hexsha
stringlengths 40
40
| size
int64 6
14.9M
| ext
stringclasses 1
value | lang
stringclasses 1
value | max_stars_repo_path
stringlengths 6
260
| max_stars_repo_name
stringlengths 6
119
| max_stars_repo_head_hexsha
stringlengths 40
41
| max_stars_repo_licenses
list | max_stars_count
int64 1
191k
⌀ | max_stars_repo_stars_event_min_datetime
stringlengths 24
24
⌀ | max_stars_repo_stars_event_max_datetime
stringlengths 24
24
⌀ | max_issues_repo_path
stringlengths 6
260
| max_issues_repo_name
stringlengths 6
119
| max_issues_repo_head_hexsha
stringlengths 40
41
| max_issues_repo_licenses
list | max_issues_count
int64 1
67k
⌀ | max_issues_repo_issues_event_min_datetime
stringlengths 24
24
⌀ | max_issues_repo_issues_event_max_datetime
stringlengths 24
24
⌀ | max_forks_repo_path
stringlengths 6
260
| max_forks_repo_name
stringlengths 6
119
| max_forks_repo_head_hexsha
stringlengths 40
41
| max_forks_repo_licenses
list | max_forks_count
int64 1
105k
⌀ | max_forks_repo_forks_event_min_datetime
stringlengths 24
24
⌀ | max_forks_repo_forks_event_max_datetime
stringlengths 24
24
⌀ | avg_line_length
float64 2
1.04M
| max_line_length
int64 2
11.2M
| alphanum_fraction
float64 0
1
| cells
list | cell_types
list | cell_type_groups
list |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
4afd2c476c4320929f9efcc48f307d6465828e31
| 51,108 |
ipynb
|
Jupyter Notebook
|
deep-learning/udacity-deeplearning/reinforcement/Q-learning-cart.ipynb
|
AadityaGupta/Artificial-Intelligence-Deep-Learning-Machine-Learning-Tutorials
|
352dd6d9a785e22fde0ce53a6b0c2e56f4964950
|
[
"Apache-2.0"
] | 3,266 |
2017-08-06T16:51:46.000Z
|
2022-03-30T07:34:24.000Z
|
deep-learning/udacity-deeplearning/reinforcement/Q-learning-cart.ipynb
|
AadityaGupta/Artificial-Intelligence-Deep-Learning-Machine-Learning-Tutorials
|
352dd6d9a785e22fde0ce53a6b0c2e56f4964950
|
[
"Apache-2.0"
] | 150 |
2017-08-28T14:59:36.000Z
|
2022-03-11T23:21:35.000Z
|
deep-learning/udacity-deeplearning/reinforcement/Q-learning-cart.ipynb
|
AadityaGupta/Artificial-Intelligence-Deep-Learning-Machine-Learning-Tutorials
|
352dd6d9a785e22fde0ce53a6b0c2e56f4964950
|
[
"Apache-2.0"
] | 1,449 |
2017-08-06T17:40:59.000Z
|
2022-03-31T12:03:24.000Z
| 85.322204 | 27,418 | 0.769508 |
[
[
[
"# Deep Q-learning\n\nIn this notebook, we'll build a neural network that can learn to play games through reinforcement learning. More specifically, we'll use Q-learning to train an agent to play a game called [Cart-Pole](https://gym.openai.com/envs/CartPole-v0). In this game, a freely swinging pole is attached to a cart. The cart can move to the left and right, and the goal is to keep the pole upright as long as possible.\n\n\n\nWe can simulate this game using [OpenAI Gym](https://gym.openai.com/). First, let's check out how OpenAI Gym works. Then, we'll get into training an agent to play the Cart-Pole game.",
"_____no_output_____"
]
],
[
[
"import gym\nimport tensorflow as tf\nimport numpy as np",
"_____no_output_____"
]
],
[
[
">**Note:** Make sure you have OpenAI Gym cloned into the same directory with this notebook. I've included `gym` as a submodule, so you can run `git submodule --init --recursive` to pull the contents into the `gym` repo.",
"_____no_output_____"
]
],
[
[
"# Create the Cart-Pole game environment\nenv = gym.make('CartPole-v0')",
"[2017-04-13 12:20:53,011] Making new env: CartPole-v0\n"
]
],
[
[
"We interact with the simulation through `env`. To show the simulation running, you can use `env.render()` to render one frame. Passing in an action as an integer to `env.step` will generate the next step in the simulation. You can see how many actions are possible from `env.action_space` and to get a random action you can use `env.action_space.sample()`. This is general to all Gym games. In the Cart-Pole game, there are two possible actions, moving the cart left or right. So there are two actions we can take, encoded as 0 and 1.\n\nRun the code below to watch the simulation run.",
"_____no_output_____"
]
],
[
[
"env.reset()\nrewards = []\nfor _ in range(100):\n env.render()\n state, reward, done, info = env.step(env.action_space.sample()) # take a random action\n rewards.append(reward)\n if done:\n rewards = []\n env.reset()",
"_____no_output_____"
]
],
[
[
"To shut the window showing the simulation, use `env.close()`.",
"_____no_output_____"
],
[
"If you ran the simulation above, we can look at the rewards:",
"_____no_output_____"
]
],
[
[
"print(rewards[-20:])",
"[1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0]\n"
]
],
[
[
"The game resets after the pole has fallen past a certain angle. For each frame while the simulation is running, it returns a reward of 1.0. The longer the game runs, the more reward we get. Then, our network's goal is to maximize the reward by keeping the pole vertical. It will do this by moving the cart to the left and the right.",
"_____no_output_____"
],
[
"## Q-Network\n\nWe train our Q-learning agent using the Bellman Equation:\n\n$$\nQ(s, a) = r + \\gamma \\max{Q(s', a')}\n$$\n\nwhere $s$ is a state, $a$ is an action, and $s'$ is the next state from state $s$ and action $a$.\n\nBefore we used this equation to learn values for a Q-_table_. However, for this game there are a huge number of states available. The state has four values: the position and velocity of the cart, and the position and velocity of the pole. These are all real-valued numbers, so ignoring floating point precisions, you practically have infinite states. Instead of using a table then, we'll replace it with a neural network that will approximate the Q-table lookup function.\n\n<img src=\"assets/deep-q-learning.png\" width=450px>\n\nNow, our Q value, $Q(s, a)$ is calculated by passing in a state to the network. The output will be Q-values for each available action, with fully connected hidden layers.\n\n<img src=\"assets/q-network.png\" width=550px>\n\n\nAs I showed before, we can define our targets for training as $\\hat{Q}(s,a) = r + \\gamma \\max{Q(s', a')}$. Then we update the weights by minimizing $(\\hat{Q}(s,a) - Q(s,a))^2$. \n\nFor this Cart-Pole game, we have four inputs, one for each value in the state, and two outputs, one for each action. To get $\\hat{Q}$, we'll first choose an action, then simulate the game using that action. This will get us the next state, $s'$, and the reward. With that, we can calculate $\\hat{Q}$ then pass it back into the $Q$ network to run the optimizer and update the weights.\n\nBelow is my implementation of the Q-network. I used two fully connected layers with ReLU activations. Two seems to be good enough, three might be better. Feel free to try it out.",
"_____no_output_____"
]
],
[
[
"class QNetwork:\n def __init__(self, learning_rate=0.01, state_size=4, \n action_size=2, hidden_size=10, \n name='QNetwork'):\n # state inputs to the Q-network\n with tf.variable_scope(name):\n self.inputs_ = tf.placeholder(tf.float32, [None, state_size], name='inputs')\n \n # One hot encode the actions to later choose the Q-value for the action\n self.actions_ = tf.placeholder(tf.int32, [None], name='actions')\n one_hot_actions = tf.one_hot(self.actions_, action_size)\n \n # Target Q values for training\n self.targetQs_ = tf.placeholder(tf.float32, [None], name='target')\n \n # ReLU hidden layers\n self.fc1 = tf.contrib.layers.fully_connected(self.inputs_, hidden_size)\n self.fc2 = tf.contrib.layers.fully_connected(self.fc1, hidden_size)\n\n # Linear output layer\n self.output = tf.contrib.layers.fully_connected(self.fc2, action_size, \n activation_fn=None)\n \n ### Train with loss (targetQ - Q)^2\n # output has length 2, for two actions. This next line chooses\n # one value from output (per row) according to the one-hot encoded actions.\n self.Q = tf.reduce_sum(tf.multiply(self.output, one_hot_actions), axis=1)\n \n self.loss = tf.reduce_mean(tf.square(self.targetQs_ - self.Q))\n self.opt = tf.train.AdamOptimizer(learning_rate).minimize(self.loss)",
"_____no_output_____"
]
],
[
[
"## Experience replay\n\nReinforcement learning algorithms can have stability issues due to correlations between states. To reduce correlations when training, we can store the agent's experiences and later draw a random mini-batch of those experiences to train on. \n\nHere, we'll create a `Memory` object that will store our experiences, our transitions $<s, a, r, s'>$. This memory will have a maxmium capacity, so we can keep newer experiences in memory while getting rid of older experiences. Then, we'll sample a random mini-batch of transitions $<s, a, r, s'>$ and train on those.\n\nBelow, I've implemented a `Memory` object. If you're unfamiliar with `deque`, this is a double-ended queue. You can think of it like a tube open on both sides. You can put objects in either side of the tube. But if it's full, adding anything more will push an object out the other side. This is a great data structure to use for the memory buffer.",
"_____no_output_____"
]
],
[
[
"from collections import deque\nclass Memory():\n def __init__(self, max_size = 1000):\n self.buffer = deque(maxlen=max_size)\n \n def add(self, experience):\n self.buffer.append(experience)\n \n def sample(self, batch_size):\n idx = np.random.choice(np.arange(len(self.buffer)), \n size=batch_size, \n replace=False)\n return [self.buffer[ii] for ii in idx]",
"_____no_output_____"
]
],
[
[
"## Exploration - Exploitation\n\nTo learn about the environment and rules of the game, the agent needs to explore by taking random actions. We'll do this by choosing a random action with some probability $\\epsilon$ (epsilon). That is, with some probability $\\epsilon$ the agent will make a random action and with probability $1 - \\epsilon$, the agent will choose an action from $Q(s,a)$. This is called an **$\\epsilon$-greedy policy**.\n\n\nAt first, the agent needs to do a lot of exploring. Later when it has learned more, the agent can favor choosing actions based on what it has learned. This is called _exploitation_. We'll set it up so the agent is more likely to explore early in training, then more likely to exploit later in training.",
"_____no_output_____"
],
[
"## Q-Learning training algorithm\n\nPutting all this together, we can list out the algorithm we'll use to train the network. We'll train the network in _episodes_. One *episode* is one simulation of the game. For this game, the goal is to keep the pole upright for 195 frames. So we can start a new episode once meeting that goal. The game ends if the pole tilts over too far, or if the cart moves too far the left or right. When a game ends, we'll start a new episode. Now, to train the agent:\n\n* Initialize the memory $D$\n* Initialize the action-value network $Q$ with random weights\n* **For** episode = 1, $M$ **do**\n * **For** $t$, $T$ **do**\n * With probability $\\epsilon$ select a random action $a_t$, otherwise select $a_t = \\mathrm{argmax}_a Q(s,a)$\n * Execute action $a_t$ in simulator and observe reward $r_{t+1}$ and new state $s_{t+1}$\n * Store transition $<s_t, a_t, r_{t+1}, s_{t+1}>$ in memory $D$\n * Sample random mini-batch from $D$: $<s_j, a_j, r_j, s'_j>$\n * Set $\\hat{Q}_j = r_j$ if the episode ends at $j+1$, otherwise set $\\hat{Q}_j = r_j + \\gamma \\max_{a'}{Q(s'_j, a')}$\n * Make a gradient descent step with loss $(\\hat{Q}_j - Q(s_j, a_j))^2$\n * **endfor**\n* **endfor**",
"_____no_output_____"
],
[
"## Hyperparameters\n\nOne of the more difficult aspects of reinforcememt learning are the large number of hyperparameters. Not only are we tuning the network, but we're tuning the simulation.",
"_____no_output_____"
]
],
[
[
"train_episodes = 1000 # max number of episodes to learn from\nmax_steps = 200 # max steps in an episode\ngamma = 0.99 # future reward discount\n\n# Exploration parameters\nexplore_start = 1.0 # exploration probability at start\nexplore_stop = 0.01 # minimum exploration probability \ndecay_rate = 0.0001 # exponential decay rate for exploration prob\n\n# Network parameters\nhidden_size = 64 # number of units in each Q-network hidden layer\nlearning_rate = 0.0001 # Q-network learning rate\n\n# Memory parameters\nmemory_size = 10000 # memory capacity\nbatch_size = 20 # experience mini-batch size\npretrain_length = batch_size # number experiences to pretrain the memory",
"_____no_output_____"
],
[
"tf.reset_default_graph()\nmainQN = QNetwork(name='main', hidden_size=hidden_size, learning_rate=learning_rate)",
"_____no_output_____"
]
],
[
[
"## Populate the experience memory\n\nHere I'm re-initializing the simulation and pre-populating the memory. The agent is taking random actions and storing the transitions in memory. This will help the agent with exploring the game.",
"_____no_output_____"
]
],
[
[
"# Initialize the simulation\nenv.reset()\n# Take one random step to get the pole and cart moving\nstate, reward, done, _ = env.step(env.action_space.sample())\n\nmemory = Memory(max_size=memory_size)\n\n# Make a bunch of random actions and store the experiences\nfor ii in range(pretrain_length):\n # Uncomment the line below to watch the simulation\n # env.render()\n\n # Make a random action\n action = env.action_space.sample()\n next_state, reward, done, _ = env.step(action)\n\n if done:\n # The simulation fails so no next state\n next_state = np.zeros(state.shape)\n # Add experience to memory\n memory.add((state, action, reward, next_state))\n \n # Start new episode\n env.reset()\n # Take one random step to get the pole and cart moving\n state, reward, done, _ = env.step(env.action_space.sample())\n else:\n # Add experience to memory\n memory.add((state, action, reward, next_state))\n state = next_state",
"_____no_output_____"
]
],
[
[
"## Training\n\nBelow we'll train our agent. If you want to watch it train, uncomment the `env.render()` line. This is slow because it's rendering the frames slower than the network can train. But, it's cool to watch the agent get better at the game.",
"_____no_output_____"
]
],
[
[
"# Now train with experiences\nsaver = tf.train.Saver()\nrewards_list = []\nwith tf.Session() as sess:\n # Initialize variables\n sess.run(tf.global_variables_initializer())\n \n step = 0\n for ep in range(1, train_episodes):\n total_reward = 0\n t = 0\n while t < max_steps:\n step += 1\n # Uncomment this next line to watch the training\n # env.render() \n \n # Explore or Exploit\n explore_p = explore_stop + (explore_start - explore_stop)*np.exp(-decay_rate*step) \n if explore_p > np.random.rand():\n # Make a random action\n action = env.action_space.sample()\n else:\n # Get action from Q-network\n feed = {mainQN.inputs_: state.reshape((1, *state.shape))}\n Qs = sess.run(mainQN.output, feed_dict=feed)\n action = np.argmax(Qs)\n \n # Take action, get new state and reward\n next_state, reward, done, _ = env.step(action)\n \n total_reward += reward\n \n if done:\n # the episode ends so no next state\n next_state = np.zeros(state.shape)\n t = max_steps\n \n print('Episode: {}'.format(ep),\n 'Total reward: {}'.format(total_reward),\n 'Training loss: {:.4f}'.format(loss),\n 'Explore P: {:.4f}'.format(explore_p))\n rewards_list.append((ep, total_reward))\n \n # Add experience to memory\n memory.add((state, action, reward, next_state))\n \n # Start new episode\n env.reset()\n # Take one random step to get the pole and cart moving\n state, reward, done, _ = env.step(env.action_space.sample())\n\n else:\n # Add experience to memory\n memory.add((state, action, reward, next_state))\n state = next_state\n t += 1\n \n # Sample mini-batch from memory\n batch = memory.sample(batch_size)\n states = np.array([each[0] for each in batch])\n actions = np.array([each[1] for each in batch])\n rewards = np.array([each[2] for each in batch])\n next_states = np.array([each[3] for each in batch])\n \n # Train network\n target_Qs = sess.run(mainQN.output, feed_dict={mainQN.inputs_: next_states})\n \n # Set target_Qs to 0 for states where episode ends\n episode_ends = (next_states == np.zeros(states[0].shape)).all(axis=1)\n target_Qs[episode_ends] = (0, 0)\n \n targets = rewards + gamma * np.max(target_Qs, axis=1)\n\n loss, _ = sess.run([mainQN.loss, mainQN.opt],\n feed_dict={mainQN.inputs_: states,\n mainQN.targetQs_: targets,\n mainQN.actions_: actions})\n \n saver.save(sess, \"checkpoints/cartpole.ckpt\")\n",
"_____no_output_____"
]
],
[
[
"## Visualizing training\n\nBelow I'll plot the total rewards for each episode. I'm plotting the rolling average too, in blue.",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nimport matplotlib.pyplot as plt\n\ndef running_mean(x, N):\n cumsum = np.cumsum(np.insert(x, 0, 0)) \n return (cumsum[N:] - cumsum[:-N]) / N ",
"_____no_output_____"
],
[
"eps, rews = np.array(rewards_list).T\nsmoothed_rews = running_mean(rews, 10)\nplt.plot(eps[-len(smoothed_rews):], smoothed_rews)\nplt.plot(eps, rews, color='grey', alpha=0.3)\nplt.xlabel('Episode')\nplt.ylabel('Total Reward')",
"_____no_output_____"
]
],
[
[
"## Testing\n\nLet's checkout how our trained agent plays the game.",
"_____no_output_____"
]
],
[
[
"test_episodes = 10\ntest_max_steps = 400\nenv.reset()\nwith tf.Session() as sess:\n saver.restore(sess, tf.train.latest_checkpoint('checkpoints'))\n \n for ep in range(1, test_episodes):\n t = 0\n while t < test_max_steps:\n env.render() \n \n # Get action from Q-network\n feed = {mainQN.inputs_: state.reshape((1, *state.shape))}\n Qs = sess.run(mainQN.output, feed_dict=feed)\n action = np.argmax(Qs)\n \n # Take action, get new state and reward\n next_state, reward, done, _ = env.step(action)\n \n if done:\n t = test_max_steps\n env.reset()\n # Take one random step to get the pole and cart moving\n state, reward, done, _ = env.step(env.action_space.sample())\n\n else:\n state = next_state\n t += 1",
"_____no_output_____"
],
[
"env.close()",
"_____no_output_____"
]
],
[
[
"## Extending this\n\nSo, Cart-Pole is a pretty simple game. However, the same model can be used to train an agent to play something much more complicated like Pong or Space Invaders. Instead of a state like we're using here though, you'd want to use convolutional layers to get the state from the screen images.\n\n\n\nI'll leave it as a challenge for you to use deep Q-learning to train an agent to play Atari games. Here's the original paper which will get you started: http://www.davidqiu.com:8888/research/nature14236.pdf.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
4afd30e2594401a9db98c46dc15d9707ce3a7b07
| 207,560 |
ipynb
|
Jupyter Notebook
|
notebooks/3_nasa_data_initial_explore.ipynb
|
douglasdaly/hurricanes
|
60d8dbf8f704c8b33d54f71950d8c9bb60c6649c
|
[
"MIT"
] | 1 |
2018-12-02T20:33:30.000Z
|
2018-12-02T20:33:30.000Z
|
notebooks/3_nasa_data_initial_explore.ipynb
|
douglasdaly/hurricanes
|
60d8dbf8f704c8b33d54f71950d8c9bb60c6649c
|
[
"MIT"
] | null | null | null |
notebooks/3_nasa_data_initial_explore.ipynb
|
douglasdaly/hurricanes
|
60d8dbf8f704c8b33d54f71950d8c9bb60c6649c
|
[
"MIT"
] | null | null | null | 801.389961 | 83,980 | 0.952245 |
[
[
[
"# NASA Data Exploration\n",
"_____no_output_____"
]
],
[
[
"raw_data_dir = '../data/raw'\nprocessed_data_dir = '../data/processed'\nfigsize_width = 12\nfigsize_height = 8\noutput_dpi = 72",
"_____no_output_____"
],
[
"# Imports\nimport os\nimport numpy as np\nimport pandas as pd\nfrom datetime import datetime\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"# Load Data\nnasa_temp_file = os.path.join(raw_data_dir, 'nasa_temperature_anomaly.txt')\nnasa_sea_file = os.path.join(raw_data_dir, 'nasa_sea_level.txt')\nnasa_co2_file = os.path.join(raw_data_dir, 'nasa_carbon_dioxide_levels.txt')",
"_____no_output_____"
],
[
"# Variable Setup\ndefault_fig_size = (figsize_width, figsize_height)",
"_____no_output_____"
],
[
"# - Process temperature data\ntemp_data = pd.read_csv(nasa_temp_file, sep='\\t', header=None)\ntemp_data.columns = ['Year', 'Annual Mean', 'Lowness Smoothing']\ntemp_data.set_index('Year', inplace=True)\n\nfig, ax = plt.subplots(figsize=default_fig_size)\n\ntemp_data.plot(ax=ax)\nax.grid(True, linestyle='--', color='grey', alpha=0.6)\n\nax.set_title('Global Temperature Anomaly Data', fontweight='bold')\nax.set_xlabel('')\nax.set_ylabel('Temperature Anomaly ($\\degree$C)')\nax.legend()\n\nplt.show();",
"_____no_output_____"
],
[
"# - Process Sea-level File\n# -- Figure out header rows\nwith open(nasa_sea_file, 'r') as fin:\n all_lines = fin.readlines()\n \nheader_lines = np.array([1 for x in all_lines if x.startswith('HDR')]).sum()\nsea_level_data = pd.read_csv(nasa_sea_file, delim_whitespace=True, \n skiprows=header_lines-1).reset_index()\n\nsea_level_data.columns = ['Altimeter Type', 'File Cycle', 'Year Fraction', \n 'N Observations', 'N Weighted Observations', 'GMSL',\n 'Std GMSL', 'GMSL (smoothed)', 'GMSL (GIA Applied)',\n 'Std GMSL (GIA Applied)', 'GMSL (GIA, smoothed)',\n 'GMSL (GIA, smoothed, filtered)']\nsea_level_data.set_index('Year Fraction', inplace=True)\n\nfig, ax = plt.subplots(figsize=default_fig_size)\n\nsea_level_var = sea_level_data.loc[:, 'GMSL (GIA, smoothed, filtered)'] \\\n - sea_level_data.loc[:, 'GMSL (GIA, smoothed, filtered)'].iloc[0]\n\nsea_level_var.plot(ax=ax)\nax.grid(True, color='grey', alpha=0.6, linestyle='--')\nax.set_title('Global Sea-Level Height Change over Time', fontweight='bold')\nax.set_xlabel('')\nax.set_ylabel('Sea Height Change (mm)')\nax.legend(loc='upper left')\n\nplt.show();",
"_____no_output_____"
],
[
"# - Process Carbon Dioxide Data\nwith open(nasa_co2_file, 'r') as fin:\n all_lines = fin.readlines()\n\nheader_lines = np.array([1 for x in all_lines if x.startswith('#')]).sum()\n\nco2_data = pd.read_csv(nasa_co2_file, skiprows=header_lines, header=None, \n delim_whitespace=True)\nco2_data[co2_data == -99.99] = np.nan\n\nco2_data.columns = ['Year', 'Month', 'Year Fraction', 'Average', 'Interpolated', \n 'Trend', 'N Days']\n\nco2_data.set_index(['Year', 'Month'], inplace=True)\nnew_idx = [datetime(x[0], x[1], 1) for x in co2_data.index]\nco2_data.index = new_idx\nco2_data.index.name = 'Date'\n\n# - Plot\nfig, ax = plt.subplots(figsize=default_fig_size)\n\nco2_data.loc[:, 'Average'].plot(ax=ax)\n\nax.grid(True, linestyle='--', color='grey', alpha=0.6)\nax.set_xlabel('')\nax.set_ylabel('$CO_2$ Level (ppm)')\nax.set_title('Global Carbon Dioxide Level over Time', fontweight='bold')\n\nplt.show();",
"_____no_output_____"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4afd45e4872e2988563b039c8df1e5d05a49017e
| 20,516 |
ipynb
|
Jupyter Notebook
|
section-04-research-and-development/titanic-assignment/03-titanic-survival-pipeline-assignment.ipynb
|
jaime-cespedes-sisniega/deploying-machine-learning-models
|
dc13180f4ee20edd96b997e847ebcd94be29dfef
|
[
"BSD-3-Clause"
] | null | null | null |
section-04-research-and-development/titanic-assignment/03-titanic-survival-pipeline-assignment.ipynb
|
jaime-cespedes-sisniega/deploying-machine-learning-models
|
dc13180f4ee20edd96b997e847ebcd94be29dfef
|
[
"BSD-3-Clause"
] | null | null | null |
section-04-research-and-development/titanic-assignment/03-titanic-survival-pipeline-assignment.ipynb
|
jaime-cespedes-sisniega/deploying-machine-learning-models
|
dc13180f4ee20edd96b997e847ebcd94be29dfef
|
[
"BSD-3-Clause"
] | null | null | null | 43.651064 | 2,612 | 0.504777 |
[
[
[
"## Predicting Survival on the Titanic\n\n### History\nPerhaps one of the most infamous shipwrecks in history, the Titanic sank after colliding with an iceberg, killing 1502 out of 2224 people on board. Interestingly, by analysing the probability of survival based on few attributes like gender, age, and social status, we can make very accurate predictions on which passengers would survive. Some groups of people were more likely to survive than others, such as women, children, and the upper-class. Therefore, we can learn about the society priorities and privileges at the time.\n\n### Assignment:\n\nBuild a Machine Learning Pipeline, to engineer the features in the data set and predict who is more likely to Survive the catastrophe.\n\nFollow the Jupyter notebook below, and complete the missing bits of code, to achieve each one of the pipeline steps.",
"_____no_output_____"
]
],
[
[
"import re\n\n# to handle datasets\nimport pandas as pd\nimport numpy as np\n\n# for visualization\nimport matplotlib.pyplot as plt\n\n# to divide train and test set\nfrom sklearn.model_selection import train_test_split\n\n# feature scaling\nfrom sklearn.preprocessing import StandardScaler\n\n# to build the models\nfrom sklearn.linear_model import LogisticRegression\n\n# to evaluate the models\nfrom sklearn.metrics import accuracy_score, roc_auc_score\n\n# to persist the model and the scaler\nimport joblib\n\n# ========== NEW IMPORTS ========\n# Respect to notebook 02-Predicting-Survival-Titanic-Solution\n\n# pipeline\nfrom sklearn.pipeline import Pipeline\n\n# for the preprocessors\nfrom sklearn.base import BaseEstimator, TransformerMixin\n\n# for imputation\nfrom feature_engine.imputation import (AddMissingIndicator,\n CategoricalImputer,\n MeanMedianImputer)\n\n# for encoding categorical variables\nfrom feature_engine.encoding import (OneHotEncoder,\n RareLabelEncoder)\n\n# typing\nfrom typing import List",
"_____no_output_____"
]
],
[
[
"## Prepare the data set",
"_____no_output_____"
]
],
[
[
"# load the data - it is available open source and online\n\ndata = pd.read_csv('https://www.openml.org/data/get_csv/16826755/phpMYEkMl')\n\n# display data\ndata.head()",
"_____no_output_____"
],
[
"# replace interrogation marks by NaN values\n\ndata = data.replace('?', np.nan)",
"_____no_output_____"
],
[
"# retain only the first cabin if more than\n# 1 are available per passenger\n\ndef get_first_cabin(row):\n try:\n return row.split()[0]\n except:\n return np.nan\n \ndata['cabin'] = data['cabin'].apply(get_first_cabin)",
"_____no_output_____"
],
[
"# extracts the title (Mr, Ms, etc) from the name variable\n\ndef get_title(passenger):\n line = passenger\n if re.search('Mrs', line):\n return 'Mrs'\n elif re.search('Mr', line):\n return 'Mr'\n elif re.search('Miss', line):\n return 'Miss'\n elif re.search('Master', line):\n return 'Master'\n else:\n return 'Other'\n \ndata['title'] = data['name'].apply(get_title)",
"_____no_output_____"
],
[
"# cast numerical variables as floats\n\ndata['fare'] = data['fare'].astype('float')\ndata['age'] = data['age'].astype('float')",
"_____no_output_____"
],
[
"# drop unnecessary variables\n\ndata.drop(labels=['name','ticket', 'boat', 'body','home.dest'], axis=1, inplace=True)\n\n# display data\ndata.head()",
"_____no_output_____"
],
[
"# # save the data set\n\n# data.to_csv('titanic.csv', index=False)",
"_____no_output_____"
]
],
[
[
"# Begin Assignment\n\n## Configuration",
"_____no_output_____"
]
],
[
[
"# list of variables to be used in the pipeline's transformers\n\nNUMERICAL_VARIABLES = ['age', 'fare']\n# The rest of the numerical variables (pclass, sibsp, parch) do not need to be transformed (add missing flag or impute values). That is why they are not included in the list.\n\nCATEGORICAL_VARIABLES = ['sex', 'cabin', 'embarked', 'title']\n\nCABIN = ['cabin']",
"_____no_output_____"
]
],
[
[
"## Separate data into train and test",
"_____no_output_____"
]
],
[
[
"X_train, X_test, y_train, y_test = train_test_split(\n data.drop('survived', axis=1), # predictors\n data['survived'], # target\n test_size=0.2, # percentage of obs in test set\n random_state=0) # seed to ensure reproducibility\n\nX_train.shape, X_test.shape",
"_____no_output_____"
]
],
[
[
"## Preprocessors\n\n### Class to extract the letter from the variable Cabin",
"_____no_output_____"
]
],
[
[
"class ExtractLetterTransformer(BaseEstimator, TransformerMixin):\n # Extract fist letter of variable\n\n def __init__(self, variables: List[str]) -> None:\n self.variables = [variables] \\\n if not isinstance(variables, list) else variables\n\n def fit(self, X: pd.DataFrame, y=None):\n return self\n\n def transform(self, X: pd.DataFrame) -> pd.DataFrame:\n X = X.copy()\n\n for var in self.variables:\n X[var] = X[var].str[0]\n\n return X",
"_____no_output_____"
]
],
[
[
"## Pipeline\n\n- Impute categorical variables with string missing\n- Add a binary missing indicator to numerical variables with missing data\n- Fill NA in original numerical variable with the median\n- Extract first letter from cabin\n- Group rare Categories\n- Perform One hot encoding\n- Scale features with standard scaler\n- Fit a Logistic regression",
"_____no_output_____"
]
],
[
[
"# set up the pipeline\ntitanic_pipe = Pipeline([\n\n # ===== IMPUTATION =====\n # impute categorical variables with string 'missing'\n ('categorical_imputation', CategoricalImputer(variables=CATEGORICAL_VARIABLES,\n imputation_method='missing')),\n\n # add missing indicator to numerical variables\n ('missing_indicator', AddMissingIndicator(variables=NUMERICAL_VARIABLES)),\n\n # impute numerical variables with the median\n ('median_imputation', MeanMedianImputer(variables=NUMERICAL_VARIABLES,\n imputation_method='median')),\n\n\n # Extract first letter from cabin\n ('extract_letter', ExtractLetterTransformer(variables=CABIN)),\n\n\n # == CATEGORICAL ENCODING ======\n # remove categories present in less than 5% of the observations (0.05)\n # group them in one category called 'Rare'\n ('rare_label_encoder', RareLabelEncoder(variables=CATEGORICAL_VARIABLES,\n tol=0.05,\n n_categories=1,\n replace_with='Other')),\n\n\n # encode categorical variables using one hot encoding into k-1 variables\n ('categorical_encoder', OneHotEncoder(variables=CATEGORICAL_VARIABLES,\n drop_last=True)),\n\n # scale using standardization\n ('scaler', StandardScaler()),\n\n # logistic regression (use C=0.0005 and random_state=0)\n ('Logit', LogisticRegression(C=5e-4,\n random_state=0)),\n])",
"_____no_output_____"
],
[
"# train the pipeline\ntitanic_pipe.fit(X_train, y_train)",
"_____no_output_____"
]
],
[
[
"## Make predictions and evaluate model performance\n\nDetermine:\n- roc-auc\n- accuracy\n\n**Important, remember that to determine the accuracy, you need the outcome 0, 1, referring to survived or not. But to determine the roc-auc you need the probability of survival.**",
"_____no_output_____"
]
],
[
[
"# make predictions for train set\nclass_ = titanic_pipe.predict(X_train)\npred = titanic_pipe.predict_proba(X_train)[:, 1]\n\n# determine mse and rmse\nprint('train roc-auc: {}'.format(roc_auc_score(y_train, pred)))\nprint('train accuracy: {}'.format(accuracy_score(y_train, class_)))\nprint()\n\n# make predictions for test set\nclass_ = titanic_pipe.predict(X_test)\npred = titanic_pipe.predict_proba(X_test)[:, 1]\n\n# determine mse and rmse\nprint('test roc-auc: {}'.format(roc_auc_score(y_test, pred)))\nprint('test accuracy: {}'.format(accuracy_score(y_test, class_)))\nprint()",
"train roc-auc: 0.8450386398763523\ntrain accuracy: 0.7220630372492837\n\ntest roc-auc: 0.8354629629629629\ntest accuracy: 0.7137404580152672\n\n"
]
],
[
[
"That's it! Well done\n\n**Keep this code safe, as we will use this notebook later on, to build production code, in our next assignement!!**",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
4afd4f93d134ef676ead3a6269be3629d985f6a4
| 13,599 |
ipynb
|
Jupyter Notebook
|
_notebooks/2021-01-04-Lock-free-data-structures.ipynb
|
abhishekSingh210193/cs_craftmanship
|
f5f4ea3691e632be0e3cfc9f19593baa5b5bb4da
|
[
"Apache-2.0"
] | null | null | null |
_notebooks/2021-01-04-Lock-free-data-structures.ipynb
|
abhishekSingh210193/cs_craftmanship
|
f5f4ea3691e632be0e3cfc9f19593baa5b5bb4da
|
[
"Apache-2.0"
] | 2 |
2021-09-28T05:46:13.000Z
|
2022-02-26T10:26:45.000Z
|
_notebooks/2021-01-04-Lock-free-data-structures.ipynb
|
abhishekSingh210193/cs_craftsmanship
|
f5f4ea3691e632be0e3cfc9f19593baa5b5bb4da
|
[
"Apache-2.0"
] | null | null | null | 47.21875 | 1,008 | 0.680712 |
[
[
[
"# Data structures for fast infinte batching or streaming requests processing \n\n> Here we dicuss one of the coolest use of a data structures to address one of the very natural use case scenario of a server processing streaming requests from clients in order.Usually processing these requests involve a pipeline of operations applied based on request and multiple threads are in charge of dealing with these satges of pipeline. The requests gets accessed by these threads and the threads performing operations in the later part of the pipeline will have to wait for the earlier threads to finish their execution.\n\nThe usual way to ensure the correctness of multiple threads handling the same data concurrently is use locks.The problem is framed as a producer / consumer problems , where one threads finishes its operation and become producer of the data to be worked upon by another thread, which is a consumer. These two threads needs to be synchronized. \n",
"_____no_output_____"
],
[
"> Note: In this blog we will discuss a \"lock-free\" circular queue data structure called disruptor. It was designed to be an efficient concurrent message passing datastructure.The official implementations and other discussions are available [here](https://lmax-exchange.github.io/disruptor/#_discussion_blogs_other_useful_links). This blog intends to summarise its use case and show the points where the design of the disruptor scores big.",
"_____no_output_____"
],
[
"# LOCKS ARE BAD\n\nWhenever we have a scenario where mutliple concurrent running threads contend on a shared data structure and you need to ensure visibility of changes (i.e. a consumer thread can only get its hand over the data after the producer has processed it and put it for further processing). The usual and most common way to ensure these two requirements is to use a lock.\nLocks need the operating system to arbitrate which thread has the responsibility on a shared piece of data. The operating system might schedule other processes and the software's thread may be waiting in a queue. Moreover, if other threads get scheduled by the CPU then the cache memory of the softwares's thread will be overwritten and when it finally gets access to the CPU, it may have to go as far as the main memory to get it's required data. All this adds a lot of overhead and is evident by the simple experiment of incrementing a single shared variable. In the experiment below we increment a shared variable in three different ways. In the first case, we have a single process incrementing the variable, in the second case we again have two threads, but they synchronize their way through the operation using locks.\nIn the third case, we have two threads which increment the variables and they synchronize their operation using atomic locks.\n",
"_____no_output_____"
],
[
"## SINGLE PROCESS INCREMENTING A SINGLE VARIABLE",
"_____no_output_____"
]
],
[
[
"import time \ndef single_thread():\n start = time.time()\n x = 0\n for i in range(500000000):\n x += 1\n end = time.time()\n return(end-start)\nprint(single_thread())",
"28.66362190246582\n"
],
[
"#another way for single threaded increment using \nclass SingleThreadedCounter():\n def __init__(self):\n self.val = 0\n def increment(self):\n self.val += 1\n",
"_____no_output_____"
]
],
[
[
"## TWO PROCESS INCREMENTING A SINGLE VARIABLE",
"_____no_output_____"
]
],
[
[
"import time\nfrom threading import Thread, Lock\nmutex = Lock()\nx = 0\ndef thread_fcn():\n global x\n mutex.acquire()\n for i in range(250000000):\n x += 1\n mutex.release()\n\ndef mutex_increment():\n start = time.time()\n t1 = Thread(target=thread_fcn)\n t2 = Thread(target=thread_fcn)\n \n t1.start()\n t2.start()\n \n t1.join()\n t2.join()\n \n end = time.time()\n return (end-start)\nprint(mutex_increment())",
"36.418396949768066\n"
]
],
[
[
"> Note: As we can see that the time for performing the increment operation has gone up substantially when we would have expected it take half the time. ",
"_____no_output_____"
],
[
"> Important: In the rest of the blog we will take in a very usual scenario we see in streaming request processing.\n\nA client sends in requests to a server in a streaming fashion. The server at its end needs to process the client's request, it may have multiple stages of processing. For example, imagine the client sends in a stream of requests and the server in JSON format. Now the probable first task that the client needs to perform is to parse the JSON request.Imagine a thread being assigned to do this parsing task. It parses requests one after another and hands over the parsed request in some form to another thread which may be responsible for performing business logic for that client. Usually the data structure to manage this message passing and flow control in screaming scenario is handled by a queue data structure. The producer threads (parser thread) puts in parsed data in this queue, from which the consumer thread (the business logic thread) will read of the parsed data. Because we have two threads working concurrently on a single data structure (the queue) we can expect contention to kick in. ",
"_____no_output_____"
],
[
"## WHY QUEUES ARE FLAWED",
"_____no_output_____"
],
[
"The queue could be an obvious choice for handling data communication between multiple threads, but the queue data structure is fundamentally flawed for communication between multiple threads. Imagine the case of the first two threads of the a system using a queue for data communication, the listener thread and the parsing thread. The listener thread listens to bytes from the wire and puts it in a queue and the parser thread will pick up bytes from the queue and parse it. Typically, a queue data structure will have a head field, a tail field and a size field (to tell an empty queue from a full one). The head field will be modified by the parser thread and the tail field by the parser thread. The size field though will be modified by both of the threads and it effectively makes the queue data structure having two writers. \n\n\nMoreover, the entire data structure will fall in the same cache line and hence when say the listener thread modifies the tail field, the head field in another core also gets invalidated and needs to be fetched from a level 2 cache.\n",
"_____no_output_____"
],
[
"## CAN WE AVOID LOCKS ?",
"_____no_output_____"
],
[
"So, using a queue structure for inter-thread communication with expensive locks could cost a lot of performance for any system. Hence, we move towards a better data structure that solves the issues of synchronization among threads.\nThe data structure we use doesn't use locks.\nThe main components of the data structure are -\nA. A circular buffer\nB. A sequence number field which has a number indicating a specific slot in the circular buffer.\nC. Each of the worker threads have their own sequence number.\nThe circular buffer is written to by the producers . The producer in each case updates the sequence number for each of the circular buffers. The worker threads (consumer thread) have their own sequence number indicating the slots they have consumed so far from the circular buffer.\n",
"_____no_output_____"
],
[
">Note: In the design, each of the elements has a SINGLE WRITER. The producer threads of the circular ring write to the ring buffer, and its sequence number. The worker consumer threads will write their own local sequence number. No field or data have more than one writer in this data structure.",
"_____no_output_____"
],
[
"## WRITE OPERATION ON THE LOCK-FREE DATA STRUCTURE",
"_____no_output_____"
],
[
" Before writing a slot in the circular buffer, the thread has to make sure that it doesn't overwrite old bytes that have not yet been processed by the consumer thread. The consumer thread also maintains a sequence number, this number indicates the slots that have been already processed. So the producer thread before writing grabs the circular buffer's sequence number, adds one to it (mod size of the circular buffer) to get the next eligible slot for writing. But before putting in the bytes in that slot it checks with the dependent consumer thread (by reading their local sequence number) if they have processed this slot. If say the consumer has not yet processed this slot, then the producer thread goes in a busy wait till the slot is available to write to. When the slot is overwritten then the circular buffer's sequence number is updated by the producer thread. This indicates to consumer threads that they have a new slot to consume.\n",
"_____no_output_____"
],
[
"Writing to the circular buffers is a 2-phase commit. In the first phase, we check out a slot from the circular buffer. We can only check out a slot if it has already been consumed. This is ensured by following the logic mentioned above. Once the slot is checked out the producer writes the next byte to it. Then it sends a commit message to commit the entry by updating the circular buffer's sequence number to its next logical value(+1 mod size of the circular buffer)\n",
"_____no_output_____"
],
[
"## READ OPERATION ON THE LOCK_FREE DATA STRUCTURE",
"_____no_output_____"
],
[
"The consumer thread reads the slots from circular buffer -1. Before reading the next slot, it checks (read) the buffer's sequence number. This number is indicative of the slots till which the buffer can read.\n",
"_____no_output_____"
],
[
"## ENSURING THAT THE READS HAPPEN IN PROGRAM ORDER\n",
"_____no_output_____"
],
[
"There is just one piece of detail that needs to be addressed for the above data structure to work. Compilers and CPU take the liberty to reorder independent instructions for optimizations. This doesn’t have any issues in the single process case where the program’s logic integrity is maintained. But this could logic breakdown in case of multiple threads.\nImagine a typical simplified read/write to the circular buffer described above—\nSay the publisher thread’s sequence of operation is indicated in black, and the consumer thread’s in brown. The publisher checks in a slot and it updates the sequence number. Then the consumer thread reads the (wrong) sequence number of the buffer and goes on to access the slot which is yet to be written.\n",
"_____no_output_____"
],
[
"The way we could solve this is by putting memory fences around the variables which tells the compiler and CPU to not reorder reads / writes before and after those shared variables. In that way programs logic integrity is maintained.\n",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
4afd631e7d3dab91bca763dbe778d0a520affd70
| 12,058 |
ipynb
|
Jupyter Notebook
|
jupyter/paddlepaddle/face_mask_detection_paddlepaddle_zh.ipynb
|
andreabrduque/djl
|
06997ce4320d656cd133a509c36f6d1a5ade4d07
|
[
"Apache-2.0"
] | null | null | null |
jupyter/paddlepaddle/face_mask_detection_paddlepaddle_zh.ipynb
|
andreabrduque/djl
|
06997ce4320d656cd133a509c36f6d1a5ade4d07
|
[
"Apache-2.0"
] | null | null | null |
jupyter/paddlepaddle/face_mask_detection_paddlepaddle_zh.ipynb
|
andreabrduque/djl
|
06997ce4320d656cd133a509c36f6d1a5ade4d07
|
[
"Apache-2.0"
] | null | null | null | 34.15864 | 202 | 0.560541 |
[
[
[
"# 用飛槳+ DJL 實作人臉口罩辨識\n在這個教學中我們將會展示利用 PaddleHub 下載預訓練好的 PaddlePaddle 模型並針對範例照片做人臉口罩辨識。這個範例總共會分成兩個步驟:\n\n- 用臉部檢測模型識別圖片中的人臉(無論是否有戴口罩) \n- 確認圖片中的臉是否有戴口罩\n\n這兩個步驟會包含使用兩個 Paddle 模型,我們會在接下來的內容介紹兩個模型對應需要做的前後處理邏輯\n\n## 導入相關環境依賴及子類別\n在這個例子中的前處理飛槳深度學習引擎需要搭配 DJL 混合模式進行深度學習推理,原因是引擎本身沒有包含 NDArray 操作,因此需要藉用其他引擎的 NDArray 操作能力來完成。這邊我們導入 PyTorch 來做協同的前處理工作:",
"_____no_output_____"
]
],
[
[
"// %mavenRepo snapshots https://oss.sonatype.org/content/repositories/snapshots/\n\n%maven ai.djl:api:0.16.0\n%maven ai.djl.paddlepaddle:paddlepaddle-model-zoo:0.16.0\n%maven org.slf4j:slf4j-simple:1.7.32\n\n// second engine to do preprocessing and postprocessing\n%maven ai.djl.pytorch:pytorch-engine:0.16.0",
"_____no_output_____"
],
[
"import ai.djl.*;\nimport ai.djl.inference.*;\nimport ai.djl.modality.*;\nimport ai.djl.modality.cv.*;\nimport ai.djl.modality.cv.output.*;\nimport ai.djl.modality.cv.transform.*;\nimport ai.djl.modality.cv.translator.*;\nimport ai.djl.modality.cv.util.*;\nimport ai.djl.ndarray.*;\nimport ai.djl.ndarray.types.Shape;\nimport ai.djl.repository.zoo.*;\nimport ai.djl.translate.*;\n\nimport java.io.*;\nimport java.nio.file.*;\nimport java.util.*;",
"_____no_output_____"
]
],
[
[
"## 臉部偵測模型\n現在我們可以開始處理第一個模型,在將圖片輸入臉部檢測模型前我們必須先做一些預處理:\n•\t調整圖片尺寸: 以特定比例縮小圖片\n•\t用一個數值對縮小後圖片正規化\n對開發者來說好消息是,DJL 提供了 Translator 介面來幫助開發做這樣的預處理. 一個比較粗略的 Translator 架構如下:\n\n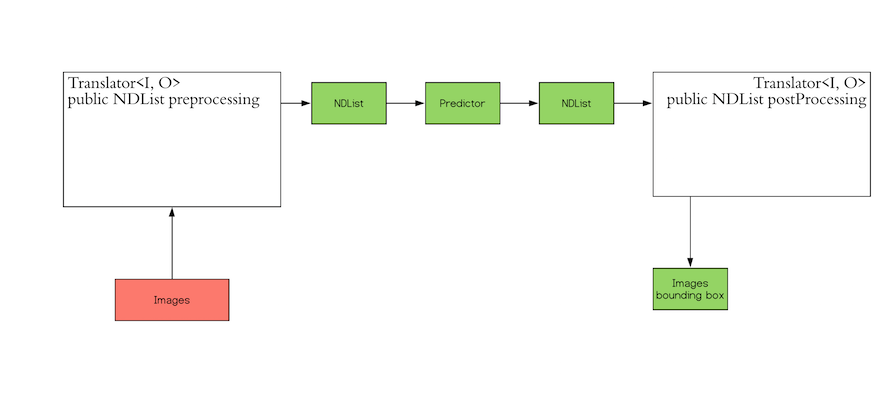\n\n在接下來的段落,我們會利用一個 FaceTranslator 子類別實作來完成工作\n### 預處理\n在這個階段我們會讀取一張圖片並且對其做一些事先的預處理,讓我們先示範讀取一張圖片:",
"_____no_output_____"
]
],
[
[
"String url = \"https://raw.githubusercontent.com/PaddlePaddle/PaddleHub/release/v1.5/demo/mask_detection/python/images/mask.jpg\";\nImage img = ImageFactory.getInstance().fromUrl(url);\nimg.getWrappedImage();",
"_____no_output_____"
]
],
[
[
"接著,讓我們試著對圖片做一些預處理的轉換:",
"_____no_output_____"
]
],
[
[
"NDList processImageInput(NDManager manager, Image input, float shrink) {\n NDArray array = input.toNDArray(manager);\n Shape shape = array.getShape();\n array = NDImageUtils.resize(\n array, (int) (shape.get(1) * shrink), (int) (shape.get(0) * shrink));\n array = array.transpose(2, 0, 1).flip(0); // HWC -> CHW BGR -> RGB\n NDArray mean = manager.create(new float[] {104f, 117f, 123f}, new Shape(3, 1, 1));\n array = array.sub(mean).mul(0.007843f); // normalization\n array = array.expandDims(0); // make batch dimension\n return new NDList(array);\n}\n\nprocessImageInput(NDManager.newBaseManager(), img, 0.5f);",
"_____no_output_____"
]
],
[
[
"如上述所見,我們已經把圖片轉成如下尺寸的 NDArray: (披量, 通道(RGB), 高度, 寬度). 這是物件檢測模型輸入的格式\n### 後處理\n當我們做後處理時, 模型輸出的格式是 (number_of_boxes, (class_id, probability, xmin, ymin, xmax, ymax)). 我們可以將其存入預先建立好的 DJL 子類別 DetectedObjects 以便做後續操作. 我們假設有一組推論後的輸出是 ((1, 0.99, 0.2, 0.4, 0.5, 0.8)) 並且試著把人像框顯示在圖片上",
"_____no_output_____"
]
],
[
[
"DetectedObjects processImageOutput(NDList list, List<String> className, float threshold) {\n NDArray result = list.singletonOrThrow();\n float[] probabilities = result.get(\":,1\").toFloatArray();\n List<String> names = new ArrayList<>();\n List<Double> prob = new ArrayList<>();\n List<BoundingBox> boxes = new ArrayList<>();\n for (int i = 0; i < probabilities.length; i++) {\n if (probabilities[i] >= threshold) {\n float[] array = result.get(i).toFloatArray();\n names.add(className.get((int) array[0]));\n prob.add((double) probabilities[i]);\n boxes.add(\n new Rectangle(\n array[2], array[3], array[4] - array[2], array[5] - array[3]));\n }\n }\n return new DetectedObjects(names, prob, boxes);\n}\n\nNDArray tempOutput = NDManager.newBaseManager().create(new float[]{1f, 0.99f, 0.1f, 0.1f, 0.2f, 0.2f}, new Shape(1, 6));\nDetectedObjects testBox = processImageOutput(new NDList(tempOutput), Arrays.asList(\"Not Face\", \"Face\"), 0.7f);\nImage newImage = img.duplicate();\nnewImage.drawBoundingBoxes(testBox);\nnewImage.getWrappedImage();",
"_____no_output_____"
]
],
[
[
"### 生成一個翻譯器並執行推理任務\n透過這個步驟,你會理解 DJL 中的前後處理如何運作,現在讓我們把前數的幾個步驟串在一起並對真實圖片進行操作:",
"_____no_output_____"
]
],
[
[
"class FaceTranslator implements NoBatchifyTranslator<Image, DetectedObjects> {\n\n private float shrink;\n private float threshold;\n private List<String> className;\n\n FaceTranslator(float shrink, float threshold) {\n this.shrink = shrink;\n this.threshold = threshold;\n className = Arrays.asList(\"Not Face\", \"Face\");\n }\n\n @Override\n public DetectedObjects processOutput(TranslatorContext ctx, NDList list) {\n return processImageOutput(list, className, threshold);\n }\n\n @Override\n public NDList processInput(TranslatorContext ctx, Image input) {\n return processImageInput(ctx.getNDManager(), input, shrink);\n }\n}",
"_____no_output_____"
]
],
[
[
"要執行這個人臉檢測推理,我們必須先從 DJL 的 Paddle Model Zoo 讀取模型,在讀取模型之前我們必須指定好 `Crieteria` . `Crieteria` 是用來確認要從哪邊讀取模型而後執行 `Translator` 來進行模型導入. 接著,我們只要利用 `Predictor` 就可以開始進行推論",
"_____no_output_____"
]
],
[
[
"Criteria<Image, DetectedObjects> criteria = Criteria.builder()\n .setTypes(Image.class, DetectedObjects.class)\n .optModelUrls(\"djl://ai.djl.paddlepaddle/face_detection/0.0.1/mask_detection\")\n .optFilter(\"flavor\", \"server\")\n .optTranslator(new FaceTranslator(0.5f, 0.7f))\n .build();\n \nvar model = criteria.loadModel();\nvar predictor = model.newPredictor();\n\nDetectedObjects inferenceResult = predictor.predict(img);\nnewImage = img.duplicate();\nnewImage.drawBoundingBoxes(inferenceResult);\nnewImage.getWrappedImage();",
"_____no_output_____"
]
],
[
[
"如圖片所示,這個推論服務已經可以正確的辨識出圖片中的三張人臉\n## 口罩分類模型\n一旦有了圖片的座標,我們就可以將圖片裁剪到適當大小並且將其傳給口罩分類模型做後續的推論\n### 圖片裁剪\n圖中方框位置的數值範圍從0到1, 只要將這個數值乘上圖片的長寬我們就可以將方框對應到圖片中的準確位置. 為了使裁剪後的圖片有更好的精確度,我們將圖片裁剪成方形,讓我們示範一下:",
"_____no_output_____"
]
],
[
[
"int[] extendSquare(\n double xmin, double ymin, double width, double height, double percentage) {\n double centerx = xmin + width / 2;\n double centery = ymin + height / 2;\n double maxDist = Math.max(width / 2, height / 2) * (1 + percentage);\n return new int[] {\n (int) (centerx - maxDist), (int) (centery - maxDist), (int) (2 * maxDist)\n };\n}\n\nImage getSubImage(Image img, BoundingBox box) {\n Rectangle rect = box.getBounds();\n int width = img.getWidth();\n int height = img.getHeight();\n int[] squareBox =\n extendSquare(\n rect.getX() * width,\n rect.getY() * height,\n rect.getWidth() * width,\n rect.getHeight() * height,\n 0.18);\n return img.getSubImage(squareBox[0], squareBox[1], squareBox[2], squareBox[2]);\n}\n\nList<DetectedObjects.DetectedObject> faces = inferenceResult.items();\ngetSubImage(img, faces.get(2).getBoundingBox()).getWrappedImage();",
"_____no_output_____"
]
],
[
[
"### 事先準備 Translator 並讀取模型\n在使用臉部檢測模型的時候,我們可以利用 DJL 預先建好的 `ImageClassificationTranslator` 並且加上一些轉換。這個 Translator 提供了一些基礎的圖片翻譯處理並且同時包含一些進階的標準化圖片處理。以這個例子來說, 我們不需要額外建立新的 `Translator` 而使用預先建立的就可以",
"_____no_output_____"
]
],
[
[
"var criteria = Criteria.builder()\n .setTypes(Image.class, Classifications.class)\n .optModelUrls(\"djl://ai.djl.paddlepaddle/mask_classification/0.0.1/mask_classification\")\n .optFilter(\"flavor\", \"server\")\n .optTranslator(\n ImageClassificationTranslator.builder()\n .addTransform(new Resize(128, 128))\n .addTransform(new ToTensor()) // HWC -> CHW div(255)\n .addTransform(\n new Normalize(\n new float[] {0.5f, 0.5f, 0.5f},\n new float[] {1.0f, 1.0f, 1.0f}))\n .addTransform(nd -> nd.flip(0)) // RGB -> GBR\n .build())\n .build();\n\nvar classifyModel = criteria.loadModel();\nvar classifier = classifyModel.newPredictor();",
"_____no_output_____"
]
],
[
[
"### 執行推論任務\n最後,要完成一個口罩識別的任務,我們只需要將上述的步驟合在一起即可。我們先將圖片做裁剪後並對其做上述的推論操作,結束之後再生成一個新的分類子類別 `DetectedObjects`:",
"_____no_output_____"
]
],
[
[
"List<String> names = new ArrayList<>();\nList<Double> prob = new ArrayList<>();\nList<BoundingBox> rect = new ArrayList<>();\nfor (DetectedObjects.DetectedObject face : faces) {\n Image subImg = getSubImage(img, face.getBoundingBox());\n Classifications classifications = classifier.predict(subImg);\n names.add(classifications.best().getClassName());\n prob.add(face.getProbability());\n rect.add(face.getBoundingBox());\n}\n\nnewImage = img.duplicate();\nnewImage.drawBoundingBoxes(new DetectedObjects(names, prob, rect));\nnewImage.getWrappedImage();",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
4afd6c40f7eeb73855f07ee7a8e6c2d2dcbf469a
| 106,571 |
ipynb
|
Jupyter Notebook
|
5 Xgboost/IrisClassification/iris_xgboost_localmode.ipynb
|
jaypeeml/AWSSagemaker
|
9ab931065e9f2af0b1c102476781a63c917e7c47
|
[
"Apache-2.0"
] | 167 |
2019-04-07T16:33:56.000Z
|
2022-03-24T12:13:13.000Z
|
5 Xgboost/IrisClassification/iris_xgboost_localmode.ipynb
|
jaypeeml/AWSSagemaker
|
9ab931065e9f2af0b1c102476781a63c917e7c47
|
[
"Apache-2.0"
] | 5 |
2019-04-13T06:39:43.000Z
|
2019-11-09T06:09:56.000Z
|
5 Xgboost/IrisClassification/iris_xgboost_localmode.ipynb
|
jaypeeml/AWSSagemaker
|
9ab931065e9f2af0b1c102476781a63c917e7c47
|
[
"Apache-2.0"
] | 317 |
2019-04-07T16:34:00.000Z
|
2022-03-31T11:20:32.000Z
| 94.0609 | 19,932 | 0.811262 |
[
[
[
"## Train a model with Iris data using XGBoost algorithm\n### Model is trained with XGBoost installed in notebook instance\n### In the later examples, we will train using SageMaker's XGBoost algorithm",
"_____no_output_____"
]
],
[
[
"# Install xgboost in notebook instance.\n#### Command to install xgboost\n!pip install xgboost==1.2",
"Collecting xgboost==1.2\n Downloading xgboost-1.2.0-py3-none-manylinux2010_x86_64.whl (148.9 MB)\n\u001b[K |████████████████████████████████| 148.9 MB 63 kB/s /s eta 0:00:01 |███████████████▊ | 73.2 MB 19.3 MB/s eta 0:00:04\n\u001b[?25hRequirement already satisfied: numpy in /home/ec2-user/anaconda3/envs/python3/lib/python3.6/site-packages (from xgboost==1.2) (1.19.5)\nRequirement already satisfied: scipy in /home/ec2-user/anaconda3/envs/python3/lib/python3.6/site-packages (from xgboost==1.2) (1.5.3)\nInstalling collected packages: xgboost\nSuccessfully installed xgboost-1.2.0\n"
],
[
"import sys\nimport numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport itertools\nimport xgboost as xgb\n\nfrom sklearn import preprocessing\nfrom sklearn.metrics import classification_report, confusion_matrix",
"_____no_output_____"
],
[
"column_list_file = 'iris_train_column_list.txt'\ntrain_file = 'iris_train.csv'\nvalidation_file = 'iris_validation.csv'",
"_____no_output_____"
],
[
"columns = ''\nwith open(column_list_file,'r') as f:\n columns = f.read().split(',')",
"_____no_output_____"
],
[
"columns",
"_____no_output_____"
],
[
"# Encode Class Labels to integers\n# Labeled Classes\nlabels=[0,1,2]\nclasses = ['Iris-setosa', 'Iris-versicolor', 'Iris-virginica']\nle = preprocessing.LabelEncoder()\nle.fit(classes)",
"_____no_output_____"
],
[
"# Specify the column names as the file does not have column header\ndf_train = pd.read_csv(train_file,names=columns)\ndf_validation = pd.read_csv(validation_file,names=columns)",
"_____no_output_____"
],
[
"df_train.head()",
"_____no_output_____"
],
[
"df_validation.head()",
"_____no_output_____"
],
[
"X_train = df_train.iloc[:,1:] # Features: 1st column onwards \ny_train = df_train.iloc[:,0].ravel() # Target: 0th column\n\nX_validation = df_validation.iloc[:,1:]\ny_validation = df_validation.iloc[:,0].ravel()",
"_____no_output_____"
],
[
"# Launch a classifier\n# XGBoost Training Parameter Reference: \n# https://xgboost.readthedocs.io/en/latest/parameter.html\n\nclassifier = xgb.XGBClassifier(objective=\"multi:softmax\",\n num_class=3,\n n_estimators=100)",
"_____no_output_____"
],
[
"classifier",
"_____no_output_____"
],
[
"classifier.fit(X_train,\n y_train,\n eval_set = [(X_train, y_train), (X_validation, y_validation)],\n eval_metric=['mlogloss'],\n early_stopping_rounds=10)\n\n# early_stopping_rounds - needs to be passed in as a hyperparameter in SageMaker XGBoost implementation\n# \"The model trains until the validation score stops improving. \n# Validation error needs to decrease at least every early_stopping_rounds to continue training.\n# Amazon SageMaker hosting uses the best model for inference.\"",
"[0]\tvalidation_0-mlogloss:0.73876\tvalidation_1-mlogloss:0.74994\nMultiple eval metrics have been passed: 'validation_1-mlogloss' will be used for early stopping.\n\nWill train until validation_1-mlogloss hasn't improved in 10 rounds.\n[1]\tvalidation_0-mlogloss:0.52787\tvalidation_1-mlogloss:0.55401\n[2]\tvalidation_0-mlogloss:0.38960\tvalidation_1-mlogloss:0.42612\n[3]\tvalidation_0-mlogloss:0.29429\tvalidation_1-mlogloss:0.34328\n[4]\tvalidation_0-mlogloss:0.22736\tvalidation_1-mlogloss:0.29000\n[5]\tvalidation_0-mlogloss:0.17920\tvalidation_1-mlogloss:0.24961\n[6]\tvalidation_0-mlogloss:0.14403\tvalidation_1-mlogloss:0.22234\n[7]\tvalidation_0-mlogloss:0.11664\tvalidation_1-mlogloss:0.20338\n[8]\tvalidation_0-mlogloss:0.09668\tvalidation_1-mlogloss:0.18999\n[9]\tvalidation_0-mlogloss:0.08128\tvalidation_1-mlogloss:0.18190\n[10]\tvalidation_0-mlogloss:0.06783\tvalidation_1-mlogloss:0.17996\n[11]\tvalidation_0-mlogloss:0.05794\tvalidation_1-mlogloss:0.18029\n[12]\tvalidation_0-mlogloss:0.05011\tvalidation_1-mlogloss:0.18306\n[13]\tvalidation_0-mlogloss:0.04428\tvalidation_1-mlogloss:0.18471\n[14]\tvalidation_0-mlogloss:0.03993\tvalidation_1-mlogloss:0.18693\n[15]\tvalidation_0-mlogloss:0.03615\tvalidation_1-mlogloss:0.18553\n[16]\tvalidation_0-mlogloss:0.03310\tvalidation_1-mlogloss:0.18571\n[17]\tvalidation_0-mlogloss:0.03065\tvalidation_1-mlogloss:0.18615\n[18]\tvalidation_0-mlogloss:0.02874\tvalidation_1-mlogloss:0.18930\n[19]\tvalidation_0-mlogloss:0.02739\tvalidation_1-mlogloss:0.18989\n[20]\tvalidation_0-mlogloss:0.02639\tvalidation_1-mlogloss:0.19251\nStopping. Best iteration:\n[10]\tvalidation_0-mlogloss:0.06783\tvalidation_1-mlogloss:0.17996\n\n"
],
[
"eval_result = classifier.evals_result()",
"_____no_output_____"
],
[
"training_rounds = range(len(eval_result['validation_0']['mlogloss']))",
"_____no_output_____"
],
[
"print(training_rounds)",
"range(0, 20)\n"
],
[
"plt.scatter(x=training_rounds,y=eval_result['validation_0']['mlogloss'],label='Training Error')\nplt.scatter(x=training_rounds,y=eval_result['validation_1']['mlogloss'],label='Validation Error')\nplt.grid(True)\nplt.xlabel('Iteration')\nplt.ylabel('LogLoss')\nplt.title('Training Vs Validation Error')\nplt.legend()\nplt.show()",
"_____no_output_____"
],
[
"xgb.plot_importance(classifier)\nplt.show()",
"_____no_output_____"
],
[
"df = pd.read_csv(validation_file,names=columns)",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
],
[
"X_test = df.iloc[:,1:]\nprint(X_test[:5])",
" sepal_length sepal_width petal_length petal_width\n0 5.8 2.7 4.1 1.0\n1 4.8 3.4 1.6 0.2\n2 6.0 2.2 4.0 1.0\n3 6.4 3.1 5.5 1.8\n4 6.7 2.5 5.8 1.8\n"
],
[
"result = classifier.predict(X_test)",
"_____no_output_____"
],
[
"result[:5]",
"_____no_output_____"
],
[
"df['predicted_class'] = result #le.inverse_transform(result)",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
],
[
"# Compare performance of Actual and Model 1 Prediction\nplt.figure()\nplt.scatter(df.index,df['encoded_class'],label='Actual')\nplt.scatter(df.index,df['predicted_class'],label='Predicted',marker='^')\nplt.legend(loc=4)\nplt.yticks([0,1,2])\nplt.xlabel('Sample')\nplt.ylabel('Class')\nplt.show()",
"_____no_output_____"
]
],
[
[
"<h2>Confusion Matrix</h2>\nConfusion Matrix is a table that summarizes performance of classification model.<br><br>",
"_____no_output_____"
]
],
[
[
"# Reference: \n# https://scikit-learn.org/stable/auto_examples/model_selection/plot_confusion_matrix.html\ndef plot_confusion_matrix(cm, classes,\n normalize=False,\n title='Confusion matrix',\n cmap=plt.cm.Blues):\n \"\"\"\n This function prints and plots the confusion matrix.\n Normalization can be applied by setting `normalize=True`.\n \"\"\"\n if normalize:\n cm = cm.astype('float') / cm.sum(axis=1)[:, np.newaxis]\n #print(\"Normalized confusion matrix\")\n #else:\n # print('Confusion matrix, without normalization')\n\n #print(cm)\n\n plt.imshow(cm, interpolation='nearest', cmap=cmap)\n plt.title(title)\n plt.colorbar()\n tick_marks = np.arange(len(classes))\n plt.xticks(tick_marks, classes, rotation=45)\n plt.yticks(tick_marks, classes)\n\n fmt = '.2f' if normalize else 'd'\n thresh = cm.max() / 2.\n for i, j in itertools.product(range(cm.shape[0]), range(cm.shape[1])):\n plt.text(j, i, format(cm[i, j], fmt),\n horizontalalignment=\"center\",\n color=\"white\" if cm[i, j] > thresh else \"black\")\n\n plt.ylabel('True label')\n plt.xlabel('Predicted label')\n plt.tight_layout()",
"_____no_output_____"
],
[
"# Compute confusion matrix\ncnf_matrix = confusion_matrix(df['encoded_class'],\n df['predicted_class'],labels=labels)",
"_____no_output_____"
],
[
"cnf_matrix",
"_____no_output_____"
],
[
"# Plot confusion matrix\nplt.figure()\nplot_confusion_matrix(cnf_matrix, classes=classes,\n title='Confusion matrix - Count')",
"_____no_output_____"
],
[
"# Plot confusion matrix\nplt.figure()\nplot_confusion_matrix(cnf_matrix, classes=classes,\n title='Confusion matrix - Count',normalize=True)",
"_____no_output_____"
],
[
"print(classification_report(\n df['encoded_class'],\n df['predicted_class'],\n labels=labels,\n target_names=classes))",
" precision recall f1-score support\n\n Iris-setosa 1.00 1.00 1.00 16\nIris-versicolor 0.91 0.91 0.91 11\n Iris-virginica 0.94 0.94 0.94 18\n\n accuracy 0.96 45\n macro avg 0.95 0.95 0.95 45\n weighted avg 0.96 0.96 0.96 45\n\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4afd8187a3c49b4bad23543f562c39ce71da1408
| 82,063 |
ipynb
|
Jupyter Notebook
|
Demonstration.ipynb
|
ashrafya/BreastCancerDetection-IDC
|
faa07398520aaad8e89120bfa2419d6017ada82b
|
[
"MIT"
] | null | null | null |
Demonstration.ipynb
|
ashrafya/BreastCancerDetection-IDC
|
faa07398520aaad8e89120bfa2419d6017ada82b
|
[
"MIT"
] | null | null | null |
Demonstration.ipynb
|
ashrafya/BreastCancerDetection-IDC
|
faa07398520aaad8e89120bfa2419d6017ada82b
|
[
"MIT"
] | null | null | null | 455.905556 | 19,540 | 0.94096 |
[
[
[
"import numpy as np\nimport time\nimport torch\nimport torch.nn as nn\nimport torch.nn.functional as F\nimport torch.optim as optim\nimport torchvision\nfrom torch.utils.data.sampler import SubsetRandomSampler\nimport torchvision.transforms as transforms\nimport matplotlib.pyplot as plt\nimport glob as glob\nimport os\n\nos.environ['KMP_DUPLICATE_LIB_OK']='True'\n\nclass CancerDetectModel(nn.Module):\n def __init__(self):\n super(CancerDetectModel, self).__init__()\n self.name = \"CancerDetectModel\"\n self.conv1 = torch.nn.Conv2d(3, 32, 5, 5)\n self.conv2 = torch.nn.Conv2d(32, 64, 3, 1)\n self.conv3 = torch.nn.Conv2d(64, 128, 4, 2)\n self.fc = nn.Linear(3 * 3 * 128, 2)\n \n def forward(self, x):\n x = F.leaky_relu(self.conv1(x))\n x = F.leaky_relu(self.conv2(x))\n x = F.leaky_relu(self.conv3(x))\n x = x.view(-1, 3 * 3 * 128)\n x = self.fc(x)\n return x",
"_____no_output_____"
],
[
"pos_image_dir = \"./demo_images/1/*.png\"\nneg_image_dir = \"./demo_images/0/*.png\"\n\nbest_model = CancerDetectModel()\nmodel_path = \"./model_files/model_CancerDetectModel_bs64_lr0.001_epoch2\"\nstate = torch.load(model_path)\nbest_model.load_state_dict(state)\n\nfor file in glob.glob(pos_image_dir):\n image = plt.imread(file)\n plt.figure()\n plt.imshow(image)\n tensor = torch.from_numpy(image)\n result = torch.argmax(best_model(torch.unsqueeze(tensor.transpose(0,2), 0))).item()\n print(\"Classification: {}\".format(result))",
"Classification: 1\nClassification: 1\n"
],
[
"for file in glob.glob(neg_image_dir):\n image = plt.imread(file)\n plt.figure()\n plt.imshow(image)\n tensor = torch.from_numpy(image)\n result = torch.argmax(best_model(torch.unsqueeze(tensor.transpose(0,2), 0))).item()\n print(\"Classification: {}\".format(result))",
"Classification: 0\nClassification: 0\n"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code"
]
] |
4afd8e54e80c7d11efc7c82052c691b3bdd2e973
| 6,303 |
ipynb
|
Jupyter Notebook
|
project-bikesharing/keyboard-shortcuts.ipynb
|
thebrightshade/Udacity-Project-Bikesharing
|
d6e83e9574575f169b7e8643c016f3adc8a23d1e
|
[
"MIT"
] | null | null | null |
project-bikesharing/keyboard-shortcuts.ipynb
|
thebrightshade/Udacity-Project-Bikesharing
|
d6e83e9574575f169b7e8643c016f3adc8a23d1e
|
[
"MIT"
] | null | null | null |
project-bikesharing/keyboard-shortcuts.ipynb
|
thebrightshade/Udacity-Project-Bikesharing
|
d6e83e9574575f169b7e8643c016f3adc8a23d1e
|
[
"MIT"
] | null | null | null | 33 | 508 | 0.61669 |
[
[
[
"# Keyboard shortcuts\n\nIn this notebook, you'll get some practice using keyboard shortcuts. These are key to becoming proficient at using notebooks and will greatly increase your work speed.\n\nFirst up, switching between edit mode and command mode. Edit mode allows you to type into cells while command mode will use key presses to execute commands such as creating new cells and openning the command palette. When you select a cell, you can tell which mode you're currently working in by the color of the box around the cell. In edit mode, the box and thick left border are colored green. In command mode, they are colored blue. Also in edit mode, you should see a cursor in the cell itself.\n\nBy default, when you create a new cell or move to the next one, you'll be in command mode. To enter edit mode, press Enter/Return. To go back from edit mode to command mode, press Escape.\n\n> **Exercise:** Click on this cell, then press Enter + Shift to get to the next cell. Switch between edit and command mode a few times.",
"_____no_output_____"
]
],
[
[
"# mode practice",
"_____no_output_____"
]
],
[
[
"## Help with commands\n\nIf you ever need to look up a command, you can bring up the list of shortcuts by pressing `H` in command mode. The keyboard shortcuts are also available above in the Help menu. Go ahead and try it now.",
"_____no_output_____"
],
[
"## Creating new cells\n\nOne of the most common commands is creating new cells. You can create a cell above the current cell by pressing `A` in command mode. Pressing `B` will create a cell below the currently selected cell.",
"_____no_output_____"
],
[
"> **Exercise:** Create a cell above this cell using the keyboard command.",
"_____no_output_____"
],
[
"> **Exercise:** Create a cell below this cell using the keyboard command.",
"_____no_output_____"
],
[
"## Switching between Markdown and code\n\nWith keyboard shortcuts, it is quick and simple to switch between Markdown and code cells. To change from Markdown to a code cell, press `Y`. To switch from code to Markdown, press `M`.\n\n> **Exercise:** Switch the cell below between Markdown and code cells.",
"_____no_output_____"
]
],
[
[
"## Practice here\n\ndef fibo(n): # Recursive Fibonacci sequence!\n if n == 0:\n return 0\n elif n == 1:\n return 1\n return fibo(n-1) + fibo(n-2)",
"_____no_output_____"
]
],
[
[
"## Line numbers\n\nA lot of times it is helpful to number the lines in your code for debugging purposes. You can turn on numbers by pressing `L` (in command mode of course) on a code cell.\n\n> **Exercise:** Turn line numbers on and off in the above code cell.",
"_____no_output_____"
],
[
"## Deleting cells\n\nDeleting cells is done by pressing `D` twice in a row so `D`, `D`. This is to prevent accidently deletions, you have to press the button twice!\n\n> **Exercise:** Delete the cell below.",
"_____no_output_____"
],
[
"## Saving the notebook\n\nNotebooks are autosaved every once in a while, but you'll often want to save your work between those times. To save the book, press `S`. So easy!",
"_____no_output_____"
],
[
"## The Command Palette\n\nYou can easily access the command palette by pressing Shift + Control/Command + `P`. \n\n> **Note:** This won't work in Firefox and Internet Explorer unfortunately. There is already a keyboard shortcut assigned to those keys in those browsers. However, it does work in Chrome and Safari.\n\nThis will bring up the command palette where you can search for commands that aren't available through the keyboard shortcuts. For instance, there are buttons on the toolbar that move cells up and down (the up and down arrows), but there aren't corresponding keyboard shortcuts. To move a cell down, you can open up the command palette and type in \"move\" which will bring up the move commands.\n\n> **Exercise:** Use the command palette to move the cell below down one position.",
"_____no_output_____"
]
],
[
[
"# below this cell",
"_____no_output_____"
],
[
"# Move this cell down",
"_____no_output_____"
]
],
[
[
"## Finishing up\n\nThere is plenty more you can do such as copying, cutting, and pasting cells. I suggest getting used to using the keyboard shortcuts, you’ll be much quicker at working in notebooks. When you become proficient with them, you'll rarely need to move your hands away from the keyboard, greatly speeding up your work.\n\nRemember, if you ever need to see the shortcuts, just press `H` in command mode.\n",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
4afd93bbdce3a8f46fd9a23fa74da87f46a7c95f
| 1,671 |
ipynb
|
Jupyter Notebook
|
Mundo01/Desafio013.ipynb
|
BrunaKuntz/PythonMundo01
|
998eb7ffff97b692de65a6ce522bae57f6582279
|
[
"Apache-2.0"
] | null | null | null |
Mundo01/Desafio013.ipynb
|
BrunaKuntz/PythonMundo01
|
998eb7ffff97b692de65a6ce522bae57f6582279
|
[
"Apache-2.0"
] | null | null | null |
Mundo01/Desafio013.ipynb
|
BrunaKuntz/PythonMundo01
|
998eb7ffff97b692de65a6ce522bae57f6582279
|
[
"Apache-2.0"
] | null | null | null | 26.52381 | 247 | 0.503291 |
[
[
[
"<a href=\"https://colab.research.google.com/github/BrunaKuntz/Python-Curso-em-Video/blob/main/Mundo01/Desafio013.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"\n\n\n# **Desafio 013**\n**Python 3 - 1º Mundo**\n\nDescrição: Faça um algoritmo que leia o salário de um funcionário e mostre seu novo salário, com 15% de aumento.\n\nLink: https://www.youtube.com/watch?v=cTkivN8XcJ0&list=TLPQMTMwNTIwMjH5tRDtoC1oig&index=14",
"_____no_output_____"
]
],
[
[
"si = float(input('Qual o seu salário? R$'))\naum = (si*15)/100\nsa = si + aum\nprint(f'Seu novo salário é de R${sa:.2f}.')",
"_____no_output_____"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
]
] |
4afdab58c80395274c66b1b756be1b687b3650fc
| 106,341 |
ipynb
|
Jupyter Notebook
|
jit_qr.ipynb
|
alewis/AdvMPS
|
3a8544a74c740db8618638acb9b01d8a7b111202
|
[
"MIT"
] | null | null | null |
jit_qr.ipynb
|
alewis/AdvMPS
|
3a8544a74c740db8618638acb9b01d8a7b111202
|
[
"MIT"
] | null | null | null |
jit_qr.ipynb
|
alewis/AdvMPS
|
3a8544a74c740db8618638acb9b01d8a7b111202
|
[
"MIT"
] | null | null | null | 106.234765 | 35,302 | 0.765387 |
[
[
[
"<a href=\"https://colab.research.google.com/github/alewis/AdvMPS/blob/master/jit_qr.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"\n\n```\n# This is formatted as code\n```\n\n# Practice writing efficient Jit code on the Householder QR algorithm.",
"_____no_output_____"
]
],
[
[
"import jax\nimport jax.numpy as jnp\nfrom jax.ops import index, index_update, index_add\nimport numpy as np\n\n# import jit\n# import scipy as sp\n# import tensorflow as tf\n# from copy import deepcopy\nimport random\nimport time\nimport functools\nimport matplotlib.pyplot as plt\nimport sys\nimport unittest\nimport cProfile\nimport pstats\nimport os\nimport getpass\nimport urllib\n%load_ext autoreload\n%autoreload 2\nrepo_loaded = False\n",
"_____no_output_____"
]
],
[
[
"\n# Credentials.\nThis block imports the git repo so that we can use the relevant libraries. \nChange the string in 'username' to your GitHub username. Input your GitHub password when requested by the dialog. Your password is not saved in the\nnotebook. Solution by Vinoj John Hosan at https://stackoverflow.com/questions/48350226/methods-for-using-git-with-google-colab.\n",
"_____no_output_____"
]
],
[
[
" def load_repo_from_colab(username, repo_name):\n password = getpass.getpass('Password: ')\n password = urllib.parse.quote(password) # your password is converted into url format\n\n cmd_string = 'git clone https://{0}:{1}@github.com/{0}/{2}.git'.format(username, password, repo_name)\n\n os.system(cmd_string)\n cmd_string, password = \"\", \"\" # removing the password from the variable\n",
"_____no_output_____"
],
[
"",
"Already up to date.\n"
],
[
"def is_local():\n return not 'google.colab' in sys.modules\n\nif not is_local():\n if not repo_loaded:\n username = \"alewis\"\n repo_name = \"dfact\"\n load_repo_from_colab(username, repo_name)\n repo_loaded = True\n else:\n !git -C /content/dfact pull\nelse:\n to_append = '/Users/adam/projects/'\n if to_append not in sys.path:\n sys.path.append(to_append)\n\n\nimport dfact.utv as utv\nimport dfact.matutils as matutils\nfrom dfact.matutils import dag\nimport dfact.utv_tests as utv_tests\n#import dfact.qr as qr",
"Password: ··········\n"
]
],
[
[
"#### Matplotlib Customizations",
"_____no_output_____"
]
],
[
[
"from matplotlib import cycler\ncolors = cycler('color',\n ['#EE6666', '#3388BB', '#9988DD',\n '#EECC55', '#88BB44', '#FFBBBB'])\n# plt.rc('axes', facecolor='#E6E6E6', edgecolor='none',\n# axisbelow=True, grid=True, prop_cycle=colors)\n# plt.rc('grid', color='w', linestyle='solid')\n# plt.rc('xtick', direction='out', color='gray')\n# plt.rc('ytick', direction='out', color='gray')\n# plt.rc('patch', edgecolor='#E6E6E6')\nplt.rc('figure', figsize=(10, 10))\nplt.rc('xtick', labelsize=18)\nplt.rc('ytick', labelsize=18)\nplt.rc('font', size=22 )\nplt.rc('lines', linewidth=2)\n",
"_____no_output_____"
]
],
[
[
"# Notebook Code",
"_____no_output_____"
]
],
[
[
"\"\"\"\nJax implementation of householder QR exposing the low-level functionality\nneeded for the UTV decomposition.\n\nAdam GM Lewis\[email protected]\[email protected]\n\"\"\"\nimport jax\nfrom jax.ops import index_update, index_add, index\nimport jax.numpy as jnp\nimport numpy as np\n\nimport dfact.matutils as matutils\nfrom dfact.matutils import dag\n\n###############################################################################\n# UTILITIES\n###############################################################################\[email protected]\ndef sign(num):\n \"\"\"\n Sign function using the standard (?) convention sign(x) = x / |x| in\n the complex case. Returns 0 with the same type as x if x == 0.\n Note the numpy implementation uses the slightly different convention\n sign(x) = x / sqrt(x * x).\n \"\"\"\n result = jax.lax.cond(num == 0,\n num, lambda num: 0*num, # 0, casted, if num is 0\n num, lambda num: num/jnp.abs(num)) # else x/|x| \n return result\n\n\n###############################################################################\n# COMPUTATION OF HOUSEHOLDER VECTORS\n###############################################################################\[email protected]\ndef house(x_in):\n \"\"\"\n Given a real or complex length-m vector x, finds the Householder vector\n v and its inverse normalization tau such that\n P = I - beta * v \\otimes dag(v)\n is the symmetric (Hermitian) and orthogonal (unitary) Householder matrix\n representing reflections about x.\n\n Returns a list [v, beta], where v is a length-m vector whose first\n component is 1, and beta = 2/(dag(v) v).\n\n x will be treated as a flattened vector whatever its shape.\n\n Parameters\n ----------\n x_in: array_like, shape(m,)\n The vector about which to compute the Householder reflector. Will\n be flattened (inside this fucntion only) to the prescribed shape.\n\n Output\n ------\n [v_out, beta]:\n v_out: array_like, shape(m,), the Householder vector including the 1.\n beta: float, the normalization 2/|v|\n \"\"\"\n x_in= x_in.ravel()\n x_2_norm = jnp.linalg.norm(x_in[1:])\n # The next two lines are logically equivalent to\n # if x_2_norm == 0:\n # v, beta = __house_zero_norm(x)\n # else:\n # v, beta = __house_nonzero_norm( (x, x_2_norm) )\n switch = (x_2_norm == 0)\n v_out, beta = jax.lax.cond(switch,\n x_in, __house_zero_norm,\n (x_in, x_2_norm), __house_nonzero_norm)\n return [v_out, beta]\n\n\[email protected]\ndef __house_zero_norm(x):\n \"\"\"\n Handles house(x) in the case that norm(x[1:])==0.\n \"\"\"\n beta = 2.\n v = x\n v = index_update(v, index[0], 1.)\n beta = jax.lax.cond(x[0] == 0,\n x, lambda x: x[0]*0,\n x, lambda x: x[0]*0 + 2\n ).real\n return [v, beta]\n\n\[email protected]\ndef __house_nonzero_norm(xtup):\n \"\"\"\n Handles house(x) in the case that norm(x[1:])!=0.\n \"\"\"\n x, x_2_norm = xtup\n x, x_2_norm = xtup\n x_norm = jnp.linalg.norm(jnp.array([x[0], x_2_norm]))\n rho = sign(x[0])*x_norm\n\n v_1p = x[0] + rho\n v_1pabs = jnp.abs(v_1p)\n v_1m = x[0] - rho\n v_1mabs = jnp.abs(v_1m)\n\n # Pick whichever of v[0] = x[0] +- sign(x[0])*||x||\n # has greater ||v[0]||, and thus leads to greater ||v||.\n # Golub and van Loan prescribes this \"for stability\".\n v_1, v_1abs = jax.lax.cond(v_1pabs >= v_1mabs,\n (v_1p, v_1pabs), lambda x: x,\n (v_1m, v_1mabs), lambda x: x)\n\n v = x\n v = index_update(v, index[1:], v[1:]/v_1)\n v = index_update(v, index[0], 1.)\n v_2_norm = x_2_norm / v_1abs\n v_norm_sqr = 1 + v_2_norm**2\n beta = (2 / v_norm_sqr).real\n return [v, beta]\n\n\n###############################################################################\n# MANIPULATION OF HOUSEHOLDER VECTORS\n###############################################################################\[email protected]\ndef form_dense_P(hlist):\n \"\"\"\n Computes the dense Householder matrix P = I - beta * (v otimes dag(v))\n from the Householder reflector stored as hlist = (v, beta). This is\n useful for testing.\n \"\"\"\n v, beta = hlist\n Id = jnp.eye(v.size, dtype=v.dtype)\n P = Id - beta * jnp.outer(v, dag(v))\n return P\n\n\[email protected]\ndef house_leftmult(A, v, beta):\n \"\"\"\n Given the m x n matrix A and the length-n vector v with normalization\n beta such that P = I - beta v otimes dag(v) is the Householder matrix that\n reflects about v, compute PA.\n\n Parameters\n ----------\n A: array_like, shape(M, N)\n Matrix to be multiplied by H.\n\n v: array_like, shape(N).\n Householder vector.\n\n beta: float\n Householder normalization.\n\n Returns\n -------\n C = PA\n \"\"\"\n C = A - jnp.outer(beta*v, jnp.dot(dag(v), A))\n return C\n\n\[email protected]\ndef house_rightmult(A, v, beta):\n \"\"\"\n Given the m x n matrix A and the length-n vector v with normalization\n beta such that P = I - beta v otimes dag(v) is the Householder matrix that\n reflects about v, compute AP.\n\n Parameters\n ----------\n A: array_like, shape(M, N)\n Matrix to be multiplied by H.\n\n v: array_like, shape(N).\n Householder vector.\n\n beta: float\n Householder normalization.\n\n Returns\n -------\n C = AP\n \"\"\"\n C = A - jnp.outer(A@v, beta*dag(v))\n return C\n\n\n###############################################################################\n# MANIPULATION OF FACTORED QR REPRESENTATION\n###############################################################################\ndef factored_rightmult_dense(A, H, betas):\n \"\"\"\n Computes C = A * Q, where Q is in the factored representation.\n With A = Hbetalist[0].shape[0], this computes Q, but less economically\n than 'factored_to_QR'.\n\n This is a dense implementation written to test 'factored_rightmult' below.\n Do not call it in production code.\n \"\"\"\n C = A\n n = C.shape[1]\n for j, beta in enumerate(betas):\n vnz = jnp.array([1.]+list(H[j+1:, j]))\n nminus = n - vnz.size\n v = jnp.array([0.]*nminus + list(vnz))\n P = form_dense_P([v, beta])\n C = index_update(C, index[:, :], C@P)\n return C\n\n\[email protected]\ndef factored_rightmult(A, H, betas):\n \"\"\"\n Computes C = A * Q, where Q is in the factored representation.\n With A = Hbetalist[0].shape[0], this computes Q, but less economically\n than 'factored_to_QR'.\n \"\"\"\n C = A\n for j, beta in enumerate(betas):\n v = jnp.array([1.]+list(H[j+1:, j]))\n C = index_update(C, index[:, j:], house_rightmult(C[:, j:], v, beta))\n return C\n\n\[email protected]\ndef factored_to_QR(h, beta):\n \"\"\"\n Computes dense matrices Q and R from the factored QR representation\n [h, tau] as computed by qr with mode == \"factored\".\n \"\"\"\n m, n = h.shape\n R = jnp.triu(h)\n Q = jnp.eye(m, dtype=h.dtype)\n for j in range(n-1, -1, -1):\n v = jnp.concatenate((jnp.array([1.]), h[j+1:, j]))\n Q = index_update(Q, index[j:, j:],\n house_leftmult(Q[j:, j:], v, beta[j]))\n out = [Q, R]\n return out\n\n###############################################################################\n# MANIPULATION OF WY QR REPRESENTATION\n###############################################################################\[email protected]\ndef times_householder_vector(A, H, j):\n \"\"\"\n Computes A * v_j where v_j is the j'th Householder vector in H.\n\n Parameters\n ----------\n A: k x M matrix to multiply by v_j.\n H: M x k matrix of Householder reflectors.\n j: The column of H from which to extract v_j.\n\n Returns\n ------\n vout: length-M vector of output.\n \"\"\"\n\n vin = jnp.array([1.]+list(H[j+1:, j]))\n vout = jnp.zeros(H.shape[0], dtype=H.dtype)\n vout = index_update(vout, index[j:], A[:, j:] @ vin)\n return vout\n\n\[email protected]\ndef factored_to_WY(hbetalist):\n \"\"\"\n Converts the 'factored' QR representation [H, beta] into the WY\n representation, Q = I - WY^H.\n\n Parameters\n ----------\n hbetalist = [H, beta] : list of array_like, shapes [M, N] and [N].\n 'factored' QR rep of a matrix A (the output from\n house_QR(A, mode='factored')).\n\n Returns\n -------\n [W, YH]: list of ndarrays of shapes [M, N].\n The matrices W and Y generating Q along with R in the 'WY'\n representation.\n -W (M x N): The matrix W.\n -YH (M x N): -Y is the lower triangular matrix with the essential parts of\n the Householder reflectors as its columns,\n obtained by setting the main diagonal of H to 1 and zeroing\n out everything above.\n -YH, the h.c. of this matrix, is thus upper triangular\n with the full Householder reflectors as its rows. This\n function returns YH, which is what one needs to compute\n C = Q @ B = (I - WY^H) @ B = B - W @ Y^H @ B.\n\n Note: it is more efficient to store W and Y^H separately\n than to precompute their product, since we will\n typically have N << M when exploiting this\n representation.\n \"\"\"\n\n H, betas = hbetalist\n m, n = matutils.matshape(H)\n W = jnp.zeros(H.shape, H.dtype)\n vj = jnp.array([1.]+list(H[1:, 0]))\n W = index_update(W, index[:, 0], betas[0] * vj)\n\n Y = jnp.zeros(H.shape, H.dtype)\n Y = index_update(Y, index[:, 0], vj)\n for j in range(1, n):\n vj = index_update(vj, index[j+1:], H[j+1:, j])\n vj = index_update(vj, index[j], 1.) # vj[j:] stores the current vector\n YHv = (dag(Y[j:, :j])) @ vj[j:]\n z = W[:, :j] @ YHv\n z = index_add(z, index[j:], -vj[j:])\n z = index_update(z, index[:], -betas[j]*z)\n\n W = index_update(W, index[:, j], z)\n Y = index_update(Y, index[j:, j], vj[j:])\n YH = dag(Y)\n return [W, YH]\n\n\[email protected]\ndef B_times_Q_WY(B, W, YH):\n \"\"\"\n Computes C(kxm) = B(kxm)@Q(mxm) with Q given as W and Y^H in\n Q = I(mxm) - W(mxr)Y^T(rxm).\n \"\"\"\n C = B - (B@W)@YH\n return C\n\n\[email protected]\ndef Qdag_WY_times_B(B, W, YH):\n \"\"\"\n Computes C(mxk) = QH(mxm)@B(mxk) with Q given as W and Y^H in\n Q = I(mxm) - W(mxr)Y^T(rxm)\n \"\"\"\n C = B - dag(YH)@(dag(W)@B)\n return C\n\n\[email protected]\ndef WY_to_Q(W, YH):\n \"\"\"\n Retrieves Q from its WY representation.\n \"\"\"\n m = W.shape[0]\n Id = jnp.eye(m, dtype=W.dtype)\n return B_times_Q_WY(Id, W, YH)\n\n\n###############################################################################\n# QR DECOMPOSITION\n###############################################################################\ndef house_qr(A, mode=\"reduced\"):\n \"\"\"\n Performs a QR decomposition of the m x n real or complex matrix A\n using the Householder algorithm.\n\n The string parameter 'mode' determines the representation of the output.\n In this way, one can retrieve various implicit representations of the\n factored matrices. This can be a significant optimization in the case\n of a highly rectangular A, which is the reason for this function's\n existence.\n\n Parameters\n ----------\n A : array_like, shape (M, N)\n Matrix to be factored.\n\n mode: {'reduced', 'complete', 'r', 'factored', 'WY'}, optional\n If K = min(M, N), then:\n - 'reduced': returns Q, R with dimensions (M, K), (K, N)\n (default)\n - 'complete': returns Q, R with dimensions (M, M), (M, N)\n - 'r': returns r only with dimensions (K, N)\n - 'factored': returns H, beta with dimensions (N, M), (K), read\n below for details.\n - 'WY' : returns W, Y with dimensions (M, K), read below for\n details.\n\n With 'reduced', 'complete', or 'r', this function simply passes to\n jnp.linalg.qr, which depending on the currect status of Jax may lead to\n NotImplemented if A is complex.\n\n With 'factored' this function returns the same H, beta as generated by\n the Lapack function dgeqrf() (but in row-major form). Thus,\n H contains the upper triangular matrix R in its upper triangle, and\n the j'th Householder reflector forming Q in the j'th column of its\n lower triangle. beta[j] contains the normalization factor of the j'th\n reflector, called 'beta' in the function 'house' in this module.\n\n The matrix Q is then represented implicitly as\n Q = H(0) H(1) ... H(K), H(i) = I - tau[i] v dag(v)\n with v[:j] = 0; v[j]=1; v[j+1:]=A[j+1:, j].\n\n Application of Q (C -> dag{Q} C) can be made directly from this implicit\n representation using the function factored_multiply(C). When\n K << max(M, N), both the QR factorization and multiplication by Q\n using factored_multiply theoretically require far fewer operations than\n would an explicit representation of Q. However, these applications\n are mostly Level-2 BLAS operations.\n\n With 'WY' this function returns (M, K) matrices W and Y such that\n Q = I - W dag(Y).\n Y is lower-triangular matrix of Householder vectors, e.g. the lower\n triangle\n of the matrix H resulting from mode='factored'. W is then computed so\n that the above identity holds.\n\n Application of Q can be made directly from the WY representation\n using the function WY_multiply(C). The WY representation is\n a bit more expensive to compute than the factored one, though still less\n expensive than the full Q. Its advantage versus 'factored' is that\n WY_multiply calls depend mostly on Level-3 BLAS operations.\n\n\n Returns\n -------\n Q: ndarray of float or complex, optional\n The column-orthonormal orthogonal/unitary matrix Q.\n\n R: ndarray of float or complex, optional.\n The upper-triangular matrix.\n\n [H, beta]: list of ndarrays of float or complex, optional.\n The matrix H and scaling factors beta generating Q along with R in the\n 'factored' representation.\n\n [W, Y, R] : list of ndarrays of float or complex, optional.\n The matrices W and Y generating Q along with R in the 'WY'\n representation.\n\n Raises\n ------\n LinAlgError\n If factoring fails.\n\n NotImplementedError\n In reduced, complete, or r mode with complex ijnp.t.\n In factored or WY mode in the case M < N.\n \"\"\"\n if mode == \"reduced\" or mode == \"complete\" or mode == \"r\":\n return jnp.linalg.qr(A, mode=mode)\n else:\n m, n = A.shape\n if n > m:\n raise NotImplementedError(\"n > m QR not implemented in factored\"\n + \"or WY mode.\")\n if mode == \"factored\":\n return __house_qr_factored(A)\n elif mode == \"WY\":\n hbetalist = __house_qr_factored(A)\n R = jnp.triu(hbetalist[0])\n WYlist = factored_to_WY(hbetalist)\n output = WYlist + [R]\n return output\n else:\n raise ValueError(\"Invalid mode: \", mode)\n\n\[email protected]\ndef __house_qr_factored(A):\n \"\"\"\n Computes the QR decomposition of A in the 'factored' representation.\n This is a workhorse function to be accessed externally by\n house_qr(A, mode=\"factored\"), and is documented more extensively in\n that function's documentation.\n\n \"\"\"\n H = A\n M, N = matutils.matshape(A)\n beta = list()\n for j in range(A.shape[1]):\n v, thisbeta = house(H[j:, j])\n beta.append(thisbeta)\n H = index_update(H, index[j:, j:], house_leftmult(H[j:, j:], v,\n thisbeta))\n if j < M:\n H = index_update(H, index[j+1:, j], v[1:])\n beta = jnp.array(beta)\n output = [H, beta]\n return output\n\n\ndef __house_qr_factored_scan(A):\n \"\"\"\n Computes the QR decomposition of A in the 'factored' representation.\n This is a workhorse function to be accessed externally by\n house_qr(A, mode=\"factored\"), and is documented more extensively in\n that function's documentation.\n\n This implementation uses jax.lax.scan to reduce the amount of emitted\n XLA code.\n\n This should work for N > M!\n \"\"\"\n H = A\n M, N = matutils.matshape(A)\n js_i = jnp.arange(0, M, 1)\n js_f = jnp.arange(M, N, 1)\n\n def house_qr_j_lt_m(H, j):\n m, n = H.shape\n Htri = jnp.tril(H)\n v, thisbeta = house_padded(Htri[:, j], j)\n # Hjj = jax.lax.dynamic_slice(H, (j, j), (m-j, n-j)) # H[j:, j:]\n # H_update = house_leftmult(Hjj, v, thisbeta)\n # H = index_update(H, index[:, :],\n # jax.lax.dynamic_update_slice(H, H_update, [j, j]))\n # H = index_update(H, index[:, :],\n # jax.lax.dynamic_update_slice(H, v[1:], [j+1, j]))\n return H, thisbeta\n\n def house_qr_j_gt_m(H, j):\n m, n = H.shape\n this_slice = jax.lax.dynamic_slice(H, (j, j), (m-j, 1)) # H[j:, j]\n v, thisbeta = house(this_slice)\n Hjj = jax.lax.dynamic_slice(H, (j, j), (m-j, n-j)) # H[j:, j:]\n H_update = house_leftmult(Hjj, v, thisbeta)\n H = index_update(H, index[:, :],\n jax.lax.dynamic_update_slice(H, H_update, [j, j]))\n return H, thisbeta\n\n # def house_qr_j_lt_m(H, j):\n # m, n = H.shape\n # H, thisbeta = house_qr_j_gt_m(H, j)\n # v = jax.lax.dynamic_slice(H, (j+1, j), (m-j-1, 1))\n # return H, thisbeta\n H, betas_i = jax.lax.scan(house_qr_j_lt_m, H, js_i)\n raise ValueError(\"Meep meep!\")\n\n #H, betas_f = jax.lax.scan(house_qr_j_gt_m, H, js_f)\n\n betas = jnp.concatenate([betas_i, betas_f])\n output = [H, betas]\n return output\n\n\n\n\n\n\n",
"_____no_output_____"
],
[
"import gc\ndef get_obj_size(obj):\n \"\"\"\n Returns the logical size of 'obj' in bytes.\n \"\"\"\n marked = {id(obj)}\n obj_q = [obj]\n sz = 0\n\n while obj_q:\n sz += sum(map(sys.getsizeof, obj_q))\n\n # Lookup all the object referred to by the object in obj_q.\n # See: https://docs.python.org/3.7/library/gc.html#gc.get_referents\n all_refr = ((id(o), o) for o in gc.get_referents(*obj_q))\n\n # Filter object that are already marked.\n # Using dict notation will prevent repeated objects.\n new_refr = {o_id: o for o_id, o in all_refr if o_id not in marked and not isinstance(o, type)}\n\n # The new obj_q will be the ones that were not marked,\n # and we will update marked with their ids so we will\n # not traverse them again.\n obj_q = new_refr.values()\n marked.update(new_refr.keys())\n\n return sz\n\n\n\nimport time\ndef time_for_jit(func, input_shape, N_med=20, N_inner=20, \n dtype=jnp.float32):\n \"\"\"\n Measures the effective bandwidth of func(A), for A an array\n of shape input_shape. The wallclock time of the first call is returned \n separately.\n\n \"\"\"\n A = matutils.gaussian_random(shape=input_shape, dtype=dtype)\n A_logical_size = get_obj_size(A) # size of A in bytes\n\n c_time_i = time.perf_counter()\n out = func(A)\n c_time_f = time.perf_counter()\n c_time = c_time_f - c_time_i\n\n e_times = []\n for _ in range(N_med):\n A = matutils.gaussian_random(shape=input_shape, dtype=dtype)\n\n e_time_i = time.perf_counter()\n for _ in range(N_inner):\n out = func(A)\n e_time_f = time.perf_counter()\n e_times.append((e_time_f-e_time_i)/N_inner)\n e_time = np.median(np.array(e_times))\n c_time = c_time - e_time\n\n e_BW = A_logical_size / (1000 * e_time) #GB / s\n c_BW = A_logical_size / (1000 * c_time)\n return (c_BW, e_BW)",
"_____no_output_____"
],
[
"f = functools.partial(house_qr, mode=\"factored\")\n\ntimes = []\nBWs = []\nns = range(2, 40, 1)\ncomp_BWs = []\nex_BWs = []\nfor n in ns:\n shape = (n, n)\n print(\"n=\", n)\n comp_BW, ex_BW = time_for_jit(f, shape)\n comp_BWs.append(comp_BW)\n ex_BWs.append(ex_BW)\n",
"n= 2\nn= 3\nn= 4\nn= 5\nn= 6\nn= 7\nn= 8\nn= 9\nn= 10\nn= 11\nn= 12\nn= 13\nn= 14\nn= 15\nn= 16\nn= 17\nn= 18\nn= 19\nn= 20\nn= 21\nn= 22\nn= 23\nn= 24\nn= 25\nn= 26\nn= 27\nn= 28\nn= 29\nn= 30\nn= 31\nn= 32\nn= 33\nn= 34\nn= 35\nn= 36\nn= 37\nn= 38\nn= 39\n"
],
[
"plt.plot(ns, ex_BWs, label=\"Execution\", marker=\"o\")\nplt.xlabel(\"n\")\nplt.ylabel(\"BW_eff\")\nplt.title(\"Execution\")\nplt.show()\n\nplt.plot(ns, comp_BWs, label=\"Compilation\", marker=\"o\")\nplt.xlabel(\"n\")\nplt.ylabel(\"BW_eff\")\nplt.title(\"Compilation\")\nplt.show()\n",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
4afdcb8354786b3a5b5bcebc0e8e5310e622a514
| 77,108 |
ipynb
|
Jupyter Notebook
|
docs/examples/quantile-regression-uncertainty.ipynb
|
maseDevelop/river
|
b4a083cf46a9ec5ab0f55f906143f72ed2436aba
|
[
"BSD-3-Clause"
] | 2 |
2021-04-13T09:19:42.000Z
|
2021-12-22T13:43:15.000Z
|
docs/examples/quantile-regression-uncertainty.ipynb
|
maseDevelop/river
|
b4a083cf46a9ec5ab0f55f906143f72ed2436aba
|
[
"BSD-3-Clause"
] | null | null | null |
docs/examples/quantile-regression-uncertainty.ipynb
|
maseDevelop/river
|
b4a083cf46a9ec5ab0f55f906143f72ed2436aba
|
[
"BSD-3-Clause"
] | null | null | null | 426.01105 | 70,420 | 0.936219 |
[
[
[
"# Handling uncertainty with quantile regression",
"_____no_output_____"
]
],
[
[
"%matplotlib inline",
"_____no_output_____"
]
],
[
[
"[Quantile regression](https://www.wikiwand.com/en/Quantile_regression) is useful when you're not so much interested in the accuracy of your model, but rather you want your model to be good at ranking observations correctly. The typical way to perform quantile regression is to use a special loss function, namely the quantile loss. The quantile loss takes a parameter, $\\alpha$ (alpha), which indicates which quantile the model should be targeting. In the case of $\\alpha = 0.5$, then this is equivalent to asking the model to predict the median value of the target, and not the most likely value which would be the mean. \n\nA nice thing we can do with quantile regression is to produce a prediction interval for each prediction. Indeed, if we predict the lower and upper quantiles of the target then we will be able to obtain a \"trust region\" in between which the true value is likely to belong. Of course, the likeliness will depend on the chosen quantiles. For a slightly more detailed explanation see [this](https://medium.com/the-artificial-impostor/quantile-regression-part-1-e25bdd8d9d43) blog post.\n\nAs an example, let us take the [simple time series model we built in another notebook](building-a-simple-time-series-model.md). Instead of predicting the mean value of the target distribution, we will predict the 5th, 50th, 95th quantiles. This will require training three separate models, so we will encapsulate the model building logic in a function called `make_model`. We also have to slightly adapt the training loop, but not by much. Finally, we will draw the prediction interval along with the predictions from for 50th quantile (i.e. the median) and the true values.",
"_____no_output_____"
]
],
[
[
"import calendar\nimport math\nimport matplotlib.pyplot as plt\nfrom river import compose\nfrom river import datasets\nfrom river import linear_model\nfrom river import metrics\nfrom river import optim\nfrom river import preprocessing\nfrom river import stats\nfrom river import time_series\n \n\ndef get_ordinal_date(x):\n return {'ordinal_date': x['month'].toordinal()} \n\n \ndef get_month_distances(x):\n return {\n calendar.month_name[month]: math.exp(-(x['month'].month - month) ** 2)\n for month in range(1, 13)\n }\n \n\ndef make_model(alpha):\n \n extract_features = compose.TransformerUnion(get_ordinal_date, get_month_distances)\n\n scale = preprocessing.StandardScaler()\n\n learn = linear_model.LinearRegression(\n intercept_lr=0,\n optimizer=optim.SGD(3),\n loss=optim.losses.Quantile(alpha=alpha)\n )\n\n model = extract_features | scale | learn\n model = time_series.Detrender(regressor=model, window_size=12)\n\n return model\n\nmetric = metrics.MAE()\n\nmodels = {\n 'lower': make_model(alpha=0.05),\n 'center': make_model(alpha=0.5),\n 'upper': make_model(alpha=0.95)\n}\n\ndates = []\ny_trues = []\ny_preds = {\n 'lower': [],\n 'center': [],\n 'upper': []\n}\n\nfor x, y in datasets.AirlinePassengers():\n y_trues.append(y)\n dates.append(x['month'])\n \n for name, model in models.items():\n y_preds[name].append(model.predict_one(x))\n model.learn_one(x, y)\n\n # Update the error metric\n metric.update(y, y_preds['center'][-1])\n\n# Plot the results\nfig, ax = plt.subplots(figsize=(10, 6))\nax.grid(alpha=0.75)\nax.plot(dates, y_trues, lw=3, color='#2ecc71', alpha=0.8, label='Truth')\nax.plot(dates, y_preds['center'], lw=3, color='#e74c3c', alpha=0.8, label='Prediction')\nax.fill_between(dates, y_preds['lower'], y_preds['upper'], color='#e74c3c', alpha=0.3, label='Prediction interval')\nax.legend()\nax.set_title(metric);",
"_____no_output_____"
]
],
[
[
"An important thing to note is that the prediction interval we obtained should not be confused with a confidence interval. Simply put, a prediction interval represents uncertainty for where the true value lies, whereas a confidence interval encapsulates the uncertainty on the prediction. You can find out more by reading [this](https://stats.stackexchange.com/questions/16493/difference-between-confidence-intervals-and-prediction-intervals) CrossValidated post.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
4afdd5912087ab9c3b439fbe8634de17ffbf05c9
| 16,865 |
ipynb
|
Jupyter Notebook
|
legacy/FaceSwap_GAN_v2_test_img.ipynb
|
EribertoO/faceswap-GAN-2
|
02e7bebc6eccffb653e30d9ffad7bbe52494fff4
|
[
"MIT"
] | 1 |
2021-09-07T09:50:58.000Z
|
2021-09-07T09:50:58.000Z
|
legacy/FaceSwap_GAN_v2_test_img.ipynb
|
EribertoO/faceswap-GAN-2
|
02e7bebc6eccffb653e30d9ffad7bbe52494fff4
|
[
"MIT"
] | null | null | null |
legacy/FaceSwap_GAN_v2_test_img.ipynb
|
EribertoO/faceswap-GAN-2
|
02e7bebc6eccffb653e30d9ffad7bbe52494fff4
|
[
"MIT"
] | null | null | null | 31.880907 | 121 | 0.547702 |
[
[
[
"<a id='1'></a>\n# 1. Import packages",
"_____no_output_____"
]
],
[
[
"from keras.models import Sequential, Model\nfrom keras.layers import *\nfrom keras.layers.advanced_activations import LeakyReLU\nfrom keras.activations import relu\nfrom keras.initializers import RandomNormal\nfrom keras.applications import *\nimport keras.backend as K\nfrom tensorflow.contrib.distributions import Beta\nimport tensorflow as tf\nfrom keras.optimizers import Adam",
"Using TensorFlow backend.\n"
],
[
"from image_augmentation import random_transform\nfrom image_augmentation import random_warp\nfrom utils import get_image_paths, load_images, stack_images\nfrom pixel_shuffler import PixelShuffler",
"_____no_output_____"
],
[
"import time\nimport numpy as np\nfrom PIL import Image\nimport cv2\nimport glob\nfrom random import randint, shuffle\nfrom IPython.display import clear_output\nfrom IPython.display import display\nimport matplotlib.pyplot as plt\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"<a id='4'></a>\n# 4. Config\n\nmixup paper: https://arxiv.org/abs/1710.09412\n\nDefault training data directories: `./faceA/` and `./faceB/`",
"_____no_output_____"
]
],
[
[
"K.set_learning_phase(0)",
"_____no_output_____"
],
[
"channel_axis=-1\nchannel_first = False",
"_____no_output_____"
],
[
"IMAGE_SHAPE = (64, 64, 3)\nnc_in = 3 # number of input channels of generators\nnc_D_inp = 6 # number of input channels of discriminators\n\nuse_self_attn = False\nw_l2 = 1e-4 # weight decay\n\nbatchSize = 8\n\n# Path of training images\nimg_dirA = './faceA/*.*'\nimg_dirB = './faceB/*.*'",
"_____no_output_____"
]
],
[
[
"<a id='5'></a>\n# 5. Define models",
"_____no_output_____"
]
],
[
[
"class Scale(Layer):\n '''\n Code borrows from https://github.com/flyyufelix/cnn_finetune\n '''\n def __init__(self, weights=None, axis=-1, gamma_init='zero', **kwargs):\n self.axis = axis\n self.gamma_init = initializers.get(gamma_init)\n self.initial_weights = weights\n super(Scale, self).__init__(**kwargs)\n\n def build(self, input_shape):\n self.input_spec = [InputSpec(shape=input_shape)]\n\n # Compatibility with TensorFlow >= 1.0.0\n self.gamma = K.variable(self.gamma_init((1,)), name='{}_gamma'.format(self.name))\n self.trainable_weights = [self.gamma]\n\n if self.initial_weights is not None:\n self.set_weights(self.initial_weights)\n del self.initial_weights\n\n def call(self, x, mask=None):\n return self.gamma * x\n\n def get_config(self):\n config = {\"axis\": self.axis}\n base_config = super(Scale, self).get_config()\n return dict(list(base_config.items()) + list(config.items()))\n\n\ndef self_attn_block(inp, nc):\n '''\n Code borrows from https://github.com/taki0112/Self-Attention-GAN-Tensorflow\n '''\n assert nc//8 > 0, f\"Input channels must be >= 8, but got nc={nc}\"\n x = inp\n shape_x = x.get_shape().as_list()\n \n f = Conv2D(nc//8, 1, kernel_initializer=conv_init)(x)\n g = Conv2D(nc//8, 1, kernel_initializer=conv_init)(x)\n h = Conv2D(nc, 1, kernel_initializer=conv_init)(x)\n \n shape_f = f.get_shape().as_list()\n shape_g = g.get_shape().as_list()\n shape_h = h.get_shape().as_list()\n flat_f = Reshape((-1, shape_f[-1]))(f)\n flat_g = Reshape((-1, shape_g[-1]))(g)\n flat_h = Reshape((-1, shape_h[-1]))(h) \n \n s = Lambda(lambda x: tf.matmul(x[0], x[1], transpose_b=True))([flat_g, flat_f])\n\n beta = Softmax(axis=-1)(s)\n o = Lambda(lambda x: tf.matmul(x[0], x[1]))([beta, flat_h])\n o = Reshape(shape_x[1:])(o)\n o = Scale()(o)\n \n out = add([o, inp])\n return out\n\ndef conv_block(input_tensor, f):\n x = input_tensor\n x = Conv2D(f, kernel_size=3, strides=2, kernel_regularizer=regularizers.l2(w_l2), \n kernel_initializer=conv_init, use_bias=False, padding=\"same\")(x)\n x = Activation(\"relu\")(x)\n return x\n\ndef conv_block_d(input_tensor, f, use_instance_norm=False):\n x = input_tensor\n x = Conv2D(f, kernel_size=4, strides=2, kernel_regularizer=regularizers.l2(w_l2), \n kernel_initializer=conv_init, use_bias=False, padding=\"same\")(x)\n x = LeakyReLU(alpha=0.2)(x)\n return x\n\ndef res_block(input_tensor, f):\n x = input_tensor\n x = Conv2D(f, kernel_size=3, kernel_regularizer=regularizers.l2(w_l2), \n kernel_initializer=conv_init, use_bias=False, padding=\"same\")(x)\n x = LeakyReLU(alpha=0.2)(x)\n x = Conv2D(f, kernel_size=3, kernel_regularizer=regularizers.l2(w_l2), \n kernel_initializer=conv_init, use_bias=False, padding=\"same\")(x)\n x = add([x, input_tensor])\n x = LeakyReLU(alpha=0.2)(x)\n return x\n\ndef upscale_ps(filters, use_norm=True):\n def block(x):\n x = Conv2D(filters*4, kernel_size=3, kernel_regularizer=regularizers.l2(w_l2), \n kernel_initializer=RandomNormal(0, 0.02), padding='same')(x)\n x = LeakyReLU(0.2)(x)\n x = PixelShuffler()(x)\n return x\n return block\n\ndef Discriminator(nc_in, input_size=64):\n inp = Input(shape=(input_size, input_size, nc_in))\n #x = GaussianNoise(0.05)(inp)\n x = conv_block_d(inp, 64, False)\n x = conv_block_d(x, 128, False)\n x = self_attn_block(x, 128) if use_self_attn else x\n x = conv_block_d(x, 256, False)\n x = self_attn_block(x, 256) if use_self_attn else x\n out = Conv2D(1, kernel_size=4, kernel_initializer=conv_init, use_bias=False, padding=\"same\")(x) \n return Model(inputs=[inp], outputs=out)\n\ndef Encoder(nc_in=3, input_size=64):\n inp = Input(shape=(input_size, input_size, nc_in))\n x = Conv2D(64, kernel_size=5, kernel_initializer=conv_init, use_bias=False, padding=\"same\")(inp)\n x = conv_block(x,128)\n x = conv_block(x,256)\n x = self_attn_block(x, 256) if use_self_attn else x\n x = conv_block(x,512) \n x = self_attn_block(x, 512) if use_self_attn else x\n x = conv_block(x,1024)\n x = Dense(1024)(Flatten()(x))\n x = Dense(4*4*1024)(x)\n x = Reshape((4, 4, 1024))(x)\n out = upscale_ps(512)(x)\n return Model(inputs=inp, outputs=out)\n\ndef Decoder_ps(nc_in=512, input_size=8):\n input_ = Input(shape=(input_size, input_size, nc_in))\n x = input_\n x = upscale_ps(256)(x)\n x = upscale_ps(128)(x)\n x = self_attn_block(x, 128) if use_self_attn else x\n x = upscale_ps(64)(x)\n x = res_block(x, 64)\n x = self_attn_block(x, 64) if use_self_attn else x\n #x = Conv2D(4, kernel_size=5, padding='same')(x) \n alpha = Conv2D(1, kernel_size=5, padding='same', activation=\"sigmoid\")(x)\n rgb = Conv2D(3, kernel_size=5, padding='same', activation=\"tanh\")(x)\n out = concatenate([alpha, rgb])\n return Model(input_, out) ",
"_____no_output_____"
],
[
"encoder = Encoder()\ndecoder_A = Decoder_ps()\ndecoder_B = Decoder_ps()\n\nx = Input(shape=IMAGE_SHAPE)\n\nnetGA = Model(x, decoder_A(encoder(x)))\nnetGB = Model(x, decoder_B(encoder(x)))",
"_____no_output_____"
],
[
"netDA = Discriminator(nc_D_inp)\nnetDB = Discriminator(nc_D_inp)",
"_____no_output_____"
]
],
[
[
"<a id='6'></a>\n# 6. Load Models",
"_____no_output_____"
]
],
[
[
"try:\n encoder.load_weights(\"models/encoder.h5\")\n decoder_A.load_weights(\"models/decoder_A.h5\")\n decoder_B.load_weights(\"models/decoder_B.h5\")\n #netDA.load_weights(\"models/netDA.h5\") \n #netDB.load_weights(\"models/netDB.h5\") \n print (\"model loaded.\")\nexcept:\n print (\"Weights file not found.\")\n pass",
"model loaded.\n"
]
],
[
[
"<a id='7'></a>\n# 7. Define Inputs/Outputs Variables\n\n distorted_A: A (batch_size, 64, 64, 3) tensor, input of generator_A (netGA).\n distorted_B: A (batch_size, 64, 64, 3) tensor, input of generator_B (netGB).\n fake_A: (batch_size, 64, 64, 3) tensor, output of generator_A (netGA).\n fake_B: (batch_size, 64, 64, 3) tensor, output of generator_B (netGB).\n mask_A: (batch_size, 64, 64, 1) tensor, mask output of generator_A (netGA).\n mask_B: (batch_size, 64, 64, 1) tensor, mask output of generator_B (netGB).\n path_A: A function that takes distorted_A as input and outputs fake_A.\n path_B: A function that takes distorted_B as input and outputs fake_B.\n path_mask_A: A function that takes distorted_A as input and outputs mask_A.\n path_mask_B: A function that takes distorted_B as input and outputs mask_B.\n path_abgr_A: A function that takes distorted_A as input and outputs concat([mask_A, fake_A]).\n path_abgr_B: A function that takes distorted_B as input and outputs concat([mask_B, fake_B]).\n real_A: A (batch_size, 64, 64, 3) tensor, target images for generator_A given input distorted_A.\n real_B: A (batch_size, 64, 64, 3) tensor, target images for generator_B given input distorted_B.",
"_____no_output_____"
]
],
[
[
"def cycle_variables(netG):\n distorted_input = netG.inputs[0]\n fake_output = netG.outputs[0]\n alpha = Lambda(lambda x: x[:,:,:, :1])(fake_output)\n rgb = Lambda(lambda x: x[:,:,:, 1:])(fake_output)\n \n masked_fake_output = alpha * rgb + (1-alpha) * distorted_input \n\n fn_generate = K.function([distorted_input], [masked_fake_output])\n fn_mask = K.function([distorted_input], [concatenate([alpha, alpha, alpha])])\n fn_abgr = K.function([distorted_input], [concatenate([alpha, rgb])])\n return distorted_input, fake_output, alpha, fn_generate, fn_mask, fn_abgr",
"_____no_output_____"
],
[
"distorted_A, fake_A, mask_A, path_A, path_mask_A, path_abgr_A = cycle_variables(netGA)\ndistorted_B, fake_B, mask_B, path_B, path_mask_B, path_abgr_B = cycle_variables(netGB)\nreal_A = Input(shape=IMAGE_SHAPE)\nreal_B = Input(shape=IMAGE_SHAPE)",
"_____no_output_____"
]
],
[
[
"<a id='11'></a>\n# 11. Helper Function: face_swap()\nThis function is provided for those who don't have enough VRAM to run dlib's CNN and GAN model at the same time.\n\n INPUTS:\n img: A RGB face image of any size.\n path_func: a function that is either path_abgr_A or path_abgr_B.\n OUPUTS:\n result_img: A RGB swapped face image after masking.\n result_mask: A single channel uint8 mask image.",
"_____no_output_____"
]
],
[
[
"def swap_face(img, path_func):\n input_size = img.shape\n img = cv2.cvtColor(img, cv2.COLOR_RGB2BGR) # generator expects BGR input \n ae_input = cv2.resize(img, (64,64))/255. * 2 - 1 \n \n result = np.squeeze(np.array([path_func([[ae_input]])]))\n result_a = result[:,:,0] * 255\n result_a = cv2.resize(result_a, (input_size[1],input_size[0]))[...,np.newaxis]\n result_bgr = np.clip( (result[:,:,1:] + 1) * 255 / 2, 0, 255)\n result_bgr = cv2.resize(result_bgr, (input_size[1],input_size[0]))\n result = (result_a/255 * result_bgr + (1 - result_a/255) * img).astype('uint8')\n \n result = cv2.cvtColor(result, cv2.COLOR_BGR2RGB) \n result = cv2.resize(result, (input_size[1],input_size[0]))\n result_a = np.expand_dims(cv2.resize(result_a, (input_size[1],input_size[0])), axis=2)\n return result, result_a",
"_____no_output_____"
],
[
"whom2whom = \"BtoA\" # default trainsforming faceB to faceA\n\nif whom2whom is \"AtoB\":\n path_func = path_abgr_B\nelif whom2whom is \"BtoA\":\n path_func = path_abgr_A\nelse:\n print (\"whom2whom should be either AtoB or BtoA\")",
"_____no_output_____"
],
[
"input_img = plt.imread(\"./IMAGE_FILENAME.jpg\")",
"_____no_output_____"
],
[
"plt.imshow(input_img)",
"_____no_output_____"
],
[
"result_img, result_mask = swap_face(input_img, path_func)",
"_____no_output_____"
],
[
"plt.imshow(result_img)",
"_____no_output_____"
],
[
"plt.imshow(result_mask[:, :, 0]) # cmap='gray'",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4afdddb0532e01d35e0078e53c49c0d83d0879b9
| 39,763 |
ipynb
|
Jupyter Notebook
|
python/example/histograms.ipynb
|
souravrhythm/opendp
|
2c576dbf98389c349ca3a3be928f0e600cd0c10b
|
[
"MIT"
] | 95 |
2021-02-17T19:50:28.000Z
|
2022-03-31T16:50:59.000Z
|
python/example/histograms.ipynb
|
souravrhythm/opendp
|
2c576dbf98389c349ca3a3be928f0e600cd0c10b
|
[
"MIT"
] | 299 |
2021-02-10T00:14:41.000Z
|
2022-03-31T16:17:33.000Z
|
python/example/histograms.ipynb
|
souravrhythm/opendp
|
2c576dbf98389c349ca3a3be928f0e600cd0c10b
|
[
"MIT"
] | 13 |
2021-04-01T14:40:56.000Z
|
2022-03-27T08:52:46.000Z
| 133.882155 | 14,655 | 0.871086 |
[
[
[
"### Privatizing Histograms\n\nSometimes we want to release the counts of individual outcomes in a dataset.\nWhen plotted, this makes a histogram.\n\nThe library currently has two approaches:\n1. Known category set `make_count_by_categories`\n2. Unknown category set `make_count_by`\n\nThe next code block imports just handles boilerplate: imports, data loading, plotting.",
"_____no_output_____"
]
],
[
[
"import os\n\nfrom opendp.meas import *\nfrom opendp.mod import enable_features, binary_search_chain, Measurement, Transformation\nfrom opendp.trans import *\nfrom opendp.typing import *\nenable_features(\"contrib\")\nmax_influence = 1\nbudget = (1., 1e-8)\n\n# public information\ncol_names = [\"age\", \"sex\", \"educ\", \"race\", \"income\", \"married\"]\ndata_path = os.path.join('.', 'data', 'PUMS_california_demographics_1000', 'data.csv')\nsize = 1000\n\nwith open(data_path) as input_data:\n data = input_data.read()\n\ndef plot_histogram(sensitive_counts, released_counts):\n \"\"\"Plot a histogram that compares true data against released data\"\"\"\n import matplotlib.pyplot as plt\n import matplotlib.ticker as ticker\n\n fig = plt.figure()\n ax = fig.add_axes([1,1,1,1])\n plt.ylim([0,225])\n tick_spacing = 1.\n ax.xaxis.set_major_locator(ticker.MultipleLocator(tick_spacing))\n plt.xlim(0,15)\n width = .4\n\n ax.bar(list([x+width for x in range(0, len(sensitive_counts))]), sensitive_counts, width=width, label='True Value')\n ax.bar(list([x+2*width for x in range(0, len(released_counts))]), released_counts, width=width, label='DP Value')\n ax.legend()\n plt.title('Histogram of Education Level')\n plt.xlabel('Years of Education')\n plt.ylabel('Count')\n plt.show()",
"_____no_output_____"
]
],
[
[
"### Private histogram via `make_count_by_categories`\n\nThis approach is only applicable if the set of potential values that the data may take on is public information.\nIf this information is not available, then use `make_count_by` instead.\nIt typically has greater utility than `make_count_by` until the size of the category set is greater than dataset size.\nIn this data, we know that the category set is public information:\nstrings consisting of the numbers between 1 and 20.\n\nThe counting aggregator computes a vector of counts in the same order as the input categories.\nIt also includes one extra count at the end of the vector,\nconsisting of the number of elements that were not members of the category set.\n\nYou'll notice that `make_base_geometric` has an additional argument that explicitly sets the type of the domain, `D`.\nIt defaults to `AllDomain[int]` which works in situations where the mechanism is noising a scalar.\nHowever, in this situation, we are noising a vector of scalars,\nand thus the appropriate domain is `VectorDomain[AllDomain[int]]`.",
"_____no_output_____"
]
],
[
[
"# public information\ncategories = list(map(str, range(1, 20)))\n\nhistogram = (\n make_split_dataframe(separator=\",\", col_names=col_names) >>\n make_select_column(key=\"educ\", TOA=str) >>\n # Compute counts for each of the categories and null\n make_count_by_categories(categories=categories)\n)\n\nnoisy_histogram = binary_search_chain(\n lambda s: histogram >> make_base_geometric(scale=s, D=VectorDomain[AllDomain[int]]),\n d_in=max_influence, d_out=budget[0])\n\nsensitive_counts = histogram(data)\nreleased_counts = noisy_histogram(data)\n\nprint(\"Educational level counts:\\n\", sensitive_counts[:-1])\nprint(\"DP Educational level counts:\\n\", released_counts[:-1])\n\nprint(\"DP estimate for the number of records that were not a member of the category set:\", released_counts[-1])\n\nplot_histogram(sensitive_counts, released_counts)",
"Educational level counts:\n [33, 14, 38, 17, 24, 21, 31, 51, 201, 60, 165, 76, 178, 54, 24, 13, 0, 0, 0]\nDP Educational level counts:\n [33, 11, 38, 17, 23, 22, 32, 50, 201, 63, 165, 77, 178, 53, 24, 10, 1, 0, 0]\nDP estimate for the number of records that were not a member of the category set: 0\n"
]
],
[
[
"### Private histogram via `make_count_by`\nThis approach is applicable when the set of categories is unknown or very large.\nAny categories with a noisy count less than a given threshold will be censored from the final release.\nThe noise scale influences the epsilon parameter of the budget, and the threshold influences the delta parameter in the budget.\n\n`ptr` stands for Propose-Test-Release, a framework for censoring queries for which the local sensitivity is greater than some threshold.\nAny category with a count sufficiently small is censored from the release.\n\nIt is sometimes referred to as a \"stability histogram\" because it only releases counts for \"stable\" categories that exist in all datasets that are considered \"neighboring\" to your private dataset.\n\nI start out by defining a function that finds the tightest noise scale and threshold for which the stability histogram is (d_in, d_out)-close.\nYou may find this useful for your application.",
"_____no_output_____"
]
],
[
[
"def make_base_ptr_budget(\n preprocess: Transformation,\n d_in, d_out,\n TK: RuntimeTypeDescriptor) -> Measurement:\n \"\"\"Make a stability histogram that respects a given d_in, d_out.\n\n :param preprocess: Transformation\n :param d_in: Input distance to satisfy\n :param d_out: Output distance to satisfy\n :param TK: Type of Key (hashable)\n \"\"\"\n from opendp.mod import binary_search_param\n def privatize(s, t=1e8):\n return preprocess >> make_base_ptr(scale=s, threshold=t, TK=TK)\n \n s = binary_search_param(lambda s: privatize(s=s), d_in, d_out)\n t = binary_search_param(lambda t: privatize(s=s, t=t), d_in, d_out)\n\n return privatize(s=s, t=t)\n",
"_____no_output_____"
]
],
[
[
"I now use the `make_count_by_ptr_budget` constructor to release a private histogram on the education data.\n\nThe stability mechanism, as currently written, samples from a continuous noise distribution.\nIf you haven't already, please read about [floating-point behavior in the docs](https://docs.opendp.org/en/latest/user/measurement-constructors.html#floating-point).\n",
"_____no_output_____"
]
],
[
[
"\nfrom opendp.mod import enable_features\nenable_features(\"floating-point\")\n\npreprocess = (\n make_split_dataframe(separator=\",\", col_names=col_names) >>\n make_select_column(key=\"educ\", TOA=str) >>\n make_count_by(MO=L1Distance[float], TK=str, TV=float)\n)\n\nnoisy_histogram = make_base_ptr_budget(\n preprocess,\n d_in=max_influence, d_out=budget,\n TK=str)\n\nsensitive_counts = histogram(data)\nreleased_counts = noisy_histogram(data)\n# postprocess to make the results easier to compare\npostprocessed_counts = {k: round(v) for k, v in released_counts.items()}\n\nprint(\"Educational level counts:\\n\", sensitive_counts)\nprint(\"DP Educational level counts:\\n\", postprocessed_counts)\n\ndef as_array(data):\n return [data.get(k, 0) for k in categories]\n\nplot_histogram(sensitive_counts, as_array(released_counts))",
"Educational level counts:\n [33, 14, 38, 17, 24, 21, 31, 51, 201, 60, 165, 76, 178, 54, 24, 13, 0, 0, 0, 0]\nDP Educational level counts:\n {'13': 180, '6': 21, '3': 38, '14': 53, '8': 51, '11': 165, '7': 32, '10': 61, '5': 25, '12': 74, '15': 24, '9': 201, '1': 32}\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
4afdde180f7755bbffcc9a335de9e668cf1a3d22
| 36,774 |
ipynb
|
Jupyter Notebook
|
Evaluate_power_vs_trip_start_end.ipynb
|
maufadel/mobilitynet-analysis-scripts
|
faf3dad8ed71b0b31363b7698c2611414c60c930
|
[
"BSD-3-Clause"
] | 1 |
2021-01-25T15:58:25.000Z
|
2021-01-25T15:58:25.000Z
|
Evaluate_power_vs_trip_start_end.ipynb
|
maufadel/mobilitynet-analysis-scripts
|
faf3dad8ed71b0b31363b7698c2611414c60c930
|
[
"BSD-3-Clause"
] | 7 |
2021-02-10T04:44:56.000Z
|
2021-03-22T06:52:15.000Z
|
Evaluate_power_vs_trip_start_end.ipynb
|
maufadel/mobilitynet-analysis-scripts
|
faf3dad8ed71b0b31363b7698c2611414c60c930
|
[
"BSD-3-Clause"
] | null | null | null | 35.912109 | 599 | 0.570512 |
[
[
[
"## Set up the dependencies",
"_____no_output_____"
]
],
[
[
"# for reading and validating data\nimport emeval.input.spec_details as eisd\nimport emeval.input.phone_view as eipv\nimport emeval.input.eval_view as eiev",
"_____no_output_____"
],
[
"# Visualization helpers\nimport emeval.viz.phone_view as ezpv\nimport emeval.viz.eval_view as ezev",
"_____no_output_____"
],
[
"# Metrics helpers\nimport emeval.metrics.segmentation as ems",
"_____no_output_____"
],
[
"# For plots\nimport matplotlib.pyplot as plt\nfrom matplotlib.collections import PatchCollection\nfrom matplotlib.patches import Rectangle\n%matplotlib inline",
"_____no_output_____"
],
[
"# For maps\nimport folium\nimport branca.element as bre",
"_____no_output_____"
],
[
"# For easier debugging while working on modules\nimport importlib",
"_____no_output_____"
],
[
"import pandas as pd\nimport numpy as np\npd.options.display.float_format = '{:.6f}'.format\nimport arrow",
"_____no_output_____"
],
[
"THIRTY_MINUTES = 30 * 60\nTIME_THRESHOLD = THIRTY_MINUTES",
"_____no_output_____"
],
[
"importlib.reload(ems)",
"_____no_output_____"
]
],
[
[
"## The spec\n\nThe spec defines what experiments were done, and over which time ranges. Once the experiment is complete, most of the structure is read back from the data, but we use the spec to validate that it all worked correctly. The spec also contains the ground truth for the legs. Here, we read the spec for the trip to UC Berkeley.",
"_____no_output_____"
]
],
[
[
"DATASTORE_URL = \"http://cardshark.cs.berkeley.edu\"\nAUTHOR_EMAIL = \"[email protected]\"\nsd_la = eisd.SpecDetails(DATASTORE_URL, AUTHOR_EMAIL, \"unimodal_trip_car_bike_mtv_la\")\nsd_sj = eisd.SpecDetails(DATASTORE_URL, AUTHOR_EMAIL, \"car_scooter_brex_san_jose\")\nsd_ucb = eisd.SpecDetails(DATASTORE_URL, AUTHOR_EMAIL, \"train_bus_ebike_mtv_ucb\")",
"_____no_output_____"
]
],
[
[
"## The views\n\nThere are two main views for the data - the phone view and the evaluation view. ",
"_____no_output_____"
],
[
"### Phone view\n\nIn the phone view, the phone is primary, and then there is a tree that you can traverse to get the data that you want. Traversing that tree typically involves nested for loops; here's an example of loading the phone view and traversing it. You can replace the print statements with real code. When you are ready to check this in, please move the function to one of the python modules so that we can invoke it more generally",
"_____no_output_____"
]
],
[
[
"importlib.reload(eipv)",
"_____no_output_____"
],
[
"pv_la = eipv.PhoneView(sd_la)",
"_____no_output_____"
],
[
"pv_sj = eipv.PhoneView(sd_sj)",
"_____no_output_____"
],
[
"pv_ucb = eipv.PhoneView(sd_ucb)",
"_____no_output_____"
]
],
[
[
"## Number of detected trips versus ground truth trips\n\nChecks to see how many spurious transitions there were",
"_____no_output_____"
]
],
[
[
"importlib.reload(ems)",
"_____no_output_____"
],
[
"ems.fill_sensed_trip_ranges(pv_la)\nems.fill_sensed_trip_ranges(pv_sj)\nems.fill_sensed_trip_ranges(pv_ucb)",
"_____no_output_____"
],
[
"pv_sj.map()[\"ios\"][\"ucb-sdb-ios-2\"][\"evaluation_ranges\"][4][\"trip_id\"]",
"_____no_output_____"
]
],
[
[
"### Start and end times mismatch",
"_____no_output_____"
]
],
[
[
"curr_run = pv_la.map()[\"android\"][\"ucb-sdb-android-2\"][\"evaluation_ranges\"][0]\nprint(curr_run.keys())\nems.find_matching_segments(curr_run[\"evaluation_trip_ranges\"], \"trip_id\", curr_run[\"sensed_trip_ranges\"])",
"_____no_output_____"
],
[
"[1,2,3][1:2]",
"_____no_output_____"
],
[
"def get_tradeoff_entries(pv):\n tradeoff_entry_list = []\n for phone_os, phone_map in pv.map().items():\n print(15 * \"=*\")\n print(phone_os, phone_map.keys())\n for phone_label, phone_detail_map in phone_map.items():\n print(4 * ' ', 15 * \"-*\")\n print(4 * ' ', phone_label, phone_detail_map.keys())\n if \"control\" in phone_detail_map[\"role\"]:\n print(\"Ignoring %s phone %s since they are always on\" % (phone_detail_map[\"role\"], phone_label))\n continue\n # this spec does not have any calibration ranges, but evaluation ranges are actually cooler\n for r in phone_detail_map[\"evaluation_ranges\"]:\n print(8 * ' ', 30 * \"=\")\n print(8 * ' ',r.keys())\n print(8 * ' ',r[\"trip_id\"], r[\"eval_common_trip_id\"], r[\"eval_role\"], len(r[\"evaluation_trip_ranges\"]))\n bcs = r[\"battery_df\"][\"battery_level_pct\"]\n delta_battery = bcs.iloc[0] - bcs.iloc[-1]\n print(\"Battery starts at %d, ends at %d, drain = %d\" % (bcs.iloc[0], bcs.iloc[-1], delta_battery))\n sensed_trips = len(r[\"sensed_trip_ranges\"])\n visit_reports = len(r[\"visit_sensed_trip_ranges\"])\n matching_trip_map = ems.find_matching_segments(r[\"evaluation_trip_ranges\"], \"trip_id\", r[\"sensed_trip_ranges\"])\n print(matching_trip_map)\n for trip in r[\"evaluation_trip_ranges\"]:\n sensed_trip_range = matching_trip_map[trip[\"trip_id\"]]\n results = ems.get_count_start_end_ts_diff(trip, sensed_trip_range)\n print(\"Got results %s\" % results)\n tradeoff_entry = {\"phone_os\": phone_os, \"phone_label\": phone_label,\n \"timeline\": pv.spec_details.curr_spec[\"id\"],\n \"range_id\": r[\"trip_id\"],\n \"run\": r[\"trip_run\"], \"duration\": r[\"duration\"],\n \"role\": r[\"eval_role_base\"], \"battery_drain\": delta_battery,\n \"trip_count\": sensed_trips, \"visit_reports\": visit_reports,\n \"trip_id\": trip[\"trip_id\"]}\n tradeoff_entry.update(results)\n tradeoff_entry_list.append(tradeoff_entry)\n return tradeoff_entry_list",
"_____no_output_____"
],
[
"# We are not going to look at battery life at the evaluation trip level; we will end with evaluation range\n# since we want to capture the overall drain for the timeline\ntradeoff_entries_list = []\ntradeoff_entries_list.extend(get_tradeoff_entries(pv_la))\ntradeoff_entries_list.extend(get_tradeoff_entries(pv_sj))\ntradeoff_entries_list.extend(get_tradeoff_entries(pv_ucb))\ntradeoff_df = pd.DataFrame(tradeoff_entries_list)",
"_____no_output_____"
],
[
"r2q_map = {\"power_control\": 0, \"HAMFDC\": 1, \"MAHFDC\": 2, \"HAHFDC\": 3, \"accuracy_control\": 4}\nq2r_map = {0: \"power\", 1: \"HAMFDC\", 2: \"MAHFDC\", 3: \"HAHFDC\", 4: \"accuracy\"}",
"_____no_output_____"
],
[
"# Make a number so that can get the plots to come out in order\ntradeoff_df[\"quality\"] = tradeoff_df.role.apply(lambda r: r2q_map[r])\ntradeoff_df[\"count_diff\"] = tradeoff_df[[\"count\"]] - 1",
"_____no_output_____"
],
[
"import itertools",
"_____no_output_____"
]
],
[
[
"## Trip count analysis",
"_____no_output_____"
],
[
"### Scatter plot",
"_____no_output_____"
]
],
[
[
"ifig, ax = plt.subplots(nrows=1, ncols=1, figsize=(12,4))\nerrorboxes = []\nfor key, df in tradeoff_df.query(\"phone_os == 'android'\").groupby([\"role\", \"timeline\"]):\n print(key, df)\n tcd = df.trip_count\n bd = df.battery_drain\n print(\"Plotting rect with params %s, %d, %d\" % (str((tcd.min(), bd.min())),\n tcd.max() - tcd.min(),\n bd.max() - bd.min()))\n print(tcd.min(), tcd.max(), tcd.std())\n xerror = np.array([[tcd.min(), tcd.max()]])\n print(xerror.shape)\n ax.errorbar(x=tcd.mean(), y=bd.mean(), xerr=[[tcd.min()], [tcd.max()]], yerr=[[bd.min()], [bd.max()]], label=key)\nplt.legend()",
"_____no_output_____"
]
],
[
[
"### Timeline + trip specific variation\n\nHow many sensed trips matched to each ground truth trip?",
"_____no_output_____"
]
],
[
[
"ifig, ax_array = plt.subplots(nrows=2,ncols=3,figsize=(9,6), sharex=False, sharey=True)\ntimeline_list = [\"train_bus_ebike_mtv_ucb\", \"car_scooter_brex_san_jose\", \"unimodal_trip_car_bike_mtv_la\"]\nfor i, tl in enumerate(timeline_list):\n tradeoff_df.query(\"timeline == @tl & phone_os == 'android'\").boxplot(ax = ax_array[0][i], column=[\"count_diff\"], by=[\"quality\"])\n ax_array[0][i].set_title(tl)\n tradeoff_df.query(\"timeline == @tl & phone_os == 'ios'\").boxplot(ax = ax_array[1][i], column=[\"count_diff\"], by=[\"quality\"])\n ax_array[1][i].set_title(\"\")\n # tradeoff_df.query(\"timeline == @tl & phone_os == 'ios'\").boxplot(ax = ax_array[2][i], column=[\"visit_reports\"], by=[\"quality\"])\n # ax_array[2][i].set_title(\"\")\n\n # print(android_ax_returned.shape, ios_ax_returned.shape)\n\nfor i, ax in enumerate(ax_array[0]):\n ax.set_xticklabels([q2r_map[int(t.get_text())] for t in ax.get_xticklabels()])\n ax.set_xlabel(\"\")\n\nfor i, ax in enumerate(ax_array[1]):\n ax.set_xticklabels([q2r_map[int(t.get_text())] for t in ax.get_xticklabels()])\n ax.set_xlabel(\"\")\n\n# for ax in ax_array[1]:\n# ax.set_xticklabels(q2r_ios_list[1:])\n# ax.set_xlabel(\"\")\n\n# for ax in ax_array[2]:\n# ax.set_xticklabels(q2r_ios_list[1:])\n# ax.set_xlabel(\"\")\n\nax_array[0][0].set_ylabel(\"Difference in trip counts (android)\")\nax_array[1][0].set_ylabel(\"Difference in trip counts (ios)\")\n# ax_array[2][0].set_ylabel(\"Difference in visit reports (ios)\")\nifig.suptitle(\"Trip count differences v/s configured quality over multiple timelines\")\n# ifig.tight_layout()",
"_____no_output_____"
]
],
[
[
"### Timeline specific variation",
"_____no_output_____"
]
],
[
[
"def plot_count_with_errors(ax_array, phone_os):\n for i, (tl, trip_gt) in enumerate(timeline_trip_gt.items()):\n ax_array[i].bar(0, trip_gt)\n for q in range(1,4):\n curr_df = tradeoff_df.query(\"timeline == @tl & phone_os == @phone_os & quality == @q\")\n print(\"%s %s %s values = %s %s %s\" % (phone_os, tl, q2r_map[q], curr_df.trip_count.min(), curr_df.trip_count.mean(), curr_df.trip_count.max()))\n lower_error = curr_df.trip_count.mean() - curr_df.trip_count.min()\n upper_error = curr_df.trip_count.max() - curr_df.trip_count.mean()\n ax_array[i].bar(x=q, height=curr_df.trip_count.mean(),\n yerr=[[lower_error], [upper_error]])\n print(\"%s %s %s errors = %s %s %s\" % (phone_os, tl, q2r_map[q], lower_error, curr_df.trip_count.mean(), upper_error))\n ax_array[i].set_title(tl)",
"_____no_output_____"
],
[
"ifig, ax_array = plt.subplots(nrows=2,ncols=3,figsize=(10,5), sharex=False, sharey=True)\ntimeline_trip_gt = {\"train_bus_ebike_mtv_ucb\": 3,\n \"car_scooter_brex_san_jose\": 2,\n \"unimodal_trip_car_bike_mtv_la\": 2}\n\nplot_count_with_errors(ax_array[0], \"android\")\nplot_count_with_errors(ax_array[1], \"ios\")\n\nfor ax in ax_array[0]:\n ax.set_xticks(range(0,4))\n ax.set_xticklabels([\"truth\"] + [q2r_map[r] for r in range(1,4)])\n ax.set_yticks(range(0,tradeoff_df.trip_count.max(),3))\n \nfor ax in ax_array[1]:\n ax.set_xticks(range(0,4))\n ax.set_xticklabels([\"truth\"] + [q2r_map[r] for r in range(1,4)])\n ax.set_yticks(range(0,tradeoff_df.trip_count.max(),3))\n \nax_array[0,0].set_ylabel(\"nTrips (android)\")\nax_array[1,0].set_ylabel(\"nTrips (ios)\")\n \nifig.tight_layout(pad=0.85)",
"_____no_output_____"
]
],
[
[
"## Start end results",
"_____no_output_____"
]
],
[
[
"for r, df in tradeoff_df.query(\"timeline == @tl & phone_os == 'android'\").groupby(\"role\"):\n print(r, df.trip_count.mean() , df.trip_count.min(), df.trip_count.max())",
"_____no_output_____"
]
],
[
[
"The HAHFDC phone ran out of battery on all three runs of the `train_bus_ebike_mtv_ucb` timeline, so the trips never ended. Let's remove those so that they don't obfuscate the values from the other runs. ",
"_____no_output_____"
]
],
[
[
"out_of_battery_phones = tradeoff_df.query(\"timeline=='train_bus_ebike_mtv_ucb' & role=='HAHFDC' & trip_id=='berkeley_to_mtv_SF_express_bus_0' & phone_os == 'android'\")\nfor i in out_of_battery_phones.index:\n tradeoff_df.loc[i,\"end_diff_mins\"] = float('nan')",
"_____no_output_____"
],
[
"tradeoff_df.query(\"timeline=='train_bus_ebike_mtv_ucb' & role=='HAHFDC' & trip_id=='berkeley_to_mtv_SF_express_bus_0' & phone_os == 'android'\")",
"_____no_output_____"
]
],
[
[
"### Overall results",
"_____no_output_____"
]
],
[
[
"ifig, ax_array = plt.subplots(nrows=1,ncols=4,figsize=(16,4), sharex=False, sharey=True)\ntradeoff_df.query(\"phone_os == 'android'\").boxplot(ax = ax_array[0], column=[\"start_diff_mins\"], by=[\"quality\"])\nax_array[0].set_title(\"start time (android)\")\ntradeoff_df.query(\"phone_os == 'android'\").boxplot(ax = ax_array[1], column=[\"end_diff_mins\"], by=[\"quality\"])\nax_array[1].set_title(\"end time (android)\")\ntradeoff_df.query(\"phone_os == 'ios'\").boxplot(ax = ax_array[2], column=[\"start_diff_mins\"], by=[\"quality\"])\nax_array[2].set_title(\"start_time (ios)\")\ntradeoff_df.query(\"phone_os == 'ios'\").boxplot(ax = ax_array[3], column=[\"end_diff_mins\"], by=[\"quality\"])\nax_array[3].set_title(\"end_time (ios)\")\n\n # print(android_ax_returned.shape, ios_ax_returned.shape)\n\nax_array[0].set_xticklabels([q2r_map[int(t.get_text())] for t in ax_array[0].get_xticklabels()])\nax_array[1].set_xticklabels([q2r_map[int(t.get_text())] for t in ax_array[1].get_xticklabels()])\nax_array[2].set_xticklabels([q2r_map[int(t.get_text())] for t in ax_array[2].get_xticklabels()])\nax_array[3].set_xticklabels([q2r_map[int(t.get_text())] for t in ax_array[3].get_xticklabels()])\n\nfor ax in ax_array:\n ax.set_xlabel(\"\")\n\nax_array[1].text(0.55,25,\"Excluding trips where battery ran out\")\n\nax_array[0].set_ylabel(\"Diff (mins)\")\nifig.suptitle(\"Trip start end accuracy v/s configured quality\")\nifig.tight_layout(pad=1.7)",
"_____no_output_____"
]
],
[
[
"### Timeline specific",
"_____no_output_____"
]
],
[
[
"ifig, ax_array = plt.subplots(nrows=4,ncols=3,figsize=(10,10), sharex=False, sharey=True)\ntimeline_list = [\"train_bus_ebike_mtv_ucb\", \"car_scooter_brex_san_jose\", \"unimodal_trip_car_bike_mtv_la\"]\nfor i, tl in enumerate(timeline_list):\n tradeoff_df.query(\"timeline == @tl & phone_os == 'android'\").boxplot(ax = ax_array[0][i], column=[\"start_diff_mins\"], by=[\"quality\"])\n ax_array[0][i].set_title(tl)\n tradeoff_df.query(\"timeline == @tl & phone_os == 'android'\").boxplot(ax = ax_array[1][i], column=[\"end_diff_mins\"], by=[\"quality\"])\n ax_array[1][i].set_title(\"\")\n tradeoff_df.query(\"timeline == @tl & phone_os == 'ios'\").boxplot(ax = ax_array[2][i], column=[\"start_diff_mins\"], by=[\"quality\"])\n ax_array[2][i].set_title(\"\")\n tradeoff_df.query(\"timeline == @tl & phone_os == 'ios'\").boxplot(ax = ax_array[3][i], column=[\"end_diff_mins\"], by=[\"quality\"])\n ax_array[3][i].set_title(\"\")\n\n # print(android_ax_returned.shape, ios_ax_returned.shape)\n\nfor ax in ax_array[0]:\n ax.set_xticklabels([q2r_map[int(t.get_text())] for t in ax.get_xticklabels()])\n ax.set_xlabel(\"\")\n \nfor ax in ax_array[1]:\n ax.set_xticklabels([q2r_map[int(t.get_text())] for t in ax.get_xticklabels()])\n ax.set_xlabel(\"\")\n\nax_array[1,0].text(0.55,25,\"Excluding trips where battery ran out\")\n\nfor ax in ax_array[2]:\n ax.set_xticklabels([q2r_map[int(t.get_text())] for t in ax.get_xticklabels()])\n ax.set_xlabel(\"\")\n\nfor ax in ax_array[3]:\n ax.set_xticklabels([q2r_map[int(t.get_text())] for t in ax.get_xticklabels()])\n ax.set_xlabel(\"\")\n\nax_array[0][0].set_ylabel(\"Start time diff (android)\")\nax_array[1][0].set_ylabel(\"End time diff (android)\")\nax_array[2][0].set_ylabel(\"Start time diff (ios)\")\nax_array[3][0].set_ylabel(\"End time diff (ios)\")\nifig.suptitle(\"Trip start end accuracy (mins) v/s configured quality over multiple timelines\")\n\n# ifig.tight_layout(pad=2.5)",
"_____no_output_____"
]
],
[
[
"## Outlier checks\n\nWe can have unexpected values for both time and count. Unfortunately, there is no overlap between the two (intersection is zero). So we will look at a random sample from both cases",
"_____no_output_____"
]
],
[
[
"expected_legs = \"&\".join([\"not (trip_id == 'bus trip with e-scooter access_0' & count == 2)\",\n \"not (trip_id == 'mtv_to_berkeley_sf_bart_0' & count == 3)\"])\ncount_outliers = tradeoff_df.query(\"count > 1 & %s\" % expected_legs)\ncount_outliers[[\"phone_os\", \"range_id\", \"trip_id\", \"run\", \"role\", \"count\", \"start_diff_mins\", \"end_diff_mins\"]].head()",
"_____no_output_____"
],
[
"tradeoff_df.query(\"count < 1 & role == 'HAHFDC'\")",
"_____no_output_____"
],
[
"time_outliers = tradeoff_df.query(\"start_diff_mins == 30 | end_diff_mins == 30\")\ntime_outliers[[\"phone_os\", \"range_id\", \"trip_id\", \"run\", \"role\", \"start_diff_mins\", \"end_diff_mins\"]].head()",
"_____no_output_____"
],
[
"print(len(time_outliers.index.union(count_outliers.index)), len(time_outliers.index.intersection(count_outliers.index)))",
"_____no_output_____"
],
[
"time_outliers.sample(n=3, random_state=1)[[\"phone_os\", \"range_id\", \"trip_id\", \"run\", \"role\", \"count\", \"start_diff_mins\", \"end_diff_mins\"]]",
"_____no_output_____"
],
[
"count_outliers.sample(n=3, random_state=1)[[\"phone_os\", \"range_id\", \"trip_id\", \"run\", \"role\", \"count\", \"start_diff_mins\", \"end_diff_mins\"]]",
"_____no_output_____"
],
[
"fmt = lambda ts: arrow.get(ts).to(\"America/Los_Angeles\")",
"_____no_output_____"
],
[
"def check_outlier(eval_range, trip_idx, mismatch_key):\n eval_trip_range = eval_range[\"evaluation_trip_ranges\"][trip_idx]\n print(\"Trip %s, ground truth experiment for metric %s, %s, trip %s\" % (eval_range[\"trip_id\"], mismatch_key, fmt(eval_range[mismatch_key]), fmt(eval_trip_range[mismatch_key])))\n print(eval_trip_range[\"transition_df\"][[\"transition\", \"fmt_time\"]])\n print(\"**** For entire experiment ***\")\n print(eval_range[\"transition_df\"][[\"transition\", \"fmt_time\"]])\n if mismatch_key == \"end_ts\":\n # print(\"Transitions after trip end\")\n # print(eval_range[\"transition_df\"].query(\"ts > %s\" % eval_trip_range[\"end_ts\"])[[\"transition\", \"fmt_time\"]])\n return ezpv.display_map_detail_from_df(eval_trip_range[\"location_df\"])\n else:\n return ezpv.display_map_detail_from_df(eval_trip_range[\"location_df\"])",
"_____no_output_____"
]
],
[
[
"##### MAHFDC is just terrible\n\nIt looks like with MAHFDC, we essentially get no trip ends on android. Let's investigate these a bit further.\n- run 0: trip never ended: trip actually ended just before next trip started `15:01:26`. And then next trip had geofence exit, but we didn't detect it because it never ended, so we didn't create a sensed range for it.\n- run 1: trip ended but after 30 mins: similar behavior; trip ended just before next trip started `15:49:39`.",
"_____no_output_____"
]
],
[
[
"tradeoff_df.query(\"phone_os == 'android' & role == 'MAHFDC' & timeline == 'car_scooter_brex_san_jose'\")[[\"range_id\", \"trip_id\", \"run\", \"role\", \"count\", \"start_diff_mins\", \"end_diff_mins\"]]",
"_____no_output_____"
],
[
"FMT_STRING = \"HH:mm:SS\"\nfor t in pv_sj.map()[\"android\"][\"ucb-sdb-android-3\"][\"evaluation_ranges\"][3][\"evaluation_trip_ranges\"]:\n print(sd_sj.fmt(t[\"start_ts\"], FMT_STRING), \"->\", sd_sj.fmt(t[\"end_ts\"], FMT_STRING))\npv_sj.map()[\"android\"][\"ucb-sdb-android-3\"][\"evaluation_ranges\"][3][\"transition_df\"]",
"_____no_output_____"
],
[
"FMT_STRING = \"HH:mm:SS\"\nfor t in pv_sj.map()[\"android\"][\"ucb-sdb-android-3\"][\"evaluation_ranges\"][4][\"evaluation_trip_ranges\"]:\n print(sd_sj.fmt(t[\"start_ts\"], FMT_STRING), \"->\", sd_sj.fmt(t[\"end_ts\"], FMT_STRING))\npv_sj.map()[\"android\"][\"ucb-sdb-android-3\"][\"evaluation_ranges\"][4][\"transition_df\"]",
"_____no_output_____"
]
],
[
[
"##### Visit detection kicked in almost at the end of the trip",
"_____no_output_____"
]
],
[
[
"# 44 \tios \tsuburb_city_driving_weekend_0 \t1 \tHAMFDC \t0 \t30.000000 \t30.000000\ncheck_outlier(pv_la.map()[\"ios\"][\"ucb-sdb-ios-3\"][\"evaluation_ranges\"][4], 0, \"start_ts\")",
"_____no_output_____"
]
],
[
[
"##### Trip end never detected\n\nTrip ended at 14:11, experiment ended at 14:45. No stopped_moving for the last trip",
"_____no_output_____"
]
],
[
[
"# 65 \tandroid \tbus trip with e-scooter access_0 \t2 \tHAMFDC \t1 \t3.632239 \t30.000000\ncheck_outlier(pv_sj.map()[\"android\"][\"ucb-sdb-android-3\"][\"evaluation_ranges\"][2], 1, \"end_ts\")",
"_____no_output_____"
]
],
[
[
"##### Trip end detection errors on iOS\nOriginal experiment, explanation for the outliers on the HAHFDC and MAHFDC first runs to San Jose\n- HAHFDC: Trip end detected 1.5 hours after real end, but before next trip start\n- MAHFDC: Trip end detected 5 hours after real end, at the end of the next trip\n- MAHFDC: Clearly this was not even detected as a separate trip, so this is correct. There was a spurious trip from `17:42:22` - `17:44:22` which ended up matching this. But clearly because of the missing trip end detection, both the previous trip and this one were incorrect. You can click on the points at the Mountain View library to confirm when the trip ended.",
"_____no_output_____"
]
],
[
[
"fig = bre.Figure()\nfig.add_subplot(1,3,1).add_child(check_outlier(pv_sj.map()[\"ios\"][\"ucb-sdb-ios-2\"][\"evaluation_ranges\"][0], 0, \"end_ts\"))\nfig.add_subplot(1,3,2).add_child(check_outlier(pv_sj.map()[\"ios\"][\"ucb-sdb-ios-3\"][\"evaluation_ranges\"][0], 0, \"end_ts\"))\nfig.add_subplot(1,3,3).add_child(check_outlier(pv_sj.map()[\"ios\"][\"ucb-sdb-ios-3\"][\"evaluation_ranges\"][0], 1, \"start_ts\"))\n# check_outlier(pv_sj.map()[\"ios\"][\"ucb-sdb-ios-2\"][\"evaluation_ranges\"][0], 0, \"end_ts\")",
"_____no_output_____"
]
],
[
[
"##### No geofence exit ever detected\n\nOn the middle trip of the second round of data collection to the San Jose library, we got no geofence exits. The entire list of transitions is \n\n```\ntransition fmt_time\n3 T_VISIT_ENDED 2019-08-06T11:29:20.573817-07:00\n6 T_VISIT_STARTED 2019-08-06T11:29:20.911773-07:00\n8 T_VISIT_ENDED 2019-08-06T11:35:38.250980-07:00\n9 T_VISIT_STARTED 2019-08-06T12:00:05.445936-07:00\n12 T_TRIP_ENDED 2019-08-06T12:00:07.093790-07:00\n15 T_VISIT_ENDED 2019-08-06T15:59:13.998068-07:00\n18 T_VISIT_STARTED 2019-08-06T17:12:38.808743-07:00\n21 T_TRIP_ENDED 2019-08-06T17:12:40.504285-07:00\n```\n\nWe did get visit notifications, so we did track location points (albeit after a long time), and we did get the trip end notifications, but we have no sensed trips. Had to handle this in the code as well",
"_____no_output_____"
]
],
[
[
"check_outlier(pv_sj.map()[\"ios\"][\"ucb-sdb-ios-2\"][\"evaluation_ranges\"][4], 0, \"start_ts\")",
"_____no_output_____"
]
],
[
[
"##### No geofence exit ever detected\n\nOn the middle trip of the second round of data collection to the San Jose library, we got no geofence exits.\nWe did get visit notifications, so we did track location points (albeit after a long time), and we did get the trip end notifications, but we have no sensed trips. Had to handle this in the code as well",
"_____no_output_____"
]
],
[
[
"# 81 \tios \tbus trip with e-scooter access_0 \t1 \tHAHFDC \t0 \t30.000000 \t30.000000\ncheck_outlier(pv_sj.map()[\"ios\"][\"ucb-sdb-ios-2\"][\"evaluation_ranges\"][4], 1, \"end_ts\")",
"_____no_output_____"
]
],
[
[
"### 7 mapped trips for one\n\nThis is essentially from the time that I wandered around looking for the bikeshare bike. This raises the question of whether I should filter out the points within the polygon in this case too. Overall, I think not. The only part within the polygon that we don't guarantee is the ground truth trajectory. We still do have the ground truth of the trip/section start end, and there really is no reason why we should have had so many \"trips\" when I was walking around. I certainly didn't wait for too long while walking and this was not semantically a \"trip\" by any stretch of the imagination.",
"_____no_output_____"
]
],
[
[
"# 113 \tandroid \tberkeley_to_mtv_SF_express_bus_0 \t2 \tHAMFDC \t7 \t2.528077 \t3.356611\ncheck_outlier(pv_ucb.map()[\"android\"][\"ucb-sdb-android-3\"][\"evaluation_ranges\"][2], 2, \"end_ts\")",
"_____no_output_____"
]
],
[
[
"### Trip split into two in medium accuracy *only*\n\nActual trip ends at `14:21`. In medium accuracy, detected trips were `14:12:15 -> 14:17:33` and `14:22:14 -> 14:24:15`. This was after we reached the destination, but there is a large gap because we basically got no points for a large part of the trip. This seems correct - it looks like iOS is just prematurely detecting the trip end in the MA case.",
"_____no_output_____"
]
],
[
[
"# 127 \tios \twalk_urban_university_0 \t1 \tMAHFDC \t2 \t4.002549 \t2.352913\nfig = bre.Figure()\n\ndef compare_med_high_accuracy():\n trip_idx = 1\n mismatch_key = \"end_ts\"\n ha_range = pv_ucb.map()[\"ios\"][\"ucb-sdb-ios-2\"][\"evaluation_ranges\"][1]\n ha_trip_range = ha_range[\"evaluation_trip_ranges\"][trip_idx]\n eval_range = pv_ucb.map()[\"ios\"][\"ucb-sdb-ios-3\"][\"evaluation_ranges\"][1]\n eval_trip_range = eval_range[\"evaluation_trip_ranges\"][trip_idx]\n print(\"Trip %s, ground truth experiment for metric %s, %s, trip %s, high accuracy %s\" %\n (eval_range[\"trip_id\"], mismatch_key,\n fmt(eval_range[mismatch_key]), fmt(eval_trip_range[mismatch_key]), fmt(ha_trip_range[mismatch_key])))\n print(eval_trip_range[\"transition_df\"][[\"transition\", \"fmt_time\"]])\n print(\"**** Expanded ***\")\n print(eval_range[\"transition_df\"].query(\"%s < ts < %s\" %\n ((eval_trip_range[\"end_ts\"] - 30*60), (eval_trip_range[\"end_ts\"] + 30*60)))[[\"transition\", \"fmt_time\"]])\n fig = bre.Figure()\n fig.add_subplot(1,2,1).add_child(ezpv.display_map_detail_from_df(ha_trip_range[\"location_df\"]))\n fig.add_subplot(1,2,2).add_child(ezpv.display_map_detail_from_df(eval_trip_range[\"location_df\"]))\n return fig\ncompare_med_high_accuracy()",
"_____no_output_____"
],
[
"[{'start_ts': fmt(1564089135.368705), 'end_ts': fmt(1564089453.8783798)},\n{'start_ts': fmt(1564089734.305933), 'end_ts': fmt(1564089855.8683748)}]",
"_____no_output_____"
]
],
[
[
"### We just didn't detect any trip ends in the middle\n\nWe only detected a trip end at the Mountain View station. This is arguably more correct than the multiple trips that we get with a dwell time.",
"_____no_output_____"
]
],
[
[
"# 120 \tios \tmtv_to_berkeley_sf_bart_0 \t2 \tHAHFDC \t2 \t3.175024 \t1.046759\ncheck_outlier(pv_ucb.map()[\"ios\"][\"ucb-sdb-ios-2\"][\"evaluation_ranges\"][2], 0, \"end_ts\")",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
4afddfe28f6e59d241c7a2b3239a4025be358b5f
| 38,850 |
ipynb
|
Jupyter Notebook
|
2021notebooks/2021_1010facial_keypoints_detection.ipynb
|
project-ccap/project-ccap.github.io
|
867e32af5459ae55d864d9d022d69eac17fbb450
|
[
"MIT"
] | null | null | null |
2021notebooks/2021_1010facial_keypoints_detection.ipynb
|
project-ccap/project-ccap.github.io
|
867e32af5459ae55d864d9d022d69eac17fbb450
|
[
"MIT"
] | 1 |
2020-12-04T11:36:15.000Z
|
2020-12-04T11:36:15.000Z
|
2021notebooks/2021_1010facial_keypoints_detection.ipynb
|
project-ccap/project-ccap.github.io
|
867e32af5459ae55d864d9d022d69eac17fbb450
|
[
"MIT"
] | 2 |
2020-07-22T02:58:14.000Z
|
2020-07-23T07:02:07.000Z
| 40.595611 | 283 | 0.520824 |
[
[
[
"<a href=\"https://colab.research.google.com/github/project-ccap/project-ccap.github.io/blob/master/2021notebooks/2021_1010facial_keypoints_detection.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"# -*- coding: utf-8 -*-",
"_____no_output_____"
]
],
[
[
"---\ntitle: Getting Started with Facial Keypoint Detection using Deep Learning and PyTorch\nauthor: Sovit Ranjan \ndate: 2020October\nsource: https://debuggercafe.com/getting-started-with-facial-keypoint-detection-using-pytorch/\n\n---\n\n- [Kaggle](https://www.kaggle.com/c/facial-keypoints-detection/data)\n- Kaggle からデータ入手\n\n```bash\nkaggle competitions download -c facial-keypoints-detection\n```",
"_____no_output_____"
]
],
[
[
"%%shell\ncurl -sc /tmp/cookie \"https://drive.google.com/uc?export=download&id=1r1XxfBOQMzfhaohgg6aq-KbsYACkiWNa\" > /dev/null\nCODE=\"$(awk '/_warning_/ {print $NF}' /tmp/cookie)\" \ncurl -Lb /tmp/cookie \"https://drive.google.com/uc?export=download&confirm=${CODE}&id=1r1XxfBOQMzfhaohgg6aq-KbsYACkiWNa\" -o kaggle_facial_keypoints_detection.tar.gz\n",
"_____no_output_____"
],
[
"!gunzip kaggle_facial_keypoints_detection.tar.gz\n!tar -xf kaggle_facial_keypoints_detection.tar",
"_____no_output_____"
]
],
[
[
"# 深層学習とPyTorchを使った顔のキーポイント検出\n\nPyTorch を使った顔のキーポイント検出デモ\n\n<center>\n<img src=\"https://debuggercafe.com/wp-content/uploads/2020/10/intro_exmp.png\" width=\"33%\"><br/>\n<p style=\"text-align:left;width:88%; background-color:cornsilk\">\nFigure 1. An example of facial keypoint detection using deep learning and PyTorch. We will try to achieve similar results after going through this tutorial.\n</p>\n</center>\n\n図 1 は濃淡画像上での顔のキーポイント検出の例です。\nこのチュートリアルの終わりまでに, 同様の結果を得ることを目標としています。\n\n* 顔のキーポイント検出の必要性を簡単に紹介します。\n* 深層学習と PyTorch を使って、顔のキーポイント検出を始めるための簡単なデータセットを使用しています。\n* シンプルな畳み込みニューラルネットワークモデルを使って、データセット上で訓練を行います。\n* 次に、訓練されたモデルを使って、テストデータセットの未見の画像の顔のキーポイントを検出します。\n* 最後に、メリット、デメリット、さらなる実験と改善のために取るべきステップにたどり着きます。\n\n# 1. なぜ顔のキーポイント検出が必要なのか?\n\n\n先に進む前に、素朴な疑問に答えてみましょう。\nなぜ、顔のキーポイント検出のような技術が必要なのか?たくさんありますが、いくつかをご紹介します。\n\nスマートフォンのアプリで, フィルターを見たことがある人も多いのではないでしょうか。\nこのようなフィルターを顔に正確に適用するためには,人の顔のキーポイント (注目点) を正しく判断する必要があります。\nそのためには, 顔のキーポイントを検出する必要があります。\nまた、顔のキーポイント検出は、人の年齢を判定するのにも利用できます。\n実際、多くの産業や企業がこの技術を利用しています。\n顔認証によるスマートフォンのロック解除にも、顔のキーポイント検出が使われています。\n上記は、実際の使用例の一部に過ぎません。\n他にもたくさんありますが、それらの詳細については今は触れません。\nもっと詳しく知りたい方は、ユースケースについてより多くのポイントを説明した[こちらの記事をお読みください](https://www.facefirst.com/blog/amazing-uses-for-face-recognition-facial-recognition-use-cases/)。\n\n上述したように、このチュートリアルでは、顔のキーポイント検出にディープラーニングを使用します。\n深層学習と畳み込みニューラルネットワークは、現在、顔認識とキーポイント検出の分野で大きな役割を果たしています。",
"_____no_output_____"
],
[
"## 1.1 データセット\n<!-- ## 1.1 The Dataset-->\n\n過去に開催された Kaggle のコンペティションのデータセットを使用します。\n競技名は Facial Keypoints Detection です。\n競技のルールに同意した後、データセットをダウンロードするように言われたら、ダウンロードしてください。\n\nデータセットは大きくありません。約 80 MBしかありません。\n訓練データセットとテストデータセットを含む CSV ファイルで構成されています。\n画像も CSV ファイルの中にピクセル値で入っています。\n画像はすべて 96×96 次元の濃淡画像です。\n濃淡画像で次元が小さいため、深層学習による顔のキーポイント検出を始めるのに適した、簡単なデータセットです。\n\nこのデータセットには (x, y) 形式の 15 個の座標特徴のキーポイントが含まれています。\nつまり、各顔画像には合計 30 個のポイント特徴があるということです。\nすべてのデータポイントは CSV ファイルの異なる列に入っており、最後の列には画像のピクセル値が入っています。\n\n<!--\nWe will use a dataset from one of the past Kaggle competitions. \nThe competition is Facial Keypoints Detection. Go ahead and download the dataset after accepting the competition rules if it asks you to do so.\n\nThe dataset is not big. It is only around 80 MB. \nIt consists of CSV files containing the training and test dataset. \nThe images are also within the CSV files with the pixel values. \nAll the images are 96×96 dimensional grayscale images. \nAs the images are grayscale and small in dimension, that is why it is a good and easy dataset to start with facial keypoint detection using deep learning.\n\nThe dataset contains the keypoints for 15 coordinate features in the form of (x, y). \nSo, there are a total of 30 point features for each face image. \nAll the data points are in different columns of the CSV file with the final column holding the image pixel values.-->\n\n次のコードスニペットは CSV ファイルのデータフォーマットを示しています。\n<!-- The following code snippet shows the data format in the CSV files. -->\n\n\n<pre style=\"background-color:powderblue\">\nleft_eye_center_x left_eye_center_y right_eye_center_x ... mouth_center_bottom_lip_x mouth_center_bottom\n_lip_y Image\n0 66.033564 39.002274 30.227008 ... 43.130707 \n 84.485774 238 236 237 238 240 240 239 241 241 243 240 23...\n1 64.332936 34.970077 29.949277 ... 45.467915 \n 85.480170 219 215 204 196 204 211 212 200 180 168 178 19...\n2 65.057053 34.909642 30.903789 ... 47.274947 \n 78.659368 144 142 159 180 188 188 184 180 167 132 84 59 ...\n3 65.225739 37.261774 32.023096 ... 51.561183 \n 78.268383 193 192 193 194 194 194 193 192 168 111 50 12 ...\n4 66.725301 39.621261 32.244810 ... 44.227141 \n 86.871166 147 148 160 196 215 214 216 217 219 220 206 18...\n... ... ... ... ... ... \n ... ...\n7044 67.402546 31.842551 29.746749 ... 50.426637 \n 79.683921 71 74 85 105 116 128 139 150 170 187 201 209 2...\n7045 66.134400 38.365501 30.478626 ... 50.287397 \n 77.983023 60 60 62 57 55 51 49 48 50 53 56 56 106 89 77 ...\n7046 66.690732 36.845221 31.666420 ... 49.462572 \n 78.117120 74 74 74 78 79 79 79 81 77 78 80 73 72 81 77 1...\n7047 70.965082 39.853666 30.543285 ... 50.065186 \n 79.586447 254 254 254 254 254 238 193 145 121 118 119 10...\n7048 66.938311 43.424510 31.096059 ... 45.900480 \n 82.773096 53 62 67 76 86 91 97 105 105 106 107 108 112 1...</pre>\n\n\nキーポイントとなる特徴的な列を見ることができます。\nこのような列は、顔の左側と右側で30個あります。\n最後の列は、ピクセル値を示す「画像」の列です。\nこれは文字列形式です。\nこのように、データセットに深層学習技術を適用する前に、少しだけ前処理を行う必要があります。\n<!-- You can see the keypoint feature columns. \nThere are 30 such columns for the left and right sides of the face. \nThe last column is the Image column with the pixel values. \nThey are in string format. \nSo, we will have to do a bit of preprocessing before we can apply our deep learning techniques to the dataset. -->\n\n\n以下は、顔にキーポイントを設定した `training.csv` ファイルのサンプル画像です。\n<!-- The following are some sample images from the training.csv file with the keypoints on the faces. -->\n\n<center>\n <img src=\"https://debuggercafe.com/wp-content/uploads/2020/10/training_samples.png\" width=\"66%\"><br/>\n<p style=\"text-align:left;width:77%;background-color:cornsilk\">\nFigure 2. Some samples from the training set with their facial keypoints. \nWe will use this dataset to train our deep neural network using PyTorch.</p>\n</center>\n\nまた、このデータセットには多くの欠損値が含まれています。\n7048 個のインスタンス (行) のうち 4909 行は 1 つ以上の列で少なくとも 1 つの NULL 値を含んでいます。\nまた、全てのキーポイントが揃っている完全なデータは 2140 行のみです。\nこのような状況は、データセットを作成する際に対処しなければなりません。\n",
"_____no_output_____"
],
[
"# 2. 深層学習とPyTorchによる顔のキーポイント検出\n<!-- # 2. Facial Keypoint Detection using Deep Learning and PyTorch -->\n\nPyTorch フレームワークを使った顔のキーポイント検出のためのコーディング作業に入っていきます。",
"_____no_output_____"
]
],
[
[
"import torch\nimport torch.optim as optim\nimport matplotlib.pyplot as plt\nimport torch.nn as nn\n\nimport matplotlib.pyplot as plt\n%matplotlib inline\nimport numpy as np\n\n!pip install japanize_matplotlib\nimport japanize_matplotlib",
"_____no_output_____"
],
[
"import torch\n\nROOT_PATH = 'kaggle_facial_keypoints_detection'\n!mkdir outputs\nOUTPUT_PATH = 'outputs'\n\n# learning parameters\nBATCH_SIZE = 256\nLR = 0.0001\nEPOCHS = 300\n\nDEVICE = torch.device('cuda' if torch.cuda.is_available() else 'cpu')\n\n# train/test split\nTEST_SPLIT = 0.2\nTEST_SPLIT = 0.1\n\n# show dataset keypoint plot\nSHOW_DATASET_PLOT = True",
"_____no_output_____"
]
],
[
[
"トレーニングと検証のための学習パラメータは以下の通りです。\n\n* バッチサイズは 256 としています。\n画像のサイズが 96×96 と小さく、また濃淡画像であるため,ミニバッチサイズを大きくしてもメモリの問題は発生しません。\nただし GPU のメモリに応じて、バッチサイズを自由に増減させてください。\n* 学習率は 0.0001 です。\n様々な学習率を試した結果、今回使用するモデルとデータセットでは、この学習率が最も安定していると思われます。\n* 顔のキーポイントのデータセットに対して 300 エポックでモデルを学習します。\n多いと思われるかもしれませんが、実際には, このように多くのエポックを行うことで, モデルは恩恵を受けます。\n* 0.2 のテスト分割を使用しています。\n データの 80 %をトレーニングに,20 %を検証に使用します。\n* SHOW_DATASET_PLOT が True の場合, 訓練直前に,いくつかの顔と,それに対応する顔のキーポイントのプロットが表示されます。\n必要であれば,これを False にしておくこともできます。\n\n## 2.1 深層学習と PyTorch による顔のキーポイント検出のためのユーティリティー関数の作成\n\nこの節では、作業を容易にするためのユーティリティー関数をいくつか書きます。\nユーティリティー関数は全部で 3 つあります。\nこの 3 つのユーティリティー関数はすべて、顔の画像上に顔のキーポイントをプロットするのに役立ちます。\nしかし 3 つとも異なるシナリオに対応しています。\n1 つずつ取り組んでいきましょう。\n\n### 2.1.1 顔に検証キーポイントをプロットする関数\n\nまず、検証用のキーポイントをプロットする関数を紹介します。\nこの関数を `valid_keypoints_plot()` と呼ぶことにします。\nこの関数は基本的に,与えられた一定のエポック数の後に,画像の顔上に検証(回帰したキーポイント)をプロットします.\n\nまずはコードを書いてみましょう。その後、重要な部分の説明に入ります。\n",
"_____no_output_____"
]
],
[
[
"def valid_keypoints_plot(image, outputs, orig_keypoints, epoch):\n \"\"\"\n This function plots the regressed (predicted) keypoints and the actual keypoints after each validation epoch for one image in the batch.\n \"\"\"\n # detach the image, keypoints, and output tensors from GPU to CPU\n image = image.detach().cpu()\n outputs = outputs.detach().cpu().numpy()\n orig_keypoints = orig_keypoints.detach().cpu().numpy()\n\n # just get a single datapoint from each batch\n img = image[0]\n output_keypoint = outputs[0]\n orig_keypoint = orig_keypoints[0]\n img = np.array(img, dtype=float)\n img = np.transpose(img, (1, 2, 0))\n img = img.reshape(96, 96)\n plt.imshow(img, cmap='gray')\n\n output_keypoint = output_keypoint.reshape(-1, 2)\n orig_keypoint = orig_keypoint.reshape(-1, 2)\n for p in range(output_keypoint.shape[0]):\n plt.plot(output_keypoint[p, 0], output_keypoint[p, 1], 'r.')\n plt.text(output_keypoint[p, 0], output_keypoint[p, 1], f\"{p}\")\n plt.plot(orig_keypoint[p, 0], orig_keypoint[p, 1], 'g.')\n plt.text(orig_keypoint[p, 0], orig_keypoint[p, 1], f\"{p}\")\n plt.savefig(f\"{OUTPUT_PATH}/val_epoch_{epoch}.png\")\n plt.close()\n\n\ndef test_keypoints_plot(images_list, outputs_list, figsize=(10,10)):\n \"\"\"\n This function plots the keypoints for the outputs and images\n in `test.csv` file.\n \"\"\"\n plt.figure(figsize=figsize)\n for i in range(len(images_list)):\n outputs = outputs_list[i]\n image = images_list[i]\n outputs = outputs.cpu().detach().numpy()\n outputs = outputs.reshape(-1, 2)\n plt.subplot(3, 3, i+1)\n #plt.imshow(image, cmap='gray')\n plt.imshow(image.reshape(96,96), cmap='gray')\n\n for p in range(outputs.shape[0]):\n plt.plot(outputs[p, 0], outputs[p, 1], 'r.')\n plt.text(outputs[p, 0], outputs[p, 1], f\"{p}\")\n plt.axis('off')\n\n plt.savefig(f\"{OUTPUT_PATH}/test_output.pdf\")\n plt.show()\n plt.close()\n\n\ndef dataset_keypoints_plot(data, figsize=(22,20), n_samples=30):\n \"\"\"\n This function shows the image faces and keypoint plots that the model will actually see. \n This is a good way to validate that our dataset is in fact corrent and the faces align wiht the keypoint features. \n The plot will be show just before training starts. Press `q` to quit the plot and start training.\n \"\"\"\n plt.figure(figsize=figsize)\n for i in range(n_samples):\n sample = data[i]\n img = sample['image']\n img = np.array(img, dtype=float)\n img = np.transpose(img, (1, 2, 0))\n img = img.reshape(96, 96)\n plt.subplot(5, 6, i+1)\n plt.imshow(img, cmap='gray')\n keypoints = sample['keypoints']\n for j in range(len(keypoints)):\n plt.plot(keypoints[j, 0], keypoints[j, 1], 'r.')\n plt.show()\n plt.close()",
"_____no_output_____"
]
],
[
[
"最初の 2 行のコメントを読めば,この関数の要点が容易に理解できると思います。\n画像テンソル(`image`),出力テンソル(`outputs`),データセットからのオリジナルキーポイント(`orig_keypoints`)を,エポック番号とともに関数に渡します.\n<!-- If you read the comment in the first two lines then you will easily get the gist of the function. \nWe provide the image tensors (`image`), the output tensors (`outputs`), and the original keypoints from the dataset (`orig_keypoints`) along with the epoch number to the function.-->\n\n* 7, 8, 9行目では GPU からデータを切り離し CPU にロードしています。\n* テンソルは,画像,予測されたキーポイント,オリジナルのキーポイントそれぞれについて 256 個のデータポイントを含むバッチの形をしています.\n12 行目から 14 行目で、それぞれの最初のデータポイントを取得します。\n* 次に,画像を NumPy の配列形式に変換し,チャンネルを最後にして転置し,元の 96×96 のサイズに整形します。\nそして,Matplotlib を使って画像をプロットします。\n* 21 行目と 22 行目では,予測されたキーポイントと元のキーポイントの形状を変更します.\n21 行目と 22 行目で,予測キーポイントとオリジナルキーポイントの形状を変更します.\n* 23 行目から 27 行目まで,顔の画像上に予測キーポイントとオリジナルキーポイントをプロットします。\n予測されたキーポイントは赤のドットで、オリジナルのキーポイントは緑のドットになります。\nまた,`plt.text()`を用いて,対応するキーポイントの番号をプロットします。\n* 最後に,画像を `outputs` フォルダに保存します。\n\n<!--\n* At lines 7, 8, and 9 we detach the data from the GPU and load them onto the CPU.\n* The tensors are in the form of a batch containing 256 datapoints each for the image, the predicted keypoints, and the original keypoints. \nWe get just the first datapoint from each from lines 12 to 14.\n* Then we convert the image to NumPy array format, transpose it make channels last, and reshape it into the original 96×96 dimensions. \nThen we plot the image using Matplotlib.\n* At lines 21 and 22, we reshape the predicted and original keypoints. \nThis will make them have 2 columns along with the respective number of rows.\n* Starting from lines 23 till 27, we plot the predicted and original keypoints on the image of the face. \nThe predicted keypoints will be red dots while the original keypoints will be green dots. \nWe also plot the corresponding keypoint numbers using `plt.text()`.\n* Finally, we save the image in the `outputs` folder.\n-->\n\nNow, we will move onto the next function for the `utils.py` file.\n\n### 2.1.2 テスト用キーポイントを顔にプロットする関数\n<!-- ### 2.1.2 Function to Plot the Test Keypoints on the Faces -->\n\nここでは、テスト時に予測するキーポイントをプロットするためのコードを書きます。\n具体的には `test.csv` ファイルにピクセル値が入っている画像が対象となります。\n<!--\nHere, we will write the code for plotting the keypoints that we will predict during testing. \nSpecifically, this is for those images whose pixel values are in the test.csv file. -->",
"_____no_output_____"
]
],
[
[
"import torch\nimport cv2\nimport pandas as pd\nimport numpy as np\n\nfrom torch.utils.data import Dataset, DataLoader\n\nresize = 96\n\ndef train_test_split(csv_path, split):\n df_data = pd.read_csv(csv_path)\n\n # drop all the rows with missing values\n df_data = df_data.dropna()\n len_data = len(df_data)\n\n # calculate the validation data sample length\n valid_split = int(len_data * split)\n\n # calculate the training data samples length\n train_split = int(len_data - valid_split)\n training_samples = df_data.iloc[:train_split][:]\n valid_samples = df_data.iloc[-valid_split:][:]\n print(f\"訓練データサンプル数: {len(training_samples)}\")\n print(f\"検証データサンプル数: {len(valid_samples)}\")\n return training_samples, valid_samples\n\n\nclass FaceKeypointDataset(Dataset):\n def __init__(self, samples):\n self.data = samples\n\n # get the image pixel column only\n self.pixel_col = self.data.Image\n self.image_pixels = []\n #for i in tqdm(range(len(self.data))):\n for i in range(len(self.data)):\n img = self.pixel_col.iloc[i].split(' ')\n self.image_pixels.append(img)\n self.images = np.array(self.image_pixels, dtype=float)\n\n def __len__(self):\n return len(self.images)\n\n def __getitem__(self, index):\n\n # reshape the images into their original 96x96 dimensions\n image = self.images[index].reshape(96, 96)\n orig_w, orig_h = image.shape\n\n # resize the image into `resize` defined above\n image = cv2.resize(image, (resize, resize))\n\n # again reshape to add grayscale channel format\n image = image.reshape(resize, resize, 1)\n image = image / 255.0\n\n # transpose for getting the channel size to index 0\n image = np.transpose(image, (2, 0, 1))\n\n # get the keypoints\n keypoints = self.data.iloc[index][:30]\n keypoints = np.array(keypoints, dtype=float)\n\n # reshape the keypoints\n keypoints = keypoints.reshape(-1, 2)\n # rescale keypoints according to image resize\n keypoints = keypoints * [resize / orig_w, resize / orig_h]\n return {\n 'image': torch.tensor(image, dtype=torch.float),\n 'keypoints': torch.tensor(keypoints, dtype=torch.float),\n }\n\n\n# get the training and validation data samples\ntraining_samples, valid_samples = train_test_split(f\"{ROOT_PATH}/training.csv\", TEST_SPLIT)\n\n# initialize the dataset - `FaceKeypointDataset()`\nprint('--- PREPARING DATA ---')\ntrain_data = FaceKeypointDataset(training_samples)\nvalid_data = FaceKeypointDataset(valid_samples)\nprint('--- DATA PREPRATION DONE ---')\n# prepare data loaders\ntrain_loader = DataLoader(train_data,\n batch_size=BATCH_SIZE,\n shuffle=True)\nvalid_loader = DataLoader(valid_data,\n batch_size=BATCH_SIZE,\n shuffle=False)\n\n\n# whether to show dataset keypoint plots\nif SHOW_DATASET_PLOT:\n dataset_keypoints_plot(valid_data)\n",
"_____no_output_____"
],
[
"#dataset_keypoints_plot(valid_data,figsize=(24,20))\n#dataset_keypoints_plot(train_data,figsize=(24,20))\n#df_data = pd.read_csv(f\"{ROOT_PATH}/training/training.csv\")\n#df_data",
"_____no_output_____"
],
[
"#from model import FaceKeypointModel\nimport torch.nn as nn\nimport torch.nn.functional as F\n\nclass FaceKeypointModel(nn.Module):\n def __init__(self):\n super(FaceKeypointModel, self).__init__()\n self.conv1 = nn.Conv2d(1, 32, kernel_size=5)\n self.conv2 = nn.Conv2d(32, 64, kernel_size=3)\n self.conv3 = nn.Conv2d(64, 128, kernel_size=3)\n self.fc1 = nn.Linear(128, 30) \n self.pool = nn.MaxPool2d(2, 2)\n self.dropout = nn.Dropout2d(p=0.2)\n\n\n def forward(self, x):\n x = F.relu(self.conv1(x))\n x = self.pool(x)\n x = F.relu(self.conv2(x))\n x = self.pool(x)\n x = F.relu(self.conv3(x))\n x = self.pool(x)\n\n bs, _, _, _ = x.shape\n x = F.adaptive_avg_pool2d(x, 1).reshape(bs, -1)\n x = self.dropout(x)\n out = self.fc1(x) \n return out",
"_____no_output_____"
]
],
[
[
"`test_keypoints_plot()` 関数の入力パラメータは, `images_list` と `outputs_list` です。\nこれらは, 指定された数の入力画像と, プロットしたい予測キーポイントを含む 2 つのリストです。\nこの関数は非常にシンプルです。\n\n* 7 行目からは,単純な for ループを実行し,2 つのリストに含まれる画像と予測されるキーポイントをループします。\n* `valid_keypoints_plot()` 関数と同じ経路をたどります。\n* しかし,今回は,すべての画像を 1 つのプロットにしたいので,Matplotlib の `subplot()` 関数を利用します。\n9 枚の画像をプロットするので,`plt.subplot(3, 3, i+1)` を使用します.\n* 最後に, プロットされた画像と予測されたキーポイントを,出力フォルダに保存します。\n\n以上でこの関数の説明は終わりです。\n\n### 2.1.3 入力データセットの顔画像とキーポイントをプロットする関数\n\nニューラルネットワークモデルにデータを入力する前に,データが正しいかどうかを確認します。\nすべてのキーポイントが顔に正しく対応しているかどうかは,わからないかもしれません。\nそのため,学習開始前に,顔画像とそれに対応するキーポイントを表示する関数を書きます。\n`SHOW_DATASET_PLOT = True` になっている場合のみ行われます。\n",
"_____no_output_____"
]
],
[
[
"import torch\nimport torch.optim as optim\nimport matplotlib.pyplot as plt\nimport torch.nn as nn\nimport matplotlib\n\n# model\nmodel = FaceKeypointModel().to(DEVICE)\n\n# optimizer\noptimizer = optim.Adam(model.parameters(), lr=LR)\n\n# we need a loss function which is good for regression like MSELoss\ncriterion = nn.MSELoss()\n\n# training function\ndef train(model, dataloader, data):\n #print('Training')\n model.train()\n train_running_loss = 0.0\n counter = 0\n # calculate the number of batches\n num_batches = int(len(data)/dataloader.batch_size)\n\n for i, data in enumerate(dataloader): #, total=num_batches):\n counter += 1\n image, keypoints = data['image'].to(DEVICE), data['keypoints'].to(DEVICE)\n # flatten the keypoints\n keypoints = keypoints.view(keypoints.size(0), -1)\n optimizer.zero_grad()\n outputs = model(image)\n loss = criterion(outputs, keypoints)\n train_running_loss += loss.item()\n loss.backward()\n optimizer.step()\n\n train_loss = train_running_loss/counter\n return train_loss\n\n\n# validatioon function\ndef validate(model, dataloader, data, epoch, print_interval=3):\n #print('Validating')\n model.eval()\n valid_running_loss = 0.0\n counter = 0\n # calculate the number of batches\n num_batches = int(len(data)/dataloader.batch_size)\n with torch.no_grad():\n for i, data in enumerate(dataloader): #, total=num_batches):\n counter += 1\n image, keypoints = data['image'].to(DEVICE), data['keypoints'].to(DEVICE)\n # flatten the keypoints\n keypoints = keypoints.view(keypoints.size(0), -1)\n outputs = model(image)\n loss = criterion(outputs, keypoints)\n valid_running_loss += loss.item()\n # plot the predicted validation keypoints after every...\n # ... print_interval epochs and from the first batch\n if (epoch+1) % print_interval == 0 and i == 0:\n valid_keypoints_plot(image, outputs, keypoints, epoch)\n\n valid_loss = valid_running_loss/counter\n return valid_loss",
"_____no_output_____"
],
[
"EPOCHS",
"_____no_output_____"
],
[
"EPOCHS = 64 # 時間の都合上 EPOCHS を少なくしています\ninterval = EPOCHS >> 3\n\ntrain_loss = []\nval_loss = []\nfor epoch in range(EPOCHS):\n train_epoch_loss = train(model, train_loader, train_data)\n val_epoch_loss = validate(model, valid_loader, valid_data, epoch, print_interval=2)\n train_loss.append(train_epoch_loss)\n val_loss.append(val_epoch_loss)\n\n if ((epoch) % interval) == 0:\n print(f\"エポック {epoch+1:<4d}/{EPOCHS:<4d}\", end=\"\")\n print(f\"訓練損失: {train_epoch_loss:.3f}\", \n f'検証損失: {val_epoch_loss:.3f}')",
"_____no_output_____"
],
[
"# loss plots\nplt.figure(figsize=(10, 7))\nplt.plot(train_loss, color='blue', label='訓練損失')\nplt.plot(val_loss, color='red', label='検証損失')\nplt.xlabel('エポック')\nplt.ylabel('損失値')\nplt.legend()\nplt.savefig(f\"{OUTPUT_PATH}/loss.pdf\")\nplt.show()",
"_____no_output_____"
],
[
"# 結果の保存\ntorch.save({\n 'epoch': EPOCHS,\n 'model_state_dict': model.state_dict(),\n 'optimizer_state_dict': optimizer.state_dict(),\n 'loss': criterion,\n }, f\"{OUTPUT_PATH}/model.pth\")\n",
"_____no_output_____"
]
],
[
[
"## 結果の再読み込みと視覚化\n\n保存した結果を再度読み込んで表示します\n",
"_____no_output_____"
]
],
[
[
"model = FaceKeypointModel().to(DEVICE)\n\n# load the model checkpoint\ncheckpoint = torch.load(f\"{OUTPUT_PATH}/model.pth\")\n\n# load model weights state_dict\nmodel.load_state_dict(checkpoint['model_state_dict'])\nmodel.eval()",
"_____no_output_____"
]
],
[
[
"## テストデータによる結果の視覚化\n\n`test.csv` を読み込んで,結果を表示します",
"_____no_output_____"
]
],
[
[
"model.eval()",
"_____no_output_____"
],
[
"# `test.csv` ファイルの読み込み\n\ncsv_file = f\"{ROOT_PATH}/test.csv\"\ndata = pd.read_csv(csv_file)\n\npixel_col = data.Image\nimage_pixels = []\nfor i in range(len(pixel_col)):\n img = pixel_col[i].split(' ')\n image_pixels.append(img)\n\n# NumPy 配列へ変換\nimages = np.array(image_pixels, dtype=float)",
"_____no_output_____"
]
],
[
[
"## キーポイント予測結果の視覚化\n\n9 つのデータを視覚化します。\n\n予測されたキーポイントを取得し, `outputs` に格納します。\n各前向き処理後に, 画像と出力をそれぞれ `images_list` と `outputs_list` に追加します。\n\n最後に,`test_keypoints_plot()` を呼び出し,予測キーポイントを顔の画像上にプロットします。\n\n検証結果と比較すると、テスト結果は良好に見えます。\n",
"_____no_output_____"
]
],
[
[
"images_list, outputs_list = [], []\nfor i in range(9):\n with torch.no_grad():\n image = images[i]\n image = image.reshape(96, 96, 1)\n image = cv2.resize(image, (resize, resize))\n image = image.reshape(resize, resize, 1)\n orig_image = image.copy()\n image = image / 255.0\n image = np.transpose(image, (2, 0, 1))\n image = torch.tensor(image, dtype=torch.float)\n image = image.unsqueeze(0).to(DEVICE)\n \n # forward pass through the model\n outputs = model(image)\n # append the current original image\n images_list.append(orig_image)\n # append the current outputs\n outputs_list.append(outputs)\n \n \ntest_keypoints_plot(images_list, outputs_list, figsize=(14,14))",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
4afde2b155efd2004aa128b5b9dd97b2e347a097
| 5,911 |
ipynb
|
Jupyter Notebook
|
src/main/python/OfficalExamples/adult.ipynb
|
xixici/AlinkLearning
|
6e0da9cc011b65e405ebbe885e0a0e60af26823c
|
[
"Apache-2.0"
] | 3 |
2020-01-26T17:26:45.000Z
|
2020-10-18T13:20:26.000Z
|
src/main/python/OfficalExamples/adult.ipynb
|
0xqq/AlinkLearning
|
a087b892526cfa7369baf717efe3f5a5fca8a335
|
[
"Apache-2.0"
] | null | null | null |
src/main/python/OfficalExamples/adult.ipynb
|
0xqq/AlinkLearning
|
a087b892526cfa7369baf717efe3f5a5fca8a335
|
[
"Apache-2.0"
] | 4 |
2020-01-26T17:26:15.000Z
|
2021-11-27T13:37:54.000Z
| 33.777143 | 707 | 0.587887 |
[
[
[
"from pyalink.alink import *\nresetEnv()\nuseLocalEnv(1, config=None)",
"_____no_output_____"
]
],
[
[
"# 准备数据",
"_____no_output_____"
]
],
[
[
"schema = \"age bigint, workclass string, fnlwgt bigint, education string, \\\n education_num bigint, marital_status string, occupation string, \\\n relationship string, race string, sex string, capital_gain bigint, \\\n capital_loss bigint, hours_per_week bigint, native_country string, label string\"\n\nadult_batch = CsvSourceBatchOp() \\\n .setFilePath(\"https://alink-release.oss-cn-beijing.aliyuncs.com/data-files/adult_train.csv\") \\\n .setSchemaStr(schema)\n\nadult_stream = CsvSourceStreamOp() \\\n .setFilePath(\"https://alink-release.oss-cn-beijing.aliyuncs.com/data-files/adult_test.csv\") \\\n .setSchemaStr(schema)",
"_____no_output_____"
]
],
[
[
"# 特征建模",
"_____no_output_____"
]
],
[
[
"categoricalColNames = [\"workclass\", \"education\", \"marital_status\", \"occupation\",\n \"relationship\", \"race\", \"sex\", \"native_country\"]\nnumerialColNames = [\"age\", \"fnlwgt\", \"education_num\", \"capital_gain\",\n \"capital_loss\", \"hours_per_week\"]\nonehot = OneHotEncoder().setSelectedCols(categoricalColNames) \\\n .setOutputCol(\"output\").setReservedCols(numerialColNames + [\"label\"])\nassembler = VectorAssembler().setSelectedCols([\"output\"] + numerialColNames) \\\n .setOutputCol(\"vec\").setReservedCols([\"label\"])\npipeline = Pipeline().add(onehot).add(assembler)",
"_____no_output_____"
]
],
[
[
"# 训练+预测+评估",
"_____no_output_____"
]
],
[
[
"logistic = LogisticRegression().setVectorCol(\"vec\").setLabelCol(\"label\") \\\n .setPredictionCol(\"pred\").setPredictionDetailCol(\"detail\")\nmodel = pipeline.add(logistic).fit(adult_batch)\n\npredictBatch = model.transform(adult_batch)\n\nmetrics = EvalBinaryClassBatchOp().setLabelCol(\"label\") \\\n .setPredictionDetailCol(\"detail\").linkFrom(predictBatch).collectMetrics()",
"_____no_output_____"
]
],
[
[
"# 输出评估结果",
"_____no_output_____"
]
],
[
[
"print(\"AUC:\", metrics.getAuc())\nprint(\"KS:\", metrics.getKs())\nprint(\"PRC:\", metrics.getPrc())\nprint(\"Precision:\", metrics.getPrecision())\nprint(\"Recall:\", metrics.getRecall())\nprint(\"F1:\", metrics.getF1())\nprint(\"ConfusionMatrix:\", metrics.getConfusionMatrix())\nprint(\"LabelArray:\", metrics.getLabelArray())\nprint(\"LogLoss:\", metrics.getLogLoss())\nprint(\"TotalSamples:\", metrics.getTotalSamples())\nprint(\"ActualLabelProportion:\", metrics.getActualLabelProportion())\nprint(\"ActualLabelFrequency:\", metrics.getActualLabelFrequency())\nprint(\"Accuracy:\", metrics.getAccuracy())\nprint(\"Kappa:\", metrics.getKappa())",
"AUC: 0.9071346253140332\nKS: 0.6508855101121852\nPRC: 0.7654668375809972\nPrecision: 0.7311696264543784\nRecall: 0.609105981379926\nF1: 0.6645794197453558\nConfusionMatrix: [[4776, 1756], [3065, 22964]]\nLabelArray: ['>50K', '<=50K']\nLogLoss: 0.31880016560096547\nTotalSamples: 32561\nActualLabelProportion: [0.2408095574460244, 0.7591904425539756]\nActualLabelFrequency: [7841, 24720]\nAccuracy: 0.8519394367494856\nKappa: 0.5705912048680206\n"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
4afdfd3ffb6516c25513d3214be3fddf3a758401
| 2,399 |
ipynb
|
Jupyter Notebook
|
ch06/overfit_dropout.ipynb
|
SSLAB-SSU/deep-learning-from-scratch
|
3609360751d67085e0963ee6d7af6d49380cd965
|
[
"MIT"
] | null | null | null |
ch06/overfit_dropout.ipynb
|
SSLAB-SSU/deep-learning-from-scratch
|
3609360751d67085e0963ee6d7af6d49380cd965
|
[
"MIT"
] | null | null | null |
ch06/overfit_dropout.ipynb
|
SSLAB-SSU/deep-learning-from-scratch
|
3609360751d67085e0963ee6d7af6d49380cd965
|
[
"MIT"
] | null | null | null | 31.986667 | 120 | 0.561901 |
[
[
[
"# coding: utf-8\nimport os\nimport sys\nsys.path.append('/Users/hxxnhxx/Documents/development/deep-learning-from-scratch') # 親ディレクトリのファイルをインポートするための設定\nimport numpy as np\nimport matplotlib.pyplot as plt\nfrom dataset.mnist import load_mnist\nfrom common.multi_layer_net_extend import MultiLayerNetExtend\nfrom common.trainer import Trainer\n\n(x_train, t_train), (x_test, t_test) = load_mnist(normalize=True)\n\n# 過学習を再現するために、学習データを削減\nx_train = x_train[:300]\nt_train = t_train[:300]\n\n# Dropuoutの有無、割り合いの設定 ========================\nuse_dropout = True # Dropoutなしのときの場合はFalseに\ndropout_ratio = 0.2\n# ====================================================\n\nnetwork = MultiLayerNetExtend(input_size=784, hidden_size_list=[100, 100, 100, 100, 100, 100],\n output_size=10, use_dropout=use_dropout, dropout_ration=dropout_ratio)\ntrainer = Trainer(network, x_train, t_train, x_test, t_test,\n epochs=301, mini_batch_size=100,\n optimizer='sgd', optimizer_param={'lr': 0.01}, verbose=True)\ntrainer.train()\n\ntrain_acc_list, test_acc_list = trainer.train_acc_list, trainer.test_acc_list\n\n# グラフの描画==========\nmarkers = {'train': 'o', 'test': 's'}\nx = np.arange(len(train_acc_list))\nplt.plot(x, train_acc_list, marker='o', label='train', markevery=10)\nplt.plot(x, test_acc_list, marker='s', label='test', markevery=10)\nplt.xlabel(\"epochs\")\nplt.ylabel(\"accuracy\")\nplt.ylim(0, 1.0)\nplt.legend(loc='lower right')\nplt.show()",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code"
]
] |
4afe00ea2be2d988f9e8ca62085d1b31d13ccabb
| 87,950 |
ipynb
|
Jupyter Notebook
|
notebooks/Revision/Revisión_4_Solver.ipynb
|
czammar/MNO_finalproject
|
d3bb2707442ec64a7f36b24358d82e6bff170323
|
[
"RSA-MD"
] | null | null | null |
notebooks/Revision/Revisión_4_Solver.ipynb
|
czammar/MNO_finalproject
|
d3bb2707442ec64a7f36b24358d82e6bff170323
|
[
"RSA-MD"
] | 61 |
2020-04-25T01:09:22.000Z
|
2020-05-29T00:18:46.000Z
|
notebooks/Revision/Revisión_4_Solver.ipynb
|
czammar/MNO_finalproject
|
d3bb2707442ec64a7f36b24358d82e6bff170323
|
[
"RSA-MD"
] | 4 |
2020-05-01T19:24:45.000Z
|
2021-01-23T01:28:44.000Z
| 31.5346 | 1,812 | 0.385139 |
[
[
[
"# Revisión Solver de Markowitz\n\n\n\n\n\n**Código revisado**",
"_____no_output_____"
],
[
"## Librerias necesarias",
"_____no_output_____"
]
],
[
[
"#Librerias necesarias\n!curl https://colab.chainer.org/install | sh -",
" % Total % Received % Xferd Average Speed Time Time Time Current\n Dload Upload Total Spent Left Speed\n\r 0 0 0 0 0 0 0 0 --:--:-- --:--:-- --:--:-- 0\r100 1580 100 1580 0 0 22571 0 --:--:-- --:--:-- --:--:-- 22571\n+ apt -y -q install cuda-libraries-dev-10-0\nReading package lists...\nBuilding dependency tree...\nReading state information...\ncuda-libraries-dev-10-0 is already the newest version (10.0.130-1).\n0 upgraded, 0 newly installed, 0 to remove and 29 not upgraded.\n+ pip install -q cupy-cuda100 chainer \n+ set +ex\nInstallation succeeded!\n"
],
[
"\nimport cupy as cp\nimport numpy as np\nimport pandas as pd\nimport fix_yahoo_finance as yf\nimport datetime\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nimport time",
"/usr/local/lib/python3.6/dist-packages/statsmodels/tools/_testing.py:19: FutureWarning: pandas.util.testing is deprecated. Use the functions in the public API at pandas.testing instead.\n import pandas.util.testing as tm\n"
]
],
[
[
"A continuación creamos un arreglo con las acciones que utilizaremos.",
"_____no_output_____"
]
],
[
[
"stocks = ['COP','AMT','LIN','LMT','AMZN','WMT','JNJ','VTI','MSFT','GOOG','XOM','CCI','BHP.AX','UNP',\n'BABA','NSRGY','RHHBY','VOO','AAPL','FB','CVX','PLD','RIO.L','HON','HD','PG','UNH','BRK-A','V','0700.HK',\n'RDSA.AS','0688.HK','AI.PA','RTX','MC.PA','KO','PFE','JPM','005930.KS','VZ','RELIANCE.NS','DLR','2010.SR',\n'UPS','7203.T','PEP','MRK','1398.HK','MA','T']",
"_____no_output_____"
],
[
"def extraer_datos_yahoo(stocks):\n '''\n Funcion para extraer datos de los portafolios de yahoo finance de 2015-01-01 a 2020-04-30\n '''\n df_c = yf.download(stocks, start='2015-01-01', end='2020-04-30').Close\n base = df_c['AAPL'].dropna().to_frame()\n for i in range(0,50):\n base = base.join(df_c.iloc[:,i].to_frame(), lsuffix='_caller', rsuffix='_other')\n base = base.drop(columns=['AAPL_caller'])\n base = base.rename(columns={\"AAPL_other\": \"AAPL\"})\n base = base.fillna(method='ffill')\n base = base.fillna(method='bfill')\n return base",
"_____no_output_____"
],
[
"datos = extraer_datos_yahoo(stocks)",
"[*********************100%***********************] 50 of 50 downloaded\n"
]
],
[
[
"Revisamos los filas de los datos",
"_____no_output_____"
]
],
[
[
"datos",
"_____no_output_____"
]
],
[
[
"## Funciones auxiliares\n",
"_____no_output_____"
]
],
[
[
"def calcular_rendimiento_vector(x):\n \"\"\"\n Función para calcular el rendimiento esperado\n\n params:\n x vector de precios\n \n return:\n r_est rendimiento esperado diario\n \"\"\"\n\n # Definimos precios iniciales y finales como arreglo alojado en la gpu\n x_o = cp.asarray(x)\n x_f = x_o[1:]\n\n # Calculamos los rendimientos diarios\n r = cp.log(x_f/x_o[:-1])\n\n return r",
"_____no_output_____"
],
[
"def calcular_rendimiento(X):\n \"\"\"\n Función para calcular el rendimiento esperado para un conjunto de acciones\n\n params:\n X matriz mxn de precios, donde:\n m es el número de observaciones y\n n el número de acciones\n \n return:\n r_est rvector de rendimientos esperados\n \"\"\"\n m,n = X.shape\n r_est = cp.zeros(n)\n X = cp.asarray(X)\n\n for i in range(n):\n r_est[i] = calcular_rendimiento_vector(X[:,i]).mean()\n\n return 264*r_est",
"_____no_output_____"
],
[
"def calcular_varianza(X):\n\n \"\"\"\n Función para calcular el la matriz de varianzas y covarianzas para un conjunto de acciones\n\n params:\n X matriz mxn de precios, donde:\n m es el número de observaciones y\n n el número de acciones\n \n return:\n S matriz de varianzas y covarianzas\n \"\"\"\n m,n=X.shape\n X = cp.asarray(X)\n\n X_m = cp.zeros((m-1,n))\n\n for i in range(n):\n X_m[:,i] = calcular_rendimiento_vector(X[:,i]) - calcular_rendimiento_vector(X[:,i]).mean()\n\n S = (cp.transpose(X_m)@X_m)/(m-2)\n\n return S",
"_____no_output_____"
]
],
[
[
"## Solución del modelo de Markowitz",
"_____no_output_____"
]
],
[
[
"def formar_vectores(mu, Sigma):\n '''\n Calcula las cantidades u = \\Sigma^{-1} \\mu y v := \\Sigma^{-1} \\cdot 1 del problema de Markowitz\n\n Args:\n mu (cupy array, vector): valores medios esperados de activos (dimension n)\n Sigma (cupy array, matriz): matriz de covarianzas asociada a activos (dimension n x n)\n\n Return:\n u (cupy array, escalar): vector dado por \\cdot Sigma^-1 \\cdot mu (dimension n)\n v (cupy array, escalar): vector dado por Sigma^-1 \\cdot 1 (dimension n)\n '''\n\n # Vector auxiliar con entradas igual a 1\n n = Sigma.shape[0]\n ones_vector = cp.ones(n)\n\n # Formamos vector \\cdot Sigma^-1 mu y Sigm^-1 1\n # Nota: \n # 1) u= Sigma^-1 \\cdot mu se obtiene resolviendo Sigma u = mu\n # 2) v= Sigma^-1 \\cdot 1 se obtiene resolviendo Sigma v = 1\n\n # Obtiene vectores de interes\n u = cp.linalg.solve(Sigma, mu)\n u = u.transpose() # correcion de expresion de array\n v = cp.linalg.solve(Sigma, ones_vector)\n\n return u , v",
"_____no_output_____"
],
[
"def formar_abc(mu, Sigma):\n '''\n Calcula las cantidades A, B y C del diagrama de flujo del problema de Markowitz\n\n Args:\n mu (cupy array, vector): valores medios esperados de activos (dimension n)\n Sigma (cupy array, matriz): matriz de covarianzas asociada a activos (dimension n x n)\n\n Return:\n A (cupy array, escalar): escalar dado por mu^t \\cdot Sigma^-1 \\cdot mu\n B (cupy array, escalar): escalar dado por 1^t \\cdot Sigma^-1 \\cdot 1\n C (cupy array, escalar): escalar dado por 1^t \\cdot Sigma^-1 \\cdot mu\n '''\n\n # Vector auxiliar con entradas igual a 1\n n = Sigma.shape[0]\n ones_vector = cp.ones(n)\n\n # Formamos vector \\cdot Sigma^-1 mu y Sigm^-1 1\n # Nota: \n # 1) u= Sigma^-1 \\cdot mu se obtiene resolviendo Sigma u = mu\n # 2) v= Sigma^-1 \\cdot 1 se obtiene resolviendo Sigma v = 1\n\n u, v = formar_vectores(mu, Sigma)\n\n # Obtiene escalares de interes\n A = mu.transpose()@u\n B = ones_vector.transpose()@v\n C = ones_vector.transpose()@u\n\n return A, B, C",
"_____no_output_____"
],
[
"def delta(A,B,C):\n '''\n Calcula las cantidad Delta = AB-C^2 del diagrama de flujo del problema de Markowitz\n\n Args:\n A (cupy array, escalar): escalar dado por mu^t \\cdot Sigma^-1 \\cdot mu\n B (cupy array, escalar): escalar dado por 1^t \\cdot Sigma^-1 \\cdot 1\n C (cupy array, escalar): escalar dado por 1^t \\cdot Sigma^-1 \\cdot mu\n\n Return:\n Delta (cupy array, escalar): escalar dado \\mu^t \\cdot \\Sigma^{-1} \\cdot \\mu\n '''\n Delta = A*B-C**2\n\n return Delta",
"_____no_output_____"
],
[
"def formar_omegas(r, mu, Sigma):\n '''\n Calcula las cantidades w_o y w_ del problema de Markowitz\n\n Args:\n mu (cupy array, vector): valores medios esperados de activos (dimension n)\n Sigma (cupy array, matriz): matriz de covarianzas asociada a activos (dimension n x n)\n\n Return:\n w_0 (cupy array, matriz): matriz dada por \n w_0 = \\frac{1}{\\Delta} (B \\Sigma^{-1} \\hat{\\mu}- C\\Sigma^{-1} 1) \n w_1 (cupy array, vector): vector dado por \n w_1 = \\frac{1}{\\Delta} (C \\Sigma^{-1} \\hat{\\mu}- A\\Sigma^{-1} 1)\n '''\n # Obtenemos u = Sigma^{-1} \\hat{\\mu}, v = \\Sigma^{-1} 1\n u, v = formar_vectores(mu, Sigma)\n # Escalares relevantes\n A, B, C = formar_abc(mu, Sigma)\n Delta = delta(A,B,C)\n # Formamos w_0 y w_1\n w_0 = (1/Delta)*(r*B-C)\n w_1 = (1/Delta)*(A-C*r)\n\n return w_0, w_1\n",
"_____no_output_____"
],
[
"def markowitz(r, mu, Sigma):\n '''\n Calcula las cantidades w_o y w_ del problema de Markowitz\n\n Args:\n mu (cupy array, vector): valores medios esperados de activos (dimension n)\n Sigma (cupy array, matriz): matriz de covarianzas asociada a activos (dimension n x n)\n\n Return:\n w_0 (cupy array, matriz): matriz dada por \n w_0 = \\frac{1}{\\Delta} (B \\Sigma^{-1} \\hat{\\mu}- C\\Sigma^{-1} 1) \n w_1 (cupy array, vector): vector dado por \n w_1 = \\frac{1}{\\Delta} (C \\Sigma^{-1} \\hat{\\mu}- A\\Sigma^{-1} 1)\n '''\n # Obtenemos u = Sigma^{-1} \\hat{\\mu}, v = \\Sigma^{-1} 1\n u, v = formar_vectores(mu, Sigma)\n\n # Formamos w_0 y w_1\n w_0, w_1 = formar_omegas(r, mu, Sigma)\n\n return w_0*u+w_1*v\n",
"_____no_output_____"
]
],
[
[
"## Revisón",
"_____no_output_____"
],
[
"**1. Documentación**\n\nLa Documentación expresa de manera clara, consica y breve lo que hace el código. De igual forma se explica de manera clara y concisa los argumentos de entrada y salida. La documentación es completa.\n\n**2. Cumplimiento de objetivos del código**\n\nLa función cumple con el objetivo devolviendo una matriz y un vector de pesos w.\n\n**3. Test**\n\nObjetivo: verificar el desempeño del código con diferentes números de activos y rendimientos, y verificar que los pesos sumen 1.\n",
"_____no_output_____"
]
],
[
[
"datos_1 = cp.random.uniform(1,1000,10**3).reshape(10**2,10)\nsigma_1 = calcular_varianza(datos_1)\nmu_1 = calcular_rendimiento(datos_1)\nr_11 = cp.mean(mu_1)**2\nr_12 = max(mu_1)",
"_____no_output_____"
],
[
"w_11 = markowitz(r_11,mu_1,sigma_1)\nw_12 = markowitz(r_12,mu_1,sigma_1)",
"_____no_output_____"
]
],
[
[
"Verificamos que la suma de las $\\sum w's=1$ y el error absoluto",
"_____no_output_____"
]
],
[
[
"sum(w_11)",
"_____no_output_____"
],
[
"abs((sum(w_11)-1)/sum(w_11))",
"_____no_output_____"
],
[
"sum(w_12)",
"_____no_output_____"
],
[
"abs((sum(w_12)-1)/sum(w_12))",
"_____no_output_____"
]
],
[
[
"Verificamos el error absoluto de $w^{t}\\mu = r$\n",
"_____no_output_____"
]
],
[
[
"error_11 = abs(r_11 - w_11@mu_1)/r_11\nprint(error_11)",
"6.155967883235464e-16\n"
],
[
"error_12 = abs(r_12 - w_12@mu_1)/r_12\nprint(error_12)",
"1.2309415527669263e-16\n"
],
[
"w_11@mu_1",
"_____no_output_____"
],
[
"r_11",
"_____no_output_____"
],
[
"w_12@mu_1",
"_____no_output_____"
],
[
"r_12",
"_____no_output_____"
]
],
[
[
"Ahora probamos con una matriz de $10^4 \\times 10^2$",
"_____no_output_____"
]
],
[
[
"datos_2 = cp.random.uniform(1,1000,10**6).reshape(10**4,10**2)\nsigma_2 = calcular_varianza(datos_2)\nmu_2 = calcular_rendimiento(datos_2)\nr_21 = cp.mean(mu_2)**2\nr_22 = max(mu_2)",
"_____no_output_____"
],
[
"w_21 = markowitz(r_21,mu_2,sigma_2)\nw_22 = markowitz(r_22,mu_2,sigma_2)",
"_____no_output_____"
]
],
[
[
"Verificamos que la suma de las $\\sum w's=1$ y el error absoluto",
"_____no_output_____"
]
],
[
[
"sum(w_21)",
"_____no_output_____"
],
[
"abs((sum(w_21)-1)/sum(w_21))",
"_____no_output_____"
],
[
"sum(w_22)",
"_____no_output_____"
],
[
"abs((sum(w_22)-1)/sum(w_22))",
"_____no_output_____"
]
],
[
[
"Verificamos el error absoluto de $w^{t}\\mu = r$",
"_____no_output_____"
]
],
[
[
"w_21@mu_2",
"_____no_output_____"
],
[
"r_21",
"_____no_output_____"
],
[
"error_21 = abs(r_21 - w_21@mu_2)/r_21\nprint(error_21)",
"1.304348104457549e-13\n"
],
[
"error_22 = abs(r_22 - w_22@mu_2)/r_22\nprint(error_22)",
"3.770752517849849e-16\n"
]
],
[
[
"Ahora probamos con una matriz de $10^5 \\times 10^3$\n\n",
"_____no_output_____"
]
],
[
[
"datos_3 = cp.random.uniform(1,1000,10**8).reshape(10**5,10**3)\nsigma_3 = calcular_varianza(datos_3)\nmu_3 = calcular_rendimiento(datos_3)\nr_31 = cp.mean(mu_3)**2\nr_32 = max(mu_3)",
"_____no_output_____"
],
[
"w_31 = markowitz(r_31,mu_3,sigma_3)\nw_32 = markowitz(r_32,mu_3,sigma_3)",
"_____no_output_____"
]
],
[
[
"Verificamos que la suma de las $\\sum w's=1$ y el error absoluto",
"_____no_output_____"
]
],
[
[
"sum(w_31)",
"_____no_output_____"
],
[
"abs(sum(w_31)-1)/sum(w_31)",
"_____no_output_____"
],
[
"sum(w_32)",
"_____no_output_____"
]
],
[
[
"Verificamos el error absoluto de $w^{t}\\mu = r$",
"_____no_output_____"
]
],
[
[
"error_31 = abs(r_31 - w_31@mu_3)/r_31\nprint(error_31)",
"5.972565684253623e-12\n"
],
[
"error_32 = abs(r_32 - w_32@mu_3)/r_32\nprint(error_32)",
"1.2765462539881994e-16\n"
]
],
[
[
"Ahora probamos con una matriz de $10^4 \\times 10^4$",
"_____no_output_____"
]
],
[
[
"datos_4 = cp.random.uniform(100,1000,10**8).reshape(10**4,10**4)\nsigma_4 = calcular_varianza(datos)\nmu_4 = calcular_rendimiento(datos)\nr_41 = cp.mean(mu_4)**2\nr_42 = max(mu_4)",
