hexsha
stringlengths 40
40
| size
int64 6
14.9M
| ext
stringclasses 1
value | lang
stringclasses 1
value | max_stars_repo_path
stringlengths 6
260
| max_stars_repo_name
stringlengths 6
119
| max_stars_repo_head_hexsha
stringlengths 40
41
| max_stars_repo_licenses
list | max_stars_count
int64 1
191k
⌀ | max_stars_repo_stars_event_min_datetime
stringlengths 24
24
⌀ | max_stars_repo_stars_event_max_datetime
stringlengths 24
24
⌀ | max_issues_repo_path
stringlengths 6
260
| max_issues_repo_name
stringlengths 6
119
| max_issues_repo_head_hexsha
stringlengths 40
41
| max_issues_repo_licenses
list | max_issues_count
int64 1
67k
⌀ | max_issues_repo_issues_event_min_datetime
stringlengths 24
24
⌀ | max_issues_repo_issues_event_max_datetime
stringlengths 24
24
⌀ | max_forks_repo_path
stringlengths 6
260
| max_forks_repo_name
stringlengths 6
119
| max_forks_repo_head_hexsha
stringlengths 40
41
| max_forks_repo_licenses
list | max_forks_count
int64 1
105k
⌀ | max_forks_repo_forks_event_min_datetime
stringlengths 24
24
⌀ | max_forks_repo_forks_event_max_datetime
stringlengths 24
24
⌀ | avg_line_length
float64 2
1.04M
| max_line_length
int64 2
11.2M
| alphanum_fraction
float64 0
1
| cells
list | cell_types
list | cell_type_groups
list |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
4a544d0af9331ebfb92912e5e46dd05a5e2b5f62
| 7,592 |
ipynb
|
Jupyter Notebook
|
09/Lab12.ipynb
|
ghostoverflow/Artificial-Intelligence-Labs
|
7e340d0115fbe79d59e2d46a0f4e3c976291ba58
|
[
"MIT"
] | null | null | null |
09/Lab12.ipynb
|
ghostoverflow/Artificial-Intelligence-Labs
|
7e340d0115fbe79d59e2d46a0f4e3c976291ba58
|
[
"MIT"
] | null | null | null |
09/Lab12.ipynb
|
ghostoverflow/Artificial-Intelligence-Labs
|
7e340d0115fbe79d59e2d46a0f4e3c976291ba58
|
[
"MIT"
] | null | null | null | 34.044843 | 106 | 0.460353 |
[
[
[
"Genetic Algorithm",
"_____no_output_____"
]
],
[
[
"import random as rnd",
"_____no_output_____"
],
[
"# returns the random array\ndef random_arr(lower, upper, size):\n return [rnd.randrange(lower, upper+1) for _ in range(size)]\n",
"_____no_output_____"
],
[
"# cross over between chromosomes\ndef reproduce(x, y):\n tmp = rnd.randint(0, len(x)-1)\n return x[:tmp]+y[tmp:]\n",
"_____no_output_____"
],
[
"# randomly change the value of index\ndef mutate(x):\n inp = rnd.randint(1, len(x))\n x[rnd.randrange(0, len(x))] = inp\n return x\n",
"_____no_output_____"
],
[
"# pick the random chromosome from population while seeing the probabilities\ndef random_pick(population, probs):\n r = rnd.uniform(0, sum(probs))\n endpoint = 0\n for pop, prob in zip(population, probs):\n if endpoint+prob >= r:\n return pop # picking random chromosome\n endpoint += prob\n print(\"Error!\")\n exit()\n",
"_____no_output_____"
],
[
"def genetic_algo(population, maxfitness):\n mutation_prob = 0.85 # mutation 85%\n new_population = []\n # all probabilites or percentages\n probs = [fitness(pop)/maxfitness for pop in population]\n for _ in range(len(population)):\n x = random_pick(population, probs) # one best chromosome\n y = random_pick(population, probs) # two best chromosome\n\n # creating child\n child = reproduce(x, y)\n if rnd.random() < mutation_prob:\n child = mutate(child) # rarely mutate\n\n new_population.append(child)\n if fitness(child) >= maxfitness:\n break\n return new_population\n ",
"_____no_output_____"
],
[
"def fitness(x): # checking the chromosome for fitness\n horizontal_collisions = sum(\n [x.count(queen)-1 for queen in x])/2\n diagonal_collisions = 0\n\n n = len(x)\n left_diagonal = [0] * 2*n\n right_diagonal = [0] * 2*n\n for i in range(n):\n left_diagonal[i + x[i] - 1] += 1\n right_diagonal[len(x) - i + x[i] - 2] += 1\n\n diagonal_collisions = 0\n for i in range(2*n-1):\n counter = 0\n if left_diagonal[i] > 1:\n counter += left_diagonal[i]-1\n if right_diagonal[i] > 1:\n counter += right_diagonal[i]-1\n diagonal_collisions += counter / (n-abs(i-n+1))\n\n # 28-(2+3)=23\n return int((n*(n-1))/2 - (horizontal_collisions + diagonal_collisions))\n",
"_____no_output_____"
],
[
"def print_chromosome(chrom):\n print(f\"Chromosome = {str(chrom)}, Fitness = {fitness(chrom)}\")\n",
"_____no_output_____"
],
[
"nq = int(input(\"Enter number of queens: \")) # number of queens\nmaxfitness = (nq*(nq-1))/2\n\npopulation = [random_arr(1, nq, nq) for _ in range(nq*nq)]\n\ngeneration = 1\n\nwhile not maxfitness in [fitness(chrom) for chrom in population]:\n population = genetic_algo(population, maxfitness)\n generation += 1\n if generation % 100 == 0:\n besttill = max([(fitness(n), n) for n in population],key=lambda x:x[0])\n print(\n f\"Generation= {generation}, Sol={besttill[1]} Maximum Fitness = {besttill[0]}\")\nprint(\"Solved!!\")\nchrom_out=[]\nfor chrom in population:\n if fitness(chrom) == maxfitness:\n chrom_out = chrom\n print(\n f\"Generation= {generation}, Sol={chrom} Maximum Fitness = {fitness(chrom)}\")\n",
"Generation= 100, Sol=[2, 3, 4, 7, 5, 2, 6, 1] Maximum Fitness = 26\nGeneration= 200, Sol=[4, 8, 3, 3, 6, 8, 1, 5] Maximum Fitness = 26\nGeneration= 300, Sol=[7, 4, 4, 2, 8, 5, 6, 3] Maximum Fitness = 26\nGeneration= 400, Sol=[3, 6, 4, 5, 1, 8, 2, 7] Maximum Fitness = 27\nGeneration= 500, Sol=[5, 8, 3, 6, 2, 7, 4, 8] Maximum Fitness = 26\nGeneration= 600, Sol=[1, 6, 5, 1, 2, 4, 7, 8] Maximum Fitness = 26\nGeneration= 700, Sol=[5, 4, 7, 2, 6, 5, 1, 3] Maximum Fitness = 26\nGeneration= 800, Sol=[2, 3, 1, 8, 5, 6, 7, 2] Maximum Fitness = 26\nGeneration= 900, Sol=[4, 1, 6, 3, 2, 7, 5, 3] Maximum Fitness = 26\nGeneration= 1000, Sol=[1, 4, 8, 2, 5, 3, 7, 6] Maximum Fitness = 27\nGeneration= 1100, Sol=[6, 4, 8, 3, 2, 5, 7, 1] Maximum Fitness = 27\nGeneration= 1200, Sol=[7, 5, 6, 8, 4, 2, 1, 3] Maximum Fitness = 27\nGeneration= 1300, Sol=[5, 2, 4, 1, 3, 8, 6, 2] Maximum Fitness = 27\nSolved!!\nGeneration= 1342, Sol=[8, 3, 1, 6, 2, 5, 7, 4] Maximum Fitness = 28\n"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a547f7cc14b720c164877a4906a4d8c3b2a2026
| 18,189 |
ipynb
|
Jupyter Notebook
|
notebooks/Lecture4.ipynb
|
quang-ha/IA-maths-Ipython
|
8ff8533d64a3d8db8e4813a7b6dfee39339fd846
|
[
"BSD-3-Clause"
] | null | null | null |
notebooks/Lecture4.ipynb
|
quang-ha/IA-maths-Ipython
|
8ff8533d64a3d8db8e4813a7b6dfee39339fd846
|
[
"BSD-3-Clause"
] | null | null | null |
notebooks/Lecture4.ipynb
|
quang-ha/IA-maths-Ipython
|
8ff8533d64a3d8db8e4813a7b6dfee39339fd846
|
[
"BSD-3-Clause"
] | null | null | null | 58.864078 | 3,643 | 0.665787 |
[
[
[
"empty"
]
]
] |
[
"empty"
] |
[
[
"empty"
]
] |
4a54952e5b7a1cf16f38d3b03282ff15631a3818
| 177,754 |
ipynb
|
Jupyter Notebook
|
Notebooks/How_to_use_the_Reactome_BQ_dataset.ipynb
|
rpatil524/Community-Notebooks
|
e87df00fefc33e6753c48b6bcdb63ab51f42cbca
|
[
"Apache-2.0"
] | null | null | null |
Notebooks/How_to_use_the_Reactome_BQ_dataset.ipynb
|
rpatil524/Community-Notebooks
|
e87df00fefc33e6753c48b6bcdb63ab51f42cbca
|
[
"Apache-2.0"
] | null | null | null |
Notebooks/How_to_use_the_Reactome_BQ_dataset.ipynb
|
rpatil524/Community-Notebooks
|
e87df00fefc33e6753c48b6bcdb63ab51f42cbca
|
[
"Apache-2.0"
] | null | null | null | 41.648079 | 527 | 0.356577 |
[
[
[
"<a href=\"https://colab.research.google.com/github/isb-cgc/Community-Notebooks/blob/master/Notebooks/How_to_use_the_Reactome_BQ_dataset.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# How to use the Reactome BigQuery dataset\nCheck out other notebooks at our [Community Notebooks Repository](https://github.com/isb-cgc/Community-Notebooks)!\n\n- **Title:** How to use the Reactome BigQuery dataset\n- **Author:** John Phan\n- **Created:** 2021-09-13\n- **Purpose:** Demonstrate basic usage of the Reactome BigQuery dataset\n- **URL:** https://github.com/isb-cgc/Community-Notebooks/blob/master/Notebooks/How_to_use_the_Reactome_BQ_dataset.ipynb\n\nThis notebook demonstrates basic usage of the Reactome BigQuery dataset. Analysis of this dataset can provide a powerful tool for identifying pathways related to cancer biomarkers. \n\nThe Reactome is an open-source, manually curated, and peer-reviewed pathway database. More information can be found here: https://reactome.org/. \n\n",
"_____no_output_____"
],
[
"# Initialize Notebook Environment\n\nBefore running the analysis, we need to load dependencies, authenticate to BigQuery, and customize notebook parameters.",
"_____no_output_____"
],
[
"## Import Dependencies",
"_____no_output_____"
]
],
[
[
"# GCP Libraries\nfrom google.cloud import bigquery\nfrom google.colab import auth\n\n# Data Analytics\nimport numpy as np\nfrom scipy import stats",
"_____no_output_____"
]
],
[
[
"## Authenticate\n\nBefore using BigQuery, we need to get authorization for access to BigQuery and the Google Cloud. For more information see ['Quick Start Guide to ISB-CGC'](https://isb-cancer-genomics-cloud.readthedocs.io/en/latest/sections/HowToGetStartedonISB-CGC.html). Alternative authentication methods can be found [here](https://googleapis.dev/python/google-api-core/latest/auth.html).",
"_____no_output_____"
]
],
[
[
"# if you're using Google Colab, authenticate to gcloud with the following\nauth.authenticate_user()\n\n# alternatively, use the gcloud SDK\n#!gcloud auth application-default login",
"_____no_output_____"
]
],
[
[
"## Parameters\n\nCustomize the following parameters based on your notebook, execution environment, or project.",
"_____no_output_____"
]
],
[
[
"# set the google project that will be billed for this notebook's computations\ngoogle_project = 'google-project' ## change me",
"_____no_output_____"
]
],
[
[
"## BigQuery Client\n\nCreate the BigQuery client.",
"_____no_output_____"
]
],
[
[
"# Create a client to access the data within BigQuery\nclient = bigquery.Client(google_project)",
"_____no_output_____"
]
],
[
[
"# Identify all Reactome Pathways Related to Genes\n\nWe can join tables from the Reactome BigQuery dataset to identify all pathways related to our genes of interest. We first have to map the gene names to Uniprot IDs in the Reactome \"physical entities\" table. These can then be mapped to Reactome pathways. We further filter the physical entity to pathway evidence codes to retain only interactions that have \"Traceable Author Statements\" (TAS) rather than just \"Inferred from Electronic Annotation\" (IEA) to avoid evidence that have not been manually curated. \n\nWe use the following genes to identify related pathways. These genes were identified in an ovarian cancer chemo-response study by [Bosquet et al](https://molecular-cancer.biomedcentral.com/articles/10.1186/s12943-016-0548-9). ",
"_____no_output_____"
]
],
[
[
"# set parameters for query\ngenes = \"'RHOT1','MYO7A','ZBTB10','MATK','ST18','RPS23','GCNT1','DROSHA','NUAK1','CCPG1',\\\n'PDGFD','KLRAP1','MTAP','RNF13','THBS1','MLX','FAP','TIMP3','PRSS1','SLC7A11',\\\n'OLFML3','RPS20','MCM5','POLE','STEAP4','LRRC8D','WBP1L','ENTPD5','SYNE1','DPT',\\\n'COPZ2','TRIO','PDPR'\"",
"_____no_output_____"
],
[
"# run query and put results in data frame\npathways = client.query((\"\"\"\n SELECT\n DISTINCT pathway.*\n\n FROM\n `isb-cgc-bq.reactome_versioned.pathway_v77` as pathway\n\n INNER JOIN `isb-cgc-bq.reactome_versioned.pe_to_pathway_v77` as pe2pathway\n -- link pathways to physical entities via intermediate table\n ON pathway.stable_id = pe2pathway.pathway_stable_id\n\n INNER JOIN `isb-cgc-bq.reactome_versioned.physical_entity_v77` AS pe\n -- link pathways to physical entities\n ON pe2pathway.pe_stable_id = pe.stable_id\n\n WHERE\n -- filter by stronger evidence: \"Traceable Author Statement\" \n pe2pathway.evidence_code = 'TAS'\n\n -- filter by pathways that are related to genes in list\n AND pe.name IN ({genes}) \n\n ORDER BY pathway.name ASC\n\"\"\").format(\n genes=genes\n)).result().to_dataframe()",
"_____no_output_____"
],
[
"# Display the data frame\npathways",
"_____no_output_____"
]
],
[
[
"If we're only interested in the lowest level pathways, i.e., pathways that are not parents of other pathways in the hierarchy, we can filter by the \"lowest_level\" field in the pathways table.",
"_____no_output_____"
]
],
[
[
"# run query and put results in data frame\nlowest_pathways = client.query((\"\"\"\n SELECT\n DISTINCT pathway.*\n\n FROM\n `isb-cgc-bq.reactome_versioned.pathway_v77` as pathway\n\n INNER JOIN `isb-cgc-bq.reactome_versioned.pe_to_pathway_v77` as pe2pathway\n -- link pathways to physical entities via intermediate table\n ON pathway.stable_id = pe2pathway.pathway_stable_id\n\n INNER JOIN `isb-cgc-bq.reactome_versioned.physical_entity_v77` AS pe\n -- link pathways to physical entities\n ON pe2pathway.pe_stable_id = pe.stable_id\n\n WHERE\n -- filter by stronger evidence: \"Traceable Author Statement\" \n pe2pathway.evidence_code = 'TAS'\n\n -- filter by pathways that are related to genes in list\n AND pe.name IN ({genes})\n\n -- filter to include just lowest level pathways\n AND pathway.lowest_level = TRUE\n\n ORDER BY pathway.name ASC\n\"\"\").format(\n genes=genes\n)).result().to_dataframe()",
"_____no_output_____"
],
[
"# display data frame\nlowest_pathways",
"_____no_output_____"
]
],
[
[
"# Pathway Enrichment Analysis\nWe can identify pathways that are \"enriched\" with the genes of interest. In other words, we can answer the question: given a set of interesting genes, which pathways contain those genes at a frequency higher than random chance? By calculating the probability that a number of target genes are contained in each pathway, we can identify pathways most likely to be related. \n\nTo do this, we can use a **chi-squared** test to determine if there is a statistically significant difference between the expected frequency of genes in a pathway compared to the observed frequency. \n",
"_____no_output_____"
]
],
[
[
"# set query parameters\nlowest_level = True # only show pathways at the lowest level",
"_____no_output_____"
]
],
[
[
"## Construct SQL Query\n\nA single query can be used to calculate the chi-squared statistic for all pathways. This query is rather lengthy, but can be broken up into a series of named sub-queries. We step through each query below.\n\nFirst, we write a query that simply gets a list of all genes in the Reactome physical entity table: ",
"_____no_output_____"
]
],
[
[
"gene_list_query = \"\"\"\n -- Table that contains a list of all distinct genes that map to Reactome\n -- physical entities \n SELECT\n DISTINCT pe.uniprot_id\n\n FROM\n `isb-cgc-bq.reactome_versioned.physical_entity_v77` AS pe\n\n WHERE\n -- filter by pathways that are related to genes in list\n pe.name IN ({genes})\n\"\"\".format(\n genes=genes\n).strip(\"\\n\")",
"_____no_output_____"
],
[
"print(gene_list_query)",
" -- Table that contains a list of all distinct genes that map to Reactome\n -- physical entities \n SELECT\n DISTINCT pe.uniprot_id\n\n FROM\n `isb-cgc-bq.reactome_versioned.physical_entity_v77` AS pe\n\n WHERE\n -- filter by pathways that are related to genes in list\n pe.name IN ('RHOT1','MYO7A','ZBTB10','MATK','ST18','RPS23','GCNT1','DROSHA','NUAK1','CCPG1','PDGFD','KLRAP1','MTAP','RNF13','THBS1','MLX','FAP','TIMP3','PRSS1','SLC7A11','OLFML3','RPS20','MCM5','POLE','STEAP4','LRRC8D','WBP1L','ENTPD5','SYNE1','DPT','COPZ2','TRIO','PDPR')\n"
]
],
[
[
"We then create a query that counts all targets associated with each Reactome pathway. This query depends on the previous `gene_list_query` sub-query. ",
"_____no_output_____"
]
],
[
[
"gene_pp_query = \"\"\"\n -- Table that maps pathways to the total number of interesting genes within\n -- that pathway\n SELECT\n COUNT(DISTINCT gene_list_query.uniprot_id) as num_genes,\n pathway.stable_id,\n pathway.name\n\n FROM\n gene_list_query\n\n INNER JOIN `isb-cgc-bq.reactome_versioned.physical_entity_v77` AS pe\n -- filter for interactions with genes that match a reactome\n -- physical entity\n ON gene_list_query.uniprot_id = pe.uniprot_id\n\n INNER JOIN `isb-cgc-bq.reactome_versioned.pe_to_pathway_v77` AS pe2pathway\n -- link physical entities to pathways via intermediate table\n ON pe.stable_id = pe2pathway.pe_stable_id\n\n INNER JOIN `isb-cgc-bq.reactome_versioned.pathway_v77` AS pathway\n -- link physical entities to pathways\n ON pe2pathway.pathway_stable_id = pathway.stable_id\n\n WHERE\n -- filter by stronger evidence: \"Traceable Author Statement\" \n pe2pathway.evidence_code = 'TAS'\n\n GROUP BY pathway.stable_id, pathway.name\n ORDER BY num_genes DESC\n\"\"\".strip(\"\\n\")",
"_____no_output_____"
],
[
"print(gene_pp_query)",
" -- Table that maps pathways to the total number of interesting genes within\n -- that pathway\n SELECT\n COUNT(DISTINCT gene_list_query.uniprot_id) as num_genes,\n pathway.stable_id,\n pathway.name\n\n FROM\n gene_list_query\n\n INNER JOIN `isb-cgc-bq.reactome_versioned.physical_entity_v77` AS pe\n -- filter for interactions with genes that match a reactome\n -- physical entity\n ON gene_list_query.uniprot_id = pe.uniprot_id\n\n INNER JOIN `isb-cgc-bq.reactome_versioned.pe_to_pathway_v77` AS pe2pathway\n -- link physical entities to pathways via intermediate table\n ON pe.stable_id = pe2pathway.pe_stable_id\n\n INNER JOIN `isb-cgc-bq.reactome_versioned.pathway_v77` AS pathway\n -- link physical entities to pathways\n ON pe2pathway.pathway_stable_id = pathway.stable_id\n\n WHERE\n -- filter by stronger evidence: \"Traceable Author Statement\" \n pe2pathway.evidence_code = 'TAS'\n\n GROUP BY pathway.stable_id, pathway.name\n ORDER BY num_genes DESC\n"
]
],
[
[
"Now we construct the same queries for genes that are NOT in the interesting gene list. These are prefixed with \"not_gene\". This query depends on the previous `gene_list_query` sub-query. ",
"_____no_output_____"
]
],
[
[
"not_gene_list_query = \"\"\"\n -- Table that contains a list of all genes that are NOT in the interest list\n -- This query depends on the previous \"gene_list_query\" sub-query.\n SELECT\n DISTINCT pe.uniprot_id\n\n FROM `isb-cgc-bq.reactome_versioned.physical_entity_v77` AS pe\n\n WHERE\n pe.uniprot_id NOT IN (\n SELECT uniprot_id FROM gene_list_query\n )\n\"\"\".strip(\"\\n\")",
"_____no_output_____"
],
[
"print(not_gene_list_query)",
" -- Table that contains a list of all genes that are NOT in the interest list\n -- This query depends on the previous \"gene_list_query\" sub-query.\n SELECT\n DISTINCT pe.uniprot_id\n\n FROM `isb-cgc-bq.reactome_versioned.physical_entity_v77` AS pe\n\n WHERE\n pe.uniprot_id NOT IN (\n SELECT uniprot_id FROM gene_list_query\n )\n"
],
[
"not_gene_pp_query = \"\"\"\n -- Table that maps pathways to the number of proteins that are NOT drug\n -- targets in that pathway.\n SELECT\n COUNT(DISTINCT not_gene_list_query.uniprot_id) AS num_not_genes,\n pathway.stable_id,\n pathway.name\n\n FROM not_gene_list_query\n\n INNER JOIN `isb-cgc-bq.reactome_versioned.physical_entity_v77` AS pe\n ON not_gene_list_query.uniprot_id = pe.uniprot_id\n\n INNER JOIN `isb-cgc-bq.reactome_versioned.pe_to_pathway_v77` AS pe2pathway\n ON pe.stable_id = pe2pathway.pe_stable_id\n \n INNER JOIN `isb-cgc-bq.reactome_versioned.pathway_v77` AS pathway\n ON pe2pathway.pathway_stable_id = pathway.stable_id\n\n WHERE\n -- filter by stronger evidence: \"Traceable Author Statement\" \n pe2pathway.evidence_code = 'TAS'\n\n GROUP BY pathway.stable_id, pathway.name\n ORDER BY num_not_genes DESC\n\"\"\".strip(\"\\n\")",
"_____no_output_____"
],
[
"print(not_gene_pp_query)",
" -- Table that maps pathways to the number of proteins that are NOT drug\n -- targets in that pathway.\n SELECT\n COUNT(DISTINCT not_gene_list_query.uniprot_id) AS num_not_genes,\n pathway.stable_id,\n pathway.name\n\n FROM not_gene_list_query\n\n INNER JOIN `isb-cgc-bq.reactome_versioned.physical_entity_v77` AS pe\n ON not_gene_list_query.uniprot_id = pe.uniprot_id\n\n INNER JOIN `isb-cgc-bq.reactome_versioned.pe_to_pathway_v77` AS pe2pathway\n ON pe.stable_id = pe2pathway.pe_stable_id\n \n INNER JOIN `isb-cgc-bq.reactome_versioned.pathway_v77` AS pathway\n ON pe2pathway.pathway_stable_id = pathway.stable_id\n\n WHERE\n -- filter by stronger evidence: \"Traceable Author Statement\" \n pe2pathway.evidence_code = 'TAS'\n\n GROUP BY pathway.stable_id, pathway.name\n ORDER BY num_not_genes DESC\n"
]
],
[
[
"For convenience, we create a sub-query to get the counts of the number of genes that are in the list and the number of genes that are not in the list.",
"_____no_output_____"
]
],
[
[
"gene_count_query = \"\"\"\n -- Table that contains the counts of # of genes that are/are not targets\n SELECT\n gene_count,\n not_gene_count,\n gene_count + not_gene_count AS total_count\n\n FROM \n (SELECT COUNT(*) AS gene_count FROM gene_list_query),\n (SELECT COUNT(*) AS not_gene_count FROM not_gene_list_query)\n\"\"\".strip(\"\\n\")",
"_____no_output_____"
]
],
[
[
"Now we can create more interesting queries for the contingency matrices that contain the observed and expected values. \n\nFirst, the observed contingency table counts for each pathway:",
"_____no_output_____"
]
],
[
[
"observed_query = \"\"\"\n -- Table with observed values per pathway in the contingency matrix\n SELECT\n gene_pp_query.num_genes AS in_gene_in_pathway,\n not_gene_pp_query.num_not_genes AS not_gene_in_pathway,\n gene_count_query.gene_count - gene_pp_query.num_genes AS in_gene_not_pathway,\n gene_count_query.not_gene_count - not_gene_pp_query.num_not_genes AS not_gene_not_pathway,\n gene_pp_query.stable_id,\n gene_pp_query.name\n\n FROM \n gene_pp_query,\n gene_count_query\n\n INNER JOIN not_gene_pp_query\n ON gene_pp_query.stable_id = not_gene_pp_query.stable_id\n\"\"\".strip(\"\\n\")",
"_____no_output_____"
]
],
[
[
"Then the observed row and column sums of the contingency table:",
"_____no_output_____"
]
],
[
[
"sum_query = \"\"\"\n -- Table with summed observed values per pathway in the contingency matrix\n SELECT\n observed_query.in_gene_in_pathway + observed_query.not_gene_in_pathway AS pathway_total,\n observed_query.in_gene_not_pathway + observed_query.not_gene_not_pathway AS not_pathway_total,\n observed_query.in_gene_in_pathway + observed_query.in_gene_not_pathway AS gene_total,\n observed_query.not_gene_in_pathway + observed_query.not_gene_not_pathway AS not_gene_total,\n observed_query.stable_id,\n observed_query.name\n\n FROM\n observed_query\n\"\"\".strip(\"\\n\")",
"_____no_output_____"
]
],
[
[
"And the expected contingency table values for each pathway:",
"_____no_output_____"
]
],
[
[
"expected_query = \"\"\" \n -- Table with the expected values per pathway in the contingency matrix\n SELECT \n sum_query.gene_total * sum_query.pathway_total / gene_count_query.total_count AS exp_in_gene_in_pathway,\n sum_query.not_gene_total * sum_query.pathway_total / gene_count_query.total_count AS exp_not_gene_in_pathway,\n sum_query.gene_total * sum_query.not_pathway_total / gene_count_query.total_count AS exp_in_gene_not_pathway,\n sum_query.not_gene_total * sum_query.not_pathway_total / gene_count_query.total_count AS exp_not_gene_not_pathway,\n sum_query.stable_id,\n sum_query.name\n\n FROM \n sum_query, gene_count_query\n\"\"\".strip(\"\\n\")",
"_____no_output_____"
]
],
[
[
"Finally, we can calculate the chi-squared statistic for each pathway:",
"_____no_output_____"
]
],
[
[
"chi_squared_query = \"\"\"\n -- Table with the chi-squared statistic for each pathway\n SELECT\n -- Chi squared statistic with Yates' correction\n POW(ABS(observed_query.in_gene_in_pathway - expected_query.exp_in_gene_in_pathway) - 0.5, 2) / expected_query.exp_in_gene_in_pathway \n + POW(ABS(observed_query.not_gene_in_pathway - expected_query.exp_not_gene_in_pathway) - 0.5, 2) / expected_query.exp_not_gene_in_pathway\n + POW(ABS(observed_query.in_gene_not_pathway - expected_query.exp_in_gene_not_pathway) - 0.5, 2) / expected_query.exp_in_gene_not_pathway\n + POW(ABS(observed_query.not_gene_not_pathway - expected_query.exp_not_gene_not_pathway) - 0.5, 2) / expected_query.exp_not_gene_not_pathway\n AS chi_squared_stat,\n observed_query.stable_id,\n observed_query.name\n\n FROM observed_query\n\n INNER JOIN expected_query\n ON observed_query.stable_id = expected_query.stable_id\n\"\"\".strip(\"\\n\")",
"_____no_output_____"
]
],
[
[
"The final piece of the query optionally adds a filter that removes all pathways that are not at the lowest level of the hierarchy. This helps remove non-specific \"pathways\" such as the generic \"Disease\" pathway.",
"_____no_output_____"
]
],
[
[
"lowest_level_filter = \"\"\"\n INNER JOIN `isb-cgc-bq.reactome_versioned.pathway_v77` AS pathway\n ON chi_squared_query.stable_id = pathway.stable_id\n\n WHERE pathway.lowest_level = TRUE\n\"\"\".strip(\"\\n\")",
"_____no_output_____"
],
[
"print(lowest_level_filter)",
" INNER JOIN `isb-cgc-bq.reactome_versioned.pathway_v77` AS pathway\n ON chi_squared_query.stable_id = pathway.stable_id\n\n WHERE pathway.lowest_level = TRUE\n"
]
],
[
[
"Now we can combine all sub-queries to create the final query:",
"_____no_output_____"
]
],
[
[
"final_query = \"\"\"\n WITH\n gene_list_query AS (\n {gene_list_query}\n ),\n\n gene_pp_query AS (\n {gene_pp_query}\n ),\n\n not_gene_list_query AS (\n {not_gene_list_query}\n ),\n\n not_gene_pp_query AS (\n {not_gene_pp_query}\n ),\n\n gene_count_query AS (\n {gene_count_query}\n ),\n\n observed_query AS (\n {observed_query}\n ),\n\n sum_query AS (\n {sum_query}\n ),\n\n expected_query AS (\n {expected_query}\n ),\n \n chi_squared_query AS (\n {chi_squared_query}\n )\n\n SELECT\n observed_query.in_gene_in_pathway,\n observed_query.in_gene_not_pathway,\n observed_query.not_gene_in_pathway,\n observed_query.not_gene_not_pathway,\n chi_squared_query.chi_squared_stat,\n chi_squared_query.stable_id,\n chi_squared_query.name\n\n FROM chi_squared_query\n\n INNER JOIN observed_query\n ON chi_squared_query.stable_id = observed_query.stable_id\n {lowest_level_filter}\n ORDER BY chi_squared_stat DESC\n\"\"\".format(\n # make final query a little easier to read by removing/adding some white space \n gene_list_query=\"\\n \".join(gene_list_query.strip().splitlines()),\n gene_pp_query=\"\\n \".join(gene_pp_query.strip().splitlines()),\n not_gene_list_query=\"\\n \".join(not_gene_list_query.strip().splitlines()),\n not_gene_pp_query=\"\\n \".join(not_gene_pp_query.strip().splitlines()),\n gene_count_query=\"\\n \".join(gene_count_query.strip().splitlines()),\n observed_query=\"\\n \".join(observed_query.strip().splitlines()),\n sum_query=\"\\n \".join(sum_query.strip().splitlines()),\n expected_query=\"\\n \".join(expected_query.strip().splitlines()),\n chi_squared_query=\"\\n \".join(chi_squared_query.strip().splitlines()),\n lowest_level_filter=(\n \"\\n \"+\"\\n\".join(lowest_level_filter.strip().splitlines())+\"\\n\" if lowest_level else \"\"\n )\n).strip(\"\\n\")",
"_____no_output_____"
]
],
[
[
"## Display Final Query",
"_____no_output_____"
]
],
[
[
"# print the formatted final query\nprint(final_query)",
" WITH\n gene_list_query AS (\n -- Table that contains a list of all distinct genes that map to Reactome\n -- physical entities \n SELECT\n DISTINCT pe.uniprot_id\n \n FROM\n `isb-cgc-bq.reactome_versioned.physical_entity_v77` AS pe\n \n WHERE\n -- filter by pathways that are related to genes in list\n pe.name IN ('RHOT1','MYO7A','ZBTB10','MATK','ST18','RPS23','GCNT1','DROSHA','NUAK1','CCPG1','PDGFD','KLRAP1','MTAP','RNF13','THBS1','MLX','FAP','TIMP3','PRSS1','SLC7A11','OLFML3','RPS20','MCM5','POLE','STEAP4','LRRC8D','WBP1L','ENTPD5','SYNE1','DPT','COPZ2','TRIO','PDPR')\n ),\n\n gene_pp_query AS (\n -- Table that maps pathways to the total number of interesting genes within\n -- that pathway\n SELECT\n COUNT(DISTINCT gene_list_query.uniprot_id) as num_genes,\n pathway.stable_id,\n pathway.name\n \n FROM\n gene_list_query\n \n INNER JOIN `isb-cgc-bq.reactome_versioned.physical_entity_v77` AS pe\n -- filter for interactions with genes that match a reactome\n -- physical entity\n ON gene_list_query.uniprot_id = pe.uniprot_id\n \n INNER JOIN `isb-cgc-bq.reactome_versioned.pe_to_pathway_v77` AS pe2pathway\n -- link physical entities to pathways via intermediate table\n ON pe.stable_id = pe2pathway.pe_stable_id\n \n INNER JOIN `isb-cgc-bq.reactome_versioned.pathway_v77` AS pathway\n -- link physical entities to pathways\n ON pe2pathway.pathway_stable_id = pathway.stable_id\n \n WHERE\n -- filter by stronger evidence: \"Traceable Author Statement\" \n pe2pathway.evidence_code = 'TAS'\n \n GROUP BY pathway.stable_id, pathway.name\n ORDER BY num_genes DESC\n ),\n\n not_gene_list_query AS (\n -- Table that contains a list of all genes that are NOT in the interest list\n -- This query depends on the previous \"gene_list_query\" sub-query.\n SELECT\n DISTINCT pe.uniprot_id\n \n FROM `isb-cgc-bq.reactome_versioned.physical_entity_v77` AS pe\n \n WHERE\n pe.uniprot_id NOT IN (\n SELECT uniprot_id FROM gene_list_query\n )\n ),\n\n not_gene_pp_query AS (\n -- Table that maps pathways to the number of proteins that are NOT drug\n -- targets in that pathway.\n SELECT\n COUNT(DISTINCT not_gene_list_query.uniprot_id) AS num_not_genes,\n pathway.stable_id,\n pathway.name\n \n FROM not_gene_list_query\n \n INNER JOIN `isb-cgc-bq.reactome_versioned.physical_entity_v77` AS pe\n ON not_gene_list_query.uniprot_id = pe.uniprot_id\n \n INNER JOIN `isb-cgc-bq.reactome_versioned.pe_to_pathway_v77` AS pe2pathway\n ON pe.stable_id = pe2pathway.pe_stable_id\n \n INNER JOIN `isb-cgc-bq.reactome_versioned.pathway_v77` AS pathway\n ON pe2pathway.pathway_stable_id = pathway.stable_id\n \n WHERE\n -- filter by stronger evidence: \"Traceable Author Statement\" \n pe2pathway.evidence_code = 'TAS'\n \n GROUP BY pathway.stable_id, pathway.name\n ORDER BY num_not_genes DESC\n ),\n\n gene_count_query AS (\n -- Table that contains the counts of # of genes that are/are not targets\n SELECT\n gene_count,\n not_gene_count,\n gene_count + not_gene_count AS total_count\n \n FROM \n (SELECT COUNT(*) AS gene_count FROM gene_list_query),\n (SELECT COUNT(*) AS not_gene_count FROM not_gene_list_query)\n ),\n\n observed_query AS (\n -- Table with observed values per pathway in the contingency matrix\n SELECT\n gene_pp_query.num_genes AS in_gene_in_pathway,\n not_gene_pp_query.num_not_genes AS not_gene_in_pathway,\n gene_count_query.gene_count - gene_pp_query.num_genes AS in_gene_not_pathway,\n gene_count_query.not_gene_count - not_gene_pp_query.num_not_genes AS not_gene_not_pathway,\n gene_pp_query.stable_id,\n gene_pp_query.name\n \n FROM \n gene_pp_query,\n gene_count_query\n \n INNER JOIN not_gene_pp_query\n ON gene_pp_query.stable_id = not_gene_pp_query.stable_id\n ),\n\n sum_query AS (\n -- Table with summed observed values per pathway in the contingency matrix\n SELECT\n observed_query.in_gene_in_pathway + observed_query.not_gene_in_pathway AS pathway_total,\n observed_query.in_gene_not_pathway + observed_query.not_gene_not_pathway AS not_pathway_total,\n observed_query.in_gene_in_pathway + observed_query.in_gene_not_pathway AS gene_total,\n observed_query.not_gene_in_pathway + observed_query.not_gene_not_pathway AS not_gene_total,\n observed_query.stable_id,\n observed_query.name\n \n FROM\n observed_query\n ),\n\n expected_query AS (\n -- Table with the expected values per pathway in the contingency matrix\n SELECT \n sum_query.gene_total * sum_query.pathway_total / gene_count_query.total_count AS exp_in_gene_in_pathway,\n sum_query.not_gene_total * sum_query.pathway_total / gene_count_query.total_count AS exp_not_gene_in_pathway,\n sum_query.gene_total * sum_query.not_pathway_total / gene_count_query.total_count AS exp_in_gene_not_pathway,\n sum_query.not_gene_total * sum_query.not_pathway_total / gene_count_query.total_count AS exp_not_gene_not_pathway,\n sum_query.stable_id,\n sum_query.name\n \n FROM \n sum_query, gene_count_query\n ),\n \n chi_squared_query AS (\n -- Table with the chi-squared statistic for each pathway\n SELECT\n -- Chi squared statistic with Yates' correction\n POW(ABS(observed_query.in_gene_in_pathway - expected_query.exp_in_gene_in_pathway) - 0.5, 2) / expected_query.exp_in_gene_in_pathway \n + POW(ABS(observed_query.not_gene_in_pathway - expected_query.exp_not_gene_in_pathway) - 0.5, 2) / expected_query.exp_not_gene_in_pathway\n + POW(ABS(observed_query.in_gene_not_pathway - expected_query.exp_in_gene_not_pathway) - 0.5, 2) / expected_query.exp_in_gene_not_pathway\n + POW(ABS(observed_query.not_gene_not_pathway - expected_query.exp_not_gene_not_pathway) - 0.5, 2) / expected_query.exp_not_gene_not_pathway\n AS chi_squared_stat,\n observed_query.stable_id,\n observed_query.name\n \n FROM observed_query\n \n INNER JOIN expected_query\n ON observed_query.stable_id = expected_query.stable_id\n )\n\n SELECT\n observed_query.in_gene_in_pathway,\n observed_query.in_gene_not_pathway,\n observed_query.not_gene_in_pathway,\n observed_query.not_gene_not_pathway,\n chi_squared_query.chi_squared_stat,\n chi_squared_query.stable_id,\n chi_squared_query.name\n\n FROM chi_squared_query\n\n INNER JOIN observed_query\n ON chi_squared_query.stable_id = observed_query.stable_id\n \n INNER JOIN `isb-cgc-bq.reactome_versioned.pathway_v77` AS pathway\n ON chi_squared_query.stable_id = pathway.stable_id\n\n WHERE pathway.lowest_level = TRUE\n\n ORDER BY chi_squared_stat DESC\n"
]
],
[
[
"## Execute the Query\n\nNow execute the query to calculate a chi-squared statistic for each pathway:",
"_____no_output_____"
]
],
[
[
"# run query and put results in data frame\nchi_squared_pathways = client.query(final_query).result().to_dataframe()",
"_____no_output_____"
],
[
"# display the data frame\nchi_squared_pathways",
"_____no_output_____"
]
],
[
[
"## Calculate P-Values\n\nBigQuery does not have statistical functions to calculate p-values, so we use the SciPy stats library and update the data frame with a new p-value column:",
"_____no_output_____"
]
],
[
[
"chi_squared_pathways['p_value'] = 1-stats.chi2.cdf(chi_squared_pathways['chi_squared_stat'], 1)\nchi_squared_pathways",
"_____no_output_____"
]
],
[
[
"## Adjust for Multiple Testing\n\nSince we're testing multiple pathways, we need to adjust the p-value threshold for significance accordingly. The number of pathways tested depends on whether or not we're considering all pathways, or just the lowest level pathways. That count can be obtained with the following query.",
"_____no_output_____"
]
],
[
[
"# run query and put results in data frame\nnum_pathways_result = client.query((\"\"\"\n SELECT\n COUNT (*) AS num_pathways\n FROM\n `isb-cgc-bq.reactome_versioned.pathway_v77` as pathway\n {lowest_level_filter}\n\"\"\").format(\n lowest_level_filter=(\"WHERE lowest_level = TRUE\" if lowest_level else \"\")\n)).result().to_dataframe()",
"_____no_output_____"
],
[
"# display data frame\nnum_pathways_result",
"_____no_output_____"
],
[
"# adjust significance threshold for multiple testing, using a p-value of 0.01\nnum_pathways = num_pathways_result['num_pathways'][0]\nsignificance_threshold = 0.01/num_pathways\nprint('Significance Threshold: {}'.format(significance_threshold))\n\n# find all pathways that meet the significance criterion after adjusting for\n# multiple testing\nsignificant_pathway_index = chi_squared_pathways['p_value']<significance_threshold\n\n# list of significant pathways\nsignificant_pathways = chi_squared_pathways[significant_pathway_index]",
"Significance Threshold: 5.640157924421884e-06\n"
]
],
[
[
"The final result is a list of all pathways in which the targeted proteins are enriched, or over-represented, at a rate higher than random chance. ",
"_____no_output_____"
]
],
[
[
"# display the final data frame\nsignificant_pathways",
"_____no_output_____"
]
],
[
[
"The results of this analysis suggest that at least three pathways may be related to the genes identified. ",
"_____no_output_____"
],
[
"## Verify Results by Comparing to SciPy Chi-Squared Function\n\nWe verify these BigQuery results by calculating the same chi-squared statistic using the SciPy package. Comparing the SciPy p-values and statistics to those of the BigQuery-derived results confirms that they are identical.",
"_____no_output_____"
]
],
[
[
"# extract observed values from bigquery result\nobserved = chi_squared_pathways[[\n 'in_gene_in_pathway',\n 'in_gene_not_pathway',\n 'not_gene_in_pathway',\n 'not_gene_not_pathway'\n]]\n\n# calculate the chi-squared statistic using the scipy stats package \nchi2_stat = []\nchi2_pvalue = []\nfor index, row in observed.iterrows():\n stat, pvalue, _, _ = stats.chi2_contingency(\n np.reshape(np.matrix(row), (2,2)), correction=True\n )\n chi2_stat.append(stat)\n chi2_pvalue.append(pvalue)\n\n# add columns to the original data frame\nchi_squared_pathways['scipy_stat'] = chi2_stat\nchi_squared_pathways['scipy_p_value'] = chi2_pvalue",
"_____no_output_____"
],
[
"# display the updated data frame\nchi_squared_pathways",
"_____no_output_____"
]
],
[
[
"# Conclusion\n\nThis notebook demonstrated usage of the Reactome BigQuery dataset for basic cancer pathway identification from a gene set, as well as a more complex pathway enrichment analysis using a chi-squared statistic.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
4a549a3a138ae7aa6ce6fe15b6219c839f66e12d
| 466,361 |
ipynb
|
Jupyter Notebook
|
week_2/arima_walkthrough.ipynb
|
technogleb/ts_summer
|
bb2c92d541a7b361232ece5a9fb5931388604275
|
[
"MIT"
] | 2 |
2021-05-17T09:45:16.000Z
|
2021-08-11T11:58:09.000Z
|
week_2/arima_walkthrough.ipynb
|
technogleb/ts_summer
|
bb2c92d541a7b361232ece5a9fb5931388604275
|
[
"MIT"
] | null | null | null |
week_2/arima_walkthrough.ipynb
|
technogleb/ts_summer
|
bb2c92d541a7b361232ece5a9fb5931388604275
|
[
"MIT"
] | 2 |
2021-05-24T18:50:59.000Z
|
2021-05-30T19:30:56.000Z
| 244.680483 | 51,608 | 0.864367 |
[
[
[
"%matplotlib inline\n%load_ext autoreload\n%autoreload 2",
"The autoreload extension is already loaded. To reload it, use:\n %reload_ext autoreload\n"
],
[
"import sys\nfrom pathlib import Path\n\nsys.path.append(str(Path.cwd().parent))",
"_____no_output_____"
],
[
"import numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport plotting\nfrom statsmodels.tsa.stattools import adfuller\nfrom statsmodels.graphics.tsaplots import plot_acf, plot_pacf\nfrom scipy.signal import detrend\n%matplotlib inline",
"_____no_output_____"
],
[
"from load_dataset import Dataset",
"_____no_output_____"
],
[
"dataset = Dataset('../data/dataset/')",
"_____no_output_____"
],
[
"# возьмем временной ряд, характеризующий продажи алкоголя по месяцам\nts = dataset[\"alcohol_sales.csv\"]",
"_____no_output_____"
],
[
"ts, ts_test = ts[:250], ts[250:]",
"_____no_output_____"
],
[
"ts.plot()",
"_____no_output_____"
]
],
[
[
"## Box-Jenkins",
"_____no_output_____"
]
],
[
[
"# как можно заметить, у него есть окололинейный тренд, гетероскедастичность, сезонный период равен 12 (месяцам)",
"_____no_output_____"
],
[
"# сначала уберем гетероскедастичность простым логарифмированием\nts_log = np.log(ts)\nplotting.plot_ts(ts_log)",
"_____no_output_____"
],
[
"# Теперь подберем порядки дифференцирования d, D\n# d малое подбирается таким образом, что d раз продифференцировав ряд, мы добьемся стационарности\n# обычно таким дифференцированием убирают тренды\n# D большое обычно подбирается так, что если d малое не дало стацинарности, мы можем D раз сезонно придифференцировать\n# ряд, пока он не станет стационарным.",
"_____no_output_____"
],
[
"# для начала просто продифференцируем один раз",
"_____no_output_____"
],
[
"ts_log.diff().plot()",
"_____no_output_____"
],
[
"# в данном случае ряд сохраняет сезонность\nplot_acf(ts_log.diff().dropna());",
"_____no_output_____"
],
[
"# попробуем применить сезонное дифференцирование \nts_log.diff(12).plot()",
"_____no_output_____"
],
[
"plot_acf(ts_log.diff(12).dropna());",
"_____no_output_____"
],
[
"# уже лучше\n# посмотрим, что скажет критерий Дики Фуллера\n# видим, что пока мы не можем отвергнуть нулевую гипотезу\nadfuller(ts_log.diff(12).dropna())[1]",
"_____no_output_____"
],
[
"# давайте тогда попробуем обьединить сезонное и простое дифференцирования\nts_log.diff(12).diff().plot()",
"_____no_output_____"
],
[
"plot_acf(ts_log.diff(12).diff().dropna(), lags=40);",
"_____no_output_____"
],
[
"adfuller(ts_log.diff(12).diff().dropna())[1]",
"_____no_output_____"
],
[
"# отлично, вердикт о стационарности подтвержден, (d, D) = (1, 1)",
"_____no_output_____"
],
[
"# теперь разберемся с параметрами q, Q, p, P.",
"_____no_output_____"
],
[
"ts_flat = ts_log.diff(12).diff().dropna()",
"_____no_output_____"
],
[
"ts_flat.plot()",
"_____no_output_____"
],
[
"# отлично, для поиска параметров q, Q, p, P нарисуем график автокорреляции и частичной автокорреляции\n# на графиках мы видим что резкое падение частичной автокорреляции, и плавное затухание полной автокорреляции,\n# следовательно, наш ряд может быть описан моделью (p, d, 0), (P, D, 0). Итак, q = 0, Q = 0.\nplot_acf(ts_flat.dropna());\nplot_pacf(ts_flat, lags=50);",
"_____no_output_____"
],
[
"# найдем теперь параметры p, P\n# p малое определяется как последний несезонный лаг, находящийся выше доверительного интервала\n# в данном случае это p = 2, аналогично с сезонными лагами мы не видим никаких сезонных всплесков, \n# значит P = 0, итак (p, P) = (2, 0)\nplot_pacf(ts_flat, lags=50);",
"_____no_output_____"
],
[
"# теперь попробуем построить SARIMA с этими параметрами",
"_____no_output_____"
],
[
"from statsmodels.tsa.statespace import sarimax",
"_____no_output_____"
],
[
"pdq = (2, 1, 0)\nPDQ = (0, 1, 0, 12)",
"_____no_output_____"
],
[
"model = sarimax.SARIMAX(ts_log, order=pdq, seasonal_order=PDQ)",
"/Users/gsinyakov/ts_summer/.env/lib/python3.8/site-packages/statsmodels/tsa/base/tsa_model.py:524: ValueWarning: No frequency information was provided, so inferred frequency MS will be used.\n warnings.warn('No frequency information was'\n/Users/gsinyakov/ts_summer/.env/lib/python3.8/site-packages/statsmodels/tsa/base/tsa_model.py:524: ValueWarning: No frequency information was provided, so inferred frequency MS will be used.\n warnings.warn('No frequency information was'\n"
],
[
"res = model.fit()",
"_____no_output_____"
],
[
"preds = res.forecast(69)",
"_____no_output_____"
],
[
"plotting.plot_ts(ts_log, preds)",
"_____no_output_____"
],
[
"# восстановим в изначальном масштабе\nplotting.plot_ts(np.exp(ts_log), np.exp(preds), ts_test)",
"_____no_output_____"
],
[
"# Видим что получилось весьма неплохо!",
"_____no_output_____"
],
[
"# чтобы убедиться еще раз, давайте проанализируем остатки",
"_____no_output_____"
],
[
"res = (np.exp(preds) - ts_test)",
"_____no_output_____"
],
[
"res.plot()",
"_____no_output_____"
],
[
"plot_acf(res, lags=40);",
"_____no_output_____"
]
],
[
[
"## Auto arima",
"_____no_output_____"
]
],
[
[
"from pmdarima import auto_arima",
"_____no_output_____"
],
[
"model = auto_arima(\n ts_log, start_p=0, start_q=0,\n max_p=3, max_q=3, m=12,\n start_P=0, start_Q=0, seasonal=True,\n d=1, D=1, trace=True,\n error_action='ignore',\n suppress_warnings=True,\n stepwise=True\n)",
"Performing stepwise search to minimize aic\n ARIMA(0,1,0)(0,1,0)[12] : AIC=-556.970, Time=0.05 sec\n ARIMA(1,1,0)(1,1,0)[12] : AIC=-647.918, Time=0.07 sec\n ARIMA(0,1,1)(0,1,1)[12] : AIC=inf, Time=0.44 sec\n ARIMA(1,1,0)(0,1,0)[12] : AIC=-649.023, Time=0.05 sec\n ARIMA(1,1,0)(0,1,1)[12] : AIC=inf, Time=0.33 sec\n ARIMA(1,1,0)(1,1,1)[12] : AIC=inf, Time=0.79 sec\n ARIMA(2,1,0)(0,1,0)[12] : AIC=-803.158, Time=0.14 sec\n ARIMA(2,1,0)(1,1,0)[12] : AIC=-806.341, Time=0.32 sec\n ARIMA(2,1,0)(2,1,0)[12] : AIC=-823.880, Time=0.74 sec\n ARIMA(2,1,0)(2,1,1)[12] : AIC=inf, Time=2.22 sec\n ARIMA(2,1,0)(1,1,1)[12] : AIC=inf, Time=0.93 sec\n ARIMA(1,1,0)(2,1,0)[12] : AIC=-679.155, Time=0.63 sec\n ARIMA(3,1,0)(2,1,0)[12] : AIC=-822.275, Time=1.03 sec\n ARIMA(2,1,1)(2,1,0)[12] : AIC=-822.639, Time=1.27 sec\n ARIMA(1,1,1)(2,1,0)[12] : AIC=-775.734, Time=0.72 sec\n ARIMA(3,1,1)(2,1,0)[12] : AIC=-827.791, Time=1.26 sec\n ARIMA(3,1,1)(1,1,0)[12] : AIC=-813.394, Time=0.63 sec\n ARIMA(3,1,1)(2,1,1)[12] : AIC=inf, Time=2.60 sec\n ARIMA(3,1,1)(1,1,1)[12] : AIC=inf, Time=2.17 sec\n ARIMA(3,1,2)(2,1,0)[12] : AIC=-829.308, Time=3.11 sec\n ARIMA(3,1,2)(1,1,0)[12] : AIC=-811.196, Time=1.28 sec\n ARIMA(3,1,2)(2,1,1)[12] : AIC=inf, Time=5.02 sec\n ARIMA(3,1,2)(1,1,1)[12] : AIC=inf, Time=1.41 sec\n ARIMA(2,1,2)(2,1,0)[12] : AIC=-829.214, Time=1.43 sec\n ARIMA(3,1,3)(2,1,0)[12] : AIC=-836.759, Time=1.44 sec\n ARIMA(3,1,3)(1,1,0)[12] : AIC=-853.056, Time=0.91 sec\n ARIMA(3,1,3)(0,1,0)[12] : AIC=-833.893, Time=0.53 sec\n ARIMA(3,1,3)(1,1,1)[12] : AIC=inf, Time=1.90 sec\n ARIMA(3,1,3)(0,1,1)[12] : AIC=inf, Time=1.74 sec\n ARIMA(3,1,3)(2,1,1)[12] : AIC=-885.282, Time=29.14 sec\n ARIMA(3,1,3)(2,1,2)[12] : AIC=-891.175, Time=4.33 sec\n ARIMA(3,1,3)(1,1,2)[12] : AIC=inf, Time=4.98 sec\n ARIMA(2,1,3)(2,1,2)[12] : AIC=-892.558, Time=3.99 sec\n ARIMA(2,1,3)(1,1,2)[12] : AIC=inf, Time=4.47 sec\n ARIMA(2,1,3)(2,1,1)[12] : AIC=-885.963, Time=3.77 sec\n ARIMA(2,1,3)(1,1,1)[12] : AIC=inf, Time=2.10 sec\n ARIMA(1,1,3)(2,1,2)[12] : AIC=-885.698, Time=4.08 sec\n ARIMA(2,1,2)(2,1,2)[12] : AIC=-893.959, Time=3.22 sec\n ARIMA(2,1,2)(1,1,2)[12] : AIC=inf, Time=3.21 sec\n ARIMA(2,1,2)(2,1,1)[12] : AIC=-884.588, Time=3.35 sec\n ARIMA(2,1,2)(1,1,1)[12] : AIC=inf, Time=1.48 sec\n ARIMA(1,1,2)(2,1,2)[12] : AIC=-867.521, Time=2.86 sec\n ARIMA(2,1,1)(2,1,2)[12] : AIC=-885.869, Time=3.14 sec\n ARIMA(3,1,2)(2,1,2)[12] : AIC=-892.614, Time=3.37 sec\n ARIMA(1,1,1)(2,1,2)[12] : AIC=-867.961, Time=3.38 sec\n ARIMA(3,1,1)(2,1,2)[12] : AIC=-890.096, Time=3.50 sec\n ARIMA(2,1,2)(2,1,2)[12] intercept : AIC=-890.223, Time=3.82 sec\n\nBest model: ARIMA(2,1,2)(2,1,2)[12] \nTotal fit time: 123.420 seconds\n"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
4a54a118bf134e111f74b23537d5132dc9551623
| 255,630 |
ipynb
|
Jupyter Notebook
|
8- How to solve Problem/A Data Science Framework for Quora/A Data Science Framework for Quora.ipynb
|
ahmed24khaled/10-steps-to-become-a-data-scientist
|
374487e92cf3a2860c2016b98251de5808fd97a2
|
[
"Apache-2.0"
] | null | null | null |
8- How to solve Problem/A Data Science Framework for Quora/A Data Science Framework for Quora.ipynb
|
ahmed24khaled/10-steps-to-become-a-data-scientist
|
374487e92cf3a2860c2016b98251de5808fd97a2
|
[
"Apache-2.0"
] | null | null | null |
8- How to solve Problem/A Data Science Framework for Quora/A Data Science Framework for Quora.ipynb
|
ahmed24khaled/10-steps-to-become-a-data-scientist
|
374487e92cf3a2860c2016b98251de5808fd97a2
|
[
"Apache-2.0"
] | 1 |
2020-01-03T13:14:33.000Z
|
2020-01-03T13:14:33.000Z
| 195.735069 | 60,273 | 0.865118 |
[
[
[
"# <div style=\"text-align: center\">A Data Science Framework for Quora </div>\n### <div align=\"center\"><b>Quite Practical and Far from any Theoretical Concepts</b></div>\n<img src='http://s9.picofile.com/file/8342477368/kq.png'>\n<div style=\"text-align:center\">last update: <b>11/15/2018</b></div>\n\nYou can Fork and Run this kernel on Github:\n> ###### [ GitHub](https://github.com/mjbahmani/10-steps-to-become-a-data-scientist)\n",
"_____no_output_____"
],
[
" <a id=\"1\"></a> <br>\n## 1- Introduction\n**Quora** has defined a competition in **Kaggle**. A realistic and attractive data set for data scientists.\non this notebook, I will provide a **comprehensive** approach to solve Quora classification problem.\n\nI am open to getting your feedback for improving this **kernel**",
"_____no_output_____"
],
[
"<a id=\"top\"></a> <br>\n## Notebook Content\n1. [Introduction](#1)\n1. [Data Science Workflow for Quora](#2)\n1. [Problem Definition](#3)\n 1. [Business View](#4)\n 1. [Real world Application Vs Competitions](#31)\n 1. [What is a insincere question?](#5)\n 1. [How can we find insincere question?](#6)\n1. [Problem feature](#7)\n 1. [Aim](#8)\n 1. [Variables](#9)\n 1. [ Inputs & Outputs](#10)\n1. [Select Framework](#11)\n 1. [Import](#12)\n 1. [Version](#13)\n 1. [Setup](#14)\n1. [Exploratory data analysis](#15)\n 1. [Data Collection](#16)\n 1. [Features](#17)\n 1. [Explorer Dataset](#18)\n 1. [Data Cleaning](#19)\n 1. [Data Preprocessing](#20)\n 1. [Is data set imbalance?](#21)\n 1. [Some Feature Engineering](#22)\n 1. [Data Visualization](#23)\n 1. [countplot](#61)\n 1. [pie plot](#62)\n 1. [Histogram](#63)\n 1. [violin plot](#64)\n 1. [kdeplot](#65)\n 1. [Data Cleaning](#24)\n1. [Model Deployment](#24)\n1. [Conclusion](#25)\n1. [References](#26)",
"_____no_output_____"
],
[
"-------------------------------------------------------------------------------------------------------------\n\n **I hope you find this kernel helpful and some <font color=\"red\"><b>UPVOTES</b></font> would be very much appreciated**\n \n -----------",
"_____no_output_____"
],
[
"<a id=\"2\"></a> <br>\n## 2- A Data Science Workflow for Quora\nOf course, the same solution can not be provided for all problems, so the best way is to create a general framework and adapt it to new problem.\n\n**You can see my workflow in the below image** :\n\n <img src=\"http://s8.picofile.com/file/8342707700/workflow2.png\" />\n\n**you should\tfeel free\tto\tadapt \tthis\tchecklist \tto\tyour needs**\n###### [Go to top](#top)",
"_____no_output_____"
],
[
"<a id=\"3\"></a> <br>\n## 3- Problem Definition\nI think one of the important things when you start a new machine learning project is Defining your problem. that means you should understand business problem.( **Problem Formalization**)\n> **we will be predicting whether a question asked on Quora is sincere or not.**\n\n## 3-1 About Quora\nQuora is a platform that empowers people to learn from each other. On Quora, people can ask questions and connect with others who contribute unique insights and quality answers. A key challenge is to weed out insincere questions -- those founded upon false premises, or that intend to make a statement rather than look for helpful answers.\n<a id=\"4\"></a> <br>\n## 3-2 Business View \nAn existential problem for any major website today is how to handle toxic and divisive content. **Quora** wants to tackle this problem head-on to keep their platform a place where users can feel safe sharing their knowledge with the world.\n\n**Quora** is a platform that empowers people to learn from each other. On Quora, people can ask questions and connect with others who contribute unique insights and quality answers. A key challenge is to weed out insincere questions -- those founded upon false premises, or that intend to make a statement rather than look for helpful answers.\n\nIn this kernel, I will develop models that identify and flag insincere questions.we Help Quora uphold their policy of “Be Nice, Be Respectful” and continue to be a place for sharing and growing the world’s knowledge.\n<a id=\"31\"></a> <br>\n### 3-2-1 Real world Application Vs Competitions\nJust a simple comparison between real-world apps with competitions:\n<img src=\"http://s9.picofile.com/file/8339956300/reallife.png\" height=\"600\" width=\"500\" />\n<a id=\"5\"></a> <br>\n## 3-3 What is a insincere question?\nis defined as a question intended to make a **statement** rather than look for **helpful answers**.\n<img src='http://s8.picofile.com/file/8342711526/Quora_moderation.png'>\n<a id=\"6\"></a> <br>\n## 3-4 How can we find insincere question?\nSome characteristics that can signify that a question is insincere:\n\n1. **Has a non-neutral tone**\n 1. Has an exaggerated tone to underscore a point about a group of people\n 1. Is rhetorical and meant to imply a statement about a group of people\n1. **Is disparaging or inflammatory**\n 1. Suggests a discriminatory idea against a protected class of people, or seeks confirmation of a stereotype\n 1. Makes disparaging attacks/insults against a specific person or group of people\n 1. Based on an outlandish premise about a group of people\n 1. Disparages against a characteristic that is not fixable and not measurable\n1. **Isn't grounded in reality**\n 1. Based on false information, or contains absurd assumptions\n 1. Uses sexual content (incest, bestiality, pedophilia) for shock value, and not to seek genuine answers\n ###### [Go to top](#top)",
"_____no_output_____"
],
[
"<a id=\"7\"></a> <br>\n## 4- Problem Feature\nProblem Definition has three steps that have illustrated in the picture below:\n\n1. Aim\n1. Variable\n1. Inputs & Outputs\n\n\n\n\n\n<a id=\"8\"></a> <br>\n### 4-1 Aim\nwe will be predicting whether a question asked on Quora is **sincere** or not.\n\n\n<a id=\"9\"></a> <br>\n### 4-2 Variables\n\n1. qid - unique question identifier\n1. question_text - Quora question text\n1. target - a question labeled \"insincere\" has a value of 1, otherwise 0\n\n<a id=\"10\"></a> <br>\n### 4-3 Inputs & Outputs\nwe use train.csv and test.csv as Input and we should upload a submission.csv as Output\n\n\n**<< Note >>**\n> You must answer the following question:\nHow does your company expect to use and benefit from **your model**.\n###### [Go to top](#top)",
"_____no_output_____"
],
[
"<a id=\"11\"></a> <br>\n## 5- Select Framework\nAfter problem definition and problem feature, we should select our framework to solve the problem.\nWhat we mean by the framework is that the programming languages you use and by what modules the problem will be solved.",
"_____no_output_____"
],
[
"<a id=\"12\"></a> <br>\n### 5-1 Import",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import train_test_split\nfrom sklearn.metrics import classification_report\nfrom sklearn.metrics import confusion_matrix\nfrom sklearn.metrics import accuracy_score\nfrom nltk.corpus import stopwords\nimport matplotlib.pylab as pylab\nimport matplotlib.pyplot as plt\nfrom pandas import get_dummies\nimport matplotlib as mpl\nimport seaborn as sns\nimport pandas as pd\nimport numpy as np\nimport matplotlib\nimport warnings\nimport sklearn\nimport string\nimport scipy\nimport numpy\nimport nltk\nimport json\nimport sys\nimport csv\nimport os",
"_____no_output_____"
]
],
[
[
"<a id=\"13\"></a> <br>\n### 5-2 version",
"_____no_output_____"
]
],
[
[
"print('matplotlib: {}'.format(matplotlib.__version__))\nprint('sklearn: {}'.format(sklearn.__version__))\nprint('scipy: {}'.format(scipy.__version__))\nprint('seaborn: {}'.format(sns.__version__))\nprint('pandas: {}'.format(pd.__version__))\nprint('numpy: {}'.format(np.__version__))\nprint('Python: {}'.format(sys.version))\n",
"matplotlib: 2.2.3\nsklearn: 0.20.0\nscipy: 1.1.0\nseaborn: 0.8.1\npandas: 0.23.4\nnumpy: 1.15.3\nPython: 3.6.6 |Anaconda, Inc.| (default, Oct 9 2018, 12:34:16) \n[GCC 7.3.0]\n"
]
],
[
[
"<a id=\"14\"></a> <br>\n### 5-3 Setup\n\nA few tiny adjustments for better **code readability**",
"_____no_output_____"
]
],
[
[
"sns.set(style='white', context='notebook', palette='deep')\npylab.rcParams['figure.figsize'] = 12,8\nwarnings.filterwarnings('ignore')\nmpl.style.use('ggplot')\nsns.set_style('white')\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"<a id=\"15\"></a> <br>\n## 6- EDA\n In this section, you'll learn how to use graphical and numerical techniques to begin uncovering the structure of your data. \n \n* Which variables suggest interesting relationships?\n* Which observations are unusual?\n* Analysis of the features!\n\nBy the end of the section, you'll be able to answer these questions and more, while generating graphics that are both insightful and beautiful. then We will review analytical and statistical operations:\n\n1. Data Collection\n1. Visualization\n1. Data Cleaning\n1. Data Preprocessing\n<img src=\"http://s9.picofile.com/file/8338476134/EDA.png\">\n\n ###### [Go to top](#top)",
"_____no_output_____"
],
[
"<a id=\"16\"></a> <br>\n## 6-1 Data Collection\n**Data collection** is the process of gathering and measuring data, information or any variables of interest in a standardized and established manner that enables the collector to answer or test hypothesis and evaluate outcomes of the particular collection.[techopedia]\n\nI start Collection Data by the training and testing datasets into **Pandas DataFrames**\n###### [Go to top](#top)",
"_____no_output_____"
]
],
[
[
"train = pd.read_csv('../input/train.csv')\ntest = pd.read_csv('../input/test.csv')",
"_____no_output_____"
]
],
[
[
"**<< Note 1 >>**\n\n* Each **row** is an observation (also known as : sample, example, instance, record)\n* Each **column** is a feature (also known as: Predictor, attribute, Independent Variable, input, regressor, Covariate)\n###### [Go to top](#top)",
"_____no_output_____"
]
],
[
[
"train.sample(1) ",
"_____no_output_____"
],
[
"test.sample(1) ",
"_____no_output_____"
]
],
[
[
"<a id=\"17\"></a> <br>\n## 6-1-1 Features\nFeatures can be from following types:\n* numeric\n* categorical\n* ordinal\n* datetime\n* coordinates\n\nFind the type of features in **Qoura dataset**?!\nfor getting some information about the dataset you can use **info()** command",
"_____no_output_____"
]
],
[
[
"print(train.info())",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 1306122 entries, 0 to 1306121\nData columns (total 3 columns):\nqid 1306122 non-null object\nquestion_text 1306122 non-null object\ntarget 1306122 non-null int64\ndtypes: int64(1), object(2)\nmemory usage: 29.9+ MB\nNone\n"
],
[
"print(test.info())",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 56370 entries, 0 to 56369\nData columns (total 2 columns):\nqid 56370 non-null object\nquestion_text 56370 non-null object\ndtypes: object(2)\nmemory usage: 880.9+ KB\nNone\n"
]
],
[
[
"<a id=\"18\"></a> <br>\n## 6-1-2 Explorer Dataset\n1- Dimensions of the dataset.\n\n2- Peek at the data itself.\n\n3- Statistical summary of all attributes.\n\n4- Breakdown of the data by the class variable.\n\nDon’t worry, each look at the data is **one command**. These are useful commands that you can use again and again on future projects.\n###### [Go to top](#top)",
"_____no_output_____"
]
],
[
[
"# shape\nprint('Shape of train:',train.shape)\nprint('Shape of train:',test.shape)",
"Shape of train: (1306122, 3)\nShape of train: (56370, 2)\n"
],
[
"#columns*rows\ntrain.size",
"_____no_output_____"
]
],
[
[
"After loading the data via **pandas**, we should checkout what the content is, description and via the following:",
"_____no_output_____"
]
],
[
[
"type(train)",
"_____no_output_____"
],
[
"type(test)",
"_____no_output_____"
]
],
[
[
"to pop up 5 random rows from the data set, we can use **sample(5)** function and find the type of features",
"_____no_output_____"
]
],
[
[
"train.sample(5) ",
"_____no_output_____"
]
],
[
[
"<a id=\"19\"></a> <br>\n## 6-2 Data Cleaning\nWhen dealing with real-world data, dirty data is the norm rather than the exception. We continuously need to predict correct values, impute missing ones, and find links between various data artefacts such as schemas and records. We need to stop treating data cleaning as a piecemeal exercise (resolving different types of errors in isolation), and instead leverage all signals and resources (such as constraints, available statistics, and dictionaries) to accurately predict corrective actions.\n\nThe primary goal of data cleaning is to detect and remove errors and **anomalies** to increase the value of data in analytics and decision making. While it has been the focus of many researchers for several years, individual problems have been addressed separately. These include missing value imputation, outliers detection, transformations, integrity constraints violations detection and repair, consistent query answering, deduplication, and many other related problems such as profiling and constraints mining.[4]\n###### [Go to top](#top)",
"_____no_output_____"
],
[
"how many NA elements in every column!!\nGood news, it is Zero!\nto check out how many null info are on the dataset, we can use **isnull().sum()**",
"_____no_output_____"
]
],
[
[
"train.isnull().sum()",
"_____no_output_____"
]
],
[
[
"But if we had , we can just use **dropna()**(be careful sometimes you should not do this!)",
"_____no_output_____"
]
],
[
[
"# remove rows that have NA's\nprint('Before Droping',train.shape)\ntrain = train.dropna()\nprint('After Droping',train.shape)",
"Before Droping (1306122, 3)\nAfter Droping (1306122, 3)\n"
]
],
[
[
"\nWe can get a quick idea of how many instances (rows) and how many attributes (columns) the data contains with the shape property.",
"_____no_output_____"
],
[
"to print dataset **columns**, we can use columns atribute",
"_____no_output_____"
]
],
[
[
"train.columns",
"_____no_output_____"
]
],
[
[
"you see number of unique item for Target with command below:",
"_____no_output_____"
]
],
[
[
"train_target = train['target'].values\n\nnp.unique(train_target)",
"_____no_output_____"
]
],
[
[
"YES, quora problem is a binary classification! :)",
"_____no_output_____"
],
[
"to check the first 5 rows of the data set, we can use head(5).",
"_____no_output_____"
]
],
[
[
"train.head(5) ",
"_____no_output_____"
]
],
[
[
"or to check out last 5 row of the data set, we use tail() function",
"_____no_output_____"
]
],
[
[
"train.tail() ",
"_____no_output_____"
]
],
[
[
"to give a **statistical summary** about the dataset, we can use **describe()",
"_____no_output_____"
]
],
[
[
"train.describe() ",
"_____no_output_____"
]
],
[
[
"**describe() is more useful for numerical data sets**",
"_____no_output_____"
],
[
"<a id=\"20\"></a> <br>\n## 6-3 Data Preprocessing\n**Data preprocessing** refers to the transformations applied to our data before feeding it to the algorithm.\n \nData Preprocessing is a technique that is used to convert the raw data into a clean data set. In other words, whenever the data is gathered from different sources it is collected in raw format which is not feasible for the analysis.\nthere are plenty of steps for data preprocessing and we just listed some of them in general(Not just for Quora) :\n* removing Target column (id)\n* Sampling (without replacement)\n* Making part of iris unbalanced and balancing (with undersampling and SMOTE)\n* Introducing missing values and treating them (replacing by average values)\n* Noise filtering\n* Data discretization\n* Normalization and standardization\n* PCA analysis\n* Feature selection (filter, embedded, wrapper)\n###### [Go to top](#top)",
"_____no_output_____"
],
[
"**<< Note 2 >>**\nin pandas's data frame you can perform some query such as \"where\"",
"_____no_output_____"
]
],
[
[
"train.where(train ['target']==1).count()",
"_____no_output_____"
]
],
[
[
"as you can see in the below in python, it is so easy perform some query on the dataframe:",
"_____no_output_____"
]
],
[
[
"train[train['target']>1]",
"_____no_output_____"
],
[
"train[train['target']==1].head(5)",
"_____no_output_____"
]
],
[
[
"<a id=\"21\"></a> <br>\n## 6-3-1 Is data set imbalance?",
"_____no_output_____"
]
],
[
[
"train_target.mean()",
"_____no_output_____"
]
],
[
[
"A large part of the data is unbalanced, but **how can we solve it?**",
"_____no_output_____"
]
],
[
[
"train[\"target\"].value_counts()\n# data is imbalance",
"_____no_output_____"
]
],
[
[
"<a id=\"22\"></a> <br>\n## 6-3-2 Some Feature Engineering",
"_____no_output_____"
],
[
"[NLTK](https://www.nltk.org/) is one of the leading platforms for working with human language data and Python, the module NLTK is used for natural language processing. NLTK is literally an acronym for Natural Language Toolkit.\n\nWe get a set of **English stop** words using the line",
"_____no_output_____"
]
],
[
[
"#from nltk.corpus import stopwords\neng_stopwords = set(stopwords.words(\"english\"))",
"_____no_output_____"
]
],
[
[
"The returned list stopWords contains **179 stop words** on my computer.\nYou can view the length or contents of this array with the lines:",
"_____no_output_____"
]
],
[
[
"print(len(eng_stopwords))\nprint(eng_stopwords)",
"179\n{'ourselves', 'most', 'here', 'after', 'up', 'over', 'll', 'our', 'ma', 'out', 'who', 'yourself', 'until', 'can', 'on', 'don', 'y', 'only', 'own', \"shouldn't\", 't', \"you're\", 'below', 'all', 'yourselves', 'm', 'now', 'a', 'i', 'are', 'too', 'o', 'wouldn', 'does', 'my', 'his', 'do', 'against', 'weren', \"hasn't\", 'were', \"haven't\", 'isn', 'myself', 'couldn', 'through', 'to', 'did', \"isn't\", 'its', 're', \"doesn't\", 'their', 'both', \"that'll\", 'where', 'no', 'what', 'themselves', 'is', \"aren't\", 'down', 'then', \"it's\", \"should've\", 'but', \"weren't\", 'as', 'them', 'mustn', 'of', 'whom', 'he', \"hadn't\", 'any', \"wouldn't\", 'we', 'have', 'has', 'off', \"didn't\", 'wasn', 'hers', 'you', 'they', 'this', 'between', 'shan', 'yours', 'doing', 'didn', \"won't\", 'be', 'in', 'very', 'theirs', 'being', 'if', 'having', 'itself', 'by', 'needn', 'and', \"she's\", 'there', 'not', 'hadn', 'ain', 'it', 'above', \"mustn't\", 'further', 'herself', 'before', 'was', 'other', 'hasn', 'or', 'why', 'aren', 'been', 'just', 'nor', \"you've\", 'her', 'which', 'an', 'each', 'once', \"wasn't\", 'how', 'about', 'won', 'd', 'had', 'than', 'haven', \"needn't\", 'under', 'into', 'she', 've', 'shouldn', 'these', 'such', 'when', 'those', 'him', 'during', 'for', 'that', 'doesn', 'same', 'at', 'some', 'with', 'your', 'am', 'will', \"mightn't\", 'himself', 's', 'so', \"you'd\", 'the', 'because', 'ours', 'from', \"couldn't\", \"you'll\", 'should', 'few', 'more', 'mightn', 'me', \"shan't\", \"don't\", 'while', 'again'}\n"
]
],
[
[
"The metafeatures that we'll create based on SRK's EDAs, [sudalairajkumar](http://http://www.kaggle.com/sudalairajkumar/simple-feature-engg-notebook-spooky-author) and [tunguz](https://www.kaggle.com/tunguz/just-some-simple-eda) are:\n* Number of words in the text\n* Number of unique words in the text\n* Number of characters in the text\n* Number of stopwords\n* Number of punctuations\n* Number of upper case words\n* Number of title case words\n* Average length of the words\n\n###### [Go to top](#top)",
"_____no_output_____"
],
[
"Number of words in the text ",
"_____no_output_____"
]
],
[
[
"train[\"num_words\"] = train[\"question_text\"].apply(lambda x: len(str(x).split()))\ntest[\"num_words\"] = test[\"question_text\"].apply(lambda x: len(str(x).split()))\nprint('maximum of num_words in train',train[\"num_words\"].max())\nprint('min of num_words in train',train[\"num_words\"].min())\nprint(\"maximum of num_words in test\",test[\"num_words\"].max())\nprint('min of num_words in train',test[\"num_words\"].min())\n",
"maximum of num_words in train 134\nmin of num_words in train 1\nmaximum of num_words in test 87\nmin of num_words in train 2\n"
]
],
[
[
"Number of unique words in the text",
"_____no_output_____"
]
],
[
[
"train[\"num_unique_words\"] = train[\"question_text\"].apply(lambda x: len(set(str(x).split())))\ntest[\"num_unique_words\"] = test[\"question_text\"].apply(lambda x: len(set(str(x).split())))\nprint('maximum of num_unique_words in train',train[\"num_unique_words\"].max())\nprint('mean of num_unique_words in train',train[\"num_unique_words\"].mean())\nprint(\"maximum of num_unique_words in test\",test[\"num_unique_words\"].max())\nprint('mean of num_unique_words in train',test[\"num_unique_words\"].mean())",
"maximum of num_unique_words in train 96\nmean of num_unique_words in train 12.135776749798257\nmaximum of num_unique_words in test 61\nmean of num_unique_words in train 12.096363313819408\n"
]
],
[
[
"Number of characters in the text ",
"_____no_output_____"
]
],
[
[
"\ntrain[\"num_chars\"] = train[\"question_text\"].apply(lambda x: len(str(x)))\ntest[\"num_chars\"] = test[\"question_text\"].apply(lambda x: len(str(x)))\nprint('maximum of num_chars in train',train[\"num_chars\"].max())\nprint(\"maximum of num_chars in test\",test[\"num_chars\"].max())",
"maximum of num_chars in train 1017\nmaximum of num_chars in test 588\n"
]
],
[
[
"Number of stopwords in the text",
"_____no_output_____"
]
],
[
[
"train[\"num_stopwords\"] = train[\"question_text\"].apply(lambda x: len([w for w in str(x).lower().split() if w in eng_stopwords]))\ntest[\"num_stopwords\"] = test[\"question_text\"].apply(lambda x: len([w for w in str(x).lower().split() if w in eng_stopwords]))\nprint('maximum of num_stopwords in train',train[\"num_stopwords\"].max())\nprint(\"maximum of num_stopwords in test\",test[\"num_stopwords\"].max())",
"maximum of num_stopwords in train 56\nmaximum of num_stopwords in test 47\n"
]
],
[
[
"Number of punctuations in the text",
"_____no_output_____"
]
],
[
[
"\ntrain[\"num_punctuations\"] =train['question_text'].apply(lambda x: len([c for c in str(x) if c in string.punctuation]) )\ntest[\"num_punctuations\"] =test['question_text'].apply(lambda x: len([c for c in str(x) if c in string.punctuation]) )\nprint('maximum of num_punctuations in train',train[\"num_punctuations\"].max())\nprint(\"maximum of num_punctuations in test\",test[\"num_punctuations\"].max())",
"maximum of num_punctuations in train 411\nmaximum of num_punctuations in test 260\n"
]
],
[
[
"Number of title case words in the text",
"_____no_output_____"
]
],
[
[
"\ntrain[\"num_words_upper\"] = train[\"question_text\"].apply(lambda x: len([w for w in str(x).split() if w.isupper()]))\ntest[\"num_words_upper\"] = test[\"question_text\"].apply(lambda x: len([w for w in str(x).split() if w.isupper()]))\nprint('maximum of num_words_upper in train',train[\"num_words_upper\"].max())\nprint(\"maximum of num_words_upper in test\",test[\"num_words_upper\"].max())",
"maximum of num_words_upper in train 37\nmaximum of num_words_upper in test 36\n"
]
],
[
[
"Number of title case words in the text",
"_____no_output_____"
]
],
[
[
"\ntrain[\"num_words_title\"] = train[\"question_text\"].apply(lambda x: len([w for w in str(x).split() if w.istitle()]))\ntest[\"num_words_title\"] = test[\"question_text\"].apply(lambda x: len([w for w in str(x).split() if w.istitle()]))\nprint('maximum of num_words_title in train',train[\"num_words_title\"].max())\nprint(\"maximum of num_words_title in test\",test[\"num_words_title\"].max())",
"maximum of num_words_title in train 37\nmaximum of num_words_title in test 24\n"
]
],
[
[
" Average length of the words in the text ",
"_____no_output_____"
]
],
[
[
"\ntrain[\"mean_word_len\"] = train[\"question_text\"].apply(lambda x: np.mean([len(w) for w in str(x).split()]))\ntest[\"mean_word_len\"] = test[\"question_text\"].apply(lambda x: np.mean([len(w) for w in str(x).split()]))\nprint('mean_word_len in train',train[\"mean_word_len\"].max())\nprint(\"mean_word_len in test\",test[\"mean_word_len\"].max())",
"mean_word_len in train 57.666666666666664\nmean_word_len in test 29.333333333333332\n"
]
],
[
[
"we add some new feature to train and test data set now, print columns agains",
"_____no_output_____"
]
],
[
[
"print(train.columns)\ntrain.head(1)",
"Index(['qid', 'question_text', 'target', 'num_words', 'num_unique_words',\n 'num_chars', 'num_stopwords', 'num_punctuations', 'num_words_upper',\n 'num_words_title', 'mean_word_len'],\n dtype='object')\n"
]
],
[
[
"**<< Note >>**\n>**Preprocessing and generation pipelines depend on a model type**",
"_____no_output_____"
],
[
"<a id=\"23\"></a> <br>\n## 6-4 Data Visualization\n**Data visualization** is the presentation of data in a pictorial or graphical format. It enables decision makers to see analytics presented visually, so they can grasp difficult concepts or identify new patterns.\n\n> * Two** important rules** for Data visualization:\n> 1. Do not put too little information\n> 1. Do not put too much information\n\n###### [Go to top](#top)",
"_____no_output_____"
],
[
"<a id=\"61\"></a> <br>\n## 6-4-1 countplot",
"_____no_output_____"
]
],
[
[
"ax=sns.countplot(x='target',hue=\"target\", data=train ,linewidth=5,edgecolor=sns.color_palette(\"dark\", 3))\nplt.title('Is data set imbalance?');",
"_____no_output_____"
],
[
"ax = sns.countplot(y=\"target\", hue=\"target\", data=train)\nplt.title('Is data set imbalance?');",
"_____no_output_____"
]
],
[
[
"<a id=\"62\"></a> <br>\n## 6-4-2 pie plot",
"_____no_output_____"
]
],
[
[
"\nax=train['target'].value_counts().plot.pie(explode=[0,0.1],autopct='%1.1f%%' ,shadow=True)\nax.set_title('target')\nax.set_ylabel('')\nplt.show()",
"_____no_output_____"
],
[
"#plt.pie(train['target'],autopct='%1.1f%%')\n \n#plt.axis('equal')\n#plt.show()",
"_____no_output_____"
]
],
[
[
"<a id=\"63\"></a> <br>\n## 6-4-3 Histogram",
"_____no_output_____"
]
],
[
[
"f,ax=plt.subplots(1,2,figsize=(20,10))\ntrain[train['target']==0].num_words.plot.hist(ax=ax[0],bins=20,edgecolor='black',color='red')\nax[0].set_title('target= 0')\nx1=list(range(0,85,5))\nax[0].set_xticks(x1)\ntrain[train['target']==1].num_words.plot.hist(ax=ax[1],color='green',bins=20,edgecolor='black')\nax[1].set_title('target= 1')\nx2=list(range(0,85,5))\nax[1].set_xticks(x2)\nplt.show()",
"_____no_output_____"
],
[
"f,ax=plt.subplots(1,2,figsize=(18,8))\ntrain[['target','num_words']].groupby(['target']).mean().plot.bar(ax=ax[0])\nax[0].set_title('num_words vs Sex')\nsns.countplot('num_words',hue='target',data=train,ax=ax[1])\nax[1].set_title('num_words:target=0 vs target=1')\nplt.show()",
"_____no_output_____"
],
[
"# histograms\ntrain.hist(figsize=(15,20))\nplt.figure()",
"_____no_output_____"
],
[
"train[\"num_words\"].hist();",
"_____no_output_____"
]
],
[
[
"<a id=\"64\"></a> <br>\n## 6-4-4 violin plot",
"_____no_output_____"
]
],
[
[
"sns.violinplot(data=train,x=\"target\", y=\"num_words\")",
"_____no_output_____"
]
],
[
[
"<a id=\"65\"></a> <br>\n## 6-4-5 kdeplot",
"_____no_output_____"
]
],
[
[
"sns.FacetGrid(train, hue=\"target\", size=5).map(sns.kdeplot, \"num_words\").add_legend()\nplt.show()",
"_____no_output_____"
]
],
[
[
"<a id=\"24\"></a> <br>\n## 7- Apply Learning\nComing soon",
"_____no_output_____"
],
[
"-----------------\n<a id=\"25\"></a> <br>\n# 8- Conclusion",
"_____no_output_____"
],
[
"This kernel is not completed yet , I have tried to cover all the parts related to the process of **Quora problem** with a variety of Python packages and I know that there are still some problems then I hope to get your feedback to improve it.\n",
"_____no_output_____"
],
[
"you can Fork and Run this kernel on Github:\n> ###### [ GitHub](https://github.com/mjbahmani/10-steps-to-become-a-data-scientist)\n\n--------------------------------------\n\n **I hope you find this kernel helpful and some <font color=\"red\"><b>UPVOTES</b></font> would be very much appreciated** ",
"_____no_output_____"
],
[
"<a id=\"26\"></a> <br>\n\n-----------\n\n# 9- References\n## 9-1 Kaggle's Kernels\n**In the end , I want to thank all the kernels I've used in this notebook**:\n1. [SRK](https://www.kaggle.com/sudalairajkumar/simple-exploration-notebook-qiqc)\n1. [mihaskalic](https://www.kaggle.com/mihaskalic/lstm-is-all-you-need-well-maybe-embeddings-also)\n1. [artgor](https://www.kaggle.com/artgor/eda-and-lstm-cnn)\n1. [tunguz](https://www.kaggle.com/tunguz/just-some-simple-eda)\n\n## 9-2 other references\n\n1. [Machine Learning Certification by Stanford University (Coursera)](https://www.coursera.org/learn/machine-learning/)\n\n1. [Machine Learning A-Z™: Hands-On Python & R In Data Science (Udemy)](https://www.udemy.com/machinelearning/)\n\n1. [Deep Learning Certification by Andrew Ng from deeplearning.ai (Coursera)](https://www.coursera.org/specializations/deep-learning)\n\n1. [Python for Data Science and Machine Learning Bootcamp (Udemy)](Python for Data Science and Machine Learning Bootcamp (Udemy))\n\n1. [Mathematics for Machine Learning by Imperial College London](https://www.coursera.org/specializations/mathematics-machine-learning)\n\n1. [Deep Learning A-Z™: Hands-On Artificial Neural Networks](https://www.udemy.com/deeplearning/)\n\n1. [Complete Guide to TensorFlow for Deep Learning Tutorial with Python](https://www.udemy.com/complete-guide-to-tensorflow-for-deep-learning-with-python/)\n\n1. [Data Science and Machine Learning Tutorial with Python – Hands On](https://www.udemy.com/data-science-and-machine-learning-with-python-hands-on/)\n-------------\n###### [Go to top](#top)",
"_____no_output_____"
],
[
"### The kernel is not complete and will be updated soon !!!",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
4a54a2821331ef3c37e5365562de1976648231b5
| 17,430 |
ipynb
|
Jupyter Notebook
|
OCR_Plagiarism_Percentage_Checker.ipynb
|
Tanishq45/OCR_Plagiarism_Percentage_Checker
|
83ff4042f7938a32fd81d968206fc00f6caf2d8c
|
[
"MIT"
] | null | null | null |
OCR_Plagiarism_Percentage_Checker.ipynb
|
Tanishq45/OCR_Plagiarism_Percentage_Checker
|
83ff4042f7938a32fd81d968206fc00f6caf2d8c
|
[
"MIT"
] | null | null | null |
OCR_Plagiarism_Percentage_Checker.ipynb
|
Tanishq45/OCR_Plagiarism_Percentage_Checker
|
83ff4042f7938a32fd81d968206fc00f6caf2d8c
|
[
"MIT"
] | null | null | null | 34.176471 | 107 | 0.593345 |
[
[
[
"import requests\nimport pytesseract\nimport os\nfrom sklearn.feature_extraction.text import TfidfVectorizer\nfrom sklearn.metrics.pairwise import cosine_similarity\nprint ('Starting to Download!')\nurl = 'https://images.sampletemplates.com/wp-content/uploads/2017/04/Technical-Paper-Example1.jpg'\nr = requests.get(url)\nfilename = '2.jpg'\nwith open(filename, 'wb') as out_file:\n out_file.write(r.content)\nprint(\"Download complete!\")",
"Starting to Download!\nDownload complete!\n"
],
[
"print ('Starting to Download!')\nurl = 'https://images.template.net/wp-content/uploads/2017/06/Technical-Support-White-Paper.jpg'\nr = requests.get(url)\nfilename_2 = '3.png'\nwith open(filename_2, 'wb') as out_file:\n out_file.write(r.content)\nprint(\"Download complete!\")",
"Starting to Download!\nDownload complete!\n"
],
[
"pytesseract.pytesseract.tesseract_cmd = r'C:\\Users\\ajayb\\AppData\\Local\\tesseract.exe'",
"_____no_output_____"
],
[
"import cv2\nimport numpy as np\nimport pytesseract\nfrom PIL import Image\ndef get_string(img_path):\n img = cv2.imread(img_path)\n img = cv2.cvtColor(img, cv2.COLOR_BGR2GRAY)\n kernel = np.ones((1, 1), np.uint8)\n img = cv2.dilate(img, kernel, iterations=1)\n img = cv2.erode(img, kernel, iterations=1)\n cv2.imwrite(\"removed_noise.png\", img)\n cv2.imwrite(img_path, img)\n result = pytesseract.image_to_string(Image.open(img_path))\n return result\nprint ('--- Start recognize text from image ---')\nprint (get_string(filename))\nprint (\"------ Done -------\")",
"--- Start recognize text from image ---\n‘Technical Research Paper\n“Microcontroller based Fault Detector”\n\nGENERAL Abstract\n\nPurpose: The aim of this research is to develop a device used to detect faults in the\nline and isolate the connected system or instrument connected to it\n\nScope: This device involves the use of microcontroller for detection and isolation of\nthe system of instrument with proper use of programming. The instrument devised is\neconomical and effective compared to other protective devices available in market.\n\nAfter the patent of the product a large scale production is also possible for consumer\n\nDesign Approach: The design methodology involves the use of microcontroller in\nconjugation with the relay circuitry with display on a LCD screen. It isa totally new\ndesign in the market and it will bea substitute to ELCB’s, MCB’s and Relays in near\nfuture\n\n \n\n \n\nPractical Implications: The circuit devised can be used in conjugation with the\n‘medical instrument, industrial instrument and even in the household application. It finds\n«alot of applications in factories and industries where costly instruments are to be saved\nfrom faults.\n\nConclusion: The device thus developed is very economical and effective in the use of\nprotection in household as well as industrial instrument protection, The added\nadvantage of this circuit is that it shows the type of fault occurred on LCD and at the\nsame time keeps the circuit in isolation until the fault is cleared.\n\nINTRODUCTION\n\n> A faut in electrical equipment is defined asa defect in its electrical circuit due to\n‘which the current is diverted from the intended path,\n\n> Faults are generally caused by mechanical failure, accidents, excessive internal and\nextemal stresses ete\n\n> The fault impedance being low, the fault currents are relatively high. During the faults,\nthe power flow is diverted towards the fault and the supply to the neighboring zone is,\naffected. Voltages become unbalanced.\n\f\n------ Done -------\n"
],
[
"def get_string_2(img_path1):\n img1= cv2.imread(img_path1)\n img1 = cv2.cvtColor(img1, cv2.COLOR_BGR2GRAY)\n kernel = np.ones((1, 1), np.uint8)\n img1 = cv2.dilate(img1, kernel, iterations=1)\n img1 = cv2.erode(img1, kernel, iterations=1)\n cv2.imwrite(\"removed_noise.png\", img1)\n cv2.imwrite(img_path1, img1)\n result = pytesseract.image_to_string(Image.open(img_path1))\n return result\nprint ('--- Start recognize text from image ---')\nprint (get_string_2(filename_2))\nprint (\"------ Done -------\")",
"--- Start recognize text from image ---\nTechnical Support White Paper\n\nDate: August 3, 2007\n\nTitle: Controlé USB Recovery documentation - using the Recovery utility to\nrestore a Control4 device to factory default condition\n\nProduct(s) affected:\n+ Control4 Media Controller, Home Theater Controller, Home Controller HC-300, Min\nTouch Screen v1 & v2, Speaker Point\n\nTopicts)\n+ How to restore a Control device to its factory default condition.\n\nSummary:\nThe Controls Recovery utit wil enable Control dealers and installers to minimize the\ntime, paperwork, inconvenience, and other costs associated with devices that fail during the\nupdate proces or at some other time, I wil quickly become a valuable tol in your\nControl arsenal\n\n(Occasionally, and most likely during an update process, a Contrld device wil become\ninoperable. At this point the Dealer / Installer cals Tech Support and troubleshoot the\nevice. if unable to restore the device using Tenet or other measures indicated by Tech\nSupport, an RMA will be issued. An RMA is inconvenient and costly both for Controld and\nforthe Dealer / Installer due to increased time on the ob, shipping charges, anda probable\nreturn trip to the customer site\n\n‘Most ofthe devices which are retumed via RMA canbe restored to a fully functional\ncondition. In order to enable Tech Support and the Dealer / Installer to have the ability to\n‘estore devices inthe field, Control has developed the Controlé Recovery utity. Now, ia\ndevice doesn't update correctly or in some other way becomes inoperable, the Dealer |\nInstaller can recover that device immediately. This reduces the time, expense, and\ninconvenience for both partes incurred through the RMA process,\n\f\n------ Done -------\n"
],
[
"if os.path.exists(\"12.txt\"):\n os.remove(\"12.txt\")\n \n # Print the statement once\n # the file is deleted \n print(\"File deleted !\") \n \nelse:\n \n # Print if file is not present \n print(\"File doesnot exist !\") ",
"File deleted !\n"
],
[
"f = open(\"12.txt\", \"x\")\nf = open(\"12.txt\", \"a\")\nf.write(get_string(filename))\nf.close()\nf = open(\"12.txt\", \"r\")\nprint(f.read())\nf.close()",
"Technical Research Paper\n“Microcontroller based Fault Detector”\n\nGENERAL Abstract\n\nPurpose: The aim of this research is to develop a device used to detect faults in the\nline and isolate the connected system or instrument connected to it\n\nSeope: This device involves the use of microcontroller for detection and isolation of\nthe system of instrument with proper use of programming. The instrument devised is\neconomical and effective compared to other protective devices available in market,\n‘After the patent of the product « large scale production is also possible for consumer\nDesign Approach: The design methodology involves the use of microcontroller in\nconjugation with the relay circuitry with display on a LCD screen. It isa totally new\ndesign in the market and it will bea substitute to ELCB’s, MCB’s and Relays in near\nfuture\n\n \n\n \n\nPractical Implications: The circuit devised can be used in conjugation with the\n‘medical instrument, industrial instrument and even in the household application. It finds\n«alot of applications in factories and industries where costly instruments are to be saved\nfrom faults.\n\nConclusion: ‘The device thus developed is very economical and effective in the use of\nprotection in household as well as industrial instrument protection, The added\nadvantage of this circuit is that it shows the type of fault occurred on LCD and at the\nsame time keeps the circuit in isolation until the fault is cleared.\n\nINTRODUCTION\n\n> A faut in electrical equipment is defined as a defect in its electrical circuit due to\n‘which the current is diverted from the intended path,\n\n> Faults are generally caused by mechanical failure, accidents, excessive internal and\nextemal stresses ete\n\n> The fault impedance being low, the fault currents are relatively high. During the faults,\nthe power flow is diverted towards the fault and the supply to the neighboring zone is,\naffected. Voltages become unbalanced.\n\f\n"
],
[
"if os.path.exists(\"13.txt\"):\n os.remove(\"13.txt\")\n \n # Print the statement once\n # the file is deleted \n print(\"File deleted !\") \n \nelse:\n \n # Print if file is not present \n print(\"File doesnot exist !\") ",
"File deleted !\n"
],
[
"g = open(\"13.txt\", \"x\")\ng = open(\"13.txt\", \"a\")\ng.write(get_string(filename_2))\ng.close()\ng = open(\"13.txt\", \"r\")\nprint(g.read())\ng.close()",
"Technical Support White Paper\n\nDate: August 3, 2007\n\nTitle: Controlé USB Recovery documentation - using the Recovery utility to\nrestore a Control4 device to factory default condition\n\nProduct(s) affected:\n+ Control4 Media Controller, Home Theater Controller, Home Controller HC-300, Min\nTouch Screen v1 & v2, Speaker Point\n\nTopicts)\n+ How to restore a Control device to its factory default condition.\n\nSummary:\nThe Controls Recovery utit wil enable Control dealers and installers to minimize the\ntime, paperwork, inconvenience, and other costs associated with devices that fail during the\nupdate proces or at some other time, I wil quickly become a valuable tol in your\nControl arsenal\n\n(Occasionally, and most likely during an update process, a Contrld device wil become\ninoperable. At this point the Dealer / Installer cals Tech Support and troubleshoot the\nevice. if unable to restore the device using Tenet or other measures indicated by Tech\nSupport, an RMA will be issued. An RMA is inconvenient and costly both for Controld and\nforthe Dealer / Installer due to increased time on the ob, shipping charges, anda probable\nreturn trip to the customer site\n\n‘Most ofthe devices which are retumed via RMA canbe restored to a fully functional\ncondition. In order to enable Tech Support and the Dealer / Installer to have the ability to\n‘estore devices inthe field, Control has developed the Controlé Recovery utity. Now, ia\ndevice doesn't update correctly or in some other way becomes inoperable, the Dealer |\nInstaller can recover that device immediately. This reduces the time, expense, and\ninconvenience for both partes incurred through the RMA process,\n\f\n"
],
[
"student_files = [doc for doc in os.listdir() if doc.endswith('.txt')]\nstudent_notes =[open(File).read() for File in student_files]",
"_____no_output_____"
],
[
"vectorize = lambda Text: TfidfVectorizer().fit_transform(Text).toarray()\nsimilarity = lambda doc1, doc2: cosine_similarity([doc1, doc2])",
"_____no_output_____"
],
[
"vectors = vectorize(student_notes)\ns_vectors = list(zip(student_files, vectors))\nplagiarism_results = set()",
"_____no_output_____"
],
[
"def check_plagiarism():\n global s_vectors\n for student_a, text_vector_a in s_vectors:\n new_vectors =s_vectors.copy()\n current_index = new_vectors.index((student_a, text_vector_a))\n del new_vectors[current_index]\n for student_b , text_vector_b in new_vectors:\n sim_score = similarity(text_vector_a, text_vector_b)[0][1]\n student_pair = sorted((student_a, student_b))\n score = (student_pair[0], student_pair[1],sim_score)\n plagiarism_results.add(score)\n return plagiarism_results",
"_____no_output_____"
],
[
"for data in check_plagiarism():\n print(data)",
"('12.txt', '13.txt', 0.46045626081270197)\n"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a54ab00094e22ae2c40952b4365ba9674d8e8bf
| 4,477 |
ipynb
|
Jupyter Notebook
|
labs/lab10.ipynb
|
Bastian951/mat281_portfolio
|
ffdb44a985e9bebf93ae3ab9c9dd4086a7860114
|
[
"MIT"
] | null | null | null |
labs/lab10.ipynb
|
Bastian951/mat281_portfolio
|
ffdb44a985e9bebf93ae3ab9c9dd4086a7860114
|
[
"MIT"
] | null | null | null |
labs/lab10.ipynb
|
Bastian951/mat281_portfolio
|
ffdb44a985e9bebf93ae3ab9c9dd4086a7860114
|
[
"MIT"
] | null | null | null | 24.2 | 212 | 0.579182 |
[
[
[
"# Laboratorio 10",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nimport numpy as np\nimport pandas as pd\nfrom sklearn.datasets import load_breast_cancer\nfrom sklearn.model_selection import train_test_split, GridSearchCV\nfrom sklearn.linear_model import LogisticRegression\nfrom sklearn.neighbors import KNeighborsClassifier\nfrom sklearn.metrics import plot_confusion_matrix, classification_report\n\n%matplotlib inline",
"_____no_output_____"
],
[
"breast_cancer = load_breast_cancer()\nX, y = breast_cancer.data, breast_cancer.target\nX_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, random_state=42)\ntarget_names = breast_cancer.target_names",
"_____no_output_____"
]
],
[
[
"## Ejercicio 1\n\n(1 pto.)\n\nAjusta una regresión logística a los datos de entrenamiento y obtén el _accuracy_ con los datos de test. Utiliza el argumento `n_jobs` igual a $-1$, si aún así no converge aumenta el valor de `max_iter`.\n\nHint: Recuerda que el _accuracy_ es el _score_ por defecto en los modelos de clasificación de scikit-learn.",
"_____no_output_____"
]
],
[
[
"lr = LogisticRegression(max_iter=3000,n_jobs=-1)\nlr.fit(X_train, y_train)\nprint(f\"Logistic Regression accuracy: {lr.score(X_test,y_test)}\")",
"Logistic Regression accuracy: 0.9707602339181286\n"
]
],
[
[
"## Ejercicio 2\n\n(1 pto.)\n\nUtiliza `GridSearchCV` con 5 _folds_ para encontrar el mejor valor de `n_neighbors` de un modelo KNN.",
"_____no_output_____"
]
],
[
[
"knn = KNeighborsClassifier(n_neighbors=5)\nknn_grid = {'n_neighbors': np.arange(2, 31)}\n\nknn_cv = GridSearchCV(\n cv=5,\n estimator=knn,\n param_grid=knn_grid\n)\n\nknn_cv.fit(X_train, y_train)",
"_____no_output_____"
],
[
"print(f\"KNN accuray: {knn_cv.fit(X_train, y_train).score(X_test,y_test)}\")",
"_____no_output_____"
]
],
[
[
"## Ejercicio 3\n\n(1 pto.)\n\n¿Cuál modelo escogerías basándote en los resultados anteriores? Justifica",
"_____no_output_____"
],
[
"__Respuesta:__ Eligiria el modelo de regresión logistica pues tiene un score mayor que el score del modelo KNN_cv",
"_____no_output_____"
],
[
"## Ejercicio 4\n\n(1 pto.)\n\nPara el modelo seleccionado en el ejercicio anterior.\n\n* Grafica la matriz de confusión (no olvides colocar los nombres originales en los _labels_).\n* Imprime el reporte de clasificación.",
"_____no_output_____"
]
],
[
[
"plot_confusion_matrix(lr, X_test, y_test, display_labels=target_names)\nplt.show()",
"_____no_output_____"
],
[
"y_pred= lr.predict(X_test)\nprint(classification_report(y_test, y_pred, target_names=breast_cancer.target_names))",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
]
] |
4a54ab680b213459f77d8737ab04c8e7baba90c1
| 46,274 |
ipynb
|
Jupyter Notebook
|
Neural Networks and Deep Learning/Logistic Regression with a Neural Network mindset.ipynb
|
Ulysses-WJL/deep-learning-coursera
|
db76fdd20a0d7aaca9f9b33c33e8c72cb4d3e021
|
[
"MIT"
] | null | null | null |
Neural Networks and Deep Learning/Logistic Regression with a Neural Network mindset.ipynb
|
Ulysses-WJL/deep-learning-coursera
|
db76fdd20a0d7aaca9f9b33c33e8c72cb4d3e021
|
[
"MIT"
] | null | null | null |
Neural Networks and Deep Learning/Logistic Regression with a Neural Network mindset.ipynb
|
Ulysses-WJL/deep-learning-coursera
|
db76fdd20a0d7aaca9f9b33c33e8c72cb4d3e021
|
[
"MIT"
] | null | null | null | 35.135915 | 434 | 0.541125 |
[
[
[
"# Logistic Regression with a Neural Network mindset\n\nWelcome to your first (required) programming assignment! You will build a logistic regression classifier to recognize cats. This assignment will step you through how to do this with a Neural Network mindset, and so will also hone your intuitions about deep learning.\n\n**Instructions:**\n- Do not use loops (for/while) in your code, unless the instructions explicitly ask you to do so.\n\n**You will learn to:**\n- Build the general architecture of a learning algorithm, including:\n - Initializing parameters\n - Calculating the cost function and its gradient\n - Using an optimization algorithm (gradient descent) \n- Gather all three functions above into a main model function, in the right order.",
"_____no_output_____"
],
[
"## 1 - Packages ##\n\nFirst, let's run the cell below to import all the packages that you will need during this assignment. \n- [numpy](www.numpy.org) is the fundamental package for scientific computing with Python.\n- [h5py](http://www.h5py.org) is a common package to interact with a dataset that is stored on an H5 file.\n- [matplotlib](http://matplotlib.org) is a famous library to plot graphs in Python.\n- [PIL](http://www.pythonware.com/products/pil/) and [scipy](https://www.scipy.org/) are used here to test your model with your own picture at the end.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\nimport h5py\nimport scipy\nfrom PIL import Image\n# from scipy import ndimage\n# from lr_utils import load_dataset\n\nnp.set_printoptions(threshold=16, suppress=True, precision=5)\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"## 2 - Overview of the Problem set ##\n\n**Problem Statement**: You are given a dataset (\"data.h5\") containing:\n - a training set of m_train images labeled as cat (y=1) or non-cat (y=0)\n - a test set of m_test images labeled as cat or non-cat\n - each image is of shape (num_px, num_px, 3) where 3 is for the 3 channels (RGB). Thus, each image is square (height = num_px) and (width = num_px).\n\nYou will build a simple image-recognition algorithm that can correctly classify pictures as cat or non-cat.\n\nLet's get more familiar with the dataset. Load the data by running the following code.",
"_____no_output_____"
]
],
[
[
"def load_dataset():\n # load dataset from h5 file\n train_dataset = h5py.File('datasets/train_catvnoncat.h5', \"r\")\n train_set_x_orig = np.array(train_dataset[\"train_set_x\"][:]) # your train set features\n train_set_y_orig = np.array(train_dataset[\"train_set_y\"][:]) # your train set labels\n\n test_dataset = h5py.File('datasets/test_catvnoncat.h5', \"r\")\n test_set_x_orig = np.array(test_dataset[\"test_set_x\"][:]) # your test set features\n test_set_y_orig = np.array(test_dataset[\"test_set_y\"][:]) # your test set labels\n\n classes = np.array(test_dataset[\"list_classes\"][:]) # the list of classes\n \n train_set_y_orig = train_set_y_orig.reshape((-1, 1))\n test_set_y_orig = test_set_y_orig.reshape((-1, 1))\n \n return train_set_x_orig, train_set_y_orig, test_set_x_orig, test_set_y_orig, classes",
"_____no_output_____"
]
],
[
[
"We added \"_orig\" at the end of image datasets (train and test) because we are going to preprocess them. After preprocessing, we will end up with train_set_x and test_set_x (the labels train_set_y and test_set_y don't need any preprocessing).\n\nEach line of your train_set_x_orig and test_set_x_orig is an array representing an image. You can visualize an example by running the following code. Feel free also to change the `index` value and re-run to see other images. ",
"_____no_output_____"
]
],
[
[
"# Loading the data (cat/non-cat)\ntrain_set_x_orig, train_set_y, test_set_x_orig, test_set_y, classes = load_dataset()",
"_____no_output_____"
],
[
"train_set_x_orig.shape # 209 张图片 大小64 × 64 RGB",
"_____no_output_____"
],
[
"train_set_y.shape",
"_____no_output_____"
],
[
"classes # 0: non-cat 1: cat",
"_____no_output_____"
],
[
"train_set_y",
"_____no_output_____"
],
[
"np.squeeze(train_set_y[2, :]) # Remove single-dimensional entries from the shape of an array.",
"_____no_output_____"
],
[
"# Example of a picture\nindex = 25\nplt.imshow(train_set_x_orig[index])\nprint(\"y = \" + str(train_set_y[index,:]) + \", it's a '\" + classes[np.squeeze(train_set_y[index, :])].decode(\"utf-8\") + \"' picture.\")",
"_____no_output_____"
]
],
[
[
"Many software bugs in deep learning come from having matrix/vector dimensions that don't fit. If you can keep your matrix/vector dimensions straight you will go a long way toward eliminating many bugs. \n\n**Exercise:** Find the values for:\n - m_train (number of training examples)\n - m_test (number of test examples)\n - num_px (= height = width of a training image)\nRemember that `train_set_x_orig` is a numpy-array of shape (m_train, num_px, num_px, 3). For instance, you can access `m_train` by writing `train_set_x_orig.shape[0]`.",
"_____no_output_____"
]
],
[
[
"### START CODE HERE ### (≈ 3 lines of code)\nm_train = train_set_y.shape[0]\nm_test = test_set_y.shape[0]\nnum_px = train_set_x_orig.shape[1]\n### END CODE HERE ###\n\nprint(\"Number of training examples: m_train = \" + str(m_train))\nprint(\"Number of testing examples: m_test = \" + str(m_test))\nprint(\"Height/Width of each image: num_px = \" + str(num_px))\nprint(\"Each image is of size: (\" + str(num_px) + \", \" + str(num_px) + \", 3)\")\nprint(\"train_set_x shape: \" + str(train_set_x_orig.shape))\nprint(\"train_set_y shape: \" + str(train_set_y.shape))\nprint(\"test_set_x shape: \" + str(test_set_x_orig.shape))\nprint(\"test_set_y shape: \" + str(test_set_y.shape))",
"_____no_output_____"
]
],
[
[
"**Expected Output for m_train, m_test and num_px**: \n<table style=\"width:15%\">\n <tr>\n <td>**m_train**</td>\n <td> 209 </td> \n </tr>\n \n <tr>\n <td>**m_test**</td>\n <td> 50 </td> \n </tr>\n \n <tr>\n <td>**num_px**</td>\n <td> 64 </td> \n </tr>\n \n</table>\n",
"_____no_output_____"
],
[
"For convenience, you should now reshape images of shape (num_px, num_px, 3) in a numpy-array of shape (num_px $*$ num_px $*$ 3, 1). After this, our training (and test) dataset is a numpy-array where each column represents a flattened image. There should be m_train (respectively m_test) columns.\n\n**Exercise:** Reshape the training and test data sets so that images of size (num_px, num_px, 3) are flattened into single vectors of shape (num\\_px $*$ num\\_px $*$ 3, 1).\n\nA trick when you want to flatten a matrix X of shape (a,b,c,d) to a matrix X_flatten of shape (b$*$c$*$d, a) is to use: \n```python\nX_flatten = X.reshape(X.shape[0], -1).T # X.T is the transpose of X\n```",
"_____no_output_____"
]
],
[
[
"# Reshape the training and test examples\n\n### START CODE HERE ### (≈ 2 lines of code)\ntrain_set_x_flatten = train_set_x_orig.reshape(train_set_x_orig.shape[0], -1)\ntest_set_x_flatten = test_set_x_orig.reshape(test_set_x_orig.shape[0], -1)\n### END CODE HERE ###\n\nprint (\"train_set_x_flatten shape: \" + str(train_set_x_flatten.shape))\nprint (\"train_set_y shape: \" + str(train_set_y.shape))\nprint (\"test_set_x_flatten shape: \" + str(test_set_x_flatten.shape))\nprint (\"test_set_y shape: \" + str(test_set_y.shape))\nprint (\"sanity check after reshaping: \" + str(train_set_x_flatten[0:5,0]))",
"_____no_output_____"
]
],
[
[
"**Expected Output**: \n\n<table style=\"width:35%\">\n <tr>\n <td>**train_set_x_flatten shape**</td>\n <td> (12288, 209)</td> \n </tr>\n <tr>\n <td>**train_set_y shape**</td>\n <td>(1, 209)</td> \n </tr>\n <tr>\n <td>**test_set_x_flatten shape**</td>\n <td>(12288, 50)</td> \n </tr>\n <tr>\n <td>**test_set_y shape**</td>\n <td>(1, 50)</td> \n </tr>\n <tr>\n <td>**sanity check after reshaping**</td>\n <td>[17 31 56 22 33]</td> \n </tr>\n</table>",
"_____no_output_____"
],
[
"To represent color images, the red, green and blue channels (RGB) must be specified for each pixel, and so the pixel value is actually a vector of three numbers ranging from 0 to 255.\n\nOne common preprocessing step in machine learning is to center and standardize your dataset, meaning that you substract the mean of the whole numpy array from each example, and then divide each example by the standard deviation of the whole numpy array. But for picture datasets, it is simpler and more convenient and works almost as well to just divide every row of the dataset by 255 (the maximum value of a pixel channel).\n\n<!-- During the training of your model, you're going to multiply weights and add biases to some initial inputs in order to observe neuron activations. Then you backpropogate with the gradients to train the model. But, it is extremely important for each feature to have a similar range such that our gradients don't explode. You will see that more in detail later in the lectures. !--> \n\nLet's standardize our dataset.",
"_____no_output_____"
]
],
[
[
"train_set_x = train_set_x_flatten / 255.\ntest_set_x = test_set_x_flatten / 255.",
"_____no_output_____"
]
],
[
[
"<font color='red'>\n**What you need to remember:**\n\nCommon steps for pre-processing a new dataset are:\n- Figure out the dimensions and shapes of the problem (m_train, m_test, num_px, ...)\n- Reshape the datasets such that each example is now a vector of size (num_px \\* num_px \\* 3, 1)\n- \"Standardize\" the data",
"_____no_output_____"
],
[
"## 3 - General Architecture of the learning algorithm ##\n\nIt's time to design a simple algorithm to distinguish cat images from non-cat images.\n\nYou will build a Logistic Regression, using a Neural Network mindset. The following Figure explains why **Logistic Regression is actually a very simple Neural Network!**\n\n<img src=\"images/LogReg_kiank.png\" style=\"width:650px;height:400px;\">\n\n**Mathematical expression of the algorithm**:\n\nFor one example $x^{(i)}$:\n$$z^{(i)} = w^T x^{(i)} + b \\tag{1}$$\n$$\\hat{y}^{(i)} = a^{(i)} = sigmoid(z^{(i)})\\tag{2}$$ \n$$ \\mathcal{L}(a^{(i)}, y^{(i)}) = - y^{(i)} \\log(a^{(i)}) - (1-y^{(i)} ) \\log(1-a^{(i)})\\tag{3}$$\n\nThe cost is then computed by summing over all training examples:\n$$ J = \\frac{1}{m} \\sum_{i=1}^m \\mathcal{L}(a^{(i)}, y^{(i)})\\tag{6}$$\n\n**Key steps**:\nIn this exercise, you will carry out the following steps: \n - Initialize the parameters of the model\n - Learn the parameters for the model by minimizing the cost \n - Use the learned parameters to make predictions (on the test set)\n - Analyse the results and conclude",
"_____no_output_____"
],
[
"## 4 - Building the parts of our algorithm ## \n\nThe main steps for building a Neural Network are:\n1. Define the model structure (such as number of input features) \n2. Initialize the model's parameters\n3. Loop:\n - Calculate current loss (forward propagation)\n - Calculate current gradient (backward propagation)\n - Update parameters (gradient descent)\n\nYou often build 1-3 separately and integrate them into one function we call `model()`.\n\n### 4.1 - Helper functions\n\n**Exercise**: Using your code from \"Python Basics\", implement `sigmoid()`. As you've seen in the figure above, you need to compute $sigmoid( w^T x + b)$ to make predictions.",
"_____no_output_____"
]
],
[
[
"# GRADED FUNCTION: sigmoid\n\ndef sigmoid(z):\n \"\"\"\n Compute the sigmoid of z\n\n Arguments:\n x -- A scalar or numpy array of any size.\n\n Return:\n s -- sigmoid(z)\n \"\"\"\n\n ### START CODE HERE ### (≈ 1 line of code)\n s = 1 / (1 + np.exp(-z))\n ### END CODE HERE ###\n \n return s",
"_____no_output_____"
],
[
"print(\"sigmoid(0) = \" + str(sigmoid(0)))\nprint(\"sigmoid(9.2) = \" + str(sigmoid(9.2)))",
"_____no_output_____"
]
],
[
[
"**Expected Output**: \n\n<table style=\"width:20%\">\n <tr>\n <td>**sigmoid(0)**</td>\n <td> 0.5</td> \n </tr>\n \n <tr>\n <td>**sigmoid(9.2)**</td>\n <td> 0.999898970806 </td> \n </tr>\n</table>",
"_____no_output_____"
],
[
"### 4.2 - Initializing parameters\n\n**Exercise:** Implement parameter initialization in the cell below. You have to initialize w as a vector of zeros. If you don't know what numpy function to use, look up np.zeros() in the Numpy library's documentation.",
"_____no_output_____"
]
],
[
[
"# GRADED FUNCTION: initialize_with_zeros\n\ndef initialize_with_zeros(dim):\n \"\"\"\n This function creates a vector of zeros of shape (dim, 1) for w and initializes b to 0.\n \n Argument:\n dim -- size of the w vector we want (or number of parameters in this case)\n \n Returns:\n w -- initialized vector of shape (dim, 1)\n b -- initialized scalar (corresponds to the bias)\n \"\"\"\n \n ### START CODE HERE ### (≈ 1 line of code)\n w = np.zeros(shape=(dim, 1))\n b = 0\n ### END CODE HERE ###\n\n assert(w.shape == (dim, 1))\n assert(isinstance(b, float) or isinstance(b, int))\n \n return w, b",
"_____no_output_____"
],
[
"dim = 2\nw, b = initialize_with_zeros(dim)\nprint (\"w = \" + str(w))\nprint (\"b = \" + str(b))",
"_____no_output_____"
]
],
[
[
"**Expected Output**: \n\n\n<table style=\"width:15%\">\n <tr>\n <td> ** w ** </td>\n <td> [[ 0.]\n [ 0.]] </td>\n </tr>\n <tr>\n <td> ** b ** </td>\n <td> 0 </td>\n </tr>\n</table>\n\nFor image inputs, w will be of shape (num_px $\\times$ num_px $\\times$ 3, 1).",
"_____no_output_____"
],
[
"### 4.3 - Forward and Backward propagation\n\nNow that your parameters are initialized, you can do the \"forward\" and \"backward\" propagation steps for learning the parameters.\n\n**Exercise:** Implement a function `propagate()` that computes the cost function and its gradient.\n\n**Hints**:\n\nForward Propagation:\n- You get X\n- You compute $A = \\sigma(w^T X + b) = (a^{(0)}, a^{(1)}, ..., a^{(m-1)}, a^{(m)})$\n- You calculate the cost function: $J = -\\frac{1}{m}\\sum_{i=1}^{m}y^{(i)}\\log(a^{(i)})+(1-y^{(i)})\\log(1-a^{(i)})$\n\nHere are the two formulas you will be using: \n\n$$ \\frac{\\partial J}{\\partial w} = \\frac{1}{m}X^T(A-Y)\\tag{7}$$\n$$ \\frac{\\partial J}{\\partial b} = \\frac{1}{m} \\sum_{i=1}^m (a^{(i)}-y^{(i)})\\tag{8}$$",
"_____no_output_____"
]
],
[
[
"# GRADED FUNCTION: propagate\n\ndef propagate(w, b, X, Y):\n \"\"\"\n Implement the cost function and its gradient for the propagation explained above\n\n Arguments:\n w -- weights, a numpy array of size (num_px * num_px * 3, 1)\n b -- bias, a scalar\n X -- data of size (number of examples, num_px * num_px * 3)\n Y -- true \"label\" vector (containing 0 if non-cat, 1 if cat) of size (number of examples, 1)\n\n Return:\n cost -- negative log-likelihood cost for logistic regression\n dw -- gradient of the loss with respect to w, thus same shape as w\n db -- gradient of the loss with respect to b, thus same shape as b\n \n Tips:\n - Write your code step by step for the propagation\n \"\"\"\n \n m = X.shape[0]\n \n # FORWARD PROPAGATION (FROM X TO COST)\n ### START CODE HERE ### (≈ 2 lines of code)\n A = sigmoid(np.dot(X, w) + b) # compute activation\n cost = (- 1 / m) * np.sum(Y * np.log(A) + (1 - Y) * (np.log(1 - A))) # compute cost\n ### END CODE HERE ###\n \n # BACKWARD PROPAGATION (TO FIND GRAD)\n ### START CODE HERE ### (≈ 2 lines of code)\n dw = (1 / m) * np.dot(X.T, (A - Y))\n db = (1 / m) * np.sum(A - Y, axis=0)\n ### END CODE HERE ###\n\n assert(dw.shape == w.shape)\n assert(db.dtype == float)\n cost = np.squeeze(cost)\n assert(cost.shape == ())\n \n grads = {\"dw\": dw,\n \"db\": db}\n \n return grads, cost",
"_____no_output_____"
],
[
"w, b, X, Y = np.array([[1], [2]]), 2, np.array([[1,2], [3,4]]), np.array([[1], [0]])\ngrads, cost = propagate(w, b, X, Y)\nprint (\"dw = \" + str(grads[\"dw\"]))\nprint (\"db = \" + str(grads[\"db\"]))\nprint (\"cost = \" + str(cost))\n",
"_____no_output_____"
]
],
[
[
"**Expected Output**:\n\n<table style=\"width:50%\">\n <tr>\n <td> ** dw ** </td>\n <td> [[ 0.99993216]\n [ 1.99980262]]</td>\n </tr>\n <tr>\n <td> ** db ** </td>\n <td> 0.499935230625 </td>\n </tr>\n <tr>\n <td> ** cost ** </td>\n <td> 6.000064773192205</td>\n </tr>\n\n</table>",
"_____no_output_____"
],
[
"### d) Optimization\n- You have initialized your parameters.\n- You are also able to compute a cost function and its gradient.\n- Now, you want to update the parameters using gradient descent.\n\n**Exercise:** Write down the optimization function. The goal is to learn $w$ and $b$ by minimizing the cost function $J$. For a parameter $\\theta$, the update rule is $ \\theta = \\theta - \\alpha \\text{ } d\\theta$, where $\\alpha$ is the learning rate.",
"_____no_output_____"
]
],
[
[
"# GRADED FUNCTION: optimize\n\ndef optimize(w, b, X, Y, num_iterations, learning_rate, print_cost = False):\n \"\"\"\n This function optimizes w and b by running a gradient descent algorithm\n \n Arguments:\n w -- weights, a numpy array of size (num_px * num_px * 3, 1)\n b -- bias, a scalar\n X -- data of shape (num_px * num_px * 3, number of examples)\n Y -- true \"label\" vector (containing 0 if non-cat, 1 if cat), of shape (1, number of examples)\n num_iterations -- number of iterations of the optimization loop\n learning_rate -- learning rate of the gradient descent update rule\n print_cost -- True to print the loss every 100 steps\n \n Returns:\n params -- dictionary containing the weights w and bias b\n grads -- dictionary containing the gradients of the weights and bias with respect to the cost function\n costs -- list of all the costs computed during the optimization, this will be used to plot the learning curve.\n \n Tips:\n You basically need to write down two steps and iterate through them:\n 1) Calculate the cost and the gradient for the current parameters. Use propagate().\n 2) Update the parameters using gradient descent rule for w and b.\n \"\"\"\n \n costs = []\n \n for i in range(num_iterations):\n \n \n # Cost and gradient calculation (≈ 1-4 lines of code)\n ### START CODE HERE ### \n grads, cost = propagate(w, b, X, Y)\n ### END CODE HERE ###\n \n # Retrieve derivatives from grads\n dw = grads[\"dw\"]\n db = grads[\"db\"]\n \n # update rule (≈ 2 lines of code)\n ### START CODE HERE ###\n w = w - learning_rate * dw # need to broadcast\n b = b - learning_rate * db\n ### END CODE HERE ###\n \n # Record the costs\n if i % 100 == 0:\n costs.append(cost)\n \n # Print the cost every 100 training examples\n if print_cost and i % 100 == 0:\n print (\"Cost after iteration %i: %f\" % (i, cost))\n \n params = {\"w\": w,\n \"b\": b}\n \n grads = {\"dw\": dw,\n \"db\": db}\n \n return params, grads, costs",
"_____no_output_____"
],
[
"params, grads, costs = optimize(w, b, X, Y, num_iterations= 100, learning_rate = 0.009, print_cost = False)\n\nprint (\"w = \" + str(params[\"w\"]))\nprint (\"b = \" + str(params[\"b\"]))\nprint (\"dw = \" + str(grads[\"dw\"]))\nprint (\"db = \" + str(grads[\"db\"]))",
"_____no_output_____"
]
],
[
[
"**Expected Output**: \n\n<table style=\"width:40%\">\n <tr>\n <td> **w** </td>\n <td>[[ 0.1124579 ]\n [ 0.23106775]] </td>\n </tr>\n \n <tr>\n <td> **b** </td>\n <td> 1.55930492484 </td>\n </tr>\n <tr>\n <td> **dw** </td>\n <td> [[ 0.90158428]\n [ 1.76250842]] </td>\n </tr>\n <tr>\n <td> **db** </td>\n <td> 0.430462071679 </td>\n </tr>\n\n</table>",
"_____no_output_____"
],
[
"**Exercise:** The previous function will output the learned w and b. We are able to use w and b to predict the labels for a dataset X. Implement the `predict()` function. There is two steps to computing predictions:\n\n1. Calculate $\\hat{Y} = A = \\sigma(w^T X + b)$\n\n2. Convert the entries of a into 0 (if activation <= 0.5) or 1 (if activation > 0.5), stores the predictions in a vector `Y_prediction`. If you wish, you can use an `if`/`else` statement in a `for` loop (though there is also a way to vectorize this). ",
"_____no_output_____"
]
],
[
[
"# GRADED FUNCTION: predict\n\ndef predict(w, b, X):\n '''\n Predict whether the label is 0 or 1 using learned logistic regression parameters (w, b)\n \n Arguments:\n w -- weights, a numpy array of size (num_px * num_px * 3, 1)\n b -- bias, a scalar\n X -- data of size (num_px * num_px * 3, number of examples)\n \n Returns:\n Y_prediction -- a numpy array (vector) containing all predictions (0/1) for the examples in X\n '''\n \n m, n = X.shape\n Y_prediction = np.zeros((m, 1))\n w = w.reshape(n, 1)\n \n # Compute vector \"A\" predicting the probabilities of a cat being present in the picture\n ### START CODE HERE ### (≈ 1 line of code)\n A = sigmoid(np.dot(X, w) + b)\n ### END CODE HERE ###\n \n Y_prediction = np.where(A > 0.5, 1, 0)\n # for i in range(A.shape[0]):\n # # Convert probabilities a[0,i] to actual predictions p[0,i]\n # ### START CODE HERE ### (≈ 4 lines of code)\n # Y_prediction[i, 0] = 1 if A[i, 0] > 0.5 else 0\n # ### END CODE HERE ###\n \n assert(Y_prediction.shape == (m, 1))\n \n return Y_prediction",
"_____no_output_____"
],
[
"print(\"predictions = \" + str(predict(w, b, X)))",
"_____no_output_____"
]
],
[
[
"**Expected Output**: \n\n<table style=\"width:30%\">\n <tr>\n <td>\n **predictions**\n </td>\n <td>\n [[ 1. 1.]]\n </td> \n </tr>\n\n</table>\n",
"_____no_output_____"
],
[
"<font color='blue'>\n**What to remember:**\nYou've implemented several functions that:\n- Initialize (w,b)\n- Optimize the loss iteratively to learn parameters (w,b):\n - computing the cost and its gradient \n - updating the parameters using gradient descent\n- Use the learned (w,b) to predict the labels for a given set of examples",
"_____no_output_____"
],
[
"## 5 - Merge all functions into a model ##\n\nYou will now see how the overall model is structured by putting together all the building blocks (functions implemented in the previous parts) together, in the right order.\n\n**Exercise:** Implement the model function. Use the following notation:\n - Y_prediction for your predictions on the test set\n - Y_prediction_train for your predictions on the train set\n - w, costs, grads for the outputs of optimize()",
"_____no_output_____"
]
],
[
[
"# GRADED FUNCTION: model\n\ndef model(X_train, Y_train, X_test, Y_test, num_iterations=2000, learning_rate=0.5, print_cost=False):\n \"\"\"\n Builds the logistic regression model by calling the function you've implemented previously\n \n Arguments:\n X_train -- training set represented by a numpy array of shape (num_px * num_px * 3, m_train)\n Y_train -- training labels represented by a numpy array (vector) of shape (1, m_train)\n X_test -- test set represented by a numpy array of shape (num_px * num_px * 3, m_test)\n Y_test -- test labels represented by a numpy array (vector) of shape (1, m_test)\n num_iterations -- hyperparameter representing the number of iterations to optimize the parameters\n learning_rate -- hyperparameter representing the learning rate used in the update rule of optimize()\n print_cost -- Set to true to print the cost every 100 iterations\n \n Returns:\n d -- dictionary containing information about the model.\n \"\"\"\n \n ### START CODE HERE ###\n # initialize parameters with zeros (≈ 1 line of code)\n w, b = initialize_with_zeros(X_train.shape[1])\n\n # Gradient descent (≈ 1 line of code)\n parameters, grads, costs = optimize(w, b, X_train, Y_train, num_iterations, learning_rate, print_cost)\n \n # Retrieve parameters w and b from dictionary \"parameters\"\n w = parameters[\"w\"]\n b = parameters[\"b\"]\n \n # Predict test/train set examples (≈ 2 lines of code)\n Y_prediction_test = predict(w, b, X_test)\n Y_prediction_train = predict(w, b, X_train)\n\n ### END CODE HERE ###\n\n # Print train/test Errors\n print(\"train accuracy: {} %\".format(100 - np.mean(np.abs(Y_prediction_train - Y_train)) * 100))\n print(\"test accuracy: {} %\".format(100 - np.mean(np.abs(Y_prediction_test - Y_test)) * 100))\n\n \n d = {\"costs\": costs,\n \"Y_prediction_test\": Y_prediction_test, \n \"Y_prediction_train\" : Y_prediction_train, \n \"w\" : w, \n \"b\" : b,\n \"learning_rate\" : learning_rate,\n \"num_iterations\": num_iterations}\n \n return d",
"_____no_output_____"
],
[
"np.zeros(1)",
"_____no_output_____"
],
[
"class LogisticRegress:\n def __init__(self, lr=0.01):\n self.w = None\n self.b = None\n self.lr = lr\n self.costs = []\n \n def activate(self, z):\n # 使用sigmoid作为激活函数\n return 1 / (1 + np.exp(-z))\n \n def loss(self, y_true, y_pred):\n # 逻辑斯蒂损失\n l = - y_true * np.log(y_pred) - (1 - y_true) * np.log(1-y_pred) \n return np.squeeze(np.mean(l, axis=0))\n \n \n def fit(self, X, y, num_iterations = 2000, verbose=1):\n m, n = X.shape\n y = y.reshape((-1, 1))\n self.init_params(n)\n for epoch in range(num_iterations):\n z = X @ self.w + self.b\n A = self.activate(z)\n l = self.loss(y, A)\n dw = (1 / m) * np.dot(X.T, (A - y))\n db = (1 / m) * np.sum(A - y, axis=0)\n self.w -= self.lr * dw\n self.b -= self.lr * db\n \n if epoch % 100 == 0:\n self.costs.append(l)\n if verbose:\n print(f'Cost after iteration {epoch}', l)\n return costs \n \n def init_params(self, n_features):\n # 逻辑斯蒂回归 全部舒适化为0\n self.w = np.zeros((n_features, 1))\n self.b = np.zeros(1)\n \n def predict(self, X):\n y_pred = X @ self.w + self.b\n return np.where(y_pred >= 0.5, 1, 0)",
"_____no_output_____"
],
[
"model = LogisticRegress(lr=0.005)\ncosts = model.fit(train_set_x, train_set_y)",
"_____no_output_____"
],
[
"y_train_pred = model.predict(train_set_x)\nprint('train accuracy: ', np.sum(train_set_y == y_train_pred) / train_set_x.shape[0])",
"_____no_output_____"
],
[
"y_test_pred = model.predict(test_set_x)\nprint('test accuracy: ', np.sum(test_set_y == y_test_pred) / test_set_x.shape[0])",
"_____no_output_____"
]
],
[
[
"Run the following cell to train your model.",
"_____no_output_____"
]
],
[
[
"# d = model(train_set_x, train_set_y, test_set_x, test_set_y, num_iterations = 2000, learning_rate = 0.005, print_cost = True)",
"_____no_output_____"
]
],
[
[
"**Expected Output**: \n\n<table style=\"width:40%\"> \n \n <tr>\n <td> **Train Accuracy** </td> \n <td> 99.04306220095694 % </td>\n </tr>\n\n <tr>\n <td>**Test Accuracy** </td> \n <td> 70.0 % </td>\n </tr>\n</table> \n\n\n",
"_____no_output_____"
],
[
"**Comment**: Training accuracy is close to 100%. This is a good sanity check: your model is working and has high enough capacity to fit the training data. Test error is 68%. It is actually not bad for this simple model, given the small dataset we used and that logistic regression is a linear classifier. But no worries, you'll build an even better classifier next week!\n\nAlso, you see that the model is clearly overfitting the training data. Later in this specialization you will learn how to reduce overfitting, for example by using regularization. Using the code below (and changing the `index` variable) you can look at predictions on pictures of the test set.",
"_____no_output_____"
]
],
[
[
"# Example of a picture that was wrongly classified.\nindex = 13\nplt.imshow(test_set_x[index, :].reshape((num_px, num_px, 3)))\nprint (\"y = \" + str(test_set_y[index, 0]) + \", you predicted that it is a \\\"\" + classes[y_test_pred[index, 0]].decode(\"utf-8\") + \"\\\" picture.\")",
"_____no_output_____"
]
],
[
[
"Let's also plot the cost function and the gradients.",
"_____no_output_____"
]
],
[
[
"# Plot learning curve (with costs)\n# costs = np.squeeze(d['costs'])\nplt.plot(costs)\nplt.ylabel('cost')\nplt.xlabel('iterations (per hundreds)')\nplt.title(\"Learning rate =\" + str(model.lr))\nplt.show()",
"_____no_output_____"
]
],
[
[
"**Interpretation**:\nYou can see the cost decreasing. It shows that the parameters are being learned. However, you see that you could train the model even more on the training set. Try to increase the number of iterations in the cell above and rerun the cells. You might see that the training set accuracy goes up, but the test set accuracy goes down. This is called overfitting. ",
"_____no_output_____"
],
[
"## 6 - Further analysis (optional/ungraded exercise) ##\n\nCongratulations on building your first image classification model. Let's analyze it further, and examine possible choices for the learning rate $\\alpha$. ",
"_____no_output_____"
],
[
"#### Choice of learning rate ####\n\n**Reminder**:\nIn order for Gradient Descent to work you must choose the learning rate wisely. The learning rate $\\alpha$ determines how rapidly we update the parameters. If the learning rate is too large we may \"overshoot\" the optimal value. Similarly, if it is too small we will need too many iterations to converge to the best values. That's why it is crucial to use a well-tuned learning rate.\n\nLet's compare the learning curve of our model with several choices of learning rates. Run the cell below. This should take about 1 minute. Feel free also to try different values than the three we have initialized the `learning_rates` variable to contain, and see what happens. ",
"_____no_output_____"
]
],
[
[
"a = 0.356\nprint(\"a = {:.2%}\".format(a))",
"_____no_output_____"
],
[
"learning_rates = [0.01, 0.001, 0.0001, 0.03]\nmodels = {}\nfor i in learning_rates:\n print (\"learning rate is: \" + str(i))\n classifier = LogisticRegress(lr=i)\n models[str(i)] = classifier\n classifier.fit(train_set_x, train_set_y, num_iterations = 1500, verbose=0)\n \n y_train_pred = classifier.predict(train_set_x)\n print('train accuracy: {:.2%}'.format(np.sum(train_set_y == y_train_pred) / train_set_x.shape[0]))\n \n y_test_pred = classifier.predict(test_set_x)\n print('test accuracy: {:.2%}'.format(np.sum(test_set_y == y_test_pred) / test_set_x.shape[0]))\n # models[str(i)] = model(train_set_x, train_set_y, test_set_x, test_set_y, num_iterations = 1500, learning_rate = i, print_cost = False)\n print ('\\n' + \"-------------------------------------------------------\" + '\\n')\n\nfor i in learning_rates:\n plt.plot(models[str(i)].costs, label= str(models[str(i)].lr))\n\nplt.ylabel('cost')\nplt.xlabel('iterations')\n\nlegend = plt.legend(loc='upper center', shadow=True)\nframe = legend.get_frame()\nframe.set_facecolor('0.90')\nplt.show()",
"_____no_output_____"
]
],
[
[
"**Interpretation**: \n- Different learning rates give different costs and thus different predictions results.\n- If the learning rate is too large (0.01), the cost may oscillate up and down. It may even diverge (though in this example, using 0.01 still eventually ends up at a good value for the cost). \n- A lower cost doesn't mean a better model. You have to check if there is possibly overfitting. It happens when the training accuracy is a lot higher than the test accuracy.\n- In deep learning, we usually recommend that you: \n - Choose the learning rate that better minimizes the cost function.\n - If your model overfits, use other techniques to reduce overfitting. (We'll talk about this in later videos.) \n",
"_____no_output_____"
],
[
"## 7 - Test with your own image (optional/ungraded exercise) ##\n\nCongratulations on finishing this assignment. You can use your own image and see the output of your model. To do that:\n 1. Click on \"File\" in the upper bar of this notebook, then click \"Open\" to go on your Coursera Hub.\n 2. Add your image to this Jupyter Notebook's directory, in the \"images\" folder\n 3. Change your image's name in the following code\n 4. Run the code and check if the algorithm is right (1 = cat, 0 = non-cat)!",
"_____no_output_____"
]
],
[
[
"## START CODE HERE ## (PUT YOUR IMAGE NAME) \nmy_image = \"my_image.jpg\" # change this to the name of your image file \n## END CODE HERE ##\n\n# We preprocess the image to fit your algorithm.\nfname = \"images/\" + my_image\nimage = np.array(plt.imread(fname))\nimage.shape",
"_____no_output_____"
],
[
"plt.imshow(image)",
"_____no_output_____"
],
[
"from skimage import transform",
"_____no_output_____"
],
[
"my_image = transform.resize(image, (64, 64, 3))\nmy_image.shape",
"_____no_output_____"
],
[
"plt.imshow(my_image)",
"_____no_output_____"
],
[
"\n# my_predicted_image = model(d[\"w\"], d[\"b\"], my_image.reshape(1, -1))\nmy_predicted_image = model.predict(my_image.reshape(1, -1))\n\nprint(\"y = \" + str(np.squeeze(my_predicted_image)) + \", your algorithm predicts a \\\"\" + classes[int(np.squeeze(my_predicted_image)),].decode(\"utf-8\") + \"\\\" picture.\")",
"_____no_output_____"
]
],
[
[
"<font color='red'>\n**What to remember from this assignment:**\n1. Preprocessing the dataset is important.\n2. You implemented each function separately: initialize(), propagate(), optimize(). Then you built a model().\n3. Tuning the learning rate (which is an example of a \"hyperparameter\") can make a big difference to the algorithm. You will see more examples of this later in this course!",
"_____no_output_____"
],
[
"Finally, if you'd like, we invite you to try different things on this Notebook. Make sure you submit before trying anything. Once you submit, things you can play with include:\n - Play with the learning rate and the number of iterations\n - Try different initialization methods and compare the results\n - Test other preprocessings (center the data, or divide each row by its standard deviation)",
"_____no_output_____"
],
[
"Bibliography:\n- http://www.wildml.com/2015/09/implementing-a-neural-network-from-scratch/\n- https://stats.stackexchange.com/questions/211436/why-do-we-normalize-images-by-subtracting-the-datasets-image-mean-and-not-the-c",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
4a54acdcf301f46c1dfa1c3ac12445b61dac788d
| 2,198 |
ipynb
|
Jupyter Notebook
|
examples/polygon.ipynb
|
kylebarron/ipycanvas
|
8c91ec4f634ff3661f594872e8050cf27d6db0c6
|
[
"BSD-3-Clause"
] | 597 |
2019-07-06T12:56:50.000Z
|
2022-03-31T06:54:37.000Z
|
examples/polygon.ipynb
|
kylebarron/ipycanvas
|
8c91ec4f634ff3661f594872e8050cf27d6db0c6
|
[
"BSD-3-Clause"
] | 142 |
2019-07-06T17:13:22.000Z
|
2022-03-15T16:41:29.000Z
|
examples/polygon.ipynb
|
kylebarron/ipycanvas
|
8c91ec4f634ff3661f594872e8050cf27d6db0c6
|
[
"BSD-3-Clause"
] | 52 |
2019-07-08T14:04:25.000Z
|
2022-03-08T23:05:04.000Z
| 21.54902 | 65 | 0.517288 |
[
[
[
"from ipycanvas import Canvas\n\ncanvas = Canvas(width=200, height=200)\n\ncanvas.fill_style = '#63934e'\ncanvas.stroke_style = '#4e6393'\ncanvas.line_width = 5\ncanvas.fill_polygon([(20, 20), (180, 20), (100, 150)])\ncanvas.stroke_polygon([(20, 20), (180, 20), (100, 150)])\n\ncanvas",
"_____no_output_____"
],
[
"from math import pi\n\nimport numpy as np\n\nfrom ipycanvas import Canvas",
"_____no_output_____"
],
[
"def polygon(canvas, x, y, radius, n_points):\n angles = (2 * pi / n_points) * np.arange(n_points)\n\n v_x = x + np.cos(angles) * radius\n v_y = y + np.sin(angles) * radius\n\n points = np.stack((v_x, v_y), axis=1)\n\n canvas.fill_polygon(points)",
"_____no_output_____"
],
[
"background_color = '#89c64f'\npolygon_color = '#c6574f'",
"_____no_output_____"
],
[
"canvas = Canvas(width=500, height=500)\n\ncanvas.fill_style = background_color\ncanvas.fill_rect(0, 0, canvas.width, canvas.height)\n\ncanvas.fill_style = polygon_color\npolygon(canvas, 250, 250, 100, 6)\n\ncanvas",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code"
]
] |
4a54af1177d394ac2b7ac829a523b24afd3d5a28
| 756,963 |
ipynb
|
Jupyter Notebook
|
data/galaxy_zoo.ipynb
|
yhyu13/Galaxy_Zoo_Capsule
|
9158a29c290d2681c9df345d7717e5abd95ba764
|
[
"MIT"
] | 2 |
2018-05-25T15:30:05.000Z
|
2018-06-26T22:18:41.000Z
|
data/galaxy_zoo.ipynb
|
joshualin24/Galaxy_Zoo_Capsule
|
fe90ff8baf79d26aafaabdaf55f4c5f3fab23462
|
[
"MIT"
] | null | null | null |
data/galaxy_zoo.ipynb
|
joshualin24/Galaxy_Zoo_Capsule
|
fe90ff8baf79d26aafaabdaf55f4c5f3fab23462
|
[
"MIT"
] | 1 |
2018-06-04T12:11:43.000Z
|
2018-06-04T12:11:43.000Z
| 1,484.241176 | 106,440 | 0.959752 |
[
[
[
"import pandas as pd\nimport matplotlib.pyplot as plt\nfrom os import listdir, mkdir\nimport numpy as np\nfrom shutil import copy2\n\n# reproducible randomness\nfrom numpy.random import RandomState",
"_____no_output_____"
],
[
"df = pd.read_csv('./training_solutions_rev1.csv')\nprint(df.count()) # show total number of samples\nprint(df.head()) # showcase data set",
"GalaxyID 61578\nClass1.1 61578\nClass1.2 61578\nClass1.3 61578\nClass2.1 61578\nClass2.2 61578\nClass3.1 61578\nClass3.2 61578\nClass4.1 61578\nClass4.2 61578\nClass5.1 61578\nClass5.2 61578\nClass5.3 61578\nClass5.4 61578\nClass6.1 61578\nClass6.2 61578\nClass7.1 61578\nClass7.2 61578\nClass7.3 61578\nClass8.1 61578\nClass8.2 61578\nClass8.3 61578\nClass8.4 61578\nClass8.5 61578\nClass8.6 61578\nClass8.7 61578\nClass9.1 61578\nClass9.2 61578\nClass9.3 61578\nClass10.1 61578\nClass10.2 61578\nClass10.3 61578\nClass11.1 61578\nClass11.2 61578\nClass11.3 61578\nClass11.4 61578\nClass11.5 61578\nClass11.6 61578\ndtype: int64\n GalaxyID Class1.1 Class1.2 Class1.3 Class2.1 Class2.2 Class3.1 \\\n0 100008 0.383147 0.616853 0.000000 0.000000 0.616853 0.038452 \n1 100023 0.327001 0.663777 0.009222 0.031178 0.632599 0.467370 \n2 100053 0.765717 0.177352 0.056931 0.000000 0.177352 0.000000 \n3 100078 0.693377 0.238564 0.068059 0.000000 0.238564 0.109493 \n4 100090 0.933839 0.000000 0.066161 0.000000 0.000000 0.000000 \n\n Class3.2 Class4.1 Class4.2 ... Class9.3 Class10.1 Class10.2 \\\n0 0.578401 0.418398 0.198455 ... 0.000000 0.279952 0.138445 \n1 0.165229 0.591328 0.041271 ... 0.018764 0.000000 0.131378 \n2 0.177352 0.000000 0.177352 ... 0.000000 0.000000 0.000000 \n3 0.129071 0.189098 0.049466 ... 0.000000 0.094549 0.000000 \n4 0.000000 0.000000 0.000000 ... 0.000000 0.000000 0.000000 \n\n Class10.3 Class11.1 Class11.2 Class11.3 Class11.4 Class11.5 Class11.6 \n0 0.000000 0.000000 0.092886 0.0 0.0 0.0 0.325512 \n1 0.459950 0.000000 0.591328 0.0 0.0 0.0 0.000000 \n2 0.000000 0.000000 0.000000 0.0 0.0 0.0 0.000000 \n3 0.094549 0.189098 0.000000 0.0 0.0 0.0 0.000000 \n4 0.000000 0.000000 0.000000 0.0 0.0 0.0 0.000000 \n\n[5 rows x 38 columns]\n"
],
[
"df_irregular = df[['GalaxyID','Class8.4']] # Irregular (1.1)",
"_____no_output_____"
],
[
"condition_irregular = df_irregular['Class8.4']>0.2 # >?% vote YES\ndf_irregular1 = df_irregular[condition_irregular]\nprint(df_irregular1.count())",
"GalaxyID 3005\nClass8.4 3005\ndtype: int64\n"
],
[
"# get irregular galaxy ID\nirregular_id = list(df_irregular1['GalaxyID'])\nprint(irregular_id[:10])",
"[100134, 100335, 100821, 101007, 101307, 101501, 101666, 101854, 102391, 102657]\n"
],
[
"GALAXY_ORIG_FOLDER = './images_training_rev1/'\n\nimport cv2\nimg = cv2.imread(GALAXY_ORIG_FOLDER+'%d.jpg' % irregular_id[0],0)\nplt.imshow(img)\nplt.show()\nimg = cv2.imread(GALAXY_ORIG_FOLDER+'%d.jpg' % irregular_id[1],0)\nplt.imshow(img)\nplt.show()\nimg = cv2.imread(GALAXY_ORIG_FOLDER+'%d.jpg' % irregular_id[2],0)\nplt.imshow(img)\nplt.show()",
"_____no_output_____"
],
[
"df_odd = df[['GalaxyID','Class6.1']] # Irregular (1.1)",
"_____no_output_____"
],
[
"condition_odd = df_odd['Class6.1']>0.78 # >?% vote YES\ndf_odd1 = df_odd[condition_odd]\nprint(df_odd1.count())",
"GalaxyID 2603\nClass6.1 2603\ndtype: int64\n"
],
[
"# get irregular galaxy ID\nodd_id = list(df_odd1['GalaxyID'])\nprint(odd_id[:10])",
"[100263, 100458, 100513, 100520, 100561, 101591, 101597, 102347, 102474, 102595]\n"
],
[
"GALAXY_ORIG_FOLDER = './images_training_rev1/'\n\nimport cv2\nimg = cv2.imread(GALAXY_ORIG_FOLDER+'%d.jpg' % odd_id[0],0)\nplt.imshow(img)\nplt.show()\nimg = cv2.imread(GALAXY_ORIG_FOLDER+'%d.jpg' % odd_id[1],0)\nplt.imshow(img)\nplt.show()\nimg = cv2.imread(GALAXY_ORIG_FOLDER+'%d.jpg' % odd_id[2],0)\nplt.imshow(img)\nplt.show()",
"_____no_output_____"
],
[
"df_elliptical = df[['GalaxyID','Class1.1']] # Smooth (1.1)",
"_____no_output_____"
],
[
"condition_elliptical = df_elliptical['Class1.1']>0.9 # >?% vote YES\ndf_elliptical1 = df_elliptical[condition_elliptical]\nprint(df_elliptical1.count())",
"GalaxyID 2502\nClass1.1 2502\ndtype: int64\n"
],
[
"df_sprial = df[['GalaxyID','Class4.1']] # Spiral Arm(4.1)",
"_____no_output_____"
],
[
"condition = df_sprial['Class4.1']>0.9 # >?% vote YES\ndf_sprial1 = df_sprial[condition]\nprint(df_sprial1.count())",
"GalaxyID 2907\nClass4.1 2907\ndtype: int64\n"
],
[
"# get ellipitical galaxy ID\nelliptical_id = list(df_elliptical1['GalaxyID'])\nprint(elliptical_id[:10])\n# get sprial galaxy ID\nsprial_id = list(df_sprial1['GalaxyID'])\nprint(sprial_id[:10])",
"[100090, 100672, 101627, 101858, 102182, 102260, 102928, 103260, 103348, 103707]\n[100380, 100673, 100765, 100813, 102243, 102402, 102407, 102422, 102433, 102735]\n"
],
[
"# select the same of samples (reproducibility ensured)\nprng = RandomState(1234567890)\nnum_samples = 2500\nnum_split = int(0.8 * num_samples)\nassert(num_samples <= len(elliptical_id) and num_samples <= len(sprial_id))\n\nelliptical_selected_idx = prng.choice(len(elliptical_id), num_samples)\nsprial_selected_idx = prng.choice(len(sprial_id), num_samples)\n\nelliptical_selected_id = []\nsprial_selected_id = []\n\nfor idx in elliptical_selected_idx:\n elliptical_selected_id.append(elliptical_id[idx])\n\nfor idx in sprial_selected_idx:\n sprial_selected_id.append(sprial_id[idx])\n \nprng.shuffle(elliptical_selected_id)\nprng.shuffle(sprial_selected_id)\n\ntrain_elliptical_id, test_elliptical_id = elliptical_selected_id[:num_split],elliptical_selected_id[num_split:]\ntrain_sprial_id, test_sprial_id = sprial_selected_id[:num_split],sprial_selected_id[num_split:]",
"_____no_output_____"
],
[
"with open('./train_simple.txt', 'w+') as f:\n for i in train_elliptical_id:\n f.write('%d,0\\n' % i)\n for i in train_sprial_id:\n f.write('%d,1\\n' % i)\nwith open('./test_simple.txt', 'w+') as f:\n for i in test_elliptical_id:\n f.write('%d,0\\n' % i)\n for i in test_sprial_id:\n f.write('%d,1\\n' % i)",
"_____no_output_____"
],
[
"GALAXY_ORIG_FOLDER = './images_training_rev1/'\n\nimport cv2\nimg = cv2.imread(GALAXY_ORIG_FOLDER+'%d.jpg' % train_elliptical_id[0],0)\nplt.imshow(img)\nplt.show()\nimg = cv2.imread(GALAXY_ORIG_FOLDER+'%d.jpg' % train_sprial_id[0],0)\nplt.imshow(img)\nplt.show()",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a54baaef4973dd0c8ef1ea21214bde081c6a3e7
| 605,643 |
ipynb
|
Jupyter Notebook
|
02_body/chapter2/images/Calcul_force_optique/Calcul optical force.ipynb
|
eXpensia/Confined-Brownian-Motion
|
bd0eb6dea929727ea081dae060a7d1aa32efafd1
|
[
"MIT"
] | null | null | null |
02_body/chapter2/images/Calcul_force_optique/Calcul optical force.ipynb
|
eXpensia/Confined-Brownian-Motion
|
bd0eb6dea929727ea081dae060a7d1aa32efafd1
|
[
"MIT"
] | null | null | null |
02_body/chapter2/images/Calcul_force_optique/Calcul optical force.ipynb
|
eXpensia/Confined-Brownian-Motion
|
bd0eb6dea929727ea081dae060a7d1aa32efafd1
|
[
"MIT"
] | null | null | null | 1,081.505357 | 88,540 | 0.956473 |
[
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\nimport miepython as mp",
"_____no_output_____"
],
[
"import numpy as np\nimport matplotlib.pyplot as plt\nimport matplotlib as mpl \nmpl.rcParams[\"xtick.direction\"] = \"in\"\nmpl.rcParams[\"ytick.direction\"] = \"in\"\nmpl.rcParams[\"lines.markeredgecolor\"] = \"k\"\nmpl.rcParams[\"lines.markeredgewidth\"] = 1\nmpl.rcParams[\"figure.dpi\"] = 200\nfrom matplotlib import rc\nrc('font', family='serif')\nrc('text', usetex=True)\nrc('xtick', labelsize='medium')\nrc('ytick', labelsize='medium')\ndef cm2inch(value):\n return value/2.54",
"_____no_output_____"
],
[
"link = r\"https://refractiveindex.info/tmp/data/organic/(C8H8)n%20-%20polystyren/Zhang.txt\"\npoly = np.genfromtxt(link, delimiter='\\t')",
"_____no_output_____"
],
[
"N = len(poly)//2\npoly_lam = poly[1:N,0][:40]\npoly_mre = poly[1:N,1][:40]\npoly_mim = poly[N+1:,1][:40]",
"_____no_output_____"
],
[
"poly_mim",
"_____no_output_____"
],
[
"plt.figure(figsize=( cm2inch(16),cm2inch(8)))\n\nplt.plot(poly_lam*1000,poly_mre,color='tab:blue')\n\n#plt.xlim(300,800)\n#plt.ylim(-,3)\n\nplt.xlabel('Wavelength (nm)')\nplt.ylabel('$n_r$')\n#plt.text(350, 1.2, '$m_{re}$', color='blue', fontsize=14)\n#plt.text(350, 2.2, '$m_{im}$', color='red', fontsize=14)\nax=plt.gca()\n\nax.spines['left'].set_color(\"red\")\n\nax2=ax.twinx()\nplt.semilogy(poly_lam*1000,poly_mim,color='tab:red')\nplt.ylabel('$n_i$', color = \"tab:red\")\n\n\nplt.tight_layout(pad=0.1)\n\nplt.savefig(\"refractive_index.pdf\")",
"_____no_output_____"
],
[
"plt.semilogy(poly_lam*1000,poly_mim,color='red')\n\n#plt.xlim(300,800)\n#plt.ylim(-,3)\n\nplt.xlabel('Wavelength (nm)')\nplt.ylabel('Refractive Index')\n#plt.text(350, 1.2, '$m_{re}$', color='blue', fontsize=14)\n#plt.text(350, 2.2, '$m_{im}$', color='red', fontsize=14)\n\nplt.title('Complex part of Refractive Index of Polystyrene')\n\nplt.show()",
"_____no_output_____"
],
[
"r = 1.5 #radius in microns\n\nx = 2*np.pi*r/poly_lam;\nm = poly_mre - 1.0j * poly_mim\nqext, qsca, qback, g = mp.mie(m,x)\nabsorb = (qext - qsca) * np.pi * r**2\nscatt = qsca * np.pi * r**2\nextinct = qext* np.pi * r**2\n\nplt.plot(poly_lam*1000,absorb, label=\"$\\sigma_{abs}$\")\nplt.plot(poly_lam*1000,scatt, label=\"$\\sigma_{sca}$\")\nplt.plot(poly_lam*1000,extinct, \"--\", label=\"$\\sigma_{ext}$\")\n#plt.text(350, 0.35,'$\\sigma_{abs}$', color='blue', fontsize=14)\n#plt.text(350, 0.54,'$\\sigma_{sca}$', color='red', fontsize=14)\n#plt.text(350, 0.84,'$\\sigma_{ext}$', color='green', fontsize=14)\n\nplt.xlabel(\"Wavelength (nm)\")\nplt.ylabel(\"Cross Section (1/microns$^2$)\")\nplt.title(\"Cross Sections for %.1f$\\mu$m Polystyrebe Spheres\" % (r*2))\n\nplt.xlim(400,800)\nplt.legend()\nplt.show()\n\n",
"_____no_output_____"
],
[
"\nx = 2*np.pi*r/poly_lam;\nm = poly_mre - 1.0j * poly_mim\nqext, qsca, qback, g = mp.mie(m,x)\nqpr = qext - g*qsca\n\nplt.plot(poly_lam*1000,qpr)\nplt.xlabel(\"Wavelength (nm)\")\nplt.ylabel(\"Efficiency $Q_{pr}$\")\nplt.title(\"Radiation Pressure Efficiency for %.1f$\\mu$m Polystyrene Spheres\" % (r*2))\n\nplt.show()\n\n",
"_____no_output_____"
],
[
"r0 = 1.5e-6\nCpr = np.pi * r0 *r0 * qpr\nE0 = 4.5e-3 / (np.pi * 1.75e-3 ** 2 )\nc = 299792458 / 1.33",
"_____no_output_____"
]
],
[
[
"## Radiation Pressure\n\nThe radiation pressure is given by [e.g., Kerker, p. 94]\n\n$$\nQ_{pr}=Q_{ext}-g Q_{sca}\n$$\n\nand is the momentum given to the scattering particle [van de Hulst, p. 13] in the direction of the incident wave. The radiation pressure cross section $C_{pr}$ is just the efficiency multiplied by the geometric cross section\n\n$$\nC_{pr} = \\pi r^2 Q_{pr}\n$$\n\nThe radiation pressure cross section $C_{pr}$ can be interpreted as the area of a black wall that would receive the same force from the same incident wave. The actual force on the particle is\nis\n\n$$\nF = E_0 \\frac{C_{pr}}{c}\n$$\n\nwhere $E_0$ is the irradiance (W/m$^2$) on the sphere and $c$ is the velocity of the radiation in the medium\n",
"_____no_output_____"
]
],
[
[
"F = E0 * Cpr / c",
"_____no_output_____"
],
[
"plt.plot(poly_lam*1000,F*1e15)\nplt.xlabel(\"Wavelength (nm)\")\nplt.ylabel(\"Force (fN)\")",
"_____no_output_____"
],
[
"Fs = []\n\nrs = np.linspace(0.5, 3, 100000)\nI = np.argmin(abs(poly_lam*1000 - 532))\n\nfor r in rs:\n x = 2*np.pi*r/poly_lam;\n m = poly_mre - 1.0j * poly_mim\n qext, qsca, qback, g = mp.mie(m,x)\n qpr = qext - g*qsca\n \n r0 = r * 1e-6\n Cpr = np.pi * r0 *r0 * qpr\n E0 = 4.5e-3 / (np.pi * 1.75e-3 ** 2 )\n c = 299792458 / 1.33\n F = E0 * Cpr / c\n \n Fs.append(F[I]*1e15)\n \n\n \n",
"_____no_output_____"
],
[
"plt.figure(figsize=(cm2inch(16),cm2inch(8)))\n\n\nplt.plot(rs, Fs)\n#plt.title(\"Optical force for a 532 $\\mu$m plane wave on Polystyrene Spheres\")\nplt.xlim(0.5,2)\nplt.ylim(0,0.022)\nplt.xlabel(\"$a$ ($\\mu$m)\")\nplt.ylabel(\"$F_\\mathrm{opt}$ (fN)\")\nplt.tight_layout(pad=0.1)\nplt.savefig(\"optical_force.pdf\")",
"_____no_output_____"
],
[
"E0",
"_____no_output_____"
],
[
"4/3 * np.pi * (0.5e-6)**3 * (-50) * 9.81 * 1e15",
"_____no_output_____"
],
[
"plt.figure(figsize=( cm2inch(16),cm2inch(8)))\n\n\nplt.plot(rs, Fs)\n\nplt.xlim(1,2)\nplt.ylim(0,0.02)",
"_____no_output_____"
],
[
"plt.figure(figsize=( cm2inch(16),cm2inch(8)))\n\n\nplt.plot(rs, Fs)\n\nplt.xlim(1.45,1.55)\nplt.ylim(0,0.02)",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a54c5fcc342d0033a505cf9d441cf25ec14b104
| 2,219 |
ipynb
|
Jupyter Notebook
|
coinTossing_.ipynb
|
avidQ/EQTransformer
|
e645189e138c320aa0ee6c461ea80160f20fef12
|
[
"MIT"
] | null | null | null |
coinTossing_.ipynb
|
avidQ/EQTransformer
|
e645189e138c320aa0ee6c461ea80160f20fef12
|
[
"MIT"
] | null | null | null |
coinTossing_.ipynb
|
avidQ/EQTransformer
|
e645189e138c320aa0ee6c461ea80160f20fef12
|
[
"MIT"
] | null | null | null | 27.395062 | 230 | 0.477693 |
[
[
[
"<a href=\"https://colab.research.google.com/github/avidQ/EQTransformer/blob/master/coinTossing_.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"# import all necessary packages. Although not necessary, \n# it is customary to have all the imports on top.\nimport random\nimport pandas as pd ",
"_____no_output_____"
],
[
"# simulate a coin flip\n# what is the sample space for a coin flip?\nn = 100000\ncount = 0;\nfor i in range(n):\n result = random.randint(0,1)\n if result == 1:\n count +=1\n\nprint (\"number of Tail =\", n-count )\n#from IPython.display import Image, display\n#Image('/blue.png', width=100, height=100)\n\n#from IPython.display import Image, display\n#Image(\"/yellow.png\", width=100, height=100) \nprint (\"number of Head =\", count )",
"number of Tail = 50001\nnumber of Head = 49999\n"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code"
]
] |
4a54d9d2f3a8986fff672e7ad971a3b6eeeeef58
| 47,837 |
ipynb
|
Jupyter Notebook
|
Assignments/Assignment04_iris.ipynb
|
meghagupta08/RIT-DSCI-633-FDS
|
8871f4452484802d1c2860ff8985566feda20b5b
|
[
"MIT"
] | null | null | null |
Assignments/Assignment04_iris.ipynb
|
meghagupta08/RIT-DSCI-633-FDS
|
8871f4452484802d1c2860ff8985566feda20b5b
|
[
"MIT"
] | null | null | null |
Assignments/Assignment04_iris.ipynb
|
meghagupta08/RIT-DSCI-633-FDS
|
8871f4452484802d1c2860ff8985566feda20b5b
|
[
"MIT"
] | null | null | null | 50.782378 | 17,276 | 0.648368 |
[
[
[
"# Get Data",
"_____no_output_____"
]
],
[
[
"import os\nimport zipfile\nimport urllib\n\nDOWNLOAD_ROOT = \"https://archive.ics.uci.edu/ml/machine-learning-databases/iris/iris.data\"\nIRIS_PATH = os.path.join(\"datasets\", \"iris\")\nIRIS_URL = DOWNLOAD_ROOT \n\ndef extract_iris_data(iris_url=IRIS_URL,iris_path=IRIS_PATH):\n if not os.path.isdir(iris_path):\n os.makedirs(iris_path)\n irisdata_path = os.path.join(iris_path,\"iris.data\")\n urllib.request.urlretrieve(iris_url, irisdata_path)\n",
"_____no_output_____"
],
[
"extract_iris_data()",
"_____no_output_____"
]
],
[
[
"## Data Manipulation and analysis",
"_____no_output_____"
]
],
[
[
"import pandas as pd\ndef load_iris_train_data(iris_path=IRIS_PATH):\n csv_path = os.path.join(iris_path, \"iris.data\")\n return pd.read_csv(csv_path,names=['SepalLength','SepalWidth','PetalLength','PetalWidth','Class'])",
"_____no_output_____"
],
[
"datasets=load_iris_train_data()\ndatasets.head()",
"_____no_output_____"
],
[
"from sklearn.preprocessing import OrdinalEncoder\n\nordinal_encoder = OrdinalEncoder()\ncat_encoded = ordinal_encoder.fit_transform(datasets[['Class']])\ndatasets['Class'] = cat_encoded\ndatasets.head()",
"_____no_output_____"
],
[
"labels = ordinal_encoder.categories_\nlabels = list(labels)\nprint(labels)",
"[array(['Iris-setosa', 'Iris-versicolor', 'Iris-virginica'], dtype=object)]\n"
],
[
"import matplotlib.pyplot as plt\nax = datasets[datasets.Class==0].plot.scatter(x='SepalLength', y='SepalWidth',\n color='red', label='Iris-setosa')\ndatasets[datasets.Class==1].plot.scatter(x='SepalLength', y='SepalWidth',\n color='green', label='Iris-versicolor', ax=ax)\ndatasets[datasets.Class==2].plot.scatter(x='SepalLength', y='SepalWidth',\n color='blue', label='Iris-virginica', ax=ax)\nax.set_title(\"scatter\")",
"_____no_output_____"
]
],
[
[
"The above scatter plot shows three classes, i.e. 'Iris-setosa', 'Iris-versicolor', 'Iris-virginica'.",
"_____no_output_____"
],
[
"## Divide dataset into Train/Test",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import train_test_split\nX=datasets[['SepalLength','SepalWidth','PetalLength','PetalWidth']]\ny=datasets['Class']\nX_train, X_test, y_train, y_test=train_test_split(X, y, test_size=0.20, random_state=42)",
"_____no_output_____"
]
],
[
[
"## Prepare data for ML",
"_____no_output_____"
],
[
"### Standard scaling",
"_____no_output_____"
]
],
[
[
"from sklearn.preprocessing import StandardScaler\nscaler = StandardScaler()\nX_train_tr = scaler.fit_transform(X_train)\nX_test_tr = scaler.fit_transform(X_test)\nX_train_tr",
"_____no_output_____"
]
],
[
[
"## Training the model",
"_____no_output_____"
],
[
"### SVC with linear kernel",
"_____no_output_____"
]
],
[
[
"from sklearn.svm import SVC\nsvc = SVC(kernel='linear')\nsvc.fit(X_train_tr, y_train)\ny_pred = svc.predict(X_test_tr)\nprint(\"C for trained model: \",svc.C)\nprint(\"gamma for trained model: \",svc.gamma)",
"C for trained model: 1.0\ngamma for trained model: scale\n"
]
],
[
[
"### RMSE for SVC linear kernel",
"_____no_output_____"
]
],
[
[
"from sklearn.metrics import mean_squared_error\nprint(\"Root mean squared error: \",mean_squared_error(y_test, y_pred, squared=False))",
"Root mean squared error: 0.31622776601683794\n"
]
],
[
[
"### Classification report",
"_____no_output_____"
]
],
[
[
"from sklearn.metrics import classification_report\nprint(\"Classification report for SVC linear kernel\")\nprint(classification_report(y_test, y_pred, target_names=['Iris-setosa', 'Iris-versicolor', 'Iris-virginica']))",
"Classification report for SVC linear kernel\n precision recall f1-score support\n\n Iris-setosa 1.00 1.00 1.00 10\nIris-versicolor 0.80 0.89 0.84 9\n Iris-virginica 0.90 0.82 0.86 11\n\n accuracy 0.90 30\n macro avg 0.90 0.90 0.90 30\n weighted avg 0.90 0.90 0.90 30\n\n"
]
],
[
[
"### SVC with rbf kernel",
"_____no_output_____"
]
],
[
[
"svc_rbf = SVC(kernel='rbf')\nsvc_rbf.fit(X_train_tr, y_train)\ny_pred_rbf = svc_rbf.predict(X_test_tr)\nprint(\"C for trained model: \",svc_rbf.C)\nprint(\"gamma for trained model: \",svc_rbf.gamma)",
"C for trained model: 1.0\ngamma for trained model: scale\n"
]
],
[
[
"### RMSE for SVC rbf kernel",
"_____no_output_____"
]
],
[
[
"print(\"Root mean squared error: \",mean_squared_error(y_test, y_pred_rbf, squared=False))",
"Root mean squared error: 0.18257418583505536\n"
]
],
[
[
"### Classification report",
"_____no_output_____"
]
],
[
[
"from sklearn.metrics import classification_report\nprint(\"Classification report for SVC linear kernel\")\nprint(classification_report(y_test, y_pred_rbf, target_names=['Iris-setosa', 'Iris-versicolor', 'Iris-virginica']))",
"Classification report for SVC linear kernel\n precision recall f1-score support\n\n Iris-setosa 1.00 1.00 1.00 10\nIris-versicolor 0.90 1.00 0.95 9\n Iris-virginica 1.00 0.91 0.95 11\n\n accuracy 0.97 30\n macro avg 0.97 0.97 0.97 30\n weighted avg 0.97 0.97 0.97 30\n\n"
]
],
[
[
"## Tuning the SVC rbf kernel model",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import GridSearchCV\nparameters_svc = {'C':[0.01, 0.1, 1, 10, 100, 1000], 'gamma':[0.01, 0.1, 1, 10, 100, 1000]}\nclf = GridSearchCV(svc_rbf, parameters_svc)\nclf.fit(X_train_tr, y_train)\nprint(\"Best parameters for C and gamma\")\nclf.best_estimator_",
"Best parameters for C and gamma\n"
]
],
[
[
"## Accuracy on test set with tuned SVC rbf kernel",
"_____no_output_____"
]
],
[
[
"from sklearn.metrics import accuracy_score\nsvc_tuned = clf.best_estimator_\ny_pred_svc_tuned = svc_tuned.predict(X_test_tr)\nprint(\"RMSE: \",mean_squared_error(y_test, y_pred_svc_tuned, squared=False))\nprint(classification_report(y_test, y_pred_svc_tuned, target_names=['Iris-setosa', 'Iris-versicolor', 'Iris-virginica']))\n\nprint(\"Acurracy: \",accuracy_score(y_test, y_pred_svc_tuned) * 100)",
"RMSE: 0.18257418583505536\n precision recall f1-score support\n\n Iris-setosa 1.00 1.00 1.00 10\nIris-versicolor 0.90 1.00 0.95 9\n Iris-virginica 1.00 0.91 0.95 11\n\n accuracy 0.97 30\n macro avg 0.97 0.97 0.97 30\n weighted avg 0.97 0.97 0.97 30\n\nAcurracy: 96.66666666666667\n"
]
],
[
[
"## KNN",
"_____no_output_____"
]
],
[
[
"from sklearn.neighbors import KNeighborsClassifier\nknn = KNeighborsClassifier(n_neighbors=9)\nknn.fit(X_train_tr, y_train)\ny_pred_knn = svc.predict(X_test_tr)\nprint(\"RMSE: \",mean_squared_error(y_test, y_pred_knn, squared=False))\nprint(\"Classification report: \",classification_report(y_test, y_pred_knn, target_names=['Iris-setosa', 'Iris-versicolor', 'Iris-virginica']))\nprint(\"Accuracy: \", accuracy_score(y_test, y_pred_knn))",
"RMSE: 0.31622776601683794\nClassification report: precision recall f1-score support\n\n Iris-setosa 1.00 1.00 1.00 10\nIris-versicolor 0.80 0.89 0.84 9\n Iris-virginica 0.90 0.82 0.86 11\n\n accuracy 0.90 30\n macro avg 0.90 0.90 0.90 30\n weighted avg 0.90 0.90 0.90 30\n\nAccuracy: 0.9\n"
]
],
[
[
"### Tuning KNN",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import RandomizedSearchCV\nparameters = {'n_neighbors': range(1,11)}\nclf_randomCV = RandomizedSearchCV(knn, parameters, random_state=0)\nsearch = clf_randomCV.fit(X_train_tr, y_train)\nprint(\"Best parameters: \", search.best_params_)",
"Best parameters: {'n_neighbors': 3}\n"
]
],
[
[
"## Accuracy on test set with tuned KNN",
"_____no_output_____"
]
],
[
[
"knn_tuned = search.best_estimator_\ny_pred_knn_tuned = knn_tuned.predict(X_test_tr)\nprint(\"RMSE: \", mean_squared_error(y_test, y_pred_knn_tuned, squared=False))\nprint(\"Classification report: \",classification_report(y_test, y_pred_svc_tuned, target_names=['Iris-setosa', 'Iris-versicolor', 'Iris-virginica']))\nprint(\"Accuracy: \",accuracy_score(y_test, y_pred_knn_tuned) * 100)",
"RMSE: 0.18257418583505536\nClassification report: precision recall f1-score support\n\n Iris-setosa 1.00 1.00 1.00 10\nIris-versicolor 0.90 1.00 0.95 9\n Iris-virginica 1.00 0.91 0.95 11\n\n accuracy 0.97 30\n macro avg 0.97 0.97 0.97 30\n weighted avg 0.97 0.97 0.97 30\n\nAccuracy: 96.66666666666667\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
4a54dd095affcc098300cf73f510fdf961156690
| 12,229 |
ipynb
|
Jupyter Notebook
|
ray-rllib/00-Ray-RLlib-Overview.ipynb
|
desktable/academy
|
6f517b1ad9f573d76a9aa2d3813dfdfe17f56008
|
[
"Apache-2.0"
] | 3 |
2020-09-29T21:55:16.000Z
|
2021-10-30T19:19:25.000Z
|
ray-rllib/00-Ray-RLlib-Overview.ipynb
|
desktable/academy
|
6f517b1ad9f573d76a9aa2d3813dfdfe17f56008
|
[
"Apache-2.0"
] | null | null | null |
ray-rllib/00-Ray-RLlib-Overview.ipynb
|
desktable/academy
|
6f517b1ad9f573d76a9aa2d3813dfdfe17f56008
|
[
"Apache-2.0"
] | null | null | null | 56.87907 | 403 | 0.678142 |
[
[
[
"# Ray RLlib - Overview\n\n© 2019-2020, Anyscale. All Rights Reserved\n\n\n\n## Join Us at Ray Summit 2020!\n\nJoin us for the [_free_ Ray Summit 2020 virtual conference](https://events.linuxfoundation.org/ray-summit/?utm_source=dean&utm_medium=embed&utm_campaign=ray_summit&utm_content=anyscale_academy), September 30 - October 1, 2020. We have an amazing lineup of luminar keynote speakers and breakout sessions on the Ray ecosystem, third-party Ray libraries, and applications of Ray in the real world.\n\n\n\n## About This Tutorial\n\nThis tutorial, part of [Anyscale Academy](https://anyscale.com/academy), introduces the broad topic of _reinforcement learning_ (RL) and [RLlib](https://ray.readthedocs.io/en/latest/rllib.html), Ray's comprehensive RL library.\n\n\n\nThe lessons in this tutorial use different _environments_ from [OpenAI Gym](https://gym.openai.com/) to illustrate how to train _policies_.\n\nSee the instructions in the [README](../README.md) for setting up your environment to use this tutorial.\n\nGo [here](../Overview.ipynb) for an overview of all tutorials.",
"_____no_output_____"
],
[
"## Tutorial Sections\n\nBecause of the breadth of RL this tutorial is divided into several sections. See below for a recommended _learning plan_.\n\n### Introduction to Reinforcement Learning and RLlib\n\n| | Lesson | Description |\n| :- | :----- | :---------- |\n| 00 | [Ray RLlib Overview](00-Ray-RLlib-Overview.ipynb) | Overview of this tutorial, including all the sections. (This file.) |\n| 01 | [Introduction to Reinforcement Learning](01-Introduction-to-Reinforcement-Learning.ipynb) | A quick introduction to the concepts of reinforcement learning. You can skim or skip this lesson if you already understand RL concepts. |\n| 02 | [Introduction to RLlib](02-Introduction-to-RLlib.ipynb) | An overview of RLlib, its goals and the capabilities it provides. |\n| | [RL References](References-Reinforcement-Learning.ipynb) | References on reinforcement learning. |\n\nExercise solutions for this introduction can be found [here](solutions/Ray-RLlib-Solutions.ipynb).",
"_____no_output_____"
],
[
"### Multi-Armed Bandits\n\n_Multi-Armed Bandits_ (MABs) are a special kind of RL problem that have broad and growing applications. They are also an excellent platform for investigating the important _exploitation vs. exploration tradeoff_ at the heart of RL. The term _multi-armed bandit_ is inspired by the slot machines in casinos, so called _one-armed bandits_, but where a machine might have more than one arm. \n\n| | Lesson | Description |\n| :- | :----- | :---------- |\n| 00 | [Multi-Armed-Bandits Overview](multi-armed-bandits/00-Multi-Armed-Bandits-Overview.ipynb) | Overview of this set of lessons. |\n| 01 | [Introduction to Multi-Armed Bandits](multi-armed-bandits/01-Introduction-to-Multi-Armed-Bandits.ipynb) | A quick introduction to the concepts of multi-armed bandits (MABs) and how they fit in the spectrum of RL problems. |\n| 02 | [Exploration vs. Exploitation Strategies](multi-armed-bandits/02-Exploration-vs-Exploitation-Strategies.ipynb) | A deeper look at algorithms that balance exploration vs. exploitation, the key challenge for efficient solutions. Much of this material is technical and can be skipped in a first reading, but skim the first part of this lesson at least. |\n| 03 | [Simple Multi-Armed Bandit](multi-armed-bandits/03-Simple-Multi-Armed-Bandit.ipynb) | A simple example of a multi-armed bandit to illustrate the core ideas. |\n| 04 | [Linear Upper Confidence Bound](multi-armed-bandits/04-Linear-Upper-Confidence-Bound.ipynb) | One popular algorithm for exploration vs. exploitation is _Upper Confidence Bound_. This lesson shows how to use a linear version in RLlib. |\n| 05 | [Linear Thompson Sampling](multi-armed-bandits/05-Linear-Thompson-Sampling.ipynb) | Another popular algorithm for exploration vs. exploitation is _Thompson Sampling_. This lesson shows how to use a linear version in RLlib. |\n| 06 | [Market Example](multi-armed-bandits/06-Market-Example.ipynb) | A simplified real-world example of MABs, finding the optimal stock and bond investment strategy. |\n\nExercise solutions for the bandits section of the tutorial can be found [here](multi-armed-bandits/solutions/Multi-Armed-Bandits-Solutions.ipynb).",
"_____no_output_____"
],
[
"### Explore Reinforcement Learning and RLlib\n\nThis section dives into more details about RL and using RLlib. It is best studied after going through the MAB material.\n\n| | Lesson | Description |\n| :- | :----- | :---------- |\n| 00 | [Explore RLlib Overview](explore-rllib/00-Explore-RLlib-Overview.ipynb) | Overview of this set of lessons. |\n| 01 | [Application - Cart Pole](explore-rllib/01-Application-Cart-Pole.ipynb) | The best starting place for learning how to use RL, in this case to train a moving car to balance a vertical pole. Based on the `CartPole-v1` environment from OpenAI Gym, combined with RLlib. |\n| 02 | [Application: Bipedal Walker](explore-rllib/02-Bipedal-Walker.ipynb) | Train a two-legged robot simulator. This is an optional lesson, due to the longer compute times required, but fun to try. |\n| 03 | [Custom Environments and Reward Shaping](explore-rllib/03-Custom-Environments-Reward-Shaping.ipynb) | How to customize environments and rewards for your applications. |\n\nSome additional examples you might explore can be found in the `extras` folder:\n\n| Lesson | Description |\n| :----- | :---------- |\n| [Extra: Application - Mountain Car](explore-rllib/extras/Extra-Application-Mountain-Car.ipynb) | Based on the `MountainCar-v0` environment from OpenAI Gym. |\n| [Extra: Application - Taxi](explore-rllib/extras/Extra-Application-Taxi.ipynb) | Based on the `Taxi-v3` environment from OpenAI Gym. |\n| [Extra: Application - Frozen Lake](explore-rllib/extras/Extra-Application-Frozen-Lake.ipynb) | Based on the `FrozenLake-v0` environment from OpenAI Gym. |\n\nIn addition, exercise solutions for this \"exploration\" section of the tutorial can be found [here](explore-rllib/solutions/Ray-RLlib-Solutions.ipynb).",
"_____no_output_____"
],
[
"### RecSys: Recommender System\n\nThis section applies RL to the problem of building a recommender system, a state-of-the-art technique that addresses many of the limitations of older approaches.\n\n| | Lesson | Description |\n| :- | :----- | :---------- |\n| 00 | [RecSys: Recommender System Overview](recsys/00-RecSys-Overview.ipynb) | Overview of this set of lessons. |\n| 01 | [Recsys: Recommender System](recsys/01-Recsys.ipynb) | An example that builds a recommender system using reinforcement learning. |\n\nThe [Custom Environments and Reward Shaping](explore-rllib/03-Custom-Environments-Reward-Shaping.ipynb) lesson from _Explore RLlib_ might be useful background for this section.",
"_____no_output_____"
],
[
"For earlier versions of some of these tutorials, see [`rllib_exercises`](https://github.com/ray-project/tutorial/blob/master/rllib_exercises/rllib_colab.ipynb) in the original [github.com/ray-project/tutorial](https://github.com/ray-project/tutorial) project.",
"_____no_output_____"
],
[
"## Learning Plan\n\nWe recommend the following _learning plan_ for working through the lessons:\n\nStart with the introduction material for RL and RLlib:\n\n* [Ray RLlib Overview](00-Ray-RLlib-Overview.ipynb) - This file\n* [Introduction to Reinforcement Learning](01-Introduction-to-Reinforcement-Learning.ipynb) \n* [Introduction to RLlib](02-Introduction-to-RLlib.ipynb)\n\nThen study several of the lessons for multi-armed bandits, starting with these lessons:\n\n* [Multi-Armed-Bandits Overview](multi-armed-bandits/00-Multi-Armed-Bandits-Overview.ipynb)\n* [Introduction to Multi-Armed Bandits](multi-armed-bandits/01-Introduction-to-Multi-Armed-Bandits.ipynb)\n* [Exploration vs. Exploitation Strategies](multi-armed-bandits/02-Exploration-vs-Exploitation-Strategies.ipynb): Skim at least the first part of this lesson. \n* [Simple Multi-Armed Bandit](multi-armed-bandits/03-Simple-Multi-Armed-Bandit.ipynb)\n\nAs time permits, study one or both of the following lessons:\n\n* [Linear Upper Confidence Bound](multi-armed-bandits/04-Linear-Upper-Confidence-Bound.ipynb)\n* [Linear Thompson Sampling](multi-armed-bandits/05-Linear-Thompson-Sampling.ipynb)\n\nThen finish with this more complete example:\n\n* [Market Example](multi-armed-bandits/06-Market-Example.ipynb)\n\nNext, return to the \"exploration\" lessons under `explore-rllib` and work through as many of the following lessons as time permits:\n\n* [Application: Cart Pole](explore-rllib/01-Application-Cart-Pole.ipynb): Further exploration of the popular `CartPole` example.\n* [Application: Bipedal Walker](explore-rllib/02-Bipedal-Walker.ipynb): A nontrivial, but simplified robot simulator.\n* [Custom Environments and Reward Shaping](explore-rllib/03-Custom-Environments-Reward-Shaping.ipynb): More about creating custom environments for your problem. Also, finetuning the rewards to ensure sufficient exploration.\n\nOther examples that use different OpenAI Gym environments are provided for your use in the `extras` directory:\n\n* [Extra: Application - Mountain Car](explore-rllib/extras/Extra-Application-Mountain-Car.ipynb)\n* [Extra: Application - Taxi](explore-rllib/extras/Extra-Application-Taxi.ipynb)\n* [Extra: Application - Frozen Lake](explore-rllib/extras/Extra-Application-Frozen-Lake.ipynb)\n\nFinally, the [references](References-Reinforcement-Learning.ipynb) collect useful books, papers, blog posts, and other available tutorial materials.",
"_____no_output_____"
],
[
"## Getting Help\n\n* The [#tutorial channel](https://ray-distributed.slack.com/archives/C011ML23W5B) on the [Ray Slack](https://ray-distributed.slack.com). [Click here](https://forms.gle/9TSdDYUgxYs8SA9e8) to join.\n* [Email](mailto:[email protected])\n\nFind an issue? Please report it!\n\n* [GitHub issues](https://github.com/anyscale/academy/issues)",
"_____no_output_____"
],
[
"## Give Us Feedback!\n\nLet us know what you like and don't like about this RL and RLlib tutorial.\n\n* [Survey](https://forms.gle/D2Lo4K5tkcqsWeKU8)",
"_____no_output_____"
]
]
] |
[
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
4a54ff89325fce99d82444c2cbd2dcb9ee71ab2d
| 39,133 |
ipynb
|
Jupyter Notebook
|
notebooks/CredentialExtraction_SSA.ipynb
|
0xFFD700/security_content
|
cac50b13166cde29bc74ca189fe1418c9675bb65
|
[
"Apache-2.0"
] | null | null | null |
notebooks/CredentialExtraction_SSA.ipynb
|
0xFFD700/security_content
|
cac50b13166cde29bc74ca189fe1418c9675bb65
|
[
"Apache-2.0"
] | null | null | null |
notebooks/CredentialExtraction_SSA.ipynb
|
0xFFD700/security_content
|
cac50b13166cde29bc74ca189fe1418c9675bb65
|
[
"Apache-2.0"
] | null | null | null | 32.049959 | 114 | 0.473667 |
[
[
[
"empty"
]
]
] |
[
"empty"
] |
[
[
"empty"
]
] |
4a5512a3902f652939f44bf950e4d26aefe45ed2
| 4,881 |
ipynb
|
Jupyter Notebook
|
notebooks/1.Conceitos-Instalacao-Ambiente.ipynb
|
fredcsilva/aprendendo-flask
|
42e943f10096d857f058b4cacc1e51e39b89b779
|
[
"Apache-2.0"
] | null | null | null |
notebooks/1.Conceitos-Instalacao-Ambiente.ipynb
|
fredcsilva/aprendendo-flask
|
42e943f10096d857f058b4cacc1e51e39b89b779
|
[
"Apache-2.0"
] | null | null | null |
notebooks/1.Conceitos-Instalacao-Ambiente.ipynb
|
fredcsilva/aprendendo-flask
|
42e943f10096d857f058b4cacc1e51e39b89b779
|
[
"Apache-2.0"
] | null | null | null | 27.268156 | 189 | 0.575907 |
[
[
[
"# Aprendendo Flask",
"_____no_output_____"
],
[
"| [Anterior](#)| [Próximo](2.Primeiros-Passos.ipynb) | \n| :------------- | :----------:|",
"_____no_output_____"
],
[
"### 1. Conceitos Básicos",
"_____no_output_____"
],
[
"- É um microframework para aplicativos Web escrito em Python.\n- Flask é baseado no kit Werkzeug WSGI (Web Server Gateway Interface) e Jinja2 (View) próprio para criar templates em Python.\n- Uma ferramenta para construção de Apis\n- Flask é uma microestrutura que mantém a simplicidade do núcleo da aplicação, porém, é bastante extensível através de módulos que podem ser agregados no decorrer do desenvolvimento. ",
"_____no_output_____"
],
[
"### 2. Instalação (ambiente Windows)",
"_____no_output_____"
],
[
"**Antes de tudo:** Baixe e Instale o Python (https://www.python.org/downloads/)\n\n**Dica:** *Crie um ambiente virtual (virtualenv) para instalar o Flask.*\n> `$ pip install virtual_env`\n<br/>\n\n**Inicie o virtualenv (ex: com o nome venv_flask)**\n<br/>\n> `$ py -3 -m venv venv_flask`\nou\n> `$ virtualenv -p python3 venv_flask`\n<br/>\n\n**Ative o virtualenv**\n<br/>\n> `$ venv_flask\\Scripts\\activate`\n<br/>\n\n**Instale as bibliotecas no ambiente virtual do projeto**\n> `$ (venv_flask) pip install Flask`\n\n**OBS:** O Flask suporta Python 3.6 e mais recente. E o suporte assíncrono no Flask requer Python 3.7+ para contextvars.ContextVar. <br/>\n**Saiba mais sobre a instalação em:** https://flask.palletsprojects.com/en/2.0.x/installation/",
"_____no_output_____"
],
[
"### 3. Confirmando a instalação do Flask",
"_____no_output_____"
],
[
"- Para confirmar que o flask foi instalado corretamente, faça o seguinte:\n\n> Acesse o seu \"prompt de comando\" <br/>\n> Digite o comando `$ python` <br/>\n> Após entrar no ambiente de shell do python, digite o comando <br/>\n> `>>> import flask` <br/>\nSe nenhum erro for apresentado é porque o flask foi instalado corretamente. Em seguida, digite o comando <br/>\n> `>>> exit()` <br/>",
"_____no_output_____"
],
[
"### 4. Gerando e Carregando as Bibliotecas Instaladas no Ambiente",
"_____no_output_____"
],
[
"**Gerando um arquivo \"requirements.txt\" contendo todas as bibliotecas do meu projeto**",
"_____no_output_____"
],
[
"> `$ pip freeze > requirements.txt`",
"_____no_output_____"
],
[
"**Importanto o arquivo \"requirements.txt\" contendo todas as bibliotecas do meu projeto**",
"_____no_output_____"
],
[
"> `$ pip install -r requirements.txt ou $ pip3 install -r requirements.txt`",
"_____no_output_____"
],
[
"### 5. Página do Flask",
"_____no_output_____"
],
[
"https://flask.palletsprojects.com/en/2.0.x/",
"_____no_output_____"
]
]
] |
[
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
4a5521d917beeedd04a3c0b010e33366026620a4
| 22,507 |
ipynb
|
Jupyter Notebook
|
data_accelerometer_gyroscope_features.ipynb
|
hontarenkoYana/driving_behaviour_prediction_model
|
3ddf9ece3a8a0529627978f5de8c71e5c48e1326
|
[
"Apache-2.0"
] | null | null | null |
data_accelerometer_gyroscope_features.ipynb
|
hontarenkoYana/driving_behaviour_prediction_model
|
3ddf9ece3a8a0529627978f5de8c71e5c48e1326
|
[
"Apache-2.0"
] | 1 |
2021-08-23T20:38:54.000Z
|
2021-08-23T20:38:54.000Z
|
data_accelerometer_gyroscope_features.ipynb
|
hontarenkoYana/driving_behaviour_prediction_model
|
3ddf9ece3a8a0529627978f5de8c71e5c48e1326
|
[
"Apache-2.0"
] | 2 |
2020-03-13T14:09:08.000Z
|
2020-03-13T14:18:24.000Z
| 39.55536 | 169 | 0.598436 |
[
[
[
"<font size=\"+0.5\">Notebook for transform data format to train the model<font>",
"_____no_output_____"
],
[
"# <center> Data transform",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nimport numpy as np\nimport pandas as pd\nimport os\n\nfrom datetime import datetime\n\nfrom scipy.signal import savgol_filter\nfrom sklearn.utils import shuffle",
"_____no_output_____"
],
[
"# Timestamp form in init data have this format\nform = \"%d/%m/%Y %H:%M:%S\"",
"_____no_output_____"
],
[
"# Dictionary to replace events\nreplacement = {\"curva_direita_agressiva\": \"Aggressive right turn\",\n \"curva_esquerda_agressiva\": \"Aggressive left turn\",\n \"evento_nao_agressivo\": \"Non-aggressive event\",\n \"troca_faixa_direita_agressiva\": \"Aggressive right lane change\",\n \"aceleracao_agressiva\": \"Aggressive acceleration\",\n \"freada_agressiva\": \"Aggressive breaking\",\n \"troca_faixa_esquerda_agressiva\": \"Aggressive left lane change\",\n \"No label\": \"No label\"}",
"_____no_output_____"
],
[
"# Function to replace events\ndef replace_event(row):\n return replacement[row['event']]",
"_____no_output_____"
],
[
"# Load and concatenate accelerometer data with its events\ndef make_labeled_data(folder_num):\n # Load events and its time\n data_label = pd.read_csv(os.path.join('data', 'data_init', str(folder_num), 'groundTruth.csv'))\n # Load accelerometer and gyroscope data\n data_acc = pd.read_csv(os.path.join('data', 'data_init', str(folder_num), 'aceleracaoLinear_terra.csv'))\n data_gyro = pd.read_csv(os.path.join('data', 'data_init', str(folder_num), 'giroscopio_terra.csv'))\n \n data = data_acc.copy()\n data = data.rename(columns={\"x\": \"x_accelerometer\", \"y\": \"y_accelerometer\", \"z\": \"z_accelerometer\"})\n data['x_gyroscope'] = data_gyro[\"x\"]\n data['y_gyroscope'] = data_gyro[\"y\"]\n data['z_gyroscope'] = data_gyro[\"z\"]\n \n # Take first time as start of the trip\n init = datetime.strptime(data.loc[0]['timestamp'], form)\n \n # Function for changing time on its duration of the time by this trip to this record\n def change_timestamp(row):\n return (datetime.strptime(row['timestamp'], form) - init).seconds\n \n \n \n data['time_duration'] = data.apply(change_timestamp, axis=1)\n \n for index, row in data_label.iterrows():\n start = row[' inicio']\n finish = row[' fim']\n data.loc[((data['time_duration'] >= start) & (data['time_duration'] < finish)), 'event'] = row['evento']\n \n data['event'] = data['event'].fillna(\"No label\")\n data['event'] = data.apply(replace_event, axis=1)\n \n return data",
"_____no_output_____"
],
[
"# Function for creating sequence of events in one dataframe\n# Each event has its own number if it is on different time interval\ndef create_events_sequence(data):\n event_num = 1\n event = data.iloc[0][\"event\"]\n sequence = []\n \n for index, row in data.iterrows():\n if row[\"event\"] != event:\n event_num += 1\n event = data.loc[index, \"event\"]\n sequence.append(event_num)\n return sequence",
"_____no_output_____"
],
[
"# Function for adding new events to the dictionary of events\ndef add_events_to_dict(data, dictionary):\n # Create events sequence in this dataframe\n data[\"event_number\"] = create_events_sequence(data)\n # Select only labeled data\n data = data[data[\"event\"] != \"No label\"]\n # Group data by unique number of event\n data_groupbed = data.groupby(\"event_number\")\n \n # For each unique event number\n for group in np.unique(data[\"event_number\"].values):\n current_group = data_groupbed.get_group(group)\n event_name = current_group[\"event\"].values[0]\n # If dictionary has this event name add dataframe to the list\n # Otherwise create list with this dataframe\n if dictionary.get(event_name):\n dictionary[event_name].append(current_group)\n else:\n dictionary[event_name] = [current_group]\n # Return updated dictionary\n return dictionary",
"_____no_output_____"
],
[
"data1 = make_labeled_data(16)\ndata2 = make_labeled_data(17)\ndata3 = make_labeled_data(20)\ndata4 = make_labeled_data(21)",
"_____no_output_____"
]
],
[
[
"# <center> Data filtering",
"_____no_output_____"
],
[
"### <center> Gyroscope data filtering",
"_____no_output_____"
],
[
"<font size=\"+0.5\">Look at both curves: initial and filtered and find those <i><b>window lenght</b></i> which filtered curva describe data in the best way.</font>",
"_____no_output_____"
]
],
[
[
"window_lengths = np.arange(11, 151, 10)\npolyorder = 3",
"_____no_output_____"
],
[
"for window_length in window_lengths:\n \n data1['x_gyroscope_fil'] = savgol_filter(data1['x_gyroscope'].values, window_length, polyorder)\n data1['y_gyroscope_fil'] = savgol_filter(data1['y_gyroscope'].values, window_length, polyorder)\n data1['z_gyroscope_fil'] = savgol_filter(data1['z_gyroscope'].values, window_length, polyorder)\n \n fig, ax = plt.subplots(1, 3, figsize=(10, 5))\n \n ax[0].plot(data1[:500]['x_gyroscope'].values, label='x gyroscope')\n ax[0].plot(data1[:500]['x_gyroscope_fil'].values, label='x gyroscope filtered')\n ax[0].legend();\n \n ax[1].plot(data1[:500]['y_gyroscope'].values, label='y gyroscope')\n ax[1].plot(data1[:500]['y_gyroscope_fil'].values, label='y gyroscope filtered')\n ax[1].legend();\n \n ax[2].plot(data1[:500]['z_gyroscope'].values, label='z gyroscope')\n ax[2].plot(data1[:500]['z_gyroscope_fil'].values, label='z gyroscope filtered')\n plt.suptitle(f\"Window length: {window_length}\", fontsize=20)\n ",
"_____no_output_____"
]
],
[
[
"<font size=\"+0.5\">Look at both curves: initial and filtered and find those <i><b>polyorder</b></i> which filtered curve describe data in the best way.</font>",
"_____no_output_____"
]
],
[
[
"polyorders = np.arange(2, 15, 1)\nwindow_length = 31",
"_____no_output_____"
],
[
"for polyorder in polyorders:\n \n data1['x_gyroscope_fil'] = savgol_filter(data1['x_gyroscope'].values, window_length, polyorder)\n data1['y_gyroscope_fil'] = savgol_filter(data1['y_gyroscope'].values, window_length, polyorder)\n data1['z_gyroscope_fil'] = savgol_filter(data1['z_gyroscope'].values, window_length, polyorder)\n \n fig, ax = plt.subplots(1, 3, figsize=(10, 5))\n \n ax[0].plot(data1[:500]['x_gyroscope'].values, label='x gyroscope')\n ax[0].plot(data1[:500]['x_gyroscope_fil'].values, label='x gyroscope filtered')\n ax[0].legend();\n \n ax[1].plot(data1[:500]['y_gyroscope'].values, label='y gyroscope')\n ax[1].plot(data1[:500]['y_gyroscope_fil'].values, label='y gyroscope filtered')\n ax[1].legend();\n \n ax[2].plot(data1[:500]['z_gyroscope'].values, label='z gyroscope')\n ax[2].plot(data1[:500]['z_gyroscope_fil'].values, label='z gyroscope filtered')\n plt.suptitle(f\"Window length: {window_length}\", fontsize=20)\n ",
"_____no_output_____"
],
[
"polyorder = 4",
"_____no_output_____"
],
[
"data1['x_gyroscope_fil'] = savgol_filter(data1['x_gyroscope'].values, window_length, polyorder)\ndata1['y_gyroscope_fil'] = savgol_filter(data1['y_gyroscope'].values, window_length, polyorder)\ndata1['z_gyroscope_fil'] = savgol_filter(data1['z_gyroscope'].values, window_length, polyorder)\n\ndata2['x_gyroscope_fil'] = savgol_filter(data2['x_gyroscope'].values, window_length, polyorder)\ndata2['y_gyroscope_fil'] = savgol_filter(data2['y_gyroscope'].values, window_length, polyorder)\ndata2['z_gyroscope_fil'] = savgol_filter(data2['z_gyroscope'].values, window_length, polyorder)\n\ndata3['x_gyroscope_fil'] = savgol_filter(data3['x_gyroscope'].values, window_length, polyorder)\ndata3['y_gyroscope_fil'] = savgol_filter(data3['y_gyroscope'].values, window_length, polyorder)\ndata3['z_gyroscope_fil'] = savgol_filter(data3['z_gyroscope'].values, window_length, polyorder)\n\ndata4['x_gyroscope_fil'] = savgol_filter(data4['x_gyroscope'].values, window_length, polyorder)\ndata4['y_gyroscope_fil'] = savgol_filter(data4['y_gyroscope'].values, window_length, polyorder)\ndata4['z_gyroscope_fil'] = savgol_filter(data4['z_gyroscope'].values, window_length, polyorder)",
"_____no_output_____"
],
[
"data1[\"mean_window_x_gyroscope\"] = data1[\"x_gyroscope_fil\"].rolling(8, min_periods=1).mean()\ndata1[\"mean_window_y_gyroscope\"] = data1[\"y_gyroscope_fil\"].rolling(8, min_periods=1).mean()\ndata1[\"mean_window_z_gyroscope\"] = data1[\"z_gyroscope_fil\"].rolling(8, min_periods=1).mean()\n\ndata2[\"mean_window_x_gyroscope\"] = data2[\"x_gyroscope_fil\"].rolling(8, min_periods=1).mean()\ndata2[\"mean_window_y_gyroscope\"] = data2[\"y_gyroscope_fil\"].rolling(8, min_periods=1).mean()\ndata2[\"mean_window_z_gyroscope\"] = data2[\"z_gyroscope_fil\"].rolling(8, min_periods=1).mean()\n\ndata3[\"mean_window_x_gyroscope\"] = data3[\"x_gyroscope_fil\"].rolling(8, min_periods=1).mean()\ndata3[\"mean_window_y_gyroscope\"] = data3[\"y_gyroscope_fil\"].rolling(8, min_periods=1).mean()\ndata3[\"mean_window_z_gyroscope\"] = data3[\"z_gyroscope_fil\"].rolling(8, min_periods=1).mean()\n\ndata4[\"mean_window_x_gyroscope\"] = data4[\"x_gyroscope_fil\"].rolling(8, min_periods=1).mean()\ndata4[\"mean_window_y_gyroscope\"] = data4[\"y_gyroscope_fil\"].rolling(8, min_periods=1).mean()\ndata4[\"mean_window_z_gyroscope\"] = data4[\"z_gyroscope_fil\"].rolling(8, min_periods=1).mean()",
"_____no_output_____"
],
[
"data1[\"std_window_x_gyroscope\"] = data1[\"x_gyroscope_fil\"].rolling(8, min_periods=1).std()\ndata1[\"std_window_y_gyroscope\"] = data1[\"y_gyroscope_fil\"].rolling(8, min_periods=1).std()\ndata1[\"std_window_z_gyroscope\"] = data1[\"z_gyroscope_fil\"].rolling(8, min_periods=1).std()\n\ndata2[\"std_window_x_gyroscope\"] = data2[\"x_gyroscope_fil\"].rolling(8, min_periods=1).std()\ndata2[\"std_window_y_gyroscope\"] = data2[\"y_gyroscope_fil\"].rolling(8, min_periods=1).std()\ndata2[\"std_window_z_gyroscope\"] = data2[\"z_gyroscope_fil\"].rolling(8, min_periods=1).std()\n\ndata3[\"std_window_x_gyroscope\"] = data3[\"x_gyroscope_fil\"].rolling(8, min_periods=1).std()\ndata3[\"std_window_y_gyroscope\"] = data3[\"y_gyroscope_fil\"].rolling(8, min_periods=1).std()\ndata3[\"std_window_z_gyroscope\"] = data3[\"z_gyroscope_fil\"].rolling(8, min_periods=1).std()\n\ndata4[\"std_window_x_gyroscope\"] = data4[\"x_gyroscope_fil\"].rolling(8, min_periods=1).std()\ndata4[\"std_window_y_gyroscope\"] = data4[\"y_gyroscope_fil\"].rolling(8, min_periods=1).std()\ndata4[\"std_window_z_gyroscope\"] = data4[\"z_gyroscope_fil\"].rolling(8, min_periods=1).std()",
"_____no_output_____"
],
[
"data1[\"median_window_x_gyroscope\"] = data1[\"x_gyroscope_fil\"].rolling(8, min_periods=1).median()\ndata1[\"median_window_y_gyroscope\"] = data1[\"y_gyroscope_fil\"].rolling(8, min_periods=1).median()\ndata1[\"median_window_z_gyroscope\"] = data1[\"z_gyroscope_fil\"].rolling(8, min_periods=1).median()\n\ndata2[\"median_window_x_gyroscope\"] = data2[\"x_gyroscope_fil\"].rolling(8, min_periods=1).median()\ndata2[\"median_window_y_gyroscope\"] = data2[\"y_gyroscope_fil\"].rolling(8, min_periods=1).median()\ndata2[\"median_window_z_gyroscope\"] = data2[\"z_gyroscope_fil\"].rolling(8, min_periods=1).median()\n\ndata3[\"median_window_x_gyroscope\"] = data3[\"x_gyroscope_fil\"].rolling(8, min_periods=1).median()\ndata3[\"median_window_y_gyroscope\"] = data3[\"y_gyroscope_fil\"].rolling(8, min_periods=1).median()\ndata3[\"median_window_z_gyroscope\"] = data3[\"z_gyroscope_fil\"].rolling(8, min_periods=1).median()\n\ndata4[\"median_window_x_gyroscope\"] = data4[\"x_gyroscope_fil\"].rolling(8, min_periods=1).median()\ndata4[\"median_window_y_gyroscope\"] = data4[\"y_gyroscope_fil\"].rolling(8, min_periods=1).median()\ndata4[\"median_window_z_gyroscope\"] = data4[\"z_gyroscope_fil\"].rolling(8, min_periods=1).median()",
"_____no_output_____"
],
[
"def roll_column_with_duplicate(column):\n result = np.roll(column, 1)\n result[0] = result[1]\n return result",
"_____no_output_____"
],
[
"data1[\"tendency_window_x_gyroscope\"] = roll_column_with_duplicate(data1[\"mean_window_x_gyroscope\"].values) / data1[\"mean_window_x_gyroscope\"]\ndata1[\"tendency_window_y_gyroscope\"] = roll_column_with_duplicate(data1[\"mean_window_y_gyroscope\"].values) / data1[\"mean_window_y_gyroscope\"]\ndata1[\"tendency_window_z_gyroscope\"] = roll_column_with_duplicate(data1[\"mean_window_z_gyroscope\"].values) / data1[\"mean_window_z_gyroscope\"]\n\ndata2[\"tendency_window_x_gyroscope\"] = roll_column_with_duplicate(data2[\"mean_window_x_gyroscope\"].values) / data2[\"mean_window_x_gyroscope\"]\ndata2[\"tendency_window_y_gyroscope\"] = roll_column_with_duplicate(data2[\"mean_window_y_gyroscope\"].values) / data2[\"mean_window_y_gyroscope\"]\ndata2[\"tendency_window_z_gyroscope\"] = roll_column_with_duplicate(data2[\"mean_window_z_gyroscope\"].values) / data2[\"mean_window_z_gyroscope\"]\n\ndata3[\"tendency_window_x_gyroscope\"] = roll_column_with_duplicate(data3[\"mean_window_x_gyroscope\"].values) / data3[\"mean_window_x_gyroscope\"]\ndata3[\"tendency_window_y_gyroscope\"] = roll_column_with_duplicate(data3[\"mean_window_y_gyroscope\"].values) / data3[\"mean_window_y_gyroscope\"]\ndata3[\"tendency_window_z_gyroscope\"] = roll_column_with_duplicate(data3[\"mean_window_z_gyroscope\"].values) / data3[\"mean_window_z_gyroscope\"]\n\ndata4[\"tendency_window_x_gyroscope\"] = roll_column_with_duplicate(data4[\"mean_window_x_gyroscope\"].values) / data4[\"mean_window_x_gyroscope\"]\ndata4[\"tendency_window_y_gyroscope\"] = roll_column_with_duplicate(data4[\"mean_window_y_gyroscope\"].values) / data4[\"mean_window_y_gyroscope\"]\ndata4[\"tendency_window_z_gyroscope\"] = roll_column_with_duplicate(data4[\"mean_window_z_gyroscope\"].values) / data4[\"mean_window_z_gyroscope\"]",
"_____no_output_____"
],
[
"# Dictionary for storing parts of dataframe by its event\nevent_dict = {}",
"_____no_output_____"
],
[
"event_dict = add_events_to_dict(data1, event_dict)\nevent_dict = add_events_to_dict(data2, event_dict)\nevent_dict = add_events_to_dict(data3, event_dict)\nevent_dict = add_events_to_dict(data4, event_dict)",
"_____no_output_____"
],
[
"train_agg_br = pd.concat([event_dict[\"Aggressive breaking\"][i] for i in [0, 2, 3, 4, 5, 6, 8, 9, 11, 7]])\nval_agg_br = pd.concat([event_dict[\"Aggressive breaking\"][i] for i in [1, 10]])\n\ntrain_agg_ac = pd.concat([event_dict[\"Aggressive acceleration\"][i] for i in [0, 2, 3, 4, 5, 6, 8, 9, 11, 7]])\nval_agg_ac = pd.concat([event_dict[\"Aggressive acceleration\"][i] for i in [1, 10]])\n\ntrain_agg_lt = pd.concat([event_dict[\"Aggressive left turn\"][i] for i in [0, 2, 3, 4, 5, 6, 8, 9, 7]])\nval_agg_lt = pd.concat([event_dict[\"Aggressive left turn\"][i] for i in [1, 10]])\n\ntrain_agg_rt = pd.concat([event_dict[\"Aggressive right turn\"][i] for i in [0, 2, 3, 4, 5, 6, 8, 9, 7]])\nval_agg_rt = pd.concat([event_dict[\"Aggressive right turn\"][i] for i in [1, 10]])\n\ntrain_agg_lc = pd.concat([event_dict[\"Aggressive left lane change\"][i] for i in [0, 2, 3]])\nval_agg_lc = pd.concat([event_dict[\"Aggressive left lane change\"][i] for i in [1]])\n\ntrain_agg_rc = pd.concat([event_dict[\"Aggressive right lane change\"][i] for i in [0, 2, 4, 3]])\nval_agg_rc = pd.concat([event_dict[\"Aggressive right lane change\"][i] for i in [1]])\n\ntrain_agg_na = pd.concat([event_dict[\"Non-aggressive event\"][i] for i in [0, 2, 3, 4, 5, 6, 8, 9, 11, 13, 10]])\nval_agg_na = pd.concat([event_dict[\"Non-aggressive event\"][i] for i in [1, 7, 12]])",
"_____no_output_____"
],
[
"train = pd.concat([train_agg_br, train_agg_ac, train_agg_lt, train_agg_rt, train_agg_lc, train_agg_rc, train_agg_na])\nval = pd.concat([val_agg_br, val_agg_ac, val_agg_lt, val_agg_rt, val_agg_lc, val_agg_rc, val_agg_na])",
"_____no_output_____"
],
[
"columns_to_save = [\"mean_window_x_gyroscope\",\n \"mean_window_y_gyroscope\",\n \"mean_window_z_gyroscope\",\n \"std_window_x_gyroscope\",\n \"std_window_y_gyroscope\",\n \"std_window_z_gyroscope\",\n \"median_window_x_gyroscope\",\n \"median_window_y_gyroscope\",\n \"median_window_z_gyroscope\",\n \"tendency_window_x_gyroscope\",\n \"tendency_window_y_gyroscope\",\n \"tendency_window_z_gyroscope\",\n \"event\"]",
"_____no_output_____"
],
[
"train = train.fillna(method=\"bfill\")\nval = val.fillna(method=\"bfill\")",
"_____no_output_____"
],
[
"train = shuffle(train)\nval = shuffle(val)",
"_____no_output_____"
],
[
"train[columns_to_save].to_csv('data/train_gyroscope_features.csv', index=False)",
"_____no_output_____"
],
[
"val[columns_to_save].to_csv('data/val_gyroscope_features.csv', index=False)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a5524f37984855c14988b4fcfde60520bb1fd2c
| 6,061 |
ipynb
|
Jupyter Notebook
|
1 - Python for Data Science/Module 5 - Working with Numpy Arrays/code/3-PY0101EN-5.2_notebook_quizz_numpy.ipynb
|
joaopaulo164/Data-Science-with-Python
|
eff0240f97c150e65e12a295e47b5dee2d1bdbf7
|
[
"MIT"
] | null | null | null |
1 - Python for Data Science/Module 5 - Working with Numpy Arrays/code/3-PY0101EN-5.2_notebook_quizz_numpy.ipynb
|
joaopaulo164/Data-Science-with-Python
|
eff0240f97c150e65e12a295e47b5dee2d1bdbf7
|
[
"MIT"
] | null | null | null |
1 - Python for Data Science/Module 5 - Working with Numpy Arrays/code/3-PY0101EN-5.2_notebook_quizz_numpy.ipynb
|
joaopaulo164/Data-Science-with-Python
|
eff0240f97c150e65e12a295e47b5dee2d1bdbf7
|
[
"MIT"
] | null | null | null | 17.931953 | 203 | 0.460155 |
[
[
[
"<center>\n <img src=\"https://cf-courses-data.s3.us.cloud-object-storage.appdomain.cloud/IBMDeveloperSkillsNetwork-PY0101EN-SkillsNetwork/IDSNlogo.png\" width=\"300\" alt=\"cognitiveclass.ai logo\" />\n</center>\n",
"_____no_output_____"
],
[
"<h3> Get to Know a Numpy Array </h3>\n",
"_____no_output_____"
],
[
"You will use the numpy array <code>A</code> for the following questions.\n",
"_____no_output_____"
]
],
[
[
"import numpy as np\nA=np.array([[11,12],[21,22],[31,32]])\n",
"_____no_output_____"
]
],
[
[
"1. Find the type of `x` using the function `type()`.\n",
"_____no_output_____"
]
],
[
[
"type(A)",
"_____no_output_____"
]
],
[
[
"<details><summary>Click here for the solution</summary>\n\n```python\ntype(A)\n```\n\n</details>\n",
"_____no_output_____"
],
[
"2. Find the shape of the array:\n",
"_____no_output_____"
]
],
[
[
"A.shape",
"_____no_output_____"
]
],
[
[
"<details><summary>Click here for the solution</summary>\n\n```python\nA.shape\n```\n\n</details>\n",
"_____no_output_____"
],
[
"3. Find the type of data in the array:\n",
"_____no_output_____"
]
],
[
[
"A.dtype",
"_____no_output_____"
]
],
[
[
"<details><summary>Click here for the solution</summary>\n\n```python\nA.dtype\n```\n\n</details>\n",
"_____no_output_____"
],
[
"4. Find the second row of the numpy array <code>A</code>:\n",
"_____no_output_____"
]
],
[
[
"A[1]",
"_____no_output_____"
]
],
[
[
"<details><summary>Click here for the solution</summary>\n\n```python\nA[1]\n```\n\n</details>\n",
"_____no_output_____"
],
[
"<h3>Two Types of Multiplication</h3>\n",
"_____no_output_____"
],
[
"You will use the following numpy arrays for the next questions:\n",
"_____no_output_____"
]
],
[
[
"A=np.array([[11,12],[21,22]])\nB=np.array([[1, 0],[0,1]])",
"_____no_output_____"
]
],
[
[
"1. Multiply array <code>A</code> and <code>B</code>.\n",
"_____no_output_____"
]
],
[
[
"C = A * B\nC",
"_____no_output_____"
]
],
[
[
"<details><summary>Click here for the solution</summary>\n\n```python\nC = A * B\nC\n```\n\n</details>\n",
"_____no_output_____"
],
[
"2. Perform matrix multiplication on array <code>A</code> and <code>B</code> (order will not matter in this case).\n",
"_____no_output_____"
]
],
[
[
"Z = np.dot(A,B)\nZ",
"_____no_output_____"
]
],
[
[
"<details><summary>Click here for the solution</summary>\n\n```python\nZ = np.dot(A,B)\nZ\n```\n\n</details>\n",
"_____no_output_____"
],
[
"<hr>\n\n<h3 align=\"center\"> © IBM Corporation 2020. All rights reserved. <h3/>\n",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
4a55339875cb2dc96955ef276a2685febe5ab352
| 4,902 |
ipynb
|
Jupyter Notebook
|
OceanPython041120.ipynb
|
franklinperseuDS/exercicios
|
752c36aac852a4e2af04547e9c6cd28ef96736c5
|
[
"MIT"
] | null | null | null |
OceanPython041120.ipynb
|
franklinperseuDS/exercicios
|
752c36aac852a4e2af04547e9c6cd28ef96736c5
|
[
"MIT"
] | null | null | null |
OceanPython041120.ipynb
|
franklinperseuDS/exercicios
|
752c36aac852a4e2af04547e9c6cd28ef96736c5
|
[
"MIT"
] | null | null | null | 22.281818 | 241 | 0.415545 |
[
[
[
"<a href=\"https://colab.research.google.com/github/franklinperseuDS/exercicios/blob/main/OceanPython041120.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"import datetime as dt",
"_____no_output_____"
],
[
"dataCriada = dt.date(2010,11,15,)",
"_____no_output_____"
],
[
"print(dataCriada)",
"2010-11-15\n"
],
[
"print(dataCriada.day)",
"15\n"
],
[
"dataHoje = dt.date.today()",
"_____no_output_____"
],
[
"print(dataHoje)",
"2020-11-04\n"
],
[
"dataHora = datetime.datetime.now()\nprint(dataHora)",
"2020-11-04 16:28:20.096560\n"
],
[
"idade = 30\nidade_maior = idade > 18\nif not idade_maior:\n print(\"teste1\")\nelse:\n print(\"teste2\")",
"teste2\n"
],
[
"\ndef jogo():\n frase = input(\"Você se considera mais medieval ou futurista?\\n\").lower().strip()\n \n if 'medieval' in frase:\n print(\"espada\")\n \n elif \"futurista\" in frase:\n print(\"sabre de luz\")\n \n else:\n print(\"Tente novamente.\")\n jogo()\n\n\njogo()\n\n",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a553a05c22132643142750e9a8d07d511d9f0a8
| 528,820 |
ipynb
|
Jupyter Notebook
|
Document similarity and clustering V1.ipynb
|
jsafyan/edav-project-4
|
0d3ad40a532da6d0b2acf70a15e55f5cd8354f88
|
[
"MIT"
] | null | null | null |
Document similarity and clustering V1.ipynb
|
jsafyan/edav-project-4
|
0d3ad40a532da6d0b2acf70a15e55f5cd8354f88
|
[
"MIT"
] | null | null | null |
Document similarity and clustering V1.ipynb
|
jsafyan/edav-project-4
|
0d3ad40a532da6d0b2acf70a15e55f5cd8354f88
|
[
"MIT"
] | null | null | null | 770.874636 | 202,116 | 0.940016 |
[
[
[
"import matplotlib\nimport matplotlib.pyplot as plt\nplt.style.use(\"ggplot\")\n%matplotlib inline\nplt.rcParams[\"figure.figsize\"] = \"15, 8\"\n\nimport pandas as pd\nimport numpy as np\nimport seaborn as sns",
"_____no_output_____"
],
[
"#http://stackoverflow.com/questions/8897593/similarity-between-two-text-documents\nimport nltk, string\nfrom sklearn.feature_extraction.text import TfidfVectorizer\n\nstemmer = nltk.stem.porter.PorterStemmer()\nremove_punctuation_map = dict((ord(char), None) for char in string.punctuation)\n\ndef stem_tokens(tokens):\n return [stemmer.stem(item) for item in tokens]\n\n'''remove punctuation, lowercase, stem'''\ndef normalize(text):\n return stem_tokens(nltk.word_tokenize(text.lower().translate(remove_punctuation_map)))\n\nvectorizer = TfidfVectorizer(tokenizer=normalize, stop_words='english')\n\ndef cosine_sim(text1, text2):\n tfidf = vectorizer.fit_transform([text1, text2])\n return ((tfidf * tfidf.T).A)[0,1]",
"_____no_output_____"
],
[
"lines = []\nwith open('sotu.txt') as f:\n for line in f:\n lines.append(line.strip())\n\nlines = [line for line in lines if line != '\\n' and line != '']\nlines.append('')\n\nstars = [i for i, x in enumerate(lines) if x == '***']\n# last line of file\nstars.append(len(lines) - 1)\n\ndef president(i):\n return ' '.join(lines[stars[i] + 2 : stars[i] + 3]).strip()\n\ndef date(i):\n return ' '.join(lines[stars[i] + 3 : stars[i] + 4]).strip()\n\ndef speech(i):\n return ' '.join(lines[stars[i] + 4 : stars[i + 1]]).strip()",
"_____no_output_____"
],
[
"def rolling_similarity(end_time):\n similarities = []\n for i in range(end_time):\n similarities.append(cosine_sim(speech(i), speech(i + 1)))\n return similarities",
"_____no_output_____"
],
[
"diff_one_series = rolling_similarity(223)",
"_____no_output_____"
],
[
"years = []\nfor i in range(224):\n years.append(date(i)[-4:])",
"_____no_output_____"
],
[
"df = pd.DataFrame()\ndfs = []\nfor i, year in enumerate(years):\n print(year)\n first = speech(i)\n temp_dist = []\n temp_df = pd.DataFrame()\n for j, _ in enumerate(years):\n if j < i:\n temp_dist.append(-1)\n else:\n temp_dist.append(cosine_sim(first, speech(j)))\n temp_df[year] = temp_dist\n dfs.append(temp_df)\ndf = pd.concat(dfs, axis=1)",
"1790\n1790\n1791\n1792\n1793\n1794\n1795\n1796\n1797\n1798\n1799\n1800\n1801\n1802\n1803\n1804\n1805\n1806\n1807\n1808\n1809\n1810\n1811\n1812\n1813\n1814\n1815\n1816\n1817\n1818\n1819\n1820\n1821\n1822\n1823\n1824\n1825\n1826\n1827\n1828\n1829\n1830\n1831\n1832\n1833\n1834\n1835\n1836\n1837\n1838\n1839\n1840\n1841\n1842\n1843\n1844\n1845\n1846\n1847\n1848\n1849\n1850\n1851\n1852\n1853\n1854\n1855\n1856\n1857\n1858\n1859\n1860\n1861\n1862\n1863\n1864\n1865\n1866\n1867\n1868\n1869\n1870\n1871\n1872\n1873\n1874\n1875\n1876\n1877\n1878\n1879\n1880\n1881\n1882\n1883\n1884\n1885\n1886\n1887\n1888\n1889\n1890\n1891\n1892\n1897\n1898\n1899\n1900\n1901\n1902\n1903\n1904\n1905\n1906\n1907\n1908\n1909\n1910\n1911\n1912\n1913\n1914\n1915\n1916\n1917\n1918\n1919\n1920\n1921\n1922\n1923\n1924\n1925\n1926\n1927\n1928\n1929\n1930\n1931\n1932\n1934\n1935\n1936\n1937\n1938\n1939\n1940\n1941\n1942\n1943\n1944\n1945\n1946\n1947\n1948\n1949\n1950\n1951\n1952\n1953\n1953\n1954\n1955\n1956\n1957\n1958\n1959\n1960\n1961\n1961\n1962\n1963\n1964\n1965\n1966\n1967\n1968\n1969\n1970\n1971\n1972\n1973\n1974\n1975\n1976\n1977\n1978\n1979\n1980\n1981\n1982\n1983\n1984\n1985\n1986\n1987\n1988\n1990\n1991\n1992\n1994\n1995\n1996\n1997\n1998\n1999\n2000\n2001\n2001\n2002\n2003\n2004\n2005\n2006\n2007\n2008\n2009\n2010\n2011\n2012\n2013\n2014\n2015\n2016\n"
],
[
"df.to_csv(\"cosine.csv\", index=False)",
"_____no_output_____"
],
[
"df.as_matrix()\n# Generate a mask for the upper triangle\nmask = np.zeros_like(df.as_matrix(), dtype=np.bool)\nmask[np.triu_indices_from(mask, k=1)] = True",
"_____no_output_____"
],
[
"m = df.as_matrix()",
"_____no_output_____"
],
[
"# copy lower triangle to upper triangle\nfor i in range(m.shape[0]):\n for j in range(i, m.shape[1]):\n m[i][j] = m[j][i]",
"_____no_output_____"
],
[
"# Set up matplotlib figure and axis\nfig, ax = plt.subplots(figsize=(16, 12))\n\n# Diverging colormap\ncmap = sns.diverging_palette(220, 10, as_cmap=True)\n\nax.set_title('Cosine Distances between tf-idf vectors of State of the Union Addresses, 1790-2016', fontsize=16)\n\ntemp = pd.DataFrame(m, columns=years)\ntemp.index = years\n# Create the seaborn heatmap with the upper triangle masked\nheatmap = sns.heatmap(temp, annot=False, cmap=cmap, vmax=1, vmin=0,xticklabels=10, yticklabels=10,\n linewidths=.5, square=True, cbar_kws={\"shrink\": .5}, ax=ax)\n# Make the axis labels legible\nplt.yticks(rotation=360)\nplt.xticks(rotation=360)",
"_____no_output_____"
],
[
"np.array(df.columns.values, dtype=np.int32)",
"_____no_output_____"
],
[
"# Set up matplotlib figure and axis\nfig, ax = plt.subplots(figsize=(16, 12))\n\n# Diverging colormap\ncmap = sns.diverging_palette(220, 10, as_cmap=True)\n\nax.set_title('Cosine Distances between tf-idf vectors of State of the Union Addresses')\n\n# Create the seaborn heatmap with the upper triangle masked\nheatmap = sns.heatmap(temp, annot=False, cmap=cmap, vmax=1, mask=mask, vmin=0,xticklabels=10, yticklabels=10,\n linewidths=.5, square=True, cbar_kws={\"shrink\": .5}, ax=ax)\n# Make the axis labels legible\nplt.yticks(rotation=360)\nplt.xticks(rotation=360)",
"_____no_output_____"
],
[
"np.sort(diff_one_series)[:-1][:10]",
"_____no_output_____"
],
[
"pd.DataFrame(m, columns=years).to_csv(\"distances.csv\", index=False)",
"_____no_output_____"
],
[
"# Set up matplotlib figure and axis\nfig, ax = plt.subplots(figsize=(16, 12))\n\n# Diverging colormap\ncmap = sns.diverging_palette(220, 10, as_cmap=True)\n\nax.set_title('Cosine Distances (Gaussian Filtered) between tf-idf vectors of State of the Union Addresses, 1790-2016', fontsize=16)\n\n# Gaussian filter applied to cosine distance matrix to smooth differences\n# order of 0 corresponds to a convolution with the Gaussian kernel\ntemp = pd.DataFrame(scipy.ndimage.filters.gaussian_filter(m, 1, order=0), columns=years)\ntemp.index = years\n# Create the seaborn heatmap with the upper triangle masked\nheatmap = sns.heatmap(temp, annot=False, cmap=cmap, vmax=1, vmin=0,xticklabels=10, yticklabels=10,\n linewidths=.5, square=True, cbar_kws={\"shrink\": .5}, ax=ax)\n# Make the axis labels legible\nplt.yticks(rotation=360)\nplt.xticks(rotation=360)",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a553af73710fc1dc9a50d2bca4f1c38ea9aa2ef
| 8,862 |
ipynb
|
Jupyter Notebook
|
guides/feature_tour_guide.ipynb
|
RelevanceAI/RelevanceAI
|
a0542f35153d9c842f3d2cd0955d6b07f6dfc07b
|
[
"Apache-2.0"
] | 21 |
2021-11-23T13:01:36.000Z
|
2022-03-23T03:45:30.000Z
|
guides/feature_tour_guide.ipynb
|
RelevanceAI/RelevanceAI
|
a0542f35153d9c842f3d2cd0955d6b07f6dfc07b
|
[
"Apache-2.0"
] | 217 |
2021-11-23T00:11:01.000Z
|
2022-03-30T08:11:49.000Z
|
guides/feature_tour_guide.ipynb
|
RelevanceAI/RelevanceAI
|
a0542f35153d9c842f3d2cd0955d6b07f6dfc07b
|
[
"Apache-2.0"
] | 4 |
2022-01-04T01:48:30.000Z
|
2022-02-11T03:19:32.000Z
| 25.761628 | 372 | 0.591063 |
[
[
[
"# 🌋 Quick Feature Tour",
"_____no_output_____"
],
[
"[](https://colab.research.google.com/github/RelevanceAI/RelevanceAI-readme-docs/blob/v2.0.0/docs/getting-started/_notebooks/RelevanceAI-ReadMe-Quick-Feature-Tour.ipynb)",
"_____no_output_____"
],
[
"### 1. Set up Relevance AI\n\nGet started using our RelevanceAI SDK and use of [Vectorhub](https://hub.getvectorai.com/)'s [CLIP model](https://hub.getvectorai.com/model/text_image%2Fclip) for encoding.",
"_____no_output_____"
]
],
[
[
"# remove `!` if running the line in a terminal\n!pip install -U RelevanceAI[notebook]==2.0.0\n# remove `!` if running the line in a terminal\n!pip install -U vectorhub[clip]",
"_____no_output_____"
]
],
[
[
"Follow the signup flow and get your credentials below otherwise, you can sign up/login and find your credentials in the settings [here](https://auth.relevance.ai/signup/?callback=https%3A%2F%2Fcloud.relevance.ai%2Flogin%3Fredirect%3Dcli-api)\n\n\n",
"_____no_output_____"
]
],
[
[
"from relevanceai import Client\n\n\"\"\"\nYou can sign up/login and find your credentials here: https://cloud.relevance.ai/sdk/api\nOnce you have signed up, click on the value under `Activation token` and paste it here\n\"\"\"\nclient = Client()",
"_____no_output_____"
]
],
[
[
"",
"_____no_output_____"
],
[
"### 2. Create a dataset and insert data\n\nUse one of our sample datasets to upload into your own project!",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nfrom relevanceai.utils.datasets import get_ecommerce_dataset_clean\n\n# Retrieve our sample dataset. - This comes in the form of a list of documents.\ndocuments = get_ecommerce_dataset_clean()\n\npd.DataFrame.from_dict(documents).head()",
"_____no_output_____"
],
[
"ds = client.Dataset(\"quickstart\")\nds.insert_documents(documents)",
"_____no_output_____"
]
],
[
[
"See your dataset in the dashboard\n\n\n\n\n",
"_____no_output_____"
],
[
"\n### 3. Encode data and upload vectors into your new dataset\n\nEncode a new product image vector using [Vectorhub's](https://hub.getvectorai.com/) `Clip2Vec` models and update your dataset with the resulting vectors. Please refer to [Vectorhub](https://github.com/RelevanceAI/vectorhub) for more details.\n",
"_____no_output_____"
]
],
[
[
"from vectorhub.bi_encoders.text_image.torch import Clip2Vec\n\nmodel = Clip2Vec()\n\n# Set the default encode to encoding an image\nmodel.encode = model.encode_image\ndocuments = model.encode_documents(fields=[\"product_image\"], documents=documents)",
"_____no_output_____"
],
[
"ds.upsert_documents(documents=documents)",
"_____no_output_____"
],
[
"ds.schema",
"_____no_output_____"
]
],
[
[
"\nMonitor your vectors in the dashboard\n\n\n\n\n\n\n",
"_____no_output_____"
],
[
"### 4. Run clustering on your vectors\n\nRun clustering on your vectors to better understand your data! \n\nYou can view your clusters in our clustering dashboard following the link which is provided after the clustering is finished! \n\n",
"_____no_output_____"
]
],
[
[
"from sklearn.cluster import KMeans\n\ncluster_model = KMeans(n_clusters=10)\nds.cluster(cluster_model, [\"product_image_clip_vector_\"])",
"_____no_output_____"
]
],
[
[
"You can see the new `_cluster_` field that is added to your document schema. \nClustering results are uploaded back to the dataset as an additional field.\nThe default `alias` of the cluster will be the `kmeans_<k>`.",
"_____no_output_____"
]
],
[
[
"ds.schema",
"_____no_output_____"
]
],
[
[
"See your cluster centers in the dashboard\n\n\n\n\n\n",
"_____no_output_____"
],
[
"### 4. Run a vector search\n\nEncode your query and find your image results!\n\nHere our query is just a simple vector query, but our search comes with out of the box support for features such as multi-vector, filters, facets and traditional keyword matching to combine with your vector search. You can read more about how to construct a multivector query with those features [here](https://docs.relevance.ai/docs/vector-search-prerequisites).\n\nSee your search results on the dashboard here https://cloud.relevance.ai/sdk/search.\n",
"_____no_output_____"
]
],
[
[
"query = \"gifts for the holidays\"\nquery_vector = model.encode(query)\nmultivector_query = [{\"vector\": query_vector, \"fields\": [\"product_image_clip_vector_\"]}]\nresults = ds.vector_search(multivector_query=multivector_query, page_size=10)",
"_____no_output_____"
]
],
[
[
"See your multi-vector search results in the dashboard\n\n\n\n",
"_____no_output_____"
],
[
"\n\nWant to quickly create some example applications with Relevance AI? Check out some other guides below!\n- [Text-to-image search with OpenAI's CLIP](https://docs.relevance.ai/docs/quickstart-text-to-image-search)\n- [Hybrid Text search with Universal Sentence Encoder using Vectorhub](https://docs.relevance.ai/docs/quickstart-text-search)\n- [Text search with Universal Sentence Encoder Question Answer from Google](https://docs.relevance.ai/docs/quickstart-question-answering)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
4a553dabed5656cde9a53f029e5f06143fe3026a
| 6,346 |
ipynb
|
Jupyter Notebook
|
4_7_SLAM/2_1. Include Landmarks, exercise.ipynb
|
gustavojoseleite/CVND_Localization_Exercises
|
7284f199bdbdf6ac2cea2539e27e21d0cf471016
|
[
"MIT"
] | null | null | null |
4_7_SLAM/2_1. Include Landmarks, exercise.ipynb
|
gustavojoseleite/CVND_Localization_Exercises
|
7284f199bdbdf6ac2cea2539e27e21d0cf471016
|
[
"MIT"
] | null | null | null |
4_7_SLAM/2_1. Include Landmarks, exercise.ipynb
|
gustavojoseleite/CVND_Localization_Exercises
|
7284f199bdbdf6ac2cea2539e27e21d0cf471016
|
[
"MIT"
] | null | null | null | 30.657005 | 399 | 0.439647 |
[
[
[
"## Updating Constraint Matrices\n\nTo implement Graph SLAM, a matrix and a vector (omega and xi, respectively) are introduced. The matrix is square and labelled with all the robot poses (xi) and all the landmarks (Li). Every time you make an observation, for example, as you move between two poses by some distance `dx` and can relate those two positions, you can represent this as a numerical relationship in these matrices.\n\nBelow you can see a matrix representation of omega and a vector representation of xi.\n\n<img src='images/omega_xi.png' width=20% height=20% />\n\n\n### Solving for x, L\n\nTo \"solve\" for all these poses and landmark positions, we can use linear algebra; all the positional values are in the vector `mu` which can be calculated as a product of the inverse of omega times xi.\n\n---\n",
"_____no_output_____"
],
[
"## Constraint Updates\n\nIn the below code, we construct `omega` and `xi` constraint matrices, and update these according to landmark sensor measurements and motion.\n\n#### Sensor Measurements\n\nWhen you sense a distance, `dl`, between a pose and a landmark, l, update the constraint matrices as follows:\n* Add `[[1, -1], [-1, 1]]` to omega at the indices for the intersection of `xt` and `l`\n* Add `-dl` and `dl` to xi at the rows for `xt` and `l`\n\nThe values 2 instead of 1 indicate the \"strength\" of the measurement.\n\nYou'll see three new `dl`'s as new inputs to our function `Z0, Z1, Z2`, below.\n\n#### Motion\nWhen your robot moves by some amount `dx` update the constraint matrices as follows:\n* Add `[[1, -1], [-1, 1]]` to omega at the indices for the intersection of `xt` and `xt+1`\n* Add `-dx` and `dx` to xi at the rows for `xt` and `xt+1`\n\n## QUIZ: Include three new sensor measurements for a single landmark, L.",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\n\ndef mu_from_positions(initial_pos, move1, move2, Z0, Z1, Z2):\n \n ## TODO: construct constraint matrices\n ## and add each position/motion constraint to them\n \n # initialize constraint matrices with 0's\n # Now these are 4x4 because of 3 poses and a landmark\n omega = np.zeros((4,4))\n xi = np.zeros((4,1))\n \n # add initial pose constraint\n omega[0][0] = 1\n xi[0] = initial_pos\n \n # account for the first motion, dx = move1\n omega += [[1., -1., 0., 0.],\n [-1., 1., 0., 0.],\n [0., 0., 0., 0.],\n [0., 0., 0., 0.]]\n xi += [[-move1],\n [move1],\n [0.],\n [0.]]\n \n # account for the second motion\n omega += [[0., 0., 0., 0.],\n [0., 1., -1., 0.],\n [0., -1., 1., 0.],\n [0., 0., 0., 0.]]\n xi += [[0.],\n [-move2],\n [move2],\n [0.]]\n \n \n ## TODO: Include three new sensor measurements for the landmark, L\n \n # Your code here\n #first sensor\n omega += [[1., 0., 0., -1.],\n [0., 0., 0., 0.],\n [0., 0., 0., 0.],\n [-1., 0., 0., 1.]]\n xi += [[-Z0],\n [0],\n [0],\n [Z0]]\n\n #second sensor\n omega += [[0., 0., 0., 0.],\n [0., 1., 0., -1.],\n [0., 0., 0., 0.],\n [0., -1., 0., 1.]]\n xi += [[0],\n [-Z1],\n [0],\n [Z1]]\n \n #third sensor\n omega += [[0., 0., 0., 0.],\n [0., 0, 0., 0],\n [0., 0., 1., -1.],\n [0., 0, -1., 1.]]\n xi += [[0],\n [0],\n [-Z2],\n [Z2]]\n # display final omega and xi\n print('Omega: \\n', omega)\n print('\\n')\n print('Xi: \\n', xi)\n print('\\n')\n \n ## TODO: calculate mu as the inverse of omega * xi\n ## recommended that you use: np.linalg.inv(np.matrix(omega)) to calculate the inverse\n omega_inv = np.linalg.inv(np.matrix(omega))\n mu = omega_inv*xi\n return mu\n",
"_____no_output_____"
],
[
"# call function and print out `mu`\nmu = mu_from_positions(-3, 5, 3, 10, 5, 2)\nprint('Mu: \\n', mu)",
"Omega: \n [[ 3. -1. 0. -1.]\n [-1. 3. -1. -1.]\n [ 0. -1. 2. -1.]\n [-1. -1. -1. 3.]]\n\n\nXi: \n [[-18.]\n [ -3.]\n [ 1.]\n [ 17.]]\n\n\nMu: \n [[-3.]\n [ 2.]\n [ 5.]\n [ 7.]]\n"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code"
]
] |
4a5540fbdf8aae4574b4ac4ac099f91aa9e31cfe
| 778,053 |
ipynb
|
Jupyter Notebook
|
UG_S17/Mba-Kalu_Onwughalu-Brazil.ipynb
|
NYUDataBootcamp/Projects
|
b456061d7b0738b3cebabaff3f91e860f32afd80
|
[
"MIT"
] | 5 |
2016-11-02T17:10:03.000Z
|
2021-06-28T02:43:51.000Z
|
UG_S17/Mba-Kalu_Onwughalu-Brazil.ipynb
|
NYUDataBootcamp/Projects
|
b456061d7b0738b3cebabaff3f91e860f32afd80
|
[
"MIT"
] | null | null | null |
UG_S17/Mba-Kalu_Onwughalu-Brazil.ipynb
|
NYUDataBootcamp/Projects
|
b456061d7b0738b3cebabaff3f91e860f32afd80
|
[
"MIT"
] | 10 |
2016-06-13T02:06:54.000Z
|
2020-03-24T08:25:47.000Z
| 581.070202 | 86,538 | 0.915823 |
[
[
[
"\n\n### <center> **Chukwuemeka Mba-Kalu** </center> <center> **Joseph Onwughalu** </center>\n### <center> **An Analysis of the Brazilian Economy between 2000 and 2012** </center>\n#### <center> Final Project In Partial Fulfillment of the Course Requirements </center> <center> [**Data Bootcamp**](http://nyu.data-bootcamp.com/) </center>\n##### <center> Stern School of Business, NYU Spring 2017 </center> <center> **May 12, 2017** </center> ",
"_____no_output_____"
],
[
"### The Brazilian Economy\nIn this project we examine in detail different complexities of Brazil’s growth between the years 2000-2012. During this period, Brazil set an example for many of the major emerging economies in Latin America, Africa, and Asia. \n\nFrom the years 2000-2012, Brazil was one of the fastest growing major economies in the world. It is the 8th largest economy in the world, with its GDP totalling 2.2 trillion dollars and GDP per Capita being at 10,308 dollars. While designing this project, we were interested to find out more about the main drivers of the Brazilian economy. Specifically, we aim to look at specific trends and indicators that directly affect economic growth, especially in fast-growing countries such as Brazil. Certain trends include household consumption and its effects on the GDP, bilateral aid and investment flows and its effects on the GDP per capita growth. We also aim to view the effects of economic growth on climate change and public health by observing the carbon emissions percentage changes and specific indicators like the mortality rate.\n\nWe will be looking at generally accepted economic concepts and trends, making some hypotheses, and comparing our hypotheses to the Brazil data we have. Did Brazil follow these trends on its path to economic growth?\n",
"_____no_output_____"
],
[
"### Methodology - Data Acquisition\nAll the data we are using in this project was acquired from the World Bank and can be accessed and downloaded from the [website](www.WorldBank.org). By going on the website and searching for “World data report” we were given access to information that has to be submitted by the respective countries on the site. By clicking “[Brazil](http://databank.worldbank.org/data/reports.aspx?source=2&country=BRA),” we’re shown the information of several economic indicators and their respective data over a time period of 2000-2012 that we downloaded as an excel file. We picked more than 20 metrics to include in our data, such as: \n* Population\n* GDP (current US Dollars)\n* Household final consumption expenditure, etc. (% of GDP)\n* General government final consumption expenditure (current US Dollars)\n* Life expectancy at birth, total (years) \n\nFor all of our analysis and data we will be looking at the 2000-2012 time period and have filtered the spreadsheets accordingly to reflect this information.\n",
"_____no_output_____"
]
],
[
[
"# Inportant Packages\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport sys\nimport datetime as dt",
"_____no_output_____"
],
[
"print('Python version is:', sys.version)\nprint('Pandas version:', pd.__version__)\nprint('Date:', dt.date.today())",
"Python version is: 3.5.2 |Anaconda 4.2.0 (64-bit)| (default, Jul 5 2016, 11:41:13) [MSC v.1900 64 bit (AMD64)]\nPandas version: 0.18.1\nDate: 2017-05-11\n"
]
],
[
[
"### Reading in and Cleaning up the Data\n\nWe downloaded our [data](http://databank.worldbank.org/data/AjaxDownload/FileDownloadHandler.ashx?filename=67fd49af-3b41-4515-b248-87b045e61886.zip&filetype=CSV&language=en&displayfile=Data_Extract_From_World_Development_Indicators.zip) in xlxs, retained and renamed the important columns, and deleted rows without enough data. We alse transposed the table to make it easier to plot diagrams.",
"_____no_output_____"
]
],
[
[
"path = 'C:\\\\Users\\\\emeka_000\\\\Desktop\\\\Bootcamp_Emeka.xlsx'\nodata = pd.read_excel(path, \n usecols = ['Series Name','2000 [YR2000]', '2001 [YR2001]', '2002 [YR2002]',\n '2003 [YR2003]', '2004 [YR2004]', '2005 [YR2005]', '2006 [YR2006]',\n '2007 [YR2007]', '2008 [YR2008]', '2009 [YR2009]', '2010 [YR2010]',\n '2011 [YR2011]', '2012 [YR2012]']\n ) #retained only the necessary columns \n\nodata.columns = ['Metric', '2000', '2001', '2002', '2003', '2004', '2005', '2006', '2007', '2008',\n '2009', '2010', '2011', '2012'] #easier column names\n\nodata = odata.drop([20, 21, 22, 23, 24]) ##delete NaN values\n\nodata = odata.transpose() #transpose to make diagram easier\n\n\nodata #data with metrics description for the chart below",
"_____no_output_____"
],
[
"data = pd.read_excel(path, \n usecols = ['2000 [YR2000]', '2001 [YR2001]', '2002 [YR2002]',\n '2003 [YR2003]', '2004 [YR2004]', '2005 [YR2005]', '2006 [YR2006]',\n '2007 [YR2007]', '2008 [YR2008]', '2009 [YR2009]', '2010 [YR2010]',\n '2011 [YR2011]', '2012 [YR2012]']\n ) #same data but modified for pandas edits \n\ndata.columns = ['2000', '2001', '2002', '2003', '2004', '2005', '2006', '2007', '2008',\n '2009', '2010', '2011', '2012'] #all columns are now string\n\ndata = data.transpose() #data used for the rest of the project",
"_____no_output_____"
]
],
[
[
"### GDP Growth and GDP Growth Rate in Brazil \nTo demonstrate Brazil's strong economic growht between 2000 and 2012, here are a few charts illustrating Brazil's GDP growth. \n\nGross domestic product (GDP) is the monetary value of all the finished goods and services produced within a country's borders in a specific time period. Though GDP is usually calculated on an annual basis, it can be calculated on a quarterly basis as well. GDP includes all private and public consumption, government outlays, investments and exports minus imports that occur within a defined territory. Put simply, GDP is a broad measurement of a nation’s overall economic activity.\n\nGDP per Capita is a measure of the total output of a country that takes gross domestic product (GDP) and divides it by the number of people in the country. \n\nRead more on [Investopedia](http://www.investopedia.com/terms/g/gdp.asp#ixzz4gjgzo4Ri)",
"_____no_output_____"
]
],
[
[
" \ndata[4].plot(kind = 'line', #line plot\n title = 'Brazil Yearly GDP (2000-2012) (current US$)', #title\n fontsize=15,\n color='Green', \n linewidth=4, #width of plot line\n figsize=(20,5),).title.set_size(20) #set figure size and title size\n\nplt.xlabel(\"Year\").set_size(15) \nplt.ylabel(\"GDP (current US$) * 1e12\").set_size(15) #set x and y axis, with their sizes\n",
"_____no_output_____"
],
[
"data[6].plot(kind = 'line',\n title = 'Brazil Yearly GDP Per Capita (2000-2012) (current US$)', \n fontsize=15, \n color='blue', \n linewidth=4, \n figsize=(20,5)).title.set_size(20)\n\nplt.xlabel(\"Year\").set_size(15)\nplt.ylabel(\"GDP per capita (current US$)\").set_size(15)",
"_____no_output_____"
],
[
"data[5].plot(kind = 'line', \n title = 'Brazil Yearly GDP Growth (2000-2012) (%)', \n fontsize=15, \n color='red', \n linewidth=4, \n figsize=(20,5)).title.set_size(20)\n\nplt.xlabel(\"Year\").set_size(15)\nplt.ylabel(\"GDP Growth (%)\").set_size(15)",
"_____no_output_____"
]
],
[
[
"#### GDP Growth vs. GDP Growth Rate\nWhile Brazil's GDP was growing quite consistently over the 12 years, its GDP growth-rate was not steady with negative growth during the 2008 financial crisis. ",
"_____no_output_____"
],
[
"### Hypothesis: Household Consumption vs. Foreign Aid\nOur hypothesis is that household consumption is a bigger driver of the Brazilian economy than foreign aid. With their rising incomes, Brazilians are expected to be empowered with larger disposable incomes to spend on goods and services. Foreign aid, on the other hand, might not filter down to the masses for spending.",
"_____no_output_____"
]
],
[
[
"fig, ax1 = plt.subplots(figsize = (20,5))\n\ny1 = data[8]\ny2 = data[4]\n\nax2 = ax1.twinx()\n\nax1.plot(y1, 'green') #household consumption\nax2.plot(y2, 'blue') #GDP growth\n\nplt.title(\"Household Consumption (% of GDP) vs. GDP\").set_size(20)\n ",
"_____no_output_____"
]
],
[
[
"#### Actual: Household Consumption\nGDP comprises of household consumption, net investments, government spending and net exports;increases or decreases in any of these areas would affect the overall GDP respectively. The data shows that despite household consumption decreasing as a % of GDP, the GDP was growing. We found this a little strange and difficult to understand. One explanation for this phenomenon could be that as emerging market economies continue to expand, there is an increased shift towards investments and government spending. \n\nThe blue line represents GDP growth and the green line represents Household Consumption. ",
"_____no_output_____"
]
],
[
[
"fig, ax1 = plt.subplots(figsize = (20, 5))\n\ny1 = data[11]\ny2 = data[4]\n\nax2 = ax1.twinx()\n\nax1.plot(y1, 'red') #Net official development assistance \nax2.plot(y2, 'blue') #GDP growth\n\nplt.title(\"Foreign Aid vs. GDP\").set_size(20)",
"_____no_output_____"
]
],
[
[
"#### Actual: Foreign Aid\nRegarding foreign aid, it should be the case that with decreases in aid there will be reduced economic growth because many developing countries do rely on that as a crucial resource. The data shows a positive corellation for Brazil. While household spending was not a major driver of the Brazil's GDP growth, foreign aid played a big role. We will now explore how foreign direct investment and government spending can affect economic growth. \n\nThe blue line represents GDP growth and the red line represents Foreign Aid. \n",
"_____no_output_____"
],
[
"### Hypothesis: Foreign Direct Investment vs. Government Spending\nFor emerging market economies, the general trend is that Governments contribute a significant proportion to the GDP. Given that Brazil experienced growth between the years 2000-2012, it is expected that a consequence was increased foreign direct investment. Naturally, we’d like to compare the increases in Government Spending versus this foreign direct investment and see who generally contributed more to the GDP growth of the country. \nOur hypothesis is that the increased foreign direct investment was a bigger contributor to the GDP growth than government spending. With increased globalisation, we expect many multinationals and investors started business operations in Brazil due to its large, fast-growing market.",
"_____no_output_____"
]
],
[
[
"fig, ax1 = plt.subplots(figsize = (20, 5))\n\ny1 = data[2]\ny2 = data[4]\n\nax2 = ax1.twinx()\n\nax1.plot(y1, 'yellow') #household consumption\nax2.plot(y2, 'blue') #GDP growth\n\nplt.title(\"Foreign Direct Investment (Inflows) (% of GDP) vs. GDP\").set_size(20)",
"_____no_output_____"
]
],
[
[
"#### Actual: Foreign Direct Investment\nContrary to popular belief and economic concepts, increased foreign direct investment did not act as a major contributor to the GDP growth Brazil experienced. There is no clear general trend or correlation between FDI and GDP growth. ",
"_____no_output_____"
]
],
[
[
"fig, ax1 = plt.subplots(figsize = (20, 5))\n\ny1 = data[14]\ny2 = data[4]\n\nax2 = ax1.twinx()\n\nax1.plot(y1, 'purple') #household consumption\nax2.plot(y2, 'blue') #GDP growth\n\nplt.title(\"Government Spending vs. GDP\").set_size(20)",
"_____no_output_____"
]
],
[
[
"#### Actual: Government Spending\nIt is clear that government spending is positively corellated with the total GDP growth Brazil experienced. We believe that this was the major driver for Brazil's growth.\n",
"_____no_output_____"
],
[
"### Hypothesis: Population Growth and GDP per capita\nBrazil’s population growth continued to increase during this time period of 2000-2012. As mentioned earlier, Brazil’s GDP growth was also growing during the same time period. Given that GDP per capita is a nice economic indicator to highlight standard of living in a country, we wanted to see if the increasing population was negating the effects of increased economic growth.\nOur hypothesis is that even though population was growing, the GDP per capita over the years generally increased at a higher rate and, all things equal, we are assured increased living standards in Brazil. This finding would prove to us that the GDP was growing at a faster rate than the population.",
"_____no_output_____"
]
],
[
[
"data.plot.scatter(x = 5, y = 0,\n title = 'Population Growth vs. GDP Growth',\n figsize=(20,5)).title.set_size(20)\n\nplt.xlabel(\"GDP Growth Rate\").set_size(15)\nplt.ylabel(\"Population Growth Rate\").set_size(15)",
"_____no_output_____"
]
],
[
[
"#### Actual: Population Growth \nThere is no correlation between the population growth rate and the overall GDP growth rate. The general GDP rate already accounts for population increases and decreases. ",
"_____no_output_____"
]
],
[
[
"data.plot.scatter(x = 6, y = 0,\n title = 'Population Growth vs. GDP per Capita',\n figsize=(20,5)).title.set_size(20)\n\nplt.xlabel(\"GDP per Capita\").set_size(15)\nplt.ylabel(\"Population Growth Rate\").set_size(15)",
"_____no_output_____"
]
],
[
[
"#### Population Growth \nThe population growth rate has a negative correlation with GDP per capita. Our explanation is that, as economies advance, the birth rate is expected to decrease. This generally causes population growth rate to fall and GDP per Capita to rise. ",
"_____no_output_____"
],
[
"### Hypothesis: Renewable Energy Expenditures and C02 Emissions\nWhat one would expect is that as a country’s economy grows, its investments in renewable energy methods would increase as well. Such actions should lead to a decrease in CO2 emissions as cleaner energy processes are being applied. Our hypothesis disagrees with this.\nWe believe that despite there being significant increases in renewable energy expenditures due to increased incomes and a larger, more diversified economy, there will still be more than proportionate increases in C02 emissions. By testing this hypothesis we will begin to understand certain explanations as to why this may be true or false.\n",
"_____no_output_____"
]
],
[
[
"data[15].plot(kind = 'bar', \n title = 'Renewable energy consumption (% of total) (2000-2012)', \n fontsize=15,\n color='green', \n linewidth=4, \n figsize=(20,5)).title.set_size(20)",
"_____no_output_____"
],
[
"data[12].plot(kind = 'bar', \n title = 'CO2 emissions from liquid fuel consumption (2000-2012)', \n fontsize=15,\n color='red', \n linewidth=4, \n figsize=(20,5)).title.set_size(20)",
"_____no_output_____"
],
[
"data[13].plot(kind = 'bar', \n title = 'CO2 emissions from gaseous fuel consumption (2000-2012)', \n fontsize=15, \n color='blue', \n linewidth=4, \n figsize=(20,5)).title.set_size(20)",
"_____no_output_____"
]
],
[
[
"#### Actual: Renewable Energy Consumption vs. CO2 Emmissions\nAs countries continue to grow their economies, it is expected that people’s incomes will continue to rise. Increased disposable incomes should cause better energy consumption methods but as our hypothesis states, C02 emissions still continue to rise. This could be due to the increase in population as more people are using carbon goods and products.",
"_____no_output_____"
],
[
"### Hypothesis: Health Expenditures and Life Expectancy\nThere should be a positive correlation between health expenditures and life expenditures. Naturally, the more a country is spending on healthcare, the higher the life expectancy ought to be. Our hypothesis agrees with this positive statement and we’d like to test it. If it turns out that health expenditure increases positively affects life expectancy, then we can attribute the increase to an improved economy that allows for more health expenditures from individuals, organisations and institutions.\n",
"_____no_output_____"
]
],
[
[
"data.plot.scatter(x = 7, y = 19, #scatter plot\n title = 'Health Expenditures vs. Life Expectancy',\n figsize=(20,5)).title.set_size(20)\n\nplt.xlabel(\"Health Expenditures\").set_size(15)\nplt.ylabel(\"Life Expectancy\").set_size(15)",
"_____no_output_____"
]
],
[
[
"#### Actual: Health Expenditures and Life Expectancy\nAs expected, there is a positive correlation between health expenditures and life expectancy in Brazil. This is the natural expectation that as a country spends more on healthcare services, products and research, the life expectancy should increase as improvements to health science will be made due to such investments. \n",
"_____no_output_____"
],
[
"### Conclusion\n\nWhen we first started working on this project, we wanted to analyze some of the generally accepted economic concepts we’ve learned in our four years at Stern. Using a previously booming emerging market economy like Brazil as a test subject, we put these economic metrics to test. Some metrics contributed to increased economic growth and some indicators also show that the economic growth played a big role in society. \n\t\nWe started with specific hypotheses of what we expected to happen before running the data. While there were some findings that met our expectations, we came across some surprising information that made us realize that economies aren’t completely systematic and will vary in functioning.\n\nAlthough household spending and foreign direct investment were generally increasing, we did not find that there was a direct correlation between its growth and the GDP growth rate. What instead was our conclusion was that the foreign aid and government spending were two of the major drivers of GDP growth during the years 2000 - 2012.\n\n",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
4a5561aaed7033d9a123999173de6772cdd0c4c7
| 2,009 |
ipynb
|
Jupyter Notebook
|
exercises/multiples_of_3_and_5/notebook.ipynb
|
hsteinshiromoto/exercises.coding
|
7a7780285016e819e846bada5be3639df492efcf
|
[
"MIT"
] | null | null | null |
exercises/multiples_of_3_and_5/notebook.ipynb
|
hsteinshiromoto/exercises.coding
|
7a7780285016e819e846bada5be3639df492efcf
|
[
"MIT"
] | null | null | null |
exercises/multiples_of_3_and_5/notebook.ipynb
|
hsteinshiromoto/exercises.coding
|
7a7780285016e819e846bada5be3639df492efcf
|
[
"MIT"
] | 1 |
2020-11-04T04:48:40.000Z
|
2020-11-04T04:48:40.000Z
| 19.133333 | 206 | 0.486809 |
[
[
[
"# Multiples of 3 and 5",
"_____no_output_____"
],
[
"**Problem**: If we list all the natural numbers below 10 that are multiples of 3 or 5, we get 3, 5, 6 and 9. The sum of these multiples is 23. Find the sum of all the multiples of 3 or 5 below 1000.\n",
"_____no_output_____"
]
],
[
[
"import numpy as np",
"_____no_output_____"
],
[
"def multiples(k: int, below_threshold: int):\n \n output = []\n for i in range(2, below_threshold):\n if i % k == 0:\n output.append(i)\n \n return output",
"_____no_output_____"
],
[
"numbers = []\nfor i in [3, 5]:\n numbers.extend(multiples(i, 1000))\n \nnumbers = set(numbers)",
"_____no_output_____"
],
[
"sum(numbers)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
4a5579fb3afe74fe4737f0f0e8db2c6978692272
| 2,347 |
ipynb
|
Jupyter Notebook
|
pcb/torque_units.ipynb
|
kreier/me2arm
|
ca2c6994b2ed854a7fac613803deee478fee8e63
|
[
"MIT"
] | 2 |
2020-05-24T18:08:20.000Z
|
2020-05-31T23:25:18.000Z
|
calculations/torque_units.ipynb
|
kreier/actuator
|
af81df83f969d024100d28b02cce60cde76b147b
|
[
"MIT"
] | null | null | null |
calculations/torque_units.ipynb
|
kreier/actuator
|
af81df83f969d024100d28b02cce60cde76b147b
|
[
"MIT"
] | null | null | null | 30.480519 | 279 | 0.493822 |
[
[
[
"# Power, Energy and Torque\n\nFor cars people talk about their max torque and power in relation to their maximum acceleration. How is that related to energy and can be converted?\n\n## Servo torque\n\nServos give their torque in $kgcm$ which is not really a unit. SG90 has $2.4 kgcm$, bigger servos have $20 kgcm$. What is that unit? Isn't torque in $Nm$?\n\n__First__ the kilogram is used as replacement for the force, using $W = mg$ with $g = 9.81ms^{-2}$ as gravitational acceleration, $W$ the weight in Newton and $m$ the mass in kilogram. With 2% error we can estimate $g \\approx 10 ms^{2}$ and then $1 kg \\equiv 10N$.\n\nIn Physics we used more the angular momentum $\\mathbf{L}$ wich relates to the torque in\n\n$$\\tau = \\frac{\\mathrm{d}\\mathbf{L}}{\\mathrm{d}t}$$",
"_____no_output_____"
]
],
[
[
"F = 2 # force in Newton\nr = 1 # radius, distance from center for force\ntau = F * r # torque in Nm if F is perpendicular to r\n\nprint(\"The torque is {:} Nm.\".format(tau))",
"The torque is 2 Nm.\n"
]
],
[
[
"Now let's got to power $P$ and energy $E = Pt$ wich is power times time.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
4a557a3123d0746ada0f464f81d9890e4c089c97
| 15,149 |
ipynb
|
Jupyter Notebook
|
pt_framework/c17e3_multi_modal_multi_task.ipynb
|
praneetheddu/LDL
|
97b81fa2e0fd8b76b354de8873dca235f81f5f17
|
[
"MIT"
] | null | null | null |
pt_framework/c17e3_multi_modal_multi_task.ipynb
|
praneetheddu/LDL
|
97b81fa2e0fd8b76b354de8873dca235f81f5f17
|
[
"MIT"
] | null | null | null |
pt_framework/c17e3_multi_modal_multi_task.ipynb
|
praneetheddu/LDL
|
97b81fa2e0fd8b76b354de8873dca235f81f5f17
|
[
"MIT"
] | null | null | null | 47.939873 | 1,105 | 0.61212 |
[
[
[
"\"\"\"\nThe MIT License (MIT)\nCopyright (c) 2021 NVIDIA\nPermission is hereby granted, free of charge, to any person obtaining a copy of\nthis software and associated documentation files (the \"Software\"), to deal in\nthe Software without restriction, including without limitation the rights to\nuse, copy, modify, merge, publish, distribute, sublicense, and/or sell copies of\nthe Software, and to permit persons to whom the Software is furnished to do so,\nsubject to the following conditions:\nThe above copyright notice and this permission notice shall be included in all\ncopies or substantial portions of the Software.\nTHE SOFTWARE IS PROVIDED \"AS IS\", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR\nIMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY, FITNESS\nFOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE AUTHORS OR\nCOPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER LIABILITY, WHETHER\nIN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM, OUT OF OR IN\nCONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE SOFTWARE.\n\"\"\"\n",
"_____no_output_____"
]
],
[
[
"This code example extends the multimodal network from c17e2_multi_modal with an additional head to build a network that does multitask learning using multimodal inputs. We teach the network to simultaneously do multiclass classification (identify the handwritten digit) and perform a simple question-answering task. The question-answering task is to provide a yes/no answer to a question about the digit in the image. The textual input will look similar to the textual input in c17e2_multi_modal ('upper half', 'lower half', 'odd number', 'even number'). However, instead of correctly describing the digit, the text is chosen randomly and represents a question. The network is then tasked with classifying the image into one of ten classes as well as with determining whether the answer to the question is yes or no (is the statement true or false). More context for this code example can be found in the section \"Programming Example: Multiclass classification and question answering with a single network\" in Chapter 17 in the book Learning Deep Learning by Magnus Ekman (ISBN: 9780137470358).\n\nAs always, we start with initialization code and loading the dataset.\n",
"_____no_output_____"
]
],
[
[
"import torch\nimport torch.nn as nn\nimport torch.nn.functional as F\nfrom torch.utils.data import TensorDataset, DataLoader\nimport torchvision\nimport torchvision.transforms as transforms\nfrom torchvision.datasets import MNIST\nfrom torch.utils.data import DataLoader\n# Using Keras Tokenizer for simplicity\nfrom tensorflow.keras.preprocessing.text import Tokenizer\nfrom tensorflow.keras.preprocessing.text \\\n import text_to_word_sequence\nfrom tensorflow.keras.preprocessing.sequence \\\n import pad_sequences\nimport numpy as np\nimport matplotlib.pyplot as plt\n\ndevice = torch.device(\"cuda:0\" if torch.cuda.is_available() else \"cpu\")\nEPOCHS = 20\nBATCH_SIZE = 64\nMAX_WORDS = 8\nEMBEDDING_WIDTH = 4\n\n# Load MNIST dataset.\ntrainset = MNIST(root='./pt_data', train=True, download=True)\ntestset = MNIST(root='./pt_data', train=False, download=True)\n\n# Convert to numpy arrays to enable us to create a richer dataset.\ntrain_images = trainset.data.numpy().astype(np.float32)\ntrain_labels = trainset.targets.numpy()\ntest_images = testset.data.numpy().astype(np.float32)\ntest_labels = testset.targets.numpy()\n\n# Standardize the data.\nmean = np.mean(train_images)\nstddev = np.std(train_images)\ntrain_images = (train_images - mean) / stddev\ntest_images = (test_images - mean) / stddev\n",
"_____no_output_____"
]
],
[
[
"The next step is to extend the MNIST dataset with questions and answers. This is done in the next code snippet. The code alternates between the four questions/statements for each training and test example. It then determines whether the answer is yes or no based on the ground truth label.\n",
"_____no_output_____"
]
],
[
[
"# Function to create question and answer text.\ndef create_question_answer(tokenizer, labels):\n text = []\n answers = np.zeros(len(labels))\n for i, label in enumerate(labels):\n question_num = i % 4\n if question_num == 0:\n text.append('lower half')\n if label < 5:\n answers[i] = 1.0\n elif question_num == 1:\n text.append('upper half')\n if label >= 5:\n answers[i] = 1.0\n elif question_num == 2:\n text.append('even number')\n if label % 2 == 0:\n answers[i] = 1.0\n elif question_num == 3:\n text.append('odd number')\n if label % 2 == 1:\n answers[i] = 1.0\n text = tokenizer.texts_to_sequences(text)\n text = pad_sequences(text).astype(np.int64)\n answers = answers.reshape((len(labels), 1))\n return text, answers\n\n# Create second modality for training and test set.\nvocabulary = ['lower', 'upper', 'half', 'even', 'odd', 'number']\ntokenizer = Tokenizer(num_words=MAX_WORDS)\ntokenizer.fit_on_texts(vocabulary)\ntrain_text, train_answers = create_question_answer(tokenizer,\n train_labels)\ntest_text, test_answers = create_question_answer(tokenizer,\n test_labels)\n\n# Create datasets.\ntrainset = TensorDataset(torch.from_numpy(train_images), \n torch.from_numpy(train_text),\n torch.from_numpy(train_labels),\n torch.from_numpy(train_answers))\n\ntestset = TensorDataset(torch.from_numpy(test_images),\n torch.from_numpy(test_text),\n torch.from_numpy(test_labels),\n torch.from_numpy(test_answers))\n",
"_____no_output_____"
]
],
[
[
"The next code snippet creates the network. Most of the network is identical to the programming example for the multimodal network. The key difference is that in parallel with the ten-unit output layer for multiclass classification, there is a one-unit output layer for binary classification. Given that there are two separate outputs, we also need to supply two separate loss functions.\n",
"_____no_output_____"
]
],
[
[
"# Define model.\nclass MultiTaskModel(nn.Module):\n def __init__(self):\n super().__init__()\n self.embedding_layer = nn.Embedding(MAX_WORDS, EMBEDDING_WIDTH)\n nn.init.uniform_(self.embedding_layer.weight, -0.05, 0.05) # Default is -1, 1.\n self.lstm_layers = nn.LSTM(EMBEDDING_WIDTH, 8, num_layers=1, batch_first=True)\n\n self.linear_layer = nn.Linear(784+8, 25)\n self.relu_layer = nn.ReLU()\n self.class_output_layer = nn.Linear(25, 10)\n self.answer_output_layer = nn.Linear(25, 1)\n\n def forward(self, inputs):\n image_input = inputs[0]\n text_input = inputs[1]\n\n # Process textual data.\n x0 = self.embedding_layer(text_input)\n x0 = self.lstm_layers(x0)\n\n # Process image data.\n # Flatten the image.\n x1 = image_input.view(-1, 784)\n\n # Concatenate input branches and build shared trunk.\n x = torch.cat((x0[1][0][0], x1), dim=1)\n x = self.linear_layer(x)\n x = self.relu_layer(x)\n\n # Define two heads.\n class_output = self.class_output_layer(x)\n answer_output = self.answer_output_layer(x)\n return [class_output, answer_output]\n\nmodel = MultiTaskModel()\n\n# Loss function and optimizer\noptimizer = torch.optim.Adam(model.parameters())\nloss_function0 = nn.CrossEntropyLoss()\nloss_function1 = nn.BCEWithLogitsLoss()\n",
"_____no_output_____"
]
],
[
[
"The training loop needs to be modified to not only handle two different inputs (the image of the digit and the text representing the question) but also the two outputs and loss functions. We need to decide on weights for these two loss functions to indicate how to weigh the two into a single loss function for training the network. The weights should be treated like any other hyperparameter. A reasonable starting point is to have the same weight for both losses, so we use 50/50 in our implementation. You can see how we compute and weigh these two losses as a part of the forward pass in the code snippet below.\n\nFinally, we also modify the print statement to print out loss and accuracy separately for the two heads. If you run this code example, you should see it achieving an accuracy above 90% for both tasks. You can experiment with tweaking the weights for the two loss functions and see how that affects the results.\n",
"_____no_output_____"
]
],
[
[
"# Training loop for multi-modal multi-task model.\n# Transfer model to GPU.\nmodel.to(device)\n\n# Create dataloaders.\ntrainloader = DataLoader(trainset, batch_size=BATCH_SIZE, shuffle=True)\ntestloader = DataLoader(testset, batch_size=BATCH_SIZE, shuffle=False)\n\nfor i in range(EPOCHS):\n model.train() # Set model in training mode.\n class_train_loss = 0.0\n class_train_correct = 0\n answer_train_loss = 0.0\n answer_train_correct = 0\n train_batches = 0\n for image_inputs, text_inputs, class_targets, answer_targets in trainloader:\n # Move data to GPU.\n image_inputs = image_inputs.to(device)\n text_inputs = text_inputs.to(device)\n class_targets = class_targets.to(device)\n answer_targets = answer_targets.to(device)\n\n # Zero the parameter gradients.\n optimizer.zero_grad()\n\n # Forward pass.\n outputs = model([image_inputs, text_inputs])\n class_loss = loss_function0(outputs[0], class_targets)\n answer_loss = loss_function1(outputs[1], answer_targets)\n loss = 0.5*class_loss + 0.5*answer_loss\n\n # Accumulate metrics.\n _, indices = torch.max(outputs[0].data, 1)\n class_train_correct += (indices == class_targets).sum().item()\n answer_train_correct += ((outputs[1].data > 0.0) == answer_targets).sum().item()\n train_batches += 1\n class_train_loss += class_loss.item()\n answer_train_loss += answer_loss.item()\n\n # Backward pass and update.\n loss.backward()\n optimizer.step()\n\n class_train_loss = class_train_loss / train_batches\n class_train_acc = class_train_correct / (train_batches * BATCH_SIZE)\n answer_train_loss = answer_train_loss / train_batches\n answer_train_acc = answer_train_correct / (train_batches * BATCH_SIZE)\n\n # Evaluate the model on the test dataset.\n model.eval() # Set model in inference mode.\n class_test_loss = 0.0\n class_test_correct = 0\n answer_test_loss = 0.0\n answer_test_correct = 0\n test_batches = 0\n for image_inputs, text_inputs, class_targets, answer_targets in testloader:\n image_inputs = image_inputs.to(device)\n text_inputs = text_inputs.to(device)\n class_targets = class_targets.to(device)\n answer_targets = answer_targets.to(device)\n outputs = model([image_inputs, text_inputs])\n class_loss = loss_function0(outputs[0], class_targets)\n answer_loss = loss_function1(outputs[1], answer_targets)\n loss = 0.5*class_loss + 0.5*answer_loss\n _, indices = torch.max(outputs[0].data, 1)\n class_test_correct += (indices == class_targets).sum().item()\n answer_test_correct += ((outputs[1].data > 0.0) == answer_targets).sum().item()\n test_batches += 1\n class_test_loss += class_loss.item()\n answer_test_loss += answer_loss.item()\n class_test_loss = class_test_loss / test_batches\n class_test_acc = class_test_correct / (test_batches * BATCH_SIZE)\n answer_test_loss = answer_test_loss / test_batches\n answer_test_acc = answer_test_correct / (test_batches * BATCH_SIZE)\n print(f'Epoch {i+1}/{EPOCHS} class loss: {class_train_loss:.4f} - answer loss: {answer_train_loss:.4f} - class acc: {class_train_acc:0.4f} - answer acc: {answer_train_acc:0.4f} - class val_loss: {class_test_loss:.4f} - answer val_loss: {answer_test_loss:.4f} - class val_acc: {class_test_acc:0.4f} - answer val_acc: {answer_test_acc:0.4f}')\n",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
4a55863aa76332ed6f99c731316604515681036e
| 31,865 |
ipynb
|
Jupyter Notebook
|
examples/Chicago/Exploring the dataset.ipynb
|
BubbleStar/PredictCode
|
1c6a5544b1d9185a4547c54fddc630a3592da3ba
|
[
"Artistic-2.0"
] | 1 |
2019-03-24T07:06:25.000Z
|
2019-03-24T07:06:25.000Z
|
examples/Chicago/Exploring the dataset.ipynb
|
BubbleStar/PredictCode
|
1c6a5544b1d9185a4547c54fddc630a3592da3ba
|
[
"Artistic-2.0"
] | null | null | null |
examples/Chicago/Exploring the dataset.ipynb
|
BubbleStar/PredictCode
|
1c6a5544b1d9185a4547c54fddc630a3592da3ba
|
[
"Artistic-2.0"
] | null | null | null | 35.723094 | 292 | 0.478613 |
[
[
[
"# Allow us to load `open_cp` without installing\nimport sys, os.path\nsys.path.insert(0, os.path.abspath(os.path.join(\"..\", \"..\")))",
"_____no_output_____"
]
],
[
[
"# Chicago data\n\nThe data can be downloaded from https://catalog.data.gov/dataset/crimes-2001-to-present-398a4 (see the module docstring of `open_cp.sources.chicago` See also https://data.cityofchicago.org/Public-Safety/Crimes-2001-to-present/ijzp-q8t2\n\nIn this notebook, we quickly look at the data, check that the data agrees between both sources, and demo some of the library features provided for loading the data.",
"_____no_output_____"
]
],
[
[
"import open_cp.sources.chicago as chicago\nimport geopandas as gpd\n\nimport sys, os, csv, lzma\nfilename = os.path.join(\"..\", \"..\", \"open_cp\", \"sources\", \"chicago.csv\")\nfilename_all = os.path.join(\"..\", \"..\", \"open_cp\", \"sources\", \"chicago_all.csv.xz\")\nfilename_all1 = os.path.join(\"..\", \"..\", \"open_cp\", \"sources\", \"chicago_all1.csv.xz\")",
"_____no_output_____"
]
],
[
[
"Let us look at the snapshot of the last year, vs the total dataset. The data appears to be the same, though the exact format changes.",
"_____no_output_____"
]
],
[
[
"with open(filename, \"rt\") as file:\n reader = csv.reader(file)\n print(next(reader))\n print(next(reader))",
"['CASE#', 'DATE OF OCCURRENCE', 'BLOCK', ' IUCR', ' PRIMARY DESCRIPTION', ' SECONDARY DESCRIPTION', ' LOCATION DESCRIPTION', 'ARREST', 'DOMESTIC', 'BEAT', 'WARD', 'FBI CD', 'X COORDINATE', 'Y COORDINATE', 'LATITUDE', 'LONGITUDE', 'LOCATION']\n['HZ560767', '12/22/2016 02:55:00 AM', '010XX N CENTRAL PARK AVE', '4387', 'OTHER OFFENSE', 'VIOLATE ORDER OF PROTECTION', 'APARTMENT', 'N', 'Y', '1112', '27', '26', '1152189', '1906649', '41.899712716', '-87.716454159', '(41.899712716, -87.716454159)']\n"
],
[
"with lzma.open(filename_all, \"rt\") as file:\n reader = csv.reader(file)\n print(next(reader))\n print(next(reader))",
"['ID', 'Case Number', 'Date', 'Block', 'IUCR', 'Primary Type', 'Description', 'Location Description', 'Arrest', 'Domestic', 'Beat', 'District', 'Ward', 'Community Area', 'FBI Code', 'X Coordinate', 'Y Coordinate', 'Year', 'Updated On', 'Latitude', 'Longitude', 'Location']\n['8651563', 'HV322174', '06/05/2012 11:00:00 AM', '022XX N CANNON DR', '0810', 'THEFT', 'OVER $500', 'STREET', 'false', 'false', '1814', '018', '43', '7', '06', '1175057', '1915111', '2012', '02/04/2016 06:33:39 AM', '41.922450893', '-87.632206293', '(41.922450893, -87.632206293)']\n"
]
],
[
[
"As well as loading data directly into a `TimedPoints` class, we can process a sub-set of the data to GeoJSON, or straight to a geopandas dataframe (if geopandas is installed).",
"_____no_output_____"
]
],
[
[
"geo_data = chicago.load_to_GeoJSON()\ngeo_data[0]",
"_____no_output_____"
],
[
"frame = chicago.load_to_geoDataFrame()\nframe.head()",
"_____no_output_____"
]
],
[
[
"## Explore with QGIS\n\nWe can save the dataframe to a shape-file which can be viewed in e.g. QGIS.\n\nTo explore the spatial-distribution, I would recommend using an interactive GIS package. Using QGIS (free and open source) you can easily add a basemap using GoogleMaps or OpenStreetMap, etc. See http://maps.cga.harvard.edu/qgis/wkshop/basemap.php\n\nI found this to be slightly buggy. On Windows, QGIS 2.18.7 I found that the following worked:\n- First open the `chicago.shp` file produced from the line above.\n- Select the Coordinate reference system \"WGS 84 / EPSG:4326\"\n- Now go to the menu \"Web\" -> \"OpenLayers plugin\" -> Whatever\n- The projection should change to EPSG:3857. The basemap will obscure the point map, so in the \"Layers Panel\" drag the basemap to the bottom.\n- Selecting EPSG:3857 at import time doesn't seem to work (which is different to the instructions..!)",
"_____no_output_____"
]
],
[
[
"# On my Windows install, if I don't do this, I get a GDAL error in\n# the Jupyter console, and the resulting \".prj\" file is empty.\n# This isn't critical, but it confuses QGIS, and you end up having to\n# choose a projection when loading the shape-file.\nimport os\nos.environ[\"GDAL_DATA\"] = \"C:\\\\Users\\\\Matthew\\\\Anaconda3\\\\Library\\\\share\\\\gdal\\\\\"\n\nframe.to_file(\"chicago\")",
"_____no_output_____"
]
],
[
[
"# A geoPandas example\n\nLet's use the \"generator of GeoJSON\" option shown above to pick out only BURGLARY crimes from the 2001-- dataset (which is too large to easily load into a dataframe in one go).",
"_____no_output_____"
]
],
[
[
"with lzma.open(filename_all, \"rt\") as file:\n features = [ event for event in chicago.generate_GeoJSON_Features(file, type=\"all\")\n if event[\"properties\"][\"crime\"] == \"THEFT\" ]\n \nframe = gpd.GeoDataFrame.from_features(features)\nframe.crs = {\"init\":\"EPSG:4326\"} # Lon/Lat native coords\nframe.head()",
"_____no_output_____"
],
[
"frame.to_file(\"chicago_all_theft\")",
"_____no_output_____"
],
[
"with lzma.open(filename_all, \"rt\") as file:\n features = [ event for event in chicago.generate_GeoJSON_Features(file, type=\"all\")\n if event[\"properties\"][\"crime\"] == \"BURGLARY\" ]\n\nframe = gpd.GeoDataFrame.from_features(features)\nframe.crs = {\"init\":\"EPSG:4326\"} # Lon/Lat native coords\nframe.head()",
"_____no_output_____"
],
[
"frame.to_file(\"chicago_all_burglary\")",
"_____no_output_____"
],
[
"frame[\"type\"].unique()",
"_____no_output_____"
],
[
"frame[\"location\"].unique()",
"_____no_output_____"
]
],
[
[
"Upon loading into QGIS to visualise, we find that the 2001 data seems to be geocoded in a different way... The events are not on the road, and the distribution looks less artificial. Let's extract the 2001 burglary data, and then the all the 2001 data, and save.",
"_____no_output_____"
]
],
[
[
"with lzma.open(filename_all, \"rt\") as file:\n features = [ event for event in chicago.generate_GeoJSON_Features(file, type=\"all\")\n if event[\"properties\"][\"timestamp\"].startswith(\"2001\") ]\n\nframe = gpd.GeoDataFrame.from_features(features)\nframe.crs = {\"init\":\"EPSG:4326\"} # Lon/Lat native coords\nframe.head()",
"_____no_output_____"
],
[
"frame.to_file(\"chicago_2001\")",
"_____no_output_____"
]
],
[
[
"# Explore rounding errors\n\nWe check the following:\n- The X and Y COORDINATES fields (which we'll see, in a different notebook, at longitude / latitude coordinates projected in EPSG:3435 in feet) are always whole numbers.\n- The longitude and latitude data contains at most 9 decimals places of accuracy.\n\nIn the other notebook, we look at map projections. The data is most consistent with the longitude / latitude coordinates being the primary source, and the X/Y projected coordinates being computed and rounded to the nearest integer.",
"_____no_output_____"
]
],
[
[
"longs, lats = [], []\nxcs, ycs = [], []\n\nwith open(filename, \"rt\") as file:\n reader = csv.reader(file)\n header = next(reader)\n print(header)\n for row in reader:\n if len(row[14]) > 0:\n longs.append(row[14])\n lats.append(row[15])\n xcs.append(row[12])\n ycs.append(row[13])",
"['CASE#', 'DATE OF OCCURRENCE', 'BLOCK', ' IUCR', ' PRIMARY DESCRIPTION', ' SECONDARY DESCRIPTION', ' LOCATION DESCRIPTION', 'ARREST', 'DOMESTIC', 'BEAT', 'WARD', 'FBI CD', 'X COORDINATE', 'Y COORDINATE', 'LATITUDE', 'LONGITUDE', 'LOCATION']\n"
],
[
"set(len(x) for x in longs), set(len(x) for x in lats)",
"_____no_output_____"
],
[
"any(x.find('.') >= 0 for x in xcs), any(y.find('.') >= 0 for y in ycs)",
"_____no_output_____"
]
],
[
[
"# Repeated data\n\nMostly the \"case\" assignment is unique, but there are a few exceptions to this.",
"_____no_output_____"
]
],
[
[
"import collections\n\nwith lzma.open(filename_all, \"rt\") as file:\n c = collections.Counter( event[\"properties\"][\"case\"] for event in\n chicago.generate_GeoJSON_Features(file, type=\"all\") )\n \nmultiples = set( key for key in c if c[key] > 1 )\nlen(multiples)",
"_____no_output_____"
],
[
"with lzma.open(file_all, \"rt\") as file:\n data = gpd.GeoDataFrame.from_features(\n event for event in chicago.generate_GeoJSON_Features(file, type=\"all\")\n if event[\"properties\"][\"case\"] in multiples\n )\n \nlen(data), len(data.case.uniques())",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
4a559b65d9bca8fe2b91079654c31a4b8cee5502
| 14,230 |
ipynb
|
Jupyter Notebook
|
chapter-7/streaming_weblogs.snb.ipynb
|
LearningSparkStreaming/notebooks
|
e3703fbf227a7de6ad54221b2a27310917204c5e
|
[
"Apache-2.0"
] | 33 |
2019-04-05T21:09:31.000Z
|
2022-02-28T01:02:29.000Z
|
chapter-7/streaming_weblogs.snb.ipynb
|
LearningSparkStreaming/notebooks
|
e3703fbf227a7de6ad54221b2a27310917204c5e
|
[
"Apache-2.0"
] | 2 |
2019-09-11T20:48:06.000Z
|
2020-11-06T14:14:31.000Z
|
chapter-7/streaming_weblogs.snb.ipynb
|
LearningSparkStreaming/notebooks
|
e3703fbf227a7de6ad54221b2a27310917204c5e
|
[
"Apache-2.0"
] | 26 |
2019-06-05T21:42:35.000Z
|
2021-12-30T15:57:02.000Z
| 55.803922 | 1,189 | 0.642656 |
[
[
[
"empty"
]
]
] |
[
"empty"
] |
[
[
"empty"
]
] |
4a559c00c0a23b8b8fc7601fd2429cab2ee7a081
| 194,409 |
ipynb
|
Jupyter Notebook
|
AI for Medicine/AI for Medical Prognosis/Part 1 - Build and Evaluate a Linear Risk model.ipynb
|
niheon/ds-ml-dl
|
16f55411d0eaeb45a952f7889eb580959e769487
|
[
"MIT"
] | 2 |
2022-03-08T19:13:01.000Z
|
2022-03-09T01:19:20.000Z
|
AI for Medicine/AI for Medical Prognosis/Part 1 - Build and Evaluate a Linear Risk model.ipynb
|
niheon/machine-learning
|
16f55411d0eaeb45a952f7889eb580959e769487
|
[
"MIT"
] | null | null | null |
AI for Medicine/AI for Medical Prognosis/Part 1 - Build and Evaluate a Linear Risk model.ipynb
|
niheon/machine-learning
|
16f55411d0eaeb45a952f7889eb580959e769487
|
[
"MIT"
] | null | null | null | 110.648264 | 18,560 | 0.852841 |
[
[
[
"# Build and Evaluate a Linear Risk model\n\nWelcome to the first assignment in Course 2!\n",
"_____no_output_____"
],
[
"## Outline\n\n- [1. Import Packages](#1)\n- [2. Load Data](#2)\n- [3. Explore the Dataset](#3)\n- [4. Mean-Normalize the Data](#4)\n - [Exercise 1](#Ex-1)\n- [5. Build the Model](#Ex-2)\n - [Exercise 2](#Ex-2)\n- [6. Evaluate the Model Using the C-Index](#6)\n - [Exercise 3](#Ex-3)\n- [7. Evaluate the Model on the Test Set](#7)\n- [8. Improve the Model](#8)\n - [Exercise 4](#Ex-4)\n- [9. Evalute the Improved Model](#9)",
"_____no_output_____"
],
[
"## Overview of the Assignment\n\nIn this assignment, you'll build a risk score model for retinopathy in diabetes patients using logistic regression.\n\nAs we develop the model, we will learn about the following topics:\n\n- Data preprocessing\n - Log transformations\n - Standardization\n- Basic Risk Models\n - Logistic Regression\n - C-index\n - Interactions Terms\n \n### Diabetic Retinopathy\nRetinopathy is an eye condition that causes changes to the blood vessels in the part of the eye called the retina.\nThis often leads to vision changes or blindness.\nDiabetic patients are known to be at high risk for retinopathy. \n \n### Logistic Regression \nLogistic regression is an appropriate analysis to use for predicting the probability of a binary outcome. In our case, this would be the probability of having or not having diabetic retinopathy.\nLogistic Regression is one of the most commonly used algorithms for binary classification. It is used to find the best fitting model to describe the relationship between a set of features (also referred to as input, independent, predictor, or explanatory variables) and a binary outcome label (also referred to as an output, dependent, or response variable). Logistic regression has the property that the output prediction is always in the range $[0,1]$. Sometimes this output is used to represent a probability from 0%-100%, but for straight binary classification, the output is converted to either $0$ or $1$ depending on whether it is below or above a certain threshold, usually $0.5$.\n\nIt may be confusing that the term regression appears in the name even though logistic regression is actually a classification algorithm, but that's just a name it was given for historical reasons.",
"_____no_output_____"
],
[
"<a name='1'></a>\n## 1. Import Packages\n\nWe'll first import all the packages that we need for this assignment. \n\n- `numpy` is the fundamental package for scientific computing in python.\n- `pandas` is what we'll use to manipulate our data.\n- `matplotlib` is a plotting library.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt",
"_____no_output_____"
]
],
[
[
"<a name='2'></a>\n## 2. Load Data\n\nFirst we will load in the dataset that we will use for training and testing our model.\n\n- Run the next cell to load the data that is stored in csv files.\n- There is a function `load_data` which randomly generates data, but for consistency, please use the data from the csv files.",
"_____no_output_____"
]
],
[
[
"from utils import load_data\n\n# This function creates randomly generated data\n# X, y = load_data(6000)\n\n# For stability, load data from files that were generated using the load_data\nX = pd.read_csv('X_data.csv',index_col=0)\ny_df = pd.read_csv('y_data.csv',index_col=0)\ny = y_df['y']",
"_____no_output_____"
]
],
[
[
"`X` and `y` are Pandas DataFrames that hold the data for 6,000 diabetic patients. ",
"_____no_output_____"
],
[
"<a name='3'></a>\n## 3. Explore the Dataset\n\nThe features (`X`) include the following fields:\n* Age: (years)\n* Systolic_BP: Systolic blood pressure (mmHg)\n* Diastolic_BP: Diastolic blood pressure (mmHg)\n* Cholesterol: (mg/DL)\n \nWe can use the `head()` method to display the first few records of each. ",
"_____no_output_____"
]
],
[
[
"X.head()",
"_____no_output_____"
]
],
[
[
"The target (`y`) is an indicator of whether or not the patient developed retinopathy.\n\n* y = 1 : patient has retinopathy.\n* y = 0 : patient does not have retinopathy.",
"_____no_output_____"
]
],
[
[
"y.head()",
"_____no_output_____"
]
],
[
[
"Before we build a model, let's take a closer look at the distribution of our training data. To do this, we will split the data into train and test sets using a 75/25 split.\n\nFor this, we can use the built in function provided by sklearn library. See the documentation for [sklearn.model_selection.train_test_split](https://scikit-learn.org/stable/modules/generated/sklearn.model_selection.train_test_split.html). ",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import train_test_split",
"_____no_output_____"
],
[
"X_train_raw, X_test_raw, y_train, y_test = train_test_split(X, y, train_size=0.75, random_state=0)",
"_____no_output_____"
]
],
[
[
"Plot the histograms of each column of `X_train` below: ",
"_____no_output_____"
]
],
[
[
"for col in X.columns:\n X_train_raw.loc[:, col].hist()\n plt.title(col)\n plt.show()",
"_____no_output_____"
]
],
[
[
"As we can see, the distributions have a generally bell shaped distribution, but with slight rightward skew.\n\nMany statistical models assume that the data is normally distributed, forming a symmetric Gaussian bell shape (with no skew) more like the example below.",
"_____no_output_____"
]
],
[
[
"from scipy.stats import norm\ndata = np.random.normal(50,12, 5000)\nfitting_params = norm.fit(data)\nnorm_dist_fitted = norm(*fitting_params)\nt = np.linspace(0,100, 100)\nplt.hist(data, bins=60, density=True)\nplt.plot(t, norm_dist_fitted.pdf(t))\nplt.title('Example of Normally Distributed Data')\nplt.show()",
"_____no_output_____"
]
],
[
[
"We can transform our data to be closer to a normal distribution by removing the skew. One way to remove the skew is by applying the log function to the data.\n\nLet's plot the log of the feature variables to see that it produces the desired effect.",
"_____no_output_____"
]
],
[
[
"for col in X_train_raw.columns:\n np.log(X_train_raw.loc[:, col]).hist()\n plt.title(col)\n plt.show()",
"_____no_output_____"
]
],
[
[
"We can see that the data is more symmetric after taking the log.",
"_____no_output_____"
],
[
"<a name='4'></a>\n## 4. Mean-Normalize the Data\n\nLet's now transform our data so that the distributions are closer to standard normal distributions.\n\nFirst we will remove some of the skew from the distribution by using the log transformation.\nThen we will \"standardize\" the distribution so that it has a mean of zero and standard deviation of 1. Recall that a standard normal distribution has mean of zero and standard deviation of 1. \n",
"_____no_output_____"
],
[
"<a name='Ex-1'></a>\n### Exercise 1\n* Write a function that first removes some of the skew in the data, and then standardizes the distribution so that for each data point $x$,\n$$\\overline{x} = \\frac{x - mean(x)}{std(x)}$$\n* Keep in mind that we want to pretend that the test data is \"unseen\" data. \n * This implies that it is unavailable to us for the purpose of preparing our data, and so we do not want to consider it when evaluating the mean and standard deviation that we use in the above equation. Instead we want to calculate these values using the training data alone, but then use them for standardizing both the training and the test data.\n * For a further discussion on the topic, see this article [\"Why do we need to re-use training parameters to transform test data\"](https://sebastianraschka.com/faq/docs/scale-training-test.html). ",
"_____no_output_____"
],
[
"#### Note\n- For the sample standard deviation, please calculate the unbiased estimator:\n$$s = \\sqrt{\\frac{\\sum_{i=1}^n(x_{i} - \\bar{x})^2}{n-1}}$$\n- In other words, if you numpy, set the degrees of freedom `ddof` to 1.\n- For pandas, the default `ddof` is already set to 1.",
"_____no_output_____"
],
[
"<details> \n<summary>\n <font size=\"3\" color=\"darkgreen\"><b>Hints</b></font>\n</summary>\n<p>\n <ul>\n <li> When working with Pandas DataFrames, you can use the aggregation functions <code>mean</code> and <code>std</code> functions. Note that in order to apply an aggregation function separately for each row or each column, you'll set the axis parameter to either <code>0</code> or <code>1</code>. One produces the aggregation along columns and the other along rows, but it is easy to get them confused. So experiment with each option below to see which one you should use to get an average for each column in the dataframe.\n<code>\navg = df.mean(axis=0)\navg = df.mean(axis=1) \n</code>\n </li>\n <br></br>\n <li>Remember to use <b>training</b> data statistics when standardizing both the training and the test data.</li>\n </ul>\n</p>\n</details> ",
"_____no_output_____"
]
],
[
[
"# UNQ_C1 (UNIQUE CELL IDENTIFIER, DO NOT EDIT)\ndef make_standard_normal(df_train, df_test):\n \"\"\"\n In order to make the data closer to a normal distribution, take log\n transforms to reduce the skew.\n Then standardize the distribution with a mean of zero and standard deviation of 1. \n \n Args:\n df_train (dataframe): unnormalized training data.\n df_test (dataframe): unnormalized test data.\n \n Returns:\n df_train_normalized (dateframe): normalized training data.\n df_test_normalized (dataframe): normalized test data.\n \"\"\"\n \n ### START CODE HERE (REPLACE INSTANCES OF 'None' with your code) ### \n # Remove skew by applying the log function to the train set, and to the test set\n df_train_unskewed = np.log(df_train)\n df_test_unskewed = np.log(df_test)\n \n #calculate the mean and standard deviation of the training set\n mean = df_train_unskewed.mean(axis=0)\n stdev = df_train_unskewed.std(axis=0)\n \n # standardize the training set\n df_train_standardized = (df_train_unskewed - mean) / stdev\n \n # standardize the test set (see instructions and hints above)\n df_test_standardized = (df_test_unskewed - mean) / stdev\n \n ### END CODE HERE ###\n return df_train_standardized, df_test_standardized",
"_____no_output_____"
]
],
[
[
"#### Test Your Work",
"_____no_output_____"
]
],
[
[
"# test\ntmp_train = pd.DataFrame({'field1': [1,2,10], 'field2': [4,5,11]})\ntmp_test = pd.DataFrame({'field1': [1,3,10], 'field2': [4,6,11]})\ntmp_train_transformed, tmp_test_transformed = make_standard_normal(tmp_train,tmp_test)\n\nprint(f\"Training set transformed field1 has mean {tmp_train_transformed['field1'].mean(axis=0):.4f} and standard deviation {tmp_train_transformed['field1'].std(axis=0):.4f} \")\nprint(f\"Test set transformed, field1 has mean {tmp_test_transformed['field1'].mean(axis=0):.4f} and standard deviation {tmp_test_transformed['field1'].std(axis=0):.4f}\")\nprint(f\"Skew of training set field1 before transformation: {tmp_train['field1'].skew(axis=0):.4f}\")\nprint(f\"Skew of training set field1 after transformation: {tmp_train_transformed['field1'].skew(axis=0):.4f}\")\nprint(f\"Skew of test set field1 before transformation: {tmp_test['field1'].skew(axis=0):.4f}\")\nprint(f\"Skew of test set field1 after transformation: {tmp_test_transformed['field1'].skew(axis=0):.4f}\")",
"Training set transformed field1 has mean -0.0000 and standard deviation 1.0000 \nTest set transformed, field1 has mean 0.1144 and standard deviation 0.9749\nSkew of training set field1 before transformation: 1.6523\nSkew of training set field1 after transformation: 1.0857\nSkew of test set field1 before transformation: 1.3896\nSkew of test set field1 after transformation: 0.1371\n"
]
],
[
[
"#### Expected Output:\n```CPP\nTraining set transformed field1 has mean -0.0000 and standard deviation 1.0000 \nTest set transformed, field1 has mean 0.1144 and standard deviation 0.9749\nSkew of training set field1 before transformation: 1.6523\nSkew of training set field1 after transformation: 1.0857\nSkew of test set field1 before transformation: 1.3896\nSkew of test set field1 after transformation: 0.1371\n```",
"_____no_output_____"
],
[
"#### Transform training and test data \nUse the function that you just implemented to make the data distribution closer to a standard normal distribution.",
"_____no_output_____"
]
],
[
[
"X_train, X_test = make_standard_normal(X_train_raw, X_test_raw)",
"_____no_output_____"
]
],
[
[
"After transforming the training and test sets, we'll expect the training set to be centered at zero with a standard deviation of $1$.\n\nWe will avoid observing the test set during model training in order to avoid biasing the model training process, but let's have a look at the distributions of the transformed training data.",
"_____no_output_____"
]
],
[
[
"for col in X_train.columns:\n X_train[col].hist()\n plt.title(col)\n plt.show()",
"_____no_output_____"
]
],
[
[
"<a name='5'></a>\n## 5. Build the Model\n\nNow we are ready to build the risk model by training logistic regression with our data.\n",
"_____no_output_____"
],
[
"<a name='Ex-2'></a>\n### Exercise 2\n\n* Implement the `lr_model` function to build a model using logistic regression with the `LogisticRegression` class from `sklearn`. \n* See the documentation for [sklearn.linear_model.LogisticRegression](https://scikit-learn.org/stable/modules/generated/sklearn.linear_model.LogisticRegression.html#sklearn.linear_model.LogisticRegression.fit).",
"_____no_output_____"
],
[
"<details> \n<summary>\n <font size=\"3\" color=\"darkgreen\"><b>Hints</b></font>\n</summary>\n<p>\n <ul>\n <li>You can leave all the parameters to their default values when constructing an instance of the <code>sklearn.linear_model.LogisticRegression</code> class. If you get a warning message regarding the <code>solver</code> parameter, however, you may want to specify that particular one explicitly with <code>solver='lbfgs'</code>. \n </li>\n <br></br>\n </ul>\n</p>\n</details> ",
"_____no_output_____"
]
],
[
[
"# UNQ_C2 (UNIQUE CELL IDENTIFIER, DO NOT EDIT)\ndef lr_model(X_train, y_train):\n \n ### START CODE HERE (REPLACE INSTANCES OF 'None' with your code) ###\n # import the LogisticRegression class\n from sklearn.linear_model import LogisticRegression\n \n # create the model object\n model = LogisticRegression()\n \n # fit the model to the training data\n model.fit(X_train,y_train)\n \n ### END CODE HERE ###\n #return the fitted model\n return model",
"_____no_output_____"
]
],
[
[
"#### Test Your Work\n\nNote: the `predict` method returns the model prediction *after* converting it from a value in the $[0,1]$ range to a $0$ or $1$ depending on whether it is below or above $0.5$.",
"_____no_output_____"
]
],
[
[
"# Test\ntmp_model = lr_model(X_train[0:3], y_train[0:3] )\nprint(tmp_model.predict(X_train[4:5]))\nprint(tmp_model.predict(X_train[5:6]))",
"[1.]\n[1.]\n"
]
],
[
[
"#### Expected Output:\n```CPP\n[1.]\n[1.]\n```",
"_____no_output_____"
],
[
"Now that we've tested our model, we can go ahead and build it. Note that the `lr_model` function also fits the model to the training data.",
"_____no_output_____"
]
],
[
[
"model_X = lr_model(X_train, y_train)",
"_____no_output_____"
]
],
[
[
"<a name='6'></a>\n## 6. Evaluate the Model Using the C-index\n\nNow that we have a model, we need to evaluate it. We'll do this using the c-index. \n* The c-index measures the discriminatory power of a risk score. \n* Intuitively, a higher c-index indicates that the model's prediction is in agreement with the actual outcomes of a pair of patients.\n* The formula for the c-index is\n\n$$ \\mbox{cindex} = \\frac{\\mbox{concordant} + 0.5 \\times \\mbox{ties}}{\\mbox{permissible}} $$\n\n* A permissible pair is a pair of patients who have different outcomes.\n* A concordant pair is a permissible pair in which the patient with the higher risk score also has the worse outcome.\n* A tie is a permissible pair where the patients have the same risk score.\n",
"_____no_output_____"
],
[
"<a name='Ex-3'></a>\n### Exercise 3\n\n* Implement the `cindex` function to compute c-index.\n* `y_true` is the array of actual patient outcomes, 0 if the patient does not eventually get the disease, and 1 if the patient eventually gets the disease.\n* `scores` is the risk score of each patient. These provide relative measures of risk, so they can be any real numbers. By convention, they are always non-negative.\n* Here is an example of input data and how to interpret it:\n```Python\ny_true = [0,1]\nscores = [0.45, 1.25]\n```\n * There are two patients. Index 0 of each array is associated with patient 0. Index 1 is associated with patient 1.\n * Patient 0 does not have the disease in the future (`y_true` is 0), and based on past information, has a risk score of 0.45.\n * Patient 1 has the disease at some point in the future (`y_true` is 1), and based on past information, has a risk score of 1.25.",
"_____no_output_____"
]
],
[
[
"# UNQ_C3 (UNIQUE CELL IDENTIFIER, DO NOT EDIT)\ndef cindex(y_true, scores):\n '''\n\n Input:\n y_true (np.array): a 1-D array of true binary outcomes (values of zero or one)\n 0: patient does not get the disease\n 1: patient does get the disease\n scores (np.array): a 1-D array of corresponding risk scores output by the model\n\n Output:\n c_index (float): (concordant pairs + 0.5*ties) / number of permissible pairs\n '''\n n = len(y_true)\n assert len(scores) == n\n\n concordant = 0\n permissible = 0\n ties = 0\n \n ### START CODE HERE (REPLACE INSTANCES OF 'None' with your code) ###\n # use two nested for loops to go through all unique pairs of patients\n for i in range(n):\n for j in range(i+1, n): #choose the range of j so that j>i\n \n # Check if the pair is permissible (the patient outcomes are different)\n if y_true[i] != y_true[j]:\n # Count the pair if it's permissible\n permissible =permissible + 1\n\n # For permissible pairs, check if they are concordant or are ties\n\n # check for ties in the score\n if scores[i] == scores[j]:\n # count the tie\n ties = ties + 1\n # if it's a tie, we don't need to check patient outcomes, continue to the top of the for loop.\n continue\n\n # case 1: patient i doesn't get the disease, patient j does\n if y_true[i] == 0 and y_true[j] == 1:\n # Check if patient i has a lower risk score than patient j\n if scores[i] < scores[j]:\n # count the concordant pair\n concordant = concordant + 1\n # Otherwise if patient i has a higher risk score, it's not a concordant pair.\n # Already checked for ties earlier\n\n # case 2: patient i gets the disease, patient j does not\n if y_true[i] == 1 and y_true[j] == 0:\n # Check if patient i has a higher risk score than patient j\n if scores[i] > scores[j]:\n #count the concordant pair\n concordant = concordant + 1\n # Otherwise if patient i has a lower risk score, it's not a concordant pair.\n # We already checked for ties earlier\n\n # calculate the c-index using the count of permissible pairs, concordant pairs, and tied pairs.\n c_index = (concordant + (0.5 * ties)) / permissible\n ### END CODE HERE ###\n \n return c_index",
"_____no_output_____"
]
],
[
[
"#### Test Your Work\n\nYou can use the following test cases to make sure your implementation is correct.",
"_____no_output_____"
]
],
[
[
"# test\ny_true = np.array([1.0, 0.0, 0.0, 1.0])\n\n# Case 1\nscores = np.array([0, 1, 1, 0])\nprint('Case 1 Output: {}'.format(cindex(y_true, scores)))\n\n# Case 2\nscores = np.array([1, 0, 0, 1])\nprint('Case 2 Output: {}'.format(cindex(y_true, scores)))\n\n# Case 3\nscores = np.array([0.5, 0.5, 0.0, 1.0])\nprint('Case 3 Output: {}'.format(cindex(y_true, scores)))\ncindex(y_true, scores)",
"Case 1 Output: 0.0\nCase 2 Output: 1.0\nCase 3 Output: 0.875\n"
]
],
[
[
"#### Expected Output:\n\n```CPP\nCase 1 Output: 0.0\nCase 2 Output: 1.0\nCase 3 Output: 0.875\n```",
"_____no_output_____"
],
[
"#### Note\nPlease check your implementation of the for loops. \n- There is way to make a mistake on the for loops that cannot be caught with unit tests.\n- Bonus: Can you think of what this error could be, and why it can't be caught by unit tests?",
"_____no_output_____"
],
[
"<a name='7'></a>\n## 7. Evaluate the Model on the Test Set\n\nNow, you can evaluate your trained model on the test set. \n\nTo get the predicted probabilities, we use the `predict_proba` method. This method will return the result from the model *before* it is converted to a binary 0 or 1. For each input case, it returns an array of two values which represent the probabilities for both the negative case (patient does not get the disease) and positive case (patient the gets the disease). ",
"_____no_output_____"
]
],
[
[
"scores = model_X.predict_proba(X_test)[:, 1]\nc_index_X_test = cindex(y_test.values, scores)\nprint(f\"c-index on test set is {c_index_X_test:.4f}\")",
"c-index on test set is 0.8182\n"
]
],
[
[
"#### Expected output:\n```CPP\nc-index on test set is 0.8182\n```",
"_____no_output_____"
],
[
"Let's plot the coefficients to see which variables (patient features) are having the most effect. You can access the model coefficients by using `model.coef_`",
"_____no_output_____"
]
],
[
[
"coeffs = pd.DataFrame(data = model_X.coef_, columns = X_train.columns)\ncoeffs.T.plot.bar(legend=None);",
"_____no_output_____"
]
],
[
[
"### Question: \n> __Which three variables have the largest impact on the model's predictions?__",
"_____no_output_____"
],
[
"<a name='8'></a>\n## 8. Improve the Model\n\nYou can try to improve your model by including interaction terms. \n* An interaction term is the product of two variables. \n * For example, if we have data \n $$ x = [x_1, x_2]$$\n * We could add the product so that:\n $$ \\hat{x} = [x_1, x_2, x_1*x_2]$$\n ",
"_____no_output_____"
],
[
"<a name='Ex-4'></a>\n### Exercise 4\n\nWrite code below to add all interactions between every pair of variables to the training and test datasets. ",
"_____no_output_____"
]
],
[
[
"# UNQ_C4 (UNIQUE CELL IDENTIFIER, DO NOT EDIT)\ndef add_interactions(X):\n \"\"\"\n Add interaction terms between columns to dataframe.\n\n Args:\n X (dataframe): Original data\n\n Returns:\n X_int (dataframe): Original data with interaction terms appended. \n \"\"\"\n features = X.columns\n m = len(features)\n X_int = X.copy(deep=True)\n\n ### START CODE HERE (REPLACE INSTANCES OF 'None' with your code) ###\n # 'i' loops through all features in the original dataframe X\n for i in range(m):\n \n # get the name of feature 'i'\n feature_i_name = features[i]\n \n # get the data for feature 'i'\n feature_i_data = X[feature_i_name]\n \n # choose the index of column 'j' to be greater than column i\n for j in range(i+1, m):\n \n # get the name of feature 'j'\n feature_j_name = features[j]\n \n # get the data for feature j'\n feature_j_data = X[feature_j_name]\n \n # create the name of the interaction feature by combining both names\n # example: \"apple\" and \"orange\" are combined to be \"apple_x_orange\"\n feature_i_j_name = feature_i_name+\"_x_\"+feature_j_name\n \n # Multiply the data for feature 'i' and feature 'j'\n # store the result as a column in dataframe X_int\n X_int[feature_i_j_name] = feature_i_data * feature_j_data\n \n ### END CODE HERE ###\n\n return X_int",
"_____no_output_____"
]
],
[
[
"#### Test Your Work\n\nRun the cell below to check your implementation. ",
"_____no_output_____"
]
],
[
[
"print(\"Original Data\")\nprint(X_train.loc[:, ['Age', 'Systolic_BP']].head())\nprint(\"Data w/ Interactions\")\nprint(add_interactions(X_train.loc[:, ['Age', 'Systolic_BP']].head()))",
"Original Data\n Age Systolic_BP\n1824 -0.912451 -0.068019\n253 -0.302039 1.719538\n1114 2.576274 0.155962\n3220 1.163621 -2.033931\n2108 -0.446238 -0.054554\nData w/ Interactions\n Age Systolic_BP Age_x_Systolic_BP\n1824 -0.912451 -0.068019 0.062064\n253 -0.302039 1.719538 -0.519367\n1114 2.576274 0.155962 0.401800\n3220 1.163621 -2.033931 -2.366725\n2108 -0.446238 -0.054554 0.024344\n"
]
],
[
[
"#### Expected Output:\n```CPP\nOriginal Data\n Age Systolic_BP\n1824 -0.912451 -0.068019\n253 -0.302039 1.719538\n1114 2.576274 0.155962\n3220 1.163621 -2.033931\n2108 -0.446238 -0.054554\nData w/ Interactions\n Age Systolic_BP Age_x_Systolic_BP\n1824 -0.912451 -0.068019 0.062064\n253 -0.302039 1.719538 -0.519367\n1114 2.576274 0.155962 0.401800\n3220 1.163621 -2.033931 -2.366725\n2108 -0.446238 -0.054554 0.024344\n```",
"_____no_output_____"
],
[
"Once you have correctly implemented `add_interactions`, use it to make transformed version of `X_train` and `X_test`.",
"_____no_output_____"
]
],
[
[
"X_train_int = add_interactions(X_train)\nX_test_int = add_interactions(X_test)",
"_____no_output_____"
]
],
[
[
"<a name='9'></a>\n## 9. Evaluate the Improved Model\n\nNow we can train the new and improved version of the model.",
"_____no_output_____"
]
],
[
[
"model_X_int = lr_model(X_train_int, y_train)",
"_____no_output_____"
]
],
[
[
"Let's evaluate our new model on the test set.",
"_____no_output_____"
]
],
[
[
"scores_X = model_X.predict_proba(X_test)[:, 1]\nc_index_X_int_test = cindex(y_test.values, scores_X)\n\nscores_X_int = model_X_int.predict_proba(X_test_int)[:, 1]\nc_index_X_int_test = cindex(y_test.values, scores_X_int)\n\nprint(f\"c-index on test set without interactions is {c_index_X_test:.4f}\")\nprint(f\"c-index on test set with interactions is {c_index_X_int_test:.4f}\")",
"c-index on test set without interactions is 0.8182\nc-index on test set with interactions is 0.8281\n"
]
],
[
[
"You should see that the model with interaction terms performs a bit better than the model without interactions.\n\nNow let's take another look at the model coefficients to try and see which variables made a difference. Plot the coefficients and report which features seem to be the most important.",
"_____no_output_____"
]
],
[
[
"int_coeffs = pd.DataFrame(data = model_X_int.coef_, columns = X_train_int.columns)\nint_coeffs.T.plot.bar();",
"_____no_output_____"
]
],
[
[
"### Questions:\n> __Which variables are most important to the model?__<br>\n> __Have the relevant variables changed?__<br>\n> __What does it mean when the coefficients are positive or negative?__<br>\n\nYou may notice that Age, Systolic_BP, and Cholesterol have a positive coefficient. This means that a higher value in these three features leads to a higher prediction probability for the disease. You also may notice that the interaction of Age x Cholesterol has a negative coefficient. This means that a higher value for the Age x Cholesterol product reduces the prediction probability for the disease.\n\nTo understand the effect of interaction terms, let's compare the output of the model we've trained on sample cases with and without the interaction. Run the cell below to choose an index and look at the features corresponding to that case in the training set. ",
"_____no_output_____"
]
],
[
[
"index = index = 3432\ncase = X_train_int.iloc[index, :]\nprint(case)",
"Age 2.502061\nSystolic_BP 1.713547\nDiastolic_BP 0.268265\nCholesterol 2.146349\nAge_x_Systolic_BP 4.287400\nAge_x_Diastolic_BP 0.671216\nAge_x_Cholesterol 5.370296\nSystolic_BP_x_Diastolic_BP 0.459685\nSystolic_BP_x_Cholesterol 3.677871\nDiastolic_BP_x_Cholesterol 0.575791\nName: 5970, dtype: float64\n"
]
],
[
[
"We can see that they have above average Age and Cholesterol. We can now see what our original model would have output by zero-ing out the value for Cholesterol and Age.",
"_____no_output_____"
]
],
[
[
"new_case = case.copy(deep=True)\nnew_case.loc[\"Age_x_Cholesterol\"] = 0\nnew_case",
"_____no_output_____"
],
[
"print(f\"Output with interaction: \\t{model_X_int.predict_proba([case.values])[:, 1][0]:.4f}\")\nprint(f\"Output without interaction: \\t{model_X_int.predict_proba([new_case.values])[:, 1][0]:.4f}\")",
"Output with interaction: \t0.9448\nOutput without interaction: \t0.9965\n"
]
],
[
[
"#### Expected output\n```CPP\nOutput with interaction: 0.9448\nOutput without interaction: 0.9965\n```",
"_____no_output_____"
],
[
"We see that the model is less confident in its prediction with the interaction term than without (the prediction value is lower when including the interaction term). With the interaction term, the model has adjusted for the fact that the effect of high cholesterol becomes less important for older patients compared to younger patients.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
]
] |
4a55a14b400d94c7a38eeb972cd6cc5051098c18
| 4,066 |
ipynb
|
Jupyter Notebook
|
plot_performance.ipynb
|
YunjaeChoi/Gain-Risk_Framework_for_Stabilization_of_Deep_Policy_Gradient_Optimization
|
68d3f8fca6c6e6b356261f568f0d8562242fa649
|
[
"MIT"
] | null | null | null |
plot_performance.ipynb
|
YunjaeChoi/Gain-Risk_Framework_for_Stabilization_of_Deep_Policy_Gradient_Optimization
|
68d3f8fca6c6e6b356261f568f0d8562242fa649
|
[
"MIT"
] | null | null | null |
plot_performance.ipynb
|
YunjaeChoi/Gain-Risk_Framework_for_Stabilization_of_Deep_Policy_Gradient_Optimization
|
68d3f8fca6c6e6b356261f568f0d8562242fa649
|
[
"MIT"
] | null | null | null | 26.232258 | 107 | 0.497541 |
[
[
[
"from utils.plot_utils import make_plots\nimport seaborn as sns\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport numpy as np\n\nimport warnings\nwarnings.filterwarnings(\"ignore\")\n\n%matplotlib inline\n\nlegend = None\nxaxis = 'TotalEnvInteracts'\n\nvalues = ['Performance', 'StopIter', 'PiLr', 'DiagNormalEntropy', 'DiagNormalKL', 'SampleEntropy']\ncount = False\nsmooth = 1\nselect = None\nexclude = ''\nest = 'mean'",
"_____no_output_____"
],
[
"logdir = ['data/sca-']\nret_values, ret_data = make_plots(logdir, legend, xaxis, values, count, \n smooth=smooth, select=select, exclude=exclude,\n estimator=est, figsize=(10, 6))",
"_____no_output_____"
],
[
"logdir = ['data/half-']\nret_values, ret_data = make_plots(logdir, legend, xaxis, values, count, \n smooth=smooth, select=select, exclude=exclude,\n estimator=est, figsize=(10, 6))",
"_____no_output_____"
],
[
"logdir = ['data/ant-']\n_ = make_plots(logdir, legend, xaxis, values, count, \n smooth=smooth, select=select, exclude=exclude,\n estimator=est, figsize=(10, 6))",
"_____no_output_____"
],
[
"logdir = ['data/hopper-']\nret_values, ret_data = make_plots(logdir, legend, xaxis, values, count, \n smooth=smooth, select=select, exclude=exclude,\n estimator=est, figsize=(10, 6))",
"_____no_output_____"
],
[
"logdir = ['data/walker-']\nret_values, ret_data = make_plots(logdir, legend, xaxis, values, count, \n smooth=smooth, select=select, exclude=exclude,\n estimator=est, figsize=(10, 6))",
"_____no_output_____"
],
[
"logdir = ['data/ipsu-']\nret_values, ret_data = make_plots(logdir, legend, xaxis, values, count, \n smooth=smooth, select=select, exclude=exclude,\n estimator=est, figsize=(10, 6))",
"_____no_output_____"
],
[
"logdir = ['data/idp-']\nret_values, ret_data = make_plots(logdir, legend, xaxis, values, count, \n smooth=smooth, select=select, exclude=exclude,\n estimator=est, figsize=(10, 6))",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a55db54bcdd6ac2456da74f523dcb5c3ef6d7d4
| 14,173 |
ipynb
|
Jupyter Notebook
|
13/13.ipynb
|
bruninmi/AoC2019
|
43579a799cb70a982eb22d65b8797f29e9b771dd
|
[
"MIT"
] | null | null | null |
13/13.ipynb
|
bruninmi/AoC2019
|
43579a799cb70a982eb22d65b8797f29e9b771dd
|
[
"MIT"
] | null | null | null |
13/13.ipynb
|
bruninmi/AoC2019
|
43579a799cb70a982eb22d65b8797f29e9b771dd
|
[
"MIT"
] | null | null | null | 40.264205 | 540 | 0.394624 |
[
[
[
"import queue\n\nclass Intcode:\n def __init__(self, intcode=None, inputs=None, outputs=None, manual=True):\n if intcode == None:\n self.intcode = [99]\n self.inputs = inputs\n if inputs == None:\n self.inputs = queue.Queue()\n if type(inputs) == list:\n q = inputs\n self.inputs = queue.Queue()\n [self.inputs.put(i) for i in q]\n self.outputs = outputs\n if outputs == None:\n self.outputs = queue.Queue()\n if type(outputs) == list:\n q = outputs\n self.outputs = queue.Queue()\n [self.outputs.put(i) for i in q]\n self.base = 0\n self.pc = 0\n self.mem = intcode.copy()\n self.manual = manual\n \n def __mem(self, p, v=None):\n if p >= len(self.mem):\n self.mem += [0] * (1 + p - len(self.mem))\n if not v == None:\n self.mem[p] = v\n return self.mem[p]\n\n def __get(self, var, mode):\n tar = self.pc + ['A', 'B', 'C'].index(var) + 1\n if mode == 0:\n return self.__mem(self.__mem(tar))\n elif mode == 1:\n return self.__mem(tar)\n elif mode == 2:\n return self.__mem(self.base + self.__mem(tar))\n return None\n\n def __set(self, var, mode, value):\n tar = self.pc + ['A', 'B', 'C'].index(var) + 1\n if mode == 0:\n return self.__mem(self.__mem(tar), value)\n elif mode == 1:\n return self.__mem(tar, value)\n elif mode == 2:\n return self.__mem(self.base + self.__mem(tar), value)\n return None\n\n def run(self):\n while not (self.__mem(self.pc) % 100 == 99):\n op = self.__mem(self.pc) % 100\n modes = []\n for i in range(3):\n modes += [int(self.__mem(self.pc) / 100 / 10**i) % 10]\n \n l = 4\n \n if op == 1:\n A = self.__get('A', modes[0])\n B = self.__get('B', modes[1])\n self.__set('C', modes[2], A + B)\n elif op == 2:\n A = self.__get('A', modes[0])\n B = self.__get('B', modes[1])\n self.__set('C', modes[2], A * B)\n elif op == 3:\n if self.manual:\n inp = input('Gief pls: ')\n if inp == 'abort':\n print(\"Aborted\")\n return \"Aborted\"\n self.inputs.put(int(inp))\n else:\n self.outputs.put(\"request\")\n self.__set('A', modes[0], self.inputs.get())\n self.inputs.task_done()\n l = 2\n elif op == 4:\n A = self.__get('A', modes[0])\n if self.manual:\n print('Take pls:', A)\n self.outputs.put(A)\n l = 2\n elif op == 5:\n A = self.__get('A', modes[0])\n B = self.__get('B', modes[1])\n if A:\n self.pc = B\n l = 0\n else:\n l = 3\n elif op == 6:\n A = self.__get('A', modes[0])\n B = self.__get('B', modes[1])\n if not A:\n self.pc = B\n l = 0\n else:\n l = 3\n elif op == 7:\n A = self.__get('A', modes[0])\n B = self.__get('B', modes[1])\n if A < B:\n self.__set('C', modes[2], 1)\n else:\n self.__set('C', modes[2], 0)\n elif op == 8:\n A = self.__get('A', modes[0])\n B = self.__get('B', modes[1])\n if A == B:\n self.__set('C', modes[2], 1)\n else:\n self.__set('C', modes[2], 0)\n elif op == 9:\n A = self.__get('A', modes[0])\n self.base += A\n l = 2\n else:\n return \"Something went wrong.\"\n self.pc += l\n return self.mem, list(self.outputs.queue)\n\ndef process_Intcode_test(params, output, message):\n prg = Intcode(*params)\n if not prg.run() == output:\n print(message)\n return False\n return True\n\nprocess_Intcode_test([[1,9,10,3,2,3,11,0,99,30,40,50]],\n ([3500,9,10,70,2,3,11,0,99,30,40,50], []), \"FAIL: process_Intcode_test 01\")\nprocess_Intcode_test([[1,0,0,0,99]], ([2,0,0,0,99], []), \"FAIL: process_Intcode_test 02\")\nprocess_Intcode_test([[2,3,0,3,99]], ([2,3,0,6,99], []), \"FAIL: process_Intcode_test 03\")\nprocess_Intcode_test([[2,4,4,5,99,0]], ([2,4,4,5,99,9801], []), \"FAIL: process_Intcode_test 04\")\nprocess_Intcode_test([[1,1,1,4,99,5,6,0,99]], ([30,1,1,4,2,5,6,0,99], []), \"FAIL: process_Intcode_test 05\")\nprocess_Intcode_test([[1002,4,3,4,33]], ([1002,4,3,4,99], []), \"FAIL: process_Intcode_test 07\")\nprocess_Intcode_test([[1101,100,-1,4,0]], ([1101,100,-1,4,99], []), \"FAIL: process_Intcode_test 08\")\n#testing with inputs\nprocess_Intcode_test([[3,0,4,0,99], [456], [], False], ([456,0,4,0,99], [456]), \"FAIL: process_Intcode_test 06\")\nprocess_Intcode_test([[3,9,8,9,10,9,4,9,99,-1,8], [8], [], False], ([3,9,8,9,10,9,4,9,99,1,8], [1]), \"FAIL: process_Intcode_test 09\")\nprocess_Intcode_test([[3,9,8,9,10,9,4,9,99,-1,8], [5], [], False], ([3,9,8,9,10,9,4,9,99,0,8], [0]), \"FAIL: process_Intcode_test 10\")\nprocess_Intcode_test([[3,9,7,9,10,9,4,9,99,-1,8], [5], [], False], ([3,9,7,9,10,9,4,9,99,1,8], [1]), \"FAIL: process_Intcode_test 11\")\nprocess_Intcode_test([[3,9,7,9,10,9,4,9,99,-1,8], [8], [], False], ([3,9,7,9,10,9,4,9,99,0,8], [0]), \"FAIL: process_Intcode_test 12\")\nprocess_Intcode_test([[3,3,1108,-1,8,3,4,3,99], [8], [], False], ([3,3,1108,1,8,3,4,3,99], [1]), \"FAIL: process_Intcode_test 13\")\nprocess_Intcode_test([[3,3,1108,-1,8,3,4,3,99], [5], [], False], ([3,3,1108,0,8,3,4,3,99], [0]), \"FAIL: process_Intcode_test 14\")\nprocess_Intcode_test([[3,3,1107,-1,8,3,4,3,99], [5], [], False], ([3,3,1107,1,8,3,4,3,99], [1]), \"FAIL: process_Intcode_test 15\")\nprocess_Intcode_test([[3,3,1107,-1,8,3,4,3,99], [8], [], False], ([3,3,1107,0,8,3,4,3,99], [0]), \"FAIL: process_Intcode_test 16\")\n#day 9 tests\nprocess_Intcode_test([[109,1,204,-1,1001,100,1,100,1008,100,16,101,1006,101,0,99], [], [], False], ([109, 1, 204, -1, 1001, 100, 1, 100, 1008, 100, 16, 101, 1006, 101, 0, 99, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 16, 1], [109,1,204,-1,1001,100,1,100,1008,100,16,101,1006,101,0,99]), \"FAIL: process_Intcode_test 17\")\nprocess_Intcode_test([[1102,34915192,34915192,7,4,7,99,0], [], [], False], ([1102, 34915192, 34915192, 7, 4, 7, 99, 1219070632396864], [1219070632396864]), \"FAIL: process_Intcode_test 18\")\nprocess_Intcode_test([[104,1125899906842624,99], [], [], False], ([104, 1125899906842624, 99], [1125899906842624]), \"FAIL: process_Intcode_test 18\")",
"FAIL: process_Intcode_test 06\nFAIL: process_Intcode_test 09\nFAIL: process_Intcode_test 10\nFAIL: process_Intcode_test 11\nFAIL: process_Intcode_test 12\nFAIL: process_Intcode_test 13\nFAIL: process_Intcode_test 14\nFAIL: process_Intcode_test 15\nFAIL: process_Intcode_test 16\n"
],
[
"with open(\"input13.txt\") as infile:\n intcode = [int(i) for i in infile.readlines()[0].split(',')]\n inp = queue.Queue()\n out = queue.Queue()\n prg = Intcode(intcode, inputs=inp, outputs=out, manual=False)\n \nout = prg.run()[1]\nscreen = []\nfor i in range(0, len(out), 3):\n (x, y, z) = (out[i], out[i+1], out[i+2])\n if y >= len(screen):\n screen += [[]] * (1 + y - len(screen))\n row = screen[y]\n if x >= len(row):\n row += [''] * (1 + x - len(row))\n row[x] = z\n \nblock = [' ', '#', 'x' ,'=' ,'o']\n \nscreen_img = '\\n'.join([''.join(block[i] for i in row) for row in screen])\nscreen_img.count('x')",
"_____no_output_____"
],
[
"from IPython.display import clear_output\n\ndef update(screen, x, y, z):\n if y >= len(screen):\n screen += [[]] * (1 + y - len(screen))\n row = screen[y]\n if x >= len(row):\n row += [''] * (1 + x - len(row))\n row[x] = z\n\n return screen\n\nwith open(\"input13.txt\") as infile:\n intcode = [int(i) for i in infile.readlines()[0].split(',')]\n intcode[0] = 2\n inp = queue.Queue()\n out = queue.Queue()\n prg = Intcode(intcode, inputs=inp, outputs=out, manual=False)\n\nimport threading\n\nt = threading.Thread(target=Intcode.run, name='arcade', args=(prg,))\nt.start()\n\nscreen = []\nscore = 0\n\nwhile t.is_alive() or not out.empty():\n x = out.get()\n if x == \"request\":\n clear_output(wait=True)\n print('\\n'.join([''.join(block[i] for i in row) for row in screen]))\n print(\"Score: \", score)\n \n target = -1\n pos = -1\n for row in screen:\n if 4 in row:\n target = row.index(4)\n if 3 in row:\n pos = row.index(3)\n if target > pos:\n inp.put(1)\n elif target < pos:\n inp.put(-1)\n else:\n inp.put(0)\n continue\n y = out.get()\n z = out.get()\n if (x, y) == (-1, 0):\n score = z\n else:\n update(screen, x, y, z)\n\nclear_output(wait=True)\nprint('\\n'.join([''.join(block[i] for i in row) for row in screen]))\nprint(\"Score: \", score)",
"###################################\n# #\n# #\n# #\n# #\n# #\n# o #\n# #\n# #\n# #\n# #\n# #\n# #\n# #\n# #\n# #\n# #\n# #\n# #\n# #\n# #\n# = #\n# #\nScore: 12952\n"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code"
]
] |
4a55e437c2946fc53d0cc773e7626a524fa6c69d
| 14,073 |
ipynb
|
Jupyter Notebook
|
lectures/01b-coordinates.ipynb
|
ghutchis/chem1000
|
07a7eac20cc04ee9a1bdb98339fbd5653a02a38d
|
[
"CC-BY-4.0"
] | 12 |
2020-06-23T18:44:37.000Z
|
2022-03-14T10:13:05.000Z
|
lectures/01b-coordinates.ipynb
|
ghutchis/chem1000
|
07a7eac20cc04ee9a1bdb98339fbd5653a02a38d
|
[
"CC-BY-4.0"
] | null | null | null |
lectures/01b-coordinates.ipynb
|
ghutchis/chem1000
|
07a7eac20cc04ee9a1bdb98339fbd5653a02a38d
|
[
"CC-BY-4.0"
] | 4 |
2021-07-29T10:45:23.000Z
|
2021-10-16T09:51:00.000Z
| 33.035211 | 240 | 0.564983 |
[
[
[
"# CHEM 1000 - Spring 2022\nProf. Geoffrey Hutchison, University of Pittsburgh\n\n## 1. Functions and Coordinate Sets\n\nChapter 1 in [*Mathematical Methods for Chemists*](http://sites.bu.edu/straub/mathematical-methods-for-molecular-science/)\n\nBy the end of this session, you should be able to:\n- Handle 2D polar and 3D spherical coordinates\n- Understand area elements in 2D polar coordinates\n- Understand volume eleements in 3D spherical coordinates",
"_____no_output_____"
],
[
"### X/Y Cartesian 2D Coordinates\n\nWe've already been using the x/y 2D Cartesian coordinate set to plot functions.\n\nBeyond `sympy`, we're going to use two new modules:\n- `numpy` which lets us create and handle arrays of numbers\n- `matplotlib` which lets us plot things\n\nIt's a little bit more complicated. For now, you can just consider these as **demos**. We'll go into code (and make our own plots) in the next recitation period.",
"_____no_output_____"
]
],
[
[
"# import numpy\n# the \"as np\" part is giving a shortcut so we can write \"np.function()\" instead of \"numpy.function()\"\n# (saving typing is nice)\nimport numpy as np\n# similarly, we import matplotlib's 'pyplot' module\n# and \"as plt\" means we can use \"plt.show\" instead of \"matplotlib.pyplot.show()\"\nimport matplotlib.pyplot as plt\n\n# insert any graphs into our notebooks directly\n%matplotlib inline\n%config InlineBackend.figure_format = 'retina'",
"_____no_output_____"
],
[
"# once we've done that import (once) - we just need to create our x/y values\nx = np.arange(0, 4*np.pi, 0.1) # start, stop, resolution\ny = np.sin(x) # creates an array with sin() of all the x values\n\nplt.plot(x,y)\nplt.show()",
"_____no_output_____"
]
],
[
[
"Sometimes, we need to get areas in the Cartesian xy system, but this is very easy - we simply multiply an increment in x ($dx$) and an increment in y ($dy$).\n\n(Image from [*Mathematical Methods for Chemists*](http://sites.bu.edu/straub/mathematical-methods-for-molecular-science/))\n\n<img src=\"../images/cartesian-area.png\" width=\"400\" />",
"_____no_output_____"
],
[
"### Polar (2D) Coordinates\n\nOf course, not all functions work well in xy Cartesian coordinates. A function should produce one y value for any x value. Thus, a circle isn't easily represented as $y = f(x)$.\n\nInstead, polar coordinates, use radius $r$ and angle $\\theta$. (Image from [*Mathematical Methods for Chemists*](http://sites.bu.edu/straub/mathematical-methods-for-molecular-science/))\n\n<img src=\"../images/cartesian-polar.png\" width=\"343\" />",
"_____no_output_____"
],
[
"As a reminder, we can interconvert x,y into r, theta:\n\n$$\nr = \\sqrt{x^2 + y^2}\n$$\n\n$$\n\\theta = \\arctan \\frac{y}{x} = \\tan^{-1} \\frac{y}{x}\n$$",
"_____no_output_____"
]
],
[
[
"x = 3.0\ny = 1.0\n\nr = np.sqrt(x**2 + y**2)\ntheta = np.arctan(y / x)\n\nprint('r =', round(r, 4), 'theta = ', round(theta, 4))",
"_____no_output_____"
]
],
[
[
"Okay, we can't express a circle as an easy $y = f(x)$ expression. Can we do that in polar coordinates? Sure. The radius will be constant, and theta will go from $0 .. 2\\pi$.",
"_____no_output_____"
]
],
[
[
"theta = np.arange(0, 2*np.pi, 0.01) # set up an array of radii from 0 to 2π with 0.01 rad\n\n# create a function r(theta) = 1.5 .. a constant\nr = np.full(theta.size, 1.5)\n\n# create a new polar plot\nax = plt.subplot(111, projection='polar')\nax.plot(theta, r, color='blue')\n\nax.set_rmax(3)\nax.set_rticks([1, 2]) # Less radial ticks\nax.set_rlabel_position(22.5) # Move radial labels away from plotted line\nax.grid(True)\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"Anything else? Sure - we can create spirals, etc. that are parametric functions in the XY Cartesian coordinates.",
"_____no_output_____"
]
],
[
[
"r = np.arange(0, 2, 0.01) # set up an array of radii from 0 to 2 with 0.01 resolution\n\n# this is a function theta(r) = 2π * r\ntheta = 2 * np.pi * r # set up an array of theta angles - spiraling outward .. from 0 to 2*2pi = 4pi\n\n# create a polar plot\nax = plt.subplot(111, projection='polar')\nax.plot(theta, r, color='red')\n\nax.set_rmax(3)\nax.set_rticks([1, 2]) # Less radial ticks\nax.set_rlabel_position(22.5) # Move radial labels away from plotted line\nax.grid(True)\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"Just like with xy Cartesian, we will eventually need to consider the area of functions in polar coordinates. (Image from [*Mathematical Methods for Chemists*](http://sites.bu.edu/straub/mathematical-methods-for-molecular-science/))\n\n<img src=\"../images/polar_area.png\" width=375 />\n\nNote that the area depends on the radius. Even if we sweep out the same $\\Delta r$ and $\\Delta \\theta$ an area further out from the center is larger:",
"_____no_output_____"
]
],
[
[
"# create a polar plot\nax = plt.subplot(111, projection='polar')\n\n# first arc at r = 1.0\nr1 = np.full(20, 1.0)\ntheta1 = np.linspace(1.0, 1.3, 20)\nax.plot(theta1, r1)\n\n# second arc at r = 1.2\nr2 = np.full(20, 1.2)\ntheta2 = np.linspace(1.0, 1.3, 20)\nax.plot(theta2, r2)\n\n# first radial line at theta = 1.0 radians\nr3 = np.linspace(1.0, 1.2, 20)\ntheta3 = np.full(20, 1.0)\nax.plot(theta3, r3)\n\n# first radial line at theta = 1.3 radians\nr4 = np.linspace(1.0, 1.2, 20)\ntheta4 = np.full(20, 1.3)\nax.plot(theta4, r4)\n\n# smaller box\n# goes from r = 0.4-> 0.6\n# sweeps out theta = 1.0-1.3 radians\nr5 = np.full(20, 0.4)\nr6 = np.full(20, 0.6)\nr7 = np.linspace(0.4, 0.6, 20)\nr8 = np.linspace(0.4, 0.6, 20)\nax.plot(theta1, r5)\nax.plot(theta2, r6)\nax.plot(theta3, r7)\nax.plot(theta4, r8)\n\nax.set_rmax(1.5)\nax.set_rticks([0.5, 1, 1.5]) # Less radial ticks\nax.set_rlabel_position(-22.5) # Move radial labels away from plotted line\nax.grid(True)\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"Thus the area element will be $r dr d\\theta$. While it's not precisely rectangular, the increments are very small and it's a reasonable approximation.",
"_____no_output_____"
],
[
"### 3D Cartesian Coordinates\n\nOf course there are many times when we need to express functions like:\n\n$$ z = f(x,y) $$\n\nThese are a standard extension of 2D Cartesian coordinates, and so the volume is simply defined as that of a rectangular solid.\n\n<img src=\"../images/cartesian-volume.png\" width=\"360\" />",
"_____no_output_____"
]
],
[
[
"from sympy import symbols\nfrom sympy.plotting import plot3d\nx, y = symbols('x y')\n\nplot3d(-0.5 * (x**2 + y**2), (x, -3, 3), (y, -3, 3))",
"_____no_output_____"
]
],
[
[
"### 3D Spherical Coordinates\n\nMuch like two dimensions we sometimes need to use spherical coordinates — atoms are spherical, after all.\n\n<div class=\"alert alert-block alert-danger\">\n\n**WARNING** Some math courses use a different [convention](https://en.wikipedia.org/wiki/Spherical_coordinate_system#Conventions) than chemistry and physics.\n \n- Physics and chemistry use $(r, \\theta, \\varphi)$ where $\\theta$ is the angle down from the z-axis (e.g., latitude)\n- Some math courses use $\\theta$ as the angle in the XY 2D plane.\n\n</div>\n\n(Image from [*Mathematical Methods for Chemists*](http://sites.bu.edu/straub/mathematical-methods-for-molecular-science/))\n\n<img src=\"../images/spherical.png\" width=\"330\" />\n\nWhere:\n- $r$ is the radius, from 0 to $\\infty$\n- $\\theta$ is the angle down from the z-axis\n - e.g., think of N/S latitude on the Earth's surface) from 0° at the N pole to 90° (π/2) at the equator and 180° (π) at the S pole\n- $\\varphi$ is the angle in the $xy$ plane\n - e.g., think of E/W longitude on the Earth), from 0 to 360° / 0..2π\n\nWe can interconvert xyz and $r\\theta\\varphi$\n\n$$x = r\\sin \\theta \\cos \\varphi$$\n$$y = r\\sin \\theta \\sin \\varphi$$\n$$z = r \\cos \\theta$$\n\nOr vice-versa:\n\n$$\n\\begin{array}{l}r=\\sqrt{x^{2}+y^{2}+z^{2}} \\\\ \\theta=\\arccos \\left(\\frac{z}{r}\\right)=\\cos ^{-1}\\left(\\frac{z}{r}\\right) \\\\ \\varphi=\\tan ^{-1}\\left(\\frac{y}{x}\\right)\\end{array}\n$$",
"_____no_output_____"
],
[
"The code below might look a little complicated. That's okay. I've added comments for the different sections and each line.\n\nYou don't need to understand all of it - it's intended to plot the function:\n\n$$ r = |\\cos(\\theta^2) | $$",
"_____no_output_____"
]
],
[
[
"# import some matplotlib modules for 3D and color scales\nimport mpl_toolkits.mplot3d.axes3d as axes3d\nimport matplotlib.colors as mcolors\ncmap = plt.get_cmap('jet') # pick a red-to-blue color map\nfig = plt.figure() # create a figure\nax = fig.add_subplot(1,1,1, projection='3d') # set up some axes for a 3D projection\n\n# We now set up the grid for evaluating our function\n# particularly the angle portion of the spherical coordinates\ntheta = np.linspace(0, np.pi, 100)\nphi = np.linspace(0, 2*np.pi, 100)\nTHETA, PHI = np.meshgrid(theta, phi)\n\n# here's the function to plot\nR = np.abs(np.cos(THETA**2))\n\n# now convert R(phi, theta) to x, y, z coordinates to plot\nX = R * np.sin(THETA) * np.cos(PHI)\nY = R * np.sin(THETA) * np.sin(PHI)\nZ = R * np.cos(THETA)\n\n# set up some colors based on the Z range .. from red to blue\nnorm = mcolors.Normalize(vmin=Z.min(), vmax=Z.max())\n# plot the surface\nplot = ax.plot_surface(X, Y, Z, rstride=1, cstride=1, facecolors=cmap(norm(Z)),\n linewidth=0, antialiased=True, alpha=0.4) # no lines, smooth graphics, semi-transparent\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"The volume element in spherical coordinates is a bit tricky, since the distances depend on the radius and angles:\n(Image from [*Mathematical Methods for Chemists*](http://sites.bu.edu/straub/mathematical-methods-for-molecular-science/))\n\n$$ dV = r^2 dr \\sin \\theta d\\theta d\\phi$$\n\n<img src=\"../images/spherical-volume.png\" width=\"414\" />\n",
"_____no_output_____"
],
[
"-------\nThis notebook is from Prof. Geoffrey Hutchison, University of Pittsburgh\nhttps://github.com/ghutchis/chem1000\n\n<a rel=\"license\" href=\"http://creativecommons.org/licenses/by/4.0/\"><img alt=\"Creative Commons License\" style=\"border-width:0\" src=\"https://i.creativecommons.org/l/by/4.0/88x31.png\" /></a>",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
4a55ef970d6e93a85f3aeaa1b7d798243c80a329
| 5,344 |
ipynb
|
Jupyter Notebook
|
student-notebooks/01.03-FAQ.ipynb
|
wrbl96/pyRosetta
|
63dccca0c7d84a04068f0d981cc0b00b138a7b9b
|
[
"MIT"
] | 226 |
2019-08-05T17:36:59.000Z
|
2022-03-27T09:30:21.000Z
|
student-notebooks/01.03-FAQ.ipynb
|
wrbl96/pyRosetta
|
63dccca0c7d84a04068f0d981cc0b00b138a7b9b
|
[
"MIT"
] | 44 |
2019-08-21T15:47:53.000Z
|
2022-03-18T03:45:07.000Z
|
student-notebooks/01.03-FAQ.ipynb
|
wrbl96/pyRosetta
|
63dccca0c7d84a04068f0d981cc0b00b138a7b9b
|
[
"MIT"
] | 86 |
2019-12-23T07:18:27.000Z
|
2022-03-31T08:33:12.000Z
| 39.294118 | 676 | 0.654753 |
[
[
[
"Before you turn this problem in, make sure everything runs as expected. First, **restart the kernel** (in the menubar, select Kernel$\\rightarrow$Restart) and then **run all cells** (in the menubar, select Cell$\\rightarrow$Run All).\n\nMake sure you fill in any place that says `YOUR CODE HERE` or \"YOUR ANSWER HERE\", as well as your name and collaborators below:",
"_____no_output_____"
]
],
[
[
"NAME = \"\"\nCOLLABORATORS = \"\"",
"_____no_output_____"
]
],
[
[
"---",
"_____no_output_____"
],
[
"<!--NOTEBOOK_HEADER-->\n*This notebook contains material from [PyRosetta](https://RosettaCommons.github.io/PyRosetta.notebooks);\ncontent is available [on Github](https://github.com/RosettaCommons/PyRosetta.notebooks.git).*",
"_____no_output_____"
],
[
"<!--NAVIGATION-->\n< [Jupyter Notebooks, Python, and Google Colaboratory](http://nbviewer.jupyter.org/github/RosettaCommons/PyRosetta.notebooks/blob/master/notebooks/01.02-Notebooks-Python-Colab.ipynb) | [Contents](toc.ipynb) | [Index](index.ipynb) | [Introduction to PyRosetta](http://nbviewer.jupyter.org/github/RosettaCommons/PyRosetta.notebooks/blob/master/notebooks/02.00-Introduction-to-PyRosetta.ipynb) ><p><a href=\"https://colab.research.google.com/github/RosettaCommons/PyRosetta.notebooks/blob/master/notebooks/01.03-FAQ.ipynb\"><img align=\"left\" src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open in Colab\" title=\"Open in Google Colaboratory\"></a>",
"_____no_output_____"
],
[
"# Frequently Asked Questions/Troubleshooting Tips",
"_____no_output_____"
],
[
"**Q1: I went through the setup instructions, but importing PyRosetta causes an error.**",
"_____no_output_____"
],
[
"Few things you could try:\n- Make sure that you ran every cell in the `PyRosetta Google Drive Setup` notebook (Chapter 1.01) before importing PyRosetta.\n\n- Make sure that you installed the correct version of PyRosetta. It has to be the Linux version or it won't work. If you did happen to use an incorrect version, just delete the wrong PyRosetta package from your Google Drive, upload the correct one, delete the `prefix` folder, and try running Chapter 1.01 again.\n\n- Make sure that the directory tree is correct. The `PyRosetta` folder should be in the _top directory_ in your Google Drive, as well as the `notebooks` or `student-notebooks` folder. Your notebooks, including Chapter 01.01 should reside in either `notebooks` or `student-notebooks`.",
"_____no_output_____"
],
[
"**Q2: The `make-student-nb.bash` script doesn't work.**\n\nThis script automatically synchronizes the `notebooks` and `student-notebooks` folders. All changes should be made in `notebooks`. The script relies on the `nbgrader` module and the `nbpages` module. Make sure you've installed these before running the script (you might even have to update these modules).\n\nIf you just want to update the Table of Contents or Keywords files, you can just use the `nbpages` command alone.\n\nIf nbgrader is the problem, you might have to run the command: `nbgrader update .`",
"_____no_output_____"
],
[
"<!--NAVIGATION-->\n< [Jupyter Notebooks, Python, and Google Colaboratory](http://nbviewer.jupyter.org/github/RosettaCommons/PyRosetta.notebooks/blob/master/notebooks/01.02-Notebooks-Python-Colab.ipynb) | [Contents](toc.ipynb) | [Index](index.ipynb) | [Introduction to PyRosetta](http://nbviewer.jupyter.org/github/RosettaCommons/PyRosetta.notebooks/blob/master/notebooks/02.00-Introduction-to-PyRosetta.ipynb) ><p><a href=\"https://colab.research.google.com/github/RosettaCommons/PyRosetta.notebooks/blob/master/notebooks/01.03-FAQ.ipynb\"><img align=\"left\" src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open in Colab\" title=\"Open in Google Colaboratory\"></a>",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
4a5612a5c0ed026b35c9c9222e2200f76e70f0c2
| 1,700 |
ipynb
|
Jupyter Notebook
|
day4/day3_tutorial1_microcircuit_modelling/Example_0_Setup.ipynb
|
SophieLafaille/kcni-school-lessons
|
5779fade25d87bb28735a5b5912418f97929f66e
|
[
"MIT"
] | null | null | null |
day4/day3_tutorial1_microcircuit_modelling/Example_0_Setup.ipynb
|
SophieLafaille/kcni-school-lessons
|
5779fade25d87bb28735a5b5912418f97929f66e
|
[
"MIT"
] | null | null | null |
day4/day3_tutorial1_microcircuit_modelling/Example_0_Setup.ipynb
|
SophieLafaille/kcni-school-lessons
|
5779fade25d87bb28735a5b5912418f97929f66e
|
[
"MIT"
] | null | null | null | 22.077922 | 132 | 0.566471 |
[
[
[
"## 1) Install Docker, clone repo\n\n- Follow link:\nhttps://github.com/krembilneuroinformatics/kcni-school-lessons/tree/master/envs\n- Complete 'Step one: Install docker on your computer'\n- Scroll down to \"Running the Jyputer Physiological Modeling Environment\" and complete\n\n",
"_____no_output_____"
],
[
"## 2) Compile mod files\n\n",
"_____no_output_____"
],
[
"Run this part once. Setup should hold for future sessions, but you can delete folder x86_64 and rerun if you encounter errors.",
"_____no_output_____"
]
],
[
[
"# Ensure you're in 'day3_tutorial1_microcircuit_modelling', if not, cd to it. \nimport os\nos.getcwd() =='/home/neuro/kcni-school-data/kcni-school-lessons/day3/day3_tutorial1_microcircuit_modelling'",
"_____no_output_____"
],
[
"%%bash\ncd mod\nnrnivmodl\ncd ..",
"_____no_output_____"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
]
] |
4a561ff44d261359229c8b2ca5a5e89ff2d86872
| 1,974 |
ipynb
|
Jupyter Notebook
|
02_NRPMHandbook/miscellaneous/models/.ipynb_checkpoints/general_methodology-checkpoint.ipynb
|
paulremo/NRPMHandbook
|
125d2947c90b1c99a11a457e0e55e06adc609ea2
|
[
"MIT"
] | null | null | null |
02_NRPMHandbook/miscellaneous/models/.ipynb_checkpoints/general_methodology-checkpoint.ipynb
|
paulremo/NRPMHandbook
|
125d2947c90b1c99a11a457e0e55e06adc609ea2
|
[
"MIT"
] | null | null | null |
02_NRPMHandbook/miscellaneous/models/.ipynb_checkpoints/general_methodology-checkpoint.ipynb
|
paulremo/NRPMHandbook
|
125d2947c90b1c99a11a457e0e55e06adc609ea2
|
[
"MIT"
] | null | null | null | 35.890909 | 490 | 0.662107 |
[
[
[
"# General Methodology\n\n## Reliability prediction for miscellaneous items\n\nFor the four different sections (System, EEE, Mechanical and Miscellaneous), the same principle will be applied with for each section a clear definition of the objective(s) and then a clear explanation of the choices. When relevant, examples will be used to illustrate.\n\nThe level of recommendation/rule or permission will be clearly indicated so that it is easy for the user to adapt its application of the method based on its needs.\n\nThis chapter deals with the miscellaneous items which are represented by all parts/item (or sets of parts) which are not fully electrical parts, nor fully mechanical parts nor fully structural parts. The current reliability models (EEE parts, mechanical parts) are not adapted and are not the preferred way to support reliability predictions for these items. Therefore, other ways to provide reliability estimates are proposed in this chapter and are detailed in the next sections.\n\nThe [following figure](figure4_1) represents examples of some miscellaneous parts/items:\n\n\n\n",
"_____no_output_____"
]
]
] |
[
"markdown"
] |
[
[
"markdown"
]
] |
4a5623506b2a1d151faa00171921c2cdd72ebaa9
| 115,972 |
ipynb
|
Jupyter Notebook
|
python/fluid_sphere_example.ipynb
|
SvenGastauer/scatmod
|
638db9b42b824deca54d8372f78fac154ff2e613
|
[
"MIT"
] | 11 |
2019-09-11T11:20:06.000Z
|
2021-11-19T18:56:39.000Z
|
python/fluid_sphere_example.ipynb
|
SvenGastauer/scatmod
|
638db9b42b824deca54d8372f78fac154ff2e613
|
[
"MIT"
] | null | null | null |
python/fluid_sphere_example.ipynb
|
SvenGastauer/scatmod
|
638db9b42b824deca54d8372f78fac154ff2e613
|
[
"MIT"
] | 3 |
2020-09-04T23:36:39.000Z
|
2021-08-05T02:41:17.000Z
| 475.295082 | 54,432 | 0.939684 |
[
[
[
"# Example for an analytical solution of weakly scattering sphere in Python\n\nIn this example the analytical solution for a weakly scattering sphere is based on Anderson, V. C., \"Sound scattering from a fluid sphere\", \n J. Acoust. Soc. America, 22 (4), pp 426-431, July 1950 is computed in it's original form and the simplified version from Jech et al. (2015).\n\nFor the original Anderson (1950) version, several variable need to be defined to compute the target strength (TS):\n- Define the sphere\n - radius (Radius) in m\n - range (Range) which is the distance from the sound emitting device to the center of the sphere (m)\n - density of the sphere (Rho_b) (kg/$m^3$)\n - sound velocity inside the sphere (c_b) (m/s)\n- Define the surrounding fluid\n - sound velocity in the surrounding fluid (c_w)(m/s)\n - density of the surrounding fluid (kg/$m^3$)\n- Define the plane wave \n - Frequency (f) (Hz)\n - Scattering angle relative to the travelling direction of the incident wave (rad)\n \nExample for a 1 cm sphere with a density and sound velcoity contrast of 1.0025, at a range of 10 m, well outside of the nearfield at a frequency of 200 kHz with an assumed sound velocity of 1500 m/s and a density of 1026 kg/m^3 for the surrounding fluid, measured at 90 degrees (i.e. 1.571 rad):",
"_____no_output_____"
]
],
[
[
"from fluid_sphere import *\nimport time #just to time the execution of the script\n#Define variables\nc_w = 1500 \nf = 200000\nc_b = 1.0025*1500\nRange = 10\nRadius = 0.01\nRho_w = 1026\nRho_b = 1.0025 * Rho_w\nTheta = 1.571\n#get TS\nTS = fluid_sphere(f=f,Radius=Radius, Range=Range,Rho_w=Rho_w,Rho_b=Rho_b,Theta=Theta,c_w=c_w,c_b=c_b)\nprint(\"TS for the sphere is %.2f dB\"%TS)",
"TS for the sphere is -98.53 dB\n"
]
],
[
[
"TS can easily becomputed for a range of frequencies, here from 1 to 300 kHz at 0.5 kHz steps:",
"_____no_output_____"
]
],
[
[
"freqs = np.arange(1,300,0.5)*1000\nstart = time.perf_counter()\nTS = [fluid_sphere(f=x,Radius=Radius, Range=Range,Rho_w=Rho_w,Rho_b=Rho_b,Theta=Theta,c_w=c_w,c_b=c_b) for x in freqs]\nend = time.perf_counter()\ntel_0 = end - start\nprint(\"Evaluating the TS took %.2f seconds\"%tel_0)",
"Evaluating the TS took 0.44 seconds\n"
]
],
[
[
"Plot the results:",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nplt.rcParams.update({'font.size': 16})\n\nfig, ax = plt.subplots(figsize=(12,6))\nax.plot(freqs/1000, TS)\nplt.xlabel(\"Frequency [kHz]\")\nplt.ylabel(\"TS [dB re 1$m^2$]\")\nplt.title(\"TS for a sphere with a=%.1f cm @ range=%.1f m\"%(Radius*100,Range))\nplt.show()",
"_____no_output_____"
]
],
[
[
"Similarly for the simplified version, the sound velocity in the external fluid (c, m/s), the frequency (f, Hz), the density and sound velocity contrasts (g and h), distance to the sphere (r, m), the radius of the sphere (a,m) and the density of the surrounding fluid (rho, kg m^-3) need to be defined.",
"_____no_output_____"
]
],
[
[
"f = 200000\nr = 10\na = 0.01\nc = 1500\nh = 1.0025\ng = 1.0025\nrho = 1026\nTS = fluid_sphere_simple(f,r,a,c,h,g,rho)\nprint(\"TS for the sphere is %.2f dB\"%TS)\n\nstart = time.perf_counter()\nTS = [fluid_sphere_simple(f=x,r=r,a=a,c=c,h=h,g=g,rho=rho) for x in freqs]\nend = time.perf_counter()\ntel_1 = end - start\nprint(\"Evaluating the TS took %.2f seconds\"%tel_1)",
"TS for the sphere is -98.53 dB\nEvaluating the TS took 0.44 seconds\n"
],
[
"plt.rcParams.update({'font.size': 16})\n\nfig, ax = plt.subplots(figsize=(12,6))\nax.plot(freqs/1000, TS)\nplt.xlabel(\"Frequency [kHz]\")\nplt.ylabel(\"TS [dB re 1$m^2$]\")\nplt.title(\"TS for a sphere with a=%.1f cm @ range=%.1f m\"%(Radius*100,Range))\nplt.show()",
"_____no_output_____"
],
[
"if tel_1 < tel_0:\n print(\"The simplified method (%.2f s) evaluated the TS %.2f s faster then \"\n \"the original Anderson (1950) method (%.2f s)\"%(tel_1,np.abs(tel_1-tel_0),tel_0))\nelse:\n print(\"The simplified method (%.2f s) evaluated the TS %.2f s slower then \"\n \"the original Anderson (1950) method (%.2f s)\"%(tel_1,np.abs(tel_1-tel_0),tel_0))",
"The simplified method (0.36 s) evaluated the TS 0.07 s slower then the original Anderson (1950) method (0.44 s)\n"
]
],
[
[
"Both methods should deliver the same results for backscatter at a 90 degrees angle, but the original Anderson (1950) is more flexible in terms of scattering angle and could easily be extended to include shear and surface pressure or be validated for a solid sphere.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
]
] |
4a56425b13fe12dd975a0f1cb2ba82facc4cc407
| 30,920 |
ipynb
|
Jupyter Notebook
|
Code/Classification1.ipynb
|
TheGadgeteer/Predicting-from-Sound-Blog
|
a7b0230c9fd3bdcd900c6b4aa160378affef1025
|
[
"Apache-2.0"
] | null | null | null |
Code/Classification1.ipynb
|
TheGadgeteer/Predicting-from-Sound-Blog
|
a7b0230c9fd3bdcd900c6b4aa160378affef1025
|
[
"Apache-2.0"
] | null | null | null |
Code/Classification1.ipynb
|
TheGadgeteer/Predicting-from-Sound-Blog
|
a7b0230c9fd3bdcd900c6b4aa160378affef1025
|
[
"Apache-2.0"
] | null | null | null | 41.171771 | 163 | 0.514618 |
[
[
[
"import os\nimport wave\nimport contextlib\nfrom pathlib import Path\nfrom google.colab import files\nfrom os import listdir\nimport sys\n\nfrom keras.preprocessing.image import load_img\nfrom keras.preprocessing.image import img_to_array\nfrom keras import backend\nfrom numpy import zeros\nfrom sklearn.metrics import fbeta_score\nfrom numpy import ones\nfrom numpy import asarray\nfrom numpy import savez_compressed\nimport pandas as pd\nfrom keras.optimizers import SGD\nimport matplotlib.pyplot as plt\nimport librosa\nimport librosa.display\nfrom scipy.io import wavfile as wav\nimport numpy as np\nfrom keras.models import Sequential\nfrom keras.layers import Dense, Dropout, Activation\nfrom keras.optimizers import Adam\nimport numpy as np\nfrom keras.layers import Input, Conv2D, BatchNormalization, MaxPooling2D, Reshape, Dense, LSTM, add, concatenate, Dropout, Lambda, Flatten, GRU, LeakyReLU\nfrom keras.models import Model\nimport keras.backend as K\nfrom keras.callbacks import ModelCheckpoint \nfrom datetime import datetime \nfrom keras import regularizers, optimizers\nfrom sklearn.model_selection import train_test_split \nfrom sklearn.preprocessing import MultiLabelBinarizer\nfrom tensorflow.keras.utils import to_categorical\nfrom matplotlib import pyplot\nfrom pandas import read_csv",
"_____no_output_____"
],
[
"sampling_rate=16000\naudio_duration=2\nn_classes=3022\nn_folds_=10\nlearning_rate=0.0001\nmax_epochs=50\nn_mfcc=20\naudio_length = sampling_rate * audio_duration\ndim = (audio_length, 1)\nbatch_size=64",
"_____no_output_____"
],
[
"#best case, the dataset is in drive/MyDrive/datasets\nfrom google.colab import drive\ndrive.mount('/content/drive')",
"_____no_output_____"
],
[
"def audio_norm(data):\n max_data = np.max(data)\n min_data = np.min(data)\n data = (data-min_data)/(max_data-min_data+1e-6)\n return data-0.5",
"_____no_output_____"
],
[
"#filter files that actually have sound by their size (not the best)\nactualFiles = []\ncounter = 0\ndirectory = r'drive/MyDrive/DISTANCE/'\nfor filename in os.listdir(directory):\n if(filename.endswith(\".wav\")):\n size = Path(directory + filename).stat().st_size\n if(size >= 59):\n actualFiles.append(directory + filename)\n counter += 1",
"_____no_output_____"
],
[
"train = pd.read_csv(\"drive/MyDrive/metadata.csv\")\nX = pd.read_csv(\"drive/MyDrive/metadata.csv\", usecols = [\"fileName\", \"distance\"])\ny = pd.read_csv(\"drive/MyDrive/metadata.csv\", usecols = [\"label\"])\n\n#test = pd.read_csv(\"drive/MyDrive/datasets/FSDKaggle2018.meta/test_post_competition_scoring_clips.csv\")\ntrain.head()",
"_____no_output_____"
],
[
"#ONLY RUN THIS ONCE, or do the second cell again\nLABELS = list(train.label.unique())\n#LABELS\nlabel_idx = {label: i for i, label in enumerate(LABELS)}\n#label_idx\n#interestingly enough, Hi-hat is 0 for some reason\ntrain.set_index(\"fileName\", inplace=True)\n#set the index row for train to the name of the file (you can create a new row instead)\n#train.head()\n#same for test\ntrain[\"label_idx\"] = train.label.apply(lambda x: label_idx[x])\n#create row where every element has the amount of times that label is used",
"_____no_output_____"
],
[
"def getModel():\n input_length = audio_length\n\n inp = Input(shape=(dim[0], dim[1],1))\n x = Conv2D(32, (3,3), padding=\"same\")(inp)\n x = BatchNormalization()(x)\n x = Activation(\"relu\")(x)\n x = MaxPooling2D()(x)\n \n x = Conv2D(32, (3,3), padding=\"same\")(x)\n x = BatchNormalization()(x)\n x = Activation(\"relu\")(x)\n x = MaxPooling2D()(x)\n \n x = Conv2D(32, (3,3), padding=\"same\")(x)\n x = BatchNormalization()(x)\n x = Activation(\"relu\")(x)\n x = MaxPooling2D()(x)\n \n x = Conv2D(32, (3,3), padding=\"same\")(x)\n x = BatchNormalization()(x)\n x = Activation(\"relu\")(x)\n x = MaxPooling2D()(x)\n\n x = Flatten()(x)\n x = Dense(64)(x)\n x = BatchNormalization()(x)\n x = Activation(\"relu\")(x)\n out = Dense(n_classes, activation='softmax')(x)\n\n model = Model(inputs=inp, outputs=out)\n opt = optimizers.Adam(learning_rate)\n\n model.compile(optimizer=opt, loss=\"categorical_crossentropy\", metrics=['acc'])\n return model",
"_____no_output_____"
],
[
"dim = (n_mfcc, 1+(int(np.floor(audio_length/512))), 1)",
"_____no_output_____"
],
[
"def prepare_data(df, data_dir):\n X = np.empty(shape=(df.shape[0], dim[0], dim[1], 1))\n input_length = audio_length\n #create librosa file\n for i, fname in enumerate(data_dir):\n file_path = fname\n data, _ = librosa.core.load(file_path, sr=sampling_rate, res_type=\"kaiser_fast\")\n\n\n # Random offset / Padding\n if len(data) > input_length:\n max_offset = len(data) - input_length\n offset = np.random.randint(max_offset)\n data = data[offset:(input_length+offset)]\n else:\n if input_length > len(data):\n max_offset = input_length - len(data)\n offset = np.random.randint(max_offset)\n else:\n offset = 0\n data = np.pad(data, (offset, input_length - len(data) - offset), \"constant\")\n #extract mfcc features\n data = librosa.feature.mfcc(data, sr=sampling_rate, n_mfcc=n_mfcc)\n data = np.expand_dims(data, axis=-1)\n #save them and do it for each file, return X\n X[i,] = data\n return X",
"_____no_output_____"
],
[
"print(train.head())\nprint(actualFiles[0])\n#this takes a lot of time\nX_train = prepare_data(train, actualFiles)\n#'drive/MyDrive/datasets/FSDKaggle2018.audio_train/'",
" distance ... label_idx\nfileName ... \n10000.wav 2 ... 0\n100005.wav 2 ... 1\n10000.wav 2 ... 0\n100005.wav 2 ... 1\n100007.wav 2 ... 1\n\n[5 rows x 3 columns]\ndrive/MyDrive/DISTANCE/119312.wav\n"
],
[
"y_train = to_categorical(train.label_idx, num_classes=3022)",
"_____no_output_____"
],
[
"mean = np.mean(X_train, axis=0)\nstd = np.std(X_train, axis=0)\n\n#some kind of normalization\nX_train = (X_train - mean)/std",
"_____no_output_____"
],
[
"print(X_train.shape)\nprint(y_train.shape)",
"(3022, 20, 63, 1)\n(3022, 3022)\n"
],
[
"PREDICTION_FOLDER = \"predictions_1d_conv\"\nif not os.path.exists(PREDICTION_FOLDER):\n os.mkdir(PREDICTION_FOLDER)",
"_____no_output_____"
],
[
"from sklearn.model_selection import KFold\nfrom keras.callbacks import (EarlyStopping, LearningRateScheduler,\n ModelCheckpoint, TensorBoard, ReduceLROnPlateau)\n#kfold this time for simplicity\n#the idea is the same as the previous one\nkf = KFold(n_splits=2)\nfor i, (train_index, test_index) in enumerate(kf.split(X_train)):\n X_t, X_te = X_train[train_index], X_train[test_index]\n y_t, y_te = y_train[train_index], y_train[test_index]\n print(\"#\"*50)\n print(\"Fold: \", i)\n model = getModel()\n checkpoint = ModelCheckpoint('best_%d.h5'%i, monitor='val_loss', verbose=1, save_best_only=True)\n early = EarlyStopping(monitor=\"val_loss\", mode=\"min\", patience=5)\n tb = TensorBoard(log_dir='./logs/' + PREDICTION_FOLDER + '/fold_%i'%i, write_graph=True)\n callbacks_list = [checkpoint, early, tb]\n history = model.fit(X_t, y_t, validation_data=(X_te, y_te), callbacks=callbacks_list, \n batch_size=64, epochs=30)\n model.load_weights('best_%d.h5'%i)\n predictions = model.predict(X_train, batch_size=64, verbose=1)\n np.save(PREDICTION_FOLDER + \"/train_predictions_%d.npy\"%i, predictions)",
"##################################################\nFold: 0\nEpoch 1/30\n24/24 [==============================] - 13s 461ms/step - loss: 7.9959 - acc: 1.6954e-04 - val_loss: 8.0074 - val_acc: 0.0000e+00\n\nEpoch 00001: val_loss improved from inf to 8.00735, saving model to best_0.h5\nEpoch 2/30\n24/24 [==============================] - 10s 429ms/step - loss: 7.8510 - acc: 0.0324 - val_loss: 7.9845 - val_acc: 0.0079\n\nEpoch 00002: val_loss improved from 8.00735 to 7.98454, saving model to best_0.h5\nEpoch 3/30\n24/24 [==============================] - 10s 432ms/step - loss: 7.7177 - acc: 0.1640 - val_loss: 7.9518 - val_acc: 0.0126\n\nEpoch 00003: val_loss improved from 7.98454 to 7.95180, saving model to best_0.h5\nEpoch 4/30\n24/24 [==============================] - 10s 430ms/step - loss: 7.5837 - acc: 0.1825 - val_loss: 7.9163 - val_acc: 0.0126\n\nEpoch 00004: val_loss improved from 7.95180 to 7.91635, saving model to best_0.h5\nEpoch 5/30\n24/24 [==============================] - 10s 430ms/step - loss: 7.4516 - acc: 0.2079 - val_loss: 7.8780 - val_acc: 0.0126\n\nEpoch 00005: val_loss improved from 7.91635 to 7.87805, saving model to best_0.h5\nEpoch 6/30\n24/24 [==============================] - 9s 390ms/step - loss: 7.3218 - acc: 0.2264 - val_loss: 7.8407 - val_acc: 0.0126\n\nEpoch 00006: val_loss improved from 7.87805 to 7.84069, saving model to best_0.h5\nEpoch 7/30\n24/24 [==============================] - 9s 391ms/step - loss: 7.1791 - acc: 0.2480 - val_loss: 7.7979 - val_acc: 0.0079\n\nEpoch 00007: val_loss improved from 7.84069 to 7.79793, saving model to best_0.h5\nEpoch 8/30\n24/24 [==============================] - 10s 432ms/step - loss: 7.0512 - acc: 0.2460 - val_loss: 7.7592 - val_acc: 0.0060\n\nEpoch 00008: val_loss improved from 7.79793 to 7.75917, saving model to best_0.h5\nEpoch 9/30\n24/24 [==============================] - 10s 431ms/step - loss: 6.9099 - acc: 0.2603 - val_loss: 7.7179 - val_acc: 0.0060\n\nEpoch 00009: val_loss improved from 7.75917 to 7.71791, saving model to best_0.h5\nEpoch 10/30\n24/24 [==============================] - 10s 433ms/step - loss: 6.7843 - acc: 0.2558 - val_loss: 7.6748 - val_acc: 0.0040\n\nEpoch 00010: val_loss improved from 7.71791 to 7.67479, saving model to best_0.h5\nEpoch 11/30\n24/24 [==============================] - 10s 432ms/step - loss: 6.6483 - acc: 0.2503 - val_loss: 7.6320 - val_acc: 0.0040\n\nEpoch 00011: val_loss improved from 7.67479 to 7.63203, saving model to best_0.h5\nEpoch 12/30\n24/24 [==============================] - 10s 432ms/step - loss: 6.4951 - acc: 0.2889 - val_loss: 7.5441 - val_acc: 0.0020\n\nEpoch 00012: val_loss improved from 7.63203 to 7.54406, saving model to best_0.h5\nEpoch 13/30\n24/24 [==============================] - 9s 390ms/step - loss: 6.3781 - acc: 0.2624 - val_loss: 7.5460 - val_acc: 0.0020\n\nEpoch 00013: val_loss did not improve from 7.54406\nEpoch 14/30\n24/24 [==============================] - 10s 428ms/step - loss: 6.2259 - acc: 0.2703 - val_loss: 7.4917 - val_acc: 0.0020\n\nEpoch 00014: val_loss improved from 7.54406 to 7.49169, saving model to best_0.h5\nEpoch 15/30\n24/24 [==============================] - 9s 387ms/step - loss: 6.0918 - acc: 0.2711 - val_loss: 7.4407 - val_acc: 0.0020\n\nEpoch 00015: val_loss improved from 7.49169 to 7.44067, saving model to best_0.h5\nEpoch 16/30\n24/24 [==============================] - 9s 386ms/step - loss: 5.9610 - acc: 0.2650 - val_loss: 7.3870 - val_acc: 0.0020\n\nEpoch 00016: val_loss improved from 7.44067 to 7.38697, saving model to best_0.h5\nEpoch 17/30\n24/24 [==============================] - 10s 430ms/step - loss: 5.8182 - acc: 0.2774 - val_loss: 7.3560 - val_acc: 0.0013\n\nEpoch 00017: val_loss improved from 7.38697 to 7.35595, saving model to best_0.h5\nEpoch 18/30\n24/24 [==============================] - 10s 438ms/step - loss: 5.6945 - acc: 0.2774 - val_loss: 7.3166 - val_acc: 0.0026\n\nEpoch 00018: val_loss improved from 7.35595 to 7.31664, saving model to best_0.h5\nEpoch 19/30\n24/24 [==============================] - 10s 430ms/step - loss: 5.5448 - acc: 0.2825 - val_loss: 7.1802 - val_acc: 0.0020\n\nEpoch 00019: val_loss improved from 7.31664 to 7.18016, saving model to best_0.h5\nEpoch 20/30\n24/24 [==============================] - 10s 432ms/step - loss: 5.4289 - acc: 0.2823 - val_loss: 7.1589 - val_acc: 0.0020\n\nEpoch 00020: val_loss improved from 7.18016 to 7.15885, saving model to best_0.h5\nEpoch 21/30\n24/24 [==============================] - 10s 433ms/step - loss: 5.2513 - acc: 0.3012 - val_loss: 7.0984 - val_acc: 0.0020\n\nEpoch 00021: val_loss improved from 7.15885 to 7.09839, saving model to best_0.h5\nEpoch 22/30\n24/24 [==============================] - 10s 431ms/step - loss: 5.1607 - acc: 0.2881 - val_loss: 7.0323 - val_acc: 0.0013\n\nEpoch 00022: val_loss improved from 7.09839 to 7.03229, saving model to best_0.h5\nEpoch 23/30\n24/24 [==============================] - 10s 432ms/step - loss: 5.0444 - acc: 0.2812 - val_loss: 7.0421 - val_acc: 0.0013\n\nEpoch 00023: val_loss did not improve from 7.03229\nEpoch 24/30\n24/24 [==============================] - 10s 431ms/step - loss: 4.9444 - acc: 0.2730 - val_loss: 6.9312 - val_acc: 0.0020\n\nEpoch 00024: val_loss improved from 7.03229 to 6.93122, saving model to best_0.h5\nEpoch 25/30\n24/24 [==============================] - 9s 390ms/step - loss: 4.8289 - acc: 0.2786 - val_loss: 6.9296 - val_acc: 0.0020\n\nEpoch 00025: val_loss improved from 6.93122 to 6.92955, saving model to best_0.h5\nEpoch 26/30\n24/24 [==============================] - 10s 431ms/step - loss: 4.6844 - acc: 0.2841 - val_loss: 6.9126 - val_acc: 0.0013\n\nEpoch 00026: val_loss improved from 6.92955 to 6.91260, saving model to best_0.h5\nEpoch 27/30\n24/24 [==============================] - 10s 433ms/step - loss: 4.5869 - acc: 0.2946 - val_loss: 6.8428 - val_acc: 0.0020\n\nEpoch 00027: val_loss improved from 6.91260 to 6.84275, saving model to best_0.h5\nEpoch 28/30\n24/24 [==============================] - 10s 433ms/step - loss: 4.4629 - acc: 0.3001 - val_loss: 6.8403 - val_acc: 0.0013\n\nEpoch 00028: val_loss improved from 6.84275 to 6.84034, saving model to best_0.h5\nEpoch 29/30\n24/24 [==============================] - 10s 433ms/step - loss: 4.3865 - acc: 0.2954 - val_loss: 6.8103 - val_acc: 0.0013\n\nEpoch 00029: val_loss improved from 6.84034 to 6.81029, saving model to best_0.h5\nEpoch 30/30\n24/24 [==============================] - 10s 432ms/step - loss: 4.3106 - acc: 0.3130 - val_loss: 6.7661 - val_acc: 0.0013\n\nEpoch 00030: val_loss improved from 6.81029 to 6.76606, saving model to best_0.h5\n48/48 [==============================] - 4s 71ms/step\n##################################################\nFold: 1\nEpoch 1/30\n24/24 [==============================] - 13s 466ms/step - loss: 8.0055 - acc: 2.3716e-04 - val_loss: 8.0078 - val_acc: 0.0073\n\nEpoch 00001: val_loss improved from inf to 8.00781, saving model to best_1.h5\nEpoch 2/30\n24/24 [==============================] - 10s 432ms/step - loss: 7.8786 - acc: 0.0062 - val_loss: 7.9796 - val_acc: 0.0073\n\nEpoch 00002: val_loss improved from 8.00781 to 7.97960, saving model to best_1.h5\nEpoch 3/30\n24/24 [==============================] - 9s 390ms/step - loss: 7.7671 - acc: 0.0352 - val_loss: 7.9401 - val_acc: 0.0073\n\nEpoch 00003: val_loss improved from 7.97960 to 7.94006, saving model to best_1.h5\nEpoch 4/30\n24/24 [==============================] - 10s 433ms/step - loss: 7.6576 - acc: 0.0758 - val_loss: 7.8863 - val_acc: 0.0073\n\nEpoch 00004: val_loss improved from 7.94006 to 7.88629, saving model to best_1.h5\nEpoch 5/30\n24/24 [==============================] - 9s 392ms/step - loss: 7.5441 - acc: 0.0933 - val_loss: 7.8310 - val_acc: 0.0066\n\nEpoch 00005: val_loss improved from 7.88629 to 7.83100, saving model to best_1.h5\nEpoch 6/30\n24/24 [==============================] - 10s 433ms/step - loss: 7.4249 - acc: 0.1440 - val_loss: 7.7642 - val_acc: 0.0079\n\nEpoch 00006: val_loss improved from 7.83100 to 7.76416, saving model to best_1.h5\nEpoch 7/30\n24/24 [==============================] - 10s 431ms/step - loss: 7.3125 - acc: 0.1658 - val_loss: 7.7030 - val_acc: 0.0073\n\nEpoch 00007: val_loss improved from 7.76416 to 7.70301, saving model to best_1.h5\nEpoch 8/30\n24/24 [==============================] - 9s 391ms/step - loss: 7.2017 - acc: 0.1859 - val_loss: 7.6371 - val_acc: 0.0086\n\nEpoch 00008: val_loss improved from 7.70301 to 7.63711, saving model to best_1.h5\nEpoch 9/30\n24/24 [==============================] - 10s 433ms/step - loss: 7.0924 - acc: 0.1921 - val_loss: 7.5748 - val_acc: 0.0033\n\nEpoch 00009: val_loss improved from 7.63711 to 7.57479, saving model to best_1.h5\nEpoch 10/30\n24/24 [==============================] - 10s 432ms/step - loss: 6.9652 - acc: 0.2224 - val_loss: 7.5301 - val_acc: 0.0033\n\nEpoch 00010: val_loss improved from 7.57479 to 7.53012, saving model to best_1.h5\nEpoch 11/30\n24/24 [==============================] - 10s 434ms/step - loss: 6.8438 - acc: 0.2429 - val_loss: 7.4810 - val_acc: 0.0040\n\nEpoch 00011: val_loss improved from 7.53012 to 7.48096, saving model to best_1.h5\nEpoch 12/30\n24/24 [==============================] - 9s 393ms/step - loss: 6.7345 - acc: 0.2367 - val_loss: 7.4347 - val_acc: 0.0033\n\nEpoch 00012: val_loss improved from 7.48096 to 7.43473, saving model to best_1.h5\nEpoch 13/30\n24/24 [==============================] - 10s 435ms/step - loss: 6.6132 - acc: 0.2604 - val_loss: 7.3774 - val_acc: 0.0026\n\nEpoch 00013: val_loss improved from 7.43473 to 7.37743, saving model to best_1.h5\nEpoch 14/30\n24/24 [==============================] - 10s 432ms/step - loss: 6.4848 - acc: 0.2666 - val_loss: 7.3118 - val_acc: 0.0020\n\nEpoch 00014: val_loss improved from 7.37743 to 7.31176, saving model to best_1.h5\nEpoch 15/30\n24/24 [==============================] - 9s 389ms/step - loss: 6.3694 - acc: 0.2732 - val_loss: 7.2560 - val_acc: 0.0026\n\nEpoch 00015: val_loss improved from 7.31176 to 7.25596, saving model to best_1.h5\nEpoch 16/30\n24/24 [==============================] - 9s 389ms/step - loss: 6.2440 - acc: 0.2733 - val_loss: 7.2403 - val_acc: 0.0053\n\nEpoch 00016: val_loss improved from 7.25596 to 7.24030, saving model to best_1.h5\nEpoch 17/30\n24/24 [==============================] - 9s 392ms/step - loss: 6.1323 - acc: 0.2764 - val_loss: 7.1575 - val_acc: 0.0053\n\nEpoch 00017: val_loss improved from 7.24030 to 7.15748, saving model to best_1.h5\nEpoch 18/30\n24/24 [==============================] - 9s 392ms/step - loss: 6.0130 - acc: 0.2821 - val_loss: 7.1310 - val_acc: 0.0053\n\nEpoch 00018: val_loss improved from 7.15748 to 7.13102, saving model to best_1.h5\nEpoch 19/30\n24/24 [==============================] - 10s 432ms/step - loss: 5.8918 - acc: 0.2936 - val_loss: 7.1074 - val_acc: 0.0013\n\nEpoch 00019: val_loss improved from 7.13102 to 7.10743, saving model to best_1.h5\nEpoch 20/30\n24/24 [==============================] - 10s 437ms/step - loss: 5.7773 - acc: 0.2816 - val_loss: 7.0649 - val_acc: 0.0046\n\nEpoch 00020: val_loss improved from 7.10743 to 7.06494, saving model to best_1.h5\nEpoch 21/30\n24/24 [==============================] - 10s 436ms/step - loss: 5.6651 - acc: 0.2728 - val_loss: 7.0027 - val_acc: 0.0046\n\nEpoch 00021: val_loss improved from 7.06494 to 7.00272, saving model to best_1.h5\nEpoch 22/30\n24/24 [==============================] - 10s 437ms/step - loss: 5.5534 - acc: 0.2717 - val_loss: 6.9465 - val_acc: 0.0053\n\nEpoch 00022: val_loss improved from 7.00272 to 6.94647, saving model to best_1.h5\nEpoch 23/30\n24/24 [==============================] - 10s 438ms/step - loss: 5.4518 - acc: 0.2737 - val_loss: 6.9152 - val_acc: 0.0046\n\nEpoch 00023: val_loss improved from 6.94647 to 6.91515, saving model to best_1.h5\nEpoch 24/30\n24/24 [==============================] - 10s 435ms/step - loss: 5.3436 - acc: 0.2838 - val_loss: 6.8856 - val_acc: 0.0060\n\nEpoch 00024: val_loss improved from 6.91515 to 6.88558, saving model to best_1.h5\nEpoch 25/30\n24/24 [==============================] - 10s 431ms/step - loss: 5.2224 - acc: 0.2990 - val_loss: 6.8097 - val_acc: 0.0040\n\nEpoch 00025: val_loss improved from 6.88558 to 6.80974, saving model to best_1.h5\nEpoch 26/30\n24/24 [==============================] - 10s 432ms/step - loss: 5.1478 - acc: 0.2634 - val_loss: 6.7992 - val_acc: 0.0040\n\nEpoch 00026: val_loss improved from 6.80974 to 6.79920, saving model to best_1.h5\nEpoch 27/30\n24/24 [==============================] - 10s 431ms/step - loss: 5.0379 - acc: 0.2852 - val_loss: 6.7847 - val_acc: 0.0046\n\nEpoch 00027: val_loss improved from 6.79920 to 6.78471, saving model to best_1.h5\nEpoch 28/30\n24/24 [==============================] - 10s 430ms/step - loss: 4.9284 - acc: 0.2737 - val_loss: 6.7453 - val_acc: 0.0040\n\nEpoch 00028: val_loss improved from 6.78471 to 6.74529, saving model to best_1.h5\nEpoch 29/30\n24/24 [==============================] - 10s 431ms/step - loss: 4.8542 - acc: 0.2773 - val_loss: 6.7046 - val_acc: 0.0053\n\nEpoch 00029: val_loss improved from 6.74529 to 6.70456, saving model to best_1.h5\nEpoch 30/30\n24/24 [==============================] - 9s 387ms/step - loss: 4.7437 - acc: 0.2828 - val_loss: 6.6670 - val_acc: 0.0046\n\nEpoch 00030: val_loss improved from 6.70456 to 6.66701, saving model to best_1.h5\n48/48 [==============================] - 4s 71ms/step\n"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a5642c0044c01af4f6de5f8b8fc056606aca5f7
| 12,305 |
ipynb
|
Jupyter Notebook
|
Level3_Patterns/Level3_Patterns.ipynb
|
amanchamola/level3_patterns
|
b3e531164ed46f27368ef9b10a906f66000fcf5e
|
[
"MIT"
] | null | null | null |
Level3_Patterns/Level3_Patterns.ipynb
|
amanchamola/level3_patterns
|
b3e531164ed46f27368ef9b10a906f66000fcf5e
|
[
"MIT"
] | null | null | null |
Level3_Patterns/Level3_Patterns.ipynb
|
amanchamola/level3_patterns
|
b3e531164ed46f27368ef9b10a906f66000fcf5e
|
[
"MIT"
] | null | null | null | 19.107143 | 45 | 0.33336 |
[
[
[
"# Patterns 2",
"_____no_output_____"
]
],
[
[
"n=int(input())\ni=1\nwhile n>=i:\n #spaces\n spaces = 1\n while n-i>=spaces:\n print(' ',end='')\n spaces+=1\n\n\n #increasing\n stars = 1\n while i>=stars:\n print('*',end='')\n stars+=1\n\n\n #decreasing\n p=i-1\n while p>=1:\n \n print('*',end='')\n p=p-1\n \n print()\n i+=1",
"5\n *\n ***\n *****\n *******\n*********\n"
],
[
"n=int(input())\ni=1\nwhile n>=i:\n #spaces\n spaces = 1\n while n-i>=spaces:\n print(' ',end='')\n spaces+=1\n\n\n #increasing\n stars = 1\n while i>=stars:\n print(i+stars-1,end='')\n stars+=1\n\n\n #decreasing\n p=i-1\n while p>=1:\n \n print(p+stars-2,end='')\n p=p-1\n \n print()\n i+=1\n",
"5\n 1\n 232\n 34543\n 4567654\n567898765\n"
],
[
"n=int(input())\ni=1\nn1 = (n+1)/2\nj=1\nn2=n1-1\n\n\nwhile n1>=i:\n \n spaces = 1\n while n1-i>=spaces:\n \n print(' ',end='')\n spaces+=1\n\n stars = 1\n while stars<=2*i-1:\n print('*',end='')\n stars+=1\n\n print() \n i+=1\n\n\nwhile n2>=j:\n spaces2 = 1\n while spaces2<=j:\n print(' ',end='')\n spaces2+=1\n\n\n stars2 = 1\n while n-2*j>=stars2:\n print('*',end='')\n stars2+=1\n\n print()\n j+=1\n",
"5\n *\n ***\n*****\n ***\n *\n"
],
[
"n=int(input())\ni=1\nwhile n>=i:\n j=1\n while i>=j:\n print(j,end='')\n j+=1\n\n #SPACES\n k=1 \n while (2*n)-(2*i)>=k:\n print(' ',end='')\n k+=1\n\n #Numbers again\n t=1\n while i>=t:\n print(i-t+1,end='')\n t+=1\n\n print()\n i+=1\n",
"7\n1 1\n12 21\n123 321\n1234 4321\n12345 54321\n123456 654321\n12345677654321\n"
],
[
"lines=int(input()) \ni=1 \nj=1 \nwhile i<=lines: \n j=1 \n while j<=lines: \n if i==j: \n print(\"*\", end='') \n else : \n print(\"0\", end='') \n j=j+1 \n j=j-1\n print(\"*\", end='') \n while j>=1: \n if i==j: \n print(\"*\", end='') \n else : \n print(\"0\", end='') \n j=j-1 \n print() \n i=i+1 \n",
"5\n*0000*0000*\n0*000*000*0\n00*00*00*00\n000*0*0*000\n0000***0000\n"
],
[
"n=int(input())\ni=1\nwhile n>=i:\n #spaces\n spaces = 1\n while n-i>=spaces:\n print(' ',end='')\n spaces+=1\n\n\n #increasing\n stars = 1\n while i>=stars:\n print(i-stars+1,end='')\n stars+=1\n\n\n #decreasing\n p=1\n while i-1>=p:\n \n print(p+1,end='')\n p=p+1\n \n print()\n i+=1\n",
"5\n 1\n 212\n 32123\n 4321234\n543212345\n"
],
[
"n=int(input())\ni=1\nn1 = (n+1)/2\nj=1\nn2=n1-1\n\nwhile n1>=i:\n \n spaces = 1\n while i-1>=spaces:\n \n print(' ',end='')\n spaces+=1\n\n stars = 1\n while stars<=i:\n print('* ',end='')\n stars+=1\n\n print() \n i+=1\n\n\n\nwhile n2>=j:\n spaces2 = 1\n while spaces2<=n2-j:\n print(' ',end='')\n spaces2+=1\n\n\n stars2 = 1\n while n1-j>=stars2:\n print('* ',end='')\n stars2+=1\n\n print()\n j+=1\n",
"11\n* \n * * \n * * * \n * * * * \n * * * * * \n * * * * * * \n * * * * * \n * * * * \n * * * \n * * \n* \n"
],
[
"n=int(input())\nfor i in range (1,n+1):\n for j in range(n,i-1,-1):\n if i%2==0:\n print('0',end='')\n\n else:\n print('1',end='')\n\n print()\n i+=1\n",
"6\n111111\n00000\n1111\n000\n11\n0\n"
],
[
"n=int(input())\nn1=n\nn2=n1-1\nfor i in range (1,n1+1):\n for s in range(1,i):\n print(' ',end='')\n\n for j in range (1,n1-i+2):\n print(i+j-1,end='')\n\n print() \n\nfor t in range (1,n2+1):\n for spaces in range (1,n2-t+1):\n print (' ',end='')\n\n for k in range (1,t+2):\n print(n-t+k-1,end='')\n \n print() \n",
"6\n123456\n 23456\n 3456\n 456\n 56\n 6\n 56\n 456\n 3456\n 23456\n123456\n"
],
[
"n=int(input())\n\nfor i in range (n,0,-1):\n for j in range (n,i,-1):\n print(j,end=\"\")\n for k in range (1,i*2,1):\n print(i,end=\"\")\n for l in range (i+1,n+1,1):\n print(l,end=\"\")\n print()\n\n \nfor m in range (1,n,1):\n for o in range (n,m,-1):\n print(o,end=\"\")\n for p in range (1,m*2,1):\n print(m+1,end=\"\")\n for q in range (m+1,n+1,1):\n print(q,end=\"\")\n print()\n",
"4\n4444444\n4333334\n4322234\n4321234\n4322234\n4333334\n4444444\n"
],
[
"n=int(input())\no=n\nimport math\nn1=math.ceil(n/2)\nn2=n-n1\ni=1\nt=1\nwhile n1>=i:\n j=1\n while n>=j:\n print(t,' ',end='')\n t=t+1\n j+=1\n print() \n t+=n \n i+=1\n\nwhile n2>=1:\n if n%2!=0:\n k=n*(o-2)\n else:\n k=n*(o-1)\n \n p=1\n while n>=p:\n print (k+p,' ',end='')\n p+=1\n\n o=o-2\n n2=n2-1\n print()\n",
"5\n1 2 3 4 5 \n11 12 13 14 15 \n21 22 23 24 25 \n16 17 18 19 20 \n6 7 8 9 10 \n"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a5642f0d4267a8722efce9d7af2a11df90f29ed
| 145,423 |
ipynb
|
Jupyter Notebook
|
code/anomaly-analysis-buoy-44014.ipynb
|
jxfischer/buoy-data-analysis
|
1a41431eb15f9547fb9040db537313f73212fd6e
|
[
"Apache-2.0"
] | 1 |
2019-05-12T18:26:48.000Z
|
2019-05-12T18:26:48.000Z
|
code/anomaly-analysis-buoy-44014.ipynb
|
jxfischer/buoy-data-analysis
|
1a41431eb15f9547fb9040db537313f73212fd6e
|
[
"Apache-2.0"
] | null | null | null |
code/anomaly-analysis-buoy-44014.ipynb
|
jxfischer/buoy-data-analysis
|
1a41431eb15f9547fb9040db537313f73212fd6e
|
[
"Apache-2.0"
] | 2 |
2019-09-17T08:14:45.000Z
|
2020-02-27T18:42:03.000Z
| 201.416898 | 66,808 | 0.873555 |
[
[
[
"import pandas as pd\nimport numpy as np\n%matplotlib inline\nimport matplotlib.pyplot as plt\nfrom utils import load_preprocessed_file,get_monthly_averages,\\\n compute_anomalies, plot_anomaly_graph",
"_____no_output_____"
],
[
"BUOYNO=44014\ndf=load_preprocessed_file(BUOYNO)\ndf = df[df.index.map(lambda x:x.year)!=1990] # drop 1990, which only has october onward\ndf.head(6)",
"_____no_output_____"
],
[
"monthly=get_monthly_averages(df)\nmonthly.head()",
"_____no_output_____"
],
[
"all_months_air = compute_anomalies(monthly, 'ATMP')\nall_months_air.head()\n",
"_____no_output_____"
],
[
"air_slope = plot_anomaly_graph(BUOYNO, 'air', all_months_air)\n",
"_____no_output_____"
],
[
"all_months_water = compute_anomalies(monthly, 'WTMP')\nall_months_water.head()",
"_____no_output_____"
],
[
"water_slope = plot_anomaly_graph(BUOYNO, 'water', all_months_water)\n",
"Unable to fit a line\nCreating a line just using the endpoint years (1991, 2018)\n"
],
[
"from dataworkspaces.kits.jupyter import NotebookLineageBuilder\nwith NotebookLineageBuilder('../results',\n run_description=\"compute air and water anomaly for buoy %s\" % BUOYNO)\\\n .with_parameters({'buoy':BUOYNO})\\\n .with_input_path('../intermediate-data/processed_%s.csv.gz'%BUOYNO)\\\n .eval() as lineage:\n lineage.write_results({'air_slope':round(air_slope,3),\n 'water_slope':round(water_slope, 3),\n 'units':'degrees C per decade'})\nprint(\"Results:\")\nprint(\" Air slope: %.3f degrees C per decade\" % air_slope)\nprint(\" Water slope: %.3f degrees C per decade\" % water_slope)",
"Wrote results to results:results.json\nResults:\n Air slope: 0.504 degrees C per decade\n Water slope: -0.183 degrees C per decade\n"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a5646db607a807f2fb38148ec54d7581ac36ec8
| 425,137 |
ipynb
|
Jupyter Notebook
|
quiz/m4_multifactor_models/Zipline_Pipeline/Zipline Pipeline.ipynb
|
masrur-ahmed/Udacity-AI-for-Trading-Nanodegree
|
bf2fe2d933f19af6e8550152d84673d5550d189a
|
[
"MIT"
] | 2 |
2020-04-29T10:07:17.000Z
|
2020-09-11T22:11:42.000Z
|
quiz/m4_multifactor_models/Zipline_Pipeline/Zipline Pipeline.ipynb
|
masrur-ahmed/Udacity-AI-for-Trading-Nanodegree
|
bf2fe2d933f19af6e8550152d84673d5550d189a
|
[
"MIT"
] | null | null | null |
quiz/m4_multifactor_models/Zipline_Pipeline/Zipline Pipeline.ipynb
|
masrur-ahmed/Udacity-AI-for-Trading-Nanodegree
|
bf2fe2d933f19af6e8550152d84673d5550d189a
|
[
"MIT"
] | null | null | null | 84.102275 | 3,812 | 0.598682 |
[
[
[
"# Zipline Pipeline",
"_____no_output_____"
],
[
"### Introduction\n\nOn any given trading day, the entire universe of stocks consists of thousands of securities. Usually, you will not be interested in investing in all the stocks in the entire universe, but rather, you will likely select only a subset of these to invest. For example, you may only want to invest in stocks that have a 10-day average closing price of \\$10.00 or less. Or you may only want to invest in the top 500 securities ranked by some factor.\n\nIn order to avoid spending a lot of time doing data wrangling to select only the securities you are interested in, people often use **pipelines**. In general, a pipeline is a placeholder for a series of data operations used to filter and rank data according to some factor or factors. \n\nIn this notebook, you will learn how to work with the **Zipline Pipeline**. Zipline is an open-source algorithmic trading simulator developed by *Quantopian*. We will learn how to use the Zipline Pipeline to filter stock data according to factors. ",
"_____no_output_____"
],
[
"### Install Packages",
"_____no_output_____"
]
],
[
[
"conda install -c Quantopian zipline",
"Collecting package metadata (current_repodata.json): done\nSolving environment: failed with initial frozen solve. Retrying with flexible solve.\nSolving environment: failed with repodata from current_repodata.json, will retry with next repodata source.\nCollecting package metadata (repodata.json): done\nSolving environment: failed with initial frozen solve. Retrying with flexible solve.\nSolving environment: - \nFound conflicts! Looking for incompatible packages.\nThis can take several minutes. Press CTRL-C to abort.\n failed\n\nUnsatisfiableError: The following specifications were found\nto be incompatible with the existing python installation in your environment:\n\nSpecifications:\n\n - zipline -> python[version='2.7.*|3.4.*|3.5.*|>=2.7,<2.8.0a0|>=3.5,<3.6.0a0|3.3.*']\n\nYour python: python=3.7\n\nIf python is on the left-most side of the chain, that's the version you've asked for.\nWhen python appears to the right, that indicates that the thing on the left is somehow\nnot available for the python version you are constrained to. Note that conda will not\nchange your python version to a different minor version unless you explicitly specify\nthat.\n\nThe following specifications were found to be incompatible with your CUDA driver:\n\n - feature:/linux-64::__cuda==10.1=0\n - feature:|@/linux-64::__cuda==10.1=0\n\nYour installed CUDA driver is: 10.1\n\n\n\nNote: you may need to restart the kernel to use updated packages.\n"
],
[
"import sys\n!{sys.executable} -m pip install -r requirements.txt",
"Requirement already satisfied: colour==0.1.5 in /home/masrur/.local/lib/python3.7/site-packages (from -r requirements.txt (line 1)) (0.1.5)\nRequirement already satisfied: cvxpy==1.0.3 in /home/masrur/.local/lib/python3.7/site-packages (from -r requirements.txt (line 2)) (1.0.3)\nRequirement already satisfied: cycler==0.10.0 in /home/masrur/.local/lib/python3.7/site-packages (from -r requirements.txt (line 3)) (0.10.0)\nCollecting numpy==1.13.3\n Using cached numpy-1.13.3.zip (5.0 MB)\nCollecting pandas==0.21.1\n Using cached pandas-0.21.1.tar.gz (11.3 MB)\n Installing build dependencies ... \u001b[?25lerror\n\u001b[31m ERROR: Command errored out with exit status 1:\n command: /home/masrur/anaconda3/bin/python /home/masrur/.local/lib/python3.7/site-packages/pip install --ignore-installed --no-user --prefix /tmp/pip-build-env-q_sd8e0o/overlay --no-warn-script-location --no-binary :none: --only-binary :none: -i https://pypi.org/simple -- wheel setuptools Cython 'numpy==1.9.3; python_version=='\"'\"'3.5'\"'\"'' 'numpy==1.12.1; python_version=='\"'\"'3.6'\"'\"'' 'numpy==1.13.1; python_version>='\"'\"'3.7'\"'\"''\n cwd: None\n Complete output (4389 lines):\n Ignoring numpy: markers 'python_version == \"3.5\"' don't match your environment\n Ignoring numpy: markers 'python_version == \"3.6\"' don't match your environment\n Collecting wheel\n Using cached wheel-0.34.2-py2.py3-none-any.whl (26 kB)\n Collecting setuptools\n Using cached setuptools-46.1.3-py3-none-any.whl (582 kB)\n Collecting Cython\n Using cached Cython-0.29.16-cp37-cp37m-manylinux1_x86_64.whl (2.1 MB)\n Collecting numpy==1.13.1\n Using cached numpy-1.13.1.zip (5.0 MB)\n Building wheels for collected packages: numpy\n Building wheel for numpy (setup.py): started\n Building wheel for numpy (setup.py): still running...\n Building wheel for numpy (setup.py): still running...\n Building wheel for numpy (setup.py): finished with status 'error'\n ERROR: Command errored out with exit status 1:\n command: /home/masrur/anaconda3/bin/python -u -c 'import sys, setuptools, tokenize; sys.argv[0] = '\"'\"'/tmp/pip-install-wkrshu4d/numpy/setup.py'\"'\"'; __file__='\"'\"'/tmp/pip-install-wkrshu4d/numpy/setup.py'\"'\"';f=getattr(tokenize, '\"'\"'open'\"'\"', open)(__file__);code=f.read().replace('\"'\"'\\r\\n'\"'\"', '\"'\"'\\n'\"'\"');f.close();exec(compile(code, __file__, '\"'\"'exec'\"'\"'))' bdist_wheel -d /tmp/pip-wheel-0oew2f6l\n cwd: /tmp/pip-install-wkrshu4d/numpy/\n Complete output (4016 lines):\n Running from numpy source directory.\n blas_opt_info:\n blas_mkl_info:\n FOUND:\n libraries = ['mkl_rt', 'pthread']\n library_dirs = ['/home/masrur/anaconda3/lib']\n define_macros = [('SCIPY_MKL_H', None), ('HAVE_CBLAS', None)]\n include_dirs = ['/usr/local/include', '/usr/include', '/home/masrur/anaconda3/include']\n \n FOUND:\n libraries = ['mkl_rt', 'pthread']\n library_dirs = ['/home/masrur/anaconda3/lib']\n define_macros = [('SCIPY_MKL_H', None), ('HAVE_CBLAS', None)]\n include_dirs = ['/usr/local/include', '/usr/include', '/home/masrur/anaconda3/include']\n \n /bin/sh: 1: svnversion: not found\n non-existing path in 'numpy/distutils': 'site.cfg'\n /bin/sh: 1: svnversion: not found\n F2PY Version 2\n lapack_opt_info:\n lapack_mkl_info:\n FOUND:\n libraries = ['mkl_rt', 'pthread']\n library_dirs = ['/home/masrur/anaconda3/lib']\n define_macros = [('SCIPY_MKL_H', None), ('HAVE_CBLAS', None)]\n include_dirs = ['/usr/local/include', '/usr/include', '/home/masrur/anaconda3/include']\n \n FOUND:\n libraries = ['mkl_rt', 'pthread']\n library_dirs = ['/home/masrur/anaconda3/lib']\n define_macros = [('SCIPY_MKL_H', None), ('HAVE_CBLAS', None)]\n include_dirs = ['/usr/local/include', '/usr/include', '/home/masrur/anaconda3/include']\n \n /home/masrur/anaconda3/lib/python3.7/distutils/dist.py:274: UserWarning: Unknown distribution option: 'define_macros'\n warnings.warn(msg)\n running bdist_wheel\n running build\n running config_cc\n unifing config_cc, config, build_clib, build_ext, build commands --compiler options\n running config_fc\n unifing config_fc, config, build_clib, build_ext, build commands --fcompiler options\n running build_src\n build_src\n building py_modules sources\n creating build\n creating build/src.linux-x86_64-3.7\n creating build/src.linux-x86_64-3.7/numpy\n creating build/src.linux-x86_64-3.7/numpy/distutils\n building library \"npymath\" sources\n customize Gnu95FCompiler\n Found executable /usr/bin/gfortran\n customize Gnu95FCompiler\n customize Gnu95FCompiler using config\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ _configtest.o -o _configtest\n success!\n removing: _configtest.c _configtest.o _configtest\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n _configtest.c:1:5: warning: conflicting types for built-in function ‘exp’; expected ‘double(double)’ [-Wbuiltin-declaration-mismatch]\n 1 | int exp (void);\n | ^~~\n _configtest.c:1:1: note: ‘exp’ is declared in header ‘<math.h>’\n +++ |+#include <math.h>\n 1 | int exp (void);\n gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ _configtest.o -o _configtest\n /home/masrur/anaconda3/compiler_compat/ld: _configtest.o: in function `main':\n /tmp/pip-install-wkrshu4d/numpy/_configtest.c:6: undefined reference to `exp'\n collect2: error: ld returned 1 exit status\n /home/masrur/anaconda3/compiler_compat/ld: _configtest.o: in function `main':\n /tmp/pip-install-wkrshu4d/numpy/_configtest.c:6: undefined reference to `exp'\n collect2: error: ld returned 1 exit status\n failure.\n removing: _configtest.c _configtest.o\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n _configtest.c:1:5: warning: conflicting types for built-in function ‘exp’; expected ‘double(double)’ [-Wbuiltin-declaration-mismatch]\n 1 | int exp (void);\n | ^~~\n _configtest.c:1:1: note: ‘exp’ is declared in header ‘<math.h>’\n +++ |+#include <math.h>\n 1 | int exp (void);\n gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ _configtest.o -lm -o _configtest\n success!\n removing: _configtest.c _configtest.o _configtest\n creating build/src.linux-x86_64-3.7/numpy/core\n creating build/src.linux-x86_64-3.7/numpy/core/src\n creating build/src.linux-x86_64-3.7/numpy/core/src/npymath\n conv_template:> build/src.linux-x86_64-3.7/numpy/core/src/npymath/npy_math_internal.h\n adding 'build/src.linux-x86_64-3.7/numpy/core/src/npymath' to include_dirs.\n conv_template:> build/src.linux-x86_64-3.7/numpy/core/src/npymath/ieee754.c\n conv_template:> build/src.linux-x86_64-3.7/numpy/core/src/npymath/npy_math_complex.c\n None - nothing done with h_files = ['build/src.linux-x86_64-3.7/numpy/core/src/npymath/npy_math_internal.h']\n building library \"npysort\" sources\n creating build/src.linux-x86_64-3.7/numpy/core/src/npysort\n conv_template:> build/src.linux-x86_64-3.7/numpy/core/src/npysort/quicksort.c\n conv_template:> build/src.linux-x86_64-3.7/numpy/core/src/npysort/mergesort.c\n conv_template:> build/src.linux-x86_64-3.7/numpy/core/src/npysort/heapsort.c\n creating build/src.linux-x86_64-3.7/numpy/core/src/private\n conv_template:> build/src.linux-x86_64-3.7/numpy/core/src/private/npy_partition.h\n adding 'build/src.linux-x86_64-3.7/numpy/core/src/private' to include_dirs.\n conv_template:> build/src.linux-x86_64-3.7/numpy/core/src/npysort/selection.c\n conv_template:> build/src.linux-x86_64-3.7/numpy/core/src/private/npy_binsearch.h\n conv_template:> build/src.linux-x86_64-3.7/numpy/core/src/npysort/binsearch.c\n None - nothing done with h_files = ['build/src.linux-x86_64-3.7/numpy/core/src/private/npy_partition.h', 'build/src.linux-x86_64-3.7/numpy/core/src/private/npy_binsearch.h']\n building extension \"numpy.core._dummy\" sources\n Generating build/src.linux-x86_64-3.7/numpy/core/include/numpy/config.h\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n success!\n removing: _configtest.c _configtest.o\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n success!\n removing: _configtest.c _configtest.o\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n _configtest.c:1:10: fatal error: sys/endian.h: No such file or directory\n 1 | #include <sys/endian.h>\n | ^~~~~~~~~~~~~~\n compilation terminated.\n _configtest.c:1:10: fatal error: sys/endian.h: No such file or directory\n 1 | #include <sys/endian.h>\n | ^~~~~~~~~~~~~~\n compilation terminated.\n failure.\n removing: _configtest.c _configtest.o\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n success!\n removing: _configtest.c _configtest.o\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n success!\n removing: _configtest.c _configtest.o\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n success!\n removing: _configtest.c _configtest.o\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n success!\n removing: _configtest.c _configtest.o\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n _configtest.c: In function ‘main’:\n _configtest.c:5:16: warning: variable ‘test_array’ set but not used [-Wunused-but-set-variable]\n 5 | static int test_array [1 - 2 * !(((long) (sizeof (npy_check_sizeof_type))) >= 0)];\n | ^~~~~~~~~~\n removing: _configtest.c _configtest.o\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n _configtest.c: In function ‘main’:\n _configtest.c:5:16: warning: variable ‘test_array’ set but not used [-Wunused-but-set-variable]\n 5 | static int test_array [1 - 2 * !(((long) (sizeof (npy_check_sizeof_type))) == 4)];\n | ^~~~~~~~~~\n removing: _configtest.c _configtest.o\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n _configtest.c: In function ‘main’:\n _configtest.c:5:16: warning: variable ‘test_array’ set but not used [-Wunused-but-set-variable]\n 5 | static int test_array [1 - 2 * !(((long) (sizeof (npy_check_sizeof_type))) >= 0)];\n | ^~~~~~~~~~\n removing: _configtest.c _configtest.o\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n _configtest.c: In function ‘main’:\n _configtest.c:5:16: warning: variable ‘test_array’ set but not used [-Wunused-but-set-variable]\n 5 | static int test_array [1 - 2 * !(((long) (sizeof (npy_check_sizeof_type))) == 8)];\n | ^~~~~~~~~~\n removing: _configtest.c _configtest.o\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n success!\n removing: _configtest.c _configtest.o\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n _configtest.c: In function ‘main’:\n _configtest.c:5:16: warning: variable ‘test_array’ set but not used [-Wunused-but-set-variable]\n 5 | static int test_array [1 - 2 * !(((long) (sizeof (npy_check_sizeof_type))) >= 0)];\n | ^~~~~~~~~~\n removing: _configtest.c _configtest.o\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n _configtest.c: In function ‘main’:\n _configtest.c:5:16: warning: variable ‘test_array’ set but not used [-Wunused-but-set-variable]\n 5 | static int test_array [1 - 2 * !(((long) (sizeof (npy_check_sizeof_type))) == 8)];\n | ^~~~~~~~~~\n removing: _configtest.c _configtest.o\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n _configtest.c: In function ‘main’:\n _configtest.c:5:16: warning: variable ‘test_array’ set but not used [-Wunused-but-set-variable]\n 5 | static int test_array [1 - 2 * !(((long) (sizeof (npy_check_sizeof_type))) >= 0)];\n | ^~~~~~~~~~\n removing: _configtest.c _configtest.o\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n _configtest.c: In function ‘main’:\n _configtest.c:5:16: warning: variable ‘test_array’ set but not used [-Wunused-but-set-variable]\n 5 | static int test_array [1 - 2 * !(((long) (sizeof (npy_check_sizeof_type))) == 16)];\n | ^~~~~~~~~~\n removing: _configtest.c _configtest.o\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n _configtest.c: In function ‘main’:\n _configtest.c:7:12: error: ‘SIZEOF_LONGDOUBLE’ undeclared (first use in this function); did you mean ‘SIZEOF_LONG_DOUBLE’?\n 7 | (void) SIZEOF_LONGDOUBLE;\n | ^~~~~~~~~~~~~~~~~\n | SIZEOF_LONG_DOUBLE\n _configtest.c:7:12: note: each undeclared identifier is reported only once for each function it appears in\n _configtest.c: In function ‘main’:\n _configtest.c:7:12: error: ‘SIZEOF_LONGDOUBLE’ undeclared (first use in this function); did you mean ‘SIZEOF_LONG_DOUBLE’?\n 7 | (void) SIZEOF_LONGDOUBLE;\n | ^~~~~~~~~~~~~~~~~\n | SIZEOF_LONG_DOUBLE\n _configtest.c:7:12: note: each undeclared identifier is reported only once for each function it appears in\n failure.\n removing: _configtest.c _configtest.o\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n _configtest.c: In function ‘main’:\n _configtest.c:5:16: warning: variable ‘test_array’ set but not used [-Wunused-but-set-variable]\n 5 | static int test_array [1 - 2 * !(((long) (sizeof (npy_check_sizeof_type))) >= 0)];\n | ^~~~~~~~~~\n removing: _configtest.c _configtest.o\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n _configtest.c: In function ‘main’:\n _configtest.c:5:16: warning: variable ‘test_array’ set but not used [-Wunused-but-set-variable]\n 5 | static int test_array [1 - 2 * !(((long) (sizeof (npy_check_sizeof_type))) == 16)];\n | ^~~~~~~~~~\n removing: _configtest.c _configtest.o\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n _configtest.c: In function ‘main’:\n _configtest.c:5:16: warning: variable ‘test_array’ set but not used [-Wunused-but-set-variable]\n 5 | static int test_array [1 - 2 * !(((long) (sizeof (npy_check_sizeof_type))) >= 0)];\n | ^~~~~~~~~~\n removing: _configtest.c _configtest.o\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n _configtest.c: In function ‘main’:\n _configtest.c:5:16: warning: variable ‘test_array’ set but not used [-Wunused-but-set-variable]\n 5 | static int test_array [1 - 2 * !(((long) (sizeof (npy_check_sizeof_type))) == 32)];\n | ^~~~~~~~~~\n removing: _configtest.c _configtest.o\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n _configtest.c: In function ‘main’:\n _configtest.c:7:16: warning: variable ‘test_array’ set but not used [-Wunused-but-set-variable]\n 7 | static int test_array [1 - 2 * !(((long) (sizeof (npy_check_sizeof_type))) >= 0)];\n | ^~~~~~~~~~\n removing: _configtest.c _configtest.o\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n _configtest.c: In function ‘main’:\n _configtest.c:7:16: warning: variable ‘test_array’ set but not used [-Wunused-but-set-variable]\n 7 | static int test_array [1 - 2 * !(((long) (sizeof (npy_check_sizeof_type))) == 8)];\n | ^~~~~~~~~~\n removing: _configtest.c _configtest.o\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n _configtest.c: In function ‘main’:\n _configtest.c:7:16: warning: variable ‘test_array’ set but not used [-Wunused-but-set-variable]\n 7 | static int test_array [1 - 2 * !(((long) (sizeof (npy_check_sizeof_type))) >= 0)];\n | ^~~~~~~~~~\n removing: _configtest.c _configtest.o\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n _configtest.c: In function ‘main’:\n _configtest.c:7:16: warning: variable ‘test_array’ set but not used [-Wunused-but-set-variable]\n 7 | static int test_array [1 - 2 * !(((long) (sizeof (npy_check_sizeof_type))) == 8)];\n | ^~~~~~~~~~\n removing: _configtest.c _configtest.o\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n success!\n removing: _configtest.c _configtest.o\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n _configtest.c: In function ‘main’:\n _configtest.c:7:16: warning: variable ‘test_array’ set but not used [-Wunused-but-set-variable]\n 7 | static int test_array [1 - 2 * !(((long) (sizeof (npy_check_sizeof_type))) >= 0)];\n | ^~~~~~~~~~\n removing: _configtest.c _configtest.o\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n _configtest.c: In function ‘main’:\n _configtest.c:7:16: warning: variable ‘test_array’ set but not used [-Wunused-but-set-variable]\n 7 | static int test_array [1 - 2 * !(((long) (sizeof (npy_check_sizeof_type))) == 8)];\n | ^~~~~~~~~~\n removing: _configtest.c _configtest.o\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n _configtest.c: In function ‘main’:\n _configtest.c:5:16: warning: variable ‘test_array’ set but not used [-Wunused-but-set-variable]\n 5 | static int test_array [1 - 2 * !(((long) (sizeof (npy_check_sizeof_type))) >= 0)];\n | ^~~~~~~~~~\n removing: _configtest.c _configtest.o\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n _configtest.c: In function ‘main’:\n _configtest.c:5:16: warning: variable ‘test_array’ set but not used [-Wunused-but-set-variable]\n 5 | static int test_array [1 - 2 * !(((long) (sizeof (npy_check_sizeof_type))) == 8)];\n | ^~~~~~~~~~\n removing: _configtest.c _configtest.o\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n success!\n removing: _configtest.c _configtest.o\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n _configtest.c:1:5: warning: conflicting types for built-in function ‘exp’; expected ‘double(double)’ [-Wbuiltin-declaration-mismatch]\n 1 | int exp (void);\n | ^~~\n _configtest.c:1:1: note: ‘exp’ is declared in header ‘<math.h>’\n +++ |+#include <math.h>\n 1 | int exp (void);\n gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ _configtest.o -o _configtest\n /home/masrur/anaconda3/compiler_compat/ld: _configtest.o: in function `main':\n /tmp/pip-install-wkrshu4d/numpy/_configtest.c:6: undefined reference to `exp'\n collect2: error: ld returned 1 exit status\n /home/masrur/anaconda3/compiler_compat/ld: _configtest.o: in function `main':\n /tmp/pip-install-wkrshu4d/numpy/_configtest.c:6: undefined reference to `exp'\n collect2: error: ld returned 1 exit status\n failure.\n removing: _configtest.c _configtest.o\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n _configtest.c:1:5: warning: conflicting types for built-in function ‘exp’; expected ‘double(double)’ [-Wbuiltin-declaration-mismatch]\n 1 | int exp (void);\n | ^~~\n _configtest.c:1:1: note: ‘exp’ is declared in header ‘<math.h>’\n +++ |+#include <math.h>\n 1 | int exp (void);\n gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ _configtest.o -lm -o _configtest\n success!\n removing: _configtest.c _configtest.o _configtest\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n _configtest.c:1:5: warning: conflicting types for built-in function ‘sin’; expected ‘double(double)’ [-Wbuiltin-declaration-mismatch]\n 1 | int sin (void);\n | ^~~\n _configtest.c:1:1: note: ‘sin’ is declared in header ‘<math.h>’\n +++ |+#include <math.h>\n 1 | int sin (void);\n _configtest.c:2:5: warning: conflicting types for built-in function ‘cos’; expected ‘double(double)’ [-Wbuiltin-declaration-mismatch]\n 2 | int cos (void);\n | ^~~\n _configtest.c:2:5: note: ‘cos’ is declared in header ‘<math.h>’\n _configtest.c:3:5: warning: conflicting types for built-in function ‘tan’; expected ‘double(double)’ [-Wbuiltin-declaration-mismatch]\n 3 | int tan (void);\n | ^~~\n _configtest.c:3:5: note: ‘tan’ is declared in header ‘<math.h>’\n _configtest.c:4:5: warning: conflicting types for built-in function ‘sinh’; expected ‘double(double)’ [-Wbuiltin-declaration-mismatch]\n 4 | int sinh (void);\n | ^~~~\n _configtest.c:4:5: note: ‘sinh’ is declared in header ‘<math.h>’\n _configtest.c:5:5: warning: conflicting types for built-in function ‘cosh’; expected ‘double(double)’ [-Wbuiltin-declaration-mismatch]\n 5 | int cosh (void);\n | ^~~~\n _configtest.c:5:5: note: ‘cosh’ is declared in header ‘<math.h>’\n _configtest.c:6:5: warning: conflicting types for built-in function ‘tanh’; expected ‘double(double)’ [-Wbuiltin-declaration-mismatch]\n 6 | int tanh (void);\n | ^~~~\n _configtest.c:6:5: note: ‘tanh’ is declared in header ‘<math.h>’\n _configtest.c:7:5: warning: conflicting types for built-in function ‘fabs’; expected ‘double(double)’ [-Wbuiltin-declaration-mismatch]\n 7 | int fabs (void);\n | ^~~~\n _configtest.c:7:5: note: ‘fabs’ is declared in header ‘<math.h>’\n _configtest.c:8:5: warning: conflicting types for built-in function ‘floor’; expected ‘double(double)’ [-Wbuiltin-declaration-mismatch]\n 8 | int floor (void);\n | ^~~~~\n _configtest.c:8:5: note: ‘floor’ is declared in header ‘<math.h>’\n _configtest.c:9:5: warning: conflicting types for built-in function ‘ceil’; expected ‘double(double)’ [-Wbuiltin-declaration-mismatch]\n 9 | int ceil (void);\n | ^~~~\n _configtest.c:9:5: note: ‘ceil’ is declared in header ‘<math.h>’\n _configtest.c:10:5: warning: conflicting types for built-in function ‘sqrt’; expected ‘double(double)’ [-Wbuiltin-declaration-mismatch]\n 10 | int sqrt (void);\n | ^~~~\n _configtest.c:10:5: note: ‘sqrt’ is declared in header ‘<math.h>’\n _configtest.c:11:5: warning: conflicting types for built-in function ‘log10’; expected ‘double(double)’ [-Wbuiltin-declaration-mismatch]\n 11 | int log10 (void);\n | ^~~~~\n _configtest.c:11:5: note: ‘log10’ is declared in header ‘<math.h>’\n _configtest.c:12:5: warning: conflicting types for built-in function ‘log’; expected ‘double(double)’ [-Wbuiltin-declaration-mismatch]\n 12 | int log (void);\n | ^~~\n _configtest.c:12:5: note: ‘log’ is declared in header ‘<math.h>’\n _configtest.c:13:5: warning: conflicting types for built-in function ‘exp’; expected ‘double(double)’ [-Wbuiltin-declaration-mismatch]\n 13 | int exp (void);\n | ^~~\n _configtest.c:13:5: note: ‘exp’ is declared in header ‘<math.h>’\n _configtest.c:14:5: warning: conflicting types for built-in function ‘asin’; expected ‘double(double)’ [-Wbuiltin-declaration-mismatch]\n 14 | int asin (void);\n | ^~~~\n _configtest.c:14:5: note: ‘asin’ is declared in header ‘<math.h>’\n _configtest.c:15:5: warning: conflicting types for built-in function ‘acos’; expected ‘double(double)’ [-Wbuiltin-declaration-mismatch]\n 15 | int acos (void);\n | ^~~~\n _configtest.c:15:5: note: ‘acos’ is declared in header ‘<math.h>’\n _configtest.c:16:5: warning: conflicting types for built-in function ‘atan’; expected ‘double(double)’ [-Wbuiltin-declaration-mismatch]\n 16 | int atan (void);\n | ^~~~\n _configtest.c:16:5: note: ‘atan’ is declared in header ‘<math.h>’\n _configtest.c:17:5: warning: conflicting types for built-in function ‘fmod’; expected ‘double(double, double)’ [-Wbuiltin-declaration-mismatch]\n 17 | int fmod (void);\n | ^~~~\n _configtest.c:17:5: note: ‘fmod’ is declared in header ‘<math.h>’\n _configtest.c:18:5: warning: conflicting types for built-in function ‘modf’; expected ‘double(double, double *)’ [-Wbuiltin-declaration-mismatch]\n 18 | int modf (void);\n | ^~~~\n _configtest.c:18:5: note: ‘modf’ is declared in header ‘<math.h>’\n _configtest.c:19:5: warning: conflicting types for built-in function ‘frexp’; expected ‘double(double, int *)’ [-Wbuiltin-declaration-mismatch]\n 19 | int frexp (void);\n | ^~~~~\n _configtest.c:19:5: note: ‘frexp’ is declared in header ‘<math.h>’\n _configtest.c:20:5: warning: conflicting types for built-in function ‘ldexp’; expected ‘double(double, int)’ [-Wbuiltin-declaration-mismatch]\n 20 | int ldexp (void);\n | ^~~~~\n _configtest.c:20:5: note: ‘ldexp’ is declared in header ‘<math.h>’\n gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ _configtest.o -lm -o _configtest\n success!\n removing: _configtest.c _configtest.o _configtest\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n success!\n removing: _configtest.c _configtest.o\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n success!\n removing: _configtest.c _configtest.o\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n success!\n removing: _configtest.c _configtest.o\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n success!\n removing: _configtest.c _configtest.o\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n success!\n removing: _configtest.c _configtest.o\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n success!\n removing: _configtest.c _configtest.o\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n success!\n removing: _configtest.c _configtest.o\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n success!\n removing: _configtest.c _configtest.o\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n success!\n removing: _configtest.c _configtest.o\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n _configtest.c:1:5: warning: conflicting types for built-in function ‘rint’; expected ‘double(double)’ [-Wbuiltin-declaration-mismatch]\n 1 | int rint (void);\n | ^~~~\n _configtest.c:1:1: note: ‘rint’ is declared in header ‘<math.h>’\n +++ |+#include <math.h>\n 1 | int rint (void);\n _configtest.c:2:5: warning: conflicting types for built-in function ‘trunc’; expected ‘double(double)’ [-Wbuiltin-declaration-mismatch]\n 2 | int trunc (void);\n | ^~~~~\n _configtest.c:2:5: note: ‘trunc’ is declared in header ‘<math.h>’\n _configtest.c:3:5: warning: conflicting types for built-in function ‘exp2’; expected ‘double(double)’ [-Wbuiltin-declaration-mismatch]\n 3 | int exp2 (void);\n | ^~~~\n _configtest.c:3:5: note: ‘exp2’ is declared in header ‘<math.h>’\n _configtest.c:4:5: warning: conflicting types for built-in function ‘log2’; expected ‘double(double)’ [-Wbuiltin-declaration-mismatch]\n 4 | int log2 (void);\n | ^~~~\n _configtest.c:4:5: note: ‘log2’ is declared in header ‘<math.h>’\n _configtest.c:5:5: warning: conflicting types for built-in function ‘atan2’; expected ‘double(double, double)’ [-Wbuiltin-declaration-mismatch]\n 5 | int atan2 (void);\n | ^~~~~\n _configtest.c:5:5: note: ‘atan2’ is declared in header ‘<math.h>’\n _configtest.c:6:5: warning: conflicting types for built-in function ‘pow’; expected ‘double(double, double)’ [-Wbuiltin-declaration-mismatch]\n 6 | int pow (void);\n | ^~~\n _configtest.c:6:5: note: ‘pow’ is declared in header ‘<math.h>’\n _configtest.c:7:5: warning: conflicting types for built-in function ‘nextafter’; expected ‘double(double, double)’ [-Wbuiltin-declaration-mismatch]\n 7 | int nextafter (void);\n | ^~~~~~~~~\n _configtest.c:7:5: note: ‘nextafter’ is declared in header ‘<math.h>’\n _configtest.c:10:5: warning: conflicting types for built-in function ‘cbrt’; expected ‘double(double)’ [-Wbuiltin-declaration-mismatch]\n 10 | int cbrt (void);\n | ^~~~\n _configtest.c:10:5: note: ‘cbrt’ is declared in header ‘<math.h>’\n gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ _configtest.o -lm -o _configtest\n success!\n removing: _configtest.c _configtest.o _configtest\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ _configtest.o -o _configtest\n success!\n removing: _configtest.c _configtest.o _configtest\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ _configtest.o -o _configtest\n success!\n removing: _configtest.c _configtest.o _configtest\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ _configtest.o -o _configtest\n success!\n removing: _configtest.c _configtest.o _configtest\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n _configtest.c:1:10: fatal error: xlocale.h: No such file or directory\n 1 | #include <xlocale.h>\n | ^~~~~~~~~~~\n compilation terminated.\n _configtest.c:1:10: fatal error: xlocale.h: No such file or directory\n 1 | #include <xlocale.h>\n | ^~~~~~~~~~~\n compilation terminated.\n failure.\n removing: _configtest.c _configtest.o\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ _configtest.o -o _configtest\n success!\n removing: _configtest.c _configtest.o _configtest\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n _configtest.c: In function ‘main’:\n _configtest.c:5:3: warning: statement with no effect [-Wunused-value]\n 5 | __builtin_isnan(5.);\n | ^~~~~~~~~~~~~~~\n gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ _configtest.o -o _configtest\n success!\n removing: _configtest.c _configtest.o _configtest\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n _configtest.c: In function ‘main’:\n _configtest.c:5:3: warning: statement with no effect [-Wunused-value]\n 5 | __builtin_isinf(5.);\n | ^~~~~~~~~~~~~~~\n gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ _configtest.o -o _configtest\n success!\n removing: _configtest.c _configtest.o _configtest\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n _configtest.c: In function ‘main’:\n _configtest.c:5:3: warning: statement with no effect [-Wunused-value]\n 5 | __builtin_isfinite(5.);\n | ^~~~~~~~~~~~~~~~~~\n gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ _configtest.o -o _configtest\n success!\n removing: _configtest.c _configtest.o _configtest\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n _configtest.c: In function ‘main’:\n _configtest.c:5:3: warning: statement with no effect [-Wunused-value]\n 5 | __builtin_bswap32(5u);\n | ^~~~~~~~~~~~~~~~~\n gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ _configtest.o -o _configtest\n success!\n removing: _configtest.c _configtest.o _configtest\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n _configtest.c: In function ‘main’:\n _configtest.c:5:3: warning: statement with no effect [-Wunused-value]\n 5 | __builtin_bswap64(5u);\n | ^~~~~~~~~~~~~~~~~\n gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ _configtest.o -o _configtest\n success!\n removing: _configtest.c _configtest.o _configtest\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n _configtest.c: In function ‘main’:\n _configtest.c:5:3: warning: statement with no effect [-Wunused-value]\n 5 | __builtin_expect(5, 0);\n | ^~~~~~~~~~~~~~~~\n gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ _configtest.o -o _configtest\n success!\n removing: _configtest.c _configtest.o _configtest\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n _configtest.c: In function ‘main’:\n _configtest.c:5:3: warning: right-hand operand of comma expression has no effect [-Wunused-value]\n 5 | __builtin_mul_overflow(5, 5, (int*)5);\n | ^~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~\n gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ _configtest.o -o _configtest\n success!\n removing: _configtest.c _configtest.o _configtest\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n _configtest.c: In function ‘main’:\n _configtest.c:7:16: warning: unused variable ‘r’ [-Wunused-variable]\n 7 | volatile int r = __builtin_cpu_supports(\"sse\");\n | ^\n gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ _configtest.o -o _configtest\n success!\n removing: _configtest.c _configtest.o _configtest\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ _configtest.o -o _configtest\n success!\n removing: _configtest.c _configtest.o _configtest\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ _configtest.o -o _configtest\n success!\n removing: _configtest.c _configtest.o _configtest\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ _configtest.o -o _configtest\n success!\n removing: _configtest.c _configtest.o _configtest\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ _configtest.o -o _configtest\n success!\n removing: _configtest.c _configtest.o _configtest\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ _configtest.o -o _configtest\n success!\n removing: _configtest.c _configtest.o _configtest\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ _configtest.o -o _configtest\n success!\n removing: _configtest.c _configtest.o _configtest\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ _configtest.o -o _configtest\n success!\n removing: _configtest.c _configtest.o _configtest\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n _configtest.c:3: warning: ignoring #pragma clang diagnostic [-Wunknown-pragmas]\n 3 | #pragma clang diagnostic error \"-Wattributes\"\n |\n _configtest.c:8:1: warning: function declaration isn’t a prototype [-Wstrict-prototypes]\n 8 | main()\n | ^~~~\n success!\n removing: _configtest.c _configtest.o\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n _configtest.c:3: warning: ignoring #pragma clang diagnostic [-Wunknown-pragmas]\n 3 | #pragma clang diagnostic error \"-Wattributes\"\n |\n _configtest.c:8:1: warning: function declaration isn’t a prototype [-Wstrict-prototypes]\n 8 | main()\n | ^~~~\n success!\n removing: _configtest.c _configtest.o\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n _configtest.c:3: warning: ignoring #pragma clang diagnostic [-Wunknown-pragmas]\n 3 | #pragma clang diagnostic error \"-Wattributes\"\n |\n _configtest.c:8:1: warning: function declaration isn’t a prototype [-Wstrict-prototypes]\n 8 | main()\n | ^~~~\n success!\n removing: _configtest.c _configtest.o\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n _configtest.c:3: warning: ignoring #pragma clang diagnostic [-Wunknown-pragmas]\n 3 | #pragma clang diagnostic error \"-Wattributes\"\n |\n _configtest.c:8:1: warning: function declaration isn’t a prototype [-Wstrict-prototypes]\n 8 | main()\n | ^~~~\n success!\n removing: _configtest.c _configtest.o\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n _configtest.c:3: warning: ignoring #pragma clang diagnostic [-Wunknown-pragmas]\n 3 | #pragma clang diagnostic error \"-Wattributes\"\n |\n _configtest.c:8:1: warning: function declaration isn’t a prototype [-Wstrict-prototypes]\n 8 | main()\n | ^~~~\n success!\n removing: _configtest.c _configtest.o\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n _configtest.c:3: warning: ignoring #pragma clang diagnostic [-Wunknown-pragmas]\n 3 | #pragma clang diagnostic error \"-Wattributes\"\n |\n _configtest.c:8:1: warning: function declaration isn’t a prototype [-Wstrict-prototypes]\n 8 | main()\n | ^~~~\n success!\n removing: _configtest.c _configtest.o\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n _configtest.c:3: warning: ignoring #pragma clang diagnostic [-Wunknown-pragmas]\n 3 | #pragma clang diagnostic error \"-Wattributes\"\n |\n _configtest.c:5:5: warning: function declaration isn’t a prototype [-Wstrict-prototypes]\n 5 | int __declspec(thread) foo;\n | ^~~~~~~~~~\n _configtest.c: In function ‘__declspec’:\n _configtest.c:5:24: error: expected declaration specifiers before ‘foo’\n 5 | int __declspec(thread) foo;\n | ^~~\n _configtest.c:9:1: error: expected ‘=’, ‘,’, ‘;’, ‘asm’ or ‘__attribute__’ before ‘{’ token\n 9 | {\n | ^\n _configtest.c:5:5: warning: type of ‘thread’ defaults to ‘int’ [-Wimplicit-int]\n 5 | int __declspec(thread) foo;\n | ^~~~~~~~~~\n _configtest.c:11: error: expected ‘{’ at end of input\n 11 | }\n |\n _configtest.c:11:1: warning: control reaches end of non-void function [-Wreturn-type]\n 11 | }\n | ^\n _configtest.c:3: warning: ignoring #pragma clang diagnostic [-Wunknown-pragmas]\n 3 | #pragma clang diagnostic error \"-Wattributes\"\n |\n _configtest.c:5:5: warning: function declaration isn’t a prototype [-Wstrict-prototypes]\n 5 | int __declspec(thread) foo;\n | ^~~~~~~~~~\n _configtest.c: In function ‘__declspec’:\n _configtest.c:5:24: error: expected declaration specifiers before ‘foo’\n 5 | int __declspec(thread) foo;\n | ^~~\n _configtest.c:9:1: error: expected ‘=’, ‘,’, ‘;’, ‘asm’ or ‘__attribute__’ before ‘{’ token\n 9 | {\n | ^\n _configtest.c:5:5: warning: type of ‘thread’ defaults to ‘int’ [-Wimplicit-int]\n 5 | int __declspec(thread) foo;\n | ^~~~~~~~~~\n _configtest.c:11: error: expected ‘{’ at end of input\n 11 | }\n |\n _configtest.c:11:1: warning: control reaches end of non-void function [-Wreturn-type]\n 11 | }\n | ^\n failure.\n removing: _configtest.c _configtest.o\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n _configtest.c:1:5: warning: conflicting types for built-in function ‘sinf’; expected ‘float(float)’ [-Wbuiltin-declaration-mismatch]\n 1 | int sinf (void);\n | ^~~~\n _configtest.c:1:1: note: ‘sinf’ is declared in header ‘<math.h>’\n +++ |+#include <math.h>\n 1 | int sinf (void);\n _configtest.c:2:5: warning: conflicting types for built-in function ‘cosf’; expected ‘float(float)’ [-Wbuiltin-declaration-mismatch]\n 2 | int cosf (void);\n | ^~~~\n _configtest.c:2:5: note: ‘cosf’ is declared in header ‘<math.h>’\n _configtest.c:3:5: warning: conflicting types for built-in function ‘tanf’; expected ‘float(float)’ [-Wbuiltin-declaration-mismatch]\n 3 | int tanf (void);\n | ^~~~\n _configtest.c:3:5: note: ‘tanf’ is declared in header ‘<math.h>’\n _configtest.c:4:5: warning: conflicting types for built-in function ‘sinhf’; expected ‘float(float)’ [-Wbuiltin-declaration-mismatch]\n 4 | int sinhf (void);\n | ^~~~~\n _configtest.c:4:5: note: ‘sinhf’ is declared in header ‘<math.h>’\n _configtest.c:5:5: warning: conflicting types for built-in function ‘coshf’; expected ‘float(float)’ [-Wbuiltin-declaration-mismatch]\n 5 | int coshf (void);\n | ^~~~~\n _configtest.c:5:5: note: ‘coshf’ is declared in header ‘<math.h>’\n _configtest.c:6:5: warning: conflicting types for built-in function ‘tanhf’; expected ‘float(float)’ [-Wbuiltin-declaration-mismatch]\n 6 | int tanhf (void);\n | ^~~~~\n _configtest.c:6:5: note: ‘tanhf’ is declared in header ‘<math.h>’\n _configtest.c:7:5: warning: conflicting types for built-in function ‘fabsf’; expected ‘float(float)’ [-Wbuiltin-declaration-mismatch]\n 7 | int fabsf (void);\n | ^~~~~\n _configtest.c:7:5: note: ‘fabsf’ is declared in header ‘<math.h>’\n _configtest.c:8:5: warning: conflicting types for built-in function ‘floorf’; expected ‘float(float)’ [-Wbuiltin-declaration-mismatch]\n 8 | int floorf (void);\n | ^~~~~~\n _configtest.c:8:5: note: ‘floorf’ is declared in header ‘<math.h>’\n _configtest.c:9:5: warning: conflicting types for built-in function ‘ceilf’; expected ‘float(float)’ [-Wbuiltin-declaration-mismatch]\n 9 | int ceilf (void);\n | ^~~~~\n _configtest.c:9:5: note: ‘ceilf’ is declared in header ‘<math.h>’\n _configtest.c:10:5: warning: conflicting types for built-in function ‘rintf’; expected ‘float(float)’ [-Wbuiltin-declaration-mismatch]\n 10 | int rintf (void);\n | ^~~~~\n _configtest.c:10:5: note: ‘rintf’ is declared in header ‘<math.h>’\n _configtest.c:11:5: warning: conflicting types for built-in function ‘truncf’; expected ‘float(float)’ [-Wbuiltin-declaration-mismatch]\n 11 | int truncf (void);\n | ^~~~~~\n _configtest.c:11:5: note: ‘truncf’ is declared in header ‘<math.h>’\n _configtest.c:12:5: warning: conflicting types for built-in function ‘sqrtf’; expected ‘float(float)’ [-Wbuiltin-declaration-mismatch]\n 12 | int sqrtf (void);\n | ^~~~~\n _configtest.c:12:5: note: ‘sqrtf’ is declared in header ‘<math.h>’\n _configtest.c:13:5: warning: conflicting types for built-in function ‘log10f’; expected ‘float(float)’ [-Wbuiltin-declaration-mismatch]\n 13 | int log10f (void);\n | ^~~~~~\n _configtest.c:13:5: note: ‘log10f’ is declared in header ‘<math.h>’\n _configtest.c:14:5: warning: conflicting types for built-in function ‘logf’; expected ‘float(float)’ [-Wbuiltin-declaration-mismatch]\n 14 | int logf (void);\n | ^~~~\n _configtest.c:14:5: note: ‘logf’ is declared in header ‘<math.h>’\n _configtest.c:15:5: warning: conflicting types for built-in function ‘log1pf’; expected ‘float(float)’ [-Wbuiltin-declaration-mismatch]\n 15 | int log1pf (void);\n | ^~~~~~\n _configtest.c:15:5: note: ‘log1pf’ is declared in header ‘<math.h>’\n _configtest.c:16:5: warning: conflicting types for built-in function ‘expf’; expected ‘float(float)’ [-Wbuiltin-declaration-mismatch]\n 16 | int expf (void);\n | ^~~~\n _configtest.c:16:5: note: ‘expf’ is declared in header ‘<math.h>’\n _configtest.c:17:5: warning: conflicting types for built-in function ‘expm1f’; expected ‘float(float)’ [-Wbuiltin-declaration-mismatch]\n 17 | int expm1f (void);\n | ^~~~~~\n _configtest.c:17:5: note: ‘expm1f’ is declared in header ‘<math.h>’\n _configtest.c:18:5: warning: conflicting types for built-in function ‘asinf’; expected ‘float(float)’ [-Wbuiltin-declaration-mismatch]\n 18 | int asinf (void);\n | ^~~~~\n _configtest.c:18:5: note: ‘asinf’ is declared in header ‘<math.h>’\n _configtest.c:19:5: warning: conflicting types for built-in function ‘acosf’; expected ‘float(float)’ [-Wbuiltin-declaration-mismatch]\n 19 | int acosf (void);\n | ^~~~~\n _configtest.c:19:5: note: ‘acosf’ is declared in header ‘<math.h>’\n _configtest.c:20:5: warning: conflicting types for built-in function ‘atanf’; expected ‘float(float)’ [-Wbuiltin-declaration-mismatch]\n 20 | int atanf (void);\n | ^~~~~\n _configtest.c:20:5: note: ‘atanf’ is declared in header ‘<math.h>’\n _configtest.c:21:5: warning: conflicting types for built-in function ‘asinhf’; expected ‘float(float)’ [-Wbuiltin-declaration-mismatch]\n 21 | int asinhf (void);\n | ^~~~~~\n _configtest.c:21:5: note: ‘asinhf’ is declared in header ‘<math.h>’\n _configtest.c:22:5: warning: conflicting types for built-in function ‘acoshf’; expected ‘float(float)’ [-Wbuiltin-declaration-mismatch]\n 22 | int acoshf (void);\n | ^~~~~~\n _configtest.c:22:5: note: ‘acoshf’ is declared in header ‘<math.h>’\n _configtest.c:23:5: warning: conflicting types for built-in function ‘atanhf’; expected ‘float(float)’ [-Wbuiltin-declaration-mismatch]\n 23 | int atanhf (void);\n | ^~~~~~\n _configtest.c:23:5: note: ‘atanhf’ is declared in header ‘<math.h>’\n _configtest.c:24:5: warning: conflicting types for built-in function ‘hypotf’; expected ‘float(float, float)’ [-Wbuiltin-declaration-mismatch]\n 24 | int hypotf (void);\n | ^~~~~~\n _configtest.c:24:5: note: ‘hypotf’ is declared in header ‘<math.h>’\n _configtest.c:25:5: warning: conflicting types for built-in function ‘atan2f’; expected ‘float(float, float)’ [-Wbuiltin-declaration-mismatch]\n 25 | int atan2f (void);\n | ^~~~~~\n _configtest.c:25:5: note: ‘atan2f’ is declared in header ‘<math.h>’\n _configtest.c:26:5: warning: conflicting types for built-in function ‘powf’; expected ‘float(float, float)’ [-Wbuiltin-declaration-mismatch]\n 26 | int powf (void);\n | ^~~~\n _configtest.c:26:5: note: ‘powf’ is declared in header ‘<math.h>’\n _configtest.c:27:5: warning: conflicting types for built-in function ‘fmodf’; expected ‘float(float, float)’ [-Wbuiltin-declaration-mismatch]\n 27 | int fmodf (void);\n | ^~~~~\n _configtest.c:27:5: note: ‘fmodf’ is declared in header ‘<math.h>’\n _configtest.c:28:5: warning: conflicting types for built-in function ‘modff’; expected ‘float(float, float *)’ [-Wbuiltin-declaration-mismatch]\n 28 | int modff (void);\n | ^~~~~\n _configtest.c:28:5: note: ‘modff’ is declared in header ‘<math.h>’\n _configtest.c:29:5: warning: conflicting types for built-in function ‘frexpf’; expected ‘float(float, int *)’ [-Wbuiltin-declaration-mismatch]\n 29 | int frexpf (void);\n | ^~~~~~\n _configtest.c:29:5: note: ‘frexpf’ is declared in header ‘<math.h>’\n _configtest.c:30:5: warning: conflicting types for built-in function ‘ldexpf’; expected ‘float(float, int)’ [-Wbuiltin-declaration-mismatch]\n 30 | int ldexpf (void);\n | ^~~~~~\n _configtest.c:30:5: note: ‘ldexpf’ is declared in header ‘<math.h>’\n _configtest.c:31:5: warning: conflicting types for built-in function ‘exp2f’; expected ‘float(float)’ [-Wbuiltin-declaration-mismatch]\n 31 | int exp2f (void);\n | ^~~~~\n _configtest.c:31:5: note: ‘exp2f’ is declared in header ‘<math.h>’\n _configtest.c:32:5: warning: conflicting types for built-in function ‘log2f’; expected ‘float(float)’ [-Wbuiltin-declaration-mismatch]\n 32 | int log2f (void);\n | ^~~~~\n _configtest.c:32:5: note: ‘log2f’ is declared in header ‘<math.h>’\n _configtest.c:33:5: warning: conflicting types for built-in function ‘copysignf’; expected ‘float(float, float)’ [-Wbuiltin-declaration-mismatch]\n 33 | int copysignf (void);\n | ^~~~~~~~~\n _configtest.c:33:5: note: ‘copysignf’ is declared in header ‘<math.h>’\n _configtest.c:34:5: warning: conflicting types for built-in function ‘nextafterf’; expected ‘float(float, float)’ [-Wbuiltin-declaration-mismatch]\n 34 | int nextafterf (void);\n | ^~~~~~~~~~\n _configtest.c:34:5: note: ‘nextafterf’ is declared in header ‘<math.h>’\n _configtest.c:35:5: warning: conflicting types for built-in function ‘cbrtf’; expected ‘float(float)’ [-Wbuiltin-declaration-mismatch]\n 35 | int cbrtf (void);\n | ^~~~~\n _configtest.c:35:5: note: ‘cbrtf’ is declared in header ‘<math.h>’\n gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ _configtest.o -lm -o _configtest\n success!\n removing: _configtest.c _configtest.o _configtest\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n _configtest.c:1:5: warning: conflicting types for built-in function ‘sinl’; expected ‘long double(long double)’ [-Wbuiltin-declaration-mismatch]\n 1 | int sinl (void);\n | ^~~~\n _configtest.c:1:1: note: ‘sinl’ is declared in header ‘<math.h>’\n +++ |+#include <math.h>\n 1 | int sinl (void);\n _configtest.c:2:5: warning: conflicting types for built-in function ‘cosl’; expected ‘long double(long double)’ [-Wbuiltin-declaration-mismatch]\n 2 | int cosl (void);\n | ^~~~\n _configtest.c:2:5: note: ‘cosl’ is declared in header ‘<math.h>’\n _configtest.c:3:5: warning: conflicting types for built-in function ‘tanl’; expected ‘long double(long double)’ [-Wbuiltin-declaration-mismatch]\n 3 | int tanl (void);\n | ^~~~\n _configtest.c:3:5: note: ‘tanl’ is declared in header ‘<math.h>’\n _configtest.c:4:5: warning: conflicting types for built-in function ‘sinhl’; expected ‘long double(long double)’ [-Wbuiltin-declaration-mismatch]\n 4 | int sinhl (void);\n | ^~~~~\n _configtest.c:4:5: note: ‘sinhl’ is declared in header ‘<math.h>’\n _configtest.c:5:5: warning: conflicting types for built-in function ‘coshl’; expected ‘long double(long double)’ [-Wbuiltin-declaration-mismatch]\n 5 | int coshl (void);\n | ^~~~~\n _configtest.c:5:5: note: ‘coshl’ is declared in header ‘<math.h>’\n _configtest.c:6:5: warning: conflicting types for built-in function ‘tanhl’; expected ‘long double(long double)’ [-Wbuiltin-declaration-mismatch]\n 6 | int tanhl (void);\n | ^~~~~\n _configtest.c:6:5: note: ‘tanhl’ is declared in header ‘<math.h>’\n _configtest.c:7:5: warning: conflicting types for built-in function ‘fabsl’; expected ‘long double(long double)’ [-Wbuiltin-declaration-mismatch]\n 7 | int fabsl (void);\n | ^~~~~\n _configtest.c:7:5: note: ‘fabsl’ is declared in header ‘<math.h>’\n _configtest.c:8:5: warning: conflicting types for built-in function ‘floorl’; expected ‘long double(long double)’ [-Wbuiltin-declaration-mismatch]\n 8 | int floorl (void);\n | ^~~~~~\n _configtest.c:8:5: note: ‘floorl’ is declared in header ‘<math.h>’\n _configtest.c:9:5: warning: conflicting types for built-in function ‘ceill’; expected ‘long double(long double)’ [-Wbuiltin-declaration-mismatch]\n 9 | int ceill (void);\n | ^~~~~\n _configtest.c:9:5: note: ‘ceill’ is declared in header ‘<math.h>’\n _configtest.c:10:5: warning: conflicting types for built-in function ‘rintl’; expected ‘long double(long double)’ [-Wbuiltin-declaration-mismatch]\n 10 | int rintl (void);\n | ^~~~~\n _configtest.c:10:5: note: ‘rintl’ is declared in header ‘<math.h>’\n _configtest.c:11:5: warning: conflicting types for built-in function ‘truncl’; expected ‘long double(long double)’ [-Wbuiltin-declaration-mismatch]\n 11 | int truncl (void);\n | ^~~~~~\n _configtest.c:11:5: note: ‘truncl’ is declared in header ‘<math.h>’\n _configtest.c:12:5: warning: conflicting types for built-in function ‘sqrtl’; expected ‘long double(long double)’ [-Wbuiltin-declaration-mismatch]\n 12 | int sqrtl (void);\n | ^~~~~\n _configtest.c:12:5: note: ‘sqrtl’ is declared in header ‘<math.h>’\n _configtest.c:13:5: warning: conflicting types for built-in function ‘log10l’; expected ‘long double(long double)’ [-Wbuiltin-declaration-mismatch]\n 13 | int log10l (void);\n | ^~~~~~\n _configtest.c:13:5: note: ‘log10l’ is declared in header ‘<math.h>’\n _configtest.c:14:5: warning: conflicting types for built-in function ‘logl’; expected ‘long double(long double)’ [-Wbuiltin-declaration-mismatch]\n 14 | int logl (void);\n | ^~~~\n _configtest.c:14:5: note: ‘logl’ is declared in header ‘<math.h>’\n _configtest.c:15:5: warning: conflicting types for built-in function ‘log1pl’; expected ‘long double(long double)’ [-Wbuiltin-declaration-mismatch]\n 15 | int log1pl (void);\n | ^~~~~~\n _configtest.c:15:5: note: ‘log1pl’ is declared in header ‘<math.h>’\n _configtest.c:16:5: warning: conflicting types for built-in function ‘expl’; expected ‘long double(long double)’ [-Wbuiltin-declaration-mismatch]\n 16 | int expl (void);\n | ^~~~\n _configtest.c:16:5: note: ‘expl’ is declared in header ‘<math.h>’\n _configtest.c:17:5: warning: conflicting types for built-in function ‘expm1l’; expected ‘long double(long double)’ [-Wbuiltin-declaration-mismatch]\n 17 | int expm1l (void);\n | ^~~~~~\n _configtest.c:17:5: note: ‘expm1l’ is declared in header ‘<math.h>’\n _configtest.c:18:5: warning: conflicting types for built-in function ‘asinl’; expected ‘long double(long double)’ [-Wbuiltin-declaration-mismatch]\n 18 | int asinl (void);\n | ^~~~~\n _configtest.c:18:5: note: ‘asinl’ is declared in header ‘<math.h>’\n _configtest.c:19:5: warning: conflicting types for built-in function ‘acosl’; expected ‘long double(long double)’ [-Wbuiltin-declaration-mismatch]\n 19 | int acosl (void);\n | ^~~~~\n _configtest.c:19:5: note: ‘acosl’ is declared in header ‘<math.h>’\n _configtest.c:20:5: warning: conflicting types for built-in function ‘atanl’; expected ‘long double(long double)’ [-Wbuiltin-declaration-mismatch]\n 20 | int atanl (void);\n | ^~~~~\n _configtest.c:20:5: note: ‘atanl’ is declared in header ‘<math.h>’\n _configtest.c:21:5: warning: conflicting types for built-in function ‘asinhl’; expected ‘long double(long double)’ [-Wbuiltin-declaration-mismatch]\n 21 | int asinhl (void);\n | ^~~~~~\n _configtest.c:21:5: note: ‘asinhl’ is declared in header ‘<math.h>’\n _configtest.c:22:5: warning: conflicting types for built-in function ‘acoshl’; expected ‘long double(long double)’ [-Wbuiltin-declaration-mismatch]\n 22 | int acoshl (void);\n | ^~~~~~\n _configtest.c:22:5: note: ‘acoshl’ is declared in header ‘<math.h>’\n _configtest.c:23:5: warning: conflicting types for built-in function ‘atanhl’; expected ‘long double(long double)’ [-Wbuiltin-declaration-mismatch]\n 23 | int atanhl (void);\n | ^~~~~~\n _configtest.c:23:5: note: ‘atanhl’ is declared in header ‘<math.h>’\n _configtest.c:24:5: warning: conflicting types for built-in function ‘hypotl’; expected ‘long double(long double, long double)’ [-Wbuiltin-declaration-mismatch]\n 24 | int hypotl (void);\n | ^~~~~~\n _configtest.c:24:5: note: ‘hypotl’ is declared in header ‘<math.h>’\n _configtest.c:25:5: warning: conflicting types for built-in function ‘atan2l’; expected ‘long double(long double, long double)’ [-Wbuiltin-declaration-mismatch]\n 25 | int atan2l (void);\n | ^~~~~~\n _configtest.c:25:5: note: ‘atan2l’ is declared in header ‘<math.h>’\n _configtest.c:26:5: warning: conflicting types for built-in function ‘powl’; expected ‘long double(long double, long double)’ [-Wbuiltin-declaration-mismatch]\n 26 | int powl (void);\n | ^~~~\n _configtest.c:26:5: note: ‘powl’ is declared in header ‘<math.h>’\n _configtest.c:27:5: warning: conflicting types for built-in function ‘fmodl’; expected ‘long double(long double, long double)’ [-Wbuiltin-declaration-mismatch]\n 27 | int fmodl (void);\n | ^~~~~\n _configtest.c:27:5: note: ‘fmodl’ is declared in header ‘<math.h>’\n _configtest.c:28:5: warning: conflicting types for built-in function ‘modfl’; expected ‘long double(long double, long double *)’ [-Wbuiltin-declaration-mismatch]\n 28 | int modfl (void);\n | ^~~~~\n _configtest.c:28:5: note: ‘modfl’ is declared in header ‘<math.h>’\n _configtest.c:29:5: warning: conflicting types for built-in function ‘frexpl’; expected ‘long double(long double, int *)’ [-Wbuiltin-declaration-mismatch]\n 29 | int frexpl (void);\n | ^~~~~~\n _configtest.c:29:5: note: ‘frexpl’ is declared in header ‘<math.h>’\n _configtest.c:30:5: warning: conflicting types for built-in function ‘ldexpl’; expected ‘long double(long double, int)’ [-Wbuiltin-declaration-mismatch]\n 30 | int ldexpl (void);\n | ^~~~~~\n _configtest.c:30:5: note: ‘ldexpl’ is declared in header ‘<math.h>’\n _configtest.c:31:5: warning: conflicting types for built-in function ‘exp2l’; expected ‘long double(long double)’ [-Wbuiltin-declaration-mismatch]\n 31 | int exp2l (void);\n | ^~~~~\n _configtest.c:31:5: note: ‘exp2l’ is declared in header ‘<math.h>’\n _configtest.c:32:5: warning: conflicting types for built-in function ‘log2l’; expected ‘long double(long double)’ [-Wbuiltin-declaration-mismatch]\n 32 | int log2l (void);\n | ^~~~~\n _configtest.c:32:5: note: ‘log2l’ is declared in header ‘<math.h>’\n _configtest.c:33:5: warning: conflicting types for built-in function ‘copysignl’; expected ‘long double(long double, long double)’ [-Wbuiltin-declaration-mismatch]\n 33 | int copysignl (void);\n | ^~~~~~~~~\n _configtest.c:33:5: note: ‘copysignl’ is declared in header ‘<math.h>’\n _configtest.c:34:5: warning: conflicting types for built-in function ‘nextafterl’; expected ‘long double(long double, long double)’ [-Wbuiltin-declaration-mismatch]\n 34 | int nextafterl (void);\n | ^~~~~~~~~~\n _configtest.c:34:5: note: ‘nextafterl’ is declared in header ‘<math.h>’\n _configtest.c:35:5: warning: conflicting types for built-in function ‘cbrtl’; expected ‘long double(long double)’ [-Wbuiltin-declaration-mismatch]\n 35 | int cbrtl (void);\n | ^~~~~\n _configtest.c:35:5: note: ‘cbrtl’ is declared in header ‘<math.h>’\n gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ _configtest.o -lm -o _configtest\n success!\n removing: _configtest.c _configtest.o _configtest\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n success!\n removing: _configtest.c _configtest.o\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n success!\n removing: _configtest.c _configtest.o\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n success!\n removing: _configtest.c _configtest.o\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n success!\n removing: _configtest.c _configtest.o\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n _configtest.c: In function ‘main’:\n _configtest.c:8:12: error: ‘HAVE_DECL_SIGNBIT’ undeclared (first use in this function); did you mean ‘HAVE_DECL_ISNAN’?\n 8 | (void) HAVE_DECL_SIGNBIT;\n | ^~~~~~~~~~~~~~~~~\n | HAVE_DECL_ISNAN\n _configtest.c:8:12: note: each undeclared identifier is reported only once for each function it appears in\n _configtest.c: In function ‘main’:\n _configtest.c:8:12: error: ‘HAVE_DECL_SIGNBIT’ undeclared (first use in this function); did you mean ‘HAVE_DECL_ISNAN’?\n 8 | (void) HAVE_DECL_SIGNBIT;\n | ^~~~~~~~~~~~~~~~~\n | HAVE_DECL_ISNAN\n _configtest.c:8:12: note: each undeclared identifier is reported only once for each function it appears in\n failure.\n removing: _configtest.c _configtest.o\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n success!\n removing: _configtest.c _configtest.o\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n success!\n removing: _configtest.c _configtest.o\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n success!\n removing: _configtest.c _configtest.o\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n success!\n removing: _configtest.c _configtest.o\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n removing: _configtest.c _configtest.o\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n removing: _configtest.c _configtest.o\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n removing: _configtest.c _configtest.o\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n _configtest.c:1:5: warning: conflicting types for built-in function ‘cabs’; expected ‘double(_Complex double)’ [-Wbuiltin-declaration-mismatch]\n 1 | int cabs (void);\n | ^~~~\n _configtest.c:1:1: note: ‘cabs’ is declared in header ‘<complex.h>’\n +++ |+#include <complex.h>\n 1 | int cabs (void);\n _configtest.c:2:5: warning: conflicting types for built-in function ‘cacos’; expected ‘_Complex double(_Complex double)’ [-Wbuiltin-declaration-mismatch]\n 2 | int cacos (void);\n | ^~~~~\n _configtest.c:2:5: note: ‘cacos’ is declared in header ‘<complex.h>’\n _configtest.c:3:5: warning: conflicting types for built-in function ‘cacosh’; expected ‘_Complex double(_Complex double)’ [-Wbuiltin-declaration-mismatch]\n 3 | int cacosh (void);\n | ^~~~~~\n _configtest.c:3:5: note: ‘cacosh’ is declared in header ‘<complex.h>’\n _configtest.c:4:5: warning: conflicting types for built-in function ‘carg’; expected ‘double(_Complex double)’ [-Wbuiltin-declaration-mismatch]\n 4 | int carg (void);\n | ^~~~\n _configtest.c:4:5: note: ‘carg’ is declared in header ‘<complex.h>’\n _configtest.c:5:5: warning: conflicting types for built-in function ‘casin’; expected ‘_Complex double(_Complex double)’ [-Wbuiltin-declaration-mismatch]\n 5 | int casin (void);\n | ^~~~~\n _configtest.c:5:5: note: ‘casin’ is declared in header ‘<complex.h>’\n _configtest.c:6:5: warning: conflicting types for built-in function ‘casinh’; expected ‘_Complex double(_Complex double)’ [-Wbuiltin-declaration-mismatch]\n 6 | int casinh (void);\n | ^~~~~~\n _configtest.c:6:5: note: ‘casinh’ is declared in header ‘<complex.h>’\n _configtest.c:7:5: warning: conflicting types for built-in function ‘catan’; expected ‘_Complex double(_Complex double)’ [-Wbuiltin-declaration-mismatch]\n 7 | int catan (void);\n | ^~~~~\n _configtest.c:7:5: note: ‘catan’ is declared in header ‘<complex.h>’\n _configtest.c:8:5: warning: conflicting types for built-in function ‘catanh’; expected ‘_Complex double(_Complex double)’ [-Wbuiltin-declaration-mismatch]\n 8 | int catanh (void);\n | ^~~~~~\n _configtest.c:8:5: note: ‘catanh’ is declared in header ‘<complex.h>’\n _configtest.c:9:5: warning: conflicting types for built-in function ‘ccos’; expected ‘_Complex double(_Complex double)’ [-Wbuiltin-declaration-mismatch]\n 9 | int ccos (void);\n | ^~~~\n _configtest.c:9:5: note: ‘ccos’ is declared in header ‘<complex.h>’\n _configtest.c:10:5: warning: conflicting types for built-in function ‘ccosh’; expected ‘_Complex double(_Complex double)’ [-Wbuiltin-declaration-mismatch]\n 10 | int ccosh (void);\n | ^~~~~\n _configtest.c:10:5: note: ‘ccosh’ is declared in header ‘<complex.h>’\n _configtest.c:11:5: warning: conflicting types for built-in function ‘cexp’; expected ‘_Complex double(_Complex double)’ [-Wbuiltin-declaration-mismatch]\n 11 | int cexp (void);\n | ^~~~\n _configtest.c:11:5: note: ‘cexp’ is declared in header ‘<complex.h>’\n _configtest.c:12:5: warning: conflicting types for built-in function ‘cimag’; expected ‘double(_Complex double)’ [-Wbuiltin-declaration-mismatch]\n 12 | int cimag (void);\n | ^~~~~\n _configtest.c:12:5: note: ‘cimag’ is declared in header ‘<complex.h>’\n _configtest.c:13:5: warning: conflicting types for built-in function ‘clog’; expected ‘_Complex double(_Complex double)’ [-Wbuiltin-declaration-mismatch]\n 13 | int clog (void);\n | ^~~~\n _configtest.c:13:5: note: ‘clog’ is declared in header ‘<complex.h>’\n _configtest.c:14:5: warning: conflicting types for built-in function ‘conj’; expected ‘_Complex double(_Complex double)’ [-Wbuiltin-declaration-mismatch]\n 14 | int conj (void);\n | ^~~~\n _configtest.c:14:5: note: ‘conj’ is declared in header ‘<complex.h>’\n _configtest.c:15:5: warning: conflicting types for built-in function ‘cpow’; expected ‘_Complex double(_Complex double, _Complex double)’ [-Wbuiltin-declaration-mismatch]\n 15 | int cpow (void);\n | ^~~~\n _configtest.c:15:5: note: ‘cpow’ is declared in header ‘<complex.h>’\n _configtest.c:16:5: warning: conflicting types for built-in function ‘cproj’; expected ‘_Complex double(_Complex double)’ [-Wbuiltin-declaration-mismatch]\n 16 | int cproj (void);\n | ^~~~~\n _configtest.c:16:5: note: ‘cproj’ is declared in header ‘<complex.h>’\n _configtest.c:17:5: warning: conflicting types for built-in function ‘creal’; expected ‘double(_Complex double)’ [-Wbuiltin-declaration-mismatch]\n 17 | int creal (void);\n | ^~~~~\n _configtest.c:17:5: note: ‘creal’ is declared in header ‘<complex.h>’\n _configtest.c:18:5: warning: conflicting types for built-in function ‘csin’; expected ‘_Complex double(_Complex double)’ [-Wbuiltin-declaration-mismatch]\n 18 | int csin (void);\n | ^~~~\n _configtest.c:18:5: note: ‘csin’ is declared in header ‘<complex.h>’\n _configtest.c:19:5: warning: conflicting types for built-in function ‘csinh’; expected ‘_Complex double(_Complex double)’ [-Wbuiltin-declaration-mismatch]\n 19 | int csinh (void);\n | ^~~~~\n _configtest.c:19:5: note: ‘csinh’ is declared in header ‘<complex.h>’\n _configtest.c:20:5: warning: conflicting types for built-in function ‘csqrt’; expected ‘_Complex double(_Complex double)’ [-Wbuiltin-declaration-mismatch]\n 20 | int csqrt (void);\n | ^~~~~\n _configtest.c:20:5: note: ‘csqrt’ is declared in header ‘<complex.h>’\n _configtest.c:21:5: warning: conflicting types for built-in function ‘ctan’; expected ‘_Complex double(_Complex double)’ [-Wbuiltin-declaration-mismatch]\n 21 | int ctan (void);\n | ^~~~\n _configtest.c:21:5: note: ‘ctan’ is declared in header ‘<complex.h>’\n _configtest.c:22:5: warning: conflicting types for built-in function ‘ctanh’; expected ‘_Complex double(_Complex double)’ [-Wbuiltin-declaration-mismatch]\n 22 | int ctanh (void);\n | ^~~~~\n _configtest.c:22:5: note: ‘ctanh’ is declared in header ‘<complex.h>’\n gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ _configtest.o -lm -o _configtest\n success!\n removing: _configtest.c _configtest.o _configtest\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n _configtest.c:1:5: warning: conflicting types for built-in function ‘cabsf’; expected ‘float(_Complex float)’ [-Wbuiltin-declaration-mismatch]\n 1 | int cabsf (void);\n | ^~~~~\n _configtest.c:1:1: note: ‘cabsf’ is declared in header ‘<complex.h>’\n +++ |+#include <complex.h>\n 1 | int cabsf (void);\n _configtest.c:2:5: warning: conflicting types for built-in function ‘cacosf’; expected ‘_Complex float(_Complex float)’ [-Wbuiltin-declaration-mismatch]\n 2 | int cacosf (void);\n | ^~~~~~\n _configtest.c:2:5: note: ‘cacosf’ is declared in header ‘<complex.h>’\n _configtest.c:3:5: warning: conflicting types for built-in function ‘cacoshf’; expected ‘_Complex float(_Complex float)’ [-Wbuiltin-declaration-mismatch]\n 3 | int cacoshf (void);\n | ^~~~~~~\n _configtest.c:3:5: note: ‘cacoshf’ is declared in header ‘<complex.h>’\n _configtest.c:4:5: warning: conflicting types for built-in function ‘cargf’; expected ‘float(_Complex float)’ [-Wbuiltin-declaration-mismatch]\n 4 | int cargf (void);\n | ^~~~~\n _configtest.c:4:5: note: ‘cargf’ is declared in header ‘<complex.h>’\n _configtest.c:5:5: warning: conflicting types for built-in function ‘casinf’; expected ‘_Complex float(_Complex float)’ [-Wbuiltin-declaration-mismatch]\n 5 | int casinf (void);\n | ^~~~~~\n _configtest.c:5:5: note: ‘casinf’ is declared in header ‘<complex.h>’\n _configtest.c:6:5: warning: conflicting types for built-in function ‘casinhf’; expected ‘_Complex float(_Complex float)’ [-Wbuiltin-declaration-mismatch]\n 6 | int casinhf (void);\n | ^~~~~~~\n _configtest.c:6:5: note: ‘casinhf’ is declared in header ‘<complex.h>’\n _configtest.c:7:5: warning: conflicting types for built-in function ‘catanf’; expected ‘_Complex float(_Complex float)’ [-Wbuiltin-declaration-mismatch]\n 7 | int catanf (void);\n | ^~~~~~\n _configtest.c:7:5: note: ‘catanf’ is declared in header ‘<complex.h>’\n _configtest.c:8:5: warning: conflicting types for built-in function ‘catanhf’; expected ‘_Complex float(_Complex float)’ [-Wbuiltin-declaration-mismatch]\n 8 | int catanhf (void);\n | ^~~~~~~\n _configtest.c:8:5: note: ‘catanhf’ is declared in header ‘<complex.h>’\n _configtest.c:9:5: warning: conflicting types for built-in function ‘ccosf’; expected ‘_Complex float(_Complex float)’ [-Wbuiltin-declaration-mismatch]\n 9 | int ccosf (void);\n | ^~~~~\n _configtest.c:9:5: note: ‘ccosf’ is declared in header ‘<complex.h>’\n _configtest.c:10:5: warning: conflicting types for built-in function ‘ccoshf’; expected ‘_Complex float(_Complex float)’ [-Wbuiltin-declaration-mismatch]\n 10 | int ccoshf (void);\n | ^~~~~~\n _configtest.c:10:5: note: ‘ccoshf’ is declared in header ‘<complex.h>’\n _configtest.c:11:5: warning: conflicting types for built-in function ‘cexpf’; expected ‘_Complex float(_Complex float)’ [-Wbuiltin-declaration-mismatch]\n 11 | int cexpf (void);\n | ^~~~~\n _configtest.c:11:5: note: ‘cexpf’ is declared in header ‘<complex.h>’\n _configtest.c:12:5: warning: conflicting types for built-in function ‘cimagf’; expected ‘float(_Complex float)’ [-Wbuiltin-declaration-mismatch]\n 12 | int cimagf (void);\n | ^~~~~~\n _configtest.c:12:5: note: ‘cimagf’ is declared in header ‘<complex.h>’\n _configtest.c:13:5: warning: conflicting types for built-in function ‘clogf’; expected ‘_Complex float(_Complex float)’ [-Wbuiltin-declaration-mismatch]\n 13 | int clogf (void);\n | ^~~~~\n _configtest.c:13:5: note: ‘clogf’ is declared in header ‘<complex.h>’\n _configtest.c:14:5: warning: conflicting types for built-in function ‘conjf’; expected ‘_Complex float(_Complex float)’ [-Wbuiltin-declaration-mismatch]\n 14 | int conjf (void);\n | ^~~~~\n _configtest.c:14:5: note: ‘conjf’ is declared in header ‘<complex.h>’\n _configtest.c:15:5: warning: conflicting types for built-in function ‘cpowf’; expected ‘_Complex float(_Complex float, _Complex float)’ [-Wbuiltin-declaration-mismatch]\n 15 | int cpowf (void);\n | ^~~~~\n _configtest.c:15:5: note: ‘cpowf’ is declared in header ‘<complex.h>’\n _configtest.c:16:5: warning: conflicting types for built-in function ‘cprojf’; expected ‘_Complex float(_Complex float)’ [-Wbuiltin-declaration-mismatch]\n 16 | int cprojf (void);\n | ^~~~~~\n _configtest.c:16:5: note: ‘cprojf’ is declared in header ‘<complex.h>’\n _configtest.c:17:5: warning: conflicting types for built-in function ‘crealf’; expected ‘float(_Complex float)’ [-Wbuiltin-declaration-mismatch]\n 17 | int crealf (void);\n | ^~~~~~\n _configtest.c:17:5: note: ‘crealf’ is declared in header ‘<complex.h>’\n _configtest.c:18:5: warning: conflicting types for built-in function ‘csinf’; expected ‘_Complex float(_Complex float)’ [-Wbuiltin-declaration-mismatch]\n 18 | int csinf (void);\n | ^~~~~\n _configtest.c:18:5: note: ‘csinf’ is declared in header ‘<complex.h>’\n _configtest.c:19:5: warning: conflicting types for built-in function ‘csinhf’; expected ‘_Complex float(_Complex float)’ [-Wbuiltin-declaration-mismatch]\n 19 | int csinhf (void);\n | ^~~~~~\n _configtest.c:19:5: note: ‘csinhf’ is declared in header ‘<complex.h>’\n _configtest.c:20:5: warning: conflicting types for built-in function ‘csqrtf’; expected ‘_Complex float(_Complex float)’ [-Wbuiltin-declaration-mismatch]\n 20 | int csqrtf (void);\n | ^~~~~~\n _configtest.c:20:5: note: ‘csqrtf’ is declared in header ‘<complex.h>’\n _configtest.c:21:5: warning: conflicting types for built-in function ‘ctanf’; expected ‘_Complex float(_Complex float)’ [-Wbuiltin-declaration-mismatch]\n 21 | int ctanf (void);\n | ^~~~~\n _configtest.c:21:5: note: ‘ctanf’ is declared in header ‘<complex.h>’\n _configtest.c:22:5: warning: conflicting types for built-in function ‘ctanhf’; expected ‘_Complex float(_Complex float)’ [-Wbuiltin-declaration-mismatch]\n 22 | int ctanhf (void);\n | ^~~~~~\n _configtest.c:22:5: note: ‘ctanhf’ is declared in header ‘<complex.h>’\n gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ _configtest.o -lm -o _configtest\n success!\n removing: _configtest.c _configtest.o _configtest\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n _configtest.c:1:5: warning: conflicting types for built-in function ‘cabsl’; expected ‘long double(_Complex long double)’ [-Wbuiltin-declaration-mismatch]\n 1 | int cabsl (void);\n | ^~~~~\n _configtest.c:1:1: note: ‘cabsl’ is declared in header ‘<complex.h>’\n +++ |+#include <complex.h>\n 1 | int cabsl (void);\n _configtest.c:2:5: warning: conflicting types for built-in function ‘cacosl’; expected ‘_Complex long double(_Complex long double)’ [-Wbuiltin-declaration-mismatch]\n 2 | int cacosl (void);\n | ^~~~~~\n _configtest.c:2:5: note: ‘cacosl’ is declared in header ‘<complex.h>’\n _configtest.c:3:5: warning: conflicting types for built-in function ‘cacoshl’; expected ‘_Complex long double(_Complex long double)’ [-Wbuiltin-declaration-mismatch]\n 3 | int cacoshl (void);\n | ^~~~~~~\n _configtest.c:3:5: note: ‘cacoshl’ is declared in header ‘<complex.h>’\n _configtest.c:4:5: warning: conflicting types for built-in function ‘cargl’; expected ‘long double(_Complex long double)’ [-Wbuiltin-declaration-mismatch]\n 4 | int cargl (void);\n | ^~~~~\n _configtest.c:4:5: note: ‘cargl’ is declared in header ‘<complex.h>’\n _configtest.c:5:5: warning: conflicting types for built-in function ‘casinl’; expected ‘_Complex long double(_Complex long double)’ [-Wbuiltin-declaration-mismatch]\n 5 | int casinl (void);\n | ^~~~~~\n _configtest.c:5:5: note: ‘casinl’ is declared in header ‘<complex.h>’\n _configtest.c:6:5: warning: conflicting types for built-in function ‘casinhl’; expected ‘_Complex long double(_Complex long double)’ [-Wbuiltin-declaration-mismatch]\n 6 | int casinhl (void);\n | ^~~~~~~\n _configtest.c:6:5: note: ‘casinhl’ is declared in header ‘<complex.h>’\n _configtest.c:7:5: warning: conflicting types for built-in function ‘catanl’; expected ‘_Complex long double(_Complex long double)’ [-Wbuiltin-declaration-mismatch]\n 7 | int catanl (void);\n | ^~~~~~\n _configtest.c:7:5: note: ‘catanl’ is declared in header ‘<complex.h>’\n _configtest.c:8:5: warning: conflicting types for built-in function ‘catanhl’; expected ‘_Complex long double(_Complex long double)’ [-Wbuiltin-declaration-mismatch]\n 8 | int catanhl (void);\n | ^~~~~~~\n _configtest.c:8:5: note: ‘catanhl’ is declared in header ‘<complex.h>’\n _configtest.c:9:5: warning: conflicting types for built-in function ‘ccosl’; expected ‘_Complex long double(_Complex long double)’ [-Wbuiltin-declaration-mismatch]\n 9 | int ccosl (void);\n | ^~~~~\n _configtest.c:9:5: note: ‘ccosl’ is declared in header ‘<complex.h>’\n _configtest.c:10:5: warning: conflicting types for built-in function ‘ccoshl’; expected ‘_Complex long double(_Complex long double)’ [-Wbuiltin-declaration-mismatch]\n 10 | int ccoshl (void);\n | ^~~~~~\n _configtest.c:10:5: note: ‘ccoshl’ is declared in header ‘<complex.h>’\n _configtest.c:11:5: warning: conflicting types for built-in function ‘cexpl’; expected ‘_Complex long double(_Complex long double)’ [-Wbuiltin-declaration-mismatch]\n 11 | int cexpl (void);\n | ^~~~~\n _configtest.c:11:5: note: ‘cexpl’ is declared in header ‘<complex.h>’\n _configtest.c:12:5: warning: conflicting types for built-in function ‘cimagl’; expected ‘long double(_Complex long double)’ [-Wbuiltin-declaration-mismatch]\n 12 | int cimagl (void);\n | ^~~~~~\n _configtest.c:12:5: note: ‘cimagl’ is declared in header ‘<complex.h>’\n _configtest.c:13:5: warning: conflicting types for built-in function ‘clogl’; expected ‘_Complex long double(_Complex long double)’ [-Wbuiltin-declaration-mismatch]\n 13 | int clogl (void);\n | ^~~~~\n _configtest.c:13:5: note: ‘clogl’ is declared in header ‘<complex.h>’\n _configtest.c:14:5: warning: conflicting types for built-in function ‘conjl’; expected ‘_Complex long double(_Complex long double)’ [-Wbuiltin-declaration-mismatch]\n 14 | int conjl (void);\n | ^~~~~\n _configtest.c:14:5: note: ‘conjl’ is declared in header ‘<complex.h>’\n _configtest.c:15:5: warning: conflicting types for built-in function ‘cpowl’; expected ‘_Complex long double(_Complex long double, _Complex long double)’ [-Wbuiltin-declaration-mismatch]\n 15 | int cpowl (void);\n | ^~~~~\n _configtest.c:15:5: note: ‘cpowl’ is declared in header ‘<complex.h>’\n _configtest.c:16:5: warning: conflicting types for built-in function ‘cprojl’; expected ‘_Complex long double(_Complex long double)’ [-Wbuiltin-declaration-mismatch]\n 16 | int cprojl (void);\n | ^~~~~~\n _configtest.c:16:5: note: ‘cprojl’ is declared in header ‘<complex.h>’\n _configtest.c:17:5: warning: conflicting types for built-in function ‘creall’; expected ‘long double(_Complex long double)’ [-Wbuiltin-declaration-mismatch]\n 17 | int creall (void);\n | ^~~~~~\n _configtest.c:17:5: note: ‘creall’ is declared in header ‘<complex.h>’\n _configtest.c:18:5: warning: conflicting types for built-in function ‘csinl’; expected ‘_Complex long double(_Complex long double)’ [-Wbuiltin-declaration-mismatch]\n 18 | int csinl (void);\n | ^~~~~\n _configtest.c:18:5: note: ‘csinl’ is declared in header ‘<complex.h>’\n _configtest.c:19:5: warning: conflicting types for built-in function ‘csinhl’; expected ‘_Complex long double(_Complex long double)’ [-Wbuiltin-declaration-mismatch]\n 19 | int csinhl (void);\n | ^~~~~~\n _configtest.c:19:5: note: ‘csinhl’ is declared in header ‘<complex.h>’\n _configtest.c:20:5: warning: conflicting types for built-in function ‘csqrtl’; expected ‘_Complex long double(_Complex long double)’ [-Wbuiltin-declaration-mismatch]\n 20 | int csqrtl (void);\n | ^~~~~~\n _configtest.c:20:5: note: ‘csqrtl’ is declared in header ‘<complex.h>’\n _configtest.c:21:5: warning: conflicting types for built-in function ‘ctanl’; expected ‘_Complex long double(_Complex long double)’ [-Wbuiltin-declaration-mismatch]\n 21 | int ctanl (void);\n | ^~~~~\n _configtest.c:21:5: note: ‘ctanl’ is declared in header ‘<complex.h>’\n _configtest.c:22:5: warning: conflicting types for built-in function ‘ctanhl’; expected ‘_Complex long double(_Complex long double)’ [-Wbuiltin-declaration-mismatch]\n 22 | int ctanhl (void);\n | ^~~~~~\n _configtest.c:22:5: note: ‘ctanhl’ is declared in header ‘<complex.h>’\n gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ _configtest.o -lm -o _configtest\n success!\n removing: _configtest.c _configtest.o _configtest\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n _configtest.c:2:12: warning: ‘static_func’ defined but not used [-Wunused-function]\n 2 | static int static_func (char * restrict a)\n | ^~~~~~~~~~~\n success!\n removing: _configtest.c _configtest.o\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n success!