"_____no_output_____"
],
[
"w_41 = markowitz(r_41,mu_4,sigma_4)\nw_42 = markowitz(r_42,mu_4,sigma_4)",
"_____no_output_____"
]
],
[
[
"Verificamos que ∑w′s=1 y el error absoluto",
"_____no_output_____"
]
],
[
[
"sum(w_41)",
"_____no_output_____"
],
[
"abs(sum(w_41)-1)/sum(w_41)",
"_____no_output_____"
],
[
"sum(w_42)",
"_____no_output_____"
],
[
"abs(sum(w_42)-1)/sum(w_42)",
"_____no_output_____"
]
],
[
[
"Verificamos el error absoluto de $w^{t}\\mu = r$",
"_____no_output_____"
]
],
[
[
"error_41 = abs(r_41 - w_41@mu_4)/r_41\nprint(error_41)",
"6.689301079114241e-14\n"
],
[
"error_42 = abs(r_42 - w_42@mu_4)/r_42\nprint(error_42)",
"1.2421353758545672e-15\n"
],
[
"r_41",
"_____no_output_____"
],
[
"w_41@mu_4",
"_____no_output_____"
],
[
"r_42",
"_____no_output_____"
],
[
"w_42@mu_4",
"_____no_output_____"
]
],
[
[
"Ahora probamos con una matriz de $10^5 \\times 10^4$",
"_____no_output_____"
]
],
[
[
"datos_5 = cp.random.uniform(1,1000,10**9).reshape(10**5,10**4)\nsigma_5 = calcular_varianza(datos_5)\nmu_5 = calcular_rendimiento(datos_5)\nr_51 = cp.mean(mu_5)**2\nr_52 = max(mu_5)",
"_____no_output_____"
]
],
[
[
"EL sistema no soporta generar matrices con 10**9 elementos.\n\nProbamos con las acciones\n\n",
"_____no_output_____"
]
],
[
[
"sigma = calcular_varianza(datos)\nmu = calcular_rendimiento(datos)\nr1 = cp.mean(mu)**2\nr2 = max(mu)",
"_____no_output_____"
],
[
"w_01 = markowitz(r1,mu,sigma)\nw_02 = markowitz(r2,mu,sigma)",
"_____no_output_____"
]
],
[
[
"Verificamos que ∑w′s=1 y el error absoluto",
"_____no_output_____"
]
],
[
[
"sum(w_01)",
"_____no_output_____"
],
[
"abs(sum(w_01)-1)/sum(w_01)",
"_____no_output_____"
],
[
"sum(w_02)",
"_____no_output_____"
],
[
"abs(sum(w_02)-1)/sum(w_02)",
"_____no_output_____"
]
],
[
[
"Verificamos el error absoluto de $w^{t}\\mu = r$",
"_____no_output_____"
]
],
[
[
"error_01 = abs(r1 - w_01@mu)/r1\nprint(error_01)",
"6.689301079114241e-14\n"
],
[
"error_02 = abs(r2 - w_02@mu)/r2\nprint(error_02)",
"1.2421353758545672e-15\n"
]
],
[
[
"# Hallazgos\n\nEl código funciona de manera correcta para distintos tamaños de matrices y para el protafolio de acciones.\n\nLas funciones generan unos valores con una exactitud de hasta 16 dígitos correctos. \n",
"_____no_output_____"
],
[
"**Nota**: Los tests fueron realizados en google colaboratory con GPU como entorno de ejecución",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
]
] |
4afe132cd8035e956c9502c2e0d204662b0f007f
| 245,873 |
ipynb
|
Jupyter Notebook
|
docs/examples/the-art-of-using-pipelines.ipynb
|
online-ml/creme
|
60872844e6052b5ef20e4075aea30f9031377136
|
[
"BSD-3-Clause"
] | 1,105 |
2019-01-24T15:15:30.000Z
|
2020-11-10T18:27:00.000Z
|
docs/examples/the-art-of-using-pipelines.ipynb
|
online-ml/creme
|
60872844e6052b5ef20e4075aea30f9031377136
|
[
"BSD-3-Clause"
] | 328 |
2019-01-25T13:48:43.000Z
|
2020-11-11T11:41:44.000Z
|
docs/examples/the-art-of-using-pipelines.ipynb
|
online-ml/creme
|
60872844e6052b5ef20e4075aea30f9031377136
|
[
"BSD-3-Clause"
] | 150 |
2019-01-29T19:05:21.000Z
|
2020-11-11T11:50:14.000Z
| 75.14456 | 770 | 0.463426 |
[
[
[
"# The art of using pipelines",
"_____no_output_____"
],
[
"Pipelines are a natural way to think about a machine learning system. Indeed with some practice a data scientist can visualise data \"flowing\" through a series of steps. The input is typically some raw data which has to be processed in some manner. The goal is to represent the data in such a way that is can be ingested by a machine learning algorithm. Along the way some steps will extract features, while others will normalize the data and remove undesirable elements. Pipelines are simple, and yet they are a powerful way of designing sophisticated machine learning systems.\n\nBoth [scikit-learn](https://stackoverflow.com/questions/33091376/python-what-is-exactly-sklearn-pipeline-pipeline) and [pandas](https://tomaugspurger.github.io/method-chaining) make it possible to use pipelines. However it's quite rare to see pipelines being used in practice (at least on Kaggle). Sometimes you get to see people using scikit-learn's `pipeline` module, however the `pipe` method from `pandas` is sadly underappreciated. A big reason why pipelines are not given much love is that it's easier to think of batch learning in terms of a script or a notebook. Indeed many people doing data science seem to prefer a procedural style to a declarative style. Moreover in practice pipelines can be a bit rigid if one wishes to do non-orthodox operations.\n\nAlthough pipelines may be a bit of an odd fit for batch learning, they make complete sense when they are used for online learning. Indeed the UNIX philosophy has advocated the use of pipelines for data processing for many decades. If you can visualise data as a stream of observations then using pipelines should make a lot of sense to you. We'll attempt to convince you by writing a machine learning algorithm in a procedural way and then converting it to a declarative pipeline in small steps. Hopefully by the end you'll be convinced, or not!\n\nIn this notebook we'll manipulate data from the [Kaggle Recruit Restaurants Visitor Forecasting competition](https://www.kaggle.com/c/recruit-restaurant-visitor-forecasting). The data is directly available through `river`'s `datasets` module.",
"_____no_output_____"
]
],
[
[
"from pprint import pprint\nfrom river import datasets\n\nfor x, y in datasets.Restaurants():\n pprint(x)\n pprint(y)\n break",
"Downloading https://maxhalford.github.io/files/datasets/kaggle_recruit_restaurants.zip (4.28 MB)\nUncompressing into /Users/max.halford/river_data/Restaurants\n{'area_name': 'Tōkyō-to Nerima-ku Toyotamakita',\n 'date': datetime.datetime(2016, 1, 1, 0, 0),\n 'genre_name': 'Izakaya',\n 'is_holiday': True,\n 'latitude': 35.7356234,\n 'longitude': 139.6516577,\n 'store_id': 'air_04341b588bde96cd'}\n10\n"
]
],
[
[
"We'll start by building and running a model using a procedural coding style. The performance of the model doesn't matter, we're simply interested in the design of the model.",
"_____no_output_____"
]
],
[
[
"from river import feature_extraction\nfrom river import linear_model\nfrom river import metrics\nfrom river import preprocessing\nfrom river import stats\n\nmeans = (\n feature_extraction.TargetAgg(by='store_id', how=stats.RollingMean(7)),\n feature_extraction.TargetAgg(by='store_id', how=stats.RollingMean(14)),\n feature_extraction.TargetAgg(by='store_id', how=stats.RollingMean(21))\n)\n\nscaler = preprocessing.StandardScaler()\nlin_reg = linear_model.LinearRegression()\nmetric = metrics.MAE()\n\nfor x, y in datasets.Restaurants():\n \n # Derive date features\n x['weekday'] = x['date'].weekday()\n x['is_weekend'] = x['date'].weekday() in (5, 6)\n \n # Process the rolling means of the target \n for mean in means:\n x = {**x, **mean.transform_one(x)}\n mean.learn_one(x, y)\n \n # Remove the key/value pairs that aren't features\n for key in ['store_id', 'date', 'genre_name', 'area_name', 'latitude', 'longitude']:\n x.pop(key)\n \n # Rescale the data\n x = scaler.learn_one(x).transform_one(x)\n \n # Fit the linear regression\n y_pred = lin_reg.predict_one(x)\n lin_reg.learn_one(x, y)\n \n # Update the metric using the out-of-fold prediction\n metric.update(y, y_pred)\n \nprint(metric)",
"MAE: 8.465114\n"
]
],
[
[
"We're not using many features. We can print the last `x` to get an idea of the features (don't forget they've been scaled!)",
"_____no_output_____"
]
],
[
[
"pprint(x)",
"{'is_holiday': -0.23103573677646685,\n 'is_weekend': 1.6249280076334165,\n 'weekday': 1.0292832579142892,\n 'y_rollingmean_14_by_store_id': -1.4125913815779154,\n 'y_rollingmean_21_by_store_id': -1.3980979075298519,\n 'y_rollingmean_7_by_store_id': -1.3502314499809096}\n"
]
],
[
[
"The above chunk of code is quite explicit but it's a bit verbose. The whole point of libraries such as `river` is to make life easier for users. Moreover there's too much space for users to mess up the order in which things are done, which increases the chance of there being target leakage. We'll now rewrite our model in a declarative fashion using a pipeline *à la sklearn*. ",
"_____no_output_____"
]
],
[
[
"from river import compose\n\n\ndef get_date_features(x):\n weekday = x['date'].weekday()\n return {'weekday': weekday, 'is_weekend': weekday in (5, 6)}\n\n\nmodel = compose.Pipeline(\n ('features', compose.TransformerUnion(\n ('date_features', compose.FuncTransformer(get_date_features)),\n ('last_7_mean', feature_extraction.TargetAgg(by='store_id', how=stats.RollingMean(7))),\n ('last_14_mean', feature_extraction.TargetAgg(by='store_id', how=stats.RollingMean(14))),\n ('last_21_mean', feature_extraction.TargetAgg(by='store_id', how=stats.RollingMean(21)))\n )),\n ('drop_non_features', compose.Discard('store_id', 'date', 'genre_name', 'area_name', 'latitude', 'longitude')),\n ('scale', preprocessing.StandardScaler()),\n ('lin_reg', linear_model.LinearRegression())\n)\n\nmetric = metrics.MAE()\n\nfor x, y in datasets.Restaurants():\n \n # Make a prediction without using the target\n y_pred = model.predict_one(x)\n \n # Update the model using the target\n model.learn_one(x, y)\n \n # Update the metric using the out-of-fold prediction\n metric.update(y, y_pred)\n \nprint(metric)",
"MAE: 8.38533\n"
]
],
[
[
"We use a `Pipeline` to arrange each step in a sequential order. A `TransformerUnion` is used to merge multiple feature extractors into a single transformer. The `for` loop is now much shorter and is thus easier to grok: we get the out-of-fold prediction, we fit the model, and finally we update the metric. This way of evaluating a model is typical of online learning, and so we put it wrapped it inside a function called `progressive_val_score` part of the `evaluate` module. We can use it to replace the `for` loop.",
"_____no_output_____"
]
],
[
[
"from river import evaluate\n\nmodel = compose.Pipeline(\n ('features', compose.TransformerUnion(\n ('date_features', compose.FuncTransformer(get_date_features)),\n ('last_7_mean', feature_extraction.TargetAgg(by='store_id', how=stats.RollingMean(7))),\n ('last_14_mean', feature_extraction.TargetAgg(by='store_id', how=stats.RollingMean(14))),\n ('last_21_mean', feature_extraction.TargetAgg(by='store_id', how=stats.RollingMean(21)))\n )),\n ('drop_non_features', compose.Discard('store_id', 'date', 'genre_name', 'area_name', 'latitude', 'longitude')),\n ('scale', preprocessing.StandardScaler()),\n ('lin_reg', linear_model.LinearRegression())\n)\n\nevaluate.progressive_val_score(dataset=datasets.Restaurants(), model=model, metric=metrics.MAE())",
"_____no_output_____"
]
],
[
[
"Notice that you couldn't have used the `progressive_val_score` method if you wrote the model in a procedural manner.\n\nOur code is getting shorter, but it's still a bit difficult on the eyes. Indeed there is a lot of boilerplate code associated with pipelines that can get tedious to write. However `river` has some special tricks up it's sleeve to save you from a lot of pain.\n\nThe first trick is that the name of each step in the pipeline can be omitted. If no name is given for a step then `river` automatically infers one.",
"_____no_output_____"
]
],
[
[
"model = compose.Pipeline(\n compose.TransformerUnion(\n compose.FuncTransformer(get_date_features),\n feature_extraction.TargetAgg(by='store_id', how=stats.RollingMean(7)),\n feature_extraction.TargetAgg(by='store_id', how=stats.RollingMean(14)),\n feature_extraction.TargetAgg(by='store_id', how=stats.RollingMean(21))\n ),\n compose.Discard('store_id', 'date', 'genre_name', 'area_name', 'latitude', 'longitude'),\n preprocessing.StandardScaler(),\n linear_model.LinearRegression()\n)\n\nevaluate.progressive_val_score(datasets.Restaurants(), model, metrics.MAE())",
"_____no_output_____"
]
],
[
[
"Under the hood a `Pipeline` inherits from `collections.OrderedDict`. Indeed this makes sense because if you think about it a `Pipeline` is simply a sequence of steps where each step has a name. The reason we mention this is because it means you can manipulate a `Pipeline` the same way you would manipulate an ordinary `dict`. For instance we can print the name of each step by using the `keys` method.",
"_____no_output_____"
]
],
[
[
"for name in model.steps:\n print(name)",
"TransformerUnion\nDiscard\nStandardScaler\nLinearRegression\n"
]
],
[
[
"The first step is a `FeatureUnion` and it's string representation contains the string representation of each of it's elements. Not having to write names saves up some time and space and is certainly less tedious.\n\nThe next trick is that we can use mathematical operators to compose our pipeline. For example we can use the `+` operator to merge `Transformer`s into a `TransformerUnion`. ",
"_____no_output_____"
]
],
[
[
"model = compose.Pipeline(\n compose.FuncTransformer(get_date_features) + \\\n feature_extraction.TargetAgg(by='store_id', how=stats.RollingMean(7)) + \\\n feature_extraction.TargetAgg(by='store_id', how=stats.RollingMean(14)) + \\\n feature_extraction.TargetAgg(by='store_id', how=stats.RollingMean(21)),\n\n compose.Discard('store_id', 'date', 'genre_name', 'area_name', 'latitude', 'longitude'),\n preprocessing.StandardScaler(),\n linear_model.LinearRegression()\n)\n\nevaluate.progressive_val_score(datasets.Restaurants(), model, metrics.MAE())",
"_____no_output_____"
]
],
[
[
"Likewhise we can use the `|` operator to assemble steps into a `Pipeline`. ",
"_____no_output_____"
]
],
[
[
"model = (\n compose.FuncTransformer(get_date_features) +\n feature_extraction.TargetAgg(by='store_id', how=stats.RollingMean(7)) +\n feature_extraction.TargetAgg(by='store_id', how=stats.RollingMean(14)) +\n feature_extraction.TargetAgg(by='store_id', how=stats.RollingMean(21))\n)\n\nto_discard = ['store_id', 'date', 'genre_name', 'area_name', 'latitude', 'longitude']\n\nmodel = model | compose.Discard(*to_discard) | preprocessing.StandardScaler()\n\nmodel |= linear_model.LinearRegression()\n\nevaluate.progressive_val_score(datasets.Restaurants(), model, metrics.MAE())",
"_____no_output_____"
]
],
[
[
"Hopefully you'll agree that this is a powerful way to express machine learning pipelines. For some people this should be quite remeniscent of the UNIX pipe operator. One final trick we want to mention is that functions are automatically wrapped with a `FuncTransformer`, which can be quite handy.",
"_____no_output_____"
]
],
[
[
"model = get_date_features\n\nfor n in [7, 14, 21]:\n model += feature_extraction.TargetAgg(by='store_id', how=stats.RollingMean(n))\n\nmodel |= compose.Discard(*to_discard)\nmodel |= preprocessing.StandardScaler()\nmodel |= linear_model.LinearRegression()\n\nevaluate.progressive_val_score(datasets.Restaurants(), model, metrics.MAE())",
"_____no_output_____"
]
],
[
[
"Naturally some may prefer the procedural style we first used because they find it easier to work with. It all depends on your style and you should use what you feel comfortable with. However we encourage you to use operators because we believe that this will increase the readability of your code, which is very important. To each their own!\n\nBefore finishing we can take an interactive look at our pipeline.",
"_____no_output_____"
]
],
[
[
"model",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
4afe16ef2eb50d7e0a314298973c62afc5c2ea99
| 85,401 |
ipynb
|
Jupyter Notebook
|
Assignments/Assignment_1/Q3/Q3_line_v3.ipynb
|
Kaustubh1Verma/CS671_Deep-Learning_2019
|
062002a1369dc962feb52d3c9561a3f1153e0f84
|
[
"MIT"
] | null | null | null |
Assignments/Assignment_1/Q3/Q3_line_v3.ipynb
|
Kaustubh1Verma/CS671_Deep-Learning_2019
|
062002a1369dc962feb52d3c9561a3f1153e0f84
|
[
"MIT"
] | null | null | null |
Assignments/Assignment_1/Q3/Q3_line_v3.ipynb
|
Kaustubh1Verma/CS671_Deep-Learning_2019
|
062002a1369dc962feb52d3c9561a3f1153e0f84
|
[
"MIT"
] | 1 |
2019-06-12T14:02:33.000Z
|
2019-06-12T14:02:33.000Z
| 188.10793 | 42,944 | 0.893221 |
[
[
[
"import tensorflow as tf\nimport numpy as np",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"# Q3 API\nfrom layers import dense ",
"_____no_output_____"
],
[
"dataset = np.load('a1_dataset.npz')\ndataset.files",
"_____no_output_____"
],
[
"# 80-10-10 split for train, validate and test images\ntrain_till = int(.8 * dataset['x'].shape[0])\nvalidate_till = int(.9 * dataset['x'].shape[0])\n\nprint(train_till, validate_till-train_till, dataset['x'].shape[0]-validate_till)\n\n# training dataset\nx_train = dataset['x'][:train_till]/255\ny_train = np.eye(96)[dataset['y'][:train_till]]\n\n# validation dataset\nx_val = dataset['x'][train_till:validate_till]/255\ny_val = np.eye(96)[dataset['y'][train_till:validate_till]]\n\n# testing dataset\nx_test = dataset['x'][validate_till:]/255\ny_test = np.eye(96)[dataset['y'][validate_till:]]",
"76800 9600 9600\n"
],
[
"n_classes = 96\nn_features = 2352\nbatch_size = 50\nepochs = 50\nlearning_rate = 0.1",
"_____no_output_____"
],
[
"# input\nx_p = tf.placeholder(tf.float32, [None, n_features])\n# output\ny_p = tf.placeholder(tf.float32, [None, n_classes])",
"_____no_output_____"
],
[
"# define architecture\nn_l1 = 256\nn_l2 = 128\n\n# set up layers\nhidden1 = dense(x=x_p, in_length=n_features, neurons=n_l1, activation=tf.nn.relu, layer_name='Layer_1')\nhidden2 = dense(x=hidden1, in_length=n_l1, neurons=n_l2, activation=tf.nn.relu, layer_name='Layer_2')\noutput = dense(x=hidden1, in_length=n_l1, neurons=n_classes, activation=tf.nn.softmax, layer_name='Layer_Output')\ny_clipped = tf.clip_by_value(output, 1e-10, 0.9999999)\ncross_entropy = -tf.reduce_mean(tf.reduce_sum(y_p * tf.log(y_clipped)+ (1 - y_p) * tf.log(1 - y_clipped), axis=1))\n\noptimiser = tf.train.GradientDescentOptimizer(learning_rate=learning_rate).minimize(cross_entropy)",
"_____no_output_____"
],
[
"prediction_vector = tf.argmax(y_p, 1)\noutput_vector = tf.argmax(output, 1)",
"_____no_output_____"
],
[
"acc, acc_op = tf.metrics.accuracy(prediction_vector, output_vector)",
"_____no_output_____"
],
[
"conmat = tf.confusion_matrix(prediction_vector, output_vector)",
"_____no_output_____"
],
[
"with tf.Session() as sess:\n sess.run(tf.global_variables_initializer())\n sess.run(tf.local_variables_initializer())\n avg_loss = []\n validate_accuracy = []\n total_batches = x_train.shape[0] // batch_size\n \n # Training\n for e in range(epochs):\n avg_loss.append(0.0)\n for b in range(total_batches):\n start = b*batch_size\n end = (b+1)*batch_size\n batch = sess.run([optimiser, cross_entropy], \n feed_dict={x_p: x_train[start:end], y_p: y_train[start:end]})\n avg_loss[e] += batch[1] / total_batches\n # validation\n accuracy = sess.run(acc_op,\n feed_dict={x_p: x_val, y_p: y_val})\n validate_accuracy.append(accuracy)\n print(\"Epoch:\",\"{:2d}\".format(e+1), \"train_loss =\", \"{:.4f}\".format(avg_loss[e]), \"validate_accuracy =\", \"{:.4f}\".format(validate_accuracy[e]))\n \n # Testing\n test_accuracy, confusion_mat = sess.run([acc_op, conmat],\n feed_dict={x_p:x_test, y_p:y_test})\n \n print('Testing Accuracy:', test_accuracy)\n print('Confusion Matrix:', confusion_mat)\n tf.io.write_graph(sess.graph_def, 'graphs/', 'line-v3.pbtxt')",
"Epoch: 1 train_loss = 32.8415 validate_accuracy = 0.0497\nEpoch: 2 train_loss = 30.6241 validate_accuracy = 0.0685\nEpoch: 3 train_loss = 28.1706 validate_accuracy = 0.0830\nEpoch: 4 train_loss = 25.8823 validate_accuracy = 0.0996\nEpoch: 5 train_loss = 23.5630 validate_accuracy = 0.1153\nEpoch: 6 train_loss = 20.9577 validate_accuracy = 0.1289\nEpoch: 7 train_loss = 17.9383 validate_accuracy = 0.1418\nEpoch: 8 train_loss = 14.9069 validate_accuracy = 0.1544\nEpoch: 9 train_loss = 12.5750 validate_accuracy = 0.1682\nEpoch: 10 train_loss = 10.4634 validate_accuracy = 0.1838\nEpoch: 11 train_loss = 8.4767 validate_accuracy = 0.2010\nEpoch: 12 train_loss = 6.8023 validate_accuracy = 0.2190\nEpoch: 13 train_loss = 5.4827 validate_accuracy = 0.2375\nEpoch: 14 train_loss = 4.5168 validate_accuracy = 0.2563\nEpoch: 15 train_loss = 3.7650 validate_accuracy = 0.2752\nEpoch: 16 train_loss = 3.1800 validate_accuracy = 0.2939\nEpoch: 17 train_loss = 2.7230 validate_accuracy = 0.3123\nEpoch: 18 train_loss = 2.3341 validate_accuracy = 0.3305\nEpoch: 19 train_loss = 1.9623 validate_accuracy = 0.3482\nEpoch: 20 train_loss = 1.7098 validate_accuracy = 0.3653\nEpoch: 21 train_loss = 1.5094 validate_accuracy = 0.3816\nEpoch: 22 train_loss = 1.3523 validate_accuracy = 0.3973\nEpoch: 23 train_loss = 1.2216 validate_accuracy = 0.4123\nEpoch: 24 train_loss = 1.1111 validate_accuracy = 0.4266\nEpoch: 25 train_loss = 1.0181 validate_accuracy = 0.4403\nEpoch: 26 train_loss = 0.9424 validate_accuracy = 0.4534\nEpoch: 27 train_loss = 0.8783 validate_accuracy = 0.4659\nEpoch: 28 train_loss = 0.8199 validate_accuracy = 0.4779\nEpoch: 29 train_loss = 0.7670 validate_accuracy = 0.4894\nEpoch: 30 train_loss = 0.7207 validate_accuracy = 0.5004\nEpoch: 31 train_loss = 0.6790 validate_accuracy = 0.5108\nEpoch: 32 train_loss = 0.6407 validate_accuracy = 0.5208\nEpoch: 33 train_loss = 0.6067 validate_accuracy = 0.5303\nEpoch: 34 train_loss = 0.5757 validate_accuracy = 0.5395\nEpoch: 35 train_loss = 0.5469 validate_accuracy = 0.5484\nEpoch: 36 train_loss = 0.5205 validate_accuracy = 0.5569\nEpoch: 37 train_loss = 0.4955 validate_accuracy = 0.5652\nEpoch: 38 train_loss = 0.4721 validate_accuracy = 0.5731\nEpoch: 39 train_loss = 0.4507 validate_accuracy = 0.5806\nEpoch: 40 train_loss = 0.4303 validate_accuracy = 0.5880\nEpoch: 41 train_loss = 0.4115 validate_accuracy = 0.5950\nEpoch: 42 train_loss = 0.3942 validate_accuracy = 0.6018\nEpoch: 43 train_loss = 0.3775 validate_accuracy = 0.6084\nEpoch: 44 train_loss = 0.3619 validate_accuracy = 0.6148\nEpoch: 45 train_loss = 0.3472 validate_accuracy = 0.6209\nEpoch: 46 train_loss = 0.3332 validate_accuracy = 0.6268\nEpoch: 47 train_loss = 0.3202 validate_accuracy = 0.6326\nEpoch: 48 train_loss = 0.3074 validate_accuracy = 0.6381\nEpoch: 49 train_loss = 0.2939 validate_accuracy = 0.6435\nEpoch: 50 train_loss = 0.2825 validate_accuracy = 0.6487\nTesting Accuracy: 0.6536152\nConfusion Matrix: [[ 87 8 1 ... 0 0 0]\n [ 6 59 7 ... 0 0 0]\n [ 2 4 77 ... 0 0 0]\n ...\n [ 0 0 0 ... 111 0 0]\n [ 0 0 0 ... 0 106 1]\n [ 0 0 0 ... 0 2 103]]\n"
],
[
"plt.xlabel('Epoch')\nplt.ylabel('Cross Entropy Loss')\nplt.plot(avg_loss[None:])\nplt.show()",
"_____no_output_____"
],
[
"plt.xlabel('Epoch')\nplt.ylabel('Validation Accuracy')\nplt.plot(validate_accuracy)\nplt.show()",
"_____no_output_____"
],
[
"True_positives = np.diag(confusion_mat)\nFalse_positives = np.sum(confusion_mat, axis=1) - True_positives\nFalse_negatives = np.sum(confusion_mat, axis=0) - True_positives\nPrecision = True_positives / (True_positives + False_positives)\nprint(\"Precision:\", Precision)\nRecall = True_positives / (True_positives + False_negatives)\nprint(\"\\nRecall:\", Recall)\nF_scores = (2*Precision*Recall) / (Recall+Precision)\nprint(\"\\nF_scores:\", F_scores)",
"Precision: [0.79090909 0.62105263 0.78571429 0.82828283 0.74025974 0.75789474\n 0.92929293 0.5952381 0.72972973 0.91836735 0.58 0.6\n 0.76699029 0.68867925 0.83333333 0.68674699 0.68807339 0.72631579\n 0.87058824 0.68224299 0.80172414 0.9 0.55555556 0.73033708\n 0.86842105 0.73863636 0.90909091 0.84615385 0.94623656 0.91964286\n 0.89108911 0.90217391 0.88333333 0.92792793 0.92929293 0.76\n 0.83018868 0.82954545 0.95798319 0.85714286 0.93939394 0.90654206\n 0.925 0.81904762 0.90291262 0.95 0.92173913 0.84375\n 0.96296296 0.98979592 0.99029126 1. 1. 1.\n 1. 1. 0.98245614 0.96330275 0.93693694 0.94845361\n 0.95555556 0.97916667 1. 1. 0.98958333 1.\n 1. 0.98989899 0.97115385 0.99029126 0.98989899 0.93693694\n 0.98 1. 1. 1. 0.99029126 1.\n 1. 1. 0.96385542 0.97 0.9 1.\n 0.98076923 1. 0.99 1. 1. 1.\n 1. 1. 0.98913043 1. 0.99065421 0.97169811]\n\nRecall: [0.83653846 0.60204082 0.79381443 0.65079365 0.73076923 0.68571429\n 0.88461538 0.80645161 0.77142857 0.72 0.79452055 0.58762887\n 0.88764045 0.81111111 0.87628866 0.57 0.81521739 0.73404255\n 0.85057471 0.74489796 0.86111111 0.62068966 0.74626866 0.51181102\n 0.76153846 0.84415584 0.87912088 0.84615385 0.93617021 0.96261682\n 0.94736842 0.8556701 0.9137931 0.91964286 0.85185185 0.82608696\n 0.88 0.85882353 0.92682927 0.87272727 0.89423077 0.92380952\n 0.88095238 0.93478261 0.88571429 0.9223301 0.90598291 0.89010989\n 1. 1. 1. 0.96774194 1. 0.99115044\n 0.97938144 0.97894737 1. 0.97222222 0.95412844 1.\n 1. 0.96907216 0.98913043 1. 0.97938144 1.\n 1. 0.98 0.97115385 0.99029126 0.98 0.98113208\n 0.98989899 1. 0.98979592 1. 0.99029126 1.\n 0.99122807 1. 0.98765432 0.98979592 0.96428571 0.97087379\n 1. 0.98958333 0.99 1. 0.99193548 1.\n 0.97142857 0.97938144 0.97849462 1. 0.98148148 0.99038462]\n\nF_scores: [0.81308411 0.61139896 0.78974359 0.72888889 0.73548387 0.72\n 0.90640394 0.68493151 0.75 0.80717489 0.67052023 0.59375\n 0.82291667 0.74489796 0.85427136 0.62295082 0.74626866 0.73015873\n 0.86046512 0.71219512 0.83035714 0.73469388 0.63694268 0.60185185\n 0.81147541 0.78787879 0.89385475 0.84615385 0.94117647 0.94063927\n 0.91836735 0.87830688 0.89830508 0.92376682 0.88888889 0.79166667\n 0.85436893 0.84393064 0.94214876 0.86486486 0.91625616 0.91509434\n 0.90243902 0.87309645 0.89423077 0.93596059 0.9137931 0.86631016\n 0.98113208 0.99487179 0.99512195 0.98360656 1. 0.99555556\n 0.98958333 0.9893617 0.99115044 0.96774194 0.94545455 0.97354497\n 0.97727273 0.97409326 0.99453552 1. 0.98445596 1.\n 1. 0.98492462 0.97115385 0.99029126 0.98492462 0.95852535\n 0.98492462 1. 0.99487179 1. 0.99029126 1.\n 0.99559471 1. 0.97560976 0.97979798 0.93103448 0.98522167\n 0.99029126 0.9947644 0.99 1. 0.99595142 1.\n 0.98550725 0.98958333 0.98378378 1. 0.98604651 0.98095238]\n"
],
[
"plt.plot(Precision, label='Precision')\nplt.plot(Recall, label='Recall')\nplt.plot(F_scores, label='F Scores')\nplt.ylabel('Score')\nplt.xlabel('Class')\nplt.legend()\nplt.show()",
"_____no_output_____"
],
[
"np.savez_compressed('linev3-conmat.npz', cmat=confusion_mat)",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4afe3a723f2f1cfa08870017d0ee787fe2c01823
| 1,029,958 |
ipynb
|
Jupyter Notebook
|
examples/explorer.ipynb
|
kratzert/EGU2017_public
|
47bcb8714f4a8744176817ee3d90e8188d2b8480
|
[
"MIT"
] | 4 |
2017-04-26T08:12:02.000Z
|
2018-10-20T17:03:45.000Z
|
examples/explorer.ipynb
|
kratzert/EGU2017_public
|
47bcb8714f4a8744176817ee3d90e8188d2b8480
|
[
"MIT"
] | null | null | null |
examples/explorer.ipynb
|
kratzert/EGU2017_public
|
47bcb8714f4a8744176817ee3d90e8188d2b8480
|
[
"MIT"
] | 7 |
2017-02-22T00:19:20.000Z
|
2021-07-08T00:12:53.000Z
| 2,555.727047 | 104,264 | 0.950877 |
[
[
[
"# Explore the classification results\n\nThis notebook will guide you through different visualizations of the test set evaluation of any of the presented models.\n\nIn a first step you can select the result file of any of the models you want to explore.",
"_____no_output_____"
]
],
[
[
"model = 'vgg_results_sample.csv' #should be placed in the /eval/ folder",
"_____no_output_____"
]
],
[
[
"Then we will import some packages and setup some basic variables",
"_____no_output_____"
]
],
[
[
"import os\nimport sys\n\nimport numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\n\nfrom skimage.io import imread\nfrom sklearn.metrics import accuracy_score, confusion_matrix\nfrom utils import plot_confusion_matrix, class_names, plots\n\n%matplotlib inline\n\n# get the path of the data and eval folder\nroot_dir = os.path.abspath(os.path.join(sys.path[0], os.pardir))\neval_dir = os.path.join(root_dir, 'eval')\ndata_dir = os.path.join(root_dir, 'data')",
"_____no_output_____"
]
],
[
[
"**Next** we will load the evaluation results and compute different **accuracy** values.\n\n1. The per-image accuracy. Should not be be confused with the per-fish accuracy. The per-image accuracy means how many of the samples (image + corresponding features) in the test set were predicted with the correct class.\n2. The per-fish accuracy if the results of the different images of one fish are combine by the `mode` of the single image predictions\n3. The per-fish accuracy if the final prediction is derived from the class with the highest overall probability if we sum up all class probabilities of the single images.\n\nTo get the different per-fish accuracies we can group the data by `video_name` and `label` because each combination of `video_name` and `label` stays for one individual fish. \nThe columns `prediction_max_prob` and `prediction_mode` contain the predicted class label for each individual fish derived through the above stated methods. ",
"_____no_output_____"
]
],
[
[
"#load the evaluation data\ndf = pd.read_csv(os.path.join(eval_dir, model), sep=';')\n\n#compute the per-image accuracy\nacc_per_img = accuracy_score(df['label'], df['pred'])\n\n# get the per-fish results by grouping the dataframe and taking first entry of each group as the columns for the \n# per-fish prediction are the same for all images of one fish\nfish = df.groupby(['video_name', 'label']).first().reset_index()\n\n# calculate the per-fish accuracies\nacc_fish_mode = accuracy_score(fish['label'], fish['prediction_mode'])\nacc_fish_max_prob = accuracy_score(fish['label'], fish['prediction_max_prob'])\n\n# print the results\nprint(\"From a total of %i images, %.2f percent of the images were classified correctly.\" %(len(df), acc_per_img*100))\nprint(\"If combined by the mode of %i fish individuals %.2f were classified correctly.\" %(len(fish), acc_fish_mode*100))\nprint(\"If derived from the max probability of the summed class probabilities, out of %i fish %.2f were classified correctly.\" %(len(fish), acc_fish_max_prob*100))",
"From a total of 1053 images, 86.32 percent of the images were classified correctly.\nIf combined by the mode of 104 fish individuals 90.38 were classified correctly.\nIf derived from the max probability of the summed class probabilities, out of 104 fish 89.42 were classified correctly.\n"
]
],
[
[
"As we are interested in the per-fish accuracy, we can see that combining the classification results of many images of one fish can help to raise the overall prediction accuracy. From the two different methods, this can be best done through the `prediction_max_prob` method.\n\n**Next** we will display the **confusion matrix**. The confusion matrix displays the true class vs. the predicted class. This might help to understand which classes the model can seperate and between which it makes the most mispredictions. \nBy changing the first line, you can select which confusion matrix should be displayed. You can chose between ['pre-img', 'mode', 'max_prob'] referring to the 3 different accuracies from above.",
"_____no_output_____"
]
],
[
[
"method = 'max_prob' # chose one of ['per-img', 'mode', 'max_prob']\n\n#compute confusion matrix\nif method == 'per-img':\n cm = confusion_matrix(df['label'], df['pred'])\nelif method == 'mode':\n cm = confusion_matrix(fish['label'], fish['prediction_mode'])\nelif method == 'max_prob':\n cm = confusion_matrix(fish['label'], fish['prediction_max_prob'])\nelse:\n raise ValueError(\"Select a valid method. Must be one of ['per-img', 'mode', 'max_prob']\")\n \n#plot confusion matrix\nplot_confusion_matrix(cm, [n.split(',')[1] for n in class_names])\n",
"_____no_output_____"
]
],
[
[
"You will see for all of the combinations you can selected (4 models and 3 different methods) that the models make the most number of misclassifications between similar (at least somehow) looking fish species. For example you can see that most misclassifications of brown trouts were made with rainbow trouts (and vice versa) and the same for chub and common nase. Through this we can conclude that the models understand the content of the images/features they are confronted with.\n\n**Next** we will look at some random images of each class the model was absolutely sure about it's prediction and the predicted class was indeed the true class.",
"_____no_output_____"
]
],
[
[
"#number of total classes and images per class we will plot\nnum_classes = 7\nn_view = 4\n\n#iterate over each class and find the images the model assigned the highest class probability to (to the true class)\nfor i in range(num_classes):\n corrects = np.where((df['label'] == i) & (df['pred'] == i))\n df_corrects = df.loc[corrects]\n df_corrects.sort_values('score'+str(i), inplace = True, ascending = False)\n \n #print number of correct images per class\n print(\"Found %i correct %s\" %(len(df_corrects), class_names[i]))\n \n #plot images\n plots(df_corrects, df_corrects.index[:n_view], data_dir)",
"Found 64 correct Lota lota, burbot, Aalrutte\nFound 160 correct Squalius cephalus, chub, Aitel\nFound 208 correct Salmo trutta, brown trout, Bachforelle\nFound 181 correct Abramis brama, bream, Brachse\nFound 65 correct Perca fluviatilis, perch, Flussbarsch\nFound 112 correct Chondrostoma nasus, common nase, Nase\nFound 119 correct Oncorhynchus mykiss, rainbow trout, Regenbogenforelle\n"
]
],
[
[
"To some degree, these images should make sense. Most of them show specific characteristics of each species that are enought to classify them correctly. Some might not make much sense for us as a human observer but might make sense to the model. \nE.g. We had plenty of rainbow trouts in turbid water, so that the model seems to have learned that a salmonide looking fish in turbid water is more likely to be a rainbow than a brown trout.\n\nUp **Next** we could visualize the opposite: We could look at the images of each class, for which the model was most uncertain about.",
"_____no_output_____"
]
],
[
[
"for i in range(num_classes):\n fish = df.loc[df['label'] == i]\n fish.is_copy = False\n fish.sort_values('score'+str(i), inplace = True, ascending = True)\n plots(fish, fish.index[:n_view], data_dir)",
"_____no_output_____"
]
],
[
[
"These results may let you wonder, why for some of these images the model predicted the wrong class, although the fish could be clearly seen. To be honest, I don't know the answer and could only guess for some of the cases. I think one key point could be the size of the dataset and one could expect that a bigger sample size per class could yield better results. \n\nOne the other hand, one could also see many images of bad quality, turbid water etc. where even for us humans it might be hard to classify the fish by this one single images.\n\nAlso note, that this are per-image prediction results. A lot of these single image misclassifications aren't influencing the per-fish predictions. ",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
4afe49687d445c4be7d9eb019d8d60cce07311e3
| 8,458 |
ipynb
|
Jupyter Notebook
|
.ipynb_checkpoints/data_conversion-checkpoint.ipynb
|
knretka/Web-Design-Challenge
|
d2faa6c27ce6bb1b9bbfab88caa897c36f36c051
|
[
"ADSL"
] | null | null | null |
.ipynb_checkpoints/data_conversion-checkpoint.ipynb
|
knretka/Web-Design-Challenge
|
d2faa6c27ce6bb1b9bbfab88caa897c36f36c051
|
[
"ADSL"
] | null | null | null |
.ipynb_checkpoints/data_conversion-checkpoint.ipynb
|
knretka/Web-Design-Challenge
|
d2faa6c27ce6bb1b9bbfab88caa897c36f36c051
|
[
"ADSL"
] | null | null | null | 120.828571 | 1,514 | 0.698037 |
[
[
[
"import pandas as pd",
"_____no_output_____"
],
[
"data=pd.read_csv(\"../Resources/cities.csv\")",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code"
]
] |
4afe614877a5a89739614eaaa467ad488d79f55c
| 9,628 |
ipynb
|
Jupyter Notebook
|
notebooks/layers/wrappers/TimeDistributed.ipynb
|
jefffriesen/keras-js
|
130cca841fc4e8fd5fc157ae57061f9576d8588f
|
[
"MIT"
] | null | null | null |
notebooks/layers/wrappers/TimeDistributed.ipynb
|
jefffriesen/keras-js
|
130cca841fc4e8fd5fc157ae57061f9576d8588f
|
[
"MIT"
] | null | null | null |
notebooks/layers/wrappers/TimeDistributed.ipynb
|
jefffriesen/keras-js
|
130cca841fc4e8fd5fc157ae57061f9576d8588f
|
[
"MIT"
] | 1 |
2020-03-12T21:03:06.000Z
|
2020-03-12T21:03:06.000Z
| 49.628866 | 1,668 | 0.594308 |
[
[
[
"import numpy as np\nfrom keras.models import Model\nfrom keras.layers import Input\nfrom keras.layers.core import Dense\nfrom keras.layers.convolutional import Convolution2D\nfrom keras.layers.wrappers import TimeDistributed\nfrom keras import backend as K",
"Using TensorFlow backend.\n"
],
[
"def format_decimal(arr, places=6):\n return [round(x * 10**places) / 10**places for x in arr]",
"_____no_output_____"
]
],
[
[
"### TimeDistributed",
"_____no_output_____"
],
[
"**[wrappers.TimeDistributed.0] wrap a Dense layer with output_dim 4 (input: 3 x 6)**",
"_____no_output_____"
]
],
[
[
"data_in_shape = (3, 6)\n\nlayer_0 = Input(shape=data_in_shape)\nlayer_1 = TimeDistributed(Dense(4))(layer_0)\nmodel = Model(input=layer_0, output=layer_1)\n\n# set weights to random (use seed for reproducibility)\nweights = []\nfor i, w in enumerate(model.get_weights()):\n np.random.seed(4000 + i)\n weights.append(2 * np.random.random(w.shape) - 1)\nmodel.set_weights(weights)\nweight_names = ['W', 'b']\nfor w_i, w_name in enumerate(weight_names):\n print('{} shape:'.format(w_name), weights[w_i].shape)\n print('{}:'.format(w_name), format_decimal(weights[w_i].ravel().tolist()))\n\ndata_in = 2 * np.random.random(data_in_shape) - 1\nprint('')\nprint('in shape:', data_in_shape)\nprint('in:', format_decimal(data_in.ravel().tolist()))\nresult = model.predict(np.array([data_in]))\nprint('out shape:', result[0].shape)\nprint('out:', format_decimal(result[0].ravel().tolist()))",
"W shape: (6, 4)\nW: [0.317596, 0.688515, -0.688309, -0.48247, 0.387223, -0.718263, 0.281673, -0.106311, 0.576861, -0.083926, 0.631691, 0.92647, 0.579655, -0.024215, -0.805793, -0.842947, -0.955415, 0.656415, 0.44667, 0.633739, 0.701525, 0.917507, -0.185671, -0.105247]\nb shape: (4,)\nb: [-0.332867, 0.650317, 0.995501, -0.458367]\n\nin shape: (3, 6)\nin: [-0.30351, 0.37881, -0.248093, 0.372204, -0.698964, -0.408058, -0.103801, 0.376217, -0.724015, 0.708616, -0.513219, -0.46074, -0.125163, -0.76111, -0.153798, 0.729255, 0.556458, -0.671966]\nout shape: (3, 4)\nout: [0.171595, -0.652137, 0.618031, -1.295817, -0.05994, -0.407387, 0.000875, -1.993142, -1.33639, 0.854801, 0.555804, -0.650907]\n"
]
],
[
[
"**[wrappers.TimeDistributed.1] wrap a Convolution2D layer with 6 3x3 filters (input: 5x4x4x2)**",
"_____no_output_____"
]
],
[
[
"data_in_shape = (5, 4, 4, 2)\n\nlayer_0 = Input(shape=data_in_shape)\nlayer_1 = TimeDistributed(Convolution2D(6, 3, 3, dim_ordering='tf'))(layer_0)\nmodel = Model(input=layer_0, output=layer_1)\n\n# set weights to random (use seed for reproducibility)\nweights = []\nfor i, w in enumerate(model.get_weights()):\n np.random.seed(4010 + i)\n weights.append(2 * np.random.random(w.shape) - 1)\nmodel.set_weights(weights)\nweight_names = ['W', 'b']\nfor w_i, w_name in enumerate(weight_names):\n print('{} shape:'.format(w_name), weights[w_i].shape)\n print('{}:'.format(w_name), format_decimal(weights[w_i].ravel().tolist()))\n\ndata_in = 2 * np.random.random(data_in_shape) - 1\nprint('')\nprint('in shape:', data_in_shape)\nprint('in:', format_decimal(data_in.ravel().tolist()))\nresult = model.predict(np.array([data_in]))\nprint('out shape:', result[0].shape)\nprint('out:', format_decimal(result[0].ravel().tolist()))",
"W shape: (3, 3, 2, 6)\nW: [0.971827, -0.898904, -0.987921, 0.529589, 0.043586, -0.541366, 0.316759, 0.351387, -0.292323, 0.445466, -0.922655, 0.437413, -0.483267, -0.478014, 0.7408, -0.595028, -0.718381, 0.349594, -0.091293, 0.14291, 0.633818, -0.686841, -0.925272, -0.740397, 0.070594, 0.67408, 0.455314, -0.402251, 0.288807, 0.001378, 0.42892, -0.251869, 0.06113, -0.703784, 0.002676, 0.965023, 0.758788, 0.1193, 0.749321, -0.017408, -0.004115, 0.18981, -0.91507, 0.132792, -0.219057, 0.19682, -0.512841, 0.954544, 0.794403, -0.663179, -0.05377, -0.855038, -0.486641, 0.625844, -0.945869, -0.474979, 0.922345, -0.334843, -0.469456, -0.394364, 0.543681, -0.817676, 0.6093, -0.77635, -0.508683, 0.22456, 0.696262, 0.079806, -0.182646, -0.718939, 0.962504, -0.386231, 0.860488, -0.918945, -0.800484, -0.590285, 0.409804, -0.822098, 0.3489, -0.4508, 0.913208, -0.414455, 0.97663, 0.956314, -0.55547, 0.594094, -0.552044, -0.137467, 0.539049, -0.320055, -0.335577, 0.974746, -0.634747, 0.085161, -0.127183, -0.061717, -0.411844, 0.774181, 0.223395, 0.163937, -0.606967, 0.178549, -0.005153, 0.452476, 0.373127, -0.726827, -0.395458, -0.769671]\nb shape: (6,)\nb: [0.180389, 0.629217, -0.656262, -0.476575, -0.36398, 0.987756]\n\nin shape: (5, 4, 4, 2)\nin: [-0.579677, 0.883193, 0.651172, -0.820251, -0.64795, 0.857328, -0.4689, 0.356044, -0.641528, -0.531973, -0.33586, -0.438823, 0.682186, 0.215781, -0.401735, 0.169171, 0.869358, -0.204078, -0.661876, -0.616139, -0.453943, -0.569439, -0.25218, 0.156473, 0.194797, -0.923921, 0.652204, -0.11765, 0.86293, 0.314218, -0.878496, -0.364761, -0.647821, 0.296841, 0.280105, 0.2753, -0.959741, -0.148037, -0.489424, -0.88939, 0.704443, 0.08354, 0.930112, -0.87023, -0.212285, 0.750133, 0.343506, -0.82568, 0.391491, 0.149626, 0.003594, -0.181464, -0.499632, 0.20694, 0.1007, 0.39826, 0.609736, -0.765775, -0.728474, -0.011711, 0.543543, 0.174309, 0.105794, -0.009876, -0.694421, -0.157031, 0.670853, -0.581331, 0.739486, -0.886014, -0.637039, 0.725753, 0.61919, 0.447635, 0.167298, 0.164242, -0.615436, -0.503061, 0.981698, -0.392795, 0.532215, 0.761817, 0.735562, -0.236234, -0.856381, 0.22419, -0.221125, 0.133757, -0.011162, -0.88018, -0.433047, -0.825617, 0.693626, -0.185243, -0.824829, 0.07932, 0.336478, 0.370138, -0.685905, -0.462037, 0.563862, 0.490274, 0.934239, -0.129323, 0.717792, -0.73658, -0.939587, 0.796637, -0.131382, -0.79957, -0.271279, 0.816961, -0.082096, 0.64553, -0.106661, 0.651369, -0.843208, -0.221077, 0.758074, 0.156006, -0.429501, 0.191698, 0.988067, -0.277344, 0.757645, -0.877824, 0.053841, 0.394075, 0.786359, 0.735302, 0.247852, -0.310899, 0.703408, -0.848404, 0.455067, 0.295289, -0.629316, 0.626332, -0.075289, -0.442735, -0.219408, -0.766048, 0.303257, 0.142211, 0.910002, -0.780858, 0.333242, -0.533434, 0.572575, 0.355883, -0.671924, 0.22028, -0.505951, -0.317892, 0.609641, -0.360548, 0.490007, 0.441024, 0.660294, 0.850007]\nout shape: (5, 2, 2, 6)\nout: [2.089554, -2.186939, -1.436176, -0.951733, -0.212962, 2.449681, 1.053569, -0.592297, -0.875753, -0.803289, -0.834779, -0.56835, -0.842922, 3.976766, -1.054281, 0.581773, 0.235047, 0.10304, -0.079684, 0.225164, -2.408352, -1.116154, 1.561833, -0.491674, 2.43274, -0.158394, -0.874487, -1.968509, -0.106465, 1.602375, 0.941225, 0.480547, 0.002478, 1.246196, -1.388929, -1.133004, 1.476556, -0.459852, -2.130519, -0.126113, -1.162246, 1.398016, -0.61384, 1.539333, -0.466156, 0.0395, 0.506595, -1.590957, -1.044266, 0.736233, 0.61792, -0.923799, 1.275832, 1.491487, 1.903215, -2.385962, -1.553725, -0.554848, -0.456638, 1.645426, 0.690055, 0.190637, -2.015925, 1.143469, -2.530135, 1.025159, -0.150503, 2.627801, -1.352068, 1.245647, 1.235627, -0.915363, 0.682647, 0.854592, -0.030856, 0.949627, 1.204568, 1.052329, -0.942961, 2.039315, 0.892454, -1.925232, 0.046332, 2.315713, -2.358421, 1.724373, -1.528506, 1.794933, 0.342617, -0.191888, -0.026605, 0.475714, -1.332559, -1.158213, 0.028725, 1.890396, -0.305622, 0.890336, -3.426138, 1.245994, -2.027975, -0.505022, 1.32001, 0.477822, -2.460816, -0.984189, 1.221664, 0.339474, 1.26535, 2.228118, 0.207158, -0.455113, -0.64988, 0.688864, 0.574933, 1.911588, -1.642422, -1.385078, 0.744757, -0.567276]\n"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
4afe65c79eb4bea948ef9876e67261a80cf68c84
| 22,148 |
ipynb
|
Jupyter Notebook
|
training/training_CAL.ipynb
|
xl-sr/CAL
|
d260c0769d536f48acd9711079d63fc11802b8e8
|
[
"MIT"
] | 71 |
2018-09-24T17:53:36.000Z
|
2022-02-04T07:26:25.000Z
|
training/training_CAL.ipynb
|
xl-sr/CAL
|
d260c0769d536f48acd9711079d63fc11802b8e8
|
[
"MIT"
] | 30 |
2018-10-30T13:37:36.000Z
|
2020-11-15T08:53:01.000Z
|
training/training_CAL.ipynb
|
xl-sr/CAL
|
d260c0769d536f48acd9711079d63fc11802b8e8
|
[
"MIT"
] | 30 |
2018-11-10T16:42:08.000Z
|
2022-01-21T23:46:08.000Z
| 45.950207 | 1,509 | 0.633375 |
[
[
[
"#### Setup",
"_____no_output_____"
]
],
[
[
"# standard imports \nimport numpy as np\nimport torch\nimport matplotlib.pyplot as plt\nfrom torch import optim\nfrom ipdb import set_trace\nfrom datetime import datetime\n\n# jupyter setup\n%matplotlib inline\n%load_ext autoreload\n%autoreload 2\n\n# own modules\nfrom dataloader import CAL_Dataset\nfrom net import get_model\nfrom dataloader import get_data, get_mini_data, load_json, save_json\nfrom train import fit, custom_loss, validate\nfrom metrics import calc_metrics\n\n# paths\ndata_path = './dataset/'",
"_____no_output_____"
]
],
[
[
"uncomment the cell below if you want your experiments to yield always the same results",
"_____no_output_____"
]
],
[
[
"# manualSeed = 42\n\n# np.random.seed(manualSeed)\n# torch.manual_seed(manualSeed)\n\n# # if you are using GPU\n# torch.cuda.manual_seed(manualSeed)\n# torch.cuda.manual_seed_all(manualSeed)\n\n# torch.backends.cudnn.enabled = False \n# torch.backends.cudnn.benchmark = False\n# torch.backends.cudnn.deterministic = True",
"_____no_output_____"
]
],
[
[
"#### Training",
"_____no_output_____"
],
[
"Initialize the model. Possible Values for the task block type: MLP, LSTM, GRU, TempConv",
"_____no_output_____"
]
],
[
[
"params = {'name': 'tempconv', 'type_': 'TempConv', 'lr': 3e-4, 'n_h': 128, 'p':0.5, 'seq_len':5}",
"_____no_output_____"
],
[
"save_json(params, f\"models/{params['name']}\")",
"_____no_output_____"
],
[
"model, opt = get_model(params)\ndevice = torch.device(\"cuda:0\" if torch.cuda.is_available() else \"cpu\")\nmodel = model.to(device)",
"_____no_output_____"
]
],
[
[
"get the data loader. get mini data gets only a subset of the training data, on which we can try if the model is able to overfit",
"_____no_output_____"
]
],
[
[
"train_dl, valid_dl = get_data(data_path, model.params.seq_len, batch_size=16)\n# train_dl, valid_dl = get_mini_data(data_path, model.params.seq_len, batch_size=16, l=4000)",
"_____no_output_____"
]
],
[
[
"Train the model. We automatically save the model with the lowest val_loss. If you want to continue the training and keep the loss history, just pass it as an additional argument as shown below.",
"_____no_output_____"
]
],
[
[
"model, val_hist = fit(1, model, custom_loss, opt, train_dl, valid_dl)",
"_____no_output_____"
],
[
"# model, val_hist = fit(1, model, custom_loss, opt, train_dl, valid_dl, val_hist=val_hist)",
"_____no_output_____"
]
],
[
[
"uncomment the following two cells if the feature extractor should also be trained",
"_____no_output_____"
]
],
[
[
"# for name,param in model.named_parameters():\n# param.requires_grad = True\n# opt = optim.Adam(model.parameters())",
"_____no_output_____"
],
[
"# model, val_hist = fit(1, model, custom_loss, opt, train_dl, valid_dl)",
"_____no_output_____"
],
[
"plt.plot(val_hist)",
"_____no_output_____"
]
],
[
[
"#### evalute the model",
"_____no_output_____"
],
[
"reload model",
"_____no_output_____"
]
],
[
[
"name = 'gru'\nparams = load_json(f\"models/{name}\")",
"_____no_output_____"
],
[
"model, _ = get_model(params)\nmodel.load_state_dict(torch.load(f\"./models/{name}.pth\"));",
"_____no_output_____"
],
[
"model.eval().to(device);\n_, valid_dl = get_data(data_path, model.params.seq_len, batch_size=16)",
"_____no_output_____"
]
],
[
[
"run evaluation on full val set",
"_____no_output_____"
]
],
[
[
"_, all_preds, all_labels = validate(model, valid_dl, custom_loss)",
"_____no_output_____"
],
[
"calc_metrics(all_preds, all_labels)",
"_____no_output_____"
]
],
[
[
"#### plot results",
"_____no_output_____"
]
],
[
[
"# for convience, we can pass an integer instead of the full string\nint2key = {0: 'red_light', 1:'hazard_stop', 2:'speed_sign', \n 3:'relative_angle', 4: 'center_distance', 5: 'veh_distance'}",
"_____no_output_____"
],
[
"def plot_preds(k, all_preds, all_labels, start=0, delta=1000):\n if isinstance(k, int): k = int2key[k]\n \n # get preds and labels\n class_labels = ['red_light', 'hazard_stop', 'speed_sign']\n pred = np.argmax(all_preds[k], axis=1) if k in class_labels else all_preds[k]\n label = all_labels[k][:, 1] if k in class_labels else all_labels[k]\n \n plt.plot(pred[start:start+delta], 'r--', label='Prediction', linewidth=2.0)\n plt.plot(label[start:start+delta], 'g', label='Ground Truth', linewidth=2.0)\n \n plt.legend()\n plt.grid()\n plt.show()",
"_____no_output_____"
],
[
"plot_preds(5, all_preds, all_labels, start=0, delta=4000)",
"_____no_output_____"
]
],
[
[
"#### param search\n\n",
"_____no_output_____"
]
],
[
[
"from numpy.random import choice\nnp.random.seed()",
"_____no_output_____"
],
[
"params = {'name': 'tempconv', 'type_': 'TempConv', 'lr': 3e-4, 'n_h': 128, 'p':0.5, 'seq_len':5}",
"_____no_output_____"
],
[
"def get_random_NN_parameters():\n params = {}\n params['type_'] = choice(['MLP', 'GRU', 'LSTM', 'TempConv'])\n params['name'] = datetime.now().strftime(\"%Y_%m_%d_%H_%M\")\n params['lr'] = np.random.uniform(1e-5, 1e-2)\n params['n_h'] = np.random.randint(5, 200)\n params['p'] = np.random.uniform(0.25, 0.75)\n params['seq_len'] = np.random.randint(1, 15)\n \n return params",
"_____no_output_____"
],
[
"while True:\n params = get_random_NN_parameters() \n print('PARAMS: {}'.format(params))\n \n # instantiate the model\n model, opt = get_model(params)\n device = torch.device(\"cuda:0\" if torch.cuda.is_available() else \"cpu\")\n model = model.to(device)\n save_json(params, f\"models/{params['name']}\")\n \n # get the data loaders\n train_dl, valid_dl = get_data(data_path, model.params.seq_len, batch_size=16)\n \n # start the training\n model, val_hist = fit(5, model, custom_loss, opt, train_dl, valid_dl)\n for name,param in model.named_parameters():\n param.requires_grad = True\n opt = optim.Adam(model.parameters())\n model, val_hist = fit(5, model, custom_loss, opt, train_dl, valid_dl, val_hist=val_hist)",
"PARAMS: {'name': '2019_06_13_19_55', 'lr': 0.007221551882651735, 'n_h': 48, 'p': 0.5985699357015684, 'seq_len': 14, 'type_': 'MLP'}\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
4afe70ab6b0f8cfc64ba6104719d15773ff40421
| 71,627 |
ipynb
|
Jupyter Notebook
|
TOA_ML_1_DataPrep_5d_APHY_Feature_Eng_&_Standardize.pkl.ipynb
|
madHatter106/TOA_ML
|
fc651f47a76a72cff2bbc516a8ee49170d78332a
|
[
"MIT"
] | null | null | null |
TOA_ML_1_DataPrep_5d_APHY_Feature_Eng_&_Standardize.pkl.ipynb
|
madHatter106/TOA_ML
|
fc651f47a76a72cff2bbc516a8ee49170d78332a
|
[
"MIT"
] | null | null | null |
TOA_ML_1_DataPrep_5d_APHY_Feature_Eng_&_Standardize.pkl.ipynb
|
madHatter106/TOA_ML
|
fc651f47a76a72cff2bbc516a8ee49170d78332a
|
[
"MIT"
] | null | null | null | 34.436058 | 113 | 0.402739 |
[
[
[
"import pickle\nimport warnings\nwarnings.filterwarnings('ignore')\nfrom sklearn.preprocessing import StandardScaler, PolynomialFeatures\nimport pandas as pd\nfrom pandas.plotting import scatter_matrix\nimport numpy as np\nimport matplotlib.pyplot as pl\nfrom matplotlib import rcParams\nfrom seaborn import PairGrid, heatmap, kdeplot\nimport cmocean.cm as cmo",
"_____no_output_____"
],
[
"% matplotlib inline\nrcParams['axes.titlesize'] = 18\nrcParams['xtick.labelsize'] = 16\nrcParams['ytick.labelsize'] = 16\nrcParams['axes.labelsize'] = 16\nrcParams['font.size'] = 16",
"_____no_output_____"
],
[
"df_pc = pd.read_pickle('./pickleJar/DevelopmentalDataSets/df_5_APHY_pc.pkl')\ndf_sat = pd.read_pickle('./pickleJar/DevelopmentalDataSets/df_5_APHY_sat')",
"_____no_output_____"
],
[
"df_pc.info()",
"<class 'pandas.core.frame.DataFrame'>\nInt64Index: 495 entries, 0 to 494\nData columns (total 36 columns):\nsin_doy 495 non-null float64\ncos_doy 495 non-null float64\nsin_minofday 495 non-null float64\ncos_minofday 495 non-null float64\nx 495 non-null float64\ny 495 non-null float64\nz 495 non-null float64\nlog10_etopo2 495 non-null float64\noisst 495 non-null float64\nsolz 495 non-null float64\nPC1 494 non-null float64\nPC2 494 non-null float64\nPC3 494 non-null float64\nPC4 494 non-null float64\nPC5 494 non-null float64\nPC6 494 non-null float64\naphy405 495 non-null float64\naphy411 495 non-null float64\naphy443 495 non-null float64\naphy455 495 non-null float64\naphy465 495 non-null float64\naphy489 495 non-null float64\naphy510 495 non-null float64\naphy520 495 non-null float64\naphy530 495 non-null float64\naphy550 495 non-null float64\naphy555 495 non-null float64\naphy560 495 non-null float64\naphy565 495 non-null float64\naphy570 495 non-null float64\naphy590 495 non-null float64\naphy619 495 non-null float64\naphy625 495 non-null float64\naphy665 495 non-null float64\naphy670 495 non-null float64\naphy683 495 non-null float64\ndtypes: float64(36)\nmemory usage: 143.1 KB\n"
],
[
"df_sat.info()",
"<class 'pandas.core.frame.DataFrame'>\nInt64Index: 495 entries, 0 to 494\nData columns (total 36 columns):\noisst 495 non-null float64\nsolz 495 non-null float64\nsat_rho_rc412 495 non-null float64\nsat_rho_rc443 495 non-null float64\nsat_rho_rc490 495 non-null float64\nsat_rho_rc510 495 non-null float64\nsat_rho_rc555 495 non-null float64\nsat_rho_rc670 495 non-null float64\naphy405 495 non-null float64\naphy411 495 non-null float64\naphy443 495 non-null float64\naphy455 495 non-null float64\naphy465 495 non-null float64\naphy489 495 non-null float64\naphy510 495 non-null float64\naphy520 495 non-null float64\naphy530 495 non-null float64\naphy550 495 non-null float64\naphy555 495 non-null float64\naphy560 495 non-null float64\naphy565 495 non-null float64\naphy570 495 non-null float64\naphy590 495 non-null float64\naphy619 495 non-null float64\naphy625 495 non-null float64\naphy665 495 non-null float64\naphy670 495 non-null float64\naphy683 495 non-null float64\nlog10_etopo2 495 non-null float64\nsin_doy 495 non-null float64\ncos_doy 495 non-null float64\nsin_minofday 495 non-null float64\ncos_minofday 495 non-null float64\nx 495 non-null float64\ny 495 non-null float64\nz 495 non-null float64\ndtypes: float64(36)\nmemory usage: 143.1 KB\n"
],
[
"df_pc = df_pc.loc[((df_pc.aphy411>0) & (df_pc.aphy443>0) & (df_pc.aphy489)\n & (df_pc.aphy510>0) & (df_pc.aphy555>0) & (df_pc.aphy670))\n ]",
"_____no_output_____"
],
[
"df_sat = df_sat.loc[((df_sat.aphy411>0) & (df_sat.aphy443>0) & (df_sat.aphy489)\n & (df_sat.aphy510>0) & (df_sat.aphy555>0) & (df_sat.aphy670))\n ]\ndf_sat = df_sat.loc[((df_sat.sat_rho_rc412>0) & (df_sat.sat_rho_rc443>0) & \n (df_sat.sat_rho_rc490>0) & (df_sat.sat_rho_rc510>0) &\n (df_sat.sat_rho_rc555>0) & (df_sat.sat_rho_rc670>0))\n ]",
"_____no_output_____"
],
[
"df_pc.loc[172:175].T",
"_____no_output_____"
],
[
"df_sat.loc[172:175].T",
"_____no_output_____"
],
[
"df_pc.dropna(inplace=True)\ndf_sat.dropna(inplace=True)",
"_____no_output_____"
],
[
"df_pc.info()",
"<class 'pandas.core.frame.DataFrame'>\nInt64Index: 163 entries, 2 to 494\nData columns (total 36 columns):\nsin_doy 163 non-null float64\ncos_doy 163 non-null float64\nsin_minofday 163 non-null float64\ncos_minofday 163 non-null float64\nx 163 non-null float64\ny 163 non-null float64\nz 163 non-null float64\nlog10_etopo2 163 non-null float64\noisst 163 non-null float64\nsolz 163 non-null float64\nPC1 162 non-null float64\nPC2 162 non-null float64\nPC3 162 non-null float64\nPC4 162 non-null float64\nPC5 162 non-null float64\nPC6 162 non-null float64\naphy405 163 non-null float64\naphy411 163 non-null float64\naphy443 163 non-null float64\naphy455 163 non-null float64\naphy465 163 non-null float64\naphy489 163 non-null float64\naphy510 163 non-null float64\naphy520 163 non-null float64\naphy530 163 non-null float64\naphy550 163 non-null float64\naphy555 163 non-null float64\naphy560 163 non-null float64\naphy565 163 non-null float64\naphy570 163 non-null float64\naphy590 163 non-null float64\naphy619 163 non-null float64\naphy625 163 non-null float64\naphy665 163 non-null float64\naphy670 163 non-null float64\naphy683 163 non-null float64\ndtypes: float64(36)\nmemory usage: 52.1 KB\n"
],
[
"sat_cols = df_sat.columns.tolist()",
"_____no_output_____"
],
[
"sat_cols_new = [col for col in sat_cols if not col.startswith('aphy')] +\\\n [col for col in sat_cols if col.startswith('aphy')]",
"_____no_output_____"
],
[
"df_sat = df_sat[sat_cols_new]",
"_____no_output_____"
],
[
"df_sat.info()",
"<class 'pandas.core.frame.DataFrame'>\nInt64Index: 162 entries, 2 to 494\nData columns (total 36 columns):\noisst 162 non-null float64\nsolz 162 non-null float64\nsat_rho_rc412 162 non-null float64\nsat_rho_rc443 162 non-null float64\nsat_rho_rc490 162 non-null float64\nsat_rho_rc510 162 non-null float64\nsat_rho_rc555 162 non-null float64\nsat_rho_rc670 162 non-null float64\nlog10_etopo2 162 non-null float64\nsin_doy 162 non-null float64\ncos_doy 162 non-null float64\nsin_minofday 162 non-null float64\ncos_minofday 162 non-null float64\nx 162 non-null float64\ny 162 non-null float64\nz 162 non-null float64\naphy405 162 non-null float64\naphy411 162 non-null float64\naphy443 162 non-null float64\naphy455 162 non-null float64\naphy465 162 non-null float64\naphy489 162 non-null float64\naphy510 162 non-null float64\naphy520 162 non-null float64\naphy530 162 non-null float64\naphy550 162 non-null float64\naphy555 162 non-null float64\naphy560 162 non-null float64\naphy565 162 non-null float64\naphy570 162 non-null float64\naphy590 162 non-null float64\naphy619 162 non-null float64\naphy625 162 non-null float64\naphy665 162 non-null float64\naphy670 162 non-null float64\naphy683 162 non-null float64\ndtypes: float64(36)\nmemory usage: 51.8 KB\n"
],
[
"df_pc.to_pickle('./pickleJar/OperationalDataSets/df_6_APHY_PC.pkl')\ndf_sat.to_pickle('./pickleJar/OperationalDataSets/df_6_APHY_SAT.pkl')",
"_____no_output_____"
],
[
"sc_pc = StandardScaler()\nsc_sat = StandardScaler()",
"_____no_output_____"
],
[
"X_s_pc = sc_pc.fit_transform(df_pc.loc[:, :'PC6'].values)\nX_s_sat = sc_sat.fit_transform(df_sat.loc[:, :'z'].values)",
"_____no_output_____"
],
[
"X_s_pc.shape, X_s_sat.shape",
"_____no_output_____"
],
[
"feat_cols_pc = df_pc.loc[:,:'PC6'].columns.tolist()\nfeat_cols_sat = df_sat.loc[:, :'z'].columns.tolist()\ndf_pc_s = pd.DataFrame(X_s_pc, columns=['%s_s' % col for col in feat_cols_pc],\n index=df_pc.index)\ndf_sat_s = pd.DataFrame(X_s_sat, columns=['%s_s' % col for col in feat_cols_sat],\n index=df_sat.index)\n",
"_____no_output_____"
],
[
"df_pc_s.head()",
"_____no_output_____"
],
[
"df_pc_s.shape, df_sat_s.shape",
"_____no_output_____"
],
[
"df_pc_s = pd.concat((df_pc_s, df_pc.filter(regex='aphy', axis=1)), axis=1)",
"_____no_output_____"
],
[
"df_pc_s.info()",
"<class 'pandas.core.frame.DataFrame'>\nInt64Index: 162 entries, 2 to 494\nData columns (total 36 columns):\nsin_doy_s 162 non-null float64\ncos_doy_s 162 non-null float64\nsin_minofday_s 162 non-null float64\ncos_minofday_s 162 non-null float64\nx_s 162 non-null float64\ny_s 162 non-null float64\nz_s 162 non-null float64\nlog10_etopo2_s 162 non-null float64\noisst_s 162 non-null float64\nsolz_s 162 non-null float64\nPC1_s 162 non-null float64\nPC2_s 162 non-null float64\nPC3_s 162 non-null float64\nPC4_s 162 non-null float64\nPC5_s 162 non-null float64\nPC6_s 162 non-null float64\naphy405 162 non-null float64\naphy411 162 non-null float64\naphy443 162 non-null float64\naphy455 162 non-null float64\naphy465 162 non-null float64\naphy489 162 non-null float64\naphy510 162 non-null float64\naphy520 162 non-null float64\naphy530 162 non-null float64\naphy550 162 non-null float64\naphy555 162 non-null float64\naphy560 162 non-null float64\naphy565 162 non-null float64\naphy570 162 non-null float64\naphy590 162 non-null float64\naphy619 162 non-null float64\naphy625 162 non-null float64\naphy665 162 non-null float64\naphy670 162 non-null float64\naphy683 162 non-null float64\ndtypes: float64(36)\nmemory usage: 51.8 KB\n"
],
[
"df_sat_s = pd.concat((df_sat_s, df_sat.filter(regex='aphy', axis=1)), axis=1)",
"_____no_output_____"
],
[
"pf_pc = PolynomialFeatures(interaction_only=True, include_bias=False)\npf_sat = PolynomialFeatures(interaction_only=True, include_bias=False)",
"_____no_output_____"
],
[
"Xsp_pc = pf_pc.fit_transform(X_s_pc)\nXsp_sat = pf_sat.fit_transform(X_s_sat)",
"_____no_output_____"
],
[
"y_aphy = df_pc.filter(regex='aphy', axis=1)",
"_____no_output_____"
],
[
"y_aphy.shape",
"_____no_output_____"
],
[
"poly_feat_nams_pc = pf_pc.get_feature_names(input_features=df_pc_s.loc[:,:'PC6_s'].columns)\npoly_df_cols_pc = poly_feat_nams_pc + y_aphy.columns.tolist()\npoly_feat_nams_sat = pf_pc.get_feature_names(input_features=df_sat_s.loc[:,:'z_s'].columns)\npoly_df_cols_sat = poly_feat_nams_sat + y_aphy.columns.tolist()",
"_____no_output_____"
],
[
"df_sp_pc = pd.DataFrame(np.c_[Xsp_pc, y_aphy], columns=poly_df_cols_pc,\n index=df_pc_s.index)",
"_____no_output_____"
],
[
"df_sp_sat = pd.DataFrame(np.c_[Xsp_sat, y_aphy], columns=poly_df_cols_sat,\n index=df_sat_s.index)",
"_____no_output_____"
],
[
"df_sp_pc.shape, df_sp_sat.shape",
"_____no_output_____"
],
[
"df_pc_s.to_pickle('./pickleJar/OperationalDataSets/df_6_APHY_Standardized_PC.pkl')\ndf_sp_pc.to_pickle('./pickleJar/OperationalDataSets/df_6_APHY_Standardized_PolyFeatures_PC.pkl')\ndf_sat_s.to_pickle('./pickleJar/OperationalDataSets/df_6_APHY_Standardized_SAT.pkl')\ndf_sp_sat.to_pickle('./pickleJar/OperationalDataSets/df_6_APHY_Standardized_PolyFeatures_SAT.pkl')",
"_____no_output_____"
],
[
"aphys = ['aphy411', 'aphy443', 'aphy489', 'aphy510', 'aphy555', 'aphy670']\ndf_s_SWF_pc = pd.concat((df_pc_s.loc[:, :'PC6_s'], df_pc_s[aphys]), axis=1)\ndf_s_SWF_sat = pd.concat((df_sat_s.loc[:, :'z_s'], df_sat_s[aphys]), axis=1)",
"_____no_output_____"
],
[
"df_sp_sat.head().T",
"_____no_output_____"
],
[
"df_sp_SWF_pc = pd.concat((df_sp_pc.loc[:, :'PC5_s PC6_s'], df_sp_pc[aphys]), axis=1)\ndf_sp_SWF_sat = pd.concat((df_sp_sat.loc[:, :'y_s z_s'], df_sp_sat[aphys]), axis=1)",
"_____no_output_____"
],
[
"df_s_SWF_pc.to_pickle('./pickleJar/OperationalDataSets/df_6_APHY_Standardized_SWF_PC.pkl')\ndf_s_SWF_sat.to_pickle('./pickleJar/OperationalDataSets/df_6_APHY_Standardized_SWF_SAT.pkl')\ndf_sp_SWF_pc.to_pickle('./pickleJar/OperationalDataSets/df_6_APHY_Standardized_PolyFeatures_SWF_PC.pkl')\ndf_sp_SWF_sat.to_pickle('./pickleJar/OperationalDataSets/df_6_APHY_Standardized_PolyFeatures_SWF_SAT.pkl')",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4afe7d9c38a2a9d22ad83e226ee3e973db4d8682
| 1,910 |
ipynb
|
Jupyter Notebook
|
Daily/Reach End.ipynb
|
jasonyum/python_go
|
910963ac0caa46cbc931b18f6b3abd10f4605f8a
|
[
"MIT"
] | 1 |
2019-04-30T23:44:35.000Z
|
2019-04-30T23:44:35.000Z
|
Daily/Reach End.ipynb
|
jasonyum/Patterns
|
910963ac0caa46cbc931b18f6b3abd10f4605f8a
|
[
"MIT"
] | null | null | null |
Daily/Reach End.ipynb
|
jasonyum/Patterns
|
910963ac0caa46cbc931b18f6b3abd10f4605f8a
|
[
"MIT"
] | null | null | null | 28.507463 | 270 | 0.563351 |
[
[
[
"# Traveling to the End\n\nYou are given an array of nonnegative integers. Let's say you start at the beginning of the array and are trying to advance to the end. You can advance at most, the number of steps that you're currently on. Determine whether you can get to the end of the array.\n\nFor example, given the array [1, 3, 1, 2, 0, 1], we can go from indices 0 -> 1 -> 3 -> 5, so return true.\n\nGiven the array [1, 2, 1, 0, 0], we can't reach the end, so return false.\n\n### solution idea...\n- keep track of the absolute furthest step we can reach. \n- then compute the furthest step we can reach and all the steps in between as well\n- each step is stateless and gets \"reset\" whenever we land on a new index\n- break if we are \"looking\" past the furthest step",
"_____no_output_____"
]
],
[
[
"def reach_end(arr): \n furthest_so_far = 0 \n for i in range(len(arr)): \n if i > furthest_so_far: \n break\n furthest_so_far = max(furthest_so_far, i + arr[i])\n return furthest_so_far >= len(arr) - 1",
"_____no_output_____"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
]
] |
4afec60a386f16f3a0e388c7918d9a83b425ebea
| 295,450 |
ipynb
|
Jupyter Notebook
|
Lambda_Labs_SP500_Analysis.ipynb
|
labs13-quake-viewer/ds-notebooks
|
3b649ff6d51ede244652b008cb49c218e9a96c54
|
[
"MIT"
] | null | null | null |
Lambda_Labs_SP500_Analysis.ipynb
|
labs13-quake-viewer/ds-notebooks
|
3b649ff6d51ede244652b008cb49c218e9a96c54
|
[
"MIT"
] | 1 |
2019-06-10T15:18:55.000Z
|
2019-06-10T15:42:05.000Z
|
Lambda_Labs_SP500_Analysis.ipynb
|
labs13-quake-viewer/ds-notebooks
|
3b649ff6d51ede244652b008cb49c218e9a96c54
|
[
"MIT"
] | 1 |
2022-01-28T19:34:19.000Z
|
2022-01-28T19:34:19.000Z
| 295,450 | 295,450 | 0.88025 |
[
[
[
"import pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nimport datetime",
"_____no_output_____"
],
[
"sp500 = pd.read_csv(\"https://raw.githubusercontent.com/labs13-quake-viewer/ds-data/master/S%26P%20500.csv\")\nsp500.shape",
"_____no_output_____"
],
[
"sp500.describe()",
"_____no_output_____"
],
[
"(sp500 == 0).sum()",
"_____no_output_____"
],
[
"sp500.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 17465 entries, 0 to 17464\nData columns (total 2 columns):\nDate 17465 non-null object\nClose 17465 non-null float64\ndtypes: float64(1), object(1)\nmemory usage: 273.0+ KB\n"
],
[
"sp500.head(12)",
"_____no_output_____"
],
[
"dates=[]\nfor i in sp500['Date']:\n dd = datetime.datetime.strptime(i,'%m/%d/%y')\n if dd.year > 2019:\n dd = dd.replace(year=dd.year-100)\n dates.append(dd.strftime( '%Y-%m-%d' ))\nsp500['Date'] = dates",
"_____no_output_____"
],
[
"sp500.plot(x=\"Date\", y=\"Close\", figsize=(20, 10));",
"_____no_output_____"
],
[
"df_sp500 = sp500.set_index('Date')",
"_____no_output_____"
],
[
"df_sp500.info()",
"<class 'pandas.core.frame.DataFrame'>\nIndex: 17465 entries, 1950-01-03 to 2019-05-31\nData columns (total 1 columns):\nClose 17465 non-null float64\ndtypes: float64(1)\nmemory usage: 272.9+ KB\n"
],
[
"df_sp500.head(12)",
"_____no_output_____"
],
[
"quakes = pd.read_csv(\"https://raw.githubusercontent.com/labs13-quake-viewer/ds-data/master/Earthquakes%205.5%201900-present.csv\")\nquakes.shape",
"_____no_output_____"
],
[
"quakes.describe()",
"_____no_output_____"
],
[
"quakes.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 30251 entries, 0 to 30250\nData columns (total 22 columns):\ntime 30251 non-null object\nlatitude 30251 non-null float64\nlongitude 30251 non-null float64\ndepth 29997 non-null float64\nmag 30251 non-null float64\nmagType 30251 non-null object\nnst 7109 non-null float64\ngap 8354 non-null float64\ndmin 2672 non-null float64\nrms 18486 non-null float64\nnet 30251 non-null object\nid 30251 non-null object\nupdated 30251 non-null object\nplace 30248 non-null object\ntype 30251 non-null object\nhorizontalError 2234 non-null float64\ndepthError 5610 non-null float64\nmagError 1369 non-null float64\nmagNst 3611 non-null float64\nstatus 30251 non-null object\nlocationSource 30251 non-null object\nmagSource 30251 non-null object\ndtypes: float64(12), object(10)\nmemory usage: 5.1+ MB\n"
],
[
"quakes.sample(12)",
"_____no_output_____"
],
[
"pd.options.mode.chained_assignment = None # default='warn'\ndf_quakes = quakes[['time', 'mag', 'latitude', 'longitude']]\ndf_quakes.time = df_quakes.time.str[:10]\ndf_quakes = df_quakes.sort_values(by=['time'])\ndf_quakes.info()",
"<class 'pandas.core.frame.DataFrame'>\nInt64Index: 30251 entries, 30250 to 0\nData columns (total 4 columns):\ntime 30251 non-null object\nmag 30251 non-null float64\nlatitude 30251 non-null float64\nlongitude 30251 non-null float64\ndtypes: float64(3), object(1)\nmemory usage: 1.2+ MB\n"
],
[
"df = df_quakes[df_quakes['time'] >= \"1950-01-03\"]\ndf_quakes = df[df['mag'] >= 6.7]\ndf_quakes = df_quakes.reset_index(drop=True)\ndf_quakes.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 1873 entries, 0 to 1872\nData columns (total 4 columns):\ntime 1873 non-null object\nmag 1873 non-null float64\nlatitude 1873 non-null float64\nlongitude 1873 non-null float64\ndtypes: float64(3), object(1)\nmemory usage: 58.6+ KB\n"
],
[
"df_quakes.head(12)",
"_____no_output_____"
],
[
"df_quakes.plot(x=\"time\", y=\"mag\", figsize=(20, 10));",
"_____no_output_____"
],
[
"def get_next_trading_day(date_in):\n t1 = date_in\n dayz = 1\n while t1 not in df_sp500.index:\n t1 = pd.to_datetime(date_in).date() + datetime.timedelta(days=dayz)\n t1 = t1.strftime('%Y-%m-%d')\n dayz += 1\n return t1 ",
"_____no_output_____"
],
[
"import datetime\ni = -1\ndata = []\nfor _, row in df_quakes.iterrows():\n\n i += 1\n \n if row.time > \"2019-04-15\":\n continue\n \n t0 = get_next_trading_day(row.time)\n \n t1 = pd.to_datetime(row.time).date() + datetime.timedelta(days=7)\n t1 = t1.strftime('%Y-%m-%d')\n t1 = get_next_trading_day(t1)\n\n t2 = pd.to_datetime(row.time).date() + datetime.timedelta(days=14)\n t2 = t2.strftime('%Y-%m-%d')\n t2 = get_next_trading_day(t2)\n \n t3 = pd.to_datetime(row.time).date() + datetime.timedelta(days=30)\n t3 = t3.strftime('%Y-%m-%d')\n t3 = get_next_trading_day(t3)\n\n x = (row.time, row.mag, df_sp500.loc[t0].Close, df_sp500.loc[t1].Close,\n df_sp500.loc[t2].Close, df_sp500.loc[t3].Close)\n \n data.append(x)",
"_____no_output_____"
],
[
"df_quake_sp500 = pd.DataFrame(data=data, columns=['Date', 'Mag', 'Price_Day_0', 'Price_Day_7', 'Price_Day_14', 'Price_Day_30'])",
"_____no_output_____"
],
[
"df_quake_sp500[\"Appr_Day_7\"] = 100 * (df_quake_sp500[\"Price_Day_7\"] - df_quake_sp500[\"Price_Day_0\"]) / df_quake_sp500[\"Price_Day_0\"]\ndf_quake_sp500[\"Appr_Day_14\"] = 100 * (df_quake_sp500[\"Price_Day_14\"] - df_quake_sp500[\"Price_Day_0\"]) / df_quake_sp500[\"Price_Day_0\"]\ndf_quake_sp500[\"Appr_Day_30\"] = 100 * (df_quake_sp500[\"Price_Day_30\"] - df_quake_sp500[\"Price_Day_0\"]) / df_quake_sp500[\"Price_Day_0\"]",
"_____no_output_____"
],
[
"df_quake_sp500.describe()",
"_____no_output_____"
],
[
"df = df_quake_sp500.groupby(['Mag'])['Appr_Day_30'].mean()\ndf.plot.bar(figsize=(20, 10));",
"_____no_output_____"
],
[
"df_quake_sp500.Mag.value_counts().sort_index()",
"_____no_output_____"
],
[
"df_quakes_usa = df_quakes.query('32.5 <= latitude < 48.75').query('-123 <= longitude < -69')\ndf_quakes_usa.info()",
"<class 'pandas.core.frame.DataFrame'>\nInt64Index: 13 entries, 53 to 1317\nData columns (total 4 columns):\ntime 13 non-null object\nmag 13 non-null float64\nlatitude 13 non-null float64\nlongitude 13 non-null float64\ndtypes: float64(3), object(1)\nmemory usage: 520.0+ bytes\n"
],
[
"df_quakes_usa",
"_____no_output_____"
],
[
"i = -1\ndata = []\nfor _, row in df_quakes_usa.iterrows():\n\n i += 1\n \n if row.time > \"2019-04-15\":\n continue\n \n t0 = get_next_trading_day(row.time)\n \n t1 = pd.to_datetime(row.time).date() + datetime.timedelta(days=7)\n t1 = t1.strftime('%Y-%m-%d')\n t1 = get_next_trading_day(t1)\n\n t2 = pd.to_datetime(row.time).date() + datetime.timedelta(days=14)\n t2 = t2.strftime('%Y-%m-%d')\n t2 = get_next_trading_day(t2)\n \n t3 = pd.to_datetime(row.time).date() + datetime.timedelta(days=30)\n t3 = t3.strftime('%Y-%m-%d')\n t3 = get_next_trading_day(t3)\n\n x = (row.time, row.mag, df_sp500.loc[t0].Close, df_sp500.loc[t1].Close,\n df_sp500.loc[t2].Close, df_sp500.loc[t3].Close)\n \n data.append(x)\ndf_quake_sp500_usa = pd.DataFrame(data=data, columns=['Date', 'Mag', 'Price_Day_0', 'Price_Day_7', 'Price_Day_14', 'Price_Day_30'])\ndf_quake_sp500_usa[\"Appr_Day_7\"] = 100 * (df_quake_sp500_usa[\"Price_Day_7\"] - df_quake_sp500_usa[\"Price_Day_0\"]) / df_quake_sp500_usa[\"Price_Day_0\"]\ndf_quake_sp500_usa[\"Appr_Day_14\"] = 100 * (df_quake_sp500_usa[\"Price_Day_14\"] - df_quake_sp500_usa[\"Price_Day_0\"]) / df_quake_sp500_usa[\"Price_Day_0\"]\ndf_quake_sp500_usa[\"Appr_Day_30\"] = 100 * (df_quake_sp500_usa[\"Price_Day_30\"] - df_quake_sp500_usa[\"Price_Day_0\"]) / df_quake_sp500_usa[\"Price_Day_0\"]",
"_____no_output_____"
],
[
"df = df_quake_sp500_usa.groupby(['Mag'])['Appr_Day_30'].mean()\ndf.plot.bar(figsize=(20, 10));",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4afec98d9e875da92ef98a5e21f386fdeb07d304
| 6,259 |
ipynb
|
Jupyter Notebook
|
notebooks/controls/python.ipynb
|
LucienZhang/website-binder
|
76148742640d77dfa33ed3af34c170fe8145c11d
|
[
"MIT"
] | null | null | null |
notebooks/controls/python.ipynb
|
LucienZhang/website-binder
|
76148742640d77dfa33ed3af34c170fe8145c11d
|
[
"MIT"
] | null | null | null |
notebooks/controls/python.ipynb
|
LucienZhang/website-binder
|
76148742640d77dfa33ed3af34c170fe8145c11d
|
[
"MIT"
] | null | null | null | 16.471053 | 50 | 0.393993 |
[
[
[
"## Version",
"_____no_output_____"
]
],
[
[
"import sys\nprint(sys.version)",
"3.8.3 (default, May 19 2020, 18:47:26) \n[GCC 7.3.0]\n"
]
],
[
[
"## Loop Structure",
"_____no_output_____"
]
],
[
[
"range(3)",
"_____no_output_____"
],
[
"list(range(3))",
"_____no_output_____"
],
[
"list(range(1, 4))",
"_____no_output_____"
],
[
"list(range(5, 2, -1))",
"_____no_output_____"
],
[
"sum(range(4))",
"_____no_output_____"
],
[
"for i in range(3):\n print(i)",
"0\n1\n2\n"
],
[
"for i in range(3):\n if i == 1:\n ...\n if i == 5:\n break\nelse:\n print('not found')",
"not found\n"
],
[
"for i in range(3):\n if i == 1:\n break\nelse:\n print('not found')",
"_____no_output_____"
],
[
"for i in range(3):\n if i == 1:\n continue\n if i == 1:\n break\nelse:\n print('not found')",
"not found\n"
],
[
"i = 0\nwhile (j := i+1) < 4:\n if i == 1:\n pass\n if i == 2:\n i += 1\n continue\n print(i, j)\n if i == 5:\n break\n i += 1\nelse:\n print('not found')",
"0 1\n1 2\nnot found\n"
],
[
"key = 'abc'\nval = [1, 2, 3]\nfor k, v in zip(key, val):\n print(f'{k = }, {v = }')",
"k = 'a', v = 1\nk = 'b', v = 2\nk = 'c', v = 3\n"
],
[
"[x for x in range(5) if x % 2 == 0]",
"_____no_output_____"
],
[
"[x if x % 2 == 0 else -x for x in range(5)]",
"_____no_output_____"
]
],
[
[
"## Branch Structure",
"_____no_output_____"
]
],
[
[
"i = 3\nif i > 5:\n print('i > 5')\nelif i > 3:\n print('i > 3')\nelse:\n print('i <= 3')",
"i <= 3\n"
],
[
"a = list(range(5))\nif (n := len(a)) > 3:\n print(f'the length is {n}')",
"the length is 5\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
4afeca75fb341748cc354c0b51abdbeb73faaec8
| 3,908 |
ipynb
|
Jupyter Notebook
|
Ear_Gait_Recognition/.ipynb_checkpoints/Untitled-checkpoint.ipynb
|
JunghyoSohn/Human-Recognition-Using-Gait-and-Ear
|
e0607db72c685ea07bb9f57343ca7ec1378007a7
|
[
"MIT"
] | 1 |
2021-06-25T10:22:56.000Z
|
2021-06-25T10:22:56.000Z
|
Ear_Gait_Recognition/.ipynb_checkpoints/Untitled-checkpoint.ipynb
|
JunghyoSohn/Human-Recognition-Using-Gait-and-Ear
|
e0607db72c685ea07bb9f57343ca7ec1378007a7
|
[
"MIT"
] | null | null | null |
Ear_Gait_Recognition/.ipynb_checkpoints/Untitled-checkpoint.ipynb
|
JunghyoSohn/Human-Recognition-Using-Gait-and-Ear
|
e0607db72c685ea07bb9f57343ca7ec1378007a7
|
[
"MIT"
] | null | null | null | 39.474747 | 1,260 | 0.635619 |
[
[
[
"import tensorflow as tf\nimport pandas as pd\nimport sys\nimport matplotlib.pyplot as plt\nimport numpy as np\nimport os",
"/home/pirl/anaconda3/lib/python3.6/site-packages/h5py/__init__.py:36: FutureWarning: Conversion of the second argument of issubdtype from `float` to `np.floating` is deprecated. In future, it will be treated as `np.float64 == np.dtype(float).type`.\n from ._conv import register_converters as _register_converters\n"
],
[
"train_data = pd.read_csv(\"./Test_Data/Ear_test_data.txt\", names=['image'])\ntrain_image = list(train_data['image'])\nprint(train_image)",
"['./Ear_image/Test/After_preprocess/0020.jpg.png', './Ear_image/Test/After_preprocess/0009.jpg.png', './Ear_image/Test/After_preprocess/0004.jpg.png', './Ear_image/Test/After_preprocess/0003.jpg.png', './Ear_image/Test/After_preprocess/0008.jpg.png', './Ear_image/Test/After_preprocess/0025.jpg.png', './Ear_image/Test/After_preprocess/0007.jpg.png', './Ear_image/Test/After_preprocess/0013.jpg.png', './Ear_image/Test/After_preprocess/0015.jpg.png', './Ear_image/Test/After_preprocess/0023.jpg.png', './Ear_image/Test/After_preprocess/0012.jpg.png', './Ear_image/Test/After_preprocess/0014.jpg.png', './Ear_image/Test/After_preprocess/0005.jpg.png', './Ear_image/Test/After_preprocess/0021.jpg.png', './Ear_image/Test/After_preprocess/0002.jpg.png', './Ear_image/Test/After_preprocess/0024.jpg.png', './Ear_image/Test/After_preprocess/0017.jpg.png', './Ear_image/Test/After_preprocess/0018.jpg.png', './Ear_image/Test/After_preprocess/0022.jpg.png', './Ear_image/Test/After_preprocess/0016.jpg.png', './Ear_image/Test/After_preprocess/0010.jpg.png', './Ear_image/Test/After_preprocess/0001.jpg.png', './Ear_image/Test/After_preprocess/0000.jpg.png', './Ear_image/Test/After_preprocess/0011.jpg.png', './Ear_image/Test/After_preprocess/0006.jpg.png']\n"
],
[
"data_queue = tf.train.slice_input_producer([train_image], num_epochs=None, shuffle=True)\n\nsess = tf.Session()\ncoord = tf.train.Coordinator()\nthreads = tf.train.start_queue_runners(sess=sess, coord=coord)\nsess.run(tf.global_variables_initializer())\ntype(sess.run(data_queue))",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code"
]
] |
4afed132f3866bdd50641f1a78f658cbf329ef13
| 3,670 |
ipynb
|
Jupyter Notebook
|
website/docs/example.ipynb
|
SamEdwardes/spaCyTextBlob
|
3ea8842c22fd6eb06f78123f25dcf59f3caddae2
|
[
"MIT"
] | 20 |
2020-11-19T13:50:45.000Z
|
2022-02-16T17:55:23.000Z
|
website/docs/example.ipynb
|
SamEdwardes/spaCyTextBlob
|
3ea8842c22fd6eb06f78123f25dcf59f3caddae2
|
[
"MIT"
] | 15 |
2020-11-12T05:42:52.000Z
|
2022-02-17T05:25:58.000Z
|
website/docs/example.ipynb
|
SamEdwardes/spaCyTextBlob
|
3ea8842c22fd6eb06f78123f25dcf59f3caddae2
|
[
"MIT"
] | 4 |
2020-11-18T07:24:45.000Z
|
2021-04-13T15:23:44.000Z
| 23.986928 | 195 | 0.550681 |
[
[
[
"---\nid: example\ntitle: Examples\n---",
"_____no_output_____"
]
],
[
[
"## Using on a single text",
"_____no_output_____"
]
],
[
[
"import spacy\nfrom spacytextblob.spacytextblob import SpacyTextBlob\n\nnlp = spacy.load('en_core_web_sm')\nnlp.add_pipe(\"spacytextblob\")\n\ntext = \"I had a really horrible day. It was the worst day ever! But every now and then I have a really good day that makes me happy.\"\ndoc = nlp(text)",
"_____no_output_____"
]
],
[
[
"By adding the pipeline, the extensions `._.polarity`, `._.subjectivity`, and `._.assessments` will be added to `Doc`, `Span`, and `Token` objects. You can assess specific details below:\n- `.polarity`: a float within the range (-1.0, 1.0)\n- `.subjectivity`: a float within the range (0.0, 1.0) where 0.0 is very objective and 1.0 is very subjective\n- `.assessments`: a list of polarity and subjectivity scores for the assessed tokens.",
"_____no_output_____"
]
],
[
[
"print(doc._.polarity)",
"_____no_output_____"
],
[
"print(doc._.subjectivity)",
"_____no_output_____"
],
[
"print(doc._.assessments)",
"_____no_output_____"
]
],
[
[
"You can identify the sentiment at the `Span` or `Token` level.",
"_____no_output_____"
]
],
[
[
"for token in doc:\n print(token.text, token._.polarity, token._.subjectivity)",
"_____no_output_____"
],
[
"for span in doc.sents:\n print(span.text, span._.polarity, span._.subjectivity)",
"_____no_output_____"
]
],
[
[
"## Using on a multiple texts",
"_____no_output_____"
]
],
[
[
"import spacy\nfrom spacytextblob.spacytextblob import SpacyTextBlob\n\nnlp = spacy.load('en_core_web_sm')\nnlp.add_pipe(\"spacytextblob\")\n\ntext1 = \"I had a really horrible day. It was the worst day ever! But every now and then I have a really good day that makes me happy.\"\ntext2 = \"Wow I had just the best day ever today! I cannot wait to tell the world.\"\ndocs = list(nlp.pipe([text1, text2]))\nfor doc in docs:\n print('=' * 64)\n print(doc.text)\n print('Polarity:', doc._.polarity)\n print('Sujectivity:', doc._.subjectivity)\n print('Assessments:', doc._.assessments)",
"_____no_output_____"
]
]
] |
[
"raw",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"raw"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
4afedde7b99bca8457084732c15f8e141400abbe
| 364,540 |
ipynb
|
Jupyter Notebook
|
02-tensorflow-data-and-deployment/04-tensorflow-deployment/02-tensorflow-hub/01_tfhub_basic.ipynb
|
pedro-abundio-wang/tensorflow-specialization
|
46ef3d342957d673143a2a17d0faf67a0c49fdb5
|
[
"Apache-2.0"
] | null | null | null |
02-tensorflow-data-and-deployment/04-tensorflow-deployment/02-tensorflow-hub/01_tfhub_basic.ipynb
|
pedro-abundio-wang/tensorflow-specialization
|
46ef3d342957d673143a2a17d0faf67a0c49fdb5
|
[
"Apache-2.0"
] | null | null | null |
02-tensorflow-data-and-deployment/04-tensorflow-deployment/02-tensorflow-hub/01_tfhub_basic.ipynb
|
pedro-abundio-wang/tensorflow-specialization
|
46ef3d342957d673143a2a17d0faf67a0c49fdb5
|
[
"Apache-2.0"
] | null | null | null | 441.331719 | 176,324 | 0.943348 |
[
[
[
"# Getting Started with TensorFlow Hub\n\n[TensorFlow Hub](https://tfhub.dev/) is a repository of reusable TensorFlow machine learning modules. A module is a self-contained piece of a TensorFlow graph, along with its weights and assets, that can be reused across different tasks. These modules can be reused to solve new tasks with less training data, diminishing training time. \n\nIn this notebook we will go over some basic examples to help you get started with TensorFlow Hub. In particular, we will cover the following topics:\n\n* Loading TensorFlow Hub Modules and Performing Inference.\n\n* Using TensorFlow Hub Modules with Keras.\n\n* Using Feature Vectors with Keras for Transfer Learning.\n\n* Saving and Running a TensorFlow Hub Module Locally.\n\n* Changing the Download Location of TensorFlow Hub Modules. ",
"_____no_output_____"
],
[
"## Setup",
"_____no_output_____"
]
],
[
[
"try:\n %tensorflow_version 2.x\nexcept:\n pass",
"_____no_output_____"
],
[
"import os\nimport numpy as np\nimport matplotlib.pyplot as plt\n\nimport tensorflow as tf\nimport tensorflow_hub as hub\n\nfrom PIL import Image\n\n\nprint(\"\\u2022 Using TensorFlow Version:\", tf.__version__)",
"• Using TensorFlow Version: 2.0.0\n"
]
],
[
[
"## Download Test Image\n\nWe will download the image of a puppy to test our TensorFlow Hub modules.",
"_____no_output_____"
]
],
[
[
"!wget -O dog.jpeg https://cdn.pixabay.com/photo/2016/12/13/05/15/puppy-1903313_960_720.jpg\n \noriginal_image = Image.open('./dog.jpeg')",
"--2021-02-06 21:10:37-- https://cdn.pixabay.com/photo/2016/12/13/05/15/puppy-1903313_960_720.jpg\nResolving cdn.pixabay.com (cdn.pixabay.com)... 104.18.21.183, 104.18.20.183, 2606:4700::6812:15b7, ...\nConnecting to cdn.pixabay.com (cdn.pixabay.com)|104.18.21.183|:443... connected.\nHTTP request sent, awaiting response... 200 OK\nLength: 85624 (84K) [image/jpeg]\nSaving to: ‘dog.jpeg’\n\ndog.jpeg 100%[===================>] 83.62K 21.6KB/s in 3.9s \n\n2021-02-06 21:10:54 (21.6 KB/s) - ‘dog.jpeg’ saved [85624/85624]\n\n"
]
],
[
[
"Let's take a look at the image we just downloaded.",
"_____no_output_____"
]
],
[
[
"plt.figure(figsize=(6,6))\nplt.imshow(original_image)\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Format Image\n\nWe will now resize and normalize our image so that is compatible with the module we are going to use. In this notebook we will use the [MobileNet](https://tfhub.dev/google/tf2-preview/mobilenet_v2/classification/4) model which was trained in ImageNet. For this module, the input images are expected to have color values in the range `[0,1]` and to have an input size of `(224,224)`.",
"_____no_output_____"
]
],
[
[
"IMAGE_SIZE = (224, 224)\n \nimg = original_image.resize(IMAGE_SIZE)\nimg = np.array(img) / 255.0",
"_____no_output_____"
]
],
[
[
"Let's now plot the reformatted image, to see what it looks like.",
"_____no_output_____"
]
],
[
[
"plt.figure(figsize=(5,5))\nplt.imshow(img)\nplt.title('New Image Size: {}'.format(img.shape), fontdict={'size': 16}, color='green')\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Get ImageNet Labels\n\nWe will now get the labels for all the 1001 classes in the ImageNet dataset.",
"_____no_output_____"
]
],
[
[
"!wget -O labels.txt --quiet https://storage.googleapis.com/download.tensorflow.org/data/ImageNetLabels.txt\n\nwith open('labels.txt', 'r') as f:\n labels = [l.strip() for l in f.readlines()]\n\n# get number of labels\nnum_classes = len(labels)\n\nprint('There are a total of {0} labels representing {0} classes.\\n'.format(num_classes))",
"There are a total of 1001 labels representing 1001 classes.\n\n"
]
],
[
[
"Let's take a look at the first 5 labels. ",
"_____no_output_____"
]
],
[
[
"for label in labels[0:5]:\n print(label)",
"background\ntench\ngoldfish\ngreat white shark\ntiger shark\n"
]
],
[
[
"## Loading a TensorFlow Hub Module\n\nTo load a module, we use its unique **module handle**, which is just a URL string. To obtain the module handle, we have to browse through the catalog of modules in the [TensorFlow Hub](https://tfhub.dev/) website. \n\nFor example, in this case, we will be using the complete **MobileNet** model. If we go to [MobileNet's webpage](https://tfhub.dev/google/tf2-preview/mobilenet_v2/classification/4) in the TensorFlow Hub website, we will see that the module handle for this module is:\n\n```\n'https://tfhub.dev/google/tf2-preview/mobilenet_v2/classification/4'\n```\n\nFinally, we'll make use of TensorFlow Hub's, `load` API to load the module into memory. ",
"_____no_output_____"
]
],
[
[
"MODULE_HANDLE = 'https://tfhub.dev/google/tf2-preview/mobilenet_v2/classification/4'\nmodule = hub.load(MODULE_HANDLE)",
"_____no_output_____"
]
],
[
[
"## Performing Inference\n\nOnce we have loaded the module, we can then start running inference on it. Note however, that the module generates the final layer's logits without any activations. Therefore, we have to apply the `softmax` activation to the module's output. The result will be a Tensor of shape `(1, 1001)`, where the first dimension refers to the batch size. In this case it is just `1` because we only passed 1 image. \n\nIn the cell below, we will pass the image of the puppy and get the top 5 predictions from our model along with their probability scores.",
"_____no_output_____"
]
],
[
[
"predictions = tf.nn.softmax(module([img]))[0]\n\ntop_k_pred_values, top_k_indices = tf.math.top_k(predictions, k=5)\n\ntop_k_pred_values = top_k_pred_values.numpy()\ntop_k_indices = top_k_indices.numpy()\n\nfor value, i in zip(top_k_pred_values, top_k_indices):\n print('{}: {:.3}'.format(labels[i], value))",
"Labrador retriever: 0.45\nkuvasz: 0.113\nGreat Pyrenees: 0.0352\nice bear: 0.0217\ndalmatian: 0.0162\n"
]
],
[
[
"## Using a TensorFlow Hub Module with Keras\n\nWe can also integrate TensorFlow Hub modules into the high level Keras API. In this case, we make use of the `hub.KerasLayer` API to load it. We can add the `hub.KerasLayer` to a Keras `sequential` model along with an activation layer. Once the model is built, all the Keras model methods can be accessed like you would normally do in Keras.",
"_____no_output_____"
]
],
[
[
"model = tf.keras.Sequential([\n hub.KerasLayer(MODULE_HANDLE, input_shape=IMAGE_SIZE + (3,)),\n tf.keras.layers.Activation('softmax')\n])",
"_____no_output_____"
]
],
[
[
"## Performing Inference\n\nTo perform inference with the Keras model, we have to add a dimension to our image to account for the batch size. Remember that our Keras model expects the input to have shape `(batch_size, image_size)`, where the `image_size` includes the number of color channels.",
"_____no_output_____"
]
],
[
[
"# Add batch dimension\nimg_arr = np.expand_dims(img, axis=0)",
"_____no_output_____"
]
],
[
[
"As we did previously, in the cell below we will pass the image of the puppy and get the top 5 predictions from our Keras model along with their probability scores.",
"_____no_output_____"
]
],
[
[
"predictions = model.predict(img_arr)[0]\n \ntop_k_pred_values, top_k_indices = tf.math.top_k(predictions, k=5)\n\ntop_k_pred_values = top_k_pred_values.numpy()\ntop_k_indices = top_k_indices.numpy()\n\nfor value, i in zip(top_k_pred_values, top_k_indices):\n print('{}: {:.3}'.format(labels[i], value))",
"_____no_output_____"
]
],
[
[
"# Using Feature Vectors with Keras\n\nWhile we can use complete models as we did in the previous section, perhaps, the most important part of TensorFlow Hub is in how it provides **Feature Vectors** that allows us to take advantage of transfer learning. Feature vectors are just complete modules that had their final classification head removed.\n\nIn the cell below we show an example of how a feature vector can be added to a Keras `sequential` model.",
"_____no_output_____"
]
],
[
[
"MODULE_HANDLE =\"https://tfhub.dev/google/tf2-preview/mobilenet_v2/feature_vector/4\"",
"_____no_output_____"
],
[
"# Number of classes in the new dataset\nNUM_CLASSES = 20\n\nmodel = tf.keras.Sequential([\n hub.KerasLayer(MODULE_HANDLE, input_shape=IMAGE_SIZE + (3,)),\n tf.keras.layers.Dense(NUM_CLASSES, activation='softmax')\n])",
"_____no_output_____"
]
],
[
[
"Now that the model is built, the next step in transfer learning will be to train the model on a new dataset with the new classifier (i.e. the last layer of the model). Remember that the number of output units in the last layer will correspond to the number of classes in your new dataset. After the model has been trained, we can perform inference in the same way as with any Keras model (see previous section).",
"_____no_output_____"
],
[
"# Saving a TensorFlow Hub Module for Local Use\n\nWe can download TensorFlow Hub modules, by explicitly downloading the module as a **SavedModel** archived as a tarball. This is useful if we want to work with the module offline.\n\nTo do this, we first have to download the Hub module by appending a query parameter to the module handled URL string. This is done by setting the TF Hub format query parameter as shown below. For now, only the compressed option is defined.",
"_____no_output_____"
]
],
[
[
"MODULE_HANDLE = 'https://tfhub.dev/google/tf2-preview/mobilenet_v2/classification/4?tf-hub-format=compressed'\n!wget -O ./saved_model.tar.gz $MODULE_HANDLE",
"_____no_output_____"
]
],
[
[
"Next, we need to decompress the tarball.",
"_____no_output_____"
]
],
[
[
"# Untar the tarball\n!mkdir -p ./saved_model\n!tar xvzf ./saved_model.tar.gz -C ./saved_model",
"./\r\n./assets/\r\n./saved_model.pb\r\n./variables/\r\n./variables/variables.index\r\n./variables/variables.data-00000-of-00001\r\n"
]
],
[
[
"# Running a TensorFlow Hub Module Locally\n\nWe can load the SavedModel containing the saved TensorFlow Hub module by using `hub.load`.",
"_____no_output_____"
]
],
[
[
"module = hub.load('./saved_model')",
"_____no_output_____"
]
],
[
[
"After the TensorFlow Hub module is loaded, we can start making inferences as shown below. As before, we will pass the image of the puppy and get the top 5 predictions from our model along with their probability scores.",
"_____no_output_____"
]
],
[
[
"predictions = tf.nn.softmax(module([img]))[0]\n\ntop_k_pred_values, top_k_indices = tf.math.top_k(predictions, k=5)\n\ntop_k_pred_values = top_k_pred_values.numpy()\ntop_k_indices = top_k_indices.numpy()\n\nfor value, i in zip(top_k_pred_values, top_k_indices):\n print('{}: {:.3}'.format(labels[i], value))",
"Labrador retriever: 0.45\nkuvasz: 0.113\nGreat Pyrenees: 0.0352\nice bear: 0.0217\ndalmatian: 0.0162\n"
]
],
[
[
"## Changing the Download Location of TensorFlow Hub Modules. \n\nFinally, we can change the download location of TensorFlow Hub modules to a more permanent location. We can do this by setting the environment variable `'TFHUB_CACHE_DIR'` to the directory we want our modules to be saved in. \n\nIn Python, we can set this environment variable in the environment dictionary that's present in the Pythons `os` module as you can see below. ",
"_____no_output_____"
]
],
[
[
"hub_cache_dir = './hub_cache_dir'\n\nos.environ['TFHUB_CACHE_DIR'] = hub_cache_dir",
"_____no_output_____"
]
],
[
[
"Once we set the new location of the TF Hub cache directory environment variable, all the subsequent modules that we request will get downloaded to that location. ",
"_____no_output_____"
]
],
[
[
"MODULE_HANDLE = 'https://tfhub.dev/google/tf2-preview/mobilenet_v2/classification/4'\nmodule = hub.load(MODULE_HANDLE)",
"_____no_output_____"
]
],
[
[
"We can take a look the contents of the new directory and all its subdirectories by using the `-R` option.",
"_____no_output_____"
]
],
[
[
"!ls -R {hub_cache_dir}",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
4afeed2c6a4136f08655759d28d7c28723789857
| 5,943 |
ipynb
|
Jupyter Notebook
|
Julia_Intro/notebooks/07.Packages.ipynb
|
hossainlab/Julia_Learning
|
47758566a53a7156a8578b3247a4f12c37aef687
|
[
"MIT"
] | null | null | null |
Julia_Intro/notebooks/07.Packages.ipynb
|
hossainlab/Julia_Learning
|
47758566a53a7156a8578b3247a4f12c37aef687
|
[
"MIT"
] | null | null | null |
Julia_Intro/notebooks/07.Packages.ipynb
|
hossainlab/Julia_Learning
|
47758566a53a7156a8578b3247a4f12c37aef687
|
[
"MIT"
] | null | null | null | 24.557851 | 259 | 0.567727 |
[
[
[
"empty"
]
]
] |
[
"empty"
] |
[
[
"empty"
]
] |
4afef2c3905acb6a9649b5b53f7c56da82772529
| 20,594 |
ipynb
|
Jupyter Notebook
|
amftrack/notebooks/analysis/check_general_stats.ipynb
|
Cocopyth/MscThesis
|
60162bc779a3a668e7447b60bb9a4b2a616b8093
|
[
"MIT"
] | 1 |
2021-06-10T02:51:53.000Z
|
2021-06-10T02:51:53.000Z
|
amftrack/notebooks/analysis/check_general_stats.ipynb
|
Cocopyth/MscThesis
|
60162bc779a3a668e7447b60bb9a4b2a616b8093
|
[
"MIT"
] | null | null | null |
amftrack/notebooks/analysis/check_general_stats.ipynb
|
Cocopyth/MscThesis
|
60162bc779a3a668e7447b60bb9a4b2a616b8093
|
[
"MIT"
] | null | null | null | 39.527831 | 1,505 | 0.467806 |
[
[
[
"%matplotlib widget\n\nimport os \nimport sys \nsys.path.insert(0, os.getenv('HOME')+'/pycode/MscThesis/')\nimport pandas as pd\nfrom amftrack.util import get_dates_datetime, get_dirname, get_plate_number, get_postion_number\n\nimport ast\nfrom amftrack.plotutil import plot_t_tp1\nfrom scipy import sparse\nfrom datetime import datetime\nfrom amftrack.pipeline.functions.node_id import orient\nimport pickle\nimport scipy.io as sio\nfrom pymatreader import read_mat\nfrom matplotlib import colors\nimport cv2\nimport imageio\nimport matplotlib.pyplot as plt\nimport numpy as np\nfrom skimage.filters import frangi\nfrom skimage import filters\nfrom random import choice\nimport scipy.sparse\nimport os\nfrom amftrack.pipeline.functions.extract_graph import from_sparse_to_graph, generate_nx_graph, sparse_to_doc\nfrom skimage.feature import hessian_matrix_det\nfrom amftrack.pipeline.functions.experiment_class_surf import Experiment\nfrom amftrack.pipeline.paths.directory import run_parallel, find_state, directory_scratch, directory_project, path_code\n\nfrom amftrack.notebooks.analysis.data_info import *\nimport matplotlib.patches as mpatches\nfrom statsmodels.stats import weightstats as stests\n",
"_____no_output_____"
],
[
"window=800\nresults={}\nfor treatment in treatments.keys():\n insts = treatments[treatment]\n for inst in insts:\n results[inst] = pickle.load(open(f'{path_code}/MscThesis/Results/straight_{window}_{inst}.pick', \"rb\"))",
"_____no_output_____"
],
[
"column_names = [\"plate\",\"inst\", \"treatment\", \"angle\", \"curvature\",\"density\",\"growth\",\"speed\",\"straightness\",\"t\",\"hyph\"]\ninfos = pd.DataFrame(columns=column_names)\nfor treatment in treatments.keys():\n insts = treatments[treatment]\n for inst in insts:\n angles, curvatures, densities,growths,speeds,tortuosities,ts,hyphs = results[inst]\n for i,angle in enumerate(angles):\n new_line = pd.DataFrame(\n { \"plate\": [plate_number[inst]],\n \"inst\": [inst],\n \"treatment\": [treatment],\n \"angle\": [angle],\n \"curvature\": [curvatures[i]],\n \"density\": [densities[i]],\n \"growth\": [growths[i]],\n \"speed\": [speeds[i]],\n \"straightness\": [tortuosities[i]],\n \"t\": [ts[i]],\n \"hyph\": [hyphs[i]],\n }\n ) # index 0 for\n # mothers need to be modified to resolve multi mother issue\n infos = infos.append(new_line, ignore_index=True)",
"_____no_output_____"
],
[
"corrected = infos.loc[infos[\"straightness\"] <= 1]",
"_____no_output_____"
],
[
"corrected",
"_____no_output_____"
],
[
"fig, ax = plt.subplots(nrows=1, ncols=1, figsize=(9, 4))\nbplot1 = corrected.boxplot(column = ['speed'],by=\"plate\",figsize =(9,8),ax =ax,patch_artist=True, showfliers=False)\ncolors = ['lightblue']+ ['pink'] +['lightgreen']\nfor i,(artist, col) in enumerate(zip(ax.artists, colors)):\n artist.set_edgecolor(col)\n artist.set_facecolor(col) \nax.set_xlabel('Plate')\nax.set_ylabel('Speed')\nax.set_ylim(0.9)\nplt.show()",
"/home/cbisot/anaconda3/envs/test/lib/python3.7/site-packages/matplotlib/__init__.py:880: MatplotlibDeprecationWarning: \nnbagg.transparent\n version, key, obj_type=\"rcparam\", alternative=alt_key)\n"
],
[
"max_speeds = []\ntotal_growth = []\nfor treatment in treatments.keys():\n insts = treatments[treatment]\n for inst in insts:\n inst_tab = corrected.loc[corrected[\"inst\"]==inst] \n for hyph in set(inst_tab['hyph']):\n max_speeds.append(np.max(inst_tab.loc[inst_tab['hyph']==hyph]['speed']))\n total_growth.append(np.sum(inst_tab.loc[inst_tab['hyph']==hyph]['growth']))",
"_____no_output_____"
],
[
"len(max_speeds)",
"_____no_output_____"
],
[
"fig, ax = plt.subplots(nrows=1, ncols=1, figsize=(9, 4))\nax.scatter(np.log(total_growth),max_speeds)\n# ax.set_xlim(100,300)",
"/home/cbisot/anaconda3/envs/test/lib/python3.7/site-packages/matplotlib/__init__.py:880: MatplotlibDeprecationWarning: \nnbagg.transparent\n version, key, obj_type=\"rcparam\", alternative=alt_key)\n"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4afef6c3ba207e49aaf816422483215a44b80c6d
| 88,570 |
ipynb
|
Jupyter Notebook
|
2018_09_11_create_seigel_data_done_3_4.ipynb
|
alistairwalsh/START_notebooks
|
115a53656c4681b9ee43c38349aac2612b9d5d3f
|
[
"MIT"
] | null | null | null |
2018_09_11_create_seigel_data_done_3_4.ipynb
|
alistairwalsh/START_notebooks
|
115a53656c4681b9ee43c38349aac2612b9d5d3f
|
[
"MIT"
] | null | null | null |
2018_09_11_create_seigel_data_done_3_4.ipynb
|
alistairwalsh/START_notebooks
|
115a53656c4681b9ee43c38349aac2612b9d5d3f
|
[
"MIT"
] | null | null | null | 37.324062 | 165 | 0.2986 |
[
[
[
"import pandas as pd\nfrom glob import glob\nfrom pypcleaner import convert_excel_time",
"_____no_output_____"
],
[
"df = pd.read_hdf('big_multi_index.h5')\nscans = pd.read_pickle('scans.p')\nimaging_id_codes = pd.read_pickle('imaging_id_codes.p')\nid_codes = imaging_id_codes.loc[scans.index,'prefix']\nscan_id_df = df.loc[scans.index]\nscan_id_df.drop('day_0', axis='columns', level=0, inplace = True)\n\ndrop_these = ['bp_three_hr','bp_three_hr_systolic','bp_three_hr_diastolic','bp_three_hr_datetime',\n 'bp_seven_hr','bp_seven_hr_systolic','bp_seven_hr_diastolic','bp_seven_hr_datetime',\n 'bp_other_hr','bp_other_hr_systolic','bp_other_hr_diastolic','bp_other_hr_datetime',\n 'hrt_seven_hr','hrt_seven_hr_rate','hrt_seven_hr_datetime','hrt_other_hr',\n 'hrt_other_hr_rate','hrt_other_hr_datetime','surgical_intervention',\n 'surgical_intervention_detail','invasive_intervention','invasive_intervention_detail',\n 'imaging_intervention','imaging_intervention_detail',\n 'other_intervention','other_intervention_detail','medication_given','medication_comment']\n\nscan_id_df.drop([('day_baseline','recent_medical_history',c) for c in drop_these], axis = 'columns',inplace = True)\n\nscan_id_df[('day_baseline','recent_medical_history','tpa_datetime')] = \\\nscan_id_df[('day_baseline','recent_medical_history','tpa_datetime')].apply(convert_excel_time).dt.round('5min')",
"_____no_output_____"
],
[
"remove_tests = \\\n[('day_baseline','physical_examination'), \n ('day_baseline', 'past_medical_history'), \n ('day_baseline', 'CCMI'), \n ('day_baseline', 'further_stroke_event_part'), \n ('day_baseline', 'bloods'), \n ('day_baseline', 'laboratory_tests'), \n ('day_baseline', 'radiological_scans'), \n ('day_baseline', 'concomitant_medications'), \n ('day_baseline', 'pre-stroke_mRS'),\n ('day_7', 'bloods') , \n ('day_7', 'history_of_depression'), \n ('day_7', 'physical_risk_factors'), \n ('day_7', 'diet_q'),\n ('day_7', 'laboratory_tests'), \n ('day_7', 'concomitant_medications'), \n ('day_7', 'concomitant_medications_plus'), \n ('day_90', 'bloods'), \n ('day_90', 'physical_examination'), \n ('day_90', 'physical_risk_factors'), \n ('day_90', 'diet_q'), \n ('day_90', 'laboratory_tests'), \n ('day_90', 'further_stroke_event_part'), \n ('day_90', 'history_of_depression'), \n ('day_90', 'concomitant_medications'), \n ('day_365', 'bloods'), \n ('day_365', 'physical_examination'), \n ('day_365', 'physical_risk_factors'), \n ('day_365', 'diet_q'), \n ('day_365', 'laboratory_tests'), \n ('day_365', 'further_stroke_event_part'), \n ('day_365', 'history_of_depression'), \n ('day_365', 'concomitant_medications'), \n ('day_1', 'vital_signs'), \n ('day_1', 'bloods'), \n ('day_1', 'laboratory_tests'), \n ('day_1', 'serious_adverse_event')]\n\nscan_id_df.drop(remove_tests, axis = 'columns',inplace = True)",
"/Users/alistair/anaconda3/lib/python3.6/site-packages/ipykernel/kernelbase.py:399: PerformanceWarning: indexing past lexsort depth may impact performance.\n user_expressions, allow_stdin)\n"
],
[
"demographics = pd.read_pickle('base_df_data.p')\n\n\nstroke_onset = demographics.loc[scans.index,['identity_stroke_onset']]\nmulti_NIHSS = scan_id_df[('day_baseline','NIHSS_multiple')]\nmulti_with_stroke_onset = stroke_onset.join(multi_NIHSS)\n\nfor c in ['nihss_three_hr_datetime', 'nihss_seven_hr_datetime', \n 'nihss_eighteen_hr_datetime', 'nihss_other_hr_datetime']:\n hrs = multi_with_stroke_onset[c] - multi_with_stroke_onset['identity_stroke_onset']\n multi_with_stroke_onset[c] = hrs.dt.round('5min')\n \nscan_id_df[('day_baseline','NIHSS_multiple')] = multi_with_stroke_onset",
"/Users/alistair/anaconda3/lib/python3.6/site-packages/ipykernel/kernelbase.py:399: PerformanceWarning: indexing past lexsort depth may impact performance.\n user_expressions, allow_stdin)\n/Users/alistair/anaconda3/lib/python3.6/site-packages/ipykernel/zmqshell.py:533: PerformanceWarning: indexing past lexsort depth may impact performance.\n return super(ZMQInteractiveShell, self).run_cell(*args, **kwargs)\n/Users/alistair/anaconda3/lib/python3.6/site-packages/ipykernel/ipkernel.py:196: PerformanceWarning: indexing past lexsort depth may impact performance.\n res = shell.run_cell(code, store_history=store_history, silent=silent)\n"
],
[
"bit_1_cols = demographics.loc[:,'identity_gender':'identity_age_at_stroke_in_years'].columns\nbit_2_cols = demographics.loc[:,'demographics_ancestry_1':].columns\nuseful_columns = bit_1_cols.append(bit_2_cols)\ndemographics = demographics.loc[scans.index,useful_columns]",
"_____no_output_____"
],
[
"demographics['identity_gender'] = demographics['identity_gender'].map({1:'female',0:'male'})",
"_____no_output_____"
],
[
"id_codes = pd.read_pickle('imaging_id_codes.p').loc[scans.index,['prefix']].join(scan_id_df[('day_1','infarct_type')])\n#infarct_type = scan_id_df[('day_1','infarct_type')]",
"/Users/alistair/anaconda3/lib/python3.6/site-packages/ipykernel/kernelbase.py:399: PerformanceWarning: indexing past lexsort depth may impact performance.\n user_expressions, allow_stdin)\n"
],
[
"id_codes.columns = pd.MultiIndex.from_product([['day_90'],['lesion_location_confirmed'], id_codes.columns])",
"_____no_output_____"
],
[
"new_bit = scan_id_df['day_90']['lesion_location_confirmed'].copy()",
"_____no_output_____"
],
[
"new_bit.columns = pd.MultiIndex.from_product([['day_90'],['lesion_location_confirmed'],\n new_bit.columns])",
"_____no_output_____"
],
[
"#scan_id_df['day_90']['lesion_location_confirmed'] = \\\nnew_bit = new_bit.join(id_codes)",
"_____no_output_____"
],
[
"scan_id_df.drop(('day_90',\n 'lesion_location_confirmed',\n 'lesion location confirmed by imaging at 3 months (BC report)'), axis = 'columns',inplace = True)",
"_____no_output_____"
],
[
"scan_id_df = scan_id_df.join(new_bit)",
"_____no_output_____"
],
[
"scan_id_df['day_90']['lesion_location_confirmed']",
"_____no_output_____"
]
],
[
[
"This doesn't work! Sort out. Probably need to create multi index for ones to join",
"_____no_output_____"
]
],
[
[
"scan_id_df[('day_90','lesion_location_confirmed')]",
"/Users/alistair/anaconda3/lib/python3.6/site-packages/ipykernel/kernelbase.py:399: PerformanceWarning: indexing past lexsort depth may impact performance.\n user_expressions, allow_stdin)\n"
],
[
"infarct_type.index",
"_____no_output_____"
],
[
"scan_id_df[('day_90','lesion_location_confirmed')].join([id_codes,infarct_type]).columns",
"_____no_output_____"
],
[
"scan_id_df[('day_90','lesion_location_confirmed')].columns",
"_____no_output_____"
]
],
[
[
"### Do it!\n\ndelete the individual columns and keep the calculated average",
"_____no_output_____"
]
],
[
[
"scan_id_df[('day_90','TUGT_average','correct_average')] = \\\nscan_id_df['day_90']['TUGT_average'][['time1', 'time2', 'time3']].mean(axis = 'columns').round(1)\nscan_id_df[('day_365','TUGT','correct_average')] = \\\nscan_id_df['day_365']['TUGT'][['time1', 'time2', 'time3']].mean(axis='columns').round(1)\n\nbad = \\\n[('day_90', 'TUGT', 'time_taken'),\n ('day_90', 'TUGT_average', 'time1'),\n ('day_90', 'TUGT_average', 'time2'),\n ('day_90', 'TUGT_average', 'time3'),\n ('day_90', 'TUGT_average', 'TUGT_average'),\n ('day_365', 'TUGT', 'time1'),\n ('day_365', 'TUGT', 'time2'),\n ('day_365', 'TUGT', 'time3'),\n ('day_365', 'TUGT', 'average')]\nscan_id_df.drop(bad,axis='columns',inplace=True)",
"_____no_output_____"
],
[
"# drop mri\nmri_drop = scan_id_df.filter(regex='MRI').columns.tolist()\nscan_id_df.drop(mri_drop,axis='columns',inplace=True)",
"_____no_output_____"
],
[
"response_drop = \\\n[('day_7', 'RAPA', 'resp'),\n ('day_7', 'RAPA', 'resp_other'),\n ('day_7', 'RAPA', 'resp_date'),\n ('day_90', 'mRS', 'resp'),\n ('day_90', 'mRS', 'resp_other'),\n ('day_90', 'BI', 'resp.1'),\n ('day_90', 'BI', 'resp_other.1'),\n ('day_90', 'SIS', 'resp.2'),\n ('day_90', 'SIS', 'resp_other.2'),\n ('day_90', 'RAPA', 'resp'),\n ('day_90', 'RAPA', 'resp_other'),\n ('day_90', 'RAPA', 'resp_date'),\n ('day_90', 'WSAS', 'resp.5'),\n ('day_90', 'WSAS', 'resp_other.5'),\n ('day_90', 'WSAS', 'resp_date.2'),\n ('day_365', 'mRS', 'resp'),\n ('day_365', 'mRS', 'resp_other'),\n ('day_365', 'BI', 'resp.1'),\n ('day_365', 'BI', 'resp_other.1'),\n ('day_365', 'SIS', 'resp.2'),\n ('day_365', 'SIS', 'resp_other.2'),\n ('day_365', 'RAPA', 'resp'),\n ('day_365', 'RAPA', 'resp_other'),\n ('day_365', 'RAPA', 'resp_date'),\n ('day_365', 'WSAS', 'resp.5'),\n ('day_365', 'WSAS', 'resp_other.5'),\n ('day_365', 'WSAS', 'resp_date.2')]\n\nscan_id_df.drop(response_drop,axis='columns',inplace=True)\nscan_id_df.columns = scan_id_df.columns.remove_unused_levels()",
"_____no_output_____"
],
[
"drop_nihss_multiple = [('day_baseline','NIHSS_multiple',c) for c in ['nihss_three_hr',\n 'nihss_seven_hr',\n 'nihss_eighteen_hr',\n 'nihss_other_hr']]\n\nscan_id_df.drop(drop_nihss_multiple,axis = 'columns',inplace= True)",
"_____no_output_____"
],
[
"# RAVENs had no data\norig_df = pd.read_excel('data/extract/START 12 months (20161121).xlsx')\nravens = orig_df.iloc[scans.index,689:762]\nravens.rename(columns = {'RAVENS_total_score_12mth':'score'}, inplace = True)\nscan_id_df[('day_365','RAVENs')] = ravens",
"_____no_output_____"
],
[
"scan_id_df.columns = scan_id_df.columns.remove_unused_levels()",
"_____no_output_____"
],
[
"scan_id_df",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4aff0df2e207fd89e2c505516b7161c7569460ab
| 9,616 |
ipynb
|
Jupyter Notebook
|
notebooks/ORG+Tagging+for+DOJ+Filings.ipynb
|
Rosster/MLFinalProject
|
521a739d4c5371db08179a54e22c76a9827136bb
|
[
"MIT"
] | null | null | null |
notebooks/ORG+Tagging+for+DOJ+Filings.ipynb
|
Rosster/MLFinalProject
|
521a739d4c5371db08179a54e22c76a9827136bb
|
[
"MIT"
] | null | null | null |
notebooks/ORG+Tagging+for+DOJ+Filings.ipynb
|
Rosster/MLFinalProject
|
521a739d4c5371db08179a54e22c76a9827136bb
|
[
"MIT"
] | 1 |
2019-03-03T16:36:00.000Z
|
2019-03-03T16:36:00.000Z
| 22.895238 | 145 | 0.551061 |
[
[
[
"import json\nimport requests\nimport pandas as pd\nfrom tqdm import tqdm\nfrom collections import Counter\nimport re\nimport Levenshtein\nfrom cleanco import cleanco",
"_____no_output_____"
],
[
"import spacy\nfrom spacy import displacy\nnlp = spacy.load('en_core_web_lg')",
"_____no_output_____"
],
[
"doj_data = pd.read_json('combined.json', lines=True)",
"_____no_output_____"
],
[
"stock_ticker_data = requests.get('https://quantquote.com/docs/symbol_map_comnam.csv').content",
"_____no_output_____"
],
[
"stock_ticker_df = stock_ticker_data.decode('utf-8').split('\\r\\n')[3:]\nstock_ticker_df = pd.DataFrame([i.split(',') for i in stock_ticker_df])\nstock_ticker_df.columns = stock_ticker_df.iloc[0]\nstock_ticker_df = stock_ticker_df[1:]\nstock_ticker_df = stock_ticker_df.dropna(subset=['COMPANY NAME'])",
"_____no_output_____"
]
],
[
[
"## Tagging Organizations with Spacy",
"_____no_output_____"
]
],
[
[
"parsed_doj_contents = [set([w.text for w in nlp(c).ents if w.label_=='ORG'])\n for c in tqdm_notebook(doj_data.contents.values)]",
"_____no_output_____"
],
[
"parsed_doj_titles = [set([w.text for w in nlp(c).ents if w.label_=='ORG'])\n for c in tqdm(doj_data.title.values)]",
"100%|██████████| 13087/13087 [02:18<00:00, 94.66it/s] \n"
],
[
"doj_data['organizations'] = parsed_doj_contents",
"_____no_output_____"
],
[
"doj_data['organizations_titles'] = parsed_doj_titles",
"_____no_output_____"
],
[
"doj_data['all_orgs'] = doj_data['organizations'].apply(list) + doj_data['organizations_titles'].apply(list)",
"_____no_output_____"
],
[
"all_orgs = [o.lower() for i in doj_data.all_orgs for o in i]",
"_____no_output_____"
],
[
"all_companies = [i.lower() for i in stock_ticker_df['COMPANY NAME']]",
"_____no_output_____"
],
[
"# doj_data.to_json('doj_data_with_orgs.json')",
"_____no_output_____"
]
],
[
[
"## Simpler Tagging :(",
"_____no_output_____"
]
],
[
[
"def process_name(nm):\n name = cleanco(nm).clean_name()\n name = re.sub(r\"[[:punct:]]+\", \"\", name)\n return name.lower()",
"_____no_output_____"
],
[
"clean_org_set_v2 = set([process_name(o) for i in tqdm(doj_data.all_orgs) for o in i])",
"100%|██████████| 13087/13087 [00:41<00:00, 318.97it/s]\n"
],
[
"clean_co_set_v2 = set([process_name(i) for i in tqdm(stock_ticker_df['COMPANY NAME']) ])",
"100%|██████████| 21190/21190 [00:02<00:00, 7236.37it/s]\n"
],
[
"clean_co_to_symbol_dict = {}\nsymbol_to_full_nm_dict = {}\nfor _,symbol,_,name in stock_ticker_df[~stock_ticker_df['QUANTQUOTE PERMTICK'].str.contains(r'\\d')].itertuples():\n if len(name.strip())>0:\n clean_co_to_symbol_dict[process_name(name)] = symbol\n symbol_to_full_nm_dict[symbol] = name",
"_____no_output_____"
],
[
"doj_data['clean_orgs'] = doj_data.all_orgs.apply(lambda st: [process_name(o) for o in st])",
"_____no_output_____"
],
[
"doj_data['tagged_symbols'] = doj_data.clean_orgs.apply(lambda st: [clean_co_to_symbol_dict[o] for o in st if o in clean_co_to_symbol_dict])",
"_____no_output_____"
],
[
"doj_data_final = doj_data[doj_data.tagged_symbols.apply(lambda x: len(x)>0)].copy()",
"_____no_output_____"
],
[
"doj_data_final['tagged_companies'] = doj_data_final['tagged_symbols'].apply(lambda li: [symbol_to_full_nm_dict[i] for i in li])",
"_____no_output_____"
],
[
"# doj_data_final.to_json('doj_data_with_tags.json')",
"_____no_output_____"
]
],
[
[
"## Industry Tagging",
"_____no_output_____"
]
],
[
[
"nyse = pd.read_csv('nyse_company_list.csv')\nnasdaq = pd.read_csv('nasdaq_company_list.csv')\n",
"_____no_output_____"
],
[
"nyse_symbol_set = set([i.lower() for i in nyse.Symbol.values])\nnasdaq_symbol_set = set([i.lower() for i in nasdaq.Symbol.values])",
"_____no_output_____"
],
[
"nyse_symbol_sector_dict = {sym.lower():sector for sym,sector in zip(nyse.Symbol,nyse.Sector)}\nnasdaq_symbol_sector_dict = {sym.lower():sector for sym,sector in zip(nasdaq.Symbol,nasdaq.Sector)}",
"_____no_output_____"
],
[
"nyse_symbol_industry_dict = {sym.lower():industry for sym,industry in zip(nyse.Symbol,nyse.Industry)}\nnasdaq_symbol_industry_dict = {sym.lower():industry for sym,industry in zip(nasdaq.Symbol,nasdaq.Industry)}",
"_____no_output_____"
],
[
"doj_data_final['sectors'] = doj_data_final.tagged_symbols.apply(\n lambda li: \n [nyse_symbol_sector_dict.get(i,nasdaq_symbol_sector_dict.get(i)) \n for i in li if (i in nyse_symbol_sector_dict) or (i in nasdaq_symbol_sector_dict)])",
"_____no_output_____"
],
[
"doj_data_final['industries'] = doj_data_final.tagged_symbols.apply(\n lambda li: \n [nyse_symbol_industry_dict.get(i,nasdaq_symbol_industry_dict.get(i)) \n for i in li if i in nyse_symbol_industry_dict or i in nasdaq_symbol_industry_dict])",
"_____no_output_____"
],
[
"doj_data_final.to_json('doj_data_with_tags_and_industries.json')",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4aff12cd87512aa586afe133bfe12f2b3068604b
| 55,569 |
ipynb
|
Jupyter Notebook
|
visual_logging_example.ipynb
|
wisrovi/pyimagesearch-buy
|
9d5682faacec4415750595d2a14eb69129ab8b52
|
[
"CC0-1.0"
] | 1 |
2021-02-05T11:24:55.000Z
|
2021-02-05T11:24:55.000Z
|
visual_logging_example.ipynb
|
wisrovi/pyimagesearch-buy
|
9d5682faacec4415750595d2a14eb69129ab8b52
|
[
"CC0-1.0"
] | null | null | null |
visual_logging_example.ipynb
|
wisrovi/pyimagesearch-buy
|
9d5682faacec4415750595d2a14eb69129ab8b52
|
[
"CC0-1.0"
] | null | null | null | 204.297794 | 45,465 | 0.895787 |
[
[
[
"<a href=\"https://colab.research.google.com/github/wisrovi/pyimagesearch-buy/blob/main/visual_logging_example.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"# Visual-logging, my new favorite tool for debugging OpenCV and Python apps\n### by [PyImageSearch.com](http://www.pyimagesearch.com)",
"_____no_output_____"
],
[
"## Welcome to **[PyImageSearch Plus](http://pyimg.co/plus)** Jupyter Notebooks!\n\nThis notebook is associated with the [Visual-logging, my new favorite tool for debugging OpenCV and Python apps](https://www.pyimagesearch.com/2014/12/22/visual-logging-new-favorite-tool-debugging-opencv-python-apps/) blog post published on 2014-12-22.\n\nOnly the code for the blog post is here. Most codeblocks have a 1:1 relationship with what you find in the blog post with two exceptions: (1) Python classes are not separate files as they are typically organized with PyImageSearch projects, and (2) Command Line Argument parsing is replaced with an `args` dictionary that you can manipulate as needed.\n\nWe recommend that you execute (press ▶️) the code block-by-block, as-is, before adjusting parameters and `args` inputs. Once you've verified that the code is working, you are welcome to hack with it and learn from manipulating inputs, settings, and parameters. For more information on using Jupyter and Colab, please refer to these resources:\n\n* [Jupyter Notebook User Interface](https://jupyter-notebook.readthedocs.io/en/stable/notebook.html#notebook-user-interface)\n* [Overview of Google Colaboratory Features](https://colab.research.google.com/notebooks/basic_features_overview.ipynb)\n\nAs a reminder, these PyImageSearch Plus Jupyter Notebooks are not for sharing; please refer to the **Copyright** directly below and **Code License Agreement** in the last cell of this notebook. \n\nHappy hacking!\n\n*Adrian*\n\n<hr>\n\n***Copyright:*** *The contents of this Jupyter Notebook, unless otherwise indicated, are Copyright 2020 Adrian Rosebrock, PyimageSearch.com. All rights reserved. Content like this is made possible by the time invested by the authors. If you received this Jupyter Notebook and did not purchase it, please consider making future content possible by joining PyImageSearch Plus at http://pyimg.co/plus/ today.*",
"_____no_output_____"
],
[
"### Install the necessary packages",
"_____no_output_____"
]
],
[
[
"!pip install visual-logging",
"_____no_output_____"
]
],
[
[
"### Download the code zip file",
"_____no_output_____"
]
],
[
[
"!wget https://www.pyimagesearch.com/wp-content/uploads/2014/12/visual-logging-example.zip\n!unzip -qq visual-logging-example.zip\n%cd visual-logging-example",
"_____no_output_____"
]
],
[
[
"## Blog Post Code",
"_____no_output_____"
],
[
"### Import Packages",
"_____no_output_____"
]
],
[
[
"# import the necessary packages\nfrom matplotlib import pyplot as plt\nfrom logging import FileHandler\nfrom vlogging import VisualRecord\nimport logging\nimport cv2",
"_____no_output_____"
]
],
[
[
"### visual-logging, my new favorite tool for debugging OpenCV and Python apps",
"_____no_output_____"
]
],
[
[
"# open the logging file\nlogger = logging.getLogger(\"visual_logging_example\")\nfh = FileHandler(\"demo.html\", mode = \"w\")\n\n# set the logger attributes\nlogger.setLevel(logging.DEBUG)\nlogger.addHandler(fh)\n\n# load our example image and convert it to grayscale\nimage = cv2.imread(\"lex.jpg\")\nimage = cv2.cvtColor(image, cv2.COLOR_BGR2GRAY)\n\n# loop over some varying sigma sizes\nfor s in range(3, 11, 2):\n\t# blur the image and detect edges\n\tblurred = cv2.GaussianBlur(image, (s, s), 0)\n\tedged = cv2.Canny(blurred, 75, 200)\n\tlogger.debug(VisualRecord((\"Detected edges using sigma = %d\" % (s)),\n\t\t[blurred, edged], fmt = \"png\"))",
"_____no_output_____"
],
[
"#@title Display `demo.html`\nimport IPython\nIPython.display.HTML(filename=\"demo.html\")",
"_____no_output_____"
]
],
[
[
"For a detailed walkthrough of the concepts and code, be sure to refer to the full tutorial, [*Visual-logging, my new favorite tool for debugging OpenCV and Python apps*](https://www.pyimagesearch.com/2014/12/22/visual-logging-new-favorite-tool-debugging-opencv-python-apps/) published on 2014-12-22.",
"_____no_output_____"
],
[
"# Code License Agreement\n```\nCopyright (c) 2020 PyImageSearch.com\n\nSIMPLE VERSION\nFeel free to use this code for your own projects, whether they are\npurely educational, for fun, or for profit. THE EXCEPTION BEING if\nyou are developing a course, book, or other educational product.\nUnder *NO CIRCUMSTANCE* may you use this code for your own paid\neducational or self-promotional ventures without written consent\nfrom Adrian Rosebrock and PyImageSearch.com.\n\nLONGER, FORMAL VERSION\nPermission is hereby granted, free of charge, to any person obtaining\na copy of this software and associated documentation files\n(the \"Software\"), to deal in the Software without restriction,\nincluding without limitation the rights to use, copy, modify, merge,\npublish, distribute, sublicense, and/or sell copies of the Software,\nand to permit persons to whom the Software is furnished to do so,\nsubject to the following conditions:\nThe above copyright notice and this permission notice shall be\nincluded in all copies or substantial portions of the Software.\nNotwithstanding the foregoing, you may not use, copy, modify, merge,\npublish, distribute, sublicense, create a derivative work, and/or\nsell copies of the Software in any work that is designed, intended,\nor marketed for pedagogical or instructional purposes related to\nprogramming, coding, application development, or information\ntechnology. Permission for such use, copying, modification, and\nmerger, publication, distribution, sub-licensing, creation of\nderivative works, or sale is expressly withheld.\nTHE SOFTWARE IS PROVIDED \"AS IS\", WITHOUT WARRANTY OF ANY KIND,\nEXPRESS OR IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES\nOF MERCHANTABILITY, FITNESS FOR A PARTICULAR PURPOSE AND\nNONINFRINGEMENT. IN NO EVENT SHALL THE AUTHORS OR COPYRIGHT HOLDERS\nBE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER LIABILITY, WHETHER IN AN\nACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM, OUT OF OR IN\nCONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE\nSOFTWARE.\n```",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
]
] |
4aff172c4041a1b584b7ab40996a555abc367ddf
| 46,882 |
ipynb
|
Jupyter Notebook
|
12_Extraction_based_Question_Answering_using_BERT.ipynb
|
bensjx/ML_repo
|
bf0ebbb8fd4717b21d6e57cb7924d668ddb73870
|
[
"MIT"
] | null | null | null |
12_Extraction_based_Question_Answering_using_BERT.ipynb
|
bensjx/ML_repo
|
bf0ebbb8fd4717b21d6e57cb7924d668ddb73870
|
[
"MIT"
] | null | null | null |
12_Extraction_based_Question_Answering_using_BERT.ipynb
|
bensjx/ML_repo
|
bf0ebbb8fd4717b21d6e57cb7924d668ddb73870
|
[
"MIT"
] | null | null | null | 37.06087 | 214 | 0.485133 |
[
[
[
"!pip install transformers",
"Collecting transformers\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/2c/4e/4f1ede0fd7a36278844a277f8d53c21f88f37f3754abf76a5d6224f76d4a/transformers-3.4.0-py3-none-any.whl (1.3MB)\n\u001b[K |████████████████████████████████| 1.3MB 3.4MB/s \n\u001b[?25hRequirement already satisfied: packaging in /usr/local/lib/python3.6/dist-packages (from transformers) (20.4)\nRequirement already satisfied: filelock in /usr/local/lib/python3.6/dist-packages (from transformers) (3.0.12)\nRequirement already satisfied: tqdm>=4.27 in /usr/local/lib/python3.6/dist-packages (from transformers) (4.41.1)\nCollecting sacremoses\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/7d/34/09d19aff26edcc8eb2a01bed8e98f13a1537005d31e95233fd48216eed10/sacremoses-0.0.43.tar.gz (883kB)\n\u001b[K |████████████████████████████████| 890kB 13.4MB/s \n\u001b[?25hRequirement already satisfied: numpy in /usr/local/lib/python3.6/dist-packages (from transformers) (1.18.5)\nCollecting sentencepiece!=0.1.92\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/e5/2d/6d4ca4bef9a67070fa1cac508606328329152b1df10bdf31fb6e4e727894/sentencepiece-0.1.94-cp36-cp36m-manylinux2014_x86_64.whl (1.1MB)\n\u001b[K |████████████████████████████████| 1.1MB 25.5MB/s \n\u001b[?25hRequirement already satisfied: regex!=2019.12.17 in /usr/local/lib/python3.6/dist-packages (from transformers) (2019.12.20)\nRequirement already satisfied: dataclasses; python_version < \"3.7\" in /usr/local/lib/python3.6/dist-packages (from transformers) (0.7)\nRequirement already satisfied: protobuf in /usr/local/lib/python3.6/dist-packages (from transformers) (3.12.4)\nRequirement already satisfied: requests in /usr/local/lib/python3.6/dist-packages (from transformers) (2.23.0)\nCollecting tokenizers==0.9.2\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/7c/a5/78be1a55b2ac8d6a956f0a211d372726e2b1dd2666bb537fea9b03abd62c/tokenizers-0.9.2-cp36-cp36m-manylinux1_x86_64.whl (2.9MB)\n\u001b[K |████████████████████████████████| 2.9MB 42.0MB/s \n\u001b[?25hRequirement already satisfied: pyparsing>=2.0.2 in /usr/local/lib/python3.6/dist-packages (from packaging->transformers) (2.4.7)\nRequirement already satisfied: six in /usr/local/lib/python3.6/dist-packages (from packaging->transformers) (1.15.0)\nRequirement already satisfied: click in /usr/local/lib/python3.6/dist-packages (from sacremoses->transformers) (7.1.2)\nRequirement already satisfied: joblib in /usr/local/lib/python3.6/dist-packages (from sacremoses->transformers) (0.17.0)\nRequirement already satisfied: setuptools in /usr/local/lib/python3.6/dist-packages (from protobuf->transformers) (50.3.2)\nRequirement already satisfied: urllib3!=1.25.0,!=1.25.1,<1.26,>=1.21.1 in /usr/local/lib/python3.6/dist-packages (from requests->transformers) (1.24.3)\nRequirement already satisfied: chardet<4,>=3.0.2 in /usr/local/lib/python3.6/dist-packages (from requests->transformers) (3.0.4)\nRequirement already satisfied: certifi>=2017.4.17 in /usr/local/lib/python3.6/dist-packages (from requests->transformers) (2020.6.20)\nRequirement already satisfied: idna<3,>=2.5 in /usr/local/lib/python3.6/dist-packages (from requests->transformers) (2.10)\nBuilding wheels for collected packages: sacremoses\n Building wheel for sacremoses (setup.py) ... \u001b[?25l\u001b[?25hdone\n Created wheel for sacremoses: filename=sacremoses-0.0.43-cp36-none-any.whl size=893257 sha256=ea71b5781458df5c26c23ff3eba4a586c5b5930bbb7417056e8560d0fa109e7c\n Stored in directory: /root/.cache/pip/wheels/29/3c/fd/7ce5c3f0666dab31a50123635e6fb5e19ceb42ce38d4e58f45\nSuccessfully built sacremoses\nInstalling collected packages: sacremoses, sentencepiece, tokenizers, transformers\nSuccessfully installed sacremoses-0.0.43 sentencepiece-0.1.94 tokenizers-0.9.2 transformers-3.4.0\n"
],
[
"import torch\nfrom transformers import BertForQuestionAnswering, BertTokenizer\n\nmodel = BertForQuestionAnswering.from_pretrained('bert-large-uncased-whole-word-masking-finetuned-squad')\ntokenizer = BertTokenizer.from_pretrained('bert-large-uncased-whole-word-masking-finetuned-squad')",
"_____no_output_____"
]
],
[
[
"### Design the question and reference text",
"_____no_output_____"
]
],
[
[
"question = \"What does NUS mean?\"\nanswer_text = \"The National University of Singapore (NUS) is the national research university of Singapore. \\\n Founded in 1905 as the Straits Settlements and Federated Malay States Government Medical School, NUS is the oldest higher education institution in Singapore. \\\n It is consistently ranked within the top 20 universities in the world and is considered to be the best university in the Asia-Pacific. \\\n NUS is a comprehensive research university, \\\n offering a wide range of disciplines, including the sciences, medicine and dentistry, design and environment, law, arts and social sciences, engineering, business, computing and music \\\n at both the undergraduate and postgraduate levels.\"",
"_____no_output_____"
],
[
"# Apply the tokenizer to the input text, treating them as a text-pair.\ninput_ids = tokenizer.encode(question, answer_text)\nprint('The input has a total of {:} tokens.'.format(len(input_ids)))",
"The input has a total of 128 tokens.\n"
],
[
"# BERT only needs the token IDs, but for the purpose of inspecting the \n# tokenizer's behavior, let's also get the token strings and display them.\ntokens = tokenizer.convert_ids_to_tokens(input_ids)\n# For each token and its id...\nfor token, id in zip(tokens, input_ids): \n # If this is the [SEP] token, add some space around it to make it stand out.\n if id == tokenizer.sep_token_id:\n print('') \n # Print the token string and its ID in two columns.\n print('{:<12} {:>6,}'.format(token, id))\n if id == tokenizer.sep_token_id:\n print('')",
"[CLS] 101\nwhat 2,054\ndoes 2,515\nnu 16,371\n##s 2,015\nmean 2,812\n? 1,029\n\n[SEP] 102\n\nthe 1,996\nnational 2,120\nuniversity 2,118\nof 1,997\nsingapore 5,264\n( 1,006\nnu 16,371\n##s 2,015\n) 1,007\nis 2,003\nthe 1,996\nnational 2,120\nresearch 2,470\nuniversity 2,118\nof 1,997\nsingapore 5,264\n. 1,012\nfounded 2,631\nin 1,999\n1905 5,497\nas 2,004\nthe 1,996\nstraits 18,849\nsettlements 7,617\nand 1,998\nfed 7,349\n##erated 16,848\nmalay 12,605\nstates 2,163\ngovernment 2,231\nmedical 2,966\nschool 2,082\n, 1,010\nnu 16,371\n##s 2,015\nis 2,003\nthe 1,996\noldest 4,587\nhigher 3,020\neducation 2,495\ninstitution 5,145\nin 1,999\nsingapore 5,264\n. 1,012\nit 2,009\nis 2,003\nconsistently 10,862\nranked 4,396\nwithin 2,306\nthe 1,996\ntop 2,327\n20 2,322\nuniversities 5,534\nin 1,999\nthe 1,996\nworld 2,088\nand 1,998\nis 2,003\nconsidered 2,641\nto 2,000\nbe 2,022\nthe 1,996\nbest 2,190\nuniversity 2,118\nin 1,999\nthe 1,996\nasia 4,021\n- 1,011\npacific 3,534\n. 1,012\nnu 16,371\n##s 2,015\nis 2,003\na 1,037\ncomprehensive 7,721\nresearch 2,470\nuniversity 2,118\n, 1,010\noffering 5,378\na 1,037\nwide 2,898\nrange 2,846\nof 1,997\ndisciplines 12,736\n, 1,010\nincluding 2,164\nthe 1,996\nsciences 4,163\n, 1,010\nmedicine 4,200\nand 1,998\ndentistry 26,556\n, 1,010\ndesign 2,640\nand 1,998\nenvironment 4,044\n, 1,010\nlaw 2,375\n, 1,010\narts 2,840\nand 1,998\nsocial 2,591\nsciences 4,163\n, 1,010\nengineering 3,330\n, 1,010\nbusiness 2,449\n, 1,010\ncomputing 9,798\nand 1,998\nmusic 2,189\nat 2,012\nboth 2,119\nthe 1,996\nundergraduate 8,324\nand 1,998\npostgraduate 15,438\nlevels 3,798\n. 1,012\n\n[SEP] 102\n\n"
]
],
[
[
"#### Split question and reference text",
"_____no_output_____"
]
],
[
[
"# Search the input_ids for the first instance of the `[SEP]` token.\nsep_index = input_ids.index(tokenizer.sep_token_id)\n# The number of segment A tokens includes the [SEP] token istelf.\nnum_seg_a = sep_index + 1\n# The remainder are segment B.\nnum_seg_b = len(input_ids) - num_seg_a\n# Construct the list of 0s and 1s.\nsegment_ids = [0]*num_seg_a + [1]*num_seg_b\n# There should be a segment_id for every input token.\nassert len(segment_ids) == len(input_ids)",
"_____no_output_____"
],
[
"start_scores, end_scores = model(torch.tensor([input_ids]), # The tokens representing our input text.\n token_type_ids=torch.tensor([segment_ids])) # The segment IDs to differentiate question from answer_text",
"_____no_output_____"
]
],
[
[
"#### Run the BERT Model",
"_____no_output_____"
]
],
[
[
"# Find the tokens with the highest `start` and `end` scores.\nanswer_start = torch.argmax(start_scores)\nanswer_end = torch.argmax(end_scores)",
"_____no_output_____"
]
],
[
[
"#### Combine the tokens in the answer and print it out.",
"_____no_output_____"
]
],
[
[
"# Start with the first token.\nanswer = tokens[answer_start]\n\n# Select the remaining answer tokens and join them with whitespace.\nfor i in range(answer_start + 1, answer_end + 1):\n \n # If it's a subword token, then recombine it with the previous token.\n if tokens[i][0:2] == '##':\n answer += tokens[i][2:]\n \n # Otherwise, add a space then the token.\n else:\n answer += ' ' + tokens[i]\n\nprint('Answer: \"' + answer + '\"')",
"Answer: \"national university of singapore\"\n"
],
[
"",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
4aff24086de77b1c1375b33d42c5ef85c72ab898
| 41,952 |
ipynb
|
Jupyter Notebook
|
11 - Tune Hyperparameters.ipynb
|
aizatrosli/mslearn-dp100
|
bd9ecc014bdaf164a36c5386d6c47e8f8c82595a
|
[
"MIT"
] | null | null | null |
11 - Tune Hyperparameters.ipynb
|
aizatrosli/mslearn-dp100
|
bd9ecc014bdaf164a36c5386d6c47e8f8c82595a
|
[
"MIT"
] | null | null | null |
11 - Tune Hyperparameters.ipynb
|
aizatrosli/mslearn-dp100
|
bd9ecc014bdaf164a36c5386d6c47e8f8c82595a
|
[
"MIT"
] | null | null | null | 69.572139 | 17,853 | 0.622926 |
[
[
[
"# Tune Hyperparameters\n\nThere are many machine learning algorithms that require *hyperparameters* (parameter values that influence training, but can't be determined from the training data itself). For example, when training a logistic regression model, you can use a *regularization rate* hyperparameter to counteract bias in the model; or when training a convolutional neural network, you can use hyperparameters like *learning rate* and *batch size* to control how weights are adjusted and how many data items are processed in a mini-batch respectively. The choice of hyperparameter values can significantly affect the performance of a trained model, or the time taken to train it; and often you need to try multiple combinations to find the optimal solution.\n\nIn this case, you'll train a classification model with two hyperparameters, but the principles apply to any kind of model you can train with Azure Machine Learning.",
"_____no_output_____"
],
[
"## Connect to your workspace\n\nTo get started, connect to your workspace.\n\n> **Note**: If you haven't already established an authenticated session with your Azure subscription, you'll be prompted to authenticate by clicking a link, entering an authentication code, and signing into Azure.",
"_____no_output_____"
]
],
[
[
"import azureml.core\nfrom azureml.core import Workspace\n\n# Load the workspace from the saved config file\nws = Workspace.from_config()\nprint('Ready to use Azure ML {} to work with {}'.format(azureml.core.VERSION, ws.name))",
"Ready to use Azure ML 1.34.0 to work with aizat\n"
]
],
[
[
"## Prepare data\n\nIn this lab, you'll use a dataset containing details of diabetes patients. Run the cell below to create this dataset (if it already exists, the existing version will be used)",
"_____no_output_____"
]
],
[
[
"from azureml.core import Dataset\n\ndefault_ds = ws.get_default_datastore()\n\nif 'diabetes dataset' not in ws.datasets:\n default_ds.upload_files(files=['./data/diabetes.csv', './data/diabetes2.csv'], # Upload the diabetes csv files in /data\n target_path='diabetes-data/', # Put it in a folder path in the datastore\n overwrite=True, # Replace existing files of the same name\n show_progress=True)\n\n #Create a tabular dataset from the path on the datastore (this may take a short while)\n tab_data_set = Dataset.Tabular.from_delimited_files(path=(default_ds, 'diabetes-data/*.csv'))\n\n # Register the tabular dataset\n try:\n tab_data_set = tab_data_set.register(workspace=ws, \n name='diabetes dataset',\n description='diabetes data',\n tags = {'format':'CSV'},\n create_new_version=True)\n print('Dataset registered.')\n except Exception as ex:\n print(ex)\nelse:\n print('Dataset already registered.')",
"Dataset already registered.\n"
]
],
[
[
"## Prepare a training script\n\nNow let's create a folder for the training script you'll use to train the model.",
"_____no_output_____"
]
],
[
[
"import os\n\nexperiment_folder = 'diabetes_training-hyperdrive'\nos.makedirs(experiment_folder, exist_ok=True)\n\nprint('Folder ready.')",
"Folder ready.\n"
]
],
[
[
"Now create the Python script to train the model. In this example, you'll use a *Gradient Boosting* algorithm to train a classification model. The script must include:\n\n- An argument for each hyperparameter you want to optimize (in this case, the learning rate and number of estimators for the Gradient Boosting algorithm)\n- Code to log the performance metric you want to optimize for (in this case, you'll log both AUC and accuracy, so you can choose to optimize the model for either of these)",
"_____no_output_____"
]
],
[
[
"%%writefile $experiment_folder/diabetes_training.py\n# Import libraries\nimport argparse, joblib, os\nfrom azureml.core import Run\nimport pandas as pd\nimport numpy as np\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.ensemble import GradientBoostingClassifier\nfrom sklearn.metrics import roc_auc_score, roc_curve\n\n# Get the experiment run context\nrun = Run.get_context()\n\n# Get script arguments\nparser = argparse.ArgumentParser()\n\n# Input dataset\nparser.add_argument(\"--input-data\", type=str, dest='input_data', help='training dataset')\n\n# Hyperparameters\nparser.add_argument('--learning_rate', type=float, dest='learning_rate', default=0.1, help='learning rate')\nparser.add_argument('--n_estimators', type=int, dest='n_estimators', default=100, help='number of estimators')\n\n# Add arguments to args collection\nargs = parser.parse_args()\n\n# Log Hyperparameter values\nrun.log('learning_rate', np.float(args.learning_rate))\nrun.log('n_estimators', np.int(args.n_estimators))\n\n# load the diabetes dataset\nprint(\"Loading Data...\")\ndiabetes = run.input_datasets['training_data'].to_pandas_dataframe() # Get the training data from the estimator input\n\n# Separate features and labels\nX, y = diabetes[['Pregnancies','PlasmaGlucose','DiastolicBloodPressure','TricepsThickness','SerumInsulin','BMI','DiabetesPedigree','Age']].values, diabetes['Diabetic'].values\n\n# Split data into training set and test set\nX_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.30, random_state=0)\n\n# Train a Gradient Boosting classification model with the specified hyperparameters\nprint('Training a classification model')\nmodel = GradientBoostingClassifier(learning_rate=args.learning_rate,\n n_estimators=args.n_estimators).fit(X_train, y_train)\n\n# calculate accuracy\ny_hat = model.predict(X_test)\nacc = np.average(y_hat == y_test)\nprint('Accuracy:', acc)\nrun.log('Accuracy', np.float(acc))\n\n# calculate AUC\ny_scores = model.predict_proba(X_test)\nauc = roc_auc_score(y_test,y_scores[:,1])\nprint('AUC: ' + str(auc))\nrun.log('AUC', np.float(auc))\n\n# Save the model in the run outputs\nos.makedirs('outputs', exist_ok=True)\njoblib.dump(value=model, filename='outputs/diabetes_model.pkl')\n\nrun.complete()",
"Overwriting diabetes_training-hyperdrive/diabetes_training.py\n"
]
],
[
[
"## Create compute\n\nHyperparameter tuning involves running multiple training iterations with different hyperparameter values and comparing the performance metrics of the resulting models. To do this efficiently, we'll take advantage of on-demand cloud compute and create a cluster - this will allow multiple training iterations to be run concurrently.\n\nUse the following code to specify an Azure Machine Learning compute cluster (it will be created if it doesn't already exist).\n\n> **Important**: Change *your-compute-cluster* to the name of your compute cluster in the code below before running it! Cluster names must be globally unique names between 2 to 16 characters in length. Valid characters are letters, digits, and the - character.",
"_____no_output_____"
]
],
[
[
"from azureml.core.compute import ComputeTarget, AmlCompute\nfrom azureml.core.compute_target import ComputeTargetException\n\ncluster_name = \"aizatcluster\"\n\ntry:\n # Check for existing compute target\n training_cluster = ComputeTarget(workspace=ws, name=cluster_name)\n print('Found existing cluster, use it.')\nexcept ComputeTargetException:\n # If it doesn't already exist, create it\n try:\n compute_config = AmlCompute.provisioning_configuration(vm_size='STANDARD_DS11_V2', max_nodes=2)\n training_cluster = ComputeTarget.create(ws, cluster_name, compute_config)\n training_cluster.wait_for_completion(show_output=True)\n except Exception as ex:\n print(ex)\n ",
"Found existing cluster, use it.\n"
]
],
[
[
"> **Note**: Compute instances and clusters are based on standard Azure virtual machine images. For this exercise, the *Standard_DS11_v2* image is recommended to achieve the optimal balance of cost and performance. If your subscription has a quota that does not include this image, choose an alternative image; but bear in mind that a larger image may incur higher cost and a smaller image may not be sufficient to complete the tasks. Alternatively, ask your Azure administrator to extend your quota.\n\nYou'll need a Python environment to be hosted on the compute, so let's define that as Conda configuration file.",
"_____no_output_____"
]
],
[
[
"%%writefile $experiment_folder/hyperdrive_env.yml\nname: batch_environment\ndependencies:\n- python=3.6.2\n- scikit-learn\n- pandas\n- numpy\n- pip\n- pip:\n - azureml-defaults\n",
"Overwriting diabetes_training-hyperdrive/hyperdrive_env.yml\n"
]
],
[
[
"## Run a hyperparameter tuning experiment\n\nAzure Machine Learning includes a hyperparameter tuning capability through *hyperdrive* experiments. These experiments launch multiple child runs, each with a different hyperparameter combination. The run producing the best model (as determined by the logged target performance metric for which you want to optimize) can be identified, and its trained model selected for registration and deployment.\n\n> **Note**: In this example, we aren't specifying an early stopping policy. Such a policy is only relevant if the training script performs multiple training iterations, logging the primary metric for each iteration. This approach is typically employed when training deep neural network models over multiple *epochs*.",
"_____no_output_____"
]
],
[
[
"from azureml.core import Experiment, ScriptRunConfig, Environment\nfrom azureml.train.hyperdrive import GridParameterSampling, HyperDriveConfig, PrimaryMetricGoal, choice\nfrom azureml.widgets import RunDetails\n\n# Create a Python environment for the experiment\nhyper_env = Environment.from_conda_specification(\"experiment_env\", experiment_folder + \"/hyperdrive_env.yml\")\n\n# Get the training dataset\ndiabetes_ds = ws.datasets.get(\"diabetes dataset\")\n\n# Create a script config\nscript_config = ScriptRunConfig(source_directory=experiment_folder,\n script='diabetes_training.py',\n # Add non-hyperparameter arguments -in this case, the training dataset\n arguments = ['--input-data', diabetes_ds.as_named_input('training_data')],\n environment=hyper_env,\n compute_target = training_cluster)\n\n# Sample a range of parameter values\nparams = GridParameterSampling(\n {\n # Hyperdrive will try 6 combinations, adding these as script arguments\n '--learning_rate': choice(0.01, 0.1, 1.0),\n '--n_estimators' : choice(10, 100)\n }\n)\n\n# Configure hyperdrive settings\nhyperdrive = HyperDriveConfig(run_config=script_config, \n hyperparameter_sampling=params, \n policy=None, # No early stopping policy\n primary_metric_name='AUC', # Find the highest AUC metric\n primary_metric_goal=PrimaryMetricGoal.MAXIMIZE, \n max_total_runs=6, # Restict the experiment to 6 iterations\n max_concurrent_runs=2) # Run up to 2 iterations in parallel\n\n# Run the experiment\nexperiment = Experiment(workspace=ws, name='mslearn-diabetes-hyperdrive')\nrun = experiment.submit(config=hyperdrive)\n\n# Show the status in the notebook as the experiment runs\nRunDetails(run).show()\nrun.wait_for_completion()",
"_____no_output_____"
]
],
[
[
"You can view the experiment run status in the widget above. You can also view the main Hyperdrive experiment run and its child runs in [Azure Machine Learning studio](https://ml.azure.com).\n\n> **Note**: If a message indicating that a non-numeric can't be visualized is displayed, you can ignore it.\n\n## Determine the best performing run\n\nWhen all of the runs have finished, you can find the best one based on the performance metric you specified (in this case, the one with the best AUC).",
"_____no_output_____"
]
],
[
[
"# Print all child runs, sorted by the primary metric\nfor child_run in run.get_children_sorted_by_primary_metric():\n print(child_run)\n\n# Get the best run, and its metrics and arguments\nbest_run = run.get_best_run_by_primary_metric()\nbest_run_metrics = best_run.get_metrics()\nscript_arguments = best_run.get_details() ['runDefinition']['arguments']\nprint('Best Run Id: ', best_run.id)\nprint(' -AUC:', best_run_metrics['AUC'])\nprint(' -Accuracy:', best_run_metrics['Accuracy'])\nprint(' -Arguments:',script_arguments)",
"{'run_id': 'HD_8e65d1c1-28b5-4baf-a835-cd42e9a147cc_3', 'hyperparameters': '{\"--learning_rate\": 0.1, \"--n_estimators\": 100}', 'best_primary_metric': 0.9885804604667666, 'status': 'Completed'}\n{'run_id': 'HD_8e65d1c1-28b5-4baf-a835-cd42e9a147cc_5', 'hyperparameters': '{\"--learning_rate\": 1.0, \"--n_estimators\": 100}', 'best_primary_metric': 0.9857517600851531, 'status': 'Completed'}\n{'run_id': 'HD_8e65d1c1-28b5-4baf-a835-cd42e9a147cc_4', 'hyperparameters': '{\"--learning_rate\": 1.0, \"--n_estimators\": 10}', 'best_primary_metric': 0.982908128731084, 'status': 'Completed'}\n{'run_id': 'HD_8e65d1c1-28b5-4baf-a835-cd42e9a147cc_1', 'hyperparameters': '{\"--learning_rate\": 0.01, \"--n_estimators\": 100}', 'best_primary_metric': 0.9559393638830617, 'status': 'Completed'}\n{'run_id': 'HD_8e65d1c1-28b5-4baf-a835-cd42e9a147cc_2', 'hyperparameters': '{\"--learning_rate\": 0.1, \"--n_estimators\": 10}', 'best_primary_metric': 0.9516323866285732, 'status': 'Completed'}\n{'run_id': 'HD_8e65d1c1-28b5-4baf-a835-cd42e9a147cc_0', 'hyperparameters': '{\"--learning_rate\": 0.01, \"--n_estimators\": 10}', 'best_primary_metric': 0.9354833786202631, 'status': 'Completed'}\nBest Run Id: HD_8e65d1c1-28b5-4baf-a835-cd42e9a147cc_3\n -AUC: 0.9885804604667666\n -Accuracy: 0.9457777777777778\n -Arguments: ['--input-data', 'DatasetConsumptionConfig:training_data', '--learning_rate', '0.1', '--n_estimators', '100']\n"
]
],
[
[
"Now that you've found the best run, you can register the model it trained.",
"_____no_output_____"
]
],
[
[
"from azureml.core import Model\n\n# Register model\nbest_run.register_model(model_path='outputs/diabetes_model.pkl', model_name='diabetes_model',\n tags={'Training context':'Hyperdrive'},\n properties={'AUC': best_run_metrics['AUC'], 'Accuracy': best_run_metrics['Accuracy']})\n\n# List registered models\nfor model in Model.list(ws):\n print(model.name, 'version:', model.version)\n for tag_name in model.tags:\n tag = model.tags[tag_name]\n print ('\\t',tag_name, ':', tag)\n for prop_name in model.properties:\n prop = model.properties[prop_name]\n print ('\\t',prop_name, ':', prop)\n print('\\n')",
"diabetes_model version: 11\n\t Training context : Hyperdrive\n\t AUC : 0.9885804604667666\n\t Accuracy : 0.9457777777777778\n\n\ndiabetes_model version: 10\n\t Training context : Inline Training\n\t AUC : 0.8798298422807104\n\t Accuracy : 0.892\n\n\ndiabetes_model version: 9\n\t Training context : Inline Training\n\t AUC : 0.8805985372406587\n\t Accuracy : 0.8936666666666667\n\n\ndiabetes_model version: 8\n\t Training context : Inline Training\n\t AUC : 0.8741031892133936\n\t Accuracy : 0.8866666666666667\n\n\ndiabetes_model version: 7\n\t Training context : Parameterized script\n\t AUC : 0.8484377332205582\n\t Accuracy : 0.774\n\n\ndiabetes_model version: 6\n\t Training context : Script\n\t AUC : 0.8483377282451863\n\t Accuracy : 0.774\n\n\ndiabetes_model version: 5\n\t Training context : Pipeline\n\t AUC : 0.8862361650715226\n\t Accuracy : 0.9004444444444445\n\n\ndiabetes_model version: 4\n\t Training context : File dataset\n\t AUC : 0.8468331741963582\n\t Accuracy : 0.7793333333333333\n\n\ndiabetes_model version: 3\n\t Training context : Tabular dataset\n\t AUC : 0.8568509052814499\n\t Accuracy : 0.7891111111111111\n\n\ndiabetes_model version: 2\n\t Training context : Parameterized script\n\t AUC : 0.8483198169063138\n\t Accuracy : 0.774\n\n\ndiabetes_model version: 1\n\t Training context : Script\n\t AUC : 0.8484929598487486\n\t Accuracy : 0.774\n\n\namlstudio-designer-predict-dia version: 1\n\t CreatedByAMLStudio : true\n\n\nAutoML7001303fd0 version: 1\n\n\n"
]
],
[
[
"> **More Information**: For more information about Hyperdrive, see the [Azure ML documentation](https://docs.microsoft.com/azure/machine-learning/how-to-tune-hyperparameters).",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
4aff2faf31e911f869181eb85ccec72b2573d91e
| 389,227 |
ipynb
|
Jupyter Notebook
|
lijin-THU:notes-python/04-scipy/04.02-interpolation-with-scipy.ipynb
|
Maecenas/python-getting-started
|
2739444e0f4aa692123dcd0c1b9a44218281f9b6
|
[
"MIT"
] | null | null | null |
lijin-THU:notes-python/04-scipy/04.02-interpolation-with-scipy.ipynb
|
Maecenas/python-getting-started
|
2739444e0f4aa692123dcd0c1b9a44218281f9b6
|
[
"MIT"
] | null | null | null |
lijin-THU:notes-python/04-scipy/04.02-interpolation-with-scipy.ipynb
|
Maecenas/python-getting-started
|
2739444e0f4aa692123dcd0c1b9a44218281f9b6
|
[
"MIT"
] | null | null | null | 382.344794 | 176,574 | 0.929733 |
[
[
[
"# 插值",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"设置 **`Numpy`** 浮点数显示格式:",
"_____no_output_____"
]
],
[
[
"np.set_printoptions(precision=2, suppress=True)",
"_____no_output_____"
]
],
[
[
"从文本中读入数据,数据来自 http://kinetics.nist.gov/janaf/html/C-067.txt ,保存为结构体数组:",
"_____no_output_____"
]
],
[
[
"data = np.genfromtxt(\"JANAF_CH4.txt\", \n delimiter=\"\\t\", # TAB 分隔\n skiprows=1, # 忽略首行\n names=True, # 读入属性\n missing_values=\"INFINITE\", # 缺失值\n filling_values=np.inf) # 填充缺失值",
"_____no_output_____"
]
],
[
[
"显示部分数据:",
"_____no_output_____"
]
],
[
[
"for row in data[:7]:\n print \"{}\\t{}\".format(row['TK'], row['Cp'])\nprint \"...\\t...\"",
"0.0\t0.0\n100.0\t33.258\n200.0\t33.473\n250.0\t34.216\n298.15\t35.639\n300.0\t35.708\n350.0\t37.874\n...\t...\n"
]
],
[
[
"绘图:",
"_____no_output_____"
]
],
[
[
"p = plt.plot(data['TK'], data['Cp'], 'kx')\nt = plt.title(\"JANAF data for Methane $CH_4$\")\na = plt.axis([0, 6000, 30, 120])\nx = plt.xlabel(\"Temperature (K)\")\ny = plt.ylabel(r\"$C_p$ ($\\frac{kJ}{kg K}$)\")",
"_____no_output_____"
]
],
[
[
"## 插值",
"_____no_output_____"
],
[
"假设我们要对这组数据进行插值。\n\n先导入一维插值函数 `interp1d`:\n\n interp1d(x, y)",
"_____no_output_____"
]
],
[
[
"from scipy.interpolate import interp1d",
"_____no_output_____"
],
[
"ch4_cp = interp1d(data['TK'], data['Cp'])",
"_____no_output_____"
]
],
[
[
"`interp1d` 的返回值可以像函数一样接受输入,并返回插值的结果。\n\n单个输入值,注意返回的是数组:",
"_____no_output_____"
]
],
[
[
"ch4_cp(382.2)",
"_____no_output_____"
]
],
[
[
"输入数组,返回的是对应的数组:",
"_____no_output_____"
]
],
[
[
"ch4_cp([32.2,323.2])",
"_____no_output_____"
]
],
[
[
"默认情况下,输入值要在插值允许的范围内,否则插值会报错:",
"_____no_output_____"
]
],
[
[
"ch4_cp(8752)",
"_____no_output_____"
]
],
[
[
"但我们可以通过参数设置允许超出范围的值存在:",
"_____no_output_____"
]
],
[
[
"ch4_cp = interp1d(data['TK'], data['Cp'], \n bounds_error=False)",
"_____no_output_____"
]
],
[
[
"不过由于超出范围,所以插值的输出是非法值:",
"_____no_output_____"
]
],
[
[
"ch4_cp(8752)",
"_____no_output_____"
]
],
[
[
"可以使用指定值替代这些非法值:",
"_____no_output_____"
]
],
[
[
"ch4_cp = interp1d(data['TK'], data['Cp'], \n bounds_error=False, fill_value=-999.25)",
"_____no_output_____"
],
[
"ch4_cp(8752)",
"_____no_output_____"
]
],
[
[
"### 线性插值",
"_____no_output_____"
],
[
"`interp1d` 默认的插值方法是线性,关于线性插值的定义,请参见:\n\n- 维基百科-线性插值: https://zh.wikipedia.org/wiki/%E7%BA%BF%E6%80%A7%E6%8F%92%E5%80%BC\n- 百度百科-线性插值: http://baike.baidu.com/view/4685624.htm\n\n其基本思想是,已知相邻两点 $x_1,x_2$ 对应的值 $y_1,y_2$ ,那么对于 $(x_1,x_2)$ 之间的某一点 $x$ ,线性插值对应的值 $y$ 满足:点 $(x,y)$ 在 $(x_1,y_1),(x_2,y_2)$ 所形成的线段上。\n\n应用线性插值:",
"_____no_output_____"
]
],
[
[
"T = np.arange(100,355,5)\nplt.plot(T, ch4_cp(T), \"+k\")\np = plt.plot(data['TK'][1:7], data['Cp'][1:7], 'ro', markersize=8)",
"_____no_output_____"
]
],
[
[
"其中红色的圆点为原来的数据点,黑色的十字点为对应的插值点,可以明显看到,相邻的数据点的插值在一条直线上。",
"_____no_output_____"
],
[
"### 多项式插值",
"_____no_output_____"
],
[
"我们可以通过 `kind` 参数来调节使用的插值方法,来得到不同的结果:\n\n- `nearest` 最近邻插值\n- `zero` 0阶插值\n- `linear` 线性插值\n- `quadratic` 二次插值\n- `cubic` 三次插值\n- `4,5,6,7` 更高阶插值\n\n最近邻插值:",
"_____no_output_____"
]
],
[
[
"cp_ch4 = interp1d(data['TK'], data['Cp'], kind=\"nearest\")\np = plt.plot(T, cp_ch4(T), \"k+\")\np = plt.plot(data['TK'][1:7], data['Cp'][1:7], 'ro', markersize=8)",
"_____no_output_____"
]
],
[
[
"0阶插值:",
"_____no_output_____"
]
],
[
[
"cp_ch4 = interp1d(data['TK'], data['Cp'], kind=\"zero\")\np = plt.plot(T, cp_ch4(T), \"k+\")\np = plt.plot(data['TK'][1:7], data['Cp'][1:7], 'ro', markersize=8)",
"_____no_output_____"
]
],
[
[
"二次插值:",
"_____no_output_____"
]
],
[
[
"cp_ch4 = interp1d(data['TK'], data['Cp'], kind=\"quadratic\")\np = plt.plot(T, cp_ch4(T), \"k+\")\np = plt.plot(data['TK'][1:7], data['Cp'][1:7], 'ro', markersize=8)",
"_____no_output_____"
]
],
[
[
"三次插值:",
"_____no_output_____"
]
],
[
[
"cp_ch4 = interp1d(data['TK'], data['Cp'], kind=\"cubic\")\np = plt.plot(T, cp_ch4(T), \"k+\")\np = plt.plot(data['TK'][1:7], data['Cp'][1:7], 'ro', markersize=8)",
"_____no_output_____"
]
],
[
[
"事实上,我们可以使用更高阶的多项式插值,只要将 `kind` 设为对应的数字即可:",
"_____no_output_____"
],
[
"四次多项式插值:",
"_____no_output_____"
]
],
[
[
"cp_ch4 = interp1d(data['TK'], data['Cp'], kind=4)\np = plt.plot(T, cp_ch4(T), \"k+\")\np = plt.plot(data['TK'][1:7], data['Cp'][1:7], 'ro', markersize=8)",
"_____no_output_____"
]
],
[
[
"可以参见:\n\n- 维基百科-多项式插值:https://zh.wikipedia.org/wiki/%E5%A4%9A%E9%A1%B9%E5%BC%8F%E6%8F%92%E5%80%BC\n- 百度百科-插值法:http://baike.baidu.com/view/754506.htm\n\n对于二维乃至更高维度的多项式插值:",
"_____no_output_____"
]
],
[
[
"from scipy.interpolate import interp2d, interpnd",
"_____no_output_____"
]
],
[
[
"其使用方法与一维类似。",
"_____no_output_____"
],
[
"### 径向基函数",
"_____no_output_____"
],
[
"关于径向基函数,可以参阅:\n- 维基百科-Radial basis fucntion:https://en.wikipedia.org/wiki/Radial_basis_function\n\n径向基函数,简单来说就是点 $x$ 处的函数值只依赖于 $x$ 与某点 $c$ 的距离:\n\n$$\\Phi(x,c) = \\Phi(\\|x-c\\|)$$",
"_____no_output_____"
]
],
[
[
"x = np.linspace(-3,3,100)",
"_____no_output_____"
]
],
[
[
"常用的径向基(`RBF`)函数有:\n\n高斯函数:",
"_____no_output_____"
]
],
[
[
"plt.plot(x, np.exp(-1 * x **2))\nt = plt.title(\"Gaussian\")",
"_____no_output_____"
]
],
[
[
"`Multiquadric` 函数:",
"_____no_output_____"
]
],
[
[
"plt.plot(x, np.sqrt(1 + x **2))\nt = plt.title(\"Multiquadric\")",
"_____no_output_____"
]
],
[
[
"`Inverse Multiquadric` 函数:",
"_____no_output_____"
]
],
[
[
"plt.plot(x, 1. / np.sqrt(1 + x **2))\nt = plt.title(\"Inverse Multiquadric\")",
"_____no_output_____"
]
],
[
[
"### 径向基函数插值",
"_____no_output_____"
],
[
"对于径向基函数,其插值的公式为:\n\n$$\nf(x) = \\sum_j n_j \\Phi(\\|x-x_j\\|)\n$$\n\n我们通过数据点 $x_j$ 来计算出 $n_j$ 的值,来计算 $x$ 处的插值结果。",
"_____no_output_____"
]
],
[
[
"from scipy.interpolate.rbf import Rbf",
"_____no_output_____"
]
],
[
[
"使用 `multiquadric` 核的:",
"_____no_output_____"
]
],
[
[
"cp_rbf = Rbf(data['TK'], data['Cp'], function = \"multiquadric\")\nplt.plot(data['TK'], data['Cp'], 'k+')\np = plt.plot(data['TK'], cp_rbf(data['TK']), 'r-')",
"_____no_output_____"
]
],
[
[
"使用 `gaussian` 核:",
"_____no_output_____"
]
],
[
[
"cp_rbf = Rbf(data['TK'], data['Cp'], function = \"gaussian\")\nplt.plot(data['TK'], data['Cp'], 'k+')\np = plt.plot(data['TK'], cp_rbf(data['TK']), 'r-')",
"_____no_output_____"
]
],
[
[
"使用 `nverse_multiquadric` 核:",
"_____no_output_____"
]
],
[
[
"cp_rbf = Rbf(data['TK'], data['Cp'], function = \"inverse_multiquadric\")\nplt.plot(data['TK'], data['Cp'], 'k+')\np = plt.plot(data['TK'], cp_rbf(data['TK']), 'r-')",
"_____no_output_____"
]
],
[
[
"不同的 `RBF` 核的结果也不同。",
"_____no_output_____"
],
[
"### 高维 `RBF` 插值",
"_____no_output_____"
]
],
[
[
"from mpl_toolkits.mplot3d import Axes3D",
"_____no_output_____"
]
],
[
[
"三维数据点:",
"_____no_output_____"
]
],
[
[
"x, y = np.mgrid[-np.pi/2:np.pi/2:5j, -np.pi/2:np.pi/2:5j]\nz = np.cos(np.sqrt(x**2 + y**2))",
"_____no_output_____"
],
[
"fig = plt.figure(figsize=(12,6))\nax = fig.gca(projection=\"3d\")\nax.scatter(x,y,z)",
"_____no_output_____"
]
],
[
[
"3维 `RBF` 插值:",
"_____no_output_____"
]
],
[
[
"zz = Rbf(x, y, z)",
"_____no_output_____"
],
[
"xx, yy = np.mgrid[-np.pi/2:np.pi/2:50j, -np.pi/2:np.pi/2:50j]\nfig = plt.figure(figsize=(12,6))\nax = fig.gca(projection=\"3d\")\nax.plot_surface(xx,yy,zz(xx,yy),rstride=1, cstride=1, cmap=plt.cm.jet)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
4aff3697e50e03c411849bc2ed13051f1104cfc2
| 217,270 |
ipynb
|
Jupyter Notebook
|
2. Data Analysis with Python Cars Dataset/Data Analysis with Python - Cars Dataset.ipynb
|
Biplob45/Python-for-Machine-Learning-and-Data-Science
|
f08cb51358b266933bc3d4f23bd5998811791ee9
|
[
"MIT"
] | 1 |
2021-05-03T18:57:24.000Z
|
2021-05-03T18:57:24.000Z
|
2. Data Analysis with Python Cars Dataset/Data Analysis with Python - Cars Dataset.ipynb
|
Biplob45/Python-for-Machine-Learning-and-Data-Science
|
f08cb51358b266933bc3d4f23bd5998811791ee9
|
[
"MIT"
] | null | null | null |
2. Data Analysis with Python Cars Dataset/Data Analysis with Python - Cars Dataset.ipynb
|
Biplob45/Python-for-Machine-Learning-and-Data-Science
|
f08cb51358b266933bc3d4f23bd5998811791ee9
|
[
"MIT"
] | 1 |
2020-08-20T07:26:04.000Z
|
2020-08-20T07:26:04.000Z
| 36.109357 | 196 | 0.292084 |
[
[
[
"import pandas as pd",
"_____no_output_____"
],
[
"car = pd.read_csv(r\"C:\\Users\\Biplob\\Desktop\\Python for Machine Learning and Data Science\\All Projects - Data Analytics\\2. Data Analysis with Python Cars Dataset\\2. Cars Data1.csv\")",
"_____no_output_____"
],
[
"car.head()",
"_____no_output_____"
],
[
"car.shape",
"_____no_output_____"
]
],
[
[
"# 1) Instruction ( For Data Cleaning)\n\t#Find all null value in the dataset. If there is any null value in any column. then fill it with the mean of the column\n",
"_____no_output_____"
]
],
[
[
"car.isnull().sum()",
"_____no_output_____"
],
[
"car['Cylinders'].fillna(car['Cylinders'].mean(), inplace = True)",
"_____no_output_____"
],
[
"car.isnull().sum()",
"_____no_output_____"
]
],
[
[
"# 2)Question ( Based on Value Counts )\n\tCheck what are the different types of Make are there in our dataset. And, what is the count(occurrence) of each make in the data ?\n",
"_____no_output_____"
]
],
[
[
"car.head(2)",
"_____no_output_____"
],
[
"car['Make'].value_counts()",
"_____no_output_____"
]
],
[
[
"# 3) Instruction (Filtering)\n\tShow all the records where Origin is Asia or Europe",
"_____no_output_____"
]
],
[
[
"car.head()",
"_____no_output_____"
],
[
"car[car['Origin'].isin(['Asia','Europe'])]",
"_____no_output_____"
],
[
"car[(car['Origin'] == 'Asia') | (car['Origin'] == 'Europe')].head(50)",
"_____no_output_____"
],
[
"car[(car['Origin'] == 'Asia') | (car['Origin'] == 'Europe')].tail(50)",
"_____no_output_____"
]
],
[
[
"# 4) Instruction (Removing unwanted records)\n\tRemove all the records(rows) where Weight is above 4000",
"_____no_output_____"
]
],
[
[
"car.head(2)",
"_____no_output_____"
],
[
"#use ~ this sign for remove..\ncar[~(car['Weight'] > 4000)]",
"_____no_output_____"
]
],
[
[
"# 5) Instruction (Applying function on a column)\n\tincrease all the values of \"MPG_City\" column by 3",
"_____no_output_____"
]
],
[
[
"car.head(2)",
"_____no_output_____"
],
[
"car['MPG_City'] = car['MPG_City'].apply(lambda x:x+3)",
"_____no_output_____"
],
[
"car.head(5)",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
4aff4d97796c5bed3aa1bf63317c28cd66d65625
| 66,077 |
ipynb
|
Jupyter Notebook
|
CNN_Pretrain/testing_stuff-checkpoint.ipynb
|
fgwy/uavSim
|
a58dc3149a52fc407c288c281eedb176738bd558
|
[
"BSD-3-Clause"
] | null | null | null |
CNN_Pretrain/testing_stuff-checkpoint.ipynb
|
fgwy/uavSim
|
a58dc3149a52fc407c288c281eedb176738bd558
|
[
"BSD-3-Clause"
] | null | null | null |
CNN_Pretrain/testing_stuff-checkpoint.ipynb
|
fgwy/uavSim
|
a58dc3149a52fc407c288c281eedb176738bd558
|
[
"BSD-3-Clause"
] | null | null | null | 54.699503 | 2,051 | 0.5506 |
[
[
[
"import tensorflow as tf\nimport numpy as np\nimport math\nimport scipy\n\n",
"_____no_output_____"
],
[
"goal_size = 4\nbatch_size = 10\nexample_goal = np.random.rand(batch_size, goal_size)\nnp.random.shuffle(example_goal)\nprint(example_goal)\narg_max = tf.argmax(example_goal, axis=1, output_type=tf.int32)\nprint(arg_max)\none_hot = tf.one_hot(arg_max, depth=1)\nprint(one_hot)\n# # example_goal[0][0] = 1\n\n# np.random.shuffle(example_goal)\n# print(example_goal, '\\n', tf.argmax(example_goal))\n\n# # np.random.shuffle(example_goal)\n# example_goal = tf.convert_to_tensor(example_goal)\n# print(tf.argmax(example_goal, 1), tf.reduce_max(example_goal))\n# highest_vals_per_col = tf.argmax(example_goal, 1)\n# print(highest_vals_per_col, highest_vals_per_col.shape)\n\n# # max_value = max(example_goal)\n# # max_index = my_list.index(max_value)",
"[[0.17570497 0.81667436 0.81373083 0.45365642]\n [0.65507156 0.9182954 0.44150857 0.9319078 ]\n [0.46075487 0.61216577 0.08352444 0.43859082]\n [0.20147648 0.34532974 0.41958065 0.19458197]\n [0.80030846 0.79636807 0.5785543 0.60433818]\n [0.39232041 0.16670067 0.46795182 0.64587489]\n [0.37149397 0.38602793 0.57724609 0.77117301]\n [0.44276678 0.61618945 0.08221358 0.92478675]\n [0.72859033 0.41533477 0.68552604 0.87852771]\n [0.53891444 0.30762412 0.18869989 0.34281981]]\ntf.Tensor([1 3 1 2 0 3 3 3 3 0], shape=(10,), dtype=int32)\ntf.Tensor(\n[[0.]\n [0.]\n [0.]\n [0.]\n [1.]\n [0.]\n [0.]\n [0.]\n [0.]\n [1.]], shape=(10, 1), dtype=float32)\n"
],
[
"place_h = []\n# for i in range(goal_size):\n# print(i, example_goal.shape)\n# a = highest_vals_per_col.data[i]\n# print[a]\n# place_h[i] = [example_goal[0][a]]\n \none_hot_idx = tf.argmax(place_h)\nprint(one_hot_idx)\none_hot = tf.one_hot(tf.argmax(example_goal), depth=4, dtype=float, on_value=1.0, off_value=0.0)\nprint(one_hot)\none_hot = tf.squeeze(one_hot)\nprint(one_hot)\n",
"_____no_output_____"
],
[
"x,y = 17,17\na = np.ones((7,7))\n# a[1][1] = 1\nmap_ = np.zeros((18,18))\n\n\n\nshape_total = map_.shape\nshape_loc = a.shape\nnp.random.shuffle(a)\nnfz = int((shape_loc[0]-1)/2)\nprint(a, \"\\n\", nfz)\n\npad_left = x\npad_right = shape_total[0] - x -1# - shape_loc[0] + nfz\npad_up = y # - nfz\npad_down = shape_total[0] - y - 1# - shape_loc[0] + nfz\n\nprint(pad_left, pad_right, pad_up, pad_down)\n\npadded = np.pad(a, ((pad_up, pad_down), (pad_left, pad_right)))\nprint(padded, \"\\n\", padded.shape)\npadded = padded[nfz:(padded.shape[0]-nfz), nfz:(padded[1]-nfz)]\nprint(padded, padded.shape)\n\n",
"289\n[[1. 1. 1. 1. 1. 1. 1.]\n [1. 1. 1. 1. 1. 1. 1.]\n [1. 1. 1. 1. 1. 1. 1.]\n [1. 1. 1. 1. 1. 1. 1.]\n [1. 1. 1. 1. 1. 1. 1.]\n [1. 1. 1. 1. 1. 1. 1.]\n [1. 1. 1. 1. 1. 1. 1.]] \n 3\n17 0 17 0\n[[0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 1. 1. 1. 1. 1. 1. 1.]\n [0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 1. 1. 1. 1. 1. 1. 1.]\n [0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 1. 1. 1. 1. 1. 1. 1.]\n [0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 1. 1. 1. 1. 1. 1. 1.]\n [0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 1. 1. 1. 1. 1. 1. 1.]\n [0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 1. 1. 1. 1. 1. 1. 1.]\n [0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 1. 1. 1. 1. 1. 1. 1.]] \n (24, 24)\n"
],
[
"# def pad_centered(state, map_in, pad_value):\nmap_in = np.zeros((10,10))\nprint(map_in.shape)\npadding_rows = math.ceil(map_in.shape[0] / 2.0)\npadding_cols = math.ceil(map_in.shape[1] / 2.0)\nposition_x, position_y = 0,0\nmap_in[position_x][position_y] = 1\npad_value = 1\n# print(\"pos\", position_x, position_y)\nposition_row_offset = padding_rows - position_y\nposition_col_offset = padding_cols - position_x\nres = np.pad(map_in,\n pad_width=[[padding_rows + position_row_offset - 1, padding_rows - position_row_offset],\n [padding_cols + position_col_offset - 1, padding_cols - position_col_offset],\n# [0, 0]\n ],\n mode='constant',\n constant_values=pad_value)\n\nprint(res,\"\\n\", res.shape)\n",
"(10, 10)\n[[1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1.]\n [1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1.]\n [1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1.]\n [1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1.]\n [1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1.]\n [1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1.]\n [1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1.]\n [1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1.]\n [1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1.]\n [1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n [1. 1. 1. 1. 1. 1. 1. 1. 1. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n [1. 1. 1. 1. 1. 1. 1. 1. 1. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n [1. 1. 1. 1. 1. 1. 1. 1. 1. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n [1. 1. 1. 1. 1. 1. 1. 1. 1. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n [1. 1. 1. 1. 1. 1. 1. 1. 1. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n [1. 1. 1. 1. 1. 1. 1. 1. 1. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n [1. 1. 1. 1. 1. 1. 1. 1. 1. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n [1. 1. 1. 1. 1. 1. 1. 1. 1. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n [1. 1. 1. 1. 1. 1. 1. 1. 1. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]] \n (19, 19)\n"
],
[
"a = np.zeros((6,6))\nb = np.zeros((6,6))\n\nfor i in range(5):\n a[i][i]=1\nnp.random.shuffle(a)\nb[0][0]=1\nprint(a, \"\\n\", bool(b.any))\n# a = not bool(a)\n\na = np.ones((6,6))\na = np.logical_not(a).astype(int)\nb = b.astype(int)\nprint(a, \"\\n\", b)\n\nc = b*a\nprint(c)\n# c = np.logical_not(c).astype(int)\nprint(c)\nprint(not np.all(c == 0))\n\n",
"[[1. 0. 0. 0. 0. 0.]\n [0. 0. 1. 0. 0. 0.]\n [0. 0. 0. 0. 1. 0.]\n [0. 1. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 1. 0. 0.]] \n True\n[[0 0 0 0 0 0]\n [0 0 0 0 0 0]\n [0 0 0 0 0 0]\n [0 0 0 0 0 0]\n [0 0 0 0 0 0]\n [0 0 0 0 0 0]] \n [[1 0 0 0 0 0]\n [0 0 0 0 0 0]\n [0 0 0 0 0 0]\n [0 0 0 0 0 0]\n [0 0 0 0 0 0]\n [0 0 0 0 0 0]]\n[[0 0 0 0 0 0]\n [0 0 0 0 0 0]\n [0 0 0 0 0 0]\n [0 0 0 0 0 0]\n [0 0 0 0 0 0]\n [0 0 0 0 0 0]]\n[[0 0 0 0 0 0]\n [0 0 0 0 0 0]\n [0 0 0 0 0 0]\n [0 0 0 0 0 0]\n [0 0 0 0 0 0]\n [0 0 0 0 0 0]]\nFalse\n"
],
[
"lm_size = (17,17)\nNT_size = (9,9)\n\nprint(17**2)\nprint(9**2)\nprint(17**2-9**2)\na = np.zeros(lm_size)\nprint(a)\n\n\n\n# NT_size[0]:(lm_size[0]-NT_size[0]), NT_size[0]:(lm_size[0]-NT_size[0])\n\na[9-5:17-4,9-5:17-4] = 1\nprint(a)",
"289\n81\n208\n[[0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]]\n[[0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 1. 1. 1. 1. 1. 1. 1. 1. 1. 0. 0. 0. 0.]\n [0. 0. 0. 0. 1. 1. 1. 1. 1. 1. 1. 1. 1. 0. 0. 0. 0.]\n [0. 0. 0. 0. 1. 1. 1. 1. 1. 1. 1. 1. 1. 0. 0. 0. 0.]\n [0. 0. 0. 0. 1. 1. 1. 1. 1. 1. 1. 1. 1. 0. 0. 0. 0.]\n [0. 0. 0. 0. 1. 1. 1. 1. 1. 1. 1. 1. 1. 0. 0. 0. 0.]\n [0. 0. 0. 0. 1. 1. 1. 1. 1. 1. 1. 1. 1. 0. 0. 0. 0.]\n [0. 0. 0. 0. 1. 1. 1. 1. 1. 1. 1. 1. 1. 0. 0. 0. 0.]\n [0. 0. 0. 0. 1. 1. 1. 1. 1. 1. 1. 1. 1. 0. 0. 0. 0.]\n [0. 0. 0. 0. 1. 1. 1. 1. 1. 1. 1. 1. 1. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]]\n"
],
[
"l1 = [10.3, 22.3, 1.1, 2.34, 0]\nl2 = [1.3, 2.3, 10.1, 20.34, 330]",
"_____no_output_____"
],
[
"def pad_lm_to_total_size(h_target, position):\n \"\"\"\n pads input of shape local_map to output of total_map_size\n \"\"\"\n\n shape_map = (32,32)\n shape_htarget = h_target.shape\n # print(shape_htarget, shape_map)\n\n x, y = position\n\n pad_left = x\n pad_right = shape_map[0] - x - 1\n pad_up = y\n pad_down = shape_map[1] - y - 1\n\n padded = np.pad(h_target, ((pad_up, pad_down), (pad_left, pad_right)))\n\n lm_as_tm_size = padded[int((shape_htarget[0] - 1) / 2):int(padded.shape[0] - (shape_htarget[0] - 1) / 2),\n int((shape_htarget[1] - 1) / 2):int(padded.shape[1] - (shape_htarget[1] - 1) / 2)]\n\n return lm_as_tm_size.astype(bool)\n\nposition = (31,31)\nh_target = np.zeros((15,15))\nh_target[7][7] = 1\n\npht = pad_lm_to_total_size(h_target, position)\nprint(pht, pht.shape)\n\nprint(pht.any()==True)",
"[[False False False ... False False False]\n [False False False ... False False False]\n [False False False ... False False False]\n ...\n [False False False ... False False False]\n [False False False ... False False False]\n [False False False ... False False True]] (32, 32)\nTrue\n"
],
[
"a = np.zeros((10,5))\n\na[9,3]=1\nprint(a)",
"[[0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0.]\n [0. 0. 0. 1. 0.]]\n"
],
[
"a = np.zeros((10,10))\na[0,1]=1\nprint(a)\n",
"[[0. 1. 0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]]\n"
],
[
"#Input \ninput_img = Input(shape=(128, 128, 3))#Encoder \ny = Conv2D(32, (3, 3), padding='same',strides =(2,2))(input_img)\ny = LeakyReLU()(y)\ny = Conv2D(64, (3, 3), padding='same',strides =(2,2))(y)\ny = LeakyReLU()(y)\ny1 = Conv2D(128, (3, 3), padding='same',strides =(2,2))(y) # skip-1\ny = LeakyReLU()(y1)\ny = Conv2D(256, (3, 3), padding='same',strides =(2,2))(y)\ny = LeakyReLU()(y)\ny2 = Conv2D(256, (3, 3), padding='same',strides =(2,2))(y)# skip-2\ny = LeakyReLU()(y2)\ny = Conv2D(512, (3, 3), padding='same',strides =(2,2))(y)\ny = LeakyReLU()(y)\ny = Conv2D(1024, (3, 3), padding='same',strides =(2,2))(y)\ny = LeakyReLU()(y)#Flattening for the bottleneck\nvol = y.shape\nx = Flatten()(y)\nlatent = Dense(128, activation='relu')(x) \n\n\n# Helper function to apply activation and batch normalization to the # output added with output of residual connection from the encoderdef lrelu_bn(inputs):\n lrelu = LeakyReLU()(inputs)\n bn = BatchNormalization()(lrelu)\n return bn#Decoder\ny = Dense(np.prod(vol[1:]), activation='relu')(latent)\ny = Reshape((vol[1], vol[2], vol[3]))(y)\ny = Conv2DTranspose(1024, (3,3), padding='same')(y)\ny = LeakyReLU()(y)\ny = Conv2DTranspose(512, (3,3), padding='same',strides=(2,2))(y)\ny = LeakyReLU()(y)\ny = Conv2DTranspose(256, (3,3), padding='same',strides=(2,2))(y)\ny= Add()([y2, y]) # second skip connection added here\ny = lrelu_bn(y)\ny = Conv2DTranspose(256, (3,3), padding='same',strides=(2,2))(y)\ny = LeakyReLU()(y)\ny = Conv2DTranspose(128, (3,3), padding='same',strides=(2,2))(y)\ny= Add()([y1, y]) # first skip connection added here\ny = lrelu_bn(y)\ny = Conv2DTranspose(64, (3,3), padding='same',strides=(2,2))(y)\ny = LeakyReLU()(y)\ny = Conv2DTranspose(32, (3,3), padding='same',strides=(2,2))(y)\ny = LeakyReLU()(y)\ny = Conv2DTranspose(3, (3,3), activation='sigmoid', padding='same',strides=(2,2))(y)",
"_____no_output_____"
],
[
"def lrelu(inputs):\n lrelu = LeakyReLU()(inputs)\n bn = BatchNormalization()(lrelu)\n return bn\n\nconv_layers = 2\nmb = 25\ncurrent_mb = 15\nhidden_layer_size = 256\nname = 'hl_model_'\nlm = np.random.rand(17,17,4)\ngm = np.random.rand(21,21,4)\nstates_proc = np.array(current_mb/mb)\n\n\ndef build_hl_model(local_map, global_map, states_proc): #local:17,17,4; global:21:21,4\n \n # local map processing layers\n# for k in range(conv_layers):\n local_map_input = tf.keras.layers.Input(shape=local_map.shape)\n global_map_input = tf.keras.layers.Input(shape=global_map.shape)\n states_proc_input = tf.keras.layers.Input(shape=states_proc.shape)\n \n local_map_1 = tf.keras.layers.Conv2D(4, 3, activation='elu',\n strides=(1, 1),\n name=name + 'local_conv_' + str(0 + 1))(local_map_input) #out:(None, 1, 15, 15, 4) 1156->\n local_map_2 = tf.keras.layers.Conv2D(8, 3, activation='elu',\n strides=(1, 1),\n name=name + 'local_conv_' + str(1 + 1))(local_map_1) #out:(None, 1, 13, 13, 8)\n local_map_3 = tf.keras.layers.Conv2D(16, 3, activation='elu',\n strides=(1, 1),\n name=name + 'local_conv_' + str(2 + 1))(local_map_2) #out:(None, 1, 11, 11, 16)\n local_map_4 = tf.keras.layers.Conv2D(16, 3, activation='elu',\n strides=(1, 1),\n name=name + 'local_conv_' + str(3 + 1))(local_map_3) #out:(None, 1, 9, 9, 16)\n flatten_local = tf.keras.layers.Flatten(name=name + 'local_flatten')(local_map_4)\n \n # global map processing layers\n\n global_map_1 = tf.keras.layers.Conv2D(4, 5, activation='elu',\n strides=(1, 1),\n name=name + 'global_conv_' + str(0 + 1))(global_map_input) #out:17\n global_map_2 = tf.keras.layers.Conv2D(8, 5, activation='elu',\n strides=(1, 1),\n name=name + 'global_map_' + str(1 + 1))(global_map_1) #out:13\n global_map_3 = tf.keras.layers.Conv2D(16, 5, activation='elu',\n strides=(1, 1),\n name=name + 'global_map_' + str(2 + 1))(global_map_2)#out:9\n\n flatten_global = tf.keras.layers.Flatten(name=name + 'global_flatten')(global_map_3)\n \n print(flatten_local.shape, flatten_global.shape)\n \n flatten_map = tf.keras.layers.Concatenate(name=name + 'concat_flatten')([flatten_global, flatten_local])\n \n layer = tf.keras.layers.Concatenate(name=name + 'concat')([flatten_map, states_proc_input])\n \n layer_1 = tf.keras.layers.Dense(256, activation='elu', name=name + 'hidden_layer_all_hl_' + str(0))(\n layer)\n layer_2 = tf.keras.layers.Dense(512, activation='elu', name=name + 'hidden_layer_all_hl_' + str(1))(\n layer_1)\n layer_3 = tf.keras.layers.Dense(256, activation='elu', name=name + 'hidden_layer_all_hl_' + str(2))(\n layer_2)\n\n output = tf.keras.layers.Dense(units=300, activation='linear', name=name + 'last_dense_layer_hl')(\n layer)\n \n reshape = tf.keras.layers.Reshape((5,5,12), name=name + 'last_dense_layer')(output)\n\n \n landing = tf.keras.layers.Dense(units=128, activation='elu', name=name + 'landing_preproc_layer_hl')(\n layer_3)\n landing = tf.keras.layers.Dense(units=1, activation='elu', name=name + 'landing_layer_hl')(landing)\n \n # deconvolutional part aiming at 17x17 \n deconv_1 = tf.keras.layers.Conv2DTranspose(filters=16, kernel_size=5, activation='elu', name=name + 'deconv_' + str(1))(reshape)\n skip_1 = tf.keras.layers.Concatenate(name=name + '1st_skip_connection_concat', axis=3)([deconv_1, tf.squeeze(local_map_4, axis=1)])\n deconv_2 = tf.keras.layers.Conv2DTranspose(filters=8, kernel_size=3, activation='elu', name=name + 'deconv_' + str(2))(skip_1)\n skip_2 = tf.keras.layers.Concatenate(name=name + '2nd_skip_connection_concat', axis=3)([deconv_2, tf.squeeze(local_map_3, axis=1)])\n deconv_2_1 = tf.keras.layers.Conv2DTranspose(filters=8, kernel_size=3, activation='elu', name=name + 'deconv_' + str(2.1))(skip_2)\n skip_3 = tf.keras.layers.Concatenate(name=name + '3rd_skip_connection_concat', axis=3)([deconv_2_1, tf.squeeze(local_map_2, axis=1)])\n deconv_3 = tf.keras.layers.Conv2DTranspose(filters=4, kernel_size=5, activation='elu', name=name + 'deconv_' + str(3))(skip_3)\n deconv_4 = tf.keras.layers.Conv2DTranspose(filters=1, kernel_size=1, activation='elu', name=name + 'deconv_' + str(4))(deconv_3)\n\n flatten_deconv = tf.keras.layers.Flatten(name=name + 'deconv_flatten')(deconv_4)\n concat_final = tf.keras.layers.Concatenate(name=name + 'concat_final')([flatten_deconv, landing])\n \n return tf.keras.Model(inputs=[local_map_input, global_map_input, states_proc_input], outputs=concat_final)\n \n\nmodel = build_hl_model(lm[tf.newaxis, ...], gm[tf.newaxis, ...], states_proc[tf.newaxis, ...]) #lm, gm, states_proc)\n\nmodel.compile(optimizer='adam', loss='mse')\n# model.build()\nmodel.summary()\n ",
"(None, 1296) (None, 1296)\nModel: \"model_18\"\n__________________________________________________________________________________________________\nLayer (type) Output Shape Param # Connected to \n==================================================================================================\ninput_157 (InputLayer) [(None, 1, 17, 17, 4 0 \n__________________________________________________________________________________________________\ninput_158 (InputLayer) [(None, 1, 21, 21, 4 0 \n__________________________________________________________________________________________________\nhl_model_local_conv_1 (Conv2D) (None, 1, 15, 15, 4) 148 input_157[0][0] \n__________________________________________________________________________________________________\nhl_model_global_conv_1 (Conv2D) (None, 1, 17, 17, 4) 404 input_158[0][0] \n__________________________________________________________________________________________________\nhl_model_local_conv_2 (Conv2D) (None, 1, 13, 13, 8) 296 hl_model_local_conv_1[0][0] \n__________________________________________________________________________________________________\nhl_model_global_map_2 (Conv2D) (None, 1, 13, 13, 8) 808 hl_model_global_conv_1[0][0] \n__________________________________________________________________________________________________\nhl_model_local_conv_3 (Conv2D) (None, 1, 11, 11, 16 1168 hl_model_local_conv_2[0][0] \n__________________________________________________________________________________________________\nhl_model_global_map_3 (Conv2D) (None, 1, 9, 9, 16) 3216 hl_model_global_map_2[0][0] \n__________________________________________________________________________________________________\nhl_model_local_conv_4 (Conv2D) (None, 1, 9, 9, 16) 2320 hl_model_local_conv_3[0][0] \n__________________________________________________________________________________________________\nhl_model_global_flatten (Flatte (None, 1296) 0 hl_model_global_map_3[0][0] \n__________________________________________________________________________________________________\nhl_model_local_flatten (Flatten (None, 1296) 0 hl_model_local_conv_4[0][0] \n__________________________________________________________________________________________________\nhl_model_concat_flatten (Concat (None, 2592) 0 hl_model_global_flatten[0][0] \n hl_model_local_flatten[0][0] \n__________________________________________________________________________________________________\ninput_159 (InputLayer) [(None, 1)] 0 \n__________________________________________________________________________________________________\nhl_model_concat (Concatenate) (None, 2593) 0 hl_model_concat_flatten[0][0] \n input_159[0][0] \n__________________________________________________________________________________________________\nhl_model_last_dense_layer_hl (D (None, 300) 778200 hl_model_concat[0][0] \n__________________________________________________________________________________________________\nhl_model_last_dense_layer (Resh (None, 5, 5, 12) 0 hl_model_last_dense_layer_hl[0][0\n__________________________________________________________________________________________________\nhl_model_deconv_1 (Conv2DTransp (None, 9, 9, 16) 4816 hl_model_last_dense_layer[0][0] \n__________________________________________________________________________________________________\ntf.compat.v1.squeeze_32 (TFOpLa (None, 9, 9, 16) 0 hl_model_local_conv_4[0][0] \n__________________________________________________________________________________________________\nhl_model_1st_skip_connection_co (None, 9, 9, 32) 0 hl_model_deconv_1[0][0] \n tf.compat.v1.squeeze_32[0][0] \n__________________________________________________________________________________________________\nhl_model_deconv_2 (Conv2DTransp (None, 11, 11, 8) 2312 hl_model_1st_skip_connection_conc\n__________________________________________________________________________________________________\ntf.compat.v1.squeeze_33 (TFOpLa (None, 11, 11, 16) 0 hl_model_local_conv_3[0][0] \n__________________________________________________________________________________________________\nhl_model_2nd_skip_connection_co (None, 11, 11, 24) 0 hl_model_deconv_2[0][0] \n tf.compat.v1.squeeze_33[0][0] \n__________________________________________________________________________________________________\nhl_model_deconv_2.1 (Conv2DTran (None, 13, 13, 8) 1736 hl_model_2nd_skip_connection_conc\n__________________________________________________________________________________________________\ntf.compat.v1.squeeze_34 (TFOpLa (None, 13, 13, 8) 0 hl_model_local_conv_2[0][0] \n__________________________________________________________________________________________________\nhl_model_hidden_layer_all_hl_0 (None, 256) 664064 hl_model_concat[0][0] \n__________________________________________________________________________________________________\nhl_model_3rd_skip_connection_co (None, 13, 13, 16) 0 hl_model_deconv_2.1[0][0] \n tf.compat.v1.squeeze_34[0][0] \n__________________________________________________________________________________________________\nhl_model_hidden_layer_all_hl_1 (None, 512) 131584 hl_model_hidden_layer_all_hl_0[0]\n__________________________________________________________________________________________________\nhl_model_deconv_3 (Conv2DTransp (None, 17, 17, 4) 1604 hl_model_3rd_skip_connection_conc\n__________________________________________________________________________________________________\nhl_model_hidden_layer_all_hl_2 (None, 256) 131328 hl_model_hidden_layer_all_hl_1[0]\n__________________________________________________________________________________________________\nhl_model_deconv_4 (Conv2DTransp (None, 17, 17, 1) 5 hl_model_deconv_3[0][0] \n__________________________________________________________________________________________________\nhl_model_landing_preproc_layer_ (None, 128) 32896 hl_model_hidden_layer_all_hl_2[0]\n__________________________________________________________________________________________________\nhl_model_deconv_flatten (Flatte (None, 289) 0 hl_model_deconv_4[0][0] \n__________________________________________________________________________________________________\nhl_model_landing_layer_hl (Dens (None, 1) 129 hl_model_landing_preproc_layer_hl\n__________________________________________________________________________________________________\nhl_model_concat_final (Concaten (None, 290) 0 hl_model_deconv_flatten[0][0] \n hl_model_landing_layer_hl[0][0] \n==================================================================================================\nTotal params: 1,757,034\nTrainable params: 1,757,034\nNon-trainable params: 0\n__________________________________________________________________________________________________\n"
],
[
"a = np.arange(10)\nprint(a)\na = a.reshape(2,5)\nprint(a)\na = tf.keras.layers.Flatten()(tf.convert_to_tensor(a))\nprint(a)",
"[0 1 2 3 4 5 6 7 8 9]\n[[0 1 2 3 4]\n [5 6 7 8 9]]\ntf.Tensor(\n[[0 1 2 3 4]\n [5 6 7 8 9]], shape=(2, 5), dtype=int64)\n"
],
[
"a, b, c = tf.stop_gradient([1,2,3])\nprint( a,b,c\n )",
"tf.Tensor(1, shape=(), dtype=int32) tf.Tensor(2, shape=(), dtype=int32) tf.Tensor(3, shape=(), dtype=int32)\n"
],
[
"local_map_in = np.zeros((4,5))\nglobal_map_in = np.ones_like(local_map_in)\nscalars_in = 1\n# local_map_in[0][4]=np.float('nan')\nif np.any(np.isnan(local_map_in)) or np.any(np.isnan(global_map_in)) or np.any(np.isnan(scalars_in)) :\n print(f'###################### Nan in act input: {np.isnan(local_map_in)}')",
"_____no_output_____"
],
[
"size = (40)\np = np.zeros(size)\np[1] = 1\na = np.random.choice(range(40), size=1, p=p)\na = tf.one_hot((1000), depth=size).numpy().reshape(5,8)\nprint(a)\na = tf.keras.layers.Flatten()(a)\nprint(a)\na = a.numpy().reshape(5,8)\nprint(a)",
"[[0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0.]]\ntf.Tensor(\n[[0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0.]], shape=(5, 8), dtype=float32)\n[[0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0.]]\n"
],
[
"b = np.array([1, 1, 0, 0, 1])\n\nb = tf.math.is_nan(tf.convert_to_tensor(b))\n\na = tf.reduce_any(b)\n\nprint(a)",
"_____no_output_____"
],
[
"sz = 11\na = np.random.rand(sz,sz)\nb = 3\nc = int((sz-1)/2 - (b-1)/2)\nprint(c)\nfor i in range(b):\n for j in range(b):\n# a[i+c][j+c]=-math.inf\n a[i+c][j+c]=0\n \n \nprint(a)\na = tf.keras.layers.Flatten()(a[tf.newaxis, ...]).numpy()\na = np.squeeze(a)\nprint(a)\na = scipy.special.softmax(a)\nprint(a)\n\n",
"4\n[[0.85903 0.45615618 0.75352573 0.28616315 0.656314 0.40986627\n 0.31044914 0.94073707 0.03260706 0.20273256 0.53378992]\n [0.56615772 0.20992389 0.21193915 0.38760111 0.7934665 0.57851711\n 0.14808255 0.07390192 0.00809374 0.86135458 0.0260759 ]\n [0.73361629 0.37659285 0.34437586 0.6400296 0.87902691 0.48436414\n 0.49118999 0.31552158 0.68295266 0.98499074 0.66384527]\n [0.65358725 0.91551074 0.35424552 0.92398924 0.2827416 0.77516516\n 0.79810265 0.21162833 0.80351687 0.90661335 0.50315294]\n [0.19520098 0.70098219 0.38246709 0.72394498 0. 0.\n 0. 0.44388019 0.18274414 0.9029086 0.77770016]\n [0.62226835 0.63678732 0.7381341 0.3879475 0. 0.\n 0. 0.69032153 0.8001331 0.8663466 0.10392525]\n [0.61907532 0.12746065 0.40523353 0.20330397 0. 0.\n 0. 0.71342267 0.3259403 0.99208458 0.24866851]\n [0.49543152 0.33516277 0.5995586 0.38818935 0.22187303 0.19782512\n 0.91076292 0.85435479 0.37687985 0.21084955 0.70612536]\n [0.80627865 0.42170152 0.43714887 0.76846941 0.23046562 0.38785737\n 0.13959836 0.92770249 0.94806577 0.58370962 0.85580163]\n [0.6092264 0.94894204 0.47589102 0.00169341 0.54790165 0.17868032\n 0.97465975 0.90827907 0.45635785 0.52106672 0.24707265]\n [0.54113419 0.81209233 0.73425428 0.22573739 0.61183313 0.23493544\n 0.19775216 0.42191449 0.131545 0.23224329 0.16233885]]\n[0.85903 0.45615616 0.75352573 0.28616315 0.656314 0.40986627\n 0.31044915 0.94073707 0.03260706 0.20273256 0.53378993 0.5661577\n 0.2099239 0.21193914 0.3876011 0.7934665 0.57851714 0.14808254\n 0.07390192 0.00809374 0.8613546 0.0260759 0.7336163 0.37659284\n 0.34437585 0.6400296 0.8790269 0.48436415 0.49119 0.31552157\n 0.68295264 0.9849907 0.66384524 0.6535873 0.9155107 0.3542455\n 0.92398924 0.2827416 0.77516514 0.7981027 0.21162833 0.80351686\n 0.90661335 0.50315297 0.19520098 0.7009822 0.3824671 0.72394496\n 0. 0. 0. 0.4438802 0.18274413 0.9029086\n 0.7777002 0.6222684 0.6367873 0.7381341 0.3879475 0.\n 0. 0. 0.6903215 0.8001331 0.8663466 0.10392525\n 0.6190753 0.12746066 0.40523353 0.20330396 0. 0.\n 0. 0.71342266 0.3259403 0.99208456 0.2486685 0.4954315\n 0.33516276 0.5995586 0.38818935 0.22187303 0.19782512 0.9107629\n 0.8543548 0.37687984 0.21084955 0.7061254 0.80627865 0.42170152\n 0.43714887 0.7684694 0.23046562 0.38785738 0.13959835 0.9277025\n 0.94806576 0.5837096 0.85580164 0.6092264 0.94894207 0.47589102\n 0.00169341 0.54790163 0.17868033 0.97465974 0.90827906 0.45635784\n 0.5210667 0.24707265 0.5411342 0.8120923 0.7342543 0.2257374\n 0.61183316 0.23493545 0.19775216 0.4219145 0.13154499 0.23224328\n 0.16233885]\n[0.01158038 0.00774029 0.01042085 0.00653026 0.0094555 0.00739016\n 0.00669079 0.01256632 0.00506771 0.00600754 0.00836514 0.00864033\n 0.0060509 0.00606311 0.00722743 0.01084549 0.00874778 0.00568804\n 0.00528137 0.004945 0.01160734 0.00503472 0.01021543 0.00714831\n 0.00692168 0.00930277 0.01181429 0.00796173 0.00801627 0.00672482\n 0.00971077 0.01313491 0.00952698 0.00942976 0.01225328 0.00699033\n 0.01235761 0.00650795 0.01064881 0.01089589 0.00606122 0.01095504\n 0.01214474 0.00811274 0.00596246 0.00988744 0.00719042 0.01011711\n 0.00490514 0.00490514 0.00490514 0.00764585 0.00588865 0.01209983\n 0.01067584 0.009139 0.00927266 0.01026168 0.00722994 0.00490514\n 0.00490514 0.00490514 0.00978259 0.01091804 0.01166542 0.00544233\n 0.00910987 0.00557194 0.007356 0.00601097 0.00490514 0.00490514\n 0.00490514 0.01001121 0.00679524 0.01322842 0.00628994 0.00805034\n 0.0068582 0.0089338 0.00723168 0.00612363 0.00597813 0.01219524\n 0.01152637 0.00715036 0.0060565 0.00993842 0.01098534 0.00747814\n 0.00759456 0.01057774 0.00617648 0.00722929 0.00563998 0.01240358\n 0.01265875 0.00879332 0.01154306 0.00902058 0.01266985 0.00789456\n 0.00491345 0.00848402 0.00586477 0.01299991 0.01216498 0.00774185\n 0.00825938 0.00627991 0.0084268 0.01104939 0.01022195 0.00614735\n 0.00904413 0.00620415 0.00597769 0.00747973 0.00559475 0.00618747\n 0.00576971]\n"