\n removing: _configtest.c _configtest.o\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n removing: _configtest.c _configtest.o\n File: build/src.linux-x86_64-3.7/numpy/core/include/numpy/config.h\n #define HAVE_ENDIAN_H 1\n #define SIZEOF_PY_INTPTR_T 8\n #define SIZEOF_OFF_T 8\n #define SIZEOF_PY_LONG_LONG 8\n #define MATHLIB m\n #define HAVE_SIN 1\n #define HAVE_COS 1\n #define HAVE_TAN 1\n #define HAVE_SINH 1\n #define HAVE_COSH 1\n #define HAVE_TANH 1\n #define HAVE_FABS 1\n #define HAVE_FLOOR 1\n #define HAVE_CEIL 1\n #define HAVE_SQRT 1\n #define HAVE_LOG10 1\n #define HAVE_LOG 1\n #define HAVE_EXP 1\n #define HAVE_ASIN 1\n #define HAVE_ACOS 1\n #define HAVE_ATAN 1\n #define HAVE_FMOD 1\n #define HAVE_MODF 1\n #define HAVE_FREXP 1\n #define HAVE_LDEXP 1\n #define HAVE_RINT 1\n #define HAVE_TRUNC 1\n #define HAVE_EXP2 1\n #define HAVE_LOG2 1\n #define HAVE_ATAN2 1\n #define HAVE_POW 1\n #define HAVE_NEXTAFTER 1\n #define HAVE_STRTOLL 1\n #define HAVE_STRTOULL 1\n #define HAVE_CBRT 1\n #define HAVE_STRTOLD_L 1\n #define HAVE_FALLOCATE 1\n #define HAVE_BACKTRACE 1\n #define HAVE_XMMINTRIN_H 1\n #define HAVE_EMMINTRIN_H 1\n #define HAVE_FEATURES_H 1\n #define HAVE_DLFCN_H 1\n #define HAVE___BUILTIN_ISNAN 1\n #define HAVE___BUILTIN_ISINF 1\n #define HAVE___BUILTIN_ISFINITE 1\n #define HAVE___BUILTIN_BSWAP32 1\n #define HAVE___BUILTIN_BSWAP64 1\n #define HAVE___BUILTIN_EXPECT 1\n #define HAVE___BUILTIN_MUL_OVERFLOW 1\n #define HAVE___BUILTIN_CPU_SUPPORTS 1\n #define HAVE__M_FROM_INT64 1\n #define HAVE__MM_LOAD_PS 1\n #define HAVE__MM_PREFETCH 1\n #define HAVE__MM_LOAD_PD 1\n #define HAVE___BUILTIN_PREFETCH 1\n #define HAVE_LINK_AVX 1\n #define HAVE_LINK_AVX2 1\n #define HAVE_ATTRIBUTE_OPTIMIZE_UNROLL_LOOPS 1\n #define HAVE_ATTRIBUTE_OPTIMIZE_OPT_3 1\n #define HAVE_ATTRIBUTE_NONNULL 1\n #define HAVE_ATTRIBUTE_TARGET_AVX 1\n #define HAVE_ATTRIBUTE_TARGET_AVX2 1\n #define HAVE___THREAD 1\n #define HAVE_SINF 1\n #define HAVE_COSF 1\n #define HAVE_TANF 1\n #define HAVE_SINHF 1\n #define HAVE_COSHF 1\n #define HAVE_TANHF 1\n #define HAVE_FABSF 1\n #define HAVE_FLOORF 1\n #define HAVE_CEILF 1\n #define HAVE_RINTF 1\n #define HAVE_TRUNCF 1\n #define HAVE_SQRTF 1\n #define HAVE_LOG10F 1\n #define HAVE_LOGF 1\n #define HAVE_LOG1PF 1\n #define HAVE_EXPF 1\n #define HAVE_EXPM1F 1\n #define HAVE_ASINF 1\n #define HAVE_ACOSF 1\n #define HAVE_ATANF 1\n #define HAVE_ASINHF 1\n #define HAVE_ACOSHF 1\n #define HAVE_ATANHF 1\n #define HAVE_HYPOTF 1\n #define HAVE_ATAN2F 1\n #define HAVE_POWF 1\n #define HAVE_FMODF 1\n #define HAVE_MODFF 1\n #define HAVE_FREXPF 1\n #define HAVE_LDEXPF 1\n #define HAVE_EXP2F 1\n #define HAVE_LOG2F 1\n #define HAVE_COPYSIGNF 1\n #define HAVE_NEXTAFTERF 1\n #define HAVE_CBRTF 1\n #define HAVE_SINL 1\n #define HAVE_COSL 1\n #define HAVE_TANL 1\n #define HAVE_SINHL 1\n #define HAVE_COSHL 1\n #define HAVE_TANHL 1\n #define HAVE_FABSL 1\n #define HAVE_FLOORL 1\n #define HAVE_CEILL 1\n #define HAVE_RINTL 1\n #define HAVE_TRUNCL 1\n #define HAVE_SQRTL 1\n #define HAVE_LOG10L 1\n #define HAVE_LOGL 1\n #define HAVE_LOG1PL 1\n #define HAVE_EXPL 1\n #define HAVE_EXPM1L 1\n #define HAVE_ASINL 1\n #define HAVE_ACOSL 1\n #define HAVE_ATANL 1\n #define HAVE_ASINHL 1\n #define HAVE_ACOSHL 1\n #define HAVE_ATANHL 1\n #define HAVE_HYPOTL 1\n #define HAVE_ATAN2L 1\n #define HAVE_POWL 1\n #define HAVE_FMODL 1\n #define HAVE_MODFL 1\n #define HAVE_FREXPL 1\n #define HAVE_LDEXPL 1\n #define HAVE_EXP2L 1\n #define HAVE_LOG2L 1\n #define HAVE_COPYSIGNL 1\n #define HAVE_NEXTAFTERL 1\n #define HAVE_CBRTL 1\n #define HAVE_DECL_SIGNBIT\n #define HAVE_COMPLEX_H 1\n #define HAVE_CABS 1\n #define HAVE_CACOS 1\n #define HAVE_CACOSH 1\n #define HAVE_CARG 1\n #define HAVE_CASIN 1\n #define HAVE_CASINH 1\n #define HAVE_CATAN 1\n #define HAVE_CATANH 1\n #define HAVE_CCOS 1\n #define HAVE_CCOSH 1\n #define HAVE_CEXP 1\n #define HAVE_CIMAG 1\n #define HAVE_CLOG 1\n #define HAVE_CONJ 1\n #define HAVE_CPOW 1\n #define HAVE_CPROJ 1\n #define HAVE_CREAL 1\n #define HAVE_CSIN 1\n #define HAVE_CSINH 1\n #define HAVE_CSQRT 1\n #define HAVE_CTAN 1\n #define HAVE_CTANH 1\n #define HAVE_CABSF 1\n #define HAVE_CACOSF 1\n #define HAVE_CACOSHF 1\n #define HAVE_CARGF 1\n #define HAVE_CASINF 1\n #define HAVE_CASINHF 1\n #define HAVE_CATANF 1\n #define HAVE_CATANHF 1\n #define HAVE_CCOSF 1\n #define HAVE_CCOSHF 1\n #define HAVE_CEXPF 1\n #define HAVE_CIMAGF 1\n #define HAVE_CLOGF 1\n #define HAVE_CONJF 1\n #define HAVE_CPOWF 1\n #define HAVE_CPROJF 1\n #define HAVE_CREALF 1\n #define HAVE_CSINF 1\n #define HAVE_CSINHF 1\n #define HAVE_CSQRTF 1\n #define HAVE_CTANF 1\n #define HAVE_CTANHF 1\n #define HAVE_CABSL 1\n #define HAVE_CACOSL 1\n #define HAVE_CACOSHL 1\n #define HAVE_CARGL 1\n #define HAVE_CASINL 1\n #define HAVE_CASINHL 1\n #define HAVE_CATANL 1\n #define HAVE_CATANHL 1\n #define HAVE_CCOSL 1\n #define HAVE_CCOSHL 1\n #define HAVE_CEXPL 1\n #define HAVE_CIMAGL 1\n #define HAVE_CLOGL 1\n #define HAVE_CONJL 1\n #define HAVE_CPOWL 1\n #define HAVE_CPROJL 1\n #define HAVE_CREALL 1\n #define HAVE_CSINL 1\n #define HAVE_CSINHL 1\n #define HAVE_CSQRTL 1\n #define HAVE_CTANL 1\n #define HAVE_CTANHL 1\n #define NPY_RESTRICT restrict\n #define NPY_RELAXED_STRIDES_CHECKING 1\n #define HAVE_LDOUBLE_INTEL_EXTENDED_16_BYTES_LE 1\n #define NPY_PY3K 1\n #ifndef __cplusplus\n /* #undef inline */\n #endif\n \n #ifndef _NPY_NPY_CONFIG_H_\n #error config.h should never be included directly, include npy_config.h instead\n #endif\n \n EOF\n adding 'build/src.linux-x86_64-3.7/numpy/core/include/numpy/config.h' to sources.\n Generating build/src.linux-x86_64-3.7/numpy/core/include/numpy/_numpyconfig.h\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n _configtest.c:1:5: warning: conflicting types for built-in function ‘exp’; expected ‘double(double)’ [-Wbuiltin-declaration-mismatch]\n 1 | int exp (void);\n | ^~~\n _configtest.c:1:1: note: ‘exp’ is declared in header ‘<math.h>’\n +++ |+#include <math.h>\n 1 | int exp (void);\n gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ _configtest.o -o _configtest\n /home/masrur/anaconda3/compiler_compat/ld: _configtest.o: in function `main':\n /tmp/pip-install-wkrshu4d/numpy/_configtest.c:6: undefined reference to `exp'\n collect2: error: ld returned 1 exit status\n /home/masrur/anaconda3/compiler_compat/ld: _configtest.o: in function `main':\n /tmp/pip-install-wkrshu4d/numpy/_configtest.c:6: undefined reference to `exp'\n collect2: error: ld returned 1 exit status\n failure.\n removing: _configtest.c _configtest.o\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n _configtest.c:1:5: warning: conflicting types for built-in function ‘exp’; expected ‘double(double)’ [-Wbuiltin-declaration-mismatch]\n 1 | int exp (void);\n | ^~~\n _configtest.c:1:1: note: ‘exp’ is declared in header ‘<math.h>’\n +++ |+#include <math.h>\n 1 | int exp (void);\n gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ _configtest.o -lm -o _configtest\n success!\n removing: _configtest.c _configtest.o _configtest\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n success!\n removing: _configtest.c _configtest.o\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n _configtest.c:3:1: warning: function declaration isn’t a prototype [-Wstrict-prototypes]\n 3 | main()\n | ^~~~\n success!\n removing: _configtest.c _configtest.o\n File: build/src.linux-x86_64-3.7/numpy/core/include/numpy/_numpyconfig.h\n #define NPY_HAVE_ENDIAN_H 1\n #define NPY_SIZEOF_SHORT SIZEOF_SHORT\n #define NPY_SIZEOF_INT SIZEOF_INT\n #define NPY_SIZEOF_LONG SIZEOF_LONG\n #define NPY_SIZEOF_FLOAT 4\n #define NPY_SIZEOF_COMPLEX_FLOAT 8\n #define NPY_SIZEOF_DOUBLE 8\n #define NPY_SIZEOF_COMPLEX_DOUBLE 16\n #define NPY_SIZEOF_LONGDOUBLE 16\n #define NPY_SIZEOF_COMPLEX_LONGDOUBLE 32\n #define NPY_SIZEOF_PY_INTPTR_T 8\n #define NPY_SIZEOF_OFF_T 8\n #define NPY_SIZEOF_PY_LONG_LONG 8\n #define NPY_SIZEOF_LONGLONG 8\n #define NPY_NO_SMP 0\n #define NPY_HAVE_DECL_ISNAN\n #define NPY_HAVE_DECL_ISINF\n #define NPY_HAVE_DECL_ISFINITE\n #define NPY_HAVE_DECL_SIGNBIT\n #define NPY_USE_C99_COMPLEX 1\n #define NPY_HAVE_COMPLEX_DOUBLE 1\n #define NPY_HAVE_COMPLEX_FLOAT 1\n #define NPY_HAVE_COMPLEX_LONG_DOUBLE 1\n #define NPY_RELAXED_STRIDES_CHECKING 1\n #define NPY_USE_C99_FORMATS 1\n #define NPY_VISIBILITY_HIDDEN __attribute__((visibility(\"hidden\")))\n #define NPY_ABI_VERSION 0x01000009\n #define NPY_API_VERSION 0x0000000B\n \n #ifndef __STDC_FORMAT_MACROS\n #define __STDC_FORMAT_MACROS 1\n #endif\n \n EOF\n adding 'build/src.linux-x86_64-3.7/numpy/core/include/numpy/_numpyconfig.h' to sources.\n executing numpy/core/code_generators/generate_numpy_api.py\n adding 'build/src.linux-x86_64-3.7/numpy/core/include/numpy/__multiarray_api.h' to sources.\n numpy.core - nothing done with h_files = ['build/src.linux-x86_64-3.7/numpy/core/include/numpy/config.h', 'build/src.linux-x86_64-3.7/numpy/core/include/numpy/_numpyconfig.h', 'build/src.linux-x86_64-3.7/numpy/core/include/numpy/__multiarray_api.h']\n building extension \"numpy.core.multiarray\" sources\n adding 'build/src.linux-x86_64-3.7/numpy/core/include/numpy/config.h' to sources.\n adding 'build/src.linux-x86_64-3.7/numpy/core/include/numpy/_numpyconfig.h' to sources.\n executing numpy/core/code_generators/generate_numpy_api.py\n adding 'build/src.linux-x86_64-3.7/numpy/core/include/numpy/__multiarray_api.h' to sources.\n creating build/src.linux-x86_64-3.7/numpy/core/src/multiarray\n conv_template:> build/src.linux-x86_64-3.7/numpy/core/src/multiarray/arraytypes.c\n conv_template:> build/src.linux-x86_64-3.7/numpy/core/src/multiarray/einsum.c\n conv_template:> build/src.linux-x86_64-3.7/numpy/core/src/multiarray/lowlevel_strided_loops.c\n conv_template:> build/src.linux-x86_64-3.7/numpy/core/src/multiarray/nditer_templ.c\n conv_template:> build/src.linux-x86_64-3.7/numpy/core/src/multiarray/scalartypes.c\n conv_template:> build/src.linux-x86_64-3.7/numpy/core/src/private/templ_common.h\n adding 'build/src.linux-x86_64-3.7/numpy/core/src/private' to include_dirs.\n numpy.core - nothing done with h_files = ['build/src.linux-x86_64-3.7/numpy/core/src/private/templ_common.h', 'build/src.linux-x86_64-3.7/numpy/core/include/numpy/config.h', 'build/src.linux-x86_64-3.7/numpy/core/include/numpy/_numpyconfig.h', 'build/src.linux-x86_64-3.7/numpy/core/include/numpy/__multiarray_api.h']\n building extension \"numpy.core.umath\" sources\n adding 'build/src.linux-x86_64-3.7/numpy/core/include/numpy/config.h' to sources.\n adding 'build/src.linux-x86_64-3.7/numpy/core/include/numpy/_numpyconfig.h' to sources.\n executing numpy/core/code_generators/generate_ufunc_api.py\n adding 'build/src.linux-x86_64-3.7/numpy/core/include/numpy/__ufunc_api.h' to sources.\n creating build/src.linux-x86_64-3.7/numpy/core/src/umath\n conv_template:> build/src.linux-x86_64-3.7/numpy/core/src/umath/funcs.inc\n adding 'build/src.linux-x86_64-3.7/numpy/core/src/umath' to include_dirs.\n conv_template:> build/src.linux-x86_64-3.7/numpy/core/src/umath/simd.inc\n conv_template:> build/src.linux-x86_64-3.7/numpy/core/src/umath/loops.h\n conv_template:> build/src.linux-x86_64-3.7/numpy/core/src/umath/loops.c\n conv_template:> build/src.linux-x86_64-3.7/numpy/core/src/umath/scalarmath.c\n numpy.core - nothing done with h_files = ['build/src.linux-x86_64-3.7/numpy/core/src/umath/funcs.inc', 'build/src.linux-x86_64-3.7/numpy/core/src/umath/simd.inc', 'build/src.linux-x86_64-3.7/numpy/core/src/umath/loops.h', 'build/src.linux-x86_64-3.7/numpy/core/include/numpy/config.h', 'build/src.linux-x86_64-3.7/numpy/core/include/numpy/_numpyconfig.h', 'build/src.linux-x86_64-3.7/numpy/core/include/numpy/__ufunc_api.h']\n building extension \"numpy.core.umath_tests\" sources\n conv_template:> build/src.linux-x86_64-3.7/numpy/core/src/umath/umath_tests.c\n building extension \"numpy.core.test_rational\" sources\n conv_template:> build/src.linux-x86_64-3.7/numpy/core/src/umath/test_rational.c\n building extension \"numpy.core.struct_ufunc_test\" sources\n conv_template:> build/src.linux-x86_64-3.7/numpy/core/src/umath/struct_ufunc_test.c\n building extension \"numpy.core.multiarray_tests\" sources\n conv_template:> build/src.linux-x86_64-3.7/numpy/core/src/multiarray/multiarray_tests.c\n building extension \"numpy.core.operand_flag_tests\" sources\n conv_template:> build/src.linux-x86_64-3.7/numpy/core/src/umath/operand_flag_tests.c\n building extension \"numpy.fft.fftpack_lite\" sources\n building extension \"numpy.linalg.lapack_lite\" sources\n creating build/src.linux-x86_64-3.7/numpy/linalg\n adding 'numpy/linalg/lapack_lite/python_xerbla.c' to sources.\n building extension \"numpy.linalg._umath_linalg\" sources\n adding 'numpy/linalg/lapack_lite/python_xerbla.c' to sources.\n conv_template:> build/src.linux-x86_64-3.7/numpy/linalg/umath_linalg.c\n building extension \"numpy.random.mtrand\" sources\n creating build/src.linux-x86_64-3.7/numpy/random\n building data_files sources\n build_src: building npy-pkg config files\n running build_py\n creating build/lib.linux-x86_64-3.7\n creating build/lib.linux-x86_64-3.7/numpy\n copying numpy/matlib.py -> build/lib.linux-x86_64-3.7/numpy\n copying numpy/__init__.py -> build/lib.linux-x86_64-3.7/numpy\n copying numpy/setup.py -> build/lib.linux-x86_64-3.7/numpy\n copying numpy/ctypeslib.py -> build/lib.linux-x86_64-3.7/numpy\n copying numpy/_globals.py -> build/lib.linux-x86_64-3.7/numpy\n copying numpy/version.py -> build/lib.linux-x86_64-3.7/numpy\n copying numpy/_distributor_init.py -> build/lib.linux-x86_64-3.7/numpy\n copying numpy/_import_tools.py -> build/lib.linux-x86_64-3.7/numpy\n copying numpy/dual.py -> build/lib.linux-x86_64-3.7/numpy\n copying numpy/add_newdocs.py -> build/lib.linux-x86_64-3.7/numpy\n copying build/src.linux-x86_64-3.7/numpy/__config__.py -> build/lib.linux-x86_64-3.7/numpy\n creating build/lib.linux-x86_64-3.7/numpy/compat\n copying numpy/compat/__init__.py -> build/lib.linux-x86_64-3.7/numpy/compat\n copying numpy/compat/setup.py -> build/lib.linux-x86_64-3.7/numpy/compat\n copying numpy/compat/_inspect.py -> build/lib.linux-x86_64-3.7/numpy/compat\n copying numpy/compat/py3k.py -> build/lib.linux-x86_64-3.7/numpy/compat\n creating build/lib.linux-x86_64-3.7/numpy/core\n copying numpy/core/_methods.py -> build/lib.linux-x86_64-3.7/numpy/core\n copying numpy/core/arrayprint.py -> build/lib.linux-x86_64-3.7/numpy/core\n copying numpy/core/numeric.py -> build/lib.linux-x86_64-3.7/numpy/core\n copying numpy/core/numerictypes.py -> build/lib.linux-x86_64-3.7/numpy/core\n copying numpy/core/__init__.py -> build/lib.linux-x86_64-3.7/numpy/core\n copying numpy/core/fromnumeric.py -> build/lib.linux-x86_64-3.7/numpy/core\n copying numpy/core/function_base.py -> build/lib.linux-x86_64-3.7/numpy/core\n copying numpy/core/setup.py -> build/lib.linux-x86_64-3.7/numpy/core\n copying numpy/core/einsumfunc.py -> build/lib.linux-x86_64-3.7/numpy/core\n copying numpy/core/defchararray.py -> build/lib.linux-x86_64-3.7/numpy/core\n copying numpy/core/machar.py -> build/lib.linux-x86_64-3.7/numpy/core\n copying numpy/core/getlimits.py -> build/lib.linux-x86_64-3.7/numpy/core\n copying numpy/core/memmap.py -> build/lib.linux-x86_64-3.7/numpy/core\n copying numpy/core/records.py -> build/lib.linux-x86_64-3.7/numpy/core\n copying numpy/core/shape_base.py -> build/lib.linux-x86_64-3.7/numpy/core\n copying numpy/core/_internal.py -> build/lib.linux-x86_64-3.7/numpy/core\n copying numpy/core/setup_common.py -> build/lib.linux-x86_64-3.7/numpy/core\n copying numpy/core/info.py -> build/lib.linux-x86_64-3.7/numpy/core\n copying numpy/core/cversions.py -> build/lib.linux-x86_64-3.7/numpy/core\n copying numpy/core/code_generators/generate_numpy_api.py -> build/lib.linux-x86_64-3.7/numpy/core\n creating build/lib.linux-x86_64-3.7/numpy/distutils\n copying numpy/distutils/compat.py -> build/lib.linux-x86_64-3.7/numpy/distutils\n copying numpy/distutils/from_template.py -> build/lib.linux-x86_64-3.7/numpy/distutils\n copying numpy/distutils/numpy_distribution.py -> build/lib.linux-x86_64-3.7/numpy/distutils\n copying numpy/distutils/system_info.py -> build/lib.linux-x86_64-3.7/numpy/distutils\n copying numpy/distutils/unixccompiler.py -> build/lib.linux-x86_64-3.7/numpy/distutils\n copying numpy/distutils/__version__.py -> build/lib.linux-x86_64-3.7/numpy/distutils\n copying numpy/distutils/log.py -> build/lib.linux-x86_64-3.7/numpy/distutils\n copying numpy/distutils/__init__.py -> build/lib.linux-x86_64-3.7/numpy/distutils\n copying numpy/distutils/setup.py -> build/lib.linux-x86_64-3.7/numpy/distutils\n copying numpy/distutils/lib2def.py -> build/lib.linux-x86_64-3.7/numpy/distutils\n copying numpy/distutils/msvc9compiler.py -> build/lib.linux-x86_64-3.7/numpy/distutils\n copying numpy/distutils/misc_util.py -> build/lib.linux-x86_64-3.7/numpy/distutils\n copying numpy/distutils/conv_template.py -> build/lib.linux-x86_64-3.7/numpy/distutils\n copying numpy/distutils/npy_pkg_config.py -> build/lib.linux-x86_64-3.7/numpy/distutils\n copying numpy/distutils/pathccompiler.py -> build/lib.linux-x86_64-3.7/numpy/distutils\n copying numpy/distutils/exec_command.py -> build/lib.linux-x86_64-3.7/numpy/distutils\n copying numpy/distutils/msvccompiler.py -> build/lib.linux-x86_64-3.7/numpy/distutils\n copying numpy/distutils/environment.py -> build/lib.linux-x86_64-3.7/numpy/distutils\n copying numpy/distutils/line_endings.py -> build/lib.linux-x86_64-3.7/numpy/distutils\n copying numpy/distutils/core.py -> build/lib.linux-x86_64-3.7/numpy/distutils\n copying numpy/distutils/ccompiler.py -> build/lib.linux-x86_64-3.7/numpy/distutils\n copying numpy/distutils/extension.py -> build/lib.linux-x86_64-3.7/numpy/distutils\n copying numpy/distutils/cpuinfo.py -> build/lib.linux-x86_64-3.7/numpy/distutils\n copying numpy/distutils/mingw32ccompiler.py -> build/lib.linux-x86_64-3.7/numpy/distutils\n copying numpy/distutils/intelccompiler.py -> build/lib.linux-x86_64-3.7/numpy/distutils\n copying numpy/distutils/info.py -> build/lib.linux-x86_64-3.7/numpy/distutils\n copying build/src.linux-x86_64-3.7/numpy/distutils/__config__.py -> build/lib.linux-x86_64-3.7/numpy/distutils\n creating build/lib.linux-x86_64-3.7/numpy/distutils/command\n copying numpy/distutils/command/bdist_rpm.py -> build/lib.linux-x86_64-3.7/numpy/distutils/command\n copying numpy/distutils/command/egg_info.py -> build/lib.linux-x86_64-3.7/numpy/distutils/command\n copying numpy/distutils/command/build_src.py -> build/lib.linux-x86_64-3.7/numpy/distutils/command\n copying numpy/distutils/command/config_compiler.py -> build/lib.linux-x86_64-3.7/numpy/distutils/command\n copying numpy/distutils/command/__init__.py -> build/lib.linux-x86_64-3.7/numpy/distutils/command\n copying numpy/distutils/command/build_py.py -> build/lib.linux-x86_64-3.7/numpy/distutils/command\n copying numpy/distutils/command/config.py -> build/lib.linux-x86_64-3.7/numpy/distutils/command\n copying numpy/distutils/command/install_headers.py -> build/lib.linux-x86_64-3.7/numpy/distutils/command\n copying numpy/distutils/command/develop.py -> build/lib.linux-x86_64-3.7/numpy/distutils/command\n copying numpy/distutils/command/sdist.py -> build/lib.linux-x86_64-3.7/numpy/distutils/command\n copying numpy/distutils/command/build_clib.py -> build/lib.linux-x86_64-3.7/numpy/distutils/command\n copying numpy/distutils/command/build.py -> build/lib.linux-x86_64-3.7/numpy/distutils/command\n copying numpy/distutils/command/build_scripts.py -> build/lib.linux-x86_64-3.7/numpy/distutils/command\n copying numpy/distutils/command/autodist.py -> build/lib.linux-x86_64-3.7/numpy/distutils/command\n copying numpy/distutils/command/install_data.py -> build/lib.linux-x86_64-3.7/numpy/distutils/command\n copying numpy/distutils/command/install.py -> build/lib.linux-x86_64-3.7/numpy/distutils/command\n copying numpy/distutils/command/build_ext.py -> build/lib.linux-x86_64-3.7/numpy/distutils/command\n copying numpy/distutils/command/install_clib.py -> build/lib.linux-x86_64-3.7/numpy/distutils/command\n creating build/lib.linux-x86_64-3.7/numpy/distutils/fcompiler\n copying numpy/distutils/fcompiler/nag.py -> build/lib.linux-x86_64-3.7/numpy/distutils/fcompiler\n copying numpy/distutils/fcompiler/lahey.py -> build/lib.linux-x86_64-3.7/numpy/distutils/fcompiler\n copying numpy/distutils/fcompiler/__init__.py -> build/lib.linux-x86_64-3.7/numpy/distutils/fcompiler\n copying numpy/distutils/fcompiler/pathf95.py -> build/lib.linux-x86_64-3.7/numpy/distutils/fcompiler\n copying numpy/distutils/fcompiler/g95.py -> build/lib.linux-x86_64-3.7/numpy/distutils/fcompiler\n copying numpy/distutils/fcompiler/mips.py -> build/lib.linux-x86_64-3.7/numpy/distutils/fcompiler\n copying numpy/distutils/fcompiler/intel.py -> build/lib.linux-x86_64-3.7/numpy/distutils/fcompiler\n copying numpy/distutils/fcompiler/compaq.py -> build/lib.linux-x86_64-3.7/numpy/distutils/fcompiler\n copying numpy/distutils/fcompiler/none.py -> build/lib.linux-x86_64-3.7/numpy/distutils/fcompiler\n copying numpy/distutils/fcompiler/sun.py -> build/lib.linux-x86_64-3.7/numpy/distutils/fcompiler\n copying numpy/distutils/fcompiler/gnu.py -> build/lib.linux-x86_64-3.7/numpy/distutils/fcompiler\n copying numpy/distutils/fcompiler/pg.py -> build/lib.linux-x86_64-3.7/numpy/distutils/fcompiler\n copying numpy/distutils/fcompiler/vast.py -> build/lib.linux-x86_64-3.7/numpy/distutils/fcompiler\n copying numpy/distutils/fcompiler/ibm.py -> build/lib.linux-x86_64-3.7/numpy/distutils/fcompiler\n copying numpy/distutils/fcompiler/hpux.py -> build/lib.linux-x86_64-3.7/numpy/distutils/fcompiler\n copying numpy/distutils/fcompiler/absoft.py -> build/lib.linux-x86_64-3.7/numpy/distutils/fcompiler\n creating build/lib.linux-x86_64-3.7/numpy/doc\n copying numpy/doc/creation.py -> build/lib.linux-x86_64-3.7/numpy/doc\n copying numpy/doc/byteswapping.py -> build/lib.linux-x86_64-3.7/numpy/doc\n copying numpy/doc/__init__.py -> build/lib.linux-x86_64-3.7/numpy/doc\n copying numpy/doc/internals.py -> build/lib.linux-x86_64-3.7/numpy/doc\n copying numpy/doc/broadcasting.py -> build/lib.linux-x86_64-3.7/numpy/doc\n copying numpy/doc/glossary.py -> build/lib.linux-x86_64-3.7/numpy/doc\n copying numpy/doc/structured_arrays.py -> build/lib.linux-x86_64-3.7/numpy/doc\n copying numpy/doc/basics.py -> build/lib.linux-x86_64-3.7/numpy/doc\n copying numpy/doc/indexing.py -> build/lib.linux-x86_64-3.7/numpy/doc\n copying numpy/doc/constants.py -> build/lib.linux-x86_64-3.7/numpy/doc\n copying numpy/doc/misc.py -> build/lib.linux-x86_64-3.7/numpy/doc\n copying numpy/doc/ufuncs.py -> build/lib.linux-x86_64-3.7/numpy/doc\n copying numpy/doc/subclassing.py -> build/lib.linux-x86_64-3.7/numpy/doc\n creating build/lib.linux-x86_64-3.7/numpy/f2py\n copying numpy/f2py/f2py2e.py -> build/lib.linux-x86_64-3.7/numpy/f2py\n copying numpy/f2py/diagnose.py -> build/lib.linux-x86_64-3.7/numpy/f2py\n copying numpy/f2py/__version__.py -> build/lib.linux-x86_64-3.7/numpy/f2py\n copying numpy/f2py/__init__.py -> build/lib.linux-x86_64-3.7/numpy/f2py\n copying numpy/f2py/use_rules.py -> build/lib.linux-x86_64-3.7/numpy/f2py\n copying numpy/f2py/f90mod_rules.py -> build/lib.linux-x86_64-3.7/numpy/f2py\n copying numpy/f2py/setup.py -> build/lib.linux-x86_64-3.7/numpy/f2py\n copying numpy/f2py/__main__.py -> build/lib.linux-x86_64-3.7/numpy/f2py\n copying numpy/f2py/func2subr.py -> build/lib.linux-x86_64-3.7/numpy/f2py\n copying numpy/f2py/f2py_testing.py -> build/lib.linux-x86_64-3.7/numpy/f2py\n copying numpy/f2py/cb_rules.py -> build/lib.linux-x86_64-3.7/numpy/f2py\n copying numpy/f2py/cfuncs.py -> build/lib.linux-x86_64-3.7/numpy/f2py\n copying numpy/f2py/common_rules.py -> build/lib.linux-x86_64-3.7/numpy/f2py\n copying numpy/f2py/crackfortran.py -> build/lib.linux-x86_64-3.7/numpy/f2py\n copying numpy/f2py/auxfuncs.py -> build/lib.linux-x86_64-3.7/numpy/f2py\n copying numpy/f2py/info.py -> build/lib.linux-x86_64-3.7/numpy/f2py\n copying numpy/f2py/capi_maps.py -> build/lib.linux-x86_64-3.7/numpy/f2py\n copying numpy/f2py/rules.py -> build/lib.linux-x86_64-3.7/numpy/f2py\n creating build/lib.linux-x86_64-3.7/numpy/fft\n copying numpy/fft/__init__.py -> build/lib.linux-x86_64-3.7/numpy/fft\n copying numpy/fft/setup.py -> build/lib.linux-x86_64-3.7/numpy/fft\n copying numpy/fft/helper.py -> build/lib.linux-x86_64-3.7/numpy/fft\n copying numpy/fft/fftpack.py -> build/lib.linux-x86_64-3.7/numpy/fft\n copying numpy/fft/info.py -> build/lib.linux-x86_64-3.7/numpy/fft\n creating build/lib.linux-x86_64-3.7/numpy/lib\n copying numpy/lib/stride_tricks.py -> build/lib.linux-x86_64-3.7/numpy/lib\n copying numpy/lib/_iotools.py -> build/lib.linux-x86_64-3.7/numpy/lib\n copying numpy/lib/financial.py -> build/lib.linux-x86_64-3.7/numpy/lib\n copying numpy/lib/type_check.py -> build/lib.linux-x86_64-3.7/numpy/lib\n copying numpy/lib/utils.py -> build/lib.linux-x86_64-3.7/numpy/lib\n copying numpy/lib/nanfunctions.py -> build/lib.linux-x86_64-3.7/numpy/lib\n copying numpy/lib/arraypad.py -> build/lib.linux-x86_64-3.7/numpy/lib\n copying numpy/lib/__init__.py -> build/lib.linux-x86_64-3.7/numpy/lib\n copying numpy/lib/npyio.py -> build/lib.linux-x86_64-3.7/numpy/lib\n copying numpy/lib/function_base.py -> build/lib.linux-x86_64-3.7/numpy/lib\n copying numpy/lib/twodim_base.py -> build/lib.linux-x86_64-3.7/numpy/lib\n copying numpy/lib/setup.py -> build/lib.linux-x86_64-3.7/numpy/lib\n copying numpy/lib/arrayterator.py -> build/lib.linux-x86_64-3.7/numpy/lib\n copying numpy/lib/ufunclike.py -> build/lib.linux-x86_64-3.7/numpy/lib\n copying numpy/lib/user_array.py -> build/lib.linux-x86_64-3.7/numpy/lib\n copying numpy/lib/index_tricks.py -> build/lib.linux-x86_64-3.7/numpy/lib\n copying numpy/lib/scimath.py -> build/lib.linux-x86_64-3.7/numpy/lib\n copying numpy/lib/format.py -> build/lib.linux-x86_64-3.7/numpy/lib\n copying numpy/lib/shape_base.py -> build/lib.linux-x86_64-3.7/numpy/lib\n copying numpy/lib/arraysetops.py -> build/lib.linux-x86_64-3.7/numpy/lib\n copying numpy/lib/_version.py -> build/lib.linux-x86_64-3.7/numpy/lib\n copying numpy/lib/mixins.py -> build/lib.linux-x86_64-3.7/numpy/lib\n copying numpy/lib/info.py -> build/lib.linux-x86_64-3.7/numpy/lib\n copying numpy/lib/polynomial.py -> build/lib.linux-x86_64-3.7/numpy/lib\n copying numpy/lib/recfunctions.py -> build/lib.linux-x86_64-3.7/numpy/lib\n copying numpy/lib/_datasource.py -> build/lib.linux-x86_64-3.7/numpy/lib\n creating build/lib.linux-x86_64-3.7/numpy/linalg\n copying numpy/linalg/__init__.py -> build/lib.linux-x86_64-3.7/numpy/linalg\n copying numpy/linalg/setup.py -> build/lib.linux-x86_64-3.7/numpy/linalg\n copying numpy/linalg/linalg.py -> build/lib.linux-x86_64-3.7/numpy/linalg\n copying numpy/linalg/info.py -> build/lib.linux-x86_64-3.7/numpy/linalg\n creating build/lib.linux-x86_64-3.7/numpy/ma\n copying numpy/ma/timer_comparison.py -> build/lib.linux-x86_64-3.7/numpy/ma\n copying numpy/ma/__init__.py -> build/lib.linux-x86_64-3.7/numpy/ma\n copying numpy/ma/setup.py -> build/lib.linux-x86_64-3.7/numpy/ma\n copying numpy/ma/extras.py -> build/lib.linux-x86_64-3.7/numpy/ma\n copying numpy/ma/mrecords.py -> build/lib.linux-x86_64-3.7/numpy/ma\n copying numpy/ma/bench.py -> build/lib.linux-x86_64-3.7/numpy/ma\n copying numpy/ma/core.py -> build/lib.linux-x86_64-3.7/numpy/ma\n copying numpy/ma/version.py -> build/lib.linux-x86_64-3.7/numpy/ma\n copying numpy/ma/testutils.py -> build/lib.linux-x86_64-3.7/numpy/ma\n creating build/lib.linux-x86_64-3.7/numpy/matrixlib\n copying numpy/matrixlib/__init__.py -> build/lib.linux-x86_64-3.7/numpy/matrixlib\n copying numpy/matrixlib/setup.py -> build/lib.linux-x86_64-3.7/numpy/matrixlib\n copying numpy/matrixlib/defmatrix.py -> build/lib.linux-x86_64-3.7/numpy/matrixlib\n creating build/lib.linux-x86_64-3.7/numpy/polynomial\n copying numpy/polynomial/_polybase.py -> build/lib.linux-x86_64-3.7/numpy/polynomial\n copying numpy/polynomial/chebyshev.py -> build/lib.linux-x86_64-3.7/numpy/polynomial\n copying numpy/polynomial/polyutils.py -> build/lib.linux-x86_64-3.7/numpy/polynomial\n copying numpy/polynomial/__init__.py -> build/lib.linux-x86_64-3.7/numpy/polynomial\n copying numpy/polynomial/setup.py -> build/lib.linux-x86_64-3.7/numpy/polynomial\n copying numpy/polynomial/laguerre.py -> build/lib.linux-x86_64-3.7/numpy/polynomial\n copying numpy/polynomial/hermite_e.py -> build/lib.linux-x86_64-3.7/numpy/polynomial\n copying numpy/polynomial/legendre.py -> build/lib.linux-x86_64-3.7/numpy/polynomial\n copying numpy/polynomial/polynomial.py -> build/lib.linux-x86_64-3.7/numpy/polynomial\n copying numpy/polynomial/hermite.py -> build/lib.linux-x86_64-3.7/numpy/polynomial\n creating build/lib.linux-x86_64-3.7/numpy/random\n copying numpy/random/__init__.py -> build/lib.linux-x86_64-3.7/numpy/random\n copying numpy/random/setup.py -> build/lib.linux-x86_64-3.7/numpy/random\n copying numpy/random/info.py -> build/lib.linux-x86_64-3.7/numpy/random\n creating build/lib.linux-x86_64-3.7/numpy/testing\n copying numpy/testing/utils.py -> build/lib.linux-x86_64-3.7/numpy/testing\n copying numpy/testing/__init__.py -> build/lib.linux-x86_64-3.7/numpy/testing\n copying numpy/testing/setup.py -> build/lib.linux-x86_64-3.7/numpy/testing\n copying numpy/testing/print_coercion_tables.py -> build/lib.linux-x86_64-3.7/numpy/testing\n copying numpy/testing/decorators.py -> build/lib.linux-x86_64-3.7/numpy/testing\n copying numpy/testing/noseclasses.py -> build/lib.linux-x86_64-3.7/numpy/testing\n copying numpy/testing/nosetester.py -> build/lib.linux-x86_64-3.7/numpy/testing\n running build_clib\n customize UnixCCompiler\n customize UnixCCompiler using build_clib\n building 'npymath' library\n compiling C sources\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n creating build/temp.linux-x86_64-3.7\n creating build/temp.linux-x86_64-3.7/numpy\n creating build/temp.linux-x86_64-3.7/numpy/core\n creating build/temp.linux-x86_64-3.7/numpy/core/src\n creating build/temp.linux-x86_64-3.7/numpy/core/src/npymath\n creating build/temp.linux-x86_64-3.7/build\n creating build/temp.linux-x86_64-3.7/build/src.linux-x86_64-3.7\n creating build/temp.linux-x86_64-3.7/build/src.linux-x86_64-3.7/numpy\n creating build/temp.linux-x86_64-3.7/build/src.linux-x86_64-3.7/numpy/core\n creating build/temp.linux-x86_64-3.7/build/src.linux-x86_64-3.7/numpy/core/src\n creating build/temp.linux-x86_64-3.7/build/src.linux-x86_64-3.7/numpy/core/src/npymath\n compile options: '-Ibuild/src.linux-x86_64-3.7/numpy/core/src/npymath -Inumpy/core/include -Ibuild/src.linux-x86_64-3.7/numpy/core/include/numpy -Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -Ibuild/src.linux-x86_64-3.7/numpy/core/src/private -Ibuild/src.linux-x86_64-3.7/numpy/core/src/npymath -Ibuild/src.linux-x86_64-3.7/numpy/core/src/private -Ibuild/src.linux-x86_64-3.7/numpy/core/src/npymath -Ibuild/src.linux-x86_64-3.7/numpy/core/src/private -Ibuild/src.linux-x86_64-3.7/numpy/core/src/npymath -c'\n gcc: numpy/core/src/npymath/npy_math.c\n gcc: build/src.linux-x86_64-3.7/numpy/core/src/npymath/ieee754.c\n gcc: build/src.linux-x86_64-3.7/numpy/core/src/npymath/npy_math_complex.c\n gcc: numpy/core/src/npymath/halffloat.c\n ar: adding 4 object files to build/temp.linux-x86_64-3.7/libnpymath.a\n building 'npysort' library\n compiling C sources\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n creating build/temp.linux-x86_64-3.7/build/src.linux-x86_64-3.7/numpy/core/src/npysort\n compile options: '-Ibuild/src.linux-x86_64-3.7/numpy/core/src/private -Inumpy/core/include -Ibuild/src.linux-x86_64-3.7/numpy/core/include/numpy -Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -Ibuild/src.linux-x86_64-3.7/numpy/core/src/private -Ibuild/src.linux-x86_64-3.7/numpy/core/src/npymath -Ibuild/src.linux-x86_64-3.7/numpy/core/src/private -Ibuild/src.linux-x86_64-3.7/numpy/core/src/npymath -Ibuild/src.linux-x86_64-3.7/numpy/core/src/private -Ibuild/src.linux-x86_64-3.7/numpy/core/src/npymath -c'\n gcc: build/src.linux-x86_64-3.7/numpy/core/src/npysort/quicksort.c\n gcc: build/src.linux-x86_64-3.7/numpy/core/src/npysort/mergesort.c\n gcc: build/src.linux-x86_64-3.7/numpy/core/src/npysort/heapsort.c\n gcc: build/src.linux-x86_64-3.7/numpy/core/src/npysort/selection.c\n In file included from numpy/core/src/npysort/selection.c.src:22:\n numpy/core/src/private/npy_partition.h.src: In function ‘get_partition_func’:\n numpy/core/src/private/npy_partition.h.src:98:19: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘long unsigned int’ [-Wsign-compare]\n 98 | for (i = 0; i < sizeof(_part_map)/sizeof(_part_map[0]); i++) {\n | ^\n numpy/core/src/private/npy_partition.h.src:99:18: warning: comparison of integer expressions of different signedness: ‘int’ and ‘enum NPY_TYPES’ [-Wsign-compare]\n 99 | if (type == _part_map[i].typenum) {\n | ^~\n numpy/core/src/private/npy_partition.h.src: In function ‘get_argpartition_func’:\n numpy/core/src/private/npy_partition.h.src:114:19: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘long unsigned int’ [-Wsign-compare]\n 114 | for (i = 0; i < sizeof(_part_map)/sizeof(_part_map[0]); i++) {\n | ^\n numpy/core/src/private/npy_partition.h.src:115:18: warning: comparison of integer expressions of different signedness: ‘int’ and ‘enum NPY_TYPES’ [-Wsign-compare]\n 115 | if (type == _part_map[i].typenum) {\n | ^~\n gcc: build/src.linux-x86_64-3.7/numpy/core/src/npysort/binsearch.c\n In file included from numpy/core/src/npysort/binsearch.c.src:6:\n numpy/core/src/private/npy_binsearch.h.src: In function ‘get_binsearch_func’:\n numpy/core/src/private/npy_binsearch.h.src:120:45: warning: comparison of integer expressions of different signedness: ‘enum NPY_TYPES’ and ‘int’ [-Wsign-compare]\n 120 | if (_@arg@binsearch_map[mid_idx].typenum < type) {\n | ^\n numpy/core/src/private/npy_binsearch.h.src:128:64: warning: comparison of integer expressions of different signedness: ‘enum NPY_TYPES’ and ‘int’ [-Wsign-compare]\n 128 | if (min_idx < num_funcs && _@arg@binsearch_map[min_idx].typenum == type) {\n | ^~\n In file included from numpy/core/src/npysort/binsearch.c.src:6:\n numpy/core/src/private/npy_binsearch.h.src: In function ‘get_argbinsearch_func’:\n numpy/core/src/private/npy_binsearch.h.src:120:48: warning: comparison of integer expressions of different signedness: ‘enum NPY_TYPES’ and ‘int’ [-Wsign-compare]\n 120 | if (_@arg@binsearch_map[mid_idx].typenum < type) {\n | ^\n numpy/core/src/private/npy_binsearch.h.src:128:67: warning: comparison of integer expressions of different signedness: ‘enum NPY_TYPES’ and ‘int’ [-Wsign-compare]\n 128 | if (min_idx < num_funcs && _@arg@binsearch_map[min_idx].typenum == type) {\n | ^~\n ar: adding 5 object files to build/temp.linux-x86_64-3.7/libnpysort.a\n running build_ext\n customize UnixCCompiler\n customize UnixCCompiler using build_ext\n building 'numpy.core._dummy' extension\n compiling C sources\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-DNPY_INTERNAL_BUILD=1 -DHAVE_NPY_CONFIG_H=1 -D_FILE_OFFSET_BITS=64 -D_LARGEFILE_SOURCE=1 -D_LARGEFILE64_SOURCE=1 -Inumpy/core/include -Ibuild/src.linux-x86_64-3.7/numpy/core/include/numpy -Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -Ibuild/src.linux-x86_64-3.7/numpy/core/src/private -Ibuild/src.linux-x86_64-3.7/numpy/core/src/npymath -Ibuild/src.linux-x86_64-3.7/numpy/core/src/private -Ibuild/src.linux-x86_64-3.7/numpy/core/src/npymath -Ibuild/src.linux-x86_64-3.7/numpy/core/src/private -Ibuild/src.linux-x86_64-3.7/numpy/core/src/npymath -c'\n gcc: numpy/core/src/dummymodule.c\n gcc -pthread -shared -B /home/masrur/anaconda3/compiler_compat -L/home/masrur/anaconda3/lib -Wl,-rpath=/home/masrur/anaconda3/lib -Wl,--no-as-needed -Wl,--sysroot=/ build/temp.linux-x86_64-3.7/numpy/core/src/dummymodule.o -Lbuild/temp.linux-x86_64-3.7 -lm -o build/lib.linux-x86_64-3.7/numpy/core/_dummy.cpython-37m-x86_64-linux-gnu.so\n building 'numpy.core.multiarray' extension\n compiling C sources\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n creating build/temp.linux-x86_64-3.7/numpy/core/src/multiarray\n creating build/temp.linux-x86_64-3.7/build/src.linux-x86_64-3.7/numpy/core/src/multiarray\n creating build/temp.linux-x86_64-3.7/numpy/core/src/private\n compile options: '-DNPY_INTERNAL_BUILD=1 -DHAVE_NPY_CONFIG_H=1 -D_FILE_OFFSET_BITS=64 -D_LARGEFILE_SOURCE=1 -D_LARGEFILE64_SOURCE=1 -DSCIPY_MKL_H -DHAVE_CBLAS -I/usr/local/include -I/usr/include -I/home/masrur/anaconda3/include -Ibuild/src.linux-x86_64-3.7/numpy/core/src/private -Inumpy/core/include -Ibuild/src.linux-x86_64-3.7/numpy/core/include/numpy -Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -Ibuild/src.linux-x86_64-3.7/numpy/core/src/private -Ibuild/src.linux-x86_64-3.7/numpy/core/src/npymath -Ibuild/src.linux-x86_64-3.7/numpy/core/src/private -Ibuild/src.linux-x86_64-3.7/numpy/core/src/npymath -Ibuild/src.linux-x86_64-3.7/numpy/core/src/private -Ibuild/src.linux-x86_64-3.7/numpy/core/src/npymath -c'\n gcc: numpy/core/src/multiarray/alloc.c\n gcc: numpy/core/src/multiarray/arrayobject.c\n gcc: build/src.linux-x86_64-3.7/numpy/core/src/multiarray/arraytypes.c\n numpy/core/src/multiarray/arraytypes.c.src: In function ‘VOID_setitem’:\n numpy/core/src/multiarray/arraytypes.c.src:874:9: warning: ‘PyObject_AsReadBuffer’ is deprecated [-Wdeprecated-declarations]\n 874 | res = PyObject_AsReadBuffer(op, &buffer, &buflen);\n | ^~~\n In file included from /home/masrur/anaconda3/include/python3.7m/Python.h:147,\n from numpy/core/src/multiarray/arraytypes.c.src:3:\n /home/masrur/anaconda3/include/python3.7m/abstract.h:489:17: note: declared here\n 489 | PyAPI_FUNC(int) PyObject_AsReadBuffer(PyObject *obj,\n | ^~~~~~~~~~~~~~~~~~~~~\n gcc: numpy/core/src/multiarray/array_assign.c\n gcc: numpy/core/src/multiarray/array_assign_scalar.c\n numpy/core/src/multiarray/array_assign_scalar.c: In function ‘PyArray_AssignRawScalar’:\n numpy/core/src/multiarray/array_assign_scalar.c:236:34: warning: comparison of integer expressions of different signedness: ‘long unsigned int’ and ‘int’ [-Wsign-compare]\n 236 | if (sizeof(scalarbuffer) >= PyArray_DESCR(dst)->elsize) {\n | ^~\n gcc: numpy/core/src/multiarray/array_assign_array.c\n gcc: numpy/core/src/multiarray/buffer.c\n gcc: numpy/core/src/multiarray/calculation.c\n gcc: numpy/core/src/multiarray/compiled_base.c\n gcc: numpy/core/src/multiarray/common.c\n numpy/core/src/multiarray/common.c: In function ‘_IsAligned’:\n numpy/core/src/multiarray/common.c:614:19: warning: comparison of integer expressions of different signedness: ‘unsigned int’ and ‘int’ [-Wsign-compare]\n 614 | for (i = 0; i < PyArray_NDIM(ap); i++) {\n | ^\n numpy/core/src/multiarray/common.c: In function ‘_IsWriteable’:\n numpy/core/src/multiarray/common.c:671:5: warning: ‘PyObject_AsWriteBuffer’ is deprecated [-Wdeprecated-declarations]\n 671 | if (PyObject_AsWriteBuffer(base, &dummy, &n) < 0) {\n | ^~\n In file included from /home/masrur/anaconda3/include/python3.7m/Python.h:147,\n from numpy/core/src/multiarray/common.c:2:\n /home/masrur/anaconda3/include/python3.7m/abstract.h:500:17: note: declared here\n 500 | PyAPI_FUNC(int) PyObject_AsWriteBuffer(PyObject *obj,\n | ^~~~~~~~~~~~~~~~~~~~~~\n gcc: numpy/core/src/multiarray/convert.c\n gcc: numpy/core/src/multiarray/convert_datatype.c\n gcc: numpy/core/src/multiarray/conversion_utils.c\n gcc: numpy/core/src/multiarray/ctors.c\n numpy/core/src/multiarray/ctors.c: In function ‘PyArray_FromInterface’:\n numpy/core/src/multiarray/ctors.c:2322:9: warning: ‘PyObject_AsWriteBuffer’ is deprecated [-Wdeprecated-declarations]\n 2322 | res = PyObject_AsWriteBuffer(base, (void **)&data, &buffer_len);\n | ^~~\n In file included from /home/masrur/anaconda3/include/python3.7m/Python.h:147,\n from numpy/core/src/multiarray/ctors.c:2:\n /home/masrur/anaconda3/include/python3.7m/abstract.h:500:17: note: declared here\n 500 | PyAPI_FUNC(int) PyObject_AsWriteBuffer(PyObject *obj,\n | ^~~~~~~~~~~~~~~~~~~~~~\n numpy/core/src/multiarray/ctors.c:2325:13: warning: ‘PyObject_AsReadBuffer’ is deprecated [-Wdeprecated-declarations]\n 2325 | res = PyObject_AsReadBuffer(\n | ^~~\n In file included from /home/masrur/anaconda3/include/python3.7m/Python.h:147,\n from numpy/core/src/multiarray/ctors.c:2:\n /home/masrur/anaconda3/include/python3.7m/abstract.h:489:17: note: declared here\n 489 | PyAPI_FUNC(int) PyObject_AsReadBuffer(PyObject *obj,\n | ^~~~~~~~~~~~~~~~~~~~~\n numpy/core/src/multiarray/ctors.c: In function ‘PyArray_FromBuffer’:\n numpy/core/src/multiarray/ctors.c:3501:5: warning: ‘PyObject_AsWriteBuffer’ is deprecated [-Wdeprecated-declarations]\n 3501 | if (PyObject_AsWriteBuffer(buf, (void *)&data, &ts) == -1) {\n | ^~\n In file included from /home/masrur/anaconda3/include/python3.7m/Python.h:147,\n from numpy/core/src/multiarray/ctors.c:2:\n /home/masrur/anaconda3/include/python3.7m/abstract.h:500:17: note: declared here\n 500 | PyAPI_FUNC(int) PyObject_AsWriteBuffer(PyObject *obj,\n | ^~~~~~~~~~~~~~~~~~~~~~\n numpy/core/src/multiarray/ctors.c:3504:9: warning: ‘PyObject_AsReadBuffer’ is deprecated [-Wdeprecated-declarations]\n 3504 | if (PyObject_AsReadBuffer(buf, (void *)&data, &ts) == -1) {\n | ^~\n In file included from /home/masrur/anaconda3/include/python3.7m/Python.h:147,\n from numpy/core/src/multiarray/ctors.c:2:\n /home/masrur/anaconda3/include/python3.7m/abstract.h:489:17: note: declared here\n 489 | PyAPI_FUNC(int) PyObject_AsReadBuffer(PyObject *obj,\n | ^~~~~~~~~~~~~~~~~~~~~\n gcc: numpy/core/src/multiarray/datetime.c\n numpy/core/src/multiarray/datetime.c: In function ‘parse_datetime_extended_unit_from_string’:\n numpy/core/src/multiarray/datetime.c:781:24: warning: comparison of integer expressions of different signedness: ‘NPY_DATETIMEUNIT’ {aka ‘enum <anonymous>’} and ‘int’ [-Wsign-compare]\n 781 | if (out_meta->base == -1) {\n | ^~\n numpy/core/src/multiarray/datetime.c: In function ‘get_datetime_units_factor’:\n numpy/core/src/multiarray/datetime.c:1076:23: warning: comparison of integer expressions of different signedness: ‘NPY_DATETIMEUNIT’ {aka ‘enum <anonymous>’} and ‘int’ [-Wsign-compare]\n 1076 | while (littlebase > unit) {\n | ^\n numpy/core/src/multiarray/datetime.c: In function ‘convert_datetime_metadata_tuple_to_datetime_metadata’:\n numpy/core/src/multiarray/datetime.c:1847:24: warning: comparison of integer expressions of different signedness: ‘NPY_DATETIMEUNIT’ {aka ‘enum <anonymous>’} and ‘int’ [-Wsign-compare]\n 1847 | if (out_meta->base == -1) {\n | ^~\n numpy/core/src/multiarray/datetime.c: In function ‘convert_pyobject_to_datetime’:\n numpy/core/src/multiarray/datetime.c:2395:24: warning: comparison of integer expressions of different signedness: ‘NPY_DATETIMEUNIT’ {aka ‘enum <anonymous>’} and ‘int’ [-Wsign-compare]\n 2395 | if (meta->base == -1) {\n | ^~\n numpy/core/src/multiarray/datetime.c:2411:24: warning: comparison of integer expressions of different signedness: ‘NPY_DATETIMEUNIT’ {aka ‘enum <anonymous>’} and ‘int’ [-Wsign-compare]\n 2411 | if (meta->base == -1 || meta->base == NPY_FR_GENERIC) {\n | ^~\n numpy/core/src/multiarray/datetime.c:2424:24: warning: comparison of integer expressions of different signedness: ‘NPY_DATETIMEUNIT’ {aka ‘enum <anonymous>’} and ‘int’ [-Wsign-compare]\n 2424 | if (meta->base == -1) {\n | ^~\n numpy/core/src/multiarray/datetime.c:2463:24: warning: comparison of integer expressions of different signedness: ‘NPY_DATETIMEUNIT’ {aka ‘enum <anonymous>’} and ‘int’ [-Wsign-compare]\n 2463 | if (meta->base == -1) {\n | ^~\n numpy/core/src/multiarray/datetime.c:2495:28: warning: comparison of integer expressions of different signedness: ‘NPY_DATETIMEUNIT’ {aka ‘enum <anonymous>’} and ‘int’ [-Wsign-compare]\n 2495 | if (meta->base == -1) {\n | ^~\n numpy/core/src/multiarray/datetime.c:2522:24: warning: comparison of integer expressions of different signedness: ‘NPY_DATETIMEUNIT’ {aka ‘enum <anonymous>’} and ‘int’ [-Wsign-compare]\n 2522 | if (meta->base == -1) {\n | ^~\n numpy/core/src/multiarray/datetime.c: In function ‘convert_pyobject_to_timedelta’:\n numpy/core/src/multiarray/datetime.c:2598:28: warning: comparison of integer expressions of different signedness: ‘NPY_DATETIMEUNIT’ {aka ‘enum <anonymous>’} and ‘int’ [-Wsign-compare]\n 2598 | if (meta->base == -1) {\n | ^~\n numpy/core/src/multiarray/datetime.c:2609:24: warning: comparison of integer expressions of different signedness: ‘NPY_DATETIMEUNIT’ {aka ‘enum <anonymous>’} and ‘int’ [-Wsign-compare]\n 2609 | if (meta->base == -1) {\n | ^~\n numpy/core/src/multiarray/datetime.c:2622:24: warning: comparison of integer expressions of different signedness: ‘NPY_DATETIMEUNIT’ {aka ‘enum <anonymous>’} and ‘int’ [-Wsign-compare]\n 2622 | if (meta->base == -1) {\n | ^~\n numpy/core/src/multiarray/datetime.c:2661:24: warning: comparison of integer expressions of different signedness: ‘NPY_DATETIMEUNIT’ {aka ‘enum <anonymous>’} and ‘int’ [-Wsign-compare]\n 2661 | if (meta->base == -1) {\n | ^~\n numpy/core/src/multiarray/datetime.c:2730:24: warning: comparison of integer expressions of different signedness: ‘NPY_DATETIMEUNIT’ {aka ‘enum <anonymous>’} and ‘int’ [-Wsign-compare]\n 2730 | if (meta->base == -1) {\n | ^~\n numpy/core/src/multiarray/datetime.c:2784:24: warning: comparison of integer expressions of different signedness: ‘NPY_DATETIMEUNIT’ {aka ‘enum <anonymous>’} and ‘int’ [-Wsign-compare]\n 2784 | if (meta->base == -1) {\n | ^~\n numpy/core/src/multiarray/datetime.c: In function ‘convert_pyobjects_to_datetimes’:\n numpy/core/src/multiarray/datetime.c:3118:26: warning: comparison of integer expressions of different signedness: ‘NPY_DATETIMEUNIT’ {aka ‘enum <anonymous>’} and ‘int’ [-Wsign-compare]\n 3118 | if (inout_meta->base == -1) {\n | ^~\n gcc: numpy/core/src/multiarray/datetime_strings.c\n numpy/core/src/multiarray/datetime_strings.c: In function ‘parse_iso_8601_datetime’:\n numpy/core/src/multiarray/datetime_strings.c:310:18: warning: comparison of integer expressions of different signedness: ‘NPY_DATETIMEUNIT’ {aka ‘enum <anonymous>’} and ‘int’ [-Wsign-compare]\n 310 | if (unit != -1 && !can_cast_datetime64_units(bestunit, unit,\n | ^~\n numpy/core/src/multiarray/datetime_strings.c:350:18: warning: comparison of integer expressions of different signedness: ‘NPY_DATETIMEUNIT’ {aka ‘enum <anonymous>’} and ‘int’ [-Wsign-compare]\n 350 | if (unit != -1 && !can_cast_datetime64_units(bestunit, unit,\n | ^~\n numpy/core/src/multiarray/datetime_strings.c:733:14: warning: comparison of integer expressions of different signedness: ‘NPY_DATETIMEUNIT’ {aka ‘enum <anonymous>’} and ‘int’ [-Wsign-compare]\n 733 | if (unit != -1 && !can_cast_datetime64_units(bestunit, unit,\n | ^~\n numpy/core/src/multiarray/datetime_strings.c: In function ‘get_datetime_iso_8601_strlen’:\n numpy/core/src/multiarray/datetime_strings.c:764:14: warning: comparison of integer expressions of different signedness: ‘NPY_DATETIMEUNIT’ {aka ‘enum <anonymous>’} and ‘int’ [-Wsign-compare]\n 764 | if (base == -1) {\n | ^~\n numpy/core/src/multiarray/datetime_strings.c: In function ‘make_iso_8601_datetime’:\n numpy/core/src/multiarray/datetime_strings.c:930:14: warning: comparison of integer expressions of different signedness: ‘NPY_DATETIMEUNIT’ {aka ‘enum <anonymous>’} and ‘int’ [-Wsign-compare]\n 930 | if (base == -1) {\n | ^~\n numpy/core/src/multiarray/datetime_strings.c: In function ‘array_datetime_as_string’:\n numpy/core/src/multiarray/datetime_strings.c:1414:22: warning: comparison of integer expressions of different signedness: ‘NPY_DATETIMEUNIT’ {aka ‘enum <anonymous>’} and ‘int’ [-Wsign-compare]\n 1414 | if (unit == -1) {\n | ^~\n numpy/core/src/multiarray/datetime_strings.c:1421:18: warning: comparison of integer expressions of different signedness: ‘NPY_DATETIMEUNIT’ {aka ‘enum <anonymous>’} and ‘int’ [-Wsign-compare]\n 1421 | if (unit != -1 && !can_cast_datetime64_units(meta->base, unit, casting)) {\n | ^~\n gcc: numpy/core/src/multiarray/datetime_busday.c\n gcc: numpy/core/src/multiarray/datetime_busdaycal.c\n gcc: numpy/core/src/multiarray/descriptor.c\n gcc: numpy/core/src/multiarray/dtype_transfer.c\n gcc: build/src.linux-x86_64-3.7/numpy/core/src/multiarray/einsum.c\n gcc: numpy/core/src/multiarray/flagsobject.c\n gcc: numpy/core/src/multiarray/getset.c\n numpy/core/src/multiarray/getset.c: In function ‘array_strides_set’:\n numpy/core/src/multiarray/getset.c:134:5: warning: ‘PyObject_AsReadBuffer’ is deprecated [-Wdeprecated-declarations]\n 134 | if (PyArray_BASE(new) && PyObject_AsReadBuffer(PyArray_BASE(new),\n | ^~\n In file included from /home/masrur/anaconda3/include/python3.7m/Python.h:147,\n from numpy/core/src/multiarray/getset.c:4:\n /home/masrur/anaconda3/include/python3.7m/abstract.h:489:17: note: declared here\n 489 | PyAPI_FUNC(int) PyObject_AsReadBuffer(PyObject *obj,\n | ^~~~~~~~~~~~~~~~~~~~~\n numpy/core/src/multiarray/getset.c: In function ‘array_data_set’:\n numpy/core/src/multiarray/getset.c:344:5: warning: ‘PyObject_AsWriteBuffer’ is deprecated [-Wdeprecated-declarations]\n 344 | if (PyObject_AsWriteBuffer(op, &buf, &buf_len) < 0) {\n | ^~\n In file included from /home/masrur/anaconda3/include/python3.7m/Python.h:147,\n from numpy/core/src/multiarray/getset.c:4:\n /home/masrur/anaconda3/include/python3.7m/abstract.h:500:17: note: declared here\n 500 | PyAPI_FUNC(int) PyObject_AsWriteBuffer(PyObject *obj,\n | ^~~~~~~~~~~~~~~~~~~~~~\n numpy/core/src/multiarray/getset.c:346:9: warning: ‘PyObject_AsReadBuffer’ is deprecated [-Wdeprecated-declarations]\n 346 | if (PyObject_AsReadBuffer(op, (const void **)&buf, &buf_len) < 0) {\n | ^~\n In file included from /home/masrur/anaconda3/include/python3.7m/Python.h:147,\n from numpy/core/src/multiarray/getset.c:4:\n /home/masrur/anaconda3/include/python3.7m/abstract.h:489:17: note: declared here\n 489 | PyAPI_FUNC(int) PyObject_AsReadBuffer(PyObject *obj,\n | ^~~~~~~~~~~~~~~~~~~~~\n gcc: numpy/core/src/multiarray/hashdescr.c\n gcc: numpy/core/src/multiarray/item_selection.c\n In file included from numpy/core/src/multiarray/item_selection.c:24:\n numpy/core/src/private/npy_partition.h.src: In function ‘get_partition_func’:\n numpy/core/src/private/npy_partition.h.src:98:19: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘long unsigned int’ [-Wsign-compare]\n 98 | for (i = 0; i < sizeof(_part_map)/sizeof(_part_map[0]); i++) {\n | ^\n numpy/core/src/private/npy_partition.h.src:99:18: warning: comparison of integer expressions of different signedness: ‘int’ and ‘enum NPY_TYPES’ [-Wsign-compare]\n 99 | if (type == _part_map[i].typenum) {\n | ^~\n numpy/core/src/private/npy_partition.h.src: In function ‘get_argpartition_func’:\n numpy/core/src/private/npy_partition.h.src:114:19: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘long unsigned int’ [-Wsign-compare]\n 114 | for (i = 0; i < sizeof(_part_map)/sizeof(_part_map[0]); i++) {\n | ^\n numpy/core/src/private/npy_partition.h.src:115:18: warning: comparison of integer expressions of different signedness: ‘int’ and ‘enum NPY_TYPES’ [-Wsign-compare]\n 115 | if (type == _part_map[i].typenum) {\n | ^~\n In file included from numpy/core/src/multiarray/item_selection.c:25:\n numpy/core/src/private/npy_binsearch.h.src: In function ‘get_binsearch_func’:\n numpy/core/src/private/npy_binsearch.h.src:120:45: warning: comparison of integer expressions of different signedness: ‘enum NPY_TYPES’ and ‘int’ [-Wsign-compare]\n 120 | if (_@arg@binsearch_map[mid_idx].typenum < type) {\n | ^\n numpy/core/src/private/npy_binsearch.h.src:128:64: warning: comparison of integer expressions of different signedness: ‘enum NPY_TYPES’ and ‘int’ [-Wsign-compare]\n 128 | if (min_idx < num_funcs && _@arg@binsearch_map[min_idx].typenum == type) {\n | ^~\n In file included from numpy/core/src/multiarray/item_selection.c:25:\n numpy/core/src/private/npy_binsearch.h.src: In function ‘get_argbinsearch_func’:\n numpy/core/src/private/npy_binsearch.h.src:120:48: warning: comparison of integer expressions of different signedness: ‘enum NPY_TYPES’ and ‘int’ [-Wsign-compare]\n 120 | if (_@arg@binsearch_map[mid_idx].typenum < type) {\n | ^\n numpy/core/src/private/npy_binsearch.h.src:128:67: warning: comparison of integer expressions of different signedness: ‘enum NPY_TYPES’ and ‘int’ [-Wsign-compare]\n 128 | if (min_idx < num_funcs && _@arg@binsearch_map[min_idx].typenum == type) {\n | ^~\n gcc: numpy/core/src/multiarray/iterators.c\n gcc: build/src.linux-x86_64-3.7/numpy/core/src/multiarray/lowlevel_strided_loops.c\n gcc: numpy/core/src/multiarray/mapping.c\n gcc: numpy/core/src/multiarray/methods.c\n gcc: numpy/core/src/multiarray/multiarraymodule.c\n gcc: build/src.linux-x86_64-3.7/numpy/core/src/multiarray/nditer_templ.c\n gcc: numpy/core/src/multiarray/nditer_api.c\n gcc: numpy/core/src/multiarray/nditer_constr.c\n gcc: numpy/core/src/multiarray/nditer_pywrap.c\n gcc: numpy/core/src/multiarray/number.c\n gcc: numpy/core/src/multiarray/numpyos.c\n gcc: numpy/core/src/multiarray/refcount.c\n numpy/core/src/multiarray/refcount.c: In function ‘_fillobject’:\n numpy/core/src/multiarray/refcount.c:279:23: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘long unsigned int’ [-Wsign-compare]\n 279 | for (i = 0; i < dtype->elsize / sizeof(obj); i++) {\n | ^\n gcc: numpy/core/src/multiarray/sequence.c\n gcc: numpy/core/src/multiarray/shape.c\n numpy/core/src/multiarray/shape.c: In function ‘_putzero’:\n numpy/core/src/multiarray/shape.c:347:23: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘long unsigned int’ [-Wsign-compare]\n 347 | for (i = 0; i < dtype->elsize / sizeof(zero); i++) {\n | ^\n gcc: numpy/core/src/multiarray/scalarapi.c\n gcc: build/src.linux-x86_64-3.7/numpy/core/src/multiarray/scalartypes.c\n numpy/core/src/multiarray/scalartypes.c.src: In function ‘gentype_reduce’:\n numpy/core/src/multiarray/scalartypes.c.src:1720:5: warning: ‘PyObject_AsReadBuffer’ is deprecated [-Wdeprecated-declarations]\n 1720 | if (PyObject_AsReadBuffer(self, (const void **)&buffer, &buflen)<0) {\n | ^~\n In file included from /home/masrur/anaconda3/include/python3.7m/Python.h:147,\n from numpy/core/src/multiarray/scalartypes.c.src:3:\n /home/masrur/anaconda3/include/python3.7m/abstract.h:489:17: note: declared here\n 489 | PyAPI_FUNC(int) PyObject_AsReadBuffer(PyObject *obj,\n | ^~~~~~~~~~~~~~~~~~~~~\n numpy/core/src/multiarray/scalartypes.c.src: In function ‘datetime_arrtype_new’:\n numpy/core/src/multiarray/scalartypes.c.src:2707:30: warning: comparison of integer expressions of different signedness: ‘NPY_DATETIMEUNIT’ {aka ‘enum <anonymous>’} and ‘int’ [-Wsign-compare]\n 2707 | if (ret->obmeta.base == -1) {\n | ^~\n numpy/core/src/multiarray/scalartypes.c.src: In function ‘timedelta_arrtype_new’:\n numpy/core/src/multiarray/scalartypes.c.src:2707:30: warning: comparison of integer expressions of different signedness: ‘NPY_DATETIMEUNIT’ {aka ‘enum <anonymous>’} and ‘int’ [-Wsign-compare]\n 2707 | if (ret->obmeta.base == -1) {\n | ^~\n gcc: numpy/core/src/multiarray/temp_elide.c\n numpy/core/src/multiarray/temp_elide.c: In function ‘check_callers’:\n numpy/core/src/multiarray/temp_elide.c:245:30: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘long unsigned int’ [-Wsign-compare]\n 245 | if (n_pyeval < sizeof(pyeval_addr) / sizeof(pyeval_addr[0])) {\n | ^\n numpy/core/src/multiarray/temp_elide.c:252:32: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘long unsigned int’ [-Wsign-compare]\n 252 | else if (n_py_addr < sizeof(py_addr) / sizeof(py_addr[0])) {\n | ^\n gcc: numpy/core/src/multiarray/usertypes.c\n gcc: numpy/core/src/multiarray/ucsnarrow.c\n gcc: numpy/core/src/multiarray/vdot.c\n gcc: numpy/core/src/private/mem_overlap.c\n numpy/core/src/private/mem_overlap.c: In function ‘diophantine_dfs’:\n numpy/core/src/private/mem_overlap.c:420:31: warning: comparison of integer expressions of different signedness: ‘int’ and ‘unsigned int’ [-Wsign-compare]\n 420 | for (j = 0; j < n; ++j) {\n | ^\n numpy/core/src/private/mem_overlap.c: In function ‘strides_to_terms’:\n numpy/core/src/private/mem_overlap.c:715:19: warning: comparison of integer expressions of different signedness: ‘unsigned int’ and ‘int’ [-Wsign-compare]\n 715 | for (i = 0; i < PyArray_NDIM(arr); ++i) {\n | ^\n numpy/core/src/private/mem_overlap.c: In function ‘solve_may_share_memory’:\n numpy/core/src/private/mem_overlap.c:801:13: warning: comparison of integer expressions of different signedness: ‘npy_int64’ {aka ‘long int’} and ‘long unsigned int’ [-Wsign-compare]\n 801 | if (rhs != (npy_uintp)rhs) {\n | ^~\n numpy/core/src/private/mem_overlap.c: In function ‘solve_may_have_internal_overlap’:\n numpy/core/src/private/mem_overlap.c:890:19: warning: comparison of integer expressions of different signedness: ‘int’ and ‘unsigned int’ [-Wsign-compare]\n 890 | for (j = 0; j < nterms; ++j) {\n | ^\n numpy/core/src/private/mem_overlap.c:908:19: warning: comparison of integer expressions of different signedness: ‘int’ and ‘unsigned int’ [-Wsign-compare]\n 908 | for (j = 0; j < nterms; ++j) {\n | ^\n gcc: numpy/core/src/private/ufunc_override.c\n gcc: numpy/core/src/multiarray/cblasfuncs.c\n gcc: numpy/core/src/multiarray/python_xerbla.c\n gcc -pthread -shared -B /home/masrur/anaconda3/compiler_compat -L/home/masrur/anaconda3/lib -Wl,-rpath=/home/masrur/anaconda3/lib -Wl,--no-as-needed -Wl,--sysroot=/ build/temp.linux-x86_64-3.7/numpy/core/src/multiarray/alloc.o build/temp.linux-x86_64-3.7/numpy/core/src/multiarray/arrayobject.o build/temp.linux-x86_64-3.7/build/src.linux-x86_64-3.7/numpy/core/src/multiarray/arraytypes.o build/temp.linux-x86_64-3.7/numpy/core/src/multiarray/array_assign.o build/temp.linux-x86_64-3.7/numpy/core/src/multiarray/array_assign_scalar.o build/temp.linux-x86_64-3.7/numpy/core/src/multiarray/array_assign_array.o build/temp.linux-x86_64-3.7/numpy/core/src/multiarray/buffer.o build/temp.linux-x86_64-3.7/numpy/core/src/multiarray/calculation.o build/temp.linux-x86_64-3.7/numpy/core/src/multiarray/compiled_base.o build/temp.linux-x86_64-3.7/numpy/core/src/multiarray/common.o build/temp.linux-x86_64-3.7/numpy/core/src/multiarray/convert.o build/temp.linux-x86_64-3.7/numpy/core/src/multiarray/convert_datatype.o build/temp.linux-x86_64-3.7/numpy/core/src/multiarray/conversion_utils.o build/temp.linux-x86_64-3.7/numpy/core/src/multiarray/ctors.o build/temp.linux-x86_64-3.7/numpy/core/src/multiarray/datetime.o build/temp.linux-x86_64-3.7/numpy/core/src/multiarray/datetime_strings.o build/temp.linux-x86_64-3.7/numpy/core/src/multiarray/datetime_busday.o build/temp.linux-x86_64-3.7/numpy/core/src/multiarray/datetime_busdaycal.o build/temp.linux-x86_64-3.7/numpy/core/src/multiarray/descriptor.o build/temp.linux-x86_64-3.7/numpy/core/src/multiarray/dtype_transfer.o build/temp.linux-x86_64-3.7/build/src.linux-x86_64-3.7/numpy/core/src/multiarray/einsum.o build/temp.linux-x86_64-3.7/numpy/core/src/multiarray/flagsobject.o build/temp.linux-x86_64-3.7/numpy/core/src/multiarray/getset.o build/temp.linux-x86_64-3.7/numpy/core/src/multiarray/hashdescr.o build/temp.linux-x86_64-3.7/numpy/core/src/multiarray/item_selection.o build/temp.linux-x86_64-3.7/numpy/core/src/multiarray/iterators.o build/temp.linux-x86_64-3.7/build/src.linux-x86_64-3.7/numpy/core/src/multiarray/lowlevel_strided_loops.o build/temp.linux-x86_64-3.7/numpy/core/src/multiarray/mapping.o build/temp.linux-x86_64-3.7/numpy/core/src/multiarray/methods.o build/temp.linux-x86_64-3.7/numpy/core/src/multiarray/multiarraymodule.o build/temp.linux-x86_64-3.7/build/src.linux-x86_64-3.7/numpy/core/src/multiarray/nditer_templ.o build/temp.linux-x86_64-3.7/numpy/core/src/multiarray/nditer_api.o build/temp.linux-x86_64-3.7/numpy/core/src/multiarray/nditer_constr.o build/temp.linux-x86_64-3.7/numpy/core/src/multiarray/nditer_pywrap.o build/temp.linux-x86_64-3.7/numpy/core/src/multiarray/number.o build/temp.linux-x86_64-3.7/numpy/core/src/multiarray/numpyos.o build/temp.linux-x86_64-3.7/numpy/core/src/multiarray/refcount.o build/temp.linux-x86_64-3.7/numpy/core/src/multiarray/sequence.o build/temp.linux-x86_64-3.7/numpy/core/src/multiarray/shape.o build/temp.linux-x86_64-3.7/numpy/core/src/multiarray/scalarapi.o build/temp.linux-x86_64-3.7/build/src.linux-x86_64-3.7/numpy/core/src/multiarray/scalartypes.o build/temp.linux-x86_64-3.7/numpy/core/src/multiarray/temp_elide.o build/temp.linux-x86_64-3.7/numpy/core/src/multiarray/usertypes.o build/temp.linux-x86_64-3.7/numpy/core/src/multiarray/ucsnarrow.o build/temp.linux-x86_64-3.7/numpy/core/src/multiarray/vdot.o build/temp.linux-x86_64-3.7/numpy/core/src/private/mem_overlap.o build/temp.linux-x86_64-3.7/numpy/core/src/private/ufunc_override.o build/temp.linux-x86_64-3.7/numpy/core/src/multiarray/cblasfuncs.o build/temp.linux-x86_64-3.7/numpy/core/src/multiarray/python_xerbla.o -L/home/masrur/anaconda3/lib -Lbuild/temp.linux-x86_64-3.7 -lnpymath -lnpysort -lmkl_rt -lpthread -lm -o build/lib.linux-x86_64-3.7/numpy/core/multiarray.cpython-37m-x86_64-linux-gnu.so\n building 'numpy.core.umath' extension\n compiling C sources\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n creating build/temp.linux-x86_64-3.7/numpy/core/src/umath\n creating build/temp.linux-x86_64-3.7/build/src.linux-x86_64-3.7/numpy/core/src/umath\n compile options: '-DNPY_INTERNAL_BUILD=1 -DHAVE_NPY_CONFIG_H=1 -D_FILE_OFFSET_BITS=64 -D_LARGEFILE_SOURCE=1 -D_LARGEFILE64_SOURCE=1 -Ibuild/src.linux-x86_64-3.7/numpy/core/src/umath -Inumpy/core/include -Ibuild/src.linux-x86_64-3.7/numpy/core/include/numpy -Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -Ibuild/src.linux-x86_64-3.7/numpy/core/src/private -Ibuild/src.linux-x86_64-3.7/numpy/core/src/npymath -Ibuild/src.linux-x86_64-3.7/numpy/core/src/private -Ibuild/src.linux-x86_64-3.7/numpy/core/src/npymath -Ibuild/src.linux-x86_64-3.7/numpy/core/src/private -Ibuild/src.linux-x86_64-3.7/numpy/core/src/npymath -c'\n gcc: numpy/core/src/umath/umathmodule.c\n gcc: numpy/core/src/umath/reduction.c\n gcc: build/src.linux-x86_64-3.7/numpy/core/src/umath/loops.c\n In file included from numpy/core/src/umath/loops.c.src:39:\n numpy/core/src/umath/simd.inc.src: In function ‘run_binary_simd_add_FLOAT’:\n numpy/core/src/umath/simd.inc.src:76:34: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘long unsigned int’ [-Wsign-compare]\n 76 | abs_intp(args[2] - args[0]) >= (esize))\n | ^~\n numpy/core/src/umath/simd.inc.src:195:9: note: in expansion of macro ‘IS_BLOCKABLE_BINARY_SCALAR1’\n 195 | if (IS_BLOCKABLE_BINARY_SCALAR1(sizeof(@type@), 16)) {\n | ^~~~~~~~~~~~~~~~~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:83:34: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘long unsigned int’ [-Wsign-compare]\n 83 | abs_intp(args[2] - args[1]) >= (esize))\n | ^~\n numpy/core/src/umath/simd.inc.src:200:14: note: in expansion of macro ‘IS_BLOCKABLE_BINARY_SCALAR2’\n 200 | else if (IS_BLOCKABLE_BINARY_SCALAR2(sizeof(@type@), 16)) {\n | ^~~~~~~~~~~~~~~~~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘run_binary_simd_subtract_FLOAT’:\n numpy/core/src/umath/simd.inc.src:76:34: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘long unsigned int’ [-Wsign-compare]\n 76 | abs_intp(args[2] - args[0]) >= (esize))\n | ^~\n numpy/core/src/umath/simd.inc.src:195:9: note: in expansion of macro ‘IS_BLOCKABLE_BINARY_SCALAR1’\n 195 | if (IS_BLOCKABLE_BINARY_SCALAR1(sizeof(@type@), 16)) {\n | ^~~~~~~~~~~~~~~~~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:83:34: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘long unsigned int’ [-Wsign-compare]\n 83 | abs_intp(args[2] - args[1]) >= (esize))\n | ^~\n numpy/core/src/umath/simd.inc.src:200:14: note: in expansion of macro ‘IS_BLOCKABLE_BINARY_SCALAR2’\n 200 | else if (IS_BLOCKABLE_BINARY_SCALAR2(sizeof(@type@), 16)) {\n | ^~~~~~~~~~~~~~~~~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘run_binary_simd_multiply_FLOAT’:\n numpy/core/src/umath/simd.inc.src:76:34: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘long unsigned int’ [-Wsign-compare]\n 76 | abs_intp(args[2] - args[0]) >= (esize))\n | ^~\n numpy/core/src/umath/simd.inc.src:195:9: note: in expansion of macro ‘IS_BLOCKABLE_BINARY_SCALAR1’\n 195 | if (IS_BLOCKABLE_BINARY_SCALAR1(sizeof(@type@), 16)) {\n | ^~~~~~~~~~~~~~~~~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:83:34: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘long unsigned int’ [-Wsign-compare]\n 83 | abs_intp(args[2] - args[1]) >= (esize))\n | ^~\n numpy/core/src/umath/simd.inc.src:200:14: note: in expansion of macro ‘IS_BLOCKABLE_BINARY_SCALAR2’\n 200 | else if (IS_BLOCKABLE_BINARY_SCALAR2(sizeof(@type@), 16)) {\n | ^~~~~~~~~~~~~~~~~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘run_binary_simd_divide_FLOAT’:\n numpy/core/src/umath/simd.inc.src:76:34: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘long unsigned int’ [-Wsign-compare]\n 76 | abs_intp(args[2] - args[0]) >= (esize))\n | ^~\n numpy/core/src/umath/simd.inc.src:195:9: note: in expansion of macro ‘IS_BLOCKABLE_BINARY_SCALAR1’\n 195 | if (IS_BLOCKABLE_BINARY_SCALAR1(sizeof(@type@), 16)) {\n | ^~~~~~~~~~~~~~~~~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:83:34: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘long unsigned int’ [-Wsign-compare]\n 83 | abs_intp(args[2] - args[1]) >= (esize))\n | ^~\n numpy/core/src/umath/simd.inc.src:200:14: note: in expansion of macro ‘IS_BLOCKABLE_BINARY_SCALAR2’\n 200 | else if (IS_BLOCKABLE_BINARY_SCALAR2(sizeof(@type@), 16)) {\n | ^~~~~~~~~~~~~~~~~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘run_binary_simd_add_DOUBLE’:\n numpy/core/src/umath/simd.inc.src:76:34: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘long unsigned int’ [-Wsign-compare]\n 76 | abs_intp(args[2] - args[0]) >= (esize))\n | ^~\n numpy/core/src/umath/simd.inc.src:195:9: note: in expansion of macro ‘IS_BLOCKABLE_BINARY_SCALAR1’\n 195 | if (IS_BLOCKABLE_BINARY_SCALAR1(sizeof(@type@), 16)) {\n | ^~~~~~~~~~~~~~~~~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:83:34: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘long unsigned int’ [-Wsign-compare]\n 83 | abs_intp(args[2] - args[1]) >= (esize))\n | ^~\n numpy/core/src/umath/simd.inc.src:200:14: note: in expansion of macro ‘IS_BLOCKABLE_BINARY_SCALAR2’\n 200 | else if (IS_BLOCKABLE_BINARY_SCALAR2(sizeof(@type@), 16)) {\n | ^~~~~~~~~~~~~~~~~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘run_binary_simd_subtract_DOUBLE’:\n numpy/core/src/umath/simd.inc.src:76:34: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘long unsigned int’ [-Wsign-compare]\n 76 | abs_intp(args[2] - args[0]) >= (esize))\n | ^~\n numpy/core/src/umath/simd.inc.src:195:9: note: in expansion of macro ‘IS_BLOCKABLE_BINARY_SCALAR1’\n 195 | if (IS_BLOCKABLE_BINARY_SCALAR1(sizeof(@type@), 16)) {\n | ^~~~~~~~~~~~~~~~~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:83:34: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘long unsigned int’ [-Wsign-compare]\n 83 | abs_intp(args[2] - args[1]) >= (esize))\n | ^~\n numpy/core/src/umath/simd.inc.src:200:14: note: in expansion of macro ‘IS_BLOCKABLE_BINARY_SCALAR2’\n 200 | else if (IS_BLOCKABLE_BINARY_SCALAR2(sizeof(@type@), 16)) {\n | ^~~~~~~~~~~~~~~~~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘run_binary_simd_multiply_DOUBLE’:\n numpy/core/src/umath/simd.inc.src:76:34: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘long unsigned int’ [-Wsign-compare]\n 76 | abs_intp(args[2] - args[0]) >= (esize))\n | ^~\n numpy/core/src/umath/simd.inc.src:195:9: note: in expansion of macro ‘IS_BLOCKABLE_BINARY_SCALAR1’\n 195 | if (IS_BLOCKABLE_BINARY_SCALAR1(sizeof(@type@), 16)) {\n | ^~~~~~~~~~~~~~~~~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:83:34: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘long unsigned int’ [-Wsign-compare]\n 83 | abs_intp(args[2] - args[1]) >= (esize))\n | ^~\n numpy/core/src/umath/simd.inc.src:200:14: note: in expansion of macro ‘IS_BLOCKABLE_BINARY_SCALAR2’\n 200 | else if (IS_BLOCKABLE_BINARY_SCALAR2(sizeof(@type@), 16)) {\n | ^~~~~~~~~~~~~~~~~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘run_binary_simd_divide_DOUBLE’:\n numpy/core/src/umath/simd.inc.src:76:34: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘long unsigned int’ [-Wsign-compare]\n 76 | abs_intp(args[2] - args[0]) >= (esize))\n | ^~\n numpy/core/src/umath/simd.inc.src:195:9: note: in expansion of macro ‘IS_BLOCKABLE_BINARY_SCALAR1’\n 195 | if (IS_BLOCKABLE_BINARY_SCALAR1(sizeof(@type@), 16)) {\n | ^~~~~~~~~~~~~~~~~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:83:34: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘long unsigned int’ [-Wsign-compare]\n 83 | abs_intp(args[2] - args[1]) >= (esize))\n | ^~\n numpy/core/src/umath/simd.inc.src:200:14: note: in expansion of macro ‘IS_BLOCKABLE_BINARY_SCALAR2’\n 200 | else if (IS_BLOCKABLE_BINARY_SCALAR2(sizeof(@type@), 16)) {\n | ^~~~~~~~~~~~~~~~~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_binary_add_FLOAT’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:428:13: note: in expansion of macro ‘LOOP_BLOCKED’\n 428 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:435:13: note: in expansion of macro ‘LOOP_BLOCKED’\n 435 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:444:9: note: in expansion of macro ‘LOOP_BLOCKED’\n 444 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:452:9: note: in expansion of macro ‘LOOP_BLOCKED’\n 452 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:461:13: note: in expansion of macro ‘LOOP_BLOCKED’\n 461 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:468:13: note: in expansion of macro ‘LOOP_BLOCKED’\n 468 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_binary_scalar1_add_FLOAT’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:489:9: note: in expansion of macro ‘LOOP_BLOCKED’\n 489 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:496:9: note: in expansion of macro ‘LOOP_BLOCKED’\n 496 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_binary_scalar2_add_FLOAT’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:515:9: note: in expansion of macro ‘LOOP_BLOCKED’\n 515 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:522:9: note: in expansion of macro ‘LOOP_BLOCKED’\n 522 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_binary_subtract_FLOAT’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:428:13: note: in expansion of macro ‘LOOP_BLOCKED’\n 428 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:435:13: note: in expansion of macro ‘LOOP_BLOCKED’\n 435 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:444:9: note: in expansion of macro ‘LOOP_BLOCKED’\n 444 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:452:9: note: in expansion of macro ‘LOOP_BLOCKED’\n 452 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:461:13: note: in expansion of macro ‘LOOP_BLOCKED’\n 461 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:468:13: note: in expansion of macro ‘LOOP_BLOCKED’\n 468 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_binary_scalar1_subtract_FLOAT’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:489:9: note: in expansion of macro ‘LOOP_BLOCKED’\n 489 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:496:9: note: in expansion of macro ‘LOOP_BLOCKED’\n 496 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_binary_scalar2_subtract_FLOAT’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:515:9: note: in expansion of macro ‘LOOP_BLOCKED’\n 515 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:522:9: note: in expansion of macro ‘LOOP_BLOCKED’\n 522 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_binary_multiply_FLOAT’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:428:13: note: in expansion of macro ‘LOOP_BLOCKED’\n 428 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:435:13: note: in expansion of macro ‘LOOP_BLOCKED’\n 435 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:444:9: note: in expansion of macro ‘LOOP_BLOCKED’\n 444 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:452:9: note: in expansion of macro ‘LOOP_BLOCKED’\n 452 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:461:13: note: in expansion of macro ‘LOOP_BLOCKED’\n 461 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:468:13: note: in expansion of macro ‘LOOP_BLOCKED’\n 468 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_binary_scalar1_multiply_FLOAT’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:489:9: note: in expansion of macro ‘LOOP_BLOCKED’\n 489 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:496:9: note: in expansion of macro ‘LOOP_BLOCKED’\n 496 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_binary_scalar2_multiply_FLOAT’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:515:9: note: in expansion of macro ‘LOOP_BLOCKED’\n 515 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:522:9: note: in expansion of macro ‘LOOP_BLOCKED’\n 522 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_binary_divide_FLOAT’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:428:13: note: in expansion of macro ‘LOOP_BLOCKED’\n 428 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:435:13: note: in expansion of macro ‘LOOP_BLOCKED’\n 435 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:444:9: note: in expansion of macro ‘LOOP_BLOCKED’\n 444 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:452:9: note: in expansion of macro ‘LOOP_BLOCKED’\n 452 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:461:13: note: in expansion of macro ‘LOOP_BLOCKED’\n 461 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:468:13: note: in expansion of macro ‘LOOP_BLOCKED’\n 468 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_binary_scalar1_divide_FLOAT’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:489:9: note: in expansion of macro ‘LOOP_BLOCKED’\n 489 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:496:9: note: in expansion of macro ‘LOOP_BLOCKED’\n 496 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_binary_scalar2_divide_FLOAT’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:515:9: note: in expansion of macro ‘LOOP_BLOCKED’\n 515 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:522:9: note: in expansion of macro ‘LOOP_BLOCKED’\n 522 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_signbit_FLOAT’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:564:5: note: in expansion of macro ‘LOOP_BLOCKED’\n 564 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_isnan_FLOAT’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:605:5: note: in expansion of macro ‘LOOP_BLOCKED’\n 605 | LOOP_BLOCKED(@type@, 64) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_isfinite_FLOAT’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:605:5: note: in expansion of macro ‘LOOP_BLOCKED’\n 605 | LOOP_BLOCKED(@type@, 64) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_isinf_FLOAT’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:605:5: note: in expansion of macro ‘LOOP_BLOCKED’\n 605 | LOOP_BLOCKED(@type@, 64) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_binary_equal_FLOAT’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:675:5: note: in expansion of macro ‘LOOP_BLOCKED’\n 675 | LOOP_BLOCKED(@type@, 64) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_binary_scalar1_equal_FLOAT’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:703:5: note: in expansion of macro ‘LOOP_BLOCKED’\n 703 | LOOP_BLOCKED(@type@, 64) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_binary_scalar2_equal_FLOAT’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:727:5: note: in expansion of macro ‘LOOP_BLOCKED’\n 727 | LOOP_BLOCKED(@type@, 64) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_binary_not_equal_FLOAT’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:675:5: note: in expansion of macro ‘LOOP_BLOCKED’\n 675 | LOOP_BLOCKED(@type@, 64) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_binary_scalar1_not_equal_FLOAT’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:703:5: note: in expansion of macro ‘LOOP_BLOCKED’\n 703 | LOOP_BLOCKED(@type@, 64) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_binary_scalar2_not_equal_FLOAT’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:727:5: note: in expansion of macro ‘LOOP_BLOCKED’\n 727 | LOOP_BLOCKED(@type@, 64) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_binary_less_FLOAT’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:675:5: note: in expansion of macro ‘LOOP_BLOCKED’\n 675 | LOOP_BLOCKED(@type@, 64) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_binary_scalar1_less_FLOAT’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:703:5: note: in expansion of macro ‘LOOP_BLOCKED’\n 703 | LOOP_BLOCKED(@type@, 64) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_binary_scalar2_less_FLOAT’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:727:5: note: in expansion of macro ‘LOOP_BLOCKED’\n 727 | LOOP_BLOCKED(@type@, 64) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_binary_less_equal_FLOAT’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:675:5: note: in expansion of macro ‘LOOP_BLOCKED’\n 675 | LOOP_BLOCKED(@type@, 64) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_binary_scalar1_less_equal_FLOAT’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:703:5: note: in expansion of macro ‘LOOP_BLOCKED’\n 703 | LOOP_BLOCKED(@type@, 64) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_binary_scalar2_less_equal_FLOAT’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:727:5: note: in expansion of macro ‘LOOP_BLOCKED’\n 727 | LOOP_BLOCKED(@type@, 64) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_binary_greater_FLOAT’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:675:5: note: in expansion of macro ‘LOOP_BLOCKED’\n 675 | LOOP_BLOCKED(@type@, 64) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_binary_scalar1_greater_FLOAT’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:703:5: note: in expansion of macro ‘LOOP_BLOCKED’\n 703 | LOOP_BLOCKED(@type@, 64) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_binary_scalar2_greater_FLOAT’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:727:5: note: in expansion of macro ‘LOOP_BLOCKED’\n 727 | LOOP_BLOCKED(@type@, 64) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_binary_greater_equal_FLOAT’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:675:5: note: in expansion of macro ‘LOOP_BLOCKED’\n 675 | LOOP_BLOCKED(@type@, 64) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_binary_scalar1_greater_equal_FLOAT’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:703:5: note: in expansion of macro ‘LOOP_BLOCKED’\n 703 | LOOP_BLOCKED(@type@, 64) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_binary_scalar2_greater_equal_FLOAT’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:727:5: note: in expansion of macro ‘LOOP_BLOCKED’\n 727 | LOOP_BLOCKED(@type@, 64) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_sqrt_FLOAT’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:753:9: note: in expansion of macro ‘LOOP_BLOCKED’\n 753 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:759:9: note: in expansion of macro ‘LOOP_BLOCKED’\n 759 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_absolute_FLOAT’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:804:9: note: in expansion of macro ‘LOOP_BLOCKED’\n 804 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:810:9: note: in expansion of macro ‘LOOP_BLOCKED’\n 810 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_negative_FLOAT’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:804:9: note: in expansion of macro ‘LOOP_BLOCKED’\n 804 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:810:9: note: in expansion of macro ‘LOOP_BLOCKED’\n 810 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n In file included from numpy/core/src/umath/loops.c.src:39:\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_maximum_FLOAT’:\n numpy/core/src/umath/simd.inc.src:836:24: warning: comparison of integer expressions of different signedness: ‘long unsigned int’ and ‘npy_intp’ {aka ‘const long int’} [-Wsign-compare]\n 836 | if (i + 3 * stride <= n) {\n | ^~\n In file included from numpy/core/src/umath/loops.c.src:39:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:844:9: note: in expansion of macro ‘LOOP_BLOCKED’\n 844 | LOOP_BLOCKED(@type@, 32) {\n | ^~~~~~~~~~~~\n In file included from numpy/core/src/umath/loops.c.src:39:\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_minimum_FLOAT’:\n numpy/core/src/umath/simd.inc.src:836:24: warning: comparison of integer expressions of different signedness: ‘long unsigned int’ and ‘npy_intp’ {aka ‘const long int’} [-Wsign-compare]\n 836 | if (i + 3 * stride <= n) {\n | ^~\n In file included from numpy/core/src/umath/loops.c.src:39:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:844:9: note: in expansion of macro ‘LOOP_BLOCKED’\n 844 | LOOP_BLOCKED(@type@, 32) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_binary_add_DOUBLE’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:428:13: note: in expansion of macro ‘LOOP_BLOCKED’\n 428 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:435:13: note: in expansion of macro ‘LOOP_BLOCKED’\n 435 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:444:9: note: in expansion of macro ‘LOOP_BLOCKED’\n 444 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:452:9: note: in expansion of macro ‘LOOP_BLOCKED’\n 452 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:461:13: note: in expansion of macro ‘LOOP_BLOCKED’\n 461 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:468:13: note: in expansion of macro ‘LOOP_BLOCKED’\n 468 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_binary_scalar1_add_DOUBLE’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:489:9: note: in expansion of macro ‘LOOP_BLOCKED’\n 489 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:496:9: note: in expansion of macro ‘LOOP_BLOCKED’\n 496 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_binary_scalar2_add_DOUBLE’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:515:9: note: in expansion of macro ‘LOOP_BLOCKED’\n 515 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:522:9: note: in expansion of macro ‘LOOP_BLOCKED’\n 522 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_binary_subtract_DOUBLE’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:428:13: note: in expansion of macro ‘LOOP_BLOCKED’\n 428 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:435:13: note: in expansion of macro ‘LOOP_BLOCKED’\n 435 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:444:9: note: in expansion of macro ‘LOOP_BLOCKED’\n 444 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:452:9: note: in expansion of macro ‘LOOP_BLOCKED’\n 452 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:461:13: note: in expansion of macro ‘LOOP_BLOCKED’\n 461 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:468:13: note: in expansion of macro ‘LOOP_BLOCKED’\n 468 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_binary_scalar1_subtract_DOUBLE’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:489:9: note: in expansion of macro ‘LOOP_BLOCKED’\n 489 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:496:9: note: in expansion of macro ‘LOOP_BLOCKED’\n 496 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_binary_scalar2_subtract_DOUBLE’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:515:9: note: in expansion of macro ‘LOOP_BLOCKED’\n 515 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:522:9: note: in expansion of macro ‘LOOP_BLOCKED’\n 522 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_binary_multiply_DOUBLE’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:428:13: note: in expansion of macro ‘LOOP_BLOCKED’\n 428 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:435:13: note: in expansion of macro ‘LOOP_BLOCKED’\n 435 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:444:9: note: in expansion of macro ‘LOOP_BLOCKED’\n 444 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:452:9: note: in expansion of macro ‘LOOP_BLOCKED’\n 452 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:461:13: note: in expansion of macro ‘LOOP_BLOCKED’\n 461 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:468:13: note: in expansion of macro ‘LOOP_BLOCKED’\n 468 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_binary_scalar1_multiply_DOUBLE’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:489:9: note: in expansion of macro ‘LOOP_BLOCKED’\n 489 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:496:9: note: in expansion of macro ‘LOOP_BLOCKED’\n 496 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_binary_scalar2_multiply_DOUBLE’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:515:9: note: in expansion of macro ‘LOOP_BLOCKED’\n 515 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:522:9: note: in expansion of macro ‘LOOP_BLOCKED’\n 522 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_binary_divide_DOUBLE’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:428:13: note: in expansion of macro ‘LOOP_BLOCKED’\n 428 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:435:13: note: in expansion of macro ‘LOOP_BLOCKED’\n 435 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:444:9: note: in expansion of macro ‘LOOP_BLOCKED’\n 444 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:452:9: note: in expansion of macro ‘LOOP_BLOCKED’\n 452 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:461:13: note: in expansion of macro ‘LOOP_BLOCKED’\n 461 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:468:13: note: in expansion of macro ‘LOOP_BLOCKED’\n 468 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_binary_scalar1_divide_DOUBLE’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:489:9: note: in expansion of macro ‘LOOP_BLOCKED’\n 489 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:496:9: note: in expansion of macro ‘LOOP_BLOCKED’\n 496 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_binary_scalar2_divide_DOUBLE’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:515:9: note: in expansion of macro ‘LOOP_BLOCKED’\n 515 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:522:9: note: in expansion of macro ‘LOOP_BLOCKED’\n 522 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_signbit_DOUBLE’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:564:5: note: in expansion of macro ‘LOOP_BLOCKED’\n 564 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_isnan_DOUBLE’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:605:5: note: in expansion of macro ‘LOOP_BLOCKED’\n 605 | LOOP_BLOCKED(@type@, 64) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_isfinite_DOUBLE’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:605:5: note: in expansion of macro ‘LOOP_BLOCKED’\n 605 | LOOP_BLOCKED(@type@, 64) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_isinf_DOUBLE’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:605:5: note: in expansion of macro ‘LOOP_BLOCKED’\n 605 | LOOP_BLOCKED(@type@, 64) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_binary_equal_DOUBLE’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:675:5: note: in expansion of macro ‘LOOP_BLOCKED’\n 675 | LOOP_BLOCKED(@type@, 64) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_binary_scalar1_equal_DOUBLE’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:703:5: note: in expansion of macro ‘LOOP_BLOCKED’\n 703 | LOOP_BLOCKED(@type@, 64) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_binary_scalar2_equal_DOUBLE’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:727:5: note: in expansion of macro ‘LOOP_BLOCKED’\n 727 | LOOP_BLOCKED(@type@, 64) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_binary_not_equal_DOUBLE’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:675:5: note: in expansion of macro ‘LOOP_BLOCKED’\n 675 | LOOP_BLOCKED(@type@, 64) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_binary_scalar1_not_equal_DOUBLE’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:703:5: note: in expansion of macro ‘LOOP_BLOCKED’\n 703 | LOOP_BLOCKED(@type@, 64) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_binary_scalar2_not_equal_DOUBLE’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:727:5: note: in expansion of macro ‘LOOP_BLOCKED’\n 727 | LOOP_BLOCKED(@type@, 64) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_binary_less_DOUBLE’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:675:5: note: in expansion of macro ‘LOOP_BLOCKED’\n 675 | LOOP_BLOCKED(@type@, 64) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_binary_scalar1_less_DOUBLE’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:703:5: note: in expansion of macro ‘LOOP_BLOCKED’\n 703 | LOOP_BLOCKED(@type@, 64) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_binary_scalar2_less_DOUBLE’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:727:5: note: in expansion of macro ‘LOOP_BLOCKED’\n 727 | LOOP_BLOCKED(@type@, 64) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_binary_less_equal_DOUBLE’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:675:5: note: in expansion of macro ‘LOOP_BLOCKED’\n 675 | LOOP_BLOCKED(@type@, 64) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_binary_scalar1_less_equal_DOUBLE’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:703:5: note: in expansion of macro ‘LOOP_BLOCKED’\n 703 | LOOP_BLOCKED(@type@, 64) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_binary_scalar2_less_equal_DOUBLE’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:727:5: note: in expansion of macro ‘LOOP_BLOCKED’\n 727 | LOOP_BLOCKED(@type@, 64) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_binary_greater_DOUBLE’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:675:5: note: in expansion of macro ‘LOOP_BLOCKED’\n 675 | LOOP_BLOCKED(@type@, 64) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_binary_scalar1_greater_DOUBLE’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:703:5: note: in expansion of macro ‘LOOP_BLOCKED’\n 703 | LOOP_BLOCKED(@type@, 64) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_binary_scalar2_greater_DOUBLE’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:727:5: note: in expansion of macro ‘LOOP_BLOCKED’\n 727 | LOOP_BLOCKED(@type@, 64) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_binary_greater_equal_DOUBLE’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:675:5: note: in expansion of macro ‘LOOP_BLOCKED’\n 675 | LOOP_BLOCKED(@type@, 64) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_binary_scalar1_greater_equal_DOUBLE’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:703:5: note: in expansion of macro ‘LOOP_BLOCKED’\n 703 | LOOP_BLOCKED(@type@, 64) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_binary_scalar2_greater_equal_DOUBLE’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:727:5: note: in expansion of macro ‘LOOP_BLOCKED’\n 727 | LOOP_BLOCKED(@type@, 64) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_sqrt_DOUBLE’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:753:9: note: in expansion of macro ‘LOOP_BLOCKED’\n 753 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:759:9: note: in expansion of macro ‘LOOP_BLOCKED’\n 759 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_absolute_DOUBLE’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:804:9: note: in expansion of macro ‘LOOP_BLOCKED’\n 804 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:810:9: note: in expansion of macro ‘LOOP_BLOCKED’\n 810 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_negative_DOUBLE’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:804:9: note: in expansion of macro ‘LOOP_BLOCKED’\n 804 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:810:9: note: in expansion of macro ‘LOOP_BLOCKED’\n 810 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n In file included from numpy/core/src/umath/loops.c.src:39:\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_maximum_DOUBLE’:\n numpy/core/src/umath/simd.inc.src:836:24: warning: comparison of integer expressions of different signedness: ‘long unsigned int’ and ‘npy_intp’ {aka ‘const long int’} [-Wsign-compare]\n 836 | if (i + 3 * stride <= n) {\n | ^~\n In file included from numpy/core/src/umath/loops.c.src:39:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:844:9: note: in expansion of macro ‘LOOP_BLOCKED’\n 844 | LOOP_BLOCKED(@type@, 32) {\n | ^~~~~~~~~~~~\n In file included from numpy/core/src/umath/loops.c.src:39:\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_minimum_DOUBLE’:\n numpy/core/src/umath/simd.inc.src:836:24: warning: comparison of integer expressions of different signedness: ‘long unsigned int’ and ‘npy_intp’ {aka ‘const long int’} [-Wsign-compare]\n 836 | if (i + 3 * stride <= n) {\n | ^~\n In file included from numpy/core/src/umath/loops.c.src:39:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:844:9: note: in expansion of macro ‘LOOP_BLOCKED’\n 844 | LOOP_BLOCKED(@type@, 32) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_binary_logical_or_BOOL’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:910:5: note: in expansion of macro ‘LOOP_BLOCKED’\n 910 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_reduce_logical_or_BOOL’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:942:5: note: in expansion of macro ‘LOOP_BLOCKED’\n 942 | LOOP_BLOCKED(npy_bool, 32) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_binary_logical_and_BOOL’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:910:5: note: in expansion of macro ‘LOOP_BLOCKED’\n 910 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_reduce_logical_and_BOOL’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:942:5: note: in expansion of macro ‘LOOP_BLOCKED’\n 942 | LOOP_BLOCKED(npy_bool, 32) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_absolute_BOOL’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:984:5: note: in expansion of macro ‘LOOP_BLOCKED’\n 984 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/simd.inc.src: In function ‘sse2_logical_not_BOOL’:\n numpy/core/src/umath/simd.inc.src:107:13: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 107 | for(; i < npy_blocked_end(peel, sizeof(type), vsize, n);\\\n | ^\n numpy/core/src/umath/simd.inc.src:984:5: note: in expansion of macro ‘LOOP_BLOCKED’\n 984 | LOOP_BLOCKED(@type@, 16) {\n | ^~~~~~~~~~~~\n numpy/core/src/umath/loops.c.src: In function ‘pairwise_sum_FLOAT’:\n numpy/core/src/umath/loops.c.src:1635:23: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 1635 | for (i = 0; i < n; i++) {\n | ^\n numpy/core/src/umath/loops.c.src:1658:23: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 1658 | for (i = 8; i < n - (n % 8); i += 8) {\n | ^\n numpy/core/src/umath/loops.c.src:1676:18: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 1676 | for (; i < n; i++) {\n | ^\n numpy/core/src/umath/loops.c.src: In function ‘pairwise_sum_DOUBLE’:\n numpy/core/src/umath/loops.c.src:1635:23: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 1635 | for (i = 0; i < n; i++) {\n | ^\n numpy/core/src/umath/loops.c.src:1658:23: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 1658 | for (i = 8; i < n - (n % 8); i += 8) {\n | ^\n numpy/core/src/umath/loops.c.src:1676:18: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 1676 | for (; i < n; i++) {\n | ^\n numpy/core/src/umath/loops.c.src: In function ‘pairwise_sum_LONGDOUBLE’:\n numpy/core/src/umath/loops.c.src:1635:23: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 1635 | for (i = 0; i < n; i++) {\n | ^\n numpy/core/src/umath/loops.c.src:1658:23: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 1658 | for (i = 8; i < n - (n % 8); i += 8) {\n | ^\n numpy/core/src/umath/loops.c.src:1676:18: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 1676 | for (; i < n; i++) {\n | ^\n numpy/core/src/umath/loops.c.src: In function ‘pairwise_sum_HALF’:\n numpy/core/src/umath/loops.c.src:1635:23: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 1635 | for (i = 0; i < n; i++) {\n | ^\n numpy/core/src/umath/loops.c.src:1658:23: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 1658 | for (i = 8; i < n - (n % 8); i += 8) {\n | ^\n numpy/core/src/umath/loops.c.src:1676:18: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 1676 | for (; i < n; i++) {\n | ^\n numpy/core/src/umath/loops.c.src: In function ‘pairwise_sum_CFLOAT’:\n numpy/core/src/umath/loops.c.src:2410:23: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 2410 | for (i = 0; i < n; i += 2) {\n | ^\n numpy/core/src/umath/loops.c.src:2434:23: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 2434 | for (i = 8; i < n - (n % 8); i += 8) {\n | ^\n numpy/core/src/umath/loops.c.src:2452:18: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 2452 | for (; i < n; i+=2) {\n | ^\n numpy/core/src/umath/loops.c.src: In function ‘pairwise_sum_CDOUBLE’:\n numpy/core/src/umath/loops.c.src:2410:23: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 2410 | for (i = 0; i < n; i += 2) {\n | ^\n numpy/core/src/umath/loops.c.src:2434:23: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 2434 | for (i = 8; i < n - (n % 8); i += 8) {\n | ^\n numpy/core/src/umath/loops.c.src:2452:18: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 2452 | for (; i < n; i+=2) {\n | ^\n numpy/core/src/umath/loops.c.src: In function ‘pairwise_sum_CLONGDOUBLE’:\n numpy/core/src/umath/loops.c.src:2410:23: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 2410 | for (i = 0; i < n; i += 2) {\n | ^\n numpy/core/src/umath/loops.c.src:2434:23: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 2434 | for (i = 8; i < n - (n % 8); i += 8) {\n | ^\n numpy/core/src/umath/loops.c.src:2452:18: warning: comparison of integer expressions of different signedness: ‘npy_intp’ {aka ‘long int’} and ‘npy_uintp’ {aka ‘long unsigned int’} [-Wsign-compare]\n 2452 | for (; i < n; i+=2) {\n | ^\n gcc: numpy/core/src/umath/ufunc_object.c\n numpy/core/src/umath/ufunc_object.c: In function ‘PyUFunc_GenericReduction’:\n numpy/core/src/umath/ufunc_object.c:3912:15: warning: unused variable ‘out_obj’ [-Wunused-variable]\n 3912 | PyObject *out_obj = NULL;\n | ^~~~~~~\n gcc: build/src.linux-x86_64-3.7/numpy/core/src/umath/scalarmath.c\n gcc: numpy/core/src/umath/ufunc_type_resolution.c\n gcc: numpy/core/src/umath/override.c\n gcc: numpy/core/src/private/mem_overlap.c\n numpy/core/src/private/mem_overlap.c: In function ‘diophantine_dfs’:\n numpy/core/src/private/mem_overlap.c:420:31: warning: comparison of integer expressions of different signedness: ‘int’ and ‘unsigned int’ [-Wsign-compare]\n 420 | for (j = 0; j < n; ++j) {\n | ^\n numpy/core/src/private/mem_overlap.c: In function ‘strides_to_terms’:\n numpy/core/src/private/mem_overlap.c:715:19: warning: comparison of integer expressions of different signedness: ‘unsigned int’ and ‘int’ [-Wsign-compare]\n 715 | for (i = 0; i < PyArray_NDIM(arr); ++i) {\n | ^\n numpy/core/src/private/mem_overlap.c: In function ‘solve_may_share_memory’:\n numpy/core/src/private/mem_overlap.c:801:13: warning: comparison of integer expressions of different signedness: ‘npy_int64’ {aka ‘long int’} and ‘long unsigned int’ [-Wsign-compare]\n 801 | if (rhs != (npy_uintp)rhs) {\n | ^~\n numpy/core/src/private/mem_overlap.c: In function ‘solve_may_have_internal_overlap’:\n numpy/core/src/private/mem_overlap.c:890:19: warning: comparison of integer expressions of different signedness: ‘int’ and ‘unsigned int’ [-Wsign-compare]\n 890 | for (j = 0; j < nterms; ++j) {\n | ^\n numpy/core/src/private/mem_overlap.c:908:19: warning: comparison of integer expressions of different signedness: ‘int’ and ‘unsigned int’ [-Wsign-compare]\n 908 | for (j = 0; j < nterms; ++j) {\n | ^\n gcc: numpy/core/src/private/ufunc_override.c\n gcc -pthread -shared -B /home/masrur/anaconda3/compiler_compat -L/home/masrur/anaconda3/lib -Wl,-rpath=/home/masrur/anaconda3/lib -Wl,--no-as-needed -Wl,--sysroot=/ build/temp.linux-x86_64-3.7/numpy/core/src/umath/umathmodule.o build/temp.linux-x86_64-3.7/numpy/core/src/umath/reduction.o build/temp.linux-x86_64-3.7/build/src.linux-x86_64-3.7/numpy/core/src/umath/loops.o build/temp.linux-x86_64-3.7/numpy/core/src/umath/ufunc_object.o build/temp.linux-x86_64-3.7/build/src.linux-x86_64-3.7/numpy/core/src/umath/scalarmath.o build/temp.linux-x86_64-3.7/numpy/core/src/umath/ufunc_type_resolution.o build/temp.linux-x86_64-3.7/numpy/core/src/umath/override.o build/temp.linux-x86_64-3.7/numpy/core/src/private/mem_overlap.o build/temp.linux-x86_64-3.7/numpy/core/src/private/ufunc_override.o -Lbuild/temp.linux-x86_64-3.7 -lnpymath -lm -o build/lib.linux-x86_64-3.7/numpy/core/umath.cpython-37m-x86_64-linux-gnu.so\n building 'numpy.core.umath_tests' extension\n compiling C sources\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-DNPY_INTERNAL_BUILD=1 -DHAVE_NPY_CONFIG_H=1 -D_FILE_OFFSET_BITS=64 -D_LARGEFILE_SOURCE=1 -D_LARGEFILE64_SOURCE=1 -Inumpy/core/include -Ibuild/src.linux-x86_64-3.7/numpy/core/include/numpy -Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -Ibuild/src.linux-x86_64-3.7/numpy/core/src/private -Ibuild/src.linux-x86_64-3.7/numpy/core/src/npymath -Ibuild/src.linux-x86_64-3.7/numpy/core/src/private -Ibuild/src.linux-x86_64-3.7/numpy/core/src/npymath -Ibuild/src.linux-x86_64-3.7/numpy/core/src/private -Ibuild/src.linux-x86_64-3.7/numpy/core/src/npymath -c'\n gcc: build/src.linux-x86_64-3.7/numpy/core/src/umath/umath_tests.c\n gcc -pthread -shared -B /home/masrur/anaconda3/compiler_compat -L/home/masrur/anaconda3/lib -Wl,-rpath=/home/masrur/anaconda3/lib -Wl,--no-as-needed -Wl,--sysroot=/ build/temp.linux-x86_64-3.7/build/src.linux-x86_64-3.7/numpy/core/src/umath/umath_tests.o -Lbuild/temp.linux-x86_64-3.7 -o build/lib.linux-x86_64-3.7/numpy/core/umath_tests.cpython-37m-x86_64-linux-gnu.so\n building 'numpy.core.test_rational' extension\n compiling C sources\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-DNPY_INTERNAL_BUILD=1 -DHAVE_NPY_CONFIG_H=1 -D_FILE_OFFSET_BITS=64 -D_LARGEFILE_SOURCE=1 -D_LARGEFILE64_SOURCE=1 -Inumpy/core/include -Ibuild/src.linux-x86_64-3.7/numpy/core/include/numpy -Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -Ibuild/src.linux-x86_64-3.7/numpy/core/src/private -Ibuild/src.linux-x86_64-3.7/numpy/core/src/npymath -Ibuild/src.linux-x86_64-3.7/numpy/core/src/private -Ibuild/src.linux-x86_64-3.7/numpy/core/src/npymath -Ibuild/src.linux-x86_64-3.7/numpy/core/src/private -Ibuild/src.linux-x86_64-3.7/numpy/core/src/npymath -c'\n gcc: build/src.linux-x86_64-3.7/numpy/core/src/umath/test_rational.c\n gcc -pthread -shared -B /home/masrur/anaconda3/compiler_compat -L/home/masrur/anaconda3/lib -Wl,-rpath=/home/masrur/anaconda3/lib -Wl,--no-as-needed -Wl,--sysroot=/ build/temp.linux-x86_64-3.7/build/src.linux-x86_64-3.7/numpy/core/src/umath/test_rational.o -Lbuild/temp.linux-x86_64-3.7 -o build/lib.linux-x86_64-3.7/numpy/core/test_rational.cpython-37m-x86_64-linux-gnu.so\n building 'numpy.core.struct_ufunc_test' extension\n compiling C sources\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-DNPY_INTERNAL_BUILD=1 -DHAVE_NPY_CONFIG_H=1 -D_FILE_OFFSET_BITS=64 -D_LARGEFILE_SOURCE=1 -D_LARGEFILE64_SOURCE=1 -Inumpy/core/include -Ibuild/src.linux-x86_64-3.7/numpy/core/include/numpy -Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -Ibuild/src.linux-x86_64-3.7/numpy/core/src/private -Ibuild/src.linux-x86_64-3.7/numpy/core/src/npymath -Ibuild/src.linux-x86_64-3.7/numpy/core/src/private -Ibuild/src.linux-x86_64-3.7/numpy/core/src/npymath -Ibuild/src.linux-x86_64-3.7/numpy/core/src/private -Ibuild/src.linux-x86_64-3.7/numpy/core/src/npymath -c'\n gcc: build/src.linux-x86_64-3.7/numpy/core/src/umath/struct_ufunc_test.c\n gcc -pthread -shared -B /home/masrur/anaconda3/compiler_compat -L/home/masrur/anaconda3/lib -Wl,-rpath=/home/masrur/anaconda3/lib -Wl,--no-as-needed -Wl,--sysroot=/ build/temp.linux-x86_64-3.7/build/src.linux-x86_64-3.7/numpy/core/src/umath/struct_ufunc_test.o -Lbuild/temp.linux-x86_64-3.7 -o build/lib.linux-x86_64-3.7/numpy/core/struct_ufunc_test.cpython-37m-x86_64-linux-gnu.so\n building 'numpy.core.multiarray_tests' extension\n compiling C sources\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-DNPY_INTERNAL_BUILD=1 -DHAVE_NPY_CONFIG_H=1 -D_FILE_OFFSET_BITS=64 -D_LARGEFILE_SOURCE=1 -D_LARGEFILE64_SOURCE=1 -Inumpy/core/include -Ibuild/src.linux-x86_64-3.7/numpy/core/include/numpy -Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -Ibuild/src.linux-x86_64-3.7/numpy/core/src/private -Ibuild/src.linux-x86_64-3.7/numpy/core/src/npymath -Ibuild/src.linux-x86_64-3.7/numpy/core/src/private -Ibuild/src.linux-x86_64-3.7/numpy/core/src/npymath -Ibuild/src.linux-x86_64-3.7/numpy/core/src/private -Ibuild/src.linux-x86_64-3.7/numpy/core/src/npymath -c'\n gcc: build/src.linux-x86_64-3.7/numpy/core/src/multiarray/multiarray_tests.c\n numpy/core/src/multiarray/multiarray_tests.c.src: In function ‘array_solve_diophantine’:\n numpy/core/src/multiarray/multiarray_tests.c.src:989:29: warning: comparison of integer expressions of different signedness: ‘Py_ssize_t’ {aka ‘long int’} and ‘long unsigned int’ [-Wsign-compare]\n 989 | if (PyTuple_GET_SIZE(A) > sizeof(terms) / sizeof(diophantine_term_t)) {\n | ^\n gcc: numpy/core/src/private/mem_overlap.c\n numpy/core/src/private/mem_overlap.c: In function ‘diophantine_dfs’:\n numpy/core/src/private/mem_overlap.c:420:31: warning: comparison of integer expressions of different signedness: ‘int’ and ‘unsigned int’ [-Wsign-compare]\n 420 | for (j = 0; j < n; ++j) {\n | ^\n numpy/core/src/private/mem_overlap.c: In function ‘strides_to_terms’:\n numpy/core/src/private/mem_overlap.c:715:19: warning: comparison of integer expressions of different signedness: ‘unsigned int’ and ‘int’ [-Wsign-compare]\n 715 | for (i = 0; i < PyArray_NDIM(arr); ++i) {\n | ^\n numpy/core/src/private/mem_overlap.c: In function ‘solve_may_share_memory’:\n numpy/core/src/private/mem_overlap.c:801:13: warning: comparison of integer expressions of different signedness: ‘npy_int64’ {aka ‘long int’} and ‘long unsigned int’ [-Wsign-compare]\n 801 | if (rhs != (npy_uintp)rhs) {\n | ^~\n numpy/core/src/private/mem_overlap.c: In function ‘solve_may_have_internal_overlap’:\n numpy/core/src/private/mem_overlap.c:890:19: warning: comparison of integer expressions of different signedness: ‘int’ and ‘unsigned int’ [-Wsign-compare]\n 890 | for (j = 0; j < nterms; ++j) {\n | ^\n numpy/core/src/private/mem_overlap.c:908:19: warning: comparison of integer expressions of different signedness: ‘int’ and ‘unsigned int’ [-Wsign-compare]\n 908 | for (j = 0; j < nterms; ++j) {\n | ^\n gcc -pthread -shared -B /home/masrur/anaconda3/compiler_compat -L/home/masrur/anaconda3/lib -Wl,-rpath=/home/masrur/anaconda3/lib -Wl,--no-as-needed -Wl,--sysroot=/ build/temp.linux-x86_64-3.7/build/src.linux-x86_64-3.7/numpy/core/src/multiarray/multiarray_tests.o build/temp.linux-x86_64-3.7/numpy/core/src/private/mem_overlap.o -Lbuild/temp.linux-x86_64-3.7 -o build/lib.linux-x86_64-3.7/numpy/core/multiarray_tests.cpython-37m-x86_64-linux-gnu.so\n building 'numpy.core.operand_flag_tests' extension\n compiling C sources\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-DNPY_INTERNAL_BUILD=1 -DHAVE_NPY_CONFIG_H=1 -D_FILE_OFFSET_BITS=64 -D_LARGEFILE_SOURCE=1 -D_LARGEFILE64_SOURCE=1 -Inumpy/core/include -Ibuild/src.linux-x86_64-3.7/numpy/core/include/numpy -Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -Ibuild/src.linux-x86_64-3.7/numpy/core/src/private -Ibuild/src.linux-x86_64-3.7/numpy/core/src/npymath -Ibuild/src.linux-x86_64-3.7/numpy/core/src/private -Ibuild/src.linux-x86_64-3.7/numpy/core/src/npymath -Ibuild/src.linux-x86_64-3.7/numpy/core/src/private -Ibuild/src.linux-x86_64-3.7/numpy/core/src/npymath -c'\n gcc: build/src.linux-x86_64-3.7/numpy/core/src/umath/operand_flag_tests.c\n gcc -pthread -shared -B /home/masrur/anaconda3/compiler_compat -L/home/masrur/anaconda3/lib -Wl,-rpath=/home/masrur/anaconda3/lib -Wl,--no-as-needed -Wl,--sysroot=/ build/temp.linux-x86_64-3.7/build/src.linux-x86_64-3.7/numpy/core/src/umath/operand_flag_tests.o -Lbuild/temp.linux-x86_64-3.7 -o build/lib.linux-x86_64-3.7/numpy/core/operand_flag_tests.cpython-37m-x86_64-linux-gnu.so\n building 'numpy.fft.fftpack_lite' extension\n compiling C sources\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n creating build/temp.linux-x86_64-3.7/numpy/fft\n compile options: '-Inumpy/core/include -Ibuild/src.linux-x86_64-3.7/numpy/core/include/numpy -Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -Ibuild/src.linux-x86_64-3.7/numpy/core/src/private -Ibuild/src.linux-x86_64-3.7/numpy/core/src/npymath -Ibuild/src.linux-x86_64-3.7/numpy/core/src/private -Ibuild/src.linux-x86_64-3.7/numpy/core/src/npymath -Ibuild/src.linux-x86_64-3.7/numpy/core/src/private -Ibuild/src.linux-x86_64-3.7/numpy/core/src/npymath -c'\n gcc: numpy/fft/fftpack_litemodule.c\n gcc: numpy/fft/fftpack.c\n gcc -pthread -shared -B /home/masrur/anaconda3/compiler_compat -L/home/masrur/anaconda3/lib -Wl,-rpath=/home/masrur/anaconda3/lib -Wl,--no-as-needed -Wl,--sysroot=/ build/temp.linux-x86_64-3.7/numpy/fft/fftpack_litemodule.o build/temp.linux-x86_64-3.7/numpy/fft/fftpack.o -Lbuild/temp.linux-x86_64-3.7 -o build/lib.linux-x86_64-3.7/numpy/fft/fftpack_lite.cpython-37m-x86_64-linux-gnu.so\n building 'numpy.linalg.lapack_lite' extension\n compiling C sources\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n creating build/temp.linux-x86_64-3.7/numpy/linalg\n creating build/temp.linux-x86_64-3.7/numpy/linalg/lapack_lite\n compile options: '-DSCIPY_MKL_H -DHAVE_CBLAS -I/usr/local/include -I/usr/include -I/home/masrur/anaconda3/include -Inumpy/core/include -Ibuild/src.linux-x86_64-3.7/numpy/core/include/numpy -Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -Ibuild/src.linux-x86_64-3.7/numpy/core/src/private -Ibuild/src.linux-x86_64-3.7/numpy/core/src/npymath -Ibuild/src.linux-x86_64-3.7/numpy/core/src/private -Ibuild/src.linux-x86_64-3.7/numpy/core/src/npymath -Ibuild/src.linux-x86_64-3.7/numpy/core/src/private -Ibuild/src.linux-x86_64-3.7/numpy/core/src/npymath -c'\n gcc: numpy/linalg/lapack_litemodule.c\n gcc: numpy/linalg/lapack_lite/python_xerbla.c\n gcc -pthread -shared -B /home/masrur/anaconda3/compiler_compat -L/home/masrur/anaconda3/lib -Wl,-rpath=/home/masrur/anaconda3/lib -Wl,--no-as-needed -Wl,--sysroot=/ build/temp.linux-x86_64-3.7/numpy/linalg/lapack_litemodule.o build/temp.linux-x86_64-3.7/numpy/linalg/lapack_lite/python_xerbla.o -L/home/masrur/anaconda3/lib -Lbuild/temp.linux-x86_64-3.7 -lmkl_rt -lpthread -o build/lib.linux-x86_64-3.7/numpy/linalg/lapack_lite.cpython-37m-x86_64-linux-gnu.so\n building 'numpy.linalg._umath_linalg' extension\n compiling C sources\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n creating build/temp.linux-x86_64-3.7/build/src.linux-x86_64-3.7/numpy/linalg\n compile options: '-DSCIPY_MKL_H -DHAVE_CBLAS -I/usr/local/include -I/usr/include -I/home/masrur/anaconda3/include -Inumpy/core/include -Ibuild/src.linux-x86_64-3.7/numpy/core/include/numpy -Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -Ibuild/src.linux-x86_64-3.7/numpy/core/src/private -Ibuild/src.linux-x86_64-3.7/numpy/core/src/npymath -Ibuild/src.linux-x86_64-3.7/numpy/core/src/private -Ibuild/src.linux-x86_64-3.7/numpy/core/src/npymath -Ibuild/src.linux-x86_64-3.7/numpy/core/src/private -Ibuild/src.linux-x86_64-3.7/numpy/core/src/npymath -c'\n gcc: build/src.linux-x86_64-3.7/numpy/linalg/umath_linalg.c\n numpy/linalg/umath_linalg.c.src: In function ‘linearize_FLOAT_matrix’:\n numpy/linalg/umath_linalg.c.src:831:23: warning: comparison of integer expressions of different signedness: ‘int’ and ‘size_t’ {aka ‘const long unsigned int’} [-Wsign-compare]\n 831 | for (i = 0; i < data->rows; i++) {\n | ^\n numpy/linalg/umath_linalg.c.src: In function ‘delinearize_FLOAT_matrix’:\n numpy/linalg/umath_linalg.c.src:877:23: warning: comparison of integer expressions of different signedness: ‘int’ and ‘size_t’ {aka ‘const long unsigned int’} [-Wsign-compare]\n 877 | for (i = 0; i < data->rows; i++) {\n | ^\n numpy/linalg/umath_linalg.c.src: In function ‘nan_FLOAT_matrix’:\n numpy/linalg/umath_linalg.c.src:917:19: warning: comparison of integer expressions of different signedness: ‘int’ and ‘size_t’ {aka ‘const long unsigned int’} [-Wsign-compare]\n 917 | for (i = 0; i < data->rows; i++) {\n | ^\n numpy/linalg/umath_linalg.c.src:920:23: warning: comparison of integer expressions of different signedness: ‘int’ and ‘size_t’ {aka ‘const long unsigned int’} [-Wsign-compare]\n 920 | for (j = 0; j < data->columns; ++j) {\n | ^\n numpy/linalg/umath_linalg.c.src: In function ‘linearize_DOUBLE_matrix’:\n numpy/linalg/umath_linalg.c.src:831:23: warning: comparison of integer expressions of different signedness: ‘int’ and ‘size_t’ {aka ‘const long unsigned int’} [-Wsign-compare]\n 831 | for (i = 0; i < data->rows; i++) {\n | ^\n numpy/linalg/umath_linalg.c.src: In function ‘delinearize_DOUBLE_matrix’:\n numpy/linalg/umath_linalg.c.src:877:23: warning: comparison of integer expressions of different signedness: ‘int’ and ‘size_t’ {aka ‘const long unsigned int’} [-Wsign-compare]\n 877 | for (i = 0; i < data->rows; i++) {\n | ^\n numpy/linalg/umath_linalg.c.src: In function ‘nan_DOUBLE_matrix’:\n numpy/linalg/umath_linalg.c.src:917:19: warning: comparison of integer expressions of different signedness: ‘int’ and ‘size_t’ {aka ‘const long unsigned int’} [-Wsign-compare]\n 917 | for (i = 0; i < data->rows; i++) {\n | ^\n numpy/linalg/umath_linalg.c.src:920:23: warning: comparison of integer expressions of different signedness: ‘int’ and ‘size_t’ {aka ‘const long unsigned int’} [-Wsign-compare]\n 920 | for (j = 0; j < data->columns; ++j) {\n | ^\n numpy/linalg/umath_linalg.c.src: In function ‘linearize_CFLOAT_matrix’:\n numpy/linalg/umath_linalg.c.src:831:23: warning: comparison of integer expressions of different signedness: ‘int’ and ‘size_t’ {aka ‘const long unsigned int’} [-Wsign-compare]\n 831 | for (i = 0; i < data->rows; i++) {\n | ^\n numpy/linalg/umath_linalg.c.src: In function ‘delinearize_CFLOAT_matrix’:\n numpy/linalg/umath_linalg.c.src:877:23: warning: comparison of integer expressions of different signedness: ‘int’ and ‘size_t’ {aka ‘const long unsigned int’} [-Wsign-compare]\n 877 | for (i = 0; i < data->rows; i++) {\n | ^\n numpy/linalg/umath_linalg.c.src: In function ‘nan_CFLOAT_matrix’:\n numpy/linalg/umath_linalg.c.src:917:19: warning: comparison of integer expressions of different signedness: ‘int’ and ‘size_t’ {aka ‘const long unsigned int’} [-Wsign-compare]\n 917 | for (i = 0; i < data->rows; i++) {\n | ^\n numpy/linalg/umath_linalg.c.src:920:23: warning: comparison of integer expressions of different signedness: ‘int’ and ‘size_t’ {aka ‘const long unsigned int’} [-Wsign-compare]\n 920 | for (j = 0; j < data->columns; ++j) {\n | ^\n numpy/linalg/umath_linalg.c.src: In function ‘linearize_CDOUBLE_matrix’:\n numpy/linalg/umath_linalg.c.src:831:23: warning: comparison of integer expressions of different signedness: ‘int’ and ‘size_t’ {aka ‘const long unsigned int’} [-Wsign-compare]\n 831 | for (i = 0; i < data->rows; i++) {\n | ^\n numpy/linalg/umath_linalg.c.src: In function ‘delinearize_CDOUBLE_matrix’:\n numpy/linalg/umath_linalg.c.src:877:23: warning: comparison of integer expressions of different signedness: ‘int’ and ‘size_t’ {aka ‘const long unsigned int’} [-Wsign-compare]\n 877 | for (i = 0; i < data->rows; i++) {\n | ^\n numpy/linalg/umath_linalg.c.src: In function ‘nan_CDOUBLE_matrix’:\n numpy/linalg/umath_linalg.c.src:917:19: warning: comparison of integer expressions of different signedness: ‘int’ and ‘size_t’ {aka ‘const long unsigned int’} [-Wsign-compare]\n 917 | for (i = 0; i < data->rows; i++) {\n | ^\n numpy/linalg/umath_linalg.c.src:920:23: warning: comparison of integer expressions of different signedness: ‘int’ and ‘size_t’ {aka ‘const long unsigned int’} [-Wsign-compare]\n 920 | for (j = 0; j < data->columns; ++j) {\n | ^\n gcc -pthread -shared -B /home/masrur/anaconda3/compiler_compat -L/home/masrur/anaconda3/lib -Wl,-rpath=/home/masrur/anaconda3/lib -Wl,--no-as-needed -Wl,--sysroot=/ build/temp.linux-x86_64-3.7/build/src.linux-x86_64-3.7/numpy/linalg/umath_linalg.o build/temp.linux-x86_64-3.7/numpy/linalg/lapack_lite/python_xerbla.o -L/home/masrur/anaconda3/lib -Lbuild/temp.linux-x86_64-3.7 -lnpymath -lmkl_rt -lpthread -o build/lib.linux-x86_64-3.7/numpy/linalg/_umath_linalg.cpython-37m-x86_64-linux-gnu.so\n building 'numpy.random.mtrand' extension\n compiling C sources\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n creating build/temp.linux-x86_64-3.7/numpy/random\n creating build/temp.linux-x86_64-3.7/numpy/random/mtrand\n compile options: '-D_FILE_OFFSET_BITS=64 -D_LARGEFILE_SOURCE=1 -D_LARGEFILE64_SOURCE=1 -Inumpy/core/include -Ibuild/src.linux-x86_64-3.7/numpy/core/include/numpy -Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -Ibuild/src.linux-x86_64-3.7/numpy/core/src/private -Ibuild/src.linux-x86_64-3.7/numpy/core/src/npymath -Ibuild/src.linux-x86_64-3.7/numpy/core/src/private -Ibuild/src.linux-x86_64-3.7/numpy/core/src/npymath -Ibuild/src.linux-x86_64-3.7/numpy/core/src/private -Ibuild/src.linux-x86_64-3.7/numpy/core/src/npymath -c'\n gcc: numpy/random/mtrand/mtrand.c\n numpy/random/mtrand/mtrand.c: In function ‘__Pyx_PyCFunction_FastCall’:\n numpy/random/mtrand/mtrand.c:44374:13: error: too many arguments to function ‘(PyObject * (*)(PyObject *, PyObject * const*, Py_ssize_t))meth’\n 44374 | return (*((__Pyx_PyCFunctionFast)meth)) (self, args, nargs, NULL);\n | ~^~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~\n numpy/random/mtrand/mtrand.c: In function ‘__Pyx__ExceptionSave’:\n numpy/random/mtrand/mtrand.c:44793:21: error: ‘PyThreadState’ {aka ‘struct _ts’} has no member named ‘exc_type’; did you mean ‘curexc_type’?\n 44793 | *type = tstate->exc_type;\n | ^~~~~~~~\n | curexc_type\n numpy/random/mtrand/mtrand.c:44794:22: error: ‘PyThreadState’ {aka ‘struct _ts’} has no member named ‘exc_value’; did you mean ‘curexc_value’?\n 44794 | *value = tstate->exc_value;\n | ^~~~~~~~~\n | curexc_value\n numpy/random/mtrand/mtrand.c:44795:19: error: ‘PyThreadState’ {aka ‘struct _ts’} has no member named ‘exc_traceback’; did you mean ‘curexc_traceback’?\n 44795 | *tb = tstate->exc_traceback;\n | ^~~~~~~~~~~~~\n | curexc_traceback\n numpy/random/mtrand/mtrand.c: In function ‘__Pyx__ExceptionReset’:\n numpy/random/mtrand/mtrand.c:44802:24: error: ‘PyThreadState’ {aka ‘struct _ts’} has no member named ‘exc_type’; did you mean ‘curexc_type’?\n 44802 | tmp_type = tstate->exc_type;\n | ^~~~~~~~\n | curexc_type\n numpy/random/mtrand/mtrand.c:44803:25: error: ‘PyThreadState’ {aka ‘struct _ts’} has no member named ‘exc_value’; did you mean ‘curexc_value’?\n 44803 | tmp_value = tstate->exc_value;\n | ^~~~~~~~~\n | curexc_value\n numpy/random/mtrand/mtrand.c:44804:22: error: ‘PyThreadState’ {aka ‘struct _ts’} has no member named ‘exc_traceback’; did you mean ‘curexc_traceback’?\n 44804 | tmp_tb = tstate->exc_traceback;\n | ^~~~~~~~~~~~~\n | curexc_traceback\n numpy/random/mtrand/mtrand.c:44805:13: error: ‘PyThreadState’ {aka ‘struct _ts’} has no member named ‘exc_type’; did you mean ‘curexc_type’?\n 44805 | tstate->exc_type = type;\n | ^~~~~~~~\n | curexc_type\n numpy/random/mtrand/mtrand.c:44806:13: error: ‘PyThreadState’ {aka ‘struct _ts’} has no member named ‘exc_value’; did you mean ‘curexc_value’?\n 44806 | tstate->exc_value = value;\n | ^~~~~~~~~\n | curexc_value\n numpy/random/mtrand/mtrand.c:44807:13: error: ‘PyThreadState’ {aka ‘struct _ts’} has no member named ‘exc_traceback’; did you mean ‘curexc_traceback’?\n 44807 | tstate->exc_traceback = tb;\n | ^~~~~~~~~~~~~\n | curexc_traceback\n numpy/random/mtrand/mtrand.c: In function ‘__Pyx__GetException’:\n numpy/random/mtrand/mtrand.c:44862:24: error: ‘PyThreadState’ {aka ‘struct _ts’} has no member named ‘exc_type’; did you mean ‘curexc_type’?\n 44862 | tmp_type = tstate->exc_type;\n | ^~~~~~~~\n | curexc_type\n numpy/random/mtrand/mtrand.c:44863:25: error: ‘PyThreadState’ {aka ‘struct _ts’} has no member named ‘exc_value’; did you mean ‘curexc_value’?\n 44863 | tmp_value = tstate->exc_value;\n | ^~~~~~~~~\n | curexc_value\n numpy/random/mtrand/mtrand.c:44864:22: error: ‘PyThreadState’ {aka ‘struct _ts’} has no member named ‘exc_traceback’; did you mean ‘curexc_traceback’?\n 44864 | tmp_tb = tstate->exc_traceback;\n | ^~~~~~~~~~~~~\n | curexc_traceback\n numpy/random/mtrand/mtrand.c:44865:13: error: ‘PyThreadState’ {aka ‘struct _ts’} has no member named ‘exc_type’; did you mean ‘curexc_type’?\n 44865 | tstate->exc_type = local_type;\n | ^~~~~~~~\n | curexc_type\n numpy/random/mtrand/mtrand.c:44866:13: error: ‘PyThreadState’ {aka ‘struct _ts’} has no member named ‘exc_value’; did you mean ‘curexc_value’?\n 44866 | tstate->exc_value = local_value;\n | ^~~~~~~~~\n | curexc_value\n numpy/random/mtrand/mtrand.c:44867:13: error: ‘PyThreadState’ {aka ‘struct _ts’} has no member named ‘exc_traceback’; did you mean ‘curexc_traceback’?\n 44867 | tstate->exc_traceback = local_tb;\n | ^~~~~~~~~~~~~\n | curexc_traceback\n numpy/random/mtrand/mtrand.c: In function ‘__Pyx_PyCFunction_FastCall’:\n numpy/random/mtrand/mtrand.c:44374:13: error: too many arguments to function ‘(PyObject * (*)(PyObject *, PyObject * const*, Py_ssize_t))meth’\n 44374 | return (*((__Pyx_PyCFunctionFast)meth)) (self, args, nargs, NULL);\n | ~^~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~\n numpy/random/mtrand/mtrand.c: In function ‘__Pyx__ExceptionSave’:\n numpy/random/mtrand/mtrand.c:44793:21: error: ‘PyThreadState’ {aka ‘struct _ts’} has no member named ‘exc_type’; did you mean ‘curexc_type’?\n 44793 | *type = tstate->exc_type;\n | ^~~~~~~~\n | curexc_type\n numpy/random/mtrand/mtrand.c:44794:22: error: ‘PyThreadState’ {aka ‘struct _ts’} has no member named ‘exc_value’; did you mean ‘curexc_value’?\n 44794 | *value = tstate->exc_value;\n | ^~~~~~~~~\n | curexc_value\n numpy/random/mtrand/mtrand.c:44795:19: error: ‘PyThreadState’ {aka ‘struct _ts’} has no member named ‘exc_traceback’; did you mean ‘curexc_traceback’?\n 44795 | *tb = tstate->exc_traceback;\n | ^~~~~~~~~~~~~\n | curexc_traceback\n numpy/random/mtrand/mtrand.c: In function ‘__Pyx__ExceptionReset’:\n numpy/random/mtrand/mtrand.c:44802:24: error: ‘PyThreadState’ {aka ‘struct _ts’} has no member named ‘exc_type’; did you mean ‘curexc_type’?\n 44802 | tmp_type = tstate->exc_type;\n | ^~~~~~~~\n | curexc_type\n numpy/random/mtrand/mtrand.c:44803:25: error: ‘PyThreadState’ {aka ‘struct _ts’} has no member named ‘exc_value’; did you mean ‘curexc_value’?\n 44803 | tmp_value = tstate->exc_value;\n | ^~~~~~~~~\n | curexc_value\n numpy/random/mtrand/mtrand.c:44804:22: error: ‘PyThreadState’ {aka ‘struct _ts’} has no member named ‘exc_traceback’; did you mean ‘curexc_traceback’?\n 44804 | tmp_tb = tstate->exc_traceback;\n | ^~~~~~~~~~~~~\n | curexc_traceback\n numpy/random/mtrand/mtrand.c:44805:13: error: ‘PyThreadState’ {aka ‘struct _ts’} has no member named ‘exc_type’; did you mean ‘curexc_type’?\n 44805 | tstate->exc_type = type;\n | ^~~~~~~~\n | curexc_type\n numpy/random/mtrand/mtrand.c:44806:13: error: ‘PyThreadState’ {aka ‘struct _ts’} has no member named ‘exc_value’; did you mean ‘curexc_value’?\n 44806 | tstate->exc_value = value;\n | ^~~~~~~~~\n | curexc_value\n numpy/random/mtrand/mtrand.c:44807:13: error: ‘PyThreadState’ {aka ‘struct _ts’} has no member named ‘exc_traceback’; did you mean ‘curexc_traceback’?\n 44807 | tstate->exc_traceback = tb;\n | ^~~~~~~~~~~~~\n | curexc_traceback\n numpy/random/mtrand/mtrand.c: In function ‘__Pyx__GetException’:\n numpy/random/mtrand/mtrand.c:44862:24: error: ‘PyThreadState’ {aka ‘struct _ts’} has no member named ‘exc_type’; did you mean ‘curexc_type’?\n 44862 | tmp_type = tstate->exc_type;\n | ^~~~~~~~\n | curexc_type\n numpy/random/mtrand/mtrand.c:44863:25: error: ‘PyThreadState’ {aka ‘struct _ts’} has no member named ‘exc_value’; did you mean ‘curexc_value’?\n 44863 | tmp_value = tstate->exc_value;\n | ^~~~~~~~~\n | curexc_value\n numpy/random/mtrand/mtrand.c:44864:22: error: ‘PyThreadState’ {aka ‘struct _ts’} has no member named ‘exc_traceback’; did you mean ‘curexc_traceback’?\n 44864 | tmp_tb = tstate->exc_traceback;\n | ^~~~~~~~~~~~~\n | curexc_traceback\n numpy/random/mtrand/mtrand.c:44865:13: error: ‘PyThreadState’ {aka ‘struct _ts’} has no member named ‘exc_type’; did you mean ‘curexc_type’?\n 44865 | tstate->exc_type = local_type;\n | ^~~~~~~~\n | curexc_type\n numpy/random/mtrand/mtrand.c:44866:13: error: ‘PyThreadState’ {aka ‘struct _ts’} has no member named ‘exc_value’; did you mean ‘curexc_value’?\n 44866 | tstate->exc_value = local_value;\n | ^~~~~~~~~\n | curexc_value\n numpy/random/mtrand/mtrand.c:44867:13: error: ‘PyThreadState’ {aka ‘struct _ts’} has no member named ‘exc_traceback’; did you mean ‘curexc_traceback’?\n 44867 | tstate->exc_traceback = local_tb;\n | ^~~~~~~~~~~~~\n | curexc_traceback\n error: Command \"gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC -D_FILE_OFFSET_BITS=64 -D_LARGEFILE_SOURCE=1 -D_LARGEFILE64_SOURCE=1 -Inumpy/core/include -Ibuild/src.linux-x86_64-3.7/numpy/core/include/numpy -Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -Ibuild/src.linux-x86_64-3.7/numpy/core/src/private -Ibuild/src.linux-x86_64-3.7/numpy/core/src/npymath -Ibuild/src.linux-x86_64-3.7/numpy/core/src/private -Ibuild/src.linux-x86_64-3.7/numpy/core/src/npymath -Ibuild/src.linux-x86_64-3.7/numpy/core/src/private -Ibuild/src.linux-x86_64-3.7/numpy/core/src/npymath -c numpy/random/mtrand/mtrand.c -o build/temp.linux-x86_64-3.7/numpy/random/mtrand/mtrand.o -MMD -MF build/temp.linux-x86_64-3.7/numpy/random/mtrand/mtrand.o.d\" failed with exit status 1\n ----------------------------------------\n ERROR: Failed building wheel for numpy\n Running setup.py clean for numpy\n ERROR: Command errored out with exit status 1:\n command: /home/masrur/anaconda3/bin/python -u -c 'import sys, setuptools, tokenize; sys.argv[0] = '\"'\"'/tmp/pip-install-wkrshu4d/numpy/setup.py'\"'\"'; __file__='\"'\"'/tmp/pip-install-wkrshu4d/numpy/setup.py'\"'\"';f=getattr(tokenize, '\"'\"'open'\"'\"', open)(__file__);code=f.read().replace('\"'\"'\\r\\n'\"'\"', '\"'\"'\\n'\"'\"');f.close();exec(compile(code, __file__, '\"'\"'exec'\"'\"'))' clean --all\n cwd: /tmp/pip-install-wkrshu4d/numpy\n Complete output (10 lines):\n Running from numpy source directory.\n \n `setup.py clean` is not supported, use one of the following instead:\n \n - `git clean -xdf` (cleans all files)\n - `git clean -Xdf` (cleans all versioned files, doesn't touch\n files that aren't checked into the git repo)\n \n Add `--force` to your command to use it anyway if you must (unsupported).\n \n ----------------------------------------\n ERROR: Failed cleaning build dir for numpy\n Failed to build numpy\n ERROR: Error checking for conflicts.\n Traceback (most recent call last):\n File \"/home/masrur/.local/lib/python3.7/site-packages/pip/_vendor/pkg_resources/__init__.py\", line 3021, in _dep_map\n return self.__dep_map\n File \"/home/masrur/.local/lib/python3.7/site-packages/pip/_vendor/pkg_resources/__init__.py\", line 2815, in __getattr__\n raise AttributeError(attr)\n AttributeError: _DistInfoDistribution__dep_map\n \n During handling of the above exception, another exception occurred:\n \n Traceback (most recent call last):\n File \"/home/masrur/.local/lib/python3.7/site-packages/pip/_vendor/pkg_resources/__init__.py\", line 3012, in _parsed_pkg_info\n return self._pkg_info\n File \"/home/masrur/.local/lib/python3.7/site-packages/pip/_vendor/pkg_resources/__init__.py\", line 2815, in __getattr__\n raise AttributeError(attr)\n AttributeError: _pkg_info\n \n During handling of the above exception, another exception occurred:\n \n Traceback (most recent call last):\n File \"/home/masrur/.local/lib/python3.7/site-packages/pip/_internal/commands/install.py\", line 517, in _warn_about_conflicts\n package_set, _dep_info = check_install_conflicts(to_install)\n File \"/home/masrur/.local/lib/python3.7/site-packages/pip/_internal/operations/check.py\", line 114, in check_install_conflicts\n package_set, _ = create_package_set_from_installed()\n File \"/home/masrur/.local/lib/python3.7/site-packages/pip/_internal/operations/check.py\", line 53, in create_package_set_from_installed\n package_set[name] = PackageDetails(dist.version, dist.requires())\n File \"/home/masrur/.local/lib/python3.7/site-packages/pip/_vendor/pkg_resources/__init__.py\", line 2736, in requires\n dm = self._dep_map\n File \"/home/masrur/.local/lib/python3.7/site-packages/pip/_vendor/pkg_resources/__init__.py\", line 3023, in _dep_map\n self.__dep_map = self._compute_dependencies()\n File \"/home/masrur/.local/lib/python3.7/site-packages/pip/_vendor/pkg_resources/__init__.py\", line 3032, in _compute_dependencies\n for req in self._parsed_pkg_info.get_all('Requires-Dist') or []:\n File \"/home/masrur/.local/lib/python3.7/site-packages/pip/_vendor/pkg_resources/__init__.py\", line 3014, in _parsed_pkg_info\n metadata = self.get_metadata(self.PKG_INFO)\n File \"/home/masrur/.local/lib/python3.7/site-packages/pip/_vendor/pkg_resources/__init__.py\", line 1420, in get_metadata\n value = self._get(path)\n File \"/home/masrur/.local/lib/python3.7/site-packages/pip/_vendor/pkg_resources/__init__.py\", line 1616, in _get\n with open(path, 'rb') as stream:\n FileNotFoundError: [Errno 2] No such file or directory: '/home/masrur/.local/lib/python3.7/site-packages/~andas-1.0.2.dist-info/METADATA'\n Installing collected packages: wheel, setuptools, Cython, numpy\n Running setup.py install for numpy: started\n Running setup.py install for numpy: finished with status 'error'\n ERROR: Command errored out with exit status 1:\n command: /home/masrur/anaconda3/bin/python -u -c 'import sys, setuptools, tokenize; sys.argv[0] = '\"'\"'/tmp/pip-install-wkrshu4d/numpy/setup.py'\"'\"'; __file__='\"'\"'/tmp/pip-install-wkrshu4d/numpy/setup.py'\"'\"';f=getattr(tokenize, '\"'\"'open'\"'\"', open)(__file__);code=f.read().replace('\"'\"'\\r\\n'\"'\"', '\"'\"'\\n'\"'\"');f.close();exec(compile(code, __file__, '\"'\"'exec'\"'\"'))' install --record /tmp/pip-record-iwi5uo6n/install-record.txt --single-version-externally-managed --prefix /tmp/pip-build-env-q_sd8e0o/overlay --compile --install-headers /tmp/pip-build-env-q_sd8e0o/overlay/include/python3.7m/numpy\n cwd: /tmp/pip-install-wkrshu4d/numpy/\n Complete output (286 lines):\n Running from numpy source directory.\n \n Note: if you need reliable uninstall behavior, then install\n with pip instead of using `setup.py install`:\n \n - `pip install .` (from a git repo or downloaded source\n release)\n - `pip install numpy` (last NumPy release on PyPi)\n \n \n blas_opt_info:\n blas_mkl_info:\n FOUND:\n libraries = ['mkl_rt', 'pthread']\n library_dirs = ['/home/masrur/anaconda3/lib']\n define_macros = [('SCIPY_MKL_H', None), ('HAVE_CBLAS', None)]\n include_dirs = ['/usr/local/include', '/usr/include', '/home/masrur/anaconda3/include']\n \n FOUND:\n libraries = ['mkl_rt', 'pthread']\n library_dirs = ['/home/masrur/anaconda3/lib']\n define_macros = [('SCIPY_MKL_H', None), ('HAVE_CBLAS', None)]\n include_dirs = ['/usr/local/include', '/usr/include', '/home/masrur/anaconda3/include']\n \n /bin/sh: 1: svnversion: not found\n non-existing path in 'numpy/distutils': 'site.cfg'\n /bin/sh: 1: svnversion: not found\n F2PY Version 2\n lapack_opt_info:\n lapack_mkl_info:\n FOUND:\n libraries = ['mkl_rt', 'pthread']\n library_dirs = ['/home/masrur/anaconda3/lib']\n define_macros = [('SCIPY_MKL_H', None), ('HAVE_CBLAS', None)]\n include_dirs = ['/usr/local/include', '/usr/include', '/home/masrur/anaconda3/include']\n \n FOUND:\n libraries = ['mkl_rt', 'pthread']\n library_dirs = ['/home/masrur/anaconda3/lib']\n define_macros = [('SCIPY_MKL_H', None), ('HAVE_CBLAS', None)]\n include_dirs = ['/usr/local/include', '/usr/include', '/home/masrur/anaconda3/include']\n \n /home/masrur/anaconda3/lib/python3.7/distutils/dist.py:274: UserWarning: Unknown distribution option: 'define_macros'\n warnings.warn(msg)\n running install\n running build\n running config_cc\n unifing config_cc, config, build_clib, build_ext, build commands --compiler options\n running config_fc\n unifing config_fc, config, build_clib, build_ext, build commands --fcompiler options\n running build_src\n build_src\n building py_modules sources\n building library \"npymath\" sources\n customize Gnu95FCompiler\n Found executable /usr/bin/gfortran\n customize Gnu95FCompiler\n customize Gnu95FCompiler using config\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ _configtest.o -o _configtest\n success!\n removing: _configtest.c _configtest.o _configtest\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n _configtest.c:1:5: warning: conflicting types for built-in function ‘exp’; expected ‘double(double)’ [-Wbuiltin-declaration-mismatch]\n 1 | int exp (void);\n | ^~~\n _configtest.c:1:1: note: ‘exp’ is declared in header ‘<math.h>’\n +++ |+#include <math.h>\n 1 | int exp (void);\n gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ _configtest.o -o _configtest\n /home/masrur/anaconda3/compiler_compat/ld: _configtest.o: in function `main':\n /tmp/pip-install-wkrshu4d/numpy/_configtest.c:6: undefined reference to `exp'\n collect2: error: ld returned 1 exit status\n /home/masrur/anaconda3/compiler_compat/ld: _configtest.o: in function `main':\n /tmp/pip-install-wkrshu4d/numpy/_configtest.c:6: undefined reference to `exp'\n collect2: error: ld returned 1 exit status\n failure.\n removing: _configtest.c _configtest.o\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -c'\n gcc: _configtest.c\n _configtest.c:1:5: warning: conflicting types for built-in function ‘exp’; expected ‘double(double)’ [-Wbuiltin-declaration-mismatch]\n 1 | int exp (void);\n | ^~~\n _configtest.c:1:1: note: ‘exp’ is declared in header ‘<math.h>’\n +++ |+#include <math.h>\n 1 | int exp (void);\n gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ _configtest.o -lm -o _configtest\n success!\n removing: _configtest.c _configtest.o _configtest\n adding 'build/src.linux-x86_64-3.7/numpy/core/src/npymath' to include_dirs.\n None - nothing done with h_files = ['build/src.linux-x86_64-3.7/numpy/core/src/npymath/npy_math_internal.h']\n building library \"npysort\" sources\n adding 'build/src.linux-x86_64-3.7/numpy/core/src/private' to include_dirs.\n None - nothing done with h_files = ['build/src.linux-x86_64-3.7/numpy/core/src/private/npy_partition.h', 'build/src.linux-x86_64-3.7/numpy/core/src/private/npy_binsearch.h']\n building extension \"numpy.core._dummy\" sources\n adding 'build/src.linux-x86_64-3.7/numpy/core/include/numpy/config.h' to sources.\n adding 'build/src.linux-x86_64-3.7/numpy/core/include/numpy/_numpyconfig.h' to sources.\n executing numpy/core/code_generators/generate_numpy_api.py\n adding 'build/src.linux-x86_64-3.7/numpy/core/include/numpy/__multiarray_api.h' to sources.\n numpy.core - nothing done with h_files = ['build/src.linux-x86_64-3.7/numpy/core/include/numpy/config.h', 'build/src.linux-x86_64-3.7/numpy/core/include/numpy/_numpyconfig.h', 'build/src.linux-x86_64-3.7/numpy/core/include/numpy/__multiarray_api.h']\n building extension \"numpy.core.multiarray\" sources\n adding 'build/src.linux-x86_64-3.7/numpy/core/include/numpy/config.h' to sources.\n adding 'build/src.linux-x86_64-3.7/numpy/core/include/numpy/_numpyconfig.h' to sources.\n executing numpy/core/code_generators/generate_numpy_api.py\n adding 'build/src.linux-x86_64-3.7/numpy/core/include/numpy/__multiarray_api.h' to sources.\n adding 'build/src.linux-x86_64-3.7/numpy/core/src/private' to include_dirs.\n numpy.core - nothing done with h_files = ['build/src.linux-x86_64-3.7/numpy/core/src/private/templ_common.h', 'build/src.linux-x86_64-3.7/numpy/core/include/numpy/config.h', 'build/src.linux-x86_64-3.7/numpy/core/include/numpy/_numpyconfig.h', 'build/src.linux-x86_64-3.7/numpy/core/include/numpy/__multiarray_api.h']\n building extension \"numpy.core.umath\" sources\n adding 'build/src.linux-x86_64-3.7/numpy/core/include/numpy/config.h' to sources.\n adding 'build/src.linux-x86_64-3.7/numpy/core/include/numpy/_numpyconfig.h' to sources.\n executing numpy/core/code_generators/generate_ufunc_api.py\n adding 'build/src.linux-x86_64-3.7/numpy/core/include/numpy/__ufunc_api.h' to sources.\n adding 'build/src.linux-x86_64-3.7/numpy/core/src/umath' to include_dirs.\n numpy.core - nothing done with h_files = ['build/src.linux-x86_64-3.7/numpy/core/src/umath/funcs.inc', 'build/src.linux-x86_64-3.7/numpy/core/src/umath/simd.inc', 'build/src.linux-x86_64-3.7/numpy/core/src/umath/loops.h', 'build/src.linux-x86_64-3.7/numpy/core/include/numpy/config.h', 'build/src.linux-x86_64-3.7/numpy/core/include/numpy/_numpyconfig.h', 'build/src.linux-x86_64-3.7/numpy/core/include/numpy/__ufunc_api.h']\n building extension \"numpy.core.umath_tests\" sources\n building extension \"numpy.core.test_rational\" sources\n building extension \"numpy.core.struct_ufunc_test\" sources\n building extension \"numpy.core.multiarray_tests\" sources\n building extension \"numpy.core.operand_flag_tests\" sources\n building extension \"numpy.fft.fftpack_lite\" sources\n building extension \"numpy.linalg.lapack_lite\" sources\n adding 'numpy/linalg/lapack_lite/python_xerbla.c' to sources.\n building extension \"numpy.linalg._umath_linalg\" sources\n adding 'numpy/linalg/lapack_lite/python_xerbla.c' to sources.\n building extension \"numpy.random.mtrand\" sources\n building data_files sources\n build_src: building npy-pkg config files\n running build_py\n copying numpy/version.py -> build/lib.linux-x86_64-3.7/numpy\n copying build/src.linux-x86_64-3.7/numpy/__config__.py -> build/lib.linux-x86_64-3.7/numpy\n copying build/src.linux-x86_64-3.7/numpy/distutils/__config__.py -> build/lib.linux-x86_64-3.7/numpy/distutils\n running build_clib\n customize UnixCCompiler\n customize UnixCCompiler using build_clib\n running build_ext\n customize UnixCCompiler\n customize UnixCCompiler using build_ext\n building 'numpy.random.mtrand' extension\n compiling C sources\n C compiler: gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC\n \n compile options: '-D_FILE_OFFSET_BITS=64 -D_LARGEFILE_SOURCE=1 -D_LARGEFILE64_SOURCE=1 -Inumpy/core/include -Ibuild/src.linux-x86_64-3.7/numpy/core/include/numpy -Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -Ibuild/src.linux-x86_64-3.7/numpy/core/src/private -Ibuild/src.linux-x86_64-3.7/numpy/core/src/npymath -Ibuild/src.linux-x86_64-3.7/numpy/core/src/private -Ibuild/src.linux-x86_64-3.7/numpy/core/src/npymath -Ibuild/src.linux-x86_64-3.7/numpy/core/src/private -Ibuild/src.linux-x86_64-3.7/numpy/core/src/npymath -c'\n gcc: numpy/random/mtrand/mtrand.c\n numpy/random/mtrand/mtrand.c: In function ‘__Pyx_PyCFunction_FastCall’:\n numpy/random/mtrand/mtrand.c:44374:13: error: too many arguments to function ‘(PyObject * (*)(PyObject *, PyObject * const*, Py_ssize_t))meth’\n 44374 | return (*((__Pyx_PyCFunctionFast)meth)) (self, args, nargs, NULL);\n | ~^~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~\n numpy/random/mtrand/mtrand.c: In function ‘__Pyx__ExceptionSave’:\n numpy/random/mtrand/mtrand.c:44793:21: error: ‘PyThreadState’ {aka ‘struct _ts’} has no member named ‘exc_type’; did you mean ‘curexc_type’?\n 44793 | *type = tstate->exc_type;\n | ^~~~~~~~\n | curexc_type\n numpy/random/mtrand/mtrand.c:44794:22: error: ‘PyThreadState’ {aka ‘struct _ts’} has no member named ‘exc_value’; did you mean ‘curexc_value’?\n 44794 | *value = tstate->exc_value;\n | ^~~~~~~~~\n | curexc_value\n numpy/random/mtrand/mtrand.c:44795:19: error: ‘PyThreadState’ {aka ‘struct _ts’} has no member named ‘exc_traceback’; did you mean ‘curexc_traceback’?\n 44795 | *tb = tstate->exc_traceback;\n | ^~~~~~~~~~~~~\n | curexc_traceback\n numpy/random/mtrand/mtrand.c: In function ‘__Pyx__ExceptionReset’:\n numpy/random/mtrand/mtrand.c:44802:24: error: ‘PyThreadState’ {aka ‘struct _ts’} has no member named ‘exc_type’; did you mean ‘curexc_type’?\n 44802 | tmp_type = tstate->exc_type;\n | ^~~~~~~~\n | curexc_type\n numpy/random/mtrand/mtrand.c:44803:25: error: ‘PyThreadState’ {aka ‘struct _ts’} has no member named ‘exc_value’; did you mean ‘curexc_value’?\n 44803 | tmp_value = tstate->exc_value;\n | ^~~~~~~~~\n | curexc_value\n numpy/random/mtrand/mtrand.c:44804:22: error: ‘PyThreadState’ {aka ‘struct _ts’} has no member named ‘exc_traceback’; did you mean ‘curexc_traceback’?\n 44804 | tmp_tb = tstate->exc_traceback;\n | ^~~~~~~~~~~~~\n | curexc_traceback\n numpy/random/mtrand/mtrand.c:44805:13: error: ‘PyThreadState’ {aka ‘struct _ts’} has no member named ‘exc_type’; did you mean ‘curexc_type’?\n 44805 | tstate->exc_type = type;\n | ^~~~~~~~\n | curexc_type\n numpy/random/mtrand/mtrand.c:44806:13: error: ‘PyThreadState’ {aka ‘struct _ts’} has no member named ‘exc_value’; did you mean ‘curexc_value’?\n 44806 | tstate->exc_value = value;\n | ^~~~~~~~~\n | curexc_value\n numpy/random/mtrand/mtrand.c:44807:13: error: ‘PyThreadState’ {aka ‘struct _ts’} has no member named ‘exc_traceback’; did you mean ‘curexc_traceback’?\n 44807 | tstate->exc_traceback = tb;\n | ^~~~~~~~~~~~~\n | curexc_traceback\n numpy/random/mtrand/mtrand.c: In function ‘__Pyx__GetException’:\n numpy/random/mtrand/mtrand.c:44862:24: error: ‘PyThreadState’ {aka ‘struct _ts’} has no member named ‘exc_type’; did you mean ‘curexc_type’?\n 44862 | tmp_type = tstate->exc_type;\n | ^~~~~~~~\n | curexc_type\n numpy/random/mtrand/mtrand.c:44863:25: error: ‘PyThreadState’ {aka ‘struct _ts’} has no member named ‘exc_value’; did you mean ‘curexc_value’?\n 44863 | tmp_value = tstate->exc_value;\n | ^~~~~~~~~\n | curexc_value\n numpy/random/mtrand/mtrand.c:44864:22: error: ‘PyThreadState’ {aka ‘struct _ts’} has no member named ‘exc_traceback’; did you mean ‘curexc_traceback’?\n 44864 | tmp_tb = tstate->exc_traceback;\n | ^~~~~~~~~~~~~\n | curexc_traceback\n numpy/random/mtrand/mtrand.c:44865:13: error: ‘PyThreadState’ {aka ‘struct _ts’} has no member named ‘exc_type’; did you mean ‘curexc_type’?\n 44865 | tstate->exc_type = local_type;\n | ^~~~~~~~\n | curexc_type\n numpy/random/mtrand/mtrand.c:44866:13: error: ‘PyThreadState’ {aka ‘struct _ts’} has no member named ‘exc_value’; did you mean ‘curexc_value’?\n 44866 | tstate->exc_value = local_value;\n | ^~~~~~~~~\n | curexc_value\n numpy/random/mtrand/mtrand.c:44867:13: error: ‘PyThreadState’ {aka ‘struct _ts’} has no member named ‘exc_traceback’; did you mean ‘curexc_traceback’?\n 44867 | tstate->exc_traceback = local_tb;\n | ^~~~~~~~~~~~~\n | curexc_traceback\n numpy/random/mtrand/mtrand.c: In function ‘__Pyx_PyCFunction_FastCall’:\n numpy/random/mtrand/mtrand.c:44374:13: error: too many arguments to function ‘(PyObject * (*)(PyObject *, PyObject * const*, Py_ssize_t))meth’\n 44374 | return (*((__Pyx_PyCFunctionFast)meth)) (self, args, nargs, NULL);\n | ~^~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~\n numpy/random/mtrand/mtrand.c: In function ‘__Pyx__ExceptionSave’:\n numpy/random/mtrand/mtrand.c:44793:21: error: ‘PyThreadState’ {aka ‘struct _ts’} has no member named ‘exc_type’; did you mean ‘curexc_type’?\n 44793 | *type = tstate->exc_type;\n | ^~~~~~~~\n | curexc_type\n numpy/random/mtrand/mtrand.c:44794:22: error: ‘PyThreadState’ {aka ‘struct _ts’} has no member named ‘exc_value’; did you mean ‘curexc_value’?\n 44794 | *value = tstate->exc_value;\n | ^~~~~~~~~\n | curexc_value\n numpy/random/mtrand/mtrand.c:44795:19: error: ‘PyThreadState’ {aka ‘struct _ts’} has no member named ‘exc_traceback’; did you mean ‘curexc_traceback’?\n 44795 | *tb = tstate->exc_traceback;\n | ^~~~~~~~~~~~~\n | curexc_traceback\n numpy/random/mtrand/mtrand.c: In function ‘__Pyx__ExceptionReset’:\n numpy/random/mtrand/mtrand.c:44802:24: error: ‘PyThreadState’ {aka ‘struct _ts’} has no member named ‘exc_type’; did you mean ‘curexc_type’?\n 44802 | tmp_type = tstate->exc_type;\n | ^~~~~~~~\n | curexc_type\n numpy/random/mtrand/mtrand.c:44803:25: error: ‘PyThreadState’ {aka ‘struct _ts’} has no member named ‘exc_value’; did you mean ‘curexc_value’?\n 44803 | tmp_value = tstate->exc_value;\n | ^~~~~~~~~\n | curexc_value\n numpy/random/mtrand/mtrand.c:44804:22: error: ‘PyThreadState’ {aka ‘struct _ts’} has no member named ‘exc_traceback’; did you mean ‘curexc_traceback’?\n 44804 | tmp_tb = tstate->exc_traceback;\n | ^~~~~~~~~~~~~\n | curexc_traceback\n numpy/random/mtrand/mtrand.c:44805:13: error: ‘PyThreadState’ {aka ‘struct _ts’} has no member named ‘exc_type’; did you mean ‘curexc_type’?\n 44805 | tstate->exc_type = type;\n | ^~~~~~~~\n | curexc_type\n numpy/random/mtrand/mtrand.c:44806:13: error: ‘PyThreadState’ {aka ‘struct _ts’} has no member named ‘exc_value’; did you mean ‘curexc_value’?\n 44806 | tstate->exc_value = value;\n | ^~~~~~~~~\n | curexc_value\n numpy/random/mtrand/mtrand.c:44807:13: error: ‘PyThreadState’ {aka ‘struct _ts’} has no member named ‘exc_traceback’; did you mean ‘curexc_traceback’?\n 44807 | tstate->exc_traceback = tb;\n | ^~~~~~~~~~~~~\n | curexc_traceback\n numpy/random/mtrand/mtrand.c: In function ‘__Pyx__GetException’:\n numpy/random/mtrand/mtrand.c:44862:24: error: ‘PyThreadState’ {aka ‘struct _ts’} has no member named ‘exc_type’; did you mean ‘curexc_type’?\n 44862 | tmp_type = tstate->exc_type;\n | ^~~~~~~~\n | curexc_type\n numpy/random/mtrand/mtrand.c:44863:25: error: ‘PyThreadState’ {aka ‘struct _ts’} has no member named ‘exc_value’; did you mean ‘curexc_value’?\n 44863 | tmp_value = tstate->exc_value;\n | ^~~~~~~~~\n | curexc_value\n numpy/random/mtrand/mtrand.c:44864:22: error: ‘PyThreadState’ {aka ‘struct _ts’} has no member named ‘exc_traceback’; did you mean ‘curexc_traceback’?\n 44864 | tmp_tb = tstate->exc_traceback;\n | ^~~~~~~~~~~~~\n | curexc_traceback\n numpy/random/mtrand/mtrand.c:44865:13: error: ‘PyThreadState’ {aka ‘struct _ts’} has no member named ‘exc_type’; did you mean ‘curexc_type’?\n 44865 | tstate->exc_type = local_type;\n | ^~~~~~~~\n | curexc_type\n numpy/random/mtrand/mtrand.c:44866:13: error: ‘PyThreadState’ {aka ‘struct _ts’} has no member named ‘exc_value’; did you mean ‘curexc_value’?\n 44866 | tstate->exc_value = local_value;\n | ^~~~~~~~~\n | curexc_value\n numpy/random/mtrand/mtrand.c:44867:13: error: ‘PyThreadState’ {aka ‘struct _ts’} has no member named ‘exc_traceback’; did you mean ‘curexc_traceback’?\n 44867 | tstate->exc_traceback = local_tb;\n | ^~~~~~~~~~~~~\n | curexc_traceback\n error: Command \"gcc -pthread -B /home/masrur/anaconda3/compiler_compat -Wl,--sysroot=/ -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC -D_FILE_OFFSET_BITS=64 -D_LARGEFILE_SOURCE=1 -D_LARGEFILE64_SOURCE=1 -Inumpy/core/include -Ibuild/src.linux-x86_64-3.7/numpy/core/include/numpy -Inumpy/core/src/private -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/home/masrur/anaconda3/include/python3.7m -Ibuild/src.linux-x86_64-3.7/numpy/core/src/private -Ibuild/src.linux-x86_64-3.7/numpy/core/src/npymath -Ibuild/src.linux-x86_64-3.7/numpy/core/src/private -Ibuild/src.linux-x86_64-3.7/numpy/core/src/npymath -Ibuild/src.linux-x86_64-3.7/numpy/core/src/private -Ibuild/src.linux-x86_64-3.7/numpy/core/src/npymath -c numpy/random/mtrand/mtrand.c -o build/temp.linux-x86_64-3.7/numpy/random/mtrand/mtrand.o -MMD -MF build/temp.linux-x86_64-3.7/numpy/random/mtrand/mtrand.o.d\" failed with exit status 1\n ----------------------------------------\n ERROR: Command errored out with exit status 1: /home/masrur/anaconda3/bin/python -u -c 'import sys, setuptools, tokenize; sys.argv[0] = '\"'\"'/tmp/pip-install-wkrshu4d/numpy/setup.py'\"'\"'; __file__='\"'\"'/tmp/pip-install-wkrshu4d/numpy/setup.py'\"'\"';f=getattr(tokenize, '\"'\"'open'\"'\"', open)(__file__);code=f.read().replace('\"'\"'\\r\\n'\"'\"', '\"'\"'\\n'\"'\"');f.close();exec(compile(code, __file__, '\"'\"'exec'\"'\"'))' install --record /tmp/pip-record-iwi5uo6n/install-record.txt --single-version-externally-managed --prefix /tmp/pip-build-env-q_sd8e0o/overlay --compile --install-headers /tmp/pip-build-env-q_sd8e0o/overlay/include/python3.7m/numpy Check the logs for full command output.\n ----------------------------------------\u001b[0m\n"
]
],
[
[
"# Loading Data with Zipline\n\nBefore we build our pipeline with Zipline, we will first see how we can load the stock data we are going to use into Zipline. Zipline uses **Data Bundles** to make it easy to use different data sources. A data bundle is a collection of pricing data, adjustment data, and an asset database. Zipline employs data bundles to preload data used to run backtests and store data for future runs. Zipline comes with a few data bundles by default but it also has the ability to ingest new bundles. The first step to using a data bundle is to ingest the data. Zipline's ingestion process will start by downloading the data or by loading data files from your local machine. It will then pass the data to a set of writer objects that converts the original data to Zipline’s internal format (`bcolz` for pricing data, and `SQLite` for split/merger/dividend data) that hs been optimized for speed. This new data is written to a standard location that Zipline can find. By default, the new data is written to a subdirectory of `ZIPLINE_ROOT/data/<bundle>`, where `<bundle>` is the name given to the bundle ingested and the subdirectory is named with the current date. This allows Zipline to look at older data and run backtests on older copies of the data. Running a backtest with an old ingestion makes it easier to reproduce backtest results later. \n\nIn this notebook, we will be using stock data from **Quotemedia**. In the Udacity Workspace you will find that the stock data from Quotemedia has already been ingested into Zipline. Therefore, in the code below we will use Zipline's `bundles.load()` function to load our previously ingested stock data from Quotemedia. In order to use the `bundles.load()` function we first need to do a couple of things. First, we need to specify the name of the bundle previously ingested. In this case, the name of the Quotemedia data bundle is `eod-quotemedia`:",
"_____no_output_____"
]
],
[
[
"# Specify the bundle name\nbundle_name = 'eod-quotemedia'",
"_____no_output_____"
]
],
[
[
"Second, we need to register the data bundle and its ingest function with Zipline, using the `bundles.register()` function. The ingest function is responsible for loading the data into memory and passing it to a set of writer objects provided by Zipline to convert the data to Zipline’s internal format. Since the original Quotemedia data was contained in `.csv` files, we will use the `csvdir_equities()` function to generate the ingest function for our Quotemedia data bundle. In addition, since Quotemedia's `.csv` files contained daily stock data, we will set the time frame for our ingest function, to `daily`.",
"_____no_output_____"
]
],
[
[
"from zipline.data import bundles\nfrom zipline.data.bundles.csvdir import csvdir_equities\n\n# Create an ingest function \ningest_func = csvdir_equities(['daily'], bundle_name)\n\n# Register the data bundle and its ingest function\nbundles.register(bundle_name, ingest_func);",
"_____no_output_____"
]
],
[
[
"Once our data bundle and ingest function are registered, we can load our data using the `bundles.load()` function. Since this function loads our previously ingested data, we need to set `ZIPLINE_ROOT` to the path of the most recent ingested data. The most recent data is located in the `cwd/../../data/project_4_eod/` directory, where `cwd` is the current working directory. We will specify this location using the `os.environ[]` command.",
"_____no_output_____"
]
],
[
[
"import os\n\n# Set environment variable 'ZIPLINE_ROOT' to the path where the most recent data is located\nos.environ['ZIPLINE_ROOT'] = os.path.join(os.getcwd(),'project_4_eod')\n\n# Load the data bundle\nbundle_data = bundles.load(bundle_name)",
"_____no_output_____"
]
],
[
[
"# Building an Empty Pipeline\n\nOnce we have loaded our data, we can start building our Zipline pipeline. We begin by creating an empty Pipeline object using Zipline's `Pipeline` class. A Pipeline object represents a collection of named expressions to be compiled and executed by a Pipeline Engine. The `Pipeline(columns=None, screen=None)` class takes two optional parameters, `columns` and `screen`. The `columns` parameter is a dictionary used to indicate the intial columns to use, and the `screen` parameter is used to setup a screen to exclude unwanted data. \n\nIn the code below we will create a `screen` for our pipeline using Zipline's built-in `.AverageDollarVolume()` class. We will use the `.AverageDollarVolume()` class to produce a 60-day Average Dollar Volume of closing prices for every stock in our universe. We then use the `.top(10)` attribute to specify that we want to filter down our universe each day to just the top 10 assets. Therefore, this screen will act as a filter to exclude data from our stock universe each day. The average dollar volume is a good first pass filter to avoid illiquid assets.",
"_____no_output_____"
]
],
[
[
"from zipline.pipeline import Pipeline\nfrom zipline.pipeline.factors import AverageDollarVolume\n\n# Create a screen for our Pipeline\nuniverse = AverageDollarVolume(window_length = 60).top(10)\n\n# Create an empty Pipeline with the given screen\npipeline = Pipeline(screen = universe)",
"_____no_output_____"
]
],
[
[
"In the code above we have named our Pipeline object `pipeline` so that we can identify it later when we make computations. Remember a Pipeline is an object that represents computations we would like to perform every day. A freshly-constructed pipeline, like the one we just created, is empty. This means it doesn’t yet know how to compute anything, and it won’t produce any values if we ask for its outputs. In the sections below, we will see how to provide our Pipeline with expressions to compute.",
"_____no_output_____"
],
[
"# Factors and Filters\n\nThe `.AverageDollarVolume()` class used above is an example of a factor. In this section we will take a look at two types of computations that can be expressed in a pipeline: **Factors** and **Filters**. In general, factors and filters represent functions that produce a value from an asset in a moment in time, but are distinguished by the types of values they produce. Let's start by looking at factors.\n\n\n### Factors\n\nIn general, a **Factor** is a function from an asset at a particular moment of time to a numerical value. A simple example of a factor is the most recent price of a security. Given a security and a specific moment in time, the most recent price is a number. Another example is the 10-day average trading volume of a security. Factors are most commonly used to assign values to securities which can then be combined with filters or other factors. The fact that you can combine multiple factors makes it easy for you to form new custom factors that can be as complex as you like. For example, constructing a Factor that computes the average of two other Factors can be simply illustrated usingthe pseudocode below:\n\n```python\nf1 = factor1(...)\nf2 = factor2(...) \naverage = (f1 + f2) / 2.0 \n```\n\n### Filters\n\nIn general, a **Filter** is a function from an asset at a particular moment in time to a boolean value (True of False). An example of a filter is a function indicating whether a security's price is below \\$5. Given a security and a specific moment in time, this evaluates to either **True** or **False**. Filters are most commonly used for selecting sets of securities to include or exclude from your stock universe. Filters are usually applied using comparison operators, such as <, <=, !=, ==, >, >=.",
"_____no_output_____"
],
[
"# Viewing the Pipeline as a Diagram\n\nZipline's Pipeline class comes with the attribute `.show_graph()` that allows you to render the Pipeline as a Directed Acyclic Graph (DAG). This graph is specified using the DOT language and consequently we need a DOT graph layout program to view the rendered image. In the code below, we will use the Graphviz pakage to render the graph produced by the `.show_graph()` attribute. Graphviz is an open-source package for drawing graphs specified in DOT language scripts.",
"_____no_output_____"
]
],
[
[
"import graphviz\n\n# Render the pipeline as a DAG\npipeline.show_graph()",
"_____no_output_____"
]
],
[
[
"Right now, our pipeline is empty and it only contains a screen. Therefore, when we rendered our `pipeline`, we only see the diagram of our `screen`:\n\n```python\nAverageDollarVolume(window_length = 60).top(10)\n```\n\nBy default, the `.AverageDollarVolume()` class uses the `USEquityPricing` dataset, containing daily trading prices and volumes, to compute the average dollar volume:\n\n```python\naverage_dollar_volume = np.nansum(close_price * volume, axis=0) / len(close_price)\n```\nThe top of the diagram reflects the fact that the `.AverageDollarVolume()` class gets its inputs (closing price and volume) from the `USEquityPricing` dataset. The bottom of the diagram shows that the output is determined by the expression `x_0 <= 10`. This expression reflects the fact that we used `.top(10)` as a filter in our `screen`. We refer to each box in the diagram as a Term. ",
"_____no_output_____"
],
[
"# Datasets and Dataloaders \n\nOne of the features of Zipline's Pipeline is that it separates the actual source of the stock data from the abstract description of that dataset. Therefore, Zipline employs **DataSets** and **Loaders** for those datasets. `DataSets` are just abstract collections of sentinel values describing the columns/types for a particular dataset. While a `loader` is an object which, given a request for a particular chunk of a dataset, can actually get the requested data. For example, the loader used for the `USEquityPricing` dataset, is the `USEquityPricingLoader` class. The `USEquityPricingLoader` class will delegate the loading of baselines and adjustments to lower-level subsystems that know how to get the pricing data in the default formats used by Zipline (`bcolz` for pricing data, and `SQLite` for split/merger/dividend data). As we saw in the beginning of this notebook, data bundles automatically convert the stock data into `bcolz` and `SQLite` formats. It is important to note that the `USEquityPricingLoader` class can also be used to load daily OHLCV data from other datasets, not just from the `USEquityPricing` dataset. Simliarly, it is also possible to write different loaders for the same dataset and use those instead of the default loader. Zipline contains lots of other loaders to allow you to load data from different datasets.\n\nIn the code below, we will use `USEquityPricingLoader(BcolzDailyBarWriter, SQLiteAdjustmentWriter)` to create a loader from a `bcolz` equity pricing directory and a `SQLite` adjustments path. Both the `BcolzDailyBarWriter` and `SQLiteAdjustmentWriter` determine the path of the pricing and adjustment data. Since we will be using the Quotemedia data bundle, we will use the `bundle_data.equity_daily_bar_reader` and the `bundle_data.adjustment_reader` as our `BcolzDailyBarWriter` and `SQLiteAdjustmentWriter`, respectively.",
"_____no_output_____"
]
],
[
[
"from zipline.pipeline.loaders import USEquityPricingLoader\n\n# Set the dataloader\npricing_loader = USEquityPricingLoader(bundle_data.equity_daily_bar_reader, bundle_data.adjustment_reader)",
"_____no_output_____"
]
],
[
[
"# Pipeline Engine\n\nZipline employs computation engines for executing Pipelines. In the code below we will use Zipline's `SimplePipelineEngine()` class as the engine to execute our pipeline. The `SimplePipelineEngine(get_loader, calendar, asset_finder)` class associates the chosen data loader with the corresponding dataset and a trading calendar. The `get_loader` parameter must be a callable function that is given a loadable term and returns a `PipelineLoader` to use to retrieve the raw data for that term in the pipeline. In our case, we will be using the `pricing_loader` defined above, we therefore, create a function called `choose_loader` that returns our `pricing_loader`. The function also checks that the data that is being requested corresponds to OHLCV data, otherwise it retunrs an error. The `calendar` parameter must be a `DatetimeIndex` array of dates to consider as trading days when computing a range between a fixed `start_date` and `end_date`. In our case, we will be using the same trading days as those used in the NYSE. We will use Zipline's `get_calendar('NYSE')` function to retrieve the trading days used by the NYSE. We then use the `.all_sessions` attribute to get the `DatetimeIndex` from our `trading_calendar` and pass it to the `calendar` parameter. Finally, the `asset_finder` parameter determines which assets are in the top-level universe of our stock data at any point in time. Since we are using the Quotemedia data bundle, we set this parameter to the `bundle_data.asset_finder`.",
"_____no_output_____"
]
],
[
[
"from zipline.utils.calendars import get_calendar\nfrom zipline.pipeline.data import USEquityPricing\nfrom zipline.pipeline.engine import SimplePipelineEngine\n\n# Define the function for the get_loader parameter\ndef choose_loader(column):\n if column not in USEquityPricing.columns:\n raise Exception('Column not in USEquityPricing')\n return pricing_loader\n\n# Set the trading calendar\ntrading_calendar = get_calendar('NYSE')\n\n# Create a Pipeline engine\nengine = SimplePipelineEngine(get_loader = choose_loader,\n calendar = trading_calendar.all_sessions,\n asset_finder = bundle_data.asset_finder)",
"_____no_output_____"
]
],
[
[
"# Running a Pipeline\n\nOnce we have chosen our engine we are ready to run or execute our pipeline. We can run our pipeline by using the `.run_pipeline()` attribute of the `SimplePipelineEngine` class. In particular, the `SimplePipelineEngine.run_pipeline(pipeline, start_date, end_date)` implements the following algorithm for executing pipelines:\n\n\n1. Build a dependency graph of all terms in the `pipeline`. In this step, the graph is sorted topologically to determine the order in which we can compute the terms.\n\n\n2. Ask our AssetFinder for a “lifetimes matrix”, which should contain, for each date between `start_date` and `end_date`, a boolean value for each known asset indicating whether the asset existed on that date.\n\n\n3. Compute each term in the dependency order determined in step 1, caching the results in a a dictionary so that they can be fed into future terms.\n\n\n4. For each date, determine the number of assets passing the `pipeline` screen. The sum, $N$, of all these values is the total number of rows in our output Pandas Dataframe, so we pre-allocate an output array of length $N$ for each factor in terms.\n\n\n5. Fill in the arrays allocated in step 4 by copying computed values from our output cache into the corresponding rows.\n\n\n6. Stick the values computed in step 5 into a Pandas DataFrame and return it.\n\nIn the code below, we run our pipeline for a single day, so our `start_date` and `end_date` will be the same. We then print some information about our `pipeline_output`.",
"_____no_output_____"
]
],
[
[
"import pandas as pd\n\n# Set the start and end dates\nstart_date = pd.Timestamp('2016-01-05', tz = 'utc')\nend_date = pd.Timestamp('2016-01-05', tz = 'utc')\n\n# Run our pipeline for the given start and end dates\npipeline_output = engine.run_pipeline(pipeline, start_date, end_date)\n\n# We print information about the pipeline output\nprint('The pipeline output has type:', type(pipeline_output), '\\n')\n\n# We print whether the pipeline output is a MultiIndex Dataframe\nprint('Is the pipeline output a MultiIndex Dataframe:', isinstance(pipeline_output.index, pd.core.index.MultiIndex), '\\n')\n\n# If the pipeline output is a MultiIndex Dataframe we print the two levels of the index\nif isinstance(pipeline_output.index, pd.core.index.MultiIndex):\n\n # We print the index level 0\n print('Index Level 0:\\n\\n', pipeline_output.index.get_level_values(0), '\\n')\n\n # We print the index level 1\n print('Index Level 1:\\n\\n', pipeline_output.index.get_level_values(1), '\\n')",
"_____no_output_____"
]
],
[
[
"We can see above that the return value of `.run_pipeline()` is a `MultiIndex` Pandas DataFrame containing a row for each asset that passed our pipeline’s screen. We can also see that the 0th level of the index contains the date and the 1st level of the index contains the tickers. In general, the returned Pandas DataFrame will also contain a column for each factor and filter we add to the pipeline using `Pipeline.add()`. At this point we haven't added any factors or filters to our pipeline, consequently, the Pandas Dataframe will have no columns. In the following sections we will see how to add factors and filters to our pipeline.",
"_____no_output_____"
],
[
"# Get Tickers\n\nWe saw in the previous section, that the tickers of the stocks that passed our pipeline’s screen are contained in the 1st level of the index. Therefore, we can use the Pandas `.get_level_values(1).values.tolist()` method to get the tickers of those stocks and save them to a list.",
"_____no_output_____"
]
],
[
[
"# Get the values in index level 1 and save them to a list\nuniverse_tickers = pipeline_output.index.get_level_values(1).values.tolist()\n\n# Display the tickers\nuniverse_tickers",
"_____no_output_____"
]
],
[
[
"# Get Data\n\nNow that we have the tickers for the stocks that passed our pipeline’s screen, we can get the historical stock data for those tickers from our data bundle. In order to get the historical data we need to use Zipline's `DataPortal` class. A `DataPortal` is an interface to all of the data that a Zipline simulation needs. In the code below, we will create a `DataPortal` and `get_pricing` function to get historical stock prices for our tickers. \n\nWe have already seen most of the parameters used below when we create the `DataPortal`, so we won't explain them again here. The only new parameter is `first_trading_day`. The `first_trading_day` parameter is a `pd.Timestamp` indicating the first trading day for the simulation. We will set the first trading day to the first trading day in the data bundle. For more information on the `DataPortal` class see the [Zipline documentation](https://www.zipline.io/appendix.html?highlight=dataportal#zipline.data.data_portal.DataPortal)",
"_____no_output_____"
]
],
[
[
"from zipline.data.data_portal import DataPortal\n\n# Create a data portal\ndata_portal = DataPortal(bundle_data.asset_finder,\n trading_calendar = trading_calendar,\n first_trading_day = bundle_data.equity_daily_bar_reader.first_trading_day,\n equity_daily_reader = bundle_data.equity_daily_bar_reader,\n adjustment_reader = bundle_data.adjustment_reader)",
"_____no_output_____"
]
],
[
[
"Now that we have created a `data_portal` we will create a helper function, `get_pricing`, that gets the historical data from the `data_portal` for a given set of `start_date` and `end_date`. The `get_pricing` function takes various parameters: \n\n```python\ndef get_pricing(data_portal, trading_calendar, assets, start_date, end_date, field='close')\n```\n\n\nThe first two parameters, `data_portal` and `trading_calendar`, have already been defined above. The third paramter, `assets`, is a list of tickers. In our case we will use the tickers from the output of our pipeline, namely, `universe_tickers`. The fourth and fifth parameters are strings specifying the `start_date` and `end_date`. The function converts these two strings into Timestamps with a Custom Business Day frequency. The last parameter, `field`, is a string used to indicate which field to return. In our case we want to get the closing price, so we set `field='close`. \n\nThe function returns the historical stock price data using the `.get_history_window()` attribute of the `DataPortal` class. This attribute returns a Pandas Dataframe containing the requested history window with the data fully adjusted. The `bar_count` parameter is an integer indicating the number of days to return. The number of days determines the number of rows of the returned dataframe. Both the `frequency` and `data_frequency` parameters are strings that indicate the frequency of the data to query, *i.e.* whether the data is in `daily` or `minute` intervals.",
"_____no_output_____"
]
],
[
[
"def get_pricing(data_portal, trading_calendar, assets, start_date, end_date, field='close'):\n \n # Set the given start and end dates to Timestamps. The frequency string C is used to\n # indicate that a CustomBusinessDay DateOffset is used\n end_dt = pd.Timestamp(end_date, tz='UTC', freq='C')\n start_dt = pd.Timestamp(start_date, tz='UTC', freq='C')\n\n # Get the locations of the start and end dates\n end_loc = trading_calendar.closes.index.get_loc(end_dt)\n start_loc = trading_calendar.closes.index.get_loc(start_dt)\n\n # return the historical data for the given window\n return data_portal.get_history_window(assets=assets, end_dt=end_dt, bar_count=end_loc - start_loc,\n frequency='1d',\n field=field,\n data_frequency='daily')\n\n# Get the historical data for the given window\nhistorical_data = get_pricing(data_portal, trading_calendar, universe_tickers,\n start_date='2011-01-05', end_date='2016-01-05')\n# Display the historical data\nhistorical_data",
"_____no_output_____"
]
],
[
[
"# Date Alignment\n\nWhen pipeline returns with a date of, e.g., `2016-01-07` this includes data that would be known as of before the **market open** on `2016-01-07`. As such, if you ask for latest known values on each day, it will return the closing price from the day before and label the date `2016-01-07`. All factor values assume to be run prior to the open on the labeled day with data known before that point in time.",
"_____no_output_____"
],
[
"# Adding Factors and Filters\n\nNow that you know how build a pipeline and execute it, in this section we will see how we can add factors and filters to our pipeline. These factors and filters will determine the computations we want our pipeline to compute each day.\n\nWe can add both factors and filters to our pipeline using the `.add(column, name)` method of the `Pipeline` class. The `column` parameter represetns the factor or filter to add to the pipeline. The `name` parameter is a string that determines the name of the column in the output Pandas Dataframe for that factor of fitler. As mentioned earlier, each factor and filter will appear as a column in the output dataframe of our pipeline. Let's start by adding a factor to our pipeline.\n\n### Factors\n\nIn the code below, we will use Zipline's built-in `SimpleMovingAverage` factor to create a factor that computes the 15-day mean closing price of securities. We will then add this factor to our pipeline and use `.show_graph()` to see a diagram of our pipeline with the factor added. ",
"_____no_output_____"
]
],
[
[
"from zipline.pipeline.factors import SimpleMovingAverage\n\n# Create a factor that computes the 15-day mean closing price of securities\nmean_close_15 = SimpleMovingAverage(inputs = [USEquityPricing.close], window_length = 15)\n\n# Add the factor to our pipeline\npipeline.add(mean_close_15, '15 Day MCP')\n\n# Render the pipeline as a DAG\npipeline.show_graph()",
"_____no_output_____"
]
],
[
[
"In the diagram above we can clearly see the factor we have added. Now, we can run our pipeline again and see its output. The pipeline is run in exactly the same way we did before. ",
"_____no_output_____"
]
],
[
[
"# Set starting and end dates\nstart_date = pd.Timestamp('2014-01-06', tz='utc')\nend_date = pd.Timestamp('2016-01-05', tz='utc')\n\n# Run our pipeline for the given start and end dates\noutput = engine.run_pipeline(pipeline, start_date, end_date)\n\n# Display the pipeline output\noutput.head()",
"_____no_output_____"
]
],
[
[
"We can see that now our output dataframe contains a column with the name `15 Day MCP`, which is the name we gave to our factor before. This ouput dataframe from our pipeline gives us the 15-day mean closing price of the securities that passed our `screen`.\n\n### Filters\n\nFilters are created and added to the pipeline in the same way as factors. In the code below, we create a filter that returns `True` whenever the 15-day average closing price is above \\$100. Remember, a filter produces a `True` or `False` value for each security every day. We will then add this filter to our pipeline and use `.show_graph()` to see a diagram of our pipeline with the filter added.",
"_____no_output_____"
]
],
[
[
"# Create a Filter that returns True whenever the 15-day average closing price is above $100\nhigh_mean = mean_close_15 > 100\n\n# Add the filter to our pipeline\npipeline.add(high_mean, 'High Mean')\n\n# Render the pipeline as a DAG\npipeline.show_graph()",
"_____no_output_____"
]
],
[
[
"In the diagram above we can clearly see the fiter we have added. Now, we can run our pipeline again and see its output. The pipeline is run in exactly the same way we did before. ",
"_____no_output_____"
]
],
[
[
"# Set starting and end dates\nstart_date = pd.Timestamp('2014-01-06', tz='utc')\nend_date = pd.Timestamp('2016-01-05', tz='utc')\n\n# Run our pipeline for the given start and end dates\noutput = engine.run_pipeline(pipeline, start_date, end_date)\n\n# Display the pipeline output\noutput.head()",
"_____no_output_____"
]
],
[
[
"We can see that now our output dataframe contains a two columns, one for the filter and one for the factor. The new column has the name `High Mean`, which is the name we gave to our filter before. Notice that the filter column only contains Boolean values, where only the securities with a 15-day average closing price above \\$100 have `True` values.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
4a567f92776d144ffee821d317fb62877815442f
| 41,388 |
ipynb
|
Jupyter Notebook
|
jupyter_notebook/legacy/debugging.ipynb
|
rsgit95/med_kg_txt_multimodal
|
80355b0cf58e0571531ad6f9728c533110ca996d
|
[
"Apache-2.0"
] | null | null | null |
jupyter_notebook/legacy/debugging.ipynb
|
rsgit95/med_kg_txt_multimodal
|
80355b0cf58e0571531ad6f9728c533110ca996d
|
[
"Apache-2.0"
] | null | null | null |
jupyter_notebook/legacy/debugging.ipynb
|
rsgit95/med_kg_txt_multimodal
|
80355b0cf58e0571531ad6f9728c533110ca996d
|
[
"Apache-2.0"
] | null | null | null | 71.358621 | 1,985 | 0.669276 |
[
[
[
"**0. Code for Colab Debugging**",
"_____no_output_____"
]
],
[
[
"from google.colab import drive\ndrive.mount('/content/gdrive')\n%cd /content/gdrive/My Drive/lxmert/src/\n!pip install transformers\nimport torch\nprint(torch.cuda.is_available())",
"Drive already mounted at /content/gdrive; to attempt to forcibly remount, call drive.mount(\"/content/gdrive\", force_remount=True).\n/content/gdrive/My Drive/lxmert/src\nRequirement already satisfied: transformers in /usr/local/lib/python3.6/dist-packages (3.4.0)\nRequirement already satisfied: regex!=2019.12.17 in /usr/local/lib/python3.6/dist-packages (from transformers) (2019.12.20)\nRequirement already satisfied: protobuf in /usr/local/lib/python3.6/dist-packages (from transformers) (3.12.4)\nRequirement already satisfied: tqdm>=4.27 in /usr/local/lib/python3.6/dist-packages (from transformers) (4.41.1)\nRequirement already satisfied: tokenizers==0.9.2 in /usr/local/lib/python3.6/dist-packages (from transformers) (0.9.2)\nRequirement already satisfied: dataclasses; python_version < \"3.7\" in /usr/local/lib/python3.6/dist-packages (from transformers) (0.7)\nRequirement already satisfied: sacremoses in /usr/local/lib/python3.6/dist-packages (from transformers) (0.0.43)\nRequirement already satisfied: sentencepiece!=0.1.92 in /usr/local/lib/python3.6/dist-packages (from transformers) (0.1.94)\nRequirement already satisfied: numpy in /usr/local/lib/python3.6/dist-packages (from transformers) (1.18.5)\nRequirement already satisfied: requests in /usr/local/lib/python3.6/dist-packages (from transformers) (2.23.0)\nRequirement already satisfied: packaging in /usr/local/lib/python3.6/dist-packages (from transformers) (20.4)\nRequirement already satisfied: filelock in /usr/local/lib/python3.6/dist-packages (from transformers) (3.0.12)\nRequirement already satisfied: setuptools in /usr/local/lib/python3.6/dist-packages (from protobuf->transformers) (50.3.2)\nRequirement already satisfied: six>=1.9 in /usr/local/lib/python3.6/dist-packages (from protobuf->transformers) (1.15.0)\nRequirement already satisfied: click in /usr/local/lib/python3.6/dist-packages (from sacremoses->transformers) (7.1.2)\nRequirement already satisfied: joblib in /usr/local/lib/python3.6/dist-packages (from sacremoses->transformers) (0.17.0)\nRequirement already satisfied: chardet<4,>=3.0.2 in /usr/local/lib/python3.6/dist-packages (from requests->transformers) (3.0.4)\nRequirement already satisfied: urllib3!=1.25.0,!=1.25.1,<1.26,>=1.21.1 in /usr/local/lib/python3.6/dist-packages (from requests->transformers) (1.24.3)\nRequirement already satisfied: idna<3,>=2.5 in /usr/local/lib/python3.6/dist-packages (from requests->transformers) (2.10)\nRequirement already satisfied: certifi>=2017.4.17 in /usr/local/lib/python3.6/dist-packages (from requests->transformers) (2020.6.20)\nRequirement already satisfied: pyparsing>=2.0.2 in /usr/local/lib/python3.6/dist-packages (from packaging->transformers) (2.4.7)\nTrue\n"
]
],
[
[
"**1. Import pckgs & Set basic configs**",
"_____no_output_____"
]
],
[
[
"# Base packages\nimport logging\nimport math\nimport os\nfrom dataclasses import dataclass, field\nfrom glob import glob\nfrom typing import Optional\nfrom torch.utils.data import ConcatDataset\n\n# Own implementation\nfrom utils.parameters import parser\nfrom utils.dataset import get_dataset\nfrom utils.data_collator import NodeMasking_DataCollator, NodeClassification_DataCollator, LiteralRegression_DataCollator\nfrom model import LxmertForPreTraining,LxmertForKGTokPredAndMaskedLM\n\n# From Huggingface transformers package\nfrom transformers import (\n CONFIG_MAPPING,\n MODEL_WITH_LM_HEAD_MAPPING,\n LxmertConfig,\n LxmertTokenizer,\n PreTrainedTokenizer,\n HfArgumentParser,\n TrainingArguments,\n Trainer,\n set_seed,\n)\n\ntrain_args = TrainingArguments(output_dir='test',\n do_train=True,\n do_eval=False,\n local_rank=-1,\n per_device_train_batch_size=4, \n learning_rate=1e-3,\n num_train_epochs=1)\nimport easydict \nPATH = '/content/gdrive/My Drive/lxmert/'\nargs = easydict.EasyDict({\n \"model_type\":\"lxmert\",\n \"model_name_or_path\":None,\n \"cache_dir\":None,\n \"config_name\":PATH+\"config/config.json\",\n \"tokenizer_name\":\"bert-base-uncased\",\n \"train_data_file\":PATH+\"data/masked_literal_prediction/train\",\n \"train_data_files\":None,\n \"eval_data_file\":PATH+\"data/masked_literal_prediction/valid\",\n \"output_dir\":PATH+\"pretrained_models/test\",\n \"mlm\":True,\n \"mlm_probability\":0.15,\n \"block_size\":512,\n})\n\nlogger = logging.getLogger(__name__)\n\nMODEL_CONFIG_CLASSES = list(MODEL_WITH_LM_HEAD_MAPPING.keys())\nMODEL_TYPES = tuple(conf.model_type for conf in MODEL_CONFIG_CLASSES)\n\n#args, args, args = parser.parse_args_into_dataclasses()\n\nif args.eval_data_file is None and args.do_eval:\n raise ValueError(\n \"Cannot do evaluation without an evaluation data file. Either supply a file to --eval_data_file \"\n \"or remove the --do_eval argument.\"\n )\nif (\n os.path.exists(args.output_dir)\n and os.listdir(args.output_dir)\n and args.do_train\n and not args.overwrite_output_dir\n):\n raise ValueError(\n f\"Output directory ({args.output_dir}) already exists and is not empty. Use --overwrite_output_dir to overcome.\"\n )\n\n# Setup logging\nlogging.basicConfig(\n format=\"%(asctime)s - %(message)s\",\n datefmt=\"%m/%d %H:%M\",\n level=logging.INFO if train_args.local_rank in [-1, 0] else logging.WARN,\n)\nlogger.warning(\n \"Process rank: %s, device: %s, n_gpu: %s, distributed training: %s, 16-bits training: %s\",\n train_args.local_rank,\n train_args.device,\n train_args.n_gpu,\n bool(train_args.local_rank != -1),\n train_args.fp16,\n)\nlogger.info(\"Training/evaluation parameters %s\", args)\n\n# Set seed\nset_seed(train_args.seed)",
"11/02 02:43 - Process rank: -1, device: cuda:0, n_gpu: 1, distributed training: False, 16-bits training: False\n11/02 02:43 - Training/evaluation parameters {'model_type': 'lxmert', 'model_name_or_path': None, 'cache_dir': None, 'config_name': '/content/gdrive/My Drive/lxmert/config/config.json', 'tokenizer_name': 'bert-base-uncased', 'train_data_file': '/content/gdrive/My Drive/lxmert/data/masked_literal_prediction/train', 'train_data_files': None, 'eval_data_file': '/content/gdrive/My Drive/lxmert/data/masked_literal_prediction/valid', 'output_dir': '/content/gdrive/My Drive/lxmert/pretrained_models/test', 'mlm': True, 'mlm_probability': 0.15, 'block_size': 512}\n"
]
],
[
[
"**2. Load model configuration**",
"_____no_output_____"
]
],
[
[
"if args.config_name:\n config = LxmertConfig.from_pretrained(args.config_name, cache_dir=args.cache_dir)\nelif args.model_name_or_path:\n config = LxmertConfig.from_pretrained(args.model_name_or_path, cache_dir=args.cache_dir)\nelse:\n config = CONFIG_MAPPING[args.model_type]()\n logger.warning(\"You are instantiating a new config instance from scratch.\")",
"_____no_output_____"
]
],
[
[
"**3. Define tokenizer (or load pretrained one)**",
"_____no_output_____"
]
],
[
[
"if args.tokenizer_name:\n tokenizer = LxmertTokenizer.from_pretrained(args.tokenizer_name, cache_dir=args.cache_dir)\nelif args.model_name_or_path:\n tokenizer = LxmertTokenizer.from_pretrained(args.model_name_or_path, cache_dir=args.cache_dir)\nelse:\n raise ValueError(\n \"You are instantiating a new tokenizer from scratch. This is not supported, but you can do it from another script, save it,\"\n \"and load it from here, using --tokenizer_name\"\n )",
"_____no_output_____"
]
],
[
[
"**4. Define model (or load pretrained one)**",
"_____no_output_____"
]
],
[
[
"if args.model_name_or_path:\n model = LxmertForKGTokPredAndMaskedLM.from_pretrained(\n args.model_name_or_path,\n from_tf=bool(\".ckpt\" in args.model_name_or_path),\n config=config,\n cache_dir=args.cache_dir,\n )\nelse:\n logger.info(\"Training new model from scratch\")\n model = LxmertForKGTokPredAndMaskedLM(config)\nif config.model_type in [\"bert\", \"roberta\", \"distilbert\", \"camembert\"] and not args.mlm:\n raise ValueError(\n \"BERT and RoBERTa-like models do not have LM heads but masked LM heads. They must be run using the\"\n \"--mlm flag (masked language modeling).\"\n )",
"11/02 02:43 - Training new model from scratch\n"
]
],
[
[
"**5. Build dataset & data loader**",
"_____no_output_____"
]
],
[
[
"if args.block_size <= 0:\n args.block_size = tokenizer.max_len\n # Our input block size will be the max possible for the model\nelse:\n args.block_size = min(args.block_size, tokenizer.max_len)\n\n# Get datasets\n\ntrain_dataset = (\n get_dataset(args, tokenizer=tokenizer,kg_pad=config.kg_special_token_ids[\"PAD\"]) if train_args.do_train else None\n)\neval_dataset = (\n get_dataset(args, tokenizer=tokenizer, kg_pad=config.kg_special_token_ids[\"PAD\"], evaluate=True)\n if train_args.do_eval\n else None\n)\ndata_collator = NodeClassification_DataCollator(tokenizer=tokenizer, kg_special_token_ids=config.kg_special_token_ids, kg_size = config.vocab_size['kg'])",
"/usr/local/lib/python3.6/dist-packages/transformers/tokenization_utils_base.py:1374: FutureWarning: The `max_len` attribute has been deprecated and will be removed in a future version, use `model_max_length` instead.\n FutureWarning,\n11/02 02:43 - Loading features from dataset file at /content/gdrive/My Drive/lxmert/data/masked_literal_prediction/train\n"
]
],
[
[
"**6. Initialize trainer & Run training**\n> Use Huggingface [trainer.py](https://github.com/huggingface/transformers/blob/master/src/transformers/trainer.py)\n\n\n\n",
"_____no_output_____"
]
],
[
[
"# Initialize our Trainer\nprint(train_args)\nprint(data_collator)\nprint(train_dataset)\ntrainer = Trainer(\n model=model,\n args=train_args,\n data_collator=data_collator,\n train_dataset=train_dataset,\n prediction_loss_only=True\n)\n\n# Training\nif train_args.do_train:\n model_path = (\n args.model_name_or_path\n if args.model_name_or_path is not None and os.path.isdir(args.model_name_or_path)\n else None\n )\n trainer.train(model_path=model_path)\n trainer.save_model()\n # For convenience, we also re-save the tokenizer to the same directory,\n # so that you can share your model easily on huggingface.co/models =)\n if trainer.is_world_master():\n tokenizer.save_pretrained(args.output_dir)",
"TrainingArguments(output_dir='test', overwrite_output_dir=False, do_train=True, do_eval=False, do_predict=False, evaluate_during_training=False, evaluation_strategy=<EvaluationStrategy.NO: 'no'>, prediction_loss_only=False, per_device_train_batch_size=4, per_device_eval_batch_size=8, per_gpu_train_batch_size=None, per_gpu_eval_batch_size=None, gradient_accumulation_steps=1, eval_accumulation_steps=None, learning_rate=0.001, weight_decay=0.0, adam_beta1=0.9, adam_beta2=0.999, adam_epsilon=1e-08, max_grad_norm=1.0, num_train_epochs=1, max_steps=-1, warmup_steps=0, logging_dir='runs/Nov02_02-43-37_71126e65e1fb', logging_first_step=False, logging_steps=500, save_steps=500, save_total_limit=None, no_cuda=False, seed=42, fp16=False, fp16_opt_level='O1', local_rank=-1, tpu_num_cores=None, tpu_metrics_debug=False, debug=False, dataloader_drop_last=False, eval_steps=500, dataloader_num_workers=0, past_index=-1, run_name='test', disable_tqdm=False, remove_unused_columns=True, label_names=None, load_best_model_at_end=False, metric_for_best_model=None, greater_is_better=None)\nNodeClassification_DataCollator(tokenizer=PreTrainedTokenizer(name_or_path='bert-base-uncased', vocab_size=30522, model_max_len=1000000000000000019884624838656, is_fast=False, padding_side='right', special_tokens={'unk_token': '[UNK]', 'sep_token': '[SEP]', 'pad_token': '[PAD]', 'cls_token': '[CLS]', 'mask_token': '[MASK]'}), kg_special_token_ids={'PAD': 0, 'MASK': 1}, kg_size=2901117, mlm=True, mlm_probability=0.15, contrastive=False)\n<utils.dataset.HeadOnlyDataset object at 0x7f76f484c128>\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
4a5688498b00dca3bbde44087a3541e5dc891d91
| 32,247 |
ipynb
|
Jupyter Notebook
|
geotweets.ipynb
|
davebshow/ftweet
|
bd8bca730ff9400d1811d5f879001d8a9e696ff8
|
[
"MIT"
] | null | null | null |
geotweets.ipynb
|
davebshow/ftweet
|
bd8bca730ff9400d1811d5f879001d8a9e696ff8
|
[
"MIT"
] | null | null | null |
geotweets.ipynb
|
davebshow/ftweet
|
bd8bca730ff9400d1811d5f879001d8a9e696ff8
|
[
"MIT"
] | null | null | null | 29.15642 | 1,283 | 0.481533 |
[
[
[
"%load_ext cypher\nimport json\nimport random",
"/home/davebshow/.virtualenvs/scientific3/lib/python3.4/site-packages/IPython/config.py:13: ShimWarning: The `IPython.config` package has been deprecated. You should import from traitlets.config instead.\n \"You should import from traitlets.config instead.\", ShimWarning)\n/home/davebshow/.virtualenvs/scientific3/lib/python3.4/site-packages/IPython/utils/traitlets.py:5: UserWarning: IPython.utils.traitlets has moved to a top-level traitlets package.\n warn(\"IPython.utils.traitlets has moved to a top-level traitlets package.\")\n"
],
[
"geotweets = %cypher match (n:tweet) where n.coordinates is not null return n.tid, n.lang, n.country, n.name, n.coordinates, n.created_at",
"55881 rows affected.\n"
],
[
"geotweets = geotweets.get_dataframe()\ngeotweets.head()",
"_____no_output_____"
],
[
"json.loads(geotweets.ix[1][\"n.coordinates\"])[0][0]",
"_____no_output_____"
],
[
"def get_random_coords(df):\n lats = []\n lons = []\n for row in df.iterrows():\n row = row[1]\n coords = json.loads(row[\"n.coordinates\"])[0]\n lat1 = coords[0][0]\n lat2 = coords[2][0]\n lon1 = coords[0][1]\n lon2 = coords[1][1]\n ran_lat = random.uniform(lat1, lat2)\n ran_lon = random.uniform(lon1, lon2)\n lats.append(ran_lat)\n lons.append(ran_lon)\n df[\"lat\"] = lats\n df[\"lon\"] = lons\n return df",
"_____no_output_____"
],
[
"df = get_random_coords(geotweets)",
"_____no_output_____"
],
[
"geotweets.columns = [\"Id\", \"Lang\", \"Country\", \"City\", \"Coords\", \"Time\", \"Lon\", \"Lat\"]",
"_____no_output_____"
],
[
"geotweets[\"Label\"] = \"tweet\"",
"_____no_output_____"
],
[
"geotweets.head()",
"_____no_output_____"
],
[
"geotweets.to_csv(\"data/geotweets.csv\")",
"_____no_output_____"
],
[
"edges_query = \"\"\"match (t:tweet)-[:USES]->(h:hashtag) where t.coordinates is not null with h.tagid as hashtag, t.tid as tweet return hashtag, tweet\n\"\"\"",
"_____no_output_____"
],
[
"geotweet_edges = %cypher match (t:tweet)-[:USES]->(h:hashtag) where t.coordinates is not null with h.tagid as hashtag, t.tid as tweet return tweet, hashtag",
"41490 rows affected.\n"
],
[
"geotweet_edges = geotweet_edges.get_dataframe()",
"_____no_output_____"
],
[
"geotweet_edges.head()",
"_____no_output_____"
],
[
"geotweet_edges.columns = [\"Source\", \"Target\"]",
"_____no_output_____"
],
[
"geotweet_edges.to_csv(\"data/geoedges.csv\")",
"_____no_output_____"
],
[
"geoedges_nohash = %cypher match (t:tweet)--(n:tweet) where t.coordinates is not null and n.coordinates is not null return t.tid as Source, n.tid as Target",
"136 rows affected.\n"
],
[
"geoedges_nohash = geoedges_nohash.get_dataframe()",
"_____no_output_____"
],
[
"len(geoedges_nohash)",
"_____no_output_____"
],
[
"geoedges_nohash.to_csv(\"data/geoedges_nohash.csv\")",
"_____no_output_____"
],
[
"geohash = %cypher match (t:tweet)-[r:USES]->(h:hashtag) where t.coordinates is not null with distinct h.tagid as Id, h.hashtag as Label, count(r) as deg return Id, Label order by deg desc limit 10",
"10 rows affected.\n"
],
[
"geohash = geohash.get_dataframe()\ngeohash.head()",
"_____no_output_____"
],
[
"labels = geohash[\"Label\"].map(lambda x: \"#\" + x)",
"_____no_output_____"
],
[
"geohash[\"Label\"] = labels",
"_____no_output_____"
],
[
"geohash.head()",
"_____no_output_____"
],
[
"geohash.to_csv(\"data/geotags.csv\")",
"_____no_output_____"
],
[
"edges = %cypher match (t:tweet)-[:USES]-(h:hashtag {hashtag: \"paris\"}) where t.coordinates is not null return h.hashtag, collect(t.tid) ",
"1 rows affected.\n"
],
[
"import itertools\nimport networkx as nx",
"_____no_output_____"
],
[
"edges = edges.get_dataframe()",
"_____no_output_____"
],
[
"edges[\"collect(t.tid)\"] = edges[\"collect(t.tid)\"].map(lambda x: list(itertools.combinations(x, 2)))",
"_____no_output_____"
],
[
"edges.head()",
"_____no_output_____"
],
[
"el = list(itertools.chain.from_iterable(edges[\"collect(t.tid)\"]))",
"_____no_output_____"
],
[
"len(el)",
"_____no_output_____"
],
[
"el[1]",
"_____no_output_____"
],
[
"len(el)",
"_____no_output_____"
],
[
"g = nx.Graph(el)",
"_____no_output_____"
],
[
"len(geotweet_edges)",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a56889c9f2cc1ae0c3bd1c3ae990d19b3c5e9cb
| 62,695 |
ipynb
|
Jupyter Notebook
|
waterfall.ipynb
|
finale80/mcpa
|
d3434ef69ea1fa525a5fe12eafd2dc27a538c481
|
[
"MIT"
] | 1 |
2019-06-22T16:05:11.000Z
|
2019-06-22T16:05:11.000Z
|
waterfall.ipynb
|
finale80/mcpa
|
d3434ef69ea1fa525a5fe12eafd2dc27a538c481
|
[
"MIT"
] | null | null | null |
waterfall.ipynb
|
finale80/mcpa
|
d3434ef69ea1fa525a5fe12eafd2dc27a538c481
|
[
"MIT"
] | null | null | null | 187.149254 | 50,896 | 0.879448 |
[
[
[
"# Network waterfall generation",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nimport matplotlib as mpl\nimport matplotlib.pyplot as plt\n%matplotlib inline\nfrom math import sqrt\nimport re, bisect\nfrom colorama import Fore",
"_____no_output_____"
]
],
[
[
"## Select input file and experiment ID (~10 experiments per file)\n- ./startup : Application startup\n- ./startup_and_click : Application startup + click (single user interaction)\n- ./multiclick : Application statup + clicks (multiple user interactions)\n\n### (*critical flows* and performance metrics available for *startup* and *startup_and_click* datasets)",
"_____no_output_____"
]
],
[
[
"##example (YouTube)\nFNAME = \"./startup/com.google.android.youtube_bursts.txt\"\nEXPID = 1",
"_____no_output_____"
]
],
[
[
"## Load experiment data and plot waterfall",
"_____no_output_____"
]
],
[
[
"##load experiment data\nd_exps = load_experiments(FNAME)\ndf = d_exps[EXPID]\nprint_head(FNAME)\n##plot waterfall\nplot_waterfall(df, fvolume=None, title=FNAME, fname_png=\"output_waterfall.png\")",
"TDT = Transport Delivery Time\nAFT = Above-the-Fold Time\n\u001b[31mCritical flows\n\u001b[34mNon-Critical flows\n"
]
],
[
[
"## A small library for plotting waterfalls, based on matplotlib",
"_____no_output_____"
]
],
[
[
"def load_experiments(fname):\n df = pd.read_csv(fname, sep = ' ', low_memory=False)\n ## split the single file in multiple dataframes based on experiment id\n d = {}\n for expid in df['expId'].unique():\n df_tmp = df[df['expId'] == expid].copy()\n df_tmp = df_tmp.sort_values(by='t_start')\n \n cat = pd.Categorical(df_tmp['flow'], ordered=False)\n cat = cat.reorder_categories(df_tmp['flow'].unique())\n df_tmp.loc[:, 'flowid'] = cat.codes\n \n d[expid] = df_tmp\n return d\n\n\n\ndef _get_reference_times(df):\n tdt = df['TDT'].values[0]\n aft = df['AFT'].values[0]\n x_max = 0.5+max(df['t_end'].max(), aft, tdt)\n return {\n 'tdt' : tdt,\n 'aft' : aft,\n 'x_max' : x_max}\n\n\ndef _get_max_time(df):\n x_max = 0.5+df['t_end'].max()\n return {'x_max' : x_max}\n \n\ndef _get_lines_burst(df, x_lim=None):\n lines_burst = []\n lines_burst_widths = []\n for flowid, x_start, x_end, burst_bytes in df[['flowid', 't_start', 't_end', 'KB']].values:\n if x_lim is None:\n lines_burst.append([(x_start, flowid), (x_end, flowid)])\n width = min(13, 2*sqrt(burst_bytes))\n width = max(1, width)\n lines_burst_widths.append(width)\n else:\n el = [(x_lim[0], flowid), (x_lim[1], flowid)]\n if el not in lines_burst:\n lines_burst.append(el)\n\n return lines_burst, lines_burst_widths\n\n\n\ndef _plot_aft_tdt_reference(ax, tdt, aft, no_legend=False):\n tdt_label = \"TDT = \" + str(tdt)[0:5]\n aft_label = \"AFT = \" + str(aft)[0:5]\n if no_legend:\n tdt_label = None\n aft_label = None\n ax.axvline(x=tdt, color=\"green\", label=tdt_label, linewidth=2) #, ax = ax)\n ax.axvline(x=aft, color=\"purple\", label=aft_label, linewidth=2) #, ax = ax)\n lgd = ax.legend(bbox_to_anchor=[1, 1])\n\n \ndef _plot_bursts(ax, df, lines_flow, \n lines_burst=None, \n lines_burst_critical=None,\n flow_kwargs={}, \n burst_kwargs={}, \n burst_critical_kwargs={},\n title=None):\n ## flow lines\n ax.add_collection(mpl.collections.LineCollection(lines_flow, **flow_kwargs)) \n\n ## burst lines\n if lines_burst is not None:\n ax.add_collection(mpl.collections.LineCollection(lines_burst, **burst_kwargs))\n\n if lines_burst_critical is not None:\n ax.add_collection(mpl.collections.LineCollection(lines_burst_critical, **burst_critical_kwargs))\n \n if 'AFT' in df and 'TDT' in df:\n d_times = _get_reference_times(df)\n ## vertical reference lines\n _plot_aft_tdt_reference(ax, tdt=d_times['tdt'], aft=d_times['aft'])\n else:\n d_times = _get_max_time(df)\n ## axis lim \n x_max = d_times['x_max']\n y_max = len(lines_flow)+1\n ax.set_ylim((-1, y_max))\n ax.set_xlim((0, x_max))\n\n chess_lines = [[(0, y),(x_max, y)] for y in range(0, y_max, 2)]\n ax.add_collection(mpl.collections.LineCollection(chess_lines, linewidths=10, color='gray', alpha=0.1))\n\n ## ticks\n ax.yaxis.set_major_locator(mpl.ticker.MultipleLocator(1))\n ax.tick_params(axis='y', length=0)\n\n ## y-labels (clipping the long ones)\n labels = df[['flow', 'flowid']].sort_values(by='flowid').drop_duplicates()['flow'].values\n ax.set_yticklabels(['',''] + list(labels))\n\n ## remove borders\n ax.spines['top'].set_visible(False)\n ax.spines['right'].set_visible(False)\n ax.spines['bottom'].set_visible(False)\n ax.spines['left'].set_visible(False)\n\n ## grid\n ax.grid(axis='x', alpha=0.3)\n\n ax.legend().remove()\n\n \n \ndef _plot_volume(ax, df, title=None, fvolume=None):\n ## get times\n if 'AFT' in df and 'TDT' in df:\n d_times = _get_reference_times(df)\n else:\n d_times = _get_max_time(df)\n if fvolume!=None:\n x=[]\n y=[]\n for line in open(fvolume):\n x.append(float(line[0:-1].split(' ')[0]))\n y.append(float(line[0:-1].split(' ')[1]))\n ax.step(x, y, color='gray', where='post', label='')\n else:\n # get volume cumulate\n df_tmp = df.copy()\n df_tmp = df_tmp.sort_values(by='t_end')\n df_tmp.loc[:, 'KB_cdf'] = df_tmp['KB'].cumsum() / df_tmp['KB'].sum()\n ax.step(x=df_tmp['t_end'], y=df_tmp['KB_cdf'], color='gray', where='post', label='')\n \n ## remove border\n ax.spines['top'].set_visible(False)\n ax.spines['right'].set_visible(False)\n ax.spines['bottom'].set_visible(False)\n ax.spines['left'].set_visible(False)\n \n if 'AFT' in df and 'TDT' in df:\n _plot_aft_tdt_reference(ax, tdt=d_times['tdt'], aft=d_times['aft'], no_legend=False)\n\n ax.tick_params(labeltop=True, labelbottom=False, length=0.1, axis='x', direction='out')\n ax.set_xlim((0, d_times['x_max']))\n \n ## grid\n ax.grid(axis='x', alpha=0.3)\n ax.yaxis.set_major_locator(mpl.ticker.MultipleLocator(0.5))\n ax.set_ylabel('CDF Volume')\n ## title\n if title is not None:\n ax.set_title(title, pad=20)\n\n \ndef print_head(fname):\n print(\"TDT = Transport Delivery Time\\nAFT = Above-the-Fold Time\")\n if 'multiclick' not in fname:\n print(Fore.RED + 'Critical flows')\n print(Fore.BLUE + 'Non-Critical flows')\n \n\ndef plot_waterfall(df, fvolume=None, title=None, fname_png=None): #, ax=None):\n ## first start and end of each flow\n df_tmp = df.groupby('flowid').agg({'t_start':'min', 't_end':'max'})\n ## ..and create lines\n lines_flow = [ [(x_start, y), (x_end, y)] for y, (x_start, x_end) in zip(df_tmp.index, df_tmp.values) ]\n \n ## lines for each burst\n lines_burst, lines_burst_widths = _get_lines_burst(df[df['KB'] > 0])\n ## lines for each critical burst (if any info on critical domains in the input file)\n if 'critical' in df:\n lines_burst_critical, lines_burst_widths_critical = _get_lines_burst(df[(df['critical']) & (df['KB'] > 0)])\n else:\n lines_burst_critical, lines_burst_widths_critical = [], []\n ######################\n \n fig_height = max(5, 0.25*len(lines_flow))\n fig = plt.figure(figsize=(8, fig_height))\n gs = mpl.gridspec.GridSpec(nrows=2, ncols=1, hspace=0.1, height_ratios=[1,3])\n \n ax0 = plt.subplot(gs[0])\n ax1 = plt.subplot(gs[1])\n \n _plot_volume(ax0, df, title, fvolume)\n \n _plot_bursts(ax1, df, lines_flow, lines_burst, lines_burst_critical,\n flow_kwargs={\n 'linewidths':2, \n 'color': 'gray', \n 'linestyle' : (0, (1, 1)), \n 'alpha':0.7},\n burst_kwargs={\n 'linewidths' : lines_burst_widths, \n 'color': 'blue'},\n burst_critical_kwargs={\n 'linewidths':lines_burst_widths_critical, \n 'color': 'red'})\n ## add click timestamps (if any)\n if 'clicks' in df:\n for click_t in df['clicks'].values[0][1:-1].split(', '):\n if float(click_t)<40 and float(click_t)>35:\n continue\n plt.axvline(x=float(click_t), color=\"grey\", linestyle=\"--\")\n if fname_png is not None:\n plt.savefig(fname_png, bbox_inches='tight')",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
4a569cd95e24e12dd1f0b0d6a72466c5787587fe
| 615,986 |
ipynb
|
Jupyter Notebook
|
Project_Airbnb_Seattle.ipynb
|
hllcbn/Seattle_Airbnb_Project
|
865ed0805a0efc6c9839eacdc86e7a80482ffe34
|
[
"CNRI-Python",
"Xnet",
"X11"
] | null | null | null |
Project_Airbnb_Seattle.ipynb
|
hllcbn/Seattle_Airbnb_Project
|
865ed0805a0efc6c9839eacdc86e7a80482ffe34
|
[
"CNRI-Python",
"Xnet",
"X11"
] | null | null | null |
Project_Airbnb_Seattle.ipynb
|
hllcbn/Seattle_Airbnb_Project
|
865ed0805a0efc6c9839eacdc86e7a80482ffe34
|
[
"CNRI-Python",
"Xnet",
"X11"
] | null | null | null | 236.28155 | 155,680 | 0.875835 |
[
[
[
"import numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nfrom sklearn.linear_model import LinearRegression\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.metrics import r2_score, mean_squared_error\nimport seaborn as sns\nimport statsmodels.api as sm\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"# 1. BUSINESS UNDERSTANDING",
"_____no_output_____"
],
[
"In this project, i used Airbnb Seatle data and focus on answering three business questions using exploratory data analysis, data visualization and machine learning algorithm. \n\n1- How are properties distributed among neighboorhood group? This will help stakeholder to understand what percent of the properties are located in what neighboorhood of Seattle.\n\n2 - How many times room types are reviewed? What is the average review score rating? Is there a relationship room type price and number of review and review score rating? This will guide if there is any correlation between the variables.\n\n3-Implement one of ML algoritims which is linear regression model to forecast price. This will help stakeholder to predict price based on independt predictors.\n\n\n\n\n",
"_____no_output_____"
],
[
"# 2. DATA UNDERSTANDING",
"_____no_output_____"
]
],
[
[
"#In this project, i will be using Seattle Airbnb dataset\n#Load Seattle listing data\ndf_listings = pd.read_csv('listings.csv')",
"_____no_output_____"
],
[
"# Check the structure of the data after it's loaded.\ndf_listings.shape",
"_____no_output_____"
],
[
"#Checking if there is abnormal among the numberical columns. lincence does not have any data so it can be dropped\ndf_listings.describe()",
"_____no_output_____"
],
[
"#Checking the data type of each column and be sure if there is any string column is required to be float value.\n# host response rate and price which will be used in the following analysis are supposed to be float \ndf_listings.dtypes",
"_____no_output_____"
]
],
[
[
"# 3 . DATA PREPARATION: CLEANING DATASET",
"_____no_output_____"
]
],
[
[
"#Redundant columns are dropped. They will not be used in the following data analysis process.\n\ndf_listings.drop(['security_deposit','weekly_price','summary','square_feet','monthly_price','space','scrape_id','notes','neighborhood_overview','transit','last_scraped','experiences_offered','thumbnail_url','medium_url','picture_url','xl_picture_url',\n 'host_id','host_thumbnail_url','host_picture_url','host_has_profile_pic','host_identity_verified',\n 'license','square_feet','cleaning_fee'],axis =1,inplace = True)\n",
"_____no_output_____"
],
[
"#There was formating issue in these two columns. Converting string column to float values \ndf_listings['host_response_rate'] = df_listings['host_response_rate'].apply(lambda x : float(x.strip('%'))/100 if pd.notna(x)==True else None)\ndf_listings['price'] = df_listings['price'].replace('[\\$,]', '', regex=True).astype(float)",
"_____no_output_____"
],
[
"nan_cols = (df_listings.isnull().sum()/df_listings.shape[0]).sort_values(ascending = False)\nax = nan_cols.hist() \nax.set_xlabel(\"% NaN\")\nax.set_ylabel(\"Column Count\")\nnan_cols.head(n=10)",
"_____no_output_____"
],
[
"#df_listings.dropna(axis = 0,how = 'any',inplace = True)",
"_____no_output_____"
],
[
"\ndef clean_data_replace(df):\n \n '''\n INPUT:\n df - pandas dataframe which is df_listings\n \n OUTPUT:\n df - returns the clean data \n df - return the data without no missing values\n '''\n#I use mean imputation here to keep same mean and the same sample size. \n for col in df:\n dt = df[col].dtype\n if dt == int or dt==float :\n df[col].fillna(df[col].mean(),inplace = True)\n else:\n df_listings[col].fillna(df[col].mode()[0],inplace=True)\n \n return df.columns[df.isnull().sum()>0]",
"_____no_output_____"
],
[
"clean_data_replace(df_listings)",
"_____no_output_____"
]
],
[
[
"# 4.EXPLORATORY DATA ANALYSIS",
"_____no_output_____"
],
[
"# Q1 : How are properties distributed among neighboorhood group?",
"_____no_output_____"
],
[
"In this question, we would like to figure out how the properties are distributed among the neighboorhood. So, this will help us to figure out which neighboorhood has most properties. ",
"_____no_output_____"
]
],
[
[
"nb_seattle = (df_listings['neighbourhood_cleansed'].value_counts()).sort_values().reset_index()\nnb_seattle",
"_____no_output_____"
],
[
"nb_seattle_percent = (df_listings['neighbourhood_cleansed'].value_counts()/df_listings.shape[0]).sort_values(ascending = False).reset_index().head()\nnb_seattle_percent",
"_____no_output_____"
],
[
"plt.figure(figsize = (20,8));\nx = nb_seattle_percent['index'];\ny = nb_seattle_percent['neighbourhood_cleansed'];\nnb_seattle_percent.plot(x = 'index',y = 'neighbourhood_cleansed',kind = 'bar',figsize = (20,10),color='green');\nplt.plot(x,y);\nplt.xlabel('Neighboorhood',fontsize = 15);\nplt.ylabel('Number of properties',fontsize = 15);\nplt.tick_params(axis='x', labelsize=15);\nplt.tick_params(axis='y', labelsize=15);",
"_____no_output_____"
]
],
[
[
"As easily seen it the graph below, Broadway has the most airbnb property in Seattle. There is a huge supply in Broadway relatively other neighboorhood",
"_____no_output_____"
]
],
[
[
"\nnb_seattle.plot(x = 'index',y = 'neighbourhood_cleansed',kind = 'bar',figsize = (25,10),color='salmon')\nplt.plot(nb_seattle['index'],nb_seattle['neighbourhood_cleansed']);\nplt.xlabel('Neighboorhood',fontsize = 12)\nplt.ylabel('Number of properties',fontsize = 12);\nplt.tick_params(axis='x', labelsize=12)\nplt.tick_params(axis='y', labelsize=12)",
"_____no_output_____"
]
],
[
[
"# Q2: How many times room types are reviewed? What is the average review score rating? Is there a relationship room type price and number of review and review score rating?",
"_____no_output_____"
],
[
"We will try to figure out most prefered room types based on number of review and review score rating. Other important point to understand if there is any relationship between room type price and review rating. We will question if price and reviews are correlated.",
"_____no_output_____"
]
],
[
[
"#Here we selected specific columns that we will use in our explotory analysis.\ndf_Seatle_price_dist = df_listings[['room_type','property_type','neighbourhood_group_cleansed','neighbourhood_cleansed','price','number_of_reviews','review_scores_rating']]",
"_____no_output_____"
],
[
"df_Seatle_price_dist.sort_values(by = ['review_scores_rating','number_of_reviews'])",
"_____no_output_____"
],
[
"#Most reviewed room type is entire home/apt\ndf_Seatle_price_dist.groupby(['room_type'])['number_of_reviews'].sum().reset_index()",
"_____no_output_____"
],
[
"#Score of the Entire home/apt and private room are almost same. Generally scores of the room types are almost same. \ndf_Seatle_price_dist.groupby(['room_type'])['review_scores_rating'].mean().reset_index()",
"_____no_output_____"
]
],
[
[
"Most reviewed neighboorhood group are Other neighborhood, Downtown and Capital Hill . In these neighboorhood , entire home/apt is received\nhigh number of reviews.",
"_____no_output_____"
]
],
[
[
"df_Seatle_price_dist.groupby(['neighbourhood_group_cleansed','room_type'])['number_of_reviews'].sum().sort_values(ascending=False).reset_index().head(5)\n",
"_____no_output_____"
],
[
"sns.catplot(x = 'neighbourhood_group_cleansed',y = 'number_of_reviews',hue = 'room_type',data =df_Seatle_price_dist, \n kind = 'swarm',height=6, aspect=2 );\nplt.xticks(rotation=90);\nplt.xlabel('Neighboorhood Group',fontsize = 12);\nplt.ylabel('Number of Reviews',fontsize = 12);\n",
"_____no_output_____"
]
],
[
[
"Lets take a look at if there is any correlation between price and number of reviews and review_score_rating given chart below.\nIt looks there is inverse correlation between price and number of reviews while there is direct relationship between price and review score rating but we cant say it is strong enough.",
"_____no_output_____"
]
],
[
[
"df_Seatle_price_dist.corr()",
"_____no_output_____"
],
[
"sns.heatmap(df_Seatle_price_dist.corr(),cmap=\"YlGnBu\",annot = True)",
"_____no_output_____"
],
[
"sns.pairplot(df_Seatle_price_dist,x_vars=['number_of_reviews','review_scores_rating'],y_vars=['price'],hue='room_type',size = 4);",
"C:\\Users\\hilal\\Anaconda3\\lib\\site-packages\\seaborn\\axisgrid.py:2065: UserWarning: The `size` parameter has been renamed to `height`; pleaes update your code.\n warnings.warn(msg, UserWarning)\n"
]
],
[
[
"Given dataframe below shows how the price are distributed between room type in the different neighboorhood group. As seen in the graph to Magnolia is the most expensive neighboorhood if entire room/apt is rented.",
"_____no_output_____"
]
],
[
[
"df_Seatle_price_dist.groupby(['neighbourhood_group_cleansed','room_type'])['price'].mean().sort_values(ascending = False).reset_index().head()",
"_____no_output_____"
],
[
"plt.figure(figsize=(20,10))\nsns.barplot(x = \"neighbourhood_group_cleansed\", y = \"price\", hue = \"room_type\", data = df_Seatle_price_dist)\nplt.xticks(rotation=90,fontsize = 14);\nplt.xlabel('Neighboorhood Group',fontsize = 15);\nplt.ylabel('Price',fontsize = 15);\nplt.show()",
"_____no_output_____"
]
],
[
[
"# 5. DATA MODELING AND EVALUATION",
"_____no_output_____"
],
[
" # Q3 : Implement linear regression model to apply ML algorithm to forecast price based on variables are selected.",
"_____no_output_____"
],
[
"We performed a ML model to forecast if price are impacted under different circumstances. In order to proceed , we selected columns as indipendent variables. Dependent variable is here price. The list of independent variables are below.\n\nSELECTED INDEPENDENT VARIABLES :::\nhost_response_time,\nhost_response_rate,\nproperty_type,\nroom_type,\naccommodates,\nbathrooms,\nbedrooms,\nbeds,\nbed_type,\nminimum_nights,\nmaximum_nights,\navailability_365,\nnumber_of_reviews,\nreview_scores_rating,\nreview_scores_accuracy,\nreview_scores_cleanliness,\nreview_scores_checkin,\nreview_scores_communication,\nreview_scores_location,\nreview_scores_value,\ninstant_bookable,\ncancellation_policy,\nrequire_guest_profile_picture,\nrequire_guest_phone_verification,\ncalculated_host_listings_count,\nreviews_per_month,\nneighbourhood_cleansed\n\nDEPENDENT VARIABLE :::\nprice",
"_____no_output_____"
]
],
[
[
"#Useful colunms for ML application has been selected\ndf_selected_vars = df_listings[['host_response_time','host_response_rate','property_type','room_type','accommodates','bathrooms',\n 'bedrooms','beds','bed_type','minimum_nights','maximum_nights','availability_365','number_of_reviews',\n 'review_scores_rating','review_scores_accuracy','review_scores_cleanliness','neighbourhood_cleansed',\n 'review_scores_checkin','review_scores_communication','review_scores_location','review_scores_value','instant_bookable',\n 'cancellation_policy','require_guest_profile_picture','require_guest_phone_verification',\n 'calculated_host_listings_count','reviews_per_month','price']]",
"_____no_output_____"
],
[
"df_selected_vars.head()",
"_____no_output_____"
]
],
[
[