],
[
"a = np.zeros((17,17))\nv = np.zeros((11,11))\n\nb = np.random.rand(11,11)\ndv = int((a.shape[0]-b.shape[0])/2)\nprint(dv)\nfor i in range(b.shape[0]):\n for j in range(b.shape[1]):\n a[i+dv][j+dv]=b[i][j]\nprint(a)\n# v = np.zeros((11,11,4))\nprint(v)\ndv = int((a.shape[0]-v.shape[0])/2)\nprint(dv)\n\nfor i in range(v.shape[0]):\n for j in range(v.shape[1]):\n v[i][j]=a[i+dv][j+dv] # [3]\n\nprint(v)",
"3\n[[0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. ]\n [0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. ]\n [0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. ]\n [0. 0. 0. 0.76181723 0.44534156 0.66537213\n 0.99453811 0.92273703 0.91531384 0.87244398 0.63392102 0.79081619\n 0.2361872 0.07628017 0. 0. 0. ]\n [0. 0. 0. 0.57719785 0.85631048 0.6116705\n 0.42807821 0.44677604 0.9827626 0.10399085 0.15583558 0.0555077\n 0.87603932 0.79392707 0. 0. 0. ]\n [0. 0. 0. 0.47938728 0.49354469 0.3395669\n 0.34023351 0.90847343 0.50912706 0.35745864 0.1611303 0.85618968\n 0.78897265 0.80688676 0. 0. 0. ]\n [0. 0. 0. 0.75091189 0.4542349 0.42034586\n 0.25246496 0.55325201 0.39630838 0.56428047 0.83955843 0.41505947\n 0.35321452 0.77643613 0. 0. 0. ]\n [0. 0. 0. 0.80781209 0.96572507 0.1031235\n 0.16607626 0.1133003 0.60075425 0.67889049 0.7119989 0.61166602\n 0.40257559 0.98249968 0. 0. 0. ]\n [0. 0. 0. 0.128886 0.85509524 0.38092101\n 0.86349268 0.18777041 0.41608138 0.83462434 0.59686333 0.53574295\n 0.14993223 0.78348061 0. 0. 0. ]\n [0. 0. 0. 0.00144689 0.02666539 0.29448727\n 0.54532638 0.44243586 0.22955977 0.09887055 0.3744612 0.0962165\n 0.2638637 0.16449845 0. 0. 0. ]\n [0. 0. 0. 0.53589767 0.27044362 0.75674756\n 0.1002469 0.38229672 0.74343444 0.08376721 0.54327714 0.92371232\n 0.70493676 0.3854846 0. 0. 0. ]\n [0. 0. 0. 0.1048669 0.97565011 0.51579935\n 0.57320343 0.38599205 0.07692458 0.95576389 0.69605442 0.0114053\n 0.40998882 0.13584043 0. 0. 0. ]\n [0. 0. 0. 0.03996253 0.20200814 0.27017422\n 0.03397632 0.62943747 0.2380568 0.32712564 0.37708691 0.2787412\n 0.83068124 0.62599562 0. 0. 0. ]\n [0. 0. 0. 0.7239421 0.35693653 0.25707462\n 0.77940229 0.07464426 0.57406134 0.00579886 0.57196461 0.55900043\n 0.52349929 0.02712629 0. 0. 0. ]\n [0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. ]\n [0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. ]\n [0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. ]]\n[[0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]]\n3\n[[0.76181723 0.44534156 0.66537213 0.99453811 0.92273703 0.91531384\n 0.87244398 0.63392102 0.79081619 0.2361872 0.07628017]\n [0.57719785 0.85631048 0.6116705 0.42807821 0.44677604 0.9827626\n 0.10399085 0.15583558 0.0555077 0.87603932 0.79392707]\n [0.47938728 0.49354469 0.3395669 0.34023351 0.90847343 0.50912706\n 0.35745864 0.1611303 0.85618968 0.78897265 0.80688676]\n [0.75091189 0.4542349 0.42034586 0.25246496 0.55325201 0.39630838\n 0.56428047 0.83955843 0.41505947 0.35321452 0.77643613]\n [0.80781209 0.96572507 0.1031235 0.16607626 0.1133003 0.60075425\n 0.67889049 0.7119989 0.61166602 0.40257559 0.98249968]\n [0.128886 0.85509524 0.38092101 0.86349268 0.18777041 0.41608138\n 0.83462434 0.59686333 0.53574295 0.14993223 0.78348061]\n [0.00144689 0.02666539 0.29448727 0.54532638 0.44243586 0.22955977\n 0.09887055 0.3744612 0.0962165 0.2638637 0.16449845]\n [0.53589767 0.27044362 0.75674756 0.1002469 0.38229672 0.74343444\n 0.08376721 0.54327714 0.92371232 0.70493676 0.3854846 ]\n [0.1048669 0.97565011 0.51579935 0.57320343 0.38599205 0.07692458\n 0.95576389 0.69605442 0.0114053 0.40998882 0.13584043]\n [0.03996253 0.20200814 0.27017422 0.03397632 0.62943747 0.2380568\n 0.32712564 0.37708691 0.2787412 0.83068124 0.62599562]\n [0.7239421 0.35693653 0.25707462 0.77940229 0.07464426 0.57406134\n 0.00579886 0.57196461 0.55900043 0.52349929 0.02712629]]\n"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4aff5954a7bc5a15e0bca2d035da29a97bcce586
| 151,704 |
ipynb
|
Jupyter Notebook
|
tools/python_ecosystem.ipynb
|
bpesquet/machine-learning-handbook
|
1a590073cf100a0473ec30c48b35054494a31fd9
|
[
"MIT"
] | 16 |
2018-12-08T17:48:47.000Z
|
2020-06-30T12:58:07.000Z
|
tools/python_ecosystem.ipynb
|
bpesquet/machine-learning-handbook
|
1a590073cf100a0473ec30c48b35054494a31fd9
|
[
"MIT"
] | null | null | null |
tools/python_ecosystem.ipynb
|
bpesquet/machine-learning-handbook
|
1a590073cf100a0473ec30c48b35054494a31fd9
|
[
"MIT"
] | 3 |
2019-04-20T23:48:07.000Z
|
2020-01-22T05:55:46.000Z
| 306.472727 | 139,076 | 0.93221 |
[
[
[
"# The Python ecosystem",
"_____no_output_____"
],
[
"## Why Python?",
"_____no_output_____"
],
[
"### Python in a nutshell\n\n[Python](https://www.python.org) is a multi-purpose programming language created in 1989 by [Guido van Rossum](https://en.wikipedia.org/wiki/Guido_van_Rossum) and developed under a open source license.\n\nIt has the following characteristics:\n\n- multi-paradigms (procedural, fonctional, object-oriented);\n- dynamic types;\n- automatic memory management;\n- and much more!",
"_____no_output_____"
],
[
"### The Python syntax\n\nFor more examples, see the [Python cheatsheet](../tools/python_cheatsheet).",
"_____no_output_____"
]
],
[
[
"def hello(name):\n print(f\"Hello, {name}\")\n\n\nfriends = [\"Lou\", \"David\", \"Iggy\"]\n\nfor friend in friends:\n hello(friend)",
"Hello, Lou\nHello, David\nHello, Iggy\n"
]
],
[
[
"### Introduction to Data Science\n\n- Main objective: extract insight from data.\n- Expression born in 1997 in the statistician community.\n- \"A Data Scientist is a statistician that lives in San Francisco\".\n- 2012 : \"Sexiest job of the 21st century\" (Harvard Business Review).\n- [Controversy](https://en.wikipedia.org/wiki/Data_science#Relationship_to_statistics) on the expression's real usefulness.",
"_____no_output_____"
],
[
"[](https://en.wikipedia.org/wiki/Data_science)",
"_____no_output_____"
],
[
"[](http://drewconway.com/zia/2013/3/26/the-data-science-venn-diagram)",
"_____no_output_____"
],
[
"[](http://drewconway.com/zia/2013/3/26/the-data-science-venn-diagram)",
"_____no_output_____"
],
[
"### Python, a standard for ML and Data Science\n\n- Language qualities (ease of use, simplicity, versatility).\n- Involvement of the scientific and academical communities.\n- Rich ecosystem of dedicated open source libraries.",
"_____no_output_____"
],
[
"## Essential Python tools",
"_____no_output_____"
],
[
"### Anaconda\n\n[Anaconda](https://www.anaconda.com/distribution/) is a scientific distribution including Python and many (1500+) specialized packages. it is the easiest way to setup a work environment for ML and Data Science with Python.\n\n[](https://www.anaconda.com/distribution/)",
"_____no_output_____"
],
[
"### Jupyter Notebook\n\nThe Jupyter Notebook is an open-source web application that allows to manage documents (_.ipynb_ files) that may contain live code, equations, visualizations and text.\n\nIt has become the *de facto* standard for sharing research results in numerical fields.\n\n[](https://jupyter.org/)",
"_____no_output_____"
],
[
"### Google Colaboratory\n\nCloud environment for executing Jupyter notebooks through CPU, GPU or TPU.\n\n[](https://colab.research.google.com)",
"_____no_output_____"
],
[
"### NumPy\n\n[NumPy](https://numpy.org/) is a Python library providing support for multi-dimensional arrays, along with a large collection of mathematical functions to operate on these arrays. \n\nIt is the fundamental package for scientific computing in Python.",
"_____no_output_____"
]
],
[
[
"# Import the NumPy package under the alias \"np\"\nimport numpy as np\n\nx = np.array([1, 4, 2, 5, 3])\nprint(x[:2])\nprint(x[2:])\nprint(np.sort(x))",
"[1 4]\n[2 5 3]\n[1 2 3 4 5]\n"
]
],
[
[
"### pandas\n\n[pandas](https://pandas.pydata.org/) is a Python library providing high-performance, easy-to-use data structures and data analysis tools.\n\nThe primary data structures in **pandas** are implemented as two classes:\n\n- **DataFrame**, which you can imagine as a relational data table, with rows and named columns.\n- **Series**, which is a single column. A DataFrame contains one or more Series and a name for each Series.\n\nThe DataFrame is a commonly used abstraction for data manipulation.",
"_____no_output_____"
]
],
[
[
"import pandas as pd\n\n# Create a DataFrame object contraining two Series\npop = pd.Series({\"CAL\": 38332521, \"TEX\": 26448193, \"NY\": 19651127})\narea = pd.Series({\"CAL\": 423967, \"TEX\": 695662, \"NY\": 141297})\npd.DataFrame({\"population\": pop, \"area\": area})",
"_____no_output_____"
]
],
[
[
"### Matplotlib and Seaborn\n\n[Matplotlib](https://matplotlib.org/) is a Python library for 2D plotting. [Seaborn](https://seaborn.pydata.org) is another visualization library that improves presentation of matplotlib-generated graphics.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\nimport seaborn as sns",
"_____no_output_____"
],
[
"# Setup plots (should be done on a separate cell for better results)\n%matplotlib inline\nplt.rcParams[\"figure.figsize\"] = 10, 8\n%config InlineBackend.figure_format = \"retina\"\nsns.set()",
"_____no_output_____"
],
[
"# Plot a single function\nx = np.linspace(0, 10, 30)\nplt.plot(x, np.cos(x), label=\"cosinus\")\nplt.plot(x, np.sin(x), '-ok', label=\"sinus\")\nplt.legend()\nplt.show()",
"_____no_output_____"
]
],
[
[
"### scikit-learn\n\n[scikit-learn](https://scikit-learn.org) is a multi-purpose library built over Numpy and Matplotlib and providing dozens of built-in ML algorithms and models.\n\nIt is the Swiss army knife of Machine Learning.\n\nFun fact: scikit-learn was originally created by [INRIA](https://www.inria.fr).",
"_____no_output_____"
],
[
"### Keras\n\n[Keras](https://keras.io/) is a high-level, user-friendly API for creating and training neural nets.\n\nOnce compatible with many back-end tools (Caffe, Theano, CNTK...), Keras is now the official high-level API of [TensorFlow](https://www.tensorflow.org/), Google's Machine Learning platform.\n\nThe [2.3.0 release](https://github.com/keras-team/keras/releases/tag/2.3.0) (Sept. 2019) was the last major release of multi-backend Keras.\n\nSee [this notebook](https://colab.research.google.com/drive/1UCJt8EYjlzCs1H1d1X0iDGYJsHKwu-NO) for a introduction to TF+Keras.",
"_____no_output_____"
],
[
"### PyTorch\n\n[PyTorch](https://pytorch.org) is a Machine Learning platform supported by Facebook and competing with [TensorFlow](https://www.tensorflow.org/) for the hearts and minds of ML practitioners worldwide. It provides:\n- a array manipulation API similar to NumPy;\n- an autodifferentiation engine for computing gradients;\n- a neural network API.\n\nIt is based on previous work, notably [Torch](http://torch.ch/) and [Chainer](https://chainer.org/).",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
4aff6c819f4b9f70f71bcf43f0e858068df2e7cc
| 139,882 |
ipynb
|
Jupyter Notebook
|
Catboost_Regression2.ipynb
|
farazattar/Python
|
b4f6705edf37f1e51d86985739b67d6aeb6586ea
|
[
"MIT"
] | null | null | null |
Catboost_Regression2.ipynb
|
farazattar/Python
|
b4f6705edf37f1e51d86985739b67d6aeb6586ea
|
[
"MIT"
] | null | null | null |
Catboost_Regression2.ipynb
|
farazattar/Python
|
b4f6705edf37f1e51d86985739b67d6aeb6586ea
|
[
"MIT"
] | null | null | null | 53.329013 | 33,824 | 0.636136 |
[
[
[
"# Catboost Regression No. 2 (With GPU)",
"_____no_output_____"
]
],
[
[
"# Import the required libraries.\nimport pandas as pd\nimport numpy as np",
"_____no_output_____"
],
[
"# Read the train data.\ndata_train = pd.read_csv('train.csv')",
"_____no_output_____"
],
[
"# Read the first lines of the train data.\ndata_train.head()",
"_____no_output_____"
],
[
"# Print the shape of the train data.\ndata_train.shape",
"_____no_output_____"
],
[
"# Count the number of null values.\ndata_train.isnull().sum()",
"_____no_output_____"
],
[
"# Calculate the sum of the null values.\ndata_train.isnull().sum().sum()",
"_____no_output_____"
],
[
"# Show information about the train data.\ndata_train.describe()",
"_____no_output_____"
],
[
"# Show additional information about the train data.\ndata_train.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 188318 entries, 0 to 188317\nColumns: 132 entries, id to loss\ndtypes: float64(15), int64(1), object(116)\nmemory usage: 189.7+ MB\n"
],
[
"# Read the test data.\ndata_test = pd.read_csv('test.csv')",
"_____no_output_____"
],
[
"# Read the first lines of the test data.\ndata_test.head()",
"_____no_output_____"
],
[
"# Print the shape of the test data.\ndata_test.shape",
"_____no_output_____"
],
[
"# Count the number of null values.\ndata_test.isnull().sum()",
"_____no_output_____"
],
[
"# Calculate the sum of the null values.\ndata_test.isnull().sum().sum()",
"_____no_output_____"
],
[
"# Show information about the test data.\ndata_test.describe()",
"_____no_output_____"
],
[
"# Show additional information about the test data.\ndata_test.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 125546 entries, 0 to 125545\nColumns: 131 entries, id to cont14\ndtypes: float64(14), int64(1), object(116)\nmemory usage: 125.5+ MB\n"
],
[
"# Data_train has an extra column, namely 'loss' column.\n# This is the column which contains the target values.\n# Let's look at it.\ndata_train.iloc[:, -1]",
"_____no_output_____"
],
[
"# For data visualization, import matplotlib and seaborn.\nimport matplotlib.pyplot as plt\nimport seaborn as sns",
"_____no_output_____"
],
[
"# Plot the 'loss' column.\nplt.figure(figsize=(20, 12))\nsns.distplot(data_train['loss'])",
"_____no_output_____"
],
[
"# Plot the 'loss' column logarithmic.\nplt.figure(figsize=(20, 12))\nsns.distplot(np.log(data_train['loss']))",
"_____no_output_____"
],
[
"# This is not a necessary step.\n# Adding a column to data_train and attribute a 'true' value to it.\ndata_train['is_train'] = True",
"_____no_output_____"
],
[
"# This is not a necessary step.\n# Adding a column to data_test and attribute a 'false' value to it.\ndata_test['is_train'] = False",
"_____no_output_____"
],
[
"# Read the first lines of the new train data.\ndata_train.head()",
"_____no_output_____"
],
[
"# Read the first lines of the new test data.\ndata_test.head()",
"_____no_output_____"
],
[
"# This is not a necessary step.\n# Concat the two data sets into one data set.\ndata_train_test = pd.concat([data_train, data_test], axis=0)",
"_____no_output_____"
],
[
"# Read the first lines of the new data set.\ndata_train_test.head()",
"_____no_output_____"
],
[
"# Prepare the train data for regression using catboost.\n# We need to execute some modification concerning column names.\n# Let's look at the column names.\ndata_train.columns",
"_____no_output_____"
],
[
"# As it was shown, generally we have two type of column:\n# 1) cat, that is an abbreviation of 'categorical',\n# 2) cont, that is an abbreviation of 'continuous'.\n# So the data set's values have two general types: categorical and continouos.\n# In order to give our data to catboost algorithm, we should seperate them.\n# Furthermore, we must only give the column indexes to this algorithm.\n# Considering this goal, we should use 'Regular Experssion (re)' pattern matching.\nimport re",
"_____no_output_____"
],
[
"# Specify our pattern for categorical column names.\ncat_pattern = re.compile(\"^cat([1-9]|[1-9][0-9]|[1-9][0-9][0-9])$\")",
"_____no_output_____"
],
[
"# Specify our pattern for continuous column names.\ncont_pattern = re.compile(\"^cont([1-9]|[1-9][0-9]|[1-9][0-9][0-9])$\")",
"_____no_output_____"
],
[
"# Devide the categorical column names.\ncat_column = [cat for cat in data_train.columns if 'cat' in cat]",
"_____no_output_____"
],
[
"# Print the categorical column names.\ncat_column",
"_____no_output_____"
],
[
"# Although the categorical column names are sorted, we should force it to be sorted.\ncat_column = sorted(cat_column, key=lambda s: int(s[3:]))",
"_____no_output_____"
],
[
"# Print the categorical column names.\ncat_column",
"_____no_output_____"
],
[
"# list the index of the categorical column names using the regual expression pattern matching the we previously compiled.\ncat_index = [i for i in range(0, len(data_train.columns)) if cat_pattern.match(data_train.columns[i])]\ncat_index",
"_____no_output_____"
],
[
"# Devide the continuous column names.\ncont_column = [cont for cont in data_train.columns if 'cont' in cont]",
"_____no_output_____"
],
[
"# Print the continuous column names.\ncont_column",
"_____no_output_____"
],
[
"# Although the continuous column names are sorted, we should force it to be sorted.\ncont_column = sorted(cont_column, key=lambda s: int(s[4:]))",
"_____no_output_____"
],
[
"# Print the continuous column names.\ncont_column",
"_____no_output_____"
],
[
"# list the index of the continuous column names using the regual expression pattern matching the we previously compiled.\ncont_index = [i for i in range(0, len(data_train.columns)) if cont_pattern.match(data_train.columns[i])]\ncont_index",
"_____no_output_____"
],
[
"# Prepare train and test datasets.\n# In order to do that, we can use a specific package in Sci-Kit library.\nfrom sklearn.model_selection import train_test_split",
"_____no_output_____"
],
[
"# Specify the inputs (X) and the target (y).\n# The input data is the data set whithout these specific columns: 'id', 'loss', 'isTrain'.\n# the target data is the logarithm of the 'loss' column.\nX = data_train.drop(['id', 'loss', 'is_train'], axis=1)\ny = np.log(data_train['loss'])",
"_____no_output_____"
],
[
"# Specify the train and test inputs and targets.\n# Note that we specify the random state.\nX_train, X_test, y_train, y_test = train_test_split(X, y, random_state=25)",
"_____no_output_____"
],
[
"# Use catboost algorithm for regression.\nfrom catboost import CatBoostRegressor",
"_____no_output_____"
],
[
"# Create a new catboost.\ncatboost_regressor_gpu = CatBoostRegressor(iterations=200, learning_rate=0.05, depth=6, eval_metric='MAE', verbose=10, task_type='GPU', save_snapshot=True, snapshot_file='faraz', snapshot_interval=10)",
"_____no_output_____"
],
[
"# For sake of the RAM capacity, we should delete the unncessary data sets from RAM.\ndel X\ndel y\ndel data_train\ndel data_test\ndel data_train_test",
"_____no_output_____"
],
[
"# Learn from the data.\n# Specify the categorical columns with the following command:\n# np.asarray(cat_index) - 1.\n# In addition, we declare the test data with setting the 'eval_set' parameter.\ncatboost_regressor_gpu.fit(X_train, y_train, np.asarray(cat_index) - 1, eval_set=(X_test, y_test))",
"_____no_output_____"
],
[
"# For further usage and reducing RAM occupation, Save the medel into the Hard Disk.\n# In order to achive this goal, use 'pickle'.\nimport pickle",
"_____no_output_____"
],
[
"# Use'Write Byte (wr)'' mode.\n# First open a file named catboost_regression,\n# Then pickle dumps the model, i.e. catboost_regressor, into it.\nwith open('catboost_regression_gpu', 'wb') as file:\n pickle.dump(catboost_regressor_gpu, file)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4aff6d3d3dc1aba38527e94139db22d58339a954
| 1,957 |
ipynb
|
Jupyter Notebook
|
HW/1/skip/json.ipynb
|
noname30081/IRR
|
c44298ad14c468eff36bc75ebc63abdc9ba24d55
|
[
"Apache-2.0"
] | null | null | null |
HW/1/skip/json.ipynb
|
noname30081/IRR
|
c44298ad14c468eff36bc75ebc63abdc9ba24d55
|
[
"Apache-2.0"
] | null | null | null |
HW/1/skip/json.ipynb
|
noname30081/IRR
|
c44298ad14c468eff36bc75ebc63abdc9ba24d55
|
[
"Apache-2.0"
] | 1 |
2022-01-16T03:40:34.000Z
|
2022-01-16T03:40:34.000Z
| 19.186275 | 77 | 0.477772 |
[
[
[
"import json\nimport re",
"_____no_output_____"
],
[
"path = 'hw1_data/test4.json'\nwith open(path, 'r') as paper:\n\n data = json.load(paper)\n pass\n",
"_____no_output_____"
],
[
"## 轉成 string 類型\nstring = []\nfor i in data:\n\n for k, v in i.items():\n\n string += [v] \n pass\n\n pass\n\nstring = \" \".join(string)\n# string.encode(encoding=\"UTF-8\").decode(\"UTF-8\")",
"_____no_output_____"
],
[
"## 關鍵字搜尋\nkeyword = \"➡️\"\nexist = bool(re.search(keyword, string))\nnumber = len(re.findall(keyword, string))",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code"
]
] |
4aff7400c1ac73bcc134cf14a085df690f69a0d6
| 39,066 |
ipynb
|
Jupyter Notebook
|
dlp_lec1.ipynb
|
adwaita-patil/Geospacial-Data-Analysis
|
94d12d432dde9050aacfb9fd8c673421b7706dae
|
[
"MIT"
] | null | null | null |
dlp_lec1.ipynb
|
adwaita-patil/Geospacial-Data-Analysis
|
94d12d432dde9050aacfb9fd8c673421b7706dae
|
[
"MIT"
] | null | null | null |
dlp_lec1.ipynb
|
adwaita-patil/Geospacial-Data-Analysis
|
94d12d432dde9050aacfb9fd8c673421b7706dae
|
[
"MIT"
] | null | null | null | 19.416501 | 210 | 0.484257 |
[
[
[
"# **Python Basics**\n**Values**\n\n* A value is the fundamental thing that a program manipulates\n* Values can be ‘Hello Python’, 1 , True\nValues have types. ",
"_____no_output_____"
],
[
"# **Variable**\n1. One of the most basic and powerful concepts is \nthat of a variable.\n2. A variable assigns a name to a value.\nVariables are nothing more than reserved memory locations that store values.\n3. Python variables does not need explicit declaration to reserve memory. \n4.Unlike C/C++ and Java, variables can change types\n",
"_____no_output_____"
]
],
[
[
"message = 'Hello Python!'",
"_____no_output_____"
],
[
"n = 10",
"_____no_output_____"
],
[
"e = 2.71",
"_____no_output_____"
],
[
"print(message)",
"Hello Python!\n"
],
[
"print(message, 'n==', n, 'e==', e)",
"Hello Python! n== 10 e== 2.71\n"
]
],
[
[
"# **Modules**\n\n\n1. module is a file containing Python definitions and statements\n2. not all functionality available comes automatically when starting Python\n1. extra functionality can be added by importing modules\n1. objects in the module can be accessed by prefixing them with the module name\n",
"_____no_output_____"
]
],
[
[
"import math",
"_____no_output_____"
],
[
"math.pow(2, 3)",
"_____no_output_____"
],
[
"import random",
"_____no_output_____"
],
[
"random.randint(1, 100)",
"_____no_output_____"
],
[
"from pathlib import Path",
"_____no_output_____"
]
],
[
[
"Comments Start with a `#`",
"_____no_output_____"
]
],
[
[
"# This is a comment",
"_____no_output_____"
]
],
[
[
"**Putting All things together**\n\n---\n\n\nNow we will write a program to calculate Simple Intrest",
"_____no_output_____"
]
],
[
[
"rate = 10 ",
"_____no_output_____"
],
[
"principle = 1000",
"_____no_output_____"
],
[
"time = 3",
"_____no_output_____"
],
[
"intrest = (rate * principle * time)/100",
"_____no_output_____"
],
[
"print('Intrest is ', intrest)",
"Intrest is 200.0\n"
]
],
[
[
"# **Data Types**\n\n\n### 1. Numeric Types\n\n\n* Integer Types: 92, 12, 0, 1\n* Floats (Floating point numbers): 3.1415\n* Complex Numbers: a + b*i (composed of real and imaginary component, both of which are floats a + b*i)\n* Booleans: True/False are a subtype of integers (0 is false, 1 is true)\n\n2. Strings\n* ‘Hello Python’\n* 'India'\n\n3. Sequence types:\n\n\n* Lists : [1,2,3,4,5]\n* Tuples: (1, 2)\n* Ranges: ",
"_____no_output_____"
],
[
"# **Indentation**\n1. In Python, blocks of code are defined using indentation\n1. The indentation within the block needs to be consistent\n1. The first line with less indentation is outside of the block\n1. The first line with more indentation starts a nested block\n1. Often a colon appears at the start of a new block",
"_____no_output_____"
]
],
[
[
"x = 9\nif x<10:\n print('x is less than 10')\n print('Hello')\nprint('Outside the If')",
"x is less than 10\nHello\nOutside the If\n"
]
],
[
[
"# **Strings**\n\n* Strings are text values like ‘Hello Anaconda’\n* Strings can be enclosed in single quote ‘ ..’ or double quotes “…”\n\n",
"_____no_output_____"
]
],
[
[
"'spam eggs' ",
"_____no_output_____"
],
[
"'doesn\\'t' # use \\' to escape the single quote...",
"_____no_output_____"
],
[
"\"doesn't\" # ...or use double quotes instead",
"_____no_output_____"
],
[
"print(\"\"\"\\\nUsage: thingy [OPTIONS]\n -h Display this usage message\n -H hostname Hostname to connect to\n\"\"\")",
"Usage: thingy [OPTIONS]\n -h Display this usage message\n -H hostname Hostname to connect to\n\n"
],
[
"3 * 'un' + 'ium'",
"_____no_output_____"
]
],
[
[
"Strings can be *indexed* (subscripted), with the first character having index 0.",
"_____no_output_____"
]
],
[
[
" word = 'Python'",
"_____no_output_____"
],
[
"word[0]",
"_____no_output_____"
],
[
"word[5]",
"_____no_output_____"
]
],
[
[
"indices may also be negative numbers, to start counting from the right. ",
"_____no_output_____"
]
],
[
[
"word[-1] # last character",
"_____no_output_____"
],
[
"word[-2] # second-last character",
"_____no_output_____"
]
],
[
[
"In addition to indexing, slicing is also supported. While indexing is used to obtain individual characters, slicing allows you to obtain substring:",
"_____no_output_____"
]
],
[
[
" word[0:2] # characters from position 0 (included) to 2 (excluded)",
"_____no_output_____"
],
[
"word[2:5] # characters from position 2 (included) to 5 (excluded)",
"_____no_output_____"
],
[
"text = 'Put several strings within parentheses '+\\\n 'to have them joined together.'",
"_____no_output_____"
],
[
"text",
"_____no_output_____"
]
],
[
[
"# **Lists**\n1. ordered sequence of information, accessible by index\n1. a list is denoted by square brackets, [ ]\n1. a list contains usually homogenous elements \n1. list elements can be changed so a list is mutable\n",
"_____no_output_____"
]
],
[
[
"squares = [1, 4, 9, 16, 25]",
"_____no_output_____"
],
[
"squares",
"_____no_output_____"
],
[
"squares[0]",
"_____no_output_____"
],
[
"squares[-1]",
"_____no_output_____"
],
[
"squares[-3:]",
"_____no_output_____"
],
[
"squares + [36, 49, 64, 81, 100]",
"_____no_output_____"
],
[
"cubes = [1, 8, 27, 65, 125]\ncubes",
"_____no_output_____"
]
],
[
[
"Unlike strings, which are immutable, lists are a mutable type, i.e. it is possible to change their content",
"_____no_output_____"
]
],
[
[
"cubes[3] = 64\ncubes",
"_____no_output_____"
]
],
[
[
"Assignment to slices is also possible, and this can even change the size of the list or clear it entirely",
"_____no_output_____"
]
],
[
[
"letters = ['a', 'b', 'c', 'd', 'e', 'f', 'g']",
"_____no_output_____"
],
[
"letters[2:5] = ['C', 'D', 'E']\nletters",
"_____no_output_____"
],
[
"letters[:] = []\nletters",
"_____no_output_____"
]
],
[
[
"# **Tuple**\n1. tuple consists of a number of values separated by commas\n1. cannot change element values, immutable\n1. tuples always enclosed in parenthesis \n1. used to return more than one value from a function\n",
"_____no_output_____"
]
],
[
[
"t = 12345, 54321, 'hello!'",
"_____no_output_____"
],
[
"t[0]",
"_____no_output_____"
],
[
"u = t, (1, 2, 3, 4, 5) # Tuples may be nested:",
"_____no_output_____"
],
[
"u",
"_____no_output_____"
],
[
"t[0] = 88888 # Tuples are immutable:",
"_____no_output_____"
],
[
"x, y, z = t\nprint(x)\nprint(y)\nprint(z)",
"12345\n54321\nhello!\n"
]
],
[
[
"# **Sets**\n1. A set is an unordered collection with no duplicate elements\n1. set objects also support mathematical operations like union, intersection, difference, and symmetric difference\n",
"_____no_output_____"
]
],
[
[
"basket = {'apple', 'orange', 'apple', 'pear', 'orange', 'banana'}",
"_____no_output_____"
],
[
"print(basket)# show that duplicates have been removed",
"{'banana', 'apple', 'pear', 'orange'}\n"
],
[
"'orange' in basket # fast membership testing",
"_____no_output_____"
],
[
"'crabgrass' in basket",
"_____no_output_____"
],
[
"a = set('abracadabra')\nb = set('alacazam')",
"_____no_output_____"
],
[
"a - b",
"_____no_output_____"
]
],
[
[
"# **Dictionary**\n1. dictionary is a set of key:value pairs\n1. dictionaries are indexed by keys, which can be any immutable type\n1. dictionary stores a value with some key and extracts the value given the key",
"_____no_output_____"
]
],
[
[
"telephone = {'jack': 4098, 'sape': 4139,'alpha':1099,'beta':1000}\ntelephone",
"_____no_output_____"
],
[
"telephone['jack']",
"_____no_output_____"
],
[
"telephone['king'] = 4127\ntelephone",
"_____no_output_____"
],
[
"del telephone['sape']",
"_____no_output_____"
],
[
"'king' in telephone",
"_____no_output_____"
]
],
[
[
"# **Control flow** \nControl flow is the concept of changing this order of code execution. Similar to the way you might use sale prices to decide which which car to buy or seeing the colour of signal decide to start or stop.",
"_____no_output_____"
]
],
[
[
"traffic_light = 'green'\nif traffic_light == 'green':\n print('Light is Green')\n print('Go Ahead ')\nelse:\n print('Light is Not Green')\n print(' Stop ')",
"Light is Green\n Go Ahead \n"
],
[
"traffic_light = 'red'\nif traffic_light == 'green':\n print('Light is Green')\nelif traffic_light=='yellow':\n print('Light is Yellow')\nelif traffic_light=='red':\n print('Light is Red')\nelse:\n print('Unknown Traffic Light')",
"Light is Red\n"
]
],
[
[
"# **While Loop**\nA while statement executes a block of code as long as a condition is `True`.This continues until the condition is `False`.",
"_____no_output_____"
]
],
[
[
"n = 0 ",
"_____no_output_____"
],
[
"while n<10:\n print('Value of n is',n)\n n=n+1",
"Value of n is 0\nValue of n is 1\nValue of n is 2\nValue of n is 3\nValue of n is 4\nValue of n is 5\nValue of n is 6\nValue of n is 7\nValue of n is 8\nValue of n is 9\n"
]
],
[
[
"# **For statements**\nPython’s for statement iterates over the items of any sequence. ",
"_____no_output_____"
]
],
[
[
"words = ['cat', 'window', 'defenestrate']\nfor w in words:\n print(w,len(w))",
"cat 3\nwindow 6\ndefenestrate 12\n"
]
],
[
[
"### To iterate over a sequence of numbers, the built-in function range() comes in handy.",
"_____no_output_____"
]
],
[
[
"for i in range(5):\n print(i,end=',')",
"_____no_output_____"
],
[
"for i in range(5,10):\n print(i,end=',')",
"_____no_output_____"
],
[
"for i in range(5,10,2):\n print(i,end=',')",
"5,7,9,"
]
],
[
[
"# **break and continue Statements**\n1. break statement breaks out of innermost enclosing for or while loop.\n1. continue statement continues with the next iteration of the loop skipping remaining statements.\n",
"_____no_output_____"
]
],
[
[
"for i in range(5):\n if i==4:\n break\n print(i)",
"0\n1\n2\n3\n"
],
[
"for i in range(5):\n if i==2:\n continue\n print(i)",
"0\n1\n3\n4\n"
]
],
[
[
"# **Write and call/invoke a function**\nIf its find that the same bits of code is reused over and over, you can create your own function and call that instead of repeating the same code.",
"_____no_output_____"
]
],
[
[
"def check_odd_even(i):\n if i%2==0:\n print(i,'is even number')\n else:\n print(i,'is odd number')",
"_____no_output_____"
],
[
"check_odd_even(10)",
"10 is even number\n"
],
[
"check_odd_even(11)",
"11 is odd number\n"
]
],
[
[
"# **Default Value Argument**",
"_____no_output_____"
]
],
[
[
"def calc_intrest(principal,rates=5,duration=5):\n intrest=(principal*rates*duration)/100\n amount=intrest+principal\n print('Principal Amount=',principal)\n print('Rate=',rates)\n print('Duration=',duration)\n print('Total Amount=',amount)",
"_____no_output_____"
],
[
"calc_intrest(10)",
"Principal Amount= 10\nRate= 5\nDuration= 5\nTotal Amount= 12.5\n"
],
[
"calc_intrest(10,rates=10)",
"Principal Amount= 10\nRate= 10\nDuration= 5\nTotal Amount= 15.0\n"
],
[
"calc_intrest(10,rates=10,duration=20)",
"Principal Amount= 10\nRate= 10\nDuration= 20\nTotal Amount= 30.0\n"
]
],
[
[
"# Reading File",
"_____no_output_____"
]
],
[
[
"f = open('workfile.txt','r')\nlines=f.read() # Entire file is read \nprint(lines)\nf.close()",
"_____no_output_____"
],
[
"f = open('workfile.txt','r')\nfor line in f:\n print(line,end='')\nf.close()",
"_____no_output_____"
],
[
"with open('workfile.txt','r')as f:\n for line in f:\n print(line,end='')",
"_____no_output_____"
]
],
[
[
"# Writing to file",
"_____no_output_____"
]
],
[
[
"value = ('the answer is ', 42)\nwith open('workfile2.txt','w') as f:\n s=str(value)\n f.write(s)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
4aff778069ee5c8aea164234c9025567491f7dbb
| 13,657 |
ipynb
|
Jupyter Notebook
|
prepare_data/expe.ipynb
|
hewu2008/mtcnn
|
6fd8ea174dcbd596f35ddd522d9a188e6c7fda85
|
[
"MIT"
] | 1,640 |
2017-08-31T08:16:49.000Z
|
2022-03-30T14:18:29.000Z
|
prepare_data/expe.ipynb
|
LebronGG/MTCNN-Tensorflow
|
3b3934d38f8d34287cc933a581537a1acfd0bb60
|
[
"MIT"
] | 299 |
2017-09-04T08:28:57.000Z
|
2022-02-08T08:28:32.000Z
|
prepare_data/expe.ipynb
|
LebronGG/MTCNN-Tensorflow
|
3b3934d38f8d34287cc933a581537a1acfd0bb60
|
[
"MIT"
] | 810 |
2017-08-31T08:58:39.000Z
|
2022-03-30T03:07:57.000Z
| 47.585366 | 1,503 | 0.597276 |
[
[
[
"#coding:utf-8\nimport sys\nimport numpy as np\n\nsys.path.append(\"..\")\nimport argparse\nfrom train_models.mtcnn_model import P_Net, R_Net, O_Net\nfrom prepare_data.loader import TestLoader\nfrom Detection.detector import Detector\nfrom Detection.fcn_detector import FcnDetector\nfrom Detection.MtcnnDetector import MtcnnDetector\nimport cv2\nimport os\n\ndata_dir = '../../DATA/WIDER_val/images'\nanno_file = 'wider_face_val.txt'\n",
"F:\\ProgramData\\Anaconda3\\lib\\site-packages\\h5py\\__init__.py:36: FutureWarning: Conversion of the second argument of issubdtype from `float` to `np.floating` is deprecated. In future, it will be treated as `np.float64 == np.dtype(float).type`.\n from ._conv import register_converters as _register_converters\n"
],
[
"def read_gt_bbox(raw_list):\n \n list_len = len(raw_list)\n bbox_num = (list_len-1)//4\n idx = 1\n bboxes = np.zeros((bbox_num,4),dtype=int)\n for i in range(4):\n for j in range(bbox_num):\n bboxes[j][i] = int(raw_list[idx])\n idx += 1\n return bboxes\n ",
"_____no_output_____"
],
[
"def get_image_info(anno_file):\n f = open(anno_file,'r')\n image_info = []\n for line in f:\n ct_list = line.strip().split(' ')\n path = ct_list[0]\n \n path_list = path.split('\\\\')\n event = path_list[0]\n name = path_list [1]\n #print(event, name )\n bboxes = read_gt_bbox(ct_list)\n image_info.append([event,name,bboxes])\n print('total number of images in validation set: ', len(image_info))\n return image_info",
"_____no_output_____"
],
[
"test_mode = \"ONet\"\nthresh = [0.6,0.5,0.4]\nmin_face_size = 24\nstride = 2\nslide_window = False\nshuffle = False\nvis = False\ndetectors = [None, None, None]\nprefix = ['../data/MTCNN_model/PNet_landmark/PNet', '../data/MTCNN_model/RNet_landmark/RNet', '../data/MTCNN_model/ONet_landmark/ONet']\nepoch = [18, 14, 16]\nbatch_size = [2048, 256, 16]\nmodel_path = ['%s-%s' % (x, y) for x, y in zip(prefix, epoch)]\n",
"_____no_output_____"
],
[
"\nif slide_window:\n PNet = Detector(P_Net, 12, batch_size[0], model_path[0])\nelse:\n PNet = FcnDetector(P_Net, model_path[0])\ndetectors[0] = PNet\n \n# load rnet model\nif test_mode in [\"RNet\", \"ONet\"]:\n RNet = Detector(R_Net, 24, batch_size[1], model_path[1])\n detectors[1] = RNet\n \n# load onet model\nif test_mode == \"ONet\":\n ONet = Detector(O_Net, 48, batch_size[2], model_path[2])\n detectors[2] = ONet\n \nmtcnn_detector = MtcnnDetector(detectors=detectors, min_face_size=min_face_size,\n stride=stride, threshold=thresh, slide_window=slide_window)\n ",
"(1, ?, ?, 3)\n(1, ?, ?, 10)\n(1, ?, ?, 10)\n(1, ?, ?, 16)\n(1, ?, ?, 32)\n(1, ?, ?, 2)\n(1, ?, ?, 4)\n(1, ?, ?, 10)\n"
],
[
"image_info = get_image_info(anno_file)",
"_____no_output_____"
],
[
"str1='aaa'\nstr2 = 'bbb'\nstr3 = 'aaa'\nprint(str1 != str2)\nprint (str1 == str3)\n",
"True\nTrue\n"
],
[
"a ='asdfasdf.jpg'\na.split('.jpg')",
"_____no_output_____"
],
[
"current_event = ''\nsave_path = ''\nfor item in image_info:\n image_file_name = os.path.join(data_dir,item[0],item[1])\n if current_event != item[0]:\n current_event = item[0]\n save_path = os.path.join('../../DATA',item[0])\n if not os.path.exists(save_path):\n os.mkdir(save_path)\n f_name= item[1].split('.jpg')[0]\n dets_file_name = os.path.join(save_path,f_name + '.txt')\n img = cv2.imread(image_file_name)\n all_boxes,_ = mtcnn_detector.detect_single_image(img)\n \n ",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4aff7b227717cdbdc5503fd30f1e75a0b6db5243
| 283,452 |
ipynb
|
Jupyter Notebook
|
docs/tutorials/Coronagraph_Basics.ipynb
|
maxwellmb/pynrc
|
0570da4df0dd9de158942630bb6cbbe356e9b282
|
[
"MIT"
] | 1 |
2021-12-10T22:30:37.000Z
|
2021-12-10T22:30:37.000Z
|
docs/tutorials/Coronagraph_Basics.ipynb
|
maxwellmb/pynrc
|
0570da4df0dd9de158942630bb6cbbe356e9b282
|
[
"MIT"
] | 1 |
2021-03-30T18:27:17.000Z
|
2021-03-30T18:27:17.000Z
|
docs/tutorials/Coronagraph_Basics.ipynb
|
mperrin/pynrc
|
f1b341162ca63f03da5fc3054d1d22ed11fbf13d
|
[
"MIT"
] | null | null | null | 317.415454 | 83,200 | 0.924301 |
[
[
[
"# Coronagraph Basics",
"_____no_output_____"
],
[
"This set of exercises guides the user through a step-by-step process of simulating NIRCam coronagraphic observations of the HR 8799 exoplanetary system. The goal is to familiarize the user with basic `pynrc` classes and functions relevant to coronagraphy.",
"_____no_output_____"
]
],
[
[
"# If running Python 2.x, makes print and division act like Python 3\nfrom __future__ import print_function, division\n\n# Import the usual libraries\nimport numpy as np\nimport matplotlib\nimport matplotlib.pyplot as plt\n\n# Enable inline plotting at lower left\n%matplotlib inline\n\nfrom IPython.display import display, Latex, clear_output",
"_____no_output_____"
]
],
[
[
"We will start by first importing `pynrc` along with the `obs_hci` (High Contrast Imaging) class, which lives in the `pynrc.obs_nircam` module. ",
"_____no_output_____"
]
],
[
[
"import pynrc\nfrom pynrc import nrc_utils # Variety of useful functions and classes\nfrom pynrc.obs_nircam import obs_hci # High-contrast imaging observation class\n\n# Disable informational messages and only include warnings and higher\npynrc.setup_logging(level='WARN')",
"pyNRC log messages of level WARN and above will be shown.\npyNRC log outputs will be directed to the screen.\n"
]
],
[
[
"## Source Definitions\n\nThe `obs_hci` class first requires two arguments describing the spectra of the science and reference sources (`sp_sci` and `sp_ref`, respectively. Each argument should be a Pysynphot spectrum already normalized to some known flux. `pynrc` includes built-in functions for generating spectra. The user may use either of these or should feel free to supply their own as long as it meets the requirements. \n\n1. The `pynrc.stellar_spectrum` function provides the simplest way to define a new spectrum:\n```python\nbp_k = pynrc.bp_2mass('k') # Define bandpass to normalize spectrum\nsp_sci = pynrc.stellar_spectrum('F0V', 5.24, 'vegamag', bp_k)\n```\nYou can also be more specific about the stellar properties with `Teff`, `metallicity`, and `log_g` keywords.\n```python\n sp_sci = pynrc.stellar_spectrum('F0V', 5.24, 'vegamag', bp_k, \n Teff=7430, metallicity=-0.47, log_g=4.35)\n```\n\n2. Alternatively, the `pynrc.source_spectrum` class ingests spectral information of a given target and generates a model fit to the known photometric SED. Two model routines can be fit. The first is a very simple scale factor that is applied to the input spectrum, while the second takes the input spectrum and adds an IR excess modeled as a modified blackbody function. The user can find the relevant photometric data at http://vizier.u-strasbg.fr/vizier/sed/ and click download data as a VOTable.",
"_____no_output_____"
]
],
[
[
"# Define 2MASS Ks bandpass and source information\nbp_k = pynrc.bp_2mass('k')\n\n# Science source, dist, age, sptype, Teff, [Fe/H], log_g, mag, band\nargs_sources = [('HR 8799', 39.0, 30, 'F0V', 7430, -0.47, 4.35, 5.24, bp_k)]\n\n# References source, sptype, Teff, [Fe/H], log_g, mag, band\nref_sources = [('HD 220657', 'F8III', 5888, -0.01, 3.22, 3.04, bp_k)]",
"_____no_output_____"
],
[
"name_sci, dist_sci, age, spt_sci, Teff_sci, feh_sci, logg_sci, mag_sci, bp_sci = args_sources[0]\nname_ref, spt_ref, Teff_ref, feh_ref, logg_ref, mag_ref, bp_ref = ref_sources[0]\n\n# For the purposes of simplicity, we will use pynrc.stellar_spectrum()\nsp_sci = pynrc.stellar_spectrum(spt_sci, mag_sci, 'vegamag', bp_sci, \n Teff=Teff_sci, metallicity=feh_sci, log_g=logg_sci)\nsp_sci.name = name_sci\n\n# And the refernece source\nsp_ref = pynrc.stellar_spectrum(spt_ref, mag_ref, 'vegamag', bp_ref, \n Teff=Teff_ref, metallicity=feh_ref, log_g=logg_ref)\nsp_ref.name = name_ref",
"_____no_output_____"
],
[
"# Plot the two spectra\nfig, ax = plt.subplots(1,1, figsize=(8,5))\n\nxr = [2.5,5.5]\n\nfor sp in [sp_sci, sp_ref]:\n w = sp.wave / 1e4\n ind = (w>=xr[0]) & (w<=xr[1])\n sp.convert('Jy')\n f = sp.flux / np.interp(4.0, w, sp.flux)\n ax.semilogy(w[ind], f[ind], lw=1.5, label=sp.name)\n ax.set_ylabel('Flux (Jy) normalized at 4 $\\mu m$')\n sp.convert('flam')\n\nax.set_xlim(xr)\nax.set_xlabel(r'Wavelength ($\\mu m$)')\nax.set_title('Spectral Sources')\n\n# Overplot Filter Bandpass\nbp = pynrc.read_filter('F444W', 'CIRCLYOT', 'MASK430R')\nax2 = ax.twinx()\nax2.plot(bp.wave/1e4, bp.throughput, color='C2', label=bp.name+' Bandpass')\nax2.set_ylim([0,0.8])\nax2.set_xlim(xr)\nax2.set_ylabel('Bandpass Throughput')\n\nax.legend(loc='upper left')\nax2.legend(loc='upper right')\n\nfig.tight_layout()",
"_____no_output_____"
]
],
[
[
"## Initialize Observation\n\nNow we will initialize the high-contrast imaging class `pynrc.obs_hci` using the spectral objects and various other settings. The `obs_hci` object is a subclass of the more generalized `NIRCam` class. It implements new settings and functions specific to high-contrast imaging observations for corongraphy and direct imaging.",
"_____no_output_____"
],
[
"For this tutorial, we want to observe these targets using the `MASK430R` coronagraph in the `F444W` filter. All circular coronagraphic masks such as the `430R` (R=round) should be paired with the `CIRCLYOT` pupil element, whereas wedge/bar masks are paired with `WEDGELYOT` pupil. Observations in the LW channel are most commonly observed in `WINDOW` mode with a 320x320 detector subarray size. Full detector sizes are also available.\n\nThe PSF simulation size (`fov_pix` keyword) should also be of similar size as the subarray window (recommend avoiding anything above `fov_pix=1024` due to computation time and memory usage). Use odd numbers to center the PSF in the middle of the pixel. If `fov_pix` is specified as even, then PSFs get centered at the corners. This distinction really only matter for unocculted observations, (ie., where the PSF flux is concentrated in a tight central core).\n\nWe also need to specify a WFE drift value (`wfe_ref_drift` parameter), which defines the anticipated drift in nm between the science and reference sources. For the moment, let's intialize with a value of 0nm. This prevents an initially long process by which `pynrc` calculates changes made to the PSF over a wide range of drift values.\n\nExtended disk models can also be specified upon initialization using the `disk_hdu` keyword.",
"_____no_output_____"
]
],
[
[
"filt, mask, pupil = ('F444W', 'MASK430R', 'CIRCLYOT')\nwind_mode, subsize = ('WINDOW', 320)\nfov_pix, oversample = (320, 2)\n\nwfe_ref_drift = 0\nobs = pynrc.obs_hci(sp_sci, sp_ref, dist_sci, filter=filt, mask=mask, pupil=pupil, \n wfe_ref_drift=wfe_ref_drift, fov_pix=fov_pix, oversample=oversample, \n wind_mode=wind_mode, xpix=subsize, ypix=subsize, verbose=True)",
"Generating background PSF coefficients...\nGenerating oversampled PSFs...\nUpdating NIRCam reference coefficients...\nCreating NIRCam reference class...\nFinished.\n"
]
],
[
[
"All information for the reference observation is stored in the attribute `obs.nrc_ref`, which is simply it's own isolated `NIRCam` (`nrc_hci`) class. After initialization, any updates made to the primary `obs` instrument configuration (e.g., filters, detector size, etc.) must also be made inside the `obs.nrc_ref` class as well. That is to say, it does not automatically propogate. In many ways, it's best to think of these as two separate classes,\n```python\nobs_sci = obs\nobs_ref = obs.nrc_ref\n```\nwith some linked references between the two.\n\nNow that we've succesffully initialized the obs_hci observations, let's specify the `wfe_ref_drift`. If this is your first time, then the `nrc_utils.wfed_coeff` function is called to determine a relationship between PSFs in the presense of WFE drift. This relationship is saved to disk in the `PYNRC_DATA` directory as a set of polynomial coefficients. Future calculations utilize these coefficients to quickly generate a new PSF for any arbitary drift value.",
"_____no_output_____"
]
],
[
[
"# WFE drift amount between rolls\n# This only gets called during gen_roll_image()\n# and temporarily updates obs.wfe_drift to create\n# a new PSF.\nobs.wfe_roll_drift = 2\n\n# Drift amount between Roll 1 and reference\n# This is simply a link to obs.nrc_ref.wfe_drift\nobs.wfe_ref_drift = 10",
"_____no_output_____"
]
],
[
[
"## Exposure Settings\n\nOptimization of exposure settings are demonstrated in another tutorial, so we will not repeat that process here. We can assume the optimization process was performed elsewhere to choose the `DEEP8` pattern with 16 groups and 5 total integrations. These settings apply to each roll position of the science observation as well as the for the reference observation.",
"_____no_output_____"
]
],
[
[
"# Update both the science and reference observations\nobs.update_detectors(read_mode='DEEP8', ngroup=16, nint=5, verbose=True)\nobs.nrc_ref.update_detectors(read_mode='DEEP8', ngroup=16, nint=5)",
"New Ramp Settings:\n read_mode : DEEP8\n nf : 8\n nd2 : 12\n ngroup : 16\n nint : 5\nNew Detector Settings\n wind_mode : WINDOW\n xpix : 320\n ypix : 320\n x0 : 914\n y0 : 1513\nNew Ramp Times\n t_group : 21.381\n t_frame : 1.069\n t_int : 329.264\n t_int_tot : 330.353\n t_exp : 1646.322\n t_acq : 1651.766\n"
]
],
[
[
"## Add Planets\n\nThere are four known giant planets orbiting HR 8799 at various locations. Ideally, we would like to position them at their predicted locations on the anticipated observation date. For this case, we choose a plausible observation date of November 1, 2019. To convert between $(x,y)$ and $(r,\\theta)$, use the `nrc_utils.xy_to_rtheta` and `nrc_utils.rtheta_to_xy` functions.\n\nWhen adding the planets, it doesn't matter too much which exoplanet model spectrum we decide to use since the spectra are still fairly unconstrained at these wavelengths. We do know roughly the planets' luminosities, so we can simply choose some reasonable model and renormalize it to the appropriate filter brightness. Currently, the only exoplanet spectral models available to `pynrc` are those from Spiegel & Burrows (2012).\n\n",
"_____no_output_____"
]
],
[
[
"# Projected locations for date 11/01/2019\n# These are prelimary positions, but within constrained orbital parameters\nloc_list = [(-1.57, 0.64), (0.42, 0.87), (0.5, -0.45), (0.35, 0.20)]\n\n# Estimated magnitudes within F444W filter\npmags = [16.0, 15.0, 14.6, 14.7]",
"_____no_output_____"
],
[
"# Add planet information to observation class.\n# These are stored in obs.planets.\n# Can be cleared using obs.kill_planets().\nobs.kill_planets()\nfor i, loc in enumerate(loc_list):\n obs.add_planet(mass=10, entropy=13, age=age, xy=loc, runits='arcsec', \n renorm_args=(pmags[i], 'vegamag', obs.bandpass))",
"_____no_output_____"
],
[
"# Generate and plot a noiseless slope image to make sure things look right\nPA1 = 85\nim_planets = obs.gen_planets_image(PA_offset=PA1)",
"_____no_output_____"
],
[
"from matplotlib.patches import Circle\nfrom pynrc.nrc_utils import (coron_ap_locs, build_mask_detid, fshift, pad_or_cut_to_size)\n\nfig, ax = plt.subplots(figsize=(6,6))\n\nxasec = obs.det_info['xpix'] * obs.pix_scale\nyasec = obs.det_info['ypix'] * obs.pix_scale\nextent = [-xasec/2, xasec/2, -yasec/2, yasec/2]\nxylim = 4\n\nvmin = 0\nvmax = 0.5*im_planets.max()\nax.imshow(im_planets, extent=extent, vmin=vmin, vmax=vmax)\n\n# Overlay the coronagraphic mask\ndetid = obs.Detectors[0].detid\nim_mask = obs.mask_images[detid]\n# Do some masked transparency overlays\nmasked = np.ma.masked_where(im_mask>0.99, im_mask)\n#ax.imshow(1-masked, extent=extent, alpha=0.5)\nax.imshow(1-masked, extent=extent, alpha=0.3, cmap='Greys_r', vmin=-0.5)\n\nxc_off = obs.bar_offset\nfor loc in loc_list:\n xc, yc = loc\n xc, yc = nrc_utils.xy_rot(xc, yc, PA1)\n xc += xc_off\n circle = Circle((xc,yc), radius=xylim/15., alpha=0.7, lw=1, edgecolor='red', facecolor='none')\n ax.add_artist(circle)\n\nxlim = ylim = np.array([-1,1])*xylim\nxlim = xlim + xc_off\nax.set_xlim(xlim)\nax.set_ylim(ylim)\n\nax.set_xlabel('Arcsec')\nax.set_ylabel('Arcsec')\n\nax.set_title('{} planets -- {} {}'.format(sp_sci.name, obs.filter, obs.mask))\n\ncolor = 'grey'\nax.tick_params(axis='both', color=color, which='both')\nfor k in ax.spines.keys():\n ax.spines[k].set_color(color)\n\nnrc_utils.plotAxes(ax, width=1, headwidth=5, alength=0.15, angle=PA1, \n position=(0.25,0.9), label1='E', label2='N')\n \nfig.tight_layout()",
"_____no_output_____"
]
],
[
[
"As we can see, even with \"perfect PSF subtraction\" and no noise, it's difficult to make out planet e. This is primarily due to its location relative to the occulting mask reducing throughput along with confusion of bright diffraction spots from nearby sources.",
"_____no_output_____"
],
[
"## Estimated Performance\n\nNow we are ready to determine contrast performance and sensitivites as a function of distance from the star. ",
"_____no_output_____"
],
[
"### 1. Roll-Subtracted Images\n\nFirst, we will create a quick simulated roll-subtracted image using the in `gen_roll_image` method. For the selected observation date of 11/1/2019, APT shows a PA range of 84$^{\\circ}$ to 96$^{\\circ}$. So, we'll assume Roll 1 has PA1=85, while Roll 2 has PA2=95. In this case, \"roll subtraction\" simply creates two science images observed at different parallactic angles, then subtracts the same reference observation from each. The two results are then de-rotated to a common PA=0 and averaged.\n\nThere is also the option to create ADI images, where the other roll position becomes the reference star by setting `no_ref=True`. ",
"_____no_output_____"
]
],
[
[
"# Cycle through a few WFE drift values\nwfe_list = [0,5,10]\n\n# PA values for each roll\nPA1, PA2 = (85, 95)\n\n# A dictionary of HDULists\nhdul_dict = {}\nfor i, wfe_drift in enumerate(wfe_list):\n print(wfe_drift)\n # Upate WFE reference drift value\n obs.wfe_ref_drift = wfe_drift\n \n # Set the final output image to be oversampled\n hdulist = obs.gen_roll_image(PA1=PA1, PA2=PA2)\n hdul_dict[wfe_drift] = hdulist\n",
"0\n5\n10\n"
],
[
"from pynrc.obs_nircam import plot_hdulist\nfrom matplotlib.patches import Circle\n\nfig, axes = plt.subplots(1,3, figsize=(14,4.3))\nxylim = 2.5\nxlim = ylim = np.array([-1,1])*xylim\n\nfor j, wfe_drift in enumerate(wfe_list):\n ax = axes[j]\n hdul = hdul_dict[wfe_drift]\n \n plot_hdulist(hdul, xr=xlim, yr=ylim, ax=ax, vmin=0, vmax=8)\n\n # Location of planet\n for loc in loc_list:\n circle = Circle(loc, radius=xylim/15., lw=1, edgecolor='red', facecolor='none')\n ax.add_artist(circle)\n\n ax.set_title('$\\Delta$WFE = {:.0f} nm'.format(wfe_drift))\n \n nrc_utils.plotAxes(ax, width=1, headwidth=5, alength=0.15, position=(0.9,0.7), label1='E', label2='N')\n\nfig.suptitle('{} -- {} {}'.format(name_sci, obs.filter, obs.mask), fontsize=14)\nfig.tight_layout()\nfig.subplots_adjust(top=0.85)",
"_____no_output_____"
]
],
[
[
"**Note:** At first glance, it appears as if the innermost Planet e is getting brighter with increased WFE drift, which would be understandably confusing. However, upon further investigation, there just happens to be a bright residual speckle that lines up well with Planet e when observed at this specific parallactic angle. This was verified by adjusting the observed PA as well as removing the planets from the simulations.",
"_____no_output_____"
],
[
"### 2. Contrast Curves\n\nNext, we will cycle through a few WFE drift values to get an idea of potential predicted sensitivity curves. The `calc_contrast` method returns a tuple of three arrays:\n1. The radius in arcsec.\n2. The n-sigma contrast.\n3. The n-sigma magnitude sensitivity limit (vega mag).",
"_____no_output_____"
]
],
[
[
"# Cycle through varying levels of WFE drift and calculate contrasts\nwfe_list = [0,5,10]\nnsig = 5\n\n# PA values for each roll\nPA1, PA2 = (85, 95)\nroll_angle = np.abs(PA2 - PA1)\n\ncurves = []\nfor i, wfe_drift in enumerate(wfe_list):\n print(wfe_drift)\n # Generate series of observations for each filter\n obs.wfe_ref_drift = wfe_drift\n \n # Generate contrast curves\n result = obs.calc_contrast(roll_angle=roll_angle, nsig=nsig)\n curves.append(result)\n",
"0\n5\n10\n"
],
[
"from pynrc.obs_nircam import plot_contrasts, plot_planet_patches, plot_contrasts_mjup\nimport matplotlib.patches as mpatches\n\n# fig, ax = plt.subplots(figsize=(8,5))\nfig, axes = plt.subplots(1,2, figsize=(14,4.5))\nxr=[0,5]\nyr=[24,8]\n\n# 1a. Plot contrast curves and set x/y limits\nax = axes[0]\nax, ax2, ax3 = plot_contrasts(curves, nsig, wfe_list, obs=obs, \n xr=xr, yr=yr, ax=ax, return_axes=True)\n# 1b. Plot the locations of exoplanet companions\nlabel = 'Companions ({})'.format(filt)\nplanet_dist = [np.sqrt(x**2+y**2) for x,y in loc_list]\nax.plot(planet_dist, pmags, marker='o', ls='None', label=label, color='k', zorder=10) \n\n# 1c. Plot Spiegel & Burrows (2012) exoplanet fluxes (Hot Start)\nplot_planet_patches(ax, obs, age=age, entropy=13, av_vals=None)\nax.legend(ncol=2)\n\n# 2. Plot in terms of MJup using COND models\nax = axes[1]\nplot_contrasts_mjup(curves, nsig, wfe_list, obs=obs, age=age,\n ax=ax, twin_ax=True, xr=xr, yr=None)\nax.set_yscale('log')\nax.set_ylim([0.08,100])\nax.legend(loc='upper right', title='COND ({:.0f} Myr)'.format(age))\n\nfig.suptitle('{} ({} + {})'.format(name_sci, obs.filter, obs.mask), fontsize=16)\n\nfig.tight_layout()\nfig.subplots_adjust(top=0.85, bottom=0.1 , left=0.05, right=0.97)",
"_____no_output_____"
]
],
[
[
"The innermost Planet e is right on the edge of the detection threshold as suggested by the simulated images.",
"_____no_output_____"
],
[
"### 3. Saturation Levels\n\nCreate an image showing level of saturation for each pixel. For NIRCam, saturation is important to track for purposes of accurate slope fits and persistence correction. In this case, we will plot the saturation levels both at `NGROUP=2` and `NGROUP=obs.det_info['ngroup']`. Saturation is defined at 80% well level, but can be modified using the `well_fill` keyword.\n\nWe want to perform this analysis for both science and reference targets.",
"_____no_output_____"
]
],
[
[
"# Saturation limits\nng_max = obs.det_info['ngroup']\nsp_flat = pynrc.stellar_spectrum('flat')\n\nprint('NGROUP=2')\n_ = obs.sat_limits(sp=sp_flat,ngroup=2,verbose=True)\n\nprint('')\nprint('NGROUP={}'.format(ng_max))\n_ = obs.sat_limits(sp=sp_flat,ngroup=ng_max,verbose=True)\n\nmag_sci = obs.star_flux('vegamag')\nmag_ref = obs.star_flux('vegamag', sp=obs.sp_ref)\nprint('')\nprint('{} flux at {}: {:0.2f} mags'.format(obs.sp_sci.name, obs.filter, mag_sci))\nprint('{} flux at {}: {:0.2f} mags'.format(obs.sp_ref.name, obs.filter, mag_ref))",
"NGROUP=2\nF444W Saturation Limit assuming Flat spectrum in photlam source: 2.35 vegamag\n\nNGROUP=16\nF444W Saturation Limit assuming Flat spectrum in photlam source: 4.95 vegamag\n\nHR 8799 flux at F444W: 5.24 mags\nHD 220657 flux at F444W: 3.03 mags\n"
]
],
[
[
"In this case, we don't expect HR 8799 to saturated. However, the reference source should have some saturated pixels before the end of an integration. ",
"_____no_output_____"
]
],
[
[
"# Well level of each pixel for science source\nsci_levels1 = obs.saturation_levels(ngroup=2)\nsci_levels2 = obs.saturation_levels(ngroup=ng_max)\n\n# Which pixels are saturated?\nsci_mask1 = sci_levels1 > 0.8\nsci_mask2 = sci_levels2 > 0.8",
"_____no_output_____"
],
[
"# Well level of each pixel for reference source\nref_levels1 = obs.saturation_levels(ngroup=2, do_ref=True)\nref_levels2 = obs.saturation_levels(ngroup=ng_max, do_ref=True)\n\n# Which pixels are saturated?\nref_mask1 = ref_levels1 > 0.8\nref_mask2 = ref_levels2 > 0.8",
"_____no_output_____"
],
[
"# How many saturated pixels?\nnsat1_sci = len(sci_levels1[sci_mask1])\nnsat2_sci = len(sci_levels2[sci_mask2])\n\nprint(obs.sp_sci.name)\nprint('{} saturated pixel at NGROUP=2'.format(nsat1_sci))\nprint('{} saturated pixel at NGROUP={}'.format(nsat2_sci,ng_max))\n\n# How many saturated pixels?\nnsat1_ref = len(ref_levels1[ref_mask1])\nnsat2_ref = len(ref_levels2[ref_mask2])\n\nprint('')\nprint(obs.sp_ref.name)\nprint('{} saturated pixel at NGROUP=2'.format(nsat1_ref))\nprint('{} saturated pixel at NGROUP={}'.format(nsat2_ref,ng_max))",
"HR 8799\n0 saturated pixel at NGROUP=2\n0 saturated pixel at NGROUP=16\n\nHD 220657\n0 saturated pixel at NGROUP=2\n719 saturated pixel at NGROUP=16\n"
],
[
"# Saturation Mask for science target\n\nnsat1, nsat2 = (nsat1_sci, nsat2_sci)\nsat_mask1, sat_mask2 = (sci_mask1, sci_mask2)\nsp = obs.sp_sci\nnrc = obs\n\n# Only display saturation masks if there are saturated pixels\nif nsat2 > 0:\n fig, axes = plt.subplots(1,2, figsize=(10,5))\n\n xasec = nrc.det_info['xpix'] * nrc.pix_scale\n yasec = nrc.det_info['ypix'] * nrc.pix_scale\n extent = [-xasec/2, xasec/2, -yasec/2, yasec/2]\n\n axes[0].imshow(sat_mask1, extent=extent)\n axes[1].imshow(sat_mask2, extent=extent)\n\n axes[0].set_title('{} Saturation (NGROUP=2)'.format(sp.name))\n axes[1].set_title('{} Saturation (NGROUP={})'.format(sp.name,ng_max))\n\n for ax in axes:\n ax.set_xlabel('Arcsec')\n ax.set_ylabel('Arcsec')\n \n ax.tick_params(axis='both', color='white', which='both')\n for k in ax.spines.keys():\n ax.spines[k].set_color('white')\n\n fig.tight_layout()\nelse:\n print('No saturation detected.')",
"No saturation detected.\n"
],
[
"# Saturation Mask for reference\n\nnsat1, nsat2 = (nsat1_ref, nsat2_ref)\nsat_mask1, sat_mask2 = (ref_mask1, ref_mask2)\nsp = obs.sp_ref\nnrc = obs.nrc_ref\n\n# Only display saturation masks if there are saturated pixels\nif nsat2 > 0:\n fig, axes = plt.subplots(1,2, figsize=(10,5))\n\n xasec = nrc.det_info['xpix'] * nrc.pix_scale\n yasec = nrc.det_info['ypix'] * nrc.pix_scale\n extent = [-xasec/2, xasec/2, -yasec/2, yasec/2]\n\n axes[0].imshow(sat_mask1, extent=extent)\n axes[1].imshow(sat_mask2, extent=extent)\n\n axes[0].set_title('{} Saturation (NGROUP=2)'.format(sp.name))\n axes[1].set_title('{} Saturation (NGROUP={})'.format(sp.name,ng_max))\n\n for ax in axes:\n ax.set_xlabel('Arcsec')\n ax.set_ylabel('Arcsec')\n \n ax.tick_params(axis='both', color='white', which='both')\n for k in ax.spines.keys():\n ax.spines[k].set_color('white')\n\n fig.tight_layout()\nelse:\n print('No saturation detected.')",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
4aff8477bcd6de2ba130caa494f8a33eb7b205eb
| 393,605 |
ipynb
|
Jupyter Notebook
|
documentation/source/usersGuide/usersGuide_15_key.ipynb
|
cuthbertLab/music21
|
1be16c255460107c10d7b4bc8eb77f0d115b5eac
|
[
"MIT"
] | 1,449 |
2015-01-09T15:53:56.000Z
|
2022-03-31T18:24:46.000Z
|
documentation/source/usersGuide/usersGuide_15_key.ipynb
|
cuthbertLab/music21
|
1be16c255460107c10d7b4bc8eb77f0d115b5eac
|
[
"MIT"
] | 1,179 |
2015-01-07T17:07:54.000Z
|
2022-03-31T16:46:02.000Z
|
documentation/source/usersGuide/usersGuide_15_key.ipynb
|
cuthbertLab/music21
|
1be16c255460107c10d7b4bc8eb77f0d115b5eac
|
[
"MIT"
] | 393 |
2015-01-03T20:38:16.000Z
|
2022-03-25T16:51:22.000Z
| 209.253057 | 93,992 | 0.917925 |
[
[
[
"# ignore this\n%matplotlib inline\n%load_ext music21.ipython21",
"_____no_output_____"
]
],
[
[
"# User's Guide, Chapter 15: Keys and KeySignatures\n\nMusic21 has two main objects for working with keys: the :class:`~music21.key.KeySignature` object, which handles the spelling of key signatures and the :class:`~music21.key.Key` object which does everything a KeySignature object does but also knows more advanced aspects of tonal harmony. We'll go through the basics of each one here.\n\nWe start, like always, by importing music21:",
"_____no_output_____"
]
],
[
[
"from music21 import *",
"_____no_output_____"
]
],
[
[
"Now let's get a couple of different key signatures, representing different numbers of sharps:",
"_____no_output_____"
]
],
[
[
"ks2 = key.KeySignature(2)\nks2.sharps",
"_____no_output_____"
],
[
"ks7 = key.KeySignature(7)\nks7",
"_____no_output_____"
]
],
[
[
"We can get a list of which pitches (as :class:`~music21.pitch.Pitch` objects) are altered by the key signature with the `.alteredPitches` property:",
"_____no_output_____"
]
],
[
[
"ks2.alteredPitches",
"_____no_output_____"
]
],
[
[
"There's also a method that lets us see what the accidental is for any given step:",
"_____no_output_____"
]
],
[
[
"ks2.accidentalByStep('C')",
"_____no_output_____"
],
[
"ks2.accidentalByStep('E') is None",
"_____no_output_____"
]
],
[
[
"Notice that we give a string of just a letter name from C-B. This won't work:",
"_____no_output_____"
]
],
[
[
"ks2.accidentalByStep('C#')",
"_____no_output_____"
]
],
[
[
"We can create key signatures with absurd numbers of sharps and get strange accidentals:",
"_____no_output_____"
]
],
[
[
"ks12 = key.KeySignature(12)\nks12.accidentalByStep('F')",
"_____no_output_____"
]
],
[
[
"These absurd key signatures display in some programs (such as Lilypond) and are exported into MusicXML but do not display in most MusicXML readers.\n\nKey Signatures transpose like Pitches and Notes, taking each of the notes and moving it:",
"_____no_output_____"
]
],
[
[
"ks4 = ks2.transpose('M2')\nks4",
"_____no_output_____"
]
],
[
[
"And the number of sharps can be changed after the fact:",
"_____no_output_____"
]
],
[
[
"ks4.sharps = 0\nks4",
"_____no_output_____"
]
],
[
[
"We can get the Major or Minor scale corresponding to the Key Signature:",
"_____no_output_____"
]
],
[
[
"ks2.getScale('major')",
"_____no_output_____"
],
[
"ks2.getScale('minor')",
"_____no_output_____"
]
],
[
[
"We'll see what we can do with scales in a bit.\n\nIf we put a KeySignature into a Stream, we can see it:",
"_____no_output_____"
]
],
[
[
"m = stream.Measure()\nm.insert(0, meter.TimeSignature('3/4'))\nm.insert(0, ks2)\nd = note.Note('D')\nc = note.Note('C')\nfis = note.Note('F#') # German name\nm.append([d, c, fis])\nm.show()",
"_____no_output_____"
]
],
[
[
"Note that the Note 'C' is treated as C-natural and thus needs the natural sign in front of it. The Note F# however does not need a natural sign to be displayed. The process of calling `.show()` on the stream made a copy of the notes and set the `.pitch.accidental.displayStatus` on the F# to `False` and created an accidental for the C note with a natural and a displayStatus of True. Then the copies were discarded, so we don't see them here:",
"_____no_output_____"
]
],
[
[
"fis.pitch.accidental.displayStatus",
"_____no_output_____"
]
],
[
[
"But we could instead call `.makeNotation(inPlace=True)` or `.makeAccidentals(inPlace=True)` on the Measure to do this manually:",
"_____no_output_____"
]
],
[
[
"m.makeAccidentals(inPlace=True)\nfis.pitch.accidental.displayStatus",
"_____no_output_____"
],
[
"c.pitch.accidental, c.pitch.accidental.displayStatus",
"_____no_output_____"
]
],
[
[
"If we have a `Measure` (not just any `Stream`) we can also set the KeySignature for the beginning of the measure with the Measure object's `.keySignature` property:",
"_____no_output_____"
]
],
[
[
"m.keySignature = key.KeySignature(4)\nm.show()",
"_____no_output_____"
]
],
[
[
"Of course life isn't all about sharps; it'd be a pretty terrible KeySignature object if we couldn't have flats. To do it, just specify the number of flats as a negative number. So -1 = one flat, -2 = two flats. Or if you have the number as a positive already, just multiply by -1.",
"_____no_output_____"
]
],
[
[
"eroicaFlats = 3\nksEroica = key.KeySignature(-1 * eroicaFlats)\nksEroica",
"_____no_output_____"
],
[
"ksEroica.sharps",
"_____no_output_____"
]
],
[
[
"There is no `.flats` routine:",
"_____no_output_____"
]
],
[
[
"ksEroica.flats",
"_____no_output_____"
]
],
[
[
"## Example: Adjusting notes to fit the Key Signature\n\nHere's a nice study, suppose you had a score like this:",
"_____no_output_____"
]
],
[
[
"m1 = stream.Measure()\nm1.timeSignature = meter.TimeSignature('2/4')\nm1.keySignature = key.KeySignature(-5)\nm1.append([note.Note('D'), note.Note('A')])\nm2 = stream.Measure()\nm2.append([note.Note('B-'), note.Note('G#')])\np = stream.Part()\np.append([m1, m2])\np.show()",
"_____no_output_____"
]
],
[
[
"Let's pretend that this was played by a young oboe player who was having trouble with the strange key signature. She got the B-flat right, and remembered to play some accidental on the G, but didn't do very well overall. Let's fix these notes so that they fit with the key signature.\n\nNow we could simply do something like this for each note:",
"_____no_output_____"
]
],
[
[
"m1.notes[0].pitch.accidental = pitch.Accidental('flat')",
"_____no_output_____"
]
],
[
[
"But that wouldn't be as great as getting the notes from the Key itself. Let's do that with the accidentalByStep routine:",
"_____no_output_____"
]
],
[
[
"ks = m1.keySignature\nfor n in p.recurse().notes: # we need to recurse because the notes are in measures...\n nStep = n.pitch.step\n rightAccidental = ks.accidentalByStep(nStep)\n n.pitch.accidental = rightAccidental\np.show()",
"_____no_output_____"
]
],
[
[
"Yep, now our student is ready to play the concert! Though wouldn't this be an easier key?",
"_____no_output_____"
]
],
[
[
"p.transpose(1).show()",
"_____no_output_____"
]
],
[
[
"## Key objects\n\nA Key is a lot like a KeySignature, but much more powerful. Unlike a KeySignature, which we initialize with the number of sharps and flats, we initialize a Key with a tonic string or Pitch:",
"_____no_output_____"
]
],
[
[
"kD = key.Key('D')\nkD",
"_____no_output_____"
],
[
"bFlat = pitch.Pitch('B-')\nkBflat = key.Key(bFlat)\nkBflat",
"_____no_output_____"
]
],
[
[
"By default, keys are major, but we can make minor keys by specifying 'minor' as the second argument:",
"_____no_output_____"
]
],
[
[
"kd = key.Key('D', 'minor')\nkd",
"_____no_output_____"
]
],
[
[
"Note that the key is represented as lowercase ('d minor' as opposed to 'D minor'). This is a clue as to a shortcut for making minor keys:",
"_____no_output_____"
]
],
[
[
"kg = key.Key('g')\nkg",
"_____no_output_____"
]
],
[
[
"We can also take KeySignatures and turn them into Keys by using the `asKey(mode)` method on them:",
"_____no_output_____"
]
],
[
[
"(ksEroica.asKey('major'), ksEroica.asKey('minor'))",
"_____no_output_____"
]
],
[
[
"(In the latter case we should probably have called the variable ksFifthSymphony...)\n\nWe can also make church modes:",
"_____no_output_____"
]
],
[
[
"amixy = key.Key('a', 'mixolydian')\namixy",
"_____no_output_____"
]
],
[
[
"If you've forgotten how many sharps or flats are in the key of A mixolydian, you'll be happy to know that all the properties and methods of KeySignatures are also available to Keys:",
"_____no_output_____"
]
],
[
[
"amixy.sharps",
"_____no_output_____"
],
[
"amixy.alteredPitches",
"_____no_output_____"
],
[
"amixy.transpose('M3')",
"_____no_output_____"
],
[
"aDarkKey = key.Key('B--', 'locrian')\naDarkKey.alteredPitches",
"_____no_output_____"
]
],
[
[
"(as a music historian and someone who specializes in history of music theory, I am contractually obliged to mention that \"locrian\" is not a historic mode and doesn't really exist in actual music before the 20th c. But it's fun to play with).\n\nKeys know their `.mode`:",
"_____no_output_____"
]
],
[
[
"kg.mode, amixy.mode",
"_____no_output_____"
]
],
[
[
"They also know their tonic pitches:",
"_____no_output_____"
]
],
[
[
"kg.tonic, amixy.tonic",
"_____no_output_____"
]
],
[
[
"For major and minor keys, we can get the relative (minor or major) and parallel (minor or major) keys simply:",
"_____no_output_____"
]
],
[
[
"kg.relative",
"_____no_output_____"
],
[
"kg.parallel",
"_____no_output_____"
]
],
[
[
"And because two keys are equal if their modes and tonics are the same, this is true:",
"_____no_output_____"
]
],
[
[
"kg.relative.relative == kg",
"_____no_output_____"
]
],
[
[
"This is pretty helpful from time to time:",
"_____no_output_____"
]
],
[
[
"kg.tonicPitchNameWithCase",
"_____no_output_____"
],
[
"kg.parallel.tonicPitchNameWithCase",
"_____no_output_____"
]
],
[
[
"Some analysis routines produce keys:",
"_____no_output_____"
]
],
[
[
"bach = corpus.parse('bwv66.6')\nbach.id = 'bach66'\nbach.analyze('key')",
"_____no_output_____"
]
],
[
[
"The keys from these routines have two extra cool features. They have a certainty measure:",
"_____no_output_____"
]
],
[
[
"fis = bach.analyze('key')\nfis.correlationCoefficient",
"_____no_output_____"
],
[
"fis.tonalCertainty()",
"_____no_output_____"
]
],
[
[
"Here are some of the other keys that the Bach piece could have been in:",
"_____no_output_____"
]
],
[
[
"fis.alternateInterpretations[0:4]",
"_____no_output_____"
]
],
[
[
"And the least likely:",
"_____no_output_____"
]
],
[
[
"fis.alternateInterpretations[-3:]",
"_____no_output_____"
],
[
"c = bach.measures(1, 4).chordify()\nfor ch in c.recurse().getElementsByClass('Chord'):\n ch.closedPosition(inPlace=True, forceOctave=4)\nc.show()",
"_____no_output_____"
]
],
[
[
"Yeah, that passes the smell test to me!",
"_____no_output_____"
],
[
"So, how does it know what the key is? The key analysis routines are a variation of the famous (well at least in the small world of computational music theory) algorithm developed by Carol Krumhansl and Mark A. Schmuckler called probe-tone key finding. The distribution of pitches used in the piece are compared to sample distributions of pitches for major and minor keys and the closest matches are reported. (see http://rnhart.net/articles/key-finding/ for more details). `Music21` can be asked to use the sample distributions of several authors, including Krumhansl and Schmuckler's original weights:",
"_____no_output_____"
]
],
[
[
"bach.analyze('key.krumhanslschmuckler')",
"_____no_output_____"
]
],
[
[
"Though the `key` returned by `.analyze('key')` and `.analyze('key.krumhanslschmuckler')` are the same, the correlationCoefficient is somewhat different. `fis` is the analysis from `.analyze('key')`.",
"_____no_output_____"
]
],
[
[
"fisNew = bach.analyze('key.krumhanslschmuckler')\nfisCC = round(fis.correlationCoefficient, 3)\nfisNewCC = round(fisNew.correlationCoefficient, 3)\n(fisCC, fisNewCC)",
"_____no_output_____"
]
],
[
[
"Calling `.analyze()` on a Stream calls :func:`music21.analysis.discrete.analyzeStream` which then calls an appropriate Class there.\n\nThere is another way of looking at the key of a piece and that is looking at differently sized windows of analysis on the piece and seeing what happens every quarter note, every half note, every measure, every two measures, etc. to the top. This plot was created by Jared Sadoian and is explained in the `analysis.windowed` module:",
"_____no_output_____"
]
],
[
[
"bach.flatten().plot('pianoroll')",
"_____no_output_____"
]
],
[
[
"A Key object is derived from a KeySignature object and also a Scale object, which we will explain more about later.",
"_____no_output_____"
]
],
[
[
"k = key.Key('E-')\nk.classes",
"_____no_output_____"
]
],
[
[
"But for now, a few methods that are present on scales that might end up being useful for Keys as well include:",
"_____no_output_____"
]
],
[
[
"k.pitchFromDegree(2)",
"_____no_output_____"
]
],
[
[
"(octaves in 4 and 5 are chosen just to give some ordering to the pitches)",
"_____no_output_____"
]
],
[
[
"k.solfeg('G')",
"_____no_output_____"
]
],
[
[
"## Key Context and Note Spelling\n\n`Key` and `KeySignature` objects affect how notes are spelled in some situations. Let's set up a simple situation of a F-natural whole note in D major and then B-flat minor.",
"_____no_output_____"
]
],
[
[
"s = stream.Stream()\ns.append(key.Key('D'))\ns.append(note.Note('F', type='whole'))\ns.append(key.Key('b-', 'minor'))\ns.append(note.Note('F', type='whole'))\ns2 = s.makeNotation()\ns2.show()",
"_____no_output_____"
]
],
[
[
"When we transpose each note up a half step (`n.transpose(1)`), music21 understands that the first F-natural should become F-sharp, while the second one will fit better as a G-flat.",
"_____no_output_____"
]
],
[
[
"for n in s2.recurse().notes:\n n.transpose(1, inPlace=True)\ns2.show()",
"_____no_output_____"
]
],
[
[
"## Example: Prepare a vocal exercise in all major keys, ascending by step.\n\nLet's create a simple exercise in playing or singing thirds. I think I remember this from the [First Division Band Method](https://www.google.com/search?q=First+Division+Band+Method&tbm=isch) \"Blue Book\":",
"_____no_output_____"
]
],
[
[
"pitchStream = stream.Part()\npitchStream.insert(0, meter.TimeSignature('4/4'))\nfor step in ('c', 'e', 'd', 'f', 'e', 'g', 'f', 'a',\n 'g', 'e', 'f', 'd', 'c', 'e', 'c'):\n n = note.Note(step, type='eighth')\n n.pitch.octave = 4\n pitchStream.append(n)\npitchStream.notes[-1].duration.type = 'quarter'\npitchStream.makeMeasures(inPlace=True)\npitchStream.show()",
"_____no_output_____"
]
],
[
[
"This melody does not have a key associated with it. Let's put a Key of C Major at the beginning of the piece:",
"_____no_output_____"
]
],
[
[
"k = key.Key('C')\npitchStream.measure(1).insert(0, k)\npitchStream.show()",
"_____no_output_____"
]
],
[
[
"Note that putting the key of C into the Stream doesn't change what it looks like when we show the Stream, since there are no sharps or flats. But what makes the difference between an instrumental and a vocal exercise is the act of transposition. When we transpose the `Key` object up 1 semitone, to D-flat major, it will show up:",
"_____no_output_____"
]
],
[
[
"k.transpose(1, inPlace=True)\npitchStream.show()",
"_____no_output_____"
]
],
[
[
"Now the key signature is D-flat, but the notes are still in C-major, so we should transpose them also:",
"_____no_output_____"
]
],
[
[
"for n in pitchStream.recurse().notes:\n n.transpose(1, inPlace=True)\n\npitchStream.show()",
"_____no_output_____"
]
],
[
[
"Notice that we choose a semitone transposition and not a diatonic transposition such as minor second (`\"m2\"`); minor second would work just as good in this case, but then to do another half-step up, we would need to remember to transpose by an augmented unison (`\"A1\"`) so that D-flat became D-natural and not E-double-flat. The semitone transposition is smart enough to make sure that the `Key` object remains between six-flats and six-sharps. Not only that, but the notes will match the best spelling for the current key signature.",
"_____no_output_____"
]
],
[
[
"k.transpose(1, inPlace=True)\nfor n in pitchStream.recurse().notes:\n n.transpose(1, inPlace=True)\npitchStream.show()",
"_____no_output_____"
],
[
"k.transpose(1, inPlace=True)\nfor n in pitchStream.recurse().notes:\n n.transpose(1, inPlace=True)\npitchStream.show()",
"_____no_output_____"
]
],
[
[
"So, we can make a nice, ascending vocal exercise by varying the transposition amount from 0 to 7 (or however high you can sing) and putting each of the two-measure excerpts together into one Part.\n\nWe will introduce the tinyNotation format here, which will be described in the next chapter:",
"_____no_output_____"
]
],
[
[
"out = stream.Part()\nfor i in range(0, 8):\n pitchStream = converter.parse(\"tinyNotation: 4/4 c8 e d f e g f a g e f d c e c4\")\n\n if i != 0:\n # remove redundant clefs and time signature\n trebleClef = pitchStream.recurse().getElementsByClass('Clef')[0]\n fourFour = pitchStream.recurse().getElementsByClass('TimeSignature')[0]\n\n pitchStream.remove(trebleClef, recurse=True)\n pitchStream.remove(fourFour, recurse=True)\n\n if i % 2 == 0:\n # add a line break at the beginning of every other line:\n pitchStream.measure(1).insert(0, layout.SystemLayout(isNew=True))\n\n k = key.Key('C')\n pitchStream.measure(1).insert(0, k)\n k.transpose(i, inPlace=True)\n for n in pitchStream.recurse().notes:\n n.transpose(i, inPlace=True)\n for el in pitchStream:\n out.append(el)\n\nout.show()",
"_____no_output_____"
]
],
[
[
"And we can listen to it as well:",
"_____no_output_____"
]
],
[
[
"out.show('midi')",
"_____no_output_____"
]
],
[
[
"That's enough about keys for now, let's move on to a fast way of getting small amounts of music into music21, with :ref:`Chapter 16, Tiny Notation <usersGuide_16_tinyNotation>`",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
4aff952f1bda4f027f0d3545893108f1330d573f
| 29,946 |
ipynb
|
Jupyter Notebook
|
Big-Data-Clusters/CU6/Public/content/cert-management/cer002-download-existing-root-ca.ipynb
|
gantz-at-incomm/tigertoolbox
|
9ea80d39a3c5e0c77553fc851c5ee787fbf9291d
|
[
"MIT"
] | 541 |
2019-05-07T11:41:25.000Z
|
2022-03-29T17:33:19.000Z
|
Big-Data-Clusters/CU6/Public/content/cert-management/cer002-download-existing-root-ca.ipynb
|
gantz-at-incomm/tigertoolbox
|
9ea80d39a3c5e0c77553fc851c5ee787fbf9291d
|
[
"MIT"
] | 89 |
2019-05-09T14:23:52.000Z
|
2022-01-13T20:21:04.000Z
|
Big-Data-Clusters/CU6/Public/content/cert-management/cer002-download-existing-root-ca.ipynb
|
gantz-at-incomm/tigertoolbox
|
9ea80d39a3c5e0c77553fc851c5ee787fbf9291d
|
[
"MIT"
] | 338 |
2019-05-08T05:45:16.000Z
|
2022-03-28T15:35:03.000Z
| 54.447273 | 408 | 0.420791 |
[
[
[
"CER002 - Download existing Root CA certificate\n==============================================\n\nUse this notebook to download a generated Root CA certificate from a\ncluster that installed one using:\n\n- [CER001 - Generate a Root CA\n certificate](../cert-management/cer001-create-root-ca.ipynb)\n\nAnd then to upload the generated Root CA to another cluster use:\n\n- [CER003 - Upload existing Root CA\n certificate](../cert-management/cer003-upload-existing-root-ca.ipynb)\n\nIf needed, use these notebooks to view and set the Kubernetes\nconfiguration context appropriately to enable downloading the Root CA\nfrom a Big Data Cluster in one Kubernetes cluster, and to upload it to a\nBig Data Cluster in another Kubernetes cluster.\n\n- [TSG010 - Get configuration\n contexts](../monitor-k8s/tsg010-get-kubernetes-contexts.ipynb)\n- [SOP011 - Set kubernetes configuration\n context](../common/sop011-set-kubernetes-context.ipynb)\n\nSteps\n-----\n\n### Parameters",
"_____no_output_____"
]
],
[
[
"local_folder_name = \"mssql-cluster-root-ca\"\n\ntest_cert_store_root = \"/var/opt/secrets/test-certificates\"",
"_____no_output_____"
]
],
[
[
"### Common functions\n\nDefine helper functions used in this notebook.",
"_____no_output_____"
]
],
[
[
"# Define `run` function for transient fault handling, suggestions on error, and scrolling updates on Windows\nimport sys\nimport os\nimport re\nimport json\nimport platform\nimport shlex\nimport shutil\nimport datetime\n\nfrom subprocess import Popen, PIPE\nfrom IPython.display import Markdown\n\nretry_hints = {} # Output in stderr known to be transient, therefore automatically retry\nerror_hints = {} # Output in stderr where a known SOP/TSG exists which will be HINTed for further help\ninstall_hint = {} # The SOP to help install the executable if it cannot be found\n\nfirst_run = True\nrules = None\ndebug_logging = False\n\ndef run(cmd, return_output=False, no_output=False, retry_count=0):\n \"\"\"Run shell command, stream stdout, print stderr and optionally return output\n\n NOTES:\n\n 1. Commands that need this kind of ' quoting on Windows e.g.:\n\n kubectl get nodes -o jsonpath={.items[?(@.metadata.annotations.pv-candidate=='data-pool')].metadata.name}\n\n Need to actually pass in as '\"':\n\n kubectl get nodes -o jsonpath={.items[?(@.metadata.annotations.pv-candidate=='\"'data-pool'\"')].metadata.name}\n\n The ' quote approach, although correct when pasting into Windows cmd, will hang at the line:\n \n `iter(p.stdout.readline, b'')`\n\n The shlex.split call does the right thing for each platform, just use the '\"' pattern for a '\n \"\"\"\n MAX_RETRIES = 5\n output = \"\"\n retry = False\n\n global first_run\n global rules\n\n if first_run:\n first_run = False\n rules = load_rules()\n\n # When running `azdata sql query` on Windows, replace any \\n in \"\"\" strings, with \" \", otherwise we see:\n #\n # ('HY090', '[HY090] [Microsoft][ODBC Driver Manager] Invalid string or buffer length (0) (SQLExecDirectW)')\n #\n if platform.system() == \"Windows\" and cmd.startswith(\"azdata sql query\"):\n cmd = cmd.replace(\"\\n\", \" \")\n\n # shlex.split is required on bash and for Windows paths with spaces\n #\n cmd_actual = shlex.split(cmd)\n\n # Store this (i.e. kubectl, python etc.) to support binary context aware error_hints and retries\n #\n user_provided_exe_name = cmd_actual[0].lower()\n\n # When running python, use the python in the ADS sandbox ({sys.executable})\n #\n if cmd.startswith(\"python \"):\n cmd_actual[0] = cmd_actual[0].replace(\"python\", sys.executable)\n\n # On Mac, when ADS is not launched from terminal, LC_ALL may not be set, which causes pip installs to fail\n # with:\n #\n # UnicodeDecodeError: 'ascii' codec can't decode byte 0xc5 in position 4969: ordinal not in range(128)\n #\n # Setting it to a default value of \"en_US.UTF-8\" enables pip install to complete\n #\n if platform.system() == \"Darwin\" and \"LC_ALL\" not in os.environ:\n os.environ[\"LC_ALL\"] = \"en_US.UTF-8\"\n\n # When running `kubectl`, if AZDATA_OPENSHIFT is set, use `oc`\n #\n if cmd.startswith(\"kubectl \") and \"AZDATA_OPENSHIFT\" in os.environ:\n cmd_actual[0] = cmd_actual[0].replace(\"kubectl\", \"oc\")\n\n # To aid supportabilty, determine which binary file will actually be executed on the machine\n #\n which_binary = None\n\n # Special case for CURL on Windows. The version of CURL in Windows System32 does not work to\n # get JWT tokens, it returns \"(56) Failure when receiving data from the peer\". If another instance\n # of CURL exists on the machine use that one. (Unfortunately the curl.exe in System32 is almost\n # always the first curl.exe in the path, and it can't be uninstalled from System32, so here we\n # look for the 2nd installation of CURL in the path)\n if platform.system() == \"Windows\" and cmd.startswith(\"curl \"):\n path = os.getenv('PATH')\n for p in path.split(os.path.pathsep):\n p = os.path.join(p, \"curl.exe\")\n if os.path.exists(p) and os.access(p, os.X_OK):\n if p.lower().find(\"system32\") == -1:\n cmd_actual[0] = p\n which_binary = p\n break\n\n # Find the path based location (shutil.which) of the executable that will be run (and display it to aid supportability), this\n # seems to be required for .msi installs of azdata.cmd/az.cmd. (otherwise Popen returns FileNotFound) \n #\n # NOTE: Bash needs cmd to be the list of the space separated values hence shlex.split.\n #\n if which_binary == None:\n which_binary = shutil.which(cmd_actual[0])\n\n if which_binary == None:\n if user_provided_exe_name in install_hint and install_hint[user_provided_exe_name] is not None:\n display(Markdown(f'HINT: Use [{install_hint[user_provided_exe_name][0]}]({install_hint[user_provided_exe_name][1]}) to resolve this issue.'))\n\n raise FileNotFoundError(f\"Executable '{cmd_actual[0]}' not found in path (where/which)\")\n else: \n cmd_actual[0] = which_binary\n\n start_time = datetime.datetime.now().replace(microsecond=0)\n\n print(f\"START: {cmd} @ {start_time} ({datetime.datetime.utcnow().replace(microsecond=0)} UTC)\")\n print(f\" using: {which_binary} ({platform.system()} {platform.release()} on {platform.machine()})\")\n print(f\" cwd: {os.getcwd()}\")\n\n # Command-line tools such as CURL and AZDATA HDFS commands output\n # scrolling progress bars, which causes Jupyter to hang forever, to\n # workaround this, use no_output=True\n #\n\n # Work around a infinite hang when a notebook generates a non-zero return code, break out, and do not wait\n #\n wait = True \n\n try:\n if no_output:\n p = Popen(cmd_actual)\n else:\n p = Popen(cmd_actual, stdout=PIPE, stderr=PIPE, bufsize=1)\n with p.stdout:\n for line in iter(p.stdout.readline, b''):\n line = line.decode()\n if return_output:\n output = output + line\n else:\n if cmd.startswith(\"azdata notebook run\"): # Hyperlink the .ipynb file\n regex = re.compile(' \"(.*)\"\\: \"(.*)\"') \n match = regex.match(line)\n if match:\n if match.group(1).find(\"HTML\") != -1:\n display(Markdown(f' - \"{match.group(1)}\": \"{match.group(2)}\"'))\n else:\n display(Markdown(f' - \"{match.group(1)}\": \"[{match.group(2)}]({match.group(2)})\"'))\n\n wait = False\n break # otherwise infinite hang, have not worked out why yet.\n else:\n print(line, end='')\n if rules is not None:\n apply_expert_rules(line)\n\n if wait:\n p.wait()\n except FileNotFoundError as e:\n if install_hint is not None:\n display(Markdown(f'HINT: Use {install_hint} to resolve this issue.'))\n\n raise FileNotFoundError(f\"Executable '{cmd_actual[0]}' not found in path (where/which)\") from e\n\n exit_code_workaround = 0 # WORKAROUND: azdata hangs on exception from notebook on p.wait()\n\n if not no_output:\n for line in iter(p.stderr.readline, b''):\n try:\n line_decoded = line.decode()\n except UnicodeDecodeError:\n # NOTE: Sometimes we get characters back that cannot be decoded(), e.g.\n #\n # \\xa0\n #\n # For example see this in the response from `az group create`:\n #\n # ERROR: Get Token request returned http error: 400 and server \n # response: {\"error\":\"invalid_grant\",# \"error_description\":\"AADSTS700082: \n # The refresh token has expired due to inactivity.\\xa0The token was \n # issued on 2018-10-25T23:35:11.9832872Z\n #\n # which generates the exception:\n #\n # UnicodeDecodeError: 'utf-8' codec can't decode byte 0xa0 in position 179: invalid start byte\n #\n print(\"WARNING: Unable to decode stderr line, printing raw bytes:\")\n print(line)\n line_decoded = \"\"\n pass\n else:\n\n # azdata emits a single empty line to stderr when doing an hdfs cp, don't\n # print this empty \"ERR:\" as it confuses.\n #\n if line_decoded == \"\":\n continue\n \n print(f\"STDERR: {line_decoded}\", end='')\n\n if line_decoded.startswith(\"An exception has occurred\") or line_decoded.startswith(\"ERROR: An error occurred while executing the following cell\"):\n exit_code_workaround = 1\n\n # inject HINTs to next TSG/SOP based on output in stderr\n #\n if user_provided_exe_name in error_hints:\n for error_hint in error_hints[user_provided_exe_name]:\n if line_decoded.find(error_hint[0]) != -1:\n display(Markdown(f'HINT: Use [{error_hint[1]}]({error_hint[2]}) to resolve this issue.'))\n\n # apply expert rules (to run follow-on notebooks), based on output\n #\n if rules is not None:\n apply_expert_rules(line_decoded)\n\n # Verify if a transient error, if so automatically retry (recursive)\n #\n if user_provided_exe_name in retry_hints:\n for retry_hint in retry_hints[user_provided_exe_name]:\n if line_decoded.find(retry_hint) != -1:\n if retry_count < MAX_RETRIES:\n print(f\"RETRY: {retry_count} (due to: {retry_hint})\")\n retry_count = retry_count + 1\n output = run(cmd, return_output=return_output, retry_count=retry_count)\n\n if return_output:\n return output\n else:\n return\n\n elapsed = datetime.datetime.now().replace(microsecond=0) - start_time\n\n # WORKAROUND: We avoid infinite hang above in the `azdata notebook run` failure case, by inferring success (from stdout output), so\n # don't wait here, if success known above\n #\n if wait: \n if p.returncode != 0:\n raise SystemExit(f'Shell command:\\n\\n\\t{cmd} ({elapsed}s elapsed)\\n\\nreturned non-zero exit code: {str(p.returncode)}.\\n')\n else:\n if exit_code_workaround !=0 :\n raise SystemExit(f'Shell command:\\n\\n\\t{cmd} ({elapsed}s elapsed)\\n\\nreturned non-zero exit code: {str(exit_code_workaround)}.\\n')\n\n print(f'\\nSUCCESS: {elapsed}s elapsed.\\n')\n\n if return_output:\n return output\n\ndef load_json(filename):\n \"\"\"Load a json file from disk and return the contents\"\"\"\n\n with open(filename, encoding=\"utf8\") as json_file:\n return json.load(json_file)\n\ndef load_rules():\n \"\"\"Load any 'expert rules' from the metadata of this notebook (.ipynb) that should be applied to the stderr of the running executable\"\"\"\n\n # Load this notebook as json to get access to the expert rules in the notebook metadata.\n #\n try:\n j = load_json(\"cer002-download-existing-root-ca.ipynb\")\n except:\n pass # If the user has renamed the book, we can't load ourself. NOTE: Is there a way in Jupyter, to know your own filename?\n else:\n if \"metadata\" in j and \\\n \"azdata\" in j[\"metadata\"] and \\\n \"expert\" in j[\"metadata\"][\"azdata\"] and \\\n \"expanded_rules\" in j[\"metadata\"][\"azdata\"][\"expert\"]:\n\n rules = j[\"metadata\"][\"azdata\"][\"expert\"][\"expanded_rules\"]\n\n rules.sort() # Sort rules, so they run in priority order (the [0] element). Lowest value first.\n\n # print (f\"EXPERT: There are {len(rules)} rules to evaluate.\")\n\n return rules\n\ndef apply_expert_rules(line):\n \"\"\"Determine if the stderr line passed in, matches the regular expressions for any of the 'expert rules', if so\n inject a 'HINT' to the follow-on SOP/TSG to run\"\"\"\n\n global rules\n\n for rule in rules:\n notebook = rule[1]\n cell_type = rule[2]\n output_type = rule[3] # i.e. stream or error\n output_type_name = rule[4] # i.e. ename or name \n output_type_value = rule[5] # i.e. SystemExit or stdout\n details_name = rule[6] # i.e. evalue or text \n expression = rule[7].replace(\"\\\\*\", \"*\") # Something escaped *, and put a \\ in front of it!\n\n if debug_logging:\n print(f\"EXPERT: If rule '{expression}' satisfied', run '{notebook}'.\")\n\n if re.match(expression, line, re.DOTALL):\n\n if debug_logging:\n print(\"EXPERT: MATCH: name = value: '{0}' = '{1}' matched expression '{2}', therefore HINT '{4}'\".format(output_type_name, output_type_value, expression, notebook))\n\n match_found = True\n\n display(Markdown(f'HINT: Use [{notebook}]({notebook}) to resolve this issue.'))\n\n\n\nprint('Common functions defined successfully.')\n\n# Hints for binary (transient fault) retry, (known) error and install guide\n#\nretry_hints = {'kubectl': ['A connection attempt failed because the connected party did not properly respond after a period of time, or established connection failed because connected host has failed to respond']}\nerror_hints = {'kubectl': [['no such host', 'TSG010 - Get configuration contexts', '../monitor-k8s/tsg010-get-kubernetes-contexts.ipynb'], ['No connection could be made because the target machine actively refused it', 'TSG056 - Kubectl fails with No connection could be made because the target machine actively refused it', '../repair/tsg056-kubectl-no-connection-could-be-made.ipynb']]}\ninstall_hint = {'kubectl': ['SOP036 - Install kubectl command line interface', '../install/sop036-install-kubectl.ipynb']}",
"_____no_output_____"
]
],
[
[
"### Get the Kubernetes namespace for the big data cluster\n\nGet the namespace of the Big Data Cluster use the kubectl command line\ninterface .\n\n**NOTE:**\n\nIf there is more than one Big Data Cluster in the target Kubernetes\ncluster, then either:\n\n- set \\[0\\] to the correct value for the big data cluster.\n- set the environment variable AZDATA\\_NAMESPACE, before starting\n Azure Data Studio.",
"_____no_output_____"
]
],
[
[
"# Place Kubernetes namespace name for BDC into 'namespace' variable\n\nif \"AZDATA_NAMESPACE\" in os.environ:\n namespace = os.environ[\"AZDATA_NAMESPACE\"]\nelse:\n try:\n namespace = run(f'kubectl get namespace --selector=MSSQL_CLUSTER -o jsonpath={{.items[0].metadata.name}}', return_output=True)\n except:\n from IPython.display import Markdown\n print(f\"ERROR: Unable to find a Kubernetes namespace with label 'MSSQL_CLUSTER'. SQL Server Big Data Cluster Kubernetes namespaces contain the label 'MSSQL_CLUSTER'.\")\n display(Markdown(f'HINT: Use [TSG081 - Get namespaces (Kubernetes)](../monitor-k8s/tsg081-get-kubernetes-namespaces.ipynb) to resolve this issue.'))\n display(Markdown(f'HINT: Use [TSG010 - Get configuration contexts](../monitor-k8s/tsg010-get-kubernetes-contexts.ipynb) to resolve this issue.'))\n display(Markdown(f'HINT: Use [SOP011 - Set kubernetes configuration context](../common/sop011-set-kubernetes-context.ipynb) to resolve this issue.'))\n raise\n\nprint(f'The SQL Server Big Data Cluster Kubernetes namespace is: {namespace}')",
"_____no_output_____"
]
],
[
[
"### Get name of the ‘Running’ `controller` `pod`",
"_____no_output_____"
]
],
[
[
"# Place the name of the 'Running' controller pod in variable `controller`\n\ncontroller = run(f'kubectl get pod --selector=app=controller -n {namespace} -o jsonpath={{.items[0].metadata.name}} --field-selector=status.phase=Running', return_output=True)\n\nprint(f\"Controller pod name: {controller}\")",
"_____no_output_____"
]
],
[
[
"### Create a temporary folder to hold Root CA certificate",
"_____no_output_____"
]
],
[
[
"import os\nimport tempfile\nimport shutil\n\npath = os.path.join(tempfile.gettempdir(), local_folder_name)\n\nif os.path.isdir(path):\n shutil.rmtree(path)\n\nos.mkdir(path)",
"_____no_output_____"
]
],
[
[
"### Copy Root CA certificate from `controller` `pod`",
"_____no_output_____"
]
],
[
[
"import os\n\ncwd = os.getcwd()\nos.chdir(path) # Workaround kubectl bug on Windows, can't put c:\\ on kubectl cp cmd line \n\nrun(f'kubectl cp {controller}:{test_cert_store_root}/cacert.pem cacert.pem -c controller -n {namespace}')\nrun(f'kubectl cp {controller}:{test_cert_store_root}/cakey.pem cakey.pem -c controller -n {namespace}')\n\nos.chdir(cwd)",
"_____no_output_____"
],
[
"print('Notebook execution complete.')",
"_____no_output_____"
]
],
[
[
"Related\n-------\n\n- [CER001 - Generate a Root CA\n certificate](../cert-management/cer001-create-root-ca.ipynb)\n\n- [CER003 - Upload existing Root CA\n certificate](../cert-management/cer003-upload-existing-root-ca.ipynb)\n\n- [CER010 - Install generated Root CA\n locally](../cert-management/cer010-install-generated-root-ca-locally.ipynb)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
4affa0488caffa1e14ca072ba10a46dfee3cb1a2
| 476,686 |
ipynb
|
Jupyter Notebook
|
notebooks/OpenCADD_Benchmark_CAMK_CMGC.ipynb
|
volkamerlab/OpenCADDSuperpositionBenchmark
|
9ee483088497f1d8666794780bbe95cf20a7d706
|
[
"MIT"
] | null | null | null |
notebooks/OpenCADD_Benchmark_CAMK_CMGC.ipynb
|
volkamerlab/OpenCADDSuperpositionBenchmark
|
9ee483088497f1d8666794780bbe95cf20a7d706
|
[
"MIT"
] | 1 |
2021-12-12T23:11:36.000Z
|
2021-12-12T23:11:36.000Z
|
notebooks/OpenCADD_Benchmark_CAMK_CMGC.ipynb
|
volkamerlab/OpenCADDSuperpositionBenchmark
|
9ee483088497f1d8666794780bbe95cf20a7d706
|
[
"MIT"
] | null | null | null | 162.138095 | 210,366 | 0.839215 |
[
[
[
"# Import used modules",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport sys\nsys.path.insert(0, '../src') \nimport benchmark_utils as bu\nimport analysis_utils as au",
"/Users/julian/opt/anaconda3/envs/OpenCADDBenchmark/lib/python3.8/site-packages/MDAnalysis/coordinates/chemfiles.py:108: DeprecationWarning: distutils Version classes are deprecated. Use packaging.version instead.\n MIN_CHEMFILES_VERSION = LooseVersion(\"0.9\")\n"
]
],
[
[
"# Run Alignments for OpenCADD.superposition for the CAMK and CMGC Structures",
"_____no_output_____"
],
[
"Perform all pairwise alignments for the given sample structures. Every method performs 2500 alignments for the 50 CAMK and 50 CMGC structures. The benchmark is done with an Intel Core i5-1038NG7 CPU and 16 GB of RAM.",
"_____no_output_____"
]
],
[
[
"#bu.run_alignments(sample1_path=\"../data/samples/CAMK_samples.txt\", \n# sample2_path=\"../data/samples/CMGC_samples.txt\", \n# output_path=\"../data/OpenCADD_results/<NAME_OF_FILE>\")",
"_____no_output_____"
]
],
[
[
"# Create a Dataframe containing the Alignments of all five Methods",
"_____no_output_____"
],
[
"The alignments for PyMol and ChimeraX MatchMaker are done in the respectively programs and are saved in seperate files. For the analysis, the DataFrames are combined.",
"_____no_output_____"
]
],
[
[
"columns = [\"reference_id\", \"mobile_id\", \"method\", \"rmsd\", \n \"coverage\", \"reference_size\", \"mobile_size\", \"time\", \n \"SI\", \"MI\", \"SAS\", \"ref_name\", \"ref_group\", \"ref_species\", \n \"ref_chain\", \"mob_name\", \"mob_group\", \"mob_species\", \"mob_chain\"]\nsuperposer_CAMK_CMGC = pd.read_csv(\"../data/OpenCADD_results/superposer_benchmark_CAMK_CMGC.csv\", names=columns)\npymol_CAMK_CMGC = pd.read_csv(\"../data/PyMol_results/pymol_benchmark_CAMK_CMGC.csv\", names=columns)\nchimerax_CAMK_CMGC = pd.read_csv(\"../data/ChimeraX_results/mmaker_benchmark_CAMK_CMGC.csv\", names=columns)\nall_CAMK_CMGC = pd.concat([superposer_CAMK_CMGC, pymol_CAMK_CMGC, chimerax_CAMK_CMGC]).reset_index(drop=True)",
"_____no_output_____"
]
],
[
[
"### Compute the relative Coverage",
"_____no_output_____"
],
[
"The relative coverage is computed the following way:\n\ncoverage / min(lenght of structure 1, lenght of structure 2)",
"_____no_output_____"
]
],
[
[
"au.compute_rel_cov(all_CAMK_CMGC)",
"_____no_output_____"
]
],
[
[
"# Analysis",
"_____no_output_____"
],
[
"## General Checks",
"_____no_output_____"
]
],
[
[
"counts, nans, times = au.general_checks(all_CAMK_CMGC)",
"_____no_output_____"
]
],
[
[
"Check if every value is present.\nIt should be 2500 for every value, because there are 2500 alignments performed per method.",
"_____no_output_____"
]
],
[
[
"counts",
"_____no_output_____"
]
],
[
[
"Next, we check for missing alignments. Some Methods have problems with some structures. \n\nIn this case, 1 alignment is missing for Theseus and 50 for MMLigner. All missing alignments for MMLigner are with the structure 4fv3 of ERK2.\nThe entries with missing alignments are removed for further analysis.",
"_____no_output_____"
]
],
[
[
"nans",
"_____no_output_____"
],
[
"all_CAMK_CMGC[all_CAMK_CMGC[\"rmsd\"].isna()]",
"_____no_output_____"
],
[
"all_CAMK_CMGC = all_CAMK_CMGC.dropna()",
"_____no_output_____"
]
],
[
[
"During the computation of the alignments, the time is measured. For all OpenCADD methods combined the CPU-time is about 11.5 hours. The time for downloading the structures is not included.\nPyMol align took less than a minute.",
"_____no_output_____"
]
],
[
[
"times",
"_____no_output_____"
]
],
[
[
"### Compute Mean and Median",
"_____no_output_____"
]
],
[
[
"mean, median = au.compute_mean_median(all_CAMK_CMGC)",
"_____no_output_____"
],
[
"mean",
"_____no_output_____"
],
[
"median",
"_____no_output_____"
]
],
[
[
"## Create basic plots",
"_____no_output_____"
],
[
"It is easy to see in both plots, that MMLigner performs the best. Besides that, Theseus and MDA perform very similar to ChimeraX MatchMaker.",
"_____no_output_____"
]
],
[
[
"au.create_scatter_plot(all_CAMK_CMGC)\nau.create_violine_plot(all_CAMK_CMGC)",
"_____no_output_____"
]
],
[
[
"## Check if data is normally distributed",
"_____no_output_____"
],
[
"The Kolmogorov-Smirnow-Test shows, that the values for RMSD, SI, MI, SAS and relative coverage are not normally distributed. The superposition methods have similar distributions for the measures except the relative coverage. MMLigner performs the best for all measures except the relative coverage.",
"_____no_output_____"
]
],
[
[
"dist_tests = au.check_distribution(all_CAMK_CMGC)",
"Results of kstest:\nKstestResult(statistic=0.9266956681911328, pvalue=0.0)\nKstestResult(statistic=0.9636117305326499, pvalue=0.0)\nKstestResult(statistic=0.7190065379254178, pvalue=0.0)\nKstestResult(statistic=0.7509352155373495, pvalue=0.0)\n"
]
],
[
[
"## Compute Correlation",
"_____no_output_____"
],
[
"Since the data is not distributed normally, the spearman correlation is used.\n\nThe three quality measures correlate very well with each other and with the rmsd. The quality measures also slightly positively correlate with the relative coverage, which means, the higher the relative coverage, the higher the quality measures. \n\nThe time negatively correlates with the quality measures, which means taking more time for an alignment produces better results. This correlation in this case is highly biased by MMLigner. It takes much more time than the other methods, but also yield overall the best results.\n\nAll three quality measures share the property, that lower values mean better alignments.",
"_____no_output_____"
]
],
[
[
"corr = au.compute_correlation(all_CAMK_CMGC, coeff=\"spearman\")\ncorr",
"_____no_output_____"
]
],
[
[
"## Check for significant differences",
"_____no_output_____"
],
[
"Because the data is not normally distributed, an ANOVA is not suitable. Therefore the Kruskal-Wallis-Test is performed. The RMSD and the three quality measures are significantly different for the groups.",
"_____no_output_____"
]
],
[
[
"kruskal = au.compute_kruskal(all_CAMK_CMGC)",
"Kruskal Wallis results for RMSD:\nKruskalResult(statistic=6202.255651831313, pvalue=0.0)\n\n\nKruskal Wallis results for Similarity Index (SI):\nKruskalResult(statistic=6246.492925252139, pvalue=0.0)\n\n\nKruskal Wallis results for Match Index (MI):\nKruskalResult(statistic=6268.9107829109325, pvalue=0.0)\n\n\nKruskal Wallis results for Structural Alignment Score (SAS):\nKruskalResult(statistic=6270.584365639992, pvalue=0.0)\n"
]
],
[
[
"## Which groups are different",
"_____no_output_____"
],
[
"The statistics show, that all groups are significantly different from each other, except PyMol and MDA. Looking at the diagrams above it is still noticable, that PyMol, ChimeraX, MDA and Theseus are in the same area.",
"_____no_output_____"
]
],
[
[
"significant, non_significant = au.compute_mannwhitneyu(all_CAMK_CMGC)",
"All significant results:\nResult for rmsd with theseus and pymol:\nMannwhitneyuResult(statistic=3514681.5, pvalue=1.8279692351988266e-14)\nResult for rmsd with theseus and mmaker:\nMannwhitneyuResult(statistic=4169232.0, pvalue=2.575930622208636e-93)\nResult for rmsd with theseus and mmligner:\nMannwhitneyuResult(statistic=6122550.0, pvalue=0.0)\nResult for rmsd with theseus and mda:\nMannwhitneyuResult(statistic=3510025.0, pvalue=3.705966767938766e-14)\nResult for rmsd with pymol and mmaker:\nMannwhitneyuResult(statistic=3779578.5, pvalue=1.1760551008640993e-37)\nResult for rmsd with pymol and mmligner:\nMannwhitneyuResult(statistic=6125000.0, pvalue=0.0)\nResult for rmsd with mmaker and mmligner:\nMannwhitneyuResult(statistic=6125000.0, pvalue=0.0)\nResult for rmsd with mmaker and mda:\nMannwhitneyuResult(statistic=2354725.0, pvalue=1.80812422997709e-51)\nResult for rmsd with mmligner and mda:\nMannwhitneyuResult(statistic=0.0, pvalue=0.0)\nResult for SI with theseus and pymol:\nMannwhitneyuResult(statistic=3514681.5, pvalue=1.8279692351988266e-14)\nResult for SI with theseus and mmaker:\nMannwhitneyuResult(statistic=4169232.0, pvalue=2.575930622208636e-93)\nResult for SI with theseus and mmligner:\nMannwhitneyuResult(statistic=6122550.0, pvalue=0.0)\nResult for SI with theseus and mda:\nMannwhitneyuResult(statistic=3510025.0, pvalue=3.705966767938766e-14)\nResult for SI with pymol and mmaker:\nMannwhitneyuResult(statistic=3779578.5, pvalue=1.1760551008640993e-37)\nResult for SI with pymol and mmligner:\nMannwhitneyuResult(statistic=6125000.0, pvalue=0.0)\nResult for SI with mmaker and mmligner:\nMannwhitneyuResult(statistic=6125000.0, pvalue=0.0)\nResult for SI with mmaker and mda:\nMannwhitneyuResult(statistic=2354725.0, pvalue=1.80812422997709e-51)\nResult for SI with mmligner and mda:\nMannwhitneyuResult(statistic=0.0, pvalue=0.0)\nResult for MI with theseus and pymol:\nMannwhitneyuResult(statistic=3514681.5, pvalue=1.8279692351988266e-14)\nResult for MI with theseus and mmaker:\nMannwhitneyuResult(statistic=4169232.0, pvalue=2.575930622208636e-93)\nResult for MI with theseus and mmligner:\nMannwhitneyuResult(statistic=6122550.0, pvalue=0.0)\nResult for MI with theseus and mda:\nMannwhitneyuResult(statistic=3510025.0, pvalue=3.705966767938766e-14)\nResult for MI with pymol and mmaker:\nMannwhitneyuResult(statistic=3779578.5, pvalue=1.1760551008640993e-37)\nResult for MI with pymol and mmligner:\nMannwhitneyuResult(statistic=6125000.0, pvalue=0.0)\nResult for MI with mmaker and mmligner:\nMannwhitneyuResult(statistic=6125000.0, pvalue=0.0)\nResult for MI with mmaker and mda:\nMannwhitneyuResult(statistic=2354725.0, pvalue=1.80812422997709e-51)\nResult for MI with mmligner and mda:\nMannwhitneyuResult(statistic=0.0, pvalue=0.0)\nResult for SAS with theseus and pymol:\nMannwhitneyuResult(statistic=3514681.5, pvalue=1.8279692351988266e-14)\nResult for SAS with theseus and mmaker:\nMannwhitneyuResult(statistic=4169232.0, pvalue=2.575930622208636e-93)\nResult for SAS with theseus and mmligner:\nMannwhitneyuResult(statistic=6122550.0, pvalue=0.0)\nResult for SAS with theseus and mda:\nMannwhitneyuResult(statistic=3510025.0, pvalue=3.705966767938766e-14)\nResult for SAS with pymol and mmaker:\nMannwhitneyuResult(statistic=3779578.5, pvalue=1.1760551008640993e-37)\nResult for SAS with pymol and mmligner:\nMannwhitneyuResult(statistic=6125000.0, pvalue=0.0)\nResult for SAS with mmaker and mmligner:\nMannwhitneyuResult(statistic=6125000.0, pvalue=0.0)\nResult for SAS with mmaker and mda:\nMannwhitneyuResult(statistic=2354725.0, pvalue=1.80812422997709e-51)\nResult for SAS with mmligner and mda:\nMannwhitneyuResult(statistic=0.0, pvalue=0.0)\n\n***********************************\n\nAll non significant results:\nResult for rmsd with pymol and mda:\nMannwhitneyuResult(statistic=3070594.0, pvalue=0.286414999728139)\nResult for SI with pymol and mda:\nMannwhitneyuResult(statistic=3070594.0, pvalue=0.286414999728139)\nResult for MI with pymol and mda:\nMannwhitneyuResult(statistic=3070594.0, pvalue=0.286414999728139)\nResult for SAS with pymol and mda:\nMannwhitneyuResult(statistic=3070594.0, pvalue=0.286414999728139)\n"
]
],
[
[
"# Count the best alignments",
"_____no_output_____"
],
[
"For every pair of structures, the method that has the best quality measure is selected. The following statistics show how often a method had the best results for the quality measures.",
"_____no_output_____"
]
],
[
[
"best_results = au.count_best_results(all_CAMK_CMGC)",
"Counts of best values for the Similarity Index (SI):\nmmligner 2450\nmda 22\nmatchmaker 16\npymol 11\ntheseus 1\nName: method, dtype: int64\n\n\nCounts of best values for the Match Index (MI):\nmmligner 2450\nmda 22\nmatchmaker 16\npymol 11\ntheseus 1\nName: method, dtype: int64\n\n\nCounts of best values for the Structural Alignment Score (SAS):\nmmligner 2450\nmda 22\nmatchmaker 16\npymol 11\ntheseus 1\nName: method, dtype: int64\n\n\nCounts of best values for the Similarity Index (SI) without MMLigner:\nmatchmaker 1344\nmda 708\npymol 446\ntheseus 2\nName: method, dtype: int64\n\n\nCounts of best values for the Match Index (MI) without MMLigner:\nmatchmaker 1363\nmda 739\npymol 396\ntheseus 2\nName: method, dtype: int64\n\n\nCounts of best values for the Structural Alignment Score (SAS) without MMLigner:\nmatchmaker 1344\nmda 708\npymol 446\ntheseus 2\nName: method, dtype: int64\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
]
] |
4affa951b217bfade6716deb2f0b08ce87e23502
| 105,125 |
ipynb
|
Jupyter Notebook
|
EDA_CaseStudy/EDA Credit Case Study.ipynb
|
nirbaaytandon/Data-Science
|
39fa79fdaf8b59202c7a2cc8e1ddb31269795e35
|
[
"MIT"
] | 1 |
2021-12-09T00:05:07.000Z
|
2021-12-09T00:05:07.000Z
|
EDA_CaseStudy/EDA Credit Case Study.ipynb
|
nirbaaytandon/Data-Science
|
39fa79fdaf8b59202c7a2cc8e1ddb31269795e35
|
[
"MIT"
] | null | null | null |
EDA_CaseStudy/EDA Credit Case Study.ipynb
|
nirbaaytandon/Data-Science
|
39fa79fdaf8b59202c7a2cc8e1ddb31269795e35
|
[
"MIT"
] | 1 |
2021-08-19T11:36:47.000Z
|
2021-08-19T11:36:47.000Z
| 36.275017 | 1,510 | 0.638364 |
[
[
[
"# Exploratory Data Analysis Case Study - \n##### Conducted by Nirbhay Tandon & Naveen Sharma",
"_____no_output_____"
],
[
"## 1.Import libraries and set required parameters",
"_____no_output_____"
]
],
[
[
"#import all the libraries and modules\nimport numpy as np\nimport pandas as pd\nimport seaborn as sns\nimport matplotlib.pyplot as plt\nimport re\nfrom scipy import stats \n\n# Supress Warnings\n#Enable autocomplete in Jupyter Notebook.\n%config IPCompleter.greedy=True\n\nimport warnings\nwarnings.filterwarnings('ignore')\nimport os",
"_____no_output_____"
],
[
"## Set the max display columns to None so that pandas doesn't sandwich the output \npd.set_option('display.max_columns', None)\npd.set_option('display.max_rows', 40)",
"_____no_output_____"
]
],
[
[
"### Reading and analysing Data",
"_____no_output_____"
]
],
[
[
"applicationData=pd.read_csv(\"./application_data.csv\")\n\napplicationData.head()",
"_____no_output_____"
]
],
[
[
"## 2. Data Inspection",
"_____no_output_____"
]
],
[
[
"#shape of application_data.csv data\n\napplicationData.shape",
"_____no_output_____"
],
[
"#take information about the data\n\napplicationData.info()",
"_____no_output_____"
],
[
"#get the information about the numerical data\n\napplicationData.describe()",
"_____no_output_____"
],
[
"## print the column names for application_data.csv\napplicationData.columns",
"_____no_output_____"
],
[
"## print the various datatypes of application_data.csv\napplicationData.dtypes",
"_____no_output_____"
]
],
[
[
"## 3. Data Cleaning & Quality Check",
"_____no_output_____"
],
[
"In this section we will perform various checks and balances on the application_data.csv file. \n\nWe will:\n* Perform a check for the number of missing/null values on each column\n* Perform a check for the percentage of missing/null values of each column\n * Drop the columns that have a high percentage of null values, i.e. over 60%\n * Print the names of the dropped columns\n * Verify that the columns were dropped by comparing the shape of the new dataframe created\n* For columns with around 13% of null values we will discuss the best way to handle the missing/null values in the columns\n * Check the data types of these columns and determine if they are categorical in nature or not\n* Check the data types for all the columns in the dataframe and convert them to numerical data types if required\n* Check for any outliers in any 3 numerical columns and treat them accordingly\n* Create a bin for continous variables and analyse them\n",
"_____no_output_____"
]
],
[
[
"### Let us create a utility function to generate a list of null values in different dataframes\n### We will utilize this function extensively througout the notebook. \ndef generateNullValuesPercentageTable(dataframe):\n totalNullValues = dataframe.isnull().sum().sort_values(ascending=False)\n percentageOfNullValues = round((dataframe.isnull().sum()*100/len(dataframe)).sort_values(ascending=False),2)\n columnNamesWithPrcntgOfNullValues = pd.concat([totalNullValues, percentageOfNullValues], axis=1, keys=['Total Null Values', 'Percentage of Null Values'])\n return columnNamesWithPrcntgOfNullValues",
"_____no_output_____"
],
[
"## Check the number of null values of each column and display them in \n## decending order along with the percentage of null values there is\n\ngenerateNullValuesPercentageTable(applicationData)",
"_____no_output_____"
],
[
"### Assess the shape of the dataframe before dropping \n### columns with a high percentage of \n### null values\nprint(\"The Initial shape of the DataFrame is: \", applicationData.shape)",
"_____no_output_____"
],
[
"#Drop all the columns where the \n## percentage of missing values is above 60% in application_data.csv\ndroppedColumns = applicationData.columns[applicationData.isnull().mean() > 0.60]\napplicationDataAfterDroppedColumns = applicationData.drop(droppedColumns, axis = 1)\n\nprint(\"The new shape of the DataFrame is: \", applicationDataAfterDroppedColumns.shape)",
"_____no_output_____"
],
[
"## analysing the dataframe is correct after dropping columns\napplicationDataAfterDroppedColumns.head()",
"_____no_output_____"
]
],
[
[
"### Observation: \n\nAs you can see, the shape of the data has changed from (307511, 122) to (307511, 105). Which mean we have dropped 17 columns that had over 60% percent null values. The dropped columns are mentioned below.",
"_____no_output_____"
]
],
[
[
"print(\"The columns that have been dropped are: \", droppedColumns)",
"_____no_output_____"
],
[
"## print the percentage of columns with null values in the \n## new data frame after the columns have been dropped\n\ngenerateNullValuesPercentageTable(applicationDataAfterDroppedColumns)\n",
"_____no_output_____"
],
[
"#### Check dataframe shape to confirm no other columns were dropped \napplicationDataAfterDroppedColumns.shape",
"_____no_output_____"
]
],
[
[
"### Observation: \n\nAs you can see above, there are still a few columns that have a above 30% of null/missing values. We can deal with those null/missing values using various methods of imputation. \n\n##### Some key points:\n- The columns with above 60% of null values have successfully been dropped\n- The column with the highest percentage of null values after the drop is \"LANDAREA_MEDI\" with 59.38% null values. Whereas earlier it was \"COMMONAREA_MEDI\" with 69.87% null values\n\n- The new shape of the dataframe is (307511, 105)",
"_____no_output_____"
],
[
"Checking the datadrame after dropping null values",
"_____no_output_____"
]
],
[
[
"applicationDataAfterDroppedColumns.head()",
"_____no_output_____"
],
[
"### Analyzing Columns with null values around 14% to determine\n### what might be the best way to impute such values\nlistOfColumnsWithLessValuesOfNull = applicationDataAfterDroppedColumns.columns[applicationDataAfterDroppedColumns.isnull().mean() < 0.14]\n\napplicationDataWithLessPrcntgOfNulls = applicationDataAfterDroppedColumns.loc[:, listOfColumnsWithLessValuesOfNull]\nprint(applicationDataWithLessPrcntgOfNulls.shape)",
"_____no_output_____"
],
[
"applicationDataWithLessPrcntgOfNulls.head(20)",
"_____no_output_____"
],
[
"### Analysing columns with around 13.5% null values\ncolumnsToDescribe = ['AMT_REQ_CREDIT_BUREAU_QRT', 'AMT_REQ_CREDIT_BUREAU_YEAR', 'AMT_REQ_CREDIT_BUREAU_MON', 'AMT_REQ_CREDIT_BUREAU_DAY', 'AMT_REQ_CREDIT_BUREAU_HOUR','AMT_REQ_CREDIT_BUREAU_WEEK', 'OBS_30_CNT_SOCIAL_CIRCLE', 'OBS_60_CNT_SOCIAL_CIRCLE', 'EXT_SOURCE_2']\napplicationDataAfterDroppedColumns[columnsToDescribe].describe()",
"_____no_output_____"
],
[
"### Let us plot a boxplot to see the various variables\nfig, axes = plt.subplots(nrows=3, ncols = 2, figsize=(40,25))\n\nsns.boxplot(data=applicationDataAfterDroppedColumns.AMT_REQ_CREDIT_BUREAU_YEAR, ax=axes[0][0])\naxes[0][0].set_title('AMT_REQ_CREDIT_BUREAU_YEAR')\n\nsns.boxplot(data=applicationDataAfterDroppedColumns.AMT_REQ_CREDIT_BUREAU_MON, ax=axes[0][1])\naxes[0][1].set_title('AMT_REQ_CREDIT_BUREAU_MON')\n\nsns.boxplot(data=applicationDataAfterDroppedColumns.AMT_REQ_CREDIT_BUREAU_DAY, ax=axes[1][0])\naxes[1][0].set_title('AMT_REQ_CREDIT_BUREAU_DAY')\n\nsns.boxplot(applicationDataAfterDroppedColumns.AMT_REQ_CREDIT_BUREAU_HOUR, ax=axes[1][1])\naxes[1][1].set_title('AMT_REQ_CREDIT_BUREAU_HOUR')\n\nsns.boxplot(applicationDataAfterDroppedColumns.AMT_REQ_CREDIT_BUREAU_WEEK, ax=axes[2][0])\naxes[2][0].set_title('AMT_REQ_CREDIT_BUREAU_WEEK')\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Observation\nAs you can see above, when we take a look at the columns that have a low number of null values, the shape of the data changes to (307511, 71) compared to (307511, 105). We lose 34 columns in the process. \n\n\nChecking columns having less no. of Null values(around 13% or so) and analysing the best metric \nto impute the missing/null values in those columns basis if the column/variable is 'Categorical' or 'Continuous''\n - AMT_REQ_CREDIT_BUREAU_HOUR (99.4% of the values are 0.0 with 4.0 and 3.0 values being outliers. Its safe to impute the missing values with 0.0) \n - AMT_REQ_CREDIT_BUREAU_DAY (99.4% of the values are 0.0 with 9.0 and 8.0 values being outliers. Its safe to impute the missing values with 0.0) \n - AMT_REQ_CREDIT_BUREAU_WEEK (96.8% of the values are 0.0 with 8.0 and 7.0 values being outliers. Its safe to impute the missing values with 0.0) \n - AMT_REQ_CREDIT_BUREAU_MON (83.6% of the values are 0.0. Its safe to impute the missing values with mode : 0.0)\n - AMT_REQ_CREDIT_BUREAU_YEAR (It seems fine to use the median value 1.0 here for imputing the missing values) ",
"_____no_output_____"
]
],
[
[
"### Checking for categorical data\ncategoricalDataColumns = applicationDataAfterDroppedColumns.nunique().sort_values()\ncategoricalDataColumns",
"_____no_output_____"
]
],
[
[
"### Observation:\nGiven the wide number of columns with a less number of unique values, we will convert all columns with upto 5 values into categorical columns",
"_____no_output_____"
]
],
[
[
"listOfColumnsWithMaxTenUniqueValues = [i for i in applicationDataAfterDroppedColumns.columns if applicationDataAfterDroppedColumns[i].nunique() <= 5]\nfor col in listOfColumnsWithMaxTenUniqueValues:\n applicationDataAfterDroppedColumns[col] = applicationDataAfterDroppedColumns[col].astype('category')\n \napplicationDataAfterDroppedColumns.shape",
"_____no_output_____"
],
[
"applicationDataAfterDroppedColumns.head()",
"_____no_output_____"
],
[
"## Check for datatypes of all columns in the new dataframe\napplicationDataAfterDroppedColumns.info()",
"_____no_output_____"
]
],
[
[
"### Observation:\n\nWe notice above that after dropping the null columns we still have:\n\n- 43 Categorical\n- 48 Float\n- 6 Integer \n- 8 Object data types",
"_____no_output_____"
]
],
[
[
"## Convert the categorical data columns into individual columns with numeric values for better analysis\n## we will do this using one-hot-encoding method\nconvertedCategoricalColumnsDataframe = pd.get_dummies(applicationDataAfterDroppedColumns, columns=listOfColumnsWithMaxTenUniqueValues, prefix=listOfColumnsWithMaxTenUniqueValues)\nconvertedCategoricalColumnsDataframe.head()",
"_____no_output_____"
],
[
"## Converting these columns has changed the shape of the data to\nprint(\"Shape of Application Data after categorical column conversion: \", convertedCategoricalColumnsDataframe.shape)",
"_____no_output_____"
]
],
[
[
"### Observation\nAs you can see above we have successfully converted the varius categorical datatypes into their own columns.\n\nThe new shape of the data is (307511, 158) compared to (307511, 105). We have introuced 53 new columns. These will help us identify the best possible method to use for imputing values.",
"_____no_output_____"
]
],
[
[
"### Count the number of missing values in the new dataframe\ngenerateNullValuesPercentageTable(convertedCategoricalColumnsDataframe)",
"_____no_output_____"
]
],
[
[
"### Observation\nLet us take the following columns - AMT_REQ_CREDIT_BUREAU_YEAR, AMT_REQ_CREDIT_BUREAU_MON, OBS_30_CNT_SOCIAL_CIRCLE, OBS_60_CNT_SOCIAL_CIRCLE, EXT_SOURCE_2.\nDetermine their datatypes and using the describe above try and identify what values can be used to impute into the null columns. \n",
"_____no_output_____"
]
],
[
[
"listOfCols = ['AMT_REQ_CREDIT_BUREAU_YEAR', 'AMT_REQ_CREDIT_BUREAU_MON', 'OBS_30_CNT_SOCIAL_CIRCLE', 'OBS_60_CNT_SOCIAL_CIRCLE', 'EXT_SOURCE_2']\nconvertedCategoricalColumnsDataframe[listOfCols].dtypes",
"_____no_output_____"
],
[
"applicationDataAfterDroppedColumns['AMT_REQ_CREDIT_BUREAU_HOUR'].fillna(0.0, inplace = True)\napplicationDataAfterDroppedColumns['AMT_REQ_CREDIT_BUREAU_HOUR'] = applicationDataAfterDroppedColumns['AMT_REQ_CREDIT_BUREAU_HOUR'].astype(int)",
"_____no_output_____"
],
[
"## convert DAYS_BIRTH to years\ndef func_age_yrs(x):\n return round(abs(x/365),0)\n\napplicationDataAfterDroppedColumns['DAYS_BIRTH'] = applicationDataAfterDroppedColumns['DAYS_BIRTH'].apply(func_age_yrs)",
"_____no_output_____"
]
],
[
[
"### Observation\nIn all the selected columns we can see that we can use the median to impute the values in the dataframe. They all correspond to 0.00 except EXT_SOURCE_2. For EXT_SOURCE_2 we observe that the mean and the median values are roughly similar at 5.143927e-01 for mean & 5.659614e-01 for median. So we could use either of those values to impute.",
"_____no_output_____"
],
[
"Let us now check for outliers on 6 numerical columns.\nFor this we can use our dataset from after we dropped the columns with over 60% null values. \n",
"_____no_output_____"
]
],
[
[
"### We will use boxplots to handle the outliers on AMT_CREDIT, AMT_ANNUITY, AMT_GOODS_PRICE\nfig, axes = plt.subplots(nrows=3, ncols = 2, figsize=(50,50))\n\nsns.boxplot(data= applicationDataAfterDroppedColumns.AMT_CREDIT.dropna(), ax=axes[0][0])\naxes[0][0].set_title('AMT_CREDIT')\n\nsns.boxplot(data= applicationDataAfterDroppedColumns.AMT_ANNUITY.dropna(), ax=axes[0][1])\naxes[0][1].set_title('AMT_ANNUITY')\n\nsns.boxplot(data= applicationDataAfterDroppedColumns.AMT_GOODS_PRICE.dropna(), ax=axes[1][0])\naxes[1][0].set_title('AMT_GOODS_PRICE')\n\nsns.boxplot(data= applicationDataAfterDroppedColumns.AMT_INCOME_TOTAL.dropna(), ax=axes[1][1])\naxes[1][1].set_title('AMT_INCOME_TOTAL')\n\nsns.boxplot(data= applicationDataAfterDroppedColumns.DAYS_BIRTH.dropna(), ax=axes[2][0])\naxes[2][0].set_title('DAYS_BIRTH')\n\nsns.boxplot(data= applicationDataAfterDroppedColumns.DAYS_EMPLOYED.dropna(), ax=axes[2][1])\naxes[2][1].set_title('DAYS_EMPLOYED')\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Observation\nWe can easily see in the box plot that there are so many outliers which has to removed for the better calculation. So, In the next part of the code we remove outliers from the function \"remove_outliers\" which accept dataframe and columns name (In which we want to remove outliers) as argument and return the outliers removed dataframe.\n\nAnalysing outliers in Numeric variables and Handling/Treating them with appropriate methods.\n\n- AMT_REQ_CREDIT_BUREAU_HOUR (99.4% of the values are 0.0 with value '4' and '3' being outliers. Should be retained) \n\nConsidering that its the number of enquiries made by the company to credit bureau, this could significantly mean that the company was extremely cautious in making a decision of whether to grant loan/credit to this particular client or not. This might imply that it could be a case of 'High Risk' client and can influence the Target variable. Its better to retain these outlier values\n\n- AMT_INCOME_TOTAL ( Clearly 117000000.0 is an outlier here.)\n\nThe above oulier can be dropped in order to not skew with the analysis. We can use IQR to remove this value. \n\n- DAYS_BIRTH ( There is no outlier in this column) \n\n- DAYS_EMPLOYED ( Clearly 1001 is an outlier here and should be deleted.18% of the column values are 1001)\n\nClearly 1001 is an outlier here. 18% of the column values are 1001. Since , this represents the no. of years of employement as on the application date, these should be deleted. Though values above 40 years till 49 years of employment seems questionable as well but lets not drop it for now considering exception cases.\nAnother way to see the distribution of is using a distribution plot.\n",
"_____no_output_____"
]
],
[
[
"fig, axes = plt.subplots(nrows=3, ncols = 2, figsize=(50,50))\n\nsns.distplot(applicationDataAfterDroppedColumns.AMT_CREDIT.dropna(), ax=axes[0][0])\naxes[0][0].set_title('AMT_CREDIT')\n\nsns.distplot(applicationDataAfterDroppedColumns.AMT_ANNUITY.dropna(), ax=axes[0][1])\naxes[0][1].set_title('AMT_ANNUITY')\n\nsns.distplot(applicationDataAfterDroppedColumns.AMT_GOODS_PRICE.dropna(), ax=axes[1][0])\naxes[1][0].set_title('AMT_GOODS_PRICE')\n\nsns.distplot(applicationDataAfterDroppedColumns.AMT_INCOME_TOTAL.dropna(), ax=axes[1][1])\naxes[1][1].set_title('AMT_INCOME_TOTAL')\n\nsns.distplot(applicationDataAfterDroppedColumns.DAYS_BIRTH.dropna(), ax=axes[2][0])\naxes[2][0].set_title('DAYS_BIRTH')\n\nsns.distplot(applicationDataAfterDroppedColumns.DAYS_EMPLOYED.dropna(), ax=axes[2][1])\naxes[2][1].set_title('DAYS_EMPLOYED')\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Observation\nAs you can see from the distplots above there are a few outliers that aren't properly normalized.\nThe 'DAYS_EMPLOYED' column is heavily skewed in the -ve side of the plot. ",
"_____no_output_____"
]
],
[
[
"#Function for removing outliers\n\ndef remove_outlier(df, col_name):\n q1 = df[col_name].quantile(0.25)\n q3 = df[col_name].quantile(0.75)\n iqr = q3-q1 #Interquartile range\n l = q1-1.5*iqr\n h = q3+1.5*iqr\n dfOutput = df.loc[(df[col_name] > l) & (df[col_name] < h)]\n return dfOutput",
"_____no_output_____"
],
[
"cols=['AMT_CREDIT','AMT_ANNUITY', 'AMT_GOODS_PRICE', 'AMT_INCOME_TOTAL', 'DAYS_EMPLOYED']\n\nfor i in cols:\n applicationDataAfterDroppedColumns=remove_outlier(applicationDataAfterDroppedColumns,i)\n \napplicationDataAfterDroppedColumns.head()",
"_____no_output_____"
],
[
"### Plot the box plot again after removing outliers\nfig, axes = plt.subplots(nrows=3, ncols = 2, figsize=(50,50))\n\nsns.boxplot(data= applicationDataAfterDroppedColumns.AMT_CREDIT.dropna(), ax=axes[0][0])\naxes[0][0].set_title('AMT_CREDIT')\n\nsns.boxplot(data= applicationDataAfterDroppedColumns.AMT_ANNUITY.dropna(), ax=axes[0][1])\naxes[0][1].set_title('AMT_ANNUITY')\n\nsns.boxplot(data= applicationDataAfterDroppedColumns.AMT_GOODS_PRICE.dropna(), ax=axes[1][0])\naxes[1][0].set_title('AMT_GOODS_PRICE')\n\nsns.boxplot(data= applicationDataAfterDroppedColumns.AMT_INCOME_TOTAL.dropna(), ax=axes[1][1])\naxes[1][1].set_title('AMT_INCOME_TOTAL')\n\nsns.boxplot(data= applicationDataAfterDroppedColumns.DAYS_BIRTH.dropna(), ax=axes[2][0])\naxes[2][0].set_title('DAYS_BIRTH')\n\nsns.boxplot(data= applicationDataAfterDroppedColumns.DAYS_EMPLOYED.dropna(), ax=axes[2][1])\naxes[2][1].set_title('DAYS_EMPLOYED')\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Observation\nAfter dropping the outliers we observe that there very few points mentioned on the box plots above for the outliers.",
"_____no_output_____"
]
],
[
[
"### Plotting the distribution plot after removing the outliers\nfig, axes = plt.subplots(nrows=3, ncols = 2, figsize=(50,50))\n\nsns.distplot(applicationDataAfterDroppedColumns.AMT_CREDIT.dropna(), ax=axes[0][0])\naxes[0][0].set_title('AMT_CREDIT')\n\nsns.distplot(applicationDataAfterDroppedColumns.AMT_ANNUITY.dropna(), ax=axes[0][1])\naxes[0][1].set_title('AMT_ANNUITY')\n\nsns.distplot(applicationDataAfterDroppedColumns.AMT_GOODS_PRICE.dropna(), ax=axes[1][0])\naxes[1][0].set_title('AMT_GOODS_PRICE')\n\nsns.distplot(applicationDataAfterDroppedColumns.AMT_INCOME_TOTAL.dropna(), ax=axes[1][1])\naxes[1][1].set_title('AMT_INCOME_TOTAL')\n\nsns.distplot(applicationDataAfterDroppedColumns.DAYS_BIRTH.dropna(), ax=axes[2][0])\naxes[2][0].set_title('DAYS_BIRTH')\n\nsns.distplot(applicationDataAfterDroppedColumns.DAYS_EMPLOYED.dropna(), ax=axes[2][1])\naxes[2][1].set_title('DAYS_EMPLOYED')\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Observation\nBased on the distplots above you can see that there is a marked difference between the minimum values for various columns, particularly the DAYS_EMPLOYED column where the minimum value increased from -7500 to -6000. This proves that the treatment of outliers was succesful",
"_____no_output_____"
]
],
[
[
"applicationDataAfterDroppedColumns.shape",
"_____no_output_____"
]
],
[
[
"### Observation\nWe observe that after removing the outliers the boxplots show a slight shift in the maximum ranges. \nThe distribution plot gives us a more significant display in changes. There is a significant reduction in the max ranges on the x-axis for all the three variables we chose. \n\n\nAs we can see above, after treating the outliers for various columns the shape of our dataset has changed significantly. The shape of the dataframe after dropping columns with high number of null values was (307511, 105) & after treating for outliers is (209624, 105).\n\nLet us now create bins for 3 different continous variables and plot them. We will use AMT_INCOME_TOTAL, AMT_CREDIT & DAYS_BIRTH to create our bins.",
"_____no_output_____"
]
],
[
[
"## Creating bins for Income range based on AMT_INCOME_TOTAL\nbins=[0,100000,200000,300000,400000,500000,600000,20000000]\nrange_period=['0-100000','100000-200000','200000-300000','300000-400000','400000-500000','500000-600000','600000 and above']\napplicationDataAfterDroppedColumns['Income_amount_range']=pd.cut(applicationDataAfterDroppedColumns['AMT_INCOME_TOTAL'],bins,labels=range_period)\nplotIncomeAmountRange = applicationDataAfterDroppedColumns['Income_amount_range'].value_counts().plot(kind='bar', title='Income Range Bins Plot')\nplotIncomeAmountRange.set_xlabel('Income Range Bins')\nplotIncomeAmountRange.set_ylabel('Count')",
"_____no_output_____"
]
],
[
[
"### Observation\nAs you can clearly see from the plot above:\n\n- The most number of people earn between 100000-200000\n- The number of people who earn between 200000-300000 is less than half of the number of people in 100000-200000 range\n- No one earns above 300000.",
"_____no_output_____"
]
],
[
[
"#create bins for credit anount\n\nbins=[0,50000,100000,150000,200000,250000,300000,400000]\nrange_period=['0-50000','50000-100000','100000-150000','150000-200000','200000-250000','250000-300000','300000-400000']\napplicationDataAfterDroppedColumns['credit_amount_range']=pd.cut(applicationDataAfterDroppedColumns['AMT_CREDIT'],bins,labels=range_period)\nplotCreditAmountRange = applicationDataAfterDroppedColumns['credit_amount_range'].value_counts().plot(kind='bar', title='Credit Amount Range Plots')\nplotCreditAmountRange.set_xlabel('Credit Amount Range Bins')\nplotCreditAmountRange.set_ylabel('Count')",
"_____no_output_____"
]
],
[
[
"### Observation\nAs you can see from the plots above\n\n- Very less number of people borrow money between 0-50000\n- Highest number of people are borrowing money between 250000-300000",
"_____no_output_____"
]
],
[
[
"##Creating bins for age range for DAYS_BIRTH in years\n\nbins = [10, 20, 30, 40, 50, 60, 70, 80]\nlabels = ['10-20','21-30','31-40','41-50','51-60','61-70','71-80']\n\napplicationDataAfterDroppedColumns['BINNED_AGE'] = pd.cut(applicationDataAfterDroppedColumns['DAYS_BIRTH'], bins=bins,labels=labels)\nplotAgeRange = applicationDataAfterDroppedColumns['BINNED_AGE'].value_counts().plot(kind='bar', title='Age Range Plot')\nplotAgeRange.set_xlabel('Age Range')\nplotAgeRange.set_ylabel('Count')",
"_____no_output_____"
]
],
[
[
"### Observation\n- People between the ages of 71-80 & 10-20 are not borrowing any money.\n- For people in the age range of 10-20, no borrowing could suggest that children/teenagers/young adults could have just opened new bank accounts with their parents or have just joined university so do not have a need of borrowing money\n- People in between the ages of 31-40 have a significantly higher number of borrowers, this could be suggestive of various personal expenses & it would be beneficial for the firm to identify the reasons why they are borrowing more so that they can introduce newer products at more competitive interest rates to these customers",
"_____no_output_____"
],
[
"# 4. Data Analysis",
"_____no_output_____"
],
[
"In this section we will perform indepth analysis on the application_data.csv file.\nThis will be achieved by:\n\n- Checking the imbalance percentage in the dataset\n- Dividing the dataset based on the \"TARGET\" column into 2 separate dataframes\n- Performing univariate analysis for categorical variables on both Target = 0 & Target = 1 columns\n- Identifying the correlation between the numerical columns for both Target = 0 & Target = 1 columns\n- Comparing the results across continous variables\n- Performing bivariate analysis for numerical variables on both Target = 0 & Target = 1 columns",
"_____no_output_____"
],
[
"## Selecting relevant columns from 'applicationDataAfterDroppedColumns' which would be used for EDA further\n\n- Selecting only the relevant columns(25 or so) from 'applicationDataAfterDroppedColumns' i.e. removing those columns which aren't relevant for analysis out of a total of 105 columns",
"_____no_output_____"
]
],
[
[
"applicationDataWithRelevantColumns = applicationDataAfterDroppedColumns.loc[:,['SK_ID_CURR',\n'TARGET',\n'NAME_CONTRACT_TYPE',\n'CODE_GENDER',\n'FLAG_OWN_CAR',\n'FLAG_OWN_REALTY',\n'CNT_CHILDREN',\n'AMT_INCOME_TOTAL',\n'AMT_CREDIT',\n'AMT_ANNUITY',\n'AMT_GOODS_PRICE',\n'NAME_INCOME_TYPE',\n'NAME_EDUCATION_TYPE',\n'NAME_FAMILY_STATUS',\n'NAME_HOUSING_TYPE',\n'REGION_POPULATION_RELATIVE',\n'BINNED_AGE',\n'DAYS_EMPLOYED',\n'DAYS_REGISTRATION',\n'DAYS_ID_PUBLISH',\n'FLAG_CONT_MOBILE',\n'OCCUPATION_TYPE',\n'CNT_FAM_MEMBERS',\n'REGION_RATING_CLIENT',\n'REGION_RATING_CLIENT_W_CITY',\n'ORGANIZATION_TYPE',\n'AMT_REQ_CREDIT_BUREAU_HOUR',\n'AMT_REQ_CREDIT_BUREAU_DAY']]",
"_____no_output_____"
]
],
[
[
"We will now use applicationDataWithRelevantColumns as our dataframe to run further analysis",
"_____no_output_____"
]
],
[
[
"### Checking shape of the new dataframe\napplicationDataWithRelevantColumns.shape",
"_____no_output_____"
],
[
"applicationDataWithRelevantColumns['CODE_GENDER'].value_counts()",
"_____no_output_____"
]
],
[
[
"Since the number of Females is higher than Males, we can safely impute XNA values with F.",
"_____no_output_____"
]
],
[
[
"applicationDataWithRelevantColumns.loc[applicationDataWithRelevantColumns['CODE_GENDER']=='XNA','CODE_GENDER']='F'\napplicationDataWithRelevantColumns['CODE_GENDER'].value_counts()",
"_____no_output_____"
],
[
"#Check the total percentage of target value as 0 and 1.\nimbalancePercentage = applicationDataWithRelevantColumns['TARGET'].value_counts()*100/len(applicationDataAfterDroppedColumns)\nimbalancePercentage",
"_____no_output_____"
],
[
"imbalancePercentage.plot(kind='bar',rot=0)",
"_____no_output_____"
]
],
[
[
"### Observation\nWe can easily see that this data is very much imbalance. Rows with target value 0 is only 90.612239% and with 1 is only 9.387761%.\nThis also means that only 9.38% of all the loan applicants default while paying back their loans.",
"_____no_output_____"
]
],
[
[
"#Splitting the data based on target values\n\none_df = applicationDataWithRelevantColumns.loc[applicationDataWithRelevantColumns['TARGET']==1]\nzero_df = applicationDataWithRelevantColumns.loc[applicationDataWithRelevantColumns['TARGET']==0]",
"_____no_output_____"
],
[
"## Inspecting data with TARGET = 1\none_df.head()",
"_____no_output_____"
],
[
"one_df.info()",
"_____no_output_____"
],
[
"one_df.shape",
"_____no_output_____"
],
[
"## Inspecting data with TARGET = 0\nzero_df.head()",
"_____no_output_____"
],
[
"zero_df.describe",
"_____no_output_____"
],
[
"zero_df.shape",
"_____no_output_____"
],
[
"zero_df.info",
"_____no_output_____"
]
],
[
[
"We will now use the following columns to perform Univariate & Bivariate analysis\n- CODE_GENDER\n- NAME_CONTRACT_TYPE\n- NAME_INCOME_TYPE\n- NAME_EDUCATION_TYPE\n- NAME_FAMILY_STATUS\n- NAME_HOUSING_TYPE\n- OCCUPATION_TYPE\n- ORGANIZATION_TYPE",
"_____no_output_____"
],
[
"### Univariate Analysis:-",
"_____no_output_____"
],
[
"Univariate Analysis on one_df dataset",
"_____no_output_____"
]
],
[
[
"#Univariate Analysis for categorical variable 'CODE_GENDER' in dataframe one_df.\n\nsns.countplot(x ='CODE_GENDER', data = one_df)\n\n\nplt.title('Number of applications by Gender')\nplt.ylabel('Number of Applications')\nplt.xlabel('Gender')\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Observation\nAs you can see above the number of Female applicants is higher than the number of Male applicants.",
"_____no_output_____"
]
],
[
[
"#Univariate Analysis for categorical variable 'NAME_EDUCATION_TYPE' in dataframe T1.\n\nsns.countplot(x ='NAME_EDUCATION_TYPE', data = one_df)\n\n\nplt.title(\"Number of applications by Client's Education Level\")\nplt.ylabel('Number of Applications')\nplt.xlabel(\"Client's Education Level\")\nplt.xticks(rotation = 90)\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Observation\nFrom the plot above we can infer that:\n\n- The highest number of applications for credit were made by people having Secondary/ secondary special education and these people defaulted on being able to pay back their loans. This could mean that they face trouble in being able to manage their money effectively or have jobs that pay less/are contractual in nature\n- People with higher education also applied for a credit and defaulted on their loans",
"_____no_output_____"
]
],
[
[
"#Univariate Analysis for categorical variable 'NAME_CONTRACT_TYPE' in dataframe one_df.\n\nsns.countplot(x ='NAME_CONTRACT_TYPE', data = one_df)\n\nplt.title('Number of applications by Contract Type')\nplt.ylabel('Number of Applications')\nplt.xlabel('Contract Type')\nplt.show()\n",
"_____no_output_____"
]
],
[
[
"### Observation\n\n- A high number of applicants who defaulted applied for cash loans",
"_____no_output_____"
]
],
[
[
"#Univariate Analysis for categorical variable 'NAME_INCOME_TYPE' in dataframe one_df.\n\nsns.countplot(x ='NAME_INCOME_TYPE', data = one_df)\n\nplt.title(\"Number of applications by Client's Income Type\")\nplt.ylabel('Number of Applications')\nplt.xlabel(\"Client's Income Type\")\nplt.xticks(rotation = 90)\nplt.show()\n",
"_____no_output_____"
]
],
[
[
"### Observation\n\n- Mostly working professionals apply for credit and are also the ones that default on being able to payback the loans on time\n- State servants have a very low number of defaulters",
"_____no_output_____"
]
],
[
[
"#Univariate Analysis for categorical variable 'NAME_FAMILY_STATUS' in dataframe one_df.\n\nsns.countplot(x ='NAME_FAMILY_STATUS', data = one_df)\n\n\nplt.title(\"Number of applications by Client's Family Status\")\nplt.ylabel('Number of Applications')\nplt.xlabel(\"Client's Family Status\")\nplt.xticks(rotation = 90)\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Observation\n\n- Married applicants make a higher number of applications as compared to other categories\n- It would be beneficial for the bank to introduce newer products for people in such a category to attract more customers",
"_____no_output_____"
]
],
[
[
"#Univariate Analysis for categorical variable 'NAME_HOUSING_TYPE' in dataframe one_df.\n\nsns.countplot(x ='NAME_HOUSING_TYPE', data = one_df)\n\n\nplt.title(\"Number of applications by Client's Housing Status\")\nplt.ylabel('Number of Applications')\nplt.xlabel(\"Client's Housing Status\")\nplt.xticks(rotation = 90)\nplt.show()\n",
"_____no_output_____"
]
],
[
[
"### Observation\n\n- People who live in their own apartment/house apply for loans almost 160 times more than those who live with their parents.\n- People living in office apartments default significantly less. This could be because their houses are rent free or they pay minimum charges to live in the house.\n",
"_____no_output_____"
]
],
[
[
"#Univariate Analysis for categorical variable 'OCCUPATION_TYPE' in dataframe one_df.\n\nsns.countplot(x ='OCCUPATION_TYPE', data = one_df)\n\n\nplt.title(\"Number of applications by Client's Occupation Type\")\nplt.ylabel('Number of Applications')\nplt.xlabel(\"Client's Occupation Type\")\nplt.xticks(rotation = 90)\nplt.show()\n",
"_____no_output_____"
]
],
[
[
"### Observation\n\n- Labourers apply for a lot of loans and default on being able to repay them. This could be because of the contractual nature of their work and the unsetady + low income they might earn from their daily jobs\n- IT & HR Staff make very few applications for credit and default the least on their loan applications. This could be, in stark contrast to the labourers, because of the stable job & salaried nature of their work. Thus enabling them to be better at handling monthly expenses. ",
"_____no_output_____"
]
],
[
[
"# Since there are subcategories like Type1,2 etc under few categories like Business Entity,Trade etc. \n# Because of this, there are a lot of categories making it difficult to analyse data\n# Its better to remove the types and just have the main category there\n\none_df.ORGANIZATION_TYPE= one_df.ORGANIZATION_TYPE.replace(\"Business Entity Type 3\", \"Business Entity\")\none_df.ORGANIZATION_TYPE= one_df.ORGANIZATION_TYPE.replace(\"Business Entity Type 2\", \"Business Entity\")\none_df.ORGANIZATION_TYPE= one_df.ORGANIZATION_TYPE.replace(\"Business Entity Type 1\", \"Business Entity\")\n\none_df.ORGANIZATION_TYPE= one_df.ORGANIZATION_TYPE.replace(\"Trade: type 7\", \"Trade\")\none_df.ORGANIZATION_TYPE= one_df.ORGANIZATION_TYPE.replace(\"Trade: type 3\", \"Trade\")\none_df.ORGANIZATION_TYPE= one_df.ORGANIZATION_TYPE.replace(\"Trade: type 2\", \"Trade\")\none_df.ORGANIZATION_TYPE= one_df.ORGANIZATION_TYPE.replace(\"Trade: type 1\", \"Trade\")\none_df.ORGANIZATION_TYPE= one_df.ORGANIZATION_TYPE.replace(\"Trade: type 6\", \"Trade\")\none_df.ORGANIZATION_TYPE= one_df.ORGANIZATION_TYPE.replace(\"Trade: type 5\", \"Trade\")\none_df.ORGANIZATION_TYPE= one_df.ORGANIZATION_TYPE.replace(\"Trade: type 4\", \"Trade\")\n\none_df.ORGANIZATION_TYPE= one_df.ORGANIZATION_TYPE.replace(\"Transport: type 4\", \"Transport\")\none_df.ORGANIZATION_TYPE= one_df.ORGANIZATION_TYPE.replace(\"Transport: type 3\", \"Transport\")\none_df.ORGANIZATION_TYPE= one_df.ORGANIZATION_TYPE.replace(\"Transport: type 2\", \"Transport\")\none_df.ORGANIZATION_TYPE= one_df.ORGANIZATION_TYPE.replace(\"Transport: type 1\", \"Transport\")\n\none_df.ORGANIZATION_TYPE= one_df.ORGANIZATION_TYPE.replace(\"Industry: type 1\", \"Industry\")\none_df.ORGANIZATION_TYPE= one_df.ORGANIZATION_TYPE.replace(\"Industry: type 2\", \"Industry\")\none_df.ORGANIZATION_TYPE= one_df.ORGANIZATION_TYPE.replace(\"Industry: type 3\", \"Industry\")\none_df.ORGANIZATION_TYPE= one_df.ORGANIZATION_TYPE.replace(\"Industry: type 4\", \"Industry\")\none_df.ORGANIZATION_TYPE= one_df.ORGANIZATION_TYPE.replace(\"Industry: type 5\", \"Industry\")\none_df.ORGANIZATION_TYPE= one_df.ORGANIZATION_TYPE.replace(\"Industry: type 6\", \"Industry\")\none_df.ORGANIZATION_TYPE= one_df.ORGANIZATION_TYPE.replace(\"Industry: type 7\", \"Industry\")\none_df.ORGANIZATION_TYPE= one_df.ORGANIZATION_TYPE.replace(\"Industry: type 8\", \"Industry\")\none_df.ORGANIZATION_TYPE= one_df.ORGANIZATION_TYPE.replace(\"Industry: type 9\", \"Industry\")\none_df.ORGANIZATION_TYPE= one_df.ORGANIZATION_TYPE.replace(\"Industry: type 10\", \"Industry\")\none_df.ORGANIZATION_TYPE= one_df.ORGANIZATION_TYPE.replace(\"Industry: type 11\", \"Industry\")\none_df.ORGANIZATION_TYPE= one_df.ORGANIZATION_TYPE.replace(\"Industry: type 12\", \"Industry\")\none_df.ORGANIZATION_TYPE= one_df.ORGANIZATION_TYPE.replace(\"Industry: type 13\", \"Industry\")\n\none_df['ORGANIZATION_TYPE'].value_counts()",
"_____no_output_____"
],
[
"#Univariate Analysis for categorical variable 'ORGANIZATION_TYPE' in dataframe one_df.\n\nplt.figure(figsize = (14,14))\nsns.countplot(x ='ORGANIZATION_TYPE', data = one_df)\n\nplt.title(\"Number of applications by Client's Organization Type\")\nplt.ylabel('Number of Applications')\nplt.xlabel(\"Client's Organization Type\")\nplt.xticks(rotation = 90)\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Observation\n\n- Based on the plot above we can see that Business Entity employees have the maximum number of loan applications\n- Religious people, priests etc dont seem to be making any credit applications at all\n- Self-employed people also make a lot of loan applications. This could be to boost their business or to repay other loans.",
"_____no_output_____"
],
[
"##### Continuous - Continuous Bivariate Analysis for one_df dataframe",
"_____no_output_____"
]
],
[
[
"## Plotting cont-cont Client Income vs Credit Amount\n\nplt.figure(figsize=(12,12))\n\n\nsns.scatterplot(x=\"AMT_INCOME_TOTAL\", y=\"AMT_CREDIT\",\n hue=\"CODE_GENDER\", style=\"CODE_GENDER\", data=one_df)\nplt.xlabel('Income of client')\nplt.ylabel('Credit Amount of loan')\nplt.title('Client Income vs Credit Amount')\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Observation\n\n- We do see some outliers here wherein Females having income less than 50000 have applied for loan with credit amount 1300000 approx\n- Most of the loans seem to be concentrated between credit amount of 200000 & 6000000 for income ranging from 50000-150000",
"_____no_output_____"
]
],
[
[
"## Plotting cont-cont Client Income vs Region population \n\nplt.figure(figsize=(12,12))\n\n\nsns.scatterplot(x=\"AMT_INCOME_TOTAL\", y=\"REGION_POPULATION_RELATIVE\",\n hue=\"CODE_GENDER\", style=\"CODE_GENDER\", data=one_df)\nplt.xlabel('Income of client')\nplt.ylabel('Population of region where client lives')\nplt.title('Client Income vs Region population')\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Observation\n\n- Very less no of people live in highly dense/populated region\n- Most of the clients live between population density of 0.00 to 0.04",
"_____no_output_____"
],
[
"##### Univariate analysis for zero_df dataframe",
"_____no_output_____"
]
],
[
[
"#Univariate Analysis for categorical variable 'CODE_GENDER' in dataframe zero_df.\n\nsns.countplot(x ='CODE_GENDER', data = zero_df)\n\n\nplt.title('Number of applications by Gender')\nplt.ylabel('Number of Applications')\nplt.xlabel('Gender')\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Observation\nAs you can see above the number of Female applicants is higher than the number of Male applicants.",
"_____no_output_____"
]
],
[
[
"#Univariate Analysis for categorical variable 'NAME_CONTRACT_TYPE' in dataframe zero_df.\n\nsns.countplot(x ='NAME_CONTRACT_TYPE', data = zero_df)\n\n\nplt.title('Number of applications by Contract Type')\nplt.ylabel('Number of Applications')\nplt.xlabel('Contract Type')\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Observation\n\nApplicants prefer to apply more for cash loans rather than revolving loans",
"_____no_output_____"
]
],
[
[
"#Univariate Analysis for categorical variable 'NAME_INCOME_TYPE' in dataframe zero_df.\n\nsns.countplot(x ='NAME_INCOME_TYPE', data = zero_df)\n\n\nplt.title(\"Number of applications by Client's Income Type\")\nplt.ylabel('Number of Applications')\nplt.xlabel(\"Client's Income Type\")\nplt.xticks(rotation = 90)\nplt.show()\n",
"_____no_output_____"
]
],
[
[
"### Observation\n\n- Working people make the most number of applications and are able to successfully repay their loans as well.\n- Students, Pensioners, Business men and Maternity leave applicants is close to 0. This could be due to a multitude of reasons.",
"_____no_output_____"
]
],
[
[
"#Univariate Analysis for categorical variable 'NAME_EDUCATION_TYPE' in dataframe zero_df.\n\nsns.countplot(x ='NAME_EDUCATION_TYPE', data = zero_df)\n\n\nplt.title(\"Number of applications by Client's Education Level\")\nplt.ylabel('Number of Applications')\nplt.xlabel(\"Client's Education Level\")\nplt.xticks(rotation = 90)\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Observation\nFrom the plot above we can infer that:\n\n- The highest number of applications for credit were made by people having Secondary/ secondary special education and these people did not default on being able to pay back their loans.\n- People with higher education also applied for a credit and were able to repay them successfully",
"_____no_output_____"
]
],
[
[
"#Univariate Analysis for categorical variable 'NAME_FAMILY_STATUS' in dataframe zero_df.\n\nsns.countplot(x ='NAME_FAMILY_STATUS', data = zero_df)\n\n\nplt.title(\"Number of applications by Client's Family Status\")\nplt.ylabel('Number of Applications')\nplt.xlabel(\"Client's Family Status\")\nplt.xticks(rotation = 90)\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Observation\nFrom the plot above we can infer that:\n\n- Married people apply for credit the most. \n- Married people are able to repay their loans without any defaults as well",
"_____no_output_____"
]
],
[
[
"#Univariate Analysis for categorical variable 'NAME_HOUSING_TYPE' in dataframe zero_df.\n\nsns.countplot(x ='NAME_HOUSING_TYPE', data = zero_df)\n\n\nplt.title(\"Number of applications by Client's Housing Status\")\nplt.ylabel('Number of Applications')\nplt.xlabel(\"Client's Housing Status\")\nplt.xticks(rotation = 90)\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Observation\n\n- People who live in their own apartment/house apply for loans almost 160 times more than those who live with their parents.\n- People living in office apartments apply for loans significantly less. This could be because their houses are rent free or they pay minimum charges to live in the house.\n- People in rented apartments apply for loans significantly less. This could be due to the added expenses of paying rent and other utility bills leaves them with not enough capital to payback their loans.",
"_____no_output_____"
]
],
[
[
"#Univariate Analysis for categorical variable 'OCCUPATION_TYPE' in dataframe zero_df.\n\nsns.countplot(x ='OCCUPATION_TYPE', data = zero_df)\n\n\nplt.title(\"Number of applications by Client's Occupation Type\")\nplt.ylabel('Number of Applications')\nplt.xlabel(\"Client's Occupation Type\")\nplt.xticks(rotation = 90)\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Observation\n- Labourers apply for a lot of loans.\n- IT & HR Staff make very few applications for credit. This could be, in stark contrast to the labourers, because of the stable job & salaried nature of their work. Thus enabling them to be better at handling monthly expenses.",
"_____no_output_____"
]
],
[
[
"# Since there are subcategories like Type1,2 etc under few categories like Business Entity,Trade etc. \n# Because of this, there are a lot of categories making it difficult to analyse data\n# Its better to remove the types and just have the main category there\n\nzero_df.ORGANIZATION_TYPE= zero_df.ORGANIZATION_TYPE.replace(\"Business Entity Type 3\", \"Business Entity\")\nzero_df.ORGANIZATION_TYPE= zero_df.ORGANIZATION_TYPE.replace(\"Business Entity Type 2\", \"Business Entity\")\nzero_df.ORGANIZATION_TYPE= zero_df.ORGANIZATION_TYPE.replace(\"Business Entity Type 1\", \"Business Entity\")\n\nzero_df.ORGANIZATION_TYPE= zero_df.ORGANIZATION_TYPE.replace(\"Trade: type 7\", \"Trade\")\nzero_df.ORGANIZATION_TYPE= zero_df.ORGANIZATION_TYPE.replace(\"Trade: type 3\", \"Trade\")\nzero_df.ORGANIZATION_TYPE= zero_df.ORGANIZATION_TYPE.replace(\"Trade: type 2\", \"Trade\")\nzero_df.ORGANIZATION_TYPE= zero_df.ORGANIZATION_TYPE.replace(\"Trade: type 1\", \"Trade\")\nzero_df.ORGANIZATION_TYPE= zero_df.ORGANIZATION_TYPE.replace(\"Trade: type 6\", \"Trade\")\nzero_df.ORGANIZATION_TYPE= zero_df.ORGANIZATION_TYPE.replace(\"Trade: type 5\", \"Trade\")\nzero_df.ORGANIZATION_TYPE= zero_df.ORGANIZATION_TYPE.replace(\"Trade: type 4\", \"Trade\")\n\nzero_df.ORGANIZATION_TYPE= zero_df.ORGANIZATION_TYPE.replace(\"Transport: type 4\", \"Transport\")\nzero_df.ORGANIZATION_TYPE= zero_df.ORGANIZATION_TYPE.replace(\"Transport: type 3\", \"Transport\")\nzero_df.ORGANIZATION_TYPE= zero_df.ORGANIZATION_TYPE.replace(\"Transport: type 2\", \"Transport\")\nzero_df.ORGANIZATION_TYPE= zero_df.ORGANIZATION_TYPE.replace(\"Transport: type 1\", \"Transport\")\n\nzero_df.ORGANIZATION_TYPE= zero_df.ORGANIZATION_TYPE.replace(\"Industry: type 1\", \"Industry\")\nzero_df.ORGANIZATION_TYPE= zero_df.ORGANIZATION_TYPE.replace(\"Industry: type 2\", \"Industry\")\nzero_df.ORGANIZATION_TYPE= zero_df.ORGANIZATION_TYPE.replace(\"Industry: type 3\", \"Industry\")\nzero_df.ORGANIZATION_TYPE= zero_df.ORGANIZATION_TYPE.replace(\"Industry: type 4\", \"Industry\")\nzero_df.ORGANIZATION_TYPE= zero_df.ORGANIZATION_TYPE.replace(\"Industry: type 5\", \"Industry\")\nzero_df.ORGANIZATION_TYPE= zero_df.ORGANIZATION_TYPE.replace(\"Industry: type 6\", \"Industry\")\nzero_df.ORGANIZATION_TYPE= zero_df.ORGANIZATION_TYPE.replace(\"Industry: type 7\", \"Industry\")\nzero_df.ORGANIZATION_TYPE= zero_df.ORGANIZATION_TYPE.replace(\"Industry: type 8\", \"Industry\")\nzero_df.ORGANIZATION_TYPE= zero_df.ORGANIZATION_TYPE.replace(\"Industry: type 9\", \"Industry\")\nzero_df.ORGANIZATION_TYPE= zero_df.ORGANIZATION_TYPE.replace(\"Industry: type 10\", \"Industry\")\nzero_df.ORGANIZATION_TYPE= zero_df.ORGANIZATION_TYPE.replace(\"Industry: type 11\", \"Industry\")\nzero_df.ORGANIZATION_TYPE= zero_df.ORGANIZATION_TYPE.replace(\"Industry: type 12\", \"Industry\")\nzero_df.ORGANIZATION_TYPE= zero_df.ORGANIZATION_TYPE.replace(\"Industry: type 13\", \"Industry\")\n\nzero_df['ORGANIZATION_TYPE'].value_counts()",
"_____no_output_____"
],
[
"#Univariate Analysis for categorical variable 'ORGANIZATION_TYPE' in dataframe zero_df.\n\nplt.figure(figsize = (14,14))\nsns.countplot(x ='ORGANIZATION_TYPE', data = zero_df)\n\n\nplt.title(\"Number of applications by Client's Organization Type\")\nplt.ylabel('Number of Applications')\nplt.xlabel(\"Client's Organization Type\")\nplt.xticks(rotation = 90)\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Observation\n\n- Based on the plot above we can see that Business Entity employees have the maximum number of loan applications\n- Religious people, priests etc dont seem to be making a lot of credit applications at all. They are able to repay their loans on time as well. \n- Self-employed people also make a lot of loan applications. This could be to boost their business or to repay other loans.",
"_____no_output_____"
],
[
"### Bivariate Analysis for zero_df",
"_____no_output_____"
]
],
[
[
"### Let us create a helper function to help with\n### plotting various graphs\n\ndef uniplot(df,col,title,hue =None):\n \n sns.set_style('whitegrid')\n sns.set_context('talk')\n plt.rcParams[\"axes.labelsize\"] = 20\n plt.rcParams['axes.titlesize'] = 22\n plt.rcParams['axes.titlepad'] = 30\n plt.figure(figsize=(40,20))\n \n \n temp = pd.Series(data = hue)\n fig, ax = plt.subplots()\n width = len(df[col].unique()) + 7 + 4*len(temp.unique())\n fig.set_size_inches(width , 8)\n plt.xticks(rotation=45)\n plt.title(title)\n ax = sns.countplot(data = df, x= col, order=df[col].value_counts().index,hue = hue,\n palette='magma') \n \n plt.show()",
"_____no_output_____"
],
[
"# PLotting for income range\nuniplot(zero_df,col='NAME_INCOME_TYPE',title='Distribution of Income type',hue='CODE_GENDER')",
"_____no_output_____"
]
],
[
[
"### Observation \n- For income type ‘working’, ’commercial associate’, and ‘State Servant’ the number of credits are higher than others.\n- For this Females are having more number of credit applications than males in all the categories.\n",
"_____no_output_____"
]
],
[
[
"uniplot(zero_df,col='NAME_CONTRACT_TYPE',title='Distribution of contract type',hue='CODE_GENDER')",
"_____no_output_____"
]
],
[
[
"### Observation\n- For contract type ‘cash loans’ is having higher number of credits than ‘Revolving loans’ contract type.\n- For this also Females are applying for credit a lot more than males.\n",
"_____no_output_____"
]
],
[
[
"uniplot(zero_df,col='NAME_FAMILY_STATUS',title='Distribution of Family status',hue='CODE_GENDER')",
"_____no_output_____"
]
],
[
[
"### Observation\n- As observed above the number of married females applying for loans is almost 3.5 times the number of single females. \n- No male widowers are applying for credit\n",
"_____no_output_____"
]
],
[
[
"uniplot(zero_df,col='NAME_EDUCATION_TYPE',title='Distribution of education level',hue='CODE_GENDER')",
"_____no_output_____"
]
],
[
[
"### Observation\n- No person with an 'Academic Degree' is applying for a loan\n- The number of females with 'Higher Education' that apply for a loan is almost double the number of males for the same category",
"_____no_output_____"
]
],
[
[
"uniplot(zero_df,col='NAME_HOUSING_TYPE',title='Distribution of Housing Type',hue='CODE_GENDER')",
"_____no_output_____"
]
],
[
[
"### Observation\n\n- Females living in their own apartments/houses apply for more loans and are able to successfully payback.\n- A very small number of females living in Co-op apartments apply for loans",
"_____no_output_____"
]
],
[
[
"uniplot(zero_df,col='OCCUPATION_TYPE',title='Distribution Occupation Type',hue='CODE_GENDER')",
"_____no_output_____"
]
],
[
[
"### Observation\n- Male Labourers & Drivers take more loans and are able to successfully payback in time.\n- Female Care staff & Sales Staff are also able to take loans and payback in time ",
"_____no_output_____"
],
[
"### Bivariate Analysis on one_df ",
"_____no_output_____"
],
[
"Perform correlation between numerical columns for finding correlation which having TARGET value as 1",
"_____no_output_____"
]
],
[
[
"uniplot(one_df,col='NAME_INCOME_TYPE',title='Distribution of Income type',hue='CODE_GENDER')",
"_____no_output_____"
]
],
[
[
"### Observation \n- For income type ‘working’, ’commercial associate’, and ‘State Servant’ the number of credits are higher than others.\n- Females have more number of credit applications than males in all the categories.",
"_____no_output_____"
]
],
[
[
"uniplot(one_df,col='NAME_CONTRACT_TYPE',title='Distribution of contract type',hue='CODE_GENDER')",
"_____no_output_____"
]
],
[
[
"### Observation\n- For contract type ‘cash loans’ is having higher number of credits than ‘Revolving loans’ contract type.\n- For this also Females are applying for credit a lot more than males.\n- Females are also able to payback their loans on time",
"_____no_output_____"
]
],
[
[
"uniplot(one_df,col='NAME_FAMILY_STATUS',title='Distribution of Family status',hue='CODE_GENDER')",
"_____no_output_____"
]
],
[
[
"### Observation\n- As observed above the number of married females applying for loans is almost 3.5 times the number of single females. \n- No male widowers are applying for credit\n- The number of males applying for loans and being able to not payback is higher if they are unmarried/single compared to females\n- A very small number of male widowers are unable to payback their loans after",
"_____no_output_____"
]
],
[
[
"uniplot(one_df,col='NAME_EDUCATION_TYPE',title='Distribution of education level',hue='CODE_GENDER')",
"_____no_output_____"
]
],
[
[
"### Observation\n\n- Males with lower secondary education make more loan applications and default more compared to females\n- There is very little difference between the number of defaulters for males and females with secondary education compared to the non-defaulters we saw above",
"_____no_output_____"
]
],
[
[
"uniplot(one_df,col='NAME_HOUSING_TYPE',title='Distribution of Housing Type',hue='CODE_GENDER')",
"_____no_output_____"
]
],
[
[
"### Observation\n\n- Males living with their parents tend to apply and default more on their loans\n- Almost an equal number of males and females default on loans if they are living in rented apartments",
"_____no_output_____"
]
],
[
[
"uniplot(one_df,col='OCCUPATION_TYPE',title='Distribution Occupation Type',hue='CODE_GENDER')",
"_____no_output_____"
]
],
[
[
"### Observations\n\n- The number of male applicants who default on paying back their loans is almost double the amount of female applicants\n- Irrespective of gender, managers seem to default on their loans equally",
"_____no_output_____"
],
[
"#### Categorical vs Numerical Analysis",
"_____no_output_____"
]
],
[
[
"# Box plotting for Credit amount for zero_df based on education type and family status\n\nplt.figure(figsize=(40,20))\nplt.xticks(rotation=45)\nsns.boxplot(data =zero_df, x='NAME_EDUCATION_TYPE',y='AMT_CREDIT', hue ='NAME_FAMILY_STATUS',orient='v')\nplt.title('Credit amount vs Education Status')\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Observation\n- Widows with secondary education have a very high median credit amount borrowing and default on paying back loans as well. It would be better to be vary of lending to them\n- Widows with an academic degree have a higher median for borrowing as compared to any other category. \n- People in civil marriages, those who are seperated and widows with secondary education have the same median values and usually borrow in around 400000",
"_____no_output_____"
]
],
[
[
"# Box plotting for Income amount for zero_df based on their education type & family status\n\nplt.figure(figsize=(40,20))\nplt.xticks(rotation=45)\nplt.yscale('log')\nsns.boxplot(data =zero_df, x='NAME_EDUCATION_TYPE',y='AMT_INCOME_TOTAL', hue ='NAME_FAMILY_STATUS',orient='v')\nplt.title('Income amount vs Education Status')\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Observation\n- Except widows, the median earning for all other family status types with an incomplete higher education is the same\n- Median income for all family status categories is the same for people with a secondary education",
"_____no_output_____"
]
],
[
[
"# Box plotting for Credit amount for one_df\n\nplt.figure(figsize=(16,12))\nplt.xticks(rotation=45)\nsns.boxplot(data =one_df, x='NAME_EDUCATION_TYPE',y='AMT_CREDIT', hue ='NAME_FAMILY_STATUS',orient='v')\nplt.title('Credit amount vs Education Status')\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Observation\n- Widows with secondary education have a very high median credit amount borrowing and default on paying back loans as well. It would be better to be vary of lending to them\n- Married people have a consistently high median across all categories of education except secondary education",
"_____no_output_____"
]
],
[
[
"# Box plotting for Income amount for one_df\n\nplt.figure(figsize=(40,20))\nplt.xticks(rotation=45)\nplt.yscale('log')\nsns.boxplot(data =one_df, x='NAME_EDUCATION_TYPE',y='AMT_INCOME_TOTAL', hue ='NAME_FAMILY_STATUS',orient='v')\nplt.title('Income amount vs Education Status')\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Observation\n- The median income for all family status types is the same for people with education type as Secondary/secondary special\n- The median income for widows is the lowest across all the education types",
"_____no_output_____"
]
],
[
[
"### Perform correlation between CNT_CHILDREN, AMT_INCOME_TOTAL, AMT_CREDIT, AMT_GOODS_PRICE, REGION_POPULATION_RELATIVE\n### and AMT_ANNUITY. Then make correlation matrix across the one_df dataframe\n\ncolumns=['CNT_CHILDREN','AMT_INCOME_TOTAL','AMT_CREDIT','AMT_GOODS_PRICE','REGION_POPULATION_RELATIVE', 'AMT_ANNUITY']\ncorr=one_df[columns].corr()\ncorr.style.background_gradient(cmap='coolwarm')",
"_____no_output_____"
]
],
[
[
"### Observation\nIn the heatmap above: The closer you are to RED there is a stronger relationship, the closer you are to blue the weaker the relationship.\n\nAs we can see from the corelation matrix above, there is a very close relationship between AMT_GOODS_PRICE & AMT_CREDIT. \n\nAMT_ANNUITY & AMT_CREDIT have a medium/strong relationship. Annuity has a similar relationship with AMT_GOODS_PRICE.\n",
"_____no_output_____"
]
],
[
[
"### Sorting based on the correlation and extracting top 10 relationships on the defaulters in one_df\ncorrOneDf = corr.where(np.triu(np.ones(corr.shape), k=1).astype(np.bool)).unstack().reset_index() \n\ncorrOneDf.columns = ['VAR1','VAR2','Correlation'] \ncorrOneDf.sort_values('Correlation', ascending = False).nlargest(10, 'Correlation')",
"_____no_output_____"
]
],
[
[
"### Observation\nIn the correlation matrix, we can identify-\n\nColumns with High Correlation:\n\n1.AMT_GOODS_PRICE and AMT_CREDIT\n\nColumns with Medium Correlation:\n\n1.REGION_POPULATION_RELATIVE and AMT_INCOME_TOTAL \n2.REGION_POPULATION_RELATIVE and AMT_GOODS_PRICE \n3.REGION_POPULATION_RELATIVE and AMT_CREDIT\n\nColumns with low correlation:\n\n1.AMT_INCOME_TOTAL and CNT_CHILDREN\n\nWe also observed that the top 10 correlation pairs are:\n- VAR1 VAR2 Correlation Value\n- AMT_GOODS_PRICE\tAMT_CREDIT\t0.981276\n- AMT_ANNUITY\tAMT_CREDIT\t0.748446\n- AMT_ANNUITY\tAMT_GOODS_PRICE\t0.747315\n- AMT_ANNUITY\tAMT_INCOME_TOTAL\t0.390809\n- AMT_GOODS_PRICE\tAMT_INCOME_TOTAL\t0.317123\n- AMT_CREDIT\tAMT_INCOME_TOTAL\t0.313347\n- REGION_POPULATION_RELATIVE\tAMT_INCOME_TOTAL\t0.141307\n- AMT_ANNUITY\tREGION_POPULATION_RELATIVE\t0.065024\n- REGION_POPULATION_RELATIVE\tAMT_GOODS_PRICE\t0.055120\n- REGION_POPULATION_RELATIVE\tAMT_CREDIT\t0.050097",
"_____no_output_____"
],
[
"Perform correlation between numerical columns for finding correlation which having TARGET value as 0",
"_____no_output_____"
]
],
[
[
"#Perform correlation between CNT_CHILDREN, AMT_INCOME_TOTAL, AMT_CREDIT, AMT_GOODS_PRICE and REGION_POPULATION_RELATIVE\n#Then make correlation matrix\n\ncorrZero=zero_df[columns].corr()\ncorrZero.style.background_gradient(cmap='coolwarm')",
"_____no_output_____"
]
],
[
[
"### Observation\nIn the heatmap above: The closer you are to RED there is a stronger relationship, the closer you are to blue the weaker the relationship.\n\nAs we can see from the corelation matrix above, there is a very close relationship between AMT_GOODS_PRICE & AMT_CREDIT.\n\nAMT_ANNUITY & AMT_CREDIT have a medium/strong relationship. Annuity has a similar relationship with AMT_GOODS_PRICE.\n\nThis relationship is consistent with the one we saw for the defaulters in the one_df dataframe. Thus confirming that the relationships are consistent across TARGET values",
"_____no_output_____"
]
],
[
[
"corrZeroDf = corrZero.where(np.triu(np.ones(corrZero.shape), k=1).astype(np.bool)).unstack().reset_index() \n\ncorrZeroDf.columns = ['VAR1','VAR2','Correlation'] \n# corrOneDf.dropna(subset - ['Correlation'],inplace = True) \ncorrZeroDf.sort_values('Correlation', ascending = False).nlargest(10, 'Correlation')",
"_____no_output_____"
]
],
[
[
"In the correlation matrix, we can identify-\n\nColumns with High Correlation:\n\n1.AMT_GOODS_PRICE and AMT_CREDIT\n\nColumns with Medium Correlation:\n\n1.AMT_INCOME_TOTAL and AMT_CREDIT \n2.AMT_INCOME_TOTAL and AMT_GOODS_PRICE \n\nColumns with low correlation:\n\n1.AMT_GOODS_PRICE and CNT_CHILDREN\n\nWe also observed that the top 10 correlation pairs are:\n\n- VAR1\tVAR2\tCorrelation\n- AMT_GOODS_PRICE\tAMT_CREDIT\t0.981276\n- AMT_ANNUITY\tAMT_CREDIT\t0.748446\n- AMT_ANNUITY\tAMT_GOODS_PRICE\t0.747315\n- AMT_ANNUITY\tAMT_INCOME_TOTAL\t0.390809\n- AMT_GOODS_PRICE\tAMT_INCOME_TOTAL\t0.317123\n- AMT_CREDIT\tAMT_INCOME_TOTAL\t0.313347\n- REGION_POPULATION_RELATIVE\tAMT_INCOME_TOTAL\t0.141307\n- AMT_ANNUITY\tREGION_POPULATION_RELATIVE\t0.065024\n- REGION_POPULATION_RELATIVE\tAMT_GOODS_PRICE\t0.055120\n- REGION_POPULATION_RELATIVE\tAMT_CREDIT\t0.050097",
"_____no_output_____"
],
[
"#### Key Obervation\nWe also observed that the top categories between both the data frames zero_df & one_df is the same:\nAMT_GOODS_PRICE\tAMT_CREDIT\t0.981276",
"_____no_output_____"
],
[
"### Analysing Numerical Data",
"_____no_output_____"
]
],
[
[
"#Box plot on the numerical columns having TARGET value as 1\n\nplt.figure(figsize=(25,25))\nplt.subplot(2,2,1)\nplt.title('CHILDREN COUNT')\nsns.boxplot(one_df['CNT_CHILDREN'])\n\n\nplt.subplot(2,2,2)\nplt.title('AMT_INCOME_TOTAL')\nsns.boxplot(one_df['AMT_INCOME_TOTAL'])\n\n\nplt.subplot(2,2,3)\nplt.title('AMT_CREDIT')\nsns.boxplot(one_df['AMT_CREDIT'])\n\nplt.subplot(2,2,4)\nplt.title('AMT_GOODS_PRICE')\nsns.boxplot(one_df['AMT_GOODS_PRICE'])\n\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Observation\n\n- From the box plots above we can safely say that having children has no impact on the reason to why someone defaults on paying back their loans\n- The amount of credit taken is roughly around 450000 by the defaulters",
"_____no_output_____"
]
],
[
[
"#Box plot on the numerical columns having TARGET value as 0\n\nplt.figure(figsize=(25,25))\nplt.subplot(2,2,1)\nplt.title('CHILDREN COUNT')\nsns.boxplot(zero_df['CNT_CHILDREN'])\n\n\nplt.subplot(2,2,2)\nplt.title('AMT_INCOME_TOTAL')\nsns.boxplot(zero_df['AMT_INCOME_TOTAL'])\n\n\nplt.subplot(2,2,3)\nplt.title('AMT_CREDIT')\nsns.boxplot(zero_df['AMT_CREDIT'])\n\nplt.subplot(2,2,4)\nplt.title('AMT_GOODS_PRICE')\nsns.boxplot(zero_df['AMT_GOODS_PRICE'])\n\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Observation\n\n- From the box plots above we can safely say that having children has no impact oa persons ability to repay their loans\n- The amount of credit taken is roughly around 450000 by the defaulters\n- There are no outliers in the amoount of goods price\n- The income median lies just below 150000",
"_____no_output_____"
],
[
"### Bivariate Analysis on zero_df for continuous - continuous (Target value =0)",
"_____no_output_____"
]
],
[
[
"## Plotting cont-cont Client Income vs Credit Amount\n\nplt.figure(figsize=(12,12))\n\n\nsns.scatterplot(x=\"AMT_INCOME_TOTAL\", y=\"AMT_CREDIT\",\n hue=\"CODE_GENDER\", style=\"CODE_GENDER\", data=zero_df)\nplt.xlabel('Income of client')\nplt.ylabel('Credit Amount of loan')\nplt.title('Client Income vs Credit Amount')\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Observation\n\n- We do see some outliers here wherein Females having income less than 50000 have applied for loan with credit amount 1300000 approx",
"_____no_output_____"
]
],
[
[
"## Plotting cont-cont Client Income vs Region population \n\nplt.figure(figsize=(12,12))\n\n\nsns.scatterplot(x=\"AMT_INCOME_TOTAL\", y=\"REGION_POPULATION_RELATIVE\",\n hue=\"CODE_GENDER\", style=\"CODE_GENDER\", data=zero_df)\nplt.xlabel('Income of client')\nplt.ylabel('Population of region where client lives')\nplt.title('Client Income vs Region population')\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Observation\n\n- Very less no of people live in highly dense/populated region >0.07\n- Most of the clients live between population density of 0.00 to 0.04",
"_____no_output_____"
],
[
"# 5 PREVIOUS DATA",
"_____no_output_____"
],
[
"Read the dataset file previous_application.csv which consist previous loan of the customer.",
"_____no_output_____"
]
],
[
[
"previousApplicationData=pd.read_csv(\"./previous_application.csv\")\n\npreviousApplicationData.head()",
"_____no_output_____"
]
],
[
[
"### Analysing previous application data ",
"_____no_output_____"
]
],
[
[
"previousApplicationData.shape",
"_____no_output_____"
],
[
"previousApplicationData.describe",
"_____no_output_____"
],
[
"previousApplicationData.columns",
"_____no_output_____"
],
[
"previousApplicationData.dtypes",
"_____no_output_____"
],
[
"### Join the previous application data and application data files using merge\n\nmergedApplicationDataAndPreviousData = pd.merge(applicationDataWithRelevantColumns, previousApplicationData, how='left', on=['SK_ID_CURR'])\nmergedApplicationDataAndPreviousData.head()",
"_____no_output_____"
]
],
[
[
"### Observation\nWe will be merging on 'SK_ID_CURR' column as we have duplicate IDs present in the SK_ID_CURR in previousApplicationData and in the application_data file all the values are unique.",
"_____no_output_____"
]
],
[
[
"mergedApplicationDataAndPreviousData.shape",
"_____no_output_____"
],
[
"mergedApplicationDataAndPreviousData.NAME_CONTRACT_STATUS.value_counts(normalize=True)",
"_____no_output_____"
]
],
[
[
"### Analysis\n\nWe will be focusing on analysing the NAME_CONTRACT_STATUS Column and the various relationships based on that.",
"_____no_output_____"
],
[
"## Univariate Analysis",
"_____no_output_____"
]
],
[
[
"uniplot(mergedApplicationDataAndPreviousData,col='NAME_CONTRACT_STATUS',title='Distribution of contract status type', hue=None)",
"_____no_output_____"
]
],
[
[
"### Observation\n- A large number of applications were approved for the clients\n- Some clients who recieved the offer did not use their loan offers\n- The number of refused & cancelled applications is roughly the same",
"_____no_output_____"
],
[
"## Bivariate Analysis",
"_____no_output_____"
]
],
[
[
"uniplot(mergedApplicationDataAndPreviousData,col='NAME_CONTRACT_STATUS',title='Distribution Occupation Type',hue='NAME_INCOME_TYPE')",
"_____no_output_____"
]
],
[
[
"### Observation\n\nBased on the plot above we can conclude that:\n\n- Working professionals have the highest number of approved loan applications.\n- Working professionals also have the highest number of refused or cancelled loan applications\n- Students, pensioners, businessmen and applicants on maternity leave have statistically low or no application status data present\n",
"_____no_output_____"
]
],
[
[
"uniplot(mergedApplicationDataAndPreviousData,col='NAME_CONTRACT_STATUS',title='Distribution based on Gender',hue='CODE_GENDER')",
"_____no_output_____"
]
],
[
[
"### Observation \n\n- Female applicants make more applications and have a higher number of applications approved\n- They also have a higher number of applications refused or canceled\n- The number of male applicant statuses is lower than female ones across the board. This could be because of low number of males present in the dataset. ",
"_____no_output_____"
]
],
[
[
"uniplot(mergedApplicationDataAndPreviousData,col='NAME_CONTRACT_STATUS',title='Distribution Target',hue='TARGET')",
"_____no_output_____"
]
],
[
[
"### Observation\n\n- Based on the target column, we see that a high number of applicants who have a history of being abe to repay their loans are approved for new loans\n- A very low number of defaulters are approved for new loans. This means that the bank is following a cautious approach to defaulters\n",
"_____no_output_____"
]
],
[
[
"uniplot(mergedApplicationDataAndPreviousData,col='NAME_CONTRACT_STATUS',title='Distribution based on Family Status',hue='NAME_FAMILY_STATUS')",
"_____no_output_____"
]
],
[
[
"### Observation\n- A large number of married people make loan applications & are approved for loans\n- Separated individuals have a very low number of applications in the unused offer\n- The number of single/not married people who apply for loans and are refused or have their applications cancelled as compared to approved is less than half.",
"_____no_output_____"
]
],
[
[
"uniplot(mergedApplicationDataAndPreviousData,col='NAME_CONTRACT_STATUS',title='Distribution based Application Start Day',hue='WEEKDAY_APPR_PROCESS_START')",
"_____no_output_____"
]
],
[
[
"### Observation\n\n- Most applicants start their loan applications on a Saturday and are successfully approved\n- Applicants who start their applications on Friday have a higher chance of getting rejected or cancelling their application compared to the other 2 weekend days, Saturday and Sunday\n- The number of cancelled applications is highest on Monday. This could suggest that after starting the application on the weekend, the client changed their mind on a workday.",
"_____no_output_____"
]
],
[
[
"uniplot(mergedApplicationDataAndPreviousData,col='NAME_CONTRACT_STATUS',title='Distribution of Age on Loans',hue='BINNED_AGE')",
"_____no_output_____"
]
],
[
[
"### Observation\n\n- People between the ages of 31-40 apply for the most number of loans and have consistently higher values across all application statuses\n\n- People above the age of 71 & below 20 dont make any loan applications\n\n- The people in the ages of 31-40 could be applying for more loans as they are married or living with a partner",
"_____no_output_____"
]
],
[
[
"plt.figure(figsize=(40,25))\n\nsns.catplot(x=\"NAME_CONTRACT_STATUS\", hue=\"TARGET\", col=\"CODE_GENDER\",\n data=mergedApplicationDataAndPreviousData, kind=\"count\")",
"_____no_output_____"
]
],
[
[
"### Observation\n\n- Female population has high chances of getting the loans approved\n\n- Cancellation of loans by females is significant across defaulters and non defaulters",
"_____no_output_____"
],
[
"### Continous & Categorical Plots",
"_____no_output_____"
]
],
[
[
"### Plotting the relationship between NAME_CONTRACT_STATUS vs AMT_CREDIT_x \n### from the merged application data and splitting on the basis of family status\nplt.figure(figsize=(40,25))\nplt.xticks(rotation=45)\nsns.boxplot(data =mergedApplicationDataAndPreviousData, x='NAME_CONTRACT_STATUS',y='AMT_CREDIT_x', hue ='NAME_FAMILY_STATUS',orient='v')\nplt.title('Income amount vs Application Status based on Family Status')\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Observation\n\n- Married people take a higher amount of credit and have a higher median chance of getting approved\n\n- People in Civil marriage, widows & separated applicants have a consistently similar median value across all the application statuses",
"_____no_output_____"
]
],
[
[
"### Plotting the relationship between NAME_CONTRACT_STATUS vs AMT_INCOME_TOTAL \n### from the merged application data and splitting on the basis of family status\nplt.figure(figsize=(40,25))\nplt.xticks(rotation=45)\nplt.yscale('log')\nsns.boxplot(data =mergedApplicationDataAndPreviousData, x='NAME_CONTRACT_STATUS',y='AMT_INCOME_TOTAL', hue ='NAME_FAMILY_STATUS',orient='v')\nplt.title('Income amount vs Application status based on Family Status')\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Observation\n\n- People who are married, live in civil marriages & single/not married earn consistently well across all application status types\n- Their median income is also the same\n\n- Widows earn less than all the other categories",
"_____no_output_____"
],
[
"### Continous & Continuous Plots",
"_____no_output_____"
]
],
[
[
"plt.figure(figsize=(30,20))\nplt.scatter(mergedApplicationDataAndPreviousData.AMT_APPLICATION, mergedApplicationDataAndPreviousData.AMT_CREDIT_y)\nplt.title(\"Final Amount Approved vs Credit Amount Applied\")\nplt.xlabel(\"Credit Amount applied by Client\")\nplt.ylabel(\"Final Amount approved by Bank\")\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Observation\n\n- The Credit Amount applied vs Final Amount approved shows a good linear relation till 2000000.\n- However post 2000000, we could see good number of outliers where the approved amount is quite less as compared to amount applied\n\n- The number of applications with credit amount > 3500000 are quite less and there are not very good chances that the same amount is going to be approved",
"_____no_output_____"
],
[
"# Conclusion",
"_____no_output_____"
],
[
"Through this case study we have made the following conclusions:\n\n- Most popular days for making applications is Saturday. The bank could focus on keeping offices open longer on Saturday to aid in completion of the applications. \n\n- Most popular age group for taking loans or credit is 31-40 with the most number of applications. The firm should focus on exploring more lucrative options for clients in that age range. They could be offered lower interest rates, longer repayment holidays etc.\n\n- Married people have the highest chance of making a loan application and being approved for a loan. \n\n- Because of the imbalance in the data, Females appear to be making the most number of loan applications. They also have a higher chance of getting approved and being able to repay the loans on time\n\n- Widows with secondary education have a very high median credit amount borrowing and default on paying back loans as well. It would be better to be vary of lending to them \n\n- Male labourers have high number of applications and also a high number of defaults as compared to females. It would be better for the bank to assess whether the person borrowing in this occupation type could be helped with staged loans or with loans on a lower interest rate than the other categories\n\n- The number of applications with credit amount > 3500000 are quite less and there are not very good chances that the same amount is going to be approved\n\n- Cancellation of loans by females is significant across defaulters and non defaulters",
"_____no_output_____"
]
],
[
[
"sns.boxplot(data= applicationData.AMT_ANNUITY.head(500000).isnull())\naxes[0][1].set_title('AMT_ANNUITY')\nplt.show()",
"_____no_output_____"
],
[
"print(applicationDataAfterDroppedColumns.AMT_ANNUITY.head(500000).isnull().sum())",
"_____no_output_____"
],
[
"print(applicationData.AMT_ANNUITY.head(500000).isnull().sum())",
"_____no_output_____"
],
[
"sns.boxplot(data= applicationDataAfterDroppedColumns.AMT_ANNUITY.dropna())\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"# END OF FILE",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
4affadbaa5f5d96c1944973b9636f0e96a419b63
| 172,335 |
ipynb
|
Jupyter Notebook
|
notebooks/intro-numpy.ipynb
|
AlJohri/DAT-DC-12
|
a98c4b68e7063c2d8f64879bc4d0630b7993ff6f
|
[
"MIT"
] | 11 |
2016-03-22T06:14:17.000Z
|
2021-11-06T16:38:00.000Z
|
notebooks/intro-numpy.ipynb
|
titoashg/DAT-DC-12
|
a98c4b68e7063c2d8f64879bc4d0630b7993ff6f
|
[
"MIT"
] | 1 |
2016-06-16T14:30:38.000Z
|
2016-06-16T15:05:29.000Z
|
notebooks/intro-numpy.ipynb
|
titoashg/DAT-DC-12
|
a98c4b68e7063c2d8f64879bc4d0630b7993ff6f
|
[
"MIT"
] | 8 |
2016-03-25T19:26:44.000Z
|
2021-10-03T21:57:29.000Z
| 34.098734 | 64,508 | 0.669922 |
[
[
[
"# Introduction to NumPy",
"_____no_output_____"
],
[
"Forked from [Lecture 2](https://github.com/jrjohansson/scientific-python-lectures/blob/master/Lecture-2-Numpy.ipynb) of [Scientific Python Lectures](http://github.com/jrjohansson/scientific-python-lectures) by [J.R. Johansson](http://jrjohansson.github.io/)",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\n\nimport traceback\nimport matplotlib.pyplot as plt\n\nimport numpy as np",
"_____no_output_____"
]
],
[
[
"### Why NumPy?",
"_____no_output_____"
]
],
[
[
"%%time\n\ntotal = 0\nfor i in range(100000):\n total += i",
"CPU times: user 17.2 ms, sys: 1.16 ms, total: 18.4 ms\nWall time: 21.5 ms\n"
],
[
"%%time\n\ntotal = np.arange(100000).sum()",
"CPU times: user 583 µs, sys: 1.11 ms, total: 1.69 ms\nWall time: 1.31 ms\n"
],
[
"%%time \n\nl = list(range(0, 1000000))\nltimes5 = [x * 5 for x in l]",
"CPU times: user 111 ms, sys: 32 ms, total: 143 ms\nWall time: 149 ms\n"
],
[
"%%time \nl = np.arange(1000000)\nltimes5 = l * 5",
"CPU times: user 23.4 ms, sys: 13.1 ms, total: 36.5 ms\nWall time: 38.2 ms\n"
]
],
[
[
"## Introduction",
"_____no_output_____"
],
[
"The `numpy` package (module) is used in almost all numerical computation using Python. It is a package that provide high-performance vector, matrix and higher-dimensional data structures for Python. It is implemented in C and Fortran so when calculations are vectorized (formulated with vectors and matrices), performance is very good. \n\nTo use `numpy` you need to import the module, using for example:",
"_____no_output_____"
]
],
[
[
"import numpy as np",
"_____no_output_____"
]
],
[
[
"In the `numpy` package the terminology used for vectors, matrices and higher-dimensional data sets is *array*. \n\n",
"_____no_output_____"
],
[
"## Creating `numpy` arrays",
"_____no_output_____"
],
[
"There are a number of ways to initialize new numpy arrays, for example from\n\n* a Python list or tuples\n* using functions that are dedicated to generating numpy arrays, such as `arange`, `linspace`, etc.\n* reading data from files",
"_____no_output_____"
],
[
"### From lists",
"_____no_output_____"
],
[
"For example, to create new vector and matrix arrays from Python lists we can use the `numpy.array` function.",
"_____no_output_____"
]
],
[
[
"# a vector: the argument to the array function is a Python list\nv = np.array([1,2,3,4])\n\nv",
"_____no_output_____"
],
[
"# a matrix: the argument to the array function is a nested Python list\nM = np.array([[1, 2], [3, 4]])\n\nM",
"_____no_output_____"
]
],
[
[
"The `v` and `M` objects are both of the type `ndarray` that the `numpy` module provides.",
"_____no_output_____"
]
],
[
[
"type(v), type(M)",
"_____no_output_____"
]
],
[
[
"The difference between the `v` and `M` arrays is only their shapes. We can get information about the shape of an array by using the `ndarray.shape` property.",
"_____no_output_____"
]
],
[
[
"v.shape",
"_____no_output_____"
],
[
"M.shape",
"_____no_output_____"
]
],
[
[
"The number of elements in the array is available through the `ndarray.size` property:",
"_____no_output_____"
]
],
[
[
"M.size",
"_____no_output_____"
]
],
[
[
"Equivalently, we could use the function `numpy.shape` and `numpy.size`",
"_____no_output_____"
]
],
[
[
"np.shape(M)",
"_____no_output_____"
],
[
"np.size(M)",
"_____no_output_____"
]
],
[
[
"So far the `numpy.ndarray` looks awefully much like a Python list (or nested list). Why not simply use Python lists for computations instead of creating a new array type? \n\nThere are several reasons:\n\n* Python lists are very general. They can contain any kind of object. They are dynamically typed. They do not support mathematical functions such as matrix and dot multiplications, etc. Implementing such functions for Python lists would not be very efficient because of the dynamic typing.\n* Numpy arrays are **statically typed** and **homogeneous**. The type of the elements is determined when the array is created.\n* Numpy arrays are memory efficient.\n* Because of the static typing, fast implementation of mathematical functions such as multiplication and addition of `numpy` arrays can be implemented in a compiled language (C and Fortran is used).\n\nUsing the `dtype` (data type) property of an `ndarray`, we can see what type the data of an array has:",
"_____no_output_____"
]
],
[
[
"M.dtype",
"_____no_output_____"
]
],
[
[
"We get an error if we try to assign a value of the wrong type to an element in a numpy array:",
"_____no_output_____"
]
],
[
[
"try:\n M[0,0] = \"hello\"\nexcept ValueError as e:\n print(traceback.format_exc())",
"Traceback (most recent call last):\n File \"<ipython-input-16-91a1594f7e08>\", line 2, in <module>\n M[0,0] = \"hello\"\nValueError: invalid literal for int() with base 10: 'hello'\n\n"
]
],
[
[
"If we want, we can explicitly define the type of the array data when we create it, using the `dtype` keyword argument: ",
"_____no_output_____"
]
],
[
[
"M = np.array([[1, 2], [3, 4]], dtype=complex)\n\nM",
"_____no_output_____"
]
],
[
[
"Common data types that can be used with `dtype` are: `int`, `float`, `complex`, `bool`, `object`, etc.\n\nWe can also explicitly define the bit size of the data types, for example: `int64`, `int16`, `float128`, `complex128`.",
"_____no_output_____"
],
[
"### Using array-generating functions",
"_____no_output_____"
],
[
"For larger arrays it is inpractical to initialize the data manually, using explicit python lists. Instead we can use one of the many functions in `numpy` that generate arrays of different forms. Some of the more common are:",
"_____no_output_____"
],
[
"#### arange",
"_____no_output_____"
]
],
[
[
"# create a range\n\nx = np.arange(0, 10, 1) # arguments: start, stop, step\n\nx",
"_____no_output_____"
],
[
"x = np.arange(-1, 1, 0.1)\n\nx",
"_____no_output_____"
]
],
[
[
"#### linspace and logspace",
"_____no_output_____"
]
],
[
[
"# using linspace, both end points ARE included\nnp.linspace(0, 10, 25)",
"_____no_output_____"
],
[
"np.logspace(0, 10, 10, base=np.e)",
"_____no_output_____"
]
],
[
[
"#### mgrid",
"_____no_output_____"
]
],
[
[
"x, y = np.mgrid[0:5, 0:5] # similar to meshgrid in MATLAB",
"_____no_output_____"
],
[
"x",
"_____no_output_____"
],
[
"y",
"_____no_output_____"
]
],
[
[
"#### random data",
"_____no_output_____"
]
],
[
[
"# uniform random numbers in [0,1]\nnp.random.rand(5,5)",
"_____no_output_____"
],
[
"# standard normal distributed random numbers\nnp.random.randn(5,5)",
"_____no_output_____"
]
],
[
[
"#### diag",
"_____no_output_____"
]
],
[
[
"# a diagonal matrix\nnp.diag([1,2,3])",
"_____no_output_____"
],
[
"# diagonal with offset from the main diagonal\nnp.diag([1,2,3], k=1) ",
"_____no_output_____"
]
],
[
[
"#### zeros and ones",
"_____no_output_____"
]
],
[
[
"np.zeros((3,3))",
"_____no_output_____"
],
[
"np.ones((3,3))",
"_____no_output_____"
]
],
[
[
"## File I/O",
"_____no_output_____"
],
[
"### Comma-separated values (CSV)",
"_____no_output_____"
],
[
"A very common file format for data files is comma-separated values (CSV), or related formats such as TSV (tab-separated values). To read data from such files into Numpy arrays we can use the `numpy.genfromtxt` function. For example, ",
"_____no_output_____"
]
],
[
[
"!head ../data/stockholm_td_adj.dat",
"1800 1 1 -6.1 -6.1 -6.1 1\r\n1800 1 2 -15.4 -15.4 -15.4 1\r\n1800 1 3 -15.0 -15.0 -15.0 1\r\n1800 1 4 -19.3 -19.3 -19.3 1\r\n1800 1 5 -16.8 -16.8 -16.8 1\r\n1800 1 6 -11.4 -11.4 -11.4 1\r\n1800 1 7 -7.6 -7.6 -7.6 1\r\n1800 1 8 -7.1 -7.1 -7.1 1\r\n1800 1 9 -10.1 -10.1 -10.1 1\r\n1800 1 10 -9.5 -9.5 -9.5 1\r\n"
],
[
"data = np.genfromtxt('../data/stockholm_td_adj.dat')",
"_____no_output_____"
],
[
"data.shape",
"_____no_output_____"
],
[
"fig, ax = plt.subplots(figsize=(14,4))\nax.plot(data[:,0]+data[:,1]/12.0+data[:,2]/365, data[:,5])\nax.axis('tight')\nax.set_title('tempeatures in Stockholm')\nax.set_xlabel('year')\nax.set_ylabel('temperature (C)');",
"_____no_output_____"
]
],
[
[
"Using `numpy.savetxt` we can store a Numpy array to a file in CSV format:",
"_____no_output_____"
]
],
[
[
"M = np.random.rand(3,3)\n\nM",
"_____no_output_____"
],
[
"np.savetxt(\"../data/random-matrix.csv\", M)",
"_____no_output_____"
],
[
"!cat ../data/random-matrix.csv",
"6.631877625544441157e-01 7.975119209766545758e-01 8.140936214893396139e-01\r\n6.414899085854186556e-02 5.510325574134632420e-01 4.226693463618673707e-01\r\n4.504356050742730488e-03 5.895893686518726140e-01 2.023804185438885517e-01\r\n"
],
[
"np.savetxt(\"../data/random-matrix.csv\", M, fmt='%.5f') # fmt specifies the format\n\n!cat ../data/random-matrix.csv",
"0.66319 0.79751 0.81409\r\n0.06415 0.55103 0.42267\r\n0.00450 0.58959 0.20238\r\n"
]
],
[
[
"### Numpy's native file format",
"_____no_output_____"
],
[
"Useful when storing and reading back numpy array data. Use the functions `numpy.save` and `numpy.load`:",
"_____no_output_____"
]
],
[
[
"np.save(\"../data/random-matrix.npy\", M)\n\n!file ../data/random-matrix.npy",
"../data/random-matrix.npy: data\r\n"
],
[
"np.load(\"../data/random-matrix.npy\")",
"_____no_output_____"
]
],
[
[
"## More properties of the numpy arrays",
"_____no_output_____"
]
],
[
[
"M.itemsize # bytes per element",
"_____no_output_____"
],
[
"M.nbytes # number of bytes",
"_____no_output_____"
],
[
"M.ndim # number of dimensions",
"_____no_output_____"
]
],
[
[
"## Manipulating arrays",
"_____no_output_____"
],
[
"### Indexing",
"_____no_output_____"
],
[
"We can index elements in an array using square brackets and indices:",
"_____no_output_____"
]
],
[
[
"# v is a vector, and has only one dimension, taking one index\nv[0]",
"_____no_output_____"
],
[
"# M is a matrix, or a 2 dimensional array, taking two indices \nM[1,1]",
"_____no_output_____"
]
],
[
[
"If we omit an index of a multidimensional array it returns the whole row (or, in general, a N-1 dimensional array) ",
"_____no_output_____"
]
],
[
[
"M",
"_____no_output_____"
],
[
"M[1]",
"_____no_output_____"
]
],
[
[
"The same thing can be achieved with using `:` instead of an index: ",
"_____no_output_____"
]
],
[
[
"M[1,:] # row 1",
"_____no_output_____"
],
[
"M[:,1] # column 1",
"_____no_output_____"
]
],
[
[
"We can assign new values to elements in an array using indexing:",
"_____no_output_____"
]
],
[
[
"M[0,0] = 1",
"_____no_output_____"
],
[
"M",
"_____no_output_____"
],
[
"# also works for rows and columns\nM[1,:] = 0\nM[:,2] = -1",
"_____no_output_____"
],
[
"M",
"_____no_output_____"
]
],
[
[
"### Index slicing",
"_____no_output_____"
],
[
"Index slicing is the technical name for the syntax `M[lower:upper:step]` to extract part of an array:",
"_____no_output_____"
]
],
[
[
"A = np.array([1,2,3,4,5])\nA",
"_____no_output_____"
],
[
"A[1:3]",
"_____no_output_____"
]
],
[
[
"Array slices are *mutable*: if they are assigned a new value the original array from which the slice was extracted is modified:",
"_____no_output_____"
]
],
[
[
"A[1:3] = [-2,-3]\n\nA",
"_____no_output_____"
]
],
[
[
"We can omit any of the three parameters in `M[lower:upper:step]`:",
"_____no_output_____"
]
],
[
[
"A[::] # lower, upper, step all take the default values",
"_____no_output_____"
],
[
"A[::2] # step is 2, lower and upper defaults to the beginning and end of the array",
"_____no_output_____"
],
[
"A[:3] # first three elements",
"_____no_output_____"
],
[
"A[3:] # elements from index 3",
"_____no_output_____"
]
],
[
[
"Negative indices counts from the end of the array (positive index from the begining):",
"_____no_output_____"
]
],
[
[
"A = np.array([1,2,3,4,5])",
"_____no_output_____"
],
[
"A[-1] # the last element in the array",
"_____no_output_____"
],
[
"A[-3:] # the last three elements",
"_____no_output_____"
]
],
[
[
"Index slicing works exactly the same way for multidimensional arrays:",
"_____no_output_____"
]
],
[
[
"A = np.array([[n+m*10 for n in range(5)] for m in range(5)])\n\nA",
"_____no_output_____"
],
[
"# a block from the original array\nA[1:4, 1:4]",
"_____no_output_____"
],
[
"# strides\nA[::2, ::2]",
"_____no_output_____"
]
],
[
[
"### Fancy indexing",
"_____no_output_____"
],
[
"Fancy indexing is the name for when an array or list is used in-place of an index: ",
"_____no_output_____"
]
],
[
[
"row_indices = [1, 2, 3]\nA[row_indices]",
"_____no_output_____"
],
[
"col_indices = [1, 2, -1] # remember, index -1 means the last element\nA[row_indices, col_indices]",
"_____no_output_____"
]
],
[
[
"We can also use index masks: If the index mask is an Numpy array of data type `bool`, then an element is selected (True) or not (False) depending on the value of the index mask at the position of each element: ",
"_____no_output_____"
]
],
[
[
"B = np.array([n for n in range(5)])\nB",
"_____no_output_____"
],
[
"row_mask = np.array([True, False, True, False, False])\nB[row_mask]",
"_____no_output_____"
],
[
"# same thing\nrow_mask = np.array([1,0,1,0,0], dtype=bool)\nB[row_mask]",
"_____no_output_____"
]
],
[
[
"This feature is very useful to conditionally select elements from an array, using for example comparison operators:",
"_____no_output_____"
]
],
[
[
"x = np.arange(0, 10, 0.5)\nx",
"_____no_output_____"
],
[
"mask = (5 < x) * (x < 7.5)\n\nmask",
"_____no_output_____"
],
[
"x[mask]",
"_____no_output_____"
]
],
[
[
"## Functions for extracting data from arrays and creating arrays",
"_____no_output_____"
],
[
"### where",
"_____no_output_____"
],
[
"The index mask can be converted to position index using the `where` function",
"_____no_output_____"
]
],
[
[
"indices = np.where(mask)\n\nindices",
"_____no_output_____"
],
[
"x[indices] # this indexing is equivalent to the fancy indexing x[mask]",
"_____no_output_____"
]
],
[
[
"### diag",
"_____no_output_____"
],
[
"With the diag function we can also extract the diagonal and subdiagonals of an array:",
"_____no_output_____"
]
],
[
[
"np.diag(A)",
"_____no_output_____"
],
[
"np.diag(A, -1)",
"_____no_output_____"
]
],
[
[
"### take",
"_____no_output_____"
],
[
"The `take` function is similar to fancy indexing described above:",
"_____no_output_____"
]
],
[
[
"v2 = np.arange(-3,3)\nv2",
"_____no_output_____"
],
[
"row_indices = [1, 3, 5]\nv2[row_indices] # fancy indexing",
"_____no_output_____"
],
[
"v2.take(row_indices)",
"_____no_output_____"
]
],
[
[
"But `take` also works on lists and other objects:",
"_____no_output_____"
]
],
[
[
"np.take([-3, -2, -1, 0, 1, 2], row_indices)",
"_____no_output_____"
]
],
[
[
"### choose",
"_____no_output_____"
],
[
"Constructs an array by picking elements from several arrays:",
"_____no_output_____"
]
],
[
[
"which = [1, 0, 1, 0]\nchoices = [[-2,-2,-2,-2], [5,5,5,5]]\n\nnp.choose(which, choices)",
"_____no_output_____"
]
],
[
[
"## Linear algebra",
"_____no_output_____"
],
[
"Vectorizing code is the key to writing efficient numerical calculation with Python/Numpy. That means that as much as possible of a program should be formulated in terms of matrix and vector operations, like matrix-matrix multiplication.",
"_____no_output_____"
],
[
"### Scalar-array operations",
"_____no_output_____"
],
[
"We can use the usual arithmetic operators to multiply, add, subtract, and divide arrays with scalar numbers.",
"_____no_output_____"
]
],
[
[
"v1 = np.arange(0, 5)",
"_____no_output_____"
],
[
"v1 * 2",
"_____no_output_____"
],
[
"v1 + 2",
"_____no_output_____"
],
[
"A * 2, A + 2",
"_____no_output_____"
]
],
[
[
"### Element-wise array-array operations",
"_____no_output_____"
],
[
"When we add, subtract, multiply and divide arrays with each other, the default behaviour is **element-wise** operations:",
"_____no_output_____"
]
],
[
[
"A * A # element-wise multiplication",
"_____no_output_____"
],
[
"v1 * v1",
"_____no_output_____"
]
],
[
[
"If we multiply arrays with compatible shapes, we get an element-wise multiplication of each row:",
"_____no_output_____"
]
],
[
[
"A.shape, v1.shape",
"_____no_output_____"
],
[
"A * v1",
"_____no_output_____"
]
],
[
[
"### Matrix algebra",
"_____no_output_____"
],
[
"What about matrix mutiplication? There are two ways. We can either use the `dot` function, which applies a matrix-matrix, matrix-vector, or inner vector multiplication to its two arguments: ",
"_____no_output_____"
]
],
[
[
"np.dot(A, A)",
"_____no_output_____"
]
],
[
[
"Python 3 has a new operator for using infix notation with matrix multiplication.",
"_____no_output_____"
]
],
[
[
"A @ A",
"_____no_output_____"
],
[
"np.dot(A, v1)",
"_____no_output_____"
],
[
"np.dot(v1, v1)",
"_____no_output_____"
]
],
[
[
"Alternatively, we can cast the array objects to the type `matrix`. This changes the behavior of the standard arithmetic operators `+, -, *` to use matrix algebra.",
"_____no_output_____"
]
],
[
[
"M = np.matrix(A)\nv = np.matrix(v1).T # make it a column vector",
"_____no_output_____"
],
[
"v",
"_____no_output_____"
],
[
"M * M",
"_____no_output_____"
],
[
"M * v",
"_____no_output_____"
],
[
"# inner product\nv.T * v",
"_____no_output_____"
],
[
"# with matrix objects, standard matrix algebra applies\nv + M*v",
"_____no_output_____"
]
],
[
[
"If we try to add, subtract or multiply objects with incomplatible shapes we get an error:",
"_____no_output_____"
]
],
[
[
"v = np.matrix([1,2,3,4,5,6]).T",
"_____no_output_____"
],
[
"M.shape, v.shape",
"_____no_output_____"
],
[
"import traceback\n\ntry:\n M * v\nexcept ValueError as e:\n print(traceback.format_exc())",
"Traceback (most recent call last):\n File \"<ipython-input-104-06fa348e920c>\", line 4, in <module>\n M * v\n File \"/Users/johria/anaconda/lib/python3.5/site-packages/numpy/matrixlib/defmatrix.py\", line 343, in __mul__\n return N.dot(self, asmatrix(other))\nValueError: shapes (5,5) and (6,1) not aligned: 5 (dim 1) != 6 (dim 0)\n\n"
]
],
[
[
"See also the related functions: `inner`, `outer`, `cross`, `kron`, `tensordot`. Try for example `help(np.kron)`.",
"_____no_output_____"
],
[
"### Array/Matrix transformations",
"_____no_output_____"
],
[
"Above we have used the `.T` to transpose the matrix object `v`. We could also have used the `transpose` function to accomplish the same thing. \n\nOther mathematical functions that transform matrix objects are:",
"_____no_output_____"
]
],
[
[
"C = np.matrix([[1j, 2j], [3j, 4j]])\nC",
"_____no_output_____"
],
[
"np.conjugate(C)",
"_____no_output_____"
]
],
[
[
"Hermitian conjugate: transpose + conjugate",
"_____no_output_____"
]
],
[
[
"C.H",
"_____no_output_____"
]
],
[
[
"We can extract the real and imaginary parts of complex-valued arrays using `real` and `imag`:",
"_____no_output_____"
]
],
[
[
"np.real(C) # same as: C.real",
"_____no_output_____"
],
[
"np.imag(C) # same as: C.imag",
"_____no_output_____"
]
],
[
[
"Or the complex argument and absolute value",
"_____no_output_____"
]
],
[
[
"np.angle(C+1) # heads up MATLAB Users, angle is used instead of arg",
"_____no_output_____"
],
[
"abs(C)",
"_____no_output_____"
]
],
[
[
"### Matrix computations",
"_____no_output_____"
],
[
"#### Inverse",
"_____no_output_____"
]
],
[
[
"np.linalg.inv(C) # equivalent to C.I ",
"_____no_output_____"
],
[
"C.I * C",
"_____no_output_____"
]
],
[
[
"#### Determinant",
"_____no_output_____"
]
],
[
[
"np.linalg.det(C)",
"_____no_output_____"
],
[
"np.linalg.det(C.I)",
"_____no_output_____"
]
],
[
[
"### Data processing",
"_____no_output_____"
],
[
"Often it is useful to store datasets in Numpy arrays. Numpy provides a number of functions to calculate statistics of datasets in arrays. \n\nFor example, let's calculate some properties from the Stockholm temperature dataset used above.",
"_____no_output_____"
]
],
[
[
"# reminder, the tempeature dataset is stored in the data variable:\nnp.shape(data)",
"_____no_output_____"
]
],
[
[
"#### mean",
"_____no_output_____"
]
],
[
[
"# the temperature data is in column 3\nnp.mean(data[:,3])",
"_____no_output_____"
]
],
[
[
"The daily mean temperature in Stockholm over the last 200 years has been about 6.2 C.",
"_____no_output_____"
],
[
"#### standard deviations and variance",
"_____no_output_____"
]
],
[
[
"np.std(data[:,3]), np.var(data[:,3])",
"_____no_output_____"
]
],
[
[
"#### min and max",
"_____no_output_____"
]
],
[
[
"# lowest daily average temperature\ndata[:,3].min()",
"_____no_output_____"
],
[
"# highest daily average temperature\ndata[:,3].max()",
"_____no_output_____"
]
],
[
[
"#### sum, prod, and trace",
"_____no_output_____"
]
],
[
[
"d = np.arange(0, 10)\nd",
"_____no_output_____"
],
[
"# sum up all elements\nnp.sum(d)",
"_____no_output_____"
],
[
"# product of all elements\nnp.prod(d+1)",
"_____no_output_____"
],
[
"# cummulative sum\nnp.cumsum(d)",
"_____no_output_____"
],
[
"# cummulative product\nnp.cumprod(d+1)",
"_____no_output_____"
],
[
"# same as: diag(A).sum()\nnp.trace(A)",
"_____no_output_____"
]
],
[
[
"### Computations on subsets of arrays",
"_____no_output_____"
],
[
"We can compute with subsets of the data in an array using indexing, fancy indexing, and the other methods of extracting data from an array (described above).\n\nFor example, let's go back to the temperature dataset:",
"_____no_output_____"
]
],
[
[
"!head -n 3 ../data/stockholm_td_adj.dat",
"1800 1 1 -6.1 -6.1 -6.1 1\r\n1800 1 2 -15.4 -15.4 -15.4 1\r\n1800 1 3 -15.0 -15.0 -15.0 1\r\n"
]
],
[
[
"The dataformat is: year, month, day, daily average temperature, low, high, location.\n\nIf we are interested in the average temperature only in a particular month, say February, then we can create a index mask and use it to select only the data for that month using:",
"_____no_output_____"
]
],
[
[
"np.unique(data[:,1]) # the month column takes values from 1 to 12",
"_____no_output_____"
],
[
"mask_feb = data[:,1] == 2",
"_____no_output_____"
],
[
"# the temperature data is in column 3\nnp.mean(data[mask_feb,3])",
"_____no_output_____"
]
],
[
[
"With these tools we have very powerful data processing capabilities at our disposal. For example, to extract the average monthly average temperatures for each month of the year only takes a few lines of code: ",
"_____no_output_____"
]
],
[
[
"months = np.arange(1,13)\nmonthly_mean = [np.mean(data[data[:,1] == month, 3]) for month in months]\n\nfig, ax = plt.subplots()\nax.bar(months, monthly_mean)\nax.set_xlabel(\"Month\")\nax.set_ylabel(\"Monthly avg. temp.\");",
"_____no_output_____"
]
],
[
[
"### Calculations with higher-dimensional data",
"_____no_output_____"
],
[
"When functions such as `min`, `max`, etc. are applied to a multidimensional arrays, it is sometimes useful to apply the calculation to the entire array, and sometimes only on a row or column basis. Using the `axis` argument we can specify how these functions should behave: ",
"_____no_output_____"
]
],
[
[
"m = np.random.rand(3,3)\nm",
"_____no_output_____"
],
[
"# global max\nm.max()",
"_____no_output_____"
],
[
"# max in each column\nm.max(axis=0)",
"_____no_output_____"
],
[
"# max in each row\nm.max(axis=1)",
"_____no_output_____"
]
],
[
[
"Many other functions and methods in the `array` and `matrix` classes accept the same (optional) `axis` keyword argument.",
"_____no_output_____"
],
[
"## Reshaping, resizing and stacking arrays",
"_____no_output_____"
],
[
"The shape of an Numpy array can be modified without copying the underlaying data, which makes it a fast operation even for large arrays.",
"_____no_output_____"
]
],
[
[
"A",
"_____no_output_____"
],
[
"n, m = A.shape",
"_____no_output_____"
],
[
"B = A.reshape((1,n*m))\nB",
"_____no_output_____"
],
[
"B[0,0:5] = 5 # modify the array\n\nB",
"_____no_output_____"
],
[
"A # and the original variable is also changed. B is only a different view of the same data",
"_____no_output_____"
]
],
[
[
"We can also use the function `flatten` to make a higher-dimensional array into a vector. But this function create a copy of the data.",
"_____no_output_____"
]
],
[
[
"B = A.flatten()\n\nB",
"_____no_output_____"
],
[
"B[0:5] = 10\n\nB",
"_____no_output_____"
],
[
"A # now A has not changed, because B's data is a copy of A's, not refering to the same data",
"_____no_output_____"
]
],
[
[
"## Adding a new dimension: newaxis",
"_____no_output_____"
],
[
"With `newaxis`, we can insert new dimensions in an array, for example converting a vector to a column or row matrix:",
"_____no_output_____"
]
],
[
[
"v = np.array([1,2,3])",
"_____no_output_____"
],
[
"v.shape",
"_____no_output_____"
],
[
"# make a column matrix of the vector v\nv[:, np.newaxis]",
"_____no_output_____"
],
[
"# column matrix\nv[:, np.newaxis].shape",
"_____no_output_____"
],
[
"# row matrix\nv[np.newaxis, :].shape",
"_____no_output_____"
]
],
[
[
"## Stacking and repeating arrays",
"_____no_output_____"
],
[
"Using function `repeat`, `tile`, `vstack`, `hstack`, and `concatenate` we can create larger vectors and matrices from smaller ones:",
"_____no_output_____"
],
[
"### tile and repeat",
"_____no_output_____"
]
],
[
[
"a = np.array([[1, 2], [3, 4]])",
"_____no_output_____"
],
[
"# repeat each element 3 times\nnp.repeat(a, 3)",
"_____no_output_____"
],
[
"# tile the matrix 3 times \nnp.tile(a, 3)",
"_____no_output_____"
]
],
[
[
"### concatenate",
"_____no_output_____"
]
],
[
[
"b = np.array([[5, 6]])",
"_____no_output_____"
],
[
"np.concatenate((a, b), axis=0)",
"_____no_output_____"
],
[
"np.concatenate((a, b.T), axis=1)",
"_____no_output_____"
]
],
[
[
"### hstack and vstack",
"_____no_output_____"
]
],
[
[
"np.vstack((a,b))",
"_____no_output_____"
],
[
"np.hstack((a,b.T))",
"_____no_output_____"
]
],
[
[
"## Copy and \"deep copy\"",
"_____no_output_____"
],
[
"To achieve high performance, assignments in Python usually do not copy the underlaying objects. This is important for example when objects are passed between functions, to avoid an excessive amount of memory copying when it is not necessary (technical term: pass by reference). ",
"_____no_output_____"
]
],
[
[
"A = np.array([[1, 2], [3, 4]])\n\nA",
"_____no_output_____"
],
[
"# now B is referring to the same array data as A \nB = A ",
"_____no_output_____"
],
[
"# changing B affects A\nB[0,0] = 10\n\nB",
"_____no_output_____"
],
[
"A",
"_____no_output_____"
]
],
[
[
"If we want to avoid this behavior, so that when we get a new completely independent object `B` copied from `A`, then we need to do a so-called \"deep copy\" using the function `copy`:",
"_____no_output_____"
]
],
[
[
"B = np.copy(A)",
"_____no_output_____"
],
[
"# now, if we modify B, A is not affected\nB[0,0] = -5\n\nB",
"_____no_output_____"
],
[
"A",
"_____no_output_____"
]
],
[
[
"## Iterating over array elements",
"_____no_output_____"
],
[
"Generally, we want to avoid iterating over the elements of arrays whenever we can (at all costs). The reason is that in a interpreted language like Python (or MATLAB/R), iterations are really slow compared to vectorized operations. \n\nHowever, sometimes iterations are unavoidable. For such cases, the Python `for` loop is the most convenient way to iterate over an array:",
"_____no_output_____"
]
],
[
[
"v = np.array([1,2,3,4])\n\nfor element in v:\n print(element)",
"1\n2\n3\n4\n"
],
[
"M = np.array([[1,2], [3,4]])\n\nfor row in M:\n print(\"row\", row)\n \n for element in row:\n print(element)",
"row [1 2]\n1\n2\nrow [3 4]\n3\n4\n"
]
],
[
[
"When we need to iterate over each element of an array and modify its elements, it is convenient to use the `enumerate` function to obtain both the element and its index in the `for` loop: ",
"_____no_output_____"
]
],
[
[
"for row_idx, row in enumerate(M):\n print(\"row_idx\", row_idx, \"row\", row)\n \n for col_idx, element in enumerate(row):\n print(\"col_idx\", col_idx, \"element\", element)\n \n # update the matrix M: square each element\n M[row_idx, col_idx] = element ** 2",
"row_idx 0 row [1 2]\ncol_idx 0 element 1\ncol_idx 1 element 2\nrow_idx 1 row [3 4]\ncol_idx 0 element 3\ncol_idx 1 element 4\n"
],
[
"# each element in M is now squared\nM",
"_____no_output_____"
]
],
[
[
"## Vectorizing functions",
"_____no_output_____"
],
[
"As mentioned several times by now, to get good performance we should try to avoid looping over elements in our vectors and matrices, and instead use vectorized algorithms. The first step in converting a scalar algorithm to a vectorized algorithm is to make sure that the functions we write work with vector inputs.",
"_____no_output_____"
]
],
[
[
"def theta(x):\n \"\"\"\n Scalar implemenation of the Heaviside step function.\n \"\"\"\n if x >= 0:\n return 1\n else:\n return 0",
"_____no_output_____"
],
[
"try:\n theta(np.array([-3,-2,-1,0,1,2,3]))\nexcept Exception as e:\n print(traceback.format_exc())",
"Traceback (most recent call last):\n File \"<ipython-input-169-c71653502221>\", line 2, in <module>\n theta(np.array([-3,-2,-1,0,1,2,3]))\n File \"<ipython-input-168-5cd8eb6ce61c>\", line 5, in theta\n if x >= 0:\nValueError: The truth value of an array with more than one element is ambiguous. Use a.any() or a.all()\n\n"
]
],
[
[
"OK, that didn't work because we didn't write the `Theta` function so that it can handle a vector input... \n\nTo get a vectorized version of Theta we can use the Numpy function `vectorize`. In many cases it can automatically vectorize a function:",
"_____no_output_____"
]
],
[
[
"theta_vec = np.vectorize(theta)",
"_____no_output_____"
],
[
"%%time \n\ntheta_vec(np.array([-3,-2,-1,0,1,2,3]))",
"CPU times: user 121 µs, sys: 12 µs, total: 133 µs\nWall time: 140 µs\n"
]
],
[
[
"We can also implement the function to accept a vector input from the beginning (requires more effort but might give better performance):",
"_____no_output_____"
]
],
[
[
"def theta(x):\n \"\"\"\n Vector-aware implemenation of the Heaviside step function.\n \"\"\"\n return 1 * (x >= 0)",
"_____no_output_____"
],
[
"%%time\n\ntheta(np.array([-3,-2,-1,0,1,2,3]))",
"CPU times: user 38 µs, sys: 9 µs, total: 47 µs\nWall time: 50.1 µs\n"
],
[
"# still works for scalars as well\ntheta(-1.2), theta(2.6)",
"_____no_output_____"
]
],
[
[
"## Using arrays in conditions",
"_____no_output_____"
],
[
"When using arrays in conditions,for example `if` statements and other boolean expressions, one needs to use `any` or `all`, which requires that any or all elements in the array evalutes to `True`:",
"_____no_output_____"
]
],
[
[
"M",
"_____no_output_____"
],
[
"if (M > 5).any():\n print(\"at least one element in M is larger than 5\")\nelse:\n print(\"no element in M is larger than 5\")",
"at least one element in M is larger than 5\n"
],
[
"if (M > 5).all():\n print(\"all elements in M are larger than 5\")\nelse:\n print(\"all elements in M are not larger than 5\")",
"all elements in M are not larger than 5\n"
]
],
[
[
"## Type casting",
"_____no_output_____"
],
[
"Since Numpy arrays are *statically typed*, the type of an array does not change once created. But we can explicitly cast an array of some type to another using the `astype` functions (see also the similar `asarray` function). This always create a new array of new type:",
"_____no_output_____"
]
],
[
[
"M.dtype",
"_____no_output_____"
],
[
"M2 = M.astype(float)\n\nM2",
"_____no_output_____"
],
[
"M2.dtype",
"_____no_output_____"
],
[
"M3 = M.astype(bool)\n\nM3",
"_____no_output_____"
]
],
[
[
"## Further reading",
"_____no_output_____"
],
[
"* http://numpy.scipy.org - Official Numpy Documentation\n* http://scipy.org/Tentative_NumPy_Tutorial - Official Numpy Quickstart Tutorial (highly recommended)\n* http://www.scipy-lectures.org/intro/numpy/index.html - Scipy Lectures: Lecture 1.3",
"_____no_output_____"
],
[
"## Versions",
"_____no_output_____"
]
],
[
[
"%reload_ext version_information\n%version_information numpy",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
]
] |
4affbbf456f441f0c176765cf57c0982ddecd318
| 153,495 |
ipynb
|
Jupyter Notebook
|
bacillusme/analysis/spore/energy_analysis.ipynb
|
jdtibochab/bacillusme
|
d7eea04ca8a8a72731f35127142a6cb05ad05754
|
[
"MIT"
] | null | null | null |
bacillusme/analysis/spore/energy_analysis.ipynb
|
jdtibochab/bacillusme
|
d7eea04ca8a8a72731f35127142a6cb05ad05754
|
[
"MIT"
] | null | null | null |
bacillusme/analysis/spore/energy_analysis.ipynb
|
jdtibochab/bacillusme
|
d7eea04ca8a8a72731f35127142a6cb05ad05754
|
[
"MIT"
] | null | null | null | 66.678975 | 23,960 | 0.623942 |
[
[
[
"# Transporter analysis of bacillus mother-spore",
"_____no_output_____"
]
],
[
[
"from __future__ import print_function, division, absolute_import\n\nimport sys\n\nimport qminospy\nfrom qminospy.me2 import ME_NLP\n\n# python imports\nfrom copy import copy\nimport re\nfrom os.path import join, dirname, abspath\nimport sys\nsys.path.append('/home/UCSD/cobra_utils')\nfrom collections import defaultdict\nimport pickle\n\n# third party imports\nimport pandas\nimport cobra\nfrom tqdm import tqdm\nimport numpy as np\nimport scipy\n\n# COBRAme\nimport cobrame\nfrom cobrame.util import building, mu, me_model_interface\nfrom cobrame.io.json import save_json_me_model, save_reduced_json_me_model\n\n# ECOLIme\nimport bacillusme\nfrom bacillusme import (transcription, translation, flat_files, generics, formulas, compartments)\nfrom cobrame.util.helper_functions import *\n\nimport copy\nfrom scipy import stats\nimport matplotlib.pyplot as plt\n%load_ext autoreload\n%autoreload 2\nprint(cobra.__file__)\nprint(cobrame.__file__)\nprint(bacillusme.__file__)\necoli_files = dirname(abspath(bacillusme.__file__))\npd.set_option('display.max_colwidth', None)",
"/home/jt/me_modeling/lib/python3.6/site-packages/cobra-0.5.11-py3.6-linux-x86_64.egg/cobra/io/sbml3.py:24: UserWarning: Install lxml for faster SBML I/O\n warn(\"Install lxml for faster SBML I/O\")\n/home/jt/me_modeling/lib/python3.6/site-packages/cobra-0.5.11-py3.6-linux-x86_64.egg/cobra/io/__init__.py:12: UserWarning: cobra.io.sbml requires libsbml\n warn(\"cobra.io.sbml requires libsbml\")\n"
],
[
"with open(\"../../me_models/solution.pickle\", \"rb\") as outfile:\n me = pickle.load(outfile)",
"_____no_output_____"
]
],
[
[
"### Closing mechanisms",
"_____no_output_____"
]
],
[
[
"with open(\"./sporeme_solution_v3.pickle\", \"rb\") as outfile:\n sporeme = pickle.load(outfile)",
"_____no_output_____"
],
[
"sporeme.solution.x_dict['biomass_dilution_s']",
"_____no_output_____"
],
[
"main_mechanisms = [ 'ACKr_REV_BSU29470-MONOMER',\n 'PGK_REV_BSU33930-MONOMER',\n 'PYK_FWD_BSU29180-MONOMER_mod_mn2_mod_k']",
"_____no_output_____"
],
[
"for r in main_mechanisms:\n sporeme.reactions.get_by_id(r).bounds = (0,0)\n sporeme.reactions.get_by_id(r+'_s').bounds = (0,0)",
"_____no_output_____"
],
[
"version = 'v5_KO_ACK_PGK_PYK'",
"_____no_output_____"
],
[
"solve_me_model(sporeme, max_mu = 0.1, min_mu = .01, using_soplex=False, precision = 1e-3,growth_key = 'sigma')",
"iter\tmuopt \ta \tb \tmu1 \tstat1\nFinished compiling expressions in 248.511381 seconds\nFinished substituting S,lb,ub in 7.094637 seconds\nFinished makeME_LP in 1.522246 seconds\nGetting MINOS parameters from ME_NLP...\n1 0.0 0.0 0.05 0.05 1\nFinished substituting S,lb,ub in 7.121589 seconds\nFinished makeME_LP in 1.508124 seconds\nGetting MINOS parameters from ME_NLP...\n2 0.0 0.0 0.025 0.025 1\nFinished substituting S,lb,ub in 7.103040 seconds\nFinished makeME_LP in 1.499345 seconds\nGetting MINOS parameters from ME_NLP...\n3 0.0 0.0 0.0125 0.0125 1\nFinished substituting S,lb,ub in 7.101150 seconds\nFinished makeME_LP in 1.501006 seconds\nGetting MINOS parameters from ME_NLP...\n4 0.00625 0.00625 0.0125 0.00625 optimal\nFinished substituting S,lb,ub in 7.148437 seconds\nFinished makeME_LP in 1.523553 seconds\nGetting MINOS parameters from ME_NLP...\n5 0.009375000000000001 0.009375000000000001 0.0125 0.009375000000000001 optimal\nFinished substituting S,lb,ub in 7.080312 seconds\nFinished makeME_LP in 1.506168 seconds\nGetting MINOS parameters from ME_NLP...\n6 0.009375000000000001 0.009375000000000001 0.010937500000000001 0.010937500000000001 1\nFinished substituting S,lb,ub in 6.898724 seconds\nFinished makeME_LP in 1.521826 seconds\nGetting MINOS parameters from ME_NLP...\n7 0.010156250000000002 0.010156250000000002 0.010937500000000001 0.010156250000000002 optimal\nBisection done in 1401.1 seconds\n"
],
[
"sporeme.solution.x_dict['biomass_dilution_s']",
"_____no_output_____"
],
[
"sporeme.reactions.get_by_id('PRPPS_REV_BSU00510-MONOMER_mod_mn2_mod_pi_s').bounds = (0,0)\nsolve_me_model(sporeme, max_mu = 0.1, min_mu = .01, using_soplex=False, precision = 1e-3,growth_key = 'sigma')",
"iter\tmuopt \ta \tb \tmu1 \tstat1\nFinished compiling expressions in 247.352470 seconds\nFinished substituting S,lb,ub in 7.186478 seconds\nFinished makeME_LP in 1.510343 seconds\nGetting MINOS parameters from ME_NLP...\n1 0.0 0.0 0.05 0.05 1\nFinished substituting S,lb,ub in 7.239314 seconds\nFinished makeME_LP in 1.509732 seconds\nGetting MINOS parameters from ME_NLP...\n2 0.0 0.0 0.025 0.025 1\nFinished substituting S,lb,ub in 7.224814 seconds\nFinished makeME_LP in 1.514063 seconds\nGetting MINOS parameters from ME_NLP...\n3 0.0 0.0 0.0125 0.0125 1\nFinished substituting S,lb,ub in 7.171512 seconds\nFinished makeME_LP in 1.501719 seconds\nGetting MINOS parameters from ME_NLP...\n4 0.0 0.0 0.00625 0.00625 1\nFinished substituting S,lb,ub in 7.181740 seconds\nFinished makeME_LP in 1.519977 seconds\nGetting MINOS parameters from ME_NLP...\n5 0.0 0.0 0.003125 0.003125 1\nFinished substituting S,lb,ub in 7.212373 seconds\nFinished makeME_LP in 1.506061 seconds\nGetting MINOS parameters from ME_NLP...\n6 0.0 0.0 0.0015625 0.0015625 1\nFinished substituting S,lb,ub in 7.172508 seconds\nFinished makeME_LP in 1.503221 seconds\nGetting MINOS parameters from ME_NLP...\n7 0.0 0.0 0.00078125 0.00078125 1\nBisection done in 2483.25 seconds\n"
],
[
"if sporeme.solution: sporeme.solution.x_dict['biomass_dilution_s']",
"_____no_output_____"
]
],
[
[
"### GK",
"_____no_output_____"
]
],
[
[
"with open(\"./sporeme_solution_v3.pickle\", \"rb\") as outfile:\n sporeme = pickle.load(outfile)",
"_____no_output_____"
],
[
"for r in sporeme.reactions.query(re.compile('BSU15680-MONOMER.*_s$')):\n print(r.id)\n r.bounds = (0,0)",
"formation_BSU15680-MONOMER_mod_mg2_s\nDGK1_REV_BSU15680-MONOMER_mod_mg2_s\nDGK1_FWD_BSU15680-MONOMER_mod_mg2_s\nGK1_REV_BSU15680-MONOMER_mod_mg2_s\nGK1_FWD_BSU15680-MONOMER_mod_mg2_s\nGK2_REV_BSU15680-MONOMER_mod_mg2_s\nGK2_FWD_BSU15680-MONOMER_mod_mg2_s\n"
],
[
"solve_me_model(sporeme, max_mu = 0.1, min_mu = .01, using_soplex=False, precision = 1e-3,growth_key = 'sigma')",
"iter\tmuopt \ta \tb \tmu1 \tstat1\nFinished compiling expressions in 249.921906 seconds\nFinished substituting S,lb,ub in 7.332601 seconds\nFinished makeME_LP in 1.530706 seconds\nGetting MINOS parameters from ME_NLP...\n1 0.0 0.0 0.05 0.05 1\nFinished substituting S,lb,ub in 7.339327 seconds\nFinished makeME_LP in 1.531912 seconds\nGetting MINOS parameters from ME_NLP...\n2 0.0 0.0 0.025 0.025 1\nFinished substituting S,lb,ub in 7.365429 seconds\nFinished makeME_LP in 1.534005 seconds\nGetting MINOS parameters from ME_NLP...\n"
],
[
"if sporeme.solution: sporeme.solution.x_dict['biomass_dilution_s']",
"_____no_output_____"
],
[
"flux_based_reactions(sporeme,'fum_c',only_types=['MetabolicReaction']).head(10)",
"_____no_output_____"
]
],
[
[
"### CYTK",
"_____no_output_____"
]
],
[
[
"with open(\"./sporeme_solution_v3.pickle\", \"rb\") as outfile:\n sporeme = pickle.load(outfile)",
"_____no_output_____"
],
[
"for r in sporeme.reactions.query('BSU22890-MONOMER'):\n print(r.id)\n r.bounds = (0,0)",
"formation_BSU22890-MONOMER_mod_mg2\nCYTK1_REV_BSU22890-MONOMER_mod_mg2\nCYTK1_FWD_BSU22890-MONOMER_mod_mg2\nCYTK2_REV_BSU22890-MONOMER_mod_mg2\nCYTK2_FWD_BSU22890-MONOMER_mod_mg2\nUMPK_FWD_BSU22890-MONOMER_mod_mg2\nURIDK2r_copy1_FWD_BSU22890-MONOMER_mod_mg2\nformation_BSU22890-MONOMER_mod_mg2_s\nCYTK1_REV_BSU22890-MONOMER_mod_mg2_s\nCYTK1_FWD_BSU22890-MONOMER_mod_mg2_s\nCYTK2_REV_BSU22890-MONOMER_mod_mg2_s\nCYTK2_FWD_BSU22890-MONOMER_mod_mg2_s\nUMPK_FWD_BSU22890-MONOMER_mod_mg2_s\nURIDK2r_copy1_FWD_BSU22890-MONOMER_mod_mg2_s\n"
],
[
"for r in sporeme.reactions.query(re.compile('BSU37150-MONOMER.*_s$')):\n print(r.id,r.reaction)\n r.bounds = (0,0)",
"formation_BSU37150-MONOMER_mod_mg2_s mg2_s + 4.0 protein_BSU37150_s --> BSU37150-MONOMER_mod_mg2_s + 0.04861 prosthetic_group_biomass_s\nCTPS1_FWD_BSU37150-MONOMER_mod_mg2_s atp_s + nh4_s + utp_s --> -4.27350427350427e-6*sigma BSU37150-MONOMER_mod_mg2_s + adp_s + ctp_s + 2.0 h_s + pi_s\nCTPS2_FWD_BSU37150-MONOMER_mod_mg2_s atp_s + gln__L_s + h2o_s + utp_s --> -3.27559071090038e-6*sigma BSU37150-MONOMER_mod_mg2_s + adp_s + ctp_s + glu__L_s + 2.0 h_s + pi_s\n"
],
[
"solve_me_model(sporeme, max_mu = 0.1, min_mu = .01, using_soplex=False, precision = 1e-3,growth_key = 'sigma')",
"iter\tmuopt \ta \tb \tmu1 \tstat1\nFinished compiling expressions in 251.678204 seconds\nFinished substituting S,lb,ub in 7.290562 seconds\nFinished makeME_LP in 1.530940 seconds\nGetting MINOS parameters from ME_NLP...\n1 0.0 0.0 0.05 0.05 1\nFinished substituting S,lb,ub in 7.328154 seconds\nFinished makeME_LP in 1.526656 seconds\nGetting MINOS parameters from ME_NLP...\n2 0.0 0.0 0.025 0.025 1\nFinished substituting S,lb,ub in 7.262715 seconds\nFinished makeME_LP in 1.528114 seconds\nGetting MINOS parameters from ME_NLP...\n3 0.0 0.0 0.0125 0.0125 1\nFinished substituting S,lb,ub in 7.295166 seconds\nFinished makeME_LP in 1.525097 seconds\nGetting MINOS parameters from ME_NLP...\n4 0.0 0.0 0.00625 0.00625 1\nFinished substituting S,lb,ub in 7.310462 seconds\nFinished makeME_LP in 1.527177 seconds\nGetting MINOS parameters from ME_NLP...\n5 0.0 0.0 0.003125 0.003125 1\nFinished substituting S,lb,ub in 7.304533 seconds\nFinished makeME_LP in 1.531065 seconds\nGetting MINOS parameters from ME_NLP...\n6 0.0 0.0 0.0015625 0.0015625 1\nFinished substituting S,lb,ub in 7.262285 seconds\nFinished makeME_LP in 1.526000 seconds\nGetting MINOS parameters from ME_NLP...\n7 0.0 0.0 0.00078125 0.00078125 1\nBisection done in 2860.59 seconds\n"
],
[
"if sporeme.solution: sporeme.solution.x_dict['biomass_dilution_s']",
"_____no_output_____"
],
[
"flux_based_reactions(sporeme,'ctp_s',only_types=['MetabolicReaction'])",
"_____no_output_____"
],
[
"flux_based_reactions(sporeme,'cbp_c',only_types=['MetabolicReaction'])",
"_____no_output_____"
]
],
[
[
"### Methionine",
"_____no_output_____"
]
],
[
[
"with open(\"./sporeme_solution_v3.pickle\", \"rb\") as outfile:\n sporeme = pickle.load(outfile)",
"_____no_output_____"
],
[
"for r in get_transport_reactions(sporeme,'met__L_s',comps=['c','s']):\n print(r.id)\n r.bounds = (0,0)",
"METabc_FWD_CPLX8J2-67_s\n"
],
[
"solve_me_model(sporeme, max_mu = 0.1, min_mu = .01, using_soplex=False, precision = 1e-3,growth_key = 'sigma')",
"iter\tmuopt \ta \tb \tmu1 \tstat1\nFinished compiling expressions in 250.193434 seconds\nFinished substituting S,lb,ub in 7.382745 seconds\nFinished makeME_LP in 1.537819 seconds\nGetting MINOS parameters from ME_NLP...\n1 0.05 0.05 0.1 0.05 optimal\nFinished substituting S,lb,ub in 7.362918 seconds\nFinished makeME_LP in 1.532017 seconds\nGetting MINOS parameters from ME_NLP...\n2 0.05 0.05 0.07500000000000001 0.07500000000000001 1\nFinished substituting S,lb,ub in 7.196329 seconds\nFinished makeME_LP in 1.528339 seconds\nGetting MINOS parameters from ME_NLP...\n3 0.05 0.05 0.0625 0.0625 1\nFinished substituting S,lb,ub in 7.299746 seconds\nFinished makeME_LP in 1.532322 seconds\nGetting MINOS parameters from ME_NLP...\n4 0.05625 0.05625 0.0625 0.05625 optimal\nFinished substituting S,lb,ub in 7.141644 seconds\nFinished makeME_LP in 1.523025 seconds\nGetting MINOS parameters from ME_NLP...\n5 0.05625 0.05625 0.059375 0.059375 1\nFinished substituting S,lb,ub in 7.171215 seconds\nFinished makeME_LP in 1.525771 seconds\nGetting MINOS parameters from ME_NLP...\n6 0.0578125 0.0578125 0.059375 0.0578125 optimal\nFinished substituting S,lb,ub in 7.133823 seconds\nFinished makeME_LP in 1.529619 seconds\nGetting MINOS parameters from ME_NLP...\n7 0.0578125 0.0578125 0.05859375 0.05859375 1\nBisection done in 620.971 seconds\n"
],
[
"if sporeme.solution: print(sporeme.solution.x_dict['biomass_dilution_s'])",
"0.0578125\n"
],
[
"met = 'suchms_s'\nprint(sporeme.metabolites.get_by_id(met).name)\nflux_based_reactions(sporeme,met,only_types=['MetabolicReaction'])",
"O-Succinyl-L-homoserine\n"
]
],
[
[
"### Mechanisms",
"_____no_output_____"
]
],
[
[
"with open(\"./sporeme_solution_v3.pickle\", \"rb\") as outfile:\n sporeme = pickle.load(outfile)",
"_____no_output_____"
],
[
"main_mechanisms = [ 'ACKr_REV_BSU29470-MONOMER_s',\n 'PGK_REV_BSU33930-MONOMER_s',\n 'PYK_FWD_BSU29180-MONOMER_mod_mn2_mod_k_s',\n 'PRPPS_REV_BSU00510-MONOMER_mod_mn2_mod_pi_s']",
"_____no_output_____"
],
[
"for r in main_mechanisms:\n sporeme.reactions.get_by_id(r).bounds = (0,0)",
"_____no_output_____"
],
[
"solve_me_model(sporeme, max_mu = 0.1, min_mu = .01, using_soplex=False, precision = 1e-6,growth_key = 'sigma')",
"_____no_output_____"
],
[
"sporeme.solution",
"_____no_output_____"
],
[
"with open(\"./sporeme_solution_{}.pickle\".format(version), \"wb\") as outfile:\n pickle.dump(sporeme, outfile)",
"_____no_output_____"
],
[
"# Previously identified essential metabolites\nexchange_list = ['4fe4s_s','2fe2s_s','udcpp_s','pydx5p_s','3fe4s_s','cl_s','sheme_s','cu_s','mn2_s',\n 'bmocogdp_s','dpm_s','thmpp_s','zn2_s','cbl1_s','cobalt2_s']\nadditional = [m.id for m in sporeme.metabolites if isinstance(m,cobrame.Metabolite)]\ntransported_metabolites = exchange_list+additional",
"_____no_output_____"
],
[
"# Get transport reactions\n\ndef get_compartments(r):\n comps = []\n if isinstance(r,cobrame.MetabolicReaction):\n for m in r.metabolites:\n if isinstance(m,cobrame.Metabolite):\n comps.append(m.id[-1])\n return list(set(comps))\n\ndef get_all_transport(model):\n transport_reactions = []\n for r in tqdm(model.reactions):\n comps = get_compartments(r)\n if len(comps) > 1 and 's' in comps:\n transport_reactions.append(r.id)\n return list(set(transport_reactions))\n\ndef get_active_transport(transport_reactions):\n active_transporters = []\n for r in tqdm(transport_reactions):\n if 'SPONT' not in r and abs(sporeme.solution.x_dict[r])>0.:\n active_transporters.append(r)\n # Include arginine transport\n arginine_transport = [r.id for r in get_transport_reactions(sporeme,'arg__L_c',comps=['c','s'])+get_transport_reactions(sporeme,'arg__L_c',comps=['s','c'])]\n [active_transporters.append(r) for r in arginine_transport]\n active_transporters = list(set(active_transporters))\n return active_transporters",
"_____no_output_____"
]
],
[
[
"## Check by group of transporters of metabolite",
"_____no_output_____"
]
],
[
[
"def get_necessary_metabolites(model,active_transporters):\n necessary_metabolites = []\n for r in tqdm(active_transporters):\n rxn = model.reactions.get_by_id(r)\n for m in rxn.products:\n if not isinstance(m,cobrame.Metabolite):\n continue\n met_root = m.id[:-2]\n for i in rxn.reactants:\n if met_root in i.id:\n necessary_metabolites.append(m.id)\n return list(set(necessary_metabolites))\n\ndef get_all_available_transport(model,necessary_metabolites):\n available_transport = []\n at_dict = {}\n for m in tqdm(necessary_metabolites):\n rxns = get_transport_reactions(model,m,comps=['c','s']) + get_transport_reactions(model,m,comps=['s','c'])\n [available_transport.append(r.id) for r in rxns]\n at_dict[m] = []\n [at_dict[m].append(r.id) for r in rxns]\n return list(set(available_transport)), at_dict",
"_____no_output_____"
],
[
"# Previously identified essential metabolites\nexchange_list = ['4fe4s_s','2fe2s_s','udcpp_s','pydx5p_s','3fe4s_s','cl_s','sheme_s','cu_s','mn2_s',\n 'bmocogdp_s','dpm_s','thmpp_s','zn2_s','cbl1_s','cobalt2_s']\nadditional = [m.id for m in sporeme.metabolites if isinstance(m,cobrame.Metabolite)]\ntransported_metabolites = exchange_list+additional",
"_____no_output_____"
],
[
"transport_reactions = get_all_transport(sporeme)\nprint('{} transport reactions identified'.format(len(transport_reactions)))",
"_____no_output_____"
],
[
"active_transporters = get_active_transport(transport_reactions)\nnecessary_metabolites = get_necessary_metabolites(sporeme,active_transporters)\nnecessary_metabolites.remove('h_s')\nnecessary_metabolites.remove('h_c')\navailable_transport, at_dict = get_all_available_transport(sporeme,necessary_metabolites)",
"_____no_output_____"
],
[
"print('{} active transport reactions identified'.format(len(active_transporters)))\nprint('{} necessary metabolites identified'.format(len(necessary_metabolites)))\nprint('{} available transport reactions identified'.format(len(available_transport)))",
"_____no_output_____"
],
[
"all_transporters_to_open = list(set(active_transporters + available_transport))\nprint('{} open transport reactions identified'.format(len(all_transporters_to_open)))\nprint('Included {}'.format(set(active_transporters)-set(available_transport)))",
"_____no_output_____"
],
[
"for r in transport_reactions:\n if r not in all_transporters_to_open and 'SPONT' not in r:\n rxn = sporeme.reactions.get_by_id(r)\n rxn.upper_bound = 0 \n rxn.lower_bound = 0",
"_____no_output_____"
],
[
"solve_me_model(sporeme, max_mu = 0.1, min_mu = .01, using_soplex=False, precision = 1e-6,growth_key = 'sigma')",
"_____no_output_____"
],
[
"from bacillusme.analysis import sensitivity as ss\nflux_results_df = ss.transporter_knockout(sporeme,necessary_metabolites, \\\n NP=20,solution=1,biomass_dilution='biomass_dilution_s',\\\n growth_key = 'sigma',single_change_function='group_knockout')",
"_____no_output_____"
],
[
"flux_results_df.to_csv('group_KO_flux_results_{}.csv'.format(version))",
"_____no_output_____"
],
[
"flux_results_df = pd.read_csv('group_KO_flux_results_{}.csv'.format(version),index_col=0)\nflux_results_df.loc['biomass_dilution_s'].sort_values().plot.bar(figsize=(12 ,4))\nplt.tight_layout()\nplt.savefig(\"group_KO_flux_results_{}.svg\".format(version), format=\"SVG\")",
"_____no_output_____"
]
],
[
[
"### Close metabolite one by one\nIncluding information about arginine being transported",
"_____no_output_____"
]
],
[
[
"with open(\"./sporeme_solution_{}.pickle\".format(version), \"rb\") as outfile:\n sporeme = pickle.load(outfile)",
"_____no_output_____"
],
[
"for r in transport_reactions:\n if r not in all_transporters_to_open and 'SPONT' not in r:\n rxn = sporeme.reactions.get_by_id(r)\n rxn.upper_bound = 0 \n rxn.lower_bound = 0",
"_____no_output_____"
],
[
"flux_results_df = pd.read_csv('group_KO_flux_results_{}.csv'.format(version),index_col=0)\nsorted_mets = flux_results_df.loc['biomass_dilution_s'].sort_values(ascending=False).drop('base').index.to_list()",
"_____no_output_____"
],
[
"sorted_mets.remove('arg__L_s')\nsorted_mets.append('arg__L_s')",
"_____no_output_____"
],
[
"from bacillusme.analysis import sensitivity as ss\nflux_results_df = ss.transporter_knockout(sporeme,sorted_mets, \\\n NP=20,solution=1,biomass_dilution='biomass_dilution_s',\\\n growth_key = 'sigma',single_change_function='group_knockout',sequential=True)\nflux_results_df.to_csv('group_1by1_KO_flux_results_{}.csv'.format(version))",
"_____no_output_____"
],
[
"flux_results_df = pd.read_csv('group_1by1_KO_flux_results_{}.csv'.format(version),index_col=0)\nflux_results_df.loc['biomass_dilution_s',sorted_mets[::-1]].plot.bar(figsize=(12,4))\nplt.tight_layout()\nplt.savefig(\"group_1by1_KO_flux_results_{}.svg\".format(version), format=\"SVG\")",
"_____no_output_____"
]
],
[
[
"# Cases",
"_____no_output_____"
]
],
[
[
"pd.set_option('display.max_colwidth', None)",
"_____no_output_____"
]
],
[
[
"### Original",
"_____no_output_____"
]
],
[
[
"# CYTK2 KO\nversion = 'v4'",
"_____no_output_____"
],
[
"flux_results_df = pd.read_csv('group_KO_flux_results_{}.csv'.format(version),index_col=0)\nflux_results_df.loc['biomass_dilution_s'].sort_values().plot.bar(figsize=(12 ,4))",
"_____no_output_____"
],
[
"flux_results_df = pd.read_csv('group_1by1_KO_flux_results_{}.csv'.format(version),index_col=0)\nsorted_mets = flux_results_df.loc['biomass_dilution_s'].sort_values(ascending=True)\nlast_met = sorted_mets.index[list(sorted_mets.index).index(sorted_mets[sorted_mets<1e-5].index[-1])+1]\nprint(last_met)\nflux_dict = flux_results_df[last_met].to_dict() # Last time before model breaks",
"_____no_output_____"
],
[
"flux_results_df = pd.read_csv('group_1by1_KO_flux_results_{}.csv'.format(version),index_col=0)\nflux_results_df.loc['biomass_dilution_s',sorted_mets.index[::-1]].plot.bar(figsize=(12,4))\nplt.tight_layout()",
"_____no_output_____"
],
[
"flux_dict['biomass_dilution']",
"_____no_output_____"
],
[
"met='atp_s' # ATP production and glucose uptake\nprod_atp_df = flux_based_reactions(sporeme,met,flux_dict=flux_dict,only_types=['MetabolicReaction'])\nprod_atp_df = prod_atp_df[prod_atp_df['met_flux']>0]\nprod_atp_df['met_flux'].sum()",
"_____no_output_____"
],
[
"prod_atp_df['met_flux'].div(prod_atp_df['met_flux'].sum())",
"_____no_output_____"
]
],
[
[
"### All mechanisms KO",
"_____no_output_____"
]
],
[
[
"version = 'v5_all_KO'",
"_____no_output_____"
],
[
"flux_results_df = pd.read_csv('group_KO_flux_results_{}.csv'.format(version),index_col=0)\nflux_results_df.loc['biomass_dilution_s'].sort_values().plot.bar(figsize=(12 ,4))",
"_____no_output_____"
],
[
"flux_results_df = pd.read_csv('group_1by1_KO_flux_results_{}.csv'.format(version),index_col=0)\nsorted_mets = flux_results_df.loc['biomass_dilution_s'].sort_values(ascending=True)\nlast_met = sorted_mets.index[list(sorted_mets.index).index(sorted_mets[sorted_mets<1e-5].index[-1])+1]\nprint(last_met)\nflux_dict = flux_results_df[last_met].to_dict() # Last time before model breaks",
"thr__L_s\n"
],
[
"flux_results_df = pd.read_csv('group_1by1_KO_flux_results_{}.csv'.format(version),index_col=0)\nflux_results_df.loc['biomass_dilution_s',sorted_mets.index[::-1]].plot.bar(figsize=(12,4))\nplt.tight_layout()",
"_____no_output_____"
],
[
"flux_dict['biomass_dilution']",
"_____no_output_____"
],
[
"met='atp_s' # ATP production and glucose uptake\nprod_atp_df = flux_based_reactions(sporeme,met,flux_dict=flux_dict,only_types=['MetabolicReaction'])\nprod_atp_df = prod_atp_df[prod_atp_df['met_flux']>0]\nprod_atp_df['met_flux'].sum()",
"_____no_output_____"
],
[
"prod_atp_df['met_flux'].div(prod_atp_df['met_flux'].sum())",
"_____no_output_____"
],
[
"flux_based_reactions(sporeme,met,flux_dict=flux_dict,only_types=['MetabolicReaction'])",
"_____no_output_____"
],
[
"flux_based_reactions(sporeme,'prpp_s',flux_dict=flux_dict,only_types=['MetabolicReaction'])",
"_____no_output_____"
],
[
"sporeme.metabolites.prpp_s.name",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4affbc4f3156ab75b69e49dd083cbc7cb2aed67c
| 42,288 |
ipynb
|
Jupyter Notebook
|
formats/formats1-lines-sol.ipynb
|
simoneb1x/softpython-en
|
4321fb033a10b57058e10c07f763d170b41a3082
|
[
"CC-BY-4.0"
] | null | null | null |
formats/formats1-lines-sol.ipynb
|
simoneb1x/softpython-en
|
4321fb033a10b57058e10c07f763d170b41a3082
|
[
"CC-BY-4.0"
] | 16 |
2020-10-24T15:16:59.000Z
|
2022-03-19T04:05:48.000Z
|
formats/formats1-lines-sol.ipynb
|
simoneb1x/softpython-en
|
4321fb033a10b57058e10c07f763d170b41a3082
|
[
"CC-BY-4.0"
] | 1 |
2021-10-30T18:09:14.000Z
|
2021-10-30T18:09:14.000Z
| 29.822285 | 653 | 0.53235 |
[
[
[
"# Data formats 1 - introduction\n\n## [Download exercises zip](../_static/generated/formats.zip)\n\n[Browse files online](https://github.com/DavidLeoni/softpython-en/tree/master/formats)\n\n## Introduction\n\nIn these tutorials we will see how to load and write tabular data such as CSV, and we will mention tree-like data such as JSON files. We will also spend a couple of words about opendata catalogs and licenses (creative commons).",
"_____no_output_____"
],
[
"\nIn these tutorials we will review main data formats:\n\nTextual formats\n\n* Line files\n* CSV (tabular data)\n* JSON (tree-like data, just mention)\n\nBinary formats (just mention)\n\n* fogli Excel\n\nWe will also mention open data catalogs and licenses (Creative Commons)",
"_____no_output_____"
],
[
"### What to do\n\n1. unzip exercises in a folder, you should get something like this: \n\n```\nformats\n formats1-lines.ipynb \n formats1-lines-sol.ipynb\n formats2-csv.ipynb \n formats2-csv-sol.ipynb \n formats3-json.ipynb \n formats3-json-sol.ipynb \n formats4-chal.ipynb\n jupman.py\n```\n\n<div class=\"alert alert-warning\">\n\n**WARNING**: to correctly visualize the notebook, it MUST be in an unzipped folder !\n</div>\n\n2. open Jupyter Notebook from that folder. Two things should open, first a console and then browser. The browser should show a file list: navigate the list and open the notebook `formats/formats1-lines.ipynb`\n3. Go on reading that notebook, and follow instuctions inside.\n\n\nShortcut keys:\n\n- to execute Python code inside a Jupyter cell, press `Control + Enter`\n- to execute Python code inside a Jupyter cell AND select next cell, press `Shift + Enter`\n- to execute Python code inside a Jupyter cell AND a create a new cell aftwerwards, press `Alt + Enter`\n- If the notebooks look stuck, try to select `Kernel -> Restart`",
"_____no_output_____"
],
[
"## Line files\n\nLine files are typically text files which contain information grouped by lines. An example using historical characters might be like the following:\n\n```\nLeonardo\nda Vinci\nSandro\nBotticelli\nNiccolò \nMacchiavelli\n```\nWe can immediately see a regularity: first two lines contain data of Leonardo da Vinci, second one the name and then the surname. Successive lines instead have data of Sandro Botticelli, with again first the name and then the surname and so on.\n\nWe might want to do a program that reads the lines and prints on the terminal names and surnames like the following:\n\n```\nLeonardo da Vinci \nSandro Botticelli\nNiccolò Macchiavelli\n```\n\nTo start having an approximation of the final result, we can open the file, read only the first line and print it:\n",
"_____no_output_____"
]
],
[
[
"with open('people-simple.txt', encoding='utf-8') as f:\n line=f.readline()\n print(line)\n",
"Leonardo\n\n"
]
],
[
[
"What happened? Let's examing first rows:\n\n\n\n### open command\n\nThe command\n\n```python\nopen('people-simple.txt', encoding='utf-8')\n```\n\nallows us to open the text file by telling PYthon the file path `'people-simple.txt'` and the encoding in which it was written (`encoding='utf-8'`). ",
"_____no_output_____"
],
[
"### The encoding\n\nThe encoding dependes on the operating system and on the editor used to write the file. When we open a file, Python is not capable to divine the encoding, and if we do not specify anything Python might open the file assuming an encoding different from the original - in other words, if we omit the encoding (or we put a wrong one) we might end up seeing weird characters (like little squares instead of accented letters).\n\nIn general, when you open a file, try first to specify the encoding `utf-8` which is the most common one. If it doesn't work try others, for example for files written in south Europe with Windows you might check `encoding='latin-1'`. If you open a file written elsewhere, you might need other encodings. For more in-depth information, you can read [Dive into Python - Chapter 4 - Strings](https://diveintopython3.problemsolving.io/strings.html), and [Dive into Python - Chapter 11 - File](https://diveintopython3.problemsolving.io/files.html), **both of which are extremely recommended readings**.",
"_____no_output_____"
],
[
"### with block\n\nThe `with` defines a block with instructions inside:\n\n```python\nwith open('people-simple.txt', encoding='utf-8') as f:\n line=f.readline()\n print(line)\n```\n\nWe used the `with` to tell PYthon that in any case, even if errors occur, we want that after having used the file, that is after having executed the instructions inside the internal block (the `line=f.readline()` and `print(line)`) Python must automatically close the file. Properly closing a file avoids to waste memory resources and creating hard to find paranormal errors. If you want to avoid hunting for never closed zombie files, always remember to open all files in `with` blocks! Furthermore, at the end of the row in the part `as f:` we assigned the file to a variable hereby called `f`, but we could have used any other name we liked.\n\n\n<div class=\"alert alert-warning\">\n\n**WARNING**: To indent the code, ALWAYS use sequences of four white spaces. Sequences of 2 spaces. Sequences of only 2 spaces even if allowed are not recommended.\n</div>\n\n<div class=\"alert alert-warning\">\n\n**WARNING**: Depending on the editor you use, by pressing TAB you might get a sequence o f white spaces like it happens in Jupyter (4 spaces which is the recommended length), or a special tabulation character (to avoid)! As much as this annoying this distinction might appear, remember it because it might generate very hard to find errors.\n\n</div>\n\n<div class=\"alert alert-warning\">\n\n**WARNING**: In the commands to create blocks such as `with`, always remember to put the character of colon `:` at the end of the line !\n</div>\n",
"_____no_output_____"
],
[
"\nThe command\n\n```\n line=f.readline()\n```\nputs in the variable `line` the entire line, like a string. Warning: the string will contain at the end the special character of line return !\n\nYou might wonder where that `readline` comes from. Like everything in Python, our variable `f` which represents the file we just opened is an object, and like any object, depending on its type, it has particular methods we can use on it. In this case the method is `readline`. \n\nThe following command prints the string content:\n\n```python\n print(line) \n```\n",
"_____no_output_____"
],
[
"**✪ 1.1 EXERCISE**: Try to rewrite here the block we've just seen, and execute the cell by pressing Control-Enter. Rewrite the code with the fingers, not with copy-paste ! Pay attention to correct indentation with spaces in the block.",
"_____no_output_____"
]
],
[
[
"# write here\n\nwith open('people-simple.txt', encoding='utf-8') as f:\n line=f.readline()\n print(line)\n",
"Leonardo\n\n"
]
],
[
[
"**✪ 1.2 EXERCISE**: you might wondering what exactly is that `f`, and what exatly the method `readlines` should be doing. When you find yourself in these situations, you might help yourself with functions `type` and `help`. This time, directly copy paste the same code here, but insert inside `with` block the commands:\n\n* `print(type(f))`\n* `help(f)`\n* `help(f.readline)` # Attention: remember the f. before the readline !!\n\nEvery time you add something, try to execute with Control+Enter and see what happens",
"_____no_output_____"
]
],
[
[
"# write here the code (copy and paste)\nwith open('people-simple.txt', encoding='utf-8') as f:\n line=f.readline()\n print(line)\n print(type(f)) \n help(f.readline)\n help(f)",
"Leonardo\n\n<class '_io.TextIOWrapper'>\nHelp on built-in function readline:\n\nreadline(size=-1, /) method of _io.TextIOWrapper instance\n Read until newline or EOF.\n \n Returns an empty string if EOF is hit immediately.\n\nHelp on TextIOWrapper object:\n\nclass TextIOWrapper(_TextIOBase)\n | TextIOWrapper(buffer, encoding=None, errors=None, newline=None, line_buffering=False, write_through=False)\n | \n | Character and line based layer over a BufferedIOBase object, buffer.\n | \n | encoding gives the name of the encoding that the stream will be\n | decoded or encoded with. It defaults to locale.getpreferredencoding(False).\n | \n | errors determines the strictness of encoding and decoding (see\n | help(codecs.Codec) or the documentation for codecs.register) and\n | defaults to \"strict\".\n | \n | newline controls how line endings are handled. It can be None, '',\n | '\\n', '\\r', and '\\r\\n'. It works as follows:\n | \n | * On input, if newline is None, universal newlines mode is\n | enabled. Lines in the input can end in '\\n', '\\r', or '\\r\\n', and\n | these are translated into '\\n' before being returned to the\n | caller. If it is '', universal newline mode is enabled, but line\n | endings are returned to the caller untranslated. If it has any of\n | the other legal values, input lines are only terminated by the given\n | string, and the line ending is returned to the caller untranslated.\n | \n | * On output, if newline is None, any '\\n' characters written are\n | translated to the system default line separator, os.linesep. If\n | newline is '' or '\\n', no translation takes place. If newline is any\n | of the other legal values, any '\\n' characters written are translated\n | to the given string.\n | \n | If line_buffering is True, a call to flush is implied when a call to\n | write contains a newline character.\n | \n | Method resolution order:\n | TextIOWrapper\n | _TextIOBase\n | _IOBase\n | builtins.object\n | \n | Methods defined here:\n | \n | __getstate__(...)\n | \n | __init__(self, /, *args, **kwargs)\n | Initialize self. See help(type(self)) for accurate signature.\n | \n | __next__(self, /)\n | Implement next(self).\n | \n | __repr__(self, /)\n | Return repr(self).\n | \n | close(self, /)\n | Flush and close the IO object.\n | \n | This method has no effect if the file is already closed.\n | \n | detach(self, /)\n | Separate the underlying buffer from the TextIOBase and return it.\n | \n | After the underlying buffer has been detached, the TextIO is in an\n | unusable state.\n | \n | fileno(self, /)\n | Returns underlying file descriptor if one exists.\n | \n | OSError is raised if the IO object does not use a file descriptor.\n | \n | flush(self, /)\n | Flush write buffers, if applicable.\n | \n | This is not implemented for read-only and non-blocking streams.\n | \n | isatty(self, /)\n | Return whether this is an 'interactive' stream.\n | \n | Return False if it can't be determined.\n | \n | read(self, size=-1, /)\n | Read at most n characters from stream.\n | \n | Read from underlying buffer until we have n characters or we hit EOF.\n | If n is negative or omitted, read until EOF.\n | \n | readable(self, /)\n | Return whether object was opened for reading.\n | \n | If False, read() will raise OSError.\n | \n | readline(self, size=-1, /)\n | Read until newline or EOF.\n | \n | Returns an empty string if EOF is hit immediately.\n | \n | reconfigure(self, /, *, encoding=None, errors=None, newline=None, line_buffering=None, write_through=None)\n | Reconfigure the text stream with new parameters.\n | \n | This also does an implicit stream flush.\n | \n | seek(self, cookie, whence=0, /)\n | Change stream position.\n | \n | Change the stream position to the given byte offset. The offset is\n | interpreted relative to the position indicated by whence. Values\n | for whence are:\n | \n | * 0 -- start of stream (the default); offset should be zero or positive\n | * 1 -- current stream position; offset may be negative\n | * 2 -- end of stream; offset is usually negative\n | \n | Return the new absolute position.\n | \n | seekable(self, /)\n | Return whether object supports random access.\n | \n | If False, seek(), tell() and truncate() will raise OSError.\n | This method may need to do a test seek().\n | \n | tell(self, /)\n | Return current stream position.\n | \n | truncate(self, pos=None, /)\n | Truncate file to size bytes.\n | \n | File pointer is left unchanged. Size defaults to the current IO\n | position as reported by tell(). Returns the new size.\n | \n | writable(self, /)\n | Return whether object was opened for writing.\n | \n | If False, write() will raise OSError.\n | \n | write(self, text, /)\n | Write string to stream.\n | Returns the number of characters written (which is always equal to\n | the length of the string).\n | \n | ----------------------------------------------------------------------\n | Static methods defined here:\n | \n | __new__(*args, **kwargs) from builtins.type\n | Create and return a new object. See help(type) for accurate signature.\n | \n | ----------------------------------------------------------------------\n | Data descriptors defined here:\n | \n | buffer\n | \n | closed\n | \n | encoding\n | Encoding of the text stream.\n | \n | Subclasses should override.\n | \n | errors\n | The error setting of the decoder or encoder.\n | \n | Subclasses should override.\n | \n | line_buffering\n | \n | name\n | \n | newlines\n | Line endings translated so far.\n | \n | Only line endings translated during reading are considered.\n | \n | Subclasses should override.\n | \n | write_through\n | \n | ----------------------------------------------------------------------\n | Methods inherited from _IOBase:\n | \n | __del__(...)\n | \n | __enter__(...)\n | \n | __exit__(...)\n | \n | __iter__(self, /)\n | Implement iter(self).\n | \n | readlines(self, hint=-1, /)\n | Return a list of lines from the stream.\n | \n | hint can be specified to control the number of lines read: no more\n | lines will be read if the total size (in bytes/characters) of all\n | lines so far exceeds hint.\n | \n | writelines(self, lines, /)\n | Write a list of lines to stream.\n | \n | Line separators are not added, so it is usual for each of the\n | lines provided to have a line separator at the end.\n | \n | ----------------------------------------------------------------------\n | Data descriptors inherited from _IOBase:\n | \n | __dict__\n\n"
]
],
[
[
"\nFirst we put the content of the first line into the variable `line`, now we might put it in a variable witha more meaningful name, like `name`. Also, we can directly read the next row into the variable `surname` and then print the concatenation of both:\n",
"_____no_output_____"
]
],
[
[
"with open('people-simple.txt', encoding='utf-8') as f:\n name=f.readline()\n surname=f.readline()\n print(name + ' ' + surname)\n",
"Leonardo\n da Vinci\n\n"
]
],
[
[
"**PROBLEM !** The printing puts a weird carriage return. Why is that? If you remember, first we said that `readline` reads the line content in a string adding to the end also the special newline character. To eliminate it, you can use the command `rstrip()`: \n",
"_____no_output_____"
]
],
[
[
"with open('people-simple.txt', encoding='utf-8') as f:\n name=f.readline().rstrip()\n surname=f.readline().rstrip()\n print(name + ' ' + surname)\n",
"Leonardo da Vinci\n"
]
],
[
[
"**✪ 1.3 EXERCISE**: Again, rewrite the block above in the cell below, ed execute the cell with Control+Enter. Question: what happens if you use `strip()` instead of `rstrip()`? What about `lstrip()`? Can you deduce the meaning of `r` and `l`? If you can't manage it, try to use python command `help` by calling `help(string.rstrip)`",
"_____no_output_____"
]
],
[
[
"# write here\n\nwith open('people-simple.txt', encoding='utf-8') as f:\n name=f.readline().rstrip()\n surname=f.readline().rstrip()\n print(name + ' ' + surname)",
"Leonardo da Vinci\n"
]
],
[
[
"Very good, we have the first line ! Now we can read all the lines in sequence. To this end, we can use a `while` cycle:",
"_____no_output_____"
]
],
[
[
"with open('people-simple.txt', encoding='utf-8') as f:\n line=f.readline()\n while line != \"\": \n name = line.rstrip()\n surname=f.readline().rstrip()\n print(name + ' ' + surname)\n line=f.readline()",
"Leonardo da Vinci\nSandro Botticelli\nNiccolò Macchiavelli\n"
]
],
[
[
"<div class=\"alert alert-info\">\n\n**NOTE**: In Python there are [shorter ways](https://thispointer.com/5-different-ways-to-read-a-file-line-by-line-in-python/) \nto read a text file line by line, we used this approach to make explicit all passages.\n\n</div>",
"_____no_output_____"
],
[
"What did we do? First, we added a `while` cycle in a new block\n\n<div class=\"alert alert-warning\">\n\n**WARNING**: In new block, since it is already within the external `with`, the instructions are indented of 8 spaces and not 4! If you use the wrong spaces, bad things happen !\n\n</div>\n\nWe first read a line, and two cases are possible: \n\na. we are the end of the file (or file is empty) : in this case `readline()` call returns an empty string\n\nb. we are not at the end of the file: the first line is put as a string inside the variable `line`. Since Python internally uses a pointer to keep track at which position we are when reading inside the file, after the read such pointer is moved at the beginning of the next line. This way the next call to `readline()` will read a line from the new position.\n\nIn `while` block we tell Python to continue the cycle as long as `line` is _not_ empty. If this is the case, inside the `while` block we parse the name from the line and put it in variable `name` (removing extra newline character with `rstrip()` as we did before), then we proceed reading the next line and parse the result inside the `surname` variable. Finally, we read again a line into the `line` variable so it will be ready for the next round of name extraction. If line is empty the cycle will terminate:\n\n\n```python\nwhile line != \"\": # enter cycle if line contains characters\n name = line.rstrip() # parses the name\n surname=f.readline().rstrip() # reads next line and parses surname\n print(name + ' ' + surname) \n line=f.readline() # read next line\n```",
"_____no_output_____"
],
[
"**✪ 1.4 EXERCISE**: As before, rewrite in the cell below the code with the `while`, paying attention to the indentation (for the external `with` line use copy-and-paste):",
"_____no_output_____"
]
],
[
[
"# write here the code of internal while\n\nwith open('people-simple.txt', encoding='utf-8') as f:\n line=f.readline()\n while line != \"\": \n name = line.rstrip()\n surname=f.readline().rstrip()\n print(name + ' ' + surname)\n line=f.readline()",
"Leonardo da Vinci\nSandro Botticelli\nNiccolò Macchiavelli\n"
]
],
[
[
"## people-complex line file\n\nLook at the file `people-complex.txt`:\n\n```\nname: Leonardo\nsurname: da Vinci\nbirthdate: 1452-04-15\nname: Sandro\nsurname: Botticelli\nbirthdate: 1445-03-01\nname: Niccolò \nsurname: Macchiavelli\nbirthdate: 1469-05-03\n```\nSupposing to read the file to print this output, how would you do it? \n\n```\nLeonardo da Vinci, 1452-04-15\nSandro Botticelli, 1445-03-01\nNiccolò Macchiavelli, 1469-05-03\n```",
"_____no_output_____"
],
[
"\n**Hint 1**: to obtain the string `'abcde'`, the substring `'cde'`, which starts at index 2, you can ue the operator square brackets, using the index followed by colon `:`\n",
"_____no_output_____"
]
],
[
[
"x = 'abcde'\nx[2:]",
"_____no_output_____"
],
[
"x[3:]",
"_____no_output_____"
]
],
[
[
"**Hint 2**: To know the length of a string, use the function `len`:",
"_____no_output_____"
]
],
[
[
"len('abcde')",
"_____no_output_____"
]
],
[
[
"**✪ 1.5 EXERCISE**: Write here the solution of the exercise 'People complex':",
"_____no_output_____"
]
],
[
[
"# write here \n \nwith open('people-complex.txt', encoding='utf-8') as f:\n line=f.readline()\n while line != \"\": \n name = line.rstrip()[len(\"name: \"):]\n surname= f.readline().rstrip()[len(\"surname: \"):]\n born = f.readline().rstrip()[len(\"birthdate: \"):]\n print(name + ' ' + surname + ', ' + born)\n line=f.readline() ",
"Leonardo da Vinci, 1452-04-15\nSandro Botticelli, 1445-03-01\nNiccolò Macchiavelli, 1469-05-03\n"
]
],
[
[
"## Exercise - line file immersione-in-python-toc\n\n✪✪✪ This exercise is more challenging, if you are a beginner you might skip it and go on to CSVs\n\nThe book Dive into Python is nice and for the italian version there is a PDF, which has a problem though: if you try to print it, you will discover that the index is missing. Without despairing, we found a program to extract titles in a file as follows, but you will discover it is not exactly nice to see. Since we are Python ninjas, we decided to transform raw titles in a [real table of contents](http://softpython.readthedocs.io/it/latest/_static/toc-immersione-in-python-3.txt). Sure enough there are smarter ways to do this, like loading the pdf in Python with an appropriate module for pdfs, still this makes for an interesting exercise.\n\nYou are given the file `immersione-in-python-toc.txt`:\n\n```\nBookmarkBegin\nBookmarkTitle: Il vostro primo programma Python\nBookmarkLevel: 1\nBookmarkPageNumber: 38\nBookmarkBegin\nBookmarkTitle: Immersione!\nBookmarkLevel: 2\nBookmarkPageNumber: 38\nBookmarkBegin\nBookmarkTitle: Dichiarare funzioni\nBookmarkLevel: 2\nBookmarkPageNumber: 41\nBookmarkBeginint\nBookmarkTitle: Argomenti opzionali e con nome\nBookmarkLevel: 3\nBookmarkPageNumber: 42\nBookmarkBegin\nBookmarkTitle: Scrivere codice leggibile\nBookmarkLevel: 2\nBookmarkPageNumber: 44\nBookmarkBegin\nBookmarkTitle: Stringhe di documentazione\nBookmarkLevel: 3\nBookmarkPageNumber: 44\nBookmarkBegin\nBookmarkTitle: Il percorso di ricerca di import\nBookmarkLevel: 2\nBookmarkPageNumber: 46\nBookmarkBegin\nBookmarkTitle: Ogni cosa è un oggetto\nBookmarkLevel: 2\nBookmarkPageNumber: 47\n```\n\nWrite a python program to print the following output:\n\n```\n Il vostro primo programma Python 38\n Immersione! 38\n Dichiarare funzioni 41\n Argomenti opzionali e con nome 42\n Scrivere codice leggibile 44\n Stringhe di documentazione 44\n Il percorso di ricerca di import 46\n Ogni cosa è un oggetto 47\n```\n\nFor this exercise, you will need to insert in the output artificial spaces, in a qunatity determined by the rows `BookmarkLevel`\n\n\n**QUESTION**: what's that weird value `è` at the end of the original file? Should we report it in the output?\n\n**HINT 1**: To convert a string into an integer number, use the function `int`:\n",
"_____no_output_____"
]
],
[
[
"x = '5'",
"_____no_output_____"
],
[
"x",
"_____no_output_____"
],
[
"int(x)",
"_____no_output_____"
]
],
[
[
"\n<div class=\"alert alert-warning\">\n\n**Warning**: `int(x)` returns a value, and never modifies the argument `x`! \n</div>\n",
"_____no_output_____"
],
[
"**HINT 2**: To substitute a substring in a string, you can use the method `.replace`:",
"_____no_output_____"
]
],
[
[
"x = 'abcde'\nx.replace('cd', 'HELLO' )",
"_____no_output_____"
]
],
[
[
"**HINT 3**: while there is only one sequence to substitute, `replace` is fine, but if we had a milion of horrible sequences like `>`, `>`, `&x3e;`, what should we do? As good data cleaners, we recognize these are [HTML escape sequences](https://corsidia.com/materia/web-design/caratterispecialihtml), so we could use methods specific to sequences like [html.escape](https://docs.python.org/3/library/html.html#html.unescape). TRy it instead of `replace` and check if it works!\n\n\nNOTE: Before using `html.unescape`, import the module `html` with the command: \n\n```python\nimport html\n```",
"_____no_output_____"
],
[
"**HINT 4**: To write _n_ copies of a character, use `*` like this:",
"_____no_output_____"
]
],
[
[
"\"b\" * 3",
"_____no_output_____"
],
[
"\"b\" * 7",
"_____no_output_____"
]
],
[
[
"**IMPLEMENTATION**: Write here the solution for the line file `immersione-in-python-toc.txt`, and try execute it by pressing Control + Enter:\n",
"_____no_output_____"
]
],
[
[
"# write here \n\nimport html\n\nwith open(\"immersione-in-python-toc.txt\", encoding='utf-8') as f:\n\n line=f.readline() \n while line != \"\":\n line = f.readline().strip()\n title = html.unescape(line[len(\"BookmarkTitle: \"):])\n line=f.readline().strip()\n level = int(line[len(\"BookmarkLevel: \"):])\n line=f.readline().strip()\n page = line[len(\"BookmarkPageNumber: \"):]\n print((\" \" * level) + title + \" \" + page)\n line=f.readline()",
" Il vostro primo programma Python 38\n Immersione! 38\n Dichiarare funzioni 41\n Argomenti opzionali e con nome 42\n Scrivere codice leggibile 44\n Stringhe di documentazione 44\n Il percorso di ricerca di import 46\n Ogni cosa è un oggetto 47\n"
]
],
[
[
"## Continue\n\nGo on with [CSV tabular files](https://en.softpython.org/formats/formats2-csv-sol.html)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
4affc86b14c2f9696c00d944eb6ae571733d9cfa
| 18,593 |
ipynb
|
Jupyter Notebook
|
site/zh-cn/tutorials/load_data/csv.ipynb
|
akalakheti/docs
|
ad602b40f8f968520d21ae81e304dde80861f745
|
[
"Apache-2.0"
] | 3 |
2020-01-09T02:58:22.000Z
|
2020-09-11T09:02:01.000Z
|
site/zh-cn/tutorials/load_data/csv.ipynb
|
akalakheti/docs
|
ad602b40f8f968520d21ae81e304dde80861f745
|
[
"Apache-2.0"
] | 1 |
2020-01-11T03:55:25.000Z
|
2020-01-11T03:55:25.000Z
|
site/zh-cn/tutorials/load_data/csv.ipynb
|
akalakheti/docs
|
ad602b40f8f968520d21ae81e304dde80861f745
|
[
"Apache-2.0"
] | 2 |
2020-01-15T21:50:31.000Z
|
2020-01-15T21:56:30.000Z
| 26.113764 | 249 | 0.469209 |
[
[
[
"##### Copyright 2019 The TensorFlow Authors.\n\n",
"_____no_output_____"
]
],
[
[
"#@title Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# https://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.",
"_____no_output_____"
]
],
[
[
"# 用 tf.data 加载 CSV 数据",
"_____no_output_____"
],
[
"<table class=\"tfo-notebook-buttons\" align=\"left\">\n <td>\n <a target=\"_blank\" href=\"https://tensorflow.google.cn/tutorials/load_data/csv\"><img src=\"https://tensorflow.google.cn/images/tf_logo_32px.png\" />在 Tensorflow.org 上查看</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://colab.research.google.com/github/tensorflow/docs/blob/master/site/zh-cn/tutorials/load_data/csv.ipynb\"><img src=\"https://tensorflow.google.cn/images/colab_logo_32px.png\" />在 Google Colab 运行</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://github.com/tensorflow/docs/blob/master/site/zh-cn/tutorials/load_data/csv.ipynb\"><img src=\"https://tensorflow.google.cn/images/GitHub-Mark-32px.png\" />在 Github 上查看源代码</a>\n </td>\n <td>\n <a href=\"https://storage.googleapis.com/tensorflow_docs/site/zh-cn/tutorials/load_data/csv.ipynb\"><img src=\"https://tensorflow.google.cn/images/download_logo_32px.png\" />下载此 notebook</a>\n </td>\n</table>",
"_____no_output_____"
],
[
"Note: 我们的 TensorFlow 社区翻译了这些文档。因为社区翻译是尽力而为, 所以无法保证它们是最准确的,并且反映了最新的\n[官方英文文档](https://www.tensorflow.org/?hl=en)。如果您有改进此翻译的建议, 请提交 pull request 到\n[tensorflow/docs](https://github.com/tensorflow/docs) GitHub 仓库。要志愿地撰写或者审核译文,请加入\n[[email protected] Google Group](https://groups.google.com/a/tensorflow.org/forum/#!forum/docs-zh-cn)。",
"_____no_output_____"
],
[
"这篇教程通过一个示例展示了怎样将 CSV 格式的数据加载进 `tf.data.Dataset`。\n\n这篇教程使用的是泰坦尼克号乘客的数据。模型会根据乘客的年龄、性别、票务舱和是否独自旅行等特征来预测乘客生还的可能性。",
"_____no_output_____"
],
[
"## 设置",
"_____no_output_____"
]
],
[
[
"try:\n # Colab only\n %tensorflow_version 2.x\nexcept Exception:\n pass\n",
"_____no_output_____"
],
[
"from __future__ import absolute_import, division, print_function, unicode_literals\nimport functools\n\nimport numpy as np\nimport tensorflow as tf\nimport tensorflow_datasets as tfds",
"_____no_output_____"
],
[
"TRAIN_DATA_URL = \"https://storage.googleapis.com/tf-datasets/titanic/train.csv\"\nTEST_DATA_URL = \"https://storage.googleapis.com/tf-datasets/titanic/eval.csv\"\n\ntrain_file_path = tf.keras.utils.get_file(\"train.csv\", TRAIN_DATA_URL)\ntest_file_path = tf.keras.utils.get_file(\"eval.csv\", TEST_DATA_URL)",
"_____no_output_____"
],
[
"# 让 numpy 数据更易读。\nnp.set_printoptions(precision=3, suppress=True)",
"_____no_output_____"
]
],
[
[
"## 加载数据\n\n开始的时候,我们通过打印 CSV 文件的前几行来了解文件的格式。",
"_____no_output_____"
]
],
[
[
"!head {train_file_path}",
"_____no_output_____"
]
],
[
[
"正如你看到的那样,CSV 文件的每列都会有一个列名。dataset 的构造函数会自动识别这些列名。如果你使用的文件的第一行不包含列名,那么需要将列名通过字符串列表传给 `make_csv_dataset` 函数的 `column_names` 参数。",
"_____no_output_____"
],
[
" \n\n\n\n```python\n\nCSV_COLUMNS = ['survived', 'sex', 'age', 'n_siblings_spouses', 'parch', 'fare', 'class', 'deck', 'embark_town', 'alone']\n\ndataset = tf.data.experimental.make_csv_dataset(\n ...,\n column_names=CSV_COLUMNS,\n ...)\n \n```\n",
"_____no_output_____"
],
[
"这个示例使用了所有的列。如果你需要忽略数据集中的某些列,创建一个包含你需要使用的列的列表,然后传给构造器的(可选)参数 `select_columns`。\n\n```python\n\ndataset = tf.data.experimental.make_csv_dataset(\n ...,\n select_columns = columns_to_use, \n ...)\n\n```",
"_____no_output_____"
],
[
"对于包含模型需要预测的值的列是你需要显式指定的。",
"_____no_output_____"
]
],
[
[
"LABEL_COLUMN = 'survived'\nLABELS = [0, 1]",
"_____no_output_____"
]
],
[
[
"现在从文件中读取 CSV 数据并且创建 dataset。\n\n(完整的文档,参考 `tf.data.experimental.make_csv_dataset`)\n",
"_____no_output_____"
]
],
[
[
"def get_dataset(file_path):\n dataset = tf.data.experimental.make_csv_dataset(\n file_path,\n batch_size=12, # 为了示例更容易展示,手动设置较小的值\n label_name=LABEL_COLUMN,\n na_value=\"?\",\n num_epochs=1,\n ignore_errors=True)\n return dataset\n\nraw_train_data = get_dataset(train_file_path)\nraw_test_data = get_dataset(test_file_path)",
"_____no_output_____"
]
],
[
[
"dataset 中的每个条目都是一个批次,用一个元组(*多个样本*,*多个标签*)表示。样本中的数据组织形式是以列为主的张量(而不是以行为主的张量),每条数据中包含的元素个数就是批次大小(这个示例中是 12)。\n\n阅读下面的示例有助于你的理解。",
"_____no_output_____"
]
],
[
[
"examples, labels = next(iter(raw_train_data)) # 第一个批次\nprint(\"EXAMPLES: \\n\", examples, \"\\n\")\nprint(\"LABELS: \\n\", labels)",
"_____no_output_____"
]
],
[
[
"## 数据预处理",
"_____no_output_____"
],
[
"### 分类数据\n\nCSV 数据中的有些列是分类的列。也就是说,这些列只能在有限的集合中取值。\n\n使用 `tf.feature_column` API 创建一个 `tf.feature_column.indicator_column` 集合,每个 `tf.feature_column.indicator_column` 对应一个分类的列。\n",
"_____no_output_____"
]
],
[
[
"CATEGORIES = {\n 'sex': ['male', 'female'],\n 'class' : ['First', 'Second', 'Third'],\n 'deck' : ['A', 'B', 'C', 'D', 'E', 'F', 'G', 'H', 'I', 'J'],\n 'embark_town' : ['Cherbourg', 'Southhampton', 'Queenstown'],\n 'alone' : ['y', 'n']\n}\n",
"_____no_output_____"
],
[
"categorical_columns = []\nfor feature, vocab in CATEGORIES.items():\n cat_col = tf.feature_column.categorical_column_with_vocabulary_list(\n key=feature, vocabulary_list=vocab)\n categorical_columns.append(tf.feature_column.indicator_column(cat_col))",
"_____no_output_____"
],
[
"# 你刚才创建的内容\ncategorical_columns",
"_____no_output_____"
]
],
[
[
"这将是后续构建模型时处理输入数据的一部分。",
"_____no_output_____"
],
[
"### 连续数据",
"_____no_output_____"
],
[
"连续数据需要标准化。\n\n写一个函数标准化这些值,然后将这些值改造成 2 维的张量。\n",
"_____no_output_____"
]
],
[
[
"def process_continuous_data(mean, data):\n # 标准化数据\n data = tf.cast(data, tf.float32) * 1/(2*mean)\n return tf.reshape(data, [-1, 1])",
"_____no_output_____"
]
],
[
[
"现在创建一个数值列的集合。`tf.feature_columns.numeric_column` API 会使用 `normalizer_fn` 参数。在传参的时候使用 [`functools.partial`](https://docs.python.org/3/library/functools.html#functools.partial),`functools.partial` 由使用每个列的均值进行标准化的函数构成。",
"_____no_output_____"
]
],
[
[
"MEANS = {\n 'age' : 29.631308,\n 'n_siblings_spouses' : 0.545455,\n 'parch' : 0.379585,\n 'fare' : 34.385399\n}\n\nnumerical_columns = []\n\nfor feature in MEANS.keys():\n num_col = tf.feature_column.numeric_column(feature, normalizer_fn=functools.partial(process_continuous_data, MEANS[feature]))\n numerical_columns.append(num_col)",
"_____no_output_____"
],
[
"# 你刚才创建的内容。\nnumerical_columns",
"_____no_output_____"
]
],
[
[
"这里使用标准化的方法需要提前知道每列的均值。如果需要计算连续的数据流的标准化的值可以使用 [TensorFlow Transform](https://www.tensorflow.org/tfx/transform/get_started)。",
"_____no_output_____"
],
[
"### 创建预处理层",
"_____no_output_____"
],
[
"将这两个特征列的集合相加,并且传给 `tf.keras.layers.DenseFeatures` 从而创建一个进行预处理的输入层。",
"_____no_output_____"
]
],
[
[
"preprocessing_layer = tf.keras.layers.DenseFeatures(categorical_columns+numerical_columns)",
"_____no_output_____"
]
],
[
[
"## 构建模型",
"_____no_output_____"
],
[
"从 `preprocessing_layer` 开始构建 `tf.keras.Sequential`。",
"_____no_output_____"
]
],
[
[
"model = tf.keras.Sequential([\n preprocessing_layer,\n tf.keras.layers.Dense(128, activation='relu'),\n tf.keras.layers.Dense(128, activation='relu'),\n tf.keras.layers.Dense(1, activation='sigmoid'),\n])\n\nmodel.compile(\n loss='binary_crossentropy',\n optimizer='adam',\n metrics=['accuracy'])",
"_____no_output_____"
]
],
[
[
"## 训练、评估和预测",
"_____no_output_____"
],
[
"现在可以实例化和训练模型。",
"_____no_output_____"
]
],
[
[
"train_data = raw_train_data.shuffle(500)\ntest_data = raw_test_data",
"_____no_output_____"
],
[
"model.fit(train_data, epochs=20)",
"_____no_output_____"
]
],
[
[
"当模型训练完成的时候,你可以在测试集 `test_data` 上检查准确性。",
"_____no_output_____"
]
],
[
[
"test_loss, test_accuracy = model.evaluate(test_data)\n\nprint('\\n\\nTest Loss {}, Test Accuracy {}'.format(test_loss, test_accuracy))",
"_____no_output_____"
]
],
[
[
"使用 `tf.keras.Model.predict` 推断一个批次或多个批次的标签。",
"_____no_output_____"
]
],
[
[
"predictions = model.predict(test_data)\n\n# 显示部分结果\nfor prediction, survived in zip(predictions[:10], list(test_data)[0][1][:10]):\n print(\"Predicted survival: {:.2%}\".format(prediction[0]),\n \" | Actual outcome: \",\n (\"SURVIVED\" if bool(survived) else \"DIED\"))\n\n",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
4affcffeac5c9ed6daccd032f4c3e43ac9913d4f
| 22,062 |
ipynb
|
Jupyter Notebook
|
notebooks/v0/methods_doc_v0_figures.ipynb
|
carbonplan/trace
|
5cf113891bdefa29c2afd4478dff099e0458c82c
|
[
"MIT"
] | 14 |
2021-02-15T22:40:52.000Z
|
2022-02-24T15:25:28.000Z
|
notebooks/v0/methods_doc_v0_figures.ipynb
|
carbonplan/trace
|
5cf113891bdefa29c2afd4478dff099e0458c82c
|
[
"MIT"
] | 75 |
2021-02-11T17:57:42.000Z
|
2022-03-22T00:47:57.000Z
|
notebooks/v0/methods_doc_v0_figures.ipynb
|
carbonplan/trace
|
5cf113891bdefa29c2afd4478dff099e0458c82c
|
[
"MIT"
] | 2 |
2021-09-28T01:51:19.000Z
|
2021-11-22T21:32:35.000Z
| 30.098226 | 146 | 0.556885 |
[
[
[
"<img width=\"100\" src=\"https://carbonplan-assets.s3.amazonaws.com/monogram/dark-small.png\" style=\"margin-left:0px;margin-top:20px\"/>\n\n# Forest Emissions Tracking - Validation\n\n_CarbonPlan ClimateTrace Team_\n\nThis notebook compares our estimates of country-level forest emissions to prior estimates from other\ngroups. The notebook currently compares againsts:\n\n- Global Forest Watch (Zarin et al. 2016)\n- Global Carbon Project (Friedlingstein et al. 2020)\n",
"_____no_output_____"
]
],
[
[
"import geopandas\nimport pandas as pd\nfrom io import StringIO\nimport matplotlib.pyplot as plt\nimport numpy as np\nfrom carbonplan_styles.mpl import set_theme\n\nset_theme()",
"_____no_output_____"
],
[
"axis_name_size = 12",
"_____no_output_____"
],
[
"# country shapes from GADM36\ncountries = geopandas.read_file(\"s3://carbonplan-climatetrace/inputs/shapes/countries.shp\")\n\n# CarbonPlan's emissions\nemissions = pd.read_csv(\"s3://carbonplan-climatetrace/v0.4/country_rollups_emissions.csv\")",
"_____no_output_____"
],
[
"agb = pd.read_csv(\"s3://carbonplan-climatetrace/v0.4/country_rollups_agb.csv\")",
"_____no_output_____"
],
[
"# Input data\n# ----------\n\n# GFW emissions\ngfw_emissions = pd.read_excel(\n \"s3://carbonplan-climatetrace/validation/gfw_global_emissions.xlsx\",\n sheet_name=\"Country co2 emissions\",\n).dropna(axis=0)\ngfw_emissions = gfw_emissions[gfw_emissions[\"threshold\"] == 10] # select threshold\n\n# rename\ngfw_emissions.loc[gfw_emissions.country == \"Republic of Congo\", \"country\"] = \"Congo\"\ngfw_emissions.loc[\n gfw_emissions.country == \"Bolivia\", \"country\"\n] = \"Bolivia (Plurinational State of)\"\ngfw_emissions.loc[gfw_emissions.country == \"Brunei\", \"country\"] = \"Brunei Darussalam\"\ngfw_emissions.loc[gfw_emissions.country == \"Côte d'Ivoire\", \"country\"] = \"Côte dIvoire\"\ngfw_emissions.loc[gfw_emissions.country == \"Laos\", \"country\"] = \"Lao Peoples Democratic Republic\"\ngfw_emissions.loc[gfw_emissions.country == \"Swaziland\", \"country\"] = \"Eswatini\"\ngfw_emissions.loc[gfw_emissions.country == \"Tanzania\", \"country\"] = \"United Republic of Tanzania\"\ngfw_emissions.loc[\n gfw_emissions.country == \"Venezuela\", \"country\"\n] = \"Venezuela (Bolivarian Republic of)\"\ngfw_emissions.loc[gfw_emissions.country == \"Vietnam\", \"country\"] = \"Viet Nam\"\ngfw_emissions.loc[\n gfw_emissions.country == \"Virgin Islands, U.S.\", \"country\"\n] = \"United States Virgin Islands\"\ngfw_emissions.loc[gfw_emissions.country == \"Zimbabwe\", \"country\"] = \"Zimbabwe)\"",
"_____no_output_____"
],
[
"emissions.groupby(\"begin_date\").sum().mean() / 1e9",
"_____no_output_____"
],
[
"# Merge emissions dataframes with countries GeoDataFrame\ngfw_countries = countries.merge(gfw_emissions.rename(columns={\"country\": \"name\"}), on=\"name\")\ntrace_countries = countries.merge(emissions.rename(columns={\"iso3_country\": \"alpha3\"}), on=\"alpha3\")\nagb_countries = countries.merge(agb.rename(columns={\"iso3_country\": \"alpha3\"}), on=\"alpha3\")",
"_____no_output_____"
],
[
"agb = pd.merge(\n left=agb_countries.rename(columns={\"agb\": \"trace_agb\"}),\n right=gfw_countries[[\"alpha3\", \"abg_co2_stock_2000__Mg\"]].rename(\n columns={\"abg_co2_stock_2000__Mg\": \"gfw_agb_co2\"}\n ),\n on=\"alpha3\",\n)\nagb[\"trace_agb_co2\"] = agb.trace_agb * 0.5 * 3.67\n\nagb[\"trace_agb_co2\"] = agb.trace_agb_co2 / 1e6\nagb[\"gfw_agb_co2\"] = agb.gfw_agb_co2 / 1e6\n\nagb = agb[[\"name\", \"alpha3\", \"geometry\", \"trace_agb_co2\", \"gfw_agb_co2\"]]",
"_____no_output_____"
],
[
"# reformat to \"wide\" format (time x country)\ntrace_wide = (\n emissions.drop(columns=[\"end_date\"])\n .pivot(index=\"begin_date\", columns=\"iso3_country\")\n .droplevel(0, axis=1)\n)\ntrace_wide.index = pd.to_datetime(trace_wide.index)\n\ngfw_wide = gfw_emissions.set_index(\"country\").filter(regex=\"whrc_aboveground_co2_emissions_Mg_.*\").T\ngfw_wide.index = [pd.to_datetime(f\"{l[-4:]}-01-01\") for l in gfw_wide.index]\n\ngfw_wide.head()",
"_____no_output_____"
],
[
"df = pd.read_csv(\"s3://carbonplan-climatetrace/v0.4/country_rollups_emissions_from_clearing.csv\")",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
],
[
"df.loc[df.iso3_country == \"AGO\"].tCO2eq / 1e6",
"_____no_output_____"
]
],
[
[
"## Part 1 - Compare time-averaged country emissions (tropics only)\n",
"_____no_output_____"
]
],
[
[
"# Create a new dataframe with average emissions\navg_emissions = countries.set_index(\"alpha3\")\navg_emissions[\"trace\"] = trace_wide.mean().transpose() / 1e6\n# avg_emissions[\"trace\"] = trace_wide.loc['2020-01-01'] / 1e6\n\navg_emissions = avg_emissions.reset_index().set_index(\"name\")\navg_emissions[\"gfw\"] = gfw_wide.mean().transpose() / 1e6\n# avg_emissions[\"gfw\"] = gfw_wide.loc['2020-01-01'] / 1e6\n\navg_emissions = avg_emissions.dropna()",
"_____no_output_____"
],
[
"len(avg_emissions)",
"_____no_output_____"
],
[
"from sklearn.metrics import r2_score",
"_____no_output_____"
],
[
"r2_score(avg_emissions.gfw, avg_emissions.trace)",
"_____no_output_____"
],
[
"avg_emissions[\"me\"] = avg_emissions.trace - avg_emissions.gfw\navg_emissions[\"mae\"] = (avg_emissions.trace - avg_emissions.gfw).abs()\navg_emissions[\"mape\"] = (avg_emissions.trace - avg_emissions.gfw).abs() / avg_emissions.gfw * 100\navg_emissions = avg_emissions.replace(np.inf, np.nan)\navg_emissions.mean().round(2)",
"_____no_output_____"
],
[
"sub = avg_emissions.loc[(avg_emissions.mape > 1) & (avg_emissions.gfw > 1)]\nsub",
"_____no_output_____"
],
[
"(avg_emissions.gfw > 1).mean()",
"_____no_output_____"
],
[
"top20 = avg_emissions.sort_values(by=\"mae\", ascending=False).head(20)",
"_____no_output_____"
],
[
"names = {\n \"Democratic Republic of the Congo\": \"DRC\",\n \"Lao Peoples Democratic Republic\": \"Laos\",\n \"Bolivia (Plurinational State of)\": \"Bolivia\",\n \"Côte dIvoire\": \"Côte d'Ivoire\",\n \"United Republic of Tanzania\": \"Tanzania\",\n \"Viet Nam\": \"Vietnam\",\n \"Venezuela (Bolivarian Republic of)\": \"Venezuela\",\n}",
"_____no_output_____"
],
[
"plt.figure(figsize=(12, 10))\n\nfor i, row in top20.reset_index()[[\"name\", \"alpha3\"]].iterrows():\n plt.subplot(5, 4, i + 1)\n name = row[\"name\"]\n alpha3 = row[\"alpha3\"]\n plt.plot(gfw_wide[name].index, gfw_wide[name].values / 1e6, label=\"Zarin et al.\")\n plt.plot(trace_wide[alpha3].index, trace_wide[alpha3].values / 1e6, label=\"CarbonPlan\")\n\n plt.xticks([\"2001-01-01\", \"2010-01-01\", \"2020-01-01\"], [2001, 2010, 2020])\n if name in names:\n name = names[name]\n plt.title(name, fontsize=axis_name_size)\n if i > 3:\n plt.ylim(0, 200)\n if i == 8:\n plt.ylabel(\"Emissions [Mt CO2 / yr]\", fontsize=axis_name_size)\n\nax = plt.gca()\nfig = plt.gcf()\nhandles, labels = ax.get_legend_handles_labels()\nfig.legend(handles, labels, loc=\"upper center\", ncol=2, bbox_to_anchor=(0.5, 1.03))\nplt.tight_layout()\nplt.savefig(\"top20_time_series.png\", bbox_inches=\"tight\")\nplt.show()\nplt.close()",
"_____no_output_____"
],
[
"# Scatter Plot\nxmin = 1e-6\nxmax = 1e4\nplt.figure(figsize=(10, 5))\nplt.subplot(1, 2, 1)\nplt.plot([xmin, xmax], [xmin, xmax], \"0.5\")\navg_emissions.plot.scatter(\"gfw\", \"trace\", ax=plt.gca())\nplt.gca().set_xscale(\"log\")\nplt.gca().set_yscale(\"log\")\nplt.ylabel(\"CarbonPlan [Mt CO$_2$ / yr]\", fontsize=axis_name_size)\nplt.xlabel(\"Zarin [Mt CO$_2$ / yr]\", fontsize=axis_name_size)\nplt.xlim(xmin, xmax)\nplt.ylim(xmin, xmax)\nplt.title(\"a) Forest related carbon emissions\", fontsize=axis_name_size)\n\nxmin = 1e-4\nxmax = 1e6\nplt.subplot(1, 2, 2)\nplt.plot([xmin, xmax], [xmin, xmax], \"0.5\")\nagb.plot.scatter(\"gfw_agb_co2\", \"trace_agb_co2\", ax=plt.gca())\nplt.gca().set_xscale(\"log\")\nplt.gca().set_yscale(\"log\")\nplt.ylabel(\"CarbonPlan [Mt CO$_2$]\", fontsize=axis_name_size)\nplt.xlabel(\"Zarin [Mt CO$_2$]\", fontsize=axis_name_size)\nplt.xlim(xmin, xmax)\nplt.ylim(xmin, xmax)\nplt.title(\"b) Forest AGB stock in 2000\", fontsize=axis_name_size)\n\nplt.tight_layout()\nplt.savefig(\"gfw_scatter.png\")",
"_____no_output_____"
]
],
[
[
"## Part 2 - Maps of Tropical Emissions\n",
"_____no_output_____"
]
],
[
[
"from mpl_toolkits.axes_grid1 import make_axes_locatable",
"_____no_output_____"
],
[
"plt.figure(figsize=(14, 8))\nplt.subplot(2, 1, 1)\nkwargs = dict(\n legend=True,\n legend_kwds={\n \"orientation\": \"vertical\",\n \"label\": \"Emissions [Mt CO$_2$ / yr]\",\n },\n lw=0.25,\n cmap=\"Reds\",\n vmin=0,\n vmax=1000,\n)\nax = plt.gca()\ndivider = make_axes_locatable(ax)\ncax = divider.append_axes(\"right\", size=\"2%\", pad=0.2)\navg_emissions.plot(\"trace\", ax=ax, cax=cax, **kwargs)\nax.set_title(\"Forest related carbon emissions from CarbonPlan\", fontsize=axis_name_size)\nax.set_xlabel(\"Longitude\", fontsize=axis_name_size)\nax.set_ylabel(\"Latitude\", fontsize=axis_name_size)\n\nplt.subplot(2, 1, 2)\nkwargs = dict(\n legend=True,\n legend_kwds={\n \"orientation\": \"vertical\",\n \"label\": \"Emissions Difference [%]\",\n },\n lw=0.25,\n cmap=\"RdBu_r\",\n vmin=-20,\n vmax=20,\n)\navg_emissions[\"pdiff\"] = (\n (avg_emissions[\"trace\"] - avg_emissions[\"gfw\"]) / avg_emissions[\"gfw\"]\n) * 100\n\nax = plt.gca()\ndivider = make_axes_locatable(ax)\ncax = divider.append_axes(\"right\", size=\"2%\", pad=0.2)\navg_emissions.plot(\"pdiff\", ax=ax, cax=cax, **kwargs)\nax.set_title(\"% difference from Zarin\", fontsize=axis_name_size)\nax.set_xlabel(\"Longitude\", fontsize=axis_name_size)\nax.set_ylabel(\"Latitude\", fontsize=axis_name_size)\n\nplt.tight_layout()\nplt.savefig(\"gfw_map.png\")",
"_____no_output_____"
]
],
[
[
"## Part 3 - Compare fire emissions\n",
"_____no_output_____"
]
],
[
[
"# CarbonPlan's emissions\nemissions = {}\nversions = [\"v0.4\"]\nfor version in versions:\n for mechanism in [\"fire\"]:\n emissions[version + \"-\" + mechanism] = pd.read_csv(\n \"s3://carbonplan-climatetrace/{}/country_rollups_emissions_from_{}.csv\".format(\n version, mechanism\n )\n )\n\n# Blue Sky Fire emissions\nemissions[\"Blue Sky\"] = pd.read_csv(\"forest-fires_bsa.csv\")",
"_____no_output_____"
],
[
"emissions[f\"{version}-fire\"]",
"_____no_output_____"
],
[
"emissions[\"Blue Sky\"]",
"_____no_output_____"
],
[
"version = \"v0.4\"\n\ncomparison = pd.merge(\n emissions[f\"{version}-fire\"].rename({\"tCO2eq\": \"CarbonPlan\"}, axis=1),\n emissions[\"Blue Sky\"].rename({\"tCO2\": \"BSA\"}, axis=1),\n how=\"inner\", # \"left\",\n left_on=[\"iso3_country\", \"begin_date\"],\n right_on=[\"iso3_country\", \"begin_date\"],\n)\n\ncomparison[\"BSA\"] /= 1e6\ncomparison[\"CarbonPlan\"] /= 1e6\n\ncomparison[\"year\"] = pd.to_datetime(comparison.begin_date).dt.year\ncomparison[\"BSA\"] = comparison.BSA.fillna(0)",
"_____no_output_____"
],
[
"r2_score(comparison.BSA, comparison.CarbonPlan)",
"_____no_output_____"
],
[
"(comparison.CarbonPlan - comparison.BSA).mean()",
"_____no_output_____"
],
[
"(comparison.CarbonPlan <= comparison.BSA).mean()",
"_____no_output_____"
],
[
"len(comparison.iso3_country.unique())",
"_____no_output_____"
],
[
"xmin = 1e-4\nxmax = 1e4\nplt.figure(figsize=(5, 5))\nplt.plot([xmin, xmax], [xmin, xmax], \"0.5\")\ncomparison.plot.scatter(\"BSA\", \"CarbonPlan\", ax=plt.gca())\nplt.gca().set_xscale(\"log\")\nplt.gca().set_yscale(\"log\")\nplt.ylabel(\"CarbonPlan [Mt CO$_2$ / yr]\", fontsize=axis_name_size)\nplt.xlabel(\"BSA [Mt CO$_2$ / yr]\", fontsize=axis_name_size)\nplt.yticks()\nplt.xlim(xmin, xmax)\nplt.ylim(xmin, xmax)\nplt.title(\"Forest fire emissions\", fontsize=axis_name_size)\nplt.savefig(\"bsa_scatter.png\", bbox_inches=\"tight\")",
"_____no_output_____"
],
[
"avg_yr = comparison.groupby(\"iso3_country\").mean()\n\nxmin = 1e-4\nxmax = 1e4\nplt.figure(figsize=(5, 5))\nplt.plot([xmin, xmax], [xmin, xmax], \"0.5\")\navg_yr.plot.scatter(\"BSA\", \"CarbonPlan\", ax=plt.gca())\nplt.gca().set_xscale(\"log\")\nplt.gca().set_yscale(\"log\")\nplt.ylabel(\"CarbonPlan [Mt CO$_2$ / yr]\", fontsize=axis_name_size)\nplt.xlabel(\"BSA [Mt CO$_2$ / yr]\", fontsize=axis_name_size)\nplt.xlim(xmin, xmax)\nplt.ylim(xmin, xmax)\nplt.title(\"Forest fire emissions\", fontsize=axis_name_size)\nplt.tight_layout()\nplt.savefig(\"bsa_scatter_avg.png\")",
"_____no_output_____"
],
[
"comparison.head()",
"_____no_output_____"
],
[
"comparison.loc[comparison.iso3_country.isin([\"RUS\", \"USA\"])]",
"_____no_output_____"
],
[
"comparison.loc[comparison.iso3_country.isin([\"BRA\"])]",
"_____no_output_____"
],
[
"emissions[\"Mt CO2\"] = emissions.tCO2eq / 1e6\nsub = emissions.loc[(emissions.iso3_country == \"LKA\"), [\"begin_date\", \"Mt CO2\", \"iso3_country\"]]\nsub[\"year\"] = pd.to_datetime(sub.begin_date).dt.year",
"_____no_output_____"
],
[
"plt.plot(sub.year, sub[\"Mt CO2\"], \"o-\")\nplt.xticks([2001, 2005, 2010, 2015, 2020], [2001, 2005, 2010, 2015, 2020])\nplt.ylabel(\"Mt CO2\")\nplt.grid()",
"_____no_output_____"
],
[
"sub[[\"iso3_country\", \"year\", \"Mt CO2\"]]",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4affd3bf59c677d3229c2cd7498eecc8b09b40b7
| 570,118 |
ipynb
|
Jupyter Notebook
|
notebooks/LasVegasStrip.ipynb
|
mherbert93/unit2-build
|
b05567cf3971f8afa4af39be1251d48a667dffd3
|
[
"MIT"
] | null | null | null |
notebooks/LasVegasStrip.ipynb
|
mherbert93/unit2-build
|
b05567cf3971f8afa4af39be1251d48a667dffd3
|
[
"MIT"
] | null | null | null |
notebooks/LasVegasStrip.ipynb
|
mherbert93/unit2-build
|
b05567cf3971f8afa4af39be1251d48a667dffd3
|
[
"MIT"
] | null | null | null | 663.699651 | 370,123 | 0.914958 |
[
[
[
"import pandas as pd\n\ndf = pd.read_csv(\"https://archive.ics.uci.edu/ml/machine-learning-databases/00397/LasVegasTripAdvisorReviews-Dataset.csv\", sep=';')",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
],
[
"df['Score'].value_counts(normalize=True)",
"_____no_output_____"
],
[
"from sklearn.model_selection import train_test_split\n\ntrain, test = train_test_split(df, train_size=0.80, test_size=0.20, stratify=df['Score'], random_state=1337)",
"_____no_output_____"
],
[
"train['Score'].value_counts(normalize=True)",
"_____no_output_____"
],
[
"def wrangle(X):\n\n X = X.copy()\n\n def country_aggregation(row): #reduce number of country categories to 4. 'USA', 'Canada', 'UK', and all else 'other'.\n if row['User country'] != \"USA\" and row['User country'] != 'Canada' and row['User country'] != 'UK':\n return \"Other\"\n else:\n return row['User country']\n\n def score_aggregation(row):\n if row['Score'] == 5:\n return \"Excellent\"\n elif row['Score'] == 4 or row['Score'] == 3:\n return \"Average\"\n else:\n return \"Bad\"\n\n X['User country'] = X.apply(country_aggregation, axis=1)\n\n #if not predict: #only modify score column if passing in training/test data. Do not run when running real predictions!\n X['Score'] = X.apply(score_aggregation, axis=1)\n X = X.drop(['Member years'], axis=1)\n\n X['Hotel stars'] = X['Hotel stars'].str.replace(\",\" , \".\").astype(str)\n X['Hotel stars'] = X['Hotel stars'].replace({\"3\": 1, \"3.5\": 2, \"4\": 3, \"4.5\": 4, \"5\": 5}).astype(int) #ordinal encoding\n\n X.loc[(X['Hotel name'] == \"Trump International Hotel Las Vegas\") | #Trump international is a hotel only, no casino.\n (X['Hotel name'] == \"Marriott's Grand Chateau\") | #Marriott's Grand Chateau is a hotel only, no casino.\n (X['Hotel name'] == \"Wyndham Grand Desert\"), 'Casino'] = \"NO\" #Wyndham Grand Desert is a hotel only, no casino.\n\n return X",
"_____no_output_____"
],
[
"train = wrangle(train)\ntest = wrangle(test)",
"_____no_output_____"
],
[
"from sklearn.metrics import accuracy_score\n\nmajority_class = train['Score'].mode()[0]\ny_pred = [majority_class] * len(train['Score'])\n\nprint(\"Train baseline accuracy is: \", accuracy_score(train['Score'], y_pred))",
"Train baseline accuracy is: 0.46898263027295284\n"
],
[
"majority_class = test['Score'].mode()[0]\ny_pred = [majority_class] * len(test['Score'])\n\nprint(\"Test baseline accuracy is: \", accuracy_score(test['Score'], y_pred))",
"Test baseline accuracy is: 0.46534653465346537\n"
],
[
"target = 'Score'\n\ntrain_features = train.drop([target], axis=1)\nnumeric_features = train_features.select_dtypes(include='number').columns.tolist()\n\ncategorical_features = train_features.select_dtypes(exclude='number').nunique().index.tolist()\n\n\nfeatures = numeric_features + categorical_features\n\nfeatures_logistic = categorical_features.copy()\nfeatures_logistic.append('Hotel stars') #add hotel stars to features list, so we can specify to onehotencoder that we want to encode it, even though it is numeric(as we already did ordinal encoding on it)",
"_____no_output_____"
],
[
"y_train = train[target]\nX_train = train[features]\nX_test = test[features]\ny_test = test[target]",
"_____no_output_____"
],
[
"X_train.shape",
"_____no_output_____"
],
[
"from sklearn.model_selection import RandomizedSearchCV, GridSearchCV\nfrom sklearn.linear_model import LogisticRegression\nfrom sklearn.ensemble import RandomForestClassifier, VotingClassifier\nfrom sklearn.preprocessing import StandardScaler\nimport category_encoders as ce\nfrom sklearn.pipeline import Pipeline\nfrom sklearn.feature_selection import SelectKBest\nfrom xgboost import XGBClassifier\nfrom imblearn.over_sampling import RandomOverSampler\n\n#When hyperparamater tuning, set tune to \"True\", and mark each model that we want to tune to \"True\".\ntune = False\nforest = False\nlogistic = False\nxgboost = False\n\nforest_distributions = {\n 'model__n_estimators': range(250, 500, 50),\n 'model__max_depth': range(3, 14),\n 'model__max_features': range(2, 14),\n 'model__min_samples_leaf': range(2, 4)\n}\nlogistic_distributions = {\n 'kbest__k': range(1, 20),\n 'model__C': [0.001, 0.01, 0.1, 1, 10, 100, 1000]\n}\n\nxgboost_distributions = {\n 'model__n_estimators': [75, 100, 125, 150, 175],\n 'model__max_depth': [6, 7, 8, 9, 10, 11, 12, 13],\n 'model__learning_rate': [0.01, 0.02, 0.03, 0.04, 0.05, 0.07, 0.10, 0.12, 0.14, 0.16],\n 'model__min_child_leaf':[1, 2, 3],\n 'model__min_child_weight': [1, 2, 3, 4],\n 'model__colsample_bytree':[0.2, 0.3, 0.4, 0.50, 0.60, 0.70],\n 'model__subsample':[0.3, 0.4, 0.5, 0.6, 0.7, 0.8],\n 'model__gamma':[0],\n 'model__scale_pos_weight': [1, 5, 10, 15, 20, 25, 30, 35, 40, 45, 50, 55, 60, 65, 70,\n 75, 80, 85, 90, 95, 100]\n }\n\nif tune: #If we are hyperparamater tuning, pass no parameters into estimators and find paramaters via GridSearchCV / RandomizedSearchCV\n\n forest_pipeline = Pipeline([('encoder', ce.OrdinalEncoder()),\n ('model', RandomForestClassifier(random_state=1337))])\n\n logistic_pipeline = Pipeline([('encoder', ce.OneHotEncoder()),\n ('scaler', StandardScaler()),\n ('kbest', SelectKBest()),\n ('model', LogisticRegression(random_state=1337))])\n\n xgboost_pipeline = Pipeline([('encoder', ce.OrdinalEncoder()),\n ('model', XGBClassifier(seed=1337))])\n\n forest_search = GridSearchCV(\n forest_pipeline,\n param_grid=forest_distributions,\n cv=3,\n scoring='neg_log_loss', #good out of the box scoring metric for multiclass hyperparameter tuning\n verbose=10,\n n_jobs=15\n )\n logistic_search = GridSearchCV(\n logistic_pipeline,\n param_grid=logistic_distributions,\n cv=3,\n scoring='neg_log_loss', #good out of the box scoring metric for multiclass hyperparameter tuning\n verbose=10,\n n_jobs=15\n )\n xgboost_search = RandomizedSearchCV(\n estimator=xgboost_pipeline,\n param_distributions=xgboost_distributions,\n n_iter=10000,\n cv=3,\n scoring='neg_log_loss', #good out of the box scoring metric for multiclass hyperparameter tuning\n verbose=10,\n random_state=1337,\n n_jobs=15\n )\n\n X_train, y_train = RandomOverSampler(sampling_strategy='not majority').fit_resample(X_train, y_train)\n\n if forest:\n forest_search.fit(X_train, y_train)\n forest_train_pred = forest_search.predict(X_train)\n forest_test_pred = forest_search.predict(X_test)\n forest_test_pred_proba = forest_search.predict_proba(X_test)\n\n if logistic:\n logistic_search.fit(X_train, y_train)\n logistic_train_pred = logistic_search.predict(X_train)\n logistic_test_pred = logistic_search.predict(X_test)\n logistic_test_pred_proba = logistic_search.predict_proba(X_test)\n\n if xgboost:\n xgboost_search.fit(X_train, y_train)\n xgboost_train_pred = xgboost_search.predict(X_train)\n xgboost_test_pred = xgboost_search.predict(X_test)\n xgboost_test_pred_proba = xgboost_search.predict_proba(X_test)\n\n #When hyperparameter tuning, pass our best estimators into votingclassifier. Only run when all 3 models are being tuned.\n if forest and logistic and xgboost:\n voting_model = VotingClassifier(estimators=[('forest', forest_search.best_estimator_), #VotingClassifier is Soft Voting/Majority Rule classifier for unfitted estimators\n ('logistic', logistic_search.best_estimator_),\n ('xgboost', xgboost_search.best_estimator_),],\n voting='soft', weights=[2, 1, 2]) #soft voting per recommendation from sklearn documentation, when used on tuned classifiers.\n voting_model.fit(X_train, y_train)\n\nelse: #If we are not hyperparameter tuning, pass in our best params(from previous tuning runs).\n forest_pipeline = Pipeline([('encoder', ce.OrdinalEncoder()),\n ('model', RandomForestClassifier(random_state=1337,\n max_depth=13,\n max_features=11,\n min_samples_leaf=2,\n n_estimators=450))])\n logistic_pipeline = Pipeline([('encoder', ce.OneHotEncoder(cols=features_logistic)), #use features list which contains \"Hotel stars\" so that it gets properly encoded.\n ('scaler', StandardScaler()),\n ('kbest', SelectKBest(k=19)),\n ('model', LogisticRegression(random_state=1337, C=0.01))])\n\n xgboost_pipeline = Pipeline([('encoder', ce.OrdinalEncoder()),\n ('model', XGBClassifier(random_state=1337, n_estimators=175, min_child_weight=1,\n min_child_leaf=2, max_depth=11, learning_rate=0.04,\n gamma=0, subsample=0.8, colsample_bytree=0.3, scale_pos_weight=100))])\n\n\n X_train, y_train = RandomOverSampler(sampling_strategy='not majority').fit_resample(X_train, y_train) #over sample all but the majority class\n\n forest_pipeline.fit(X_train, y_train)\n forest_train_pred = forest_pipeline.predict(X_train)\n forest_test_pred = forest_pipeline.predict(X_test)\n forest_test_pred_proba = forest_pipeline.predict_proba(X_test)\n\n logistic_pipeline.fit(X_train, y_train)\n logistic_train_pred = logistic_pipeline.predict(X_train)\n logistic_test_pred = logistic_pipeline.predict(X_test)\n logistic_test_pred_proba = logistic_pipeline.predict_proba(X_test)\n\n xgboost_pipeline.fit(X_train, y_train)\n xgboost_train_pred = xgboost_pipeline.predict(X_train)\n xgboost_test_pred = xgboost_pipeline.predict(X_test)\n xgboost_test_pred_proba = xgboost_pipeline.predict_proba(X_test)\n\n voting_model = VotingClassifier(estimators=[('forest', forest_pipeline), #VotingClassifier is Soft Voting/Majority Rule classifier for unfitted estimators\n ('logistic', logistic_pipeline),\n ('xgboost', xgboost_pipeline),],\n voting='soft') #soft voting per recommendation from sklearn documentation, when used on tuned classifiers.\n voting_model.fit(X_train, y_train)",
"_____no_output_____"
],
[
"#pickle the model\nfrom joblib import dump\ndump(voting_model, 'voting_model.joblib', compress=True)",
"_____no_output_____"
],
[
"#print package versions for pipenv\nimport joblib\nimport sklearn\nimport category_encoders as ce\nimport xgboost\nimport imblearn\nprint(f'joblib=={joblib.__version__}')\nprint(f'scikit-learn=={sklearn.__version__}')\nprint(f'category_encoders=={ce.__version__}')\nprint(f'xgboost=={xgboost.__version__}')\nprint(f'imblearn=={imblearn.__version__}')\n\n",
"joblib==0.14.1\nscikit-learn==0.22.2.post1\ncategory_encoders==2.1.0\nxgboost==1.0.2\nimblearn==0.6.2\n"
],
[
"from sklearn.metrics import accuracy_score\nfrom sklearn.metrics import classification_report\nfrom sklearn.metrics import roc_auc_score, plot_roc_curve\nif tune:\n target_names = ['Average', 'Bad', 'Excellent']\n\n if forest:\n print(forest_search.best_params_, '\\n')\n print(\"Best Random Forest CV score: \", forest_search.best_score_, '\\n')\n print(\"Random forest ROC-AUC: \", roc_auc_score(y_test, forest_test_pred_proba, multi_class='ovr', labels=target_names), '\\n')\n print(classification_report(y_test, forest_test_pred, target_names=target_names), '\\n')\n\n print('\\n')\n\n if logistic:\n print(logistic_search.best_params_, '\\n')\n print(\"Best logistic regression CV score: \", logistic_search.best_score_, '\\n')\n print(\"Logistic regression ROC-AUC: \", roc_auc_score(y_test, logistic_test_pred_proba, multi_class='ovr', labels=target_names), '\\n')\n print(classification_report(y_test, logistic_test_pred, target_names=target_names), '\\n')\n\n print('\\n')\n\n if xgboost:\n print(xgboost_search.best_params_, '\\n')\n print(\"Best xgboost CV score: \", xgboost_search.best_score_, '\\n')\n print(\"Xgboost ROC-AUC: \", roc_auc_score(y_test, xgboost_test_pred_proba, multi_class='ovr', labels=target_names), '\\n')\n print(classification_report(y_test, xgboost_test_pred, target_names=target_names), '\\n')\n\n print('\\n')\n\n if forest and logistic and xgboost:\n print(\"Voting classifier, final accuracy score on test set: \", voting_model.score(X_test, y_test))\n\n combined_model = voting_model.predict_proba(X_test)\n print(\"Voting classifier, final ROC AUC on test set: \", roc_auc_score(y_test, combined_model, multi_class='ovr', labels=target_names))\nelse:\n target_names = ['Average', 'Bad', 'Excellent']\n print(\"Random forest ROC-AUC: \", roc_auc_score(y_test, forest_test_pred_proba, multi_class='ovr', labels=target_names), '\\n')\n print(classification_report(y_test, forest_test_pred, target_names=target_names))\n\n print('\\n')\n\n print(\"Logistic regression ROC-AUC: \", roc_auc_score(y_test, logistic_test_pred_proba, multi_class='ovr', labels=target_names), '\\n')\n print(classification_report(y_test, logistic_test_pred, target_names=target_names))\n\n print('\\n')\n\n print(\"Xgboost ROC-AUC: \", roc_auc_score(y_test, xgboost_test_pred_proba, multi_class='ovr', labels=target_names), '\\n')\n print(classification_report(y_test, xgboost_test_pred, target_names=target_names))\n\n print('\\n')\n\n print(\"Voting classifier, final accuracy score on test set: \", voting_model.score(X_test, y_test))\n\n combined_model = voting_model.predict_proba(X_test)\n print(\"Voting classifier, final ROC AUC on test set: \", roc_auc_score(y_test, combined_model, multi_class='ovr', labels=target_names))",
"Random forest ROC-AUC: 0.7010360544292188 \n\n precision recall f1-score support\n\n Average 0.57 0.55 0.56 47\n Bad 0.31 0.50 0.38 8\n Excellent 0.48 0.43 0.45 46\n\n accuracy 0.50 101\n macro avg 0.45 0.50 0.46 101\nweighted avg 0.50 0.50 0.50 101\n\n\n\nLogistic regression ROC-AUC: 0.6103605551769339 \n\n precision recall f1-score support\n\n Average 0.59 0.43 0.49 47\n Bad 0.16 0.38 0.22 8\n Excellent 0.50 0.52 0.51 46\n\n accuracy 0.47 101\n macro avg 0.42 0.44 0.41 101\nweighted avg 0.51 0.47 0.48 101\n\n\n\nXgboost ROC-AUC: 0.6821405168030156 \n\n precision recall f1-score support\n\n Average 0.59 0.64 0.61 47\n Bad 0.50 0.25 0.33 8\n Excellent 0.52 0.52 0.52 46\n\n accuracy 0.55 101\n macro avg 0.54 0.47 0.49 101\nweighted avg 0.55 0.55 0.55 101\n\n\n\nVoting classifier, final accuracy score on test set: 0.5445544554455446\nVoting classifier, final ROC AUC on test set: 0.6850820555730883\n"
],
[
"###While this is a good way to evaluate the estimator we are passing in, unless using a 3 way train/validate/test split, this should not be used for feature selection.\n###Instead, passing in an unfit estimator and specifying the amount of CV rounds will allow us to see more generalized permutation importance.\nimport eli5\nfrom eli5.sklearn import PermutationImportance\n\npermuter = PermutationImportance(\n xgboost_pipeline.named_steps.model, #prefit estimator\n scoring='roc_auc_ovo',\n n_iter=300,\n random_state=1337\n)\n\npermuter.fit(xgboost_pipeline.named_steps.encoder.transform(X_test), y_test)\nfeature_names = X_test.columns.tolist()\n\neli5.show_weights(\n permuter,\n top=None,\n feature_names=feature_names\n)",
"c:\\users\\user\\venv\\lib\\site-packages\\sklearn\\utils\\deprecation.py:144: FutureWarning: The sklearn.metrics.scorer module is deprecated in version 0.22 and will be removed in version 0.24. The corresponding classes / functions should instead be imported from sklearn.metrics. Anything that cannot be imported from sklearn.metrics is now part of the private API.\n warnings.warn(message, FutureWarning)\nc:\\users\\user\\venv\\lib\\site-packages\\sklearn\\utils\\deprecation.py:144: FutureWarning: The sklearn.feature_selection.base module is deprecated in version 0.22 and will be removed in version 0.24. The corresponding classes / functions should instead be imported from sklearn.feature_selection. Anything that cannot be imported from sklearn.feature_selection is now part of the private API.\n warnings.warn(message, FutureWarning)\n"
],
[
"import eli5\nfrom eli5.sklearn import PermutationImportance\n\npermuter = PermutationImportance(\n forest_pipeline.named_steps.model, #prefit estimator\n scoring='roc_auc_ovo',\n n_iter=300,\n random_state=1337\n)\n\npermuter.fit(forest_pipeline.named_steps.encoder.transform(X_test), y_test)\nfeature_names = X_test.columns.tolist()\n\neli5.show_weights(\n permuter,\n top=None,\n feature_names=feature_names\n)",
"_____no_output_____"
],
[
"permuter = PermutationImportance(\n logistic_pipeline.named_steps.model, #prefit estimator\n scoring='roc_auc_ovo',\n n_iter=300,\n random_state=1337\n)\n\nX_test_encoded = logistic_pipeline.named_steps.encoder.transform(X_test)\nX_test_scaled = logistic_pipeline.named_steps.scaler.transform(X_test_encoded)\nX_test_final = logistic_pipeline.named_steps.kbest.transform(X_test_scaled)\n\nselected_mask = logistic_pipeline.named_steps.kbest.get_support()\nall_names = X_test_encoded.columns\nselected_names = all_names[selected_mask]\n\n\npermuter.fit(X_test_final, y_test)\nfeature_names = selected_names.tolist()\n\neli5.show_weights(\n permuter,\n top=None,\n feature_names=feature_names\n)",
"_____no_output_____"
],
[
"from pdpbox.pdp import pdp_isolate, pdp_plot\n\nfeature = 'Hotel stars'\n\nisolated = pdp_isolate(\n model=forest_pipeline,\n dataset=X_test,\n model_features=X_test.columns,\n feature=feature\n )\n\npdp_plot(isolated,feature_name=feature, plot_lines=True);",
"_____no_output_____"
],
[
"from pdpbox.pdp import pdp_interact, pdp_interact_plot\n\nfeatures_interact = ['Nr. reviews', 'Hotel stars']\n\ninteraction = pdp_interact(\n model=forest_pipeline,\n dataset=X_test,\n model_features=X_test.columns,\n features=features_interact\n)\n\npdp_interact_plot(interaction, plot_type='grid', feature_names=features_interact);\n\n\n",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4affdf34dcd9061a9b08b4d3afb01568176c5f2f
| 20,272 |
ipynb
|
Jupyter Notebook
|
lecture_21_python_oop/python_oop.ipynb
|
marcel-goldschen-ohm/CompNeuroSpring2019
|
1b982c86c5db336a63daf3868a3577c7d0fbb156
|
[
"MIT"
] | 3 |
2019-06-14T01:46:33.000Z
|
2020-12-10T20:13:21.000Z
|
lecture_21_python_oop/python_oop.ipynb
|
marcel-goldschen-ohm/NEU-Spring-2019
|
1b982c86c5db336a63daf3868a3577c7d0fbb156
|
[
"MIT"
] | null | null | null |
lecture_21_python_oop/python_oop.ipynb
|
marcel-goldschen-ohm/NEU-Spring-2019
|
1b982c86c5db336a63daf3868a3577c7d0fbb156
|
[
"MIT"
] | 7 |
2019-01-24T20:24:53.000Z
|
2020-12-10T23:44:58.000Z
| 22.010858 | 375 | 0.444653 |
[
[
[
"# Object Oriented Programming (OOP)",
"_____no_output_____"
],
[
"### classes and attributes",
"_____no_output_____"
]
],
[
[
"# definition of a class object\nclass vec3:\n pass\n\n# instance of the vec3 class object\na = vec3()\n\n# add some attributes to the v instance\na.x = 1\na.y = 2\na.z = 2.5\n\nprint(a)\nprint(a.z)\nprint(a.__dict__)",
"<__main__.vec3 object at 0x10dcac160>\n2.5\n{'x': 1, 'y': 2, 'z': 2.5}\n"
],
[
"# another instance of the vec3 class object\nb = vec3()\n\nprint(b)\nprint(b.__dict__)",
"<__main__.vec3 object at 0x10dc88e10>\n{}\n"
],
[
"class vec2:\n pass\n\nprint(isinstance(a, vec3))\nprint(isinstance(b, vec3))\nprint(isinstance(a, vec2))",
"True\nTrue\nFalse\n"
],
[
"# all vec3 instances should have the attributes x, y, z\nclass vec3:\n # attributes\n x = 1\n y = 2\n z = 2.5\n\na = vec3()\nb = vec3()\n\nprint(a, a.__dict__)\nprint(b, b.__dict__)\n\n# !!! Neither a nor b has x, y, or z! Huh?",
"<__main__.vec3 object at 0x10dcac358> {}\n<__main__.vec3 object at 0x10dcac320> {}\n"
],
[
"# the class vec3 owns x, y and z!\nprint(vec3.__dict__)",
"{'__module__': '__main__', 'x': 1, 'y': 2, 'z': 2.5, '__dict__': <attribute '__dict__' of 'vec3' objects>, '__weakref__': <attribute '__weakref__' of 'vec3' objects>, '__doc__': None}\n"
],
[
"# but a and b still have access to x, y and z\nprint(a.x, a.y, a.z)\nprint(b.x, b.y, b.z)",
"1 2 2.5\n1 2 2.5\n"
],
[
"# this changes z for all vec3 instances\nvec3.z = 3\n\nprint(vec3.__dict__)\nprint(a.x, a.y, a.z)\nprint(b.x, b.y, b.z)",
"{'__module__': '__main__', 'x': 1, 'y': 2, 'z': 3, '__dict__': <attribute '__dict__' of 'vec3' objects>, '__weakref__': <attribute '__weakref__' of 'vec3' objects>, '__doc__': None}\n1 2 3\n1 2 3\n"
],
[
"# what if we change z only for a?\na.z = 7\n\nprint(vec3.__dict__)\nprint(a.x, a.y, a.z)\nprint(b.x, b.y, b.z)",
"{'__module__': '__main__', 'x': 1, 'y': 2, 'z': 3, '__dict__': <attribute '__dict__' of 'vec3' objects>, '__weakref__': <attribute '__weakref__' of 'vec3' objects>, '__doc__': None}\n1 2 7\n1 2 3\n"
],
[
"# a now has both a class level attribute z and it's own attribute z!\nprint(a.__dict__)",
"{'z': 7}\n"
],
[
"# if we get rid of a.z, a will default back to vec3.z\ndel a.__dict__['z']\n\nprint(a.__dict__)\nprint(a.x, a.y, a.z)",
"{}\n1 2 3\n"
]
],
[
[
"### initialization",
"_____no_output_____"
]
],
[
[
"# class initialization\nclass vec3:\n \"\"\" __init__() is a method of vec3 (i.e. a function belonging to the class vec3).\n It is called whenever we create a new instance of a vec3 object.\n \"\"\"\n def __init__(self):\n self.x = 10\n self.y = 20\n self.z = 30\n\na = vec3()\n\nprint(vec3.__dict__)\nprint(a.__dict__)",
"{'__module__': '__main__', '__doc__': ' __init__() is a method of vec3 (i.e. a function belonging to the class vec3).\\n It is called whenever we create a new instance of a vec3 object.\\n ', '__init__': <function vec3.__init__ at 0x10dc91840>, '__dict__': <attribute '__dict__' of 'vec3' objects>, '__weakref__': <attribute '__weakref__' of 'vec3' objects>}\n{'x': 10, 'y': 20, 'z': 30}\n"
],
[
"# a and b are two separate instances of the vec3 object\nb = vec3()\n\nprint(a)\nprint(b)",
"<__main__.vec3 object at 0x10dcac550>\n<__main__.vec3 object at 0x10dcac5c0>\n"
],
[
"a.x = 5\n\nprint(a.__dict__)\nprint(b.__dict__)",
"{'x': 5, 'y': 20, 'z': 30}\n{'x': 10, 'y': 20, 'z': 30}\n"
],
[
"# passing arguments during class instantiation\nclass vec3:\n def __init__(self, x, y, z):\n self.x = x\n self.y = y\n self.z = z\n\na = vec3(2, 4, 6)\n\nprint(a.__dict__)",
"{'x': 2, 'y': 4, 'z': 6}\n"
],
[
"class vec3:\n def __init__(self, x=0, y=0, z=0):\n self.x = x\n self.y = y\n self.z = z\n\na = vec3()\nb = vec3(1, 2, 3)\n\nprint(a.__dict__)\nprint(b.__dict__)",
"{'x': 0, 'y': 0, 'z': 0}\n{'x': 1, 'y': 2, 'z': 3}\n"
]
],
[
[
"### methods",
"_____no_output_____"
]
],
[
[
"# class methods are just functions (e.g. __init__) that are wrapped up into the class\nclass vec3:\n def __init__(self, x=0, y=0, z=0):\n self.x = x\n self.y = y\n self.z = z\n \n def translate(self, dx, dy, dz):\n self.x += dx\n self.y += dy\n self.z += dz\n\na = vec3(1, 2, 3)\nprint(a.__dict__)\n\na.translate(10, 10, -10)\nprint(a.__dict__)",
"{'x': 1, 'y': 2, 'z': 3}\n{'x': 11, 'y': 12, 'z': -7}\n"
],
[
"# two ways to call a class method\na = vec3(1, 2, 3)\nvec3.translate(a, 10, 10, -10)\nprint(a.__dict__)\n\na = vec3(1, 2, 3)\na.translate(10, 10, -10)\nprint(a.__dict__)",
"{'x': 11, 'y': 12, 'z': -7}\n{'x': 11, 'y': 12, 'z': -7}\n"
]
],
[
[
"### special methods",
"_____no_output_____"
]
],
[
[
"class vec3:\n def __init__(self, x=0, y=0, z=0):\n self.x = x\n self.y = y\n self.z = z\n \n def __repr__(self):\n return f\"({self.x}, {self.y}, {self.z})\"\n \n def translate(self, dx, dy, dz):\n self.x += dx\n self.y += dy\n self.z += dz\n\na = vec3(1, 2, 3)\n\nprint(a)",
"(1, 2, 3)\n"
]
],
[
[
"### inheritance",
"_____no_output_____"
]
],
[
[
"# vec4 inherits all of vec3's functionality\nclass vec4(vec3):\n pass\n\na = vec4()\nprint(a.__dict__)\n\na = vec4(1, 2, 3)\nprint(a.__dict__)\n\na.translate(10, 10, -10)\nprint(a.__dict__)",
"{'x': 0, 'y': 0, 'z': 0}\n{'x': 1, 'y': 2, 'z': 3}\n{'x': 11, 'y': 12, 'z': -7}\n"
],
[
"print(issubclass(vec4, vec2))\nprint(issubclass(vec4, vec3))",
"False\nTrue\n"
],
[
"# vec4 extends vec3's functionality\nclass vec4(vec3):\n \"\"\" vec4 instantiation will use this __init__ instead of vec3's\n \"\"\"\n def __init__(self):\n self.w = 1\n\na = vec4()\n\nprint(a.__dict__)",
"{'w': 1}\n"
],
[
"class vec4(vec3):\n def __init__(self, x=0, y=0, z=0, w=1):\n vec3.__init__(self, x, y, z) # or you could use `super().__init__(x, y, z)`\n self.w = w\n\na = vec4()\nprint(a.__dict__)\n\na = vec4(1, 2, 3)\nprint(a.__dict__)\n\na = vec4(1, 2, 3, 0)\nprint(a.__dict__)",
"{'x': 0, 'y': 0, 'z': 0, 'w': 1}\n{'x': 1, 'y': 2, 'z': 3, 'w': 1}\n{'x': 1, 'y': 2, 'z': 3, 'w': 0}\n"
],
[
"print(help(vec4))",
"Help on class vec4 in module __main__:\n\nclass vec4(vec3)\n | vec4(x=0, y=0, z=0, w=1)\n | \n | Method resolution order:\n | vec4\n | vec3\n | builtins.object\n | \n | Methods defined here:\n | \n | __init__(self, x=0, y=0, z=0, w=1)\n | Initialize self. See help(type(self)) for accurate signature.\n | \n | ----------------------------------------------------------------------\n | Methods inherited from vec3:\n | \n | __repr__(self)\n | Return repr(self).\n | \n | translate(self, dx, dy, dz)\n | \n | ----------------------------------------------------------------------\n | Data descriptors inherited from vec3:\n | \n | __dict__\n | dictionary for instance variables (if defined)\n | \n | __weakref__\n | list of weak references to the object (if defined)\n\nNone\n"
],
[
"print(help(vec3))",
"Help on class vec3 in module __main__:\n\nclass vec3(builtins.object)\n | vec3(x=0, y=0, z=0)\n | \n | Methods defined here:\n | \n | __init__(self, x=0, y=0, z=0)\n | Initialize self. See help(type(self)) for accurate signature.\n | \n | __repr__(self)\n | Return repr(self).\n | \n | translate(self, dx, dy, dz)\n | \n | ----------------------------------------------------------------------\n | Data descriptors defined here:\n | \n | __dict__\n | dictionary for instance variables (if defined)\n | \n | __weakref__\n | list of weak references to the object (if defined)\n\nNone\n"
],
[
"# all classes inherit from builtins.object by default\nclass tmp1(object):\n pass\n\nclass tmp2():\n pass\n\nprint(help(tmp1))\nprint(help(tmp2))",
"Help on class tmp1 in module __main__:\n\nclass tmp1(builtins.object)\n | Data descriptors defined here:\n | \n | __dict__\n | dictionary for instance variables (if defined)\n | \n | __weakref__\n | list of weak references to the object (if defined)\n\nNone\nHelp on class tmp2 in module __main__:\n\nclass tmp2(builtins.object)\n | Data descriptors defined here:\n | \n | __dict__\n | dictionary for instance variables (if defined)\n | \n | __weakref__\n | list of weak references to the object (if defined)\n\nNone\n"
]
],
[
[
"### Exercise: Create a class ",
"_____no_output_____"
],
[
"### Diabetes dataset",
"_____no_output_____"
]
],
[
[
"import numpy as np\nfrom sklearn import datasets\n\ndiabetes = datasets.load_diabetes()\nX = diabetes.data\ny = diabetes.target\ny -= y.mean()\nfeatures = \"age sex bmi map tc ldl hdl tch ltg glu\".split()",
"_____no_output_____"
]
],
[
[
"### OLS regression class",
"_____no_output_____"
]
],
[
[
"from sklearn.linear_model import LinearRegression\n\nclass MyLinearRegression:\n def __init__(self, X, y):\n self.data = X\n self.target = y\n self.model = LinearRegression()\n self.fit()\n \n def fit(self):\n self.model.fit(self.data, self.target)\n return self.predict(self.data)\n \n def predict(self, X):\n return self.model.predict(X)\n \n def params(self):\n return self.model.coef_\n \n def getMSE(self, X, y):\n return np.mean((y - self.predict(X))**2)\n \n def getR2(self, X, y):\n return self.model.score(X, y)",
"_____no_output_____"
],
[
"mymodel = MyLinearRegression(X, y)\n\nprint(mymodel.params())\nprint(f\"MSE = {mymodel.getMSE(X, y)}\")\nprint(f\"R^2 = {mymodel.getR2(X, y)}\")",
"[ -10.01219782 -239.81908937 519.83978679 324.39042769 -792.18416163\n 476.74583782 101.04457032 177.06417623 751.27932109 67.62538639]\nMSE = 2859.6903987680657\nR^2 = 0.5177494254132934\n"
]
],
[
[
"### Exercise: Add a method to MyLinearRegression that automatically loads the dibaetes data set.",
"_____no_output_____"
],
[
"### Exercise: Add a method to MyLinearRegression that plots the slope factors in a bar graph.",
"_____no_output_____"
],
[
"### Exercise: Ridge regression class",
"_____no_output_____"
]
],
[
[
"from sklearn.linear_model import Ridge\nfrom sklearn.model_selection import cross_validate, GridSearchCV\n\nclass MyRigeRegression:\n def __init__(self, X, y, alphas):",
"_____no_output_____"
]
],
[
[
"### Exercise: KNN regression class",
"_____no_output_____"
]
],
[
[
"from sklearn import neighbors\n\nclass MyRigeRegression:\n def __init__(self, X, y, K):",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
4affe952c420544960efcfd4e7e5ac19f1d83c1c
| 57,328 |
ipynb
|
Jupyter Notebook
|
python/pprint_module.ipynb
|
RenZorRUS/apollo-lib
|
497af7714b0374c3242eccc38db1a55300e02294
|
[
"MIT"
] | null | null | null |
python/pprint_module.ipynb
|
RenZorRUS/apollo-lib
|
497af7714b0374c3242eccc38db1a55300e02294
|
[
"MIT"
] | null | null | null |
python/pprint_module.ipynb
|
RenZorRUS/apollo-lib
|
497af7714b0374c3242eccc38db1a55300e02294
|
[
"MIT"
] | null | null | null | 48.09396 | 4,433 | 0.540382 |
[
[
[
"## Prettify Your Data Structures With Pretty Print in Python\n\nDealing with data is essential for any Pythonista, but sometimes that data is just not very pretty. Computers don’t care about formatting, but without good formatting, humans may find something hard to read. The output isn’t pretty when you use `print()` on large dictionaries or long lists—it’s efficient, but not pretty.\n\nThe `pprint` module in Python is a utility module that you can use to print data structures in a readable, pretty way. It’s a part of the standard library that’s especially useful for debugging code dealing with API requests, large JSON files, and data in general.\n\n## Understanding the Need for Python’s Pretty Print\n\nThe Python `pprint` module is helpful in many situations. It comes in handy when making API requests, dealing with JSON files, or handling complicated and nested data. You’ll probably find that using the normal `print()` function isn’t adequate to efficiently explore your data and debug your application. When you use `print()` with dictionaries and lists, the output doesn’t contain any newlines.\n\nExample: You’ll make a request to {JSON} Placeholder for some mock user information. The first thing to do is to make the HTTP `GET` request and put the response into a dictionary:",
"_____no_output_____"
]
],
[
[
"from urllib import request\nimport json\n\n# Here, you make a basic GET request and then parse the response\n# Into a dictionary with `json.loads()`. With the dictionary \n# Now in a variable, a common next step is to print the contents with print():\nresponse = request.urlopen(\"https://jsonplaceholder.typicode.com/users\")\njson_response = response.read()\n\n# `json.loads(str)` - decoding JSON\nusers = json.loads(json_response)\n\nprint(users)",
"[{'id': 1, 'name': 'Leanne Graham', 'username': 'Bret', 'email': '[email protected]', 'address': {'street': 'Kulas Light', 'suite': 'Apt. 556', 'city': 'Gwenborough', 'zipcode': '92998-3874', 'geo': {'lat': '-37.3159', 'lng': '81.1496'}}, 'phone': '1-770-736-8031 x56442', 'website': 'hildegard.org', 'company': {'name': 'Romaguera-Crona', 'catchPhrase': 'Multi-layered client-server neural-net', 'bs': 'harness real-time e-markets'}}, {'id': 2, 'name': 'Ervin Howell', 'username': 'Antonette', 'email': '[email protected]', 'address': {'street': 'Victor Plains', 'suite': 'Suite 879', 'city': 'Wisokyburgh', 'zipcode': '90566-7771', 'geo': {'lat': '-43.9509', 'lng': '-34.4618'}}, 'phone': '010-692-6593 x09125', 'website': 'anastasia.net', 'company': {'name': 'Deckow-Crist', 'catchPhrase': 'Proactive didactic contingency', 'bs': 'synergize scalable supply-chains'}}, {'id': 3, 'name': 'Clementine Bauch', 'username': 'Samantha', 'email': '[email protected]', 'address': {'street': 'Douglas Extension', 'suite': 'Suite 847', 'city': 'McKenziehaven', 'zipcode': '59590-4157', 'geo': {'lat': '-68.6102', 'lng': '-47.0653'}}, 'phone': '1-463-123-4447', 'website': 'ramiro.info', 'company': {'name': 'Romaguera-Jacobson', 'catchPhrase': 'Face to face bifurcated interface', 'bs': 'e-enable strategic applications'}}, {'id': 4, 'name': 'Patricia Lebsack', 'username': 'Karianne', 'email': '[email protected]', 'address': {'street': 'Hoeger Mall', 'suite': 'Apt. 692', 'city': 'South Elvis', 'zipcode': '53919-4257', 'geo': {'lat': '29.4572', 'lng': '-164.2990'}}, 'phone': '493-170-9623 x156', 'website': 'kale.biz', 'company': {'name': 'Robel-Corkery', 'catchPhrase': 'Multi-tiered zero tolerance productivity', 'bs': 'transition cutting-edge web services'}}, {'id': 5, 'name': 'Chelsey Dietrich', 'username': 'Kamren', 'email': '[email protected]', 'address': {'street': 'Skiles Walks', 'suite': 'Suite 351', 'city': 'Roscoeview', 'zipcode': '33263', 'geo': {'lat': '-31.8129', 'lng': '62.5342'}}, 'phone': '(254)954-1289', 'website': 'demarco.info', 'company': {'name': 'Keebler LLC', 'catchPhrase': 'User-centric fault-tolerant solution', 'bs': 'revolutionize end-to-end systems'}}, {'id': 6, 'name': 'Mrs. Dennis Schulist', 'username': 'Leopoldo_Corkery', 'email': '[email protected]', 'address': {'street': 'Norberto Crossing', 'suite': 'Apt. 950', 'city': 'South Christy', 'zipcode': '23505-1337', 'geo': {'lat': '-71.4197', 'lng': '71.7478'}}, 'phone': '1-477-935-8478 x6430', 'website': 'ola.org', 'company': {'name': 'Considine-Lockman', 'catchPhrase': 'Synchronised bottom-line interface', 'bs': 'e-enable innovative applications'}}, {'id': 7, 'name': 'Kurtis Weissnat', 'username': 'Elwyn.Skiles', 'email': '[email protected]', 'address': {'street': 'Rex Trail', 'suite': 'Suite 280', 'city': 'Howemouth', 'zipcode': '58804-1099', 'geo': {'lat': '24.8918', 'lng': '21.8984'}}, 'phone': '210.067.6132', 'website': 'elvis.io', 'company': {'name': 'Johns Group', 'catchPhrase': 'Configurable multimedia task-force', 'bs': 'generate enterprise e-tailers'}}, {'id': 8, 'name': 'Nicholas Runolfsdottir V', 'username': 'Maxime_Nienow', 'email': '[email protected]', 'address': {'street': 'Ellsworth Summit', 'suite': 'Suite 729', 'city': 'Aliyaview', 'zipcode': '45169', 'geo': {'lat': '-14.3990', 'lng': '-120.7677'}}, 'phone': '586.493.6943 x140', 'website': 'jacynthe.com', 'company': {'name': 'Abernathy Group', 'catchPhrase': 'Implemented secondary concept', 'bs': 'e-enable extensible e-tailers'}}, {'id': 9, 'name': 'Glenna Reichert', 'username': 'Delphine', 'email': '[email protected]', 'address': {'street': 'Dayna Park', 'suite': 'Suite 449', 'city': 'Bartholomebury', 'zipcode': '76495-3109', 'geo': {'lat': '24.6463', 'lng': '-168.8889'}}, 'phone': '(775)976-6794 x41206', 'website': 'conrad.com', 'company': {'name': 'Yost and Sons', 'catchPhrase': 'Switchable contextually-based project', 'bs': 'aggregate real-time technologies'}}, {'id': 10, 'name': 'Clementina DuBuque', 'username': 'Moriah.Stanton', 'email': '[email protected]', 'address': {'street': 'Kattie Turnpike', 'suite': 'Suite 198', 'city': 'Lebsackbury', 'zipcode': '31428-2261', 'geo': {'lat': '-38.2386', 'lng': '57.2232'}}, 'phone': '024-648-3804', 'website': 'ambrose.net', 'company': {'name': 'Hoeger LLC', 'catchPhrase': 'Centralized empowering task-force', 'bs': 'target end-to-end models'}}]\n"
]
],
[
[
"One huge line with no newlines. Depending on your console settings, this might appear as one very long line. Alternatively, your console output might have its word-wrapping mode on, which is the most common situation. Unfortunately, that doesn’t make the output much friendlier!\n\nIf you look at the 1st and last characters, you can see that this appears to be a `list`. You might be tempted to start writing a loop to print the items:",
"_____no_output_____"
]
],
[
[
"for user in users:\n print(user)",
"{'id': 1, 'name': 'Leanne Graham', 'username': 'Bret', 'email': '[email protected]', 'address': {'street': 'Kulas Light', 'suite': 'Apt. 556', 'city': 'Gwenborough', 'zipcode': '92998-3874', 'geo': {'lat': '-37.3159', 'lng': '81.1496'}}, 'phone': '1-770-736-8031 x56442', 'website': 'hildegard.org', 'company': {'name': 'Romaguera-Crona', 'catchPhrase': 'Multi-layered client-server neural-net', 'bs': 'harness real-time e-markets'}}\n{'id': 2, 'name': 'Ervin Howell', 'username': 'Antonette', 'email': '[email protected]', 'address': {'street': 'Victor Plains', 'suite': 'Suite 879', 'city': 'Wisokyburgh', 'zipcode': '90566-7771', 'geo': {'lat': '-43.9509', 'lng': '-34.4618'}}, 'phone': '010-692-6593 x09125', 'website': 'anastasia.net', 'company': {'name': 'Deckow-Crist', 'catchPhrase': 'Proactive didactic contingency', 'bs': 'synergize scalable supply-chains'}}\n{'id': 3, 'name': 'Clementine Bauch', 'username': 'Samantha', 'email': '[email protected]', 'address': {'street': 'Douglas Extension', 'suite': 'Suite 847', 'city': 'McKenziehaven', 'zipcode': '59590-4157', 'geo': {'lat': '-68.6102', 'lng': '-47.0653'}}, 'phone': '1-463-123-4447', 'website': 'ramiro.info', 'company': {'name': 'Romaguera-Jacobson', 'catchPhrase': 'Face to face bifurcated interface', 'bs': 'e-enable strategic applications'}}\n{'id': 4, 'name': 'Patricia Lebsack', 'username': 'Karianne', 'email': '[email protected]', 'address': {'street': 'Hoeger Mall', 'suite': 'Apt. 692', 'city': 'South Elvis', 'zipcode': '53919-4257', 'geo': {'lat': '29.4572', 'lng': '-164.2990'}}, 'phone': '493-170-9623 x156', 'website': 'kale.biz', 'company': {'name': 'Robel-Corkery', 'catchPhrase': 'Multi-tiered zero tolerance productivity', 'bs': 'transition cutting-edge web services'}}\n{'id': 5, 'name': 'Chelsey Dietrich', 'username': 'Kamren', 'email': '[email protected]', 'address': {'street': 'Skiles Walks', 'suite': 'Suite 351', 'city': 'Roscoeview', 'zipcode': '33263', 'geo': {'lat': '-31.8129', 'lng': '62.5342'}}, 'phone': '(254)954-1289', 'website': 'demarco.info', 'company': {'name': 'Keebler LLC', 'catchPhrase': 'User-centric fault-tolerant solution', 'bs': 'revolutionize end-to-end systems'}}\n{'id': 6, 'name': 'Mrs. Dennis Schulist', 'username': 'Leopoldo_Corkery', 'email': '[email protected]', 'address': {'street': 'Norberto Crossing', 'suite': 'Apt. 950', 'city': 'South Christy', 'zipcode': '23505-1337', 'geo': {'lat': '-71.4197', 'lng': '71.7478'}}, 'phone': '1-477-935-8478 x6430', 'website': 'ola.org', 'company': {'name': 'Considine-Lockman', 'catchPhrase': 'Synchronised bottom-line interface', 'bs': 'e-enable innovative applications'}}\n{'id': 7, 'name': 'Kurtis Weissnat', 'username': 'Elwyn.Skiles', 'email': '[email protected]', 'address': {'street': 'Rex Trail', 'suite': 'Suite 280', 'city': 'Howemouth', 'zipcode': '58804-1099', 'geo': {'lat': '24.8918', 'lng': '21.8984'}}, 'phone': '210.067.6132', 'website': 'elvis.io', 'company': {'name': 'Johns Group', 'catchPhrase': 'Configurable multimedia task-force', 'bs': 'generate enterprise e-tailers'}}\n{'id': 8, 'name': 'Nicholas Runolfsdottir V', 'username': 'Maxime_Nienow', 'email': '[email protected]', 'address': {'street': 'Ellsworth Summit', 'suite': 'Suite 729', 'city': 'Aliyaview', 'zipcode': '45169', 'geo': {'lat': '-14.3990', 'lng': '-120.7677'}}, 'phone': '586.493.6943 x140', 'website': 'jacynthe.com', 'company': {'name': 'Abernathy Group', 'catchPhrase': 'Implemented secondary concept', 'bs': 'e-enable extensible e-tailers'}}\n{'id': 9, 'name': 'Glenna Reichert', 'username': 'Delphine', 'email': '[email protected]', 'address': {'street': 'Dayna Park', 'suite': 'Suite 449', 'city': 'Bartholomebury', 'zipcode': '76495-3109', 'geo': {'lat': '24.6463', 'lng': '-168.8889'}}, 'phone': '(775)976-6794 x41206', 'website': 'conrad.com', 'company': {'name': 'Yost and Sons', 'catchPhrase': 'Switchable contextually-based project', 'bs': 'aggregate real-time technologies'}}\n{'id': 10, 'name': 'Clementina DuBuque', 'username': 'Moriah.Stanton', 'email': '[email protected]', 'address': {'street': 'Kattie Turnpike', 'suite': 'Suite 198', 'city': 'Lebsackbury', 'zipcode': '31428-2261', 'geo': {'lat': '-38.2386', 'lng': '57.2232'}}, 'phone': '024-648-3804', 'website': 'ambrose.net', 'company': {'name': 'Hoeger LLC', 'catchPhrase': 'Centralized empowering task-force', 'bs': 'target end-to-end models'}}\n"
]
],
[
[
"This `for` loop would print each object on a separate line, but even then, each object takes up way more space than can fit on a single line. Printing in this way does make things a bit better, but it’s by no means ideal. The above example is a relatively simple data structure, but what would you do with a deeply nested dictionary 100 times the size?\n\nSure, you could write a function that uses recursion to find a way to print everything. Unfortunately, you’ll likely run into some edge cases where this won’t work. You might even find yourself writing a whole module of functions just to get to grips with the structure of the data!\n\nEnter the `pprint` module!\n\n## Working With pprint\n\n`pprint` is a Python module made to print data structures in a pretty way. It has long been part of the Python standard library, so installing it separately isn’t necessary. All you need to do is to import its `pprint()` function:",
"_____no_output_____"
]
],
[
[
"from pprint import pprint\n\n# Then, instead of going with the normal `print(users)` approach \n# As you did in the example above, you can call your new\n# Favorite function to make the output pretty\n\npprint(users)",
"[{'address': {'city': 'Gwenborough',\n 'geo': {'lat': '-37.3159', 'lng': '81.1496'},\n 'street': 'Kulas Light',\n 'suite': 'Apt. 556',\n 'zipcode': '92998-3874'},\n 'company': {'bs': 'harness real-time e-markets',\n 'catchPhrase': 'Multi-layered client-server neural-net',\n 'name': 'Romaguera-Crona'},\n 'email': '[email protected]',\n 'id': 1,\n 'name': 'Leanne Graham',\n 'phone': '1-770-736-8031 x56442',\n 'username': 'Bret',\n 'website': 'hildegard.org'},\n {'address': {'city': 'Wisokyburgh',\n 'geo': {'lat': '-43.9509', 'lng': '-34.4618'},\n 'street': 'Victor Plains',\n 'suite': 'Suite 879',\n 'zipcode': '90566-7771'},\n 'company': {'bs': 'synergize scalable supply-chains',\n 'catchPhrase': 'Proactive didactic contingency',\n 'name': 'Deckow-Crist'},\n 'email': '[email protected]',\n 'id': 2,\n 'name': 'Ervin Howell',\n 'phone': '010-692-6593 x09125',\n 'username': 'Antonette',\n 'website': 'anastasia.net'},\n {'address': {'city': 'McKenziehaven',\n 'geo': {'lat': '-68.6102', 'lng': '-47.0653'},\n 'street': 'Douglas Extension',\n 'suite': 'Suite 847',\n 'zipcode': '59590-4157'},\n 'company': {'bs': 'e-enable strategic applications',\n 'catchPhrase': 'Face to face bifurcated interface',\n 'name': 'Romaguera-Jacobson'},\n 'email': '[email protected]',\n 'id': 3,\n 'name': 'Clementine Bauch',\n 'phone': '1-463-123-4447',\n 'username': 'Samantha',\n 'website': 'ramiro.info'},\n {'address': {'city': 'South Elvis',\n 'geo': {'lat': '29.4572', 'lng': '-164.2990'},\n 'street': 'Hoeger Mall',\n 'suite': 'Apt. 692',\n 'zipcode': '53919-4257'},\n 'company': {'bs': 'transition cutting-edge web services',\n 'catchPhrase': 'Multi-tiered zero tolerance productivity',\n 'name': 'Robel-Corkery'},\n 'email': '[email protected]',\n 'id': 4,\n 'name': 'Patricia Lebsack',\n 'phone': '493-170-9623 x156',\n 'username': 'Karianne',\n 'website': 'kale.biz'},\n {'address': {'city': 'Roscoeview',\n 'geo': {'lat': '-31.8129', 'lng': '62.5342'},\n 'street': 'Skiles Walks',\n 'suite': 'Suite 351',\n 'zipcode': '33263'},\n 'company': {'bs': 'revolutionize end-to-end systems',\n 'catchPhrase': 'User-centric fault-tolerant solution',\n 'name': 'Keebler LLC'},\n 'email': '[email protected]',\n 'id': 5,\n 'name': 'Chelsey Dietrich',\n 'phone': '(254)954-1289',\n 'username': 'Kamren',\n 'website': 'demarco.info'},\n {'address': {'city': 'South Christy',\n 'geo': {'lat': '-71.4197', 'lng': '71.7478'},\n 'street': 'Norberto Crossing',\n 'suite': 'Apt. 950',\n 'zipcode': '23505-1337'},\n 'company': {'bs': 'e-enable innovative applications',\n 'catchPhrase': 'Synchronised bottom-line interface',\n 'name': 'Considine-Lockman'},\n 'email': '[email protected]',\n 'id': 6,\n 'name': 'Mrs. Dennis Schulist',\n 'phone': '1-477-935-8478 x6430',\n 'username': 'Leopoldo_Corkery',\n 'website': 'ola.org'},\n {'address': {'city': 'Howemouth',\n 'geo': {'lat': '24.8918', 'lng': '21.8984'},\n 'street': 'Rex Trail',\n 'suite': 'Suite 280',\n 'zipcode': '58804-1099'},\n 'company': {'bs': 'generate enterprise e-tailers',\n 'catchPhrase': 'Configurable multimedia task-force',\n 'name': 'Johns Group'},\n 'email': '[email protected]',\n 'id': 7,\n 'name': 'Kurtis Weissnat',\n 'phone': '210.067.6132',\n 'username': 'Elwyn.Skiles',\n 'website': 'elvis.io'},\n {'address': {'city': 'Aliyaview',\n 'geo': {'lat': '-14.3990', 'lng': '-120.7677'},\n 'street': 'Ellsworth Summit',\n 'suite': 'Suite 729',\n 'zipcode': '45169'},\n 'company': {'bs': 'e-enable extensible e-tailers',\n 'catchPhrase': 'Implemented secondary concept',\n 'name': 'Abernathy Group'},\n 'email': '[email protected]',\n 'id': 8,\n 'name': 'Nicholas Runolfsdottir V',\n 'phone': '586.493.6943 x140',\n 'username': 'Maxime_Nienow',\n 'website': 'jacynthe.com'},\n {'address': {'city': 'Bartholomebury',\n 'geo': {'lat': '24.6463', 'lng': '-168.8889'},\n 'street': 'Dayna Park',\n 'suite': 'Suite 449',\n 'zipcode': '76495-3109'},\n 'company': {'bs': 'aggregate real-time technologies',\n 'catchPhrase': 'Switchable contextually-based project',\n 'name': 'Yost and Sons'},\n 'email': '[email protected]',\n 'id': 9,\n 'name': 'Glenna Reichert',\n 'phone': '(775)976-6794 x41206',\n 'username': 'Delphine',\n 'website': 'conrad.com'},\n {'address': {'city': 'Lebsackbury',\n 'geo': {'lat': '-38.2386', 'lng': '57.2232'},\n 'street': 'Kattie Turnpike',\n 'suite': 'Suite 198',\n 'zipcode': '31428-2261'},\n 'company': {'bs': 'target end-to-end models',\n 'catchPhrase': 'Centralized empowering task-force',\n 'name': 'Hoeger LLC'},\n 'email': '[email protected]',\n 'id': 10,\n 'name': 'Clementina DuBuque',\n 'phone': '024-648-3804',\n 'username': 'Moriah.Stanton',\n 'website': 'ambrose.net'}]\n"
]
],
[
[
"How pretty! The keys of the dictionaries are even visually indented! This output makes it so much more straightforward to scan and visually analyze data structures. \n\nIf you’re a fan of typing as little as possible, then you’ll be pleased to know that `pprint()` has an alias, `pp()`:",
"_____no_output_____"
]
],
[
[
"from pprint import pp\n\n# `pp()` is just a wrapper around `pprint()`, \n# and it’ll behave exactly the same way.\n\npp(users)",
"[{'id': 1,\n 'name': 'Leanne Graham',\n 'username': 'Bret',\n 'email': '[email protected]',\n 'address': {'street': 'Kulas Light',\n 'suite': 'Apt. 556',\n 'city': 'Gwenborough',\n 'zipcode': '92998-3874',\n 'geo': {'lat': '-37.3159', 'lng': '81.1496'}},\n 'phone': '1-770-736-8031 x56442',\n 'website': 'hildegard.org',\n 'company': {'name': 'Romaguera-Crona',\n 'catchPhrase': 'Multi-layered client-server neural-net',\n 'bs': 'harness real-time e-markets'}},\n {'id': 2,\n 'name': 'Ervin Howell',\n 'username': 'Antonette',\n 'email': '[email protected]',\n 'address': {'street': 'Victor Plains',\n 'suite': 'Suite 879',\n 'city': 'Wisokyburgh',\n 'zipcode': '90566-7771',\n 'geo': {'lat': '-43.9509', 'lng': '-34.4618'}},\n 'phone': '010-692-6593 x09125',\n 'website': 'anastasia.net',\n 'company': {'name': 'Deckow-Crist',\n 'catchPhrase': 'Proactive didactic contingency',\n 'bs': 'synergize scalable supply-chains'}},\n {'id': 3,\n 'name': 'Clementine Bauch',\n 'username': 'Samantha',\n 'email': '[email protected]',\n 'address': {'street': 'Douglas Extension',\n 'suite': 'Suite 847',\n 'city': 'McKenziehaven',\n 'zipcode': '59590-4157',\n 'geo': {'lat': '-68.6102', 'lng': '-47.0653'}},\n 'phone': '1-463-123-4447',\n 'website': 'ramiro.info',\n 'company': {'name': 'Romaguera-Jacobson',\n 'catchPhrase': 'Face to face bifurcated interface',\n 'bs': 'e-enable strategic applications'}},\n {'id': 4,\n 'name': 'Patricia Lebsack',\n 'username': 'Karianne',\n 'email': '[email protected]',\n 'address': {'street': 'Hoeger Mall',\n 'suite': 'Apt. 692',\n 'city': 'South Elvis',\n 'zipcode': '53919-4257',\n 'geo': {'lat': '29.4572', 'lng': '-164.2990'}},\n 'phone': '493-170-9623 x156',\n 'website': 'kale.biz',\n 'company': {'name': 'Robel-Corkery',\n 'catchPhrase': 'Multi-tiered zero tolerance productivity',\n 'bs': 'transition cutting-edge web services'}},\n {'id': 5,\n 'name': 'Chelsey Dietrich',\n 'username': 'Kamren',\n 'email': '[email protected]',\n 'address': {'street': 'Skiles Walks',\n 'suite': 'Suite 351',\n 'city': 'Roscoeview',\n 'zipcode': '33263',\n 'geo': {'lat': '-31.8129', 'lng': '62.5342'}},\n 'phone': '(254)954-1289',\n 'website': 'demarco.info',\n 'company': {'name': 'Keebler LLC',\n 'catchPhrase': 'User-centric fault-tolerant solution',\n 'bs': 'revolutionize end-to-end systems'}},\n {'id': 6,\n 'name': 'Mrs. Dennis Schulist',\n 'username': 'Leopoldo_Corkery',\n 'email': '[email protected]',\n 'address': {'street': 'Norberto Crossing',\n 'suite': 'Apt. 950',\n 'city': 'South Christy',\n 'zipcode': '23505-1337',\n 'geo': {'lat': '-71.4197', 'lng': '71.7478'}},\n 'phone': '1-477-935-8478 x6430',\n 'website': 'ola.org',\n 'company': {'name': 'Considine-Lockman',\n 'catchPhrase': 'Synchronised bottom-line interface',\n 'bs': 'e-enable innovative applications'}},\n {'id': 7,\n 'name': 'Kurtis Weissnat',\n 'username': 'Elwyn.Skiles',\n 'email': '[email protected]',\n 'address': {'street': 'Rex Trail',\n 'suite': 'Suite 280',\n 'city': 'Howemouth',\n 'zipcode': '58804-1099',\n 'geo': {'lat': '24.8918', 'lng': '21.8984'}},\n 'phone': '210.067.6132',\n 'website': 'elvis.io',\n 'company': {'name': 'Johns Group',\n 'catchPhrase': 'Configurable multimedia task-force',\n 'bs': 'generate enterprise e-tailers'}},\n {'id': 8,\n 'name': 'Nicholas Runolfsdottir V',\n 'username': 'Maxime_Nienow',\n 'email': '[email protected]',\n 'address': {'street': 'Ellsworth Summit',\n 'suite': 'Suite 729',\n 'city': 'Aliyaview',\n 'zipcode': '45169',\n 'geo': {'lat': '-14.3990', 'lng': '-120.7677'}},\n 'phone': '586.493.6943 x140',\n 'website': 'jacynthe.com',\n 'company': {'name': 'Abernathy Group',\n 'catchPhrase': 'Implemented secondary concept',\n 'bs': 'e-enable extensible e-tailers'}},\n {'id': 9,\n 'name': 'Glenna Reichert',\n 'username': 'Delphine',\n 'email': '[email protected]',\n 'address': {'street': 'Dayna Park',\n 'suite': 'Suite 449',\n 'city': 'Bartholomebury',\n 'zipcode': '76495-3109',\n 'geo': {'lat': '24.6463', 'lng': '-168.8889'}},\n 'phone': '(775)976-6794 x41206',\n 'website': 'conrad.com',\n 'company': {'name': 'Yost and Sons',\n 'catchPhrase': 'Switchable contextually-based project',\n 'bs': 'aggregate real-time technologies'}},\n {'id': 10,\n 'name': 'Clementina DuBuque',\n 'username': 'Moriah.Stanton',\n 'email': '[email protected]',\n 'address': {'street': 'Kattie Turnpike',\n 'suite': 'Suite 198',\n 'city': 'Lebsackbury',\n 'zipcode': '31428-2261',\n 'geo': {'lat': '-38.2386', 'lng': '57.2232'}},\n 'phone': '024-648-3804',\n 'website': 'ambrose.net',\n 'company': {'name': 'Hoeger LLC',\n 'catchPhrase': 'Centralized empowering task-force',\n 'bs': 'target end-to-end models'}}]\n"
]
],
[
[
"**NOTE:** Python has included this alias since version 3.8.0 alpha 2.\n\nHowever, even the default output may be too much information to scan at first. Maybe all you really want is to verify that you’re dealing with a list of plain objects. For that, you’ll want to tweak the output a little.\n\nFor these situations, there are various parameters you can pass to `pprint()` to make even the tersest data structures pretty.\n\n## Exploring Optional Parameters of pprint()\n\nThere are 7 parameters that you can use to configure your Pythonic pretty printer. You don’t need to use them all, and some will be more useful than others. The one you’ll find most valuable will probably be depth.\n\n## Summarizing Your Data: depth\n\nOne of the handiest parameters to play around with is `depth`. The following Python command will only print the full contents of users if the data structure is at or lower than the specified `depth`—all while keeping things pretty, of course. The contents of deeper data structures are replaced with three dots `...`:",
"_____no_output_____"
]
],
[
[
"pprint(users, depth=1)\nprint('-------------------')\n\n# Now you can immediately see that this is indeed a list of dictionaries. \n# To explore the data structure further, you can increase the depth by one level,\n# Which will print all the top-level keys of the dictionaries in users:\npprint(users, depth=2)",
"[{...}, {...}, {...}, {...}, {...}, {...}, {...}, {...}, {...}, {...}]\n-------------------\n[{'address': {...},\n 'company': {...},\n 'email': '[email protected]',\n 'id': 1,\n 'name': 'Leanne Graham',\n 'phone': '1-770-736-8031 x56442',\n 'username': 'Bret',\n 'website': 'hildegard.org'},\n {'address': {...},\n 'company': {...},\n 'email': '[email protected]',\n 'id': 2,\n 'name': 'Ervin Howell',\n 'phone': '010-692-6593 x09125',\n 'username': 'Antonette',\n 'website': 'anastasia.net'},\n {'address': {...},\n 'company': {...},\n 'email': '[email protected]',\n 'id': 3,\n 'name': 'Clementine Bauch',\n 'phone': '1-463-123-4447',\n 'username': 'Samantha',\n 'website': 'ramiro.info'},\n {'address': {...},\n 'company': {...},\n 'email': '[email protected]',\n 'id': 4,\n 'name': 'Patricia Lebsack',\n 'phone': '493-170-9623 x156',\n 'username': 'Karianne',\n 'website': 'kale.biz'},\n {'address': {...},\n 'company': {...},\n 'email': '[email protected]',\n 'id': 5,\n 'name': 'Chelsey Dietrich',\n 'phone': '(254)954-1289',\n 'username': 'Kamren',\n 'website': 'demarco.info'},\n {'address': {...},\n 'company': {...},\n 'email': '[email protected]',\n 'id': 6,\n 'name': 'Mrs. Dennis Schulist',\n 'phone': '1-477-935-8478 x6430',\n 'username': 'Leopoldo_Corkery',\n 'website': 'ola.org'},\n {'address': {...},\n 'company': {...},\n 'email': '[email protected]',\n 'id': 7,\n 'name': 'Kurtis Weissnat',\n 'phone': '210.067.6132',\n 'username': 'Elwyn.Skiles',\n 'website': 'elvis.io'},\n {'address': {...},\n 'company': {...},\n 'email': '[email protected]',\n 'id': 8,\n 'name': 'Nicholas Runolfsdottir V',\n 'phone': '586.493.6943 x140',\n 'username': 'Maxime_Nienow',\n 'website': 'jacynthe.com'},\n {'address': {...},\n 'company': {...},\n 'email': '[email protected]',\n 'id': 9,\n 'name': 'Glenna Reichert',\n 'phone': '(775)976-6794 x41206',\n 'username': 'Delphine',\n 'website': 'conrad.com'},\n {'address': {...},\n 'company': {...},\n 'email': '[email protected]',\n 'id': 10,\n 'name': 'Clementina DuBuque',\n 'phone': '024-648-3804',\n 'username': 'Moriah.Stanton',\n 'website': 'ambrose.net'}]\n"
]
],
[
[
"Now you can quickly check whether all the dictionaries share their top-level keys. This is a valuable observation to make, especially if you’re tasked with developing an application that consumes data like this.\n\n## Giving Your Data Space: indent\n\nThe `indent` parameter controls how indented each level of the pretty-printed representation will be in the output. The default `indent` is just `1`, which translates to **one space character**:",
"_____no_output_____"
]
],
[
[
"pprint(users[0], depth=1)\nprint('------------')\npprint(users[0], depth=1, indent=4)",
"{'address': {...},\n 'company': {...},\n 'email': '[email protected]',\n 'id': 1,\n 'name': 'Leanne Graham',\n 'phone': '1-770-736-8031 x56442',\n 'username': 'Bret',\n 'website': 'hildegard.org'}\n------------\n{ 'address': {...},\n 'company': {...},\n 'email': '[email protected]',\n 'id': 1,\n 'name': 'Leanne Graham',\n 'phone': '1-770-736-8031 x56442',\n 'username': 'Bret',\n 'website': 'hildegard.org'}\n"
]
],
[
[
"The most important part of the indenting behavior of `pprint()` is keeping all the keys aligned visually. How much indentation is applied depends on both the `indent` parameter and where the key is.\n\nSince there’s no nesting in the examples above, the amount of indentation is based completely on the `indent` parameter. In both examples, note how the opening curly bracket (`{`) is counted as a unit of indentation for the 1st key. In the 1st example, the opening single quote `'` for the 1st key comes right after `{` without any spaces in between because the indent is set to `1`.\n\nWhen there is nesting, however, the indentation is applied to the 1st element in-line, and `pprint()` then keeps all following elements aligned with the 1st one. So if you set your `indent` to `4` when printing users, the 1st element will be indented by four characters, while the nested elements will be indented by more than 8 characters because the indentation starts from the end of the 1st key:",
"_____no_output_____"
]
],
[
[
"pprint(users[0], depth=2, indent=4)",
"{ 'address': { 'city': 'Gwenborough',\n 'geo': {...},\n 'street': 'Kulas Light',\n 'suite': 'Apt. 556',\n 'zipcode': '92998-3874'},\n 'company': { 'bs': 'harness real-time e-markets',\n 'catchPhrase': 'Multi-layered client-server neural-net',\n 'name': 'Romaguera-Crona'},\n 'email': '[email protected]',\n 'id': 1,\n 'name': 'Leanne Graham',\n 'phone': '1-770-736-8031 x56442',\n 'username': 'Bret',\n 'website': 'hildegard.org'}\n"
]
],
[
[
"## Limiting Your Line Lengths: width\n\nBy default, `pprint()` will only output up to 80 characters per line. You can customize this value by passing in a `width` argument. `pprint()` will make an effort to fit the contents on one line. If the contents of a data structure go over this limit, then it’ll print every element of the current data structure on a new line:",
"_____no_output_____"
]
],
[
[
"pprint(users[0])",
"{'address': {'city': 'Gwenborough',\n 'geo': {'lat': '-37.3159', 'lng': '81.1496'},\n 'street': 'Kulas Light',\n 'suite': 'Apt. 556',\n 'zipcode': '92998-3874'},\n 'company': {'bs': 'harness real-time e-markets',\n 'catchPhrase': 'Multi-layered client-server neural-net',\n 'name': 'Romaguera-Crona'},\n 'email': '[email protected]',\n 'id': 1,\n 'name': 'Leanne Graham',\n 'phone': '1-770-736-8031 x56442',\n 'username': 'Bret',\n 'website': 'hildegard.org'}\n"
]
],
[
[
"When you leave the `width` at the default of 80 characters, the dictionary at `users[0]['address']['geo']` only contains a '`lat`' and a '`lng`' attribute. This means that taking the sum of the indent and the number of characters needed to print out the dictionary, including the spaces in between, comes to less than 80 characters. Since it’s less than 80 characters, the default `width`, `pprint()` puts it all on one line.\n\nHowever, the dictionary at `users[0]['company']` would go over the default `width`, so `pprint()` puts each key on a new line. This is true of dictionaries, lists, tuples, and sets:",
"_____no_output_____"
]
],
[
[
"pprint(users[0], width=160)",
"{'address': {'city': 'Gwenborough', 'geo': {'lat': '-37.3159', 'lng': '81.1496'}, 'street': 'Kulas Light', 'suite': 'Apt. 556', 'zipcode': '92998-3874'},\n 'company': {'bs': 'harness real-time e-markets', 'catchPhrase': 'Multi-layered client-server neural-net', 'name': 'Romaguera-Crona'},\n 'email': '[email protected]',\n 'id': 1,\n 'name': 'Leanne Graham',\n 'phone': '1-770-736-8031 x56442',\n 'username': 'Bret',\n 'website': 'hildegard.org'}\n"
]
],
[
[
"If you set the `width` to a large value like `160`, then all the nested dictionaries fit on one line. You can even take it to extremes and use a huge value like `500`, which, for this example, prints the whole dictionary on one line:",
"_____no_output_____"
]
],
[
[
"pprint(users[0], width=500)",
"{'address': {'city': 'Gwenborough', 'geo': {'lat': '-37.3159', 'lng': '81.1496'}, 'street': 'Kulas Light', 'suite': 'Apt. 556', 'zipcode': '92998-3874'}, 'company': {'bs': 'harness real-time e-markets', 'catchPhrase': 'Multi-layered client-server neural-net', 'name': 'Romaguera-Crona'}, 'email': '[email protected]', 'id': 1, 'name': 'Leanne Graham', 'phone': '1-770-736-8031 x56442', 'username': 'Bret', 'website': 'hildegard.org'}\n"
]
],
[
[
"Here, you get the effects of setting width to a relatively large value. You can go the other way and set width to a low value such as `1`. However, the main effect that this will have is making sure every data structure will display its components on separate lines. You’ll still get the visual indentation that lines up the components:",
"_____no_output_____"
]
],
[
[
"pprint(users[0], width=5)",
"{'address': {'city': 'Gwenborough',\n 'geo': {'lat': '-37.3159',\n 'lng': '81.1496'},\n 'street': 'Kulas '\n 'Light',\n 'suite': 'Apt. '\n '556',\n 'zipcode': '92998-3874'},\n 'company': {'bs': 'harness '\n 'real-time '\n 'e-markets',\n 'catchPhrase': 'Multi-layered '\n 'client-server '\n 'neural-net',\n 'name': 'Romaguera-Crona'},\n 'email': '[email protected]',\n 'id': 1,\n 'name': 'Leanne '\n 'Graham',\n 'phone': '1-770-736-8031 '\n 'x56442',\n 'username': 'Bret',\n 'website': 'hildegard.org'}\n"
]
],
[
[
"## Squeezing Your Long Sequences: compact\n\nYou might think that compact refers to the behavior you explored in the section about `width` — that is, whether `compact` makes data structures appear on one line or separate lines. However, `compact` only affects the output once a line goes over the `width`.\n\n**NOTE:** `compact` only affects the output of sequences: lists, sets, and tuples, and NOT dictionaries. This is intentional, though it’s not clear why this decision was taken.\n\nIf `compact` is `True`, then the output will wrap onto the next line. The default behavior is for each element to appear on its own line if the data structure is longer than the `width`:",
"_____no_output_____"
]
],
[
[
"pprint(users, depth=1)\nprint('-------------')\npprint(users, depth=1, width=40)\nprint('-------------')\npprint(users, depth=1, width=40, compact=True)",
"[{...}, {...}, {...}, {...}, {...}, {...}, {...}, {...}, {...}, {...}]\n-------------\n[{...},\n {...},\n {...},\n {...},\n {...},\n {...},\n {...},\n {...},\n {...},\n {...}]\n-------------\n[{...}, {...}, {...}, {...}, {...},\n {...}, {...}, {...}, {...}, {...}]\n"
]
],
[
[
"Pretty-printing this list using the default settings prints out the abbreviated version on one line. Limiting `width` to 40 characters, you force `pprint()` to output all the list’s elements on separate lines. If you then set `compact=True`, then the list will wrap at 40 characters and be more compact than it would typically look.\n\n**NOTE:** Beware that setting the `width` to less than `7` characters — which, in this case, is equivalent to the `[{...},` output — seems to bypass the depth argument completely, and `pprint()` ends up printing everything without any folding. This has been reported as bug #45611.\n\n`compact` is useful for long sequences with short elements that would otherwise take up many lines and make the output less readable.\n\n## Directing Your Output: stream\n\nThe `stream` parameter refers to the output of `pprint()`. By default, it goes to the same place that `print()` goes to. Specifically, it goes to `sys.stdout`, which is actually a file object in Python. However, you can redirect this to any file object, just like you can with `print()`:",
"_____no_output_____"
]
],
[
[
"from urllib import request\nfrom pprint import pprint\nimport json\n\nresponse = request.urlopen(\"https://jsonplaceholder.typicode.com/users\")\nprint('type:', type(response), '\\n')\n\njson_response = response.read()\nusers = json.loads(json_response)\n\nwith open(\"output.txt\", mode=\"w\") as file_object:\n pprint(users, stream=file_object)",
"type: <class 'http.client.HTTPResponse'> \n\n"
]
],
[
[
"Here you create a `file_object` with `open()`, and then you set the stream parameter in `pprint()` to that `file_object`. If you then open the `output.txt` file, you should see that you’ve pretty-printed everything in users there.\n\nPython does have its own `logging` module. However, you can also use `pprint()` to send pretty outputs to files and have these act as logs if you prefer.\n\n## Preventing Dictionary Sorting: sort_dicts\n\nAlthough dictionaries are generally considered unordered data structures, since Python 3.6, dictionaries are ordered by insertion.\n\n`pprint()` orders the keys alphabetically for printing:",
"_____no_output_____"
]
],
[
[
"pprint(users[0], depth=1)\nprint()\npprint(users[0], depth=1, sort_dicts=False)",
"{'address': {...},\n 'company': {...},\n 'email': '[email protected]',\n 'id': 1,\n 'name': 'Leanne Graham',\n 'phone': '1-770-736-8031 x56442',\n 'username': 'Bret',\n 'website': 'hildegard.org'}\n\n{'id': 1,\n 'name': 'Leanne Graham',\n 'username': 'Bret',\n 'email': '[email protected]',\n 'address': {...},\n 'phone': '1-770-736-8031 x56442',\n 'website': 'hildegard.org',\n 'company': {...}}\n"
]
],
[
[
"Unless you set `sort_dicts` to `False`, Python’s `pprint()` sorts the keys alphabetically. It keeps the output for dictionaries consistent, readable, and—well—pretty!\n\nWhen `pprint()` was first implemented, dictionaries were unordered. Without alphabetically ordering the keys, a dictionary’s keys could have theoretically differed at each print.\n\n## Prettifying Your Numbers: underscore_numbers\n\nThe `underscore_numbers` parameter is a feature introduced in Python 3.10 that makes long numbers more readable. Considering that the example you’ve been using so far doesn’t contain any long numbers, you’ll need a new example to try it out:",
"_____no_output_____"
]
],
[
[
"number_list = [123456789, 10000000000000]\npprint(number_list, underscore_numbers=True)",
"[123456789, 10000000000000]\n"
]
],
[
[
"If you tried running this call to `pprint()` and got an error, you’re not alone. As of October 2021, this argument doesn’t work when calling `pprint()` directly. The Python community noticed this quickly, and it’s been fixed in the December 2021 3.10.1 bug fix release. The folks at Python care about their pretty printer! \n\nIf `underscore_numbers` doesn’t work when you call `pprint()` directly and you really want pretty numbers, there is a workaround: When you create your own `PrettyPrinter` object, this parameter should work just like it does in the example above.\n\n## Creating a Custom PrettyPrinter Object\n\nIt’s possible to create an instance of `PrettyPrinter` that has defaults you’ve defined. Once you have this new instance of your custom `PrettyPrinter` object, you can use it by calling the `.pprint()` method on the `PrettyPrinter` instance:",
"_____no_output_____"
]
],
[
[
"from pprint import PrettyPrinter\n\ncustom_printer = PrettyPrinter(\n indent=4,\n width=100,\n depth=2,\n compact=True,\n sort_dicts=False,\n underscore_numbers=True\n)\n\ncustom_printer.pprint(users[0])\n\nnumber_list = [123456789, 10000000000000]\ncustom_printer.pprint(number_list)",
"{ 'id': 1,\n 'name': 'Leanne Graham',\n 'username': 'Bret',\n 'email': '[email protected]',\n 'address': { 'street': 'Kulas Light',\n 'suite': 'Apt. 556',\n 'city': 'Gwenborough',\n 'zipcode': '92998-3874',\n 'geo': {...}},\n 'phone': '1-770-736-8031 x56442',\n 'website': 'hildegard.org',\n 'company': { 'name': 'Romaguera-Crona',\n 'catchPhrase': 'Multi-layered client-server neural-net',\n 'bs': 'harness real-time e-markets'}}\n[123_456_789, 10_000_000_000_000]\n"
]
],
[
[
"With these commands, you:\n\n1. Imported `PrettyPrinter`, which is a class definition\n2. Created a new instance of that class with certain parameters\n3. Printed the first user in users\n4. Defined a list of a couple of long numbers\n5. Printed `number_list`, which also demonstrates `underscore_numbers` in action\n\n**NOTE:** that the arguments you passed to `PrettyPrinter` are exactly the same as the default `pprint()` arguments, except that you skipped the 1st parameter. In `pprint()`, this is the object you want to print.\n\nThis way, you can have various printer presets—perhaps some going to different streams—and call them when you need them.\n\n## Getting a Pretty String With pformat()\n\nWhat if you don’t want to send the pretty output of `pprint()` to a stream? Perhaps you want to do some **regex matching** and replace certain keys. For plain dictionaries, you might find yourself wanting to remove the brackets and quotes to make them look even more human-readable.\n\nWhatever it is that you might want to do with the string pre-output, you can get the string by using `pformat()`:",
"_____no_output_____"
]
],
[
[
"from pprint import pformat\n\naddress = pformat(users[0][\"address\"])\nprint('type:', type(address))\nprint(address, '\\n------------')\n\nchars_to_remove = [\"{\", \"}\", \"'\"]\n\nfor char in chars_to_remove:\n address = address.replace(char, \"\")\n\nprint(address)",
"type: <class 'str'>\n{'city': 'Gwenborough',\n 'geo': {'lat': '-37.3159', 'lng': '81.1496'},\n 'street': 'Kulas Light',\n 'suite': 'Apt. 556',\n 'zipcode': '92998-3874'} \n------------\ncity: Gwenborough,\n geo: lat: -37.3159, lng: 81.1496,\n street: Kulas Light,\n suite: Apt. 556,\n zipcode: 92998-3874\n"
]
],
[
[
"`pformat()` is a tool you can use to get between the pretty printer and the output stream.\n\nAnother use case for this might be if you’re building an API and want to send a pretty string representation of the JSON string. Your end users would probably appreciate it!\n\n## Handling Recursive Data Structures\n\nPython’s `pprint()` is recursive, meaning it’ll pretty-print all the contents of a dictionary, all the contents of any child dictionaries, and so on.\n\nAsk yourself what happens when a recursive function runs into a recursive data structure. Imagine that you have dictionary `A` and dictionary `B`:\n\n* `A` has one attribute, `.link`, which points to `B`\n* `B` has one attribute, `.link`, which points to `A`\n\nIf your imaginary recursive function has no way to handle this circular reference, it’ll never finish printing! It would print `A` and then its child, `B`. But `B` also has `A` as a child, so it would go on into infinity.\n\nLuckily, both the normal `print()` function and the `pprint()` function handle this gracefully:",
"_____no_output_____"
]
],
[
[
"from pprint import pprint\n\nA = {}\nB = {\"link\": A}\nA[\"link\"] = B\n\nprint(A, '\\n----------')\npprint(A)",
"{'link': {'link': {...}}} \n----------\n{'link': {'link': <Recursion on dict with id=2228667041920>}}\n"
]
],
[
[
"While Python’s regular `print()` just abbreviates the output, `pprint()` explicitly notifies you of recursion and also adds the ID of the dictionary.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
4affe9631f036944a8a52aabed5731d24fe1f9f0
| 41,847 |
ipynb
|
Jupyter Notebook
|
tutorials/transformations_01_coordinate_systems.ipynb
|
marscher/weldx
|
a5debd8af957009b12fd366589fed1aa41f78176
|
[
"BSD-3-Clause"
] | null | null | null |
tutorials/transformations_01_coordinate_systems.ipynb
|
marscher/weldx
|
a5debd8af957009b12fd366589fed1aa41f78176
|
[
"BSD-3-Clause"
] | null | null | null |
tutorials/transformations_01_coordinate_systems.ipynb
|
marscher/weldx
|
a5debd8af957009b12fd366589fed1aa41f78176
|
[
"BSD-3-Clause"
] | null | null | null | 47.178129 | 911 | 0.602241 |
[
[
[
"# Transformations Tutorial #1: Coordinate Systems\n## Introduction\n\nThis tutorial is about the transformation packages `LocalCoordinateSystem` class which describes the orientation and position of a Cartesian coordinate system towards another reference coordinate system. The reference coordinate systems origin is always at $(0, 0, 0)$ and its orientation is described by the basis: $e_x = (1, 0, 0)$, $e_y = (0, 1, 0)$, $e_z = (0, 0, 1)$.\n\n## Imports\nThe packages required in this tutorial are:",
"_____no_output_____"
]
],
[
[
"# if the package is not installed in your python environment, run this to execute the notebook directly from inside the GitHub repository\n%cd -q ..",
"_____no_output_____"
],
[
"# enable interactive plots on Jupyterlab with ipympl and jupyterlab-matplotlib installed\n# %matplotlib widget",
"_____no_output_____"
],
[
"# plotting\nfrom mpl_toolkits.mplot3d import Axes3D # noqa: F401 unused import\nimport matplotlib.pyplot as plt\n\n# interactive plots\nimport ipywidgets as widgets\nfrom ipywidgets import VBox, HBox, IntSlider, Checkbox, interactive_output, FloatSlider\nfrom IPython.display import display\n\nimport numpy as np\nimport pandas as pd\nimport xarray as xr\n\nimport weldx.visualization as vs\nimport weldx.transformations as tf",
"_____no_output_____"
]
],
[
[
"## Construction\n\nThe constructor of the `LocalCoordinateSystem` class takes 2 parameters, the `orientation` and the `coordinates`. `orientation` is a 3x3 matrix. It can either be viewed as a rotation/reflection matrix or a set of normalized column vectors that represent the 3 basis vectors of the coordinate system. The matrix needs to be orthogonal, otherwise, an exception is raised. `coordinates` is the position of the local coordinate systems origin inside the reference coordinate system. The default parameters are the identity matrix and the zero vector. Hence, we get a system that is identical to the reference system if no parameter is passed to the constructor.",
"_____no_output_____"
]
],
[
[
"lcs_ref = tf.LocalCoordinateSystem()",
"_____no_output_____"
]
],
[
[
"We create some coordinate systems and visualize them using the `visualization` package. The coordinate axes are colored as follows:\n\n- x = red\n- y = green\n- z = blue",
"_____no_output_____"
]
],
[
[
"# create a translated coordinate system\nlcs_01 = tf.LocalCoordinateSystem(coordinates=[2, 4, -1])\n\n# create a rotated coordinate system using a rotation matrix as basis\nrotation_matrix = tf.WXRotation.from_euler(\"z\",np.pi / 3).as_matrix()\nlcs_02 = tf.LocalCoordinateSystem(orientation=rotation_matrix, coordinates=[0, 0, 3])",
"_____no_output_____"
]
],
[
[
"**Plot:**",
"_____no_output_____"
]
],
[
[
"vs.plot_coordinate_systems([(lcs_ref, {\"color\":\"r\", \"label\":\"reference_system\"}), \n (lcs_01, {\"color\":\"g\", \"label\":\"system 1\"}), \n (lcs_02, {\"color\":\"b\", \"label\":\"system 2\"}) ])",
"_____no_output_____"
]
],
[
[
"> **HINT:** In the jupyter notebook version of this tutorial, you can rotate the plot by pressing the left mouse button and moving the mouse. This helps to understand how the different coordinate systems are positioned in the 3d space.\n\nApart from the class constructor, there are some factory functions implemented to create a coordinate system. The `from_orientation` provides the same functionality as the class constructor. The `from_xyz` takes 3 basis vectors instead of a matrix. `from_xy_and_orientation`, `from_xz_and_orientation` and `from_yz_and_orientation` create a coordinate system with 2 basis vectors and a `bool` which specifies if the coordinate system should have a positive or negative orientation. Here are some examples:",
"_____no_output_____"
]
],
[
[
"# coordinate system using 3 basis vectors\ne_x = [1, 2, 0]\ne_y = [-2, 1, 0]\ne_z = [0, 0, 5]\nlcs_03 = tf.LocalCoordinateSystem.from_xyz(e_x, e_y, e_z, coordinates=[1, 1, 0])\n\n# create a negatively oriented coordinate system with 2 vectors\nlcs_04 = tf.LocalCoordinateSystem.from_yz_and_orientation(\n e_y, e_z, positive_orientation=False, coordinates=[1, 1, 2]\n)",
"_____no_output_____"
]
],
[
[
"**Plot:**",
"_____no_output_____"
]
],
[
[
"vs.plot_coordinate_systems([(lcs_ref, {\"color\":\"r\", \"label\":\"reference_system\"}), \n (lcs_03, {\"color\":\"g\", \"label\":\"system 3\"}), \n (lcs_04, {\"color\":\"b\", \"label\":\"system 4\"}) ])",
"_____no_output_____"
]
],
[
[
"As you can see, the y- and z-axis of system 3 and 4 are pointing into the same direction, since we used the same basis vectors. The automatically determined x axis of system 4 points into the opposite direction, since we wanted a system with negative orientation.\n\nAnother method to create a `LocalCoordinateSystem` is `from_euler`. It utilizes the `scipy.spatial.transform.Rotation.from_euler` function to calculate a rotation matrix from Euler sequences and uses it to describe the orientation of the coordinate system. The parameters `sequence`, `angles`, and `degrees` of the `from_euler` method are directly passed to the SciPy function. `sequence` expects a string that determines the rotation sequence around the coordinate axes. For example `\"xyz\"`. `angles` is a scalar or list of the corresponding number of angles and `degrees` a `bool` that specifies if the angles are provided in degrees (`degrees=True`) or radians (`degrees=False`). For further details, have a look at the [SciPy documentation](https://docs.scipy.org/doc/scipy/reference/generated/scipy.spatial.transform.Rotation.from_euler.html) of the `from_euler` function. Here is a short example:",
"_____no_output_____"
]
],
[
[
"# create a coordinate system by a 90° rotation around the x axis and subsequent 45° rotation around the y axis\nlcs_05 = tf.LocalCoordinateSystem.from_euler(sequence=\"x\", angles=90, degrees=True, coordinates=[1, -1, 0])\nlcs_06 = tf.LocalCoordinateSystem.from_euler(\n sequence=\"xy\", angles=[90, 45], degrees=True, coordinates=[2, -2, 0]\n)",
"_____no_output_____"
]
],
[
[
"**Plot:**",
"_____no_output_____"
]
],
[
[
"# create 3d plot\nvs.plot_coordinate_systems([(lcs_ref, {\"color\":\"r\", \"label\":\"reference_system\"}), \n (lcs_05, {\"color\":\"g\", \"label\":\"system 5\"}), \n (lcs_06, {\"color\":\"b\", \"label\":\"system 6\"}) ])",
"_____no_output_____"
]
],
[
[
"## Coordinate transformations\n\nIt is quite common that there exists a chain or tree-like dependency between coordinate systems. We might have a moving object with a local coordinate system that describes its position and orientation towards a fixed reference coordinate system. This object can have another object attached to it, with its position and orientation given in relation to its parent objects coordinate system. If we want to know the attached object coordinate system in relation to the reference coordinate system, we have to perform a coordinate transformation.\n\nTo avoid confusion about the reference systems of each coordinate system, we will use the following naming convention for the coordinate systems: `lcs_NAME_in_REFERENCE`. This is a coordinate system with the name \"NAME\" and its reference system has the name \"REFERENCE\". The only exception to this convention will be the reference coordinate system \"lcs_ref\", which has no reference system.\n\nThe `LocalCoordinateSystem` class provides the `+` and `-` operators to change the reference system easily. The `+` operator will transform a coordinate system to the reference coordinate system of its current reference system:\n\n~~~ python\nlcs_child_in_ref = lcs_child_in_parent + lcs_parent_in_ref\n~~~\nAs the naming of the variables already implies, the `+` operator should only be used if there exists a **child-parent relation** between the left-hand side and right-hand side system.\nIf two coordinate systems share a **common reference system**, the `-` operator transforms one of those systems into the other:\n\n~~~ python\nlcs_child_in_parent = lcs_child_in_ref - lcs_parent_in_ref\n~~~\n\nIt is important to remember that this operation is in general not commutative since it involves matrix multiplication which is also not commutative. During those operations, the local system that should be transformed into another coordinate system is always located to the left of the `+` or `-` operator. You can also chain multiple transformations, like this:\n\n~~~ python\nlcs_A_in_C = lcs_A_in_B + lcs_B_in_ref - lcs_C_in_ref\n~~~\n\nPythons operator associativity ([link](https://www.faceprep.in/python/python-operator-precedence-associativity/)) for the `+` and `-` operator ensures, that all operations are performed from left to right. So in the previously shown example, we first calculate an intermediate coordinate system `lcs_A_in_ref` (`lcs_A_in_B + lcs_B_in_ref`) without actually storing it to a variable and subsequently transform it to the reference coordinate system C (`lcs_A_in_ref - lcs_C_in_ref`). Keep in mind, that the intermediate results and the coordinate system on the right-hand side of the next operator must either have a child-parent relation (`+` operator) or share a common coordinate system (`-` operator), otherwise the transformation chain produces invalid results.\n\nYou can think about both operators in the context of a tree-like graph structure where all dependency chains lead to a common root coordinate system. The `+` operator moves a coordinate system 1 level higher and closer to the root. Since its way to the root leads over its parent coordinate system, the parent is the only valid system than can be used on the right-hand side of the `+` operator. The `-` operator pushes a coordinate system one level lower and further away from the root. It can only be pushed one level deeper if there is another coordinate system connected to its parent system.\n\n> TODO: Add pictures\n\n### Interactive examples for the + and - operator\n\nThe following small interactive examples should give you a better understanding of how the `+` and `-` operators work. The examples provide several sliders to modify the orientations and positions of 2 coordinate systems. From those, a third coordinate system is calculated using the `+` and `-` operator. Subsequently, the coordinate systems are plotted in relation to each other. The relevant lines of code, which generate the coordinate systems are:\n\n> **Hint:** The interactive plots need the `%matplotlib widget` magic command. Make sure you have the corresponding extensions installed and executed the command at the beginning of the tutorial",
"_____no_output_____"
]
],
[
[
"def coordinate_system_addition(parent_orientation, parent_coordinates, child_orientation, child_coordinates):\n lcs_parent_in_ref = tf.LocalCoordinateSystem(orientation=parent_orientation, coordinates=parent_coordinates)\n lcs_child_in_parent = tf.LocalCoordinateSystem(orientation=child_orientation, coordinates=child_coordinates)\n\n lcs_child_in_ref = lcs_child_in_parent + lcs_parent_in_ref\n\n return [lcs_parent_in_ref, lcs_child_in_parent, lcs_child_in_ref]\n\n\ndef coordinate_system_subtraction(\n sys1_in_ref_orientation, sys1_in_ref_coordinates, sys2_in_ref_orientation, sys2_in_ref_coordinates\n):\n lcs_sys1_in_ref = tf.LocalCoordinateSystem(orientation=sys1_in_ref_orientation, coordinates=sys1_in_ref_coordinates)\n lcs_sys2_in_ref = tf.LocalCoordinateSystem(orientation=sys2_in_ref_orientation, coordinates=sys2_in_ref_coordinates)\n\n lcs_sys2_in_sys1 = lcs_sys2_in_ref - lcs_sys1_in_ref\n lcs_sys1_in_sys2 = lcs_sys1_in_ref - lcs_sys2_in_ref\n\n return [lcs_sys1_in_ref, lcs_sys2_in_ref, lcs_sys1_in_sys2, lcs_sys2_in_sys1]",
"_____no_output_____"
]
],
[
[
"Now just execute the following code cells. You don't need to understand them since they just create the sliders and plots:",
"_____no_output_____"
]
],
[
[
"def create_output_widget(window_size=900):\n # create output widget that will hold the figure\n out = widgets.Output(layout={\"border\": \"2px solid black\"})\n\n # create figure inside output widget\n with out:\n fig = plt.figure()\n try:\n fig.canvas.layout.height = str(window_size) + \"px\"\n fig.canvas.layout.width = str(window_size) + \"px\"\n except:\n pass\n gs = fig.add_gridspec(3, 2)\n ax_0 = fig.add_subplot(gs[0, 0], projection=\"3d\")\n ax_1 = fig.add_subplot(gs[0, 1], projection=\"3d\")\n ax_2 = fig.add_subplot(gs[1:, 0:], projection=\"3d\")\n return [out, fig, ax_0, ax_1, ax_2]\n\n\ndef setup_axes(axes, limit, title=\"\"):\n axes.set_xlim([-limit, limit])\n axes.set_ylim([-limit, limit])\n axes.set_zlim([-limit, limit])\n axes.set_xlabel(\"x\")\n axes.set_ylabel(\"y\")\n axes.set_zlabel(\"z\")\n axes.set_title(title)\n axes.legend(loc=\"lower left\")\n\n\ndef get_orientation_and_location(t_x, t_y, t_z, r_x, r_y, r_z):\n rot_angles = np.array([r_x, r_y, r_z], float) / 180 * np.pi\n\n rot_x = tf.WXRotation.from_euler(\"x\", rot_angles[0]).as_matrix()\n rot_y = tf.WXRotation.from_euler(\"y\", rot_angles[1]).as_matrix()\n rot_z = tf.WXRotation.from_euler(\"z\", rot_angles[2]).as_matrix()\n\n orientation = np.matmul(rot_z, np.matmul(rot_y, rot_x))\n location = [t_x, t_y, t_z]\n return [orientation, location]\n\n\ndef create_slider(limit, step, label):\n layout = widgets.Layout(width=\"200px\", height=\"40px\")\n style = {\"description_width\": \"initial\"}\n return FloatSlider(\n min=-limit, max=limit, step=step, description=label, continuous_update=True, layout=layout, style=style\n )\n\n\ndef create_interactive_plot(function, limit_loc=3, name_sys1=\"system 1\", name_sys2=\"system 2\"):\n step_loc = 0.25\n\n w_s1_l = dict(\n s1_x=create_slider(limit_loc, step_loc, \"x\"),\n s1_y=create_slider(limit_loc, step_loc, \"y\"),\n s1_z=create_slider(limit_loc, step_loc, \"z\"),\n )\n\n w_s1_r = dict(\n s1_rx=create_slider(180, 10, \"x\"), s1_ry=create_slider(180, 10, \"y\"), s1_rz=create_slider(180, 10, \"z\")\n )\n\n w_s2_l = dict(\n s2_x=create_slider(limit_loc, step_loc, \"x\"),\n s2_y=create_slider(limit_loc, step_loc, \"y\"),\n s2_z=create_slider(limit_loc, step_loc, \"z\"),\n )\n\n w_s2_r = dict(\n s2_rx=create_slider(180, 10, \"x\"), s2_ry=create_slider(180, 10, \"y\"), s2_rz=create_slider(180, 10, \"z\")\n )\n\n w = {**w_s1_l, **w_s1_r, **w_s2_l, **w_s2_r}\n\n output = interactive_output(function, w)\n box_0 = VBox([widgets.Label(name_sys1 + \" coordinates\"), *w_s1_l.values()])\n box_1 = VBox([widgets.Label(name_sys1 + \" rotation (deg)\"), *w_s1_r.values()])\n box_2 = VBox([widgets.Label(name_sys2 + \" coordinates\"), *w_s2_l.values()])\n box_3 = VBox([widgets.Label(name_sys2 + \" rotation (deg)\"), *w_s2_r.values()])\n box = HBox([box_0, box_1, box_2, box_3])\n display(box)",
"_____no_output_____"
],
[