"In this part of the modeling, we created Dummy variables. Right after that,dummy variables are merged with numerical columns.",
"_____no_output_____"
]
],
[
[
"#CREATE DUMMY VARIABLES\ncat_df = df_selected_vars.select_dtypes(include=['object'])\ncat_df_copy = cat_df.copy()\ncat_cols_lst = cat_df.columns\n\ndef create_dummy_df(df,cat_cols):\n \n '''\n INPUT:\n df - pandas dataframe with categorical variables you want to dummy\n cat_cols - list of strings that are associated with names of the categorical columns\n \n OUTPUT:\n df - a new dataframe that has the following characteristics:\n 1. contains all columns that were not specified as categorical\n 2. removes all the original columns in cat_cols\n 3. dummy columns for each of the categorical columns in cat_cols\n 4. Use a prefix of the column name with an underscore (_) for separating \n '''\n for var in cat_cols:\n try:\n df = pd.concat([df.drop(var, axis=1), pd.get_dummies(df[var], prefix=var, prefix_sep='_', drop_first=True)], axis=1)\n except:\n continue\n return df",
"_____no_output_____"
],
[
"\n#Pull a list of the column names of the categorical variables\ncat_df = df_selected_vars.select_dtypes(include=['object'])\ncat_cols_lst = cat_df.columns\n\ndf_new = create_dummy_df(df_selected_vars, cat_df) #Use your newly created function\n\n# Show a header of df_new to check\n\ndf_new.head()\n\n",
"_____no_output_____"
],
[
"def fit_linear_mod(df,response_col,test_size=.3, rand_state=42):\n \n '''\n INPUT:\n df - a dataframe holding all the variables of interest\n response_col - a string holding the name of the column \n test_size - a float between [0,1] about what proportion of data should be in the test dataset\n rand_state - an int that is provided as the random state for splitting the data into training and test \n \n OUTPUT:\n test_score - float - r2 score on the test data\n train_score - float - r2 score on the test data\n lm_model - model object from sklearn\n X_train, X_test, y_train, y_test - output from sklearn train test split used for optimal model\n \n\n '''\n #Split your data into an X matrix and a response vector y\n X = df.drop(response_col,axis =1)\n y = df[response_col]\n \n #Create training and test set of data\n X_train,X_test,y_train,y_test = train_test_split(X,y,test_size = test_size, random_state = rand_state)\n \n #Instantiate a Linear Regression model with normalized data\n lm_model = LinearRegression(normalize = True)\n \n #Fit your model to thre training data\n lm_model.fit(X_train,y_train)\n \n #Predict the response for the training data and the test data\n y_test_preds = lm_model.predict(X_test)\n y_train_preds = lm_model.predict(X_train)\n \n #Obtain an rsquared value for both the training and test data\n test_score = r2_score(y_test,y_test_preds)\n train_score = r2_score(y_train,y_train_preds)\n \n return test_score, train_score, lm_model, X_train, X_test, y_train, y_test,y_test_preds,y_train_preds\n \n \n \n\n#Test your function with the above dataset\ntest_score, train_score, lm_model, X_train, X_test, y_train, y_test,y_test_preds,y_train_preds = fit_linear_mod(df_new, 'price')\n ",
"_____no_output_____"
]
],
[
[
"\nOur linear regression model explains around 60% of the variation of pricing in the training set, and 60% of variation of pricing in test set",
"_____no_output_____"
]
],
[
[
"#Print training and testing score . R square measures the strength of the relationship between mode and dependent variables \n#Our model is fitted 60% our observations.\nprint(\"The rsquared on the training data was {}. The rsquared on the test data was {}.\".format(train_score, test_score))",
"The rsquared on the training data was 0.5973214647874412. The rsquared on the test data was 0.5992518123850483.\n"
],
[
"lm_model.intercept_",
"_____no_output_____"
],
[
"#plotting y_test, y_test_prediction\nplt.figure(figsize=(10,5))\nsns.regplot(y_test,y_test_preds)\nplt.xlabel('actual_price')\nplt.ylabel('predicted_price')",
"_____no_output_____"
],
[
"plt.figure(figsize=(10,5))\nsns.scatterplot(y_test, y_test_preds)",
"_____no_output_____"
]
],
[
[
"We also look at p-values and coefficients of the model. if pvalues is less than 0.05 for each independent variable,there is a correlation between X vars and Y var. For example, p-value of 'accommodates','bathrooms','bedrooms','reviews_per_month' less than 0.05, we can say change in in the independent variables are associated with price. This variable is statistically significant and probably a worthwhile addition to your regression model. Otherwise, it is accepted that there is no significant relationship between X and y variables. \n\nThe sign of a regression coefficient tells you whether there is a positive or negative correlation between each independent variable the dependent variable. For example, while ‘accommodates’,’bathrooms’,’bedrooms’,’beds’ have positive correlation with price, ‘number of review’,’review score checkin’ has negative correlation. \n",
"_____no_output_____"
]
],
[
[
"import statsmodels.api as sm\n\nX_train_Sm= sm.add_constant(X_train)\nX_train_Sm= sm.add_constant(X_train)\nls=sm.OLS(y_train,X_train_Sm).fit()\nprint(ls.summary())\n",
"C:\\Users\\hilal\\Anaconda3\\lib\\site-packages\\numpy\\core\\fromnumeric.py:2389: FutureWarning: Method .ptp is deprecated and will be removed in a future version. Use numpy.ptp instead.\n return ptp(axis=axis, out=out, **kwargs)\n"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
4a56a1018db227711d97c744d44688ef01e3aa05
| 397,459 |
ipynb
|
Jupyter Notebook
|
Taller.ipynb
|
algoruz7/diplomado2017
|
cb5b659ae99a123a3d337e14c4c75c4c168083c9
|
[
"MIT"
] | null | null | null |
Taller.ipynb
|
algoruz7/diplomado2017
|
cb5b659ae99a123a3d337e14c4c75c4c168083c9
|
[
"MIT"
] | null | null | null |
Taller.ipynb
|
algoruz7/diplomado2017
|
cb5b659ae99a123a3d337e14c4c75c4c168083c9
|
[
"MIT"
] | null | null | null | 302.020517 | 120,394 | 0.895056 |
[
[
[
"# Taller evaluable sobre la extracción, transformación y visualización de datos usando IPython",
"_____no_output_____"
],
[
"**Juan David Velásquez Henao** \[email protected] \nUniversidad Nacional de Colombia, Sede Medellín \nFacultad de Minas \nMedellín, Colombia ",
"_____no_output_____"
],
[
"# Instrucciones",
"_____no_output_____"
],
[
"En la carpeta 'Taller' del repositorio 'ETVL-IPython' se encuentran los archivos 'Precio_Bolsa_Nacional_($kwh)_'*'.xls' en formato de Microsoft Excel, los cuales contienen los precios históricos horarios de la electricidad para el mercado eléctrico Colombiano entre los años 1995 y 2017 en COL-PESOS/kWh. A partir de la información suministrada resuelva los siguientes puntos usando el lenguaje de programación Python. ",
"_____no_output_____"
],
[
"# Preguntas",
"_____no_output_____"
],
[
"**1.--** Lea los archivos y cree una tabla única concatenando la información para cada uno de los años. Debe transformar la tabla de tal forma que quede con las columnas `Fecha`, `Hora` y `Precio` (únicamente tres columnas). Imprima el encabezamiento de la tabla usando `head()`. ",
"_____no_output_____"
]
],
[
[
"import os, pandas, numpy, matplotlib, matplotlib.pyplot\ndireccion = \"C:/Users/Asus/Downloads/ETVL-IPython-master (2)/ETVL-IPython-master/Taller\"\nlistaArchivos=[]\nlistaInicial=os.walk(direccion)\n\n# ESTA PARTE LISTA LOS ARCHIVOS DE EXCEL INCLUIDOS EN UNA CARPETA ESPECIFICADA. SE DESCARTAN LOS ARCHIVOS TEMPORALES Y/O \n# ARCHIVOS QUE SE ENCUENTREN EN SUBCARPETAS.\n\nfor Dire, CarpetasDentroDire, ArchivosDentroDire in listaInicial:\n if Dire == direccion:\n for nombres in ArchivosDentroDire:\n# print('Nombres: %s' % nombres)\n (nombreArchivo, extArchivo) = os.path.splitext(nombres)\n if nombreArchivo[0]=='~':\n# print('No Almacenado')\n pass\n elif(extArchivo == \".xlsx\"):\n listaArchivos.append(nombreArchivo+extArchivo)\n# print('Almacenado')\n elif(extArchivo == \".xls\"):\n listaArchivos.append(nombreArchivo+extArchivo)\n# print('Almacenado')\n else:\n# print('No Almacenado')\n pass\n else:\n# print('Carpeta Excluida')\n pass\nprint('Archivos a cargador:')\nprint(pandas.Series(listaArchivos).values)",
"Archivos a cargador:\n['Precio_Bolsa_Nacional_($kwh)_1995.xlsx'\n 'Precio_Bolsa_Nacional_($kwh)_1996.xlsx'\n 'Precio_Bolsa_Nacional_($kwh)_1997.xlsx'\n 'Precio_Bolsa_Nacional_($kwh)_1998.xlsx'\n 'Precio_Bolsa_Nacional_($kwh)_1999.xlsx'\n 'Precio_Bolsa_Nacional_($kwh)_2000.xlsx'\n 'Precio_Bolsa_Nacional_($kwh)_2001.xlsx'\n 'Precio_Bolsa_Nacional_($kwh)_2002.xlsx'\n 'Precio_Bolsa_Nacional_($kwh)_2003.xlsx'\n 'Precio_Bolsa_Nacional_($kwh)_2004.xlsx'\n 'Precio_Bolsa_Nacional_($kwh)_2005.xlsx'\n 'Precio_Bolsa_Nacional_($kwh)_2006.xlsx'\n 'Precio_Bolsa_Nacional_($kwh)_2007.xlsx'\n 'Precio_Bolsa_Nacional_($kwh)_2008.xlsx'\n 'Precio_Bolsa_Nacional_($kwh)_2009.xlsx'\n 'Precio_Bolsa_Nacional_($kwh)_2010.xlsx'\n 'Precio_Bolsa_Nacional_($kwh)_2011.xlsx'\n 'Precio_Bolsa_Nacional_($kwh)_2012.xlsx'\n 'Precio_Bolsa_Nacional_($kwh)_2013.xlsx'\n 'Precio_Bolsa_Nacional_($kwh)_2014.xlsx'\n 'Precio_Bolsa_Nacional_($kwh)_2015.xlsx'\n 'Precio_Bolsa_Nacional_($kwh)_2016.xls'\n 'Precio_Bolsa_Nacional_($kwh)_2017.xls']\n"
],
[
"# ESTA PARTE CARGA LOS ARCHIVOS Y CARGA LOS DATOS EN UNA SOLA TABLA. NO SE CARGAN DATOS DE LAS HOJAS CUYOS NOMBRES SEAN \"Hoja XX\"\ndataF=pandas.DataFrame()\nfor Archivo in listaArchivos:\n# print('Archivo=',Archivo)\n xl = pandas.ExcelFile(Archivo)\n hojas=xl.sheet_names\n skip=-1\n for hj in hojas:\n if hj[0:4] == 'Hoja':\n print( hj, 'del archivo', Archivo, 'descartada')\n else:\n dataini= xl.parse(hj)\n for i in range(0,len(dataini)):\n datamod= xl.parse(hj,skiprows=i)\n encabezado=datamod.columns\n if (encabezado[0])=='Fecha':\n skip=i\n break\n if skip>0:\n break\n datax=xl.parse(hj,skiprows=skip,parse_cols=24)\n dataF=dataF.append(datax,ignore_index=True)\n# print(dataF.shape)\ndataF_sin_NA=dataF.dropna()\ndataF_sin_dupli=dataF.drop_duplicates()\ndatos_limpios=dataF_sin_NA.drop_duplicates().reset_index(drop=True)\nprint('El tamaño tabla cargada es de', dataF.shape,'y tiene la siguiente forma:')\n(dataF.head(3))\n",
"El tamaño tabla cargada es de (7962, 25) y tiene la siguiente forma:\n"
],
[
"#ESTA PARTE DEPURA Y REORGANIZA LOS DATOS\nlista_horasDia=['00:00:00','01:00:00', '02:00:00', '03:00:00', '04:00:00', '05:00:00'\n , '06:00:00', '07:00:00', '08:00:00', '09:00:00', '10:00:00', '11:00:00'\n ,'12:00:00', '13:00:00', '14:00:00', '15:00:00', '16:00:00', '17:00:00'\n , '18:00:00', '19:00:00', '20:00:00', '21:00:00', '22:00:00', '23:00:00']\n\n\nlista_horas=['0','1', '2', '3', '4', '5', '6', '7', '8', '9', '10', '11','12', '13', '14', '15', '16', '17', '18', '19', \n '20', '21', '22', '23']\nfechas=pandas.to_datetime(datos_limpios.Fecha)\nfor iorg in range(len(datos_limpios)):\n asd1=datos_limpios.ix[iorg,lista_horas].T.values\n asd2=pandas.DataFrame(asd1)\n temp1=datos_limpios.ix[iorg,'Fecha']\n temp2=[temp1]*24\n temp3=pandas.DataFrame(temp2) \n horatemp1=pandas.DataFrame(lista_horasDia) \n if iorg == 0: \n dataxorg=asd2\n tempp=temp3\n horatemp=horatemp1\n else:\n datawass=[dataxorg,asd2]\n temppwas=[tempp,temp3] \n horatempwas=[horatemp,horatemp1]\n dataxorg = pandas.concat(datawass, ignore_index = True)\n tempp=pandas.concat(temppwas, ignore_index = True)\n horatemp=pandas.concat(horatempwas,ignore_index = True)\nhoora=[]\npreec=[]\nañomes1=[]\nfechatotal=[]\nsolofecha=[]\nfor tg in range(len(tempp)):\n hoora.append(str(horatemp.ix[tg,0])[0:8])\n preec.append(str(dataxorg.ix[tg,0])[0:100])\n añomes1.append(str(tempp.ix[tg,0])[0:7]+'-01')\n solofecha.append(str(tempp.ix[tg,0])[0:10])\n fechatotal.append(str(tempp.ix[tg,0])[0:10]+' '+str(horatemp.ix[tg,0])[0:8])\nsolofecha=pandas.to_datetime(pandas.Series((solofecha)))\nfechatotal=pandas.to_datetime(pandas.Series((fechatotal)))\ndf2 = pandas.DataFrame({'Fecha': pandas.Series(solofecha),'Hora': pandas.Series(hoora), 'Precio': preec})\ndf2['Precio'] = df2['Precio'].convert_objects(convert_numeric=True)\n\ndf4 = pandas.DataFrame({'Año': pandas.Series(fechatotal.dt.year),'Mes': pandas.Series(fechatotal.dt.month),\n 'Dia calendario': pandas.Series(fechatotal.dt.day), 'Dia': pandas.Series(fechatotal.dt.weekday_name),\n 'Hora': pandas.Series(fechatotal.dt.hour),'Precio': preec, 'Fecha Completa': fechatotal})\ndf4['Precio'] = df4['Precio'].convert_objects(convert_numeric=True)\nañomes2=pandas.Series(añomes1)\nañomes2=pandas.DataFrame(añomes2)\nañomes=añomes2.drop_duplicates().reset_index(drop=True)\nañomes3=[]\nfor tg in range(len(añomes)):\n añomes3.append(str(añomes.ix[tg,0]))\nañosymeses=pandas.to_datetime(pandas.Series((añomes3)))\ndias=fechas\nprint('El tamaño tabla depurada es de', df2.shape,'y tiene la siguiente forma:')\ndf2.head(3)",
"C:\\Users\\Asus\\Anaconda3\\lib\\site-packages\\ipykernel\\__main__.py:43: FutureWarning: convert_objects is deprecated. Use the data-type specific converters pd.to_datetime, pd.to_timedelta and pd.to_numeric.\nC:\\Users\\Asus\\Anaconda3\\lib\\site-packages\\ipykernel\\__main__.py:48: FutureWarning: convert_objects is deprecated. Use the data-type specific converters pd.to_datetime, pd.to_timedelta and pd.to_numeric.\n"
]
],
[
[
"**2.--** Compute e imprima el número de registros con datos faltantes.",
"_____no_output_____"
]
],
[
[
"print('Registros iniciales con Datos Faltantes =',len(dataF)-len(dataF_sin_NA))",
"Registros iniciales con Datos Faltantes = 28\n"
]
],
[
[
"**3.--** Compute e imprima el número de registros duplicados.",
"_____no_output_____"
]
],
[
[
"print('Registros iniciales Duplicados =',len(dataF)-len(dataF_sin_dupli))",
"Registros iniciales Duplicados = 67\n"
]
],
[
[
"**4.--** Elimine los registros con datos duplicados o datos faltantes, e imprima la cantidad de registros que quedan (registros completos).",
"_____no_output_____"
]
],
[
[
"print('Registros iniciales completos =',len(datos_limpios))",
"Registros iniciales completos = 7875\n"
]
],
[
[
"**5.--** Compute y grafique el precio primedio diario.",
"_____no_output_____"
]
],
[
[
"#ESTA PARTE COMPUTA PROMEDIOS Y GRAFICA EL PRECIO PROMEDIO DE CADA DÍA\npromDia=df4.groupby(['Año', 'Mes', 'Dia calendario'])['Precio'].mean().values#['Precio'].mean()#.values\n# print(promDia.shape)\npromMes=df4.groupby(['Año', 'Mes'])['Precio'].mean().values\n# print(promMes.shape)\nmatplotlib.pyplot.plot(dias, promDia)\nmatplotlib.pyplot.title(\"Precio Promedio por Dia\")\nmatplotlib.pyplot.ylabel('$/kwh')\nmatplotlib.pyplot.xticks(rotation=70)\nmatplotlib.pyplot.show()",
"_____no_output_____"
]
],
[
[
"**6.--** Compute y grafique el precio máximo por mes.",
"_____no_output_____"
]
],
[
[
"#ESTA PARTE EXTRÁE Y GRAFICA EL PRECIO MÁXIMO MENSUAL\nmaxMes=pandas.Series(df4.groupby(['Año', 'Mes'])['Precio'].max().values)\nmatplotlib.pyplot.plot(añosymeses,maxMes)\nmatplotlib.pyplot.title(\"Precio Máximo por Mes\")\nmatplotlib.pyplot.ylabel('$/kwh')\nmatplotlib.pyplot.xticks(rotation=70)\nmatplotlib.pyplot.show()",
"_____no_output_____"
]
],
[
[
"**7.--** Compute y grafique el precio mínimo mensual.",
"_____no_output_____"
]
],
[
[
"#ESTA PARTE EXTRÁE Y GRAFICA EL PRECIO MÍNIMO MENSUAL\nminMes=pandas.Series(df4.groupby(['Año', 'Mes'])['Precio'].min().values)\nmatplotlib.pyplot.plot(añosymeses,minMes)\nmatplotlib.pyplot.title(\"Precio Mínimo por Mes\") \nmatplotlib.pyplot.ylabel('$/kwh')\nmatplotlib.pyplot.xticks(rotation=70)\nmatplotlib.pyplot.show()",
"_____no_output_____"
]
],
[
[
"**8.--** Haga un gráfico para comparar el precio máximo del mes (para cada mes) y el precio promedio mensual.",
"_____no_output_____"
]
],
[
[
"\nmatplotlib.pyplot.plot(añosymeses, maxMes, linestyle=':', color='r',label='Precio Máximo Mensual')#marker='x', linestyle=':', color='b',label='Precio Máximo Mensual')\nmatplotlib.pyplot.ion() \nmatplotlib.pyplot.plot(añosymeses, promMes, linestyle='--', color='b',label='Precio Promedio Mensual')#marker='o', linestyle='--', color='r',label='Precio Promedio Mensual')\nmatplotlib.pyplot.legend(loc=\"best\")\nmatplotlib.pyplot.title(\"Precios Máximos y Promedio por Mes\") \nmatplotlib.pyplot.ylabel('$/kwh')\nmatplotlib.pyplot.xticks(rotation=70)\nmatplotlib.pyplot.show()",
"_____no_output_____"
]
],
[
[
"**9.--** Haga un histograma que muestre a que horas se produce el máximo precio diario para los días laborales.",
"_____no_output_____"
]
],
[
[
"\nokm=df4.groupby(['Año','Dia', 'Hora'],as_index = False)['Precio'].max()\nokm2=okm.groupby(['Año','Dia'],as_index = False)['Precio'].max()\ncolunasencomun=list(set(okm.columns) & set(okm2.columns))\nokm3=pandas.merge(okm,okm2, on=colunasencomun, how='inner')\nokm4=okm3.groupby(['Año','Dia'],as_index = False).max()\nfechagrafhabil=[]\nhoragrafhabil=[]\nfechagrafSAB=[]\nhoragrafSAB=[]\nfechagrafDOM=[]\nhoragrafDOM=[]\nfor vbg in range(len(okm4)):\n if okm4.ix[vbg,'Dia']== 'Saturday':\n fechagrafSAB.append(str(okm4.ix[vbg,'Año'])+'-'+str(okm4.ix[vbg,'Dia']))\n horagrafSAB.append(str(okm4.ix[vbg,'Hora']))\n elif okm4.ix[vbg,'Dia']== 'Sunday':\n fechagrafDOM.append(str(okm4.ix[vbg,'Año'])+'-'+str(okm4.ix[vbg,'Dia']))\n horagrafDOM.append(str(okm4.ix[vbg,'Hora']))\n else: \n fechagrafhabil.append(str(okm4.ix[vbg,'Año'])+'-'+str(okm4.ix[vbg,'Dia']))\n horagrafhabil.append(str(okm4.ix[vbg,'Hora']))\n\nhoragrafhabil=pandas.Series(horagrafhabil).convert_objects(convert_numeric=True)\nhoragrafSAB=pandas.Series(horagrafSAB).convert_objects(convert_numeric=True)\nhoragrafDOM=pandas.Series(horagrafDOM).convert_objects(convert_numeric=True)\nindixx=numpy.arange(len(horagrafhabil))\nmatplotlib.pyplot.barh(indixx,horagrafhabil,align = \"center\")\nmatplotlib.pyplot.yticks(indixx, fechagrafhabil)\nmatplotlib.pyplot.xlabel('Horas')\nmatplotlib.pyplot.ylabel('Año-Dia')\nmatplotlib.pyplot.title('Hora de precio máximo para los días laborales de cada año.')\nmatplotlib.pyplot.xticks(numpy.arange(0,24,1))\nmatplotlib.pyplot.xlim(0,23.05)\nmatplotlib.pyplot.ylim(-1,len(horagrafhabil))\nmatplotlib.pyplot.grid(True)\nmatplotlib.pyplot.show()",
"C:\\Users\\Asus\\Anaconda3\\lib\\site-packages\\ipykernel\\__main__.py:24: FutureWarning: convert_objects is deprecated. Use the data-type specific converters pd.to_datetime, pd.to_timedelta and pd.to_numeric.\nC:\\Users\\Asus\\Anaconda3\\lib\\site-packages\\ipykernel\\__main__.py:25: FutureWarning: convert_objects is deprecated. Use the data-type specific converters pd.to_datetime, pd.to_timedelta and pd.to_numeric.\nC:\\Users\\Asus\\Anaconda3\\lib\\site-packages\\ipykernel\\__main__.py:26: FutureWarning: convert_objects is deprecated. Use the data-type specific converters pd.to_datetime, pd.to_timedelta and pd.to_numeric.\n"
]
],
[
[
"**10.--** Haga un histograma que muestre a que horas se produce el máximo precio diario para los días sabado.",
"_____no_output_____"
]
],
[
[
"indixx=numpy.arange(len(horagrafSAB))\nmatplotlib.pyplot.barh(indixx,horagrafSAB,align = \"center\")\nmatplotlib.pyplot.yticks(indixx, fechagrafSAB)\nmatplotlib.pyplot.xlabel('Horas')\nmatplotlib.pyplot.ylabel('Año-Dia')\nmatplotlib.pyplot.title('Hora de precio máximo para los Sábados de cada año.')\nmatplotlib.pyplot.xticks(numpy.arange(0,24,1))\nmatplotlib.pyplot.xlim(0,23.05)\nmatplotlib.pyplot.ylim(-1,len(horagrafSAB))\nmatplotlib.pyplot.grid(True)\nmatplotlib.pyplot.show()",
"_____no_output_____"
]
],
[
[
"**11.--** Haga un histograma que muestre a que horas se produce el máximo precio diario para los días domingo.",
"_____no_output_____"
]
],
[
[
"indixx=numpy.arange(len(horagrafDOM))\nmatplotlib.pyplot.barh(indixx,horagrafSAB,align = \"center\")\nmatplotlib.pyplot.yticks(indixx, fechagrafDOM)\nmatplotlib.pyplot.xlabel('Horas')\nmatplotlib.pyplot.ylabel('Año-Dia')\nmatplotlib.pyplot.title('Hora de precio máximo para los Sábados de cada año.')\nmatplotlib.pyplot.xticks(numpy.arange(0,24,1))\nmatplotlib.pyplot.xlim(0,23.05)\nmatplotlib.pyplot.ylim(-1,len(horagrafDOM))\nmatplotlib.pyplot.grid(True)\nmatplotlib.pyplot.show()",
"_____no_output_____"
]
],
[
[
"**12.--** Imprima una tabla con la fecha y el valor más bajo por año del precio de bolsa.",
"_____no_output_____"
]
],
[
[
"# print(df4)\n\nokm=df4.groupby(['Año','Dia', 'Hora'],as_index = False)['Precio'].min()\nprint('okm=',okm)\nokm2=okm.groupby(['Año','Dia'],as_index = False)['Precio'].min()\nprint('okm2=',okm2)\ncolunasencomun=list(set(okm.columns) & set(okm2.columns))\nokm3=pandas.merge(okm,okm2, on=colunasencomun, how='inner')\nprint('okm3=',okm3)\nokm4=okm3.groupby(['Año','Dia'],as_index = False).min()\nprint('okm4=',okm4)\ncolunasencomun2=list(set(okm.columns) & set(okm2.columns))",
"okm= Año Dia Hora Precio\n0 1995 Friday 0 0.000000\n1 1995 Friday 1 0.000000\n2 1995 Friday 2 0.000000\n3 1995 Friday 3 0.000000\n4 1995 Friday 4 0.000000\n5 1995 Friday 5 0.463000\n6 1995 Friday 6 0.463000\n7 1995 Friday 7 1.000000\n8 1995 Friday 8 0.463000\n9 1995 Friday 9 0.463000\n10 1995 Friday 10 1.073000\n11 1995 Friday 11 1.073000\n12 1995 Friday 12 0.463000\n13 1995 Friday 13 0.463000\n14 1995 Friday 14 0.463000\n15 1995 Friday 15 0.463000\n16 1995 Friday 16 0.463000\n17 1995 Friday 17 0.463000\n18 1995 Friday 18 1.073000\n19 1995 Friday 19 1.073000\n20 1995 Friday 20 1.073000\n21 1995 Friday 21 1.000000\n22 1995 Friday 22 0.000000\n23 1995 Friday 23 0.000000\n24 1995 Monday 0 0.000000\n25 1995 Monday 1 0.000000\n26 1995 Monday 2 0.000000\n27 1995 Monday 3 0.000000\n28 1995 Monday 4 0.000000\n29 1995 Monday 5 1.000000\n... ... ... ... ...\n3834 2017 Tuesday 18 84.472945\n3835 2017 Tuesday 19 114.472945\n3836 2017 Tuesday 20 84.472945\n3837 2017 Tuesday 21 84.472945\n3838 2017 Tuesday 22 84.472945\n3839 2017 Tuesday 23 63.000923\n3840 2017 Wednesday 0 62.306708\n3841 2017 Wednesday 1 62.306708\n3842 2017 Wednesday 2 61.356315\n3843 2017 Wednesday 3 62.306708\n3844 2017 Wednesday 4 61.356315\n3845 2017 Wednesday 5 62.306708\n3846 2017 Wednesday 6 62.306708\n3847 2017 Wednesday 7 73.279315\n3848 2017 Wednesday 8 74.279315\n3849 2017 Wednesday 9 89.279315\n3850 2017 Wednesday 10 89.279315\n3851 2017 Wednesday 11 114.279315\n3852 2017 Wednesday 12 114.279315\n3853 2017 Wednesday 13 114.279315\n3854 2017 Wednesday 14 114.279315\n3855 2017 Wednesday 15 89.279315\n3856 2017 Wednesday 16 89.279315\n3857 2017 Wednesday 17 89.279315\n3858 2017 Wednesday 18 120.355594\n3859 2017 Wednesday 19 126.655594\n3860 2017 Wednesday 20 114.279315\n3861 2017 Wednesday 21 89.279315\n3862 2017 Wednesday 22 74.279315\n3863 2017 Wednesday 23 62.306708\n\n[3864 rows x 4 columns]\nokm2= Año Dia Precio\n0 1995 Friday 0.000000\n1 1995 Monday 0.000000\n2 1995 Saturday 0.000000\n3 1995 Sunday 0.000000\n4 1995 Thursday 0.400000\n5 1995 Tuesday 0.000000\n6 1995 Wednesday 0.000000\n7 1996 Friday 0.000000\n8 1996 Monday 0.000000\n9 1996 Saturday 1.000000\n10 1996 Sunday 0.000000\n11 1996 Thursday 0.000000\n12 1996 Tuesday 0.000000\n13 1996 Wednesday 0.000000\n14 1997 Friday 13.226270\n15 1997 Monday 13.169270\n16 1997 Saturday 13.166270\n17 1997 Sunday 12.221270\n18 1997 Thursday 13.170270\n19 1997 Tuesday 10.882310\n20 1997 Wednesday 13.226270\n21 1998 Friday 13.847330\n22 1998 Monday 13.847330\n23 1998 Saturday 13.847330\n24 1998 Sunday 13.847330\n25 1998 Thursday 13.847330\n26 1998 Tuesday 13.847330\n27 1998 Wednesday 13.847330\n28 1999 Friday 18.409530\n29 1999 Monday 18.409530\n.. ... ... ...\n131 2013 Tuesday 41.586302\n132 2013 Wednesday 40.415346\n133 2014 Friday 39.929670\n134 2014 Monday 44.904704\n135 2014 Saturday 38.941951\n136 2014 Sunday 44.268975\n137 2014 Thursday 40.830458\n138 2014 Tuesday 39.560203\n139 2014 Wednesday 41.236232\n140 2015 Friday 84.480854\n141 2015 Monday 51.116576\n142 2015 Saturday 83.776752\n143 2015 Sunday 51.649498\n144 2015 Thursday 66.170951\n145 2015 Tuesday 46.791501\n146 2015 Wednesday 52.928171\n147 2016 Friday 61.100689\n148 2016 Monday 61.243079\n149 2016 Saturday 61.913119\n150 2016 Sunday 61.172545\n151 2016 Thursday 61.609287\n152 2016 Tuesday 62.098851\n153 2016 Wednesday 61.822375\n154 2017 Friday 61.812631\n155 2017 Monday 62.699828\n156 2017 Saturday 61.516733\n157 2017 Sunday 61.493259\n158 2017 Thursday 65.440433\n159 2017 Tuesday 62.934251\n160 2017 Wednesday 61.356315\n\n[161 rows x 3 columns]\nokm3= Año Dia Hora Precio\n0 1995 Friday 0 0.000000\n1 1995 Friday 1 0.000000\n2 1995 Friday 2 0.000000\n3 1995 Friday 3 0.000000\n4 1995 Friday 4 0.000000\n5 1995 Friday 22 0.000000\n6 1995 Friday 23 0.000000\n7 1995 Monday 0 0.000000\n8 1995 Monday 1 0.000000\n9 1995 Monday 2 0.000000\n10 1995 Monday 3 0.000000\n11 1995 Monday 4 0.000000\n12 1995 Monday 7 0.000000\n13 1995 Monday 8 0.000000\n14 1995 Monday 10 0.000000\n15 1995 Monday 11 0.000000\n16 1995 Monday 13 0.000000\n17 1995 Monday 14 0.000000\n18 1995 Monday 15 0.000000\n19 1995 Monday 16 0.000000\n20 1995 Monday 17 0.000000\n21 1995 Monday 23 0.000000\n22 1995 Saturday 2 0.000000\n23 1995 Saturday 3 0.000000\n24 1995 Saturday 5 0.000000\n25 1995 Sunday 2 0.000000\n26 1995 Sunday 3 0.000000\n27 1995 Sunday 4 0.000000\n28 1995 Sunday 5 0.000000\n29 1995 Sunday 15 0.000000\n.. ... ... ... ...\n861 2017 Saturday 5 61.516733\n862 2017 Saturday 6 61.516733\n863 2017 Saturday 23 61.516733\n864 2017 Sunday 0 61.493259\n865 2017 Sunday 1 61.493259\n866 2017 Sunday 2 61.493259\n867 2017 Sunday 3 61.493259\n868 2017 Sunday 4 61.493259\n869 2017 Sunday 5 61.493259\n870 2017 Sunday 6 61.493259\n871 2017 Sunday 7 61.493259\n872 2017 Sunday 8 61.493259\n873 2017 Sunday 9 61.493259\n874 2017 Sunday 10 61.493259\n875 2017 Sunday 13 61.493259\n876 2017 Sunday 14 61.493259\n877 2017 Sunday 15 61.493259\n878 2017 Sunday 16 61.493259\n879 2017 Sunday 17 61.493259\n880 2017 Sunday 23 61.493259\n881 2017 Thursday 0 65.440433\n882 2017 Thursday 1 65.440433\n883 2017 Thursday 2 65.440433\n884 2017 Thursday 3 65.440433\n885 2017 Thursday 4 65.440433\n886 2017 Thursday 6 65.440433\n887 2017 Thursday 23 65.440433\n888 2017 Tuesday 3 62.934251\n889 2017 Wednesday 2 61.356315\n890 2017 Wednesday 4 61.356315\n\n[891 rows x 4 columns]\nokm4= Año Dia Hora Precio\n0 1995 Friday 0 0.000000\n1 1995 Monday 0 0.000000\n2 1995 Saturday 2 0.000000\n3 1995 Sunday 2 0.000000\n4 1995 Thursday 17 0.400000\n5 1995 Tuesday 0 0.000000\n6 1995 Wednesday 23 0.000000\n7 1996 Friday 0 0.000000\n8 1996 Monday 0 0.000000\n9 1996 Saturday 0 1.000000\n10 1996 Sunday 0 0.000000\n11 1996 Thursday 2 0.000000\n12 1996 Tuesday 0 0.000000\n13 1996 Wednesday 0 0.000000\n14 1997 Friday 0 13.226270\n15 1997 Monday 0 13.169270\n16 1997 Saturday 0 13.166270\n17 1997 Sunday 1 12.221270\n18 1997 Thursday 1 13.170270\n19 1997 Tuesday 2 10.882310\n20 1997 Wednesday 1 13.226270\n21 1998 Friday 0 13.847330\n22 1998 Monday 0 13.847330\n23 1998 Saturday 0 13.847330\n24 1998 Sunday 0 13.847330\n25 1998 Thursday 0 13.847330\n26 1998 Tuesday 0 13.847330\n27 1998 Wednesday 0 13.847330\n28 1999 Friday 23 18.409530\n29 1999 Monday 18 18.409530\n.. ... ... ... ...\n131 2013 Tuesday 3 41.586302\n132 2013 Wednesday 0 40.415346\n133 2014 Friday 0 39.929670\n134 2014 Monday 0 44.904704\n135 2014 Saturday 2 38.941951\n136 2014 Sunday 3 44.268975\n137 2014 Thursday 0 40.830458\n138 2014 Tuesday 2 39.560203\n139 2014 Wednesday 1 41.236232\n140 2015 Friday 2 84.480854\n141 2015 Monday 2 51.116576\n142 2015 Saturday 2 83.776752\n143 2015 Sunday 0 51.649498\n144 2015 Thursday 2 66.170951\n145 2015 Tuesday 2 46.791501\n146 2015 Wednesday 0 52.928171\n147 2016 Friday 0 61.100689\n148 2016 Monday 0 61.243079\n149 2016 Saturday 3 61.913119\n150 2016 Sunday 0 61.172545\n151 2016 Thursday 0 61.609287\n152 2016 Tuesday 2 62.098851\n153 2016 Wednesday 3 61.822375\n154 2017 Friday 0 61.812631\n155 2017 Monday 0 62.699828\n156 2017 Saturday 1 61.516733\n157 2017 Sunday 0 61.493259\n158 2017 Thursday 0 65.440433\n159 2017 Tuesday 3 62.934251\n160 2017 Wednesday 2 61.356315\n\n[161 rows x 4 columns]\n"
]
],
[
[
"**13.--** Haga una gráfica en que se muestre el precio promedio diario y el precio promedio mensual.",
"_____no_output_____"
]
],
[
[
"promDia=df4.groupby(['Año', 'Mes', 'Dia calendario'],as_index = False)['Precio'].mean()#['Precio'].mean()#.values\nprint(promDia)\npromMes=df4.groupby(['Año', 'Mes'],as_index = False)['Precio'].mean()\nprint(promMes)\npromMescompara=[]\nfor thnm in range(len(promDia)):\n for ijn in range(len(promMes)):\n if ((promDia.ix(thnm,'Año')==promMes.ix(ijn,'Año'))):\n if (promDia.ix(thnm,'Mes')==promMes.ix(ijn,'Mes')):\n promMescompara.append(str(promMes.ix[ijn,'Precio']))\npromMescompara=pandas.Series(promMescompara)\npromMescompara= promMescompara.convert_objects(convert_numeric=True)\nprint(promMescompara)\n \n \n \n ",
" Año Mes Dia calendario Precio\n0 1995 7 21 4.924333\n1 1995 7 22 1.269500\n2 1995 7 23 0.953083\n3 1995 7 24 4.305917\n4 1995 7 25 1.149167\n5 1995 7 26 1.108625\n6 1995 7 27 0.585958\n7 1995 7 28 0.499792\n8 1995 7 29 0.927667\n9 1995 7 30 0.669458\n10 1995 7 31 0.679458\n11 1995 8 1 1.106917\n12 1995 8 2 1.077500\n13 1995 8 3 1.517917\n14 1995 8 4 1.786042\n15 1995 8 5 2.383792\n16 1995 8 6 2.387583\n17 1995 8 7 3.707208\n18 1995 8 8 6.650000\n19 1995 8 9 13.991958\n20 1995 8 10 20.041667\n21 1995 8 11 21.191667\n22 1995 8 12 30.710750\n23 1995 8 13 34.327833\n24 1995 8 14 33.190250\n25 1995 8 15 14.513250\n26 1995 8 16 8.664333\n27 1995 8 17 1.813250\n28 1995 8 18 1.941667\n29 1995 8 19 1.101833\n... ... ... ... ...\n7845 2017 1 14 69.696400\n7846 2017 1 15 84.918995\n7847 2017 1 16 89.348661\n7848 2017 1 17 96.043590\n7849 2017 1 18 111.541249\n7850 2017 1 19 105.999284\n7851 2017 1 20 110.215487\n7852 2017 1 21 101.231979\n7853 2017 1 22 75.254842\n7854 2017 1 23 115.965815\n7855 2017 1 24 148.968501\n7856 2017 1 25 130.411958\n7857 2017 1 26 144.226841\n7858 2017 1 27 132.357035\n7859 2017 1 28 122.145329\n7860 2017 1 29 132.564871\n7861 2017 1 30 133.677754\n7862 2017 1 31 122.424193\n7863 2017 2 1 103.459761\n7864 2017 2 2 105.866599\n7865 2017 2 3 110.059089\n7866 2017 2 4 127.794467\n7867 2017 2 5 90.072541\n7868 2017 2 6 119.555657\n7869 2017 2 7 146.880526\n7870 2017 2 8 131.514214\n7871 2017 2 9 133.727413\n7872 2017 2 10 167.128157\n7873 2017 2 11 151.908170\n7874 2017 2 12 142.414385\n\n[7875 rows x 4 columns]\n Año Mes Precio\n0 1995 7 1.552087\n1 1995 8 7.086462\n2 1995 9 10.955819\n3 1995 10 10.445442\n4 1995 11 27.534782\n5 1995 12 68.876234\n6 1996 1 23.008071\n7 1996 2 8.044093\n8 1996 3 3.460622\n9 1996 4 3.040025\n10 1996 5 1.809852\n11 1996 6 1.889136\n12 1996 7 1.649266\n13 1996 8 3.717004\n14 1996 9 15.894114\n15 1996 10 5.523819\n16 1996 11 20.601340\n17 1996 12 14.869731\n18 1997 1 21.623433\n19 1997 2 26.916226\n20 1997 3 25.705822\n21 1997 4 25.252256\n22 1997 5 35.462962\n23 1997 6 33.356151\n24 1997 7 31.958213\n25 1997 8 38.467690\n26 1997 9 138.986062\n27 1997 10 137.127820\n28 1997 11 132.435171\n29 1997 12 128.869191\n.. ... ... ...\n230 2014 9 177.318519\n231 2014 10 207.026344\n232 2014 11 166.568139\n233 2014 12 175.054653\n234 2015 1 187.595911\n235 2015 2 166.231229\n236 2015 3 205.502951\n237 2015 4 161.304436\n238 2015 5 259.198931\n239 2015 6 186.416549\n240 2015 7 206.158070\n241 2015 8 183.708405\n242 2015 9 458.834686\n243 2015 10 1106.619160\n244 2015 11 766.991982\n245 2015 12 630.193550\n246 2016 1 554.158982\n247 2016 2 645.999911\n248 2016 3 830.018197\n249 2016 4 332.493264\n250 2016 5 140.310381\n251 2016 6 167.971917\n252 2016 7 129.214374\n253 2016 8 198.012720\n254 2016 9 150.592068\n255 2016 10 184.907520\n256 2016 11 166.532592\n257 2016 12 107.439280\n258 2017 1 111.560500\n259 2017 2 127.531748\n\n[260 rows x 3 columns]\n"
]
],
[
[
"---",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
4a56af2795a4a39713b6fa4e6958f60c73c54aa5
| 10,612 |
ipynb
|
Jupyter Notebook
|
docs/advanced_usage.ipynb
|
johanvdw/niche_vlaanderen
|
8dee32b21e81146c6c0ea39a050930ee9a4f5216
|
[
"MIT"
] | null | null | null |
docs/advanced_usage.ipynb
|
johanvdw/niche_vlaanderen
|
8dee32b21e81146c6c0ea39a050930ee9a4f5216
|
[
"MIT"
] | null | null | null |
docs/advanced_usage.ipynb
|
johanvdw/niche_vlaanderen
|
8dee32b21e81146c6c0ea39a050930ee9a4f5216
|
[
"MIT"
] | null | null | null | 34.679739 | 508 | 0.636638 |
[
[
[
"## Advanced usage\n### Using config files\n\nInstead of specifying all inputs using [set_input](https://inbo.github.io/niche_vlaanderen/lowlevel.html#niche_vlaanderen.Niche.set_input), it is possible to use a config file. A config file can be loaded using [read_config_file](https://inbo.github.io/niche_vlaanderen/lowlevel.html#niche_vlaanderen.Niche.read_config_file) or it can be read and executed immediately by using [run_config_file](https://inbo.github.io/niche_vlaanderen/lowlevel.html#niche_vlaanderen.Niche.run_config_file).\n\nThe syntax of the config file is explained more in detail in [Niche Configuration file](https://inbo.github.io/niche_vlaanderen/cli.html), but is already introduced here because it will be used in the next examples.\n\nIf you want to recreate the examples below, the config files can be found under the `docs` folder, so if you [extract all the data](https://inbo.github.io/niche_vlaanderen/getting_started.html#Interactive-usage) the you should be able to run the examples from the notebook. \n",
"_____no_output_____"
],
[
"\n### Comparing Niche classes\n\nNiche models can be compared using a [NicheDelta](lowlevel.rst#niche_vlaanderen.NicheDelta) class. This can be used to compare different scenario's. \n\nIn our example, we will compare the results of the running Niche two times, once using a simple model and once using a full model. ",
"_____no_output_____"
]
],
[
[
"import niche_vlaanderen as nv\nimport matplotlib.pyplot as plt\n\nsimple = nv.Niche()\nsimple.run_config_file(\"simple.yml\")\n\nfull = nv.Niche()\nfull.run_config_file(\"full.yml\")\n\ndelta = nv.NicheDelta(simple, full)\nax = delta.plot(7)\nplt.show()",
"_____no_output_____"
]
],
[
[
"It is also possible to show the areas in a dataframe by using the [table](lowlevel.rst#niche_vlaanderen.NicheDelta.table) attribute.",
"_____no_output_____"
]
],
[
[
"delta.table.head()",
"_____no_output_____"
]
],
[
[
"Like Niche, NicheDelta also has a write method, which takes a directory as an argument.",
"_____no_output_____"
]
],
[
[
"delta.write(\"comparison_output\", overwrite_files=True)",
"_____no_output_____"
]
],
[
[
"### Creating deviation maps\n\nIn many cases, it is not only important to find out which vegetation types are possible given the different input files, but also to find out how much change would be required to `mhw` or `mlw` to allow a certain vegetation type.\n\nTo create deviation maps, it is necessary to [run](lowlevel.rst#niche_vlaanderen.Niche.run) a model with the `deviation` option.",
"_____no_output_____"
]
],
[
[
"dev = nv.Niche()\ndev.set_input(\"mhw\",\"../testcase/zwarte_beek/input/mhw.asc\")\ndev.set_input(\"mlw\",\"../testcase/zwarte_beek/input/mhw.asc\")\ndev.set_input(\"soil_code\",\"../testcase/zwarte_beek/input/soil_code.asc\")\ndev.run(deviation=True, full_model=False)",
"_____no_output_____"
]
],
[
[
"The deviation maps can be plotted by specifying either mhw or mlw with the vegetation type, eg mhw_14 (to show the deviation between mhw and the required mhw for vegetation type 14).\nPositive values indicate that the actual condition is too dry for the vegetation type. Negative values indicate that the actual condition is too wet for the vegetation type. ",
"_____no_output_____"
]
],
[
[
"dev.plot(\"mlw\")\ndev.plot(\"mlw_14\")\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Creating statistics per shape object\n\nNiche also contains a helper function that allows one to calculate the possible vegetation by using a vector dataset, such as a .geojson or .shp file.\n\nThe vegetation is returned as a pandas dataframe, where shapes are identified by their id and the area not covered by a shape gets `shape_id` -1.",
"_____no_output_____"
]
],
[
[
"df = full.zonal_stats(\"../testcase/zwarte_beek/input/study_area_l72.geojson\")\ndf",
"_____no_output_____"
]
],
[
[
"### Using abiotic grids\n\nIn certain cases the intermediary grids of Acidity or NutrientLevel need changes, to compensate for specific circumstances.\n\nIn that case it is possible to run a Niche model and make some adjustments to the grid and then using an abiotic grid as an input.\n\n",
"_____no_output_____"
]
],
[
[
"import niche_vlaanderen as nv\nimport matplotlib.pyplot as plt\n\nfull = nv.Niche()\nfull.run_config_file(\"full.yml\")\nfull.write(\"output_abiotic\", overwrite_files=True)",
"_____no_output_____"
]
],
[
[
"Now it is possible to adapt the `acidity` and `nutrient_level` grids outside niche. For this demo, we will use some Python magic to make all nutrient levels one level lower. Note that there is no need to do this in Python, any other tool could be used as well. So if you don't understand this code - don't panic (and ignore the warning)!",
"_____no_output_____"
]
],
[
[
"import rasterio\nwith rasterio.open(\"output_abiotic/full_nutrient_level.tif\") as src:\n nutrient_level = src.read(1)\n profile = src.profile\n nodata = src.nodatavals[0]\n \nnutrient_level[nutrient_level != nodata] = nutrient_level[nutrient_level != nodata] -1\n\n# we can not have nutrient level 0, so we set all places where this occurs to 1\nnutrient_level[nutrient_level ==0 ] = 1\n\nwith rasterio.open(\"output_abiotic/adjusted_nutrient.tif\", 'w', **profile) as dst:\n dst.write(nutrient_level, 1)",
"_____no_output_____"
]
],
[
[
"Next we will create a new niche model using the same options as our previous full models, but we will also add the previously calculated acidity and nutrient level values as input, and run with the `abiotic=True` option. Note that we use the `read_config_file` method (and not `run_config_file`) because we still want to edit the configuration before running.",
"_____no_output_____"
]
],
[
[
"adjusted = nv.Niche()\nadjusted.read_config_file(\"full.yml\")\nadjusted.set_input(\"acidity\", \"output_abiotic/full_acidity.tif\")\nadjusted.set_input(\"nutrient_level\", \"output_abiotic/adjusted_nutrient.tif\")\nadjusted.name = \"adjusted\"\nadjusted.run(abiotic=True)\n\nadjusted.plot(7)\nfull.plot(7)\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Overwriting standard code tables\n\nOne is free to adapt the [standard code tables](https://inbo.github.io/niche_vlaanderen/codetables.html) that are used by NICHE. By specifying the paths to the adapted code tables in a NICHE class object, the standard code tables can be overwritten. In this way, standard model functioning can be tweaked. However, it is strongly advised to use ecological data that is reviewed by experts and to have in-depth knowledge of the [model functioning](https://inbo.github.io/niche_vlaanderen/model.html).\n\nThe possible code tables that can be adapted and set within a [NICHE object](https://inbo.github.io/niche_vlaanderen/lowlevel.html) are:\n\nct_acidity, ct_soil_mlw_class, ct_soil_codes, lnk_acidity, ct_seepage, ct_vegetation, ct_management, ct_nutrient_level and ct_mineralisation",
"_____no_output_____"
],
[
"After adapting the vegetation code table for type 7 (Caricion gracilis) on peaty soil (V) by randomly altering the maximum ``mhw`` and ``mlw`` to 5 and 4 cm resp. (i.e. below ground, instead of standard values of -28 and -29 cm) and saving the file to ct_vegetation_adj7.csv, the adjusted model can be built and run.",
"_____no_output_____"
]
],
[
[
"adjusted_ct = nv.Niche(ct_vegetation = \"ct_vegetation_adj7.csv\")\nadjusted_ct.read_config_file(\"full.yml\")\nadjusted_ct.run()",
"_____no_output_____"
]
],
[
[
"Example of changed potential area of Caricion gracilis vegetation type because of the changes set in the vegetation code table:",
"_____no_output_____"
]
],
[
[
"adjusted_ct.plot(7)\nfull.plot(7)\nplt.show()",
"_____no_output_____"
]
],
[
[
"Potential area is shrinking because of the range of grondwater levels that have become more narrow (excluding the wettest places).",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
4a56d459e23ceaee9bd65984c2b263e7fa48887f
| 30,592 |
ipynb
|
Jupyter Notebook
|
Benchmarks.ipynb
|
csdai0324/benchmarks
|
dea8ef6373a0e5a7990f024be85696fb468cf19b
|
[
"MIT"
] | null | null | null |
Benchmarks.ipynb
|
csdai0324/benchmarks
|
dea8ef6373a0e5a7990f024be85696fb468cf19b
|
[
"MIT"
] | null | null | null |
Benchmarks.ipynb
|
csdai0324/benchmarks
|
dea8ef6373a0e5a7990f024be85696fb468cf19b
|
[
"MIT"
] | null | null | null | 44.594752 | 1,775 | 0.50085 |
[
[
[
"### Requirements\n\n#### Jupyter Nbextensions \n- Python Markdown\n- Load Tex Macros\n\n#### Python & Libs\n\n- Python version $\\geq$ 3.4\n- Numpy version $\\geq$ 1.17\n- Pandas version $\\geq$ 1.0.3",
"_____no_output_____"
]
],
[
[
"import string\nimport operator \nimport functools \nimport numpy as np\nimport pandas as pd\n\nfrom collections import Counter\nfrom IPython.display import display, Math, Latex, Markdown",
"_____no_output_____"
],
[
"ref_1 = 'The cat sat on the mat.'\ncand_1 = 'The cat is on the mat.'\nref_2 = 'There is a cat on the mat.'\ncand_2 = 'The the the the the the the the.'\n\npreprocess = lambda x: x.lower().translate(str.maketrans('', '', string.punctuation))\n\ndef extract(sentence):\n sentence = preprocess(sentence)\n uni_gram = sentence.split()\n bi_gram = [' '.join(words) for words in zip(uni_gram[::], uni_gram[1::])]\n tri_gram = [' '.join(words) for words in zip(uni_gram[::], uni_gram[1::], uni_gram[2::])]\n quad_gram = [' '.join(words) for words in zip(uni_gram[::], uni_gram[1::], uni_gram[2::], uni_gram[3::])]\n return uni_gram, bi_gram, tri_gram, quad_gram",
"_____no_output_____"
]
],
[
[
"## N-gram Evaluation",
"_____no_output_____"
],
[
"### Example \n\nReference\n\n`{{ref_1}}` \n\n$\\xrightarrow[\\text{}]{\\text{ Preprocessing }}$ `{{preprocess(ref_1)}}` \n \n$\\xrightarrow[\\text{}]{\\text{Extract 1-gram}} $ `{{extract(ref_1)[0]}}`\n \n$\\xrightarrow[\\text{}]{\\text{Extract 2-gram}} $ `{{extract(ref_1)[1]}}` \n\n$\\xrightarrow[\\text{}]{\\text{Extract 3-gram}} $ `{{extract(ref_1)[2]}}` \n\nCandidate\n\n`{{cand_1}}`\n\n$\\xrightarrow[\\text{}]{\\text{ Preprocessing }}$ `{{preprocess(cand_1)}}` \n \n$\\xrightarrow[\\text{}]{\\text{Extract 1-gram}} $ `{{extract(cand_1)[0]}}` \n \n$\\xrightarrow[\\text{}]{\\text{Extract 2-gram}} $ `{{extract(cand_1)[1]}}` \n\n$\\xrightarrow[\\text{}]{\\text{Extract 3-gram}} $ `{{extract(cand_1)[2]}}` ",
"_____no_output_____"
],
[
"## Considering Recall %\n\n### Modified Precision - Clipping",
"_____no_output_____"
],
[
"### Example \n\nCandidate\n\n`{{cand_2}}` \n\n$\\xrightarrow[\\text{}]{\\text{ Preprocessing }}$ `{{preprocess(cand_2)}}` \n \n$\\xrightarrow[\\text{}]{\\text{Extract 1-gram}} $ `{{extract(cand_2)[0]}}`\n \n$\\xrightarrow[\\text{}]{\\text{Extract 2-gram}} $ `{{extract(cand_2)[1]}}` \n\n$\\xrightarrow[\\text{}]{\\text{Extract 3-gram}} $ `{{extract(cand_2)[2][:2] + ['...']}}` ",
"_____no_output_____"
],
[
"## [BLEU - Bilingual Evaluation Understudy](https://www.aclweb.org/anthology/P02-1040.pdf)\n\n### BLEU_n Formula\n\n$\n\\begin{align}\n \\quad\n BLEU = BP \\cdot exp(\\sum_{n=1}^{N} w_n\\log_{}{P_n}) \\cr\n\\end{align}\n$\n \n$\n\\begin{align}\n \\quad\n BP \\quad\\,\\ = \\begin{cases} \n 1 &, \\ c > r \\cr\n exp(1-\\frac{r}{c}) &, \\ c \\leq r \\cr\n \\end{cases} \n\\end{align}\n$",
"_____no_output_____"
]
],
[
[
"def BLEU_n(candidate, reference):\n candidate = extract(candidate)\n reference = extract(reference)\n BLEU = 0\n W_n = 1. / len(candidate)\n for cand, ref in zip(candidate, reference):\n BLEU += W_n * np.log(P_n(cand, ref))\n BLEU = np.exp(BLEU) * BP(candidate[0], reference[0])\n return BLEU\n\ndef P_n(cand, ref):\n count = 0\n for c in cand:\n if c in ref:\n count += 1\n ref.remove(c)\n return 1 if count == 0 else count / len(cand)\n \ndef BP(candidate, reference):\n c, r = len(candidate), len(reference)\n return 1 if c > r else np.exp(1 - r / c)\n\n#BLEU_n(cand_1, ref_1)",
"_____no_output_____"
]
],
[
[
"## [ROUGE - Recall-Oriented Understudy for Gisting Evaluation](https://www.aclweb.org/anthology/W04-1013.pdf)\n\n### Rouge-N Formula\n\n$\n\\begin{align}\n \\quad\n \\textit{Rouge-N}\\; = \\frac{\\sum\\limits_{S \\in \\{\\textit{ReferenceSummaries}\\}} \n \\sum\\limits_{gram_n \\in S} Count_{macth}(gram_n)}\n {\\sum\\limits_{S \\in \\{\\textit{ReferenceSummaries}\\}} \n \\sum\\limits_{gram_n \\in S} Count(gram_n)}\n\\end{align}\n$\n ",
"_____no_output_____"
]
],
[
[
"ref_3 = 'The cat was under the bed.'\ncand_3 = 'The cat was found under the bed.'\n\ndef Rouge_n(candidate, reference, n=1):\n cand, ref = extract(candidate)[n-1], extract(reference)[n-1]\n cand = list(map(lambda x: 1 if x in ref else 0, cand))\n return functools.reduce(operator.add, cand) / len(ref)\n\n#Rouge_n(cand_3, ref_3)",
"_____no_output_____"
]
],
[
[
"### LCS(Longest Common Subsequence)\n\n#### Example \n\nReference\n\n`{{ref_1}}` \n\n$\\xrightarrow[\\text{}]{\\text{ Preprocessing }}$ `{{preprocess(ref_1)}}` \n\nCandidate\n\n`{{cand_1}}`\n\n$\\xrightarrow[\\text{}]{\\text{ Preprocessing }}$ `{{preprocess(cand_1)}}` \n\nLCS\n\n`the cat on the mat`\n\n### Rouge-L Formula\n\n$\n\\begin{align}\n \\cr\n \\quad\n R_{lcs} = \\frac{LCS(\\textit{Reference}, \\textit{Candidate})}{m}, \n \\; m \\;\\text{for }\\textit{Reference} \\text{ length} \\cr \n P_{lcs} = \\frac{LCS(\\textit{Reference}, \\textit{Candidate})}{n}, \n \\; n \\;\\text{for }\\textit{Candidate} \\text{ length} \\cr \n\\end{align}\n$\n\n$\n\\begin{align}\n \\quad \\;\n F_{lcs} = \\frac{(1+\\beta^2)R_{lcs}P_{lcs}}{R_{lcs} + \\beta^2P_{lcs}} \n\\end{align}\n$",
"_____no_output_____"
]
],
[
[
"def Rouge_l(candidate, reference, beta=1.2):\n cand, ref = extract(candidate)[0], extract(reference)[0]\n lcs = LCS(cand, ref)\n r_lcs, p_lcs = lcs / len(ref), lcs / len(cand)\n return ((1 + beta**2)*r_lcs*p_lcs) / (r_lcs + beta**2*p_lcs)\n \ndef LCS(cand, ref):\n l_c, l_r = len(cand), len(ref)\n dp = np.zeros(shape=(l_c + 1, l_r + 1))\n for i in range(l_c):\n for j in range(l_r):\n if cand[i] == ref[j]: \n dp[i + 1][j + 1] = dp[i][j] + 1\n elif dp[i + 1][j] > dp[i][j + 1]:\n dp[i + 1][j + 1] = dp[i + 1][j]\n else: \n dp[i + 1][j + 1] = dp[i][j + 1]\n return int(dp[-1][-1])\n\n#Rouge_l(cand_1, ref_1)",
"_____no_output_____"
]
],
[
[
"### Rouge-W\n \n - WLCS\n - Weighted LCS-based statistics that favors consecutive LCSes.\n\n### Rouge-S\n\n - Skip-gram\n - Skip-bigram based co-occurrence statistics. \n - Skip-bigram is any pair of words in their sentence order.\n \n### Rouge-SU\n\n - Skip-bigram plus unigram-based co-occurrence statistics.",
"_____no_output_____"
],
[
"## [CIDEr - Consensus-based Image Description Evaluation](https://www.cv-foundation.org/openaccess/content_cvpr_2015/papers/Vedantam_CIDEr_Consensus-Based_Image_2015_CVPR_paper.pdf)\n\n### TF-IDF\n\n - Term Frequency\n \n $ \n \\begin{align}\n \\quad \n \\textit{TF}(𝑥)\n \\;\\text{for count of term } x \\;\\text{ in the document}\n \\cr\n \\end{align}\n $\n \n \n - Inverse Document Frequency\n \n $ \n \\begin{align}\n \\quad \n \\textit{IDF}(𝑥) = \\log \\frac{N + 1}{N(x) + 1} +1\n ,\\; N\\;\\text{for total document count and }N(x) \\text{ for document which includes term } x\n \\cr\n \\end{align}\n $\n \n \n - TF-IDF\n \n $\n \\begin{align}\n \\quad\n \\textit{TF-IDF}(x) = \\textit{TF}(x)\\;\\times\\;\\textit{IDF}(x) \\cr \n \\end{align}\n $",
"_____no_output_____"
],
[
"### Cosine Similiraty\n\n - The cosine of two non-zero vectors can be derived by using the Euclidean dot product formula\n \n $\n \\begin{align}\n \\quad\n A \\cdot B = \\lVert A \\rVert \\lVert B \\rVert \\cos{\\theta}\n \\cr\n \\end{align}\n $\n \n \n - Similiraty \n \n $\n \\begin{align}\n \\quad\n \\textit{Similiraty } = \n \\cos{(\\theta)} = \n \\frac{A \\cdot B}\n {\\lVert A \\rVert \\lVert B \\rVert} = \n \\frac{\\sum\\limits_{i=1}^{N}A_i B_i}\n {\\sqrt{\\sum\\limits_{i=1}^{N}A_i^2} \\sqrt{\\sum\\limits_{i=1}^{N}B_i^2}}, \n \\text{ where } A_i \\text{ and } B_i \\text{are components of vector } A \\text{ and } B \\text{ respectively.}\n \\end{align}\n $",
"_____no_output_____"
]
],
[
[
"@np.vectorize\ndef nonzero(x):\n return 1 if x > 0 else 0\n\[email protected]\ndef TFIDF(x, IDF):\n return x * IDF\n\ndef gen_matrix(documents):\n d = Counter()\n for doc in documents:\n d += Counter(doc)\n d = dict(d.most_common()).fromkeys(d, 0)\n matrix = []\n for doc in documents:\n dest = d.copy() \n dest.update(dict(Counter(doc)))\n matrix.append(list(dest.values()))\n return d, matrix\n\ndef gen_table(d, matrix, fn):\n columns = {}\n index = ['Candidate'] + ['Reference_{}'.format(i+1) for i in range(len(matrix) - 1)]\n if fn.__name__ == 'CIDEr':\n for idx, key in enumerate(list(d.keys()) +['CIDEr']):\n columns.update({idx:key})\n N = [len(matrix)]*len(matrix[0])\n N_x = np.matrix.sum(nonzero(np.matrix(matrix)), axis=0).tolist()[0]\n N = np.array(N) + np.array(1)\n N_x = np.array(N_x) + np.array(1)\n IDF = (np.log(N / N_x) / np.log(10) + (np.array(1)))\n matrix = TFIDF(matrix, IDF).tolist()\n else:\n for idx, key in enumerate(list(d.keys()) +['Cos_Sim']):\n columns.update({idx:key})\n temp = []\n for i in range(len(matrix)):\n temp.append(matrix[i] + [fn(matrix[0], matrix[i])])\n df = pd.DataFrame(temp).rename(columns=columns)\n df.reset_index(drop=True, inplace=True)\n df.index = index\n return Markdown(df.to_markdown())\n\ndef cosine_similiraty(cand, ref):\n fn = lambda x: (np.sqrt(np.sum(np.power(x, 2))))\n return np.dot(cand, ref) / (fn(cand) * fn(ref))",
"_____no_output_____"
],
[
"# Cosine similiraty\n\ntable = []\nfor i in range(0, 4):\n table.append(gen_table(*gen_matrix([extract(cand_1)[i], \n extract(ref_1)[i], extract(ref_2)[i]]), cosine_similiraty))",
"_____no_output_____"
]
],
[
[
"### Example \n\nCandidate\n\n`{{cand_1}}`\n\n$\\xrightarrow[\\text{}]{\\text{ Preprocessing }}$ `{{preprocess(cand_1)}}` \n$\\xrightarrow[\\text{}]{\\text{Extract 2-gram}} $ `{{extract(cand_1)[1]}}` \n\nReference_1\n\n`{{ref_1}}` \n\n$\\xrightarrow[\\text{}]{\\text{ Preprocessing }}$ `{{preprocess(ref_1)}}` \n$\\xrightarrow[\\text{}]{\\text{Extract 2-gram}}$ `{{extract(ref_1)[1]}}` \n\nReference_2\n\n`{{ref_2}}` \n\n$\\xrightarrow[\\text{}]{\\text{ Preprocessing }}$ `{{preprocess(ref_2)}}` \n$\\xrightarrow[\\text{}]{\\text{Extract 2-gram}}$ `{{extract(ref_2)[1]}}`",
"_____no_output_____"
],
[
"### Doc-Term Matrix with Cosine Similiraty\n\n`{{table[0]}}`\n\n`{{table[1]}}`\n\n`{{table[2]}}`",
"_____no_output_____"
],
[
"### CIDEr_n Formula\n\n\n$\n\\begin{align}\n\\cr \\quad\n\\textit{CIDEr_n}(\\textit{candidate}, \n \\textit{references}) = \n \\frac{1}{M}\\sum\\limits_{i=1}^{M}\n \\frac{ g^n(\\textit{candidate}) \\cdot g^n(\\textit{references}) }\n {\\lVert g^n(\\textit{candidate}) \\rVert \\times \\lVert g^n(\\textit{references}) \\rVert},\n\\text{ where } g^n(x) \\text{ is } \\textit{TF-IDF} \\text{ weight of n-gram in sentence } x \\text{.}\n\\end{align}\n$\n\n$ \n$",
"_____no_output_____"
]
],
[
[
"def CIDEr(cand, ref):\n fn = lambda x: (np.sqrt(np.sum(np.power(x, 2))))\n return np.dot(cand, ref) / (fn(cand) * fn(ref))\n\ntable = []\nfor i in range(0, 3):\n table.append(gen_table(*gen_matrix([extract(cand_1)[i], \n extract(ref_1)[i], extract(ref_2)[i]]), CIDEr))",
"_____no_output_____"
]
],
[
[
"### Doc-Term TF-IDF Matrix with CIDEr-n\n\n`{{table[0]}}`\n\n`{{table[1]}}`\n\n`{{table[2]}}`",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
4a56d75993aa6736d8cb0eb4c2be2e36e9eed7c9
| 124,672 |
ipynb
|
Jupyter Notebook
|
forPaper2/paperFigures/fig_transport.ipynb
|
UBC-MOAD/outputanalysisnotebooks
|
50839cde3832d26bac6641427fed03c818fbe170
|
[
"Apache-2.0"
] | null | null | null |
forPaper2/paperFigures/fig_transport.ipynb
|
UBC-MOAD/outputanalysisnotebooks
|
50839cde3832d26bac6641427fed03c818fbe170
|
[
"Apache-2.0"
] | null | null | null |
forPaper2/paperFigures/fig_transport.ipynb
|
UBC-MOAD/outputanalysisnotebooks
|
50839cde3832d26bac6641427fed03c818fbe170
|
[
"Apache-2.0"
] | null | null | null | 271.026087 | 105,142 | 0.89016 |
[
[
[
"## Figure tracer transport ",
"_____no_output_____"
]
],
[
[
"#import gsw as sw # Gibbs seawater package\nimport cmocean as cmo\nimport matplotlib.pyplot as plt\nimport matplotlib.colors as mcolors\nimport matplotlib.gridspec as gspec\n%matplotlib inline\nfrom netCDF4 import Dataset\nimport numpy as np\nimport pandas as pd\nimport seaborn as sns\nimport sys\nimport xarray as xr\nimport canyon_tools.readout_tools as rout \nimport canyon_tools.metrics_tools as mpt",
"_____no_output_____"
],
[
"from IPython.display import HTML\n\nHTML('''<script>\ncode_show=true; \nfunction code_toggle() {\n if (code_show){\n $('div.input').hide();\n } else {\n $('div.input').show();\n }\n code_show = !code_show\n} \n$( document ).ready(code_toggle);\n</script>\n<form action=\"javascript:code_toggle()\"><input type=\"submit\" value=\"Click here to toggle on/off the raw code.\"></form>''')\n",
"_____no_output_____"
],
[
"sns.set_context('paper')\nsns.set_style('white')\nplt.rcParams.update({'font.size': 11})",
"_____no_output_____"
],
[
"def plot_transports_CS(g0,g1,g2,g3,g4,g5, dfcan, dfdif, color, lab):\n \n ax0 = plt.subplot(g0)\n ax1 = plt.subplot(g1)\n ax2 = plt.subplot(g2)\n ax3 = plt.subplot(g3)\n ax4 = plt.subplot(g4)\n ax5 = plt.subplot(g5)\n \n axs = [ax0,ax1,ax2,ax3,ax4,ax5]\n \n for ax in axs:\n ax.axhline(0, color='gold')\n ax.tick_params(axis='y', pad=0)\n ax.tick_params(axis='x', pad=0.05)\n ax.grid(which='both',color='0.9', linestyle='-')\n ax.set_ylim(-5, 5)\n ax.set_xlabel('Days', labelpad=0)\n ax.set_xticks([0,3,6,9])\n for ax in axs[1:]:\n #ax.set_yticks([-200,-100,0,100,200])\n ax.set_yticklabels(['','','','','',''])\n \n # Tracers\n vertical = dfcan.Vert_adv_trans_sb # only advective parts, ignoring diffusve for now \n ax0.plot(np.arange(1,19,1)/2.0,(vertical)/1E6,color=color, label=lab)\n ax1.plot(np.arange(1,19,1)/2.0,(dfcan.CS1_adv_trans+dfcan.CS2_adv_trans)/1E6,color=color, label=lab)\n ax2.plot(np.arange(1,19,1)/2.0,(dfcan.CS3_adv_trans )/1E6,color=color, label=lab)\n ax3.plot(np.arange(1,19,1)/2.0,(dfcan.CS4_adv_trans+dfcan.CS5_adv_trans)/1E6,color=color, label=lab)\n ax4.plot(np.arange(1,19,1)/2.0,(dfcan.CS6_adv_trans )/1E6,color=color, label=lab)\n\n total = ( (dfcan.CS1_adv_trans ) +\n (dfcan.CS2_adv_trans ) +\n (dfcan.CS3_adv_trans ) +\n (dfcan.CS4_adv_trans ) +\n (dfcan.CS5_adv_trans ) +\n (dfcan.CS6_adv_trans ) +\n vertical)\n\n ax5.plot(np.arange(1,19,1)/2.0,total/1E6,color=color, label=lab)\n return(ax0,ax1,ax2,ax3,ax4,ax5) \ndef plot_can_effect(gs_c, dfcan, dfdif, dfcanNoC, dfdifNoC, color, lab, id_sup):\n ax = plt.subplot(gs_c, xticks=[])\n ax.axhline(0, color='gold')\n canyon = tot_trans(dfcan, dfdif)\n no_canyon = tot_trans(dfcanNoC, dfdifNoC)\n ax.plot(np.arange(1,19,1)/2.0,(canyon-no_canyon)/1E5,color=color, label=lab)\n ax.tick_params(axis='y', pad=0.5)\n ax.grid(which='both',color='0.9', linestyle='-')\n ax.yaxis.tick_right()\n if lab =='ARGO':\n ax.text(0.8,0.8,id_sup,transform=ax.transAxes)\n\n return(ax)\ndef tot_trans(dfcan, dfdif):\n vertical = (dfdif.Vert_dif_trans_sb + dfcan.Vert_adv_trans_sb) \n total = ( (dfcan.CS1_adv_trans ) +\n (dfcan.CS2_adv_trans ) +\n (dfcan.CS3_adv_trans ) +\n (dfcan.CS4_adv_trans ) +\n (dfcan.CS5_adv_trans ) +\n (dfcan.CS6_adv_trans ) +\n vertical)\n return(total)\n\ndef plotCSPos(ax,CS1,CS2,CS3,CS4,CS5,CS6):\n ax.axvline(CS1,color='k',linestyle=':')\n ax.axvline(CS2,color='k',linestyle=':')\n ax.axvline(CS3,color='k',linestyle=':')\n ax.axvline(CS4,color='k',linestyle=':')\n ax.axvline(CS5,color='k',linestyle=':')\n ax.axvline(CS6,color='k',linestyle=':')\n\ndef plot_CS_slice(fig, gs_a, gs_b, t_slice, x_slice, x_slice_vert, y_slice_vert, z_slice, z_slice_zoom, y_ind, z_ind,\n grid,Flux,FluxV,unit):\n \n ax_a = plt.subplot(gs_a)#,xticks=[])\n ax_b = plt.subplot(gs_b)#,xticks=[])\n \n areas = (np.expand_dims(grid.dxF.isel(X=x_slice,Y=y_ind).data,0))*(np.expand_dims(grid.drF.isel(Z=z_slice).data,1))\n\n # Zoom shelf ---------------------------------------------------------------------------\n cnt = ax_a.contourf(grid.X.isel(X=x_slice)/1000,\n grid.Z.isel(Z=z_slice_zoom),\n Flux.isel(Zmd000104=z_slice_zoom, X=x_slice)/areas[z_slice_zoom,:],\n 16,cmap=cmo.cm.tarn,\n vmax=np.max(Flux.isel(Zmd000104=z_slice_zoom,X=x_slice)/areas[z_slice_zoom,:]), \n vmin=-np.max(Flux.isel(Zmd000104=z_slice_zoom,X=x_slice)/areas[z_slice_zoom,:]))\n ax_a.contourf(grid.X.isel(X=x_slice)/1000,\n grid.Z.isel(Z=z_slice_zoom),\n grid.HFacC.isel(Z=z_slice_zoom,Y=y_ind,X=x_slice),\n [0,0.1], colors='#a99582')\n\n cb_a = fig.colorbar(cnt, ax=ax_a)\n cb_a.ax.yaxis.set_tick_params(pad=1.5)\n\n ax_a.set_ylabel('Depth / m',labelpad=0.0)\n ax_a.text(0.001,0.05,'%s' %unit,transform=ax_a.transAxes, fontsize=8, color='k',fontweight='bold') \n\n # Vertical section ---------------------------------------------------------------------------\n cnt=ax_b.contourf(grid.X.isel(X=x_slice_vert)/1000,\n grid.Y.isel(Y=y_slice_vert)/1000,\n 100*(FluxV.isel(X=x_slice_vert,Y=y_slice_vert).data)/(grid.rA[y_slice_vert,x_slice_vert]),\n 16,cmap=cmo.cm.tarn,\n vmax= np.max(100*(FluxV.isel(X=x_slice_vert,Y=y_slice_vert).data)/(grid.rA[y_slice_vert,x_slice_vert])),\n vmin=-np.max(100*(FluxV.isel(X=x_slice_vert,Y=y_slice_vert).data)/(grid.rA[y_slice_vert,x_slice_vert])))\n ax_b.contourf(grid.X.isel(X=x_slice_vert)/1000, \n grid.Y.isel(Y=y_slice_vert)/1000,\n grid.HFacC.isel(Z=z_ind,X=x_slice_vert,Y=y_slice_vert),\n [0,0.1], colors='#a99582')\n \n cb_b=fig.colorbar(cnt, ax=ax_b)#,ticks=[-2,-1,0,1,2,3,4])\n cb_b.ax.yaxis.set_tick_params(pad=1.5)\n ax_b.set_ylabel('C-S distance / km', labelpad=0)\n ax_b.set_aspect(1)\n \n return(ax_a,ax_b)",
"_____no_output_____"
],
[
"#Exp\nGrid = '/data/kramosmu/results/TracerExperiments/UPW_10TR_BF2_AST/01_Ast03/gridGlob.nc' \nGridOut = Dataset(Grid)\n\nGridNoC = '/data/kramosmu/results/TracerExperiments/UPW_10TR_BF2_AST/02_Ast03_No_Cny/gridGlob.nc' \nGridNoCOut = Dataset(GridNoC)\n\nState = '/data/kramosmu/results/TracerExperiments/UPW_10TR_BF2_AST/01_Ast03/stateGlob.nc' \nStateNoC = '/data/kramosmu/results/TracerExperiments/UPW_10TR_BF2_AST/02_Ast03_No_Cny/stateGlob.nc' ",
"_____no_output_____"
],
[
"units = ['$10^5$ $\\mu$mol kg$^{-1}$ m$^3$s$^{-1}$',\n '$10^5$ nM m$^3$s$^{-1}$',\n '$10^5$ $\\mu$mol kg$^{-1}$ m$^3$s$^{-1}$']\ntracers = ['TR03','TR08','TR09']\ntr_labels = ['Oxygen','Methane','DIC']\n\nexps = ['UPW_10TR_BF2_AST/01_Ast03',\n 'UPW_10TR_BF2_AST/03_Ast03_Argo',\n 'UPW_10TR_BF4_BAR/01_Bar03',\n 'UPW_10TR_BF4_BAR/03_Bar03_Path']\n\nexpsNoC = ['UPW_10TR_BF2_AST/02_Ast03_No_Cny',\n 'UPW_10TR_BF2_AST/04_Ast03_No_Cny_Argo',\n 'UPW_10TR_BF4_BAR/02_Bar03_No_Cny',\n 'UPW_10TR_BF4_BAR/04_Bar03_No_Cny_Path']\n\ncolors = ['steelblue', 'skyblue', 'orangered', 'lightsalmon']\nlabels = ['Astoria','ARGO', 'Barkley', 'Pathways']\nsubplots_id = ['a3', 'b3', 'c3',]",
"_____no_output_____"
],
[
"t_slice = slice(10,20)\nx_slice = slice(0,400)\nx_slice_vert = slice(120,240)\ny_slice_vert = slice(130,230)\nz_slice = slice(0,80)\nz_slice_zoom = slice(0,31)\ny_ind = 130 # sb index\nz_ind = 30 # sb index",
"_____no_output_____"
],
[
"fig = plt.figure(figsize = (7.48,7.05))\n\ngg = gspec.GridSpec(2, 1, hspace=0.12, height_ratios=[3,1])\ngs = gspec.GridSpecFromSubplotSpec(3, 1, subplot_spec=gg[0])\ngs1 = gspec.GridSpecFromSubplotSpec(1, 3, subplot_spec=gs[0],hspace=0.15,wspace=0.1,width_ratios=[1,0.43,0.38])\ngs3 = gspec.GridSpecFromSubplotSpec(1, 3, subplot_spec=gs[1],hspace=0.15,wspace=0.1,width_ratios=[1,0.43,0.38])\ngs4 = gspec.GridSpecFromSubplotSpec(1, 3, subplot_spec=gs[2],hspace=0.15,wspace=0.1,width_ratios=[1,0.43,0.38])\ngs5 = gspec.GridSpecFromSubplotSpec(1, 6, subplot_spec=gg[1])\n\nggs = [gs1,gs3,gs4]\ngrid = xr.open_dataset(Grid)\n\n# This is horrible ------------------------------------------------------------------------------------\n# - Oxygen\nflux_file = ('/data/kramosmu/results/TracerExperiments/UPW_10TR_BF2_AST/01_Ast03/Flux%sGlob.nc' %tracers[0])\nflux = xr.open_dataset(flux_file)\n\nadv_flux_AP = (flux.ADVyTr03[t_slice,:,y_ind,:]).mean(dim='T')\ndif_flux_AP = (flux.DFyETr03[t_slice,:,y_ind,:]).mean(dim='T')\nFlux = adv_flux_AP + dif_flux_AP\n\nadv_fluxV_AP = (flux.ADVrTr03[t_slice,z_ind,:,:]).mean(dim='T')\ndif_fluxV_AP = (flux.DFrITr03[t_slice,z_ind,:,:]+flux.DFrETr03[t_slice,z_ind,:,:]).mean(dim='T')\nFluxV = adv_fluxV_AP + dif_fluxV_AP \n\nax3,ax4 = plot_CS_slice(fig, gs1[0], gs1[1],t_slice, x_slice, x_slice_vert, y_slice_vert, \n z_slice, z_slice_zoom, y_ind, z_ind, grid,Flux,FluxV, units[0])\n\nax3.text(0.05,0.85,tr_labels[0],fontweight='bold',transform=ax3.transAxes)\nax3.text(0.85,0.8,'a1',transform=ax3.transAxes)\nax4.text(0.7,0.8,'a2',transform=ax4.transAxes)\nax3.tick_params(axis='y', pad=0.5)\nax4.tick_params(axis='y', pad=0.5)\n\n# - Methane\nflux_file = ('/data/kramosmu/results/TracerExperiments/UPW_10TR_BF2_AST/01_Ast03/Flux%sGlob.nc' %tracers[1])\nflux = xr.open_dataset(flux_file)\n\nadv_flux_AP = (flux.ADVyTr08[t_slice,:,y_ind,:]).mean(dim='T')\ndif_flux_AP = (flux.DFyETr08[t_slice,:,y_ind,:]).mean(dim='T')\nFlux = adv_flux_AP + dif_flux_AP\n\nadv_fluxV_AP = (flux.ADVrTr08[t_slice,z_ind,:,:]).mean(dim='T')\ndif_fluxV_AP = (flux.DFrITr08[t_slice,z_ind,:,:]+flux.DFrETr08[t_slice,z_ind,:,:]).mean(dim='T')\nFluxV = adv_fluxV_AP + dif_fluxV_AP \n\nax7,ax8 = plot_CS_slice(fig, gs3[0], gs3[1],t_slice, x_slice, x_slice_vert, y_slice_vert, \n z_slice, z_slice_zoom, y_ind, z_ind, grid,Flux,FluxV, units[1])\nax7.text(0.05,0.85,tr_labels[1],fontweight='bold',transform=ax7.transAxes)\nax7.text(0.85,0.8,'b1',transform=ax7.transAxes)\nax8.text(0.7,0.8,'b2',transform=ax8.transAxes)\nax7.tick_params(axis='y', pad=0.5)\nax8.tick_params(axis='y', pad=0.5)\n\n# - DIC\nflux_file = ('/data/kramosmu/results/TracerExperiments/UPW_10TR_BF2_AST/01_Ast03/Flux%sGlob.nc' %tracers[2])\nflux = xr.open_dataset(flux_file)\n\nadv_flux_AP = (flux.ADVyTr09[t_slice,:,y_ind,:]).mean(dim='T')\ndif_flux_AP = (flux.DFyETr09[t_slice,:,y_ind,:]).mean(dim='T')\nFlux = adv_flux_AP + dif_flux_AP\n\nadv_fluxV_AP = (flux.ADVrTr09[t_slice,z_ind,:,:]).mean(dim='T')\ndif_fluxV_AP = (flux.DFrITr09[t_slice,z_ind,:,:]+flux.DFrETr09[t_slice,z_ind,:,:]).mean(dim='T')\nFluxV = adv_fluxV_AP + dif_fluxV_AP \n\nax9,ax10 = plot_CS_slice(fig, gs4[0], gs4[1],t_slice, x_slice, x_slice_vert, y_slice_vert, \n z_slice, z_slice_zoom, y_ind, z_ind, grid,Flux,FluxV, units[2])\nax9.text(0.05,0.85,tr_labels[2],fontweight='bold',transform=ax9.transAxes)\nax9.text(0.85,0.8,'c1',transform=ax9.transAxes)\nax10.text(0.7,0.8,'c2',transform=ax10.transAxes)\nax9.tick_params(axis='y', pad=0.5)\nax10.tick_params(axis='y', pad=0.5)\n#ax9.set_xticks([20,40,60,80,100,120,140])\n#ax10.set_xticks([60,80,100])\nax9.set_xlabel('Alongshelf distance / km', labelpad=0)\nax10.set_xlabel('Alongshelf dist. / km', labelpad=0)\n#------------------------------------------------------------------------------------------------------------\n# - Canyon Effect\nfor tr, unit, tr_lab, gss, id_sup in zip(tracers, units, tr_labels, ggs,subplots_id):\n for exp,expNoC, color, lab in zip(exps,expsNoC, colors, labels):\n # net canyon effect\n file = ('/data/kramosmu/results/TracerExperiments/%s/adv%s_CS_transports.nc' %(exp,tr))\n filedif = ('/data/kramosmu/results/TracerExperiments/%s/dif%s_CS_transports.nc' %(exp,tr))\n fileNoC = ('/data/kramosmu/results/TracerExperiments/%s/adv%s_CS_transports.nc' %(expNoC,tr))\n filedifNoC = ('/data/kramosmu/results/TracerExperiments/%s/dif%s_CS_transports.nc' %(expNoC,tr))\n\n dfcan = xr.open_dataset(file)\n dfdif = xr.open_dataset(filedif)\n dfcanNoC = xr.open_dataset(fileNoC)\n dfdifNoC = xr.open_dataset(filedifNoC)\n axx = plot_can_effect(gss[2], dfcan, dfdif, dfcanNoC, dfdifNoC, color, lab, id_sup)\n axx.set_xticks([0,2,4,6,8])\n axx.set_xticklabels(['','','','',''])\n if tr_lab == 'DIC':\n axx.set_xticklabels([0,2,4,6,8])\n axx.set_xlabel('Days', labelpad=0)\n \n#-------------------------------------------------------------------------------------------------------------\n#- Linear profile cross-shelf transport through CS sections\ntr = tracers[1]\nunit = units[1]\ntr_lab = tr_labels[1]\nfor exp, color, lab in zip(exps, colors, labels):\n # net canyon effect\n file = ('/data/kramosmu/results/TracerExperiments/%s/adv%s_CS_transports.nc' %(exp,tr))\n filedif = ('/data/kramosmu/results/TracerExperiments/%s/dif%s_CS_transports.nc' %(exp,tr))\n\n dfcan = xr.open_dataset(file)\n dfdif = xr.open_dataset(filedif)\n axa,axb,axc,axd,axe,axf,= plot_transports_CS(gs5[0],gs5[1],gs5[2],gs5[3],gs5[4],gs5[5], dfcan, dfdif, color, lab)\n\n axf.legend(ncol=1,handletextpad=0 , labelspacing=0.1, handlelength=1.5)\n#---------------------------------------------------------------------------------------------------------\n# - aesthetics\n\naxa.set_ylabel('Transport \\n / $10^{6}$$\\mu$mol kg$^{-1}$ m$^3$s$^{-1}$', labelpad=0)\naxa.text(0.75,0.05,'d1',transform=axa.transAxes)\naxb.text(0.75,0.05,'d2',transform=axb.transAxes)\naxc.text(0.75,0.05,'d3',transform=axc.transAxes)\naxd.text(0.75,0.05,'d4',transform=axd.transAxes)\naxe.text(0.75,0.05,'d5',transform=axe.transAxes)\naxf.text(0.75,0.85,'d6',transform=axf.transAxes)\n\nplt.savefig('tracer_transport_rev01.eps',format='eps', bbox_inches='tight')",
"/home/kramosmu/anaconda3/lib/python3.5/site-packages/matplotlib/cbook/deprecation.py:107: MatplotlibDeprecationWarning: Adding an axes using the same arguments as a previous axes currently reuses the earlier instance. In a future version, a new instance will always be created and returned. Meanwhile, this warning can be suppressed, and the future behavior ensured, by passing a unique label to each axes instance.\n warnings.warn(message, mplDeprecation, stacklevel=1)\n"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a56e4d90c02c3c057d18146729f48b99bf83378
| 31,658 |
ipynb
|
Jupyter Notebook
|
code/transformer_201102/prepare_data.ipynb
|
steveyu323/motor_embedding
|
65b05e024ca5a0aa339330eff6b63927af5ce4aa
|
[
"MIT"
] | null | null | null |
code/transformer_201102/prepare_data.ipynb
|
steveyu323/motor_embedding
|
65b05e024ca5a0aa339330eff6b63927af5ce4aa
|
[
"MIT"
] | null | null | null |
code/transformer_201102/prepare_data.ipynb
|
steveyu323/motor_embedding
|
65b05e024ca5a0aa339330eff6b63927af5ce4aa
|
[
"MIT"
] | null | null | null | 31.500498 | 162 | 0.431234 |
[
[
[
"# Documentation\n- Generate the datasets used for evotuning the esm model\n- for each dataset, filter out those sequence longer than 1024\n- pfamA_balanced: 18000 entries for 4 clans related to motors\n- motor_toolkit: motor toolkit\n- kinesin_labelled: kinesin labelled dataset\n- pfamA_target_shuffled: pfamA_target\n- pfamA_target_sub: 396 of each protein family, for embedding visualization only",
"_____no_output_____"
]
],
[
[
"import torch\nimport torch.nn as nn \nimport torch.optim as optim \n\nimport torchvision \nimport torchvision.transforms as transforms \nimport torch.nn.functional as F\nfrom torch.autograd import Variable\nfrom torch.nn.utils.rnn import pad_sequence\nfrom torch.utils.data import Dataset, IterableDataset, DataLoader\n# import tqdm\nimport numpy as np\nimport pandas as pd\n\nimport math\n\nseed = 7\ntorch.manual_seed(seed)\nnp.random.seed(seed)",
"_____no_output_____"
],
[
"pfamA_motors = pd.read_csv(\"../../data/pfamA_motors_named.csv\")\npfamA_motors.head()",
"_____no_output_____"
],
[
"sum(np.array([len(a) for a in pfamA_motors[\"seq\"]])<1025)",
"_____no_output_____"
],
[
"sum(np.array([len(a) for a in pfamA_motors[\"seq\"]])>=1025)",
"_____no_output_____"
],
[
"7502/1907329",
"_____no_output_____"
],
[
"pfamA_motors = pfamA_motors.loc[np.array([len(a) for a in pfamA_motors[\"seq\"]])<1025,:]",
"_____no_output_____"
],
[
"motor_toolkit = pd.read_csv(\"../../data/motor_tookits.csv\")\nmotor_toolkit.head()",
"_____no_output_____"
],
[
"# truncate motor_toolkit to be <=1024 \nsum(motor_toolkit[\"Length\"]<=1024)",
"_____no_output_____"
],
[
"motor_toolkit.loc[motor_toolkit[\"Length\"]>1024,\"seq\"] = motor_toolkit.loc[motor_toolkit[\"Length\"]>1024,\"seq\"].apply(lambda s: s[0:1024])",
"_____no_output_____"
],
[
"motor_toolkit[\"Length\"] = motor_toolkit.loc[:,\"seq\"].apply(lambda s: len(s))",
"_____no_output_____"
],
[
"sum(motor_toolkit[\"Length\"]>1024)",
"_____no_output_____"
],
[
"kinesin_labelled = pd.read_csv(\"../../data/kinesin_labelled.csv\")",
"_____no_output_____"
],
[
"kinesin_labelled.head()",
"_____no_output_____"
],
[
"kinesin_labelled.loc[kinesin_labelled[\"Length\"]>1024,\"seq\"] = kinesin_labelled.loc[kinesin_labelled[\"Length\"]>1024,\"seq\"].apply(lambda s: s[0:1024])",
"_____no_output_____"
],
[
"kinesin_labelled[\"Length\"] = kinesin_labelled.loc[:,\"seq\"].apply(lambda s: len(s))",
"_____no_output_____"
],
[
"sum(kinesin_labelled[\"Length\"]>1024)",
"_____no_output_____"
],
[
"pfamA_motors_balanced = pfamA_motors.groupby('clan_x').apply(lambda _df: _df.sample(4500,random_state=1))\npfamA_motors_balanced = pfamA_motors_balanced.apply(lambda x: x.reset_index(drop = True))",
"_____no_output_____"
],
[
"pfamA_motors_balanced.shape",
"_____no_output_____"
],
[
"sum(np.array([len(a) for a in pfamA_motors_balanced[\"seq\"]])>=1025)",
"_____no_output_____"
],
[
"pfamA_target_name = [\"PF00349\",\"PF00022\",\"PF03727\",\"PF06723\",\\\n \"PF14450\",\"PF03953\",\"PF12327\",\"PF00091\",\"PF10644\",\\\n \"PF13809\",\"PF14881\",\"PF00063\",\"PF00225\",\"PF03028\"]\n\npfamA_target = pfamA_motors.loc[pfamA_motors[\"pfamA_acc\"].isin(pfamA_target_name),:].reset_index()",
"_____no_output_____"
],
[
"pfamA_target = pfamA_target.iloc[:,1:]",
"_____no_output_____"
],
[
"pfamA_target_sub = pfamA_target.sample(frac = 1).groupby(\"pfamA_acc\").head(396)",
"_____no_output_____"
],
[
"pfamA_target_sub.groupby(\"pfamA_acc\").count()",
"_____no_output_____"
],
[
"sum(np.array([len(a) for a in pfamA_target_sub[\"seq\"]])>=1025)",
"_____no_output_____"
],
[
"pfamA_target_sub.to_csv(\"../../data/esm/pfamA_target_sub.csv\",index = False)\npfamA_target.to_csv(\"../../data/esm/pfamA_target.csv\",index = False)\nkinesin_labelled.to_csv(\"../../data/esm/kinesin_labelled.csv\",index = False)\nmotor_toolkit.to_csv(\"../../data/esm/motor_toolkit.csv\",index = False)\npfamA_motors_balanced.to_csv(\"../../data/esm/pfamA_motors_balanced.csv\",index = False)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a56e4f4a60c31e9d8aa2b3552f1ebe5150a3ebc
| 607,955 |
ipynb
|
Jupyter Notebook
|
face_detection/08_CompareNetworks.ipynb
|
qAp/kgl_deepfake
|
d3ee36d704d82d5d72068ea16276a88b5746c8de
|
[
"Apache-2.0"
] | null | null | null |
face_detection/08_CompareNetworks.ipynb
|
qAp/kgl_deepfake
|
d3ee36d704d82d5d72068ea16276a88b5746c8de
|
[
"Apache-2.0"
] | null | null | null |
face_detection/08_CompareNetworks.ipynb
|
qAp/kgl_deepfake
|
d3ee36d704d82d5d72068ea16276a88b5746c8de
|
[
"Apache-2.0"
] | null | null | null | 137.39096 | 320,500 | 0.833822 |
[
[
[
"## Compare Network Architectures",
"_____no_output_____"
],
[
"Now that we have a reasonable baseline for our EasyDeepFakes dataset, let's try to improve performance. For starters, let's just compare how a variety of networks perform on this dataset. We will try:\n\n - ResNet\n - XResNet\n - EfficientNet\n - MesoNet\n - XceptionNet",
"_____no_output_____"
]
],
[
[
"from fastai.core import *\nfrom fastai.vision import *",
"_____no_output_____"
],
[
"path = Path('../data/EasyDeepFakes')\nsrc = ImageList.from_folder(path).split_by_folder(train='train', valid='val')",
"_____no_output_____"
],
[
"def get_data(bs,size):\n data = (src.label_from_re('([A-Z]+).png$')\n .transform(get_transforms(max_warp=0, max_zoom=1), size=size)\n .databunch(bs=bs).normalize(imagenet_stats))\n return data",
"_____no_output_____"
],
[
"bs, sz = 32, 256\ndata = get_data(bs, sz)\ndata.show_batch(rows=4, figsize=(10,7))",
"_____no_output_____"
]
],
[
[
"# ResNet",
"_____no_output_____"
],
[
"The ResNet architecture is one of the most common and trusted baseline architectures. We will use it to establish a reasonable baseline for performance and compare our other networks to it.",
"_____no_output_____"
],
[
"### ResNet18",
"_____no_output_____"
]
],
[
[
"from fastai.vision.models import resnet18\nlearner = cnn_learner(data, resnet18, metrics=[accuracy])\nlearner.lr_find()\nlearner.recorder.plot()",
"_____no_output_____"
],
[
"# Train only the head of the network\nlearner.fit_one_cycle(5, 1e-3)\n# Unfreeze other layers and train the entire network\nlearner.unfreeze()\nlearner.fit_one_cycle(20, max_lr=slice(1e-5, 1e-3))",
"_____no_output_____"
]
],
[
[
"ResNet18 gets a final accuracy of **85.6%** and a peak accuracy of **86.8%**",
"_____no_output_____"
],
[
"### ResNet34",
"_____no_output_____"
]
],
[
[
"from fastai.vision.models import resnet34\nlearner = cnn_learner(data, resnet34, metrics=[accuracy])\nlearner.lr_find()\nlearner.recorder.plot()",
"_____no_output_____"
],
[
"# Train only the head of the network\nlearner.fit_one_cycle(5, 1e-3)\n# Unfreeze other layers and train the entire network\nlearner.unfreeze()\nlearner.fit_one_cycle(20, max_lr=slice(1e-5, 1e-3))",
"_____no_output_____"
]
],
[
[
"ResNet34 has a final accuracy of **87.6%** and a peak accuracy of **89.1%**",
"_____no_output_____"
],
[
"### ResNet50",
"_____no_output_____"
]
],
[
[
"from fastai.vision.models import resnet50\nlearner = cnn_learner(data, resnet50, metrics=[accuracy])\nlearner.lr_find()\nlearner.recorder.plot()",
"_____no_output_____"
],
[
"# Train only the head of the network\nlearner.fit_one_cycle(5, 5e-3)\n# Unfreeze other layers and train the entire network\nlearner.unfreeze()\nlearner.fit_one_cycle(20, max_lr=slice(1e-5, 1e-3))",
"_____no_output_____"
]
],
[
[
"ResNet50 has a final accuracy of **91.1%** and a peak accuracy of **91.1%**.",
"_____no_output_____"
],
[
"# XResNet",
"_____no_output_____"
],
[
"`xresnet` is modified resnet architecture developed by fast.ai in according with the paper [Bag of Tricks for Image Classification with Convolutional Neural Networks](https://arxiv.org/abs/1812.01187).\n\nNotably the initial 7x7 conv is replaced by a series of 3x3 convolutions. I believe they have also changed some of the 1x1 convolutions in the bottleneck layers.\n\n\n**NOTE:** In fastai v1, there is no pretrained model for xresnet.",
"_____no_output_____"
],
[
"### XResNet18",
"_____no_output_____"
]
],
[
[
"from fastai.vision.models import xresnet18\nlearner = cnn_learner(data, xresnet18, metrics=[accuracy], pretrained=False)\nlearner.lr_find()\nlearner.recorder.plot()",
"_____no_output_____"
],
[
"# Train only the head of the network\nlearner.fit_one_cycle(5, 1e-3)\n# Unfreeze other layers and train the entire network\nlearner.unfreeze()\nlearner.fit_one_cycle(20, max_lr=slice(1e-5, 1e-3))",
"_____no_output_____"
]
],
[
[
"(Non-pretrained) XResNet18 gets a final accuracy of **65.0%** and a peak accuracy of **68.4%**.",
"_____no_output_____"
],
[
"### XResNet34",
"_____no_output_____"
]
],
[
[
"from fastai.vision.models import xresnet34\nlearner = cnn_learner(data, xresnet34, metrics=[accuracy], pretrained=False)\nlearner.lr_find()\nlearner.recorder.plot()",
"_____no_output_____"
],
[
"# Train only the head of the network\nlearner.fit_one_cycle(5, 1e-3)\n# Unfreeze other layers and train the entire network\nlearner.unfreeze()\nlearner.fit_one_cycle(20, max_lr=slice(1e-5, 1e-3))",
"_____no_output_____"
]
],
[
[
"`xresnet34` has a final accuracy of **68.9%** and a peak accuracy of **73.4%**.",
"_____no_output_____"
],
[
"### XResNet50",
"_____no_output_____"
]
],
[
[
"from fastai.vision.models import xresnet50\nlearner = cnn_learner(data, xresnet50, metrics=[accuracy], pretrained=False)\nlearner.lr_find()\nlearner.recorder.plot()",
"_____no_output_____"
],
[
"# Train only the head of the network\nlearner.fit_one_cycle(5, 1e-3)\n# Unfreeze other layers and train the entire network\nlearner.unfreeze()\nlearner.fit_one_cycle(20, max_lr=slice(1e-5, 1e-3))",
"_____no_output_____"
]
],
[
[
"`xresnet50` has a final accuracy of **71.1%** and a peak accuracy of **72.7%**.",
"_____no_output_____"
],
[
"# EfficientNet",
"_____no_output_____"
],
[
"EfficientNet is an architecture released by Google with the intention of reducing the number of parameters while maintaining good performance. There are 8 versions of Efficient with increasing capacity from `efficientb0` to `efficientnetb7`.\n\nI haven't figured out how to set up layer groups so I'm unable to do discriminitive learning with EfficientNet, but we'll give it a shot anyways.",
"_____no_output_____"
]
],
[
[
"# !pip install efficientnet-pytorch\nfrom efficientnet_pytorch import EfficientNet",
"_____no_output_____"
]
],
[
[
"### EfficientNetB0",
"_____no_output_____"
]
],
[
[
"model = EfficientNet.from_pretrained('efficientnet-b0', num_classes=data.c)\nlearner = Learner(data, model, metrics=[accuracy])\n\nlearner.lr_find()\nlearner.recorder.plot()",
"Loaded pretrained weights for efficientnet-b0\n"
],
[
"# Train only the head of the network\nlearner.fit_one_cycle(5, 1e-3)\n# Unfreeze other layers and train the entire network\nlearner.unfreeze()\n\n#NOTE: Not using discriminitive learning rates!\nlearner.fit_one_cycle(20, max_lr=1e-4)",
"_____no_output_____"
]
],
[
[
"`efficientnetb0` has a final accuracy of **91.1%** and a peak accuracy of **92.7%**.",
"_____no_output_____"
],
[
"### EfficientNetB1",
"_____no_output_____"
]
],
[
[
"model = EfficientNet.from_pretrained('efficientnet-b1', num_classes=data.c)\nlearner = Learner(data, model, metrics=[accuracy])\n\nlearner.lr_find()\nlearner.recorder.plot()",
"Loaded pretrained weights for efficientnet-b1\n"
],
[
"# Train only the head of the network\nlearner.fit_one_cycle(5, 1e-3)\n# Unfreeze other layers and train the entire network\nlearner.unfreeze()\n\n#NOTE: Not using discriminitive learning rates!\nlearner.fit_one_cycle(20, max_lr=1e-4)",
"_____no_output_____"
]
],
[
[
"`efficientnetb1` hsa an final accuracy of **91.9%** and a peak accuracy of **93.7%**",
"_____no_output_____"
],
[
"### EfficientNetB2",
"_____no_output_____"
]
],
[
[
"model = EfficientNet.from_pretrained('efficientnet-b2', num_classes=data.c)\nlearner = Learner(data, model, metrics=[accuracy])\n\nlearner.lr_find()\nlearner.recorder.plot()",
"Downloading: \"https://publicmodels.blob.core.windows.net/container/aa/efficientnet-b2-8bb594d6.pth\" to /home/josh/.cache/torch/checkpoints/efficientnet-b2-8bb594d6.pth\n"
],
[
"# Train only the head of the network\nlearner.fit_one_cycle(5, 1e-3)\n# Unfreeze other layers and train the entire network\nlearner.unfreeze()\n\n#NOTE: Not using discriminitive learning rates!\nlearner.fit_one_cycle(20, max_lr=1e-4)",
"_____no_output_____"
]
],
[
[
"`efficientnetb2` get an final accuracy of **89.4%** and a peak accuracy of **90.1%**.",
"_____no_output_____"
],
[
"## MesoNet",
"_____no_output_____"
],
[
"MesoNet was developed to detect deep fakes in the [MesoNet: a Compact Facial Video Forgery Detection Network](https://arxiv.org/abs/1809.00888) paper.\n\nLike EfficientNet, I'm unsure how to build layer groups, so we will not use discriminitive fine tuning on this network.",
"_____no_output_____"
]
],
[
[
"#export\n# By Nathan Hubens.\n# Paper implementation does not use Adaptive Average Pooling. To get the exact same implementation, \n# comment the avg_pool and uncomment the final max_pool layer.\nclass MesoNet(nn.Module):\n def __init__(self):\n super().__init__()\n \n self.conv1 = nn.Conv2d(3, 8, 3, 1,1) # 8 x 256 x 256\n self.bn1 = nn.BatchNorm2d(8)\n self.conv2 = nn.Conv2d(8, 8, 5, 1,2) # 8 x 128 x 128\n self.bn2 = nn.BatchNorm2d(8)\n self.conv3 = nn.Conv2d(8, 16, 5, 1,2) # 8 x 64 x 64\n self.bn3 = nn.BatchNorm2d(16)\n self.conv4 = nn.Conv2d(16,16,5,1,2) # 8 x 32 x 32\n self.bn4 = nn.BatchNorm2d(16)\n self.avg_pool = nn.AdaptiveAvgPool2d((8))\n self.fc1 = nn.Linear(1024, 16)\n self.fc2 = nn.Linear(16, 2)\n \n def forward(self, x):\n\n x = F.relu(self.conv1(x))\n x = self.bn1(x)\n x = F.max_pool2d(x, 2, 2)\n \n x = F.relu(self.conv2(x))\n x = self.bn2(x)\n x = F.max_pool2d(x, 2, 2)\n \n x = F.relu(self.conv3(x)) \n x = self.bn3(x)\n x = F.max_pool2d(x, 2, 2)\n \n x = F.relu(self.conv4(x))\n x = self.bn4(x)\n #x = F.max_pool2d(x, 4, 4)\n \n x = self.avg_pool(x)\n\n x = x.reshape(x.shape[0], -1)\n\n x = F.dropout(x, 0.5)\n x = F.relu(self.fc1(x))\n x = F.dropout(x,0.5)\n x = self.fc2(x)\n return x",
"_____no_output_____"
],
[
"model = MesoNet()\nlearner = Learner(data, model, metrics=[accuracy])\n\nlearner.lr_find()\nlearner.recorder.plot()",
"_____no_output_____"
],
[
"# Train only the head of the network\nlearner.fit_one_cycle(5, 1e-3)\n# Unfreeze other layers and train the entire network\nlearner.unfreeze()\n\n#NOTE: Not using discriminitive learning rates!\nlearner.fit_one_cycle(20, max_lr=1e-4)",
"_____no_output_____"
]
],
[
[
"`mesonet` has a final accuracy of **65.6%** and a peak accuracy of **67.6%**.",
"_____no_output_____"
],
[
"## XceptionNet",
"_____no_output_____"
],
[
"[XceptionNet](https://arxiv.org/abs/1610.02357) was developed to be a more performant version of Google's InceptionNet. They replace Inception modules with depthwise separable convolution modules.\n\nIt was also the best performing model used in [FaceForensics++: Learning to Detect Manipulated Facial Images](https://arxiv.org/abs/1901.08971).",
"_____no_output_____"
]
],
[
[
"## xception.py\n\"\"\"\nPorted to pytorch thanks to [tstandley](https://github.com/tstandley/Xception-PyTorch)\n@author: tstandley\nAdapted by cadene\nCreates an Xception Model as defined in:\nFrancois Chollet\nXception: Deep Learning with Depthwise Separable Convolutions\nhttps://arxiv.org/pdf/1610.02357.pdf\nThis weights ported from the Keras implementation. Achieves the following performance on the validation set:\nLoss:0.9173 Prec@1:78.892 Prec@5:94.292\nREMEMBER to set your image size to 3x299x299 for both test and validation\nnormalize = transforms.Normalize(mean=[0.5, 0.5, 0.5],\n std=[0.5, 0.5, 0.5])\nThe resize parameter of the validation transform should be 333, and make sure to center crop at 299x299\n\"\"\"\nimport math\nimport torch\nimport torch.nn as nn\nimport torch.nn.functional as F\nimport torch.utils.model_zoo as model_zoo\nfrom torch.nn import init\n\npretrained_settings = {\n 'xception': {\n 'imagenet': {\n 'url': 'http://data.lip6.fr/cadene/pretrainedmodels/xception-b5690688.pth',\n 'input_space': 'RGB',\n 'input_size': [3, 299, 299],\n 'input_range': [0, 1],\n 'mean': [0.5, 0.5, 0.5],\n 'std': [0.5, 0.5, 0.5],\n 'num_classes': 1000,\n 'scale': 0.8975 # The resize parameter of the validation transform should be 333, and make sure to center crop at 299x299\n }\n }\n}\n\n\nclass SeparableConv2d(nn.Module):\n def __init__(self,in_channels,out_channels,kernel_size=1,stride=1,padding=0,dilation=1,bias=False):\n super(SeparableConv2d,self).__init__()\n\n self.conv1 = nn.Conv2d(in_channels,in_channels,kernel_size,stride,padding,dilation,groups=in_channels,bias=bias)\n self.pointwise = nn.Conv2d(in_channels,out_channels,1,1,0,1,1,bias=bias)\n\n def forward(self,x):\n x = self.conv1(x)\n x = self.pointwise(x)\n return x\n\n\nclass Block(nn.Module):\n def __init__(self,in_filters,out_filters,reps,strides=1,start_with_relu=True,grow_first=True):\n super(Block, self).__init__()\n\n if out_filters != in_filters or strides!=1:\n self.skip = nn.Conv2d(in_filters,out_filters,1,stride=strides, bias=False)\n self.skipbn = nn.BatchNorm2d(out_filters)\n else:\n self.skip=None\n\n self.relu = nn.ReLU(inplace=True)\n rep=[]\n\n filters=in_filters\n if grow_first:\n rep.append(self.relu)\n rep.append(SeparableConv2d(in_filters,out_filters,3,stride=1,padding=1,bias=False))\n rep.append(nn.BatchNorm2d(out_filters))\n filters = out_filters\n\n for i in range(reps-1):\n rep.append(self.relu)\n rep.append(SeparableConv2d(filters,filters,3,stride=1,padding=1,bias=False))\n rep.append(nn.BatchNorm2d(filters))\n\n if not grow_first:\n rep.append(self.relu)\n rep.append(SeparableConv2d(in_filters,out_filters,3,stride=1,padding=1,bias=False))\n rep.append(nn.BatchNorm2d(out_filters))\n\n if not start_with_relu:\n rep = rep[1:]\n else:\n rep[0] = nn.ReLU(inplace=False)\n\n if strides != 1:\n rep.append(nn.MaxPool2d(3,strides,1))\n self.rep = nn.Sequential(*rep)\n\n def forward(self,inp):\n x = self.rep(inp)\n\n if self.skip is not None:\n skip = self.skip(inp)\n skip = self.skipbn(skip)\n else:\n skip = inp\n\n x+=skip\n return x\n\n\nclass Xception(nn.Module):\n \"\"\"\n Xception optimized for the ImageNet dataset, as specified in\n https://arxiv.org/pdf/1610.02357.pdf\n \"\"\"\n def __init__(self, num_classes=1000):\n \"\"\" Constructor\n Args:\n num_classes: number of classes\n \"\"\"\n super(Xception, self).__init__()\n self.num_classes = num_classes\n\n self.conv1 = nn.Conv2d(3, 32, 3,2, 0, bias=False)\n self.bn1 = nn.BatchNorm2d(32)\n self.relu = nn.ReLU(inplace=True)\n\n self.conv2 = nn.Conv2d(32,64,3,bias=False)\n self.bn2 = nn.BatchNorm2d(64)\n #do relu here\n\n self.block1=Block(64,128,2,2,start_with_relu=False,grow_first=True)\n self.block2=Block(128,256,2,2,start_with_relu=True,grow_first=True)\n self.block3=Block(256,728,2,2,start_with_relu=True,grow_first=True)\n\n self.block4=Block(728,728,3,1,start_with_relu=True,grow_first=True)\n self.block5=Block(728,728,3,1,start_with_relu=True,grow_first=True)\n self.block6=Block(728,728,3,1,start_with_relu=True,grow_first=True)\n self.block7=Block(728,728,3,1,start_with_relu=True,grow_first=True)\n\n self.block8=Block(728,728,3,1,start_with_relu=True,grow_first=True)\n self.block9=Block(728,728,3,1,start_with_relu=True,grow_first=True)\n self.block10=Block(728,728,3,1,start_with_relu=True,grow_first=True)\n self.block11=Block(728,728,3,1,start_with_relu=True,grow_first=True)\n\n self.block12=Block(728,1024,2,2,start_with_relu=True,grow_first=False)\n\n self.conv3 = SeparableConv2d(1024,1536,3,1,1)\n self.bn3 = nn.BatchNorm2d(1536)\n\n #do relu here\n self.conv4 = SeparableConv2d(1536,2048,3,1,1)\n self.bn4 = nn.BatchNorm2d(2048)\n\n self.fc = nn.Linear(2048, num_classes)\n\n # #------- init weights --------\n # for m in self.modules():\n # if isinstance(m, nn.Conv2d):\n # n = m.kernel_size[0] * m.kernel_size[1] * m.out_channels\n # m.weight.data.normal_(0, math.sqrt(2. / n))\n # elif isinstance(m, nn.BatchNorm2d):\n # m.weight.data.fill_(1)\n # m.bias.data.zero_()\n # #-----------------------------\n\n def features(self, input):\n x = self.conv1(input)\n x = self.bn1(x)\n x = self.relu(x)\n\n x = self.conv2(x)\n x = self.bn2(x)\n x = self.relu(x)\n\n x = self.block1(x)\n x = self.block2(x)\n x = self.block3(x)\n x = self.block4(x)\n x = self.block5(x)\n x = self.block6(x)\n x = self.block7(x)\n x = self.block8(x)\n x = self.block9(x)\n x = self.block10(x)\n x = self.block11(x)\n x = self.block12(x)\n\n x = self.conv3(x)\n x = self.bn3(x)\n x = self.relu(x)\n\n x = self.conv4(x)\n x = self.bn4(x)\n return x\n\n def logits(self, features):\n x = self.relu(features)\n\n x = F.adaptive_avg_pool2d(x, (1, 1))\n x = x.view(x.size(0), -1)\n x = self.last_linear(x)\n return x\n\n def forward(self, input):\n x = self.features(input)\n x = self.logits(x)\n return x\n\n\ndef xception(num_classes=1000, pretrained='imagenet'):\n model = Xception(num_classes=num_classes)\n if pretrained:\n settings = pretrained_settings['xception'][pretrained]\n assert num_classes == settings['num_classes'], \\\n \"num_classes should be {}, but is {}\".format(settings['num_classes'], num_classes)\n\n model = Xception(num_classes=num_classes)\n model.load_state_dict(model_zoo.load_url(settings['url']))\n\n model.input_space = settings['input_space']\n model.input_size = settings['input_size']\n model.input_range = settings['input_range']\n model.mean = settings['mean']\n model.std = settings['std']\n\n # TODO: ugly\n model.last_linear = model.fc\n del model.fc\n return model",
"_____no_output_____"
],
[
"XCEPTION_MODEL = 'xception/xception-b5690688.pth'\n\ndef return_pytorch04_xception(pretrained=True):\n # Raises warning \"src not broadcastable to dst\" but thats fine\n model = xception(pretrained=False)\n if pretrained:\n # Load model in torch 0.4+\n model.fc = model.last_linear\n del model.last_linear\n state_dict = torch.load(\n XCEPTION_MODEL)\n for name, weights in state_dict.items():\n if 'pointwise' in name:\n state_dict[name] = weights.unsqueeze(-1).unsqueeze(-1)\n model.load_state_dict(state_dict)\n model.last_linear = model.fc\n del model.fc\n return model",
"_____no_output_____"
],
[
"model = return_pytorch04_xception()\nmodel.last_linear = torch.nn.Linear(in_features=2048, out_features=2, bias=True)",
"_____no_output_____"
],
[
"learner = Learner(data, model, metrics=[accuracy])\n\nlearner.lr_find()\nlearner.recorder.plot()",
"_____no_output_____"
],
[
"# Train only the head of the network\nlearner.fit_one_cycle(5, 1e-3)\n# Unfreeze other layers and train the entire network\nlearner.unfreeze()\n\n#NOTE: Not using discriminitive learning rates!\nlearner.fit_one_cycle(20, max_lr=1e-4)",
"_____no_output_____"
]
],
[
[
"XceptionNet has a final accuracy of **83.5%** and a peak accuracy of **86.8%**.",
"_____no_output_____"
],
[
"## Results",
"_____no_output_____"
],
[
"|Network | Pretrained | Discriminitive | Final Accuracy % | Peak Accuracy %| Time for 1 Epoch (s) |\n|----------------|----------------|----------------|------------------|----------------|----------------------|\n|`resnet18` | True | True | 85.6 | 86.8 | **5** |\n|`resnet34` | True | True | 87.6 | 89.1 | 7 |\n|`resnet50` | True | True | 91.1 | 91.1 | 13 |\n|`xresnet18` | False | True | 65.0 | 68.4 | 6 |\n|`xresnet34` | False | True | 68.9 | 73.4 | 8 |\n|`xresnet50` | False | True | 71.1 | 72.7 | 14 |\n|`efficientnetb0`| True | False | 91.1 | 92.7 | 12 |\n|`efficientnetb1`| True | False | **91.9** | **93.7** | 15 |\n|`efficientnetb2`| True | False | 89.4 | 90.1 | 16 |\n| `mesonet` | False | False | 65.6 | 67.6 | **5** |\n| `xceptionnet` | True | False | 83.5 | 86.8 | 21 |",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
4a56e85d42b1f58752e6a1105bc9ed7d7f1f8d5e
| 317,966 |
ipynb
|
Jupyter Notebook
|
Image Classifier Project-zh.ipynb
|
jiashuyu/Image-Classifier-APP
|
9c3423c9ccbc977433e03477cf6340a1c09737a8
|
[
"MIT"
] | null | null | null |
Image Classifier Project-zh.ipynb
|
jiashuyu/Image-Classifier-APP
|
9c3423c9ccbc977433e03477cf6340a1c09737a8
|
[
"MIT"
] | null | null | null |
Image Classifier Project-zh.ipynb
|
jiashuyu/Image-Classifier-APP
|
9c3423c9ccbc977433e03477cf6340a1c09737a8
|
[
"MIT"
] | null | null | null | 415.099217 | 145,204 | 0.934471 |
[
[
[
"# 开发 AI 应用\n\n未来,AI 算法在日常生活中的应用将越来越广泛。例如,你可能想要在智能手机应用中包含图像分类器。为此,在整个应用架构中,你将使用一个用成百上千个图像训练过的深度学习模型。未来的软件开发很大一部分将是使用这些模型作为应用的常用部分。\n\n在此项目中,你将训练一个图像分类器来识别不同的花卉品种。可以想象有这么一款手机应用,当你对着花卉拍摄时,它能够告诉你这朵花的名称。在实际操作中,你会训练此分类器,然后导出它以用在你的应用中。我们将使用[此数据集](http://www.robots.ox.ac.uk/~vgg/data/flowers/102/index.html),其中包含 102 个花卉类别。你可以在下面查看几个示例。 \n\n<img src='assets/Flowers.png' width=500px>\n\n该项目分为多个步骤:\n\n* 加载和预处理图像数据集\n* 用数据集训练图像分类器\n* 使用训练的分类器预测图像内容\n\n我们将指导你完成每一步,你将用 Python 实现这些步骤。\n\n完成此项目后,你将拥有一个可以用任何带标签图像的数据集进行训练的应用。你的网络将学习花卉,并成为一个命令行应用。但是,你对新技能的应用取决于你的想象力和构建数据集的精力。例如,想象有一款应用能够拍摄汽车,告诉你汽车的制造商和型号,然后查询关于该汽车的信息。构建你自己的数据集并开发一款新型应用吧。\n\n首先,导入你所需的软件包。建议在代码开头导入所有软件包。当你创建此 notebook 时,如果发现你需要导入某个软件包,确保在开头导入该软件包。",
"_____no_output_____"
]
],
[
[
"# Imports here\n% matplotlib inline\n\nimport matplotlib.pyplot as plt\nimport numpy as np\nimport time\n\nimport json\n\nimport torch\nfrom torch import nn\nfrom torch import optim\nimport torch.nn.functional as F\nfrom torch.autograd import Variable\nfrom torchvision import datasets, transforms, models\n\nfrom collections import OrderedDict\nfrom PIL import Image",
"_____no_output_____"
]
],
[
[
"## 加载数据\n\n在此项目中,你将使用 `torchvision` 加载数据([文档](http://pytorch.org/docs/master/torchvision/transforms.html#))。数据应该和此 notebook 一起包含在内,否则你可以[在此处下载数据](https://s3.amazonaws.com/content.udacity-data.com/nd089/flower_data.tar.gz)。数据集分成了三部分:训练集、验证集和测试集。对于训练集,你需要变换数据,例如随机缩放、剪裁和翻转。这样有助于网络泛化,并带来更好的效果。你还需要确保将输入数据的大小调整为 224x224 像素,因为预训练的网络需要这么做。\n\n验证集和测试集用于衡量模型对尚未见过的数据的预测效果。对此步骤,你不需要进行任何缩放或旋转变换,但是需要将图像剪裁到合适的大小。\n\n对于所有三个数据集,你都需要将均值和标准差标准化到网络期望的结果。均值为 `[0.485, 0.456, 0.406]`,标准差为 `[0.229, 0.224, 0.225]`。这样使得每个颜色通道的值位于 -1 到 1 之间,而不是 0 到 1 之间。",
"_____no_output_____"
]
],
[
[
"train_dir = 'flowers/train'\nvalid_dir = 'flowers/valid'\ntest_dir = 'flowers/test'",
"_____no_output_____"
],
[
"# TODO: Define your transforms for the training, validation, and testing sets\ntrain_transforms = transforms.Compose([transforms.RandomRotation(30),\n transforms.RandomResizedCrop(224),\n transforms.RandomHorizontalFlip(),\n transforms.ToTensor(),\n transforms.Normalize([0.485, 0.456, 0.406], \n [0.229, 0.224, 0.225])])\n\n\nvalid_transforms = transforms.Compose([transforms.Resize(256),\n transforms.CenterCrop(224),\n transforms.ToTensor(),\n transforms.Normalize([0.485, 0.456, 0.406], \n [0.229, 0.224, 0.225])])\n\ntransforms.RandomRotation(30),\n\n# TODO: Load the datasets with ImageFolder\ntrain_dataset = datasets.ImageFolder(train_dir, transform=train_transforms)\nvalid_dataset = datasets.ImageFolder(valid_dir, transform=valid_transforms)\ntest_dataset = datasets.ImageFolder(test_dir, transform=valid_transforms)\n\n# TODO: Using the image datasets and the trainforms, define the dataloaders\ntrain_loader = torch.utils.data.DataLoader(train_dataset, batch_size=32, shuffle=True)\nvalid_loader = torch.utils.data.DataLoader(valid_dataset, batch_size=32, shuffle=True)\ntest_loader = torch.utils.data.DataLoader(test_dataset, batch_size=32, shuffle=True)",
"_____no_output_____"
]
],
[
[
"### 标签映射\n\n你还需要加载从类别标签到类别名称的映射。你可以在文件 `cat_to_name.json` 中找到此映射。它是一个 JSON 对象,可以使用 [`json` 模块](https://docs.python.org/2/library/json.html)读取它。这样可以获得一个从整数编码的类别到实际花卉名称的映射字典。",
"_____no_output_____"
]
],
[
[
"with open('cat_to_name.json', 'r') as f:\n cat_to_name = json.load(f)",
"_____no_output_____"
]
],
[
[
"# 构建和训练分类器\n\n数据准备好后,就开始构建和训练分类器了。和往常一样,你应该使用 `torchvision.models` 中的某个预训练模型获取图像特征。使用这些特征构建和训练新的前馈分类器。\n\n这部分将由你来完成。如果你想与他人讨论这部分,欢迎与你的同学讨论!你还可以在论坛上提问或在工作时间内咨询我们的课程经理和助教导师。\n\n请参阅[审阅标准](https://review.udacity.com/#!/rubrics/1663/view),了解如何成功地完成此部分。你需要执行以下操作:\n\n* 加载[预训练的网络](http://pytorch.org/docs/master/torchvision/models.html)(如果你需要一个起点,推荐使用 VGG 网络,它简单易用)\n* 使用 ReLU 激活函数和丢弃定义新的未训练前馈网络作为分类器\n* 使用反向传播训练分类器层,并使用预训练的网络获取特征\n* 跟踪验证集的损失和准确率,以确定最佳超参数\n\n我们在下面为你留了一个空的单元格,但是你可以使用多个单元格。建议将问题拆分为更小的部分,并单独运行。检查确保每部分都达到预期效果,然后再完成下个部分。你可能会发现,当你实现每部分时,可能需要回去修改之前的代码,这很正常!\n\n训练时,确保仅更新前馈网络的权重。如果一切构建正确的话,验证准确率应该能够超过 70%。确保尝试不同的超参数(学习速率、分类器中的单元、周期等),寻找最佳模型。保存这些超参数并用作项目下个部分的默认值。",
"_____no_output_____"
]
],
[
[
"# Load AlexNet as my model\nmodel = models.alexnet(pretrained=True)\nmodel",
"Downloading: \"https://download.pytorch.org/models/alexnet-owt-4df8aa71.pth\" to /root/.torch/models/alexnet-owt-4df8aa71.pth\n100%|██████████| 244418560/244418560 [00:01<00:00, 126626907.37it/s]\n"
],
[
"# Freeze parameters so we don't backprop through them\nfor p in model.parameters():\n p.requires_grad = False\n \nclassifier = nn.Sequential(OrderedDict([\n ('fc1', nn.Linear(9216, 1000)),\n ('relu', nn.ReLU()),\n ('fc2', nn.Linear(1000, 102)),\n ('output', nn.LogSoftmax(dim=1))\n ]))\n\nmodel.classifier = classifier",
"_____no_output_____"
],
[
"# Train a model with a pre-trained network\ncriterion = nn.NLLLoss()\noptimizer = optim.Adam(model.classifier.parameters(), lr=0.001)",
"_____no_output_____"
],
[
"epochs = 3\nprint_every = 40\nsteps = 0\n\n# change to cuda\nmodel.to('cuda')\n\nfor e in range(epochs):\n model.train()\n running_loss = 0\n for ii, (inputs, labels) in enumerate(train_loader):\n steps += 1\n \n inputs, labels = inputs.to('cuda'), labels.to('cuda')\n \n optimizer.zero_grad()\n \n # Forward and backward passes\n outputs = model.forward(inputs)\n loss = criterion(outputs, labels)\n loss.backward()\n optimizer.step()\n \n running_loss += loss.item()\n \n if steps % print_every == 0:\n model.eval()\n correct = 0\n total = 0\n test_loss = 0\n \n with torch.no_grad():\n for data in valid_loader:\n images, labels = data\n images = images.to('cuda')\n labels = labels.to('cuda')\n outputs = model.forward(images)\n test_loss += criterion(outputs, labels).item()\n _, predicted = torch.max(outputs.data, 1)\n total += labels.size(0)\n correct += (predicted == labels).sum().item()\n \n print(\"Epoch: {}/{}... \".format(e+1, epochs),\n \"Training Loss: {:.4f}\".format(running_loss/print_every),\n \" Test Loss: {:.4f}\".format(test_loss/len(valid_loader)),\n \" Accuracy: {:.4f}\".format(correct / total))\n running_loss = 0\n model.train()",
"Epoch: 1/3... Training Loss: 4.5910 Test Loss: 2.7071 Accuracy: 0.3741\nEpoch: 1/3... Training Loss: 2.3924 Test Loss: 1.5787 Accuracy: 0.5941\nEpoch: 1/3... Training Loss: 1.8183 Test Loss: 1.3634 Accuracy: 0.6540\nEpoch: 1/3... Training Loss: 1.6817 Test Loss: 1.2887 Accuracy: 0.6369\nEpoch: 1/3... Training Loss: 1.5187 Test Loss: 0.9631 Accuracy: 0.7200\nEpoch: 2/3... Training Loss: 1.0291 Test Loss: 0.8121 Accuracy: 0.7567\nEpoch: 2/3... Training Loss: 1.2033 Test Loss: 0.9343 Accuracy: 0.7335\nEpoch: 2/3... Training Loss: 1.1127 Test Loss: 0.7286 Accuracy: 0.7738\nEpoch: 2/3... Training Loss: 1.2338 Test Loss: 0.7340 Accuracy: 0.7812\nEpoch: 2/3... Training Loss: 1.1414 Test Loss: 0.7432 Accuracy: 0.7861\nEpoch: 3/3... Training Loss: 0.7024 Test Loss: 0.6860 Accuracy: 0.8252\nEpoch: 3/3... Training Loss: 0.9885 Test Loss: 0.7302 Accuracy: 0.8020\nEpoch: 3/3... Training Loss: 1.0018 Test Loss: 0.7333 Accuracy: 0.8142\nEpoch: 3/3... Training Loss: 1.0665 Test Loss: 0.8263 Accuracy: 0.7885\nEpoch: 3/3... Training Loss: 0.9734 Test Loss: 0.6461 Accuracy: 0.8166\n"
]
],
[
[
"## 测试网络\n\n建议使用网络在训练或验证过程中从未见过的测试数据测试训练的网络。这样,可以很好地判断模型预测全新图像的效果。用网络预测测试图像,并测量准确率,就像验证过程一样。如果模型训练良好的话,你应该能够达到大约 70% 的准确率。",
"_____no_output_____"
]
],
[
[
"correct = 0\ntotal = 0\nwith torch.no_grad():\n for data in test_loader:\n images, labels = data\n images = images.to('cuda')\n labels = labels.to('cuda')\n outputs = model(images)\n _, predicted = torch.max(outputs.data, 1)\n total += labels.size(0)\n correct += (predicted == labels).sum().item()\n\nprint('accuracy on the test dataset=', correct / total)",
"accuracy on the test dataset= 0.7814407814407814\n"
]
],
[
[
"## 保存检查点\n\n训练好网络后,保存模型,以便稍后加载它并进行预测。你可能还需要保存其他内容,例如从类别到索引的映射,索引是从某个图像数据集中获取的:`image_datasets['train'].class_to_idx`。你可以将其作为属性附加到模型上,这样稍后推理会更轻松。",
"_____no_output_____"
],
[
"注意,稍后你需要完全重新构建模型,以便用模型进行推理。确保在检查点中包含你所需的任何信息。如果你想加载模型并继续训练,则需要保存周期数量和优化器状态 `optimizer.state_dict`。你可能需要在下面的下个部分使用训练的模型,因此建议立即保存它。",
"_____no_output_____"
]
],
[
[
"# TODO: Save the checkpoint \ncheckpoint = {'class_to_idx': train_dataset.class_to_idx,\n 'classifier_input_size': 9216,\n 'output_size': 102,\n 'classifier_hidden_layers': 1000,\n 'state_dict': model.state_dict()}\n\ntorch.save(checkpoint, 'checkpoint.pth')",
"_____no_output_____"
]
],
[
[
"## 加载检查点\n\n此刻,建议写一个可以加载检查点并重新构建模型的函数。这样的话,你可以回到此项目并继续完善它,而不用重新训练网络。",
"_____no_output_____"
]
],
[
[
"# TODO: Write a function that loads a checkpoint and rebuilds the model\ndef load_checkpoint(filepath):\n checkpoint = torch.load(filepath)\n model = models.alexnet(pretrained=True)\n classifier = nn.Sequential(OrderedDict([\n ('fc1', nn.Linear(checkpoint['classifier_input_size'], checkpoint['classifier_hidden_layers'])),\n ('relu', nn.ReLU()),\n ('fc2', nn.Linear(checkpoint['classifier_hidden_layers'], checkpoint['output_size'])),\n ('output', nn.LogSoftmax(dim=1))\n ]))\n model.classifier = classifier\n model.load_state_dict(checkpoint['state_dict'])\n model.class_to_idx = checkpoint['class_to_idx']\n return model",
"_____no_output_____"
],
[
"m = load_checkpoint('checkpoint.pth')\nm",
"_____no_output_____"
]
],
[
[
"# 类别推理\n\n现在,你需要写一个使用训练的网络进行推理的函数。即你将向网络中传入一个图像,并预测图像中的花卉类别。写一个叫做 `predict` 的函数,该函数会接受图像和模型,然后返回概率在前 $K$ 的类别及其概率。应该如下所示:",
"_____no_output_____"
]
],
[
[
"probs, classes = predict(image_path, model)\nprint(probs)\nprint(classes)\n> [ 0.01558163 0.01541934 0.01452626 0.01443549 0.01407339]\n> ['70', '3', '45', '62', '55']",
"_____no_output_____"
]
],
[
[
"首先,你需要处理输入图像,使其可以用于你的网络。\n\n## 图像处理\n\n你需要使用 `PIL` 加载图像([文档](https://pillow.readthedocs.io/en/latest/reference/Image.html))。建议写一个函数来处理图像,使图像可以作为模型的输入。该函数应该按照训练的相同方式处理图像。\n\n首先,调整图像大小,使最小的边为 256 像素,并保持宽高比。为此,可以使用 [`thumbnail`](http://pillow.readthedocs.io/en/3.1.x/reference/Image.html#PIL.Image.Image.thumbnail) 或 [`resize`](http://pillow.readthedocs.io/en/3.1.x/reference/Image.html#PIL.Image.Image.thumbnail) 方法。然后,你需要从图像的中心裁剪出 224x224 的部分。\n\n图像的颜色通道通常编码为整数 0-255,但是该模型要求值为浮点数 0-1。你需要变换值。使用 Numpy 数组最简单,你可以从 PIL 图像中获取,例如 `np_image = np.array(pil_image)`。\n\n和之前一样,网络要求图像按照特定的方式标准化。均值应标准化为 `[0.485, 0.456, 0.406]`,标准差应标准化为 `[0.229, 0.224, 0.225]`。你需要用每个颜色通道减去均值,然后除以标准差。\n\n最后,PyTorch 要求颜色通道为第一个维度,但是在 PIL 图像和 Numpy 数组中是第三个维度。你可以使用 [`ndarray.transpose`](https://docs.scipy.org/doc/numpy-1.13.0/reference/generated/numpy.ndarray.transpose.html)对维度重新排序。颜色通道必须是第一个维度,并保持另外两个维度的顺序。",
"_____no_output_____"
]
],
[
[
"def process_image(image):\n im = Image.open(image)\n image = valid_transforms(im)\n return image.numpy()",
"_____no_output_____"
]
],
[
[
"要检查你的项目,可以使用以下函数来转换 PyTorch 张量并将其显示在 notebook 中。如果 `process_image` 函数可行,用该函数运行输出应该会返回原始图像(但是剪裁掉的部分除外)。",
"_____no_output_____"
]
],
[
[
"def imshow(image, ax=None, title=None):\n \"\"\"Imshow for Tensor.\"\"\"\n if ax is None:\n fig, ax = plt.subplots()\n \n # PyTorch tensors assume the color channel is the first dimension\n # but matplotlib assumes is the third dimension\n image = image.numpy().transpose((1, 2, 0))\n \n # Undo preprocessing\n mean = np.array([0.485, 0.456, 0.406])\n std = np.array([0.229, 0.224, 0.225])\n image = std * image + mean\n \n # Image needs to be clipped between 0 and 1 or it looks like noise when displayed\n image = np.clip(image, 0, 1)\n \n ax.imshow(image)\n \n return ax",
"_____no_output_____"
],
[
"np_image = process_image('flowers/test/1/image_06743.jpg')\nimage = torch.from_numpy(np_image)\nimshow(image)",
"_____no_output_____"
]
],
[
[
"## 类别预测\n\n可以获得格式正确的图像后 \n\n要获得前 $K$ 个值,在张量中使用 [`x.topk(k)`](http://pytorch.org/docs/master/torch.html#torch.topk)。该函数会返回前 `k` 个概率和对应的类别索引。你需要使用 `class_to_idx`(希望你将其添加到了模型中)将这些索引转换为实际类别标签,或者从用来加载数据的[ `ImageFolder`](https://pytorch.org/docs/master/torchvision/datasets.html?highlight=imagefolder#torchvision.datasets.ImageFolder)进行转换。确保颠倒字典\n\n同样,此方法应该接受图像路径和模型检查点,并返回概率和类别。",
"_____no_output_____"
]
],
[
[
"def predict(image_path, model, topk=5):\n ''' Predict the class (or classes) of an image using a trained deep learning model.\n '''\n \n # TODO: Implement the code to predict the class from an image file\n np_image = process_image(image_path)\n image = torch.from_numpy(np_image)\n image.unsqueeze_(0)\n model.eval()\n output = model(image)\n x = torch.topk(output, topk)\n list_of_class = {}\n np_log_probs = x[0][0].detach().numpy()\n tags = x[1][0].detach().numpy()\n for i in range(topk):\n for classes, idx in model.class_to_idx.items():\n if idx == tags[i]:\n list_of_class[classes] = np.exp(np_log_probs[i])\n return list_of_class",
"_____no_output_____"
],
[
"predict('flowers/valid/99/image_08063.jpg', m, 5)",
"_____no_output_____"
]
],
[
[
"## 检查运行状况\n\n你已经可以使用训练的模型做出预测,现在检查模型的性能如何。即使测试准确率很高,始终有必要检查是否存在明显的错误。使用 `matplotlib` 将前 5 个类别的概率以及输入图像绘制为条形图,应该如下所示:\n\n<img src='assets/inference_example.png' width=300px>\n\n你可以使用 `cat_to_name.json` 文件(应该之前已经在 notebook 中加载该文件)将类别整数编码转换为实际花卉名称。要将 PyTorch 张量显示为图像,请使用定义如下的 `imshow` 函数。",
"_____no_output_____"
]
],
[
[
"# TODO: Display an image along with the top 5 classes\npath = 'flowers/valid/99/image_08063.jpg' \nim = torch.from_numpy(process_image(path))\nimshow(im)",
"_____no_output_____"
],
[
"def showbarplot(cat_to_name, dictionary):\n name_of_class = []\n probs = []\n for classes, prob in dictionary.items():\n name_of_class.append(cat_to_name[classes])\n probs.append(prob)\n plt.bar(name_of_class, probs);\n plt.xticks(rotation=30);\n plt.xlabel('type of flowers');\n plt.ylabel('probabilities');\n plt.title('Top guesses of flower categories and their probabilities');",
"_____no_output_____"
],
[
"list_of_class = predict('flowers/valid/99/image_08063.jpg', m, 5)\nshowbarplot(cat_to_name, list_of_class)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
4a56f0fd4661516a46f525ee35259861233e0289
| 7,458 |
ipynb
|
Jupyter Notebook
|
cifar10_classification.ipynb
|
ganeshpc/Cifar10_Classification
|
df38f598e610c4bc78ab05cca65c386c0e02ae7c
|
[
"MIT"
] | null | null | null |
cifar10_classification.ipynb
|
ganeshpc/Cifar10_Classification
|
df38f598e610c4bc78ab05cca65c386c0e02ae7c
|
[
"MIT"
] | null | null | null |
cifar10_classification.ipynb
|
ganeshpc/Cifar10_Classification
|
df38f598e610c4bc78ab05cca65c386c0e02ae7c
|
[
"MIT"
] | null | null | null | 24.29316 | 100 | 0.438455 |
[
[
[
"## Cifar10 image classification\n\nLoading data...",
"_____no_output_____"
]
],
[
[
"from keras.datasets import cifar10\n\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"(x_train, y_train), (x_test, y_test) = cifar10.load_data()\n",
"_____no_output_____"
]
],
[
[
"### Visualizing data",
"_____no_output_____"
]
],
[
[
"n = 6\n\nplt.figure(figsize=(20, 10))\n\nfor i in range(10):\n plt.imshow(x_train[i])\n plt.show()\n",
"_____no_output_____"
]
],
[
[
"### Required Imports",
"_____no_output_____"
]
],
[
[
"from keras.models import Sequential\nfrom keras.layers import Dense, Dropout, Flatten\nfrom keras.constraints import maxnorm\nfrom keras.optimizers import SGD\nfrom keras.layers.convolutional import Conv2D, MaxPooling2D\nfrom keras.utils import np_utils",
"_____no_output_____"
]
],
[
[
"### Data preprocessing",
"_____no_output_____"
]
],
[
[
"x_train = x_train.astype('float32')\nx_test = x_test.astype('float32')\n\nx_train = x_train / 255\nx_test = x_test / 255",
"_____no_output_____"
],
[
"y_train = np_utils.to_categorical(y_train)\ny_test = np_utils.to_categorical(y_test)\n\nnum_classes = y_train.shape[1]",
"_____no_output_____"
]
],
[
[
"### Model Creation",
"_____no_output_____"
]
],
[
[
"model = Sequential()\nmodel.add(Conv2D(32, (3, 3), input_shape=(32, 32, 3), padding='same',\n activation='relu', kernel_constraint=maxnorm(3)))\nmodel.add(Dropout(0.2))\nmodel.add(Conv2D(32, (3, 3), activation='relu', padding='same',\n kernel_constraint=maxnorm(3)))\nmodel.add(MaxPooling2D(pool_size=(2, 2)))\nmodel.add(Flatten())\nmodel.add(Dense(512, activation='relu', kernel_constraint=maxnorm(3)))\nmodel.add(Dropout(0.5))\nmodel.add(Dense(num_classes, activation='softmax'))",
"_____no_output_____"
],
[
"sgd = SGD(lr=0.01, momentum=0.9, decay=(0.01/25), nesterov=False)\nmodel.compile(loss='categorical_crossentropy', optimizer=sgd,\n metrics=['accuracy'])",
"_____no_output_____"
],
[
"model.summary()",
"_____no_output_____"
]
],
[
[
"### Training the model",
"_____no_output_____"
]
],
[
[
"model.fit(x_train, y_train, validation_data=(x_test, y_test), epochs=50, batch_size=32)\n\nmodel.save('/content/drive/My Drive/Colab Notebooks/Cifar10/cifar_model_50_epochs.h5')",
"_____no_output_____"
]
],
[
[
"### Testing model",
"_____no_output_____"
]
],
[
[
"results={\n 0:'aeroplane',\n 1:'automobile',\n 2:'bird',\n 3:'cat',\n 4:'deer',\n 5:'dog',\n 6:'frog',\n 7:'horse',\n 8:'ship',\n 9:'truck'\n}\nfrom PIL import Image\nimport numpy as np\n\nfor i in range(9):\n path = \"/content/img\" + str(i) + \".jpg\"\n im=Image.open(path)\n # the input image is required to be in the shape of dataset, i.e (32,32,3)\n \n im=im.resize((32,32))\n plt.imshow(im)\n plt.show()\n im=np.expand_dims(im,axis=0)\n im=np.array(im)\n pred=model.predict_classes([im])[0]\n print(pred,results[pred])",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
4a56f9416af8225be8dd949764b216ebed3ed94a
| 150,794 |
ipynb
|
Jupyter Notebook
|
source/industry/telecom/notebooks/Ml-Telecom-NaiveBayes.ipynb
|
Devesh929/Telecom-Data-insight-project
|
a89bc9d5f4cc5dd5e11d221bd2cd7b34e5288213
|
[
"Apache-2.0"
] | 20 |
2018-11-08T13:58:16.000Z
|
2021-08-21T04:34:35.000Z
|
source/industry/telecom/notebooks/Ml-Telecom-NaiveBayes.ipynb
|
Devesh929/Telecom-Data-insight-project
|
a89bc9d5f4cc5dd5e11d221bd2cd7b34e5288213
|
[
"Apache-2.0"
] | 2 |
2019-09-18T01:10:00.000Z
|
2020-04-29T14:33:22.000Z
|
source/industry/telecom/notebooks/Ml-Telecom-NaiveBayes.ipynb
|
Devesh929/Telecom-Data-insight-project
|
a89bc9d5f4cc5dd5e11d221bd2cd7b34e5288213
|
[
"Apache-2.0"
] | 26 |
2018-11-09T13:16:12.000Z
|
2021-09-16T21:15:30.000Z
| 73.4148 | 17,052 | 0.665285 |
[
[
[
"# Machine Learning for Telecom with Naive Bayes",
"_____no_output_____"
],
[
"# Introduction",
"_____no_output_____"
],
[
"Machine Learning for CallDisconnectReason is a notebook which demonstrates exploration of dataset and CallDisconnectReason classification with Spark ml Naive Bayes Algorithm.\n",
"_____no_output_____"
]
],
[
[
"from pyspark.sql.types import *\nfrom pyspark.sql import SparkSession\nfrom sagemaker import get_execution_role\nimport sagemaker_pyspark\n\n\nrole = get_execution_role()\n\n# Configure Spark to use the SageMaker Spark dependency jars\njars = sagemaker_pyspark.classpath_jars()\n\nclasspath = \":\".join(sagemaker_pyspark.classpath_jars())\n\nspark = SparkSession.builder.config(\"spark.driver.extraClassPath\", classpath)\\\n .master(\"local[*]\").getOrCreate()",
"_____no_output_____"
]
],
[
[
"Using S3 Select, enables applications to retrieve only a subset of data from an object by using simple SQL expressions. By using S3 Select to retrieve only the data, you can achieve drastic performance increases – in many cases you can get as much as a 400% improvement.\n\n- _We first read a parquet compressed format of CDR dataset using s3select which has already been processed by Glue._\n",
"_____no_output_____"
]
],
[
[
"cdr_start_loc = \"<%CDRStartFile%>\"\ncdr_stop_loc = \"<%CDRStopFile%>\"\ncdr_start_sample_loc = \"<%CDRStartSampleFile%>\"\ncdr_stop_sample_loc = \"<%CDRStopSampleFile%>\"\n\ndf = spark.read.format(\"s3select\").parquet(cdr_stop_sample_loc)\ndf.createOrReplaceTempView(\"cdr\")",
"_____no_output_____"
],
[
"durationDF = spark.sql(\"SELECT _c13 as CallServiceDuration FROM cdr where _c0 = 'STOP'\")\ndurationDF.count()",
"_____no_output_____"
]
],
[
[
"# Exploration of Data",
"_____no_output_____"
],
[
"- _We see how we can explore and visualize the dataset used for processing. Here we create a bar chart representation of CallServiceDuration from CDR dataset._",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\ndurationpd = durationDF.toPandas().astype(int) ",
"_____no_output_____"
],
[
"durationpd.plot(kind='bar',stacked=True,width=1)",
"_____no_output_____"
]
],
[
[
"- _We can represent the data and visualize with a box plot. The box extends from the lower to upper quartile values of the data, with a line at the median._",
"_____no_output_____"
]
],
[
[
"color = dict(boxes='DarkGreen', whiskers='DarkOrange',\n medians='DarkBlue', caps='Gray')\n\ndurationpd.plot.box(color=color, sym='r+')",
"_____no_output_____"
],
[
"from pyspark.sql.functions import col\ndurationDF = durationDF.withColumn(\"CallServiceDuration\", col(\"CallServiceDuration\").cast(DoubleType())) ",
"_____no_output_____"
]
],
[
[
"- _We can represent the data and visualize the data with histograms partitioned in different bins._",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\n\nbins, counts = durationDF.select('CallServiceDuration').rdd.flatMap(lambda x: x).histogram(durationDF.count())\nplt.hist(bins[:-1], bins=bins, weights=counts,color=['green'])",
"_____no_output_____"
],
[
"sqlDF = spark.sql(\"SELECT _c2 as Accounting_ID, _c19 as Calling_Number,_c20 as Called_Number, _c14 as CallDisconnectReason FROM cdr where _c0 = 'STOP'\")\nsqlDF.show()",
"+------------------+--------------+-------------+--------------------+\n| Accounting_ID|Calling_Number|Called_Number|CallDisconnectReason|\n+------------------+--------------+-------------+--------------------+\n|0x00016E0F5BDACAF7| 9645000046| 3512000046| 16|\n|0x00016E0F36A4A836| 9645000048| 3512000048| 16|\n|0x00016E0F4C261126| 9645000050| 3512000050| 16|\n|0x00016E0F4A446638| 9645000052| 3512000052| 16|\n|0x00016E0F4040CE81| 9645000054| 3512000054| 16|\n|0x00016E0F4D522D63| 9645000055| 3512000055| 16|\n|0x00016E0F5854A088| 9645000057| 3512000057| 16|\n|0x00016E0F7DFDA482| 9645000060| 3512000060| 16|\n|0x00016E0F65D65F76| 9645000062| 3512000062| 16|\n|0x00016E0F2378A4AE| 9645000064| 3512000064| 16|\n|0x00016E0F5003BC72| 9645000066| 3512000066| 16|\n| 0x00016E0F44702AB| 9645000067| 3512000067| 16|\n|0x00016E0F500EED75| 9645000069| 3512000069| 16|\n|0x00016E0F38D99C7D| 9645000071| 3512000071| 16|\n|0x00016E0F4D14C078| 9645000074| 3512000074| 16|\n|0x00016E0F4116E96C| 9645000075| 3512000075| 16|\n|0x00016E0F1F5CDE40| 9645000077| 3512000077| 16|\n|0x00016E0F1BFE3E2A| 9645000079| 3512000079| 16|\n|0x00016E0F7E203CC9| 9645000081| 3512000081| 16|\n| 0x00016E0F5B43F12| 9645000084| 3512000084| 16|\n+------------------+--------------+-------------+--------------------+\nonly showing top 20 rows\n\n"
]
],
[
[
"# Featurization ",
"_____no_output_____"
]
],
[
[
"from pyspark.ml.feature import StringIndexer\n\naccountIndexer = StringIndexer(inputCol=\"Accounting_ID\", outputCol=\"AccountingIDIndex\")\naccountIndexer.setHandleInvalid(\"skip\")\ntempdf1 = accountIndexer.fit(sqlDF).transform(sqlDF)\n\ncallingNumberIndexer = StringIndexer(inputCol=\"Calling_Number\", outputCol=\"Calling_NumberIndex\")\ncallingNumberIndexer.setHandleInvalid(\"skip\")\ntempdf2 = callingNumberIndexer.fit(tempdf1).transform(tempdf1)\n\ncalledNumberIndexer = StringIndexer(inputCol=\"Called_Number\", outputCol=\"Called_NumberIndex\")\ncalledNumberIndexer.setHandleInvalid(\"skip\")\ntempdf3 = calledNumberIndexer.fit(tempdf2).transform(tempdf2)",
"_____no_output_____"
],
[
"from pyspark.ml.feature import StringIndexer\n# Convert target into numerical categories\nlabelIndexer = StringIndexer(inputCol=\"CallDisconnectReason\", outputCol=\"label\")\nlabelIndexer.setHandleInvalid(\"skip\")",
"_____no_output_____"
],
[
"from pyspark.sql.functions import rand\n\ntrainingFraction = 0.75; \ntestingFraction = (1-trainingFraction);\nseed = 1234;\n\ntrainData, testData = tempdf3.randomSplit([trainingFraction, testingFraction], seed=seed);\n\n# CACHE TRAIN AND TEST DATA\ntrainData.cache()\ntestData.cache()\ntrainData.count(),testData.count()",
"_____no_output_____"
]
],
[
[
"# Analyzing the label distribution\n\n- We analyze the distribution of our target labels using a histogram where 16 represents Normal_Call_Clearing.",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\n\nnegcount = trainData.filter(\"CallDisconnectReason != 16\").count()\nposcount = trainData.filter(\"CallDisconnectReason == 16\").count()\n\nnegfrac = 100*float(negcount)/float(negcount+poscount)\nposfrac = 100*float(poscount)/float(poscount+negcount)\nind = [0.0,1.0]\nfrac = [negfrac,posfrac]\nwidth = 0.35\n\nplt.title('Label Distribution')\nplt.bar(ind, frac, width, color='r')\nplt.xlabel(\"CallDisconnectReason\")\nplt.ylabel('Percentage share')\nplt.xticks(ind,['0.0','1.0'])\nplt.show()",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt\n\nnegcount = testData.filter(\"CallDisconnectReason != 16\").count()\nposcount = testData.filter(\"CallDisconnectReason == 16\").count()\n\nnegfrac = 100*float(negcount)/float(negcount+poscount)\nposfrac = 100*float(poscount)/float(poscount+negcount)\nind = [0.0,1.0]\nfrac = [negfrac,posfrac]\nwidth = 0.35\n\nplt.title('Label Distribution')\nplt.bar(ind, frac, width, color='r')\nplt.xlabel(\"CallDisconnectReason\")\nplt.ylabel('Percentage share')\nplt.xticks(ind,['0.0','1.0'])\nplt.show()",
"_____no_output_____"
],
[
"from pyspark.ml.feature import VectorAssembler\n\nfrom pyspark.ml.feature import VectorAssembler\n\nvecAssembler = VectorAssembler(inputCols=[\"AccountingIDIndex\",\"Calling_NumberIndex\", \"Called_NumberIndex\"], outputCol=\"features\")\n",
"_____no_output_____"
]
],
[
[
"__Spark ML Naive Bayes__: \n Naive Bayes is a simple multiclass classification algorithm with the assumption of independence between every pair of features. Naive Bayes can be trained very efficiently. Within a single pass to the training data, it computes the conditional probability distribution of each feature given label, and then it applies Bayes’ theorem to compute the conditional probability distribution of label given an observation and use it for prediction.\n\n\n\n- _We use Spark ML Naive Bayes Algorithm and spark Pipeline to train the data set._",
"_____no_output_____"
]
],
[
[
"from pyspark.ml.classification import NaiveBayes\nfrom pyspark.ml.clustering import KMeans\nfrom pyspark.ml import Pipeline\n\n# Train a NaiveBayes model\nnb = NaiveBayes(smoothing=1.0, modelType=\"multinomial\")\n\n# Chain labelIndexer, vecAssembler and NBmodel in a \npipeline = Pipeline(stages=[labelIndexer,vecAssembler, nb])\n\n# Run stages in pipeline and train model\nmodel = pipeline.fit(trainData)",
"_____no_output_____"
],
[
" # Run inference on the test data and show some results\npredictions = model.transform(testData)\npredictions.printSchema()\npredictions.show()",
"root\n |-- Accounting_ID: string (nullable = true)\n |-- Calling_Number: string (nullable = true)\n |-- Called_Number: string (nullable = true)\n |-- CallDisconnectReason: string (nullable = true)\n |-- AccountingIDIndex: double (nullable = false)\n |-- Calling_NumberIndex: double (nullable = false)\n |-- Called_NumberIndex: double (nullable = false)\n |-- label: double (nullable = false)\n |-- features: vector (nullable = true)\n |-- rawPrediction: vector (nullable = true)\n |-- probability: vector (nullable = true)\n |-- prediction: double (nullable = false)\n\n+------------------+--------------+-------------+--------------------+-----------------+-------------------+------------------+-----+-------------------+--------------------+-----------+----------+\n| Accounting_ID|Calling_Number|Called_Number|CallDisconnectReason|AccountingIDIndex|Calling_NumberIndex|Called_NumberIndex|label| features| rawPrediction|probability|prediction|\n+------------------+--------------+-------------+--------------------+-----------------+-------------------+------------------+-----+-------------------+--------------------+-----------+----------+\n| 0x00016E0F1005CE4| 9645000075| 3512000075| 16| 2577.0| 38.0| 39.0| 0.0| [2577.0,38.0,39.0]|[-440.84691368645...| [1.0]| 0.0|\n|0x00016E0F100A017D| 9645000010| 3512000010| 16| 21710.0| 33.0| 35.0| 0.0|[21710.0,33.0,35.0]|[-560.2845426594596]| [1.0]| 0.0|\n|0x00016E0F100AF60A| 9645000077| 3512000077| 16| 6832.0| 34.0| 45.0| 0.0| [6832.0,34.0,45.0]|[-489.13697166313...| [1.0]| 0.0|\n|0x00016E0F104511E6| 9645000059| 3512000059| 16| 9768.0| 25.0| 21.0| 0.0| [9768.0,25.0,21.0]|[-335.75198901002...| [1.0]| 0.0|\n|0x00016E0F107E0142| 9645000038| 3512000038| 16| 13013.0| 12.0| 11.0| 0.0|[13013.0,12.0,11.0]| [-239.387649472332]| [1.0]| 0.0|\n|0x00016E0F107F6253| 9645000093| 3512000093| 16| 13936.0| 97.0| 98.0| 0.0|[13936.0,97.0,98.0]|[-1181.6164141280...| [1.0]| 0.0|\n|0x00016E0F109060DA| 9645000068| 3512000068| 16| 13255.0| 21.0| 13.0| 0.0|[13255.0,21.0,13.0]| [-301.258614162017]| [1.0]| 0.0|\n|0x00016E0F109962EA| 9645000014| 3512000014| 16| 12198.0| 57.0| 56.0| 0.0|[12198.0,57.0,56.0]|[-720.9963093850602]| [1.0]| 0.0|\n|0x00016E0F10AD5AA3| 9645000077| 3512000077| 16| 17980.0| 34.0| 45.0| 0.0|[17980.0,34.0,45.0]|[-587.2075683476078]| [1.0]| 0.0|\n|0x00016E0F10B7685D| 9645000080| 3512000080| 16| 19118.0| 51.0| 62.0| 0.0|[19118.0,51.0,62.0]|[-781.8683149993153]| [1.0]| 0.0|\n|0x00016E0F10BF7B70| 9645000092| 3512000092| 16| 13766.0| 18.0| 20.0| 0.0|[13766.0,18.0,20.0]|[-327.4738938976721]| [1.0]| 0.0|\n|0x00016E0F10C18111| 9645000037| 3512000037| 16| 4031.0| 83.0| 85.0| 0.0| [4031.0,83.0,85.0]|[-947.8468165201091]| [1.0]| 0.0|\n|0x00016E0F11135782| 9645000025| 3512000025| 16| 4836.0| 35.0| 32.0| 0.0| [4836.0,35.0,32.0]|[-406.41238288465...| [1.0]| 0.0|\n|0x00016E0F113C9E41| 9645000074| 3512000074| 16| 15726.0| 85.0| 83.0| 0.0|[15726.0,85.0,83.0]|[-1050.7308703158...| [1.0]| 0.0|\n|0x00016E0F11525D52| 9645000023| 3512000023| 16| 15228.0| 62.0| 63.0| 0.0|[15228.0,62.0,63.0]|[-812.8214011632385]| [1.0]| 0.0|\n|0x00016E0F11533B86| 9645000023| 3512000023| 16| 899.0| 62.0| 63.0| 0.0| [899.0,62.0,63.0]|[-686.7670793214941]| [1.0]| 0.0|\n|0x00016E0F1156E519| 9645000047| 3512000047| 16| 14638.0| 42.0| 34.0| 0.0|[14638.0,42.0,34.0]|[-541.5216249696589]| [1.0]| 0.0|\n|0x00016E0F1159A234| 9645000005| 3512000005| 16| 11407.0| 16.0| 26.0| 0.0|[11407.0,16.0,26.0]|[-328.44207005065...| [1.0]| 0.0|\n|0x00016E0F11780902| 9645000099| 3512000099| 16| 19361.0| 27.0| 27.0| 0.0|[19361.0,27.0,27.0]|[-463.58856733144...| [1.0]| 0.0|\n|0x00016E0F117D2932| 9645000080| 3512000080| 16| 13710.0| 51.0| 62.0| 0.0|[13710.0,51.0,62.0]|[-734.2933430877965]| [1.0]| 0.0|\n+------------------+--------------+-------------+--------------------+-----------------+-------------------+------------------+-----+-------------------+--------------------+-----------+----------+\nonly showing top 20 rows\n\n"
],
[
"predictiondf = predictions.select(\"label\", \"prediction\", \"probability\")",
"_____no_output_____"
],
[
"pddf_pred = predictions.toPandas()",
"_____no_output_____"
],
[
"pddf_pred",
"_____no_output_____"
]
],
[
[
"- _We use Scatter plot for visualization and represent the dataset._",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nimport numpy as np\n# Set the size of the plot\nplt.figure(figsize=(14,7))\n \n# Create a colormap\ncolormap = np.array(['red', 'lime', 'black'])\n \n# Plot CDR\nplt.subplot(1, 2, 1)\nplt.scatter(pddf_pred.Calling_NumberIndex, pddf_pred.Called_NumberIndex, c=pddf_pred.prediction)\nplt.title('CallDetailRecord')\nplt.show()",
"_____no_output_____"
]
],
[
[
"# Evaluation",
"_____no_output_____"
]
],
[
[
"from pyspark.ml.evaluation import MulticlassClassificationEvaluator\n\nevaluator = MulticlassClassificationEvaluator(labelCol=\"label\", predictionCol=\"prediction\",\n metricName=\"accuracy\")\naccuracy = evaluator.evaluate(predictiondf)\nprint(accuracy)",
"1.0\n"
]
],
[
[
"# Confusion Matrix",
"_____no_output_____"
]
],
[
[
"from sklearn.metrics import confusion_matrix\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport seaborn as sn\n\noutdataframe = predictiondf.select(\"prediction\", \"label\")\npandadf = outdataframe.toPandas()\nnpmat = pandadf.values\nlabels = npmat[:,0]\npredicted_label = npmat[:,1]\n\ncnf_matrix = confusion_matrix(labels, predicted_label)\n",
"_____no_output_____"
],
[
"import numpy as np\n\ndef plot_confusion_matrix(cm,\n target_names,\n title='Confusion matrix',\n cmap=None,\n normalize=True):\n\n import matplotlib.pyplot as plt\n import numpy as np\n import itertools\n\n accuracy = np.trace(cm) / float(np.sum(cm))\n misclass = 1 - accuracy\n\n if cmap is None:\n cmap = plt.get_cmap('Blues')\n\n plt.figure(figsize=(8, 6))\n plt.imshow(cm, interpolation='nearest', cmap=cmap)\n plt.title(title)\n plt.colorbar()\n\n if target_names is not None:\n tick_marks = np.arange(len(target_names))\n plt.xticks(tick_marks, target_names, rotation=45)\n plt.yticks(tick_marks, target_names)\n\n if normalize:\n cm = cm.astype('float') / cm.sum(axis=1)[:, np.newaxis]\n\n thresh = cm.max() / 1.5 if normalize else cm.max() / 2\n for i, j in itertools.product(range(cm.shape[0]), range(cm.shape[1])):\n if normalize:\n plt.text(j, i, \"{:0.4f}\".format(cm[i, j]),\n horizontalalignment=\"center\",\n color=\"white\" if cm[i, j] > thresh else \"black\")\n else:\n plt.text(j, i, \"{:,}\".format(cm[i, j]),\n horizontalalignment=\"center\",\n color=\"white\" if cm[i, j] > thresh else \"black\")\n\n\n plt.tight_layout()\n plt.ylabel('label')\n plt.xlabel('Predicted \\naccuracy={:0.4f}; misclass={:0.4f}'.format(accuracy, misclass))\n plt.show()",
"_____no_output_____"
],
[
"plot_confusion_matrix(cnf_matrix,\n normalize = False,\n target_names = ['Positive', 'Negative'],\n title = \"Confusion Matrix\")",
"_____no_output_____"
],
[
"from pyspark.mllib.evaluation import MulticlassMetrics\n# Create (prediction, label) pairs\npredictionAndLabel = predictiondf.select(\"prediction\", \"label\").rdd\n\n# Generate confusion matrix\nmetrics = MulticlassMetrics(predictionAndLabel)\nprint(metrics.confusionMatrix())",
"DenseMatrix([[5469.]])\n\n"
]
],
[
[
"# Cross Validation",
"_____no_output_____"
]
],
[
[
"from pyspark.ml.tuning import ParamGridBuilder, CrossValidator\n\n# Create ParamGrid and Evaluator for Cross Validation\nparamGrid = ParamGridBuilder().addGrid(nb.smoothing, [0.0, 0.2, 0.4, 0.6, 0.8, 1.0]).build()\ncvEvaluator = MulticlassClassificationEvaluator(metricName=\"accuracy\")",
"_____no_output_____"
],
[
"# Run Cross-validation\ncv = CrossValidator(estimator=pipeline, estimatorParamMaps=paramGrid, evaluator=cvEvaluator)\ncvModel = cv.fit(trainData)",
"_____no_output_____"
],
[
"# Make predictions on testData. cvModel uses the bestModel.\ncvPredictions = cvModel.transform(testData)",
"_____no_output_____"
],
[
"cvPredictions.select(\"label\", \"prediction\", \"probability\").show()",
"+-----+----------+-----------+\n|label|prediction|probability|\n+-----+----------+-----------+\n| 0.0| 0.0| [1.0]|\n| 0.0| 0.0| [1.0]|\n| 0.0| 0.0| [1.0]|\n| 0.0| 0.0| [1.0]|\n| 0.0| 0.0| [1.0]|\n| 0.0| 0.0| [1.0]|\n| 0.0| 0.0| [1.0]|\n| 0.0| 0.0| [1.0]|\n| 0.0| 0.0| [1.0]|\n| 0.0| 0.0| [1.0]|\n| 0.0| 0.0| [1.0]|\n| 0.0| 0.0| [1.0]|\n| 0.0| 0.0| [1.0]|\n| 0.0| 0.0| [1.0]|\n| 0.0| 0.0| [1.0]|\n| 0.0| 0.0| [1.0]|\n| 0.0| 0.0| [1.0]|\n| 0.0| 0.0| [1.0]|\n| 0.0| 0.0| [1.0]|\n| 0.0| 0.0| [1.0]|\n+-----+----------+-----------+\nonly showing top 20 rows\n\n"
],
[
"# Evaluate bestModel found from Cross Validation\nevaluator.evaluate(cvPredictions)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
4a56ff09ec1c69a146bc897fc7892cab7bcc5340
| 293,077 |
ipynb
|
Jupyter Notebook
|
DataCamp_spark/Feature Engineering with PySpark/Feature Engineering with PySpark.ipynb
|
obirgul/SPARK
|
e3b4fcf32b2c2811c87af47dcc87cd2768eb2bad
|
[
"Apache-2.0"
] | null | null | null |
DataCamp_spark/Feature Engineering with PySpark/Feature Engineering with PySpark.ipynb
|
obirgul/SPARK
|
e3b4fcf32b2c2811c87af47dcc87cd2768eb2bad
|
[
"Apache-2.0"
] | null | null | null |
DataCamp_spark/Feature Engineering with PySpark/Feature Engineering with PySpark.ipynb
|
obirgul/SPARK
|
e3b4fcf32b2c2811c87af47dcc87cd2768eb2bad
|
[
"Apache-2.0"
] | null | null | null | 91.07427 | 51,012 | 0.794661 |
[
[
[
"# Feature Engineering with PySpark",
"_____no_output_____"
],
[
"## Exploratory Data Analysis",
"_____no_output_____"
]
],
[
[
"import pyspark as sp",
"_____no_output_____"
],
[
"sp.version",
"_____no_output_____"
],
[
"import sys\nprint(sys.version_info)",
"sys.version_info(major=3, minor=6, micro=7, releaselevel='final', serial=0)\n"
],
[
"sys.version",
"_____no_output_____"
]
],
[
[
"import os\nos.environ[\"JAVA_HOME\"] = \"/Library/Java/JavaVirtualMachines/jdk1.8.0_151.jdk/Contents/Home\"",
"_____no_output_____"
]
],
[
[
"sc = sp.SparkContext.getOrCreate()",
"_____no_output_____"
],
[
"sc.version",
"_____no_output_____"
],
[
"# spark session\n# Import SparkSession from pyspark.sql\nfrom pyspark.sql import SparkSession \n\n# Create a session as spark \nspark = SparkSession.builder.getOrCreate()",
"_____no_output_____"
],
[
"df = spark.read.csv('2017_StPaul_MN_Real_Estate.csv', header=True)\ndf.columns",
"_____no_output_____"
],
[
"df.count()",
"_____no_output_____"
],
[
"df.dtypes",
"_____no_output_____"
]
],
[
[
"### What are we predicting?",
"_____no_output_____"
]
],
[
[
"# Select our dependent variable\nY_df = df.select(['SalesClosePrice'])\n\n# Display summary statistics\nY_df.describe().show()",
"+-------+------------------+\n|summary| SalesClosePrice|\n+-------+------------------+\n| count| 5000|\n| mean| 262804.4668|\n| stddev|140559.82591998563|\n| min| 100000|\n| max| 99900|\n+-------+------------------+\n\n"
]
],
[
[
"Looks like we need to convert the data type of SalesClosePrice:",
"_____no_output_____"
]
],
[
[
"# convert the data type of SalesClosePrice to integer\ndf = df.withColumn(\"SalesClosePrice\", df.SalesClosePrice.cast(\"integer\"))",
"_____no_output_____"
],
[
"df.select('SalesClosePrice').describe().show()",
"+-------+------------------+\n|summary| SalesClosePrice|\n+-------+------------------+\n| count| 5000|\n| mean| 262804.4668|\n| stddev|140559.82591998563|\n| min| 48000|\n| max| 1700000|\n+-------+------------------+\n\n"
],
[
"df = df.withColumn(\"AssessedValuation\", df.AssessedValuation.cast(\"double\"))\ndf = df.withColumn(\"AssociationFee\", df.AssociationFee.cast(\"bigint\"))\ndf = df.withColumn(\"SQFTBELOWGROUND\", df.SQFTBELOWGROUND.cast(\"bigint\"))",
"_____no_output_____"
],
[
"required_dtypes = [('NO', 'bigint'),\n ('MLSID', 'string'),\n ('STREETNUMBERNUMERIC', 'bigint'),\n ('STREETADDRESS', 'string'),\n ('STREETNAME', 'string'),\n ('POSTALCODE', 'bigint'),\n ('STATEORPROVINCE', 'string'),\n ('CITY', 'string'),\n ('SALESCLOSEPRICE', 'bigint'),\n ('LISTDATE', 'string'),\n ('LISTPRICE', 'bigint'),\n ('LISTTYPE', 'string'),\n ('ORIGINALLISTPRICE', 'bigint'),\n ('PRICEPERTSFT', 'double'),\n ('FOUNDATIONSIZE', 'bigint'),\n ('FENCE', 'string'),\n ('MAPLETTER', 'string'),\n ('LOTSIZEDIMENSIONS', 'string'),\n ('SCHOOLDISTRICTNUMBER', 'string'),\n ('DAYSONMARKET', 'bigint'),\n ('OFFMARKETDATE', 'string'),\n ('FIREPLACES', 'bigint'),\n ('ROOMAREA4', 'string'),\n ('ROOMTYPE', 'string'),\n ('ROOF', 'string'),\n ('ROOMFLOOR4', 'string'),\n ('POTENTIALSHORTSALE', 'string'),\n ('POOLDESCRIPTION', 'string'),\n ('PDOM', 'bigint'),\n ('GARAGEDESCRIPTION', 'string'),\n ('SQFTABOVEGROUND', 'bigint'),\n ('TAXES', 'bigint'),\n ('ROOMFLOOR1', 'string'),\n ('ROOMAREA1', 'string'),\n ('TAXWITHASSESSMENTS', 'double'),\n ('TAXYEAR', 'bigint'),\n ('LIVINGAREA', 'bigint'),\n ('UNITNUMBER', 'string'),\n ('YEARBUILT', 'bigint'),\n ('ZONING', 'string'),\n ('STYLE', 'string'),\n ('ACRES', 'double'),\n ('COOLINGDESCRIPTION', 'string'),\n ('APPLIANCES', 'string'),\n ('BACKONMARKETDATE', 'double'),\n ('ROOMFAMILYCHAR', 'string'),\n ('ROOMAREA3', 'string'),\n ('EXTERIOR', 'string'),\n ('ROOMFLOOR3', 'string'),\n ('ROOMFLOOR2', 'string'),\n ('ROOMAREA2', 'string'),\n ('DININGROOMDESCRIPTION', 'string'),\n ('BASEMENT', 'string'),\n ('BATHSFULL', 'bigint'),\n ('BATHSHALF', 'bigint'),\n ('BATHQUARTER', 'bigint'),\n ('BATHSTHREEQUARTER', 'double'),\n ('CLASS', 'string'),\n ('BATHSTOTAL', 'bigint'),\n ('BATHDESC', 'string'),\n ('ROOMAREA5', 'string'),\n ('ROOMFLOOR5', 'string'),\n ('ROOMAREA6', 'string'),\n ('ROOMFLOOR6', 'string'),\n ('ROOMAREA7', 'string'),\n ('ROOMFLOOR7', 'string'),\n ('ROOMAREA8', 'string'),\n ('ROOMFLOOR8', 'string'),\n ('BEDROOMS', 'bigint'),\n ('SQFTBELOWGROUND', 'bigint'),\n ('ASSUMABLEMORTGAGE', 'string'),\n ('ASSOCIATIONFEE', 'bigint'),\n ('ASSESSMENTPENDING', 'string'),\n ('ASSESSEDVALUATION', 'double')]",
"_____no_output_____"
],
[
"old_columns = df.columns",
"_____no_output_____"
],
[
"new_columns = [c for c, d in required_dtypes]",
"_____no_output_____"
],
[
"for n, o in zip(new_columns, old_columns): \n df = df.withColumnRenamed(o, n)",
"_____no_output_____"
]
],
[
[
"### Verifying Data Load",
"_____no_output_____"
]
],
[
[
"def check_load(df, num_records, num_columns):\n # Takes a dataframe and compares record and column counts to input\n # Message to return if the critera below aren't met\n message = 'Validation Failed'\n # Check number of records\n if num_records == df.count():\n # Check number of columns\n if num_columns == len(df.columns):\n # Success message\n message = 'Validation Passed'\n return message\n\n# Print the data validation message\nprint(check_load(df, 5000, 74))",
"Validation Passed\n"
]
],
[
[
"### Verifying DataTypes",
"_____no_output_____"
]
],
[
[
"validation_dict = {'ASSESSMENTPENDING': 'string',\n 'ASSESSEDVALUATION': 'double',\n 'ASSOCIATIONFEE': 'bigint',\n 'ASSUMABLEMORTGAGE': 'string',\n 'SQFTBELOWGROUND': 'bigint'}",
"_____no_output_____"
],
[
"# create list of actual dtypes to check\nactual_dtypes_list = df.dtypes\n\n# Iterate through the list of actual dtypes tuples\nfor attribute_tuple in actual_dtypes_list:\n \n # Check if column name is dictionary of expected dtypes\n col_name = attribute_tuple[0]\n if col_name in validation_dict:\n\n # Compare attribute types\n col_type = attribute_tuple[1]\n if col_type == validation_dict[col_name]:\n print(col_name + ' has expected dtype.')",
"SQFTBELOWGROUND has expected dtype.\nASSUMABLEMORTGAGE has expected dtype.\nASSOCIATIONFEE has expected dtype.\nASSESSMENTPENDING has expected dtype.\nASSESSEDVALUATION has expected dtype.\n"
]
],
[
[
"### Using `Corr()`",
"_____no_output_____"
]
],
[
[
"for required_type, current_column in zip(required_dtypes, df.columns):\n # since the required and current column names are the exact order we can do:\n if required_type[1] != 'string':\n# df = df.withColumn(\"{:}\".format(current_column), df[\"`{:}`\".format(current_column)].cast(req[1]))\n df = df.withColumn(current_column, df[\"{:}\".format(current_column)].cast(required_type[1]))",
"_____no_output_____"
],
[
"check_columns = ['FOUNDATIONSIZE',\n 'DAYSONMARKET',\n 'FIREPLACES',\n 'PDOM',\n 'SQFTABOVEGROUND',\n 'TAXES',\n 'TAXWITHASSESSMENTS',\n 'TAXYEAR',\n 'LIVINGAREA',\n 'YEARBUILT',\n 'ACRES',\n 'BACKONMARKETDATE',\n 'BATHSFULL',\n 'BATHSHALF',\n 'BATHQUARTER',\n 'BATHSTHREEQUARTER',\n 'BATHSTOTAL',\n 'BEDROOMS',\n 'SQFTBELOWGROUND',\n 'ASSOCIATIONFEE',\n 'ASSESSEDVALUATION']",
"_____no_output_____"
],
[
"# Name and value of col with max corr\ncorr_max = 0\ncorr_max_col = check_columns[0]\n\n# Loop to check all columns contained in list\nfor col in check_columns:\n # Check the correlation of a pair of columns\n corr_val = df.corr(col, 'SALESCLOSEPRICE')\n # Logic to compare corr_max with current corr_val\n if corr_val > corr_max:\n # Update the column name and corr value\n corr_max = corr_val\n corr_max_col = col\n\nprint(corr_max_col)",
"LIVINGAREA\n"
]
],
[
[
"### Using Visualizations: distplot",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\n\nimport matplotlib.pyplot as plt\nimport seaborn as sns",
"_____no_output_____"
],
[
"# Select a single column and sample and convert to pandas\n# sample 50% and not use replacement and setting the random seed to 42.\nsample_df = df.select(['LISTPRICE']).sample(False, .5, 42)\npandas_df = sample_df.toPandas()\n\n# Plot distribution of pandas_df and display plot\nsns.distplot(pandas_df)\nplt.show()\n\n# Import skewness function\nfrom pyspark.sql.functions import skewness\n\n# Compute and print skewness of LISTPRICE\nprint(df.agg({'LISTPRICE': 'skewness'}).collect())",
"_____no_output_____"
]
],
[
[
"We can use the skewness function to verify this numerically rather than visually.",
"_____no_output_____"
],
[
"### Using Visualizations: lmplot",
"_____no_output_____"
]
],
[
[
"# Select a the relevant columns and sample\nsample_df = df.select(['SALESCLOSEPRICE','LIVINGAREA']).sample(False, .5, 42)\n\n# Convert to pandas dataframe\npandas_df = sample_df.toPandas()\n\n# Linear model plot of pandas_df\nsns.lmplot(x='LIVINGAREA', y='SALESCLOSEPRICE', data=pandas_df)",
"_____no_output_____"
]
],
[
[
"we can see that as LivingArea increases, the price of the home increases at a relatively steady rate",
"_____no_output_____"
],
[
"## Wrangling with Spark Functions",
"_____no_output_____"
],
[
"### Dropping a list of columns",
"_____no_output_____"
]
],
[
[
"# List of columns to remove from dataset\ncols_to_drop = ['STREETNUMBERNUMERIC', 'LOTSIZEDIMENSIONS']\n\n# Drop columns in list\ndf = df.drop(*cols_to_drop)",
"_____no_output_____"
]
],
[
[
"We can always come back to these after our intial model if we need more information.",
"_____no_output_____"
],
[
"### Using text filters to remove records",
"_____no_output_____"
]
],
[
[
"# Inspect unique values in the column 'ASSUMABLEMORTGAGE'\ndf.select(['ASSUMABLEMORTGAGE']).distinct().show()\n\n# List of possible values containing 'yes'\nyes_values = ['Yes w/ Qualifying', 'Yes w/No Qualifying']\n\n# Filter the text values out of df but keep null values\ntext_filter = ~df['ASSUMABLEMORTGAGE'].isin(yes_values) | df['ASSUMABLEMORTGAGE'].isNull()\ndf = df.where(text_filter)\n\n# Print count of remaining records\nprint(df.count())",
"+-------------------+\n| ASSUMABLEMORTGAGE|\n+-------------------+\n| Yes w/ Qualifying|\n| Information Coming|\n| null|\n|Yes w/No Qualifying|\n| Not Assumable|\n+-------------------+\n\n4976\n"
]
],
[
[
"### Filtering numeric fields conditionally",
"_____no_output_____"
]
],
[
[
"from pyspark.sql.functions import log\n\ndf = df.withColumn('log_SalesClosePrice', log('SalesClosePrice'))",
"_____no_output_____"
],
[
"from pyspark.sql.functions import mean, stddev\n\n# Calculate values used for outlier filtering\nmean_val = df.agg({'log_SalesClosePrice': 'mean'}).collect()[0][0]\nstddev_val = df.agg({'log_SalesClosePrice': 'stddev'}).collect()[0][0]\n\n# Create three standard deviation (μ ± 3σ) lower and upper bounds for data\nlow_bound = mean_val - (3 * stddev_val)\nhi_bound = mean_val + (3 * stddev_val)\n\n# Filter the data to fit between the lower and upper bounds\ndf = df.where((df['log_SalesClosePrice'] < hi_bound) & (df['log_SalesClosePrice'] > low_bound))",
"_____no_output_____"
]
],
[
[
"### Custom Percentage Scaling",
"_____no_output_____"
]
],
[
[
"from pyspark.sql.functions import round",
"_____no_output_____"
],
[
"# Define max and min values and collect them\nmax_days = df.agg({'DAYSONMARKET': 'max'}).collect()[0][0]\nmin_days = df.agg({'DAYSONMARKET': 'min'}).collect()[0][0]\n\n# Create a new column based off the scaled data\ndf = df.withColumn('percentage_scaled_days', \n round((df['DAYSONMARKET'] - min_days) / (max_days - min_days)) * 100)\n\n# Calc max and min for new column\nprint(df.agg({'percentage_scaled_days': 'max'}).collect())\nprint(df.agg({'percentage_scaled_days': 'min'}).collect())",
"[Row(max(percentage_scaled_days)=100.0)]\n[Row(min(percentage_scaled_days)=0.0)]\n"
]
],
[
[
"### Scaling your scalers",
"_____no_output_____"
]
],
[
[
"def min_max_scaler(df, cols_to_scale):\n # Takes a dataframe and list of columns to minmax scale. Returns a dataframe.\n for col in cols_to_scale:\n # Define min and max values and collect them\n max_days = df.agg({col: 'max'}).collect()[0][0]\n min_days = df.agg({col: 'min'}).collect()[0][0]\n new_column_name = 'scaled_' + col\n # Create a new column based off the scaled data\n df = df.withColumn(new_column_name, \n (df[col] - min_days) / (max_days - min_days))\n return df\n \ndf = min_max_scaler(df, ['FOUNDATIONSIZE', 'DAYSONMARKET', 'FIREPLACES'])\n# Show that our data is now between 0 and 1\ndf[['DAYSONMARKET', 'scaled_DAYSONMARKET']].show()",
"+------------+--------------------+\n|DAYSONMARKET| scaled_DAYSONMARKET|\n+------------+--------------------+\n| 10|0.044444444444444446|\n| 4|0.017777777777777778|\n| 28| 0.12444444444444444|\n| 19| 0.08444444444444445|\n| 21| 0.09333333333333334|\n| 17| 0.07555555555555556|\n| 32| 0.14222222222222222|\n| 5|0.022222222222222223|\n| 23| 0.10222222222222223|\n| 73| 0.3244444444444444|\n| 80| 0.35555555555555557|\n| 79| 0.3511111111111111|\n| 12| 0.05333333333333334|\n| 1|0.004444444444444...|\n| 18| 0.08|\n| 2|0.008888888888888889|\n| 12| 0.05333333333333334|\n| 45| 0.2|\n| 31| 0.13777777777777778|\n| 16| 0.07111111111111111|\n+------------+--------------------+\nonly showing top 20 rows\n\n"
]
],
[
[
"### Correcting Right Skew Data",
"_____no_output_____"
]
],
[
[
"# Compute the skewness\nprint(df.agg({'YEARBUILT': 'skewness'}).collect())\n\n# Calculate the max year\nmax_year = df.agg({'YEARBUILT': 'max'}).collect()[0][0]\n\n# Create a new column of reflected data\ndf = df.withColumn('Reflect_YearBuilt', (max_year + 1) - df['YEARBUILT'])\n\n# Create a new column based reflected data\ndf = df.withColumn('adj_yearbuilt', 1 / log(df['Reflect_YearBuilt']))",
"[Row(skewness(YEARBUILT)=-0.25317374723020336)]\n"
]
],
[
[
"What you've seen here are only a few of the ways that you might try to make your data fit a normal distribution.",
"_____no_output_____"
],
[
"### Visualizing Missing Data",
"_____no_output_____"
]
],
[
[
"columns = ['APPLIANCES',\n 'BACKONMARKETDATE',\n 'ROOMFAMILYCHAR',\n 'BASEMENT',\n 'DININGROOMDESCRIPTION']",
"_____no_output_____"
],
[
"df.select(columns).show()",
"+--------------------+----------------+--------------------+--------------------+---------------------+\n| APPLIANCES|BACKONMARKETDATE| ROOMFAMILYCHAR| BASEMENT|DININGROOMDESCRIPTION|\n+--------------------+----------------+--------------------+--------------------+---------------------+\n|Range, Dishwasher...| null| null| Full| Eat In Kitchen|\n|Range, Microwave,...| null| Lower Level|Full, Partial Fin...| Informal Dining R...|\n|Range, Microwave,...| null| null|Full, Crawl Space...| Informal Dining Room|\n|Range, Washer, Dryer| null| Main Level|Full, Partial Fin...| null|\n|Range, Exhaust Fa...| null| Lower Level|Walkout, Full, Fi...| Breakfast Area, K...|\n|Range, Microwave,...| null| null|Full, Sump Pump, ...| Separate/Formal D...|\n|Range, Microwave,...| null| Loft| Slab| Informal Dining R...|\n|Range, Microwave,...| null|Main Level, Famil...| None| Informal Dining R...|\n|Range, Microwave,...| null| null| None| Informal Dining R...|\n|Range, Microwave,...| null|Main Level, Famil...| None| Informal Dining R...|\n|Range, Microwave,...| null|Main Level, Famil...| None| Informal Dining R...|\n|Range, Microwave,...| null| null| None| Informal Dining R...|\n|Range, Microwave,...| null|Main Level, Famil...| None| Informal Dining R...|\n|Range, Microwave,...| null|Main Level, Great...| None| Informal Dining R...|\n|Cooktop, Wall Ove...| null| null|Full, Concrete Bl...| Informal Dining R...|\n|Range, Microwave,...| null| Loft| None| Informal Dining R...|\n|Range, Microwave,...| null| Loft| None| Informal Dining R...|\n|Range, Microwave,...| null|Main Level, Great...| None| Informal Dining R...|\n|Range, Microwave,...| null| Loft| None| Informal Dining R...|\n|Range, Microwave,...| null| Loft| None| Informal Dining R...|\n+--------------------+----------------+--------------------+--------------------+---------------------+\nonly showing top 20 rows\n\n"
],
[
"# Sample the dataframe and convert to Pandas\nsample_df = df.select(columns).sample(False, 0.5, 42)\npandas_df = sample_df.toPandas()\n\n# Convert all values to T/F\ntf_df = pandas_df.isnull()\n\n# Plot it\nsns.heatmap(data=tf_df)\nplt.xticks(rotation=30, fontsize=10)\nplt.yticks(rotation=0, fontsize=10)\nplt.show()\n\n# Set the answer to the column with the most missing data\nanswer = 'BACKONMARKETDATE'\nanswer",
"_____no_output_____"
]
],
[
[
"### Imputing Missing Data",
"_____no_output_____"
]
],
[
[
"# Count missing rows\nmissing = df.where(df['PDOM'].isNull()).count()\n\n# Calculate the mean value\ncol_mean = df.agg({'PDOM': 'mean'}).collect()[0][0]\n\n# Replacing with the mean value for that column\ndf.fillna(col_mean, subset=['PDOM'])",
"_____no_output_____"
]
],
[
[
"Make sure to spend time considering the appropriate ways to handle missing data in your problems.",
"_____no_output_____"
],
[
"### Calculate Missing Percents",
"_____no_output_____"
]
],
[
[
"def column_dropper(df, threshold):\n # Takes a dataframe and threshold for missing values. Returns a dataframe.\n total_records = df.count()\n for col in df.columns:\n # Calculate the percentage of missing values\n missing = df.where(df[col].isNull()).count()\n missing_percent = missing / total_records\n # Drop column if percent of missing is more than threshold\n if missing_percent > threshold:\n df = df.drop(col)\n return df\n\n# Drop columns that are more than 60% missing\ndf = column_dropper(df, .6)",
"_____no_output_____"
]
],
[
[
"### A Dangerous Join",
"_____no_output_____"
]
],
[
[
"# Cast data types\nwalk_df = walk_df.withColumn('longitude', walk_df.longitude.cast('double'))\nwalk_df = walk_df.withColumn('latitude', walk_df.latitude.cast('double'))\n\n# Round precision\ndf = df.withColumn('longitude', round(df['longitude'], 5))\ndf = df.withColumn('latitude', round(df['latitude'], 5))\n\n# Create join condition\ncondition = [walk_df['latitude'] == df['latitude'], walk_df['longitude'] == df['longitude']]\n\n# Join the dataframes together\njoin_df = df.join(walk_df, on=condition, how='left')\n# Count non-null records from new field\nprint(join_df.where(~join_df['walkscore'].isNull()).count())",
"_____no_output_____"
]
],
[
[
"### Spark SQL Join",
"_____no_output_____"
]
],
[
[
"# Register dataframes as tables\ndf.createOrReplaceTempView(\"df\")\nwalk_df.createOrReplaceTempView(\"walk_df\")\n\n# SQL to join dataframes\njoin_sql = \t\"\"\"\n\t\t\tSELECT \n\t\t\t\t*\n\t\t\tFROM df\n\t\t\tLEFT JOIN walk_df\n\t\t\tON df.longitude = walk_df.longitude\n\t\t\tAND df.latitude = walk_df.latitude\n\t\t\t\"\"\"\n# Perform sql join\njoined_df = spark.sql(join_sql)",
"_____no_output_____"
]
],
[
[
"### Checking for Bad Joins",
"_____no_output_____"
]
],
[
[
"# Join on mismatched keys precision \nwrong_prec_cond = [walk_df['latitude'] == df_orig['latitude'], walk_df['longitude'] == df_orig['longitude']]\nwrong_prec_df = df_orig.join(walk_df, on=wrong_prec_cond, how='left')\n\n# Compare bad join to the correct one\nprint(wrong_prec_df.where(wrong_prec_df['walkscore'].isNull()).count())\nprint(correct_join_df.where(correct_join_df['walkscore'].isNull()).count())\n\n# Create a join on too few keys\nfew_keys_cond = [walk_df['longitude'] == df['longitude']]\nfew_keys_df = df.join(walk_df, on=few_keys_cond, how='left')\n\n# Compare bad join to the correct one\nprint(\"Record Count of the Too Few Keys Join Example: \" + str(few_keys_df.count()))\nprint(\"Record Count of the Correct Join Example: \" + str(correct_join_df.count()))",
"_____no_output_____"
]
],
[
[
"## Feature Engineering",
"_____no_output_____"
],
[
"### Differences",
"_____no_output_____"
]
],
[
[
"# Lot size in square feet\nacres_to_sqfeet = 43560\ndf = df.withColumn('LOT_SIZE_SQFT', df['ACRES'] * acres_to_sqfeet)\n\n# Create new column YARD_SIZE\ndf = df.withColumn('YARD_SIZE', df['LOT_SIZE_SQFT'] - df['FOUNDATIONSIZE'])\n\n# Corr of ACRES vs SALESCLOSEPRICE\nprint(\"Corr of ACRES vs SALESCLOSEPRICE: \" + str(df.corr('ACRES', 'SALESCLOSEPRICE')))\n# Corr of FOUNDATIONSIZE vs SALESCLOSEPRICE\nprint(\"Corr of FOUNDATIONSIZE vs SALESCLOSEPRICE: \" + str(df.corr('FOUNDATIONSIZE', 'SALESCLOSEPRICE')))\n# Corr of YARD_SIZE vs SALESCLOSEPRICE\nprint(\"Corr of YARD_SIZE vs SALESCLOSEPRICE: \" + str(df.corr('YARD_SIZE', 'SALESCLOSEPRICE')))",
"Corr of ACRES vs SALESCLOSEPRICE: 0.2130819260750225\nCorr of FOUNDATIONSIZE vs SALESCLOSEPRICE: 0.5976957088401492\nCorr of YARD_SIZE vs SALESCLOSEPRICE: 0.20032633979612804\n"
]
],
[
[
"### Ratios",
"_____no_output_____"
]
],
[
[
"# ASSESSED_TO_LIST\ndf = df.withColumn('ASSESSED_TO_LIST', df['ASSESSEDVALUATION'] / df['LISTPRICE'])\ndf[['ASSESSEDVALUATION', 'LISTPRICE', 'ASSESSED_TO_LIST']].show(5)\n# TAX_TO_LIST\ndf = df.withColumn('TAX_TO_LIST', df['TAXES'] / df['LISTPRICE'])\ndf[['TAX_TO_LIST', 'TAXES', 'LISTPRICE']].show(5)\n# BED_TO_BATHS\ndf = df.withColumn('BED_TO_BATHS', df['BEDROOMS'] / df['BATHSTOTAL'])\ndf[['BED_TO_BATHS', 'BEDROOMS', 'BATHSTOTAL']].show(5)",
"+-----------------+---------+----------------+\n|ASSESSEDVALUATION|LISTPRICE|ASSESSED_TO_LIST|\n+-----------------+---------+----------------+\n| 0.0| 139900| 0.0|\n| 0.0| 210000| 0.0|\n| 0.0| 225000| 0.0|\n| 0.0| 230000| 0.0|\n| 0.0| 239900| 0.0|\n+-----------------+---------+----------------+\nonly showing top 5 rows\n\n+--------------------+-----+---------+\n| TAX_TO_LIST|TAXES|LISTPRICE|\n+--------------------+-----+---------+\n|0.013280914939242315| 1858| 139900|\n| 0.00780952380952381| 1640| 210000|\n|0.010622222222222222| 2390| 225000|\n|0.009330434782608695| 2146| 230000|\n|0.008378491037932471| 2010| 239900|\n+--------------------+-----+---------+\nonly showing top 5 rows\n\n+------------------+--------+----------+\n| BED_TO_BATHS|BEDROOMS|BATHSTOTAL|\n+------------------+--------+----------+\n| 1.5| 3| 2|\n|1.3333333333333333| 4| 3|\n| 2.0| 2| 1|\n| 1.0| 2| 2|\n| 1.5| 3| 2|\n+------------------+--------+----------+\nonly showing top 5 rows\n\n"
]
],
[
[
"### Deeper Features",
"_____no_output_____"
]
],
[
[
"from scipy import stats\n\ndef r2(x, y):\n return stats.pearsonr(x, y)[0] ** 2",
"_____no_output_____"
],
[
"# Create new feature by adding two features together\ndf = df.withColumn('Total_SQFT', df['SQFTBELOWGROUND'] + df['SQFTABOVEGROUND'])\n\n# Create additional new feature using previously created feature\ndf = df.withColumn('BATHS_PER_1000SQFT', df['BATHSTOTAL'] / (df['Total_SQFT'] / 1000))\ndf[['BATHS_PER_1000SQFT']].describe().show()\n\n# Sample and create pandas dataframe\npandas_df = df.sample(False, 0.5, 0).toPandas()\n\n# Linear model plots\nsns.jointplot(x='Total_SQFT', y='SALESCLOSEPRICE', data=pandas_df, kind=\"reg\", stat_func=r2)\nsns.jointplot(x='BATHS_PER_1000SQFT', y='SALESCLOSEPRICE', data=pandas_df, kind=\"reg\", stat_func=r2)",
"+-------+-------------------+\n|summary| BATHS_PER_1000SQFT|\n+-------+-------------------+\n| count| 4946|\n| mean| 1.4339460530042523|\n| stddev| 14.205761576339642|\n| min|0.39123630672926446|\n| max| 1000.0|\n+-------+-------------------+\n\n"
]
],
[
[
"### Time Components",
"_____no_output_____"
]
],
[
[
"# Import needed functions\nfrom pyspark.sql.functions import to_date, dayofweek\n\n# Convert to date type\ndf = df.withColumn('LISTDATE', to_date(df['LISTDATE'], format='MM/dd/yyyy HH:mm'))\n\n# Get the day of the week\ndf = df.withColumn('List_Day_of_Week', dayofweek(df['LISTDATE']))\n\n# Sample and convert to pandas dataframe\nsample_df = df.sample(False, .5, 42).toPandas()",
"_____no_output_____"
],
[
"# Plot count plot of of day of week\nsns.countplot(x=\"List_Day_of_Week\", data=sample_df)\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Joining On Time Components",
"_____no_output_____"
]
],
[
[
"import pandas as pd",
"_____no_output_____"
],
[
"data = dict(City=['LELM - Lake Elmo', 'MAPW - Maplewood','STP - Saint Paul','WB - Woodbury', \\\n 'OAKD - Oakdale', 'LELM - Lake Elmo', 'MAPW - Maplewood', \\\n 'STP - Saint Paul', 'WB - Woodbury', 'OAKD - Oakdale'],\n MedianHomeValue=[401000, 193000, 172000, 291000, 210000, 385000, 187000, 162000, 277000, 192000],\n Year= [2016,2016,2016,2016,2016,2015,2015,2015,2015, 2015])\n\ndf_price = pd.DataFrame(data)\nprice_df = spark.createDataFrame(df_price)",
"_____no_output_____"
],
[
"price_df.show()",
"+----------------+---------------+----+\n| City|MedianHomeValue|Year|\n+----------------+---------------+----+\n|LELM - Lake Elmo| 401000|2016|\n|MAPW - Maplewood| 193000|2016|\n|STP - Saint Paul| 172000|2016|\n| WB - Woodbury| 291000|2016|\n| OAKD - Oakdale| 210000|2016|\n|LELM - Lake Elmo| 385000|2015|\n|MAPW - Maplewood| 187000|2015|\n|STP - Saint Paul| 162000|2015|\n| WB - Woodbury| 277000|2015|\n| OAKD - Oakdale| 192000|2015|\n+----------------+---------------+----+\n\n"
],
[
"from pyspark.sql.functions import year\n\n# Create year column\ndf = df.withColumn('list_year', year(df['LISTDATE']))\n\n# Adjust year to match\ndf = df.withColumn('report_year', (df['list_year'] - 1))\n\n# Create join condition\ncondition = [df['CITY'] == price_df['City'], df['report_year'] == price_df['year']]\n\n# Join the dataframes together\ndf = df.join(price_df, on=condition, how='left')\n# Inspect that new columns are available\ndf[['MedianHomeValue']].show()",
"+---------------+\n|MedianHomeValue|\n+---------------+\n| 172000|\n| 172000|\n| 172000|\n| 172000|\n| 172000|\n| 172000|\n| 172000|\n| 172000|\n| 172000|\n| 172000|\n| 172000|\n| 172000|\n| 172000|\n| 172000|\n| 172000|\n| 172000|\n| 172000|\n| 172000|\n| 172000|\n| 172000|\n+---------------+\nonly showing top 20 rows\n\n"
]
],
[
[
"### Date Math",
"_____no_output_____"
]
],
[
[
"from pyspark.sql.functions import lag, datediff, to_date\nfrom pyspark.sql.window import Window\n\n# Cast data type\nmort_df = mort_df.withColumn('DATE', to_date(mort_df['DATE']))\n\n# Create window\nw = Window().orderBy(mort_df['DATE'])\n# Create lag column\nmort_df = mort_df.withColumn('DATE-1', lag(mort_df['DATE'], count=1).over(w))\n\n# Calculate difference between date columns\nmort_df = mort_df.withColumn('Days_Between_Report', datediff(mort_df['DATE'], mort_df['DATE-1']))\n# Print results\nmort_df.select('Days_Between_Report').distinct().show()",
"_____no_output_____"
]
],
[
[
"### Extracting Text to New Features",
"_____no_output_____"
]
],
[
[
"# Import needed functions\nfrom pyspark.sql.functions import when\n\n# Create boolean conditions for string matches\nhas_attached_garage = df['GARAGEDESCRIPTION'].like('%Attached%')\nhas_detached_garage = df['GARAGEDESCRIPTION'].like('%Detached%')\n\n# Conditional value assignment \ndf = df.withColumn('has_attached_garage', (when(has_attached_garage, 1)\n .when(has_detached_garage, 0)\n .otherwise(None)))\n\n# Inspect results\ndf[['GARAGEDESCRIPTION', 'has_attached_garage']].show(truncate=100)",
"+------------------------------------------------------------------------------+-------------------+\n| GARAGEDESCRIPTION|has_attached_garage|\n+------------------------------------------------------------------------------+-------------------+\n| Driveway - Concrete| null|\n| Driveway - Gravel| null|\n| Other| null|\n| Detached Garage| 0|\n| Other| null|\n| Detached Garage| 0|\n| Detached Garage| 0|\n| None| null|\n| Detached Garage, Driveway - Concrete| 0|\n| Underground Garage, Secured, Assigned, Garage Door Opener| null|\n|Underground Garage, Driveway - Concrete, Secured, Assigned, Garage Door Opener| null|\n| Uncovered/Open| null|\n| Detached Garage| 0|\n| Underground Garage| null|\n| Other| null|\n| None| null|\n| Detached Garage| 0|\n| Detached Garage, Driveway - Concrete| 0|\n| Contract Pkg Required| null|\n| Detached Garage| 0|\n+------------------------------------------------------------------------------+-------------------+\nonly showing top 20 rows\n\n"
]
],
[
[
"### Splitting & Exploding",
"_____no_output_____"
]
],
[
[
"df.select(['GARAGEDESCRIPTION']).show(truncate=100)",
"+------------------------------------------------------------------------------+\n| GARAGEDESCRIPTION|\n+------------------------------------------------------------------------------+\n| Driveway - Concrete|\n| Driveway - Gravel|\n| Other|\n| Detached Garage|\n| Other|\n| Detached Garage|\n| Detached Garage|\n| None|\n| Detached Garage, Driveway - Concrete|\n| Underground Garage, Secured, Assigned, Garage Door Opener|\n|Underground Garage, Driveway - Concrete, Secured, Assigned, Garage Door Opener|\n| Uncovered/Open|\n| Detached Garage|\n| Underground Garage|\n| Other|\n| None|\n| Detached Garage|\n| Detached Garage, Driveway - Concrete|\n| Contract Pkg Required|\n| Detached Garage|\n+------------------------------------------------------------------------------+\nonly showing top 20 rows\n\n"
],
[
"# Import needed functions\nfrom pyspark.sql.functions import split, explode\n\n# Convert string to list-like array\ndf = df.withColumn('garage_list', split(df['GARAGEDESCRIPTION'], ', '))\n\n# Explode the values into new records\nex_df = df.withColumn('ex_garage_list', explode(df['garage_list']))\n\n# Inspect the values\nex_df[['ex_garage_list']].distinct().show(100, truncate=50)",
"+----------------------------+\n| ex_garage_list|\n+----------------------------+\n| Attached Garage|\n| On-Street Parking Only|\n| None|\n| More Parking Onsite for Fee|\n| Garage Door Opener|\n| No Int Access to Dwelling|\n| Driveway - Gravel|\n| Valet Parking for Fee|\n| Uncovered/Open|\n| Heated Garage|\n| Underground Garage|\n| Other|\n| Unassigned|\n|More Parking Offsite for Fee|\n| Driveway - Other Surface|\n| Contract Pkg Required|\n| Carport|\n| Secured|\n| Detached Garage|\n| Driveway - Asphalt|\n| Units Vary|\n| Assigned|\n| Tuckunder|\n| Covered|\n| Insulated Garage|\n| Driveway - Concrete|\n| Tandem|\n| Driveway - Shared|\n+----------------------------+\n\n"
]
],
[
[
"### Pivot & Join",
"_____no_output_____"
]
],
[
[
"from pyspark.sql.functions import coalesce, first\n\n# Pivot \npiv_df = ex_df.groupBy('NO').pivot('ex_garage_list').agg(coalesce(first('constant_val')))\n\n# Join the dataframes together and fill null\njoined_df = df.join(piv_df, on='NO', how='left')\n\n# Columns to zero fill\nzfill_cols = piv_df.columns\n\n# Zero fill the pivoted values\nzfilled_df = joined_df.fillna(0, subset=zfill_cols)",
"_____no_output_____"
]
],
[
[
"### Binarizing Day of Week",
"_____no_output_____"
]
],
[
[
"df = df.withColumn('List_Day_of_Week', df['List_Day_of_Week'].cast('double'))",
"_____no_output_____"
],
[
"# Import transformer\nfrom pyspark.ml.feature import Binarizer\n\n# Create the transformer\nbinarizer = Binarizer(threshold=5, inputCol='List_Day_of_Week', outputCol='Listed_On_Weekend')\n\n# Apply the transformation to df\ndf = binarizer.transform(df)\n\n# Verify transformation\ndf[['List_Day_of_Week', 'Listed_On_Weekend']].show()",
"+----------------+-----------------+\n|List_Day_of_Week|Listed_On_Weekend|\n+----------------+-----------------+\n| 6.0| 1.0|\n| 5.0| 0.0|\n| 6.0| 1.0|\n| 5.0| 0.0|\n| 6.0| 1.0|\n| 5.0| 0.0|\n| 5.0| 0.0|\n| 2.0| 0.0|\n| 7.0| 1.0|\n| 6.0| 1.0|\n| 6.0| 1.0|\n| 4.0| 0.0|\n| 3.0| 0.0|\n| 4.0| 0.0|\n| 5.0| 0.0|\n| 7.0| 1.0|\n| 4.0| 0.0|\n| 3.0| 0.0|\n| 5.0| 0.0|\n| 4.0| 0.0|\n+----------------+-----------------+\nonly showing top 20 rows\n\n"
]
],
[
[
"### Bucketing",
"_____no_output_____"
]
],
[
[
"sample_df.head()",
"_____no_output_____"
],
[
"sample_df.BEDROOMS.dtype",
"_____no_output_____"
],
[
"from pyspark.ml.feature import Bucketizer\n\n# Plot distribution of sample_df\nsns.distplot(sample_df.BEDROOMS, axlabel='BEDROOMS')\nplt.show()\n\n# Create the bucket splits and bucketizer\nsplits = [0, 1, 2, 3, 4, 5, float('Inf')]\nbuck = Bucketizer(splits=splits, inputCol='BEDROOMS', outputCol='bedrooms')\n\n# Apply the transformation to df\ndf = buck.transform(df)\n\n# Display results\ndf[['BEDROOMS', 'bedrooms']].show()",
"_____no_output_____"
]
],
[
[
"### One Hot Encoding",
"_____no_output_____"
]
],
[
[
"df.select(['SCHOOLDISTRICTNUMBER']).show()",
"+--------------------+\n|SCHOOLDISTRICTNUMBER|\n+--------------------+\n| 625 - St. Paul|\n| 625 - St. Paul|\n| 625 - St. Paul|\n| 625 - St. Paul|\n| 625 - St. Paul|\n| 625 - St. Paul|\n| 625 - St. Paul|\n| 625 - St. Paul|\n| 625 - St. Paul|\n| 625 - St. Paul|\n| 625 - St. Paul|\n| 625 - St. Paul|\n| 625 - St. Paul|\n| 625 - St. Paul|\n| 625 - St. Paul|\n| 625 - St. Paul|\n| 625 - St. Paul|\n| 625 - St. Paul|\n| 625 - St. Paul|\n| 625 - St. Paul|\n+--------------------+\nonly showing top 20 rows\n\n"
],
[
"from pyspark.ml.feature import OneHotEncoder, StringIndexer\n\n# Map strings to numbers with string indexer\nstring_indexer = StringIndexer(inputCol='SCHOOLDISTRICTNUMBER', outputCol='School_Index')\nindexed_df = string_indexer.fit(df).transform(df)\n\n# Onehot encode indexed values\nencoder = OneHotEncoder(inputCol='School_Index', outputCol='School_Vec')\nencoded_df = encoder.transform(indexed_df)\n\n# Inspect the transformation steps\nencoded_df[['SCHOOLDISTRICTNUMBER', 'School_Index', 'School_Vec']].show(truncate=100)",
"+--------------------+------------+-------------+\n|SCHOOLDISTRICTNUMBER|School_Index| School_Vec|\n+--------------------+------------+-------------+\n| 625 - St. Paul| 0.0|(7,[0],[1.0])|\n| 625 - St. Paul| 0.0|(7,[0],[1.0])|\n| 625 - St. Paul| 0.0|(7,[0],[1.0])|\n| 625 - St. Paul| 0.0|(7,[0],[1.0])|\n| 625 - St. Paul| 0.0|(7,[0],[1.0])|\n| 625 - St. Paul| 0.0|(7,[0],[1.0])|\n| 625 - St. Paul| 0.0|(7,[0],[1.0])|\n| 625 - St. Paul| 0.0|(7,[0],[1.0])|\n| 625 - St. Paul| 0.0|(7,[0],[1.0])|\n| 625 - St. Paul| 0.0|(7,[0],[1.0])|\n| 625 - St. Paul| 0.0|(7,[0],[1.0])|\n| 625 - St. Paul| 0.0|(7,[0],[1.0])|\n| 625 - St. Paul| 0.0|(7,[0],[1.0])|\n| 625 - St. Paul| 0.0|(7,[0],[1.0])|\n| 625 - St. Paul| 0.0|(7,[0],[1.0])|\n| 625 - St. Paul| 0.0|(7,[0],[1.0])|\n| 625 - St. Paul| 0.0|(7,[0],[1.0])|\n| 625 - St. Paul| 0.0|(7,[0],[1.0])|\n| 625 - St. Paul| 0.0|(7,[0],[1.0])|\n| 625 - St. Paul| 0.0|(7,[0],[1.0])|\n+--------------------+------------+-------------+\nonly showing top 20 rows\n\n"
]
],
[
[
"notice that the implementation in PySpark is different than Pandas get_dummies() as it puts everything into a single column of type vector rather than a new column for each value. It's also different from sklearn's OneHotEncoder in that the last categorical value is captured by a vector of all zeros",
"_____no_output_____"
],
[
"### Building a Model",
"_____no_output_____"
]
],
[
[
"df.select(['OFFMARKETDATE']).show()",
"+---------------+\n| OFFMARKETDATE|\n+---------------+\n| 9/6/2017 0:00|\n| 8/12/2017 0:00|\n| 10/4/2017 0:00|\n| 8/22/2017 0:00|\n| 3/27/2017 0:00|\n| 8/18/2017 0:00|\n| 5/10/2017 0:00|\n| 9/2/2017 0:00|\n| 9/1/2017 0:00|\n| 7/26/2017 0:00|\n| 5/18/2017 0:00|\n| 9/20/2017 0:00|\n| 8/29/2017 0:00|\n| 3/29/2017 0:00|\n| 3/13/2017 0:00|\n| 9/6/2017 0:00|\n| 6/25/2017 0:00|\n| 9/15/2017 0:00|\n| 6/14/2017 0:00|\n|11/20/2017 0:00|\n+---------------+\nonly showing top 20 rows\n\n"
],
[
"from datetime import timedelta\n\ndf = df.withColumn('OFFMARKETDATE', to_date(df['OFFMARKETDATE'], format='MM/dd/yyyy HH:mm'))",
"_____no_output_____"
],
[
"def train_test_split_date(df, split_col, test_days=45):\n \"\"\"Calculate the date to split test and training sets\"\"\"\n # Find how many days our data spans\n max_date = df.agg({split_col: 'max'}).collect()[0][0]\n min_date = df.agg({split_col: 'min'}).collect()[0][0]\n # Subtract an integer number of days from the last date in dataset\n split_date = max_date - timedelta(days=test_days)\n return split_date\n\n# Find the date to use in spitting test and train\nsplit_date = train_test_split_date(df, 'OFFMARKETDATE')\n\n# Create Sequential Test and Training Sets\ntrain_df = df.where(df['OFFMARKETDATE'] < split_date) \ntest_df = df.where(df['OFFMARKETDATE'] >= split_date).where(df['LISTDATE'] <= split_date) ",
"_____no_output_____"
],
[
"split_date",
"_____no_output_____"
],
[
"train_df.count(), test_df.count()",
"_____no_output_____"
]
],
[
[
"### Adjusting Time Features",
"_____no_output_____"
]
],
[
[
"from pyspark.sql.functions import datediff, to_date, lit\n\nsplit_date = to_date(lit('2017-12-10'))\n\n# Create a copy of DAYSONMARKET to review later\ntest_df = test_df.withColumn('DAYSONMARKET_Original', test_df['DAYSONMARKET'])\n\n# Recalculate DAYSONMARKET from what we know on our split date\ntest_df = test_df.withColumn('DAYSONMARKET', datediff(split_date, test_df['LISTDATE']))\n\n# Review the difference\ntest_df[['LISTDATE', 'OFFMARKETDATE', 'DAYSONMARKET_Original', 'DAYSONMARKET']].show()",
"+----------+-------------+---------------------+------------+\n| LISTDATE|OFFMARKETDATE|DAYSONMARKET_Original|DAYSONMARKET|\n+----------+-------------+---------------------+------------+\n|2017-10-07| 2017-12-17| 71| 64|\n|2017-08-14| 2018-01-09| 114| 118|\n|2017-10-24| 2017-12-19| 56| 47|\n|2017-09-21| 2018-01-03| 100| 80|\n|2017-12-08| 2017-12-13| 4| 2|\n|2017-11-03| 2017-12-19| 46| 37|\n|2017-10-14| 2017-12-15| 6| 57|\n|2017-11-27| 2017-12-27| 3| 13|\n|2017-12-10| 2017-12-14| 4| 0|\n|2017-11-14| 2018-01-02| 49| 26|\n|2017-11-21| 2017-12-29| 38| 19|\n|2017-12-08| 2017-12-12| 4| 2|\n|2017-11-13| 2017-12-21| 38| 27|\n|2017-12-03| 2018-01-15| 43| 7|\n|2017-07-27| 2017-12-14| 138| 136|\n|2017-12-04| 2018-01-04| 31| 6|\n|2017-12-07| 2017-12-27| 20| 3|\n|2017-12-04| 2017-12-17| 13| 6|\n|2017-11-10| 2017-12-31| 50| 30|\n|2017-11-07| 2017-12-22| 7| 33|\n+----------+-------------+---------------------+------------+\nonly showing top 20 rows\n\n"
]
],
[
[
"if the house is still on the market, we don't know how many more days it will stay on the market. We need to adjust our test_df to reflect what information we currently have as of 2017-12-10.",
"_____no_output_____"
],
[
"Missing values are handled by Random Forests internally where they partition on missing values. As long as you replace them with something outside of the range of normal values, they will be handled correctly. Likewise, categorical features only need to be mapped to numbers, they are fine to stay all in one column by using a StringIndexer as we saw in chapter 3. OneHot encoding which converts each possible value to its own boolean feature is not needed.",
"_____no_output_____"
],
[
"### Dropping Columns with Low Observations",
"_____no_output_____"
]
],
[
[
"df.select('FENCE').show()",
"+--------------------+\n| FENCE|\n+--------------------+\n| null|\n| null|\n| Other|\n| Chain Link|\n| Chain Link|\n| None|\n| Wire, Partial|\n| null|\n| Chain Link, Partial|\n| null|\n| None|\n| Chain Link, Partial|\n| null|\n| null|\n|Wood, Chain Link,...|\n| Chain Link|\n| null|\n| Chain Link, Partial|\n| None|\n| null|\n+--------------------+\nonly showing top 20 rows\n\n"
],
[
"binary_cols = ['FENCE_WIRE',\n 'FENCE_ELECTRIC',\n 'FENCE_NAN',\n 'FENCE_PARTIAL',\n 'FENCE_RAIL',\n 'FENCE_OTHER',\n 'FENCE_CHAIN LINK',\n 'FENCE_FULL',\n 'FENCE_NONE',\n 'FENCE_PRIVACY',\n 'FENCE_WOOD',\n 'FENCE_INVISIBLE', # e.g. one hot = fence columns \n 'ROOF_ASPHALT SHINGLES',\n 'ROOF_SHAKES',\n 'ROOF_NAN',\n 'ROOF_UNSPECIFIED SHINGLE',\n 'ROOF_SLATE',\n 'ROOF_PITCHED',\n 'ROOF_FLAT',\n 'ROOF_TAR/GRAVEL',\n 'ROOF_OTHER',\n 'ROOF_METAL',\n 'ROOF_TILE',\n 'ROOF_RUBBER',\n 'ROOF_WOOD SHINGLES',\n 'ROOF_AGE OVER 8 YEARS',\n 'ROOF_AGE 8 YEARS OR LESS',\n 'POOLDESCRIPTION_NAN',\n 'POOLDESCRIPTION_HEATED',\n 'POOLDESCRIPTION_NONE',\n 'POOLDESCRIPTION_SHARED',\n 'POOLDESCRIPTION_INDOOR',\n 'POOLDESCRIPTION_OUTDOOR',\n 'POOLDESCRIPTION_ABOVE GROUND',\n 'POOLDESCRIPTION_BELOW GROUND',\n 'GARAGEDESCRIPTION_ASSIGNED',\n 'GARAGEDESCRIPTION_TANDEM',\n 'GARAGEDESCRIPTION_UNCOVERED/OPEN',\n 'GARAGEDESCRIPTION_TUCKUNDER',\n 'GARAGEDESCRIPTION_DRIVEWAY - ASPHALT',\n 'GARAGEDESCRIPTION_HEATED GARAGE',\n 'GARAGEDESCRIPTION_UNDERGROUND GARAGE',\n 'GARAGEDESCRIPTION_DRIVEWAY - SHARED',\n 'GARAGEDESCRIPTION_CONTRACT PKG REQUIRED',\n 'GARAGEDESCRIPTION_GARAGE DOOR OPENER',\n 'GARAGEDESCRIPTION_MORE PARKING OFFSITE FOR FEE',\n 'GARAGEDESCRIPTION_VALET PARKING FOR FEE',\n 'GARAGEDESCRIPTION_OTHER',\n 'GARAGEDESCRIPTION_MORE PARKING ONSITE FOR FEE',\n 'GARAGEDESCRIPTION_DRIVEWAY - OTHER SURFACE',\n 'GARAGEDESCRIPTION_DETACHED GARAGE',\n 'GARAGEDESCRIPTION_SECURED',\n 'GARAGEDESCRIPTION_CARPORT',\n 'GARAGEDESCRIPTION_DRIVEWAY - CONCRETE',\n 'GARAGEDESCRIPTION_ON-STREET PARKING ONLY',\n 'GARAGEDESCRIPTION_COVERED',\n 'GARAGEDESCRIPTION_INSULATED GARAGE',\n 'GARAGEDESCRIPTION_UNASSIGNED',\n 'GARAGEDESCRIPTION_NONE',\n 'GARAGEDESCRIPTION_DRIVEWAY - GRAVEL',\n 'GARAGEDESCRIPTION_NO INT ACCESS TO DWELLING',\n 'GARAGEDESCRIPTION_UNITS VARY',\n 'GARAGEDESCRIPTION_ATTACHED GARAGE',\n 'APPLIANCES_NAN',\n 'APPLIANCES_COOKTOP',\n 'APPLIANCES_WALL OVEN',\n 'APPLIANCES_WATER SOFTENER - OWNED',\n 'APPLIANCES_DISPOSAL',\n 'APPLIANCES_DISHWASHER',\n 'APPLIANCES_OTHER',\n 'APPLIANCES_INDOOR GRILL',\n 'APPLIANCES_WASHER',\n 'APPLIANCES_RANGE',\n 'APPLIANCES_REFRIGERATOR',\n 'APPLIANCES_FURNACE HUMIDIFIER',\n 'APPLIANCES_TANKLESS WATER HEATER',\n 'APPLIANCES_ELECTRONIC AIR FILTER',\n 'APPLIANCES_MICROWAVE',\n 'APPLIANCES_EXHAUST FAN/HOOD',\n 'APPLIANCES_NONE',\n 'APPLIANCES_CENTRAL VACUUM',\n 'APPLIANCES_TRASH COMPACTOR',\n 'APPLIANCES_AIR-TO-AIR EXCHANGER',\n 'APPLIANCES_DRYER',\n 'APPLIANCES_FREEZER',\n 'APPLIANCES_WATER SOFTENER - RENTED',\n 'EXTERIOR_SHAKES',\n 'EXTERIOR_CEMENT BOARD',\n 'EXTERIOR_BLOCK',\n 'EXTERIOR_VINYL',\n 'EXTERIOR_FIBER BOARD',\n 'EXTERIOR_OTHER',\n 'EXTERIOR_METAL',\n 'EXTERIOR_BRICK/STONE',\n 'EXTERIOR_STUCCO',\n 'EXTERIOR_ENGINEERED WOOD',\n 'EXTERIOR_WOOD',\n 'DININGROOMDESCRIPTION_EAT IN KITCHEN',\n 'DININGROOMDESCRIPTION_NAN',\n 'DININGROOMDESCRIPTION_OTHER',\n 'DININGROOMDESCRIPTION_LIVING/DINING ROOM',\n 'DININGROOMDESCRIPTION_SEPARATE/FORMAL DINING ROOM',\n 'DININGROOMDESCRIPTION_KITCHEN/DINING ROOM',\n 'DININGROOMDESCRIPTION_INFORMAL DINING ROOM',\n 'DININGROOMDESCRIPTION_BREAKFAST AREA',\n 'BASEMENT_FINISHED (LIVABLE)',\n 'BASEMENT_PARTIAL',\n 'BASEMENT_SUMP PUMP',\n 'BASEMENT_INSULATING CONCRETE FORMS',\n 'BASEMENT_CRAWL SPACE',\n 'BASEMENT_PARTIAL FINISHED',\n 'BASEMENT_CONCRETE BLOCK',\n 'BASEMENT_DRAINAGE SYSTEM',\n 'BASEMENT_POURED CONCRETE',\n 'BASEMENT_UNFINISHED',\n 'BASEMENT_DRAIN TILED',\n 'BASEMENT_WOOD',\n 'BASEMENT_FULL',\n 'BASEMENT_EGRESS WINDOWS',\n 'BASEMENT_DAY/LOOKOUT WINDOWS',\n 'BASEMENT_SLAB',\n 'BASEMENT_STONE',\n 'BASEMENT_NONE',\n 'BASEMENT_WALKOUT',\n 'BATHDESC_MAIN FLOOR 1/2 BATH',\n 'BATHDESC_TWO MASTER BATHS',\n 'BATHDESC_MASTER WALK-THRU',\n 'BATHDESC_WHIRLPOOL',\n 'BATHDESC_NAN',\n 'BATHDESC_3/4 BASEMENT',\n 'BATHDESC_TWO BASEMENT BATHS',\n 'BATHDESC_OTHER',\n 'BATHDESC_3/4 MASTER',\n 'BATHDESC_MAIN FLOOR 3/4 BATH',\n 'BATHDESC_FULL MASTER',\n 'BATHDESC_MAIN FLOOR FULL BATH',\n 'BATHDESC_WALK-IN SHOWER',\n 'BATHDESC_SEPARATE TUB & SHOWER',\n 'BATHDESC_FULL BASEMENT',\n 'BATHDESC_BASEMENT',\n 'BATHDESC_WALK THRU',\n 'BATHDESC_BATHROOM ENSUITE',\n 'BATHDESC_PRIVATE MASTER',\n 'BATHDESC_JACK & JILL 3/4',\n 'BATHDESC_UPPER LEVEL 1/2 BATH',\n 'BATHDESC_ROUGH IN',\n 'BATHDESC_UPPER LEVEL FULL BATH',\n 'BATHDESC_1/2 MASTER',\n 'BATHDESC_1/2 BASEMENT',\n 'BATHDESC_JACK AND JILL',\n 'BATHDESC_UPPER LEVEL 3/4 BATH',\n 'ZONING_INDUSTRIAL',\n 'ZONING_BUSINESS/COMMERCIAL',\n 'ZONING_OTHER',\n 'ZONING_RESIDENTIAL-SINGLE',\n 'ZONING_RESIDENTIAL-MULTI-FAMILY',\n 'COOLINGDESCRIPTION_WINDOW',\n 'COOLINGDESCRIPTION_WALL',\n 'COOLINGDESCRIPTION_DUCTLESS MINI-SPLIT',\n 'COOLINGDESCRIPTION_NONE',\n 'COOLINGDESCRIPTION_GEOTHERMAL',\n 'COOLINGDESCRIPTION_CENTRAL',\n 'CITY:LELM - LAKE ELMO',\n 'CITY:MAPW - MAPLEWOOD',\n 'CITY:OAKD - OAKDALE',\n 'CITY:STP - SAINT PAUL',\n 'CITY:WB - WOODBURY',\n 'LISTTYPE:EXCLUSIVE AGENCY',\n 'LISTTYPE:EXCLUSIVE RIGHT',\n 'LISTTYPE:EXCLUSIVE RIGHT WITH EXCLUSIONS',\n 'LISTTYPE:OTHER',\n 'LISTTYPE:SERVICE AGREEMENT',\n 'SCHOOLDISTRICTNUMBER:6 - SOUTH ST. PAUL',\n 'SCHOOLDISTRICTNUMBER:622 - NORTH ST PAUL-MAPLEWOOD',\n 'SCHOOLDISTRICTNUMBER:623 - ROSEVILLE',\n 'SCHOOLDISTRICTNUMBER:624 - WHITE BEAR LAKE',\n 'SCHOOLDISTRICTNUMBER:625 - ST. PAUL',\n 'SCHOOLDISTRICTNUMBER:832 - MAHTOMEDI',\n 'SCHOOLDISTRICTNUMBER:833 - SOUTH WASHINGTON COUNTY',\n 'SCHOOLDISTRICTNUMBER:834 - STILLWATER',\n 'POTENTIALSHORTSALE:NO',\n 'POTENTIALSHORTSALE:NOT DISCLOSED',\n 'STYLE:(CC) CONVERTED MANSION',\n 'STYLE:(CC) HIGH RISE (4+ LEVELS)',\n 'STYLE:(CC) LOW RISE (3- LEVELS)',\n 'STYLE:(CC) MANOR/VILLAGE',\n 'STYLE:(CC) TWO UNIT',\n 'STYLE:(SF) FOUR OR MORE LEVEL SPLIT',\n 'STYLE:(SF) MODIFIED TWO STORY',\n 'STYLE:(SF) MORE THAN TWO STORIES',\n 'STYLE:(SF) ONE 1/2 STORIES',\n 'STYLE:(SF) ONE STORY',\n 'STYLE:(SF) OTHER',\n 'STYLE:(SF) SPLIT ENTRY (BI-LEVEL)',\n 'STYLE:(SF) THREE LEVEL SPLIT',\n 'STYLE:(SF) TWO STORIES',\n 'STYLE:(TH) DETACHED',\n 'STYLE:(TH) QUAD/4 CORNERS',\n 'STYLE:(TH) SIDE X SIDE',\n 'STYLE:(TW) TWIN HOME',\n 'ASSUMABLEMORTGAGE:INFORMATION COMING',\n 'ASSUMABLEMORTGAGE:NOT ASSUMABLE',\n 'ASSUMABLEMORTGAGE:YES W/ QUALIFYING',\n 'ASSUMABLEMORTGAGE:YES W/NO QUALIFYING',\n 'ASSESSMENTPENDING:NO',\n 'ASSESSMENTPENDING:UNKNOWN',\n 'ASSESSMENTPENDING:YES']",
"_____no_output_____"
],
[
"len(binary_cols)",
"_____no_output_____"
],
[
"obs_threshold = 30\ncols_to_remove = list()\n# Inspect first 10 binary columns in list\nfor col in binary_cols[0:10]:\n # Count the number of 1 values in the binary column\n obs_count = df.agg({col: 'sum'}).collect()[0][0]\n # If less than our observation threshold, remove\n if obs_count < obs_threshold:\n cols_to_remove.append(col)\n \n# Drop columns and print starting and ending dataframe shapes\nnew_df = df.drop(*cols_to_remove)\n\nprint('Rows: ' + str(df.count()) + ' Columns: ' + str(len(df.columns)))\nprint('Rows: ' + str(new_df.count()) + ' Columns: ' + str(len(new_df.columns)))",
"_____no_output_____"
]
],
[
[
" Rows: 5000 Columns: 253\n Rows: 5000 Columns: 250",
"_____no_output_____"
],
[
"### Naively Handling Missing and Categorical Values",
"_____no_output_____"
],
[
"For missing values since our data is strictly positive, we will assign -1. The random forest will split on this value and handle it differently than the rest of the values in the same feature.",
"_____no_output_____"
]
],
[
[
"categorical_cols = ['CITY', 'LISTTYPE',\n'SCHOOLDISTRICTNUMBER',\n'POTENTIALSHORTSALE',\n'STYLE',\n'ASSUMABLEMORTGAGE',\n'ASSESSMENTPENDING']",
"_____no_output_____"
],
[
"from pyspark.ml import Pipeline",
"_____no_output_____"
],
[
"# Replace missing values\ndf = df.fillna(-1, subset=['WALKSCORE', 'BIKESCORE'])\n\n# Create list of StringIndexers using list comprehension\nindexers = [StringIndexer(inputCol=col, outputCol=col+\"_IDX\")\\\n .setHandleInvalid(\"keep\") for col in categorical_cols]\n\n# Create pipeline of indexers\nindexer_pipeline = Pipeline(stages=indexers)\n# Fit and Transform the pipeline to the original data\ndf_indexed = indexer_pipeline.fit(df).transform(df)\n\n# Clean up redundant columns\ndf_indexed = df_indexed.drop(*categorical_cols)\n# Inspect data transformations\nprint(df_indexed.dtypes)",
"_____no_output_____"
]
],
[
[
"### Building a Regression Model",
"_____no_output_____"
]
],
[
[
"from pyspark.ml.regression import GBTRegressor\n\n# Train a Gradient Boosted Trees (GBT) model.\ngbt = GBTRegressor(featuresCol='features',\n labelCol='SALESCLOSEPRICE',\n predictionCol=\"Prediction_Price\",\n seed=42\n )\n\n# Train model.\nmodel = gbt.fit(train_df)",
"_____no_output_____"
]
],
[
[
"### Evaluating & Comparing Algorithms",
"_____no_output_____"
]
],
[
[
"from pyspark.ml.evaluation import RegressionEvaluator\n\n# Select columns to compute test error\nevaluator = RegressionEvaluator(labelCol='SALESCLOSEPRICE', \n predictionCol='Prediction_Price')\n# Dictionary of model predictions to loop over\nmodels = {'Gradient Boosted Trees': gbt_predictions, 'Random Forest Regression': rfr_predictions}\nfor key, preds in models.items():\n # Create evaluation metrics\n rmse = evaluator.evaluate(preds, {evaluator.metricName: 'rmse'})\n r2 = evaluator.evaluate(preds, {evaluator.metricName: 'r2'})\n\n # Print Model Metrics\n print(key + ' RMSE: ' + str(rmse))\n print(key + ' R^2: ' + str(r2))",
"_____no_output_____"
]
],
[
[
" Gradient Boosted Trees RMSE: 74380.63652512032\n Gradient Boosted Trees R^2: 0.6482244200795505\n Random Forest Regression RMSE: 22898.84041072095\n Random Forest Regression R^2: 0.9666594402208077",
"_____no_output_____"
],
[
"### Interpreting Results",
"_____no_output_____"
]
],
[
[
"# Convert feature importances to a pandas column\nfi_df = pd.DataFrame(importances, columns=['importance'])\n\n# Convert list of feature names to pandas column\nfi_df['feature'] = pd.Series(feature_cols)\n\n# Sort the data based on feature importance\nfi_df.sort_values(by=['importance'], ascending=False, inplace=True)\n\n# Inspect Results\nfi_df.head(10)",
"_____no_output_____"
]
],
[
[
"### Saving & Loading Models",
"_____no_output_____"
]
],
[
[
"from pyspark.ml.regression import RandomForestRegressionModel\n\n# Save model\nmodel.save('rfr_no_listprice')\n\n# Load model\nloaded_model = RandomForestRegressionModel.load('rfr_no_listprice')",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
4a57017a9e3e504e4dac9379deae992a903e216d
| 71,315 |
ipynb
|
Jupyter Notebook
|
example/ctpfrec_retailrocket.ipynb
|
k-tahiro/ctpfrec
|
973daa964ef85b000783f6e54ef8cb959d426e7c
|
[
"BSD-2-Clause"
] | 32 |
2018-11-23T02:17:56.000Z
|
2022-01-01T14:09:23.000Z
|
example/ctpfrec_retailrocket.ipynb
|
k-tahiro/ctpfrec
|
973daa964ef85b000783f6e54ef8cb959d426e7c
|
[
"BSD-2-Clause"
] | 5 |
2020-02-27T16:21:00.000Z
|
2021-03-06T21:04:25.000Z
|
example/ctpfrec_retailrocket.ipynb
|
k-tahiro/ctpfrec
|
973daa964ef85b000783f6e54ef8cb959d426e7c
|
[
"BSD-2-Clause"
] | 9 |
2018-12-07T23:13:43.000Z
|
2021-03-21T12:17:30.000Z
| 49.148863 | 7,128 | 0.658529 |
[
[
[
"# Recommending products with RetailRocket event logs\n\nThis IPython notebook illustrates the usage of the [ctpfrec](https://github.com/david-cortes/ctpfrec/) Python package for _Collaborative Topic Poisson Factorization_ in recommender systems based on sparse count data using the [RetailRocket](https://www.kaggle.com/retailrocket/ecommerce-dataset) dataset, consisting of event logs (view, add to cart, purchase) from an online catalog of products plus anonymized text descriptions of items.\n\nCollaborative Topic Poisson Factorization is a probabilistic model that tries to jointly factorize the user-item interaction matrix along with item-word text descriptions (as bag-of-words) of the items by the product of lower dimensional matrices. The package can also extend this model to add user attributes in the same format as the items’.\n\nCompared to competing methods such as BPR (Bayesian Personalized Ranking) or weighted-implicit NMF (non-negative matrix factorization of the non-probabilistic type that uses squared loss), it only requires iterating over the data for which an interaction was observed and not over data for which no interaction was observed (i.e. it doesn’t iterate over items not clicked by a user), thus being more scalable, and at the same time producing better results when fit to sparse count data (in general). Same for the word counts of items.\n\nThe implementation here is based on the paper _Content-based recommendations with poisson factorization (Gopalan, P.K., Charlin, L. and Blei, D., 2014)_.\n\nFor a similar package for explicit feedback data see also [cmfrec](https://github.com/david-cortes/cmfrec/). For Poisson factorization without side information see [hpfrec](https://github.com/david-cortes/hpfrec/).\n\n**Small note: if the TOC here is not clickable or the math symbols don't show properly, try visualizing this same notebook from nbviewer following [this link](http://nbviewer.jupyter.org/github/david-cortes/ctpfrec/blob/master/example/ctpfrec_retailrocket.ipynb).**\n\n** *\n## Sections\n* [1. Model description](#p1)\n* [2. Loading and processing the dataset](#p2)\n* [3. Fitting the model](#p3)\n* [4. Common sense checks](#p4)\n* [5. Comparison to model without item information](#p5)\n* [6. Making recommendations](#p6)\n* [7. References](#p7)\n** *\n<a id=\"p1\"></a>\n## 1. Model description\n\nThe model consists in producing a low-rank non-negative matrix factorization of the item-word matrix (a.k.a. bag-of-words, a matrix where each row represents an item and each column a word, with entries containing the number of times each word appeared in an item’s text, ideally with some pre-processing on the words such as stemming or lemmatization) by the product of two lower-rank matrices\n\n$$ W_{iw} \\approx \\Theta_{ik} \\beta_{wk}^T $$\n\nalong with another low-rank matrix factorization of the user-item activity matrix (a matrix where each entry corresponds to how many times each user interacted with each item) that shares the same item-factor matrix above plus an offset based on user activity and not based on items’ words\n\n$$ Y_{ui} \\approx \\eta_{uk} (\\Theta_{ik} + \\epsilon_{ik})^T $$\n\nThese matrices are assumed to come from a generative process as follows:\n\n* Items:\n\n$$ \\beta_{wk} \\sim Gamma(a,b) $$\n$$ \\Theta_{ik} \\sim Gamma(c,d)$$\n$$ W_{iw} \\sim Poisson(\\Theta_{ik} \\beta_{wk}^T) $$\n_(Where $W$ is the item-word count matrix, $k$ is the number of latent factors, $i$ is the number of items, $w$ is the number of words)_\n\n* User-Item interactions\n$$ \\eta_{uk} \\sim Gamma(e,f) $$\n$$ \\epsilon_{ik} \\sim Gamma(g,h) $$\n$$ Y_{ui} \\sim Poisson(\\eta_{uk} (\\Theta_{ik} + \\epsilon_{ik})^T) $$\n_(Where $u$ is the number of users, $Y$ is the user-item interaction matrix)_\n\nThe model is fit using mean-field variational inference with coordinate ascent. For more details see the paper in the references.\n** *\n<a id=\"p2\"></a>\n## 2. Loading and processing the data\n\nReading and concatenating the data. First the event logs:",
"_____no_output_____"
]
],
[
[
"import numpy as np, pandas as pd\n\nevents = pd.read_csv(\"events.csv\")\nevents.head()",
"_____no_output_____"
],
[
"events.event.value_counts()",
"_____no_output_____"
]
],
[
[
"In order to put all user-item interactions in one scale, I will arbitrarily assign values as follows:\n* View: +1\n* Add to basket: +3\n* Purchase: +3\n\nThus, if a user clicks an item, that `(user, item)` pair will have `value=1`, if she later adds it to cart and purchases it, will have `value=7` (plus any other views of the same item), and so on.\n\nThe reasoning behind this scale is because the distributions of counts and sums of counts seem to still follow a nice exponential distribution with these values, but different values might give better results in terms of models fit to them.",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\n\nequiv = {\n 'view':1,\n 'addtocart':3,\n 'transaction':3\n}\nevents['count']=events.event.map(equiv)\nevents.groupby('visitorid')['count'].sum().value_counts().hist(bins=200)",
"_____no_output_____"
],
[
"events = events.groupby(['visitorid','itemid'])['count'].sum().to_frame().reset_index()\nevents.rename(columns={'visitorid':'UserId', 'itemid':'ItemId', 'count':'Count'}, inplace=True)\nevents.head()",
"_____no_output_____"
]
],
[
[
"Now creating a train and test split. For simplicity purposes and in order to be able to make a fair comparison with a model that doesn't use item descriptions, I will try to only take users that had >= 3 items in the training data, and items that had >= 3 users.\n\nGiven the lack of user attributes and the fact that it will be compared later to a model without side information, the test set will only have users from the training data, but it's also possible to use user attributes if they follow the same format as the items', in which case the model can also recommend items to new users.\n\nIn order to compare it later to a model without items' text, I will also filter out the test set to have only items that were in the training set. **This is however not a model limitation, as it can also recommend items that have descriptions but no user interactions**.",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import train_test_split\n\nevents_train, events_test = train_test_split(events, test_size=.2, random_state=1)\ndel events\n\n## In order to find users and items with at least 3 interactions each,\n## it's easier and faster to use a simple heuristic that first filters according to one criteria,\n## then, according to the other, and repeats.\n## Finding a real subset of the data in which each item has strictly >= 3 users,\n## and each user has strictly >= 3 items, is a harder graph partitioning or optimization\n## problem. For a similar example of finding such subsets see also:\n## http://nbviewer.ipython.org/github/david-cortes/datascienceprojects/blob/master/optimization/dataset_splitting.ipynb\nusers_filter_out = events_train.groupby('UserId')['ItemId'].agg(lambda x: len(tuple(x)))\nusers_filter_out = np.array(users_filter_out.index[users_filter_out < 3])\n\nitems_filter_out = events_train.loc[~np.in1d(events_train.UserId, users_filter_out)].groupby('ItemId')['UserId'].agg(lambda x: len(tuple(x)))\nitems_filter_out = np.array(items_filter_out.index[items_filter_out < 3])\n\nusers_filter_out = events_train.loc[~np.in1d(events_train.ItemId, items_filter_out)].groupby('UserId')['ItemId'].agg(lambda x: len(tuple(x)))\nusers_filter_out = np.array(users_filter_out.index[users_filter_out < 3])\n\nevents_train = events_train.loc[~np.in1d(events_train.UserId.values, users_filter_out)]\nevents_train = events_train.loc[~np.in1d(events_train.ItemId.values, items_filter_out)]\nevents_test = events_test.loc[np.in1d(events_test.UserId.values, events_train.UserId.values)]\nevents_test = events_test.loc[np.in1d(events_test.ItemId.values, events_train.ItemId.values)]\n\nprint(events_train.shape)\nprint(events_test.shape)",
"(381963, 3)\n(68490, 3)\n"
]
],
[
[
"Now processing the text descriptions of the items:",
"_____no_output_____"
]
],
[
[
"iteminfo = pd.read_csv(\"item_properties_part1.csv\")\niteminfo2 = pd.read_csv(\"item_properties_part2.csv\")\niteminfo = iteminfo.append(iteminfo2, ignore_index=True)\niteminfo.head()",
"_____no_output_____"
]
],
[
[
"The item's description contain many fields and have a mixture of words and numbers. The numeric variables, as per the documentation, are prefixed with an \"n\" and have three digits decimal precision - I will exclude them here since this model is insensitive to numeric attributes such as price. The words are already lemmazed, and since we only have their IDs, it's not possible to do any other pre-processing on them.\n\nAlthough the descriptions don't say anything about it, looking at the contents and the lengths of the different fields, here I will assume that the field $283$ is the product title and the field $888$ is the product description. I will just concatenate them to obtain an overall item text, but there might be better ways of doing this (such as having different IDs for the same word when it appears in the title or the body, or multiplying those in the title by some number, etc.)\n\nAs the descriptions vary over time, I will only take the most recent version for each item:",
"_____no_output_____"
]
],
[
[
"iteminfo = iteminfo.loc[iteminfo.property.isin(('888','283'))]\niteminfo = iteminfo.loc[iteminfo.groupby(['itemid','property'])['timestamp'].idxmax()]\niteminfo.reset_index(drop=True, inplace=True)\niteminfo.head()",
"_____no_output_____"
]
],
[
[
"**Note that for simplicity I am completely ignoring the categories (these are easily incorporated e.g. by adding a count of +1 for each category to which an item belongs) and important factors such as the price. I am also completely ignoring all the other fields.**",
"_____no_output_____"
]
],
[
[
"from sklearn.feature_extraction.text import CountVectorizer\nfrom scipy.sparse import coo_matrix\nimport re\n\ndef concat_fields(x):\n x = list(x)\n out = x[0]\n for i in x[1:]:\n out += \" \" + i\n return out\n\nclass NonNumberTokenizer(object):\n def __init__(self):\n pass\n def __call__(self, txt):\n return [i for i in txt.split(\" \") if bool(re.search(\"^\\d\", i))]\n\niteminfo = iteminfo.groupby('itemid')['value'].agg(lambda x: concat_fields(x))\n\nt = CountVectorizer(tokenizer=NonNumberTokenizer(), stop_words=None,\n dtype=np.int32, strip_accents=None, lowercase=False)\nbag_of_words = t.fit_transform(iteminfo)\n\nbag_of_words = coo_matrix(bag_of_words)\nbag_of_words = pd.DataFrame({\n 'ItemId' : iteminfo.index[bag_of_words.row],\n 'WordId' : bag_of_words.col,\n 'Count' : bag_of_words.data\n})\ndel iteminfo\nbag_of_words.head()",
"_____no_output_____"
]
],
[
[
"In this case, I will not filter it out by only items that were in the training set, as other items can still be used to get better latent factors.",
"_____no_output_____"
],
[
"** *\n<a id=\"p3\"></a>\n## 3. Fitting the model\n\nFitting the model - note that I'm using some enhancements (passed as arguments to the class constructor) over the original version in the paper:\n* Standardizing item counts so as not to favor items with longer descriptions.\n* Initializing $\\Theta$ and $\\beta$ through hierarchical Poisson factorization instead of latent Dirichlet allocation.\n* Using a small step size for the updates for the parameters obtained from hierarchical Poisson factorization at the beginning, which then grows to one with increasing iteration numbers (informally, this achieves to somehwat \"preserve\" these fits while the user parameters are adjusted to these already-fit item parameters - then as the user parameters are already defined towards them, the item and word parameters start changing too).\n\nI'll be also fitting two slightly different models: one that takes (and can make recommendations for) all the items for which there are either descriptions or user clicks, and another that uses all the items for which there are descriptions to initialize the item-related parameters but discards the ones without clicks (can only make recommendations for items that users have clicked).\n\nFor more information about the parameters and what they do, see the online documentation:\n\n[http://ctpfrec.readthedocs.io](http://ctpfrec.readthedocs.io)",
"_____no_output_____"
]
],
[
[
"print(events_train.shape)\nprint(events_test.shape)\nprint(bag_of_words.shape)",
"(381963, 3)\n(68490, 3)\n(7676561, 3)\n"
],
[
"%%time\nfrom ctpfrec import CTPF\n\nrecommender_all_items = CTPF(k=70, step_size=lambda x: 1-1/np.sqrt(x+1),\n standardize_items=True, initialize_hpf=True, reindex=True,\n missing_items='include', allow_inconsistent_math=True, random_seed=1)\nrecommender_all_items.fit(counts_df=events_train.copy(), words_df=bag_of_words.copy())",
"*****************************************\nCollaborative Topic Poisson Factorization\n*****************************************\n\nNumber of users: 65913\nNumber of items: 418301\nNumber of words: 342260\nLatent factors to use: 70\n\nInitializing parameters...\nInitializing Theta and Beta through HPF...\n\n**********************************\nHierarchical Poisson Factorization\n**********************************\n\nNumber of users: 417053\nNumber of items: 342260\nLatent factors to use: 70\n\nInitializing parameters...\nAllocating Phi matrix...\nInitializing optimization procedure...\nIteration 10 | Norm(Theta_{10} - Theta_{0}): 3373.40234\nIteration 20 | Norm(Theta_{20} - Theta_{10}): 13.27755\nIteration 30 | Norm(Theta_{30} - Theta_{20}): 11.13662\nIteration 40 | Norm(Theta_{40} - Theta_{30}): 5.30947\nIteration 50 | Norm(Theta_{50} - Theta_{40}): 3.23760\nIteration 60 | Norm(Theta_{60} - Theta_{50}): 2.57951\nIteration 70 | Norm(Theta_{70} - Theta_{60}): 1.99546\nIteration 80 | Norm(Theta_{80} - Theta_{70}): 1.91506\nIteration 90 | Norm(Theta_{90} - Theta_{80}): 1.49374\nIteration 100 | Norm(Theta_{100} - Theta_{90}): 1.17536\n\n\nOptimization finished\nFinal log-likelihood: -54256333\nFinal RMSE: 2.4187\nMinutes taken (optimization part): 23.7\n\n**********************************\n\nAllocating intermediate matrices...\nInitializing optimization procedure...\nIteration 10 | train llk: -6305341 | train rmse: 2.8694\nIteration 20 | train llk: -6248204 | train rmse: 2.8681\nIteration 30 | train llk: -6228858 | train rmse: 2.8675\nIteration 40 | train llk: -6220805 | train rmse: 2.8672\nIteration 50 | train llk: -6212324 | train rmse: 2.8670\nIteration 60 | train llk: -6212101 | train rmse: 2.8670\n\n\nOptimization finished\nFinal log-likelihood: -6212101\nFinal RMSE: 2.8670\nMinutes taken (optimization part): 15.4\n\nProducing Python dictionaries...\nCPU times: user 5h 10min 38s, sys: 1min 39s, total: 5h 12min 18s\nWall time: 39min 46s\n"
],
[
"%%time\nrecommender_clicked_items_only = CTPF(k=70, step_size=lambda x: 1-1/np.sqrt(x+1),\n standardize_items=True, initialize_hpf=True, reindex=True,\n missing_items='exclude', allow_inconsistent_math=True, random_seed=1)\nrecommender_clicked_items_only.fit(counts_df=events_train.copy(), words_df=bag_of_words.copy())",
"*****************************************\nCollaborative Topic Poisson Factorization\n*****************************************\n\n"
]
],
[
[
"Most of the time here was spent in fitting the model to items that no user in the training set had clicked. If using instead a random initialization, it would have taken a lot less time to fit this model (there would be only a fraction of the items - see above time spent in each procedure), but the results are slightly worse.\n\n_Disclaimer: this notebook was run on a Google cloud server with Skylake CPU using 8 cores, and memory usage tops at around 6GB of RAM for the first model (including all the objects loaded before). In a desktop computer, it would take a bit longer to fit._\n** *\n<a id=\"p4\"></a>\n## 4. Common sense checks\n\nThere are many different metrics to evaluate recommendation quality in implicit datasets, but all of them have their drawbacks. The idea of this notebook is to illustrate the package usage and not to introduce and compare evaluation metrics, so I will only perform some common sense checks on the test data.\n\nFor implementations of evaluation metrics for implicit recommendations see other packages such as [lightFM](https://github.com/lyst/lightfm).\n\nAs some common sense checks, the predictions should:\n* Be higher for this non-zero hold-out sample than for random items.\n* Produce a good discrimination between random items and those in the hold-out sample (very related to the first point).\n* Be correlated with the numer of events per user-item pair in the hold-out sample.\n* Follow an exponential distribution rather than a normal or some other symmetric distribution.\n\nHere I'll check these four conditions:\n\n#### Model with all items",
"_____no_output_____"
]
],
[
[
"events_test['Predicted'] = recommender_all_items.predict(user=events_test.UserId, item=events_test.ItemId)\nevents_test['RandomItem'] = np.random.choice(events_train.ItemId.unique(), size=events_test.shape[0])\nevents_test['PredictedRandom'] = recommender_all_items.predict(user=events_test.UserId,\n item=events_test.RandomItem)\nprint(\"Average prediction for combinations in test set: \", events_test.Predicted.mean())\nprint(\"Average prediction for random combinations: \", events_test.PredictedRandom.mean())",
"Average prediction for combinations in test set: 0.017780766\nAverage prediction for random combinations: 0.0047758827\n"
],
[
"from sklearn.metrics import roc_auc_score\n\nwas_clicked = np.r_[np.ones(events_test.shape[0]), np.zeros(events_test.shape[0])]\nscore_model = np.r_[events_test.Predicted.values, events_test.PredictedRandom.values]\nroc_auc_score(was_clicked[~np.isnan(score_model)], score_model[~np.isnan(score_model)])",
"_____no_output_____"
],
[
"np.corrcoef(events_test.Count[~events_test.Predicted.isnull()], events_test.Predicted[~events_test.Predicted.isnull()])[0,1]",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt\n%matplotlib inline\n\n_ = plt.hist(events_test.Predicted, bins=200)\nplt.xlim(0,5)\nplt.show()",
"_____no_output_____"
]
],
[
[
"#### Model with clicked items only",
"_____no_output_____"
]
],
[
[
"events_test['Predicted'] = recommender_clicked_items_only.predict(user=events_test.UserId, item=events_test.ItemId)\nevents_test['PredictedRandom'] = recommender_clicked_items_only.predict(user=events_test.UserId,\n item=events_test.RandomItem)\nprint(\"Average prediction for combinations in test set: \", events_test.Predicted.mean())\nprint(\"Average prediction for random combinations: \", events_test.PredictedRandom.mean())",
"Average prediction for combinations in test set: 0.025673132\nAverage prediction for random combinations: 0.008127485\n"
],
[
"was_clicked = np.r_[np.ones(events_test.shape[0]), np.zeros(events_test.shape[0])]\nscore_model = np.r_[events_test.Predicted.values, events_test.PredictedRandom.values]\nroc_auc_score(was_clicked, score_model)",
"_____no_output_____"
],
[
"np.corrcoef(events_test.Count, events_test.Predicted)[0,1]",
"_____no_output_____"
],
[
"_ = plt.hist(events_test.Predicted, bins=200)\nplt.xlim(0,5)\nplt.show()",
"_____no_output_____"
]
],
[
[
"** *\n<a id=\"p5\"></a>\n## 5. Comparison to model without item information\n\nA natural benchmark to compare this model is to is a Poisson factorization model without any item side information - here I'll do the comparison with a _Hierarchical Poisson factorization_ model with the same metrics as above:",
"_____no_output_____"
]
],
[
[
"%%time\nfrom hpfrec import HPF\n\nrecommender_no_sideinfo = HPF(k=70)\nrecommender_no_sideinfo.fit(events_train.copy())",
"**********************************\nHierarchical Poisson Factorization\n**********************************\n\nNumber of users: 65913\nNumber of items: 39578\nLatent factors to use: 70\n\nInitializing parameters...\nAllocating Phi matrix...\nInitializing optimization procedure...\nIteration 10 | train llk: -4635584 | train rmse: 2.8502\nIteration 20 | train llk: -4548912 | train rmse: 2.8397\nIteration 30 | train llk: -4512693 | train rmse: 2.8336\nIteration 40 | train llk: -4492286 | train rmse: 2.8297\nIteration 50 | train llk: -4476969 | train rmse: 2.8287\nIteration 60 | train llk: -4464443 | train rmse: 2.8282\nIteration 70 | train llk: -4454397 | train rmse: 2.8282\nIteration 80 | train llk: -4448200 | train rmse: 2.8280\nIteration 90 | train llk: -4442528 | train rmse: 2.8275\nIteration 100 | train llk: -4437068 | train rmse: 2.8272\n\n\nOptimization finished\nFinal log-likelihood: -4437068\nFinal RMSE: 2.8272\nMinutes taken (optimization part): 1.5\n\nCPU times: user 12min 22s, sys: 2.19 s, total: 12min 24s\nWall time: 1min 34s\n"
],
[
"events_test_comp = events_test.copy()\nevents_test_comp['Predicted'] = recommender_no_sideinfo.predict(user=events_test_comp.UserId, item=events_test_comp.ItemId)\nevents_test_comp['PredictedRandom'] = recommender_no_sideinfo.predict(user=events_test_comp.UserId,\n item=events_test_comp.RandomItem)\nprint(\"Average prediction for combinations in test set: \", events_test_comp.Predicted.mean())\nprint(\"Average prediction for random combinations: \", events_test_comp.PredictedRandom.mean())",
"Average prediction for combinations in test set: 0.023392139\nAverage prediction for random combinations: 0.0063642794\n"
],
[
"was_clicked = np.r_[np.ones(events_test_comp.shape[0]), np.zeros(events_test_comp.shape[0])]\nscore_model = np.r_[events_test_comp.Predicted.values, events_test_comp.PredictedRandom.values]\nroc_auc_score(was_clicked, score_model)",
"_____no_output_____"
],
[
"np.corrcoef(events_test_comp.Count, events_test_comp.Predicted)[0,1]",
"_____no_output_____"
]
],
[
[
"As can be seen, adding the side information and widening the catalog to include more items using only their text descriptions (no clicks) results in an improvemnet over all 3 metrics, especially correlation with number of clicks.\n\nMore important than that however, is its ability to make recommendations from a far wider catalog of items, which in practice can make a much larger difference in recommendation quality than improvement in typicall offline metrics.\n** *\n<a id=\"p6\"></a>\n## 6. Making recommendations\n\nThe package provides a simple API for making predictions and Top-N recommended lists. These Top-N lists can be made among all items, or across some user-provided subset only, and you can choose to discard items with which the user had already interacted in the training set.\n\nHere I will:\n* Pick a random user with a reasonably long event history.\n* See which items would the model recommend to them among those which he has not yet clicked.\n* Compare it with the recommended list from the model without item side information.\n\nUnfortunately, since all the data is anonymized, it's not possible to make a qualitative evaluation of the results by looking at the recommended lists as it is in other datasets.",
"_____no_output_____"
]
],
[
[
"users_many_events = events_train.groupby('UserId')['ItemId'].agg(lambda x: len(tuple(x)))\nusers_many_events = np.array(users_many_events.index[users_many_events > 20])\n\nnp.random.seed(1)\nchosen_user = np.random.choice(users_many_events)\nchosen_user",
"_____no_output_____"
],
[
"%%time\nrecommender_all_items.topN(chosen_user, n=20)",
"CPU times: user 44 ms, sys: 0 ns, total: 44 ms\nWall time: 52 ms\n"
]
],
[
[
"*(These numbers represent the IDs of the items being recommended as they appeared in the `events_train` data frame)*",
"_____no_output_____"
]
],
[
[
"%%time\nrecommender_clicked_items_only.topN(chosen_user, n=20)",
"CPU times: user 8 ms, sys: 0 ns, total: 8 ms\nWall time: 1.65 ms\n"
],
[
"%%time\nrecommender_no_sideinfo.topN(chosen_user, n=20)",
"CPU times: user 4 ms, sys: 0 ns, total: 4 ms\nWall time: 1.48 ms\n"
]
],
[
[
"** *\n<a id=\"p7\"></a>\n## 7. References\n* Gopalan, Prem K., Laurent Charlin, and David Blei. \"Content-based recommendations with poisson factorization.\" Advances in Neural Information Processing Systems. 2014.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
4a5706831986cf48e84d6d6d8e015e09e782da70
| 4,819 |
ipynb
|
Jupyter Notebook
|
book/_build/jupyter_execute/fundamentals/modeling_process.ipynb
|
bradleyboehmke/PyML
|
51be5b340f3f5460d20c92cfe2ec9e02405d552e
|
[
"MIT"
] | 2 |
2021-11-21T02:31:41.000Z
|
2022-01-20T03:19:29.000Z
|
book/_build/jupyter_execute/fundamentals/modeling_process.ipynb
|
bradleyboehmke/PyML
|
51be5b340f3f5460d20c92cfe2ec9e02405d552e
|
[
"MIT"
] | null | null | null |
book/_build/jupyter_execute/fundamentals/modeling_process.ipynb
|
bradleyboehmke/PyML
|
51be5b340f3f5460d20c92cfe2ec9e02405d552e
|
[
"MIT"
] | null | null | null | 35.962687 | 1,003 | 0.586636 |
[
[
[
"# Content with notebooks\n\nYou can also create content with Jupyter Notebooks. This means that you can include\ncode blocks and their outputs in your book.\n\n## Markdown + notebooks\n\nAs it is markdown, you can embed images, HTML, etc into your posts!\n\n\n\nYou an also $add_{math}$ and\n\n$$\nmath^{blocks}\n$$\n\nor\n\n$$\n\\begin{aligned}\n\\mbox{mean} la_{tex} \\\\ \\\\\nmath blocks\n\\end{aligned}\n$$\n\nBut make sure you \\$Escape \\$your \\$dollar signs \\$you want to keep!\n\n## MyST markdown\n\nMyST markdown works in Jupyter Notebooks as well. For more information about MyST markdown, check\nout [the MyST guide in Jupyter Book](https://jupyterbook.org/content/myst.html),\nor see [the MyST markdown documentation](https://myst-parser.readthedocs.io/en/latest/).\n\n## Code blocks and outputs\n\nJupyter Book will also embed your code blocks and output in your book.\nFor example, here's some sample Matplotlib code:",
"_____no_output_____"
]
],
[
[
"from matplotlib import rcParams, cycler\nimport matplotlib.pyplot as plt\nimport numpy as np\nplt.ion()",
"_____no_output_____"
],
[
"# Fixing random state for reproducibility\nnp.random.seed(19680801)\n\nN = 10\ndata = [np.logspace(0, 1, 100) + np.random.randn(100) + ii for ii in range(N)]\ndata = np.array(data).T\ncmap = plt.cm.coolwarm\nrcParams['axes.prop_cycle'] = cycler(color=cmap(np.linspace(0, 1, N)))\n\n\nfrom matplotlib.lines import Line2D\ncustom_lines = [Line2D([0], [0], color=cmap(0.), lw=4),\n Line2D([0], [0], color=cmap(.5), lw=4),\n Line2D([0], [0], color=cmap(1.), lw=4)]\n\nfig, ax = plt.subplots(figsize=(10, 5))\nlines = ax.plot(data)\nax.legend(custom_lines, ['Cold', 'Medium', 'Hot']);",
"_____no_output_____"
]
],
[
[
"There is a lot more that you can do with outputs (such as including interactive outputs)\nwith your book. For more information about this, see [the Jupyter Book documentation](https://jupyterbook.org)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
4a570bf6d0edb2f7b3b1dbd0b77be028fd964984
| 582,163 |
ipynb
|
Jupyter Notebook
|
01_Python_Assignments/Haberman_cancer_survival_dataset.ipynb
|
VijayRameshkumar/AppliedAI
|
2e5012c18d6754f99cb81e2d23f0e6391bb0162d
|
[
"Apache-2.0"
] | 2 |
2021-01-26T09:29:13.000Z
|
2021-06-30T02:01:56.000Z
|
01_Python_Assignments/Haberman_cancer_survival_dataset.ipynb
|
VijayRameshkumar/AppliedAI
|
2e5012c18d6754f99cb81e2d23f0e6391bb0162d
|
[
"Apache-2.0"
] | null | null | null |
01_Python_Assignments/Haberman_cancer_survival_dataset.ipynb
|
VijayRameshkumar/AppliedAI
|
2e5012c18d6754f99cb81e2d23f0e6391bb0162d
|
[
"Apache-2.0"
] | 1 |
2021-03-30T09:47:59.000Z
|
2021-03-30T09:47:59.000Z
| 598.933128 | 140,612 | 0.943629 |
[
[
[
"import numpy as np\nimport seaborn as sns\nimport pandas as pd\nimport matplotlib.pyplot as plt\n\nplt.rcParams['axes.titlesize'] = 20\nplt.rcParams['axes.titleweight'] = 10",
"_____no_output_____"
]
],
[
[
"## 1. Dataset Read",
"_____no_output_____"
]
],
[
[
"df = pd.read_csv(\"haberman.csv\")\ndf.head()",
"_____no_output_____"
]
],
[
[
"## 2. Basic Analysis",
"_____no_output_____"
]
],
[
[
"print(\"No. of features are in given dataset : {} \\n\".format(len(df.columns[:-1])))\n\nprint(\"Features are : {} \\n\".format(list(df.columns)[:-1]))\n\nprint(\"Target Feature is : {}\".format(df.columns[-1]))",
"No. of features are in given dataset : 3 \n\nFeatures are : ['age', 'year', 'nodes'] \n\nTarget Feature is : status\n"
],
[
"df.info() # Note: No null entries",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 306 entries, 0 to 305\nData columns (total 4 columns):\nage 306 non-null int64\nyear 306 non-null int64\nnodes 306 non-null int64\nstatus 306 non-null int64\ndtypes: int64(4)\nmemory usage: 9.7 KB\n"
],
[
"df.describe() # basic stats about df",
"_____no_output_____"
],
[
"# !pip install statsmodels\n\n#Median, Quantiles, Percentiles, IQR.\nprint(\"\\nMedians:\")\nprint(np.median(df[\"nodes\"]))\n#Median with an outlier\nprint(np.median(np.append(df[\"age\"],50)));\nprint(np.median(df[\"year\"]))\n\n\nprint(\"\\nQuantiles:\")\nprint(np.percentile(df[\"nodes\"],np.arange(0, 100, 25)))\nprint(np.percentile(df[\"age\"],np.arange(0, 100, 25)))\nprint(np.percentile(df[\"year\"], np.arange(0, 100, 25)))\n\nprint(\"\\n90th Percentiles:\")\nprint(np.percentile(df[\"nodes\"],90))\nprint(np.percentile(df[\"age\"],90))\nprint(np.percentile(df[\"year\"], 90))\n\nfrom statsmodels import robust\nprint (\"\\nMedian Absolute Deviation\")\nprint(robust.mad(df[\"nodes\"]))\nprint(robust.mad(df[\"age\"]))\nprint(robust.mad(df[\"year\"]))\n",
"\nMedians:\n1.0\n52.0\n63.0\n\nQuantiles:\n[0. 0. 1. 4.]\n[30. 44. 52. 60.75]\n[58. 60. 63. 65.75]\n\n90th Percentiles:\n13.0\n67.0\n67.0\n\nMedian Absolute Deviation\n1.482602218505602\n11.860817748044816\n4.447806655516806\n"
],
[
"print(\"No of datapoint is in each feature : {} \\n\".format(df.size / 4))\n\nprint(\"No of classes and datapoint is in dataset :\\n{}\\n\".format(df.status.value_counts()))",
"No of datapoint is in each feature : 306.0 \n\nNo of classes and datapoint is in dataset :\n1 225\n2 81\nName: status, dtype: int64\n\n"
]
],
[
[
"## 3. Insights of cancer patients survival rate of haberman hospital before/after surgery",
"_____no_output_____"
]
],
[
[
"plt.figure(figsize=(15,8));\nsns.set(style='whitegrid');\nplt.rcParams['axes.titlesize'] = 15\nplt.rcParams['axes.titleweight'] = 50\n\nsns.FacetGrid(df, hue='status', size=4) \\\n.map(plt.scatter, 'age', 'nodes') \\\n.add_legend();\n\nplt.title(\"AGE - NODES SCATTER PLOT\");\n\nplt.show();",
"/home/vijay/.conda/envs/pose/lib/python3.6/site-packages/seaborn/axisgrid.py:230: UserWarning: The `size` paramter has been renamed to `height`; please update your code.\n warnings.warn(msg, UserWarning)\n"
]
],
[
[
"`Observation :`\n 1. Features are tightly merged with each other\n 2. Higher lyph nodes causes higher risks \n ",
"_____no_output_____"
],
[
"### 3.1 Multivariate Analysis",
"_____no_output_____"
]
],
[
[
"plt.close();\nsns.set_style(\"whitegrid\");\nsns.pairplot(df, hue=\"status\", size=4, vars=['year','age', 'nodes'], diag_kind='kde', kind='scatter');\nplt.show()\n",
"/home/vijay/.conda/envs/pose/lib/python3.6/site-packages/seaborn/axisgrid.py:2065: UserWarning: The `size` parameter has been renamed to `height`; pleaes update your code.\n warnings.warn(msg, UserWarning)\n/home/vijay/.conda/envs/pose/lib/python3.6/site-packages/scipy-1.1.0-py3.6-linux-x86_64.egg/scipy/stats/stats.py:1713: FutureWarning: Using a non-tuple sequence for multidimensional indexing is deprecated; use `arr[tuple(seq)]` instead of `arr[seq]`. In the future this will be interpreted as an array index, `arr[np.array(seq)]`, which will result either in an error or a different result.\n return np.add.reduce(sorted[indexer] * weights, axis=axis) / sumval\n"
]
],
[
[
"`observation : `\n\n 1. All features are tightly overlapped\n 2. aged peoples with higher lyph nodes is having higher possibility of survival risk",
"_____no_output_____"
],
[
"### 3.2 Univariate Analysis\n\n#### Histogram, CDF, PDF",
"_____no_output_____"
]
],
[
[
"sns.set_style(\"whitegrid\");\nplt.figure(figsize=(15,8))\nplt.rcParams['axes.titlesize'] = 20\nplt.rcParams['axes.titleweight'] = 10\n\nsns.distplot(df.loc[df['status'] == 1].nodes , bins=20, label='survived', color='Green');\nsns.distplot(df.loc[df['status'] == 2].nodes , bins=20, label='unsurvived', color='Red');\nplt.legend();\nplt.title(\"NODES DISTRIBUTION OVER TARGET\");",
"_____no_output_____"
]
],
[
[
"`observation :`\n\n 1. The person who has nodes > 10, is having higher survival risk;\n 2. if the person who has < 10, is having low survival risk",
"_____no_output_____"
]
],
[
[
"sns.set_style(\"whitegrid\");\nplt.figure(figsize=(15,8))\n\nplt.rcParams['axes.titlesize'] = 20\nplt.rcParams['axes.titleweight'] = 10\n\nsns.distplot(df.loc[df['status'] == 1].age , bins=20, label='survived', color='Green');\nsns.distplot(df.loc[df['status'] == 2].age , bins=20, label='unsurvived', color='Red');\nplt.legend();\nplt.title(\"AGE DISTRIBUTION OVER TARGET\");",
"_____no_output_____"
]
],
[
[
"`observation : `\n\n 1. people whose age lies between 42-55, they slightly have higher survival risk possibility;",
"_____no_output_____"
]
],
[
[
"sns.set_style(\"whitegrid\");\nplt.figure(figsize=(15,8))\n\nplt.rcParams['axes.titlesize'] = 20\nplt.rcParams['axes.titleweight'] = 10\n\nsns.distplot(df.loc[df['status'] == 1].year , bins=20, label='survived', color='Green')\nsns.distplot(df.loc[df['status'] == 2].year , bins=20, label='unsurvived', color='Red')\nplt.legend()\nplt.title(\"YEAR DISTRIBUTION OVER TARGET\");",