"axes_lim = 3\nwindow_size = 1000\n\n[out, fig_iadd, ax_iadd_0, ax_iadd_1, ax_iadd_2] = create_output_widget(window_size)\n\n\ndef update_output(s1_x, s1_y, s1_z, s1_rx, s1_ry, s1_rz, s2_x, s2_y, s2_z, s2_rx, s2_ry, s2_rz):\n\n [parent_orientation, parent_coordinates] = get_orientation_and_location(s1_x, s1_y, s1_z, s1_rx, s1_ry, s1_rz)\n [child_orientation, child_coordinates] = get_orientation_and_location(s2_x, s2_y, s2_z, s2_rx, s2_ry, s2_rz)\n\n [lcs_parent, lcs_child, lcs_child_ref] = coordinate_system_addition(\n parent_orientation, parent_coordinates, child_orientation, child_coordinates\n )\n\n coordinates_cr = lcs_child_ref.coordinates\n cr_x = coordinates_cr[0]\n cr_y = coordinates_cr[1]\n cr_z = coordinates_cr[2]\n\n ax_iadd_0.clear()\n vs.draw_coordinate_system_matplotlib(lcs_ref, ax_iadd_0, color=\"r\", label=\"reference\")\n vs.draw_coordinate_system_matplotlib(lcs_parent, ax_iadd_0, color=\"g\", label=\"parent\")\n ax_iadd_0.plot([0, s1_x], [0, s1_y], [0, s1_z], \"c--\", label=\"ref -> parent\")\n setup_axes(ax_iadd_0, axes_lim, \"'parent' in reference coordinate system\")\n\n ax_iadd_1.clear()\n vs.draw_coordinate_system_matplotlib(lcs_ref, ax_iadd_1, color=\"g\", label=\"parent\")\n vs.draw_coordinate_system_matplotlib(lcs_child, ax_iadd_1, color=\"y\", label=\"child\")\n ax_iadd_1.plot([0, s2_x], [0, s2_y], [0, s2_z], \"m--\", label=\"parent -> child\")\n setup_axes(ax_iadd_1, axes_lim, \"'child' in 'parent' coordinate system\")\n\n ax_iadd_2.clear()\n vs.draw_coordinate_system_matplotlib(lcs_ref, ax_iadd_2, color=\"r\", label=\"reference\")\n vs.draw_coordinate_system_matplotlib(lcs_parent, ax_iadd_2, color=\"g\", label=\"parent\")\n vs.draw_coordinate_system_matplotlib(lcs_child_ref, ax_iadd_2, color=\"y\", label=\"parent + child\")\n ax_iadd_2.plot([0, s1_x], [0, s1_y], [0, s1_z], \"c--\", label=\"ref -> parent\")\n ax_iadd_2.plot([s1_x, cr_x], [s1_y, cr_y], [s1_z, cr_z], \"m--\", label=\"parent -> child\")\n setup_axes(ax_iadd_2, axes_lim * 2, \"'parent' and 'child' in reference coordinate system\")\n\ncreate_interactive_plot(update_output, limit_loc=axes_lim, name_sys1=\"parent\", name_sys2=\"child\")\nout",
"_____no_output_____"
],
[
"axes_lim = 1.5\nwindow_size = 1000\n\n[out_2, fig2, ax_isub_0, ax_isub_1, ax_isub_2] = create_output_widget(window_size)\n\n\ndef update_output2(s1_x, s1_y, s1_z, s1_rx, s1_ry, s1_rz, s2_x, s2_y, s2_z, s2_rx, s2_ry, s2_rz):\n\n [sys1_orientation, sys1_coordinates] = get_orientation_and_location(s1_x, s1_y, s1_z, s1_rx, s1_ry, s1_rz)\n [sys2_orientation, sys2_coordinates] = get_orientation_and_location(s2_x, s2_y, s2_z, s2_rx, s2_ry, s2_rz)\n\n [lcs_sys1_in_ref, lcs_sys2_in_ref, lcs_sys1_in_sys2, lcs_sys2_in_sys1] = coordinate_system_subtraction(\n sys1_orientation, sys1_coordinates, sys2_orientation, sys2_coordinates\n )\n sys12_o = lcs_sys1_in_sys2.coordinates\n sys12_x = sys12_o[0]\n sys12_y = sys12_o[1]\n sys12_z = sys12_o[2]\n\n sys21_o = lcs_sys2_in_sys1.coordinates\n sys21_x = sys21_o[0]\n sys21_y = sys21_o[1]\n sys21_z = sys21_o[2]\n\n ax_isub_1.clear()\n vs.draw_coordinate_system_matplotlib(lcs_ref, ax_isub_1, color=\"g\", label=\"system 1 (reference)\")\n vs.draw_coordinate_system_matplotlib(lcs_sys2_in_sys1, ax_isub_1, color=\"b\", label=\"system 2 - system 1\")\n ax_isub_1.plot([0, sys21_x], [0, sys21_y], [0, sys21_z], \"y--\", label=\"system 1 -> system 2\")\n setup_axes(ax_isub_1, axes_lim * 2, \"'system 2' in 'system 1'\")\n\n ax_isub_0.clear()\n vs.draw_coordinate_system_matplotlib(lcs_ref, ax_isub_0, color=\"b\", label=\"system 2 (reference)\")\n vs.draw_coordinate_system_matplotlib(lcs_sys1_in_sys2, ax_isub_0, color=\"g\", label=\"system_1 - system 2\")\n ax_isub_0.plot([0, sys12_x], [0, sys12_y], [0, sys12_z], \"y--\", label=\"system 1 -> system 2\")\n setup_axes(ax_isub_0, axes_lim * 2, \"'system 1' in 'system 2'\")\n\n ax_isub_2.clear()\n vs.draw_coordinate_system_matplotlib(lcs_ref, ax_isub_2, color=\"r\", label=\"reference\")\n vs.draw_coordinate_system_matplotlib(lcs_sys1_in_ref, ax_isub_2, color=\"g\", label=\"system 1\")\n vs.draw_coordinate_system_matplotlib(lcs_sys2_in_ref, ax_isub_2, color=\"b\", label=\"system 2\")\n ax_isub_2.plot([0, s1_x], [0, s1_y], [0, s1_z], \"g--\", label=\"ref -> system 1\")\n ax_isub_2.plot([0, s2_x], [0, s2_y], [0, s2_z], \"b--\", label=\"ref -> system 2\")\n ax_isub_2.plot([s1_x, s2_x], [s1_y, s2_y], [s1_z, s2_z], \"y--\", label=\"system 1 <-> system 2\")\n setup_axes(ax_isub_2, axes_lim, \"'system 1' and 'system 2' in reference coordinate system\")\n\n\ncreate_interactive_plot(update_output2, limit_loc=axes_lim)\nout_2",
"_____no_output_____"
]
],
[
[
"## Invert method\n\nThe `invert` method calculates how a parent coordinate system is positioned and oriented in its child coordinate system:\n\n~~~ python\nlcs_child_in_parent = lcs_parent_in_child.invert()\n~~~\n\nHere is a short example with visualization:",
"_____no_output_____"
]
],
[
[
"lcs_child_in_parent = tf.LocalCoordinateSystem.from_euler(\n sequence=\"xy\", angles=[90, 45], degrees=True, coordinates=[2, 3, 0]\n)\nlcs_parent_in_child = lcs_child_in_parent.invert()",
"_____no_output_____"
]
],
[
[
"**Plot:**",
"_____no_output_____"
]
],
[
[
"_, (ax_invert_0, ax_invert_1) = vs.new_3d_figure_and_axes(num_subplots=2, width=1000)\n\n# left plot\nvs.plot_coordinate_systems([(lcs_ref, {\"color\":\"r\", \"label\":\"parent\"}), \n (lcs_child_in_parent, {\"color\":\"g\", \"label\":\"child\"})],\n axes=ax_invert_0,\n limits=(-3, 3),\n title=\"child in parent\")\n \n# right plot\nvs.plot_coordinate_systems([(lcs_parent_in_child, {\"color\":\"r\", \"label\":\"parent\"}), \n (lcs_ref, {\"color\":\"g\", \"label\":\"child\"})],\n axes=ax_invert_1,\n limits=(-3, 3),\n title=\"parent in child\")",
"_____no_output_____"
]
],
[
[
"## Time dependency\n\nThe orientation and position of a local coordinate system towards their reference system might vary in time. For example, in a welding process, the position of the torch towards the specimen is changing constantly. The `LocalCoordinateSystem` provides an interface for such cases. All previously shown construction methods also provide the option to pass a `time` parameter.\n\n> **IMPORTANT HINT:** In the current package version, only the constructors `time` parameter is actually working\n\n> **TODO:** Rewrite examples if factory methods also support time dependency\n\nTo create a time-dependent system, you have to provide a list of timestamps that can be converted to a `pandas.DateTimeIndex`. If you do, you also need to provide the extra data for the `orientation` and/or `coordinates` to the constructor or construction method. You do this by adding an extra outer dimension to the corresponding time-dependent function parameter. For example: If you want to create a moving coordinate system with 2 timestamps, you can do it by like this:\n\n~~~ python\ntime = [\"2010-02-01\", \"2010-02-02\"]\n\n # fixed orientation (no extra dimension)\norientation_mov = [[0, -1, 0], [1, 0, 0], [0, 0, 1]]\n\n# time dependent coordinates (extra dimension)\ncoordinates_mov = [[-3, 0, 0], [0, 0, 2]]\n\nlcs_mov_in_ref = tf.LocalCoordinateSystem(orientation=orientation_mov, coordinates=coordinates_mov, time=time)\n~~~\n\nA coordinate system with varying orientation between 2 timestamps is defined by:\n\n~~~ python\ntime = [\"2010-02-01\", \"2010-02-02\"]\n\n# time dependent orientation (extra coordinate)\norientation_rot = [[[1, 0, 0], [0, 1, 0], [0, 0, 1]], [[0, -1, 0], [1, 0, 0], [0, 0, 1]]]\n\n# fixed coordinates (no extra dimension)\ncoordinates_rot = [1, 0, 2]\n\nlcs_rot_in_ref = tf.LocalCoordinateSystem(orientation=orientation_rot, coordinates=coordinates_rot, time=time)\n~~~\n\nYou can also create a rotating and moving coordinate system:\n\n~~~ python\ntime = [\"2010-02-01\", \"2010-02-02\"]\n\ncoordinates_movrot = [[0, 3, 0], [-2, 3, 2]]\norientation_movrot = [[[1, 0, 0], [0, 1, 0], [0, 0, 1]], [[0, -1, 0], [1, 0, 0], [0, 0, 1]]]\n\nlcs_movrot_in_ref = tf.LocalCoordinateSystem(orientation=orientation_movrot, coordinates=coordinates_movrot, time=time)\n~~~\n\nYou have to ensure, that the extra outer dimension has always the same number of entries as a number of timestamps you provided. Otherwise, an exception is raised. In case your coordinate system data is not always providing orientation and coordinates at the same time, you need to interpolate the missing values first.\n\nHere is a visualization of the discussed coordinate systems at the two different times:",
"_____no_output_____"
]
],
[
[
"# define coordinate systems\ntime = [\"2010-02-01\", \"2010-02-02\"]\n\norientation_mov = [[0, -1, 0], [1, 0, 0], [0, 0, 1]]\ncoordinates_mov = [[-3, 0, 0], [0, 0, 2]]\nlcs_mov_in_ref = tf.LocalCoordinateSystem(orientation=orientation_mov, coordinates=coordinates_mov, time=time)\n\norientation_rot = [[[1, 0, 0], [0, 1, 0], [0, 0, 1]], [[0, -1, 0], [1, 0, 0], [0, 0, 1]]]\ncoordinates_rot = [1, 0, 2]\nlcs_rot_in_ref = tf.LocalCoordinateSystem(orientation=orientation_rot, coordinates=coordinates_rot, time=time)\n\ncoordinates_movrot = [[0, 3, 0], [-2, 3, 2]]\norientation_movrot = [[[1, 0, 0], [0, 1, 0], [0, 0, 1]], [[0, -1, 0], [1, 0, 0], [0, 0, 1]]]\nlcs_movrot_in_ref = tf.LocalCoordinateSystem(orientation=orientation_movrot, coordinates=coordinates_movrot, time=time)",
"_____no_output_____"
]
],
[
[
"**Plot:**",
"_____no_output_____"
]
],
[
[
"# plot coordinate systems\n_, axes_array = vs.new_3d_figure_and_axes(num_subplots=2, width=1000)\n\nfor i, ax in enumerate(axes_array):\n vs.plot_coordinate_systems([(lcs_ref, {\"color\":\"r\", \"label\":\"reference\"}),\n (lcs_mov_in_ref, {\"color\":\"g\", \"label\":\"lcs_mov_in_ref\"}),\n (lcs_rot_in_ref, {\"color\":\"b\", \"label\":\"lcs_rot_in_ref\"}),\n (lcs_movrot_in_ref, {\"color\":\"c\", \"label\":\"lcs_movrot_in_ref\"})],\n axes=ax,\n limits=(-3, 3),\n title=f\"timestep {i}\",\n time_index=i)\n",
"_____no_output_____"
]
],
[
[
"## Time interpolation\n\nIt is also possible, to interpolate a coordinate system's orientations and coordinates in time by using the `interp_time` function. You have to pass it a single or multiple target times for the interpolation. As for the constructor, you can pass any type that is convertible to a `pandas.DatetimeIndex`. Alternatively, you can pass another `LocalCoordinateSystem` which provides the target timestamps. The return value of this function is a new `LocalCoordinateSystem` with interpolated orientations and coordinates.\n\nIn case that a target time for the interpolation lies outside of the `LocalCoordinateSystem`s' time range, the boundary value is broadcasted.\n\nHere is an example:",
"_____no_output_____"
]
],
[
[
"# original coordinate system\ntime = [\"2010-02-02\", \"2010-02-07\"]\n\ncoordinates_tdp = [[0, 3, 0], [-2, 3, 2]]\norientation_tdp = [[[1, 0, 0], [0, 1, 0], [0, 0, 1]], [[0, -1, 0], [1, 0, 0], [0, 0, 1]]]\nlcs_tdp_in_ref = tf.LocalCoordinateSystem(orientation=orientation_movrot, coordinates=coordinates_movrot, time=time)\n\ntime_interp = pd.DatetimeIndex([\"2010-02-01\", \"2010-02-03\", \"2010-02-04\", \"2010-02-05\", \"2010-02-06\", \"2010-02-11\"])\nlcs_interp_in_ref = lcs_tdp_in_ref.interp_time(time_interp)",
"_____no_output_____"
]
],
[
[
"**Plot:**",
"_____no_output_____"
]
],
[
[
"# plot coordinate systems\n_, (ax_0_interp, ax_1_interp) = vs.new_3d_figure_and_axes(num_subplots=2, width=1000)\n\n# original systems\nvs.plot_coordinate_systems([(lcs_ref, {\"color\":\"r\", \"label\":\"reference\"}),\n (lcs_tdp_in_ref, {\"color\":\"g\", \"label\":\"2010-02-01\", \"time_index\":0}),\n (lcs_tdp_in_ref, {\"color\":\"b\", \"label\":\"2010-02-07\", \"time_index\":1})],\n axes=ax_0_interp,\n limits=(-3, 3),\n title=\"original system\")\n\n\n# interpolated systems\ncolors = [[0, 1, 0], [0, 1, 0.5], [0, 1, 1], [0, 0.5, 1], [0, 0, 1], [0.5, 0, 1]]\nplot_data = [\n (lcs_interp_in_ref, {\"color\":colors[i], \"label\":time_interp[i], \"time_index\":i}) \n for i in range(len(time_interp))\n]\nplot_data = [(lcs_ref, {\"color\":\"r\", \"label\":\"reference\"}), *plot_data]\n\nvs.plot_coordinate_systems(plot_data,\n axes=ax_1_interp,\n limits=(-3, 3),\n title=\"interpolated system\")",
"_____no_output_____"
]
],
[
[
"As you can see, the time values `\"2010-02-01\"` and `\"2010-02-11\"`, which lie outside the original range from `\"2010-02-02\"` and `\"2010-02-07\"` still get valid values due to the broadcasting across time range boundaries. The intermediate coordinates and orientations are interpolated as expected.\n\n> **TODO:** Mention interpolation behavior of `+` and `*`\n\n## Transformation of spatial data\n\nThe `LocalCoordinateSystem` only defines how the different coordinate systems are oriented towards each other. If you want to transform spatial data which is defined in one coordinate system (for example specimen geometry/point cloud) you have to use the `CoordinateSystemManager`, which is discussed in the next tutorial or do the transformation manually.\nFor the manual transformation, you can get all you need from the `LocalCoordinateSystem` using its accessor properties:\n\n~~~\norientation = lcs_a_in_b.orientation\ncoordinates = lcs_a_in_b.coordinates\n~~~\n\nThe returned data is an `xarray.DataFrame`. In case you are not used to work with this data type, you can get a `numpy.ndarray` by simply using their `data` property:\n\n~~~\norientation_numpy = lcs_a_in_b.orientation.data\ncoordinates_numpy = lcs_a_in_b.coordinates.data\n~~~\n\nKeep in mind, that you actually get an array of matrices (`orientation`) and vectors (`coordinates`) if the corresponding component is time dependent. The transformation itself is done by the equation:\n\n$$v_b = O_{ab} \\cdot v_a + c_{ab}$$\n\nwhere $v_a$ is a data point defined in coordinate system `a`, $O_{ab}$ is the orientation matrix of `a` in `b`, $c_{ab}$ the coordinates of `a` in `b` and $v_b$ the transformed data point.\n\n\nHere is a short example that transforms the points of a square from one coordinate system to another:",
"_____no_output_____"
]
],
[
[
"lcs_target_in_ref = tf.LocalCoordinateSystem.from_euler(\n sequence=\"zy\", angles=[90, 45], degrees=True, coordinates=[2, -2, 0]\n)\nlcs_ref_in_target = lcs_target_in_ref.invert()\n\npoints_in_ref = np.array([[-1, 1, 0], [1, 1, 0], [1, -1, 0], [-1, -1, 0], [-1, 1, 0]], dtype=float).transpose()\n\n# Transform points to target system\npoints_in_target = (\n np.matmul(lcs_ref_in_target.orientation.data, points_in_ref) + lcs_ref_in_target.coordinates.data[:, np.newaxis]\n)",
"_____no_output_____"
]
],
[
[
"**Plot:**",
"_____no_output_____"
]
],
[
[
"# plot coordinate systems\n_, (ax_0_trans, ax_1_trans) = vs.new_3d_figure_and_axes(num_subplots=2, width=1000)\n\n# first plot\nvs.plot_coordinate_systems([(lcs_ref, {\"color\":\"r\", \"label\":\"original system\"}),\n (lcs_target_in_ref, {\"color\":\"g\", \"label\":\"target system\"})],\n axes=ax_0_trans,\n limits=(-3, 3),\n title=\"Data in original system\")\nax_0_trans.plot(points_in_ref[0], points_in_ref[1], points_in_ref[2])\n\n\n# second plot\nvs.plot_coordinate_systems([(lcs_ref_in_target, {\"color\":\"r\", \"label\":\"original system\"}),\n (lcs_ref, {\"color\":\"g\", \"label\":\"target system\"})],\n axes=ax_1_trans,\n limits=(-3, 3),\n title=\"Data in original system\")\nax_1_trans.plot(points_in_target[0], points_in_target[1], points_in_target[2])",
"_____no_output_____"
]
],
[
[
"## Internal xarray structure\n\nThe local coordinate system and many other components of the WeldX package use xarray data frames internally. So it is also possible to pass xarray data frames to a lot of functions and the constructor. However, they need a certain structure which will be described here. If you are not familiar with the xarray package, you should first read the [documentation](http://xarray.pydata.org/en/stable/).\n\nTo pass a xarray data frame as coordinate to a `LocalCoordinateSystem`, it must at least have a dimension `c`. It represents the location in 3d space of the coordinate system and must always be of length 3. Those components must be named coordinates of the data frame (`coords={\"c\": [\"x\", \"y\", \"z\"]}`). An optional dimension is `time`. It can be of arbitrary length, but the timestamps must be added as coordinates.\n\nThe same conventions that are used for the coordinates also apply to the orientations. Additionally, they must have another dimension `v` of length 3, which are enumerated (`\"v\": [0, 1, 2]`). `c` and `v` are the rows and columns of the orientation matrix.\n\n**Example:**",
"_____no_output_____"
]
],
[
[
"time = pd.TimedeltaIndex([0,5],\"D\")\ncoordinates_tdp = [[0, 3, 0], [-2, 3, 2]]\norientation_tdp = [[[1, 0, 0], [0, 1, 0], [0, 0, 1]], [[0, -1, 0], [1, 0, 0], [0, 0, 1]]]\n\ndsx_coordinates = xr.DataArray(data=coordinates_tdp, dims=[\"time\", \"c\"], coords={\"time\": time, \"c\": [\"x\", \"y\", \"z\"]})\ndsx_orientation = xr.DataArray(\n data=orientation_tdp, dims=[\"time\", \"c\", \"v\"], coords={\"time\": time, \"c\": [\"x\", \"y\", \"z\"], \"v\": [0, 1, 2]},\n)\n\nlcs_xr = tf.LocalCoordinateSystem(orientation=dsx_orientation, coordinates=dsx_coordinates)",
"_____no_output_____"
]
],
[
[
"**Plot:**",
"_____no_output_____"
]
],
[
[
"# plot coordinate systems\n_, ax_dsx = vs.new_3d_figure_and_axes()\n\n# first timestep\nvs.plot_coordinate_systems([(lcs_ref, {\"color\":\"r\", \"label\":\"reference\"}),\n (lcs_xr, {\"color\":\"g\", \"label\":str(lcs_xr.time[0]), \"time_index\":0}),\n (lcs_xr, {\"color\":\"b\", \"label\":str(lcs_xr.time[1]), \"time_index\":1})],\n axes=ax_dsx,\n limits=(-3, 3))",
"_____no_output_____"
]
],
[
[
"The `weldx.utility` package contains two utility functions to create xarray data frames that can be passed as `orientation` and `coordinates` to an `LocalCoordinateSystem`. They are named `xr_3d_vector` and `xr_3d_matrix`. Please read the API documentation for further information.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
4affec30f6fa5ce9470b0b46c3ef8711562a58ce
| 64,833 |
ipynb
|
Jupyter Notebook
|
tdx/Untitled2.ipynb
|
dizzy21c/easyqtrs
|
4704674d2175d40afdc306afd8a002a486c83220
|
[
"MIT"
] | 2 |
2021-12-30T13:43:13.000Z
|
2022-01-23T13:39:54.000Z
|
tdx/Untitled2.ipynb
|
dizzy21c/easyqtrs
|
4704674d2175d40afdc306afd8a002a486c83220
|
[
"MIT"
] | null | null | null |
tdx/Untitled2.ipynb
|
dizzy21c/easyqtrs
|
4704674d2175d40afdc306afd8a002a486c83220
|
[
"MIT"
] | 4 |
2021-10-22T01:44:47.000Z
|
2022-01-05T05:49:20.000Z
| 35.447239 | 180 | 0.346058 |
[
[
[
"from py2neo import Graph,Node,Relationship",
"_____no_output_____"
],
[
"import pandas as pd\nimport os\nimport QUANTAXIS as QA\nimport datetime\nimport numpy as np \nimport statsmodels.formula.api as sml\nfrom QAStrategy.qastockbase import QAStrategyStockBase\n\nimport matplotlib.pyplot as plt\nimport scipy.stats as scs\nimport matplotlib.mlab as mlab\nfrom easyquant.indicator.base import *\n\nimport json\nfrom easyquant import MongoIo\nimport statsmodels.api as sm\nfrom multiprocessing import Process, Pool, cpu_count, Manager\n",
"_____no_output_____"
],
[
"mongo = MongoIo()\n",
"_____no_output_____"
],
[
"def tdx_base_func(data, code_list = None):\n \"\"\"\n 准备数据\n \"\"\"\n # highs = data.high\n # start_t = datetime.datetime.now()\n # print(\"begin-tdx_base_func:\", start_t)\n if len(data) < 10:\n data = data.copy()\n data['bflg'] = 0\n data['sflg'] = 0\n return data\n\n CLOSE=data.close\n C=data.close\n # df_macd = MACD(C,12,26,9)\n # mtj1 = IFAND(df_macd.DIFF < 0, df_macd.DEA < 0, 1, 0)\n # mtj2 = IFAND(mtj1, df_macd.MACD < 0, 1, 0)\n 花 = SLOPE(EMA(C, 3), 3)\n 神 = SLOPE(EMA(C, 7), 7)\n 买 = IFAND(COUNT(花 < 神, 5)==4 , 花 >= 神,1,0)\n 卖 = IFAND(COUNT(花 >= 神, 5)==4, 花 < 神,1,0)\n 钻石 = IFAND(CROSS(花, 神), CLOSE / REF(CLOSE, 1) > 1.03, 1, 0)\n 买股 = IFAND(买, 钻石,1,0)\n # 买股 = IFAND(mtj2, 买股1, 1, 0)\n # AND(CROSS(花, 神)\n # AND\n # CLOSE / REF(CLOSE, 1) > 1.03);\n\n # return pd.DataFrame({'FLG': 后炮}).iloc[-1]['FLG']\n # return 后炮.iloc[-1]\n\n # 斜率\n data = data.copy()\n # data['bflg'] = IF(REF(后炮,1) > 0, 1, 0)\n data['bflg'] = 买股\n data['sflg'] = 卖\n # print(\"code=%s, bflg=%s\" % (code, data['bflg'].iloc[-1]))\n # data['beta'] = 0\n # data['R2'] = 0\n # beta_rsquared = np.zeros((len(data), 2),)\n #\n # for i in range(N - 1, len(highs) - 1):\n # #for i in range(len(highs))[N:]:\n # df_ne = data.iloc[i - N + 1:i + 1, :]\n # model = sml.ols(formula='high~low', data = df_ne)\n # result = model.fit()\n #\n # # beta = low\n # beta_rsquared[i + 1, 0] = result.params[1]\n # beta_rsquared[i + 1, 1] = result.rsquared\n #\n # data[['beta', 'R2']] = beta_rsquared\n\n # 日收益率\n data['ret'] = data.close.pct_change(1)\n\n # 标准分\n # data['beta_norm'] = (data['beta'] - data.beta.rolling(M).mean().shift(1)) / data.beta.rolling(M).std().shift(1)\n #\n # beta_norm = data.columns.get_loc('beta_norm')\n # beta = data.columns.get_loc('beta')\n # for i in range(min(M, len(highs))):\n # data.iat[i, beta_norm] = (data.iat[i, beta] - data.iloc[:i - 1, beta].mean()) / data.iloc[:i - 1, beta].std() if (data.iloc[:i - 1, beta].std() != 0) else np.nan\n\n # data.iat[2, beta_norm] = 0\n # data['RSRS_R2'] = data.beta_norm * data.R2\n # data = data.fillna(0)\n #\n # # 右偏标准分\n # data['beta_right'] = data.RSRS_R2 * data.beta\n # if code == '000732':\n # print(data.tail(22))\n\n return data\n",
"_____no_output_____"
],
[
"def buy_sell_fun(price, S1=1.0, S2=0.8):\n \"\"\"\n 斜率指标交易策略标准分策略\n \"\"\"\n data = price.copy()\n data['flag'] = 0 # 买卖标记\n data['position'] = 0 # 持仓标记\n data['hold_price'] = 0 # 持仓价格\n bflag = data.columns.get_loc('bflg')\n sflag = data.columns.get_loc('sflg')\n # beta = data.columns.get_loc('beta')\n flag = data.columns.get_loc('flag')\n position_col = data.columns.get_loc('position')\n close_col = data.columns.get_loc('close')\n high_col = data.columns.get_loc('high')\n open_col = data.columns.get_loc('open')\n hold_price_col = data.columns.get_loc('hold_price')\n position = 0 # 是否持仓,持仓:1,不持仓:0\n for i in range(1,data.shape[0] - 1):\n # 开仓\n if data.iat[i, bflag] > 0 and position == 0:\n data.iat[i, flag] = 1\n data.iat[i, position_col] = 1\n data.iat[i, hold_price_col] = data.iat[i, open_col]\n data.iat[i + 1, position_col] = 1\n data.iat[i + 1, hold_price_col] = data.iat[i, open_col]\n\n position = 1\n\n print(\"buy : date=%s code=%s price=%.2f\" % (data.iloc[i].name[0], data.iloc[i].name[1], data.iloc[i].close))\n code = data.iloc[i].name[1]\n price = data.iloc[i].close\n# qa_order.send_order('BUY', 'OPEN', code=code, price=price, volume=1000)\n # order.send_order('SELL', 'CLOSE', code=code, price=price, volume=1000)\n # 平仓\n # elif data.iat[i, bflag] == S2 and position == 1:\n elif data.iat[i, position_col] > 0 and position == 1:\n cprice = data.iat[i, close_col]\n # oprice = data.iat[i, open_col]\n hole_price = data.iat[i, hold_price_col]\n high_price = data.iat[i, high_col]\n if cprice < hole_price * 0.95:# or cprice > hprice * 1.2:\n data.iat[i, flag] = -1\n data.iat[i + 1, position_col] = 0\n data.iat[i + 1, hold_price_col] = 0\n position = 0\n print(\"sell : code=%s date=%s price=%.2f\" % (data.iloc[i].name[0], data.iloc[i].name[1], data.iloc[i].close))\n code = data.iloc[i].name[1]\n price = data.iloc[i].close\n # order.send_order('BUY', 'OPEN', code=code, price=price, volume=1000)\n# qa_order.send_order('SELL', 'CLOSE', code=code, price=price, volume=1000)\n\n elif cprice > hole_price * 1.1 and high_price / cprice > 1.05:\n data.iat[i, flag] = -1\n data.iat[i + 1, position_col] = 0\n data.iat[i + 1, hold_price_col] = 0\n position = 0\n print(\"sell : code=%s date=%s price=%.2f\" % (data.iloc[i].name[0], data.iloc[i].name[1], data.iloc[i].close))\n code = data.iloc[i].name[1]\n price = data.iloc[i].close\n # order.send_order('BUY', 'OPEN', code=code, price=price, volume=1000)\n# qa_order.send_order('SELL', 'CLOSE', code=code, price=price, volume=1000)\n\n elif cprice > hole_price * 1.2 and high_price / cprice > 1.06:\n data.iat[i, flag] = -1\n data.iat[i + 1, position_col] = 0\n data.iat[i + 1, hold_price_col] = 0\n position = 0\n print(\"sell : code=%s date=%s price=%.2f\" % (data.iloc[i].name[0], data.iloc[i].name[1], data.iloc[i].close))\n code = data.iloc[i].name[1]\n price = data.iloc[i].close\n # order.send_order('BUY', 'OPEN', code=code, price=price, volume=1000)\n# qa_order.send_order('SELL', 'CLOSE', code=code, price=price, volume=1000)\n\n elif data.iat[i, sflag] > 0:\n data.iat[i, flag] = -1\n data.iat[i + 1, position_col] = 0\n data.iat[i + 1, hold_price_col] = 0\n position = 0\n print(\"sell : code=%s date=%s price=%.2f\" % (data.iloc[i].name[0], data.iloc[i].name[1], data.iloc[i].close))\n code = data.iloc[i].name[1]\n price = data.iloc[i].close\n # order.send_order('BUY', 'OPEN', code=code, price=price, volume=1000)\n# qa_order.send_order('SELL', 'CLOSE', code=code, price=price, volume=1000)\n else:\n data.iat[i + 1, position_col] = data.iat[i, position_col]\n data.iat[i + 1, hold_price_col] = data.iat[i, hold_price_col]\n # 保持\n else:\n data.iat[i + 1, position_col] = data.iat[i, position_col]\n data.iat[i + 1, hold_price_col] = data.iat[i, hold_price_col]\n\n data['nav'] = (1+data.close.pct_change(1).fillna(0) * data.position).cumprod()\n data['nav1'] = data.close * data.position\n return data\n",
"_____no_output_____"
],
[
"df=mongo.get_stock_day('600718')",
"_____no_output_____"
],
[
"df.tail()",
"_____no_output_____"
],
[
"data1=buy_sell_fun(df1)",
"buy : date=2018-04-16 00:00:00 code=600718 price=15.35\nsell : code=2018-04-23 00:00:00 date=600718 price=14.97\nbuy : date=2018-10-17 00:00:00 code=600718 price=9.61\nsell : code=2018-10-25 00:00:00 date=600718 price=9.78\nbuy : date=2019-04-01 00:00:00 code=600718 price=15.58\nsell : code=2019-04-08 00:00:00 date=600718 price=15.02\nbuy : date=2020-02-05 00:00:00 code=600718 price=11.96\nsell : code=2020-02-11 00:00:00 date=600718 price=12.65\nbuy : date=2020-07-14 00:00:00 code=600718 price=13.78\nsell : code=2020-07-16 00:00:00 date=600718 price=12.28\n"
],
[
"df1=tdx_base_func(df)",
"_____no_output_____"
],
[
"df1.tail()",
"_____no_output_____"
],
[
"data1.loc['2018-04-10':]",
"_____no_output_____"
],
[
"a = np.array([10,11,13,15,12,7,14])\nap = np.array([1,1,1,1,1,0,0])\nb = np.array([1.2,1.1,1.8,1.5,1.2,0.7,1.4])\n# a = np.array([[10,11,13,15,12,7,14],[10,11,18,15,12,7,14]])",
"_____no_output_____"
],
[
"dfn=pd.Series(a)\ndfb=pd.Series(b)\ndf=pd.DataFrame()\ndf['a']=pd.Series(a)\ndf['ap']=pd.Series(ap)\ndf",
"_____no_output_____"
],
[
"bc1=(1+dfn.pct_change(1).fillna(0)).cumprod()\nbc2=(1+dfb.pct_change(1).fillna(0)).cumprod()",
"_____no_output_____"
],
[
"bc2",
"_____no_output_____"
],
[
"result01=bc2\nround(float(max([(result01.iloc[idx] - result01.iloc[idx::].min()) / result01.iloc[idx] for idx in range(len(result01))])), 2)",
"_____no_output_____"
],
[
"bc",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4afff7e1ec5944e4398adb865288ecf9fcce9411
| 139,058 |
ipynb
|
Jupyter Notebook
|
examples/_debug/style.ipynb
|
CristianPachacama/cartoframes
|
3dc4e10d175069a7d7b734db3d9526127aad9dec
|
[
"BSD-3-Clause"
] | 1 |
2020-11-23T23:44:32.000Z
|
2020-11-23T23:44:32.000Z
|
examples/_debug/style.ipynb
|
CristianPachacama/cartoframes
|
3dc4e10d175069a7d7b734db3d9526127aad9dec
|
[
"BSD-3-Clause"
] | null | null | null |
examples/_debug/style.ipynb
|
CristianPachacama/cartoframes
|
3dc4e10d175069a7d7b734db3d9526127aad9dec
|
[
"BSD-3-Clause"
] | null | null | null | 40.541691 | 793 | 0.421975 |
[
[
[
"from cartoframes.auth import set_default_credentials\nfrom cartoframes.viz import Map, Layer, Source, Style\n\nset_default_credentials('cartovl')",
"_____no_output_____"
],
[
"# Style: simple string\nMap(\n Layer(\n Source('populated_places'),\n Style('color: blue')\n )\n)\n\n# Style: simple string + sugar\nMap(Layer('populated_places', 'color: blue'))",
"_____no_output_____"
],
[
"# Style: simple object\nMap(\n Layer(\n Source('populated_places'),\n Style({\n 'color': 'blue' \n })\n )\n)\n\n# Style: simple object + sugar\nMap(Layer('populated_places', { 'color': 'blue' }))",
"_____no_output_____"
],
[
"# Style: complex string\nMap(\n Layer(\n Source('populated_places'),\n Style('''\n @sum: sqrt($pop_max) / 100\n @grad: [red, blue, green]\n color: ramp(globalEqIntervals($pop_min, 3), @grad)\n filter: @sum > 20\n ''')\n )\n)\n\n# Style: complex string + sugar\nMap(\n Layer(\n 'populated_places',\n '''\n @sum: sqrt($pop_max) / 100\n @grad: [red, blue, green]\n color: ramp(globalEqIntervals($pop_min, 3), @grad)\n filter: @sum > 20\n '''\n )\n)",
"_____no_output_____"
],
[
"# Style: complex object\nMap(\n Layer(\n Source('populated_places'),\n Style({\n 'vars': {\n 'sum': 'sqrt($pop_max) / 100',\n 'grad': '[red, blue, green]'\n },\n 'color': 'ramp(globalEqIntervals($pop_min, 3), @grad)',\n 'filter': '@sum > 20'\n })\n )\n)\n\n# Style: complex object + sugar\nMap(\n Layer(\n 'populated_places',\n {\n 'vars': {\n 'sum': 'sqrt($pop_max) / 100',\n 'grad': '[red, blue, green]'\n },\n 'color': 'ramp(globalEqIntervals($pop_min, 3), @grad)',\n 'filter': '@sum > 20'\n }\n )\n)",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code"
]
] |
c5001578cadfd0e51948dc8f2bfc19dc2ade00b8
| 1,510 |
ipynb
|
Jupyter Notebook
|
Array2BitMap.ipynb
|
llewyn-jh/python-practice
|
1c3e16076cb611516fc38da936ce930e2ebe8f88
|
[
"Unlicense"
] | null | null | null |
Array2BitMap.ipynb
|
llewyn-jh/python-practice
|
1c3e16076cb611516fc38da936ce930e2ebe8f88
|
[
"Unlicense"
] | null | null | null |
Array2BitMap.ipynb
|
llewyn-jh/python-practice
|
1c3e16076cb611516fc38da936ce930e2ebe8f88
|
[
"Unlicense"
] | null | null | null | 24.754098 | 103 | 0.433775 |
[
[
[
"height = input('Enter an height: ')\nwidth = input('Enter an width: ')\n\narrays = []\nfor n in range(0, int(height)):\n arr = input(\"Enter integer array{} with space ' ': \".format(n + 1))\n arr = arr.split(' ')\n arr = list(map(int, arr))\n assert sum(arr) <= int(width), 'The sum of array can not be greater than {}'.format(width)\n arrays.append(arr)\n \nfor arr in arrays:\n arr2bit = []\n for i, n in enumerate(arr):\n if i == 0 and n == 0:\n pass\n else:\n if i % 2 == 0:\n arr2bit.append(n * '□')\n else:\n arr2bit.append(n * '■')\n \n m = ''\n for b in arr2bit:\n m += b\n print('\\n', m)\n ",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code"
]
] |
c50027f3ff878901cea099537073bed1b559a1d4
| 137,688 |
ipynb
|
Jupyter Notebook
|
notebooks/TAO_Integration_pgie.ipynb
|
MesaTech/deepstream_tao_integration
|
aa07e9c6c6007775c0f9f14fb43171278795a752
|
[
"CC0-1.0"
] | 4 |
2022-01-27T04:57:37.000Z
|
2022-02-17T21:21:37.000Z
|
notebooks/TAO_Integration_pgie.ipynb
|
MesaTech/deepstream_tao_integration
|
aa07e9c6c6007775c0f9f14fb43171278795a752
|
[
"CC0-1.0"
] | null | null | null |
notebooks/TAO_Integration_pgie.ipynb
|
MesaTech/deepstream_tao_integration
|
aa07e9c6c6007775c0f9f14fb43171278795a752
|
[
"CC0-1.0"
] | 3 |
2022-02-10T17:19:53.000Z
|
2022-02-17T21:21:49.000Z
| 65.784998 | 401 | 0.714666 |
[
[
[
"# Integrating TAO Models in DeepStream",
"_____no_output_____"
],
[
"In the first of two notebooks, we will be building a 4-class object detection pipeline as shown in the illustration below using Nvidia's TrafficCamNet pretrained model, directly downloaded from NGC. \n\nNote: This notebook has code inspired from a sample application provided by NVIDIA in a GitHub repository. You can find this repository [here](https://github.com/NVIDIA-AI-IOT/deepstream_python_apps).\n\n## The Pipeline\n\n\n",
"_____no_output_____"
],
[
"We notice there are multiple DeepStream plugins used in the pipeline , Let us have a look at them and try to understand them. \n\n## NVIDIA DeepStream Plugins\n\n### Nvinfer\n\nThe nvinfer plugin provides [TensorRT](https://docs.nvidia.com/deeplearning/tensorrt/developer-guide/index.html)-based inference for detection and tracking. The lowlevel library (libnvds_infer) operates either on float RGB or BGR planar data with dimensions of Network Height and Network Width. The plugin accepts NV12/RGBA data from upstream components like the decoder, muxer, and dewarper.\nThe Gst-nvinfer plugin also performs preprocessing operations like format conversion, scaling, mean subtraction, and produces final float RGB/BGR planar data which is passed to the low-level library. The low-level library uses the TensorRT engine for inferencing. It outputs each classified object’s class and each detected object’s bounding boxes (Bboxes) after clustering.\n\n\n\n### Nvvidconv \n\nWe create the nvvidconv plugin that performs color format conversions, which is required to make data ready for the nvosd plugin.\n\n\n\n\n### Nvosd\n\nThe nvosd plugin draws bounding boxes, text, and RoI (Regions of Interest) polygons (Polygons are presented as a set of lines). The plugin accepts an RGBA buffer with attached metadata from the upstream component. It\ndraws bounding boxes, which may be shaded depending on the configuration (e.g. width, color, and opacity) of a given bounding box. It also draws text and RoI polygons at specified locations in the frame. Text and polygon parameters are configurable through metadata.\n\n\n\n\nNow with this idea , let us get started into building the pipeline.",
"_____no_output_____"
],
[
"# Building the pipeline \n\n",
"_____no_output_____"
]
],
[
[
"# Import Required Libraries \nimport sys\nsys.path.append('../source_code')\nimport gi\nimport time\ngi.require_version('Gst', '1.0')\nfrom gi.repository import GObject, Gst, GLib\nfrom common.bus_call import bus_call\nimport pyds\n\n# Defining the Class Labels\nPGIE_CLASS_ID_VEHICLE = 0\nPGIE_CLASS_ID_BICYCLE = 1\nPGIE_CLASS_ID_PERSON = 2\nPGIE_CLASS_ID_ROADSIGN = 3\n\n# Defining the input output video file \nINPUT_VIDEO_NAME = '../videos/sample_720p.h264'\nOUTPUT_VIDEO_NAME = \"../videos/out.mp4\"",
"_____no_output_____"
]
],
[
[
"We define a function `make_elm_or_print_err()` to create our elements and report any errors if the creation fails.\n\nElements are created using the `Gst.ElementFactory.make()` function as part of Gstreamer library.",
"_____no_output_____"
]
],
[
[
"## Make Element or Print Error and any other detail\ndef make_elm_or_print_err(factoryname, name, printedname, detail=\"\"):\n print(\"Creating\", printedname)\n elm = Gst.ElementFactory.make(factoryname, name)\n if not elm:\n sys.stderr.write(\"Unable to create \" + printedname + \" \\n\")\n if detail:\n sys.stderr.write(detail)\n return elm",
"_____no_output_____"
]
],
[
[
"#### Initialise GStreamer and Create an Empty Pipeline",
"_____no_output_____"
]
],
[
[
"# Standard GStreamer initialization\nGst.init(None)\n\n\n# Create Gstreamer elements\n# Create Pipeline element that will form a connection of other elements\nprint(\"Creating Pipeline \\n \")\npipeline = Gst.Pipeline()\n\nif not pipeline:\n sys.stderr.write(\" Unable to create Pipeline \\n\")",
"Creating Pipeline \n \n"
]
],
[
[
"#### Create Elements that are required for our pipeline ",
"_____no_output_____"
]
],
[
[
"# Creating elements required for the pipeline\n# Source element for reading from file\nsource = make_elm_or_print_err(\"filesrc\", \"file-source\",\"Source\")\n# Parse the data since the input is an elementary .h264 stream\nh264parser = make_elm_or_print_err(\"h264parse\", \"h264-parser\",\"h264 parse\")\n# For hardware accelerated decoding of the stream\ndecoder = make_elm_or_print_err(\"nvv4l2decoder\", \"nvv4l2-decoder\",\"Nvv4l2 Decoder\")\n# Form batches from one or more sources\nstreammux = make_elm_or_print_err(\"nvstreammux\", \"Stream-muxer\",'NvStreamMux')\n# Run inference on the decoded stream, this property is set through a configuration file later\npgie = make_elm_or_print_err(\"nvinfer\", \"primary-inference\" ,\"pgie\")\n# Convert output stream to formatted buffer accepted by Nvosd\nnvvidconv = make_elm_or_print_err(\"nvvideoconvert\", \"convertor\",\"nvvidconv\")\n# Draw on the buffer\nnvosd = make_elm_or_print_err(\"nvdsosd\", \"onscreendisplay\",\"nvosd\")\n# Encode and save the OSD output\nqueue = make_elm_or_print_err(\"queue\", \"queue\", \"Queue\")\n# Convert output for saving\nnvvidconv2 = make_elm_or_print_err(\"nvvideoconvert\", \"convertor2\",\"nvvidconv2\")\n# Save as video file\nencoder = make_elm_or_print_err(\"avenc_mpeg4\", \"encoder\", \"Encoder\")\n# Parse output from encoder\ncodeparser = make_elm_or_print_err(\"mpeg4videoparse\", \"mpeg4-parser\", 'Code Parser')\n# Create a container\ncontainer = make_elm_or_print_err(\"qtmux\", \"qtmux\", \"Container\")\n# Create sink for string the output\nsink = make_elm_or_print_err(\"filesink\", \"filesink\", \"Sink\")",
"Creating Source\nCreating h264 parse\nCreating Nvv4l2 Decoder\nCreating NvStreamMux\nCreating pgie\nCreating nvvidconv\nCreating nvosd\nCreating Queue\nCreating nvvidconv2\nCreating Encoder\nCreating Code Parser\nCreating Container\nCreating Sink\n"
]
],
[
[
"Now that we have created the elements ,we can now set various properties for out pipeline at this point. \n\n### Understanding the configuration file \n\nWe set an `config-file-path` for our nvinfer ( Interference plugin ) and it points to the file `config_infer_primary_trafficcamnet.txt`\n\nYou can have a have a look at the [file](../configs/config_infer_primary_trafficcamnet.txt)\n\nHere are some parts of the configuration file : \n\n```\n# Copyright (c) 2020 NVIDIA Corporation. All rights reserved.\n#\n# NVIDIA Corporation and its licensors retain all intellectual property\n# and proprietary rights in and to this software, related documentation\n# and any modifications thereto. Any use, reproduction, disclosure or\n# distribution of this software and related documentation without an express\n# license agreement from NVIDIA Corporation is strictly prohibited.\n\n[property]\ngpu-id=0\nnet-scale-factor=0.0039215697906911373\ntlt-model-key=tlt_encode\ntlt-encoded-model=../models/trafficcamnet/resnet18_trafficcamnet_pruned.etlt\nlabelfile-path=labels_trafficnet.txt\nint8-calib-file=../models/trafficcamnet/trafficnet_int8.bin\nmodel-engine-file=../models/trafficcamnet/resnet18_trafficcamnet_pruned.etlt_b1_gpu0_int8.engine\ninput-dims=3;544;960;0\nuff-input-blob-name=input_1\nbatch-size=1\nprocess-mode=1\nmodel-color-format=0\n## 0=FP32, 1=INT8, 2=FP16 mode\nnetwork-mode=2\nnum-detected-classes=4\ninterval=0\ngie-unique-id=1\noutput-blob-names=output_bbox/BiasAdd;output_cov/Sigmoid\n\n[class-attrs-all]\npre-cluster-threshold=0.2\ngroup-threshold=1\n## Set eps=0.7 and minBoxes for cluster-mode=1(DBSCAN)\neps=0.2\n#minBoxes=3\n```\n\nHere we define all the parameters of our model. In this example we use model-file `resnet18_trafficcamnet_pruned`. `Nvinfer` creates an TensorRT Engine specific to the Host GPU to accelerate it's inference performance.",
"_____no_output_____"
]
],
[
[
"# Set properties for elements\nprint(\"Playing file %s\" %INPUT_VIDEO_NAME)\n# Set input file\nsource.set_property('location', INPUT_VIDEO_NAME)\n# Set input height, width, and batch size\nstreammux.set_property('width', 1920)\nstreammux.set_property('height', 1080)\nstreammux.set_property('batch-size', 1)\n# Set timer (in microseconds) to wait after the first buffer is available\n# to push the batch even if batch is never completely formed\nstreammux.set_property('batched-push-timeout', 4000000)\n# Set configuration files for Nvinfer\npgie.set_property('config-file-path', \"../configs/config_infer_primary_trafficcamnet.txt\")\n# Set encoder bitrate for output video\nencoder.set_property(\"bitrate\", 2000000)\n# Set output file location, disable sync and async\nsink.set_property(\"location\", OUTPUT_VIDEO_NAME)\nsink.set_property(\"sync\", 0)\nsink.set_property(\"async\", 0)",
"Playing file ../videos/sample_720p.h264\n"
]
],
[
[
"We now link all the elements in the order we prefer and create Gstreamer bus to feed all messages through it. ",
"_____no_output_____"
]
],
[
[
"# Add and link all elements to the pipeline\n# Adding elements\nprint(\"Adding elements to Pipeline \\n\")\n\npipeline.add(source)\npipeline.add(h264parser)\npipeline.add(decoder)\npipeline.add(streammux)\npipeline.add(pgie)\npipeline.add(nvvidconv)\npipeline.add(nvosd)\npipeline.add(queue)\npipeline.add(nvvidconv2)\npipeline.add(encoder)\npipeline.add(codeparser)\npipeline.add(container)\npipeline.add(sink)\n\n# Linking elements\n# Order: source -> h264parser -> decoder -> streammux -> pgie ->\n# -> vidconv -> osd -> queue -> vidconv2 -> encoder -> parser ->\n# -> container -> sink\n\nprint(\"Linking elements in the Pipeline \\n\")\nsource.link(h264parser)\nh264parser.link(decoder)\n\n\nsinkpad = streammux.get_request_pad(\"sink_0\")\nif not sinkpad:\n sys.stderr.write(\" Unable to get the sink pad of streammux \\n\")\n# Create source pad from Decoder \nsrcpad = decoder.get_static_pad(\"src\")\nif not srcpad:\n sys.stderr.write(\" Unable to get source pad of decoder \\n\")\n \nsrcpad.link(sinkpad)\nstreammux.link(pgie)\npgie.link(nvvidconv)\nnvvidconv.link(nvosd)\nnvosd.link(queue)\nqueue.link(nvvidconv2)\nnvvidconv2.link(encoder)\nencoder.link(codeparser)\ncodeparser.link(container)\ncontainer.link(sink)",
"Adding elements to Pipeline \n\nLinking elements in the Pipeline \n\n"
],
[
"# Create an event loop and feed GStreamer bus messages to it\nloop = GLib.MainLoop()\nbus = pipeline.get_bus()\nbus.add_signal_watch()\nbus.connect (\"message\", bus_call, loop)",
"_____no_output_____"
]
],
[
[
"## Working with the Metadata \n\nOur pipeline now carries the metadata forward but we have not done anything with it until now, but as mentoioned in the above pipeline diagram , we will now create a callback function to write relevant data on the frame once called and create a sink pad in the `nvosd` element to call the function.",
"_____no_output_____"
]
],
[
[
"# Working with metadata\ndef osd_sink_pad_buffer_probe(pad,info,u_data):\n \n obj_counter = {\n PGIE_CLASS_ID_VEHICLE:0,\n PGIE_CLASS_ID_PERSON:0,\n PGIE_CLASS_ID_BICYCLE:0,\n PGIE_CLASS_ID_ROADSIGN:0\n }\n # Reset frame number and number of rectanges to zero\n frame_number=0\n num_rects=0\n \n gst_buffer = info.get_buffer()\n if not gst_buffer:\n print(\"Unable to get GstBuffer \")\n return\n\n # Retrieve metadata from gst_buffer\n # Note: since we use the pyds shared object library,\n # the input is the C address of gst_buffer\n batch_meta = pyds.gst_buffer_get_nvds_batch_meta(hash(gst_buffer))\n l_frame = batch_meta.frame_meta_list\n while l_frame is not None:\n try:\n frame_meta = pyds.NvDsFrameMeta.cast(l_frame.data)\n except StopIteration:\n break\n \n # Get frame number, number of rectangles to draw and object metadata\n frame_number=frame_meta.frame_num\n num_rects = frame_meta.num_obj_meta\n l_obj=frame_meta.obj_meta_list\n \n while l_obj is not None:\n try:\n obj_meta=pyds.NvDsObjectMeta.cast(l_obj.data)\n except StopIteration:\n break\n # Increment object class by 1 and set box border color to red\n obj_counter[obj_meta.class_id] += 1\n obj_meta.rect_params.border_color.set(0.0, 0.0, 1.0, 0.0)\n try: \n l_obj=l_obj.next\n except StopIteration:\n break\n \n # Setting metadata display configuration\n # Acquire display meta object\n display_meta=pyds.nvds_acquire_display_meta_from_pool(batch_meta)\n display_meta.num_labels = 1\n py_nvosd_text_params = display_meta.text_params[0]\n # Set display text to be shown on screen\n py_nvosd_text_params.display_text = \"Frame Number={} Number of Objects={} Vehicle_count={} Person_count={}\".format(frame_number, num_rects, obj_counter[PGIE_CLASS_ID_VEHICLE], obj_counter[PGIE_CLASS_ID_PERSON])\n # Set where the string will appear\n py_nvosd_text_params.x_offset = 10\n py_nvosd_text_params.y_offset = 12\n # Font, font colour and font size\n py_nvosd_text_params.font_params.font_name = \"Serif\"\n py_nvosd_text_params.font_params.font_size = 10\n # Set color (We used white)\n py_nvosd_text_params.font_params.font_color.set(1.0, 1.0, 1.0, 1.0)\n # Set text background colour (We used black)\n py_nvosd_text_params.set_bg_clr = 1\n py_nvosd_text_params.text_bg_clr.set(0.0, 0.0, 0.0, 1.0)\n # Print the display text in the console as well\n print(pyds.get_string(py_nvosd_text_params.display_text))\n pyds.nvds_add_display_meta_to_frame(frame_meta, display_meta)\n \n \n try:\n l_frame=l_frame.next\n except StopIteration:\n break\n return Gst.PadProbeReturn.OK",
"_____no_output_____"
],
[
"# Adding probe to sinkpad of the OSD element\nosdsinkpad = nvosd.get_static_pad(\"sink\")\nif not osdsinkpad:\n sys.stderr.write(\" Unable to get sink pad of nvosd \\n\")\n \nosdsinkpad.add_probe(Gst.PadProbeType.BUFFER, osd_sink_pad_buffer_probe, 0)",
"_____no_output_____"
]
],
[
[
"Now with everything defined , we can start the playback and listen the events.",
"_____no_output_____"
]
],
[
[
"# Start the pipeline\nprint(\"Starting pipeline \\n\")\nstart_time = time.time()\npipeline.set_state(Gst.State.PLAYING)\ntry:\n loop.run()\nexcept:\n pass\n# Cleanup\npipeline.set_state(Gst.State.NULL)\nprint(\"--- %s seconds ---\" % (time.time() - start_time))",
"Starting pipeline \n\nFrame Number=0 Number of Objects=6 Vehicle_count=3 Person_count=3\nFrame Number=1 Number of Objects=7 Vehicle_count=2 Person_count=5\nFrame Number=2 Number of Objects=5 Vehicle_count=3 Person_count=2\nFrame Number=3 Number of Objects=6 Vehicle_count=4 Person_count=2\nFrame Number=4 Number of Objects=6 Vehicle_count=3 Person_count=3\nFrame Number=5 Number of Objects=7 Vehicle_count=5 Person_count=2\nFrame Number=6 Number of Objects=7 Vehicle_count=5 Person_count=2\nFrame Number=7 Number of Objects=6 Vehicle_count=4 Person_count=2\nFrame Number=8 Number of Objects=8 Vehicle_count=6 Person_count=2\nFrame Number=9 Number of Objects=7 Vehicle_count=5 Person_count=2\nFrame Number=10 Number of Objects=6 Vehicle_count=4 Person_count=2\nFrame Number=11 Number of Objects=8 Vehicle_count=6 Person_count=2\nFrame Number=12 Number of Objects=7 Vehicle_count=5 Person_count=2\nFrame Number=13 Number of Objects=3 Vehicle_count=1 Person_count=2\nFrame Number=14 Number of Objects=5 Vehicle_count=3 Person_count=2\nFrame Number=15 Number of Objects=5 Vehicle_count=3 Person_count=2\nFrame Number=16 Number of Objects=6 Vehicle_count=4 Person_count=2\nFrame Number=17 Number of Objects=6 Vehicle_count=4 Person_count=2\nFrame Number=18 Number of Objects=7 Vehicle_count=5 Person_count=2\nFrame Number=19 Number of Objects=8 Vehicle_count=6 Person_count=2\nFrame Number=20 Number of Objects=9 Vehicle_count=7 Person_count=2\nFrame Number=21 Number of Objects=7 Vehicle_count=5 Person_count=2\nFrame Number=22 Number of Objects=10 Vehicle_count=8 Person_count=2\nFrame Number=23 Number of Objects=8 Vehicle_count=6 Person_count=2\nFrame Number=24 Number of Objects=11 Vehicle_count=9 Person_count=2\nFrame Number=25 Number of Objects=9 Vehicle_count=6 Person_count=3\nFrame Number=26 Number of Objects=8 Vehicle_count=6 Person_count=2\nFrame Number=27 Number of Objects=8 Vehicle_count=6 Person_count=2\nFrame Number=28 Number of Objects=8 Vehicle_count=6 Person_count=2\nFrame Number=29 Number of Objects=9 Vehicle_count=7 Person_count=2\nFrame Number=30 Number of Objects=10 Vehicle_count=8 Person_count=2\nFrame Number=31 Number of Objects=11 Vehicle_count=8 Person_count=3\nFrame Number=32 Number of Objects=10 Vehicle_count=8 Person_count=2\nFrame Number=33 Number of Objects=11 Vehicle_count=9 Person_count=2\nFrame Number=34 Number of Objects=8 Vehicle_count=6 Person_count=2\nFrame Number=35 Number of Objects=11 Vehicle_count=9 Person_count=2\nFrame Number=36 Number of Objects=11 Vehicle_count=8 Person_count=3\nFrame Number=37 Number of Objects=10 Vehicle_count=7 Person_count=3\nFrame Number=38 Number of Objects=12 Vehicle_count=9 Person_count=3\nFrame Number=39 Number of Objects=14 Vehicle_count=10 Person_count=4\nFrame Number=40 Number of Objects=10 Vehicle_count=6 Person_count=4\nFrame Number=41 Number of Objects=9 Vehicle_count=6 Person_count=3\nFrame Number=42 Number of Objects=13 Vehicle_count=9 Person_count=4\nFrame Number=43 Number of Objects=12 Vehicle_count=9 Person_count=3\nFrame Number=44 Number of Objects=12 Vehicle_count=9 Person_count=3\nFrame Number=45 Number of Objects=9 Vehicle_count=6 Person_count=3\nFrame Number=46 Number of Objects=10 Vehicle_count=7 Person_count=3\nFrame Number=47 Number of Objects=11 Vehicle_count=8 Person_count=3\nFrame Number=48 Number of Objects=10 Vehicle_count=7 Person_count=3\nFrame Number=49 Number of Objects=9 Vehicle_count=6 Person_count=3\nFrame Number=50 Number of Objects=9 Vehicle_count=6 Person_count=3\nFrame Number=51 Number of Objects=8 Vehicle_count=5 Person_count=3\nFrame Number=52 Number of Objects=9 Vehicle_count=6 Person_count=3\nFrame Number=53 Number of Objects=8 Vehicle_count=5 Person_count=3\nFrame Number=54 Number of Objects=8 Vehicle_count=5 Person_count=3\nFrame Number=55 Number of Objects=9 Vehicle_count=6 Person_count=3\nFrame Number=56 Number of Objects=10 Vehicle_count=7 Person_count=3\nFrame Number=57 Number of Objects=9 Vehicle_count=5 Person_count=4\nFrame Number=58 Number of Objects=11 Vehicle_count=7 Person_count=4\nFrame Number=59 Number of Objects=11 Vehicle_count=7 Person_count=4\nFrame Number=60 Number of Objects=9 Vehicle_count=6 Person_count=3\nFrame Number=61 Number of Objects=8 Vehicle_count=5 Person_count=3\nFrame Number=62 Number of Objects=8 Vehicle_count=5 Person_count=3\nFrame Number=63 Number of Objects=7 Vehicle_count=4 Person_count=3\nFrame Number=64 Number of Objects=9 Vehicle_count=5 Person_count=4\nFrame Number=65 Number of Objects=12 Vehicle_count=8 Person_count=4\nFrame Number=66 Number of Objects=12 Vehicle_count=8 Person_count=4\nFrame Number=67 Number of Objects=13 Vehicle_count=9 Person_count=4\nFrame Number=68 Number of Objects=10 Vehicle_count=7 Person_count=3\nFrame Number=69 Number of Objects=11 Vehicle_count=7 Person_count=4\nFrame Number=70 Number of Objects=12 Vehicle_count=8 Person_count=4\nFrame Number=71 Number of Objects=14 Vehicle_count=10 Person_count=4\nFrame Number=72 Number of Objects=14 Vehicle_count=10 Person_count=4\nFrame Number=73 Number of Objects=13 Vehicle_count=9 Person_count=4\nFrame Number=74 Number of Objects=14 Vehicle_count=10 Person_count=4\nFrame Number=75 Number of Objects=14 Vehicle_count=10 Person_count=4\nFrame Number=76 Number of Objects=14 Vehicle_count=10 Person_count=4\nFrame Number=77 Number of Objects=16 Vehicle_count=12 Person_count=4\nFrame Number=78 Number of Objects=16 Vehicle_count=12 Person_count=4\nFrame Number=79 Number of Objects=14 Vehicle_count=10 Person_count=4\nFrame Number=80 Number of Objects=12 Vehicle_count=8 Person_count=4\nFrame Number=81 Number of Objects=14 Vehicle_count=10 Person_count=4\nFrame Number=82 Number of Objects=14 Vehicle_count=10 Person_count=4\nFrame Number=83 Number of Objects=13 Vehicle_count=9 Person_count=4\nFrame Number=84 Number of Objects=14 Vehicle_count=10 Person_count=4\nFrame Number=85 Number of Objects=13 Vehicle_count=9 Person_count=4\nFrame Number=86 Number of Objects=14 Vehicle_count=10 Person_count=4\nFrame Number=87 Number of Objects=13 Vehicle_count=8 Person_count=5\nFrame Number=88 Number of Objects=13 Vehicle_count=8 Person_count=5\nFrame Number=89 Number of Objects=12 Vehicle_count=7 Person_count=5\nFrame Number=90 Number of Objects=11 Vehicle_count=7 Person_count=4\nFrame Number=91 Number of Objects=13 Vehicle_count=9 Person_count=4\nFrame Number=92 Number of Objects=12 Vehicle_count=8 Person_count=4\nFrame Number=93 Number of Objects=15 Vehicle_count=11 Person_count=4\nFrame Number=94 Number of Objects=15 Vehicle_count=11 Person_count=4\nFrame Number=95 Number of Objects=13 Vehicle_count=9 Person_count=4\nFrame Number=96 Number of Objects=13 Vehicle_count=9 Person_count=4\nFrame Number=97 Number of Objects=12 Vehicle_count=8 Person_count=4\nFrame Number=98 Number of Objects=12 Vehicle_count=8 Person_count=4\nFrame Number=99 Number of Objects=13 Vehicle_count=9 Person_count=4\nFrame Number=100 Number of Objects=13 Vehicle_count=8 Person_count=5\nFrame Number=101 Number of Objects=12 Vehicle_count=7 Person_count=5\nFrame Number=102 Number of Objects=13 Vehicle_count=7 Person_count=6\nFrame Number=103 Number of Objects=12 Vehicle_count=6 Person_count=6\nFrame Number=104 Number of Objects=13 Vehicle_count=7 Person_count=6\nFrame Number=105 Number of Objects=10 Vehicle_count=6 Person_count=4\nFrame Number=106 Number of Objects=14 Vehicle_count=8 Person_count=6\nFrame Number=107 Number of Objects=10 Vehicle_count=5 Person_count=5\nFrame Number=108 Number of Objects=11 Vehicle_count=6 Person_count=5\nFrame Number=109 Number of Objects=11 Vehicle_count=5 Person_count=6\nFrame Number=110 Number of Objects=8 Vehicle_count=4 Person_count=4\nFrame Number=111 Number of Objects=8 Vehicle_count=4 Person_count=4\nFrame Number=112 Number of Objects=7 Vehicle_count=3 Person_count=4\nFrame Number=113 Number of Objects=7 Vehicle_count=3 Person_count=4\nFrame Number=114 Number of Objects=8 Vehicle_count=4 Person_count=4\nFrame Number=115 Number of Objects=7 Vehicle_count=3 Person_count=4\nFrame Number=116 Number of Objects=7 Vehicle_count=3 Person_count=4\nFrame Number=117 Number of Objects=7 Vehicle_count=3 Person_count=4\nFrame Number=118 Number of Objects=7 Vehicle_count=3 Person_count=4\nFrame Number=119 Number of Objects=7 Vehicle_count=3 Person_count=4\nFrame Number=120 Number of Objects=8 Vehicle_count=3 Person_count=5\nFrame Number=121 Number of Objects=8 Vehicle_count=3 Person_count=5\nFrame Number=122 Number of Objects=8 Vehicle_count=3 Person_count=5\nFrame Number=123 Number of Objects=8 Vehicle_count=3 Person_count=5\nFrame Number=124 Number of Objects=8 Vehicle_count=3 Person_count=5\nFrame Number=125 Number of Objects=9 Vehicle_count=3 Person_count=6\nFrame Number=126 Number of Objects=9 Vehicle_count=3 Person_count=6\nFrame Number=127 Number of Objects=8 Vehicle_count=3 Person_count=5\nFrame Number=128 Number of Objects=8 Vehicle_count=3 Person_count=5\nFrame Number=129 Number of Objects=9 Vehicle_count=4 Person_count=5\nFrame Number=130 Number of Objects=8 Vehicle_count=3 Person_count=5\nFrame Number=131 Number of Objects=9 Vehicle_count=4 Person_count=5\nFrame Number=132 Number of Objects=8 Vehicle_count=3 Person_count=5\nFrame Number=133 Number of Objects=7 Vehicle_count=2 Person_count=5\nFrame Number=134 Number of Objects=7 Vehicle_count=2 Person_count=5\nFrame Number=135 Number of Objects=8 Vehicle_count=3 Person_count=5\nFrame Number=136 Number of Objects=8 Vehicle_count=3 Person_count=5\nFrame Number=137 Number of Objects=8 Vehicle_count=3 Person_count=5\nFrame Number=138 Number of Objects=9 Vehicle_count=4 Person_count=5\nFrame Number=139 Number of Objects=9 Vehicle_count=4 Person_count=5\nFrame Number=140 Number of Objects=10 Vehicle_count=4 Person_count=6\nFrame Number=141 Number of Objects=12 Vehicle_count=7 Person_count=5\nFrame Number=142 Number of Objects=11 Vehicle_count=6 Person_count=5\nFrame Number=143 Number of Objects=10 Vehicle_count=5 Person_count=5\nFrame Number=144 Number of Objects=10 Vehicle_count=5 Person_count=5\nFrame Number=145 Number of Objects=12 Vehicle_count=7 Person_count=5\nFrame Number=146 Number of Objects=13 Vehicle_count=8 Person_count=5\nFrame Number=147 Number of Objects=13 Vehicle_count=8 Person_count=5\nFrame Number=148 Number of Objects=12 Vehicle_count=7 Person_count=5\nFrame Number=149 Number of Objects=12 Vehicle_count=7 Person_count=5\nFrame Number=150 Number of Objects=13 Vehicle_count=7 Person_count=6\nFrame Number=151 Number of Objects=13 Vehicle_count=7 Person_count=6\nFrame Number=152 Number of Objects=11 Vehicle_count=6 Person_count=5\nFrame Number=153 Number of Objects=10 Vehicle_count=6 Person_count=4\nFrame Number=154 Number of Objects=12 Vehicle_count=7 Person_count=5\nFrame Number=155 Number of Objects=11 Vehicle_count=6 Person_count=5\nFrame Number=156 Number of Objects=11 Vehicle_count=5 Person_count=6\nFrame Number=157 Number of Objects=11 Vehicle_count=5 Person_count=6\nFrame Number=158 Number of Objects=9 Vehicle_count=5 Person_count=4\nFrame Number=159 Number of Objects=9 Vehicle_count=5 Person_count=4\nFrame Number=160 Number of Objects=9 Vehicle_count=4 Person_count=4\nFrame Number=161 Number of Objects=10 Vehicle_count=5 Person_count=5\nFrame Number=162 Number of Objects=9 Vehicle_count=3 Person_count=5\nFrame Number=163 Number of Objects=7 Vehicle_count=3 Person_count=4\nFrame Number=164 Number of Objects=9 Vehicle_count=3 Person_count=5\nFrame Number=165 Number of Objects=8 Vehicle_count=2 Person_count=5\nFrame Number=166 Number of Objects=9 Vehicle_count=2 Person_count=6\nFrame Number=167 Number of Objects=8 Vehicle_count=2 Person_count=5\nFrame Number=168 Number of Objects=8 Vehicle_count=2 Person_count=5\nFrame Number=169 Number of Objects=8 Vehicle_count=3 Person_count=4\nFrame Number=170 Number of Objects=9 Vehicle_count=2 Person_count=6\nFrame Number=171 Number of Objects=8 Vehicle_count=2 Person_count=5\nFrame Number=172 Number of Objects=7 Vehicle_count=2 Person_count=5\nFrame Number=173 Number of Objects=8 Vehicle_count=2 Person_count=5\nFrame Number=174 Number of Objects=8 Vehicle_count=2 Person_count=5\nFrame Number=175 Number of Objects=7 Vehicle_count=2 Person_count=4\nFrame Number=176 Number of Objects=9 Vehicle_count=3 Person_count=5\nFrame Number=177 Number of Objects=12 Vehicle_count=6 Person_count=5\nFrame Number=178 Number of Objects=8 Vehicle_count=4 Person_count=3\nFrame Number=179 Number of Objects=9 Vehicle_count=4 Person_count=4\nFrame Number=180 Number of Objects=8 Vehicle_count=4 Person_count=3\nFrame Number=181 Number of Objects=9 Vehicle_count=4 Person_count=4\nFrame Number=182 Number of Objects=7 Vehicle_count=3 Person_count=3\nFrame Number=183 Number of Objects=6 Vehicle_count=2 Person_count=3\nFrame Number=184 Number of Objects=7 Vehicle_count=2 Person_count=3\nFrame Number=185 Number of Objects=7 Vehicle_count=2 Person_count=4\nFrame Number=186 Number of Objects=7 Vehicle_count=3 Person_count=3\nFrame Number=187 Number of Objects=8 Vehicle_count=4 Person_count=3\nFrame Number=188 Number of Objects=10 Vehicle_count=6 Person_count=2\nFrame Number=189 Number of Objects=6 Vehicle_count=3 Person_count=3\nFrame Number=190 Number of Objects=8 Vehicle_count=5 Person_count=3\nFrame Number=191 Number of Objects=9 Vehicle_count=6 Person_count=2\nFrame Number=192 Number of Objects=12 Vehicle_count=7 Person_count=3\nFrame Number=193 Number of Objects=11 Vehicle_count=7 Person_count=3\nFrame Number=194 Number of Objects=8 Vehicle_count=5 Person_count=3\nFrame Number=195 Number of Objects=9 Vehicle_count=5 Person_count=4\nFrame Number=196 Number of Objects=10 Vehicle_count=5 Person_count=4\nFrame Number=197 Number of Objects=8 Vehicle_count=4 Person_count=3\nFrame Number=198 Number of Objects=9 Vehicle_count=5 Person_count=3\nFrame Number=199 Number of Objects=13 Vehicle_count=9 Person_count=4\nFrame Number=200 Number of Objects=10 Vehicle_count=7 Person_count=3\nFrame Number=201 Number of Objects=8 Vehicle_count=5 Person_count=3\nFrame Number=202 Number of Objects=11 Vehicle_count=7 Person_count=3\nFrame Number=203 Number of Objects=11 Vehicle_count=7 Person_count=4\nFrame Number=204 Number of Objects=9 Vehicle_count=6 Person_count=3\nFrame Number=205 Number of Objects=12 Vehicle_count=8 Person_count=4\nFrame Number=206 Number of Objects=14 Vehicle_count=8 Person_count=5\nFrame Number=207 Number of Objects=12 Vehicle_count=8 Person_count=4\nFrame Number=208 Number of Objects=11 Vehicle_count=7 Person_count=3\nFrame Number=209 Number of Objects=9 Vehicle_count=6 Person_count=3\nFrame Number=210 Number of Objects=10 Vehicle_count=7 Person_count=3\nFrame Number=211 Number of Objects=13 Vehicle_count=10 Person_count=3\nFrame Number=212 Number of Objects=13 Vehicle_count=10 Person_count=3\nFrame Number=213 Number of Objects=12 Vehicle_count=9 Person_count=3\nFrame Number=214 Number of Objects=13 Vehicle_count=9 Person_count=4\nFrame Number=215 Number of Objects=15 Vehicle_count=11 Person_count=4\nFrame Number=216 Number of Objects=13 Vehicle_count=9 Person_count=4\nFrame Number=217 Number of Objects=12 Vehicle_count=9 Person_count=3\nFrame Number=218 Number of Objects=12 Vehicle_count=9 Person_count=3\nFrame Number=219 Number of Objects=13 Vehicle_count=10 Person_count=3\nFrame Number=220 Number of Objects=10 Vehicle_count=7 Person_count=3\nFrame Number=221 Number of Objects=10 Vehicle_count=7 Person_count=3\nFrame Number=222 Number of Objects=9 Vehicle_count=6 Person_count=3\nFrame Number=223 Number of Objects=7 Vehicle_count=4 Person_count=3\nFrame Number=224 Number of Objects=10 Vehicle_count=7 Person_count=3\nFrame Number=225 Number of Objects=9 Vehicle_count=6 Person_count=3\nFrame Number=226 Number of Objects=8 Vehicle_count=6 Person_count=2\nFrame Number=227 Number of Objects=7 Vehicle_count=5 Person_count=2\nFrame Number=228 Number of Objects=10 Vehicle_count=7 Person_count=3\nFrame Number=229 Number of Objects=9 Vehicle_count=7 Person_count=2\nFrame Number=230 Number of Objects=11 Vehicle_count=8 Person_count=3\nFrame Number=231 Number of Objects=11 Vehicle_count=9 Person_count=2\nFrame Number=232 Number of Objects=10 Vehicle_count=7 Person_count=3\nFrame Number=233 Number of Objects=9 Vehicle_count=7 Person_count=2\nFrame Number=234 Number of Objects=9 Vehicle_count=7 Person_count=2\nFrame Number=235 Number of Objects=10 Vehicle_count=7 Person_count=3\nFrame Number=236 Number of Objects=9 Vehicle_count=7 Person_count=2\nFrame Number=237 Number of Objects=9 Vehicle_count=6 Person_count=3\nFrame Number=238 Number of Objects=11 Vehicle_count=7 Person_count=4\nFrame Number=239 Number of Objects=11 Vehicle_count=8 Person_count=3\nFrame Number=240 Number of Objects=10 Vehicle_count=7 Person_count=3\nFrame Number=241 Number of Objects=9 Vehicle_count=6 Person_count=3\nFrame Number=242 Number of Objects=10 Vehicle_count=7 Person_count=3\nFrame Number=243 Number of Objects=8 Vehicle_count=6 Person_count=2\nFrame Number=244 Number of Objects=9 Vehicle_count=6 Person_count=3\nFrame Number=245 Number of Objects=9 Vehicle_count=6 Person_count=3\nFrame Number=246 Number of Objects=8 Vehicle_count=5 Person_count=3\nFrame Number=247 Number of Objects=9 Vehicle_count=6 Person_count=3\nFrame Number=248 Number of Objects=9 Vehicle_count=6 Person_count=3\nFrame Number=249 Number of Objects=9 Vehicle_count=6 Person_count=3\nFrame Number=250 Number of Objects=10 Vehicle_count=7 Person_count=3\nFrame Number=251 Number of Objects=10 Vehicle_count=7 Person_count=3\nFrame Number=252 Number of Objects=8 Vehicle_count=5 Person_count=3\nFrame Number=253 Number of Objects=9 Vehicle_count=6 Person_count=3\nFrame Number=254 Number of Objects=9 Vehicle_count=6 Person_count=3\nFrame Number=255 Number of Objects=8 Vehicle_count=5 Person_count=3\nFrame Number=256 Number of Objects=7 Vehicle_count=4 Person_count=3\nFrame Number=257 Number of Objects=6 Vehicle_count=3 Person_count=3\nFrame Number=258 Number of Objects=6 Vehicle_count=3 Person_count=3\nFrame Number=259 Number of Objects=8 Vehicle_count=5 Person_count=3\nFrame Number=260 Number of Objects=7 Vehicle_count=4 Person_count=3\nFrame Number=261 Number of Objects=8 Vehicle_count=5 Person_count=3\nFrame Number=262 Number of Objects=8 Vehicle_count=5 Person_count=3\nFrame Number=263 Number of Objects=8 Vehicle_count=4 Person_count=4\nFrame Number=264 Number of Objects=8 Vehicle_count=5 Person_count=3\nFrame Number=265 Number of Objects=10 Vehicle_count=5 Person_count=5\nFrame Number=266 Number of Objects=10 Vehicle_count=7 Person_count=3\nFrame Number=267 Number of Objects=11 Vehicle_count=7 Person_count=4\nFrame Number=268 Number of Objects=12 Vehicle_count=8 Person_count=4\nFrame Number=269 Number of Objects=12 Vehicle_count=9 Person_count=3\nFrame Number=270 Number of Objects=12 Vehicle_count=8 Person_count=4\nFrame Number=271 Number of Objects=10 Vehicle_count=7 Person_count=3\nFrame Number=272 Number of Objects=10 Vehicle_count=7 Person_count=3\nFrame Number=273 Number of Objects=11 Vehicle_count=8 Person_count=3\nFrame Number=274 Number of Objects=11 Vehicle_count=8 Person_count=3\nFrame Number=275 Number of Objects=10 Vehicle_count=6 Person_count=4\nFrame Number=276 Number of Objects=12 Vehicle_count=8 Person_count=4\nFrame Number=277 Number of Objects=10 Vehicle_count=6 Person_count=4\nFrame Number=278 Number of Objects=11 Vehicle_count=7 Person_count=4\nFrame Number=279 Number of Objects=13 Vehicle_count=9 Person_count=4\nFrame Number=280 Number of Objects=12 Vehicle_count=8 Person_count=4\nFrame Number=281 Number of Objects=12 Vehicle_count=8 Person_count=4\nFrame Number=282 Number of Objects=14 Vehicle_count=8 Person_count=6\nFrame Number=283 Number of Objects=12 Vehicle_count=7 Person_count=5\nFrame Number=284 Number of Objects=11 Vehicle_count=6 Person_count=5\nFrame Number=285 Number of Objects=11 Vehicle_count=5 Person_count=6\nFrame Number=286 Number of Objects=9 Vehicle_count=5 Person_count=4\nFrame Number=287 Number of Objects=11 Vehicle_count=6 Person_count=5\nFrame Number=288 Number of Objects=11 Vehicle_count=6 Person_count=5\nFrame Number=289 Number of Objects=10 Vehicle_count=5 Person_count=5\nFrame Number=290 Number of Objects=9 Vehicle_count=5 Person_count=4\nFrame Number=291 Number of Objects=9 Vehicle_count=5 Person_count=4\nFrame Number=292 Number of Objects=11 Vehicle_count=5 Person_count=6\nFrame Number=293 Number of Objects=8 Vehicle_count=4 Person_count=4\nFrame Number=294 Number of Objects=9 Vehicle_count=4 Person_count=5\nFrame Number=295 Number of Objects=9 Vehicle_count=4 Person_count=5\nFrame Number=296 Number of Objects=9 Vehicle_count=4 Person_count=5\nFrame Number=297 Number of Objects=11 Vehicle_count=5 Person_count=6\nFrame Number=298 Number of Objects=9 Vehicle_count=4 Person_count=5\nFrame Number=299 Number of Objects=10 Vehicle_count=4 Person_count=6\nFrame Number=300 Number of Objects=10 Vehicle_count=4 Person_count=6\nFrame Number=301 Number of Objects=10 Vehicle_count=5 Person_count=5\nFrame Number=302 Number of Objects=9 Vehicle_count=4 Person_count=5\nFrame Number=303 Number of Objects=10 Vehicle_count=4 Person_count=6\nFrame Number=304 Number of Objects=11 Vehicle_count=6 Person_count=5\nFrame Number=305 Number of Objects=16 Vehicle_count=10 Person_count=6\nFrame Number=306 Number of Objects=14 Vehicle_count=9 Person_count=5\nFrame Number=307 Number of Objects=15 Vehicle_count=10 Person_count=5\nFrame Number=308 Number of Objects=16 Vehicle_count=10 Person_count=6\nFrame Number=309 Number of Objects=14 Vehicle_count=10 Person_count=4\nFrame Number=310 Number of Objects=15 Vehicle_count=10 Person_count=5\nFrame Number=311 Number of Objects=13 Vehicle_count=9 Person_count=4\nFrame Number=312 Number of Objects=14 Vehicle_count=10 Person_count=4\nFrame Number=313 Number of Objects=14 Vehicle_count=10 Person_count=4\nFrame Number=314 Number of Objects=14 Vehicle_count=10 Person_count=4\nFrame Number=315 Number of Objects=11 Vehicle_count=7 Person_count=4\nFrame Number=316 Number of Objects=12 Vehicle_count=8 Person_count=4\nFrame Number=317 Number of Objects=12 Vehicle_count=8 Person_count=4\nFrame Number=318 Number of Objects=10 Vehicle_count=6 Person_count=4\nFrame Number=319 Number of Objects=9 Vehicle_count=5 Person_count=4\nFrame Number=320 Number of Objects=9 Vehicle_count=5 Person_count=4\nFrame Number=321 Number of Objects=11 Vehicle_count=7 Person_count=4\nFrame Number=322 Number of Objects=10 Vehicle_count=6 Person_count=4\nFrame Number=323 Number of Objects=11 Vehicle_count=7 Person_count=4\nFrame Number=324 Number of Objects=12 Vehicle_count=8 Person_count=4\nFrame Number=325 Number of Objects=12 Vehicle_count=8 Person_count=4\nFrame Number=326 Number of Objects=12 Vehicle_count=8 Person_count=4\nFrame Number=327 Number of Objects=12 Vehicle_count=8 Person_count=4\nFrame Number=328 Number of Objects=13 Vehicle_count=9 Person_count=4\nFrame Number=329 Number of Objects=14 Vehicle_count=10 Person_count=4\nFrame Number=330 Number of Objects=12 Vehicle_count=8 Person_count=4\nFrame Number=331 Number of Objects=16 Vehicle_count=12 Person_count=4\nFrame Number=332 Number of Objects=16 Vehicle_count=12 Person_count=4\nFrame Number=333 Number of Objects=15 Vehicle_count=11 Person_count=4\nFrame Number=334 Number of Objects=18 Vehicle_count=13 Person_count=5\nFrame Number=335 Number of Objects=18 Vehicle_count=13 Person_count=5\nFrame Number=336 Number of Objects=17 Vehicle_count=12 Person_count=5\nFrame Number=337 Number of Objects=16 Vehicle_count=12 Person_count=4\nFrame Number=338 Number of Objects=16 Vehicle_count=12 Person_count=4\nFrame Number=339 Number of Objects=15 Vehicle_count=11 Person_count=4\nFrame Number=340 Number of Objects=17 Vehicle_count=12 Person_count=5\nFrame Number=341 Number of Objects=17 Vehicle_count=12 Person_count=5\nFrame Number=342 Number of Objects=16 Vehicle_count=11 Person_count=5\nFrame Number=343 Number of Objects=17 Vehicle_count=12 Person_count=5\nFrame Number=344 Number of Objects=18 Vehicle_count=13 Person_count=5\nFrame Number=345 Number of Objects=16 Vehicle_count=11 Person_count=5\nFrame Number=346 Number of Objects=15 Vehicle_count=10 Person_count=5\nFrame Number=347 Number of Objects=14 Vehicle_count=9 Person_count=5\nFrame Number=348 Number of Objects=13 Vehicle_count=8 Person_count=5\nFrame Number=349 Number of Objects=13 Vehicle_count=8 Person_count=5\nFrame Number=350 Number of Objects=15 Vehicle_count=10 Person_count=5\nFrame Number=351 Number of Objects=12 Vehicle_count=7 Person_count=5\nFrame Number=352 Number of Objects=13 Vehicle_count=8 Person_count=5\nFrame Number=353 Number of Objects=14 Vehicle_count=9 Person_count=5\nFrame Number=354 Number of Objects=13 Vehicle_count=8 Person_count=5\nFrame Number=355 Number of Objects=15 Vehicle_count=10 Person_count=5\nFrame Number=356 Number of Objects=14 Vehicle_count=9 Person_count=5\nFrame Number=357 Number of Objects=13 Vehicle_count=8 Person_count=5\nFrame Number=358 Number of Objects=13 Vehicle_count=8 Person_count=5\nFrame Number=359 Number of Objects=11 Vehicle_count=6 Person_count=5\nFrame Number=360 Number of Objects=13 Vehicle_count=8 Person_count=5\nFrame Number=361 Number of Objects=12 Vehicle_count=7 Person_count=5\nFrame Number=362 Number of Objects=11 Vehicle_count=6 Person_count=5\nFrame Number=363 Number of Objects=11 Vehicle_count=6 Person_count=5\nFrame Number=364 Number of Objects=10 Vehicle_count=5 Person_count=5\nFrame Number=365 Number of Objects=9 Vehicle_count=5 Person_count=4\nFrame Number=366 Number of Objects=8 Vehicle_count=5 Person_count=3\nFrame Number=367 Number of Objects=10 Vehicle_count=5 Person_count=5\nFrame Number=368 Number of Objects=8 Vehicle_count=4 Person_count=4\nFrame Number=369 Number of Objects=8 Vehicle_count=4 Person_count=4\nFrame Number=370 Number of Objects=8 Vehicle_count=4 Person_count=4\nFrame Number=371 Number of Objects=8 Vehicle_count=4 Person_count=4\nFrame Number=372 Number of Objects=8 Vehicle_count=3 Person_count=5\nFrame Number=373 Number of Objects=7 Vehicle_count=3 Person_count=4\nFrame Number=374 Number of Objects=9 Vehicle_count=5 Person_count=4\nFrame Number=375 Number of Objects=8 Vehicle_count=3 Person_count=5\nFrame Number=376 Number of Objects=7 Vehicle_count=3 Person_count=4\nFrame Number=377 Number of Objects=7 Vehicle_count=3 Person_count=4\nFrame Number=378 Number of Objects=7 Vehicle_count=3 Person_count=4\nFrame Number=379 Number of Objects=9 Vehicle_count=5 Person_count=4\nFrame Number=380 Number of Objects=10 Vehicle_count=6 Person_count=4\nFrame Number=381 Number of Objects=12 Vehicle_count=7 Person_count=5\nFrame Number=382 Number of Objects=14 Vehicle_count=9 Person_count=5\nFrame Number=383 Number of Objects=9 Vehicle_count=4 Person_count=5\nFrame Number=384 Number of Objects=12 Vehicle_count=7 Person_count=5\nFrame Number=385 Number of Objects=12 Vehicle_count=7 Person_count=5\nFrame Number=386 Number of Objects=12 Vehicle_count=7 Person_count=5\nFrame Number=387 Number of Objects=13 Vehicle_count=8 Person_count=5\nFrame Number=388 Number of Objects=13 Vehicle_count=9 Person_count=4\nFrame Number=389 Number of Objects=11 Vehicle_count=7 Person_count=4\nFrame Number=390 Number of Objects=11 Vehicle_count=7 Person_count=4\nFrame Number=391 Number of Objects=13 Vehicle_count=9 Person_count=4\nFrame Number=392 Number of Objects=13 Vehicle_count=9 Person_count=4\nFrame Number=393 Number of Objects=9 Vehicle_count=5 Person_count=4\nFrame Number=394 Number of Objects=9 Vehicle_count=5 Person_count=4\nFrame Number=395 Number of Objects=9 Vehicle_count=5 Person_count=4\nFrame Number=396 Number of Objects=10 Vehicle_count=6 Person_count=4\nFrame Number=397 Number of Objects=12 Vehicle_count=8 Person_count=4\nFrame Number=398 Number of Objects=13 Vehicle_count=8 Person_count=5\nFrame Number=399 Number of Objects=12 Vehicle_count=7 Person_count=5\nFrame Number=400 Number of Objects=10 Vehicle_count=6 Person_count=4\nFrame Number=401 Number of Objects=10 Vehicle_count=6 Person_count=4\nFrame Number=402 Number of Objects=8 Vehicle_count=4 Person_count=4\nFrame Number=403 Number of Objects=6 Vehicle_count=3 Person_count=3\nFrame Number=404 Number of Objects=8 Vehicle_count=3 Person_count=5\nFrame Number=405 Number of Objects=8 Vehicle_count=3 Person_count=5\nFrame Number=406 Number of Objects=8 Vehicle_count=3 Person_count=5\nFrame Number=407 Number of Objects=8 Vehicle_count=3 Person_count=5\nFrame Number=408 Number of Objects=10 Vehicle_count=5 Person_count=5\nFrame Number=409 Number of Objects=8 Vehicle_count=3 Person_count=5\nFrame Number=410 Number of Objects=9 Vehicle_count=4 Person_count=5\nFrame Number=411 Number of Objects=10 Vehicle_count=5 Person_count=5\nFrame Number=412 Number of Objects=10 Vehicle_count=5 Person_count=5\nFrame Number=413 Number of Objects=10 Vehicle_count=5 Person_count=5\nFrame Number=414 Number of Objects=10 Vehicle_count=5 Person_count=5\nFrame Number=415 Number of Objects=10 Vehicle_count=5 Person_count=5\nFrame Number=416 Number of Objects=11 Vehicle_count=6 Person_count=5\nFrame Number=417 Number of Objects=9 Vehicle_count=5 Person_count=4\nFrame Number=418 Number of Objects=8 Vehicle_count=4 Person_count=4\nFrame Number=419 Number of Objects=9 Vehicle_count=5 Person_count=4\nFrame Number=420 Number of Objects=10 Vehicle_count=6 Person_count=4\nFrame Number=421 Number of Objects=10 Vehicle_count=6 Person_count=4\nFrame Number=422 Number of Objects=12 Vehicle_count=8 Person_count=4\nFrame Number=423 Number of Objects=11 Vehicle_count=7 Person_count=4\nFrame Number=424 Number of Objects=13 Vehicle_count=9 Person_count=4\nFrame Number=425 Number of Objects=10 Vehicle_count=6 Person_count=4\nFrame Number=426 Number of Objects=15 Vehicle_count=11 Person_count=4\nFrame Number=427 Number of Objects=14 Vehicle_count=10 Person_count=4\nFrame Number=428 Number of Objects=12 Vehicle_count=8 Person_count=4\nFrame Number=429 Number of Objects=14 Vehicle_count=10 Person_count=4\nFrame Number=430 Number of Objects=12 Vehicle_count=8 Person_count=4\nFrame Number=431 Number of Objects=13 Vehicle_count=9 Person_count=4\nFrame Number=432 Number of Objects=15 Vehicle_count=10 Person_count=5\nFrame Number=433 Number of Objects=15 Vehicle_count=10 Person_count=5\nFrame Number=434 Number of Objects=11 Vehicle_count=6 Person_count=5\nFrame Number=435 Number of Objects=12 Vehicle_count=8 Person_count=4\nFrame Number=436 Number of Objects=12 Vehicle_count=7 Person_count=5\nFrame Number=437 Number of Objects=14 Vehicle_count=9 Person_count=5\nFrame Number=438 Number of Objects=13 Vehicle_count=9 Person_count=4\nFrame Number=439 Number of Objects=15 Vehicle_count=10 Person_count=5\nFrame Number=440 Number of Objects=13 Vehicle_count=9 Person_count=4\nFrame Number=441 Number of Objects=14 Vehicle_count=9 Person_count=5\nFrame Number=442 Number of Objects=12 Vehicle_count=8 Person_count=4\nFrame Number=443 Number of Objects=13 Vehicle_count=8 Person_count=5\nFrame Number=444 Number of Objects=14 Vehicle_count=9 Person_count=5\nFrame Number=445 Number of Objects=14 Vehicle_count=9 Person_count=5\nFrame Number=446 Number of Objects=13 Vehicle_count=9 Person_count=4\nFrame Number=447 Number of Objects=16 Vehicle_count=11 Person_count=5\nFrame Number=448 Number of Objects=15 Vehicle_count=10 Person_count=5\nFrame Number=449 Number of Objects=14 Vehicle_count=9 Person_count=5\nFrame Number=450 Number of Objects=15 Vehicle_count=10 Person_count=5\nFrame Number=451 Number of Objects=14 Vehicle_count=10 Person_count=4\nFrame Number=452 Number of Objects=15 Vehicle_count=10 Person_count=5\nFrame Number=453 Number of Objects=16 Vehicle_count=12 Person_count=4\nFrame Number=454 Number of Objects=16 Vehicle_count=11 Person_count=5\nFrame Number=455 Number of Objects=18 Vehicle_count=13 Person_count=5\nFrame Number=456 Number of Objects=19 Vehicle_count=14 Person_count=5\nFrame Number=457 Number of Objects=15 Vehicle_count=10 Person_count=5\nFrame Number=458 Number of Objects=17 Vehicle_count=12 Person_count=5\nFrame Number=459 Number of Objects=18 Vehicle_count=13 Person_count=5\nFrame Number=460 Number of Objects=19 Vehicle_count=14 Person_count=5\nFrame Number=461 Number of Objects=19 Vehicle_count=14 Person_count=5\nFrame Number=462 Number of Objects=19 Vehicle_count=14 Person_count=5\nFrame Number=463 Number of Objects=17 Vehicle_count=12 Person_count=5\nFrame Number=464 Number of Objects=17 Vehicle_count=12 Person_count=5\nFrame Number=465 Number of Objects=19 Vehicle_count=14 Person_count=5\nFrame Number=466 Number of Objects=15 Vehicle_count=11 Person_count=4\nFrame Number=467 Number of Objects=17 Vehicle_count=12 Person_count=5\nFrame Number=468 Number of Objects=15 Vehicle_count=10 Person_count=5\nFrame Number=469 Number of Objects=17 Vehicle_count=12 Person_count=5\nFrame Number=470 Number of Objects=17 Vehicle_count=12 Person_count=5\nFrame Number=471 Number of Objects=17 Vehicle_count=12 Person_count=5\nFrame Number=472 Number of Objects=15 Vehicle_count=10 Person_count=5\nFrame Number=473 Number of Objects=15 Vehicle_count=10 Person_count=5\nFrame Number=474 Number of Objects=17 Vehicle_count=12 Person_count=5\nFrame Number=475 Number of Objects=16 Vehicle_count=11 Person_count=5\nFrame Number=476 Number of Objects=16 Vehicle_count=11 Person_count=5\nFrame Number=477 Number of Objects=15 Vehicle_count=10 Person_count=5\nFrame Number=478 Number of Objects=16 Vehicle_count=10 Person_count=5\nFrame Number=479 Number of Objects=17 Vehicle_count=11 Person_count=5\nFrame Number=480 Number of Objects=17 Vehicle_count=11 Person_count=5\nFrame Number=481 Number of Objects=17 Vehicle_count=11 Person_count=5\nFrame Number=482 Number of Objects=18 Vehicle_count=12 Person_count=5\nFrame Number=483 Number of Objects=16 Vehicle_count=10 Person_count=5\nFrame Number=484 Number of Objects=17 Vehicle_count=11 Person_count=5\nFrame Number=485 Number of Objects=15 Vehicle_count=10 Person_count=5\nFrame Number=486 Number of Objects=16 Vehicle_count=10 Person_count=5\nFrame Number=487 Number of Objects=14 Vehicle_count=9 Person_count=5\nFrame Number=488 Number of Objects=13 Vehicle_count=8 Person_count=5\nFrame Number=489 Number of Objects=13 Vehicle_count=8 Person_count=5\nFrame Number=490 Number of Objects=12 Vehicle_count=7 Person_count=5\nFrame Number=491 Number of Objects=13 Vehicle_count=7 Person_count=5\nFrame Number=492 Number of Objects=13 Vehicle_count=7 Person_count=5\nFrame Number=493 Number of Objects=14 Vehicle_count=7 Person_count=5\nFrame Number=494 Number of Objects=12 Vehicle_count=6 Person_count=5\nFrame Number=495 Number of Objects=13 Vehicle_count=6 Person_count=5\nFrame Number=496 Number of Objects=11 Vehicle_count=5 Person_count=5\nFrame Number=497 Number of Objects=11 Vehicle_count=5 Person_count=5\nFrame Number=498 Number of Objects=12 Vehicle_count=7 Person_count=5\nFrame Number=499 Number of Objects=13 Vehicle_count=8 Person_count=5\nFrame Number=500 Number of Objects=12 Vehicle_count=7 Person_count=5\nFrame Number=501 Number of Objects=13 Vehicle_count=8 Person_count=5\nFrame Number=502 Number of Objects=13 Vehicle_count=8 Person_count=5\nFrame Number=503 Number of Objects=11 Vehicle_count=6 Person_count=5\nFrame Number=504 Number of Objects=13 Vehicle_count=7 Person_count=5\nFrame Number=505 Number of Objects=11 Vehicle_count=5 Person_count=5\nFrame Number=506 Number of Objects=10 Vehicle_count=5 Person_count=5\nFrame Number=507 Number of Objects=10 Vehicle_count=5 Person_count=5\nFrame Number=508 Number of Objects=12 Vehicle_count=6 Person_count=5\nFrame Number=509 Number of Objects=11 Vehicle_count=5 Person_count=5\nFrame Number=510 Number of Objects=10 Vehicle_count=5 Person_count=5\nFrame Number=511 Number of Objects=11 Vehicle_count=5 Person_count=5\nFrame Number=512 Number of Objects=12 Vehicle_count=6 Person_count=5\nFrame Number=513 Number of Objects=12 Vehicle_count=7 Person_count=5\nFrame Number=514 Number of Objects=13 Vehicle_count=7 Person_count=6\nFrame Number=515 Number of Objects=12 Vehicle_count=6 Person_count=6\nFrame Number=516 Number of Objects=12 Vehicle_count=6 Person_count=5\nFrame Number=517 Number of Objects=10 Vehicle_count=5 Person_count=5\nFrame Number=518 Number of Objects=9 Vehicle_count=3 Person_count=5\nFrame Number=519 Number of Objects=9 Vehicle_count=4 Person_count=5\nFrame Number=520 Number of Objects=9 Vehicle_count=5 Person_count=4\nFrame Number=521 Number of Objects=10 Vehicle_count=5 Person_count=5\nFrame Number=522 Number of Objects=11 Vehicle_count=5 Person_count=5\nFrame Number=523 Number of Objects=11 Vehicle_count=5 Person_count=5\nFrame Number=524 Number of Objects=11 Vehicle_count=5 Person_count=5\nFrame Number=525 Number of Objects=10 Vehicle_count=5 Person_count=5\nFrame Number=526 Number of Objects=11 Vehicle_count=5 Person_count=5\nFrame Number=527 Number of Objects=10 Vehicle_count=5 Person_count=5\nFrame Number=528 Number of Objects=12 Vehicle_count=6 Person_count=5\nFrame Number=529 Number of Objects=11 Vehicle_count=5 Person_count=6\nFrame Number=530 Number of Objects=12 Vehicle_count=5 Person_count=6\nFrame Number=531 Number of Objects=12 Vehicle_count=5 Person_count=6\nFrame Number=532 Number of Objects=13 Vehicle_count=6 Person_count=6\nFrame Number=533 Number of Objects=10 Vehicle_count=5 Person_count=5\nFrame Number=534 Number of Objects=10 Vehicle_count=5 Person_count=5\nFrame Number=535 Number of Objects=12 Vehicle_count=7 Person_count=5\nFrame Number=536 Number of Objects=15 Vehicle_count=8 Person_count=5\nFrame Number=537 Number of Objects=15 Vehicle_count=8 Person_count=5\nFrame Number=538 Number of Objects=13 Vehicle_count=6 Person_count=5\nFrame Number=539 Number of Objects=12 Vehicle_count=6 Person_count=5\nFrame Number=540 Number of Objects=11 Vehicle_count=6 Person_count=4\nFrame Number=541 Number of Objects=11 Vehicle_count=6 Person_count=5\nFrame Number=542 Number of Objects=12 Vehicle_count=6 Person_count=5\nFrame Number=543 Number of Objects=13 Vehicle_count=6 Person_count=6\nFrame Number=544 Number of Objects=15 Vehicle_count=7 Person_count=6\nFrame Number=545 Number of Objects=16 Vehicle_count=7 Person_count=6\nFrame Number=546 Number of Objects=14 Vehicle_count=7 Person_count=6\nFrame Number=547 Number of Objects=14 Vehicle_count=8 Person_count=6\nFrame Number=548 Number of Objects=14 Vehicle_count=8 Person_count=5\nFrame Number=549 Number of Objects=13 Vehicle_count=8 Person_count=5\nFrame Number=550 Number of Objects=14 Vehicle_count=8 Person_count=5\nFrame Number=551 Number of Objects=15 Vehicle_count=7 Person_count=6\nFrame Number=552 Number of Objects=16 Vehicle_count=9 Person_count=5\nFrame Number=553 Number of Objects=14 Vehicle_count=9 Person_count=5\nFrame Number=554 Number of Objects=14 Vehicle_count=9 Person_count=5\nFrame Number=555 Number of Objects=13 Vehicle_count=8 Person_count=5\nFrame Number=556 Number of Objects=12 Vehicle_count=8 Person_count=4\nFrame Number=557 Number of Objects=13 Vehicle_count=8 Person_count=5\nFrame Number=558 Number of Objects=14 Vehicle_count=9 Person_count=5\nFrame Number=559 Number of Objects=13 Vehicle_count=7 Person_count=6\nFrame Number=560 Number of Objects=15 Vehicle_count=9 Person_count=6\nFrame Number=561 Number of Objects=14 Vehicle_count=8 Person_count=5\nFrame Number=562 Number of Objects=12 Vehicle_count=7 Person_count=5\nFrame Number=563 Number of Objects=13 Vehicle_count=8 Person_count=5\nFrame Number=564 Number of Objects=11 Vehicle_count=7 Person_count=4\nFrame Number=565 Number of Objects=12 Vehicle_count=7 Person_count=4\nFrame Number=566 Number of Objects=11 Vehicle_count=6 Person_count=5\nFrame Number=567 Number of Objects=11 Vehicle_count=7 Person_count=4\nFrame Number=568 Number of Objects=12 Vehicle_count=7 Person_count=5\nFrame Number=569 Number of Objects=13 Vehicle_count=8 Person_count=5\nFrame Number=570 Number of Objects=12 Vehicle_count=6 Person_count=6\nFrame Number=571 Number of Objects=11 Vehicle_count=6 Person_count=5\nFrame Number=572 Number of Objects=10 Vehicle_count=5 Person_count=5\nFrame Number=573 Number of Objects=11 Vehicle_count=6 Person_count=5\nFrame Number=574 Number of Objects=11 Vehicle_count=6 Person_count=5\nFrame Number=575 Number of Objects=10 Vehicle_count=6 Person_count=4\nFrame Number=576 Number of Objects=11 Vehicle_count=7 Person_count=4\nFrame Number=577 Number of Objects=11 Vehicle_count=7 Person_count=4\nFrame Number=578 Number of Objects=11 Vehicle_count=7 Person_count=4\nFrame Number=579 Number of Objects=11 Vehicle_count=7 Person_count=4\nFrame Number=580 Number of Objects=12 Vehicle_count=7 Person_count=5\nFrame Number=581 Number of Objects=12 Vehicle_count=8 Person_count=4\nFrame Number=582 Number of Objects=12 Vehicle_count=7 Person_count=5\nFrame Number=583 Number of Objects=11 Vehicle_count=7 Person_count=4\nFrame Number=584 Number of Objects=13 Vehicle_count=9 Person_count=4\nFrame Number=585 Number of Objects=14 Vehicle_count=9 Person_count=5\nFrame Number=586 Number of Objects=13 Vehicle_count=8 Person_count=5\nFrame Number=587 Number of Objects=14 Vehicle_count=10 Person_count=4\nFrame Number=588 Number of Objects=14 Vehicle_count=10 Person_count=4\nFrame Number=589 Number of Objects=14 Vehicle_count=10 Person_count=4\nFrame Number=590 Number of Objects=16 Vehicle_count=12 Person_count=4\nFrame Number=591 Number of Objects=15 Vehicle_count=10 Person_count=5\nFrame Number=592 Number of Objects=13 Vehicle_count=9 Person_count=4\nFrame Number=593 Number of Objects=15 Vehicle_count=10 Person_count=5\nFrame Number=594 Number of Objects=12 Vehicle_count=7 Person_count=5\nFrame Number=595 Number of Objects=10 Vehicle_count=6 Person_count=4\nFrame Number=596 Number of Objects=10 Vehicle_count=6 Person_count=4\nFrame Number=597 Number of Objects=11 Vehicle_count=6 Person_count=5\nFrame Number=598 Number of Objects=10 Vehicle_count=5 Person_count=5\nFrame Number=599 Number of Objects=10 Vehicle_count=5 Person_count=5\nFrame Number=600 Number of Objects=12 Vehicle_count=7 Person_count=5\nFrame Number=601 Number of Objects=12 Vehicle_count=7 Person_count=5\nFrame Number=602 Number of Objects=14 Vehicle_count=8 Person_count=5\nFrame Number=603 Number of Objects=11 Vehicle_count=7 Person_count=4\nFrame Number=604 Number of Objects=11 Vehicle_count=6 Person_count=5\nFrame Number=605 Number of Objects=12 Vehicle_count=7 Person_count=5\nFrame Number=606 Number of Objects=13 Vehicle_count=7 Person_count=5\nFrame Number=607 Number of Objects=14 Vehicle_count=9 Person_count=5\nFrame Number=608 Number of Objects=11 Vehicle_count=6 Person_count=5\nFrame Number=609 Number of Objects=13 Vehicle_count=8 Person_count=5\nFrame Number=610 Number of Objects=12 Vehicle_count=7 Person_count=5\nFrame Number=611 Number of Objects=13 Vehicle_count=8 Person_count=5\nFrame Number=612 Number of Objects=11 Vehicle_count=7 Person_count=4\nFrame Number=613 Number of Objects=11 Vehicle_count=7 Person_count=4\nFrame Number=614 Number of Objects=12 Vehicle_count=6 Person_count=6\nFrame Number=615 Number of Objects=16 Vehicle_count=11 Person_count=5\nFrame Number=616 Number of Objects=14 Vehicle_count=9 Person_count=5\nFrame Number=617 Number of Objects=14 Vehicle_count=9 Person_count=5\nFrame Number=618 Number of Objects=11 Vehicle_count=7 Person_count=4\nFrame Number=619 Number of Objects=12 Vehicle_count=7 Person_count=5\nFrame Number=620 Number of Objects=12 Vehicle_count=7 Person_count=5\nFrame Number=621 Number of Objects=13 Vehicle_count=8 Person_count=5\nFrame Number=622 Number of Objects=9 Vehicle_count=5 Person_count=4\nFrame Number=623 Number of Objects=10 Vehicle_count=6 Person_count=4\nFrame Number=624 Number of Objects=11 Vehicle_count=7 Person_count=4\nFrame Number=625 Number of Objects=12 Vehicle_count=8 Person_count=4\nFrame Number=626 Number of Objects=14 Vehicle_count=10 Person_count=4\nFrame Number=627 Number of Objects=15 Vehicle_count=10 Person_count=5\nFrame Number=628 Number of Objects=13 Vehicle_count=9 Person_count=4\nFrame Number=629 Number of Objects=12 Vehicle_count=8 Person_count=4\nFrame Number=630 Number of Objects=14 Vehicle_count=10 Person_count=4\nFrame Number=631 Number of Objects=15 Vehicle_count=11 Person_count=4\nFrame Number=632 Number of Objects=15 Vehicle_count=11 Person_count=4\nFrame Number=633 Number of Objects=16 Vehicle_count=12 Person_count=4\nFrame Number=634 Number of Objects=14 Vehicle_count=10 Person_count=4\nFrame Number=635 Number of Objects=12 Vehicle_count=8 Person_count=4\nFrame Number=636 Number of Objects=14 Vehicle_count=10 Person_count=4\nFrame Number=637 Number of Objects=14 Vehicle_count=10 Person_count=4\nFrame Number=638 Number of Objects=13 Vehicle_count=9 Person_count=4\nFrame Number=639 Number of Objects=12 Vehicle_count=7 Person_count=5\nFrame Number=640 Number of Objects=13 Vehicle_count=8 Person_count=5\nFrame Number=641 Number of Objects=12 Vehicle_count=7 Person_count=5\nFrame Number=642 Number of Objects=11 Vehicle_count=6 Person_count=5\nFrame Number=643 Number of Objects=11 Vehicle_count=6 Person_count=5\nFrame Number=644 Number of Objects=11 Vehicle_count=6 Person_count=5\nFrame Number=645 Number of Objects=12 Vehicle_count=7 Person_count=5\nFrame Number=646 Number of Objects=10 Vehicle_count=5 Person_count=5\nFrame Number=647 Number of Objects=9 Vehicle_count=4 Person_count=5\nFrame Number=648 Number of Objects=11 Vehicle_count=6 Person_count=5\nFrame Number=649 Number of Objects=8 Vehicle_count=4 Person_count=4\nFrame Number=650 Number of Objects=9 Vehicle_count=4 Person_count=5\nFrame Number=651 Number of Objects=7 Vehicle_count=4 Person_count=3\nFrame Number=652 Number of Objects=6 Vehicle_count=3 Person_count=3\nFrame Number=653 Number of Objects=7 Vehicle_count=3 Person_count=4\nFrame Number=654 Number of Objects=8 Vehicle_count=4 Person_count=4\nFrame Number=655 Number of Objects=7 Vehicle_count=4 Person_count=3\nFrame Number=656 Number of Objects=7 Vehicle_count=3 Person_count=4\nFrame Number=657 Number of Objects=5 Vehicle_count=2 Person_count=3\nFrame Number=658 Number of Objects=7 Vehicle_count=3 Person_count=4\nFrame Number=659 Number of Objects=8 Vehicle_count=4 Person_count=4\nFrame Number=660 Number of Objects=7 Vehicle_count=3 Person_count=4\nFrame Number=661 Number of Objects=7 Vehicle_count=4 Person_count=3\nFrame Number=662 Number of Objects=9 Vehicle_count=5 Person_count=4\nFrame Number=663 Number of Objects=10 Vehicle_count=6 Person_count=4\nFrame Number=664 Number of Objects=11 Vehicle_count=7 Person_count=4\nFrame Number=665 Number of Objects=11 Vehicle_count=7 Person_count=4\nFrame Number=666 Number of Objects=10 Vehicle_count=6 Person_count=4\nFrame Number=667 Number of Objects=12 Vehicle_count=8 Person_count=4\nFrame Number=668 Number of Objects=11 Vehicle_count=7 Person_count=4\nFrame Number=669 Number of Objects=11 Vehicle_count=7 Person_count=4\nFrame Number=670 Number of Objects=12 Vehicle_count=8 Person_count=4\nFrame Number=671 Number of Objects=12 Vehicle_count=7 Person_count=5\nFrame Number=672 Number of Objects=11 Vehicle_count=7 Person_count=4\nFrame Number=673 Number of Objects=13 Vehicle_count=9 Person_count=4\nFrame Number=674 Number of Objects=11 Vehicle_count=7 Person_count=4\nFrame Number=675 Number of Objects=10 Vehicle_count=6 Person_count=4\nFrame Number=676 Number of Objects=10 Vehicle_count=5 Person_count=4\nFrame Number=677 Number of Objects=12 Vehicle_count=7 Person_count=5\nFrame Number=678 Number of Objects=10 Vehicle_count=6 Person_count=4\nFrame Number=679 Number of Objects=9 Vehicle_count=5 Person_count=4\nFrame Number=680 Number of Objects=11 Vehicle_count=7 Person_count=4\nFrame Number=681 Number of Objects=14 Vehicle_count=9 Person_count=5\nFrame Number=682 Number of Objects=15 Vehicle_count=10 Person_count=5\nFrame Number=683 Number of Objects=15 Vehicle_count=10 Person_count=4\nFrame Number=684 Number of Objects=14 Vehicle_count=9 Person_count=5\nFrame Number=685 Number of Objects=14 Vehicle_count=9 Person_count=4\nFrame Number=686 Number of Objects=14 Vehicle_count=9 Person_count=4\nFrame Number=687 Number of Objects=14 Vehicle_count=8 Person_count=5\nFrame Number=688 Number of Objects=12 Vehicle_count=7 Person_count=4\nFrame Number=689 Number of Objects=13 Vehicle_count=7 Person_count=5\nFrame Number=690 Number of Objects=14 Vehicle_count=7 Person_count=5\nFrame Number=691 Number of Objects=14 Vehicle_count=7 Person_count=5\nFrame Number=692 Number of Objects=14 Vehicle_count=7 Person_count=5\nFrame Number=693 Number of Objects=11 Vehicle_count=6 Person_count=4\nFrame Number=694 Number of Objects=11 Vehicle_count=6 Person_count=4\nFrame Number=695 Number of Objects=11 Vehicle_count=6 Person_count=4\nFrame Number=696 Number of Objects=11 Vehicle_count=6 Person_count=4\nFrame Number=697 Number of Objects=9 Vehicle_count=4 Person_count=4\nFrame Number=698 Number of Objects=9 Vehicle_count=4 Person_count=4\nFrame Number=699 Number of Objects=7 Vehicle_count=3 Person_count=4\nFrame Number=700 Number of Objects=8 Vehicle_count=3 Person_count=4\nFrame Number=701 Number of Objects=9 Vehicle_count=4 Person_count=4\nFrame Number=702 Number of Objects=7 Vehicle_count=2 Person_count=4\nFrame Number=703 Number of Objects=7 Vehicle_count=2 Person_count=4\nFrame Number=704 Number of Objects=7 Vehicle_count=2 Person_count=4\nFrame Number=705 Number of Objects=6 Vehicle_count=1 Person_count=4\nFrame Number=706 Number of Objects=6 Vehicle_count=1 Person_count=4\nFrame Number=707 Number of Objects=7 Vehicle_count=2 Person_count=4\nFrame Number=708 Number of Objects=9 Vehicle_count=3 Person_count=5\nFrame Number=709 Number of Objects=7 Vehicle_count=2 Person_count=4\nFrame Number=710 Number of Objects=9 Vehicle_count=4 Person_count=4\nFrame Number=711 Number of Objects=9 Vehicle_count=3 Person_count=5\nFrame Number=712 Number of Objects=8 Vehicle_count=3 Person_count=4\nFrame Number=713 Number of Objects=9 Vehicle_count=3 Person_count=5\nFrame Number=714 Number of Objects=8 Vehicle_count=3 Person_count=4\nFrame Number=715 Number of Objects=7 Vehicle_count=1 Person_count=5\nFrame Number=716 Number of Objects=7 Vehicle_count=1 Person_count=5\nFrame Number=717 Number of Objects=7 Vehicle_count=1 Person_count=5\nFrame Number=718 Number of Objects=8 Vehicle_count=2 Person_count=5\nFrame Number=719 Number of Objects=8 Vehicle_count=2 Person_count=5\nFrame Number=720 Number of Objects=8 Vehicle_count=2 Person_count=5\nFrame Number=721 Number of Objects=8 Vehicle_count=2 Person_count=5\nFrame Number=722 Number of Objects=6 Vehicle_count=2 Person_count=4\nFrame Number=723 Number of Objects=7 Vehicle_count=2 Person_count=5\nFrame Number=724 Number of Objects=6 Vehicle_count=2 Person_count=4\nFrame Number=725 Number of Objects=9 Vehicle_count=3 Person_count=6\nFrame Number=726 Number of Objects=7 Vehicle_count=2 Person_count=5\nFrame Number=727 Number of Objects=7 Vehicle_count=3 Person_count=4\nFrame Number=728 Number of Objects=8 Vehicle_count=3 Person_count=5\nFrame Number=729 Number of Objects=8 Vehicle_count=3 Person_count=5\nFrame Number=730 Number of Objects=8 Vehicle_count=3 Person_count=5\nFrame Number=731 Number of Objects=8 Vehicle_count=3 Person_count=5\nFrame Number=732 Number of Objects=9 Vehicle_count=3 Person_count=5\nFrame Number=733 Number of Objects=8 Vehicle_count=3 Person_count=5\nFrame Number=734 Number of Objects=8 Vehicle_count=3 Person_count=5\nFrame Number=735 Number of Objects=8 Vehicle_count=3 Person_count=5\nFrame Number=736 Number of Objects=8 Vehicle_count=3 Person_count=5\nFrame Number=737 Number of Objects=8 Vehicle_count=3 Person_count=5\nFrame Number=738 Number of Objects=9 Vehicle_count=3 Person_count=6\nFrame Number=739 Number of Objects=8 Vehicle_count=3 Person_count=5\nFrame Number=740 Number of Objects=7 Vehicle_count=2 Person_count=5\nFrame Number=741 Number of Objects=10 Vehicle_count=3 Person_count=5\nFrame Number=742 Number of Objects=10 Vehicle_count=3 Person_count=5\nFrame Number=743 Number of Objects=11 Vehicle_count=4 Person_count=5\nFrame Number=744 Number of Objects=12 Vehicle_count=6 Person_count=5\nFrame Number=745 Number of Objects=14 Vehicle_count=5 Person_count=7\nFrame Number=746 Number of Objects=13 Vehicle_count=6 Person_count=5\nFrame Number=747 Number of Objects=13 Vehicle_count=7 Person_count=5\nFrame Number=748 Number of Objects=12 Vehicle_count=7 Person_count=5\nFrame Number=749 Number of Objects=10 Vehicle_count=5 Person_count=5\nFrame Number=750 Number of Objects=11 Vehicle_count=6 Person_count=5\nFrame Number=751 Number of Objects=14 Vehicle_count=9 Person_count=5\nFrame Number=752 Number of Objects=13 Vehicle_count=8 Person_count=5\nFrame Number=753 Number of Objects=13 Vehicle_count=8 Person_count=5\nFrame Number=754 Number of Objects=11 Vehicle_count=7 Person_count=4\nFrame Number=755 Number of Objects=13 Vehicle_count=8 Person_count=5\nFrame Number=756 Number of Objects=10 Vehicle_count=6 Person_count=4\nFrame Number=757 Number of Objects=10 Vehicle_count=6 Person_count=4\nFrame Number=758 Number of Objects=11 Vehicle_count=7 Person_count=4\nFrame Number=759 Number of Objects=10 Vehicle_count=6 Person_count=4\nFrame Number=760 Number of Objects=12 Vehicle_count=8 Person_count=4\nFrame Number=761 Number of Objects=12 Vehicle_count=8 Person_count=4\nFrame Number=762 Number of Objects=11 Vehicle_count=8 Person_count=3\nFrame Number=763 Number of Objects=10 Vehicle_count=7 Person_count=3\nFrame Number=764 Number of Objects=10 Vehicle_count=6 Person_count=4\nFrame Number=765 Number of Objects=10 Vehicle_count=6 Person_count=4\nFrame Number=766 Number of Objects=9 Vehicle_count=5 Person_count=4\nFrame Number=767 Number of Objects=10 Vehicle_count=6 Person_count=4\nFrame Number=768 Number of Objects=9 Vehicle_count=5 Person_count=4\nFrame Number=769 Number of Objects=9 Vehicle_count=5 Person_count=4\nFrame Number=770 Number of Objects=12 Vehicle_count=8 Person_count=4\nFrame Number=771 Number of Objects=11 Vehicle_count=7 Person_count=4\nFrame Number=772 Number of Objects=11 Vehicle_count=7 Person_count=4\nFrame Number=773 Number of Objects=12 Vehicle_count=8 Person_count=4\nFrame Number=774 Number of Objects=13 Vehicle_count=10 Person_count=3\nFrame Number=775 Number of Objects=15 Vehicle_count=10 Person_count=4\nFrame Number=776 Number of Objects=13 Vehicle_count=10 Person_count=3\nFrame Number=777 Number of Objects=13 Vehicle_count=9 Person_count=4\nFrame Number=778 Number of Objects=14 Vehicle_count=10 Person_count=3\nFrame Number=779 Number of Objects=18 Vehicle_count=12 Person_count=4\nFrame Number=780 Number of Objects=12 Vehicle_count=9 Person_count=3\nFrame Number=781 Number of Objects=11 Vehicle_count=8 Person_count=3\nFrame Number=782 Number of Objects=12 Vehicle_count=9 Person_count=3\nFrame Number=783 Number of Objects=11 Vehicle_count=7 Person_count=4\nFrame Number=784 Number of Objects=11 Vehicle_count=7 Person_count=4\nFrame Number=785 Number of Objects=10 Vehicle_count=6 Person_count=4\nFrame Number=786 Number of Objects=13 Vehicle_count=9 Person_count=4\nFrame Number=787 Number of Objects=11 Vehicle_count=7 Person_count=4\nFrame Number=788 Number of Objects=9 Vehicle_count=5 Person_count=4\nFrame Number=789 Number of Objects=11 Vehicle_count=7 Person_count=4\nFrame Number=790 Number of Objects=9 Vehicle_count=5 Person_count=4\nFrame Number=791 Number of Objects=8 Vehicle_count=4 Person_count=4\nFrame Number=792 Number of Objects=8 Vehicle_count=4 Person_count=4\nFrame Number=793 Number of Objects=9 Vehicle_count=5 Person_count=4\nFrame Number=794 Number of Objects=9 Vehicle_count=5 Person_count=4\nFrame Number=795 Number of Objects=8 Vehicle_count=4 Person_count=4\nFrame Number=796 Number of Objects=8 Vehicle_count=4 Person_count=4\nFrame Number=797 Number of Objects=8 Vehicle_count=4 Person_count=4\nFrame Number=798 Number of Objects=8 Vehicle_count=4 Person_count=4\nFrame Number=799 Number of Objects=9 Vehicle_count=3 Person_count=4\nFrame Number=800 Number of Objects=9 Vehicle_count=5 Person_count=4\nFrame Number=801 Number of Objects=8 Vehicle_count=4 Person_count=4\nFrame Number=802 Number of Objects=10 Vehicle_count=6 Person_count=4\nFrame Number=803 Number of Objects=13 Vehicle_count=7 Person_count=4\nFrame Number=804 Number of Objects=10 Vehicle_count=5 Person_count=4\nFrame Number=805 Number of Objects=12 Vehicle_count=6 Person_count=4\nFrame Number=806 Number of Objects=12 Vehicle_count=6 Person_count=4\nFrame Number=807 Number of Objects=13 Vehicle_count=7 Person_count=4\nFrame Number=808 Number of Objects=13 Vehicle_count=7 Person_count=4\nFrame Number=809 Number of Objects=10 Vehicle_count=6 Person_count=4\nFrame Number=810 Number of Objects=10 Vehicle_count=6 Person_count=4\nFrame Number=811 Number of Objects=10 Vehicle_count=6 Person_count=4\nFrame Number=812 Number of Objects=14 Vehicle_count=9 Person_count=4\nFrame Number=813 Number of Objects=14 Vehicle_count=9 Person_count=4\nFrame Number=814 Number of Objects=13 Vehicle_count=7 Person_count=4\nFrame Number=815 Number of Objects=15 Vehicle_count=10 Person_count=4\nFrame Number=816 Number of Objects=15 Vehicle_count=10 Person_count=4\nFrame Number=817 Number of Objects=15 Vehicle_count=9 Person_count=4\nFrame Number=818 Number of Objects=15 Vehicle_count=9 Person_count=4\nFrame Number=819 Number of Objects=14 Vehicle_count=8 Person_count=4\nFrame Number=820 Number of Objects=12 Vehicle_count=7 Person_count=4\nFrame Number=821 Number of Objects=12 Vehicle_count=8 Person_count=4\nFrame Number=822 Number of Objects=12 Vehicle_count=7 Person_count=4\nFrame Number=823 Number of Objects=9 Vehicle_count=5 Person_count=4\nFrame Number=824 Number of Objects=14 Vehicle_count=9 Person_count=4\nFrame Number=825 Number of Objects=15 Vehicle_count=10 Person_count=4\nFrame Number=826 Number of Objects=12 Vehicle_count=8 Person_count=4\nFrame Number=827 Number of Objects=12 Vehicle_count=8 Person_count=4\nFrame Number=828 Number of Objects=14 Vehicle_count=10 Person_count=4\nFrame Number=829 Number of Objects=12 Vehicle_count=8 Person_count=4\nFrame Number=830 Number of Objects=13 Vehicle_count=9 Person_count=4\nFrame Number=831 Number of Objects=13 Vehicle_count=9 Person_count=4\nFrame Number=832 Number of Objects=13 Vehicle_count=8 Person_count=4\nFrame Number=833 Number of Objects=15 Vehicle_count=10 Person_count=4\nFrame Number=834 Number of Objects=15 Vehicle_count=10 Person_count=4\nFrame Number=835 Number of Objects=13 Vehicle_count=8 Person_count=4\nFrame Number=836 Number of Objects=13 Vehicle_count=8 Person_count=4\nFrame Number=837 Number of Objects=11 Vehicle_count=6 Person_count=4\nFrame Number=838 Number of Objects=12 Vehicle_count=7 Person_count=4\nFrame Number=839 Number of Objects=12 Vehicle_count=7 Person_count=4\nFrame Number=840 Number of Objects=11 Vehicle_count=7 Person_count=4\nFrame Number=841 Number of Objects=12 Vehicle_count=8 Person_count=4\nFrame Number=842 Number of Objects=11 Vehicle_count=7 Person_count=4\nFrame Number=843 Number of Objects=10 Vehicle_count=6 Person_count=4\nFrame Number=844 Number of Objects=10 Vehicle_count=6 Person_count=4\nFrame Number=845 Number of Objects=10 Vehicle_count=6 Person_count=4\nFrame Number=846 Number of Objects=12 Vehicle_count=8 Person_count=4\nFrame Number=847 Number of Objects=11 Vehicle_count=7 Person_count=4\nFrame Number=848 Number of Objects=10 Vehicle_count=6 Person_count=4\nFrame Number=849 Number of Objects=8 Vehicle_count=5 Person_count=3\nFrame Number=850 Number of Objects=8 Vehicle_count=5 Person_count=3\nFrame Number=851 Number of Objects=7 Vehicle_count=4 Person_count=3\nFrame Number=852 Number of Objects=12 Vehicle_count=7 Person_count=4\nFrame Number=853 Number of Objects=11 Vehicle_count=6 Person_count=4\nFrame Number=854 Number of Objects=12 Vehicle_count=7 Person_count=4\nFrame Number=855 Number of Objects=12 Vehicle_count=6 Person_count=4\nFrame Number=856 Number of Objects=15 Vehicle_count=8 Person_count=4\nFrame Number=857 Number of Objects=12 Vehicle_count=6 Person_count=4\nFrame Number=858 Number of Objects=12 Vehicle_count=7 Person_count=4\nFrame Number=859 Number of Objects=13 Vehicle_count=8 Person_count=4\nFrame Number=860 Number of Objects=13 Vehicle_count=8 Person_count=4\nFrame Number=861 Number of Objects=14 Vehicle_count=9 Person_count=4\nFrame Number=862 Number of Objects=13 Vehicle_count=8 Person_count=4\nFrame Number=863 Number of Objects=12 Vehicle_count=8 Person_count=4\nFrame Number=864 Number of Objects=14 Vehicle_count=10 Person_count=4\nFrame Number=865 Number of Objects=12 Vehicle_count=8 Person_count=4\nFrame Number=866 Number of Objects=14 Vehicle_count=10 Person_count=4\nFrame Number=867 Number of Objects=13 Vehicle_count=9 Person_count=3\nFrame Number=868 Number of Objects=12 Vehicle_count=7 Person_count=5\nFrame Number=869 Number of Objects=11 Vehicle_count=7 Person_count=4\nFrame Number=870 Number of Objects=13 Vehicle_count=9 Person_count=4\nFrame Number=871 Number of Objects=14 Vehicle_count=10 Person_count=4\nFrame Number=872 Number of Objects=12 Vehicle_count=8 Person_count=4\nFrame Number=873 Number of Objects=12 Vehicle_count=8 Person_count=4\nFrame Number=874 Number of Objects=12 Vehicle_count=8 Person_count=4\nFrame Number=875 Number of Objects=12 Vehicle_count=7 Person_count=5\nFrame Number=876 Number of Objects=14 Vehicle_count=10 Person_count=4\nFrame Number=877 Number of Objects=14 Vehicle_count=9 Person_count=5\nFrame Number=878 Number of Objects=11 Vehicle_count=7 Person_count=4\nFrame Number=879 Number of Objects=13 Vehicle_count=9 Person_count=4\nFrame Number=880 Number of Objects=12 Vehicle_count=9 Person_count=3\nFrame Number=881 Number of Objects=10 Vehicle_count=7 Person_count=3\nFrame Number=882 Number of Objects=11 Vehicle_count=8 Person_count=3\nFrame Number=883 Number of Objects=12 Vehicle_count=9 Person_count=3\nFrame Number=884 Number of Objects=11 Vehicle_count=8 Person_count=3\nFrame Number=885 Number of Objects=11 Vehicle_count=7 Person_count=3\nFrame Number=886 Number of Objects=13 Vehicle_count=9 Person_count=3\nFrame Number=887 Number of Objects=11 Vehicle_count=7 Person_count=3\nFrame Number=888 Number of Objects=11 Vehicle_count=7 Person_count=3\nFrame Number=889 Number of Objects=8 Vehicle_count=5 Person_count=3\nFrame Number=890 Number of Objects=10 Vehicle_count=7 Person_count=3\nFrame Number=891 Number of Objects=10 Vehicle_count=7 Person_count=3\nFrame Number=892 Number of Objects=12 Vehicle_count=8 Person_count=4\nFrame Number=893 Number of Objects=10 Vehicle_count=7 Person_count=3\nFrame Number=894 Number of Objects=12 Vehicle_count=9 Person_count=3\nFrame Number=895 Number of Objects=14 Vehicle_count=9 Person_count=4\nFrame Number=896 Number of Objects=14 Vehicle_count=9 Person_count=4\nFrame Number=897 Number of Objects=13 Vehicle_count=8 Person_count=4\nFrame Number=898 Number of Objects=12 Vehicle_count=7 Person_count=4\nFrame Number=899 Number of Objects=12 Vehicle_count=7 Person_count=4\nFrame Number=900 Number of Objects=12 Vehicle_count=7 Person_count=4\nFrame Number=901 Number of Objects=11 Vehicle_count=6 Person_count=4\nFrame Number=902 Number of Objects=11 Vehicle_count=6 Person_count=4\nFrame Number=903 Number of Objects=11 Vehicle_count=6 Person_count=4\nFrame Number=904 Number of Objects=11 Vehicle_count=6 Person_count=4\nFrame Number=905 Number of Objects=11 Vehicle_count=6 Person_count=4\nFrame Number=906 Number of Objects=10 Vehicle_count=5 Person_count=4\nFrame Number=907 Number of Objects=12 Vehicle_count=7 Person_count=4\nFrame Number=908 Number of Objects=13 Vehicle_count=8 Person_count=4\nFrame Number=909 Number of Objects=14 Vehicle_count=9 Person_count=4\nFrame Number=910 Number of Objects=15 Vehicle_count=9 Person_count=4\nFrame Number=911 Number of Objects=11 Vehicle_count=7 Person_count=3\nFrame Number=912 Number of Objects=16 Vehicle_count=11 Person_count=3\nFrame Number=913 Number of Objects=16 Vehicle_count=13 Person_count=3\nFrame Number=914 Number of Objects=17 Vehicle_count=12 Person_count=4\nFrame Number=915 Number of Objects=16 Vehicle_count=12 Person_count=4\nFrame Number=916 Number of Objects=15 Vehicle_count=11 Person_count=4\nFrame Number=917 Number of Objects=14 Vehicle_count=10 Person_count=4\nFrame Number=918 Number of Objects=14 Vehicle_count=10 Person_count=4\nFrame Number=919 Number of Objects=14 Vehicle_count=10 Person_count=4\nFrame Number=920 Number of Objects=14 Vehicle_count=10 Person_count=4\nFrame Number=921 Number of Objects=14 Vehicle_count=10 Person_count=4\nFrame Number=922 Number of Objects=15 Vehicle_count=11 Person_count=4\nFrame Number=923 Number of Objects=15 Vehicle_count=11 Person_count=4\nFrame Number=924 Number of Objects=14 Vehicle_count=10 Person_count=4\nFrame Number=925 Number of Objects=14 Vehicle_count=10 Person_count=4\nFrame Number=926 Number of Objects=13 Vehicle_count=9 Person_count=4\nFrame Number=927 Number of Objects=14 Vehicle_count=9 Person_count=5\nFrame Number=928 Number of Objects=14 Vehicle_count=9 Person_count=5\nFrame Number=929 Number of Objects=14 Vehicle_count=9 Person_count=5\nFrame Number=930 Number of Objects=14 Vehicle_count=9 Person_count=5\nFrame Number=931 Number of Objects=14 Vehicle_count=9 Person_count=5\nFrame Number=932 Number of Objects=14 Vehicle_count=8 Person_count=6\nFrame Number=933 Number of Objects=15 Vehicle_count=9 Person_count=6\nFrame Number=934 Number of Objects=16 Vehicle_count=10 Person_count=6\nFrame Number=935 Number of Objects=15 Vehicle_count=10 Person_count=5\nFrame Number=936 Number of Objects=15 Vehicle_count=10 Person_count=5\nFrame Number=937 Number of Objects=16 Vehicle_count=11 Person_count=5\nFrame Number=938 Number of Objects=14 Vehicle_count=10 Person_count=4\nFrame Number=939 Number of Objects=14 Vehicle_count=11 Person_count=3\nFrame Number=940 Number of Objects=12 Vehicle_count=9 Person_count=3\nFrame Number=941 Number of Objects=13 Vehicle_count=10 Person_count=3\nFrame Number=942 Number of Objects=15 Vehicle_count=10 Person_count=5\nFrame Number=943 Number of Objects=13 Vehicle_count=9 Person_count=4\nFrame Number=944 Number of Objects=14 Vehicle_count=10 Person_count=4\nFrame Number=945 Number of Objects=13 Vehicle_count=9 Person_count=4\nFrame Number=946 Number of Objects=12 Vehicle_count=8 Person_count=4\nFrame Number=947 Number of Objects=13 Vehicle_count=8 Person_count=5\nFrame Number=948 Number of Objects=13 Vehicle_count=9 Person_count=4\nFrame Number=949 Number of Objects=12 Vehicle_count=7 Person_count=5\nFrame Number=950 Number of Objects=12 Vehicle_count=7 Person_count=5\nFrame Number=951 Number of Objects=11 Vehicle_count=7 Person_count=4\nFrame Number=952 Number of Objects=12 Vehicle_count=7 Person_count=5\nFrame Number=953 Number of Objects=12 Vehicle_count=7 Person_count=5\nFrame Number=954 Number of Objects=10 Vehicle_count=5 Person_count=5\nFrame Number=955 Number of Objects=11 Vehicle_count=6 Person_count=5\nFrame Number=956 Number of Objects=11 Vehicle_count=6 Person_count=5\nFrame Number=957 Number of Objects=11 Vehicle_count=6 Person_count=5\nFrame Number=958 Number of Objects=10 Vehicle_count=5 Person_count=5\nFrame Number=959 Number of Objects=11 Vehicle_count=6 Person_count=5\nFrame Number=960 Number of Objects=12 Vehicle_count=6 Person_count=5\nFrame Number=961 Number of Objects=12 Vehicle_count=6 Person_count=5\nFrame Number=962 Number of Objects=9 Vehicle_count=5 Person_count=4\nFrame Number=963 Number of Objects=9 Vehicle_count=4 Person_count=5\nFrame Number=964 Number of Objects=9 Vehicle_count=5 Person_count=4\nFrame Number=965 Number of Objects=9 Vehicle_count=5 Person_count=4\nFrame Number=966 Number of Objects=9 Vehicle_count=5 Person_count=4\nFrame Number=967 Number of Objects=10 Vehicle_count=5 Person_count=5\nFrame Number=968 Number of Objects=12 Vehicle_count=7 Person_count=4\nFrame Number=969 Number of Objects=13 Vehicle_count=6 Person_count=5\nFrame Number=970 Number of Objects=12 Vehicle_count=5 Person_count=6\nFrame Number=971 Number of Objects=15 Vehicle_count=10 Person_count=4\nFrame Number=972 Number of Objects=16 Vehicle_count=11 Person_count=4\nFrame Number=973 Number of Objects=15 Vehicle_count=10 Person_count=4\nFrame Number=974 Number of Objects=15 Vehicle_count=10 Person_count=4\nFrame Number=975 Number of Objects=16 Vehicle_count=11 Person_count=4\nFrame Number=976 Number of Objects=17 Vehicle_count=12 Person_count=4\nFrame Number=977 Number of Objects=16 Vehicle_count=11 Person_count=4\nFrame Number=978 Number of Objects=14 Vehicle_count=10 Person_count=4\nFrame Number=979 Number of Objects=13 Vehicle_count=8 Person_count=4\nFrame Number=980 Number of Objects=15 Vehicle_count=10 Person_count=4\nFrame Number=981 Number of Objects=14 Vehicle_count=9 Person_count=4\nFrame Number=982 Number of Objects=16 Vehicle_count=11 Person_count=4\nFrame Number=983 Number of Objects=15 Vehicle_count=11 Person_count=3\nFrame Number=984 Number of Objects=16 Vehicle_count=12 Person_count=3\nFrame Number=985 Number of Objects=13 Vehicle_count=10 Person_count=3\nFrame Number=986 Number of Objects=13 Vehicle_count=10 Person_count=2\nFrame Number=987 Number of Objects=17 Vehicle_count=12 Person_count=4\nFrame Number=988 Number of Objects=12 Vehicle_count=8 Person_count=3\nFrame Number=989 Number of Objects=14 Vehicle_count=10 Person_count=3\nFrame Number=990 Number of Objects=12 Vehicle_count=8 Person_count=3\nFrame Number=991 Number of Objects=13 Vehicle_count=9 Person_count=3\nFrame Number=992 Number of Objects=13 Vehicle_count=9 Person_count=3\nFrame Number=993 Number of Objects=14 Vehicle_count=9 Person_count=4\nFrame Number=994 Number of Objects=15 Vehicle_count=10 Person_count=4\nFrame Number=995 Number of Objects=14 Vehicle_count=10 Person_count=3\nFrame Number=996 Number of Objects=13 Vehicle_count=10 Person_count=3\nFrame Number=997 Number of Objects=14 Vehicle_count=11 Person_count=3\nFrame Number=998 Number of Objects=14 Vehicle_count=11 Person_count=3\nFrame Number=999 Number of Objects=12 Vehicle_count=9 Person_count=3\nFrame Number=1000 Number of Objects=12 Vehicle_count=9 Person_count=3\nFrame Number=1001 Number of Objects=10 Vehicle_count=7 Person_count=3\nFrame Number=1002 Number of Objects=11 Vehicle_count=8 Person_count=3\nFrame Number=1003 Number of Objects=11 Vehicle_count=8 Person_count=3\nFrame Number=1004 Number of Objects=11 Vehicle_count=8 Person_count=3\nFrame Number=1005 Number of Objects=11 Vehicle_count=8 Person_count=3\nFrame Number=1006 Number of Objects=11 Vehicle_count=8 Person_count=3\nFrame Number=1007 Number of Objects=12 Vehicle_count=8 Person_count=4\nFrame Number=1008 Number of Objects=14 Vehicle_count=10 Person_count=4\nFrame Number=1009 Number of Objects=13 Vehicle_count=9 Person_count=4\nFrame Number=1010 Number of Objects=11 Vehicle_count=7 Person_count=4\nFrame Number=1011 Number of Objects=11 Vehicle_count=7 Person_count=4\nFrame Number=1012 Number of Objects=12 Vehicle_count=8 Person_count=4\nFrame Number=1013 Number of Objects=11 Vehicle_count=7 Person_count=4\nFrame Number=1014 Number of Objects=10 Vehicle_count=6 Person_count=4\nFrame Number=1015 Number of Objects=10 Vehicle_count=6 Person_count=4\nFrame Number=1016 Number of Objects=14 Vehicle_count=10 Person_count=4\nFrame Number=1017 Number of Objects=12 Vehicle_count=8 Person_count=4\nFrame Number=1018 Number of Objects=13 Vehicle_count=9 Person_count=4\nFrame Number=1019 Number of Objects=11 Vehicle_count=8 Person_count=3\nFrame Number=1020 Number of Objects=11 Vehicle_count=8 Person_count=3\nFrame Number=1021 Number of Objects=11 Vehicle_count=7 Person_count=4\nFrame Number=1022 Number of Objects=8 Vehicle_count=5 Person_count=3\nFrame Number=1023 Number of Objects=9 Vehicle_count=6 Person_count=3\nFrame Number=1024 Number of Objects=9 Vehicle_count=6 Person_count=3\nFrame Number=1025 Number of Objects=8 Vehicle_count=5 Person_count=3\nFrame Number=1026 Number of Objects=11 Vehicle_count=8 Person_count=3\nFrame Number=1027 Number of Objects=8 Vehicle_count=5 Person_count=3\nFrame Number=1028 Number of Objects=8 Vehicle_count=5 Person_count=3\nFrame Number=1029 Number of Objects=14 Vehicle_count=10 Person_count=4\nFrame Number=1030 Number of Objects=10 Vehicle_count=6 Person_count=4\nFrame Number=1031 Number of Objects=11 Vehicle_count=7 Person_count=4\nFrame Number=1032 Number of Objects=14 Vehicle_count=10 Person_count=4\nFrame Number=1033 Number of Objects=13 Vehicle_count=9 Person_count=4\nFrame Number=1034 Number of Objects=17 Vehicle_count=13 Person_count=4\nFrame Number=1035 Number of Objects=14 Vehicle_count=10 Person_count=4\nFrame Number=1036 Number of Objects=16 Vehicle_count=12 Person_count=4\nFrame Number=1037 Number of Objects=16 Vehicle_count=12 Person_count=4\nFrame Number=1038 Number of Objects=15 Vehicle_count=11 Person_count=4\nFrame Number=1039 Number of Objects=17 Vehicle_count=13 Person_count=4\nFrame Number=1040 Number of Objects=16 Vehicle_count=12 Person_count=4\nFrame Number=1041 Number of Objects=14 Vehicle_count=10 Person_count=4\nFrame Number=1042 Number of Objects=15 Vehicle_count=11 Person_count=4\nFrame Number=1043 Number of Objects=15 Vehicle_count=11 Person_count=4\nFrame Number=1044 Number of Objects=14 Vehicle_count=10 Person_count=4\nFrame Number=1045 Number of Objects=15 Vehicle_count=11 Person_count=4\nFrame Number=1046 Number of Objects=15 Vehicle_count=11 Person_count=4\nFrame Number=1047 Number of Objects=15 Vehicle_count=11 Person_count=4\nFrame Number=1048 Number of Objects=15 Vehicle_count=11 Person_count=4\nFrame Number=1049 Number of Objects=14 Vehicle_count=10 Person_count=4\nFrame Number=1050 Number of Objects=15 Vehicle_count=11 Person_count=4\nFrame Number=1051 Number of Objects=13 Vehicle_count=9 Person_count=4\nFrame Number=1052 Number of Objects=14 Vehicle_count=10 Person_count=4\nFrame Number=1053 Number of Objects=14 Vehicle_count=10 Person_count=4\nFrame Number=1054 Number of Objects=16 Vehicle_count=12 Person_count=4\nFrame Number=1055 Number of Objects=16 Vehicle_count=12 Person_count=4\nFrame Number=1056 Number of Objects=17 Vehicle_count=13 Person_count=4\nFrame Number=1057 Number of Objects=16 Vehicle_count=12 Person_count=4\nFrame Number=1058 Number of Objects=18 Vehicle_count=14 Person_count=4\nFrame Number=1059 Number of Objects=15 Vehicle_count=11 Person_count=4\nFrame Number=1060 Number of Objects=14 Vehicle_count=10 Person_count=4\nFrame Number=1061 Number of Objects=13 Vehicle_count=9 Person_count=4\nFrame Number=1062 Number of Objects=13 Vehicle_count=9 Person_count=4\nFrame Number=1063 Number of Objects=13 Vehicle_count=9 Person_count=4\nFrame Number=1064 Number of Objects=14 Vehicle_count=10 Person_count=4\nFrame Number=1065 Number of Objects=13 Vehicle_count=9 Person_count=4\nFrame Number=1066 Number of Objects=13 Vehicle_count=9 Person_count=4\nFrame Number=1067 Number of Objects=12 Vehicle_count=8 Person_count=4\nFrame Number=1068 Number of Objects=11 Vehicle_count=7 Person_count=4\nFrame Number=1069 Number of Objects=12 Vehicle_count=9 Person_count=3\nFrame Number=1070 Number of Objects=13 Vehicle_count=9 Person_count=4\nFrame Number=1071 Number of Objects=13 Vehicle_count=8 Person_count=5\nFrame Number=1072 Number of Objects=12 Vehicle_count=8 Person_count=4\nFrame Number=1073 Number of Objects=13 Vehicle_count=9 Person_count=4\nFrame Number=1074 Number of Objects=13 Vehicle_count=8 Person_count=5\nFrame Number=1075 Number of Objects=10 Vehicle_count=6 Person_count=4\nFrame Number=1076 Number of Objects=11 Vehicle_count=8 Person_count=3\nFrame Number=1077 Number of Objects=9 Vehicle_count=6 Person_count=3\nFrame Number=1078 Number of Objects=12 Vehicle_count=8 Person_count=4\nFrame Number=1079 Number of Objects=11 Vehicle_count=7 Person_count=4\nFrame Number=1080 Number of Objects=16 Vehicle_count=12 Person_count=4\nFrame Number=1081 Number of Objects=14 Vehicle_count=11 Person_count=3\nFrame Number=1082 Number of Objects=13 Vehicle_count=10 Person_count=3\nFrame Number=1083 Number of Objects=16 Vehicle_count=12 Person_count=4\nFrame Number=1084 Number of Objects=14 Vehicle_count=11 Person_count=3\nFrame Number=1085 Number of Objects=14 Vehicle_count=11 Person_count=3\nFrame Number=1086 Number of Objects=14 Vehicle_count=11 Person_count=3\nFrame Number=1087 Number of Objects=14 Vehicle_count=11 Person_count=3\nFrame Number=1088 Number of Objects=14 Vehicle_count=11 Person_count=3\nFrame Number=1089 Number of Objects=13 Vehicle_count=10 Person_count=3\nFrame Number=1090 Number of Objects=12 Vehicle_count=9 Person_count=3\nFrame Number=1091 Number of Objects=13 Vehicle_count=10 Person_count=3\nFrame Number=1092 Number of Objects=10 Vehicle_count=7 Person_count=3\nFrame Number=1093 Number of Objects=11 Vehicle_count=8 Person_count=3\nFrame Number=1094 Number of Objects=9 Vehicle_count=7 Person_count=2\nFrame Number=1095 Number of Objects=10 Vehicle_count=7 Person_count=3\nFrame Number=1096 Number of Objects=10 Vehicle_count=7 Person_count=3\nFrame Number=1097 Number of Objects=10 Vehicle_count=7 Person_count=3\nFrame Number=1098 Number of Objects=9 Vehicle_count=6 Person_count=3\nFrame Number=1099 Number of Objects=9 Vehicle_count=6 Person_count=3\nFrame Number=1100 Number of Objects=9 Vehicle_count=6 Person_count=3\nFrame Number=1101 Number of Objects=9 Vehicle_count=6 Person_count=3\nFrame Number=1102 Number of Objects=9 Vehicle_count=6 Person_count=3\nFrame Number=1103 Number of Objects=7 Vehicle_count=4 Person_count=3\nFrame Number=1104 Number of Objects=7 Vehicle_count=4 Person_count=3\nFrame Number=1105 Number of Objects=8 Vehicle_count=5 Person_count=3\nFrame Number=1106 Number of Objects=7 Vehicle_count=4 Person_count=3\nFrame Number=1107 Number of Objects=7 Vehicle_count=4 Person_count=3\nFrame Number=1108 Number of Objects=7 Vehicle_count=4 Person_count=3\nFrame Number=1109 Number of Objects=6 Vehicle_count=3 Person_count=3\nFrame Number=1110 Number of Objects=6 Vehicle_count=3 Person_count=3\nFrame Number=1111 Number of Objects=8 Vehicle_count=5 Person_count=3\nFrame Number=1112 Number of Objects=7 Vehicle_count=4 Person_count=3\nFrame Number=1113 Number of Objects=7 Vehicle_count=4 Person_count=3\nFrame Number=1114 Number of Objects=8 Vehicle_count=5 Person_count=3\nFrame Number=1115 Number of Objects=9 Vehicle_count=6 Person_count=3\nFrame Number=1116 Number of Objects=9 Vehicle_count=6 Person_count=3\nFrame Number=1117 Number of Objects=8 Vehicle_count=5 Person_count=3\nFrame Number=1118 Number of Objects=7 Vehicle_count=4 Person_count=3\nFrame Number=1119 Number of Objects=8 Vehicle_count=5 Person_count=3\nFrame Number=1120 Number of Objects=8 Vehicle_count=5 Person_count=3\nFrame Number=1121 Number of Objects=10 Vehicle_count=7 Person_count=3\nFrame Number=1122 Number of Objects=11 Vehicle_count=8 Person_count=3\nFrame Number=1123 Number of Objects=12 Vehicle_count=9 Person_count=3\nFrame Number=1124 Number of Objects=15 Vehicle_count=12 Person_count=3\nFrame Number=1125 Number of Objects=12 Vehicle_count=9 Person_count=3\nFrame Number=1126 Number of Objects=14 Vehicle_count=11 Person_count=3\nFrame Number=1127 Number of Objects=14 Vehicle_count=11 Person_count=3\nFrame Number=1128 Number of Objects=13 Vehicle_count=10 Person_count=3\nFrame Number=1129 Number of Objects=13 Vehicle_count=10 Person_count=3\nFrame Number=1130 Number of Objects=13 Vehicle_count=10 Person_count=3\nFrame Number=1131 Number of Objects=15 Vehicle_count=12 Person_count=3\nFrame Number=1132 Number of Objects=14 Vehicle_count=11 Person_count=3\nFrame Number=1133 Number of Objects=13 Vehicle_count=10 Person_count=3\nFrame Number=1134 Number of Objects=16 Vehicle_count=13 Person_count=3\nFrame Number=1135 Number of Objects=16 Vehicle_count=13 Person_count=3\nFrame Number=1136 Number of Objects=16 Vehicle_count=13 Person_count=3\nFrame Number=1137 Number of Objects=16 Vehicle_count=13 Person_count=3\nFrame Number=1138 Number of Objects=15 Vehicle_count=12 Person_count=3\nFrame Number=1139 Number of Objects=15 Vehicle_count=12 Person_count=3\nFrame Number=1140 Number of Objects=15 Vehicle_count=12 Person_count=3\nFrame Number=1141 Number of Objects=14 Vehicle_count=11 Person_count=3\nFrame Number=1142 Number of Objects=14 Vehicle_count=11 Person_count=3\nFrame Number=1143 Number of Objects=15 Vehicle_count=12 Person_count=3\nFrame Number=1144 Number of Objects=14 Vehicle_count=11 Person_count=3\nFrame Number=1145 Number of Objects=13 Vehicle_count=10 Person_count=3\nFrame Number=1146 Number of Objects=13 Vehicle_count=10 Person_count=3\nFrame Number=1147 Number of Objects=12 Vehicle_count=9 Person_count=3\nFrame Number=1148 Number of Objects=12 Vehicle_count=9 Person_count=3\nFrame Number=1149 Number of Objects=11 Vehicle_count=8 Person_count=3\nFrame Number=1150 Number of Objects=9 Vehicle_count=6 Person_count=3\nFrame Number=1151 Number of Objects=10 Vehicle_count=7 Person_count=3\nFrame Number=1152 Number of Objects=9 Vehicle_count=6 Person_count=3\nFrame Number=1153 Number of Objects=11 Vehicle_count=8 Person_count=3\nFrame Number=1154 Number of Objects=9 Vehicle_count=6 Person_count=3\nFrame Number=1155 Number of Objects=10 Vehicle_count=7 Person_count=3\nFrame Number=1156 Number of Objects=10 Vehicle_count=7 Person_count=3\nFrame Number=1157 Number of Objects=10 Vehicle_count=8 Person_count=2\nFrame Number=1158 Number of Objects=8 Vehicle_count=5 Person_count=3\nFrame Number=1159 Number of Objects=9 Vehicle_count=6 Person_count=3\nFrame Number=1160 Number of Objects=8 Vehicle_count=5 Person_count=3\nFrame Number=1161 Number of Objects=11 Vehicle_count=8 Person_count=3\nFrame Number=1162 Number of Objects=12 Vehicle_count=9 Person_count=3\nFrame Number=1163 Number of Objects=12 Vehicle_count=9 Person_count=3\nFrame Number=1164 Number of Objects=10 Vehicle_count=7 Person_count=3\nFrame Number=1165 Number of Objects=11 Vehicle_count=8 Person_count=3\nFrame Number=1166 Number of Objects=11 Vehicle_count=8 Person_count=3\nFrame Number=1167 Number of Objects=11 Vehicle_count=8 Person_count=3\nFrame Number=1168 Number of Objects=10 Vehicle_count=7 Person_count=3\nFrame Number=1169 Number of Objects=10 Vehicle_count=7 Person_count=3\nFrame Number=1170 Number of Objects=10 Vehicle_count=7 Person_count=3\nFrame Number=1171 Number of Objects=10 Vehicle_count=7 Person_count=3\nFrame Number=1172 Number of Objects=12 Vehicle_count=9 Person_count=3\nFrame Number=1173 Number of Objects=10 Vehicle_count=8 Person_count=2\nFrame Number=1174 Number of Objects=14 Vehicle_count=11 Person_count=3\nFrame Number=1175 Number of Objects=14 Vehicle_count=12 Person_count=2\nFrame Number=1176 Number of Objects=15 Vehicle_count=12 Person_count=3\nFrame Number=1177 Number of Objects=13 Vehicle_count=11 Person_count=2\nFrame Number=1178 Number of Objects=12 Vehicle_count=10 Person_count=2\nFrame Number=1179 Number of Objects=13 Vehicle_count=11 Person_count=2\nFrame Number=1180 Number of Objects=12 Vehicle_count=10 Person_count=2\nFrame Number=1181 Number of Objects=13 Vehicle_count=10 Person_count=3\nFrame Number=1182 Number of Objects=12 Vehicle_count=10 Person_count=2\nFrame Number=1183 Number of Objects=10 Vehicle_count=8 Person_count=2\nFrame Number=1184 Number of Objects=10 Vehicle_count=8 Person_count=2\nFrame Number=1185 Number of Objects=10 Vehicle_count=8 Person_count=2\nFrame Number=1186 Number of Objects=10 Vehicle_count=8 Person_count=2\nFrame Number=1187 Number of Objects=11 Vehicle_count=9 Person_count=2\nFrame Number=1188 Number of Objects=10 Vehicle_count=8 Person_count=2\nFrame Number=1189 Number of Objects=10 Vehicle_count=7 Person_count=3\nFrame Number=1190 Number of Objects=9 Vehicle_count=7 Person_count=2\nFrame Number=1191 Number of Objects=9 Vehicle_count=6 Person_count=3\nFrame Number=1192 Number of Objects=9 Vehicle_count=6 Person_count=3\nFrame Number=1193 Number of Objects=8 Vehicle_count=6 Person_count=2\nFrame Number=1194 Number of Objects=8 Vehicle_count=5 Person_count=3\nFrame Number=1195 Number of Objects=9 Vehicle_count=6 Person_count=3\nFrame Number=1196 Number of Objects=9 Vehicle_count=6 Person_count=3\nFrame Number=1197 Number of Objects=8 Vehicle_count=5 Person_count=3\nFrame Number=1198 Number of Objects=9 Vehicle_count=6 Person_count=3\nFrame Number=1199 Number of Objects=8 Vehicle_count=5 Person_count=3\nFrame Number=1200 Number of Objects=9 Vehicle_count=6 Person_count=3\nFrame Number=1201 Number of Objects=8 Vehicle_count=5 Person_count=3\nFrame Number=1202 Number of Objects=8 Vehicle_count=5 Person_count=3\nFrame Number=1203 Number of Objects=8 Vehicle_count=5 Person_count=3\nFrame Number=1204 Number of Objects=9 Vehicle_count=6 Person_count=3\nFrame Number=1205 Number of Objects=7 Vehicle_count=4 Person_count=3\nFrame Number=1206 Number of Objects=9 Vehicle_count=5 Person_count=4\nFrame Number=1207 Number of Objects=8 Vehicle_count=5 Person_count=3\nFrame Number=1208 Number of Objects=9 Vehicle_count=5 Person_count=4\nFrame Number=1209 Number of Objects=10 Vehicle_count=6 Person_count=4\nFrame Number=1210 Number of Objects=10 Vehicle_count=7 Person_count=3\nFrame Number=1211 Number of Objects=12 Vehicle_count=8 Person_count=4\nFrame Number=1212 Number of Objects=15 Vehicle_count=11 Person_count=4\nFrame Number=1213 Number of Objects=17 Vehicle_count=14 Person_count=3\nFrame Number=1214 Number of Objects=13 Vehicle_count=10 Person_count=3\nFrame Number=1215 Number of Objects=14 Vehicle_count=11 Person_count=3\nFrame Number=1216 Number of Objects=15 Vehicle_count=12 Person_count=3\nFrame Number=1217 Number of Objects=14 Vehicle_count=11 Person_count=3\nFrame Number=1218 Number of Objects=15 Vehicle_count=11 Person_count=4\nFrame Number=1219 Number of Objects=14 Vehicle_count=11 Person_count=3\nFrame Number=1220 Number of Objects=13 Vehicle_count=10 Person_count=3\nFrame Number=1221 Number of Objects=16 Vehicle_count=13 Person_count=3\nFrame Number=1222 Number of Objects=15 Vehicle_count=12 Person_count=3\nFrame Number=1223 Number of Objects=14 Vehicle_count=11 Person_count=3\nFrame Number=1224 Number of Objects=12 Vehicle_count=9 Person_count=3\nFrame Number=1225 Number of Objects=14 Vehicle_count=11 Person_count=3\nFrame Number=1226 Number of Objects=12 Vehicle_count=9 Person_count=3\nFrame Number=1227 Number of Objects=11 Vehicle_count=8 Person_count=3\nFrame Number=1228 Number of Objects=10 Vehicle_count=7 Person_count=3\nFrame Number=1229 Number of Objects=9 Vehicle_count=6 Person_count=3\nFrame Number=1230 Number of Objects=9 Vehicle_count=6 Person_count=3\nFrame Number=1231 Number of Objects=7 Vehicle_count=4 Person_count=3\nFrame Number=1232 Number of Objects=9 Vehicle_count=6 Person_count=3\nFrame Number=1233 Number of Objects=8 Vehicle_count=5 Person_count=3\nFrame Number=1234 Number of Objects=10 Vehicle_count=6 Person_count=4\nFrame Number=1235 Number of Objects=9 Vehicle_count=6 Person_count=3\nFrame Number=1236 Number of Objects=10 Vehicle_count=6 Person_count=4\nFrame Number=1237 Number of Objects=11 Vehicle_count=7 Person_count=4\nFrame Number=1238 Number of Objects=9 Vehicle_count=5 Person_count=4\nFrame Number=1239 Number of Objects=8 Vehicle_count=4 Person_count=4\nFrame Number=1240 Number of Objects=8 Vehicle_count=4 Person_count=4\nFrame Number=1241 Number of Objects=8 Vehicle_count=4 Person_count=4\nFrame Number=1242 Number of Objects=8 Vehicle_count=5 Person_count=3\nFrame Number=1243 Number of Objects=7 Vehicle_count=4 Person_count=3\nFrame Number=1244 Number of Objects=8 Vehicle_count=4 Person_count=4\nFrame Number=1245 Number of Objects=12 Vehicle_count=8 Person_count=4\nFrame Number=1246 Number of Objects=15 Vehicle_count=11 Person_count=4\nFrame Number=1247 Number of Objects=13 Vehicle_count=9 Person_count=4\nFrame Number=1248 Number of Objects=11 Vehicle_count=7 Person_count=4\nFrame Number=1249 Number of Objects=12 Vehicle_count=8 Person_count=4\nFrame Number=1250 Number of Objects=13 Vehicle_count=9 Person_count=4\nFrame Number=1251 Number of Objects=13 Vehicle_count=9 Person_count=4\nFrame Number=1252 Number of Objects=13 Vehicle_count=9 Person_count=4\nFrame Number=1253 Number of Objects=13 Vehicle_count=9 Person_count=4\nFrame Number=1254 Number of Objects=13 Vehicle_count=9 Person_count=4\nFrame Number=1255 Number of Objects=14 Vehicle_count=10 Person_count=4\nFrame Number=1256 Number of Objects=13 Vehicle_count=9 Person_count=4\nFrame Number=1257 Number of Objects=12 Vehicle_count=8 Person_count=4\nFrame Number=1258 Number of Objects=12 Vehicle_count=8 Person_count=4\nFrame Number=1259 Number of Objects=12 Vehicle_count=8 Person_count=4\nFrame Number=1260 Number of Objects=13 Vehicle_count=8 Person_count=5\nFrame Number=1261 Number of Objects=12 Vehicle_count=7 Person_count=5\nFrame Number=1262 Number of Objects=13 Vehicle_count=8 Person_count=5\nFrame Number=1263 Number of Objects=12 Vehicle_count=8 Person_count=4\nFrame Number=1264 Number of Objects=13 Vehicle_count=9 Person_count=4\nFrame Number=1265 Number of Objects=14 Vehicle_count=9 Person_count=5\nFrame Number=1266 Number of Objects=14 Vehicle_count=10 Person_count=4\nFrame Number=1267 Number of Objects=13 Vehicle_count=9 Person_count=4\nFrame Number=1268 Number of Objects=13 Vehicle_count=10 Person_count=3\nFrame Number=1269 Number of Objects=14 Vehicle_count=10 Person_count=4\nFrame Number=1270 Number of Objects=13 Vehicle_count=10 Person_count=3\nFrame Number=1271 Number of Objects=12 Vehicle_count=9 Person_count=3\nFrame Number=1272 Number of Objects=14 Vehicle_count=11 Person_count=3\nFrame Number=1273 Number of Objects=13 Vehicle_count=10 Person_count=3\nFrame Number=1274 Number of Objects=12 Vehicle_count=9 Person_count=3\nFrame Number=1275 Number of Objects=11 Vehicle_count=8 Person_count=3\nFrame Number=1276 Number of Objects=12 Vehicle_count=9 Person_count=3\nFrame Number=1277 Number of Objects=13 Vehicle_count=9 Person_count=4\nFrame Number=1278 Number of Objects=13 Vehicle_count=9 Person_count=4\nFrame Number=1279 Number of Objects=12 Vehicle_count=9 Person_count=3\nFrame Number=1280 Number of Objects=12 Vehicle_count=8 Person_count=4\nFrame Number=1281 Number of Objects=14 Vehicle_count=10 Person_count=4\nFrame Number=1282 Number of Objects=12 Vehicle_count=8 Person_count=4\nFrame Number=1283 Number of Objects=14 Vehicle_count=10 Person_count=4\nFrame Number=1284 Number of Objects=13 Vehicle_count=9 Person_count=4\nFrame Number=1285 Number of Objects=12 Vehicle_count=8 Person_count=4\nFrame Number=1286 Number of Objects=11 Vehicle_count=8 Person_count=3\nFrame Number=1287 Number of Objects=11 Vehicle_count=8 Person_count=3\nFrame Number=1288 Number of Objects=10 Vehicle_count=7 Person_count=3\nFrame Number=1289 Number of Objects=10 Vehicle_count=7 Person_count=3\nFrame Number=1290 Number of Objects=10 Vehicle_count=7 Person_count=3\nFrame Number=1291 Number of Objects=10 Vehicle_count=7 Person_count=3\nFrame Number=1292 Number of Objects=10 Vehicle_count=7 Person_count=3\nFrame Number=1293 Number of Objects=9 Vehicle_count=5 Person_count=4\nFrame Number=1294 Number of Objects=10 Vehicle_count=5 Person_count=5\nFrame Number=1295 Number of Objects=10 Vehicle_count=5 Person_count=5\nFrame Number=1296 Number of Objects=7 Vehicle_count=4 Person_count=3\nFrame Number=1297 Number of Objects=7 Vehicle_count=4 Person_count=3\nFrame Number=1298 Number of Objects=7 Vehicle_count=4 Person_count=3\nFrame Number=1299 Number of Objects=6 Vehicle_count=3 Person_count=3\nFrame Number=1300 Number of Objects=9 Vehicle_count=5 Person_count=4\nFrame Number=1301 Number of Objects=7 Vehicle_count=4 Person_count=3\nFrame Number=1302 Number of Objects=8 Vehicle_count=4 Person_count=4\nFrame Number=1303 Number of Objects=8 Vehicle_count=4 Person_count=4\nFrame Number=1304 Number of Objects=7 Vehicle_count=3 Person_count=4\nFrame Number=1305 Number of Objects=6 Vehicle_count=2 Person_count=4\nFrame Number=1306 Number of Objects=6 Vehicle_count=2 Person_count=4\nFrame Number=1307 Number of Objects=8 Vehicle_count=5 Person_count=3\nFrame Number=1308 Number of Objects=9 Vehicle_count=5 Person_count=4\nFrame Number=1309 Number of Objects=10 Vehicle_count=6 Person_count=4\nFrame Number=1310 Number of Objects=10 Vehicle_count=6 Person_count=4\nFrame Number=1311 Number of Objects=9 Vehicle_count=5 Person_count=4\nFrame Number=1312 Number of Objects=10 Vehicle_count=6 Person_count=4\nFrame Number=1313 Number of Objects=10 Vehicle_count=6 Person_count=4\nFrame Number=1314 Number of Objects=12 Vehicle_count=9 Person_count=3\nFrame Number=1315 Number of Objects=11 Vehicle_count=8 Person_count=3\nFrame Number=1316 Number of Objects=11 Vehicle_count=8 Person_count=3\nFrame Number=1317 Number of Objects=14 Vehicle_count=11 Person_count=3\nFrame Number=1318 Number of Objects=12 Vehicle_count=9 Person_count=3\nFrame Number=1319 Number of Objects=13 Vehicle_count=10 Person_count=3\nFrame Number=1320 Number of Objects=12 Vehicle_count=9 Person_count=3\nFrame Number=1321 Number of Objects=13 Vehicle_count=10 Person_count=3\nFrame Number=1322 Number of Objects=14 Vehicle_count=11 Person_count=3\nFrame Number=1323 Number of Objects=15 Vehicle_count=12 Person_count=3\nFrame Number=1324 Number of Objects=17 Vehicle_count=14 Person_count=3\nFrame Number=1325 Number of Objects=15 Vehicle_count=12 Person_count=3\nFrame Number=1326 Number of Objects=15 Vehicle_count=12 Person_count=3\nFrame Number=1327 Number of Objects=13 Vehicle_count=10 Person_count=3\nFrame Number=1328 Number of Objects=13 Vehicle_count=10 Person_count=3\nFrame Number=1329 Number of Objects=14 Vehicle_count=11 Person_count=3\nFrame Number=1330 Number of Objects=14 Vehicle_count=11 Person_count=3\nFrame Number=1331 Number of Objects=15 Vehicle_count=12 Person_count=3\nFrame Number=1332 Number of Objects=14 Vehicle_count=11 Person_count=3\nFrame Number=1333 Number of Objects=14 Vehicle_count=11 Person_count=3\nFrame Number=1334 Number of Objects=15 Vehicle_count=12 Person_count=3\nFrame Number=1335 Number of Objects=14 Vehicle_count=11 Person_count=3\nFrame Number=1336 Number of Objects=13 Vehicle_count=10 Person_count=3\nFrame Number=1337 Number of Objects=14 Vehicle_count=11 Person_count=3\nFrame Number=1338 Number of Objects=13 Vehicle_count=10 Person_count=3\nFrame Number=1339 Number of Objects=12 Vehicle_count=9 Person_count=3\nFrame Number=1340 Number of Objects=12 Vehicle_count=9 Person_count=3\nFrame Number=1341 Number of Objects=10 Vehicle_count=7 Person_count=3\nFrame Number=1342 Number of Objects=8 Vehicle_count=5 Person_count=3\nFrame Number=1343 Number of Objects=9 Vehicle_count=6 Person_count=3\nFrame Number=1344 Number of Objects=9 Vehicle_count=6 Person_count=3\nFrame Number=1345 Number of Objects=9 Vehicle_count=6 Person_count=3\nFrame Number=1346 Number of Objects=9 Vehicle_count=6 Person_count=3\nFrame Number=1347 Number of Objects=8 Vehicle_count=5 Person_count=3\nFrame Number=1348 Number of Objects=8 Vehicle_count=5 Person_count=3\nFrame Number=1349 Number of Objects=7 Vehicle_count=4 Person_count=3\nFrame Number=1350 Number of Objects=7 Vehicle_count=4 Person_count=3\nFrame Number=1351 Number of Objects=7 Vehicle_count=4 Person_count=3\nFrame Number=1352 Number of Objects=7 Vehicle_count=4 Person_count=3\nFrame Number=1353 Number of Objects=7 Vehicle_count=4 Person_count=3\nFrame Number=1354 Number of Objects=8 Vehicle_count=5 Person_count=3\nFrame Number=1355 Number of Objects=7 Vehicle_count=4 Person_count=3\nFrame Number=1356 Number of Objects=6 Vehicle_count=4 Person_count=2\nFrame Number=1357 Number of Objects=7 Vehicle_count=5 Person_count=2\nFrame Number=1358 Number of Objects=8 Vehicle_count=6 Person_count=2\nFrame Number=1359 Number of Objects=7 Vehicle_count=5 Person_count=2\nFrame Number=1360 Number of Objects=8 Vehicle_count=6 Person_count=2\nFrame Number=1361 Number of Objects=11 Vehicle_count=9 Person_count=2\nFrame Number=1362 Number of Objects=11 Vehicle_count=9 Person_count=2\nFrame Number=1363 Number of Objects=10 Vehicle_count=8 Person_count=2\nFrame Number=1364 Number of Objects=13 Vehicle_count=11 Person_count=2\nFrame Number=1365 Number of Objects=11 Vehicle_count=9 Person_count=2\nFrame Number=1366 Number of Objects=11 Vehicle_count=9 Person_count=2\nFrame Number=1367 Number of Objects=13 Vehicle_count=11 Person_count=2\nFrame Number=1368 Number of Objects=11 Vehicle_count=9 Person_count=2\nFrame Number=1369 Number of Objects=11 Vehicle_count=9 Person_count=2\nFrame Number=1370 Number of Objects=11 Vehicle_count=9 Person_count=2\nFrame Number=1371 Number of Objects=12 Vehicle_count=10 Person_count=2\nFrame Number=1372 Number of Objects=12 Vehicle_count=10 Person_count=2\nFrame Number=1373 Number of Objects=12 Vehicle_count=9 Person_count=3\nFrame Number=1374 Number of Objects=14 Vehicle_count=11 Person_count=3\nFrame Number=1375 Number of Objects=13 Vehicle_count=10 Person_count=3\nFrame Number=1376 Number of Objects=12 Vehicle_count=10 Person_count=2\nFrame Number=1377 Number of Objects=13 Vehicle_count=11 Person_count=2\nFrame Number=1378 Number of Objects=15 Vehicle_count=13 Person_count=2\nFrame Number=1379 Number of Objects=13 Vehicle_count=10 Person_count=3\nFrame Number=1380 Number of Objects=13 Vehicle_count=10 Person_count=3\nFrame Number=1381 Number of Objects=14 Vehicle_count=11 Person_count=3\nFrame Number=1382 Number of Objects=13 Vehicle_count=10 Person_count=3\nFrame Number=1383 Number of Objects=13 Vehicle_count=10 Person_count=3\nFrame Number=1384 Number of Objects=11 Vehicle_count=8 Person_count=3\nFrame Number=1385 Number of Objects=11 Vehicle_count=9 Person_count=2\nFrame Number=1386 Number of Objects=10 Vehicle_count=8 Person_count=2\nFrame Number=1387 Number of Objects=12 Vehicle_count=10 Person_count=2\nFrame Number=1388 Number of Objects=10 Vehicle_count=8 Person_count=2\nFrame Number=1389 Number of Objects=10 Vehicle_count=8 Person_count=2\nFrame Number=1390 Number of Objects=10 Vehicle_count=8 Person_count=2\nFrame Number=1391 Number of Objects=11 Vehicle_count=9 Person_count=2\nFrame Number=1392 Number of Objects=11 Vehicle_count=9 Person_count=2\nFrame Number=1393 Number of Objects=11 Vehicle_count=9 Person_count=2\nFrame Number=1394 Number of Objects=11 Vehicle_count=8 Person_count=3\nFrame Number=1395 Number of Objects=11 Vehicle_count=8 Person_count=3\nFrame Number=1396 Number of Objects=11 Vehicle_count=8 Person_count=3\nFrame Number=1397 Number of Objects=12 Vehicle_count=9 Person_count=3\nFrame Number=1398 Number of Objects=11 Vehicle_count=8 Person_count=3\nFrame Number=1399 Number of Objects=11 Vehicle_count=8 Person_count=3\nFrame Number=1400 Number of Objects=10 Vehicle_count=8 Person_count=2\nFrame Number=1401 Number of Objects=12 Vehicle_count=9 Person_count=3\nFrame Number=1402 Number of Objects=10 Vehicle_count=7 Person_count=3\nFrame Number=1403 Number of Objects=10 Vehicle_count=8 Person_count=2\nFrame Number=1404 Number of Objects=10 Vehicle_count=8 Person_count=2\nFrame Number=1405 Number of Objects=11 Vehicle_count=9 Person_count=2\nFrame Number=1406 Number of Objects=10 Vehicle_count=8 Person_count=2\nFrame Number=1407 Number of Objects=10 Vehicle_count=8 Person_count=2\nFrame Number=1408 Number of Objects=8 Vehicle_count=6 Person_count=2\nFrame Number=1409 Number of Objects=7 Vehicle_count=5 Person_count=2\nFrame Number=1410 Number of Objects=8 Vehicle_count=6 Person_count=2\nFrame Number=1411 Number of Objects=7 Vehicle_count=5 Person_count=2\nFrame Number=1412 Number of Objects=9 Vehicle_count=7 Person_count=2\nFrame Number=1413 Number of Objects=8 Vehicle_count=6 Person_count=2\nFrame Number=1414 Number of Objects=6 Vehicle_count=4 Person_count=2\nFrame Number=1415 Number of Objects=7 Vehicle_count=5 Person_count=2\nFrame Number=1416 Number of Objects=8 Vehicle_count=5 Person_count=3\nFrame Number=1417 Number of Objects=6 Vehicle_count=3 Person_count=3\nFrame Number=1418 Number of Objects=6 Vehicle_count=3 Person_count=3\nFrame Number=1419 Number of Objects=8 Vehicle_count=5 Person_count=3\nFrame Number=1420 Number of Objects=8 Vehicle_count=5 Person_count=3\nFrame Number=1421 Number of Objects=6 Vehicle_count=3 Person_count=3\nFrame Number=1422 Number of Objects=8 Vehicle_count=5 Person_count=3\nFrame Number=1423 Number of Objects=8 Vehicle_count=5 Person_count=3\nFrame Number=1424 Number of Objects=7 Vehicle_count=4 Person_count=3\nFrame Number=1425 Number of Objects=8 Vehicle_count=5 Person_count=3\nFrame Number=1426 Number of Objects=8 Vehicle_count=5 Person_count=3\nFrame Number=1427 Number of Objects=11 Vehicle_count=8 Person_count=3\nFrame Number=1428 Number of Objects=10 Vehicle_count=7 Person_count=3\nFrame Number=1429 Number of Objects=11 Vehicle_count=8 Person_count=3\nFrame Number=1430 Number of Objects=9 Vehicle_count=6 Person_count=3\nFrame Number=1431 Number of Objects=10 Vehicle_count=7 Person_count=3\nFrame Number=1432 Number of Objects=12 Vehicle_count=9 Person_count=3\nFrame Number=1433 Number of Objects=12 Vehicle_count=9 Person_count=3\nFrame Number=1434 Number of Objects=12 Vehicle_count=9 Person_count=3\nFrame Number=1435 Number of Objects=11 Vehicle_count=8 Person_count=3\nFrame Number=1436 Number of Objects=11 Vehicle_count=8 Person_count=3\nFrame Number=1437 Number of Objects=10 Vehicle_count=7 Person_count=3\nFrame Number=1438 Number of Objects=10 Vehicle_count=7 Person_count=3\nFrame Number=1439 Number of Objects=12 Vehicle_count=9 Person_count=3\nFrame Number=1440 Number of Objects=12 Vehicle_count=9 Person_count=3\nFrame Number=1441 Number of Objects=0 Vehicle_count=0 Person_count=0\nEnd-of-stream\n--- 24.878137350082397 seconds ---\n"
]
],
[
[
"This video output is not compatible to be shown in this Notebook. To circumvent this, we convert the output in a Jupyter Notebook-readable format. For this we use the shell command `ffmpeg`.",
"_____no_output_____"
]
],
[
[
"# Convert video profile to be compatible with the Notebook\n!ffmpeg -loglevel panic -y -an -i ../videos/out.mp4 -vcodec libx264 -pix_fmt yuv420p -profile:v baseline -level 3 ../videos/output.mp4",
"_____no_output_____"
]
],
[
[
"Finally, we display the output in the notbook by creating an HTML video element.",
"_____no_output_____"
]
],
[
[
"# Display the Output\nfrom IPython.display import HTML\nHTML(\"\"\"\n <video width=\"640\" height=\"480\" controls>\n <source src=\"../videos/output.mp4\"\n </video>\n\"\"\".format())",
"_____no_output_____"
]
],
[
[
"\nIn the next notebook , we will learn about object tracking and build an attribute classification pipeline along with the primary inference built in this notebook.\n",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
c5003024b999b2392cdad609d6117a27002b4721
| 139,618 |
ipynb
|
Jupyter Notebook
|
docs/source/use_cases/customer_segmentation.ipynb
|
zhxt95/featuretools
|
4fd90031e2b525ddaacddf46ff06a58103dbca35
|
[
"BSD-3-Clause"
] | null | null | null |
docs/source/use_cases/customer_segmentation.ipynb
|
zhxt95/featuretools
|
4fd90031e2b525ddaacddf46ff06a58103dbca35
|
[
"BSD-3-Clause"
] | null | null | null |
docs/source/use_cases/customer_segmentation.ipynb
|
zhxt95/featuretools
|
4fd90031e2b525ddaacddf46ff06a58103dbca35
|
[
"BSD-3-Clause"
] | null | null | null | 750.634409 | 122,418 | 0.937028 |
[
[
[
"# Customer segmentation using k-means clustering\n\nBelow is a demo applying automated feature engineering to a retail dataset to automatically segment customers based on historical behavior",
"_____no_output_____"
]
],
[
[
"import featuretools as ft\nimport pandas as pd",
"_____no_output_____"
],
[
"retail_es = ft.demo.load_retail()",
"_____no_output_____"
]
],
[
[
"## Use Deep Feature Synthesis\n\nThe input to DFS is a set of entities and a list of relationships (defined by our EntitySet) and the \"target_entity\" to calculate features for. We can supply \"cutoff times\" to specify the that we want to calculate features one year after a customers first invoices.\n\nThe ouput of DFS is a feature matrix and the corresponding list of feature defintions",
"_____no_output_____"
]
],
[
[
"from featuretools.primitives import AvgTimeBetween, Mean, Sum, Count, Day\n\nfeature_matrix, features = ft.dfs(entityset=retail_es, target_entity=\"customers\",\n agg_primitives=[AvgTimeBetween, Mean, Sum, Count],\n trans_primitives=[Day], max_depth=5, verbose=True)\n\nfeature_matrix, features = ft.encode_features(feature_matrix, features)",
"calulate_feature_matrix: 100%|██████████| 2/2 [01:23<00:00, 41.92s/it]\n"
],
[
"from sklearn import decomposition\nfrom sklearn.preprocessing import Imputer, StandardScaler\n# Create our imputer to replace missing values with the mean e.g.\nimp = Imputer(missing_values='NaN', strategy='mean', axis=0)\nX = imp.fit_transform(feature_matrix.values)\nscale = StandardScaler()\nX = scale.fit_transform(X)\npca = decomposition.PCA(n_components=10)\nX_pca = pca.fit_transform(X)\n\nfrom sklearn import preprocessing, manifold\n# Reduce dimension to 2-D\ntsne = manifold.TSNE(n_components=2)\nX_2_dim = tsne.fit_transform(X_pca)",
"_____no_output_____"
],
[
"from sklearn import preprocessing, decomposition, cluster\nimport matplotlib.pyplot as plt\nplt.style.use('ggplot')\nimport seaborn as sns\nimport matplotlib.pyplot as plt\nimport matplotlib as mpl\n%matplotlib inline\n\n# do kmeans clustering\nn_clusters=10\nclust = cluster.MiniBatchKMeans(n_clusters=n_clusters,\n reassignment_ratio=.03,\n batch_size=1000,\n n_init=100)\ncluster_labels = clust.fit_predict(X_pca)\n\n# plot cluster sizes\nplt.hist(cluster_labels, bins=range(n_clusters+1))\nplt.title('# Customers per Cluster')\nplt.xlabel('Cluster')\nplt.ylabel('# Customers')\nplt.show()",
"_____no_output_____"
],
[
"df = pd.DataFrame(X_2_dim, columns=[\"x\", \"y\"])\ndf['color'] = cluster_labels\n\nsns.set(font_scale=1.2) \ng = sns.lmplot(\"x\", \"y\", data=df, hue='color',\n fit_reg=False, size=8, palette=\"hls\", legend=False)\n\ng.set(yticks=[], xticks=[], xlabel=\"\", ylabel=\"\")\nplt.title(\"Customer Clusters\")\nplt.show()",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
c5003525ec4172f3ad57b3c63465b76ecde5dbca
| 15,785 |
ipynb
|
Jupyter Notebook
|
demo/FineTuning.ipynb
|
KirillVladimirov/pytorch-lifestream
|
83005b950d41de8afc11711fc955ffafb5ff7a9e
|
[
"Apache-2.0"
] | null | null | null |
demo/FineTuning.ipynb
|
KirillVladimirov/pytorch-lifestream
|
83005b950d41de8afc11711fc955ffafb5ff7a9e
|
[
"Apache-2.0"
] | null | null | null |
demo/FineTuning.ipynb
|
KirillVladimirov/pytorch-lifestream
|
83005b950d41de8afc11711fc955ffafb5ff7a9e
|
[
"Apache-2.0"
] | null | null | null | 26.134106 | 181 | 0.489199 |
[
[
[
"from IPython.core.display import HTML, display\ndisplay(HTML(\"<style>.container { width:100% !important; }</style>\"))",
"/tmp/ipykernel_17014/1410932729.py:1: DeprecationWarning: Importing display from IPython.core.display is deprecated since IPython 7.14, please import from IPython display\n from IPython.core.display import HTML, display\n"
],
[
"%load_ext autoreload\n%autoreload 2\n\nimport sys\nsys.path.append('..')\n\nimport logging\nimport pytorch_lightning as pl\nimport warnings\n\nwarnings.filterwarnings('ignore')\nlogging.getLogger(\"pytorch_lightning\").setLevel(logging.ERROR)",
"_____no_output_____"
]
],
[
[
"## Data load",
"_____no_output_____"
]
],
[
[
"! mkdir ../../data\n! curl -OL https://storage.googleapis.com/di-datasets/age-prediction-nti-sbebank-2019.zip\n! unzip -j -o age-prediction-nti-sbebank-2019.zip 'data/*.csv' -d ../../data\n! mv age-prediction-nti-sbebank-2019.zip ../../data/",
"mkdir: cannot create directory ‘../../data’: File exists\n % Total % Received % Xferd Average Speed Time Time Time Current\n Dload Upload Total Spent Left Speed\n100 239M 100 239M 0 0 198M 0 0:00:01 0:00:01 --:--:-- 197M\nArchive: age-prediction-nti-sbebank-2019.zip\n inflating: ../../data/test.csv \n inflating: ../../data/small_group_description.csv \n inflating: ../../data/train_target.csv \n inflating: ../../data/transactions_train.csv \n inflating: ../../data/transactions_test.csv \n"
]
],
[
[
"## Data Preproccessing",
"_____no_output_____"
]
],
[
[
"import os\nimport pandas as pd\n\ndata_path = '../../data/'\n\nsource_data = pd.read_csv(os.path.join(data_path, 'transactions_train.csv'))\nsource_data.head(2)",
"_____no_output_____"
],
[
"from dltranz.data_preprocessing import PandasDataPreprocessor\n\npreprocessor = PandasDataPreprocessor(\n col_id='client_id',\n cols_event_time='trans_date',\n time_transformation='float',\n cols_category=[\"trans_date\", \"small_group\"],\n cols_log_norm=[\"amount_rur\"],\n print_dataset_info=False,\n)",
"_____no_output_____"
],
[
"# Split data into train and finetuning parts:\n\nmetric_learn_data, finetune_data = train_test_split(source_data, test_size=0.5, random_state=42)",
"_____no_output_____"
],
[
"%%time\n\nimport pickle\n\npreproc_fitted = preprocessor.fit(metric_learn_data)\n\n# Save preprocessor:\n# with open('preproc_fitted.pickle', 'wb') as handle:\n# pickle.dump(preproc_fitted, handle, protocol=pickle.HIGHEST_PROTOCOL)\n \ndataset = preproc_fitted.transform(metric_learn_data)\n",
"CPU times: user 42.7 s, sys: 8.74 s, total: 51.4 s\nWall time: 51.3 s\n"
],
[
"from sklearn.model_selection import train_test_split\n\ntrain, test = train_test_split(dataset, test_size=0.2, random_state=42)\n\nprint(len(train), len(test))",
"24000 6000\n"
]
],
[
[
"## Embedding training",
"_____no_output_____"
],
[
"Model training in our framework organised via pytorch-lightning (pl) framework.\nThe key parts of neural networks training in pl are: \n\n * model (pl.LightningModule)\n * data_module (pl.LightningDataModule)\n * pl.trainer (pl.trainer)\n \nFor futher details check https://www.pytorchlightning.ai/",
"_____no_output_____"
],
[
"### model ",
"_____no_output_____"
]
],
[
[
"from dltranz.seq_encoder import SequenceEncoder\nfrom dltranz.models import Head\nfrom dltranz.lightning_modules.emb_module import EmbModule\n\nseq_encoder = SequenceEncoder(\n category_features=preprocessor.get_category_sizes(),\n numeric_features=[\"amount_rur\"],\n trx_embedding_noize=0.003\n)\n\nhead = Head(input_size=seq_encoder.embedding_size, use_norm_encoder=True)\n\nmodel = EmbModule(seq_encoder=seq_encoder, head=head)",
"_____no_output_____"
]
],
[
[
"### Data module",
"_____no_output_____"
]
],
[
[
"from dltranz.data_load.data_module.emb_data_module import EmbeddingTrainDataModule\n\ndm = EmbeddingTrainDataModule(\n dataset=train,\n pl_module=model,\n min_seq_len=25,\n seq_split_strategy='SampleSlices',\n category_names = model.seq_encoder.category_names,\n category_max_size = model.seq_encoder.category_max_size,\n split_count=5,\n split_cnt_min=25,\n split_cnt_max=200,\n train_num_workers=16,\n train_batch_size=256,\n valid_num_workers=16,\n valid_batch_size=256\n)",
"_____no_output_____"
]
],
[
[
"### Trainer",
"_____no_output_____"
]
],
[
[
"import torch\nimport pytorch_lightning as pl\n\nimport logging\n# logging.getLogger(\"lightning\").addHandler(logging.NullHandler())\n# logging.getLogger(\"lightning\").propagate = False\n\ntrainer = pl.Trainer(\n# progress_bar_refresh_rate=0,\n max_epochs=10,\n gpus=1 if torch.cuda.is_available() else 0\n)",
"_____no_output_____"
]
],
[
[
"### Training ",
"_____no_output_____"
]
],
[
[
"%%time\n\ntrainer.fit(model, dm)",
"_____no_output_____"
]
],
[
[
"## FineTuning",
"_____no_output_____"
]
],
[
[
"from pyhocon import ConfigFactory\nfrom dltranz.seq_to_target import SequenceToTarget\n\n\nclass SeqToTargetDemo(SequenceToTarget):\n def __init__(self,\n seq_encoder = None,\n encoder_lr: float = 0.0001,\n in_features: int = 256,\n out_features: int = 1,\n head_lr: float = 0.005,\n weight_decay: float = 0.0,\n lr_step_size: int = 1,\n lr_step_gamma: float = 0.60): \n \n params = {\n 'score_metric': ['auroc', 'accuracy'],\n\n 'encoder_type': 'pretrained',\n 'pretrained': {\n 'pl_module_class': 'dltranz.lightning_modules.coles_module.CoLESModule',\n 'lr': encoder_lr\n },\n\n 'head_layers': [\n ['BatchNorm1d', {'num_features': in_features}],\n ['Linear', {\"in_features\": in_features, \"out_features\": out_features}],\n ['Sigmoid', {}],\n ['Squeeze', {}]\n ],\n\n 'train': {\n 'random_neg': 'false',\n 'loss': 'bce',\n 'lr': head_lr,\n 'weight_decay': weight_decay,\n },\n 'lr_scheduler': {\n 'step_size': lr_step_size,\n 'step_gamma': lr_step_gamma\n }\n }\n super().__init__(ConfigFactory.from_dict(params), seq_encoder)\n\n\npretrained_encoder = model.seq_encoder\ndownstream_model = SeqToTargetDemo(pretrained_encoder,\n encoder_lr=0.0001,\n in_features=model.seq_encoder.embedding_size,\n out_features=1,\n head_lr=0.05,\n weight_decay=0.0,\n lr_step_size=1,\n lr_step_gamma=0.60)\n",
"_____no_output_____"
],
[
"finetune_dataset = preproc_fitted.transform(finetune_data)\n\nfinetune_dm = EmbeddingTrainDataModule(\n dataset=finetune_dataset,\n pl_module=downstream_model,\n min_seq_len=25,\n seq_split_strategy='SampleSlices',\n category_names = model.seq_encoder.category_names,\n category_max_size = model.seq_encoder.category_max_size,\n split_count=5,\n split_cnt_min=25,\n split_cnt_max=200,\n train_num_workers=16,\n train_batch_size=256,\n valid_num_workers=16,\n valid_batch_size=256\n)",
"_____no_output_____"
],
[
"# trainer.fit(downstream_model, dm)",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
c50035a39518878470fdfaba8cd080de45c51471
| 65,715 |
ipynb
|
Jupyter Notebook
|
.ipynb_checkpoints/Radar_analysis-checkpoint.ipynb
|
guilistocco/Udacity-Car_Radar_Lanes
|
b9b43ffe5032c8e0d84b5f855a953cde3c6f6bc8
|
[
"MIT"
] | 1 |
2021-01-05T14:31:16.000Z
|
2021-01-05T14:31:16.000Z
|
.ipynb_checkpoints/Radar_analysis-checkpoint.ipynb
|
guilistocco/Udacity-Car_Radar_Lanes
|
b9b43ffe5032c8e0d84b5f855a953cde3c6f6bc8
|
[
"MIT"
] | null | null | null |
.ipynb_checkpoints/Radar_analysis-checkpoint.ipynb
|
guilistocco/Udacity-Car_Radar_Lanes
|
b9b43ffe5032c8e0d84b5f855a953cde3c6f6bc8
|
[
"MIT"
] | null | null | null | 55.362258 | 18,252 | 0.634847 |
[
[
[
"# Importing libaries",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nfrom sklearn.linear_model import LinearRegression, BayesianRidge\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.metrics import mean_squared_error, r2_score\nfrom sklearn import linear_model\n\n",
"_____no_output_____"
]
],
[
[
"# Importing first databases\n\nRadars.csv contains all cars, trucks, motorcycles and buses that comes thru São Paulo's radar system \n",
"_____no_output_____"
]
],
[
[
"df_base = pd.read_csv(r\"D:\\\\Users\\\\guilh\\\\Documents\\\\GitHub\\\\Dados_CET\\\\Marco_2018_nAg\\\\2_nAg.csv\", index_col= \"Data\")\n",
"_____no_output_____"
],
[
"df_base.head()",
"_____no_output_____"
]
],
[
[
"### Reading the columns\n\n* **Radar** is the number of identification of a street section\n* **Lane** goes from 1 to 6 in most radars, low lane number are closer to the center of the freeway, high lane numbers are \"local\" lanes, to the right\n* **Register** represents each vehicle\n* **Types** are: motorcycle = 0, car = 1, bus = 2 ou truck = 3\n* **Classes** are: *light* (motorcycle and car) = 0 ou *heavy* (bus and truck) = 1\n* **Speeds** are in kilometer per hour\n* **Radar_Lane** comes to identify each lane on a single radar (will be usefull to merge dataframes)\n",
"_____no_output_____"
]
],
[
[
"# Preprocessing\n\ndf = df_base[[\"Numero Agrupado\", \"Faixa\", \"Registro\", \"Especie\", \"Classe\", \"Velocidade\"]]\n# turns speed from dm/s to km/h\ndf[\"Velocidade\"] = df[\"Velocidade\"] * 0.36\n\ndf.index.names = [\"Date\"]\n\ndf[\"Radar_Lane\"] = df[\"Numero Agrupado\"].astype(str) + df[\"Faixa\"].astype(str)\n\n# renaming columns to english\ndf.columns = [\"Radar\", \"Lane\", \"Register\", \"Type\", \"Class\", \"Speed [km/h]\", \"Radar_Lane\"]\n\ndf.head()",
"<ipython-input-4-35ba3737c1f8>:5: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\n df[\"Velocidade\"] = df[\"Velocidade\"] * 0.36\n<ipython-input-4-35ba3737c1f8>:9: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\n df[\"Radar_Lane\"] = df[\"Numero Agrupado\"].astype(str) + df[\"Faixa\"].astype(str)\n"
]
],
[
[
"### Lane types database\n\nHelps to tell the **use of each lane** . \n\n\"Tipo\" contains the information of lanes where all types of vehicycles can use ( *mix_use* ) and other that are for buses only ( *exclusive_bus* )",
"_____no_output_____"
]
],
[
[
"lane_types = pd.read_excel(r\"D:\\Users\\guilh\\Documents\\[POLI]_6_Semestre\\IC\\2021\\codigos olimpio\\\\Faixa Tipo.xlsx\", usecols = [\"Num_agrupado\",\"faixa\", \"Num_fx\",\"tipo\"],engine='openpyxl')\n",
"_____no_output_____"
],
[
"lane_types.head()",
"_____no_output_____"
]
],
[
[
"### Merge dataframes\n\nTo identify the type of the lane, if it is exclusive for buses, or multipurpose",
"_____no_output_____"
]
],
[
[
"df_merged = lane_types[[\"Num_fx\", \"tipo\"]].merge(df, left_on = \"Num_fx\", right_on = \"Radar_Lane\", how=\"right\")\n\ndf_merged[\"Lane_use\"] = df_merged[\"tipo\"].map({\"mista\":\"mix_use\", \"onibus\": \"exclusive_bus\"})\n\ndf_merged = df_merged[[\"Radar\", \"Lane\", \"Register\", \"Type\", \"Class\", \"Speed [km/h]\", \"Lane_use\"]]\n\ndf_merged.head()",
"_____no_output_____"
]
],
[
[
"### Looking for NaNs\n\nAs shown below, NaNs are less than 1% (actually, less than 0,2%)\n\nWith this information, there will be low loss in dropping NaNs\n\n",
"_____no_output_____"
]
],
[
[
"print(df_merged.isna().mean() *100)\n\ndf_merged.dropna(inplace=True)",
"Radar 0.000000\nLane 0.000000\nRegister 0.000000\nType 0.185455\nClass 0.185455\nSpeed [km/h] 0.185455\nLane_use 0.000000\ndtype: float64\n"
]
],
[
[
"### Selection of Lanes\n\nUsing only the data from mix_use lanes, select for each lane to create comparison\n\nThe max numper of lanes is 6, but only few roads have all 6, so it can be excluded from the analysis",
"_____no_output_____"
]
],
[
[
"lanes = df_merged.loc[df_merged[\"Lane_use\"] == \"mix_use\"]\n\nlane_1 = lanes.loc[lanes[\"Lane\"] == 1]\n\nlane_2 = lanes.loc[lanes[\"Lane\"] == 2]\n\nlane_3 = lanes.loc[lanes[\"Lane\"] == 3]\n\nlane_4 = lanes.loc[lanes[\"Lane\"] == 4]\n\nlane_5 = lanes.loc[lanes[\"Lane\"] == 5]\n\nlane_6 = lanes.loc[lanes[\"Lane\"] == 6]\n\nprint(lane_1.shape, lane_2.shape, lane_3.shape, lane_4.shape, lane_5.shape, lane_6.shape)",
"(364148, 7) (586260, 7) (436164, 7) (167320, 7) (22417, 7) (6075, 7)\n"
]
],
[
[
"### Plotting the means\n\n",
"_____no_output_____"
]
],
[
[
"means = []\n\nfor lane in [lane_1,lane_2,lane_3,lane_4,lane_5]:\n means.append(lane[\"Speed [km/h]\"].mean())\n\nmeans = [ round(elem, 2) for elem in means ]\n\n\nfig, ax = plt.subplots()\n\nrects = ax.bar([1,2,3,4,5],means, width= 0.5)\n\nax.set_ylabel(\"Speed [km/h]\")\nax.set_xlabel(\"Lanes\")\nax.set_title('Speeds per lane')\n\n\ndef autolabel(rects):\n \"\"\"Attach a text label above each bar in *rects*, displaying its height.\"\"\"\n for rect in rects:\n height = rect.get_height()\n ax.annotate('{}'.format(height),\n xy=(rect.get_x() + rect.get_width() / 2, height),\n xytext=(0, 3), # 3 points vertical offset\n textcoords=\"offset points\",\n ha='center', va='bottom')\n\nautolabel(rects)\n\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"# How can we predicti a new car?",
"_____no_output_____"
]
],
[
[
"df_regression = df_base[[\"Numero Agrupado\", \"Faixa\", \"Registro\", \"Especie\", \"Classe\", \"Velocidade\", \"Comprimento\"]]\n\ndf_regression.loc[:,\"Comprimento\"] = df_regression.loc[:,\"Comprimento\"] /10\n\ndf_regression.loc[:,\"Velocidade\"] = df_regression.loc[:,\"Velocidade\"] * 0.36\n",
"_____no_output_____"
]
],
[
[
"### Reading the columns refazerrrrrrrrrrrrrrrr\n\n\n* **Radar** is the number of identification of a street section\n* **Lane** goes from 1 to 6 in most radars, low lane number are closer to the center of the freeway, high lane numbers are \"local\" lanes, to the right\n* **Register** represents each vehicle\n* **Types** are: motorcycle = 0, car = 1, bus = 2 ou truck = 3\n* **Classes** are: *light* (motorcycle and car) = 0 ou *heavy* (bus and truck) = 1\n* **Speeds** are in kilometer per hour\n* **Radar_Lane** comes to identify each lane on a single radar (will be usefull to merge dataframes)\n",
"_____no_output_____"
]
],
[
[
"df_regression.columns = [\"Radar\", \"Lane\", \"Register\", \"Type\", \"Class\", \"Speed [km/h]\", \"Length\"]\n\n\nValidation = df_regression.loc[df_regression[\"Speed [km/h]\"].isna()]\nX = df_regression[[\"Lane\", \"Type\", \"Class\", \"Length\"]].dropna()\nX = pd.concat([pd.get_dummies(X[[\"Lane\", \"Type\", \"Class\"]].astype(\"object\")),X[\"Length\"]], axis=1)\n\ny = df_regression[\"Speed [km/h]\"].dropna()\n\n\nX.head()",
"_____no_output_____"
],
[
"X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=42)",
"_____no_output_____"
],
[
"lr = LinearRegression(normalize=True)\n\nlr.fit(X_train, y_train)\n\npred = lr.predict(X_test)\n\nprint(lr.score(X_train, y_train),lr.score(X_test, y_test))",
"0.037942678575454525 0.03946052183332238\n"
],
[
"corrMatrix = df_regression[[\"Lane\", \"Type\", \"Speed [km/h]\",\"Length\"]].corr()\ndisplay (corrMatrix)",
"_____no_output_____"
],
[
"sns.heatmap(corrMatrix, annot=True)\nplt.show()",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
c50038532de0e2866c14d85591049f10a68e78f7
| 946,863 |
ipynb
|
Jupyter Notebook
|
notebooks/01.01_bb_EDA_augmented_emotion_classification_2020_05_13.ipynb
|
bhattbhuwan13/fuseai-training
|
2739c8bd51f5ab6ad8e765184ecae1d1c2881945
|
[
"FTL"
] | 1 |
2020-08-13T04:27:08.000Z
|
2020-08-13T04:27:08.000Z
|
notebooks/01.01_bb_EDA_augmented_emotion_classification_2020_05_13.ipynb
|
prajwollamichhane11/fuseai-training
|
2739c8bd51f5ab6ad8e765184ecae1d1c2881945
|
[
"FTL"
] | 3 |
2021-04-30T21:15:06.000Z
|
2021-09-08T02:02:12.000Z
|
notebooks/01.01_bb_EDA_augmented_emotion_classification_2020_05_13.ipynb
|
prajwollamichhane11/fuseai-training
|
2739c8bd51f5ab6ad8e765184ecae1d1c2881945
|
[
"FTL"
] | 1 |
2020-08-13T04:27:03.000Z
|
2020-08-13T04:27:03.000Z
| 500.191759 | 121,560 | 0.936528 |
[
[
[
"import matplotlib as mpl\nimport numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\n%matplotlib inline\nimport seaborn as sns\nimport nltk\nfrom wordcloud import WordCloud, STOPWORDS, ImageColorGenerator\n\nfrom sklearn.metrics import accuracy_score",
"_____no_output_____"
],
[
"from sklearn.feature_extraction.text import CountVectorizer, TfidfTransformer",
"_____no_output_____"
],
[
"# configuring matplotlib\nplt.axes.titlesize : 24\nplt.axes.labelsize : 20\nplt.figsize = (15, 10)\n# plt.cmap.",
"_____no_output_____"
],
[
"RANDOM_STATE = 42\nnp.random.seed(RANDOM_STATE)",
"_____no_output_____"
]
],
[
[
"## For augmenting the dataset",
"_____no_output_____"
],
[
"### Random Deletion",
"_____no_output_____"
]
],
[
[
"# random deletion use list.pop()b\nimport random\np = [1, 23,4 ,5, 34, 35, 23, 54, 645, 53]\nrandom.randrange(len(p))\n\ndef delete_random(text):\n \n text = text.split(\" \")\n random_index = random.randrange(len(text))\n \n text.pop(random_index)\n text = \" \".join(text)\n return text",
"_____no_output_____"
],
[
"delete_random('I feel guilty when when I realize that I consider material things more important than caring for my relatives. I feel very self-centered.')",
"_____no_output_____"
]
],
[
[
"### Random swap",
"_____no_output_____"
]
],
[
[
"# Random swap\ndef swap_random(text):\n text = text.split(\" \")\n idx = range(len(text))\n i1, i2 = random.sample(idx, 2)\n text[i1], text[i2] = text[i2], text[i1]\n text = \" \".join(text)\n return text",
"_____no_output_____"
],
[
"swap_random(\"I feel guilty when when I realize that I consider material things more important than caring for my relatives. I feel very self-centered.\")",
"_____no_output_____"
]
],
[
[
"### Lemmatization",
"_____no_output_____"
]
],
[
[
"from nltk.tokenize import sent_tokenize, word_tokenize\nfrom nltk.stem import PorterStemmer\nporter = PorterStemmer()\ndef lemmatize(text):\n \n sentences = sent_tokenize(text)\n stem_sentence=[]\n for sent in sentences:\n token_words=word_tokenize(sent)\n \n for word in token_words:\n stem_sentence.append(porter.stem(word))\n stem_sentence.append(\" \")\n return \"\".join(stem_sentence)",
"_____no_output_____"
],
[
"lemmatize(\"I feel guilty when when I realize that I consider material things more important than caring for my relatives. I feel very self-centered.\")",
"_____no_output_____"
],
[
"raw_data = pd.read_csv('../data/raw/ISEAR.csv', header=None)\nraw_data.head(15)",
"_____no_output_____"
],
[
"raw_data.columns = ['index', 'sentiment', 'text']\nraw_data.set_index('index')\nraw_data.head()",
"_____no_output_____"
],
[
"raw_data['text'][6]",
"_____no_output_____"
]
],
[
[
"### Remove newline character",
"_____no_output_____"
]
],
[
[
"raw_data['text'] = raw_data['text'].apply(lambda x: x.replace('\\n', ''))",
"_____no_output_____"
],
[
"raw_data['text'][6]",
"_____no_output_____"
]
],
[
[
"## Convert text to lowercase",
"_____no_output_____"
]
],
[
[
"raw_data['text'] = raw_data['text'].apply( lambda x: x.lower())\nraw_data.head()",
"_____no_output_____"
]
],
[
[
"### Dividing into train and test set",
"_____no_output_____"
]
],
[
[
"# Diving data into train, validation and test set\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.naive_bayes import GaussianNB\n\nX, y = raw_data['text'], raw_data['sentiment']\nX_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=RANDOM_STATE, stratify=y)\n\n# X_train, X_test = list(X_train), list(y_train)",
"_____no_output_____"
],
[
"X_train.head()",
"_____no_output_____"
],
[
"# Lemmatize X_train\nX_train = X_train.apply(lemmatize)",
"_____no_output_____"
],
[
"# Apply swap random and delet random to X\nX_train_original = X_train\ny_train_original = y_train\n\nX_train_swapped = X_train.apply(swap_random)\ny_train_swapped = y_train\n\n\nX_train_deleted = X_train.apply(delete_random)\ny_train_deleted = y_train",
"_____no_output_____"
],
[
"y_train_original.shape, X_train_swapped.shape, X_train_deleted.shape",
"_____no_output_____"
],
[
"X_train_combined = X_train_original.append(X_train_swapped)\nX_train_combined = X_train_combined.append(X_train_deleted)\n\ny_train_combined = y_train_original.append(y_train_swapped)\ny_train_combined = y_train_combined.append(y_train_deleted)\n\nX_train_combined.shape, y_train_combined.shape",
"_____no_output_____"
]
],
[
[
"### Vectorizing the training and testing features separately",
"_____no_output_____"
]
],
[
[
"vectorizer = CountVectorizer(\n analyzer = 'word',\n stop_words = 'english', # removes common english words\n ngram_range = (2, 2), # extracting bigrams\n lowercase = True,\n)\n\ntfidf_transformer = TfidfTransformer()\n\nfeatures_train = vectorizer.fit_transform(\n X_train_combined\n)\n\n\n\nfeatures_train = tfidf_transformer.fit_transform(features_train)\n\nfeatures_train = features_train.toarray() # for easy usage",
"_____no_output_____"
],
[
"# for testing features\nfeatures_test = vectorizer.transform(\n X_test\n)\n\nfeatures_test = tfidf_transformer.transform(features_test)\n\nfeatures_test = features_test.toarray() # for easy usage",
"_____no_output_____"
]
],
[
[
"## Encoding the training and testing labels separately using the same label encoder",
"_____no_output_____"
]
],
[
[
"from sklearn import preprocessing\nle = preprocessing.LabelEncoder()\ny_train = le.fit_transform(y_train_combined)",
"_____no_output_____"
],
[
"# encodeing the labels of test set\ny_test = le.transform(y_test)",
"_____no_output_____"
],
[
"y_test, y_train",
"_____no_output_____"
]
],
[
[
"## making the classifier",
"_____no_output_____"
]
],
[
[
"from sklearn.linear_model import SGDClassifier\n\nclassifier = SGDClassifier(random_state=RANDOM_STATE)\ny_pred = classifier.fit(features_train, y_train).predict(features_test)\n\naccuracy = accuracy_score(y_test, y_pred)\naccuracy",
"_____no_output_____"
]
],
[
[
"### MNB",
"_____no_output_____"
]
],
[
[
"from sklearn.naive_bayes import MultinomialNB\n\n\nclassifier = MultinomialNB()\ny_pred = classifier.fit(features_train, y_train).predict(features_test)\n\naccuracy = accuracy_score(y_test, y_pred)\naccuracy",
"_____no_output_____"
],
[
"my_colors = [(0.5,0.4,0.5), (0.75, 0.75, 0.25)]*7 # <-- make two custom RGBs and repeat/alternate them over all the bar elements.\n\nraw_data['sentiment'].value_counts().plot(kind='bar', stacked=True, color=my_colors)\nplt.savefig('../images/sentiment_distribution.png')",
"_____no_output_____"
]
],
[
[
"From above graph it is clear that all classes of sentiment have almost equal number of instances",
"_____no_output_____"
]
],
[
[
"def make_wordcloud(texts, stopwords=STOPWORDS):\n texts = texts.lower()\n sw = set(stopwords)\n wordcloud = WordCloud(stopwords=stopwords, background_color=\"white\").generate(texts)\n return wordcloud",
"_____no_output_____"
],
[
"# def plot_wordclouds(dataframe, subplot_rows, subplot_columns):\n\nrows = 4\ncolumns = 3\nfig = plt.figure()\np = 0\n\nfor col in raw_data['sentiment'].unique():\n temp_df = raw_data[raw_data['sentiment']==col]\n temp_df_texts = \" \".join(text for text in temp_df['text'])\n \n wordcloud = make_wordcloud(temp_df_texts)\n \n \n plt.imshow(wordcloud, interpolation='bilinear')\n plt.axis(\"off\")\n plt.title(col)\n image_name = '../images/'+ col+ '_wordcloud.png'\n plt.savefig(image_name)\n plt.show()\n \n",
"_____no_output_____"
]
],
[
[
"From above plots it is common that words like friend, mother, felt is common in all the texts. So we will need to remove them.",
"_____no_output_____"
],
[
"## Creating a new column that will hold the text as a list of words",
"_____no_output_____"
]
],
[
[
"frequent_words = []\ndef get_most_common_words(dataframe):\n for col in dataframe['sentiment'].unique():\n temp_df = dataframe[raw_data['sentiment']==col]\n temp_df_texts = \" \".join(text for text in temp_df['text'])\n temp_df_texts = temp_df_texts.lower()\n\n wordcloud = make_wordcloud(temp_df_texts)\n frequent_words.append(list(wordcloud.words_.keys())[:50])\n \n return frequent_words\n \n ",
"_____no_output_____"
],
[
"most_frequent_words = get_most_common_words(raw_data)",
"_____no_output_____"
],
[
"print(len(most_frequent_words))\np =set(most_frequent_words[0])",
"7\n"
],
[
"for i in range(1, len(most_frequent_words)):\n print(i)\n p.intersection_update(set(most_frequent_words[i]))\nprint(p)",
"1\n2\n3\n4\n5\n6\n{'my friend', 'felt', 'time', 'day', 'one', 'got', 'friend'}\n"
]
],
[
[
"The words present above are the most frequent words so they can also be removed from the text.",
"_____no_output_____"
]
],
[
[
"p = \" \".join(list(p))\nmost_frequent_wordcloud = make_wordcloud(p)\n\n\nplt.imshow(most_frequent_wordcloud, interpolation='bilinear')\nplt.axis(\"off\")\nplt.title('Most frequent words')\n\nimage_name = '../images/'+ 'most_frequent_words'+ '_wordcloud.png'\nplt.savefig(image_name)\nplt.show()",
"_____no_output_____"
],
[
"raw_data['text_length'] = raw_data['text'].apply(lambda x: len(x.split(' ')))\nraw_data.head()",
"_____no_output_____"
],
[
"raw_data['text_length'].plot.hist()\nplt.title('Distribution of text length')\nplt.savefig('../images/distribution_of_text_length.png')",
"_____no_output_____"
],
[
"stopwords = list(STOPWORDS) + list(p)",
"_____no_output_____"
]
],
[
[
"## Converting all the text to lowercase",
"_____no_output_____"
]
],
[
[
"raw_data['text'] = raw_data['text'].apply( lambda x: x.lower())\nraw_data.head()",
"_____no_output_____"
],
[
"vectorizer = CountVectorizer(\n analyzer = 'word',\n stop_words = 'english', # removes common english words\n ngram_range = (2, 2), # extracting bigrams\n lowercase = True,\n)\n\nfeatures = vectorizer.fit_transform(\n raw_data['text']\n)\n\ntfidf_transformer = TfidfTransformer()\n\nfeatures = tfidf_transformer.fit_transform(features)",
"_____no_output_____"
]
],
[
[
"## Saving countvectorizer",
"_____no_output_____"
]
],
[
[
"import pickle\n# Save the label encoder as pickel object\noutput = open('../models/encoder_and_vectorizer/tf_idf_transformer.pkl', 'wb')\npickle.dump(tfidf_transformer, output)\noutput.close()",
"_____no_output_____"
],
[
"features_nd = features.toarray() # for easy usage\n# print(features_nd.shape)\n# raw_data['text_vectorized'] = list(features_nd)\n# print(raw_data['text_vectorized'].shape)\n# raw_data.head()",
"_____no_output_____"
],
[
"output = open('../models/encoder_and_vectorizer/vectorizer.pkl', 'wb')\npickle.dump(vectorizer, output)\noutput.close()",
"_____no_output_____"
]
],
[
[
"The vectorizer will also need to be saved. Because we will need to use the same vectorizer for making new predictions",
"_____no_output_____"
]
],
[
[
"from sklearn import preprocessing\nle = preprocessing.LabelEncoder()\nraw_data['sentiment_encoded'] = le.fit_transform(raw_data['sentiment'])\n\n",
"_____no_output_____"
],
[
"# raw_data = raw_data[['sentiment_encoded','text_vectorized']]",
"_____no_output_____"
]
],
[
[
"Save the label encoder as a pickle or in some form. Make a function that takes column names as input, converts the column, saves the label encoder and then returns the new column values.",
"_____no_output_____"
],
[
"## Saving label encoder to a file",
"_____no_output_____"
]
],
[
[
"# Save the label encoder as pickel object\noutput = open('../models/encoder_and_vectorizer/label_encoder.pkl', 'wb')\npickle.dump(le, output)\noutput.close()",
"_____no_output_____"
],
[
"# Saving the processed data\n# raw_data.to_csv('../data/processed/sentiment_features.csv')",
"_____no_output_____"
]
],
[
[
"## Making the actual model",
"_____no_output_____"
]
],
[
[
"# Diving data into train, validation and test set\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.naive_bayes import GaussianNB\n\nX, y = features_nd, raw_data['sentiment_encoded']\nX_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=RANDOM_STATE, stratify=y)\n\n# X_train, X_test = list(X_train), list(y_train)",
"_____no_output_____"
]
],
[
[
"### knn model",
"_____no_output_____"
]
],
[
[
"from sklearn import neighbors\nknn=neighbors.KNeighborsClassifier()\n# we create an instance of Neighbours Classifier and fit the data.\nknn.fit(X_train, y_train)",
"_____no_output_____"
],
[
"\n\npredicted_results = knn.predict(X_test)\n\naccuracy = accuracy_score(y_test, predicted_results)\naccuracy",
"_____no_output_____"
]
],
[
[
"### naive bayes'",
"_____no_output_____"
]
],
[
[
"gnb = GaussianNB()\ny_pred = gnb.fit(X_train, y_train).predict(X_test)\n\naccuracy = accuracy_score(y_test, y_pred)\naccuracy",
"_____no_output_____"
]
],
[
[
"### Random Forest",
"_____no_output_____"
]
],
[
[
"from sklearn.ensemble import RandomForestClassifier\nclassifier = RandomForestClassifier(n_estimators=100, random_state=RANDOM_STATE)\ny_pred = classifier.fit(X_train, y_train).predict(X_test)\n\naccuracy = accuracy_score(y_test, y_pred)\naccuracy",
"_____no_output_____"
]
],
[
[
"### SGD",
"_____no_output_____"
]
],
[
[
"from sklearn.linear_model import SGDClassifier\n\nclassifier = SGDClassifier(random_state=RANDOM_STATE)\ny_pred = classifier.fit(X_train, y_train).predict(X_test)\n\naccuracy = accuracy_score(y_test, y_pred)\naccuracy",
"_____no_output_____"
]
],
[
[
"## Random Search with SGD",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import RandomizedSearchCV\nfrom scipy.stats import uniform\n\nclf = SGDClassifier()\n\ndistributions = dict(\n loss=['hinge', 'log', 'modified_huber', 'squared_hinge', 'perceptron'],\n learning_rate=['optimal', 'invscaling', 'adaptive'],\n eta0=uniform(loc=1e-7, scale=1e-2)\n)\n\nrandom_search_cv = RandomizedSearchCV(\n estimator=clf,\n param_distributions=distributions,\n cv=5,\n n_iter=50\n)\n\nrandom_search_cv.fit(X_train, y_train)\n",
"_____no_output_____"
],
[
"! ls",
"01.01_bb_EDA_2020_05_11.ipynb\r\n01.01_bb_EDA_emotion_classification_2020_05_13.ipynb\r\n01.01_pl_EDA_2020_05_11.ipynb\r\ncarads-tp-data_exploration.ipynb\r\n"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
c5004b4e71418e34fa53425b3d7599916f84e4a8
| 609,912 |
ipynb
|
Jupyter Notebook
|
notebooks/trial_specification_demo.ipynb
|
deflaux/metis
|
f85e78110bd3449878116cdd2ef21c3b34cdd492
|
[
"BSD-Source-Code"
] | 10 |
2020-10-20T02:52:26.000Z
|
2021-01-12T04:18:54.000Z
|
notebooks/trial_specification_demo.ipynb
|
deflaux/metis
|
f85e78110bd3449878116cdd2ef21c3b34cdd492
|
[
"BSD-Source-Code"
] | 8 |
2020-10-21T16:07:05.000Z
|
2021-03-02T21:47:42.000Z
|
notebooks/trial_specification_demo.ipynb
|
deflaux/metis
|
f85e78110bd3449878116cdd2ef21c3b34cdd492
|
[
"BSD-Source-Code"
] | 3 |
2020-10-20T16:05:54.000Z
|
2020-10-30T21:07:31.000Z
| 534.54163 | 91,704 | 0.934486 |
[
[
[
"Copyright 2020 Verily Life Sciences LLC\n\nUse of this source code is governed by a BSD-style\nlicense that can be found in the LICENSE file or at\nhttps://developers.google.com/open-source/licenses/bsd",
"_____no_output_____"
],
[
"# Trial Specification Demo\n\nThe first step to use the Baseline Site Selection Tool is to specify your trial.\n\nAll data in the Baseline Site Selection Tool is stored in [xarray.DataArray](http://xarray.pydata.org/en/stable/generated/xarray.DataArray.html) datasets. This is a [convenient datastructure](http://xarray.pydata.org/en/stable/why-xarray.html) for storing multidimensional arrays with different labels, coordinates or attributes. You don't need to have any expertise with xr.Datasets to use the Baseline Site Selection Tool. The goal of this notebook is to walk you through the construction of the dataset that contains the specification of your trial.\n\nThis notebook has several sections:\n1. **Define the Trial**. In this section you will load all aspects of your trial, including the trial sites, the expected recruitment demographics for each trial site (e.g. from a census) as well as the rules for how the trial will be carried out.\n2. **Load Incidence Forecasts**. In this section you will load forecasts for covid incidence at the locations of your trial. We highly recommend using forecasts that are as local as possible for the sites of the trial. There is significant variation in covid incidence among counties in the same state, and taking the state (province) average can be highly misleading. Here we include code to preload forecasts for county level forecasts from the US Center for Disease Control. The trial planner should include whatever forecasts they find most compelling. \n3. **Simulate the Trial** Given the incidence forecasts and the trial rules, the third section will simulate the trial.\n4. **Optimize the Trial** Given the parameters of the trial within our control, the next section asks whether we can set those parameters to make the trial meet our objective criteria, for example most likely to succeed or to succeed as quickly as possible. We have written a set of optimization routines for optimizing different types of trials.\n\nWe write out different trial plans, which you can then examine interactively in the second notebook in the Baseline Site Selection Tool. That notebook lets you visualize how the trial is proceeding at a per site level and experiment with what will happen when you turn up or down different sites.\n\nIf you have questions about how to implement these steps for your clinical trial, or there are variations in the trial specification that are not captured with this framework, please contact [email protected] for additional help.",
"_____no_output_____"
],
[
"## Imports",
"_____no_output_____"
]
],
[
[
"import matplotlib as mpl\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nsns.set_style('ticks')\n\nimport functools\nimport importlib.resources\nimport numpy as np\nimport os\nimport pandas as pd\npd.plotting.register_matplotlib_converters()\nimport xarray as xr\n\nfrom IPython.display import display\n\n# bsst imports\nfrom bsst import demo_data\nfrom bsst import io as bsst_io\nfrom bsst import util\nfrom bsst import optimization\nfrom bsst import sim\nfrom bsst import sim_scenarios\nfrom bsst import public_data",
"_____no_output_____"
]
],
[
[
"## Helper methods for visualization",
"_____no_output_____"
]
],
[
[
"def plot_participants(participants):\n time = participants.time.values\n util.sum_all_but_dims(['time'], participants).cumsum('time').plot()\n plt.title('Participants recruited (both control and treatment arm)')\n plt.xlim(time[0], time[-1])\n plt.ylim(bottom=0)\n plt.show()\n\ndef plot_events(events):\n time = events.time.values\n events.cumsum('time').plot.line(x='time', color='k', alpha=.02, add_legend=False)\n for analysis, num_events in c.needed_control_arm_events.to_series().items():\n plt.axhline(num_events, linestyle='--')\n plt.text(time[0], num_events, analysis, ha='left', va='bottom')\n plt.ylim(0, 120)\n plt.xlim(time[0], time[-1])\n plt.title(f'Control arm events\\n{events.scenario.size} simulated scenarios')\n plt.show()\n\ndef plot_success(c, events):\n time = c.time.values\n success_day = xr.DataArray(util.success_day(c.needed_control_arm_events, events),\n coords=(events.scenario, c.analysis))\n\n fig, axes = plt.subplots(c.analysis.size, 1, sharex=True)\n step = max(1, int(np.timedelta64(3, 'D') / (time[1] - time[0])))\n bins = mpl.units.registry[np.datetime64].convert(time[::step], None, None)\n\n for analysis, ax in zip(c.analysis.values, axes):\n success_days = success_day.sel(analysis=analysis).values\n np.where(np.isnat(success_days), np.datetime64('2050-06-01'), success_days)\n ax.hist(success_days, bins=bins, density=True)\n ax.yaxis.set_visible(False)\n # subtract time[0] to make into timedelta64s so that we can take a mean/median\n median = np.median(success_days - time[0]) + time[0]\n median = pd.to_datetime(median).date()\n ax.axvline(median, color='r')\n ax.text(time[0], 0, f'{analysis}\\n{median} median', ha='left', va='bottom')\n\n plt.xlabel('Date when sufficient statistical power is achieved')\n plt.xlim(time[0], time[-1])\n plt.xticks(rotation=35)\n plt.show()",
"_____no_output_____"
]
],
[
[
"# 1. Define the trial\n\n## Choose the sites\nA trial specification consists a list of sites, together with various properties of the sites. \n\nFor this demo, we read demonstration data embedded in the Baseline Site Selection Tool Python package. Specifically, this information is loaded from the file `demo_data/site_list1.csv`. Each row of this file contains the name of a site, as well as the detailed information about the trial. In this illustrative example, we pick sites in real US counties. Each column contains the following information:\n\n* `opencovid_key` . This is a key that specifies location within [COVID-19 Open Data](https://github.com/GoogleCloudPlatform/covid-19-open-data). It is required by this schema because it is the way we join the incidence forecasts to the site locations. \n* `capacity`, the number of participants the site can recruit each week, including both control arm and treatment arms. For simplicity, we assume this is constant over time, but variable recruitment rates are also supported. (See the construction of the `site_capacity` array below).\n* `start_date`. This is the first date on which the site can recruit participants.\n* The proportion of the population in various demographic categories. For this example, we consider categories for age (`over_60`), ethnicity (`black`, `hisp_lat`), and comorbidities (`smokers`, `diabetes`, `obese`). **Here we just fill in demographic information with random numbers.** We assume different categories are independent, but the data structure supports complex beliefs about how different categories intersect, how much each site can enrich for different categories, and different infection risks for different categories. These are represented in the factors `population_fraction`, `participant_fraction`, `incidence_scaler`, and `incidence_to_event_factor` below. In a practical situation, we recommend that the trial planner uses accurate estimates of the populations for the different sites they are drawing from.",
"_____no_output_____"
]
],
[
[
"with importlib.resources.path(demo_data, 'site_list1.csv') as p:\n demo_data_file_path = os.fspath(p)\n site_df = pd.read_csv(demo_data_file_path, index_col=0)\n\nsite_df.index.name = 'location'\nsite_df['start_date'] = pd.to_datetime(site_df['start_date'])\ndisplay(site_df)\n\n# Add in information we have about each county.\nsite_df = pd.concat([site_df, public_data.us_county_data().loc[site_df.opencovid_key].set_index(site_df.index)], axis=1)",
"_____no_output_____"
]
],
[
[
"## Choose trial parameters\nThe trial requires a number of parameters that have to be specified to be able to simulate what will happen in the trial: These include:\n\n\n* `trial_size_cap`: the maximum number of participants in the trial (includes both control and treatment arms)\n* `start_day` and `end_day`: the boundaries of the time period we will simulate.\n* `proportion_control_arm`: what proportion of participants are in the control arm. It's assumed that the control arm is as uniformly distributed across locations and time (e.g. at each location on each day, half of the recruited participants are assigned to the control arm).\n* `needed_control_arm_events`: the number of events required in the *control* arm of the trial at various intermediate analysis points. For this example we assume intermediate analyses which would demonstrate a vaccine efficacy of about 55%, 65%, 75%, 85%, or 95%.\n* `observation_delay`: how long after a participant is recruited before they contribute an event. This is measured in the same time units as your incidence forecasts. Here we assume 28 days.\n* `site_capacity` and `site_activation`: the number of participants each site could recruit *if* it were activated, and whether each site is activated at any given time. Here we assume each site as a constant weekly capacity, but time dependence can be included (e.g. to model ramp up of recruitment).\n* `population_fraction`, `participant_fraction`, and `incidence_scaler`: the proportion of the general population and the proportion of participants who fall into different demographic categories at each location, and the infection risk factor for each category. These three are required to translate an overall incidence forecast for the population into the incidence forecast for your control arm.\n* `incidence_to_event_factor`: what proportion of infections lead to a clinical event. We assume a constant 0.6, but you can specify different values for different demographic categories.\n\nThese factors are specified in the datastructure below.\n",
"_____no_output_____"
]
],
[
[
"start_day = np.datetime64('2021-05-15')\nend_day = np.datetime64('2021-10-01')\ntime_resolution = np.timedelta64(1, 'D')\n\ntime = np.arange(start_day, end_day + time_resolution, time_resolution)\nc = xr.Dataset(coords=dict(time=time))\nc['proportion_control_arm'] = 0.5\n\n# Assume some intermediate analyses.\nfrac_control = float(c.proportion_control_arm)\nefficacy = np.array([.55, .65, .75, .85, .95])\nctrl_events = util.needed_control_arm_events(efficacy, frac_control)\nvaccine_events = (1 - efficacy) * ctrl_events * (1 - frac_control) / frac_control\nctrl_events, vaccine_events = np.round(ctrl_events), np.round(vaccine_events)\nefficacy = 1 - (vaccine_events / ctrl_events)\ntotal_events = ctrl_events + vaccine_events\nanalysis_names = [\n f'{int(t)} total events @{int(100 * e)}% VE' for t, e in zip(total_events, efficacy)\n]\nc['needed_control_arm_events'] = xr.DataArray(\n ctrl_events, dims=('analysis',)).assign_coords(analysis=analysis_names)\n\nc['recruitment_type'] = 'default'\nc['observation_delay'] = int(np.timedelta64(28, 'D') / time_resolution) # 28 days\nc['trial_size_cap'] = 30000\n\n# convert weekly capacity to capacity per time step\nsite_capacity = site_df.capacity.to_xarray() * time_resolution / np.timedelta64(7, 'D')\nsite_capacity = site_capacity.broadcast_like(c.time).astype('float')\n# Can't recruit before the activation date\nactivation_date = site_df.start_date.to_xarray()\nfor l in activation_date.location.values:\n date = activation_date.loc[l]\n site_capacity.loc[site_capacity.time < date, l] = 0.0\nc['site_capacity'] = site_capacity.transpose('location', 'time')\n\nc['site_activation'] = xr.ones_like(c.site_capacity)\n\n# For the sake of simplicity, this code assumes black and hisp_lat are\n# non-overlapping, and that obese/smokers/diabetes are non-overlapping.\nfrac_and_scalar = util.fraction_and_incidence_scaler\nfraction_scalers = [\n frac_and_scalar(site_df, 'age', ['over_60'], [1], 'under_60'),\n frac_and_scalar(site_df, 'ethnicity', ['black', 'hisp_lat'], [1, 1],\n 'other'),\n frac_and_scalar(site_df, 'comorbidity', ['smokers', 'diabetes', 'obese'],\n [1, 1, 1], 'none')\n]\nfractions, incidence_scalers = zip(*fraction_scalers)\n\n# We assume that different categories are independent (e.g. the proportion of\n# smokers over 60 is the same as the proportion of smokers under 60)\nc['population_fraction'] = functools.reduce(lambda x, y: x * y, fractions)\n# We assume the participants are drawn uniformly from the population.\nc['participant_fraction'] = c['population_fraction']\n# Assume some boosted incidence risk for subpopulations. We pick random numbers\n# here, but in actual use you'd put your best estimate for the incidence risk\n# of each demographic category.\n# Since we assume participants are uniformly drawn from the county population,\n# this actually doesn't end up affecting the estimated number of clinical events.\nc['incidence_scaler'] = functools.reduce(lambda x, y: x * y,\n incidence_scalers)\nc.incidence_scaler.loc[dict(age='over_60')] = 1 + 2 * np.random.random()\nc.incidence_scaler.loc[dict(comorbidity=['smokers', 'diabetes', 'obese'])] = 1 + 2 * np.random.random()\nc.incidence_scaler.loc[dict(ethnicity=['black', 'hisp_lat'])] = 1 + 2 * np.random.random()\n\n# We assume a constant incidence_to_event_factor.\nc['incidence_to_event_factor'] = 0.6 * xr.ones_like(c.incidence_scaler)\n\nutil.add_empty_history(c)",
"_____no_output_____"
]
],
[
[
"# 2. Load incidence forecasts\n\nWe load historical incidence data from [COVID-19 Open Data](https://github.com/GoogleCloudPlatform/covid-19-open-data) and forecasts from [COVID-19 Forecast Hub](https://github.com/reichlab/covid19-forecast-hub).\n\nWe note that there are a set of caveats when using the CDC models that should be considered when using these for trial planning:\n* Forecasts are only available for US counties. Hence, these forecasts will only work for US-only trials. Trials with sites outside the US will need to supplement these forecasts.\n* Forecasts only go out for four weeks. Trials take much longer than four weeks to complete, when measured from site selection to logging the required number of cases in the control arm. For simplicity, here we extrapolate incidence as *constant* after the last point of the forecast. Here we extrapolate out to October 1, 2021. \n* The forecasts from the CDC are provided with quantile estimates. Our method depends on getting *representative forecasts* from the model: we need a set of sample forecasts for each site which represent the set of scenarios that can occur. Ideally these scenarios will be equally probable so that we can compute probabilities by averaging over samples. To get samples from quantiles, we interpolate/extrapolate to get 100 evenly spaced quantile estimates, which we treat as representative samples.\n\nYou can of course replace these forecasts with whatever represents your beliefs and uncertainty about what will happen.",
"_____no_output_____"
]
],
[
[
"# Extrapolate out a bit extra to ensure we're within bounds when we interpolate later.\nfull_pred = public_data.fetch_cdc_forecasts([('COVIDhub-ensemble', '2021-05-10'),\n ('COVIDhub-baseline', '2021-05-10')],\n end_date=c.time.values[-1] + np.timedelta64(15, 'D'),\n num_samples=50)\nfull_gt = public_data.fetch_opencovid_incidence()\n\n# Suppose we only have ground truth through 2021-05-09.\nfull_gt = full_gt.sel(time=slice(None, np.datetime64('2021-05-09')))\n\n# Include more historical incidence here for context. It will be trimmed off when\n# we construct scenarios to simulate. The funny backwards range is to ensure that if\n# we use weekly instead of daily resolution, we use the same day of the week as c.\ntime = np.arange(c.time.values[-1], np.datetime64('2021-04-01'), -time_resolution)[::-1]\nincidence_model = public_data.assemble_forecast(full_gt, full_pred, site_df, time)",
"_____no_output_____"
],
[
"locs = np.random.choice(c.location.values, size=5, replace=False)\nincidence_model.sel(location=locs).plot.line(x='time', color='k', alpha=.1, add_legend=False, col='location', row='model')\nplt.ylim(0.0, 1e-3)\nplt.suptitle('Forecast incidence at a sampling of sites', y=1.0)\npass",
"_____no_output_____"
]
],
[
[
"# 3. Simulate the trial\n\nNow that we've specified how the trial works, we can compute how the trial will turn out given the incidence forecasts you've specified. We do this by first imagining what sampling what incidence will be at all locations simultaneously. For any given fully-specified scenario, we compute how many participants will be under observation at any given time in any given location (in any given combination of demographic buckets), then based on the specified local incidence we compute how many will become infected, and how many will produce clinical events.\n\nHere we assume that the incidence trajectories of different locations are drawn at random from the available forecasts. Other scenario-generation methods in `sim_scenarios` support more complex approaches. For example, we may be highly uncertain about the incidence at each site, but believe that if incidence is high at a site, then it will also be high at geographically nearby sites. If this is the case then the simulation should not choose forecasts independently at each site but instead should take these correlations into account. The code scenario-generating methods in `sim_scenarios` allows us to do that.",
"_____no_output_____"
]
],
[
[
"# incidence_flattened: rolls together all the models you've included in your ensemble, treating them as independent samples.\nincidence_flattened = sim_scenarios.get_incidence_flattened(incidence_model, c)\n\n# incidence_scenarios: chooses scenarios given the incidence curves and your chosen method of scenario-generation.\nincidence_scenarios = sim_scenarios.generate_scenarios_independently(incidence_flattened, num_scenarios=100)",
"_____no_output_____"
],
[
"# compute the number of participants recruited under your trial rule\nparticipants = sim.recruitment(c)\n# compute the number of control arm events under your trial rules and incidence_scenarios.\nevents = sim.control_arm_events(c, participants, incidence_scenarios)\n\nplot_participants(participants)\n# plot events and label different vaccine efficacies\nplot_events(events)\n# plot histograms of time to success\nplot_success(c, events)",
"_____no_output_____"
],
[
"sim.add_stuff_to_ville(c, incidence_model, site_df, num_scenarios=100)\n!mkdir -p demo_data\nbsst_io.write_ville_to_netcdf(c, 'demo_data/site_list1_all_site_on.nc')",
"Populating participants.\nPopulating control_arm_events based on independent scenarios.\n"
]
],
[
[
"# 4. Optimize the trial\n\nThe simulations above supposed that all sites are activated as soon as possible (i.e. `site_activation` is identically 1). Now that we have shown the ability to simulate the outcome of the trial, we can turn it into a mathematical optimization problem. \n\n**Given the parameters of the trial within our control, how can we set those parameters to make the trial most likely to succeed or to succeed as quickly as possible?**\n\nWe imagine the main levers of control are which sites to activate or which sites to prioritize activating, and this is what is implemented here. \n\nHowever, the framework we have developed is very general and could be extended to just about anything you control which you can predict the impact of. For example,\n* If you can estimate the impact of money spent boosting recruitment of high-risk participants, we could use those estimates to help figure out how to best allocate a fixed budget.\n* If you had requirements for the number of people infected in different demographic groups, we could use those to help figure out how to best allocate doses between sites with different population characteristics.\n\nThe optimization algorithms are implemented in [JAX](https://github.com/google/jax), a python library that makes it possible to differentiate through native python and numpy functions. The flexibility of the language makes it possible to compose a variety of trial optimization scenarios and then to write algorithms that find optima. There are a number of technical details in how the optimization algorithms are written that will be discussed elsewhere.\n",
"_____no_output_____"
],
[
"### Example: Optimizing Static site activations\n\nSuppose that the only variable we can control is which sites should be activated, and we have to make this decision at the beginning of the trial. This decision is then set in stone for the duration of the trial. To calculate this we proceed as follows:\n\nThe optimizer takes in the trial plan, encoded in the xarray `c` as well as the `incidence_scenarios`, and then calls the optimizer to find the sites that should be activated to minimize the time to success of the trial. The algorithm modifies `c` *in place*, so that after the algorithm runs, it returns the trial plan `c` but with the site activations chosen to be on or off in accordance with the optimizion.",
"_____no_output_____"
]
],
[
[
"%time optimization.optimize_static_activation(c, incidence_scenarios)",
"WARNING:absl:No GPU/TPU found, falling back to CPU. (Set TF_CPP_MIN_LOG_LEVEL=0 and rerun for more info.)\n"
]
],
[
[
"#### Plot the resulting sites\n\nNow we can plot the activations for the resulting sites. Only a subset of the original sites are activated in the optimized plan. Comparing the distributions for the time to success for the optimized sites to those in the original trial plan (all sites activated), the optimized plan will save a bit of time if the vaccine efficacy is low. If the vaccine efficacy is high, then just getting as many participants as possible as quickly as possible is optimal.",
"_____no_output_____"
]
],
[
[
"all_sites = c.location.values\nactivated_sites = c.location.values[c.site_activation.mean('time') == 1]\n\n# Simulate the results with this activation scheme.\nprint(f'\\n\\n{len(activated_sites)} of {len(all_sites)} activated')\nparticipants = sim.recruitment(c)\nevents = sim.control_arm_events(c, participants, incidence_scenarios)\nplot_participants(participants)\nplot_events(events)\nplot_success(c, events)\n\ndf = (participants.sum(['location', 'time', 'comorbidity']) / participants.sum()).to_pandas()\ndisplay(df.style.set_caption('Proportion of participants by age and ethnicity'))",
"\n\n118 of 146 activated\n"
],
[
"sim.add_stuff_to_ville(c, incidence_model, site_df, num_scenarios=100)\n!mkdir -p demo_data\nbsst_io.write_ville_to_netcdf(c, 'demo_data/site_list1_optimized_static.nc')",
"_____no_output_____"
]
],
[
[
"### Example: Custom loss penalizing site activation and promoting diverse participants\n\nSuppose we want to factor in considerations aside from how quickly the trial succeeds. In this example, we assume that activating sites is expensive, so we'd like to activate as few of them as possible, so long as it doesn't delay the success of the trial too much. Similarly, we assume that it's valuable to have a larger proportion of elderly, black, or hispanic participants, and we're willing to activate sites which can recruit from these demographic groups, even if doing so delays success a bit.",
"_____no_output_____"
]
],
[
[
"def loss_fn(c):\n # sum over location, time, comorbidity\n # remaining dimensions are [age, ethnicity]\n participants = c.participants.sum(axis=0).sum(axis=0).sum(axis=-1)\n total_participants = participants.sum()\n\n return (\n optimization.negative_mean_successiness(c) # demonstrate efficacy fast\n + 0.2 * c.site_activation.mean() # turning on sites is costly\n - 0.5 * participants[1:, :].sum() / total_participants # we want people over 60\n - 0.5 * participants[:, 1:].sum() / total_participants # we want blacks and hispanics\n )\n\n%time optimization.optimize_static_activation(c, incidence_scenarios, loss_fn)",
"Skipping jax conversion of subregion1_name (dtype is \"object\")\nSkipping jax conversion of subregion2_name (dtype is \"object\")\nSkipping jax conversion of opencovid_key (dtype is \"object\")\nstep 10, loss value -0.657703697681427\nstep 20, loss value -0.6844117641448975\nstep 30, loss value -0.6997547149658203\nstep 40, loss value -0.7068135142326355\nstep 50, loss value -0.7099326848983765\nstep 60, loss value -0.7114608883857727\nstep 70, loss value -0.7123234272003174\nstep 80, loss value -0.7128790020942688\nSkipping jax conversion of subregion1_name (dtype is \"object\")\nSkipping jax conversion of subregion2_name (dtype is \"object\")\nSkipping jax conversion of opencovid_key (dtype is \"object\")\nCPU times: user 21.9 s, sys: 608 ms, total: 22.5 s\nWall time: 13.9 s\n"
]
],
[
[
"#### Plot the resulting sites\n\nThis time only 53 of 146 sites are activated. The slower recruitment costs us 1-2 weeks until the trial succeeds (depending on vaccine efficacy). In exchange, we don't need to activate as many sites, and we end up with a greater proportion of participants who are elderly, black, or hispanic (dropping from 55.7% to 45.6% young white).",
"_____no_output_____"
]
],
[
[
"all_sites = c.location.values\nactivated_sites = c.location.values[c.site_activation.mean('time') == 1]\n\n# Simulate the results with this activation scheme.\nprint(f'\\n\\n{len(activated_sites)} of {len(all_sites)} activated')\nparticipants = sim.recruitment(c)\nevents = sim.control_arm_events(c, participants, incidence_scenarios)\nplot_participants(participants)\nplot_events(events)\nplot_success(c, events)\n\ndf = (participants.sum(['location', 'time', 'comorbidity']) / participants.sum()).to_pandas()\ndisplay(df.style.set_caption('Proportion of participants by age and ethnicity'))",
"\n\n53 of 146 activated\n"
]
],
[
[
"### Example: prioritizing sites\nSuppose we can activate up to 20 sites each week for 10 weeks. How do we prioritize them?",
"_____no_output_____"
]
],
[
[
"# We put all sites in on group. We also support prioritizing sites within groupings.\n# For example, if you can activate 2 sites per state per week, sites would be grouped\n# according to the state they're in.\nsite_to_group = pd.Series(['all_sites'] * len(site_df), index=site_df.index)\ndecision_dates = c.time.values[:70:7]\nallowed_activations = pd.DataFrame([[20] * len(decision_dates)], index=['all_sites'], columns=decision_dates)\nparameterizer = optimization.PivotTableActivation(c, site_to_group, allowed_activations, can_deactivate=False)",
"_____no_output_____"
],
[
"optimization.optimize_params(c, incidence_scenarios, parameterizer)\nc['site_activation'] = c.site_activation.round() # each site has to be on or off at each time",
"Skipping jax conversion of subregion1_name (dtype is \"object\")\nSkipping jax conversion of subregion2_name (dtype is \"object\")\nSkipping jax conversion of opencovid_key (dtype is \"object\")\nstep 10, loss value -0.4285351634025574\nstep 20, loss value -0.4439670145511627\nstep 30, loss value -0.4540953040122986\nstep 40, loss value -0.45961153507232666\nstep 50, loss value -0.4631063938140869\nstep 60, loss value -0.465656578540802\nstep 70, loss value -0.4669983685016632\nstep 80, loss value -0.46810150146484375\nstep 90, loss value -0.4691193103790283\nstep 100, loss value -0.4694053530693054\nSkipping jax conversion of subregion1_name (dtype is \"object\")\nSkipping jax conversion of subregion2_name (dtype is \"object\")\nSkipping jax conversion of opencovid_key (dtype is \"object\")\n"
],
[
"df = c.site_activation.to_pandas()\ndf.columns = [pd.to_datetime(x).date() for x in df.columns]\nsns.heatmap(df, cbar=False)\nplt.title('Which sites are activated when')\nplt.show()",
"_____no_output_____"
],
[
"participants = sim.recruitment(c)\nevents = sim.control_arm_events(c, participants, incidence_scenarios)\nplot_participants(participants)\nplot_events(events)\nplot_success(c, events)",
"_____no_output_____"
],
[
"sim.add_stuff_to_ville(c, incidence_model, site_df, num_scenarios=100)\n!mkdir -p demo_data\nbsst_io.write_ville_to_netcdf(c, 'demo_data/site_list1_prioritized.nc')",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
c5005c5639fcadc86f5be9cccc5b416752f48087
| 253,705 |
ipynb
|
Jupyter Notebook
|
Advanced Level/PD_Monitoring/PD_monitoring.ipynb
|
zubairfarahi/Data-Science--Machine-Learning-
|
803aee3decf673774f88e088e5d319ea69005751
|
[
"MIT"
] | null | null | null |
Advanced Level/PD_Monitoring/PD_monitoring.ipynb
|
zubairfarahi/Data-Science--Machine-Learning-
|
803aee3decf673774f88e088e5d319ea69005751
|
[
"MIT"
] | null | null | null |
Advanced Level/PD_Monitoring/PD_monitoring.ipynb
|
zubairfarahi/Data-Science--Machine-Learning-
|
803aee3decf673774f88e088e5d319ea69005751
|
[
"MIT"
] | null | null | null | 129.971824 | 44,630 | 0.80704 |
[
[
[
"import pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport seaborn as sns",
"_____no_output_____"
],
[
"dataset = pd.read_csv(\"data/EMA_data.csv\")\ndataset.head()",
"_____no_output_____"
],
[
"dataset.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 1539 entries, 0 to 1538\nData columns (total 46 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 ID 1539 non-null int64 \n 1 beep_time_start 1539 non-null object \n 2 beep_time_end 1539 non-null object \n 3 mood_well 1539 non-null int64 \n 4 mood_down 1539 non-null int64 \n 5 mood_fright 1539 non-null int64 \n 6 mood_tense 1539 non-null int64 \n 7 phy_sleepy 1539 non-null int64 \n 8 phy_tired 1539 non-null int64 \n 9 mood_cheerf 1538 non-null float64\n 10 mood_relax 1538 non-null float64\n 11 thou_concent 1538 non-null float64\n 12 pat_hallu 1538 non-null float64\n 13 loc_where 1538 non-null float64\n 14 soc_who 1537 non-null float64\n 15 soc_who02 168 non-null float64\n 16 soc_who03 12 non-null float64\n 17 act_what 1536 non-null float64\n 18 act_what02 77 non-null float64\n 19 act_what03 7 non-null float64\n 20 act_problemless 1536 non-null float64\n 21 mobility_well 1536 non-null float64\n 22 sit_still 1535 non-null float64\n 23 speech_well 1535 non-null float64\n 24 walk_well 1535 non-null float64\n 25 tremor 1534 non-null float64\n 26 slowness 1534 non-null float64\n 27 stiffness 1534 non-null float64\n 28 muscle_tension 1534 non-null float64\n 29 dyskinesia 1534 non-null float64\n 30 sanpar_onoff 1533 non-null float64\n 31 sanpar_medic 1533 non-null float64\n 32 beep_disturb 1530 non-null float64\n 33 mor_sleptwell 1514 non-null float64\n 34 mor_often_awake 1513 non-null float64\n 35 mor_rested 1513 non-null float64\n 36 mor_tired_phys 1514 non-null float64\n 37 mor_tired_ment 1514 non-null float64\n 38 eve_many_offs 1471 non-null float64\n 39 eve_long_offs 1471 non-null float64\n 40 eve_walk_well 1471 non-null float64\n 41 eve_clothing 1471 non-null float64\n 42 eve_eat_well 1471 non-null float64\n 43 eve_personalcare 1471 non-null float64\n 44 eve_household 1471 non-null float64\n 45 eve_tired 1471 non-null float64\ndtypes: float64(37), int64(7), object(2)\nmemory usage: 553.2+ KB\n"
],
[
"dataset.columns",
"_____no_output_____"
],
[
"pd.set_option(\"display.max_columns\", None)\ndataset.head()",
"_____no_output_____"
],
[
"dataset.describe()",
"_____no_output_____"
],
[
"smaler_dataset = dataset[[\"ID\",\"tremor\",\"dyskinesia\"]]\nsmaler_dataset.tremor.value_counts(normalize=True)\n",
"_____no_output_____"
],
[
"smaler_dataset.tremor.dropna()\nsmaler_dataset.tremor = smaler_dataset.tremor.fillna(1.0)\nsmaler_dataset.tremor.value_counts(normalize=True)",
"C:\\Users\\zubair\\anaconda3\\lib\\site-packages\\pandas\\core\\generic.py:5494: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\n self[name] = value\n"
],
[
"plt.figure(figsize=(15,8))\nsns.boxplot(x=\"ID\", y=\"tremor\", data=dataset, palette=\"Set1\")\nplt.show()",
"_____no_output_____"
],
[
"plt.figure(figsize=(15,8))\nsns.boxplot(x=\"ID\", y=\"tremor\", data=dataset)\nsns.swarmplot(x=\"ID\", y=\"tremor\", data=dataset, color=\".25\")\nplt.show()",
"C:\\Users\\zubair\\anaconda3\\lib\\site-packages\\seaborn\\categorical.py:1296: UserWarning: 85.7% of the points cannot be placed; you may want to decrease the size of the markers or use stripplot.\n warnings.warn(msg, UserWarning)\nC:\\Users\\zubair\\anaconda3\\lib\\site-packages\\seaborn\\categorical.py:1296: UserWarning: 76.0% of the points cannot be placed; you may want to decrease the size of the markers or use stripplot.\n warnings.warn(msg, UserWarning)\nC:\\Users\\zubair\\anaconda3\\lib\\site-packages\\seaborn\\categorical.py:1296: UserWarning: 90.5% of the points cannot be placed; you may want to decrease the size of the markers or use stripplot.\n warnings.warn(msg, UserWarning)\nC:\\Users\\zubair\\anaconda3\\lib\\site-packages\\seaborn\\categorical.py:1296: UserWarning: 84.6% of the points cannot be placed; you may want to decrease the size of the markers or use stripplot.\n warnings.warn(msg, UserWarning)\nC:\\Users\\zubair\\anaconda3\\lib\\site-packages\\seaborn\\categorical.py:1296: UserWarning: 63.0% of the points cannot be placed; you may want to decrease the size of the markers or use stripplot.\n warnings.warn(msg, UserWarning)\nC:\\Users\\zubair\\anaconda3\\lib\\site-packages\\seaborn\\categorical.py:1296: UserWarning: 71.8% of the points cannot be placed; you may want to decrease the size of the markers or use stripplot.\n warnings.warn(msg, UserWarning)\nC:\\Users\\zubair\\anaconda3\\lib\\site-packages\\seaborn\\categorical.py:1296: UserWarning: 32.1% of the points cannot be placed; you may want to decrease the size of the markers or use stripplot.\n warnings.warn(msg, UserWarning)\nC:\\Users\\zubair\\anaconda3\\lib\\site-packages\\seaborn\\categorical.py:1296: UserWarning: 82.0% of the points cannot be placed; you may want to decrease the size of the markers or use stripplot.\n warnings.warn(msg, UserWarning)\nC:\\Users\\zubair\\anaconda3\\lib\\site-packages\\seaborn\\categorical.py:1296: UserWarning: 88.7% of the points cannot be placed; you may want to decrease the size of the markers or use stripplot.\n warnings.warn(msg, UserWarning)\nC:\\Users\\zubair\\anaconda3\\lib\\site-packages\\seaborn\\categorical.py:1296: UserWarning: 87.2% of the points cannot be placed; you may want to decrease the size of the markers or use stripplot.\n warnings.warn(msg, UserWarning)\nC:\\Users\\zubair\\anaconda3\\lib\\site-packages\\seaborn\\categorical.py:1296: UserWarning: 68.5% of the points cannot be placed; you may want to decrease the size of the markers or use stripplot.\n warnings.warn(msg, UserWarning)\nC:\\Users\\zubair\\anaconda3\\lib\\site-packages\\seaborn\\categorical.py:1296: UserWarning: 68.3% of the points cannot be placed; you may want to decrease the size of the markers or use stripplot.\n warnings.warn(msg, UserWarning)\nC:\\Users\\zubair\\anaconda3\\lib\\site-packages\\seaborn\\categorical.py:1296: UserWarning: 64.8% of the points cannot be placed; you may want to decrease the size of the markers or use stripplot.\n warnings.warn(msg, UserWarning)\nC:\\Users\\zubair\\anaconda3\\lib\\site-packages\\seaborn\\categorical.py:1296: UserWarning: 52.5% of the points cannot be placed; you may want to decrease the size of the markers or use stripplot.\n warnings.warn(msg, UserWarning)\nC:\\Users\\zubair\\anaconda3\\lib\\site-packages\\seaborn\\categorical.py:1296: UserWarning: 63.7% of the points cannot be placed; you may want to decrease the size of the markers or use stripplot.\n warnings.warn(msg, UserWarning)\nC:\\Users\\zubair\\anaconda3\\lib\\site-packages\\seaborn\\categorical.py:1296: UserWarning: 76.1% of the points cannot be placed; you may want to decrease the size of the markers or use stripplot.\n warnings.warn(msg, UserWarning)\nC:\\Users\\zubair\\anaconda3\\lib\\site-packages\\seaborn\\categorical.py:1296: UserWarning: 48.0% of the points cannot be placed; you may want to decrease the size of the markers or use stripplot.\n warnings.warn(msg, UserWarning)\nC:\\Users\\zubair\\anaconda3\\lib\\site-packages\\seaborn\\categorical.py:1296: UserWarning: 53.8% of the points cannot be placed; you may want to decrease the size of the markers or use stripplot.\n warnings.warn(msg, UserWarning)\nC:\\Users\\zubair\\anaconda3\\lib\\site-packages\\seaborn\\categorical.py:1296: UserWarning: 90.3% of the points cannot be placed; you may want to decrease the size of the markers or use stripplot.\n warnings.warn(msg, UserWarning)\nC:\\Users\\zubair\\anaconda3\\lib\\site-packages\\seaborn\\categorical.py:1296: UserWarning: 70.6% of the points cannot be placed; you may want to decrease the size of the markers or use stripplot.\n warnings.warn(msg, UserWarning)\n"
],
[
"plt.figure(figsize=(15,8))\nsns.boxplot(x=\"ID\", y=\"dyskinesia\", data=dataset, palette=\"Set1\")\nplt.show()",
"_____no_output_____"
],
[
"plt.figure(figsize=(15,8))\nsns.boxplot(x=\"ID\", y=\"mor_tired_phys\", data=dataset, palette=\"Set1\")\nplt.show()",
"_____no_output_____"
],
[
"smaler_dataset.groupby('ID').tremor.mean().sort_values(ascending=False)\n",
"_____no_output_____"
],
[
"smaler_dataset.groupby('ID').dyskinesia.mean().sort_values(ascending=False)",
"_____no_output_____"
],
[
"smaler_dataset.groupby('ID').mor_tired_phys.mean()\nsmaler_dataset.groupby('ID').mor_tired_phys.mean().sort_values(ascending=False)",
"_____no_output_____"
],
[
"plt.figure(figsize=(15,8))\nsmaler_dataset.groupby('ID').tremor.mean().sort_values(ascending=False).plot(kind='bar')",
"_____no_output_____"
],
[
"plt.figure(figsize=(15,8))\nsmaler_dataset.groupby('ID').dyskinesia.mean().sort_values(ascending=False).plot(kind='bar')\n",
"_____no_output_____"
],
[
"plt.figure(figsize=(15,8))\nsmaler_dataset.groupby('ID').mor_tired_phys.mean().sort_values(ascending=False).plot(kind='bar')",
"_____no_output_____"
],
[
"sns.set_style(\"whitegrid\")\nsns.FacetGrid(smaler_dataset, hue=\"ID\", height=5).map(sns.histplot, \"mor_tired_phys\").add_legend()\nplt.show()",
"_____no_output_____"
],
[
"sns.set_style(\"whitegrid\")\nsns.FacetGrid(smaler_dataset, hue=\"ID\", height=5).map(sns.histplot, \"tremor\").add_legend()\nplt.show()",
"_____no_output_____"
],
[
"\nsns.set_style(\"whitegrid\")\nsns.FacetGrid(smaler_dataset, hue=\"ID\", height=5).map(sns.histplot, \"dyskinesia\").add_legend()\nplt.show()",
"_____no_output_____"
],
[
"smaler_dataset.describe()",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
c50067d09aa6bc27a69a9fac626c11b1822217a2
| 5,314 |
ipynb
|
Jupyter Notebook
|
examples/neb_path_mapping.ipynb
|
cajfisher/pymatgen-diffusion
|
a4265d232b21804ba397b2b193ffc65c9f5665e8
|
[
"BSD-3-Clause"
] | null | null | null |
examples/neb_path_mapping.ipynb
|
cajfisher/pymatgen-diffusion
|
a4265d232b21804ba397b2b193ffc65c9f5665e8
|
[
"BSD-3-Clause"
] | null | null | null |
examples/neb_path_mapping.ipynb
|
cajfisher/pymatgen-diffusion
|
a4265d232b21804ba397b2b193ffc65c9f5665e8
|
[
"BSD-3-Clause"
] | null | null | null | 33.2125 | 140 | 0.439029 |
[
[
[
"# Using the FullPathMapper for Migration For Graph based Migration Barrier Analysis",
"_____no_output_____"
]
],
[
[
"# Start with a lithiated structure\nfrom pymatgen import MPRester, Structure\nstruct = MPRester().get_structure_by_material_id(\"mp-770533\")",
"_____no_output_____"
],
[
"# BASIC usage of the Full \nfrom pymatgen_diffusion.neb.full_path_mapper import FullPathMapper\n# instanciate the full path mapper, all hops with distance below a cutoff of 4 Angstroms will be considered.\nfpm = FullPathMapper(struct, 'Li', max_path_length=4) \n# populate the edges with the MigratationPath objects\nfpm.populate_edges_with_migration_paths() \n# group the edges together based on equivalence of the MigratationPath objects\nfpm.group_and_label_hops()",
"_____no_output_____"
]
],
[
[
"The result is a dicationary in the form of:\n```\n{\n (start_index, end_index, edges_index) : {'hop_label', unique_hop_label}\n}\n```\n\nThe `edge_index` only increments for hop with the same starting and ending site indices which represent hops to different unit cells. ",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
c5006db5c6c20aedb09998d80392765e12a7aff8
| 10,142 |
ipynb
|
Jupyter Notebook
|
LabWork/02_formatsandlibraries.ipynb
|
engr-rudn/pyee01
|
831a780a8285e5650a55e1286d7565656dc089dc
|
[
"MIT"
] | null | null | null |
LabWork/02_formatsandlibraries.ipynb
|
engr-rudn/pyee01
|
831a780a8285e5650a55e1286d7565656dc089dc
|
[
"MIT"
] | null | null | null |
LabWork/02_formatsandlibraries.ipynb
|
engr-rudn/pyee01
|
831a780a8285e5650a55e1286d7565656dc089dc
|
[
"MIT"
] | null | null | null | 50.71 | 211 | 0.624137 |
[
[
[
"<h1 class=\"maintitle\">Raster Formats and Libraries</h1>\n\n<blockquote class=\"objectives\">\n<h2>Overview</h2>\n\n<div class=\"row\">\n<div class=\"col-md-3\">\n<strong>Teaching:</strong> 5 min\n <br>\n<strong>Exercises:</strong> 0 min\n</div>\n<div class=\"col-md-9\">\n<strong>Questions</strong>\n<ul>\n<li><p>What sorts of formats are available for representing raster datasets?</p>\n</li>\n</ul>\n</div>\n</div>\n<div class=\"row\">\n<div class=\"col-md-3\">\n</div>\n<div class=\"col-md-9\">\n<strong>Objectives</strong>\n<ul>\n<li><p>Understand the high-level data interchange formats for raster datasets.</p>\n</li>\n</ul>\n</div>\n</div>\n</blockquote>\n\n<h1 id=\"libraries-and-file-formats-for-raster-datasets\">Libraries and file formats for raster datasets</h1>\n\n<p><a href=\"http://gdal.org\">GDAL</a> (Geospatial Data Abstraction Library) is the de facto standard library for\ninteraction and manipulation of geospatial raster data. The primary purpose of GDAL or a\nGDAL-enabled library is to read, write and transform geospatial datasets in a\nway that makes sense in the context of its spatial metadata. GDAL also includes\na set of <a href=\"http://www.gdal.org/gdal_utilities.html\">command-line utilities</a> (e.g., gdalinfo, gdal_translate)\nfor convenient inspection and manipulation of raster data.</p>\n\n<p>Other libraries also exist (we’ll introduce rasterio in the next section of this\ntutorial, and even more exist in the fields of geoprocessing (which would\ninclude hydrological routing and other routines needed for Earth Systems\nSciences) and digital signal processing (including image classification,\npattern recognition, and feature extraction).</p>\n\n<p>GDAL’s support for different file formats depends on the format drivers that\nhave been implemented, and the libraries that are available at compile time.\nTo find the available formats for your current install of GDAL:</p>\n\n<div class=\"shell highlighter-rouge\"><div class=\"highlight\"><pre class=\"highlight\"><code>$ gdalinfo --formats\n</code></pre></div></div>\n\n<div class=\"highlighter-rouge\"><div class=\"highlight\"><pre class=\"highlight\"><code>Supported Formats:\n VRT -raster- (rw+v): Virtual Raster\n GTiff -raster- (rw+vs): GeoTIFF\n NITF -raster- (rw+vs): National Imagery Transmission Format\n RPFTOC -raster- (rovs): Raster Product Format TOC format\n ...\n # There are lots more, results depend on your build\n</code></pre></div></div>\n\n<p>Details about a specific format can be found with the <code class=\"highlighter-rouge\">--format</code> parameter,\nor by taking a look at the\n<a href=\"http://www.gdal.org/formats_list.html\">formats list on their website</a>.</p>\n\n<div class=\"highlighter-rouge\"><div class=\"highlight\"><pre class=\"highlight\"><code>$ gdalinfo --format GTiff\n</code></pre></div></div>\n\n<p>GDAL can operate on local files or even read files from the web like so:</p>\n<div class=\"shell highlighter-rouge\"><div class=\"highlight\"><pre class=\"highlight\"><code>$ SERVER='http://landsat-pds.s3.amazonaws.com/c1/L8/042/034/LC08_L1TP_042034_20170616_20170629_01_T1'\n$ IMAGE='LC08_L1TP_042034_20170616_20170629_01_T1_B4.TIF'\n$ gdalinfo /vsicurl/$SERVER/$IMAGE\n</code></pre></div></div>\n\n<div class=\"highlighter-rouge\"><div class=\"highlight\"><pre class=\"highlight\"><code>Driver: GTiff/GeoTIFF\nFiles: /vsicurl/http://landsat-pds.s3.amazonaws.com/c1/L8/042/034/LC08_L1TP_042034_20170616_20170629_01_T1/LC08_L1TP_042034_20170616_20170629_01_T1_B4.TIF\n /vsicurl/http://landsat-pds.s3.amazonaws.com/c1/L8/042/034/LC08_L1TP_042034_20170616_20170629_01_T1/LC08_L1TP_042034_20170616_20170629_01_T1_B4.TIF.ovr\n /vsicurl/http://landsat-pds.s3.amazonaws.com/c1/L8/042/034/LC08_L1TP_042034_20170616_20170629_01_T1/LC08_L1TP_042034_20170616_20170629_01_T1_MTL.txt\nSize is 7821, 7951\nCoordinate System is:\nPROJCS[\"WGS 84 / UTM zone 11N\",\n GEOGCS[\"WGS 84\",\n DATUM[\"WGS_1984\",\n SPHEROID[\"WGS 84\",6378137,298.257223563,\n AUTHORITY[\"EPSG\",\"7030\"]],\n AUTHORITY[\"EPSG\",\"6326\"]],\n PRIMEM[\"Greenwich\",0,\n AUTHORITY[\"EPSG\",\"8901\"]],\n UNIT[\"degree\",0.0174532925199433,\n AUTHORITY[\"EPSG\",\"9122\"]],\n AUTHORITY[\"EPSG\",\"4326\"]],\n PROJECTION[\"Transverse_Mercator\"],\n PARAMETER[\"latitude_of_origin\",0],\n PARAMETER[\"central_meridian\",-117],\n PARAMETER[\"scale_factor\",0.9996],\n PARAMETER[\"false_easting\",500000],\n PARAMETER[\"false_northing\",0],\n UNIT[\"metre\",1,\n AUTHORITY[\"EPSG\",\"9001\"]],\n AXIS[\"Easting\",EAST],\n AXIS[\"Northing\",NORTH],\n AUTHORITY[\"EPSG\",\"32611\"]]\nOrigin = (204285.000000000000000,4268115.000000000000000)\nPixel Size = (30.000000000000000,-30.000000000000000)\nMetadata:\n AREA_OR_POINT=Point\n METADATATYPE=ODL\nImage Structure Metadata:\n COMPRESSION=DEFLATE\n INTERLEAVE=BAND\nCorner Coordinates:\nUpper Left ( 204285.000, 4268115.000) (120d23'29.18\"W, 38d30'44.39\"N)\nLower Left ( 204285.000, 4029585.000) (120d17'44.96\"W, 36d21'57.41\"N)\nUpper Right ( 438915.000, 4268115.000) (117d42' 3.98\"W, 38d33'33.76\"N)\nLower Right ( 438915.000, 4029585.000) (117d40'52.67\"W, 36d24'34.20\"N)\nCenter ( 321600.000, 4148850.000) (119d 1' 2.61\"W, 37d28' 9.59\"N)\nBand 1 Block=512x512 Type=UInt16, ColorInterp=Gray\n Overviews: 2607x2651, 869x884, 290x295, 97x99\n</code></pre></div></div>\n\n<p>Often you want files in a specific format. GDAL is great for format conversions,\nhere is an example that saves a reference to a remote file to your local disk\nin the <a href=\"https://www.gdal.org/gdal_vrttut.html\">VRT format</a>.</p>\n<div class=\"highlighter-rouge\"><div class=\"highlight\"><pre class=\"highlight\"><code>$ gdal_translate -of VRT /vsicurl/$SERVER/$IMAGE LC08_L1TP_042034_20170616_20170629_01_T1_B4.vrt\n</code></pre></div></div>\n\n<p>Now you can forget about the strange ‘/vsicurl/’ syntax and just work directly\nwith the local file. The command below should give you the same print-out as\nearlier.</p>\n<div class=\"highlighter-rouge\"><div class=\"highlight\"><pre class=\"highlight\"><code>$ gdalinfo LC08_L1TP_042034_20170616_20170629_01_T1_B4.vrt\n</code></pre></div></div>\n\n<p>Another common task is warping an image to a different coordinate system. The\nexample command below warps the image from UTM Coordinates to WGS84 lat/lon\ncoordinates:</p>\n<div class=\"highlighter-rouge\"><div class=\"highlight\"><pre class=\"highlight\"><code>$ gdalwarp -t_srs EPSG:4326 -of VRT /vsicurl/$SERVER/$IMAGE LC08_L1TP_042034_20170616_20170629_01_T1_B4-wgs84.vrt\n</code></pre></div></div>\n\n<p>Confirm by looking at the new coordinates:</p>\n<div class=\"highlighter-rouge\"><div class=\"highlight\"><pre class=\"highlight\"><code>$ gdalinfo LC08_L1TP_042034_20170616_20170629_01_T1_B4-wgs84.vrt\n</code></pre></div></div>\n\n<p>As you can see, there is a lot you can do with GDAL command line utilities!</p>\n\n<h1 id=\"programming-model-numpy-arrays\">Programming model: NumPy arrays</h1>\n\n<p>Because rasters are images, they are best thought of as 2-dimensional arrays. If we\nhave multiple bands, we could think of an image as a 3-dimensional array.\nEither way, we are working with arrays (matrices) of pixel values, which in the\npython programming language are best represented by <a href=\"http://numpy.org\">NumPy</a> arrays.</p>\n\n<p>For this tutorial, we’ll perform basic operations with NumPy arrays extracted\nfrom geospatial rasters. For more information about multidimensional array\nanalysis, take a look at Thursday’s tutorial on\n<a href=\"https://geohackweek.github.io/nDarrays\">N-Dimensional Arrays</a>.</p>\n<blockquote class=\"keypoints\">\n<h2>Key Points</h2>\n<ul>\n<li><p>Geospatial libraries such as GDAL are very useful for reading, writing and transforming rasters</p>\n</li>\n<li><p>Once a raster’s pixel values have been extracted to a NumPy array, they can be processed with more specialized libraries and routines.</p>\n</li> \n</ul>\n</blockquote>\n<hr>\nThis notebook is inspired by the material in the website <a href=\"https://geohackweewebsite k.github.io/raster/04-workingwithrasters/\"> GeoHackWeek </a>",
"_____no_output_____"
]
]
] |
[
"markdown"
] |
[
[
"markdown"
]
] |
c5008a316ebbfcef2da43a84ba9d0b6080dc9976
| 2,811 |
ipynb
|
Jupyter Notebook
|
.ipynb_checkpoints/Roots-checkpoint.ipynb
|
smithrockmaker/PhysicsTools
|
d55c311009ac3e6552d18a179bbbc82f41349130
|
[
"MIT"
] | null | null | null |
.ipynb_checkpoints/Roots-checkpoint.ipynb
|
smithrockmaker/PhysicsTools
|
d55c311009ac3e6552d18a179bbbc82f41349130
|
[
"MIT"
] | null | null | null |
.ipynb_checkpoints/Roots-checkpoint.ipynb
|
smithrockmaker/PhysicsTools
|
d55c311009ac3e6552d18a179bbbc82f41349130
|
[
"MIT"
] | null | null | null | 24.657895 | 445 | 0.569192 |
[
[
[
"### Roots Finder Using Numpy\n\nJust because I am always using my phone as a calculator and it doesn't do this:) I originally consider a direct quadratic solver from the ```math``` library and then realized is was useful to have a more general solver available. [This model](https://nkrvavica.github.io/post/on_computing_roots/) from a computational engineer in Croatia ([Nino Krvavica](https://nkrvavica.github.io/#about)) totally got me so that's what this is based on.",
"_____no_output_____"
]
],
[
[
"import numpy as np",
"_____no_output_____"
]
],
[
[
"### numpy.roots\n\nDocumentation for [numpy.roots](https://numpy.org/doc/stable/reference/generated/numpy.roots.html) suggests that it will report complex roots when they occur. Since speed is not my issue many of Nino's concerns are not mine. As long as I provide a coefficient vector numpt.roots will find all appropriate roots and return them. \n\nThe coefficients are entered from highest order down to the constant!",
"_____no_output_____"
],
[
"### Quadratic:",
"_____no_output_____"
]
],
[
[
"coeffsQuad = [.83,-6.66, 7.5]\nquadRoots = np.roots(coeffsQuad)\nprint(\"quadratic roots: \",quadRoots)",
"quadratic roots: [6.6691869 1.35490948]\n"
]
],
[
[
"### Cubic:",
"_____no_output_____"
]
],
[
[
"coeffsCubic = [4.9, -6.8, 1.82, 3.]\nquadCubic = np.roots(coeffsCubic)\nprint(\"cubic roots: \",quadCubic)",
"quadratic roots: [ 0.93463374+0.63084369j 0.93463374-0.63084369j -0.48151237+0.j ]\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
c5008ce38db25f96d69b5ef81190da3d88c0405c
| 47,535 |
ipynb
|
Jupyter Notebook
|
notebooks/community/migration/UJ6 legacy AutoML Natural Language Text Classification.ipynb
|
nayaknishant/vertex-ai-samples
|
3ce120b953f1cdc2ec2c5a3f4509cfeab106b7d0
|
[
"Apache-2.0"
] | 418 |
2019-06-26T05:55:42.000Z
|
2022-03-31T10:46:57.000Z
|
notebooks/community/migration/UJ6 legacy AutoML Natural Language Text Classification.ipynb
|
nayaknishant/vertex-ai-samples
|
3ce120b953f1cdc2ec2c5a3f4509cfeab106b7d0
|
[
"Apache-2.0"
] | 362 |
2019-06-26T20:41:17.000Z
|
2022-02-10T16:02:16.000Z
|
notebooks/community/migration/UJ6 legacy AutoML Natural Language Text Classification.ipynb
|
nayaknishant/vertex-ai-samples
|
3ce120b953f1cdc2ec2c5a3f4509cfeab106b7d0
|
[
"Apache-2.0"
] | 229 |
2019-06-29T17:55:33.000Z
|
2022-03-14T15:52:58.000Z
| 28.093972 | 967 | 0.471211 |
[
[
[
"# Copyright 2021 Google LLC\n#\n# Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# https://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.",
"_____no_output_____"
]
],
[
[
"# AutoML natural language text classification model\n",
"_____no_output_____"
],
[
"## Installation\n\nInstall the latest version of AutoML SDK.",
"_____no_output_____"
]
],
[
[
"! pip3 install google-cloud-automl",
"_____no_output_____"
]
],
[
[
"Install the Google *cloud-storage* library as well.",
"_____no_output_____"
]
],
[
[
"! pip3 install google-cloud-storage",
"_____no_output_____"
]
],
[
[
"### Restart the Kernel\n\nOnce you've installed the AutoML SDK and Google *cloud-storage*, you need to restart the notebook kernel so it can find the packages.",
"_____no_output_____"
]
],
[
[
"import os\n\nif not os.getenv(\"AUTORUN\"):\n # Automatically restart kernel after installs\n import IPython\n\n app = IPython.Application.instance()\n app.kernel.do_shutdown(True)",
"_____no_output_____"
]
],
[
[
"## Before you begin\r\n\r\n### GPU run-time\r\n\r\n*Make sure you're running this notebook in a GPU runtime if you have that option. In Colab, select* **Runtime > Change Runtime Type > GPU**\r\n\r\n### Set up your GCP project\r\n\r\n**The following steps are required, regardless of your notebook environment.**\r\n\r\n1. [Select or create a GCP project](https://console.cloud.google.com/cloud-resource-manager). When you first create an account, you get a $300 free credit towards your compute/storage costs.\r\n\r\n2. [Make sure that billing is enabled for your project.](https://cloud.google.com/billing/docs/how-to/modify-project)\r\n\r\n3. [Enable the AutoML APIs and Compute Engine APIs.](https://console.cloud.google.com/flows/enableapi?apiid=ml.googleapis.com,compute_component)\r\n\r\n4. [Google Cloud SDK](https://cloud.google.com/sdk) is already installed in AutoML Notebooks.\r\n\r\n5. Enter your project ID in the cell below. Then run the cell to make sure the\r\nCloud SDK uses the right project for all the commands in this notebook.\r\n\r\n**Note**: Jupyter runs lines prefixed with `!` as shell commands, and it interpolates Python variables prefixed with `$` into these commands.",
"_____no_output_____"
],
[
"#### Project ID\n\n**If you don't know your project ID**, try to get your project ID using `gcloud` command by executing the second cell below.",
"_____no_output_____"
]
],
[
[
"PROJECT_ID = \"[your-project-id]\" # @param {type:\"string\"}",
"_____no_output_____"
],
[
"if PROJECT_ID == \"\" or PROJECT_ID is None or PROJECT_ID == \"[your-project-id]\":\n # Get your GCP project id from gcloud\n shell_output = !gcloud config list --format 'value(core.project)' 2>/dev/null\n PROJECT_ID = shell_output[0]\n print(\"Project ID:\", PROJECT_ID)",
"_____no_output_____"
],
[
"! gcloud config set project $PROJECT_ID",
"_____no_output_____"
]
],
[
[
"#### Region\n\nYou can also change the `REGION` variable, which is used for operations\nthroughout the rest of this notebook. Below are regions supported for AutoML. We recommend when possible, to choose the region closest to you.\n\n- Americas: `us-central1`\n- Europe: `europe-west4`\n- Asia Pacific: `asia-east1`\n\nYou cannot use a Multi-Regional Storage bucket for training with AutoML. Not all regions provide support for all AutoML services. For the latest support per region, see [Region support for AutoML services]()",
"_____no_output_____"
]
],
[
[
"REGION = \"us-central1\" # @param {type: \"string\"}",
"_____no_output_____"
]
],
[
[
"#### Timestamp\n\nIf you are in a live tutorial session, you might be using a shared test account or project. To avoid name collisions between users on resources created, you create a timestamp for each instance session, and append onto the name of resources which will be created in this tutorial.",
"_____no_output_____"
]
],
[
[
"from datetime import datetime\n\nTIMESTAMP = datetime.now().strftime(\"%Y%m%d%H%M%S\")",
"_____no_output_____"
]
],
[
[
"### Authenticate your GCP account\r\n\r\n**If you are using AutoML Notebooks**, your environment is already\r\nauthenticated. Skip this step.\r\n\r\n*Note: If you are on an AutoML notebook and run the cell, the cell knows to skip executing the authentication steps.*",
"_____no_output_____"
]
],
[
[
"import os\nimport sys\n\n# If you are running this notebook in Colab, run this cell and follow the\n# instructions to authenticate your Google Cloud account. This provides access\n# to your Cloud Storage bucket and lets you submit training jobs and prediction\n# requests.\n\n# If on Vertex, then don't execute this code\nif not os.path.exists(\"/opt/deeplearning/metadata/env_version\"):\n if \"google.colab\" in sys.modules:\n from google.colab import auth as google_auth\n\n google_auth.authenticate_user()\n\n # If you are running this tutorial in a notebook locally, replace the string\n # below with the path to your service account key and run this cell to\n # authenticate your Google Cloud account.\n else:\n %env GOOGLE_APPLICATION_CREDENTIALS your_path_to_credentials.json\n\n # Log in to your account on Google Cloud\n ! gcloud auth login",
"_____no_output_____"
]
],
[
[
"### Create a Cloud Storage bucket\n\n**The following steps are required, regardless of your notebook environment.**\n\nThis tutorial is designed to use training data that is in a public Cloud Storage bucket and a local Cloud Storage bucket for your batch predictions. You may alternatively use your own training data that you have stored in a local Cloud Storage bucket.\n\nSet the name of your Cloud Storage bucket below. It must be unique across all Cloud Storage buckets. ",
"_____no_output_____"
]
],
[
[
"BUCKET_NAME = \"[your-bucket-name]\" # @param {type:\"string\"}",
"_____no_output_____"
],
[
"if BUCKET_NAME == \"\" or BUCKET_NAME is None or BUCKET_NAME == \"[your-bucket-name]\":\n BUCKET_NAME = PROJECT_ID + \"aip-\" + TIMESTAMP",
"_____no_output_____"
]
],
[
[
"**Only if your bucket doesn't already exist**: Run the following cell to create your Cloud Storage bucket.",
"_____no_output_____"
]
],
[
[
"! gsutil mb -l $REGION gs://$BUCKET_NAME",
"_____no_output_____"
]
],
[
[
"Finally, validate access to your Cloud Storage bucket by examining its contents:",
"_____no_output_____"
]
],
[
[
"! gsutil ls -al gs://$BUCKET_NAME",
"_____no_output_____"
]
],
[
[
"### Set up variables\n\nNext, set up some variables used throughout the tutorial.\n### Import libraries and define constants",
"_____no_output_____"
],
[
"#### Import AutoML SDK\n\nImport the AutoM SDK into our Python environment.",
"_____no_output_____"
]
],
[
[
"import json\nimport time\n\nfrom google.cloud import automl\nfrom google.protobuf.json_format import MessageToJson",
"_____no_output_____"
]
],
[
[
"#### AutoML constants\n\nSetup up the following constants for AutoML:\n\n- `PARENT`: The AutoM location root path for dataset, model and endpoint resources.",
"_____no_output_____"
]
],
[
[
"# AutoM location root path for your dataset, model and endpoint resources\nPARENT = \"projects/\" + PROJECT_ID + \"/locations/\" + REGION",
"_____no_output_____"
]
],
[
[
"## Clients\n\nThe AutoML SDK works as a client/server model. On your side (the Python script) you will create a client that sends requests and receives responses from the server (AutoML).\n\nYou will use several clients in this tutorial, so set them all up upfront.\n",
"_____no_output_____"
]
],
[
[
"def automl_client():\n return automl.AutoMlClient()\n\n\ndef prediction_client():\n return automl.PredictionServiceClient()\n\n\ndef operations_client():\n return automl.AutoMlClient()._transport.operations_client\n\n\nclients = {}\nclients[\"automl\"] = automl_client()\nclients[\"prediction\"] = prediction_client()\nclients[\"operations\"] = operations_client()\n\nfor client in clients.items():\n print(client)",
"_____no_output_____"
],
[
"IMPORT_FILE = \"gs://cloud-ml-data/NL-classification/happiness.csv\"",
"_____no_output_____"
],
[
"! gsutil cat $IMPORT_FILE | head -n 10",
"_____no_output_____"
]
],
[
[
"*Example output*:\n```\nI went on a successful date with someone I felt sympathy and connection with.,affection\nI was happy when my son got 90% marks in his examination,affection\nI went to the gym this morning and did yoga.,exercise\nWe had a serious talk with some friends of ours who have been flaky lately. They understood and we had a good evening hanging out.,bonding\nI went with grandchildren to butterfly display at Crohn Conservatory,affection\nI meditated last night.,leisure\n\"I made a new recipe for peasant bread, and it came out spectacular!\",achievement\nI got gift from my elder brother which was really surprising me,affection\nYESTERDAY MY MOMS BIRTHDAY SO I ENJOYED,enjoy_the_moment\nWatching cupcake wars with my three teen children,affection\n```\n",
"_____no_output_____"
],
[
"## Create a dataset",
"_____no_output_____"
],
[
"### [projects.locations.datasets.create](https://cloud.google.com/automl/docs/reference/rest/v1beta1/projects.locations.datasets/create)",
"_____no_output_____"
],
[
"#### Request",
"_____no_output_____"
]
],
[
[
"dataset = {\n \"display_name\": \"happiness_\" + TIMESTAMP,\n \"text_classification_dataset_metadata\": {\"classification_type\": \"MULTICLASS\"},\n}\n\nprint(\n MessageToJson(\n automl.CreateDatasetRequest(parent=PARENT, dataset=dataset).__dict__[\"_pb\"]\n )\n)",
"_____no_output_____"
]
],
[
[
"*Example output*:\n```\n{\n \"parent\": \"projects/migration-ucaip-training/locations/us-central1\",\n \"dataset\": {\n \"displayName\": \"happiness_20210228224317\",\n \"textClassificationDatasetMetadata\": {\n \"classificationType\": \"MULTICLASS\"\n }\n }\n}\n```\n",
"_____no_output_____"
],
[
"#### Call",
"_____no_output_____"
]
],
[
[
"request = clients[\"automl\"].create_dataset(parent=PARENT, dataset=dataset)",
"_____no_output_____"
]
],
[
[
"#### Response",
"_____no_output_____"
]
],
[
[
"result = request.result()\n\nprint(MessageToJson(result.__dict__[\"_pb\"]))",
"_____no_output_____"
]
],
[
[
"*Example output*:\n```\n{\n \"name\": \"projects/116273516712/locations/us-central1/datasets/TCN2705019056410329088\"\n}\n```\n",
"_____no_output_____"
]
],
[
[
"# The full unique ID for the dataset\ndataset_id = result.name\n# The short numeric ID for the dataset\ndataset_short_id = dataset_id.split(\"/\")[-1]\n\nprint(dataset_id)",
"_____no_output_____"
]
],
[
[
"### [projects.locations.datasets.importData](https://cloud.google.com/automl/docs/reference/rest/v1beta1/projects.locations.datasets/importData)",
"_____no_output_____"
],
[
"#### Request",
"_____no_output_____"
]
],
[
[
"input_config = {\"gcs_source\": {\"input_uris\": [IMPORT_FILE]}}\n\nprint(\n MessageToJson(\n automl.ImportDataRequest(name=dataset_id, input_config=input_config).__dict__[\n \"_pb\"\n ]\n )\n)",
"_____no_output_____"
]
],
[
[
"*Example output*:\n```\n{\n \"name\": \"projects/116273516712/locations/us-central1/datasets/TCN2705019056410329088\",\n \"inputConfig\": {\n \"gcsSource\": {\n \"inputUris\": [\n \"gs://cloud-ml-data/NL-classification/happiness.csv\"\n ]\n }\n }\n}\n```\n",
"_____no_output_____"
],
[
"#### Call",
"_____no_output_____"
]
],
[
[
"request = clients[\"automl\"].import_data(name=dataset_id, input_config=input_config)",
"_____no_output_____"
]
],
[
[
"#### Response",
"_____no_output_____"
]
],
[
[
"result = request.result()\n\nprint(MessageToJson(result))",
"_____no_output_____"
]
],
[
[
"*Example output*:\n```\n{}\n```\n",
"_____no_output_____"
],
[
"## Train a model",
"_____no_output_____"
],
[
"### [projects.locations.models.create](https://cloud.google.com/automl/docs/reference/rest/v1beta1/projects.locations.models/create)",
"_____no_output_____"
],
[
"#### Request",
"_____no_output_____"
]
],
[
[
"model = automl.Model(\n display_name=\"happiness_\" + TIMESTAMP,\n dataset_id=dataset_short_id,\n text_classification_model_metadata=automl.TextClassificationModelMetadata(),\n)\n\nprint(\n MessageToJson(automl.CreateModelRequest(parent=PARENT, model=model).__dict__[\"_pb\"])\n)",
"_____no_output_____"
]
],
[
[
"*Example output*:\n```\n{\n \"parent\": \"projects/migration-ucaip-training/locations/us-central1\",\n \"model\": {\n \"displayName\": \"happiness_20210228224317\",\n \"datasetId\": \"TCN2705019056410329088\",\n \"textClassificationModelMetadata\": {}\n }\n}\n```\n",
"_____no_output_____"
],
[
"#### Call",
"_____no_output_____"
]
],
[
[
"request = clients[\"automl\"].create_model(parent=PARENT, model=model)",
"_____no_output_____"
]
],
[
[
"#### Response",
"_____no_output_____"
]
],
[
[
"result = request.result()\n\nprint(MessageToJson(result.__dict__[\"_pb\"]))",
"_____no_output_____"
]
],
[
[
"*Example output*:\n```\n{\n \"name\": \"projects/116273516712/locations/us-central1/models/TCN5333697920992542720\"\n}\n```\n",
"_____no_output_____"
]
],
[
[
"# The full unique ID for the training pipeline\nmodel_id = result.name\n# The short numeric ID for the training pipeline\nmodel_short_id = model_id.split(\"/\")[-1]\n\nprint(model_short_id)",
"_____no_output_____"
]
],
[
[
"## Evaluate the model",
"_____no_output_____"
],
[
"### [projects.locations.models.modelEvaluations.list](https://cloud.google.com/automl/docs/reference/rest/v1beta1/projects.locations.models.modelEvaluations/list)",
"_____no_output_____"
],
[
"#### Call",
"_____no_output_____"
]
],
[
[
"request = clients[\"automl\"].list_model_evaluations(parent=model_id, filter=\"\")",
"_____no_output_____"
]
],
[
[
"#### Response",
"_____no_output_____"
]
],
[
[
"evaluations_list = [\n json.loads(MessageToJson(me.__dict__[\"_pb\"])) for me in request.model_evaluation\n]\n\nprint(json.dumps(evaluations_list, indent=2))\n# The evaluation slice\nevaluation_slice = request.model_evaluation[0].name",
"_____no_output_____"
]
],
[
[
"*Example output*:\n```\n[\n {\n \"name\": \"projects/116273516712/locations/us-central1/models/TCN5333697920992542720/modelEvaluations/1436745357261371663\",\n \"annotationSpecId\": \"3130761503557287936\",\n \"createTime\": \"2021-03-01T02:56:28.878044Z\",\n \"evaluatedExampleCount\": 1193,\n \"classificationEvaluationMetrics\": {\n \"auPrc\": 0.99065405,\n \"confidenceMetricsEntry\": [\n {\n \"recall\": 1.0,\n \"precision\": 0.01424979,\n \"f1Score\": 0.028099174\n },\n {\n \"confidenceThreshold\": 0.05,\n \"recall\": 1.0,\n \"precision\": 0.5862069,\n \"f1Score\": 0.73913044\n },\n {\n \"confidenceThreshold\": 0.94,\n \"recall\": 0.64705884,\n \"precision\": 1.0,\n \"f1Score\": 0.7857143\n },\n \n # REMOVED FOR BREVITY\n \n {\n \"confidenceThreshold\": 0.999,\n \"recall\": 0.21372032,\n \"precision\": 1.0,\n \"f1Score\": 0.35217392\n },\n {\n \"confidenceThreshold\": 1.0,\n \"recall\": 0.0026385225,\n \"precision\": 1.0,\n \"f1Score\": 0.005263158\n }\n ],\n \"logLoss\": 0.14686257\n },\n \"displayName\": \"achievement\"\n }\n]\n```\n",
"_____no_output_____"
],
[
"### [projects.locations.models.modelEvaluations.get](https://cloud.google.com/automl/docs/reference/rest/v1beta1/projects.locations.models.modelEvaluations/get)",
"_____no_output_____"
],
[
"#### Call",
"_____no_output_____"
]
],
[
[
"request = clients[\"automl\"].get_model_evaluation(name=evaluation_slice)",
"_____no_output_____"
]
],
[
[
"#### Response",
"_____no_output_____"
]
],
[
[
"print(MessageToJson(request.__dict__[\"_pb\"]))",
"_____no_output_____"
]
],
[
[
"*Example output*:\n```\n{\n \"name\": \"projects/116273516712/locations/us-central1/models/TCN5333697920992542720/modelEvaluations/1436745357261371663\",\n \"annotationSpecId\": \"3130761503557287936\",\n \"createTime\": \"2021-03-01T02:56:28.878044Z\",\n \"evaluatedExampleCount\": 1193,\n \"classificationEvaluationMetrics\": {\n \"auPrc\": 0.99065405,\n \"confidenceMetricsEntry\": [\n {\n \"recall\": 1.0,\n \"precision\": 0.01424979,\n \"f1Score\": 0.028099174\n },\n {\n \"confidenceThreshold\": 0.05,\n \"recall\": 1.0,\n \"precision\": 0.5862069,\n \"f1Score\": 0.73913044\n },\n \n # REMOVED FOR BREVITY\n \n {\n \"confidenceThreshold\": 0.999,\n \"recall\": 0.23529412,\n \"precision\": 1.0,\n \"f1Score\": 0.3809524\n },\n {\n \"confidenceThreshold\": 1.0,\n \"precision\": 1.0\n }\n ],\n \"logLoss\": 0.005436425\n },\n \"displayName\": \"exercise\"\n}\n```\n",
"_____no_output_____"
],
[
"## Make batch predictions",
"_____no_output_____"
],
[
"### Prepare files for batch prediction",
"_____no_output_____"
]
],
[
[
"test_item = ! gsutil cat $IMPORT_FILE | head -n1\ntest_item, test_label = str(test_item[0]).split(\",\")\n\nprint(test_item, test_label)",
"_____no_output_____"
],
[
"import json\n\nimport tensorflow as tf\n\ntest_item_uri = \"gs://\" + BUCKET_NAME + \"/test.txt\"\nwith tf.io.gfile.GFile(test_item_uri, \"w\") as f:\n f.write(test_item + \"\\n\")\n\ngcs_input_uri = \"gs://\" + BUCKET_NAME + \"/batch.csv\"\nwith tf.io.gfile.GFile(gcs_input_uri, \"w\") as f:\n f.write(test_item_uri + \"\\n\")",
"_____no_output_____"
],
[
"! gsutil cat $gcs_input_uri\n! gsutil cat $test_item_uri",
"_____no_output_____"
]
],
[
[
"*Example output*:\n```\ngs://migration-ucaip-trainingaip-20210228224317/test.txt\nI went on a successful date with someone I felt sympathy and connection with.\n```\n",
"_____no_output_____"
],
[
"### [projects.locations.models.batchPredict](https://cloud.google.com/automl/docs/reference/rest/v1beta1/projects.locations.models/batchPredict)",
"_____no_output_____"
],
[
"#### Request",
"_____no_output_____"
]
],
[
[
"input_config = {\"gcs_source\": {\"input_uris\": [gcs_input_uri]}}\n\noutput_config = {\n \"gcs_destination\": {\"output_uri_prefix\": \"gs://\" + f\"{BUCKET_NAME}/batch_output/\"}\n}\n\nprint(\n MessageToJson(\n automl.BatchPredictRequest(\n name=model_id, input_config=input_config, output_config=output_config\n ).__dict__[\"_pb\"]\n )\n)",
"_____no_output_____"
]
],
[
[
"*Example output*:\n```\n{\n \"name\": \"projects/116273516712/locations/us-central1/models/TCN5333697920992542720\",\n \"inputConfig\": {\n \"gcsSource\": {\n \"inputUris\": [\n \"gs://migration-ucaip-trainingaip-20210228224317/batch.csv\"\n ]\n }\n },\n \"outputConfig\": {\n \"gcsDestination\": {\n \"outputUriPrefix\": \"gs://migration-ucaip-trainingaip-20210228224317/batch_output/\"\n }\n }\n}\n```\n",
"_____no_output_____"
],
[
"#### Call",
"_____no_output_____"
]
],
[
[
"request = clients[\"prediction\"].batch_predict(\n name=model_id, input_config=input_config, output_config=output_config\n)",
"_____no_output_____"
]
],
[
[
"#### Response",
"_____no_output_____"
]
],
[
[
"result = request.result()\n\nprint(MessageToJson(result.__dict__[\"_pb\"]))",
"_____no_output_____"
]
],
[
[
"*Example output*:\r\n```\r\n{}\r\n```",
"_____no_output_____"
]
],
[
[
"destination_uri = output_config[\"gcs_destination\"][\"output_uri_prefix\"][:-1]\n\n! gsutil ls $destination_uri/*\n! gsutil cat $destination_uri/prediction*/*.jsonl",
"_____no_output_____"
]
],
[
[
"*Example output*:\n```\ngs://migration-ucaip-trainingaip-20210228224317/batch_output/prediction-happiness_20210228224317-2021-03-01T02:57:02.004934Z/text_classification_1.jsonl\ngs://migration-ucaip-trainingaip-20210228224317/batch_output/prediction-happiness_20210228224317-2021-03-01T02:57:02.004934Z/text_classification_2.jsonl\n{\"textSnippet\":{\"contentUri\":\"gs://migration-ucaip-trainingaip-20210228224317/test.txt\"},\"annotations\":[{\"annotationSpecId\":\"5436604512770981888\",\"classification\":{\"score\":0.93047273},\"displayName\":\"affection\"},{\"annotationSpecId\":\"3707222255860711424\",\"classification\":{\"score\":0.002518793},\"displayName\":\"achievement\"},{\"annotationSpecId\":\"7742447521984675840\",\"classification\":{\"score\":1.3182563E-4},\"displayName\":\"enjoy_the_moment\"},{\"annotationSpecId\":\"824918494343593984\",\"classification\":{\"score\":0.06613126},\"displayName\":\"bonding\"},{\"annotationSpecId\":\"1977839998950440960\",\"classification\":{\"score\":1.5267624E-5},\"displayName\":\"leisure\"},{\"annotationSpecId\":\"8318908274288099328\",\"classification\":{\"score\":8.887557E-6},\"displayName\":\"nature\"},{\"annotationSpecId\":\"3130761503557287936\",\"classification\":{\"score\":7.2130124E-4},\"displayName\":\"exercise\"}]}\n```",
"_____no_output_____"
],
[
"## Make online predictions",
"_____no_output_____"
],
[
"### [projects.locations.models.deploy](https://cloud.google.com/automl/docs/reference/rest/v1beta1/projects.locations.models/deploy)",
"_____no_output_____"
],
[
"#### Call",
"_____no_output_____"
]
],
[
[
"request = clients[\"automl\"].deploy_model(name=model_id)",
"_____no_output_____"
]
],
[
[
"#### Response",
"_____no_output_____"
]
],
[
[
"result = request.result()\n\nprint(MessageToJson(result))",
"_____no_output_____"
]
],
[
[
"*Example output*:\r\n```\r\n{}\r\n```",
"_____no_output_____"
],
[
"### [projects.locations.models.predict](https://cloud.google.com/automl/docs/reference/rest/v1beta1/projects.locations.models/predict)",
"_____no_output_____"
],
[
"### Prepare data item for online prediction",
"_____no_output_____"
]
],
[
[
"test_item = ! gsutil cat $IMPORT_FILE | head -n1\ntest_item, test_label = str(test_item[0]).split(\",\")",
"_____no_output_____"
]
],
[
[
"#### Request",
"_____no_output_____"
]
],
[
[
"payload = {\"text_snippet\": {\"content\": test_item, \"mime_type\": \"text/plain\"}}\n\nrequest = automl.PredictRequest(name=model_id, payload=payload)\n\nprint(MessageToJson(request.__dict__[\"_pb\"]))",
"_____no_output_____"
]
],
[
[
"*Example output*:\n```\n{\n \"name\": \"projects/116273516712/locations/us-central1/models/TCN5333697920992542720\",\n \"payload\": {\n \"textSnippet\": {\n \"content\": \"I went on a successful date with someone I felt sympathy and connection with.\",\n \"mimeType\": \"text/plain\"\n }\n }\n}\n```\n",
"_____no_output_____"
],
[
"#### Call",
"_____no_output_____"
]
],
[
[
"request = clients[\"prediction\"].predict(request=request)",
"_____no_output_____"
]
],
[
[
"#### Response",
"_____no_output_____"
]
],
[
[
"print(MessageToJson(request.__dict__[\"_pb\"]))",
"_____no_output_____"
]
],
[
[
"*Example output*:\n```\n{\n \"payload\": [\n {\n \"annotationSpecId\": \"5436604512770981888\",\n \"classification\": {\n \"score\": 0.9272586\n },\n \"displayName\": \"affection\"\n },\n {\n \"annotationSpecId\": \"824918494343593984\",\n \"classification\": {\n \"score\": 0.068884976\n },\n \"displayName\": \"bonding\"\n },\n {\n \"annotationSpecId\": \"3707222255860711424\",\n \"classification\": {\n \"score\": 0.0028119811\n },\n \"displayName\": \"achievement\"\n },\n {\n \"annotationSpecId\": \"3130761503557287936\",\n \"classification\": {\n \"score\": 0.0008869726\n },\n \"displayName\": \"exercise\"\n },\n {\n \"annotationSpecId\": \"7742447521984675840\",\n \"classification\": {\n \"score\": 0.00013229548\n },\n \"displayName\": \"enjoy_the_moment\"\n },\n {\n \"annotationSpecId\": \"1977839998950440960\",\n \"classification\": {\n \"score\": 1.5584701e-05\n },\n \"displayName\": \"leisure\"\n },\n {\n \"annotationSpecId\": \"8318908274288099328\",\n \"classification\": {\n \"score\": 9.5975e-06\n },\n \"displayName\": \"nature\"\n }\n ]\n}\n```\n",
"_____no_output_____"
],
[
"# Cleaning up\r\n\r\nTo clean up all GCP resources used in this project, you can [delete the GCP\r\nproject](https://cloud.google.com/resource-manager/docs/creating-managing-projects#shutting_down_projects) you used for the tutorial.\r\n\r\nOtherwise, you can delete the individual resources you created in this tutorial.",
"_____no_output_____"
]
],
[
[
"delete_dataset = True\ndelete_model = True\ndelete_bucket = True\n\n# Delete the dataset using the AutoML fully qualified identifier for the dataset\ntry:\n if delete_dataset:\n clients[\"automl\"].delete_dataset(name=dataset_id)\nexcept Exception as e:\n print(e)\n\n# Delete the model using the AutoML fully qualified identifier for the model\ntry:\n if delete_model:\n clients[\"automl\"].delete_model(name=model_id)\nexcept Exception as e:\n print(e)\n\n\nif delete_bucket and \"BUCKET_NAME\" in globals():\n ! gsutil rm -r gs://$BUCKET_NAME",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
]
] |
c50098527143a05c8f84c1a661f1019a8e2b0fa8
| 29,570 |
ipynb
|
Jupyter Notebook
|
ml/cc/exercises/ko/intro_to_neural_nets.ipynb
|
ananci/eng-edu
|
ac907dab212d4232f5fbbd69ba75272d8f3110e5
|
[
"Apache-2.0"
] | null | null | null |
ml/cc/exercises/ko/intro_to_neural_nets.ipynb
|
ananci/eng-edu
|
ac907dab212d4232f5fbbd69ba75272d8f3110e5
|
[
"Apache-2.0"
] | null | null | null |
ml/cc/exercises/ko/intro_to_neural_nets.ipynb
|
ananci/eng-edu
|
ac907dab212d4232f5fbbd69ba75272d8f3110e5
|
[
"Apache-2.0"
] | null | null | null | 44.466165 | 919 | 0.566994 |
[
[
[
"#### Copyright 2017 Google LLC.",
"_____no_output_____"
]
],
[
[
"# Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# https://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.",
"_____no_output_____"
]
],
[
[
" # 신경망 소개",
"_____no_output_____"
],
[
" **학습 목표:**\n * 텐서플로우의 `DNNRegressor` 클래스를 사용하여 신경망(NN) 및 히든 레이어를 정의한다\n * 비선형성을 갖는 데이터 세트를 신경망에 학습시켜 선형 회귀 모델보다 우수한 성능을 달성한다",
"_____no_output_____"
],
[
" 이전 실습에서는 모델에 비선형성을 통합하는 데 도움이 되는 합성 특성을 사용했습니다.\n\n비선형성을 갖는 대표적인 세트는 위도와 경도였지만 다른 특성도 있을 수 있습니다.\n\n일단 이전 실습의 로지스틱 회귀 작업이 아닌 표준 회귀 작업으로 돌아가겠습니다. 즉, `median_house_value`를 직접 예측할 것입니다.",
"_____no_output_____"
],
[
" ## 설정\n\n우선 데이터를 로드하고 준비하겠습니다.",
"_____no_output_____"
]
],
[
[
"from __future__ import print_function\n\nimport math\n\nfrom IPython import display\nfrom matplotlib import cm\nfrom matplotlib import gridspec\nfrom matplotlib import pyplot as plt\nimport numpy as np\nimport pandas as pd\nfrom sklearn import metrics\nimport tensorflow as tf\nfrom tensorflow.python.data import Dataset\n\ntf.logging.set_verbosity(tf.logging.ERROR)\npd.options.display.max_rows = 10\npd.options.display.float_format = '{:.1f}'.format\n\ncalifornia_housing_dataframe = pd.read_csv(\"https://download.mlcc.google.com/mledu-datasets/california_housing_train.csv\", sep=\",\")\n\ncalifornia_housing_dataframe = california_housing_dataframe.reindex(\n np.random.permutation(california_housing_dataframe.index))",
"_____no_output_____"
],
[
"def preprocess_features(california_housing_dataframe):\n \"\"\"Prepares input features from California housing data set.\n\n Args:\n california_housing_dataframe: A Pandas DataFrame expected to contain data\n from the California housing data set.\n Returns:\n A DataFrame that contains the features to be used for the model, including\n synthetic features.\n \"\"\"\n selected_features = california_housing_dataframe[\n [\"latitude\",\n \"longitude\",\n \"housing_median_age\",\n \"total_rooms\",\n \"total_bedrooms\",\n \"population\",\n \"households\",\n \"median_income\"]]\n processed_features = selected_features.copy()\n # Create a synthetic feature.\n processed_features[\"rooms_per_person\"] = (\n california_housing_dataframe[\"total_rooms\"] /\n california_housing_dataframe[\"population\"])\n return processed_features\n\ndef preprocess_targets(california_housing_dataframe):\n \"\"\"Prepares target features (i.e., labels) from California housing data set.\n\n Args:\n california_housing_dataframe: A Pandas DataFrame expected to contain data\n from the California housing data set.\n Returns:\n A DataFrame that contains the target feature.\n \"\"\"\n output_targets = pd.DataFrame()\n # Scale the target to be in units of thousands of dollars.\n output_targets[\"median_house_value\"] = (\n california_housing_dataframe[\"median_house_value\"] / 1000.0)\n return output_targets",
"_____no_output_____"
],
[
"# Choose the first 12000 (out of 17000) examples for training.\ntraining_examples = preprocess_features(california_housing_dataframe.head(12000))\ntraining_targets = preprocess_targets(california_housing_dataframe.head(12000))\n\n# Choose the last 5000 (out of 17000) examples for validation.\nvalidation_examples = preprocess_features(california_housing_dataframe.tail(5000))\nvalidation_targets = preprocess_targets(california_housing_dataframe.tail(5000))\n\n# Double-check that we've done the right thing.\nprint(\"Training examples summary:\")\ndisplay.display(training_examples.describe())\nprint(\"Validation examples summary:\")\ndisplay.display(validation_examples.describe())\n\nprint(\"Training targets summary:\")\ndisplay.display(training_targets.describe())\nprint(\"Validation targets summary:\")\ndisplay.display(validation_targets.describe())",
"_____no_output_____"
]
],
[
[
" ## 신경망 구축\n\nNN은 [DNNRegressor](https://www.tensorflow.org/api_docs/python/tf/estimator/DNNRegressor) 클래스에 의해 정의됩니다.\n\n**`hidden_units`**를 사용하여 NN의 구조를 정의합니다. `hidden_units` 인수는 정수의 목록을 제공하며, 각 정수는 히든 레이어에 해당하고 포함된 노드의 수를 나타냅니다. 예를 들어 아래 대입식을 살펴보세요.\n\n`hidden_units=[3,10]`\n\n위 대입식은 히든 레이어 2개를 갖는 신경망을 지정합니다.\n\n* 1번 히든 레이어는 노드 3개를 포함합니다.\n* 2번 히든 레이어는 노드 10개를 포함합니다.\n\n레이어를 늘리려면 목록에 정수를 더 추가하면 됩니다. 예를 들어 `hidden_units=[10,20,30,40]`은 각각 10개, 20개, 30개, 40개의 유닛을 갖는 4개의 레이어를 만듭니다.\n\n기본적으로 모든 히든 레이어는 ReLu 활성화를 사용하며 완전 연결성을 갖습니다.",
"_____no_output_____"
]
],
[
[
"def construct_feature_columns(input_features):\n \"\"\"Construct the TensorFlow Feature Columns.\n\n Args:\n input_features: The names of the numerical input features to use.\n Returns:\n A set of feature columns\n \"\"\" \n return set([tf.feature_column.numeric_column(my_feature)\n for my_feature in input_features])",
"_____no_output_____"
],
[
"def my_input_fn(features, targets, batch_size=1, shuffle=True, num_epochs=None):\n \"\"\"Trains a neural net regression model.\n \n Args:\n features: pandas DataFrame of features\n targets: pandas DataFrame of targets\n batch_size: Size of batches to be passed to the model\n shuffle: True or False. Whether to shuffle the data.\n num_epochs: Number of epochs for which data should be repeated. None = repeat indefinitely\n Returns:\n Tuple of (features, labels) for next data batch\n \"\"\"\n \n # Convert pandas data into a dict of np arrays.\n features = {key:np.array(value) for key,value in dict(features).items()} \n \n # Construct a dataset, and configure batching/repeating.\n ds = Dataset.from_tensor_slices((features,targets)) # warning: 2GB limit\n ds = ds.batch(batch_size).repeat(num_epochs)\n \n # Shuffle the data, if specified.\n if shuffle:\n ds = ds.shuffle(10000)\n \n # Return the next batch of data.\n features, labels = ds.make_one_shot_iterator().get_next()\n return features, labels",
"_____no_output_____"
],
[
"def train_nn_regression_model(\n learning_rate,\n steps,\n batch_size,\n hidden_units,\n training_examples,\n training_targets,\n validation_examples,\n validation_targets):\n \"\"\"Trains a neural network regression model.\n \n In addition to training, this function also prints training progress information,\n as well as a plot of the training and validation loss over time.\n \n Args:\n learning_rate: A `float`, the learning rate.\n steps: A non-zero `int`, the total number of training steps. A training step\n consists of a forward and backward pass using a single batch.\n batch_size: A non-zero `int`, the batch size.\n hidden_units: A `list` of int values, specifying the number of neurons in each layer.\n training_examples: A `DataFrame` containing one or more columns from\n `california_housing_dataframe` to use as input features for training.\n training_targets: A `DataFrame` containing exactly one column from\n `california_housing_dataframe` to use as target for training.\n validation_examples: A `DataFrame` containing one or more columns from\n `california_housing_dataframe` to use as input features for validation.\n validation_targets: A `DataFrame` containing exactly one column from\n `california_housing_dataframe` to use as target for validation.\n \n Returns:\n A `DNNRegressor` object trained on the training data.\n \"\"\"\n\n periods = 10\n steps_per_period = steps / periods\n \n # Create a DNNRegressor object.\n my_optimizer = tf.train.GradientDescentOptimizer(learning_rate=learning_rate)\n my_optimizer = tf.contrib.estimator.clip_gradients_by_norm(my_optimizer, 5.0)\n dnn_regressor = tf.estimator.DNNRegressor(\n feature_columns=construct_feature_columns(training_examples),\n hidden_units=hidden_units,\n optimizer=my_optimizer\n )\n \n # Create input functions.\n training_input_fn = lambda: my_input_fn(training_examples, \n training_targets[\"median_house_value\"], \n batch_size=batch_size)\n predict_training_input_fn = lambda: my_input_fn(training_examples, \n training_targets[\"median_house_value\"], \n num_epochs=1, \n shuffle=False)\n predict_validation_input_fn = lambda: my_input_fn(validation_examples, \n validation_targets[\"median_house_value\"], \n num_epochs=1, \n shuffle=False)\n\n # Train the model, but do so inside a loop so that we can periodically assess\n # loss metrics.\n print(\"Training model...\")\n print(\"RMSE (on training data):\")\n training_rmse = []\n validation_rmse = []\n for period in range (0, periods):\n # Train the model, starting from the prior state.\n dnn_regressor.train(\n input_fn=training_input_fn,\n steps=steps_per_period\n )\n # Take a break and compute predictions.\n training_predictions = dnn_regressor.predict(input_fn=predict_training_input_fn)\n training_predictions = np.array([item['predictions'][0] for item in training_predictions])\n \n validation_predictions = dnn_regressor.predict(input_fn=predict_validation_input_fn)\n validation_predictions = np.array([item['predictions'][0] for item in validation_predictions])\n \n # Compute training and validation loss.\n training_root_mean_squared_error = math.sqrt(\n metrics.mean_squared_error(training_predictions, training_targets))\n validation_root_mean_squared_error = math.sqrt(\n metrics.mean_squared_error(validation_predictions, validation_targets))\n # Occasionally print the current loss.\n print(\" period %02d : %0.2f\" % (period, training_root_mean_squared_error))\n # Add the loss metrics from this period to our list.\n training_rmse.append(training_root_mean_squared_error)\n validation_rmse.append(validation_root_mean_squared_error)\n print(\"Model training finished.\")\n\n # Output a graph of loss metrics over periods.\n plt.ylabel(\"RMSE\")\n plt.xlabel(\"Periods\")\n plt.title(\"Root Mean Squared Error vs. Periods\")\n plt.tight_layout()\n plt.plot(training_rmse, label=\"training\")\n plt.plot(validation_rmse, label=\"validation\")\n plt.legend()\n\n print(\"Final RMSE (on training data): %0.2f\" % training_root_mean_squared_error)\n print(\"Final RMSE (on validation data): %0.2f\" % validation_root_mean_squared_error)\n\n return dnn_regressor",
"_____no_output_____"
]
],
[
[
" ## 작업 1: NN 모델 학습\n\n**RMSE를 110 미만으로 낮추는 것을 목표로 초매개변수를 조정합니다.**\n\n다음 블록을 실행하여 NN 모델을 학습시킵니다. \n\n많은 특성을 사용한 선형 회귀 실습에서 RMSE이 110 정도면 상당히 양호하다고 설명한 바 있습니다. 더 우수한 모델을 목표로 해 보겠습니다.\n\n이번에 수행할 작업은 다양한 학습 설정을 수정하여 검증 데이터에 대한 정확성을 높이는 것입니다.\n\nNN에는 과적합이라는 위험이 도사리고 있습니다. 학습 데이터에 대한 손실과 검증 데이터에 대한 손실의 격차를 조사하면 모델에서 과적합이 시작되고 있는지를 판단하는 데 도움이 됩니다. 일반적으로 격차가 증가하기 시작하면 과적합의 확실한 증거가 됩니다.\n\n매우 다양한 설정이 가능하므로, 각 시도에서 설정을 잘 기록하여 개발 방향을 잡는 데 참고하는 것이 좋습니다.\n\n또한 괜찮은 설정을 발견했다면 여러 번 실행하여 결과의 재현성을 확인하시기 바랍니다. NN 가중치는 일반적으로 작은 무작위 값으로 초기화되므로 실행 시마다 약간의 차이를 보입니다.\n",
"_____no_output_____"
]
],
[
[
"dnn_regressor = train_nn_regression_model(\n learning_rate=0.01,\n steps=500,\n batch_size=10,\n hidden_units=[10, 2],\n training_examples=training_examples,\n training_targets=training_targets,\n validation_examples=validation_examples,\n validation_targets=validation_targets)",
"_____no_output_____"
]
],
[
[
" ### 해결 방법\n\n가능한 해결 방법을 보려면 아래를 클릭하세요.",
"_____no_output_____"
],
[
" **참고:** 이 매개변수 선택은 어느 정도 임의적인 것입니다. 여기에서는 오차가 목표치 아래로 떨어질 때까지 점점 복잡한 조합을 시도하면서 학습 시간을 늘렸습니다. 이 조합은 결코 최선의 조합이 아니며, 다른 조합이 더 낮은 RMSE를 달성할 수도 있습니다. 오차를 최소화하는 모델을 찾는 것이 목표라면 매개변수 검색과 같은 보다 엄밀한 절차를 사용해야 합니다.",
"_____no_output_____"
]
],
[
[
"dnn_regressor = train_nn_regression_model(\n learning_rate=0.001,\n steps=2000,\n batch_size=100,\n hidden_units=[10, 10],\n training_examples=training_examples,\n training_targets=training_targets,\n validation_examples=validation_examples,\n validation_targets=validation_targets)",
"_____no_output_____"
]
],
[
[
" ## 작업 2: 테스트 데이터로 평가\n\n**검증 성능 결과가 테스트 데이터에 대해서도 유지되는지 확인합니다.**\n\n만족할 만한 모델이 만들어졌으면 테스트 데이터로 평가하고 검증 성능과 비교해 봅니다.\n\n테스트 데이터 세트는 [여기](https://download.mlcc.google.com/mledu-datasets/california_housing_test.csv)에 있습니다.",
"_____no_output_____"
]
],
[
[
"california_housing_test_data = pd.read_csv(\"https://download.mlcc.google.com/mledu-datasets/california_housing_test.csv\", sep=\",\")\n\n# YOUR CODE HERE",
"_____no_output_____"
]
],
[
[
" ### 해결 방법\n\n가능한 해결 방법을 보려면 아래를 클릭하세요.",
"_____no_output_____"
],
[
" 위 코드에서 수행하는 작업과 마찬가지로 적절한 데이터 파일을 로드하고 전처리한 후 predict 및 mean_squared_error를 호출해야 합니다.\n\n모든 레코드를 사용할 것이므로 테스트 데이터를 무작위로 추출할 필요는 없습니다.",
"_____no_output_____"
]
],
[
[
"california_housing_test_data = pd.read_csv(\"https://download.mlcc.google.com/mledu-datasets/california_housing_test.csv\", sep=\",\")\n\ntest_examples = preprocess_features(california_housing_test_data)\ntest_targets = preprocess_targets(california_housing_test_data)\n\npredict_testing_input_fn = lambda: my_input_fn(test_examples, \n test_targets[\"median_house_value\"], \n num_epochs=1, \n shuffle=False)\n\ntest_predictions = dnn_regressor.predict(input_fn=predict_testing_input_fn)\ntest_predictions = np.array([item['predictions'][0] for item in test_predictions])\n\nroot_mean_squared_error = math.sqrt(\n metrics.mean_squared_error(test_predictions, test_targets))\n\nprint(\"Final RMSE (on test data): %0.2f\" % root_mean_squared_error)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
]
] |
c5009c193255e45676d836ab83d1a9197341d782
| 110,171 |
ipynb
|
Jupyter Notebook
|
Job Search - On-the-Job Search.ipynb
|
DiogoRibeiro7/Finance
|
6babc706bd523fc83e1dd1fda7f57aef969c5347
|
[
"Apache-2.0"
] | null | null | null |
Job Search - On-the-Job Search.ipynb
|
DiogoRibeiro7/Finance
|
6babc706bd523fc83e1dd1fda7f57aef969c5347
|
[
"Apache-2.0"
] | null | null | null |
Job Search - On-the-Job Search.ipynb
|
DiogoRibeiro7/Finance
|
6babc706bd523fc83e1dd1fda7f57aef969c5347
|
[
"Apache-2.0"
] | null | null | null | 278.209596 | 39,060 | 0.915686 |
[
[
[
"# Job Search - On-the-Job Search\n\n\n<a id='index-1'></a>",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport scipy.stats as stats\nfrom interpolation import interp\nfrom numba import njit, prange\nimport matplotlib.pyplot as plt\n%matplotlib inline\nfrom math import gamma",
"_____no_output_____"
],
[
"class JVWorker:\n r\"\"\"\n A Jovanovic-type model of employment with on-the-job search.\n\n \"\"\"\n\n def __init__(self,\n A=1.4,\n α=0.6,\n β=0.96, # Discount factor\n π=np.sqrt, # Search effort function\n a=2, # Parameter of f\n b=2, # Parameter of f\n grid_size=50,\n mc_size=100,\n ɛ=1e-4):\n\n self.A, self.α, self.β, self.π = A, α, β, π\n self.mc_size, self.ɛ = mc_size, ɛ\n\n self.g = njit(lambda x, ϕ: A * (x * ϕ)**α) # Transition function\n self.f_rvs = np.random.beta(a, b, mc_size)\n\n # Max of grid is the max of a large quantile value for f and the\n # fixed point y = g(y, 1)\n ɛ = 1e-4\n grid_max = max(A**(1 / (1 - α)), stats.beta(a, b).ppf(1 - ɛ))\n\n # Human capital\n self.x_grid = np.linspace(ɛ, grid_max, grid_size)",
"_____no_output_____"
],
[
"def operator_factory(jv, parallel_flag=True):\n\n \"\"\"\n Returns a jitted version of the Bellman operator T\n\n jv is an instance of JVWorker\n\n \"\"\"\n\n π, β = jv.π, jv.β\n x_grid, ɛ, mc_size = jv.x_grid, jv.ɛ, jv.mc_size\n f_rvs, g = jv.f_rvs, jv.g\n\n @njit\n def state_action_values(z, x, v):\n s, ϕ = z\n v_func = lambda x: interp(x_grid, v, x)\n\n integral = 0\n for m in range(mc_size):\n u = f_rvs[m]\n integral += v_func(max(g(x, ϕ), u))\n integral = integral / mc_size\n\n q = π(s) * integral + (1 - π(s)) * v_func(g(x, ϕ))\n return x * (1 - ϕ - s) + β * q\n\n @njit(parallel=parallel_flag)\n def T(v):\n \"\"\"\n The Bellman operator\n \"\"\"\n\n v_new = np.empty_like(v)\n for i in prange(len(x_grid)):\n x = x_grid[i]\n\n # Search on a grid\n search_grid = np.linspace(ɛ, 1, 15)\n max_val = -1\n for s in search_grid:\n for ϕ in search_grid:\n current_val = state_action_values((s, ϕ), x, v) if s + ϕ <= 1 else -1\n if current_val > max_val:\n max_val = current_val\n v_new[i] = max_val\n\n return v_new\n\n @njit\n def get_greedy(v):\n \"\"\"\n Computes the v-greedy policy of a given function v\n \"\"\"\n s_policy, ϕ_policy = np.empty_like(v), np.empty_like(v)\n\n for i in range(len(x_grid)):\n x = x_grid[i]\n # Search on a grid\n search_grid = np.linspace(ɛ, 1, 15)\n max_val = -1\n for s in search_grid:\n for ϕ in search_grid:\n current_val = state_action_values((s, ϕ), x, v) if s + ϕ <= 1 else -1\n if current_val > max_val:\n max_val = current_val\n max_s, max_ϕ = s, ϕ\n s_policy[i], ϕ_policy[i] = max_s, max_ϕ\n return s_policy, ϕ_policy\n\n return T, get_greedy",
"_____no_output_____"
],
[
"def solve_model(jv,\n use_parallel=True,\n tol=1e-4,\n max_iter=1000,\n verbose=True,\n print_skip=25):\n\n \"\"\"\n Solves the model by value function iteration\n\n * jv is an instance of JVWorker\n\n \"\"\"\n\n T, _ = operator_factory(jv, parallel_flag=use_parallel)\n\n # Set up loop\n v = jv.x_grid * 0.5 # Initial condition\n i = 0\n error = tol + 1\n\n while i < max_iter and error > tol:\n v_new = T(v)\n error = np.max(np.abs(v - v_new))\n i += 1\n if verbose and i % print_skip == 0:\n print(f\"Error at iteration {i} is {error}.\")\n v = v_new\n\n if i == max_iter:\n print(\"Failed to converge!\")\n\n if verbose and i < max_iter:\n print(f\"\\nConverged in {i} iterations.\")\n\n return v_new",
"_____no_output_____"
],
[
"jv = JVWorker()\nT, get_greedy = operator_factory(jv)\nv_star = solve_model(jv)\ns_star, ϕ_star = get_greedy(v_star)",
"C:\\Users\\Diogo\\Anaconda3\\lib\\site-packages\\numba\\np\\ufunc\\parallel.py:355: NumbaWarning: \u001b[1mThe TBB threading layer requires TBB version 2019.5 or later i.e., TBB_INTERFACE_VERSION >= 11005. Found TBB_INTERFACE_VERSION = 10005. The TBB threading layer is disabled.\u001b[0m\n warnings.warn(problem)\n"
],
[
"plots = [s_star, ϕ_star, v_star]\ntitles = [\"s policy\", \"ϕ policy\", \"value function\"]\n\nfig, axes = plt.subplots(3, 1, figsize=(12, 12))\n\nfor ax, plot, title in zip(axes, plots, titles):\n ax.plot(jv.x_grid, plot)\n ax.set(title=title)\n ax.grid()\n\naxes[-1].set_xlabel(\"x\")\nplt.show()",
"_____no_output_____"
],
[
"jv = JVWorker(grid_size=25, mc_size=50)\nπ, g, f_rvs, x_grid = jv.π, jv.g, jv.f_rvs, jv.x_grid\nT, get_greedy = operator_factory(jv)\nv_star = solve_model(jv, verbose=False)\ns_policy, ϕ_policy = get_greedy(v_star)\n\n# Turn the policy function arrays into actual functions\ns = lambda y: interp(x_grid, s_policy, y)\nϕ = lambda y: interp(x_grid, ϕ_policy, y)\n\ndef h(x, b, u):\n return (1 - b) * g(x, ϕ(x)) + b * max(g(x, ϕ(x)), u)\n\n\nplot_grid_max, plot_grid_size = 1.2, 100\nplot_grid = np.linspace(0, plot_grid_max, plot_grid_size)\nfig, ax = plt.subplots(figsize=(8, 8))\nticks = (0.25, 0.5, 0.75, 1.0)\nax.set(xticks=ticks, yticks=ticks,\n xlim=(0, plot_grid_max),\n ylim=(0, plot_grid_max),\n xlabel='$x_t$', ylabel='$x_{t+1}$')\n\nax.plot(plot_grid, plot_grid, 'k--', alpha=0.6) # 45 degree line\nfor x in plot_grid:\n for i in range(jv.mc_size):\n b = 1 if np.random.uniform(0, 1) < π(s(x)) else 0\n u = f_rvs[i]\n y = h(x, b, u)\n ax.plot(x, y, 'go', alpha=0.25)\n\nplt.show()",
"_____no_output_____"
],
[
"jv = JVWorker()\n\ndef xbar(ϕ):\n A, α = jv.A, jv.α\n return (A * ϕ**α)**(1 / (1 - α))\n\nϕ_grid = np.linspace(0, 1, 100)\nfig, ax = plt.subplots(figsize=(9, 7))\nax.set(xlabel='$\\phi$')\nax.plot(ϕ_grid, [xbar(ϕ) * (1 - ϕ) for ϕ in ϕ_grid], label='$w^*(\\phi)$')\nax.legend()\n\nplt.show()",
"_____no_output_____"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
c500a9782a4a7cc2da96fb22e98079ae3947b6c7
| 24,019 |
ipynb
|
Jupyter Notebook
|
site/en-snapshot/agents/tutorials/6_reinforce_tutorial.ipynb
|
leeyspaul/docs-l10n
|
0e2f1a4d1c507b8a71be6b506b275ab2c83ca359
|
[
"Apache-2.0"
] | null | null | null |
site/en-snapshot/agents/tutorials/6_reinforce_tutorial.ipynb
|
leeyspaul/docs-l10n
|
0e2f1a4d1c507b8a71be6b506b275ab2c83ca359
|
[
"Apache-2.0"
] | null | null | null |
site/en-snapshot/agents/tutorials/6_reinforce_tutorial.ipynb
|
leeyspaul/docs-l10n
|
0e2f1a4d1c507b8a71be6b506b275ab2c83ca359
|
[
"Apache-2.0"
] | null | null | null | 34.411175 | 504 | 0.548524 |
[
[
[
"##### Copyright 2018 The TF-Agents Authors.",
"_____no_output_____"
]
],
[
[
"#@title Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# https://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.",
"_____no_output_____"
]
],
[
[
"# REINFORCE agent\n\n<table class=\"tfo-notebook-buttons\" align=\"left\">\n <td>\n <a target=\"_blank\" href=\"https://www.tensorflow.org/agents/tutorials/6_reinforce_tutorial\">\n <img src=\"https://www.tensorflow.org/images/tf_logo_32px.png\" />\n View on TensorFlow.org</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://colab.research.google.com/github/tensorflow/agents/blob/master/docs/tutorials/6_reinforce_tutorial.ipynb\">\n <img src=\"https://www.tensorflow.org/images/colab_logo_32px.png\" />\n Run in Google Colab</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://github.com/tensorflow/agents/blob/master/docs/tutorials/6_reinforce_tutorial.ipynb\">\n <img src=\"https://www.tensorflow.org/images/GitHub-Mark-32px.png\" />\n View source on GitHub</a>\n </td>\n <td>\n <a href=\"https://storage.googleapis.com/tensorflow_docs/agents/docs/tutorials/6_reinforce_tutorial.ipynb\"><img src=\"https://www.tensorflow.org/images/download_logo_32px.png\" />Download notebook</a>\n </td>\n</table>",
"_____no_output_____"
],
[
"## Introduction",
"_____no_output_____"
],
[
"This example shows how to train a [REINFORCE](http://www-anw.cs.umass.edu/~barto/courses/cs687/williams92simple.pdf) agent on the Cartpole environment using the TF-Agents library, similar to the [DQN tutorial](1_dqn_tutorial.ipynb).\n\n\n\nWe will walk you through all the components in a Reinforcement Learning (RL) pipeline for training, evaluation and data collection.\n",
"_____no_output_____"
],
[
"## Setup",
"_____no_output_____"
],
[
"If you haven't installed the following dependencies, run:",
"_____no_output_____"
]
],
[
[
"!sudo apt-get install -y xvfb ffmpeg\n!pip install gym\n!pip install 'imageio==2.4.0'\n!pip install PILLOW\n!pip install pyvirtualdisplay\n!pip install tf-agents",
"_____no_output_____"
],
[
"from __future__ import absolute_import\nfrom __future__ import division\nfrom __future__ import print_function\n\nimport base64\nimport imageio\nimport IPython\nimport matplotlib\nimport matplotlib.pyplot as plt\nimport numpy as np\nimport PIL.Image\nimport pyvirtualdisplay\n\nimport tensorflow as tf\n\nfrom tf_agents.agents.reinforce import reinforce_agent\nfrom tf_agents.drivers import dynamic_step_driver\nfrom tf_agents.environments import suite_gym\nfrom tf_agents.environments import tf_py_environment\nfrom tf_agents.eval import metric_utils\nfrom tf_agents.metrics import tf_metrics\nfrom tf_agents.networks import actor_distribution_network\nfrom tf_agents.replay_buffers import tf_uniform_replay_buffer\nfrom tf_agents.trajectories import trajectory\nfrom tf_agents.utils import common\n\ntf.compat.v1.enable_v2_behavior()\n\n\n# Set up a virtual display for rendering OpenAI gym environments.\ndisplay = pyvirtualdisplay.Display(visible=0, size=(1400, 900)).start()",
"_____no_output_____"
]
],
[
[
"## Hyperparameters",
"_____no_output_____"
]
],
[
[
"env_name = \"CartPole-v0\" # @param {type:\"string\"}\nnum_iterations = 250 # @param {type:\"integer\"}\ncollect_episodes_per_iteration = 2 # @param {type:\"integer\"}\nreplay_buffer_capacity = 2000 # @param {type:\"integer\"}\n\nfc_layer_params = (100,)\n\nlearning_rate = 1e-3 # @param {type:\"number\"}\nlog_interval = 25 # @param {type:\"integer\"}\nnum_eval_episodes = 10 # @param {type:\"integer\"}\neval_interval = 50 # @param {type:\"integer\"}",
"_____no_output_____"
]
],
[
[
"## Environment\n\nEnvironments in RL represent the task or problem that we are trying to solve. Standard environments can be easily created in TF-Agents using `suites`. We have different `suites` for loading environments from sources such as the OpenAI Gym, Atari, DM Control, etc., given a string environment name.\n\nNow let us load the CartPole environment from the OpenAI Gym suite.",
"_____no_output_____"
]
],
[
[
"env = suite_gym.load(env_name)",
"_____no_output_____"
]
],
[
[
"We can render this environment to see how it looks. A free-swinging pole is attached to a cart. The goal is to move the cart right or left in order to keep the pole pointing up.",
"_____no_output_____"
]
],
[
[
"#@test {\"skip\": true}\nenv.reset()\nPIL.Image.fromarray(env.render())",
"_____no_output_____"
]
],
[
[
"The `time_step = environment.step(action)` statement takes `action` in the environment. The `TimeStep` tuple returned contains the environment's next observation and reward for that action. The `time_step_spec()` and `action_spec()` methods in the environment return the specifications (types, shapes, bounds) of the `time_step` and `action` respectively.",
"_____no_output_____"
]
],
[
[
"print('Observation Spec:')\nprint(env.time_step_spec().observation)\nprint('Action Spec:')\nprint(env.action_spec())",
"_____no_output_____"
]
],
[
[
"So, we see that observation is an array of 4 floats: the position and velocity of the cart, and the angular position and velocity of the pole. Since only two actions are possible (move left or move right), the `action_spec` is a scalar where 0 means \"move left\" and 1 means \"move right.\"",
"_____no_output_____"
]
],
[
[
"time_step = env.reset()\nprint('Time step:')\nprint(time_step)\n\naction = np.array(1, dtype=np.int32)\n\nnext_time_step = env.step(action)\nprint('Next time step:')\nprint(next_time_step)",
"_____no_output_____"
]
],
[
[
"Usually we create two environments: one for training and one for evaluation. Most environments are written in pure python, but they can be easily converted to TensorFlow using the `TFPyEnvironment` wrapper. The original environment's API uses numpy arrays, the `TFPyEnvironment` converts these to/from `Tensors` for you to more easily interact with TensorFlow policies and agents.\n",
"_____no_output_____"
]
],
[
[
"train_py_env = suite_gym.load(env_name)\neval_py_env = suite_gym.load(env_name)\n\ntrain_env = tf_py_environment.TFPyEnvironment(train_py_env)\neval_env = tf_py_environment.TFPyEnvironment(eval_py_env)",
"_____no_output_____"
]
],
[
[
"## Agent\n\nThe algorithm that we use to solve an RL problem is represented as an `Agent`. In addition to the REINFORCE agent, TF-Agents provides standard implementations of a variety of `Agents` such as [DQN](https://storage.googleapis.com/deepmind-media/dqn/DQNNaturePaper.pdf), [DDPG](https://arxiv.org/pdf/1509.02971.pdf), [TD3](https://arxiv.org/pdf/1802.09477.pdf), [PPO](https://arxiv.org/abs/1707.06347) and [SAC](https://arxiv.org/abs/1801.01290).\n\nTo create a REINFORCE Agent, we first need an `Actor Network` that can learn to predict the action given an observation from the environment.\n\nWe can easily create an `Actor Network` using the specs of the observations and actions. We can specify the layers in the network which, in this example, is the `fc_layer_params` argument set to a tuple of `ints` representing the sizes of each hidden layer (see the Hyperparameters section above).\n",
"_____no_output_____"
]
],
[
[
"actor_net = actor_distribution_network.ActorDistributionNetwork(\n train_env.observation_spec(),\n train_env.action_spec(),\n fc_layer_params=fc_layer_params)",
"_____no_output_____"
]
],
[
[
"We also need an `optimizer` to train the network we just created, and a `train_step_counter` variable to keep track of how many times the network was updated.\n",
"_____no_output_____"
]
],
[
[
"optimizer = tf.compat.v1.train.AdamOptimizer(learning_rate=learning_rate)\n\ntrain_step_counter = tf.compat.v2.Variable(0)\n\ntf_agent = reinforce_agent.ReinforceAgent(\n train_env.time_step_spec(),\n train_env.action_spec(),\n actor_network=actor_net,\n optimizer=optimizer,\n normalize_returns=True,\n train_step_counter=train_step_counter)\ntf_agent.initialize()",
"_____no_output_____"
]
],
[
[
"## Policies\n\nIn TF-Agents, policies represent the standard notion of policies in RL: given a `time_step` produce an action or a distribution over actions. The main method is `policy_step = policy.step(time_step)` where `policy_step` is a named tuple `PolicyStep(action, state, info)`. The `policy_step.action` is the `action` to be applied to the environment, `state` represents the state for stateful (RNN) policies and `info` may contain auxiliary information such as log probabilities of the actions.\n\nAgents contain two policies: the main policy that is used for evaluation/deployment (agent.policy) and another policy that is used for data collection (agent.collect_policy).",
"_____no_output_____"
]
],
[
[
"eval_policy = tf_agent.policy\ncollect_policy = tf_agent.collect_policy",
"_____no_output_____"
]
],
[
[
"## Metrics and Evaluation\n\nThe most common metric used to evaluate a policy is the average return. The return is the sum of rewards obtained while running a policy in an environment for an episode, and we usually average this over a few episodes. We can compute the average return metric as follows.\n",
"_____no_output_____"
]
],
[
[
"#@test {\"skip\": true}\ndef compute_avg_return(environment, policy, num_episodes=10):\n\n total_return = 0.0\n for _ in range(num_episodes):\n\n time_step = environment.reset()\n episode_return = 0.0\n\n while not time_step.is_last():\n action_step = policy.action(time_step)\n time_step = environment.step(action_step.action)\n episode_return += time_step.reward\n total_return += episode_return\n\n avg_return = total_return / num_episodes\n return avg_return.numpy()[0]\n\n\n# Please also see the metrics module for standard implementations of different\n# metrics.",
"_____no_output_____"
]
],
[
[
"## Replay Buffer\n\nIn order to keep track of the data collected from the environment, we will use the TFUniformReplayBuffer. This replay buffer is constructed using specs describing the tensors that are to be stored, which can be obtained from the agent using `tf_agent.collect_data_spec`.",
"_____no_output_____"
]
],
[
[
"replay_buffer = tf_uniform_replay_buffer.TFUniformReplayBuffer(\n data_spec=tf_agent.collect_data_spec,\n batch_size=train_env.batch_size,\n max_length=replay_buffer_capacity)",
"_____no_output_____"
]
],
[
[
"For most agents, the `collect_data_spec` is a `Trajectory` named tuple containing the observation, action, reward etc.",
"_____no_output_____"
],
[
"## Data Collection\n\nAs REINFORCE learns from whole episodes, we define a function to collect an episode using the given data collection policy and save the data (observations, actions, rewards etc.) as trajectories in the replay buffer.",
"_____no_output_____"
]
],
[
[
"#@test {\"skip\": true}\n\ndef collect_episode(environment, policy, num_episodes):\n\n episode_counter = 0\n environment.reset()\n\n while episode_counter < num_episodes:\n time_step = environment.current_time_step()\n action_step = policy.action(time_step)\n next_time_step = environment.step(action_step.action)\n traj = trajectory.from_transition(time_step, action_step, next_time_step)\n\n # Add trajectory to the replay buffer\n replay_buffer.add_batch(traj)\n\n if traj.is_boundary():\n episode_counter += 1\n\n\n# This loop is so common in RL, that we provide standard implementations of\n# these. For more details see the drivers module.",
"_____no_output_____"
]
],
[
[
"## Training the agent\n\nThe training loop involves both collecting data from the environment and optimizing the agent's networks. Along the way, we will occasionally evaluate the agent's policy to see how we are doing.\n\nThe following will take ~3 minutes to run.",
"_____no_output_____"
]
],
[
[
"#@test {\"skip\": true}\ntry:\n %%time\nexcept:\n pass\n\n# (Optional) Optimize by wrapping some of the code in a graph using TF function.\ntf_agent.train = common.function(tf_agent.train)\n\n# Reset the train step\ntf_agent.train_step_counter.assign(0)\n\n# Evaluate the agent's policy once before training.\navg_return = compute_avg_return(eval_env, tf_agent.policy, num_eval_episodes)\nreturns = [avg_return]\n\nfor _ in range(num_iterations):\n\n # Collect a few episodes using collect_policy and save to the replay buffer.\n collect_episode(\n train_env, tf_agent.collect_policy, collect_episodes_per_iteration)\n\n # Use data from the buffer and update the agent's network.\n experience = replay_buffer.gather_all()\n train_loss = tf_agent.train(experience)\n replay_buffer.clear()\n\n step = tf_agent.train_step_counter.numpy()\n\n if step % log_interval == 0:\n print('step = {0}: loss = {1}'.format(step, train_loss.loss))\n\n if step % eval_interval == 0:\n avg_return = compute_avg_return(eval_env, tf_agent.policy, num_eval_episodes)\n print('step = {0}: Average Return = {1}'.format(step, avg_return))\n returns.append(avg_return)",
"_____no_output_____"
]
],
[
[
"## Visualization\n",
"_____no_output_____"
],
[
"### Plots\n\nWe can plot return vs global steps to see the performance of our agent. In `Cartpole-v0`, the environment gives a reward of +1 for every time step the pole stays up, and since the maximum number of steps is 200, the maximum possible return is also 200.",
"_____no_output_____"
]
],
[
[
"#@test {\"skip\": true}\n\nsteps = range(0, num_iterations + 1, eval_interval)\nplt.plot(steps, returns)\nplt.ylabel('Average Return')\nplt.xlabel('Step')\nplt.ylim(top=250)",
"_____no_output_____"
]
],
[
[
"### Videos",
"_____no_output_____"
],
[
"It is helpful to visualize the performance of an agent by rendering the environment at each step. Before we do that, let us first create a function to embed videos in this colab.",
"_____no_output_____"
]
],
[
[
"def embed_mp4(filename):\n \"\"\"Embeds an mp4 file in the notebook.\"\"\"\n video = open(filename,'rb').read()\n b64 = base64.b64encode(video)\n tag = '''\n <video width=\"640\" height=\"480\" controls>\n <source src=\"data:video/mp4;base64,{0}\" type=\"video/mp4\">\n Your browser does not support the video tag.\n </video>'''.format(b64.decode())\n\n return IPython.display.HTML(tag)",
"_____no_output_____"
]
],
[
[
"The following code visualizes the agent's policy for a few episodes:",
"_____no_output_____"
]
],
[
[
"num_episodes = 3\nvideo_filename = 'imageio.mp4'\nwith imageio.get_writer(video_filename, fps=60) as video:\n for _ in range(num_episodes):\n time_step = eval_env.reset()\n video.append_data(eval_py_env.render())\n while not time_step.is_last():\n action_step = tf_agent.policy.action(time_step)\n time_step = eval_env.step(action_step.action)\n video.append_data(eval_py_env.render())\n\nembed_mp4(video_filename)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
c500b55aba0eb9bbc97a55cefd248c54829759ea
| 741,492 |
ipynb
|
Jupyter Notebook
|
notebooks/3-4.0.2-intel-ordering_and_retraining-configuration_2.ipynb
|
fjdurlop/guided-retraining
|
1eba5175019ecac9fd02b7529f913c4750bf5a28
|
[
"MIT"
] | null | null | null |
notebooks/3-4.0.2-intel-ordering_and_retraining-configuration_2.ipynb
|
fjdurlop/guided-retraining
|
1eba5175019ecac9fd02b7529f913c4750bf5a28
|
[
"MIT"
] | null | null | null |
notebooks/3-4.0.2-intel-ordering_and_retraining-configuration_2.ipynb
|
fjdurlop/guided-retraining
|
1eba5175019ecac9fd02b7529f913c4750bf5a28
|
[
"MIT"
] | null | null | null | 91.002946 | 41,100 | 0.70279 |
[
[
[
"# Ordering by metrics and retraining phase\n\n## Dataset: Intel",
"_____no_output_____"
],
[
"## Configuration 2\n\t2. Incremental guided retraining starting from the original model using the new adversarial inputs and original training set.\n ",
"_____no_output_____"
]
],
[
[
"pip install --user tensorflow==2.5",
"_____no_output_____"
],
[
"import argparse\n\nimport numpy as np\nimport tensorflow as tf\nimport keras.backend as K\n\nimport matplotlib.pyplot as plt\n\n\nfrom keras.utils import np_utils\nfrom keras.models import Sequential\nfrom keras.layers import Dense, Dropout, Activation, Flatten, Conv2D, MaxPooling2D\nfrom keras.regularizers import l2\n\nimport glob\nimport os\nimport cv2\nimport keras\n\n\nimport time\nimport argparse\n\nfrom tqdm import tqdm\n\nfrom keras.models import load_model, Model\n",
"_____no_output_____"
],
[
"cd '../utils'",
"_____no_output_____"
],
[
"# utils for project\nimport utils_guided_retraining as utils",
"_____no_output_____"
],
[
"cd '../notebooks/'",
"_____no_output_____"
],
[
"x_train,y_train = utils.get_data(\"intel\",\"Train\")\nx_val,y_val = utils.get_data(\"intel\",\"Val\")\nx_test,y_test = utils.get_data(\"intel\",\"Test\")",
"_____no_output_____"
],
[
"x_train_and_adversary,y_train_and_adversary = utils.get_data(\"intel\",\"Train_and_adversary\")",
"_____no_output_____"
],
[
"x_adversary_training = x_train_and_adversary[len(x_train):]\nprint(len(x_adversary_training))\ny_adversary_training = y_train_and_adversary[len(y_train):]\n\nprint(len(y_adversary_training))",
"3000\n3000\n"
],
[
"# Obtaining adversarial examples for testing \nx_test_and_adversary,y_test_and_adversary = utils.get_adversarial_data(\"intel\",'Test_fgsm') ",
"_____no_output_____"
],
[
"x_adversary_test_fgsm = x_test_and_adversary[len(x_test):]\nprint(len(x_adversary_test_fgsm))\ny_adversary_test_fgsm = y_test_and_adversary[len(y_test):]\n\nprint(len(y_adversary_test_fgsm))",
"3000\n3000\n"
]
],
[
[
"## ----",
"_____no_output_____"
]
],
[
[
"# Original model \nmodel_dir = \"C:/Users/fjdur/Desktop/upc/project_notebooks/github_project/DL_notebooks/models/intel_model_21_10/\"\n",
"_____no_output_____"
],
[
"model_original = utils.My_model(\"intel\",True, model_dir)",
"Model loaded correctly\n"
],
[
"model_original.evaluate(x_test,y_test)",
"94/94 [==============================] - 12s 22ms/step - loss: 0.5392 - accuracy: 0.8037\n"
],
[
"model_original.evaluate(x_adversary_test_fgsm,y_adversary_test_fgsm)",
"94/94 [==============================] - 2s 23ms/step - loss: 3.1384 - accuracy: 0.1497\n"
],
[
"model_original.evaluate(x_adversary_test_fgsm,y_adversary_test_fgsm)",
"_____no_output_____"
]
],
[
[
"## Obtaining new LSA and DSA values ",
"_____no_output_____"
]
],
[
[
"save_dir_lsa = \"C:/Users/fjdur/Desktop/upc/project_notebooks/github_project/DL_notebooks/SA_values/intel_lsa_values_2.npy\"\n\n\nsave_dir_dsa = \"C:/Users/fjdur/Desktop/upc/project_notebooks/github_project/DL_notebooks/SA_values/intel_dsa_values_2.npy\"\n\ntarget_lsa = np.load(save_dir_lsa)\ntarget_dsa = np.load(save_dir_dsa)",
"_____no_output_____"
],
[
"lsa_values = target_lsa\ndsa_values = target_dsa",
"_____no_output_____"
],
[
"\n# Obtaining top n images by LSA values\ntop_images_by_lsa = utils.get_x_of_indexes(list(np.flip(np.argsort(lsa_values))),x_train_and_adversary)\ntop_labels_by_lsa = utils.get_x_of_indexes(list(np.flip(np.argsort(lsa_values))),y_train_and_adversary)\n",
"_____no_output_____"
],
[
"\ntop_images_by_dsa = utils.get_x_of_indexes(list(np.flip(np.argsort(dsa_values))),x_train_and_adversary)\ntop_labels_by_dsa = utils.get_x_of_indexes(list(np.flip(np.argsort(dsa_values))),y_train_and_adversary)\n",
"_____no_output_____"
],
[
"len(top_images_by_lsa)//20",
"_____no_output_____"
],
[
"top_images_by_lsa_5000 = np.array(top_images_by_lsa[:5000])\ntop_labels_by_lsa_5000 = np.array(top_labels_by_lsa[:5000])\n\n",
"_____no_output_____"
],
[
"m = 700\nn = 0\nimage_sets_lsa = []\nlabel_sets_lsa = []\n\n\nfor i in range(len(top_images_by_lsa)//m):\n print(i,\":\")\n if (i+1 >= len(top_images_by_lsa)//m):\n print(\"Last\")\n print(0,\" -> \",n+m+(len(top_images_by_lsa)%m))\n top_images_by_lsa_n = np.array(top_images_by_lsa[:n+m+(len(top_images_by_lsa)%m)])\n top_labels_by_lsa_n = np.array(top_labels_by_lsa[:n+m+(len(top_images_by_lsa)%m)])\n else:\n print(0,\" -> \",m+n)\n top_images_by_lsa_n = np.array(top_images_by_lsa[:n+m])\n top_labels_by_lsa_n = np.array(top_labels_by_lsa[:n+m])\n image_sets_lsa.append(top_images_by_lsa_n)\n label_sets_lsa.append(top_labels_by_lsa_n)\n print(len(top_images_by_lsa_n))\n n += m\n\n",
"0 :\n0 -> 700\n700\n1 :\n0 -> 1400\n1400\n2 :\n0 -> 2100\n2100\n3 :\n0 -> 2800\n2800\n4 :\n0 -> 3500\n3500\n5 :\n0 -> 4200\n4200\n6 :\n0 -> 4900\n4900\n7 :\n0 -> 5600\n5600\n8 :\n0 -> 6300\n6300\n9 :\n0 -> 7000\n7000\n10 :\n0 -> 7700\n7700\n11 :\n0 -> 8400\n8400\n12 :\n0 -> 9100\n9100\n13 :\n0 -> 9800\n9800\n14 :\n0 -> 10500\n10500\n15 :\n0 -> 11200\n11200\n16 :\n0 -> 11900\n11900\n17 :\n0 -> 12600\n12600\n18 :\n0 -> 13300\n13300\n19 :\nLast\n0 -> 14224\n14224\n"
]
],
[
[
"## Training guided by LSA",
"_____no_output_____"
]
],
[
[
"dataset = 'intel'",
"_____no_output_____"
],
[
"model_lsa_5000 = utils.My_model(dataset,True,model_dir)\nmodel_lsa_5000.compile_model()",
"Model loaded correctly\nModel compiled\n"
],
[
"model_lsa_5000.fit_model(top_images_by_lsa_5000,top_labels_by_lsa_5000,x_val,y_val)",
"Epoch 1/10\n79/79 [==============================] - 29s 176ms/step - loss: 1.1935 - accuracy: 0.5353 - val_loss: 0.8302 - val_accuracy: 0.6701\nEpoch 2/10\n79/79 [==============================] - 13s 169ms/step - loss: 0.9267 - accuracy: 0.6339 - val_loss: 1.0896 - val_accuracy: 0.5899\nEpoch 3/10\n79/79 [==============================] - 14s 172ms/step - loss: 0.8681 - accuracy: 0.6547 - val_loss: 1.0818 - val_accuracy: 0.5913\nEpoch 4/10\n79/79 [==============================] - 13s 171ms/step - loss: 0.8098 - accuracy: 0.6864 - val_loss: 0.9624 - val_accuracy: 0.6316\nEpoch 5/10\n79/79 [==============================] - 13s 170ms/step - loss: 0.8086 - accuracy: 0.6873 - val_loss: 1.4199 - val_accuracy: 0.4583\nEpoch 6/10\n79/79 [==============================] - 13s 169ms/step - loss: 0.7126 - accuracy: 0.7243 - val_loss: 1.0978 - val_accuracy: 0.6031\nEpoch 7/10\n79/79 [==============================] - 13s 167ms/step - loss: 0.6787 - accuracy: 0.7435 - val_loss: 0.9577 - val_accuracy: 0.6337\nEpoch 8/10\n79/79 [==============================] - 13s 167ms/step - loss: 0.6863 - accuracy: 0.7380 - val_loss: 1.0050 - val_accuracy: 0.6123\nEpoch 9/10\n79/79 [==============================] - 14s 173ms/step - loss: 0.6714 - accuracy: 0.7454 - val_loss: 0.9199 - val_accuracy: 0.6227\nEpoch 10/10\n79/79 [==============================] - 13s 169ms/step - loss: 0.5977 - accuracy: 0.7755 - val_loss: 1.0140 - val_accuracy: 0.5920\nDuration: 0:02:30.110022\n"
],
[
"for i in range(7):\n print(i)\n models_lsa[i] = utils.My_model('intel',True,model_dir)\n models_lsa[i].compile_model()",
"0\nModel loaded correctly\nModel compiled\n1\nModel loaded correctly\nModel compiled\n2\nModel loaded correctly\nModel compiled\n3\nModel loaded correctly\nModel compiled\n4\nModel loaded correctly\nModel compiled\n5\nModel loaded correctly\nModel compiled\n6\nModel loaded correctly\nModel compiled\n"
],
[
"models_lsa[1].evaluate(x_test,y_test)",
"94/94 [==============================] - 3s 24ms/step - loss: 0.4941 - accuracy: 0.8294\n"
],
[
"print(model_dir)\n\nmodels_lsa = []\nfor i in range(len(label_sets_lsa)):\n print(i,\":\")\n model = utils.My_model('intel',True,model_dir)\n model.compile_model()\n models_lsa.append(model)\n",
"C:/Users/fjdur/Desktop/upc/project_notebooks/github_project/DL_notebooks/models/intel_model_21_10/\n0 :\nModel loaded correctly\nModel compiled\n1 :\nModel loaded correctly\nModel compiled\n2 :\nModel loaded correctly\nModel compiled\n3 :\nModel loaded correctly\nModel compiled\n4 :\nModel loaded correctly\nModel compiled\n5 :\nModel loaded correctly\nModel compiled\n6 :\nModel loaded correctly\nModel compiled\n7 :\nModel loaded correctly\nModel compiled\n8 :\nModel loaded correctly\nModel compiled\n9 :\nModel loaded correctly\nModel compiled\n10 :\nModel loaded correctly\nModel compiled\n11 :\nModel loaded correctly\nModel compiled\n12 :\nModel loaded correctly\nModel compiled\n13 :\nModel loaded correctly\nModel compiled\n14 :\nModel loaded correctly\nModel compiled\n15 :\nModel loaded correctly\nModel compiled\n16 :\nModel loaded correctly\nModel compiled\n17 :\nModel loaded correctly\nModel compiled\n18 :\nModel loaded correctly\nModel compiled\n19 :\nModel loaded correctly\nModel compiled\n"
],
[
"print(len(models_lsa))\n",
"20\n"
],
[
"n = 0\nmodels_lsa[n].fit_model(image_sets_lsa[n],label_sets_lsa[n],x_val,y_val,epochs=20,batch_size = 128)",
"Epoch 1/20\n6/6 [==============================] - 5s 636ms/step - loss: 2.0641 - accuracy: 0.2457 - val_loss: 1.1151 - val_accuracy: 0.5603\nEpoch 2/20\n6/6 [==============================] - 4s 639ms/step - loss: 1.4756 - accuracy: 0.3757 - val_loss: 0.9574 - val_accuracy: 0.7036\nEpoch 3/20\n6/6 [==============================] - 4s 638ms/step - loss: 1.3338 - accuracy: 0.4529 - val_loss: 0.8454 - val_accuracy: 0.7304\nEpoch 4/20\n6/6 [==============================] - 4s 678ms/step - loss: 1.4258 - accuracy: 0.4214 - val_loss: 0.9247 - val_accuracy: 0.6740\nEpoch 5/20\n6/6 [==============================] - 4s 673ms/step - loss: 1.1768 - accuracy: 0.5457 - val_loss: 0.8539 - val_accuracy: 0.6994\nEpoch 6/20\n6/6 [==============================] - 4s 635ms/step - loss: 1.1639 - accuracy: 0.5343 - val_loss: 0.8525 - val_accuracy: 0.7072\nEpoch 7/20\n6/6 [==============================] - 3s 636ms/step - loss: 1.1793 - accuracy: 0.5486 - val_loss: 0.9379 - val_accuracy: 0.6883\nEpoch 8/20\n6/6 [==============================] - 4s 649ms/step - loss: 1.0407 - accuracy: 0.6157 - val_loss: 0.9068 - val_accuracy: 0.6501\nEpoch 9/20\n6/6 [==============================] - 4s 663ms/step - loss: 1.0513 - accuracy: 0.5957 - val_loss: 0.9039 - val_accuracy: 0.6558\nEpoch 10/20\n6/6 [==============================] - 3s 627ms/step - loss: 1.0279 - accuracy: 0.5943 - val_loss: 0.8752 - val_accuracy: 0.6534\nEpoch 11/20\n6/6 [==============================] - 3s 627ms/step - loss: 0.9793 - accuracy: 0.6286 - val_loss: 0.8517 - val_accuracy: 0.6794\nEpoch 12/20\n6/6 [==============================] - 3s 626ms/step - loss: 0.9016 - accuracy: 0.6486 - val_loss: 0.8642 - val_accuracy: 0.6944\nEpoch 13/20\n6/6 [==============================] - 3s 624ms/step - loss: 0.8665 - accuracy: 0.6500 - val_loss: 0.9294 - val_accuracy: 0.6009\nEpoch 14/20\n6/6 [==============================] - 3s 629ms/step - loss: 0.9954 - accuracy: 0.6200 - val_loss: 0.8286 - val_accuracy: 0.6887\nEpoch 15/20\n6/6 [==============================] - 3s 626ms/step - loss: 0.8422 - accuracy: 0.6714 - val_loss: 0.8491 - val_accuracy: 0.6819\nEpoch 16/20\n6/6 [==============================] - 3s 633ms/step - loss: 0.7811 - accuracy: 0.7043 - val_loss: 0.8510 - val_accuracy: 0.6961\nEpoch 17/20\n6/6 [==============================] - 3s 629ms/step - loss: 0.7561 - accuracy: 0.7214 - val_loss: 0.8482 - val_accuracy: 0.6986\nEpoch 18/20\n6/6 [==============================] - 3s 632ms/step - loss: 0.6925 - accuracy: 0.7314 - val_loss: 0.8442 - val_accuracy: 0.6801\nEpoch 19/20\n6/6 [==============================] - 3s 628ms/step - loss: 0.9227 - accuracy: 0.6714 - val_loss: 0.9800 - val_accuracy: 0.6534\nEpoch 20/20\n6/6 [==============================] - 3s 625ms/step - loss: 0.7154 - accuracy: 0.7343 - val_loss: 0.8701 - val_accuracy: 0.6673\nDuration: 0:01:11.779573\n"
],
[
"n = 1\nmodels_lsa[n].fit_model(image_sets_lsa[n],label_sets_lsa[n],x_val,y_val,epochs=20,batch_size = 128)",
"Epoch 1/20\n11/11 [==============================] - 7s 482ms/step - loss: 1.8946 - accuracy: 0.3236 - val_loss: 1.1215 - val_accuracy: 0.6462\nEpoch 2/20\n11/11 [==============================] - 5s 484ms/step - loss: 1.2982 - accuracy: 0.4771 - val_loss: 0.9860 - val_accuracy: 0.6480\nEpoch 3/20\n11/11 [==============================] - 5s 480ms/step - loss: 1.1976 - accuracy: 0.5364 - val_loss: 0.9669 - val_accuracy: 0.6558\nEpoch 4/20\n11/11 [==============================] - 5s 481ms/step - loss: 1.1207 - accuracy: 0.5643 - val_loss: 0.8854 - val_accuracy: 0.6819\nEpoch 5/20\n11/11 [==============================] - 5s 490ms/step - loss: 1.1931 - accuracy: 0.5471 - val_loss: 0.9150 - val_accuracy: 0.6626\nEpoch 6/20\n11/11 [==============================] - 5s 478ms/step - loss: 1.0171 - accuracy: 0.6157 - val_loss: 1.1086 - val_accuracy: 0.5913\nEpoch 7/20\n11/11 [==============================] - 5s 475ms/step - loss: 0.9804 - accuracy: 0.6400 - val_loss: 0.8846 - val_accuracy: 0.6612\nEpoch 8/20\n11/11 [==============================] - 5s 502ms/step - loss: 0.9827 - accuracy: 0.6193 - val_loss: 0.8754 - val_accuracy: 0.6644\nEpoch 9/20\n11/11 [==============================] - 6s 529ms/step - loss: 0.8869 - accuracy: 0.6721 - val_loss: 0.9599 - val_accuracy: 0.6266\nEpoch 10/20\n11/11 [==============================] - 5s 498ms/step - loss: 0.9265 - accuracy: 0.6621 - val_loss: 0.8709 - val_accuracy: 0.6598\nEpoch 11/20\n11/11 [==============================] - 6s 523ms/step - loss: 0.8521 - accuracy: 0.6836 - val_loss: 0.9033 - val_accuracy: 0.6516\nEpoch 12/20\n11/11 [==============================] - 6s 516ms/step - loss: 0.8134 - accuracy: 0.6979 - val_loss: 0.9116 - val_accuracy: 0.6551\nEpoch 13/20\n11/11 [==============================] - 6s 546ms/step - loss: 0.8364 - accuracy: 0.6814 - val_loss: 0.8297 - val_accuracy: 0.6723\nEpoch 14/20\n11/11 [==============================] - 6s 574ms/step - loss: 0.7585 - accuracy: 0.7150 - val_loss: 0.8616 - val_accuracy: 0.6655\nEpoch 15/20\n11/11 [==============================] - 6s 566ms/step - loss: 0.7321 - accuracy: 0.7371 - val_loss: 0.8710 - val_accuracy: 0.6605\nEpoch 16/20\n11/11 [==============================] - 6s 528ms/step - loss: 0.6922 - accuracy: 0.7336 - val_loss: 0.9260 - val_accuracy: 0.6391\nEpoch 17/20\n11/11 [==============================] - 6s 554ms/step - loss: 0.6282 - accuracy: 0.7650 - val_loss: 0.9375 - val_accuracy: 0.6184\nEpoch 18/20\n11/11 [==============================] - 5s 498ms/step - loss: 0.7511 - accuracy: 0.7300 - val_loss: 0.9233 - val_accuracy: 0.6637\nEpoch 19/20\n11/11 [==============================] - 5s 495ms/step - loss: 0.6333 - accuracy: 0.7636 - val_loss: 0.8548 - val_accuracy: 0.6765\nEpoch 20/20\n11/11 [==============================] - 5s 490ms/step - loss: 0.5589 - accuracy: 0.7886 - val_loss: 1.1196 - val_accuracy: 0.5678\nDuration: 0:01:50.326372\n"
],
[
"n = 2\nmodels_lsa[n].fit_model(image_sets_lsa[n],label_sets_lsa[n],x_val,y_val,epochs=20,batch_size = 128)",
"Epoch 1/20\n17/17 [==============================] - 9s 454ms/step - loss: 1.8661 - accuracy: 0.3703 - val_loss: 0.8873 - val_accuracy: 0.6826\nEpoch 2/20\n17/17 [==============================] - 7s 404ms/step - loss: 1.1814 - accuracy: 0.5126 - val_loss: 0.9045 - val_accuracy: 0.7008\nEpoch 3/20\n17/17 [==============================] - 7s 407ms/step - loss: 1.0810 - accuracy: 0.5562 - val_loss: 1.2201 - val_accuracy: 0.5535\nEpoch 4/20\n17/17 [==============================] - 7s 426ms/step - loss: 1.1343 - accuracy: 0.5553 - val_loss: 0.9419 - val_accuracy: 0.6612\nEpoch 5/20\n17/17 [==============================] - 8s 468ms/step - loss: 1.0833 - accuracy: 0.5747 - val_loss: 1.0173 - val_accuracy: 0.6312\nEpoch 6/20\n17/17 [==============================] - 8s 453ms/step - loss: 0.9682 - accuracy: 0.6209 - val_loss: 0.8684 - val_accuracy: 0.6944\nEpoch 7/20\n17/17 [==============================] - 8s 461ms/step - loss: 0.9501 - accuracy: 0.6100 - val_loss: 1.0084 - val_accuracy: 0.6059\nEpoch 8/20\n17/17 [==============================] - 7s 437ms/step - loss: 0.8573 - accuracy: 0.6614 - val_loss: 0.9154 - val_accuracy: 0.6815\nEpoch 9/20\n17/17 [==============================] - 7s 436ms/step - loss: 0.8818 - accuracy: 0.6571 - val_loss: 0.9704 - val_accuracy: 0.6234\nEpoch 10/20\n17/17 [==============================] - 8s 464ms/step - loss: 0.7790 - accuracy: 0.6898 - val_loss: 0.9311 - val_accuracy: 0.6459\nEpoch 11/20\n17/17 [==============================] - 8s 458ms/step - loss: 0.7683 - accuracy: 0.7089 - val_loss: 0.9931 - val_accuracy: 0.6291\nEpoch 12/20\n17/17 [==============================] - 8s 471ms/step - loss: 0.7478 - accuracy: 0.6990 - val_loss: 0.9532 - val_accuracy: 0.6544\nEpoch 13/20\n17/17 [==============================] - 8s 451ms/step - loss: 0.7483 - accuracy: 0.7061 - val_loss: 0.9334 - val_accuracy: 0.6430\nEpoch 14/20\n17/17 [==============================] - 7s 419ms/step - loss: 0.7301 - accuracy: 0.7036 - val_loss: 0.9408 - val_accuracy: 0.6641\nEpoch 15/20\n17/17 [==============================] - 7s 430ms/step - loss: 0.7776 - accuracy: 0.7198 - val_loss: 1.3140 - val_accuracy: 0.5118\nEpoch 16/20\n17/17 [==============================] - 8s 454ms/step - loss: 0.6392 - accuracy: 0.7404 - val_loss: 1.0015 - val_accuracy: 0.6287\nEpoch 17/20\n17/17 [==============================] - 7s 439ms/step - loss: 0.5989 - accuracy: 0.7705 - val_loss: 0.9678 - val_accuracy: 0.6466\nEpoch 18/20\n17/17 [==============================] - 7s 423ms/step - loss: 0.5836 - accuracy: 0.7830 - val_loss: 0.9590 - val_accuracy: 0.6623\nEpoch 19/20\n17/17 [==============================] - 7s 421ms/step - loss: 0.5588 - accuracy: 0.7888 - val_loss: 0.9917 - val_accuracy: 0.6320\nEpoch 20/20\n17/17 [==============================] - 7s 412ms/step - loss: 0.6361 - accuracy: 0.7617 - val_loss: 1.0590 - val_accuracy: 0.6013\nDuration: 0:02:29.278780\n"
],
[
"n = 3\nmodels_lsa[n].fit_model(image_sets_lsa[n],label_sets_lsa[n],x_val,y_val,epochs=20,batch_size = 128)",
"Epoch 1/20\n22/22 [==============================] - 11s 428ms/step - loss: 1.5531 - accuracy: 0.4740 - val_loss: 0.8377 - val_accuracy: 0.7068\nEpoch 2/20\n22/22 [==============================] - 9s 417ms/step - loss: 0.9646 - accuracy: 0.6090 - val_loss: 0.8973 - val_accuracy: 0.6858\nEpoch 3/20\n22/22 [==============================] - 9s 428ms/step - loss: 0.9248 - accuracy: 0.6363 - val_loss: 0.8556 - val_accuracy: 0.6865\nEpoch 4/20\n22/22 [==============================] - 9s 409ms/step - loss: 0.8847 - accuracy: 0.6510 - val_loss: 0.8897 - val_accuracy: 0.6858\nEpoch 5/20\n22/22 [==============================] - 9s 410ms/step - loss: 0.8457 - accuracy: 0.6793 - val_loss: 0.9218 - val_accuracy: 0.6455\nEpoch 6/20\n22/22 [==============================] - 9s 412ms/step - loss: 0.8002 - accuracy: 0.6896 - val_loss: 1.1707 - val_accuracy: 0.5271\nEpoch 7/20\n22/22 [==============================] - 9s 409ms/step - loss: 0.8258 - accuracy: 0.6888 - val_loss: 1.0025 - val_accuracy: 0.5956\nEpoch 8/20\n22/22 [==============================] - 9s 434ms/step - loss: 0.7404 - accuracy: 0.7080 - val_loss: 1.3450 - val_accuracy: 0.4383\nEpoch 9/20\n22/22 [==============================] - 9s 418ms/step - loss: 0.7097 - accuracy: 0.7180 - val_loss: 0.9532 - val_accuracy: 0.6323\nEpoch 10/20\n22/22 [==============================] - 9s 420ms/step - loss: 0.7233 - accuracy: 0.7317 - val_loss: 0.9749 - val_accuracy: 0.6045\nEpoch 11/20\n22/22 [==============================] - 9s 425ms/step - loss: 0.6510 - accuracy: 0.7637 - val_loss: 1.0011 - val_accuracy: 0.6127\nEpoch 12/20\n22/22 [==============================] - 9s 432ms/step - loss: 0.6149 - accuracy: 0.7756 - val_loss: 0.9478 - val_accuracy: 0.6473\nEpoch 13/20\n22/22 [==============================] - 10s 449ms/step - loss: 0.6201 - accuracy: 0.7642 - val_loss: 0.8858 - val_accuracy: 0.6651\nEpoch 14/20\n22/22 [==============================] - 10s 443ms/step - loss: 0.5953 - accuracy: 0.7722 - val_loss: 1.0934 - val_accuracy: 0.5852\nEpoch 15/20\n22/22 [==============================] - 9s 431ms/step - loss: 0.5604 - accuracy: 0.7853 - val_loss: 1.1574 - val_accuracy: 0.5603\nEpoch 16/20\n22/22 [==============================] - 9s 402ms/step - loss: 0.5685 - accuracy: 0.7805 - val_loss: 1.2570 - val_accuracy: 0.5218\nEpoch 17/20\n22/22 [==============================] - 9s 404ms/step - loss: 0.5401 - accuracy: 0.7968 - val_loss: 0.9041 - val_accuracy: 0.6530\nEpoch 18/20\n22/22 [==============================] - 9s 410ms/step - loss: 0.4967 - accuracy: 0.8127 - val_loss: 1.3973 - val_accuracy: 0.4832\nEpoch 19/20\n22/22 [==============================] - 10s 456ms/step - loss: 0.4896 - accuracy: 0.8130 - val_loss: 1.1225 - val_accuracy: 0.5781\nEpoch 20/20\n22/22 [==============================] - 9s 405ms/step - loss: 0.4411 - accuracy: 0.8366 - val_loss: 1.2082 - val_accuracy: 0.5742\nDuration: 0:03:05.790499\n"
],
[
"n = 4\nmodels_lsa[n].fit_model(image_sets_lsa[n],label_sets_lsa[n],x_val,y_val,epochs=20,batch_size = 128)",
"Epoch 1/20\n28/28 [==============================] - 15s 416ms/step - loss: 1.3594 - accuracy: 0.5072 - val_loss: 1.0544 - val_accuracy: 0.6031\nEpoch 2/20\n28/28 [==============================] - 12s 420ms/step - loss: 0.9471 - accuracy: 0.6356 - val_loss: 0.8675 - val_accuracy: 0.7165\nEpoch 3/20\n28/28 [==============================] - 11s 407ms/step - loss: 0.8981 - accuracy: 0.6602 - val_loss: 1.1100 - val_accuracy: 0.5681\nEpoch 4/20\n28/28 [==============================] - 11s 390ms/step - loss: 0.8470 - accuracy: 0.6838 - val_loss: 0.9087 - val_accuracy: 0.6869\nEpoch 5/20\n28/28 [==============================] - 12s 416ms/step - loss: 0.8293 - accuracy: 0.6881 - val_loss: 0.8904 - val_accuracy: 0.6555\nEpoch 6/20\n28/28 [==============================] - 10s 374ms/step - loss: 0.7493 - accuracy: 0.7252 - val_loss: 1.1010 - val_accuracy: 0.5414\nEpoch 7/20\n28/28 [==============================] - 10s 373ms/step - loss: 0.7674 - accuracy: 0.7181 - val_loss: 1.0420 - val_accuracy: 0.5642\nEpoch 8/20\n28/28 [==============================] - 11s 384ms/step - loss: 0.7359 - accuracy: 0.7210 - val_loss: 1.0661 - val_accuracy: 0.5728\nEpoch 9/20\n28/28 [==============================] - 11s 390ms/step - loss: 0.6449 - accuracy: 0.7643 - val_loss: 1.4586 - val_accuracy: 0.4597\nEpoch 10/20\n28/28 [==============================] - 12s 434ms/step - loss: 0.7111 - accuracy: 0.7421 - val_loss: 1.0048 - val_accuracy: 0.5852\nEpoch 11/20\n28/28 [==============================] - 12s 419ms/step - loss: 0.6203 - accuracy: 0.7722 - val_loss: 0.9508 - val_accuracy: 0.6280\nEpoch 12/20\n28/28 [==============================] - 11s 398ms/step - loss: 0.5815 - accuracy: 0.7779 - val_loss: 0.8788 - val_accuracy: 0.6801\nEpoch 13/20\n28/28 [==============================] - 11s 402ms/step - loss: 0.5672 - accuracy: 0.7976 - val_loss: 0.9882 - val_accuracy: 0.6180\nEpoch 14/20\n28/28 [==============================] - 11s 402ms/step - loss: 0.5770 - accuracy: 0.7896 - val_loss: 1.0442 - val_accuracy: 0.5842\nEpoch 15/20\n28/28 [==============================] - 11s 401ms/step - loss: 0.5412 - accuracy: 0.7962 - val_loss: 0.9656 - val_accuracy: 0.6295\nEpoch 16/20\n28/28 [==============================] - 11s 391ms/step - loss: 0.5530 - accuracy: 0.7997 - val_loss: 0.9658 - val_accuracy: 0.6441\nEpoch 17/20\n28/28 [==============================] - 10s 376ms/step - loss: 0.4846 - accuracy: 0.8268 - val_loss: 0.9189 - val_accuracy: 0.6405\nEpoch 18/20\n28/28 [==============================] - 11s 384ms/step - loss: 0.5108 - accuracy: 0.8134 - val_loss: 0.9830 - val_accuracy: 0.6234\nEpoch 19/20\n28/28 [==============================] - 11s 395ms/step - loss: 0.5626 - accuracy: 0.7928 - val_loss: 0.9988 - val_accuracy: 0.6159\nEpoch 20/20\n28/28 [==============================] - 11s 387ms/step - loss: 0.4240 - accuracy: 0.8483 - val_loss: 1.5242 - val_accuracy: 0.5128\nDuration: 0:03:45.424812\n"
],
[
"n = 5\nmodels_lsa[n].fit_model(image_sets_lsa[n],label_sets_lsa[n],x_val,y_val,epochs=20,batch_size = 128)",
"Epoch 1/20\n33/33 [==============================] - 15s 396ms/step - loss: 1.5245 - accuracy: 0.4783 - val_loss: 0.8874 - val_accuracy: 0.7019\nEpoch 2/20\n33/33 [==============================] - 13s 385ms/step - loss: 0.9869 - accuracy: 0.6223 - val_loss: 0.9243 - val_accuracy: 0.6669\nEpoch 3/20\n33/33 [==============================] - 13s 380ms/step - loss: 0.9366 - accuracy: 0.6428 - val_loss: 0.9287 - val_accuracy: 0.6501\nEpoch 4/20\n33/33 [==============================] - 13s 391ms/step - loss: 0.9052 - accuracy: 0.6534 - val_loss: 0.9730 - val_accuracy: 0.6041\nEpoch 5/20\n33/33 [==============================] - 13s 384ms/step - loss: 0.8564 - accuracy: 0.6722 - val_loss: 0.8483 - val_accuracy: 0.6805\nEpoch 6/20\n33/33 [==============================] - 12s 369ms/step - loss: 0.8124 - accuracy: 0.6891 - val_loss: 1.0211 - val_accuracy: 0.5852\nEpoch 7/20\n33/33 [==============================] - 2494s 78s/step - loss: 0.7682 - accuracy: 0.7008 - val_loss: 0.9547 - val_accuracy: 0.6180\nEpoch 8/20\n33/33 [==============================] - 14s 422ms/step - loss: 0.7100 - accuracy: 0.7323 - val_loss: 1.3337 - val_accuracy: 0.4700\nEpoch 9/20\n33/33 [==============================] - 13s 384ms/step - loss: 0.7394 - accuracy: 0.7204 - val_loss: 1.1667 - val_accuracy: 0.5328\nEpoch 10/20\n33/33 [==============================] - 12s 371ms/step - loss: 0.6853 - accuracy: 0.7256 - val_loss: 1.3475 - val_accuracy: 0.4971\nEpoch 11/20\n33/33 [==============================] - 12s 367ms/step - loss: 0.6472 - accuracy: 0.7535 - val_loss: 0.9886 - val_accuracy: 0.6241\nEpoch 12/20\n33/33 [==============================] - 12s 368ms/step - loss: 0.6398 - accuracy: 0.7525 - val_loss: 0.9352 - val_accuracy: 0.6402\nEpoch 13/20\n33/33 [==============================] - 12s 364ms/step - loss: 0.6172 - accuracy: 0.7687 - val_loss: 1.2990 - val_accuracy: 0.5121\nEpoch 14/20\n33/33 [==============================] - 12s 377ms/step - loss: 0.5755 - accuracy: 0.7917 - val_loss: 1.0118 - val_accuracy: 0.5934\nEpoch 15/20\n33/33 [==============================] - 12s 376ms/step - loss: 0.5533 - accuracy: 0.7957 - val_loss: 0.9680 - val_accuracy: 0.6352\nEpoch 16/20\n33/33 [==============================] - 12s 375ms/step - loss: 0.5513 - accuracy: 0.7880 - val_loss: 0.9857 - val_accuracy: 0.6341\nEpoch 17/20\n33/33 [==============================] - 12s 374ms/step - loss: 0.4900 - accuracy: 0.8160 - val_loss: 1.1406 - val_accuracy: 0.5813\nEpoch 18/20\n33/33 [==============================] - 12s 368ms/step - loss: 0.4646 - accuracy: 0.8376 - val_loss: 1.9233 - val_accuracy: 0.4016\nEpoch 19/20\n33/33 [==============================] - 12s 366ms/step - loss: 0.4829 - accuracy: 0.8204 - val_loss: 1.1202 - val_accuracy: 0.5781\nEpoch 20/20\n33/33 [==============================] - 12s 365ms/step - loss: 0.4253 - accuracy: 0.8420 - val_loss: 1.2650 - val_accuracy: 0.5300\nDuration: 0:45:32.496735\n"
],
[
"n = 6\nmodels_lsa[n].fit_model(image_sets_lsa[n],label_sets_lsa[n],x_val,y_val,epochs=20,batch_size = 128)",
"Epoch 1/20\n39/39 [==============================] - 16s 368ms/step - loss: 1.2959 - accuracy: 0.5116 - val_loss: 1.0358 - val_accuracy: 0.6220\nEpoch 2/20\n39/39 [==============================] - 14s 353ms/step - loss: 0.9867 - accuracy: 0.6167 - val_loss: 1.1220 - val_accuracy: 0.5806\nEpoch 3/20\n39/39 [==============================] - 14s 356ms/step - loss: 0.9196 - accuracy: 0.6397 - val_loss: 0.9736 - val_accuracy: 0.6637\nEpoch 4/20\n39/39 [==============================] - 14s 361ms/step - loss: 0.8998 - accuracy: 0.6487 - val_loss: 0.8598 - val_accuracy: 0.6737\nEpoch 5/20\n39/39 [==============================] - 14s 360ms/step - loss: 0.8201 - accuracy: 0.6734 - val_loss: 0.9242 - val_accuracy: 0.6701\nEpoch 6/20\n39/39 [==============================] - 14s 353ms/step - loss: 0.7887 - accuracy: 0.6901 - val_loss: 1.0069 - val_accuracy: 0.6580\nEpoch 7/20\n39/39 [==============================] - 14s 354ms/step - loss: 0.7982 - accuracy: 0.6908 - val_loss: 0.8798 - val_accuracy: 0.6887\nEpoch 8/20\n39/39 [==============================] - 14s 356ms/step - loss: 0.7056 - accuracy: 0.7252 - val_loss: 1.4547 - val_accuracy: 0.5292\nEpoch 9/20\n39/39 [==============================] - 14s 355ms/step - loss: 0.7537 - accuracy: 0.7045 - val_loss: 0.9854 - val_accuracy: 0.6377\nEpoch 10/20\n39/39 [==============================] - 15s 374ms/step - loss: 0.6520 - accuracy: 0.7478 - val_loss: 0.8058 - val_accuracy: 0.7154\nEpoch 11/20\n39/39 [==============================] - 14s 356ms/step - loss: 0.5992 - accuracy: 0.7755 - val_loss: 0.9326 - val_accuracy: 0.6373\nEpoch 12/20\n39/39 [==============================] - 15s 378ms/step - loss: 0.6000 - accuracy: 0.7779 - val_loss: 0.9292 - val_accuracy: 0.6295\nEpoch 13/20\n39/39 [==============================] - 15s 376ms/step - loss: 0.5964 - accuracy: 0.7706 - val_loss: 1.0841 - val_accuracy: 0.5849\nEpoch 14/20\n39/39 [==============================] - 16s 401ms/step - loss: 0.5826 - accuracy: 0.7834 - val_loss: 1.2264 - val_accuracy: 0.5756\nEpoch 15/20\n39/39 [==============================] - 14s 359ms/step - loss: 0.5488 - accuracy: 0.7913 - val_loss: 1.1725 - val_accuracy: 0.5899\nEpoch 16/20\n39/39 [==============================] - 14s 367ms/step - loss: 0.5405 - accuracy: 0.7955 - val_loss: 0.9635 - val_accuracy: 0.6348\nEpoch 17/20\n39/39 [==============================] - 15s 380ms/step - loss: 0.4669 - accuracy: 0.8281 - val_loss: 1.1636 - val_accuracy: 0.5959\nEpoch 18/20\n39/39 [==============================] - 15s 374ms/step - loss: 0.4972 - accuracy: 0.8157 - val_loss: 1.0279 - val_accuracy: 0.6009\nEpoch 19/20\n39/39 [==============================] - 15s 386ms/step - loss: 0.5155 - accuracy: 0.8132 - val_loss: 1.0452 - val_accuracy: 0.6316\nEpoch 20/20\n39/39 [==============================] - 14s 360ms/step - loss: 0.4348 - accuracy: 0.8424 - val_loss: 1.1446 - val_accuracy: 0.5981\nDuration: 0:04:46.913209\n"
],
[
"n = 7\nmodels_lsa[n].fit_model(image_sets_lsa[n],label_sets_lsa[n],x_val,y_val,epochs=20,batch_size = 128)",
"Epoch 1/20\n44/44 [==============================] - 16s 338ms/step - loss: 1.0141 - accuracy: 0.6048 - val_loss: 0.7934 - val_accuracy: 0.7322\nEpoch 2/20\n44/44 [==============================] - 14s 322ms/step - loss: 0.8564 - accuracy: 0.6668 - val_loss: 0.8025 - val_accuracy: 0.7161\nEpoch 3/20\n44/44 [==============================] - 14s 324ms/step - loss: 0.8062 - accuracy: 0.6920 - val_loss: 0.7807 - val_accuracy: 0.7200\nEpoch 4/20\n44/44 [==============================] - 15s 334ms/step - loss: 0.7598 - accuracy: 0.7039 - val_loss: 0.9071 - val_accuracy: 0.6587\nEpoch 5/20\n44/44 [==============================] - 14s 326ms/step - loss: 0.7213 - accuracy: 0.7175 - val_loss: 1.0161 - val_accuracy: 0.5956\nEpoch 6/20\n44/44 [==============================] - 14s 328ms/step - loss: 0.6876 - accuracy: 0.7311 - val_loss: 2.0429 - val_accuracy: 0.4198\nEpoch 7/20\n44/44 [==============================] - 14s 325ms/step - loss: 0.6943 - accuracy: 0.7312 - val_loss: 1.3482 - val_accuracy: 0.5011\nEpoch 8/20\n44/44 [==============================] - 14s 324ms/step - loss: 0.6565 - accuracy: 0.7520 - val_loss: 0.7806 - val_accuracy: 0.6983\nEpoch 9/20\n44/44 [==============================] - 14s 325ms/step - loss: 0.6137 - accuracy: 0.7657 - val_loss: 0.8346 - val_accuracy: 0.6976\nEpoch 10/20\n44/44 [==============================] - 14s 324ms/step - loss: 0.6187 - accuracy: 0.7652 - val_loss: 1.1757 - val_accuracy: 0.5599\nEpoch 11/20\n44/44 [==============================] - 14s 325ms/step - loss: 0.5809 - accuracy: 0.7805 - val_loss: 0.8818 - val_accuracy: 0.6644\nEpoch 12/20\n44/44 [==============================] - 14s 324ms/step - loss: 0.5500 - accuracy: 0.7968 - val_loss: 0.9220 - val_accuracy: 0.6352\nEpoch 13/20\n44/44 [==============================] - 14s 325ms/step - loss: 0.5498 - accuracy: 0.7900 - val_loss: 1.5051 - val_accuracy: 0.4904\nEpoch 14/20\n44/44 [==============================] - 14s 324ms/step - loss: 0.5182 - accuracy: 0.8054 - val_loss: 1.3590 - val_accuracy: 0.5392\nEpoch 15/20\n44/44 [==============================] - 14s 326ms/step - loss: 0.4916 - accuracy: 0.8175 - val_loss: 1.2730 - val_accuracy: 0.5556\nEpoch 16/20\n44/44 [==============================] - 14s 324ms/step - loss: 0.4617 - accuracy: 0.8298 - val_loss: 1.1493 - val_accuracy: 0.6009\nEpoch 17/20\n44/44 [==============================] - 14s 324ms/step - loss: 0.4553 - accuracy: 0.8329 - val_loss: 1.0280 - val_accuracy: 0.6002\nEpoch 18/20\n44/44 [==============================] - 14s 326ms/step - loss: 0.4439 - accuracy: 0.8330 - val_loss: 0.9410 - val_accuracy: 0.6330\nEpoch 19/20\n44/44 [==============================] - 14s 324ms/step - loss: 0.4008 - accuracy: 0.8512 - val_loss: 0.9906 - val_accuracy: 0.6238\nEpoch 20/20\n44/44 [==============================] - 14s 324ms/step - loss: 0.4084 - accuracy: 0.8570 - val_loss: 1.0077 - val_accuracy: 0.6180\nDuration: 0:04:47.674746\n"
],
[
"n = 8\nmodels_lsa[n].fit_model(image_sets_lsa[n],label_sets_lsa[n],x_val,y_val,epochs=20,batch_size = 128)",
"Epoch 1/20\n50/50 [==============================] - 18s 333ms/step - loss: 1.0302 - accuracy: 0.6033 - val_loss: 0.8136 - val_accuracy: 0.7468\nEpoch 2/20\n50/50 [==============================] - 16s 316ms/step - loss: 0.8675 - accuracy: 0.6563 - val_loss: 1.5622 - val_accuracy: 0.4490\nEpoch 3/20\n50/50 [==============================] - 16s 319ms/step - loss: 0.7999 - accuracy: 0.6937 - val_loss: 0.9258 - val_accuracy: 0.6473\nEpoch 4/20\n50/50 [==============================] - 16s 318ms/step - loss: 0.7712 - accuracy: 0.6997 - val_loss: 0.7783 - val_accuracy: 0.7165\nEpoch 5/20\n50/50 [==============================] - 16s 318ms/step - loss: 0.7258 - accuracy: 0.7156 - val_loss: 0.9879 - val_accuracy: 0.6230\nEpoch 6/20\n50/50 [==============================] - 16s 317ms/step - loss: 0.6963 - accuracy: 0.7286 - val_loss: 1.0448 - val_accuracy: 0.6284\nEpoch 7/20\n50/50 [==============================] - 16s 318ms/step - loss: 0.6679 - accuracy: 0.7468 - val_loss: 0.9360 - val_accuracy: 0.6327\nEpoch 8/20\n50/50 [==============================] - 16s 317ms/step - loss: 0.6393 - accuracy: 0.7489 - val_loss: 0.7726 - val_accuracy: 0.7015\nEpoch 9/20\n50/50 [==============================] - 16s 318ms/step - loss: 0.6100 - accuracy: 0.7716 - val_loss: 1.0814 - val_accuracy: 0.5774\nEpoch 10/20\n50/50 [==============================] - 16s 319ms/step - loss: 0.5979 - accuracy: 0.7763 - val_loss: 1.0184 - val_accuracy: 0.6127\nEpoch 11/20\n50/50 [==============================] - 17s 334ms/step - loss: 0.5711 - accuracy: 0.7879 - val_loss: 0.8104 - val_accuracy: 0.7068\nEpoch 12/20\n50/50 [==============================] - 16s 319ms/step - loss: 0.5292 - accuracy: 0.8070 - val_loss: 1.2299 - val_accuracy: 0.5931\nEpoch 13/20\n50/50 [==============================] - 16s 320ms/step - loss: 0.5266 - accuracy: 0.8006 - val_loss: 0.8266 - val_accuracy: 0.6844\nEpoch 14/20\n50/50 [==============================] - 16s 319ms/step - loss: 0.4951 - accuracy: 0.8170 - val_loss: 0.9707 - val_accuracy: 0.6334\nEpoch 15/20\n50/50 [==============================] - 16s 319ms/step - loss: 0.4693 - accuracy: 0.8284 - val_loss: 1.6673 - val_accuracy: 0.4772\nEpoch 16/20\n50/50 [==============================] - 16s 319ms/step - loss: 0.4733 - accuracy: 0.8263 - val_loss: 1.0761 - val_accuracy: 0.5788\nEpoch 17/20\n50/50 [==============================] - 16s 320ms/step - loss: 0.4267 - accuracy: 0.8395 - val_loss: 1.0778 - val_accuracy: 0.5874\nEpoch 18/20\n50/50 [==============================] - 16s 320ms/step - loss: 0.4450 - accuracy: 0.8379 - val_loss: 1.2456 - val_accuracy: 0.5995\nEpoch 19/20\n50/50 [==============================] - 16s 319ms/step - loss: 0.4104 - accuracy: 0.8516 - val_loss: 1.7010 - val_accuracy: 0.5032\nEpoch 20/20\n50/50 [==============================] - 16s 320ms/step - loss: 0.3833 - accuracy: 0.8597 - val_loss: 1.0084 - val_accuracy: 0.6255\nDuration: 0:05:21.520598\n"
],
[
"n = 9\nmodels_lsa[n].fit_model(image_sets_lsa[n],label_sets_lsa[n],x_val,y_val,epochs=20,batch_size = 128)",
"Epoch 1/20\n55/55 [==============================] - 19s 324ms/step - loss: 1.0165 - accuracy: 0.5970 - val_loss: 0.7971 - val_accuracy: 0.7372\nEpoch 2/20\n55/55 [==============================] - 17s 318ms/step - loss: 0.8632 - accuracy: 0.6559 - val_loss: 0.9980 - val_accuracy: 0.6555\nEpoch 3/20\n55/55 [==============================] - 18s 331ms/step - loss: 0.8279 - accuracy: 0.6780 - val_loss: 0.7247 - val_accuracy: 0.7447\nEpoch 4/20\n55/55 [==============================] - 17s 319ms/step - loss: 0.8013 - accuracy: 0.6854 - val_loss: 0.9289 - val_accuracy: 0.6448\nEpoch 5/20\n55/55 [==============================] - 18s 320ms/step - loss: 0.7616 - accuracy: 0.7109 - val_loss: 0.7194 - val_accuracy: 0.7536\nEpoch 6/20\n55/55 [==============================] - 18s 322ms/step - loss: 0.7236 - accuracy: 0.7193 - val_loss: 0.7984 - val_accuracy: 0.6879\nEpoch 7/20\n55/55 [==============================] - 18s 323ms/step - loss: 0.6860 - accuracy: 0.7327 - val_loss: 0.7932 - val_accuracy: 0.6851\nEpoch 8/20\n55/55 [==============================] - 18s 320ms/step - loss: 0.6826 - accuracy: 0.7371 - val_loss: 0.7292 - val_accuracy: 0.7225\nEpoch 9/20\n55/55 [==============================] - 17s 318ms/step - loss: 0.6618 - accuracy: 0.7487 - val_loss: 0.7265 - val_accuracy: 0.7272\nEpoch 10/20\n55/55 [==============================] - 17s 318ms/step - loss: 0.6325 - accuracy: 0.7581 - val_loss: 0.7766 - val_accuracy: 0.7154\nEpoch 11/20\n55/55 [==============================] - 17s 318ms/step - loss: 0.6040 - accuracy: 0.7696 - val_loss: 0.7070 - val_accuracy: 0.7325\nEpoch 12/20\n55/55 [==============================] - 18s 319ms/step - loss: 0.5811 - accuracy: 0.7809 - val_loss: 0.7249 - val_accuracy: 0.7322\nEpoch 13/20\n55/55 [==============================] - 20s 365ms/step - loss: 0.5691 - accuracy: 0.7821 - val_loss: 0.8235 - val_accuracy: 0.6815\nEpoch 14/20\n55/55 [==============================] - 19s 337ms/step - loss: 0.5448 - accuracy: 0.7933 - val_loss: 0.7487 - val_accuracy: 0.7129\nEpoch 15/20\n55/55 [==============================] - 18s 321ms/step - loss: 0.5257 - accuracy: 0.8054 - val_loss: 0.8158 - val_accuracy: 0.6979\nEpoch 16/20\n55/55 [==============================] - 18s 320ms/step - loss: 0.5074 - accuracy: 0.8091 - val_loss: 0.8471 - val_accuracy: 0.6833\nEpoch 17/20\n55/55 [==============================] - 18s 321ms/step - loss: 0.4983 - accuracy: 0.8114 - val_loss: 0.7642 - val_accuracy: 0.7172\nEpoch 18/20\n55/55 [==============================] - 18s 321ms/step - loss: 0.4684 - accuracy: 0.8274 - val_loss: 0.8835 - val_accuracy: 0.6894\nEpoch 19/20\n55/55 [==============================] - 18s 328ms/step - loss: 0.4443 - accuracy: 0.8411 - val_loss: 0.8346 - val_accuracy: 0.6833\nEpoch 20/20\n55/55 [==============================] - 18s 323ms/step - loss: 0.4287 - accuracy: 0.8447 - val_loss: 1.1799 - val_accuracy: 0.5917\nDuration: 0:05:57.855470\n"
],
[
"n = 10\nmodels_lsa[n].fit_model(image_sets_lsa[n],label_sets_lsa[n],x_val,y_val,epochs=20,batch_size = 128)",
"Epoch 1/20\n61/61 [==============================] - 22s 328ms/step - loss: 0.9818 - accuracy: 0.6234 - val_loss: 0.8760 - val_accuracy: 0.7040\nEpoch 2/20\n61/61 [==============================] - 19s 315ms/step - loss: 0.8478 - accuracy: 0.6725 - val_loss: 0.8838 - val_accuracy: 0.6737\nEpoch 3/20\n61/61 [==============================] - 19s 315ms/step - loss: 0.8165 - accuracy: 0.6843 - val_loss: 0.8286 - val_accuracy: 0.6648\nEpoch 4/20\n61/61 [==============================] - 19s 315ms/step - loss: 0.7676 - accuracy: 0.7048 - val_loss: 0.9776 - val_accuracy: 0.6316\nEpoch 5/20\n61/61 [==============================] - 19s 317ms/step - loss: 0.7404 - accuracy: 0.7144 - val_loss: 1.0183 - val_accuracy: 0.5981\nEpoch 6/20\n61/61 [==============================] - 19s 318ms/step - loss: 0.7205 - accuracy: 0.7239 - val_loss: 0.7239 - val_accuracy: 0.7290\nEpoch 7/20\n61/61 [==============================] - 19s 317ms/step - loss: 0.6916 - accuracy: 0.7312 - val_loss: 0.9580 - val_accuracy: 0.6202\nEpoch 8/20\n61/61 [==============================] - 19s 315ms/step - loss: 0.6731 - accuracy: 0.7414 - val_loss: 0.8468 - val_accuracy: 0.6698\nEpoch 9/20\n61/61 [==============================] - 20s 327ms/step - loss: 0.6428 - accuracy: 0.7513 - val_loss: 0.9564 - val_accuracy: 0.6384\nEpoch 10/20\n61/61 [==============================] - 19s 317ms/step - loss: 0.6183 - accuracy: 0.7684 - val_loss: 0.7993 - val_accuracy: 0.7019\nEpoch 11/20\n61/61 [==============================] - 19s 315ms/step - loss: 0.5842 - accuracy: 0.7777 - val_loss: 0.7108 - val_accuracy: 0.7400\nEpoch 12/20\n61/61 [==============================] - 19s 314ms/step - loss: 0.5809 - accuracy: 0.7805 - val_loss: 1.0145 - val_accuracy: 0.5909\nEpoch 13/20\n61/61 [==============================] - 19s 317ms/step - loss: 0.5649 - accuracy: 0.7871 - val_loss: 0.7320 - val_accuracy: 0.7215\nEpoch 14/20\n61/61 [==============================] - 20s 320ms/step - loss: 0.5305 - accuracy: 0.7935 - val_loss: 0.9582 - val_accuracy: 0.6312\nEpoch 15/20\n61/61 [==============================] - 21s 345ms/step - loss: 0.5214 - accuracy: 0.8048 - val_loss: 0.8126 - val_accuracy: 0.7061\nEpoch 16/20\n61/61 [==============================] - 19s 314ms/step - loss: 0.4982 - accuracy: 0.8162 - val_loss: 0.9771 - val_accuracy: 0.6608\nEpoch 17/20\n61/61 [==============================] - 19s 316ms/step - loss: 0.4816 - accuracy: 0.8183 - val_loss: 0.8599 - val_accuracy: 0.6979\nEpoch 18/20\n61/61 [==============================] - 19s 313ms/step - loss: 0.4671 - accuracy: 0.8205 - val_loss: 0.8884 - val_accuracy: 0.6580\nEpoch 19/20\n61/61 [==============================] - 19s 316ms/step - loss: 0.4463 - accuracy: 0.8353 - val_loss: 1.0291 - val_accuracy: 0.6437\nEpoch 20/20\n61/61 [==============================] - 19s 318ms/step - loss: 0.4467 - accuracy: 0.8357 - val_loss: 1.0232 - val_accuracy: 0.6427\nDuration: 0:06:29.986582\n"
],
[
"n = 11\nmodels_lsa[n].fit_model(image_sets_lsa[n],label_sets_lsa[n],x_val,y_val,epochs=20,batch_size = 128)",
"Epoch 1/20\n66/66 [==============================] - 23s 319ms/step - loss: 0.9548 - accuracy: 0.6389 - val_loss: 0.8210 - val_accuracy: 0.7154\nEpoch 2/20\n66/66 [==============================] - 21s 313ms/step - loss: 0.8450 - accuracy: 0.6792 - val_loss: 0.8767 - val_accuracy: 0.6537\nEpoch 3/20\n66/66 [==============================] - 21s 313ms/step - loss: 0.7912 - accuracy: 0.6932 - val_loss: 0.7726 - val_accuracy: 0.7133\nEpoch 4/20\n66/66 [==============================] - 21s 311ms/step - loss: 0.7552 - accuracy: 0.7118 - val_loss: 0.7189 - val_accuracy: 0.7536\nEpoch 5/20\n66/66 [==============================] - 21s 312ms/step - loss: 0.7471 - accuracy: 0.7175 - val_loss: 0.8621 - val_accuracy: 0.6665\nEpoch 6/20\n66/66 [==============================] - 20s 311ms/step - loss: 0.6959 - accuracy: 0.7344 - val_loss: 0.7927 - val_accuracy: 0.6819\nEpoch 7/20\n66/66 [==============================] - 21s 311ms/step - loss: 0.6828 - accuracy: 0.7392 - val_loss: 0.7152 - val_accuracy: 0.7225\nEpoch 8/20\n66/66 [==============================] - 21s 311ms/step - loss: 0.6529 - accuracy: 0.7513 - val_loss: 0.7717 - val_accuracy: 0.7022\nEpoch 9/20\n66/66 [==============================] - 21s 312ms/step - loss: 0.6426 - accuracy: 0.7585 - val_loss: 0.8844 - val_accuracy: 0.6391\nEpoch 10/20\n66/66 [==============================] - 21s 316ms/step - loss: 0.6073 - accuracy: 0.7699 - val_loss: 0.6602 - val_accuracy: 0.7671\nEpoch 11/20\n66/66 [==============================] - 21s 313ms/step - loss: 0.5829 - accuracy: 0.7812 - val_loss: 0.7941 - val_accuracy: 0.7022\nEpoch 12/20\n66/66 [==============================] - 21s 315ms/step - loss: 0.5786 - accuracy: 0.7849 - val_loss: 0.7342 - val_accuracy: 0.7222\nEpoch 13/20\n66/66 [==============================] - 21s 311ms/step - loss: 0.5437 - accuracy: 0.7992 - val_loss: 0.7949 - val_accuracy: 0.6947\nEpoch 14/20\n66/66 [==============================] - 21s 312ms/step - loss: 0.5331 - accuracy: 0.8021 - val_loss: 0.7667 - val_accuracy: 0.7108\nEpoch 15/20\n66/66 [==============================] - 20s 311ms/step - loss: 0.5034 - accuracy: 0.8098 - val_loss: 0.9727 - val_accuracy: 0.6434\nEpoch 16/20\n66/66 [==============================] - 21s 312ms/step - loss: 0.5098 - accuracy: 0.8125 - val_loss: 0.8025 - val_accuracy: 0.6958\nEpoch 17/20\n66/66 [==============================] - 21s 313ms/step - loss: 0.4842 - accuracy: 0.8212 - val_loss: 0.8357 - val_accuracy: 0.6644\nEpoch 18/20\n66/66 [==============================] - 21s 324ms/step - loss: 0.4674 - accuracy: 0.8270 - val_loss: 0.8785 - val_accuracy: 0.6680\nEpoch 19/20\n66/66 [==============================] - 21s 320ms/step - loss: 0.4677 - accuracy: 0.8275 - val_loss: 0.9145 - val_accuracy: 0.6865\nEpoch 20/20\n66/66 [==============================] - 21s 322ms/step - loss: 0.4308 - accuracy: 0.8414 - val_loss: 0.9848 - val_accuracy: 0.6573\nDuration: 0:06:56.196394\n"
],
[
"n = 12\nmodels_lsa[n].fit_model(image_sets_lsa[n],label_sets_lsa[n],x_val,y_val,epochs=20,batch_size = 128)",
"Epoch 1/20\n72/72 [==============================] - 24s 307ms/step - loss: 0.9401 - accuracy: 0.6445 - val_loss: 0.7893 - val_accuracy: 0.7072\nEpoch 2/20\n72/72 [==============================] - 22s 307ms/step - loss: 0.8291 - accuracy: 0.6824 - val_loss: 0.8862 - val_accuracy: 0.6751\nEpoch 3/20\n72/72 [==============================] - 23s 317ms/step - loss: 0.7826 - accuracy: 0.7012 - val_loss: 0.8630 - val_accuracy: 0.6673\nEpoch 4/20\n72/72 [==============================] - 22s 301ms/step - loss: 0.7521 - accuracy: 0.7115 - val_loss: 0.7412 - val_accuracy: 0.7193\nEpoch 5/20\n72/72 [==============================] - 22s 302ms/step - loss: 0.7211 - accuracy: 0.7263 - val_loss: 0.7178 - val_accuracy: 0.7504\nEpoch 6/20\n72/72 [==============================] - 22s 301ms/step - loss: 0.7102 - accuracy: 0.7326 - val_loss: 0.7650 - val_accuracy: 0.7004\nEpoch 7/20\n72/72 [==============================] - 22s 302ms/step - loss: 0.6792 - accuracy: 0.7409 - val_loss: 0.8653 - val_accuracy: 0.6865\nEpoch 8/20\n72/72 [==============================] - 22s 302ms/step - loss: 0.6674 - accuracy: 0.7451 - val_loss: 0.6719 - val_accuracy: 0.7575\nEpoch 9/20\n72/72 [==============================] - 22s 302ms/step - loss: 0.6289 - accuracy: 0.7598 - val_loss: 1.2657 - val_accuracy: 0.5624\nEpoch 10/20\n72/72 [==============================] - 22s 301ms/step - loss: 0.6141 - accuracy: 0.7649 - val_loss: 0.7711 - val_accuracy: 0.7126\nEpoch 11/20\n72/72 [==============================] - 22s 303ms/step - loss: 0.6041 - accuracy: 0.7664 - val_loss: 0.9243 - val_accuracy: 0.6651\nEpoch 12/20\n72/72 [==============================] - 22s 302ms/step - loss: 0.5900 - accuracy: 0.7777 - val_loss: 1.0059 - val_accuracy: 0.6494\nEpoch 13/20\n72/72 [==============================] - 22s 301ms/step - loss: 0.5598 - accuracy: 0.7838 - val_loss: 0.9711 - val_accuracy: 0.6919\nEpoch 14/20\n72/72 [==============================] - 22s 311ms/step - loss: 0.5433 - accuracy: 0.7931 - val_loss: 0.7862 - val_accuracy: 0.6947\nEpoch 15/20\n72/72 [==============================] - 22s 312ms/step - loss: 0.5328 - accuracy: 0.7989 - val_loss: 0.7691 - val_accuracy: 0.7229\nEpoch 16/20\n72/72 [==============================] - 23s 326ms/step - loss: 0.5070 - accuracy: 0.8111 - val_loss: 1.1300 - val_accuracy: 0.6106\nEpoch 17/20\n72/72 [==============================] - 23s 314ms/step - loss: 0.5076 - accuracy: 0.8145 - val_loss: 0.8347 - val_accuracy: 0.7111\nEpoch 18/20\n72/72 [==============================] - 22s 307ms/step - loss: 0.4831 - accuracy: 0.8187 - val_loss: 0.7321 - val_accuracy: 0.7350\nEpoch 19/20\n72/72 [==============================] - 22s 304ms/step - loss: 0.4681 - accuracy: 0.8249 - val_loss: 0.9948 - val_accuracy: 0.7076\nEpoch 20/20\n72/72 [==============================] - 22s 303ms/step - loss: 0.4431 - accuracy: 0.8351 - val_loss: 0.8727 - val_accuracy: 0.7101\nDuration: 0:07:22.767589\n"
],
[
"models_lsa[13] = utils.My_model('intel',True,model_dir)\nmodels_lsa[13].compile_model()\nmodels_lsa[14] = utils.My_model('intel',True,model_dir)\nmodels_lsa[14].compile_model()",
"Model loaded correctly\nModel compiled\nModel loaded correctly\nModel compiled\n"
],
[
"n = 13\nmodels_lsa[n].fit_model(image_sets_lsa[n],label_sets_lsa[n],x_val,y_val,epochs=20,batch_size = 128)",
"Epoch 1/20\n77/77 [==============================] - 25s 303ms/step - loss: 1.0739 - accuracy: 0.6188 - val_loss: 0.8010 - val_accuracy: 0.7150\nEpoch 2/20\n77/77 [==============================] - 23s 297ms/step - loss: 0.8105 - accuracy: 0.6902 - val_loss: 0.6996 - val_accuracy: 0.7361\nEpoch 3/20\n77/77 [==============================] - 23s 298ms/step - loss: 0.7501 - accuracy: 0.7111 - val_loss: 0.6619 - val_accuracy: 0.7461\nEpoch 4/20\n77/77 [==============================] - 23s 297ms/step - loss: 0.7336 - accuracy: 0.7264 - val_loss: 0.7042 - val_accuracy: 0.7293\nEpoch 5/20\n77/77 [==============================] - 23s 302ms/step - loss: 0.7178 - accuracy: 0.7375 - val_loss: 0.7076 - val_accuracy: 0.7386\nEpoch 6/20\n77/77 [==============================] - 23s 300ms/step - loss: 0.6841 - accuracy: 0.7435 - val_loss: 0.8195 - val_accuracy: 0.7090\nEpoch 7/20\n77/77 [==============================] - 23s 300ms/step - loss: 0.6774 - accuracy: 0.7502 - val_loss: 0.7132 - val_accuracy: 0.7343\nEpoch 8/20\n77/77 [==============================] - 23s 300ms/step - loss: 0.6296 - accuracy: 0.7672 - val_loss: 0.6941 - val_accuracy: 0.7411\nEpoch 9/20\n77/77 [==============================] - 23s 300ms/step - loss: 0.6345 - accuracy: 0.7650 - val_loss: 0.8884 - val_accuracy: 0.6701\nEpoch 10/20\n77/77 [==============================] - 23s 304ms/step - loss: 0.5743 - accuracy: 0.7900 - val_loss: 0.7081 - val_accuracy: 0.7382\nEpoch 11/20\n77/77 [==============================] - 24s 309ms/step - loss: 0.5951 - accuracy: 0.7761 - val_loss: 0.6866 - val_accuracy: 0.7479\nEpoch 12/20\n77/77 [==============================] - 23s 304ms/step - loss: 0.5808 - accuracy: 0.7827 - val_loss: 0.6869 - val_accuracy: 0.7414\nEpoch 13/20\n77/77 [==============================] - 23s 300ms/step - loss: 0.5275 - accuracy: 0.8058 - val_loss: 0.7015 - val_accuracy: 0.7343\nEpoch 14/20\n77/77 [==============================] - 23s 305ms/step - loss: 0.5109 - accuracy: 0.8047 - val_loss: 0.7457 - val_accuracy: 0.7343\nEpoch 15/20\n77/77 [==============================] - 23s 300ms/step - loss: 0.5065 - accuracy: 0.8182 - val_loss: 0.7865 - val_accuracy: 0.7329\nEpoch 16/20\n77/77 [==============================] - 24s 306ms/step - loss: 0.5004 - accuracy: 0.8094 - val_loss: 0.8880 - val_accuracy: 0.7047\nEpoch 17/20\n77/77 [==============================] - 23s 301ms/step - loss: 0.4674 - accuracy: 0.8325 - val_loss: 0.8937 - val_accuracy: 0.6983\nEpoch 18/20\n77/77 [==============================] - 23s 301ms/step - loss: 0.4776 - accuracy: 0.8241 - val_loss: 0.9107 - val_accuracy: 0.7154\nEpoch 19/20\n77/77 [==============================] - 23s 299ms/step - loss: 0.4562 - accuracy: 0.8317 - val_loss: 0.7944 - val_accuracy: 0.7247\nEpoch 20/20\n77/77 [==============================] - 23s 299ms/step - loss: 0.4305 - accuracy: 0.8440 - val_loss: 1.0069 - val_accuracy: 0.6890\nDuration: 0:07:45.186916\n"
],
[
"n = 14\nmodels_lsa[n].fit_model(image_sets_lsa[n],label_sets_lsa[n],x_val,y_val,epochs=20,batch_size = 128)",
"Epoch 1/20\n83/83 [==============================] - 26s 299ms/step - loss: 0.9787 - accuracy: 0.6390 - val_loss: 1.0858 - val_accuracy: 0.6159\nEpoch 2/20\n83/83 [==============================] - 24s 293ms/step - loss: 0.8370 - accuracy: 0.6831 - val_loss: 0.7888 - val_accuracy: 0.7297\nEpoch 3/20\n83/83 [==============================] - 25s 295ms/step - loss: 0.8074 - accuracy: 0.6955 - val_loss: 0.8777 - val_accuracy: 0.6776\nEpoch 4/20\n83/83 [==============================] - 24s 294ms/step - loss: 0.7663 - accuracy: 0.7121 - val_loss: 0.8881 - val_accuracy: 0.6904\nEpoch 5/20\n83/83 [==============================] - 25s 296ms/step - loss: 0.7507 - accuracy: 0.7155 - val_loss: 0.8028 - val_accuracy: 0.7186\nEpoch 6/20\n83/83 [==============================] - 25s 296ms/step - loss: 0.7325 - accuracy: 0.7294 - val_loss: 1.7042 - val_accuracy: 0.4786\nEpoch 7/20\n83/83 [==============================] - 24s 294ms/step - loss: 0.7233 - accuracy: 0.7327 - val_loss: 0.7150 - val_accuracy: 0.7489\nEpoch 8/20\n83/83 [==============================] - 24s 295ms/step - loss: 0.6633 - accuracy: 0.7476 - val_loss: 1.0785 - val_accuracy: 0.6262\nEpoch 9/20\n83/83 [==============================] - 25s 307ms/step - loss: 0.6621 - accuracy: 0.7548 - val_loss: 1.0198 - val_accuracy: 0.6544\nEpoch 10/20\n83/83 [==============================] - 25s 302ms/step - loss: 0.6573 - accuracy: 0.7578 - val_loss: 0.7187 - val_accuracy: 0.7257\nEpoch 11/20\n83/83 [==============================] - 25s 297ms/step - loss: 0.6141 - accuracy: 0.7669 - val_loss: 0.6917 - val_accuracy: 0.7600\nEpoch 12/20\n83/83 [==============================] - 26s 313ms/step - loss: 0.5857 - accuracy: 0.7832 - val_loss: 0.8734 - val_accuracy: 0.6715\nEpoch 13/20\n83/83 [==============================] - 30s 361ms/step - loss: 0.5884 - accuracy: 0.7771 - val_loss: 0.6749 - val_accuracy: 0.7564\nEpoch 14/20\n83/83 [==============================] - 29s 347ms/step - loss: 0.5498 - accuracy: 0.7909 - val_loss: 0.8043 - val_accuracy: 0.7093\nEpoch 15/20\n83/83 [==============================] - 27s 332ms/step - loss: 0.5506 - accuracy: 0.7918 - val_loss: 0.9964 - val_accuracy: 0.6516\nEpoch 16/20\n83/83 [==============================] - 28s 332ms/step - loss: 0.5479 - accuracy: 0.7937 - val_loss: 0.6900 - val_accuracy: 0.7557\nEpoch 17/20\n83/83 [==============================] - 27s 321ms/step - loss: 0.5188 - accuracy: 0.8117 - val_loss: 0.9152 - val_accuracy: 0.7033\nEpoch 18/20\n83/83 [==============================] - 26s 318ms/step - loss: 0.5052 - accuracy: 0.8134 - val_loss: 0.6837 - val_accuracy: 0.7589\nEpoch 19/20\n83/83 [==============================] - 26s 315ms/step - loss: 0.4771 - accuracy: 0.8185 - val_loss: 0.8107 - val_accuracy: 0.7033\nEpoch 20/20\n83/83 [==============================] - 26s 312ms/step - loss: 0.4839 - accuracy: 0.8226 - val_loss: 0.8726 - val_accuracy: 0.7133\nDuration: 0:08:37.667555\n"
],
[
"n = 15\nmodels_lsa[n].fit_model(image_sets_lsa[n],label_sets_lsa[n],x_val,y_val,epochs=20,batch_size = 128)",
"Epoch 1/20\n88/88 [==============================] - 28s 298ms/step - loss: 0.9273 - accuracy: 0.6525 - val_loss: 0.6116 - val_accuracy: 0.7846\nEpoch 2/20\n88/88 [==============================] - 26s 299ms/step - loss: 0.8230 - accuracy: 0.6817 - val_loss: 0.7093 - val_accuracy: 0.7361\nEpoch 3/20\n88/88 [==============================] - 26s 295ms/step - loss: 0.7700 - accuracy: 0.7038 - val_loss: 0.6230 - val_accuracy: 0.7832\nEpoch 4/20\n88/88 [==============================] - 27s 302ms/step - loss: 0.7625 - accuracy: 0.7092 - val_loss: 0.6244 - val_accuracy: 0.7743\nEpoch 5/20\n88/88 [==============================] - 26s 298ms/step - loss: 0.7274 - accuracy: 0.7221 - val_loss: 0.6227 - val_accuracy: 0.7743\nEpoch 6/20\n88/88 [==============================] - 26s 297ms/step - loss: 0.7131 - accuracy: 0.7278 - val_loss: 0.7245 - val_accuracy: 0.7364\nEpoch 7/20\n88/88 [==============================] - 26s 296ms/step - loss: 0.6896 - accuracy: 0.7377 - val_loss: 0.5607 - val_accuracy: 0.7971\nEpoch 8/20\n88/88 [==============================] - 26s 296ms/step - loss: 0.6662 - accuracy: 0.7477 - val_loss: 0.5727 - val_accuracy: 0.7964\nEpoch 9/20\n88/88 [==============================] - 26s 295ms/step - loss: 0.6445 - accuracy: 0.7544 - val_loss: 0.7508 - val_accuracy: 0.7282\nEpoch 10/20\n88/88 [==============================] - 26s 301ms/step - loss: 0.6208 - accuracy: 0.7618 - val_loss: 0.5680 - val_accuracy: 0.7989\nEpoch 11/20\n88/88 [==============================] - 26s 297ms/step - loss: 0.6101 - accuracy: 0.7638 - val_loss: 0.6879 - val_accuracy: 0.7482\nEpoch 12/20\n88/88 [==============================] - 26s 295ms/step - loss: 0.5974 - accuracy: 0.7754 - val_loss: 0.7721 - val_accuracy: 0.7275\nEpoch 13/20\n88/88 [==============================] - 26s 301ms/step - loss: 0.5695 - accuracy: 0.7873 - val_loss: 0.6459 - val_accuracy: 0.7807\nEpoch 14/20\n88/88 [==============================] - 27s 311ms/step - loss: 0.5698 - accuracy: 0.7831 - val_loss: 0.6375 - val_accuracy: 0.7746\nEpoch 15/20\n88/88 [==============================] - 30s 337ms/step - loss: 0.5560 - accuracy: 0.7921 - val_loss: 0.6152 - val_accuracy: 0.7807\nEpoch 16/20\n88/88 [==============================] - 27s 307ms/step - loss: 0.5351 - accuracy: 0.7998 - val_loss: 0.6096 - val_accuracy: 0.7746\nEpoch 17/20\n88/88 [==============================] - 27s 306ms/step - loss: 0.5106 - accuracy: 0.8052 - val_loss: 0.5741 - val_accuracy: 0.7989\nEpoch 18/20\n88/88 [==============================] - 26s 297ms/step - loss: 0.4980 - accuracy: 0.8141 - val_loss: 0.7270 - val_accuracy: 0.7511\nEpoch 19/20\n88/88 [==============================] - 28s 318ms/step - loss: 0.4876 - accuracy: 0.8163 - val_loss: 0.7024 - val_accuracy: 0.7571\nEpoch 20/20\n88/88 [==============================] - 27s 308ms/step - loss: 0.4721 - accuracy: 0.8275 - val_loss: 0.6239 - val_accuracy: 0.7867\nDuration: 0:08:53.923035\n"
],
[
"n = 16\nmodels_lsa[n].fit_model(image_sets_lsa[n],label_sets_lsa[n],x_val,y_val,epochs=20,batch_size = 128)",
"Epoch 1/20\n93/93 [==============================] - 32s 325ms/step - loss: 0.8866 - accuracy: 0.6546 - val_loss: 0.5979 - val_accuracy: 0.7946\nEpoch 2/20\n93/93 [==============================] - 28s 297ms/step - loss: 0.7942 - accuracy: 0.6920 - val_loss: 0.5786 - val_accuracy: 0.7939\nEpoch 3/20\n93/93 [==============================] - 28s 300ms/step - loss: 0.7695 - accuracy: 0.7046 - val_loss: 0.6349 - val_accuracy: 0.7728\nEpoch 4/20\n93/93 [==============================] - 28s 302ms/step - loss: 0.7495 - accuracy: 0.7088 - val_loss: 0.6135 - val_accuracy: 0.7867\nEpoch 5/20\n93/93 [==============================] - 28s 296ms/step - loss: 0.7141 - accuracy: 0.7229 - val_loss: 0.5786 - val_accuracy: 0.7960\nEpoch 6/20\n93/93 [==============================] - 28s 296ms/step - loss: 0.7122 - accuracy: 0.7282 - val_loss: 0.7061 - val_accuracy: 0.7571\nEpoch 7/20\n93/93 [==============================] - 27s 295ms/step - loss: 0.6698 - accuracy: 0.7424 - val_loss: 0.6325 - val_accuracy: 0.7921\nEpoch 8/20\n93/93 [==============================] - 28s 296ms/step - loss: 0.6544 - accuracy: 0.7504 - val_loss: 0.6262 - val_accuracy: 0.7889\nEpoch 9/20\n93/93 [==============================] - 27s 294ms/step - loss: 0.6428 - accuracy: 0.7571 - val_loss: 0.5638 - val_accuracy: 0.8049\nEpoch 10/20\n93/93 [==============================] - 27s 295ms/step - loss: 0.6203 - accuracy: 0.7643 - val_loss: 0.5686 - val_accuracy: 0.7989\nEpoch 11/20\n93/93 [==============================] - 27s 294ms/step - loss: 0.6001 - accuracy: 0.7725 - val_loss: 0.5473 - val_accuracy: 0.8113\nEpoch 12/20\n93/93 [==============================] - 28s 296ms/step - loss: 0.5898 - accuracy: 0.7775 - val_loss: 0.7342 - val_accuracy: 0.7364\nEpoch 13/20\n93/93 [==============================] - 28s 296ms/step - loss: 0.5629 - accuracy: 0.7845 - val_loss: 0.5657 - val_accuracy: 0.8060\nEpoch 14/20\n93/93 [==============================] - 28s 298ms/step - loss: 0.5516 - accuracy: 0.7915 - val_loss: 0.5663 - val_accuracy: 0.8063\nEpoch 15/20\n93/93 [==============================] - 27s 295ms/step - loss: 0.5368 - accuracy: 0.7998 - val_loss: 0.6395 - val_accuracy: 0.7871\nEpoch 16/20\n93/93 [==============================] - 27s 295ms/step - loss: 0.5274 - accuracy: 0.8016 - val_loss: 0.6700 - val_accuracy: 0.7718\nEpoch 17/20\n93/93 [==============================] - 27s 295ms/step - loss: 0.5096 - accuracy: 0.8079 - val_loss: 0.6590 - val_accuracy: 0.7767\nEpoch 18/20\n93/93 [==============================] - 27s 295ms/step - loss: 0.5038 - accuracy: 0.8119 - val_loss: 0.6921 - val_accuracy: 0.7571\nEpoch 19/20\n93/93 [==============================] - 28s 297ms/step - loss: 0.4899 - accuracy: 0.8163 - val_loss: 0.5883 - val_accuracy: 0.7999\nEpoch 20/20\n93/93 [==============================] - 29s 317ms/step - loss: 0.4816 - accuracy: 0.8186 - val_loss: 0.7103 - val_accuracy: 0.7657\nDuration: 0:09:17.186845\n"
],
[
"n = 17\nmodels_lsa[n].fit_model(image_sets_lsa[n],label_sets_lsa[n],x_val,y_val,epochs=20,batch_size = 128)",
"Epoch 1/20\n99/99 [==============================] - 39s 363ms/step - loss: 0.8981 - accuracy: 0.6487 - val_loss: 0.7695 - val_accuracy: 0.7183\nEpoch 2/20\n99/99 [==============================] - 31s 314ms/step - loss: 0.8266 - accuracy: 0.6824 - val_loss: 0.7250 - val_accuracy: 0.7657\nEpoch 3/20\n99/99 [==============================] - 30s 303ms/step - loss: 0.7979 - accuracy: 0.6928 - val_loss: 0.8440 - val_accuracy: 0.7197\nEpoch 4/20\n99/99 [==============================] - 30s 303ms/step - loss: 0.7620 - accuracy: 0.7057 - val_loss: 0.6930 - val_accuracy: 0.7557\nEpoch 5/20\n99/99 [==============================] - 30s 301ms/step - loss: 0.7410 - accuracy: 0.7173 - val_loss: 0.6677 - val_accuracy: 0.7874\nEpoch 6/20\n99/99 [==============================] - 30s 306ms/step - loss: 0.7115 - accuracy: 0.7278 - val_loss: 0.7550 - val_accuracy: 0.7204\nEpoch 7/20\n99/99 [==============================] - 31s 313ms/step - loss: 0.6987 - accuracy: 0.7317 - val_loss: 0.7945 - val_accuracy: 0.7350\nEpoch 8/20\n99/99 [==============================] - 29s 295ms/step - loss: 0.6788 - accuracy: 0.7446 - val_loss: 0.7411 - val_accuracy: 0.7564\nEpoch 9/20\n99/99 [==============================] - 29s 294ms/step - loss: 0.6536 - accuracy: 0.7502 - val_loss: 0.6457 - val_accuracy: 0.7767\nEpoch 10/20\n99/99 [==============================] - 29s 292ms/step - loss: 0.6356 - accuracy: 0.7603 - val_loss: 0.8298 - val_accuracy: 0.7097\nEpoch 11/20\n99/99 [==============================] - 29s 294ms/step - loss: 0.6143 - accuracy: 0.7664 - val_loss: 0.6321 - val_accuracy: 0.7917\nEpoch 12/20\n99/99 [==============================] - 29s 295ms/step - loss: 0.6045 - accuracy: 0.7694 - val_loss: 0.6097 - val_accuracy: 0.7960\nEpoch 13/20\n99/99 [==============================] - 30s 305ms/step - loss: 0.5913 - accuracy: 0.7739 - val_loss: 0.5732 - val_accuracy: 0.8078\nEpoch 14/20\n99/99 [==============================] - 32s 322ms/step - loss: 0.5599 - accuracy: 0.7868 - val_loss: 0.7784 - val_accuracy: 0.7140\nEpoch 15/20\n99/99 [==============================] - 29s 295ms/step - loss: 0.5554 - accuracy: 0.7912 - val_loss: 0.6599 - val_accuracy: 0.7718\nEpoch 16/20\n99/99 [==============================] - 29s 292ms/step - loss: 0.5463 - accuracy: 0.7955 - val_loss: 0.6533 - val_accuracy: 0.7853\nEpoch 17/20\n99/99 [==============================] - 29s 294ms/step - loss: 0.5180 - accuracy: 0.8034 - val_loss: 0.7128 - val_accuracy: 0.7354\nEpoch 18/20\n99/99 [==============================] - 29s 293ms/step - loss: 0.5105 - accuracy: 0.8109 - val_loss: 0.6191 - val_accuracy: 0.7803\nEpoch 19/20\n99/99 [==============================] - 29s 298ms/step - loss: 0.4925 - accuracy: 0.8152 - val_loss: 0.7036 - val_accuracy: 0.7643\nEpoch 20/20\n99/99 [==============================] - 29s 293ms/step - loss: 0.4897 - accuracy: 0.8140 - val_loss: 0.7342 - val_accuracy: 0.7468\nDuration: 0:10:03.315815\n"
],
[
"n = 18\nmodels_lsa[n].fit_model(image_sets_lsa[n],label_sets_lsa[n],x_val,y_val,epochs=20,batch_size = 128)",
"Epoch 1/20\n104/104 [==============================] - 32s 297ms/step - loss: 0.8969 - accuracy: 0.6632 - val_loss: 0.6320 - val_accuracy: 0.7807\nEpoch 2/20\n104/104 [==============================] - 30s 291ms/step - loss: 0.8038 - accuracy: 0.6947 - val_loss: 0.6904 - val_accuracy: 0.7582\nEpoch 3/20\n104/104 [==============================] - 30s 293ms/step - loss: 0.7664 - accuracy: 0.7074 - val_loss: 0.5988 - val_accuracy: 0.7953\nEpoch 4/20\n104/104 [==============================] - 30s 292ms/step - loss: 0.7500 - accuracy: 0.7135 - val_loss: 0.5814 - val_accuracy: 0.7996\nEpoch 5/20\n104/104 [==============================] - 30s 292ms/step - loss: 0.7146 - accuracy: 0.7235 - val_loss: 0.5815 - val_accuracy: 0.7853\nEpoch 6/20\n104/104 [==============================] - 30s 292ms/step - loss: 0.7050 - accuracy: 0.7332 - val_loss: 0.5399 - val_accuracy: 0.8092\nEpoch 7/20\n104/104 [==============================] - 30s 292ms/step - loss: 0.6797 - accuracy: 0.7403 - val_loss: 0.6413 - val_accuracy: 0.7653\nEpoch 8/20\n104/104 [==============================] - 30s 293ms/step - loss: 0.6594 - accuracy: 0.7483 - val_loss: 0.5775 - val_accuracy: 0.7999\nEpoch 9/20\n104/104 [==============================] - 30s 291ms/step - loss: 0.6330 - accuracy: 0.7614 - val_loss: 0.5925 - val_accuracy: 0.7971\nEpoch 10/20\n104/104 [==============================] - 30s 292ms/step - loss: 0.6187 - accuracy: 0.7677 - val_loss: 0.5904 - val_accuracy: 0.7835\nEpoch 11/20\n104/104 [==============================] - 31s 297ms/step - loss: 0.5903 - accuracy: 0.7782 - val_loss: 0.6443 - val_accuracy: 0.7743\nEpoch 12/20\n104/104 [==============================] - 34s 332ms/step - loss: 0.5947 - accuracy: 0.7798 - val_loss: 0.5508 - val_accuracy: 0.8042\nEpoch 13/20\n104/104 [==============================] - 31s 295ms/step - loss: 0.5667 - accuracy: 0.7850 - val_loss: 0.5646 - val_accuracy: 0.8010\nEpoch 14/20\n104/104 [==============================] - 31s 294ms/step - loss: 0.5641 - accuracy: 0.7880 - val_loss: 0.6007 - val_accuracy: 0.7821\nEpoch 15/20\n104/104 [==============================] - 31s 295ms/step - loss: 0.5383 - accuracy: 0.7998 - val_loss: 0.5739 - val_accuracy: 0.8017\nEpoch 16/20\n104/104 [==============================] - 30s 293ms/step - loss: 0.5293 - accuracy: 0.7999 - val_loss: 0.5960 - val_accuracy: 0.7921\nEpoch 17/20\n104/104 [==============================] - 30s 292ms/step - loss: 0.5136 - accuracy: 0.8078 - val_loss: 0.5468 - val_accuracy: 0.8153\nEpoch 18/20\n104/104 [==============================] - 32s 307ms/step - loss: 0.4975 - accuracy: 0.8203 - val_loss: 0.5551 - val_accuracy: 0.8092\nEpoch 19/20\n104/104 [==============================] - 31s 294ms/step - loss: 0.4807 - accuracy: 0.8194 - val_loss: 0.5949 - val_accuracy: 0.7924\nEpoch 20/20\n104/104 [==============================] - 31s 295ms/step - loss: 0.4720 - accuracy: 0.8246 - val_loss: 0.7685 - val_accuracy: 0.7546\nDuration: 0:10:16.869981\n"
],
[
"n = 19\nmodels_lsa[n].fit_model(image_sets_lsa[n],label_sets_lsa[n],x_val,y_val,epochs=20,batch_size = 128)",
"Epoch 1/20\n112/112 [==============================] - 34s 290ms/step - loss: 0.8577 - accuracy: 0.6857 - val_loss: 0.5769 - val_accuracy: 0.7924\nEpoch 2/20\n112/112 [==============================] - 32s 289ms/step - loss: 0.7639 - accuracy: 0.7145 - val_loss: 0.6809 - val_accuracy: 0.7821\nEpoch 3/20\n112/112 [==============================] - 32s 289ms/step - loss: 0.7349 - accuracy: 0.7188 - val_loss: 1.0961 - val_accuracy: 0.5424\nEpoch 4/20\n112/112 [==============================] - 32s 289ms/step - loss: 0.7127 - accuracy: 0.7308 - val_loss: 0.7965 - val_accuracy: 0.7222\nEpoch 5/20\n112/112 [==============================] - 33s 295ms/step - loss: 0.6877 - accuracy: 0.7395 - val_loss: 0.6855 - val_accuracy: 0.7561\nEpoch 6/20\n112/112 [==============================] - 33s 290ms/step - loss: 0.6663 - accuracy: 0.7496 - val_loss: 1.1202 - val_accuracy: 0.5795\nEpoch 7/20\n112/112 [==============================] - 32s 288ms/step - loss: 0.6573 - accuracy: 0.7537 - val_loss: 0.7033 - val_accuracy: 0.7514\nEpoch 8/20\n112/112 [==============================] - 33s 295ms/step - loss: 0.6214 - accuracy: 0.7649 - val_loss: 0.6092 - val_accuracy: 0.7885\nEpoch 9/20\n112/112 [==============================] - 35s 308ms/step - loss: 0.6084 - accuracy: 0.7701 - val_loss: 0.7119 - val_accuracy: 0.7504\nEpoch 10/20\n112/112 [==============================] - 32s 290ms/step - loss: 0.5916 - accuracy: 0.7762 - val_loss: 0.6675 - val_accuracy: 0.7643\nEpoch 11/20\n112/112 [==============================] - 32s 289ms/step - loss: 0.5826 - accuracy: 0.7814 - val_loss: 0.6600 - val_accuracy: 0.7636\nEpoch 12/20\n112/112 [==============================] - 32s 289ms/step - loss: 0.5585 - accuracy: 0.7903 - val_loss: 0.7380 - val_accuracy: 0.7211\nEpoch 13/20\n112/112 [==============================] - 32s 289ms/step - loss: 0.5378 - accuracy: 0.8015 - val_loss: 0.6118 - val_accuracy: 0.8024\nEpoch 14/20\n112/112 [==============================] - 32s 289ms/step - loss: 0.5307 - accuracy: 0.8031 - val_loss: 0.8139 - val_accuracy: 0.7136\nEpoch 15/20\n112/112 [==============================] - 33s 291ms/step - loss: 0.5137 - accuracy: 0.8099 - val_loss: 0.6391 - val_accuracy: 0.7842\nEpoch 16/20\n112/112 [==============================] - 32s 290ms/step - loss: 0.5006 - accuracy: 0.8174 - val_loss: 0.5803 - val_accuracy: 0.7992\nEpoch 17/20\n112/112 [==============================] - 33s 293ms/step - loss: 0.4889 - accuracy: 0.8176 - val_loss: 0.6892 - val_accuracy: 0.7589\nEpoch 18/20\n112/112 [==============================] - 33s 295ms/step - loss: 0.4730 - accuracy: 0.8247 - val_loss: 0.6746 - val_accuracy: 0.7785\nEpoch 19/20\n112/112 [==============================] - 32s 290ms/step - loss: 0.4691 - accuracy: 0.8261 - val_loss: 0.5809 - val_accuracy: 0.8021\nEpoch 20/20\n112/112 [==============================] - 33s 292ms/step - loss: 0.4430 - accuracy: 0.8371 - val_loss: 0.6540 - val_accuracy: 0.7785\nDuration: 0:10:54.431207\n"
],
[
"loading = True\n\nmodels_lsa = []\n\nif loading:\n for i in range(20):\n model_lsa_dir = \"D:/models/intel_models/C2/intel_model_c2_sep_lsa_e2_\"+str(i)\n print(model_lsa_dir)\n model =utils.My_model('intel',True,model_lsa_dir)\n model.model.compile(loss= 'categorical_crossentropy', optimizer = 'rmsprop', metrics = ['accuracy',tf.keras.metrics.Precision(), tf.keras.metrics.Recall()])\n models_lsa.append(model)\n \n ",
"D:/models/intel_models/C2/intel_model_c2_sep_lsa_e2_0\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_lsa_e2_1\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_lsa_e2_2\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_lsa_e2_3\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_lsa_e2_4\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_lsa_e2_5\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_lsa_e2_6\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_lsa_e2_7\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_lsa_e2_8\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_lsa_e2_9\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_lsa_e2_10\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_lsa_e2_11\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_lsa_e2_12\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_lsa_e2_13\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_lsa_e2_14\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_lsa_e2_15\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_lsa_e2_16\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_lsa_e2_17\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_lsa_e2_18\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_lsa_e2_19\nModel loaded correctly\n"
]
],
[
[
"## Training guided by DSA",
"_____no_output_____"
]
],
[
[
"top_images_by_dsa_5000 = np.array(top_images_by_dsa[:5000])\ntop_labels_by_dsa_5000 = np.array(top_labels_by_dsa[:5000])\nmodel_dsa_5000 = utils.My_model(dataset,True,model_dir)\nmodel_dsa_5000.compile_model()\nmodel_dsa_5000.fit_model(top_images_by_dsa_5000,top_labels_by_dsa_5000,x_val,y_val)",
"Model loaded correctly\nModel compiled\nEpoch 1/10\n79/79 [==============================] - 16s 183ms/step - loss: 1.2097 - accuracy: 0.5704 - val_loss: 1.4648 - val_accuracy: 0.4975\nEpoch 2/10\n79/79 [==============================] - 13s 168ms/step - loss: 0.9588 - accuracy: 0.6459 - val_loss: 2.6739 - val_accuracy: 0.3627\nEpoch 3/10\n79/79 [==============================] - 14s 175ms/step - loss: 0.9534 - accuracy: 0.6621 - val_loss: 1.0395 - val_accuracy: 0.5977\nEpoch 4/10\n79/79 [==============================] - 14s 177ms/step - loss: 0.8596 - accuracy: 0.6855 - val_loss: 1.3329 - val_accuracy: 0.4971\nEpoch 5/10\n79/79 [==============================] - 14s 180ms/step - loss: 0.7888 - accuracy: 0.7085 - val_loss: 1.9018 - val_accuracy: 0.5178\nEpoch 6/10\n79/79 [==============================] - 15s 194ms/step - loss: 0.7790 - accuracy: 0.7204 - val_loss: 1.2383 - val_accuracy: 0.5788\nEpoch 7/10\n79/79 [==============================] - 15s 195ms/step - loss: 0.7200 - accuracy: 0.7398 - val_loss: 1.0920 - val_accuracy: 0.6195\nEpoch 8/10\n79/79 [==============================] - 15s 186ms/step - loss: 0.6766 - accuracy: 0.7547 - val_loss: 1.5629 - val_accuracy: 0.5032\nEpoch 9/10\n79/79 [==============================] - 16s 207ms/step - loss: 0.6354 - accuracy: 0.7698 - val_loss: 1.3361 - val_accuracy: 0.5895\nEpoch 10/10\n79/79 [==============================] - 14s 183ms/step - loss: 0.6132 - accuracy: 0.7894 - val_loss: 1.7874 - val_accuracy: 0.5164\nDuration: 0:02:27.422643\n"
],
[
"m = 700\nn = 0\nimage_sets_dsa = []\nlabel_sets_dsa = []\n\n\nfor i in range(len(top_images_by_dsa)//m):\n print(i,\":\")\n if (i+1 >= len(top_images_by_dsa)//m):\n print(\"Last\")\n print(0,\" -> \",n+m+(len(top_images_by_dsa)%m))\n top_images_by_dsa_n = np.array(top_images_by_dsa[:n+m+(len(top_images_by_dsa)%m)])\n top_labels_by_dsa_n = np.array(top_labels_by_dsa[:n+m+(len(top_images_by_dsa)%m)])\n else:\n print(0,\" -> \",m+n)\n top_images_by_dsa_n = np.array(top_images_by_dsa[:n+m])\n top_labels_by_dsa_n = np.array(top_labels_by_dsa[:n+m])\n image_sets_dsa.append(top_images_by_dsa_n)\n label_sets_dsa.append(top_labels_by_dsa_n)\n print(len(top_images_by_dsa_n))\n n += m\n\n",
"0 :\n0 -> 700\n700\n1 :\n0 -> 1400\n1400\n2 :\n0 -> 2100\n2100\n3 :\n0 -> 2800\n2800\n4 :\n0 -> 3500\n3500\n5 :\n0 -> 4200\n4200\n6 :\n0 -> 4900\n4900\n7 :\n0 -> 5600\n5600\n8 :\n0 -> 6300\n6300\n9 :\n0 -> 7000\n7000\n10 :\n0 -> 7700\n7700\n11 :\n0 -> 8400\n8400\n12 :\n0 -> 9100\n9100\n13 :\n0 -> 9800\n9800\n14 :\n0 -> 10500\n10500\n15 :\n0 -> 11200\n11200\n16 :\n0 -> 11900\n11900\n17 :\n0 -> 12600\n12600\n18 :\n0 -> 13300\n13300\n19 :\nLast\n0 -> 14224\n14224\n"
],
[
"print(model_dir)\n\nmodels_dsa = []\nfor i in range(len(label_sets_dsa)):\n print(i,\":\")\n model = utils.My_model('intel',True,model_dir)\n model.compile_model()\n models_dsa.append(model)\n",
"C:/Users/fjdur/Desktop/upc/project_notebooks/github_project/DL_notebooks/models/intel_model_21_10/\n0 :\nModel loaded correctly\nModel compiled\n1 :\nModel loaded correctly\nModel compiled\n2 :\nModel loaded correctly\nModel compiled\n3 :\nModel loaded correctly\nModel compiled\n4 :\nModel loaded correctly\nModel compiled\n5 :\nModel loaded correctly\nModel compiled\n6 :\nModel loaded correctly\nModel compiled\n7 :\nModel loaded correctly\nModel compiled\n8 :\nModel loaded correctly\nModel compiled\n9 :\nModel loaded correctly\nModel compiled\n10 :\nModel loaded correctly\nModel compiled\n11 :\nModel loaded correctly\nModel compiled\n12 :\nModel loaded correctly\nModel compiled\n13 :\nModel loaded correctly\nModel compiled\n14 :\nModel loaded correctly\nModel compiled\n15 :\nModel loaded correctly\nModel compiled\n16 :\nModel loaded correctly\nModel compiled\n17 :\nModel loaded correctly\nModel compiled\n18 :\nModel loaded correctly\nModel compiled\n19 :\nModel loaded correctly\nModel compiled\n"
],
[
"n=0",
"_____no_output_____"
],
[
"for i in range(7):\n print(i)\n models_dsa[i] = utils.My_model('intel',True,model_dir)\n models_dsa[i].compile_model()",
"0\nModel loaded correctly\nModel compiled\n1\nModel loaded correctly\nModel compiled\n2\nModel loaded correctly\nModel compiled\n3\nModel loaded correctly\nModel compiled\n4\nModel loaded correctly\nModel compiled\n5\nModel loaded correctly\nModel compiled\n6\nModel loaded correctly\nModel compiled\n"
],
[
"print(models_dsa[0].evaluate(x_test,y_test))\nprint(models_dsa[1].evaluate(x_test,y_test))",
"94/94 [==============================] - 2s 25ms/step - loss: 0.5392 - accuracy: 0.8037 - precision_21: 0.5778 - recall_21: 0.5040\n[0.5391883850097656, 0.8036666512489319, 0.5778048038482666, 0.5040168166160583]\n94/94 [==============================] - 2s 25ms/step - loss: 0.5392 - accuracy: 0.8037 - precision_22: 0.5778 - recall_22: 0.5040\n[0.5391883850097656, 0.8036666512489319, 0.5778048038482666, 0.5040168166160583]\n"
],
[
"n=0\nprint(n)\n\nmodels_dsa[n].fit_model(image_sets_dsa[n],label_sets_dsa[n],x_val,y_val,epochs=20,batch_size = 128)\nn = n+1",