"_____no_output_____"
]
],
[
[
"`observation : `\n\n 1. patient who had surgery inbetween [1958 - mid of 1963] and [1966 - 1968] years had highly survived;\n 2. patient who had surgery inbetween [1963 - 1966] years had higher survival risk.",
"_____no_output_____"
]
],
[
[
"sns.set_style(\"whitegrid\");\nplt.figure(figsize=(10,6))\n\nplt.rcParams['axes.titlesize'] = 20\nplt.rcParams['axes.titleweight'] = 10\n\ncount, bin_edges = np.histogram(df.loc[df['status'] == 1].nodes, bins=10, density=True)\nnodes_pdf = count / sum(count)\nnodes_cdf = np.cumsum(nodes_pdf)\nplt.plot(bin_edges[1:],nodes_pdf, color='green', marker='o', linestyle='dashed')\nplt.plot(bin_edges[1:],nodes_cdf, color='black', marker='o', linestyle='dashed')\n\ncount, bin_edges = np.histogram(df.loc[df['status'] == 1].age, bins=10, density=True)\nage_pdf = count / sum(count)\nage_cdf = np.cumsum(age_pdf)\nplt.plot(bin_edges[1:],age_pdf, color='red', marker='o', linestyle='dotted')\nplt.plot(bin_edges[1:],age_cdf, color='black', marker='o', linestyle='dotted')\n\ncount, bin_edges = np.histogram(df.loc[df['status'] == 1].year, bins=10, density=True)\nyear_pdf = count / sum(count)\nyear_cdf = np.cumsum(year_pdf)\nplt.plot(bin_edges[1:],year_pdf, color='blue', marker='o', linestyle='solid')\nplt.plot(bin_edges[1:],year_cdf, color='black', marker='o', linestyle='solid')\n\nplt.title(\"SURVIVED PATIENTS PDF & CDF\")\n\nplt.legend([\"nodes_pdf\",\"nodes_cdf\", \"age_pdf\", \"age_cdf\", \"year_pdf\", \"year_cdf\"])\n\nplt.show();",
"_____no_output_____"
]
],
[
[
"`observation : `\n\n 1. if nodes < 10, 82% patient survived, else: 10% chances of survival\n 2. if age 45-65, 18% patient survived, than among other patients;\n ",
"_____no_output_____"
]
],
[
[
"sns.set_style(\"whitegrid\");\nplt.figure(figsize=(10,6))\nplt.rcParams['axes.titlesize'] = 20\nplt.rcParams['axes.titleweight'] = 10\n\ncount, bin_edges = np.histogram(df.loc[df['status'] == 2].nodes, bins=10, density=True)\nnodes_pdf = count / sum(count)\nnodes_cdf = np.cumsum(nodes_pdf)\nplt.plot(bin_edges[1:],nodes_pdf, color='green', marker='o', linestyle='dashed')\nplt.plot(bin_edges[1:],nodes_cdf, color='black', marker='o', linestyle='dashed')\n\ncount, bin_edges = np.histogram(df.loc[df['status'] == 2].age, bins=10, density=True)\nage_pdf = count / sum(count)\nage_cdf = np.cumsum(age_pdf)\nplt.plot(bin_edges[1:],age_pdf, color='red', marker='o', linestyle='dotted')\nplt.plot(bin_edges[1:],age_cdf, color='black', marker='o', linestyle='dotted')\n\ncount, bin_edges = np.histogram(df.loc[df['status'] == 2].year, bins=10, density=True)\nyear_pdf = count / sum(count)\nyear_cdf = np.cumsum(year_pdf)\nplt.plot(bin_edges[1:],year_pdf, color='blue', marker='o', linestyle='solid')\nplt.plot(bin_edges[1:],year_cdf, color='black', marker='o', linestyle='solid')\n\nplt.title(\"UNSURVIVED PATIENTS PDF & CDF\")\n\nplt.legend([\"nodes_pdf\",\"nodes_cdf\", \"age_pdf\", \"age_cdf\", \"year_pdf\", \"year_cdf\"])\n\nplt.show();",
"_____no_output_____"
]
],
[
[
"`observation : `\n\n 1. nodes > 20, 97% of unsurvived rate.\n 2. age inbetween 38 - 48 has 20% of unsurvived rate.",
"_____no_output_____"
]
],
[
[
"sns.set_style(\"whitegrid\");\n\ng = sns.catplot(x=\"status\", y=\"nodes\", hue=\"status\", data=df, kind=\"box\", height=4, aspect=.7)\n\ng.fig.set_figwidth(10)\ng.fig.set_figheight(5)\ng.fig.suptitle('[BOX PLOT] NODES OVER STATUS', fontsize=20)\ng.add_legend()\nplt.show()\n",
"_____no_output_____"
]
],
[
[
"`observation :`\n\n 1. 75 percentile of patient who has survived had lyph node less than 5.\n 2. Threshold for unsurvival is 25 percentile, 75 percentile is 12, 25 percentile is 2",
"_____no_output_____"
]
],
[
[
"sns.set_style(\"whitegrid\");\n\ng = sns.catplot(x=\"status\", y=\"nodes\", hue=\"status\", data=df, kind=\"violin\", height=4, aspect=.7);\n\ng.fig.set_figwidth(10)\ng.fig.set_figheight(5)\ng.add_legend()\ng.fig.suptitle('[VIOLIN PLOT] NODES OVER STATUS', fontsize=20)\nplt.show()",
"_____no_output_____"
]
],
[
[
"`observation : `\n\n 1. plot 1 clearly is showing lymph nodes closer to zero has highely survived, whiskers also 0-7.\n 2. plot 2 is showing lymph nodes far away from zero has highly unsurvived, whiskers 0-20 has threshold 0-12 short survival chance.",
"_____no_output_____"
]
],
[
[
"plt.rcParams['axes.titlesize'] = 20\nplt.rcParams['axes.titleweight'] = 10\n\nsns.jointplot(x='age',y='nodes',data=df,kind='kde')\n\nplt.suptitle(\"JOINT_PLOT FOR NODES - AGE\",fontsize=20)\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"`observation : `\n\n 1. long survival is more from age range 47–60 and axillary nodes from 0–3.",
"_____no_output_____"
],
[
"### 3.3 BAR_PLOTS [SUMMARIZATION]",
"_____no_output_____"
]
],
[
[
"plt.figure(figsize=(15,8));\n\nsns.set(style=\"whitegrid\");\n# sns.FacetGrid(df, hue='status')\n\n# Draw a nested barplot to show survival for class and sex\ng = sns.catplot(x=\"nodes\", y=\"status\", data=df,\n height=6, kind=\"bar\", palette=\"muted\");\n\n# g.despine(left=True)\ng.set_ylabels(\"survival probability\");\n\ng.fig.set_figwidth(15);\ng.fig.set_figheight(8.27);\ng.fig.suptitle(\"Survival rate for node vise [0-2]\", fontsize=20);",
"_____no_output_____"
],
[
"plt.figure(figsize=(15,8))\n\nsns.set(style=\"whitegrid\");\n\ng = sns.catplot(x=\"age\", y=\"status\", data=df,\n height=6, kind=\"bar\", palette=\"muted\");\n\ng.set_ylabels(\"survival probability\");\n\ng.fig.set_figwidth(15)\ng.fig.set_figheight(8.27)\n\ng.fig.suptitle(\"Survival rate for age vise [0-2]\", fontsize=20);",
"_____no_output_____"
]
],
[
[
"## 4. Conclusion :",
"_____no_output_____"
],
[
"\n`1. Patient’s age and operation year alone are not deciding factors for his/her survival. Yet, people less than 35 years have more chance of survival.`\n\n`2. Survival chance is inversely proportional to the number of positive axillary nodes. We also saw that the absence of positive axillary nodes cannot always guarantee survival.`\n\n`3. The objective of classifying the survival status of a new patient based on the given features is a difficult task as the data is imbalanced.`",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
]
] |
4a573fa32bfe209c9217873ccb2220f9dc0fb1f3
| 2,245 |
ipynb
|
Jupyter Notebook
|
examples/protyping/dicom/create-ct-dicom/create_dosecheck_comm.ipynb
|
lipteck/pymedphys
|
6e8e2b5db8173eafa6006481ceeca4f4341789e0
|
[
"Apache-2.0"
] | 2 |
2020-02-04T03:21:20.000Z
|
2020-04-11T14:17:53.000Z
|
prototyping/dicom/create-ct-dicom/create_dosecheck_comm.ipynb
|
SimonBiggs/pymedphys
|
83f02eac6549ac155c6963e0a8d1f9284359b652
|
[
"Apache-2.0"
] | 6 |
2020-10-06T15:36:46.000Z
|
2022-02-27T05:15:17.000Z
|
prototyping/dicom/create-ct-dicom/create_dosecheck_comm.ipynb
|
SimonBiggs/pymedphys
|
83f02eac6549ac155c6963e0a8d1f9284359b652
|
[
"Apache-2.0"
] | 1 |
2020-12-20T14:14:00.000Z
|
2020-12-20T14:14:00.000Z
| 21.380952 | 74 | 0.523831 |
[
[
[
"import numpy as np\nfrom dicomutils.builders import StudyBuilder",
"_____no_output_____"
],
[
"import shutil\nimport os",
"_____no_output_____"
],
[
"shutil.rmtree('results')\nos.mkdir('results')",
"_____no_output_____"
],
[
"study_builder = StudyBuilder()\n\nstudy_builder.current_study['PatientPosition'] = 'HFS'\nstudy_builder.current_study['PatientID'] = 'DOSECHECK'\nstudy_builder.current_study['PatientsName'] = 'PHANTOM^DOSECHECK'\n\nct_builder = study_builder.build_ct(\n num_voxels=[303, 252, 283],\n voxel_size=[2.0, 2.0, 2.0],\n pixel_representation=1,\n rescale_slope=1.0,\n rescale_intercept=0,\n center=np.array([0,150,0])\n)\n\nx, y, z = ct_builder.mgrid()\nprint np.min(x), np.max(x)\nprint np.min(y), np.max(y)\nprint np.min(z), np.max(z)\n\nct_builder.pixel_array[:,:,:] = -1024.0\nct_builder.pixel_array[1:-1, 1:-1, 1:-1] = 0.0\n\nct_builder.build()\n\nstudy_builder.write('results')",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code"
]
] |
4a574cdc2f1ae046762bdd75f3049716ada1c021
| 55,965 |
ipynb
|
Jupyter Notebook
|
tutorials/asr/ASR_with_NeMo.ipynb
|
pasandi20/NeMo
|
5a00bc43bd2d520670c6e37d2e1218d08b0189ed
|
[
"Apache-2.0"
] | null | null | null |
tutorials/asr/ASR_with_NeMo.ipynb
|
pasandi20/NeMo
|
5a00bc43bd2d520670c6e37d2e1218d08b0189ed
|
[
"Apache-2.0"
] | null | null | null |
tutorials/asr/ASR_with_NeMo.ipynb
|
pasandi20/NeMo
|
5a00bc43bd2d520670c6e37d2e1218d08b0189ed
|
[
"Apache-2.0"
] | null | null | null | 47.589286 | 984 | 0.648727 |
[
[
[
"\"\"\"\nYou can run either this notebook locally (if you have all the dependencies and a GPU) or on Google Colab.\n\nInstructions for setting up Colab are as follows:\n1. Open a new Python 3 notebook.\n2. Import this notebook from GitHub (File -> Upload Notebook -> \"GITHUB\" tab -> copy/paste GitHub URL)\n3. Connect to an instance with a GPU (Runtime -> Change runtime type -> select \"GPU\" for hardware accelerator)\n4. Run this cell to set up dependencies.\n5. Restart the runtime (Runtime -> Restart Runtime) for any upgraded packages to take effect\n\"\"\"\n# If you're using Google Colab and not running locally, run this cell.\n\n## Install dependencies\n!pip install wget\n!apt-get install sox libsndfile1 ffmpeg\n!pip install unidecode\n!pip install matplotlib>=3.3.2\n\n## Install NeMo\nBRANCH = 'r1.3.0'\n!python -m pip install git+https://github.com/NVIDIA/NeMo.git@$BRANCH#egg=nemo_toolkit[all]\n\n## Grab the config we'll use in this example\n!mkdir configs\n!wget -P configs/ https://raw.githubusercontent.com/NVIDIA/NeMo/$BRANCH/examples/asr/conf/config.yaml\n\n\"\"\"\nRemember to restart the runtime for the kernel to pick up any upgraded packages (e.g. matplotlib)!\nAlternatively, you can uncomment the exit() below to crash and restart the kernel, in the case\nthat you want to use the \"Run All Cells\" (or similar) option.\n\"\"\"\n# exit()",
"_____no_output_____"
]
],
[
[
"# Introduction to End-To-End Automatic Speech Recognition\n\nThis notebook contains a basic tutorial of Automatic Speech Recognition (ASR) concepts, introduced with code snippets using the [NeMo framework](https://github.com/NVIDIA/NeMo).\nWe will first introduce the basics of the main concepts behind speech recognition, then explore concrete examples of what the data looks like and walk through putting together a simple end-to-end ASR pipeline.\n\nWe assume that you are familiar with general machine learning concepts and can follow Python code, and we'll be using the [AN4 dataset from CMU](http://www.speech.cs.cmu.edu/databases/an4/) (with processing using `sox`).",
"_____no_output_____"
],
[
"## Conceptual Overview: What is ASR?\n\nASR, or **Automatic Speech Recognition**, refers to the problem of getting a program to automatically transcribe spoken language (speech-to-text). Our goal is usually to have a model that minimizes the **Word Error Rate (WER)** metric when transcribing speech input. In other words, given some audio file (e.g. a WAV file) containing speech, how do we transform this into the corresponding text with as few errors as possible?\n\nTraditional speech recognition takes a generative approach, modeling the full pipeline of how speech sounds are produced in order to evaluate a speech sample. We would start from a **language model** that encapsulates the most likely orderings of words that are generated (e.g. an n-gram model), to a **pronunciation model** for each word in that ordering (e.g. a pronunciation table), to an **acoustic model** that translates those pronunciations to audio waveforms (e.g. a Gaussian Mixture Model).\n\nThen, if we receive some spoken input, our goal would be to find the most likely sequence of text that would result in the given audio according to our generative pipeline of models. Overall, with traditional speech recognition, we try to model `Pr(audio|transcript)*Pr(transcript)`, and take the argmax of this over possible transcripts.\n\nOver time, neural nets advanced to the point where each component of the traditional speech recognition model could be replaced by a neural model that had better performance and that had a greater potential for generalization. For example, we could replace an n-gram model with a neural language model, and replace a pronunciation table with a neural pronunciation model, and so on. However, each of these neural models need to be trained individually on different tasks, and errors in any model in the pipeline could throw off the whole prediction.\n\nThus, we can see the appeal of **end-to-end ASR architectures**: discriminative models that simply take an audio input and give a textual output, and in which all components of the architecture are trained together towards the same goal. The model's encoder would be akin to an acoustic model for extracting speech features, which can then be directly piped to a decoder which outputs text. If desired, we could integrate a language model that would improve our predictions, as well.\n\nAnd the entire end-to-end ASR model can be trained at once--a much easier pipeline to handle! ",
"_____no_output_____"
],
[
"### End-To-End ASR\n\nWith an end-to-end model, we want to directly learn `Pr(transcript|audio)` in order to predict the transcripts from the original audio. Since we are dealing with sequential information--audio data over time that corresponds to a sequence of letters--RNNs are the obvious choice. But now we have a pressing problem to deal with: since our input sequence (number of audio timesteps) is not the same length as our desired output (transcript length), how do we match each time step from the audio data to the correct output characters?\n\nEarlier speech recognition approaches relied on **temporally-aligned data**, in which each segment of time in an audio file was matched up to a corresponding speech sound such as a phoneme or word. However, if we would like to have the flexibility to predict letter-by-letter to prevent OOV (out of vocabulary) issues, then each time step in the data would have to be labeled with the letter sound that the speaker is making at that point in the audio file. With that information, it seems like we should simply be able to try to predict the correct letter for each time step and then collapse the repeated letters (e.g. the prediction output `LLLAAAAPPTOOOPPPP` would become `LAPTOP`). It turns out that this idea has some problems: not only does alignment make the dataset incredibly labor-intensive to label, but also, what do we do with words like \"book\" that contain consecutive repeated letters? Simply squashing repeated letters together would not work in that case!\n\n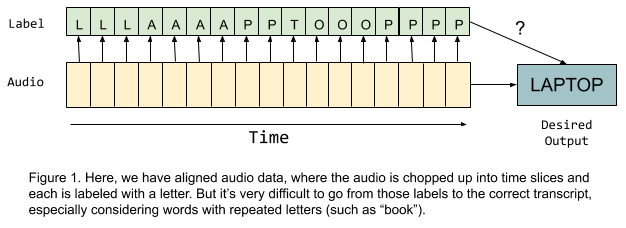\n\nModern end-to-end approaches get around this using methods that don't require manual alignment at all, so that the input-output pairs are really just the raw audio and the transcript--no extra data or labeling required. Let's briefly go over two popular approaches that allow us to do this, Connectionist Temporal Classification (CTC) and sequence-to-sequence models with attention.\n\n#### Connectionist Temporal Classification (CTC)\n\nIn normal speech recognition prediction output, we would expect to have characters such as the letters from A through Z, numbers 0 through 9, spaces (\"\\_\"), and so on. CTC introduces a new intermediate output token called the **blank token** (\"-\") that is useful for getting around the alignment issue.\n\nWith CTC, we still predict one token per time segment of speech, but we use the blank token to figure out where we can and can't collapse the predictions. The appearance of a blank token helps separate repeating letters that should not be collapsed. For instance, with an audio snippet segmented into `T=11` time steps, we could get predictions that look like `BOO-OOO--KK`, which would then collapse to `\"BO-O-K\"`, and then we would remove the blank tokens to get our final output, `BOOK`.\n\nNow, we can predict one output token per time step, then collapse and clean to get sensible output without any fear of ambiguity from repeating letters! A simple way of getting predictions like this would be to apply a bidirectional RNN to the audio input, apply softmax over each time step's output, and then take the token with the highest probability. The method of always taking the best token at each time step is called **greedy decoding, or max decoding**.\n\nTo calculate our loss for backprop, we would like to know the log probability of the model producing the correct transcript, `log(Pr(transcript|audio))`. We can get the log probability of a single intermediate output sequence (e.g. `BOO-OOO--KK`) by summing over the log probabilities we get from each token's softmax value, but note that the resulting sum is different from the log probability of the transcript itself (`BOOK`). This is because there are multiple possible output sequences of the same length that can be collapsed to get the same transcript (e.g. `BBO--OO-KKK` also results in `BOOK`), and so we need to **marginalize over every valid sequence of length `T` that collapses to the transcript**.\n\nTherefore, to get our transcript's log probability given our audio input, we must sum the log probabilities of every sequence of length `T` that collapses to the transcript (e.g. `log(Pr(output: \"BOOK\"|audio)) = log(Pr(BOO-OOO--KK|audio)) + log(Pr(BBO--OO-KKK|audio)) + ...`). In practice, we can use a dynamic programming approach to calculate this, accumulating our log probabilities over different \"paths\" through the softmax outputs at each time step.\n\nIf you would like a more in-depth explanation of how CTC works, or how we can improve our results by using a modified beam search algorithm, feel free to check out the Further Reading section at the end of this notebook for more resources.\n\n#### Sequence-to-Sequence with Attention\n\nOne problem with CTC is that predictions at different time steps are conditionally independent, which is an issue because the words in a continuous utterance tend to be related to each other in some sensible way. With this conditional independence assumption, we can't learn a language model that can represent such dependencies, though we can add a language model on top of the CTC output to mitigate this to some degree.\n\nA popular alternative is to use a sequence-to-sequence model with attention. A typical seq2seq model for ASR consists of some sort of **bidirectional RNN encoder** that consumes the audio sequence timestep-by-timestep, and where the outputs are then passed to an **attention-based decoder**. Each prediction from the decoder is based on attending to some parts of the entire encoded input, as well as the previously outputted tokens.\n\nThe outputs of the decoder can be anything from word pieces to phonemes to letters, and since predictions are not directly tied to time steps of the input, we can just continue producing tokens one-by-one until an end token is given (or we reach a specified max output length). This way, we do not need to deal with audio alignment, and our predicted transcript is just the sequence of outputs given by our decoder.\n\nNow that we have an idea of what some popular end-to-end ASR models look like, let's take a look at the audio data we'll be working with for our example.",
"_____no_output_____"
],
[
"## Taking a Look at Our Data (AN4)\n\nThe AN4 dataset, also known as the Alphanumeric dataset, was collected and published by Carnegie Mellon University. It consists of recordings of people spelling out addresses, names, telephone numbers, etc., one letter or number at a time, as well as their corresponding transcripts. We choose to use AN4 for this tutorial because it is relatively small, with 948 training and 130 test utterances, and so it trains quickly.\n\nBefore we get started, let's download and prepare the dataset. The utterances are available as `.sph` files, so we will need to convert them to `.wav` for later processing. If you are not using Google Colab, please make sure you have [Sox](http://sox.sourceforge.net/) installed for this step--see the \"Downloads\" section of the linked Sox homepage. (If you are using Google Colab, Sox should have already been installed in the setup cell at the beginning.)",
"_____no_output_____"
]
],
[
[
"# This is where the an4/ directory will be placed.\n# Change this if you don't want the data to be extracted in the current directory.\ndata_dir = '.'",
"_____no_output_____"
],
[
"import glob\nimport os\nimport subprocess\nimport tarfile\nimport wget\n\n# Download the dataset. This will take a few moments...\nprint(\"******\")\nif not os.path.exists(data_dir + '/an4_sphere.tar.gz'):\n an4_url = 'http://www.speech.cs.cmu.edu/databases/an4/an4_sphere.tar.gz'\n an4_path = wget.download(an4_url, data_dir)\n print(f\"Dataset downloaded at: {an4_path}\")\nelse:\n print(\"Tarfile already exists.\")\n an4_path = data_dir + '/an4_sphere.tar.gz'\n\nif not os.path.exists(data_dir + '/an4/'):\n # Untar and convert .sph to .wav (using sox)\n tar = tarfile.open(an4_path)\n tar.extractall(path=data_dir)\n\n print(\"Converting .sph to .wav...\")\n sph_list = glob.glob(data_dir + '/an4/**/*.sph', recursive=True)\n for sph_path in sph_list:\n wav_path = sph_path[:-4] + '.wav'\n cmd = [\"sox\", sph_path, wav_path]\n subprocess.run(cmd)\nprint(\"Finished conversion.\\n******\")",
"_____no_output_____"
]
],
[
[
"You should now have a folder called `an4` that contains `etc/an4_train.transcription`, `etc/an4_test.transcription`, audio files in `wav/an4_clstk` and `wav/an4test_clstk`, along with some other files we will not need.\n\nNow we can load and take a look at the data. As an example, file `cen2-mgah-b.wav` is a 2.6 second-long audio recording of a man saying the letters \"G L E N N\" one-by-one. To confirm this, we can listen to the file:",
"_____no_output_____"
]
],
[
[
"import librosa\nimport IPython.display as ipd\n\n# Load and listen to the audio file\nexample_file = data_dir + '/an4/wav/an4_clstk/mgah/cen2-mgah-b.wav'\naudio, sample_rate = librosa.load(example_file)\n\nipd.Audio(example_file, rate=sample_rate)",
"_____no_output_____"
]
],
[
[
"In an ASR task, if this WAV file was our input, then \"G L E N N\" would be our desired output.\n\nLet's plot the waveform, which is simply a line plot of the sequence of values that we read from the file. This is a format of viewing audio that you are likely to be familiar with seeing in many audio editors and visualizers:",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nimport librosa.display\nimport matplotlib.pyplot as plt\n\n# Plot our example audio file's waveform\nplt.rcParams['figure.figsize'] = (15,7)\nplt.title('Waveform of Audio Example')\nplt.ylabel('Amplitude')\n\n_ = librosa.display.waveplot(audio)",
"_____no_output_____"
]
],
[
[
"We can see the activity in the waveform that corresponds to each letter in the audio, as our speaker here enunciates quite clearly!\nYou can kind of tell that each spoken letter has a different \"shape,\" and it's interesting to note that last two blobs look relatively similar, which is expected because they are both the letter \"N.\"\n\n### Spectrograms and Mel Spectrograms\n\nHowever, since audio information is more useful in the context of frequencies of sound over time, we can get a better representation than this raw sequence of 57,330 values.\nWe can apply a [Fourier Transform](https://en.wikipedia.org/wiki/Fourier_transform) on our audio signal to get something more useful: a **spectrogram**, which is a representation of the energy levels (i.e. amplitude, or \"loudness\") of each frequency (i.e. pitch) of the signal over the duration of the file.\nA spectrogram (which can be viewed as a heat map) is a good way of seeing how the *strengths of various frequencies in the audio vary over time*, and is obtained by breaking up the signal into smaller, usually overlapping chunks and performing a Short-Time Fourier Transform (STFT) on each.\n\nLet's examine what the spectrogram of our sample looks like.",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\n# Get spectrogram using Librosa's Short-Time Fourier Transform (stft)\nspec = np.abs(librosa.stft(audio))\nspec_db = librosa.amplitude_to_db(spec, ref=np.max) # Decibels\n\n# Use log scale to view frequencies\nlibrosa.display.specshow(spec_db, y_axis='log', x_axis='time')\nplt.colorbar()\nplt.title('Audio Spectrogram');",
"_____no_output_____"
]
],
[
[
"Again, we are able to see each letter being pronounced, and that the last two blobs that correspond to the \"N\"s are pretty similar-looking. But how do we interpret these shapes and colors? Just as in the waveform plot before, we see time passing on the x-axis (all 2.6s of audio). But now, the y-axis represents different frequencies (on a log scale), and *the color on the plot shows the strength of a frequency at a particular point in time*.\n\nWe're still not done yet, as we can make one more potentially useful tweak: using the **Mel Spectrogram** instead of the normal spectrogram. This is simply a change in the frequency scale that we use from linear (or logarithmic) to the mel scale, which is \"a perceptual scale of pitches judged by listeners to be equal in distance from one another\" (from [Wikipedia](https://en.wikipedia.org/wiki/Mel_scale)).\n\nIn other words, it's a transformation of the frequencies to be more aligned to what humans perceive; a change of +1000Hz from 2000Hz->3000Hz sounds like a larger difference to us than 9000Hz->10000Hz does, so the mel scale normalizes this such that equal distances sound like equal differences to the human ear. Intuitively, we use the mel spectrogram because in this case we are processing and transcribing human speech, such that transforming the scale to better match what we hear is a useful procedure.",
"_____no_output_____"
]
],
[
[
"# Plot the mel spectrogram of our sample\nmel_spec = librosa.feature.melspectrogram(audio, sr=sample_rate)\nmel_spec_db = librosa.power_to_db(mel_spec, ref=np.max)\n\nlibrosa.display.specshow(\n mel_spec_db, x_axis='time', y_axis='mel')\nplt.colorbar()\nplt.title('Mel Spectrogram');",
"_____no_output_____"
]
],
[
[
"## Convolutional ASR Models\n\nLet's take a look at the model that we will be building, and how we specify its parameters.\n\n### The Jasper Model\n\nWe will be training a small [Jasper (Just Another SPeech Recognizer) model](https://arxiv.org/abs/1904.03288) from scratch (e.g. initialized randomly). \nIn brief, Jasper architectures consist of a repeated block structure that utilizes 1D convolutions.\nIn a Jasper_KxR model, `R` sub-blocks (consisting of a 1D convolution, batch norm, ReLU, and dropout) are grouped into a single block, which is then repeated `K` times.\nWe also have a one extra block at the beginning and a few more at the end that are invariant of `K` and `R`, and we use CTC loss.\n\n### The QuartzNet Model\n\nThe QuartzNet is better variant of Jasper with a key difference that it uses time-channel separable 1D convolutions. This allows it to dramatically reduce number of weights while keeping similar accuracy.\n\nA Jasper/QuartzNet models look like this (QuartzNet model is pictured):\n\n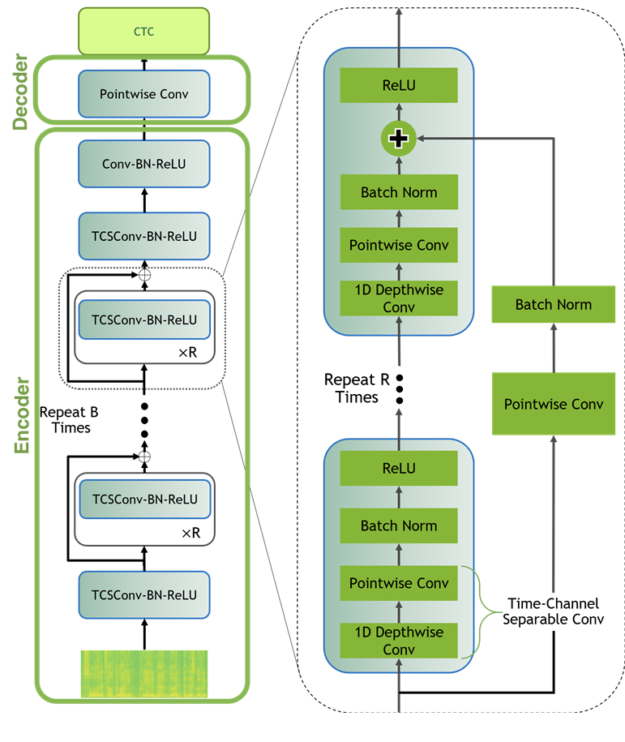",
"_____no_output_____"
],
[
"# Using NeMo for Automatic Speech Recognition\n\nNow that we have an idea of what ASR is and how the audio data looks like, we can start using NeMo to do some ASR!\n\nWe'll be using the **Neural Modules (NeMo) toolkit** for this part, so if you haven't already, you should download and install NeMo and its dependencies. To do so, just follow the directions on the [GitHub page](https://github.com/NVIDIA/NeMo), or in the [documentation](https://docs.nvidia.com/deeplearning/nemo/user-guide/docs/en/v1.0.2/).\n\nNeMo lets us easily hook together the components (modules) of our model, such as the data layer, intermediate layers, and various losses, without worrying too much about implementation details of individual parts or connections between modules. NeMo also comes with complete models which only require your data and hyperparameters for training.",
"_____no_output_____"
]
],
[
[
"# NeMo's \"core\" package\nimport nemo\n# NeMo's ASR collection - this collections contains complete ASR models and\n# building blocks (modules) for ASR\nimport nemo.collections.asr as nemo_asr",
"_____no_output_____"
]
],
[
[
"## Using an Out-of-the-Box Model\n\nNeMo's ASR collection comes with many building blocks and even complete models that we can use for training and evaluation. Moreover, several models come with pre-trained weights. Let's instantiate a complete QuartzNet15x5 model.",
"_____no_output_____"
]
],
[
[
"# This line will download pre-trained QuartzNet15x5 model from NVIDIA's NGC cloud and instantiate it for you\nquartznet = nemo_asr.models.EncDecCTCModel.from_pretrained(model_name=\"QuartzNet15x5Base-En\")",
"_____no_output_____"
]
],
[
[
"Next, we'll simply add paths to files we want to transcribe into the list and pass it to our model. Note that it will work for relatively short (<25 seconds) files. ",
"_____no_output_____"
]
],
[
[
"files = ['./an4/wav/an4_clstk/mgah/cen2-mgah-b.wav']\nfor fname, transcription in zip(files, quartznet.transcribe(paths2audio_files=files)):\n print(f\"Audio in {fname} was recognized as: {transcription}\")",
"_____no_output_____"
]
],
[
[
"That was easy! But there are plenty of scenarios where you would want to fine-tune the model on your own data or even train from scratch. For example, this out-of-the box model will obviously not work for Spanish and would likely perform poorly for telephone audio. So if you have collected your own data, you certainly should attempt to fine-tune or train on it!",
"_____no_output_____"
],
[
"## Training from Scratch\n\nTo train from scratch, you need to prepare your training data in the right format and specify your models architecture.",
"_____no_output_____"
],
[
"### Creating Data Manifests\n\nThe first thing we need to do now is to create manifests for our training and evaluation data, which will contain the metadata of our audio files. NeMo data sets take in a standardized manifest format where each line corresponds to one sample of audio, such that the number of lines in a manifest is equal to the number of samples that are represented by that manifest. A line must contain the path to an audio file, the corresponding transcript (or path to a transcript file), and the duration of the audio sample.\n\nHere's an example of what one line in a NeMo-compatible manifest might look like:\n```\n{\"audio_filepath\": \"path/to/audio.wav\", \"duration\": 3.45, \"text\": \"this is a nemo tutorial\"}\n```\n\nWe can build our training and evaluation manifests using `an4/etc/an4_train.transcription` and `an4/etc/an4_test.transcription`, which have lines containing transcripts and their corresponding audio file IDs:\n```\n...\n<s> P I T T S B U R G H </s> (cen5-fash-b)\n<s> TWO SIX EIGHT FOUR FOUR ONE EIGHT </s> (cen7-fash-b)\n...\n```",
"_____no_output_____"
]
],
[
[
"# --- Building Manifest Files --- #\nimport json\n\n# Function to build a manifest\ndef build_manifest(transcripts_path, manifest_path, wav_path):\n with open(transcripts_path, 'r') as fin:\n with open(manifest_path, 'w') as fout:\n for line in fin:\n # Lines look like this:\n # <s> transcript </s> (fileID)\n transcript = line[: line.find('(')-1].lower()\n transcript = transcript.replace('<s>', '').replace('</s>', '')\n transcript = transcript.strip()\n\n file_id = line[line.find('(')+1 : -2] # e.g. \"cen4-fash-b\"\n audio_path = os.path.join(\n data_dir, wav_path,\n file_id[file_id.find('-')+1 : file_id.rfind('-')],\n file_id + '.wav')\n\n duration = librosa.core.get_duration(filename=audio_path)\n\n # Write the metadata to the manifest\n metadata = {\n \"audio_filepath\": audio_path,\n \"duration\": duration,\n \"text\": transcript\n }\n json.dump(metadata, fout)\n fout.write('\\n')\n \n# Building Manifests\nprint(\"******\")\ntrain_transcripts = data_dir + '/an4/etc/an4_train.transcription'\ntrain_manifest = data_dir + '/an4/train_manifest.json'\nif not os.path.isfile(train_manifest):\n build_manifest(train_transcripts, train_manifest, 'an4/wav/an4_clstk')\n print(\"Training manifest created.\")\n\ntest_transcripts = data_dir + '/an4/etc/an4_test.transcription'\ntest_manifest = data_dir + '/an4/test_manifest.json'\nif not os.path.isfile(test_manifest):\n build_manifest(test_transcripts, test_manifest, 'an4/wav/an4test_clstk')\n print(\"Test manifest created.\")\nprint(\"***Done***\")",
"_____no_output_____"
]
],
[
[
"### Specifying Our Model with a YAML Config File\n\nFor this tutorial, we'll build a *Jasper_4x1 model*, with `K=4` blocks of single (`R=1`) sub-blocks and a *greedy CTC decoder*, using the configuration found in `./configs/config.yaml`.\n\nIf we open up this config file, we find model section which describes architecture of our model. A model contains an entry labeled `encoder`, with a field called `jasper` that contains a list with multiple entries. Each of the members in this list specifies one block in our model, and looks something like this:\n```\n- filters: 128\n repeat: 1\n kernel: [11]\n stride: [2]\n dilation: [1]\n dropout: 0.2\n residual: false\n separable: true\n se: true\n se_context_size: -1\n```\nThe first member of the list corresponds to the first block in the Jasper architecture diagram, which appears regardless of `K` and `R`.\nNext, we have four entries that correspond to the `K=4` blocks, and each has `repeat: 1` since we are using `R=1`.\nThese are followed by two more entries for the blocks that appear at the end of our Jasper model before the CTC loss.\n\nThere are also some entries at the top of the file that specify how we will handle training (`train_ds`) and validation (`validation_ds`) data.\n\nUsing a YAML config such as this is helpful for getting a quick and human-readable overview of what your architecture looks like, and allows you to swap out model and run configurations easily without needing to change your code.",
"_____no_output_____"
]
],
[
[
"# --- Config Information ---#\ntry:\n from ruamel.yaml import YAML\nexcept ModuleNotFoundError:\n from ruamel_yaml import YAML\nconfig_path = './configs/config.yaml'\n\nyaml = YAML(typ='safe')\nwith open(config_path) as f:\n params = yaml.load(f)\nprint(params)",
"_____no_output_____"
]
],
[
[
"### Training with PyTorch Lightning\n\nNeMo models and modules can be used in any PyTorch code where torch.nn.Module is expected.\n\nHowever, NeMo's models are based on [PytorchLightning's](https://github.com/PyTorchLightning/pytorch-lightning) LightningModule and we recommend you use PytorchLightning for training and fine-tuning as it makes using mixed precision and distributed training very easy. So to start, let's create Trainer instance for training on GPU for 50 epochs",
"_____no_output_____"
]
],
[
[
"import pytorch_lightning as pl\ntrainer = pl.Trainer(gpus=1, max_epochs=50)",
"_____no_output_____"
]
],
[
[
"Next, we instantiate and ASR model based on our ``config.yaml`` file from the previous section.\nNote that this is a stage during which we also tell the model where our training and validation manifests are.",
"_____no_output_____"
]
],
[
[
"from omegaconf import DictConfig\nparams['model']['train_ds']['manifest_filepath'] = train_manifest\nparams['model']['validation_ds']['manifest_filepath'] = test_manifest\nfirst_asr_model = nemo_asr.models.EncDecCTCModel(cfg=DictConfig(params['model']), trainer=trainer)",
"_____no_output_____"
]
],
[
[
"With that, we can start training with just one line!",
"_____no_output_____"
]
],
[
[
"# Start training!!!\ntrainer.fit(first_asr_model)",
"_____no_output_____"
]
],
[
[
"There we go! We've put together a full training pipeline for the model and trained it for 50 epochs.\n\nIf you'd like to save this model checkpoint for loading later (e.g. for fine-tuning, or for continuing training), you can simply call `first_asr_model.save_to(<checkpoint_path>)`. Then, to restore your weights, you can rebuild the model using the config (let's say you call it `first_asr_model_continued` this time) and call `first_asr_model_continued.restore_from(<checkpoint_path>)`.\n\n### After Training: Monitoring Progress and Changing Hyperparameters\nWe can now start Tensorboard to see how training went. Recall that WER stands for Word Error Rate and so the lower it is, the better.",
"_____no_output_____"
]
],
[
[
"try:\n from google import colab\n COLAB_ENV = True\nexcept (ImportError, ModuleNotFoundError):\n COLAB_ENV = False\n\n# Load the TensorBoard notebook extension\nif COLAB_ENV:\n %load_ext tensorboard\n %tensorboard --logdir lightning_logs/\nelse:\n print(\"To use tensorboard, please use this notebook in a Google Colab environment.\")",
"_____no_output_____"
]
],
[
[
"We could improve this model by playing with hyperparameters. We can look at the current hyperparameters with the following:",
"_____no_output_____"
]
],
[
[
"print(params['model']['optim'])",
"_____no_output_____"
]
],
[
[
"Let's say we wanted to change the learning rate. To do so, we can create a `new_opt` dict and set our desired learning rate, then call `<model>.setup_optimization()` with the new optimization parameters.",
"_____no_output_____"
]
],
[
[
"import copy\nnew_opt = copy.deepcopy(params['model']['optim'])\nnew_opt['lr'] = 0.001\nfirst_asr_model.setup_optimization(optim_config=DictConfig(new_opt))\n# And then you can invoke trainer.fit(first_asr_model)",
"_____no_output_____"
]
],
[
[
"## Inference\n\nLet's have a quick look at how one could run inference with NeMo's ASR model.\n\nFirst, ``EncDecCTCModel`` and its subclasses contain a handy ``transcribe`` method which can be used to simply obtain audio files' transcriptions. It also has batch_size argument to improve performance.",
"_____no_output_____"
]
],
[
[
"print(first_asr_model.transcribe(paths2audio_files=['./an4/wav/an4_clstk/mgah/cen2-mgah-b.wav',\n './an4/wav/an4_clstk/fmjd/cen7-fmjd-b.wav',\n './an4/wav/an4_clstk/fmjd/cen8-fmjd-b.wav',\n './an4/wav/an4_clstk/fkai/cen8-fkai-b.wav'],\n batch_size=4))",
"_____no_output_____"
]
],
[
[
"Below is an example of a simple inference loop in pure PyTorch. It also shows how one can compute Word Error Rate (WER) metric between predictions and references.",
"_____no_output_____"
]
],
[
[
"# Bigger batch-size = bigger throughput\nparams['model']['validation_ds']['batch_size'] = 16\n\n# Setup the test data loader and make sure the model is on GPU\nfirst_asr_model.setup_test_data(test_data_config=params['model']['validation_ds'])\nfirst_asr_model.cuda()\n\n# We will be computing Word Error Rate (WER) metric between our hypothesis and predictions.\n# WER is computed as numerator/denominator.\n# We'll gather all the test batches' numerators and denominators.\nwer_nums = []\nwer_denoms = []\n\n# Loop over all test batches.\n# Iterating over the model's `test_dataloader` will give us:\n# (audio_signal, audio_signal_length, transcript_tokens, transcript_length)\n# See the AudioToCharDataset for more details.\nfor test_batch in first_asr_model.test_dataloader():\n test_batch = [x.cuda() for x in test_batch]\n targets = test_batch[2]\n targets_lengths = test_batch[3] \n log_probs, encoded_len, greedy_predictions = first_asr_model(\n input_signal=test_batch[0], input_signal_length=test_batch[1]\n )\n # Notice the model has a helper object to compute WER\n first_asr_model._wer.update(greedy_predictions, targets, targets_lengths)\n _, wer_num, wer_denom = first_asr_model._wer.compute()\n first_asr_model._wer.reset()\n wer_nums.append(wer_num.detach().cpu().numpy())\n wer_denoms.append(wer_denom.detach().cpu().numpy())\n\n # Release tensors from GPU memory\n del test_batch, log_probs, targets, targets_lengths, encoded_len, greedy_predictions\n\n# We need to sum all numerators and denominators first. Then divide.\nprint(f\"WER = {sum(wer_nums)/sum(wer_denoms)}\")",
"_____no_output_____"
]
],
[
[
"This WER is not particularly impressive and could be significantly improved. You could train longer (try 100 epochs) to get a better number. Check out the next section on how to improve it further.",
"_____no_output_____"
],
[
"## Model Improvements\n\nYou already have all you need to create your own ASR model in NeMo, but there are a few more tricks that you can employ if you so desire. In this section, we'll briefly cover a few possibilities for improving an ASR model.\n\n### Data Augmentation\n\nThere exist several ASR data augmentation methods that can increase the size of our training set.\n\nFor example, we can perform augmentation on the spectrograms by zeroing out specific frequency segments (\"frequency masking\") or time segments (\"time masking\") as described by [SpecAugment](https://arxiv.org/abs/1904.08779), or zero out rectangles on the spectrogram as in [Cutout](https://arxiv.org/pdf/1708.04552.pdf). In NeMo, we can do all three of these by simply adding in a `SpectrogramAugmentation` neural module. (As of now, it does not perform the time warping from the SpecAugment paper.)\n\nOur toy model does not do spectrogram augmentation. But the real one we got from cloud does:",
"_____no_output_____"
]
],
[
[
"print(quartznet._cfg['spec_augment'])",
"_____no_output_____"
]
],
[
[
"If you want to enable SpecAugment in your model, make sure your .yaml config file contains 'model/spec_augment' section which looks like the one above.",
"_____no_output_____"
],
[
"### Transfer learning\n\nTransfer learning is an important machine learning technique that uses a model’s knowledge of one task to make it perform better on another. Fine-tuning is one of the techniques to perform transfer learning. It is an essential part of the recipe for many state-of-the-art results where a base model is first pretrained on a task with abundant training data and then fine-tuned on different tasks of interest where the training data is less abundant or even scarce.\n\nIn ASR you might want to do fine-tuning in multiple scenarios, for example, when you want to improve your model's performance on a particular domain (medical, financial, etc.) or on accented speech. You can even transfer learn from one language to another! Check out [this paper](https://arxiv.org/abs/2005.04290) for examples.\n\nTransfer learning with NeMo is simple. Let's demonstrate how the model we got from the cloud could be fine-tuned on AN4 data. (NOTE: this is a toy example). And, while we are at it, we will change model's vocabulary, just to demonstrate how it's done.",
"_____no_output_____"
]
],
[
[
"# Check what kind of vocabulary/alphabet the model has right now\nprint(quartznet.decoder.vocabulary)\n\n# Let's add \"!\" symbol there. Note that you can (and should!) change the vocabulary\n# entirely when fine-tuning using a different language.\nquartznet.change_vocabulary(\n new_vocabulary=[\n ' ', 'a', 'b', 'c', 'd', 'e', 'f', 'g', 'h', 'i', 'j', 'k', 'l', 'm', 'n',\n 'o', 'p', 'q', 'r', 's', 't', 'u', 'v', 'w', 'x', 'y', 'z', \"'\", \"!\"\n ]\n)",
"_____no_output_____"
]
],
[
[
"After this, our decoder has completely changed, but our encoder (which is where most of the weights are) remained intact. Let's fine tune-this model for 2 epochs on AN4 dataset. We will also use the smaller learning rate from ``new_opt` (see the \"After Training\" section)`.",
"_____no_output_____"
]
],
[
[
"# Use the smaller learning rate we set before\nquartznet.setup_optimization(optim_config=DictConfig(new_opt))\n\n# Point to the data we'll use for fine-tuning as the training set\nquartznet.setup_training_data(train_data_config=params['model']['train_ds'])\n\n# Point to the new validation data for fine-tuning\nquartznet.setup_validation_data(val_data_config=params['model']['validation_ds'])\n\n# And now we can create a PyTorch Lightning trainer and call `fit` again.\ntrainer = pl.Trainer(gpus=[1], max_epochs=2)\ntrainer.fit(quartznet)",
"_____no_output_____"
]
],
[
[
"### Fast Training\n\nLast but not least, we could simply speed up training our model! If you have the resources, you can speed up training by splitting the workload across multiple GPUs. Otherwise (or in addition), there's always mixed precision training, which allows you to increase your batch size.\n\nYou can use [PyTorch Lightning's Trainer object](https://pytorch-lightning.readthedocs.io/en/latest/common/trainer.html?highlight=Trainer) to handle mixed-precision and distributed training for you. Below are some examples of flags you would pass to the `Trainer` to use these features:\n\n```python\n# Mixed precision:\ntrainer = pl.Trainer(amp_level='O1', precision=16)\n\n# Trainer with a distributed backend:\ntrainer = pl.Trainer(gpus=2, num_nodes=2, accelerator='ddp')\n\n# Of course, you can combine these flags as well.\n```\n\nFinally, have a look at [example scripts in NeMo repository](https://github.com/NVIDIA/NeMo/blob/stable/examples/asr/speech_to_text.py) which can handle mixed precision and distributed training using command-line arguments.",
"_____no_output_____"
],
[
"### Deployment\n\nNote: It is recommended to run the deployment code from the NVIDIA PyTorch container.\n\nLet's get back to our pre-trained model and see how easy it can be exported to an ONNX file\nin order to run it in an inference engine like TensorRT or ONNXRuntime.\n\nIf you are running in an environment outside of the NVIDIA PyTorch container (like Google Colab for example) then you will have to build the onnxruntime and onnxruntime-gpu. The cell below gives an example of how to build those runtimes but the example may have to be adapted depending on your environment.",
"_____no_output_____"
]
],
[
[
"!pip install --upgrade onnxruntime onnxruntime-gpu\n#!mkdir -p ort\n#%cd ort\n#!git clean -xfd\n#!git clone --depth 1 --branch v1.8.0 https://github.com/microsoft/onnxruntime.git .\n#!./build.sh --skip_tests --config Release --build_shared_lib --parallel --use_cuda --cuda_home /usr/local/cuda --cudnn_home /usr/lib/#x86_64-linux-gnu --build_wheel\n#!pip uninstall -y onnxruntime\n#!pip uninstall -y onnxruntime-gpu\n#!pip install --upgrade --force-reinstall ./build/Linux/Release/dist/onnxruntime*.whl\n#%cd ..",
"_____no_output_____"
]
],
[
[
"Then run",
"_____no_output_____"
]
],
[
[
"import json\nimport os\nimport tempfile\nimport onnxruntime\nimport torch\n\nimport numpy as np\nimport nemo.collections.asr as nemo_asr\nfrom nemo.collections.asr.data.audio_to_text import AudioToCharDataset\nfrom nemo.collections.asr.metrics.wer import WER\n\ndef to_numpy(tensor):\n return tensor.detach().cpu().numpy() if tensor.requires_grad else tensor.cpu().numpy()\n\ndef setup_transcribe_dataloader(cfg, vocabulary):\n config = {\n 'manifest_filepath': os.path.join(cfg['temp_dir'], 'manifest.json'),\n 'sample_rate': 16000,\n 'labels': vocabulary,\n 'batch_size': min(cfg['batch_size'], len(cfg['paths2audio_files'])),\n 'trim_silence': True,\n 'shuffle': False,\n }\n dataset = AudioToCharDataset(\n manifest_filepath=config['manifest_filepath'],\n labels=config['labels'],\n sample_rate=config['sample_rate'],\n int_values=config.get('int_values', False),\n augmentor=None,\n max_duration=config.get('max_duration', None),\n min_duration=config.get('min_duration', None),\n max_utts=config.get('max_utts', 0),\n blank_index=config.get('blank_index', -1),\n unk_index=config.get('unk_index', -1),\n normalize=config.get('normalize_transcripts', False),\n trim=config.get('trim_silence', True),\n parser=config.get('parser', 'en'),\n )\n return torch.utils.data.DataLoader(\n dataset=dataset,\n batch_size=config['batch_size'],\n collate_fn=dataset.collate_fn,\n drop_last=config.get('drop_last', False),\n shuffle=False,\n num_workers=config.get('num_workers', 0),\n pin_memory=config.get('pin_memory', False),\n )\n\nquartznet = nemo_asr.models.EncDecCTCModel.from_pretrained(model_name=\"QuartzNet15x5Base-En\")\n\nquartznet.export('qn.onnx')\n\nort_session = onnxruntime.InferenceSession('qn.onnx')\n\nwith tempfile.TemporaryDirectory() as tmpdir:\n with open(os.path.join(tmpdir, 'manifest.json'), 'w') as fp:\n for audio_file in files:\n entry = {'audio_filepath': audio_file, 'duration': 100000, 'text': 'nothing'}\n fp.write(json.dumps(entry) + '\\n')\n\n config = {'paths2audio_files': files, 'batch_size': 4, 'temp_dir': tmpdir}\n temporary_datalayer = setup_transcribe_dataloader(config, quartznet.decoder.vocabulary)\n for test_batch in temporary_datalayer:\n processed_signal, processed_signal_len = quartznet.preprocessor(\n input_signal=test_batch[0].to(quartznet.device), length=test_batch[1].to(quartznet.device)\n )\n ort_inputs = {ort_session.get_inputs()[0].name: to_numpy(processed_signal),}\n ologits = ort_session.run(None, ort_inputs)\n alogits = np.asarray(ologits)\n logits = torch.from_numpy(alogits[0])\n greedy_predictions = logits.argmax(dim=-1, keepdim=False)\n wer = WER(vocabulary=quartznet.decoder.vocabulary, batch_dim_index=0, use_cer=False, ctc_decode=True)\n hypotheses = wer.ctc_decoder_predictions_tensor(greedy_predictions)\n print(hypotheses)\n break\n",
"_____no_output_____"
]
],
[
[
"## Under the Hood\n\nNeMo is open-source and we do all our model development in the open, so you can inspect our code if you wish.\n\nIn particular, ``nemo_asr.model.EncDecCTCModel`` is an encoder-decoder model which is constructed using several ``Neural Modules`` taken from ``nemo_asr.modules.`` Here is what its forward pass looks like:\n```python\ndef forward(self, input_signal, input_signal_length):\n processed_signal, processed_signal_len = self.preprocessor(\n input_signal=input_signal, length=input_signal_length,\n )\n # Spec augment is not applied during evaluation/testing\n if self.spec_augmentation is not None and self.training:\n processed_signal = self.spec_augmentation(input_spec=processed_signal)\n encoded, encoded_len = self.encoder(audio_signal=processed_signal, length=processed_signal_len)\n log_probs = self.decoder(encoder_output=encoded)\n greedy_predictions = log_probs.argmax(dim=-1, keepdim=False)\n return log_probs, encoded_len, greedy_predictions\n```\nHere:\n\n* ``self.preprocessor`` is an instance of ``nemo_asr.modules.AudioToMelSpectrogramPreprocessor``, which is a neural module that takes audio signal and converts it into a Mel-Spectrogram\n* ``self.spec_augmentation`` - is a neural module of type ```nemo_asr.modules.SpectrogramAugmentation``, which implements data augmentation. \n* ``self.encoder`` - is a convolutional Jasper/QuartzNet-like encoder of type ``nemo_asr.modules.ConvASREncoder``\n* ``self.decoder`` - is a ``nemo_asr.modules.ConvASRDecoder`` which simply projects into the target alphabet (vocabulary).\n\nAlso, ``EncDecCTCModel`` uses the audio dataset class ``nemo_asr.data.AudioToCharDataset`` and CTC loss implemented in ``nemo_asr.losses.CTCLoss``.\n\nYou can use these and other neural modules (or create new ones yourself!) to construct new ASR models.",
"_____no_output_____"
],
[
"# Further Reading/Watching:\n\nThat's all for now! If you'd like to learn more about the topics covered in this tutorial, here are some resources that may interest you:\n- [Stanford Lecture on ASR](https://www.youtube.com/watch?v=3MjIkWxXigM)\n- [\"An Intuitive Explanation of Connectionist Temporal Classification\"](https://towardsdatascience.com/intuitively-understanding-connectionist-temporal-classification-3797e43a86c)\n- [Explanation of CTC with Prefix Beam Search](https://medium.com/corti-ai/ctc-networks-and-language-models-prefix-beam-search-explained-c11d1ee23306)\n- [Listen Attend and Spell Paper (seq2seq ASR model)](https://arxiv.org/abs/1508.01211)\n- [Explanation of the mel spectrogram in more depth](https://towardsdatascience.com/getting-to-know-the-mel-spectrogram-31bca3e2d9d0)\n- [Jasper Paper](https://arxiv.org/abs/1904.03288)\n- [QuartzNet paper](https://arxiv.org/abs/1910.10261)\n- [SpecAugment Paper](https://arxiv.org/abs/1904.08779)\n- [Explanation and visualization of SpecAugment](https://towardsdatascience.com/state-of-the-art-audio-data-augmentation-with-google-brains-specaugment-and-pytorch-d3d1a3ce291e)\n- [Cutout Paper](https://arxiv.org/pdf/1708.04552.pdf)\n- [Transfer Learning Blogpost](https://developer.nvidia.com/blog/jump-start-training-for-speech-recognition-models-with-nemo/)",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
4a5750d542cefe0382c3d5f31cb62c6bbea50872
| 402,167 |
ipynb
|
Jupyter Notebook
|
work/neptune_notebook.ipynb
|
freddy887/neptune
|
2d7b510d7500bb379b44a33789139b4293fadf30
|
[
"Apache-2.0"
] | null | null | null |
work/neptune_notebook.ipynb
|
freddy887/neptune
|
2d7b510d7500bb379b44a33789139b4293fadf30
|
[
"Apache-2.0"
] | null | null | null |
work/neptune_notebook.ipynb
|
freddy887/neptune
|
2d7b510d7500bb379b44a33789139b4293fadf30
|
[
"Apache-2.0"
] | null | null | null | 435.717226 | 49,290 | 0.922532 |
[
[
[
"from os import fsdecode\nimport subprocess\nimport math\nimport json\nfrom numpy import linalg as la, ma\nimport numpy as np\nimport time\nimport os\nimport julian\nimport matplotlib.pyplot as plt\nimport pandas as pd\nfrom numpy.linalg import linalg\nfrom scipy.spatial.transform import Rotation as R\nfrom scipy.spatial import distance\nfrom scipy.stats import burr\nfrom datetime import datetime as dt\nimport uuid\nimport sys\nfrom pprint import pprint\nimport shutil\nimport astropy\nimport numpy as np\nimport sys\nfrom astropy.utils import iers\nfrom astropy.time import Time",
"_____no_output_____"
],
[
"#Define the function that generates/modifies the neptune.inp file and then executes neptune\ndef propagate(\n inputTypeStateVector = 2,\n inputTypeCovarianceMatrix = ' 2',\n beginDate = '2016 07 20 00 31 50.00',\n endDate = '2016 07 27 00 31 50.00',\n radiusX = 615.119526,\n radiusY = -7095.644839,\n radiusZ = -678.668352,\n velocityX = 0.390367,\n velocityY = 0.741902,\n velocityZ = -7.396980,\n semiMajorAxis = 6800.59176,\n eccentricity = 0.0012347,\n inclination = 98.4076293,\n rightAscensionOfAscendingNode = 30.3309997,\n argumentOfPerigee = 68.5606724,\n trueAnomaly = 91.5725696,\n variancePositionX = 10.,\n variancePositionY = 100.,\n variancePositionZ = 30.,\n varianceVelocityX = 2.,\n varianceVelocityY = 1.,\n varianceVelocityZ = 1.,\n covMatrix2row = '0.d0',\n covMatrix3row = '0.d0 0.d0',\n covMatrix4row = '0.d0 0.d0 0.d0',\n covMatrix5row = '0.d0 0.d0 0.d0 0.d0',\n covMatrix6row = '0.d0 0.d0 0.d0 0.d0 0.d0',\n geopotential = 6,\n atmosphericDrag = 1,\n sunGravity = 1,\n moonGravity = 1,\n solarRadiationPressure = 1,\n earthAlbedo = 1,\n solidEarthTides = 1,\n oceanTides = 0,\n orbitalManeuvers = 0,\n geopotentialModel = 3,\n atmosphericModel = 2,\n geopotentialHarmonicSwitch = 0,\n geopotentialHarmonic = '20 30',\n shadowModelSwitch = 1,\n shadowBoundaryCorrection = 0,\n covariancePropagationSwitch = 1,\n covariancePropagationDegGeopotential = 36,\n covariancePropagationAtmosDrag = 1,\n covariancePropagationSun = 0,\n covariancePropagationMoon = 0,\n covariancePropagationSolarRadPressure = 0,\n noiseMatrixComputation = 0,\n fapDayFile = 'fap_day.dat'):\n \n runId = str(uuid.uuid4())\n\n with open(\"input/neptune.inp\", 'r', encoding=\"utf-8\") as f:\n lines = f.readlines()\n\n lines[23] = str(runId) + '\\n'\n lines[44] = str(inputTypeStateVector) + '\\n'\n lines[51] = str(inputTypeCovarianceMatrix) + '\\n'\n lines[59] = str(beginDate) + '\\n'\n lines[60] = str(endDate) + '\\n'\n lines[66] = str(radiusX) + 'd0 \\n'\n lines[67] = str(radiusY) + 'd0 \\n'\n lines[68] = str(radiusZ) + 'd0 \\n'\n lines[69] = str(velocityX) + 'd0 \\n'\n lines[70] = str(velocityY) + 'd0 \\n'\n lines[71] = str(velocityZ) + 'd0 \\n'\n lines[75] = str(semiMajorAxis) + '\\n'\n lines[76] = str(eccentricity) + '\\n'\n lines[77] = str(inclination) + '\\n'\n lines[78] = str(rightAscensionOfAscendingNode) + '\\n'\n lines[79] = str(argumentOfPerigee) + '\\n'\n lines[80] = str(trueAnomaly) + '\\n'\n lines[84] = str(variancePositionX) + 'd0 \\n'\n lines[85] = str(variancePositionY) + 'd0 \\n'\n lines[86] = str(variancePositionZ) + 'd0 \\n'\n lines[87] = str(varianceVelocityX) + 'd-4 \\n'\n lines[88] = str(varianceVelocityY) + 'd-4 \\n'\n lines[89] = str(varianceVelocityZ) + 'd-4 \\n'\n lines[91] = str(covMatrix2row) + '\\n'\n lines[92] = str(covMatrix3row) + '\\n'\n lines[93] = str(covMatrix4row) + '\\n'\n lines[94] = str(covMatrix5row) + '\\n'\n lines[95] = str(covMatrix6row) + '\\n'\n lines[105] = str(geopotential) + '\\n'\n lines[106] = str(atmosphericDrag) + '\\n'\n lines[107] = str(sunGravity) + '\\n'\n lines[108] = str(moonGravity) + '\\n'\n lines[109] = str(solarRadiationPressure) + '\\n'\n lines[110] = str(earthAlbedo) + '\\n'\n lines[111] = str(solidEarthTides) + '\\n'\n lines[112] = str(oceanTides) + '\\n'\n lines[113] = str(orbitalManeuvers) + '\\n'\n lines[120] = str(geopotentialModel) + '\\n'\n lines[127] = str(atmosphericModel) + '\\n'\n lines[135] = str(geopotentialHarmonicSwitch) + '\\n'\n lines[136] = str(geopotentialHarmonic) + '\\n'\n lines[140] = str(shadowModelSwitch) + '\\n'\n lines[141] = str(shadowBoundaryCorrection) + '\\n'\n lines[145] = str(covariancePropagationSwitch) + '\\n'\n lines[146] = str(covariancePropagationDegGeopotential) + '\\n'\n lines[147] = str(covariancePropagationAtmosDrag) + '\\n'\n lines[148] = str(covariancePropagationSun) + '\\n'\n lines[149] = str(covariancePropagationMoon) + '\\n'\n lines[150] = str(covariancePropagationSolarRadPressure) + '\\n'\n lines[157] = str(noiseMatrixComputation) + '\\n'\n lines[246] = str(fapDayFile) + '\\n'\n\n with open(\"input/neptune.inp\", 'w', encoding=\"utf-8\") as file:\n file.writelines(lines)\n\n input_dict = {\n 'runId': runId,\n 'inputTypeStateVector': inputTypeStateVector,\n 'inputTypeCovarianceMatrix': inputTypeCovarianceMatrix,\n 'beginDate': beginDate,\n 'endDate': endDate,\n 'radiusX': radiusX, \n 'radiusY': radiusY, \n 'radiusZ': radiusZ, \n 'velocityX': velocityX, \n 'velocityY': velocityY, \n 'velocityZ': velocityZ, \n 'semiMajorAxis': semiMajorAxis,\n 'eccentricity': eccentricity,\n 'inclination': inclination,\n 'rightAscensionOfAscendingNode': rightAscensionOfAscendingNode,\n 'argumentOfPerigee': argumentOfPerigee,\n 'trueAnomaly': trueAnomaly,\n 'variancePositionX': variancePositionX, \n 'variancePositionY': variancePositionY, \n 'variancePositionZ': variancePositionZ, \n 'varianceVelocityX': varianceVelocityX, \n 'varianceVelocityY': varianceVelocityY, \n 'varianceVelocityZ': varianceVelocityZ, \n 'covMatrix2row': covMatrix2row,\n 'covMatrix3row': covMatrix3row,\n 'covMatrix4row': covMatrix4row,\n 'covMatrix5row': covMatrix5row,\n 'covMatrix6row': covMatrix6row,\n 'geopotential': geopotential,\n 'atmosphericDrag': atmosphericDrag,\n 'sunGravity': sunGravity,\n 'moonGravity': moonGravity,\n 'solarRadiationPressure': solarRadiationPressure,\n 'earthAlbedo': earthAlbedo,\n 'solidEarthTides': solidEarthTides,\n 'oceanTides': oceanTides,\n 'orbitalManeuvers': orbitalManeuvers,\n 'geopotentialModel': geopotentialModel,\n 'atmosphericModel': atmosphericModel,\n 'geopotentialHarmonicSwitch': geopotentialHarmonicSwitch,\n 'geopotentialHarmonic': geopotentialHarmonic,\n 'shadowModelSwitch': shadowModelSwitch,\n 'shadowBoundaryCorrection': shadowBoundaryCorrection,\n 'covariancePropagationSwitch': covariancePropagationSwitch,\n 'covariancePropagationDegGeopotential': covariancePropagationDegGeopotential,\n 'covariancePropagationAtmosDrag': covariancePropagationAtmosDrag,\n 'covariancePropagationSun' : covariancePropagationSun,\n 'covariancePropagationMoon': covariancePropagationMoon,\n 'covariancePropagationSolarRadPressure': covariancePropagationSolarRadPressure,\n 'noiseMatrixComputation': noiseMatrixComputation,\n 'fapDayFile': fapDayFile\n }\n\n subprocess.call(\"../bin/neptune-sa\", stdout=subprocess.DEVNULL, stderr=subprocess.DEVNULL)\n\n output_dir = os.path.join(\"output\",str(input_dict['runId']))\n os.mkdir(output_dir)\n\n for filetype in ['.acc', '.csv', '.cvu', '.osc', '.vru']:\n filename = str(input_dict['runId']) + str(filetype)\n src = os.path.join(\"output\", filename)\n dst = os.path.join(\"output\",str(input_dict['runId']))\n shutil.move(src,dst)\n\n filename = str(input_dict['runId']) + '.json'\n filepath = os.path.join(\"output\",str(input_dict['runId']), filename)\n\n with open(filepath, 'w') as f:\n json.dump(input_dict, f)\n \n return runId\n",
"_____no_output_____"
],
[
"# Override runID\n# a81d2b74-9cfb-4590-a5b8-84088184112e geopotential = 36\n# 9783a265-62c7-4f40-9c40-0bd5463a24a2 geopotential = 6\ndef plot_rundId(runId):\n filename = str(runId) + '.json'\n input_json_path = os.path.join(\"output\", str(runId), filename)\n\n with open(input_json_path) as json_file:\n input_json = json.load(json_file)\n\n\n print('Geopotential: ' + str(input_json['geopotential']))\n ######################################################\n # Plot the covariances \n ######################################################\n\n filename = str(runId) + \".vru\"\n output_file_path = os.path.join(\"output\", str(runId), filename)\n\n # read file to pandas data frame\n data = pd.read_table(\n output_file_path, \n comment='#', \n header=None, \n sep='\\s+', \n names=['date','time','mjd','rx','ry','rz','vx','vy','vz'], parse_dates=[[0,1]]\n )\n\n data_labels = ['rx','ry','rz','vx','vy','vz']\n\n data[data_labels] = data[data_labels].apply(np.sqrt)\n data[data_labels] = data[data_labels].multiply(1000.0)\n\n # strip MJD\n data = data[['date_time', 'rx', 'ry', 'rz', 'vx', 'vy', 'vz']]\n\n ######################################################\n # Plot the kepler elements\n ######################################################\n\n filename = str(runId) + \".osc\"\n output_file_path = os.path.join(\"output\", str(runId), filename)\n\n # now plot\n data.plot(x='date_time', subplots=True, sharex=True, title='$1\\sigma$ errors (r in m, v in m/s)')\n plt.show()\n data = pd.read_table(\n output_file_path, \n comment='#', \n header=None, \n sep='\\s+', \n names=['date','time','mjd','sma','ecc','inc','raan','aop','tran','mean'], parse_dates=[[0,1]]\n )\n\n # strip MJD\n sma = data[['date_time', 'sma']]\n # now plot\n sma.plot( \n x='date_time',\n subplots=True, \n sharex=True, \n title='SMA (km, deg)',\n color='c'\n )\n # set the \n #plt.xlim([dt(2016, 7, 21), dt(2016, 7, 23)])\n plt.show()\n\n # strip MJD\n ecc = data[['date_time', 'ecc']]\n # now plot\n ecc.plot( \n x='date_time',\n subplots=True, \n sharex=True, \n title='ecc (km, deg)',\n color='r'\n )\n # set the \n #plt.xlim([dt(2016, 7, 21), dt(2016, 7, 23)])\n plt.show()\n\n # strip MJD\n inc = data[['date_time', 'inc']]\n # now plot\n inc.plot( \n x='date_time',\n subplots=True, \n sharex=True, \n title='inc (km, deg)',\n color='b'\n )\n # set the \n #plt.xlim([dt(2016, 7, 21), dt(2016, 7, 23)])\n plt.show()\n\n # strip MJD\n raan = data[['date_time', 'raan']]\n # now plot\n raan.plot( \n x='date_time',\n subplots=True, \n sharex=True, \n title='Raan (km, deg)',\n color='y'\n )\n # set the \n #plt.xlim([dt(2016, 7, 21), dt(2016, 7, 23)])\n plt.show()\n\n # strip MJD\n data = data[['date_time', 'aop']]\n data['aop'] = data['aop'].apply(lambda x: math.radians(x))\n data['aop'] = np.unwrap(data['aop'].tolist())\n # now plot\n data.plot( \n x='date_time',\n subplots=True, \n sharex=True, \n title='aop (km, deg)',\n color = 'k'\n )\n # set the \n #plt.xlim([dt(2016, 7, 21), dt(2016, 7, 23)])\n plt.show()\n",
"_____no_output_____"
],
[
"def fap_day_modifier(f10Mod = 0, f3mMod = 0, ssnMod = 0, apMod = 0):\n with open(\"data/fap_day.dat\", 'r') as f:\n lines = f.readlines()\n\n for i in range(2, len(lines)):\n splitLine = lines[i].split()\n\n f10 = int(splitLine[1]) + int(f10Mod)\n splitLine[1] = str(f10).zfill(3)\n\n f3m = int(splitLine[2]) + int(f3mMod)\n splitLine[2] = str(f3m).zfill(3)\n\n ssn = int(splitLine[3]) + int(ssnMod)\n splitLine[3] = str(ssn).zfill(3)\n\n ap = int(splitLine[4]) + int(apMod)\n splitLine[4] = str(ap).zfill(3)\n\n splitLine.append('\\n')\n lines[i] = ' '.join(splitLine)\n\n with open(\"data/fap_day_modified.dat\", 'w') as file:\n file.writelines(lines)\n\nfap_day_modifier()\n ",
"_____no_output_____"
],
[
"def get_rtn_matrix(state_vector):\n r = state_vector[0:3]\n v = state_vector[3:6]\n rxv = np.cross(r, v)\n vecRTN = np.empty([3,3],float)\n # process vector R\n vecRTN[0,:] = np.divide(r,np.linalg.norm(r))\n\n # process vector W\n vecRTN[2,:] = np.divide(rxv,np.linalg.norm(rxv))\n\n # process vector S\n vecRTN[1,:] = np.cross(vecRTN[2,:], vecRTN[0,:])\n\n return vecRTN",
"_____no_output_____"
],
[
"# Run propagation with burr distribution on f10.7 values\nnumberIterations = 100\n# Generate f10.7 modifier list using normal distribution\nf10ModList = np.random.normal(0.0, 20, numberIterations)\napModList = np.random.normal(0.0, 7, numberIterations)\n\n# Initialise the runIdList IF no additional \nrunIdList = []\n\n#Set variables\nnoiseMatrixComputation = 0\ncovariancePropagationSwitch = 0\nbeginDate = '2016 07 20 00 31 50.00',\nendDate = '2016 07 27 00 31 50.00',\n\n# Create a initial \"unmodified\" reference propagation\nrunIdList.append(propagate(beginDate = beginDate, endDate = endDate,noiseMatrixComputation=noiseMatrixComputation, covariancePropagationSwitch=covariancePropagationSwitch))\n\nfor i in range(0, len(f10ModList)):\n fap_day_modifier(f10Mod=f10ModList[i])\n\n runIdList.append(propagate(beginDate = beginDate, endDate = endDate,noiseMatrixComputation=noiseMatrixComputation, covariancePropagationSwitch=covariancePropagationSwitch, fapDayFile='fap_day_modified.dat',))\n",
"_____no_output_____"
],
[
"\nplt.hist(f10ModList)\nplt.show()",
"_____no_output_____"
],
[
"# x = []\n# y = []\n# z = []\n# u = []\n# v = []\n# w = []\nr = []\nt = []\nn = []\n\n# Calculate required data from the reference propagation\nfilename = str(runIdList[0]) + \".csv\"\noutput_file_path = os.path.join(\"output\", str(runIdList[0]), filename)\n\ndata = pd.read_table(\n output_file_path, \n comment='#', \n header=None, \n sep='\\s+', \n names=['date','time','mjd','x','y','z','u','v','w'], parse_dates=[[0,1]]\n )\n\nstateVector = [\n data.tail(1)['x'].values[0],\n data.tail(1)['y'].values[0],\n data.tail(1)['z'].values[0],\n data.tail(1)['u'].values[0],\n data.tail(1)['v'].values[0],\n data.tail(1)['w'].values[0]]\n\nrtnMatrix = get_rtn_matrix(state_vector=stateVector)\nrtnMatrix = np.array(rtnMatrix)\nstateVector = np.array([\n data.tail(1)['x'].values[0],\n data.tail(1)['y'].values[0],\n data.tail(1)['z'].values[0]\n])\nRTN1 = np.dot(rtnMatrix, stateVector)\n\n# filename = str(runIdList[0]) + \".vru\"\n# output_file_path = os.path.join(\"output\", str(runIdList[0]), filename)\n\n# # read file to pandas data frame\n# data = pd.read_table(\n# output_file_path, \n# comment='#', \n# header=None, \n# sep='\\s+', \n# names=['date','time','mjd','rx','ry','rz','vx','vy','vz'], parse_dates=[[0,1]]\n# )\n\n# data_labels = ['rx','ry','rz','vx','vy','vz']\n\n# data[data_labels] = data[data_labels].apply(np.sqrt)\n# #data[data_labels] = data[data_labels].multiply(1000.0)\n\n# # strip MJD\n# data = data[['date_time', 'rx', 'ry', 'rz', 'vx', 'vy', 'vz']]\n\n# covarianceVector = np.array([data.tail(1)['rx'].values[0], data.tail(1)['ry'].values[0], data.tail(1)['ry'].values[0]])\n# covarianceVectorRTN = np.dot(rtnMatrix, covarianceVector)\n\nprint(\"Number of Propagations: \" + str(len(runIdList)))\nfor i in range(1, len(runIdList)):\n filename = str(runIdList[i]) + \".csv\"\n output_file_path = os.path.join(\"output\", str(runIdList[i]), filename)\n\n data = pd.read_table(\n output_file_path, \n comment='#', \n header=None, \n sep='\\s+', \n names=['date','time','mjd','x','y','z','u','v','w'], parse_dates=[[0,1]]\n )\n \n stateVector = np.array([\n data.tail(1)['x'].values[0],\n data.tail(1)['y'].values[0],\n data.tail(1)['z'].values[0]\n ])\n RTN2 = np.dot(rtnMatrix, stateVector)\n\n r.append(RTN1[0]-RTN2[0])\n t.append(RTN1[1]-RTN2[1])\n n.append(RTN1[2]-RTN2[2])\n\n\nplt.hist(r, bins=40)\nplt.xlabel(\"km\")\nplt.title(\"Radial Standard Deviation\")\nplt.axvline(0, color='k', linestyle='dashed', linewidth=2)\n#plt.axvline(covarianceVectorRTN[0], color='r', linestyle='dashed', linewidth=1)\n#plt.axvline(-covarianceVectorRTN[0], color='r', linestyle='dashed', linewidth=1)\nplt.show()\n\nplt.hist(t, bins=25)\nplt.xlabel(\"km\")\nplt.title(\"Tangential Standard Deviation\")\nplt.axvline(0, color='k', linestyle='dashed', linewidth=2)\n# plt.axvline(covarianceVectorRTN[1], color='r', linestyle='dashed', linewidth=1)\n# plt.axvline(-covarianceVectorRTN[1], color='r', linestyle='dashed', linewidth=1)\nplt.show()\n\nplt.hist(n, bins=25)\nplt.xlabel(\"km\")\nplt.title(\"Normal Standard Deviation\")\nplt.axvline(0, color='k', linestyle='dashed', linewidth=2)\n#plt.axvline(-covarianceVectorRTN[2], color='r', linestyle='dashed', linewidth=1)\nplt.show()\n\n\n",
"Number of Propagations: 101\n"
],
[
"# Override runID\n# a81d2b74-9cfb-4590-a5b8-84088184112e geopotential = 36\n# 9783a265-62c7-4f40-9c40-0bd5463a24a2 geopotential = 6\n\nfor id in ['9783a265-62c7-4f40-9c40-0bd5463a24a2','a81d2b74-9cfb-4590-a5b8-84088184112e']:\n plot_rundId(id)",
"Geopotential: 6\n"
],
[
"filename = \"fap_day.dat\"\noutput_file_path = os.path.join(\"data\", filename)\n\n# read file to pandas data frame\ndata = pd.read_table(\n output_file_path, \n comment='#', \n header=None, \n sep='\\s+', \n names=['date', 'F10', 'F3M', 'SSN', 'Ap'], parse_dates=[1]\n )\n\n\nprint(data)\n# now plot\ndata.plot(x='date', subplots=True, sharex=True, title='Space Weather')\nplt.show()\n# set the \n#plt.xlim([dt(2016, 7, 21), dt(2016, 7, 23)])",
"_____no_output_____"
],
[
"print(len(runIdList))",
"52\n"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a5759d08d7f6f105fe7eb414c63935d8db86e7f
| 76,707 |
ipynb
|
Jupyter Notebook
|
p2_continuous-control/Continuous_Control.ipynb
|
salviosage/Deep_R_Learning
|
be10c90f551ae812435549196fc09e948ac50691
|
[
"MIT"
] | 1 |
2021-07-01T17:12:20.000Z
|
2021-07-01T17:12:20.000Z
|
p2_continuous-control/Continuous_Control.ipynb
|
salviosage/Deep_R_Learning
|
be10c90f551ae812435549196fc09e948ac50691
|
[
"MIT"
] | null | null | null |
p2_continuous-control/Continuous_Control.ipynb
|
salviosage/Deep_R_Learning
|
be10c90f551ae812435549196fc09e948ac50691
|
[
"MIT"
] | null | null | null | 187.547677 | 22,864 | 0.888667 |
[
[
[
"# Continuous Control\n\n---\n\nIn this notebook, you will learn how to use the Unity ML-Agents environment for the second project of the [Deep Reinforcement Learning Nanodegree](https://www.udacity.com/course/deep-reinforcement-learning-nanodegree--nd893) program.\n\n### 1. Start the Environment\n\nWe begin by importing the necessary packages. If the code cell below returns an error, please revisit the project instructions to double-check that you have installed [Unity ML-Agents](https://github.com/Unity-Technologies/ml-agents/blob/master/docs/Installation.md) and [NumPy](http://www.numpy.org/).",
"_____no_output_____"
]
],
[
[
"\nimport torch\nimport numpy as np\nimport pandas as pd\nfrom collections import deque\nfrom unityagents import UnityEnvironment\nimport random\nimport matplotlib.pyplot as plt\n%matplotlib inline\n\nfrom ddpg_agent import Agent",
"_____no_output_____"
]
],
[
[
"Next, we will start the environment! **_Before running the code cell below_**, change the `file_name` parameter to match the location of the Unity environment that you downloaded.\n\n- **Mac**: `\"path/to/Reacher.app\"`\n- **Windows** (x86): `\"path/to/Reacher_Windows_x86/Reacher.exe\"`\n- **Windows** (x86_64): `\"path/to/Reacher_Windows_x86_64/Reacher.exe\"`\n- **Linux** (x86): `\"path/to/Reacher_Linux/Reacher.x86\"`\n- **Linux** (x86_64): `\"path/to/Reacher_Linux/Reacher.x86_64\"`\n- **Linux** (x86, headless): `\"path/to/Reacher_Linux_NoVis/Reacher.x86\"`\n- **Linux** (x86_64, headless): `\"path/to/Reacher_Linux_NoVis/Reacher.x86_64\"`\n\nFor instance, if you are using a Mac, then you downloaded `Reacher.app`. If this file is in the same folder as the notebook, then the line below should appear as follows:\n```\nenv = UnityEnvironment(file_name=\"Reacher.app\")\n```",
"_____no_output_____"
]
],
[
[
"env = UnityEnvironment(file_name=\"Reacher1.app\")",
"INFO:unityagents:\n'Academy' started successfully!\nUnity Academy name: Academy\n Number of Brains: 1\n Number of External Brains : 1\n Lesson number : 0\n Reset Parameters :\n\t\tgoal_speed -> 1.0\n\t\tgoal_size -> 5.0\nUnity brain name: ReacherBrain\n Number of Visual Observations (per agent): 0\n Vector Observation space type: continuous\n Vector Observation space size (per agent): 33\n Number of stacked Vector Observation: 1\n Vector Action space type: continuous\n Vector Action space size (per agent): 4\n Vector Action descriptions: , , , \n"
]
],
[
[
"Environments contain **_brains_** which are responsible for deciding the actions of their associated agents. Here we check for the first brain available, and set it as the default brain we will be controlling from Python.",
"_____no_output_____"
]
],
[
[
"# get the default brain\nbrain_name = env.brain_names[0]\nbrain = env.brains[brain_name]",
"_____no_output_____"
]
],
[
[
"### 2. Examine the State and Action Spaces\n\nIn this environment, a double-jointed arm can move to target locations. A reward of `+0.1` is provided for each step that the agent's hand is in the goal location. Thus, the goal of your agent is to maintain its position at the target location for as many time steps as possible.\n\nThe observation space consists of `33` variables corresponding to position, rotation, velocity, and angular velocities of the arm. Each action is a vector with four numbers, corresponding to torque applicable to two joints. Every entry in the action vector must be a number between `-1` and `1`.\n\nRun the code cell below to print some information about the environment.",
"_____no_output_____"
]
],
[
[
"# reset the environment\nenv_info = env.reset(train_mode=True)[brain_name]\n\n# number of agents\nnum_agents = len(env_info.agents)\nprint('Number of agents:', num_agents)\n\n# size of each action\naction_size = brain.vector_action_space_size\nprint('Size of each action:', action_size)\n\n# examine the state space \nstates = env_info.vector_observations\nstate_size = states.shape[1]\nprint('There are {} agents. Each observes a state with length: {}'.format(states.shape[0], state_size))\nprint('The state for the first agent looks like:', states[0])",
"Number of agents: 20\nSize of each action: 4\nThere are 20 agents. Each observes a state with length: 33\nThe state for the first agent looks like: [ 0.00000000e+00 -4.00000000e+00 0.00000000e+00 1.00000000e+00\n -0.00000000e+00 -0.00000000e+00 -4.37113883e-08 0.00000000e+00\n 0.00000000e+00 0.00000000e+00 0.00000000e+00 0.00000000e+00\n 0.00000000e+00 0.00000000e+00 -1.00000000e+01 0.00000000e+00\n 1.00000000e+00 -0.00000000e+00 -0.00000000e+00 -4.37113883e-08\n 0.00000000e+00 0.00000000e+00 0.00000000e+00 0.00000000e+00\n 0.00000000e+00 0.00000000e+00 5.75471878e+00 -1.00000000e+00\n 5.55726624e+00 0.00000000e+00 1.00000000e+00 0.00000000e+00\n -1.68164849e-01]\n"
]
],
[
[
"### 3. Take Random Actions in the Environment\n\nIn the next code cell, you will learn how to use the Python API to control the agent and receive feedback from the environment.\n\nOnce this cell is executed, you will watch the agent's performance, if it selects an action at random with each time step. A window should pop up that allows you to observe the agent, as it moves through the environment. \n\nOf course, as part of the project, you'll have to change the code so that the agent is able to use its experience to gradually choose better actions when interacting with the environment!",
"_____no_output_____"
]
],
[
[
"env_info = env.reset(train_mode=False)[brain_name] # reset the environment \nstates = env_info.vector_observations # get the current state (for each agent)\nscores = np.zeros(num_agents) # initialize the score (for each agent)\nwhile True:\n actions = np.random.randn(num_agents, action_size) # select an action (for each agent)\n actions = np.clip(actions, -1, 1) # all actions between -1 and 1\n env_info = env.step(actions)[brain_name] # send all actions to tne environment\n next_states = env_info.vector_observations # get next state (for each agent)\n rewards = env_info.rewards # get reward (for each agent)\n dones = env_info.local_done # see if episode finished\n scores += env_info.rewards # update the score (for each agent)\n states = next_states # roll over states to next time step\n if np.any(dones): # exit loop if episode finished\n break\nprint('Total score (averaged over agents) this episode: {}'.format(np.mean(scores)))",
"Total score (averaged over agents) this episode: 0.11949999732896685\n"
]
],
[
[
"### 4. It's Your Turn!\n\nNow it's your turn to train your own agent to solve the environment! When training the environment, set `train_mode=True`, so that the line for resetting the environment looks like the following:\n```python\nenv_info = env.reset(train_mode=True)[brain_name]\n```",
"_____no_output_____"
]
],
[
[
"agent = Agent(state_size=state_size, action_size=action_size,\n n_agents=num_agents, random_seed=42)",
"_____no_output_____"
],
[
"def plot_scores(scores, rolling_window=10, save_fig=False):\n \"\"\"Plot scores and optional rolling mean using specified window.\"\"\"\n fig = plt.figure()\n ax = fig.add_subplot(111)\n plt.plot(np.arange(len(scores)), scores)\n plt.ylabel('Score')\n plt.xlabel('Episode #')\n plt.title(f'scores')\n rolling_mean = pd.Series(scores).rolling(rolling_window).mean()\n plt.plot(rolling_mean);\n\n if save_fig:\n plt.savefig(f'figures_scores.png', bbox_inches='tight', pad_inches=0)",
"_____no_output_____"
],
[
"\ndef ddpg(n_episodes=10000, max_t=1000, print_every=100):\n scores_deque = deque(maxlen=print_every)\n scores = []\n for i_episode in range(1, n_episodes+1):\n env_info = env.reset(train_mode=True)[brain_name]\n states = env_info.vector_observations \n agent.reset()\n score = np.zeros(num_agents)\n for t in range(max_t):\n actions = agent.act(states)\n \n env_info = env.step(actions)[brain_name] \n next_states = env_info.vector_observations # get next state (for each agent)\n rewards = env_info.rewards # get reward (for each agent)\n dones = env_info.local_done # see if episode finished\n\n agent.step(states, actions, rewards, next_states, dones)\n states = next_states\n score += rewards\n if any(dones):\n break \n scores_deque.append(np.mean(score))\n scores.append(np.mean(score))\n \n print('\\rEpisode {}\\tAverage Score: {:.2f}'.format(i_episode, np.mean(scores_deque)), end=\"\")\n torch.save(agent.actor_local.state_dict(), './weights/checkpoint_actor.pth')\n torch.save(agent.critic_local.state_dict(), './weights/checkpoint_critic.pth')\n if i_episode % print_every == 0:\n print('\\rEpisode {}\\tAverage Score: {:.2f}'.format(i_episode, np.mean(scores_deque)))\n plot_scores(scores)\n if np.mean(scores_deque) >= 30.0:\n print('\\nEnvironment solved in {:d} episodes!\\tAverage Score: {:.2f}'.format(i_episode - print_every, np.mean(scores_deque)))\n torch.save(agent.actor_local.state_dict(), './weights/checkpoint_actor.pth')\n torch.save(agent.critic_local.state_dict(), './weights/checkpoint_critic.pth')\n break\n \n return scores\n\nscores = ddpg()\n\nfig = plt.figure()\nax = fig.add_subplot(111)\nplt.plot(np.arange(1, len(scores)+1), scores)\nplt.ylabel('Score')\nplt.xlabel('Episode #')\nplt.show()",
"Episode 100\tAverage Score: 16.49\nEpisode 146\tAverage Score: 30.08\nEnvironment solved in 46 episodes!\tAverage Score: 30.08\n"
],
[
"\nplot_scores(scores)",
"_____no_output_____"
]
],
[
[
"When finished, you can close the environment.",
"_____no_output_____"
]
],
[
[
"env.close()",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
4a57619da70474101b7835e5b71643b4a37e8f3c
| 33,485 |
ipynb
|
Jupyter Notebook
|
Lecture 6 Plotting Graphs/Pie Graph/Pie Graphs-checkpoint.ipynb
|
Paraskk/Data-Science-and-Machine-Leaning-
|
b29223a82ea39f7860d3729d7297bac2a4724c8f
|
[
"MIT"
] | 1 |
2021-12-13T12:37:25.000Z
|
2021-12-13T12:37:25.000Z
|
Lecture 6 Plotting Graphs/Pie Graph/Pie Graphs-checkpoint.ipynb
|
Udaysonu/Coding-Ninjas-Machine-Learning
|
4fd6b4b62f07b28dbe80c084ad820630f2351a76
|
[
"MIT"
] | null | null | null |
Lecture 6 Plotting Graphs/Pie Graph/Pie Graphs-checkpoint.ipynb
|
Udaysonu/Coding-Ninjas-Machine-Learning
|
4fd6b4b62f07b28dbe80c084ad820630f2351a76
|
[
"MIT"
] | 2 |
2020-08-27T13:03:33.000Z
|
2020-09-01T17:34:23.000Z
| 321.971154 | 15,672 | 0.924265 |
[
[
[
"## Plot function x^3 ",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nimport numpy as np\n#x=np.array([1,2,3,4])\nx=np.arange(0,5,0.1)\n#color,marker,line width for axis,dataset and title,axis and grid\ny=x**3\ny2=x**2\n#plt.plot(x,y,color=\"black\",marker=\"0)\nplt.plot(x,y,color=\"black\",linewidth=5,label=\"x^3\")\nplt.plot(x,y2,color=\"red\",linewidth=5,label=\"x^2\")\nplt.axis([0,10,0,200])\nplt.ylabel(\"y\")\nplt.xlabel(\"x\")\nplt.grid()\nplt.text(2,80,\"text\",fontsize=12)\nplt.legend()\nplt.title(\"Matplotlib Demo\")\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Pie Graph",
"_____no_output_____"
]
],
[
[
"labels=[\"A\",\"B\",\"C\",\"D\"]\nsize=[3,4,6,2]\ncolors=[\"blue\",\"purple\",\"red\",\"pink\"]\nexplode=[0.1,0,0,0]\nplt.title(\"Split among classes\")\nplt.pie(size,colors=colors,explode=explode,labels=labels,autopct=\"%.2f%%\",counterclock=False,startangle=100)\nplt.axis(\"equal\")\nplt.show()",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
4a5765780c0e6886f9558dc6463a8f392f852e26
| 46,642 |
ipynb
|
Jupyter Notebook
|
Notebooks/investment_dist.ipynb
|
TessPalan/stock-research
|
86bb48d8f5d5d67a60af0d3ade9e3a1791e80863
|
[
"Apache-2.0"
] | 1 |
2021-03-13T06:18:31.000Z
|
2021-03-13T06:18:31.000Z
|
Notebooks/investment_dist.ipynb
|
RickyDoubles/stock-research
|
3a98e6e756ab29f76938b567ddec19802856dadc
|
[
"Apache-2.0"
] | null | null | null |
Notebooks/investment_dist.ipynb
|
RickyDoubles/stock-research
|
3a98e6e756ab29f76938b567ddec19802856dadc
|
[
"Apache-2.0"
] | 2 |
2021-02-13T22:16:03.000Z
|
2021-03-13T06:01:31.000Z
| 58.229713 | 22,368 | 0.637323 |
[
[
[
"## Investment Distribution",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nimport pandas as pd\nimport numpy as np",
"_____no_output_____"
],
[
"# read top stocks csv\ntop_stocks = pd.read_csv('../data/top_stocks.csv').head()\ntop_stocks",
"_____no_output_____"
]
],
[
[
"## Stock Distribution\n- Stock YHOO = 35 % \n- Stock FTR = 30 % \n- Stock AKAM = 15 %\n- Stock EW = 10 %\n- Stock EBAY = 10 %\n- Pie chart = 100 % \n",
"_____no_output_____"
]
],
[
[
"ranking = len(top_stocks['Ranking'])",
"_____no_output_____"
],
[
"# pie chart with our 5 stocks distribution\ntop_stocks = plt.pie(\n x=[35,30,15,10,10], \n labels=[\"YHOO\", \"FTR\",\"AKAM\",\"EW\",\"EBAY\"], \n explode=[0,0,0,0,0], \n colors=[\"darksalmon\", \"royalblue\", \"darkseagreen\",\"slategray\",\"peachpuff\"],\n autopct=\"%1.1f%%\", \n shadow=True, \n startangle=140\n)\nplt.title('Stock Distribution')\n\nplt.axis(\"equal\")\n\nplt.savefig(\"../visuals/top_stocks.png\")\n\nplt.show()",
"_____no_output_____"
],
[
"# investor \nbudget = input(\"How much money do you want to invest today?\")",
"How muh money do you want to invest today? 500\n"
],
[
"alocation = {\n 'YHOO': 0.35,\n 'FTR': 0.30,\n 'AKAM': 0.15,\n 'EW': 0.10,\n 'EBAY': 0.10\n}",
"_____no_output_____"
],
[
"for stock, pct in alocation.items():\n print(f'You should invest ${int(budget)* pct} in the {stock} stock')\n ",
"You should invest $175.0 in the YHOO stock\nYou should invest $150.0 in the FTR stock\nYou should invest $75.0 in the AKAM stock\nYou should invest $50.0 in the EW stock\nYou should invest $50.0 in the EBAY stock\n"
],
[
"# API call data frame to here \ntop_stocks_final = pd.read_csv('../data/top_stocks_final.csv').head()\ntop_stocks_final",
"_____no_output_____"
],
[
"# ROI \nroi = ((top_stocks_final['Current Closing Price'] - top_stocks_final['Stock Price'])/top_stocks_final['Stock Price'])*100\nroi",
"_____no_output_____"
],
[
"top_stocks_final['ROI %'] = roi\ntop_stocks_final_roi = top_stocks_final.round(2)\ntop_stocks_final_roi",
"_____no_output_____"
],
[
"# Tell the investor how much money they made \ntop_stocks_final_roi['Investment Return'] = (top_stocks_final_roi['ROI %']/100) * (top_stocks_final_roi['Ticker Symbol'].apply(alocation.get)) * int(budget)\ntop_stocks_final_roi",
"_____no_output_____"
],
[
"total_return = top_stocks_final_roi['Investment Return'].sum()\ntotal_return",
"_____no_output_____"
],
[
"top_stocks_final_roi.to_csv(\"../data/top_stocks_final_roi.csv\")",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a5765a7d2a097319c0d3224c12e08cf78195d8b
| 273,096 |
ipynb
|
Jupyter Notebook
|
Projet_Gomoku.ipynb
|
GMTAccount/Projets-scolaires
|
b48f0fb0a7573ad8198abce350d8889047198bfd
|
[
"CC0-1.0"
] | null | null | null |
Projet_Gomoku.ipynb
|
GMTAccount/Projets-scolaires
|
b48f0fb0a7573ad8198abce350d8889047198bfd
|
[
"CC0-1.0"
] | null | null | null |
Projet_Gomoku.ipynb
|
GMTAccount/Projets-scolaires
|
b48f0fb0a7573ad8198abce350d8889047198bfd
|
[
"CC0-1.0"
] | null | null | null | 33.402153 | 1,335 | 0.305134 |
[
[
[
"<a href=\"https://colab.research.google.com/github/GMTAccount/Projets-scolaires/blob/main/Projet_Gomoku.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"<h1>\n Liste des erreurs\n</h1>\n<ul>\n <li style=\"color:green;\">\n Recentrer le premier point (fait)\n </li>\n <li>\n Problème de l'emplacement des valeurs\n </li>\n <li>\n Problème des valeurs placées sur la matrice\n </li>\n <li>\n Problème de valeurs infinies\n </li>\n <li>\n Manque Utility\n </li>\n</ul>",
"_____no_output_____"
]
],
[
[
"print ( \"hello\" , \" test \")\nprint(\"test2\")\n#fonction 1: Méthod einitialisation (creation plateau de jeu) V\n#fonction 2: Attribution Joueur V\n#fonction 3: Affichage du plateau + avec coordonées V\n#fonction 4: Saisie sécurisée de placement + affichage placement chosii par l'IA (en cours)\n#fonction 5: menu V\n#fonction 5,5: Saisie 1ers pions (en cours)\n#fonction 6: timer V\n#fonction 7: Fonction minimax V\n#fonction 8: Terminal Test return 0 (égalité) 1(on a gagné) 2 (aversaire a gagné) -1 (partie non finie) V\n#fonction 9: Actions (récapitule toutes les acttions) V\n#fonction 10: utility (détermine quelle est la meilleure action) (en cours)\n#Fonction 11:décision,/result (applique l'action) V\n\n#class Gomoko V\n#attributs: matrice de 0 1 2\n #joueur: booleen\n#méthodes: Affichage\n #saisie etc\n #init timer\n\n#MAIN: menu\n\n#0 : pas occupé, 1 : occupé par nous, 2 : occupé par l'adversaire",
"hello test \ntest2\n"
],
[
"import time\n\n \ndef AfficherPlateau(plateau):\n rep =\"\" \n lettres= {\"A\" : 1 , \"B\" : 2 , \"C\" : 3, \"D\" : 4, \"E\" : 5, \"F\": 6 , \"G\" : 7 , \"H\" :8 , \"I\" :9 , \"J\" : 10 , \"K\" :11 , \"L\" :12 , \"M\" :13 , \"N\":14 , \"O\": 15 }\n rep += \"\\t \"\n rep += \" \".join(lettres) \n rep += \"\\n\" \n for i in range(15):\n for j in range(15):\n if plateau[i][j]==0:\n if j==0:\n if i<9: \n rep += \"\\t \" +str(i+1) + \" . \"\n else:\n rep += \"\\t\" + str(i+1) +\" . \"\n else:\n rep += \". \"\n \n elif plateau[i][j]==1:\n if j==0:\n if i<9:\n rep += \"\\t \" + str(i+1) + \" O \"\n else:\n rep += \"\\t\" + str(i+1) + \" O \"\n else:\n rep += \"O \"\n else:\n if j==0:\n if i<9:\n rep += \"\\t \" + str(i+1) + \" X \"\n else:\n rep += \"\\t\" + str(i+1) +\" X \"\n else:\n rep += \"X \"\n rep += \"\\n\"\n print (rep)\n return rep\n \nplateau= [[ 0 for x in range (15)] for y in range(15)]\n\ndef Jouable(ligne,col, plateau): #ok\n\tif (col < 0 or col > 14 or ligne >14 or ligne < 0):\n\t\treturn 0\n\telif (plateau[ligne][col] == 0):\n\t\treturn 1\n\telse:\n\t\treturn 0\n \ndef EstNul(plateau): #ok\n for i in range(15):\n for j in range(15):\n if(Jouable(i,j,plateau)==1):\n return False\n return True\n\ndef TerminalTest(plateau): #ok\n\tfor i in range(15):\n\t\tfor j in range(15):\n\t\t\tif(j>= 4):\n\t\t\t\tif(pow(plateau[i][j]+plateau[i][j-1]+plateau[i][j-2]+plateau[i][j-3]+plateau[i][j-4],2) == 25):\n\t\t\t\t\treturn True\n\t\t\t\telif(i >= 4):\n\t\t\t\t\tif(pow(plateau[i][j]+plateau[i-1][j-1]+plateau[i-2][j-2]+plateau[i-3][j-3]+plateau[i-4][j-4],2) == 25):\n\t\t\t\t\t\treturn True\n\t\t\t\telif(i<11):\n\t\t\t\t\tif(pow(plateau[i][j]+plateau[i+1][j-1]+plateau[i+2][j-2]+plateau[i+3][j-3]+plateau[i+4][j-4],2) == 25):\n\t\t\t\t\t\treturn True\t\t\t\t\n\t\t\tif(j< 11):\n\t\t\t\tif(pow(plateau[i][j]+plateau[i][j+1]+plateau[i][j+2]+plateau[i][j+3]+plateau[i][j+4],2) == 25):\n\t\t\t\t\treturn True\n\t\t\t\telif(i >= 4):\n\t\t\t\t\tif(pow(plateau[i][j]+plateau[i-1][j+1]+plateau[i-2][j+2]+plateau[i-3][j+3]+plateau[i-4][j+4],2) == 25):\n\t\t\t\t\t\treturn True\n\t\t\t\telif(i<11):\n\t\t\t\t\tif(pow(plateau[i][j]+plateau[i+1][j+1]+plateau[i+2][j+2]+plateau[i+3][j+3]+plateau[i+4][j+4],2) == 25):\n\t\t\t\t\t\treturn True\t\t\t\t\n\t\t\tif(i >= 4):\n\t\t\t\tif(pow(plateau[i][j]+plateau[i-1][j]+plateau[i-2][j]+plateau[i-3][j]+plateau[i-4][j],2) == 25):\n\t\t\t\t\treturn True\n\t\t\tif(i<11):\n\t\t\t\tif(pow(plateau[i][j]+plateau[i+1][j]+plateau[i+2][j]+plateau[i+3][j]+plateau[i+4][j],2) == 25):\n\t\t\t\t\treturn True\n\treturn False\n \t\ndef Saisie(plateau, Joueur): #ok\n secu = False\n lettres= {\"A\" : 1 , \"B\" : 2 , \"C\" : 3 , \"D\" : 4, \"E\" : 5, \"F\": 6 , \"G\" : 7 , \"H\" :8 , \"I\" :9 , \"J\" : 10 , \"K\" :11 , \"L\" :12 , \"M\" :13 , \"N\":14 , \"O\": 15 }\n while secu == False : \n print(\"Veuillez saisir les coordonnées de la case que vous voulez jouer\")\n print(\"(Séparez les axes par un point virgule, par exemple D;7)\")\n saisie = input()\n try:\n saisieTab = saisie.split(\";\")\n \n if len (saisieTab) == 2 and int(saisieTab[1]) > 0 and int(saisieTab[1]) <= len(lettres) and \"\".join(saisieTab[0].split() ).upper() in lettres : \n secu = True\n saisiex = int(saisieTab[1])\n saisiey = \"\".join(saisieTab[0].split() ).upper() \n \n if plateau[saisiex-1][lettres[saisiey]-1] == 1 or plateau[saisiex-1][lettres[saisiey]-1] == -1 :\n secu = False \n except:\n print(\"Saisie invalide, veuillez réessayer\")\n return (lettres[saisiey]-1,saisiex-1)\n print('\\nC\\'est au tour du joueur', Joueur, ' de jouer !')\n \n print('\\nChoisissez la colonne où vous voulez mettre votre pion ', end ='')\n col=int(input())-1\n \n print('\\nChoisissez la ligne ou vous voulez mettre votre pion', end='')\n ligne=int(input())-1\n \n if(col < 0 or col > 14 or Jouable(ligne,col, plateau) == 0):\n while(col < 0 or col > 14 or Jouable(ligne, col, plateau) == 0):\n print('Vous ne pouvez pas choisir cette colonne, choisissez une colonne valide !')\n col = int(input())\n \n if(ligne<0 or ligne>14 or Jouable(ligne,col,plateau)==0):\n while(ligne<0 or ligne>14 or Jouable(ligne,col,plateau)==0):\n print('Vous ne pouvez pas choisir cette ligne, choisissez une ligne valide !')\n ligne=int(input())\n \n print('Vous avez joué la colonne', col+1, 'et la ligne', ligne+1, '\\n') \n return col, ligne\t \n\ndef MinMax(plateau,Joueur):\n Fin=False\n J=1\n while(Fin==False):\n if(Joueur==J):\n col,ligne=Saisie(plateau,Joueur)\n #print(ligne,col)\n plateau[ligne][col]= 1 \n else:\n print('\\nC\\'est au tour de l\\'IA de jouer')\n start = time.time()\n [ligne,col] = JeuxIA(plateau,3,-float(\"inf\"),float(\"inf\"),True)[1]\n print(ligne+1,col+1)\n end = time.time()\n print('Temps de réponse: {}s'.format(round(end - start, 7)))\n plateau[ligne][col]= -1\n AfficherPlateau(plateau)\n if(TerminalTest(plateau)):\n print('\\nLe joueur', J, 'a gagné ! \\nVoulez-vous rejouer ? (y/n) ', end = '')\n rejouer=str(input)\n Fin=True\n elif(EstNul(plateau)):\n print('\\nMatch nul, voulez-vous rejouer ? (y/n) ', end = '')\n rejouer=str(input())\n Fin=True\n if(J==1):\n J=2\n else:\n J=1\n return rejouer\n\ndef JeuxIA(M, profondeur,alpha,beta,maximizingPlayer): \n actions=Actions(M)\n if profondeur == 0 or TerminalTest(M) == True or EstNul(M) == True:\n cout = Utility(M,maximizingPlayer)\n return [cout,[0,0]]\n elif maximizingPlayer == True:\n maxEval = [-1000000,[0,0]]\n for i in range(len(actions)):\n evale = JeuxIA(Result(M,actions[i][0],actions[i][1],True),profondeur-1,alpha,beta,False)\n evale[1] = [actions[i][0],actions[i][1]]\n if evale[0] > maxEval[0]:\n maxEval[0] = evale[0]\n maxEval[1] = evale[1]\n alpha = max(alpha,evale[0])\n if beta <= alpha :\n break\n return maxEval\n else:\n minEval = [1000000,[0,0]]\n for i in range(len(actions)):\n evale = JeuxIA(Result(M,actions[i][0],actions[i][1],False),profondeur-1,alpha,beta,True)\n evale[1] = [actions[i][0],actions[i][1]]\n if evale[0] < minEval[0]:\n minEval[0] = evale[0]\n minEval[1] = evale[1]\n beta = min(beta,evale[0])\n if beta <= alpha:\n break\n return minEval \n\ndef Actions(state): # ok ù\n actions=[]\n for a in range(15):\n for b in range(15):\n if Jouable(a,b,state)==0:\n if Jouable(a-1,b-1,state) == 1 and actions.count([a-1,b-1])== 0 :\n actions.append([a-1,b-1])\n if Jouable(a-1,b,state) == 1 and actions.count([a-1,b]) == 0:\n actions.append([a-1,b])\n if Jouable(a-1,b+1,state) == 1 and actions.count([a-1,b+1]) == 0:\n actions.append([a-1,b+1])\n if Jouable(a,b-1,state) == 1 and actions.count([a,b-1]) == 0:\n actions.append([a,b-1])\n if Jouable(a,b+1,state) == 1 and actions.count([a,b+1]) == 0:\n actions.append([a,b+1])\n if Jouable(a+1,b-1,state) == 1 and actions.count([a+1,b-1]) == 0:\n actions.append([a+1,b-1])\n if Jouable(a+1,b,state) == 1 and actions.count([a+1,b]) == 0:\n actions.append([a+1,b])\n if Jouable(a+1,b+1,state) == 1 and actions.count([a+1,b+1]) == 0:\n actions.append([a+1,b+1])\n if len(actions) == 0:\n actions.append([6,6])\n return actions\n \ndef Result(state, a, b, IA):\n\tnewState = [[],[],[],[],[],[],[],[],[],[],[],[],[],[],[]]\n\tfor i in range(15):\n\t\tnewState[i] = state[i][:]\n\tif IA == True : \n\t\tnewState[a][b] = -1\n\telse :\n\t newState[a][b] = 1\n\treturn newState\n\ndef Utility(state,J): \n\t\t\t\t\t\n somme = 0\n if TerminalTest(state) == True:\n if J == True :\n return -float(\"inf\")\n else :\n return float(\"inf\")\n else :\n for i in range(1,14):\n for j in range(1,14):\n if(j>= 3):\n if (state[i][j]+state[i][j-1]+state[i][j-2] == -3) and (state[i][j-3] == 0 or state[i][j+1] == 0):\n somme += 500\n elif(i >= 3):\n if(state[i][j]+state[i-1][j-1]+state[i-2][j-2] == -3) and (state[i-3][j-3] == 0 or state[i+1][j+1] == 0):\n somme += 1000\n elif(i<9):\n if(state[i][j]+state[i+1][j-1]+state[i+2][j-2] == -3)and (state[i+3][j-3] == 0 or state[i-1][j+1] == 0):\n somme += 1000\t\t\t\t\n if(j< 9):\n if(state[i][j]+state[i][j+1]+state[i][j+2] == -3)and (state[i][j+3] == 0 or state[i][j-1] == 0):\n somme += 500\n elif(i >= 3):\n if(state[i][j]+state[i-1][j+1]+state[i-2][j+2] == -3)and (state[i-3][j+3] == 0 or state[i+1][j-1] == 0):\n somme += 1000\n elif(i<9):\n if(state[i][j]+state[i+1][j+1]+state[i+2][j+2] == -3)and (state[i+3][j+3] == 0 or state[i-1][j-1] == 0):\n somme += 1000\t\t\t\t\n if(i >= 3):\n if(state[i][j]+state[i-1][j]+state[i-2][j] == -3)and (state[i-3][j] == 0 or state[i+1][j] == 0):\n somme += 500\n if(i<9):\n if(state[i][j]+state[i+1][j]+state[i+2][j] == -3)and (state[i+3][j] == 0 or state[i-1][j] == 0):\n somme += 500\n for i in range(1,14):\n for j in range(1,14):\n if(j>= 3):\n if (state[i][j]+state[i][j-1]+state[i][j-2] == -3) and (state[i][j-3] == 0 and state[i][j+1] == 0):\n somme += 50000\n elif(i >= 3):\n if(state[i][j]+state[i-1][j-1]+state[i-2][j-2] == -3) and (state[i-3][j-3] == 0 and state[i+1][j+1] == 0):\n somme += 100000\n elif(i<9):\n if(state[i][j]+state[i+1][j-1]+state[i+2][j-2] == -3)and (state[i+3][j-3] == 0 and state[i-1][j+1] == 0):\n somme += 100000\t\t\t\t\n if(j< 9):\n if(state[i][j]+state[i][j+1]+state[i][j+2] == -3)and (state[i][j+3] == 0 and state[i][j-1] == 0):\n somme += 50000\n elif(i >= 3):\n if(state[i][j]+state[i-1][j+1]+state[i-2][j+2] == -3)and (state[i-3][j+3] == 0 and state[i+1][j-1] == 0):\n somme += 100000\n elif(i<9):\n if(state[i][j]+state[i+1][j+1]+state[i+2][j+2] == -3)and (state[i+3][j+3] == 0 and state[i-1][j-1] == 0):\n somme += 100000\t\t\t\t\n if(i >= 3):\n if(state[i][j]+state[i-1][j]+state[i-2][j] == -3)and (state[i-3][j] == 0 and state[i+1][j] == 0):\n somme += 50000\n if(i<9):\n if(state[i][j]+state[i+1][j]+state[i+2][j] == -3)and (state[i+3][j] == 0 and state[i-1][j] == 0):\n somme += 50000\n for i in range(1,14):\n for j in range(1,14):\n if(j>= 3):\n if (state[i][j]+state[i][j-1]+plateau[i][j-2] == 3) and (state[i][j-3] == 0 or state[i][j+1] == 0):\n somme -= 50000\n elif(i >= 3):\n if(state[i][j]+state[i-1][j-1]+state[i-2][j-2] == 3) and (state[i-3][j-3] == 0 or state[i+1][j+1] == 0):\n somme -= 100000\n elif(i<9):\n if(state[i][j]+state[i+1][j-1]+state[i+2][j-2] == 3)and (state[i+3][j-3] == 0 or state[i-1][j+1] == 0):\n somme -= 100000\t\t\t\t\n if(j< 9):\n if(state[i][j]+state[i][j+1]+state[i][j+2] == 3)and (state[i][j+3] == 0 or state[i][j-1] == 0):\n somme -= 50000\n elif(i >= 3):\n if(state[i][j]+state[i-1][j+1]+state[i-2][j+2] == 3)and (state[i-3][j+3] == 0 or state[i+1][j-1] == 0):\n somme -= 100000\n elif(i<9):\n if(state[i][j]+state[i+1][j+1]+state[i+2][j+2] == 3)and (state[i+3][j+3] == 0 or state[i-1][j-1] == 0):\n somme -= 100000\t\t\t\t\n if(i >= 3):\n if(state[i][j]+state[i-1][j]+state[i-2][j] == 3)and (state[i-3][j] == 0 or state[i+1][j] == 0):\n somme -= 50000\n if(i<9):\n if(state[i][j]+state[i+1][j]+state[i+2][j] == 3)and (state[i+3][j] == 0 or state[i-1][j] == 0):\n somme -= 50000\n for i in range(1,14):\n for j in range(1,14):\n if(j>= 3):\n if (state[i][j]+state[i][j-1]+plateau[i][j-2] == 3) and (state[i][j-3] == 0 and state[i][j+1] == 0):\n somme -= 500000\n elif(i >= 3):\n if(state[i][j]+state[i-1][j-1]+state[i-2][j-2] == 3) and (state[i-3][j-3] == 0 and state[i+1][j+1] == 0):\n somme -= 1000000\n elif(i<9):\n if(state[i][j]+state[i+1][j-1]+state[i+2][j-2] == 3)and (state[i+3][j-3] == 0 and state[i-1][j+1] == 0):\n somme -= 1000000\t\t\t\t\n if(j< 9):\n if(state[i][j]+state[i][j+1]+state[i][j+2] == 3)and (state[i][j+3] == 0 and state[i][j-1] == 0):\n somme -= 500000\n elif(i >= 3):\n if(state[i][j]+state[i-1][j+1]+state[i-2][j+2] == 3)and (state[i-3][j+3] == 0 and state[i+1][j-1] == 0):\n somme -= 1000000\n elif(i<9):\n if(state[i][j]+state[i+1][j+1]+state[i+2][j+2] == 3)and (state[i+3][j+3] == 0 and state[i-1][j-1] == 0):\n somme -= 1000000\t\t\t\t\n if(i >= 3):\n if(state[i][j]+state[i-1][j]+state[i-2][j] == 3)and (state[i-3][j] == 0 and state[i+1][j] == 0):\n somme -= 500000\n if(i<9):\n if(state[i][j]+state[i+1][j]+state[i+2][j] == 3)and (state[i+3][j] == 0 and state[i-1][j] == 0):\n somme -= 500000\n for i in range(1,14):\n for j in range(1,14):\n if(j>= 2):\n if (state[i][j]+state[i][j-1] == 2) and (state[i][j-2] == 0 and state[i][j+1] == 0):\n somme -= 50000\n elif(i >= 2):\n if(state[i][j]+state[i-1][j-1] == 2) and (state[i-2][j-2] == 0 and state[i+1][j+1] == 0):\n somme -= 100000\n elif(i<10):\n if(state[i][j]+state[i+1][j-1] == 2)and (state[i+2][j-2] == 0 and state[i-1][j+1] == 0):\n somme -= 100000\t\t\t\t\n if(j< 10):\n if(state[i][j]+state[i][j+1] == 2)and (state[i][j+2] == 0 and state[i][j-1] == 0):\n somme -= 50000\n elif(i >= 2):\n if(state[i][j]+state[i-1][j+1] == 2)and (state[i-2][j+2] == 0 and state[i+1][j-1] == 0):\n somme -= 100000\n elif(i<10):\n if(state[i][j]+state[i+1][j+1] == 2)and (state[i+2][j+2] == 0 and state[i-1][j-1] == 0):\n somme -= 100000\t\t\t\t\n if(i >= 2):\n if(state[i][j]+state[i-1][j] == 2)and (state[i-2][j] == 0 and state[i+1][j] == 0):\n somme -= 50000\n if(i<10):\n if(state[i][j]+state[i+1][j] == 2)and (state[i+2][j] == 0 and state[i-1][j] == 0):\n somme -= 50000\n for i in range(1,14):\n for j in range(1,14):\n if(j>= 2):\n if (state[i][j]+state[i][j-1] == -2) and (state[i][j-2] == 0 and state[i][j+1] == 0):\n #print('2 cercles en ligne')\n somme += 5000\n elif(i >= 2):\n if(state[i][j]+state[i-1][j-1] == -2) and (state[i-2][j-2] == 0 and state[i+1][j+1] == 0):\n #print('2 cercles en diagonales')\n somme += 10000\n elif(i<10):\n if(state[i][j]+state[i+1][j-1] == -2)and (state[i+2][j-2] == 0 and state[i-1][j+1] == 0):\n #print('2 cercles en diagonales')\n somme += 10000\t\t\t\t\n if(j< 10):\n if(state[i][j]+state[i][j+1] == -2)and (state[i][j+2] ==0 and state[i][j-1] == 0):\n #print('2 cercles en ligne')\n somme += 5000\n elif(i >= 2):\n if(state[i][j]+state[i-1][j+1] == -2)and (state[i-2][j+2] == 0 and state[i+1][j-1] == 0):\n #print('2 cercles en diagonales')\n somme += 10000\n elif(i<10):\n if(state[i][j]+state[i+1][j+1] == -2)and (state[i+2][j+2] == 0 and state[i-1][j-1] == 0):\n #print('2 cercles en diagonales')\n somme += 10000\t\t\t\t\n if(i >= 2):\n if(state[i][j]+state[i-1][j] == -2)and (state[i-2][j] == 0 and state[i+1][j] == 0):\n #print('2 cercles en colonne')\n somme += 5000\n if(i<10):\n if(state[i][j]+state[i+1][j] == -2)and (state[i+2][j] == 0 and state[i-1][j] == 0):\n #print('2 cercles en colonne')\n somme += 5000\n for i in range(1,14):\n for j in range(1,14):\n if(j>= 2):\n if (state[i][j]+state[i][j-1] == -2) and (state[i][j-2] == 0 or state[i][j+1] == 0):\n #print('2 cercles en ligne')\n somme += 500\n elif(i >= 2):\n if(state[i][j]+state[i-1][j-1] == -2) and (state[i-2][j-2] == 0 or state[i+1][j+1] == 0):\n #print('2 cercles en diagonales')\n somme += 1000\n elif(i<10):\n if(state[i][j]+state[i+1][j-1] == -2)and (state[i+2][j-2] == 0 or state[i-1][j+1] == 0):\n #print('2 cercles en diagonales')\n somme += 1000\t\t\t\t\n if(j< 10):\n if(state[i][j]+state[i][j+1] == -2)and (state[i][j+2] == 0 or state[i][j-1] == 0):\n #print('2 cercles en ligne')\n somme += 500\n elif(i >= 2):\n if(state[i][j]+state[i-1][j+1] == -2)and (state[i-2][j+2] == 0 or state[i+1][j-1] == 0):\n #print('2 cercles en diagonales')\n somme += 1000\n elif(i<10):\n if(state[i][j]+state[i+1][j+1] == -2)and (state[i+2][j+2] == 0 or state[i-1][j-1] == 0):\n #print('2 cercles en diagonales')\n somme += 1000\t\t\t\t\n if(i >= 2):\n if(state[i][j]+state[i-1][j] == -2)and (state[i-2][j] == 0 or state[i+1][j] == 0):\n #print('2 cercles en colonne')\n somme += 500\n if(i<10):\n if(state[i][j]+state[i+1][j] == -2)and (state[i+2][j] == 0 or state[i-1][j] == 0):\n #print('2 cercles en colonne')\n somme += 500\n \n \n for i in range(1,14):\n for j in range(1,14):\n if(j>= 2):\n if (state[i][j]+state[i][j-1] == 2) and (state[i][j-2] == 0 or state[i][j+1] == 0):\n #print('2 cercles en ligne')\n somme -= 500\n elif(i >= 2):\n if(state[i][j]+state[i-1][j-1] == 2) and (state[i-2][j-2] == 0 or state[i+1][j+1] == 0):\n #print('2 cercles en diagonales')\n somme -= 1000\n elif(i<10):\n if(state[i][j]+state[i+1][j-1] == 2)and (state[i+2][j-2] == 0 or state[i-1][j+1] == 0):\n #print('2 cercles en diagonales')\n somme -= 1000\t\t\t\t\n if(j< 10):\n if(state[i][j]+state[i][j+1] == 2)and (state[i][j+2] == 0 or state[i][j-1] == 0):\n #print('2 cercles en ligne')\n somme -= 500\n elif(i >= 2):\n if(state[i][j]+state[i-1][j+1] == 2)and (state[i-2][j+2] == 0 or state[i+1][j-1] == 0):\n #print('2 cercles en diagonales')\n somme -= 1000\n elif(i<10):\n if(state[i][j]+state[i+1][j+1] == 2)and (state[i+2][j+2] == 0 or state[i-1][j-1] == 0):\n #print('2 cercles en diagonales')\n somme -= 1000\t\t\t\t\n if(i >= 2):\n if(state[i][j]+state[i-1][j] == 2)and (state[i-2][j] == 0 or state[i+1][j] == 0):\n #print('2 cercles en colonne')\n somme -= 500\n if(i<10):\n if(state[i][j]+state[i+1][j] == 2)and (state[i+2][j] == 0 or state[i-1][j] == 0):\n #print('2 cercles en colonne')\n somme -= 500\n return somme\n \n \n\n\n\n\n\nAfficherPlateau([[ 0 for x in range (15)] for y in range(15)])\nrejouer = 'y'\nwhile(rejouer == 'y' or rejouer == 'Y'):\n print(\"\\n\\n***** Bonjour et bienvenue dans le jeux Gomoko ***\")\n mat = [[ 0 for x in range (15)] for y in range(15)]\n AfficherPlateau(mat)\n \n Joueur=0\n niveau=0\n while(Joueur!=1 and Joueur!=2):\n \n saisieValide=False\n while(saisieValide==False):\n print(\"\\nVeuillez choisir un joueur\\n\")\n print(\"\\t1 - Joueur 1\\n\\t2 - Joueur 2\")\n Joueur=input(\"Votre choix : \")\n try:\n Joueur=int(Joueur)\n saisieValide=(Joueur==1) or (Joueur==2)\n if(not saisieValide):\n print(\"Saisie invalide, veuillez réessayer\")\n except:\n print(\"Saisie invalide, veuillez réessayer\")\n if Joueur==1:\n col,ligne=Saisie(mat,Joueur)\n mat[ligne][col]= 1\n AfficherPlateau(mat)\n rejouer=MinMax(mat,2)\n \n rejouer=input()",
"\t A B C D E F G H I J K L M N O\n\t 1 . . . . . . . . . . . . . . . \n\t 2 . . . . . . . . . . . . . . . \n\t 3 . . . . . . . . . . . . . . . \n\t 4 . . . . . . . . . . . . . . . \n\t 5 . . . . . . . . . . . . . . . \n\t 6 . . . . . . . . . . . . . . . \n\t 7 . . . . . . . . . . . . . . . \n\t 8 . . . . . . . . . . . . . . . \n\t 9 . . . . . . . . . . . . . . . \n\t10 . . . . . . . . . . . . . . . \n\t11 . . . . . . . . . . . . . . . \n\t12 . . . . . . . . . . . . . . . \n\t13 . . . . . . . . . . . . . . . \n\t14 . . . . . . . . . . . . . . . \n\t15 . . . . . . . . . . . . . . . \n\n\n\n***** Bonjour et bienvenue dans le jeux Gomoko ***\n\t A B C D E F G H I J K L M N O\n\t 1 . . . . . . . . . . . . . . . \n\t 2 . . . . . . . . . . . . . . . \n\t 3 . . . . . . . . . . . . . . . \n\t 4 . . . . . . . . . . . . . . . \n\t 5 . . . . . . . . . . . . . . . \n\t 6 . . . . . . . . . . . . . . . \n\t 7 . . . . . . . . . . . . . . . \n\t 8 . . . . . . . . . . . . . . . \n\t 9 . . . . . . . . . . . . . . . \n\t10 . . . . . . . . . . . . . . . \n\t11 . . . . . . . . . . . . . . . \n\t12 . . . . . . . . . . . . . . . \n\t13 . . . . . . . . . . . . . . . \n\t14 . . . . . . . . . . . . . . . \n\t15 . . . . . . . . . . . . . . . \n\n\nVeuillez choisir un joueur\n\n\t1 - Joueur 1\n\t2 - Joueur 2\n"
],
[
"",
"_____no_output_____"
]
],
[
[
"<h1>\n <em>Projet IA - Gomoku<em>\n</h1>\n<ul>\n <li style=\"color: green;\">Fonction 1 : Méthode initialisation (creation plateau de jeu) V</li>\n <li style=\"color=green;\">Fonction 2: Attribution Joueur V</li>\n <li style=\"color=green;\">Fonction 3: Affichage du plateau + avec coordonées V</li>\n <li style=\"color=green;\">\nFonction 4: Saisie sécurisée de placement + affichage placement chosii par l'IA (en cours)</li>\n <li style=\"color=green;\">Fonction 5: menu V</li>\n <li style=\"color=green;\">Fonction 5,5: Saisie 1ers pions (à compléter)</li>\n <li style=\"color=green;\">Fonction 6: timer V</li>\n <li style=\"color=green;\">Fonction 7: Fonction minimax</li>\n <li style=\"color=green;\">Fonction 8: Terminal Test return 0 (égalité) 1(on a gagné) 2 (aversaire a gagné) -1 (partie non finie) V</li>\n <li style=\"color=green;\">Fonction 9: Actions (récapitule toutes les acttions) (en cours)</li>\n <li style=\"color=green;\">Fonction 10: utility (détermine quelle est la meilleure action)</li>\n <li style=\"color=green;\">Fonction 11:décision,/result (applique l'action) V</li>\n <li style=\"color=green;\">class Gomoko) V</li>\n</ul>",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport time",
"_____no_output_____"
],
[
"#FONCTION 1: INITIALISATION\n\ndef init_Game():\n mat = [[0]*15 for _ in range(15)]\n return mat",
"_____no_output_____"
],
[
"#Fonction 3: AFFICHAGE PLATEAU\ndef Affichage(plateau): \n print(\" A B C D E F G H I J K L M N O\")\n for i in range(15):\n for j in range(15):\n if plateau[i][j]==0:\n if j==0:\n if i<10:\n print(i, \" . \",end=\"\")\n else:\n print(i,\". \",end=\"\")\n else:\n print(\". \", end=\"\")\n\n elif plateau[i][j]==1:\n if j==0:\n if i<10:\n print(i, \" O \",end=\"\")\n else:\n print(i,\"O \",end=\"\")\n else:\n print(\"O \", end=\"\")\n else:\n if j==0:\n if i<10:\n print(i, \" X \",end=\"\")\n else:\n print(i,\"X \",end=\"\")\n else:\n print(\"X \", end=\"\")\n print(\"\")",
"_____no_output_____"
],
[
"#TAB_ACTIONS_POSSIBLES 9\ndef Actions(plateau):\n tab_actions=[]\n nbr_cases=0\n\n for i in range(15):\n for j in range(15):\n if plateau[i][j]==0:\n tab_actions.append([i,j])\n nbr_cases+=1\n \n return tab_actions",
"_____no_output_____"
],
[
"#Fonction 5 : MENU\n\ndef Menu():\n print(\"Bonjour et bienvenue dans le Gomoko\")\n saisieValide=False\n while(saisieValide==False):\n print(\"Veuillez choisir un joueur\")\n print(\"1 - Joueur 1\\n2 - Joueur 2\")\n joueur=input(\"Votre choix : \")\n try:\n joueur=int(joueur)\n saisieValide=(joueur==1) or (joueur==2)\n if(not saisieValide):\n print(\"Saisie invalide, veuillez réessayer\")\n except:\n print(\"Saisie invalide, veuillez réessayer\")\n print()\n print(\"C'est bon\")\n estTermine=False\n gomoko=Gomoko(joueur)\n t=-1\n gomoko.Initialisation() #A modifier lorsque la méthode sera crée\n while(not estTermine):\n gomoko.Affichage() # A modifier si besoin est\n gomoko.Saisie()\n gomoko.MinMax() # Ajouter d'autres fonctions si y a lieu\n t=gomoko.TerminalTest()\n if(t!=-1):\n estTermine=True\n if(t==0):\n print(\"Et nous avons une égalité\")\n elif(t==1):\n print(\"Et notre IA vous a battu.e\")\n elif(t==2):\n print(\"Et c'est gagné\")\n print(\"Merci d'avoir joué\")",
"_____no_output_____"
],
[
"#Fonction 6 : timer\n#L'idée, c'est qu'il soit appliqué à une autre méthode, via un @timer juste avant\ndef timer(fonction):\n \"\"\"\n Méthode qui chronomètre le temps d'exécution d'une méthode\n Sera utilisé pour mesurer le temps de traitement du MinMax\n\n Parameters\n ----------\n fonction : Fonction sur laquelle appliquer le timer\n\n Returns\n -------\n None.\n\n \"\"\"\n \n def inner(*args,**kwargs):\n #Coeur de la méthode de mesure du temps\n t=time.time()#Temps en entrée\n f=fonction(*args,**kwargs)\n print(time.time()-t)#Affichage du temps finel (en secondes)\n return f\n return inner",
"_____no_output_____"
],
[
"#Fonction 7 : MinMax\n#Celle-ci est divisée en 3 fonctions différentes, pour simplifier le traitement\n@timer\ndef MaxValue(state):\n if TerminalTest(state):\n return Utility(state)\n v=-float(\"inf\")\n for a in Actions(state):\n v=max(v,MinValue(application_result(state,a)))\n return v\n\ndef MinValue(state):\n if TerminalTest(state):\n return Utility(state)\n v=float(\"inf\")\n for a in Actions(state):\n v=min(v,MaxValue(application_result(state,a)))\n return v\n\ndef MinMax():\n #Réaliser un classement des différentes valeurs trouvées\n a=Actions(self.plateau)\n b=(MinValue(Result(action,a)) for action in a)\n dico=dict(a,b) #Attention, cette ligne pourrait crash\n dico = sorted(dico.items(), key=lambda x: x[1], reverse=True)\n actionValide=dico.keys()[0]\n application_result(self.plateau,actionValide[0],actionValide[1],self.joueur)",
"_____no_output_____"
],
[
"#Fonction 8 : determination du gagnant\n\nCOL_COUNT=15\nROW_COUNT=15\ndef gagnant(plateau, pion):\n for c in range(COL_COUNT - 4):\n for r in range(ROW_COUNT):\n if plateau[r][c] == pion and plateau[r][c+1] == pion and plateau[r][c+2] == pion and plateau[r][c+3] == pion and plateau[r][c+4] == pion:\n return True\n \n for c in range(COL_COUNT):\n for r in range(ROW_COUNT-4):\n if plateau[r][c] == pion and plateau[r+1][c] == pion and plateau[r+2][c] == pion and plateau[r+3][c] == pion and plateau[r+4][c] == pion:\n return True\n \n for c in range(COL_COUNT-4):\n for r in range(4,ROW_COUNT):\n if plateau[r][c] == pion and plateau[r-1][c+1] == pion and plateau[r-2][c+2] == pion and plateau[r-3][c+3] == pion and plateau[r-4][c+4] == pion:\n return True\n \n for c in range(COL_COUNT-4):\n for r in range(ROW_COUNT-4):\n if plateau[r][c] == pion and plateau[r+1][c+1] == pion and plateau[r+2][c+2] == pion and plateau[r+3][c+3] == pion and plateau[r+4][c+4] == pion:\n return True",
"_____no_output_____"
],
[
"#FONCTION 11 RESULT/Application\ndef application_result(plateau,i,j,joueur):\n plateau[i][j]=joueur\n return plateau",
"_____no_output_____"
],
[
"class Gomoko () :\n def __init__ ( self):\n self.plateau = [[0,0,0,0,0,0,0,0,0,0,0,0,0,0,0],\n [0,0,0,0,0,0,0,0,0,0,0,0,0,0,0],\n [0,0,0,0,0,0,0,0,0,0,0,0,0,0,0],\n [0,0,0,0,0,0,0,0,0,0,0,0,0,0,0],\n [0,0,0,0,0,0,0,0,0,0,0,0,0,0,0],\n [0,0,0,0,0,0,0,0,0,0,0,0,0,0,0],\n [0,0,0,0,0,0,0,0,0,0,0,0,0,0,0],\n [0,0,0,0,0,0,0,0,0,0,0,0,0,0,0],\n [0,0,0,0,0,0,0,0,0,0,0,0,0,0,0],\n [0,0,0,0,0,0,0,0,0,0,0,0,0,0,0],\n [0,0,0,0,0,0,0,0,0,0,0,0,0,0,0],\n [0,0,0,0,0,0,0,0,0,0,0,0,0,0,0],\n [0,0,0,0,0,0,0,0,0,0,0,0,0,0,0],\n [0,0,0,0,0,0,0,0,0,0,0,0,0,0,0],\n [0,0,0,0,0,0,0,0,0,0,0,0,0,0,0]]\n self.joueur \n \n def __str__ (self):\n for i in range( len(self.plateau) ) :\n for j in range (len(self.plateau) ) : \n print(self.plateau[i][j], end = \"\")\n print( )\n def Saisie (self) : \n \n \n \n lettre = {\"A\" : 1 , \"B\" : 2 , \"C\" : 3 , \"D\" : 4, \"E\" : 5,\n \"F\": 6 , \"G\" : 7 , \"H\" :8 , \"I\" :9 , \"J\" : 10 , \"K\" :11 , \"L\" :12\n , \"M\" :13 , \"N\":14 , \"O\": 15 }\n \n \n \n \n\n \n \n secu3 = False\n while secu3 == False :\n \n \n secu1 = False\n while secu1 == False : \n saisiex =input(\"Saisir la ligne ou vous voulez jouer : (chiffre) \")\n try:\n saisiex = int(saisiex)\n \n if saisiex > 0 and saisiex <= 15 :\n secu1 = True\n \n \n if(not secu1):\n print(\"Saisie invalide, veuillez réessayer\")\n except:\n print(\"Saisie invalide, veuillez réessayer\")\n \n secu2 = False\n while secu2 == False : \n saisiey =input(\"Saisir la colonnes ou vous voulez jouer : (lettre)\")\n \n try:\n\n if saisiey in lettre :\n secu2 = True\n \n if(not secu2):\n print(\"Saisie invalide, veuillez réessayer\")\n except:\n print(\"Saisie invalide, veuillez réessayer\")\n \n \n \n try:\n\n if self.plateau[saisiex-1][lettre[saisiey]-1] == 1 or self.plateau[saisiex-1][lettre[saisiey]-1] == 2 :\n print( \"pas bien \")\n secu3 = False\n else :\n secu3 = True\n \n if(not secu3):\n print(\"Position non valide, veuillez réessayer\")\n \n except:\n print(\"Saisie invalide, veuillez réessayer\") \ndef Start(joueur):\n if(joueur==1):\n #Placement du joueur 1 au centre du plateau\n self.plateau[8][8]=1\n #Placement selon l'IA de notre point\n self.MinMax() # A modifier, si elle ne fait pas le placement\n #Saisie des valeurs\n lettre = {\"A\" : 1 , \"B\" : 2 , \"C\" : 3 , \"D\" : 4, \"E\" : 5,\n \"L\" :12\n , \"M\" :13 , \"N\":14 , \"O\": 15 }\n secu3 = False\n while secu3 == False :\n secu1 = False\n while secu1 == False : \n saisiex =input(\"Saisir la ligne ou vous voulez jouer : (chiffre) \")\n try:\n saisiex = int(saisiex)\n \n if saisiex > 0 and saisiex <= 15 and (saisiex<5 or saisiex>11) :\n secu1 = True\n \n \n if(not secu1):\n print(\"Saisie invalide, veuillez réessayer\")\n except:\n print(\"Saisie invalide, veuillez réessayer\")\n \n secu2 = False\n while secu2 == False : \n saisiey =input(\"Saisir la colonnes ou vous voulez jouer : (lettre)\")\n \n try:\n \n if saisiey in lettre:\n secu2 = True\n \n if(not secu2):\n print(\"Saisie invalide, veuillez réessayer\")\n except:\n print(\"Saisie invalide, veuillez réessayer\")\n try:\n \n if self.plateau[saisiex-1][lettre[saisiey]-1] == 1 or self.plateau[saisiex-1][lettre[saisiey]-1] == 2 :\n print( \"pas bien \")\n secu3 = False\n else :\n secu3 = True\n \n if(not secu3):\n print(\"Position non valide, veuillez réessayer\")\n \n except:\n print(\"Saisie invalide, veuillez réessayer\") \n \n else:\n self.plateau[8][8]=2\n Saisie() # A voir en fonction de ce qui est souhaité\n #Réaliser un classement des différentes valeurs trouvées\n a=Actions(self.plateau)\n cpt=0\n for i in range(len(a)):\n if (a[i][0]>5 or a[i][0]<11) or (a[i][1]>5 or a[i][1]<11):\n a.pop(i)\n i-=1\n b=(MinValue(Result(action,a)) for action in a)\n dico=dict(a,b) #Attention, cette ligne pourrait crash\n dico = sorted(dico.items(), key=lambda x: x[1], reverse=True)\n actionValide=dico.keys()[0]\n application_result(self.plateau,actionValide[0],actionValide[1],self.joueur)\n\n ",
"_____no_output_____"
],
[
" def MaxValue(self, state):\n if self.TerminalTest(state):\n return self.Utility(state)\n v=-float(\"inf\")\n for a in self.Actions(state):\n v=max(v,self.MinValue(self.application_result(state,a[0],a[1],self.joueur)))\n return v\n \ndef MinValue(self, state):\n if self.TerminalTest(state):\n return self.Utility(state)\n v=float(\"inf\")\n for a in self.Actions(state):\n \n self.plateau , action[0] , action[1]\n \n v=min(v,self.MaxValue(self.application_result(state,a[0],a[1],self.joueur)))\n return v\n",
"_____no_output_____"
]
],
[
[
"### VERSION 1\n",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport time\n\ndef Menu():\n \n \n \n #%% Accueil\n print(\"\\n\\nBonjour et bienvenue dans le Gomoko\")\n saisieValide=False\n joueur = 0\n while(saisieValide==False):\n print(\"\\nVeuillez choisir un joueur\\n\")\n print(\"\\t1 - Joueur 1\\n\\t2 - Joueur 2\")\n joueur=input(\"Votre choix : \")\n try:\n joueur=int(joueur)\n saisieValide=(joueur==1) or (joueur==2)\n if(not saisieValide):\n print(\"Saisie invalide, veuillez réessayer\")\n except:\n print(\"Saisie invalide, veuillez réessayer\")\n print()\n print(\"C'est bon\")\n \n \n #%% Debut jeux\n estTermine=False \n gomoko=Gomoko(joueur)\n t=-1\n gomoko.Start(joueur) #A modifier lorsque la méthode sera crée\n while(not estTermine):\n gomoko.Affichage() # A modifier si besoin est\n gomoko.Saisie()\n gomoko.MinMax() # Ajouter d'autres fonctions si y a lieu\n t=gomoko.TerminalTest()\n if(t!=-1):\n estTermine=True\n if(t==0):\n print(\"Et nous avons une égalité\")\n elif(t==1):\n print(\"Et notre IA vous a battu.e\")\n elif(t==2):\n print(\"Et c'est gagné\")\n print(\"Merci d'avoir joué\")\n \n\n \n\n\n\n\nclass Gomoko () :\n def __init__ ( self, joueur ):\n self.plateau = [[0,0,0,0,0,0,0,0,0,0,0,0,0,0,0],\n [0,0,0,0,0,0,0,0,0,0,0,0,0,0,0],\n [0,0,0,0,0,0,0,0,0,0,0,0,0,0,0],\n [0,0,0,0,0,0,0,0,0,0,0,0,0,0,0],\n [0,0,0,0,0,0,0,0,0,0,0,0,0,0,0],\n [0,0,0,0,0,0,0,0,0,0,0,0,0,0,0],\n [0,0,0,0,0,0,0,0,0,0,0,0,0,0,0],\n [0,0,0,0,0,0,0,0,0,0,0,0,0,0,0],\n [0,0,0,0,0,0,0,0,0,0,0,0,0,0,0],\n [0,0,0,0,0,0,0,0,0,0,0,0,0,0,0],\n [0,0,0,0,0,0,0,0,0,0,0,0,0,0,0],\n [0,0,0,0,0,0,0,0,0,0,0,0,0,0,0],\n [0,0,0,0,0,0,0,0,0,0,0,0,0,0,0],\n [0,0,0,0,0,0,0,0,0,0,0,0,0,0,0],\n [0,0,0,0,0,0,0,0,0,0,0,0,0,0,0]]\n self.joueur = joueur \n \n def __str__ (self):\n for i in range( len(self.plateau) ) :\n for j in range (len(self.plateau) ) : \n print(self.plateau[i][j], end = \"\")\n print( )\n \n \n def application_result(self, plateau,i,j,joueur):\n plateau[i][j]=joueur\n return plateau\n \n def MaxValue(self, state):\n if self.TerminalTest(state):\n return self.Utility(state)\n v=-float(\"inf\")\n for a in self.Actions(state):\n v=max(v,self.MinValue(self.application_result(state,a[0],a[1],self.joueur)))\n return v\n \n def MinValue(self, state):\n if self.TerminalTest(state):\n return self.Utility(state)\n v=float(\"inf\")\n for a in self.Actions(state): \n v=min(v,self.MaxValue(self.application_result(state,a[0],a[1],self.joueur)))\n return v\n\n \n def MinMax( self):\n #Réaliser un classement des différentes valeurs trouvées\n a=self.Actions(self.plateau)\n b=(self.MinValue(self.application_result(self.plateau , action[0] , action[1] , self.joueur)) for action in a)\n print ( a)\n a2=[]\n for b in a:\n print(b)\n a2.append(str(b[0])+\" \"+str(b[1]))\n zip_iterator = zip(a2 ,b )\n dico=dict(zip_iterator)\n\n dico = dict(sorted(dico.items(), key=lambda x: x[1], reverse=True))\n print(dico.keys())\n actionValide=dico.keys()[0].split(sep=\" \")\n actionValide[0]=int(actionValide[0])\n actionValide[1]=int(actionValide[1])\n self.application_result(self.plateau,actionValide[0],actionValide[1],self.joueur)\n\n \n def Actions(self, plateau):\n tab_actions=[]\n nbr_cases=0\n for i in range(15):\n for j in range(15):\n if plateau[i][j]==0:\n tab_actions.append([i,j])\n nbr_cases+=1\n \n return tab_actions\n\n def timer(self, fonction):\n \"\"\"\n Méthode qui chronomètre le temps d'exécution d'une méthode\n Sera utilisé pour mesurer le temps de traitement du MinMax\n \n Parameters\n ----------\n fonction : Fonction sur laquelle appliquer le timer\n \n Returns\n -------\n None.\n \n \"\"\"\n \n def inner(*args,**kwargs):\n #Coeur de la méthode de mesure du temps\n t=time.time()#Temps en entrée\n f=fonction(*args,**kwargs)\n print(time.time()-t)#Affichage du temps finel (en secondes)\n return f\n return inner\n\n def TerminalTest(self,pion):\n COL_COUNT=15\n ROW_COUNT=15\n for c in range(COL_COUNT - 4):\n for r in range(ROW_COUNT):\n if self.plateau[r][c] == pion and self.plateau[r][c+1] == pion and self.plateau[r][c+2] == pion and self.plateau[r][c+3] == pion and self.plateau[r][c+4] == pion:\n return True\n \n for c in range(COL_COUNT):\n for r in range(ROW_COUNT-4):\n if self.plateau[r][c] == pion and self.plateau[r+1][c] == pion and self.plateau[r+2][c] == pion and self.plateau[r+3][c] == pion and self.plateau[r+4][c] == pion:\n return True\n \n for c in range(COL_COUNT-4):\n for r in range(4,ROW_COUNT):\n if self.plateau[r][c] == pion and self.plateau[r-1][c+1] == pion and self.plateau[r-2][c+2] == pion and self.plateau[r-3][c+3] == pion and self.plateau[r-4][c+4] == pion:\n return True\n \n for c in range(COL_COUNT-4):\n for r in range(ROW_COUNT-4):\n if self.plateau[r][c] == pion and self.plateau[r+1][c+1] == pion and self.plateau[r+2][c+2] == pion and self.plateau[r+3][c+3] == pion and self.plateau[r+4][c+4] == pion:\n return True\n \n def Saisie (self) : \n \n \n \n lettre = {\"A\" : 1 , \"B\" : 2 , \"C\" : 3 , \"D\" : 4, \"E\" : 5,\n \"F\": 6 , \"G\" : 7 , \"H\" :8 , \"I\" :9 , \"J\" : 10 , \"K\" :11 , \"L\" :12\n , \"M\" :13 , \"N\":14 , \"O\": 15 }\n \n \n \n \n\n \n \n secu3 = False\n while secu3 == False :\n \n \n secu1 = False\n while secu1 == False : \n saisiex =input(\"Saisir la ligne ou vous voulez jouer : (chiffre) \")\n try:\n saisiex = int(saisiex)\n \n if saisiex > 0 and saisiex <= 15 :\n secu1 = True\n \n \n if(not secu1):\n print(\"Saisie invalide, veuillez réessayer\")\n except:\n print(\"Saisie invalide, veuillez réessayer\")\n \n secu2 = False\n while secu2 == False : \n saisiey =input(\"Saisir la colonnes ou vous voulez jouer : (lettre)\")\n \n try:\n\n if saisiey in lettre :\n secu2 = True\n \n if(not secu2):\n print(\"Saisie invalide, veuillez réessayer\")\n except:\n print(\"Saisie invalide, veuillez réessayer\")\n \n \n \n try:\n\n if self.plateau[saisiex-1][lettre[saisiey]-1] == 1 or self.plateau[saisiex-1][lettre[saisiey]-1] == 2 :\n print( \"pas bien \")\n secu3 = False\n else :\n secu3 = True\n \n if(not secu3):\n print(\"Position non valide, veuillez réessayer\")\n \n except:\n print(\"Saisie invalide, veuillez réessayer\") \n #Fonction 5,5 : saisie des premiers points\n #Ira dans la classe Gomoko\n def Start(self, joueur):\n if(joueur==1):\n #Placement du joueur 1 au centre du plateau\n self.plateau[8][8]=1\n #Placement selon l'IA de notre point\n self.MinMax() # A modifier, si elle ne fait pas le placement\n #Saisie des valeurs\n lettre = {\"A\" : 1 , \"B\" : 2 , \"C\" : 3 , \"D\" : 4, \"E\" : 5,\n \"L\" :12\n , \"M\" :13 , \"N\":14 , \"O\": 15 }\n secu3 = False\n while secu3 == False :\n secu1 = False\n while secu1 == False : \n saisiex =input(\"Saisir la ligne ou vous voulez jouer : (chiffre) \")\n try:\n saisiex = int(saisiex)\n \n if saisiex > 0 and saisiex <= 15 and (saisiex<5 or saisiex>11) :\n secu1 = True\n \n \n if(not secu1):\n print(\"Saisie invalide, veuillez réessayer\")\n except:\n print(\"Saisie invalide, veuillez réessayer\")\n \n secu2 = False\n while secu2 == False : \n saisiey =input(\"Saisir la colonnes ou vous voulez jouer : (lettre)\")\n \n try:\n \n if saisiey in lettre:\n secu2 = True\n \n if(not secu2):\n print(\"Saisie invalide, veuillez réessayer\")\n except:\n print(\"Saisie invalide, veuillez réessayer\")\n try:\n \n if self.plateau[saisiex-1][lettre[saisiey]-1] == 1 or self.plateau[saisiex-1][lettre[saisiey]-1] == 2 :\n print( \"pas bien \")\n secu3 = False\n else :\n secu3 = True\n \n if(not secu3):\n print(\"Position non valide, veuillez réessayer\")\n \n except:\n print(\"Saisie invalide, veuillez réessayer\") \n \n else:\n self.plateau[8][8]=2\n self.Saisie() # A voir en fonction de ce qui est souhaité\n #Réaliser un classement des différentes valeurs trouvées\n a=self.Actions(self.plateau)\n cpt=0\n for i in range(len(a)):\n if (a[i][0]>5 or a[i][0]<11) or (a[i][1]>5 or a[i][1]<11):\n a.pop(i)\n i-=1\n b=(self.MinValue(self.Result(action,a)) for action in a)\n dico=dict(a,b) #Attention, cette ligne pourrait crash\n dico = sorted(dico.items(), key=lambda x: x[1], reverse=True)\n actionValide=dico.keys()[0]\n self.application_result(self.plateau,actionValide[0],actionValide[1],self.joueur)\n\n \n\n\nMenu()",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
],
[
[
"### **VERSION JEUX A DEUX ( sans IA) --> pour faire des tests**",
"_____no_output_____"
]
],
[
[
"class Gomoko () :\n def __init__ ( self ): #ok \n self.plateau = [[ 0 for x in range ( 15)] for y in range(15)]\n self.nbPions = 0\n self.lettres= {\"A\" : 1 , \"B\" : 2 , \"C\" : 3 , \"D\" : 4, \"E\" : 5, \"F\": 6 , \"G\" : 7 , \"H\" :8 , \"I\" :9 , \"J\" : 10 , \"K\" :11 \n , \"L\" :12 , \"M\" :13 , \"N\":14 , \"O\": 15 }\n\n \n \n def __str__ (self): ## ok\n rep = \"\"\n rep += \"\\n\\t \"\n rep += \" \".join(self.lettres) \n if self.nbPions % 2 == 0 :\n rep += \"\\t\\t\\t Joueur 1 : O (noir) \\n\"\n elif self.nbPions % 2 != 0 :\n rep += \"\\t\\t\\t Joueur 2 : X (blanc) \\n\"\n else :\n rep += \"\\n\"\n \n for i in range(len(self.lettres)):\n for j in range(len(self.lettres)):\n if self.plateau[i][j]==0:\n if j==0:\n if i<9:\n rep += \"\\t \" +str(i+1) + \" . \"\n else:\n rep += \"\\t\" + str(i+1) +\" . \"\n else:\n rep += \". \"\n \n elif self.plateau[i][j]==1:\n if j==0:\n if i<9:\n rep += \"\\t \" + str(i+1) + \" O \"\n else:\n rep += \"\\t\" + str(i+1) + \" O \"\n else:\n rep += \"O \"\n else:\n if j==0:\n if i<9:\n rep += \"\\t \" + str(i+1) + \" X \"\n else:\n rep += \"\\t\" + str(i+1) +\" X \"\n else:\n rep += \"X \"\n rep += \"\\n\"\n return rep\n \n \n def Saisie (self) : ## ok \n \n secu3 = False\n while secu3 == False :\n \n \n secu1 = False\n while secu1 == False : \n saisiex =input(\"Saisir la ligne ou vous voulez jouer : (chiffre) \")\n try:\n saisiex = int(saisiex)\n \n if saisiex > 0 and saisiex <= len(self.lettres) :\n secu1 = True\n \n \n if(not secu1):\n print(\"Saisie invalide, veuillez réessayer\")\n except:\n print(\"Saisie invalide, veuillez réessayer\")\n \n secu2 = False\n while secu2 == False : \n saisiey =input(\"Saisir la colonnes ou vous voulez jouer : (lettres)\")\n \n try:\n saisiey = \"\".join(saisiey.split() ).upper()\n if saisiey in self.lettres :\n secu2 = True\n \n \n if(not secu2):\n print(\"Saisie invalide, veuillez réessayer\")\n except:\n print(\"Saisie invalide, veuillez réessayer\")\n \n try:\n\n if self.plateau[saisiex-1][self.lettres[saisiey]-1] == 1 or self.plateau[saisiex-1][self.lettres[saisiey]-1] == 2 :\n print( \"pas bien \")\n secu3 = False\n else :\n secu3 = True\n \n if(not secu3):\n print(\"Position non valide, veuillez réessayer\")\n \n except:\n print(\"Saisie invalide, veuillez réessayer\") \n return (saisiex-1 , self.lettres[saisiey]-1)\n \n \n \n def Appliquer(self,i,j):\n if self.nbPions % 2 == 0 :\n joueur = 0\n else :\n joueur = 1\n \n self.plateau[i][j]=joueur + 1 \n self.nbPions += 1 \n print ( \"nb pions \" , self.nbPions)\n \n def Jouer (self) :\n boucle = True\n while ( boucle ) :\n print ( \"terminal \" ,self.TerminalTest() )\n if self.nbPions % 2 == 0 :\n print ( gomoko)\n x, y = self.Saisie()\n self.Appliquer( x , y )\n \n elif self.nbPions % 2 != 0: \n print ( gomoko)\n x, y = self.Saisie()\n self.Appliquer( x , y )\n else : \n print ( \"Erreur\")\n \n if self.TerminalTest() or self.nbPions >= 120 :\n boucle = False\n print ( \"fin de la partie\")\n\n\ngomoko=Gomoko()\ngomoko.Jouer()\n",
"_____no_output_____"
]
],
[
[
"## version 2 ( by romu ) ",
"_____no_output_____"
]
],
[
[
"def Menu():\n print(\"\\n\\n***** Bonjour et bienvenue dans le jeux Gomoko *****\")\n saisieValide=False\n joueur = 0\n while(saisieValide==False):\n print(\"\\nVeuillez choisir un joueur\\n\")\n print(\"\\t1 - Joueur 1\\n\\t2 - Joueur 2\")\n joueur=input(\"Votre choix : \")\n try:\n joueur=int(joueur)\n saisieValide=(joueur==1) or (joueur==2)\n if(not saisieValide):\n print(\"Saisie invalide, veuillez réessayer\")\n except:\n print(\"Saisie invalide, veuillez réessayer\")\n print(\"\\nC'est bon\")\n \n \n \n\n gomoko = Gomoko(joueur-1)\n gomoko.Jouer()\n \n\n\nclass Gomoko () :\n def __init__ ( self , joueur): #ok \n self.plateau = [[ 0 for x in range ( 15)] for y in range(15)]\n self.nbPions = 0\n self.joueur = joueur\n self.lettres= {\"A\" : 1 , \"B\" : 2 , \"C\" : 3 , \"D\" : 4, \"E\" : 5, \"F\": 6 , \"G\" : 7 , \"H\" :8 , \"I\" :9 , \"J\" : 10 , \"K\" :11 \n , \"L\" :12 , \"M\" :13 , \"N\":14 , \"O\": 15 }\n\n \n def __str__ (self): ## ok\n rep = \"\\t\\t\\t\\t\\t\\t\\t\\t\\t\\t\\t\\t\\t\\t\\t Pions\\t : \" + str(self.nbPions) \n rep += \"\\n\\t \"\n rep += \" \".join(self.lettres) \n if self.joueur == 0 :\n rep += \"\\t\\t\\t joueur 1 : O (noir) \\n\"\n elif self.joueur == 1 :\n rep += \"\\t\\t\\t joueur 2 : X (blanc) \\n\"\n else :\n rep += \"\\n\"\n \n for i in range(len(self.lettres)):\n for j in range(len(self.lettres)):\n if self.plateau[i][j]==0:\n if j==0:\n if i<9: \n rep += \"\\t \" +str(i+1) + \" . \"\n else:\n rep += \"\\t\" + str(i+1) +\" . \"\n else:\n rep += \". \"\n \n elif self.plateau[i][j]==1:\n if j==0:\n if i<9:\n rep += \"\\t \" + str(i+1) + \" O \"\n else:\n rep += \"\\t\" + str(i+1) + \" O \"\n else:\n rep += \"O \"\n else:\n if j==0:\n if i<9:\n rep += \"\\t \" + str(i+1) + \" X \"\n else:\n rep += \"\\t\" + str(i+1) +\" X \"\n else:\n rep += \"X \"\n rep += \"\\n\"\n return rep\n \n \n def MaxValue(self, state):\n if self.TerminalTest(state):\n return self.Utility(state)\n v=-float(\"inf\")\n for a in self.Actions(state):\n v=max(v,self.MinValue(self.application_result(state.copy(),a[0],a[1],self.joueur)))\n return v\n \n \n def application_result(self, state ,i,j,joueur ):\n state[i][j]=joueur + 1 \n return state\n\n def MinValue(self, state):\n if self.TerminalTest(state):\n return self.Utility(state)\n v=float(\"inf\")\n for a in self.Actions(state): \n v=min(v,self.MaxValue(self.application_result(state.copy(),a[0],a[1],self.joueur)))\n return v\n\n \n \n #@timer\n def MinMax(self):\n #Réaliser un classement des différentes valeurs trouvées\n a=self.Actions(self.plateau.copy())\n print (\"actions \" , a)\n print ( \"taille actionc \" , len(a))\n \n t=self.plateau.copy()\n b=[self.MinValue(self.application_result(t, action[0] , action[1] , self.joueur)) for action in a]\n print (\"b = \" , b)\n \n a2=[]\n for t in a:\n c=str(t[0])+\" \"+str(t[1])\n a2.append(c)\n\n zip_iterator = zip(a2 ,b)\n dico=dict(zip_iterator)\n \n\n dico = dict(sorted(dico.items(), key=lambda x: x[1], reverse=True))\n \n print ( dico)\n actionValide=list( dico.keys()) [0].split(sep=\" \")\n actionValide[0]=int(actionValide[0])\n actionValide[1]=int(actionValide[1])\n \n self.application_result(self.plateau,actionValide[0],actionValide[1],self.joueur)\n\n \n\n def Actions(self, state):\n tab_actions = []\n [tab_actions.append([i,j]) if state[i][j]==0 else \"\" for i in range ( len(self.lettres)) for j in range(len(self.lettres))]\n return tab_actions\n \n \n def Utility(self,state):\n \n if(self.TerminalTest(state)):\n return -1\n t=self.joueur\n if(t==1):\n t=2\n else:\n t=1\n if(self.TerminalTest(state)):\n return 1\n return 0 \n \n def TerminalTest(self , state):\n for c in range(len(self.lettres) - 4):\n for r in range(len(self.lettres)):\n if state[r][c]== state[r][c+1]== state[r][c+2] == state[r][c+3]== state[r][c+4] and state[r][c]!=0:\n return True\n \n for c in range(len(self.lettres)):\n for r in range(len(self.lettres)-4):\n if state[r][c] == state[r+1][c] == state[r+2][c] == state[r+3][c] == state[r+4][c] and state[r][c]!=0:\n return True\n \n for c in range(len(self.lettres)-4):\n for r in range(4,len(self.lettres)):\n if state[r][c] == state[r-1][c+1] == state[r-2][c+2] == state[r-3][c+3] == state[r-4][c+4] and state[r][c]!=0:\n return True\n \n for c in range(len(self.lettres)-4):\n for r in range(len(self.lettres)-4):\n if state[r][c] == state[r+1][c+1] == state[r+2][c+2] == state[r+3][c+3] == state[r+4][c+4] and state[r][c]!=0:\n return True \n return False\n \n \n def Saisie (self) : ## ok \n secu = False\n while secu == False : \n saisie =input(\"\\t Coordonnées (D;7) \\t : \")\n try:\n saisieTab = saisie.split(\";\")\n \n if len (saisieTab) == 2 and int(saisieTab[1]) > 0 and int(saisieTab[1]) <= len(self.lettres) and \"\".join(saisieTab[0].split() ).upper() in self.lettres : \n secu = True\n saisiex = int(saisieTab[1])\n saisiey = \"\".join(saisieTab[0].split() ).upper() \n \n if self.plateau[saisiex-1][self.lettres[saisiey]-1] == 1 or self.plateau[saisiex-1][self.lettres[saisiey]-1] == 2 :\n secu = False \n except:\n print(\"Saisie invalide, veuillez réessayer\")\n \n return (saisiex-1 , self.lettres[saisiey]-1)\n \n \n \n \n def Appliquer(self,i,j):\n \n self.plateau[i][j]=self.joueur + 1 \n self.nbPions += 1\n if self.joueur == 1 : self.joueur = 0\n elif self.joueur == 0 : self.joueur = 1\n \n \n \"\"\" Long Pro\n \n \n \n def JouerLongPro(self):\n ## ##\n ## Si Joueur 1 commance\n ## ##\n if(self.joueur == 0):\n #Placement du joueur 1 au centre du plateau (\"humain\")\n self.Appliquer(8,8)\n #Placement selon l'IA de notre point (\"IA\")\n self.MinMax() # A modifier, si elle ne fait pas le placement\n #Saisie des valeurs carré de 7 (\"humain\")\n \n \n \n secu = False\n while secu == False : \n print (\" MERCI DE SAISIR DES VALEURS ENTRE 5 & 10 ET E & K\")\n print ( self)\n x, y = self.Saisie()\n if ( x >4 and x <9 and y >4 and y <9 ) :\n secu = True\n self.Appliquer( x , y )\n # (suite classique) \n \n else:\n ## ##\n ## Si Joueur 2 commance\n ## ##\n #Placement du joueur 2 au centre du plateau (IA)\n self.Appliquer(8,8)\n # PLacement\n print ( self) # (Humaine)\n x, y = self.Saisie()\n self.Appliquer( x , y )\n #Réaliser un classement des différentes valeurs trouvées (IA)\n a=self.Actions(self.plateau)\n cpt=0\n for i in range(len(a)):\n if (a[i][0]>5 or a[i][0]<11) or (a[i][1]>5 or a[i][1]<11):\n a.pop(i)\n i-=1\n b=(MinValue(Result(action,a)) for action in a)\n dico=dict(a,b) #Attention, cette ligne pourrait crash\n dico = sorted(dico.items(), key=lambda x: x[1], reverse=True)\n actionValide=dico.keys()[0]\n application_result(self.plateau,actionValide[0],actionValide[1],self.joueur) \n \n \"\"\"\n def JouerADeux (self) :\n boucle = True\n while ( boucle ) :\n if self.joueur == 0 :\n print ( self)\n x, y = self.Saisie()\n self.Appliquer( x , y )\n \n elif self.joueur == 1: \n print ( self)\n x, y = self.Saisie()\n self.Appliquer( x , y )\n else : \n print ( \"Erreur\")\n \n if self.TerminalTest(self.plateau) or self.nbPions >= 120 :\n boucle = False\n \n \n print ( \"\\n\\n************************************************************\")\n print ( \"*** FIN DE LA PARTIE ***\")\n print ( \"*** ***\")\n if self.joueur == 0 :\n print ( \"*** Le joueur 1 à gagné la partie! (pion noir) ***\")\n elif self.joueur == 1 :\n print ( \"*** Le joueur 2 à gagné la partie! (pion blanc) ***\")\n else: \n print ( \"*** ERREUR GAGNANT ***\")\n print ( \"*** ***\")\n print ( \"*** Vous avez utilisé\", self.nbPions, \"pions. ***\")\n print ( \"*** ***\")\n print ( \"************************************************************\")\n\n def JouerIA (self) :\n boucle = True\n while ( boucle ) :\n if self.joueur == 0 :\n print ( self)\n x, y = self.Saisie()\n self.Appliquer( x , y )\n \n elif self.joueur == 1: \n print ( self)\n print(len (self.Actions(self.plateau)))\n \n self.MinMax()\n\n \n \n \n else : \n print ( \"Erreur\")\n \n if self.TerminalTest(self.plateau) or self.nbPions >= 120 :\n boucle = True ## attention\n print ( \"fin de la partie\")\n \n \n \n \n \n \n\ngomoko=Gomoko(0)\ngomoko.JouerADeux ()\n\n#Menu()\n\n\n\n\n\n\n\n\n\n",
"\t\t\t\t\t\t\t\t\t\t\t\t\t\t\t Pions\t : 0\n\t A B C D E F G H I J K L M N O\t\t\t joueur 1 : O (noir) \n\t 1 . . . . . . . . . . . . . . . \n\t 2 . . . . . . . . . . . . . . . \n\t 3 . . . . . . . . . . . . . . . \n\t 4 . . . . . . . . . . . . . . . \n\t 5 . . . . . . . . . . . . . . . \n\t 6 . . . . . . . . . . . . . . . \n\t 7 . . . . . . . . . . . . . . . \n\t 8 . . . . . . . . . . . . . . . \n\t 9 . . . . . . . . . . . . . . . \n\t10 . . . . . . . . . . . . . . . \n\t11 . . . . . . . . . . . . . . . \n\t12 . . . . . . . . . . . . . . . \n\t13 . . . . . . . . . . . . . . . \n\t14 . . . . . . . . . . . . . . . \n\t15 . . . . . . . . . . . . . . . \n\n\t Coordonnées (D;7) \t : A;1\n\t\t\t\t\t\t\t\t\t\t\t\t\t\t\t Pions\t : 1\n\t A B C D E F G H I J K L M N O\t\t\t joueur 2 : X (blanc) \n\t 1 O . . . . . . . . . . . . . . \n\t 2 . . . . . . . . . . . . . . . \n\t 3 . . . . . . . . . . . . . . . \n\t 4 . . . . . . . . . . . . . . . \n\t 5 . . . . . . . . . . . . . . . \n\t 6 . . . . . . . . . . . . . . . \n\t 7 . . . . . . . . . . . . . . . \n\t 8 . . . . . . . . . . . . . . . \n\t 9 . . . . . . . . . . . . . . . \n\t10 . . . . . . . . . . . . . . . \n\t11 . . . . . . . . . . . . . . . \n\t12 . . . . . . . . . . . . . . . \n\t13 . . . . . . . . . . . . . . . \n\t14 . . . . . . . . . . . . . . . \n\t15 . . . . . . . . . . . . . . . \n\n\t Coordonnées (D;7) \t : B;2\n\t\t\t\t\t\t\t\t\t\t\t\t\t\t\t Pions\t : 2\n\t A B C D E F G H I J K L M N O\t\t\t joueur 1 : O (noir) \n\t 1 O . . . . . . . . . . . . . . \n\t 2 . X . . . . . . . . . . . . . \n\t 3 . . . . . . . . . . . . . . . \n\t 4 . . . . . . . . . . . . . . . \n\t 5 . . . . . . . . . . . . . . . \n\t 6 . . . . . . . . . . . . . . . \n\t 7 . . . . . . . . . . . . . . . \n\t 8 . . . . . . . . . . . . . . . \n\t 9 . . . . . . . . . . . . . . . \n\t10 . . . . . . . . . . . . . . . \n\t11 . . . . . . . . . . . . . . . \n\t12 . . . . . . . . . . . . . . . \n\t13 . . . . . . . . . . . . . . . \n\t14 . . . . . . . . . . . . . . . \n\t15 . . . . . . . . . . . . . . . \n\n\t Coordonnées (D;7) \t : c;\nSaisie invalide, veuillez réessayer\n\t Coordonnées (D;7) \t : c;1\n\t\t\t\t\t\t\t\t\t\t\t\t\t\t\t Pions\t : 3\n\t A B C D E F G H I J K L M N O\t\t\t joueur 2 : X (blanc) \n\t 1 O . O . . . . . . . . . . . . \n\t 2 . X . . . . . . . . . . . . . \n\t 3 . . . . . . . . . . . . . . . \n\t 4 . . . . . . . . . . . . . . . \n\t 5 . . . . . . . . . . . . . . . \n\t 6 . . . . . . . . . . . . . . . \n\t 7 . . . . . . . . . . . . . . . \n\t 8 . . . . . . . . . . . . . . . \n\t 9 . . . . . . . . . . . . . . . \n\t10 . . . . . . . . . . . . . . . \n\t11 . . . . . . . . . . . . . . . \n\t12 . . . . . . . . . . . . . . . \n\t13 . . . . . . . . . . . . . . . \n\t14 . . . . . . . . . . . . . . . \n\t15 . . . . . . . . . . . . . . . \n\n\t Coordonnées (D;7) \t : d;10\n\t\t\t\t\t\t\t\t\t\t\t\t\t\t\t Pions\t : 4\n\t A B C D E F G H I J K L M N O\t\t\t joueur 1 : O (noir) \n\t 1 O . O . . . . . . . . . . . . \n\t 2 . X . . . . . . . . . . . . . \n\t 3 . . . . . . . . . . . . . . . \n\t 4 . . . . . . . . . . . . . . . \n\t 5 . . . . . . . . . . . . . . . \n\t 6 . . . . . . . . . . . . . . . \n\t 7 . . . . . . . . . . . . . . . \n\t 8 . . . . . . . . . . . . . . . \n\t 9 . . . . . . . . . . . . . . . \n\t10 . . . X . . . . . . . . . . . \n\t11 . . . . . . . . . . . . . . . \n\t12 . . . . . . . . . . . . . . . \n\t13 . . . . . . . . . . . . . . . \n\t14 . . . . . . . . . . . . . . . \n\t15 . . . . . . . . . . . . . . . \n\n\t Coordonnées (D;7) \t : d;1\n\t\t\t\t\t\t\t\t\t\t\t\t\t\t\t Pions\t : 5\n\t A B C D E F G H I J K L M N O\t\t\t joueur 2 : X (blanc) \n\t 1 O . O O . . . . . . . . . . . \n\t 2 . X . . . . . . . . . . . . . \n\t 3 . . . . . . . . . . . . . . . \n\t 4 . . . . . . . . . . . . . . . \n\t 5 . . . . . . . . . . . . . . . \n\t 6 . . . . . . . . . . . . . . . \n\t 7 . . . . . . . . . . . . . . . \n\t 8 . . . . . . . . . . . . . . . \n\t 9 . . . . . . . . . . . . . . . \n\t10 . . . X . . . . . . . . . . . \n\t11 . . . . . . . . . . . . . . . \n\t12 . . . . . . . . . . . . . . . \n\t13 . . . . . . . . . . . . . . . \n\t14 . . . . . . . . . . . . . . . \n\t15 . . . . . . . . . . . . . . . \n\n\t Coordonnées (D;7) \t : m;1\n\t\t\t\t\t\t\t\t\t\t\t\t\t\t\t Pions\t : 6\n\t A B C D E F G H I J K L M N O\t\t\t joueur 1 : O (noir) \n\t 1 O . O O . . . . . . . . X . . \n\t 2 . X . . . . . . . . . . . . . \n\t 3 . . . . . . . . . . . . . . . \n\t 4 . . . . . . . . . . . . . . . \n\t 5 . . . . . . . . . . . . . . . \n\t 6 . . . . . . . . . . . . . . . \n\t 7 . . . . . . . . . . . . . . . \n\t 8 . . . . . . . . . . . . . . . \n\t 9 . . . . . . . . . . . . . . . \n\t10 . . . X . . . . . . . . . . . \n\t11 . . . . . . . . . . . . . . . \n\t12 . . . . . . . . . . . . . . . \n\t13 . . . . . . . . . . . . . . . \n\t14 . . . . . . . . . . . . . . . \n\t15 . . . . . . . . . . . . . . . \n\n\t Coordonnées (D;7) \t : b;1\n\t\t\t\t\t\t\t\t\t\t\t\t\t\t\t Pions\t : 7\n\t A B C D E F G H I J K L M N O\t\t\t joueur 2 : X (blanc) \n\t 1 O O O O . . . . . . . . X . . \n\t 2 . X . . . . . . . . . . . . . \n\t 3 . . . . . . . . . . . . . . . \n\t 4 . . . . . . . . . . . . . . . \n\t 5 . . . . . . . . . . . . . . . \n\t 6 . . . . . . . . . . . . . . . \n\t 7 . . . . . . . . . . . . . . . \n\t 8 . . . . . . . . . . . . . . . \n\t 9 . . . . . . . . . . . . . . . \n\t10 . . . X . . . . . . . . . . . \n\t11 . . . . . . . . . . . . . . . \n\t12 . . . . . . . . . . . . . . . \n\t13 . . . . . . . . . . . . . . . \n\t14 . . . . . . . . . . . . . . . \n\t15 . . . . . . . . . . . . . . . \n\n\t Coordonnées (D;7) \t : f;8\n\t\t\t\t\t\t\t\t\t\t\t\t\t\t\t Pions\t : 8\n\t A B C D E F G H I J K L M N O\t\t\t joueur 1 : O (noir) \n\t 1 O O O O . . . . . . . . X . . \n\t 2 . X . . . . . . . . . . . . . \n\t 3 . . . . . . . . . . . . . . . \n\t 4 . . . . . . . . . . . . . . . \n\t 5 . . . . . . . . . . . . . . . \n\t 6 . . . . . . . . . . . . . . . \n\t 7 . . . . . . . . . . . . . . . \n\t 8 . . . . . X . . . . . . . . . \n\t 9 . . . . . . . . . . . . . . . \n\t10 . . . X . . . . . . . . . . . \n\t11 . . . . . . . . . . . . . . . \n\t12 . . . . . . . . . . . . . . . \n\t13 . . . . . . . . . . . . . . . \n\t14 . . . . . . . . . . . . . . . \n\t15 . . . . . . . . . . . . . . . \n\n\t Coordonnées (D;7) \t : e;1\n\n\n************************************************************\n*** FIN DE LA PARTIE ***\n*** ***\n*** Le joueur 2 à gagné la partie! (pion blanc) ***\n*** ***\n*** Vous avez utilisé 9 pions. ***\n*** ***\n************************************************************\n"
],
[
"# -*- coding: utf-8 -*-\n\"\"\"\nCreated on Fri May 6 17:18:09 2022\n\n@author: Romuald\n\"\"\"\nimport time\n\n\n\ndef Menu():\n print(\"\\n\\n***** Bonjour et bienvenue dans le jeux Gomoko *****\")\n saisieValide=False\n joueur = 0\n while(saisieValide==False):\n print(\"\\nVeuillez choisir un joueur\\n\")\n print(\"\\t1 - Joueur 1\\n\\t2 - Joueur 2\")\n joueur=input(\"Votre choix : \")\n try:\n joueur=int(joueur)\n saisieValide=(joueur==1) or (joueur==2)\n if(not saisieValide):\n print(\"Saisie invalide, veuillez réessayer\")\n except:\n print(\"Saisie invalide, veuillez réessayer\")\n print(\"\\nC'est bon\")\n \n \n \n\n gomoko = Gomoko(joueur-1)\n gomoko.Jouer()\n \n\n\nclass Gomoko () :\n def __init__ ( self , joueur): #ok \n self.plateau = [[ 0 for x in range ( 15)] for y in range(15)]\n self.nbPions = 0\n self.joueur = joueur\n \n self.player_turn = 2\n \n \n self.lettres= {\"A\" : 1 , \"B\" : 2 , \"C\" : 3 , \"D\" : 4, \"E\" : 5, \"F\": 6 }\n #, \"G\" : 7 , \"H\" :8 , \"I\" :9 , \"J\" : 10 , \"K\" :11 \n # , \"L\" :12 , \"M\" :13 , \"N\":14 , \"O\": 15 }\n\n \n def __str__ (self): ## ok\n rep = \"\\t\\t\\t\\t\\t\\t\\t\\t\\t\\t\\t\\t\\t\\t\\t Pions\\t : \" + str(self.nbPions) \n rep += \"\\n\\t \"\n rep += \" \".join(self.lettres) \n if self.joueur == 0 :\n rep += \"\\t\\t\\t joueur 1 : O (noir) \\n\"\n elif self.joueur == 1 :\n rep += \"\\t\\t\\t joueur 2 : X (blanc) \\n\"\n else :\n rep += \"\\n\"\n \n for i in range(len(self.lettres)):\n for j in range(len(self.lettres)):\n if self.plateau[i][j]==0:\n if j==0:\n if i<9: \n rep += \"\\t \" +str(i+1) + \" . \"\n else:\n rep += \"\\t\" + str(i+1) +\" . \"\n else:\n rep += \". \"\n \n elif self.plateau[i][j]==1:\n if j==0:\n if i<9:\n rep += \"\\t \" + str(i+1) + \" O \"\n else:\n rep += \"\\t\" + str(i+1) + \" O \"\n else:\n rep += \"O \"\n else:\n if j==0:\n if i<9:\n rep += \"\\t \" + str(i+1) + \" X \"\n else:\n rep += \"\\t\" + str(i+1) +\" X \"\n else:\n rep += \"X \"\n rep += \"\\n\"\n return rep\n \n \n def MaxValue(self, state):\n if self.TerminalTest(state):\n return self.Utility(state)\n v=-float(\"inf\")\n for a in self.Actions(state):\n v=max(v,self.MinValue(self.application_result(state.copy(),a[0],a[1],self.joueur)))\n return v\n \n \n def application_result(self, state ,i,j,joueur ):\n state[i][j]=joueur + 1 \n return state\n\n def MinValue(self, state):\n if self.TerminalTest(state):\n return self.Utility(state)\n v=float(\"inf\")\n for a in self.Actions(state): \n v=min(v,self.MaxValue(self.application_result(state.copy(),a[0],a[1],self.joueur)))\n return v\n\n \n \n # @ timer\n def MinMax(self):\n #Réaliser un classement des différentes valeurs trouvées\n a=self.Actions(self.plateau.copy())\n print (\"actions \" , a)\n print ( \"taille actionc \" , len(a))\n \n t=self.plateau.copy()\n b=[self.MinValue(self.application_result(t, action[0] , action[1] , self.joueur)) for action in a]\n print (\"b = \" , b)\n \n a2=[]\n for t in a:\n c=str(t[0])+\" \"+str(t[1])\n a2.append(c)\n\n zip_iterator = zip(a2 ,b)\n dico=dict(zip_iterator)\n \n\n dico = dict(sorted(dico.items(), key=lambda x: x[1], reverse=True))\n \n print ( dico)\n actionValide=list( dico.keys()) [0].split(sep=\" \") \n actionValide[0]=int(actionValide[0])\n actionValide[1]=int(actionValide[1])\n \n self.application_result(self.plateau,actionValide[0],actionValide[1],self.joueur)\n\n \n\n def Actions(self, state):\n tab_actions = []\n [tab_actions.append([i,j]) if state[i][j]==0 else \"\" for i in range ( len(self.lettres)) for j in range(len(self.lettres))]\n return tab_actions\n \n \n def Utility(self,state):\n \n if(self.TerminalTest(state)):\n return -1\n t=self.joueur\n if(t==1):\n t=2\n else:\n t=1\n if(self.TerminalTest(state)):\n return 1\n return 0 \n \n \n def is_end(self ): # internet \n \n state = self.plateau \n for c in range(len(self.lettres) - 4):\n for r in range(len(self.lettres)):\n if state[r][c]== state[r][c+1]== state[r][c+2] == state[r][c+3]== state[r][c+4] and state[r][c]!=0:\n return state[r][c]\n \n for c in range(len(self.lettres)):\n for r in range(len(self.lettres)-4):\n if state[r][c] == state[r+1][c] == state[r+2][c] == state[r+3][c] == state[r+4][c] and state[r][c]!=0:\n return state[r][c]\n \n for c in range(len(self.lettres)-4):\n for r in range(4,len(self.lettres)):\n if state[r][c] == state[r-1][c+1] == state[r-2][c+2] == state[r-3][c+3] == state[r-4][c+4] and state[r][c]!=0:\n return state[r][c]\n \n for c in range(len(self.lettres)-4):\n for r in range(len(self.lettres)-4):\n if state[r][c] == state[r+1][c+1] == state[r+2][c+2] == state[r+3][c+3] == state[r+4][c+4] and state[r][c]!=0:\n return state[r][c] \n return \n \n def min(self):\n \n \n # Possible values for minv are:\n # -1 - win\n # 0 - a tie\n # 1 - loss\n \n # We're initially setting it to 2 as worse than the worst case:\n minv = 2\n \n qx = None\n qy = None\n \n result = self.is_end()\n \n if result == 0:\n return (-1, 0, 0)\n elif result == 1:\n return (1, 0, 0)\n elif result == False:\n return (0, 0, 0)\n \n for i in range(0, len(self.lettres)):\n for j in range(0, len(self.lettres)):\n if self.plateau[i][j] == 0:\n\n self.plateau[i][j] = 1\n (m, max_i, max_j) = self.max()\n if m < minv:\n minv = m\n qx = i\n qy = j\n\n self.plateau[i][j] = 0\n \n return (minv, qx, qy)\n \n \n def play(self):\n while True:\n print(self)\n self.result = self.is_end()\n \n # Printing the appropriate message if the game has ended\n if self.result != None:\n if self.result == 2:\n print('The winner is X!')\n elif self.result == 1:\n print('The winner is O!')\n elif self.result == '.':\n print(\"It's a tie!\")\n \n self\n return\n \n # If it's player's turn\n if self.player_turn == 2:\n \n while True:\n \n start = time.time()\n print ( \"cocou\")\n (m, qx, qy) = self.min()\n \n end = time.time()\n print('Evaluation time: {}s'.format(round(end - start, 7)))\n print('Recommended move: X = {}, Y = {}'.format(qx, qy))\n \n px = int(input('Insert the X coordinate: '))\n py = int(input('Insert the Y coordinate: '))\n \n (qx, qy) = (px, py)\n \n if self.is_valid(px, py):\n self.current_state[px][py] = 2\n self.player_turn = 1\n break\n else:\n print('The move is not valid! Try again.')\n \n # If it's AI's turn\n else:\n (m, px, py) = self.max()\n self.plateau[px][py] = 1\n self.player_turn = 2 \n \n \n \n \n\n \n def max(self):\n \n # Possible values for maxv are:\n # -1 - loss\n # 0 - a tie\n # 1 - win\n \n # We're initially setting it to -2 as worse than the worst case:\n maxv = -2\n \n px = None\n py = None\n \n result = self.is_end()\n \n # If the game came to an end, the function needs to return\n # the evaluation function of the end. That can be:\n # -1 - loss\n # 0 - a tie\n # 1 - win\n if result == 0:\n return (-1, 0, 0)\n elif result == 1:\n return (1, 0, 0)\n elif result == False:\n return (0, 0, 0)\n \n for i in range(0, len(self.lettres)):\n print ( \"coucou\")\n for j in range(0, len(self.lettres)):\n if self.plateau[i][j] == 0:\n # On the empty field player 'O' makes a move and calls Min\n # That's one branch of the game tree.\n self.plateau[i][j] = 2\n (m, min_i, min_j) = self.min()\n # Fixing the maxv value if needed\n if m > maxv:\n maxv = m\n px = i\n py = j\n # Setting back the field to empty\n self.plateau[i][j] = 0\n return (maxv, px, py)\n\n def TerminalTest(self , state ):\n for c in range(len(self.lettres) - 4):\n for r in range(len(self.lettres)):\n if state[r][c]== state[r][c+1]== state[r][c+2] == state[r][c+3]== state[r][c+4] and state[r][c]!=0:\n return True\n \n for c in range(len(self.lettres)):\n for r in range(len(self.lettres)-4):\n if state[r][c] == state[r+1][c] == state[r+2][c] == state[r+3][c] == state[r+4][c] and state[r][c]!=0:\n return True\n \n for c in range(len(self.lettres)-4):\n for r in range(4,len(self.lettres)):\n if state[r][c] == state[r-1][c+1] == state[r-2][c+2] == state[r-3][c+3] == state[r-4][c+4] and state[r][c]!=0:\n return True\n \n for c in range(len(self.lettres)-4):\n for r in range(len(self.lettres)-4):\n if state[r][c] == state[r+1][c+1] == state[r+2][c+2] == state[r+3][c+3] == state[r+4][c+4] and state[r][c]!=0:\n return True \n return False\n \n \n def Saisie (self) : ## ok \n secu = False\n while secu == False : \n saisie =input(\"\\t Coordonnées (D;7) \\t : \")\n try:\n saisieTab = saisie.split(\";\")\n \n if len (saisieTab) == 2 and int(saisieTab[1]) > 0 and int(saisieTab[1]) <= len(self.lettres) and \"\".join(saisieTab[0].split() ).upper() in self.lettres : \n secu = True\n saisiex = int(saisieTab[1])\n saisiey = \"\".join(saisieTab[0].split() ).upper() \n \n if self.plateau[saisiex-1][self.lettres[saisiey]-1] == 1 or self.plateau[saisiex-1][self.lettres[saisiey]-1] == 2 :\n secu = False \n except:\n print(\"Saisie invalide, veuillez réessayer\")\n \n return (saisiex-1 , self.lettres[saisiey]-1)\n \n \n \n \n def Appliquer(self,i,j):\n \n self.plateau[i][j]=self.joueur + 1 \n self.nbPions += 1\n if self.joueur == 1 : self.joueur = 0\n elif self.joueur == 0 : self.joueur = 1\n \n \n \"\"\" Long Pro\n \n \n \n def JouerLongPro(self):\n ## ##\n ## Si Joueur 1 commance\n ## ##\n if(self.joueur == 0):\n #Placement du joueur 1 au centre du plateau (\"humain\")\n self.Appliquer(8,8)\n #Placement selon l'IA de notre point (\"IA\")\n self.MinMax() # A modifier, si elle ne fait pas le placement\n #Saisie des valeurs carré de 7 (\"humain\")\n \n \n \n secu = False\n while secu == False : \n print (\" MERCI DE SAISIR DES VALEURS ENTRE 5 & 10 ET E & K\")\n print ( self)\n x, y = self.Saisie()\n if ( x >4 and x <9 and y >4 and y <9 ) :\n secu = True\n self.Appliquer( x , y )\n # (suite classique) \n \n else:\n ## ##\n ## Si Joueur 2 commance\n ## ##\n #Placement du joueur 2 au centre du plateau (IA)\n self.Appliquer(8,8)\n # PLacement\n print ( self) # (Humaine)\n x, y = self.Saisie()\n self.Appliquer( x , y )\n #Réaliser un classement des différentes valeurs trouvées (IA)\n a=self.Actions(self.plateau)\n cpt=0\n for i in range(len(a)):\n if (a[i][0]>5 or a[i][0]<11) or (a[i][1]>5 or a[i][1]<11):\n a.pop(i)\n i-=1\n b=(MinValue(Result(action,a)) for action in a)\n dico=dict(a,b) #Attention, cette ligne pourrait crash\n dico = sorted(dico.items(), key=lambda x: x[1], reverse=True)\n actionValide=dico.keys()[0]\n application_result(self.plateau,actionValide[0],actionValide[1],self.joueur) \n \n \"\"\"\n def JouerADeux (self) :\n boucle = True\n while ( boucle ) :\n if self.joueur == 0 :\n print ( self)\n x, y = self.Saisie()\n self.Appliquer( x , y )\n \n elif self.joueur == 1: \n print ( self)\n x, y = self.Saisie()\n self.Appliquer( x , y )\n else : \n print ( \"Erreur\")\n \n if self.TerminalTest(self.plateau) or self.nbPions >= 120 :\n boucle = False\n \n \n print ( \"\\n\\n************************************************************\")\n print ( \"*** FIN DE LA PARTIE ***\")\n print ( \"*** ***\")\n if self.joueur == 0 :\n print ( \"*** Le joueur 1 à gagné la partie! (pion noir) ***\")\n elif self.joueur == 1 :\n print ( \"*** Le joueur 2 à gagné la partie! (pion blanc) ***\")\n else: \n print ( \"*** ERREUR GAGNANT ***\")\n print ( \"*** ***\")\n print ( \"*** Vous avez utilisé\", self.nbPions, \"pions. ***\")\n print ( \"*** ***\")\n print ( \"************************************************************\")\n\n def JouerIA (self) :\n boucle = True\n while ( boucle ) :\n if self.joueur == 0 :\n print ( self)\n x, y = self.Saisie()\n self.Appliquer( x , y )\n \n elif self.joueur == 1: \n print ( self)\n print(len (self.Actions(self.plateau)))\n \n self.MinMax()\n\n \n \n \n else : \n print ( \"Erreur\")\n \n if self.TerminalTest(self.plateau) or self.nbPions >= 120 :\n boucle = True ## attention\n print ( \"fin de la partie\")\n \n \n \n \n \n \n\n#gomoko=Gomoko(0)\n#gomoko.JouerADeux()\n\ng = Gomoko(3)\n\ng.play()\n\n\n\n#Menu()\n\n\n\n\n\n",
"\u001b[1;30;43mLe flux de sortie a été tronqué et ne contient que les 5000 dernières lignes.\u001b[0m\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\ncoucou\n"
],
[
"import time\n\ndef Menu():\n print(\"\\n\\n***** Bonjour et bienvenue dans le jeux Gomoko *****\")\n saisieValide=False\n joueur = 0\n while(saisieValide==False):\n print(\"\\nVeuillez choisir un joueur\\n\")\n print(\"\\t1 - Joueur 1\\n\\t2 - Joueur 2\")\n joueur=input(\"Votre choix : \")\n try:\n joueur=int(joueur)\n saisieValide=(joueur==1) or (joueur==2)\n if(not saisieValide):\n print(\"Saisie invalide, veuillez réessayer\")\n except:\n print(\"Saisie invalide, veuillez réessayer\")\n gomoko = Gomoko(joueur)\n gomoko.Jouer()\n\ndef timer(fonction):\n \"\"\"\n Méthode qui chronomètre le temps d'exécution d'une méthode\n Sera utilisé pour mesurer le temps de traitement du MinMax\n\n Parameters\n ----------\n fonction : Fonction sur laquelle appliquer le timer\n\n Returns\n -------\n None.\n\n \"\"\"\n \n def inner(*args,**kwargs):\n #Coeur de la méthode de mesure du temps\n t=time.time()#Temps en entrée\n f=fonction(*args,**kwargs)\n print(time.time()-t)#Affichage du temps finel (en secondes)\n return f\n return inner \n \ndef Duplicate(t2):\n t=[]\n for a in t2:\n k=[]\n for b in a:\n if(b>=3):\n print(\"Y a un 3\")\n k.append(b)\n t.append(k)\n return t\n\n\n\n\nclass Gomoko () :\n iteration=0\n def __init__ ( self , joueur): #ok \n self.plateau = [[ 0 for x in range ( 15)] for y in range(15)]\n self.nbPions = 0\n self.joueur = joueur\n self.lettres= {\"A\" : 1 , \"B\" : 2 , \"C\" : 3 , \"D\" : 4, \"E\" : 5, \"F\": 6 , \"G\" : 7 , \"H\" :8 , \"I\" :9 , \"J\" : 10 , \"K\" :11 , \"L\" :12 , \"M\" :13 , \"N\":14 , \"O\": 15 }\n #self.lettres= {\"A\" : 1 , \"B\" : 2 , \"C\" : 3 , \"D\" : 4, \"E\" : 5}\n \n def __str__ (self): ## ok\n rep = \"\\t\\t\\t\\t\\t\\t\\t\\t\\t\\t\\t\\t\\t\\t\\t Pions\\t : \" + str(self.nbPions) \n rep += \"\\n\\t \"\n rep += \" \".join(self.lettres) \n if self.joueur == 0 :\n rep += \"\\t\\t\\t joueur 1 : O (noir) \\n\"\n elif self.joueur == 1 :\n rep += \"\\t\\t\\t joueur 2 : X (blanc) \\n\"\n else :\n rep += \"\\n\"\n \n for i in range(len(self.lettres)):\n for j in range(len(self.lettres)):\n if self.plateau[i][j]==0:\n if j==0:\n if i<9: \n rep += \"\\t \" +str(i+1) + \" . \"\n else:\n rep += \"\\t\" + str(i+1) +\" . \"\n else:\n rep += \". \"\n \n elif self.plateau[i][j]==1:\n if j==0:\n if i<9:\n rep += \"\\t \" + str(i+1) + \" O \"\n else:\n rep += \"\\t\" + str(i+1) + \" O \"\n else:\n rep += \"O \"\n else:\n if j==0:\n if i<9:\n rep += \"\\t \" + str(i+1) + \" X \"\n else:\n rep += \"\\t\" + str(i+1) +\" X \"\n else:\n rep += \"X \"\n rep += \"\\n\"\n return rep\n \n \n def MaxValue(self, state):\n print(\"Max\")\n t=Duplicate(state)\n if self.TerminalTest(t)!=0:\n return self.Utility(t)\n v=-float(\"inf\")\n k=0\n for a in self.Actions(t,self.CasesPossibles(t)):\n v=max(v,self.MinValue(self.application_result(t,a[0],a[1],self.joueur)))\n return v\n \n \n def application_result(self, state ,i,j,joueur):\n state[i][j]=joueur + 1\n return state\n\n def MinValue(self, state):\n print(\"Min\")\n t=Duplicate(state)\n if self.TerminalTest(t)!=0:\n print(\"Ici2\")\n return self.Utility(t)\n print(\"Ici\")\n v=float(\"inf\")\n if(self.joueur==1):\n k=0\n else:\n k=1\n for a in self.Actions(t,self.CasesPossibles(t)): \n v=min(v,self.MaxValue(self.application_result(t,a[0],a[1],k)))\n return v\n\n \n \n @timer\n def MinMax(self):\n #Réaliser un classement des différentes valeurs trouvées\n #print(\"Plateau de base :\",self.plateau)\n t=Duplicate(self.plateau)\n cas=self.CasesPossibles(t)\n a=self.Actions(t,cas)\n #print (\"actions \" , a)\n #print ( \"taille actionc \" , len(a))\n b=[]\n #print(\"Actions : \",a)\n if(self.joueur==1):\n k=0\n else:\n k=1\n for action in a:\n # Le passage à \n b.append(self.MinValue(self.application_result(t, action[0] , action[1] , k)))\n #print (\"b = \" , b)\n a2=[]\n for t in a:\n c=str(t[0])+\" \"+str(t[1])\n a2.append(c)\n zip_iterator = zip(a2 ,b)\n dico=dict(zip_iterator)\n \n \n dico = dict(sorted(dico.items(), key=lambda x: x[1], reverse=True))\n \n #print(dico)\n #print(dico.keys())\n actionValide=list( dico.keys()) [0].split(sep=\" \")\n actionValide[0]=int(actionValide[0])\n actionValide[1]=int(actionValide[1])\n \n self.application_result(self.plateau,actionValide[0],actionValide[1],self.joueur)\n\n\n def CasesPossibles(self,state):\n b=[]\n for i in range(len(state)):\n for j in range(len(state)):\n cpt=0\n for k in range(-2,3):\n for l in range(-2,3):\n if((i+k>=0 and i+k<len(state)) and (j+l>=0 and j+l<len(state))):\n if(state[i+k][j+l]!=0):\n b.append([i,j])\n cpt=1\n break\n return b\n\n def Actions(self, state,cas):\n tab_actions = []\n casO=[]\n casI=[]\n for a in cas:\n casO.append(a[0])\n casI.append(a[1])\n print(casO)\n print(casI)\n [tab_actions.append([i,j]) if state[i][j]==0 else \"\" for i in casO for j in casI]\n return tab_actions\n \n \n def Utility2(self,state):\n if(self.TerminalTest(state)==self.joueur+1):\n print(\"-1\")\n return -1\n t=self.joueur+1\n if(t==1):\n t=2\n else:\n t=1\n if(self.TerminalTest(state)==t):\n print(\"1\")\n return 1\n print(\"0\")\n return 0 \n \n def Utility(self,state):\n if(self.TerminalTest(state)==1):\n print(-1)\n return -1\n elif(self.TerminalTest(state)==2):\n print(1)\n return 1\n else:\n print(0)\n return 0\n \n def TerminalTest(self , state ):\n for c in range(len(self.lettres) - 4):\n for r in range(len(self.lettres)):\n if state[r][c]== state[r][c+1]== state[r][c+2] == state[r][c+3]== state[r][c+4] and state[r][c]!=0:\n return state[r][c]\n \n for c in range(len(self.lettres)):\n for r in range(len(self.lettres)-4):\n if state[r][c] == state[r+1][c] == state[r+2][c] == state[r+3][c] == state[r+4][c] and state[r][c]!=0:\n return state[r][c]\n \n for c in range(len(self.lettres)-4):\n for r in range(4,len(self.lettres)):\n if state[r][c] == state[r-1][c+1] == state[r-2][c+2] == state[r-3][c+3] == state[r-4][c+4] and state[r][c]!=0:\n return state[r][c]\n \n for c in range(len(self.lettres)-4):\n for r in range(len(self.lettres)-4):\n if state[r][c] == state[r+1][c+1] == state[r+2][c+2] == state[r+3][c+3] == state[r+4][c+4] and state[r][c]!=0:\n return state[r][c] \n return 0\n \n \n def Saisie (self) : ## ok \n secu = False\n while secu == False : \n saisie =input(\"\\t Coordonnées (D;7) \\t : \")\n try:\n saisieTab = saisie.split(\";\")\n \n if len (saisieTab) == 2 and int(saisieTab[1]) > 0 and int(saisieTab[1]) <= len(self.lettres) and \"\".join(saisieTab[0].split() ).upper() in self.lettres : \n secu = True\n saisiex = int(saisieTab[1])\n saisiey = \"\".join(saisieTab[0].split() ).upper() \n \n if self.plateau[saisiex-1][self.lettres[saisiey]-1] == 1 or self.plateau[saisiex-1][self.lettres[saisiey]-1] == 2 :\n secu = False \n except:\n print(\"Saisie invalide, veuillez réessayer\")\n \n return (saisiex-1 , self.lettres[saisiey]-1)\n \n \n \n \n def Appliquer(self,i,j):\n \n self.plateau[i][j]=self.joueur + 1 \n self.nbPions += 1\n if self.joueur == 1 : self.joueur = 0\n elif self.joueur == 0 : self.joueur = 1\n \n \n \"\"\" Long Pro\n \n \n \n def JouerLongPro(self):\n ## ##\n ## Si Joueur 1 commance\n ## ##\n if(self.joueur == 0):\n #Placement du joueur 1 au centre du plateau (\"humain\")\n self.Appliquer(8,8)\n #Placement selon l'IA de notre point (\"IA\")\n self.MinMax() # A modifier, si elle ne fait pas le placement\n #Saisie des valeurs carré de 7 (\"humain\")\n \n \n \n secu = False\n while secu == False : \n print (\" MERCI DE SAISIR DES VALEURS ENTRE 5 & 10 ET E & K\")\n print ( self)\n x, y = self.Saisie()\n if ( x >4 and x <9 and y >4 and y <9 ) :\n secu = True\n self.Appliquer( x , y )\n # (suite classique) \n \n else:\n ## ##\n ## Si Joueur 2 commance\n ## ##\n #Placement du joueur 2 au centre du plateau (IA)\n self.Appliquer(8,8)\n # PLacement\n print ( self) # (Humaine)\n x, y = self.Saisie()\n self.Appliquer( x , y )\n #Réaliser un classement des différentes valeurs trouvées (IA)\n a=self.Actions(self.plateau)\n cpt=0\n for i in range(len(a)):\n if (a[i][0]>5 or a[i][0]<11) or (a[i][1]>5 or a[i][1]<11):\n a.pop(i)\n i-=1\n b=(MinValue(Result(action,a)) for action in a)\n dico=dict(a,b) #Attention, cette ligne pourrait crash\n dico = sorted(dico.items(), key=lambda x: x[1], reverse=True)\n actionValide=dico.keys()[0]\n application_result(self.plateau,actionValide[0],actionValide[1],self.joueur) \n \n \"\"\"\n def JouerADeux (self) :\n boucle = True\n while ( boucle ) :\n if self.joueur == 0 :\n print ( self)\n x, y = self.Saisie()\n self.Appliquer( x , y )\n \n elif self.joueur == 1: \n print ( self)\n x, y = self.Saisie()\n self.Appliquer( x , y )\n else : \n print ( \"Erreur\")\n \n if self.TerminalTest(self.plateau)!=0 or self.nbPions >= 120 :\n boucle = False\n \n \n print ( \"\\n\\n************************************************************\")\n print ( \"*** FIN DE LA PARTIE ***\")\n print ( \"*** ***\")\n if self.joueur == 0 :\n print ( \"*** Le joueur 1 à gagné la partie! (pion noir) ***\")\n elif self.joueur == 1 :\n print ( \"*** Le joueur 2 à gagné la partie! (pion blanc) ***\")\n else: \n print ( \"*** ERREUR GAGNANT ***\")\n print ( \"*** ***\")\n print ( \"*** Vous avez utilisé\", self.nbPions, \"pions. ***\")\n print ( \"*** ***\")\n print ( \"************************************************************\")\n\n def JouerIA (self) :\n boucle = True\n while ( boucle ) :\n if self.joueur == 0 :\n print ( self)\n x, y = self.Saisie()\n self.Appliquer( x , y )\n \n elif self.joueur == 1: \n print ( self)\n print(len (self.Actions(self.plateau,self.CasesPossibles(self.plateau))))\n \n self.MinMax()\n print(\"Ici\")\n self.joueur=0\n \n \n \n else : \n print ( \"Erreur\")\n \n if self.TerminalTest(self.plateau) or self.nbPions >= 120 :\n boucle = True ## attention\n print ( \"fin de la partie\")\n \n \n \n \n \n \n\ngomoko=Gomoko(0)\ngomoko.JouerIA()\n\n#Menu()\n\n\n\n\n\n\n\n\n\n",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
4a576767bb7269dde0c63d117e510143cfc419b6
| 7,998 |
ipynb
|
Jupyter Notebook
|
teaching/stat_775_2021_fall/activities/activity-2021-09-01.ipynb
|
cgrudz/cgrudz.github.io
|
b6c2c0fcbe2b988f8827dfcece92075594037c4a
|
[
"MIT"
] | 3 |
2019-08-29T17:53:52.000Z
|
2021-12-28T02:53:29.000Z
|
teaching/stat_775_2021_fall/activities/activity-2021-09-01.ipynb
|
cgrudz/cgrudz.github.io
|
b6c2c0fcbe2b988f8827dfcece92075594037c4a
|
[
"MIT"
] | 5 |
2020-02-25T01:02:27.000Z
|
2021-09-27T20:36:01.000Z
|
teaching/stat_775_2021_fall/activities/activity-2021-09-01.ipynb
|
cgrudz/cgrudz.github.io
|
b6c2c0fcbe2b988f8827dfcece92075594037c4a
|
[
"MIT"
] | 1 |
2021-05-16T00:40:03.000Z
|
2021-05-16T00:40:03.000Z
| 28.564286 | 358 | 0.586522 |
[
[
[
"# Introduction to Python part IV (And a discussion of linear transformations)",
"_____no_output_____"
],
[
"## Activity 1: Discussion of linear transformations\n\n\n* Orthogonality also plays a key role in understanding linear transformations. How can we understand linear transformations in terms of a composition of rotations and diagonal matrices? There are two specific matrix factorizations that arise this way, can you name them and describe the conditions in which they are applicable?\n\n* What is a linear inverse problem? What conditions guarantee a solution?\n\n* What is a pseudo-inverse? How is this related to an orthogonal projection? How is this related to the linear inverse problem?\n\n* What is a weighted norm and what is a weighted pseudo-norm?",
"_____no_output_____"
],
[
"## Activity 2: Basic data analysis and manipulation",
"_____no_output_____"
]
],
[
[
"import numpy as np",
"_____no_output_____"
]
],
[
[
"### Exercise 1:\n\nArrays can be concatenated and stacked on top of one another, using NumPy’s `vstack` and `hstack` functions for vertical and horizontal stacking, respectively.\n",
"_____no_output_____"
]
],
[
[
"A = np.array([[1,2,3], [4,5,6], [7, 8, 9]])\nprint('A = ')\nprint(A)\n\nB = np.hstack([A, A])\nprint('B = ')\nprint(B)\n\nC = np.vstack([A, A])\nprint('C = ')\nprint(C)",
"_____no_output_____"
]
],
[
[
"Write some additional code that slices the first and last columns of A, and stacks them into a 3x2 array. Make sure to print the results to verify your solution.\n\nNote a ‘gotcha’ with array indexing is that singleton dimensions are dropped by default. That means `A[:, 0]` is a one dimensional array, which won’t stack as desired. To preserve singleton dimensions, the index itself can be a slice or array. For example, `A[:, :1]` returns a two dimensional array with one singleton dimension (i.e. a column vector).",
"_____no_output_____"
]
],
[
[
"D = np.hstack((A[:, :1], A[:, -1:]))\nprint('D = ')\nprint(D)",
"_____no_output_____"
]
],
[
[
"An alternative way to achieve the same result is to use Numpy’s delete function to remove the second column of A. Use the search function for the documentation on the `np.delete` function to find the syntax for constructing such an array.\n",
"_____no_output_____"
],
[
"### Exercise 2:\n\nThe patient data is longitudinal in the sense that each row represents a series of observations relating to one individual. This means that the change in inflammation over time is a meaningful concept. Let’s find out how to calculate changes in the data contained in an array with NumPy.\n\nThe `np.diff` function takes an array and returns the differences between two successive values. Let’s use it to examine the changes each day across the first week of patient 3 from our inflammation dataset.",
"_____no_output_____"
]
],
[
[
"patient3_week1 = data[3, :7]\nprint(patient3_week1)",
"_____no_output_____"
]
],
[
[
"Calling `np.diff(patient3_week1)` would do the following calculations",
"_____no_output_____"
],
[
"`[ 0 - 0, 2 - 0, 0 - 2, 4 - 0, 2 - 4, 2 - 2 ]`",
"_____no_output_____"
],
[
"and return the 6 difference values in a new array.",
"_____no_output_____"
]
],
[
[
"np.diff(patient3_week1)",
"_____no_output_____"
]
],
[
[
"Note that the array of differences is shorter by one element (length 6).",
"_____no_output_____"
],
[
"When calling `np.diff` with a multi-dimensional array, an axis argument may be passed to the function to specify which axis to process. When applying `np.diff` to our 2D inflammation array data, which axis would we specify? Take the differences in the appropriate axis and compute a basic summary of the differences with our standard statistics above.",
"_____no_output_____"
],
[
"If the shape of an individual data file is (60, 40) (60 rows and 40 columns), what is the shape of the array after you run the `np.diff` function and why?",
"_____no_output_____"
],
[
"How would you find the largest change in inflammation for each patient? Does it matter if the change in inflammation is an increase or a decrease?",
"_____no_output_____"
],
[
"## Summary of key points\n\nSome of the key takeaways from this activity are the following:\n\n * Import a library into a program using import libraryname.\n\n * Use the numpy library to work with arrays in Python.\n\n * The expression `array.shape` gives the shape of an array.\n\n * Use `array[x, y]` to select a single element from a 2D array.\n\n * Array indices start at 0, not 1.\n\n * Use `low:high` to specify a slice that includes the indices from `low` to `high-1`.\n\n * Use `# some kind of explanation` to add comments to programs.\n\n * Use `np.mean(array)`, `np.std(array)`, `np.quantile(array)`, `np.max(array)`, and `np.min(array)` to calculate simple statistics.\n \n * Use `sp.mode(array)` to compute additional statistics.\n \n * Use `np.mean(array, axis=0)` or `np.mean(array, axis=1)` to calculate statistics across the specified axis.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
4a576a3e8deedefd8cb0b4d94a2a200a3de07126
| 191,772 |
ipynb
|
Jupyter Notebook
|
content/learning_curves/machine_learning/pytorch_tute/MorvanZhou_PYTORCH_cnn.ipynb
|
anuragvvworkspace/anuragvvworkspace.github.io
|
2aa5d6f3ac700ca12150b8cf3a4b9d2f73515c34
|
[
"MIT"
] | null | null | null |
content/learning_curves/machine_learning/pytorch_tute/MorvanZhou_PYTORCH_cnn.ipynb
|
anuragvvworkspace/anuragvvworkspace.github.io
|
2aa5d6f3ac700ca12150b8cf3a4b9d2f73515c34
|
[
"MIT"
] | null | null | null |
content/learning_curves/machine_learning/pytorch_tute/MorvanZhou_PYTORCH_cnn.ipynb
|
anuragvvworkspace/anuragvvworkspace.github.io
|
2aa5d6f3ac700ca12150b8cf3a4b9d2f73515c34
|
[
"MIT"
] | null | null | null | 465.466019 | 25,920 | 0.923154 |
[
[
[
"View more, visit my tutorial page: https://morvanzhou.github.io/tutorials/\nMy Youtube Channel: https://www.youtube.com/user/MorvanZhou\n\nDependencies:\n* torch: 0.1.11\n* torchvision\n* matplotlib",
"_____no_output_____"
]
],
[
[
"import torch\nimport torch.nn as nn\nfrom torch.autograd import Variable\nimport torch.utils.data as Data\nimport torchvision\nimport matplotlib.pyplot as plt\n%matplotlib inline",
"_____no_output_____"
],
[
"torch.manual_seed(1) # reproducible",
"_____no_output_____"
],
[
"# Hyper Parameters\nEPOCH = 1 # train the training data n times, to save time, we just train 1 epoch\nBATCH_SIZE = 50\nLR = 0.001 # learning rate\nDOWNLOAD_MNIST = True # set to False if you have downloaded",
"_____no_output_____"
],
[
"# Mnist digits dataset\ntrain_data = torchvision.datasets.MNIST(\n root='./mnist/',\n train=True, # this is training data\n transform=torchvision.transforms.ToTensor(), # Converts a PIL.Image or numpy.ndarray to\n # torch.FloatTensor of shape (C x H x W) and normalize in the range [0.0, 1.0]\n download=DOWNLOAD_MNIST, # download it if you don't have it\n)",
"_____no_output_____"
],
[
"# plot one example\nprint(train_data.train_data.size()) # (60000, 28, 28)\nprint(train_data.train_labels.size()) # (60000)\nplt.imshow(train_data.train_data[0].numpy(), cmap='gray')\nplt.title('%i' % train_data.train_labels[0])\nplt.show()",
"torch.Size([60000, 28, 28])\ntorch.Size([60000])\n"
],
[
"# Data Loader for easy mini-batch return in training, the image batch shape will be (50, 1, 28, 28)\ntrain_loader = Data.DataLoader(dataset=train_data, batch_size=BATCH_SIZE, shuffle=True)",
"_____no_output_____"
],
[
"# convert test data into Variable, pick 2000 samples to speed up testing\ntest_data = torchvision.datasets.MNIST(root='./mnist/', train=False)\ntest_x = Variable(torch.unsqueeze(test_data.test_data, dim=1)).type(torch.FloatTensor)[:2000]/255. # shape from (2000, 28, 28) to (2000, 1, 28, 28), value in range(0,1)\ntest_y = test_data.test_labels[:2000]",
"_____no_output_____"
],
[
"class CNN(nn.Module):\n def __init__(self):\n super(CNN, self).__init__()\n self.conv1 = nn.Sequential( # input shape (1, 28, 28)\n nn.Conv2d(\n in_channels=1, # input height\n out_channels=16, # n_filters\n kernel_size=5, # filter size\n stride=1, # filter movement/step\n padding=2, # if want same width and length of this image after con2d, padding=(kernel_size-1)/2 if stride=1\n ), # output shape (16, 28, 28)\n nn.ReLU(), # activation\n nn.MaxPool2d(kernel_size=2), # choose max value in 2x2 area, output shape (16, 14, 14)\n )\n self.conv2 = nn.Sequential( # input shape (1, 28, 28)\n nn.Conv2d(16, 32, 5, 1, 2), # output shape (32, 14, 14)\n nn.ReLU(), # activation\n nn.MaxPool2d(2), # output shape (32, 7, 7)\n )\n self.out = nn.Linear(32 * 7 * 7, 10) # fully connected layer, output 10 classes\n\n def forward(self, x):\n x = self.conv1(x)\n x = self.conv2(x)\n x = x.view(x.size(0), -1) # flatten the output of conv2 to (batch_size, 32 * 7 * 7)\n output = self.out(x)\n return output, x # return x for visualization",
"_____no_output_____"
],
[
"cnn = CNN()\nprint(cnn) # net architecture",
"CNN(\n (conv1): Sequential(\n (0): Conv2d(1, 16, kernel_size=(5, 5), stride=(1, 1), padding=(2, 2))\n (1): ReLU()\n (2): MaxPool2d(kernel_size=2, stride=2, padding=0, dilation=1, ceil_mode=False)\n )\n (conv2): Sequential(\n (0): Conv2d(16, 32, kernel_size=(5, 5), stride=(1, 1), padding=(2, 2))\n (1): ReLU()\n (2): MaxPool2d(kernel_size=2, stride=2, padding=0, dilation=1, ceil_mode=False)\n )\n (out): Linear(in_features=1568, out_features=10, bias=True)\n)\n"
],
[
"optimizer = torch.optim.Adam(cnn.parameters(), lr=LR) # optimize all cnn parameters\nloss_func = nn.CrossEntropyLoss() # the target label is not one-hotted",
"_____no_output_____"
],
[
"# following function (plot_with_labels) is for visualization, can be ignored if not interested\nfrom matplotlib import cm\ntry: from sklearn.manifold import TSNE; HAS_SK = True\nexcept: HAS_SK = False; print('Please install sklearn for layer visualization')\ndef plot_with_labels(lowDWeights, labels):\n plt.cla()\n X, Y = lowDWeights[:, 0], lowDWeights[:, 1]\n for x, y, s in zip(X, Y, labels):\n c = cm.rainbow(int(255 * s / 9)); plt.text(x, y, s, backgroundcolor=c, fontsize=9)\n plt.xlim(X.min(), X.max()); plt.ylim(Y.min(), Y.max()); plt.title('Visualize last layer'); plt.show(); plt.pause(0.01)\n\nplt.ion()\n# training and testing\nfor epoch in range(EPOCH):\n for step, (x, y) in enumerate(train_loader): # gives batch data, normalize x when iterate train_loader\n b_x = Variable(x) # batch x\n b_y = Variable(y) # batch y\n\n output = cnn(b_x)[0] # cnn output\n loss = loss_func(output, b_y) # cross entropy loss\n optimizer.zero_grad() # clear gradients for this training step\n loss.backward() # backpropagation, compute gradients\n optimizer.step() # apply gradients\n\n if step % 100 == 0:\n test_output, last_layer = cnn(test_x)\n pred_y = torch.max(test_output, 1)[1].data.squeeze()\n accuracy = (pred_y == test_y).sum().item() / float(test_y.size(0))\n #print('Epoch: ', epoch, '| train loss: %.4f' % loss.data[0], '| test accuracy: %.2f' % accuracy)\n if HAS_SK:\n # Visualization of trained flatten layer (T-SNE)\n tsne = TSNE(perplexity=30, n_components=2, init='pca', n_iter=5000)\n plot_only = 500\n low_dim_embs = tsne.fit_transform(last_layer.data.numpy()[:plot_only, :])\n labels = test_y.numpy()[:plot_only]\n plot_with_labels(low_dim_embs, labels)\nplt.ioff()",
"_____no_output_____"
],
[
"# print 10 predictions from test data\ntest_output, _ = cnn(test_x[:10])\npred_y = torch.max(test_output, 1)[1].data.numpy().squeeze()\nprint(pred_y, 'prediction number')\nprint(test_y[:10].numpy(), 'real number')",
"[7 2 1 0 4 1 4 9 5 9] prediction number\n[7 2 1 0 4 1 4 9 5 9] real number\n"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a57ba7501291f6c1b8cdfbe8860535d39ef06fd
| 14,850 |
ipynb
|
Jupyter Notebook
|
Pytorch basics/linear_regression.ipynb
|
niloysh/machine-learning
|
165b32647d7d2fbaedbea5916c60fa2dfc96e181
|
[
"MIT"
] | null | null | null |
Pytorch basics/linear_regression.ipynb
|
niloysh/machine-learning
|
165b32647d7d2fbaedbea5916c60fa2dfc96e181
|
[
"MIT"
] | null | null | null |
Pytorch basics/linear_regression.ipynb
|
niloysh/machine-learning
|
165b32647d7d2fbaedbea5916c60fa2dfc96e181
|
[
"MIT"
] | null | null | null | 23.571429 | 386 | 0.492458 |
[
[
[
"import torch",
"_____no_output_____"
],
[
"torch.cuda.is_available() # check if GPU is available",
"/home/niloy/anaconda3/envs/tf/lib/python3.8/site-packages/torch/cuda/__init__.py:52: UserWarning: CUDA initialization: Found no NVIDIA driver on your system. Please check that you have an NVIDIA GPU and installed a driver from http://www.nvidia.com/Download/index.aspx (Triggered internally at /opt/conda/conda-bld/pytorch_1603729062494/work/c10/cuda/CUDAFunctions.cpp:100.)\n return torch._C._cuda_getDeviceCount() > 0\n"
]
],
[
[
"# Automatic differentiation using Autograd",
"_____no_output_____"
]
],
[
[
"x = torch.ones(2, 2, requires_grad=True)\nprint(x)",
"tensor([[1., 1.],\n [1., 1.]], requires_grad=True)\n"
],
[
"y = x + 2\nprint(y)",
"tensor([[3., 3.],\n [3., 3.]], grad_fn=<AddBackward0>)\n"
],
[
"z = y * y * 3\nout = z.mean()\nprint(z, out)",
"tensor([[27., 27.],\n [27., 27.]], grad_fn=<MulBackward0>) tensor(27., grad_fn=<MeanBackward0>)\n"
],
[
"a = torch.randn(2, 2)\na = ((a * 3) / (a - 1))\nprint(a.requires_grad)",
"False\n"
],
[
"a.requires_grad_(True)\nprint(a.requires_grad)\nb = (a * a).sum()\nprint(b.grad_fn)",
"True\n<SumBackward0 object at 0x7f04643d4dc0>\n"
],
[
"out.backward()\nprint(x.grad) # d(out) / dx",
"tensor([[4.5000, 4.5000],\n [4.5000, 4.5000]])\n"
],
[
"x = torch.randn(3, requires_grad=True)\ny = x * 2\nwhile y.data.norm() < 1000:\n y = y * 2\nprint(x)\nprint(y)",
"tensor([-0.1443, -2.1702, 0.5571], requires_grad=True)\ntensor([ -73.8692, -1111.1251, 285.2525], grad_fn=<MulBackward0>)\n"
]
],
[
[
"# Linear regression manually",
"_____no_output_____"
]
],
[
[
"import numpy as np",
"_____no_output_____"
]
],
[
[
"Try to learn the function $f(x) = 2x$ ",
"_____no_output_____"
]
],
[
[
"X = np.array([1, 2, 3, 4], dtype=np.float32)\nY = np.array([2, 4, 6, 8], dtype=np.float32)\nw = 0.0",
"_____no_output_____"
],
[
"# model prediction\ndef forward(x):\n return w * x\n\n# MSE loss\ndef loss(y, y_pred):\n return ((y - y_pred) ** 2).mean()\n\n# gradient\ndef gradient(x, y, y_pred):\n return (np.dot(2*x, y_pred - y)).mean()",
"_____no_output_____"
],
[
"print(f'Prediction before training: f(5) = {forward(5):.3f}')",
"Prediction before training: f(5) = 0.000\n"
],
[
"# training\nlearning_rate = 0.01\nn_iters = 20\nfor epoch in range(n_iters):\n y_pred = forward(X)\n l = loss(Y, y_pred)\n dw = gradient(X, Y, y_pred)\n w -= learning_rate * dw\n if epoch % 2 == 0:\n print(f'Epoch {epoch+1}: weights = {w:.3f}, loss = {l:.8f}')\n \nprint(f'Prediction after training: f(5) = {forward(5):.3f}')",
"Epoch 1: weights = 1.200, loss = 30.00000000\nEpoch 3: weights = 1.872, loss = 0.76800019\nEpoch 5: weights = 1.980, loss = 0.01966083\nEpoch 7: weights = 1.997, loss = 0.00050332\nEpoch 9: weights = 1.999, loss = 0.00001288\nEpoch 11: weights = 2.000, loss = 0.00000033\nEpoch 13: weights = 2.000, loss = 0.00000001\nEpoch 15: weights = 2.000, loss = 0.00000000\nEpoch 17: weights = 2.000, loss = 0.00000000\nEpoch 19: weights = 2.000, loss = 0.00000000\nPrediction after training: f(5) = 10.000\n"
]
],
[
[
"# Linear regression using pytorch (only gradient computation)",
"_____no_output_____"
]
],
[
[
"import torch",
"_____no_output_____"
],
[
"X = torch.tensor([1, 2, 3, 4], dtype=torch.float32)\nY = torch.tensor([2, 4, 6, 8], dtype=torch.float32)\nw = torch.tensor(0.0, dtype=torch.float32, requires_grad=True)",
"_____no_output_____"
],
[
"# training\nlearning_rate = 0.01\nn_iters = 50\nfor epoch in range(n_iters):\n y_pred = forward(X)\n l = loss(Y, y_pred)\n l.backward() # compute dl/dw\n \n # update gradients\n with torch.no_grad():\n w -= learning_rate * w.grad\n \n # zero gradients in-place (very imp!)\n # otherwise gradients will accumulate over iterations\n w.grad.zero_()\n \n \n if epoch % 5 == 0:\n print(f'Epoch {epoch+1}: weights = {w:.3f}, loss = {l:.8f}')\n \nprint(f'Prediction after training: f(5) = {forward(5):.3f}')",
"Epoch 1: weights = 0.300, loss = 30.00000000\nEpoch 6: weights = 1.246, loss = 5.90623236\nEpoch 11: weights = 1.665, loss = 1.16278565\nEpoch 16: weights = 1.851, loss = 0.22892261\nEpoch 21: weights = 1.934, loss = 0.04506890\nEpoch 26: weights = 1.971, loss = 0.00887291\nEpoch 31: weights = 1.987, loss = 0.00174685\nEpoch 36: weights = 1.994, loss = 0.00034392\nEpoch 41: weights = 1.997, loss = 0.00006770\nEpoch 46: weights = 1.999, loss = 0.00001333\nPrediction after training: f(5) = 9.997\n"
]
],
[
[
"**Observation**: The autograd function is not as exact as the numerically computed gradient, so it needs more iterations.",
"_____no_output_____"
],
[
"# Linear regression using pytorch",
"_____no_output_____"
]
],
[
[
"import torch\nimport torch.nn as nn",
"_____no_output_____"
],
[
"X = torch.tensor([1, 2, 3, 4], dtype=torch.float32)\nY = torch.tensor([2, 4, 6, 8], dtype=torch.float32)",
"_____no_output_____"
],
[
"# pytorch expects data in format n_samples, n_features\n# therefore we need to reshape the data\nX = X.view(X.shape[0], 1)\nY = Y.view(X.shape[0], 1)\nprint(X)\nprint(Y)",
"tensor([[1.],\n [2.],\n [3.],\n [4.]])\ntensor([[2.],\n [4.],\n [6.],\n [8.]])\n"
],
[
"# define the model\nclass LinearRegression(nn.Module):\n def __init__(self, input_size, output_size):\n super(LinearRegression, self).__init__()\n self.linear = nn.Linear(input_size, output_size)\n \n def forward(self, x):\n return self.linear(x)",
"_____no_output_____"
],
[
"n_samples, n_features = X.shape\ninput_size = n_features\noutput_size = 1",
"_____no_output_____"
],
[
"model = LinearRegression(input_size, output_size)",
"_____no_output_____"
],
[
"learning_rate = 0.01\n\n# define loss\ncriterion = nn.MSELoss()\n\n# define optimizer\n# model.parameters() are the weights\noptimizer = torch.optim.SGD(model.parameters(), lr=learning_rate) ",
"_____no_output_____"
],
[
"# training loop\nn_iters = 100\nfor epoch in range(n_iters):\n y_pred = model(X) # forward pass\n l = criterion(y_pred, Y) # compute loss\n l.backward() # backcward pass\n \n optimizer.step() # update weights\n optimizer.zero_grad()\n \n w, b = model.parameters()\n if epoch % 5 == 0:\n print(f'Epoch {epoch+1}: weights = {w[0][0]:.3f}, loss = {l:.8f}')\n\nX_test = torch.tensor([5], dtype=torch.float32) # test point must also be a tensor\nprint(f'Prediction after training: f(5) = {model(X_test).item():.3f}')",
"Epoch 1: weights = -0.001, loss = 46.26792526\nEpoch 6: weights = 1.112, loss = 7.46706581\nEpoch 11: weights = 1.560, loss = 1.22525179\nEpoch 16: weights = 1.741, loss = 0.22054875\nEpoch 21: weights = 1.815, loss = 0.05825001\nEpoch 26: weights = 1.846, loss = 0.03147167\nEpoch 31: weights = 1.859, loss = 0.02651152\nEpoch 36: weights = 1.866, loss = 0.02508005\nEpoch 41: weights = 1.870, loss = 0.02423492\nEpoch 46: weights = 1.872, loss = 0.02350228\nEpoch 51: weights = 1.874, loss = 0.02280535\nEpoch 56: weights = 1.876, loss = 0.02213126\nEpoch 61: weights = 1.878, loss = 0.02147749\nEpoch 66: weights = 1.880, loss = 0.02084307\nEpoch 71: weights = 1.882, loss = 0.02022736\nEpoch 76: weights = 1.884, loss = 0.01962989\nEpoch 81: weights = 1.885, loss = 0.01905004\nEpoch 86: weights = 1.887, loss = 0.01848734\nEpoch 91: weights = 1.889, loss = 0.01794124\nEpoch 96: weights = 1.891, loss = 0.01741129\nPrediction after training: f(5) = 9.777\n"
],
[
"model(X_test).detach()",
"_____no_output_____"
],
[
"for parameter in model.parameters():\n print(parameter)",
"Parameter containing:\ntensor([[1.8918]], requires_grad=True)\nParameter containing:\ntensor([0.3181], requires_grad=True)\n"
],
[
"print(model)",
"LinearRegression(\n (linear): Linear(in_features=1, out_features=1, bias=True)\n)\n"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a57c362787ab0fe7662417a8e4291e81286ad7d
| 27,877 |
ipynb
|
Jupyter Notebook
|
day4.ipynb
|
saycon1/dw_matrix
|
c5e1480d1c3af29ed6e2001ea06fcae5f8d09f7f
|
[
"MIT"
] | null | null | null |
day4.ipynb
|
saycon1/dw_matrix
|
c5e1480d1c3af29ed6e2001ea06fcae5f8d09f7f
|
[
"MIT"
] | null | null | null |
day4.ipynb
|
saycon1/dw_matrix
|
c5e1480d1c3af29ed6e2001ea06fcae5f8d09f7f
|
[
"MIT"
] | null | null | null | 27,877 | 27,877 | 0.828066 |
[
[
[
"import pandas as pd\nimport numpy as np\nfrom sklearn.tree import DecisionTreeRegressor\nfrom sklearn.metrics import mean_absolute_error\nfrom sklearn.model_selection import cross_val_score\n",
"_____no_output_____"
],
[
"cd \"/content/drive/My Drive/Colab Notebooks/dw_matrix\"",
"/content/drive/My Drive/Colab Notebooks/dw_matrix\n"
],
[
"df = pd.read_csv('data/men_shoes.csv', low_memory=False)",
"_____no_output_____"
],
[
"df.shape",
"_____no_output_____"
],
[
"df.columns",
"_____no_output_____"
],
[
"mean_price = np.mean(df['prices_amountmin'])\nmean_price",
"_____no_output_____"
],
[
"[4] * 5",
"_____no_output_____"
],
[
"y_true = df['prices_amountmin']\ny_pred = [mean_price] * y_true.shape[0]\nmean_absolute_error(y_true, y_pred)",
"_____no_output_____"
],
[
"df['prices_amountmin'].hist(bins=100)",
"_____no_output_____"
],
[
"np.log1p(df['prices_amountmin']).hist(bins=100)",
"_____no_output_____"
],
[
"y_true = df['prices_amountmin']\ny_pred = [np.median(y_true)] * y_true.shape[0]\nmean_absolute_error(y_true, y_pred)",
"_____no_output_____"
],
[
"y_true = df['prices_amountmin']\nprice_log_mean = np.mean(np.log1p(y_true))\ny_pred = [price_log_mean] * y_true.shape[0]\nmean_absolute_error(y_true, y_pred)",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"df['brand_cat'] = df['brand'].factorize()[0]\ndf['manufacturer_cat'] = df['manufacturer'].factorize()[0]\ndf['colors_cat'] = df['colors'].factorize()[0] ",
"_____no_output_____"
],
[
"def run_model(feats):\n X = df[feats].values\n y = df['prices_amountmin'].values\n model = DecisionTreeRegressor(max_depth=5)\n scores = cross_val_score(model, X, y, scoring='neg_mean_absolute_error')\n return np.mean(scores), np.std(scores)",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"run_model(['brand_cat'])",
"_____no_output_____"
],
[
"colors",
"_____no_output_____"
],
[
"run_model(['brand_cat','manufacturer_cat'])",
"_____no_output_____"
],
[
"colors",
"_____no_output_____"
],
[
"run_model(['colors_cat'])\n",
"_____no_output_____"
],
[
"run_model(['brand_cat','colors_cat'])",
"_____no_output_____"
],
[
"!git add day4.ipymb",
"fatal: pathspec 'day4.ipymb' did not match any files\n"
],
[
"ls",
"\u001b[0m\u001b[01;34mdata\u001b[0m/ HelloGithub.ipynb LICENSE \u001b[01;34mmatrix_one\u001b[0m/ \u001b[01;34mmx\u001b[0m/ README.md\n"
],
[
"cd ",
"/root\n"
],
[
"cd \"/content/drive/My Drive/Colab Notebooks/dw_matrix\"",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a57dfa60430e26285dc9da7f5e76137d6ee581b
| 103,076 |
ipynb
|
Jupyter Notebook
|
Seminar1.ipynb
|
AashinShazar/SFSU-PythonSeminar
|
4788cd41590b33f0c79d19d072feaf60c1253d46
|
[
"BSD-3-Clause"
] | null | null | null |
Seminar1.ipynb
|
AashinShazar/SFSU-PythonSeminar
|
4788cd41590b33f0c79d19d072feaf60c1253d46
|
[
"BSD-3-Clause"
] | null | null | null |
Seminar1.ipynb
|
AashinShazar/SFSU-PythonSeminar
|
4788cd41590b33f0c79d19d072feaf60c1253d46
|
[
"BSD-3-Clause"
] | null | null | null | 103,076 | 103,076 | 0.862752 |
[
[
[
"# **Welcome to the Python Workshop! Here is your starter code**\n",
"_____no_output_____"
]
],
[
[
"%matplotlib inline",
"_____no_output_____"
]
],
[
[
"### Try out hello world below! It's easy I promise.",
"_____no_output_____"
]
],
[
[
"print(\"Hello World\")",
"Hello World\n"
]
],
[
[
"## What are we analyzing? \n\nWe're going to be looking at the sales numbers over a 5 year period of batmobiles for Wayne Enterprises. We can see that data below here, press ctrl+enter in the code block to run it!",
"_____no_output_____"
]
],
[
[
"import pandas as pd\n\nsales = [455, 495, 516, 570, 575]\nyears = [1,2,3,4,5]\n\ndf = pd.DataFrame({'Total Sales($)':sales, 'Year':years})\ndf",
"_____no_output_____"
]
],
[
[
"## Let's do some simple math! In the blocks below, try your hand at calculating the mean, variance and standard deviation.\n\nFeel free to comment out the code and see how easy it is to work in python.",
"_____no_output_____"
]
],
[
[
"def calcMean(data):\n totalSum = sum(sales)\n numTotal = len(sales)\n mean = totalSum/numTotal\n return mean\n\ncalcMean(sales)",
"_____no_output_____"
],
[
"def calcVariance(data):\n mean = calcMean(data)\n var = (sum(pow(x-mean,2) for x in sales)) / len(sales) \n return var\n\nfoundVar = calcVariance(sales)\nprint(foundVar)",
"2073.36\n"
],
[
"import math\n\ndef calcStd(varianceData):\n std = math.sqrt(calcVariance(varianceData)) \n return std\n\nprint(calcStd(foundVar))",
"45.53416299878587\n"
]
],
[
[
"## We can make it easier than that! \n\nThe above was the beauty of python, easy to work with and follow along. But we can make the calculations easier than that!\n\nEnter numpy!",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\nnp.mean(sales)",
"_____no_output_____"
],
[
"np.var(sales)",
"_____no_output_____"
],
[
"np.std(sales)",
"_____no_output_____"
]
],
[
[
"## What about linear regression?\n\nRefresher: https://www.statisticshowto.com/probability-and-statistics/regression-analysis/find-a-linear-regression-equation/\n\nLittle bit more dicey right? Yeah it sucks.\n\n\n\n",
"_____no_output_____"
],
[
"## Here's the quick and easy alternative...",
"_____no_output_____"
]
],
[
[
"linregressModel = np.polyfit(years, sales, 1)\nslope = linregressModel[0]\nintercept = linregressModel[1]\nprint(\"slope: %f intercept: %f\" % (linregressModel[0], linregressModel[1]))",
"slope: 31.500000 intercept: 427.700000\n"
],
[
"modelPredictions = np.polyval(linregressModel, years)\nabsError = modelPredictions - sales\n\nSE = np.square(absError) # squared errors\nMSE = np.mean(SE) # mean squared errors\nRMSE = np.sqrt(MSE) # Root Mean Squared Error, RMSE\nRsquared = 1.0 - (np.var(absError) / np.var(sales)) \n\nprint('RMSE:', RMSE)\nprint('R-squared:', Rsquared)",
"RMSE: 9.426558226627588\nR-squared: 0.957142030327584\n"
],
[
"import matplotlib.pyplot as plt\n\nx = np.array(years)\ny = np.array(sales)\n\nplt.plot(x, y, 'o', label='original data')\nplt.plot(x, intercept + slope*x, 'r', label='fitted line')\nplt.xlabel('Years', fontsize=15)\nplt.ylabel('Sales', fontsize=15)\nplt.legend()\nplt.show()",
"_____no_output_____"
]
],
[
[
"## What if we wanted to predict the sales in the future?\n\nWell that's easy! We can just use the linear regression model we have above to predict for year 6 for example. \n\nFeel free to change the yearToPredict!",
"_____no_output_____"
]
],
[
[
"yearToPredict = 6\n\npredict = np.poly1d(linregressModel)\npredictedSale = predict(yearToPredict)\n\nx_new = np.array([yearToPredict])\ny_new = np.array([predictedSale])\n\nprint(predictedSale)",
"616.6999999999999\n"
],
[
"xFull = np.append(x, x_new)\n\nplt.plot(x_new, y_new, 'X', label='new data')\nplt.plot(x, y, 'o', label='original data')\nplt.plot(xFull, intercept + slope*xFull, 'g', label='fitted line')\nplt.xlabel('Years', fontsize=15)\nplt.ylabel('Sales', fontsize=15)\nplt.legend()\nplt.show()",
"_____no_output_____"
]
],
[
[
"## What about a real life scenario?\n\nIn real life, data is often a lot larger than just 5 sets of data. They could be in excel files, databases or you have to go hunting for them. \n\nLet's look at the sample dataset below and see what magic we can do with that with everything we learned above!\n\nBegin by reading in the sampleDataset.csv file :)",
"_____no_output_____"
]
],
[
[
"dfFull = pd.read_csv(\"sampleDataset.csv\") #read in csv file",
"_____no_output_____"
]
],
[
[
"It's always good practice to explore what kind of data we're working with. Sure you can open the file and explore it for yourself but it is often the case that datasets are huge and impractical to manually inspect. \n\nThankfully, we have the pandas library to explore the data for us and give us an idea of what we're working with. ",
"_____no_output_____"
]
],
[
[
"dfFull.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 21613 entries, 0 to 21612\nData columns (total 21 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 id 21613 non-null int64 \n 1 date 21613 non-null object \n 2 price 21613 non-null float64\n 3 bedrooms 21613 non-null int64 \n 4 bathrooms 21613 non-null float64\n 5 sqft_living 21613 non-null int64 \n 6 sqft_lot 21613 non-null int64 \n 7 floors 21613 non-null float64\n 8 waterfront 21613 non-null int64 \n 9 view 21613 non-null int64 \n 10 condition 21613 non-null int64 \n 11 grade 21613 non-null int64 \n 12 sqft_above 21613 non-null int64 \n 13 sqft_basement 21613 non-null int64 \n 14 yr_built 21613 non-null int64 \n 15 yr_renovated 21613 non-null int64 \n 16 zipcode 21613 non-null int64 \n 17 lat 21613 non-null float64\n 18 long 21613 non-null float64\n 19 sqft_living15 21613 non-null int64 \n 20 sqft_lot15 21613 non-null int64 \ndtypes: float64(5), int64(15), object(1)\nmemory usage: 3.5+ MB\n"
]
],
[
[
"Looks like it's some kind of housing dataset...",
"_____no_output_____"
],
[
"## What about common statistics?\n\nWell we can just use the .describe() command and it'll give us this beautiful chart of all the meaningful stats you'd ever need to get into data analysis with just one line of code for the entire dataset!",
"_____no_output_____"
]
],
[
[
"dfFull.describe()",
"_____no_output_____"
]
],
[
[
"Now let's actually look at the data! .head() gives us the first 5 columns but you can easily view more than that by adding a number in the brackets!",
"_____no_output_____"
]
],
[
[
"dfFull.head()",
"_____no_output_____"
]
],
[
[
"## Enter Linear Regression (again)!\n\nNow that we have the data, let's just analyze it like we did earlier with the smaller data! \n\nLet's apply this to analyze the relationship between price and the sqft_living.",
"_____no_output_____"
]
],
[
[
"xData = np.array(dfFull['sqft_living'])\nyData = np.array(dfFull['price'])\n\nlinregressModelFull = np.polyfit(xData, yData, 1)\nslope = linregressModelFull[0]\nintercept = linregressModelFull[1]\nprint(\"slope: %f intercept: %f\" % (linregressModelFull[0], linregressModelFull[1]))",
"slope: 280.623568 intercept: -43580.743094\n"
],
[
"modelPredictions = np.polyval(linregressModelFull, xData)\nabsError = modelPredictions - yData\n\nSE = np.square(absError) # squared errors\nMSE = np.mean(SE) # mean squared errors\nRMSE = np.sqrt(MSE) # Root Mean Squared Error, RMSE\nRsquared = 1.0 - (np.var(absError) / np.var(yData)) \n\nprint('RMSE:', RMSE)\nprint('R-squared:', Rsquared)",
"RMSE: 261440.79030067174\nR-squared: 0.4928532179037932\n"
],
[
"plt.plot(xData, yData, 'o', label='original data')\nplt.plot(xData, intercept + slope*xData, 'r', label='fitted line')\nplt.xlabel('Sqft Living', fontsize=15)\nplt.ylabel('Price', fontsize=15)\nplt.legend()\nplt.show()",
"_____no_output_____"
],
[
"sqftToPredict = 20000\n\npredict = np.poly1d(linregressModelFull)\npredictedVal = predict(sqftToPredict)\n\nx_new = np.array([sqftToPredict])\ny_new = np.array([predictedVal])\n\nprint(predictedVal)",
"5568890.614854492\n"
],
[
"xFull = np.append(xData, x_new)\n\nplt.plot(x_new, y_new, 'X', label='new data')\nplt.plot(xData, yData, 'o', label='original data')\nplt.plot(xFull, intercept + slope*xFull, 'g', label='fitted line')\nplt.xlabel('Sqft Living', fontsize=15)\nplt.ylabel('Price', fontsize=15)\nplt.legend()\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Report Building\n\nNow that you have all this work done, you might want to show it to someone or share it with other people. All you have to do is go to File in the top left corner and click Download as .pdf!",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
4a57f2db53d62777e4379410ced1d44758470125
| 14,813 |
ipynb
|
Jupyter Notebook
|
testWalkerDetection.ipynb
|
davidruffner/cv-people-detector
|
65f6087ab00db204235496c269641720c6344336
|
[
"MIT"
] | null | null | null |
testWalkerDetection.ipynb
|
davidruffner/cv-people-detector
|
65f6087ab00db204235496c269641720c6344336
|
[
"MIT"
] | 1 |
2018-05-30T13:14:12.000Z
|
2018-05-30T13:20:17.000Z
|
testWalkerDetection.ipynb
|
davidruffner/cv-people-detector
|
65f6087ab00db204235496c269641720c6344336
|
[
"MIT"
] | null | null | null | 17.365768 | 151 | 0.343617 |
[
[
[
"import cv2\nimport matplotlib.pyplot as plt\nimport time\nimport cProfile\nimport numpy as np\n",
"_____no_output_____"
]
],
[
[
"# Walker detection with openCV",
"_____no_output_____"
],
[
"## Open video and get video info",
"_____no_output_____"
]
],
[
[
"video_capture = cv2.VideoCapture('resources/TestWalker.mp4')",
"_____no_output_____"
],
[
"# From https://www.learnopencv.com/how-to-find-frame-rate-or-frames-per-second-fps-in-opencv-python-cpp/\n# Find OpenCV version\n(major_ver, minor_ver, subminor_ver) = (cv2.__version__).split('.')\nprint major_ver, minor_ver, subminor_ver\n\n# With webcam get(CV_CAP_PROP_FPS) does not work.\n# Let's see for ourselves.\n\nif int(major_ver) < 3 :\n fps = video_capture.get(cv2.cv.CV_CAP_PROP_FPS)\n print \"Frames per second using video.get(cv2.cv.CV_CAP_PROP_FPS): {0}\".format(fps)\nelse :\n fps = video_capture.get(cv2.CAP_PROP_FPS)\n print \"Frames per second using video.get(cv2.CAP_PROP_FPS) : {0}\".format(fps)\n\n\n# Number of frames to capture\nnum_frames = 120;\n\n\nprint \"Capturing {0} frames\".format(num_frames)\n\n# Start time\nstart = time.time()\n\n# Grab a few frames\nfor i in xrange(0, num_frames) :\n ret, frame = video_capture.read()\n\n\n# End time\nend = time.time()\n\n# Time elapsed\nseconds = end - start\nprint \"Time taken : {0} seconds\".format(seconds)\n\n# Calculate frames per second\nfps = num_frames / seconds;\nprint \"Estimated frames per second : {0}\".format(fps);\n",
"2 4 8\nFrames per second using video.get(cv2.cv.CV_CAP_PROP_FPS): 20.8333333333\nCapturing 120 frames\nTime taken : 13.8059420586 seconds\nEstimated frames per second : 8.69190957712\n"
],
[
"# cProfile.runctx('video_capture.read()', globals(), locals(), 'profile.prof')\n# use snakeviz to read the output of the profiling",
"_____no_output_____"
]
],
[
[
"## Track walker using difference between frames",
"_____no_output_____"
],
[
"Following http://www.codeproject.com/Articles/10248/Motion-Detection-Algorithms",
"_____no_output_____"
]
],
[
[
"def getSmallGrayFrame(video):\n ret, frame = video.read()\n if not ret:\n return ret, frame\n frameSmall = frame[::4, ::-4]\n gray = cv2.cvtColor(frameSmall, cv2.COLOR_BGR2GRAY)\n return ret, gray\n",
"_____no_output_____"
],
[
"#cv2.startWindowThread()\ncount = 0\nfor x in range(200):\n count = count + 1\n print count\n ret1, gray1 = getSmallGrayFrame(video_capture)\n ret2, gray2 = getSmallGrayFrame(video_capture)\n diff = cv2.absdiff(gray1, gray2)\n print np.amax(diff), np.amin(diff)\n print \n diffThresh = cv2.threshold(diff, 15, 255, cv2.THRESH_BINARY)\n \n kernel = np.ones((3,3),np.uint8)\n erosion = cv2.erode(diffThresh[1],kernel,iterations = 1)\n dilation = cv2.dilate(erosion,kernel,iterations = 1)\n \n color1 = cv2.cvtColor(gray1, cv2.COLOR_GRAY2RGB)\n color1[:,:,0:1] = color1[:,:,0:1]\n \n colorDil = cv2.cvtColor(dilation, cv2.COLOR_GRAY2RGB)\n colorDil[:,:,1:2] = colorDil[:,:,1:2]*0\n \n total = cv2.add(color1, colorDil)\n if not ret1 or not ret2:\n break\n cv2.imshow('Video', total)\n cv2.imwrite('resources/frame{}.png'.format(x), total)\n if cv2.waitKey(1) & 0xFF == ord('q'): # Need the cv2.waitKey to update plot\n break",
"1\n193 0\n\n2\n201 0\n\n3\n214 0\n\n4\n212 0\n\n5\n233 0\n\n6\n220 0\n\n7\n219 0\n\n8\n197 0\n\n9\n193 0\n\n10\n195 0\n\n11\n197 0\n\n12\n159 0\n\n13\n197 0\n\n14\n131 0\n\n15\n157 0\n\n16\n96 0\n\n17\n81 0\n\n18\n77 0\n\n19\n59 0\n\n20\n27 0\n\n21\n30 0\n\n22\n28 0\n\n23\n27 0\n\n24\n26 0\n\n25\n27 0\n\n26\n23 0\n\n27\n33 0\n\n28\n24 0\n\n29\n30 0\n\n30\n31 0\n\n31\n30 0\n\n32\n32 0\n\n33\n28 0\n\n34\n27 0\n\n35\n34 0\n\n36\n32 0\n\n37\n30 0\n\n38\n26 0\n\n39\n23 0\n\n40\n65 0\n\n41\n85 0\n\n42\n91 0\n\n43\n110 0\n\n44\n119 0\n\n45\n136 0\n\n46\n134 0\n\n47\n160 0\n\n48\n163 0\n\n49\n183 0\n\n50\n198 0\n\n51\n198 0\n\n52\n183 0\n\n53\n185 0\n\n54\n188 0\n\n55\n223 0\n\n56\n197 0\n\n57\n218 0\n\n58\n242 0\n\n59\n217 0\n\n60\n224 0\n\n61\n203 0\n\n62\n193 0\n\n63\n208 0\n\n64\n196 0\n\n65\n203 0\n\n66\n198 0\n\n67\n188 0\n\n68\n194 0\n\n69\n197 0\n\n70\n197 0\n\n71\n163 0\n\n72\n185 0\n\n73\n168 0\n\n74\n165 0\n\n75\n161 0\n\n76\n160 0\n\n77\n156 0\n\n78\n108 0\n\n79\n112 0\n\n80\n132 0\n\n81\n105 0\n\n82\n113 0\n\n83\n116 0\n\n84\n126 0\n\n85\n143 0\n\n86\n156 0\n\n87\n141 0\n\n88\n151 0\n\n89\n123 0\n\n90\n133 0\n\n91\n157 0\n\n92\n156 0\n\n93\n85 0\n\n94\n117 0\n\n95\n142 0\n\n96\n144 0\n\n97\n105 0\n\n98\n94 0\n\n99\n77 0\n\n100\n93 0\n\n101\n64 0\n\n102\n72 0\n\n103\n84 0\n\n104\n75 0\n\n105\n78 0\n\n106\n81 0\n\n107\n67 0\n\n108\n63 0\n\n109\n64 0\n\n110\n88 0\n\n111\n112 0\n\n112\n78 0\n\n113\n88 0\n\n114\n78 0\n\n115\n91 0\n\n116\n91 0\n\n117\n112 0\n\n118\n102 0\n\n119\n88 0\n\n120\n90 0\n\n121\n112 0\n\n122\n89 0\n\n123\n87 0\n\n124\n93 0\n\n125\n86 0\n\n126\n91 0\n\n127\n100 0\n\n128\n100 0\n\n129\n97 0\n\n130\n98 0\n\n131\n96 0\n\n132\n99 0\n\n133\n108 0\n\n134\n104 0\n\n135\n92 0\n\n136\n109 0\n\n137\n86 0\n\n138\n100 0\n\n139\n89 0\n\n140\n89 0\n\n141\n88 0\n\n142\n87 0\n\n143\n88 0\n\n144\n91 0\n\n145\n51 0\n\n146\n26 0\n\n147\n22 0\n\n148\n28 0\n\n149\n28 0\n\n150\n27 0\n\n151\n25 0\n\n152\n27 0\n\n153\n26 0\n\n154\n31 0\n\n155\n28 0\n\n156\n23 0\n\n157\n27 0\n\n158\n26 0\n\n159\n39 0\n\n160\n30 0\n\n161\n24 0\n\n162\n23 0\n\n163\n25 0\n\n164\n31 0\n\n165\n30 0\n\n166\n26 0\n\n167\n28 0\n\n168\n29 0\n\n169\n25 0\n\n170\n31 0\n\n171\n30 0\n\n172\n24 0\n\n173\n25 0\n\n174\n32 0\n\n175\n29 0\n\n176\n25 0\n\n177\n30 0\n\n178\n23 0\n\n179\n27 0\n\n180\n34 0\n\n181\n24 0\n\n182\n24 0\n\n183\n26 0\n\n184\n22 0\n\n185\n23 0\n\n186\n23 0\n\n187\n34 0\n\n188\n22 0\n\n189\n27 0\n\n190\n28 0\n\n191\n22 0\n\n192\n26 0\n\n193\n24 0\n\n194\n25 0\n\n195\n32 0\n\n196\n28 0\n\n197\n24 0\n\n198\n25 0\n\n199\n31 0\n\n200\n30 0\n\n"
],
[
"# To close the windows: http://stackoverflow.com/questions/6116564/destroywindow-does-not-close-window-on-mac-using-python-and-opencv#15058451\ncv2.waitKey(1000)\n\ncv2.waitKey(1)\ncv2.destroyAllWindows()\ncv2.waitKey(1)",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
]
] |
4a580ae0e0af33e63906c56397da32304981c98d
| 10,579 |
ipynb
|
Jupyter Notebook
|
Video_Lecture_NBs/NB_03_Advanced_Topics.ipynb
|
jonayed-i/OANADA-Forex-algorithms
|
18cc6d0fb0093e968827409f8d277190eeed4e1b
|
[
"MIT"
] | null | null | null |
Video_Lecture_NBs/NB_03_Advanced_Topics.ipynb
|
jonayed-i/OANADA-Forex-algorithms
|
18cc6d0fb0093e968827409f8d277190eeed4e1b
|
[
"MIT"
] | null | null | null |
Video_Lecture_NBs/NB_03_Advanced_Topics.ipynb
|
jonayed-i/OANADA-Forex-algorithms
|
18cc6d0fb0093e968827409f8d277190eeed4e1b
|
[
"MIT"
] | null | null | null | 16.792063 | 93 | 0.485112 |
[
[
[
"# Advanced Topics",
"_____no_output_____"
],
[
"## Helpful DatetimeIndex Attributes and Methods",
"_____no_output_____"
]
],
[
[
"import pandas as pd",
"_____no_output_____"
],
[
"stocks = pd.read_csv(\"stocks.csv\", header = [0,1], index_col= [0], parse_dates= [0])",
"_____no_output_____"
],
[
"stocks.head()",
"_____no_output_____"
],
[
"close = stocks.loc[:, \"Close\"].copy()",
"_____no_output_____"
],
[
"close.head()",
"_____no_output_____"
],
[
"close.info()",
"_____no_output_____"
],
[
"close.index",
"_____no_output_____"
],
[
"close.index.day",
"_____no_output_____"
],
[
"close.index.month",
"_____no_output_____"
],
[
"close.index.year",
"_____no_output_____"
],
[
"close.index.day_name()",
"_____no_output_____"
],
[
"#close.index.weekday_name",
"_____no_output_____"
],
[
"close.index.month_name()",
"_____no_output_____"
],
[
" close.index.weekday",
"_____no_output_____"
],
[
"close.index.quarter",
"_____no_output_____"
],
[
"close.index.days_in_month",
"_____no_output_____"
],
[
"close.index.week",
"_____no_output_____"
],
[
"close.index.weekofyear",
"_____no_output_____"
],
[
"close.index.is_month_end",
"_____no_output_____"
],
[
"close[\"Day\"] = stocks.index.day_name()\nclose[\"Quarter\"] = stocks.index.quarter",
"_____no_output_____"
],
[
"close.head()",
"_____no_output_____"
]
],
[
[
"## Filling NA Values with bfill, ffill and interpolation",
"_____no_output_____"
]
],
[
[
"close.head()",
"_____no_output_____"
],
[
"close.tail()",
"_____no_output_____"
],
[
"all_days = pd.date_range(start = \"2009-12-31\", end = \"2019-02-06\", freq = \"D\")\nall_days",
"_____no_output_____"
],
[
"close = close.reindex(all_days)",
"_____no_output_____"
],
[
"close.head(20)",
"_____no_output_____"
],
[
"close.Day = close.index.day_name()\nclose.Quarter = close.index.quarter",
"_____no_output_____"
],
[
"close.fillna(method = \"ffill\", inplace= True)",
"_____no_output_____"
],
[
"close.head(15)",
"_____no_output_____"
],
[
"temp = pd.read_csv(\"temp.csv\", parse_dates=[\"datetime\"], index_col = \"datetime\")",
"_____no_output_____"
],
[
"temp.head(10)",
"_____no_output_____"
],
[
"temp = temp.resample(\"30 Min\").mean()\ntemp.head(10)",
"_____no_output_____"
],
[
"temp.interpolate()",
"_____no_output_____"
]
],
[
[
"## Timezones and Converting (Part 1)",
"_____no_output_____"
]
],
[
[
"import pandas as pd",
"_____no_output_____"
],
[
"ge = pd.read_csv(\"GE_prices.csv\", parse_dates= [\"date\"], index_col= \"date\")",
"_____no_output_____"
],
[
"ge.head(30)",
"_____no_output_____"
],
[
"ge.info()",
"_____no_output_____"
],
[
"ge.index",
"_____no_output_____"
],
[
"print(ge.index.tz)",
"_____no_output_____"
],
[
"ge.tz_localize(\"UTC\")",
"_____no_output_____"
],
[
"ge.tz_localize(\"America/New_York\")",
"_____no_output_____"
],
[
"ge = ge.tz_localize(\"America/New_York\")",
"_____no_output_____"
],
[
"ge.head()",
"_____no_output_____"
]
],
[
[
"## Timezones and Converting (Part 2)",
"_____no_output_____"
]
],
[
[
"ge.index.tz",
"_____no_output_____"
],
[
"ge.tz_convert(\"UTC\")",
"_____no_output_____"
],
[
"ge.tz_convert(\"America/Los_Angeles\")",
"_____no_output_____"
],
[
"ge_la = ge.tz_convert(\"America/Los_Angeles\")",
"_____no_output_____"
],
[
"ge_la.head()",
"_____no_output_____"
],
[
"ge.head()",
"_____no_output_____"
],
[
"comb = pd.concat([ge, ge_la], axis = 1)",
"_____no_output_____"
],
[
"comb.head()",
"_____no_output_____"
],
[
"comb.index",
"_____no_output_____"
],
[
"comb[\"NY_time\"] = comb.index.tz_convert(\"America/New_York\")\ncomb[\"LA_time\"] = comb.index.tz_convert(\"America/Los_Angeles\")",
"_____no_output_____"
],
[
"comb.head()",
"_____no_output_____"
],
[
"import pytz",
"_____no_output_____"
],
[
"len(pytz.all_timezones)",
"_____no_output_____"
],
[
"pytz.common_timezones",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a58124b2273395cb466f04353508990db4e53d4
| 597,788 |
ipynb
|
Jupyter Notebook
|
Face_Detection/face_detection.ipynb
|
yinguobing/Playground
|
8637c0da3d46e799afd29a3bdffce719a1fdc430
|
[
"Apache-2.0"
] | 3 |
2021-03-14T14:03:15.000Z
|
2022-01-19T03:33:18.000Z
|
Face_Detection/face_detection.ipynb
|
yinguobing/Playground
|
8637c0da3d46e799afd29a3bdffce719a1fdc430
|
[
"Apache-2.0"
] | 1 |
2021-11-30T12:51:15.000Z
|
2021-12-01T09:29:16.000Z
|
Face_Detection/face_detection.ipynb
|
yinguobing/Playground
|
8637c0da3d46e799afd29a3bdffce719a1fdc430
|
[
"Apache-2.0"
] | 2 |
2021-03-14T14:03:18.000Z
|
2022-01-19T03:33:25.000Z
| 1,688.666667 | 295,362 | 0.957744 |
[
[
[
"# 人脸检测",
"_____no_output_____"
],
[
"人脸检测,顾名思义,从图像中找到人脸。这是计算机视觉中一个非常经典的物体检测问题。经典人脸检测算法如Viola-Jones算法已经内置在OpenCV中,一度是使用OpenCV实现人脸检测的默认方案。不过OpenCV最新发布的4.5.4版本中提供了一个全新的基于神经网络的人脸检测器。这篇笔记展示了该检测器的使用方法。",
"_____no_output_____"
],
[
"## 准备工作\n\n首先载入必要的包,并检查OpenCV版本。\n\n如果你还没有安装OpenCV,可以通过如下命令安装:\n```bash \npip install opencv-python\n```",
"_____no_output_____"
]
],
[
[