"0\nEpoch 1/20\n6/6 [==============================] - 6s 739ms/step - loss: 1.8341 - accuracy: 0.3353 - val_loss: 0.9751 - val_accuracy: 0.6516\nEpoch 2/20\n6/6 [==============================] - 4s 665ms/step - loss: 1.3965 - accuracy: 0.4720 - val_loss: 0.8531 - val_accuracy: 0.6926\nEpoch 3/20\n6/6 [==============================] - 4s 638ms/step - loss: 1.4183 - accuracy: 0.5115 - val_loss: 0.7926 - val_accuracy: 0.7072\nEpoch 4/20\n6/6 [==============================] - 4s 674ms/step - loss: 1.1483 - accuracy: 0.5770 - val_loss: 0.9252 - val_accuracy: 0.6673\nEpoch 5/20\n6/6 [==============================] - 4s 656ms/step - loss: 1.1505 - accuracy: 0.5826 - val_loss: 0.8574 - val_accuracy: 0.6569\nEpoch 6/20\n6/6 [==============================] - 3s 629ms/step - loss: 1.0109 - accuracy: 0.6365 - val_loss: 0.9656 - val_accuracy: 0.6027\nEpoch 7/20\n6/6 [==============================] - 4s 665ms/step - loss: 1.2730 - accuracy: 0.5258 - val_loss: 0.8445 - val_accuracy: 0.6865\nEpoch 8/20\n6/6 [==============================] - 4s 661ms/step - loss: 0.9185 - accuracy: 0.6569 - val_loss: 0.9039 - val_accuracy: 0.6501\nEpoch 9/20\n6/6 [==============================] - 4s 677ms/step - loss: 0.9251 - accuracy: 0.6681 - val_loss: 0.9411 - val_accuracy: 0.6280\nEpoch 10/20\n6/6 [==============================] - 4s 663ms/step - loss: 0.9292 - accuracy: 0.6439 - val_loss: 0.8679 - val_accuracy: 0.6698\nEpoch 11/20\n6/6 [==============================] - 3s 631ms/step - loss: 0.9371 - accuracy: 0.6636 - val_loss: 0.8041 - val_accuracy: 0.7022\nEpoch 12/20\n6/6 [==============================] - 4s 685ms/step - loss: 0.8334 - accuracy: 0.6899 - val_loss: 0.9045 - val_accuracy: 0.6530\nEpoch 13/20\n6/6 [==============================] - 4s 678ms/step - loss: 0.7448 - accuracy: 0.7415 - val_loss: 0.8841 - val_accuracy: 0.6897\nEpoch 14/20\n6/6 [==============================] - 4s 658ms/step - loss: 0.7744 - accuracy: 0.7244 - val_loss: 0.7985 - val_accuracy: 0.7122\nEpoch 15/20\n6/6 [==============================] - 4s 717ms/step - loss: 0.6400 - accuracy: 0.7632 - val_loss: 0.8573 - val_accuracy: 0.6969\nEpoch 16/20\n6/6 [==============================] - 4s 707ms/step - loss: 0.7734 - accuracy: 0.7033 - val_loss: 1.0267 - val_accuracy: 0.5777\nEpoch 17/20\n6/6 [==============================] - 4s 737ms/step - loss: 0.7321 - accuracy: 0.7348 - val_loss: 0.8320 - val_accuracy: 0.6961\nEpoch 18/20\n6/6 [==============================] - 4s 762ms/step - loss: 0.6417 - accuracy: 0.7860 - val_loss: 0.9154 - val_accuracy: 0.6790\nEpoch 19/20\n6/6 [==============================] - 4s 680ms/step - loss: 0.6835 - accuracy: 0.7539 - val_loss: 1.0281 - val_accuracy: 0.6494\nEpoch 20/20\n6/6 [==============================] - 4s 730ms/step - loss: 0.6023 - accuracy: 0.7837 - val_loss: 1.0922 - val_accuracy: 0.6177\nDuration: 0:01:16.887977\n"
],
[
"print(n)\n\nmodels_dsa[n].fit_model(image_sets_dsa[n],label_sets_dsa[n],x_val,y_val,epochs=20,batch_size = 128)\nn = n+1",
"1\nEpoch 1/20\n11/11 [==============================] - 9s 610ms/step - loss: 1.8194 - accuracy: 0.3002 - val_loss: 0.9017 - val_accuracy: 0.7029\nEpoch 2/20\n11/11 [==============================] - 5s 473ms/step - loss: 1.2239 - accuracy: 0.5101 - val_loss: 0.8405 - val_accuracy: 0.7175\nEpoch 3/20\n11/11 [==============================] - 5s 480ms/step - loss: 1.1612 - accuracy: 0.5530 - val_loss: 0.9563 - val_accuracy: 0.6398\nEpoch 4/20\n11/11 [==============================] - 5s 478ms/step - loss: 1.2435 - accuracy: 0.5126 - val_loss: 0.8654 - val_accuracy: 0.6758\nEpoch 5/20\n11/11 [==============================] - 5s 476ms/step - loss: 1.0440 - accuracy: 0.5929 - val_loss: 0.8110 - val_accuracy: 0.6990\nEpoch 6/20\n11/11 [==============================] - 5s 494ms/step - loss: 1.0054 - accuracy: 0.6294 - val_loss: 0.8614 - val_accuracy: 0.6772\nEpoch 7/20\n11/11 [==============================] - 8s 735ms/step - loss: 0.9600 - accuracy: 0.6227 - val_loss: 0.8509 - val_accuracy: 0.6787\nEpoch 8/20\n11/11 [==============================] - 6s 522ms/step - loss: 0.8875 - accuracy: 0.6600 - val_loss: 0.8111 - val_accuracy: 0.7204\nEpoch 9/20\n11/11 [==============================] - 6s 607ms/step - loss: 0.8617 - accuracy: 0.6725 - val_loss: 0.8915 - val_accuracy: 0.6708\nEpoch 10/20\n11/11 [==============================] - 7s 626ms/step - loss: 0.8220 - accuracy: 0.6956 - val_loss: 0.7906 - val_accuracy: 0.7086\nEpoch 11/20\n11/11 [==============================] - 6s 543ms/step - loss: 0.8136 - accuracy: 0.6908 - val_loss: 0.8658 - val_accuracy: 0.6837\nEpoch 12/20\n11/11 [==============================] - 6s 546ms/step - loss: 0.8191 - accuracy: 0.6964 - val_loss: 0.9055 - val_accuracy: 0.6805\nEpoch 13/20\n11/11 [==============================] - 6s 532ms/step - loss: 0.7934 - accuracy: 0.7059 - val_loss: 0.8202 - val_accuracy: 0.7022\nEpoch 14/20\n11/11 [==============================] - 6s 552ms/step - loss: 0.6845 - accuracy: 0.7331 - val_loss: 0.9004 - val_accuracy: 0.6790\nEpoch 15/20\n11/11 [==============================] - 6s 573ms/step - loss: 0.7093 - accuracy: 0.7254 - val_loss: 0.8263 - val_accuracy: 0.6951\nEpoch 16/20\n11/11 [==============================] - 6s 547ms/step - loss: 0.6929 - accuracy: 0.7505 - val_loss: 0.8238 - val_accuracy: 0.7019\nEpoch 17/20\n11/11 [==============================] - 6s 545ms/step - loss: 0.5916 - accuracy: 0.7856 - val_loss: 0.8612 - val_accuracy: 0.6929\nEpoch 18/20\n11/11 [==============================] - 5s 482ms/step - loss: 0.6932 - accuracy: 0.7652 - val_loss: 0.8412 - val_accuracy: 0.6976\nEpoch 19/20\n11/11 [==============================] - 5s 477ms/step - loss: 0.5770 - accuracy: 0.7769 - val_loss: 0.8414 - val_accuracy: 0.6933\nEpoch 20/20\n11/11 [==============================] - 5s 488ms/step - loss: 0.5750 - accuracy: 0.7913 - val_loss: 0.8690 - val_accuracy: 0.7065\nDuration: 0:01:57.110130\n"
],
[
"print(n)\n\nmodels_dsa[n].fit_model(image_sets_dsa[n],label_sets_dsa[n],x_val,y_val,epochs=20,batch_size = 128)\nn = n+1",
"2\nEpoch 1/20\n17/17 [==============================] - 9s 412ms/step - loss: 2.0086 - accuracy: 0.2918 - val_loss: 0.9769 - val_accuracy: 0.6787\nEpoch 2/20\n17/17 [==============================] - 7s 390ms/step - loss: 1.2672 - accuracy: 0.4997 - val_loss: 0.9330 - val_accuracy: 0.6801\nEpoch 3/20\n17/17 [==============================] - 7s 403ms/step - loss: 1.1926 - accuracy: 0.5458 - val_loss: 1.0821 - val_accuracy: 0.5863\nEpoch 4/20\n17/17 [==============================] - 7s 412ms/step - loss: 1.1949 - accuracy: 0.5529 - val_loss: 0.8693 - val_accuracy: 0.6787\nEpoch 5/20\n17/17 [==============================] - 7s 396ms/step - loss: 0.9911 - accuracy: 0.6169 - val_loss: 1.0233 - val_accuracy: 0.6145\nEpoch 6/20\n17/17 [==============================] - 7s 398ms/step - loss: 1.0158 - accuracy: 0.6031 - val_loss: 0.8416 - val_accuracy: 0.6940\nEpoch 7/20\n17/17 [==============================] - 7s 388ms/step - loss: 1.0038 - accuracy: 0.6094 - val_loss: 0.8555 - val_accuracy: 0.6758\nEpoch 8/20\n17/17 [==============================] - 7s 398ms/step - loss: 0.8955 - accuracy: 0.6390 - val_loss: 0.9352 - val_accuracy: 0.6444\nEpoch 9/20\n17/17 [==============================] - 7s 401ms/step - loss: 0.8899 - accuracy: 0.6673 - val_loss: 1.0283 - val_accuracy: 0.6302\nEpoch 10/20\n17/17 [==============================] - 7s 394ms/step - loss: 0.9324 - accuracy: 0.6583 - val_loss: 0.9051 - val_accuracy: 0.6566\nEpoch 11/20\n17/17 [==============================] - 7s 393ms/step - loss: 0.7944 - accuracy: 0.6942 - val_loss: 0.8947 - val_accuracy: 0.6573\nEpoch 12/20\n17/17 [==============================] - 7s 405ms/step - loss: 0.8194 - accuracy: 0.6805 - val_loss: 0.8506 - val_accuracy: 0.6872\nEpoch 13/20\n17/17 [==============================] - 7s 401ms/step - loss: 0.7370 - accuracy: 0.7348 - val_loss: 1.0124 - val_accuracy: 0.6270\nEpoch 14/20\n17/17 [==============================] - 7s 394ms/step - loss: 0.7714 - accuracy: 0.7055 - val_loss: 0.8245 - val_accuracy: 0.6901\nEpoch 15/20\n17/17 [==============================] - 7s 403ms/step - loss: 0.7265 - accuracy: 0.7234 - val_loss: 0.9594 - val_accuracy: 0.6573\nEpoch 16/20\n17/17 [==============================] - 7s 429ms/step - loss: 0.6861 - accuracy: 0.7505 - val_loss: 0.9974 - val_accuracy: 0.6501\nEpoch 17/20\n17/17 [==============================] - 7s 430ms/step - loss: 0.6697 - accuracy: 0.7611 - val_loss: 0.9034 - val_accuracy: 0.6755\nEpoch 18/20\n17/17 [==============================] - 7s 423ms/step - loss: 0.5738 - accuracy: 0.7717 - val_loss: 0.8751 - val_accuracy: 0.6737\nEpoch 19/20\n17/17 [==============================] - 7s 405ms/step - loss: 0.5954 - accuracy: 0.7837 - val_loss: 0.9253 - val_accuracy: 0.6701\nEpoch 20/20\n17/17 [==============================] - 7s 398ms/step - loss: 0.5815 - accuracy: 0.7943 - val_loss: 0.9785 - val_accuracy: 0.6623\nDuration: 0:02:17.028100\n"
],
[
"print(n)\n\nmodels_dsa[n].fit_model(image_sets_dsa[n],label_sets_dsa[n],x_val,y_val,epochs=20,batch_size = 128)\nn = n+1",
"3\nEpoch 1/20\n22/22 [==============================] - 10s 405ms/step - loss: 1.7335 - accuracy: 0.3105 - val_loss: 0.9461 - val_accuracy: 0.6758\nEpoch 2/20\n22/22 [==============================] - 8s 381ms/step - loss: 1.2345 - accuracy: 0.4819 - val_loss: 0.9555 - val_accuracy: 0.6673\nEpoch 3/20\n22/22 [==============================] - 8s 386ms/step - loss: 1.1619 - accuracy: 0.5276 - val_loss: 0.8928 - val_accuracy: 0.6715\nEpoch 4/20\n22/22 [==============================] - 9s 403ms/step - loss: 1.1328 - accuracy: 0.5508 - val_loss: 0.9218 - val_accuracy: 0.6487\nEpoch 5/20\n22/22 [==============================] - 9s 428ms/step - loss: 1.0466 - accuracy: 0.5857 - val_loss: 0.8869 - val_accuracy: 0.6619\nEpoch 6/20\n22/22 [==============================] - 8s 386ms/step - loss: 0.9508 - accuracy: 0.6252 - val_loss: 0.8624 - val_accuracy: 0.6901\nEpoch 7/20\n22/22 [==============================] - 8s 386ms/step - loss: 0.9559 - accuracy: 0.6215 - val_loss: 0.9122 - val_accuracy: 0.6769\nEpoch 8/20\n22/22 [==============================] - 9s 406ms/step - loss: 0.9360 - accuracy: 0.6322 - val_loss: 0.9013 - val_accuracy: 0.6598\nEpoch 9/20\n22/22 [==============================] - 8s 380ms/step - loss: 0.8595 - accuracy: 0.6696 - val_loss: 0.8827 - val_accuracy: 0.6601\nEpoch 10/20\n22/22 [==============================] - 8s 387ms/step - loss: 0.8431 - accuracy: 0.6746 - val_loss: 0.8460 - val_accuracy: 0.6912\nEpoch 11/20\n22/22 [==============================] - 9s 412ms/step - loss: 0.8225 - accuracy: 0.6880 - val_loss: 0.9007 - val_accuracy: 0.6412\nEpoch 12/20\n22/22 [==============================] - 9s 393ms/step - loss: 0.8293 - accuracy: 0.6820 - val_loss: 0.9221 - val_accuracy: 0.6580\nEpoch 13/20\n22/22 [==============================] - 8s 388ms/step - loss: 0.7603 - accuracy: 0.7225 - val_loss: 1.0540 - val_accuracy: 0.5909\nEpoch 14/20\n22/22 [==============================] - 9s 392ms/step - loss: 0.7515 - accuracy: 0.7180 - val_loss: 0.9530 - val_accuracy: 0.6284\nEpoch 15/20\n22/22 [==============================] - 8s 385ms/step - loss: 0.6536 - accuracy: 0.7557 - val_loss: 1.0044 - val_accuracy: 0.6227\nEpoch 16/20\n22/22 [==============================] - 8s 384ms/step - loss: 0.6150 - accuracy: 0.7746 - val_loss: 1.1464 - val_accuracy: 0.5799\nEpoch 17/20\n22/22 [==============================] - 9s 395ms/step - loss: 0.5872 - accuracy: 0.7954 - val_loss: 1.2692 - val_accuracy: 0.5485\nEpoch 18/20\n22/22 [==============================] - 9s 406ms/step - loss: 0.6588 - accuracy: 0.7715 - val_loss: 1.1609 - val_accuracy: 0.5792\nEpoch 19/20\n22/22 [==============================] - 9s 395ms/step - loss: 0.5457 - accuracy: 0.7938 - val_loss: 1.3381 - val_accuracy: 0.5606\nEpoch 20/20\n22/22 [==============================] - 9s 405ms/step - loss: 0.5105 - accuracy: 0.8126 - val_loss: 1.3212 - val_accuracy: 0.5360\nDuration: 0:02:53.924100\n"
],
[
"print(n)#4\n\nmodels_dsa[n].fit_model(image_sets_dsa[n],label_sets_dsa[n],x_val,y_val,epochs=20,batch_size = 128)\nn = n+1",
"4\nEpoch 1/20\n28/28 [==============================] - 12s 386ms/step - loss: 1.6714 - accuracy: 0.3680 - val_loss: 1.2927 - val_accuracy: 0.5160\nEpoch 2/20\n28/28 [==============================] - 10s 361ms/step - loss: 1.2143 - accuracy: 0.5088 - val_loss: 1.1113 - val_accuracy: 0.5506\nEpoch 3/20\n28/28 [==============================] - 11s 382ms/step - loss: 1.1633 - accuracy: 0.5645 - val_loss: 1.2252 - val_accuracy: 0.5210\nEpoch 4/20\n28/28 [==============================] - 10s 364ms/step - loss: 1.0421 - accuracy: 0.5703 - val_loss: 1.0424 - val_accuracy: 0.5642\nEpoch 5/20\n28/28 [==============================] - 10s 371ms/step - loss: 1.0320 - accuracy: 0.5956 - val_loss: 0.9483 - val_accuracy: 0.6373\nEpoch 6/20\n28/28 [==============================] - 11s 383ms/step - loss: 0.9840 - accuracy: 0.6240 - val_loss: 2.0712 - val_accuracy: 0.5118\nEpoch 7/20\n28/28 [==============================] - 11s 392ms/step - loss: 0.9702 - accuracy: 0.6242 - val_loss: 1.3188 - val_accuracy: 0.4936\nEpoch 8/20\n28/28 [==============================] - 11s 385ms/step - loss: 0.9481 - accuracy: 0.6356 - val_loss: 2.1468 - val_accuracy: 0.5460\nEpoch 9/20\n28/28 [==============================] - 10s 370ms/step - loss: 0.8849 - accuracy: 0.6680 - val_loss: 1.3288 - val_accuracy: 0.5849\nEpoch 10/20\n28/28 [==============================] - 11s 384ms/step - loss: 0.8105 - accuracy: 0.6938 - val_loss: 3.4350 - val_accuracy: 0.4868\nEpoch 11/20\n28/28 [==============================] - 10s 369ms/step - loss: 0.7748 - accuracy: 0.7097 - val_loss: 2.1538 - val_accuracy: 0.5267\nEpoch 12/20\n28/28 [==============================] - 11s 379ms/step - loss: 0.7545 - accuracy: 0.7292 - val_loss: 2.4403 - val_accuracy: 0.5004\nEpoch 13/20\n28/28 [==============================] - 10s 360ms/step - loss: 0.6967 - accuracy: 0.7388 - val_loss: 2.5034 - val_accuracy: 0.4829\nEpoch 14/20\n28/28 [==============================] - 10s 362ms/step - loss: 0.6377 - accuracy: 0.7667 - val_loss: 2.6990 - val_accuracy: 0.4772\nEpoch 15/20\n28/28 [==============================] - 10s 363ms/step - loss: 0.5980 - accuracy: 0.7834 - val_loss: 3.1646 - val_accuracy: 0.4779\nEpoch 16/20\n28/28 [==============================] - 10s 369ms/step - loss: 0.5928 - accuracy: 0.7814 - val_loss: 3.5012 - val_accuracy: 0.5118\nEpoch 17/20\n28/28 [==============================] - 10s 355ms/step - loss: 0.5713 - accuracy: 0.7864 - val_loss: 1.6059 - val_accuracy: 0.5432\nEpoch 18/20\n28/28 [==============================] - 10s 363ms/step - loss: 0.5441 - accuracy: 0.8055 - val_loss: 4.0505 - val_accuracy: 0.4643\nEpoch 19/20\n28/28 [==============================] - 10s 364ms/step - loss: 0.4674 - accuracy: 0.8319 - val_loss: 2.5596 - val_accuracy: 0.5378\nEpoch 20/20\n28/28 [==============================] - 10s 365ms/step - loss: 0.4841 - accuracy: 0.8270 - val_loss: 1.3531 - val_accuracy: 0.5014\nDuration: 0:03:28.592317\n"
],
[
"print(n)\n\nmodels_dsa[n].fit_model(image_sets_dsa[n],label_sets_dsa[n],x_val,y_val,epochs=20,batch_size = 128)\nn = n+1",
"5\nEpoch 1/20\n33/33 [==============================] - 15s 389ms/step - loss: 1.4727 - accuracy: 0.4627 - val_loss: 1.1130 - val_accuracy: 0.5456\nEpoch 2/20\n33/33 [==============================] - 12s 370ms/step - loss: 1.0792 - accuracy: 0.5815 - val_loss: 1.0811 - val_accuracy: 0.5770\nEpoch 3/20\n33/33 [==============================] - 14s 413ms/step - loss: 1.0184 - accuracy: 0.6002 - val_loss: 1.6744 - val_accuracy: 0.5200\nEpoch 4/20\n33/33 [==============================] - 13s 394ms/step - loss: 0.9497 - accuracy: 0.6292 - val_loss: 1.3306 - val_accuracy: 0.5292\nEpoch 5/20\n33/33 [==============================] - 13s 393ms/step - loss: 0.8901 - accuracy: 0.6738 - val_loss: 1.3546 - val_accuracy: 0.5728\nEpoch 6/20\n33/33 [==============================] - 13s 408ms/step - loss: 0.8357 - accuracy: 0.6902 - val_loss: 1.8005 - val_accuracy: 0.5310\nEpoch 7/20\n33/33 [==============================] - 14s 415ms/step - loss: 0.8049 - accuracy: 0.6932 - val_loss: 1.1787 - val_accuracy: 0.5934\nEpoch 8/20\n33/33 [==============================] - 13s 397ms/step - loss: 0.7679 - accuracy: 0.7195 - val_loss: 2.4720 - val_accuracy: 0.4932\nEpoch 9/20\n33/33 [==============================] - 12s 377ms/step - loss: 0.7700 - accuracy: 0.7223 - val_loss: 1.6558 - val_accuracy: 0.5346\nEpoch 10/20\n33/33 [==============================] - 12s 362ms/step - loss: 0.6732 - accuracy: 0.7563 - val_loss: 1.4414 - val_accuracy: 0.5670\nEpoch 11/20\n33/33 [==============================] - 12s 367ms/step - loss: 0.6581 - accuracy: 0.7579 - val_loss: 1.8973 - val_accuracy: 0.5296\nEpoch 12/20\n33/33 [==============================] - 12s 367ms/step - loss: 0.5977 - accuracy: 0.7851 - val_loss: 2.5465 - val_accuracy: 0.4907\nEpoch 13/20\n33/33 [==============================] - 12s 361ms/step - loss: 0.6074 - accuracy: 0.7872 - val_loss: 2.9372 - val_accuracy: 0.4975\nEpoch 14/20\n33/33 [==============================] - 12s 369ms/step - loss: 0.5845 - accuracy: 0.7911 - val_loss: 4.1378 - val_accuracy: 0.4173\nEpoch 15/20\n33/33 [==============================] - 12s 372ms/step - loss: 0.5431 - accuracy: 0.8113 - val_loss: 3.0619 - val_accuracy: 0.5200\nEpoch 16/20\n33/33 [==============================] - 14s 435ms/step - loss: 0.5193 - accuracy: 0.8175 - val_loss: 1.9752 - val_accuracy: 0.5471\nEpoch 17/20\n33/33 [==============================] - 14s 428ms/step - loss: 0.4760 - accuracy: 0.8266 - val_loss: 2.2246 - val_accuracy: 0.5367\nEpoch 18/20\n33/33 [==============================] - 13s 406ms/step - loss: 0.4429 - accuracy: 0.8450 - val_loss: 3.9415 - val_accuracy: 0.4519\nEpoch 19/20\n33/33 [==============================] - 13s 402ms/step - loss: 0.4359 - accuracy: 0.8495 - val_loss: 3.6743 - val_accuracy: 0.4843\nEpoch 20/20\n33/33 [==============================] - 14s 414ms/step - loss: 0.4046 - accuracy: 0.8537 - val_loss: 4.8601 - val_accuracy: 0.4497\nDuration: 0:04:19.634906\n"
],
[
"print(n)\n\nmodels_dsa[n].fit_model(image_sets_dsa[n],label_sets_dsa[n],x_val,y_val,epochs=20,batch_size = 128)\nn = n+1",
"6\nEpoch 1/20\n39/39 [==============================] - 17s 385ms/step - loss: 1.4967 - accuracy: 0.5053 - val_loss: 1.0042 - val_accuracy: 0.5617\nEpoch 2/20\n39/39 [==============================] - 15s 386ms/step - loss: 0.9886 - accuracy: 0.6371 - val_loss: 1.1294 - val_accuracy: 0.5674\nEpoch 3/20\n39/39 [==============================] - 14s 369ms/step - loss: 0.9068 - accuracy: 0.6551 - val_loss: 1.3311 - val_accuracy: 0.5328\nEpoch 4/20\n39/39 [==============================] - 15s 388ms/step - loss: 0.8781 - accuracy: 0.6723 - val_loss: 1.2314 - val_accuracy: 0.5631\nEpoch 5/20\n39/39 [==============================] - 15s 388ms/step - loss: 0.8109 - accuracy: 0.6923 - val_loss: 1.0422 - val_accuracy: 0.5556\nEpoch 6/20\n39/39 [==============================] - 16s 400ms/step - loss: 0.7864 - accuracy: 0.7136 - val_loss: 1.7174 - val_accuracy: 0.5018\nEpoch 7/20\n39/39 [==============================] - 14s 367ms/step - loss: 0.7551 - accuracy: 0.7109 - val_loss: 2.2118 - val_accuracy: 0.5235\nEpoch 8/20\n39/39 [==============================] - 14s 353ms/step - loss: 0.7116 - accuracy: 0.7309 - val_loss: 2.4543 - val_accuracy: 0.4522\nEpoch 9/20\n39/39 [==============================] - 14s 351ms/step - loss: 0.6979 - accuracy: 0.7359 - val_loss: 2.1299 - val_accuracy: 0.4957\nEpoch 10/20\n39/39 [==============================] - 13s 341ms/step - loss: 0.6354 - accuracy: 0.7679 - val_loss: 2.3614 - val_accuracy: 0.4932\nEpoch 11/20\n39/39 [==============================] - 15s 391ms/step - loss: 0.6400 - accuracy: 0.7739 - val_loss: 1.1451 - val_accuracy: 0.5959\nEpoch 12/20\n39/39 [==============================] - 15s 396ms/step - loss: 0.5907 - accuracy: 0.7941 - val_loss: 2.2039 - val_accuracy: 0.4711\nEpoch 13/20\n39/39 [==============================] - 16s 415ms/step - loss: 0.5326 - accuracy: 0.8075 - val_loss: 2.4222 - val_accuracy: 0.4576\nEpoch 14/20\n39/39 [==============================] - 15s 385ms/step - loss: 0.6641 - accuracy: 0.7653 - val_loss: 1.0891 - val_accuracy: 0.6209\nEpoch 15/20\n39/39 [==============================] - 15s 378ms/step - loss: 0.4983 - accuracy: 0.8175 - val_loss: 4.3358 - val_accuracy: 0.4536\nEpoch 16/20\n39/39 [==============================] - 14s 367ms/step - loss: 0.5225 - accuracy: 0.8260 - val_loss: 1.3370 - val_accuracy: 0.5571\nEpoch 17/20\n39/39 [==============================] - 16s 402ms/step - loss: 0.4549 - accuracy: 0.8329 - val_loss: 2.9284 - val_accuracy: 0.4729\nEpoch 18/20\n39/39 [==============================] - 14s 366ms/step - loss: 0.4663 - accuracy: 0.8372 - val_loss: 3.9989 - val_accuracy: 0.4558\nEpoch 19/20\n39/39 [==============================] - 13s 347ms/step - loss: 0.3890 - accuracy: 0.8711 - val_loss: 3.6470 - val_accuracy: 0.4961\nEpoch 20/20\n39/39 [==============================] - 15s 379ms/step - loss: 0.4240 - accuracy: 0.8478 - val_loss: 3.0149 - val_accuracy: 0.4807\nDuration: 0:04:55.783409\n"
],
[
"for i in range(7):\n print(i)",
"0\n1\n2\n3\n4\n5\n6\n"
],
[
"loading = True\n\nmodels_dsa = []\n\nif loading:\n for i in range(7):\n model_dsa_dir = \"D:/models/intel/C3/intel_model_c3_sep_dsa_e2_\"+str(i)\n print(model_dsa_dir)\n model =utils.My_model('intel',True,model_dsa_dir)\n model.model.compile(loss= 'categorical_crossentropy', optimizer = 'rmsprop', metrics = ['accuracy',tf.keras.metrics.Precision(), tf.keras.metrics.Recall()])\n models_lsa.append(model)\n ",
"_____no_output_____"
],
[
"n=7\nprint(n)\n\nmodels_dsa[n].fit_model(image_sets_dsa[n],label_sets_dsa[n],x_val,y_val,epochs=20,batch_size = 128)\nn = n+1",
"7\nEpoch 1/20\n44/44 [==============================] - 20s 323ms/step - loss: 1.3139 - accuracy: 0.5490 - val_loss: 1.0318 - val_accuracy: 0.6352\nEpoch 2/20\n44/44 [==============================] - 13s 300ms/step - loss: 0.9326 - accuracy: 0.6607 - val_loss: 1.5681 - val_accuracy: 0.4904\nEpoch 3/20\n44/44 [==============================] - 13s 297ms/step - loss: 0.8626 - accuracy: 0.6820 - val_loss: 1.1357 - val_accuracy: 0.5642\nEpoch 4/20\n44/44 [==============================] - 13s 300ms/step - loss: 0.8330 - accuracy: 0.6876 - val_loss: 1.1728 - val_accuracy: 0.5720\nEpoch 5/20\n44/44 [==============================] - 13s 296ms/step - loss: 0.7329 - accuracy: 0.7380 - val_loss: 1.0324 - val_accuracy: 0.5892\nEpoch 6/20\n44/44 [==============================] - 13s 295ms/step - loss: 0.7097 - accuracy: 0.7382 - val_loss: 1.5976 - val_accuracy: 0.5221\nEpoch 7/20\n44/44 [==============================] - 13s 296ms/step - loss: 0.6782 - accuracy: 0.7512 - val_loss: 1.1679 - val_accuracy: 0.5756\nEpoch 8/20\n44/44 [==============================] - 13s 299ms/step - loss: 0.6536 - accuracy: 0.7551 - val_loss: 2.3692 - val_accuracy: 0.4943\nEpoch 9/20\n44/44 [==============================] - 13s 301ms/step - loss: 0.5884 - accuracy: 0.7893 - val_loss: 2.5407 - val_accuracy: 0.4344\nEpoch 10/20\n44/44 [==============================] - 13s 299ms/step - loss: 0.6002 - accuracy: 0.7775 - val_loss: 2.2317 - val_accuracy: 0.4750\nEpoch 11/20\n44/44 [==============================] - 13s 303ms/step - loss: 0.5723 - accuracy: 0.7932 - val_loss: 1.6865 - val_accuracy: 0.5332\nEpoch 12/20\n44/44 [==============================] - 14s 312ms/step - loss: 0.5212 - accuracy: 0.8152 - val_loss: 1.4755 - val_accuracy: 0.5414\nEpoch 13/20\n44/44 [==============================] - 14s 313ms/step - loss: 0.5050 - accuracy: 0.8224 - val_loss: 1.7552 - val_accuracy: 0.5296\nEpoch 14/20\n44/44 [==============================] - 16s 367ms/step - loss: 0.4372 - accuracy: 0.8447 - val_loss: 1.4731 - val_accuracy: 0.5863\nEpoch 15/20\n44/44 [==============================] - 14s 319ms/step - loss: 0.4556 - accuracy: 0.8379 - val_loss: 1.8662 - val_accuracy: 0.5114\nEpoch 16/20\n44/44 [==============================] - 14s 312ms/step - loss: 0.4750 - accuracy: 0.8363 - val_loss: 2.6916 - val_accuracy: 0.4879\nEpoch 17/20\n44/44 [==============================] - 13s 301ms/step - loss: 0.3990 - accuracy: 0.8574 - val_loss: 1.2868 - val_accuracy: 0.6066\nEpoch 18/20\n44/44 [==============================] - 13s 302ms/step - loss: 0.3973 - accuracy: 0.8672 - val_loss: 1.0835 - val_accuracy: 0.5892\nEpoch 19/20\n44/44 [==============================] - 13s 299ms/step - loss: 0.3987 - accuracy: 0.8612 - val_loss: 1.8301 - val_accuracy: 0.5182\nEpoch 20/20\n44/44 [==============================] - 13s 298ms/step - loss: 0.3392 - accuracy: 0.8876 - val_loss: 2.1462 - val_accuracy: 0.4925\nDuration: 0:04:35.402243\n"
],
[
"print(n)\n\nmodels_dsa[n].fit_model(image_sets_dsa[n],label_sets_dsa[n],x_val,y_val,epochs=20,batch_size = 128)\nn = n+1",
"8\nEpoch 1/20\n50/50 [==============================] - 16s 296ms/step - loss: 1.1786 - accuracy: 0.6038 - val_loss: 0.9140 - val_accuracy: 0.6680\nEpoch 2/20\n50/50 [==============================] - 15s 292ms/step - loss: 0.8841 - accuracy: 0.6683 - val_loss: 1.8508 - val_accuracy: 0.5057\nEpoch 3/20\n50/50 [==============================] - 15s 291ms/step - loss: 0.8622 - accuracy: 0.6847 - val_loss: 1.2368 - val_accuracy: 0.5767\nEpoch 4/20\n50/50 [==============================] - 15s 299ms/step - loss: 0.7787 - accuracy: 0.7153 - val_loss: 1.5844 - val_accuracy: 0.4964\nEpoch 5/20\n50/50 [==============================] - 15s 293ms/step - loss: 0.7483 - accuracy: 0.7227 - val_loss: 1.2816 - val_accuracy: 0.5806\nEpoch 6/20\n50/50 [==============================] - 15s 292ms/step - loss: 0.7209 - accuracy: 0.7277 - val_loss: 1.4454 - val_accuracy: 0.5089\nEpoch 7/20\n50/50 [==============================] - 15s 291ms/step - loss: 0.6938 - accuracy: 0.7507 - val_loss: 2.0445 - val_accuracy: 0.5196\nEpoch 8/20\n50/50 [==============================] - 15s 296ms/step - loss: 0.7009 - accuracy: 0.7518 - val_loss: 1.5727 - val_accuracy: 0.5489\nEpoch 9/20\n50/50 [==============================] - 15s 293ms/step - loss: 0.6169 - accuracy: 0.7715 - val_loss: 1.8273 - val_accuracy: 0.5239\nEpoch 10/20\n50/50 [==============================] - 15s 294ms/step - loss: 0.6070 - accuracy: 0.7795 - val_loss: 1.3299 - val_accuracy: 0.5695\nEpoch 11/20\n50/50 [==============================] - 15s 292ms/step - loss: 0.5581 - accuracy: 0.8013 - val_loss: 1.2393 - val_accuracy: 0.5999\nEpoch 12/20\n50/50 [==============================] - 15s 292ms/step - loss: 0.5287 - accuracy: 0.8106 - val_loss: 1.3451 - val_accuracy: 0.5585\nEpoch 13/20\n50/50 [==============================] - 15s 297ms/step - loss: 0.4894 - accuracy: 0.8224 - val_loss: 2.2404 - val_accuracy: 0.5303\nEpoch 14/20\n50/50 [==============================] - 15s 293ms/step - loss: 0.5212 - accuracy: 0.8134 - val_loss: 1.1923 - val_accuracy: 0.6198\nEpoch 15/20\n50/50 [==============================] - 15s 294ms/step - loss: 0.4717 - accuracy: 0.8307 - val_loss: 1.7635 - val_accuracy: 0.6113\nEpoch 16/20\n50/50 [==============================] - 15s 293ms/step - loss: 0.4531 - accuracy: 0.8445 - val_loss: 2.1762 - val_accuracy: 0.5246\nEpoch 17/20\n50/50 [==============================] - 15s 293ms/step - loss: 0.4621 - accuracy: 0.8413 - val_loss: 1.3012 - val_accuracy: 0.5884\nEpoch 18/20\n50/50 [==============================] - 15s 297ms/step - loss: 0.4499 - accuracy: 0.8394 - val_loss: 2.4453 - val_accuracy: 0.5432\nEpoch 19/20\n50/50 [==============================] - 15s 292ms/step - loss: 0.4218 - accuracy: 0.8549 - val_loss: 1.3633 - val_accuracy: 0.5674\nEpoch 20/20\n50/50 [==============================] - 15s 311ms/step - loss: 0.4215 - accuracy: 0.8504 - val_loss: 2.1619 - val_accuracy: 0.5350\nDuration: 0:04:55.624787\n"
],
[
"print(n)#9\n\nmodels_dsa[n].fit_model(image_sets_dsa[n],label_sets_dsa[n],x_val,y_val,epochs=20,batch_size = 128)\nn = n+1",
"9\nEpoch 1/20\n55/55 [==============================] - 18s 299ms/step - loss: 1.2194 - accuracy: 0.5793 - val_loss: 1.4030 - val_accuracy: 0.5432\nEpoch 2/20\n55/55 [==============================] - 16s 292ms/step - loss: 0.8515 - accuracy: 0.6864 - val_loss: 1.7573 - val_accuracy: 0.5556\nEpoch 3/20\n55/55 [==============================] - 16s 285ms/step - loss: 0.7792 - accuracy: 0.7096 - val_loss: 0.9715 - val_accuracy: 0.6273\nEpoch 4/20\n55/55 [==============================] - 16s 291ms/step - loss: 0.7214 - accuracy: 0.7310 - val_loss: 1.4171 - val_accuracy: 0.5724\nEpoch 5/20\n55/55 [==============================] - 16s 298ms/step - loss: 0.7025 - accuracy: 0.7310 - val_loss: 1.3097 - val_accuracy: 0.5610\nEpoch 6/20\n55/55 [==============================] - 16s 288ms/step - loss: 0.7078 - accuracy: 0.7384 - val_loss: 0.9445 - val_accuracy: 0.6551\nEpoch 7/20\n55/55 [==============================] - 17s 305ms/step - loss: 0.6242 - accuracy: 0.7658 - val_loss: 1.0727 - val_accuracy: 0.6134\nEpoch 8/20\n55/55 [==============================] - 16s 295ms/step - loss: 0.6252 - accuracy: 0.7750 - val_loss: 1.3155 - val_accuracy: 0.5752\nEpoch 9/20\n55/55 [==============================] - 17s 311ms/step - loss: 0.5633 - accuracy: 0.8001 - val_loss: 1.4355 - val_accuracy: 0.5852\nEpoch 10/20\n55/55 [==============================] - 16s 284ms/step - loss: 0.5542 - accuracy: 0.7981 - val_loss: 1.2442 - val_accuracy: 0.5874\nEpoch 11/20\n55/55 [==============================] - 16s 283ms/step - loss: 0.5282 - accuracy: 0.8133 - val_loss: 1.4388 - val_accuracy: 0.5439\nEpoch 12/20\n55/55 [==============================] - 16s 285ms/step - loss: 0.5081 - accuracy: 0.8131 - val_loss: 1.6776 - val_accuracy: 0.5510\nEpoch 13/20\n55/55 [==============================] - 16s 289ms/step - loss: 0.4916 - accuracy: 0.8289 - val_loss: 1.6443 - val_accuracy: 0.5353\nEpoch 14/20\n55/55 [==============================] - 18s 322ms/step - loss: 0.4579 - accuracy: 0.8280 - val_loss: 2.2269 - val_accuracy: 0.5499\nEpoch 15/20\n55/55 [==============================] - 18s 325ms/step - loss: 0.4349 - accuracy: 0.8452 - val_loss: 2.0284 - val_accuracy: 0.5432\nEpoch 16/20\n55/55 [==============================] - 17s 316ms/step - loss: 0.4185 - accuracy: 0.8498 - val_loss: 2.3254 - val_accuracy: 0.5496\nEpoch 17/20\n55/55 [==============================] - 17s 314ms/step - loss: 0.4834 - accuracy: 0.8341 - val_loss: 1.7106 - val_accuracy: 0.5781\nEpoch 18/20\n55/55 [==============================] - 17s 302ms/step - loss: 0.3868 - accuracy: 0.8710 - val_loss: 1.6543 - val_accuracy: 0.5653\nEpoch 19/20\n55/55 [==============================] - 17s 316ms/step - loss: 0.3853 - accuracy: 0.8632 - val_loss: 1.5551 - val_accuracy: 0.5489\nEpoch 20/20\n55/55 [==============================] - 18s 325ms/step - loss: 0.3248 - accuracy: 0.8818 - val_loss: 1.0686 - val_accuracy: 0.6031\nDuration: 0:05:32.412901\n"
],
[
"print(n)\n\nmodels_dsa[n].fit_model(image_sets_dsa[n],label_sets_dsa[n],x_val,y_val,epochs=20,batch_size = 128)\nn = n+1",
"10\nEpoch 1/20\n61/61 [==============================] - 22s 336ms/step - loss: 1.1040 - accuracy: 0.6195 - val_loss: 1.5230 - val_accuracy: 0.5235\nEpoch 2/20\n61/61 [==============================] - 18s 298ms/step - loss: 0.8082 - accuracy: 0.7131 - val_loss: 1.2927 - val_accuracy: 0.5428\nEpoch 3/20\n61/61 [==============================] - 20s 332ms/step - loss: 0.7644 - accuracy: 0.7204 - val_loss: 1.2494 - val_accuracy: 0.5770\nEpoch 4/20\n61/61 [==============================] - 21s 350ms/step - loss: 0.7030 - accuracy: 0.7447 - val_loss: 1.1273 - val_accuracy: 0.5959\nEpoch 5/20\n61/61 [==============================] - 20s 329ms/step - loss: 0.6739 - accuracy: 0.7635 - val_loss: 1.7665 - val_accuracy: 0.5399\nEpoch 6/20\n61/61 [==============================] - 19s 319ms/step - loss: 0.6408 - accuracy: 0.7653 - val_loss: 1.7034 - val_accuracy: 0.5321\nEpoch 7/20\n61/61 [==============================] - 18s 290ms/step - loss: 0.5969 - accuracy: 0.7843 - val_loss: 1.7040 - val_accuracy: 0.4861\nEpoch 8/20\n61/61 [==============================] - 18s 288ms/step - loss: 0.5917 - accuracy: 0.7872 - val_loss: 2.3044 - val_accuracy: 0.4929\nEpoch 9/20\n61/61 [==============================] - 17s 284ms/step - loss: 0.5530 - accuracy: 0.8022 - val_loss: 1.8371 - val_accuracy: 0.5168\nEpoch 10/20\n61/61 [==============================] - 17s 282ms/step - loss: 0.5501 - accuracy: 0.8024 - val_loss: 1.5767 - val_accuracy: 0.5514\nEpoch 11/20\n61/61 [==============================] - 18s 290ms/step - loss: 0.4994 - accuracy: 0.8234 - val_loss: 1.7483 - val_accuracy: 0.5503\nEpoch 12/20\n61/61 [==============================] - 18s 299ms/step - loss: 0.4837 - accuracy: 0.8225 - val_loss: 1.1356 - val_accuracy: 0.6038\nEpoch 13/20\n61/61 [==============================] - 17s 286ms/step - loss: 0.4715 - accuracy: 0.8364 - val_loss: 1.2563 - val_accuracy: 0.5906\nEpoch 14/20\n61/61 [==============================] - 17s 283ms/step - loss: 0.4625 - accuracy: 0.8340 - val_loss: 1.7464 - val_accuracy: 0.5300\nEpoch 15/20\n61/61 [==============================] - 17s 284ms/step - loss: 0.4285 - accuracy: 0.8594 - val_loss: 1.6004 - val_accuracy: 0.5599\nEpoch 16/20\n61/61 [==============================] - 17s 285ms/step - loss: 0.4108 - accuracy: 0.8569 - val_loss: 1.7552 - val_accuracy: 0.5407\nEpoch 17/20\n61/61 [==============================] - 17s 283ms/step - loss: 0.3935 - accuracy: 0.8638 - val_loss: 1.7824 - val_accuracy: 0.5624\nEpoch 18/20\n61/61 [==============================] - 17s 285ms/step - loss: 0.3929 - accuracy: 0.8589 - val_loss: 1.2477 - val_accuracy: 0.5720\nEpoch 19/20\n61/61 [==============================] - 17s 284ms/step - loss: 0.3768 - accuracy: 0.8713 - val_loss: 1.7906 - val_accuracy: 0.5535\nEpoch 20/20\n61/61 [==============================] - 17s 285ms/step - loss: 0.3590 - accuracy: 0.8739 - val_loss: 2.1418 - val_accuracy: 0.5321\nDuration: 0:06:05.606389\n"
],
[
"print(n)\n\nmodels_dsa[n].fit_model(image_sets_dsa[n],label_sets_dsa[n],x_val,y_val,epochs=20,batch_size = 128)\nn = n+1",
"11\nEpoch 1/20\n66/66 [==============================] - 21s 295ms/step - loss: 1.0265 - accuracy: 0.6546 - val_loss: 1.1369 - val_accuracy: 0.5774\nEpoch 2/20\n66/66 [==============================] - 19s 288ms/step - loss: 0.7898 - accuracy: 0.7142 - val_loss: 1.1505 - val_accuracy: 0.5399\nEpoch 3/20\n66/66 [==============================] - 19s 286ms/step - loss: 0.6944 - accuracy: 0.7488 - val_loss: 1.1718 - val_accuracy: 0.5863\nEpoch 4/20\n66/66 [==============================] - 19s 286ms/step - loss: 0.6881 - accuracy: 0.7557 - val_loss: 0.8664 - val_accuracy: 0.6733\nEpoch 5/20\n66/66 [==============================] - 19s 287ms/step - loss: 0.6315 - accuracy: 0.7726 - val_loss: 1.3921 - val_accuracy: 0.5364\nEpoch 6/20\n66/66 [==============================] - 19s 295ms/step - loss: 0.5790 - accuracy: 0.7921 - val_loss: 1.9832 - val_accuracy: 0.5164\nEpoch 7/20\n66/66 [==============================] - 19s 286ms/step - loss: 0.5793 - accuracy: 0.7900 - val_loss: 1.3164 - val_accuracy: 0.5624\nEpoch 8/20\n66/66 [==============================] - 19s 293ms/step - loss: 0.5696 - accuracy: 0.7936 - val_loss: 1.3092 - val_accuracy: 0.5713\nEpoch 9/20\n66/66 [==============================] - 19s 288ms/step - loss: 0.5208 - accuracy: 0.8121 - val_loss: 1.3003 - val_accuracy: 0.5849\nEpoch 10/20\n66/66 [==============================] - 19s 285ms/step - loss: 0.4916 - accuracy: 0.8249 - val_loss: 1.5522 - val_accuracy: 0.5731\nEpoch 11/20\n66/66 [==============================] - 19s 284ms/step - loss: 0.4690 - accuracy: 0.8278 - val_loss: 1.2364 - val_accuracy: 0.5795\nEpoch 12/20\n66/66 [==============================] - 19s 284ms/step - loss: 0.4443 - accuracy: 0.8446 - val_loss: 1.5829 - val_accuracy: 0.5464\nEpoch 13/20\n66/66 [==============================] - 19s 284ms/step - loss: 0.4387 - accuracy: 0.8482 - val_loss: 1.5690 - val_accuracy: 0.5243\nEpoch 14/20\n66/66 [==============================] - 19s 285ms/step - loss: 0.3887 - accuracy: 0.8650 - val_loss: 2.0296 - val_accuracy: 0.5300\nEpoch 15/20\n66/66 [==============================] - 19s 286ms/step - loss: 0.4316 - accuracy: 0.8466 - val_loss: 1.5617 - val_accuracy: 0.5360\nEpoch 16/20\n66/66 [==============================] - 19s 284ms/step - loss: 0.4015 - accuracy: 0.8622 - val_loss: 1.6134 - val_accuracy: 0.5310\nEpoch 17/20\n66/66 [==============================] - 19s 286ms/step - loss: 0.3477 - accuracy: 0.8811 - val_loss: 1.6304 - val_accuracy: 0.5524\nEpoch 18/20\n66/66 [==============================] - 19s 285ms/step - loss: 0.3311 - accuracy: 0.8833 - val_loss: 1.5870 - val_accuracy: 0.5563\nEpoch 19/20\n66/66 [==============================] - 19s 284ms/step - loss: 0.3620 - accuracy: 0.8702 - val_loss: 1.5225 - val_accuracy: 0.5717\nEpoch 20/20\n66/66 [==============================] - 19s 284ms/step - loss: 0.3409 - accuracy: 0.8849 - val_loss: 1.6933 - val_accuracy: 0.5403\nDuration: 0:06:19.754355\n"
],
[
"print(n)\n\nmodels_dsa[n].fit_model(image_sets_dsa[n],label_sets_dsa[n],x_val,y_val,epochs=20,batch_size = 128)\nn = n+1",
"12\nEpoch 1/20\n72/72 [==============================] - 22s 285ms/step - loss: 0.9956 - accuracy: 0.6508 - val_loss: 0.8741 - val_accuracy: 0.6733\nEpoch 2/20\n72/72 [==============================] - 20s 278ms/step - loss: 0.7771 - accuracy: 0.7218 - val_loss: 0.8981 - val_accuracy: 0.6772\nEpoch 3/20\n72/72 [==============================] - 20s 279ms/step - loss: 0.7401 - accuracy: 0.7294 - val_loss: 1.3747 - val_accuracy: 0.5510\nEpoch 4/20\n72/72 [==============================] - 21s 292ms/step - loss: 0.7305 - accuracy: 0.7343 - val_loss: 1.1316 - val_accuracy: 0.6302\nEpoch 5/20\n72/72 [==============================] - 20s 279ms/step - loss: 0.6831 - accuracy: 0.7472 - val_loss: 1.0581 - val_accuracy: 0.6177\nEpoch 6/20\n72/72 [==============================] - 20s 280ms/step - loss: 0.6482 - accuracy: 0.7680 - val_loss: 0.9766 - val_accuracy: 0.6494\nEpoch 7/20\n72/72 [==============================] - 20s 282ms/step - loss: 0.6140 - accuracy: 0.7751 - val_loss: 1.1395 - val_accuracy: 0.5906\nEpoch 8/20\n72/72 [==============================] - 20s 280ms/step - loss: 0.6054 - accuracy: 0.7802 - val_loss: 1.0921 - val_accuracy: 0.5710\nEpoch 9/20\n72/72 [==============================] - 20s 281ms/step - loss: 0.5835 - accuracy: 0.7878 - val_loss: 1.0704 - val_accuracy: 0.5660\nEpoch 10/20\n72/72 [==============================] - 20s 281ms/step - loss: 0.5311 - accuracy: 0.8072 - val_loss: 0.8753 - val_accuracy: 0.6701\nEpoch 11/20\n72/72 [==============================] - 20s 279ms/step - loss: 0.5114 - accuracy: 0.8111 - val_loss: 1.1351 - val_accuracy: 0.5949\nEpoch 12/20\n72/72 [==============================] - 20s 279ms/step - loss: 0.5468 - accuracy: 0.8103 - val_loss: 1.0202 - val_accuracy: 0.6305\nEpoch 13/20\n72/72 [==============================] - 20s 281ms/step - loss: 0.4899 - accuracy: 0.8251 - val_loss: 1.0013 - val_accuracy: 0.6555\nEpoch 14/20\n72/72 [==============================] - 21s 291ms/step - loss: 0.4493 - accuracy: 0.8384 - val_loss: 0.8588 - val_accuracy: 0.6805\nEpoch 15/20\n72/72 [==============================] - 20s 284ms/step - loss: 0.4406 - accuracy: 0.8461 - val_loss: 0.9382 - val_accuracy: 0.6683\nEpoch 16/20\n72/72 [==============================] - 21s 291ms/step - loss: 0.4381 - accuracy: 0.8456 - val_loss: 1.0086 - val_accuracy: 0.6220\nEpoch 17/20\n72/72 [==============================] - 20s 283ms/step - loss: 0.4308 - accuracy: 0.8459 - val_loss: 1.0197 - val_accuracy: 0.6890\nEpoch 18/20\n72/72 [==============================] - 21s 288ms/step - loss: 0.3754 - accuracy: 0.8666 - val_loss: 1.1711 - val_accuracy: 0.6320\nEpoch 19/20\n72/72 [==============================] - 20s 283ms/step - loss: 0.3958 - accuracy: 0.8630 - val_loss: 0.8553 - val_accuracy: 0.6680\nEpoch 20/20\n72/72 [==============================] - 20s 280ms/step - loss: 0.3706 - accuracy: 0.8710 - val_loss: 0.9553 - val_accuracy: 0.6826\nDuration: 0:06:48.721765\n"
],
[
"print(n)\n\nmodels_dsa[n].fit_model(image_sets_dsa[n],label_sets_dsa[n],x_val,y_val,epochs=20,batch_size = 128)\nn = n+1",
"13\nEpoch 1/20\n77/77 [==============================] - 25s 303ms/step - loss: 0.9526 - accuracy: 0.6602 - val_loss: 0.7496 - val_accuracy: 0.7168\nEpoch 2/20\n77/77 [==============================] - 22s 280ms/step - loss: 0.7868 - accuracy: 0.7117 - val_loss: 0.7901 - val_accuracy: 0.7133\nEpoch 3/20\n77/77 [==============================] - 23s 293ms/step - loss: 0.7276 - accuracy: 0.7344 - val_loss: 0.8348 - val_accuracy: 0.7004\nEpoch 4/20\n77/77 [==============================] - 22s 283ms/step - loss: 0.7051 - accuracy: 0.7407 - val_loss: 0.8352 - val_accuracy: 0.7040\nEpoch 5/20\n77/77 [==============================] - 22s 284ms/step - loss: 0.6659 - accuracy: 0.7621 - val_loss: 0.9804 - val_accuracy: 0.6783\nEpoch 6/20\n77/77 [==============================] - 22s 282ms/step - loss: 0.6421 - accuracy: 0.7651 - val_loss: 0.9201 - val_accuracy: 0.6623\nEpoch 7/20\n77/77 [==============================] - 22s 284ms/step - loss: 0.6189 - accuracy: 0.7735 - val_loss: 0.8604 - val_accuracy: 0.6937\nEpoch 8/20\n77/77 [==============================] - 22s 282ms/step - loss: 0.5951 - accuracy: 0.7796 - val_loss: 0.9165 - val_accuracy: 0.6755\nEpoch 9/20\n77/77 [==============================] - 22s 287ms/step - loss: 0.5727 - accuracy: 0.7909 - val_loss: 0.7476 - val_accuracy: 0.7186\nEpoch 10/20\n77/77 [==============================] - 22s 290ms/step - loss: 0.5339 - accuracy: 0.7996 - val_loss: 0.8550 - val_accuracy: 0.6944\nEpoch 11/20\n77/77 [==============================] - 22s 281ms/step - loss: 0.5172 - accuracy: 0.8117 - val_loss: 0.8523 - val_accuracy: 0.7061\nEpoch 12/20\n77/77 [==============================] - 22s 282ms/step - loss: 0.5032 - accuracy: 0.8181 - val_loss: 0.8448 - val_accuracy: 0.7090\nEpoch 13/20\n77/77 [==============================] - 22s 281ms/step - loss: 0.4823 - accuracy: 0.8215 - val_loss: 1.1084 - val_accuracy: 0.6427\nEpoch 14/20\n77/77 [==============================] - 24s 311ms/step - loss: 0.4726 - accuracy: 0.8273 - val_loss: 0.7012 - val_accuracy: 0.7361\nEpoch 15/20\n77/77 [==============================] - 22s 286ms/step - loss: 0.4411 - accuracy: 0.8418 - val_loss: 0.8860 - val_accuracy: 0.7061\nEpoch 16/20\n77/77 [==============================] - 22s 288ms/step - loss: 0.4502 - accuracy: 0.8399 - val_loss: 0.8918 - val_accuracy: 0.7133\nEpoch 17/20\n77/77 [==============================] - 22s 281ms/step - loss: 0.4185 - accuracy: 0.8507 - val_loss: 0.7738 - val_accuracy: 0.7140\nEpoch 18/20\n77/77 [==============================] - 23s 299ms/step - loss: 0.3966 - accuracy: 0.8620 - val_loss: 1.3686 - val_accuracy: 0.6345\nEpoch 19/20\n77/77 [==============================] - 28s 365ms/step - loss: 0.3976 - accuracy: 0.8609 - val_loss: 1.1240 - val_accuracy: 0.6865\nEpoch 20/20\n77/77 [==============================] - 33s 426ms/step - loss: 0.3879 - accuracy: 0.8623 - val_loss: 0.9530 - val_accuracy: 0.6983\nDuration: 0:07:40.879816\n"
],
[
"print(n)#14\n\nmodels_dsa[n].fit_model(image_sets_dsa[n],label_sets_dsa[n],x_val,y_val,epochs=20,batch_size = 128)\nn = n+1",
"14\nEpoch 1/20\n83/83 [==============================] - 31s 318ms/step - loss: 1.0091 - accuracy: 0.6414 - val_loss: 1.1078 - val_accuracy: 0.6184\nEpoch 2/20\n83/83 [==============================] - 23s 281ms/step - loss: 0.8259 - accuracy: 0.7058 - val_loss: 1.3660 - val_accuracy: 0.5136\nEpoch 3/20\n83/83 [==============================] - 23s 283ms/step - loss: 0.7882 - accuracy: 0.7124 - val_loss: 0.8392 - val_accuracy: 0.6972\nEpoch 4/20\n83/83 [==============================] - 24s 289ms/step - loss: 0.7018 - accuracy: 0.7371 - val_loss: 0.7954 - val_accuracy: 0.7108\nEpoch 5/20\n83/83 [==============================] - 25s 302ms/step - loss: 0.6815 - accuracy: 0.7436 - val_loss: 0.9211 - val_accuracy: 0.6790\nEpoch 6/20\n83/83 [==============================] - 25s 298ms/step - loss: 0.6689 - accuracy: 0.7573 - val_loss: 0.9311 - val_accuracy: 0.6705\nEpoch 7/20\n83/83 [==============================] - 24s 290ms/step - loss: 0.6173 - accuracy: 0.7684 - val_loss: 0.8598 - val_accuracy: 0.6951\nEpoch 8/20\n83/83 [==============================] - 27s 324ms/step - loss: 0.6124 - accuracy: 0.7742 - val_loss: 0.6578 - val_accuracy: 0.7525\nEpoch 9/20\n83/83 [==============================] - 25s 307ms/step - loss: 0.5715 - accuracy: 0.7924 - val_loss: 0.7497 - val_accuracy: 0.7150\nEpoch 10/20\n83/83 [==============================] - 27s 327ms/step - loss: 0.5415 - accuracy: 0.8008 - val_loss: 1.1043 - val_accuracy: 0.6459\nEpoch 11/20\n83/83 [==============================] - 26s 312ms/step - loss: 0.5953 - accuracy: 0.7916 - val_loss: 1.0647 - val_accuracy: 0.6648\nEpoch 12/20\n83/83 [==============================] - 24s 288ms/step - loss: 0.5201 - accuracy: 0.8112 - val_loss: 1.0952 - val_accuracy: 0.6576\nEpoch 13/20\n83/83 [==============================] - 25s 296ms/step - loss: 0.5255 - accuracy: 0.8077 - val_loss: 0.9114 - val_accuracy: 0.7072\nEpoch 14/20\n83/83 [==============================] - 24s 288ms/step - loss: 0.5077 - accuracy: 0.8184 - val_loss: 0.9886 - val_accuracy: 0.6837\nEpoch 15/20\n83/83 [==============================] - 26s 312ms/step - loss: 0.4731 - accuracy: 0.8254 - val_loss: 0.8011 - val_accuracy: 0.7175\nEpoch 16/20\n83/83 [==============================] - 23s 282ms/step - loss: 0.4319 - accuracy: 0.8448 - val_loss: 0.9333 - val_accuracy: 0.6366\nEpoch 17/20\n83/83 [==============================] - 23s 282ms/step - loss: 0.4532 - accuracy: 0.8355 - val_loss: 0.7355 - val_accuracy: 0.7315\nEpoch 18/20\n83/83 [==============================] - 26s 309ms/step - loss: 0.4193 - accuracy: 0.8478 - val_loss: 1.5233 - val_accuracy: 0.5795\nEpoch 19/20\n83/83 [==============================] - 25s 307ms/step - loss: 0.5280 - accuracy: 0.8321 - val_loss: 0.8900 - val_accuracy: 0.7183\nEpoch 20/20\n83/83 [==============================] - 27s 326ms/step - loss: 0.3915 - accuracy: 0.8614 - val_loss: 1.4148 - val_accuracy: 0.5924\nDuration: 0:08:23.994035\n"
],
[
"print(n)\n\nmodels_dsa[n].fit_model(image_sets_dsa[n],label_sets_dsa[n],x_val,y_val,epochs=20,batch_size = 128)\nn = n+1",
"15\nEpoch 1/20\n88/88 [==============================] - 27s 286ms/step - loss: 0.9539 - accuracy: 0.6649 - val_loss: 0.7131 - val_accuracy: 0.7368\nEpoch 2/20\n88/88 [==============================] - 25s 287ms/step - loss: 0.7550 - accuracy: 0.7226 - val_loss: 0.7933 - val_accuracy: 0.7261\nEpoch 3/20\n88/88 [==============================] - 27s 302ms/step - loss: 0.7200 - accuracy: 0.7385 - val_loss: 0.7096 - val_accuracy: 0.7257\nEpoch 4/20\n88/88 [==============================] - 27s 312ms/step - loss: 0.6833 - accuracy: 0.7476 - val_loss: 1.0486 - val_accuracy: 0.6334\nEpoch 5/20\n88/88 [==============================] - 25s 281ms/step - loss: 0.6510 - accuracy: 0.7627 - val_loss: 0.7480 - val_accuracy: 0.7293\nEpoch 6/20\n88/88 [==============================] - 25s 282ms/step - loss: 0.6375 - accuracy: 0.7628 - val_loss: 0.7179 - val_accuracy: 0.7293\nEpoch 7/20\n88/88 [==============================] - 28s 316ms/step - loss: 0.6041 - accuracy: 0.7796 - val_loss: 0.7775 - val_accuracy: 0.7254\nEpoch 8/20\n88/88 [==============================] - 28s 320ms/step - loss: 0.5610 - accuracy: 0.7940 - val_loss: 0.7494 - val_accuracy: 0.7165\nEpoch 9/20\n88/88 [==============================] - 27s 309ms/step - loss: 0.5590 - accuracy: 0.7920 - val_loss: 0.9400 - val_accuracy: 0.6972\nEpoch 10/20\n88/88 [==============================] - 25s 280ms/step - loss: 0.5306 - accuracy: 0.8032 - val_loss: 0.9634 - val_accuracy: 0.7079\nEpoch 11/20\n88/88 [==============================] - 26s 291ms/step - loss: 0.5164 - accuracy: 0.8172 - val_loss: 0.7137 - val_accuracy: 0.7357\nEpoch 12/20\n88/88 [==============================] - 27s 307ms/step - loss: 0.5157 - accuracy: 0.8120 - val_loss: 0.8025 - val_accuracy: 0.7172\nEpoch 13/20\n88/88 [==============================] - 27s 312ms/step - loss: 0.4775 - accuracy: 0.8324 - val_loss: 0.8861 - val_accuracy: 0.7172\nEpoch 14/20\n88/88 [==============================] - 25s 285ms/step - loss: 0.4750 - accuracy: 0.8271 - val_loss: 0.8914 - val_accuracy: 0.6676\nEpoch 15/20\n88/88 [==============================] - 25s 280ms/step - loss: 0.4541 - accuracy: 0.8343 - val_loss: 0.8957 - val_accuracy: 0.7001\nEpoch 16/20\n88/88 [==============================] - 28s 321ms/step - loss: 0.4291 - accuracy: 0.8434 - val_loss: 0.7985 - val_accuracy: 0.7261\nEpoch 17/20\n88/88 [==============================] - 24s 278ms/step - loss: 0.4031 - accuracy: 0.8554 - val_loss: 0.8211 - val_accuracy: 0.7154\nEpoch 18/20\n88/88 [==============================] - 27s 310ms/step - loss: 0.4010 - accuracy: 0.8598 - val_loss: 0.7700 - val_accuracy: 0.7129\nEpoch 19/20\n88/88 [==============================] - 24s 278ms/step - loss: 0.3821 - accuracy: 0.8630 - val_loss: 1.0756 - val_accuracy: 0.7008\nEpoch 20/20\n88/88 [==============================] - 24s 276ms/step - loss: 0.3597 - accuracy: 0.8766 - val_loss: 0.9164 - val_accuracy: 0.7008\nDuration: 0:08:41.622655\n"
],
[
"print(n)\n\nmodels_dsa[n].fit_model(image_sets_dsa[n],label_sets_dsa[n],x_val,y_val,epochs=20,batch_size = 128)\nn = n+1",
"16\nEpoch 1/20\n93/93 [==============================] - 28s 281ms/step - loss: 0.9355 - accuracy: 0.6643 - val_loss: 0.7606 - val_accuracy: 0.7272\nEpoch 2/20\n93/93 [==============================] - 28s 297ms/step - loss: 0.7602 - accuracy: 0.7136 - val_loss: 0.6578 - val_accuracy: 0.7632\nEpoch 3/20\n93/93 [==============================] - 27s 287ms/step - loss: 0.7165 - accuracy: 0.7329 - val_loss: 0.6528 - val_accuracy: 0.7739\nEpoch 4/20\n93/93 [==============================] - 26s 275ms/step - loss: 0.7030 - accuracy: 0.7408 - val_loss: 0.6222 - val_accuracy: 0.7693\nEpoch 5/20\n93/93 [==============================] - 26s 276ms/step - loss: 0.6607 - accuracy: 0.7491 - val_loss: 0.6787 - val_accuracy: 0.7675\nEpoch 6/20\n93/93 [==============================] - 26s 275ms/step - loss: 0.6515 - accuracy: 0.7585 - val_loss: 0.6856 - val_accuracy: 0.7546\nEpoch 7/20\n93/93 [==============================] - 26s 278ms/step - loss: 0.6325 - accuracy: 0.7717 - val_loss: 0.6051 - val_accuracy: 0.7792\nEpoch 8/20\n93/93 [==============================] - 26s 281ms/step - loss: 0.5805 - accuracy: 0.7849 - val_loss: 0.6058 - val_accuracy: 0.7882\nEpoch 9/20\n93/93 [==============================] - 26s 278ms/step - loss: 0.5846 - accuracy: 0.7774 - val_loss: 0.6405 - val_accuracy: 0.7564\nEpoch 10/20\n93/93 [==============================] - 26s 278ms/step - loss: 0.5538 - accuracy: 0.7863 - val_loss: 0.5923 - val_accuracy: 0.7889\nEpoch 11/20\n93/93 [==============================] - 26s 281ms/step - loss: 0.5208 - accuracy: 0.8069 - val_loss: 0.6070 - val_accuracy: 0.7775\nEpoch 12/20\n93/93 [==============================] - 26s 278ms/step - loss: 0.5327 - accuracy: 0.7998 - val_loss: 0.6543 - val_accuracy: 0.7653\nEpoch 13/20\n93/93 [==============================] - 26s 279ms/step - loss: 0.5043 - accuracy: 0.8123 - val_loss: 0.6746 - val_accuracy: 0.7714\nEpoch 14/20\n93/93 [==============================] - 26s 278ms/step - loss: 0.4918 - accuracy: 0.8198 - val_loss: 0.6396 - val_accuracy: 0.7753\nEpoch 15/20\n93/93 [==============================] - 26s 278ms/step - loss: 0.5074 - accuracy: 0.8137 - val_loss: 0.6255 - val_accuracy: 0.7732\nEpoch 16/20\n93/93 [==============================] - 26s 279ms/step - loss: 0.4432 - accuracy: 0.8354 - val_loss: 0.6454 - val_accuracy: 0.7703\nEpoch 17/20\n93/93 [==============================] - 26s 277ms/step - loss: 0.4327 - accuracy: 0.8399 - val_loss: 0.6499 - val_accuracy: 0.7671\nEpoch 18/20\n93/93 [==============================] - 26s 278ms/step - loss: 0.4220 - accuracy: 0.8457 - val_loss: 0.6929 - val_accuracy: 0.7468\nEpoch 19/20\n93/93 [==============================] - 26s 278ms/step - loss: 0.4104 - accuracy: 0.8511 - val_loss: 0.7404 - val_accuracy: 0.7646\nEpoch 20/20\n93/93 [==============================] - 26s 285ms/step - loss: 0.4006 - accuracy: 0.8574 - val_loss: 0.6595 - val_accuracy: 0.7628\nDuration: 0:08:41.913704\n"
],
[
"print(n)\n\nmodels_dsa[n].fit_model(image_sets_dsa[n],label_sets_dsa[n],x_val,y_val,epochs=20,batch_size = 128)\nn = n+1",
"17\nEpoch 1/20\n99/99 [==============================] - 31s 294ms/step - loss: 0.9186 - accuracy: 0.6660 - val_loss: 0.6461 - val_accuracy: 0.7689\nEpoch 2/20\n99/99 [==============================] - 30s 304ms/step - loss: 0.7797 - accuracy: 0.7138 - val_loss: 0.7021 - val_accuracy: 0.7600\nEpoch 3/20\n99/99 [==============================] - 31s 312ms/step - loss: 0.7419 - accuracy: 0.7216 - val_loss: 0.7270 - val_accuracy: 0.7275\nEpoch 4/20\n99/99 [==============================] - 30s 304ms/step - loss: 0.7022 - accuracy: 0.7345 - val_loss: 0.6678 - val_accuracy: 0.7571\nEpoch 5/20\n99/99 [==============================] - 30s 304ms/step - loss: 0.6597 - accuracy: 0.7592 - val_loss: 0.6579 - val_accuracy: 0.7593\nEpoch 6/20\n99/99 [==============================] - 30s 302ms/step - loss: 0.6483 - accuracy: 0.7594 - val_loss: 0.6492 - val_accuracy: 0.7696\nEpoch 7/20\n99/99 [==============================] - 28s 284ms/step - loss: 0.6345 - accuracy: 0.7609 - val_loss: 0.5867 - val_accuracy: 0.7892\nEpoch 8/20\n99/99 [==============================] - 27s 275ms/step - loss: 0.6163 - accuracy: 0.7711 - val_loss: 0.5891 - val_accuracy: 0.7878\nEpoch 9/20\n99/99 [==============================] - 29s 296ms/step - loss: 0.5956 - accuracy: 0.7756 - val_loss: 0.5672 - val_accuracy: 0.8003\nEpoch 10/20\n99/99 [==============================] - 29s 295ms/step - loss: 0.5765 - accuracy: 0.7867 - val_loss: 0.6265 - val_accuracy: 0.7860\nEpoch 11/20\n99/99 [==============================] - 29s 291ms/step - loss: 0.5821 - accuracy: 0.7860 - val_loss: 0.5974 - val_accuracy: 0.7964\nEpoch 12/20\n99/99 [==============================] - 28s 288ms/step - loss: 0.5390 - accuracy: 0.7920 - val_loss: 0.5793 - val_accuracy: 0.7978\nEpoch 13/20\n99/99 [==============================] - 28s 278ms/step - loss: 0.5309 - accuracy: 0.8043 - val_loss: 0.6784 - val_accuracy: 0.7718\nEpoch 14/20\n99/99 [==============================] - 27s 276ms/step - loss: 0.5251 - accuracy: 0.8080 - val_loss: 0.5877 - val_accuracy: 0.7949\nEpoch 15/20\n99/99 [==============================] - 27s 276ms/step - loss: 0.5133 - accuracy: 0.8116 - val_loss: 0.6169 - val_accuracy: 0.7839\nEpoch 16/20\n99/99 [==============================] - 28s 280ms/step - loss: 0.4759 - accuracy: 0.8281 - val_loss: 0.6405 - val_accuracy: 0.7832\nEpoch 17/20\n99/99 [==============================] - 29s 298ms/step - loss: 0.4688 - accuracy: 0.8315 - val_loss: 0.5818 - val_accuracy: 0.7946\nEpoch 18/20\n99/99 [==============================] - 27s 274ms/step - loss: 0.4417 - accuracy: 0.8383 - val_loss: 0.6759 - val_accuracy: 0.7700\nEpoch 19/20\n99/99 [==============================] - 27s 278ms/step - loss: 0.4530 - accuracy: 0.8388 - val_loss: 0.7065 - val_accuracy: 0.7764\nEpoch 20/20\n99/99 [==============================] - 27s 274ms/step - loss: 0.4249 - accuracy: 0.8478 - val_loss: 0.6294 - val_accuracy: 0.7817\nDuration: 0:09:33.955572\n"
],
[
"print(n)\n\nmodels_dsa[n].fit_model(image_sets_dsa[n],label_sets_dsa[n],x_val,y_val,epochs=20,batch_size = 128)\nn = n+1",
"18\nEpoch 1/20\n104/104 [==============================] - 32s 289ms/step - loss: 0.9410 - accuracy: 0.6561 - val_loss: 0.6836 - val_accuracy: 0.7696\nEpoch 2/20\n104/104 [==============================] - 29s 275ms/step - loss: 0.7681 - accuracy: 0.7180 - val_loss: 0.6620 - val_accuracy: 0.7689\nEpoch 3/20\n104/104 [==============================] - 29s 275ms/step - loss: 0.7424 - accuracy: 0.7254 - val_loss: 0.7090 - val_accuracy: 0.7500\nEpoch 4/20\n104/104 [==============================] - 29s 275ms/step - loss: 0.7052 - accuracy: 0.7328 - val_loss: 0.5789 - val_accuracy: 0.7921\nEpoch 5/20\n104/104 [==============================] - 29s 277ms/step - loss: 0.6981 - accuracy: 0.7340 - val_loss: 0.7064 - val_accuracy: 0.7464\nEpoch 6/20\n104/104 [==============================] - 30s 288ms/step - loss: 0.6690 - accuracy: 0.7421 - val_loss: 0.6033 - val_accuracy: 0.7828\nEpoch 7/20\n104/104 [==============================] - 39s 373ms/step - loss: 0.6350 - accuracy: 0.7650 - val_loss: 0.5657 - val_accuracy: 0.7967\nEpoch 8/20\n104/104 [==============================] - 33s 321ms/step - loss: 0.6405 - accuracy: 0.7654 - val_loss: 0.6266 - val_accuracy: 0.7721\nEpoch 9/20\n104/104 [==============================] - 30s 293ms/step - loss: 0.5852 - accuracy: 0.7745 - val_loss: 0.6317 - val_accuracy: 0.7689\nEpoch 10/20\n104/104 [==============================] - 31s 296ms/step - loss: 0.5758 - accuracy: 0.7845 - val_loss: 0.5860 - val_accuracy: 0.7989\nEpoch 11/20\n104/104 [==============================] - 31s 300ms/step - loss: 0.5745 - accuracy: 0.7849 - val_loss: 0.6346 - val_accuracy: 0.7764\nEpoch 12/20\n104/104 [==============================] - 30s 293ms/step - loss: 0.5543 - accuracy: 0.7968 - val_loss: 0.6058 - val_accuracy: 0.7928\nEpoch 13/20\n104/104 [==============================] - 29s 276ms/step - loss: 0.5200 - accuracy: 0.8073 - val_loss: 0.5869 - val_accuracy: 0.7917\nEpoch 14/20\n104/104 [==============================] - 29s 281ms/step - loss: 0.5196 - accuracy: 0.8054 - val_loss: 0.5670 - val_accuracy: 0.8060\nEpoch 15/20\n104/104 [==============================] - 29s 277ms/step - loss: 0.5072 - accuracy: 0.8125 - val_loss: 0.6344 - val_accuracy: 0.7696\nEpoch 16/20\n104/104 [==============================] - 29s 278ms/step - loss: 0.4858 - accuracy: 0.8243 - val_loss: 0.9626 - val_accuracy: 0.6926\nEpoch 17/20\n104/104 [==============================] - 29s 277ms/step - loss: 0.5049 - accuracy: 0.8174 - val_loss: 0.5640 - val_accuracy: 0.8106\nEpoch 18/20\n104/104 [==============================] - 29s 278ms/step - loss: 0.4791 - accuracy: 0.8290 - val_loss: 0.6171 - val_accuracy: 0.7864\nEpoch 19/20\n104/104 [==============================] - 29s 282ms/step - loss: 0.4446 - accuracy: 0.8328 - val_loss: 0.5598 - val_accuracy: 0.8042\nEpoch 20/20\n104/104 [==============================] - 29s 278ms/step - loss: 0.4364 - accuracy: 0.8368 - val_loss: 0.6002 - val_accuracy: 0.7956\nDuration: 0:10:02.768448\n"
],
[
"print(n)#19\n\nmodels_dsa[n].fit_model(image_sets_dsa[n],label_sets_dsa[n],x_val,y_val,epochs=20,batch_size = 128)\nn = n+1",
"19\nEpoch 1/20\n112/112 [==============================] - 39s 328ms/step - loss: 0.9049 - accuracy: 0.6743 - val_loss: 0.6402 - val_accuracy: 0.7732\nEpoch 2/20\n112/112 [==============================] - 30s 272ms/step - loss: 0.7729 - accuracy: 0.7142 - val_loss: 0.9989 - val_accuracy: 0.6530\nEpoch 3/20\n112/112 [==============================] - 31s 274ms/step - loss: 0.7468 - accuracy: 0.7206 - val_loss: 0.9305 - val_accuracy: 0.6690\nEpoch 4/20\n112/112 [==============================] - 31s 273ms/step - loss: 0.7256 - accuracy: 0.7314 - val_loss: 0.7152 - val_accuracy: 0.7407\nEpoch 5/20\n112/112 [==============================] - 31s 278ms/step - loss: 0.6799 - accuracy: 0.7414 - val_loss: 0.7996 - val_accuracy: 0.7179\nEpoch 6/20\n112/112 [==============================] - 31s 279ms/step - loss: 0.6719 - accuracy: 0.7494 - val_loss: 0.6102 - val_accuracy: 0.7775\nEpoch 7/20\n112/112 [==============================] - 35s 310ms/step - loss: 0.6392 - accuracy: 0.7585 - val_loss: 0.6540 - val_accuracy: 0.7796\nEpoch 8/20\n112/112 [==============================] - 30s 272ms/step - loss: 0.6462 - accuracy: 0.7548 - val_loss: 0.8461 - val_accuracy: 0.6822\nEpoch 9/20\n112/112 [==============================] - 31s 273ms/step - loss: 0.6215 - accuracy: 0.7640 - val_loss: 0.6285 - val_accuracy: 0.7796\nEpoch 10/20\n112/112 [==============================] - 31s 273ms/step - loss: 0.5805 - accuracy: 0.7790 - val_loss: 0.6018 - val_accuracy: 0.7924\nEpoch 11/20\n112/112 [==============================] - 31s 274ms/step - loss: 0.5576 - accuracy: 0.7903 - val_loss: 0.5697 - val_accuracy: 0.8071\nEpoch 12/20\n112/112 [==============================] - 31s 279ms/step - loss: 0.5507 - accuracy: 0.7933 - val_loss: 0.8532 - val_accuracy: 0.7150\nEpoch 13/20\n112/112 [==============================] - 34s 300ms/step - loss: 0.5488 - accuracy: 0.7961 - val_loss: 0.5912 - val_accuracy: 0.7942\nEpoch 14/20\n112/112 [==============================] - 33s 297ms/step - loss: 0.5271 - accuracy: 0.8030 - val_loss: 0.6484 - val_accuracy: 0.7743\nEpoch 15/20\n112/112 [==============================] - 33s 290ms/step - loss: 0.5040 - accuracy: 0.8138 - val_loss: 0.6683 - val_accuracy: 0.7539\nEpoch 16/20\n112/112 [==============================] - 31s 281ms/step - loss: 0.4907 - accuracy: 0.8185 - val_loss: 0.5995 - val_accuracy: 0.7928\nEpoch 17/20\n112/112 [==============================] - 31s 273ms/step - loss: 0.4941 - accuracy: 0.8168 - val_loss: 0.5791 - val_accuracy: 0.8024\nEpoch 18/20\n112/112 [==============================] - 30s 272ms/step - loss: 0.4779 - accuracy: 0.8247 - val_loss: 0.9117 - val_accuracy: 0.6933\nEpoch 19/20\n112/112 [==============================] - 31s 278ms/step - loss: 0.4834 - accuracy: 0.8234 - val_loss: 0.6478 - val_accuracy: 0.7682\nEpoch 20/20\n112/112 [==============================] - 31s 280ms/step - loss: 0.4422 - accuracy: 0.8342 - val_loss: 0.5668 - val_accuracy: 0.8024\nDuration: 0:10:35.550820\n"
],
[
"loading = True\n\nmodels_dsa = []\n\nif loading:\n for i in range(20):\n model_dsa_dir = \"D:/models/intel_models/C2/intel_model_c2_sep_dsa_e2_\"+str(i)\n print(model_dsa_dir)\n model =utils.My_model('intel',True,model_dsa_dir)\n model.model.compile(loss= 'categorical_crossentropy', optimizer = 'rmsprop', metrics = ['accuracy',tf.keras.metrics.Precision(), tf.keras.metrics.Recall()])\n models_dsa.append(model)\n \n ",
"D:/models/intel_models/C2/intel_model_c2_sep_dsa_e2_0\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_dsa_e2_1\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_dsa_e2_2\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_dsa_e2_3\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_dsa_e2_4\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_dsa_e2_5\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_dsa_e2_6\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_dsa_e2_7\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_dsa_e2_8\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_dsa_e2_9\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_dsa_e2_10\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_dsa_e2_11\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_dsa_e2_12\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_dsa_e2_13\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_dsa_e2_14\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_dsa_e2_15\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_dsa_e2_16\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_dsa_e2_17\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_dsa_e2_18\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_dsa_e2_19\nModel loaded correctly\n"
]
],
[
[
"## Training guided by Random",
"_____no_output_____"
]
],
[
[
"import random\nrandom_indexes =list(range(len(x_train_and_adversary)))\nrandom.shuffle(random_indexes)\nprint(random_indexes[:10])\nprint(len(random_indexes))",
"_____no_output_____"
],
[
"save_dir = \"C:/Users/fjdur/Desktop/upc/project_notebooks/github_project/DL_notebooks/SA_values/intel_random_values_e2.npy\"\n\n#np.save(save_dir,np.array(random_indexes))\n\nrandom_indexes = np.load(save_dir)",
"_____no_output_____"
],
[
"# Obtaining top n images by LSA values\ntop_images_by_random = utils.get_x_of_indexes(list(np.flip(np.argsort(random_indexes))),x_train_and_adversary)\ntop_labels_by_random = utils.get_x_of_indexes(list(np.flip(np.argsort(random_indexes))),y_train_and_adversary)",
"_____no_output_____"
],
[
"top_images_by_random_5000 = np.array(top_images_by_random[:5000])\ntop_labels_by_random_5000 = np.array(top_labels_by_random[:5000])\n\nmodel_random_5000 = utils.My_model(dataset,True,model_dir)\nmodel_random_5000.compile_model()\n\nmodel_random_5000.fit_model(top_images_by_random_5000,top_labels_by_random_5000,x_val,y_val)",
"Model loaded correctly\nModel compiled\nEpoch 1/10\n79/79 [==============================] - 15s 174ms/step - loss: 0.9726 - accuracy: 0.6593 - val_loss: 0.8246 - val_accuracy: 0.7140\nEpoch 2/10\n79/79 [==============================] - 13s 166ms/step - loss: 0.7901 - accuracy: 0.7008 - val_loss: 0.8300 - val_accuracy: 0.7286\nEpoch 3/10\n79/79 [==============================] - 13s 164ms/step - loss: 0.7689 - accuracy: 0.7128 - val_loss: 0.6727 - val_accuracy: 0.7707\nEpoch 4/10\n79/79 [==============================] - 13s 165ms/step - loss: 0.7580 - accuracy: 0.7095 - val_loss: 0.6485 - val_accuracy: 0.7718\nEpoch 5/10\n79/79 [==============================] - 13s 163ms/step - loss: 0.7001 - accuracy: 0.7294 - val_loss: 0.9690 - val_accuracy: 0.6619\nEpoch 6/10\n79/79 [==============================] - 13s 163ms/step - loss: 0.7165 - accuracy: 0.7479 - val_loss: 0.6327 - val_accuracy: 0.7871\nEpoch 7/10\n79/79 [==============================] - 13s 168ms/step - loss: 0.6617 - accuracy: 0.7437 - val_loss: 1.0373 - val_accuracy: 0.6305\nEpoch 8/10\n79/79 [==============================] - 13s 164ms/step - loss: 0.6948 - accuracy: 0.7491 - val_loss: 0.6284 - val_accuracy: 0.7800\nEpoch 9/10\n79/79 [==============================] - 13s 165ms/step - loss: 0.5997 - accuracy: 0.7709 - val_loss: 1.2105 - val_accuracy: 0.6152\nEpoch 10/10\n79/79 [==============================] - 13s 163ms/step - loss: 0.6949 - accuracy: 0.7554 - val_loss: 0.7108 - val_accuracy: 0.7511\nDuration: 0:02:12.184972\n"
],
[
"m = 700\nn = 0\nimage_sets_random = []\nlabel_sets_random = []\n\n\nfor i in range(len(top_images_by_random)//m):\n print(i,\":\")\n if (i+1 >= len(top_images_by_random)//m):\n print(\"Last\")\n print(0,\" -> \",n+m+(len(top_images_by_random)%m))\n top_images_by_random_n = np.array(top_images_by_random[:n+m+(len(top_images_by_random)%m)])\n top_labels_by_random_n = np.array(top_labels_by_random[:n+m+(len(top_images_by_random)%m)])\n else:\n print(0,\" -> \",m+n)\n top_images_by_random_n = np.array(top_images_by_random[:n+m])\n top_labels_by_random_n = np.array(top_labels_by_random[:n+m])\n image_sets_random.append(top_images_by_random_n)\n label_sets_random.append(top_labels_by_random_n)\n print(len(top_images_by_random_n))\n n += m\n\n",
"0 :\n0 -> 700\n700\n1 :\n0 -> 1400\n1400\n2 :\n0 -> 2100\n2100\n3 :\n0 -> 2800\n2800\n4 :\n0 -> 3500\n3500\n5 :\n0 -> 4200\n4200\n6 :\n0 -> 4900\n4900\n7 :\n0 -> 5600\n5600\n8 :\n0 -> 6300\n6300\n9 :\n0 -> 7000\n7000\n10 :\n0 -> 7700\n7700\n11 :\n0 -> 8400\n8400\n12 :\n0 -> 9100\n9100\n13 :\n0 -> 9800\n9800\n14 :\n0 -> 10500\n10500\n15 :\n0 -> 11200\n11200\n16 :\n0 -> 11900\n11900\n17 :\n0 -> 12600\n12600\n18 :\n0 -> 13300\n13300\n19 :\nLast\n0 -> 14224\n14224\n"
],
[
"print(model_dir)\n\nmodels_random = []\nfor i in range(len(label_sets_random)):\n print(i,\":\")\n model = utils.My_model('intel',True,model_dir)\n model.compile_model()\n models_random.append(model)\n",
"C:/Users/fjdur/Desktop/upc/project_notebooks/github_project/DL_notebooks/models/intel_model_21_10/\n0 :\nModel loaded correctly\nModel compiled\n1 :\nModel loaded correctly\nModel compiled\n2 :\nModel loaded correctly\nModel compiled\n3 :\nModel loaded correctly\nModel compiled\n4 :\nModel loaded correctly\nModel compiled\n5 :\nModel loaded correctly\nModel compiled\n6 :\nModel loaded correctly\nModel compiled\n7 :\nModel loaded correctly\nModel compiled\n8 :\nModel loaded correctly\nModel compiled\n9 :\nModel loaded correctly\nModel compiled\n10 :\nModel loaded correctly\nModel compiled\n11 :\nModel loaded correctly\nModel compiled\n12 :\nModel loaded correctly\nModel compiled\n13 :\nModel loaded correctly\nModel compiled\n14 :\nModel loaded correctly\nModel compiled\n15 :\nModel loaded correctly\nModel compiled\n16 :\nModel loaded correctly\nModel compiled\n17 :\nModel loaded correctly\nModel compiled\n18 :\nModel loaded correctly\nModel compiled\n19 :\nModel loaded correctly\nModel compiled\n"
],
[
"n=0\nprint(n)\n\nmodels_random[n].fit_model(image_sets_random[n],label_sets_random[n],x_val,y_val,epochs=20,batch_size = 128)\nn = n+1",
"0\nEpoch 1/20\n6/6 [==============================] - 5s 646ms/step - loss: 1.2836 - accuracy: 0.5596 - val_loss: 0.7477 - val_accuracy: 0.7693\nEpoch 2/20\n6/6 [==============================] - 3s 569ms/step - loss: 0.8344 - accuracy: 0.6906 - val_loss: 0.9677 - val_accuracy: 0.6223\nEpoch 3/20\n6/6 [==============================] - 3s 571ms/step - loss: 0.9391 - accuracy: 0.6581 - val_loss: 0.6466 - val_accuracy: 0.7743\nEpoch 4/20\n6/6 [==============================] - 3s 571ms/step - loss: 0.7012 - accuracy: 0.7308 - val_loss: 0.6058 - val_accuracy: 0.7924\nEpoch 5/20\n6/6 [==============================] - 3s 571ms/step - loss: 0.6889 - accuracy: 0.7357 - val_loss: 0.7124 - val_accuracy: 0.7493\nEpoch 6/20\n6/6 [==============================] - 3s 576ms/step - loss: 0.7906 - accuracy: 0.6990 - val_loss: 0.6390 - val_accuracy: 0.7646\nEpoch 7/20\n6/6 [==============================] - 3s 573ms/step - loss: 0.6094 - accuracy: 0.7674 - val_loss: 0.6596 - val_accuracy: 0.7710\nEpoch 8/20\n6/6 [==============================] - 3s 574ms/step - loss: 0.5809 - accuracy: 0.7752 - val_loss: 0.6909 - val_accuracy: 0.7479\nEpoch 9/20\n6/6 [==============================] - 3s 570ms/step - loss: 0.5552 - accuracy: 0.7976 - val_loss: 0.7084 - val_accuracy: 0.7418\nEpoch 10/20\n6/6 [==============================] - 3s 571ms/step - loss: 0.5527 - accuracy: 0.7857 - val_loss: 0.6716 - val_accuracy: 0.7664\nEpoch 11/20\n6/6 [==============================] - 3s 568ms/step - loss: 0.5287 - accuracy: 0.8183 - val_loss: 0.5976 - val_accuracy: 0.7953\nEpoch 12/20\n6/6 [==============================] - 3s 586ms/step - loss: 0.4524 - accuracy: 0.8271 - val_loss: 0.7186 - val_accuracy: 0.7511\nEpoch 13/20\n6/6 [==============================] - 3s 617ms/step - loss: 0.5057 - accuracy: 0.8090 - val_loss: 0.6217 - val_accuracy: 0.7889\nEpoch 14/20\n6/6 [==============================] - 3s 603ms/step - loss: 0.4590 - accuracy: 0.8091 - val_loss: 0.7193 - val_accuracy: 0.7521\nEpoch 15/20\n6/6 [==============================] - 3s 605ms/step - loss: 0.4326 - accuracy: 0.8437 - val_loss: 0.8408 - val_accuracy: 0.7215\nEpoch 16/20\n6/6 [==============================] - 3s 582ms/step - loss: 0.5379 - accuracy: 0.7814 - val_loss: 0.6200 - val_accuracy: 0.7907\nEpoch 17/20\n6/6 [==============================] - 3s 578ms/step - loss: 0.3596 - accuracy: 0.8594 - val_loss: 0.8593 - val_accuracy: 0.7557\nEpoch 18/20\n6/6 [==============================] - 3s 590ms/step - loss: 0.5159 - accuracy: 0.7991 - val_loss: 0.7335 - val_accuracy: 0.7653\nEpoch 19/20\n6/6 [==============================] - 3s 612ms/step - loss: 0.3904 - accuracy: 0.8421 - val_loss: 0.7290 - val_accuracy: 0.7614\nEpoch 20/20\n6/6 [==============================] - 3s 621ms/step - loss: 0.4363 - accuracy: 0.8313 - val_loss: 0.7180 - val_accuracy: 0.7689\nDuration: 0:01:05.703367\n"
],
[
"print(n)\n\nmodels_random[n].fit_model(image_sets_random[n],label_sets_random[n],x_val,y_val,epochs=20,batch_size = 128)\nn = n+1",
"1\nEpoch 1/20\n11/11 [==============================] - 8s 566ms/step - loss: 1.1568 - accuracy: 0.6029 - val_loss: 0.6663 - val_accuracy: 0.7750\nEpoch 2/20\n11/11 [==============================] - 5s 460ms/step - loss: 0.7539 - accuracy: 0.7294 - val_loss: 0.5747 - val_accuracy: 0.8042\nEpoch 3/20\n11/11 [==============================] - 5s 444ms/step - loss: 0.7137 - accuracy: 0.7274 - val_loss: 0.6230 - val_accuracy: 0.7778\nEpoch 4/20\n11/11 [==============================] - 5s 427ms/step - loss: 0.7936 - accuracy: 0.7025 - val_loss: 0.5934 - val_accuracy: 0.7924\nEpoch 5/20\n11/11 [==============================] - 5s 429ms/step - loss: 0.6864 - accuracy: 0.7392 - val_loss: 0.7229 - val_accuracy: 0.7539\nEpoch 6/20\n11/11 [==============================] - 5s 429ms/step - loss: 0.6617 - accuracy: 0.7610 - val_loss: 0.6210 - val_accuracy: 0.7832\nEpoch 7/20\n11/11 [==============================] - 5s 444ms/step - loss: 0.6829 - accuracy: 0.7401 - val_loss: 0.5699 - val_accuracy: 0.8056\nEpoch 8/20\n11/11 [==============================] - 5s 460ms/step - loss: 0.5852 - accuracy: 0.7889 - val_loss: 0.6748 - val_accuracy: 0.7650\nEpoch 9/20\n11/11 [==============================] - 5s 455ms/step - loss: 0.6723 - accuracy: 0.7438 - val_loss: 0.6276 - val_accuracy: 0.7817\nEpoch 10/20\n11/11 [==============================] - 5s 443ms/step - loss: 0.5466 - accuracy: 0.7824 - val_loss: 0.6282 - val_accuracy: 0.7878\nEpoch 11/20\n11/11 [==============================] - 5s 434ms/step - loss: 0.5270 - accuracy: 0.8060 - val_loss: 0.6706 - val_accuracy: 0.7746\nEpoch 12/20\n11/11 [==============================] - 5s 463ms/step - loss: 0.5464 - accuracy: 0.7799 - val_loss: 0.6433 - val_accuracy: 0.7782\nEpoch 13/20\n11/11 [==============================] - 5s 449ms/step - loss: 0.5490 - accuracy: 0.7958 - val_loss: 0.6190 - val_accuracy: 0.7924\nEpoch 14/20\n11/11 [==============================] - 5s 431ms/step - loss: 0.5410 - accuracy: 0.7926 - val_loss: 0.6666 - val_accuracy: 0.7668\nEpoch 15/20\n11/11 [==============================] - 5s 439ms/step - loss: 0.4970 - accuracy: 0.8153 - val_loss: 0.8250 - val_accuracy: 0.7215\nEpoch 16/20\n11/11 [==============================] - 5s 432ms/step - loss: 0.5332 - accuracy: 0.7801 - val_loss: 0.6860 - val_accuracy: 0.7700\nEpoch 17/20\n11/11 [==============================] - 5s 434ms/step - loss: 0.4318 - accuracy: 0.8206 - val_loss: 0.6194 - val_accuracy: 0.7853\nEpoch 18/20\n11/11 [==============================] - 5s 434ms/step - loss: 0.4331 - accuracy: 0.8368 - val_loss: 0.6396 - val_accuracy: 0.7846\nEpoch 19/20\n11/11 [==============================] - 5s 446ms/step - loss: 0.4062 - accuracy: 0.8397 - val_loss: 0.7764 - val_accuracy: 0.7475\nEpoch 20/20\n11/11 [==============================] - 5s 451ms/step - loss: 0.5113 - accuracy: 0.7980 - val_loss: 0.6892 - val_accuracy: 0.7628\nDuration: 0:01:37.472121\n"
],
[
"print(n)\nn=2\n\nmodels_random[n].fit_model(image_sets_random[n],label_sets_random[n],x_val,y_val,epochs=20,batch_size = 128)\nn = n+1",
"2\nEpoch 1/20\n17/17 [==============================] - 8s 384ms/step - loss: 1.1232 - accuracy: 0.6054 - val_loss: 0.7775 - val_accuracy: 0.7290\nEpoch 2/20\n17/17 [==============================] - 6s 361ms/step - loss: 0.8419 - accuracy: 0.6732 - val_loss: 0.6292 - val_accuracy: 0.7767\nEpoch 3/20\n17/17 [==============================] - 6s 361ms/step - loss: 0.7205 - accuracy: 0.7310 - val_loss: 0.8380 - val_accuracy: 0.6961\nEpoch 4/20\n17/17 [==============================] - 6s 361ms/step - loss: 0.7453 - accuracy: 0.7146 - val_loss: 0.7557 - val_accuracy: 0.7250\nEpoch 5/20\n17/17 [==============================] - 6s 370ms/step - loss: 0.7455 - accuracy: 0.7322 - val_loss: 0.6529 - val_accuracy: 0.7764\nEpoch 6/20\n17/17 [==============================] - 6s 373ms/step - loss: 0.6692 - accuracy: 0.7375 - val_loss: 0.7017 - val_accuracy: 0.7471\nEpoch 7/20\n17/17 [==============================] - 6s 364ms/step - loss: 0.6529 - accuracy: 0.7550 - val_loss: 0.5874 - val_accuracy: 0.7899\nEpoch 8/20\n17/17 [==============================] - 6s 364ms/step - loss: 0.5886 - accuracy: 0.7748 - val_loss: 0.9301 - val_accuracy: 0.6530\nEpoch 9/20\n17/17 [==============================] - 6s 364ms/step - loss: 0.6043 - accuracy: 0.7745 - val_loss: 0.5947 - val_accuracy: 0.7932\nEpoch 10/20\n17/17 [==============================] - 6s 365ms/step - loss: 0.5768 - accuracy: 0.7843 - val_loss: 0.6344 - val_accuracy: 0.7860\nEpoch 11/20\n17/17 [==============================] - 6s 362ms/step - loss: 0.5694 - accuracy: 0.7901 - val_loss: 0.7511 - val_accuracy: 0.7254\nEpoch 12/20\n17/17 [==============================] - 6s 367ms/step - loss: 0.5429 - accuracy: 0.7842 - val_loss: 0.7002 - val_accuracy: 0.7500\nEpoch 13/20\n17/17 [==============================] - 6s 364ms/step - loss: 0.6047 - accuracy: 0.7729 - val_loss: 0.6150 - val_accuracy: 0.7850\nEpoch 14/20\n17/17 [==============================] - 6s 365ms/step - loss: 0.4427 - accuracy: 0.8380 - val_loss: 0.6417 - val_accuracy: 0.7835\nEpoch 15/20\n17/17 [==============================] - 6s 365ms/step - loss: 0.4553 - accuracy: 0.8169 - val_loss: 0.6678 - val_accuracy: 0.7778\nEpoch 16/20\n17/17 [==============================] - 6s 365ms/step - loss: 0.4586 - accuracy: 0.8251 - val_loss: 0.7906 - val_accuracy: 0.7300\nEpoch 17/20\n17/17 [==============================] - 6s 361ms/step - loss: 0.5287 - accuracy: 0.7966 - val_loss: 0.6944 - val_accuracy: 0.7725\nEpoch 18/20\n17/17 [==============================] - 6s 365ms/step - loss: 0.4188 - accuracy: 0.8397 - val_loss: 0.7060 - val_accuracy: 0.7682\nEpoch 19/20\n17/17 [==============================] - 6s 379ms/step - loss: 0.4618 - accuracy: 0.8238 - val_loss: 0.7058 - val_accuracy: 0.7743\nEpoch 20/20\n17/17 [==============================] - 6s 369ms/step - loss: 0.4138 - accuracy: 0.8436 - val_loss: 0.6704 - val_accuracy: 0.7775\nDuration: 0:02:04.288846\n"
],
[
"print(n)\n\nmodels_random[n].fit_model(image_sets_random[n],label_sets_random[n],x_val,y_val,epochs=20,batch_size = 128)\nn = n+1",
"3\nEpoch 1/20\n22/22 [==============================] - 9s 363ms/step - loss: 1.1172 - accuracy: 0.6257 - val_loss: 0.6357 - val_accuracy: 0.7928\nEpoch 2/20\n22/22 [==============================] - 7s 342ms/step - loss: 0.7337 - accuracy: 0.7295 - val_loss: 0.6205 - val_accuracy: 0.7953\nEpoch 3/20\n22/22 [==============================] - 7s 343ms/step - loss: 0.7633 - accuracy: 0.7121 - val_loss: 0.6636 - val_accuracy: 0.7693\nEpoch 4/20\n22/22 [==============================] - 9s 415ms/step - loss: 0.6845 - accuracy: 0.7413 - val_loss: 0.5830 - val_accuracy: 0.7989\nEpoch 5/20\n22/22 [==============================] - 33709s 1605s/step - loss: 0.6694 - accuracy: 0.7435 - val_loss: 0.6399 - val_accuracy: 0.7789\nEpoch 6/20\n22/22 [==============================] - 9s 421ms/step - loss: 0.6500 - accuracy: 0.7578 - val_loss: 0.6795 - val_accuracy: 0.7593\nEpoch 7/20\n22/22 [==============================] - 9s 392ms/step - loss: 0.6623 - accuracy: 0.7571 - val_loss: 0.6887 - val_accuracy: 0.7561\nEpoch 8/20\n22/22 [==============================] - 8s 388ms/step - loss: 0.5964 - accuracy: 0.7748 - val_loss: 0.6213 - val_accuracy: 0.7864\nEpoch 9/20\n22/22 [==============================] - 9s 403ms/step - loss: 0.5781 - accuracy: 0.7714 - val_loss: 0.6572 - val_accuracy: 0.7682\nEpoch 10/20\n22/22 [==============================] - 9s 393ms/step - loss: 0.5793 - accuracy: 0.7770 - val_loss: 0.5981 - val_accuracy: 0.7946\nEpoch 11/20\n22/22 [==============================] - 8s 387ms/step - loss: 0.5636 - accuracy: 0.7741 - val_loss: 0.7534 - val_accuracy: 0.7357\nEpoch 12/20\n22/22 [==============================] - 8s 386ms/step - loss: 0.5970 - accuracy: 0.7801 - val_loss: 0.6564 - val_accuracy: 0.7732\nEpoch 13/20\n22/22 [==============================] - 8s 383ms/step - loss: 0.5250 - accuracy: 0.8068 - val_loss: 0.6113 - val_accuracy: 0.7867\nEpoch 14/20\n22/22 [==============================] - 8s 369ms/step - loss: 0.4673 - accuracy: 0.8258 - val_loss: 0.6702 - val_accuracy: 0.7725\nEpoch 15/20\n22/22 [==============================] - 8s 376ms/step - loss: 0.5128 - accuracy: 0.8033 - val_loss: 0.7151 - val_accuracy: 0.7757\nEpoch 16/20\n22/22 [==============================] - 8s 355ms/step - loss: 0.4689 - accuracy: 0.8168 - val_loss: 0.7640 - val_accuracy: 0.7411\nEpoch 17/20\n22/22 [==============================] - 8s 354ms/step - loss: 0.4973 - accuracy: 0.8209 - val_loss: 0.6434 - val_accuracy: 0.7839\nEpoch 18/20\n22/22 [==============================] - 8s 352ms/step - loss: 0.4416 - accuracy: 0.8335 - val_loss: 0.6861 - val_accuracy: 0.7803\nEpoch 19/20\n22/22 [==============================] - 8s 369ms/step - loss: 0.4328 - accuracy: 0.8318 - val_loss: 0.6661 - val_accuracy: 0.7932\nEpoch 20/20\n22/22 [==============================] - 8s 379ms/step - loss: 0.4272 - accuracy: 0.8374 - val_loss: 0.6832 - val_accuracy: 0.7878\nDuration: 9:24:27.089724\n"
],
[
"print(n)#4\n\nmodels_random[n].fit_model(image_sets_random[n],label_sets_random[n],x_val,y_val,epochs=20,batch_size = 128)\nn = n+1",
"4\nEpoch 1/20\n28/28 [==============================] - 13s 378ms/step - loss: 1.0184 - accuracy: 0.6396 - val_loss: 0.6994 - val_accuracy: 0.7529\nEpoch 2/20\n28/28 [==============================] - 10s 344ms/step - loss: 0.8102 - accuracy: 0.6894 - val_loss: 0.6423 - val_accuracy: 0.7743\nEpoch 3/20\n28/28 [==============================] - 10s 343ms/step - loss: 0.7515 - accuracy: 0.7264 - val_loss: 0.7493 - val_accuracy: 0.7386\nEpoch 4/20\n28/28 [==============================] - 10s 350ms/step - loss: 0.7618 - accuracy: 0.7163 - val_loss: 0.6176 - val_accuracy: 0.7864\nEpoch 5/20\n28/28 [==============================] - 10s 368ms/step - loss: 0.6883 - accuracy: 0.7397 - val_loss: 0.8094 - val_accuracy: 0.7247\nEpoch 6/20\n28/28 [==============================] - 11s 389ms/step - loss: 0.6541 - accuracy: 0.7486 - val_loss: 0.6457 - val_accuracy: 0.7803\nEpoch 7/20\n28/28 [==============================] - 10s 369ms/step - loss: 0.6578 - accuracy: 0.7445 - val_loss: 0.7541 - val_accuracy: 0.7379\nEpoch 8/20\n28/28 [==============================] - 10s 364ms/step - loss: 0.6201 - accuracy: 0.7596 - val_loss: 0.5911 - val_accuracy: 0.7989\nEpoch 9/20\n28/28 [==============================] - 10s 366ms/step - loss: 0.5973 - accuracy: 0.7853 - val_loss: 0.6607 - val_accuracy: 0.7750\nEpoch 10/20\n28/28 [==============================] - 10s 361ms/step - loss: 0.5901 - accuracy: 0.7723 - val_loss: 0.7022 - val_accuracy: 0.7653\nEpoch 11/20\n28/28 [==============================] - 10s 366ms/step - loss: 0.5898 - accuracy: 0.7762 - val_loss: 0.7318 - val_accuracy: 0.7589\nEpoch 12/20\n28/28 [==============================] - 10s 350ms/step - loss: 0.5872 - accuracy: 0.7748 - val_loss: 0.8419 - val_accuracy: 0.7211\nEpoch 13/20\n28/28 [==============================] - 10s 356ms/step - loss: 0.5794 - accuracy: 0.7810 - val_loss: 0.7392 - val_accuracy: 0.7760\nEpoch 14/20\n28/28 [==============================] - 10s 357ms/step - loss: 0.5411 - accuracy: 0.7945 - val_loss: 0.6259 - val_accuracy: 0.7867\nEpoch 15/20\n28/28 [==============================] - 10s 352ms/step - loss: 0.4951 - accuracy: 0.8120 - val_loss: 0.6685 - val_accuracy: 0.7871\nEpoch 16/20\n28/28 [==============================] - 10s 348ms/step - loss: 0.5116 - accuracy: 0.8078 - val_loss: 0.6229 - val_accuracy: 0.7960\nEpoch 17/20\n28/28 [==============================] - 10s 354ms/step - loss: 0.4921 - accuracy: 0.8175 - val_loss: 0.7814 - val_accuracy: 0.7479\nEpoch 18/20\n28/28 [==============================] - 9s 338ms/step - loss: 0.4774 - accuracy: 0.8125 - val_loss: 0.9852 - val_accuracy: 0.7044\nEpoch 19/20\n28/28 [==============================] - 9s 340ms/step - loss: 0.5010 - accuracy: 0.8157 - val_loss: 0.7702 - val_accuracy: 0.7689\nEpoch 20/20\n28/28 [==============================] - 9s 337ms/step - loss: 0.4529 - accuracy: 0.8308 - val_loss: 0.7939 - val_accuracy: 0.7311\nDuration: 0:03:20.558046\n"
],
[
"print(n)\n\nmodels_random[n].fit_model(image_sets_random[n],label_sets_random[n],x_val,y_val,epochs=20,batch_size = 128)\nn = n+1",
"5\nEpoch 1/20\n33/33 [==============================] - 15s 361ms/step - loss: 0.9801 - accuracy: 0.6526 - val_loss: 0.5929 - val_accuracy: 0.7842\nEpoch 2/20\n33/33 [==============================] - 11s 336ms/step - loss: 0.8031 - accuracy: 0.6996 - val_loss: 0.5824 - val_accuracy: 0.7917\nEpoch 3/20\n33/33 [==============================] - 11s 336ms/step - loss: 0.7910 - accuracy: 0.7078 - val_loss: 0.5693 - val_accuracy: 0.8003\nEpoch 4/20\n33/33 [==============================] - 11s 337ms/step - loss: 0.7335 - accuracy: 0.7197 - val_loss: 0.6506 - val_accuracy: 0.7675\nEpoch 5/20\n33/33 [==============================] - 12s 355ms/step - loss: 0.6739 - accuracy: 0.7424 - val_loss: 0.6634 - val_accuracy: 0.7678\nEpoch 6/20\n33/33 [==============================] - 11s 340ms/step - loss: 0.6594 - accuracy: 0.7491 - val_loss: 0.6797 - val_accuracy: 0.7707\nEpoch 7/20\n33/33 [==============================] - 11s 346ms/step - loss: 0.6566 - accuracy: 0.7458 - val_loss: 0.6241 - val_accuracy: 0.7878\nEpoch 8/20\n33/33 [==============================] - 11s 343ms/step - loss: 0.6017 - accuracy: 0.7783 - val_loss: 0.6171 - val_accuracy: 0.7924\nEpoch 9/20\n33/33 [==============================] - 11s 337ms/step - loss: 0.6170 - accuracy: 0.7624 - val_loss: 0.6487 - val_accuracy: 0.7874\nEpoch 10/20\n33/33 [==============================] - 11s 332ms/step - loss: 0.6005 - accuracy: 0.7624 - val_loss: 0.6585 - val_accuracy: 0.7767\nEpoch 11/20\n33/33 [==============================] - 10s 318ms/step - loss: 0.5624 - accuracy: 0.7813 - val_loss: 0.6241 - val_accuracy: 0.7860\nEpoch 12/20\n33/33 [==============================] - 10s 319ms/step - loss: 0.5793 - accuracy: 0.7802 - val_loss: 0.6677 - val_accuracy: 0.7725\nEpoch 13/20\n33/33 [==============================] - 11s 322ms/step - loss: 0.5318 - accuracy: 0.7974 - val_loss: 0.6323 - val_accuracy: 0.7896\nEpoch 14/20\n33/33 [==============================] - 11s 321ms/step - loss: 0.5131 - accuracy: 0.8087 - val_loss: 0.6301 - val_accuracy: 0.7889\nEpoch 15/20\n33/33 [==============================] - 10s 319ms/step - loss: 0.5172 - accuracy: 0.8073 - val_loss: 0.6581 - val_accuracy: 0.7767\nEpoch 16/20\n33/33 [==============================] - 10s 319ms/step - loss: 0.4628 - accuracy: 0.8288 - val_loss: 0.7716 - val_accuracy: 0.7208\nEpoch 17/20\n33/33 [==============================] - 11s 320ms/step - loss: 0.4685 - accuracy: 0.8229 - val_loss: 0.6851 - val_accuracy: 0.7735\nEpoch 18/20\n33/33 [==============================] - 10s 318ms/step - loss: 0.4486 - accuracy: 0.8181 - val_loss: 0.6446 - val_accuracy: 0.7892\nEpoch 19/20\n33/33 [==============================] - 11s 320ms/step - loss: 0.4471 - accuracy: 0.8232 - val_loss: 1.0927 - val_accuracy: 0.6790\nEpoch 20/20\n33/33 [==============================] - 10s 319ms/step - loss: 0.4698 - accuracy: 0.8311 - val_loss: 0.7058 - val_accuracy: 0.7700\nDuration: 0:03:41.056583\n"
],
[
"print(n)\n\nmodels_random[n].fit_model(image_sets_random[n],label_sets_random[n],x_val,y_val,epochs=20,batch_size = 128)\nn = n+1",
"6\nEpoch 1/20\n39/39 [==============================] - 14s 317ms/step - loss: 0.9925 - accuracy: 0.6421 - val_loss: 0.7643 - val_accuracy: 0.7514\nEpoch 2/20\n39/39 [==============================] - 12s 303ms/step - loss: 0.7966 - accuracy: 0.6910 - val_loss: 0.6480 - val_accuracy: 0.7671\nEpoch 3/20\n39/39 [==============================] - 12s 307ms/step - loss: 0.7562 - accuracy: 0.7091 - val_loss: 0.6302 - val_accuracy: 0.7671\nEpoch 4/20\n39/39 [==============================] - 12s 319ms/step - loss: 0.7007 - accuracy: 0.7348 - val_loss: 0.6919 - val_accuracy: 0.7571\nEpoch 5/20\n39/39 [==============================] - 12s 310ms/step - loss: 0.6747 - accuracy: 0.7391 - val_loss: 0.6682 - val_accuracy: 0.7507\nEpoch 6/20\n39/39 [==============================] - 12s 309ms/step - loss: 0.6784 - accuracy: 0.7366 - val_loss: 0.6659 - val_accuracy: 0.7614\nEpoch 7/20\n39/39 [==============================] - 12s 306ms/step - loss: 0.6271 - accuracy: 0.7623 - val_loss: 0.6574 - val_accuracy: 0.7639\nEpoch 8/20\n39/39 [==============================] - 12s 306ms/step - loss: 0.6078 - accuracy: 0.7660 - val_loss: 0.5975 - val_accuracy: 0.7903\nEpoch 9/20\n39/39 [==============================] - 12s 306ms/step - loss: 0.5796 - accuracy: 0.7767 - val_loss: 0.7341 - val_accuracy: 0.7389\nEpoch 10/20\n39/39 [==============================] - 12s 306ms/step - loss: 0.5976 - accuracy: 0.7730 - val_loss: 0.6770 - val_accuracy: 0.7596\nEpoch 11/20\n39/39 [==============================] - 12s 309ms/step - loss: 0.5668 - accuracy: 0.7870 - val_loss: 0.6381 - val_accuracy: 0.7871\nEpoch 12/20\n39/39 [==============================] - 12s 306ms/step - loss: 0.5590 - accuracy: 0.7903 - val_loss: 0.6980 - val_accuracy: 0.7660\nEpoch 13/20\n39/39 [==============================] - 12s 307ms/step - loss: 0.5673 - accuracy: 0.7838 - val_loss: 0.7369 - val_accuracy: 0.7461\nEpoch 14/20\n39/39 [==============================] - 12s 307ms/step - loss: 0.5507 - accuracy: 0.7941 - val_loss: 0.8793 - val_accuracy: 0.7072\nEpoch 15/20\n39/39 [==============================] - 12s 308ms/step - loss: 0.5140 - accuracy: 0.8115 - val_loss: 0.6669 - val_accuracy: 0.7657\nEpoch 16/20\n39/39 [==============================] - 12s 307ms/step - loss: 0.5096 - accuracy: 0.8086 - val_loss: 0.9175 - val_accuracy: 0.6822\nEpoch 17/20\n39/39 [==============================] - 12s 306ms/step - loss: 0.5055 - accuracy: 0.8027 - val_loss: 0.7660 - val_accuracy: 0.7593\nEpoch 18/20\n39/39 [==============================] - 12s 306ms/step - loss: 0.4708 - accuracy: 0.8227 - val_loss: 0.7598 - val_accuracy: 0.7561\nEpoch 19/20\n39/39 [==============================] - 12s 305ms/step - loss: 0.4646 - accuracy: 0.8200 - val_loss: 0.9009 - val_accuracy: 0.7129\nEpoch 20/20\n39/39 [==============================] - 13s 328ms/step - loss: 0.5050 - accuracy: 0.8123 - val_loss: 0.6884 - val_accuracy: 0.7682\nDuration: 0:04:02.253475\n"
],
[
"print(n)\n\nmodels_random[n].fit_model(image_sets_random[n],label_sets_random[n],x_val,y_val,epochs=20,batch_size = 128)\nn = n+1",
"7\nEpoch 1/20\n44/44 [==============================] - 17s 332ms/step - loss: 0.9714 - accuracy: 0.6588 - val_loss: 0.7163 - val_accuracy: 0.7354\nEpoch 2/20\n44/44 [==============================] - 13s 301ms/step - loss: 0.7789 - accuracy: 0.7098 - val_loss: 0.6274 - val_accuracy: 0.7814\nEpoch 3/20\n44/44 [==============================] - 13s 304ms/step - loss: 0.7541 - accuracy: 0.7109 - val_loss: 0.5996 - val_accuracy: 0.7921\nEpoch 4/20\n44/44 [==============================] - 13s 305ms/step - loss: 0.7040 - accuracy: 0.7375 - val_loss: 0.6209 - val_accuracy: 0.7860\nEpoch 5/20\n44/44 [==============================] - 13s 305ms/step - loss: 0.6792 - accuracy: 0.7482 - val_loss: 0.5889 - val_accuracy: 0.8017\nEpoch 6/20\n44/44 [==============================] - 13s 305ms/step - loss: 0.6562 - accuracy: 0.7574 - val_loss: 0.6563 - val_accuracy: 0.7760\nEpoch 7/20\n44/44 [==============================] - 13s 304ms/step - loss: 0.6564 - accuracy: 0.7518 - val_loss: 0.6256 - val_accuracy: 0.7860\nEpoch 8/20\n44/44 [==============================] - 14s 310ms/step - loss: 0.6122 - accuracy: 0.7741 - val_loss: 0.6062 - val_accuracy: 0.7921\nEpoch 9/20\n44/44 [==============================] - 14s 325ms/step - loss: 0.6233 - accuracy: 0.7676 - val_loss: 0.6130 - val_accuracy: 0.7889\nEpoch 10/20\n44/44 [==============================] - 14s 315ms/step - loss: 0.6073 - accuracy: 0.7791 - val_loss: 0.6687 - val_accuracy: 0.7878\nEpoch 11/20\n44/44 [==============================] - 13s 304ms/step - loss: 0.5808 - accuracy: 0.7867 - val_loss: 0.6284 - val_accuracy: 0.7953\nEpoch 12/20\n44/44 [==============================] - 13s 304ms/step - loss: 0.5486 - accuracy: 0.7876 - val_loss: 0.6094 - val_accuracy: 0.7928\nEpoch 13/20\n44/44 [==============================] - 13s 306ms/step - loss: 0.5376 - accuracy: 0.7962 - val_loss: 0.5893 - val_accuracy: 0.8003\nEpoch 14/20\n44/44 [==============================] - 13s 305ms/step - loss: 0.5195 - accuracy: 0.8065 - val_loss: 0.7218 - val_accuracy: 0.7464\nEpoch 15/20\n44/44 [==============================] - 13s 305ms/step - loss: 0.5081 - accuracy: 0.8111 - val_loss: 0.5820 - val_accuracy: 0.7971\nEpoch 16/20\n44/44 [==============================] - 13s 304ms/step - loss: 0.4942 - accuracy: 0.8101 - val_loss: 0.6132 - val_accuracy: 0.7956\nEpoch 17/20\n44/44 [==============================] - 13s 304ms/step - loss: 0.4993 - accuracy: 0.8162 - val_loss: 0.7414 - val_accuracy: 0.7521\nEpoch 18/20\n44/44 [==============================] - 13s 305ms/step - loss: 0.5028 - accuracy: 0.8146 - val_loss: 0.6981 - val_accuracy: 0.7539\nEpoch 19/20\n44/44 [==============================] - 13s 302ms/step - loss: 0.4482 - accuracy: 0.8329 - val_loss: 0.6172 - val_accuracy: 0.7953\nEpoch 20/20\n44/44 [==============================] - 13s 302ms/step - loss: 0.4709 - accuracy: 0.8264 - val_loss: 0.7145 - val_accuracy: 0.7675\nDuration: 0:04:31.783805\n"
],
[
"print(n)\n\nmodels_random[n].fit_model(image_sets_random[n],label_sets_random[n],x_val,y_val,epochs=20,batch_size = 128)\nn = n+1",
"8\nEpoch 1/20\n50/50 [==============================] - 17s 301ms/step - loss: 0.9425 - accuracy: 0.6607 - val_loss: 1.0058 - val_accuracy: 0.6690\nEpoch 2/20\n50/50 [==============================] - 15s 297ms/step - loss: 0.8023 - accuracy: 0.7037 - val_loss: 0.7386 - val_accuracy: 0.7414\nEpoch 3/20\n50/50 [==============================] - 15s 297ms/step - loss: 0.7640 - accuracy: 0.7203 - val_loss: 0.6345 - val_accuracy: 0.7646\nEpoch 4/20\n50/50 [==============================] - 15s 298ms/step - loss: 0.7240 - accuracy: 0.7328 - val_loss: 0.7860 - val_accuracy: 0.7343\nEpoch 5/20\n50/50 [==============================] - 15s 296ms/step - loss: 0.7075 - accuracy: 0.7346 - val_loss: 0.6318 - val_accuracy: 0.7764\nEpoch 6/20\n50/50 [==============================] - 15s 297ms/step - loss: 0.6563 - accuracy: 0.7508 - val_loss: 0.6442 - val_accuracy: 0.7614\nEpoch 7/20\n50/50 [==============================] - 15s 295ms/step - loss: 0.6531 - accuracy: 0.7476 - val_loss: 0.7450 - val_accuracy: 0.7486\nEpoch 8/20\n50/50 [==============================] - 15s 295ms/step - loss: 0.6676 - accuracy: 0.7521 - val_loss: 0.6805 - val_accuracy: 0.7746\nEpoch 9/20\n50/50 [==============================] - 15s 296ms/step - loss: 0.6089 - accuracy: 0.7651 - val_loss: 0.7569 - val_accuracy: 0.7329\nEpoch 10/20\n50/50 [==============================] - 15s 298ms/step - loss: 0.6327 - accuracy: 0.7560 - val_loss: 0.6924 - val_accuracy: 0.7511\nEpoch 11/20\n50/50 [==============================] - 15s 296ms/step - loss: 0.5690 - accuracy: 0.7806 - val_loss: 0.7142 - val_accuracy: 0.7443\nEpoch 12/20\n50/50 [==============================] - 15s 295ms/step - loss: 0.5998 - accuracy: 0.7726 - val_loss: 0.8013 - val_accuracy: 0.7297\nEpoch 13/20\n50/50 [==============================] - 15s 296ms/step - loss: 0.5671 - accuracy: 0.7792 - val_loss: 0.6602 - val_accuracy: 0.7896\nEpoch 14/20\n50/50 [==============================] - 15s 295ms/step - loss: 0.5699 - accuracy: 0.7852 - val_loss: 0.6073 - val_accuracy: 0.7985\nEpoch 15/20\n50/50 [==============================] - 15s 297ms/step - loss: 0.4917 - accuracy: 0.8117 - val_loss: 0.6440 - val_accuracy: 0.7871\nEpoch 16/20\n50/50 [==============================] - 15s 294ms/step - loss: 0.4960 - accuracy: 0.8161 - val_loss: 0.9031 - val_accuracy: 0.6912\nEpoch 17/20\n50/50 [==============================] - 15s 296ms/step - loss: 0.5190 - accuracy: 0.8144 - val_loss: 0.7167 - val_accuracy: 0.7611\nEpoch 18/20\n50/50 [==============================] - 15s 298ms/step - loss: 0.4978 - accuracy: 0.8079 - val_loss: 0.6716 - val_accuracy: 0.7753\nEpoch 19/20\n50/50 [==============================] - 15s 300ms/step - loss: 0.4786 - accuracy: 0.8155 - val_loss: 1.4874 - val_accuracy: 0.5934\nEpoch 20/20\n50/50 [==============================] - 15s 299ms/step - loss: 0.5674 - accuracy: 0.8090 - val_loss: 0.6553 - val_accuracy: 0.7850\nDuration: 0:04:57.759505\n"
],
[
"print(n)#9\n\nmodels_random[n].fit_model(image_sets_random[n],label_sets_random[n],x_val,y_val,epochs=20,batch_size = 128)\nn = n+1",
"9\nEpoch 1/20\n55/55 [==============================] - 18s 300ms/step - loss: 0.9626 - accuracy: 0.6475 - val_loss: 0.7462 - val_accuracy: 0.7329\nEpoch 2/20\n55/55 [==============================] - 16s 297ms/step - loss: 0.7756 - accuracy: 0.7133 - val_loss: 0.8295 - val_accuracy: 0.6947\nEpoch 3/20\n55/55 [==============================] - 16s 297ms/step - loss: 0.7588 - accuracy: 0.7133 - val_loss: 0.5684 - val_accuracy: 0.7985\nEpoch 4/20\n55/55 [==============================] - 16s 295ms/step - loss: 0.7083 - accuracy: 0.7316 - val_loss: 0.6397 - val_accuracy: 0.7839\nEpoch 5/20\n55/55 [==============================] - 16s 295ms/step - loss: 0.6716 - accuracy: 0.7480 - val_loss: 0.6019 - val_accuracy: 0.7871\nEpoch 6/20\n55/55 [==============================] - 16s 299ms/step - loss: 0.6757 - accuracy: 0.7437 - val_loss: 0.5716 - val_accuracy: 0.7981\nEpoch 7/20\n55/55 [==============================] - 16s 295ms/step - loss: 0.6566 - accuracy: 0.7571 - val_loss: 0.5975 - val_accuracy: 0.7896\nEpoch 8/20\n55/55 [==============================] - 16s 295ms/step - loss: 0.6150 - accuracy: 0.7641 - val_loss: 0.5672 - val_accuracy: 0.8028\nEpoch 9/20\n55/55 [==============================] - 16s 296ms/step - loss: 0.5871 - accuracy: 0.7733 - val_loss: 0.6234 - val_accuracy: 0.7850\nEpoch 10/20\n55/55 [==============================] - 16s 297ms/step - loss: 0.6176 - accuracy: 0.7749 - val_loss: 0.6169 - val_accuracy: 0.7817\nEpoch 11/20\n55/55 [==============================] - 16s 295ms/step - loss: 0.5907 - accuracy: 0.7801 - val_loss: 0.6076 - val_accuracy: 0.7939\nEpoch 12/20\n55/55 [==============================] - 16s 294ms/step - loss: 0.5490 - accuracy: 0.7906 - val_loss: 0.6464 - val_accuracy: 0.7771\nEpoch 13/20\n55/55 [==============================] - 16s 297ms/step - loss: 0.5404 - accuracy: 0.7932 - val_loss: 0.6215 - val_accuracy: 0.7842\nEpoch 14/20\n55/55 [==============================] - 16s 295ms/step - loss: 0.5341 - accuracy: 0.7979 - val_loss: 0.5708 - val_accuracy: 0.8131\nEpoch 15/20\n55/55 [==============================] - 16s 295ms/step - loss: 0.5122 - accuracy: 0.8016 - val_loss: 0.5874 - val_accuracy: 0.8042\nEpoch 16/20\n55/55 [==============================] - 16s 297ms/step - loss: 0.5048 - accuracy: 0.8080 - val_loss: 0.6048 - val_accuracy: 0.8006\nEpoch 17/20\n55/55 [==============================] - 16s 297ms/step - loss: 0.4993 - accuracy: 0.8155 - val_loss: 0.5898 - val_accuracy: 0.7996\nEpoch 18/20\n55/55 [==============================] - 16s 295ms/step - loss: 0.4797 - accuracy: 0.8208 - val_loss: 0.7111 - val_accuracy: 0.7596\nEpoch 19/20\n55/55 [==============================] - 16s 296ms/step - loss: 0.4714 - accuracy: 0.8229 - val_loss: 0.6421 - val_accuracy: 0.7832\nEpoch 20/20\n55/55 [==============================] - 16s 295ms/step - loss: 0.4650 - accuracy: 0.8269 - val_loss: 0.6310 - val_accuracy: 0.7899\nDuration: 0:05:26.650157\n"
],
[
"print(n)\n\nmodels_random[n].fit_model(image_sets_random[n],label_sets_random[n],x_val,y_val,epochs=20,batch_size = 128)\nn = n+1",
"10\nEpoch 1/20\n61/61 [==============================] - 20s 300ms/step - loss: 1.1486 - accuracy: 0.6263 - val_loss: 0.6493 - val_accuracy: 0.7682\nEpoch 2/20\n61/61 [==============================] - 17s 287ms/step - loss: 0.7537 - accuracy: 0.7155 - val_loss: 0.6974 - val_accuracy: 0.7532\nEpoch 3/20\n61/61 [==============================] - 18s 289ms/step - loss: 0.7237 - accuracy: 0.7294 - val_loss: 0.7426 - val_accuracy: 0.7432\nEpoch 4/20\n61/61 [==============================] - 18s 290ms/step - loss: 0.7435 - accuracy: 0.7224 - val_loss: 0.5717 - val_accuracy: 0.7960\nEpoch 5/20\n61/61 [==============================] - 18s 290ms/step - loss: 0.6851 - accuracy: 0.7370 - val_loss: 0.6195 - val_accuracy: 0.7771\nEpoch 6/20\n61/61 [==============================] - 18s 297ms/step - loss: 0.6755 - accuracy: 0.7501 - val_loss: 0.6598 - val_accuracy: 0.7764\nEpoch 7/20\n61/61 [==============================] - 18s 298ms/step - loss: 0.6459 - accuracy: 0.7620 - val_loss: 0.7511 - val_accuracy: 0.7347\nEpoch 8/20\n61/61 [==============================] - 18s 297ms/step - loss: 0.6460 - accuracy: 0.7581 - val_loss: 0.7104 - val_accuracy: 0.7379\nEpoch 9/20\n61/61 [==============================] - 18s 288ms/step - loss: 0.5943 - accuracy: 0.7730 - val_loss: 0.7314 - val_accuracy: 0.7311\nEpoch 10/20\n61/61 [==============================] - 18s 290ms/step - loss: 0.6169 - accuracy: 0.7718 - val_loss: 0.6450 - val_accuracy: 0.7846\nEpoch 11/20\n61/61 [==============================] - 18s 289ms/step - loss: 0.5808 - accuracy: 0.7785 - val_loss: 0.7093 - val_accuracy: 0.7532\nEpoch 12/20\n61/61 [==============================] - 18s 288ms/step - loss: 0.5735 - accuracy: 0.7821 - val_loss: 0.6727 - val_accuracy: 0.7721\nEpoch 13/20\n61/61 [==============================] - 18s 288ms/step - loss: 0.5588 - accuracy: 0.7853 - val_loss: 0.9150 - val_accuracy: 0.6655\nEpoch 14/20\n61/61 [==============================] - 18s 291ms/step - loss: 0.5610 - accuracy: 0.7888 - val_loss: 1.0720 - val_accuracy: 0.6623\nEpoch 15/20\n61/61 [==============================] - 18s 289ms/step - loss: 0.5883 - accuracy: 0.7816 - val_loss: 0.8357 - val_accuracy: 0.7315\nEpoch 16/20\n61/61 [==============================] - 17s 287ms/step - loss: 0.5136 - accuracy: 0.8066 - val_loss: 0.6555 - val_accuracy: 0.7850\nEpoch 17/20\n61/61 [==============================] - 18s 289ms/step - loss: 0.5279 - accuracy: 0.8059 - val_loss: 0.7749 - val_accuracy: 0.7525\nEpoch 18/20\n61/61 [==============================] - 18s 290ms/step - loss: 0.4903 - accuracy: 0.8182 - val_loss: 0.7591 - val_accuracy: 0.7564\nEpoch 19/20\n61/61 [==============================] - 18s 289ms/step - loss: 0.4805 - accuracy: 0.8238 - val_loss: 0.7241 - val_accuracy: 0.7814\nEpoch 20/20\n61/61 [==============================] - 18s 288ms/step - loss: 0.4727 - accuracy: 0.8234 - val_loss: 0.7584 - val_accuracy: 0.7578\nDuration: 0:05:55.645484\n"
],
[
"print(n)\n\nmodels_random[n].fit_model(image_sets_random[n],label_sets_random[n],x_val,y_val,epochs=20,batch_size = 128)\nn = n+1",
"11\nEpoch 1/20\n66/66 [==============================] - 21s 297ms/step - loss: 0.9442 - accuracy: 0.6608 - val_loss: 0.6492 - val_accuracy: 0.7757\nEpoch 2/20\n66/66 [==============================] - 19s 287ms/step - loss: 0.7707 - accuracy: 0.7137 - val_loss: 0.5978 - val_accuracy: 0.7885\nEpoch 3/20\n66/66 [==============================] - 19s 291ms/step - loss: 0.7445 - accuracy: 0.7231 - val_loss: 0.6068 - val_accuracy: 0.7850\nEpoch 4/20\n66/66 [==============================] - 19s 290ms/step - loss: 0.7450 - accuracy: 0.7229 - val_loss: 0.5875 - val_accuracy: 0.7924\nEpoch 5/20\n66/66 [==============================] - 19s 289ms/step - loss: 0.6852 - accuracy: 0.7472 - val_loss: 0.6207 - val_accuracy: 0.7828\nEpoch 6/20\n66/66 [==============================] - 19s 289ms/step - loss: 0.6662 - accuracy: 0.7523 - val_loss: 0.6375 - val_accuracy: 0.7842\nEpoch 7/20\n66/66 [==============================] - 19s 290ms/step - loss: 0.6510 - accuracy: 0.7526 - val_loss: 0.6425 - val_accuracy: 0.7771\nEpoch 8/20\n66/66 [==============================] - 19s 291ms/step - loss: 0.6299 - accuracy: 0.7580 - val_loss: 0.6420 - val_accuracy: 0.7785\nEpoch 9/20\n66/66 [==============================] - 19s 289ms/step - loss: 0.6065 - accuracy: 0.7698 - val_loss: 0.5958 - val_accuracy: 0.8006\nEpoch 10/20\n66/66 [==============================] - 19s 290ms/step - loss: 0.5874 - accuracy: 0.7808 - val_loss: 0.6890 - val_accuracy: 0.7650\nEpoch 11/20\n66/66 [==============================] - 19s 290ms/step - loss: 0.5897 - accuracy: 0.7765 - val_loss: 0.5816 - val_accuracy: 0.8021\nEpoch 12/20\n66/66 [==============================] - 19s 291ms/step - loss: 0.5379 - accuracy: 0.8005 - val_loss: 0.5844 - val_accuracy: 0.7960\nEpoch 13/20\n66/66 [==============================] - 19s 289ms/step - loss: 0.5792 - accuracy: 0.7895 - val_loss: 0.6622 - val_accuracy: 0.7792\nEpoch 14/20\n66/66 [==============================] - 19s 292ms/step - loss: 0.5240 - accuracy: 0.8065 - val_loss: 0.6240 - val_accuracy: 0.7853\nEpoch 15/20\n66/66 [==============================] - 19s 295ms/step - loss: 0.5087 - accuracy: 0.8111 - val_loss: 0.6264 - val_accuracy: 0.7857\nEpoch 16/20\n66/66 [==============================] - 19s 290ms/step - loss: 0.5164 - accuracy: 0.8085 - val_loss: 0.7176 - val_accuracy: 0.7792\nEpoch 17/20\n66/66 [==============================] - 19s 288ms/step - loss: 0.4901 - accuracy: 0.8223 - val_loss: 0.5777 - val_accuracy: 0.8039\nEpoch 18/20\n66/66 [==============================] - 19s 289ms/step - loss: 0.4596 - accuracy: 0.8305 - val_loss: 0.6363 - val_accuracy: 0.7860\nEpoch 19/20\n66/66 [==============================] - 19s 290ms/step - loss: 0.4333 - accuracy: 0.8356 - val_loss: 0.6924 - val_accuracy: 0.7668\nEpoch 20/20\n66/66 [==============================] - 19s 291ms/step - loss: 0.4565 - accuracy: 0.8287 - val_loss: 0.6664 - val_accuracy: 0.7703\nDuration: 0:06:24.509251\n"
],
[
"print(n)\n\nmodels_random[n].fit_model(image_sets_random[n],label_sets_random[n],x_val,y_val,epochs=20,batch_size = 128)\nn = n+1",
"12\nEpoch 1/20\n72/72 [==============================] - 22s 290ms/step - loss: 0.9299 - accuracy: 0.6647 - val_loss: 0.9709 - val_accuracy: 0.6476\nEpoch 2/20\n72/72 [==============================] - 20s 283ms/step - loss: 0.7771 - accuracy: 0.7071 - val_loss: 0.7329 - val_accuracy: 0.7183\nEpoch 3/20\n72/72 [==============================] - 21s 286ms/step - loss: 0.7302 - accuracy: 0.7256 - val_loss: 0.7229 - val_accuracy: 0.7482\nEpoch 4/20\n72/72 [==============================] - 20s 285ms/step - loss: 0.7126 - accuracy: 0.7373 - val_loss: 0.7188 - val_accuracy: 0.7575\nEpoch 5/20\n72/72 [==============================] - 21s 285ms/step - loss: 0.6985 - accuracy: 0.7330 - val_loss: 0.6376 - val_accuracy: 0.7750\nEpoch 6/20\n72/72 [==============================] - 21s 286ms/step - loss: 0.6718 - accuracy: 0.7452 - val_loss: 0.7154 - val_accuracy: 0.7532\nEpoch 7/20\n72/72 [==============================] - 20s 284ms/step - loss: 0.6625 - accuracy: 0.7447 - val_loss: 0.6516 - val_accuracy: 0.7710\nEpoch 8/20\n72/72 [==============================] - 20s 285ms/step - loss: 0.6457 - accuracy: 0.7618 - val_loss: 0.8809 - val_accuracy: 0.6826\nEpoch 9/20\n72/72 [==============================] - 21s 285ms/step - loss: 0.6264 - accuracy: 0.7658 - val_loss: 0.6338 - val_accuracy: 0.7832\nEpoch 10/20\n72/72 [==============================] - 20s 284ms/step - loss: 0.6105 - accuracy: 0.7775 - val_loss: 0.7146 - val_accuracy: 0.7521\nEpoch 11/20\n72/72 [==============================] - 21s 285ms/step - loss: 0.5728 - accuracy: 0.7846 - val_loss: 0.7252 - val_accuracy: 0.7571\nEpoch 12/20\n72/72 [==============================] - 21s 286ms/step - loss: 0.5784 - accuracy: 0.7834 - val_loss: 0.6735 - val_accuracy: 0.7703\nEpoch 13/20\n72/72 [==============================] - 21s 287ms/step - loss: 0.5616 - accuracy: 0.7922 - val_loss: 0.7952 - val_accuracy: 0.7172\nEpoch 14/20\n72/72 [==============================] - 21s 286ms/step - loss: 0.5532 - accuracy: 0.7975 - val_loss: 1.3495 - val_accuracy: 0.6630\nEpoch 15/20\n72/72 [==============================] - 21s 285ms/step - loss: 0.6080 - accuracy: 0.7885 - val_loss: 0.6718 - val_accuracy: 0.7593\nEpoch 16/20\n72/72 [==============================] - 21s 285ms/step - loss: 0.5185 - accuracy: 0.8037 - val_loss: 0.6624 - val_accuracy: 0.7725\nEpoch 17/20\n72/72 [==============================] - 21s 285ms/step - loss: 0.4871 - accuracy: 0.8171 - val_loss: 0.6328 - val_accuracy: 0.7996\nEpoch 18/20\n72/72 [==============================] - 21s 286ms/step - loss: 0.4632 - accuracy: 0.8261 - val_loss: 0.8117 - val_accuracy: 0.7250\nEpoch 19/20\n72/72 [==============================] - 20s 285ms/step - loss: 0.4700 - accuracy: 0.8210 - val_loss: 0.7308 - val_accuracy: 0.7743\nEpoch 20/20\n72/72 [==============================] - 21s 285ms/step - loss: 0.4822 - accuracy: 0.8196 - val_loss: 0.5955 - val_accuracy: 0.7999\nDuration: 0:06:52.145383\n"
],
[
"print(n)\n\nmodels_random[n].fit_model(image_sets_random[n],label_sets_random[n],x_val,y_val,epochs=20,batch_size = 128)\nn = n+1",
"13\nEpoch 1/20\n77/77 [==============================] - 24s 289ms/step - loss: 0.9826 - accuracy: 0.6545 - val_loss: 0.6198 - val_accuracy: 0.7981\nEpoch 2/20\n77/77 [==============================] - 22s 284ms/step - loss: 0.7657 - accuracy: 0.7163 - val_loss: 0.6749 - val_accuracy: 0.7718\nEpoch 3/20\n77/77 [==============================] - 22s 287ms/step - loss: 0.7578 - accuracy: 0.7203 - val_loss: 0.6234 - val_accuracy: 0.7882\nEpoch 4/20\n77/77 [==============================] - 22s 285ms/step - loss: 0.7029 - accuracy: 0.7407 - val_loss: 0.6949 - val_accuracy: 0.7486\nEpoch 5/20\n77/77 [==============================] - 22s 284ms/step - loss: 0.6788 - accuracy: 0.7421 - val_loss: 0.6958 - val_accuracy: 0.7361\nEpoch 6/20\n77/77 [==============================] - 22s 286ms/step - loss: 0.6893 - accuracy: 0.7434 - val_loss: 0.5533 - val_accuracy: 0.8117\nEpoch 7/20\n77/77 [==============================] - 22s 287ms/step - loss: 0.6429 - accuracy: 0.7503 - val_loss: 0.6026 - val_accuracy: 0.7907\nEpoch 8/20\n77/77 [==============================] - 22s 288ms/step - loss: 0.6068 - accuracy: 0.7818 - val_loss: 0.5948 - val_accuracy: 0.7896\nEpoch 9/20\n77/77 [==============================] - 22s 286ms/step - loss: 0.6126 - accuracy: 0.7709 - val_loss: 0.5795 - val_accuracy: 0.7967\nEpoch 10/20\n77/77 [==============================] - 22s 286ms/step - loss: 0.5942 - accuracy: 0.7723 - val_loss: 0.5717 - val_accuracy: 0.7942\nEpoch 11/20\n77/77 [==============================] - 22s 288ms/step - loss: 0.5783 - accuracy: 0.7847 - val_loss: 0.5746 - val_accuracy: 0.7992\nEpoch 12/20\n77/77 [==============================] - 22s 286ms/step - loss: 0.5404 - accuracy: 0.7949 - val_loss: 0.6473 - val_accuracy: 0.7792\nEpoch 13/20\n77/77 [==============================] - 22s 285ms/step - loss: 0.5409 - accuracy: 0.7989 - val_loss: 0.6579 - val_accuracy: 0.7660\nEpoch 14/20\n77/77 [==============================] - 22s 287ms/step - loss: 0.5258 - accuracy: 0.8011 - val_loss: 0.6457 - val_accuracy: 0.7817\nEpoch 15/20\n77/77 [==============================] - 22s 290ms/step - loss: 0.5222 - accuracy: 0.8088 - val_loss: 0.5976 - val_accuracy: 0.7910\nEpoch 16/20\n77/77 [==============================] - 22s 287ms/step - loss: 0.5011 - accuracy: 0.8132 - val_loss: 0.6257 - val_accuracy: 0.7910\nEpoch 17/20\n77/77 [==============================] - 22s 286ms/step - loss: 0.4562 - accuracy: 0.8286 - val_loss: 0.5906 - val_accuracy: 0.7935\nEpoch 18/20\n77/77 [==============================] - 22s 286ms/step - loss: 0.4610 - accuracy: 0.8279 - val_loss: 0.5903 - val_accuracy: 0.8021\nEpoch 19/20\n77/77 [==============================] - 22s 286ms/step - loss: 0.4750 - accuracy: 0.8253 - val_loss: 0.5595 - val_accuracy: 0.8085\nEpoch 20/20\n77/77 [==============================] - 22s 287ms/step - loss: 0.4438 - accuracy: 0.8384 - val_loss: 0.6123 - val_accuracy: 0.7964\nDuration: 0:07:22.554227\n"
],
[
"print(n)#14\n\nmodels_random[n].fit_model(image_sets_random[n],label_sets_random[n],x_val,y_val,epochs=20,batch_size = 128)\nn = n+1",
"14\nEpoch 1/20\n83/83 [==============================] - 25s 285ms/step - loss: 0.9105 - accuracy: 0.6650 - val_loss: 0.9335 - val_accuracy: 0.5952\nEpoch 2/20\n83/83 [==============================] - 23s 281ms/step - loss: 0.7910 - accuracy: 0.7040 - val_loss: 1.0065 - val_accuracy: 0.6341\nEpoch 3/20\n83/83 [==============================] - 23s 281ms/step - loss: 0.7785 - accuracy: 0.7082 - val_loss: 0.7396 - val_accuracy: 0.7653\nEpoch 4/20\n83/83 [==============================] - 23s 283ms/step - loss: 0.7226 - accuracy: 0.7297 - val_loss: 0.8295 - val_accuracy: 0.7061\nEpoch 5/20\n83/83 [==============================] - 23s 283ms/step - loss: 0.6912 - accuracy: 0.7417 - val_loss: 0.8473 - val_accuracy: 0.7193\nEpoch 6/20\n83/83 [==============================] - 23s 282ms/step - loss: 0.6796 - accuracy: 0.7439 - val_loss: 1.0960 - val_accuracy: 0.6337\nEpoch 7/20\n83/83 [==============================] - 23s 283ms/step - loss: 0.7013 - accuracy: 0.7394 - val_loss: 0.6161 - val_accuracy: 0.7750\nEpoch 8/20\n83/83 [==============================] - 23s 282ms/step - loss: 0.6647 - accuracy: 0.7522 - val_loss: 0.6614 - val_accuracy: 0.7710\nEpoch 9/20\n83/83 [==============================] - 23s 283ms/step - loss: 0.6332 - accuracy: 0.7638 - val_loss: 0.6873 - val_accuracy: 0.7689\nEpoch 10/20\n83/83 [==============================] - 24s 283ms/step - loss: 0.6138 - accuracy: 0.7631 - val_loss: 0.7459 - val_accuracy: 0.7307\nEpoch 11/20\n83/83 [==============================] - 23s 282ms/step - loss: 0.5803 - accuracy: 0.7839 - val_loss: 0.6562 - val_accuracy: 0.7628\nEpoch 12/20\n83/83 [==============================] - 24s 284ms/step - loss: 0.5720 - accuracy: 0.7848 - val_loss: 0.6571 - val_accuracy: 0.7721\nEpoch 13/20\n83/83 [==============================] - 23s 282ms/step - loss: 0.5494 - accuracy: 0.7921 - val_loss: 0.7519 - val_accuracy: 0.7546\nEpoch 14/20\n83/83 [==============================] - 24s 286ms/step - loss: 0.5432 - accuracy: 0.7954 - val_loss: 0.7955 - val_accuracy: 0.7122\nEpoch 15/20\n83/83 [==============================] - 24s 284ms/step - loss: 0.5465 - accuracy: 0.7943 - val_loss: 0.7420 - val_accuracy: 0.7650\nEpoch 16/20\n83/83 [==============================] - 24s 287ms/step - loss: 0.5824 - accuracy: 0.7921 - val_loss: 0.6797 - val_accuracy: 0.7703\nEpoch 17/20\n83/83 [==============================] - 24s 285ms/step - loss: 0.4844 - accuracy: 0.8257 - val_loss: 1.0106 - val_accuracy: 0.6979\nEpoch 18/20\n83/83 [==============================] - 23s 282ms/step - loss: 0.4997 - accuracy: 0.8126 - val_loss: 0.6346 - val_accuracy: 0.7871\nEpoch 19/20\n83/83 [==============================] - 23s 282ms/step - loss: 0.4471 - accuracy: 0.8348 - val_loss: 0.8381 - val_accuracy: 0.7429\nEpoch 20/20\n83/83 [==============================] - 24s 284ms/step - loss: 0.4620 - accuracy: 0.8340 - val_loss: 0.6726 - val_accuracy: 0.7621\nDuration: 0:07:51.398111\n"
],
[
"print(n)\n\nmodels_random[n].fit_model(image_sets_random[n],label_sets_random[n],x_val,y_val,epochs=20,batch_size = 128)\nn = n+1",
"15\nEpoch 1/20\n88/88 [==============================] - 27s 286ms/step - loss: 0.9266 - accuracy: 0.6664 - val_loss: 0.6982 - val_accuracy: 0.7578\nEpoch 2/20\n88/88 [==============================] - 25s 283ms/step - loss: 0.7619 - accuracy: 0.7166 - val_loss: 0.6365 - val_accuracy: 0.7857\nEpoch 3/20\n88/88 [==============================] - 25s 282ms/step - loss: 0.7351 - accuracy: 0.7234 - val_loss: 0.5855 - val_accuracy: 0.7914\nEpoch 4/20\n88/88 [==============================] - 25s 283ms/step - loss: 0.6964 - accuracy: 0.7351 - val_loss: 0.5955 - val_accuracy: 0.7910\nEpoch 5/20\n88/88 [==============================] - 25s 283ms/step - loss: 0.6787 - accuracy: 0.7408 - val_loss: 0.6450 - val_accuracy: 0.7721\nEpoch 6/20\n88/88 [==============================] - 25s 281ms/step - loss: 0.6764 - accuracy: 0.7452 - val_loss: 0.5585 - val_accuracy: 0.7999\nEpoch 7/20\n88/88 [==============================] - 25s 283ms/step - loss: 0.6189 - accuracy: 0.7644 - val_loss: 0.6958 - val_accuracy: 0.7543\nEpoch 8/20\n88/88 [==============================] - 25s 282ms/step - loss: 0.6086 - accuracy: 0.7734 - val_loss: 0.5762 - val_accuracy: 0.7939\nEpoch 9/20\n88/88 [==============================] - 25s 284ms/step - loss: 0.6116 - accuracy: 0.7695 - val_loss: 0.5720 - val_accuracy: 0.7996\nEpoch 10/20\n88/88 [==============================] - 27s 306ms/step - loss: 0.5941 - accuracy: 0.7816 - val_loss: 0.6740 - val_accuracy: 0.7789\nEpoch 11/20\n88/88 [==============================] - 28s 314ms/step - loss: 0.5904 - accuracy: 0.7769 - val_loss: 0.7650 - val_accuracy: 0.7215\nEpoch 12/20\n88/88 [==============================] - 28s 321ms/step - loss: 0.5589 - accuracy: 0.7899 - val_loss: 0.5628 - val_accuracy: 0.8031\nEpoch 13/20\n88/88 [==============================] - 25s 289ms/step - loss: 0.5332 - accuracy: 0.8008 - val_loss: 0.5698 - val_accuracy: 0.8014\nEpoch 14/20\n88/88 [==============================] - 25s 284ms/step - loss: 0.5369 - accuracy: 0.7980 - val_loss: 0.5764 - val_accuracy: 0.7939\nEpoch 15/20\n88/88 [==============================] - 25s 283ms/step - loss: 0.5085 - accuracy: 0.8059 - val_loss: 0.5582 - val_accuracy: 0.8067\nEpoch 16/20\n88/88 [==============================] - 25s 283ms/step - loss: 0.4748 - accuracy: 0.8176 - val_loss: 0.5604 - val_accuracy: 0.8071\nEpoch 17/20\n88/88 [==============================] - 25s 283ms/step - loss: 0.4895 - accuracy: 0.8168 - val_loss: 0.6480 - val_accuracy: 0.7800\nEpoch 18/20\n88/88 [==============================] - 25s 283ms/step - loss: 0.4616 - accuracy: 0.8289 - val_loss: 0.6401 - val_accuracy: 0.7846\nEpoch 19/20\n88/88 [==============================] - 25s 283ms/step - loss: 0.4513 - accuracy: 0.8283 - val_loss: 0.6234 - val_accuracy: 0.7803\nEpoch 20/20\n88/88 [==============================] - 25s 283ms/step - loss: 0.4430 - accuracy: 0.8410 - val_loss: 0.6245 - val_accuracy: 0.7835\nDuration: 0:08:28.119134\n"
],
[
"print(n)\n\nmodels_random[n].fit_model(image_sets_random[n],label_sets_random[n],x_val,y_val,epochs=20,batch_size = 128)\nn = n+1",
"16\nEpoch 1/20\n93/93 [==============================] - 28s 287ms/step - loss: 0.8911 - accuracy: 0.6747 - val_loss: 0.6219 - val_accuracy: 0.7825\nEpoch 2/20\n93/93 [==============================] - 26s 283ms/step - loss: 0.7857 - accuracy: 0.7057 - val_loss: 0.6123 - val_accuracy: 0.7964\nEpoch 3/20\n93/93 [==============================] - 26s 283ms/step - loss: 0.7162 - accuracy: 0.7270 - val_loss: 0.6007 - val_accuracy: 0.7921\nEpoch 4/20\n93/93 [==============================] - 26s 284ms/step - loss: 0.7165 - accuracy: 0.7275 - val_loss: 0.5962 - val_accuracy: 0.7921\nEpoch 5/20\n93/93 [==============================] - 26s 283ms/step - loss: 0.6910 - accuracy: 0.7402 - val_loss: 0.6245 - val_accuracy: 0.7792\nEpoch 6/20\n93/93 [==============================] - 26s 283ms/step - loss: 0.6570 - accuracy: 0.7512 - val_loss: 0.6028 - val_accuracy: 0.7935\nEpoch 7/20\n93/93 [==============================] - 26s 284ms/step - loss: 0.6312 - accuracy: 0.7666 - val_loss: 0.6589 - val_accuracy: 0.7689\nEpoch 8/20\n93/93 [==============================] - 26s 284ms/step - loss: 0.6125 - accuracy: 0.7725 - val_loss: 0.5927 - val_accuracy: 0.7942\nEpoch 9/20\n93/93 [==============================] - 27s 286ms/step - loss: 0.5951 - accuracy: 0.7775 - val_loss: 0.6000 - val_accuracy: 0.7871\nEpoch 10/20\n93/93 [==============================] - 26s 283ms/step - loss: 0.5926 - accuracy: 0.7786 - val_loss: 0.5967 - val_accuracy: 0.7935\nEpoch 11/20\n93/93 [==============================] - 26s 285ms/step - loss: 0.5561 - accuracy: 0.7953 - val_loss: 0.5448 - val_accuracy: 0.8117\nEpoch 12/20\n93/93 [==============================] - 26s 283ms/step - loss: 0.5399 - accuracy: 0.7991 - val_loss: 0.5667 - val_accuracy: 0.8010\nEpoch 13/20\n93/93 [==============================] - 26s 285ms/step - loss: 0.5328 - accuracy: 0.8043 - val_loss: 0.5526 - val_accuracy: 0.8081\nEpoch 14/20\n93/93 [==============================] - 26s 285ms/step - loss: 0.5426 - accuracy: 0.7938 - val_loss: 0.6198 - val_accuracy: 0.7882\nEpoch 15/20\n93/93 [==============================] - 26s 283ms/step - loss: 0.5061 - accuracy: 0.8099 - val_loss: 0.6753 - val_accuracy: 0.7611\nEpoch 16/20\n93/93 [==============================] - 26s 282ms/step - loss: 0.4967 - accuracy: 0.8149 - val_loss: 0.5537 - val_accuracy: 0.8092\nEpoch 17/20\n93/93 [==============================] - 26s 283ms/step - loss: 0.4704 - accuracy: 0.8224 - val_loss: 0.6306 - val_accuracy: 0.7967\nEpoch 18/20\n93/93 [==============================] - 26s 285ms/step - loss: 0.4595 - accuracy: 0.8304 - val_loss: 0.5519 - val_accuracy: 0.8117\nEpoch 19/20\n93/93 [==============================] - 26s 283ms/step - loss: 0.4343 - accuracy: 0.8413 - val_loss: 0.5671 - val_accuracy: 0.8071\nEpoch 20/20\n93/93 [==============================] - 26s 282ms/step - loss: 0.4385 - accuracy: 0.8365 - val_loss: 0.5990 - val_accuracy: 0.7981\nDuration: 0:08:49.057053\n"
],
[
"print(n)\n\nmodels_random[n].fit_model(image_sets_random[n],label_sets_random[n],x_val,y_val,epochs=20,batch_size = 128)\nn = n+1",
"17\nEpoch 1/20\n99/99 [==============================] - 30s 285ms/step - loss: 0.9402 - accuracy: 0.6659 - val_loss: 0.8094 - val_accuracy: 0.7058\nEpoch 2/20\n99/99 [==============================] - 28s 281ms/step - loss: 0.7897 - accuracy: 0.7044 - val_loss: 0.5902 - val_accuracy: 0.7921\nEpoch 3/20\n99/99 [==============================] - 28s 281ms/step - loss: 0.7265 - accuracy: 0.7239 - val_loss: 0.5526 - val_accuracy: 0.7989\nEpoch 4/20\n99/99 [==============================] - 28s 281ms/step - loss: 0.6906 - accuracy: 0.7411 - val_loss: 0.6186 - val_accuracy: 0.7810\nEpoch 5/20\n99/99 [==============================] - 28s 281ms/step - loss: 0.6963 - accuracy: 0.7384 - val_loss: 0.6224 - val_accuracy: 0.7767\nEpoch 6/20\n99/99 [==============================] - 28s 282ms/step - loss: 0.6577 - accuracy: 0.7542 - val_loss: 0.6352 - val_accuracy: 0.7778\nEpoch 7/20\n99/99 [==============================] - 28s 285ms/step - loss: 0.6340 - accuracy: 0.7616 - val_loss: 0.6577 - val_accuracy: 0.7650\nEpoch 8/20\n99/99 [==============================] - 28s 283ms/step - loss: 0.6438 - accuracy: 0.7549 - val_loss: 0.5554 - val_accuracy: 0.8067\nEpoch 9/20\n99/99 [==============================] - 28s 282ms/step - loss: 0.6095 - accuracy: 0.7726 - val_loss: 0.6698 - val_accuracy: 0.7757\nEpoch 10/20\n99/99 [==============================] - 28s 281ms/step - loss: 0.5941 - accuracy: 0.7729 - val_loss: 0.5715 - val_accuracy: 0.8046\nEpoch 11/20\n99/99 [==============================] - 28s 282ms/step - loss: 0.5537 - accuracy: 0.7946 - val_loss: 0.5939 - val_accuracy: 0.7917\nEpoch 12/20\n99/99 [==============================] - 28s 284ms/step - loss: 0.5669 - accuracy: 0.7904 - val_loss: 0.6006 - val_accuracy: 0.7910\nEpoch 13/20\n99/99 [==============================] - 28s 280ms/step - loss: 0.5329 - accuracy: 0.7993 - val_loss: 0.6652 - val_accuracy: 0.7914\nEpoch 14/20\n99/99 [==============================] - 28s 281ms/step - loss: 0.5402 - accuracy: 0.7991 - val_loss: 0.6532 - val_accuracy: 0.7935\nEpoch 15/20\n99/99 [==============================] - 28s 282ms/step - loss: 0.5153 - accuracy: 0.8104 - val_loss: 0.6148 - val_accuracy: 0.7899\nEpoch 16/20\n99/99 [==============================] - 28s 282ms/step - loss: 0.4947 - accuracy: 0.8212 - val_loss: 0.5974 - val_accuracy: 0.7992\nEpoch 17/20\n99/99 [==============================] - 28s 280ms/step - loss: 0.4879 - accuracy: 0.8203 - val_loss: 0.5773 - val_accuracy: 0.7978\nEpoch 18/20\n99/99 [==============================] - 28s 282ms/step - loss: 0.4834 - accuracy: 0.8248 - val_loss: 0.6907 - val_accuracy: 0.7468\nEpoch 19/20\n99/99 [==============================] - 28s 281ms/step - loss: 0.4536 - accuracy: 0.8347 - val_loss: 1.0069 - val_accuracy: 0.6929\nEpoch 20/20\n99/99 [==============================] - 28s 282ms/step - loss: 0.4965 - accuracy: 0.8215 - val_loss: 0.6033 - val_accuracy: 0.7942\nDuration: 0:09:19.312522\n"
],
[
"print(n)\n\nmodels_random[n].fit_model(image_sets_random[n],label_sets_random[n],x_val,y_val,epochs=20,batch_size = 128)\nn = n+1",
"18\nEpoch 1/20\n104/104 [==============================] - 31s 286ms/step - loss: 0.9320 - accuracy: 0.6638 - val_loss: 0.6721 - val_accuracy: 0.7603\nEpoch 2/20\n104/104 [==============================] - 29s 281ms/step - loss: 0.7762 - accuracy: 0.7051 - val_loss: 0.6299 - val_accuracy: 0.7792\nEpoch 3/20\n104/104 [==============================] - 30s 284ms/step - loss: 0.7352 - accuracy: 0.7184 - val_loss: 0.6903 - val_accuracy: 0.7675\nEpoch 4/20\n104/104 [==============================] - 29s 280ms/step - loss: 0.7230 - accuracy: 0.7283 - val_loss: 0.5774 - val_accuracy: 0.7910\nEpoch 5/20\n104/104 [==============================] - 29s 280ms/step - loss: 0.6714 - accuracy: 0.7498 - val_loss: 0.6998 - val_accuracy: 0.7636\nEpoch 6/20\n104/104 [==============================] - 29s 280ms/step - loss: 0.6593 - accuracy: 0.7492 - val_loss: 0.5941 - val_accuracy: 0.7853\nEpoch 7/20\n104/104 [==============================] - 29s 282ms/step - loss: 0.6377 - accuracy: 0.7638 - val_loss: 0.5929 - val_accuracy: 0.7860\nEpoch 8/20\n104/104 [==============================] - 31s 294ms/step - loss: 0.6230 - accuracy: 0.7675 - val_loss: 0.6073 - val_accuracy: 0.7878\nEpoch 9/20\n104/104 [==============================] - 29s 284ms/step - loss: 0.5844 - accuracy: 0.7857 - val_loss: 0.5634 - val_accuracy: 0.8053\nEpoch 10/20\n104/104 [==============================] - 29s 283ms/step - loss: 0.6009 - accuracy: 0.7791 - val_loss: 0.8679 - val_accuracy: 0.6883\nEpoch 11/20\n104/104 [==============================] - 29s 283ms/step - loss: 0.5735 - accuracy: 0.7848 - val_loss: 0.5370 - val_accuracy: 0.8142\nEpoch 12/20\n104/104 [==============================] - 30s 285ms/step - loss: 0.5440 - accuracy: 0.7944 - val_loss: 0.5801 - val_accuracy: 0.8003\nEpoch 13/20\n104/104 [==============================] - 29s 283ms/step - loss: 0.5229 - accuracy: 0.8041 - val_loss: 0.5877 - val_accuracy: 0.8003\nEpoch 14/20\n104/104 [==============================] - 29s 283ms/step - loss: 0.5204 - accuracy: 0.8023 - val_loss: 0.5459 - val_accuracy: 0.8121\nEpoch 15/20\n104/104 [==============================] - 29s 283ms/step - loss: 0.5088 - accuracy: 0.8091 - val_loss: 0.6607 - val_accuracy: 0.7760\nEpoch 16/20\n104/104 [==============================] - 30s 287ms/step - loss: 0.4901 - accuracy: 0.8240 - val_loss: 0.5818 - val_accuracy: 0.8053\nEpoch 17/20\n104/104 [==============================] - 29s 283ms/step - loss: 0.4742 - accuracy: 0.8242 - val_loss: 0.6280 - val_accuracy: 0.7810\nEpoch 18/20\n104/104 [==============================] - 30s 284ms/step - loss: 0.4679 - accuracy: 0.8263 - val_loss: 0.6345 - val_accuracy: 0.7899\nEpoch 19/20\n104/104 [==============================] - 30s 288ms/step - loss: 0.4634 - accuracy: 0.8275 - val_loss: 0.5402 - val_accuracy: 0.8142\nEpoch 20/20\n104/104 [==============================] - 30s 286ms/step - loss: 0.4321 - accuracy: 0.8410 - val_loss: 0.6297 - val_accuracy: 0.7885\nDuration: 0:09:52.042785\n"
],
[
"print(n)#19\n\nmodels_random[n].fit_model(image_sets_random[n],label_sets_random[n],x_val,y_val,epochs=20,batch_size = 128)\nn = n+1",
"19\nEpoch 1/20\n112/112 [==============================] - 33s 283ms/step - loss: 0.8993 - accuracy: 0.6757 - val_loss: 0.7548 - val_accuracy: 0.7165\nEpoch 2/20\n112/112 [==============================] - 32s 283ms/step - loss: 0.7655 - accuracy: 0.7149 - val_loss: 0.7105 - val_accuracy: 0.7322\nEpoch 3/20\n112/112 [==============================] - 31s 280ms/step - loss: 0.7296 - accuracy: 0.7308 - val_loss: 0.6598 - val_accuracy: 0.7714\nEpoch 4/20\n112/112 [==============================] - 31s 280ms/step - loss: 0.7175 - accuracy: 0.7319 - val_loss: 0.6476 - val_accuracy: 0.7678\nEpoch 5/20\n112/112 [==============================] - 31s 280ms/step - loss: 0.6740 - accuracy: 0.7437 - val_loss: 0.6966 - val_accuracy: 0.7293\nEpoch 6/20\n112/112 [==============================] - 31s 280ms/step - loss: 0.6739 - accuracy: 0.7470 - val_loss: 0.6575 - val_accuracy: 0.7600\nEpoch 7/20\n112/112 [==============================] - 32s 282ms/step - loss: 0.6545 - accuracy: 0.7554 - val_loss: 0.7266 - val_accuracy: 0.7414\nEpoch 8/20\n112/112 [==============================] - 31s 281ms/step - loss: 0.6322 - accuracy: 0.7662 - val_loss: 0.6360 - val_accuracy: 0.7960\nEpoch 9/20\n112/112 [==============================] - 31s 280ms/step - loss: 0.6066 - accuracy: 0.7723 - val_loss: 0.9037 - val_accuracy: 0.6872\nEpoch 10/20\n112/112 [==============================] - 32s 281ms/step - loss: 0.6024 - accuracy: 0.7753 - val_loss: 0.6069 - val_accuracy: 0.7942\nEpoch 11/20\n112/112 [==============================] - 31s 280ms/step - loss: 0.5795 - accuracy: 0.7819 - val_loss: 0.6925 - val_accuracy: 0.7696\nEpoch 12/20\n112/112 [==============================] - 32s 282ms/step - loss: 0.5685 - accuracy: 0.7890 - val_loss: 0.8871 - val_accuracy: 0.6748\nEpoch 13/20\n112/112 [==============================] - 31s 280ms/step - loss: 0.5568 - accuracy: 0.7930 - val_loss: 0.6966 - val_accuracy: 0.7618\nEpoch 14/20\n112/112 [==============================] - 31s 280ms/step - loss: 0.5231 - accuracy: 0.8012 - val_loss: 0.7061 - val_accuracy: 0.7543\nEpoch 15/20\n112/112 [==============================] - 31s 281ms/step - loss: 0.5132 - accuracy: 0.8078 - val_loss: 0.7167 - val_accuracy: 0.7336\nEpoch 16/20\n112/112 [==============================] - 31s 280ms/step - loss: 0.5119 - accuracy: 0.8112 - val_loss: 0.7189 - val_accuracy: 0.7571\nEpoch 17/20\n112/112 [==============================] - 32s 282ms/step - loss: 0.5186 - accuracy: 0.8122 - val_loss: 0.7239 - val_accuracy: 0.7910\nEpoch 18/20\n112/112 [==============================] - 31s 279ms/step - loss: 0.4813 - accuracy: 0.8204 - val_loss: 0.6690 - val_accuracy: 0.7800\nEpoch 19/20\n112/112 [==============================] - 31s 279ms/step - loss: 0.4921 - accuracy: 0.8209 - val_loss: 0.7433 - val_accuracy: 0.7443\nEpoch 20/20\n112/112 [==============================] - 31s 278ms/step - loss: 0.4532 - accuracy: 0.8347 - val_loss: 0.7976 - val_accuracy: 0.7257\nDuration: 0:10:30.011363\n"
],
[
"loading = True\n\nmodels_random = []\n\nif loading:\n for i in range(20):\n model_random_dir = \"D:/models/intel_models/C2/intel_model_c2_sep_random_e2_\"+str(i)\n print(model_random_dir)\n model =utils.My_model('intel',True,model_random_dir)\n model.model.compile(loss= 'categorical_crossentropy', optimizer = 'rmsprop', metrics = ['accuracy',tf.keras.metrics.Precision(), tf.keras.metrics.Recall()])\n models_random.append(model)\n \n ",
"D:/models/intel_models/C2/intel_model_c2_sep_random_e2_0\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_random_e2_1\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_random_e2_2\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_random_e2_3\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_random_e2_4\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_random_e2_5\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_random_e2_6\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_random_e2_7\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_random_e2_8\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_random_e2_9\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_random_e2_10\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_random_e2_11\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_random_e2_12\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_random_e2_13\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_random_e2_14\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_random_e2_15\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_random_e2_16\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_random_e2_17\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_random_e2_18\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_random_e2_19\nModel loaded correctly\n"
]
],
[
[
"## Training guided by NC",
"_____no_output_____"
]
],
[
[
"# NC\nnc_values = []\nfor i in range(1,15):\n save_dir_rand = \"C:/Users/fjdur/Desktop/upc/project_notebooks/github_project/DL_notebooks/NC_values/intel_nc_values_\"+str(i)+\".npy\"\n #print(save_dir_rand)\n tmp_values = np.load(save_dir_rand)\n #print(tmp_values.shape)\n nc_values = np.append(nc_values,tmp_values)",
"_____no_output_____"
],
[
"\ntop_images_by_nc = utils.get_x_of_indexes(list(np.flip(np.argsort(nc_values))),x_train_and_adversary)\ntop_labels_by_nc = utils.get_x_of_indexes(list(np.flip(np.argsort(nc_values))),y_train_and_adversary)\n",
"_____no_output_____"
],
[
"top_images_by_nc_5000 = np.array(top_images_by_nc[:5000])\ntop_labels_by_nc_5000 = np.array(top_labels_by_nc[:5000])\n\nmodel_nc_5000 = utils.My_model(dataset,True,model_dir)\nmodel_nc_5000.compile_model()\n\nmodel_nc_5000.fit_model(top_images_by_nc_5000,top_labels_by_nc_5000,x_val,y_val)",
"Model loaded correctly\nModel compiled\nEpoch 1/10\n79/79 [==============================] - 15s 167ms/step - loss: 0.9155 - accuracy: 0.6827 - val_loss: 0.9797 - val_accuracy: 0.6252\nEpoch 2/10\n79/79 [==============================] - 13s 162ms/step - loss: 0.7808 - accuracy: 0.7188 - val_loss: 0.8830 - val_accuracy: 0.6976\nEpoch 3/10\n79/79 [==============================] - 13s 161ms/step - loss: 0.7079 - accuracy: 0.7482 - val_loss: 0.7339 - val_accuracy: 0.7479\nEpoch 4/10\n79/79 [==============================] - 13s 164ms/step - loss: 0.6930 - accuracy: 0.7453 - val_loss: 0.6557 - val_accuracy: 0.7735\nEpoch 5/10\n79/79 [==============================] - 13s 168ms/step - loss: 0.6461 - accuracy: 0.7686 - val_loss: 0.8490 - val_accuracy: 0.7101\nEpoch 6/10\n79/79 [==============================] - 13s 168ms/step - loss: 0.6403 - accuracy: 0.7628 - val_loss: 0.8018 - val_accuracy: 0.7200\nEpoch 7/10\n79/79 [==============================] - 13s 171ms/step - loss: 0.6455 - accuracy: 0.7668 - val_loss: 0.6711 - val_accuracy: 0.7721\nEpoch 8/10\n79/79 [==============================] - 13s 169ms/step - loss: 0.5780 - accuracy: 0.7916 - val_loss: 0.7400 - val_accuracy: 0.7732\nEpoch 9/10\n79/79 [==============================] - 14s 173ms/step - loss: 0.5996 - accuracy: 0.7915 - val_loss: 0.8316 - val_accuracy: 0.7407\nEpoch 10/10\n79/79 [==============================] - 15s 187ms/step - loss: 0.5942 - accuracy: 0.7781 - val_loss: 0.8322 - val_accuracy: 0.7233\nDuration: 0:02:14.828512\n"
],
[
"m = 700\nn = 0\nimage_sets_nc = []\nlabel_sets_nc = []\n\n\nfor i in range(len(top_images_by_nc)//m):\n print(i,\":\")\n if (i+1 >= len(top_images_by_nc)//m):\n print(\"Last\")\n print(0,\" -> \",n+m+(len(top_images_by_nc)%m))\n top_images_by_nc_n = np.array(top_images_by_nc[:n+m+(len(top_images_by_nc)%m)])\n top_labels_by_nc_n = np.array(top_labels_by_nc[:n+m+(len(top_images_by_nc)%m)])\n else:\n print(0,\" -> \",m+n)\n top_images_by_nc_n = np.array(top_images_by_nc[:n+m])\n top_labels_by_nc_n = np.array(top_labels_by_nc[:n+m])\n image_sets_nc.append(top_images_by_nc_n)\n label_sets_nc.append(top_labels_by_nc_n)\n print(len(top_images_by_nc_n))\n n += m\n\n",
"0 :\n0 -> 700\n700\n1 :\n0 -> 1400\n1400\n2 :\n0 -> 2100\n2100\n3 :\n0 -> 2800\n2800\n4 :\n0 -> 3500\n3500\n5 :\n0 -> 4200\n4200\n6 :\n0 -> 4900\n4900\n7 :\n0 -> 5600\n5600\n8 :\n0 -> 6300\n6300\n9 :\n0 -> 7000\n7000\n10 :\n0 -> 7700\n7700\n11 :\n0 -> 8400\n8400\n12 :\n0 -> 9100\n9100\n13 :\n0 -> 9800\n9800\n14 :\n0 -> 10500\n10500\n15 :\n0 -> 11200\n11200\n16 :\n0 -> 11900\n11900\n17 :\n0 -> 12600\n12600\n18 :\n0 -> 13300\n13300\n19 :\nLast\n0 -> 14224\n14224\n"
],
[
"print(model_dir)\n\nmodels_nc = []\nfor i in range(len(label_sets_nc)):\n print(i,\":\")\n model = utils.My_model('intel',True,model_dir)\n model.compile_model()\n models_nc.append(model)\n",
"C:/Users/fjdur/Desktop/upc/project_notebooks/github_project/DL_notebooks/models/intel_model_21_10/\n0 :\nModel loaded correctly\nModel compiled\n1 :\nModel loaded correctly\nModel compiled\n2 :\nModel loaded correctly\nModel compiled\n3 :\nModel loaded correctly\nModel compiled\n4 :\nModel loaded correctly\nModel compiled\n5 :\nModel loaded correctly\nModel compiled\n6 :\nModel loaded correctly\nModel compiled\n7 :\nModel loaded correctly\nModel compiled\n8 :\nModel loaded correctly\nModel compiled\n9 :\nModel loaded correctly\nModel compiled\n10 :\nModel loaded correctly\nModel compiled\n11 :\nModel loaded correctly\nModel compiled\n12 :\nModel loaded correctly\nModel compiled\n13 :\nModel loaded correctly\nModel compiled\n14 :\nModel loaded correctly\nModel compiled\n15 :\nModel loaded correctly\nModel compiled\n16 :\nModel loaded correctly\nModel compiled\n17 :\nModel loaded correctly\nModel compiled\n18 :\nModel loaded correctly\nModel compiled\n19 :\nModel loaded correctly\nModel compiled\n"
],
[
"n=0\nprint(n)\n\nmodels_nc[n].fit_model(image_sets_nc[n],label_sets_nc[n],x_val,y_val,epochs=20,batch_size = 128)\nn = n+1",
"0\nEpoch 1/20\n6/6 [==============================] - 5s 638ms/step - loss: 1.1656 - accuracy: 0.6301 - val_loss: 0.7497 - val_accuracy: 0.7518\nEpoch 2/20\n6/6 [==============================] - 3s 558ms/step - loss: 0.7278 - accuracy: 0.7486 - val_loss: 0.7630 - val_accuracy: 0.7364\nEpoch 3/20\n6/6 [==============================] - 3s 572ms/step - loss: 0.7037 - accuracy: 0.7520 - val_loss: 1.0144 - val_accuracy: 0.7008\nEpoch 4/20\n6/6 [==============================] - 3s 584ms/step - loss: 0.7348 - accuracy: 0.7799 - val_loss: 1.0723 - val_accuracy: 0.6509\nEpoch 5/20\n6/6 [==============================] - 3s 565ms/step - loss: 0.7431 - accuracy: 0.7504 - val_loss: 0.8908 - val_accuracy: 0.7115\nEpoch 6/20\n6/6 [==============================] - 3s 561ms/step - loss: 0.6569 - accuracy: 0.7524 - val_loss: 0.8974 - val_accuracy: 0.7343\nEpoch 7/20\n6/6 [==============================] - 3s 564ms/step - loss: 0.6345 - accuracy: 0.7772 - val_loss: 0.7422 - val_accuracy: 0.7607\nEpoch 8/20\n6/6 [==============================] - 3s 557ms/step - loss: 0.5366 - accuracy: 0.8009 - val_loss: 0.7693 - val_accuracy: 0.7614\nEpoch 9/20\n6/6 [==============================] - 3s 558ms/step - loss: 0.4615 - accuracy: 0.8355 - val_loss: 0.6596 - val_accuracy: 0.7639\nEpoch 10/20\n6/6 [==============================] - 3s 559ms/step - loss: 0.5987 - accuracy: 0.7839 - val_loss: 0.8201 - val_accuracy: 0.7582\nEpoch 11/20\n6/6 [==============================] - 3s 559ms/step - loss: 0.4105 - accuracy: 0.8420 - val_loss: 0.7837 - val_accuracy: 0.7468\nEpoch 12/20\n6/6 [==============================] - 3s 557ms/step - loss: 0.5909 - accuracy: 0.7806 - val_loss: 0.7388 - val_accuracy: 0.7586\nEpoch 13/20\n6/6 [==============================] - 3s 564ms/step - loss: 0.3808 - accuracy: 0.8608 - val_loss: 0.8320 - val_accuracy: 0.7233\nEpoch 14/20\n6/6 [==============================] - 3s 572ms/step - loss: 0.3942 - accuracy: 0.8602 - val_loss: 0.9021 - val_accuracy: 0.7450\nEpoch 15/20\n6/6 [==============================] - 3s 566ms/step - loss: 0.3784 - accuracy: 0.8787 - val_loss: 1.7438 - val_accuracy: 0.5624\nEpoch 16/20\n6/6 [==============================] - 3s 568ms/step - loss: 0.5234 - accuracy: 0.8172 - val_loss: 0.9946 - val_accuracy: 0.7161\nEpoch 17/20\n6/6 [==============================] - 3s 571ms/step - loss: 0.2935 - accuracy: 0.8883 - val_loss: 1.2260 - val_accuracy: 0.7065\nEpoch 18/20\n6/6 [==============================] - 3s 565ms/step - loss: 0.3565 - accuracy: 0.8730 - val_loss: 0.9174 - val_accuracy: 0.7311\nEpoch 19/20\n6/6 [==============================] - 3s 573ms/step - loss: 0.3892 - accuracy: 0.8600 - val_loss: 1.0645 - val_accuracy: 0.7290\nEpoch 20/20\n6/6 [==============================] - 3s 569ms/step - loss: 0.2707 - accuracy: 0.9069 - val_loss: 0.9103 - val_accuracy: 0.7504\nDuration: 0:01:03.823340\n"
],
[
"n=1\nprint(n)\n\nmodels_nc[n].fit_model(image_sets_nc[n],label_sets_nc[n],x_val,y_val,epochs=20,batch_size = 128)\nn = n+1",
"1\nEpoch 1/20\n11/11 [==============================] - 7s 478ms/step - loss: 1.1393 - accuracy: 0.6472 - val_loss: 0.7696 - val_accuracy: 0.7165\nEpoch 2/20\n11/11 [==============================] - 5s 424ms/step - loss: 0.7460 - accuracy: 0.7373 - val_loss: 0.9574 - val_accuracy: 0.7115\nEpoch 3/20\n11/11 [==============================] - 5s 425ms/step - loss: 0.7890 - accuracy: 0.7303 - val_loss: 0.6407 - val_accuracy: 0.7703\nEpoch 4/20\n11/11 [==============================] - 5s 428ms/step - loss: 0.6637 - accuracy: 0.7588 - val_loss: 0.6624 - val_accuracy: 0.7668\nEpoch 5/20\n11/11 [==============================] - 4s 423ms/step - loss: 0.6424 - accuracy: 0.7558 - val_loss: 0.6970 - val_accuracy: 0.7657\nEpoch 6/20\n11/11 [==============================] - 5s 426ms/step - loss: 0.6284 - accuracy: 0.7733 - val_loss: 0.8015 - val_accuracy: 0.7493\nEpoch 7/20\n11/11 [==============================] - 5s 425ms/step - loss: 0.5909 - accuracy: 0.7978 - val_loss: 0.6744 - val_accuracy: 0.7743\nEpoch 8/20\n11/11 [==============================] - 5s 428ms/step - loss: 0.6008 - accuracy: 0.7935 - val_loss: 1.0083 - val_accuracy: 0.6986\nEpoch 9/20\n11/11 [==============================] - 5s 429ms/step - loss: 0.5628 - accuracy: 0.7919 - val_loss: 0.8660 - val_accuracy: 0.7479\nEpoch 10/20\n11/11 [==============================] - 5s 430ms/step - loss: 0.5425 - accuracy: 0.8138 - val_loss: 0.8085 - val_accuracy: 0.7529\nEpoch 11/20\n11/11 [==============================] - 5s 433ms/step - loss: 0.5266 - accuracy: 0.8062 - val_loss: 0.8134 - val_accuracy: 0.7468\nEpoch 12/20\n11/11 [==============================] - 5s 435ms/step - loss: 0.4904 - accuracy: 0.8141 - val_loss: 0.8588 - val_accuracy: 0.7511\nEpoch 13/20\n11/11 [==============================] - 5s 436ms/step - loss: 0.4517 - accuracy: 0.8397 - val_loss: 0.8175 - val_accuracy: 0.7575\nEpoch 14/20\n11/11 [==============================] - 5s 434ms/step - loss: 0.4548 - accuracy: 0.8275 - val_loss: 0.7460 - val_accuracy: 0.7771\nEpoch 15/20\n11/11 [==============================] - 5s 427ms/step - loss: 0.4202 - accuracy: 0.8511 - val_loss: 0.9028 - val_accuracy: 0.7514\nEpoch 16/20\n11/11 [==============================] - 5s 430ms/step - loss: 0.4555 - accuracy: 0.8344 - val_loss: 0.9665 - val_accuracy: 0.6990\nEpoch 17/20\n11/11 [==============================] - 5s 429ms/step - loss: 0.3751 - accuracy: 0.8670 - val_loss: 0.8282 - val_accuracy: 0.7675\nEpoch 18/20\n11/11 [==============================] - 5s 430ms/step - loss: 0.3496 - accuracy: 0.8767 - val_loss: 0.8170 - val_accuracy: 0.7785\nEpoch 19/20\n11/11 [==============================] - 5s 430ms/step - loss: 0.3609 - accuracy: 0.8689 - val_loss: 0.9303 - val_accuracy: 0.7482\nEpoch 20/20\n11/11 [==============================] - 5s 433ms/step - loss: 0.3822 - accuracy: 0.8548 - val_loss: 0.9944 - val_accuracy: 0.7468\nDuration: 0:01:33.352888\n"
],
[
"print(n)\n\nmodels_nc[n].fit_model(image_sets_nc[n],label_sets_nc[n],x_val,y_val,epochs=20,batch_size = 128)\nn = n+1",
"2\nEpoch 1/20\n17/17 [==============================] - 8s 382ms/step - loss: 0.9874 - accuracy: 0.6564 - val_loss: 1.0628 - val_accuracy: 0.6298\nEpoch 2/20\n17/17 [==============================] - 6s 357ms/step - loss: 0.8326 - accuracy: 0.7149 - val_loss: 0.9070 - val_accuracy: 0.6887\nEpoch 3/20\n17/17 [==============================] - 6s 357ms/step - loss: 0.7121 - accuracy: 0.7479 - val_loss: 0.7244 - val_accuracy: 0.7596\nEpoch 4/20\n17/17 [==============================] - 6s 363ms/step - loss: 0.7380 - accuracy: 0.7522 - val_loss: 0.8415 - val_accuracy: 0.7051\nEpoch 5/20\n17/17 [==============================] - 6s 363ms/step - loss: 0.7053 - accuracy: 0.7411 - val_loss: 0.7267 - val_accuracy: 0.7693\nEpoch 6/20\n17/17 [==============================] - 6s 368ms/step - loss: 0.6252 - accuracy: 0.7671 - val_loss: 0.6720 - val_accuracy: 0.7792\nEpoch 7/20\n17/17 [==============================] - 6s 360ms/step - loss: 0.5394 - accuracy: 0.8065 - val_loss: 0.6830 - val_accuracy: 0.7632\nEpoch 8/20\n17/17 [==============================] - 6s 360ms/step - loss: 0.5278 - accuracy: 0.7963 - val_loss: 0.6957 - val_accuracy: 0.7732\nEpoch 9/20\n17/17 [==============================] - 6s 359ms/step - loss: 0.5933 - accuracy: 0.7984 - val_loss: 0.8562 - val_accuracy: 0.7539\nEpoch 10/20\n17/17 [==============================] - 6s 359ms/step - loss: 0.5288 - accuracy: 0.8186 - val_loss: 0.7168 - val_accuracy: 0.7618\nEpoch 11/20\n17/17 [==============================] - 6s 380ms/step - loss: 0.5274 - accuracy: 0.8017 - val_loss: 0.6911 - val_accuracy: 0.7703\nEpoch 12/20\n17/17 [==============================] - 6s 377ms/step - loss: 0.5218 - accuracy: 0.8099 - val_loss: 0.9649 - val_accuracy: 0.7147\nEpoch 13/20\n17/17 [==============================] - 6s 366ms/step - loss: 0.5756 - accuracy: 0.7766 - val_loss: 0.7967 - val_accuracy: 0.7650\nEpoch 14/20\n17/17 [==============================] - 6s 361ms/step - loss: 0.4253 - accuracy: 0.8385 - val_loss: 0.8442 - val_accuracy: 0.7447\nEpoch 15/20\n17/17 [==============================] - 6s 360ms/step - loss: 0.4763 - accuracy: 0.8279 - val_loss: 0.7163 - val_accuracy: 0.7814\nEpoch 16/20\n17/17 [==============================] - 6s 360ms/step - loss: 0.4022 - accuracy: 0.8537 - val_loss: 0.8627 - val_accuracy: 0.7514\nEpoch 17/20\n17/17 [==============================] - 6s 360ms/step - loss: 0.4740 - accuracy: 0.8330 - val_loss: 0.8853 - val_accuracy: 0.7204\nEpoch 18/20\n17/17 [==============================] - 6s 362ms/step - loss: 0.4587 - accuracy: 0.8279 - val_loss: 0.8689 - val_accuracy: 0.7778\nEpoch 19/20\n17/17 [==============================] - 6s 361ms/step - loss: 0.4285 - accuracy: 0.8423 - val_loss: 1.1165 - val_accuracy: 0.7450\nEpoch 20/20\n17/17 [==============================] - 6s 359ms/step - loss: 0.4245 - accuracy: 0.8570 - val_loss: 1.1453 - val_accuracy: 0.7190\nDuration: 0:02:03.414520\n"
],
[
"print(n)\n\nmodels_nc[n].fit_model(image_sets_nc[n],label_sets_nc[n],x_val,y_val,epochs=20,batch_size = 128)\nn = n+1",
"3\nEpoch 1/20\n22/22 [==============================] - 9s 359ms/step - loss: 0.9905 - accuracy: 0.6645 - val_loss: 0.5942 - val_accuracy: 0.7917\nEpoch 2/20\n22/22 [==============================] - 7s 339ms/step - loss: 0.7359 - accuracy: 0.7496 - val_loss: 0.6275 - val_accuracy: 0.7842\nEpoch 3/20\n22/22 [==============================] - 7s 340ms/step - loss: 0.6394 - accuracy: 0.7627 - val_loss: 0.6526 - val_accuracy: 0.7778\nEpoch 4/20\n22/22 [==============================] - 8s 349ms/step - loss: 0.6522 - accuracy: 0.7674 - val_loss: 0.6431 - val_accuracy: 0.7750\nEpoch 5/20\n22/22 [==============================] - 7s 344ms/step - loss: 0.6051 - accuracy: 0.7799 - val_loss: 0.7205 - val_accuracy: 0.7607\nEpoch 6/20\n22/22 [==============================] - 8s 345ms/step - loss: 0.5834 - accuracy: 0.7818 - val_loss: 0.8318 - val_accuracy: 0.7215\nEpoch 7/20\n22/22 [==============================] - 7s 342ms/step - loss: 0.5997 - accuracy: 0.7764 - val_loss: 0.6581 - val_accuracy: 0.7668\nEpoch 8/20\n22/22 [==============================] - 7s 344ms/step - loss: 0.5906 - accuracy: 0.7886 - val_loss: 0.6494 - val_accuracy: 0.7803\nEpoch 9/20\n22/22 [==============================] - 7s 341ms/step - loss: 0.5364 - accuracy: 0.8035 - val_loss: 0.7541 - val_accuracy: 0.7357\nEpoch 10/20\n22/22 [==============================] - 7s 343ms/step - loss: 0.5402 - accuracy: 0.8077 - val_loss: 1.1234 - val_accuracy: 0.6434\nEpoch 11/20\n22/22 [==============================] - 8s 346ms/step - loss: 0.7027 - accuracy: 0.7520 - val_loss: 0.9367 - val_accuracy: 0.7272\nEpoch 12/20\n22/22 [==============================] - 7s 343ms/step - loss: 0.4991 - accuracy: 0.8243 - val_loss: 0.7177 - val_accuracy: 0.7735\nEpoch 13/20\n22/22 [==============================] - 7s 344ms/step - loss: 0.4733 - accuracy: 0.8227 - val_loss: 0.9378 - val_accuracy: 0.7211\nEpoch 14/20\n22/22 [==============================] - 8s 346ms/step - loss: 0.4755 - accuracy: 0.8252 - val_loss: 0.7187 - val_accuracy: 0.7832\nEpoch 15/20\n22/22 [==============================] - 7s 344ms/step - loss: 0.4713 - accuracy: 0.8377 - val_loss: 0.7750 - val_accuracy: 0.7682\nEpoch 16/20\n22/22 [==============================] - 7s 344ms/step - loss: 0.4217 - accuracy: 0.8510 - val_loss: 0.8884 - val_accuracy: 0.7297\nEpoch 17/20\n22/22 [==============================] - 7s 343ms/step - loss: 0.4386 - accuracy: 0.8331 - val_loss: 0.7781 - val_accuracy: 0.7750\nEpoch 18/20\n22/22 [==============================] - 8s 345ms/step - loss: 0.3769 - accuracy: 0.8542 - val_loss: 0.8606 - val_accuracy: 0.7728\nEpoch 19/20\n22/22 [==============================] - 8s 344ms/step - loss: 0.3944 - accuracy: 0.8687 - val_loss: 0.8162 - val_accuracy: 0.7418\nEpoch 20/20\n22/22 [==============================] - 8s 349ms/step - loss: 0.3662 - accuracy: 0.8616 - val_loss: 0.7797 - val_accuracy: 0.7636\nDuration: 0:02:31.751599\n"
],
[
"print(n)#4\n\nmodels_nc[n].fit_model(image_sets_nc[n],label_sets_nc[n],x_val,y_val,epochs=20,batch_size = 128)\nn = n+1",
"4\nEpoch 1/20\n28/28 [==============================] - 11s 335ms/step - loss: 1.0233 - accuracy: 0.6629 - val_loss: 0.6750 - val_accuracy: 0.7700\nEpoch 2/20\n28/28 [==============================] - 9s 319ms/step - loss: 0.7411 - accuracy: 0.7348 - val_loss: 0.7149 - val_accuracy: 0.7710\nEpoch 3/20\n28/28 [==============================] - 9s 321ms/step - loss: 0.6960 - accuracy: 0.7490 - val_loss: 0.7056 - val_accuracy: 0.7728\nEpoch 4/20\n28/28 [==============================] - 9s 321ms/step - loss: 0.6702 - accuracy: 0.7662 - val_loss: 0.9546 - val_accuracy: 0.6819\nEpoch 5/20\n28/28 [==============================] - 9s 322ms/step - loss: 0.7089 - accuracy: 0.7460 - val_loss: 0.6958 - val_accuracy: 0.7639\nEpoch 6/20\n28/28 [==============================] - 9s 321ms/step - loss: 0.6241 - accuracy: 0.7763 - val_loss: 0.7223 - val_accuracy: 0.7600\nEpoch 7/20\n28/28 [==============================] - 9s 320ms/step - loss: 0.6255 - accuracy: 0.7758 - val_loss: 0.7074 - val_accuracy: 0.7718\nEpoch 8/20\n28/28 [==============================] - 9s 321ms/step - loss: 0.5433 - accuracy: 0.8018 - val_loss: 0.8692 - val_accuracy: 0.7175\nEpoch 9/20\n28/28 [==============================] - 9s 323ms/step - loss: 0.5644 - accuracy: 0.7935 - val_loss: 0.7240 - val_accuracy: 0.7746\nEpoch 10/20\n28/28 [==============================] - 9s 322ms/step - loss: 0.5296 - accuracy: 0.8019 - val_loss: 0.7986 - val_accuracy: 0.7668\nEpoch 11/20\n28/28 [==============================] - 9s 322ms/step - loss: 0.5308 - accuracy: 0.8020 - val_loss: 0.7834 - val_accuracy: 0.7625\nEpoch 12/20\n28/28 [==============================] - 9s 320ms/step - loss: 0.5218 - accuracy: 0.8009 - val_loss: 0.8154 - val_accuracy: 0.7735\nEpoch 13/20\n28/28 [==============================] - 9s 325ms/step - loss: 0.5030 - accuracy: 0.8242 - val_loss: 0.6960 - val_accuracy: 0.7796\nEpoch 14/20\n28/28 [==============================] - 9s 323ms/step - loss: 0.4447 - accuracy: 0.8367 - val_loss: 0.7128 - val_accuracy: 0.7664\nEpoch 15/20\n28/28 [==============================] - 9s 320ms/step - loss: 0.4815 - accuracy: 0.8272 - val_loss: 0.8803 - val_accuracy: 0.7568\nEpoch 16/20\n28/28 [==============================] - 9s 321ms/step - loss: 0.4752 - accuracy: 0.8275 - val_loss: 0.8112 - val_accuracy: 0.7743\nEpoch 17/20\n28/28 [==============================] - 9s 322ms/step - loss: 0.4462 - accuracy: 0.8312 - val_loss: 0.8825 - val_accuracy: 0.7486\nEpoch 18/20\n28/28 [==============================] - 9s 319ms/step - loss: 0.4261 - accuracy: 0.8380 - val_loss: 0.8815 - val_accuracy: 0.7521\nEpoch 19/20\n28/28 [==============================] - 9s 321ms/step - loss: 0.4433 - accuracy: 0.8347 - val_loss: 0.9838 - val_accuracy: 0.7447\nEpoch 20/20\n28/28 [==============================] - 9s 334ms/step - loss: 0.3937 - accuracy: 0.8588 - val_loss: 0.8487 - val_accuracy: 0.7625\nDuration: 0:03:01.148893\n"
],
[
"print(n)\n\nmodels_nc[n].fit_model(image_sets_nc[n],label_sets_nc[n],x_val,y_val,epochs=20,batch_size = 128)\nn = n+1",
"5\nEpoch 1/20\n33/33 [==============================] - 12s 332ms/step - loss: 1.0076 - accuracy: 0.6437 - val_loss: 0.7114 - val_accuracy: 0.7507\nEpoch 2/20\n33/33 [==============================] - 10s 315ms/step - loss: 0.7331 - accuracy: 0.7331 - val_loss: 0.6486 - val_accuracy: 0.7700\nEpoch 3/20\n33/33 [==============================] - 10s 314ms/step - loss: 0.7026 - accuracy: 0.7520 - val_loss: 0.6941 - val_accuracy: 0.7564\nEpoch 4/20\n33/33 [==============================] - 10s 316ms/step - loss: 0.7044 - accuracy: 0.7383 - val_loss: 0.6545 - val_accuracy: 0.7796\nEpoch 5/20\n33/33 [==============================] - 11s 324ms/step - loss: 0.6319 - accuracy: 0.7658 - val_loss: 0.7690 - val_accuracy: 0.7393\nEpoch 6/20\n33/33 [==============================] - 10s 319ms/step - loss: 0.6154 - accuracy: 0.7724 - val_loss: 0.6158 - val_accuracy: 0.7867\nEpoch 7/20\n33/33 [==============================] - 10s 314ms/step - loss: 0.5668 - accuracy: 0.7925 - val_loss: 0.7058 - val_accuracy: 0.7486\nEpoch 8/20\n33/33 [==============================] - 10s 316ms/step - loss: 0.6922 - accuracy: 0.7662 - val_loss: 0.7131 - val_accuracy: 0.7618\nEpoch 9/20\n33/33 [==============================] - 10s 316ms/step - loss: 0.5640 - accuracy: 0.7952 - val_loss: 0.6370 - val_accuracy: 0.7899\nEpoch 10/20\n33/33 [==============================] - 10s 314ms/step - loss: 0.5262 - accuracy: 0.8012 - val_loss: 0.6488 - val_accuracy: 0.7821\nEpoch 11/20\n33/33 [==============================] - 10s 315ms/step - loss: 0.5605 - accuracy: 0.7975 - val_loss: 0.7415 - val_accuracy: 0.7885\nEpoch 12/20\n33/33 [==============================] - 10s 316ms/step - loss: 0.5367 - accuracy: 0.8026 - val_loss: 0.7867 - val_accuracy: 0.7611\nEpoch 13/20\n33/33 [==============================] - 10s 317ms/step - loss: 0.5051 - accuracy: 0.8139 - val_loss: 0.7288 - val_accuracy: 0.7782\nEpoch 14/20\n33/33 [==============================] - 10s 316ms/step - loss: 0.4759 - accuracy: 0.8292 - val_loss: 0.6978 - val_accuracy: 0.7828\nEpoch 15/20\n33/33 [==============================] - 10s 315ms/step - loss: 0.4720 - accuracy: 0.8308 - val_loss: 0.9283 - val_accuracy: 0.7058\nEpoch 16/20\n33/33 [==============================] - 10s 316ms/step - loss: 0.4686 - accuracy: 0.8263 - val_loss: 0.7202 - val_accuracy: 0.7892\nEpoch 17/20\n33/33 [==============================] - 10s 317ms/step - loss: 0.4217 - accuracy: 0.8496 - val_loss: 0.8221 - val_accuracy: 0.7653\nEpoch 18/20\n33/33 [==============================] - 10s 317ms/step - loss: 0.4861 - accuracy: 0.8203 - val_loss: 0.9748 - val_accuracy: 0.7051\nEpoch 19/20\n33/33 [==============================] - 10s 317ms/step - loss: 0.4372 - accuracy: 0.8386 - val_loss: 1.0414 - val_accuracy: 0.7275\nEpoch 20/20\n33/33 [==============================] - 10s 317ms/step - loss: 0.4234 - accuracy: 0.8493 - val_loss: 0.7397 - val_accuracy: 0.7810\nDuration: 0:03:29.906213\n"
],
[
"print(n)\n\nmodels_nc[n].fit_model(image_sets_nc[n],label_sets_nc[n],x_val,y_val,epochs=20,batch_size = 128)\nn = n+1",
"6\nEpoch 1/20\n39/39 [==============================] - 14s 313ms/step - loss: 0.9042 - accuracy: 0.6954 - val_loss: 0.6776 - val_accuracy: 0.7771\nEpoch 2/20\n39/39 [==============================] - 12s 301ms/step - loss: 0.7103 - accuracy: 0.7370 - val_loss: 0.6164 - val_accuracy: 0.7785\nEpoch 3/20\n39/39 [==============================] - 12s 303ms/step - loss: 0.7048 - accuracy: 0.7468 - val_loss: 0.6044 - val_accuracy: 0.7885\nEpoch 4/20\n39/39 [==============================] - 12s 304ms/step - loss: 0.7002 - accuracy: 0.7418 - val_loss: 0.7152 - val_accuracy: 0.7471\nEpoch 5/20\n39/39 [==============================] - 12s 303ms/step - loss: 0.6593 - accuracy: 0.7654 - val_loss: 0.7492 - val_accuracy: 0.7568\nEpoch 6/20\n39/39 [==============================] - 12s 303ms/step - loss: 0.6312 - accuracy: 0.7667 - val_loss: 0.8804 - val_accuracy: 0.6912\nEpoch 7/20\n39/39 [==============================] - 12s 303ms/step - loss: 0.6191 - accuracy: 0.7668 - val_loss: 1.0128 - val_accuracy: 0.6865\nEpoch 8/20\n39/39 [==============================] - 12s 307ms/step - loss: 0.5975 - accuracy: 0.7876 - val_loss: 0.7041 - val_accuracy: 0.7796\nEpoch 9/20\n39/39 [==============================] - 12s 305ms/step - loss: 0.5803 - accuracy: 0.7855 - val_loss: 0.6564 - val_accuracy: 0.7946\nEpoch 10/20\n39/39 [==============================] - 12s 303ms/step - loss: 0.5440 - accuracy: 0.7987 - val_loss: 0.8012 - val_accuracy: 0.7447\nEpoch 11/20\n39/39 [==============================] - 12s 304ms/step - loss: 0.6093 - accuracy: 0.7856 - val_loss: 0.6799 - val_accuracy: 0.7835\nEpoch 12/20\n39/39 [==============================] - 12s 304ms/step - loss: 0.5042 - accuracy: 0.8120 - val_loss: 0.7000 - val_accuracy: 0.7525\nEpoch 13/20\n39/39 [==============================] - 12s 305ms/step - loss: 0.5018 - accuracy: 0.8211 - val_loss: 0.7326 - val_accuracy: 0.7792\nEpoch 14/20\n39/39 [==============================] - 12s 305ms/step - loss: 0.4763 - accuracy: 0.8278 - val_loss: 0.7021 - val_accuracy: 0.7653\nEpoch 15/20\n39/39 [==============================] - 12s 305ms/step - loss: 0.4528 - accuracy: 0.8364 - val_loss: 0.8894 - val_accuracy: 0.6961\nEpoch 16/20\n39/39 [==============================] - 12s 304ms/step - loss: 0.4797 - accuracy: 0.8219 - val_loss: 0.9069 - val_accuracy: 0.7703\nEpoch 17/20\n39/39 [==============================] - 12s 303ms/step - loss: 0.4466 - accuracy: 0.8311 - val_loss: 0.7112 - val_accuracy: 0.7885\nEpoch 18/20\n39/39 [==============================] - 12s 306ms/step - loss: 0.4018 - accuracy: 0.8562 - val_loss: 0.7362 - val_accuracy: 0.7600\nEpoch 19/20\n39/39 [==============================] - 13s 334ms/step - loss: 0.4159 - accuracy: 0.8580 - val_loss: 0.8168 - val_accuracy: 0.7700\nEpoch 20/20\n39/39 [==============================] - 13s 334ms/step - loss: 0.4601 - accuracy: 0.8331 - val_loss: 1.2481 - val_accuracy: 0.6830\nDuration: 0:04:00.590220\n"
],
[
"print(n)\n\nmodels_nc[n].fit_model(image_sets_nc[n],label_sets_nc[n],x_val,y_val,epochs=20,batch_size = 128)\nn = n+1",
"7\nEpoch 1/20\n44/44 [==============================] - 16s 323ms/step - loss: 0.9002 - accuracy: 0.6857 - val_loss: 0.7370 - val_accuracy: 0.7279\nEpoch 2/20\n44/44 [==============================] - 5120s 119s/step - loss: 0.7426 - accuracy: 0.7226 - val_loss: 0.6289 - val_accuracy: 0.7835\nEpoch 3/20\n44/44 [==============================] - 16s 356ms/step - loss: 0.6855 - accuracy: 0.7557 - val_loss: 0.6996 - val_accuracy: 0.7518\nEpoch 4/20\n44/44 [==============================] - 15s 353ms/step - loss: 0.6767 - accuracy: 0.7492 - val_loss: 0.6475 - val_accuracy: 0.7853\nEpoch 5/20\n44/44 [==============================] - 14s 313ms/step - loss: 0.6401 - accuracy: 0.7683 - val_loss: 0.7150 - val_accuracy: 0.7618\nEpoch 6/20\n44/44 [==============================] - 14s 311ms/step - loss: 0.6420 - accuracy: 0.7724 - val_loss: 0.6166 - val_accuracy: 0.7917\nEpoch 7/20\n44/44 [==============================] - 14s 330ms/step - loss: 0.6187 - accuracy: 0.7750 - val_loss: 0.7452 - val_accuracy: 0.7596\nEpoch 8/20\n44/44 [==============================] - 14s 316ms/step - loss: 0.6032 - accuracy: 0.7820 - val_loss: 0.6769 - val_accuracy: 0.7710\nEpoch 9/20\n44/44 [==============================] - 17s 388ms/step - loss: 0.5585 - accuracy: 0.7899 - val_loss: 0.7841 - val_accuracy: 0.7386\nEpoch 10/20\n44/44 [==============================] - 17s 379ms/step - loss: 0.5826 - accuracy: 0.7881 - val_loss: 0.6916 - val_accuracy: 0.7853\nEpoch 11/20\n44/44 [==============================] - 17s 385ms/step - loss: 0.5246 - accuracy: 0.8102 - val_loss: 0.6455 - val_accuracy: 0.7860\nEpoch 12/20\n44/44 [==============================] - 18s 400ms/step - loss: 0.4985 - accuracy: 0.8196 - val_loss: 0.7129 - val_accuracy: 0.7832\nEpoch 13/20\n44/44 [==============================] - 17s 388ms/step - loss: 0.4972 - accuracy: 0.8227 - val_loss: 0.7374 - val_accuracy: 0.7721\nEpoch 14/20\n44/44 [==============================] - 16s 371ms/step - loss: 0.5037 - accuracy: 0.8248 - val_loss: 0.8742 - val_accuracy: 0.7461\nEpoch 15/20\n44/44 [==============================] - 15s 339ms/step - loss: 0.5086 - accuracy: 0.8230 - val_loss: 0.8567 - val_accuracy: 0.7721\nEpoch 16/20\n44/44 [==============================] - 15s 346ms/step - loss: 0.4737 - accuracy: 0.8293 - val_loss: 0.9519 - val_accuracy: 0.7543\nEpoch 17/20\n44/44 [==============================] - 14s 312ms/step - loss: 0.4511 - accuracy: 0.8347 - val_loss: 0.7585 - val_accuracy: 0.7693\nEpoch 18/20\n44/44 [==============================] - 17s 385ms/step - loss: 0.4227 - accuracy: 0.8470 - val_loss: 0.7117 - val_accuracy: 0.7874\nEpoch 19/20\n44/44 [==============================] - 14s 314ms/step - loss: 0.4108 - accuracy: 0.8430 - val_loss: 0.9171 - val_accuracy: 0.7743\nEpoch 20/20\n44/44 [==============================] - 14s 316ms/step - loss: 0.4224 - accuracy: 0.8501 - val_loss: 0.7697 - val_accuracy: 0.7700\nDuration: 1:30:13.001859\n"
],
[
"print(n)\n\nmodels_nc[n].fit_model(image_sets_nc[n],label_sets_nc[n],x_val,y_val,epochs=20,batch_size = 128)\nn = n+1",
"8\nEpoch 1/20\n50/50 [==============================] - 17s 312ms/step - loss: 0.9197 - accuracy: 0.6747 - val_loss: 0.6257 - val_accuracy: 0.7678\nEpoch 2/20\n50/50 [==============================] - 16s 315ms/step - loss: 0.7358 - accuracy: 0.7325 - val_loss: 0.7556 - val_accuracy: 0.7536\nEpoch 3/20\n50/50 [==============================] - 15s 310ms/step - loss: 0.7071 - accuracy: 0.7431 - val_loss: 1.0012 - val_accuracy: 0.7097\nEpoch 4/20\n50/50 [==============================] - 16s 325ms/step - loss: 0.7703 - accuracy: 0.7371 - val_loss: 0.8014 - val_accuracy: 0.7193\nEpoch 5/20\n50/50 [==============================] - 15s 311ms/step - loss: 0.6754 - accuracy: 0.7563 - val_loss: 0.7185 - val_accuracy: 0.7471\nEpoch 6/20\n50/50 [==============================] - 15s 305ms/step - loss: 0.6500 - accuracy: 0.7550 - val_loss: 0.7427 - val_accuracy: 0.7511\nEpoch 7/20\n50/50 [==============================] - 15s 305ms/step - loss: 0.6377 - accuracy: 0.7698 - val_loss: 0.6642 - val_accuracy: 0.7807\nEpoch 8/20\n50/50 [==============================] - 15s 305ms/step - loss: 0.5898 - accuracy: 0.7795 - val_loss: 0.7031 - val_accuracy: 0.7668\nEpoch 9/20\n50/50 [==============================] - 15s 305ms/step - loss: 0.5817 - accuracy: 0.7872 - val_loss: 0.7465 - val_accuracy: 0.7375\nEpoch 10/20\n50/50 [==============================] - 16s 312ms/step - loss: 0.5754 - accuracy: 0.7943 - val_loss: 0.6917 - val_accuracy: 0.7675\nEpoch 11/20\n50/50 [==============================] - 16s 314ms/step - loss: 0.5413 - accuracy: 0.8036 - val_loss: 0.7134 - val_accuracy: 0.7650\nEpoch 12/20\n50/50 [==============================] - 16s 330ms/step - loss: 0.5109 - accuracy: 0.8138 - val_loss: 0.7163 - val_accuracy: 0.7707\nEpoch 13/20\n50/50 [==============================] - 19s 376ms/step - loss: 0.5266 - accuracy: 0.8085 - val_loss: 0.7585 - val_accuracy: 0.7800\nEpoch 14/20\n50/50 [==============================] - 17s 347ms/step - loss: 0.4876 - accuracy: 0.8184 - val_loss: 1.1353 - val_accuracy: 0.7222\nEpoch 15/20\n50/50 [==============================] - 16s 313ms/step - loss: 0.4992 - accuracy: 0.8119 - val_loss: 0.9427 - val_accuracy: 0.7611\nEpoch 16/20\n50/50 [==============================] - 17s 340ms/step - loss: 0.5282 - accuracy: 0.8108 - val_loss: 0.9353 - val_accuracy: 0.7186\nEpoch 17/20\n50/50 [==============================] - 17s 332ms/step - loss: 0.4999 - accuracy: 0.8259 - val_loss: 0.6802 - val_accuracy: 0.7803\nEpoch 18/20\n50/50 [==============================] - 18s 355ms/step - loss: 0.4578 - accuracy: 0.8395 - val_loss: 0.8432 - val_accuracy: 0.7636\nEpoch 19/20\n50/50 [==============================] - 19s 373ms/step - loss: 0.4552 - accuracy: 0.8391 - val_loss: 0.7463 - val_accuracy: 0.7439\nEpoch 20/20\n50/50 [==============================] - 16s 312ms/step - loss: 0.4397 - accuracy: 0.8383 - val_loss: 0.9218 - val_accuracy: 0.7183\nDuration: 0:05:25.834807\n"
],
[
"print(n)#9\n\nmodels_nc[n].fit_model(image_sets_nc[n],label_sets_nc[n],x_val,y_val,epochs=20,batch_size = 128)\nn = n+1",
"9\nEpoch 1/20\n55/55 [==============================] - 21s 347ms/step - loss: 0.9754 - accuracy: 0.6676 - val_loss: 0.5735 - val_accuracy: 0.7956\nEpoch 2/20\n55/55 [==============================] - 20s 371ms/step - loss: 0.7849 - accuracy: 0.7096 - val_loss: 0.6698 - val_accuracy: 0.7636\nEpoch 3/20\n55/55 [==============================] - 19s 350ms/step - loss: 0.7105 - accuracy: 0.7402 - val_loss: 0.7166 - val_accuracy: 0.7618\nEpoch 4/20\n55/55 [==============================] - 19s 337ms/step - loss: 0.6968 - accuracy: 0.7455 - val_loss: 0.6185 - val_accuracy: 0.7871\nEpoch 5/20\n55/55 [==============================] - 18s 330ms/step - loss: 0.6454 - accuracy: 0.7541 - val_loss: 0.6676 - val_accuracy: 0.7746\nEpoch 6/20\n55/55 [==============================] - 18s 328ms/step - loss: 0.6610 - accuracy: 0.7571 - val_loss: 0.7348 - val_accuracy: 0.7660\nEpoch 7/20\n55/55 [==============================] - 19s 347ms/step - loss: 0.6397 - accuracy: 0.7612 - val_loss: 0.5957 - val_accuracy: 0.8006\nEpoch 8/20\n55/55 [==============================] - 18s 332ms/step - loss: 0.6164 - accuracy: 0.7745 - val_loss: 0.6446 - val_accuracy: 0.7874\nEpoch 9/20\n55/55 [==============================] - 18s 324ms/step - loss: 0.5725 - accuracy: 0.7804 - val_loss: 0.8787 - val_accuracy: 0.7208\nEpoch 10/20\n55/55 [==============================] - 18s 322ms/step - loss: 0.5525 - accuracy: 0.7930 - val_loss: 0.6970 - val_accuracy: 0.7846\nEpoch 11/20\n55/55 [==============================] - 20s 363ms/step - loss: 0.5860 - accuracy: 0.7852 - val_loss: 0.6936 - val_accuracy: 0.7832\nEpoch 12/20\n55/55 [==============================] - 19s 337ms/step - loss: 0.5479 - accuracy: 0.8019 - val_loss: 0.7029 - val_accuracy: 0.7878\nEpoch 13/20\n55/55 [==============================] - 19s 340ms/step - loss: 0.5300 - accuracy: 0.8028 - val_loss: 0.6920 - val_accuracy: 0.7757\nEpoch 14/20\n55/55 [==============================] - 17s 309ms/step - loss: 0.5004 - accuracy: 0.8176 - val_loss: 0.7095 - val_accuracy: 0.7707\nEpoch 15/20\n55/55 [==============================] - 18s 322ms/step - loss: 0.4807 - accuracy: 0.8247 - val_loss: 0.7817 - val_accuracy: 0.7600\nEpoch 16/20\n55/55 [==============================] - 17s 310ms/step - loss: 0.4806 - accuracy: 0.8231 - val_loss: 0.7263 - val_accuracy: 0.7700\nEpoch 17/20\n55/55 [==============================] - 17s 312ms/step - loss: 0.4565 - accuracy: 0.8395 - val_loss: 0.7678 - val_accuracy: 0.7817\nEpoch 18/20\n55/55 [==============================] - 17s 314ms/step - loss: 0.4864 - accuracy: 0.8255 - val_loss: 0.8624 - val_accuracy: 0.7689\nEpoch 19/20\n55/55 [==============================] - 17s 310ms/step - loss: 0.4570 - accuracy: 0.8359 - val_loss: 0.7889 - val_accuracy: 0.7871\nEpoch 20/20\n55/55 [==============================] - 18s 326ms/step - loss: 0.4345 - accuracy: 0.8410 - val_loss: 0.7966 - val_accuracy: 0.7850\nDuration: 0:06:05.802695\n"
],
[
"print(n)\n\nmodels_nc[n].fit_model(image_sets_nc[n],label_sets_nc[n],x_val,y_val,epochs=20,batch_size = 128)\nn = n+1",
"10\nEpoch 1/20\n61/61 [==============================] - 22s 325ms/step - loss: 0.9880 - accuracy: 0.6588 - val_loss: 0.8487 - val_accuracy: 0.6673\nEpoch 2/20\n61/61 [==============================] - 19s 314ms/step - loss: 0.7937 - accuracy: 0.6956 - val_loss: 0.6868 - val_accuracy: 0.7603\nEpoch 3/20\n61/61 [==============================] - 19s 309ms/step - loss: 0.7298 - accuracy: 0.7254 - val_loss: 0.6146 - val_accuracy: 0.7728\nEpoch 4/20\n61/61 [==============================] - 19s 313ms/step - loss: 0.6865 - accuracy: 0.7453 - val_loss: 0.7555 - val_accuracy: 0.7411\nEpoch 5/20\n61/61 [==============================] - 19s 313ms/step - loss: 0.6703 - accuracy: 0.7520 - val_loss: 1.0765 - val_accuracy: 0.6740\nEpoch 6/20\n61/61 [==============================] - 19s 304ms/step - loss: 0.7311 - accuracy: 0.7424 - val_loss: 0.6852 - val_accuracy: 0.7614\nEpoch 7/20\n61/61 [==============================] - 18s 301ms/step - loss: 0.6248 - accuracy: 0.7641 - val_loss: 1.0415 - val_accuracy: 0.6648\nEpoch 8/20\n61/61 [==============================] - 18s 301ms/step - loss: 0.6482 - accuracy: 0.7641 - val_loss: 0.6234 - val_accuracy: 0.7882\nEpoch 9/20\n61/61 [==============================] - 19s 314ms/step - loss: 0.6039 - accuracy: 0.7738 - val_loss: 0.9409 - val_accuracy: 0.7561\nEpoch 10/20\n61/61 [==============================] - 19s 305ms/step - loss: 0.6074 - accuracy: 0.7722 - val_loss: 0.6868 - val_accuracy: 0.7703\nEpoch 11/20\n61/61 [==============================] - 20s 326ms/step - loss: 0.5665 - accuracy: 0.7912 - val_loss: 0.8233 - val_accuracy: 0.7675\nEpoch 12/20\n61/61 [==============================] - 19s 317ms/step - loss: 0.5688 - accuracy: 0.7860 - val_loss: 0.8214 - val_accuracy: 0.7553\nEpoch 13/20\n61/61 [==============================] - 19s 305ms/step - loss: 0.5443 - accuracy: 0.7962 - val_loss: 0.7688 - val_accuracy: 0.7725\nEpoch 14/20\n61/61 [==============================] - 19s 319ms/step - loss: 0.5309 - accuracy: 0.8074 - val_loss: 0.6919 - val_accuracy: 0.7896\nEpoch 15/20\n61/61 [==============================] - 18s 301ms/step - loss: 0.5019 - accuracy: 0.8137 - val_loss: 0.7262 - val_accuracy: 0.7832\nEpoch 16/20\n61/61 [==============================] - 18s 301ms/step - loss: 0.4592 - accuracy: 0.8320 - val_loss: 1.0055 - val_accuracy: 0.7304\nEpoch 17/20\n61/61 [==============================] - 19s 308ms/step - loss: 0.5269 - accuracy: 0.8212 - val_loss: 0.8181 - val_accuracy: 0.7386\nEpoch 18/20\n61/61 [==============================] - 19s 305ms/step - loss: 0.4679 - accuracy: 0.8237 - val_loss: 0.8004 - val_accuracy: 0.7682\nEpoch 19/20\n61/61 [==============================] - 19s 306ms/step - loss: 0.4539 - accuracy: 0.8330 - val_loss: 0.7543 - val_accuracy: 0.7632\nEpoch 20/20\n61/61 [==============================] - 21s 338ms/step - loss: 0.4566 - accuracy: 0.8281 - val_loss: 0.8863 - val_accuracy: 0.7782\nDuration: 0:06:21.124306\n"
],
[
"print(n)\n\nmodels_nc[n].fit_model(image_sets_nc[n],label_sets_nc[n],x_val,y_val,epochs=20,batch_size = 128)\nn = n+1",
"11\nEpoch 1/20\n66/66 [==============================] - 26s 356ms/step - loss: 0.8878 - accuracy: 0.6736 - val_loss: 0.7346 - val_accuracy: 0.7482\nEpoch 2/20\n66/66 [==============================] - 22s 336ms/step - loss: 0.7644 - accuracy: 0.7174 - val_loss: 0.6456 - val_accuracy: 0.7817\nEpoch 3/20\n66/66 [==============================] - 22s 332ms/step - loss: 0.7159 - accuracy: 0.7337 - val_loss: 0.6096 - val_accuracy: 0.7885\nEpoch 4/20\n66/66 [==============================] - 21s 313ms/step - loss: 0.7062 - accuracy: 0.7359 - val_loss: 0.6286 - val_accuracy: 0.7835\nEpoch 5/20\n66/66 [==============================] - 20s 308ms/step - loss: 0.6621 - accuracy: 0.7530 - val_loss: 0.8252 - val_accuracy: 0.7279\nEpoch 6/20\n66/66 [==============================] - 21s 319ms/step - loss: 0.6782 - accuracy: 0.7568 - val_loss: 0.6438 - val_accuracy: 0.7817\nEpoch 7/20\n66/66 [==============================] - 22s 326ms/step - loss: 0.6212 - accuracy: 0.7663 - val_loss: 0.9280 - val_accuracy: 0.7218\nEpoch 8/20\n66/66 [==============================] - 21s 319ms/step - loss: 0.6154 - accuracy: 0.7738 - val_loss: 0.6831 - val_accuracy: 0.7850\nEpoch 9/20\n66/66 [==============================] - 22s 328ms/step - loss: 0.5995 - accuracy: 0.7775 - val_loss: 0.7335 - val_accuracy: 0.7628\nEpoch 10/20\n66/66 [==============================] - 22s 331ms/step - loss: 0.5722 - accuracy: 0.7863 - val_loss: 0.7133 - val_accuracy: 0.7714\nEpoch 11/20\n66/66 [==============================] - 22s 333ms/step - loss: 0.5428 - accuracy: 0.7974 - val_loss: 0.7517 - val_accuracy: 0.7450\nEpoch 12/20\n66/66 [==============================] - 21s 312ms/step - loss: 0.5296 - accuracy: 0.8017 - val_loss: 0.7957 - val_accuracy: 0.7639\nEpoch 13/20\n66/66 [==============================] - 21s 326ms/step - loss: 0.5421 - accuracy: 0.8025 - val_loss: 0.6812 - val_accuracy: 0.7871\nEpoch 14/20\n66/66 [==============================] - 22s 331ms/step - loss: 0.5256 - accuracy: 0.8051 - val_loss: 0.7506 - val_accuracy: 0.7735\nEpoch 15/20\n66/66 [==============================] - 22s 327ms/step - loss: 0.5172 - accuracy: 0.7982 - val_loss: 0.8808 - val_accuracy: 0.7368\nEpoch 16/20\n66/66 [==============================] - 21s 320ms/step - loss: 0.5140 - accuracy: 0.8137 - val_loss: 0.7975 - val_accuracy: 0.7596\nEpoch 17/20\n66/66 [==============================] - 21s 325ms/step - loss: 0.4669 - accuracy: 0.8301 - val_loss: 0.7681 - val_accuracy: 0.7728\nEpoch 18/20\n66/66 [==============================] - 22s 335ms/step - loss: 0.4785 - accuracy: 0.8241 - val_loss: 0.7486 - val_accuracy: 0.7614\nEpoch 19/20\n66/66 [==============================] - 22s 336ms/step - loss: 0.4621 - accuracy: 0.8355 - val_loss: 0.7771 - val_accuracy: 0.7707\nEpoch 20/20\n66/66 [==============================] - 21s 321ms/step - loss: 0.4480 - accuracy: 0.8359 - val_loss: 0.7162 - val_accuracy: 0.7817\nDuration: 0:07:13.530899\n"
],
[
"print(n)\n\nmodels_nc[n].fit_model(image_sets_nc[n],label_sets_nc[n],x_val,y_val,epochs=20,batch_size = 128)\nn = n+1",
"12\nEpoch 1/20\n72/72 [==============================] - 27s 341ms/step - loss: 0.9053 - accuracy: 0.6713 - val_loss: 0.8258 - val_accuracy: 0.6908\nEpoch 2/20\n72/72 [==============================] - 25s 344ms/step - loss: 0.7928 - accuracy: 0.7051 - val_loss: 1.1022 - val_accuracy: 0.6259\nEpoch 3/20\n72/72 [==============================] - 22s 312ms/step - loss: 0.7678 - accuracy: 0.7143 - val_loss: 0.7916 - val_accuracy: 0.7218\nEpoch 4/20\n72/72 [==============================] - 23s 315ms/step - loss: 0.7286 - accuracy: 0.7311 - val_loss: 0.7726 - val_accuracy: 0.7389\nEpoch 5/20\n72/72 [==============================] - 24s 338ms/step - loss: 0.6901 - accuracy: 0.7392 - val_loss: 0.7844 - val_accuracy: 0.7628\nEpoch 6/20\n72/72 [==============================] - 22s 304ms/step - loss: 0.6656 - accuracy: 0.7499 - val_loss: 0.7866 - val_accuracy: 0.7511\nEpoch 7/20\n72/72 [==============================] - 22s 302ms/step - loss: 0.6463 - accuracy: 0.7592 - val_loss: 0.7559 - val_accuracy: 0.7625\nEpoch 8/20\n72/72 [==============================] - 22s 303ms/step - loss: 0.6502 - accuracy: 0.7624 - val_loss: 0.6964 - val_accuracy: 0.7664\nEpoch 9/20\n72/72 [==============================] - 22s 303ms/step - loss: 0.6261 - accuracy: 0.7714 - val_loss: 0.7785 - val_accuracy: 0.7450\nEpoch 10/20\n72/72 [==============================] - 22s 302ms/step - loss: 0.6020 - accuracy: 0.7770 - val_loss: 0.6736 - val_accuracy: 0.7828\nEpoch 11/20\n72/72 [==============================] - 22s 302ms/step - loss: 0.5833 - accuracy: 0.7795 - val_loss: 0.7668 - val_accuracy: 0.7350\nEpoch 12/20\n72/72 [==============================] - 22s 306ms/step - loss: 0.5559 - accuracy: 0.7919 - val_loss: 0.6892 - val_accuracy: 0.7960\nEpoch 13/20\n72/72 [==============================] - 22s 313ms/step - loss: 0.5343 - accuracy: 0.8023 - val_loss: 0.8827 - val_accuracy: 0.7372\nEpoch 14/20\n72/72 [==============================] - 24s 333ms/step - loss: 0.5667 - accuracy: 0.7910 - val_loss: 0.9472 - val_accuracy: 0.6894\nEpoch 15/20\n72/72 [==============================] - 24s 337ms/step - loss: 0.5550 - accuracy: 0.7931 - val_loss: 0.7279 - val_accuracy: 0.7696\nEpoch 16/20\n72/72 [==============================] - 22s 308ms/step - loss: 0.5316 - accuracy: 0.7988 - val_loss: 0.8619 - val_accuracy: 0.7129\nEpoch 17/20\n72/72 [==============================] - 22s 307ms/step - loss: 0.5343 - accuracy: 0.8046 - val_loss: 0.7651 - val_accuracy: 0.7721\nEpoch 18/20\n72/72 [==============================] - 22s 304ms/step - loss: 0.5069 - accuracy: 0.8158 - val_loss: 1.1289 - val_accuracy: 0.7168\nEpoch 19/20\n72/72 [==============================] - 22s 302ms/step - loss: 0.4718 - accuracy: 0.8306 - val_loss: 0.8796 - val_accuracy: 0.7083\nEpoch 20/20\n72/72 [==============================] - 22s 307ms/step - loss: 0.4835 - accuracy: 0.8232 - val_loss: 0.8053 - val_accuracy: 0.7375\nDuration: 0:07:34.369681\n"
],
[
"print(n)\n\nmodels_nc[n].fit_model(image_sets_nc[n],label_sets_nc[n],x_val,y_val,epochs=20,batch_size = 128)\nn = n+1",
"13\nEpoch 1/20\n77/77 [==============================] - 26s 311ms/step - loss: 0.9006 - accuracy: 0.6747 - val_loss: 0.6444 - val_accuracy: 0.7807\nEpoch 2/20\n77/77 [==============================] - 23s 303ms/step - loss: 0.7671 - accuracy: 0.7196 - val_loss: 0.7991 - val_accuracy: 0.6972\nEpoch 3/20\n77/77 [==============================] - 23s 302ms/step - loss: 0.7320 - accuracy: 0.7321 - val_loss: 0.5837 - val_accuracy: 0.7996\nEpoch 4/20\n77/77 [==============================] - 23s 302ms/step - loss: 0.6824 - accuracy: 0.7358 - val_loss: 0.9115 - val_accuracy: 0.6937\nEpoch 5/20\n77/77 [==============================] - 23s 303ms/step - loss: 0.6776 - accuracy: 0.7431 - val_loss: 0.6454 - val_accuracy: 0.7767\nEpoch 6/20\n77/77 [==============================] - 23s 302ms/step - loss: 0.6691 - accuracy: 0.7525 - val_loss: 1.0005 - val_accuracy: 0.6705\nEpoch 7/20\n77/77 [==============================] - 25s 319ms/step - loss: 0.6239 - accuracy: 0.7682 - val_loss: 0.6233 - val_accuracy: 0.7889\nEpoch 8/20\n77/77 [==============================] - 24s 307ms/step - loss: 0.6190 - accuracy: 0.7700 - val_loss: 0.6211 - val_accuracy: 0.7907\nEpoch 9/20\n77/77 [==============================] - 23s 304ms/step - loss: 0.5864 - accuracy: 0.7830 - val_loss: 0.7127 - val_accuracy: 0.7553\nEpoch 10/20\n77/77 [==============================] - 23s 303ms/step - loss: 0.5827 - accuracy: 0.7751 - val_loss: 0.6727 - val_accuracy: 0.7714\nEpoch 11/20\n77/77 [==============================] - 23s 304ms/step - loss: 0.5702 - accuracy: 0.7883 - val_loss: 0.6689 - val_accuracy: 0.7718\nEpoch 12/20\n77/77 [==============================] - 23s 305ms/step - loss: 0.5573 - accuracy: 0.7882 - val_loss: 0.7513 - val_accuracy: 0.7364\nEpoch 13/20\n77/77 [==============================] - 23s 304ms/step - loss: 0.5414 - accuracy: 0.7928 - val_loss: 0.6263 - val_accuracy: 0.7939\nEpoch 14/20\n77/77 [==============================] - 23s 303ms/step - loss: 0.5361 - accuracy: 0.7991 - val_loss: 0.6489 - val_accuracy: 0.7857\nEpoch 15/20\n77/77 [==============================] - 23s 303ms/step - loss: 0.5123 - accuracy: 0.8090 - val_loss: 0.7006 - val_accuracy: 0.7835\nEpoch 16/20\n77/77 [==============================] - 23s 302ms/step - loss: 0.4912 - accuracy: 0.8181 - val_loss: 0.6379 - val_accuracy: 0.7981\nEpoch 17/20\n77/77 [==============================] - 23s 304ms/step - loss: 0.4734 - accuracy: 0.8219 - val_loss: 0.7092 - val_accuracy: 0.7685\nEpoch 18/20\n77/77 [==============================] - 23s 303ms/step - loss: 0.4881 - accuracy: 0.8207 - val_loss: 0.7114 - val_accuracy: 0.7660\nEpoch 19/20\n77/77 [==============================] - 23s 303ms/step - loss: 0.4515 - accuracy: 0.8320 - val_loss: 0.6752 - val_accuracy: 0.7785\nEpoch 20/20\n77/77 [==============================] - 24s 309ms/step - loss: 0.4185 - accuracy: 0.8474 - val_loss: 0.7552 - val_accuracy: 0.7504\nDuration: 0:07:50.755710\n"
],
[
"print(n)#14\n\nmodels_nc[n].fit_model(image_sets_nc[n],label_sets_nc[n],x_val,y_val,epochs=20,batch_size = 128)\nn = n+1",
"14\nEpoch 1/20\n83/83 [==============================] - 27s 303ms/step - loss: 0.8818 - accuracy: 0.6831 - val_loss: 0.7968 - val_accuracy: 0.6997\nEpoch 2/20\n83/83 [==============================] - 25s 299ms/step - loss: 0.7660 - accuracy: 0.7161 - val_loss: 0.8203 - val_accuracy: 0.7236\nEpoch 3/20\n83/83 [==============================] - 25s 299ms/step - loss: 0.7508 - accuracy: 0.7227 - val_loss: 0.6854 - val_accuracy: 0.7660\nEpoch 4/20\n83/83 [==============================] - 25s 299ms/step - loss: 0.7048 - accuracy: 0.7380 - val_loss: 0.9760 - val_accuracy: 0.6498\nEpoch 5/20\n83/83 [==============================] - 25s 303ms/step - loss: 0.6890 - accuracy: 0.7453 - val_loss: 0.7398 - val_accuracy: 0.7454\nEpoch 6/20\n83/83 [==============================] - 25s 301ms/step - loss: 0.6604 - accuracy: 0.7514 - val_loss: 0.7243 - val_accuracy: 0.7832\nEpoch 7/20\n83/83 [==============================] - 25s 301ms/step - loss: 0.6683 - accuracy: 0.7547 - val_loss: 1.3760 - val_accuracy: 0.5631\nEpoch 8/20\n83/83 [==============================] - 25s 298ms/step - loss: 0.6955 - accuracy: 0.7501 - val_loss: 1.1584 - val_accuracy: 0.6155\nEpoch 9/20\n83/83 [==============================] - 25s 299ms/step - loss: 0.6839 - accuracy: 0.7525 - val_loss: 1.0667 - val_accuracy: 0.6287\nEpoch 10/20\n83/83 [==============================] - 25s 300ms/step - loss: 0.6173 - accuracy: 0.7697 - val_loss: 0.9503 - val_accuracy: 0.6683\nEpoch 11/20\n83/83 [==============================] - 25s 298ms/step - loss: 0.5820 - accuracy: 0.7805 - val_loss: 0.7903 - val_accuracy: 0.7529\nEpoch 12/20\n83/83 [==============================] - 25s 303ms/step - loss: 0.5879 - accuracy: 0.7845 - val_loss: 0.6203 - val_accuracy: 0.8010\nEpoch 13/20\n83/83 [==============================] - 25s 300ms/step - loss: 0.5593 - accuracy: 0.7935 - val_loss: 0.7111 - val_accuracy: 0.7389\nEpoch 14/20\n83/83 [==============================] - 25s 298ms/step - loss: 0.5399 - accuracy: 0.8013 - val_loss: 0.6700 - val_accuracy: 0.7732\nEpoch 15/20\n83/83 [==============================] - 25s 305ms/step - loss: 0.5281 - accuracy: 0.8036 - val_loss: 0.7644 - val_accuracy: 0.7197\nEpoch 16/20\n83/83 [==============================] - 26s 316ms/step - loss: 0.5129 - accuracy: 0.8129 - val_loss: 0.7488 - val_accuracy: 0.7379\nEpoch 17/20\n83/83 [==============================] - 25s 299ms/step - loss: 0.5185 - accuracy: 0.8138 - val_loss: 0.6317 - val_accuracy: 0.7942\nEpoch 18/20\n83/83 [==============================] - 25s 303ms/step - loss: 0.4954 - accuracy: 0.8161 - val_loss: 0.8336 - val_accuracy: 0.7450\nEpoch 19/20\n83/83 [==============================] - 26s 310ms/step - loss: 0.4692 - accuracy: 0.8248 - val_loss: 1.0662 - val_accuracy: 0.6430\nEpoch 20/20\n83/83 [==============================] - 27s 324ms/step - loss: 0.4750 - accuracy: 0.8267 - val_loss: 0.7093 - val_accuracy: 0.7967\nDuration: 0:08:24.320607\n"
],
[
"print(n)\n\nmodels_nc[n].fit_model(image_sets_nc[n],label_sets_nc[n],x_val,y_val,epochs=20,batch_size = 128)\nn = n+1",
"15\nEpoch 1/20\n88/88 [==============================] - 33s 354ms/step - loss: 0.9361 - accuracy: 0.6765 - val_loss: 0.6157 - val_accuracy: 0.7760\nEpoch 2/20\n88/88 [==============================] - 29s 329ms/step - loss: 0.7561 - accuracy: 0.7174 - val_loss: 0.7046 - val_accuracy: 0.7511\nEpoch 3/20\n88/88 [==============================] - 29s 331ms/step - loss: 0.7541 - accuracy: 0.7204 - val_loss: 0.6416 - val_accuracy: 0.7685\nEpoch 4/20\n88/88 [==============================] - 30s 336ms/step - loss: 0.6982 - accuracy: 0.7406 - val_loss: 0.5963 - val_accuracy: 0.7864\nEpoch 5/20\n88/88 [==============================] - 29s 331ms/step - loss: 0.6808 - accuracy: 0.7405 - val_loss: 0.5996 - val_accuracy: 0.8006\nEpoch 6/20\n88/88 [==============================] - 28s 322ms/step - loss: 0.6608 - accuracy: 0.7502 - val_loss: 0.6859 - val_accuracy: 0.7660\nEpoch 7/20\n88/88 [==============================] - 30s 336ms/step - loss: 0.6243 - accuracy: 0.7658 - val_loss: 0.5623 - val_accuracy: 0.8010\nEpoch 8/20\n88/88 [==============================] - 30s 337ms/step - loss: 0.6325 - accuracy: 0.7628 - val_loss: 0.6536 - val_accuracy: 0.7696\nEpoch 9/20\n88/88 [==============================] - 28s 324ms/step - loss: 0.6139 - accuracy: 0.7739 - val_loss: 0.6075 - val_accuracy: 0.7935\nEpoch 10/20\n88/88 [==============================] - 31s 350ms/step - loss: 0.5833 - accuracy: 0.7822 - val_loss: 0.5856 - val_accuracy: 0.7960\nEpoch 11/20\n88/88 [==============================] - 29s 334ms/step - loss: 0.5509 - accuracy: 0.7961 - val_loss: 0.6019 - val_accuracy: 0.8003\nEpoch 12/20\n88/88 [==============================] - 29s 329ms/step - loss: 0.5656 - accuracy: 0.7944 - val_loss: 0.8565 - val_accuracy: 0.6901\nEpoch 13/20\n88/88 [==============================] - 29s 335ms/step - loss: 0.5830 - accuracy: 0.7886 - val_loss: 0.6579 - val_accuracy: 0.7789\nEpoch 14/20\n88/88 [==============================] - 30s 339ms/step - loss: 0.5105 - accuracy: 0.8095 - val_loss: 0.7098 - val_accuracy: 0.7643\nEpoch 15/20\n88/88 [==============================] - 29s 328ms/step - loss: 0.5132 - accuracy: 0.8066 - val_loss: 0.6066 - val_accuracy: 0.7942\nEpoch 16/20\n88/88 [==============================] - 32s 363ms/step - loss: 0.4814 - accuracy: 0.8205 - val_loss: 0.6201 - val_accuracy: 0.7942\nEpoch 17/20\n88/88 [==============================] - 30s 336ms/step - loss: 0.4772 - accuracy: 0.8210 - val_loss: 0.6149 - val_accuracy: 0.7956\nEpoch 18/20\n88/88 [==============================] - 29s 332ms/step - loss: 0.4719 - accuracy: 0.8273 - val_loss: 0.7119 - val_accuracy: 0.7628\nEpoch 19/20\n88/88 [==============================] - 29s 326ms/step - loss: 0.4652 - accuracy: 0.8288 - val_loss: 0.6452 - val_accuracy: 0.7825\nEpoch 20/20\n88/88 [==============================] - 29s 328ms/step - loss: 0.4536 - accuracy: 0.8316 - val_loss: 0.6915 - val_accuracy: 0.8039\nDuration: 0:09:50.655437\n"