"import cv2\nfrom PIL import Image\n\nprint(f\"你需要OpenCV 4.5.4或者更高版本。当前版本为:{cv2.__version__}\")\n",
"你需要OpenCV 4.5.4或者更高版本。当前版本为:4.5.4\n"
]
],
[
[
"请从下方地址下载模型文件,并将模型文件放置在当前目录下。\n\n模型下载地址:https://github.com/ShiqiYu/libfacedetection.train/tree/master/tasks/task1/onnx\n\n当前目录为:",
"_____no_output_____"
]
],
[
[
"\n!pwd\n",
"/Users/Robin/Developer/Playground/Face_Detection\n"
]
],
[
[
"## 构建检测器",
"_____no_output_____"
],
[
"检测器的构建函数为`FaceDetectorYN_create`,必选参数有三个:\n- `model` ONNX模型路径\n- `config` 配置(使用ONNX时为可选项)\n- `input_size` 输入图像的尺寸。如果构建时输入尺寸未知,可以在执行前指定。",
"_____no_output_____"
]
],
[
[
"face_detector = cv2.FaceDetectorYN_create(\"yunet.onnx\", \"\", (0, 0))\nprint(\"检测器构建完成。\")\n",
"检测器构建完成。\n"
]
],
[
[
"## 执行检测\n\n一旦检测器构建完成,便可以使用`detect`方法检测人脸。注意,如果在构建时未指定输入大小,可以在调用前通过`setInputSzie`方法指定。",
"_____no_output_____"
]
],
[
[
"# 读入待检测图像。图像作者:@anyataylorjoy on Instagram\nimage = cv2.imread(\"queen.jpg\")\n\n# 获取图像大小并设定检测器\nheight, width, _ = image.shape\nface_detector.setInputSize((width, height))\n\n# 执行检测\nresult, faces = face_detector.detect(image)\n\nprint(\"检测完成。\")\n",
"检测完成。\n"
]
],
[
[
"## 绘制检测结果\n\n首先将检测结果打印出来。",
"_____no_output_____"
]
],
[
[
"print(faces)\n",
"[[136.21078 77.48543 118.0509 143.32883 173.94562 147.02783\n 222.2979 139.54152 205.95425 172.52359 185.9506 192.57774\n 226.84074 186.21704 0.9998491]]\n"
]
],
[
[
"检测结果为一个嵌套列表。最外层代表了检测结果的数量,即检测到几个人脸。每一个检测结果包含15个数。其含义如下。\n\n| 序号 | 含义 |\n| --- | --- |\n| 0 | 人脸框坐标x | \n| 1 | 人脸框坐标y |\n| 2 | 人脸框的宽度 |\n| 3 | 人脸框的高度 |\n| 4 | 左眼瞳孔坐标x |\n| 5 | 左眼瞳孔坐标y |\n| 6 | 右眼瞳孔坐标x |\n| 7 | 右眼瞳孔坐标y |\n| 8 | 鼻尖坐标x |\n| 9 | 鼻尖坐标y |\n| 10 | 左侧嘴角坐标x |\n| 11 | 左侧嘴角坐标y |\n| 12 | 右侧嘴角坐标x |\n| 13 | 右侧嘴角坐标y |\n| 14 | 人脸置信度分值 |\n\n接下来依次在图中绘制这些坐标。",
"_____no_output_____"
],
[
"### 绘制人脸框\n\nOpenCV提供了`rectangle`与`cirle`函数用于在图像中绘制方框与圆点。首先使用`rectangle`绘制人脸框。",
"_____no_output_____"
]
],
[
[
"# 提取第一个检测结果,并将坐标转换为整数,用于绘制。\nface = faces[0].astype(int)\n\n# 获得人脸框的位置与宽高。\nx, y, w, h = face[:4]\n\n# 在图像中绘制结果。\nimage_with_marks = cv2.rectangle(image, (x, y), (x+w, y+h), (255, 255, 255))\n\n# 显示绘制结果\ndisplay(Image.fromarray(cv2.cvtColor(image_with_marks, cv2.COLOR_BGR2RGB)))\n",
"_____no_output_____"
]
],
[
[
"# 绘制五官坐标",
"_____no_output_____"
]
],
[
[
"# 绘制瞳孔位置\nleft_eye_x, left_eye_y, right_eye_x, right_eye_y = face[4:8]\ncv2.circle(image_with_marks, (left_eye_x, left_eye_y), 2, (0, 255, 0), -1)\ncv2.circle(image_with_marks, (right_eye_x, right_eye_y), 2, (0, 255, 0), -1)\n\n# 绘制鼻尖\nnose_x, nose_y = face[8:10]\ncv2.circle(image_with_marks, (nose_x, nose_y), 2, (0, 255, 0), -1)\n\n# 绘制嘴角\nmouth_left_x, mouth_left_y, mouth_right_x, mouth_right_y = face[10:14]\ncv2.circle(image_with_marks, (mouth_left_x, mouth_left_y), 2, (0, 255, 0), -1)\ncv2.circle(image_with_marks, (mouth_right_x, mouth_right_y), 2, (0, 255, 0), -1)\n\n# 显示绘制结果\ndisplay(Image.fromarray(cv2.cvtColor(image_with_marks, cv2.COLOR_BGR2RGB)))",
"_____no_output_____"
]
],
[
[
"## 性能测试\n\n人脸检测器很有可能用在一些实时运算场景。此时的性能便是一个不可忽略的因素。下边这段代码展示了新版人脸检测器在当前计算设备上的运行速度。",
"_____no_output_____"
]
],
[
[
"tm = cv2.TickMeter()\nfor _ in range(1000):\n tm.start()\n _ = face_detector.detect(image)\n tm.stop()\nprint(f\"检测速度:{tm.getFPS():.0f} FPS\")",
"检测速度:61 FPS\n"
]
],
[
[
"## 总结\n\nOpenCV 4.5.4提供的人脸检测器采用了基于神经网络的方案。与先前基于Viola-Jones算法的方案相比还可以提供面部五官的位置。可以考虑作为默认检测方案使用。",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
4a582aab61c54a03e704d0540200396d9ab5324f
| 407,262 |
ipynb
|
Jupyter Notebook
|
nbs_gil/severstal/Severstal Competition 10.ipynb
|
gilfernandes/fastai-exercises
|
d506db42c62785044cc41c5ae76ea62e53c60ec0
|
[
"Apache-2.0"
] | null | null | null |
nbs_gil/severstal/Severstal Competition 10.ipynb
|
gilfernandes/fastai-exercises
|
d506db42c62785044cc41c5ae76ea62e53c60ec0
|
[
"Apache-2.0"
] | 3 |
2020-03-09T14:00:13.000Z
|
2022-02-26T05:29:47.000Z
|
nbs_gil/severstal/Severstal Competition 10.ipynb
|
gilfernandes/fastai-exercises
|
d506db42c62785044cc41c5ae76ea62e53c60ec0
|
[
"Apache-2.0"
] | null | null | null | 219.311793 | 300,076 | 0.898102 |
[
[
[
"# This Python 3 environment comes with many helpful analytics libraries installed\n# It is defined by the kaggle/python docker image: https://github.com/kaggle/docker-python\n# For example, here's several helpful packages to load in \n\nimport numpy as np # linear algebra\nimport pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)\nfrom fastai.vision import *\nfrom fastai import *\nimport os\nfrom collections import defaultdict",
"_____no_output_____"
]
],
[
[
"### Set up paths",
"_____no_output_____"
]
],
[
[
"train_pd = pd.read_csv('/root/.fastai/data/severstal/train.csv')",
"_____no_output_____"
],
[
"train_pd.head(5)",
"_____no_output_____"
],
[
"path = Path('/root/.fastai/data/severstal')",
"_____no_output_____"
],
[
"path.ls()",
"_____no_output_____"
],
[
"train_images = get_image_files(path/'train_images')\ntrain_images[:3]",
"_____no_output_____"
]
],
[
[
"### Check maximum size of images",
"_____no_output_____"
]
],
[
[
"def check_img_max_size(folder):\n max_height = 0\n max_width = 0\n for train_image in train_images:\n img = open_image(train_image)\n if max_height < img.shape[1]:\n max_height = img.shape[1]\n if max_width < img.shape[2]:\n max_width = img.shape[2]\n return max_height, max_width\n\ndef show_image(images, index):\n img_f = images[index]\n print(type(img_f))\n img = open_image(img_f)\n print(img)\n img.show(figsize=(5,5))",
"_____no_output_____"
],
[
"mask_path = Path('/kaggle/mask')\nif not os.path.exists(mask_path):\n os.makedirs(str(mask_path))",
"_____no_output_____"
],
[
"def convert_encoded_to_array(encoded_pixels):\n pos_array = []\n len_array = []\n splits = encoded_pixels.split()\n pos_array = [int(n) - 1 for i, n in enumerate(splits) if i % 2 == 0]\n len_array = [int(n) for i, n in enumerate(splits) if i % 2 == 1]\n return pos_array, len_array\n \ndef convert_to_pair(pos_array, rows):\n return [(p % rows, p // rows) for p in pos_array]\n\ndef create_positions(single_pos, size):\n return [i for i in range(single_pos, single_pos + size)]\n\ndef create_positions_pairs(single_pos, size, row_size):\n return convert_to_pair(create_positions(single_pos, size), row_size)\n\ndef convert_to_mask(encoded_pixels, row_size, col_size, category):\n pos_array, len_array = convert_encoded_to_array(encoded_pixels)\n mask = np.zeros([row_size, col_size])\n for(p, l) in zip(pos_array, len_array):\n for row, col in create_positions_pairs(p, l, row_size):\n mask[row][col] = category\n return mask\n\ndef save_to_image(masked, image_name):\n im = PIL.Image.fromarray(masked)\n im = im.convert(\"L\")\n image_name = re.sub(r'(.+)\\.jpg', r'\\1', image_name) + \".png\"\n real_path = mask_path/image_name\n im.save(real_path)\n return real_path\n\ndef open_single_image(path):\n img = open_image(path)\n img.show(figsize=(20,20))\n \ndef get_y_fn(x):\n return mask_path/(x.stem + '.png')\n\ndef group_by(train_images, train_pd):\n tran_dict = {image.name:[] for image in train_images}\n pattern = re.compile('(.+)_(\\d+)')\n for index, image_path in train_pd.iterrows():\n m = pattern.match(image_path['ImageId_ClassId'])\n file_name = m.group(1)\n category = m.group(2)\n tran_dict[file_name].append((int(category), image_path['EncodedPixels']))\n return tran_dict\n\ndef display_image_with_mask(img_name):\n full_image = path/'train_images'/img_name\n print(full_image)\n open_single_image(full_image)\n mask_image = get_y_fn(full_image)\n mask = open_mask(mask_image)\n print(full_image)\n mask.show(figsize=(20, 20), alpha=0.5)",
"_____no_output_____"
],
[
"grouped_categories_mask = group_by(train_images, train_pd)",
"_____no_output_____"
]
],
[
[
"### Create mask files and save these to kaggle/mask/",
"_____no_output_____"
]
],
[
[
"image_height = 256\nimage_width = 1600\nfor image_name, cat_list in grouped_categories_mask.items():\n masked = np.zeros([image_height, image_width])\n for cat_mask in cat_list:\n encoded_pixels = cat_mask[1]\n if pd.notna(cat_mask[1]):\n masked += convert_to_mask(encoded_pixels, image_height, image_width, cat_mask[0])\n if np.amax(masked) > 4:\n print(f'Check {image_name} for max category {np.amax(masked)}')\n save_to_image(masked, image_name)",
"_____no_output_____"
]
],
[
[
"### Prepare Transforms",
"_____no_output_____"
]
],
[
[
"def limited_dihedral_affine(k:partial(uniform_int,0,3)):\n \"Randomly flip `x` image based on `k`.\"\n x = -1 if k&1 else 1\n y = -1 if k&2 else 1\n if k&4: return [[0, x, 0.],\n [y, 0, 0],\n [0, 0, 1.]]\n return [[x, 0, 0.],\n [0, y, 0],\n [0, 0, 1.]]\n\ndihedral_affine = TfmAffine(limited_dihedral_affine)\n\ndef get_extra_transforms(max_rotate:float=3., max_zoom:float=1.1,\n max_lighting:float=0.2, max_warp:float=0.2, p_affine:float=0.75,\n p_lighting:float=0.75, xtra_tfms:Optional[Collection[Transform]]=None)->Collection[Transform]:\n \"Utility func to easily create a list of flip, rotate, `zoom`, warp, lighting transforms.\"\n p_lightings = [p_lighting, p_lighting + 0.2, p_lighting + 0.4, p_lighting + 0.6, p_lighting + 0.7]\n max_lightings = [max_lighting, max_lighting + 0.2, max_lighting + 0.4, max_lighting + 0.6, max_lighting + 0.7]\n res = [rand_crop(), dihedral_affine(), \n symmetric_warp(magnitude=(-max_warp,max_warp), p=p_affine),\n rotate(degrees=(-max_rotate,max_rotate), p=p_affine),\n rand_zoom(scale=(1., max_zoom), p=p_affine)]\n res.extend([brightness(change=(0.5*(1-mp[0]), 0.5*(1+mp[0])), p=mp[1]) for mp in zip(max_lightings, p_lightings)])\n res.extend([contrast(scale=(1-mp[0], 1/(1-mp[0])), p=mp[1]) for mp in zip(max_lightings, p_lightings)])\n # train , valid\n return (res, [crop_pad()])",
"_____no_output_____"
]
],
[
[
"### Prepare data bunch",
"_____no_output_____"
]
],
[
[
"train_images = (path/'train_images').ls()\nsrc_size = np.array(open_image(str(train_images[0])).shape[1:])\nvalid_pct = 0.10",
"_____no_output_____"
],
[
"codes = array(['0', '1', '2', '3', '4'])",
"_____no_output_____"
],
[
"src = (SegmentationItemList.from_folder(path/'train_images')\n .split_by_rand_pct(valid_pct=valid_pct)\n .label_from_func(get_y_fn, classes=codes))",
"_____no_output_____"
],
[
"bs = 4\nsize = src_size//2",
"_____no_output_____"
],
[
"data = (src.transform(get_extra_transforms(), size=size, tfm_y=True)\n .add_test(ImageList.from_folder(path/'test_images'), tfms=None, tfm_y=False)\n .databunch(bs=bs)\n .normalize(imagenet_stats))",
"_____no_output_____"
]
],
[
[
"### Create learner and training\nStarting with low resolution training",
"_____no_output_____"
],
[
"##### Some metrics functions",
"_____no_output_____"
]
],
[
[
"name2id = {v:k for k,v in enumerate(codes)}\nvoid_code = name2id['0']\n\ndef acc_camvid(input, target):\n target = target.squeeze(1)\n mask = target != void_code\n argmax = (input.argmax(dim=1))\n comparison = argmax[mask]==target[mask]\n return torch.tensor(0.) if comparison.numel() == 0 else comparison.float().mean()\n\ndef acc_camvid_with_zero_check(input, target):\n target = target.squeeze(1)\n argmax = (input.argmax(dim=1))\n batch_size = input.shape[0]\n total = torch.empty([batch_size])\n for b in range(batch_size):\n if(torch.sum(argmax[b]).item() == 0.0 and torch.sum(target[b]).item() == 0.0):\n total[b] = 1\n else:\n mask = target[b] != void_code\n comparison = argmax[b][mask]==target[b][mask]\n total[b] = torch.tensor(0.) if comparison.numel() == 0 else comparison.float().mean()\n return total.mean()\n\n\ndef calc_dice_coefficients(argmax, target, cats):\n def calc_dice_coefficient(seg, gt, cat: int):\n mask_seg = seg == cat\n mask_gt = gt == cat\n sum_seg = torch.sum(mask_seg.float())\n sum_gt = torch.sum(mask_gt.float())\n if sum_seg + sum_gt == 0:\n return torch.tensor(1.0)\n return (torch.sum((seg[gt == cat] / cat).float()) * 2.0) / (sum_seg + sum_gt)\n\n total_avg = torch.empty([len(cats)])\n for i, c in enumerate(cats):\n total_avg[i] = calc_dice_coefficient(argmax, target, c)\n return total_avg.mean()\n\n\ndef dice_coefficient(input, target):\n target = target.squeeze(1)\n argmax = (input.argmax(dim=1))\n batch_size = input.shape[0]\n cats = [1, 2, 3, 4]\n total = torch.empty([batch_size])\n for b in range(batch_size):\n total[b] = calc_dice_coefficients(argmax[b], target[b], cats)\n return total.mean()\n\ndef calc_dice_coefficients_2(argmax, target, cats):\n def calc_dice_coefficient(seg, gt, cat: int):\n mask_seg = seg == cat\n mask_gt = gt == cat\n sum_seg = torch.sum(mask_seg.float())\n sum_gt = torch.sum(mask_gt.float())\n return (torch.sum((seg[gt == cat] / cat).float())), (sum_seg + sum_gt)\n\n total_avg = torch.empty([len(cats), 2])\n for i, c in enumerate(cats):\n total_avg[i][0], total_avg[i][1] = calc_dice_coefficient(argmax, target, c)\n total_sum = total_avg.sum(axis=0)\n if (total_sum[1] == 0.0):\n return torch.tensor(1.0)\n return total_sum[0] * 2.0 / total_sum[1]\n\n\ndef dice_coefficient_2(input, target):\n target = target.squeeze(1)\n argmax = (input.argmax(dim=1))\n batch_size = input.shape[0]\n cats = [1, 2, 3, 4]\n total = torch.empty([batch_size])\n for b in range(batch_size):\n total[b] = calc_dice_coefficients_2(argmax[b], target[b], cats)\n return total.mean()\n\n\ndef accuracy_simple(input, target):\n target = target.squeeze(1)\n return (input.argmax(dim=1)==target).float().mean()\n\n\ndef dice_coeff(pred, target):\n smooth = 1.\n num = pred.size(0)\n m1 = pred.view(num, -1) # Flatten\n m2 = target.view(num, -1) # Flatten\n intersection = (m1 * m2).sum()\n return (2. * intersection + smooth) / (m1.sum() + m2.sum() + smooth)",
"_____no_output_____"
]
],
[
[
"##### The main training function",
"_____no_output_____"
]
],
[
[
"from fastai import callbacks\n\ndef train_learner(learn, slice_lr, epochs=10, pct_start=0.8, best_model_name='best_model', \n patience_early_stop=4, patience_reduce_lr = 3):\n learn.fit_one_cycle(epochs, slice_lr, pct_start=pct_start, \n callbacks=[callbacks.SaveModelCallback(learn, monitor='dice_coefficient',mode='max', name=best_model_name),\n callbacks.EarlyStoppingCallback(learn=learn, monitor='dice_coefficient', patience=patience_early_stop),\n callbacks.ReduceLROnPlateauCallback(learn=learn, monitor='dice_coefficient', patience=patience_reduce_lr),\n callbacks.TerminateOnNaNCallback()])",
"_____no_output_____"
],
[
"metrics=accuracy_simple, acc_camvid_with_zero_check, dice_coefficient, dice_coefficient_2\nwd=1e-2",
"_____no_output_____"
],
[
"learn = unet_learner(data, models.resnet34, metrics=metrics, wd=wd, bottle=True)\nlearn.loss_func = CrossEntropyFlat(axis=1, weight=torch.tensor([1.5, .5, .5, .5, .5]).cuda())\nlearn.loss_func",
"_____no_output_____"
],
[
"learn.model_dir = Path('/kaggle/model')",
"_____no_output_____"
],
[
"learn = to_fp16(learn, loss_scale=4.0)",
"_____no_output_____"
],
[
"lr_find(learn, num_it=300)\nlearn.recorder.plot()",
"_____no_output_____"
],
[
"lr=1e-04",
"_____no_output_____"
],
[
"train_learner(learn, slice(lr), epochs=12, pct_start=0.8, best_model_name='bestmodel-frozen-1', \n patience_early_stop=4, patience_reduce_lr = 3)",
"_____no_output_____"
],
[
"learn.save('stage-1')",
"_____no_output_____"
],
[
"learn.load('bestmodel');",
"_____no_output_____"
],
[
"learn.export(file='/kaggle/model/export-1.pkl')",
"_____no_output_____"
],
[
"learn.unfreeze()",
"_____no_output_____"
],
[
"lrs = slice(lr/100,lr)",
"_____no_output_____"
],
[
"train_learner(learn, lrs, epochs=10, pct_start=0.8, best_model_name='bestmodel-unfrozen-1', \n patience_early_stop=4, patience_reduce_lr = 3)",
"_____no_output_____"
],
[
"learn.save('stage-2');",
"_____no_output_____"
],
[
"learn.load('bestmodel-unfrozen-1-mini');",
"_____no_output_____"
],
[
"learn.save('stage-2');",
"_____no_output_____"
],
[
"learn.export(file='/kaggle/model/export-2.pkl')",
"_____no_output_____"
]
],
[
[
"### Go Large",
"_____no_output_____"
]
],
[
[
"src = (SegmentationItemList.from_folder(path/'train_images')\n .split_by_rand_pct(valid_pct=valid_pct)\n .label_from_func(get_y_fn, classes=codes))",
"_____no_output_____"
],
[
"data = (src.transform(get_extra_transforms(), size=src_size, tfm_y=True)\n .add_test(ImageList.from_folder(path/'test_images'), tfms=None, tfm_y=False)\n .databunch(bs=bs)\n .normalize(imagenet_stats))",
"_____no_output_____"
],
[
"learn = unet_learner(data, models.resnet34, metrics=metrics, wd=wd, bottle=True)\nlearn.model_dir = Path('/kaggle/model')\nlearn.loss_func = CrossEntropyFlat(axis=1, weight=torch.tensor([2.0, .5, .5, .5, .5]).cuda())\nlearn = to_fp16(learn, loss_scale=4.0)\nlearn.load('stage-2');",
"_____no_output_____"
],
[
"lr_find(learn, num_it=400)\nlearn.recorder.plot()",
"_____no_output_____"
],
[
"lr=1e-05",
"_____no_output_____"
],
[
"train_learner(learn, slice(lr), epochs=10, pct_start=0.8, best_model_name='bestmodel-frozen-3', \n patience_early_stop=4, patience_reduce_lr = 3)",
"_____no_output_____"
],
[
"learn.save('stage-3');",
"_____no_output_____"
],
[
"learn.load('bestmodel-3');",
"_____no_output_____"
],
[
"learn.export(file='/kaggle/model/export-3.pkl')",
"_____no_output_____"
],
[
"learn.unfreeze()",
"_____no_output_____"
],
[
"lrs = slice(lr/1000,lr/10)",
"_____no_output_____"
],
[
"train_learner(learn, lrs, epochs=10, pct_start=0.8, best_model_name='bestmodel-4', \n patience_early_stop=4, patience_reduce_lr = 3)",
"_____no_output_____"
],
[
"learn.save('stage-4');",
"_____no_output_____"
],
[
"learn.load('bestmodel-4');",
"_____no_output_____"
],
[
"learn.export(file='/kaggle/model/export-4.pkl')",
"_____no_output_____"
],
[
"!pwd\n!cp /kaggle/model/export.pkl /opt/fastai/fastai-exercises/nbs_gil\nfrom IPython.display import FileLink\nFileLink(r'export-4.pkl')",
"/opt/fastai/fastai-exercises/nbs_gil\r\n"
]
],
[
[
"### Inference",
"_____no_output_____"
]
],
[
[
"learn=None\ngc.collect()",
"_____no_output_____"
],
[
"test_images = (path/'test_images').ls()",
"_____no_output_____"
],
[
"!mv /kaggle/model/export-4.pkl /kaggle/model/export.pkl ",
"_____no_output_____"
],
[
"inference_learn = load_learner('/kaggle/model/')",
"_____no_output_____"
],
[
"def predict(img_path):\n pred_class, pred_idx, outputs = inference_learn.predict(open_image(str(img_path)))\n return pred_class, pred_idx, outputs\n\ndef encode_classes(pred_class_data):\n pixels = np.concatenate([[0], torch.transpose(pred_class_data.squeeze(), 0, 1).flatten(), [0]])\n classes_dict = {1: [], 2: [], 3: [], 4: []}\n count = 0\n previous = pixels[0]\n for i, val in enumerate(pixels):\n if val != previous:\n if previous in classes_dict:\n classes_dict[previous].append((i - count, count))\n count = 0\n previous = val\n count += 1\n return classes_dict\n\n\ndef convert_classes_to_text(classes_dict, clazz):\n return ' '.join([f'{v[0]} {v[1]}' for v in classes_dict[clazz]])",
"_____no_output_____"
],
[
"image_to_predict = train_images[16].name\ndisplay_image_with_mask(image_to_predict)\npred_class, pred_idx, outputs = predict(path/f'train_images/{image_to_predict}')\npred_class",
"/root/.fastai/data/severstal/train_images/f21b0be46.jpg\n/root/.fastai/data/severstal/train_images/f21b0be46.jpg\n"
],
[
"torch.transpose(pred_class.data.squeeze(), 0, 1).shape",
"_____no_output_____"
]
],
[
[
"#### Checking encoding methods",
"_____no_output_____"
]
],
[
[
"encoded_all = encode_classes(pred_class.data)\nprint(convert_classes_to_text(encoded_all, 3))",
"221319 18 221543 2 221571 22 221747 2 221751 2 221754 4 221759 15 221775 3 221779 7 221787 69 221905 3 221910 1 221912 1 221971 2 221974 4 221979 1 221981 1 221983 1 221985 7 221993 132 222129 9 222139 2 222143 3 222147 3 222161 12 222222 215 222477 218 222696 1 222728 226 222984 226 223240 227 223479 2 223496 1 223498 225 223757 16 223774 208 223985 9 224016 1 224020 7 224028 1 224032 207 224240 11 224279 6 224289 204 224496 11 224538 1 224540 1 224546 192 224739 8 224748 1 224753 10 224791 212 225005 1 225011 1 225013 4 225048 1 225051 197 225303 200 225563 172 225736 19 225815 182 226001 1 226004 4 226071 22 226094 2 226104 1 226110 133 226245 2 226249 3 226326 177 226505 2 226582 177 226761 2 226817 2 226823 192 227017 2 227073 2 227077 1 227079 192 227329 202 227585 204 227841 2 227844 203 228049 2 228097 2 228100 208 228353 2 228357 210 228569 1 228609 2 228613 214 228865 1 228869 4 228875 222 229098 1 229121 1 229139 217 229395 216 229653 214 229910 1 229913 207 230217 11 230233 5 230239 64 230305 61 230501 52 230601 3 230605 1 230761 6 230773 24 230798 5 231033 8 296189 2 296446 1 296700 5 296893 6 296903 2 296949 12 297143 22 297181 36 297396 77 297645 1 297649 80 297893 1 297899 1 297901 84 298145 8 298155 86 298403 6 298411 86 298658 3 298662 1 298669 84 298914 5 298929 80 299209 56 299474 1 299476 3 299499 1 299501 18 299520 1 299764 5 299770 1 367016 1 367270 1 367526 1\n"
],
[
"image_name = train_images[16]\nprint(get_y_fn(image_name))\nimg = open_mask(get_y_fn(image_name))\nimg_data = img.data\nprint(convert_classes_to_text(encode_classes(img_data), 3))\nimg_data.shape",
"_____no_output_____"
]
],
[
[
"### Loop through the test images and create submission csv",
"_____no_output_____"
]
],
[
[
"import time\nstart_time = time.time()\n\ndefect_classes = [1, 2, 3, 4]\nwith open('submission.csv', 'w') as submission_file:\n submission_file.write('ImageId_ClassId,EncodedPixels\\n')\n for i, test_image in enumerate(test_images):\n pred_class, pred_idx, outputs = predict(test_image)\n encoded_all = encode_classes(pred_class.data)\n for defect_class in defect_classes:\n submission_file.write(f'{test_image.name}_{defect_class},{convert_classes_to_text(encoded_all, defect_class)}\\n')\n if i % 5 == 0:\n print(f'Processed {i} images\\r', end='')\n \nprint(f\"--- {time.time() - start_time} seconds ---\")",
"_____no_output_____"
]
],
[
[
"### Alternative prediction methods",
"_____no_output_____"
]
],
[
[
"preds,y = learn.get_preds(ds_type=DatasetType.Test, with_loss=False)",
"_____no_output_____"
],
[
"preds.shape",
"_____no_output_____"
],
[
"pred_class_data = preds.argmax(dim=1)",
"_____no_output_____"
],
[
"len((path/'test_images').ls())",
"_____no_output_____"
],
[
"data.test_ds.x",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
4a582df6af897f0359082aff1e44416a8ba0feae
| 68,718 |
ipynb
|
Jupyter Notebook
|
tutorials/02-intermediate/Training_a_Classifier.ipynb
|
awesome-archive/LIS-YNP
|
0cb434e209cee1aa83a7f73ddc7c81b6983ab7d7
|
[
"MIT"
] | 129 |
2019-07-05T05:22:15.000Z
|
2022-03-31T12:21:20.000Z
|
tutorials/02-intermediate/Training_a_Classifier.ipynb
|
awesome-archive/LIS-YNP
|
0cb434e209cee1aa83a7f73ddc7c81b6983ab7d7
|
[
"MIT"
] | null | null | null |
tutorials/02-intermediate/Training_a_Classifier.ipynb
|
awesome-archive/LIS-YNP
|
0cb434e209cee1aa83a7f73ddc7c81b6983ab7d7
|
[
"MIT"
] | 29 |
2019-07-07T00:53:51.000Z
|
2021-10-16T03:40:32.000Z
| 90.896825 | 23,220 | 0.787567 |
[
[
[
"<a href=\"https://colab.research.google.com/github/Eurus-Holmes/PyTorch-Tutorials/blob/master/Training_a__Classifier.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"%matplotlib inline",
"_____no_output_____"
]
],
[
[
"\nTraining a Classifier\n=====================\n\nThis is it. You have seen how to define neural networks, compute loss and make\nupdates to the weights of the network.\n\nNow you might be thinking,\n\nWhat about data?\n----------------\n\nGenerally, when you have to deal with image, text, audio or video data,\nyou can use standard python packages that load data into a numpy array.\nThen you can convert this array into a ``torch.*Tensor``.\n\n- For images, packages such as Pillow, OpenCV are useful\n- For audio, packages such as scipy and librosa\n- For text, either raw Python or Cython based loading, or NLTK and\n SpaCy are useful\n\nSpecifically for vision, we have created a package called\n``torchvision``, that has data loaders for common datasets such as\nImagenet, CIFAR10, MNIST, etc. and data transformers for images, viz.,\n``torchvision.datasets`` and ``torch.utils.data.DataLoader``.\n\nThis provides a huge convenience and avoids writing boilerplate code.\n\nFor this tutorial, we will use the CIFAR10 dataset.\nIt has the classes: ‘airplane’, ‘automobile’, ‘bird’, ‘cat’, ‘deer’,\n‘dog’, ‘frog’, ‘horse’, ‘ship’, ‘truck’. The images in CIFAR-10 are of\nsize 3x32x32, i.e. 3-channel color images of 32x32 pixels in size.\n\n\nTraining an image classifier\n----------------------------\n\nWe will do the following steps in order:\n\n1. Load and normalizing the CIFAR10 training and test datasets using\n ``torchvision``\n2. Define a Convolution Neural Network\n3. Define a loss function\n4. Train the network on the training data\n5. Test the network on the test data\n\n1. Loading and normalizing CIFAR10\n----------------------------\n\n# 1. Loading and normalizing CIFAR10\n\nUsing ``torchvision``, it’s extremely easy to load CIFAR10.\n\n",
"_____no_output_____"
]
],
[
[
"import torch\nimport torchvision\nimport torchvision.transforms as transforms",
"_____no_output_____"
]
],
[
[
"The output of torchvision datasets are PILImage images of range [0, 1].\nWe transform them to Tensors of normalized range [-1, 1].\n\n",
"_____no_output_____"
]
],
[
[
"transform = transforms.Compose(\n [transforms.ToTensor(),\n transforms.Normalize((0.5, 0.5, 0.5), (0.5, 0.5, 0.5))])\n\ntrainset = torchvision.datasets.CIFAR10(root='./data', train=True,\n download=True, transform=transform)\ntrainloader = torch.utils.data.DataLoader(trainset, batch_size=4,\n shuffle=True, num_workers=2)\n\ntestset = torchvision.datasets.CIFAR10(root='./data', train=False,\n download=True, transform=transform)\ntestloader = torch.utils.data.DataLoader(testset, batch_size=4,\n shuffle=False, num_workers=2)\n\nclasses = ('plane', 'car', 'bird', 'cat',\n 'deer', 'dog', 'frog', 'horse', 'ship', 'truck')",
"Files already downloaded and verified\nFiles already downloaded and verified\n"
]
],
[
[
"Let us show some of the training images, for fun.\n\n",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nimport numpy as np\n\n# functions to show an image\n\n\ndef imshow(img):\n img = img / 2 + 0.5 # unnormalize\n npimg = img.numpy()\n plt.imshow(np.transpose(npimg, (1, 2, 0)))\n\n\n# get some random training images\ndataiter = iter(trainloader)\nimages, labels = dataiter.next()\n\n# show images\nimshow(torchvision.utils.make_grid(images))\n# print labels\nprint(' '.join('%5s' % classes[labels[j]] for j in range(4)))",
" ship bird ship deer\n"
]
],
[
[
"# 2. Define a Convolution Neural Network\n----\n\nCopy the neural network from the Neural Networks section before and modify it to\ntake 3-channel images (instead of 1-channel images as it was defined).\n\n",
"_____no_output_____"
]
],
[
[
"import torch.nn as nn\nimport torch.nn.functional as F\n\n\nclass Net(nn.Module):\n def __init__(self):\n super(Net, self).__init__()\n self.conv1 = nn.Conv2d(3, 6, 5)\n self.pool = nn.MaxPool2d(2, 2)\n self.conv2 = nn.Conv2d(6, 16, 5)\n self.fc1 = nn.Linear(16 * 5 * 5, 120)\n self.fc2 = nn.Linear(120, 84)\n self.fc3 = nn.Linear(84, 10)\n\n def forward(self, x):\n x = self.pool(F.relu(self.conv1(x)))\n x = self.pool(F.relu(self.conv2(x)))\n x = x.view(-1, 16 * 5 * 5)\n x = F.relu(self.fc1(x))\n x = F.relu(self.fc2(x))\n x = self.fc3(x)\n return x\n\n\nnet = Net()",
"_____no_output_____"
]
],
[
[
"# 3. Define a Loss function and optimizer\n----\n\nLet's use a Classification Cross-Entropy loss and SGD with momentum.\n\n",
"_____no_output_____"
]
],
[
[
"import torch.optim as optim\n\ncriterion = nn.CrossEntropyLoss()\noptimizer = optim.SGD(net.parameters(), lr=0.001, momentum=0.9)",
"_____no_output_____"
]
],
[
[
"# 4. Train the network\n----\n\nThis is when things start to get interesting.\nWe simply have to loop over our data iterator, and feed the inputs to the\nnetwork and optimize.\n\n",
"_____no_output_____"
]
],
[
[
"for epoch in range(2): # loop over the dataset multiple times\n\n running_loss = 0.0\n for i, data in enumerate(trainloader, 0):\n # get the inputs\n inputs, labels = data\n\n # zero the parameter gradients\n optimizer.zero_grad()\n\n # forward + backward + optimize\n outputs = net(inputs)\n loss = criterion(outputs, labels)\n loss.backward()\n optimizer.step()\n\n # print statistics\n running_loss += loss.item()\n if i % 2000 == 1999: # print every 2000 mini-batches\n print('[%d, %5d] loss: %.3f' %\n (epoch + 1, i + 1, running_loss / 2000))\n running_loss = 0.0\n\nprint('Finished Training')",
"[1, 2000] loss: 2.148\n[1, 4000] loss: 1.803\n[1, 6000] loss: 1.658\n[1, 8000] loss: 1.551\n[1, 10000] loss: 1.493\n[1, 12000] loss: 1.458\n[2, 2000] loss: 1.383\n[2, 4000] loss: 1.367\n[2, 6000] loss: 1.341\n[2, 8000] loss: 1.314\n[2, 10000] loss: 1.301\n[2, 12000] loss: 1.264\nFinished Training\n"
]
],
[
[
"# 5. Test the network on the test data\n----\n\nWe have trained the network for 2 passes over the training dataset.\nBut we need to check if the network has learnt anything at all.\n\nWe will check this by predicting the class label that the neural network\noutputs, and checking it against the ground-truth. If the prediction is\ncorrect, we add the sample to the list of correct predictions.\n\nOkay, first step. Let us display an image from the test set to get familiar.\n\n",
"_____no_output_____"
]
],
[
[
"dataiter = iter(testloader)\nimages, labels = dataiter.next()\n\n# print images\nimshow(torchvision.utils.make_grid(images))\nprint('GroundTruth: ', ' '.join('%5s' % classes[labels[j]] for j in range(4)))",
"GroundTruth: cat ship ship plane\n"
]
],
[
[
"Okay, now let us see what the neural network thinks these examples above are:\n\n",
"_____no_output_____"
]
],
[
[
"outputs = net(images)",
"_____no_output_____"
]
],
[
[
"The outputs are energies for the 10 classes.\nHigher the energy for a class, the more the network\nthinks that the image is of the particular class.\nSo, let's get the index of the highest energy:\n\n",
"_____no_output_____"
]
],
[
[
"_, predicted = torch.max(outputs, 1)\n\nprint('Predicted: ', ' '.join('%5s' % classes[predicted[j]]\n for j in range(4)))",
"Predicted: ship ship ship ship\n"
]
],
[
[
"The results seem pretty good.\n\nLet us look at how the network performs on the whole dataset.\n\n",
"_____no_output_____"
]
],
[
[
"correct = 0\ntotal = 0\nwith torch.no_grad():\n for data in testloader:\n images, labels = data\n outputs = net(images)\n _, predicted = torch.max(outputs.data, 1)\n total += labels.size(0)\n correct += (predicted == labels).sum().item()\n\nprint('Accuracy of the network on the 10000 test images: %d %%' % (\n 100 * correct / total))",
"Accuracy of the network on the 10000 test images: 55 %\n"
]
],
[
[
"That looks waaay better than chance, which is 10% accuracy (randomly picking\na class out of 10 classes).\nSeems like the network learnt something.\n\nHmmm, what are the classes that performed well, and the classes that did\nnot perform well:\n\n",
"_____no_output_____"
]
],
[
[
"class_correct = list(0. for i in range(10))\nclass_total = list(0. for i in range(10))\nwith torch.no_grad():\n for data in testloader:\n images, labels = data\n outputs = net(images)\n _, predicted = torch.max(outputs, 1)\n c = (predicted == labels).squeeze()\n for i in range(4):\n label = labels[i]\n class_correct[label] += c[i].item()\n class_total[label] += 1\n\n\nfor i in range(10):\n print('Accuracy of %5s : %2d %%' % (\n classes[i], 100 * class_correct[i] / class_total[i]))",
"Accuracy of plane : 53 %\nAccuracy of car : 60 %\nAccuracy of bird : 34 %\nAccuracy of cat : 33 %\nAccuracy of deer : 47 %\nAccuracy of dog : 53 %\nAccuracy of frog : 62 %\nAccuracy of horse : 61 %\nAccuracy of ship : 81 %\nAccuracy of truck : 63 %\n"
]
],
[
[
"Okay, so what next?\n\nHow do we run these neural networks on the GPU?\n\nTraining on GPU\n----------------\nJust like how you transfer a Tensor on to the GPU, you transfer the neural\nnet onto the GPU.\n\nLet's first define our device as the first visible cuda device if we have\nCUDA available:\n\n",
"_____no_output_____"
]
],
[
[
"device = torch.device(\"cuda:0\" if torch.cuda.is_available() else \"cpu\")\n\n# Assume that we are on a CUDA machine, then this should print a CUDA device:\n\nprint(device)",
"cuda:0\n"
]
],
[
[
"The rest of this section assumes that `device` is a CUDA device.\n\nThen these methods will recursively go over all modules and convert their\nparameters and buffers to CUDA tensors:\n\n\n `net.to(device)`\n\n\nRemember that you will have to send the inputs and targets at every step\nto the GPU too:\n\n\n`inputs, labels = inputs.to(device), labels.to(device)`\n\nWhy dont I notice MASSIVE speedup compared to CPU? Because your network\nis realllly small.\n\n**Exercise:** Try increasing the width of your network (argument 2 of\nthe first ``nn.Conv2d``, and argument 1 of the second ``nn.Conv2d`` –\nthey need to be the same number), see what kind of speedup you get.\n\n**Goals achieved**:\n\n- Understanding PyTorch's Tensor library and neural networks at a high level.\n- Train a small neural network to classify images\n\nTraining on multiple GPUs\n-------------------------\nIf you want to see even more MASSIVE speedup using all of your GPUs,\nplease check out :doc:`data_parallel_tutorial`.\n\nWhere do I go next?\n-------------------\n\n- `Train neural nets to play video games`\n- `Train a state-of-the-art ResNet network on imagenet`\n- `Train a face generator using Generative Adversarial Networks`\n- `Train a word-level language model using Recurrent LSTM networks`\n- `More examples`\n- `More tutorials`\n- `Discuss PyTorch on the Forums`\n- `Chat with other users on Slack`\n\n",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
4a58431db0db98797e788fed1be0ada26a2df1f0
| 819,785 |
ipynb
|
Jupyter Notebook
|
examples/standardization.ipynb
|
arita37/causallib
|
e5ec1afafa26d0e04fedb75ee5ac95256f5b98a8
|
[
"Apache-2.0"
] | 1 |
2020-01-26T08:17:11.000Z
|
2020-01-26T08:17:11.000Z
|
examples/standardization.ipynb
|
arita37/causallib
|
e5ec1afafa26d0e04fedb75ee5ac95256f5b98a8
|
[
"Apache-2.0"
] | null | null | null |
examples/standardization.ipynb
|
arita37/causallib
|
e5ec1afafa26d0e04fedb75ee5ac95256f5b98a8
|
[
"Apache-2.0"
] | null | null | null | 518.523087 | 304,644 | 0.935572 |
[
[
[
"# Direct Outcome Prediction Model\nAlso known as standardization",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nfrom sklearn.linear_model import LinearRegression\nfrom sklearn.ensemble import GradientBoostingRegressor\n\nfrom causallib.datasets import load_smoking_weight\nfrom causallib.estimation import Standardization, StratifiedStandardization\nfrom causallib.evaluation import OutcomeEvaluator",
"_____no_output_____"
]
],
[
[
"#### Data:\nThe effect of quitting to smoke on weight loss. \nData example is taken from [Hernan and Robins Causal Inference Book](https://www.hsph.harvard.edu/miguel-hernan/causal-inference-book/)",
"_____no_output_____"
]
],
[
[
"data = load_smoking_weight()\ndata.X.join(data.a).join(data.y).head()",
"_____no_output_____"
]
],
[
[
"## \"Standard\" Standardization\nA single model is trained with the treatment assignment as an additional feature. \nDuring inference, the model assigns a treatment value for all samples, \nthus predicting the potential outcome of all samples.",
"_____no_output_____"
]
],
[
[
"std = Standardization(LinearRegression())\nstd.fit(data.X, data.a, data.y)",
"_____no_output_____"
]
],
[
[
"##### Outcome Prediction\nThe model can be used to predict individual outcomes: \nThe potential outcome under each intervention",
"_____no_output_____"
]
],
[
[
"ind_outcomes = std.estimate_individual_outcome(data.X, data.a)\nind_outcomes.head()",
"_____no_output_____"
]
],
[
[
"The model can be used to predict population outcomes, \nBy aggregating the individual outcome prediction (e.g., mean or median). \nProviding `agg_func` which is defaulted to `'mean'`",
"_____no_output_____"
]
],
[
[
"median_pop_outcomes = std.estimate_population_outcome(data.X, data.a, agg_func=\"median\")\nmedian_pop_outcomes.rename(\"median\", inplace=True)\n\nmean_pop_outcomes = std.estimate_population_outcome(data.X, data.a, agg_func=\"mean\")\nmean_pop_outcomes.rename(\"mean\", inplace=True)\n\npop_outcomes = mean_pop_outcomes.to_frame().join(median_pop_outcomes)\npop_outcomes",
"_____no_output_____"
]
],
[
[
"##### Effect Estimation\nSimilarly, Effect estimation can be done on either individual or population level, depending on the outcomes provided.",
"_____no_output_____"
],
[
"Population level effect using population outcomes:",
"_____no_output_____"
]
],
[
[
"std.estimate_effect(mean_pop_outcomes[1], mean_pop_outcomes[0])",
"_____no_output_____"
]
],
[
[
"Population level effect using individual outcome, but asking for aggregation (default behaviour):",
"_____no_output_____"
]
],
[
[
"std.estimate_effect(ind_outcomes[1], ind_outcomes[0], agg=\"population\")",
"_____no_output_____"
]
],
[
[
"Individual level effect using inidiviual outcomes: \nSince we're using a binary treatment with logistic regression on a standard model, \nthe difference is same for all individuals, and is equal to the coefficient of the treatment varaible",
"_____no_output_____"
]
],
[
[
"print(std.learner.coef_[0])\nstd.estimate_effect(ind_outcomes[1], ind_outcomes[0], agg=\"individual\").head()",
"3.4626218292258635\n"
]
],
[
[
"Multiple types of effect are also supported:",
"_____no_output_____"
]
],
[
[
"std.estimate_effect(ind_outcomes[1], ind_outcomes[0], \n agg=\"individual\", effect_types=[\"diff\", \"ratio\"]).head()",
"_____no_output_____"
]
],
[
[
"### Treament one-hot encoded\nFor multi-treatment cases, where treatments are coded as 0, 1, 2, ... but have no ordinal interpretation, \nIt is possible to make the model encode the treatment assignment vector as one hot matrix.",
"_____no_output_____"
]
],
[
[
"std = Standardization(LinearRegression(), encode_treatment=True)\nstd.fit(data.X, data.a, data.y)\npop_outcomes = std.estimate_population_outcome(data.X, data.a, agg_func=\"mean\")\nstd.estimate_effect(mean_pop_outcomes[1], mean_pop_outcomes[0])",
"_____no_output_____"
]
],
[
[
"## Stratified Standarziation\nWhile standardization can be viewed as a **\"complete pooled\"** estimator, \nas it includes both treatment groups together, \nStratified Standardization can viewed as **\"complete unpooled\"** one, \nas it completly stratifies the dataset by treatment values and learns a different model for each treatment group.",
"_____no_output_____"
]
],
[
[
"std = StratifiedStandardization(LinearRegression())\nstd.fit(data.X, data.a, data.y)",
"_____no_output_____"
]
],
[
[
"Checking the core `learner` we can see that it actually has two models, indexed by the treatment value:",
"_____no_output_____"
]
],
[
[
"std.learner",
"_____no_output_____"
]
],
[
[
"We can apply same analysis as above.",
"_____no_output_____"
]
],
[
[
"pop_outcomes = std.estimate_population_outcome(data.X, data.a, agg_func=\"mean\")\nstd.estimate_effect(mean_pop_outcomes[1], mean_pop_outcomes[0])",
"_____no_output_____"
]
],
[
[
"We can see that internally, when asking for some potential outcome, \nthe model simply applies the model trained on the group of that treatment:",
"_____no_output_____"
]
],
[
[
"potential_outcome = std.estimate_individual_outcome(data.X, data.a)[1]\ndirect_prediction = std.learner[1].predict(data.X)\n(potential_outcome == direct_prediction).all()",
"_____no_output_____"
]
],
[
[
"#### Providing complex scheme of learners\nWhen supplying a single learner to the standardization above, \nthe model simply duplicates it for each treatment value. \nHowever, it is possible to specify a different model for each treatment value explicitly. \nFor example, in cases where the treated are more complex than the untreated \n(because, say, background of those choosed to be treated), \nit is possible to specify them with a more expressive model:",
"_____no_output_____"
]
],
[
[
"learner = {0: LinearRegression(),\n 1: GradientBoostingRegressor()}\nstd = StratifiedStandardization(learner)\nstd.fit(data.X, data.a, data.y)\nstd.learner",
"_____no_output_____"
],
[
"ind_outcomes = std.estimate_individual_outcome(data.X, data.a)\nind_outcomes.head()",
"_____no_output_____"
],
[
"std.estimate_effect(ind_outcomes[1], ind_outcomes[0])",
"_____no_output_____"
]
],
[
[
"## Evaluation\n#### Simple evaluation",
"_____no_output_____"
]
],
[
[
"plots = [\"common_support\", \"continuous_accuracy\"]\nevaluator = OutcomeEvaluator(std)\nevaluator._regression_metrics.pop(\"msle\") # We have negative values and this is log transforms\nresults = evaluator.evaluate_simple(data.X, data.a, data.y, plots=plots)",
"_____no_output_____"
]
],
[
[
"Results show the results for each treatment group separetly and also combined:",
"_____no_output_____"
]
],
[
[
"results.scores",
"_____no_output_____"
]
],
[
[
"#### Thorough evaluation",
"_____no_output_____"
]
],
[
[
"plots=[\"common_support\", \"continuous_accuracy\", \"residuals\"]\nevaluator = OutcomeEvaluator(Standardization(LinearRegression()))\nresults = evaluator.evaluate_cv(data.X, data.a, data.y,\n plots=plots)",
"_____no_output_____"
],
[
"results.scores",
"_____no_output_____"
],
[
"results.models",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
4a584b86bc4b7e2785dbd1afacec6c6125b4cbd3
| 842,817 |
ipynb
|
Jupyter Notebook
|
Jessicas_Run01.ipynb
|
UBC-MOAD/outputanalysisnotebooks
|
50839cde3832d26bac6641427fed03c818fbe170
|
[
"Apache-2.0"
] | null | null | null |
Jessicas_Run01.ipynb
|
UBC-MOAD/outputanalysisnotebooks
|
50839cde3832d26bac6641427fed03c818fbe170
|
[
"Apache-2.0"
] | null | null | null |
Jessicas_Run01.ipynb
|
UBC-MOAD/outputanalysisnotebooks
|
50839cde3832d26bac6641427fed03c818fbe170
|
[
"Apache-2.0"
] | null | null | null | 647.324885 | 434,478 | 0.930317 |
[
[
[
"Getting rid of bottom bands - Jessica's run (run01)\n===================================================\n\nRun01 Jessica's runs (360x360x90, her bathymetry and stratification initial files)\n--------------------------------------------------------------",
"_____no_output_____"
],
[
"Initial stratifications, Depths 162, 315, 705 m, Across-shelf slice 40; T, NO3, S, and velocity plots\nRun01 and run03 from 180x180x35_BodyForcing_6Tr_LinProfiles",
"_____no_output_____"
]
],
[
[
"#KRM\n\nimport numpy as np\n\nimport matplotlib.pyplot as plt\n\nimport matplotlib.colors as mcolors\n\nfrom math import *\n\nimport scipy.io\n\nimport scipy as spy\n\n%matplotlib inline\n\nfrom netCDF4 import Dataset\n\nimport pylab as pl\n\n",
"_____no_output_____"
],
[
"#'''\n#NAME\n# Custom Colormaps for Matplotlib\n#PURPOSE\n# This program shows how to implement make_cmap which is a function that\n# generates a colorbar. If you want to look at different color schemes,\n# check out https://kuler.adobe.com/create.\n#PROGRAMMER(S)\n# Chris Slocum\n#REVISION HISTORY\n# 20130411 -- Initial version created\n# 20140313 -- Small changes made and code posted online\n# 20140320 -- Added the ability to set the position of each color\n#'''\n\ndef make_cmap(colors, position=None, bit=False):\n #'''\n #make_cmap takes a list of tuples which contain RGB values. The RGB\n #values may either be in 8-bit [0 to 255] (in which bit must be set to\n #rue when called) or arithmetic [0 to 1] (default). make_cmap returns\n #a cmap with equally spaced colors.\n #Arrange your tuples so that the first color is the lowest value for the\n #colorbar and the last is the highest.\n #position contains values from 0 to 1 to dictate the location of each color.\n #'''\n import matplotlib as mpl\n import numpy as np\n bit_rgb = np.linspace(0,1,256)\n if position == None:\n position = np.linspace(0,1,len(colors))\n else:\n if len(position) != len(colors):\n sys.exit(\"position length must be the same as colors\")\n elif position[0] != 0 or position[-1] != 1:\n sys.exit(\"position must start with 0 and end with 1\")\n if bit:\n for i in range(len(colors)):\n colors[i] = (bit_rgb[colors[i][0]],\n bit_rgb[colors[i][1]],\n bit_rgb[colors[i][2]])\n cdict = {'red':[], 'green':[], 'blue':[]}\n for pos, color in zip(position, colors):\n cdict['red'].append((pos, color[0], color[0]))\n cdict['green'].append((pos, color[1], color[1]))\n cdict['blue'].append((pos, color[2], color[2]))\n\n cmap = mpl.colors.LinearSegmentedColormap('my_colormap',cdict,256)\n return cmap",
"_____no_output_____"
],
[
"def unstagger(ugrid, vgrid):\n \"\"\"Interpolate u and v component values to values at grid cell centres.\n\n The shapes of the returned arrays are 1 less than those of\n the input arrays in the y and x dimensions.\n\n :arg ugrid: u velocity component values with axes (..., y, x)\n :type ugrid: :py:class:`numpy.ndarray`\n\n :arg vgrid: v velocity component values with axes (..., y, x)\n :type vgrid: :py:class:`numpy.ndarray`\n\n :returns u, v: u and v component values at grid cell centres\n :rtype: 2-tuple of :py:class:`numpy.ndarray`\n \"\"\"\n u = np.add(ugrid[..., :-1], ugrid[..., 1:]) / 2\n v = np.add(vgrid[..., :-1, :], vgrid[..., 1:, :]) / 2\n return u[..., 1:, :], v[..., 1:]\n",
"_____no_output_____"
],
[
"# Get field from MITgcm netCDF output\n#\n''' :statefile : string with /path/to/state.0000000000.t001.nc\n :fieldname : string with the variable name as written on the netCDF file ('Temp', 'S','Eta', etc.)'''\n\ndef getField(statefile, fieldname):\n \n StateOut = Dataset(statefile)\n \n Fld = StateOut.variables[fieldname][:]\n \n shFld = np.shape(Fld)\n \n \n if len(shFld) == 2:\n \n Fld2 = np.reshape(Fld,(shFld[0],shFld[1])) # reshape to pcolor order\n return Fld2 \n \n elif len(shFld) == 3:\n \n Fld2 = np.zeros((shFld[0],shFld[1],shFld[2])) \n Fld2 = np.reshape(Fld,(shFld[0],shFld[1],shFld[2])) # reshape to pcolor order\n return Fld2\n \n elif len(shFld) == 4:\n \n Fld2 = np.zeros((shFld[0],shFld[1],shFld[2],shFld[3])) \n Fld2 = np.reshape(Fld,(shFld[0],shFld[1],shFld[2],shFld[3])) # reshape to pcolor order\n return Fld2\n \n else:\n \n print (' Check size of field ')\n \n \n\n",
"_____no_output_____"
]
],
[
[
"Inquire variable from NetCDF - RUN01",
"_____no_output_____"
]
],
[
[
"filenameb='/ocean/kramosmu/MITgcm/CanyonUpwelling/360x360x90_BodyForcing_1Tr/run01/mnc_0001/state.0000000000.t001.nc'\nStateOutb = Dataset(filenameb)\n\nfor dimobj in StateOutb.variables.values():\n print dimobj\n\n\nfilename2b='/ocean/kramosmu/MITgcm/CanyonUpwelling/360x360x90_BodyForcing_1Tr/run01/mnc_0001/grid.t001.nc'\nGridOutb = Dataset(filename2b)\n\nfor dimobj in GridOutb.variables.values():\n print dimobj\n\n\nfilename3b='/ocean/kramosmu/MITgcm/CanyonUpwelling/360x360x90_BodyForcing_1Tr/run01/mnc_0001/ptracers.0000000000.t001.nc'\nPtracersOutb = Dataset(filename3b)\n\nfor dimobj in PtracersOutb.variables.values():\n print dimobj\n",
"<type 'netCDF4.Variable'>\nfloat64 T(T)\n long_name: model_time\n units: s\nunlimited dimensions: T\ncurrent shape = (5,)\n\n<type 'netCDF4.Variable'>\nint32 iter(T)\n long_name: iteration_count\nunlimited dimensions: T\ncurrent shape = (5,)\n\n<type 'netCDF4.Variable'>\nfloat64 Xp1(Xp1)\n long_name: X-Coordinate of cell corner\n units: meters\nunlimited dimensions: \ncurrent shape = (361,)\n\n<type 'netCDF4.Variable'>\nfloat64 Y(Y)\n long_name: Y-Coordinate of cell center\n units: meters\nunlimited dimensions: \ncurrent shape = (360,)\n\n<type 'netCDF4.Variable'>\nfloat64 Z(Z)\n long_name: vertical coordinate of cell center\n units: meters\n positive: up\nunlimited dimensions: \ncurrent shape = (90,)\n\n<type 'netCDF4.Variable'>\nfloat32 U(T, Z, Y, Xp1)\n units: m/s\n coordinates: XU YU RC iter\nunlimited dimensions: T\ncurrent shape = (5, 90, 360, 361)\n\n<type 'netCDF4.Variable'>\nfloat64 X(X)\n long_name: X-coordinate of cell center\n units: meters\nunlimited dimensions: \ncurrent shape = (360,)\n\n<type 'netCDF4.Variable'>\nfloat64 Yp1(Yp1)\n long_name: Y-Coordinate of cell corner\n units: meters\nunlimited dimensions: \ncurrent shape = (361,)\n\n<type 'netCDF4.Variable'>\nfloat32 V(T, Z, Yp1, X)\n units: m/s\n coordinates: XV YV RC iter\nunlimited dimensions: T\ncurrent shape = (5, 90, 361, 360)\n\n<type 'netCDF4.Variable'>\nfloat32 Temp(T, Z, Y, X)\n units: degC\n long_name: potential_temperature\n coordinates: XC YC RC iter\nunlimited dimensions: T\ncurrent shape = (5, 90, 360, 360)\n\n<type 'netCDF4.Variable'>\nfloat32 S(T, Z, Y, X)\n long_name: salinity\n coordinates: XC YC RC iter\nunlimited dimensions: T\ncurrent shape = (5, 90, 360, 360)\n\n<type 'netCDF4.Variable'>\nfloat32 Eta(T, Y, X)\n long_name: free-surface_r-anomaly\n units: m\n coordinates: XC YC iter\nunlimited dimensions: T\ncurrent shape = (5, 360, 360)\n\n<type 'netCDF4.Variable'>\nfloat64 Zl(Zl)\n long_name: vertical coordinate of upper cell interface\n units: meters\n positive: up\nunlimited dimensions: \ncurrent shape = (90,)\n\n<type 'netCDF4.Variable'>\nfloat32 W(T, Zl, Y, X)\n units: m/s\n coordinates: XC YC RC iter\nunlimited dimensions: T\ncurrent shape = (5, 90, 360, 360)\n\n<type 'netCDF4.Variable'>\nfloat32 phi_nh(T, Z, Y, X)\nunlimited dimensions: T\ncurrent shape = (5, 90, 360, 360)\n\n<type 'netCDF4.Variable'>\nfloat64 Z(Z)\n long_name: vertical coordinate of cell center\n units: meters\n positive: up\nunlimited dimensions: \ncurrent shape = (90,)\n\n<type 'netCDF4.Variable'>\nfloat64 RC(Z)\n description: R coordinate of cell center\n units: m\nunlimited dimensions: \ncurrent shape = (90,)\n\n<type 'netCDF4.Variable'>\nfloat64 Zp1(Zp1)\n long_name: vertical coordinate of cell interface\n units: meters\n positive: up\nunlimited dimensions: \ncurrent shape = (91,)\n\n<type 'netCDF4.Variable'>\nfloat64 RF(Zp1)\n description: R coordinate of cell interface\n units: m\nunlimited dimensions: \ncurrent shape = (91,)\n\n<type 'netCDF4.Variable'>\nfloat64 Zu(Zu)\n long_name: vertical coordinate of lower cell interface\n units: meters\n positive: up\nunlimited dimensions: \ncurrent shape = (90,)\n\n<type 'netCDF4.Variable'>\nfloat64 RU(Zu)\n description: R coordinate of upper interface\n units: m\nunlimited dimensions: \ncurrent shape = (90,)\n\n<type 'netCDF4.Variable'>\nfloat64 Zl(Zl)\n long_name: vertical coordinate of upper cell interface\n units: meters\n positive: up\nunlimited dimensions: \ncurrent shape = (90,)\n\n<type 'netCDF4.Variable'>\nfloat64 RL(Zl)\n description: R coordinate of lower interface\n units: m\nunlimited dimensions: \ncurrent shape = (90,)\n\n<type 'netCDF4.Variable'>\nfloat64 drC(Zp1)\n description: r cell center separation\nunlimited dimensions: \ncurrent shape = (91,)\n\n<type 'netCDF4.Variable'>\nfloat64 drF(Z)\n description: r cell face separation\nunlimited dimensions: \ncurrent shape = (90,)\n\n<type 'netCDF4.Variable'>\nfloat64 X(X)\n long_name: X-coordinate of cell center\n units: meters\nunlimited dimensions: \ncurrent shape = (360,)\n\n<type 'netCDF4.Variable'>\nfloat64 Y(Y)\n long_name: Y-Coordinate of cell center\n units: meters\nunlimited dimensions: \ncurrent shape = (360,)\n\n<type 'netCDF4.Variable'>\nfloat64 XC(Y, X)\n description: X coordinate of cell center (T-P point)\n units: degree_east\nunlimited dimensions: \ncurrent shape = (360, 360)\n\n<type 'netCDF4.Variable'>\nfloat64 YC(Y, X)\n description: Y coordinate of cell center (T-P point)\n units: degree_north\nunlimited dimensions: \ncurrent shape = (360, 360)\n\n<type 'netCDF4.Variable'>\nfloat64 Xp1(Xp1)\n long_name: X-Coordinate of cell corner\n units: meters\nunlimited dimensions: \ncurrent shape = (361,)\n\n<type 'netCDF4.Variable'>\nfloat64 Yp1(Yp1)\n long_name: Y-Coordinate of cell corner\n units: meters\nunlimited dimensions: \ncurrent shape = (361,)\n\n<type 'netCDF4.Variable'>\nfloat64 XG(Yp1, Xp1)\n description: X coordinate of cell corner (Vorticity point)\n units: degree_east\nunlimited dimensions: \ncurrent shape = (361, 361)\n\n<type 'netCDF4.Variable'>\nfloat64 YG(Yp1, Xp1)\n description: Y coordinate of cell corner (Vorticity point)\n units: degree_north\nunlimited dimensions: \ncurrent shape = (361, 361)\n\n<type 'netCDF4.Variable'>\nfloat64 dxC(Y, Xp1)\n description: x cell center separation\nunlimited dimensions: \ncurrent shape = (360, 361)\n\n<type 'netCDF4.Variable'>\nfloat64 dyC(Yp1, X)\n description: y cell center separation\nunlimited dimensions: \ncurrent shape = (361, 360)\n\n<type 'netCDF4.Variable'>\nfloat64 dxF(Y, X)\n description: x cell face separation\nunlimited dimensions: \ncurrent shape = (360, 360)\n\n<type 'netCDF4.Variable'>\nfloat64 dyF(Y, X)\n description: y cell face separation\nunlimited dimensions: \ncurrent shape = (360, 360)\n\n<type 'netCDF4.Variable'>\nfloat64 dxG(Yp1, X)\n description: x cell corner separation\nunlimited dimensions: \ncurrent shape = (361, 360)\n\n<type 'netCDF4.Variable'>\nfloat64 dyG(Y, Xp1)\n description: y cell corner separation\nunlimited dimensions: \ncurrent shape = (360, 361)\n\n<type 'netCDF4.Variable'>\nfloat64 dxV(Yp1, Xp1)\n description: x v-velocity separation\nunlimited dimensions: \ncurrent shape = (361, 361)\n\n<type 'netCDF4.Variable'>\nfloat64 dyU(Yp1, Xp1)\n description: y u-velocity separation\nunlimited dimensions: \ncurrent shape = (361, 361)\n\n<type 'netCDF4.Variable'>\nfloat64 rA(Y, X)\n description: r-face area at cell center\nunlimited dimensions: \ncurrent shape = (360, 360)\n\n<type 'netCDF4.Variable'>\nfloat64 rAw(Y, Xp1)\n description: r-face area at U point\nunlimited dimensions: \ncurrent shape = (360, 361)\n\n<type 'netCDF4.Variable'>\nfloat64 rAs(Yp1, X)\n description: r-face area at V point\nunlimited dimensions: \ncurrent shape = (361, 360)\n\n<type 'netCDF4.Variable'>\nfloat64 rAz(Yp1, Xp1)\n description: r-face area at cell corner\nunlimited dimensions: \ncurrent shape = (361, 361)\n\n<type 'netCDF4.Variable'>\nfloat64 fCori(Y, X)\n description: Coriolis f at cell center\nunlimited dimensions: \ncurrent shape = (360, 360)\n\n<type 'netCDF4.Variable'>\nfloat64 fCoriG(Yp1, Xp1)\n description: Coriolis f at cell corner\nunlimited dimensions: \ncurrent shape = (361, 361)\n\n<type 'netCDF4.Variable'>\nfloat64 R_low(Y, X)\n description: base of fluid in r-units\nunlimited dimensions: \ncurrent shape = (360, 360)\n\n<type 'netCDF4.Variable'>\nfloat64 Ro_surf(Y, X)\n description: surface reference (at rest) position\nunlimited dimensions: \ncurrent shape = (360, 360)\n\n<type 'netCDF4.Variable'>\nfloat64 Depth(Y, X)\n description: fluid thickness in r coordinates (at rest)\nunlimited dimensions: \ncurrent shape = (360, 360)\n\n<type 'netCDF4.Variable'>\nfloat64 HFacC(Z, Y, X)\n description: vertical fraction of open cell at cell center\nunlimited dimensions: \ncurrent shape = (90, 360, 360)\n\n<type 'netCDF4.Variable'>\nfloat64 HFacW(Z, Y, Xp1)\n description: vertical fraction of open cell at West face\nunlimited dimensions: \ncurrent shape = (90, 360, 361)\n\n<type 'netCDF4.Variable'>\nfloat64 HFacS(Z, Yp1, X)\n description: vertical fraction of open cell at South face\nunlimited dimensions: \ncurrent shape = (90, 361, 360)\n\n<type 'netCDF4.Variable'>\nfloat64 T(T)\n long_name: model_time\n units: s\nunlimited dimensions: T\ncurrent shape = (5,)\n\n<type 'netCDF4.Variable'>\nint32 iter(T)\n long_name: iteration_count\nunlimited dimensions: T\ncurrent shape = (5,)\n\n<type 'netCDF4.Variable'>\nfloat64 X(X)\n long_name: X-coordinate of cell center\n units: meters\nunlimited dimensions: \ncurrent shape = (360,)\n\n<type 'netCDF4.Variable'>\nfloat64 Y(Y)\n long_name: Y-Coordinate of cell center\n units: meters\nunlimited dimensions: \ncurrent shape = (360,)\n\n<type 'netCDF4.Variable'>\nfloat64 Z(Z)\n long_name: vertical coordinate of cell center\n units: meters\n positive: up\nunlimited dimensions: \ncurrent shape = (90,)\n\n<type 'netCDF4.Variable'>\nfloat32 NO3(T, Z, Y, X)\n units: microM\nunlimited dimensions: T\ncurrent shape = (5, 90, 360, 360)\n\n"
],
[
"# General input\n\nnx = 360\nny = 360\nnz = 90\nnta = 10 # t dimension size run 04 and 05 (output every 2 hr for 4.5 days)\nntc = 10 # t dimension size run 06 (output every half-day for 4.5 days)\n\nz = StateOutb.variables['Z']\nprint(z[:])\nTime = StateOutb.variables['T']\nprint(Time[:])\n\nxc = getField(filenameb, 'XC') # x coords tracer cells\n\n\nyc = getField(filenameb, 'YC') # y coords tracer cells\n\nprint(z[65])",
"[ -2.5 -7.5 -12.5 -17.5 -22.5 -27.5 -32.5 -37.5 -42.5\n -47.5 -52.5 -57.5 -62.5 -67.5 -72.5 -77.5 -82.5 -87.5\n -92.5 -97.5 -102.5 -107.5 -112.5 -117.5 -122.5 -127.5 -132.5\n -137.5 -142.5 -147.5 -152.5 -157.5 -162.5 -167.5 -175. -185.\n -195. -205. -215. -225. -235. -245. -255. -270. -290.\n -310. -330. -350. -370. -390. -410. -430. -450. -470.\n -490. -510. -530. -550. -570. -590. -610. -630. -650.\n -670. -690. -710. -730. -750. -770. -790. -810. -830.\n -850. -870. -890. -910. -930. -950. -970. -990. -1010.\n -1030. -1050. -1070. -1090. -1110. -1130. -1150. -1170. -1190. ]\n[ 0. 86400. 172800. 259200. 345600.]\n Check size of field \n Check size of field \n-710.0\n"
],
[
"#bathy = getField(filename2, 'Depth')\n\n\n#plt.rcParams.update({'font.size': 14})\n\n#fig = plt.figure(figsize=(20,15))\n\n#CS = plt.contour(xc,yc,bathy,30,colors='k' )\n\n#plt.clabel(CS, \n# inline=1,\n# fmt='%1.1f',\n# fontsize=14)\n\n#plt.plot(xc[:,:],yc[:,:],linewidth=0.75, linestyle='-', color='0.75')\n\n#plt.xlabel('m',fontsize=14)\n#plt.ylabel('m',fontsize=14)\n\n#plt.title('Bathymetry (m) 180x180',fontsize=16)\n\n\n\n#plt.show\n",
"_____no_output_____"
]
],
[
[
"Depth 705 m\n============",
"_____no_output_____"
]
],
[
[
"zlev = 65 # 65 corresponds to 710m\ntimesc = [0,1,2,3,4] # These correspond to 1,2,4,6,8,10 days\n\n\n",
"_____no_output_____"
],
[
"ugridb = getField(filenameb,'U')\nvgridb = getField(filenameb,'V')\n\n\n\nprint(np.shape(ugridb))\n\n\nprint(np.shape(vgridb))\n\n",
"(5, 90, 360, 361)\n(5, 90, 361, 360)\n"
]
],
[
[
"Get mask from T field (not the best, I know)",
"_____no_output_____"
]
],
[
[
"tempb = getField(filenameb, 'Temp')\n\n\ntemp0b = np.ma.masked_values(tempb, 0)\n\nMASKb = np.ma.getmask(temp0b)\n",
"_____no_output_____"
],
[
"#### T controls for plot ####\n\nplt.rcParams.update({'font.size':13})\n\ncolorsTemp = [(245.0/255.0,245/255.0,245./255.0), (255/255.0,20/255.0,0)] #(khaki 1246/255.0,143./255.0 ,orangered2)\n\nposTemp = [0, 1] \n \nNumLev = 30 # number of levels for contour\n\n",
"_____no_output_____"
],
[
"#### PLOT ####\n\nplt.rcParams.update({'font.size':14})\n\nkk=1\n\nfig45=plt.figure(figsize=(18,48))\n\nfor tt in timesc :\n \n ### Temperature run01\n plt.subplot(6,2,kk)\n \n ax = plt.gca()\n ax.set_axis_bgcolor((205/255.0, 201/255.0, 201/255.0))\n\n plt.contourf(xc,yc,temp0b[tt,zlev,:,:],NumLev,cmap=make_cmap(colorsTemp, position=posTemp))\n \n plt.ticklabel_format(style='sci', axis='x', scilimits=(0,0))\n plt.ticklabel_format(style='sci', axis='y', scilimits=(0,0))\n \n plt.xlabel('m')\n plt.ylabel('m')\n \n cb = plt.colorbar()\n \n cb.set_label(r'$^{\\circ}$C',position=(1, 0),rotation=0)\n \n plt.title(\" depth=%1.1f m,%1.1f days \" % (z[zlev],tt))\n \n kk=kk+1\n ",
"_____no_output_____"
]
],
[
[
"NO3 PLOTS ",
"_____no_output_____"
]
],
[
[
"#### NO3 controls for plot ####\n\n\nNO3b = getField(filename3b, 'NO3')\n\n\n\nNO3Maskb = np.ma.array(NO3b,mask=MASKb)\n\n\ncolorsNO3 = [(245.0/255.0,245/255.0,245./255.0), (0./255.0,139.0/255.0,69.0/255.0)] #(white-ish, forest green)\n\n\nposNO3 = [0, 1] \n \nNumLev = 30 # number of levels for contour\n\n",
"_____no_output_____"
],
[
"#### PLOT ####\n\nplt.rcParams.update({'font.size':14})\n\nkk=1\n\nfig45=plt.figure(figsize=(18,48))\n\nfor tt in timesc :\n \n \n ### Temperature run06\n plt.subplot(6,2,kk)\n ax = plt.gca()\n ax.set_axis_bgcolor((205/255.0, 201/255.0, 201/255.0))\n\n plt.contourf(xc,yc,NO3Maskb[tt,zlev,:,:],NumLev,cmap=make_cmap(colorsNO3, position=posNO3))\n \n plt.ticklabel_format(style='sci', axis='x', scilimits=(0,0))\n plt.ticklabel_format(style='sci', axis='y', scilimits=(0,0))\n \n plt.xlabel('m')\n plt.ylabel('m')\n \n cb = plt.colorbar()\n \n cb.set_label(r'$Mol/m^3$',position=(1, 0),rotation=0)\n \n plt.title(\" depth=%1.1f m,%1.1f hr \" % (z[zlev],tt))\n \n \n \n \n kk=kk+1\n ",
"_____no_output_____"
]
],
[
[
"Velocity plots",
"_____no_output_____"
]
],
[
[
"#### PLOT ####\n\nplt.rcParams.update({'font.size':14})\n\nkk=1\n\nfig45=plt.figure(figsize=(18,48))\n\nfor tt in timesc :\n \n ### Speed and vel vectors, run01\n \n plt.subplot(6,2,kk)\n ax = plt.gca()\n ax.set_axis_bgcolor((205/255.0, 201/255.0, 201/255.0))\n\n u2,v2 = unstagger(ugridb[tt,zlev,:,:-1],vgridb[tt,zlev,:-1,:])\n \n umaskb=np.ma.array(u2,mask=MASKb[tt,zlev,:-1,:-1])\n vmaskb=np.ma.array(v2,mask=MASKb[tt,zlev,:-1,:-1])\n \n y_slice = yc[:]#np.arange(0, ny-1)\n x_slice = xc[:]#np.arange(0, nx-1)\n \n arrow_step = 6\n y_slice_a = y_slice[::arrow_step,::arrow_step]\n x_slice_a = x_slice[::arrow_step,::arrow_step]\n \n Usliceb = umaskb[::arrow_step,::arrow_step]\n Vsliceb = vmaskb[::arrow_step,::arrow_step]\n \n #print(np.shape(Uslice))\n #print(np.shape(Vslice))\n #print(np.shape(x_slice_a))\n #print(np.shape(y_slice_a))\n \n spdb = np.sqrt(umaskb**2 + vmaskb**2)\n \n \n pos = [0, 1] # to keep white color on zero\n \n colorsSpd = [(245.0/255.0,245/255.0,245./255.0), (71./255.0,60.0/255.0,139.0/255.0)] #(white-ish, Slate blue 4)\n \n plt.contourf(xc[:-1,:-1],yc[:-1,:-1],spdb,NumLev,cmap=make_cmap(colorsSpd, position=pos))\n \n cb = plt.colorbar()\n \n cb.set_label('m/s', position=(1, 0),rotation=0)\n \n plt.quiver(y_slice_a,x_slice_a,Usliceb,Vsliceb,pivot='middle')\n \n plt.xlabel('m')\n \n plt.ylabel('m')\n \n \n kk=kk+1\n ",
"/home/kramosmu/anaconda/lib/python2.7/site-packages/matplotlib/quiver.py:570: RuntimeWarning: divide by zero encountered in double_scalars\n length = a * (widthu_per_lenu / (self.scale * self.width))\n/home/kramosmu/anaconda/lib/python2.7/site-packages/matplotlib/quiver.py:570: RuntimeWarning: invalid value encountered in multiply\n length = a * (widthu_per_lenu / (self.scale * self.width))\n"
]
],
[
[
"Line plots across-shelf slice at x = 39.37 km (for T)",
"_____no_output_____"
]
],
[
[
"\nplt.rcParams.update({'font.size':14})\n\nalongshpos = 40\n\nkk=1\n\nfig45=plt.figure(figsize=(27,10))\n\n\nfor ii in timesc:\n \n posTemp = [0, 1] \n \n NumLev = 30 \n \n plt.subplot(1,3,kk)\n ax=plt.gca()\n plt.plot(yc[:,0],temp0b[ii,zlev,:,alongshpos],linewidth = 2)\n \n \n plt.ylabel('Temperature ($^{\\circ}C$)')\n plt.xlabel('m')\n \n plt.show\n plt.title(\"z=%1.1f m, x=%1.1f m \" % (z[zlev],xc[1,alongshpos]))\n \n\nplt.legend(('1 day','2 days','3 days','4 days','5 days'),loc=3)\n\nkk=2\n\n\n",
"_____no_output_____"
]
],
[
[
"Plot depth vs salinity/temperature\n",
"_____no_output_____"
]
],
[
[
"z = StateOutb.variables['Z']\nprint(z[:])\n\nzl = GridOutb.variables['Zl']\nprint(zl[:])\n\nzp1 = GridOutb.variables['Zp1']\nprint(zp1[:])\n\nzu = GridOutb.variables['Zu']\nprint(zu[:])\n\n\ndepth= GridOutb.variables['Depth']\n\ndrc = GridOutb.variables['drC']\nprint(drc[:])\n\ndrf = GridOutb.variables['drF']\nprint(drf[:])\n\n\n\nT = getField(filenameb, 'Temp')\nS = getField(filenameb,'S')\n\n\n\n",
"[ -2.5 -7.5 -12.5 -17.5 -22.5 -27.5 -32.5 -37.5 -42.5\n -47.5 -52.5 -57.5 -62.5 -67.5 -72.5 -77.5 -82.5 -87.5\n -92.5 -97.5 -102.5 -107.5 -112.5 -117.5 -122.5 -127.5 -132.5\n -137.5 -142.5 -147.5 -152.5 -157.5 -162.5 -167.5 -175. -185.\n -195. -205. -215. -225. -235. -245. -255. -270. -290.\n -310. -330. -350. -370. -390. -410. -430. -450. -470.\n -490. -510. -530. -550. -570. -590. -610. -630. -650.\n -670. -690. -710. -730. -750. -770. -790. -810. -830.\n -850. -870. -890. -910. -930. -950. -970. -990. -1010.\n -1030. -1050. -1070. -1090. -1110. -1130. -1150. -1170. -1190. ]\n[ 0. -5. -10. -15. -20. -25. -30. -35. -40. -45.\n -50. -55. -60. -65. -70. -75. -80. -85. -90. -95.\n -100. -105. -110. -115. -120. -125. -130. -135. -140. -145.\n -150. -155. -160. -165. -170. -180. -190. -200. -210. -220.\n -230. -240. -250. -260. -280. -300. -320. -340. -360. -380.\n -400. -420. -440. -460. -480. -500. -520. -540. -560. -580.\n -600. -620. -640. -660. -680. -700. -720. -740. -760. -780.\n -800. -820. -840. -860. -880. -900. -920. -940. -960. -980.\n -1000. -1020. -1040. -1060. -1080. -1100. -1120. -1140. -1160. -1180.]\n[ 0. -5. -10. -15. -20. -25. -30. -35. -40. -45.\n -50. -55. -60. -65. -70. -75. -80. -85. -90. -95.\n -100. -105. -110. -115. -120. -125. -130. -135. -140. -145.\n -150. -155. -160. -165. -170. -180. -190. -200. -210. -220.\n -230. -240. -250. -260. -280. -300. -320. -340. -360. -380.\n -400. -420. -440. -460. -480. -500. -520. -540. -560. -580.\n -600. -620. -640. -660. -680. -700. -720. -740. -760. -780.\n -800. -820. -840. -860. -880. -900. -920. -940. -960. -980.\n -1000. -1020. -1040. -1060. -1080. -1100. -1120. -1140. -1160. -1180.\n -1200.]\n[ -5. -10. -15. -20. -25. -30. -35. -40. -45. -50.\n -55. -60. -65. -70. -75. -80. -85. -90. -95. -100.\n -105. -110. -115. -120. -125. -130. -135. -140. -145. -150.\n -155. -160. -165. -170. -180. -190. -200. -210. -220. -230.\n -240. -250. -260. -280. -300. -320. -340. -360. -380. -400.\n -420. -440. -460. -480. -500. -520. -540. -560. -580. -600.\n -620. -640. -660. -680. -700. -720. -740. -760. -780. -800.\n -820. -840. -860. -880. -900. -920. -940. -960. -980. -1000.\n -1020. -1040. -1060. -1080. -1100. -1120. -1140. -1160. -1180. -1200.]\n[ 2.5 5. 5. 5. 5. 5. 5. 5. 5. 5. 5. 5.\n 5. 5. 5. 5. 5. 5. 5. 5. 5. 5. 5. 5.\n 5. 5. 5. 5. 5. 5. 5. 5. 5. 5. 7.5 10.\n 10. 10. 10. 10. 10. 10. 10. 15. 20. 20. 20. 20.\n 20. 20. 20. 20. 20. 20. 20. 20. 20. 20. 20. 20.\n 20. 20. 20. 20. 20. 20. 20. 20. 20. 20. 20. 20.\n 20. 20. 20. 20. 20. 20. 20. 20. 20. 20. 20. 20.\n 20. 20. 20. 20. 20. 20. 10. ]\n[ 5. 5. 5. 5. 5. 5. 5. 5. 5. 5. 5. 5. 5. 5. 5.\n 5. 5. 5. 5. 5. 5. 5. 5. 5. 5. 5. 5. 5. 5. 5.\n 5. 5. 5. 5. 10. 10. 10. 10. 10. 10. 10. 10. 10. 20. 20.\n 20. 20. 20. 20. 20. 20. 20. 20. 20. 20. 20. 20. 20. 20. 20.\n 20. 20. 20. 20. 20. 20. 20. 20. 20. 20. 20. 20. 20. 20. 20.\n 20. 20. 20. 20. 20. 20. 20. 20. 20. 20. 20. 20. 20. 20. 20.]\n"
],
[
"fig46 = plt.figure(figsize=(10,10))\n\nplt.plot(T[0,:,200,180],z[:],'ro')\n\n",
"_____no_output_____"
],
[
"\n\n",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
4a585ea4178f4ee2a065117281fe2579f9a83e6c
| 37,955 |
ipynb
|
Jupyter Notebook
|
intro-to-pytorch/Part 4 - Fashion-MNIST (Exercises).ipynb
|
Arush0113/deep-learning-v2-pytorch
|
d5c8d3a1b40bab99672259dc0a2136ee6d8b23f2
|
[
"MIT"
] | null | null | null |
intro-to-pytorch/Part 4 - Fashion-MNIST (Exercises).ipynb
|
Arush0113/deep-learning-v2-pytorch
|
d5c8d3a1b40bab99672259dc0a2136ee6d8b23f2
|
[
"MIT"
] | null | null | null |
intro-to-pytorch/Part 4 - Fashion-MNIST (Exercises).ipynb
|
Arush0113/deep-learning-v2-pytorch
|
d5c8d3a1b40bab99672259dc0a2136ee6d8b23f2
|
[
"MIT"
] | null | null | null | 139.540441 | 24,948 | 0.869609 |
[
[
[
"# Classifying Fashion-MNIST\n\nNow it's your turn to build and train a neural network. You'll be using the [Fashion-MNIST dataset](https://github.com/zalandoresearch/fashion-mnist), a drop-in replacement for the MNIST dataset. MNIST is actually quite trivial with neural networks where you can easily achieve better than 97% accuracy. Fashion-MNIST is a set of 28x28 greyscale images of clothes. It's more complex than MNIST, so it's a better representation of the actual performance of your network, and a better representation of datasets you'll use in the real world.\n\n<img src='assets/fashion-mnist-sprite.png' width=500px>\n\nIn this notebook, you'll build your own neural network. For the most part, you could just copy and paste the code from Part 3, but you wouldn't be learning. It's important for you to write the code yourself and get it to work. Feel free to consult the previous notebooks though as you work through this.\n\nFirst off, let's load the dataset through torchvision.",
"_____no_output_____"
]
],
[
[
"import torch\nfrom torchvision import datasets, transforms\nimport helper\n\n# Define a transform to normalize the data\ntransform = transforms.Compose([transforms.ToTensor(),\n transforms.Normalize((0.5,), (0.5,))])\n# Download and load the training data\ntrainset = datasets.FashionMNIST('~/.pytorch/F_MNIST_data/', download=True, train=True, transform=transform)\ntrainloader = torch.utils.data.DataLoader(trainset, batch_size=64, shuffle=True)\n\n# Download and load the test data\ntestset = datasets.FashionMNIST('~/.pytorch/F_MNIST_data/', download=True, train=False, transform=transform)\ntestloader = torch.utils.data.DataLoader(testset, batch_size=64, shuffle=True)",
"_____no_output_____"
]
],
[
[
"Here we can see one of the images.",
"_____no_output_____"
]
],
[
[
"image, label = next(iter(trainloader))\nhelper.imshow(image[0,:]);",
"_____no_output_____"
]
],
[
[
"## Building the network\n\nHere you should define your network. As with MNIST, each image is 28x28 which is a total of 784 pixels, and there are 10 classes. You should include at least one hidden layer. We suggest you use ReLU activations for the layers and to return the logits or log-softmax from the forward pass. It's up to you how many layers you add and the size of those layers.",
"_____no_output_____"
]
],
[
[
"# TODO: Define your network architecture here\nfrom torch import nn\nimport torch.nn.functional as F\n# model = nn.Sequential(nn.Linear(784, 256),\n# nn.ReLU(),\n# nn.Linear(256, 64),\n# nn.ReLU(),\n# nn.Linear(64, 10),\n# # nn.LogSoftmax(dim = 1),\n# )\n\nclass Network(nn.Module):\n def __init__(self):\n super().__init__()\n self.hidden1 = nn.Linear(784, 256)\n self.hidden2 = nn.Linear(256, 128)\n self.hidden3 = nn.Linear(128, 64)\n self.output = nn.Linear(64, 10)\n \n def forward(self, x):\n x = F.relu(self.hidden1(x))\n x = F.relu(self.hidden2(x))\n x = F.relu(self.hidden3(x))\n x = F.log_softmax(self.output(x), dim = 1)\n \n return x",
"_____no_output_____"
]
],
[
[
"# Train the network\n\nNow you should create your network and train it. First you'll want to define [the criterion](http://pytorch.org/docs/master/nn.html#loss-functions) ( something like `nn.CrossEntropyLoss`) and [the optimizer](http://pytorch.org/docs/master/optim.html) (typically `optim.SGD` or `optim.Adam`).\n\nThen write the training code. Remember the training pass is a fairly straightforward process:\n\n* Make a forward pass through the network to get the logits \n* Use the logits to calculate the loss\n* Perform a backward pass through the network with `loss.backward()` to calculate the gradients\n* Take a step with the optimizer to update the weights\n\nBy adjusting the hyperparameters (hidden units, learning rate, etc), you should be able to get the training loss below 0.4.",
"_____no_output_____"
]
],
[
[
"# TODO: Create the network, define the criterion and optimizer\nfrom torch import optim\nmodel = Network()\ncriterion = nn.CrossEntropyLoss()\noptimizer = optim.Adam(model.parameters(), lr = 0.01)\n",
"_____no_output_____"
],
[
"# TODO: Train the network here\n# dataiter = iter(trainloader)\n# images, labels = dataiter.next()\nprint(images.shape)\n\n# print(images.shape)\n\nepochs = 5\nfor i in range(epochs):\n running_loss = 0\n for images, labels in trainloader:\n images = images.view(images.shape[0], -1)\n output = model(images)\n loss = criterion(output, labels)\n optimizer.zero_grad()\n loss.backward()\n # optimizer.zero_grad()\n optimizer.step()\n running_loss += loss.item()\n# print(loss.item())\n else:\n# print(len(trainloader))\n print(f\"The training loss of {i+1}th epoch is: {running_loss}\")",
"torch.Size([64, 784])\nThe training loss of 1th epoch is: 542.2710281908512\nThe training loss of 2th epoch is: 439.61831034719944\nThe training loss of 3th epoch is: 402.98729169368744\nThe training loss of 4th epoch is: 402.6209799796343\nThe training loss of 5th epoch is: 377.83217369019985\n"
],
[
"%matplotlib inline\n%config InlineBackend.figure_format = 'retina'\n\nimport helper\n\n# Test out your network!\n# model = Network()\n\ndataiter = iter(testloader)\nimages, labels = dataiter.next()\nimg = images[0]\n# Convert 2D image to 1D vector\nimg = img.resize_(1, 784)\n\n# TODO: Calculate the class probabilities (softmax) for img\nps = torch.exp(model(img))\nprint(ps)\n\n# Plot the image and probabilities\nhelper.view_classify(img.resize_(1, 28, 28), ps, version='Fashion')",
"tensor([[5.5068e-05, 1.3520e-10, 3.6068e-07, 5.9116e-07, 2.0650e-07, 8.7346e-03,\n 5.4973e-05, 3.1731e-01, 5.1315e-03, 6.6871e-01]],\n grad_fn=<ExpBackward>)\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
4a588e309ca1c6175dcd8eaf71722d1ea8258240
| 379,847 |
ipynb
|
Jupyter Notebook
|
Ejercicio_3,_Perceptrón.ipynb
|
HenrryCordovillo/Redes_Neuronales_con_Python
|
a2d99aaaa317f4ea15cdbee38a61532ceb899dcc
|
[
"Apache-2.0"
] | null | null | null |
Ejercicio_3,_Perceptrón.ipynb
|
HenrryCordovillo/Redes_Neuronales_con_Python
|
a2d99aaaa317f4ea15cdbee38a61532ceb899dcc
|
[
"Apache-2.0"
] | null | null | null |
Ejercicio_3,_Perceptrón.ipynb
|
HenrryCordovillo/Redes_Neuronales_con_Python
|
a2d99aaaa317f4ea15cdbee38a61532ceb899dcc
|
[
"Apache-2.0"
] | null | null | null | 72.670174 | 9,286 | 0.42822 |
[
[
[
"<a href=\"https://colab.research.google.com/github/HenrryCordovillo/Redes_Neuronales_con_Python/blob/main/Ejercicio_3%2C_Perceptr%C3%B3n.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"import numpy as np\r\nfrom keras.models import Sequential\r\nfrom keras.layers.core import Dense\r\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"# cargamos las 4 combinaciones de las compuertas AND\r\ndatos_entrenamiento = np.array([[0,0],[0,1],[1,0],[1,1]], \"float32\")\r\n \r\n# y estos son los resultados que se obtienen, en el mismo orden\r\ndatos_etiquetas = np.array([[0],[0],[0],[1]], \"float32\")",
"_____no_output_____"
],
[
"x=datos_entrenamiento[:,0]\r\ny=datos_entrenamiento[:,1]\r\ncolors = datos_etiquetas\r\nplt.scatter(x,y,s=100,c=colors)\r\nplt.xlabel(\"Eje x\")\r\nplt.ylabel(\"Eje y\")\r\nplt.title(\"Grafica Datos a clasificar\")\r\nplt.show()",
"_____no_output_____"
],
[
"#Modelo para AND\r\nmodelo = Sequential()\r\nmodelo.add(Dense(1, input_dim=2, activation='relu'))\r\n\r\nmodelo.compile(loss='mean_squared_error',\r\n optimizer='adam',\r\n metrics=['binary_accuracy'])\r\n \r\nmodelo.fit(datos_entrenamiento, datos_etiquetas, epochs=800)",
"Epoch 1/800\n1/1 [==============================] - 0s 290ms/step - loss: 0.5812 - binary_accuracy: 0.7500\nEpoch 2/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5785 - binary_accuracy: 0.7500\nEpoch 3/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5759 - binary_accuracy: 0.7500\nEpoch 4/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5732 - binary_accuracy: 0.7500\nEpoch 5/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5706 - binary_accuracy: 0.7500\nEpoch 6/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5679 - binary_accuracy: 0.7500\nEpoch 7/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5653 - binary_accuracy: 0.7500\nEpoch 8/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5627 - binary_accuracy: 0.7500\nEpoch 9/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5601 - binary_accuracy: 0.7500\nEpoch 10/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5575 - binary_accuracy: 0.7500\nEpoch 11/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5549 - binary_accuracy: 0.7500\nEpoch 12/800\n1/1 [==============================] - 0s 3ms/step - loss: 0.5523 - binary_accuracy: 0.7500\nEpoch 13/800\n1/1 [==============================] - 0s 3ms/step - loss: 0.5498 - binary_accuracy: 0.7500\nEpoch 14/800\n1/1 [==============================] - 0s 3ms/step - loss: 0.5472 - binary_accuracy: 0.7500\nEpoch 15/800\n1/1 [==============================] - 0s 3ms/step - loss: 0.5447 - binary_accuracy: 0.7500\nEpoch 16/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5421 - binary_accuracy: 0.7500\nEpoch 17/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5396 - binary_accuracy: 0.7500\nEpoch 18/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5371 - binary_accuracy: 0.7500\nEpoch 19/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5346 - binary_accuracy: 0.7500\nEpoch 20/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5321 - binary_accuracy: 0.7500\nEpoch 21/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5296 - binary_accuracy: 0.7500\nEpoch 22/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5271 - binary_accuracy: 0.7500\nEpoch 23/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5247 - binary_accuracy: 0.7500\nEpoch 24/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5222 - binary_accuracy: 0.7500\nEpoch 25/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5198 - binary_accuracy: 0.7500\nEpoch 26/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5173 - binary_accuracy: 0.7500\nEpoch 27/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5149 - binary_accuracy: 0.7500\nEpoch 28/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5125 - binary_accuracy: 0.7500\nEpoch 29/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5101 - binary_accuracy: 0.7500\nEpoch 30/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5077 - binary_accuracy: 0.7500\nEpoch 31/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.5054 - binary_accuracy: 0.7500\nEpoch 32/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5030 - binary_accuracy: 0.7500\nEpoch 33/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5007 - binary_accuracy: 0.7500\nEpoch 34/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.4983 - binary_accuracy: 0.7500\nEpoch 35/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.4960 - binary_accuracy: 0.7500\nEpoch 36/800\n1/1 [==============================] - 0s 12ms/step - loss: 0.4937 - binary_accuracy: 0.7500\nEpoch 37/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.4914 - binary_accuracy: 0.7500\nEpoch 38/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.4891 - binary_accuracy: 0.7500\nEpoch 39/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.4868 - binary_accuracy: 0.7500\nEpoch 40/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.4845 - binary_accuracy: 0.7500\nEpoch 41/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.4823 - binary_accuracy: 0.7500\nEpoch 42/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.4800 - binary_accuracy: 0.7500\nEpoch 43/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.4778 - binary_accuracy: 0.7500\nEpoch 44/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.4756 - binary_accuracy: 0.7500\nEpoch 45/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.4734 - binary_accuracy: 0.7500\nEpoch 46/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.4712 - binary_accuracy: 0.7500\nEpoch 47/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.4690 - binary_accuracy: 0.7500\nEpoch 48/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.4668 - binary_accuracy: 0.7500\nEpoch 49/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.4646 - binary_accuracy: 0.7500\nEpoch 50/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.4625 - binary_accuracy: 0.7500\nEpoch 51/800\n1/1 [==============================] - 0s 3ms/step - loss: 0.4603 - binary_accuracy: 0.7500\nEpoch 52/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.4582 - binary_accuracy: 0.7500\nEpoch 53/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.4561 - binary_accuracy: 0.7500\nEpoch 54/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.4540 - binary_accuracy: 0.7500\nEpoch 55/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.4519 - binary_accuracy: 0.7500\nEpoch 56/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.4498 - binary_accuracy: 0.7500\nEpoch 57/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.4477 - binary_accuracy: 0.7500\nEpoch 58/800\n1/1 [==============================] - 0s 3ms/step - loss: 0.4456 - binary_accuracy: 0.7500\nEpoch 59/800\n1/1 [==============================] - 0s 3ms/step - loss: 0.4436 - binary_accuracy: 0.7500\nEpoch 60/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.4415 - binary_accuracy: 0.7500\nEpoch 61/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.4395 - binary_accuracy: 0.7500\nEpoch 62/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.4375 - binary_accuracy: 0.7500\nEpoch 63/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.4355 - binary_accuracy: 0.7500\nEpoch 64/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.4335 - binary_accuracy: 0.7500\nEpoch 65/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.4315 - binary_accuracy: 0.7500\nEpoch 66/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.4295 - binary_accuracy: 0.7500\nEpoch 67/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.4276 - binary_accuracy: 0.7500\nEpoch 68/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.4256 - binary_accuracy: 0.7500\nEpoch 69/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.4236 - binary_accuracy: 0.7500\nEpoch 70/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.4217 - binary_accuracy: 0.7500\nEpoch 71/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.4198 - binary_accuracy: 0.7500\nEpoch 72/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.4179 - binary_accuracy: 0.7500\nEpoch 73/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.4160 - binary_accuracy: 0.7500\nEpoch 74/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.4141 - binary_accuracy: 0.7500\nEpoch 75/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.4122 - binary_accuracy: 0.7500\nEpoch 76/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.4103 - binary_accuracy: 0.7500\nEpoch 77/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.4085 - binary_accuracy: 0.7500\nEpoch 78/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.4066 - binary_accuracy: 0.7500\nEpoch 79/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.4048 - binary_accuracy: 0.7500\nEpoch 80/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.4029 - binary_accuracy: 0.7500\nEpoch 81/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.4011 - binary_accuracy: 0.7500\nEpoch 82/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3993 - binary_accuracy: 0.7500\nEpoch 83/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.3975 - binary_accuracy: 0.7500\nEpoch 84/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.3957 - binary_accuracy: 0.7500\nEpoch 85/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.3939 - binary_accuracy: 0.7500\nEpoch 86/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3922 - binary_accuracy: 0.7500\nEpoch 87/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.3904 - binary_accuracy: 0.7500\nEpoch 88/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3887 - binary_accuracy: 0.7500\nEpoch 89/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3869 - binary_accuracy: 0.7500\nEpoch 90/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3852 - binary_accuracy: 0.7500\nEpoch 91/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3835 - binary_accuracy: 0.7500\nEpoch 92/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.3818 - binary_accuracy: 0.7500\nEpoch 93/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3801 - binary_accuracy: 0.7500\nEpoch 94/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.3784 - binary_accuracy: 0.7500\nEpoch 95/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3767 - binary_accuracy: 0.7500\nEpoch 96/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.3750 - binary_accuracy: 0.7500\nEpoch 97/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3734 - binary_accuracy: 0.7500\nEpoch 98/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3717 - binary_accuracy: 0.7500\nEpoch 99/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3701 - binary_accuracy: 0.7500\nEpoch 100/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3684 - binary_accuracy: 0.7500\nEpoch 101/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3668 - binary_accuracy: 0.7500\nEpoch 102/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.3652 - binary_accuracy: 0.7500\nEpoch 103/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3636 - binary_accuracy: 0.7500\nEpoch 104/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.3620 - binary_accuracy: 0.7500\nEpoch 105/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.3604 - binary_accuracy: 0.7500\nEpoch 106/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3588 - binary_accuracy: 0.7500\nEpoch 107/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.3573 - binary_accuracy: 0.7500\nEpoch 108/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3557 - binary_accuracy: 0.7500\nEpoch 109/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3542 - binary_accuracy: 0.7500\nEpoch 110/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3526 - binary_accuracy: 0.7500\nEpoch 111/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3511 - binary_accuracy: 0.7500\nEpoch 112/800\n1/1 [==============================] - 0s 3ms/step - loss: 0.3496 - binary_accuracy: 0.7500\nEpoch 113/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.3481 - binary_accuracy: 0.7500\nEpoch 114/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3466 - binary_accuracy: 0.7500\nEpoch 115/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.3451 - binary_accuracy: 0.7500\nEpoch 116/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3436 - binary_accuracy: 0.7500\nEpoch 117/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3421 - binary_accuracy: 0.7500\nEpoch 118/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.3406 - binary_accuracy: 0.7500\nEpoch 119/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3392 - binary_accuracy: 0.7500\nEpoch 120/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.3377 - binary_accuracy: 0.7500\nEpoch 121/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.3363 - binary_accuracy: 0.7500\nEpoch 122/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3348 - binary_accuracy: 0.7500\nEpoch 123/800\n1/1 [==============================] - 0s 3ms/step - loss: 0.3334 - binary_accuracy: 0.7500\nEpoch 124/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.3320 - binary_accuracy: 0.7500\nEpoch 125/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.3306 - binary_accuracy: 0.7500\nEpoch 126/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.3292 - binary_accuracy: 0.7500\nEpoch 127/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.3278 - binary_accuracy: 0.7500\nEpoch 128/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3264 - binary_accuracy: 0.7500\nEpoch 129/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.3250 - binary_accuracy: 0.7500\nEpoch 130/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.3236 - binary_accuracy: 0.7500\nEpoch 131/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.3223 - binary_accuracy: 0.7500\nEpoch 132/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.3209 - binary_accuracy: 0.7500\nEpoch 133/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.3196 - binary_accuracy: 0.7500\nEpoch 134/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.3183 - binary_accuracy: 0.7500\nEpoch 135/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3169 - binary_accuracy: 0.7500\nEpoch 136/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3156 - binary_accuracy: 0.7500\nEpoch 137/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.3143 - binary_accuracy: 0.7500\nEpoch 138/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3130 - binary_accuracy: 0.7500\nEpoch 139/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.3117 - binary_accuracy: 0.7500\nEpoch 140/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3104 - binary_accuracy: 0.7500\nEpoch 141/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3091 - binary_accuracy: 0.7500\nEpoch 142/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.3078 - binary_accuracy: 0.7500\nEpoch 143/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.3066 - binary_accuracy: 0.7500\nEpoch 144/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.3053 - binary_accuracy: 0.7500\nEpoch 145/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.3041 - binary_accuracy: 0.7500\nEpoch 146/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3028 - binary_accuracy: 0.7500\nEpoch 147/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3016 - binary_accuracy: 0.7500\nEpoch 148/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.3004 - binary_accuracy: 0.7500\nEpoch 149/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.2992 - binary_accuracy: 0.7500\nEpoch 150/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.2979 - binary_accuracy: 0.7500\nEpoch 151/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.2967 - binary_accuracy: 0.7500\nEpoch 152/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.2955 - binary_accuracy: 0.7500\nEpoch 153/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.2943 - binary_accuracy: 0.7500\nEpoch 154/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.2932 - binary_accuracy: 0.7500\nEpoch 155/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.2920 - binary_accuracy: 0.7500\nEpoch 156/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.2908 - binary_accuracy: 0.7500\nEpoch 157/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.2897 - binary_accuracy: 0.7500\nEpoch 158/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.2885 - binary_accuracy: 0.7500\nEpoch 159/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.2873 - binary_accuracy: 0.7500\nEpoch 160/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.2862 - binary_accuracy: 0.7500\nEpoch 161/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.2851 - binary_accuracy: 0.7500\nEpoch 162/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.2839 - binary_accuracy: 0.7500\nEpoch 163/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.2828 - binary_accuracy: 0.7500\nEpoch 164/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.2817 - binary_accuracy: 0.7500\nEpoch 165/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.2806 - binary_accuracy: 0.7500\nEpoch 166/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.2795 - binary_accuracy: 0.7500\nEpoch 167/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.2784 - binary_accuracy: 0.7500\nEpoch 168/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.2773 - binary_accuracy: 0.7500\nEpoch 169/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.2762 - binary_accuracy: 0.7500\nEpoch 170/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.2752 - binary_accuracy: 0.7500\nEpoch 171/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.2741 - binary_accuracy: 0.7500\nEpoch 172/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.2730 - binary_accuracy: 0.7500\nEpoch 173/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.2720 - binary_accuracy: 0.7500\nEpoch 174/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.2709 - binary_accuracy: 0.7500\nEpoch 175/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.2699 - binary_accuracy: 0.7500\nEpoch 176/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.2689 - binary_accuracy: 0.7500\nEpoch 177/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.2678 - binary_accuracy: 0.7500\nEpoch 178/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.2668 - binary_accuracy: 0.7500\nEpoch 179/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.2658 - binary_accuracy: 0.7500\nEpoch 180/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.2648 - binary_accuracy: 0.7500\nEpoch 181/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.2638 - binary_accuracy: 0.7500\nEpoch 182/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.2628 - binary_accuracy: 0.7500\nEpoch 183/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.2618 - binary_accuracy: 0.7500\nEpoch 184/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.2608 - binary_accuracy: 0.7500\nEpoch 185/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.2598 - binary_accuracy: 0.7500\nEpoch 186/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.2589 - binary_accuracy: 0.7500\nEpoch 187/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.2579 - binary_accuracy: 0.7500\nEpoch 188/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.2569 - binary_accuracy: 0.7500\nEpoch 189/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.2560 - binary_accuracy: 0.7500\nEpoch 190/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.2550 - binary_accuracy: 0.7500\nEpoch 191/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.2541 - binary_accuracy: 0.7500\nEpoch 192/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.2531 - binary_accuracy: 0.7500\nEpoch 193/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.2522 - binary_accuracy: 0.7500\nEpoch 194/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.2513 - binary_accuracy: 0.7500\nEpoch 195/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.2504 - binary_accuracy: 0.7500\nEpoch 196/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.2494 - binary_accuracy: 0.7500\nEpoch 197/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.2485 - binary_accuracy: 0.7500\nEpoch 198/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.2476 - binary_accuracy: 0.7500\nEpoch 199/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.2467 - binary_accuracy: 0.7500\nEpoch 200/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.2458 - binary_accuracy: 0.7500\nEpoch 201/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.2450 - binary_accuracy: 0.7500\nEpoch 202/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.2441 - binary_accuracy: 0.7500\nEpoch 203/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.2432 - binary_accuracy: 0.7500\nEpoch 204/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.2423 - binary_accuracy: 0.7500\nEpoch 205/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.2415 - binary_accuracy: 0.7500\nEpoch 206/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.2406 - binary_accuracy: 0.7500\nEpoch 207/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.2397 - binary_accuracy: 0.7500\nEpoch 208/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.2389 - binary_accuracy: 0.7500\nEpoch 209/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.2380 - binary_accuracy: 0.7500\nEpoch 210/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.2372 - binary_accuracy: 0.7500\nEpoch 211/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.2364 - binary_accuracy: 0.7500\nEpoch 212/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.2355 - binary_accuracy: 0.7500\nEpoch 213/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.2347 - binary_accuracy: 0.7500\nEpoch 214/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.2339 - binary_accuracy: 0.7500\nEpoch 215/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.2331 - binary_accuracy: 0.7500\nEpoch 216/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.2323 - binary_accuracy: 0.7500\nEpoch 217/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.2315 - binary_accuracy: 0.7500\nEpoch 218/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.2307 - binary_accuracy: 0.7500\nEpoch 219/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.2299 - binary_accuracy: 0.7500\nEpoch 220/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.2291 - binary_accuracy: 0.7500\nEpoch 221/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.2283 - binary_accuracy: 0.7500\nEpoch 222/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.2275 - binary_accuracy: 0.7500\nEpoch 223/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.2267 - binary_accuracy: 0.7500\nEpoch 224/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.2259 - binary_accuracy: 0.7500\nEpoch 225/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.2252 - binary_accuracy: 0.7500\nEpoch 226/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.2244 - binary_accuracy: 0.7500\nEpoch 227/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.2237 - binary_accuracy: 0.7500\nEpoch 228/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.2229 - binary_accuracy: 0.7500\nEpoch 229/800\n1/1 [==============================] - 0s 15ms/step - loss: 0.2221 - binary_accuracy: 0.7500\nEpoch 230/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.2214 - binary_accuracy: 0.7500\nEpoch 231/800\n1/1 [==============================] - 0s 11ms/step - loss: 0.2207 - binary_accuracy: 0.7500\nEpoch 232/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.2199 - binary_accuracy: 0.7500\nEpoch 233/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.2192 - binary_accuracy: 0.7500\nEpoch 234/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.2184 - binary_accuracy: 0.7500\nEpoch 235/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.2177 - binary_accuracy: 0.7500\nEpoch 236/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.2170 - binary_accuracy: 0.7500\nEpoch 237/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.2163 - binary_accuracy: 0.7500\nEpoch 238/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.2155 - binary_accuracy: 0.7500\nEpoch 239/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.2148 - binary_accuracy: 0.7500\nEpoch 240/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.2141 - binary_accuracy: 0.7500\nEpoch 241/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.2134 - binary_accuracy: 0.7500\nEpoch 242/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.2127 - binary_accuracy: 0.7500\nEpoch 243/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.2120 - binary_accuracy: 0.7500\nEpoch 244/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.2113 - binary_accuracy: 0.7500\nEpoch 245/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.2106 - binary_accuracy: 0.7500\nEpoch 246/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.2099 - binary_accuracy: 0.7500\nEpoch 247/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.2092 - binary_accuracy: 0.7500\nEpoch 248/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.2086 - binary_accuracy: 0.7500\nEpoch 249/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.2079 - binary_accuracy: 0.7500\nEpoch 250/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.2072 - binary_accuracy: 0.7500\nEpoch 251/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.2065 - binary_accuracy: 0.7500\nEpoch 252/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.2058 - binary_accuracy: 0.7500\nEpoch 253/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.2052 - binary_accuracy: 0.7500\nEpoch 254/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.2045 - binary_accuracy: 0.7500\nEpoch 255/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.2039 - binary_accuracy: 0.7500\nEpoch 256/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.2032 - binary_accuracy: 0.7500\nEpoch 257/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.2025 - binary_accuracy: 0.7500\nEpoch 258/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.2019 - binary_accuracy: 0.7500\nEpoch 259/800\n1/1 [==============================] - 0s 12ms/step - loss: 0.2012 - binary_accuracy: 0.7500\nEpoch 260/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.2006 - binary_accuracy: 0.7500\nEpoch 261/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.2000 - binary_accuracy: 0.7500\nEpoch 262/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1993 - binary_accuracy: 0.7500\nEpoch 263/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1987 - binary_accuracy: 0.7500\nEpoch 264/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1980 - binary_accuracy: 0.7500\nEpoch 265/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1974 - binary_accuracy: 0.7500\nEpoch 266/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1968 - binary_accuracy: 0.7500\nEpoch 267/800\n1/1 [==============================] - 0s 15ms/step - loss: 0.1962 - binary_accuracy: 0.7500\nEpoch 268/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.1955 - binary_accuracy: 0.7500\nEpoch 269/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.1949 - binary_accuracy: 0.7500\nEpoch 270/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.1943 - binary_accuracy: 0.7500\nEpoch 271/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1937 - binary_accuracy: 0.7500\nEpoch 272/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.1931 - binary_accuracy: 0.7500\nEpoch 273/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.1925 - binary_accuracy: 0.7500\nEpoch 274/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1919 - binary_accuracy: 0.7500\nEpoch 275/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1913 - binary_accuracy: 0.7500\nEpoch 276/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1907 - binary_accuracy: 0.7500\nEpoch 277/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1901 - binary_accuracy: 0.7500\nEpoch 278/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1895 - binary_accuracy: 0.7500\nEpoch 279/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1889 - binary_accuracy: 0.7500\nEpoch 280/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.1883 - binary_accuracy: 0.7500\nEpoch 281/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.1877 - binary_accuracy: 0.7500\nEpoch 282/800\n1/1 [==============================] - 0s 14ms/step - loss: 0.1871 - binary_accuracy: 0.7500\nEpoch 283/800\n1/1 [==============================] - 0s 17ms/step - loss: 0.1866 - binary_accuracy: 0.7500\nEpoch 284/800\n1/1 [==============================] - 0s 13ms/step - loss: 0.1860 - binary_accuracy: 0.7500\nEpoch 285/800\n1/1 [==============================] - 0s 14ms/step - loss: 0.1854 - binary_accuracy: 0.7500\nEpoch 286/800\n1/1 [==============================] - 0s 15ms/step - loss: 0.1849 - binary_accuracy: 0.7500\nEpoch 287/800\n1/1 [==============================] - 0s 14ms/step - loss: 0.1843 - binary_accuracy: 0.7500\nEpoch 288/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.1837 - binary_accuracy: 0.7500\nEpoch 289/800\n1/1 [==============================] - 0s 12ms/step - loss: 0.1832 - binary_accuracy: 0.7500\nEpoch 290/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1826 - binary_accuracy: 0.7500\nEpoch 291/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1820 - binary_accuracy: 0.7500\nEpoch 292/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1815 - binary_accuracy: 0.7500\nEpoch 293/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.1809 - binary_accuracy: 0.7500\nEpoch 294/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1804 - binary_accuracy: 0.7500\nEpoch 295/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.1798 - binary_accuracy: 0.7500\nEpoch 296/800\n1/1 [==============================] - 0s 3ms/step - loss: 0.1793 - binary_accuracy: 0.7500\nEpoch 297/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.1788 - binary_accuracy: 0.7500\nEpoch 298/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.1782 - binary_accuracy: 0.7500\nEpoch 299/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.1777 - binary_accuracy: 0.7500\nEpoch 300/800\n1/1 [==============================] - 0s 16ms/step - loss: 0.1772 - binary_accuracy: 0.7500\nEpoch 301/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1766 - binary_accuracy: 0.7500\nEpoch 302/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1761 - binary_accuracy: 0.7500\nEpoch 303/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1756 - binary_accuracy: 0.7500\nEpoch 304/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1751 - binary_accuracy: 0.7500\nEpoch 305/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.1745 - binary_accuracy: 0.7500\nEpoch 306/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1740 - binary_accuracy: 0.7500\nEpoch 307/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1735 - binary_accuracy: 0.7500\nEpoch 308/800\n1/1 [==============================] - 0s 16ms/step - loss: 0.1730 - binary_accuracy: 0.7500\nEpoch 309/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1725 - binary_accuracy: 0.7500\nEpoch 310/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.1720 - binary_accuracy: 0.7500\nEpoch 311/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.1715 - binary_accuracy: 0.7500\nEpoch 312/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1710 - binary_accuracy: 0.7500\nEpoch 313/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1705 - binary_accuracy: 0.7500\nEpoch 314/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1700 - binary_accuracy: 0.7500\nEpoch 315/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1695 - binary_accuracy: 0.7500\nEpoch 316/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1690 - binary_accuracy: 0.7500\nEpoch 317/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1685 - binary_accuracy: 0.7500\nEpoch 318/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1680 - binary_accuracy: 0.7500\nEpoch 319/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1675 - binary_accuracy: 0.7500\nEpoch 320/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.1670 - binary_accuracy: 0.7500\nEpoch 321/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1665 - binary_accuracy: 0.7500\nEpoch 322/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1661 - binary_accuracy: 0.7500\nEpoch 323/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.1656 - binary_accuracy: 0.7500\nEpoch 324/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1651 - binary_accuracy: 0.7500\nEpoch 325/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1646 - binary_accuracy: 0.7500\nEpoch 326/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1642 - binary_accuracy: 0.7500\nEpoch 327/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1637 - binary_accuracy: 0.7500\nEpoch 328/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1632 - binary_accuracy: 0.7500\nEpoch 329/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.1628 - binary_accuracy: 0.7500\nEpoch 330/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.1623 - binary_accuracy: 0.7500\nEpoch 331/800\n1/1 [==============================] - 0s 13ms/step - loss: 0.1618 - binary_accuracy: 0.7500\nEpoch 332/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.1614 - binary_accuracy: 0.7500\nEpoch 333/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1609 - binary_accuracy: 0.7500\nEpoch 334/800\n1/1 [==============================] - 0s 11ms/step - loss: 0.1605 - binary_accuracy: 0.7500\nEpoch 335/800\n1/1 [==============================] - 0s 12ms/step - loss: 0.1600 - binary_accuracy: 0.7500\nEpoch 336/800\n1/1 [==============================] - 0s 17ms/step - loss: 0.1596 - binary_accuracy: 0.7500\nEpoch 337/800\n1/1 [==============================] - 0s 11ms/step - loss: 0.1591 - binary_accuracy: 0.7500\nEpoch 338/800\n1/1 [==============================] - 0s 11ms/step - loss: 0.1587 - binary_accuracy: 0.7500\nEpoch 339/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.1582 - binary_accuracy: 0.7500\nEpoch 340/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.1578 - binary_accuracy: 0.7500\nEpoch 341/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.1573 - binary_accuracy: 0.7500\nEpoch 342/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.1569 - binary_accuracy: 0.7500\nEpoch 343/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.1565 - binary_accuracy: 0.7500\nEpoch 344/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.1560 - binary_accuracy: 0.7500\nEpoch 345/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1556 - binary_accuracy: 0.7500\nEpoch 346/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1552 - binary_accuracy: 0.7500\nEpoch 347/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1547 - binary_accuracy: 0.7500\nEpoch 348/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1543 - binary_accuracy: 0.7500\nEpoch 349/800\n1/1 [==============================] - 0s 11ms/step - loss: 0.1539 - binary_accuracy: 0.7500\nEpoch 350/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.1535 - binary_accuracy: 0.7500\nEpoch 351/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1531 - binary_accuracy: 0.7500\nEpoch 352/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1526 - binary_accuracy: 0.7500\nEpoch 353/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1522 - binary_accuracy: 0.7500\nEpoch 354/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1518 - binary_accuracy: 0.7500\nEpoch 355/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1514 - binary_accuracy: 0.7500\nEpoch 356/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.1510 - binary_accuracy: 0.7500\nEpoch 357/800\n1/1 [==============================] - 0s 3ms/step - loss: 0.1506 - binary_accuracy: 0.7500\nEpoch 358/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.1502 - binary_accuracy: 0.7500\nEpoch 359/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1498 - binary_accuracy: 0.7500\nEpoch 360/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1494 - binary_accuracy: 0.7500\nEpoch 361/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1490 - binary_accuracy: 0.7500\nEpoch 362/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1486 - binary_accuracy: 0.7500\nEpoch 363/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.1482 - binary_accuracy: 0.7500\nEpoch 364/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.1478 - binary_accuracy: 0.7500\nEpoch 365/800\n1/1 [==============================] - 0s 12ms/step - loss: 0.1474 - binary_accuracy: 0.7500\nEpoch 366/800\n1/1 [==============================] - 0s 13ms/step - loss: 0.1470 - binary_accuracy: 0.7500\nEpoch 367/800\n1/1 [==============================] - 0s 11ms/step - loss: 0.1466 - binary_accuracy: 0.7500\nEpoch 368/800\n1/1 [==============================] - 0s 12ms/step - loss: 0.1462 - binary_accuracy: 0.7500\nEpoch 369/800\n1/1 [==============================] - 0s 11ms/step - loss: 0.1458 - binary_accuracy: 0.7500\nEpoch 370/800\n1/1 [==============================] - 0s 13ms/step - loss: 0.1454 - binary_accuracy: 0.7500\nEpoch 371/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.1450 - binary_accuracy: 0.7500\nEpoch 372/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.1446 - binary_accuracy: 0.7500\nEpoch 373/800\n1/1 [==============================] - 0s 12ms/step - loss: 0.1443 - binary_accuracy: 0.7500\nEpoch 374/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.1439 - binary_accuracy: 0.7500\nEpoch 375/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.1435 - binary_accuracy: 0.7500\nEpoch 376/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1431 - binary_accuracy: 0.7500\nEpoch 377/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1428 - binary_accuracy: 0.7500\nEpoch 378/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.1424 - binary_accuracy: 0.7500\nEpoch 379/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.1420 - binary_accuracy: 0.7500\nEpoch 380/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1416 - binary_accuracy: 0.7500\nEpoch 381/800\n1/1 [==============================] - 0s 3ms/step - loss: 0.1413 - binary_accuracy: 0.7500\nEpoch 382/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.1409 - binary_accuracy: 0.7500\nEpoch 383/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.1405 - binary_accuracy: 0.7500\nEpoch 384/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.1402 - binary_accuracy: 0.7500\nEpoch 385/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1398 - binary_accuracy: 0.7500\nEpoch 386/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.1395 - binary_accuracy: 0.7500\nEpoch 387/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1391 - binary_accuracy: 0.7500\nEpoch 388/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.1387 - binary_accuracy: 0.7500\nEpoch 389/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1384 - binary_accuracy: 0.7500\nEpoch 390/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1380 - binary_accuracy: 0.7500\nEpoch 391/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1377 - binary_accuracy: 0.7500\nEpoch 392/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.1373 - binary_accuracy: 0.7500\nEpoch 393/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1370 - binary_accuracy: 0.7500\nEpoch 394/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.1366 - binary_accuracy: 0.7500\nEpoch 395/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.1363 - binary_accuracy: 0.7500\nEpoch 396/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1359 - binary_accuracy: 0.7500\nEpoch 397/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1356 - binary_accuracy: 0.7500\nEpoch 398/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1352 - binary_accuracy: 0.7500\nEpoch 399/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1349 - binary_accuracy: 0.7500\nEpoch 400/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1346 - binary_accuracy: 0.7500\nEpoch 401/800\n1/1 [==============================] - 0s 12ms/step - loss: 0.1342 - binary_accuracy: 0.7500\nEpoch 402/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1339 - binary_accuracy: 0.7500\nEpoch 403/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1335 - binary_accuracy: 0.7500\nEpoch 404/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1332 - binary_accuracy: 0.7500\nEpoch 405/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1329 - binary_accuracy: 0.7500\nEpoch 406/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1325 - binary_accuracy: 0.7500\nEpoch 407/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1322 - binary_accuracy: 0.7500\nEpoch 408/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1319 - binary_accuracy: 0.7500\nEpoch 409/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1316 - binary_accuracy: 0.7500\nEpoch 410/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1312 - binary_accuracy: 0.7500\nEpoch 411/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1309 - binary_accuracy: 0.7500\nEpoch 412/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1306 - binary_accuracy: 0.7500\nEpoch 413/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1303 - binary_accuracy: 0.7500\nEpoch 414/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1299 - binary_accuracy: 0.7500\nEpoch 415/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1296 - binary_accuracy: 0.7500\nEpoch 416/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1293 - binary_accuracy: 0.7500\nEpoch 417/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1290 - binary_accuracy: 0.7500\nEpoch 418/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1287 - binary_accuracy: 0.7500\nEpoch 419/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.1283 - binary_accuracy: 0.7500\nEpoch 420/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1280 - binary_accuracy: 0.7500\nEpoch 421/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1277 - binary_accuracy: 0.7500\nEpoch 422/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1274 - binary_accuracy: 0.7500\nEpoch 423/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1271 - binary_accuracy: 0.7500\nEpoch 424/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1268 - binary_accuracy: 0.7500\nEpoch 425/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1265 - binary_accuracy: 0.7500\nEpoch 426/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1262 - binary_accuracy: 0.7500\nEpoch 427/800\n1/1 [==============================] - 0s 11ms/step - loss: 0.1259 - binary_accuracy: 0.7500\nEpoch 428/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.1256 - binary_accuracy: 0.7500\nEpoch 429/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1253 - binary_accuracy: 0.7500\nEpoch 430/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1250 - binary_accuracy: 0.7500\nEpoch 431/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1246 - binary_accuracy: 0.7500\nEpoch 432/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1243 - binary_accuracy: 0.7500\nEpoch 433/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.1240 - binary_accuracy: 0.7500\nEpoch 434/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1238 - binary_accuracy: 0.7500\nEpoch 435/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1235 - binary_accuracy: 0.7500\nEpoch 436/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1232 - binary_accuracy: 0.7500\nEpoch 437/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1229 - binary_accuracy: 0.7500\nEpoch 438/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1226 - binary_accuracy: 0.7500\nEpoch 439/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1223 - binary_accuracy: 0.7500\nEpoch 440/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1220 - binary_accuracy: 0.7500\nEpoch 441/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1217 - binary_accuracy: 0.7500\nEpoch 442/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.1214 - binary_accuracy: 0.7500\nEpoch 443/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.1211 - binary_accuracy: 0.7500\nEpoch 444/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1208 - binary_accuracy: 0.7500\nEpoch 445/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.1205 - binary_accuracy: 0.7500\nEpoch 446/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.1203 - binary_accuracy: 0.7500\nEpoch 447/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1200 - binary_accuracy: 0.7500\nEpoch 448/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.1197 - binary_accuracy: 0.7500\nEpoch 449/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.1194 - binary_accuracy: 0.7500\nEpoch 450/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.1191 - binary_accuracy: 0.7500\nEpoch 451/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1188 - binary_accuracy: 0.7500\nEpoch 452/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1186 - binary_accuracy: 0.7500\nEpoch 453/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1183 - binary_accuracy: 0.7500\nEpoch 454/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1180 - binary_accuracy: 0.7500\nEpoch 455/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1177 - binary_accuracy: 0.7500\nEpoch 456/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1175 - binary_accuracy: 0.7500\nEpoch 457/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.1172 - binary_accuracy: 0.7500\nEpoch 458/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.1169 - binary_accuracy: 0.7500\nEpoch 459/800\n1/1 [==============================] - 0s 13ms/step - loss: 0.1166 - binary_accuracy: 0.7500\nEpoch 460/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.1164 - binary_accuracy: 0.7500\nEpoch 461/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.1161 - binary_accuracy: 0.7500\nEpoch 462/800\n1/1 [==============================] - 0s 11ms/step - loss: 0.1158 - binary_accuracy: 0.7500\nEpoch 463/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1155 - binary_accuracy: 0.7500\nEpoch 464/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1153 - binary_accuracy: 0.7500\nEpoch 465/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1150 - binary_accuracy: 0.7500\nEpoch 466/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1147 - binary_accuracy: 0.7500\nEpoch 467/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.1145 - binary_accuracy: 0.7500\nEpoch 468/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.1142 - binary_accuracy: 0.7500\nEpoch 469/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.1140 - binary_accuracy: 0.7500\nEpoch 470/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1137 - binary_accuracy: 0.7500\nEpoch 471/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1134 - binary_accuracy: 0.7500\nEpoch 472/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1132 - binary_accuracy: 0.7500\nEpoch 473/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1129 - binary_accuracy: 0.7500\nEpoch 474/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1126 - binary_accuracy: 0.7500\nEpoch 475/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1124 - binary_accuracy: 0.7500\nEpoch 476/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1121 - binary_accuracy: 0.7500\nEpoch 477/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1119 - binary_accuracy: 0.7500\nEpoch 478/800\n1/1 [==============================] - 0s 3ms/step - loss: 0.1116 - binary_accuracy: 0.7500\nEpoch 479/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1114 - binary_accuracy: 0.7500\nEpoch 480/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1111 - binary_accuracy: 0.7500\nEpoch 481/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1108 - binary_accuracy: 0.7500\nEpoch 482/800\n1/1 [==============================] - 0s 3ms/step - loss: 0.1106 - binary_accuracy: 0.7500\nEpoch 483/800\n1/1 [==============================] - 0s 3ms/step - loss: 0.1103 - binary_accuracy: 0.7500\nEpoch 484/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1101 - binary_accuracy: 0.7500\nEpoch 485/800\n1/1 [==============================] - 0s 3ms/step - loss: 0.1098 - binary_accuracy: 0.7500\nEpoch 486/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.1096 - binary_accuracy: 0.7500\nEpoch 487/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.1093 - binary_accuracy: 0.7500\nEpoch 488/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.1091 - binary_accuracy: 0.7500\nEpoch 489/800\n1/1 [==============================] - 0s 11ms/step - loss: 0.1088 - binary_accuracy: 0.7500\nEpoch 490/800\n1/1 [==============================] - 0s 13ms/step - loss: 0.1086 - binary_accuracy: 0.7500\nEpoch 491/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1084 - binary_accuracy: 0.7500\nEpoch 492/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.1081 - binary_accuracy: 0.7500\nEpoch 493/800\n1/1 [==============================] - 0s 13ms/step - loss: 0.1079 - binary_accuracy: 0.7500\nEpoch 494/800\n1/1 [==============================] - 0s 13ms/step - loss: 0.1076 - binary_accuracy: 0.7500\nEpoch 495/800\n1/1 [==============================] - 0s 14ms/step - loss: 0.1074 - binary_accuracy: 0.7500\nEpoch 496/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1071 - binary_accuracy: 0.7500\nEpoch 497/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.1069 - binary_accuracy: 0.7500\nEpoch 498/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.1066 - binary_accuracy: 0.7500\nEpoch 499/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.1064 - binary_accuracy: 0.7500\nEpoch 500/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.1062 - binary_accuracy: 0.7500\nEpoch 501/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.1059 - binary_accuracy: 0.7500\nEpoch 502/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.1057 - binary_accuracy: 0.7500\nEpoch 503/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.1055 - binary_accuracy: 0.7500\nEpoch 504/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1052 - binary_accuracy: 0.7500\nEpoch 505/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1050 - binary_accuracy: 0.7500\nEpoch 506/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1047 - binary_accuracy: 0.7500\nEpoch 507/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1045 - binary_accuracy: 0.7500\nEpoch 508/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1043 - binary_accuracy: 0.7500\nEpoch 509/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.1040 - binary_accuracy: 0.7500\nEpoch 510/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1038 - binary_accuracy: 0.7500\nEpoch 511/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1036 - binary_accuracy: 0.7500\nEpoch 512/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1033 - binary_accuracy: 0.7500\nEpoch 513/800\n1/1 [==============================] - 0s 3ms/step - loss: 0.1031 - binary_accuracy: 0.7500\nEpoch 514/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1029 - binary_accuracy: 0.7500\nEpoch 515/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.1027 - binary_accuracy: 0.7500\nEpoch 516/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.1024 - binary_accuracy: 0.7500\nEpoch 517/800\n1/1 [==============================] - 0s 3ms/step - loss: 0.1022 - binary_accuracy: 0.7500\nEpoch 518/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.1020 - binary_accuracy: 0.7500\nEpoch 519/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.1017 - binary_accuracy: 0.7500\nEpoch 520/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.1015 - binary_accuracy: 0.7500\nEpoch 521/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.1013 - binary_accuracy: 0.7500\nEpoch 522/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.1011 - binary_accuracy: 0.7500\nEpoch 523/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.1008 - binary_accuracy: 0.7500\nEpoch 524/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.1006 - binary_accuracy: 0.7500\nEpoch 525/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.1004 - binary_accuracy: 0.7500\nEpoch 526/800\n1/1 [==============================] - 0s 12ms/step - loss: 0.1002 - binary_accuracy: 0.7500\nEpoch 527/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.0999 - binary_accuracy: 0.7500\nEpoch 528/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.0997 - binary_accuracy: 0.7500\nEpoch 529/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0995 - binary_accuracy: 0.7500\nEpoch 530/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0993 - binary_accuracy: 0.7500\nEpoch 531/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.0991 - binary_accuracy: 0.7500\nEpoch 532/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0988 - binary_accuracy: 0.7500\nEpoch 533/800\n1/1 [==============================] - 0s 11ms/step - loss: 0.0986 - binary_accuracy: 0.7500\nEpoch 534/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.0984 - binary_accuracy: 0.7500\nEpoch 535/800\n1/1 [==============================] - 0s 16ms/step - loss: 0.0982 - binary_accuracy: 0.7500\nEpoch 536/800\n1/1 [==============================] - 0s 13ms/step - loss: 0.0980 - binary_accuracy: 0.7500\nEpoch 537/800\n1/1 [==============================] - 0s 12ms/step - loss: 0.0978 - binary_accuracy: 0.7500\nEpoch 538/800\n1/1 [==============================] - 0s 11ms/step - loss: 0.0975 - binary_accuracy: 0.7500\nEpoch 539/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0973 - binary_accuracy: 0.7500\nEpoch 540/800\n1/1 [==============================] - 0s 11ms/step - loss: 0.0971 - binary_accuracy: 0.7500\nEpoch 541/800\n1/1 [==============================] - 0s 13ms/step - loss: 0.0969 - binary_accuracy: 0.7500\nEpoch 542/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.0967 - binary_accuracy: 0.7500\nEpoch 543/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0965 - binary_accuracy: 0.7500\nEpoch 544/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0963 - binary_accuracy: 0.7500\nEpoch 545/800\n1/1 [==============================] - 0s 16ms/step - loss: 0.0960 - binary_accuracy: 0.7500\nEpoch 546/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.0958 - binary_accuracy: 0.7500\nEpoch 547/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0956 - binary_accuracy: 0.7500\nEpoch 548/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0954 - binary_accuracy: 0.7500\nEpoch 549/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0952 - binary_accuracy: 0.7500\nEpoch 550/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0950 - binary_accuracy: 0.7500\nEpoch 551/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0948 - binary_accuracy: 0.7500\nEpoch 552/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0946 - binary_accuracy: 0.7500\nEpoch 553/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0944 - binary_accuracy: 0.7500\nEpoch 554/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0942 - binary_accuracy: 0.7500\nEpoch 555/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0940 - binary_accuracy: 0.7500\nEpoch 556/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0938 - binary_accuracy: 0.7500\nEpoch 557/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0935 - binary_accuracy: 0.7500\nEpoch 558/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0933 - binary_accuracy: 0.7500\nEpoch 559/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0931 - binary_accuracy: 0.7500\nEpoch 560/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.0929 - binary_accuracy: 0.7500\nEpoch 561/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0927 - binary_accuracy: 0.7500\nEpoch 562/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0925 - binary_accuracy: 0.7500\nEpoch 563/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0923 - binary_accuracy: 0.7500\nEpoch 564/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0921 - binary_accuracy: 0.7500\nEpoch 565/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.0919 - binary_accuracy: 0.7500\nEpoch 566/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.0917 - binary_accuracy: 0.7500\nEpoch 567/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.0915 - binary_accuracy: 0.7500\nEpoch 568/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.0913 - binary_accuracy: 0.7500\nEpoch 569/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.0911 - binary_accuracy: 0.7500\nEpoch 570/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.0909 - binary_accuracy: 0.7500\nEpoch 571/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.0907 - binary_accuracy: 0.7500\nEpoch 572/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.0905 - binary_accuracy: 0.7500\nEpoch 573/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.0903 - binary_accuracy: 0.7500\nEpoch 574/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.0901 - binary_accuracy: 0.7500\nEpoch 575/800\n1/1 [==============================] - 0s 14ms/step - loss: 0.0899 - binary_accuracy: 0.7500\nEpoch 576/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.0897 - binary_accuracy: 0.7500\nEpoch 577/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.0895 - binary_accuracy: 0.7500\nEpoch 578/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.0893 - binary_accuracy: 0.7500\nEpoch 579/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.0891 - binary_accuracy: 0.7500\nEpoch 580/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.0889 - binary_accuracy: 0.7500\nEpoch 581/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.0888 - binary_accuracy: 0.7500\nEpoch 582/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.0886 - binary_accuracy: 0.7500\nEpoch 583/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.0884 - binary_accuracy: 0.7500\nEpoch 584/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0882 - binary_accuracy: 0.7500\nEpoch 585/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.0880 - binary_accuracy: 0.7500\nEpoch 586/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0878 - binary_accuracy: 0.7500\nEpoch 587/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0876 - binary_accuracy: 0.7500\nEpoch 588/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.0874 - binary_accuracy: 0.7500\nEpoch 589/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.0872 - binary_accuracy: 0.7500\nEpoch 590/800\n1/1 [==============================] - 0s 3ms/step - loss: 0.0870 - binary_accuracy: 0.7500\nEpoch 591/800\n1/1 [==============================] - 0s 15ms/step - loss: 0.0868 - binary_accuracy: 0.7500\nEpoch 592/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0866 - binary_accuracy: 0.7500\nEpoch 593/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0865 - binary_accuracy: 0.7500\nEpoch 594/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0863 - binary_accuracy: 0.7500\nEpoch 595/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.0861 - binary_accuracy: 0.7500\nEpoch 596/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0859 - binary_accuracy: 0.7500\nEpoch 597/800\n1/1 [==============================] - 0s 3ms/step - loss: 0.0857 - binary_accuracy: 0.7500\nEpoch 598/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0855 - binary_accuracy: 0.7500\nEpoch 599/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.0853 - binary_accuracy: 0.7500\nEpoch 600/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0851 - binary_accuracy: 0.7500\nEpoch 601/800\n1/1 [==============================] - 0s 11ms/step - loss: 0.0850 - binary_accuracy: 0.7500\nEpoch 602/800\n1/1 [==============================] - 0s 11ms/step - loss: 0.0848 - binary_accuracy: 0.7500\nEpoch 603/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.0846 - binary_accuracy: 0.7500\nEpoch 604/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0844 - binary_accuracy: 0.7500\nEpoch 605/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0842 - binary_accuracy: 0.7500\nEpoch 606/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0840 - binary_accuracy: 0.7500\nEpoch 607/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0838 - binary_accuracy: 0.7500\nEpoch 608/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0837 - binary_accuracy: 0.7500\nEpoch 609/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0835 - binary_accuracy: 0.7500\nEpoch 610/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0833 - binary_accuracy: 0.7500\nEpoch 611/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.0831 - binary_accuracy: 0.7500\nEpoch 612/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.0829 - binary_accuracy: 0.7500\nEpoch 613/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0828 - binary_accuracy: 0.7500\nEpoch 614/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.0826 - binary_accuracy: 0.7500\nEpoch 615/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.0824 - binary_accuracy: 0.7500\nEpoch 616/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0822 - binary_accuracy: 0.7500\nEpoch 617/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.0820 - binary_accuracy: 0.7500\nEpoch 618/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0819 - binary_accuracy: 0.7500\nEpoch 619/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0817 - binary_accuracy: 0.7500\nEpoch 620/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0815 - binary_accuracy: 0.7500\nEpoch 621/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.0813 - binary_accuracy: 0.7500\nEpoch 622/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0811 - binary_accuracy: 0.7500\nEpoch 623/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0810 - binary_accuracy: 0.7500\nEpoch 624/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.0808 - binary_accuracy: 0.7500\nEpoch 625/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0806 - binary_accuracy: 0.7500\nEpoch 626/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.0804 - binary_accuracy: 0.7500\nEpoch 627/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0803 - binary_accuracy: 0.7500\nEpoch 628/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.0801 - binary_accuracy: 0.7500\nEpoch 629/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0799 - binary_accuracy: 0.7500\nEpoch 630/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.0797 - binary_accuracy: 0.7500\nEpoch 631/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.0796 - binary_accuracy: 0.7500\nEpoch 632/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.0794 - binary_accuracy: 0.7500\nEpoch 633/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.0792 - binary_accuracy: 0.7500\nEpoch 634/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0790 - binary_accuracy: 0.7500\nEpoch 635/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.0789 - binary_accuracy: 0.7500\nEpoch 636/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.0787 - binary_accuracy: 0.7500\nEpoch 637/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0785 - binary_accuracy: 0.7500\nEpoch 638/800\n1/1 [==============================] - 0s 11ms/step - loss: 0.0783 - binary_accuracy: 0.7500\nEpoch 639/800\n1/1 [==============================] - 0s 18ms/step - loss: 0.0782 - binary_accuracy: 0.7500\nEpoch 640/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.0780 - binary_accuracy: 0.7500\nEpoch 641/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.0778 - binary_accuracy: 0.7500\nEpoch 642/800\n1/1 [==============================] - 0s 11ms/step - loss: 0.0777 - binary_accuracy: 0.7500\nEpoch 643/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0775 - binary_accuracy: 0.7500\nEpoch 644/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.0773 - binary_accuracy: 0.7500\nEpoch 645/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0771 - binary_accuracy: 0.7500\nEpoch 646/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0770 - binary_accuracy: 0.7500\nEpoch 647/800\n1/1 [==============================] - 0s 14ms/step - loss: 0.0768 - binary_accuracy: 0.7500\nEpoch 648/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.0766 - binary_accuracy: 0.7500\nEpoch 649/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0765 - binary_accuracy: 0.7500\nEpoch 650/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.0763 - binary_accuracy: 0.7500\nEpoch 651/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.0761 - binary_accuracy: 0.7500\nEpoch 652/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.0760 - binary_accuracy: 0.7500\nEpoch 653/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0758 - binary_accuracy: 0.7500\nEpoch 654/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0756 - binary_accuracy: 0.7500\nEpoch 655/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0755 - binary_accuracy: 0.7500\nEpoch 656/800\n1/1 [==============================] - 0s 17ms/step - loss: 0.0753 - binary_accuracy: 0.7500\nEpoch 657/800\n1/1 [==============================] - 0s 14ms/step - loss: 0.0751 - binary_accuracy: 0.7500\nEpoch 658/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.0750 - binary_accuracy: 0.7500\nEpoch 659/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0748 - binary_accuracy: 0.7500\nEpoch 660/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0746 - binary_accuracy: 0.7500\nEpoch 661/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0745 - binary_accuracy: 0.7500\nEpoch 662/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0743 - binary_accuracy: 0.7500\nEpoch 663/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0741 - binary_accuracy: 0.7500\nEpoch 664/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0740 - binary_accuracy: 0.7500\nEpoch 665/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.0738 - binary_accuracy: 0.7500\nEpoch 666/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0736 - binary_accuracy: 0.7500\nEpoch 667/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0735 - binary_accuracy: 0.7500\nEpoch 668/800\n1/1 [==============================] - 0s 3ms/step - loss: 0.0733 - binary_accuracy: 0.7500\nEpoch 669/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0732 - binary_accuracy: 0.7500\nEpoch 670/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.0730 - binary_accuracy: 0.7500\nEpoch 671/800\n1/1 [==============================] - 0s 14ms/step - loss: 0.0728 - binary_accuracy: 0.7500\nEpoch 672/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.0727 - binary_accuracy: 0.7500\nEpoch 673/800\n1/1 [==============================] - 0s 17ms/step - loss: 0.0725 - binary_accuracy: 0.7500\nEpoch 674/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0724 - binary_accuracy: 0.7500\nEpoch 675/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0722 - binary_accuracy: 0.7500\nEpoch 676/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.0720 - binary_accuracy: 0.7500\nEpoch 677/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0719 - binary_accuracy: 0.7500\nEpoch 678/800\n1/1 [==============================] - 0s 13ms/step - loss: 0.0717 - binary_accuracy: 1.0000\nEpoch 679/800\n1/1 [==============================] - 0s 19ms/step - loss: 0.0716 - binary_accuracy: 1.0000\nEpoch 680/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0714 - binary_accuracy: 1.0000\nEpoch 681/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0712 - binary_accuracy: 1.0000\nEpoch 682/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0711 - binary_accuracy: 1.0000\nEpoch 683/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0709 - binary_accuracy: 1.0000\nEpoch 684/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0708 - binary_accuracy: 1.0000\nEpoch 685/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0706 - binary_accuracy: 1.0000\nEpoch 686/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0704 - binary_accuracy: 1.0000\nEpoch 687/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0703 - binary_accuracy: 1.0000\nEpoch 688/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0701 - binary_accuracy: 1.0000\nEpoch 689/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0700 - binary_accuracy: 1.0000\nEpoch 690/800\n1/1 [==============================] - 0s 16ms/step - loss: 0.0698 - binary_accuracy: 1.0000\nEpoch 691/800\n1/1 [==============================] - 0s 21ms/step - loss: 0.0697 - binary_accuracy: 1.0000\nEpoch 692/800\n1/1 [==============================] - 0s 21ms/step - loss: 0.0695 - binary_accuracy: 1.0000\nEpoch 693/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0694 - binary_accuracy: 1.0000\nEpoch 694/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.0692 - binary_accuracy: 1.0000\nEpoch 695/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.0690 - binary_accuracy: 1.0000\nEpoch 696/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.0689 - binary_accuracy: 1.0000\nEpoch 697/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.0687 - binary_accuracy: 1.0000\nEpoch 698/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0686 - binary_accuracy: 1.0000\nEpoch 699/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.0684 - binary_accuracy: 1.0000\nEpoch 700/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.0683 - binary_accuracy: 1.0000\nEpoch 701/800\n1/1 [==============================] - 0s 14ms/step - loss: 0.0681 - binary_accuracy: 1.0000\nEpoch 702/800\n1/1 [==============================] - 0s 14ms/step - loss: 0.0680 - binary_accuracy: 1.0000\nEpoch 703/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.0678 - binary_accuracy: 1.0000\nEpoch 704/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.0677 - binary_accuracy: 1.0000\nEpoch 705/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0675 - binary_accuracy: 1.0000\nEpoch 706/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0674 - binary_accuracy: 1.0000\nEpoch 707/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0672 - binary_accuracy: 1.0000\nEpoch 708/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.0671 - binary_accuracy: 1.0000\nEpoch 709/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.0669 - binary_accuracy: 1.0000\nEpoch 710/800\n1/1 [==============================] - 0s 12ms/step - loss: 0.0668 - binary_accuracy: 1.0000\nEpoch 711/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.0666 - binary_accuracy: 1.0000\nEpoch 712/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.0665 - binary_accuracy: 1.0000\nEpoch 713/800\n1/1 [==============================] - 0s 23ms/step - loss: 0.0663 - binary_accuracy: 1.0000\nEpoch 714/800\n1/1 [==============================] - 0s 13ms/step - loss: 0.0662 - binary_accuracy: 1.0000\nEpoch 715/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.0660 - binary_accuracy: 1.0000\nEpoch 716/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.0659 - binary_accuracy: 1.0000\nEpoch 717/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0657 - binary_accuracy: 1.0000\nEpoch 718/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0656 - binary_accuracy: 1.0000\nEpoch 719/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0654 - binary_accuracy: 1.0000\nEpoch 720/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0653 - binary_accuracy: 1.0000\nEpoch 721/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0651 - binary_accuracy: 1.0000\nEpoch 722/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0650 - binary_accuracy: 1.0000\nEpoch 723/800\n1/1 [==============================] - 0s 13ms/step - loss: 0.0648 - binary_accuracy: 1.0000\nEpoch 724/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0647 - binary_accuracy: 1.0000\nEpoch 725/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0645 - binary_accuracy: 1.0000\nEpoch 726/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0644 - binary_accuracy: 1.0000\nEpoch 727/800\n1/1 [==============================] - 0s 3ms/step - loss: 0.0642 - binary_accuracy: 1.0000\nEpoch 728/800\n1/1 [==============================] - 0s 2ms/step - loss: 0.0641 - binary_accuracy: 1.0000\nEpoch 729/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0640 - binary_accuracy: 1.0000\nEpoch 730/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.0638 - binary_accuracy: 1.0000\nEpoch 731/800\n1/1 [==============================] - 0s 3ms/step - loss: 0.0637 - binary_accuracy: 1.0000\nEpoch 732/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0635 - binary_accuracy: 1.0000\nEpoch 733/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.0634 - binary_accuracy: 1.0000\nEpoch 734/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.0632 - binary_accuracy: 1.0000\nEpoch 735/800\n1/1 [==============================] - 0s 11ms/step - loss: 0.0631 - binary_accuracy: 1.0000\nEpoch 736/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.0629 - binary_accuracy: 1.0000\nEpoch 737/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0628 - binary_accuracy: 1.0000\nEpoch 738/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0627 - binary_accuracy: 1.0000\nEpoch 739/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0625 - binary_accuracy: 1.0000\nEpoch 740/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0624 - binary_accuracy: 1.0000\nEpoch 741/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.0622 - binary_accuracy: 1.0000\nEpoch 742/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0621 - binary_accuracy: 1.0000\nEpoch 743/800\n1/1 [==============================] - 0s 15ms/step - loss: 0.0620 - binary_accuracy: 1.0000\nEpoch 744/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.0618 - binary_accuracy: 1.0000\nEpoch 745/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0617 - binary_accuracy: 1.0000\nEpoch 746/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0615 - binary_accuracy: 1.0000\nEpoch 747/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0614 - binary_accuracy: 1.0000\nEpoch 748/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0612 - binary_accuracy: 1.0000\nEpoch 749/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0611 - binary_accuracy: 1.0000\nEpoch 750/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0610 - binary_accuracy: 1.0000\nEpoch 751/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0608 - binary_accuracy: 1.0000\nEpoch 752/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0607 - binary_accuracy: 1.0000\nEpoch 753/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.0606 - binary_accuracy: 1.0000\nEpoch 754/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.0604 - binary_accuracy: 1.0000\nEpoch 755/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.0603 - binary_accuracy: 1.0000\nEpoch 756/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.0601 - binary_accuracy: 1.0000\nEpoch 757/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.0600 - binary_accuracy: 1.0000\nEpoch 758/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.0599 - binary_accuracy: 1.0000\nEpoch 759/800\n1/1 [==============================] - 0s 2ms/step - loss: 0.0597 - binary_accuracy: 1.0000\nEpoch 760/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0596 - binary_accuracy: 1.0000\nEpoch 761/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0594 - binary_accuracy: 1.0000\nEpoch 762/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.0593 - binary_accuracy: 1.0000\nEpoch 763/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.0592 - binary_accuracy: 1.0000\nEpoch 764/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.0590 - binary_accuracy: 1.0000\nEpoch 765/800\n1/1 [==============================] - 0s 13ms/step - loss: 0.0589 - binary_accuracy: 1.0000\nEpoch 766/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0588 - binary_accuracy: 1.0000\nEpoch 767/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0586 - binary_accuracy: 1.0000\nEpoch 768/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0585 - binary_accuracy: 1.0000\nEpoch 769/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0584 - binary_accuracy: 1.0000\nEpoch 770/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.0582 - binary_accuracy: 1.0000\nEpoch 771/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0581 - binary_accuracy: 1.0000\nEpoch 772/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0580 - binary_accuracy: 1.0000\nEpoch 773/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0578 - binary_accuracy: 1.0000\nEpoch 774/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0577 - binary_accuracy: 1.0000\nEpoch 775/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0576 - binary_accuracy: 1.0000\nEpoch 776/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0574 - binary_accuracy: 1.0000\nEpoch 777/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0573 - binary_accuracy: 1.0000\nEpoch 778/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.0572 - binary_accuracy: 1.0000\nEpoch 779/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0570 - binary_accuracy: 1.0000\nEpoch 780/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0569 - binary_accuracy: 1.0000\nEpoch 781/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.0568 - binary_accuracy: 1.0000\nEpoch 782/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.0566 - binary_accuracy: 1.0000\nEpoch 783/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0565 - binary_accuracy: 1.0000\nEpoch 784/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.0564 - binary_accuracy: 1.0000\nEpoch 785/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0562 - binary_accuracy: 1.0000\nEpoch 786/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.0561 - binary_accuracy: 1.0000\nEpoch 787/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0560 - binary_accuracy: 1.0000\nEpoch 788/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.0558 - binary_accuracy: 1.0000\nEpoch 789/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.0557 - binary_accuracy: 1.0000\nEpoch 790/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0556 - binary_accuracy: 1.0000\nEpoch 791/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0555 - binary_accuracy: 1.0000\nEpoch 792/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0553 - binary_accuracy: 1.0000\nEpoch 793/800\n1/1 [==============================] - 0s 14ms/step - loss: 0.0552 - binary_accuracy: 1.0000\nEpoch 794/800\n1/1 [==============================] - 0s 12ms/step - loss: 0.0551 - binary_accuracy: 1.0000\nEpoch 795/800\n1/1 [==============================] - 0s 16ms/step - loss: 0.0549 - binary_accuracy: 1.0000\nEpoch 796/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.0548 - binary_accuracy: 1.0000\nEpoch 797/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0547 - binary_accuracy: 1.0000\nEpoch 798/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0546 - binary_accuracy: 1.0000\nEpoch 799/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0544 - binary_accuracy: 1.0000\nEpoch 800/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.0543 - binary_accuracy: 1.0000\n"
],
[
"#AND\r\nscores = modelo.evaluate(datos_entrenamiento, datos_etiquetas)\r\n \r\nprint(\"\\n%s: %.2f%%\" % (modelo.metrics_names[1], scores[1]*100))\r\nprint (modelo.predict(datos_entrenamiento).round())",
"WARNING:tensorflow:5 out of the last 6 calls to <function Model.make_test_function.<locals>.test_function at 0x7f8c57824378> triggered tf.function retracing. Tracing is expensive and the excessive number of tracings could be due to (1) creating @tf.function repeatedly in a loop, (2) passing tensors with different shapes, (3) passing Python objects instead of tensors. For (1), please define your @tf.function outside of the loop. For (2), @tf.function has experimental_relax_shapes=True option that relaxes argument shapes that can avoid unnecessary retracing. For (3), please refer to https://www.tensorflow.org/guide/function#controlling_retracing and https://www.tensorflow.org/api_docs/python/tf/function for more details.\n1/1 [==============================] - 0s 91ms/step - loss: 0.0542 - binary_accuracy: 1.0000\n\nbinary_accuracy: 100.00%\nWARNING:tensorflow:5 out of the last 5 calls to <function Model.make_predict_function.<locals>.predict_function at 0x7f8c57824620> triggered tf.function retracing. Tracing is expensive and the excessive number of tracings could be due to (1) creating @tf.function repeatedly in a loop, (2) passing tensors with different shapes, (3) passing Python objects instead of tensors. For (1), please define your @tf.function outside of the loop. For (2), @tf.function has experimental_relax_shapes=True option that relaxes argument shapes that can avoid unnecessary retracing. For (3), please refer to https://www.tensorflow.org/guide/function#controlling_retracing and https://www.tensorflow.org/api_docs/python/tf/function for more details.\n[[0.]\n [0.]\n [0.]\n [1.]]\n"
],
[
"# cargamos las 4 combinaciones de las compuertas OR\r\ndatos_entrenamiento = np.array([[0,0],[0,1],[1,0],[1,1]], \"float32\")\r\n \r\n# y estos son los resultados que se obtienen, en el mismo orden\r\ndatos_etiquetas = np.array([[0],[1],[1],[1]], \"float32\")",
"_____no_output_____"
],
[
"x=datos_entrenamiento[:,0]\r\ny=datos_entrenamiento[:,1]\r\ncolors = datos_etiquetas\r\nplt.scatter(x,y,s=100,c=colors)\r\nplt.xlabel(\"Eje x\")\r\nplt.ylabel(\"Eje y\")\r\nplt.title(\"Grafica Datos a clasificar\")\r\nplt.show()",
"_____no_output_____"
],
[
"#Modelo para OR\r\nmodelo = Sequential()\r\nmodelo.add(Dense(1, input_dim=2, activation='relu'))\r\nmodelo.compile(loss='mean_squared_error',\r\n optimizer='adam',\r\n metrics=['binary_accuracy'])\r\n \r\nmodelo.fit(datos_entrenamiento, datos_etiquetas, epochs=800)",
"Epoch 1/800\n1/1 [==============================] - 0s 289ms/step - loss: 0.5277 - binary_accuracy: 0.2500\nEpoch 2/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5258 - binary_accuracy: 0.2500\nEpoch 3/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5239 - binary_accuracy: 0.2500\nEpoch 4/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5220 - binary_accuracy: 0.2500\nEpoch 5/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5201 - binary_accuracy: 0.2500\nEpoch 6/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5183 - binary_accuracy: 0.2500\nEpoch 7/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5164 - binary_accuracy: 0.2500\nEpoch 8/800\n1/1 [==============================] - 0s 3ms/step - loss: 0.5146 - binary_accuracy: 0.2500\nEpoch 9/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5127 - binary_accuracy: 0.2500\nEpoch 10/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5109 - binary_accuracy: 0.2500\nEpoch 11/800\n1/1 [==============================] - 0s 3ms/step - loss: 0.5091 - binary_accuracy: 0.2500\nEpoch 12/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5072 - binary_accuracy: 0.2500\nEpoch 13/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5054 - binary_accuracy: 0.2500\nEpoch 14/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5036 - binary_accuracy: 0.2500\nEpoch 15/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5018 - binary_accuracy: 0.2500\nEpoch 16/800\n1/1 [==============================] - 0s 3ms/step - loss: 0.5000 - binary_accuracy: 0.2500\nEpoch 17/800\n1/1 [==============================] - 0s 3ms/step - loss: 0.4983 - binary_accuracy: 0.2500\nEpoch 18/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.4965 - binary_accuracy: 0.2500\nEpoch 19/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.4947 - binary_accuracy: 0.2500\nEpoch 20/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.4930 - binary_accuracy: 0.2500\nEpoch 21/800\n1/1 [==============================] - 0s 15ms/step - loss: 0.4912 - binary_accuracy: 0.2500\nEpoch 22/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.4895 - binary_accuracy: 0.2500\nEpoch 23/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.4878 - binary_accuracy: 0.2500\nEpoch 24/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.4861 - binary_accuracy: 0.2500\nEpoch 25/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.4844 - binary_accuracy: 0.2500\nEpoch 26/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.4827 - binary_accuracy: 0.2500\nEpoch 27/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.4810 - binary_accuracy: 0.2500\nEpoch 28/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.4793 - binary_accuracy: 0.2500\nEpoch 29/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.4776 - binary_accuracy: 0.2500\nEpoch 30/800\n1/1 [==============================] - 0s 11ms/step - loss: 0.4760 - binary_accuracy: 0.2500\nEpoch 31/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.4743 - binary_accuracy: 0.2500\nEpoch 32/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.4727 - binary_accuracy: 0.2500\nEpoch 33/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.4711 - binary_accuracy: 0.2500\nEpoch 34/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.4694 - binary_accuracy: 0.2500\nEpoch 35/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.4678 - binary_accuracy: 0.2500\nEpoch 36/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.4662 - binary_accuracy: 0.2500\nEpoch 37/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.4646 - binary_accuracy: 0.2500\nEpoch 38/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.4630 - binary_accuracy: 0.2500\nEpoch 39/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.4615 - binary_accuracy: 0.2500\nEpoch 40/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.4599 - binary_accuracy: 0.2500\nEpoch 41/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.4583 - binary_accuracy: 0.2500\nEpoch 42/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.4568 - binary_accuracy: 0.2500\nEpoch 43/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.4553 - binary_accuracy: 0.2500\nEpoch 44/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.4537 - binary_accuracy: 0.2500\nEpoch 45/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.4522 - binary_accuracy: 0.2500\nEpoch 46/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.4507 - binary_accuracy: 0.2500\nEpoch 47/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.4492 - binary_accuracy: 0.2500\nEpoch 48/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.4477 - binary_accuracy: 0.5000\nEpoch 49/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.4462 - binary_accuracy: 0.5000\nEpoch 50/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.4448 - binary_accuracy: 0.5000\nEpoch 51/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.4433 - binary_accuracy: 0.5000\nEpoch 52/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.4418 - binary_accuracy: 0.5000\nEpoch 53/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.4404 - binary_accuracy: 0.5000\nEpoch 54/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.4390 - binary_accuracy: 0.5000\nEpoch 55/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.4375 - binary_accuracy: 0.5000\nEpoch 56/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.4361 - binary_accuracy: 0.5000\nEpoch 57/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.4347 - binary_accuracy: 0.5000\nEpoch 58/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.4333 - binary_accuracy: 0.5000\nEpoch 59/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.4319 - binary_accuracy: 0.5000\nEpoch 60/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.4305 - binary_accuracy: 0.5000\nEpoch 61/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.4292 - binary_accuracy: 0.5000\nEpoch 62/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.4278 - binary_accuracy: 0.5000\nEpoch 63/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.4264 - binary_accuracy: 0.5000\nEpoch 64/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.4251 - binary_accuracy: 0.5000\nEpoch 65/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.4237 - binary_accuracy: 0.5000\nEpoch 66/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.4224 - binary_accuracy: 0.5000\nEpoch 67/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.4211 - binary_accuracy: 0.5000\nEpoch 68/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.4198 - binary_accuracy: 0.5000\nEpoch 69/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.4185 - binary_accuracy: 0.5000\nEpoch 70/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.4172 - binary_accuracy: 0.5000\nEpoch 71/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.4159 - binary_accuracy: 0.5000\nEpoch 72/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.4146 - binary_accuracy: 0.5000\nEpoch 73/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.4134 - binary_accuracy: 0.5000\nEpoch 74/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.4121 - binary_accuracy: 0.5000\nEpoch 75/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.4109 - binary_accuracy: 0.5000\nEpoch 76/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.4096 - binary_accuracy: 0.5000\nEpoch 77/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.4084 - binary_accuracy: 0.5000\nEpoch 78/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.4072 - binary_accuracy: 0.5000\nEpoch 79/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.4059 - binary_accuracy: 0.5000\nEpoch 80/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.4047 - binary_accuracy: 0.5000\nEpoch 81/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.4035 - binary_accuracy: 0.5000\nEpoch 82/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.4023 - binary_accuracy: 0.5000\nEpoch 83/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.4012 - binary_accuracy: 0.5000\nEpoch 84/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.4000 - binary_accuracy: 0.5000\nEpoch 85/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3988 - binary_accuracy: 0.5000\nEpoch 86/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.3976 - binary_accuracy: 0.5000\nEpoch 87/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.3965 - binary_accuracy: 0.5000\nEpoch 88/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.3954 - binary_accuracy: 0.5000\nEpoch 89/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.3942 - binary_accuracy: 0.5000\nEpoch 90/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.3931 - binary_accuracy: 0.5000\nEpoch 91/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3920 - binary_accuracy: 0.5000\nEpoch 92/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.3909 - binary_accuracy: 0.5000\nEpoch 93/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3898 - binary_accuracy: 0.5000\nEpoch 94/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.3887 - binary_accuracy: 0.5000\nEpoch 95/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3876 - binary_accuracy: 0.5000\nEpoch 96/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3865 - binary_accuracy: 0.5000\nEpoch 97/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3854 - binary_accuracy: 0.5000\nEpoch 98/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3844 - binary_accuracy: 0.5000\nEpoch 99/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.3833 - binary_accuracy: 0.5000\nEpoch 100/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.3822 - binary_accuracy: 0.5000\nEpoch 101/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.3812 - binary_accuracy: 0.5000\nEpoch 102/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.3802 - binary_accuracy: 0.5000\nEpoch 103/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3791 - binary_accuracy: 0.5000\nEpoch 104/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3781 - binary_accuracy: 0.5000\nEpoch 105/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3771 - binary_accuracy: 0.5000\nEpoch 106/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3761 - binary_accuracy: 0.5000\nEpoch 107/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3751 - binary_accuracy: 0.5000\nEpoch 108/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3741 - binary_accuracy: 0.5000\nEpoch 109/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3731 - binary_accuracy: 0.5000\nEpoch 110/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.3722 - binary_accuracy: 0.5000\nEpoch 111/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.3712 - binary_accuracy: 0.5000\nEpoch 112/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3702 - binary_accuracy: 0.5000\nEpoch 113/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3693 - binary_accuracy: 0.5000\nEpoch 114/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3683 - binary_accuracy: 0.5000\nEpoch 115/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3674 - binary_accuracy: 0.5000\nEpoch 116/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3665 - binary_accuracy: 0.5000\nEpoch 117/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3655 - binary_accuracy: 0.5000\nEpoch 118/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.3646 - binary_accuracy: 0.5000\nEpoch 119/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3637 - binary_accuracy: 0.5000\nEpoch 120/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3628 - binary_accuracy: 0.5000\nEpoch 121/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3619 - binary_accuracy: 0.5000\nEpoch 122/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3610 - binary_accuracy: 0.5000\nEpoch 123/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3601 - binary_accuracy: 0.5000\nEpoch 124/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3592 - binary_accuracy: 0.5000\nEpoch 125/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.3584 - binary_accuracy: 0.5000\nEpoch 126/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3575 - binary_accuracy: 0.5000\nEpoch 127/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.3566 - binary_accuracy: 0.5000\nEpoch 128/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3558 - binary_accuracy: 0.5000\nEpoch 129/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3549 - binary_accuracy: 0.5000\nEpoch 130/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3541 - binary_accuracy: 0.5000\nEpoch 131/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3533 - binary_accuracy: 0.5000\nEpoch 132/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3524 - binary_accuracy: 0.5000\nEpoch 133/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.3516 - binary_accuracy: 0.5000\nEpoch 134/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.3508 - binary_accuracy: 0.5000\nEpoch 135/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3500 - binary_accuracy: 0.5000\nEpoch 136/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3492 - binary_accuracy: 0.5000\nEpoch 137/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3484 - binary_accuracy: 0.5000\nEpoch 138/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3476 - binary_accuracy: 0.5000\nEpoch 139/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3468 - binary_accuracy: 0.5000\nEpoch 140/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3461 - binary_accuracy: 0.5000\nEpoch 141/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.3453 - binary_accuracy: 0.7500\nEpoch 142/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.3445 - binary_accuracy: 0.7500\nEpoch 143/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3438 - binary_accuracy: 0.7500\nEpoch 144/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.3430 - binary_accuracy: 0.7500\nEpoch 145/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.3423 - binary_accuracy: 0.7500\nEpoch 146/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3416 - binary_accuracy: 0.7500\nEpoch 147/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3408 - binary_accuracy: 0.7500\nEpoch 148/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3401 - binary_accuracy: 0.7500\nEpoch 149/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.3394 - binary_accuracy: 0.7500\nEpoch 150/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.3387 - binary_accuracy: 0.7500\nEpoch 151/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.3379 - binary_accuracy: 0.7500\nEpoch 152/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.3372 - binary_accuracy: 0.7500\nEpoch 153/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3365 - binary_accuracy: 0.7500\nEpoch 154/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3359 - binary_accuracy: 0.7500\nEpoch 155/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3352 - binary_accuracy: 0.7500\nEpoch 156/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3345 - binary_accuracy: 0.7500\nEpoch 157/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3338 - binary_accuracy: 0.7500\nEpoch 158/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.3331 - binary_accuracy: 0.7500\nEpoch 159/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3325 - binary_accuracy: 0.7500\nEpoch 160/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.3318 - binary_accuracy: 0.7500\nEpoch 161/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.3312 - binary_accuracy: 0.7500\nEpoch 162/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.3305 - binary_accuracy: 0.7500\nEpoch 163/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.3299 - binary_accuracy: 0.7500\nEpoch 164/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.3292 - binary_accuracy: 0.7500\nEpoch 165/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.3281 - binary_accuracy: 0.7500\nEpoch 166/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.3267 - binary_accuracy: 0.7500\nEpoch 167/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3250 - binary_accuracy: 0.7500\nEpoch 168/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3233 - binary_accuracy: 0.7500\nEpoch 169/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3214 - binary_accuracy: 0.7500\nEpoch 170/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.3195 - binary_accuracy: 0.7500\nEpoch 171/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.3175 - binary_accuracy: 0.7500\nEpoch 172/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3154 - binary_accuracy: 0.7500\nEpoch 173/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.3133 - binary_accuracy: 0.7500\nEpoch 174/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3111 - binary_accuracy: 0.7500\nEpoch 175/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3089 - binary_accuracy: 0.7500\nEpoch 176/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3067 - binary_accuracy: 0.7500\nEpoch 177/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.3045 - binary_accuracy: 0.7500\nEpoch 178/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3022 - binary_accuracy: 0.7500\nEpoch 179/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.3000 - binary_accuracy: 0.7500\nEpoch 180/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.2977 - binary_accuracy: 0.7500\nEpoch 181/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.2954 - binary_accuracy: 0.7500\nEpoch 182/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.2932 - binary_accuracy: 0.7500\nEpoch 183/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.2909 - binary_accuracy: 0.7500\nEpoch 184/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.2887 - binary_accuracy: 0.7500\nEpoch 185/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.2865 - binary_accuracy: 0.7500\nEpoch 186/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.2843 - binary_accuracy: 0.7500\nEpoch 187/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.2821 - binary_accuracy: 0.7500\nEpoch 188/800\n1/1 [==============================] - 0s 41ms/step - loss: 0.2799 - binary_accuracy: 0.7500\nEpoch 189/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.2777 - binary_accuracy: 0.7500\nEpoch 190/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.2756 - binary_accuracy: 0.7500\nEpoch 191/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.2735 - binary_accuracy: 0.7500\nEpoch 192/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.2714 - binary_accuracy: 0.7500\nEpoch 193/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.2693 - binary_accuracy: 0.7500\nEpoch 194/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.2672 - binary_accuracy: 0.7500\nEpoch 195/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.2652 - binary_accuracy: 0.7500\nEpoch 196/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.2632 - binary_accuracy: 0.7500\nEpoch 197/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.2612 - binary_accuracy: 0.7500\nEpoch 198/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.2593 - binary_accuracy: 0.7500\nEpoch 199/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.2573 - binary_accuracy: 0.7500\nEpoch 200/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.2554 - binary_accuracy: 0.7500\nEpoch 201/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.2535 - binary_accuracy: 0.7500\nEpoch 202/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.2517 - binary_accuracy: 0.7500\nEpoch 203/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.2498 - binary_accuracy: 0.7500\nEpoch 204/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.2480 - binary_accuracy: 0.7500\nEpoch 205/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.2462 - binary_accuracy: 0.7500\nEpoch 206/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.2444 - binary_accuracy: 0.7500\nEpoch 207/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.2427 - binary_accuracy: 0.7500\nEpoch 208/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.2409 - binary_accuracy: 0.7500\nEpoch 209/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.2392 - binary_accuracy: 0.7500\nEpoch 210/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.2375 - binary_accuracy: 0.7500\nEpoch 211/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.2359 - binary_accuracy: 0.7500\nEpoch 212/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.2342 - binary_accuracy: 0.7500\nEpoch 213/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.2326 - binary_accuracy: 0.7500\nEpoch 214/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.2310 - binary_accuracy: 0.7500\nEpoch 215/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.2294 - binary_accuracy: 0.7500\nEpoch 216/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.2278 - binary_accuracy: 0.7500\nEpoch 217/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.2263 - binary_accuracy: 0.7500\nEpoch 218/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.2248 - binary_accuracy: 0.7500\nEpoch 219/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.2233 - binary_accuracy: 0.7500\nEpoch 220/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.2218 - binary_accuracy: 0.7500\nEpoch 221/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.2203 - binary_accuracy: 0.7500\nEpoch 222/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.2188 - binary_accuracy: 0.7500\nEpoch 223/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.2174 - binary_accuracy: 0.7500\nEpoch 224/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.2160 - binary_accuracy: 0.7500\nEpoch 225/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.2146 - binary_accuracy: 0.7500\nEpoch 226/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.2132 - binary_accuracy: 0.7500\nEpoch 227/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.2119 - binary_accuracy: 0.7500\nEpoch 228/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.2105 - binary_accuracy: 0.7500\nEpoch 229/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.2092 - binary_accuracy: 0.7500\nEpoch 230/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.2079 - binary_accuracy: 0.7500\nEpoch 231/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.2066 - binary_accuracy: 0.7500\nEpoch 232/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.2053 - binary_accuracy: 0.7500\nEpoch 233/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.2040 - binary_accuracy: 0.7500\nEpoch 234/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.2028 - binary_accuracy: 0.7500\nEpoch 235/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.2015 - binary_accuracy: 0.7500\nEpoch 236/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.2003 - binary_accuracy: 0.7500\nEpoch 237/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1991 - binary_accuracy: 0.7500\nEpoch 238/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1979 - binary_accuracy: 0.7500\nEpoch 239/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1967 - binary_accuracy: 0.7500\nEpoch 240/800\n1/1 [==============================] - 0s 22ms/step - loss: 0.1956 - binary_accuracy: 0.7500\nEpoch 241/800\n1/1 [==============================] - 0s 18ms/step - loss: 0.1944 - binary_accuracy: 0.7500\nEpoch 242/800\n1/1 [==============================] - 0s 18ms/step - loss: 0.1933 - binary_accuracy: 0.7500\nEpoch 243/800\n1/1 [==============================] - 0s 20ms/step - loss: 0.1922 - binary_accuracy: 0.7500\nEpoch 244/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.1911 - binary_accuracy: 0.7500\nEpoch 245/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1900 - binary_accuracy: 0.7500\nEpoch 246/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1889 - binary_accuracy: 0.7500\nEpoch 247/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1878 - binary_accuracy: 0.7500\nEpoch 248/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1868 - binary_accuracy: 0.7500\nEpoch 249/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1857 - binary_accuracy: 0.7500\nEpoch 250/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1847 - binary_accuracy: 0.7500\nEpoch 251/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1837 - binary_accuracy: 0.7500\nEpoch 252/800\n1/1 [==============================] - 0s 19ms/step - loss: 0.1827 - binary_accuracy: 0.7500\nEpoch 253/800\n1/1 [==============================] - 0s 13ms/step - loss: 0.1817 - binary_accuracy: 0.7500\nEpoch 254/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1807 - binary_accuracy: 0.7500\nEpoch 255/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.1797 - binary_accuracy: 0.7500\nEpoch 256/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1787 - binary_accuracy: 0.7500\nEpoch 257/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1778 - binary_accuracy: 0.7500\nEpoch 258/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.1768 - binary_accuracy: 0.7500\nEpoch 259/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1759 - binary_accuracy: 0.7500\nEpoch 260/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1750 - binary_accuracy: 0.7500\nEpoch 261/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1741 - binary_accuracy: 0.7500\nEpoch 262/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1732 - binary_accuracy: 0.7500\nEpoch 263/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1723 - binary_accuracy: 0.7500\nEpoch 264/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1714 - binary_accuracy: 0.7500\nEpoch 265/800\n1/1 [==============================] - 0s 21ms/step - loss: 0.1706 - binary_accuracy: 0.7500\nEpoch 266/800\n1/1 [==============================] - 0s 16ms/step - loss: 0.1697 - binary_accuracy: 0.7500\nEpoch 267/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.1689 - binary_accuracy: 0.7500\nEpoch 268/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1680 - binary_accuracy: 0.7500\nEpoch 269/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1672 - binary_accuracy: 0.7500\nEpoch 270/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1664 - binary_accuracy: 0.7500\nEpoch 271/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1656 - binary_accuracy: 0.7500\nEpoch 272/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1648 - binary_accuracy: 0.7500\nEpoch 273/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1640 - binary_accuracy: 0.7500\nEpoch 274/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1632 - binary_accuracy: 0.7500\nEpoch 275/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.1624 - binary_accuracy: 0.7500\nEpoch 276/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1616 - binary_accuracy: 0.7500\nEpoch 277/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1609 - binary_accuracy: 0.7500\nEpoch 278/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1601 - binary_accuracy: 0.7500\nEpoch 279/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1594 - binary_accuracy: 0.7500\nEpoch 280/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1587 - binary_accuracy: 0.7500\nEpoch 281/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1579 - binary_accuracy: 0.7500\nEpoch 282/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1572 - binary_accuracy: 0.7500\nEpoch 283/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1565 - binary_accuracy: 0.7500\nEpoch 284/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1558 - binary_accuracy: 0.7500\nEpoch 285/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1551 - binary_accuracy: 0.7500\nEpoch 286/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.1544 - binary_accuracy: 0.7500\nEpoch 287/800\n1/1 [==============================] - 0s 3ms/step - loss: 0.1538 - binary_accuracy: 0.7500\nEpoch 288/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1531 - binary_accuracy: 0.7500\nEpoch 289/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1524 - binary_accuracy: 0.7500\nEpoch 290/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1518 - binary_accuracy: 0.7500\nEpoch 291/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1511 - binary_accuracy: 0.7500\nEpoch 292/800\n1/1 [==============================] - 0s 3ms/step - loss: 0.1505 - binary_accuracy: 0.7500\nEpoch 293/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1499 - binary_accuracy: 0.7500\nEpoch 294/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1492 - binary_accuracy: 0.7500\nEpoch 295/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1486 - binary_accuracy: 0.7500\nEpoch 296/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1480 - binary_accuracy: 0.7500\nEpoch 297/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1474 - binary_accuracy: 0.7500\nEpoch 298/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1468 - binary_accuracy: 0.7500\nEpoch 299/800\n1/1 [==============================] - 0s 3ms/step - loss: 0.1462 - binary_accuracy: 0.7500\nEpoch 300/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1456 - binary_accuracy: 0.7500\nEpoch 301/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.1450 - binary_accuracy: 0.7500\nEpoch 302/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.1445 - binary_accuracy: 0.7500\nEpoch 303/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1439 - binary_accuracy: 0.7500\nEpoch 304/800\n1/1 [==============================] - 0s 3ms/step - loss: 0.1433 - binary_accuracy: 0.7500\nEpoch 305/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1428 - binary_accuracy: 0.7500\nEpoch 306/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.1422 - binary_accuracy: 0.7500\nEpoch 307/800\n1/1 [==============================] - 0s 3ms/step - loss: 0.1417 - binary_accuracy: 0.7500\nEpoch 308/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1412 - binary_accuracy: 0.7500\nEpoch 309/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1406 - binary_accuracy: 0.7500\nEpoch 310/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1401 - binary_accuracy: 0.7500\nEpoch 311/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.1396 - binary_accuracy: 0.7500\nEpoch 312/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1391 - binary_accuracy: 0.7500\nEpoch 313/800\n1/1 [==============================] - 0s 3ms/step - loss: 0.1386 - binary_accuracy: 0.7500\nEpoch 314/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1381 - binary_accuracy: 0.7500\nEpoch 315/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1376 - binary_accuracy: 0.7500\nEpoch 316/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1371 - binary_accuracy: 0.7500\nEpoch 317/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1366 - binary_accuracy: 0.7500\nEpoch 318/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1361 - binary_accuracy: 0.7500\nEpoch 319/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1356 - binary_accuracy: 0.7500\nEpoch 320/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1352 - binary_accuracy: 0.7500\nEpoch 321/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1347 - binary_accuracy: 0.7500\nEpoch 322/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1342 - binary_accuracy: 0.7500\nEpoch 323/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1338 - binary_accuracy: 0.7500\nEpoch 324/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1333 - binary_accuracy: 0.7500\nEpoch 325/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1329 - binary_accuracy: 0.7500\nEpoch 326/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1324 - binary_accuracy: 0.7500\nEpoch 327/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1320 - binary_accuracy: 0.7500\nEpoch 328/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1316 - binary_accuracy: 0.7500\nEpoch 329/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1311 - binary_accuracy: 0.7500\nEpoch 330/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1307 - binary_accuracy: 0.7500\nEpoch 331/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1303 - binary_accuracy: 0.7500\nEpoch 332/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1299 - binary_accuracy: 0.7500\nEpoch 333/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1295 - binary_accuracy: 0.7500\nEpoch 334/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1291 - binary_accuracy: 0.7500\nEpoch 335/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1287 - binary_accuracy: 0.7500\nEpoch 336/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1283 - binary_accuracy: 0.7500\nEpoch 337/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1279 - binary_accuracy: 0.7500\nEpoch 338/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1275 - binary_accuracy: 0.7500\nEpoch 339/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1271 - binary_accuracy: 0.7500\nEpoch 340/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1267 - binary_accuracy: 0.7500\nEpoch 341/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1263 - binary_accuracy: 0.7500\nEpoch 342/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1260 - binary_accuracy: 0.7500\nEpoch 343/800\n1/1 [==============================] - 0s 3ms/step - loss: 0.1256 - binary_accuracy: 0.7500\nEpoch 344/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1252 - binary_accuracy: 0.7500\nEpoch 345/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1249 - binary_accuracy: 0.7500\nEpoch 346/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1245 - binary_accuracy: 0.7500\nEpoch 347/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1241 - binary_accuracy: 0.7500\nEpoch 348/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1238 - binary_accuracy: 0.7500\nEpoch 349/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1234 - binary_accuracy: 0.7500\nEpoch 350/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1231 - binary_accuracy: 0.7500\nEpoch 351/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1228 - binary_accuracy: 0.7500\nEpoch 352/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1224 - binary_accuracy: 0.7500\nEpoch 353/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1221 - binary_accuracy: 0.7500\nEpoch 354/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1218 - binary_accuracy: 0.7500\nEpoch 355/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1214 - binary_accuracy: 0.7500\nEpoch 356/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1211 - binary_accuracy: 0.7500\nEpoch 357/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.1208 - binary_accuracy: 0.7500\nEpoch 358/800\n1/1 [==============================] - 0s 13ms/step - loss: 0.1205 - binary_accuracy: 0.7500\nEpoch 359/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.1201 - binary_accuracy: 0.7500\nEpoch 360/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.1198 - binary_accuracy: 0.7500\nEpoch 361/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.1195 - binary_accuracy: 0.7500\nEpoch 362/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1192 - binary_accuracy: 0.7500\nEpoch 363/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1189 - binary_accuracy: 0.7500\nEpoch 364/800\n1/1 [==============================] - 0s 3ms/step - loss: 0.1186 - binary_accuracy: 0.7500\nEpoch 365/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1183 - binary_accuracy: 0.7500\nEpoch 366/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1180 - binary_accuracy: 0.7500\nEpoch 367/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1177 - binary_accuracy: 0.7500\nEpoch 368/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1175 - binary_accuracy: 0.7500\nEpoch 369/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1172 - binary_accuracy: 0.7500\nEpoch 370/800\n1/1 [==============================] - 0s 19ms/step - loss: 0.1169 - binary_accuracy: 0.7500\nEpoch 371/800\n1/1 [==============================] - 0s 3ms/step - loss: 0.1166 - binary_accuracy: 0.7500\nEpoch 372/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1163 - binary_accuracy: 0.7500\nEpoch 373/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.1161 - binary_accuracy: 0.7500\nEpoch 374/800\n1/1 [==============================] - 0s 15ms/step - loss: 0.1158 - binary_accuracy: 0.7500\nEpoch 375/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.1155 - binary_accuracy: 0.7500\nEpoch 376/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.1153 - binary_accuracy: 0.7500\nEpoch 377/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.1150 - binary_accuracy: 0.7500\nEpoch 378/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1147 - binary_accuracy: 0.7500\nEpoch 379/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1145 - binary_accuracy: 0.7500\nEpoch 380/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1142 - binary_accuracy: 0.7500\nEpoch 381/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1140 - binary_accuracy: 0.7500\nEpoch 382/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.1137 - binary_accuracy: 0.7500\nEpoch 383/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.1135 - binary_accuracy: 0.7500\nEpoch 384/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1132 - binary_accuracy: 0.7500\nEpoch 385/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1130 - binary_accuracy: 0.7500\nEpoch 386/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1127 - binary_accuracy: 0.7500\nEpoch 387/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1125 - binary_accuracy: 0.7500\nEpoch 388/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1122 - binary_accuracy: 0.7500\nEpoch 389/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.1120 - binary_accuracy: 0.7500\nEpoch 390/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1118 - binary_accuracy: 0.7500\nEpoch 391/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1115 - binary_accuracy: 0.7500\nEpoch 392/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1113 - binary_accuracy: 0.7500\nEpoch 393/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1111 - binary_accuracy: 0.7500\nEpoch 394/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1109 - binary_accuracy: 0.7500\nEpoch 395/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.1106 - binary_accuracy: 0.7500\nEpoch 396/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.1104 - binary_accuracy: 0.7500\nEpoch 397/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1102 - binary_accuracy: 0.7500\nEpoch 398/800\n1/1 [==============================] - 0s 3ms/step - loss: 0.1100 - binary_accuracy: 0.7500\nEpoch 399/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1098 - binary_accuracy: 0.7500\nEpoch 400/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.1096 - binary_accuracy: 0.7500\nEpoch 401/800\n1/1 [==============================] - 0s 3ms/step - loss: 0.1093 - binary_accuracy: 0.7500\nEpoch 402/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1091 - binary_accuracy: 0.7500\nEpoch 403/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1089 - binary_accuracy: 0.7500\nEpoch 404/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1087 - binary_accuracy: 0.7500\nEpoch 405/800\n1/1 [==============================] - 0s 14ms/step - loss: 0.1085 - binary_accuracy: 0.7500\nEpoch 406/800\n1/1 [==============================] - 0s 18ms/step - loss: 0.1083 - binary_accuracy: 0.7500\nEpoch 407/800\n1/1 [==============================] - 0s 15ms/step - loss: 0.1081 - binary_accuracy: 0.7500\nEpoch 408/800\n1/1 [==============================] - 0s 17ms/step - loss: 0.1079 - binary_accuracy: 0.7500\nEpoch 409/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.1077 - binary_accuracy: 0.7500\nEpoch 410/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.1075 - binary_accuracy: 0.7500\nEpoch 411/800\n1/1 [==============================] - 0s 14ms/step - loss: 0.1073 - binary_accuracy: 0.7500\nEpoch 412/800\n1/1 [==============================] - 0s 13ms/step - loss: 0.1071 - binary_accuracy: 0.7500\nEpoch 413/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1069 - binary_accuracy: 0.7500\nEpoch 414/800\n1/1 [==============================] - 0s 12ms/step - loss: 0.1068 - binary_accuracy: 0.7500\nEpoch 415/800\n1/1 [==============================] - 0s 27ms/step - loss: 0.1066 - binary_accuracy: 0.7500\nEpoch 416/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.1064 - binary_accuracy: 0.7500\nEpoch 417/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.1062 - binary_accuracy: 0.7500\nEpoch 418/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.1060 - binary_accuracy: 0.7500\nEpoch 419/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.1058 - binary_accuracy: 0.7500\nEpoch 420/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.1057 - binary_accuracy: 0.7500\nEpoch 421/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.1055 - binary_accuracy: 0.7500\nEpoch 422/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.1053 - binary_accuracy: 0.7500\nEpoch 423/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.1051 - binary_accuracy: 0.7500\nEpoch 424/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1050 - binary_accuracy: 0.7500\nEpoch 425/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.1048 - binary_accuracy: 0.7500\nEpoch 426/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.1046 - binary_accuracy: 0.7500\nEpoch 427/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.1044 - binary_accuracy: 0.7500\nEpoch 428/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.1043 - binary_accuracy: 0.7500\nEpoch 429/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.1041 - binary_accuracy: 0.7500\nEpoch 430/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1039 - binary_accuracy: 0.7500\nEpoch 431/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1038 - binary_accuracy: 0.7500\nEpoch 432/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1036 - binary_accuracy: 0.7500\nEpoch 433/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1034 - binary_accuracy: 0.7500\nEpoch 434/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1033 - binary_accuracy: 0.7500\nEpoch 435/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1031 - binary_accuracy: 0.7500\nEpoch 436/800\n1/1 [==============================] - 0s 3ms/step - loss: 0.1030 - binary_accuracy: 0.7500\nEpoch 437/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1028 - binary_accuracy: 0.7500\nEpoch 438/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1027 - binary_accuracy: 0.7500\nEpoch 439/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1025 - binary_accuracy: 0.7500\nEpoch 440/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1023 - binary_accuracy: 0.7500\nEpoch 441/800\n1/1 [==============================] - 0s 3ms/step - loss: 0.1022 - binary_accuracy: 0.7500\nEpoch 442/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1020 - binary_accuracy: 0.7500\nEpoch 443/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1019 - binary_accuracy: 0.7500\nEpoch 444/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1017 - binary_accuracy: 0.7500\nEpoch 445/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.1016 - binary_accuracy: 0.7500\nEpoch 446/800\n1/1 [==============================] - 0s 12ms/step - loss: 0.1015 - binary_accuracy: 0.7500\nEpoch 447/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.1013 - binary_accuracy: 0.7500\nEpoch 448/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.1012 - binary_accuracy: 0.7500\nEpoch 449/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.1010 - binary_accuracy: 0.7500\nEpoch 450/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.1009 - binary_accuracy: 0.7500\nEpoch 451/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.1007 - binary_accuracy: 0.7500\nEpoch 452/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.1006 - binary_accuracy: 0.7500\nEpoch 453/800\n1/1 [==============================] - 0s 20ms/step - loss: 0.1004 - binary_accuracy: 0.7500\nEpoch 454/800\n1/1 [==============================] - 0s 11ms/step - loss: 0.1003 - binary_accuracy: 0.7500\nEpoch 455/800\n1/1 [==============================] - 0s 15ms/step - loss: 0.1002 - binary_accuracy: 0.7500\nEpoch 456/800\n1/1 [==============================] - 0s 18ms/step - loss: 0.1000 - binary_accuracy: 0.7500\nEpoch 457/800\n1/1 [==============================] - 0s 16ms/step - loss: 0.0999 - binary_accuracy: 0.7500\nEpoch 458/800\n1/1 [==============================] - 0s 14ms/step - loss: 0.0998 - binary_accuracy: 0.7500\nEpoch 459/800\n1/1 [==============================] - 0s 15ms/step - loss: 0.0996 - binary_accuracy: 0.7500\nEpoch 460/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.0995 - binary_accuracy: 0.7500\nEpoch 461/800\n1/1 [==============================] - 0s 11ms/step - loss: 0.0994 - binary_accuracy: 0.7500\nEpoch 462/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0992 - binary_accuracy: 0.7500\nEpoch 463/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.0991 - binary_accuracy: 0.7500\nEpoch 464/800\n1/1 [==============================] - 0s 13ms/step - loss: 0.0990 - binary_accuracy: 0.7500\nEpoch 465/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.0988 - binary_accuracy: 0.7500\nEpoch 466/800\n1/1 [==============================] - 0s 13ms/step - loss: 0.0987 - binary_accuracy: 0.7500\nEpoch 467/800\n1/1 [==============================] - 0s 11ms/step - loss: 0.0986 - binary_accuracy: 0.7500\nEpoch 468/800\n1/1 [==============================] - 0s 13ms/step - loss: 0.0985 - binary_accuracy: 0.7500\nEpoch 469/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.0983 - binary_accuracy: 0.7500\nEpoch 470/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.0982 - binary_accuracy: 0.7500\nEpoch 471/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0981 - binary_accuracy: 0.7500\nEpoch 472/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0980 - binary_accuracy: 0.7500\nEpoch 473/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0978 - binary_accuracy: 0.7500\nEpoch 474/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0977 - binary_accuracy: 0.7500\nEpoch 475/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0976 - binary_accuracy: 0.7500\nEpoch 476/800\n1/1 [==============================] - 0s 13ms/step - loss: 0.0975 - binary_accuracy: 0.7500\nEpoch 477/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0974 - binary_accuracy: 0.7500\nEpoch 478/800\n1/1 [==============================] - 0s 3ms/step - loss: 0.0972 - binary_accuracy: 0.7500\nEpoch 479/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0971 - binary_accuracy: 0.7500\nEpoch 480/800\n1/1 [==============================] - 0s 12ms/step - loss: 0.0970 - binary_accuracy: 0.7500\nEpoch 481/800\n1/1 [==============================] - 0s 12ms/step - loss: 0.0969 - binary_accuracy: 0.7500\nEpoch 482/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.0968 - binary_accuracy: 0.7500\nEpoch 483/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.0966 - binary_accuracy: 0.7500\nEpoch 484/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0965 - binary_accuracy: 0.7500\nEpoch 485/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0964 - binary_accuracy: 0.7500\nEpoch 486/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0963 - binary_accuracy: 0.7500\nEpoch 487/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0962 - binary_accuracy: 0.7500\nEpoch 488/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0961 - binary_accuracy: 0.7500\nEpoch 489/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0960 - binary_accuracy: 0.7500\nEpoch 490/800\n1/1 [==============================] - 0s 3ms/step - loss: 0.0959 - binary_accuracy: 0.7500\nEpoch 491/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0957 - binary_accuracy: 0.7500\nEpoch 492/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0956 - binary_accuracy: 0.7500\nEpoch 493/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0955 - binary_accuracy: 0.7500\nEpoch 494/800\n1/1 [==============================] - 0s 3ms/step - loss: 0.0954 - binary_accuracy: 0.7500\nEpoch 495/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.0953 - binary_accuracy: 0.7500\nEpoch 496/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0952 - binary_accuracy: 0.7500\nEpoch 497/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0951 - binary_accuracy: 0.7500\nEpoch 498/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0950 - binary_accuracy: 0.7500\nEpoch 499/800\n1/1 [==============================] - 0s 3ms/step - loss: 0.0949 - binary_accuracy: 0.7500\nEpoch 500/800\n1/1 [==============================] - 0s 3ms/step - loss: 0.0948 - binary_accuracy: 1.0000\nEpoch 501/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0947 - binary_accuracy: 1.0000\nEpoch 502/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0946 - binary_accuracy: 1.0000\nEpoch 503/800\n1/1 [==============================] - 0s 11ms/step - loss: 0.0945 - binary_accuracy: 1.0000\nEpoch 504/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0944 - binary_accuracy: 1.0000\nEpoch 505/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0942 - binary_accuracy: 1.0000\nEpoch 506/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0941 - binary_accuracy: 1.0000\nEpoch 507/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.0940 - binary_accuracy: 1.0000\nEpoch 508/800\n1/1 [==============================] - 0s 11ms/step - loss: 0.0939 - binary_accuracy: 1.0000\nEpoch 509/800\n1/1 [==============================] - 0s 16ms/step - loss: 0.0938 - binary_accuracy: 1.0000\nEpoch 510/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.0937 - binary_accuracy: 1.0000\nEpoch 511/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0936 - binary_accuracy: 1.0000\nEpoch 512/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0935 - binary_accuracy: 1.0000\nEpoch 513/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0934 - binary_accuracy: 1.0000\nEpoch 514/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.0933 - binary_accuracy: 1.0000\nEpoch 515/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.0932 - binary_accuracy: 1.0000\nEpoch 516/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0931 - binary_accuracy: 1.0000\nEpoch 517/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0931 - binary_accuracy: 1.0000\nEpoch 518/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0930 - binary_accuracy: 1.0000\nEpoch 519/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0929 - binary_accuracy: 1.0000\nEpoch 520/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0928 - binary_accuracy: 1.0000\nEpoch 521/800\n1/1 [==============================] - 0s 15ms/step - loss: 0.0927 - binary_accuracy: 1.0000\nEpoch 522/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0926 - binary_accuracy: 1.0000\nEpoch 523/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0925 - binary_accuracy: 1.0000\nEpoch 524/800\n1/1 [==============================] - 0s 3ms/step - loss: 0.0924 - binary_accuracy: 1.0000\nEpoch 525/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0923 - binary_accuracy: 1.0000\nEpoch 526/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0922 - binary_accuracy: 1.0000\nEpoch 527/800\n1/1 [==============================] - 0s 19ms/step - loss: 0.0921 - binary_accuracy: 1.0000\nEpoch 528/800\n1/1 [==============================] - 0s 13ms/step - loss: 0.0920 - binary_accuracy: 1.0000\nEpoch 529/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0919 - binary_accuracy: 1.0000\nEpoch 530/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.0918 - binary_accuracy: 1.0000\nEpoch 531/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0917 - binary_accuracy: 1.0000\nEpoch 532/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0916 - binary_accuracy: 1.0000\nEpoch 533/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0916 - binary_accuracy: 1.0000\nEpoch 534/800\n1/1 [==============================] - 0s 3ms/step - loss: 0.0915 - binary_accuracy: 1.0000\nEpoch 535/800\n1/1 [==============================] - 0s 16ms/step - loss: 0.0914 - binary_accuracy: 1.0000\nEpoch 536/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0913 - binary_accuracy: 1.0000\nEpoch 537/800\n1/1 [==============================] - 0s 16ms/step - loss: 0.0912 - binary_accuracy: 1.0000\nEpoch 538/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0911 - binary_accuracy: 1.0000\nEpoch 539/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.0910 - binary_accuracy: 1.0000\nEpoch 540/800\n1/1 [==============================] - 0s 15ms/step - loss: 0.0909 - binary_accuracy: 1.0000\nEpoch 541/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.0908 - binary_accuracy: 1.0000\nEpoch 542/800\n1/1 [==============================] - 0s 12ms/step - loss: 0.0908 - binary_accuracy: 1.0000\nEpoch 543/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.0907 - binary_accuracy: 1.0000\nEpoch 544/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0906 - binary_accuracy: 1.0000\nEpoch 545/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.0905 - binary_accuracy: 1.0000\nEpoch 546/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.0904 - binary_accuracy: 1.0000\nEpoch 547/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0903 - binary_accuracy: 1.0000\nEpoch 548/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0902 - binary_accuracy: 1.0000\nEpoch 549/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0901 - binary_accuracy: 1.0000\nEpoch 550/800\n1/1 [==============================] - 0s 16ms/step - loss: 0.0901 - binary_accuracy: 1.0000\nEpoch 551/800\n1/1 [==============================] - 0s 13ms/step - loss: 0.0900 - binary_accuracy: 1.0000\nEpoch 552/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0899 - binary_accuracy: 1.0000\nEpoch 553/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.0898 - binary_accuracy: 1.0000\nEpoch 554/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0897 - binary_accuracy: 1.0000\nEpoch 555/800\n1/1 [==============================] - 0s 11ms/step - loss: 0.0896 - binary_accuracy: 1.0000\nEpoch 556/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.0896 - binary_accuracy: 1.0000\nEpoch 557/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.0895 - binary_accuracy: 1.0000\nEpoch 558/800\n1/1 [==============================] - 0s 13ms/step - loss: 0.0894 - binary_accuracy: 1.0000\nEpoch 559/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0893 - binary_accuracy: 1.0000\nEpoch 560/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.0892 - binary_accuracy: 1.0000\nEpoch 561/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.0891 - binary_accuracy: 1.0000\nEpoch 562/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.0891 - binary_accuracy: 1.0000\nEpoch 563/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0890 - binary_accuracy: 1.0000\nEpoch 564/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0889 - binary_accuracy: 1.0000\nEpoch 565/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0888 - binary_accuracy: 1.0000\nEpoch 566/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0887 - binary_accuracy: 1.0000\nEpoch 567/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.0887 - binary_accuracy: 1.0000\nEpoch 568/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0886 - binary_accuracy: 1.0000\nEpoch 569/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0885 - binary_accuracy: 1.0000\nEpoch 570/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0884 - binary_accuracy: 1.0000\nEpoch 571/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0883 - binary_accuracy: 1.0000\nEpoch 572/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.0883 - binary_accuracy: 1.0000\nEpoch 573/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0882 - binary_accuracy: 1.0000\nEpoch 574/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0881 - binary_accuracy: 1.0000\nEpoch 575/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0880 - binary_accuracy: 1.0000\nEpoch 576/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.0880 - binary_accuracy: 1.0000\nEpoch 577/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.0879 - binary_accuracy: 1.0000\nEpoch 578/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0878 - binary_accuracy: 1.0000\nEpoch 579/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.0877 - binary_accuracy: 1.0000\nEpoch 580/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.0876 - binary_accuracy: 1.0000\nEpoch 581/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.0876 - binary_accuracy: 1.0000\nEpoch 582/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.0875 - binary_accuracy: 1.0000\nEpoch 583/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.0874 - binary_accuracy: 1.0000\nEpoch 584/800\n1/1 [==============================] - 0s 12ms/step - loss: 0.0873 - binary_accuracy: 1.0000\nEpoch 585/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.0873 - binary_accuracy: 1.0000\nEpoch 586/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.0872 - binary_accuracy: 1.0000\nEpoch 587/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.0871 - binary_accuracy: 1.0000\nEpoch 588/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.0870 - binary_accuracy: 1.0000\nEpoch 589/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.0870 - binary_accuracy: 1.0000\nEpoch 590/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.0869 - binary_accuracy: 1.0000\nEpoch 591/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0868 - binary_accuracy: 1.0000\nEpoch 592/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0867 - binary_accuracy: 1.0000\nEpoch 593/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.0867 - binary_accuracy: 1.0000\nEpoch 594/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0866 - binary_accuracy: 1.0000\nEpoch 595/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.0865 - binary_accuracy: 1.0000\nEpoch 596/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.0865 - binary_accuracy: 1.0000\nEpoch 597/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.0864 - binary_accuracy: 1.0000\nEpoch 598/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.0863 - binary_accuracy: 1.0000\nEpoch 599/800\n1/1 [==============================] - 0s 11ms/step - loss: 0.0862 - binary_accuracy: 1.0000\nEpoch 600/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0862 - binary_accuracy: 1.0000\nEpoch 601/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.0861 - binary_accuracy: 1.0000\nEpoch 602/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0860 - binary_accuracy: 1.0000\nEpoch 603/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0859 - binary_accuracy: 1.0000\nEpoch 604/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0859 - binary_accuracy: 1.0000\nEpoch 605/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0858 - binary_accuracy: 1.0000\nEpoch 606/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0857 - binary_accuracy: 1.0000\nEpoch 607/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.0857 - binary_accuracy: 1.0000\nEpoch 608/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.0856 - binary_accuracy: 1.0000\nEpoch 609/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.0855 - binary_accuracy: 1.0000\nEpoch 610/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0855 - binary_accuracy: 1.0000\nEpoch 611/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.0854 - binary_accuracy: 1.0000\nEpoch 612/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.0853 - binary_accuracy: 1.0000\nEpoch 613/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.0852 - binary_accuracy: 1.0000\nEpoch 614/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0852 - binary_accuracy: 1.0000\nEpoch 615/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.0851 - binary_accuracy: 1.0000\nEpoch 616/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0850 - binary_accuracy: 1.0000\nEpoch 617/800\n1/1 [==============================] - 0s 17ms/step - loss: 0.0850 - binary_accuracy: 1.0000\nEpoch 618/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.0849 - binary_accuracy: 1.0000\nEpoch 619/800\n1/1 [==============================] - 0s 11ms/step - loss: 0.0848 - binary_accuracy: 1.0000\nEpoch 620/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.0848 - binary_accuracy: 1.0000\nEpoch 621/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.0847 - binary_accuracy: 1.0000\nEpoch 622/800\n1/1 [==============================] - 0s 11ms/step - loss: 0.0846 - binary_accuracy: 1.0000\nEpoch 623/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0846 - binary_accuracy: 1.0000\nEpoch 624/800\n1/1 [==============================] - 0s 11ms/step - loss: 0.0845 - binary_accuracy: 1.0000\nEpoch 625/800\n1/1 [==============================] - 0s 11ms/step - loss: 0.0844 - binary_accuracy: 1.0000\nEpoch 626/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.0844 - binary_accuracy: 1.0000\nEpoch 627/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.0843 - binary_accuracy: 1.0000\nEpoch 628/800\n1/1 [==============================] - 0s 12ms/step - loss: 0.0842 - binary_accuracy: 1.0000\nEpoch 629/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.0842 - binary_accuracy: 1.0000\nEpoch 630/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.0841 - binary_accuracy: 1.0000\nEpoch 631/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0840 - binary_accuracy: 1.0000\nEpoch 632/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0840 - binary_accuracy: 1.0000\nEpoch 633/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0839 - binary_accuracy: 1.0000\nEpoch 634/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0838 - binary_accuracy: 1.0000\nEpoch 635/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0838 - binary_accuracy: 1.0000\nEpoch 636/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0837 - binary_accuracy: 1.0000\nEpoch 637/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0836 - binary_accuracy: 1.0000\nEpoch 638/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0836 - binary_accuracy: 1.0000\nEpoch 639/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0835 - binary_accuracy: 1.0000\nEpoch 640/800\n1/1 [==============================] - 0s 12ms/step - loss: 0.0834 - binary_accuracy: 1.0000\nEpoch 641/800\n1/1 [==============================] - 0s 13ms/step - loss: 0.0834 - binary_accuracy: 1.0000\nEpoch 642/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0833 - binary_accuracy: 1.0000\nEpoch 643/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0832 - binary_accuracy: 1.0000\nEpoch 644/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0832 - binary_accuracy: 1.0000\nEpoch 645/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.0831 - binary_accuracy: 1.0000\nEpoch 646/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0831 - binary_accuracy: 1.0000\nEpoch 647/800\n1/1 [==============================] - 0s 3ms/step - loss: 0.0830 - binary_accuracy: 1.0000\nEpoch 648/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.0829 - binary_accuracy: 1.0000\nEpoch 649/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0829 - binary_accuracy: 1.0000\nEpoch 650/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.0828 - binary_accuracy: 1.0000\nEpoch 651/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.0827 - binary_accuracy: 1.0000\nEpoch 652/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.0827 - binary_accuracy: 1.0000\nEpoch 653/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0826 - binary_accuracy: 1.0000\nEpoch 654/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.0826 - binary_accuracy: 1.0000\nEpoch 655/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.0825 - binary_accuracy: 1.0000\nEpoch 656/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0824 - binary_accuracy: 1.0000\nEpoch 657/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.0824 - binary_accuracy: 1.0000\nEpoch 658/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.0823 - binary_accuracy: 1.0000\nEpoch 659/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.0822 - binary_accuracy: 1.0000\nEpoch 660/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0822 - binary_accuracy: 1.0000\nEpoch 661/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.0821 - binary_accuracy: 1.0000\nEpoch 662/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.0821 - binary_accuracy: 1.0000\nEpoch 663/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0820 - binary_accuracy: 1.0000\nEpoch 664/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.0819 - binary_accuracy: 1.0000\nEpoch 665/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0819 - binary_accuracy: 1.0000\nEpoch 666/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0818 - binary_accuracy: 1.0000\nEpoch 667/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0818 - binary_accuracy: 1.0000\nEpoch 668/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0817 - binary_accuracy: 1.0000\nEpoch 669/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0816 - binary_accuracy: 1.0000\nEpoch 670/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.0816 - binary_accuracy: 1.0000\nEpoch 671/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0815 - binary_accuracy: 1.0000\nEpoch 672/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0815 - binary_accuracy: 1.0000\nEpoch 673/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0814 - binary_accuracy: 1.0000\nEpoch 674/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0813 - binary_accuracy: 1.0000\nEpoch 675/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0813 - binary_accuracy: 1.0000\nEpoch 676/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0812 - binary_accuracy: 1.0000\nEpoch 677/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0812 - binary_accuracy: 1.0000\nEpoch 678/800\n1/1 [==============================] - 0s 14ms/step - loss: 0.0811 - binary_accuracy: 1.0000\nEpoch 679/800\n1/1 [==============================] - 0s 12ms/step - loss: 0.0811 - binary_accuracy: 1.0000\nEpoch 680/800\n1/1 [==============================] - 0s 14ms/step - loss: 0.0810 - binary_accuracy: 1.0000\nEpoch 681/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0809 - binary_accuracy: 1.0000\nEpoch 682/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0809 - binary_accuracy: 1.0000\nEpoch 683/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0808 - binary_accuracy: 1.0000\nEpoch 684/800\n1/1 [==============================] - 0s 12ms/step - loss: 0.0808 - binary_accuracy: 1.0000\nEpoch 685/800\n1/1 [==============================] - 0s 13ms/step - loss: 0.0807 - binary_accuracy: 1.0000\nEpoch 686/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0806 - binary_accuracy: 1.0000\nEpoch 687/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0806 - binary_accuracy: 1.0000\nEpoch 688/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0805 - binary_accuracy: 1.0000\nEpoch 689/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0805 - binary_accuracy: 1.0000\nEpoch 690/800\n1/1 [==============================] - 0s 23ms/step - loss: 0.0804 - binary_accuracy: 1.0000\nEpoch 691/800\n1/1 [==============================] - 0s 11ms/step - loss: 0.0804 - binary_accuracy: 1.0000\nEpoch 692/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0803 - binary_accuracy: 1.0000\nEpoch 693/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0803 - binary_accuracy: 1.0000\nEpoch 694/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0802 - binary_accuracy: 1.0000\nEpoch 695/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0801 - binary_accuracy: 1.0000\nEpoch 696/800\n1/1 [==============================] - 0s 16ms/step - loss: 0.0801 - binary_accuracy: 1.0000\nEpoch 697/800\n1/1 [==============================] - 0s 16ms/step - loss: 0.0800 - binary_accuracy: 1.0000\nEpoch 698/800\n1/1 [==============================] - 0s 17ms/step - loss: 0.0800 - binary_accuracy: 1.0000\nEpoch 699/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.0799 - binary_accuracy: 1.0000\nEpoch 700/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0799 - binary_accuracy: 1.0000\nEpoch 701/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0798 - binary_accuracy: 1.0000\nEpoch 702/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0798 - binary_accuracy: 1.0000\nEpoch 703/800\n1/1 [==============================] - 0s 13ms/step - loss: 0.0797 - binary_accuracy: 1.0000\nEpoch 704/800\n1/1 [==============================] - 0s 16ms/step - loss: 0.0796 - binary_accuracy: 1.0000\nEpoch 705/800\n1/1 [==============================] - 0s 17ms/step - loss: 0.0796 - binary_accuracy: 1.0000\nEpoch 706/800\n1/1 [==============================] - 0s 19ms/step - loss: 0.0795 - binary_accuracy: 1.0000\nEpoch 707/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0795 - binary_accuracy: 1.0000\nEpoch 708/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0794 - binary_accuracy: 1.0000\nEpoch 709/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0794 - binary_accuracy: 1.0000\nEpoch 710/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0793 - binary_accuracy: 1.0000\nEpoch 711/800\n1/1 [==============================] - 0s 11ms/step - loss: 0.0793 - binary_accuracy: 1.0000\nEpoch 712/800\n1/1 [==============================] - 0s 16ms/step - loss: 0.0792 - binary_accuracy: 1.0000\nEpoch 713/800\n1/1 [==============================] - 0s 22ms/step - loss: 0.0792 - binary_accuracy: 1.0000\nEpoch 714/800\n1/1 [==============================] - 0s 26ms/step - loss: 0.0791 - binary_accuracy: 1.0000\nEpoch 715/800\n1/1 [==============================] - 0s 12ms/step - loss: 0.0790 - binary_accuracy: 1.0000\nEpoch 716/800\n1/1 [==============================] - 0s 24ms/step - loss: 0.0790 - binary_accuracy: 1.0000\nEpoch 717/800\n1/1 [==============================] - 0s 17ms/step - loss: 0.0789 - binary_accuracy: 1.0000\nEpoch 718/800\n1/1 [==============================] - 0s 19ms/step - loss: 0.0789 - binary_accuracy: 1.0000\nEpoch 719/800\n1/1 [==============================] - 0s 19ms/step - loss: 0.0788 - binary_accuracy: 1.0000\nEpoch 720/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.0788 - binary_accuracy: 1.0000\nEpoch 721/800\n1/1 [==============================] - 0s 12ms/step - loss: 0.0787 - binary_accuracy: 1.0000\nEpoch 722/800\n1/1 [==============================] - 0s 17ms/step - loss: 0.0787 - binary_accuracy: 1.0000\nEpoch 723/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0786 - binary_accuracy: 1.0000\nEpoch 724/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0786 - binary_accuracy: 1.0000\nEpoch 725/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.0785 - binary_accuracy: 1.0000\nEpoch 726/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0785 - binary_accuracy: 1.0000\nEpoch 727/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0784 - binary_accuracy: 1.0000\nEpoch 728/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0784 - binary_accuracy: 1.0000\nEpoch 729/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0783 - binary_accuracy: 1.0000\nEpoch 730/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0783 - binary_accuracy: 1.0000\nEpoch 731/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0782 - binary_accuracy: 1.0000\nEpoch 732/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.0782 - binary_accuracy: 1.0000\nEpoch 733/800\n1/1 [==============================] - 0s 12ms/step - loss: 0.0781 - binary_accuracy: 1.0000\nEpoch 734/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.0781 - binary_accuracy: 1.0000\nEpoch 735/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0780 - binary_accuracy: 1.0000\nEpoch 736/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0780 - binary_accuracy: 1.0000\nEpoch 737/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0779 - binary_accuracy: 1.0000\nEpoch 738/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0779 - binary_accuracy: 1.0000\nEpoch 739/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0778 - binary_accuracy: 1.0000\nEpoch 740/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0778 - binary_accuracy: 1.0000\nEpoch 741/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0777 - binary_accuracy: 1.0000\nEpoch 742/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0777 - binary_accuracy: 1.0000\nEpoch 743/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.0776 - binary_accuracy: 1.0000\nEpoch 744/800\n1/1 [==============================] - 0s 12ms/step - loss: 0.0776 - binary_accuracy: 1.0000\nEpoch 745/800\n1/1 [==============================] - 0s 14ms/step - loss: 0.0775 - binary_accuracy: 1.0000\nEpoch 746/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.0775 - binary_accuracy: 1.0000\nEpoch 747/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0774 - binary_accuracy: 1.0000\nEpoch 748/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0774 - binary_accuracy: 1.0000\nEpoch 749/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0773 - binary_accuracy: 1.0000\nEpoch 750/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0773 - binary_accuracy: 1.0000\nEpoch 751/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.0772 - binary_accuracy: 1.0000\nEpoch 752/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.0772 - binary_accuracy: 1.0000\nEpoch 753/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0771 - binary_accuracy: 1.0000\nEpoch 754/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0771 - binary_accuracy: 1.0000\nEpoch 755/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0770 - binary_accuracy: 1.0000\nEpoch 756/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0770 - binary_accuracy: 1.0000\nEpoch 757/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0769 - binary_accuracy: 1.0000\nEpoch 758/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.0769 - binary_accuracy: 1.0000\nEpoch 759/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0768 - binary_accuracy: 1.0000\nEpoch 760/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.0768 - binary_accuracy: 1.0000\nEpoch 761/800\n1/1 [==============================] - 0s 13ms/step - loss: 0.0767 - binary_accuracy: 1.0000\nEpoch 762/800\n1/1 [==============================] - 0s 22ms/step - loss: 0.0767 - binary_accuracy: 1.0000\nEpoch 763/800\n1/1 [==============================] - 0s 12ms/step - loss: 0.0767 - binary_accuracy: 1.0000\nEpoch 764/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.0766 - binary_accuracy: 1.0000\nEpoch 765/800\n1/1 [==============================] - 0s 15ms/step - loss: 0.0766 - binary_accuracy: 1.0000\nEpoch 766/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.0765 - binary_accuracy: 1.0000\nEpoch 767/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0765 - binary_accuracy: 1.0000\nEpoch 768/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.0764 - binary_accuracy: 1.0000\nEpoch 769/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.0764 - binary_accuracy: 1.0000\nEpoch 770/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.0763 - binary_accuracy: 1.0000\nEpoch 771/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.0763 - binary_accuracy: 1.0000\nEpoch 772/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.0762 - binary_accuracy: 1.0000\nEpoch 773/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.0762 - binary_accuracy: 1.0000\nEpoch 774/800\n1/1 [==============================] - 0s 16ms/step - loss: 0.0761 - binary_accuracy: 1.0000\nEpoch 775/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.0761 - binary_accuracy: 1.0000\nEpoch 776/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0761 - binary_accuracy: 1.0000\nEpoch 777/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.0760 - binary_accuracy: 1.0000\nEpoch 778/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.0760 - binary_accuracy: 1.0000\nEpoch 779/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.0759 - binary_accuracy: 1.0000\nEpoch 780/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.0759 - binary_accuracy: 1.0000\nEpoch 781/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.0758 - binary_accuracy: 1.0000\nEpoch 782/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0758 - binary_accuracy: 1.0000\nEpoch 783/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.0757 - binary_accuracy: 1.0000\nEpoch 784/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.0757 - binary_accuracy: 1.0000\nEpoch 785/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.0757 - binary_accuracy: 1.0000\nEpoch 786/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.0756 - binary_accuracy: 1.0000\nEpoch 787/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.0756 - binary_accuracy: 1.0000\nEpoch 788/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.0755 - binary_accuracy: 1.0000\nEpoch 789/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.0755 - binary_accuracy: 1.0000\nEpoch 790/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.0754 - binary_accuracy: 1.0000\nEpoch 791/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.0754 - binary_accuracy: 1.0000\nEpoch 792/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.0753 - binary_accuracy: 1.0000\nEpoch 793/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.0753 - binary_accuracy: 1.0000\nEpoch 794/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.0753 - binary_accuracy: 1.0000\nEpoch 795/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.0752 - binary_accuracy: 1.0000\nEpoch 796/800\n1/1 [==============================] - 0s 11ms/step - loss: 0.0752 - binary_accuracy: 1.0000\nEpoch 797/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.0751 - binary_accuracy: 1.0000\nEpoch 798/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.0751 - binary_accuracy: 1.0000\nEpoch 799/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.0750 - binary_accuracy: 1.0000\nEpoch 800/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.0750 - binary_accuracy: 1.0000\n"
],
[
"#OR\r\nscores = modelo.evaluate(datos_entrenamiento, datos_etiquetas)\r\n \r\nprint(\"\\n%s: %.2f%%\" % (modelo.metrics_names[1], scores[1]*100))\r\nprint (modelo.predict(datos_entrenamiento).round())",
"WARNING:tensorflow:6 out of the last 7 calls to <function Model.make_test_function.<locals>.test_function at 0x7f8c55662510> triggered tf.function retracing. Tracing is expensive and the excessive number of tracings could be due to (1) creating @tf.function repeatedly in a loop, (2) passing tensors with different shapes, (3) passing Python objects instead of tensors. For (1), please define your @tf.function outside of the loop. For (2), @tf.function has experimental_relax_shapes=True option that relaxes argument shapes that can avoid unnecessary retracing. For (3), please refer to https://www.tensorflow.org/guide/function#controlling_retracing and https://www.tensorflow.org/api_docs/python/tf/function for more details.\n1/1 [==============================] - 0s 92ms/step - loss: 0.0750 - binary_accuracy: 1.0000\n\nbinary_accuracy: 100.00%\nWARNING:tensorflow:6 out of the last 6 calls to <function Model.make_predict_function.<locals>.predict_function at 0x7f8c54d4b2f0> triggered tf.function retracing. Tracing is expensive and the excessive number of tracings could be due to (1) creating @tf.function repeatedly in a loop, (2) passing tensors with different shapes, (3) passing Python objects instead of tensors. For (1), please define your @tf.function outside of the loop. For (2), @tf.function has experimental_relax_shapes=True option that relaxes argument shapes that can avoid unnecessary retracing. For (3), please refer to https://www.tensorflow.org/guide/function#controlling_retracing and https://www.tensorflow.org/api_docs/python/tf/function for more details.\n[[0.]\n [1.]\n [1.]\n [1.]]\n"
],
[
"# cargamos las 4 combinaciones de las compuertas XOR\r\ndatos_entrenamiento = np.array([[0,0],[0,1],[1,0],[1,1]], \"float32\")\r\n \r\n# y estos son los resultados que se obtienen, en el mismo orden\r\ndatos_etiquetas = np.array([[0],[1],[1],[0]], \"float32\")",
"_____no_output_____"
],
[
"x=datos_entrenamiento[:,0]\r\ny=datos_entrenamiento[:,1]\r\ncolors = datos_etiquetas\r\nplt.scatter(x,y,s=100,c=colors)\r\nplt.xlabel(\"Eje x\")\r\nplt.ylabel(\"Eje y\")\r\nplt.title(\"Grafica Datos a clasificar\")\r\nplt.show()",
"_____no_output_____"
],
[
"#Modelo para XOR\r\nmodelo = Sequential()\r\nmodelo.add(Dense(1, input_dim=2, activation='relu'))\r\nmodelo.compile(loss='mean_squared_error',\r\n optimizer='adam',\r\n metrics=['binary_accuracy'])\r\n \r\nmodelo.fit(datos_entrenamiento, datos_etiquetas, epochs=800)",
"Epoch 1/800\n1/1 [==============================] - 0s 312ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 2/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 3/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 4/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 5/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 6/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 7/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 8/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 9/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 10/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 11/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 12/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 13/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 14/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 15/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 16/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 17/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 18/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 19/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 20/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 21/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 22/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 23/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 24/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 25/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 26/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 27/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 28/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 29/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 30/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 31/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 32/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 33/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 34/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 35/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 36/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 37/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 38/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 39/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 40/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 41/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 42/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 43/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 44/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 45/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 46/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 47/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 48/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 49/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 50/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 51/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 52/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 53/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 54/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 55/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 56/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 57/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 58/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 59/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 60/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 61/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 62/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 63/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 64/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 65/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 66/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 67/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 68/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 69/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 70/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 71/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 72/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 73/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 74/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 75/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 76/800\n1/1 [==============================] - 0s 12ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 77/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 78/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 79/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 80/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 81/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 82/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 83/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 84/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 85/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 86/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 87/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 88/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 89/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 90/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 91/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 92/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 93/800\n1/1 [==============================] - 0s 12ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 94/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 95/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 96/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 97/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 98/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 99/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 100/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 101/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 102/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 103/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 104/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 105/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 106/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 107/800\n1/1 [==============================] - 0s 12ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 108/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 109/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 110/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 111/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 112/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 113/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 114/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 115/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 116/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 117/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 118/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 119/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 120/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 121/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 122/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 123/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 124/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 125/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 126/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 127/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 128/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 129/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 130/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 131/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 132/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 133/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 134/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 135/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 136/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 137/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 138/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 139/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 140/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 141/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 142/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 143/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 144/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 145/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 146/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 147/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 148/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 149/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 150/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 151/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 152/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 153/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 154/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 155/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 156/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 157/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 158/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 159/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 160/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 161/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 162/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 163/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 164/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 165/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 166/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 167/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 168/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 169/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 170/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 171/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 172/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 173/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 174/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 175/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 176/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 177/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 178/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 179/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 180/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 181/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 182/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 183/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 184/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 185/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 186/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 187/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 188/800\n1/1 [==============================] - 0s 3ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 189/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 190/800\n1/1 [==============================] - 0s 3ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 191/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 192/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 193/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 194/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 195/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 196/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 197/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 198/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 199/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 200/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 201/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 202/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 203/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 204/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 205/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 206/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 207/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 208/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 209/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 210/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 211/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 212/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 213/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 214/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 215/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 216/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 217/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 218/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 219/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 220/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 221/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 222/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 223/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 224/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 225/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 226/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 227/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 228/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 229/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 230/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 231/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 232/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 233/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 234/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 235/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 236/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 237/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 238/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 239/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 240/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 241/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 242/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 243/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 244/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 245/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 246/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 247/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 248/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 249/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 250/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 251/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 252/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 253/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 254/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 255/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 256/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 257/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 258/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 259/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 260/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 261/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 262/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 263/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 264/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 265/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 266/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 267/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 268/800\n1/1 [==============================] - 0s 14ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 269/800\n1/1 [==============================] - 0s 32ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 270/800\n1/1 [==============================] - 0s 11ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 271/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 272/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 273/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 274/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 275/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 276/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 277/800\n1/1 [==============================] - 0s 11ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 278/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 279/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 280/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 281/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 282/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 283/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 284/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 285/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 286/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 287/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 288/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 289/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 290/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 291/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 292/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 293/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 294/800\n1/1 [==============================] - 0s 13ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 295/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 296/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 297/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 298/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 299/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 300/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 301/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 302/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 303/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 304/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 305/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 306/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 307/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 308/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 309/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 310/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 311/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 312/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 313/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 314/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 315/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 316/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 317/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 318/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 319/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 320/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 321/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 322/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 323/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 324/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 325/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 326/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 327/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 328/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 329/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 330/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 331/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 332/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 333/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 334/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 335/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 336/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 337/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 338/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 339/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 340/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 341/800\n1/1 [==============================] - 0s 40ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 342/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 343/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 344/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 345/800\n1/1 [==============================] - 0s 21ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 346/800\n1/1 [==============================] - 0s 16ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 347/800\n1/1 [==============================] - 0s 22ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 348/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 349/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 350/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 351/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 352/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 353/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 354/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 355/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 356/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 357/800\n1/1 [==============================] - 0s 14ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 358/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 359/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 360/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 361/800\n1/1 [==============================] - 0s 11ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 362/800\n1/1 [==============================] - 0s 17ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 363/800\n1/1 [==============================] - 0s 19ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 364/800\n1/1 [==============================] - 0s 19ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 365/800\n1/1 [==============================] - 0s 15ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 366/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 367/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 368/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 369/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 370/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 371/800\n1/1 [==============================] - 0s 18ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 372/800\n1/1 [==============================] - 0s 15ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 373/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 374/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 375/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 376/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 377/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 378/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 379/800\n1/1 [==============================] - 0s 12ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 380/800\n1/1 [==============================] - 0s 17ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 381/800\n1/1 [==============================] - 0s 16ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 382/800\n1/1 [==============================] - 0s 19ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 383/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 384/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 385/800\n1/1 [==============================] - 0s 16ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 386/800\n1/1 [==============================] - 0s 27ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 387/800\n1/1 [==============================] - 0s 20ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 388/800\n1/1 [==============================] - 0s 17ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 389/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 390/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 391/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 392/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 393/800\n1/1 [==============================] - 0s 11ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 394/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 395/800\n1/1 [==============================] - 0s 13ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 396/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 397/800\n1/1 [==============================] - 0s 19ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 398/800\n1/1 [==============================] - 0s 14ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 399/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 400/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 401/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 402/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 403/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 404/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 405/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 406/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 407/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 408/800\n1/1 [==============================] - 0s 14ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 409/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 410/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 411/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 412/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 413/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 414/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 415/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 416/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 417/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 418/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 419/800\n1/1 [==============================] - 0s 15ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 420/800\n1/1 [==============================] - 0s 16ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 421/800\n1/1 [==============================] - 0s 16ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 422/800\n1/1 [==============================] - 0s 16ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 423/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 424/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 425/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 426/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 427/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 428/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 429/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 430/800\n1/1 [==============================] - 0s 11ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 431/800\n1/1 [==============================] - 0s 16ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 432/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 433/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 434/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 435/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 436/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 437/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 438/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 439/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 440/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 441/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 442/800\n1/1 [==============================] - 0s 11ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 443/800\n1/1 [==============================] - 0s 14ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 444/800\n1/1 [==============================] - 0s 12ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 445/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 446/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 447/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 448/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 449/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 450/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 451/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 452/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 453/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 454/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 455/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 456/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 457/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 458/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 459/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 460/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 461/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 462/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 463/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 464/800\n1/1 [==============================] - 0s 3ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 465/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 466/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 467/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 468/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 469/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 470/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 471/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 472/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 473/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 474/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 475/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 476/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 477/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 478/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 479/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 480/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 481/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 482/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 483/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 484/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 485/800\n1/1 [==============================] - 0s 3ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 486/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 487/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 488/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 489/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 490/800\n1/1 [==============================] - 0s 11ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 491/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 492/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 493/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 494/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 495/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 496/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 497/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 498/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 499/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 500/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 501/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 502/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 503/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 504/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 505/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 506/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 507/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 508/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 509/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 510/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 511/800\n1/1 [==============================] - 0s 3ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 512/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 513/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 514/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 515/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 516/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 517/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 518/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 519/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 520/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 521/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 522/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 523/800\n1/1 [==============================] - 0s 11ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 524/800\n1/1 [==============================] - 0s 15ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 525/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 526/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 527/800\n1/1 [==============================] - 0s 3ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 528/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 529/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 530/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 531/800\n1/1 [==============================] - 0s 2ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 532/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 533/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 534/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 535/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 536/800\n1/1 [==============================] - 0s 16ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 537/800\n1/1 [==============================] - 0s 14ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 538/800\n1/1 [==============================] - 0s 11ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 539/800\n1/1 [==============================] - 0s 11ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 540/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 541/800\n1/1 [==============================] - 0s 11ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 542/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 543/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 544/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 545/800\n1/1 [==============================] - 0s 11ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 546/800\n1/1 [==============================] - 0s 15ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 547/800\n1/1 [==============================] - 0s 12ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 548/800\n1/1 [==============================] - 0s 15ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 549/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 550/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 551/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 552/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 553/800\n1/1 [==============================] - 0s 11ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 554/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 555/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 556/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 557/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 558/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 559/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 560/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 561/800\n1/1 [==============================] - 0s 13ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 562/800\n1/1 [==============================] - 0s 13ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 563/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 564/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 565/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 566/800\n1/1 [==============================] - 0s 19ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 567/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 568/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 569/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 570/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 571/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 572/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 573/800\n1/1 [==============================] - 0s 14ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 574/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 575/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 576/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 577/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 578/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 579/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 580/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 581/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 582/800\n1/1 [==============================] - 0s 11ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 583/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 584/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 585/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 586/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 587/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 588/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 589/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 590/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 591/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 592/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 593/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 594/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 595/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 596/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 597/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 598/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 599/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 600/800\n1/1 [==============================] - 0s 15ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 601/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 602/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 603/800\n1/1 [==============================] - 0s 14ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 604/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 605/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 606/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 607/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 608/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 609/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 610/800\n1/1 [==============================] - 0s 17ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 611/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 612/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 613/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 614/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 615/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 616/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 617/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 618/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 619/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 620/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 621/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 622/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 623/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 624/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 625/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 626/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 627/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 628/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 629/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 630/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 631/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 632/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 633/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 634/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 635/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 636/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 637/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 638/800\n1/1 [==============================] - 0s 3ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 639/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 640/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 641/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 642/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 643/800\n1/1 [==============================] - 0s 20ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 644/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 645/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 646/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 647/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 648/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 649/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 650/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 651/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 652/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 653/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 654/800\n1/1 [==============================] - 0s 17ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 655/800\n1/1 [==============================] - 0s 12ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 656/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 657/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 658/800\n1/1 [==============================] - 0s 11ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 659/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 660/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 661/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 662/800\n1/1 [==============================] - 0s 11ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 663/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 664/800\n1/1 [==============================] - 0s 11ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 665/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 666/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 667/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 668/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 669/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 670/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 671/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 672/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 673/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 674/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 675/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 676/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 677/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 678/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 679/800\n1/1 [==============================] - 0s 12ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 680/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 681/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 682/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 683/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 684/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 685/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 686/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 687/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 688/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 689/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 690/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 691/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 692/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 693/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 694/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 695/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 696/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 697/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 698/800\n1/1 [==============================] - 0s 11ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 699/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 700/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 701/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 702/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 703/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 704/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 705/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 706/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 707/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 708/800\n1/1 [==============================] - 0s 11ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 709/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 710/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 711/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 712/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 713/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 714/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 715/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 716/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 717/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 718/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 719/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 720/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 721/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 722/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 723/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 724/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 725/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 726/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 727/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 728/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 729/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 730/800\n1/1 [==============================] - 0s 13ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 731/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 732/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 733/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 734/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 735/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 736/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 737/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 738/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 739/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 740/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 741/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 742/800\n1/1 [==============================] - 0s 15ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 743/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 744/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 745/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 746/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 747/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 748/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 749/800\n1/1 [==============================] - 0s 11ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 750/800\n1/1 [==============================] - 0s 12ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 751/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 752/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 753/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 754/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 755/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 756/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 757/800\n1/1 [==============================] - 0s 11ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 758/800\n1/1 [==============================] - 0s 10ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 759/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 760/800\n1/1 [==============================] - 0s 11ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 761/800\n1/1 [==============================] - 0s 11ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 762/800\n1/1 [==============================] - 0s 14ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 763/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 764/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 765/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 766/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 767/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 768/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 769/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 770/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 771/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 772/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 773/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 774/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 775/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 776/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 777/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 778/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 779/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 780/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 781/800\n1/1 [==============================] - 0s 8ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 782/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 783/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 784/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 785/800\n1/1 [==============================] - 0s 3ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 786/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 787/800\n1/1 [==============================] - 0s 7ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 788/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 789/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 790/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 791/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 792/800\n1/1 [==============================] - 0s 6ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 793/800\n1/1 [==============================] - 0s 9ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 794/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 795/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 796/800\n1/1 [==============================] - 0s 15ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 797/800\n1/1 [==============================] - 0s 13ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 798/800\n1/1 [==============================] - 0s 12ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 799/800\n1/1 [==============================] - 0s 4ms/step - loss: 0.5000 - binary_accuracy: 0.5000\nEpoch 800/800\n1/1 [==============================] - 0s 5ms/step - loss: 0.5000 - binary_accuracy: 0.5000\n"
],
[
"#XOR\r\nscores = modelo.evaluate(datos_entrenamiento, datos_etiquetas)\r\n \r\nprint(\"\\n%s: %.2f%%\" % (modelo.metrics_names[1], scores[1]*100))\r\nprint (modelo.predict(datos_entrenamiento).round())",
"WARNING:tensorflow:7 out of the last 8 calls to <function Model.make_test_function.<locals>.test_function at 0x7f8c533fe598> triggered tf.function retracing. Tracing is expensive and the excessive number of tracings could be due to (1) creating @tf.function repeatedly in a loop, (2) passing tensors with different shapes, (3) passing Python objects instead of tensors. For (1), please define your @tf.function outside of the loop. For (2), @tf.function has experimental_relax_shapes=True option that relaxes argument shapes that can avoid unnecessary retracing. For (3), please refer to https://www.tensorflow.org/guide/function#controlling_retracing and https://www.tensorflow.org/api_docs/python/tf/function for more details.\n1/1 [==============================] - 0s 99ms/step - loss: 0.5000 - binary_accuracy: 0.5000\n\nbinary_accuracy: 50.00%\nWARNING:tensorflow:7 out of the last 7 calls to <function Model.make_predict_function.<locals>.predict_function at 0x7f8c533fe730> triggered tf.function retracing. Tracing is expensive and the excessive number of tracings could be due to (1) creating @tf.function repeatedly in a loop, (2) passing tensors with different shapes, (3) passing Python objects instead of tensors. For (1), please define your @tf.function outside of the loop. For (2), @tf.function has experimental_relax_shapes=True option that relaxes argument shapes that can avoid unnecessary retracing. For (3), please refer to https://www.tensorflow.org/guide/function#controlling_retracing and https://www.tensorflow.org/api_docs/python/tf/function for more details.\n[[0.]\n [0.]\n [0.]\n [0.]]\n"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a589925d7ae75cfcf021bf2f607b82a4debcd05
| 96,575 |
ipynb
|
Jupyter Notebook
|
Pymaceuticals.ipynb
|
rchiovaro/matplotlib-challenge
|
993ef4b9d85dcad6817e45e19d6c5c0bd52c7d3c
|
[
"ADSL"
] | null | null | null |
Pymaceuticals.ipynb
|
rchiovaro/matplotlib-challenge
|
993ef4b9d85dcad6817e45e19d6c5c0bd52c7d3c
|
[
"ADSL"
] | null | null | null |
Pymaceuticals.ipynb
|
rchiovaro/matplotlib-challenge
|
993ef4b9d85dcad6817e45e19d6c5c0bd52c7d3c
|
[
"ADSL"
] | null | null | null | 116.918886 | 10,752 | 0.83649 |
[
[
[
"## Observations and Insights",
"_____no_output_____"
],
[
"## Dependencies and starter code",
"_____no_output_____"
]
],
[
[
"# Dependencies and Setup\nimport matplotlib.pyplot as plt\nimport pandas as pd\nimport scipy.stats as st\nimport numpy as np\n\n# Study data files\nmouse_metadata = \"data/Mouse_metadata.csv\"\nstudy_results = \"data/Study_results.csv\"\n\n# Read the mouse data and the study results\nmouse_metadata = pd.read_csv(mouse_metadata)\nstudy_results = pd.read_csv(study_results)\n\n# Combine the data into a single dataset\ncombined_df = pd.merge(mouse_metadata, study_results, how='outer', on='Mouse ID')\ncombined_df.head()",
"_____no_output_____"
]
],
[
[
"## Summary statistics",
"_____no_output_____"
]
],
[
[
"# Generate a summary statistics table of mean, median, variance, standard deviation, and SEM of the tumor volume for each regimen\ngroup_by_regimen = combined_df.groupby(\"Drug Regimen\")\n\nmean = group_by_regimen['Tumor Volume (mm3)'].mean()\nmedian = group_by_regimen['Tumor Volume (mm3)'].median()\nvar = group_by_regimen['Tumor Volume (mm3)'].var()\nsd = group_by_regimen['Tumor Volume (mm3)'].std()\nsem = group_by_regimen['Tumor Volume (mm3)'].sem()\n\nsummary_statistics = pd.DataFrame({\"Mean\": mean,\n \"Median\": median,\n \"Variance\": var,\n \"Standard Deviation\": sd,\n \"SEM\": sem\n })\n\nsummary_statistics",
"_____no_output_____"
]
],
[
[
"## Bar plots",
"_____no_output_____"
]
],
[
[
"# Generate a bar plot showing number of data points for each treatment regimen using pandas\ncombined_df['Drug Regimen'].value_counts().plot(kind='bar')",
"_____no_output_____"
],
[
"# Generate a bar plot showing number of data points for each treatment regimen using pyplot\nval_counts = combined_df['Drug Regimen'].value_counts()\n\nplt.bar(val_counts.index.values, val_counts.values)\nplt.xticks(rotation=90)",
"_____no_output_____"
]
],
[
[
"## Pie plots",
"_____no_output_____"
]
],
[
[
"# Generate a pie plot showing the distribution of female versus male mice using pandas\ncombined_df['Sex'].value_counts().plot(kind='pie')",
"_____no_output_____"
],
[
"# Generate a pie plot showing the distribution of female versus male mice using pyplot\n\nsex_val_counts = combined_df['Sex'].value_counts()\n\nplt.pie(sex_val_counts.values, labels = sex_val_counts.index.values)",
"_____no_output_____"
]
],
[
[
"## Quartiles, outliers and boxplots",
"_____no_output_____"
]
],
[
[
"# Calculate the final tumor volume of each mouse across four of the most promising treatment regimens. Calculate the IQR and quantitatively determine if there are any potential outliers. \noutlier_index = combined_df.set_index('Drug Regimen')\ntop4 = outlier_index.loc[['Capomulin', 'Infubinol', 'Ceftamin', 'Ketapril'], ['Tumor Volume (mm3)']]\nquartiles = top4['Tumor Volume (mm3)'].quantile([.25,.5,.75])\nlowerq = quartiles[0.25]\nupperq = quartiles[0.75]\niqr = upperq-lowerq\n\nprint(f\"The lower quartile of Tumor Volume (mm3) is: {lowerq}\")\nprint(f\"The upper quartile of Tumor Volume (mm3) is: {upperq}\")\nprint(f\"The interquartile range of Tumor Volume (mm3) is: {iqr}\")\nprint(f\"The the median of Tumor Volume (mm3) is: {quartiles[0.5]} \")\n\nlower_bound = lowerq - (1.5*iqr)\nupper_bound = upperq + (1.5*iqr)\nprint(f\"Values below {lower_bound} could be outliers.\")\nprint(f\"Values above {upper_bound} could be outliers.\")\n\ntumor_outlier = top4.loc[(top4['Tumor Volume (mm3)'] < lower_bound) | (top4['Tumor Volume (mm3)'] > upper_bound)]\ntumor_outlier",
"The lower quartile of Tumor Volume (mm3) is: 45.0\nThe upper quartile of Tumor Volume (mm3) is: 55.650798762499996\nThe interquartile range of Tumor Volume (mm3) is: 10.650798762499996\nThe the median of Tumor Volume (mm3) is: 48.31576031 \nValues below 29.023801856250007 could be outliers.\nValues above 71.62699690624999 could be outliers.\n"
],
[
"# Generate a box plot of the final tumor volume of each mouse across four regimens of interest\ncap = outlier_index.loc['Capomulin','Tumor Volume (mm3)']\nram = outlier_index.loc['Ramicane','Tumor Volume (mm3)']\ninf = outlier_index.loc['Infubinol','Tumor Volume (mm3)']\ncef = outlier_index.loc['Ceftamin','Tumor Volume (mm3)']\nvar = [cap, ram,inf, cef]\nnames = ['Capomulin','Ramicane','Infubinol','Ceftamin']\n\nfig1, ax1 = plt.subplots()\nax1.set_title('Top 4 Drug Regimens')\nax1.set_ylabel('Tumor Volume (mm3)')\nax1.boxplot(var)\nx_axis = (np.arange(len(var))) + 1\ntick_locations = [value for value in x_axis]\nplt.xticks(tick_locations, names, rotation = 'horizontal')\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Line and scatter plots",
"_____no_output_____"
]
],
[
[
"# Generate a line plot of time point versus tumor volume for a mouse treated with Capomulin\n\ncap_table = combined_df.loc[combined_df['Drug Regimen'] == 'Capomulin']\nmouse = cap_table.loc[cap_table['Mouse ID'] == 's185']\nplt.plot(mouse['Timepoint'], mouse['Tumor Volume (mm3)'])",
"_____no_output_____"
],
[
"# Generate a scatter plot of mouse weight versus average tumor volume for the Capomulin regimen\n\naverage = cap_table.groupby(['Mouse ID']).mean()\nplt.scatter(average['Weight (g)'],average['Tumor Volume (mm3)'])",
"_____no_output_____"
],
[
"# Calculate the correlation coefficient and linear regression model for mouse weight and average tumor volume for the Capomulin regimen\n\nreg_line = st.linregress(average['Weight (g)'],average['Tumor Volume (mm3)'])\ny_value = average['Weight (g)']*reg_line[0]+reg_line[1]\nplt.scatter(average['Weight (g)'],average['Tumor Volume (mm3)'])\nplt.plot(average['Weight (g)'], y_value, color = 'green')",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
4a589ea54a5bd704af6ca26d87c7e4d36ce9a949
| 73,482 |
ipynb
|
Jupyter Notebook
|
docs/examples/DataSet/Accessing-data-in-DataSet.ipynb
|
qnl/Qcodes
|
ea2c5188f04828c6a76c9cfd9a66509277d7c09f
|
[
"MIT"
] | 1 |
2019-12-07T01:25:49.000Z
|
2019-12-07T01:25:49.000Z
|
docs/examples/DataSet/Accessing-data-in-DataSet.ipynb
|
Dominik-Vogel/Qcodes
|
b4cf7d58bc1bf3be97af6bf48f57cb6b87d588bb
|
[
"MIT"
] | null | null | null |
docs/examples/DataSet/Accessing-data-in-DataSet.ipynb
|
Dominik-Vogel/Qcodes
|
b4cf7d58bc1bf3be97af6bf48f57cb6b87d588bb
|
[
"MIT"
] | 1 |
2020-05-03T22:47:40.000Z
|
2020-05-03T22:47:40.000Z
| 69.84981 | 20,616 | 0.786805 |
[
[
[
"# Accessing data in a DataSet\n\nAfter a measurement is completed all the acquired data and metadata around it is accessible via a `DataSet` object. This notebook presents the useful methods and properties of the `DataSet` object which enable convenient access to the data, parameters information, and more. For general overview of the `DataSet` class, refer to [DataSet class walkthrough](DataSet-class-walkthrough.ipynb).",
"_____no_output_____"
],
[
"## Preparation: a DataSet from a dummy Measurement\n\nIn order to obtain a `DataSet` object, we are going to run a `Measurement` storing some dummy data (see [Dataset Context Manager](Dataset%20Context%20Manager.ipynb) notebook for more details).",
"_____no_output_____"
]
],
[
[
"import tempfile\nimport os\n\nimport numpy as np\n\nimport qcodes\nfrom qcodes import initialise_or_create_database_at, \\\n load_or_create_experiment, Measurement, Parameter, \\\n Station\nfrom qcodes.dataset.plotting import plot_dataset",
"_____no_output_____"
],
[
"db_path = os.path.join(tempfile.gettempdir(), 'data_access_example.db')\ninitialise_or_create_database_at(db_path)\n\nexp = load_or_create_experiment(experiment_name='greco', sample_name='draco')",
"_____no_output_____"
],
[
"x = Parameter(name='x', label='Voltage', unit='V',\n set_cmd=None, get_cmd=None)\nt = Parameter(name='t', label='Time', unit='s',\n set_cmd=None, get_cmd=None)\ny = Parameter(name='y', label='Voltage', unit='V',\n set_cmd=None, get_cmd=None)\ny2 = Parameter(name='y2', label='Current', unit='A',\n set_cmd=None, get_cmd=None)\nq = Parameter(name='q', label='Qredibility', unit='$',\n set_cmd=None, get_cmd=None)",
"_____no_output_____"
],
[
"meas = Measurement(exp=exp, name='fresco')\n\nmeas.register_parameter(x)\nmeas.register_parameter(t)\nmeas.register_parameter(y, setpoints=(x, t))\nmeas.register_parameter(y2, setpoints=(x, t))\nmeas.register_parameter(q) # a standalone parameter\n\nx_vals = np.linspace(-4, 5, 50)\nt_vals = np.linspace(-500, 1500, 25)\n\nwith meas.run() as datasaver:\n for xv in x_vals:\n for tv in t_vals:\n yv = np.sin(2*np.pi*xv)*np.cos(2*np.pi*0.001*tv) + 0.001*tv\n y2v = np.sin(2*np.pi*xv)*np.cos(2*np.pi*0.001*tv + 0.5*np.pi) - 0.001*tv\n datasaver.add_result((x, xv), (t, tv), (y, yv), (y2, y2v))\n q_val = np.max(yv) - np.min(y2v) # a meaningless value\n datasaver.add_result((q, q_val))\n\ndataset = datasaver.dataset",
"Starting experimental run with id: 5\n"
]
],
[
[
"For the sake of demonstrating what kind of data we've produced, let's use `plot_dataset` to make some default plots of the data.",
"_____no_output_____"
]
],
[
[
"plot_dataset(dataset)",
"_____no_output_____"
]
],
[
[
"## DataSet indentification\n\nBefore we dive into what's in the `DataSet`, let's briefly note how a `DataSet` is identified.",
"_____no_output_____"
]
],
[
[
"dataset.captured_run_id",
"_____no_output_____"
],
[
"dataset.exp_name",
"_____no_output_____"
],
[
"dataset.sample_name",
"_____no_output_____"
],
[
"dataset.name",
"_____no_output_____"
]
],
[
[
"## Parameters in the DataSet\n\nIn this section we are getting information about the parameters stored in the given `DataSet`.\n\n> Why is that important? Let's jump into *data*!\n\nAs it turns out, just \"arrays of numbers\" are not enough to reason about a given `DataSet`. Even comping up with a reasonable deafult plot, which is what `plot_dataset` does, requires information on `DataSet`'s parameters. In this notebook, we first have a detailed look at what is stored about parameters and how to work with this information. After that, we will cover data access methods.",
"_____no_output_____"
],
[
"### Run description\n\nEvery dataset comes with a \"description\" (aka \"run description\"):",
"_____no_output_____"
]
],
[
[
"dataset.description",
"_____no_output_____"
]
],
[
[
"The description, an instance of `RunDescriber` object, is intended to describe the details of a dataset. In the future releases of QCoDeS it will likely be expanded. At the moment, it only contains an `InterDependencies_` object under its `interdeps` attribute - which stores all the information about the parameters of the `DataSet`.\n\nLet's look into this `InterDependencies_` object.",
"_____no_output_____"
],
[
"### Interdependencies\n\n`Interdependencies_` object inside the run description contains information about all the parameters that are stored in the `DataSet`. Subsections below explain how the individual information about the parameters as well as their relationships are captured in the `Interdependencies_` object.",
"_____no_output_____"
]
],
[
[
"interdeps = dataset.description.interdeps\ninterdeps",
"_____no_output_____"
]
],
[
[
"#### Dependencies, inferences, standalones",
"_____no_output_____"
],
[
"Information about every parameter is stored in the form of `ParamSpecBase` objects, and the releationship between parameters is captured via `dependencies`, `inferences`, and `standalones` attributes.\n\nFor example, the dataset that we are inspecting contains no inferences, and one standalone parameter `q`, and two dependent parameters `y` and `y2`, which both depend on independent `x` and `t` parameters:",
"_____no_output_____"
]
],
[
[
"interdeps.inferences",
"_____no_output_____"
],
[
"interdeps.standalones",
"_____no_output_____"
],
[
"interdeps.dependencies",
"_____no_output_____"
]
],
[
[
"`dependencies` is a dictionary of `ParamSpecBase` objects. The keys are dependent parameters (those which depend on other parameters), and the corresponding values in the dictionary are tuples of independent parameters that the dependent parameter in the key depends on. Coloquially, each key-value pair of the `dependencies` dictionary is sometimes referred to as \"parameter tree\".\n\n`inferences` follows the same structure as `dependencies`.\n\n`standalones` is a set - an unordered collection of `ParamSpecBase` objects representing \"standalone\" parameters, the ones which do not depend on other parameters, and no other parameter depends on them.",
"_____no_output_____"
],
[
"#### ParamSpecBase objects",
"_____no_output_____"
],
[
"`ParamSpecBase` object contains all the necessary information about a given parameter, for example, its `name` and `unit`:",
"_____no_output_____"
]
],
[
[
"ps = list(interdeps.dependencies.keys())[0]\nprint(f'Parameter {ps.name!r} is in {ps.unit!r}')",
"Parameter 'y' is in 'V'\n"
]
],
[
[
"`paramspecs` property returns a tuple of `ParamSpecBase`s for all the parameters contained in the `Interdependencies_` object:",
"_____no_output_____"
]
],
[
[
"interdeps.paramspecs",
"_____no_output_____"
]
],
[
[
"Here's a trivial example of iterating through dependent parameters of the `Interdependencies_` object and extracting information about them from the `ParamSpecBase` objects:",
"_____no_output_____"
]
],
[
[
"for d in interdeps.dependencies.keys():\n print(f'Parameter {d.name!r} ({d.label}, {d.unit}) depends on:')\n for i in interdeps.dependencies[d]:\n print(f'- {i.name!r} ({i.label}, {i.unit})')",
"Parameter 'y' (Voltage, V) depends on:\n- 'x' (Voltage, V)\n- 't' (Time, s)\nParameter 'y2' (Current, A) depends on:\n- 'x' (Voltage, V)\n- 't' (Time, s)\n"
]
],
[
[
"#### Other useful methods and properties",
"_____no_output_____"
],
[
"`Interdependencies_` object has a few useful properties and methods which make it easy to work it and with other `Interdependencies_` and `ParamSpecBase` objects.\n\nFor example, `non_dependencies` returns a tuple of all dependent parameters together with standalone parameters:",
"_____no_output_____"
]
],
[
[
"interdeps.non_dependencies",
"_____no_output_____"
]
],
[
[
"`what_depends_on` method allows to find what parameters depend on a given parameter:",
"_____no_output_____"
]
],
[
[
"t_ps = interdeps.paramspecs[2]\nt_deps = interdeps.what_depends_on(t_ps)\n\nprint(f'Following parameters depend on {t_ps.name!r} ({t_ps.label}, {t_ps.unit}):')\nfor t_dep in t_deps:\n print(f'- {t_dep.name!r} ({t_dep.label}, {t_dep.unit})')",
"Following parameters depend on 't' (Time, s):\n- 'y' (Voltage, V)\n- 'y2' (Current, A)\n"
]
],
[
[
"### Shortcuts to important parameters\n\nFor the frequently needed groups of parameters, `DataSet` object itself provides convenient methods and properties.\n\nFor example, use `dependent_parameters` property to get only dependent parameters of a given `DataSet`:",
"_____no_output_____"
]
],
[
[
"dataset.dependent_parameters",
"_____no_output_____"
]
],
[
[
"This is equivalent to:",
"_____no_output_____"
]
],
[
[
"tuple(dataset.description.interdeps.dependencies.keys())",
"_____no_output_____"
]
],
[
[
"### Note on inferences\n\nInferences between parameters is a feature that has not been used yet within QCoDeS. The initial concepts around `DataSet` included it in order to link parameters that are not directly dependent on each other as \"dependencies\" are. It is very likely that \"inferences\" will be eventually deprecated and removed.",
"_____no_output_____"
],
[
"### Note on ParamSpec's\n\n> `ParamSpec`s originate from QCoDeS versions prior to `0.2.0` and for now are kept for backwards compatibility. `ParamSpec`s are completely superseded by `InterDependencies_`/`ParamSpecBase` bundle and will likely be deprecated in future versions of QCoDeS together with the `DataSet` methods/properties that return `ParamSpec`s objects.\n\nIn addition to the `Interdependencies_` object, `DataSet` also holds `ParamSpec` objects (not to be confused with `ParamSpecBase` objects from above). Similar to `Interdependencies_` object, the `ParamSpec` objects hold information about parameters and their interdependencies but in a different way: for a given parameter, `ParamSpec` object itself contains information on names of parameters that it depends on, while for the `InterDependencies_`/`ParamSpecBase`s this information is stored only in the `InterDependencies_` object.",
"_____no_output_____"
],
[
"`DataSet` exposes `paramspecs` property and `get_parameters()` method, both of which return `ParamSpec` objects of all the parameters of the dataset, and are not recommended for use:",
"_____no_output_____"
]
],
[
[
"dataset.paramspecs",
"_____no_output_____"
],
[
"dataset.get_parameters()",
"_____no_output_____"
],
[
"dataset.parameters",
"_____no_output_____"
]
],
[
[
"To give an example of what it takes to work with `ParamSpec` objects as opposed to `Interdependencies_` object, here's a function that one needs to write in order to find standalone `ParamSpec`s from a given list of `ParamSpec`s:",
"_____no_output_____"
]
],
[
[
"def get_standalone_parameters(paramspecs):\n all_independents = set(spec.name\n for spec in paramspecs\n if len(spec.depends_on_) == 0)\n used_independents = set(d for spec in paramspecs for d in spec.depends_on_)\n standalones = all_independents.difference(used_independents)\n return tuple(ps for ps in paramspecs if ps.name in standalones)\n\nall_parameters = dataset.get_parameters()\nstandalone_parameters = get_standalone_parameters(all_parameters)\nstandalone_parameters",
"_____no_output_____"
]
],
[
[
"## Getting data from DataSet\n\nIn this section methods for retrieving the actual data from the `DataSet` are discussed.\n\n### `get_parameter_data` - the powerhorse\n\n`DataSet` provides one main method of accessing data - `get_parameter_data`. It returns data for groups of dependent-parameter-and-its-independent-parameters in a form of a nested dictionary of `numpy` arrays:",
"_____no_output_____"
]
],
[
[
"dataset.get_parameter_data()",
"_____no_output_____"
]
],
[
[
"#### Avoid excessive calls to loading data\n\nNote that this call actually reads the data of the `DataSet` and in case of a `DataSet` with a lot of data can take noticable amount of time. Hence, it is recommended to limit the number of times the same data gets loaded in order to speed up the user's code.",
"_____no_output_____"
],
[
"#### Loading data of selected parameters\n\nSometimes data only for a particular parameter or parameters needs to be loaded. For example, let's assume that after inspecting the `InterDependencies_` object from `dataset.description.interdeps`, we concluded that we want to load data of the `q` parameter and the `y2` parameter. In order to do that, we just pass the names of these parameters, or their `ParamSpecBase`s to `get_parameter_data` call:",
"_____no_output_____"
]
],
[
[
"q_param_spec = list(interdeps.standalones)[0]\nq_param_spec",
"_____no_output_____"
],
[
"y2_param_spec = interdeps.non_dependencies[-1]\ny2_param_spec",
"_____no_output_____"
],
[
"dataset.get_parameter_data(q_param_spec, y2_param_spec)",
"_____no_output_____"
]
],
[
[
"### `get_data_as_pandas_dataframe` - for `pandas` fans\n\n`DataSet` provides one main method of accessing data - `get_data_as_pandas_dataframe`. It returns data for groups of dependent-parameter-and-its-independent-parameters in a form of a dictionary of `pandas.DataFrame` s:",
"_____no_output_____"
]
],
[
[
"dfs = dataset.get_data_as_pandas_dataframe()\n\n# For the sake of making this article more readable,\n# we will print the contents of the `dfs` dictionary\n# manually by calling `.head()` on each of the DataFrames\nfor parameter_name, df in dfs.items():\n print(f\"DataFrame for parameter {parameter_name}\")\n print(\"-----------------------------\")\n print(f\"{df.head()!r}\")\n print(\"\")",
"DataFrame for parameter q\n-----------------------------\n q\n0 3.000000\n1 2.085587\n2 2.259722\n3 3.315108\n4 3.995379\n\nDataFrame for parameter y\n-----------------------------\n y\nx t \n-4.0 -500.000000 -0.500000\n -416.666667 -0.416667\n -333.333333 -0.333333\n -250.000000 -0.250000\n -166.666667 -0.166667\n\nDataFrame for parameter y2\n-----------------------------\n y2\nx t \n-4.0 -500.000000 0.500000\n -416.666667 0.416667\n -333.333333 0.333333\n -250.000000 0.250000\n -166.666667 0.166667\n\n"
]
],
[
[
"Similar to `get_parameter_data`, `get_data_as_pandas_dataframe` also supports retrieving data for a given parameter(s), as well as `start`/`stop` arguments.\n\n`get_data_as_pandas_dataframe` is implemented based on `get_parameter_data`, hence the performance considerations mentioned above for `get_parameter_data` apply to `get_data_as_pandas_dataframe` as well.\n\nFor more details on `get_data_as_pandas_dataframe` refer to [Working with pandas and xarray article](Working-With-Pandas-and-XArray.ipynb).",
"_____no_output_____"
],
[
"### Data extraction into \"other\" formats\n\nIf the user desires to export a QCoDeS `DataSet` into a format that is not readily supported by `DataSet` methods, we recommend to use `get_data_as_pandas_dataframe` first, and then convert the resulting `DataFrame` s into a the desired format. This is becuase `pandas` package already implements converting `DataFrame` to various popular formats including comma-separated text file (`.csv`), HDF (`.hdf5`), xarray, Excel (`.xls`, `.xlsx`), and more; refer to [Working with pandas and xarray article](Working-With-Pandas-and-XArray.ipynb), and [`pandas` documentation](https://pandas.pydata.org/pandas-docs/stable/reference/frame.html#serialization-io-conversion) for more information.\n\nNevertheless, `DataSet` also provides the following convenient methods:\n\n* `DataSet.write_data_to_text_file`\n\nRefer to the docstrings of those methods for more information on how to use them.",
"_____no_output_____"
],
[
"### Not recommended data access methods",
"_____no_output_____"
],
[
"The following tree methods of accessing data in a dataset are not recommended for use, and will be deprecated soon:\n\n* `DataSet.get_data`\n* `DataSet.get_values`\n* `DataSet.get_setpoints`",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
4a589eb845ba8039f9a61d44ae444a18645a3af3
| 215,971 |
ipynb
|
Jupyter Notebook
|
.ipynb_checkpoints/Naruse_2020_ESURF_Figs-checkpoint.ipynb
|
narusehajime/nninv1d
|
697743346c7e24a8f06d676e2e9f3330aee93afe
|
[
"MIT"
] | null | null | null |
.ipynb_checkpoints/Naruse_2020_ESURF_Figs-checkpoint.ipynb
|
narusehajime/nninv1d
|
697743346c7e24a8f06d676e2e9f3330aee93afe
|
[
"MIT"
] | null | null | null |
.ipynb_checkpoints/Naruse_2020_ESURF_Figs-checkpoint.ipynb
|
narusehajime/nninv1d
|
697743346c7e24a8f06d676e2e9f3330aee93afe
|
[
"MIT"
] | null | null | null | 153.935139 | 45,560 | 0.853439 |
[
[
[
"# Inverse Analysis of Turbidites by Machine Learning Technique",
"_____no_output_____"
],
[
"# Preprocessing of training and test data sets",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport os\nimport ipdb\n\ndef connect_dataset(dist_start, dist_end, file_list, outputdir,\n topodx=5, offset=5000,gclass_num=4,test_data_num=100):\n \"\"\"\n Connect multiple raw data to produce the training and test data sets\n \"\"\"\n \n # Define start and end points in the data sets\n prox = np.round((dist_start+offset)/topodx).astype(np.int32)\n dist = np.round((dist_end+offset)/topodx).astype(np.int32)\n H = np.zeros([0,(dist-prox)* (gclass_num) ])\n icond = np.zeros([0,gclass_num + 3])\n \n # Read files and combine them\n for i in range(len(file_list)):\n H_temp = np.loadtxt(file_list[i] + '/H1.txt', delimiter = ',')[:,prox:dist]\n for j in range(2, gclass_num + 1):\n H_next = np.loadtxt(file_list[i] + '/H{}.txt'.format(j), delimiter = ',')[:,prox:dist]\n H_temp = np.concatenate([H_temp, H_next], axis = 1)\n icond_temp = np.loadtxt(file_list[i] + '/initial_conditions.txt', delimiter = ',')\n if icond_temp.shape[0] != H_temp.shape[0]:\n icond_temp = icond_temp[:-1,:]\n H = np.concatenate((H,H_temp),axis=0)\n icond = np.concatenate((icond,icond_temp),axis = 0)\n \n # Detect the maximum and minimum values in data sets\n max_x = np.max(H)\n min_x = np.min(H)\n icond_max = np.max(icond, axis=0)\n icond_min = np.min(icond, axis=0)\n \n # Split data for test and training sets\n H_train = H[0:-test_data_num,:]\n H_test = H[H.shape[0] - test_data_num:,:]\n icond_train = icond[0:-test_data_num,:]\n icond_test = icond[H.shape[0] - test_data_num:,:]\n \n # Save data sets\n if not os.path.exists(outputdir):\n os.mkdir(outputdir)\n np.save(os.path.join(outputdir, 'H_train.npy'), H_train)\n np.save(os.path.join(outputdir, 'H_test.npy'),H_test)\n np.save(os.path.join(outputdir, 'icond_train.npy'),icond_train)\n np.save(os.path.join(outputdir, 'icond_test.npy'),icond_test)\n np.save(os.path.join(outputdir, 'icond_min.npy'),icond_min)\n np.save(os.path.join(outputdir, 'icond_max.npy'),icond_max)\n np.save(os.path.join(outputdir, 'x_minmax.npy'),[min_x, max_x])\n \nif __name__==\"__main__\":\n\n # dist_end = 30000\n original_data_dir = \"/home/naruse/public/naruse/TC_training_data_4\"\n # parent_dir = \"/home/naruse/antidune/Documents/PythonScripts/DeepLearningTurbidite/20201018_30km\"\n parent_prefix = \"/home/naruse/public/naruse/DeepLearningTurbidite/distance\"\n if not os.path.exists(parent_prefix):\n os.mkdir(parent_prefix)\n \n output_dir = []\n \n test_distance = [1, 2, 3, 4, 5, 10, 15, 20, 25, 30]\n dist_start = [0]\n # test_distance = [95]\n for i in range(len(test_distance)):\n parent_dir = os.path.join(parent_prefix, str(test_distance[i]))\n if not os.path.exists(parent_dir):\n os.mkdir(parent_dir)\n output_dir.append(os.path.join(parent_dir, \"data\"))\n \n file_list = []\n for j in range(1,23):\n dirname = os.path.join(original_data_dir, \"TCModel_for_ML{0:02d}\".format(j), \"output\")\n if os.path.exists(dirname):\n file_list.append(dirname)\n # connect_dataset(dist_start, dist_end, file_list, outputdir, test_data_num=300)\n\n for k in range(len(test_distance)):\n connect_dataset(dist_start[0] * 1000, (test_distance[k] + dist_start[0]) * 1000,\n file_list, output_dir[k], test_data_num=300) \n \n ",
"_____no_output_____"
]
],
[
[
"# Common settings for plotting",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\n\n# settings for plotting\nlinewidth = 0.5\nlinestyle = ['-', '--', ':', '-.']\nlinecolor = [\"r\", \"g\", \"b\", \"c\", \"m\", \"y\", \"k\"]\nlc_id = 0\nparams = {'legend.fontsize': 5,\n 'legend.handlelength': 1.,\n 'legend.frameon': False,\n 'font.size' : 7,\n 'font.family': ['sans-serif'],\n 'font.sans-serif': ['Arial'],\n 'legend.labelspacing' : 0.5,\n 'legend.handletextpad' : 0.5,\n 'legend.markerscale' : 1.,\n }\nplt.rcParams.update(params)\n",
"_____no_output_____"
]
],
[
[
"# Check basic properties of training data sets",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nimport numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\n\nthick_file = '/home/naruse/public/naruse/DeepLearningTurbidite/fulldata/95/data/H_test.npy'\ngclass_num = 4\ndx = 5.0\ngclass_value = np.array([1.5, 2.5, 3.5, 4.5])\ngclass_name = []\nfor i in range(gclass_num):\n gclass_name.append('{}$\\phi$'.format(gclass_value[i]))\nH_test = np.load(thick_file) # data sets for values of volume-per-unit-area of all grain size classes\nnum_grids = int(H_test.shape[1]/gclass_num)\nnum_data = H_test.shape[0]\n\n# split data sets for every grain size classes\nvolume_unit_area = np.empty([gclass_num, num_data, num_grids]) # array for volume-per-unit-area for each grain size classes\n\nfor i in range(gclass_num):\n volume_unit_area[i, :, :] = H_test[:,i*num_grids:(i+1)*num_grids]\n\nthickness = np.sum(volume_unit_area, axis=0) # total thickness",
"_____no_output_____"
],
[
"# Calculate longitudinal variation of mean grain size\nmean_grain_size = np.zeros([num_data, num_grids])\nsignificant_thick = np.where(thickness > 0.01)\nfor i in range(gclass_num):\n mean_grain_size[significant_thick] += gclass_value[i] * volume_unit_area[i][significant_thick]\nmean_grain_size[significant_thick] /= thickness[significant_thick]\n\n# Calculate mean and standard deviation of thickness and maximum reach of beds\nmean_max_thick = np.average(np.max(thickness, axis=1))\nstd_max_thick = np.std(np.max(thickness, axis=1), ddof=1)\nx = np.tile(np.arange(0, num_grids * dx, dx), num_data).reshape(num_data, num_grids)\nx[thickness < 0.01] = 0\nmean_max_reach = np.average(np.max(x, axis=1))\nstd_max_reach = np.std(np.max(x, axis=1), ddof=1)\n\nprint('Mean of maximum thickness of beds: {} m'.format(mean_max_thick))\nprint('Standard deviation of maximum thickness of beds: {} m'.format(std_max_thick))\nprint('Mean of maximum reach of bed (> 1cm): {}'.format(mean_max_reach))\nprint('Standard deviation of maximum reach of bed (> 1cm): {}'.format(std_max_reach))\n\n# plot data sets\nxrange=np.array([0, 50000])\nxrange_grid = (xrange / dx).astype(np.int32)\nx = np.arange(xrange[0], xrange[1], dx)\nstart_id = 6\nnum_beds = 4\n\n# settings for plotting\nlinewidth = 0.5\nlinestyle = ['-', '--', ':', '-.']\nlinecolor = [\"r\", \"g\", \"b\", \"c\", \"m\", \"y\", \"k\"]\nlc_id = 0\nparams = {'legend.fontsize': 5,\n 'legend.handlelength': 3,\n 'legend.frameon': False,\n 'font.size' : 7,\n 'font.family': ['sans-serif'],\n 'font.sans-serif': ['Arial'],\n }\nplt.rcParams.update(params)\n\n# Plot results\nfig, ax = plt.subplots(2, 1, figsize=(8/2.54,8/2.54))\nplt.subplots_adjust(bottom=0.3, wspace=0.4)\nfor i in range(start_id, start_id + num_beds):\n ax[0].plot(x / 1000, thickness[i,xrange_grid[0]:xrange_grid[1]],\n lw=linewidth, linestyle=linestyle[(i - start_id)%4],\n color=linecolor[lc_id%7], label='bed {}'.format(i - start_id + 1))\n lc_id += 1\nax[0].set_xlabel('Distance (km)', fontsize=7)\nax[0].set_ylabel('Thickness (m)', fontsize=7)\nax[0].legend()\nylim = ax[0].get_ylim()\nxlim = ax[0].get_xlim()\nax[0].text(xlim[0] - 0.1 * xlim[1], ylim[0] + (ylim[1] - ylim[0])*1.05, 'a.', fontweight='bold', fontsize=9)\n\n# for k in range(start_id, start_id + num_beds):\n# ax[0,1].plot(x, mean_grain_size[k, xrange_grid[0]:xrange_grid[1]],label='bed{}'.format(k))\n# ax[0,1].legend()\n# ax[0,1].set_ylim([1.5, 4.5])\n\n# for j in range(gclass_num):\nfor j in range(gclass_num):\n ax[1].plot(x / 1000, volume_unit_area[j, start_id, xrange_grid[0]:xrange_grid[1]],\n lw=linewidth, color=linecolor[lc_id%7], label=gclass_name[j])\n lc_id += 1\nax[1].set_xlabel('Distance (km)', fontsize=7)\nax[1].set_ylabel('Volume per Unit Area (m)', fontsize=7)\n# ax[1].set_xlim(0,)\n# ax[1].set_ylim(0,)\nax[1].legend()\nylim = ax[1].get_ylim()\nxlim = ax[1].get_xlim()\nax[1].text(xlim[0] - 0.1 * xlim[1], ylim[0] + (ylim[1] - ylim[0])*1.05, 'b.', fontweight='bold', fontsize=9)\n\n# for j in range(gclass_num):\n# ax[1,1].plot(x, volume_unit_area[j, start_id + 1, xrange_grid[0]:xrange_grid[1]],label=gclass_name[j])\n# ax[1,1].legend()\n#plt.tight_layout()\nplt.tight_layout()\nplt.savefig('tex/fig04.eps')\nplt.show()",
"Mean of maximum thickness of beds: 1.2715092481333332 m\nStandard deviation of maximum thickness of beds: 1.6545673324702501 m\nMean of maximum reach of bed (> 1cm): 41976.46666666667\nStandard deviation of maximum reach of bed (> 1cm): 15697.077168089567\n"
]
],
[
[
"# Show training results depending on number of training data sets and length of sampling window",
"_____no_output_____"
]
],
[
[
"import os\nfrom os.path import join\nimport numpy as np\nimport matplotlib.pyplot as plt\n%matplotlib inline\n\ndatadir = '/home/naruse/public/naruse/DeepLearningTurbidite/distance'\nresdir_train_num = '/home/naruse/public/naruse/DeepLearningTurbidite/result_training_num_10'\nresdir_distance = '/home/naruse/public/naruse/DeepLearningTurbidite/result_distance_3500'\nbase_distance = 10\nbase_train_num = 3500\ncase_train_num = [500, 1000, 1500, 2000, 2500, 3000, 3500]\ncase_distance = [1, 2, 3, 4, 5, 10, 15, 20, 25, 30]\n\n# settings for plotting\nlinewidth = 0.5\nlinestyle = ['-', '--', ':', '-.']\nlinecolor = [\"r\", \"g\", \"b\", \"c\", \"m\", \"y\", \"k\"]\nlc_id = 0\nparams = {'legend.fontsize': 5,\n 'legend.handlelength': 1.,\n 'legend.frameon': False,\n 'font.size' : 7,\n 'font.family': ['sans-serif'],\n 'font.sans-serif': ['Arial'],\n 'legend.labelspacing' : 0.5,\n 'legend.handletextpad' : 0.5,\n 'legend.markerscale' : 1.,\n }\nplt.rcParams.update(params)\n\n# Plot results\nfig, ax = plt.subplots(2, 1, figsize=(8/2.54,8/2.54))\nplt.subplots_adjust(bottom=0.3, wspace=0.5)\n\n# Plot results depending on number of training data sets\nloss_train_num = []\nval_loss_train_num = []\nfor train_num in case_train_num:\n loss_train_num.append(\n np.loadtxt(join(resdir_train_num, '{}'.format(train_num),\n 'loss.txt'), delimiter=',')[-1])\n val_loss_train_num.append(\n np.loadtxt(join(resdir_train_num, '{}'.format(train_num),\n 'val_loss.txt'), delimiter=',')[-1])\nax[0].plot(case_train_num, loss_train_num, 'bo', markerfacecolor='w',\n label='Training', markersize=3)\nax[0].plot(case_train_num, val_loss_train_num, 'ro', markerfacecolor='r',\n label='Validation', markersize=3)\nax[0].set_xlabel('Number of Data Sets', fontsize=7)\nax[0].set_ylabel('Loss function (MSE)', fontsize=7)\nax[0].legend()\nylim = ax[0].get_ylim()\nxlim = ax[0].get_xlim()\nax[0].text(xlim[0] - 0.1 * xlim[1], ylim[0] + (ylim[1] - ylim[0])*1.05, 'a.', fontweight='bold', fontsize=9)\n\n# Plot results depending on lengths of sampling window\nloss_distance = []\nval_loss_distance = []\nfor distance in case_distance:\n loss_distance.append(\n np.loadtxt(join(resdir_distance, '{}'.format(distance),\n 'loss.txt'), delimiter=',')[-1])\n val_loss_distance.append(\n np.loadtxt(join(resdir_distance, '{}'.format(distance),\n 'val_loss.txt'), delimiter=',')[-1])\nax[1].plot(case_distance, loss_distance, 'go', markerfacecolor='w',\n label='Training', markersize=3)\nax[1].plot(case_distance, val_loss_distance, 'mo', markerfacecolor='m',\n label='Validation', markersize=3)\nax[1].set_xlabel('Length of Sampling Window (km)', fontsize=7)\nax[1].set_ylabel('Loss function (MSE)', fontsize=7)\nax[1].legend()\nylim = ax[1].get_ylim()\nxlim = ax[1].get_xlim()\nax[1].text(xlim[0] - 0.1 * xlim[1], ylim[0] + (ylim[1] - ylim[0])*1.05, 'b.', fontweight='bold', fontsize=9)\n\n# Save figures\nplt.tight_layout()\nplt.savefig('tex/fig05.eps')\n\n",
"_____no_output_____"
]
],
[
[
"# Show test results",
"_____no_output_____"
]
],
[
[
"import os\nimport numpy as np\nimport matplotlib.pyplot as plt\nfrom sklearn.metrics import r2_score\nfrom scipy import stats\nfrom sklearn.utils import resample\nimport pandas as pd\n%matplotlib inline\n\n# datadir = '/home/naruse/antidune/Documents/PythonScripts/DeepLearningTurbidite/20180419/data/'\n# resdir = '/home/naruse/antidune/Documents/PythonScripts/DeepLearningTurbidite/20180419/result_testGPU_4layers/2670/'\ndatadir = '/home/naruse/public/naruse/DeepLearningTurbidite/distance/10/data/'\nresdir = '/home/naruse/public/naruse/DeepLearningTurbidite/result_training_num_10/3500/'\n\ntest_result = np.loadtxt(os.path.join(resdir, 'test_result.txt'),delimiter=',')\nicond = np.load(os.path.join(datadir, 'icond_test.npy'))\nloss = np.loadtxt(os.path.join(resdir, 'loss.txt'), delimiter=',')\nvloss = np.loadtxt(os.path.join(resdir, 'val_loss.txt'), delimiter=',')\nepoch = range(0,loss.shape[0])\n\n# Calculate statistics\nresi_ratio = (test_result - icond) / icond\nresi = test_result - icond\nr2value = []\nfor i in range(icond.shape[1]):\n r2value.append(r2_score(icond[:, i], test_result[:, i]))\n\nmean_bias = np.average(resi,axis=0)\nstd_bias = np.std(resi,axis=0, ddof=1)\nrmse = np.sqrt(np.sum(resi ** 2, axis=0) / resi.shape[0])\nmae = np.sum(np.abs(resi), axis=0) / resi.shape[0]\n\nmean_bias_ratio = np.average(resi_ratio,axis=0)\nstd_bias_ratio = np.std(resi_ratio,axis=0, ddof=1)\nrmse_ratio = np.sqrt(np.sum(resi_ratio ** 2, axis=0) / resi_ratio.shape[0])\nmae_ratio = np.sum(np.abs(resi_ratio), axis=0) / resi.shape[0]\n\n# make a table for exhibiting statistics\ndf_stats = pd.DataFrame(\n {\n \"R^2\" : r2value,\n \"RMSE\" : rmse,\n \"RMSE (normalized)\" : rmse_ratio * 100,\n \"MAE\" : mae,\n \"MAE (normalized)\" : mae_ratio * 100,\n \"Mean bias\" : mean_bias,\n \"Mean bias (normalized)\" : mean_bias_ratio * 100,\n },\n index = [\n 'Initial height',\n 'Initial length',\n 'C_1',\n 'C_2',\n 'C_3',\n 'C_4',\n 'S_l']\n)\ndf_stats.loc['C_1':'S_l' ,['RMSE', 'MAE', 'Mean bias']] *= 100\nprint(df_stats.to_latex(float_format='%.2f'))\n\n\n# Boostrap resampling\n# n = 10000\n# resampled_resi = np.empty(resi.shape)\n# resampled_mean = np.zeros([n, resi.shape[1]])\n# for i in range(resi.shape[1]):\n# for j in range(n):\n# resampled_resi[:,i] = resample(resi_ratio[:,i])\n# resampled_mean[j, i] = np.average(resampled_resi[:,i])\n\n# Bootstrap mean and error range\n# mean_bias_bootstrap = np.average(resampled_mean, axis=0)\n# lowerbounds_bias_bootstrap = np.percentile(resampled_mean, 2.5, axis=0)\n# upperbounds_bias_bootstrap = np.percentile(resampled_mean, 97.5, axis=0)\n\n# settings for plotting\nlinewidth = 0.5\nlinestyle = ['-', '--', ':', '-.']\nlinecolor = [\"r\", \"g\", \"b\", \"c\", \"m\", \"y\", \"k\"]\nlc_id = 0\nparams = {'legend.fontsize': 5,\n 'legend.handlelength': 1.,\n 'legend.frameon': False,\n 'font.size' : 7,\n 'font.family': ['sans-serif'],\n 'font.sans-serif': ['Arial'],\n 'legend.labelspacing' : 0.5,\n 'legend.handletextpad' : 0.5,\n 'legend.markerscale' : 1.,\n }\nplt.rcParams.update(params)\n\n# plot training history\nfig, ax = plt.subplots(1,1, figsize=(8/2.54,4/2.54))\nax.plot(epoch, loss, 'b-',label='Loss', lw=0.5)\nax.plot(epoch, vloss, 'y-',label='Validation', lw=0.5)\nax.set_xlabel('Epoch')\nax.set_ylabel('Loss function (MSE)')\nax.legend(loc=\"upper right\")\nplt.savefig('tex/fig06.eps')\n\nprint('Training loss: {}'.format(loss[-1]))\nprint('Validation loss: {}'.format(vloss[-1]))\n\nhfont = {'fontname':'Century Gothic'}\ntextcol = 'k'\ntitlelabel = ['Initial Length\\n(m)', 'Initial Height\\n(m)', '$C_1$', '$C_2$', '$C_3$', '$C_4$', '$S_L$']\n\n# Scattered plots to compare the predicted values with the true values\nfig2, ax2 = plt.subplots(int(len(titlelabel)/2) + 1, 2, figsize=(12/2.54, 19/2.54))\nplt.subplots_adjust(wspace=0.1, hspace=0.6)\nfor i in range(len(titlelabel)):\n x_fig = int(i/2)\n y_fig = i%2 \n ax2[x_fig, y_fig].plot(icond[:,i],test_result[:,i],\"o\", markersize=1)\n ax2[x_fig, y_fig].plot([0,np.max(test_result[:,i])], [0, np.max(test_result[:,i])],\n \"-\", lw=linewidth*2)\n ax2[x_fig, y_fig].set_xlabel('True Value',color=textcol,fontsize=7)\n ax2[x_fig, y_fig].set_ylabel('Estimated Value',color=textcol,fontsize=7)\n ax2[x_fig, y_fig].set_title(titlelabel[i],color=textcol,fontsize=9)\n ax2[x_fig, y_fig].tick_params(colors=textcol,length=2,labelsize=5)\n ax2[x_fig, y_fig].set_aspect('equal')\n xlim = ax2[x_fig, y_fig].get_xlim()\n ylim = ax2[x_fig, y_fig].get_ylim()\n xloc = xlim[0] + (xlim[1] - xlim[0]) * 0.1\n yloc = ylim[0] + (ylim[1] - ylim[0]) * 0.85\n ax2[x_fig, y_fig].text(xloc, yloc, '$R^2 = ${:.3f}'.format(r2value[i]))\n# fig.tight_layout()\nplt.savefig('tex/fig07.eps')\n #plt.show()\n\n# Histograms for prediction errors\nfig3, ax3 = plt.subplots(int(len(titlelabel)/2) + 1, 2, figsize=(12/2.54, 16/2.54))\nplt.subplots_adjust(wspace=0.5, hspace=0.7)\nfor i in range(len(titlelabel)):\n x_fig = int(i/2)\n y_fig = i%2\n ax3[x_fig, y_fig].hist(resi[:,i],bins=20)\n ax3[x_fig, y_fig].set_title(titlelabel[i],color=textcol)\n ax3[x_fig, y_fig].set_xlabel('Deviation from true value',color=textcol, fontsize=7)\n ax3[x_fig, y_fig].set_ylabel('Frequency',color=textcol, fontsize=7)\n ax3[x_fig, y_fig].tick_params(colors=textcol, length=2, labelsize=5)\n # xlim = ax3[x_fig, y_fig].get_xlim()\n # ylim = ax3[x_fig, y_fig].get_ylim()\n # xloc = xlim[0] + (xlim[1] - xlim[0]) * 0.1\n # yloc = ylim[0] + (ylim[1] - ylim[0]) * 0.7\n ax3[x_fig, y_fig].text(0.99, 0.95,\n 'RMSE = {0:.1f} %\\n Mean Bias = {1:.1f}'.format(\n rmse[i] * 100, mean_bias_ratio[i] * 100),\n # lowerbounds_bias_bootstrap[i] * 100,\n # upperbounds_bias_bootstrap[i] * 100),\n horizontalalignment='right', verticalalignment='top',\n transform=ax3[x_fig, y_fig].transAxes, fontsize=5)\n\nfig.tight_layout()\nplt.savefig('tex/fig08.eps')\n #plt.show()\n",
"\\begin{tabular}{lrrrrrrr}\n\\toprule\n{} & R\\textasciicircum 2 & RMSE & RMSE (normalized) & MAE & MAE (normalized) & Mean bias & Mean bias (normalized) \\\\\n\\midrule\nInitial height & 0.99 & 18.97 & 8.55 & 14.81 & 5.96 & -12.93 & -5.18 \\\\\nInitial length & 0.99 & 15.82 & 7.53 & 12.09 & 4.92 & -2.33 & -2.06 \\\\\nC\\_1 & 0.99 & 0.02 & 12.91 & 0.02 & 6.00 & -0.01 & -4.44 \\\\\nC\\_2 & 0.99 & 0.02 & 15.57 & 0.02 & 7.67 & -0.01 & -4.29 \\\\\nC\\_3 & 0.99 & 0.02 & 13.03 & 0.02 & 6.39 & -0.00 & -2.49 \\\\\nC\\_4 & 0.99 & 0.03 & 13.71 & 0.02 & 6.67 & -0.01 & -4.21 \\\\\nS\\_l & 0.98 & 0.03 & 19.56 & 0.03 & 11.67 & 0.03 & 11.45 \\\\\n\\bottomrule\n\\end{tabular}\n\nTraining loss: 0.0037841848097741604\nValidation loss: 0.0010345189366489649\n"
]
],
[
[
"# Check bias and errors of predicted values",
"_____no_output_____"
]
],
[
[
"from scipy import stats\nimport numpy as np\nfrom sklearn.utils import resample\nimport ipdb\n\nresi_ratio = (test_result - icond) / icond\nresi = test_result - icond\n\nprint(\"mean bias\")\nprint(np.average(resi,axis=0))\nprint(\"2σ of bias\")\nprint(np.std(resi,axis=0, ddof=1)*2)\nprint(\"RMSE\")\nprint(np.sqrt(np.sum(resi**2)/resi.shape[0]/resi.shape[1]))\n\nprint(\"mean bias (ratio)\")\nprint(np.average(resi_ratio,axis=0))\nprint(\"2σ of bias (ratio)\")\nprint(np.std(resi_ratio,axis=0, ddof=1)*2)\nprint(\"RMSE (ratio)\")\nprint(np.sqrt(np.sum(resi_ratio**2)/resi_ratio.shape[0]/resi_ratio.shape[1]))\n\nprint(\"p-values of the Shapiro-Wilk test for normality\")\nfor i in range(resi.shape[1]):\n print(stats.shapiro(resi[:,i])[1])\n\n\n# Bootstrap mean and error range\nprint(\"mean bias (bootstrap samples)\")\nprint(np.average(resampled_mean, axis=0))\nprint(\"2.5 percentile of biases (bootstrap samples)\")\nprint(np.percentile(resampled_mean, 2.5, axis=0))\nprint(\"97.5 percentile of biases (bootstrap samples)\")\nprint(np.percentile(resampled_mean, 97.5, axis=0))\n\n# Histograms of bootstrap samples\nhfont = {'fontname':'Century Gothic'}\ntextcol = 'k'\ntitlelabel = ['Initial Length', 'Initial Height', '$C_1$', '$C_2$', '$C_3$', '$C_4$', '$S_L$']\nfig4, ax4 = plt.subplots(int(len(titlelabel)/2) + 1, 2, figsize=(8, 4 * np.ceil(len(titlelabel) / 2)))\nplt.subplots_adjust(wspace=0.6, hspace=0.4)\nfor i in range(len(titlelabel)):\n ax4[int(i/2), i%2].hist(resampled_mean[:,i],bins=20)\n ax4[int(i/2), i%2].set_title(titlelabel[i],color=textcol,size=14,**hfont)\n ax4[int(i/2), i%2].set_xlabel('Bias in Bootstrap sample',color=textcol,size=14,**hfont)\n ax4[int(i/2), i%2].set_ylabel('Frequency',color=textcol,size=14,**hfont)\n ax4[int(i/2), i%2].tick_params(labelsize=14,colors=textcol)\nfig.tight_layout()\nplt.savefig('hist_bootstrap.pdf')\n\n",
"mean bias\n[-1.29266316e+01 -2.32764039e+00 -1.42043143e-04 -5.58904978e-05\n -3.05209377e-05 -9.54839696e-05 2.89349487e-04]\n2σ of bias\n[2.78196676e+01 3.13524848e+01 4.04460461e-04 4.01557574e-04\n 4.80633580e-04 5.31698802e-04 3.78284572e-04]\nRMSE\n9.33717357851972\nmean bias (ratio)\n[-0.05178297 -0.02058269 -0.04440971 -0.0429332 -0.02491598 -0.04206466\n 0.11448568]\n2σ of bias (ratio)\n[0.13624629 0.14501837 0.24293689 0.29978833 0.2562501 0.26138126\n 0.31784317]\nRMSE (ratio)\n0.13519205042019936\np-values of the Shapiro-Wilk test for normality\n7.849864286152072e-15\n0.00040593656012788415\n8.476071343466174e-06\n2.2713582126243637e-09\n5.597754790080678e-14\n1.5576918959347638e-14\n3.69856643374078e-05\nmean bias (bootstrap samples)\n[-0.05177063 -0.02051589 -0.04449836 -0.04274484 -0.02480929 -0.04208389\n 0.11432631]\n2.5 percentile of biases (bootstrap samples)\n[-0.05933748 -0.02881048 -0.05900583 -0.06028632 -0.03990595 -0.05753963\n 0.09736634]\n97.5 percentile of biases (bootstrap samples)\n[-0.04408708 -0.01256159 -0.03179467 -0.02651334 -0.01097467 -0.02810994\n 0.13281014]\n"
]
],
[
[
"# Compare time evolution of reconstructed parameters with original ones",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\nfrom os.path import join\nfrom os import mkdir\nfrom scipy.interpolate import interp1d\nimport pandas as pd\n%matplotlib\n\noriginal_dir = '/home/naruse/antidune/Documents/MATLAB/TCtrainData_forML/TCModel_for_MLTEST/test_output_original5'\nestimated_dir = '/home/naruse/antidune/Documents/MATLAB/TCtrainData_forML/TCModel_for_MLTEST/test_output_reconst5'\ndist_offset = 5000.\ndist_max = 30000.\ntopodx = 5\ngrid_origin = int(dist_offset / topodx)\ngrid_end = int((dist_max + dist_offset)/topodx)\nsnapshot_time = np.array([2000, 3500, 5000])\ntime_interval = 200.\ntime_frame = (snapshot_time / time_interval).astype(np.int64)\n\nicond_estimated = np.loadtxt(join(estimated_dir, 'icond.txt'),delimiter=',')\nHt_estimated = np.loadtxt(join(estimated_dir, 'Ht.txt'),delimiter=',')\nCt_estimated = np.loadtxt(join(estimated_dir, 'Ct.txt'),delimiter=',')\nU_estimated = np.loadtxt(join(estimated_dir, 'U.txt'),delimiter=',')\nx_estimated = np.loadtxt(join(estimated_dir, 'x.txt'),delimiter=',')\nx_bed = np.loadtxt(join(estimated_dir, 'x_init.txt'),delimiter=',')\ntime_estimated = np.loadtxt(join(estimated_dir, 'time.txt'),delimiter=',') \n\nicond_original = np.loadtxt(join(original_dir, 'icond.txt'),delimiter=',')\nHt_original = np.loadtxt(join(original_dir, 'Ht.txt'),delimiter=',')\nCt_original = np.loadtxt(join(original_dir, 'Ct.txt'),delimiter=',')\nU_original = np.loadtxt(join(original_dir, 'U.txt'),delimiter=',')\nx_original = np.loadtxt(join(original_dir, 'x.txt'),delimiter=',')\ntime_original = np.loadtxt(join(original_dir, 'time.txt'),delimiter=',')\n\n\nprint('Reconstructed values: {}'.format(icond_estimated))\nprint('True values: {}'.format(icond_original))\nprint('RMSE: {}'.format(np.sqrt(np.sum(((icond_estimated - icond_original)/icond_original)**2)/icond_estimated.shape[0])))\n\n# Make a table to exhibit true and predicted values of model input parameters\ndf = pd.DataFrame(np.array([[icond_original[:]], [icond_estimated[:]]]).reshape(2, 7),\n columns=[\n 'Initial height (m)',\n 'Initial length (m)',\n 'C_1 (%)',\n 'C_2 (%)',\n 'C_3 (%)',\n 'C_4 (%)',\n 'S_l (%)'\n ],\n index=[\n 'True input parameters',\n 'Estimated parameters'\n ])\ndf.loc[:, 'C_1 (%)':'S_l (%)'] *= 100\nprint(df.to_latex(float_format='%.2f'))\n\n\n# settings for plotting\nlinewidth = 0.5\nlinestyle = ['-', '--', ':', '-.']\nlinecolor = [\"r\", \"g\", \"b\", \"c\", \"m\", \"y\", \"k\"]\nlc_id = 0\nparams = {'legend.fontsize': 5,\n 'legend.handlelength': 1.,\n 'legend.frameon': False,\n 'font.size' : 7,\n 'font.family': ['sans-serif'],\n 'font.sans-serif': ['Arial'],\n 'legend.labelspacing' : 0.5,\n 'legend.handletextpad' : 0.5,\n 'legend.markerscale' : 1.,\n }\nplt.rcParams.update(params)\n\n# Plot results\nfig1, ax1 = plt.subplots(3, 1, figsize=(8/2.54, 12/2.54))\nplt.subplots_adjust(bottom=0.3, wspace=0.5)\n\n# plot flow velocity\nfor tframe, col in zip(time_frame, linecolor):\n ax1[0].plot(x_estimated[tframe,:]/1000, U_estimated[tframe,:],\n '-', color=col, lw=linewidth, label='{} sec.'.format(tframe*time_interval))\n ax1[0].plot(x_original[tframe,:]/1000, U_original[tframe,:],'--',\n color=col, lw=linewidth, label=None)\n# ax1[0].set_title('Flow Velocity', fontsize=9)\nax1[0].set_xlabel('Distance (km)', fontsize = 7)\nax1[0].set_ylabel('Velocity (m/s)', fontsize = 7)\nax1[0].legend()\nxlim = ax1[0].get_xlim()\nylim = ax1[0].get_ylim()\nax1[0].text(xlim[0] - 0.1 * xlim[1], ylim[0] + (ylim[1] - ylim[0])*1.05, 'a.', fontweight='bold', fontsize=9)\n\n# plot sediment concentration\nfor tframe, col in zip(time_frame, linecolor):\n ax1[1].plot(x_estimated[tframe,:]/1000, Ct_estimated[tframe,:] * 100, '-',\n color=col, lw=linewidth, label='{} sec.'.format(tframe*time_interval))\n ax1[1].plot(x_original[tframe,:]/1000, Ct_original[tframe,:] * 100, '--',\n color=col, lw=linewidth, label=None)\n# ax1[1].set_title('Total Concentration', fontsize = 9)\nax1[1].set_xlabel('Distance (km)', fontsize = 7)\nax1[1].set_ylabel('Concentration (%)', fontsize = 7)\nax1[1].legend()\nxlim = ax1[1].get_xlim()\nylim = ax1[1].get_ylim()\nax1[1].text(xlim[0] - 0.1 * xlim[1], ylim[0] + (ylim[1] - ylim[0])*1.05, 'b.', fontweight='bold', fontsize=9)\n\n# plot thickness \nax1[2].plot(x_bed[grid_origin:grid_end]/1000, Ht_estimated[-1,grid_origin:grid_end],'k--',\n lw=linewidth, label='Estimated')\nax1[2].plot(x_bed[grid_origin:grid_end]/1000, Ht_original[-1,grid_origin:grid_end],'k-',\n lw=linewidth, label='Original')\n# ax1[2].set_title('Bed thickness', size = 9, **hfont)\nax1[2].set_xlabel('Distance (km)', fontsize = 7)\nax1[2].set_ylabel('Thickness (m)', fontsize = 7)\nxlim = ax1[2].get_xlim()\nylim = ax1[2].get_ylim()\nax1[2].legend()\nax1[2].text(xlim[0] - 0.1 * xlim[1], ylim[0] + (ylim[1] - ylim[0])*1.05, 'c.', fontweight='bold', fontsize=9)\n\n# save figure\nplt.tight_layout()\nplt.savefig('tex/fig09.eps')\n\n# Time evolution at fixed location\nstart = 0.0\nendtime = 5000.0\nstart_d = int(start / time_interval)\nendtime_d = int(endtime / time_interval)\n\noutcrop = np.array([5*1000, 8 * 1000, 10 * 1000])\nlinecolor = ['r', 'g', 'b']\n\nU_original_loc = np.zeros([len(time_original),len(outcrop)])\nU_estimated_loc = np.zeros([len(time_original),len(outcrop)])\nif len(time_original) > len(time_estimated):\n time_length = len(time_estimated)\nelse:\n time_length = len(time_original)\n\nfor j in range(time_length):\n f_original = interp1d(x_original[j,:], U_original[j,:], kind=\"linear\", bounds_error=False, fill_value=0)\n U_original_loc[j,:] = f_original(outcrop)\n f_estimated = interp1d(x_estimated[j,:], U_estimated[j,:], kind=\"linear\", bounds_error=False, fill_value=0)\n U_estimated_loc[j,:] = f_estimated(outcrop)\n\n#図にプロットする\nfig2, ax2 = plt.subplots(1, 1, figsize=(8, 4))\nplt.subplots_adjust(wspace=0.6, hspace=0.4)\nfor k in range(len(outcrop)):\n ax2.plot(time_original[start_d:endtime_d], U_original_loc[start_d:endtime_d,k], '--',\n color= linecolor[k], label=None)\n ax2.plot(time_estimated[start_d:endtime_d], U_estimated_loc[start_d:endtime_d,k], '-',\n color= linecolor[k], label='{} km'.format(outcrop[k] / 1000))\nax2.legend()\nax2.set_xlabel('Time (s.)')\nax2.set_ylabel('Velocity (m/s)')\n# ax2.set_title('Velocity')\nplt.savefig('compare_result_fixedloc.svg')\n\n\n",
"Using matplotlib backend: agg\nReconstructed values: [4.5467e+02 3.0173e+02 1.7977e-03 1.5926e-04 9.2914e-03 7.2649e-03\n 2.5254e-03]\nTrue values: [4.8441e+02 3.1818e+02 1.6565e-03 5.4632e-04 9.5318e-03 7.4443e-03\n 2.2881e-03]\nRMSE: 0.27454715314370537\n\\begin{tabular}{lrrrrrrr}\n\\toprule\n{} & Initial height (m) & Initial length (m) & C\\_1 (\\%) & C\\_2 (\\%) & C\\_3 (\\%) & C\\_4 (\\%) & S\\_l (\\%) \\\\\n\\midrule\nTrue input parameters & 484.41 & 318.18 & 0.17 & 0.05 & 0.95 & 0.74 & 0.23 \\\\\nEstimated parameters & 454.67 & 301.73 & 0.18 & 0.02 & 0.93 & 0.73 & 0.25 \\\\\n\\bottomrule\n\\end{tabular}\n\n"
]
],
[
[
"# tests with normal random numbers",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\nfrom tensorflow.keras.models import load_model\nfrom scipy import stats\nfrom scipy.stats import sem\nimport os\n%matplotlib inline\n\ndef check_noise(model=None,\n X_test=None,\n y_test=None,\n y_min=None,\n y_max=None,\n min_x=None,\n max_x=None,\n err_rate=0.10,\n datadir = None,\n resdir = None,\n gclass = 4,\n topodx = 5,\n plot_fig = True,\n ):\n \n # Obtain the original data sets\n if X_test is None: X_test = np.load(os.path.join(datadir, 'H_test.npy'))\n if y_test is None: y_test = np.load(os.path.join(datadir, 'icond_test.npy'))\n if y_min is None: y_min = np.load(os.path.join(datadir, 'icond_min.npy'))\n if y_max is None: y_max = np.load(os.path.join(datadir, 'icond_max.npy'))\n\n \n # normalization\n if min_x is None or max_x is None: min_x, max_x = np.load(os.path.join(datadir, 'x_minmax.npy'))\n X_test_norm = (X_test - min_x) / (max_x - min_x)\n\n # add noise\n # 2 sigma = true parameter times err_rate\n err = np.random.normal(size=X_test_norm.shape)\n x_test_norm_w_error = X_test_norm + err * 0.5 * err_rate * X_test_norm\n num_node_per_gclass = int(X_test_norm.shape[1] / gclass)\n dist = np.arange(0,num_node_per_gclass)* topodx\n \n\n #print(X_test_norm[1,1000:1010])\n #print(x_test_norm_w_error[1,1000:1010])\n #print(err[1,1000:1010])\n\n # load the model if the model is None\n # model = load_model(resdir+'model.hdf5')\n test_result = model.predict(X_test_norm)\n test_result = test_result * (y_max - y_min) + y_min\n test_result_w_error = model.predict(x_test_norm_w_error)\n test_result_w_error = test_result_w_error * (y_max - y_min) + y_min\n\n # Load true parameters\n icond = np.load(os.path.join(datadir, 'icond_test.npy'))\n loss = np.loadtxt(os.path.join(resdir, 'loss.txt'), delimiter=',')\n epoch = range(0,len(loss))\n vloss = np.loadtxt(resdir+'val_loss.txt',delimiter=',')\n \n # Calculate residuals\n resi = (test_result - icond)\n resi_w_error = (test_result_w_error - icond)\n resi_w_error_ratio = (test_result_w_error - icond) / icond\n\n # Plot figure of each test\n if plot_fig:\n plt.figure()\n plt.plot(x_test_norm_w_error[1,0:num_node_per_gclass], label='With Error')\n plt.plot(X_test_norm[1,0:num_node_per_gclass], label='Original')\n plt.xlabel('Distance')\n plt.ylabel('Normalized thickness')\n plt.legend()\n\n \n titlelabel = ['Initial Length', 'Initial Height', '$C_1$', '$C_2$', '$C_3$', '$C_4$', '$S_1$']\n hfont = {'fontname':'Century Gothic'}\n textcol = 'k'\n \n for i in range(len(titlelabel)):\n plt.figure()\n plt.plot(icond[:,i],test_result[:,i],\"bo\",label='without error')\n plt.plot(icond[:,i],test_result_w_error[:,i],\"ro\",label='with error ({:.0f}%)'.format(err_rate*100))\n plt.title(titlelabel[i],color=textcol,size=14,**hfont)\n plt.xlabel('True values',color=textcol,size=14,**hfont)\n plt.ylabel('Estimated values',color=textcol,size=14,**hfont)\n plt.legend()\n plt.tick_params(labelsize=14,colors=textcol)\n plt.savefig(titlelabel[i] + 'err{:.0f}'.format(err_rate*100) + '.pdf')\n #plt.show()\n\n for i in range(len(titlelabel)):\n plt.figure()\n plt.hist(resi_w_error[:,i],bins=20)\n plt.title(titlelabel[i])\n plt.xlabel('Deviation from true value')\n plt.ylabel('Frequency')\n #plt.show()\n\n# print(\"Mean Square error\")\n# print(np.average(resi**2,axis=0))\n# print(\"MSE with noise\")\n# print(np.average(resi_w_error**2,axis=0))\n# print(\"Mean error\")\n# print(np.average(resi,axis=0))\n# print(\"Mean error with noise)\n# print(np.average(resi_w_error,axis=0))\n# print(\"2 sigma of residuals\")\n# print(np.std(resi,axis=0)*2)\n# print(\"2 sigma of residuals with noise\")\n# print(np.std(resi_w_error,axis=0)*2)\n# print(\"ratio of residuals to true value\")\n# print(np.average(np.abs(resi)/icond,axis=0))\n# print(\"ratio of residuals to true value with noise\")\n# print(np.average(np.abs(resi_w_error)/icond,axis=0))\n# print(\"p-values of the Shapiro-Wilk test for normality\")\n# for i in range(resi.shape[1]):\n # print(stats.shapiro(resi[:,i])[1])\n # print(\"p-values of the Shapiro-Wilk test for normality (with error)\")\n # for i in range(resi_w_error.shape[1]):\n # print(stats.shapiro(resi_w_error[:,i])[1])\n \n # Return normalized RMSE\n RMS = np.sqrt(np.sum(resi_w_error_ratio ** 2) / resi_w_error_ratio.shape[0] / resi_w_error_ratio.shape[1])\n \n return RMS\n \nif __name__ == \"__main__\":\n datadir = '/home/naruse/public/naruse/DeepLearningTurbidite/distance/10/data/'\n resdir = '/home/naruse/public/naruse/DeepLearningTurbidite/result_training_num_10/3500/'\n model = load_model(os.path.join(resdir, 'model.hdf5'))\n \n noisetest_err_rate = np.linspace(0,2.0,40)\n result_noise = np.zeros(len(noisetest_err_rate))\n result_noise_stderr = np.zeros(len(noisetest_err_rate))\n num_tests = 20\n for i in range(len(noisetest_err_rate)):\n testres = np.zeros([num_tests])\n for j in range(num_tests):\n testres[j] = check_noise(model, datadir=datadir, resdir=resdir, err_rate=noisetest_err_rate[i], plot_fig=False)\n result_noise[i] = np.average(testres)\n result_noise_stderr[i] = sem(testres)\n \n np.savetxt(\"result_w_error.csv\",result_noise,delimiter=',')\n \n ",
"_____no_output_____"
],
[
"%matplotlib inline\n\n# plot result of noise tests\n\nfig1, ax1 = plt.subplots(1, 1, figsize=(8/2.54, 5/2.54))\nplt.subplots_adjust(bottom=0.3, wspace=0.5, hspace=0.3)\n\nax1.errorbar(noisetest_err_rate*100, result_noise, color='g', yerr=result_noise_stderr, ecolor='k', capsize=1.)\n# ax1.title(\"$S_L$\")\nax1.set_xlabel('Ratio of standard deviation of\\n random noise to original value (%)')\nax1.set_ylabel('RMS error')\nax1.set_ylim([0,0.5])\n# ax1.legend()\n# plt.tick_params(labelsize=14,colors=textcol)\nplt.tight_layout()\nplt.savefig(\"tex/fig10.eps\")\n \n ",
"_____no_output_____"
]
],
[
[
"# Subsampling tests",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\nfrom tensorflow.keras.models import load_model\nfrom scipy import stats\nfrom scipy.interpolate import interp1d \nfrom scipy.stats import sem\nfrom os.path import join\n%matplotlib inline\n\ndef check_interp(model=None,\n X_test=None,\n y_test=None,\n y_min=None,\n y_max=None,\n frac = 0.005,\n datadir = None,\n resdir = None,\n plot_fig = True,\n ):\n\n # Obtain the original data sets\n if X_test is None: X_test = np.load(join(datadir, 'H_test.npy'))\n if y_test is None: y_test = np.load(join(datadir, 'icond_test.npy'))\n if y_min is None: y_min = np.load(join(datadir, 'icond_min.npy'))\n if y_max is None: y_max = np.load(join(datadir, 'icond_max.npy'))\n\n \n # normalization\n min_x, max_x = np.load(join(datadir, 'x_minmax.npy'))\n X_test_norm = (X_test - min_x) / (max_x - min_x)\n\n # Subsampling\n #frac = 0.005 # ratio of subsampling\n gclass = 4 # number of grain size classes\n coord_num = X_test_norm.shape[1] / gclass # number of grids\n sam_coord_num = np.round(frac * coord_num) # number of subsampled grids\n x_coord = np.arange(X_test_norm.shape[1]/ gclass) # Index number of grids\n sampleid = np.sort(np.random.choice(x_coord,int(sam_coord_num),replace=False)) # subsampled id of grids\n\n thick_interp = np.zeros(X_test.shape) # interpolated thickness data\n for j in range(gclass):\n sid = sampleid + coord_num * j\n #print(sid)\n sindex = sid.astype(np.int32)\n f = interp1d(sid,X_test_norm[:,sindex], kind=\"linear\", fill_value='extrapolate') # interpolation funciton for the jth grain size class\n coord_range = np.arange(coord_num*j, coord_num*(j+1)) # range to interpolate\n thick_interp[:,coord_range.astype(np.int32)] = f(coord_range) # interpolated data\n\n\n # Load the model and predict from subsampled data\n if model is None: model = load_model(join(resdir, 'model.hdf5'))\n test_result = model.predict(X_test_norm)\n test_result = test_result * (y_max - y_min) + y_min\n test_result_sample = model.predict(thick_interp)\n test_result_sample = test_result_sample * (y_max - y_min) + y_min\n\n # calculate residuals\n icond = np.load(join(datadir, 'icond_test.npy'))\n resi = test_result - icond\n resi_sample = test_result_sample - icond\n resi_sample_ratio = (test_result_sample - icond) / icond\n\n # comparison with original reconstruction\n titlelabel = ['Initial Length', 'Initial Height', '$C_1$', '$C_2$', '$C_3$', '$C_4$','$S_1$']\n hfont = {'fontname':'Century Gothic'}\n textcol = 'w'\n \n if plot_fig:\n for i in range(len(titlelabel)):\n plt.figure()\n plt.plot(icond[:,i],test_result[:,i],\"bo\",label='Original')\n plt.plot(icond[:,i],test_result_sample[:,i],\"ro\",label='Resampled data ({:.1f}%)'.format(frac*100))\n plt.title(titlelabel[i],color=textcol,size=14,**hfont)\n plt.xlabel('True values',color=textcol,size=14,**hfont)\n plt.ylabel('Estimated values',color=textcol,size=14,**hfont)\n plt.legend()\n plt.tick_params(labelsize=14,colors=textcol)\n plt.savefig(titlelabel[i] + 'resample{:.1f})'.format(frac*100) + '.pdf')\n plt.show()\n\n for i in range(len(titlelabel)):\n plt.figure()\n plt.hist(resi_sample[:,i],bins=20)\n plt.title(titlelabel[i])\n plt.xlabel('Deviation from true value')\n plt.ylabel('Frequency')\n plt.show()\n\n print(\"mean residuals\")\n print(np.average(resi,axis=0))\n print(\"mean residuals (subsampled)\")\n print(np.average(resi_sample,axis=0))\n print(\"2 sigma of residuals\")\n print(np.std(resi,axis=0)*2)\n print(\"2 sigma of residuals (subsampled)\")\n print(np.std(resi_sample,axis=0)*2)\n print()\n print(\"p-values of the Shapiro-Wilk test for normality\")\n for i in range(resi.shape[1]):\n print(stats.shapiro(resi[:,i])[1])\n print(\"p-values of the Shapiro-Wilk test for normality (with error)\")\n for i in range(resi_sample.shape[1]):\n print(stats.shapiro(resi_sample[:,i])[1])\n \n # Return normalized RMSE\n RMS = np.sqrt(np.sum(resi_sample_ratio ** 2) / resi_sample_ratio.shape[0] / resi_sample_ratio.shape[1])\n \n return RMS\n\nif __name__ == \"__main__\":\n datadir = '/home/naruse/public/naruse/DeepLearningTurbidite/distance/10/data/'\n resdir = '/home/naruse/public/naruse/DeepLearningTurbidite/result_training_num_10/3500/'\n subsampling_result_file = join(resdir, 'subsampling_result.npy')\n subsampling_result_error_file = join(resdir, 'subsampling_result_error.npy')\n model = load_model(join(resdir, 'model.hdf5'))\n \n subsampling_test_err_rate = np.linspace(0.05,0.001,50)\n result_subsampling = np.zeros([len(subsampling_test_err_rate)])\n result_subsampling_error = np.zeros([len(subsampling_test_err_rate)])\n num_tests = 20\n for i in range(len(subsampling_test_err_rate)):\n testres = np.zeros([num_tests])\n for j in range(num_tests):\n testres[j] = check_interp(model, datadir=datadir, resdir=resdir, frac=subsampling_test_err_rate[i], plot_fig=False)\n result_subsampling[i] = np.average(testres)\n result_subsampling_error[i] = sem(testres)\n \n np.save(subsampling_result_file, result_subsampling)\n np.save(subsampling_result_error_file, result_subsampling_error)\n\n \n \n ",
"_____no_output_____"
],
[
"%matplotlib inline\n\nfig1, ax1 = plt.subplots(1, 1, figsize=(8/2.54, 5/2.54))\nplt.subplots_adjust(bottom=0.3, wspace=0.5, hspace=0.3)\n\n\nplt.errorbar(subsampling_test_err_rate*100, result_subsampling, yerr=result_subsampling_error, ecolor='k', capsize=1.)\nax1.set_xlabel('Ratio of Subsampled Grids (%)') \nax1.set_ylabel('RMS error')\nax1.set_xticks(np.arange(0, 5, 0.5))\n# ax1.legend()\n# plt.tick_params(labelsize=14,colors=textcol)\nplt.tight_layout()\nplt.savefig(\"tex/fig11.eps\")\n",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
4a58b5b26856833f944774f7f2f7356676540518
| 3,412 |
ipynb
|
Jupyter Notebook
|
trial.ipynb
|
pranavgirish/Performance-Analysis-of-ML-Algorithms
|
eb839b9473d71b92f0960b30cfb0582c51d7c2f0
|
[
"MIT"
] | null | null | null |
trial.ipynb
|
pranavgirish/Performance-Analysis-of-ML-Algorithms
|
eb839b9473d71b92f0960b30cfb0582c51d7c2f0
|
[
"MIT"
] | null | null | null |
trial.ipynb
|
pranavgirish/Performance-Analysis-of-ML-Algorithms
|
eb839b9473d71b92f0960b30cfb0582c51d7c2f0
|
[
"MIT"
] | null | null | null | 19.953216 | 71 | 0.498535 |
[
[
[
"import numpy as np\nfrom sklearn import svm, datasets\nfrom sklearn.model_selection import cross_val_score, KFold\nfrom sklearn.metrics import accuracy_score, f1_score",
"_____no_output_____"
],
[
"iris = datasets.load_iris()\nX, y = iris.data, iris.target\nclf = svm.SVC(probability=True, random_state=0)\ncross_val_score(clf, X, y, scoring='neg_log_loss') ",
"_____no_output_____"
],
[
"model = svm.SVC()\ncross_val_score(model, X, y, scoring='accuracy')",
"_____no_output_____"
],
[
"\nfolds = 10\nkf = KFold(n_splits=folds, shuffle=True, random_state=3245236)\n\naccs = np.zeros(folds)\n\ni = 0\nfor index_train, index_test in kf.split(X):\n model = svm.SVC()\n model.fit(X[index_train], y[index_train])\n \n y_true = y[index_test]\n y_pred = model.predict(X[index_test])\n \n accs[i] = f1_score(y_true, y_pred, average='micro')\n i = i + 1\n \nprint('mean ', np.mean(accs))\nprint('std ', np.std(accs))\n\n\n\n\n",
"mean 0.973333333333\nstd 0.0326598632371\n"
],
[
"model = svm.SVC()\ncross_val_score(model, X, y, scoring='f1_macro')",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code"
]
] |
4a58cdabc623140837fac9019364082f3b625c90
| 15,508 |
ipynb
|
Jupyter Notebook
|
lessons/3A.ipynb
|
microbial-pangenomes-lab/2022_python_course
|
94ebb0a4dc2ff7a5d23bbf684d2f49e6b51c46ec
|
[
"Apache-2.0"
] | null | null | null |
lessons/3A.ipynb
|
microbial-pangenomes-lab/2022_python_course
|
94ebb0a4dc2ff7a5d23bbf684d2f49e6b51c46ec
|
[
"Apache-2.0"
] | null | null | null |
lessons/3A.ipynb
|
microbial-pangenomes-lab/2022_python_course
|
94ebb0a4dc2ff7a5d23bbf684d2f49e6b51c46ec
|
[
"Apache-2.0"
] | null | null | null | 24.23125 | 243 | 0.483428 |
[
[
[
"Plotting with matplotlib - 1\n========================",
"_____no_output_____"
]
],
[
[
"# plotting imports\nimport matplotlib.pyplot as plt\nimport seaborn as sns",
"_____no_output_____"
],
[
"# other imports\nimport numpy as np\nimport pandas as pd\nfrom scipy import stats",
"_____no_output_____"
]
],
[
[
"Hello world\n---\n\nUsing the `pyplot` notation, very similar to how MATLAB works",
"_____no_output_____"
]
],
[
[
"plt.plot([0, 1, 2, 3, 4],\n [0, 1, 2, 5, 10], 'bo-')\nplt.text(1.5, 5, 'Hello world', size=14)\nplt.xlabel('X axis\\n($\\mu g/mL$)')\nplt.ylabel('y axis\\n($X^2$)');",
"_____no_output_____"
]
],
[
[
"Hello world, reprise\n---\n\nUsing the reccommended \"object-oriented\" (OO) style",
"_____no_output_____"
]
],
[
[
"fig, ax = plt.subplots()\nax.plot([0, 1, 2, 3, 4],\n [0, 1, 2, 5, 10], 'bo-')\nax.text(1.5, 5, 'Hello world', size=14)\nax.set_xlabel('X axis\\n($\\mu g/mL$)')\nax.set_ylabel('y axis\\n($X^2$)');",
"_____no_output_____"
],
[
"# create some data\nx = np.linspace(0, 2, 100)",
"_____no_output_____"
],
[
"fig, ax = plt.subplots()\n\nax.plot(x, x, label='linear')\nax.plot(x, x**2, label='quadratic')\nax.plot(x, x**3, label='cubic')\n\nax.set_xlabel('x label')\nax.set_ylabel('y label')\nax.set_title('Simple Plot')\nax.legend()",
"_____no_output_____"
]
],
[
[
"Controlling a figure aspect\n---",
"_____no_output_____"
]
],
[
[
"# figure size\n# width / height\nfig, ax = plt.subplots(figsize=(9, 4))\n\nax.plot(x, x, label='linear')\nax.plot(x, x**2, label='quadratic')\nax.plot(x, x**3, label='cubic')\n\nax.set_xlabel('x label')\nax.set_ylabel('y label')\nax.set_title('Simple Plot')\nax.legend();",
"_____no_output_____"
],
[
"fig, ax = plt.subplots(figsize=(9, 4))\n\n# change markers\nax.plot(x, x, '--', color='grey', label='linear')\nax.plot(x, x**2, '.-', color='red', label='quadratic')\nax.plot(x, x**3, '*', color='#3bb44a', label='cubic')\n\nax.set_xlabel('x label')\nax.set_ylabel('y label')\nax.set_title('Simple Plot')\n\n# move the legend\nax.legend(loc='upper right');\n# alternative ways to move it\n# ax.legend(loc='center left',\n# bbox_to_anchor=(1, 0.5),\n# ncol=3);",
"_____no_output_____"
]
],
[
[
"Multiple panels\n---",
"_____no_output_____"
]
],
[
[
"x1 = np.linspace(0.0, 5.0)\nx2 = np.linspace(0.0, 2.0)\n\ny1 = np.cos(2 * np.pi * x1) * np.exp(-x1)\ny2 = np.cos(2 * np.pi * x2)\n\n# rows, columns\nfig, axes = plt.subplots(2, 1, figsize=(6, 4))\n\n# axes is a list of \"panels\"\nprint(axes)\n\nax = axes[0]\nax.plot(x1, y1, 'o-')\nax.set_title('A tale of 2 subplots')\nax.set_ylabel('Damped oscillation')\n\nax = axes[1]\nax.plot(x2, y2, '.-')\nax.set_xlabel('time (s)')\nax.set_ylabel('Undamped');",
"_____no_output_____"
]
],
[
[
"Automagically adjust panels so that they fit in the figure\n---",
"_____no_output_____"
]
],
[
[
"def example_plot(ax, fontsize=12):\n ax.plot([1, 2])\n\n ax.set_xlabel('x-label', fontsize=fontsize)\n ax.set_ylabel('y-label', fontsize=fontsize)\n ax.set_title('Title', fontsize=fontsize)",
"_____no_output_____"
],
[
"fig, axs = plt.subplots(2, 2, figsize=(4, 4),\n constrained_layout=False)\nprint(axs)\nfor ax in axs.flat:\n example_plot(ax)",
"_____no_output_____"
],
[
"# warning: \"constrained_layout\" is an experimental feature\nfig, axs = plt.subplots(2, 2, figsize=(4, 4),\n constrained_layout=True)\n\nfor ax in axs.flat:\n example_plot(ax)",
"_____no_output_____"
],
[
"# alternative way\nfig, axs = plt.subplots(2, 2, figsize=(4, 4), constrained_layout=False)\n\nfor ax in axs.flat:\n example_plot(ax)\n \n# alternative to constrained_layout\nplt.tight_layout();",
"_____no_output_____"
]
],
[
[
"Example of manipulating axes limits\n---\n\nExtra: a look at ways to choose colors\nand manipulating transparency",
"_____no_output_____"
]
],
[
[
"fig, axes = plt.subplots(1, 2, figsize=(9, 4))\n\n# same plot for both panels\n# we are just gonna change the axes' limits\nfor ax in axes:\n # more color choices\n # (see here for a full list: https://matplotlib.org/tutorials/colors/colors.html)\n \n # xkcd rgb color survey: https://xkcd.com/color/rgb/\n ax.plot(x, x, '--', color='xkcd:olive green', label='linear')\n # RGBA (red, green, blue, alpha)\n ax.plot(x, x**2, '.-', color=(0.1, 0.2, 0.5, 0.3), label='quadratic')\n # one of {'b', 'g', 'r', 'c', 'm', 'y', 'k', 'w'}\n # they are the single character short-hand notations for:\n # blue, green, red, cyan, magenta, yellow, black, and white\n ax.plot(x, x**3, '*', color='m', label='cubic')\n # transparency can be manipulated with the \"alpha\" kwarg (= keyword argument)\n ax.plot(x, x**4, '-', color='b', linewidth=4, alpha=0.3, label='white house')\n\n ax.set_xlabel('x label')\n ax.set_ylabel('y label')\n ax.set_title('Simple Plot')\n\n# only manipulate last axes\nax.set_ylim(1, 16.4)\nax.set_xlim(1.65, 2.03)\n\nax.legend(loc='center left',\n bbox_to_anchor=(1, 0.5),\n title='Fit');",
"_____no_output_____"
]
],
[
[
"Other sample plots using \"vanilla\" matplotlib\n---",
"_____no_output_____"
]
],
[
[
"# scatter plot\nfig, ax = plt.subplots(figsize=(6, 4))\n\nN = 10\nx = np.linspace(0, 1, N)\ny = x ** 2\n# colors is a list of colors\n# in the same format as shown before\ncolors = np.linspace(0, 1, N)\n# alternative\n# colors = ['b', 'b', 'b',\n# 'k', 'k', 'k',\n# 'r', 'r', 'r',\n# 'xkcd:jade']\narea = 5 + (20 * x) ** 3\n\nprint(f'x: {x}')\nprint(f'y: {y}')\nprint(f'colors: {colors}')\nprint(f'area: {area}')\n\nax.scatter(x, y, s=area, c=colors,\n alpha=0.9,\n edgecolors='w', linewidths=3,\n label='Data')\nax.legend(loc='upper left');",
"_____no_output_____"
],
[
"# generate 2d random data\ndata = np.random.randn(2, 100)\ndata",
"_____no_output_____"
],
[
"# histogram\nfig, axs = plt.subplots(1, 2, figsize=(6, 3))\n\nbins = 25\n\naxs[0].hist(data[0], bins=bins)\naxs[1].hist2d(data[0], data[1], bins=bins);",
"_____no_output_____"
]
],
[
[
"Other useful tips\n---",
"_____no_output_____"
]
],
[
[
"# scatter plot with log axes\nfig, ax = plt.subplots(figsize=(6, 4))\n\nN = 10\nx = np.linspace(0, 10, N)\ny = 2 ** x\ncolors = np.linspace(0, 1, N)\narea = 500\n\nax.scatter(x, y, s=area, c=colors,\n alpha=0.9,\n edgecolors='w', linewidths=3,\n label='Data')\nax.set_yscale('log', base=10);",
"_____no_output_____"
],
[
"# scatter plot with log axes\nfig, ax = plt.subplots(figsize=(6, 4))\n\nN = 10\nx = 10 ** np.linspace(1, 4, N)\ny = x ** 2\ncolors = np.linspace(0, 1, N)\narea = 500\n\nax.scatter(x, y, s=area, c=colors,\n alpha=0.9,\n edgecolors='w', linewidths=3,\n label='Data')\nax.set_yscale('log', base=2)\nax.set_xscale('log', base=10);",
"_____no_output_____"
],
[
"# changing colormap\n# find an exhaustive list here:\n# https://matplotlib.org/3.1.0/tutorials/colors/colormaps.html\nfig, ax = plt.subplots(figsize=(6, 4))\n\nN = 10\nx = 10 ** np.linspace(1, 4, N)\ny = x ** 2\ncolors = np.linspace(0, 1, N)\narea = 500\n\nax.scatter(x, y, s=area, c=colors,\n alpha=0.9,\n edgecolors='w', linewidths=3,\n label='Data',\n# cmap='plasma',\n# cmap='jet',\n# cmap='Blues',\n# cmap='Blues_r',\n cmap='tab20',\n )\nax.set_yscale('log', base=2)\nax.set_xscale('log', base=10);",
"_____no_output_____"
]
],
[
[
"Saving your plot\n---",
"_____no_output_____"
]
],
[
[
"fig, ax = plt.subplots(figsize=(3, 2))\n\nN = 10\nx = 10 ** np.linspace(1, 4, N)\ny = x ** 2\ncolors = np.linspace(0, 1, N)\narea = 500\n\nax.scatter(x, y, s=area, c=colors,\n alpha=0.9,\n edgecolors='w', linewidths=3,\n cmap='tab20',\n label='My awesome data is the best thing ever',\n# rasterized=True\n )\n\nax.legend(bbox_to_anchor=(1, 0.5),\n loc='center left')\n\nax.set_yscale('log', basey=2)\nax.set_xscale('log', basex=10)\n\nplt.savefig('the_awesomest_plot_ever.png',\n dpi=300,\n bbox_inches='tight',\n transparent=True\n )\nplt.savefig('the_awesomest_plot_ever.svg',\n dpi=300, bbox_inches='tight',\n transparent=True);",
"_____no_output_____"
]
],
[
[
"---\n",
"_____no_output_____"
],
[
"Exercises\n---------",
"_____no_output_____"
],
[
"Using the data from this URL: https://evocellnet.github.io/ecoref/data/phenotypic_data.tsv",
"_____no_output_____"
],
[
"Can you make a scatterplot for the relationship between s-scores and the corrected p-value?",
"_____no_output_____"
],
[
"Can you make a scatterplot for the relationship between s-scores and the corrected p-value, but only considering two strains plotted with different colors?",
"_____no_output_____"
],
[
"Select four conditions and create a multipanel figure with the same scatterplot for each condition. Experiment with different layouts",
"_____no_output_____"
],
[
"Using the [Iris dataset](https://en.wikipedia.org/wiki/Iris_flower_data_set) (which you can find at `../data/iris.csv`), prepare the following plot: for each pair of variables, prepare a scatterplot with each species having its own color",
"_____no_output_____"
],
[
"Make the same series of plots as before but in a single figure",
"_____no_output_____"
],
[
"Make a single panel now, changing the dots' sizes according to the third variable",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
4a58df1e3a4cfa1b8836fa83ad1af7c8825d93d8
| 90,268 |
ipynb
|
Jupyter Notebook
|
Logistic-Reg/Analysis.ipynb
|
nickedes/Baby-science
|
3d9f1baf09591da3afaade0f70dabe007d2df0c7
|
[
"MIT"
] | null | null | null |
Logistic-Reg/Analysis.ipynb
|
nickedes/Baby-science
|
3d9f1baf09591da3afaade0f70dabe007d2df0c7
|
[
"MIT"
] | null | null | null |
Logistic-Reg/Analysis.ipynb
|
nickedes/Baby-science
|
3d9f1baf09591da3afaade0f70dabe007d2df0c7
|
[
"MIT"
] | null | null | null | 150.446667 | 53,666 | 0.854733 |
[
[
[
"%pylab inline\nimport pandas as pd\ndfr = pd.read_csv('../datasets/loanf.csv')\n# inspect, sanity check\ndfr.head()",
"Populating the interactive namespace from numpy and matplotlib\n"
],
[
"# we add a column which indicates (True/False) whether the interest rate is <= 12 \ndfr['TF']=dfr['Interest.Rate']<=12\n# inspect again\ndfr.head()\n# we see that the TF values are False as Interest.Rate is higher than 12 in all these cases",
"_____no_output_____"
],
[
"# now we check the rows that have interest rate == 10 (just some number < 12)\n# this is just to confirm that the TF value is True where we expect it to be\nd = dfr[dfr['Interest.Rate']==10]\nd.head()\n# all is well",
"_____no_output_____"
],
[
"import statsmodels.api as sm\n# statsmodels requires us to add a constant column representing the intercept\ndfr['intercept']=1.0\n# identify the independent variables \nind_cols=['FICO.Score','Loan.Amount','intercept']\nlogit = sm.Logit(dfr['TF'], dfr[ind_cols])\nresult=logit.fit()",
"Optimization terminated successfully.\n Current function value: 0.319503\n Iterations 8\n"
],
[
"# get the fitted coefficients from the results\ncoeff = result.params\nprint(coeff)",
"FICO.Score 0.087423\nLoan.Amount -0.000174\nintercept -60.125045\ndtype: float64\n"
],
[
"def pz(fico,amt,coeff):\n # compute the linear expression by multipyling the inputs by their respective coefficients.\n # note that the coefficient array has the intercept coefficient at the end\n z = coeff[0]*fico + coeff[1]*amt + coeff[2]\n return 1/(1+exp(-1*z))",
"_____no_output_____"
],
[
"pz(720,10000,coeff)",
"_____no_output_____"
],
[
"print(\"Trying multiple FICO Loan Amount combinations: \")\nprint('----')\nprint(\"fico=720, amt=10,000\")\nprint(pz(720,10000,coeff))\nprint(\"fico=720, amt=20,000\")\nprint(pz(720,20000,coeff))\nprint(\"fico=720, amt=30,000\")\nprint(pz(720,30000,coeff))\nprint(\"fico=820, amt=10,000\")\nprint(pz(820,10000,coeff))\nprint(\"fico=820, amt=20,000\")\nprint(pz(820,20000,coeff))\nprint(\"fico=820, amt=30,000\")\nprint(pz(820,30000,coeff))\n",
"Trying multiple FICO Loan Amount combinations: \n----\nfico=720, amt=10,000\n0.746378588952\nfico=720, amt=20,000\n0.340539857688\nfico=720, amt=30,000\n0.083083595237\nfico=820, amt=10,000\n0.999945742327\nfico=820, amt=20,000\n0.999690867752\nfico=820, amt=30,000\n0.998240830138\n"
],
[
"pz(820,63000,coeff)",
"_____no_output_____"
],
[
"print(\"Trying multiple FICO Loan Amount combinations: \")\nprint('----')\nprint(\"fico=820, amt=50,000\")\nprint(pz(820,50000,coeff))\nprint(\"fico=820, amt=60,000\")\nprint(pz(820,60000,coeff))\nprint(\"fico=820, amt=70,000\")\nprint(pz(820,70000,coeff))\nprint(\"fico=820, amt=63,000\")\nprint(pz(820,63000,coeff))\nprint(\"fico=820, amt=65,000\")\nprint(pz(820,65000,coeff))\nprint(\"fico=820, amt=67,000\")\nprint(pz(820,67000,coeff))",
"Trying multiple FICO Loan Amount combinations: \n----\nfico=820, amt=50,000\n0.945863681761\nfico=820, amt=60,000\n0.754046864085\nfico=820, amt=70,000\n0.349789145957\nfico=820, amt=63,000\n0.645251163193\nfico=820, amt=65,000\n0.562219246329\nfico=820, amt=67,000\n0.475548952428\n"
],
[
"import matplotlib.pyplot as plt\nloans = dfr[dfr['FICO.Score'] == 720]\nx = loans['Loan.Amount']\ny = pz(720,x,coeff)\nplt.plot(x,y)",
"_____no_output_____"
],
[
"z = dfr['FICO.Score']\nx = dfr['Loan.Amount']\ny = pz(z,x,coeff)\nplt.plot(x,y)\n# this doesn't looks fine!",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a58e041d05ebb83206f59757cba4d237e6ae7cb
| 157,884 |
ipynb
|
Jupyter Notebook
|
examples/import_and_analyse_data.ipynb
|
MD2Korg/CerebralCortex-Kernel
|
206e9419fecf587ad1eb69a2e2c605db5ee3eb05
|
[
"BSD-2-Clause"
] | 1 |
2020-10-19T14:36:59.000Z
|
2020-10-19T14:36:59.000Z
|
examples/import_and_analyse_data.ipynb
|
MD2Korg/CerebralCortex-Kernel
|
206e9419fecf587ad1eb69a2e2c605db5ee3eb05
|
[
"BSD-2-Clause"
] | 4 |
2019-05-16T15:41:06.000Z
|
2020-04-07T06:41:39.000Z
|
examples/import_and_analyse_data.ipynb
|
MD2Korg/CerebralCortex-Kernel
|
206e9419fecf587ad1eb69a2e2c605db5ee3eb05
|
[
"BSD-2-Clause"
] | 10 |
2019-01-25T20:16:54.000Z
|
2021-05-04T16:53:46.000Z
| 157,884 | 157,884 | 0.677225 |
[
[
[
"## Interacting with CerebralCortex Data",
"_____no_output_____"
],
[
"Cerebral Cortex is MD2K's big data cloud tool designed to support population-scale data analysis, visualization, model development, and intervention design for mobile-sensor data. It provides the ability to do machine learning model development on population scale datasets and provides interoperable interfaces for aggregation of diverse data sources.\n\nThis page provides an overview of the core Cerebral Cortex operations to familiarilze you with how to discover and interact with different sources of data that could be contained within the system.\n\n_Note:_ While some of these examples are showing generated data, they are designed to function on real-world mCerebrum data and the signal generators were built to facilitate the testing and evaluation of the Cerebral Cortex platform by those individuals that are unable to see those original datasets or do not wish to collect data before evaluating the system.",
"_____no_output_____"
],
[
"## Setting Up Environment\n\nNotebook does not contain the necessary runtime enviornments necessary to run Cerebral Cortex. The following commands will download and install these tools, framework, and datasets.",
"_____no_output_____"
]
],
[
[
"import importlib, sys, os\nfrom os.path import expanduser\nsys.path.insert(0, os.path.abspath('..'))\n\nDOWNLOAD_USER_DATA=False\nALL_USERS=False #this will only work if DOWNLOAD_USER_DATA=True\nIN_COLAB = 'google.colab' in sys.modules\nMD2K_JUPYTER_NOTEBOOK = \"MD2K_JUPYTER_NOTEBOOK\" in os.environ\nif (get_ipython().__class__.__name__==\"ZMQInteractiveShell\"): IN_JUPYTER_NOTEBOOK = True\nJAVA_HOME_DEFINED = \"JAVA_HOME\" in os.environ\nSPARK_HOME_DEFINED = \"SPARK_HOME\" in os.environ\nPYSPARK_PYTHON_DEFINED = \"PYSPARK_PYTHON\" in os.environ\nPYSPARK_DRIVER_PYTHON_DEFINED = \"PYSPARK_DRIVER_PYTHON\" in os.environ\nHAVE_CEREBRALCORTEX_KERNEL = importlib.util.find_spec(\"cerebralcortex\") is not None\nSPARK_VERSION = \"3.1.2\"\nSPARK_URL = \"https://archive.apache.org/dist/spark/spark-\"+SPARK_VERSION+\"/spark-\"+SPARK_VERSION+\"-bin-hadoop2.7.tgz\"\nSPARK_FILE_NAME = \"spark-\"+SPARK_VERSION+\"-bin-hadoop2.7.tgz\"\nCEREBRALCORTEX_KERNEL_VERSION = \"3.3.14\"\n\nDATA_PATH = expanduser(\"~\")\nif DATA_PATH[:-1]!=\"/\":\n DATA_PATH+=\"/\"\nUSER_DATA_PATH = DATA_PATH+\"cc_data/\"\n\nif MD2K_JUPYTER_NOTEBOOK:\n print(\"Java, Spark, and CerebralCortex-Kernel are installed and paths are already setup.\")\nelse:\n\n SPARK_PATH = DATA_PATH+\"spark-\"+SPARK_VERSION+\"-bin-hadoop2.7/\"\n \n\n if(not HAVE_CEREBRALCORTEX_KERNEL):\n print(\"Installing CerebralCortex-Kernel\")\n !pip -q install cerebralcortex-kernel==$CEREBRALCORTEX_KERNEL_VERSION\n else:\n print(\"CerebralCortex-Kernel is already installed.\")\n\n if not JAVA_HOME_DEFINED:\n if not os.path.exists(\"/usr/lib/jvm/java-8-openjdk-amd64/\") and not os.path.exists(\"/usr/lib/jvm/java-11-openjdk-amd64/\"):\n print(\"\\nInstalling/Configuring Java\")\n !sudo apt update\n !sudo apt-get install -y openjdk-8-jdk-headless\n os.environ[\"JAVA_HOME\"] = \"/usr/lib/jvm/java-8-openjdk-amd64/\"\n elif os.path.exists(\"/usr/lib/jvm/java-8-openjdk-amd64/\"):\n print(\"\\nSetting up Java path\")\n os.environ[\"JAVA_HOME\"] = \"/usr/lib/jvm/java-8-openjdk-amd64/\"\n elif os.path.exists(\"/usr/lib/jvm/java-11-openjdk-amd64/\"):\n print(\"\\nSetting up Java path\")\n os.environ[\"JAVA_HOME\"] = \"/usr/lib/jvm/java-11-openjdk-amd64/\"\n else:\n print(\"JAVA is already installed.\")\n\n if (IN_COLAB or IN_JUPYTER_NOTEBOOK) and not MD2K_JUPYTER_NOTEBOOK:\n if SPARK_HOME_DEFINED:\n print(\"SPARK is already installed.\")\n elif not os.path.exists(SPARK_PATH):\n print(\"\\nSetting up Apache Spark \", SPARK_VERSION)\n !pip -q install findspark\n import pyspark\n spark_installation_path = os.path.dirname(pyspark.__file__)\n import findspark\n findspark.init(spark_installation_path)\n if not os.getenv(\"PYSPARK_PYTHON\"):\n os.environ[\"PYSPARK_PYTHON\"] = os.popen('which python3').read().replace(\"\\n\",\"\")\n if not os.getenv(\"PYSPARK_DRIVER_PYTHON\"):\n os.environ[\"PYSPARK_DRIVER_PYTHON\"] = os.popen('which python3').read().replace(\"\\n\",\"\")\n else:\n print(\"SPARK is already installed.\")\n else:\n raise SystemExit(\"Please check your environment configuration at: https://github.com/MD2Korg/CerebralCortex-Kernel/\")\n\nif DOWNLOAD_USER_DATA:\n if not os.path.exists(USER_DATA_PATH):\n if ALL_USERS:\n print(\"\\nDownloading all users' data.\")\n !rm -rf $USER_DATA_PATH\n !wget -q http://mhealth.md2k.org/images/datasets/cc_data.tar.bz2 && tar -xf cc_data.tar.bz2 -C $DATA_PATH && rm cc_data.tar.bz2\n else:\n print(\"\\nDownloading a user's data.\")\n !rm -rf $USER_DATA_PATH\n !wget -q http://mhealth.md2k.org/images/datasets/s2_data.tar.bz2 && tar -xf s2_data.tar.bz2 -C $DATA_PATH && rm s2_data.tar.bz2\n else:\n print(\"Data already exist. Please remove folder\", USER_DATA_PATH, \"if you want to download the data again\")",
"Installing CerebralCortex-Kernel\n\u001b[K |████████████████████████████████| 194 kB 5.2 MB/s \n\u001b[K |████████████████████████████████| 1.3 MB 9.3 MB/s \n\u001b[K |████████████████████████████████| 100 kB 9.1 MB/s \n\u001b[K |████████████████████████████████| 105 kB 25.9 MB/s \n\u001b[K |████████████████████████████████| 21.8 MB 6.5 MB/s \n\u001b[K |████████████████████████████████| 20.6 MB 1.3 MB/s \n\u001b[K |████████████████████████████████| 721 kB 40.9 MB/s \n\u001b[K |████████████████████████████████| 636 kB 37.2 MB/s \n\u001b[K |████████████████████████████████| 212.4 MB 61 kB/s \n\u001b[K |████████████████████████████████| 77 kB 7.4 MB/s \n\u001b[K |████████████████████████████████| 44 kB 3.0 MB/s \n\u001b[K |████████████████████████████████| 94 kB 3.9 MB/s \n\u001b[K |████████████████████████████████| 198 kB 61.3 MB/s \n\u001b[K |████████████████████████████████| 554 kB 42.9 MB/s \n\u001b[?25h Building wheel for datascience (setup.py) ... \u001b[?25l\u001b[?25hdone\n Building wheel for hdfs3 (setup.py) ... \u001b[?25l\u001b[?25hdone\n Building wheel for pyspark (setup.py) ... \u001b[?25l\u001b[?25hdone\n Building wheel for gatspy (setup.py) ... \u001b[?25l\u001b[?25hdone\n\nSetting up Java path\n\nSetting up Apache Spark 3.1.2\n\nDownloading a user's data.\n"
]
],
[
[
"# Import Your Own Data\nmCerebrum is not the only way to collect and load data into *Cerebral Cortex*. It is possible to import your own structured datasets into the platform. This example will demonstrate how to load existing data and subsequently how to read it back from Cerebral Cortex through the same mechanisms you have been utilizing. Additionally, it demonstrates how to write a custom data transformation fuction to manipulate data and produce a smoothed result which can then be visualized.",
"_____no_output_____"
],
[
"## Initialize the system",
"_____no_output_____"
]
],
[
[
"from cerebralcortex.kernel import Kernel\nCC = Kernel(cc_configs=\"default\", study_name=\"default\", new_study=True)",
"/usr/local/lib/python3.7/dist-packages/psycopg2/__init__.py:144: UserWarning: The psycopg2 wheel package will be renamed from release 2.8; in order to keep installing from binary please use \"pip install psycopg2-binary\" instead. For details see: <http://initd.org/psycopg/docs/install.html#binary-install-from-pypi>.\n \"\"\")\n"
]
],
[
[
"# Import Data\nCerebral Cortex provides a set of predefined data import routines that fit typical use cases. The most common is CSV data parser, `csv_data_parser`. These parsers are easy to write and can be extended to support most types of data. Additionally, the data importer, `import_data`, needs to be brought into this notebook so that we can start the data import process.\n\nThe `import_data` method requires several parameters that are discussed below.\n- `cc_config`: The path to the configuration files for Cerebral Cortex; this is the same folder that you would utilize for the `Kernel` initialization\n- `input_data_dir`: The path to where the data to be imported is located; in this example, `sample_data` is available in the file/folder browser on the left and you should explore the files located inside of it\n- `user_id`: The universally unique identifier (UUID) that owns the data to be imported into the system\n- `data_file_extension`: The type of files to be considered for import\n- `data_parser`: The import method or another that defines how to interpret the data samples on a per-line basis\n- `gen_report`: A simple True/False value that controls if a report is printed to the screen when complete",
"_____no_output_____"
],
[
"### Download sample data",
"_____no_output_____"
]
],
[
[
"sample_file = DATA_PATH+\"data.csv\"\n!wget -q https://raw.githubusercontent.com/MD2Korg/CerebralCortex/master/jupyter_demo/sample_data/data.csv -O $sample_file",
"_____no_output_____"
],
[
"iot_stream = CC.read_csv(file_path=sample_file, stream_name=\"some-sample-iot-stream\", column_names=[\"timestamp\", \"some_vals\", \"version\", \"user\"])",
"_____no_output_____"
]
],
[
[
"## View Imported Data",
"_____no_output_____"
]
],
[
[
"iot_stream.show(4)",
"+-------------------+-----------+-------+--------------------+\n| timestamp| some_vals|version| user|\n+-------------------+-----------+-------+--------------------+\n|2019-01-09 17:35:00|0.085188727| 1|00000000-afb8-476...|\n|2019-01-09 17:35:01|0.168675497| 1|00000000-afb8-476...|\n|2019-01-09 17:35:02|0.740485082| 1|00000000-afb8-476...|\n|2019-01-09 17:35:03|0.713160997| 1|00000000-afb8-476...|\n+-------------------+-----------+-------+--------------------+\nonly showing top 4 rows\n\n"
]
],
[
[
"## Document Data",
"_____no_output_____"
]
],
[
[
"from cerebralcortex.core.metadata_manager.stream.metadata import Metadata, DataDescriptor, ModuleMetadata\n\nstream_metadata = Metadata()\nstream_metadata.set_name(\"iot-data-stream\").set_description(\"This is randomly generated data for demo purposes.\") \\\n .add_dataDescriptor(\n DataDescriptor().set_name(\"timestamp\").set_type(\"datetime\").set_attribute(\"description\", \"UTC timestamp of data point collection.\")) \\\n .add_dataDescriptor(\n DataDescriptor().set_name(\"some_vals\").set_type(\"float\").set_attribute(\"description\", \\\n \"Random values\").set_attribute(\"range\", \\\n \"Data is between 0 and 1.\")) \\\n .add_dataDescriptor(\n DataDescriptor().set_name(\"version\").set_type(\"int\").set_attribute(\"description\", \"version of the data\")) \\\n .add_dataDescriptor(\n DataDescriptor().set_name(\"user\").set_type(\"string\").set_attribute(\"description\", \"user id\")) \\\n .add_module(ModuleMetadata().set_name(\"cerebralcortex.data_importer\").set_attribute(\"url\", \"hhtps://md2k.org\").set_author(\n \"Nasir Ali\", \"[email protected]\"))\niot_stream.metadata = stream_metadata",
"_____no_output_____"
]
],
[
[
"## View Metadata",
"_____no_output_____"
]
],
[
[
"iot_stream.metadata",
"_____no_output_____"
]
],
[
[
"## How to write an algorithm\nThis section provides an example of how to write a simple smoothing algorithm and apply it to the data that was just imported.",
"_____no_output_____"
],
[
"### Import the necessary modules",
"_____no_output_____"
]
],
[
[
"from pyspark.sql.functions import pandas_udf, PandasUDFType\nfrom pyspark.sql.types import StructField, StructType, StringType, FloatType, TimestampType, IntegerType\nfrom pyspark.sql.functions import minute, second, mean, window\nfrom pyspark.sql import functions as F\nimport numpy as np",
"_____no_output_____"
]
],
[
[
"### Define the Schema\nThis schema defines what the computation module will return to the execution context for each row or window in the datastream.",
"_____no_output_____"
]
],
[
[
"# column name and return data type\n# acceptable data types for schem are - \"null\", \"string\", \"binary\", \"boolean\",\n# \"date\", \"timestamp\", \"decimal\", \"double\", \"float\", \"byte\", \"integer\",\n# \"long\", \"short\", \"array\", \"map\", \"structfield\", \"struct\"\nschema=\"timestamp timestamp, some_vals double, version int, user string, vals_avg double\"\n",
"_____no_output_____"
]
],
[
[
"### Write a user defined function\nThe user-defined function (UDF) is one of two mechanisms available for distributed data processing within the Apache Spark framework. In this case, we are computing a simple windowed average.",
"_____no_output_____"
]
],
[
[
"def smooth_algo(key, df):\n # key contains all the grouped column values\n # In this example, grouped columns are (userID, version, window{start, end})\n # For example, if you want to get the start and end time of a window, you can\n # get both values by calling key[2][\"start\"] and key[2][\"end\"]\n some_vals_mean = df[\"some_vals\"].mean()\n df[\"vals_avg\"] = some_vals_mean\n return df",
"_____no_output_____"
]
],
[
[
"## Run the smoothing algorithm on imported data\nThe smoothing algorithm is applied to the datastream by calling the `run_algorithm` method and passing the method as a parameter along with which columns, `some_vals`, that should be sent. Finally, the `windowDuration` parameter specified the size of the time windows on which to segment the data before applying the algorithm. Notice that when the next cell is run, the operation completes nearly instantaneously. This is due to the lazy evaluation aspects of the Spark framework. When you run the next cell to show the data, the algorithm will be applied to the whole dataset before displaying the results on the screen. ",
"_____no_output_____"
]
],
[
[
"smooth_stream = iot_stream.compute(smooth_algo, schema=schema, windowDuration=10)",
"_____no_output_____"
],
[
"smooth_stream.show(truncate=False)",
"+-------------------+-----------+-------+------------------------------------+-------------------+\n|timestamp |some_vals |version|user |vals_avg |\n+-------------------+-----------+-------+------------------------------------+-------------------+\n|2019-01-09 17:46:30|0.070952751|1 |00000000-afb8-476e-9872-6472b4e66b68|0.37378515760000003|\n|2019-01-09 17:46:31|0.279759975|1 |00000000-afb8-476e-9872-6472b4e66b68|0.37378515760000003|\n|2019-01-09 17:46:32|0.096120952|1 |00000000-afb8-476e-9872-6472b4e66b68|0.37378515760000003|\n|2019-01-09 17:46:33|0.121091841|1 |00000000-afb8-476e-9872-6472b4e66b68|0.37378515760000003|\n|2019-01-09 17:46:34|0.356470355|1 |00000000-afb8-476e-9872-6472b4e66b68|0.37378515760000003|\n|2019-01-09 17:46:35|0.800499717|1 |00000000-afb8-476e-9872-6472b4e66b68|0.37378515760000003|\n|2019-01-09 17:46:36|0.799160143|1 |00000000-afb8-476e-9872-6472b4e66b68|0.37378515760000003|\n|2019-01-09 17:46:37|0.372062031|1 |00000000-afb8-476e-9872-6472b4e66b68|0.37378515760000003|\n|2019-01-09 17:46:38|0.601158405|1 |00000000-afb8-476e-9872-6472b4e66b68|0.37378515760000003|\n|2019-01-09 17:46:39|0.240575406|1 |00000000-afb8-476e-9872-6472b4e66b68|0.37378515760000003|\n|2019-01-09 17:39:00|0.073887651|1 |00000000-afb8-476e-9872-6472b4e66b68|0.3879962564 |\n|2019-01-09 17:39:01|0.45542365 |1 |00000000-afb8-476e-9872-6472b4e66b68|0.3879962564 |\n|2019-01-09 17:39:02|0.629757033|1 |00000000-afb8-476e-9872-6472b4e66b68|0.3879962564 |\n|2019-01-09 17:39:03|0.67459103 |1 |00000000-afb8-476e-9872-6472b4e66b68|0.3879962564 |\n|2019-01-09 17:39:04|0.513277101|1 |00000000-afb8-476e-9872-6472b4e66b68|0.3879962564 |\n|2019-01-09 17:39:05|0.131369076|1 |00000000-afb8-476e-9872-6472b4e66b68|0.3879962564 |\n|2019-01-09 17:39:06|0.604344202|1 |00000000-afb8-476e-9872-6472b4e66b68|0.3879962564 |\n|2019-01-09 17:39:07|0.12731815 |1 |00000000-afb8-476e-9872-6472b4e66b68|0.3879962564 |\n|2019-01-09 17:39:08|0.424741964|1 |00000000-afb8-476e-9872-6472b4e66b68|0.3879962564 |\n|2019-01-09 17:39:09|0.245252707|1 |00000000-afb8-476e-9872-6472b4e66b68|0.3879962564 |\n+-------------------+-----------+-------+------------------------------------+-------------------+\nonly showing top 20 rows\n\n"
]
],
[
[
"## Visualize data\nThese are two plots that show the original and smoothed data to visually check how the algorithm transformed the data.",
"_____no_output_____"
]
],
[
[
"from cerebralcortex.plotting.basic.plots import plot_timeseries",
"_____no_output_____"
],
[
"plot_timeseries(iot_stream)",
"/usr/local/lib/python3.7/dist-packages/jsonschema/compat.py:6: DeprecationWarning:\n\nUsing or importing the ABCs from 'collections' instead of from 'collections.abc' is deprecated since Python 3.3,and in 3.9 it will stop working\n\n/usr/local/lib/python3.7/dist-packages/jsonschema/compat.py:6: DeprecationWarning:\n\nUsing or importing the ABCs from 'collections' instead of from 'collections.abc' is deprecated since Python 3.3,and in 3.9 it will stop working\n\n"
],
[
"plot_timeseries(smooth_stream)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
4a58f2656c9d4b968d4f9c70a240b25ef976dab8
| 5,975 |
ipynb
|
Jupyter Notebook
|
python_algorithm/pyPractice/p071to080.ipynb
|
theVelopr/algorithm
|
1d19e5ab6e8cb05242625df7af749e22f7bfb6a7
|
[
"MIT"
] | null | null | null |
python_algorithm/pyPractice/p071to080.ipynb
|
theVelopr/algorithm
|
1d19e5ab6e8cb05242625df7af749e22f7bfb6a7
|
[
"MIT"
] | null | null | null |
python_algorithm/pyPractice/p071to080.ipynb
|
theVelopr/algorithm
|
1d19e5ab6e8cb05242625df7af749e22f7bfb6a7
|
[
"MIT"
] | null | null | null | 17.942943 | 202 | 0.431632 |
[
[
[
"### 071\nmy_variable 이름의 비어있는 튜플을 만들라.",
"_____no_output_____"
]
],
[
[
"my_variable = ()\nprint(type(my_variable))",
"<class 'tuple'>\n"
]
],
[
[
"### 072\n2016년 11월 영화 예매 순위 기준 top3는 다음과 같다. 영화 제목을 movie_rank 이름의 튜플에 저장하라. (순위 정보는 저장하지 않는다.)\n\n순위\t영화\n<br>1\t닥터 스트레인지\n<br>2\t스플릿\n<br>3\t럭키",
"_____no_output_____"
]
],
[
[
"movie_rank = ('닥터 스트레인지', '스플릿', '럭키')\n\nprint(movie_rank)",
"('닥터 스트레인지', '스플릿', '럭키')\n"
]
],
[
[
"### 073\n숫자 1 이 저장된 튜플을 생성하라.",
"_____no_output_____"
]
],
[
[
"var = (1,)\n\nprint(type(var))",
"<class 'tuple'>\n"
]
],
[
[
"하나의 데이터가 튜플에 저장되는 경우 ,를 넣어줘야 됨.",
"_____no_output_____"
],
[
"### 075\n아래와 같이 t에는 1, 2, 3, 4 데이터가 바인딩되어 있다. t가 바인딩하는 데이터 타입은 무엇인가?\n\nt = 1, 2, 3, 4",
"_____no_output_____"
]
],
[
[
"t = 1, 2, 3, 4\n\nprint(type(t))",
"<class 'tuple'>\n"
]
],
[
[
"원칙적으로 튜플은 괄호와 함께 데이터를 정의해야 하지만, 사용자 편의를 위해 괄호 없이도 동작합니다. ",
"_____no_output_____"
],
[
"### 076\n변수 t에는 아래와 같은 값이 저장되어 있다. 변수 t가 ('A', 'b', 'c') 튜플을 가리키도록 수정 하라.\n\nt = ('a', 'b', 'c')",
"_____no_output_____"
]
],
[
[
"t = ('a', 'b', 'c')\n\n# 튜플은 수정불가. 따라서, 새로 재정의 해줘야함.\nt = ('A', 'b', 'c')\n\nprint(t)",
"('A', 'b', 'c')\n"
]
],
[
[
"### 077\n다음 튜플을 리스트로 변환하라.\n\ninterest = ('삼성전자', 'LG전자', 'SK Hynix')",
"_____no_output_____"
]
],
[
[
"interest = ('삼성전자', 'LG전자', 'SK Hynix')\ndata = list(interest)\n\nprint(type(data))",
"<class 'list'>\n"
]
],
[
[
"### 078\n다음 리스트를 튜플로 변경하라.\n\ninterest = ['삼성전자', 'LG전자', 'SK Hynix']",
"_____no_output_____"
]
],
[
[
"interest = ['삼성전자', 'LG전자', 'SK Hynix']\ndata = tuple(interest)\n\nprint(type(data))",
"<class 'tuple'>\n"
]
],
[
[
"### 079 튜플 언팩킹\n다음 코드의 실행 결과를 예상하라.\n\ntemp = ('apple', 'banana', 'cake')\n<br>a, b, c = temp\n<br>print(a, b, c)",
"_____no_output_____"
]
],
[
[
"temp = ('apple', 'banana', 'cake')\na, b, c = temp\nprint(a, b, c)",
"apple banana cake\n"
]
],
[
[
"### 080 range 함수\n1 부터 99까지의 정수 중 짝수만 저장된 튜플을 생성하라.\n\n(2, 4, 6, 8 ... 98)",
"_____no_output_____"
]
],
[
[
"data = tuple(range(2, 100, 2))\nprint(data)",
"(2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26, 28, 30, 32, 34, 36, 38, 40, 42, 44, 46, 48, 50, 52, 54, 56, 58, 60, 62, 64, 66, 68, 70, 72, 74, 76, 78, 80, 82, 84, 86, 88, 90, 92, 94, 96, 98)\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
4a58f5f3541e7a5e683689b650aac45f38aa4a29
| 245,970 |
ipynb
|
Jupyter Notebook
|
4_synthetic_data_attention/toy_problem_mosaic/toy_problem_Mosaic_type4.ipynb
|
lnpandey/DL_explore_synth_data
|
0a5d8b417091897f4c7f358377d5198a155f3f24
|
[
"MIT"
] | 2 |
2019-08-24T07:20:35.000Z
|
2020-03-27T08:16:59.000Z
|
4_synthetic_data_attention/toy_problem_mosaic/toy_problem_Mosaic_type4.ipynb
|
lnpandey/DL_explore_synth_data
|
0a5d8b417091897f4c7f358377d5198a155f3f24
|
[
"MIT"
] | null | null | null |
4_synthetic_data_attention/toy_problem_mosaic/toy_problem_Mosaic_type4.ipynb
|
lnpandey/DL_explore_synth_data
|
0a5d8b417091897f4c7f358377d5198a155f3f24
|
[
"MIT"
] | 3 |
2019-06-21T09:34:32.000Z
|
2019-09-19T10:43:07.000Z
| 125.111902 | 51,722 | 0.796117 |
[
[
[
"import numpy as np\nimport pandas as pd\nfrom matplotlib import pyplot as plt\nfrom tqdm import tqdm\n%matplotlib inline\nfrom torch.utils.data import Dataset, DataLoader\nimport torch\nimport torchvision\n\nimport torch.nn as nn\nimport torch.optim as optim\nfrom torch.nn import functional as F\ndevice = torch.device(\"cuda\" if torch.cuda.is_available() else \"cpu\")\nprint(device)",
"cuda\n"
]
],
[
[
"# Generate dataset",
"_____no_output_____"
]
],
[
[
"y = np.random.randint(0,10,5000)\nidx= []\nfor i in range(10):\n print(i,sum(y==i))\n idx.append(y==i)",
"0 495\n1 477\n2 514\n3 478\n4 528\n5 522\n6 488\n7 467\n8 507\n9 524\n"
],
[
"x = np.zeros((5000,2))",
"_____no_output_____"
],
[
"x[idx[0],:] = np.random.multivariate_normal(mean = [4,6.5],cov=[[0.01,0],[0,0.01]],size=sum(idx[0]))\n\nx[idx[1],:] = np.random.multivariate_normal(mean = [5.5,6],cov=[[0.01,0],[0,0.01]],size=sum(idx[1]))\n\nx[idx[2],:] = np.random.multivariate_normal(mean = [4.5,4.5],cov=[[0.01,0],[0,0.01]],size=sum(idx[2]))\n\n# x[idx[0],:] = np.random.multivariate_normal(mean = [5,5],cov=[[0.1,0],[0,0.1]],size=sum(idx[0]))\n\n# x[idx[1],:] = np.random.multivariate_normal(mean = [6,6],cov=[[0.1,0],[0,0.1]],size=sum(idx[1]))\n\n# x[idx[2],:] = np.random.multivariate_normal(mean = [5.5,6.5],cov=[[0.1,0],[0,0.1]],size=sum(idx[2]))\n\nx[idx[3],:] = np.random.multivariate_normal(mean = [3,3.5],cov=[[0.01,0],[0,0.01]],size=sum(idx[3]))\n\n\nx[idx[4],:] = np.random.multivariate_normal(mean = [2.5,5.5],cov=[[0.01,0],[0,0.01]],size=sum(idx[4]))\n\nx[idx[5],:] = np.random.multivariate_normal(mean = [3.5,8],cov=[[0.01,0],[0,0.01]],size=sum(idx[5]))\n\nx[idx[6],:] = np.random.multivariate_normal(mean = [5.5,8],cov=[[0.01,0],[0,0.01]],size=sum(idx[6]))\n\nx[idx[7],:] = np.random.multivariate_normal(mean = [7,6.5],cov=[[0.01,0],[0,0.01]],size=sum(idx[7]))\n\nx[idx[8],:] = np.random.multivariate_normal(mean = [6.5,4.5],cov=[[0.01,0],[0,0.01]],size=sum(idx[8]))\n\nx[idx[9],:] = np.random.multivariate_normal(mean = [5,3],cov=[[0.01,0],[0,0.01]],size=sum(idx[9]))",
"_____no_output_____"
],
[
"for i in range(10):\n plt.scatter(x[idx[i],0],x[idx[i],1],label=\"class_\"+str(i))\nplt.legend(loc='center left', bbox_to_anchor=(1, 0.5))",
"_____no_output_____"
],
[
"foreground_classes = {'class_0','class_1', 'class_2'}\n\nbackground_classes = {'class_3','class_4', 'class_5', 'class_6','class_7', 'class_8', 'class_9'}",
"_____no_output_____"
],
[
"fg_class = np.random.randint(0,3)\nfg_idx = np.random.randint(0,9)\n\na = []\nfor i in range(9):\n if i == fg_idx:\n b = np.random.choice(np.where(idx[fg_class]==True)[0],size=1)\n a.append(x[b])\n print(\"foreground \"+str(fg_class)+\" present at \" + str(fg_idx))\n else:\n bg_class = np.random.randint(3,10)\n b = np.random.choice(np.where(idx[bg_class]==True)[0],size=1)\n a.append(x[b])\n print(\"background \"+str(bg_class)+\" present at \" + str(i))\na = np.concatenate(a,axis=0)\nprint(a.shape)\n\nprint(fg_class , fg_idx)",
"background 3 present at 0\nbackground 5 present at 1\nbackground 3 present at 2\nbackground 3 present at 3\nbackground 3 present at 4\nbackground 4 present at 5\nbackground 9 present at 6\nforeground 2 present at 7\nbackground 8 present at 8\n(9, 2)\n2 7\n"
],
[
"a.shape",
"_____no_output_____"
],
[
"np.reshape(a,(18,1))",
"_____no_output_____"
],
[
"a=np.reshape(a,(3,6))",
"_____no_output_____"
],
[
"plt.imshow(a)",
"_____no_output_____"
],
[
"desired_num = 3000\nmosaic_list =[]\nmosaic_label = []\nfore_idx=[]\nfor j in range(desired_num):\n fg_class = np.random.randint(0,3)\n fg_idx = np.random.randint(0,9)\n a = []\n for i in range(9):\n if i == fg_idx:\n b = np.random.choice(np.where(idx[fg_class]==True)[0],size=1)\n a.append(x[b])\n# print(\"foreground \"+str(fg_class)+\" present at \" + str(fg_idx))\n else:\n bg_class = np.random.randint(3,10)\n b = np.random.choice(np.where(idx[bg_class]==True)[0],size=1)\n a.append(x[b])\n# print(\"background \"+str(bg_class)+\" present at \" + str(i))\n a = np.concatenate(a,axis=0)\n mosaic_list.append(np.reshape(a,(18,1)))\n mosaic_label.append(fg_class)\n fore_idx.append(fg_idx)",
"_____no_output_____"
],
[
"mosaic_list = np.concatenate(mosaic_list,axis=1).T\n# print(mosaic_list)",
"_____no_output_____"
],
[
"print(np.shape(mosaic_label))\nprint(np.shape(fore_idx))",
"(3000,)\n(3000,)\n"
],
[
"class MosaicDataset(Dataset):\n \"\"\"MosaicDataset dataset.\"\"\"\n\n def __init__(self, mosaic_list, mosaic_label, fore_idx):\n \"\"\"\n Args:\n csv_file (string): Path to the csv file with annotations.\n root_dir (string): Directory with all the images.\n transform (callable, optional): Optional transform to be applied\n on a sample.\n \"\"\"\n self.mosaic = mosaic_list\n self.label = mosaic_label\n self.fore_idx = fore_idx\n\n def __len__(self):\n return len(self.label)\n\n def __getitem__(self, idx):\n return self.mosaic[idx] , self.label[idx], self.fore_idx[idx]\n\nbatch = 250\nmsd = MosaicDataset(mosaic_list, mosaic_label , fore_idx)\ntrain_loader = DataLoader( msd,batch_size= batch ,shuffle=True)",
"_____no_output_____"
],
[
"class Wherenet(nn.Module):\n def __init__(self):\n super(Wherenet,self).__init__()\n self.linear1 = nn.Linear(2,50)\n self.linear2 = nn.Linear(50,50)\n self.linear3 = nn.Linear(50,1)\n def forward(self,z):\n x = torch.zeros([batch,9],dtype=torch.float64)\n y = torch.zeros([batch,2], dtype=torch.float64)\n #x,y = x.to(\"cuda\"),y.to(\"cuda\")\n for i in range(9):\n x[:,i] = self.helper(z[:,2*i:2*i+2])[:,0]\n #print(k[:,0].shape,x[:,i].shape)\n x = F.softmax(x,dim=1) # alphas\n x1 = x[:,0]\n for i in range(9):\n x1 = x[:,i] \n #print()\n y = y+torch.mul(x1[:,None],z[:,2*i:2*i+2])\n return y , x \n\n \n def helper(self,x):\n x = F.relu(self.linear1(x))\n x = F.relu(self.linear2(x))\n x = self.linear3(x)\n return x\n\n ",
"_____no_output_____"
],
[
"trainiter = iter(train_loader)\ninput1,labels1,index1 = trainiter.next()\n\n",
"_____no_output_____"
],
[
"where = Wherenet().double()\nwhere = where\nout_where,alphas = where(input1)\nout_where.shape,alphas.shape",
"_____no_output_____"
],
[
"class Whatnet(nn.Module):\n def __init__(self):\n super(Whatnet,self).__init__()\n self.linear1 = nn.Linear(2,50)\n self.linear2 = nn.Linear(50,3)\n# self.linear3 = nn.Linear(8,3)\n def forward(self,x):\n x = F.relu(self.linear1(x))\n #x = F.relu(self.linear2(x))\n x = self.linear2(x)\n return x",
"_____no_output_____"
],
[
"what = Whatnet().double()\n# what(out_where)",
"_____no_output_____"
],
[
"test_data_required = 1000\nmosaic_list_test =[]\nmosaic_label_test = []\nfore_idx_test=[]\nfor j in range(test_data_required):\n fg_class = np.random.randint(0,3)\n fg_idx = np.random.randint(0,9)\n a = []\n for i in range(9):\n if i == fg_idx:\n b = np.random.choice(np.where(idx[fg_class]==True)[0],size=1)\n a.append(x[b])\n# print(\"foreground \"+str(fg_class)+\" present at \" + str(fg_idx))\n else:\n bg_class = np.random.randint(3,10)\n b = np.random.choice(np.where(idx[bg_class]==True)[0],size=1)\n a.append(x[b])\n# print(\"background \"+str(bg_class)+\" present at \" + str(i))\n a = np.concatenate(a,axis=0)\n mosaic_list_test.append(np.reshape(a,(18,1)))\n mosaic_label_test.append(fg_class)\n fore_idx_test.append(fg_idx)",
"_____no_output_____"
],
[
"mosaic_list_test = np.concatenate(mosaic_list_test,axis=1).T\nprint(mosaic_list_test.shape)",
"(1000, 18)\n"
],
[
"test_data = MosaicDataset(mosaic_list_test,mosaic_label_test,fore_idx_test)\ntest_loader = DataLoader( test_data,batch_size= batch ,shuffle=False)",
"_____no_output_____"
],
[
"focus_true_pred_true =0\nfocus_false_pred_true =0\nfocus_true_pred_false =0\nfocus_false_pred_false =0\n\nargmax_more_than_half = 0\nargmax_less_than_half =0\n\ncol1=[]\ncol2=[]\ncol3=[]\ncol4=[]\ncol5=[]\ncol6=[]\ncol7=[]\ncol8=[]\ncol9=[]\ncol10=[]\ncol11=[]\ncol12=[]\ncol13=[]\n\ncriterion = nn.CrossEntropyLoss()\noptimizer_where = optim.SGD(where.parameters(), lr=0.01, momentum=0.9)\noptimizer_what = optim.SGD(what.parameters(), lr=0.01, momentum=0.9)\n\nnos_epochs = 250\n\ntrain_loss=[]\ntest_loss =[]\ntrain_acc = []\ntest_acc = []\nloss_curi = []\nfor epoch in range(nos_epochs): # loop over the dataset multiple times\n\n focus_true_pred_true =0\n focus_false_pred_true =0\n focus_true_pred_false =0\n focus_false_pred_false =0\n\n argmax_more_than_half = 0\n argmax_less_than_half =0\n\n running_loss = 0.0\n cnt=0\n ep_lossi = []\n iteration = desired_num // batch\n\n #training data set\n\n for i, data in enumerate(train_loader):\n inputs , labels , fore_idx = data\n #inputs,labels,fore_idx = inputs.to(device),labels.to(device),fore_idx.to(device)\n # zero the parameter gradients\n\n optimizer_what.zero_grad()\n optimizer_where.zero_grad()\n \n \n avg_inp,alphas = where(inputs)\n \n outputs = what(avg_inp)\n \n _, predicted = torch.max(outputs.data, 1)\n \n loss = criterion(outputs, labels) \n loss.backward() \n \n optimizer_what.step()\n optimizer_where.step() \n \n\n running_loss += loss.item()\n if cnt % 6 == 5: # print every 6 mini-batches\n print('[%d, %5d] loss: %.3f' %(epoch + 1, cnt + 1, running_loss / 6))\n ep_lossi.append(running_loss/6)\n running_loss = 0.0\n cnt=cnt+1\n \n \n if epoch % 1 == 0:\n for j in range (batch):\n focus = torch.argmax(alphas[j])\n\n if(alphas[j][focus] >= 0.5):\n argmax_more_than_half +=1\n else:\n argmax_less_than_half +=1\n\n if(focus == fore_idx[j] and predicted[j] == labels[j]):\n focus_true_pred_true += 1\n\n elif(focus != fore_idx[j] and predicted[j] == labels[j]):\n focus_false_pred_true +=1\n\n elif(focus == fore_idx[j] and predicted[j] != labels[j]):\n focus_true_pred_false +=1\n\n elif(focus != fore_idx[j] and predicted[j] != labels[j]):\n focus_false_pred_false +=1\n\n loss_curi.append(np.mean(ep_lossi)) #loss per epoch\n if (np.mean(ep_lossi) <= 0.01):\n break\n \n if epoch % 1 == 0:\n col1.append(epoch)\n col2.append(argmax_more_than_half)\n col3.append(argmax_less_than_half)\n col4.append(focus_true_pred_true)\n col5.append(focus_false_pred_true)\n col6.append(focus_true_pred_false)\n col7.append(focus_false_pred_false)\n\n #************************************************************************\n #testing data set \n with torch.no_grad():\n focus_true_pred_true =0\n focus_false_pred_true =0\n focus_true_pred_false =0\n focus_false_pred_false =0\n\n argmax_more_than_half = 0\n argmax_less_than_half =0\n for data in test_loader:\n inputs, labels , fore_idx = data\n #inputs,labels,fore_idx = inputs.to(device),labels.to(device),fore_idx.to(device) \n# print(inputs.shtorch.save(where.state_dict(),\"model_epoch\"+str(epoch)+\".pt\")ape,labels.shape)\n avg_inp,alphas = where(inputs)\n outputs = what(avg_inp)\n _, predicted = torch.max(outputs.data, 1)\n\n for j in range (batch):\n focus = torch.argmax(alphas[j])\n\n if(alphas[j][focus] >= 0.5):\n argmax_more_than_half +=1\n else:\n argmax_less_than_half +=1\n\n if(focus == fore_idx[j] and predicted[j] == labels[j]):\n focus_true_pred_true += 1\n\n elif(focus != fore_idx[j] and predicted[j] == labels[j]):\n focus_false_pred_true +=1\n\n elif(focus == fore_idx[j] and predicted[j] != labels[j]):\n focus_true_pred_false +=1\n\n elif(focus != fore_idx[j] and predicted[j] != labels[j]):\n focus_false_pred_false +=1\n\n col8.append(argmax_more_than_half)\n col9.append(argmax_less_than_half)\n col10.append(focus_true_pred_true)\n col11.append(focus_false_pred_true)\n col12.append(focus_true_pred_false)\n col13.append(focus_false_pred_false)\n torch.save(where.state_dict(),\"where_model_epoch\"+str(epoch)+\".pt\")\n torch.save(what.state_dict(),\"what_model_epoch\"+str(epoch)+\".pt\")\n \nprint('Finished Training')\n# torch.save(where.state_dict(),\"where_model_epoch\"+str(nos_epochs)+\".pt\")\n# torch.save(what.state_dict(),\"what_model_epoch\"+str(epoch)+\".pt\")\n",
"[1, 6] loss: 1.096\n[1, 12] loss: 1.106\n[2, 6] loss: 1.109\n[2, 12] loss: 1.126\n[3, 6] loss: 1.110\n[3, 12] loss: 1.155\n[4, 6] loss: 1.130\n[4, 12] loss: 1.108\n[5, 6] loss: 1.111\n[5, 12] loss: 1.112\n[6, 6] loss: 1.094\n[6, 12] loss: 1.090\n[7, 6] loss: 1.107\n[7, 12] loss: 1.111\n[8, 6] loss: 1.109\n[8, 12] loss: 1.087\n[9, 6] loss: 1.081\n[9, 12] loss: 1.087\n[10, 6] loss: 1.099\n[10, 12] loss: 1.093\n[11, 6] loss: 1.086\n[11, 12] loss: 1.089\n[12, 6] loss: 1.081\n[12, 12] loss: 1.093\n[13, 6] loss: 1.088\n[13, 12] loss: 1.072\n[14, 6] loss: 1.072\n[14, 12] loss: 1.084\n[15, 6] loss: 1.076\n[15, 12] loss: 1.078\n[16, 6] loss: 1.083\n[16, 12] loss: 1.120\n[17, 6] loss: 1.122\n[17, 12] loss: 1.103\n[18, 6] loss: 1.097\n[18, 12] loss: 1.091\n[19, 6] loss: 1.062\n[19, 12] loss: 1.066\n[20, 6] loss: 1.071\n[20, 12] loss: 1.070\n[21, 6] loss: 1.060\n[21, 12] loss: 1.058\n[22, 6] loss: 1.054\n[22, 12] loss: 1.062\n[23, 6] loss: 1.041\n[23, 12] loss: 1.042\n[24, 6] loss: 1.016\n[24, 12] loss: 1.010\n[25, 6] loss: 1.000\n[25, 12] loss: 1.010\n[26, 6] loss: 0.973\n[26, 12] loss: 0.989\n[27, 6] loss: 0.949\n[27, 12] loss: 0.941\n[28, 6] loss: 0.918\n[28, 12] loss: 0.907\n[29, 6] loss: 0.892\n[29, 12] loss: 0.872\n[30, 6] loss: 0.849\n[30, 12] loss: 0.836\n[31, 6] loss: 0.812\n[31, 12] loss: 0.824\n[32, 6] loss: 0.807\n[32, 12] loss: 0.800\n[33, 6] loss: 0.776\n[33, 12] loss: 0.768\n[34, 6] loss: 0.756\n[34, 12] loss: 0.754\n[35, 6] loss: 0.740\n[35, 12] loss: 0.735\n[36, 6] loss: 0.720\n[36, 12] loss: 0.716\n[37, 6] loss: 0.691\n[37, 12] loss: 0.707\n[38, 6] loss: 0.706\n[38, 12] loss: 0.685\n[39, 6] loss: 0.652\n[39, 12] loss: 0.625\n[40, 6] loss: 0.630\n[40, 12] loss: 0.644\n[41, 6] loss: 0.579\n[41, 12] loss: 0.600\n[42, 6] loss: 0.727\n[42, 12] loss: 0.677\n[43, 6] loss: 0.585\n[43, 12] loss: 0.625\n[44, 6] loss: 0.645\n[44, 12] loss: 0.580\n[45, 6] loss: 0.779\n[45, 12] loss: 0.987\n[46, 6] loss: 0.869\n[46, 12] loss: 0.919\n[47, 6] loss: 0.724\n[47, 12] loss: 0.548\n[48, 6] loss: 0.506\n[48, 12] loss: 0.554\n[49, 6] loss: 0.669\n[49, 12] loss: 0.517\n[50, 6] loss: 0.463\n[50, 12] loss: 0.445\n[51, 6] loss: 0.435\n[51, 12] loss: 0.418\n[52, 6] loss: 0.440\n[52, 12] loss: 0.517\n[53, 6] loss: 1.473\n[53, 12] loss: 1.222\n[54, 6] loss: 0.806\n[54, 12] loss: 0.792\n[55, 6] loss: 0.751\n[55, 12] loss: 0.622\n[56, 6] loss: 0.417\n[56, 12] loss: 0.362\n[57, 6] loss: 0.368\n[57, 12] loss: 0.316\n[58, 6] loss: 0.297\n[58, 12] loss: 0.283\n[59, 6] loss: 0.284\n[59, 12] loss: 0.255\n[60, 6] loss: 0.246\n[60, 12] loss: 0.232\n[61, 6] loss: 0.228\n[61, 12] loss: 0.225\n[62, 6] loss: 0.211\n[62, 12] loss: 0.194\n[63, 6] loss: 0.188\n[63, 12] loss: 0.180\n[64, 6] loss: 0.172\n[64, 12] loss: 0.173\n[65, 6] loss: 0.172\n[65, 12] loss: 0.154\n[66, 6] loss: 0.174\n[66, 12] loss: 0.170\n[67, 6] loss: 0.213\n[67, 12] loss: 0.160\n[68, 6] loss: 0.144\n[68, 12] loss: 0.155\n[69, 6] loss: 0.129\n[69, 12] loss: 0.128\n[70, 6] loss: 0.112\n[70, 12] loss: 0.116\n[71, 6] loss: 0.111\n[71, 12] loss: 0.097\n[72, 6] loss: 0.210\n[72, 12] loss: 1.357\n[73, 6] loss: 0.955\n[73, 12] loss: 0.430\n[74, 6] loss: 0.226\n[74, 12] loss: 0.210\n[75, 6] loss: 0.171\n[75, 12] loss: 0.124\n[76, 6] loss: 0.132\n[76, 12] loss: 0.159\n[77, 6] loss: 0.174\n[77, 12] loss: 0.123\n[78, 6] loss: 0.104\n[78, 12] loss: 0.094\n[79, 6] loss: 0.090\n[79, 12] loss: 0.079\n[80, 6] loss: 0.071\n[80, 12] loss: 0.069\n[81, 6] loss: 0.064\n[81, 12] loss: 0.065\n[82, 6] loss: 0.061\n[82, 12] loss: 0.061\n[83, 6] loss: 0.056\n[83, 12] loss: 0.059\n[84, 6] loss: 0.058\n[84, 12] loss: 0.052\n[85, 6] loss: 0.053\n[85, 12] loss: 0.053\n[86, 6] loss: 0.061\n[86, 12] loss: 0.058\n[87, 6] loss: 0.060\n[87, 12] loss: 0.063\n[88, 6] loss: 0.058\n[88, 12] loss: 0.052\n[89, 6] loss: 0.048\n[89, 12] loss: 0.044\n[90, 6] loss: 0.043\n[90, 12] loss: 0.044\n[91, 6] loss: 0.042\n[91, 12] loss: 0.042\n[92, 6] loss: 0.040\n[92, 12] loss: 0.041\n[93, 6] loss: 0.038\n[93, 12] loss: 0.040\n[94, 6] loss: 0.038\n[94, 12] loss: 0.037\n[95, 6] loss: 0.037\n[95, 12] loss: 0.037\n[96, 6] loss: 0.036\n[96, 12] loss: 0.036\n[97, 6] loss: 0.036\n[97, 12] loss: 0.036\n[98, 6] loss: 0.032\n[98, 12] loss: 0.033\n[99, 6] loss: 0.033\n[99, 12] loss: 0.030\n[100, 6] loss: 0.030\n[100, 12] loss: 0.030\n[101, 6] loss: 0.031\n[101, 12] loss: 0.029\n[102, 6] loss: 0.030\n[102, 12] loss: 0.027\n[103, 6] loss: 0.028\n[103, 12] loss: 0.029\n[104, 6] loss: 0.028\n[104, 12] loss: 0.028\n[105, 6] loss: 0.026\n[105, 12] loss: 0.028\n[106, 6] loss: 0.025\n[106, 12] loss: 0.027\n[107, 6] loss: 0.025\n[107, 12] loss: 0.026\n[108, 6] loss: 0.026\n[108, 12] loss: 0.024\n[109, 6] loss: 0.025\n[109, 12] loss: 0.024\n[110, 6] loss: 0.024\n[110, 12] loss: 0.023\n[111, 6] loss: 0.023\n[111, 12] loss: 0.022\n[112, 6] loss: 0.024\n[112, 12] loss: 0.021\n[113, 6] loss: 0.023\n[113, 12] loss: 0.021\n[114, 6] loss: 0.022\n[114, 12] loss: 0.021\n[115, 6] loss: 0.021\n[115, 12] loss: 0.021\n[116, 6] loss: 0.020\n[116, 12] loss: 0.021\n[117, 6] loss: 0.021\n[117, 12] loss: 0.019\n[118, 6] loss: 0.018\n[118, 12] loss: 0.021\n[119, 6] loss: 0.019\n[119, 12] loss: 0.020\n[120, 6] loss: 0.018\n[120, 12] loss: 0.020\n[121, 6] loss: 0.020\n[121, 12] loss: 0.018\n[122, 6] loss: 0.020\n[122, 12] loss: 0.017\n[123, 6] loss: 0.017\n[123, 12] loss: 0.019\n[124, 6] loss: 0.019\n[124, 12] loss: 0.016\n[125, 6] loss: 0.017\n[125, 12] loss: 0.018\n[126, 6] loss: 0.018\n[126, 12] loss: 0.016\n[127, 6] loss: 0.017\n[127, 12] loss: 0.016\n[128, 6] loss: 0.016\n[128, 12] loss: 0.017\n[129, 6] loss: 0.016\n[129, 12] loss: 0.017\n[130, 6] loss: 0.016\n[130, 12] loss: 0.017\n[131, 6] loss: 0.015\n[131, 12] loss: 0.017\n[132, 6] loss: 0.015\n[132, 12] loss: 0.016\n[133, 6] loss: 0.015\n[133, 12] loss: 0.015\n[134, 6] loss: 0.015\n[134, 12] loss: 0.015\n[135, 6] loss: 0.015\n[135, 12] loss: 0.014\n[136, 6] loss: 0.015\n[136, 12] loss: 0.014\n[137, 6] loss: 0.015\n[137, 12] loss: 0.014\n[138, 6] loss: 0.014\n[138, 12] loss: 0.014\n[139, 6] loss: 0.015\n[139, 12] loss: 0.013\n[140, 6] loss: 0.014\n[140, 12] loss: 0.013\n[141, 6] loss: 0.013\n[141, 12] loss: 0.014\n[142, 6] loss: 0.013\n[142, 12] loss: 0.013\n[143, 6] loss: 0.013\n[143, 12] loss: 0.014\n[144, 6] loss: 0.013\n[144, 12] loss: 0.013\n[145, 6] loss: 0.013\n[145, 12] loss: 0.012\n[146, 6] loss: 0.012\n[146, 12] loss: 0.013\n[147, 6] loss: 0.013\n[147, 12] loss: 0.012\n[148, 6] loss: 0.012\n[148, 12] loss: 0.013\n[149, 6] loss: 0.013\n[149, 12] loss: 0.011\n[150, 6] loss: 0.012\n[150, 12] loss: 0.012\n[151, 6] loss: 0.012\n[151, 12] loss: 0.012\n[152, 6] loss: 0.011\n[152, 12] loss: 0.012\n[153, 6] loss: 0.010\n[153, 12] loss: 0.012\n[154, 6] loss: 0.012\n[154, 12] loss: 0.010\n[155, 6] loss: 0.011\n[155, 12] loss: 0.011\n[156, 6] loss: 0.011\n[156, 12] loss: 0.011\n[157, 6] loss: 0.011\n[157, 12] loss: 0.011\n[158, 6] loss: 0.011\n[158, 12] loss: 0.011\n[159, 6] loss: 0.010\n[159, 12] loss: 0.012\n[160, 6] loss: 0.011\n[160, 12] loss: 0.010\n[161, 6] loss: 0.011\n[161, 12] loss: 0.010\n[162, 6] loss: 0.010\n[162, 12] loss: 0.010\n[163, 6] loss: 0.011\n[163, 12] loss: 0.010\n[164, 6] loss: 0.009\n[164, 12] loss: 0.011\nFinished Training\n"
],
[
"columns = [\"epochs\", \"argmax > 0.5\" ,\"argmax < 0.5\", \"focus_true_pred_true\", \"focus_false_pred_true\", \"focus_true_pred_false\", \"focus_false_pred_false\" ]\ndf_train = pd.DataFrame()\ndf_test = pd.DataFrame()\ndf_train[columns[0]] = col1\ndf_train[columns[1]] = col2\ndf_train[columns[2]] = col3\ndf_train[columns[3]] = col4\ndf_train[columns[4]] = col5\ndf_train[columns[5]] = col6\ndf_train[columns[6]] = col7\n\ndf_test[columns[0]] = col1\ndf_test[columns[1]] = col8\ndf_test[columns[2]] = col9\ndf_test[columns[3]] = col10\ndf_test[columns[4]] = col11\ndf_test[columns[5]] = col12\ndf_test[columns[6]] = col13",
"_____no_output_____"
],
[
"df_train",
"_____no_output_____"
],
[
"plt.plot(col1,col2, label='argmax > 0.5')\nplt.plot(col1,col3, label='argmax < 0.5')\n\nplt.legend(loc='center left', bbox_to_anchor=(1, 0.5))\nplt.xlabel(\"epochs\")\nplt.ylabel(\"training data\")\nplt.title(\"On Training set\")\nplt.show()\n\nplt.plot(col1,col4, label =\"focus_true_pred_true \")\nplt.plot(col1,col5, label =\"focus_false_pred_true \")\nplt.plot(col1,col6, label =\"focus_true_pred_false \")\nplt.plot(col1,col7, label =\"focus_false_pred_false \")\nplt.title(\"On Training set\")\nplt.legend(loc='center left', bbox_to_anchor=(1, 0.5))\nplt.xlabel(\"epochs\")\nplt.ylabel(\"training data\")\nplt.show()",
"_____no_output_____"
],
[
"df_test",
"_____no_output_____"
],
[
"plt.plot(col1,col8, label='argmax > 0.5')\nplt.plot(col1,col9, label='argmax < 0.5')\n\nplt.legend(loc='center left', bbox_to_anchor=(1, 0.5))\nplt.xlabel(\"epochs\")\nplt.ylabel(\"Testing data\")\nplt.title(\"On Testing set\")\nplt.show()\n\nplt.plot(col1,col10, label =\"focus_true_pred_true \")\nplt.plot(col1,col11, label =\"focus_false_pred_true \")\nplt.plot(col1,col12, label =\"focus_true_pred_false \")\nplt.plot(col1,col13, label =\"focus_false_pred_false \")\nplt.title(\"On Testing set\")\nplt.legend(loc='center left', bbox_to_anchor=(1, 0.5))\nplt.xlabel(\"epochs\")\nplt.ylabel(\"Testing data\")\nplt.show()",
"_____no_output_____"
],
[
"print(x[0])",
"[4.37668763 4.55057721]\n"
],
[
"for i in range(9):\n print(x[0,2*i:2*i+2])",
"[4.37668763 4.55057721]\n[]\n[]\n[]\n[]\n[]\n[]\n[]\n[]\n"
],
[
"correct = 0\ntotal = 0\ncount = 0\nflag = 1\nfocus_true_pred_true =0\nfocus_false_pred_true =0\nfocus_true_pred_false =0\nfocus_false_pred_false =0\n\nargmax_more_than_half = 0\nargmax_less_than_half =0\n\nwith torch.no_grad():\n for data in train_loader:\n inputs , labels , fore_idx = data\n #inputs,labels,fore_idx = inputs.to(device),labels.to(device),fore_idx.to(device)\n # zero the parameter gradients\n\n optimizer_what.zero_grad()\n optimizer_where.zero_grad()\n \n \n avg_inp,alphas = where(inputs)\n \n outputs = what(avg_inp)\n \n _, predicted = torch.max(outputs.data, 1)\n\n\n for j in range(labels.size(0)):\n count += 1\n focus = torch.argmax(alphas[j])\n if alphas[j][focus] >= 0.5 :\n argmax_more_than_half += 1\n else:\n argmax_less_than_half += 1\n\n if(focus == fore_idx[j] and predicted[j] == labels[j]):\n focus_true_pred_true += 1\n elif(focus != fore_idx[j] and predicted[j] == labels[j]):\n focus_false_pred_true += 1\n elif(focus == fore_idx[j] and predicted[j] != labels[j]):\n focus_true_pred_false += 1\n elif(focus != fore_idx[j] and predicted[j] != labels[j]):\n focus_false_pred_false += 1\n\n total += labels.size(0)\n correct += (predicted == labels).sum().item()\n\nprint('Accuracy of the network on the 3000 train images: %d %%' % ( 100 * correct / total))\nprint(\"total correct\", correct)\nprint(\"total train set images\", total)\n\nprint(\"focus_true_pred_true %d =============> FTPT : %d %%\" % (focus_true_pred_true , (100 * focus_true_pred_true / total) ) )\nprint(\"focus_false_pred_true %d =============> FFPT : %d %%\" % (focus_false_pred_true, (100 * focus_false_pred_true / total) ) )\nprint(\"focus_true_pred_false %d =============> FTPF : %d %%\" %( focus_true_pred_false , ( 100 * focus_true_pred_false / total) ) )\nprint(\"focus_false_pred_false %d =============> FFPF : %d %%\" % (focus_false_pred_false, ( 100 * focus_false_pred_false / total) ) )\n\nprint(\"argmax_more_than_half ==================> \",argmax_more_than_half)\nprint(\"argmax_less_than_half ==================> \",argmax_less_than_half)\nprint(count)\n\nprint(\"=\"*100)",
"Accuracy of the network on the 3000 train images: 99 %\ntotal correct 2999\ntotal train set images 3000\nfocus_true_pred_true 2076 =============> FTPT : 69 %\nfocus_false_pred_true 923 =============> FFPT : 30 %\nfocus_true_pred_false 0 =============> FTPF : 0 %\nfocus_false_pred_false 1 =============> FFPF : 0 %\nargmax_more_than_half ==================> 2095\nargmax_less_than_half ==================> 905\n3000\n====================================================================================================\n"
],
[
"correct = 0\ntotal = 0\ncount = 0\nflag = 1\nfocus_true_pred_true =0\nfocus_false_pred_true =0\nfocus_true_pred_false =0\nfocus_false_pred_false =0\n\nargmax_more_than_half = 0\nargmax_less_than_half =0\n\nwith torch.no_grad():\n for data in test_loader:\n inputs , labels , fore_idx = data\n #inputs,labels,fore_idx = inputs.to(device),labels.to(device),fore_idx.to(device)\n # zero the parameter gradients\n\n optimizer_what.zero_grad()\n optimizer_where.zero_grad()\n \n \n avg_inp,alphas = where(inputs)\n \n outputs = what(avg_inp)\n \n _, predicted = torch.max(outputs.data, 1)\n\n for j in range(labels.size(0)):\n focus = torch.argmax(alphas[j])\n if alphas[j][focus] >= 0.5 :\n argmax_more_than_half += 1\n else:\n argmax_less_than_half += 1\n\n if(focus == fore_idx[j] and predicted[j] == labels[j]):\n focus_true_pred_true += 1\n elif(focus != fore_idx[j] and predicted[j] == labels[j]):\n focus_false_pred_true += 1\n elif(focus == fore_idx[j] and predicted[j] != labels[j]):\n focus_true_pred_false += 1\n elif(focus != fore_idx[j] and predicted[j] != labels[j]):\n focus_false_pred_false += 1\n\n total += labels.size(0)\n correct += (predicted == labels).sum().item()\n\nprint('Accuracy of the network on the 1000 test images: %d %%' % (\n 100 * correct / total))\nprint(\"total correct\", correct)\nprint(\"total train set images\", total)\n\nprint(\"focus_true_pred_true %d =============> FTPT : %d %%\" % (focus_true_pred_true , (100 * focus_true_pred_true / total) ) )\nprint(\"focus_false_pred_true %d =============> FFPT : %d %%\" % (focus_false_pred_true, (100 * focus_false_pred_true / total) ) )\nprint(\"focus_true_pred_false %d =============> FTPF : %d %%\" %( focus_true_pred_false , ( 100 * focus_true_pred_false / total) ) )\nprint(\"focus_false_pred_false %d =============> FFPF : %d %%\" % (focus_false_pred_false, ( 100 * focus_false_pred_false / total) ) )\n\nprint(\"argmax_more_than_half ==================> \",argmax_more_than_half)\nprint(\"argmax_less_than_half ==================> \",argmax_less_than_half)",
"Accuracy of the network on the 1000 test images: 99 %\ntotal correct 999\ntotal train set images 1000\nfocus_true_pred_true 686 =============> FTPT : 68 %\nfocus_false_pred_true 313 =============> FFPT : 31 %\nfocus_true_pred_false 0 =============> FTPF : 0 %\nfocus_false_pred_false 1 =============> FFPF : 0 %\nargmax_more_than_half ==================> 686\nargmax_less_than_half ==================> 314\n"
],
[
"",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code"
] |
[
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a58fdcc3ff11007fce88f1f4408cb376d055838
| 776,810 |
ipynb
|
Jupyter Notebook
|
UKIRTdataload.ipynb
|
dibyajit30/ExoplanetHunting
|
7dfbd8e40d1ed57d06c167a9010231a877a21073
|
[
"MIT"
] | null | null | null |
UKIRTdataload.ipynb
|
dibyajit30/ExoplanetHunting
|
7dfbd8e40d1ed57d06c167a9010231a877a21073
|
[
"MIT"
] | null | null | null |
UKIRTdataload.ipynb
|
dibyajit30/ExoplanetHunting
|
7dfbd8e40d1ed57d06c167a9010231a877a21073
|
[
"MIT"
] | null | null | null | 624.947707 | 55,512 | 0.937192 |
[
[
[
"import pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport tarfile",
"_____no_output_____"
],
[
"# # extract tarfile \n# tar = tarfile.open('UKIRT/CASU_16_11_1.tar')\n# tar.extractall()\n# tar.close()",
"_____no_output_____"
],
[
"import os\nimport re\nfrom tqdm import tqdm\ndirectory = 'CASU_16_11_1'\n\nmag = []\nemag = []\nfor filename in tqdm(sorted(os.listdir(directory))):\n if filename.endswith(\".tbl\"): \n# print(os.path.join(directory, filename))\n tmp=pd.read_table(os.path.join(directory, filename), skiprows=21, \n delim_whitespace=True, \n header=None,\n index_col=0, \n names=['hjd', 'mag', 'emag']\n )\n\n # Use Regex to get Index ID\n idx = re.findall(\"(\\d{7})\", filename)[0]\n # print(idx)\n mag.append(tmp['mag'].rename(idx))\n emag.append(tmp['emag'].rename(idx))\n else:\n continue\n\npd.concat(mag,axis=1).T.to_pickle(directory+'_mag.pkl')\npd.concat(emag,axis=1).T.to_pickle(directory+'_emag.pkl')",
"100%|██████████| 89962/89962 [02:20<00:00, 638.96it/s]\n"
],
[
"pd.concat(mag,axis=1).T",
"_____no_output_____"
],
[
"k2=pd.read_table('ukirt_c_2016_s_61_3_0049492_h_lc.tbl', skiprows=21, \n delim_whitespace=True, \n header=None,\n names=['hjd', 'mag', 'emag']\n )",
"_____no_output_____"
],
[
"plt.plot(k2.mag)",
"_____no_output_____"
],
[
"plt.plot(k2.emag)",
"_____no_output_____"
]
],
[
[
"# ",
"_____no_output_____"
]
],
[
[
"kplr1=pd.read_table('kplr002304168-2009131105131_llc_lc.tbl', \n skiprows=(lambda x: x in np.concatenate((np.arange(0,203), np.array([204,205,206])))), \n delim_whitespace=True, \n header=[0])\n\nkplr1",
"_____no_output_____"
],
[
"for i in range(22):\n plt.plot(kplr1.iloc[:,i])\n plt.title(kplr1.iloc[:,i].name)\n plt.show()",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
4a590f855005779cc011d8ef9bf183494dec2321
| 621,740 |
ipynb
|
Jupyter Notebook
|
wrangle_act.ipynb
|
nakaokacynthia/WeRateDogs
|
9fec1eede9917b8a91a481c3a675e7e8257a604c
|
[
"MIT"
] | null | null | null |
wrangle_act.ipynb
|
nakaokacynthia/WeRateDogs
|
9fec1eede9917b8a91a481c3a675e7e8257a604c
|
[
"MIT"
] | null | null | null |
wrangle_act.ipynb
|
nakaokacynthia/WeRateDogs
|
9fec1eede9917b8a91a481c3a675e7e8257a604c
|
[
"MIT"
] | null | null | null | 89.087262 | 150,540 | 0.734252 |
[
[
[
"# Project: Wrangling and Analyze Data",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nfrom twython import Twython\nimport requests\nimport json\nimport time\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nfrom wordcloud import WordCloud, STOPWORDS\nfrom PIL import Image\nimport urllib\n",
"_____no_output_____"
]
],
[
[
"## Data Gathering\nIn the cells below was gathered **all** three pieces of data for this project and loaded them in the notebook. \n1. Directly download the WeRateDogs Twitter archive data (twitter_archive_enhanced.csv)",
"_____no_output_____"
]
],
[
[
"# Supplied file\narchive = pd.read_csv('twitter-archive-enhanced.csv')",
"_____no_output_____"
]
],
[
[
"2. Use the Requests library to download the tweet image prediction (image_predictions.tsv)",
"_____no_output_____"
]
],
[
[
"# Requesting tweet image predictions\n\nwith open('image_predictions.tsv' , 'wb') as file:\n image_predictions = requests.get('https://d17h27t6h515a5.cloudfront.net/topher/2017/August/599fd2ad_image-predictions/image-predictions.tsv', auth=('user', 'pass'))\n file.write(image_predictions.content)\n \n# Reading image predictions\npredictions = pd.read_csv('image_predictions.tsv', sep='\\t')",
"_____no_output_____"
]
],
[
[
"3. Use the Tweepy library to query additional data via the Twitter API (tweet_json.txt)",
"_____no_output_____"
]
],
[
[
"#Use Python's Tweepy library and store each tweet's entire set of JSON data in a file called tweet_json.txt file.\nimport tweepy\n\nconsumer_key = '--'\nconsumer_secret = '--'\naccess_token = '--'\naccess_secret = '--'\n\nauth = tweepy.OAuthHandler(consumer_key, consumer_secret)\nauth.set_access_token(access_token, access_secret)\n\napi = tweepy.API(auth, wait_on_rate_limit=True, wait_on_rate_limit_notify=True)\n",
"_____no_output_____"
],
[
"collected =[]\nnot_collected = [] \n\nwith open('tweet_json.txt', 'w') as file:\n for tweet_id in list(archive['tweet_id']):\n try:\n tweet_status = api.get_status(tweet_id,tweet_mode='extended')\n json.dump(tweet_status._json, file)\n file.write('\\n')\n collected.append(tweet_id)\n except Exception as e:\n not_collected.append(tweet_id)",
"Rate limit reached. Sleeping for: 622\nRate limit reached. Sleeping for: 625\n"
],
[
"#Reading JSON content as pandas dataframe\ntweets = pd.read_json('tweet_json.txt', lines = True,encoding='utf-8')",
"_____no_output_____"
]
],
[
[
"## Assessing Data\nIn this section was detect and documented **nine (9) quality issues and five (5) tidiness issues**. Were use **both** visual assessment programmatic assessement to assess the data.\n",
"_____no_output_____"
]
],
[
[
"# Load the data gathered data files\narchive.head()",
"_____no_output_____"
],
[
"archive.tail()",
"_____no_output_____"
],
[
"archive.shape",
"_____no_output_____"
],
[
"archive.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 2356 entries, 0 to 2355\nData columns (total 17 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 tweet_id 2356 non-null int64 \n 1 in_reply_to_status_id 78 non-null float64\n 2 in_reply_to_user_id 78 non-null float64\n 3 timestamp 2356 non-null object \n 4 source 2356 non-null object \n 5 text 2356 non-null object \n 6 retweeted_status_id 181 non-null float64\n 7 retweeted_status_user_id 181 non-null float64\n 8 retweeted_status_timestamp 181 non-null object \n 9 expanded_urls 2297 non-null object \n 10 rating_numerator 2356 non-null int64 \n 11 rating_denominator 2356 non-null int64 \n 12 name 2356 non-null object \n 13 doggo 2356 non-null object \n 14 floofer 2356 non-null object \n 15 pupper 2356 non-null object \n 16 puppo 2356 non-null object \ndtypes: float64(4), int64(3), object(10)\nmemory usage: 313.0+ KB\n"
],
[
"archive.describe()",
"_____no_output_____"
],
[
"# Load the data gathered data files\npredictions.head()",
"_____no_output_____"
],
[
"predictions.tail()",
"_____no_output_____"
],
[
"predictions.shape",
"_____no_output_____"
],
[
"predictions.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 2075 entries, 0 to 2074\nData columns (total 12 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 tweet_id 2075 non-null int64 \n 1 jpg_url 2075 non-null object \n 2 img_num 2075 non-null int64 \n 3 p1 2075 non-null object \n 4 p1_conf 2075 non-null float64\n 5 p1_dog 2075 non-null bool \n 6 p2 2075 non-null object \n 7 p2_conf 2075 non-null float64\n 8 p2_dog 2075 non-null bool \n 9 p3 2075 non-null object \n 10 p3_conf 2075 non-null float64\n 11 p3_dog 2075 non-null bool \ndtypes: bool(3), float64(3), int64(2), object(4)\nmemory usage: 152.1+ KB\n"
],
[
"predictions.describe()",
"_____no_output_____"
],
[
"# Load the data gathered data files\ntweets.head()",
"_____no_output_____"
],
[
"tweets.tail()",
"_____no_output_____"
],
[
"tweets.shape",
"_____no_output_____"
],
[
"tweets.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 2327 entries, 0 to 2326\nData columns (total 32 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 created_at 2327 non-null datetime64[ns, UTC]\n 1 id 2327 non-null int64 \n 2 id_str 2327 non-null int64 \n 3 full_text 2327 non-null object \n 4 truncated 2327 non-null bool \n 5 display_text_range 2327 non-null object \n 6 entities 2327 non-null object \n 7 extended_entities 2057 non-null object \n 8 source 2327 non-null object \n 9 in_reply_to_status_id 77 non-null float64 \n 10 in_reply_to_status_id_str 77 non-null float64 \n 11 in_reply_to_user_id 77 non-null float64 \n 12 in_reply_to_user_id_str 77 non-null float64 \n 13 in_reply_to_screen_name 77 non-null object \n 14 user 2327 non-null object \n 15 geo 0 non-null float64 \n 16 coordinates 0 non-null float64 \n 17 place 1 non-null object \n 18 contributors 0 non-null float64 \n 19 is_quote_status 2327 non-null bool \n 20 retweet_count 2327 non-null int64 \n 21 favorite_count 2327 non-null int64 \n 22 favorited 2327 non-null bool \n 23 retweeted 2327 non-null bool \n 24 possibly_sensitive 2195 non-null float64 \n 25 possibly_sensitive_appealable 2195 non-null float64 \n 26 lang 2327 non-null object \n 27 retweeted_status 160 non-null object \n 28 quoted_status_id 26 non-null float64 \n 29 quoted_status_id_str 26 non-null float64 \n 30 quoted_status_permalink 26 non-null object \n 31 quoted_status 24 non-null object \ndtypes: bool(4), datetime64[ns, UTC](1), float64(11), int64(4), object(12)\nmemory usage: 518.2+ KB\n"
],
[
"tweets.describe()",
"_____no_output_____"
]
],
[
[
"### Quality issues\n#### Archive\n\n1. [The timestamp field is in string format (object) and tweet_id is in int64](#1)\n\n2. [There are only 181 retweets (retweeted_status_id, retweeted_status_user_id, retweeted_status_timestamp)](#2)\n\n3. [There are only 78 replies (in_reply_to_status_id, in_reply_to_user_id)](#3)\n\n4. [There are missing values in the column expanded_urls](#4)\n\n5. [Column name floofer should be spelled 'floof'](#5)\n\n6. [Dogs with no name in the description have given names of \"a\", \"an\" and \"None\" instead of \"NaN\"](#6)\n\n7. [In the column rating_denominator there are votes greater than 10](#7)\n\n8. [Drop unnecessary columns](#8)\n\n#### Predictions\n\n9. [The types of dogs in columns p1, p2, and p3 have some lowercase and uppercase letters](#9)\n\n10. [The tweet_id field is in int64, should be in string format](#10)\n\n#### Tweets\n\n11. [Rename the column 'id' to 'tweet_id' to facilitate merging](#11)\n\n12. [Clean up text column to show only the text](#12)\n",
"_____no_output_____"
],
[
"### Tidiness issues\n\n#### Archive\n\n1. [Several columns representing the same category, which is divided into \"doggo\", \"flooter\", \"pupper\", \"puppo\" columns, but we need only one column to represent this classifications](#a)\n\n2. [Merge all tables to realize any analysis](#b)\n",
"_____no_output_____"
],
[
"## Cleaning Data\nIn this section were clean up **all** of the issues you documented while assessing. ",
"_____no_output_____"
]
],
[
[
"# Make copies of original pieces of data\narchive_clean = archive.copy()\npredictions_clean = predictions.copy()\ntweets_clean = tweets.copy()",
"_____no_output_____"
]
],
[
[
"### Quality issues",
"_____no_output_____"
],
[
"### Issue #1: Erroneous data types\n<a id=\"1\"></a>",
"_____no_output_____"
],
[
"#### Define: The timestamp field is in string format (object) and tweet_id is in int64",
"_____no_output_____"
],
[
"#### Code",
"_____no_output_____"
]
],
[
[
"#change the dtype of column timestamp from object to datetime\narchive_clean.timestamp = archive_clean.timestamp.astype('datetime64')\narchive_clean.tweet_id = archive_clean.tweet_id.astype(str)",
"_____no_output_____"
]
],
[
[
"#### Test",
"_____no_output_____"
]
],
[
[
"#Check for changes\narchive_clean.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 2356 entries, 0 to 2355\nData columns (total 17 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 tweet_id 2356 non-null object \n 1 in_reply_to_status_id 78 non-null float64 \n 2 in_reply_to_user_id 78 non-null float64 \n 3 timestamp 2356 non-null datetime64[ns]\n 4 source 2356 non-null object \n 5 text 2356 non-null object \n 6 retweeted_status_id 181 non-null float64 \n 7 retweeted_status_user_id 181 non-null float64 \n 8 retweeted_status_timestamp 181 non-null object \n 9 expanded_urls 2297 non-null object \n 10 rating_numerator 2356 non-null int64 \n 11 rating_denominator 2356 non-null int64 \n 12 name 2356 non-null object \n 13 doggo 2356 non-null object \n 14 floofer 2356 non-null object \n 15 pupper 2356 non-null object \n 16 puppo 2356 non-null object \ndtypes: datetime64[ns](1), float64(4), int64(2), object(10)\nmemory usage: 313.0+ KB\n"
]
],
[
[
"### Issue #2: Missing records \n<a id=\"2\"></a>",
"_____no_output_____"
],
[
"#### Define: There are only 181 retweets (retweeted_status_id, retweeted_status_user_id, retweeted_status_timestamp)",
"_____no_output_____"
],
[
"#### Code",
"_____no_output_____"
]
],
[
[
"#Use drop function to drop the non necessary columns\narchive_clean = archive_clean.drop(['retweeted_status_id', 'retweeted_status_user_id', 'retweeted_status_timestamp'], axis=1)",
"_____no_output_____"
]
],
[
[
"#### Test",
"_____no_output_____"
]
],
[
[
"#Check for changes\narchive_clean.head()",
"_____no_output_____"
]
],
[
[
"### Issue #3: Missing records \n<a id=\"3\"></a>",
"_____no_output_____"
],
[
"#### Define: There are only 78 replies (in_reply_to_status_id, in_reply_to_user_id)",
"_____no_output_____"
],
[
"#### Code",
"_____no_output_____"
]
],
[
[
"#Use drop function to drop the non necessary columns\narchive_clean = archive_clean.drop(['in_reply_to_status_id', 'in_reply_to_user_id'], axis=1)",
"_____no_output_____"
]
],
[
[
"#### Test",
"_____no_output_____"
]
],
[
[
"#Check for changes\narchive_clean.head()",
"_____no_output_____"
]
],
[
[
"### Issue #4: Missing records \n<a id=\"4\"></a>",
"_____no_output_____"
],
[
"#### Define: There are missing values in the column expanded_urls",
"_____no_output_____"
],
[
"#### Code",
"_____no_output_____"
]
],
[
[
"#Use drop function to drop the expanded_urls. We wont use this column for the analysis\narchive_clean = archive_clean.drop(['expanded_urls'], axis=1)",
"_____no_output_____"
]
],
[
[
"#### Test",
"_____no_output_____"
]
],
[
[
"#Check for changes\narchive_clean.head()",
"_____no_output_____"
]
],
[
[
"### Issue #5: Correct the column name\n<a id=\"5\"></a>",
"_____no_output_____"
],
[
"#### Define: Column name floofer should be spelled 'floof'",
"_____no_output_____"
],
[
"#### Code",
"_____no_output_____"
]
],
[
[
"# Rename the column 'floofer'\narchive_clean = archive_clean.rename(columns={'floofer':'floof'})",
"_____no_output_____"
]
],
[
[
"#### Test",
"_____no_output_____"
]
],
[
[
"#Check for changes\narchive_clean.head()",
"_____no_output_____"
],
[
"archive_clean.floof = archive_clean['floof'].map({'floofer':'floof'},\n na_action=None)",
"_____no_output_____"
],
[
"archive_clean",
"_____no_output_____"
]
],
[
[
"### Issue #6: Differents inputs for the same categories \n<a id=\"6\"></a>",
"_____no_output_____"
],
[
"#### Define: Dogs with no name in the description have given names of \"a\", \"an\" and \"None\" instead of \"NaN\"",
"_____no_output_____"
],
[
"#### Code",
"_____no_output_____"
]
],
[
[
"# Replace the value 'None' with NaN \narchive_clean = archive_clean.replace('None', np.nan)\narchive_clean = archive_clean.replace('a', np.nan)\narchive_clean = archive_clean.replace('an', np.nan)",
"_____no_output_____"
]
],
[
[
"#### Test",
"_____no_output_____"
]
],
[
[
"#Check for changes\narchive_clean.name.value_counts()",
"_____no_output_____"
]
],
[
[
"### Issue #7: There are no delimitations for the rating demonimator\n<a id=\"7\"></a>",
"_____no_output_____"
],
[
"#### Define: In the column rating_denominator there are votes greater than 10",
"_____no_output_____"
],
[
"#### Code",
"_____no_output_____"
]
],
[
[
"#Select only the values in the column rating_denominator that should only be \"10\"\narchive_clean.rating_denominator = archive_clean[archive_clean.rating_denominator == 10]",
"_____no_output_____"
]
],
[
[
"#### Test",
"_____no_output_____"
]
],
[
[
"#Check for changes\narchive_clean",
"_____no_output_____"
]
],
[
[
"### Issue #8: Unnecessary columns\n<a id=\"8\"></a>",
"_____no_output_____"
],
[
"#### Define: Drop unnecessary columns",
"_____no_output_____"
],
[
"#### Code",
"_____no_output_____"
]
],
[
[
"#Use drop function to drop source column\narchive_clean.drop(columns='source', inplace=True)",
"_____no_output_____"
]
],
[
[
"#### Test",
"_____no_output_____"
]
],
[
[
"#Check for change \narchive_clean.head()",
"_____no_output_____"
]
],
[
[
"### Issue #9: Differents letter cases\n<a id=\"9\"></a>",
"_____no_output_____"
],
[
"#### Define: The types of dogs in columns p1, p2, and p3 have some lowercase and uppercase letters",
"_____no_output_____"
],
[
"#### Code",
"_____no_output_____"
]
],
[
[
"#Convert all the dogs names to lowercase letters\npredictions_clean['p1'] = predictions_clean['p1'].str.lower()\npredictions_clean['p2'] = predictions_clean['p2'].str.lower()\npredictions_clean['p3'] = predictions_clean['p3'].str.lower()",
"_____no_output_____"
]
],
[
[
"#### Test",
"_____no_output_____"
]
],
[
[
"#Check for changes\npredictions_clean.p1.head()",
"_____no_output_____"
]
],
[
[
"### Issue #10: Differents data type format\n<a id=\"10\"></a>",
"_____no_output_____"
],
[
"#### Define: The tweet_id field is in int64, should be in string format",
"_____no_output_____"
],
[
"#### Code",
"_____no_output_____"
]
],
[
[
"#change the dtype of column tweed_id from int64 to string format\npredictions_clean.tweet_id = predictions_clean.tweet_id.astype(str)\ntweets_clean.id = tweets_clean.id.astype(str)",
"_____no_output_____"
]
],
[
[
"#### Test",
"_____no_output_____"
]
],
[
[
"#Check for changes\npredictions_clean.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 2075 entries, 0 to 2074\nData columns (total 12 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 tweet_id 2075 non-null object \n 1 jpg_url 2075 non-null object \n 2 img_num 2075 non-null int64 \n 3 p1 2075 non-null object \n 4 p1_conf 2075 non-null float64\n 5 p1_dog 2075 non-null bool \n 6 p2 2075 non-null object \n 7 p2_conf 2075 non-null float64\n 8 p2_dog 2075 non-null bool \n 9 p3 2075 non-null object \n 10 p3_conf 2075 non-null float64\n 11 p3_dog 2075 non-null bool \ndtypes: bool(3), float64(3), int64(1), object(5)\nmemory usage: 152.1+ KB\n"
],
[
"#Check for changes\ntweets_clean.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 2327 entries, 0 to 2326\nData columns (total 32 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 created_at 2327 non-null datetime64[ns, UTC]\n 1 id 2327 non-null object \n 2 id_str 2327 non-null int64 \n 3 full_text 2327 non-null object \n 4 truncated 2327 non-null bool \n 5 display_text_range 2327 non-null object \n 6 entities 2327 non-null object \n 7 extended_entities 2057 non-null object \n 8 source 2327 non-null object \n 9 in_reply_to_status_id 77 non-null float64 \n 10 in_reply_to_status_id_str 77 non-null float64 \n 11 in_reply_to_user_id 77 non-null float64 \n 12 in_reply_to_user_id_str 77 non-null float64 \n 13 in_reply_to_screen_name 77 non-null object \n 14 user 2327 non-null object \n 15 geo 0 non-null float64 \n 16 coordinates 0 non-null float64 \n 17 place 1 non-null object \n 18 contributors 0 non-null float64 \n 19 is_quote_status 2327 non-null bool \n 20 retweet_count 2327 non-null int64 \n 21 favorite_count 2327 non-null int64 \n 22 favorited 2327 non-null bool \n 23 retweeted 2327 non-null bool \n 24 possibly_sensitive 2195 non-null float64 \n 25 possibly_sensitive_appealable 2195 non-null float64 \n 26 lang 2327 non-null object \n 27 retweeted_status 160 non-null object \n 28 quoted_status_id 26 non-null float64 \n 29 quoted_status_id_str 26 non-null float64 \n 30 quoted_status_permalink 26 non-null object \n 31 quoted_status 24 non-null object \ndtypes: bool(4), datetime64[ns, UTC](1), float64(11), int64(3), object(13)\nmemory usage: 518.2+ KB\n"
]
],
[
[
"### Issue #11: Differents columns names for the same content\n<a id=\"11\"></a>",
"_____no_output_____"
],
[
"#### Define: Rename the column 'id' to 'tweet_id' to facilitate merging",
"_____no_output_____"
],
[
"#### Code",
"_____no_output_____"
]
],
[
[
"#Use rename() function to rename the column\ntweets_clean = tweets_clean.rename(columns={'id':'tweet_id'})",
"_____no_output_____"
]
],
[
[
"#### Test",
"_____no_output_____"
]
],
[
[
"#Check for changes\ntweets_clean.head()",
"_____no_output_____"
]
],
[
[
"### Issue #12: Column Text has multiples variables\n<a id=\"12\"></a>",
"_____no_output_____"
],
[
"#### Define: Clean up text column to show only the text",
"_____no_output_____"
],
[
"#### Code",
"_____no_output_____"
]
],
[
[
"#Remove url link\narchive_clean['text'] = archive_clean.text.str.replace(r\"http\\S+\", \"\")\narchive_clean['text'] = archive_clean.text.str.strip()",
"<ipython-input-326-1a7db23b574b>:2: FutureWarning: The default value of regex will change from True to False in a future version.\n archive_clean['text'] = archive_clean.text.str.replace(r\"http\\S+\", \"\")\n"
]
],
[
[
"#### Test",
"_____no_output_____"
]
],
[
[
"archive_clean['text'][0]",
"_____no_output_____"
]
],
[
[
"### Tidiness issues",
"_____no_output_____"
],
[
"### Issue #1: Unify the dogs classes\n<a id=\"a\"></a>",
"_____no_output_____"
],
[
"#### Define: Several columns representing the same category, which is divided into \"doggo\", \"flooter\", \"pupper\", \"puppo\" columns, but we need only one column to represent this classifications",
"_____no_output_____"
],
[
"#### Code",
"_____no_output_____"
]
],
[
[
"#Use loc function to add a new column to represent the dog stage \narchive_clean.loc[archive_clean['doggo'] == 'doggo', 'stage'] = 'doggo'\narchive_clean.loc[archive_clean['floof'] == 'floof', 'stage'] = 'floof'\narchive_clean.loc[archive_clean['pupper'] == 'pupper', 'stage'] = 'pupper'\narchive_clean.loc[archive_clean['puppo'] == 'puppo', 'stage'] = 'puppo'",
"_____no_output_____"
]
],
[
[
"#### Test",
"_____no_output_____"
]
],
[
[
"#Check for changes\narchive_clean.head()",
"_____no_output_____"
]
],
[
[
"#### Code",
"_____no_output_____"
]
],
[
[
"#Dropping the columns: doggo, floofer, pupper and poppo\narchive_clean = archive_clean.drop(['doggo', 'floof', 'pupper', 'puppo'], axis = 1)",
"_____no_output_____"
]
],
[
[
"#### Test",
"_____no_output_____"
]
],
[
[
"#Check the final change in the dogs stages\narchive_clean.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 2356 entries, 0 to 2355\nData columns (total 7 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 tweet_id 2356 non-null object \n 1 timestamp 2356 non-null datetime64[ns]\n 2 text 2356 non-null object \n 3 rating_numerator 2356 non-null int64 \n 4 rating_denominator 2333 non-null object \n 5 name 1549 non-null object \n 6 stage 380 non-null object \ndtypes: datetime64[ns](1), int64(1), object(5)\nmemory usage: 129.0+ KB\n"
]
],
[
[
"### Issue #2: Separated tables\n<a id=\"b\"></a>",
"_____no_output_____"
],
[
"#### Define: Merge all tables to realize any analysis",
"_____no_output_____"
],
[
"#### Code",
"_____no_output_____"
]
],
[
[
"#Merge the archive_clean and tweets_clean table\nmerge_df = archive_clean.join(tweets_clean.set_index('tweet_id'), on='tweet_id')\nmerge_df.head()",
"_____no_output_____"
]
],
[
[
"#### Test",
"_____no_output_____"
]
],
[
[
"#Check the new df\nmerge_df.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 2356 entries, 0 to 2355\nData columns (total 38 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 tweet_id 2356 non-null object \n 1 timestamp 2356 non-null datetime64[ns] \n 2 text 2356 non-null object \n 3 rating_numerator 2356 non-null int64 \n 4 rating_denominator 2333 non-null object \n 5 name 1549 non-null object \n 6 stage 380 non-null object \n 7 created_at 2327 non-null datetime64[ns, UTC]\n 8 id_str 2327 non-null float64 \n 9 full_text 2327 non-null object \n 10 truncated 2327 non-null object \n 11 display_text_range 2327 non-null object \n 12 entities 2327 non-null object \n 13 extended_entities 2057 non-null object \n 14 source 2327 non-null object \n 15 in_reply_to_status_id 77 non-null float64 \n 16 in_reply_to_status_id_str 77 non-null float64 \n 17 in_reply_to_user_id 77 non-null float64 \n 18 in_reply_to_user_id_str 77 non-null float64 \n 19 in_reply_to_screen_name 77 non-null object \n 20 user 2327 non-null object \n 21 geo 0 non-null float64 \n 22 coordinates 0 non-null float64 \n 23 place 1 non-null object \n 24 contributors 0 non-null float64 \n 25 is_quote_status 2327 non-null object \n 26 retweet_count 2327 non-null float64 \n 27 favorite_count 2327 non-null float64 \n 28 favorited 2327 non-null object \n 29 retweeted 2327 non-null object \n 30 possibly_sensitive 2195 non-null float64 \n 31 possibly_sensitive_appealable 2195 non-null float64 \n 32 lang 2327 non-null object \n 33 retweeted_status 160 non-null object \n 34 quoted_status_id 26 non-null float64 \n 35 quoted_status_id_str 26 non-null float64 \n 36 quoted_status_permalink 26 non-null object \n 37 quoted_status 24 non-null object \ndtypes: datetime64[ns, UTC](1), datetime64[ns](1), float64(14), int64(1), object(21)\nmemory usage: 699.6+ KB\n"
],
[
"#Join the merge_df to the predictions_clean table\ntwitter_master = merge_df.join(predictions_clean.set_index('tweet_id'), on='tweet_id')\ntwitter_master.head()",
"_____no_output_____"
],
[
"twitter_master.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 2356 entries, 0 to 2355\nData columns (total 49 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 tweet_id 2356 non-null object \n 1 timestamp 2356 non-null datetime64[ns] \n 2 text 2356 non-null object \n 3 rating_numerator 2356 non-null int64 \n 4 rating_denominator 2333 non-null object \n 5 name 1549 non-null object \n 6 stage 380 non-null object \n 7 created_at 2327 non-null datetime64[ns, UTC]\n 8 id_str 2327 non-null float64 \n 9 full_text 2327 non-null object \n 10 truncated 2327 non-null object \n 11 display_text_range 2327 non-null object \n 12 entities 2327 non-null object \n 13 extended_entities 2057 non-null object \n 14 source 2327 non-null object \n 15 in_reply_to_status_id 77 non-null float64 \n 16 in_reply_to_status_id_str 77 non-null float64 \n 17 in_reply_to_user_id 77 non-null float64 \n 18 in_reply_to_user_id_str 77 non-null float64 \n 19 in_reply_to_screen_name 77 non-null object \n 20 user 2327 non-null object \n 21 geo 0 non-null float64 \n 22 coordinates 0 non-null float64 \n 23 place 1 non-null object \n 24 contributors 0 non-null float64 \n 25 is_quote_status 2327 non-null object \n 26 retweet_count 2327 non-null float64 \n 27 favorite_count 2327 non-null float64 \n 28 favorited 2327 non-null object \n 29 retweeted 2327 non-null object \n 30 possibly_sensitive 2195 non-null float64 \n 31 possibly_sensitive_appealable 2195 non-null float64 \n 32 lang 2327 non-null object \n 33 retweeted_status 160 non-null object \n 34 quoted_status_id 26 non-null float64 \n 35 quoted_status_id_str 26 non-null float64 \n 36 quoted_status_permalink 26 non-null object \n 37 quoted_status 24 non-null object \n 38 jpg_url 2075 non-null object \n 39 img_num 2075 non-null float64 \n 40 p1 2075 non-null object \n 41 p1_conf 2075 non-null float64 \n 42 p1_dog 2075 non-null object \n 43 p2 2075 non-null object \n 44 p2_conf 2075 non-null float64 \n 45 p2_dog 2075 non-null object \n 46 p3 2075 non-null object \n 47 p3_conf 2075 non-null float64 \n 48 p3_dog 2075 non-null object \ndtypes: datetime64[ns, UTC](1), datetime64[ns](1), float64(18), int64(1), object(28)\nmemory usage: 902.0+ KB\n"
]
],
[
[
"#### Code",
"_____no_output_____"
]
],
[
[
"#Filter the columns to further analisys\ntwitter_master_clean = twitter_master.filter(['tweet_id','timestamp','text', 'rating_numerator', 'rating_denominator','name','stage','retweet_count', 'favorite_count', 'jpg_url','img_num', 'p1', 'p1_conf','p1_dog', 'p2', 'p2_conf', 'p2_dog', 'p3', 'p3_conf', 'p3_dog'])\ntwitter_master_clean.head()",
"_____no_output_____"
]
],
[
[
"## Storing Data\nSave gathered, assessed, and cleaned master dataset to a CSV file named \"twitter_archive_master.csv\".",
"_____no_output_____"
]
],
[
[
"#store data with to_csv function\ntwitter_master_clean.to_csv('twitter_archive_master.csv', index = False)",
"_____no_output_____"
]
],
[
[
"## Analyzing and Visualizing Data\nIn this section, analyze and visualize your wrangled data. You must produce at least **three (3) insights and one (1) visualization.**",
"_____no_output_____"
]
],
[
[
"#Make a copy\nrate_dogs = twitter_master_clean.copy()",
"_____no_output_____"
],
[
"rate_dogs.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 2356 entries, 0 to 2355\nData columns (total 20 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 tweet_id 2356 non-null object \n 1 timestamp 2356 non-null datetime64[ns]\n 2 text 2356 non-null object \n 3 rating_numerator 2356 non-null int64 \n 4 rating_denominator 2333 non-null object \n 5 name 1549 non-null object \n 6 stage 380 non-null object \n 7 retweet_count 2327 non-null float64 \n 8 favorite_count 2327 non-null float64 \n 9 jpg_url 2075 non-null object \n 10 img_num 2075 non-null float64 \n 11 p1 2075 non-null object \n 12 p1_conf 2075 non-null float64 \n 13 p1_dog 2075 non-null object \n 14 p2 2075 non-null object \n 15 p2_conf 2075 non-null float64 \n 16 p2_dog 2075 non-null object \n 17 p3 2075 non-null object \n 18 p3_conf 2075 non-null float64 \n 19 p3_dog 2075 non-null object \ndtypes: datetime64[ns](1), float64(6), int64(1), object(12)\nmemory usage: 368.2+ KB\n"
],
[
"#Select missing values from table merged to drop later\ndrop = rate_dogs[pd.isnull(rate_dogs['retweet_count'])].index\n\ndrop",
"_____no_output_____"
],
[
"#Drop missing data from merged table\nrate_dogs.drop(index=drop, inplace=True)",
"_____no_output_____"
],
[
"#Check the changes\nrate_dogs.info()",
"<class 'pandas.core.frame.DataFrame'>\nInt64Index: 2327 entries, 0 to 2355\nData columns (total 20 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 tweet_id 2327 non-null object \n 1 timestamp 2327 non-null datetime64[ns]\n 2 text 2327 non-null object \n 3 rating_numerator 2327 non-null int64 \n 4 rating_denominator 2305 non-null object \n 5 name 1533 non-null object \n 6 stage 374 non-null object \n 7 retweet_count 2327 non-null float64 \n 8 favorite_count 2327 non-null float64 \n 9 jpg_url 2057 non-null object \n 10 img_num 2057 non-null float64 \n 11 p1 2057 non-null object \n 12 p1_conf 2057 non-null float64 \n 13 p1_dog 2057 non-null object \n 14 p2 2057 non-null object \n 15 p2_conf 2057 non-null float64 \n 16 p2_dog 2057 non-null object \n 17 p3 2057 non-null object \n 18 p3_conf 2057 non-null float64 \n 19 p3_dog 2057 non-null object \ndtypes: datetime64[ns](1), float64(6), int64(1), object(12)\nmemory usage: 381.8+ KB\n"
],
[
"#Investigating the time of the tweets\nrate_dogs.timestamp.min(), rate_dogs.timestamp.max()",
"_____no_output_____"
],
[
"#Set the index to the datatime \nrate_dogs = rate_dogs.set_index('timestamp')",
"_____no_output_____"
],
[
"#Look for informations\nrate_dogs.describe()",
"_____no_output_____"
],
[
"rate_dogs.favorite_count.max()/rate_dogs.retweet_count.max()",
"_____no_output_____"
],
[
"#See if there is any correlations\nrate_dogs.corr()",
"_____no_output_____"
],
[
"#Plot the correlations\nsns.pairplot(rate_dogs, \n vars = ['rating_numerator', 'retweet_count', 'favorite_count', 'p1_conf'], \n diag_kind = 'kde', plot_kws = {'alpha': 0.9});",
"_____no_output_____"
],
[
"#Check the most favorited tweet\nrate_dogs.sort_values(by = 'favorite_count', ascending = False).head(3)",
"_____no_output_____"
],
[
"#Check the most retweeted tweet\nrate_dogs.sort_values(by = 'retweet_count', ascending = False).head(3)",
"_____no_output_____"
],
[
"#Check fot the most common dogs stages\nrate_dogs.stage.value_counts()",
"_____no_output_____"
],
[
"#Check for the most common dogs breeds \nrate_dogs.p1.value_counts().head(10)",
"_____no_output_____"
],
[
"#Plot the most common dogs breeds \nplt.barh(rate_dogs.p1.value_counts().head(10).index, rate_dogs.p1.value_counts().head(10), color = 'g', alpha=0.9)\nplt.xlabel('Number of tweets', fontsize = 10)\nplt.title('Top 10 dog breeds by tweet count', fontsize = 14)\nplt.gca().invert_yaxis()\nplt.show();",
"_____no_output_____"
],
[
"#Group favorite count with dogs breeds and see what are the most favorites\ntop10 = rate_dogs.favorite_count.groupby(rate_dogs['p1']).sum().sort_values(ascending = False)\ntop10.head(10)",
"_____no_output_____"
],
[
"#Plot the most favorites dogs breeds \nplt.barh(top10.head(10).index, top10.head(10), color = 'g', alpha=0.9)\nplt.xlabel('Favorite count', fontsize = 10)\nplt.title('Top 10 favorite dog breeds', fontsize = 14)\nplt.gca().invert_yaxis()\nplt.show();",
"_____no_output_____"
],
[
"#Plot the most favorites dogs stages\nfavorite_count_stages = rate_dogs.groupby('stage').favorite_count.mean().sort_values()\nfavorite_count_stages.plot(x=\"stage\",y='favorite_count',kind='barh',color='g', alpha=0.9)\nplt.xlabel('Favorite count', fontsize = 10)\nplt.ylabel('Dogs stages', fontsize = 10)\nplt.title('Average favorite counts by dogs stages', fontsize = 14)",
"_____no_output_____"
]
],
[
[
"### Insights:\n1. The quantity of people who favorite the posts is 2.039 time higher than people that retweet the posts. This shows a preference of just favorite the posts than retweet.\n\n2. There are a strong correlation between the favorites counts and retweets. To be more precised the correlation is 0.801345.\nTo evidenciate better, the most retweeted and favorited dog is a doggo labrador retriever who received 72474 retweets and 147742 favorites votes. His ID is 744234799360020481.\n\n3. The most common dogs breeds are golden retriever, labrador retriever and pembroke, respectively. They receive the most favorite counts too.\n",
"_____no_output_____"
],
[
"### Visualizations",
"_____no_output_____"
]
],
[
[
"#Plot a scatter plot to verify a possible trend in the amount os favorite count over time\nplt.scatter(rate_dogs.index, rate_dogs['favorite_count'])\nplt.title('Daily tweets by favorite count', fontsize = 14)\nplt.xlabel('Days', fontsize = 14)\nplt.ylabel('Favorite count', fontsize = 14)\nplt.show();",
"_____no_output_____"
],
[
"#Plot a Word Cloud with the texts written\n\ntweets = np.array(rate_dogs.text)\nlist1 = []\nfor tweet in tweets:\n list1.append(tweet.replace(\"\\n\",\"\"))\n \n \n \nmask = np.array(Image.open(requests.get('https://img.favpng.com/23/21/16/dog-vector-graphics-bengal-cat-illustration-clip-art-png-favpng-RWmY6zWcLaCxWurMaPEpZpARA.jpg', stream=True).raw))\ntext = list1\n\n\n\n\ndef gen_wc(text, mask):\n word_cloud = WordCloud(width = 700, height = 400, background_color='white', mask=mask).generate(str(text))\n plt.figure(figsize=(16,10),facecolor = 'white', edgecolor='red')\n plt.imshow(word_cloud)\n plt.axis('off')\n plt.tight_layout(pad=0)\n plt.show()\n \n \n \n \ngen_wc(text, mask)\n\n# The code used above was modeled from this blog on how to generate a word cloud in python.\n#https://blog.goodaudience.com/how-to-generate-a-word-cloud-of-any-shape-in-python-7bce27a55f6e",
"_____no_output_____"
]
],
[
[
"### Insights:\n\n1. We can have a visual look in the Daily tweets by favorite count chart and verify a positive trend in the amount of favorite tweets over time.\n\n2. In the cloud chart we can see that the word pooper, dog, pup and meet are the most frequently written.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
4a5913b2fa63c90c3042d13a9a55afa0eff80418
| 776 |
ipynb
|
Jupyter Notebook
|
Concepts/Fundamentals/roscore.ipynb
|
RobInLabUJI/ROSIN-Tutorials
|
8886865aab83c32cafa73b0ae0e54b27fa4dc210
|
[
"Apache-2.0"
] | 11 |
2019-08-16T08:34:22.000Z
|
2021-09-21T20:34:18.000Z
|
Concepts/Fundamentals/roscore.ipynb
|
RobInLabUJI/ROSIN-Tutorials
|
8886865aab83c32cafa73b0ae0e54b27fa4dc210
|
[
"Apache-2.0"
] | null | null | null |
Concepts/Fundamentals/roscore.ipynb
|
RobInLabUJI/ROSIN-Tutorials
|
8886865aab83c32cafa73b0ae0e54b27fa4dc210
|
[
"Apache-2.0"
] | 2 |
2020-09-16T12:17:56.000Z
|
2021-04-16T03:54:05.000Z
| 16.869565 | 84 | 0.515464 |
[
[
[
"empty"
]
]
] |
[
"empty"
] |
[
[
"empty"
]
] |
4a591d495c6b737277d4971ddf330ba2538f9a13
| 20,591 |
ipynb
|
Jupyter Notebook
|
sites_positionsWihinProteins.ipynb
|
dblyon/mod_sites_and_prob
|
ce6668828ae55708ea00876cb20de0ec4fd77e63
|
[
"MIT"
] | null | null | null |
sites_positionsWihinProteins.ipynb
|
dblyon/mod_sites_and_prob
|
ce6668828ae55708ea00876cb20de0ec4fd77e63
|
[
"MIT"
] | null | null | null |
sites_positionsWihinProteins.ipynb
|
dblyon/mod_sites_and_prob
|
ce6668828ae55708ea00876cb20de0ec4fd77e63
|
[
"MIT"
] | null | null | null | 29.798842 | 225 | 0.419552 |
[
[
[
"import sites_positionWithinProteins as pos\nreload(pos)",
"_____no_output_____"
],
[
"fn_fasta = r\"/Volumes/Speedy/FASTA/HUMAN20150706.fasta\"\nfn_evidence = r\"/Users/dblyon/CloudStation/CPR/BTW_sites/sites_positionsWithinProteins_input_v2.txt\"",
"_____no_output_____"
],
[
"fn_fasta\nfa = pos.Fasta()\nfa.set_file(fn_fasta)\nfa.parse_fasta()",
"_____no_output_____"
],
[
"COLUMN_MODSEQ = \"Modified sequence\"\nCOLUMN_PROTEINS = \"Proteins\"\nCOLUMN_MODPROB = \"Acetyl (K) Probabilities\"\nCOLUMN_ID = \"id\"\nMODTYPE = \"(ac)\"\n\n##### new columns\nCOLUMN_SITES = \"Sites\"\nCOLUMN_PROB = \"Probability\"\nremove_n_terminal_acetylation = True\n\ndf = pd.read_csv(fn_evidence, sep='\\t', low_memory=False)\ndf.dropna(axis=0, how=\"all\", inplace=True)\n\ndf[\"pepseq\"] = df[COLUMN_MODSEQ].apply(lambda aaseq: aaseq.replace(\"_\", \"\").replace(MODTYPE, \"\"))\ndf[\"pepseq\"] = df[\"pepseq\"].apply(pos.remove_modification_in_parentheses)\ndf[\"start_pos\"] = df.apply(pos.get_start_position_of_sequence_proteinGroups, args=(fa, ), axis=1)",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
],
[
"df = df.head()",
"_____no_output_____"
],
[
"df",
"_____no_output_____"
],
[
"df.loc[1, \"start_pos\"] = \"413;nan;45\"",
"_____no_output_____"
],
[
"pepseq_mod = \"_IVEM(ox)STSK(ac)TGK(ac)_\"\nsites_list = pos.parse_sites_within_pepseq(pepseq_mod, [])\nsites_list",
"_____no_output_____"
],
[
"COLUMN_MODSEQ_index = df.columns.tolist().index(COLUMN_MODSEQ) + 1 # due to index\nstart_pos_index = df.columns.tolist().index(\"start_pos\") + 1 # due to index\nSites_pos_within_pep_list = []\nfor row in df.itertuples():\n mod_seq = row[COLUMN_MODSEQ_index] \n start_pos_list = row[start_pos_index].split(\";\") # string for every protein in proteinGroups the start position of the peptide\n sites_list = pos.parse_sites_within_pepseq(mod_seq, [])\n \n sites_per_row = \"\"\n for protein_start_pos in start_pos_list:\n try:\n protein_start_pos = int(float(protein_start_pos))\n except ValueError: \n sites_per_row += \"(nan)\" + \";\" \n continue\n sites_per_protein = \"(\" + \"+\".join([str(site + protein_start_pos) for site in sites_list]) + \")\"\n sites_per_row += sites_per_protein + \";\"\n Sites_pos_within_pep_list.append(sites_per_row[:-1])\ndf[\"Sites_pos_within_pep\"] = Sites_pos_within_pep_list",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
],
[
"def add_length_of_peptide_2_start_pos(row):\n pepseq = row[\"Modified sequence\"]\n \ndf[\"Positions within Proteins\"] = df.apply(add_length_of_peptide_2_start_pos, axis=1)",
"_____no_output_____"
],
[
"aaseq = fa.an2aaseq_dict[\"I3L397\"]\naaseq",
"_____no_output_____"
],
[
"aaseq[39:39+11]",
"_____no_output_____"
],
[
"len(\"IVEMSTSKTGK\")",
"_____no_output_____"
],
[
"# import re\n# my_regex = re.compile(r\"(\\(\\w+\\))\")\n# df[\"pepseq_mod\"] = df[COLUMN_MODSEQ].apply(pos.remove_modifications_not_MODTYPE, args=(my_regex, remove_n_terminal_acetylation, ))\n# df = pos.add_COLUMN_SITES_and_PROB_2_df(df)",
"_____no_output_____"
],
[
"l = [1, 3, np.nan]",
"_____no_output_____"
],
[
"\";\".join([str(ele) for ele in l])",
"_____no_output_____"
],
[
"# add sites and probabilities\nmy_regex = re.compile(r\"(\\(\\w+\\))\")\ndf[\"pepseq_mod\"] = df[COLUMN_MODSEQ].apply(remove_modifications_not_MODTYPE, args=(my_regex, remove_n_terminal_acetylation, ))\ndf = add_COLUMN_SITES_and_PROB_2_df(df)\ndf = df[df[COLUMN_SITES].notnull()]\n\nif probability_threshold > 0:\n df = df[df[COLUMN_PROB].apply(is_any_above_threshold, args=(probability_threshold,))]\n\nif conventional_counting > 0:\n df[COLUMN_SITES] = df[COLUMN_SITES].apply(start_counting_from_num, args=(conventional_counting, )) #lambda num_string: \";\".join([str(int(float(num))) + conventional_counting for num in num_string.split(\";\")])\n\n# keep only relevant columns and write to file\ndf2write = df[[COLUMN_ID, COLUMN_MODSEQ, COLUMN_MODPROB, COLUMN_LEADRAZPROT, COLUMN_SITES, COLUMN_PROB]]\ndf2write[COLUMN_SITES] = df2write[COLUMN_SITES].apply(lambda ele: ele.replace(\".0\", \"\"))\ndf2write.to_csv(fn_output, sep='\\t', header=True, index=False)",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a592e335f0ac41fd10b50c7826316e9ac455123
| 116,476 |
ipynb
|
Jupyter Notebook
|
intro-neural-networks/student-admissions/StudentAdmissions.ipynb
|
cshreyastech/deep-learning-v2-pytorch
|
ec74f21a184ed0e3eaff5ae22fea5eb5731fb065
|
[
"MIT"
] | null | null | null |
intro-neural-networks/student-admissions/StudentAdmissions.ipynb
|
cshreyastech/deep-learning-v2-pytorch
|
ec74f21a184ed0e3eaff5ae22fea5eb5731fb065
|
[
"MIT"
] | 1 |
2020-01-28T22:31:47.000Z
|
2020-01-28T22:31:47.000Z
|
intro-neural-networks/student-admissions/StudentAdmissions.ipynb
|
cshreyastech/deep-learning-v2-pytorch
|
ec74f21a184ed0e3eaff5ae22fea5eb5731fb065
|
[
"MIT"
] | null | null | null | 151.859192 | 27,884 | 0.861044 |
[
[
[
"# Predicting Student Admissions with Neural Networks\nIn this notebook, we predict student admissions to graduate school at UCLA based on three pieces of data:\n- GRE Scores (Test)\n- GPA Scores (Grades)\n- Class rank (1-4)\n\nThe dataset originally came from here: http://www.ats.ucla.edu/\n\n## Loading the data\nTo load the data and format it nicely, we will use two very useful packages called Pandas and Numpy. You can read on the documentation here:\n- https://pandas.pydata.org/pandas-docs/stable/\n- https://docs.scipy.org/",
"_____no_output_____"
]
],
[
[
"# Importing pandas and numpy\nimport pandas as pd\nimport numpy as np\n\n# Reading the csv file into a pandas DataFrame\ndata = pd.read_csv('student_data.csv')\n\n# Printing out the first 10 rows of our data\ndata.head()",
"_____no_output_____"
]
],
[
[
"## Plotting the data\n\nFirst let's make a plot of our data to see how it looks. In order to have a 2D plot, let's ingore the rank.",
"_____no_output_____"
]
],
[
[
"# %matplotlib inline\nimport matplotlib.pyplot as plt\n\n# Function to help us plot\ndef plot_points(data):\n X = np.array(data[[\"gre\",\"gpa\"]])\n y = np.array(data[\"admit\"])\n admitted = X[np.argwhere(y==1)]\n rejected = X[np.argwhere(y==0)]\n plt.scatter([s[0][0] for s in rejected], [s[0][1] for s in rejected], s = 25, color = 'red', edgecolor = 'k')\n plt.scatter([s[0][0] for s in admitted], [s[0][1] for s in admitted], s = 25, color = 'cyan', edgecolor = 'k')\n plt.xlabel('Test (GRE)')\n plt.ylabel('Grades (GPA)')\n \n# Plotting the points\nplot_points(data)\nplt.show()",
"_____no_output_____"
]
],
[
[
"Roughly, it looks like the students with high scores in the grades and test passed, while the ones with low scores didn't, but the data is not as nicely separable as we hoped it would. Maybe it would help to take the rank into account? Let's make 4 plots, each one for each rank.",
"_____no_output_____"
]
],
[
[
"# Separating the ranks\ndata_rank1 = data[data[\"rank\"]==1]\ndata_rank2 = data[data[\"rank\"]==2]\ndata_rank3 = data[data[\"rank\"]==3]\ndata_rank4 = data[data[\"rank\"]==4]\n\n# Plotting the graphs\nplot_points(data_rank1)\nplt.title(\"Rank 1\")\nplt.show()\nplot_points(data_rank2)\nplt.title(\"Rank 2\")\nplt.show()\nplot_points(data_rank3)\nplt.title(\"Rank 3\")\nplt.show()\nplot_points(data_rank4)\nplt.title(\"Rank 4\")\nplt.show()",
"_____no_output_____"
]
],
[
[
"This looks more promising, as it seems that the lower the rank, the higher the acceptance rate. Let's use the rank as one of our inputs. In order to do this, we should one-hot encode it.\n\n## TODO: One-hot encoding the rank\nUse the `get_dummies` function in pandas in order to one-hot encode the data.\n\nHint: To drop a column, it's suggested that you use `one_hot_data`[.drop( )](https://pandas.pydata.org/pandas-docs/stable/generated/pandas.DataFrame.drop.html).",
"_____no_output_____"
]
],
[
[
"# TODO: Make dummy variables for rank and concat existing columns\none_hot_data = pd.get_dummies(data, columns=['rank'])\n\n# # TODO: Drop the previous rank column\n# one_hot_data = pass\n\n# # Print the first 10 rows of our data\n# one_hot_data[:10]",
"_____no_output_____"
]
],
[
[
"## TODO: Scaling the data\nThe next step is to scale the data. We notice that the range for grades is 1.0-4.0, whereas the range for test scores is roughly 200-800, which is much larger. This means our data is skewed, and that makes it hard for a neural network to handle. Let's fit our two features into a range of 0-1, by dividing the grades by 4.0, and the test score by 800.",
"_____no_output_____"
]
],
[
[
"# Making a copy of our data\nprocessed_data = one_hot_data[:]\n\n# TODO: Scale the columns\nprocessed_data['gre'] = processed_data['gre']/800\nprocessed_data['gpa'] = processed_data['gpa']/4.0\n# Printing the first 10 rows of our procesed data\nprocessed_data[:10]",
"_____no_output_____"
]
],
[
[
"## Splitting the data into Training and Testing",
"_____no_output_____"
],
[
"In order to test our algorithm, we'll split the data into a Training and a Testing set. The size of the testing set will be 10% of the total data.",
"_____no_output_____"
]
],
[
[
"sample = np.random.choice(processed_data.index, size=int(len(processed_data)*0.9), replace=False)\ntrain_data, test_data = processed_data.iloc[sample], processed_data.drop(sample)\n\nprint(\"Number of training samples is\", len(train_data))\nprint(\"Number of testing samples is\", len(test_data))\nprint(train_data[:10])\nprint(test_data[:10])",
"Number of training samples is 360\nNumber of testing samples is 40\n admit gre gpa rank_1 rank_2 rank_3 rank_4\n330 0 0.925 1.0000 0 0 1 0\n97 0 0.600 0.8925 0 1 0 0\n3 1 0.800 0.7975 0 0 0 1\n138 0 0.775 0.8500 0 1 0 0\n261 0 0.550 0.7875 0 1 0 0\n102 0 0.475 0.8325 0 0 0 1\n216 0 0.425 0.7250 1 0 0 0\n270 1 0.800 0.9875 0 1 0 0\n281 0 0.450 0.8175 0 0 1 0\n95 0 0.825 0.8325 0 1 0 0\n admit gre gpa rank_1 rank_2 rank_3 rank_4\n15 0 0.600 0.8600 0 0 1 0\n36 0 0.725 0.8125 1 0 0 0\n41 1 0.725 0.8300 0 1 0 0\n49 0 0.500 0.8375 0 0 1 0\n55 1 0.925 1.0000 0 0 1 0\n59 0 0.750 0.7050 0 0 0 1\n72 0 0.600 0.8475 0 0 0 1\n85 0 0.650 0.7450 0 1 0 0\n90 0 0.875 0.9575 0 1 0 0\n112 0 0.450 0.7500 0 0 1 0\n"
]
],
[
[
"## Splitting the data into features and targets (labels)\nNow, as a final step before the training, we'll split the data into features (X) and targets (y).",
"_____no_output_____"
]
],
[
[
"features = train_data.drop('admit', axis=1)\ntargets = train_data['admit']\nfeatures_test = test_data.drop('admit', axis=1)\ntargets_test = test_data['admit']\n\nprint(features[:10])\nprint(targets[:10])",
" gre gpa rank_1 rank_2 rank_3 rank_4\n330 0.925 1.0000 0 0 1 0\n97 0.600 0.8925 0 1 0 0\n3 0.800 0.7975 0 0 0 1\n138 0.775 0.8500 0 1 0 0\n261 0.550 0.7875 0 1 0 0\n102 0.475 0.8325 0 0 0 1\n216 0.425 0.7250 1 0 0 0\n270 0.800 0.9875 0 1 0 0\n281 0.450 0.8175 0 0 1 0\n95 0.825 0.8325 0 1 0 0\n330 0\n97 0\n3 1\n138 0\n261 0\n102 0\n216 0\n270 1\n281 0\n95 0\nName: admit, dtype: int64\n"
]
],
[
[
"## Training the 1-layer Neural Network\nThe following function trains the 1-layer neural network. \nFirst, we'll write some helper functions.",
"_____no_output_____"
]
],
[
[
"# Activation (sigmoid) function\ndef sigmoid(x):\n return 1 / (1 + np.exp(-x))\ndef sigmoid_prime(x):\n return sigmoid(x) * (1-sigmoid(x))\ndef error_formula(y, output):\n return - y*np.log(output) - (1 - y) * np.log(1-output)",
"_____no_output_____"
]
],
[
[
"# TODO: Backpropagate the error\nNow it's your turn to shine. Write the error term. Remember that this is given by the equation $$ (y-\\hat{y})x $$ for binary cross entropy loss function and \n$$ (y-\\hat{y})\\sigma'(x)x $$ for mean square error. ",
"_____no_output_____"
]
],
[
[
"# TODO: Write the error term formula\ndef error_term_formula(x, y, output):\n return (y - output) * sigmoid_prime(x) * x",
"_____no_output_____"
],
[
"# Neural Network hyperparameters\nepochs = 1000\nlearnrate = 0.0001\n\n# Training function\ndef train_nn(features, targets, epochs, learnrate):\n \n # Use to same seed to make debugging easier\n np.random.seed(42)\n\n n_records, n_features = features.shape\n last_loss = None\n\n # Initialize weights\n weights = np.random.normal(scale=1 / n_features**.5, size=n_features)\n\n for e in range(epochs):\n del_w = np.zeros(weights.shape)\n for x, y in zip(features.values, targets):\n # Loop through all records, x is the input, y is the target\n\n # Activation of the output unit\n # Notice we multiply the inputs and the weights here \n # rather than storing h as a separate variable \n output = sigmoid(np.dot(x, weights))\n\n # The error term\n error_term = error_term_formula(x, y, output)\n\n # The gradient descent step, the error times the gradient times the inputs\n del_w += error_term\n\n # Update the weights here. The learning rate times the \n # change in weights\n # don't have to divide by n_records since it is compensated by the learning rate\n weights += learnrate * del_w #/ n_records \n\n # Printing out the mean square error on the training set\n if e % (epochs / 10) == 0:\n out = sigmoid(np.dot(features, weights))\n loss = np.mean(error_formula(targets, out))\n print(\"Epoch:\", e)\n if last_loss and last_loss < loss:\n print(\"Train loss: \", loss, \" WARNING - Loss Increasing\")\n else:\n print(\"Train loss: \", loss)\n last_loss = loss\n print(\"=========\")\n print(\"Finished training!\")\n return weights\n \nweights = train_nn(features, targets, epochs, learnrate)",
"Epoch: 0\nTrain loss: 0.7511513235400568\n=========\nEpoch: 100\nTrain loss: 0.6917623454581121\n=========\nEpoch: 200\nTrain loss: 0.6578355244458959\n=========\nEpoch: 300\nTrain loss: 0.6383530414219198\n=========\nEpoch: 400\nTrain loss: 0.6269635542193962\n=========\nEpoch: 500\nTrain loss: 0.6201209674986199\n=========\nEpoch: 600\nTrain loss: 0.6158577887845291\n=========\nEpoch: 700\nTrain loss: 0.613076807257163\n=========\nEpoch: 800\nTrain loss: 0.6111604106266163\n=========\nEpoch: 900\nTrain loss: 0.6097573740684256\n=========\nFinished training!\n"
]
],
[
[
"## Calculating the Accuracy on the Test Data",
"_____no_output_____"
]
],
[
[
"# Calculate accuracy on test data\ntest_out = sigmoid(np.dot(features_test, weights))\npredictions = test_out > 0.5\naccuracy = np.mean(predictions == targets_test)\nprint(\"Prediction accuracy: {:.3f}\".format(accuracy))",
"Prediction accuracy: 0.700\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
4a593b5631306cbef3a7b733eb1412fd2ecefe44
| 762,941 |
ipynb
|
Jupyter Notebook
|
src/assets/data/.ipynb_checkpoints/job_retraining-checkpoint.ipynb
|
polygraph-cool/job_retraining
|
42e638f9797f0b2bd503bcd74f94e78c6c33bac5
|
[
"MIT"
] | null | null | null |
src/assets/data/.ipynb_checkpoints/job_retraining-checkpoint.ipynb
|
polygraph-cool/job_retraining
|
42e638f9797f0b2bd503bcd74f94e78c6c33bac5
|
[
"MIT"
] | 4 |
2018-05-28T23:41:50.000Z
|
2018-05-30T02:12:32.000Z
|
src/assets/data/.ipynb_checkpoints/job_retraining-checkpoint.ipynb
|
polygraph-cool/job_retraining
|
42e638f9797f0b2bd503bcd74f94e78c6c33bac5
|
[
"MIT"
] | null | null | null | 26.707075 | 1,451 | 0.458492 |
[
[
[
"import pandas as pd",
"_____no_output_____"
],
[
"auto_df = pd.read_csv(\"automatability.csv\") #to transpose\nrelative_emp_df = pd.read_csv(\"relativeEmployment.csv\") #to transpose\nsimilar_df = pd.read_csv(\"newsimilarity.csv\")\nwagechange_df = pd.read_csv(\"wageChange.csv\")\n\n# all_csvs = [auto_df, relative_emp_df, similar_df, wagechange_df]\n# for csv in all_csvs:\n# auto_df.columns.values[0]='Job_Compared'\n# relative_emp_df.columns.values[0]='Job_Compared'\n# similar_df.columns.values[0]='Job_Compared'\n# wagechange_df.columns.values[0]='Job_Compared'\n ",
"_____no_output_____"
],
[
"auto_df = auto_df.transpose()\nauto_df.columns = auto_df.iloc[0]\nauto_df.drop(auto_df.index[0], inplace=True)\n\nrelative_emp_df = relative_emp_df.transpose()\nrelative_emp_df.columns = relative_emp_df.iloc[0]\nrelative_emp_df.drop(relative_emp_df.index[0], inplace=True)\n\n\n\n# # auto_df=auto_df.set_index('Job Compared')\n# # relative_emp_df=relative_emp_df.set_index('Job Compared')\n# # similar_df=similar_df.set_index('Job Compared')\n# # wagechange_df = wagechange_df.set_index('Job Compared')",
"_____no_output_____"
],
[
"# Creating an id/job name hash table\n\n# temp_sim = pd.melt(similar_df, id_vars=['Job_Compared'], var_name='job_selected', value_name='similarity')\n# temp_sim.columns =['job_compared','job_selected','similarity']\n# temp_rec_therapist = temp_sim[temp_sim['job_selected']=='Recreational Therapists']\n# temp_pre_export = temp_rec_therapist.reset_index()\n# temp_pre_export = temp_pre_export[['index', 'Job_Compared']]\n# temp_pre_export.columns=['id','job_name']\n# temp_pre_export.to_csv(\"crosswalk.csv\", index=False)",
"_____no_output_____"
],
[
"# Similarity - setting up data struture\ntemp_sim = pd.melt(similar_df, id_vars=['Job_Compared'], var_name='job_selected', value_name='similarity')\ntemp_sim.columns =['job_compared','job_selected','similarity']\n\n\n# Intermediate step of setting up hash table for job name/id\n# First we create the job hash table\ntemp_rec_therapist = temp_sim[temp_sim['job_selected']=='Recreational Therapists']\ntemp_pre_export = temp_rec_therapist.reset_index()\ntemp_pre_export = temp_pre_export[['index', 'job_compared']]\ntemp_pre_export.columns=['id','job_name']\n# Next we load in the job descriptive stats and load join these to the hash table\njobs_descriptive = pd.read_csv(\"jobs.csv\")\njobs_descriptive_hashed = pd.merge(jobs_descriptive, temp_pre_export, left_on = \"Occupation\", right_on = \"job_name\")\njobs_descriptive_hashed.columns=['to_delete', 'auto','wage','number','id','job_name']\njobs_descriptive_hashed = jobs_descriptive_hashed[['auto','wage','number','id','job_name']]\n# Exporting crosswalk table\njobs_descriptive_hashed.to_csv(\"crosswalk.csv\", index=False)\n\n\n# Creating melted csv of selected job vs. compared job similarities, with IDs hashed\ntemp_sim = pd.merge(temp_sim,temp_pre_export, left_on='job_selected',right_on='job_name')\ntemp_sim.columns=['job_compared','to_delete_0','similarity', 'id_selected','to_delete_1']\ntemp_sim = temp_sim[['job_compared', 'similarity', 'id_selected']]\n\ntemp_sim = pd.merge(temp_sim,temp_pre_export, left_on='job_compared',right_on='job_name')\ntemp_sim.columns = ['to_delete_0','similarity','id_compared','id_selected','to_delete_1']\ntemp_sim = temp_sim[['similarity','id_compared','id_selected']]\ntemp_sim.to_csv(\"similarity.csv\", index=False)",
"_____no_output_____"
],
[
"# skills_raw.columns[451:500]\n# Additional Jobs\n\n# Old additional job to include in jobs stacked chart\n# 'Choreographers',\n# 'Dentists, General',\n# 'Registered Nurses',\n# 'Chiropractors',\n# 'Farmers, Ranchers, and Other Agricultural Managers',\n# 'Construction Managers',\n# 'Firefighters',\n# 'Geographers',\n# 'Heavy and Tractor-Trailer Truck Drivers',\n# 'Embalmers',\n# 'Pipelayers'\n\n\n\n# Old column names: ['choreographers','dentists','nurses','chiropractors','farmers',\n# 'construction_managers','firefighters','geographers','truckers','embalmers','pipelayers','skills']\n\njobs_to_keep=[]\njobs_to_keep_renamed=[]\n\njobs_to_keep = [\n'Choreographers',\n\"Dentists, General\", \n\"Registered Nurses\",\\\n\"Chiropractors\",\\\n'Farmers, Ranchers, and Other Agricultural Managers',\n'Construction Managers',\n\"Firefighters\",\\\n'Geographers',\n'Heavy and Tractor-Trailer Truck Drivers',\n'Embalmers',\n'Pipelayers',\n\"Podiatrists\", \n\"Fabric and Apparel Patternmakers\",\\\n\"Clergy\",\\\n\"Makeup Artists, Theatrical and Performance\",\\\n\"Marriage and Family Therapists\",\\\n\"Chief Executives\",\\\n\"Art Directors\",\\\n\"Interior Designers\",\\\n\"Craft Artists\",\\\n\"Meeting, Convention, and Event Planners\",\\\n\"Veterinarians\",\\\n\"Writers and Authors\",\\\n\"Political Scientists\",\\\n\"Ship Engineers\",\\\n\"Emergency Medical Technicians and Paramedics\",\\\n\"Mathematicians\",\\\n\"Floral Designers\",\\\n\"Travel Guides\",\\\n\"Broadcast News Analysts\",\\\n\"Musicians and Singers\",\\\n\"Fitness Trainers and Aerobics Instructors\",\\\n\"Graphic Designers\",\\\n\"Childcare Workers\",\\\n\"Police and Sheriff's Patrol Officers\",\\\n\"Hairdressers, Hairstylists, and Cosmetologists\",\\\n\"Reporters and Correspondents\",\\\n\"Air Traffic Controllers\",\\\n\"Dancers\",\\\n\"Optometrists\",\\\n\"Physician Assistants\",\\\n\"Electricians\",\\\n\"Ambulance Drivers and Attendants, Except Emergency Medical Technicians\",\\\n\"Athletes and Sports Competitors\",\\\n\"Skincare Specialists\",\\\n\"Cooks, Private Household\",\\\n\"Funeral Attendants\",\\\n\"Actors\",\\\n\"Judges, Magistrate Judges, and Magistrates\",\\\n\"Economists\",\\\n\"Historians\",\\\n\"Dental Assistants\",\\\n\"Shoe and Leather Workers and Repairers\",\\\n\"Massage Therapists\",\\\n\"Millwrights\",\\\n\"Librarians\",\\\n\"Maids and Housekeeping Cleaners\",\n\"Bartenders\",\n\"Dishwashers\",\n\"Cooks, Fast Food\",\n\"Barbers\",\n\"Real Estate Sales Agents\",\n\"Proofreaders and Copy Markers\"]\n\n\n\n\n# New column names for jobs to keep\njobs_to_keep_renamed=[\"Choreographers\",\n\"Dentists\",\n\"Nurses\",\n\"Chiropractors\",\n\"Farmers\",\n\"Construction_Managers\",\n\"Firefighters\",\n\"Geographers\",\n\"Truck_drivers\",\n\"Embalmers\",\n\"Piplayers\",\n\"Podiatrists\",\n\"Fabric_Patternmakers\",\n\"Clergy\",\n\"Makeup_Artists\",\n\"Family_Therapists\",\n\"CEOs\",\n\"Art_Directors\",\n\"Interrior_Designers\",\n\"Craft_Artists\",\n\"Event_Planners\",\n\"Veterinarians\",\n\"Writers\",\n\"Political_Scientists\",\n\"Ship_Engineers\",\n\"Paramedics\",\n\"Mathematicians\",\n\"Florists\",\n\"Travel_Guides\",\n\"News_Analysts\",\n\"Musicians\",\n\"Fitness_Trainers\",\n\"Graphic_Designers\",\n\"Childcare_Workers\",\n\"Police_Officers\",\n\"Hairdressers\",\n\"Journalists\",\n\"Air_Traffic_Controllers\",\n\"Dancers\",\n\"Optometrists\",\n\"Physician_Assistants\",\n\"Electricians\",\n\"Ambulance_Drivers\",\n\"Athletes\",\n\"Skincare_Specialists\",\n\"Private_Cooks\",\n\"Funeral_Attendants\",\n\"Actors\",\n\"Judges\",\n\"Economists\",\n\"historians\",\n\"Dental_Assistants\",\n\"Cobblers\",\n\"Massage_Therapists\",\n\"Millwrights\",\n\"Librarians\",\n\"Maids\",\n\"Bartenders\",\n\"Dishwashers\",\n\"Fast_Food_Cooks\",\n\"Barbers\",\n\"Real_Estate_Agents\",\n\"Proofreaders\",\n\"skills\"] #Note we're adding this because there will be a skill columns added to the relevant data frame\n",
"_____no_output_____"
],
[
"# Adding in skills crosswalk\nskills_crosswalk = pd.read_csv(\"skills_raw.csv\")\nskills_crosswalk.columns.values[0]='skill_type'\nskills_crosswalk.columns.values[1]='skill_name'\n\nskills_crosswalk = pd.melt(skills_crosswalk, id_vars=['skill_type','skill_name'], var_name='job', value_name='imp')\nskills_crosswalk = skills_crosswalk[skills_crosswalk['skill_type']!='ability']\n\nall_skills = list(set(skills_crosswalk['skill_name'].tolist()))\n\nskills_crosswalk = pd.DataFrame(all_skills)\nskills_crosswalk['skill_id'] = range(0,68)\nskills_crosswalk.columns=['skill','skill_id']\nskills_crosswalk.to_csv(\"crosswalk_skills.csv\", index=False)",
"_____no_output_____"
],
[
"# Adding in skills for each job\nskills_raw = pd.read_csv(\"skills_raw.csv\")\nskills_raw.columns.values[0]='skill_type'\nskills_raw.columns.values[1]='skill_name'\n\n# Creating a skill name and type data frame\nskills_name_and_type=skills_raw.iloc[:, [0,1]]\n\n# Melting the data frame into a d3-friendly format\nskills_edited = pd.melt(skills_raw, id_vars=['skill_type','skill_name'], var_name='job', value_name='imp')\nskills_edited = skills_edited[skills_edited['skill_type']!='ability']\nskills_edited = pd.merge(skills_edited, jobs_descriptive_hashed, left_on='job', right_on='job_name')\n\nskills_edited = skills_edited[['id','skill_name','imp']]\nskills_edited[\"rank\"] = skills_edited.groupby(\"id\")[\"imp\"].rank(\"dense\", ascending=False)\nskills_edited=skills_edited[skills_edited['rank']<6]\n\n# Sorting the skills df to get rid of the tied ranks\nskills_edited = skills_edited.sort_values(by=['id','rank','skill_name'])\nskills_edited['rank_final']=skills_edited.groupby(\"id\")[\"imp\"].rank(\"first\", ascending=False)\nskills_edited=skills_edited[skills_edited['rank_final']<6]\n\nskills_edited.columns=['id_selected','skill','imp','to_delete','rank']\nskills_edited=skills_edited[['id_selected','skill','imp','rank']]\n\n\n# Joining skills for each profession to skills crosswalk\nskills = pd.merge(skills_edited, skills_crosswalk, how='inner', left_on='skill', right_on='skill')\nskills=skills[['id_selected','imp','skill_id','rank']]\nskills = skills.sort_values(by=['id_selected','imp'])\nskills.to_csv(\"skills.csv\", index=False)",
"_____no_output_____"
],
[
"# Getting full skill list and importance for devs and truck trivers\n\n# (Getting a column after it's been made into an index/can't be accessed in another way)\nskills_list = pd.DataFrame(skills_raw.ix[:,1])\n\n# Creating dev + trucker dF\ndev_and_trucker_skills=skills_raw[['Software Developers, Applications','Heavy and Tractor-Trailer Truck Drivers']]\ndev_and_trucker_skills['skill']=skills_list\n# Renaming the columns in the data frame to reduce size\ndev_and_trucker_skills.columns=['devs','truckers','skills']\n\n# Filtering out all abilities\ndev_and_trucker_skills = pd.merge(dev_and_trucker_skills, skills_name_and_type, left_on ='skills', right_on='skill_name')\ndev_and_trucker_skills = dev_and_trucker_skills[dev_and_trucker_skills['skill_type']!='ability']\n\n# Devs and truckers skills to CSV\ndev_and_trucker_skills.to_csv(\"devs_and_truckers_skills.csv\", index=False)\n\n",
"/Users/iblinderman/anaconda/lib/python2.7/site-packages/ipykernel_launcher.py:8: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n \n"
],
[
"# ten_profession_skills=skills_raw[jobs_to_keep]\n# ten_profession_skills['skill']=skills_list\n# ten_profession_skills.columns=jobs_to_keep_renamed\n\n# # Getting difference scores\n# ten_profession_skills['choreographers_difference']=abs(ten_profession_skills['truckers']-ten_profession_skills['choreographers'])\n# ten_profession_skills['dentists_difference']=abs(ten_profession_skills['truckers']-ten_profession_skills['dentists'])\n# ten_profession_skills['nurses_difference']=abs(ten_profession_skills['truckers']-ten_profession_skills['nurses'])\n# ten_profession_skills['chiropractors_difference']=abs(ten_profession_skills['truckers']-ten_profession_skills['chiropractors'])\n# ten_profession_skills['farmers_difference']=abs(ten_profession_skills['truckers']-ten_profession_skills['farmers'])\n# ten_profession_skills['construction_managers_difference']=abs(ten_profession_skills['truckers']-ten_profession_skills['construction_managers'])\n# ten_profession_skills['firefighters_difference']=abs(ten_profession_skills['truckers']-ten_profession_skills['firefighters'])\n# ten_profession_skills['geographers_difference']=abs(ten_profession_skills['truckers']-ten_profession_skills['geographers'])\n# ten_profession_skills['embalmers_difference']=abs(ten_profession_skills['truckers']-ten_profession_skills['embalmers'])\n# ten_profession_skills['pipelayers_difference']=abs(ten_profession_skills['truckers']-ten_profession_skills['pipelayers'])\n# ten_profession_skills.to_csv(\"ten_profession_skills.csv\", index=False)",
"/Users/iblinderman/anaconda/lib/python2.7/site-packages/ipykernel_launcher.py:2: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n \n"
],
[
"# choreographer = ten_profession_skills[['choreographers','truckers','skills','choreographers_difference']]\n# dentists = ten_profession_skills[['dentists','truckers','skills','dentists_difference']]\n# nurses = ten_profession_skills[['nurses','truckers','skills','nurses_difference']]\n# chiropractors = ten_profession_skills[['chiropractors','truckers','skills','chiropractors_difference']]\n# farmers = ten_profession_skills[['farmers','truckers','skills','farmers_difference']]\n# construction_managers = ten_profession_skills[['construction_managers','truckers','skills','construction_managers_difference']]\n# firefighters = ten_profession_skills[['firefighters','truckers','skills','firefighters_difference']]\n# geographers = ten_profession_skills[['geographers','truckers','skills','geographers_difference']]\n# embalmers = ten_profession_skills[['embalmers','truckers','skills','embalmers_difference']]\n# pipelayers = ten_profession_skills[['pipelayers','truckers','skills','pipelayers_difference']]\n\n# choreographer.columns=['job_compared','job_selected','skills','difference']\n# dentists.columns=['job_compared','job_selected','skills','difference']\n# nurses.columns=['job_compared','job_selected','skills','difference']\n# chiropractors.columns=['job_compared','job_selected','skills','difference']\n# farmers.columns=['job_compared','job_selected','skills','difference']\n# construction_managers.columns=['job_compared','job_selected','skills','difference']\n# firefighters.columns=['job_compared','job_selected','skills','difference']\n# geographers.columns=['job_compared','job_selected','skills','difference']\n# embalmers.columns=['job_compared','job_selected','skills','difference']\n# pipelayers.columns=['job_compared','job_selected','skills','difference']\n\n# list_of_dfs=[choreographer,dentists,nurses,chiropractors,farmers,construction_managers,\n# firefighters,geographers,embalmers,pipelayers]\n\n# for df in list_of_dfs:\n# difference_list=[]\n# difference_list = df['difference'].tolist()\n# i=0\n# for skill in difference_list:\n# print \"i :\"+str(i)\n# print \"skill :\"+str(skill)\n# skill+=i\n# print \"skill :\"+str(i) +\" updated: \"+str(skill)\n# difference_list[i]=skill\n# i+=1\n# df['difference']=pd.Series(difference_list).values\n \n \n# choreographer['job_compared_name']='choreographer'\n# dentists['job_compared_name']='dentists'\n# nurses['job_compared_name']='nurses'\n# chiropractors['job_compared_name']='chiropractors'\n# farmers['job_compared_name']='farmers'\n# construction_managers['job_compared_name']='construction_managers'\n# firefighters['job_compared_name']='firefighters'\n# geographers['job_compared_name']='geographers'\n# embalmers['job_compared_name']='embalmers'\n# pipelayers['job_compared_name']='pipelayers'\n\n# choreographer.to_csv('choreographer.csv', index=False)\n# dentists.to_csv('dentists.csv', index=False)\n# nurses.to_csv('nurses.csv', index=False)\n# chiropractors.to_csv('chiropractors.csv', index=False)\n# farmers.to_csv('farmers.csv', index=False)\n# construction_managers.to_csv('construction_managers.csv', index=False)\n# firefighters.to_csv('firefighters.csv', index=False)\n# geographers.to_csv('geographers.csv', index=False)\n# embalmers.to_csv('embalmers.csv', index=False)\n# pipelayers.to_csv('pipelayers.csv', index=False)\n\n",
"i :0\nskill :3\nskill :0 updated: 3\ni :1\nskill :13\nskill :1 updated: 14\ni :2\nskill :14\nskill :2 updated: 16\ni :3\nskill :13\nskill :3 updated: 16\ni :4\nskill :10\nskill :4 updated: 14\ni :5\nskill :13\nskill :5 updated: 18\ni :6\nskill :24\nskill :6 updated: 30\ni :7\nskill :22\nskill :7 updated: 29\ni :8\nskill :41\nskill :8 updated: 49\ni :9\nskill :3\nskill :9 updated: 12\ni :10\nskill :12\nskill :10 updated: 22\ni :11\nskill :11\nskill :11 updated: 22\ni :12\nskill :47\nskill :12 updated: 59\ni :13\nskill :6\nskill :13 updated: 19\ni :14\nskill :19\nskill :14 updated: 33\ni :15\nskill :12\nskill :15 updated: 27\ni :16\nskill :10\nskill :16 updated: 26\ni :17\nskill :34\nskill :17 updated: 51\ni :18\nskill :9\nskill :18 updated: 27\ni :19\nskill :6\nskill :19 updated: 25\ni :20\nskill :35\nskill :20 updated: 55\ni :21\nskill :25\nskill :21 updated: 46\ni :22\nskill :3\nskill :22 updated: 25\ni :23\nskill :16\nskill :23 updated: 39\ni :24\nskill :0\nskill :24 updated: 24\ni :25\nskill :22\nskill :25 updated: 47\ni :26\nskill :22\nskill :26 updated: 48\ni :27\nskill :25\nskill :27 updated: 52\ni :28\nskill :22\nskill :28 updated: 50\ni :29\nskill :6\nskill :29 updated: 35\ni :30\nskill :0\nskill :30 updated: 30\ni :31\nskill :9\nskill :31 updated: 40\ni :32\nskill :7\nskill :32 updated: 39\ni :33\nskill :37\nskill :33 updated: 70\ni :34\nskill :8\nskill :34 updated: 42\ni :35\nskill :22\nskill :35 updated: 57\ni :36\nskill :31\nskill :36 updated: 67\ni :37\nskill :16\nskill :37 updated: 53\ni :38\nskill :28\nskill :38 updated: 66\ni :39\nskill :31\nskill :39 updated: 70\ni :40\nskill :16\nskill :40 updated: 56\ni :41\nskill :6\nskill :41 updated: 47\ni :42\nskill :38\nskill :42 updated: 80\ni :43\nskill :19\nskill :43 updated: 62\ni :44\nskill :16\nskill :44 updated: 60\ni :45\nskill :28\nskill :45 updated: 73\ni :46\nskill :12\nskill :46 updated: 58\ni :47\nskill :25\nskill :47 updated: 72\ni :48\nskill :16\nskill :48 updated: 64\ni :49\nskill :7\nskill :49 updated: 56\ni :50\nskill :9\nskill :50 updated: 59\ni :51\nskill :21\nskill :51 updated: 72\ni :52\nskill :22\nskill :52 updated: 74\ni :53\nskill :9\nskill :53 updated: 62\ni :54\nskill :9\nskill :54 updated: 63\ni :55\nskill :18\nskill :55 updated: 73\ni :56\nskill :96\nskill :56 updated: 152\ni :57\nskill :19\nskill :57 updated: 76\ni :58\nskill :50\nskill :58 updated: 108\ni :59\nskill :15\nskill :59 updated: 74\ni :60\nskill :46\nskill :60 updated: 106\ni :61\nskill :37\nskill :61 updated: 98\ni :62\nskill :3\nskill :62 updated: 65\ni :63\nskill :22\nskill :63 updated: 85\ni :64\nskill :19\nskill :64 updated: 83\ni :65\nskill :42\nskill :65 updated: 107\ni :66\nskill :14\nskill :66 updated: 80\ni :67\nskill :1\nskill :67 updated: 68\ni :68\nskill :3\nskill :68 updated: 71\ni :69\nskill :3\nskill :69 updated: 72\ni :70\nskill :16\nskill :70 updated: 86\ni :71\nskill :0\nskill :71 updated: 71\ni :72\nskill :28\nskill :72 updated: 100\ni :73\nskill :28\nskill :73 updated: 101\ni :74\nskill :19\nskill :74 updated: 93\ni :75\nskill :3\nskill :75 updated: 78\ni :76\nskill :11\nskill :76 updated: 87\ni :77\nskill :4\nskill :77 updated: 81\ni :78\nskill :5\nskill :78 updated: 83\ni :79\nskill :39\nskill :79 updated: 118\ni :80\nskill :16\nskill :80 updated: 96\ni :81\nskill :22\nskill :81 updated: 103\ni :82\nskill :22\nskill :82 updated: 104\ni :83\nskill :12\nskill :83 updated: 95\ni :84\nskill :32\nskill :84 updated: 116\ni :85\nskill :22\nskill :85 updated: 107\ni :86\nskill :5\nskill :86 updated: 91\ni :87\nskill :23\nskill :87 updated: 110\ni :88\nskill :25\nskill :88 updated: 113\ni :89\nskill :56\nskill :89 updated: 145\ni :90\nskill :3\nskill :90 updated: 93\ni :91\nskill :28\nskill :91 updated: 119\ni :92\nskill :38\nskill :92 updated: 130\ni :93\nskill :16\nskill :93 updated: 109\ni :94\nskill :31\nskill :94 updated: 125\ni :95\nskill :16\nskill :95 updated: 111\ni :96\nskill :31\nskill :96 updated: 127\ni :97\nskill :22\nskill :97 updated: 119\ni :98\nskill :47\nskill :98 updated: 145\ni :99\nskill :31\nskill :99 updated: 130\ni :100\nskill :22\nskill :100 updated: 122\ni :101\nskill :41\nskill :101 updated: 142\ni :102\nskill :44\nskill :102 updated: 146\ni :103\nskill :0\nskill :103 updated: 103\ni :104\nskill :47\nskill :104 updated: 151\ni :105\nskill :31\nskill :105 updated: 136\ni :106\nskill :72\nskill :106 updated: 178\ni :107\nskill :75\nskill :107 updated: 182\ni :108\nskill :47\nskill :108 updated: 155\ni :109\nskill :50\nskill :109 updated: 159\ni :110\nskill :40\nskill :110 updated: 150\ni :111\nskill :12\nskill :111 updated: 123\ni :112\nskill :47\nskill :112 updated: 159\ni :113\nskill :55\nskill :113 updated: 168\ni :114\nskill :55\nskill :114 updated: 169\ni :115\nskill :66\nskill :115 updated: 181\ni :116\nskill :44\nskill :116 updated: 160\ni :117\nskill :6\nskill :117 updated: 123\ni :118\nskill :62\nskill :118 updated: 180\ni :119\nskill :56\nskill :119 updated: 175\ni :0\nskill :13\nskill :0 updated: 13\ni :1\nskill :14\nskill :1 updated: 15\ni :2\nskill :21\nskill :2 updated: 23\ni :3\nskill :22\nskill :3 updated: 25\ni :4\nskill :69\nskill :4 updated: 73\ni :5\nskill :16\nskill :5 updated: 21\ni :6\nskill :23\nskill :6 updated: 29\ni :7\nskill :13\nskill :7 updated: 20\ni :8\nskill :25\nskill :8 updated: 33\ni :9\nskill :9\nskill :9 updated: 18\ni :10\nskill :22\nskill :10 updated: 32\ni :11\nskill :5\nskill :11 updated: 16\ni :12\nskill :22\nskill :12 updated: 34\ni :13\nskill :28\nskill :13 updated: 41\ni :14\nskill :31\nskill :14 updated: 45\ni :15\nskill :19\nskill :15 updated: 34\ni :16\nskill :25\nskill :16 updated: 41\ni :17\nskill :28\nskill :17 updated: 45\ni :18\nskill :19\nskill :18 updated: 37\ni :19\nskill :28\nskill :19 updated: 47\ni :20\nskill :35\nskill :20 updated: 55\ni :21\nskill :41\nskill :21 updated: 62\ni :22\nskill :19\nskill :22 updated: 41\ni :23\nskill :28\nskill :23 updated: 51\ni :24\nskill :19\nskill :24 updated: 43\ni :25\nskill :28\nskill :25 updated: 53\ni :26\nskill :19\nskill :26 updated: 45\ni :27\nskill :22\nskill :27 updated: 49\ni :28\nskill :15\nskill :28 updated: 43\ni :29\nskill :18\nskill :29 updated: 47\ni :30\nskill :19\nskill :30 updated: 49\ni :31\nskill :16\nskill :31 updated: 47\ni :32\nskill :12\nskill :32 updated: 44\ni :33\nskill :18\nskill :33 updated: 51\ni :34\nskill :11\nskill :34 updated: 45\ni :35\nskill :19\nskill :35 updated: 54\ni :36\nskill :25\nskill :36 updated: 61\ni :37\nskill :25\nskill :37 updated: 62\ni :38\nskill :25\nskill :38 updated: 63\ni :39\nskill :28\nskill :39 updated: 67\ni :40\nskill :28\nskill :40 updated: 68\ni :41\nskill :12\nskill :41 updated: 53\ni :42\nskill :3\nskill :42 updated: 45\ni :43\nskill :25\nskill :43 updated: 68\ni :44\nskill :25\nskill :44 updated: 69\ni :45\nskill :22\nskill :45 updated: 67\ni :46\nskill :28\nskill :46 updated: 74\ni :47\nskill :28\nskill :47 updated: 75\ni :48\nskill :10\nskill :48 updated: 58\ni :49\nskill :19\nskill :49 updated: 68\ni :50\nskill :3\nskill :50 updated: 53\ni :51\nskill :5\nskill :51 updated: 56\ni :52\nskill :25\nskill :52 updated: 77\ni :53\nskill :25\nskill :53 updated: 78\ni :54\nskill :25\nskill :54 updated: 79\ni :55\nskill :17\nskill :55 updated: 72\ni :56\n"
],
[
"ten_profession_skills=skills_raw[jobs_to_keep]\nten_profession_skills['skill']=skills_list\nten_profession_skills.columns=jobs_to_keep_renamed",
"_____no_output_____"
],
[
"ten_profession_skills['Choreographers_difference']=abs(ten_profession_skills['Truck_drivers']-ten_profession_skills['Choreographers'])\nten_profession_skills['Dentists_difference']=abs(ten_profession_skills['Truck_drivers']-ten_profession_skills['Dentists'])\nten_profession_skills['Nurses_difference']=abs(ten_profession_skills['Truck_drivers']-ten_profession_skills['Nurses'])\nten_profession_skills['Chiropractors_difference']=abs(ten_profession_skills['Truck_drivers']-ten_profession_skills['Chiropractors'])\nten_profession_skills['Farmers_difference']=abs(ten_profession_skills['Truck_drivers']-ten_profession_skills['Farmers'])\nten_profession_skills['Construction_Managers_difference']=abs(ten_profession_skills['Truck_drivers']-ten_profession_skills['Construction_Managers'])\nten_profession_skills['Firefighters_difference']=abs(ten_profession_skills['Truck_drivers']-ten_profession_skills['Firefighters'])\nten_profession_skills['Geographers_difference']=abs(ten_profession_skills['Truck_drivers']-ten_profession_skills['Geographers'])\nten_profession_skills['Truck_drivers_difference']=abs(ten_profession_skills['Truck_drivers']-ten_profession_skills['Truck_drivers'])\nten_profession_skills['Embalmers_difference']=abs(ten_profession_skills['Truck_drivers']-ten_profession_skills['Embalmers'])\nten_profession_skills['Piplayers_difference']=abs(ten_profession_skills['Truck_drivers']-ten_profession_skills['Piplayers'])\nten_profession_skills['Podiatrists_difference']=abs(ten_profession_skills['Truck_drivers']-ten_profession_skills['Podiatrists'])\nten_profession_skills['Fabric_Patternmakers_difference']=abs(ten_profession_skills['Truck_drivers']-ten_profession_skills['Fabric_Patternmakers'])\nten_profession_skills['Clergy_difference']=abs(ten_profession_skills['Truck_drivers']-ten_profession_skills['Clergy'])\nten_profession_skills['Makeup_Artists_difference']=abs(ten_profession_skills['Truck_drivers']-ten_profession_skills['Makeup_Artists'])\nten_profession_skills['Family_Therapists_difference']=abs(ten_profession_skills['Truck_drivers']-ten_profession_skills['Family_Therapists'])\nten_profession_skills['CEOs_difference']=abs(ten_profession_skills['Truck_drivers']-ten_profession_skills['CEOs'])\nten_profession_skills['Art_Directors_difference']=abs(ten_profession_skills['Truck_drivers']-ten_profession_skills['Art_Directors'])\nten_profession_skills['Interrior_Designers_difference']=abs(ten_profession_skills['Truck_drivers']-ten_profession_skills['Interrior_Designers'])\nten_profession_skills['Craft_Artists_difference']=abs(ten_profession_skills['Truck_drivers']-ten_profession_skills['Craft_Artists'])\nten_profession_skills['Event_Planners_difference']=abs(ten_profession_skills['Truck_drivers']-ten_profession_skills['Event_Planners'])\nten_profession_skills['Veterinarians_difference']=abs(ten_profession_skills['Truck_drivers']-ten_profession_skills['Veterinarians'])\nten_profession_skills['Writers_difference']=abs(ten_profession_skills['Truck_drivers']-ten_profession_skills['Writers'])\nten_profession_skills['Political_Scientists_difference']=abs(ten_profession_skills['Truck_drivers']-ten_profession_skills['Political_Scientists'])\nten_profession_skills['Ship_Engineers_difference']=abs(ten_profession_skills['Truck_drivers']-ten_profession_skills['Ship_Engineers'])\nten_profession_skills['Paramedics_difference']=abs(ten_profession_skills['Truck_drivers']-ten_profession_skills['Paramedics'])\nten_profession_skills['Mathematicians_difference']=abs(ten_profession_skills['Truck_drivers']-ten_profession_skills['Mathematicians'])\nten_profession_skills['Florists_difference']=abs(ten_profession_skills['Truck_drivers']-ten_profession_skills['Florists'])\nten_profession_skills['Travel_Guides_difference']=abs(ten_profession_skills['Truck_drivers']-ten_profession_skills['Travel_Guides'])\nten_profession_skills['News_Analysts_difference']=abs(ten_profession_skills['Truck_drivers']-ten_profession_skills['News_Analysts'])\nten_profession_skills['Musicians_difference']=abs(ten_profession_skills['Truck_drivers']-ten_profession_skills['Musicians'])\nten_profession_skills['Fitness_Trainers_difference']=abs(ten_profession_skills['Truck_drivers']-ten_profession_skills['Fitness_Trainers'])\nten_profession_skills['Graphic_Designers_difference']=abs(ten_profession_skills['Truck_drivers']-ten_profession_skills['Graphic_Designers'])\nten_profession_skills['Childcare_Workers_difference']=abs(ten_profession_skills['Truck_drivers']-ten_profession_skills['Childcare_Workers'])\nten_profession_skills['Police_Officers_difference']=abs(ten_profession_skills['Truck_drivers']-ten_profession_skills['Police_Officers'])\nten_profession_skills['Hairdressers_difference']=abs(ten_profession_skills['Truck_drivers']-ten_profession_skills['Hairdressers'])\nten_profession_skills['Journalists_difference']=abs(ten_profession_skills['Truck_drivers']-ten_profession_skills['Journalists'])\nten_profession_skills['Air_Traffic_Controllers_difference']=abs(ten_profession_skills['Truck_drivers']-ten_profession_skills['Air_Traffic_Controllers'])\nten_profession_skills['Dancers_difference']=abs(ten_profession_skills['Truck_drivers']-ten_profession_skills['Dancers'])\nten_profession_skills['Optometrists_difference']=abs(ten_profession_skills['Truck_drivers']-ten_profession_skills['Optometrists'])\nten_profession_skills['Physician_Assistants_difference']=abs(ten_profession_skills['Truck_drivers']-ten_profession_skills['Physician_Assistants'])\nten_profession_skills['Electricians_difference']=abs(ten_profession_skills['Truck_drivers']-ten_profession_skills['Electricians'])\nten_profession_skills['Ambulance_Drivers_difference']=abs(ten_profession_skills['Truck_drivers']-ten_profession_skills['Ambulance_Drivers'])\nten_profession_skills['Athletes_difference']=abs(ten_profession_skills['Truck_drivers']-ten_profession_skills['Athletes'])\nten_profession_skills['Skincare_Specialists_difference']=abs(ten_profession_skills['Truck_drivers']-ten_profession_skills['Skincare_Specialists'])\nten_profession_skills['Private_Cooks_difference']=abs(ten_profession_skills['Truck_drivers']-ten_profession_skills['Private_Cooks'])\nten_profession_skills['Funeral_Attendants_difference']=abs(ten_profession_skills['Truck_drivers']-ten_profession_skills['Funeral_Attendants'])\nten_profession_skills['Actors_difference']=abs(ten_profession_skills['Truck_drivers']-ten_profession_skills['Actors'])\nten_profession_skills['Judges_difference']=abs(ten_profession_skills['Truck_drivers']-ten_profession_skills['Judges'])\nten_profession_skills['Economists_difference']=abs(ten_profession_skills['Truck_drivers']-ten_profession_skills['Economists'])\nten_profession_skills['historians_difference']=abs(ten_profession_skills['Truck_drivers']-ten_profession_skills['historians'])\nten_profession_skills['Dental_Assistants_difference']=abs(ten_profession_skills['Truck_drivers']-ten_profession_skills['Dental_Assistants'])\nten_profession_skills['Cobblers_difference']=abs(ten_profession_skills['Truck_drivers']-ten_profession_skills['Cobblers'])\nten_profession_skills['Massage_Therapists_difference']=abs(ten_profession_skills['Truck_drivers']-ten_profession_skills['Massage_Therapists'])\nten_profession_skills['Millwrights_difference']=abs(ten_profession_skills['Truck_drivers']-ten_profession_skills['Millwrights'])\nten_profession_skills['Librarians_difference']=abs(ten_profession_skills['Truck_drivers']-ten_profession_skills['Librarians'])\nten_profession_skills['Maids_difference']=abs(ten_profession_skills['Truck_drivers']-ten_profession_skills['Maids'])\nten_profession_skills['Bartenders_difference']=abs(ten_profession_skills['Truck_drivers']-ten_profession_skills['Bartenders'])\nten_profession_skills['Dishwashers_difference']=abs(ten_profession_skills['Truck_drivers']-ten_profession_skills['Dishwashers'])\nten_profession_skills['Fast_Food_Cooks_difference']=abs(ten_profession_skills['Truck_drivers']-ten_profession_skills['Fast_Food_Cooks'])\nten_profession_skills['Barbers_difference']=abs(ten_profession_skills['Truck_drivers']-ten_profession_skills['Barbers'])\nten_profession_skills['Real_Estate_Agents_difference']=abs(ten_profession_skills['Truck_drivers']-ten_profession_skills['Real_Estate_Agents'])\nten_profession_skills['Proofreaders_difference']=abs(ten_profession_skills['Truck_drivers']-ten_profession_skills['Proofreaders'])",
"/Users/iblinderman/anaconda/lib/python2.7/site-packages/ipykernel_launcher.py:1: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n \"\"\"Entry point for launching an IPython kernel.\n/Users/iblinderman/anaconda/lib/python2.7/site-packages/ipykernel_launcher.py:2: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n \n/Users/iblinderman/anaconda/lib/python2.7/site-packages/ipykernel_launcher.py:3: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n This is separate from the ipykernel package so we can avoid doing imports until\n/Users/iblinderman/anaconda/lib/python2.7/site-packages/ipykernel_launcher.py:4: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n after removing the cwd from sys.path.\n/Users/iblinderman/anaconda/lib/python2.7/site-packages/ipykernel_launcher.py:5: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n \"\"\"\n/Users/iblinderman/anaconda/lib/python2.7/site-packages/ipykernel_launcher.py:6: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n \n/Users/iblinderman/anaconda/lib/python2.7/site-packages/ipykernel_launcher.py:7: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n import sys\n/Users/iblinderman/anaconda/lib/python2.7/site-packages/ipykernel_launcher.py:8: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n \n/Users/iblinderman/anaconda/lib/python2.7/site-packages/ipykernel_launcher.py:9: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n if __name__ == '__main__':\n/Users/iblinderman/anaconda/lib/python2.7/site-packages/ipykernel_launcher.py:10: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n # Remove the CWD from sys.path while we load stuff.\n/Users/iblinderman/anaconda/lib/python2.7/site-packages/ipykernel_launcher.py:11: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n # This is added back by InteractiveShellApp.init_path()\n/Users/iblinderman/anaconda/lib/python2.7/site-packages/ipykernel_launcher.py:12: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n if sys.path[0] == '':\n/Users/iblinderman/anaconda/lib/python2.7/site-packages/ipykernel_launcher.py:13: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n del sys.path[0]\n/Users/iblinderman/anaconda/lib/python2.7/site-packages/ipykernel_launcher.py:14: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n \n/Users/iblinderman/anaconda/lib/python2.7/site-packages/ipykernel_launcher.py:15: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n from ipykernel import kernelapp as app\n/Users/iblinderman/anaconda/lib/python2.7/site-packages/ipykernel_launcher.py:16: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n app.launch_new_instance()\n/Users/iblinderman/anaconda/lib/python2.7/site-packages/ipykernel_launcher.py:17: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n/Users/iblinderman/anaconda/lib/python2.7/site-packages/ipykernel_launcher.py:18: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n/Users/iblinderman/anaconda/lib/python2.7/site-packages/ipykernel_launcher.py:19: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n/Users/iblinderman/anaconda/lib/python2.7/site-packages/ipykernel_launcher.py:20: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n/Users/iblinderman/anaconda/lib/python2.7/site-packages/ipykernel_launcher.py:21: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n/Users/iblinderman/anaconda/lib/python2.7/site-packages/ipykernel_launcher.py:22: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n/Users/iblinderman/anaconda/lib/python2.7/site-packages/ipykernel_launcher.py:23: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n/Users/iblinderman/anaconda/lib/python2.7/site-packages/ipykernel_launcher.py:24: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n/Users/iblinderman/anaconda/lib/python2.7/site-packages/ipykernel_launcher.py:25: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n/Users/iblinderman/anaconda/lib/python2.7/site-packages/ipykernel_launcher.py:26: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n/Users/iblinderman/anaconda/lib/python2.7/site-packages/ipykernel_launcher.py:27: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n"
],
[
"Choreographers = ten_profession_skills[['Choreographers','Truck_drivers','skills','Choreographers_difference']]\nDentists = ten_profession_skills[['Dentists','Truck_drivers','skills','Dentists_difference']]\nNurses = ten_profession_skills[['Nurses','Truck_drivers','skills','Nurses_difference']]\nChiropractors = ten_profession_skills[['Chiropractors','Truck_drivers','skills','Chiropractors_difference']]\nFarmers = ten_profession_skills[['Farmers','Truck_drivers','skills','Farmers_difference']]\nConstruction_Managers = ten_profession_skills[['Construction_Managers','Truck_drivers','skills','Construction_Managers_difference']]\nFirefighters = ten_profession_skills[['Firefighters','Truck_drivers','skills','Firefighters_difference']]\nGeographers = ten_profession_skills[['Geographers','Truck_drivers','skills','Geographers_difference']]\nTruck_drivers = ten_profession_skills[['Truck_drivers','Truck_drivers','skills','Truck_drivers_difference']]\nEmbalmers = ten_profession_skills[['Embalmers','Truck_drivers','skills','Embalmers_difference']]\nPiplayers = ten_profession_skills[['Piplayers','Truck_drivers','skills','Piplayers_difference']]\nPodiatrists = ten_profession_skills[['Podiatrists','Truck_drivers','skills','Podiatrists_difference']]\nFabric_Patternmakers = ten_profession_skills[['Fabric_Patternmakers','Truck_drivers','skills','Fabric_Patternmakers_difference']]\nClergy = ten_profession_skills[['Clergy','Truck_drivers','skills','Clergy_difference']]\nMakeup_Artists = ten_profession_skills[['Makeup_Artists','Truck_drivers','skills','Makeup_Artists_difference']]\nFamily_Therapists = ten_profession_skills[['Family_Therapists','Truck_drivers','skills','Family_Therapists_difference']]\nCEOs = ten_profession_skills[['CEOs','Truck_drivers','skills','CEOs_difference']]\nArt_Directors = ten_profession_skills[['Art_Directors','Truck_drivers','skills','Art_Directors_difference']]\nInterrior_Designers = ten_profession_skills[['Interrior_Designers','Truck_drivers','skills','Interrior_Designers_difference']]\nCraft_Artists = ten_profession_skills[['Craft_Artists','Truck_drivers','skills','Craft_Artists_difference']]\nEvent_Planners = ten_profession_skills[['Event_Planners','Truck_drivers','skills','Event_Planners_difference']]\nVeterinarians = ten_profession_skills[['Veterinarians','Truck_drivers','skills','Veterinarians_difference']]\nWriters = ten_profession_skills[['Writers','Truck_drivers','skills','Writers_difference']]\nPolitical_Scientists = ten_profession_skills[['Political_Scientists','Truck_drivers','skills','Political_Scientists_difference']]\nShip_Engineers = ten_profession_skills[['Ship_Engineers','Truck_drivers','skills','Ship_Engineers_difference']]\nParamedics = ten_profession_skills[['Paramedics','Truck_drivers','skills','Paramedics_difference']]\nMathematicians = ten_profession_skills[['Mathematicians','Truck_drivers','skills','Mathematicians_difference']]\nFlorists = ten_profession_skills[['Florists','Truck_drivers','skills','Florists_difference']]\nTravel_Guides = ten_profession_skills[['Travel_Guides','Truck_drivers','skills','Travel_Guides_difference']]\nNews_Analysts = ten_profession_skills[['News_Analysts','Truck_drivers','skills','News_Analysts_difference']]\nMusicians = ten_profession_skills[['Musicians','Truck_drivers','skills','Musicians_difference']]\nFitness_Trainers = ten_profession_skills[['Fitness_Trainers','Truck_drivers','skills','Fitness_Trainers_difference']]\nGraphic_Designers = ten_profession_skills[['Graphic_Designers','Truck_drivers','skills','Graphic_Designers_difference']]\nChildcare_Workers = ten_profession_skills[['Childcare_Workers','Truck_drivers','skills','Childcare_Workers_difference']]\nPolice_Officers = ten_profession_skills[['Police_Officers','Truck_drivers','skills','Police_Officers_difference']]\nHairdressers = ten_profession_skills[['Hairdressers','Truck_drivers','skills','Hairdressers_difference']]\nJournalists = ten_profession_skills[['Journalists','Truck_drivers','skills','Journalists_difference']]\nAir_Traffic_Controllers = ten_profession_skills[['Air_Traffic_Controllers','Truck_drivers','skills','Air_Traffic_Controllers_difference']]\nDancers = ten_profession_skills[['Dancers','Truck_drivers','skills','Dancers_difference']]\nOptometrists = ten_profession_skills[['Optometrists','Truck_drivers','skills','Optometrists_difference']]\nPhysician_Assistants = ten_profession_skills[['Physician_Assistants','Truck_drivers','skills','Physician_Assistants_difference']]\nElectricians = ten_profession_skills[['Electricians','Truck_drivers','skills','Electricians_difference']]\nAmbulance_Drivers = ten_profession_skills[['Ambulance_Drivers','Truck_drivers','skills','Ambulance_Drivers_difference']]\nAthletes = ten_profession_skills[['Athletes','Truck_drivers','skills','Athletes_difference']]\nSkincare_Specialists = ten_profession_skills[['Skincare_Specialists','Truck_drivers','skills','Skincare_Specialists_difference']]\nPrivate_Cooks = ten_profession_skills[['Private_Cooks','Truck_drivers','skills','Private_Cooks_difference']]\nFuneral_Attendants = ten_profession_skills[['Funeral_Attendants','Truck_drivers','skills','Funeral_Attendants_difference']]\nActors = ten_profession_skills[['Actors','Truck_drivers','skills','Actors_difference']]\nJudges = ten_profession_skills[['Judges','Truck_drivers','skills','Judges_difference']]\nEconomists = ten_profession_skills[['Economists','Truck_drivers','skills','Economists_difference']]\nhistorians = ten_profession_skills[['historians','Truck_drivers','skills','historians_difference']]\nDental_Assistants = ten_profession_skills[['Dental_Assistants','Truck_drivers','skills','Dental_Assistants_difference']]\nCobblers = ten_profession_skills[['Cobblers','Truck_drivers','skills','Cobblers_difference']]\nMassage_Therapists = ten_profession_skills[['Massage_Therapists','Truck_drivers','skills','Massage_Therapists_difference']]\nMillwrights = ten_profession_skills[['Millwrights','Truck_drivers','skills','Millwrights_difference']]\nLibrarians = ten_profession_skills[['Librarians','Truck_drivers','skills','Librarians_difference']]\nMaids = ten_profession_skills[['Maids','Truck_drivers','skills','Maids_difference']]\nBartenders = ten_profession_skills[['Bartenders','Truck_drivers','skills','Bartenders_difference']]\nDishwashers = ten_profession_skills[['Dishwashers','Truck_drivers','skills','Dishwashers_difference']]\nFast_Food_Cooks = ten_profession_skills[['Fast_Food_Cooks','Truck_drivers','skills','Fast_Food_Cooks_difference']]\nBarbers = ten_profession_skills[['Barbers','Truck_drivers','skills','Barbers_difference']]\nReal_Estate_Agents = ten_profession_skills[['Real_Estate_Agents','Truck_drivers','skills','Real_Estate_Agents_difference']]\nProofreaders = ten_profession_skills[['Proofreaders','Truck_drivers','skills','Proofreaders_difference']]",
"_____no_output_____"
],
[
"Choreographers.columns=['job_compared','job_selected','skills','difference']\nDentists.columns=['job_compared','job_selected','skills','difference']\nNurses.columns=['job_compared','job_selected','skills','difference']\nChiropractors.columns=['job_compared','job_selected','skills','difference']\nFarmers.columns=['job_compared','job_selected','skills','difference']\nConstruction_Managers.columns=['job_compared','job_selected','skills','difference']\nFirefighters.columns=['job_compared','job_selected','skills','difference']\nGeographers.columns=['job_compared','job_selected','skills','difference']\nTruck_drivers.columns=['job_compared','job_selected','skills','difference']\nEmbalmers.columns=['job_compared','job_selected','skills','difference']\nPiplayers.columns=['job_compared','job_selected','skills','difference']\nPodiatrists.columns=['job_compared','job_selected','skills','difference']\nFabric_Patternmakers.columns=['job_compared','job_selected','skills','difference']\nClergy.columns=['job_compared','job_selected','skills','difference']\nMakeup_Artists.columns=['job_compared','job_selected','skills','difference']\nFamily_Therapists.columns=['job_compared','job_selected','skills','difference']\nCEOs.columns=['job_compared','job_selected','skills','difference']\nArt_Directors.columns=['job_compared','job_selected','skills','difference']\nInterrior_Designers.columns=['job_compared','job_selected','skills','difference']\nCraft_Artists.columns=['job_compared','job_selected','skills','difference']\nEvent_Planners.columns=['job_compared','job_selected','skills','difference']\nVeterinarians.columns=['job_compared','job_selected','skills','difference']\nWriters.columns=['job_compared','job_selected','skills','difference']\nPolitical_Scientists.columns=['job_compared','job_selected','skills','difference']\nShip_Engineers.columns=['job_compared','job_selected','skills','difference']\nParamedics.columns=['job_compared','job_selected','skills','difference']\nMathematicians.columns=['job_compared','job_selected','skills','difference']\nFlorists.columns=['job_compared','job_selected','skills','difference']\nTravel_Guides.columns=['job_compared','job_selected','skills','difference']\nNews_Analysts.columns=['job_compared','job_selected','skills','difference']\nMusicians.columns=['job_compared','job_selected','skills','difference']\nFitness_Trainers.columns=['job_compared','job_selected','skills','difference']\nGraphic_Designers.columns=['job_compared','job_selected','skills','difference']\nChildcare_Workers.columns=['job_compared','job_selected','skills','difference']\nPolice_Officers.columns=['job_compared','job_selected','skills','difference']\nHairdressers.columns=['job_compared','job_selected','skills','difference']\nJournalists.columns=['job_compared','job_selected','skills','difference']\nAir_Traffic_Controllers.columns=['job_compared','job_selected','skills','difference']\nDancers.columns=['job_compared','job_selected','skills','difference']\nOptometrists.columns=['job_compared','job_selected','skills','difference']\nPhysician_Assistants.columns=['job_compared','job_selected','skills','difference']\nElectricians.columns=['job_compared','job_selected','skills','difference']\nAmbulance_Drivers.columns=['job_compared','job_selected','skills','difference']\nAthletes.columns=['job_compared','job_selected','skills','difference']\nSkincare_Specialists.columns=['job_compared','job_selected','skills','difference']\nPrivate_Cooks.columns=['job_compared','job_selected','skills','difference']\nFuneral_Attendants.columns=['job_compared','job_selected','skills','difference']\nActors.columns=['job_compared','job_selected','skills','difference']\nJudges.columns=['job_compared','job_selected','skills','difference']\nEconomists.columns=['job_compared','job_selected','skills','difference']\nhistorians.columns=['job_compared','job_selected','skills','difference']\nDental_Assistants.columns=['job_compared','job_selected','skills','difference']\nCobblers.columns=['job_compared','job_selected','skills','difference']\nMassage_Therapists.columns=['job_compared','job_selected','skills','difference']\nMillwrights.columns=['job_compared','job_selected','skills','difference']\nLibrarians.columns=['job_compared','job_selected','skills','difference']\nMaids.columns=['job_compared','job_selected','skills','difference']\nBartenders.columns=['job_compared','job_selected','skills','difference']\nDishwashers.columns=['job_compared','job_selected','skills','difference']\nFast_Food_Cooks.columns=['job_compared','job_selected','skills','difference']\nBarbers.columns=['job_compared','job_selected','skills','difference']\nReal_Estate_Agents.columns=['job_compared','job_selected','skills','difference']\nProofreaders.columns=['job_compared','job_selected','skills','difference']",
"_____no_output_____"
],
[
"list_of_dfs=[\nChoreographers,\nDentists,\nNurses,\nChiropractors,\nFarmers,\nConstruction_Managers,\nFirefighters,\nGeographers,\nTruck_drivers,\nEmbalmers,\nPiplayers,\nPodiatrists,\nFabric_Patternmakers,\nClergy,\nMakeup_Artists,\nFamily_Therapists,\nCEOs,\nArt_Directors,\nInterrior_Designers,\nCraft_Artists,\nEvent_Planners,\nVeterinarians,\nWriters,\nPolitical_Scientists,\nShip_Engineers,\nParamedics,\nMathematicians,\nFlorists,\nTravel_Guides,\nNews_Analysts,\nMusicians,\nFitness_Trainers,\nGraphic_Designers,\nChildcare_Workers,\nPolice_Officers,\nHairdressers,\nJournalists,\nAir_Traffic_Controllers,\nDancers,\nOptometrists,\nPhysician_Assistants,\nElectricians,\nAmbulance_Drivers,\nAthletes,\nSkincare_Specialists,\nPrivate_Cooks,\nFuneral_Attendants,\nActors,\nJudges,\nEconomists,\nhistorians,\nDental_Assistants,\nCobblers,\nMassage_Therapists,\nMillwrights,\nLibrarians,\nMaids,\nBartenders,\nDishwashers,\nFast_Food_Cooks,\nBarbers,\nReal_Estate_Agents,\nProofreaders]",
"_____no_output_____"
],
[
"for df in list_of_dfs:\n difference_list=[]\n difference_list = df['difference'].tolist()\n i=0\n for skill in difference_list:\n print \"i :\"+str(i)\n print \"skill :\"+str(skill)\n skill+=i\n print \"skill :\"+str(i) +\" updated: \"+str(skill)\n difference_list[i]=skill\n i+=1\n df['difference']=pd.Series(difference_list).values\n \n ",
"i :0\nskill :3\nskill :0 updated: 3\ni :1\nskill :13\nskill :1 updated: 14\ni :2\nskill :14\nskill :2 updated: 16\ni :3\nskill :13\nskill :3 updated: 16\ni :4\nskill :10\nskill :4 updated: 14\ni :5\nskill :13\nskill :5 updated: 18\ni :6\nskill :24\nskill :6 updated: 30\ni :7\nskill :22\nskill :7 updated: 29\ni :8\nskill :41\nskill :8 updated: 49\ni :9\nskill :3\nskill :9 updated: 12\ni :10\nskill :12\nskill :10 updated: 22\ni :11\nskill :11\nskill :11 updated: 22\ni :12\nskill :47\nskill :12 updated: 59\ni :13\nskill :6\nskill :13 updated: 19\ni :14\nskill :19\nskill :14 updated: 33\ni :15\nskill :12\nskill :15 updated: 27\ni :16\nskill :10\nskill :16 updated: 26\ni :17\nskill :34\nskill :17 updated: 51\ni :18\nskill :9\nskill :18 updated: 27\ni :19\nskill :6\nskill :19 updated: 25\ni :20\nskill :35\nskill :20 updated: 55\ni :21\nskill :25\nskill :21 updated: 46\ni :22\nskill :3\nskill :22 updated: 25\ni :23\nskill :16\nskill :23 updated: 39\ni :24\nskill :0\nskill :24 updated: 24\ni :25\nskill :22\nskill :25 updated: 47\ni :26\nskill :22\nskill :26 updated: 48\ni :27\nskill :25\nskill :27 updated: 52\ni :28\nskill :22\nskill :28 updated: 50\ni :29\nskill :6\nskill :29 updated: 35\ni :30\nskill :0\nskill :30 updated: 30\ni :31\nskill :9\nskill :31 updated: 40\ni :32\nskill :7\nskill :32 updated: 39\ni :33\nskill :37\nskill :33 updated: 70\ni :34\nskill :8\nskill :34 updated: 42\ni :35\nskill :22\nskill :35 updated: 57\ni :36\nskill :31\nskill :36 updated: 67\ni :37\nskill :16\nskill :37 updated: 53\ni :38\nskill :28\nskill :38 updated: 66\ni :39\nskill :31\nskill :39 updated: 70\ni :40\nskill :16\nskill :40 updated: 56\ni :41\nskill :6\nskill :41 updated: 47\ni :42\nskill :38\nskill :42 updated: 80\ni :43\nskill :19\nskill :43 updated: 62\ni :44\nskill :16\nskill :44 updated: 60\ni :45\nskill :28\nskill :45 updated: 73\ni :46\nskill :12\nskill :46 updated: 58\ni :47\nskill :25\nskill :47 updated: 72\ni :48\nskill :16\nskill :48 updated: 64\ni :49\nskill :7\nskill :49 updated: 56\ni :50\nskill :9\nskill :50 updated: 59\ni :51\nskill :21\nskill :51 updated: 72\ni :52\nskill :22\nskill :52 updated: 74\ni :53\nskill :9\nskill :53 updated: 62\ni :54\nskill :9\nskill :54 updated: 63\ni :55\nskill :18\nskill :55 updated: 73\ni :56\nskill :96\nskill :56 updated: 152\ni :57\nskill :19\nskill :57 updated: 76\ni :58\nskill :50\nskill :58 updated: 108\ni :59\nskill :15\nskill :59 updated: 74\ni :60\nskill :46\nskill :60 updated: 106\ni :61\nskill :37\nskill :61 updated: 98\ni :62\nskill :3\nskill :62 updated: 65\ni :63\nskill :22\nskill :63 updated: 85\ni :64\nskill :19\nskill :64 updated: 83\ni :65\nskill :42\nskill :65 updated: 107\ni :66\nskill :14\nskill :66 updated: 80\ni :67\nskill :1\nskill :67 updated: 68\ni :68\nskill :3\nskill :68 updated: 71\ni :69\nskill :3\nskill :69 updated: 72\ni :70\nskill :16\nskill :70 updated: 86\ni :71\nskill :0\nskill :71 updated: 71\ni :72\nskill :28\nskill :72 updated: 100\ni :73\nskill :28\nskill :73 updated: 101\ni :74\nskill :19\nskill :74 updated: 93\ni :75\nskill :3\nskill :75 updated: 78\ni :76\nskill :11\nskill :76 updated: 87\ni :77\nskill :4\nskill :77 updated: 81\ni :78\nskill :5\nskill :78 updated: 83\ni :79\nskill :39\nskill :79 updated: 118\ni :80\nskill :16\nskill :80 updated: 96\ni :81\nskill :22\nskill :81 updated: 103\ni :82\nskill :22\nskill :82 updated: 104\ni :83\nskill :12\nskill :83 updated: 95\ni :84\nskill :32\nskill :84 updated: 116\ni :85\nskill :22\nskill :85 updated: 107\ni :86\nskill :5\nskill :86 updated: 91\ni :87\nskill :23\nskill :87 updated: 110\ni :88\nskill :25\nskill :88 updated: 113\ni :89\nskill :56\nskill :89 updated: 145\ni :90\nskill :3\nskill :90 updated: 93\ni :91\nskill :28\nskill :91 updated: 119\ni :92\nskill :38\nskill :92 updated: 130\ni :93\nskill :16\nskill :93 updated: 109\ni :94\nskill :31\nskill :94 updated: 125\ni :95\nskill :16\nskill :95 updated: 111\ni :96\nskill :31\nskill :96 updated: 127\ni :97\nskill :22\nskill :97 updated: 119\ni :98\nskill :47\nskill :98 updated: 145\ni :99\nskill :31\nskill :99 updated: 130\ni :100\nskill :22\nskill :100 updated: 122\ni :101\nskill :41\nskill :101 updated: 142\ni :102\nskill :44\nskill :102 updated: 146\ni :103\nskill :0\nskill :103 updated: 103\ni :104\nskill :47\nskill :104 updated: 151\ni :105\nskill :31\nskill :105 updated: 136\ni :106\nskill :72\nskill :106 updated: 178\ni :107\nskill :75\nskill :107 updated: 182\ni :108\nskill :47\nskill :108 updated: 155\ni :109\nskill :50\nskill :109 updated: 159\ni :110\nskill :40\nskill :110 updated: 150\ni :111\nskill :12\nskill :111 updated: 123\ni :112\nskill :47\nskill :112 updated: 159\ni :113\nskill :55\nskill :113 updated: 168\ni :114\nskill :55\nskill :114 updated: 169\ni :115\nskill :66\nskill :115 updated: 181\ni :116\nskill :44\nskill :116 updated: 160\ni :117\nskill :6\nskill :117 updated: 123\ni :118\nskill :62\nskill :118 updated: 180\ni :119\nskill :56\nskill :119 updated: 175\ni :0\nskill :13\nskill :0 updated: 13\ni :1\nskill :14\nskill :1 updated: 15\ni :2\nskill :21\nskill :2 updated: 23\ni :3\nskill :22\nskill :3 updated: 25\ni :4\nskill :69\nskill :4 updated: 73\ni :5\nskill :16\nskill :5 updated: 21\ni :6\nskill :23\nskill :6 updated: 29\ni :7\nskill :13\nskill :7 updated: 20\ni :8\nskill :25\nskill :8 updated: 33\ni :9\nskill :9\nskill :9 updated: 18\ni :10\nskill :22\nskill :10 updated: 32\ni :11\nskill :5\nskill :11 updated: 16\ni :12\nskill :22\nskill :12 updated: 34\ni :13\nskill :28\nskill :13 updated: 41\ni :14\nskill :31\nskill :14 updated: 45\ni :15\nskill :19\nskill :15 updated: 34\ni :16\nskill :25\nskill :16 updated: 41\ni :17\nskill :28\nskill :17 updated: 45\ni :18\nskill :19\nskill :18 updated: 37\ni :19\nskill :28\nskill :19 updated: 47\ni :20\nskill :35\nskill :20 updated: 55\ni :21\nskill :41\nskill :21 updated: 62\ni :22\nskill :19\nskill :22 updated: 41\ni :23\nskill :28\nskill :23 updated: 51\ni :24\nskill :19\nskill :24 updated: 43\ni :25\nskill :28\nskill :25 updated: 53\ni :26\nskill :19\nskill :26 updated: 45\ni :27\nskill :22\nskill :27 updated: 49\ni :28\nskill :15\nskill :28 updated: 43\ni :29\nskill :18\nskill :29 updated: 47\ni :30\nskill :19\nskill :30 updated: 49\ni :31\nskill :16\nskill :31 updated: 47\ni :32\nskill :12\nskill :32 updated: 44\ni :33\nskill :18\nskill :33 updated: 51\ni :34\nskill :11\nskill :34 updated: 45\ni :35\nskill :19\nskill :35 updated: 54\ni :36\nskill :25\nskill :36 updated: 61\ni :37\nskill :25\nskill :37 updated: 62\ni :38\nskill :25\nskill :38 updated: 63\ni :39\nskill :28\nskill :39 updated: 67\ni :40\nskill :28\nskill :40 updated: 68\ni :41\nskill :12\nskill :41 updated: 53\ni :42\nskill :3\nskill :42 updated: 45\ni :43\nskill :25\nskill :43 updated: 68\ni :44\nskill :25\nskill :44 updated: 69\ni :45\nskill :22\nskill :45 updated: 67\ni :46\nskill :28\nskill :46 updated: 74\ni :47\nskill :28\nskill :47 updated: 75\ni :48\nskill :10\nskill :48 updated: 58\ni :49\nskill :19\nskill :49 updated: 68\ni :50\nskill :3\nskill :50 updated: 53\ni :51\nskill :5\nskill :51 updated: 56\ni :52\nskill :25\nskill :52 updated: 77\ni :53\nskill :25\nskill :53 updated: 78\ni :54\nskill :25\nskill :54 updated: 79\ni :55\nskill :17\nskill :55 updated: 72\ni :56\nskill :10\nskill :56 updated: 66\ni :57\nskill :6\nskill :57 updated: 63\ni :58\nskill :25\nskill :58 updated: 83\ni :59\nskill :31\nskill :59 updated: 90\ni :60\nskill :2\nskill :60 updated: 62\ni :61\nskill :3\nskill :61 updated: 64\ni :62\nskill :22\nskill :62 updated: 84\ni :63\nskill :26\nskill :63 updated: 89\ni :64\nskill :19\nskill :64 updated: 83\ni :65\nskill :10\nskill :65 updated: 75\ni :66\nskill :10\nskill :66 updated: 76"
],
[
"Choreographers['job_compared_name']='Choreographers'\nDentists['job_compared_name']='Dentists'\nNurses['job_compared_name']='Nurses'\nChiropractors['job_compared_name']='Chiropractors'\nFarmers['job_compared_name']='Farmers'\nConstruction_Managers['job_compared_name']='Construction_Managers'\nFirefighters['job_compared_name']='Firefighters'\nGeographers['job_compared_name']='Geographers'\nTruck_drivers['job_compared_name']='Truck_drivers'\nEmbalmers['job_compared_name']='Embalmers'\nPiplayers['job_compared_name']='Piplayers'\nPodiatrists['job_compared_name']='Podiatrists'\nFabric_Patternmakers['job_compared_name']='Fabric_Patternmakers'\nClergy['job_compared_name']='Clergy'\nMakeup_Artists['job_compared_name']='Makeup_Artists'\nFamily_Therapists['job_compared_name']='Family_Therapists'\nCEOs['job_compared_name']='CEOs'\nArt_Directors['job_compared_name']='Art_Directors'\nInterrior_Designers['job_compared_name']='Interrior_Designers'\nCraft_Artists['job_compared_name']='Craft_Artists'\nEvent_Planners['job_compared_name']='Event_Planners'\nVeterinarians['job_compared_name']='Veterinarians'\nWriters['job_compared_name']='Writers'\nPolitical_Scientists['job_compared_name']='Political_Scientists'\nShip_Engineers['job_compared_name']='Ship_Engineers'\nParamedics['job_compared_name']='Paramedics'\nMathematicians['job_compared_name']='Mathematicians'\nFlorists['job_compared_name']='Florists'\nTravel_Guides['job_compared_name']='Travel_Guides'\nNews_Analysts['job_compared_name']='News_Analysts'\nMusicians['job_compared_name']='Musicians'\nFitness_Trainers['job_compared_name']='Fitness_Trainers'\nGraphic_Designers['job_compared_name']='Graphic_Designers'\nChildcare_Workers['job_compared_name']='Childcare_Workers'\nPolice_Officers['job_compared_name']='Police_Officers'\nHairdressers['job_compared_name']='Hairdressers'\nJournalists['job_compared_name']='Journalists'\nAir_Traffic_Controllers['job_compared_name']='Air_Traffic_Controllers'\nDancers['job_compared_name']='Dancers'\nOptometrists['job_compared_name']='Optometrists'\nPhysician_Assistants['job_compared_name']='Physician_Assistants'\nElectricians['job_compared_name']='Electricians'\nAmbulance_Drivers['job_compared_name']='Ambulance_Drivers'\nAthletes['job_compared_name']='Athletes'\nSkincare_Specialists['job_compared_name']='Skincare_Specialists'\nPrivate_Cooks['job_compared_name']='Private_Cooks'\nFuneral_Attendants['job_compared_name']='Funeral_Attendants'\nActors['job_compared_name']='Actors'\nJudges['job_compared_name']='Judges'\nEconomists['job_compared_name']='Economists'\nhistorians['job_compared_name']='historians'\nDental_Assistants['job_compared_name']='Dental_Assistants'\nCobblers['job_compared_name']='Cobblers'\nMassage_Therapists['job_compared_name']='Massage_Therapists'\nMillwrights['job_compared_name']='Millwrights'\nLibrarians['job_compared_name']='Librarians'\nMaids['job_compared_name']='Maids'\nBartenders['job_compared_name']='Bartenders'\nDishwashers['job_compared_name']='Dishwashers'\nFast_Food_Cooks['job_compared_name']='Fast_Food_Cooks'\nBarbers['job_compared_name']='Barbers'\nReal_Estate_Agents['job_compared_name']='Real_Estate_Agents'\nProofreaders['job_compared_name']='Proofreaders'",
"/Users/iblinderman/anaconda/lib/python2.7/site-packages/ipykernel_launcher.py:1: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n \"\"\"Entry point for launching an IPython kernel.\n/Users/iblinderman/anaconda/lib/python2.7/site-packages/ipykernel_launcher.py:2: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n \n/Users/iblinderman/anaconda/lib/python2.7/site-packages/ipykernel_launcher.py:3: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n This is separate from the ipykernel package so we can avoid doing imports until\n/Users/iblinderman/anaconda/lib/python2.7/site-packages/ipykernel_launcher.py:4: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n after removing the cwd from sys.path.\n/Users/iblinderman/anaconda/lib/python2.7/site-packages/ipykernel_launcher.py:5: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n \"\"\"\n/Users/iblinderman/anaconda/lib/python2.7/site-packages/ipykernel_launcher.py:6: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n \n/Users/iblinderman/anaconda/lib/python2.7/site-packages/ipykernel_launcher.py:7: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n import sys\n/Users/iblinderman/anaconda/lib/python2.7/site-packages/ipykernel_launcher.py:8: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n \n/Users/iblinderman/anaconda/lib/python2.7/site-packages/ipykernel_launcher.py:9: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n if __name__ == '__main__':\n/Users/iblinderman/anaconda/lib/python2.7/site-packages/ipykernel_launcher.py:10: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n # Remove the CWD from sys.path while we load stuff.\n/Users/iblinderman/anaconda/lib/python2.7/site-packages/ipykernel_launcher.py:11: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n # This is added back by InteractiveShellApp.init_path()\n/Users/iblinderman/anaconda/lib/python2.7/site-packages/ipykernel_launcher.py:12: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n if sys.path[0] == '':\n/Users/iblinderman/anaconda/lib/python2.7/site-packages/ipykernel_launcher.py:13: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n del sys.path[0]\n/Users/iblinderman/anaconda/lib/python2.7/site-packages/ipykernel_launcher.py:14: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n \n/Users/iblinderman/anaconda/lib/python2.7/site-packages/ipykernel_launcher.py:15: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n from ipykernel import kernelapp as app\n/Users/iblinderman/anaconda/lib/python2.7/site-packages/ipykernel_launcher.py:16: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n app.launch_new_instance()\n/Users/iblinderman/anaconda/lib/python2.7/site-packages/ipykernel_launcher.py:17: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n/Users/iblinderman/anaconda/lib/python2.7/site-packages/ipykernel_launcher.py:18: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n/Users/iblinderman/anaconda/lib/python2.7/site-packages/ipykernel_launcher.py:19: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n/Users/iblinderman/anaconda/lib/python2.7/site-packages/ipykernel_launcher.py:20: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n/Users/iblinderman/anaconda/lib/python2.7/site-packages/ipykernel_launcher.py:21: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n/Users/iblinderman/anaconda/lib/python2.7/site-packages/ipykernel_launcher.py:22: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n/Users/iblinderman/anaconda/lib/python2.7/site-packages/ipykernel_launcher.py:23: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n/Users/iblinderman/anaconda/lib/python2.7/site-packages/ipykernel_launcher.py:24: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n/Users/iblinderman/anaconda/lib/python2.7/site-packages/ipykernel_launcher.py:25: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n/Users/iblinderman/anaconda/lib/python2.7/site-packages/ipykernel_launcher.py:26: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n/Users/iblinderman/anaconda/lib/python2.7/site-packages/ipykernel_launcher.py:27: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n/Users/iblinderman/anaconda/lib/python2.7/site-packages/ipykernel_launcher.py:28: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n/Users/iblinderman/anaconda/lib/python2.7/site-packages/ipykernel_launcher.py:29: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n"
],
[
"Choreographers.to_csv('Choreographers.csv', index=False)\nDentists.to_csv('Dentists.csv', index=False)\nNurses.to_csv('Nurses.csv', index=False)\nChiropractors.to_csv('Chiropractors.csv', index=False)\nFarmers.to_csv('Farmers.csv', index=False)\nConstruction_Managers.to_csv('Construction_Managers.csv', index=False)\nFirefighters.to_csv('Firefighters.csv', index=False)\nGeographers.to_csv('Geographers.csv', index=False)\nTruck_drivers.to_csv('Truck_drivers.csv', index=False)\nEmbalmers.to_csv('Embalmers.csv', index=False)\nPiplayers.to_csv('Piplayers.csv', index=False)\nPodiatrists.to_csv('Podiatrists.csv', index=False)\nFabric_Patternmakers.to_csv('Fabric_Patternmakers.csv', index=False)\nClergy.to_csv('Clergy.csv', index=False)\nMakeup_Artists.to_csv('Makeup_Artists.csv', index=False)\nFamily_Therapists.to_csv('Family_Therapists.csv', index=False)\nCEOs.to_csv('CEOs.csv', index=False)\nArt_Directors.to_csv('Art_Directors.csv', index=False)\nInterrior_Designers.to_csv('Interrior_Designers.csv', index=False)\nCraft_Artists.to_csv('Craft_Artists.csv', index=False)\nEvent_Planners.to_csv('Event_Planners.csv', index=False)\nVeterinarians.to_csv('Veterinarians.csv', index=False)\nWriters.to_csv('Writers.csv', index=False)\nPolitical_Scientists.to_csv('Political_Scientists.csv', index=False)\nShip_Engineers.to_csv('Ship_Engineers.csv', index=False)\nParamedics.to_csv('Paramedics.csv', index=False)\nMathematicians.to_csv('Mathematicians.csv', index=False)\nFlorists.to_csv('Florists.csv', index=False)\nTravel_Guides.to_csv('Travel_Guides.csv', index=False)\nNews_Analysts.to_csv('News_Analysts.csv', index=False)\nMusicians.to_csv('Musicians.csv', index=False)\nFitness_Trainers.to_csv('Fitness_Trainers.csv', index=False)\nGraphic_Designers.to_csv('Graphic_Designers.csv', index=False)\nChildcare_Workers.to_csv('Childcare_Workers.csv', index=False)\nPolice_Officers.to_csv('Police_Officers.csv', index=False)\nHairdressers.to_csv('Hairdressers.csv', index=False)\nJournalists.to_csv('Journalists.csv', index=False)\nAir_Traffic_Controllers.to_csv('Air_Traffic_Controllers.csv', index=False)\nDancers.to_csv('Dancers.csv', index=False)\nOptometrists.to_csv('Optometrists.csv', index=False)\nPhysician_Assistants.to_csv('Physician_Assistants.csv', index=False)\nElectricians.to_csv('Electricians.csv', index=False)\nAmbulance_Drivers.to_csv('Ambulance_Drivers.csv', index=False)\nAthletes.to_csv('Athletes.csv', index=False)\nSkincare_Specialists.to_csv('Skincare_Specialists.csv', index=False)\nPrivate_Cooks.to_csv('Private_Cooks.csv', index=False)\nFuneral_Attendants.to_csv('Funeral_Attendants.csv', index=False)\nActors.to_csv('Actors.csv', index=False)\nJudges.to_csv('Judges.csv', index=False)\nEconomists.to_csv('Economists.csv', index=False)\nhistorians.to_csv('historians.csv', index=False)\nDental_Assistants.to_csv('Dental_Assistants.csv', index=False)\nCobblers.to_csv('Cobblers.csv', index=False)\nMassage_Therapists.to_csv('Massage_Therapists.csv', index=False)\nMillwrights.to_csv('Millwrights.csv', index=False)\nLibrarians.to_csv('Librarians.csv', index=False)\nMaids.to_csv('Maids.csv', index=False)\nBartenders.to_csv('Bartenders.csv', index=False)\nDishwashers.to_csv('Dishwashers.csv', index=False)\nFast_Food_Cooks.to_csv('Fast_Food_Cooks.csv', index=False)\nBarbers.to_csv('Barbers.csv', index=False)\nReal_Estate_Agents.to_csv('Real_Estate_Agents.csv', index=False)\nProofreaders.to_csv('Proofreaders.csv', index=False)",
"_____no_output_____"
]
],
[
[
"# OLD",
"_____no_output_____"
]
],
[
[
"auto_df = auto_df[['Job_Compared','Heavy and Tractor-Trailer Truck Drivers','Athletes and Sports Competitors','Compensation and Benefits Managers','Construction Laborers']]\nrelative_emp_df = relative_emp_df[['Job_Compared','Heavy and Tractor-Trailer Truck Drivers','Athletes and Sports Competitors','Compensation and Benefits Managers','Construction Laborers']]\nsimilar_df = similar_df[['Job_Compared','Heavy and Tractor-Trailer Truck Drivers','Athletes and Sports Competitors','Compensation and Benefits Managers','Construction Laborers']]\nwagechange_df = wagechange_df[['Job_Compared','Heavy and Tractor-Trailer Truck Drivers','Athletes and Sports Competitors','Compensation and Benefits Managers','Construction Laborers']]",
"_____no_output_____"
],
[
"auto_df=auto_df.set_index('Job_Compared')\nrelative_emp_df=relative_emp_df.set_index('Job_Compared')\nsimilar_df=similar_df.set_index('Job_Compared')\nwagechange_df = wagechange_df.set_index('Job_Compared')",
"_____no_output_____"
],
[
"auto_df = auto_df.add_prefix('auto_')\nrelative_emp_df = relative_emp_df.add_prefix('empl_')\nsimilar_df = similar_df.add_prefix('similar_')\nwagechange_df = wagechange_df.add_prefix('wage_')\n",
"_____no_output_____"
],
[
"auto_df= auto_df= auto_df.rename(columns=lambda x: x.replace(' ','_'))\nrelative_emp_df= relative_emp_df.rename(columns=lambda x: x.replace(' ','_'))\nsimilar_df= similar_df.rename(columns=lambda x: x.replace(' ','_'))\nwagechange_df= wagechange_df.rename(columns=lambda x: x.replace(' ','_'))",
"_____no_output_____"
],
[
"auto_df= auto_df= auto_df.rename(columns=lambda x: x.replace(' ','_'))\nrelative_emp_df= relative_emp_df.rename(columns=lambda x: x.replace(' ','_'))\nsimilar_df= similar_df.rename(columns=lambda x: x.replace(' ','_'))\nwagechange_df= wagechange_df.rename(columns=lambda x: x.replace(' ','_'))",
"_____no_output_____"
],
[
"truckers_wage = wagechange_df[wagechange_df.columns[0]].reset_index()\nathletes_wage = wagechange_df[wagechange_df.columns[1]].reset_index()\ncomp_managers_wage = wagechange_df[wagechange_df.columns[2]].reset_index()\nlaborers_wage = wagechange_df[wagechange_df.columns[3]].reset_index()\n\n \ntruckers_similarity = similar_df[similar_df.columns[0]].reset_index()\nathletes_similarity = similar_df[similar_df.columns[1]].reset_index()\ncomp_managers_similarity = similar_df[similar_df.columns[2]].reset_index()\nlaborers_similarity = similar_df[similar_df.columns[3]].reset_index()\n\n\ntruckers_relative_emp = relative_emp_df[relative_emp_df.columns[0]].reset_index()\nathletes_relative_emp = relative_emp_df[relative_emp_df.columns[1]].reset_index()\ncomp_managers_relative_emp = relative_emp_df[relative_emp_df.columns[2]].reset_index()\nlaborers_relative_emp = relative_emp_df[relative_emp_df.columns[3]].reset_index()\n\n\ntruckers_automatability = auto_df[auto_df.columns[0]].reset_index()\nathletes_automatability = auto_df[auto_df.columns[1]].reset_index()\ncomp_managers_automatability = auto_df[auto_df.columns[2]].reset_index()\nlaborers_automatability = auto_df[auto_df.columns[3]].reset_index()",
"_____no_output_____"
],
[
"dfs_trucker = [truckers_wage, truckers_similarity, truckers_relative_emp, truckers_automatability]\n\ndfs_athlete = [athletes_wage,athletes_similarity,athletes_relative_emp,athletes_automatability]\n\ndfs_comp_managers = [comp_managers_wage,comp_managers_similarity,comp_managers_relative_emp,comp_managers_automatability]\n\ndfs_laborers = [laborers_wage,laborers_similarity,laborers_relative_emp,laborers_automatability]\n\n\ndf_trucker_final =reduce(lambda left,right: pd.merge(left,right,on='Job_Compared'), dfs_trucker)\ndf_athlete_final =reduce(lambda left,right: pd.merge(left,right,on='Job_Compared'), dfs_athlete)\ndf_comp_managers_final=reduce(lambda left,right: pd.merge(left,right,on='Job_Compared'), dfs_comp_managers)\ndf_laborers_final =reduce(lambda left,right: pd.merge(left,right,on='Job_Compared'), dfs_laborers)\n",
"_____no_output_____"
],
[
"df_trucker_final = df_trucker_final.reset_index()\ndf_athlete_final = df_athlete_final.reset_index(\"Job_Compared\")\ndf_comp_managers_final = df_comp_managers_final.reset_index(\"Job_Compared\")\ndf_laborers_final = df_laborers_final.reset_index(\"Job_Compared\")\n",
"_____no_output_____"
],
[
"raw_wage_auto_jobs = pd.read_csv(\"jobs.csv\")\nraw_wage_auto_jobs= raw_wage_auto_jobs\n# .set_index(\"Occupation\")\n",
"_____no_output_____"
],
[
"laborers = pd.merge(raw_wage_auto_jobs,df_laborers_final, right_on='Job_Compared',left_on='Occupation')\ntruckers = pd.merge(raw_wage_auto_jobs,df_trucker_final, right_on='Job_Compared',left_on='Occupation')\nathletes = pd.merge(raw_wage_auto_jobs,df_athlete_final, right_on='Job_Compared',left_on='Occupation')\ncomp_managers = pd.merge(raw_wage_auto_jobs,df_comp_managers_final, right_on='Job_Compared',left_on='Occupation')",
"_____no_output_____"
],
[
"laborers.to_csv(\"laborers.csv\")\ntruckers.to_csv(\"truckers.csv\")\nathletes.to_csv(\"athletes.csv\")\ncomp_managers.to_csv(\"comp_managers.csv\")",
"_____no_output_____"
],
[
"# auto_df.to_csv(\"auto.csv\")\n# relative_emp.to_csv(\"relative_emp.csv\")\n# similar_df.to_csv(\"simlar.csv\")\n# wagechange.to_csv(\"wage.csv\")",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a59474144197071d85535e28fc1d786c614225b
| 393,443 |
ipynb
|
Jupyter Notebook
|
nbs/dl1/rossman_data_clean-attempt.ipynb
|
rforgione/course-v3
|
f496c2eea82153e23c20b3fcb2bd2c3409677cf7
|
[
"Apache-2.0"
] | null | null | null |
nbs/dl1/rossman_data_clean-attempt.ipynb
|
rforgione/course-v3
|
f496c2eea82153e23c20b3fcb2bd2c3409677cf7
|
[
"Apache-2.0"
] | null | null | null |
nbs/dl1/rossman_data_clean-attempt.ipynb
|
rforgione/course-v3
|
f496c2eea82153e23c20b3fcb2bd2c3409677cf7
|
[
"Apache-2.0"
] | null | null | null | 34.385859 | 360 | 0.31021 |
[
[
[
"%reload_ext autoreload\n%autoreload 2",
"_____no_output_____"
],
[
"from fastai.basics import *",
"_____no_output_____"
],
[
"from pathlib import Path",
"_____no_output_____"
],
[
"import pandas as pd",
"_____no_output_____"
]
],
[
[
"# Rossmann",
"_____no_output_____"
],
[
"## Data preparation / Feature engineering",
"_____no_output_____"
],
[
"Set `PATH` to the path `~/data/rossmann/`. Create a list of table names, with one entry for each CSV that you'll be loading: \n- train\n- store\n- store_states\n- state_names\n- googletrend\n- weather\n- test\n\nFor each csv, read it in using pandas (with `low_memory=False`), and assign it to a variable corresponding with its name. Print out the lengths of the `train` and `test` tables.",
"_____no_output_____"
]
],
[
[
"PATH = Path(\"~/.fastai/data/rossmann/\")",
"_____no_output_____"
],
[
"csvs = ['train', 'store', 'store_states', 'state_names', 'googletrend', 'weather', 'test']",
"_____no_output_____"
],
[
"tables = [pd.read_csv(f\"{PATH}/{csv}.csv\", low_memory=False) for csv in csvs]",
"_____no_output_____"
],
[
"train, store, store_states, state_names, googletrend, weather, test = tables",
"_____no_output_____"
]
],
[
[
"Turn the `StateHoliday` column into a boolean indicating whether or not the day was a holiday.",
"_____no_output_____"
]
],
[
[
"train['StateHoliday'] = train['StateHoliday'] != '0'\ntest['StateHoliday'] = test['StateHoliday'] != '0'\ntrain['SchoolHoliday'] = train['SchoolHoliday'] != 0\ntest['SchoolHoliday'] = test['SchoolHoliday'] != 0",
"_____no_output_____"
],
[
"train['StateHoliday'].value_counts()",
"_____no_output_____"
],
[
"train['SchoolHoliday'].value_counts()",
"_____no_output_____"
]
],
[
[
"Print out the head of the dataframe.",
"_____no_output_____"
]
],
[
[
"train.head()",
"_____no_output_____"
]
],
[
[
"Create a function `join_df` that joins two dataframes together. It should take the following arguments:\n- left (the df on the lft)\n- right (the df on the right)\n- left_on (the left table join key)\n- right_on (the right table join key, defaulting to None; if nothing passed, default to the same as the left join key)\n- suffix (default to '_y'; a suffix to give to duplicate columns)",
"_____no_output_____"
]
],
[
[
"def join_df(left, right, left_on, right_on=None, suffix='_y'):\n return left.merge(right, left_on=left_on, right_on=right_on if right_on is not None else left_on, \n how='left', suffixes=('', suffix))",
"_____no_output_____"
],
[
"import pandas as pd\ndf1 = pd.DataFrame({'a': [1,2], 'b': [100.0, 1000.0]})\ndf2 = pd.DataFrame({'a': [1,1,1,3,3,3], 'b': [10.0, 10.0, 10.0, 20.0, 20.0, 20.0]})\njoin_df(df1, df2, 'a')",
"_____no_output_____"
]
],
[
[
"Join the weather and state names tables together, and reassign them to the variable `weather`.",
"_____no_output_____"
]
],
[
[
"weather.head()",
"_____no_output_____"
],
[
"state_names",
"_____no_output_____"
],
[
"weather = join_df(weather, state_names, left_on='file', right_on='StateName')",
"_____no_output_____"
]
],
[
[
"Show the first few rows of the weather df.",
"_____no_output_____"
]
],
[
[
"weather.head()",
"_____no_output_____"
]
],
[
[
"In the `googletrend` table, set the `Date` variable to the first date in the hyphen-separated date string in the `week` field. Set the `State` field to the third element in the underscore-separated string from the `file` field. In all rows where `State == NI`, make it instead equal `HB,NI` which is how it's referred to throughout the reset of the data.",
"_____no_output_____"
]
],
[
[
"googletrend['Date'] = googletrend['week'].str.split(' - ', expand=True)[0]",
"_____no_output_____"
],
[
"googletrend['State'] = googletrend['file'].str.split('_', expand=True)[2]",
"_____no_output_____"
],
[
"googletrend['State'][googletrend['State'] == 'NI'] = 'HB,NI'",
"/home/paperspace/anaconda3/lib/python3.6/site-packages/ipykernel_launcher.py:1: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n \"\"\"Entry point for launching an IPython kernel.\n"
],
[
"googletrend.head()",
"_____no_output_____"
]
],
[
[
"Write a function `add_datepart` that takes a date field and adds a bunch of numeric columns containing information about the date. It should take the following arguments:\n- df (the dataframe you'll be modifying)\n- fldname (the date field you'll be splitting into new columns)\n- drop (whether or not to drop the old date field; defaults to True)\n- time (whether or not to add time fields -- Hour, Minute, Second; defaults to False)\n\nIt should append ```\n['Year', 'Month', 'Week', 'Day', 'Dayofweek', 'Dayofyear','Is_month_end', 'Is_month_start', 'Is_quarter_end', 'Is_quarter_start', 'Is_year_end', 'Is_year_start']```\n\nRemember the edge cases around the dtype of the field. Specifically, if it's of type DatetimeTZDtype, cast it instead to np.datetime64. If it's not a subtype of datetime64 already, infer it (see `pd.to_datetime`).",
"_____no_output_____"
]
],
[
[
"type(np.datetime64)",
"_____no_output_____"
],
[
"Series(np.array([1,2,3], dtype='int64')).dtype",
"_____no_output_____"
],
[
"ex_date = pd.to_datetime('2019-01-01')",
"_____no_output_____"
],
[
"ex_date",
"_____no_output_____"
],
[
"ex_date",
"_____no_output_____"
],
[
"def add_datepart(df, fldname, drop=True, time=False):\n fld = df[fldname]\n \n if fld.dtype == pd.DatetimeTZDtype:\n fld = fld.astype(np.datetime64)\n \n if not np.issubdtype(fld.dtype, np.datetime64):\n fld = pd.to_datetime(fld)\n \n fields = [\n 'Year', 'Month', 'Week', \n 'Day', 'Dayofweek', 'Dayofyear',\n 'Is_month_end', 'Is_month_start', 'Is_quarter_end', \n 'Is_quarter_start', 'Is_year_end', 'Is_year_start'\n ]\n \n for i in fields:\n df[i] = getattr(fld.dt, i.lower())\n \n if drop:\n df.drop(fldname, inplace=True)\n \n return df",
"_____no_output_____"
],
[
"googletrend = add_datepart(googletrend, 'Date', False)",
"_____no_output_____"
]
],
[
[
"Use `add_datepart` to add date fields to the weather, googletrend, train and test tables.",
"_____no_output_____"
]
],
[
[
"weather.head()",
"_____no_output_____"
],
[
"weather = add_datepart(weather, 'Date', False)",
"_____no_output_____"
],
[
"train.head()",
"_____no_output_____"
],
[
"train = add_datepart(train, 'Date', False)",
"_____no_output_____"
],
[
"test = add_datepart(test, 'Date', False)",
"_____no_output_____"
]
],
[
[
"Print out the head of the weather table.",
"_____no_output_____"
]
],
[
[
"weather.head()",
"_____no_output_____"
]
],
[
[
"In the `googletrend` table, the `file` column has an entry `Rossmann_DE` that represents the whole of germany; we'll want to break that out into its own separate table, since we'll need to join it on `Date` alone rather than both `Date` and `Store`.",
"_____no_output_____"
]
],
[
[
"googletrend",
"_____no_output_____"
],
[
"rossmann_full = googletrend[googletrend['file'] == 'Rossmann_DE']",
"_____no_output_____"
]
],
[
[
"Now let's do a bunch of joins to build our entire dataset! Remember after each one to check if the right-side data is null. This is the benefit of left-joining; it's easy to debug by checking for null rows. Let's start by joining `store` and `store_states` in a new table called `store`.",
"_____no_output_____"
]
],
[
[
"store.head()",
"_____no_output_____"
],
[
"store_states.head()",
"_____no_output_____"
],
[
"store = join_df(store, store_states, 'Store')",
"_____no_output_____"
],
[
"store",
"_____no_output_____"
],
[
"store['State'].isna().sum()",
"_____no_output_____"
]
],
[
[
"Next let's join `train` and `store` in a table called `joined`. Do the same for `test` and `store` in a table called `joined_test`.",
"_____no_output_____"
]
],
[
[
"joined = join_df(train, store, 'Store')",
"_____no_output_____"
],
[
"joined.head()",
"_____no_output_____"
],
[
"joined_test = join_df(test, store, 'Store')",
"_____no_output_____"
],
[
"joined_test.head()",
"_____no_output_____"
]
],
[
[
"Next join `joined` and `googletrend` on the columns `[\"State\", \"Year\", \"Week\"]`. Again, do the same for the test data.",
"_____no_output_____"
]
],
[
[
"joined = join_df(joined, googletrend[[\"State\", \"Year\", \"Week\", \"trend\"]], [\"State\", \"Year\", \"Week\"])\njoined_test = join_df(joined_test, googletrend[[\"State\", \"Year\", \"Week\", \"trend\"]], [\"State\", \"Year\", \"Week\"])",
"_____no_output_____"
]
],
[
[
"Join `joined` and `trend_de` on `[\"Year\", \"Week\"]` with suffix `_DE`. Same for test.",
"_____no_output_____"
]
],
[
[
"rossmann_full.head()",
"_____no_output_____"
],
[
"joined = join_df(joined, rossmann_full[['Date', 'trend']], 'Date', suffix='_DE')",
"_____no_output_____"
],
[
"joined_test = join_df(joined_test, rossmann_full[['Date', 'trend']], 'Date', suffix='_DE')",
"_____no_output_____"
]
],
[
[
"Join `joined` and `weather` on `[\"State\", \"Date\"]`. Same for test.",
"_____no_output_____"
]
],
[
[
"weather.head()",
"_____no_output_____"
],
[
"joined = join_df(joined, weather, [\"State\", \"Date\"])\njoined_test = join_df(joined_test, weather, [\"State\", \"Date\"])",
"_____no_output_____"
],
[
"joined.columns",
"_____no_output_____"
],
[
"joined['Min_DewpointC'].head()",
"_____no_output_____"
]
],
[
[
"Now for every column in both `joined` and `joined_test`, check to see if it has the `_y` suffix, and if so, drop it. Warning: a data frame can have duplicate column names, but calling `df.drop` will drop _all_ instances with the passed-in column name! This could lead to calling drop a second time on a column that no longer exists!",
"_____no_output_____"
]
],
[
[
"for i in (joined, joined_test):\n for j in i.columns:\n if j in i.columns and '_y' in j: i.drop(j, axis=1, inplace=True)",
"_____no_output_____"
],
[
"joined.columns",
"_____no_output_____"
],
[
"joined_test.columns",
"_____no_output_____"
],
[
"len(joined.columns)",
"_____no_output_____"
],
[
"len(joined_test.columns)",
"_____no_output_____"
]
],
[
[
"For the columns `CompetitionOpenSinceYear`, `CompetitionOpenSinceMonth`, `Promo2SinceYear`, and `Promo2SinceMonth`, replace `NA` values with the following values (respectively):\n- 1900\n- 1\n- 1900\n- 1",
"_____no_output_____"
]
],
[
[
"for i in (joined, joined_test):\n i.loc[i['CompetitionOpenSinceYear'].isna(), 'CompetitionOpenSinceYear'] = 1900\n i.loc[i['CompetitionOpenSinceMonth'].isna(), 'CompetitionOpenSinceMonth'] = 1\n i.loc[i['Promo2SinceYear'].isna(), 'Promo2SinceYear'] = 1900\n i.loc[i['Promo2SinceWeek'].isna(), 'Promo2SinceWeek'] = 1",
"_____no_output_____"
],
[
"joined.head()['Promo2SinceYear']",
"_____no_output_____"
]
],
[
[
"Create a new field `CompetitionOpenSince` that converts `CompetitionOpenSinceYear` and `CompetitionOpenSinceMonth` and maps them to a specific date. Then create a new field `CompetitionDaysOpen` that subtracts `CompetitionOpenSince` from `Date`. ",
"_____no_output_____"
]
],
[
[
"pd.to_datetime({'year': [2019], 'month': [1], 'day': [1]})",
"_____no_output_____"
],
[
"joined['Date'].dtype",
"_____no_output_____"
],
[
"import datetime\nfor i in (joined, joined_test):\n i['CompetitionOpenSince'] = pd.to_datetime({'year': i['CompetitionOpenSinceYear'], \n 'month': i['CompetitionOpenSinceMonth'],\n 'day': 15})\n i['CompetitionDaysOpen'] = (pd.to_datetime(i['Date']) - i['CompetitionOpenSince']) / datetime.timedelta(days=1)\n ",
"_____no_output_____"
],
[
"joined[['CompetitionOpenSince', 'CompetitionDaysOpen']].head()",
"_____no_output_____"
]
],
[
[
"For `CompetitionDaysOpen`, replace values where `CompetitionDaysOpen < 0` with 0, and cases where `CompetitionOpenSinceYear < 1990` with 0.",
"_____no_output_____"
]
],
[
[
"joined['CompetitionOpenSinceYear'].dtype, joined['CompetitionDaysOpen'].dtype",
"_____no_output_____"
],
[
"for i in (joined, joined_test):\n i['CompetitionDaysOpen'][i['CompetitionDaysOpen'] < 0] = 0\n i['CompetitionOpenSinceYear'][i['CompetitionOpenSinceYear'] < 1990] = 1990",
"/home/paperspace/anaconda3/lib/python3.6/site-packages/ipykernel_launcher.py:2: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n \n/home/paperspace/anaconda3/lib/python3.6/site-packages/ipykernel_launcher.py:3: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n This is separate from the ipykernel package so we can avoid doing imports until\n"
],
[
"joined[['CompetitionDaysOpen', 'CompetitionOpenSinceYear']]",
"_____no_output_____"
]
],
[
[
"We add \"CompetitionMonthsOpen\" field, limiting the maximum to 2 years to limit number of unique categories.",
"_____no_output_____"
]
],
[
[
"for i in (joined, joined_test):\n i['CompetitionMonthsOpen'] = i['CompetitionDaysOpen'] // 30\n i['CompetitionMonthsOpen'] = i.apply(lambda x: 24 if x['CompetitionMonthsOpen'] > 24 else x['CompetitionMonthsOpen'], axis=1)",
"_____no_output_____"
],
[
"joined['CompetitionMonthsOpen'].value_counts()",
"_____no_output_____"
]
],
[
[
"Same process for Promo dates. You may need to install the `isoweek` package first.",
"_____no_output_____"
]
],
[
[
"# If needed, uncomment:\n# ! pip install isoweek",
"_____no_output_____"
]
],
[
[
"Use the `isoweek` package to turn `Promo2Since` to a specific date -- the Monday of the week specified in the column. Compute a field `Promo2SinceDays` that subtracts the current date from the `Promo2Since` date.",
"_____no_output_____"
]
],
[
[
"joined['Date'].dtype",
"_____no_output_____"
],
[
"from isoweek import Week\nfor i in (joined, joined_test):\n i['Promo2Since'] = i.apply(lambda x: Week(int(x['Promo2SinceYear']), int(x['Promo2SinceWeek'])).monday(), axis=1)\n i['Promo2Since'] = pd.to_datetime(i['Promo2Since'])\n i['Promo2Days'] = (pd.to_datetime(i['Date']) - i['Promo2Since']) / datetime.timedelta(days=1)\n i['Promo2Weeks'] = i['Promo2Days'] // 7",
"_____no_output_____"
],
[
"joined['Promo2Since'].head(25)",
"_____no_output_____"
],
[
"joined.columns",
"_____no_output_____"
]
],
[
[
"Perform the following modifications on both the train and test set:\n- For cases where `Promo2Days` is negative or `Promo2SinceYear` is before 1990, set `Promo2Days` to 0\n- Create `Promo2Weeks\n- For cases where `Promo2Weeks` is negative, set `Promo2Weeks` to 0\n- For cases where `Promo2Weeks` is above 25, set `Promo2Weeks` to 25\n\nPrint the number of unique values for `Promo2Weeks` in training and test df's.",
"_____no_output_____"
]
],
[
[
"for i in (joined, joined_test):\n i['Promo2Days'][i['Promo2Days'] < 0] = 0\n i['Promo2Days'][i['Promo2SinceYear'] < 1990] = 0\n i['Promo2Weeks'][i['Promo2Weeks'] < 0] = 0 \n i['Promo2Weeks'][i['Promo2Weeks'] > 25] = 25\n ",
"/home/paperspace/anaconda3/lib/python3.6/site-packages/ipykernel_launcher.py:2: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n \n/home/paperspace/anaconda3/lib/python3.6/site-packages/ipykernel_launcher.py:3: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n This is separate from the ipykernel package so we can avoid doing imports until\n/home/paperspace/anaconda3/lib/python3.6/site-packages/ipykernel_launcher.py:4: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n after removing the cwd from sys.path.\n/home/paperspace/anaconda3/lib/python3.6/site-packages/ipykernel_launcher.py:5: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n \"\"\"\n"
],
[
"len(joined['Promo2Weeks'].unique())",
"_____no_output_____"
]
],
[
[
"Pickle `joined` to `PATH/'joined'` and `joined_test` to `PATH/'joined_test'`.",
"_____no_output_____"
]
],
[
[
"joined.to_pickle(PATH/'joined')",
"_____no_output_____"
],
[
"joined_test.to_pickle(PATH/'joined_test')",
"_____no_output_____"
]
],
[
[
"## Durations",
"_____no_output_____"
],
[
"Write a function `get_elapsed` that takes arguments `fld` (a boolean field) and `pre` (a prefix to be appended to `fld` in a new column representing the days until/since the event in `fld`), and adds a column `pre+fld` representing the date-diff (in days) between the current date and the last date `fld` was true.",
"_____no_output_____"
]
],
[
[
"joined = pd.read_pickle(PATH/'joined')\njoined_test = pd.read_pickle(PATH/'joined_test')",
"_____no_output_____"
],
[
"joined['Date'] = pd.to_datetime(joined['Date'])",
"_____no_output_____"
],
[
"joined_test['Date'] = pd.to_datetime(joined_test['Date'])",
"_____no_output_____"
],
[
"def get_elapsed(df, fld, pre):\n day1 = np.timedelta64(1, 'D')\n store = 0\n last_date = np.timedelta64()\n vals = []\n for s,v,d in zip(df['Store'].values, df[fld].values, df['Date'].values):\n if s != store: \n store = s\n last_date = np.datetime64()\n if v: last_date = d\n vals.append(((d - last_date).astype('timedelta64[D]') / day1))\n df[pre+fld] = vals\n return df",
"_____no_output_____"
],
[
"joined.sort_values(['Store', 'Date'], ascending=[True, True], inplace=True)\nget_elapsed(joined, 'SchoolHoliday', 'After')",
"_____no_output_____"
]
],
[
[
"We'll be applying this to a subset of columns:",
"_____no_output_____"
],
[
"Create a variable `columns` containing the strings: \n- Date\n- Store\n- Promo\n- StateHoliday\n- SchoolHoliday\n\nThese will be the fields on which we'll be computing elapsed days since/until.",
"_____no_output_____"
]
],
[
[
"columns = ['Date', 'Store', 'Promo', 'StateHoliday', 'SchoolHoliday']",
"_____no_output_____"
]
],
[
[
"Create one big dataframe with both the train and test sets called `df`.",
"_____no_output_____"
]
],
[
[
"df = pd.concat([joined[columns], joined_test[columns]], axis=0)",
"_____no_output_____"
],
[
"df.shape",
"_____no_output_____"
]
],
[
[
"Sort by `Store` and `Date` ascending, and use `add_elapsed` to get the days since the last `SchoolHoliday` on each daya. Reorder by `Store` ascending and `Date` descending to get the days _until_ the next `SchoolHoliday`.",
"_____no_output_____"
]
],
[
[
"df.sort_values(['Store', 'Date'], inplace=True)\nget_elapsed(df, 'SchoolHoliday', 'After')\ndf.sort_values(['Store', 'Date'], ascending=[True, False], inplace=True)\nget_elapsed(df, 'SchoolHoliday', 'Before').head()",
"_____no_output_____"
]
],
[
[
"Do the same for `StateHoliday`.",
"_____no_output_____"
]
],
[
[
"df.sort_values(['Store', 'Date'], inplace=True)\nget_elapsed(df, 'StateHoliday', 'After')\ndf.sort_values(['Store', 'Date'], ascending=[True, False], inplace=True)\nget_elapsed(df, 'StateHoliday', 'Before').head()",
"_____no_output_____"
]
],
[
[
"Do the same for `Promo`.",
"_____no_output_____"
]
],
[
[
"df.sort_values(['Store', 'Date'], inplace=True)\nget_elapsed(df, 'Promo', 'After')\ndf.sort_values(['Store', 'Date'], ascending=[True, False], inplace=True)\nget_elapsed(df, 'Promo', 'Before').head()",
"_____no_output_____"
]
],
[
[
"Set the index on `df` to `Date`.",
"_____no_output_____"
]
],
[
[
"df.set_index('Date', inplace=True)",
"_____no_output_____"
]
],
[
[
"Reassign `columns` to `['SchoolHoliday', 'StateHoliday', 'Promo']`.",
"_____no_output_____"
]
],
[
[
"columns = ['SchoolHoliday', 'StateHoliday', 'Promo']",
"_____no_output_____"
]
],
[
[
"For columns `Before/AfterSchoolHoliday`, `Before/AfterStateHoliday`, and `Before/AfterPromo`, fill null values with 0.",
"_____no_output_____"
]
],
[
[
"df.columns",
"_____no_output_____"
],
[
"for column in columns:\n for prefix in ['Before', 'After']:\n df[prefix+column].fillna(0, inplace=True) ",
"_____no_output_____"
]
],
[
[
"Create a dataframe `bwd` that gets 7-day backward-rolling sums of the columns in `columns`, grouped by `Store`.",
"_____no_output_____"
]
],
[
[
"print(\"hello world\")",
"hello world\n"
],
[
"bwd = (df[columns+['Store']]\n .sort_index(ascending=True)\n .groupby('Store')\n .rolling(window=7, min_periods=1)\n .sum()).drop('Store', axis=1).reset_index()",
"_____no_output_____"
],
[
"bwd.head(25)",
"_____no_output_____"
],
[
"fwd = (df[columns+['Store']]\n .sort_index(ascending=False)\n .groupby('Store')\n .rolling(window=7, min_periods=1)\n .sum()).drop('Store', axis=1).reset_index()",
"_____no_output_____"
],
[
"fwd.head(25)",
"_____no_output_____"
]
],
[
[
"Create a dataframe `fwd` that gets 7-day forward-rolling sums of the columns in `columns`, grouped by `Store`.",
"_____no_output_____"
],
[
"Show the head of `bwd`.",
"_____no_output_____"
],
[
"Show the head of `fwd`.",
"_____no_output_____"
],
[
"Drop the `Store` column from `fwd` and `bwd` inplace, and reset the index inplace on each.",
"_____no_output_____"
],
[
"Reset the index on `df`.",
"_____no_output_____"
],
[
"Merge `df` with `bwd` and `fwd`.",
"_____no_output_____"
]
],
[
[
"df = join_df(df, bwd, ['Date', 'Store'], ['Date', 'Store'], '_bwd')",
"_____no_output_____"
],
[
"df = join_df(df, fwd, ['Date', 'Store'], ['Date', 'Store'], '_fwd')",
"_____no_output_____"
]
],
[
[
"Drop `columns` from df inplace -- we don't need them anymore, since we've captured their information in columns with types more suitable for machine learning.",
"_____no_output_____"
]
],
[
[
"df.columns",
"_____no_output_____"
],
[
"columns",
"_____no_output_____"
],
[
"df.drop(columns, axis=1, inplace=True)",
"_____no_output_____"
]
],
[
[
"Print out the head of `df`.",
"_____no_output_____"
]
],
[
[
"df.head()",
"_____no_output_____"
]
],
[
[
"Pickle `df` to `PATH/'df'`.",
"_____no_output_____"
]
],
[
[
"df.to_pickle(PATH/'attempt_df')",
"_____no_output_____"
]
],
[
[
"Cast the `Date` column to a datetime column.",
"_____no_output_____"
]
],
[
[
"df['Date'] = pd.to_datetime(df['Date'])",
"_____no_output_____"
]
],
[
[
"Join `joined` with `df` on `['Store', 'Date']`.",
"_____no_output_____"
]
],
[
[
"train = join_df(joined, df, left_on=['Date', 'Store'])",
"_____no_output_____"
],
[
"test = join_df(joined_test, df, left_on=['Date', 'Store'])",
"_____no_output_____"
],
[
"train.head(25)",
"_____no_output_____"
]
],
[
[
"This is not necessarily the best idea, but the authors removed all examples for which sales were equal to zero. If you're trying to stay true to what the authors did, do that now.",
"_____no_output_____"
]
],
[
[
"train_clean = train.loc[train['Sales'] != 0, :]",
"_____no_output_____"
]
],
[
[
"Reset the indices, and pickle train and test to `train_clean` and `test_clean`.",
"_____no_output_____"
]
],
[
[
"train_clean.head()",
"_____no_output_____"
],
[
"train_clean.to_pickle(PATH/'train_clean')\ntrain.to_pickle(PATH/'train_full')\ntest.to_pickle(PATH/'test')",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
4a594814df4be8768137da2baf7aecdf067a265c
| 3,431 |
ipynb
|
Jupyter Notebook
|
competitions/titanic/test.ipynb
|
yellowzp/algorithm_model_examples
|
4c8c2e29f4724fe565e73bb243709d7c302178c9
|
[
"MIT"
] | null | null | null |
competitions/titanic/test.ipynb
|
yellowzp/algorithm_model_examples
|
4c8c2e29f4724fe565e73bb243709d7c302178c9
|
[
"MIT"
] | null | null | null |
competitions/titanic/test.ipynb
|
yellowzp/algorithm_model_examples
|
4c8c2e29f4724fe565e73bb243709d7c302178c9
|
[
"MIT"
] | null | null | null | 37.293478 | 735 | 0.436607 |
[
[
[
"# https://www.kaggle.com/c/titanic/overview\nimport numpy as np\nimport pandas as pd\ntrain_data = pd.read_csv(\"./tmp/data/train.csv\")\ntest_data = pd.read_csv(\"./tmp/data/test.csv\")\nprint(train_data.head())",
" PassengerId Survived Pclass \\\n0 1 0 3 \n1 2 1 1 \n2 3 1 3 \n3 4 1 1 \n4 5 0 3 \n\n Name Sex Age SibSp \\\n0 Braund, Mr. Owen Harris male 22.0 1 \n1 Cumings, Mrs. John Bradley (Florence Briggs Th... female 38.0 1 \n2 Heikkinen, Miss. Laina female 26.0 0 \n3 Futrelle, Mrs. Jacques Heath (Lily May Peel) female 35.0 1 \n4 Allen, Mr. William Henry male 35.0 0 \n\n Parch Ticket Fare Cabin Embarked \n0 0 A/5 21171 7.2500 NaN S \n1 0 PC 17599 71.2833 C85 C \n2 0 STON/O2. 3101282 7.9250 NaN S \n3 0 113803 53.1000 C123 S \n4 0 373450 8.0500 NaN S \n"
],
[
"train_data[['Pclass', 'Survived']].groupby(['Pclass'], as_index=False).mean().sort_values(by='Survived', ascending=False)",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code"
]
] |
4a5948c957daa74110fb15b54fe54caf4f729172
| 232,632 |
ipynb
|
Jupyter Notebook
|
01_train_example.ipynb
|
tcapelle/moving_mnist
|
406eb0cebd47efce1fb93c45a9b43808394075ee
|
[
"Apache-2.0"
] | 22 |
2020-07-10T15:25:51.000Z
|
2022-03-16T07:51:52.000Z
|
01_train_example.ipynb
|
tcapelle/moving_mnist
|
406eb0cebd47efce1fb93c45a9b43808394075ee
|
[
"Apache-2.0"
] | 1 |
2021-07-22T16:43:10.000Z
|
2021-07-22T16:43:10.000Z
|
01_train_example.ipynb
|
tcapelle/moving_mnist
|
406eb0cebd47efce1fb93c45a9b43808394075ee
|
[
"Apache-2.0"
] | 5 |
2020-11-03T04:24:32.000Z
|
2021-11-23T08:35:32.000Z
| 323.549374 | 144,644 | 0.930366 |
[
[
[
"from fastai.vision.all import *\nfrom moving_mnist.models.conv_rnn import *\nfrom moving_mnist.data import *",
"_____no_output_____"
],
[
"if torch.cuda.is_available():\n torch.cuda.set_device(1)\n print(torch.cuda.get_device_name())",
"GeForce RTX 2070 SUPER\n"
]
],
[
[
"# Train Example:",
"_____no_output_____"
],
[
"We wil predict:\n- `n_in`: 5 images\n- `n_out`: 5 images \n- `n_obj`: up to 3 objects",
"_____no_output_____"
]
],
[
[
"DATA_PATH = Path.cwd()/'data'",
"_____no_output_____"
],
[
"ds = MovingMNIST(DATA_PATH, n_in=5, n_out=5, n_obj=[1,2,3])",
"_____no_output_____"
],
[
"train_tl = TfmdLists(range(7500), ImageTupleTransform(ds))\nvalid_tl = TfmdLists(range(100), ImageTupleTransform(ds))",
"_____no_output_____"
],
[
"dls = DataLoaders.from_dsets(train_tl, valid_tl, bs=32,\n after_batch=[Normalize.from_stats(imagenet_stats[0][0], \n imagenet_stats[1][0])]).cuda()",
"_____no_output_____"
],
[
"loss_func = StackLoss(MSELossFlat())",
"_____no_output_____"
]
],
[
[
"Left: Input, Right: Target",
"_____no_output_____"
]
],
[
[
"dls.show_batch()",
"_____no_output_____"
],
[
"b = dls.one_batch()\nexplode_types(b)",
"_____no_output_____"
]
],
[
[
"`StackUnstack` takes cares of stacking the list of images into a fat tensor, and unstacking them at the end, we will need to modify our loss function to take a list of tensors as input and target.",
"_____no_output_____"
],
[
"## Simple model",
"_____no_output_____"
]
],
[
[
"model = StackUnstack(SimpleModel())",
"_____no_output_____"
]
],
[
[
"As the `ImageSeq` is a `tuple` of images, we will need to stack them to compute loss.",
"_____no_output_____"
]
],
[
[
"learn = Learner(dls, model, loss_func=loss_func, cbs=[]).to_fp16()",
"_____no_output_____"
]
],
[
[
"I have a weird bug that if I use `nn.LeakyReLU` after doing `learn.lr_find()` the model does not train (the loss get stucked).",
"_____no_output_____"
]
],
[
[
"x,y = dls.one_batch()",
"_____no_output_____"
],
[
"learn.lr_find()",
"_____no_output_____"
],
[
"learn.fit_one_cycle(10, 1e-4)",
"_____no_output_____"
],
[
"p,t = learn.get_preds()",
"_____no_output_____"
]
],
[
[
"As you can see, the results is a list of 5 tensors with 100 samples each.",
"_____no_output_____"
]
],
[
[
"len(p), p[0].shape",
"_____no_output_____"
],
[
"def show_res(t, idx):\n im_seq = ImageSeq.create([t[i][idx] for i in range(5)])\n im_seq.show(figsize=(8,4));",
"_____no_output_____"
],
[
"k = random.randint(0,100)\nshow_res(t,k)\nshow_res(p,k)",
"_____no_output_____"
]
],
[
[
"## A bigger Decoder",
"_____no_output_____"
],
[
"We will pass:\n- `blur`: to use blur on the upsampling path (this is done by using and a poolling layer and a replication)\n- `attn`: to include a self attention layer on the decoder",
"_____no_output_____"
]
],
[
[
"model2 = StackUnstack(SimpleModel(szs=[16,64,96], act=partial(nn.LeakyReLU, 0.2, inplace=True),blur=True, attn=True))",
"_____no_output_____"
]
],
[
[
"We have to reduce batch size as the self attention layer is heavy.",
"_____no_output_____"
]
],
[
[
"dls = DataLoaders.from_dsets(train_tl, valid_tl, bs=8,\n after_batch=[Normalize.from_stats(imagenet_stats[0][0], \n imagenet_stats[1][0])]).cuda()",
"_____no_output_____"
],
[
"learn2 = Learner(dls, model2, loss_func=loss_func, cbs=[]).to_fp16()",
"_____no_output_____"
],
[
"learn2.lr_find()",
"_____no_output_____"
],
[
"learn2.fit_one_cycle(10, 1e-4)",
"_____no_output_____"
],
[
"p,t = learn2.get_preds()",
"_____no_output_____"
]
],
[
[
"As you can see, the results is a list of 5 tensors with 100 samples each.",
"_____no_output_____"
]
],
[
[
"len(p), p[0].shape",
"_____no_output_____"
],
[
"def show_res(t, idx):\n im_seq = ImageSeq.create([t[i][idx] for i in range(5)])\n im_seq.show(figsize=(8,4));",
"_____no_output_____"
],
[
"k = random.randint(0,100)\nshow_res(t,k)\nshow_res(p,k)",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
4a5949d39fed60199e430a0380b1167df18b1017
| 8,468 |
ipynb
|
Jupyter Notebook
|
skin_cancer.ipynb
|
yunjung-lee/class_python_data
|
67ceab73e67ec63d408894a6ab016a8d25a4e30b
|
[
"MIT"
] | null | null | null |
skin_cancer.ipynb
|
yunjung-lee/class_python_data
|
67ceab73e67ec63d408894a6ab016a8d25a4e30b
|
[
"MIT"
] | null | null | null |
skin_cancer.ipynb
|
yunjung-lee/class_python_data
|
67ceab73e67ec63d408894a6ab016a8d25a4e30b
|
[
"MIT"
] | null | null | null | 28.13289 | 239 | 0.467052 |
[
[
[
"<a href=\"https://colab.research.google.com/github/yunjung-lee/class_python_data/blob/master/skin_cancer.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"########## skin cancer in kaggle dataset\n\n#### used knn ,dropout,save file",
"_____no_output_____"
]
],
[
[
"!git clone https://github.com/yunjung-lee/python_mini_project_skin_cancer.git",
"_____no_output_____"
]
],
[
[
"!git clone : 깃을 접속함\nhttps://github.com/yunjung-lee/python_mini_project_skin_cancer.git : 깃의 주소\n",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport tensorflow as tf\nfrom sklearn.model_selection import train_test_split\nimport sklearn.preprocessing",
"_____no_output_____"
]
],
[
[
"opne data(data name : hmnist_28_28_RGB.csv)",
"_____no_output_____"
]
],
[
[
"data = np.loadtxt('drive/Colab Notebooks/dataset/hmnist_28_28_RGB.csv', dtype = np.float32, delimiter = \",\", skiprows = 1,encoding = \"utf-8\")\n# print(data)\nxdata = data[:,:-1]\nydata = data[:,[-1]]\n\n",
"_____no_output_____"
]
],
[
[
"one_hot encoding",
"_____no_output_____"
]
],
[
[
"one_hot = sklearn.preprocessing.LabelBinarizer()\nydata = one_hot.fit_transform(ydata)",
"_____no_output_____"
]
],
[
[
"train_ test_split",
"_____no_output_____"
]
],
[
[
"X_train, X_test, y_train, y_test = train_test_split(xdata, ydata, test_size=0.33, random_state=42)",
"_____no_output_____"
]
],
[
[
"nn-learning&drop-out 선언부\noutput에서 sigmoid 사용에 softmax_cross_entropy_with_logits를 사용",
"_____no_output_____"
]
],
[
[
"x = tf.placeholder(tf.float32, [None,28*28*3])\ny = tf.placeholder(tf.float32, [None,7])\nkeep_prob=tf.placeholder(tf.float32)\n\nw1 = tf.Variable(tf.random_normal([28*28*3,256],stddev=0.01))\nL1 = tf.nn.relu(tf.matmul(x,w1))\nL1=tf.nn.dropout(L1,keep_prob)\n\nw2 = tf.Variable(tf.random_normal([256,256],stddev=0.01))\nL2 = tf.nn.relu(tf.matmul(L1,w2))\nL2=tf.nn.dropout(L2,keep_prob)\n\nw3 = tf.Variable(tf.random_normal([256,7],stddev=0.01))\nmodel = tf.matmul(L2,w3)\n\ncost = tf.reduce_mean(tf.nn.softmax_cross_entropy_with_logits(logits=model,labels=y))\noptimizer = tf.train.AdamOptimizer(0.001).minimize(cost)\n",
"_____no_output_____"
]
],
[
[
"신경망 모델 학습\n\n batch size를 100으로 두고 drop-out을 0.8로 설정하여 진행",
"_____no_output_____"
]
],
[
[
"init = tf.global_variables_initializer()\nsaver = tf.train.Saver()\nsess = tf.Session()\nsess.run(init)\n\nbatch_size = 100\ntotal_batch = int(len(X_train[0])/batch_size)\nfor epoch in range(20):\n total_cost = 0\n\n for i in range(total_batch):\n _, cv = sess.run([optimizer,cost],feed_dict={x:X_train,y:y_train,keep_prob:0.8})\n total_cost += cv\n# print(\"epoch :\", \" %d\" % (epoch+1), \"avg cost : \",\"{:.3f}\".format(total_cost/total_batch))\n is_correct = tf.equal(tf.argmax(model,1), tf.argmax(y,1))\n accuracy = tf.reduce_mean(tf.cast(is_correct, dtype=tf.float32))\nprint(\"정확도 : \", sess.run(accuracy,feed_dict={x:X_test,y:y_test,keep_prob:1.0}))\n\nsaver.save(sess, 'drive/Colab Notebooks/datasetcnn_session')\nsess.close()\n",
"정확도 : 0.66596067\n"
]
],
[
[
"결과 avg cost : 2.989\n정확도 : 0.66596067\nepoch : 2 avg cost : 1.084\n정확도 : 0.66596067\nepoch : 3 avg cost : 1.006\n정확도 : 0.66596067\nepoch : 4 avg cost : 0.992\n정확도 : 0.66596067\nepoch : 5 avg cost : 0.959\n정확도 : 0.66596067\nepoch : 6 avg cost : 0.940\n정확도 : 0.66596067\nepoch : 7 avg cost : 0.927\n정확도 : 0.66596067\nepoch : 8 avg cost : 0.921\n정확도 : 0.66596067\nepoch : 9 avg cost : 1.014\n정확도 : 0.66596067\nepoch : 10 avg cost : 0.955\n정확도 : 0.66596067\nepoch : 11 avg cost : 0.938\n정확도 : 0.66596067\nepoch : 12 avg cost : 0.935\n정확도 : 0.66596067\nepoch : 13 avg cost : 0.921\n정확도 : 0.66596067\nepoch : 14 avg cost : 0.921\n정확도 : 0.66596067\nepoch : 15 avg cost : 0.920\n정확도 : 0.66596067\nepoch : 16 avg cost : 0.952\n정확도 : 0.66596067\nepoch : 17 avg cost : 0.916\n정확도 : 0.66656584\nepoch : 18 avg cost : 0.911\n정확도 : 0.66807866\nepoch : 19 avg cost : 0.917\n정확도 : 0.66717094\nepoch : 20 avg cost : 0.903\n정확도 : 0.66747355",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
4a59557fd3393f38169d345f4ababd282ec34a1b
| 39,458 |
ipynb
|
Jupyter Notebook
|
Encoder/train_encoder_stylegan3.ipynb
|
TeamTechArt/HanGAN
|
45f1f4fa81a62b47a8224f50aad1bab03624a3f5
|
[
"MIT"
] | 2 |
2022-03-18T03:51:51.000Z
|
2022-03-19T12:03:10.000Z
|
Encoder/train_encoder_stylegan3.ipynb
|
TeamTechArt/AKMA
|
45f1f4fa81a62b47a8224f50aad1bab03624a3f5
|
[
"MIT"
] | null | null | null |
Encoder/train_encoder_stylegan3.ipynb
|
TeamTechArt/AKMA
|
45f1f4fa81a62b47a8224f50aad1bab03624a3f5
|
[
"MIT"
] | null | null | null | 36.43398 | 474 | 0.431649 |
[
[
[
"https://github.com/soushirou/stylegan3-encoder",
"_____no_output_____"
]
],
[
[
"!pip install -q condacolab\nimport condacolab\n#condacolab.install_from_url('https://repo.anaconda.com/miniconda/Miniconda3-py39_4.10.3-Linux-x86_64.sh'\ncondacolab.install_from_url('https://repo.anaconda.com/miniconda/Miniconda3-py37_4.10.3-Linux-x86_64.sh')",
"\u001b[33mWARNING: Running pip as the 'root' user can result in broken permissions and conflicting behaviour with the system package manager. It is recommended to use a virtual environment instead: https://pip.pypa.io/warnings/venv\u001b[0m\n✨🍰✨ Everything looks OK!\n"
],
[
"!git clone https://github.com/NVlabs/stylegan3.git\n!cp -r /content/drive/MyDrive/colabdata/project/teamtechart/encoder_train/code_encoder_0223/pixel2style2pixel /content/pixel2style2pixel ",
"Cloning into 'stylegan3'...\nremote: Enumerating objects: 193, done.\u001b[K\nremote: Total 193 (delta 0), reused 0 (delta 0), pack-reused 193\u001b[K\nReceiving objects: 100% (193/193), 4.18 MiB | 7.02 MiB/s, done.\nResolving deltas: 100% (86/86), done.\n"
],
[
"#!cp -r /content/drive/MyDrive/colabdata/project/teamtechart/encoder_train/code_encoder_022400/stylegan3-encoder /content/stylegan3-encoder04\n!cp -r /content/drive/MyDrive/colabdata/project/teamtechart/encoder_train/code_encoder_0223/stylegan3-encoder/stylegan3-encoder /content/stylegan3-encoder03",
"_____no_output_____"
],
[
"!pip install lpips",
"Collecting lpips\n Downloading lpips-0.1.4-py3-none-any.whl (53 kB)\n\u001b[?25l\r\u001b[K |██████ | 10 kB 30.8 MB/s eta 0:00:01\r\u001b[K |████████████▏ | 20 kB 3.6 MB/s eta 0:00:01\r\u001b[K |██████████████████▎ | 30 kB 4.1 MB/s eta 0:00:01\r\u001b[K |████████████████████████▍ | 40 kB 3.5 MB/s eta 0:00:01\r\u001b[K |██████████████████████████████▌ | 51 kB 4.0 MB/s eta 0:00:01\r\u001b[K |████████████████████████████████| 53 kB 1.6 MB/s \n\u001b[?25hRequirement already satisfied: torchvision>=0.2.1 in /usr/local/lib/python3.7/dist-packages (from lpips) (0.11.1+cu111)\nRequirement already satisfied: numpy>=1.14.3 in /usr/local/lib/python3.7/dist-packages (from lpips) (1.21.5)\nRequirement already satisfied: tqdm>=4.28.1 in /usr/local/lib/python3.7/dist-packages (from lpips) (4.63.0)\nRequirement already satisfied: torch>=0.4.0 in /usr/local/lib/python3.7/dist-packages (from lpips) (1.10.0+cu111)\nRequirement already satisfied: scipy>=1.0.1 in /usr/local/lib/python3.7/dist-packages (from lpips) (1.4.1)\nRequirement already satisfied: typing-extensions in /usr/local/lib/python3.7/dist-packages (from torch>=0.4.0->lpips) (3.10.0.2)\nRequirement already satisfied: pillow!=8.3.0,>=5.3.0 in /usr/local/lib/python3.7/dist-packages (from torchvision>=0.2.1->lpips) (7.1.2)\nInstalling collected packages: lpips\nSuccessfully installed lpips-0.1.4\n"
],
[
"import torch\nimport pickle\nimport sys\nimport matplotlib.pyplot as plt\nimport torchvision",
"_____no_output_____"
],
[
"if torch.cuda.is_available():\n print(torch.cuda.get_device_name())",
"Tesla V100-SXM2-16GB\n"
],
[
"sys.path.append('/content/stylegan3')\nsys.path.append('/content/stylegan3-encoder')\n",
"_____no_output_____"
],
[
"!pip install ninja",
"Collecting ninja\n Downloading ninja-1.10.2.3-py2.py3-none-manylinux_2_5_x86_64.manylinux1_x86_64.whl (108 kB)\n\u001b[K |████████████████████████████████| 108 kB 5.5 MB/s \n\u001b[?25hInstalling collected packages: ninja\nSuccessfully installed ninja-1.10.2.3\n"
],
[
"!cp -r /content/drive/Shareddrives/\\ TeamTechArt_Dataset/YOUTUBE-WAVE/wave44_high /content/wave44_high",
"_____no_output_____"
],
[
"!cp -r /content/drive/MyDrive/colabdata/project/teamtechart/encoder/encoder_ire /content/pretrained",
"_____no_output_____"
],
[
"%cd stylegan3-encoder03",
"/content/stylegan3-encoder03\n"
],
[
"!cp -r stylegan3-encoder stylegan3-encoder_cp",
"_____no_output_____"
],
[
"!python /content/stylegan3-encoder03/train.py \\\n --outdir /content/result_cp \\ # Saved path\n --encoder base \\\n --data /content/wave44_high \\ # Used data\n --gpus 1 \\ # The number of gpu used\n --batch 4 \\ # The number of batchsize\n --training_steps 100000\\\n --val_steps 5000\\\n --workers 2 \\ # The number of cpu to call data\n --enc_layers 2\\ # The number of layers to use Transformer network.\n --lr 0.001 \\ # learning rate\n --lpips_lambda 0.65\\ # Additional learning rate for lpips loss\n --moco_lambda 0.2\\ # Additional learning rate for moco_lambda\n --d_lambda 0.0007\\ # Additional learning rate for Discriminator\n --generator /content/drive/MyDrive/colabdata/project/teamtechart/stylegan3model/wave-44-network-snapshot-002000.pkl \\\n --img_snshot_steps 100\\\n --net_snshot_steps 1500\\\n #--g_reg\n #--resume_pkl /content/result/00001-base-wave-256x256-gpus1-batch16/network_snapshots/network-snapshot-180000.pkl \\",
"\nTraining options:\n{\n \"model_architecture\": \"base\",\n \"dataset_dir\": \"/content/wave44_high\",\n \"num_gpus\": 1,\n \"batch_size\": 4,\n \"batch_gpu\": 4,\n \"generator_pkl\": \"/content/drive/MyDrive/colabdata/project/teamtechart/stylegan3model/wave-44-network-snapshot-002000.pkl\",\n \"w_avg\": false,\n \"num_encoder_layers\": 2,\n \"val_dataset_dir\": null,\n \"training_steps\": 100000,\n \"val_steps\": 5000,\n \"print_steps\": 50,\n \"tensorboard_steps\": 50,\n \"image_snapshot_steps\": 100,\n \"network_snapshot_steps\": 3000,\n \"learning_rate\": 0.001,\n \"l2_lambda\": 1.0,\n \"lpips_lambda\": 0.65,\n \"moco_lambda\": 0.2,\n \"reg_lambda\": 0.0,\n \"gan_lambda\": 0.0,\n \"edit_lambda\": 0.0,\n \"d_lambda\": 0.001,\n \"random_seed\": 0,\n \"num_workers\": 2,\n \"resume_pkl\": null,\n \"run_dir\": \"/content/result_cp/00001-base-wave44_high-gpus1-batch4\"\n}\n\nCreating output directory...\nLaunching processes...\nLoading training set...\n\nNum images: 1000\nImage shape: torch.Size([3, 256, 256])\n\nConstructing networks...\nInitialize loss...\nLoading MOCO model from path: /content/pretrained/moco_v2_800ep_pretrain.pt\nInitialize optimizer...\nInitialize tensorboard logs...\nSetting up PyTorch plugin \"filtered_lrelu_plugin\"... Done.\nSetting up PyTorch plugin \"bias_act_plugin\"... Done.\n\nCurrent batch step: 0\n16.90 sec\n{'d_loss': 0.001436164602637291,\n 'g_loss': 0.0006570785772055387,\n 'l2': 0.579058051109314,\n 'loss': 1.2591338157653809,\n 'lpips': 0.8187288045883179,\n 'moco': 0.7395098209381104,\n 'moco_improve': -0.7395098209381104}\nSaving image snapshot at step 0...\nSaving netowrk snapshot at step 0...\n\nCurrent batch step: 50\n62.41 sec\n{'d_loss': 0.0015441415598616004,\n 'g_loss': 0.013267168775200844,\n 'l2': 0.26337289810180664,\n 'loss': 0.8666693568229675,\n 'lpips': 0.7201738357543945,\n 'moco': 0.6759175658226013,\n 'moco_improve': -0.6759176105260849}\n\nCurrent batch step: 100\n58.84 sec\n{'d_loss': 0.0029490163215086795,\n 'g_loss': 0.0002818876237142831,\n 'l2': 0.3965882956981659,\n 'loss': 0.944462239742279,\n 'lpips': 0.6628456115722656,\n 'moco': 0.5851215720176697,\n 'moco_improve': -0.5851215720176697}\nSaving image snapshot at step 100...\n\nCurrent batch step: 150\n59.06 sec\n{'d_loss': 0.0014462385152000934,\n 'g_loss': 0.00034434316330589354,\n 'l2': 0.13948166370391846,\n 'loss': 0.6258171796798706,\n 'lpips': 0.5934539437294006,\n 'moco': 0.5029522776603699,\n 'moco_improve': -0.5029523074626923}\n\nCurrent batch step: 200\n58.85 sec\n{'d_loss': 0.0016946368268691003,\n 'g_loss': 0.0002484219439793378,\n 'l2': 0.3269107937812805,\n 'loss': 0.8139296770095825,\n 'lpips': 0.593259334564209,\n 'moco': 0.507001519203186,\n 'moco_improve': -0.5070014894008636}\nSaving image snapshot at step 200...\n\nCurrent batch step: 250\n62.94 sec\n{'d_loss': 0.0011001931852661073,\n 'g_loss': 0.012012398801743984,\n 'l2': 0.4265762269496918,\n 'loss': 0.898638904094696,\n 'lpips': 0.6023520231246948,\n 'moco': 0.4026692509651184,\n 'moco_improve': -0.4026692062616348}\n\nCurrent batch step: 300\n58.84 sec\n{'d_loss': 0.002134419948561117,\n 'g_loss': 0.0003700070083141327,\n 'l2': 0.17885112762451172,\n 'loss': 0.6900913119316101,\n 'lpips': 0.6060736179351807,\n 'moco': 0.5864618420600891,\n 'moco_improve': -0.5864618867635727}\nSaving image snapshot at step 300...\n\nCurrent batch step: 350\n59.05 sec\n{'d_loss': 0.0017904429842019454,\n 'g_loss': 0.00020692931138910353,\n 'l2': 0.22743293642997742,\n 'loss': 0.7933739423751831,\n 'lpips': 0.6754913926124573,\n 'moco': 0.6343579888343811,\n 'moco_improve': -0.6343580186367035}\n\nCurrent batch step: 400\n58.80 sec\n{'d_loss': 0.0020843629972659983,\n 'g_loss': 0.00019416552095208317,\n 'l2': 0.16738931834697723,\n 'loss': 0.6098400950431824,\n 'lpips': 0.5579326748847961,\n 'moco': 0.3989727199077606,\n 'moco_improve': -0.3989727348089218}\nSaving image snapshot at step 400...\n\nCurrent batch step: 450\n59.08 sec\n{'d_loss': 0.0031515100126853213,\n 'g_loss': 8.764439553488046e-05,\n 'l2': 0.4040870666503906,\n 'loss': 0.8982693552970886,\n 'lpips': 0.6070798635482788,\n 'moco': 0.4979017972946167,\n 'moco_improve': -0.4979018047451973}\n\nCurrent batch step: 500\n62.77 sec\n{'d_loss': 0.0012481075682444498,\n 'g_loss': 0.0016332785598933697,\n 'l2': 14.4644193649292,\n 'loss': 15.044516563415527,\n 'lpips': 0.7418944835662842,\n 'moco': 0.48932939767837524,\n 'moco_improve': -0.48932936415076256}\nSaving image snapshot at step 500...\n\nCurrent batch step: 550\n59.04 sec\n{'d_loss': 0.002033252181718126,\n 'g_loss': 0.00022489906405098736,\n 'l2': 0.3738184869289398,\n 'loss': 0.8308008909225464,\n 'lpips': 0.5926247239112854,\n 'moco': 0.3588818311691284,\n 'moco_improve': -0.3588818460702896}\n\nCurrent batch step: 600\n58.90 sec\n{'d_loss': 0.0017812805526773445,\n 'g_loss': 0.0002370347356190905,\n 'l2': 0.17057105898857117,\n 'loss': 0.6188242435455322,\n 'lpips': 0.5699413418769836,\n 'moco': 0.3889567255973816,\n 'moco_improve': -0.3889567106962204}\nSaving image snapshot at step 600...\n\nCurrent batch step: 650\n59.13 sec\n{'d_loss': 0.0022238825040403754,\n 'g_loss': 0.0001464279630454257,\n 'l2': 0.2709035575389862,\n 'loss': 0.7544607520103455,\n 'lpips': 0.6067314743995667,\n 'moco': 0.4459086060523987,\n 'moco_improve': -0.4459085986018181}\n\nCurrent batch step: 700\n58.91 sec\n{'d_loss': 0.001706369497696869,\n 'g_loss': 0.00028789270436391234,\n 'l2': 0.22087368369102478,\n 'loss': 0.7088747620582581,\n 'lpips': 0.6158475875854492,\n 'moco': 0.4385009706020355,\n 'moco_improve': -0.4385010525584221}\nSaving image snapshot at step 700...\n\nCurrent batch step: 750\n63.00 sec\n{'d_loss': 0.001483709886088036,\n 'g_loss': 0.0003770859620999545,\n 'l2': 0.20758625864982605,\n 'loss': 0.6420116424560547,\n 'lpips': 0.5560145378112793,\n 'moco': 0.365079790353775,\n 'moco_improve': -0.3650798052549362}\n\nCurrent batch step: 800\n58.84 sec\n{'d_loss': 0.002397095537162386,\n 'g_loss': 0.00012507857172749937,\n 'l2': 0.22545157372951508,\n 'loss': 0.6622551083564758,\n 'lpips': 0.5706579089164734,\n 'moco': 0.32937943935394287,\n 'moco_improve': -0.32937954366207123}\nSaving image snapshot at step 800...\n\nCurrent batch step: 850\n59.11 sec\n{'d_loss': 0.0019449359679128975,\n 'g_loss': 0.00025972267030738294,\n 'l2': 0.3472118079662323,\n 'loss': 0.8088719844818115,\n 'lpips': 0.5979439616203308,\n 'moco': 0.3649832010269165,\n 'moco_improve': -0.3649832010269165}\n\nCurrent batch step: 900\n58.84 sec\n{'d_loss': 0.0016533204761799425,\n 'g_loss': 0.00033778854412958026,\n 'l2': 0.25154584646224976,\n 'loss': 0.7330566644668579,\n 'lpips': 0.6409695148468018,\n 'moco': 0.3244030475616455,\n 'moco_improve': -0.32440315186977386}\nSaving image snapshot at step 900...\n\nCurrent batch step: 950\n59.10 sec\n{'d_loss': 0.0019446029109531082,\n 'g_loss': 0.0002379280631430447,\n 'l2': 0.3705904483795166,\n 'loss': 0.8328309059143066,\n 'lpips': 0.5939850807189941,\n 'moco': 0.38075077533721924,\n 'moco_improve': -0.380750834941864}\n\nCurrent batch step: 1000\n62.64 sec\n{'d_loss': 0.0021957182325422764,\n 'g_loss': 0.0001953242754098028,\n 'l2': 0.17528849840164185,\n 'loss': 0.5954281091690063,\n 'lpips': 0.5428906679153442,\n 'moco': 0.33630338311195374,\n 'moco_improve': -0.33630336821079254}\nSaving image snapshot at step 1000...\n\nCurrent batch step: 1050\n59.07 sec\n{'d_loss': 0.002517346787499264,\n 'g_loss': 0.0001646963064558804,\n 'l2': 0.15129798650741577,\n 'loss': 0.5683995485305786,\n 'lpips': 0.5242962837219238,\n 'moco': 0.38154491782188416,\n 'moco_improve': -0.38154496252536774}\n\nCurrent batch step: 1100\n58.81 sec\n{'d_loss': 0.0016310800274368376,\n 'g_loss': 0.00041159294778481126,\n 'l2': 0.24249742925167084,\n 'loss': 0.758398175239563,\n 'lpips': 0.6543539762496948,\n 'moco': 0.45285338163375854,\n 'moco_improve': -0.4528534412384033}\nSaving image snapshot at step 1100...\n\nCurrent batch step: 1150\n59.22 sec\n{'d_loss': 0.0024401706032222137,\n 'g_loss': 0.00011260298197157681,\n 'l2': 0.13720300793647766,\n 'loss': 0.5573955774307251,\n 'lpips': 0.544468879699707,\n 'moco': 0.33143913745880127,\n 'moco_improve': -0.33143918216228485}\n\nCurrent batch step: 1200\n58.85 sec\n{'d_loss': 0.00182701155426912,\n 'g_loss': 0.00023728315136395395,\n 'l2': 0.23926222324371338,\n 'loss': 0.6656973958015442,\n 'lpips': 0.5442364811897278,\n 'moco': 0.3634074926376343,\n 'moco_improve': -0.36340755224227905}\nSaving image snapshot at step 1200...\n\nCurrent batch step: 1250\n62.95 sec\n{'d_loss': 0.001754128075845074,\n 'g_loss': 0.00027127619250677526,\n 'l2': 0.17531105875968933,\n 'loss': 0.5632606148719788,\n 'lpips': 0.5273750424385071,\n 'moco': 0.22577901184558868,\n 'moco_improve': -0.2257789969444275}\n\nCurrent batch step: 1300\n58.81 sec\n{'d_loss': 0.0022228085072129034,\n 'g_loss': 0.00016076842439360917,\n 'l2': 0.1420900672674179,\n 'loss': 0.532794713973999,\n 'lpips': 0.5242006182670593,\n 'moco': 0.2498713880777359,\n 'moco_improve': -0.2498714178800583}\nSaving image snapshot at step 1300...\n\nCurrent batch step: 1350\n59.11 sec\n{'d_loss': 0.0015328187437262386,\n 'g_loss': 0.0003547806409187615,\n 'l2': 0.210394024848938,\n 'loss': 0.6186249256134033,\n 'lpips': 0.5358916521072388,\n 'moco': 0.2995068430900574,\n 'moco_improve': -0.2995068430900574}\n\nCurrent batch step: 1400\n58.81 sec\n{'d_loss': 0.002267789706820622,\n 'g_loss': 0.00013354624388739467,\n 'l2': 0.1870475709438324,\n 'loss': 0.6011337637901306,\n 'lpips': 0.5334151983261108,\n 'moco': 0.33683153986930847,\n 'moco_improve': -0.3368315249681473}\nSaving image snapshot at step 1400...\n\nCurrent batch step: 1450\n59.09 sec\n{'d_loss': 0.008840371898259036,\n 'g_loss': 0.03661893308162689,\n 'l2': 0.1865495890378952,\n 'loss': 0.6231440305709839,\n 'lpips': 0.5590264797210693,\n 'moco': 0.36613619327545166,\n 'moco_improve': -0.36613619327545166}\n\nCurrent batch step: 1500\n62.64 sec\n{'d_loss': nan,\n 'g_loss': nan,\n 'l2': nan,\n 'loss': nan,\n 'lpips': nan,\n 'moco': nan,\n 'moco_improve': nan}\nSaving image snapshot at step 1500...\n\nCurrent batch step: 1550\n58.50 sec\n{'d_loss': nan,\n 'g_loss': nan,\n 'l2': nan,\n 'loss': nan,\n 'lpips': nan,\n 'moco': nan,\n 'moco_improve': nan}\n\nCurrent batch step: 1600\n58.41 sec\n{'d_loss': nan,\n 'g_loss': nan,\n 'l2': nan,\n 'loss': nan,\n 'lpips': nan,\n 'moco': nan,\n 'moco_improve': nan}\nSaving image snapshot at step 1600...\n\nCurrent batch step: 1650\n58.50 sec\n{'d_loss': nan,\n 'g_loss': nan,\n 'l2': nan,\n 'loss': nan,\n 'lpips': nan,\n 'moco': nan,\n 'moco_improve': nan}\n\nCurrent batch step: 1700\n58.40 sec\n{'d_loss': nan,\n 'g_loss': nan,\n 'l2': nan,\n 'loss': nan,\n 'lpips': nan,\n 'moco': nan,\n 'moco_improve': nan}\nSaving image snapshot at step 1700...\n\nCurrent batch step: 1750\n62.42 sec\n{'d_loss': nan,\n 'g_loss': nan,\n 'l2': nan,\n 'loss': nan,\n 'lpips': nan,\n 'moco': nan,\n 'moco_improve': nan}\n\nCurrent batch step: 1800\n58.39 sec\n{'d_loss': nan,\n 'g_loss': nan,\n 'l2': nan,\n 'loss': nan,\n 'lpips': nan,\n 'moco': nan,\n 'moco_improve': nan}\nSaving image snapshot at step 1800...\n\nCurrent batch step: 1850\n58.54 sec\n{'d_loss': nan,\n 'g_loss': nan,\n 'l2': nan,\n 'loss': nan,\n 'lpips': nan,\n 'moco': nan,\n 'moco_improve': nan}\n\nCurrent batch step: 1900\n58.35 sec\n{'d_loss': nan,\n 'g_loss': nan,\n 'l2': nan,\n 'loss': nan,\n 'lpips': nan,\n 'moco': nan,\n 'moco_improve': nan}\nSaving image snapshot at step 1900...\n\nCurrent batch step: 1950\n58.50 sec\n{'d_loss': nan,\n 'g_loss': nan,\n 'l2': nan,\n 'loss': nan,\n 'lpips': nan,\n 'moco': nan,\n 'moco_improve': nan}\n\nCurrent batch step: 2000\n62.24 sec\n{'d_loss': nan,\n 'g_loss': nan,\n 'l2': nan,\n 'loss': nan,\n 'lpips': nan,\n 'moco': nan,\n 'moco_improve': nan}\nSaving image snapshot at step 2000...\n\nCurrent batch step: 2050\n58.47 sec\n{'d_loss': nan,\n 'g_loss': nan,\n 'l2': nan,\n 'loss': nan,\n 'lpips': nan,\n 'moco': nan,\n 'moco_improve': nan}\n\nCurrent batch step: 2100\n58.40 sec\n{'d_loss': nan,\n 'g_loss': nan,\n 'l2': nan,\n 'loss': nan,\n 'lpips': nan,\n 'moco': nan,\n 'moco_improve': nan}\nSaving image snapshot at step 2100...\n\nCurrent batch step: 2150\n58.47 sec\n{'d_loss': nan,\n 'g_loss': nan,\n 'l2': nan,\n 'loss': nan,\n 'lpips': nan,\n 'moco': nan,\n 'moco_improve': nan}\n\nCurrent batch step: 2200\n58.37 sec\n{'d_loss': nan,\n 'g_loss': nan,\n 'l2': nan,\n 'loss': nan,\n 'lpips': nan,\n 'moco': nan,\n 'moco_improve': nan}\nSaving image snapshot at step 2200...\n\nCurrent batch step: 2250\n62.33 sec\n{'d_loss': nan,\n 'g_loss': nan,\n 'l2': nan,\n 'loss': nan,\n 'lpips': nan,\n 'moco': nan,\n 'moco_improve': nan}\n\nCurrent batch step: 2300\n58.30 sec\n{'d_loss': nan,\n 'g_loss': nan,\n 'l2': nan,\n 'loss': nan,\n 'lpips': nan,\n 'moco': nan,\n 'moco_improve': nan}\nSaving image snapshot at step 2300...\n\nCurrent batch step: 2350\n58.49 sec\n{'d_loss': nan,\n 'g_loss': nan,\n 'l2': nan,\n 'loss': nan,\n 'lpips': nan,\n 'moco': nan,\n 'moco_improve': nan}\n\nCurrent batch step: 2400\n58.52 sec\n{'d_loss': nan,\n 'g_loss': nan,\n 'l2': nan,\n 'loss': nan,\n 'lpips': nan,\n 'moco': nan,\n 'moco_improve': nan}\nSaving image snapshot at step 2400...\n\nCurrent batch step: 2450\n58.42 sec\n{'d_loss': nan,\n 'g_loss': nan,\n 'l2': nan,\n 'loss': nan,\n 'lpips': nan,\n 'moco': nan,\n 'moco_improve': nan}\n\nCurrent batch step: 2500\n62.20 sec\n{'d_loss': nan,\n 'g_loss': nan,\n 'l2': nan,\n 'loss': nan,\n 'lpips': nan,\n 'moco': nan,\n 'moco_improve': nan}\nSaving image snapshot at step 2500...\n\nCurrent batch step: 2550\n58.55 sec\n{'d_loss': nan,\n 'g_loss': nan,\n 'l2': nan,\n 'loss': nan,\n 'lpips': nan,\n 'moco': nan,\n 'moco_improve': nan}\n\nCurrent batch step: 2600\n58.31 sec\n{'d_loss': nan,\n 'g_loss': nan,\n 'l2': nan,\n 'loss': nan,\n 'lpips': nan,\n 'moco': nan,\n 'moco_improve': nan}\nSaving image snapshot at step 2600...\n\nCurrent batch step: 2650\n58.46 sec\n{'d_loss': nan,\n 'g_loss': nan,\n 'l2': nan,\n 'loss': nan,\n 'lpips': nan,\n 'moco': nan,\n 'moco_improve': nan}\n\nCurrent batch step: 2700\n58.40 sec\n{'d_loss': nan,\n 'g_loss': nan,\n 'l2': nan,\n 'loss': nan,\n 'lpips': nan,\n 'moco': nan,\n 'moco_improve': nan}\nSaving image snapshot at step 2700...\n\nCurrent batch step: 2750\n62.35 sec\n{'d_loss': nan,\n 'g_loss': nan,\n 'l2': nan,\n 'loss': nan,\n 'lpips': nan,\n 'moco': nan,\n 'moco_improve': nan}\n\nCurrent batch step: 2800\n58.33 sec\n{'d_loss': nan,\n 'g_loss': nan,\n 'l2': nan,\n 'loss': nan,\n 'lpips': nan,\n 'moco': nan,\n 'moco_improve': nan}\nSaving image snapshot at step 2800...\n\nCurrent batch step: 2850\n58.48 sec\n{'d_loss': nan,\n 'g_loss': nan,\n 'l2': nan,\n 'loss': nan,\n 'lpips': nan,\n 'moco': nan,\n 'moco_improve': nan}\n\nCurrent batch step: 2900\n58.33 sec\n{'d_loss': nan,\n 'g_loss': nan,\n 'l2': nan,\n 'loss': nan,\n 'lpips': nan,\n 'moco': nan,\n 'moco_improve': nan}\nSaving image snapshot at step 2900...\n\nCurrent batch step: 2950\n58.50 sec\n{'d_loss': nan,\n 'g_loss': nan,\n 'l2': nan,\n 'loss': nan,\n 'lpips': nan,\n 'moco': nan,\n 'moco_improve': nan}\n\nCurrent batch step: 3000\n62.22 sec\n{'d_loss': nan,\n 'g_loss': nan,\n 'l2': nan,\n 'loss': nan,\n 'lpips': nan,\n 'moco': nan,\n 'moco_improve': nan}\nSaving image snapshot at step 3000...\nSaving netowrk snapshot at step 3000...\n\nCurrent batch step: 3050\n61.98 sec\n{'d_loss': nan,\n 'g_loss': nan,\n 'l2': nan,\n 'loss': nan,\n 'lpips': nan,\n 'moco': nan,\n 'moco_improve': nan}\n\nCurrent batch step: 3100\n58.29 sec\n{'d_loss': nan,\n 'g_loss': nan,\n 'l2': nan,\n 'loss': nan,\n 'lpips': nan,\n 'moco': nan,\n 'moco_improve': nan}\nSaving image snapshot at step 3100...\n\nCurrent batch step: 3150\n58.56 sec\n{'d_loss': nan,\n 'g_loss': nan,\n 'l2': nan,\n 'loss': nan,\n 'lpips': nan,\n 'moco': nan,\n 'moco_improve': nan}\n\nCurrent batch step: 3200\n58.34 sec\n{'d_loss': nan,\n 'g_loss': nan,\n 'l2': nan,\n 'loss': nan,\n 'lpips': nan,\n 'moco': nan,\n 'moco_improve': nan}\nSaving image snapshot at step 3200...\n\nCurrent batch step: 3250\n62.39 sec\n{'d_loss': nan,\n 'g_loss': nan,\n 'l2': nan,\n 'loss': nan,\n 'lpips': nan,\n 'moco': nan,\n 'moco_improve': nan}\n\nCurrent batch step: 3300\n58.29 sec\n{'d_loss': nan,\n 'g_loss': nan,\n 'l2': nan,\n 'loss': nan,\n 'lpips': nan,\n 'moco': nan,\n 'moco_improve': nan}\nSaving image snapshot at step 3300...\n\nCurrent batch step: 3350\n58.39 sec\n{'d_loss': nan,\n 'g_loss': nan,\n 'l2': nan,\n 'loss': nan,\n 'lpips': nan,\n 'moco': nan,\n 'moco_improve': nan}\n\nCurrent batch step: 3400\n58.28 sec\n{'d_loss': nan,\n 'g_loss': nan,\n 'l2': nan,\n 'loss': nan,\n 'lpips': nan,\n 'moco': nan,\n 'moco_improve': nan}\nSaving image snapshot at step 3400...\n\nAborted!\n^C\n"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a59590bbbb754d4d6eb8aaf6f12a6b48e55038f
| 577,478 |
ipynb
|
Jupyter Notebook
|
module4-rf-gb/LS_DS_234_Random_Forests_Gradient_Boosting.ipynb
|
will-cotton4/DS-Unit-2-Sprint-3-Classification-Validation
|
5adbe4fc3e3603cae139a2c1362246ac58935d57
|
[
"MIT"
] | null | null | null |
module4-rf-gb/LS_DS_234_Random_Forests_Gradient_Boosting.ipynb
|
will-cotton4/DS-Unit-2-Sprint-3-Classification-Validation
|
5adbe4fc3e3603cae139a2c1362246ac58935d57
|
[
"MIT"
] | null | null | null |
module4-rf-gb/LS_DS_234_Random_Forests_Gradient_Boosting.ipynb
|
will-cotton4/DS-Unit-2-Sprint-3-Classification-Validation
|
5adbe4fc3e3603cae139a2c1362246ac58935d57
|
[
"MIT"
] | null | null | null | 403.548567 | 167,157 | 0.947039 |
[
[
[
"<a href=\"https://colab.research.google.com/github/will-cotton4/DS-Unit-2-Sprint-3-Classification-Validation/blob/master/module4-rf-gb/LS_DS_234_Random_Forests_Gradient_Boosting.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"_Lambda School Data Science — Classification & Validation_ \n\n# Random Forests & Gradient Boosting",
"_____no_output_____"
],
[
"#### Gradient Boosting and Random Forest are often the best choice for “Spreadsheet Machine Learning.”\n- Meaning, [Tree Ensembles often have the best predictive accuracy](https://arxiv.org/abs/1708.05070) for supervised learning with structured, tabular data.\n- Because trees can fit non-linear, non-monotonic relationships, and interactions between features.\n- A single decision tree, grown to unlimited depth, will overfit. We solve this problem by ensembling trees, with bagging or boosting.\n- One-hot encoding isn’t the only way, and may not be the best way, of categorical encoding for tree ensembles.\n\n",
"_____no_output_____"
],
[
"### Links\n\n#### Decision Trees\n- A Visual Introduction to Machine Learning, [Part 1: A Decision Tree](http://www.r2d3.us/visual-intro-to-machine-learning-part-1/), and [Part 2: Bias and Variance](http://www.r2d3.us/visual-intro-to-machine-learning-part-2/)\n- [Decision Trees: Advantages & Disadvantages](https://christophm.github.io/interpretable-ml-book/tree.html#advantages-2)\n- [How decision trees work](https://brohrer.github.io/how_decision_trees_work.html)\n- [How a Russian mathematician constructed a decision tree - by hand - to solve a medical problem](http://fastml.com/how-a-russian-mathematician-constructed-a-decision-tree-by-hand-to-solve-a-medical-problem/)\n- [Let’s Write a Decision Tree Classifier from Scratch](https://www.youtube.com/watch?v=LDRbO9a6XPU)\n\n#### Random Forests\n- [Random Forests for Complete Beginners: The definitive guide to Random Forests and Decision Trees](https://victorzhou.com/blog/intro-to-random-forests/)\n- [Coloring with Random Forests](http://structuringtheunstructured.blogspot.com/2017/11/coloring-with-random-forests.html)\n- [Beware Default Random Forest Importances](https://explained.ai/rf-importance/index.html)\n\n#### Gradient Boosting\n- [A Gentle Introduction to the Gradient Boosting Algorithm for Machine Learning](https://machinelearningmastery.com/gentle-introduction-gradient-boosting-algorithm-machine-learning/)\n- [A Kaggle Master Explains Gradient Boosting](http://blog.kaggle.com/2017/01/23/a-kaggle-master-explains-gradient-boosting/)\n- [How to explain gradient boosting](https://explained.ai/gradient-boosting/index.html)\n\n#### Python libraries for Gradient Boosting\n- [scikit-learn Gradient Tree Boosting](https://scikit-learn.org/stable/modules/ensemble.html#gradient-boosting) — slower than other libraries, but [the next version may be better](https://twitter.com/amuellerml/status/1123613520426426368)\n - Anaconda: already installed\n - Google Colab: already installed\n- [xgboost](https://xgboost.readthedocs.io/en/latest/) — can accept missing values and enforce [monotonic constraints](https://xiaoxiaowang87.github.io/monotonicity_constraint/)\n - Anaconda, Mac/Linux: `conda install -c conda-forge xgboost`\n - Windows: `pip install xgboost`\n - Google Colab: already installed\n- [LightGBM](https://lightgbm.readthedocs.io/en/latest/) — can accept missing values and enforce [monotonic constraints](https://blog.datadive.net/monotonicity-constraints-in-machine-learning/)\n - Anaconda: `conda install -c conda-forge lightgbm`\n - Google Colab: already installed\n- [CatBoost](https://catboost.ai/) — can accept missing values and use [categorical features](https://catboost.ai/docs/concepts/algorithm-main-stages_cat-to-numberic.html) without preprocessing\n - Anaconda: `conda install -c conda-forge catboost`\n - Google Colab: `pip install catboost`\n\n#### Categorical encoding for trees\n- [Are categorical variables getting lost in your random forests?](https://roamanalytics.com/2016/10/28/are-categorical-variables-getting-lost-in-your-random-forests/)\n- [Beyond One-Hot: An Exploration of Categorical Variables](http://www.willmcginnis.com/2015/11/29/beyond-one-hot-an-exploration-of-categorical-variables/)\n- [Categorical Features and Encoding in Decision Trees](https://medium.com/data-design/visiting-categorical-features-and-encoding-in-decision-trees-53400fa65931)\n- [Coursera — How to Win a Data Science Competition: Learn from Top Kagglers — Concept of mean encoding](https://www.coursera.org/lecture/competitive-data-science/concept-of-mean-encoding-b5Gxv)\n- [Mean (likelihood) encodings: a comprehensive study](https://www.kaggle.com/vprokopev/mean-likelihood-encodings-a-comprehensive-study)\n- [The Mechanics of Machine Learning, Chapter 6: Categorically Speaking](https://mlbook.explained.ai/catvars.html)",
"_____no_output_____"
]
],
[
[
"!pip install catboost\n!pip install category_encoders\n!pip install ipywidgets",
"Requirement already satisfied: catboost in /usr/local/lib/python3.6/dist-packages (0.14.2)\nRequirement already satisfied: pandas>=0.19.1 in /usr/local/lib/python3.6/dist-packages (from catboost) (0.24.2)\nRequirement already satisfied: graphviz in /usr/local/lib/python3.6/dist-packages (from catboost) (0.10.1)\nRequirement already satisfied: enum34 in /usr/local/lib/python3.6/dist-packages (from catboost) (1.1.6)\nRequirement already satisfied: numpy>=1.11.1 in /usr/local/lib/python3.6/dist-packages (from catboost) (1.16.3)\nRequirement already satisfied: six in /usr/local/lib/python3.6/dist-packages (from catboost) (1.12.0)\nRequirement already satisfied: python-dateutil>=2.5.0 in /usr/local/lib/python3.6/dist-packages (from pandas>=0.19.1->catboost) (2.5.3)\nRequirement already satisfied: pytz>=2011k in /usr/local/lib/python3.6/dist-packages (from pandas>=0.19.1->catboost) (2018.9)\nCollecting category_encoders\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/6e/a1/f7a22f144f33be78afeb06bfa78478e8284a64263a3c09b1ef54e673841e/category_encoders-2.0.0-py2.py3-none-any.whl (87kB)\n\u001b[K |████████████████████████████████| 92kB 3.5MB/s \n\u001b[?25hRequirement already satisfied: scikit-learn>=0.20.0 in /usr/local/lib/python3.6/dist-packages (from category_encoders) (0.20.3)\nRequirement already satisfied: numpy>=1.11.3 in /usr/local/lib/python3.6/dist-packages (from category_encoders) (1.16.3)\nRequirement already satisfied: scipy>=0.19.0 in /usr/local/lib/python3.6/dist-packages (from category_encoders) (1.2.1)\nRequirement already satisfied: statsmodels>=0.6.1 in /usr/local/lib/python3.6/dist-packages (from category_encoders) (0.9.0)\nRequirement already satisfied: pandas>=0.21.1 in /usr/local/lib/python3.6/dist-packages (from category_encoders) (0.24.2)\nRequirement already satisfied: patsy>=0.4.1 in /usr/local/lib/python3.6/dist-packages (from category_encoders) (0.5.1)\nRequirement already satisfied: python-dateutil>=2.5.0 in /usr/local/lib/python3.6/dist-packages (from pandas>=0.21.1->category_encoders) (2.5.3)\nRequirement already satisfied: pytz>=2011k in /usr/local/lib/python3.6/dist-packages (from pandas>=0.21.1->category_encoders) (2018.9)\nRequirement already satisfied: six in /usr/local/lib/python3.6/dist-packages (from patsy>=0.4.1->category_encoders) (1.12.0)\nInstalling collected packages: category-encoders\nSuccessfully installed category-encoders-2.0.0\nRequirement already satisfied: ipywidgets in /usr/local/lib/python3.6/dist-packages (7.4.2)\nRequirement already satisfied: traitlets>=4.3.1 in /usr/local/lib/python3.6/dist-packages (from ipywidgets) (4.3.2)\nRequirement already satisfied: widgetsnbextension~=3.4.0 in /usr/local/lib/python3.6/dist-packages (from ipywidgets) (3.4.2)\nRequirement already satisfied: nbformat>=4.2.0 in /usr/local/lib/python3.6/dist-packages (from ipywidgets) (4.4.0)\nRequirement already satisfied: ipython>=4.0.0; python_version >= \"3.3\" in /usr/local/lib/python3.6/dist-packages (from ipywidgets) (5.5.0)\nRequirement already satisfied: ipykernel>=4.5.1 in /usr/local/lib/python3.6/dist-packages (from ipywidgets) (4.6.1)\nRequirement already satisfied: ipython-genutils in /usr/local/lib/python3.6/dist-packages (from traitlets>=4.3.1->ipywidgets) (0.2.0)\nRequirement already satisfied: decorator in /usr/local/lib/python3.6/dist-packages (from traitlets>=4.3.1->ipywidgets) (4.4.0)\nRequirement already satisfied: six in /usr/local/lib/python3.6/dist-packages (from traitlets>=4.3.1->ipywidgets) (1.12.0)\nRequirement already satisfied: notebook>=4.4.1 in /usr/local/lib/python3.6/dist-packages (from widgetsnbextension~=3.4.0->ipywidgets) (5.2.2)\nRequirement already satisfied: jupyter-core in /usr/local/lib/python3.6/dist-packages (from nbformat>=4.2.0->ipywidgets) (4.4.0)\nRequirement already satisfied: jsonschema!=2.5.0,>=2.4 in /usr/local/lib/python3.6/dist-packages (from nbformat>=4.2.0->ipywidgets) (2.6.0)\nRequirement already satisfied: pickleshare in /usr/local/lib/python3.6/dist-packages (from ipython>=4.0.0; python_version >= \"3.3\"->ipywidgets) (0.7.5)\nRequirement already satisfied: prompt-toolkit<2.0.0,>=1.0.4 in /usr/local/lib/python3.6/dist-packages (from ipython>=4.0.0; python_version >= \"3.3\"->ipywidgets) (1.0.16)\nRequirement already satisfied: setuptools>=18.5 in /usr/local/lib/python3.6/dist-packages (from ipython>=4.0.0; python_version >= \"3.3\"->ipywidgets) (41.0.1)\nRequirement already satisfied: pygments in /usr/local/lib/python3.6/dist-packages (from ipython>=4.0.0; python_version >= \"3.3\"->ipywidgets) (2.1.3)\nRequirement already satisfied: simplegeneric>0.8 in /usr/local/lib/python3.6/dist-packages (from ipython>=4.0.0; python_version >= \"3.3\"->ipywidgets) (0.8.1)\nRequirement already satisfied: pexpect; sys_platform != \"win32\" in /usr/local/lib/python3.6/dist-packages (from ipython>=4.0.0; python_version >= \"3.3\"->ipywidgets) (4.7.0)\nRequirement already satisfied: jupyter-client in /usr/local/lib/python3.6/dist-packages (from ipykernel>=4.5.1->ipywidgets) (5.2.4)\nRequirement already satisfied: tornado>=4.0 in /usr/local/lib/python3.6/dist-packages (from ipykernel>=4.5.1->ipywidgets) (4.5.3)\nRequirement already satisfied: jinja2 in /usr/local/lib/python3.6/dist-packages (from notebook>=4.4.1->widgetsnbextension~=3.4.0->ipywidgets) (2.10.1)\nRequirement already satisfied: terminado>=0.3.3; sys_platform != \"win32\" in /usr/local/lib/python3.6/dist-packages (from notebook>=4.4.1->widgetsnbextension~=3.4.0->ipywidgets) (0.8.2)\nRequirement already satisfied: nbconvert in /usr/local/lib/python3.6/dist-packages (from notebook>=4.4.1->widgetsnbextension~=3.4.0->ipywidgets) (5.5.0)\nRequirement already satisfied: wcwidth in /usr/local/lib/python3.6/dist-packages (from prompt-toolkit<2.0.0,>=1.0.4->ipython>=4.0.0; python_version >= \"3.3\"->ipywidgets) (0.1.7)\nRequirement already satisfied: ptyprocess>=0.5 in /usr/local/lib/python3.6/dist-packages (from pexpect; sys_platform != \"win32\"->ipython>=4.0.0; python_version >= \"3.3\"->ipywidgets) (0.6.0)\nRequirement already satisfied: pyzmq>=13 in /usr/local/lib/python3.6/dist-packages (from jupyter-client->ipykernel>=4.5.1->ipywidgets) (17.0.0)\nRequirement already satisfied: python-dateutil>=2.1 in /usr/local/lib/python3.6/dist-packages (from jupyter-client->ipykernel>=4.5.1->ipywidgets) (2.5.3)\nRequirement already satisfied: MarkupSafe>=0.23 in /usr/local/lib/python3.6/dist-packages (from jinja2->notebook>=4.4.1->widgetsnbextension~=3.4.0->ipywidgets) (1.1.1)\nRequirement already satisfied: defusedxml in /usr/local/lib/python3.6/dist-packages (from nbconvert->notebook>=4.4.1->widgetsnbextension~=3.4.0->ipywidgets) (0.6.0)\nRequirement already satisfied: testpath in /usr/local/lib/python3.6/dist-packages (from nbconvert->notebook>=4.4.1->widgetsnbextension~=3.4.0->ipywidgets) (0.4.2)\nRequirement already satisfied: mistune>=0.8.1 in /usr/local/lib/python3.6/dist-packages (from nbconvert->notebook>=4.4.1->widgetsnbextension~=3.4.0->ipywidgets) (0.8.4)\nRequirement already satisfied: pandocfilters>=1.4.1 in /usr/local/lib/python3.6/dist-packages (from nbconvert->notebook>=4.4.1->widgetsnbextension~=3.4.0->ipywidgets) (1.4.2)\nRequirement already satisfied: bleach in /usr/local/lib/python3.6/dist-packages (from nbconvert->notebook>=4.4.1->widgetsnbextension~=3.4.0->ipywidgets) (3.1.0)\nRequirement already satisfied: entrypoints>=0.2.2 in /usr/local/lib/python3.6/dist-packages (from nbconvert->notebook>=4.4.1->widgetsnbextension~=3.4.0->ipywidgets) (0.3)\nRequirement already satisfied: webencodings in /usr/local/lib/python3.6/dist-packages (from bleach->nbconvert->notebook>=4.4.1->widgetsnbextension~=3.4.0->ipywidgets) (0.5.1)\n"
],
[
"import warnings\nwarnings.simplefilter(action='ignore', category=FutureWarning)",
"_____no_output_____"
],
[
"import xgboost\nxgboost.__version__",
"_____no_output_____"
]
],
[
[
"# Golf Putts (regression, 1 feature, non-linear)\n\nhttps://statmodeling.stat.columbia.edu/2008/12/04/the_golf_puttin/",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nfrom ipywidgets import interact\nimport category_encoders\nimport matplotlib.pyplot as plt\nimport numpy as np\nimport pandas as pd\n\nputts = pd.DataFrame(\n columns=['distance', 'tries', 'successes'], \n data = [[2, 1443, 1346],\n [3, 694, 577],\n [4, 455, 337],\n [5, 353, 208],\n [6, 272, 149],\n [7, 256, 136],\n [8, 240, 111],\n [9, 217, 69],\n [10, 200, 67],\n [11, 237, 75],\n [12, 202, 52],\n [13, 192, 46],\n [14, 174, 54],\n [15, 167, 28],\n [16, 201, 27],\n [17, 195, 31],\n [18, 191, 33],\n [19, 147, 20],\n [20, 152, 24]]\n)\n\nputts['rate of success'] = putts['successes'] / putts['tries']\nputts_X = putts[['distance']]\nputts_y = putts['rate of success']",
"_____no_output_____"
]
],
[
[
"#### Docs\n- [Scikit-Learn User Guide: Random Forests](https://scikit-learn.org/stable/modules/ensemble.html#random-forests) (`from sklearn.ensemble import RandomForestRegressor, RandomForestClassifier`)\n- [XGBoost Python API Reference: Scikit-Learn API](https://xgboost.readthedocs.io/en/latest/python/python_api.html#module-xgboost.sklearn) (`from xgboost import XGBRegressor, XGBClassifier`)",
"_____no_output_____"
]
],
[
[
"from sklearn.ensemble import RandomForestRegressor\nfrom sklearn.tree import DecisionTreeRegressor\nfrom xgboost import XGBRegressor\n\ndef putt_trees(max_depth=1, n_estimators=1):\n models = [DecisionTreeRegressor(max_depth=max_depth), \n RandomForestRegressor(max_depth=max_depth, n_estimators=n_estimators), \n XGBRegressor(max_depth=max_depth, n_estimators=n_estimators)]\n \n for model in models:\n name = model.__class__.__name__\n model.fit(putts_X, putts_y)\n ax = putts.plot('distance', 'rate of success', kind='scatter', title=name)\n ax.step(putts_X, model.predict(putts_X), where='mid')\n plt.show()\n \ninteract(putt_trees, max_depth=(1,6,1), n_estimators=(10,40,10));",
"_____no_output_____"
]
],
[
[
"### Bagging\nhttps://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.DataFrame.sample.html",
"_____no_output_____"
]
],
[
[
"# Do-it-yourself Bagging Ensemble of Decision Trees (like a Random Forest)\ndef diy_bagging(max_depth=1, n_estimators=1):\n y_preds = []\n for i in range(n_estimators):\n title = f'Tree {i+1}'\n bootstrap_sample = putts.sample(n=len(putts), replace=True).sort_values(by='distance')\n bootstrap_X = bootstrap_sample[['distance']]\n bootstrap_y = bootstrap_sample['rate of success']\n tree = DecisionTreeRegressor(max_depth=max_depth)\n tree.fit(bootstrap_X, bootstrap_y)\n y_pred = tree.predict(bootstrap_X)\n y_preds.append(y_pred)\n ax = bootstrap_sample.plot('distance', 'rate of success', kind='scatter', title=title)\n ax.step(bootstrap_X, y_pred, where='mid')\n plt.show()\n \n ensembled = np.vstack(y_preds).mean(axis=0)\n title = f'Ensemble of {n_estimators} trees, with max_depth={max_depth}'\n ax = putts.plot('distance', 'rate of success', kind='scatter', title=title)\n ax.step(putts_X, ensembled, where='mid')\n plt.show()\n \ninteract(diy_bagging, max_depth=(1,6,1), n_estimators=(2,5,1));",
"_____no_output_____"
]
],
[
[
"### What's \"random\" about random forests?\n1. Each tree trains on a random bootstrap sample of the data. (In scikit-learn, for `RandomForestRegressor` and `RandomForestClassifier`, the `bootstrap` parameter's default is `True`.) This type of ensembling is called Bagging.\n2. Each split considers a random subset of the features. (In scikit-learn, when the `max_features` parameter is not `None`.) \n\nFor extra randomness, you can try [\"extremely randomized trees\"](https://scikit-learn.org/stable/modules/ensemble.html#extremely-randomized-trees)!\n\n>In extremely randomized trees (see [ExtraTreesClassifier](https://scikit-learn.org/stable/modules/generated/sklearn.ensemble.ExtraTreesClassifier.html) and [ExtraTreesRegressor](https://scikit-learn.org/stable/modules/generated/sklearn.ensemble.ExtraTreesRegressor.html) classes), randomness goes one step further in the way splits are computed. As in random forests, a random subset of candidate features is used, but instead of looking for the most discriminative thresholds, thresholds are drawn at random for each candidate feature and the best of these randomly-generated thresholds is picked as the splitting rule. This usually allows to reduce the variance of the model a bit more, at the expense of a slightly greater increase in bias",
"_____no_output_____"
],
[
"### Boosting \n\nBoosting (used by Gradient Boosting) is different than Bagging (used by Random Forests). \n\n[_An Introduction to Statistical Learning_](http://www-bcf.usc.edu/~gareth/ISL/ISLR%20Seventh%20Printing.pdf) Chapter 8.2.3, Boosting:\n\n>Recall that bagging involves creating multiple copies of the original training data set using the bootstrap, fitting a separate decision tree to each copy, and then combining all of the trees in order to create a single predictive model.\n\n>**Boosting works in a similar way, except that the trees are grown _sequentially_: each tree is grown using information from previously grown trees.**\n\n>Unlike fitting a single large decision tree to the data, which amounts to _fitting the data hard_ and potentially overfitting, the boosting approach instead _learns slowly._ Given the current model, we fit a decision tree to the residuals from the model.\n\n>We then add this new decision tree into the fitted function in order to update the residuals. Each of these trees can be rather small, with just a few terminal nodes. **By fitting small trees to the residuals, we slowly improve fˆ in areas where it does not perform well.**\n\n>Note that in boosting, unlike in bagging, the construction of each tree depends strongly on the trees that have already been grown.",
"_____no_output_____"
],
[
"# Wave (regression, 1 feature, non-monotonic, train/test split)\n\nhttp://scikit-learn.org/stable/auto_examples/tree/plot_tree_regression.html",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import train_test_split\n\ndef make_data():\n import numpy as np\n rng = np.random.RandomState(1)\n X = np.sort(5 * rng.rand(80, 1), axis=0)\n y = np.sin(X).ravel()\n y[::5] += 2 * (0.5 - rng.rand(16))\n return X, y\n\nwave_X, wave_y = make_data()\nwave_X_train, wave_X_test, wave_y_train, wave_y_test = train_test_split(\n wave_X, wave_y, test_size=0.25, random_state=42)",
"_____no_output_____"
],
[
"def wave_trees(max_depth=1, n_estimators=10):\n models = [DecisionTreeRegressor(max_depth=max_depth), \n RandomForestRegressor(max_depth=max_depth, n_estimators=n_estimators), \n XGBRegressor(max_depth=max_depth, n_estimators=n_estimators)]\n \n for model in models:\n name = model.__class__.__name__\n model.fit(wave_X_train, wave_y_train)\n print(f'{name} Train R^2 score:', model.score(wave_X_train, wave_y_train))\n print(f'{name} Test R^2 score:', model.score(wave_X_test, wave_y_test))\n plt.scatter(wave_X_train, wave_y_train)\n plt.scatter(wave_X_test, wave_y_test)\n plt.step(wave_X, model.predict(wave_X), where='mid')\n plt.show()\n \ninteract(wave_trees, max_depth=(1,8,1), n_estimators=(10,40,10));",
"_____no_output_____"
]
],
[
[
"# Titanic (classification, 2 features, interactions, non-linear / non-monotonic)\n\n#### viz2D helper function",
"_____no_output_____"
]
],
[
[
"def viz2D(fitted_model, X, feature1, feature2, num=100, title=''):\n \"\"\"\n Visualize model predictions as a 2D heatmap\n For regression or binary classification models, fitted on 2 features\n \n Parameters\n ----------\n fitted_model : scikit-learn model, already fitted\n df : pandas dataframe, which was used to fit model\n feature1 : string, name of feature 1\n feature2 : string, name of feature 2\n target : string, name of target\n num : int, number of grid points for each feature\n \n Returns\n -------\n predictions: numpy array, predictions/predicted probabilities at each grid point\n \n References\n ----------\n https://scikit-learn.org/stable/auto_examples/classification/plot_classification_probability.html\n https://jakevdp.github.io/PythonDataScienceHandbook/04.04-density-and-contour-plots.html\n \"\"\"\n x1 = np.linspace(X[feature1].min(), X[feature1].max(), num)\n x2 = np.linspace(X[feature2].min(), X[feature2].max(), num)\n X1, X2 = np.meshgrid(x1, x2)\n X = np.c_[X1.flatten(), X2.flatten()]\n if hasattr(fitted_model, 'predict_proba'):\n predicted = fitted_model.predict_proba(X)[:,1]\n else:\n predicted = fitted_model.predict(X)\n \n plt.imshow(predicted.reshape(num, num), cmap='viridis')\n plt.title(title)\n plt.xlabel(feature1)\n plt.ylabel(feature2)\n plt.xticks([])\n plt.yticks([])\n plt.colorbar()\n plt.show()\n return predicted",
"_____no_output_____"
]
],
[
[
"### Read data, encode categorical feature, impute missing values",
"_____no_output_____"
]
],
[
[
"import category_encoders as ce\nimport seaborn as sns\nfrom sklearn.impute import SimpleImputer\nfrom sklearn.pipeline import make_pipeline\n\ntitanic = sns.load_dataset('titanic')\nfeatures = ['age', 'sex']\ntarget = 'survived'\n\npreprocessor = make_pipeline(ce.OrdinalEncoder(), SimpleImputer())\nX = preprocessor.fit_transform(titanic[features])\nX = pd.DataFrame(X, columns=features)\ny = titanic[target]\n\nX.head()",
"_____no_output_____"
]
],
[
[
"### Logistic Regression",
"_____no_output_____"
]
],
[
[
"from sklearn.linear_model import LogisticRegression\nlr = LogisticRegression(solver='lbfgs')\nlr.fit(X, y)\nviz2D(lr, X, feature1='age', feature2='sex', title='Logistic Regression');",
"_____no_output_____"
]
],
[
[
"### Decision Tree, Random Forest, Gradient Boosting\n\n#### Docs\n- [Scikit-Learn User Guide: Random Forests](https://scikit-learn.org/stable/modules/ensemble.html#random-forests) (`from sklearn.ensemble import RandomForestRegressor, RandomForestClassifier`)\n- [XGBoost Python API Reference: Scikit-Learn API](https://xgboost.readthedocs.io/en/latest/python/python_api.html#module-xgboost.sklearn) (`from xgboost import XGBRegressor, XGBClassifier`)",
"_____no_output_____"
]
],
[
[
"from sklearn.ensemble import RandomForestClassifier\nfrom sklearn.tree import DecisionTreeClassifier\nfrom xgboost import XGBClassifier\n\ndef titanic_trees(max_depth=1, n_estimators=1):\n models = [DecisionTreeClassifier(max_depth=max_depth), \n RandomForestClassifier(max_depth=max_depth, n_estimators=n_estimators), \n XGBClassifier(max_depth=max_depth, n_estimators=n_estimators)]\n \n for model in models:\n name = model.__class__.__name__\n model.fit(X.values, y.values)\n viz2D(model, X, feature1='age', feature2='sex', title=name)\n \ninteract(titanic_trees, max_depth=(1,6,1), n_estimators=(10,40,10));",
"_____no_output_____"
]
],
[
[
"### Bagging",
"_____no_output_____"
]
],
[
[
"# Do-it-yourself Bagging Ensemble of Decision Trees (like a Random Forest)\n\ndef titanic_bagging(max_depth=1, n_estimators=1):\n predicteds = []\n for i in range(n_estimators):\n title = f'Tree {i+1}'\n bootstrap_sample = titanic.sample(n=len(titanic), replace=True)\n preprocessor = make_pipeline(ce.OrdinalEncoder(), SimpleImputer())\n bootstrap_X = preprocessor.fit_transform(bootstrap_sample[['age', 'sex']])\n bootstrap_y = bootstrap_sample['survived']\n tree = DecisionTreeClassifier(max_depth=max_depth)\n tree.fit(bootstrap_X, bootstrap_y)\n predicted = viz2D(tree, X, feature1='age', feature2='sex', title=title)\n predicteds.append(predicted)\n \n ensembled = np.vstack(predicteds).mean(axis=0)\n title = f'Ensemble of {n_estimators} trees, with max_depth={max_depth}'\n plt.imshow(ensembled.reshape(100, 100), cmap='viridis')\n plt.title(title)\n plt.xlabel('age')\n plt.ylabel('sex')\n plt.xticks([])\n plt.yticks([])\n plt.colorbar()\n plt.show()\n \ninteract(titanic_bagging, max_depth=(1,6,1), n_estimators=(2,5,1));",
"_____no_output_____"
]
],
[
[
"### Select more features, compare models",
"_____no_output_____"
]
],
[
[
"from sklearn.preprocessing import MinMaxScaler\n\ntitanic['deck'] = titanic['deck'].astype(str)\nfeatures = ['age', 'sex', 'pclass', 'sibsp', 'parch', 'fare', 'deck', 'embark_town']\ntarget = 'survived'\n\npreprocessor = make_pipeline(ce.OrdinalEncoder(), SimpleImputer(), MinMaxScaler())\ntitanic_X = preprocessor.fit_transform(titanic[features])\ntitanic_X = pd.DataFrame(titanic_X, columns=features)\ntitanic_y = titanic[target]\n\ntitanic_X.head()",
"_____no_output_____"
],
[
"from sklearn.model_selection import cross_val_score\n\nmodels = [LogisticRegression(solver='lbfgs', max_iter=1000), \n DecisionTreeClassifier(max_depth=3), \n DecisionTreeClassifier(max_depth=None), \n RandomForestClassifier(max_depth=3, n_estimators=100, n_jobs=-1, random_state=42), \n RandomForestClassifier(max_depth=None, n_estimators=100, n_jobs=-1, random_state=42), \n XGBClassifier(max_depth=3, n_estimators=100, n_jobs=-1, random_state=42)]\n\nfor model in models:\n print(model, '\\n')\n score = cross_val_score(model, titanic_X, titanic_y, scoring='accuracy', cv=5).mean()\n print('Cross-Validation Accuracy:', score, '\\n', '\\n')",
"_____no_output_____"
]
],
[
[
"### Feature importances",
"_____no_output_____"
]
],
[
[
"for model in models:\n name = model.__class__.__name__\n model.fit(titanic_X, titanic_y)\n if name == 'LogisticRegression':\n coefficients = pd.Series(model.coef_[0], titanic_X.columns)\n coefficients.sort_values().plot.barh(color='grey', title=name)\n plt.show()\n else:\n importances = pd.Series(model.feature_importances_, titanic_X.columns)\n title = f'{name}, max_depth={model.max_depth}'\n importances.sort_values().plot.barh(color='grey', title=title)\n plt.show()",
"_____no_output_____"
]
],
[
[
"# ASSIGNMENT\n\n**Train Random Forest and Gradient Boosting models**, on the Bank Marketing dataset. (Or another dataset of your choice, not used during this lesson.) You may use any Python libraries for Gradient Boosting.\n\nThen, you have many options!\n\n#### Keep improving your model\n- **Try new categorical encodings.**\n- Explore and visualize your data. \n- Wrangle [bad data](https://github.com/Quartz/bad-data-guide), outliers, and missing values.\n- Try engineering more features. You can transform, bin, and combine features. \n- Try selecting fewer features.\n",
"_____no_output_____"
]
],
[
[
"!wget https://archive.ics.uci.edu/ml/machine-learning-databases/00222/bank-additional.zip\n!unzip bank-additional.zip",
"--2019-05-09 19:39:55-- https://archive.ics.uci.edu/ml/machine-learning-databases/00222/bank-additional.zip\nResolving archive.ics.uci.edu (archive.ics.uci.edu)... 128.195.10.252\nConnecting to archive.ics.uci.edu (archive.ics.uci.edu)|128.195.10.252|:443... connected.\nHTTP request sent, awaiting response... 200 OK\nLength: 444572 (434K) [application/x-httpd-php]\nSaving to: ‘bank-additional.zip’\n\nbank-additional.zip 100%[===================>] 434.15K 1.08MB/s in 0.4s \n\n2019-05-09 19:39:56 (1.08 MB/s) - ‘bank-additional.zip’ saved [444572/444572]\n\nArchive: bank-additional.zip\n creating: bank-additional/\n inflating: bank-additional/.DS_Store \n creating: __MACOSX/\n creating: __MACOSX/bank-additional/\n inflating: __MACOSX/bank-additional/._.DS_Store \n inflating: bank-additional/.Rhistory \n inflating: bank-additional/bank-additional-full.csv \n inflating: bank-additional/bank-additional-names.txt \n inflating: bank-additional/bank-additional.csv \n inflating: __MACOSX/._bank-additional \n"
],
[
"#The core of this code comes from the lecture notebook earlier this week, but\n#I've modified it to include:\n# - A different CE\n# - Newly engineered features\n\n# Imports\n%matplotlib inline\nimport warnings\nimport category_encoders as ce\nimport matplotlib.pyplot as plt\nimport pandas as pd\nfrom sklearn.linear_model import LogisticRegression\nfrom sklearn.model_selection import cross_val_score\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.pipeline import make_pipeline\nfrom sklearn.exceptions import DataConversionWarning\nfrom sklearn.preprocessing import StandardScaler\nwarnings.filterwarnings(action='ignore', category=DataConversionWarning)\n\n# Load data\nbank = pd.read_csv('bank-additional/bank-additional-full.csv', sep=';')\n\n# Assign to X, y\nX = bank.drop(columns='y')\ny = bank['y'] == 'yes'\n\n# Drop leaky & random features\nX = X.drop(columns='duration')\n\n# Split Train, Test\nX_train, X_test, y_train, y_test = train_test_split(\n X, y, test_size=0.2, random_state=42, stratify=y)\n\n# Make pipeline\npipeline = make_pipeline(\n ce.OrdinalEncoder(), \n StandardScaler(), \n RandomForestClassifier(max_depth=4, n_estimators=100, n_jobs=-1, random_state=42)\n)\n\n# Cross-validate with training data\nscores = cross_val_score(pipeline, X_train, y_train, scoring='roc_auc', cv=10, n_jobs=-1, verbose=10)",
"[Parallel(n_jobs=-1)]: Using backend LokyBackend with 2 concurrent workers.\n[Parallel(n_jobs=-1)]: Done 1 tasks | elapsed: 2.4s\n[Parallel(n_jobs=-1)]: Done 4 tasks | elapsed: 5.0s\n[Parallel(n_jobs=-1)]: Done 10 out of 10 | elapsed: 12.3s finished\n"
],
[
"X.describe()",
"_____no_output_____"
],
[
"scores.mean()",
"_____no_output_____"
],
[
"#Now we'll try it again with gradient boosting:\npipeline = make_pipeline(\n ce.OrdinalEncoder(), \n StandardScaler(), \n XGBClassifier(max_depth=3, n_estimators=100, n_jobs=-1, random_state=42)\n)\n\n# Cross-validate with training data\nscores = cross_val_score(pipeline, X_train, y_train, scoring='roc_auc', cv=10, n_jobs=-1, verbose=10)\n\n",
"[Parallel(n_jobs=-1)]: Using backend LokyBackend with 2 concurrent workers.\n[Parallel(n_jobs=-1)]: Done 1 tasks | elapsed: 2.9s\n[Parallel(n_jobs=-1)]: Done 4 tasks | elapsed: 6.7s\n[Parallel(n_jobs=-1)]: Done 10 out of 10 | elapsed: 17.4s finished\n"
],
[
"scores.mean()",
"_____no_output_____"
]
],
[
[
"\n#### Follow the links — learn by reading & doing\n- Links at the top of this notebook\n- Links in previous notebooks\n- Extra notebook for today, about **\"monotonic constraints\"** and \"early stopping\" with xgboost",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
4a5978e3796c1465279459546897edf5329696a7
| 15,534 |
ipynb
|
Jupyter Notebook
|
Fall-19/Lecture_notebooks/L19/L19_Randomization.ipynb
|
sungsy12345/ENVECON-118
|
b2b62f49115f37e3a3c1f13b7ac6a3550c4c0600
|
[
"BSD-3-Clause"
] | 2 |
2020-09-10T13:45:34.000Z
|
2021-11-01T21:41:59.000Z
|
Fall-19/Lecture_notebooks/L19/L19_Randomization.ipynb
|
sungsy12345/ENVECON-118
|
b2b62f49115f37e3a3c1f13b7ac6a3550c4c0600
|
[
"BSD-3-Clause"
] | 1 |
2021-01-22T17:02:59.000Z
|
2021-06-20T14:03:54.000Z
|
Fall-19/Lecture_notebooks/L19/L19_Randomization.ipynb
|
sungsy12345/ENVECON-118
|
b2b62f49115f37e3a3c1f13b7ac6a3550c4c0600
|
[
"BSD-3-Clause"
] | 7 |
2019-11-06T20:55:27.000Z
|
2021-06-20T08:14:41.000Z
| 48.849057 | 659 | 0.457577 |
[
[
[
"library(haven)\njobdata<-read_dta(\"jip_dataset_forclass.dta\")\nhead(jobdata)",
"_____no_output_____"
],
[
"jobdata$geo_stata_zone <-as.factor(jobdata$geo_stata_zone) \njobdata$trade_strata<- as.factor(jobdata$trade_strata)\njobdata$caste<-as.factor(jobdata$caste)",
"_____no_output_____"
],
[
"#run simple regression\nreg1<- lm(emp~access + priority, data = jobdata, subset = survey_round==3)\nsummary(reg1)",
"_____no_output_____"
],
[
"#add controls\nreg2<- lm(emp~access + priority + geo_stata_zone + trade_strata + a_sex + age + educ + caste, data = jobdata, subset = survey_round==3)\nsummary(reg2)",
"_____no_output_____"
],
[
"#include both survey waves\njobdata$survey_round<- as.factor(jobdata$survey_round)\nreg3<- lm(emp~access + priority + geo_stata_zone + trade_strata + a_sex + age + educ + caste + survey_round, data = jobdata, subset = survey_round==2|survey_round==3)\nsummary(reg3)",
"_____no_output_____"
],
[
"#test unaffectable variable\nreg4<-lm(age~access + priority, data = jobdata, subset = survey_round==2)\nsummary(reg4)",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.