],
[
"print(n)\n\nmodels_nc[n].fit_model(image_sets_nc[n],label_sets_nc[n],x_val,y_val,epochs=20,batch_size = 128)\nn = n+1",
"16\nEpoch 1/20\n93/93 [==============================] - 33s 335ms/step - loss: 0.8748 - accuracy: 0.6891 - val_loss: 0.6865 - val_accuracy: 0.7899\nEpoch 2/20\n93/93 [==============================] - 31s 335ms/step - loss: 0.7378 - accuracy: 0.7282 - val_loss: 0.5928 - val_accuracy: 0.7917\nEpoch 3/20\n93/93 [==============================] - 29s 310ms/step - loss: 0.7099 - accuracy: 0.7342 - val_loss: 0.5822 - val_accuracy: 0.8049\nEpoch 4/20\n93/93 [==============================] - 30s 320ms/step - loss: 0.6988 - accuracy: 0.7371 - val_loss: 0.6082 - val_accuracy: 0.7867\nEpoch 5/20\n93/93 [==============================] - 31s 330ms/step - loss: 0.6626 - accuracy: 0.7532 - val_loss: 0.5697 - val_accuracy: 0.7928\nEpoch 6/20\n93/93 [==============================] - 32s 340ms/step - loss: 0.6319 - accuracy: 0.7645 - val_loss: 0.6369 - val_accuracy: 0.7825\nEpoch 7/20\n93/93 [==============================] - 31s 335ms/step - loss: 0.6292 - accuracy: 0.7653 - val_loss: 0.5772 - val_accuracy: 0.7949\nEpoch 8/20\n93/93 [==============================] - 30s 327ms/step - loss: 0.6112 - accuracy: 0.7742 - val_loss: 0.6204 - val_accuracy: 0.7821\nEpoch 9/20\n93/93 [==============================] - 30s 325ms/step - loss: 0.5860 - accuracy: 0.7784 - val_loss: 0.5842 - val_accuracy: 0.7946\nEpoch 10/20\n93/93 [==============================] - 32s 338ms/step - loss: 0.5790 - accuracy: 0.7814 - val_loss: 0.5721 - val_accuracy: 0.7992\nEpoch 11/20\n93/93 [==============================] - 31s 338ms/step - loss: 0.5490 - accuracy: 0.7982 - val_loss: 0.5553 - val_accuracy: 0.7978\nEpoch 12/20\n93/93 [==============================] - 32s 345ms/step - loss: 0.5517 - accuracy: 0.7967 - val_loss: 0.5626 - val_accuracy: 0.8060\nEpoch 13/20\n93/93 [==============================] - 30s 328ms/step - loss: 0.5496 - accuracy: 0.7915 - val_loss: 0.6467 - val_accuracy: 0.7632\nEpoch 14/20\n93/93 [==============================] - 31s 338ms/step - loss: 0.5263 - accuracy: 0.7999 - val_loss: 0.6754 - val_accuracy: 0.7582\nEpoch 15/20\n93/93 [==============================] - 35s 379ms/step - loss: 0.5007 - accuracy: 0.8116 - val_loss: 0.5647 - val_accuracy: 0.8021\nEpoch 16/20\n93/93 [==============================] - 32s 341ms/step - loss: 0.4906 - accuracy: 0.8190 - val_loss: 0.6313 - val_accuracy: 0.7685\nEpoch 17/20\n93/93 [==============================] - 31s 330ms/step - loss: 0.4786 - accuracy: 0.8209 - val_loss: 0.6436 - val_accuracy: 0.7867\nEpoch 18/20\n93/93 [==============================] - 31s 331ms/step - loss: 0.4720 - accuracy: 0.8274 - val_loss: 0.5937 - val_accuracy: 0.8071\nEpoch 19/20\n93/93 [==============================] - 30s 321ms/step - loss: 0.4487 - accuracy: 0.8316 - val_loss: 0.6092 - val_accuracy: 0.7946\nEpoch 20/20\n93/93 [==============================] - 30s 325ms/step - loss: 0.4390 - accuracy: 0.8377 - val_loss: 0.6178 - val_accuracy: 0.8031\nDuration: 0:10:21.933504\n"
],
[
"print(n)\n\nmodels_nc[n].fit_model(image_sets_nc[n],label_sets_nc[n],x_val,y_val,epochs=20,batch_size = 128)\nn = n+1",
"17\nEpoch 1/20\n99/99 [==============================] - 36s 338ms/step - loss: 0.9040 - accuracy: 0.6817 - val_loss: 0.6053 - val_accuracy: 0.7871\nEpoch 2/20\n99/99 [==============================] - 33s 332ms/step - loss: 0.7538 - accuracy: 0.7266 - val_loss: 0.6326 - val_accuracy: 0.7668\nEpoch 3/20\n99/99 [==============================] - 32s 323ms/step - loss: 0.7327 - accuracy: 0.7283 - val_loss: 0.6089 - val_accuracy: 0.7903\nEpoch 4/20\n99/99 [==============================] - 32s 326ms/step - loss: 0.6935 - accuracy: 0.7477 - val_loss: 0.7835 - val_accuracy: 0.7200\nEpoch 5/20\n99/99 [==============================] - 32s 323ms/step - loss: 0.6986 - accuracy: 0.7413 - val_loss: 0.7709 - val_accuracy: 0.7650\nEpoch 6/20\n99/99 [==============================] - 32s 326ms/step - loss: 0.6578 - accuracy: 0.7573 - val_loss: 0.6213 - val_accuracy: 0.7803\nEpoch 7/20\n99/99 [==============================] - 31s 315ms/step - loss: 0.6281 - accuracy: 0.7663 - val_loss: 0.5922 - val_accuracy: 0.7967\nEpoch 8/20\n99/99 [==============================] - 32s 324ms/step - loss: 0.6340 - accuracy: 0.7607 - val_loss: 0.6096 - val_accuracy: 0.7924\nEpoch 9/20\n99/99 [==============================] - 33s 329ms/step - loss: 0.6290 - accuracy: 0.7665 - val_loss: 0.6373 - val_accuracy: 0.7707\nEpoch 10/20\n99/99 [==============================] - 32s 322ms/step - loss: 0.5740 - accuracy: 0.7876 - val_loss: 0.5580 - val_accuracy: 0.8056\nEpoch 11/20\n99/99 [==============================] - 32s 325ms/step - loss: 0.5651 - accuracy: 0.7864 - val_loss: 0.5485 - val_accuracy: 0.8067\nEpoch 12/20\n99/99 [==============================] - 33s 332ms/step - loss: 0.5354 - accuracy: 0.7990 - val_loss: 0.5904 - val_accuracy: 0.7971\nEpoch 13/20\n99/99 [==============================] - 34s 342ms/step - loss: 0.5250 - accuracy: 0.8023 - val_loss: 0.5815 - val_accuracy: 0.7949\nEpoch 14/20\n99/99 [==============================] - 32s 324ms/step - loss: 0.5397 - accuracy: 0.8021 - val_loss: 0.6593 - val_accuracy: 0.7653\nEpoch 15/20\n99/99 [==============================] - 32s 325ms/step - loss: 0.5115 - accuracy: 0.8079 - val_loss: 0.6640 - val_accuracy: 0.7860\nEpoch 16/20\n99/99 [==============================] - 32s 323ms/step - loss: 0.5033 - accuracy: 0.8160 - val_loss: 0.6453 - val_accuracy: 0.7853\nEpoch 17/20\n99/99 [==============================] - 32s 322ms/step - loss: 0.4686 - accuracy: 0.8279 - val_loss: 0.5877 - val_accuracy: 0.8031\nEpoch 18/20\n99/99 [==============================] - 32s 321ms/step - loss: 0.4561 - accuracy: 0.8293 - val_loss: 0.6229 - val_accuracy: 0.7999\nEpoch 19/20\n99/99 [==============================] - 32s 323ms/step - loss: 0.4657 - accuracy: 0.8292 - val_loss: 0.6023 - val_accuracy: 0.7985\nEpoch 20/20\n99/99 [==============================] - 33s 331ms/step - loss: 0.4407 - accuracy: 0.8407 - val_loss: 0.6564 - val_accuracy: 0.7800\nDuration: 0:10:47.680401\n"
],
[
"print(n)\n\nmodels_nc[n].fit_model(image_sets_nc[n],label_sets_nc[n],x_val,y_val,epochs=20,batch_size = 128)\nn = n+1",
"18\nEpoch 1/20\n104/104 [==============================] - 37s 335ms/step - loss: 0.9316 - accuracy: 0.6613 - val_loss: 0.5931 - val_accuracy: 0.7914\nEpoch 2/20\n104/104 [==============================] - 34s 328ms/step - loss: 0.7570 - accuracy: 0.7177 - val_loss: 0.7490 - val_accuracy: 0.7158\nEpoch 3/20\n104/104 [==============================] - 32s 309ms/step - loss: 0.7062 - accuracy: 0.7405 - val_loss: 0.7483 - val_accuracy: 0.7511\nEpoch 4/20\n104/104 [==============================] - 33s 321ms/step - loss: 0.7167 - accuracy: 0.7302 - val_loss: 0.5809 - val_accuracy: 0.7914\nEpoch 5/20\n104/104 [==============================] - 31s 303ms/step - loss: 0.6815 - accuracy: 0.7435 - val_loss: 0.6486 - val_accuracy: 0.7664\nEpoch 6/20\n104/104 [==============================] - 31s 300ms/step - loss: 0.6424 - accuracy: 0.7581 - val_loss: 0.7047 - val_accuracy: 0.7432\nEpoch 7/20\n104/104 [==============================] - 31s 302ms/step - loss: 0.6437 - accuracy: 0.7556 - val_loss: 0.5616 - val_accuracy: 0.8024\nEpoch 8/20\n104/104 [==============================] - 34s 331ms/step - loss: 0.6234 - accuracy: 0.7678 - val_loss: 0.5434 - val_accuracy: 0.8103\nEpoch 9/20\n104/104 [==============================] - 34s 327ms/step - loss: 0.5993 - accuracy: 0.7760 - val_loss: 0.5861 - val_accuracy: 0.7896\nEpoch 10/20\n104/104 [==============================] - 32s 312ms/step - loss: 0.5844 - accuracy: 0.7836 - val_loss: 0.6920 - val_accuracy: 0.7475\nEpoch 11/20\n104/104 [==============================] - 32s 308ms/step - loss: 0.5698 - accuracy: 0.7785 - val_loss: 0.6352 - val_accuracy: 0.7899\nEpoch 12/20\n104/104 [==============================] - 34s 325ms/step - loss: 0.5484 - accuracy: 0.7897 - val_loss: 0.6225 - val_accuracy: 0.7785\nEpoch 13/20\n104/104 [==============================] - 34s 329ms/step - loss: 0.5426 - accuracy: 0.7960 - val_loss: 0.5542 - val_accuracy: 0.8106\nEpoch 14/20\n104/104 [==============================] - 34s 322ms/step - loss: 0.5086 - accuracy: 0.8105 - val_loss: 0.6020 - val_accuracy: 0.7878\nEpoch 15/20\n104/104 [==============================] - 35s 334ms/step - loss: 0.5080 - accuracy: 0.8130 - val_loss: 0.5571 - val_accuracy: 0.8035\nEpoch 16/20\n104/104 [==============================] - 33s 318ms/step - loss: 0.4858 - accuracy: 0.8176 - val_loss: 0.7793 - val_accuracy: 0.7329\nEpoch 17/20\n104/104 [==============================] - 34s 329ms/step - loss: 0.4862 - accuracy: 0.8200 - val_loss: 0.5849 - val_accuracy: 0.7985\nEpoch 18/20\n104/104 [==============================] - 35s 333ms/step - loss: 0.4666 - accuracy: 0.8290 - val_loss: 0.5695 - val_accuracy: 0.8035\nEpoch 19/20\n104/104 [==============================] - 32s 304ms/step - loss: 0.4406 - accuracy: 0.8340 - val_loss: 0.5887 - val_accuracy: 0.8039\nEpoch 20/20\n104/104 [==============================] - 34s 323ms/step - loss: 0.4540 - accuracy: 0.8359 - val_loss: 0.5946 - val_accuracy: 0.8035\nDuration: 0:11:06.981311\n"
],
[
"print(n)#19\n\nmodels_nc[n].fit_model(image_sets_nc[n],label_sets_nc[n],x_val,y_val,epochs=20,batch_size = 128)\nn = n+1",
"19\nEpoch 1/20\n112/112 [==============================] - 38s 325ms/step - loss: 0.8897 - accuracy: 0.6740 - val_loss: 0.8277 - val_accuracy: 0.7179\nEpoch 2/20\n112/112 [==============================] - 37s 328ms/step - loss: 0.7749 - accuracy: 0.7114 - val_loss: 0.7148 - val_accuracy: 0.7589\nEpoch 3/20\n112/112 [==============================] - 36s 325ms/step - loss: 0.7272 - accuracy: 0.7278 - val_loss: 0.8833 - val_accuracy: 0.6548\nEpoch 4/20\n112/112 [==============================] - 37s 330ms/step - loss: 0.7178 - accuracy: 0.7306 - val_loss: 0.7772 - val_accuracy: 0.7439\nEpoch 5/20\n112/112 [==============================] - 37s 327ms/step - loss: 0.6683 - accuracy: 0.7487 - val_loss: 0.7488 - val_accuracy: 0.7108\nEpoch 6/20\n112/112 [==============================] - 37s 330ms/step - loss: 0.6591 - accuracy: 0.7521 - val_loss: 0.7600 - val_accuracy: 0.7372\nEpoch 7/20\n112/112 [==============================] - 37s 331ms/step - loss: 0.6410 - accuracy: 0.7610 - val_loss: 0.6140 - val_accuracy: 0.7935\nEpoch 8/20\n112/112 [==============================] - 36s 324ms/step - loss: 0.6216 - accuracy: 0.7673 - val_loss: 0.6759 - val_accuracy: 0.7714\nEpoch 9/20\n112/112 [==============================] - 36s 321ms/step - loss: 0.6156 - accuracy: 0.7693 - val_loss: 0.7038 - val_accuracy: 0.7507\nEpoch 10/20\n112/112 [==============================] - 37s 328ms/step - loss: 0.5583 - accuracy: 0.7890 - val_loss: 0.6179 - val_accuracy: 0.7825\nEpoch 11/20\n112/112 [==============================] - 37s 329ms/step - loss: 0.5548 - accuracy: 0.7918 - val_loss: 0.8918 - val_accuracy: 0.6969\nEpoch 12/20\n112/112 [==============================] - 36s 323ms/step - loss: 0.5671 - accuracy: 0.7910 - val_loss: 0.7114 - val_accuracy: 0.7675\nEpoch 13/20\n112/112 [==============================] - 37s 327ms/step - loss: 0.5384 - accuracy: 0.8000 - val_loss: 0.6374 - val_accuracy: 0.7828\nEpoch 14/20\n112/112 [==============================] - 35s 316ms/step - loss: 0.5184 - accuracy: 0.8097 - val_loss: 0.6308 - val_accuracy: 0.7757\nEpoch 15/20\n112/112 [==============================] - 38s 342ms/step - loss: 0.5203 - accuracy: 0.8065 - val_loss: 0.6664 - val_accuracy: 0.7721\nEpoch 16/20\n112/112 [==============================] - 38s 336ms/step - loss: 0.4998 - accuracy: 0.8123 - val_loss: 0.7446 - val_accuracy: 0.7368\nEpoch 17/20\n112/112 [==============================] - 35s 308ms/step - loss: 0.4964 - accuracy: 0.8202 - val_loss: 0.6398 - val_accuracy: 0.7899\nEpoch 18/20\n112/112 [==============================] - 36s 318ms/step - loss: 0.4563 - accuracy: 0.8361 - val_loss: 0.7487 - val_accuracy: 0.7364\nEpoch 19/20\n112/112 [==============================] - 38s 340ms/step - loss: 0.4697 - accuracy: 0.8237 - val_loss: 0.6118 - val_accuracy: 0.7946\nEpoch 20/20\n112/112 [==============================] - 37s 333ms/step - loss: 0.4544 - accuracy: 0.8331 - val_loss: 0.6699 - val_accuracy: 0.7582\nDuration: 0:12:14.403769\n"
],
[
"loading = True\n\nmodels_nc = []\n\nif loading:\n for i in range(20):\n model_nc_dir = \"D:/models/intel_models/C2/intel_model_c2_sep_nc_e2_\"+str(i)\n print(model_nc_dir)\n model =utils.My_model('intel',True,model_nc_dir)\n model.model.compile(loss= 'categorical_crossentropy', optimizer = 'rmsprop', metrics = ['accuracy',tf.keras.metrics.Precision(), tf.keras.metrics.Recall()])\n models_nc.append(model)\n \n ",
"D:/models/intel_models/C2/intel_model_c2_sep_nc_e2_0\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_nc_e2_1\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_nc_e2_2\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_nc_e2_3\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_nc_e2_4\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_nc_e2_5\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_nc_e2_6\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_nc_e2_7\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_nc_e2_8\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_nc_e2_9\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_nc_e2_10\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_nc_e2_11\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_nc_e2_12\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_nc_e2_13\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_nc_e2_14\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_nc_e2_15\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_nc_e2_16\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_nc_e2_17\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_nc_e2_18\nModel loaded correctly\nD:/models/intel_models/C2/intel_model_c2_sep_nc_e2_19\nModel loaded correctly\n"
]
],
[
[
"## Evaluating",
"_____no_output_____"
]
],
[
[
"model_lsa_5000.model.compile(loss= 'categorical_crossentropy', optimizer = 'rmsprop', metrics = ['accuracy',tf.keras.metrics.Precision(), tf.keras.metrics.Recall()])\nmodel_dsa_5000.model.compile(loss= 'categorical_crossentropy', optimizer = 'rmsprop', metrics = ['accuracy',tf.keras.metrics.Precision(), tf.keras.metrics.Recall()])\nmodel_random_5000.model.compile(loss= 'categorical_crossentropy', optimizer = 'rmsprop', metrics = ['accuracy',tf.keras.metrics.Precision(), tf.keras.metrics.Recall()])\nmodel_nc_5000.model.compile(loss= 'categorical_crossentropy', optimizer = 'rmsprop', metrics = ['accuracy',tf.keras.metrics.Precision(), tf.keras.metrics.Recall()])",
"_____no_output_____"
],
[
"model_original.model.compile(loss= 'categorical_crossentropy', optimizer = 'rmsprop', metrics = ['accuracy',tf.keras.metrics.Precision(), tf.keras.metrics.Recall()])",
"_____no_output_____"
],
[
"evaluate_lsa_5k_0 = []\nevaluate_dsa_5k_0 = []\nevaluate_nc_5k_0 = []\nevaluate_random_5k_0 = []\n\nevaluate_lsa_5k_0.append(model_lsa_5000.evaluate(x_adversary_test_fgsm,y_adversary_test_fgsm))\nevaluate_dsa_5k_0.append(model_dsa_5000.evaluate(x_adversary_test_fgsm,y_adversary_test_fgsm))\nevaluate_random_5k_0.append(model_random_5000.evaluate(x_adversary_test_fgsm,y_adversary_test_fgsm))\nevaluate_nc_5k_0.append(model_nc_5000.evaluate(x_adversary_test_fgsm,y_adversary_test_fgsm))",
"94/94 [==============================] - 3s 23ms/step - loss: 1.1961 - accuracy: 0.5823 - precision: 0.6831 - recall: 0.3655\n94/94 [==============================] - 3s 23ms/step - loss: 1.1526 - accuracy: 0.6261 - precision_1: 0.7158 - recall_1: 0.5604\n94/94 [==============================] - 3s 24ms/step - loss: 1.5592 - accuracy: 0.4125 - precision_2: 0.4003 - recall_2: 0.2362\n94/94 [==============================] - 3s 23ms/step - loss: 2.1211 - accuracy: 0.4166 - precision_3: 0.3631 - recall_3: 0.2446\n"
],
[
"# Metrics using adversarial test\nevaluate_lsa_0 = []\nevaluate_dsa_0 = []\nevaluate_nc_0 = []\nevaluate_random_0 = []\n\nevaluate_lsa_0.append(model_original.evaluate(x_adversary_test_fgsm,y_adversary_test_fgsm))\nevaluate_dsa_0.append(model_original.evaluate(x_adversary_test_fgsm,y_adversary_test_fgsm))\nevaluate_nc_0.append(model_original.evaluate(x_adversary_test_fgsm,y_adversary_test_fgsm))\nevaluate_random_0.append(model_original.evaluate(x_adversary_test_fgsm,y_adversary_test_fgsm))\n\nfor model in models_lsa:\n evaluate_lsa_0.append(model.evaluate(x_adversary_test_fgsm,y_adversary_test_fgsm))\n \nfor model in models_dsa:\n evaluate_dsa_0.append(model.evaluate(x_adversary_test_fgsm,y_adversary_test_fgsm))\n \nfor model in models_random:\n evaluate_random_0.append(model.evaluate(x_adversary_test_fgsm,y_adversary_test_fgsm))\n \nfor model in models_nc:\n evaluate_nc_0.append(model.evaluate(x_adversary_test_fgsm,y_adversary_test_fgsm))",
"94/94 [==============================] - 9s 34ms/step - loss: 3.1689 - accuracy: 0.1456 - precision_140: 0.1084 - recall_140: 0.0858\n94/94 [==============================] - 3s 34ms/step - loss: 3.1384 - accuracy: 0.1497 - precision_140: 0.1314 - recall_140: 0.1048\n94/94 [==============================] - 3s 32ms/step - loss: 3.1384 - accuracy: 0.1497 - precision_140: 0.1312 - recall_140: 0.1048\n94/94 [==============================] - 3s 32ms/step - loss: 3.1384 - accuracy: 0.1497 - precision_140: 0.1311 - recall_140: 0.1048\n94/94 [==============================] - 4s 29ms/step - loss: 1.6743 - accuracy: 0.5013 - precision: 0.4590 - recall: 0.3547\n94/94 [==============================] - 4s 29ms/step - loss: 1.4151 - accuracy: 0.5177 - precision_1: 0.4891 - recall_1: 0.3837\n94/94 [==============================] - 4s 29ms/step - loss: 1.6931 - accuracy: 0.4457 - precision_2: 0.3649 - recall_2: 0.2791\n94/94 [==============================] - 4s 29ms/step - loss: 1.4600 - accuracy: 0.5361 - precision_3: 0.5075 - recall_3: 0.4196\n94/94 [==============================] - 4s 28ms/step - loss: 1.3591 - accuracy: 0.5651 - precision_4: 0.5298 - recall_4: 0.4251\n94/94 [==============================] - 4s 28ms/step - loss: 0.9693 - accuracy: 0.6619 - precision_5: 0.7101 - recall_5: 0.5601\n94/94 [==============================] - 4s 28ms/step - loss: 1.0831 - accuracy: 0.6287 - precision_6: 0.6898 - recall_6: 0.5801\n94/94 [==============================] - 4s 29ms/step - loss: 1.2635 - accuracy: 0.5665 - precision_7: 0.5802 - recall_7: 0.4346\n94/94 [==============================] - 4s 27ms/step - loss: 1.4714 - accuracy: 0.5315 - precision_8: 0.5095 - recall_8: 0.4081\n94/94 [==============================] - 4s 25ms/step - loss: 1.3267 - accuracy: 0.5676 - precision_9: 0.6573 - recall_9: 0.5366\n94/94 [==============================] - 5s 25ms/step - loss: 1.1856 - accuracy: 0.5828 - precision_10: 0.5685 - recall_10: 0.4143\n94/94 [==============================] - 4s 25ms/step - loss: 1.2884 - accuracy: 0.5883 - precision_11: 0.5769 - recall_11: 0.4632: 0s - loss: 1.3441 - accuracy: 0.5796 - precision_11: 0.5569 - recall\n94/94 [==============================] - 4s 27ms/step - loss: 1.2392 - accuracy: 0.5734 - precision_12: 0.5916 - recall_12: 0.4644\n94/94 [==============================] - 5s 36ms/step - loss: 1.2549 - accuracy: 0.5934 - precision_13: 0.6508 - recall_13: 0.5546\n94/94 [==============================] - 5s 30ms/step - loss: 1.6943 - accuracy: 0.4877 - precision_14: 0.4448 - recall_14: 0.3387\n94/94 [==============================] - 4s 24ms/step - loss: 1.3173 - accuracy: 0.5421 - precision_15: 0.5349 - recall_15: 0.4124\n94/94 [==============================] - 3s 23ms/step - loss: 1.4854 - accuracy: 0.5259 - precision_16: 0.5687 - recall_16: 0.4783\n94/94 [==============================] - 3s 23ms/step - loss: 1.3583 - accuracy: 0.5389 - precision_17: 0.5165 - recall_17: 0.3571\n94/94 [==============================] - 3s 23ms/step - loss: 1.3332 - accuracy: 0.6125 - precision_18: 0.5888 - recall_18: 0.5202\n94/94 [==============================] - 3s 24ms/step - loss: 1.3747 - accuracy: 0.5359 - precision_19: 0.5849 - recall_19: 0.4766\n94/94 [==============================] - 3s 23ms/step - loss: 2.0880 - accuracy: 0.4147 - precision_80: 0.4582 - recall_80: 0.3694\n94/94 [==============================] - 3s 24ms/step - loss: 1.4194 - accuracy: 0.5949 - precision_81: 0.6089 - recall_81: 0.5256\n94/94 [==============================] - 3s 23ms/step - loss: 1.1814 - accuracy: 0.6432 - precision_82: 0.6566 - recall_82: 0.5588\n94/94 [==============================] - 3s 23ms/step - loss: 1.0547 - accuracy: 0.6525 - precision_83: 0.6381 - recall_83: 0.5320: 0s - loss: 1.1402 - accuracy: 0.6326 - precision_83: 0.6087 - recall\n94/94 [==============================] - 3s 24ms/step - loss: 0.8778 - accuracy: 0.6849 - precision_84: 0.7775 - recall_84: 0.6114\n94/94 [==============================] - 3s 24ms/step - loss: 0.9425 - accuracy: 0.6586 - precision_85: 0.7042 - recall_85: 0.5743\n94/94 [==============================] - 3s 24ms/step - loss: 1.3228 - accuracy: 0.6129 - precision_86: 0.6157 - recall_86: 0.5618\n94/94 [==============================] - 3s 24ms/step - loss: 0.8627 - accuracy: 0.7068 - precision_87: 0.7024 - recall_87: 0.5880\n94/94 [==============================] - 3s 25ms/step - loss: 0.9757 - accuracy: 0.7004 - precision_88: 0.7091 - recall_88: 0.6264\n94/94 [==============================] - 4s 25ms/step - loss: 0.9178 - accuracy: 0.6872 - precision_89: 0.6941 - recall_89: 0.5843\n94/94 [==============================] - 4s 26ms/step - loss: 1.1188 - accuracy: 0.6934 - precision_90: 0.6636 - recall_90: 0.5945\n94/94 [==============================] - 4s 26ms/step - loss: 0.7465 - accuracy: 0.7466 - precision_91: 0.8023 - recall_91: 0.6842\n94/94 [==============================] - 4s 28ms/step - loss: 1.1000 - accuracy: 0.6304 - precision_92: 0.6284 - recall_92: 0.5139\n94/94 [==============================] - 4s 29ms/step - loss: 0.9916 - accuracy: 0.6697 - precision_93: 0.7232 - recall_93: 0.5973\n94/94 [==============================] - 4s 29ms/step - loss: 2.1638 - accuracy: 0.4322 - precision_94: 0.4165 - recall_94: 0.3222\n94/94 [==============================] - 4s 30ms/step - loss: 1.0600 - accuracy: 0.6493 - precision_95: 0.6235 - recall_95: 0.5238\n94/94 [==============================] - 4s 31ms/step - loss: 1.0726 - accuracy: 0.5988 - precision_96: 0.6645 - recall_96: 0.4399\n94/94 [==============================] - 4s 31ms/step - loss: 1.2326 - accuracy: 0.5604 - precision_97: 0.5891 - recall_97: 0.4362\n94/94 [==============================] - 4s 34ms/step - loss: 1.2616 - accuracy: 0.5651 - precision_98: 0.5912 - recall_98: 0.4762\n94/94 [==============================] - 5s 38ms/step - loss: 1.2880 - accuracy: 0.5678 - precision_99: 0.5694 - recall_99: 0.4683\n94/94 [==============================] - 5s 39ms/step - loss: 2.0499 - accuracy: 0.3648 - precision_100: 0.3323 - recall_100: 0.2538\n94/94 [==============================] - 5s 40ms/step - loss: 1.8827 - accuracy: 0.3727 - precision_101: 0.3447 - recall_101: 0.2411\n94/94 [==============================] - 5s 40ms/step - loss: 1.8549 - accuracy: 0.3890 - precision_102: 0.3788 - recall_102: 0.2590\n94/94 [==============================] - 5s 41ms/step - loss: 1.9452 - accuracy: 0.4205 - precision_103: 0.3620 - recall_103: 0.2917\n94/94 [==============================] - 5s 42ms/step - loss: 1.6859 - accuracy: 0.4122 - precision_104: 0.3813 - recall_104: 0.2421\n94/94 [==============================] - 5s 42ms/step - loss: 1.6781 - accuracy: 0.4199 - precision_105: 0.3580 - recall_105: 0.2550\n94/94 [==============================] - 5s 42ms/step - loss: 1.5373 - accuracy: 0.4806 - precision_106: 0.5020 - recall_106: 0.3434\n94/94 [==============================] - 5s 43ms/step - loss: 1.6613 - accuracy: 0.4322 - precision_107: 0.3798 - recall_107: 0.2453\n94/94 [==============================] - 5s 44ms/step - loss: 1.5076 - accuracy: 0.4874 - precision_108: 0.4434 - recall_108: 0.3328\n94/94 [==============================] - 5s 43ms/step - loss: 1.4788 - accuracy: 0.4682 - precision_109: 0.4127 - recall_109: 0.2785\n94/94 [==============================] - 5s 45ms/step - loss: 1.6406 - accuracy: 0.4933 - precision_110: 0.4824 - recall_110: 0.3752\n94/94 [==============================] - 5s 45ms/step - loss: 1.3865 - accuracy: 0.5372 - precision_111: 0.6102 - recall_111: 0.4999\n94/94 [==============================] - 5s 45ms/step - loss: 1.4061 - accuracy: 0.5169 - precision_112: 0.5096 - recall_112: 0.3858\n94/94 [==============================] - 6s 46ms/step - loss: 1.4202 - accuracy: 0.5183 - precision_113: 0.4907 - recall_113: 0.3768\n94/94 [==============================] - 6s 48ms/step - loss: 1.3247 - accuracy: 0.5500 - precision_114: 0.5866 - recall_114: 0.4666\n94/94 [==============================] - 6s 47ms/step - loss: 1.2444 - accuracy: 0.5462 - precision_115: 0.5359 - recall_115: 0.3935\n94/94 [==============================] - 6s 49ms/step - loss: 1.1189 - accuracy: 0.6030 - precision_116: 0.6340 - recall_116: 0.5034\n94/94 [==============================] - 6s 49ms/step - loss: 1.3281 - accuracy: 0.5412 - precision_117: 0.5171 - recall_117: 0.4009\n94/94 [==============================] - 6s 50ms/step - loss: 1.1154 - accuracy: 0.6190 - precision_118: 0.6457 - recall_118: 0.5367\n94/94 [==============================] - 6s 51ms/step - loss: 1.2651 - accuracy: 0.5805 - precision_119: 0.5834 - recall_119: 0.4602\n94/94 [==============================] - 7s 51ms/step - loss: 3.0393 - accuracy: 0.2747 - precision_120: 0.2587 - recall_120: 0.2146\n94/94 [==============================] - 6s 53ms/step - loss: 2.8886 - accuracy: 0.3384 - precision_121: 0.3458 - recall_121: 0.2972\n94/94 [==============================] - 6s 53ms/step - loss: 3.0231 - accuracy: 0.3354 - precision_122: 0.3538 - recall_122: 0.2716\n94/94 [==============================] - 6s 53ms/step - loss: 2.4298 - accuracy: 0.3788 - precision_123: 0.4226 - recall_123: 0.3131\n94/94 [==============================] - 6s 54ms/step - loss: 2.5234 - accuracy: 0.4269 - precision_124: 0.4605 - recall_124: 0.3618\n94/94 [==============================] - 6s 54ms/step - loss: 2.6160 - accuracy: 0.4027 - precision_125: 0.3593 - recall_125: 0.2899\n94/94 [==============================] - 6s 55ms/step - loss: 3.0732 - accuracy: 0.2759 - precision_126: 0.2588 - recall_126: 0.2227\n94/94 [==============================] - 7s 61ms/step - loss: 2.2346 - accuracy: 0.4495 - precision_127: 0.3900 - recall_127: 0.2831\n94/94 [==============================] - 7s 57ms/step - loss: 2.4547 - accuracy: 0.4099 - precision_128: 0.4152 - recall_128: 0.3131\n94/94 [==============================] - 7s 59ms/step - loss: 2.7150 - accuracy: 0.4318 - precision_129: 0.4099 - recall_129: 0.3368\n94/94 [==============================] - 7s 55ms/step - loss: 2.5278 - accuracy: 0.4905 - precision_130: 0.4176 - recall_130: 0.3689\n94/94 [==============================] - 7s 55ms/step - loss: 1.8514 - accuracy: 0.5109 - precision_131: 0.4868 - recall_131: 0.3826\n94/94 [==============================] - 7s 55ms/step - loss: 1.8326 - accuracy: 0.5223 - precision_132: 0.5235 - recall_132: 0.3949\n94/94 [==============================] - 6s 55ms/step - loss: 1.8023 - accuracy: 0.4881 - precision_133: 0.4807 - recall_133: 0.3451\n94/94 [==============================] - 6s 55ms/step - loss: 1.8004 - accuracy: 0.5207 - precision_134: 0.4772 - recall_134: 0.4130\n94/94 [==============================] - 7s 55ms/step - loss: 1.7998 - accuracy: 0.5344 - precision_135: 0.4802 - recall_135: 0.4256\n94/94 [==============================] - 6s 54ms/step - loss: 1.5619 - accuracy: 0.5446 - precision_136: 0.5172 - recall_136: 0.4399\n94/94 [==============================] - 6s 54ms/step - loss: 1.5091 - accuracy: 0.5142 - precision_137: 0.4743 - recall_137: 0.3832\n94/94 [==============================] - 6s 54ms/step - loss: 1.2455 - accuracy: 0.5798 - precision_138: 0.5771 - recall_138: 0.4750\n94/94 [==============================] - 6s 54ms/step - loss: 1.3515 - accuracy: 0.5318 - precision_139: 0.5143 - recall_139: 0.3749\n"
],
[
"print(len(evaluate_lsa_0))\nprint(len(evaluate_dsa_0))\nprint(len(evaluate_random_0))\nprint(len(evaluate_nc_0))",
"21\n21\n21\n21\n"
],
[
"evaluate_lsa_5k_1 = []\nevaluate_dsa_5k_1 = []\nevaluate_nc_5k_1 = []\nevaluate_random_5k_1 = []\n\nevaluate_lsa_5k_1.append(model_lsa_5000.evaluate(x_test,y_test))\nevaluate_dsa_5k_1.append(model_dsa_5000.evaluate(x_test,y_test))\nevaluate_nc_5k_1.append(model_random_5000.evaluate(x_test,y_test))\nevaluate_random_5k_1.append(model_nc_5000.evaluate(x_test,y_test))",
"94/94 [==============================] - 2s 23ms/step - loss: 1.0256 - accuracy: 0.5807 - precision: 0.7100 - recall: 0.4094\n94/94 [==============================] - 2s 23ms/step - loss: 1.8615 - accuracy: 0.5073 - precision_1: 0.6286 - recall_1: 0.4814\n94/94 [==============================] - 2s 24ms/step - loss: 0.7475 - accuracy: 0.7393 - precision_2: 0.6064 - recall_2: 0.4190\n94/94 [==============================] - 2s 22ms/step - loss: 0.8464 - accuracy: 0.7127 - precision_3: 0.6006 - recall_3: 0.4322\n"
],
[
"# Metrics using original test\nevaluate_lsa_1 = []\nevaluate_dsa_1 = []\nevaluate_nc_1 = []\nevaluate_random_1 = []\n\nevaluate_lsa_1.append(model_original.evaluate(x_test,y_test))\nevaluate_dsa_1.append(model_original.evaluate(x_test,y_test))\nevaluate_nc_1.append(model_original.evaluate(x_test,y_test))\nevaluate_random_1.append(model_original.evaluate(x_test,y_test))\nprint(\"lsa---\")\nfor model in models_lsa:\n evaluate_lsa_1.append(model.evaluate(x_test,y_test))\n\nprint(\"dsa---\")\n \nfor model in models_dsa:\n evaluate_dsa_1.append(model.evaluate(x_test,y_test))\n \nprint(\"random---\")\n\nfor model in models_random:\n evaluate_random_1.append(model.evaluate(x_test,y_test))\n \nprint(\"nc---\")\n\nfor model in models_nc:\n evaluate_nc_1.append(model.evaluate(x_test,y_test))",
"94/94 [==============================] - 5s 54ms/step - loss: 0.5392 - accuracy: 0.8037 - precision_140: 0.2196 - recall_140: 0.1788\n94/94 [==============================] - 5s 54ms/step - loss: 0.5392 - accuracy: 0.8037 - precision_140: 0.3442 - recall_140: 0.2867\n94/94 [==============================] - 5s 54ms/step - loss: 0.5392 - accuracy: 0.8037 - precision_140: 0.4268 - recall_140: 0.3612\n94/94 [==============================] - 5s 54ms/step - loss: 0.5392 - accuracy: 0.8037 - precision_140: 0.4857 - recall_140: 0.4157\nlsa---\n94/94 [==============================] - 5s 54ms/step - loss: 0.9717 - accuracy: 0.6690 - precision: 0.6133 - recall: 0.4625\n94/94 [==============================] - 5s 54ms/step - loss: 0.9240 - accuracy: 0.6720 - precision_1: 0.6341 - recall_1: 0.5143\n94/94 [==============================] - 5s 54ms/step - loss: 1.0626 - accuracy: 0.6007 - precision_2: 0.5901 - recall_2: 0.4816\n94/94 [==============================] - 5s 56ms/step - loss: 1.1786 - accuracy: 0.5970 - precision_3: 0.6422 - recall_3: 0.5294\n94/94 [==============================] - 5s 53ms/step - loss: 0.9930 - accuracy: 0.6337 - precision_4: 0.6582 - recall_4: 0.5046\n94/94 [==============================] - 5s 53ms/step - loss: 1.3059 - accuracy: 0.5360 - precision_5: 0.7220 - recall_5: 0.5853\n94/94 [==============================] - 5s 53ms/step - loss: 1.3436 - accuracy: 0.5480 - precision_6: 0.6526 - recall_6: 0.5210\n94/94 [==============================] - 5s 54ms/step - loss: 1.0106 - accuracy: 0.6257 - precision_7: 0.6804 - recall_7: 0.5474\n94/94 [==============================] - 5s 53ms/step - loss: 1.0561 - accuracy: 0.6150 - precision_8: 0.6617 - recall_8: 0.5414\n94/94 [==============================] - 5s 53ms/step - loss: 1.1601 - accuracy: 0.5897 - precision_9: 0.6216 - recall_9: 0.4638\n94/94 [==============================] - 5s 53ms/step - loss: 1.0207 - accuracy: 0.6390 - precision_10: 0.6677 - recall_10: 0.5501\n94/94 [==============================] - 5s 53ms/step - loss: 0.9552 - accuracy: 0.6750 - precision_11: 0.6846 - recall_11: 0.5995\n94/94 [==============================] - 5s 53ms/step - loss: 0.8972 - accuracy: 0.6907 - precision_12: 0.6910 - recall_12: 0.5798\n94/94 [==============================] - 5s 53ms/step - loss: 1.0010 - accuracy: 0.6690 - precision_13: 0.6420 - recall_13: 0.5633\n94/94 [==============================] - 5s 53ms/step - loss: 0.9112 - accuracy: 0.6973 - precision_14: 0.6127 - recall_14: 0.5240\n94/94 [==============================] - 5s 53ms/step - loss: 0.6607 - accuracy: 0.7820 - precision_15: 0.6724 - recall_15: 0.5507\n94/94 [==============================] - 5s 53ms/step - loss: 0.7470 - accuracy: 0.7563 - precision_16: 0.6365 - recall_16: 0.5319\n94/94 [==============================] - 5s 53ms/step - loss: 0.7497 - accuracy: 0.7403 - precision_17: 0.6918 - recall_17: 0.5235\n94/94 [==============================] - 5s 53ms/step - loss: 0.7872 - accuracy: 0.7447 - precision_18: 0.6672 - recall_18: 0.6039\n94/94 [==============================] - 5s 53ms/step - loss: 0.6976 - accuracy: 0.7647 - precision_19: 0.6656 - recall_19: 0.5051\ndsa---\n94/94 [==============================] - 5s 53ms/step - loss: 1.1960 - accuracy: 0.6047 - precision_80: 0.5224 - recall_80: 0.4202\n94/94 [==============================] - 5s 53ms/step - loss: 0.9296 - accuracy: 0.6993 - precision_81: 0.6491 - recall_81: 0.5490\n94/94 [==============================] - 5s 53ms/step - loss: 1.0476 - accuracy: 0.6553 - precision_82: 0.6818 - recall_82: 0.5918\n94/94 [==============================] - 5s 53ms/step - loss: 1.3665 - accuracy: 0.5277 - precision_83: 0.6912 - recall_83: 0.5911\n94/94 [==============================] - 5s 54ms/step - loss: 1.3481 - accuracy: 0.5143 - precision_84: 0.6942 - recall_84: 0.4664\n94/94 [==============================] - 5s 53ms/step - loss: 4.8531 - accuracy: 0.4577 - precision_85: 0.6422 - recall_85: 0.5465\n94/94 [==============================] - 5s 53ms/step - loss: 3.0725 - accuracy: 0.4777 - precision_86: 0.6326 - recall_86: 0.5912\n94/94 [==============================] - 5s 53ms/step - loss: 2.1315 - accuracy: 0.5020 - precision_87: 0.7036 - recall_87: 0.6279\n94/94 [==============================] - 5s 53ms/step - loss: 2.2134 - accuracy: 0.5370 - precision_88: 0.7025 - recall_88: 0.6456\n94/94 [==============================] - 5s 53ms/step - loss: 1.0841 - accuracy: 0.6030 - precision_89: 0.7351 - recall_89: 0.6087\n94/94 [==============================] - 5s 53ms/step - loss: 2.1392 - accuracy: 0.5320 - precision_90: 0.7038 - recall_90: 0.6561\n94/94 [==============================] - 5s 53ms/step - loss: 1.6590 - accuracy: 0.5433 - precision_91: 0.7691 - recall_91: 0.6681\n94/94 [==============================] - 5s 53ms/step - loss: 0.9740 - accuracy: 0.6837 - precision_92: 0.7141 - recall_92: 0.6304\n94/94 [==============================] - 5s 53ms/step - loss: 0.9656 - accuracy: 0.6913 - precision_93: 0.7150 - recall_93: 0.6197\n94/94 [==============================] - 5s 53ms/step - loss: 1.4496 - accuracy: 0.5953 - precision_94: 0.5789 - recall_94: 0.4843\n94/94 [==============================] - 5s 53ms/step - loss: 0.8886 - accuracy: 0.7093 - precision_95: 0.7295 - recall_95: 0.6298\n94/94 [==============================] - 5s 53ms/step - loss: 0.6726 - accuracy: 0.7603 - precision_96: 0.7648 - recall_96: 0.5643\n94/94 [==============================] - 5s 53ms/step - loss: 0.6538 - accuracy: 0.7713 - precision_97: 0.6922 - recall_97: 0.5205\n94/94 [==============================] - 5s 53ms/step - loss: 0.6260 - accuracy: 0.7853 - precision_98: 0.6840 - recall_98: 0.5679\n94/94 [==============================] - 5s 53ms/step - loss: 0.5897 - accuracy: 0.8037 - precision_99: 0.6862 - recall_99: 0.5701\nrandom---\n94/94 [==============================] - 5s 52ms/step - loss: 0.7516 - accuracy: 0.7577 - precision_100: 0.5192 - recall_100: 0.4190\n94/94 [==============================] - 5s 53ms/step - loss: 0.7243 - accuracy: 0.7600 - precision_101: 0.5599 - recall_101: 0.4011\n94/94 [==============================] - 5s 53ms/step - loss: 0.7209 - accuracy: 0.7643 - precision_102: 0.5577 - recall_102: 0.4097\n94/94 [==============================] - 5s 52ms/step - loss: 0.7175 - accuracy: 0.7743 - precision_103: 0.5661 - recall_103: 0.4619\n94/94 [==============================] - 5s 53ms/step - loss: 0.7979 - accuracy: 0.7207 - precision_104: 0.5970 - recall_104: 0.4263\n94/94 [==============================] - 5s 53ms/step - loss: 0.7218 - accuracy: 0.7533 - precision_105: 0.6109 - recall_105: 0.4410\n94/94 [==============================] - 5s 58ms/step - loss: 0.7227 - accuracy: 0.7510 - precision_106: 0.6374 - recall_106: 0.4180\n94/94 [==============================] - 5s 57ms/step - loss: 0.7435 - accuracy: 0.7543 - precision_107: 0.6222 - recall_107: 0.4504\n94/94 [==============================] - 6s 60ms/step - loss: 0.6848 - accuracy: 0.7627 - precision_108: 0.6270 - recall_108: 0.4818\n94/94 [==============================] - 5s 55ms/step - loss: 0.6760 - accuracy: 0.7650 - precision_109: 0.6561 - recall_109: 0.4693\n94/94 [==============================] - 5s 54ms/step - loss: 0.7861 - accuracy: 0.7447 - precision_110: 0.6159 - recall_110: 0.4957\n94/94 [==============================] - 5s 54ms/step - loss: 0.6942 - accuracy: 0.7613 - precision_111: 0.6251 - recall_111: 0.5189\n94/94 [==============================] - 5s 55ms/step - loss: 0.6260 - accuracy: 0.7907 - precision_112: 0.6567 - recall_112: 0.5377\n94/94 [==============================] - 5s 53ms/step - loss: 0.6471 - accuracy: 0.7730 - precision_113: 0.6554 - recall_113: 0.5283\n94/94 [==============================] - 5s 53ms/step - loss: 0.7010 - accuracy: 0.7553 - precision_114: 0.6302 - recall_114: 0.5280\n94/94 [==============================] - 5s 54ms/step - loss: 0.6473 - accuracy: 0.7840 - precision_115: 0.7010 - recall_115: 0.5126\n94/94 [==============================] - 6s 62ms/step - loss: 0.6176 - accuracy: 0.7810 - precision_116: 0.7071 - recall_116: 0.5686\n94/94 [==============================] - 6s 61ms/step - loss: 0.6239 - accuracy: 0.7863 - precision_117: 0.6659 - recall_117: 0.5531\n94/94 [==============================] - 5s 55ms/step - loss: 0.6336 - accuracy: 0.7813 - precision_118: 0.7020 - recall_118: 0.6093\n94/94 [==============================] - 5s 54ms/step - loss: 0.8006 - accuracy: 0.7163 - precision_119: 0.6718 - recall_119: 0.5644\nnc---\n94/94 [==============================] - 5s 54ms/step - loss: 1.0090 - accuracy: 0.7360 - precision_120: 0.4766 - recall_120: 0.4154\n94/94 [==============================] - 5s 56ms/step - loss: 1.0655 - accuracy: 0.7333 - precision_121: 0.4951 - recall_121: 0.4354\n94/94 [==============================] - 5s 58ms/step - loss: 1.2105 - accuracy: 0.7023 - precision_122: 0.5081 - recall_122: 0.4006\n94/94 [==============================] - 5s 54ms/step - loss: 0.8371 - accuracy: 0.7480 - precision_123: 0.5519 - recall_123: 0.4111\n94/94 [==============================] - 5s 53ms/step - loss: 0.9104 - accuracy: 0.7547 - precision_124: 0.5624 - recall_124: 0.4606\n94/94 [==============================] - 5s 53ms/step - loss: 0.7820 - accuracy: 0.7833 - precision_125: 0.5988 - recall_125: 0.5085\n94/94 [==============================] - 5s 53ms/step - loss: 1.3974 - accuracy: 0.6707 - precision_126: 0.4773 - recall_126: 0.4251\n94/94 [==============================] - 6s 61ms/step - loss: 0.8129 - accuracy: 0.7570 - precision_127: 0.6401 - recall_127: 0.4995\n94/94 [==============================] - 5s 54ms/step - loss: 0.9983 - accuracy: 0.6960 - precision_128: 0.5742 - recall_128: 0.4537\n94/94 [==============================] - 5s 54ms/step - loss: 0.8504 - accuracy: 0.7730 - precision_129: 0.6122 - recall_129: 0.5050\n94/94 [==============================] - 5s 55ms/step - loss: 0.9097 - accuracy: 0.7747 - precision_130: 0.6382 - recall_130: 0.5836\n94/94 [==============================] - 5s 54ms/step - loss: 0.7171 - accuracy: 0.7867 - precision_131: 0.6622 - recall_131: 0.5560\n94/94 [==============================] - 5s 55ms/step - loss: 0.8478 - accuracy: 0.7343 - precision_132: 0.6698 - recall_132: 0.4742\n94/94 [==============================] - 5s 55ms/step - loss: 0.7993 - accuracy: 0.7520 - precision_133: 0.6477 - recall_133: 0.5042\n94/94 [==============================] - 5s 54ms/step - loss: 0.7365 - accuracy: 0.8007 - precision_134: 0.6594 - recall_134: 0.5815\n94/94 [==============================] - 5s 55ms/step - loss: 0.7250 - accuracy: 0.7970 - precision_135: 0.6618 - recall_135: 0.6022\n94/94 [==============================] - 5s 54ms/step - loss: 0.6395 - accuracy: 0.8017 - precision_136: 0.6620 - recall_136: 0.5797\n94/94 [==============================] - 5s 54ms/step - loss: 0.6719 - accuracy: 0.7700 - precision_137: 0.6686 - recall_137: 0.5645\n94/94 [==============================] - 5s 54ms/step - loss: 0.6022 - accuracy: 0.7960 - precision_138: 0.6822 - recall_138: 0.5682\n94/94 [==============================] - 5s 53ms/step - loss: 0.7159 - accuracy: 0.7537 - precision_139: 0.6694 - recall_139: 0.5318\n"
],
[
"print(len(evaluate_lsa_1))\nprint(len(evaluate_dsa_1))\nprint(len(evaluate_random_1))\nprint(len(evaluate_nc_1))",
"21\n21\n21\n21\n"
],
[
"import pandas as pd\n\ndf_evaluate_lsa_5k_0 = pd.DataFrame(np.array(evaluate_lsa_5k_0),columns=[\"loss\",\"accuracy\",\"precision\",\"recall\"])\ndf_evaluate_lsa_5k_1 = pd.DataFrame(np.array(evaluate_lsa_5k_1),columns=[\"loss\",\"accuracy\",\"precision\",\"recall\"])\ndf_evaluate_dsa_5k_0 = pd.DataFrame(np.array(evaluate_dsa_5k_0),columns=[\"loss\",\"accuracy\",\"precision\",\"recall\"])\ndf_evaluate_dsa_5k_1 = pd.DataFrame(np.array(evaluate_dsa_5k_1),columns=[\"loss\",\"accuracy\",\"precision\",\"recall\"])\n\ndf_evaluate_random_5k_0 = pd.DataFrame(np.array(evaluate_random_5k_0),columns=[\"loss\",\"accuracy\",\"precision\",\"recall\"])\ndf_evaluate_random_5k_1 = pd.DataFrame(np.array(evaluate_random_5k_1),columns=[\"loss\",\"accuracy\",\"precision\",\"recall\"])\ndf_evaluate_nc_5k_0 = pd.DataFrame(np.array(evaluate_nc_5k_0),columns=[\"loss\",\"accuracy\",\"precision\",\"recall\"])\ndf_evaluate_nc_5k_1 = pd.DataFrame(np.array(evaluate_nc_5k_1),columns=[\"loss\",\"accuracy\",\"precision\",\"recall\"])\n",
"_____no_output_____"
],
[
"import pandas as pd\n\ndf_evaluate_lsa_0 = pd.DataFrame(np.array(evaluate_lsa_0),columns=[\"loss\",\"accuracy\",\"precision\",\"recall\"])\ndf_evaluate_lsa_1 = pd.DataFrame(np.array(evaluate_lsa_1),columns=[\"loss\",\"accuracy\",\"precision\",\"recall\"])\ndf_evaluate_dsa_0 = pd.DataFrame(np.array(evaluate_dsa_0),columns=[\"loss\",\"accuracy\",\"precision\",\"recall\"])\ndf_evaluate_dsa_1 = pd.DataFrame(np.array(evaluate_dsa_1),columns=[\"loss\",\"accuracy\",\"precision\",\"recall\"])\ndf_evaluate_random_0 = pd.DataFrame(np.array(evaluate_random_0),columns=[\"loss\",\"accuracy\",\"precision\",\"recall\"])\ndf_evaluate_random_1 = pd.DataFrame(np.array(evaluate_random_1),columns=[\"loss\",\"accuracy\",\"precision\",\"recall\"])\ndf_evaluate_nc_0 = pd.DataFrame(np.array(evaluate_nc_0),columns=[\"loss\",\"accuracy\",\"precision\",\"recall\"])\ndf_evaluate_nc_1 = pd.DataFrame(np.array(evaluate_nc_1),columns=[\"loss\",\"accuracy\",\"precision\",\"recall\"])",
"_____no_output_____"
],
[
"# Original test set + adversarial set\n\nmetric =\"accuracy\"\naccuracy_c3_lsa_5k_3 = (np.array(df_evaluate_lsa_5k_0[metric])+np.array(df_evaluate_lsa_5k_1[metric]))/2\naccuracy_c3_dsa_5k_3 = (np.array(df_evaluate_dsa_5k_0[metric])+np.array(df_evaluate_dsa_5k_1[metric]))/2\naccuracy_c3_nc_5k_3 = (np.array(df_evaluate_nc_5k_0[metric])+np.array(df_evaluate_nc_5k_1[metric]))/2\naccuracy_c3_random_5k_3 = (np.array(df_evaluate_random_5k_0[metric])+np.array(df_evaluate_random_5k_1[metric]))/2\n",
"_____no_output_____"
],
[
"# Original test set + adversarial set\n\nmetric =\"accuracy\"\naccuracy_c3_lsa_3 = (np.array(df_evaluate_lsa_0[metric])+np.array(df_evaluate_lsa_1[metric]))/2\naccuracy_c3_dsa_3 = (np.array(df_evaluate_dsa_0[metric])+np.array(df_evaluate_dsa_1[metric]))/2\naccuracy_c3_nc_3 = (np.array(df_evaluate_nc_0[metric])+np.array(df_evaluate_nc_1[metric]))/2\naccuracy_c3_random_3 = (np.array(df_evaluate_random_0[metric])+np.array(df_evaluate_random_1[metric]))/2\n",
"_____no_output_____"
]
],
[
[
"## Charts",
"_____no_output_____"
]
],
[
[
"n_inputs = [700*i for i in range(20)]\nn_inputs.append(len(x_train_and_adversary))",
"_____no_output_____"
],
[
"cd \"C:/Users/fjdur/Desktop/intel_graphs/e2_graphs_dec/\"",
"C:\\Users\\fjdur\\Desktop\\intel_graphs\\e2_graphs_dec\n"
],
[
"linestyles = ['solid','dotted','dashed','dashdot']\ncolors =['k','k','k','k']",
"_____no_output_____"
],
[
"\nimport matplotlib.pyplot as plt\nimport matplotlib.patches as mpatches\nfrom matplotlib.patches import Patch\nfrom matplotlib.lines import Line2D\n\n#metric = \"accuracy\" # accuracy loss\nmy_metrics =[\"accuracy\",\"loss\",\"precision\",\"recall\"]\n\nfor metric in my_metrics:\n plt.clf()\n plt.plot(n_inputs,df_evaluate_lsa_0[metric],colors[0],linestyle=linestyles[0])\n plt.plot(n_inputs,df_evaluate_dsa_0[metric],colors[1],linestyle=linestyles[1])\n plt.plot(n_inputs,df_evaluate_random_0[metric],colors[2],linestyle=linestyles[2])\n plt.plot(n_inputs,df_evaluate_nc_0[metric],colors[3],linestyle=linestyles[3])\n\n\n legend_elements = [Line2D([0], [0], color='k', label='LSA',ls = linestyles[0]),\n Line2D([0], [0], color='k', label='DSA',ls = linestyles[1]),\n Line2D([0], [0], color='k', label='Random',ls = linestyles[2]),\n Line2D([0], [0], color='k', label='NC',ls = linestyles[3])]\n\n\n\n plt.legend(handles=legend_elements)#\n plt.title(metric + \" with test set of adversarial examples FGSM\")\n plt.xlim([0, 15000])\n #plt.ylim([0, 1])\n plt.xlabel('number of inputs')\n plt.ylabel(metric)\n plt.savefig(\"intel_c2_\"+metric + \"_0.svg\")\n plt.show()",
"_____no_output_____"
],
[
"\nimport matplotlib.pyplot as plt\nimport matplotlib.patches as mpatches\n\nmy_metrics =[\"accuracy\",\"loss\",\"precision\",\"recall\"]\n\nfor metric in my_metrics:\n \n#metric = \"accuracy\" # accuracy loss\n plt.clf()\n plt.plot(n_inputs,df_evaluate_lsa_1[metric],colors[0],linestyle=linestyles[0])\n plt.plot(n_inputs,df_evaluate_dsa_1[metric],colors[1],linestyle=linestyles[1])\n plt.plot(n_inputs,df_evaluate_random_1[metric],colors[2],linestyle=linestyles[2])\n plt.plot(n_inputs,df_evaluate_nc_1[metric],colors[3],linestyle=linestyles[3])\n\n legend_elements = [Line2D([0], [0], color='k', label='LSA',ls = linestyles[0]),\n Line2D([0], [0], color='k', label='DSA',ls = linestyles[1]),\n Line2D([0], [0], color='k', label='Random',ls = linestyles[2]),\n Line2D([0], [0], color='k', label='NC',ls = linestyles[3])]\n\n\n\n plt.legend(handles=legend_elements)#\n plt.title(metric + \" with original set\")\n plt.xlim([0, 15000])\n #plt.ylim([0, 1])\n plt.xlabel('number of inputs')\n plt.ylabel(metric)\n plt.savefig(\"intel_c2_\"+metric + \"_1.svg\")\n plt.show()",
"_____no_output_____"
],
[
"#adversarial jsma test set\n#configuration 3\n\"\"\"\n6) Incremental guided retraining starting from the original model using only the new adversarial inputs.\nIncremental training, starting with the previous trained model. Using at each iteration a subset of the new inputs.\n\"\"\"\nimport matplotlib.pyplot as plt\nimport matplotlib.patches as mpatches\n\n\nplt.plot(n_inputs,accuracy_c3_lsa_3,colors[0],linestyle=linestyles[0])\nplt.plot(n_inputs,accuracy_c3_dsa_3,colors[1],linestyle=linestyles[1])\nplt.plot(n_inputs,accuracy_c3_random_3,colors[2],linestyle=linestyles[2])\nplt.plot(n_inputs,accuracy_c3_nc_3,colors[3],linestyle=linestyles[3])\n\n\nlegend_elements = [Line2D([0], [0], color='k', label='LSA',ls = linestyles[0]),\n Line2D([0], [0], color='k', label='DSA',ls = linestyles[1]),\n Line2D([0], [0], color='k', label='Random',ls = linestyles[2]),\n Line2D([0], [0], color='k', label='NC',ls = linestyles[3])]\n\n\n\nplt.legend(handles=legend_elements)#\n\nplt.title(\"accuracy with both sets\")\n\nprint(n_inputs[np.argmax(accuracy_c3_lsa_3)],accuracy_c3_lsa_3.max())\nprint(n_inputs[np.argmax(accuracy_c3_dsa_3)],accuracy_c3_dsa_3.max())\nprint(n_inputs[np.argmax(accuracy_c3_random_3)],accuracy_c3_random_3.max())\nprint(n_inputs[np.argmax(accuracy_c3_nc_3)],accuracy_c3_nc_3.max())\n\nplt.plot(n_inputs[np.argmax(accuracy_c3_lsa_3)],accuracy_c3_lsa_3.max(),'-kD')\nplt.plot(n_inputs[np.argmax(accuracy_c3_dsa_3)],accuracy_c3_dsa_3.max(),'-ko')\nplt.plot(n_inputs[np.argmax(accuracy_c3_random_3)],accuracy_c3_random_3.max(),'-kv')\nplt.plot(n_inputs[np.argmax(accuracy_c3_nc_3)],accuracy_c3_nc_3.max(),'-kp')\n\nplt.xlabel('number of inputs')\nplt.ylabel('accuracy')\nplt.xlim([0, 15000])\n#plt.ylim([0, 1])\n\nplt.savefig(\"intel_c2_\"+\"accuracy\" + \"_both.svg\")\n\nplt.show()",
"11200 0.6689999997615814\n11900 0.695000022649765\n13300 0.6859999895095825\n11200 0.6759999990463257\n"
],
[
"print(accuracy_c3_lsa_5k_3)\nprint(accuracy_c3_dsa_5k_3)\nprint(accuracy_c3_random_5k_3)\nprint(accuracy_c3_nc_5k_3)\n",
"[0.55466667]\n[0.54416665]\n[0.5795]\n[0.6015]\n"
]
],
[
[
"## Saving models",
"_____no_output_____"
]
],
[
[
"new_model_lsa_dir = \"D:/models/intel_models/C2/intel_model_c2_sep_lsa_e2\"\ni=0\n\nfor model in models_lsa:\n model.save(new_model_lsa_dir+\"_\"+str(i))\n i+=1",
"INFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_lsa_e2_0\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_lsa_e2_1\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_lsa_e2_2\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_lsa_e2_3\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_lsa_e2_4\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_lsa_e2_5\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_lsa_e2_6\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_lsa_e2_7\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_lsa_e2_8\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_lsa_e2_9\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_lsa_e2_10\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_lsa_e2_11\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_lsa_e2_12\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_lsa_e2_13\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_lsa_e2_14\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_lsa_e2_15\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_lsa_e2_16\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_lsa_e2_17\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_lsa_e2_18\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_lsa_e2_19\\assets\nModel has been saved\n"
],
[
"new_model_dsa_dir = \"D:/models/intel_models/C2/intel_model_c2_sep_dsa_e2\"\ni=0\nfor model in models_dsa:\n model.save(new_model_dsa_dir+\"_\"+str(i))\n i+=1",
"INFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_dsa_e2_0\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_dsa_e2_1\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_dsa_e2_2\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_dsa_e2_3\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_dsa_e2_4\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_dsa_e2_5\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_dsa_e2_6\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_dsa_e2_7\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_dsa_e2_8\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_dsa_e2_9\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_dsa_e2_10\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_dsa_e2_11\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_dsa_e2_12\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_dsa_e2_13\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_dsa_e2_14\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_dsa_e2_15\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_dsa_e2_16\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_dsa_e2_17\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_dsa_e2_18\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_dsa_e2_19\\assets\nModel has been saved\n"
],
[
"new_model_random_dir = \"D:/models/intel_models/C2/intel_model_c2_sep_random_e2\"\n\ni=0\nfor model in models_random:\n model.save(new_model_random_dir+\"_\"+str(i))\n i+=1",
"INFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_random_e2_0\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_random_e2_1\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_random_e2_2\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_random_e2_3\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_random_e2_4\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_random_e2_5\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_random_e2_6\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_random_e2_7\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_random_e2_8\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_random_e2_9\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_random_e2_10\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_random_e2_11\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_random_e2_12\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_random_e2_13\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_random_e2_14\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_random_e2_15\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_random_e2_16\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_random_e2_17\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_random_e2_18\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_random_e2_19\\assets\nModel has been saved\n"
],
[
"new_model_nc_dir = \"D:/models/intel_models/C2/intel_model_c2_sep_nc_e2\"\n\ni=0\nfor model in models_nc:\n model.save(new_model_nc_dir+\"_\"+str(i))\n i+=1",
"INFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_nc_e2_0\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_nc_e2_1\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_nc_e2_2\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_nc_e2_3\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_nc_e2_4\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_nc_e2_5\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_nc_e2_6\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_nc_e2_7\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_nc_e2_8\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_nc_e2_9\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_nc_e2_10\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_nc_e2_11\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_nc_e2_12\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_nc_e2_13\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_nc_e2_14\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_nc_e2_15\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_nc_e2_16\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_nc_e2_17\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_nc_e2_18\\assets\nModel has been saved\nINFO:tensorflow:Assets written to: D:/models/intel_models/C2/intel_model_c2_sep_nc_e2_19\\assets\nModel has been saved\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
c500b59df85d96f6f3bebb77c9c5c44025db3a0a
| 4,147 |
ipynb
|
Jupyter Notebook
|
notebooks/04_Script_CLI.ipynb
|
marcocaggioni/seq2video
|
53de4e7b6ae90c4544da590575d559bbae82ccee
|
[
"MIT"
] | null | null | null |
notebooks/04_Script_CLI.ipynb
|
marcocaggioni/seq2video
|
53de4e7b6ae90c4544da590575d559bbae82ccee
|
[
"MIT"
] | null | null | null |
notebooks/04_Script_CLI.ipynb
|
marcocaggioni/seq2video
|
53de4e7b6ae90c4544da590575d559bbae82ccee
|
[
"MIT"
] | null | null | null | 28.798611 | 156 | 0.545696 |
[
[
[
"%%writefile dissolution_video.py\n\nimport click\n\nfrom pathlib import Path\nimport datetime\nfrom moviepy.editor import *\nfrom PIL import Image\nfrom PIL import ImageFont\nfrom PIL import ImageDraw\nimport numpy as np\n\nfont = ImageFont.truetype(\"arial.ttf\", 40)\n\[email protected]()\[email protected]('--exp_folder', default='./', help='Path to experiment folder with images. Exp_folder will be also used to name the timelapse video')\ndef make_timelapse_video_from_folder(exp_folder):\n '''Read images in exp_folder, find date created, \n stamps it on the image and generate mp4 video \n named as exp_folder in current folder'''\n \n exp_folder=Path(exp_folder)\n \n print(f'Processing images in folder: {exp_folder.resolve()}')\n \n p = Path(exp_folder).glob('**/*')\n \n files = [item for item in p if item.is_file() and item.suffix=='.jpg']\n times = [datetime.datetime.fromtimestamp(file.stat().st_mtime) for file in files]\n times=list(map(lambda x: x-times[0],times))\n\n clips=[]\n for file, time in zip(files, times):\n img=Image.open(file)\n ImageDraw.Draw(img).text((0,0), str(time),font=font)\n clip =ImageClip(np.array(img)).set_duration(0.1)\n clips.append(clip)\n\n concat_clip = concatenate_videoclips(clips, method=\"compose\")\n concat_clip.write_videofile(f\"{exp_folder.name}.mp4\", fps=24)\n\nif __name__ == '__main__':\n make_timelapse_video_from_folder()",
"Overwriting dissolution_video.py\n"
],
[
"!python dissolution_video.py --help",
"Usage: dissolution_video.py [OPTIONS]\n\n Read images in exp_folder, find date created, stamps it on the image and\n generate mp4 video named as exp_folder in current folder\n\nOptions:\n --exp_folder TEXT Path to experiment folder with images. Exp_folder will be\n also used to name the timelapse video\n\n --help Show this message and exit.\n"
],
[
"!python dissolution_video.py --exp_folder cf90r1-041321-80LAS-diwater",
"Processing images in folder: /home/jovyan/work/Documents/timelapse/cf90r1-041321-80LAS-diwater\nMoviepy - Building video cf90r1-041321-80LAS-diwater.mp4.\nMoviepy - Writing video cf90r1-041321-80LAS-diwater.mp4\n\nMoviepy - Done ! \nMoviepy - video ready cf90r1-041321-80LAS-diwater.mp4\n"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code"
]
] |
c500c41d930af9e1d362e0349ae68cf7cde97c69
| 459,364 |
ipynb
|
Jupyter Notebook
|
notebooks/plot_cifar100-vgg19bn-full-runs.ipynb
|
awwong1/ml-research
|
6f0bb585fef0c4567a5f02937fea62726b9c88dd
|
[
"MIT"
] | null | null | null |
notebooks/plot_cifar100-vgg19bn-full-runs.ipynb
|
awwong1/ml-research
|
6f0bb585fef0c4567a5f02937fea62726b9c88dd
|
[
"MIT"
] | null | null | null |
notebooks/plot_cifar100-vgg19bn-full-runs.ipynb
|
awwong1/ml-research
|
6f0bb585fef0c4567a5f02937fea62726b9c88dd
|
[
"MIT"
] | null | null | null | 2,126.685185 | 233,296 | 0.961242 |
[
[
[
"%matplotlib inline\nimport sys\nsys.path.append(\"..\")",
"_____no_output_____"
],
[
"import os\nimport pandas as pd\nimport seaborn as sns\nimport matplotlib.pyplot as plt\n\nfrom util.tablogger import TabLogger",
"_____no_output_____"
],
[
"# load the file from the experiments directories\n\ndf = pd.DataFrame({})\nfor run in range(10): \n fpath = os.path.join(\n \"..\",\n \"experiments\",\n \"cifar100-vgg19_bn-run{}\".format(run),\n \"out\",\n \"epoch.out\"\n )\n tlog = TabLogger(fpath, resume=True)\n tlog.close()\n\n # add the run column to the data dictionary\n dd = tlog.numbers\n dd[\"Run\"] = [\"Run{}\".format(run)] * len(list(dd.values())[0])\n\n # concat the dataframe\n df = pd.concat([df, pd.DataFrame(dd)])",
"_____no_output_____"
],
[
"print(df)",
" Epoch Train Task Loss Train Acc Eval Task Loss Eval Acc LR Run\n0 0.0 4.653552 1.488 4.505377 2.34 0.100 Run0\n1 1.0 4.201894 4.332 4.076854 5.68 0.100 Run0\n2 2.0 3.866523 8.080 3.768365 9.97 0.100 Run0\n3 3.0 3.601185 12.004 3.546763 13.97 0.100 Run0\n4 4.0 3.304286 16.884 3.462945 16.08 0.100 Run0\n.. ... ... ... ... ... ... ...\n159 159.0 0.009764 99.860 1.478205 73.11 0.001 Run9\n160 160.0 0.009133 99.838 1.479901 73.16 0.001 Run9\n161 161.0 0.010223 99.796 1.481426 73.09 0.001 Run9\n162 162.0 0.009247 99.832 1.476014 73.19 0.001 Run9\n163 163.0 0.009659 99.822 1.477621 73.17 0.001 Run9\n\n[1640 rows x 7 columns]\n"
],
[
"# plot in Seaborn\n# plt.figure(figsize=(15, 6))\n\n\ntl = sns.relplot(\n x=\"Epoch\", y=\"Train Task Loss\", hue=\"Run\", data=df,\n col_order=[0.1, 0.01, 0.001], kind=\"scatter\", palette=\"colorblind\",\n col=\"LR\", facet_kws=dict(sharex=False, sharey=False),\n)\nplt.savefig(\"train_loss-cifar100-vgg19_bn-full.png\", dpi=200)\nplt.show()\nta = sns.relplot(\n x=\"Epoch\", y=\"Train Acc\", hue=\"Run\", data=df,\n col_order=[0.1, 0.01, 0.001], kind=\"line\", palette=\"colorblind\",\n col=\"LR\", facet_kws=dict(sharex=False, sharey=False),\n)\nplt.savefig(\"train_acc-cifar100-vgg19_bn-full.png\", dpi=200)\nplt.show()\nea = sns.relplot(\n x=\"Epoch\", y=\"Eval Acc\", hue=\"Run\", data=df,\n col_order=[0.1, 0.01, 0.001], kind=\"line\", palette=\"colorblind\",\n col=\"LR\", facet_kws=dict(sharex=False, sharey=False),\n)\n# g.set_yscale(\"log\")\nplt.savefig(\"eval_acc-cifar100-vgg19_bn-full.png\", dpi=200)\nplt.show()",
"_____no_output_____"
],
[
"ea = sns.lineplot(\n x=\"Epoch\", y=\"Eval Acc\", hue=\"Run\", data=df,\n palette=\"colorblind\"\n)\nea.set(title=\"VGG19_BN Full Evaluation Acuracy over Epoch\")\nplt.savefig(\"eval_acc-cifar100-vgg19_bn-full-nofacet.png\", dpi=200)\n# g.set_yscale(\"log\")\nplt.show()",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
c500c67e3edf48ee34bcc191cdb0add69fce6dfd
| 27,423 |
ipynb
|
Jupyter Notebook
|
Python_ReadingData.ipynb
|
UWashington-Astro300/Astro300-W21
|
bb23395e9823ea5eb482a45c8ff5626c7ed8d59c
|
[
"MIT"
] | null | null | null |
Python_ReadingData.ipynb
|
UWashington-Astro300/Astro300-W21
|
bb23395e9823ea5eb482a45c8ff5626c7ed8d59c
|
[
"MIT"
] | null | null | null |
Python_ReadingData.ipynb
|
UWashington-Astro300/Astro300-W21
|
bb23395e9823ea5eb482a45c8ff5626c7ed8d59c
|
[
"MIT"
] | 2 |
2021-04-13T22:28:47.000Z
|
2022-01-21T20:52:57.000Z
| 20.541573 | 140 | 0.492834 |
[
[
[
"<img style=\"float: right;\" src=\"./images/DataReading.png\" width=\"120\"/>\n\n# Reading Data\n\n* Python has a large number of different ways to read data from external files. \n* Python supports almost any type of file you can think of, from simple text files to complex binary formats.\n* In this class we are going to mainly use the package **`pandas`** to load external files into `DataFrames`.\n* Most of our datafiles will be `csv` files (comma separated values)",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np",
"_____no_output_____"
]
],
[
[
"##### Let us read-in the file: `./Data/Planets.csv`\n\n```\nName,a,\nMercury,0.3871,0.2056\nEarth,0.9991,0.0166\nJupiter,5.2016,0.0490\nNeptune,29.9769,0.0088\n```",
"_____no_output_____"
]
],
[
[
"planet_table = pd.read_csv('./Data/Planets.csv')",
"_____no_output_____"
],
[
"planet_table",
"_____no_output_____"
]
],
[
[
"## Renaming columns",
"_____no_output_____"
]
],
[
[
"planet_table.rename(columns={'Unnamed: 2': 'ecc'}, inplace=True)\n\nplanet_table",
"_____no_output_____"
],
[
"planet_table['ecc']",
"_____no_output_____"
]
],
[
[
"## Adding a column - `insert`\n\n`.insert(loc, column, value, allow_duplicates = False)`",
"_____no_output_____"
],
[
"#### perihelion distance [AU] = `semi_major axis * ( 1 - eccentricity )`",
"_____no_output_____"
]
],
[
[
"def find_perihelion(semi_major, eccentricity):\n result = semi_major * (1.0 - eccentricity)\n return result",
"_____no_output_____"
]
],
[
[
"#### Use `DataFrame` columns as arguments to the `find_perihelion` function",
"_____no_output_____"
]
],
[
[
"my_perihelion = find_perihelion(planet_table['a'], planet_table['ecc'])",
"_____no_output_____"
],
[
"my_perihelion",
"_____no_output_____"
],
[
"# Add column in position 1 (2nd column)\n\nplanet_table.insert(1, 'Perihelion', my_perihelion, allow_duplicates = False)",
"_____no_output_____"
],
[
"planet_table",
"_____no_output_____"
]
],
[
[
"## Removing a column - `drop`",
"_____no_output_____"
]
],
[
[
"planet_table.drop(columns='Perihelion', inplace = True)",
"_____no_output_____"
],
[
"planet_table",
"_____no_output_____"
]
],
[
[
"## Adding a column (quick) - always to the end of the table",
"_____no_output_____"
]
],
[
[
"planet_table['Perihelion'] = my_perihelion",
"_____no_output_____"
],
[
"planet_table",
"_____no_output_____"
]
],
[
[
"## Rearranging columns",
"_____no_output_____"
]
],
[
[
"planet_table.columns",
"_____no_output_____"
],
[
"my_new_order = ['a', 'Perihelion', 'Name', 'ecc']",
"_____no_output_____"
],
[
"planet_table = planet_table[my_new_order]",
"_____no_output_____"
],
[
"planet_table",
"_____no_output_____"
]
],
[
[
"## Adding a row `.append`\n\n* The new row has to be a `dictionary` or another `DataFrame`\n* Almost always need to use: `ignore_index=True`",
"_____no_output_____"
]
],
[
[
"my_new_row = {'Name': 'Venus',\n 'a': 0.723,\n 'ecc': 0.007}",
"_____no_output_____"
],
[
"my_new_row",
"_____no_output_____"
],
[
"planet_table.append(my_new_row, ignore_index=True)",
"_____no_output_____"
],
[
"planet_table",
"_____no_output_____"
],
[
"planet_table = planet_table.append(my_new_row, ignore_index=True)",
"_____no_output_____"
],
[
"planet_table",
"_____no_output_____"
]
],
[
[
"#### `NaN` = Not_A_Number, python's null value",
"_____no_output_____"
],
[
"----\n\n<img style=\"float: right;\" src=\"./images/Lore.jpg\" width=\"200\"/>\n\n# Reading (bad) Data\n\n## Different Delimiters\n\nSome people just want to watch the world burn, so they create datasets where the columns are separted by something other than a comma.",
"_____no_output_____"
],
[
"#### Bad - Using another delimiter like `:`\n\n##### `./Data/Planets_Ver2.txt`\n\n```\nName:a:\nMercury:0.3871:0.2056\nEarth:0.9991:0.0166\nJupiter:5.2016:0.0490\nNeptune:29.9769:0.0088\n```",
"_____no_output_____"
]
],
[
[
"planet_table_2 = pd.read_csv('./Data/Planets_Ver2.txt', delimiter = \":\")",
"_____no_output_____"
],
[
"planet_table_2",
"_____no_output_____"
]
],
[
[
"#### Worse - Using whitespace as a delimiter\n\n##### `./Data/Planets_Ver3.txt`\n\n```\nName a \nMercury 0.3871 0.2056\nEarth 0.9991 0.0166\nJupiter 5.2016 0.0490\nNeptune 29.9769 0.0088\n```",
"_____no_output_____"
]
],
[
[
"planet_table_3 = pd.read_csv('./Data/Planets_Ver3.txt', delimiter = \" \")",
"_____no_output_____"
],
[
"planet_table_3",
"_____no_output_____"
]
],
[
[
"#### WORST! - Using inconsistent whitespace as a delimiter!\n\n##### `./Data/Planets_Ver4.txt`\n\n```\n Name a \n Mercury 0.3871 0.2056\n Earth 0.9991 0.0166\n Jupiter 5.2016 0.0490\n Neptune 29.9769 0.0088\n```",
"_____no_output_____"
]
],
[
[
"planet_table_4 = pd.read_csv('./Data/Planets_Ver4.txt', delimiter = \" \", skipinitialspace=True)",
"_____no_output_____"
],
[
"planet_table_4",
"_____no_output_____"
]
],
[
[
"---\n\n<img style=\"float: right;\" src=\"./images/MessyData.jpg\" width=\"230\"/>\n\n# Messy Data\n\n* `pandas` is a good choice when working with messy data files.\n* In the \"real world\" all data is messy.",
"_____no_output_____"
],
[
"##### Let us read-in the file: `./Data/Mess.csv`\n\n```\n#######################################################\n#\n# Col 1 - Name\n# Col 2 - Size (km)\n#\n#######################################################\n\"Sample 1\",10\n\"\",23\n,\n\"Another Sample\",\n```",
"_____no_output_____"
],
[
"### This is not going to end well ... (errors galore!)",
"_____no_output_____"
]
],
[
[
"messy_table = pd.read_csv('./Data/Mess.csv')",
"_____no_output_____"
]
],
[
[
"### Tell `pandas` about the comments:",
"_____no_output_____"
]
],
[
[
"messy_table = pd.read_csv('./Data/Mess.csv', comment = \"#\")\n\nmessy_table",
"_____no_output_____"
]
],
[
[
"## Not quite correct ...",
"_____no_output_____"
],
[
"### Turn off the header",
"_____no_output_____"
]
],
[
[
"messy_table = pd.read_csv('./Data/Mess.csv', comment = \"#\", header= None)\n\nmessy_table",
"_____no_output_____"
]
],
[
[
"### Add the column names",
"_____no_output_____"
]
],
[
[
"my_column_name = ['Name', 'Size']\n\nmessy_table = pd.read_csv('./Data/Mess.csv', comment = \"#\", header= None, names = my_column_name)\n\nmessy_table",
"_____no_output_____"
]
],
[
[
"### Deal with the missing data with `.fillna()`",
"_____no_output_____"
]
],
[
[
"messy_table['Name'].fillna(\"unknown\", inplace=True)\nmessy_table['Size'].fillna(999.0, inplace=True)\n\nmessy_table",
"_____no_output_____"
]
],
[
[
"----\n\n# Fixed-Width Data Tables\n\n* These types of data tables are **VERY** common in astronomy\n* The columns have a fixed-widths\n* Whitespace is used to seperate columns **AND** used within columns\n",
"_____no_output_____"
]
],
[
[
"TPHE A 00 30 09\nPG0029+024 00 31 50\n92 309 00 53 14",
"_____no_output_____"
]
],
[
[
"#### Trying to read this in with `pd.read_csv()` is a mess!",
"_____no_output_____"
]
],
[
[
"standard_table = pd.read_csv('./Data/StdStars.dat', header= None, delimiter = \" \", skipinitialspace=True)",
"_____no_output_____"
],
[
"standard_table",
"_____no_output_____"
]
],
[
[
"|--- 11 ---|2 |2 |2 |\n\nTPHE A 00 30 09\nPG0029+024 00 31 50\n92 309 00 53 14",
"_____no_output_____"
]
],
[
[
"## `pd.read_fwf()` - fixed-width-format\n\n* You can set the beginning to end of a column: `colspecs`\n* Or you can set the width of the columns: `widths`",
"_____no_output_____"
],
[
"`colspecs` uses the same format as a slice\n\n* 0-based indexing\n* First number is the first element you want\n* Second number is the first element you DON'T want",
"_____no_output_____"
]
],
[
[
"my_colspecs = [(0,12), (12,14), (15,17), (18,20)]",
"_____no_output_____"
],
[
"standard_table = pd.read_fwf('./Data/StdStars.dat', colspecs = my_colspecs)",
"_____no_output_____"
],
[
"standard_table",
"_____no_output_____"
],
[
"my_column_name = ['Star', 'RAh', 'RAm', 'RAs']",
"_____no_output_____"
],
[
"standard_table = pd.read_fwf('./Data/StdStars.dat', colspecs = my_colspecs, header= None, names = my_column_name)",
"_____no_output_____"
],
[
"standard_table",
"_____no_output_____"
]
],
[
[
"----\n# Real World Example\n\n",
"_____no_output_____"
],
[
"[Landolt Paper SIMBAD page](http://simbad.u-strasbg.fr/simbad/sim-basic?Ident=1992AJ....104..340L)\n\n[Mirror](http://simbad.cfa.harvard.edu/simbad/sim-basic?Ident=1992AJ....104..340L)",
"_____no_output_____"
]
],
[
[
"table2.dat",
"_____no_output_____"
],
[
"TPHE A 00 30 09 -46 31 22 14.651 0.793 0.380 0.435 0.405 0.841 29 12 0.0028 0.0046 0.0071 0.0019 0.0035 0.0032\nTPHE B 00 30 16 -46 27 55 12.334 0.405 0.156 0.262 0.271 0.535 29 17 0.0115 0.0026 0.0039 0.0020 0.0019 0.0035\nTPHE C 00 30 17 -46 32 34 14.376 -0.298 -1.217 -0.148 -0.211 -0.360 39 23 0.0022 0.0024 0.0043 0.0038 0.0133 0.0149",
"_____no_output_____"
],
[
"ReadMe",
"_____no_output_____"
],
[
"Byte-by-byte Description of file: table2.dat\n--------------------------------------------------------------------------------\n Bytes Format Units Label Explanations\n--------------------------------------------------------------------------------\n 1- 11 A11 --- Star *Star Designation\n 13- 14 I2 h RAh Right Ascension J2000 (hours)\n 16- 17 I2 min RAm Right Ascension J2000 (minutes)\n 19- 20 I2 s RAs Right Ascension J2000 (seconds)\n 22 A1 --- DE- Declination J2000 (sign)\n 23- 24 I2 deg DEd Declination J2000 (degrees)\n 26- 27 I2 arcmin DEm Declination J2000 (minutes)\n 29- 30 I2 arcsec DEs Declination J2000 (seconds)\n 33- 38 F6.3 mag Vmag V magnitude\n 40- 45 F6.3 mag B-V B-V color\n 47- 52 F6.3 mag U-B U-B color\n 54- 59 F6.3 mag V-R V-R color\n 61- 66 F6.3 mag R-I R-I color\n 68- 73 F6.3 mag V-I V-I color\n 75- 77 I3 --- o_Vmag Number of observations\n 79- 81 I3 --- Nd Number of nights\n 84- 89 F6.4 mag e_Vmag Mean error of the Mean Vmag\n 91- 96 F6.4 mag e_B-V Mean error of the Mean (B-V)\n 98-103 F6.4 mag e_U-B Mean error of the Mean (U-B)\n 105-110 F6.4 mag e_V-R Mean error of the Mean (V-R)\n 112-117 F6.4 mag e_R-I Mean error of the Mean (R-I)\n 119-124 F6.4 mag e_V-I Mean error of the Mean (V-I)\n--------------------------------------------------------------------------------",
"_____no_output_____"
]
],
[
[
"#### Set the column names",
"_____no_output_____"
]
],
[
[
"my_column_name = ['Star', \n 'RAh', 'RAm','RAs',\n 'DEd', 'DEm','DEs',\n 'Vmag', 'B-V', 'U-B', 'V-R', 'R-I', 'V-I',\n 'o_Vmag', 'Nd',\n 'e_Vmag', 'e_B-V', 'e_U-B', 'e_V-R', 'e_R-I', 'e_V-I']",
"_____no_output_____"
]
],
[
[
"#### Set the column widths\n\n* include leading whitespace\n* pandas will trim off surrounding whitespace",
"_____no_output_____"
]
],
[
[
"my_column_width = [12, 2, 3, 3, 4, 3, 3, 8, 7, 7, 7, 7, 7, 5, 4, 8, 7, 7, 7, 7, 7]",
"_____no_output_____"
]
],
[
[
"#### Set the URL for the data",
"_____no_output_____"
]
],
[
[
"my_url = 'https://cdsarc.unistra.fr/ftp/II/183A/table2.dat'\n\n#my_url = 'Data/table2.dat'",
"_____no_output_____"
],
[
"standard_table = pd.read_fwf(my_url, header=None, widths=my_column_width, names = my_column_name)",
"_____no_output_____"
],
[
"standard_table.head(10)",
"_____no_output_____"
]
],
[
[
"----\n\n<img style=\"float: right;\" src=\"./images/LotsData.jpg\" width=\"230\"/>\n\n# Lots of Data\n\n* `pandas` will cutoff the display of really long tables\n* You can change this with:\n * `pd.set_option('display.max_rows', # of rows)`\n * `pd.set_option('display.max_columns', # of columns)`",
"_____no_output_____"
]
],
[
[
"standard_table.info()",
"_____no_output_____"
],
[
"pd.set_option('display.max_columns', 21)",
"_____no_output_____"
],
[
"standard_table.head(10)",
"_____no_output_____"
],
[
"pd.set_option('display.max_rows', 526)",
"_____no_output_____"
],
[
"standard_table",
"_____no_output_____"
]
],
[
[
"----\n\n# Bonus Content: Reading HTML tables (Wikipedia)\n\n* `pandas` can (sort-of) easily import HTML tables - `read_html()`\n* This is great for pulling in data from Wikipedia\n* The results are often far from perfect",
"_____no_output_____"
],
[
"# [List of impact craters in North America](https://en.wikipedia.org/wiki/List_of_impact_craters_in_North_America)\n\n* There are 4 tables on this page\n* Plus a bunch of other table-ish-looking content\n* Let us see how this works out ....",
"_____no_output_____"
]
],
[
[
"crater_wiki = 'https://en.wikipedia.org/wiki/List_of_impact_craters_in_North_America'",
"_____no_output_____"
],
[
"crater_table = pd.read_html(crater_wiki)",
"_____no_output_____"
]
],
[
[
"### What did we get?",
"_____no_output_____"
]
],
[
[
"type(crater_table)",
"_____no_output_____"
]
],
[
[
"### A list (array), close but not a `pandas` DataFrame",
"_____no_output_____"
]
],
[
[
"len(crater_table)",
"_____no_output_____"
]
],
[
[
"### And 7 of them. OK, what do we have at index 0?",
"_____no_output_____"
]
],
[
[
"crater_table[0]",
"_____no_output_____"
]
],
[
[
"### Garbage",
"_____no_output_____"
]
],
[
[
"type(crater_table[0])",
"_____no_output_____"
]
],
[
[
"### ... But is it a DataFrame!",
"_____no_output_____"
],
[
"### And at index 1?",
"_____no_output_____"
]
],
[
[
"crater_table[1]",
"_____no_output_____"
]
],
[
[
"### Sweet! A DataFrame of the first table (Impact Craters in Canada)",
"_____no_output_____"
],
[
"### Index 2 is the Mexican table",
"_____no_output_____"
]
],
[
[
"crater_table[2]",
"_____no_output_____"
]
],
[
[
"### Index 3 is the US Table",
"_____no_output_____"
]
],
[
[
"crater_table[3]",
"_____no_output_____"
]
],
[
[
"### Index 4 is the Unconfirmed impact craters",
"_____no_output_____"
]
],
[
[
"crater_table[4]",
"_____no_output_____"
]
],
[
[
"### The last two [5 and 6] are garbage",
"_____no_output_____"
],
[
"### The DataFrames are not perfect, and will need some cleaning, but is great starting point",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"raw",
"markdown",
"code",
"raw",
"markdown",
"code",
"markdown",
"raw",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"raw"
],
[
"markdown"
],
[
"code",
"code"
],
[
"raw"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"raw",
"raw",
"raw",
"raw"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
c500c83402e545e55cb4213ba3fb06e2a5a0cf26
| 152,922 |
ipynb
|
Jupyter Notebook
|
jupyterlab/graphs/MaintainerReach.ipynb
|
markuszm/npm-analysis
|
b7fba0ccf0eb6bfd9ac4a739cdf3c9aee5be5a9f
|
[
"MIT"
] | 2 |
2019-09-30T07:50:31.000Z
|
2020-05-08T10:56:56.000Z
|
jupyterlab/graphs/MaintainerReach.ipynb
|
markuszm/npm-analysis
|
b7fba0ccf0eb6bfd9ac4a739cdf3c9aee5be5a9f
|
[
"MIT"
] | 2 |
2021-05-07T22:04:20.000Z
|
2021-05-07T22:07:14.000Z
|
jupyterlab/graphs/MaintainerReach.ipynb
|
markuszm/npm-analysis
|
b7fba0ccf0eb6bfd9ac4a739cdf3c9aee5be5a9f
|
[
"MIT"
] | 2 |
2019-09-30T08:30:19.000Z
|
2019-11-18T19:26:48.000Z
| 749.617647 | 44,028 | 0.951511 |
[
[
[
"from matplotlib_venn import venn2, venn3\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport numpy as np\n\nisaacs = pd.read_json('isaacs-reach.json', typ='series')\nsetA = set(isaacs)\n\nmathias = pd.read_json('mathias-reach.json', typ='series')\nsetB = set(mathias)\n\npackages = pd.read_json('latestPackages.json', typ='series')\nsetC = set(packages)\n\n# print(setA,setB,setC)\n\nplt.figure(figsize=(8,6), dpi=100)\nvenn3([setA, setB, setC], ('isaacs', 'mathias', 'All packages'))\nplt.savefig(\"top2_reach_maintainer.png\")",
"_____no_output_____"
],
[
"import matplotlib.ticker as mtick\nimport pandas as pd\nimport matplotlib.pyplot as plt\n\nranking = pd.read_json('optimal_ranking_2018_all.json', typ='series')\n\n# for i, x in to100Percent.iteritems():\n# if i > 5:\n# amountTop5 = x\n# break\n\n# print(amountTop5, amountTop5 / 667224)\n \n\n# max = 0\n# index = 0\n# for i, x in to100Percent.iteritems():\n# if x > max:\n# max = x\n# index = i\n \n# print('full reach at maintainer count {} with {} packages'.format(index, max))\n\nplt.rcParams.update({'font.size': 14})\n\nprint(len(ranking[\"IntersectionCounts\"]))\n\nplt.figure(figsize=(10,6), dpi=100)\nplt.plot(ranking[\"IntersectionCounts\"])\n# plt.axvline(index, color='r', linestyle='--')\nplt.ylim(0,500000)\nplt.xlabel('Number of maintainers ordered by optimal reach to reach all packages with a dependency')\nplt.ylabel('Reached packages')\n\n# plt.annotate(\"Full reach\", (index, max), xytext=(+10, +30), textcoords='offset points', fontsize=10,\n# arrowprops=dict(arrowstyle=\"->\", connectionstyle=\"arc3,rad=.2\"))\n\nplt.gca().set_yticklabels(['{:.0f}%'.format(x/667224*100) for x in plt.gca().get_yticks()]) \n\nplt.savefig(\"complete_reach_maintainer.png\")",
"7387\n"
],
[
"import matplotlib.ticker as mtick\nimport pandas as pd\nimport matplotlib.pyplot as plt\n\nranking = pd.read_json('optimalRanking_2018_top100.json', typ='series')\n\nplt.figure(figsize=(8,6), dpi=100)\nplt.plot(ranking[\"IntersectionCounts\"], '.')\nplt.ylim(0,400000)\nplt.xlabel('Number of maintainers ordered by optimal reach')\nplt.ylabel('Reached packages')\n\nplt.gca().set_yticklabels(['{:.0f}%'.format(x/667224*100) for x in plt.gca().get_yticks()]) \n\nplt.savefig(\"top100_reach_maintainer.png\")",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt\n\nx = [2012,2013,2014,2015,2016,2017,2018]\npackageCount = [5165, 18574, 50393, 112373, 218744, 390190, 605079]\nreachCount = [2261, 9462, 27216, 63555, 124638, 210433, 322578]\n\npercentage = []\n\nfor i in range(7):\n percentage.append(reachCount[i] / packageCount[i])\n \nplt.figure(figsize=(8,6), dpi=100)\nplt.plot(x, percentage, '--bo')\nplt.xlabel('Year')\nplt.ylabel('Reach of Top 20 Maintainers (% of All Packages)')\n\nplt.rcParams.update({'font.size': 14})\n\nplt.gca().set_yticklabels(['{:.2f}%'.format(x*100) for x in plt.gca().get_yticks()]) \n\nplt.savefig(\"top20_maintainer_reach_evolution.png\")",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code"
]
] |
c500de9ee8102ec28b8503a9f0761d2ace4359a2
| 2,892 |
ipynb
|
Jupyter Notebook
|
lectures/RootLocus.ipynb
|
DhruvJ22/aae497-f19
|
2942f2b58874c18d41f3441460c794f6a3478d07
|
[
"BSD-3-Clause"
] | null | null | null |
lectures/RootLocus.ipynb
|
DhruvJ22/aae497-f19
|
2942f2b58874c18d41f3441460c794f6a3478d07
|
[
"BSD-3-Clause"
] | null | null | null |
lectures/RootLocus.ipynb
|
DhruvJ22/aae497-f19
|
2942f2b58874c18d41f3441460c794f6a3478d07
|
[
"BSD-3-Clause"
] | null | null | null | 28.352941 | 120 | 0.505533 |
[
[
[
"%matplotlib widget\nimport control\nimport numpy as np\nimport matplotlib.pyplot as plt\nfrom matplotlib.widgets import Slider, Button, RadioButtons\n\n\ndef root_locus(G):\n\n fig, ax = plt.subplots()\n plt.subplots_adjust(bottom=0.25)\n\n roots = control.rlocus(G, kvect=np.logspace(-3, 3, 1000), Plot=False)[0].T\n l_locus = [ ax.plot(np.real(root), np.imag(root), '-')[0] for root in roots]\n\n a0 = 2\n poles = control.pole(control.feedback(G*a0, 1))\n l_poles, = ax.plot(np.real(poles), np.imag(poles), 'rx')\n h_text = [ax.text(np.real(pole), np.imag(pole), '') for pole in poles]\n \n ax.grid()\n ax.set_title('root locus')\n ax.margins(x=0)\n\n axcolor = 'lightgoldenrodyellow'\n axamp = plt.axes([0.2, 0.15, 0.65, 0.03], facecolor=axcolor)\n\n samp = Slider(axamp, 'Gain', 0.1, 10.0, valinit=a0)\n\n def update(jnk):\n amp = samp.val\n wns, zetas, poles = control.damp(control.feedback(G*amp, 1), doprint=False)\n l_poles.set_xdata(np.real(poles))\n l_poles.set_ydata(np.imag(poles))\n #for h, pole, zeta, wn in zip(h_text, poles, zetas, wns):\n # h.set_x(np.real(pole))\n # h.set_y(np.imag(pole))\n # h.set_text(r'{:0.2f}, {:0.2f}'.format(zeta, wn))\n fig.canvas.draw_idle()\n\n samp.on_changed(update)\n update(0)\n ax.axis([-10, 10, -10, 10]);\n\nG = control.tf([1, 3, 4], [1, 2, 3, 4])\nroot_locus(G)",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code"
]
] |
c500fc6c74efb4d0ef2fe10932b86da5218842ee
| 205,267 |
ipynb
|
Jupyter Notebook
|
examples/si_cross_entropy_loss.ipynb
|
microsoft/BackwardCompatibilityML
|
5910e485453f07fd5c85114d15c423c5db521122
|
[
"MIT"
] | 54 |
2020-09-11T18:36:59.000Z
|
2022-03-29T00:47:55.000Z
|
examples/si_cross_entropy_loss.ipynb
|
microsoft/BackwardCompatibilityML
|
5910e485453f07fd5c85114d15c423c5db521122
|
[
"MIT"
] | 115 |
2020-10-08T16:55:34.000Z
|
2022-03-12T00:50:21.000Z
|
examples/si_cross_entropy_loss.ipynb
|
microsoft/BackwardCompatibilityML
|
5910e485453f07fd5c85114d15c423c5db521122
|
[
"MIT"
] | 11 |
2020-10-04T09:40:11.000Z
|
2021-12-21T21:03:33.000Z
| 226.064978 | 30,532 | 0.896311 |
[
[
[
"import torch\nimport torchvision\nimport torch.nn as nn\nimport torch.nn.functional as F\nimport torch.optim as optim\nimport matplotlib.pyplot as plt\nimport random\n\nimport backwardcompatibilityml.loss as bcloss\nimport backwardcompatibilityml.scores as scores\n\n# Initialize random seed\nrandom.seed(123)\ntorch.manual_seed(456)\ntorch.backends.cudnn.deterministic = True\ntorch.backends.cudnn.benchmark = False\n\n%matplotlib inline",
"_____no_output_____"
],
[
"n_epochs = 3\nbatch_size_train = 64\nbatch_size_test = 1000\nlearning_rate = 0.01\nmomentum = 0.5\nlog_interval = 10\n\ntorch.backends.cudnn.enabled = False",
"_____no_output_____"
],
[
"train_loader = list(torch.utils.data.DataLoader(\n torchvision.datasets.MNIST('datasets/', train=True, download=True,\n transform=torchvision.transforms.Compose([\n torchvision.transforms.ToTensor(),\n torchvision.transforms.Normalize(\n (0.1307,), (0.3081,))\n ])),\n batch_size=batch_size_train, shuffle=True))\n\ntest_loader = list(torch.utils.data.DataLoader(\n torchvision.datasets.MNIST('datasets/', train=False, download=True,\n transform=torchvision.transforms.Compose([\n torchvision.transforms.ToTensor(),\n torchvision.transforms.Normalize(\n (0.1307,), (0.3081,))\n ])),\n batch_size=batch_size_test, shuffle=True))",
"_____no_output_____"
],
[
"train_loader_a = train_loader[:int(len(train_loader)/2)]\ntrain_loader_b = train_loader[int(len(train_loader)/2):]",
"_____no_output_____"
],
[
"fig = plt.figure()\nfor i in range(6):\n plt.subplot(2,3,i+1)\n plt.tight_layout()\n plt.imshow(train_loader_a[0][0][i][0], cmap='gray', interpolation='none')\n plt.title(\"Ground Truth: {}\".format(train_loader_a[0][1][i]))\n plt.xticks([])\n plt.yticks([])\nfig",
"_____no_output_____"
],
[
"class Net(nn.Module):\n def __init__(self):\n super(Net, self).__init__()\n self.conv1 = nn.Conv2d(1, 10, kernel_size=5)\n self.conv2 = nn.Conv2d(10, 20, kernel_size=5)\n self.conv2_drop = nn.Dropout2d()\n self.fc1 = nn.Linear(320, 50)\n self.fc2 = nn.Linear(50, 10)\n\n def forward(self, x):\n x = F.relu(F.max_pool2d(self.conv1(x), 2))\n x = F.relu(F.max_pool2d(self.conv2_drop(self.conv2(x)), 2))\n x = x.view(-1, 320)\n x = F.relu(self.fc1(x))\n x = F.dropout(x, training=self.training)\n x = self.fc2(x)\n return x, F.softmax(x, dim=1), F.log_softmax(x, dim=1)",
"_____no_output_____"
],
[
"network = Net()\noptimizer = optim.SGD(network.parameters(), lr=learning_rate, momentum=momentum)",
"_____no_output_____"
],
[
"train_losses = []\ntrain_counter = []\ntest_losses = []\ntest_counter = [i*len(train_loader_a)*batch_size_train for i in range(n_epochs + 1)]",
"_____no_output_____"
],
[
"def train(epoch):\n network.train()\n for batch_idx, (data, target) in enumerate(train_loader_a):\n optimizer.zero_grad()\n _, _, output = network(data)\n loss = F.nll_loss(output, target)\n loss.backward()\n optimizer.step()\n if batch_idx % log_interval == 0:\n print('Train Epoch: {} [{}/{} ({:.0f}%)]\\tLoss: {:.6f}'.format(\n epoch, batch_idx * len(data), len(train_loader_a)*batch_size_train,\n 100. * batch_idx / len(train_loader_a), loss.item()))\n train_losses.append(loss.item())\n train_counter.append(\n (batch_idx*64) + ((epoch-1)*len(train_loader_a)*batch_size_train))",
"_____no_output_____"
],
[
"def test():\n network.eval()\n test_loss = 0\n correct = 0\n with torch.no_grad():\n for data, target in test_loader:\n _, _, output = network(data)\n test_loss += F.nll_loss(output, target, reduction=\"sum\").item()\n pred = output.data.max(1, keepdim=True)[1]\n correct += pred.eq(target.data.view_as(pred)).sum()\n test_loss /= len(train_loader_a)*batch_size_train\n test_losses.append(test_loss)\n print('\\nTest set: Avg. loss: {:.4f}, Accuracy: {}/{} ({:.0f}%)\\n'.format(\n test_loss, correct, len(train_loader_a)*batch_size_train,\n 100. * correct / (len(train_loader_a)*batch_size_train)))",
"_____no_output_____"
],
[
"test()\nfor epoch in range(1, n_epochs + 1):\n train(epoch)\n test()",
"\nTest set: Avg. loss: 0.7645, Accuracy: 1359/30016 (5%)\n\nTrain Epoch: 1 [0/30016 (0%)]\tLoss: 2.338004\nTrain Epoch: 1 [640/30016 (2%)]\tLoss: 2.281939\nTrain Epoch: 1 [1280/30016 (4%)]\tLoss: 2.274189\nTrain Epoch: 1 [1920/30016 (6%)]\tLoss: 2.249804\nTrain Epoch: 1 [2560/30016 (9%)]\tLoss: 2.218965\nTrain Epoch: 1 [3200/30016 (11%)]\tLoss: 2.199637\nTrain Epoch: 1 [3840/30016 (13%)]\tLoss: 2.167684\nTrain Epoch: 1 [4480/30016 (15%)]\tLoss: 2.119798\nTrain Epoch: 1 [5120/30016 (17%)]\tLoss: 2.025222\nTrain Epoch: 1 [5760/30016 (19%)]\tLoss: 1.932501\nTrain Epoch: 1 [6400/30016 (21%)]\tLoss: 1.671945\nTrain Epoch: 1 [7040/30016 (23%)]\tLoss: 1.561831\nTrain Epoch: 1 [7680/30016 (26%)]\tLoss: 1.626387\nTrain Epoch: 1 [8320/30016 (28%)]\tLoss: 1.685211\nTrain Epoch: 1 [8960/30016 (30%)]\tLoss: 1.400331\nTrain Epoch: 1 [9600/30016 (32%)]\tLoss: 1.427622\nTrain Epoch: 1 [10240/30016 (34%)]\tLoss: 0.989379\nTrain Epoch: 1 [10880/30016 (36%)]\tLoss: 1.192472\nTrain Epoch: 1 [11520/30016 (38%)]\tLoss: 1.240278\nTrain Epoch: 1 [12160/30016 (41%)]\tLoss: 1.094558\nTrain Epoch: 1 [12800/30016 (43%)]\tLoss: 1.126665\nTrain Epoch: 1 [13440/30016 (45%)]\tLoss: 0.786467\nTrain Epoch: 1 [14080/30016 (47%)]\tLoss: 0.961599\nTrain Epoch: 1 [14720/30016 (49%)]\tLoss: 0.717606\nTrain Epoch: 1 [15360/30016 (51%)]\tLoss: 0.923883\nTrain Epoch: 1 [16000/30016 (53%)]\tLoss: 0.835599\nTrain Epoch: 1 [16640/30016 (55%)]\tLoss: 0.820179\nTrain Epoch: 1 [17280/30016 (58%)]\tLoss: 1.214673\nTrain Epoch: 1 [17920/30016 (60%)]\tLoss: 0.545910\nTrain Epoch: 1 [18560/30016 (62%)]\tLoss: 0.864912\nTrain Epoch: 1 [19200/30016 (64%)]\tLoss: 1.045161\nTrain Epoch: 1 [19840/30016 (66%)]\tLoss: 0.737489\nTrain Epoch: 1 [20480/30016 (68%)]\tLoss: 0.576639\nTrain Epoch: 1 [21120/30016 (70%)]\tLoss: 0.706978\nTrain Epoch: 1 [21760/30016 (72%)]\tLoss: 0.708799\nTrain Epoch: 1 [22400/30016 (75%)]\tLoss: 0.598541\nTrain Epoch: 1 [23040/30016 (77%)]\tLoss: 0.870604\nTrain Epoch: 1 [23680/30016 (79%)]\tLoss: 0.716791\nTrain Epoch: 1 [24320/30016 (81%)]\tLoss: 0.696189\nTrain Epoch: 1 [24960/30016 (83%)]\tLoss: 0.651915\nTrain Epoch: 1 [25600/30016 (85%)]\tLoss: 0.746274\nTrain Epoch: 1 [26240/30016 (87%)]\tLoss: 0.463703\nTrain Epoch: 1 [26880/30016 (90%)]\tLoss: 0.636440\nTrain Epoch: 1 [27520/30016 (92%)]\tLoss: 0.778516\nTrain Epoch: 1 [28160/30016 (94%)]\tLoss: 0.637036\nTrain Epoch: 1 [28800/30016 (96%)]\tLoss: 0.829187\nTrain Epoch: 1 [29440/30016 (98%)]\tLoss: 0.561778\n\nTest set: Avg. loss: 0.0993, Accuracy: 9171/30016 (31%)\n\nTrain Epoch: 2 [0/30016 (0%)]\tLoss: 0.611573\nTrain Epoch: 2 [640/30016 (2%)]\tLoss: 0.685103\nTrain Epoch: 2 [1280/30016 (4%)]\tLoss: 0.856729\nTrain Epoch: 2 [1920/30016 (6%)]\tLoss: 0.519098\nTrain Epoch: 2 [2560/30016 (9%)]\tLoss: 0.601045\nTrain Epoch: 2 [3200/30016 (11%)]\tLoss: 0.542169\nTrain Epoch: 2 [3840/30016 (13%)]\tLoss: 0.590203\nTrain Epoch: 2 [4480/30016 (15%)]\tLoss: 0.535176\nTrain Epoch: 2 [5120/30016 (17%)]\tLoss: 0.604264\nTrain Epoch: 2 [5760/30016 (19%)]\tLoss: 0.699412\nTrain Epoch: 2 [6400/30016 (21%)]\tLoss: 0.469525\nTrain Epoch: 2 [7040/30016 (23%)]\tLoss: 0.458269\nTrain Epoch: 2 [7680/30016 (26%)]\tLoss: 0.676925\nTrain Epoch: 2 [8320/30016 (28%)]\tLoss: 0.671360\nTrain Epoch: 2 [8960/30016 (30%)]\tLoss: 0.663215\nTrain Epoch: 2 [9600/30016 (32%)]\tLoss: 0.678063\nTrain Epoch: 2 [10240/30016 (34%)]\tLoss: 0.329252\nTrain Epoch: 2 [10880/30016 (36%)]\tLoss: 0.597346\nTrain Epoch: 2 [11520/30016 (38%)]\tLoss: 0.701631\nTrain Epoch: 2 [12160/30016 (41%)]\tLoss: 0.436469\nTrain Epoch: 2 [12800/30016 (43%)]\tLoss: 0.484635\nTrain Epoch: 2 [13440/30016 (45%)]\tLoss: 0.334166\nTrain Epoch: 2 [14080/30016 (47%)]\tLoss: 0.391909\nTrain Epoch: 2 [14720/30016 (49%)]\tLoss: 0.508151\nTrain Epoch: 2 [15360/30016 (51%)]\tLoss: 0.416259\nTrain Epoch: 2 [16000/30016 (53%)]\tLoss: 0.356686\nTrain Epoch: 2 [16640/30016 (55%)]\tLoss: 0.406188\nTrain Epoch: 2 [17280/30016 (58%)]\tLoss: 0.717582\nTrain Epoch: 2 [17920/30016 (60%)]\tLoss: 0.322463\nTrain Epoch: 2 [18560/30016 (62%)]\tLoss: 0.713308\nTrain Epoch: 2 [19200/30016 (64%)]\tLoss: 0.486474\nTrain Epoch: 2 [19840/30016 (66%)]\tLoss: 0.370916\nTrain Epoch: 2 [20480/30016 (68%)]\tLoss: 0.453681\nTrain Epoch: 2 [21120/30016 (70%)]\tLoss: 0.339929\nTrain Epoch: 2 [21760/30016 (72%)]\tLoss: 0.503270\nTrain Epoch: 2 [22400/30016 (75%)]\tLoss: 0.363914\nTrain Epoch: 2 [23040/30016 (77%)]\tLoss: 0.569946\nTrain Epoch: 2 [23680/30016 (79%)]\tLoss: 0.385622\nTrain Epoch: 2 [24320/30016 (81%)]\tLoss: 0.560251\nTrain Epoch: 2 [24960/30016 (83%)]\tLoss: 0.700014\nTrain Epoch: 2 [25600/30016 (85%)]\tLoss: 0.483964\nTrain Epoch: 2 [26240/30016 (87%)]\tLoss: 0.375324\nTrain Epoch: 2 [26880/30016 (90%)]\tLoss: 0.397195\nTrain Epoch: 2 [27520/30016 (92%)]\tLoss: 0.387728\nTrain Epoch: 2 [28160/30016 (94%)]\tLoss: 0.463883\nTrain Epoch: 2 [28800/30016 (96%)]\tLoss: 0.671692\nTrain Epoch: 2 [29440/30016 (98%)]\tLoss: 0.200388\n\nTest set: Avg. loss: 0.0606, Accuracy: 9467/30016 (32%)\n\nTrain Epoch: 3 [0/30016 (0%)]\tLoss: 0.433216\nTrain Epoch: 3 [640/30016 (2%)]\tLoss: 0.626047\nTrain Epoch: 3 [1280/30016 (4%)]\tLoss: 0.614806\nTrain Epoch: 3 [1920/30016 (6%)]\tLoss: 0.351351\nTrain Epoch: 3 [2560/30016 (9%)]\tLoss: 0.372902\nTrain Epoch: 3 [3200/30016 (11%)]\tLoss: 0.315961\nTrain Epoch: 3 [3840/30016 (13%)]\tLoss: 0.629242\nTrain Epoch: 3 [4480/30016 (15%)]\tLoss: 0.325787\nTrain Epoch: 3 [5120/30016 (17%)]\tLoss: 0.332519\nTrain Epoch: 3 [5760/30016 (19%)]\tLoss: 0.428229\nTrain Epoch: 3 [6400/30016 (21%)]\tLoss: 0.375956\nTrain Epoch: 3 [7040/30016 (23%)]\tLoss: 0.369950\nTrain Epoch: 3 [7680/30016 (26%)]\tLoss: 0.362622\nTrain Epoch: 3 [8320/30016 (28%)]\tLoss: 0.520664\nTrain Epoch: 3 [8960/30016 (30%)]\tLoss: 0.621075\nTrain Epoch: 3 [9600/30016 (32%)]\tLoss: 0.418881\nTrain Epoch: 3 [10240/30016 (34%)]\tLoss: 0.219880\nTrain Epoch: 3 [10880/30016 (36%)]\tLoss: 0.499471\nTrain Epoch: 3 [11520/30016 (38%)]\tLoss: 0.484748\nTrain Epoch: 3 [12160/30016 (41%)]\tLoss: 0.372163\nTrain Epoch: 3 [12800/30016 (43%)]\tLoss: 0.318991\nTrain Epoch: 3 [13440/30016 (45%)]\tLoss: 0.298097\nTrain Epoch: 3 [14080/30016 (47%)]\tLoss: 0.514588\nTrain Epoch: 3 [14720/30016 (49%)]\tLoss: 0.333074\nTrain Epoch: 3 [15360/30016 (51%)]\tLoss: 0.374257\nTrain Epoch: 3 [16000/30016 (53%)]\tLoss: 0.249366\nTrain Epoch: 3 [16640/30016 (55%)]\tLoss: 0.413403\nTrain Epoch: 3 [17280/30016 (58%)]\tLoss: 0.644515\nTrain Epoch: 3 [17920/30016 (60%)]\tLoss: 0.345536\nTrain Epoch: 3 [18560/30016 (62%)]\tLoss: 0.406339\nTrain Epoch: 3 [19200/30016 (64%)]\tLoss: 0.398300\nTrain Epoch: 3 [19840/30016 (66%)]\tLoss: 0.237821\nTrain Epoch: 3 [20480/30016 (68%)]\tLoss: 0.485557\nTrain Epoch: 3 [21120/30016 (70%)]\tLoss: 0.381991\nTrain Epoch: 3 [21760/30016 (72%)]\tLoss: 0.561565\nTrain Epoch: 3 [22400/30016 (75%)]\tLoss: 0.319745\nTrain Epoch: 3 [23040/30016 (77%)]\tLoss: 0.636540\nTrain Epoch: 3 [23680/30016 (79%)]\tLoss: 0.355421\nTrain Epoch: 3 [24320/30016 (81%)]\tLoss: 0.471419\nTrain Epoch: 3 [24960/30016 (83%)]\tLoss: 0.373985\nTrain Epoch: 3 [25600/30016 (85%)]\tLoss: 0.348540\nTrain Epoch: 3 [26240/30016 (87%)]\tLoss: 0.196206\nTrain Epoch: 3 [26880/30016 (90%)]\tLoss: 0.278208\nTrain Epoch: 3 [27520/30016 (92%)]\tLoss: 0.475131\nTrain Epoch: 3 [28160/30016 (94%)]\tLoss: 0.369040\nTrain Epoch: 3 [28800/30016 (96%)]\tLoss: 0.359578\nTrain Epoch: 3 [29440/30016 (98%)]\tLoss: 0.251359\n\nTest set: Avg. loss: 0.0484, Accuracy: 9571/30016 (32%)\n\n"
],
[
"fig = plt.figure()\nplt.plot(train_counter, train_losses, color='blue')\nplt.scatter(test_counter, test_losses, color='red')\nplt.legend(['Train Loss', 'Test Loss'], loc='upper right')\nplt.xlabel('number of training examples seen')\nplt.ylabel('negative log likelihood loss')\nfig",
"_____no_output_____"
],
[
"with torch.no_grad():\n _, _, output = network(test_loader[0][0])",
"_____no_output_____"
],
[
"fig = plt.figure()\nfor i in range(6):\n plt.subplot(2,3,i+1)\n plt.tight_layout()\n plt.imshow(test_loader[0][0][i][0], cmap='gray', interpolation='none')\n plt.title(\"Prediction: {}\".format(\n output.data.max(1, keepdim=True)[1][i].item()))\n plt.xticks([])\n plt.yticks([])\nfig",
"_____no_output_____"
],
[
"import copy\nimport importlib\nimportlib.reload(bcloss)\n\nh1 = copy.deepcopy(network)\nh2 = copy.deepcopy(network)\nh1.eval()\nnew_optimizer = optim.SGD(h2.parameters(), lr=learning_rate, momentum=momentum)\nlambda_c = 1.0\nsi_loss = bcloss.StrictImitationCrossEntropyLoss(h1, h2, lambda_c)",
"_____no_output_____"
],
[
"update_train_losses = []\nupdate_train_counter = []\nupdate_test_losses = []\nupdate_test_counter = [i*len(train_loader_b)*batch_size_train for i in range(n_epochs + 1)]",
"_____no_output_____"
],
[
"def train_update(epoch):\n for batch_idx, (data, target) in enumerate(train_loader_b):\n new_optimizer.zero_grad()\n loss = si_loss(data, target)\n loss.backward()\n new_optimizer.step()\n if batch_idx % log_interval == 0:\n print('Train Epoch: {} [{}/{} ({:.0f}%)]\\tLoss: {:.6f}'.format(\n epoch, batch_idx * len(data), len(train_loader_b)*batch_size_train,\n 100. * batch_idx / len(train_loader_b), loss.item()))\n update_train_losses.append(loss.item())\n update_train_counter.append(\n (batch_idx*64) + ((epoch-1)*len(train_loader_b)*batch_size_train))",
"_____no_output_____"
],
[
"def test_update():\n h2.eval()\n test_loss = 0\n correct = 0\n with torch.no_grad():\n for data, target in test_loader:\n _, _, output = h2(data)\n test_loss += F.nll_loss(output, target, reduction=\"sum\").item()\n pred = output.data.max(1, keepdim=True)[1]\n correct += pred.eq(target.data.view_as(pred)).sum()\n test_loss /= len(train_loader_b)*batch_size_train\n update_test_losses.append(test_loss)\n print('\\nTest set: Avg. loss: {:.4f}, Accuracy: {}/{} ({:.0f}%)\\n'.format(\n test_loss, correct, len(train_loader_b)*batch_size_train,\n 100. * correct / (len(train_loader_b)*batch_size_train)))",
"_____no_output_____"
],
[
"test_update()\nfor epoch in range(1, n_epochs + 1):\n train_update(epoch)\n test_update()",
"\nTest set: Avg. loss: 0.0484, Accuracy: 9571/30016 (32%)\n\nTrain Epoch: 1 [0/30016 (0%)]\tLoss: 0.187793\nTrain Epoch: 1 [640/30016 (2%)]\tLoss: 0.314032\nTrain Epoch: 1 [1280/30016 (4%)]\tLoss: 0.157210\nTrain Epoch: 1 [1920/30016 (6%)]\tLoss: 0.271693\nTrain Epoch: 1 [2560/30016 (9%)]\tLoss: 0.172420\nTrain Epoch: 1 [3200/30016 (11%)]\tLoss: 0.154311\nTrain Epoch: 1 [3840/30016 (13%)]\tLoss: 0.053996\nTrain Epoch: 1 [4480/30016 (15%)]\tLoss: 0.195913\nTrain Epoch: 1 [5120/30016 (17%)]\tLoss: 0.060688\nTrain Epoch: 1 [5760/30016 (19%)]\tLoss: 0.115756\nTrain Epoch: 1 [6400/30016 (21%)]\tLoss: 0.144222\nTrain Epoch: 1 [7040/30016 (23%)]\tLoss: 0.127553\nTrain Epoch: 1 [7680/30016 (26%)]\tLoss: 0.203719\nTrain Epoch: 1 [8320/30016 (28%)]\tLoss: 0.158996\nTrain Epoch: 1 [8960/30016 (30%)]\tLoss: 0.206717\nTrain Epoch: 1 [9600/30016 (32%)]\tLoss: 0.083080\nTrain Epoch: 1 [10240/30016 (34%)]\tLoss: 0.071567\nTrain Epoch: 1 [10880/30016 (36%)]\tLoss: 0.206395\nTrain Epoch: 1 [11520/30016 (38%)]\tLoss: 0.061903\nTrain Epoch: 1 [12160/30016 (41%)]\tLoss: 0.063054\nTrain Epoch: 1 [12800/30016 (43%)]\tLoss: 0.182085\nTrain Epoch: 1 [13440/30016 (45%)]\tLoss: 0.154939\nTrain Epoch: 1 [14080/30016 (47%)]\tLoss: 0.141559\nTrain Epoch: 1 [14720/30016 (49%)]\tLoss: 0.216437\nTrain Epoch: 1 [15360/30016 (51%)]\tLoss: 0.147652\nTrain Epoch: 1 [16000/30016 (53%)]\tLoss: 0.025959\nTrain Epoch: 1 [16640/30016 (55%)]\tLoss: 0.455275\nTrain Epoch: 1 [17280/30016 (58%)]\tLoss: 0.099623\nTrain Epoch: 1 [17920/30016 (60%)]\tLoss: 0.271166\nTrain Epoch: 1 [18560/30016 (62%)]\tLoss: 0.170574\nTrain Epoch: 1 [19200/30016 (64%)]\tLoss: 0.067997\nTrain Epoch: 1 [19840/30016 (66%)]\tLoss: 0.141764\nTrain Epoch: 1 [20480/30016 (68%)]\tLoss: 0.152821\nTrain Epoch: 1 [21120/30016 (70%)]\tLoss: 0.107717\nTrain Epoch: 1 [21760/30016 (72%)]\tLoss: 0.233322\nTrain Epoch: 1 [22400/30016 (75%)]\tLoss: 0.221370\nTrain Epoch: 1 [23040/30016 (77%)]\tLoss: 0.093659\nTrain Epoch: 1 [23680/30016 (79%)]\tLoss: 0.084277\nTrain Epoch: 1 [24320/30016 (81%)]\tLoss: 0.053813\nTrain Epoch: 1 [24960/30016 (83%)]\tLoss: 0.182935\nTrain Epoch: 1 [25600/30016 (85%)]\tLoss: 0.036450\nTrain Epoch: 1 [26240/30016 (87%)]\tLoss: 0.137724\nTrain Epoch: 1 [26880/30016 (90%)]\tLoss: 0.175103\nTrain Epoch: 1 [27520/30016 (92%)]\tLoss: 0.064044\nTrain Epoch: 1 [28160/30016 (94%)]\tLoss: 0.070072\nTrain Epoch: 1 [28800/30016 (96%)]\tLoss: 0.132992\nTrain Epoch: 1 [29440/30016 (98%)]\tLoss: 0.125697\n\nTest set: Avg. loss: 0.0280, Accuracy: 9748/30016 (32%)\n\nTrain Epoch: 2 [0/30016 (0%)]\tLoss: 0.064774\nTrain Epoch: 2 [640/30016 (2%)]\tLoss: 0.173727\nTrain Epoch: 2 [1280/30016 (4%)]\tLoss: 0.120233\nTrain Epoch: 2 [1920/30016 (6%)]\tLoss: 0.156266\nTrain Epoch: 2 [2560/30016 (9%)]\tLoss: 0.112449\nTrain Epoch: 2 [3200/30016 (11%)]\tLoss: 0.091798\nTrain Epoch: 2 [3840/30016 (13%)]\tLoss: 0.024527\nTrain Epoch: 2 [4480/30016 (15%)]\tLoss: 0.144569\nTrain Epoch: 2 [5120/30016 (17%)]\tLoss: 0.037021\nTrain Epoch: 2 [5760/30016 (19%)]\tLoss: 0.055271\nTrain Epoch: 2 [6400/30016 (21%)]\tLoss: 0.071087\nTrain Epoch: 2 [7040/30016 (23%)]\tLoss: 0.077232\nTrain Epoch: 2 [7680/30016 (26%)]\tLoss: 0.113851\nTrain Epoch: 2 [8320/30016 (28%)]\tLoss: 0.086933\nTrain Epoch: 2 [8960/30016 (30%)]\tLoss: 0.126416\nTrain Epoch: 2 [9600/30016 (32%)]\tLoss: 0.060134\nTrain Epoch: 2 [10240/30016 (34%)]\tLoss: 0.045356\nTrain Epoch: 2 [10880/30016 (36%)]\tLoss: 0.180100\nTrain Epoch: 2 [11520/30016 (38%)]\tLoss: 0.067510\nTrain Epoch: 2 [12160/30016 (41%)]\tLoss: 0.057529\nTrain Epoch: 2 [12800/30016 (43%)]\tLoss: 0.116505\nTrain Epoch: 2 [13440/30016 (45%)]\tLoss: 0.079541\nTrain Epoch: 2 [14080/30016 (47%)]\tLoss: 0.090319\nTrain Epoch: 2 [14720/30016 (49%)]\tLoss: 0.164027\nTrain Epoch: 2 [15360/30016 (51%)]\tLoss: 0.084880\nTrain Epoch: 2 [16000/30016 (53%)]\tLoss: 0.011989\nTrain Epoch: 2 [16640/30016 (55%)]\tLoss: 0.373237\nTrain Epoch: 2 [17280/30016 (58%)]\tLoss: 0.067686\nTrain Epoch: 2 [17920/30016 (60%)]\tLoss: 0.210395\nTrain Epoch: 2 [18560/30016 (62%)]\tLoss: 0.143066\nTrain Epoch: 2 [19200/30016 (64%)]\tLoss: 0.037438\nTrain Epoch: 2 [19840/30016 (66%)]\tLoss: 0.096844\nTrain Epoch: 2 [20480/30016 (68%)]\tLoss: 0.121761\nTrain Epoch: 2 [21120/30016 (70%)]\tLoss: 0.090457\nTrain Epoch: 2 [21760/30016 (72%)]\tLoss: 0.234806\nTrain Epoch: 2 [22400/30016 (75%)]\tLoss: 0.126038\nTrain Epoch: 2 [23040/30016 (77%)]\tLoss: 0.072192\nTrain Epoch: 2 [23680/30016 (79%)]\tLoss: 0.058036\nTrain Epoch: 2 [24320/30016 (81%)]\tLoss: 0.036218\nTrain Epoch: 2 [24960/30016 (83%)]\tLoss: 0.144304\nTrain Epoch: 2 [25600/30016 (85%)]\tLoss: 0.024255\nTrain Epoch: 2 [26240/30016 (87%)]\tLoss: 0.135833\nTrain Epoch: 2 [26880/30016 (90%)]\tLoss: 0.126680\nTrain Epoch: 2 [27520/30016 (92%)]\tLoss: 0.031605\nTrain Epoch: 2 [28160/30016 (94%)]\tLoss: 0.049616\nTrain Epoch: 2 [28800/30016 (96%)]\tLoss: 0.113463\nTrain Epoch: 2 [29440/30016 (98%)]\tLoss: 0.067239\n\nTest set: Avg. loss: 0.0233, Accuracy: 9791/30016 (33%)\n\nTrain Epoch: 3 [0/30016 (0%)]\tLoss: 0.043380\nTrain Epoch: 3 [640/30016 (2%)]\tLoss: 0.127507\nTrain Epoch: 3 [1280/30016 (4%)]\tLoss: 0.101459\nTrain Epoch: 3 [1920/30016 (6%)]\tLoss: 0.115581\nTrain Epoch: 3 [2560/30016 (9%)]\tLoss: 0.083779\nTrain Epoch: 3 [3200/30016 (11%)]\tLoss: 0.074234\nTrain Epoch: 3 [3840/30016 (13%)]\tLoss: 0.015534\nTrain Epoch: 3 [4480/30016 (15%)]\tLoss: 0.119388\nTrain Epoch: 3 [5120/30016 (17%)]\tLoss: 0.026608\nTrain Epoch: 3 [5760/30016 (19%)]\tLoss: 0.043011\nTrain Epoch: 3 [6400/30016 (21%)]\tLoss: 0.051572\nTrain Epoch: 3 [7040/30016 (23%)]\tLoss: 0.063801\nTrain Epoch: 3 [7680/30016 (26%)]\tLoss: 0.065052\nTrain Epoch: 3 [8320/30016 (28%)]\tLoss: 0.063846\nTrain Epoch: 3 [8960/30016 (30%)]\tLoss: 0.113324\nTrain Epoch: 3 [9600/30016 (32%)]\tLoss: 0.063456\nTrain Epoch: 3 [10240/30016 (34%)]\tLoss: 0.033673\nTrain Epoch: 3 [10880/30016 (36%)]\tLoss: 0.158807\nTrain Epoch: 3 [11520/30016 (38%)]\tLoss: 0.072090\nTrain Epoch: 3 [12160/30016 (41%)]\tLoss: 0.057743\nTrain Epoch: 3 [12800/30016 (43%)]\tLoss: 0.087218\nTrain Epoch: 3 [13440/30016 (45%)]\tLoss: 0.055660\nTrain Epoch: 3 [14080/30016 (47%)]\tLoss: 0.070236\nTrain Epoch: 3 [14720/30016 (49%)]\tLoss: 0.137697\nTrain Epoch: 3 [15360/30016 (51%)]\tLoss: 0.062384\nTrain Epoch: 3 [16000/30016 (53%)]\tLoss: 0.007788\nTrain Epoch: 3 [16640/30016 (55%)]\tLoss: 0.298692\nTrain Epoch: 3 [17280/30016 (58%)]\tLoss: 0.055365\nTrain Epoch: 3 [17920/30016 (60%)]\tLoss: 0.161824\nTrain Epoch: 3 [18560/30016 (62%)]\tLoss: 0.137108\nTrain Epoch: 3 [19200/30016 (64%)]\tLoss: 0.026783\nTrain Epoch: 3 [19840/30016 (66%)]\tLoss: 0.081656\nTrain Epoch: 3 [20480/30016 (68%)]\tLoss: 0.100504\nTrain Epoch: 3 [21120/30016 (70%)]\tLoss: 0.072531\nTrain Epoch: 3 [21760/30016 (72%)]\tLoss: 0.229085\nTrain Epoch: 3 [22400/30016 (75%)]\tLoss: 0.087314\nTrain Epoch: 3 [23040/30016 (77%)]\tLoss: 0.058883\nTrain Epoch: 3 [23680/30016 (79%)]\tLoss: 0.045408\nTrain Epoch: 3 [24320/30016 (81%)]\tLoss: 0.026812\nTrain Epoch: 3 [24960/30016 (83%)]\tLoss: 0.112134\nTrain Epoch: 3 [25600/30016 (85%)]\tLoss: 0.015544\nTrain Epoch: 3 [26240/30016 (87%)]\tLoss: 0.135921\nTrain Epoch: 3 [26880/30016 (90%)]\tLoss: 0.104146\nTrain Epoch: 3 [27520/30016 (92%)]\tLoss: 0.020319\nTrain Epoch: 3 [28160/30016 (94%)]\tLoss: 0.044700\nTrain Epoch: 3 [28800/30016 (96%)]\tLoss: 0.101523\nTrain Epoch: 3 [29440/30016 (98%)]\tLoss: 0.049281\n\nTest set: Avg. loss: 0.0209, Accuracy: 9814/30016 (33%)\n\n"
],
[
"fig = plt.figure()\nplt.plot(update_train_counter, update_train_losses, color='blue')\nplt.scatter(update_test_counter, update_test_losses, color='red')\nplt.legend(['Train Loss', 'Test Loss'], loc='upper right')\nplt.xlabel('number of training examples seen')\nplt.ylabel('negative log likelihood loss')\nfig",
"_____no_output_____"
],
[
"h2.eval()\nh1.eval()",
"_____no_output_____"
],
[
"test_index = 5",
"_____no_output_____"
],
[
"with torch.no_grad():\n _, _, h1_output = h1(test_loader[test_index][0])\n _, _, h2_output = h2(test_loader[test_index][0])",
"_____no_output_____"
],
[
"h1_labels = h1_output.data.max(1)[1]\nh2_labels = h2_output.data.max(1)[1]\nexpected_labels = test_loader[test_index][1]",
"_____no_output_____"
],
[
"fig = plt.figure()\nfor i in range(6):\n plt.subplot(2,3,i+1)\n plt.tight_layout()\n plt.imshow(test_loader[test_index][0][i][0], cmap='gray', interpolation='none')\n plt.title(\"Prediction: {}\".format(\n h2_labels[i].item()))\n plt.xticks([])\n plt.yticks([])\nfig",
"_____no_output_____"
],
[
"trust_compatibility = scores.trust_compatibility_score(h1_labels, h2_labels, expected_labels)\nerror_compatibility = scores.error_compatibility_score(h1_labels, h2_labels, expected_labels)\n\nprint(f\"Error Compatibility Score: {error_compatibility}\")\nprint(f\"Trust Compatibility Score: {trust_compatibility}\")",
"Error Compatibility Score: 0.47619047619047616\nTrust Compatibility Score: 0.9989561586638831\n"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
c50102d19aa4a85e5c0c75d0b4cf85460703d7b7
| 30,034 |
ipynb
|
Jupyter Notebook
|
FluentPython/Chapter05_1class_func.ipynb
|
Liuzkai/PythonScript
|
fb21ad80e085f6390ae970b81404f7e5c7923f4e
|
[
"MIT"
] | 1 |
2021-01-16T16:09:33.000Z
|
2021-01-16T16:09:33.000Z
|
FluentPython/Chapter05_1class_func.ipynb
|
Liuzkai/PythonScript
|
fb21ad80e085f6390ae970b81404f7e5c7923f4e
|
[
"MIT"
] | null | null | null |
FluentPython/Chapter05_1class_func.ipynb
|
Liuzkai/PythonScript
|
fb21ad80e085f6390ae970b81404f7e5c7923f4e
|
[
"MIT"
] | 1 |
2021-01-16T16:09:36.000Z
|
2021-01-16T16:09:36.000Z
| 20.543092 | 106 | 0.445429 |
[
[
[
"# 一等函数\n\n函数是一等对象。\n\n## 一等对象\n\n一等对象:\n- 在运行时创建\n- 能赋值给变量或数据结构中的元素\n- 能作为参数传给函数\n- 能作为函数的返回结果\n\n",
"_____no_output_____"
]
],
[
[
"def factorial(n):\n '''return n!'''\n return 1 if n < 2 else n * factorial(n-1)\n\n# 将函数看作是对象传入方法中:\nlist(map(factorial, range(11)))",
"_____no_output_____"
],
[
"dir(factorial)",
"_____no_output_____"
]
],
[
[
"\n## 可调用对象 Callable Object\n\n### 一共7种:\n\n- 用户定义的函数 : def或lambda\n- 内置函数\n- 内置方法\n- 方法:在类的定义体内定义的函数\n- 类:\\_\\_new\\_\\_:创建一个实例;\\_\\_init\\_\\_初始化实例。\\_\\_call\\_\\_实例可以作为函数调用\n- 类的实例:\\_\\_call\\_\\_实例可以作为函数调用\n- 生成器函数: yield的函数或方法,返回生成器对象(14章)",
"_____no_output_____"
]
],
[
[
"# callable\nimport random\n\nclass BingoCage:\n \n def __init__(self, items):\n self._items = list(items)\n random.shuffle(self._items)\n \n def pick(self):\n try:\n return self._items.pop()\n except IndexError:\n raise LookupError('pick from an empty BingCage')\n \n def __call__(self):\n '''\n The class is callable only on defined the __call__ function\n '''\n return self.pick()\n \n \nbingo = BingoCage(range(3))\nbingo.pick()",
"_____no_output_____"
],
[
"bingo()",
"_____no_output_____"
],
[
"callable(bingo)",
"_____no_output_____"
],
[
"dir(BingoCage)",
"_____no_output_____"
]
],
[
[
"## 函数内省",
"_____no_output_____"
]
],
[
[
"class C:pass #自定义类\nobj = C() #自定义类实例\n\ndef func():pass #自定义函数\n\nsorted( set(dir(func)) - set(dir(obj))) #求差集",
"_____no_output_____"
]
],
[
[
"### 函数专有的属性\n| 名称 | 类型 | 说明 |\n|:--------------------|:----|:----|\n| \\_\\_annotations\\_\\_ | dict |参数和返回值的注解 |\n|\\_\\_call\\_\\_ |method-wrapper|实现()运算符;即可调用对象协议|\n|\\_\\_closure\\_\\_ |tuple|函数闭包,即自由变量的绑定(通常是None)|\n|\\_\\_code\\_\\_ |code|编译成字节码的函数元数据和函数定义体\n|\\_\\_defaults\\_\\_ |tuple|形式参数的默认值\n|\\_\\_get\\_\\_ |method-wrapper|实现只读描述符协议(20章)\n|\\_\\_globals\\_\\_ |dict|函数所在模块中的全局变量\n|\\_\\_kwdefaults\\_\\_ |dict|仅限关键字形式参数的默认值\n|\\_\\_name\\_\\_ |str|函数名称\n|\\_\\_qualname\\_\\_ |str|函数的限定名称",
"_____no_output_____"
],
[
"## 函数参数\n\n- 仅限关键字参数\n- 使用\\*存无变量名的一个或以上个参数到元组中\n- 使用\\**存有变量名的一个或以上个参数到字典中\n\n\n\n\n",
"_____no_output_____"
]
],
[
[
"def tag(name, *content, cls=None, **attrs):\n \"\"\"生成一个或多个HTML标签\"\"\"\n if cls is not None:\n attrs['class'] = cls\n \n if attrs:\n attr_str = ''.join(' %s=\"%s\"'%(attr, value) \n for attr, value \n in sorted(attrs.items()))\n else:\n attr_str = ''\n \n if content:\n return '\\n'.join('<%s%s>%s</%s>'%(name, attr_str, c, name)\n for c in content)\n \n else:\n return '<%s%s />'%(name, attr_str)\n ",
"_____no_output_____"
],
[
"# 传入单个定位参数,生成一个指定名称的空标签。\n# name = 'br'\ntag('br')",
"_____no_output_____"
],
[
"# 第一个参数后面的任意个参数会被 *content 捕获,存入一个元组。\n# name = 'p'\n# content = ('hello')\ntag('p', 'hello')",
"_____no_output_____"
],
[
"# name = 'p'\n# content = ('hello', 'world')\ntag('p', 'hello', 'world')",
"_____no_output_____"
],
[
"# tag 函数签名中没有明确指定名称的关键字参数会被 **attrs 捕 获,存入一个字典。\n\n# name = 'p'\n# content = ('hello')\n# attrs['id'] = 33\ntag('p', 'hello', id=33)",
"_____no_output_____"
],
[
"# cls 参数只能作为关键字参数传入。\n\n# name = 'p'\n# content = ('hello', 'world')\n# cls = 'sidebar'\nprint(tag('p', 'hello', 'world', cls='sidebar'))",
"<p class=\"sidebar\">hello</p>\n<p class=\"sidebar\">world</p>\n"
],
[
"# 调用 tag 函数时,即便第一个定位参数也能作为关键字参数传入。\n\n# name = 'img'\n# attrs['content'] = 'testing' \ntag(content='testing', name=\"img\")",
"_____no_output_____"
],
[
"# 在 my_tag 前面加上 **,字典中的所有元素作为单个参数传入,\n# 同名键会绑定到对应的具名参数上,余下的则被 **attrs 捕获。\n\n# name = 'img'\n# attrs['title'] = 'Sunset Buolevard'\n# attrs['src'] = 'sunset.jpg'\n# cls = 'framed'\n\nmy_tag = {'name':'img', 'title':'Sunset Buolevard',\n 'src':'sunset.jpg', 'cls':'framed'}\ntag(**my_tag)",
"_____no_output_____"
]
],
[
[
"定义函数 时若想指定仅限关键字参数(如上面参数中的 *cls*),要把它们放到前面有 \\* 的参数后面。如果不想支持数量不定的定位参数,但是想支持仅限关键字参数,在签名中 放一个 \\*,如下所示:",
"_____no_output_____"
]
],
[
[
"def f(a, *, b):\n return a,b\n\nf(1, b=2) # b参数必须指定设置了",
"_____no_output_____"
]
],
[
[
"# 获取关于参数的信息\n",
"_____no_output_____"
]
],
[
[
"def clip(text, max_len = 80):\n \"\"\"\n 在max_len前面或后面的第一个空格处截断文本\n \"\"\"\n end = None\n if len(text) > max_len:\n space_before = text.rfind(' ', 0, max_len)\n if space_before >= 0:\n end = space_before\n else:\n space_after = text.rfind(' ', max_len)\n if space_after >= 0:\n end = space_after\n if end == None:\n end = len(text)\n return text[:end].rstrip()\n\nclip('I will learn python by myself every night.', 18)",
"_____no_output_____"
],
[
"'''\n函数对象:\n__defaults__: 定位参数和关键字参数的默认值元组\n__kwdefaults__:仅限关键字参数默认值元组\n\n参数的默认值只能通过它们在 __defaults__ 元组中的位置确定,\n因此要从后向前扫描才能把参数和默认值对应起来。 \n在这个示例中 clip 函数有两个参数,text 和 max_len,\n其中一个有默认值,即 80,因此它必然属于最后一个参数,即max_len。这有违常理。\n'''\nclip.__defaults__",
"_____no_output_____"
],
[
"\"\"\"\n__code__.varname:函数参数名称,但也包含了局部变量名\n\"\"\"\nclip.__code__.co_varnames",
"_____no_output_____"
],
[
"\"\"\"\n__code__.co_argcount:结合上面的变量,可以获得参数变量\n\n需要注意,这里返回的数量是不包括前缀*或**参数的。\n\"\"\"\nclip.__code__.co_argcount",
"_____no_output_____"
]
],
[
[
"## 使用inspect模块提取函数签名",
"_____no_output_____"
]
],
[
[
"from inspect import signature\n\nsig = signature(clip)\nsig",
"_____no_output_____"
],
[
"for name, param in sig.parameters.items():\n print (param.kind, \":\", name, \"=\", param.default)",
"POSITIONAL_OR_KEYWORD : text = <class 'inspect._empty'>\nPOSITIONAL_OR_KEYWORD : max_len = 80\n"
]
],
[
[
"如果没有默认值,返回的是inspect.\\_empty,因为None本身也是一个值。",
"_____no_output_____"
],
[
"| kind | meaning |\n| :--- | ------: |\n|POSITIONAL_OR_KEYWORD | 可以通过定位参数和关键字参数传入的形参(多数) |\n|VAR_POSITIONAL| 定位参数元组 |\n|VAR_KEYWORD| 关键字参数字典 |\n|KEYWORD_ONLY| 仅限关键字参数(python3新增) |\n|POSITIONAL_ONLY| 仅限定位参数,python声明函数的句法不支持 |",
"_____no_output_____"
],
[
"# 函数注解",
"_____no_output_____"
]
],
[
[
"\"\"\"\n没有设置函数注解时,返回一个空字典\n\"\"\"\nclip.__annotations__",
"_____no_output_____"
],
[
"def clip_ann(text:str, max_len:'int > 0' = 80) -> str:\n \"\"\"\n 在max_len前面或后面的第一个空格处截断文本\n \"\"\"\n end = None\n if len(text) > max_len:\n space_before = text.rfind(' ', 0, max_len)\n if space_before >= 0:\n end = space_before\n else:\n space_after = text.rfind(' ', max_len)\n if space_after >= 0:\n end = space_after\n if end == None:\n end = len(text)\n return text[:end].rstrip()",
"_____no_output_____"
],
[
"clip_ann.__annotations__",
"_____no_output_____"
]
],
[
[
"Python 对注解所做的唯一的事情是,把它们存储在函数的\n__annotations__ 属性里。仅此而已,Python 不做检查、不做强制、\n不做验证,什么操作都不做。换句话说,注解对 Python 解释器没有任何 意义。注解只是元数据,可以供 IDE、框架和装饰器等工具使用。",
"_____no_output_____"
],
[
"# 函数式编程\n\n涉及到两个包:\n- operator\n- functools",
"_____no_output_____"
],
[
"## operator",
"_____no_output_____"
],
[
"### 使用mul替代* (相乘)运算符",
"_____no_output_____"
]
],
[
[
"# 如何将运算符当作函数使用\n\n# 传统做法:\n\nfrom functools import reduce\n\ndef fact(n):\n return reduce(lambda a,b: a*b, range(1,n+1))\n\nfact(5)",
"_____no_output_____"
],
[
"# 如果使用operator库,就可以避免使用匿名函数\nfrom functools import reduce\nfrom operator import mul\n\ndef fact_op(n):\n return reduce(mul, range(1,n+1))\n\nfact_op(5)",
"_____no_output_____"
]
],
[
[
"### 使用itemgetter来代表[ ]序号运算符",
"_____no_output_____"
]
],
[
[
"metro_data = [\n ('Tokyo', 'JP', 36.933, (35.689722, 139.691667)),\n ('Delhi NCR', 'IN', 21.935, (28.613889, 77.208889)),\n ('Mexico City', 'MX', 20.142, (19.433333, -99.133333)),\n ('New York-Newark', 'US', 20.104, (40.808611, -74.020386)),\n ('Sao Paulo', 'BR', 19.649, (-23.547778, -46.635833)),\n ]\n\nfrom operator import itemgetter\nfor city in sorted(metro_data, key = itemgetter(1)):\n print(city)\n \n \n# 这里的itemgetter(1)等价于 lambda fields:fields[1]",
"('Sao Paulo', 'BR', 19.649, (-23.547778, -46.635833))\n('Delhi NCR', 'IN', 21.935, (28.613889, 77.208889))\n('Tokyo', 'JP', 36.933, (35.689722, 139.691667))\n('Mexico City', 'MX', 20.142, (19.433333, -99.133333))\n('New York-Newark', 'US', 20.104, (40.808611, -74.020386))\n"
],
[
"cc_name = itemgetter(1,0) # 将返回提取的值构成的元组\nfor city in metro_data:\n print( cc_name(city) )",
"('JP', 'Tokyo')\n('IN', 'Delhi NCR')\n('MX', 'Mexico City')\n('US', 'New York-Newark')\n('BR', 'Sao Paulo')\n"
]
],
[
[
"### 使用attrgetter获取指定的属性,类似ver.attr(点运算符)",
"_____no_output_____"
]
],
[
[
"from collections import namedtuple\nmetro_data = [\n ('Tokyo', 'JP', 36.933, (35.689722, 139.691667)),\n ('Delhi NCR', 'IN', 21.935, (28.613889, 77.208889)),\n ('Mexico City', 'MX', 20.142, (19.433333, -99.133333)),\n ('New York-Newark', 'US', 20.104, (40.808611, -74.020386)),\n ('Sao Paulo', 'BR', 19.649, (-23.547778, -46.635833)),\n ]\n\nLatLog = namedtuple('LatLong','lat long')\nMetropolis = namedtuple('Metropolis', 'name cc pop coord')\nmetro_areas = [Metropolis(name, cc, pop, LatLog(lat, long)) for name, cc\n , pop, (lat, long) in metro_data]\n\n# 点运算符\nmetro_areas[0].coord.lat",
"_____no_output_____"
],
[
"# 使用operator函数来代替操作符\n\nfrom operator import attrgetter\nname_lat = attrgetter('name', 'coord.lat')\n\n# 用坐标lat排序\nfor city in sorted(metro_areas, key = attrgetter('coord.lat')):\n print(name_lat(city))",
"('Sao Paulo', -23.547778)\n('Mexico City', 19.433333)\n('Delhi NCR', 28.613889)\n('Tokyo', 35.689722)\n('New York-Newark', 40.808611)\n"
]
],
[
[
"### methodcaller为参数调用指定的方法",
"_____no_output_____"
]
],
[
[
"from operator import methodcaller\ns = 'The time has come'\n\n# 可以把upcase看作是一个创建出来的函数\nupcase = methodcaller('upper')# 指定方法\n\nupcase(s)",
"_____no_output_____"
],
[
"# 冻结参数:replace(str, ' ', '-')--->部分应用\nhiphenate = methodcaller('replace', ' ','-')\n\nhiphenate(s)",
"_____no_output_____"
]
],
[
[
"### operator 中的函数列表",
"_____no_output_____"
]
],
[
[
"import operator\nfuncs = [name for name in dir(operator) if not name.startswith('_')]\nfor func in funcs:\n print(func)",
"abs\nadd\nand_\nattrgetter\nconcat\ncontains\ncountOf\ndelitem\neq\nfloordiv\nge\ngetitem\ngt\niadd\niand\niconcat\nifloordiv\nilshift\nimatmul\nimod\nimul\nindex\nindexOf\ninv\ninvert\nior\nipow\nirshift\nis_\nis_not\nisub\nitemgetter\nitruediv\nixor\nle\nlength_hint\nlshift\nlt\nmatmul\nmethodcaller\nmod\nmul\nne\nneg\nnot_\nor_\npos\npow\nrshift\nsetitem\nsub\ntruediv\ntruth\nxor\n"
]
],
[
[
"## functools\n### functools.partial 冻结参数\n\npartial 的第一个参数是一个可调用对象,后面跟着任意个要绑定的 定位参数和关键字参数。",
"_____no_output_____"
]
],
[
[
"from operator import mul\nfrom functools import partial\n\n# 将mul(a,b)的一个参数冻结为3\ntriple = partial(mul, 3)\n\ntriple(7)",
"_____no_output_____"
],
[
"# 当参数只接受只有一个参数的函数时\nlist(map(triple,range(1,10)))",
"_____no_output_____"
]
],
[
[
"### functools.partial 规范化函数",
"_____no_output_____"
]
],
[
[
"# 在需要经常使用的一些函数时,我们可以将其作为一个冻结变量,会更加方便\nimport unicodedata, functools\n# 提炼常用函数 unicodedata.normalize('NFC',s)\nnfc = functools.partial(unicodedata.normalize, 'NFC') \n\ns1 ='café'\ns2 = 'cafe\\u0301'\n\ns1 == s2",
"_____no_output_____"
],
[
"nfc(s1) == nfc(s2)",
"_____no_output_____"
],
[
"def tag(name, *content, cls=None, **attrs):\n \"\"\"生成一个或多个HTML标签\"\"\"\n if cls is not None:\n attrs['class'] = cls\n \n if attrs:\n attr_str = ''.join(' %s=\"%s\"'%(attr, value) \n for attr, value \n in sorted(attrs.items()))\n else:\n attr_str = ''\n \n if content:\n return '\\n'.join('<%s%s>%s</%s>'%(name, attr_str, c, name)\n for c in content)\n \n else:\n return '<%s%s />'%(name, attr_str)\n \n \n# 将tag函数参数进行部分冻结,使用将更方便:\nfrom functools import partial\npicture = partial(tag, 'img', cls='pic-frame')\npicture(src = 'wum.jpeg')",
"_____no_output_____"
],
[
"picture",
"_____no_output_____"
],
[
"picture.func",
"_____no_output_____"
],
[
"tag",
"_____no_output_____"
],
[
"picture.args",
"_____no_output_____"
],
[
"picture.keywords",
"_____no_output_____"
],
[
"def clip",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
c50109042d4104e6a3ab229325f6197dcf55665b
| 173,877 |
ipynb
|
Jupyter Notebook
|
Model backlog/Train/107-jigsaw-fold1-xlm-roberta-large-best6.ipynb
|
dimitreOliveira/Jigsaw-Multilingual-Toxic-Comment-Classification
|
44422e6aeeff227e22dbb5c05101322e9d4aabbe
|
[
"MIT"
] | 4 |
2020-06-23T02:31:07.000Z
|
2020-07-04T11:50:08.000Z
|
Model backlog/Train/107-jigsaw-fold1-xlm-roberta-large-best6.ipynb
|
dimitreOliveira/Jigsaw-Multilingual-Toxic-Comment-Classification
|
44422e6aeeff227e22dbb5c05101322e9d4aabbe
|
[
"MIT"
] | null | null | null |
Model backlog/Train/107-jigsaw-fold1-xlm-roberta-large-best6.ipynb
|
dimitreOliveira/Jigsaw-Multilingual-Toxic-Comment-Classification
|
44422e6aeeff227e22dbb5c05101322e9d4aabbe
|
[
"MIT"
] | null | null | null | 120.330104 | 62,072 | 0.804747 |
[
[
[
"## Dependencies",
"_____no_output_____"
]
],
[
[
"import json, warnings, shutil, glob\nfrom jigsaw_utility_scripts import *\nfrom scripts_step_lr_schedulers import *\nfrom transformers import TFXLMRobertaModel, XLMRobertaConfig\nfrom tensorflow.keras.models import Model\nfrom tensorflow.keras import optimizers, metrics, losses, layers\n\nSEED = 0\nseed_everything(SEED)\nwarnings.filterwarnings(\"ignore\")",
"_____no_output_____"
]
],
[
[
"## TPU configuration",
"_____no_output_____"
]
],
[
[
"strategy, tpu = set_up_strategy()\nprint(\"REPLICAS: \", strategy.num_replicas_in_sync)\nAUTO = tf.data.experimental.AUTOTUNE",
"Running on TPU grpc://10.0.0.2:8470\nREPLICAS: 8\n"
]
],
[
[
"# Load data",
"_____no_output_____"
]
],
[
[
"database_base_path = '/kaggle/input/jigsaw-data-split-roberta-192-ratio-2-clean-tail6/'\nk_fold = pd.read_csv(database_base_path + '5-fold.csv')\nvalid_df = pd.read_csv(\"/kaggle/input/jigsaw-multilingual-toxic-comment-classification/validation.csv\", \n usecols=['comment_text', 'toxic', 'lang'])\n\nprint('Train samples: %d' % len(k_fold))\ndisplay(k_fold.head())\nprint('Validation samples: %d' % len(valid_df))\ndisplay(valid_df.head())\n\nbase_data_path = 'fold_1/'\nfold_n = 1\n# Unzip files\n!tar -xf /kaggle/input/jigsaw-data-split-roberta-192-ratio-2-clean-tail6/fold_1.tar.gz",
"Train samples: 400830\n"
]
],
[
[
"# Model parameters",
"_____no_output_____"
]
],
[
[
"base_path = '/kaggle/input/jigsaw-transformers/XLM-RoBERTa/'\n\nconfig = {\n \"MAX_LEN\": 192,\n \"BATCH_SIZE\": 128,\n \"EPOCHS\": 4,\n \"LEARNING_RATE\": 1e-5, \n \"ES_PATIENCE\": None,\n \"base_model_path\": base_path + 'tf-xlm-roberta-large-tf_model.h5',\n \"config_path\": base_path + 'xlm-roberta-large-config.json'\n}\n\nwith open('config.json', 'w') as json_file:\n json.dump(json.loads(json.dumps(config)), json_file)",
"_____no_output_____"
]
],
[
[
"## Learning rate schedule",
"_____no_output_____"
]
],
[
[
"lr_min = 1e-7\nlr_start = 1e-7\nlr_max = config['LEARNING_RATE']\nstep_size = len(k_fold[k_fold[f'fold_{fold_n}'] == 'train']) // config['BATCH_SIZE']\ntotal_steps = config['EPOCHS'] * step_size\nhold_max_steps = 0\nwarmup_steps = step_size * 1\ndecay = .9997\n\nrng = [i for i in range(0, total_steps, config['BATCH_SIZE'])]\ny = [exponential_schedule_with_warmup(tf.cast(x, tf.float32), warmup_steps, hold_max_steps, \n lr_start, lr_max, lr_min, decay) for x in rng]\n\nsns.set(style=\"whitegrid\")\nfig, ax = plt.subplots(figsize=(20, 6))\nplt.plot(rng, y)\nprint(\"Learning rate schedule: {:.3g} to {:.3g} to {:.3g}\".format(y[0], max(y), y[-1]))",
"Learning rate schedule: 1e-07 to 9.84e-06 to 1.06e-06\n"
]
],
[
[
"# Model",
"_____no_output_____"
]
],
[
[
"module_config = XLMRobertaConfig.from_pretrained(config['config_path'], output_hidden_states=False)\n\ndef model_fn(MAX_LEN):\n input_ids = layers.Input(shape=(MAX_LEN,), dtype=tf.int32, name='input_ids')\n attention_mask = layers.Input(shape=(MAX_LEN,), dtype=tf.int32, name='attention_mask')\n \n base_model = TFXLMRobertaModel.from_pretrained(config['base_model_path'], config=module_config)\n last_hidden_state, _ = base_model({'input_ids': input_ids, 'attention_mask': attention_mask})\n cls_token = last_hidden_state[:, 0, :]\n \n output = layers.Dense(1, activation='sigmoid', name='output')(cls_token)\n \n model = Model(inputs=[input_ids, attention_mask], outputs=output)\n \n return model",
"_____no_output_____"
]
],
[
[
"# Train",
"_____no_output_____"
]
],
[
[
"# Load data\nx_train = np.load(base_data_path + 'x_train.npy')\ny_train = np.load(base_data_path + 'y_train_int.npy').reshape(x_train.shape[1], 1).astype(np.float32)\nx_valid_ml = np.load(database_base_path + 'x_valid.npy')\ny_valid_ml = np.load(database_base_path + 'y_valid.npy').reshape(x_valid_ml.shape[1], 1).astype(np.float32)\n\n#################### ADD TAIL ####################\nx_train_tail = np.load(base_data_path + 'x_train_tail.npy')\ny_train_tail = np.load(base_data_path + 'y_train_int_tail.npy').reshape(x_train_tail.shape[1], 1).astype(np.float32)\nx_train = np.hstack([x_train, x_train_tail])\ny_train = np.vstack([y_train, y_train_tail])\n\nstep_size = x_train.shape[1] // config['BATCH_SIZE']\nvalid_step_size = x_valid_ml.shape[1] // config['BATCH_SIZE']\n\n# Build TF datasets\ntrain_dist_ds = strategy.experimental_distribute_dataset(get_training_dataset(x_train, y_train, config['BATCH_SIZE'], AUTO, seed=SEED))\nvalid_dist_ds = strategy.experimental_distribute_dataset(get_validation_dataset(x_valid_ml, y_valid_ml, config['BATCH_SIZE'], AUTO, repeated=True, seed=SEED))\ntrain_data_iter = iter(train_dist_ds)\nvalid_data_iter = iter(valid_dist_ds)\n\n# Step functions\[email protected]\ndef train_step(data_iter):\n def train_step_fn(x, y):\n with tf.GradientTape() as tape:\n probabilities = model(x, training=True)\n loss = loss_fn(y, probabilities)\n grads = tape.gradient(loss, model.trainable_variables)\n optimizer.apply_gradients(zip(grads, model.trainable_variables))\n train_auc.update_state(y, probabilities)\n train_loss.update_state(loss)\n for _ in tf.range(step_size):\n strategy.experimental_run_v2(train_step_fn, next(data_iter))\n\[email protected]\ndef valid_step(data_iter):\n def valid_step_fn(x, y):\n probabilities = model(x, training=False)\n loss = loss_fn(y, probabilities)\n valid_auc.update_state(y, probabilities)\n valid_loss.update_state(loss)\n for _ in tf.range(valid_step_size):\n strategy.experimental_run_v2(valid_step_fn, next(data_iter))\n\n# Train model\nwith strategy.scope():\n model = model_fn(config['MAX_LEN'])\n optimizer = optimizers.Adam(learning_rate=lambda: \n exponential_schedule_with_warmup(tf.cast(optimizer.iterations, tf.float32), \n warmup_steps, hold_max_steps, lr_start, \n lr_max, lr_min, decay))\n loss_fn = losses.binary_crossentropy\n train_auc = metrics.AUC()\n valid_auc = metrics.AUC()\n train_loss = metrics.Sum()\n valid_loss = metrics.Sum()\n\nmetrics_dict = {'loss': train_loss, 'auc': train_auc, \n 'val_loss': valid_loss, 'val_auc': valid_auc}\n\nhistory = custom_fit(model, metrics_dict, train_step, valid_step, train_data_iter, valid_data_iter, \n step_size, valid_step_size, config['BATCH_SIZE'], config['EPOCHS'], \n config['ES_PATIENCE'], save_last=False)\n# model.save_weights('model.h5')\n\n# Make predictions\nx_train = np.load(base_data_path + 'x_train.npy')\nx_valid = np.load(base_data_path + 'x_valid.npy')\nx_valid_ml_eval = np.load(database_base_path + 'x_valid.npy')\n\ntrain_preds = model.predict(get_test_dataset(x_train, config['BATCH_SIZE'], AUTO))\nvalid_preds = model.predict(get_test_dataset(x_valid, config['BATCH_SIZE'], AUTO))\nvalid_ml_preds = model.predict(get_test_dataset(x_valid_ml_eval, config['BATCH_SIZE'], AUTO))\n\nk_fold.loc[k_fold[f'fold_{fold_n}'] == 'train', f'pred_{fold_n}'] = np.round(train_preds)\nk_fold.loc[k_fold[f'fold_{fold_n}'] == 'validation', f'pred_{fold_n}'] = np.round(valid_preds)\nvalid_df[f'pred_{fold_n}'] = valid_ml_preds\n\n\n# Fine-tune on validation set\n#################### ADD TAIL ####################\nx_valid_ml_tail = np.hstack([x_valid_ml, np.load(database_base_path + 'x_valid_tail.npy')])\ny_valid_ml_tail = np.vstack([y_valid_ml, y_valid_ml])\n\nvalid_step_size_tail = x_valid_ml_tail.shape[1] // config['BATCH_SIZE']\n\n# Build TF datasets\ntrain_ml_dist_ds = strategy.experimental_distribute_dataset(get_training_dataset(x_valid_ml_tail, y_valid_ml_tail, \n config['BATCH_SIZE'], AUTO, seed=SEED))\ntrain_ml_data_iter = iter(train_ml_dist_ds)\n\nhistory_ml = custom_fit(model, metrics_dict, train_step, valid_step, train_ml_data_iter, valid_data_iter, \n valid_step_size_tail, valid_step_size, config['BATCH_SIZE'], 1, \n config['ES_PATIENCE'], save_last=False)\n\n# Join history\nfor key in history_ml.keys():\n history[key] += history_ml[key]\n \nmodel.save_weights('model_ml.h5')\n\n# Make predictions\nvalid_ml_preds = model.predict(get_test_dataset(x_valid_ml_eval, config['BATCH_SIZE'], AUTO))\nvalid_df[f'pred_ml_{fold_n}'] = valid_ml_preds\n\n### Delete data dir\nshutil.rmtree(base_data_path)",
"Train for 5010 steps, validate for 62 steps\n\nEPOCH 1/4\ntime: 1709.3s loss: 0.2442 auc: 0.9587 val_loss: 0.2794 val_auc: 0.9214\n\nEPOCH 2/4\ntime: 1519.4s loss: 0.1701 auc: 0.9797 val_loss: 0.3026 val_auc: 0.9108\n\nEPOCH 3/4\ntime: 1519.5s loss: 0.1526 auc: 0.9836 val_loss: 0.3036 val_auc: 0.9137\n\nEPOCH 4/4\ntime: 1519.4s loss: 0.1478 auc: 0.9846 val_loss: 0.3073 val_auc: 0.9118\nTraining finished\nTrain for 125 steps, validate for 62 steps\n\nEPOCH 1/1\ntime: 1621.1s loss: 7.3961 auc: 0.9550 val_loss: 0.1309 val_auc: 0.9786\nTraining finished\n"
]
],
[
[
"## Model loss graph",
"_____no_output_____"
]
],
[
[
"plot_metrics(history)",
"_____no_output_____"
]
],
[
[
"# Model evaluation",
"_____no_output_____"
]
],
[
[
"display(evaluate_model_single_fold(k_fold, fold_n, label_col='toxic_int').style.applymap(color_map))",
"_____no_output_____"
]
],
[
[
"# Confusion matrix",
"_____no_output_____"
]
],
[
[
"train_set = k_fold[k_fold[f'fold_{fold_n}'] == 'train']\nvalidation_set = k_fold[k_fold[f'fold_{fold_n}'] == 'validation'] \nplot_confusion_matrix(train_set['toxic_int'], train_set[f'pred_{fold_n}'], \n validation_set['toxic_int'], validation_set[f'pred_{fold_n}'])",
"_____no_output_____"
]
],
[
[
"# Model evaluation by language",
"_____no_output_____"
]
],
[
[
"display(evaluate_model_single_fold_lang(valid_df, fold_n).style.applymap(color_map))\n# ML fine-tunned preds\ndisplay(evaluate_model_single_fold_lang(valid_df, fold_n, pred_col='pred_ml').style.applymap(color_map))",
"_____no_output_____"
]
],
[
[
"# Visualize predictions",
"_____no_output_____"
]
],
[
[
"pd.set_option('max_colwidth', 120)\nprint('English validation set')\ndisplay(k_fold[['comment_text', 'toxic'] + [c for c in k_fold.columns if c.startswith('pred')]].head(10))\n\nprint('Multilingual validation set')\ndisplay(valid_df[['comment_text', 'toxic'] + [c for c in valid_df.columns if c.startswith('pred')]].head(10))",
"English validation set\n"
]
],
[
[
"# Test set predictions",
"_____no_output_____"
]
],
[
[
"x_test = np.load(database_base_path + 'x_test.npy')\ntest_preds = model.predict(get_test_dataset(x_test, config['BATCH_SIZE'], AUTO))",
"_____no_output_____"
],
[
"submission = pd.read_csv('/kaggle/input/jigsaw-multilingual-toxic-comment-classification/sample_submission.csv')\nsubmission['toxic'] = test_preds\nsubmission.to_csv('submission.csv', index=False)\n\ndisplay(submission.describe())\ndisplay(submission.head(10))",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
c5011aaa5521344a1c730ba041ec04c236310eb1
| 12,465 |
ipynb
|
Jupyter Notebook
|
how-to-use-azureml/machine-learning-pipelines/intro-to-pipelines/aml-pipelines-how-to-use-estimatorstep.ipynb
|
skaarthik/MachineLearningNotebooks
|
6622a6c5f28839bb8387f3fe779dfcf19095dd14
|
[
"MIT"
] | 3 |
2020-08-04T18:37:21.000Z
|
2020-09-21T20:09:31.000Z
|
how-to-use-azureml/machine-learning-pipelines/intro-to-pipelines/aml-pipelines-how-to-use-estimatorstep.ipynb
|
skaarthik/MachineLearningNotebooks
|
6622a6c5f28839bb8387f3fe779dfcf19095dd14
|
[
"MIT"
] | null | null | null |
how-to-use-azureml/machine-learning-pipelines/intro-to-pipelines/aml-pipelines-how-to-use-estimatorstep.ipynb
|
skaarthik/MachineLearningNotebooks
|
6622a6c5f28839bb8387f3fe779dfcf19095dd14
|
[
"MIT"
] | 1 |
2020-06-13T07:46:05.000Z
|
2020-06-13T07:46:05.000Z
| 42.688356 | 476 | 0.594625 |
[
[
[
"Copyright (c) Microsoft Corporation. All rights reserved.\n\nLicensed under the MIT License.",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"# How to use EstimatorStep in AML Pipeline\n\nThis notebook shows how to use the EstimatorStep with Azure Machine Learning Pipelines. Estimator is a convenient object in Azure Machine Learning that wraps run configuration information to help simplify the tasks of specifying how a script is executed.\n\n\n## Prerequisite:\n* Understand the [architecture and terms](https://docs.microsoft.com/azure/machine-learning/service/concept-azure-machine-learning-architecture) introduced by Azure Machine Learning\n* If you are using an Azure Machine Learning Notebook VM, you are all set. Otherwise, go through the [configuration notebook](https://aka.ms/pl-config) to:\n * install the AML SDK\n * create a workspace and its configuration file (`config.json`)",
"_____no_output_____"
],
[
"Let's get started. First let's import some Python libraries.",
"_____no_output_____"
]
],
[
[
"import azureml.core\n# check core SDK version number\nprint(\"Azure ML SDK Version: \", azureml.core.VERSION)",
"_____no_output_____"
]
],
[
[
"## Initialize workspace\nInitialize a [Workspace](https://docs.microsoft.com/azure/machine-learning/service/concept-azure-machine-learning-architecture#workspace) object from the existing workspace you created in the Prerequisites step. `Workspace.from_config()` creates a workspace object from the details stored in `config.json`.",
"_____no_output_____"
]
],
[
[
"from azureml.core import Workspace\nws = Workspace.from_config()\nprint('Workspace name: ' + ws.name, \n 'Azure region: ' + ws.location, \n 'Subscription id: ' + ws.subscription_id, \n 'Resource group: ' + ws.resource_group, sep = '\\n')",
"_____no_output_____"
]
],
[
[
"## Create or Attach existing AmlCompute\nYou will need to create a [compute target](https://docs.microsoft.com/azure/machine-learning/service/concept-azure-machine-learning-architecture#compute-target) for training your model. In this tutorial, you create `AmlCompute` as your training compute resource.",
"_____no_output_____"
],
[
"If we could not find the cluster with the given name, then we will create a new cluster here. We will create an `AmlCompute` cluster of `STANDARD_NC6` GPU VMs. This process is broken down into 3 steps:\n1. create the configuration (this step is local and only takes a second)\n2. create the cluster (this step will take about **20 seconds**)\n3. provision the VMs to bring the cluster to the initial size (of 1 in this case). This step will take about **3-5 minutes** and is providing only sparse output in the process. Please make sure to wait until the call returns before moving to the next cell",
"_____no_output_____"
]
],
[
[
"from azureml.core.compute import ComputeTarget, AmlCompute\nfrom azureml.core.compute_target import ComputeTargetException\n\n# choose a name for your cluster\ncluster_name = \"cpu-cluster\"\n\ntry:\n cpu_cluster = ComputeTarget(workspace=ws, name=cluster_name)\n print('Found existing compute target')\nexcept ComputeTargetException:\n print('Creating a new compute target...')\n compute_config = AmlCompute.provisioning_configuration(vm_size='STANDARD_NC6', max_nodes=4)\n\n # create the cluster\n cpu_cluster = ComputeTarget.create(ws, cluster_name, compute_config)\n\n # can poll for a minimum number of nodes and for a specific timeout. \n # if no min node count is provided it uses the scale settings for the cluster\n cpu_cluster.wait_for_completion(show_output=True, min_node_count=None, timeout_in_minutes=20)\n\n# use get_status() to get a detailed status for the current cluster. \nprint(cpu_cluster.get_status().serialize())",
"_____no_output_____"
]
],
[
[
"Now that you have created the compute target, let's see what the workspace's `compute_targets` property returns. You should now see one entry named 'cpu-cluster' of type `AmlCompute`.",
"_____no_output_____"
],
[
"## Use a simple script\nWe have already created a simple \"hello world\" script. This is the script that we will submit through the estimator pattern. It prints a hello-world message, and if Azure ML SDK is installed, it will also logs an array of values ([Fibonacci numbers](https://en.wikipedia.org/wiki/Fibonacci_number)).",
"_____no_output_____"
],
[
"## Build an Estimator object\nEstimator by default will attempt to use Docker-based execution. You can also enable Docker and let estimator pick the default CPU image supplied by Azure ML for execution. You can target an AmlCompute cluster (or any other supported compute target types). You can also customize the conda environment by adding conda and/or pip packages.\n\n> Note: The arguments to the entry script used in the Estimator object should be specified as *list* using\n 'estimator_entry_script_arguments' parameter when instantiating EstimatorStep. Estimator object's parameter\n 'script_params' accepts a dictionary. However 'estimator_entry_script_arguments' parameter expects arguments as\n a list.\n\n> Estimator object initialization involves specifying a list of DataReference objects in its 'inputs' parameter.\n In Pipelines, a step can take another step's output or DataReferences as input. So when creating an EstimatorStep,\n the parameters 'inputs' and 'outputs' need to be set explicitly and that will override 'inputs' parameter\n specified in the Estimator object.\n \n> The best practice is to use separate folders for scripts and its dependent files for each step and specify that folder as the `source_directory` for the step. This helps reduce the size of the snapshot created for the step (only the specific folder is snapshotted). Since changes in any files in the `source_directory` would trigger a re-upload of the snapshot, this helps keep the reuse of the step when there are no changes in the `source_directory` of the step.",
"_____no_output_____"
]
],
[
[
"from azureml.core import Datastore\nfrom azureml.data.data_reference import DataReference\nfrom azureml.pipeline.core import PipelineData\n\ndef_blob_store = Datastore(ws, \"workspaceblobstore\")\n\ninput_data = DataReference(\n datastore=def_blob_store,\n data_reference_name=\"input_data\",\n path_on_datastore=\"20newsgroups/20news.pkl\")\n\noutput = PipelineData(\"output\", datastore=def_blob_store)\n\nsource_directory = 'estimator_train'",
"_____no_output_____"
],
[
"from azureml.train.estimator import Estimator\n\nest = Estimator(source_directory=source_directory, \n compute_target=cpu_cluster, \n entry_script='dummy_train.py', \n conda_packages=['scikit-learn'])",
"_____no_output_____"
]
],
[
[
"## Create an EstimatorStep\n[EstimatorStep](https://docs.microsoft.com/en-us/python/api/azureml-pipeline-steps/azureml.pipeline.steps.estimator_step.estimatorstep?view=azure-ml-py) adds a step to run Estimator in a Pipeline.\n\n- **name:** Name of the step\n- **estimator:** Estimator object\n- **estimator_entry_script_arguments:** \n- **runconfig_pipeline_params:** Override runconfig properties at runtime using key-value pairs each with name of the runconfig property and PipelineParameter for that property\n- **inputs:** Inputs\n- **outputs:** Output is list of PipelineData\n- **compute_target:** Compute target to use \n- **allow_reuse:** Whether the step should reuse previous results when run with the same settings/inputs. If this is false, a new run will always be generated for this step during pipeline execution.\n- **version:** Optional version tag to denote a change in functionality for the step",
"_____no_output_____"
]
],
[
[
"from azureml.pipeline.steps import EstimatorStep\n\nest_step = EstimatorStep(name=\"Estimator_Train\", \n estimator=est, \n estimator_entry_script_arguments=[\"--datadir\", input_data, \"--output\", output],\n runconfig_pipeline_params=None, \n inputs=[input_data], \n outputs=[output], \n compute_target=cpu_cluster)",
"_____no_output_____"
]
],
[
[
"## Build and Submit the Experiment",
"_____no_output_____"
]
],
[
[
"from azureml.pipeline.core import Pipeline\nfrom azureml.core import Experiment\npipeline = Pipeline(workspace=ws, steps=[est_step])\npipeline_run = Experiment(ws, 'Estimator_sample').submit(pipeline)",
"_____no_output_____"
]
],
[
[
"## View Run Details",
"_____no_output_____"
]
],
[
[
"from azureml.widgets import RunDetails\nRunDetails(pipeline_run).show()",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
c5012d7842b155b306de0e7d70ffc415de44ffee
| 140,520 |
ipynb
|
Jupyter Notebook
|
main.ipynb
|
mehrdadhz-77/ADM-HW3
|
bc25949411576f6df6cb6fb84195bab8d7ea8fb9
|
[
"MIT"
] | null | null | null |
main.ipynb
|
mehrdadhz-77/ADM-HW3
|
bc25949411576f6df6cb6fb84195bab8d7ea8fb9
|
[
"MIT"
] | null | null | null |
main.ipynb
|
mehrdadhz-77/ADM-HW3
|
bc25949411576f6df6cb6fb84195bab8d7ea8fb9
|
[
"MIT"
] | null | null | null | 31.185087 | 459 | 0.536137 |
[
[
[
"# 3rd Homework of ADM",
"_____no_output_____"
],
[
"### Group members",
"_____no_output_____"
],
[
"#### 1. Mehrdad Hassanzadeh, 1961575, [email protected]\n#### 2. Vahid Ghanbarizadeh, 2002527, [email protected]\n#### 3. Andrea Giordano , 1871786, [email protected]",
"_____no_output_____"
],
[
"# Importing useful packages",
"_____no_output_____"
]
],
[
[
"import requests\nfrom bs4 import BeautifulSoup\nfrom collections import defaultdict, Counter\nimport os\nfrom pathlib import Path\nimport re\nimport multiprocessing as mp\nfrom math import ceil\nimport nltk\nfrom nltk.corpus import stopwords\nfrom nltk.stem import PorterStemmer\nimport json\nimport pandas as pd\nfrom math import log\nfrom copy import deepcopy\nfrom heapq import heappop, heappush ",
"_____no_output_____"
]
],
[
[
"# 1. Data collection",
"_____no_output_____"
],
[
"We are asked to collect some information about the anime in a spcific website. Here is the link to that site. 'https://myanimelist.net/topanime.php'",
"_____no_output_____"
],
[
"Here we send a request to get the content of a specific webpage. We wanted the content of the main page of the TopAnime website. ",
"_____no_output_____"
]
],
[
[
"TopAnime = requests.get('https://myanimelist.net/topanime.php')",
"_____no_output_____"
]
],
[
[
"Now we check if we have successfully received the content of the desired webpage or not. ",
"_____no_output_____"
]
],
[
[
"TopAnime",
"_____no_output_____"
]
],
[
[
"As we have the code '200' as the status of the reponse, so it means that we have successfully received the desired page. ",
"_____no_output_____"
],
[
"Then we take a brief look at the content of this page. ",
"_____no_output_____"
]
],
[
[
"#TopAnime.content",
"_____no_output_____"
]
],
[
[
"As we can see in the output, we have the HTML code of the webpage. Now we should parse this HTML code in order to get the data we want. To do so, we will be using **BeautifulSoap** library which is designed to parse HTML codes. ",
"_____no_output_____"
]
],
[
[
"TopAnimeSoup = BeautifulSoup(TopAnime.content, 'html.parser')",
"_____no_output_____"
]
],
[
[
"Now we look at the produced HTML code of the webpage in nice and more readable format using BeautifulSoup. ",
"_____no_output_____"
]
],
[
[
"#print(TopAnimeSoup.prettify())",
"_____no_output_____"
]
],
[
[
"# 1.1. Get the list of animes",
"_____no_output_____"
],
[
"### Steps to extract the url for just one anime. ",
"_____no_output_____"
],
[
"Here we will go through the required steps to extract the url and the name of just the first anime in the list. We can then expand this idea to all the anime in the page. ",
"_____no_output_____"
],
[
"### 1.1.1 Getting all the rows (anime) in a page",
"_____no_output_____"
],
[
"After chekcing the HTML code, we saw that the information related to each anime is stored in a table which its class is \"top-ranking-table\" and the animes' information are stored in trs of this table. ",
"_____no_output_____"
],
[
"Let's take a look at how many tables we have in the webpage. ",
"_____no_output_____"
]
],
[
[
"len(list(TopAnimeSoup.find_all('table')))",
"_____no_output_____"
]
],
[
[
"As we have only one table in the webpage, so every tr that we have in the webpage belongs to this table. ",
"_____no_output_____"
],
[
"Here we will take a look at how many tr tags we have in the webpage. ",
"_____no_output_____"
]
],
[
[
"#print(list(TopAnimeSoup.find_all('tr')))\n#len(list(TopAnimeSoup.find_all('tr')))",
"_____no_output_____"
]
],
[
[
"We know that in each page, we have the information related to 50 animes. But why we have 51 tr tags here? <br/>\nBecause the first row corresponds to the name of table's column and the rest store the inramtion related to each anime. ",
"_____no_output_____"
],
[
"So in order to get the rows which contain the information of each anime, we should go through the tr tags that we have in the webpage except the first one which contains the information of the columns' name of of the table. ",
"_____no_output_____"
]
],
[
[
"Rows = list(TopAnimeSoup.find_all('tr'))[1:]\nlen(Rows)",
"_____no_output_____"
]
],
[
[
"Here we have all the information of the anime in a specific page. As we know the number of anime in each page is 50. ",
"_____no_output_____"
],
[
"### 1.1.2 Extracting the name and url of an anime",
"_____no_output_____"
],
[
"Now we should get the name and the url correspond to each anime. We found out that this information can be found in 'a' tag of each row which its class name is \"hoverinfo_trigger fl-l ml12 mr8\" and is included in the second 'td' tag of each 'tr'. ",
"_____no_output_____"
],
[
"We get all the 'td' tags of the second 'tr' the first 'tr' tag contains the columns' name. ",
"_____no_output_____"
]
],
[
[
"Tds = Rows[0].find_all('td')",
"_____no_output_____"
]
],
[
[
"Then we will go to the second 'td' tag's information. The first one contains just the ranking number. ",
"_____no_output_____"
]
],
[
[
"SecondTD = Tds[1]",
"_____no_output_____"
]
],
[
[
"Then inside this 'td' tag we look for the 'a' tag which its class is \"hoverinfo_trigger fl-l ml12 mr8\"",
"_____no_output_____"
]
],
[
[
"TagA = SecondTD.find('class' == \"hoverinfo_trigger fl-l ml12 mr8\")\nTagA",
"_____no_output_____"
]
],
[
[
"The url of an anime is the value of 'href' property of this tag. Let's take a look at it. ",
"_____no_output_____"
]
],
[
[
"URL = TagA['href']\nURL",
"_____no_output_____"
]
],
[
[
"Here to extract the name of the anime we have two options. 1. To split the 'href' and get the last value of it. 2. Get the value of 'alt' property of the 'img' tag in the 'a' tag. Here we will go for the second approach. ",
"_____no_output_____"
]
],
[
[
"Image = TagA.find('img')\nAnimeName = Image['alt'].replace('Anime: ', '')\nAnimeName",
"_____no_output_____"
]
],
[
[
"### 1.1.3 Getting the information for all anime in a page",
"_____no_output_____"
],
[
"Now here we want to get the name and url of all the animes in this specific page. ",
"_____no_output_____"
]
],
[
[
"# Dictionary to store the url or each anime in the page \nMyDict = defaultdict(str)\n\n# Going through all the rows in the page\nfor Row in Rows: \n \n # Take the second column of the row\n TDs = Row.find_all('td')\n \n # Take the tag a which consists the anime name and the url of that anime\n TagA = TDs[1].find('class' == \"hoverinfo_trigger fl-l ml12 mr8\")\n \n # Extract the anime name and url of the anime\n AnimeName, URL = TagA.find('img')['alt'].replace('Anime: ', ''), TagA['href']\n \n # Store the url of the anime in the dictionary\n MyDict[AnimeName] = URL",
"_____no_output_____"
]
],
[
[
"Now we will check the information for five animes. ",
"_____no_output_____"
]
],
[
[
"for anime in list(MyDict.keys())[:5]:\n print('Name of anime: ' + anime+'\\nURL: ' + MyDict[anime], end = '\\n\\n')",
"Name of anime: Fullmetal Alchemist: Brotherhood\nURL: https://myanimelist.net/anime/5114/Fullmetal_Alchemist__Brotherhood\n\nName of anime: Gintama°\nURL: https://myanimelist.net/anime/28977/Gintama°\n\nName of anime: Shingeki no Kyojin Season 3 Part 2\nURL: https://myanimelist.net/anime/38524/Shingeki_no_Kyojin_Season_3_Part_2\n\nName of anime: Steins;Gate\nURL: https://myanimelist.net/anime/9253/Steins_Gate\n\nName of anime: Fruits Basket: The Final\nURL: https://myanimelist.net/anime/42938/Fruits_Basket__The_Final\n\n"
]
],
[
[
"Here we want to check if we have all the 50 animes' URL in the dictionay. ",
"_____no_output_____"
]
],
[
[
"print('Number of animes ', len(MyDict))",
"Number of animes 50\n"
]
],
[
[
"# 1.2 Crawl animes",
"_____no_output_____"
],
[
"### 1.2.1 Function Crawl urls in a webpage",
"_____no_output_____"
],
[
"In order to have more readable implementation we will write a function which receives the URL of the webpage that we want to scrap and writes the urls to the URLs.txt file. ",
"_____no_output_____"
]
],
[
[
"def GetAnimeInfo(webpage):\n \n # Opening the file that we want to write our information in \n File = open('URLs.txt', mode = 'a')\n \n # Send a request to get the webpage\n Request = requests.get(webpage)\n \n # Parse the HTML code of the webpage\n TopAnimeSoup = BeautifulSoup(Request.content, 'html.parser')\n \n # Take all the tr tags in the webpage which contain all the anime information in the given webpage\n Rows = TopAnimeSoup.find_all('tr')\n \n # Going throguh all the anime \n for Row in Rows[1:]: \n \n # Get the td tags of each anime \n TDs = Row.find_all('td')\n \n # Take the tag A which its class is equal to 'hoverinfo_trigger fl-l ml12 mr8'\n TagA = TDs[1].find('class' == \"hoverinfo_trigger fl-l ml12 mr8\")\n \n # Extract the URL of the anime\n URL = TagA['href']\n \n # Write the URL to the file\n File.write(URL+'\\n')\n \n # Close the file we have just written it\n File.close()",
"_____no_output_____"
]
],
[
[
"Now at each time we should pass the function the URL of the webpage that we want to scrap. ",
"_____no_output_____"
],
[
"After checking the URL of the next pages, we understood that there is a pattern in URL of the pages. <br/>\nFor example the 2nd webpage's URL is 'https://myanimelist.net/topanime.php?limit=50' and we can see the only difference between this URL and the main page URL (which is 'https://myanimelist.net/topanime.php') is that there is '?limit=50' string at the end. <br/><br/>\n* So we can use this pattern the find the next pages URL. ",
"_____no_output_____"
],
[
"### 1.2.2 Main Function to extract all the URLs",
"_____no_output_____"
]
],
[
[
"# This is the root webpage of the website\nMainPageURL = 'https://myanimelist.net/topanime.php'\n\n# We go through first 400 pages \nfor i in range(400):\n \n # This is the address that we want for the first page\n if i == 0:\n GetAnimeInfo(MainPageURL)\n \n # For the next pages, we should use the address of the webpage and put the limit \n # at the end to reach to the next pages\n else:\n GetAnimeInfo(MainPageURL+'?limit='+str(50*i))\n ",
"_____no_output_____"
]
],
[
[
"### 1.2.3 Function Get HTML code",
"_____no_output_____"
],
[
"Now we want to download the HTML file for each anime. <br/> \nIn order to do this we are going to write a function that passes a number of URLs which their HTML file should be downloaded and stored in a folder.<br/>\nThese URLS are given in a list of list which correspond to a number of webpages in the website and a number of anime's URL which are in a specific webpage. ",
"_____no_output_____"
],
[
"* **Note:** We will download the HTML codes in parallel by using all the processors in the system. ",
"_____no_output_____"
],
[
"As this function will be called from all the processors in the system, we should find a way so that all the processors can download their own given pages' HTML code. ",
"_____no_output_____"
]
],
[
[
"# The parameters of the function: \n\n# URLS: A list of a list which each element in the first list is the pages and each element in the list \n# of each page is the anime's URL in that page that we should get their HTML code. \n\n# GroupIndex: Specifies which processor is calling this function \n\n# NumPageGroup: Number of the pages that was given to each processor\n\n# Start: Start the downloading from which page? Start = 50 -> Starting extracting the HTML code of the anime \n# in the page 50\n\ndef GetHTML(URLS, GroupIndex, NumPageGroup, Start):\n \n # Going through each page\n for index, ListURLs in enumerate(URLS):\n \n # If the page number that we are considering at this moment lower than Start, we will ignore it\n if index < Start:\n continue\n \n # For each page we create a new folder to store its anime HTML code in it. \n FolderName = 'page' + str(GroupIndex*NumPageGroup + index +1)\n Path = os.path.join(os.getcwd(),'HTMLS', FolderName)\n os.mkdir(Path)\n \n # Iterating over all the anime URL in a specific page\n for i, url in enumerate(ListURLs):\n \n # Request to get the webpage of that anime's URL\n AnimePage = requests.get(url)\n \n # Get the HTML code of that webpage\n AnimePageSoup = BeautifulSoup(AnimePage.content, 'html.parser')\n \n # Create a html file to store the HTML code of the webpage in it\n file = open(Path + '/' + 'anime_' + str(i) + '.html',\"w\")\n file.write(str(AnimePageSoup))\n file.close() ",
"_____no_output_____"
]
],
[
[
"### 1.2.4 Parallelizing the Crawling process",
"_____no_output_____"
],
[
"So first we check how many processors we have in the system:",
"_____no_output_____"
]
],
[
[
"print(\"The number of processors:\", mp.cpu_count())",
"The number of processors: 4\n"
]
],
[
[
"So in this case we should distribute our URLS over these processors. We read all the URLs that we have stored in the 'URLs.txt' file.",
"_____no_output_____"
]
],
[
[
"AllURLs = []\nwith open('URLs.txt') as f:\n for row in f:\n AllURLs.append(row.strip())",
"_____no_output_____"
]
],
[
[
"Now we want to check how many URLs we have:",
"_____no_output_____"
]
],
[
[
"print('The total number of URLs is:', len(AllURLs))",
"The total number of URLs is: 19128\n"
]
],
[
[
"* **Note:** We know that we should store all the anime's webpage HTML code in a single folder for the ones that have been appeared in the same wepbage, so all of these URLs should be entirely given to a processor. ",
"_____no_output_____"
],
[
"* **Note:** We know that we extracted the URLs in order, so if we take the URLs 50 by 50 we will have the anime URLs that are in the same page. ",
"_____no_output_____"
],
[
"Now we group each 50 URLs to construct the pages. ",
"_____no_output_____"
]
],
[
[
"EachPageURLs = [AllURLs[i*50:(i+1)*50] for i in range(len(AllURLs)//50+1)]",
"_____no_output_____"
]
],
[
[
"Here we will take a look at how many pages that we have here: ",
"_____no_output_____"
]
],
[
[
"print(\"Number of webpages in the website:\", len(EachPageURLs))",
"Number of webpages in the website: 383\n"
]
],
[
[
"Now we check how many pages should be given to each of the processors. ",
"_____no_output_____"
]
],
[
[
"NumOfPage = ceil(ceil(len(AllURLs)/50)/mp.cpu_count())\nprint('Number of pages to be given to each processor: ', NumOfPage)",
"Number of pages to be given to each processor: 96\n"
]
],
[
[
"* Each processor is responsible to download the anime's HTML code that are in 96 pages. ",
"_____no_output_____"
],
[
"Now we group the pages that should be given to each of the processors. ",
"_____no_output_____"
]
],
[
[
"GroupPage = [EachPageURLs[i*NumOfPage:(i+1)*NumOfPage] for i in range(mp.cpu_count())]",
"_____no_output_____"
]
],
[
[
"Let's check the number of pages in each group:",
"_____no_output_____"
]
],
[
[
"print(list(map(len , GroupPage)))",
"[96, 96, 96, 95]\n"
]
],
[
[
"As we can see all the processors have been assigned 96 pages to download except the last one. We remind that we have 383 page which couldn't be distributed evenly between all the processors. ",
"_____no_output_____"
],
[
"### Downloading HTMLs in parallel",
"_____no_output_____"
],
[
"As we saw before, we have 4 processors in the system and we want to distribute the work among these 4 processors.<br/>\nWe will give a subsets of pages to each of these processors to download their anime HTMl code. ",
"_____no_output_____"
]
],
[
[
"# How many processors we want to pool handle \npool = mp.Pool(mp.cpu_count())\n\n# Call the GetHTML function for each of the processors given their assigned pages\nresults = [pool.apply_async(GetHTML, args = (GroupPage[i],i, NumOfPage, 5)) for i in range(mp.cpu_count())]\npool.close()",
"_____no_output_____"
]
],
[
[
"# 1.3 Parse downloaded pages",
"_____no_output_____"
],
[
"Now in this question we should produce a .tsv file for each anime which contains some information extracted from their webpage. ",
"_____no_output_____"
],
[
"In order to do this, first we check how we can get the information for just one anime and then we will expand the idea to other anime that we have. ",
"_____no_output_____"
],
[
"The information that we should extract for each anime are: ",
"_____no_output_____"
],
[
"1- **Anime Name** (to save as animeTitle): String <br/>\n2- **Anime Type** (to save as animeType): String<br/>\n3- **Number of episode** (to save as animeNumEpisode): Integer<br/>\n4- **Release and End Dates of anime** (to save as releaseDate and endDate): Convert both release and end date into datetime format.<br/>\n5- **Number of members** (to save as animeNumMembers): Integer<br/>\n6- **Score** (to save as animeScore): Float<br/>\n7- **Users** (to save as animeUsers): Integer<br/>\n8- **Rank** (to save as animeRank): Integer<br/>\n9- **Popularity** (to save as animePopularity): Integer<br/>\n10- **Synopsis** (to save as animeDescription): String<br/>\n11- **Related Anime** (to save as animeRelated): Extract all the related animes, but only keep unique values and those that have a hyperlink associated to them. List of strings.<br/>\n12- **Characters** (to save as animeCharacters): List of strings.<br/>\n13- **Voices** (to save as animeVoices): List of strings<br/>\n14- **Staff** (to save as animeStaff): Include the staff name and their responsibility/task in a list of lists.<br/>",
"_____no_output_____"
],
[
"* For getting each of the information that we need, we write a function to have a better understanding of how we should extract each of these information. ",
"_____no_output_____"
],
[
"### 1.3.1 Function AnimeName",
"_____no_output_____"
],
[
"We know that the name of the anime can be extracted from the title of the webpage. So we send the html code of the webpage to this function, and we will return the name of the anime extracted from the title of the page. ",
"_____no_output_____"
]
],
[
[
"def GetAnimeName(webpage):\n return webpage.title.getText().strip().replace(' - MyAnimeList.net', '')",
"_____no_output_____"
]
],
[
[
"### 1.3.2 Function GetAnimeType",
"_____no_output_____"
],
[
"This information can be extracted from the values stored in a specific 'div' tag. In order do this we will send that specific tag to this function and receive the type of the anime. ",
"_____no_output_____"
]
],
[
[
"def GetAnimeType(Tag):\n Temp = Tag.getText().split()\n return Temp[-1] if len(Temp) >1 else \"\"",
"_____no_output_____"
]
],
[
[
"### 1.3.3 Function Episode",
"_____no_output_____"
],
[
"This information can be extracted from the values stored in a specific 'div' tag. In order do this we will send that specific tag to this function and receive the type of the anime. ",
"_____no_output_____"
]
],
[
[
"def GetNumOfEpisode(Tag):\n Temp = Tag.getText().strip().split()[-1]\n return int(Temp) if Temp.isdigit() else ''",
"_____no_output_____"
]
],
[
[
"### 1.3.4 Function DateTime",
"_____no_output_____"
],
[
"This information can be extracted from the values stored in a specific 'div' tag. In order do this we will send that specific tag to this function and receive the type of the anime. <br/><br/>\n* Some of the animes may have just the release date. So we should be careful about this. ",
"_____no_output_____"
]
],
[
[
"def GetDates(Tag):\n Release, End = '', ''\n Temp = Tag.getText().strip().replace('Aired:\\n ', '').split('to')\n Release = Temp[0] if len(Temp) else ''\n End = Temp[1] if len(Temp) == 2 else ''\n return Release, End",
"_____no_output_____"
]
],
[
[
"### 1.3.5 Function Members",
"_____no_output_____"
],
[
"This information can be extracted from the values stored in a specific 'div' tag. In order do this we will send that specific tag to this function and receive the type of the anime.",
"_____no_output_____"
]
],
[
[
"def GetMembers(Tag):\n return int(Tag.getText().replace('\\n', '').split()[1].replace(',', ''))",
"_____no_output_____"
]
],
[
[
"### 1.3.6 Function ScoreAndUsers",
"_____no_output_____"
],
[
"This information can be extracted from the values stored in a specific 'div' tag. In order do this we will send that specific tag to this function and receive the type of the anime.",
"_____no_output_____"
]
],
[
[
"def GetScoreAndUsers(Tag):\n Rank = re.findall('[0-9|,]+ users', str(Tag))\n RankValue = int(Rank[0].split()[0].replace(',', '')) if len(Rank) else ''\n Score = re.findall('Score:[0-9|.]+', Tag.getText().replace(\"\\n\", ''))\n ScoreValue = float(Score[0].split(\":\")[1]) if len(Score) else ''\n return ScoreValue, RankValue",
"_____no_output_____"
]
],
[
[
"### 1.3.7 Function Rank",
"_____no_output_____"
],
[
"This information can be extracted from the values stored in a specific 'div' tag. In order do this we will send that specific tag to this function and receive the type of the anime.",
"_____no_output_____"
]
],
[
[
"def GetRank(Tag):\n Temp = re.findall('#[0-9|,]+', str(Tag))\n return int(Temp[0][1:]) if len(Temp) else ''",
"_____no_output_____"
]
],
[
[
"### 1.3.8 Function Popularity",
"_____no_output_____"
],
[
"This information can be extracted from the values stored in a specific 'div' tag. In order do this we will send that specific tag to this function and receive the type of the anime.",
"_____no_output_____"
]
],
[
[
"def GetPopularity(Tag):\n Temp = re.findall('#[0-9|,]+', str(Tag))\n return int(Temp[0][1:]) if len(Temp) else ''",
"_____no_output_____"
]
],
[
[
"### 1.3.9 Function Synopsis",
"_____no_output_____"
],
[
"In order to get the description of the anime, there is a specific tag which can be easily identified by its 'itemprop' property. So we will give the whole html to this function and get back the description. ",
"_____no_output_____"
]
],
[
[
"def GetSynopsis(webpage):\n Temp = webpage.find('p', {'itemprop': \"description\"})\n if not Temp:\n return ''\n Temp = Temp.getText().replace('\\n', '')\n return Temp",
"_____no_output_____"
]
],
[
[
"### 1.3.10 Function Related Anime",
"_____no_output_____"
],
[
"To get the related animes, there is a table which its 'class' is equal to 'anime_detail_related_anime'. So to get the table we should give the function the whole html file to this function. ",
"_____no_output_____"
]
],
[
[
"def GetRelatedAnime(webpage):\n Related = webpage.find('table', {'class': \"anime_detail_related_anime\"})\n if not Related:\n return \"\"\n Related = Related.find_all('a')\n UniqueRelated = set(i.getText() for i in Related)\n return list(UniqueRelated)",
"_____no_output_____"
]
],
[
[
"### 1.3.11 Function Characters",
"_____no_output_____"
],
[
"We can find this information in tag 'a' that are included in a div which is its class is equal to 'detail-characters-list clearfix'. When we go for the first div that have this feature we will be received the table which is the characters and their original voices is there. ",
"_____no_output_____"
]
],
[
[
"def GetCharacters(webpage):\n Characters = []\n Tags = webpage.find_all('div', {'class': \"detail-characters-list clearfix\"})\n for tag in Tags:\n if str(tag).count('character') >1:\n \n # We can get the name of the character in the 'href' value of the tag 'a' property\n AllTagA = tag.find_all('a')\n \n # Filter the hrefs to of the characters\n CharactersHrefs = list(set([i['href'] for i in AllTagA if 'character' in i['href']]))\n \n # Filter the names of the characters\n Characters = [i.split('/')[-1].replace('_', ' ') for i in CharactersHrefs]\n return Characters",
"_____no_output_____"
]
],
[
[
"### 1.3.12 Function Voices ",
"_____no_output_____"
],
[
"With the same approach as previous, now we just extract the voices. ",
"_____no_output_____"
]
],
[
[
"def GetVoices(webpage):\n Voices = []\n Tags = webpage.find_all('div', {'class': \"detail-characters-list clearfix\"})\n for tag in Tags:\n if str(tag).count('character') > 1:\n \n # We can get the name of the person in charge for the voice in the 'href' value in tag 'a' property\n AllTagA = tag.find_all('a')\n \n # Filter the hrefs for voices\n VoicesHrefs = list(set([i['href'] for i in AllTagA if 'people' in i['href']]))\n \n # Filter the names of the people\n Voices = [i.split('/')[-1].replace('_', ' ') for i in VoicesHrefs]\n return Voices",
"_____no_output_____"
]
],
[
[
"### 1.3.13 Function Staff",
"_____no_output_____"
],
[
"Here in this function we will look for the other div tha its 'class' is equal to 'detail-characters-list clearfix'. Then the name of the staff can be extracted from 'img' tags and also their duties can be found in the 'small' tags which are in the same 'row' as their image. ",
"_____no_output_____"
]
],
[
[
"def GetStaff(webpage):\n StaffDuty = []\n Tags = webpage.find_all('div', {'class': \"detail-characters-list clearfix\"})\n for tag in Tags:\n if str(tag).count('character') == 1:\n Staff = tag.find_all('tr')\n for i in Staff:\n NewStaff = [i.find('a')['href'].split('/')[-1].replace('_', ' ')] # Extracting the name of the staff\n Duties = list(i.find('small').getText().split(',')) # Getting the duties of the staff\n NewStaff += [i.strip() for i in Duties]\n StaffDuty.append(NewStaff)\n return StaffDuty",
"_____no_output_____"
]
],
[
[
"### 1.3.14 Function Write to TSV",
"_____no_output_____"
],
[
"With the help of this function, we can write the information that we extracted from a page to its corresponded .tsv file. ",
"_____no_output_____"
]
],
[
[
"def WriteToTSV(AnimeInfo, Path):\n \n # Open the corresponded file \n File = open(Path, mode = 'w')\n \n # Writing the header. \n File.write(\"\\t\".join(Keys))\n File.write('\\n')\n \n # Going through all the information for that specific anime\n for i in AnimeInfo:\n \n # Get the string version of that value\n STR = str(AnimeInfo[i])\n \n # In case an information was missing, we should put '' in the file\n if STR == '[]':\n STR = ''\n \n # Write the value of the information into the file\n File.write(STR + ('\\t' if i!='animeStaff' else \"\" ))\n File.close() ",
"_____no_output_____"
]
],
[
[
"### 1.3.15 Function Extract",
"_____no_output_____"
],
[
"With the help of this function we want to extract the desired information from all the .html files that we have. ",
"_____no_output_____"
],
[
"Here we used some key words so we can easily get the needed information which are in some tags that share the same class. ",
"_____no_output_____"
]
],
[
[
"Info = ['Type:', 'Episodes:', 'Aired:', 'Members:','Score:','Ranked:', 'Popularity:']",
"_____no_output_____"
]
],
[
[
"Here we define the default datastructure which is a dictionary to store the inforamtion of each page. ",
"_____no_output_____"
]
],
[
[
"Keys = ['animeTitle', 'animeType', 'animeNumEpisode', 'releaseDate', 'endDate', 'animeNumMembers'\n , 'animeScore', 'animeUsers', 'animeRank', 'animePopularity', 'animeDescription', 'animeRelated',\n 'animeCharacters', 'animeVoices', 'animeStaff']\n\n# Empty instance for storing the information for each anime\nMyAnimeInfo = {i:\"\" for i in Keys} ",
"_____no_output_____"
]
],
[
[
"Here we will go through each HTML file that we have just downloaded. Then we extract the infomration that we need from the HTML file and then store them in a .tsv file. ",
"_____no_output_____"
],
[
"* **Note:** as we wanted to avoid each processor override the datastructure of another processor we gave each processor its own private datastructure. That is the usage of exec functions. ",
"_____no_output_____"
],
[
"* exec function: Given a string it will turn that string into a python code and run the code. In this case we gave this possibility to each processor to work on its own datastructure. ",
"_____no_output_____"
]
],
[
[
"def FunctionExtract(ListNumPage, ProcesserNum):\n \n # Going through all the pages\n for i in ListNumPage:\n \n # Going through all the anime in each page\n for j in range((50 if i!= 383 else 28)):\n \n # Get the copy of the default datastructures\n exec('NewAnime'+str(ProcesserNum)+'= MyAnimeInfo.copy()')\n \n # Reading the .html file of a specific anime\n Path = '/home/mehrdad/ADM-HW3/HTMLS/page'\n File = open(Path + str(i) + '/anime_' + str(j) +'.html', mode = 'r')\n \n # Get the parsed HTML code of the webpage of the anime \n Webpage = BeautifulSoup(File.read(), 'html.parser')\n \n # Extracting some of the information that have been stored in the tags that have the \n # same value as the class\n for div in Webpage.find_all('div', {'class':\"spaceit_pad\"}):\n if Info[0] in str(div):\n exec('NewAnime'+ str(ProcesserNum) +\"['animeType'] = GetAnimeType(div)\")\n elif Info[1] in str(div):\n exec('NewAnime'+ str(ProcesserNum) +\"['animeNumEpisode'] = GetNumOfEpisode(div)\")\n elif Info[2] in str(div):\n exec('NewAnime'+ str(ProcesserNum) +\"['releaseDate'] = GetDates(div)[0]\")\n exec('NewAnime'+ str(ProcesserNum) +\"['endDate'] = GetDates(div)[1]\")\n elif Info[3] in str(div):\n exec('NewAnime'+ str(ProcesserNum) +\"['animeNumMembers'] = GetMembers(div)\")\n elif Info[4] in str(div):\n exec('NewAnime'+ str(ProcesserNum) +\"['animeScore'] = GetScoreAndUsers(div)[0]\")\n exec('NewAnime'+ str(ProcesserNum) +\"['animeUsers'] = GetScoreAndUsers(div)[1]\")\n elif Info[5] in str(div):\n exec('NewAnime'+ str(ProcesserNum) +\"['animeRank'] = GetRank(div)\")\n elif Info[6] in str(div):\n exec('NewAnime'+ str(ProcesserNum) +\"['animePopularity'] = GetPopularity(div)\")\n \n # Extracting the other information \n exec('NewAnime'+ str(ProcesserNum) +\"['animeTitle'] = GetAnimeName(Webpage)\")\n exec('NewAnime'+ str(ProcesserNum) +\"['animeDescription'] = GetSynopsis(Webpage)\")\n exec('NewAnime'+ str(ProcesserNum) +\"['animeRelated'] = GetRelatedAnime(Webpage)\")\n exec('NewAnime'+ str(ProcesserNum) +\"['animeCharacters'] = GetCharacters(Webpage)\")\n exec('NewAnime'+ str(ProcesserNum) +\"['animeVoices'] = GetVoices(Webpage)\")\n exec('NewAnime'+ str(ProcesserNum) +\"['animeStaff'] = GetStaff(Webpage)\")\n \n TSVPath = Path + str(i) + '/anime_' + str(j) +'.tsv'\n exec('WriteToTSV(NewAnime'+ str(ProcesserNum)+',TSVPath)')",
"_____no_output_____"
]
],
[
[
"### Creating the .tsv files in parallel. ",
"_____no_output_____"
],
[
"* **In order to speed up the process, we will distribute the work among the available CPUs.**",
"_____no_output_____"
],
[
"We should give a subset of pages to each CPU to make its animes' .tsv file. <br/>\nHere we will group the page numbers that should be given to each processor. ",
"_____no_output_____"
]
],
[
[
"RangePage = list(range(1, len(EachPageURLs) + 1))\nPageNums = [RangePage[i * NumOfPage:(i+1) * NumOfPage] for i in range(mp.cpu_count())]",
"_____no_output_____"
]
],
[
[
"Call the function for each CPU give a susbset of .html files. ",
"_____no_output_____"
]
],
[
[
"pool = mp.Pool(mp.cpu_count())\nresults = [pool.apply_async(FunctionExtract, args = (PageNums[i],i)) for i in range(mp.cpu_count())]\npool.close()",
"_____no_output_____"
]
],
[
[
"# 2. Search Engine\n",
"_____no_output_____"
],
[
"Here we are asked to build a search engine which given a query, will give back the documents that are similar to the given query. ",
"_____no_output_____"
],
[
"### 2.0 Pre-processing the information",
"_____no_output_____"
],
[
"Here we will pre-process all the information of an anime and store it to another .tsv file which will be name SynopsisPrepAnime_(i).csv which i corresponds to the index number of the page in its page. ",
"_____no_output_____"
],
[
"To do pre-processing we have 5 stages and for each stage we will write a function. ",
"_____no_output_____"
],
[
"### 2.0.1 Tokenization ",
"_____no_output_____"
],
[
"Given a string, we will return the words that are in the given string. ",
"_____no_output_____"
]
],
[
[
"# Using this functions to extract the terms in each given sentence \ndef Tokenization(Sentence):\n return nltk.word_tokenize(Sentence)",
"_____no_output_____"
]
],
[
[
"### 2.0.2 Lowercasing",
"_____no_output_____"
],
[
"Given a list of strings, we will return the same strings but in lower case. ",
"_____no_output_____"
]
],
[
[
"# Turn all the words that we have in the given list of words to their lowercase\ndef Lowercasing(Words):\n return [w.lower() for w in Words]",
"_____no_output_____"
]
],
[
[
"### 2.0.3 StopWordsRemoval",
"_____no_output_____"
],
[
"Here we will define a list which contains all the stopwords in English language. ",
"_____no_output_____"
]
],
[
[
"StopWords = stopwords.words('english')",
"_____no_output_____"
]
],
[
[
"Given a list of strings, we will remove the stopwords from that strings. ",
"_____no_output_____"
]
],
[
[
"# We will consider just those terms that are not in stopwords\ndef StopWordsRemoval(Words):\n return [w for w in Words if w not in StopWords]",
"_____no_output_____"
]
],
[
[
"### 2.0.4 PunctuationsRemoval",
"_____no_output_____"
],
[
"In this function given a list of strings, we will remove the punctuations from those strings. ",
"_____no_output_____"
]
],
[
[
"# To remove the punctuations, we only will consider the alphabetic letters\ndef PunctuationsRemoval(Words):\n return [w for w in Words if w.isalpha()]",
"_____no_output_____"
]
],
[
[
"### 2.0.5 Stemming",
"_____no_output_____"
],
[
"In this function, given a list of strings, we try to return back the stem of each string that we have in the list. <br/>\nHere we will use 'PorterStemmer' algorithm to do stemming. ",
"_____no_output_____"
]
],
[
[
"# The stem of each word will be returned \ndef Stemming(Words):\n return [PorterStemmer().stem(w) for w in Words]",
"_____no_output_____"
]
],
[
[
"* Here we are asked to work with the 'Synopsis' of each anime, so we decided pre-process only the synopsis of each anime and not all the information that we extracted. ",
"_____no_output_____"
],
[
"* **Note:** For your information, we want to do the pre-process we will distribute the work among the available number of CPUs in the system. Each CPU will be given a subset of pages to do the pre-process for the contained anime information. ",
"_____no_output_____"
],
[
"### 2.0.6 Main Function ",
"_____no_output_____"
]
],
[
[
"def MainFunction(Pages):\n \n # For each page \n for page in Pages:\n \n # For each anime in a specific page\n for anime in range((50 if page != 383 else 28)):\n \n # Read the '.tsv' file of that specific anime\n Path = '/home/mehrdad/ADM-HW3/HTMLS/page'\n File = open(Path + str(page) + '/anime_' + str(anime) +'.tsv', mode = 'r')\n \n # For each anime we want to extract the synopsis \n Data = File.read().split('\\n')[1].split('\\t')[10]\n File.close()\n \n # Given the synopsis of a specific anime to pre-processing stage\n Tokens = Tokenization(Data)\n Lowercase = Lowercasing(Tokens)\n WithoutStop = StopWordsRemoval(Lowercase)\n WithoutPuncs = PunctuationsRemoval(WithoutStop)\n Stems = Stemming(WithoutPuncs)\n \n # Store the result of the pre-processing stage in a '.csv' file for each specific anime\n PreProcessed = open(Path + str(page) + '/anime_' + str(anime) +'_synopsisPrep.csv', mode = 'w')\n PreProcessed.write(\",\".join(Stems))\n PreProcessed.close()",
"_____no_output_____"
]
],
[
[
"## 2.1 Conjunctive query",
"_____no_output_____"
],
[
"Here we will pre-process the synopsis of each anime. ",
"_____no_output_____"
],
[
"Group the pages to be given to the CPUs. ",
"_____no_output_____"
]
],
[
[
"RangePage = list(range(1, len(EachPageURLs) + 1))\nPageNums = [RangePage[i * NumOfPage:(i+1) * NumOfPage] for i in range(mp.cpu_count())]",
"_____no_output_____"
]
],
[
[
"Pre-processing all the synopsis of animes. ",
"_____no_output_____"
]
],
[
[
"pool = mp.Pool(mp.cpu_count())\nresults = [pool.apply_async(MainFunction, args = (PageNums[i],)) for i in range(mp.cpu_count())]\npool.close()",
"_____no_output_____"
]
],
[
[
"## 2.1.1 Create your index!",
"_____no_output_____"
],
[
"Here we will create a file name **'vocabulary.json'** which will map each word that we want to take into account for our analysis to a specific ID. ",
"_____no_output_____"
],
[
"We will go through all the pre-processed files that we have created for each anime and extract all the words in them and give each word a specific ID. ",
"_____no_output_____"
]
],
[
[
"# Dictionary which stores each words in all anime \nMyDict = dict()\n\n# For each page\nfor i in range(1, 384):\n \n # For each anime\n for j in range((50 if i != 383 else 28)):\n \n # Open the '.csv' file of each specific anime\n Path = '/home/mehrdad/ADM-HW3/HTMLS/page'\n File = open(Path + str(i) + '/anime_' + str(j) +'_synopsisPrep.csv', mode = 'r')\n \n # Get the words in the synopsis of each anime\n DescriptionWord = File.read().split(',')\n \n # For each word in the synopsis we will have an entry in the dictionary\n for term in DescriptionWord:\n MyDict[term] = 0 \n \n# Assign each word a unique id\nfor term_id,term in enumerate(MyDict.keys()):\n MyDict[term] = term_id\n\n# Writing the resulted information into a .json file so we can easily retrieve them\nwith open(\"Vocabulary.json\", \"w\") as write_file:\n json.dump(MyDict, write_file)",
"_____no_output_____"
]
],
[
[
"### 2.1.1.1 Creating Inverted Indexes",
"_____no_output_____"
],
[
"Here we will go through all the preprocessed files and add the documents id to the values of the terms we find in each document. ",
"_____no_output_____"
],
[
"We will store the document ids as a combination of page number of anime id (#page, #anime_id).",
"_____no_output_____"
],
[
"* We will store our created inverted index into a file named **'InvertedIndex_1'**. ",
"_____no_output_____"
]
],
[
[
"# A dictionary which have all the terms in the corpus and their term_id\nVocabularies = json.load(open(\"Vocabulary.json\", \"r\"))\n\n# We will store our inverted index in this dictionary\n# Keys: term\n# Value: a list of document_id (which is a pair of page and anime index) \nInvertedIndexes = defaultdict(set)\n\n# For each page \nfor i in range(1, 384):\n # For each anime \n for j in range((50 if i != 383 else 28)):\n \n # Read the '.csv' file for each specific anime\n Path = '/home/mehrdad/ADM-HW3/HTMLS/page'\n File = open(Path + str(i) + '/anime_' + str(j) +'_synopsisPrep.csv', mode = 'r')\n \n # Extract the words in the synopsis for each anime\n DescriptionWord = File.read().split(',')\n \n # For all the terms that are in this anime, we will add the document_id to its list\n for term in DescriptionWord:\n InvertedIndexes[Vocabularies[term]].add((i,j))\n \n# Sorting the document_ids\nfor term in InvertedIndexes:\n InvertedIndexes[term] = sorted(InvertedIndexes[term])\n \n# We will store these information in a '.json' file so we can easily retrieve them\nwith open(\"InvertedIndex_1.json\", \"w\") as write_file:\n json.dump(InvertedIndexes, write_file)",
"_____no_output_____"
]
],
[
[
"## 2.1.2 Execute the query",
"_____no_output_____"
],
[
"Here given a query we want to return the documents that are containing all the words in the given query. ",
"_____no_output_____"
],
[
"In order to answer the query, first we should pre-process the query as well. We need to do this as we have pre-processed all the information that we have extracted from the htmls. ",
"_____no_output_____"
],
[
"* Here we will write a function which given a string, it will pre-process the string and return the results. ",
"_____no_output_____"
],
[
"### 2.1.2.1 Function String Pre-process",
"_____no_output_____"
]
],
[
[
"# Given a stirng, the string will go to the pre-process stage\ndef StringPreProcess(string):\n Tokens = Tokenization(string)\n LowerCased = Lowercasing(Tokens)\n WithoutStop = StopWordsRemoval(LowerCased)\n WithoutPunc = PunctuationsRemoval(WithoutStop)\n Stemmed = Stemming(WithoutPunc)\n return Stemmed",
"_____no_output_____"
]
],
[
[
"### 2.1.2.2 Function token to TermID",
"_____no_output_____"
],
[
"This function given a token and the Vocabulary data structure will return back the term_id of that token. If the token is not in the Vocabulary, this function will return None. ",
"_____no_output_____"
]
],
[
[
"def WordToTermID(Token, Vocabulary):\n return Vocabulary.get(Token)",
"_____no_output_____"
]
],
[
[
"### 2.1.2.3 Function Extracting Documents",
"_____no_output_____"
],
[
"Given a TermID and inverted indexes, this function will return the documents which are containing this specific token. If TermID is equal to 'None', this function will return empty list. ",
"_____no_output_____"
]
],
[
[
"def ExtractDocs(TermId, InvertedIndex):\n return InvertedIndex.get(str(TermId)) if TermId else []",
"_____no_output_____"
]
],
[
[
"### 2.1.2.4 Function Query to Documents",
"_____no_output_____"
],
[
"This function will process the given query and return back the documents that are containing all the words in the query. ",
"_____no_output_____"
]
],
[
[
"def QueryToDocuments(Query, Vocabulary, InvertedIndex):\n PreProcessedQuery = StringPreProcess(Query)\n \n # If the query after the pre-process stage was empty, return an empty list\n if not len(PreProcessedQuery):\n return []\n \n # This list will store all the documents in which each term was there. \n Documents = []\n\n # Convert each term to its termid\n TermIDs = [WordToTermID(term, Vocabulary) for term in PreProcessedQuery]\n \n # All the documents for each Term\n Documents = [set(map(tuple, ExtractDocs(TermID, InvertedIndex))) for TermID in TermIDs]\n \n # Take the intersection of the documents\n Result = sorted(Documents[0].intersection(*Documents))\n return Result ",
"_____no_output_____"
]
],
[
[
"### 2.1.2.5 Function Data to Dataframe",
"_____no_output_____"
],
[
"This function given the name of the columns and the values for each column will create a dataframe with those information which help to visualize the results in tabular format. ",
"_____no_output_____"
]
],
[
[
"# Given the column name and the records, will produce a dataframe\ndef DataToDataFrame(ColumnsName, Records):\n \n # Declare a datafram with specific column name\n Table = pd.DataFrame(columns = ColumnsName)\n \n # For each records \n for rec in Records:\n \n # We will store the values in each records in a dictionary as for adding a record to the dataframe\n # we should pass a dictionary\n TempDict = dict()\n \n # For each value of the columns of the records\n for col in range(len(ColumnsName)):\n \n # Give the value of that column to the dictionary\n TempDict[ColumnsName[col]] = rec[col]\n \n # Add this new record to the dataframe\n Table = Table.append(TempDict,ignore_index = True )\n display(Table)",
"_____no_output_____"
]
],
[
[
"### 2.1.2.6 Function Main Answer Query",
"_____no_output_____"
]
],
[
[
"# Loading Vocabulary\nVocabulary = json.load(open(\"Vocabulary.json\", \"r\"))\n\n# Loading Inverted Indexes\nInvertedIndex = json.load(open(\"InvertedIndex_1.json\", 'r'))\n\nQuery = input('Please enter your query here: ')\n\n# Results \nResults = QueryToDocuments(Query, Vocabulary, InvertedIndex)\n\n# To pass the data to show in tabular format \nColumns, Data = ['animeTitle', 'animeDescription', 'URL'], []\n\n# Now here we should iterate over all the results and retrieve the information for each of these anime\nfor Res in Results:\n Path = '/home/mehrdad/ADM-HW3/HTMLS/page'\n FileURL = open('URLs.txt', mode = 'r')\n FileInfo = open(Path + str(Res[0]) + '/anime_' + str(Res[1]) +'.tsv', mode = 'r')\n\n Information = FileInfo.read().split('\\n')[1].split('\\t')\n Title = Information[0]\n Description = Information[10]\n URL = FileURL.read().split('\\n')[(Res[0]-1)*50 + Res[1]]\n Data.append([Title, Description, URL])\n\n# Showing the results\nDataToDataFrame(Columns, Data)",
"Please enter your query here: saiyan race\n"
]
],
[
[
"## 2.2. Conjunctive query & Ranking score\n",
"_____no_output_____"
],
[
"For this part we are asking to given a query, show the top k documents that are more closer to this query using scoring schema.",
"_____no_output_____"
],
[
"To answer this question, first we should go through some definitions. ",
"_____no_output_____"
],
[
"### - What is tf?",
"_____no_output_____"
],
[
"We define tf(t, d) of term t in documnet d as the **division** of the **number of occurance of term t in document d** by **the total number of occurance of term t in all documents** in the corpus. You can see the formula here: ",
"_____no_output_____"
],
[
"### - What is idf?",
"_____no_output_____"
],
[
"Idf (inverse document frequency) is a measure of how much information the word provides. This value will be obtained by **dividing** the **total number of documents** by the **number of documents containing the term**, and then taking the **logarithm** of that quotient. You can see the formula here: ",
"_____no_output_____"
],
[
"### - What is tfidf?",
"_____no_output_____"
],
[
"Tfidf for each term and document can be obtained by the **multiplication** of **tf(t,d) and idf(t,D)**. The higher this value is, the more important that term is in that specific document. ",
"_____no_output_____"
],
[
"## 2.2.1 Creating the inverted indexes",
"_____no_output_____"
],
[
"Here we should build an inverted index in the format that each term corresponds to the documents in which that term is appearing and also the tfidf of that term in that specific document. ",
"_____no_output_____"
],
[
"Here we will define some functions to help us to do this process. ",
"_____no_output_____"
],
[
"### 2.2.1.1 Function tfidf",
"_____no_output_____"
],
[
"This function given the list of documents and the number of occurance of each specific term T in each of these documents and also the number of all the documents in the corpus, will return the terms and its tfidf for each of the documents that are containing that term. ",
"_____no_output_____"
]
],
[
[
"def GetTfIdf(invertedIndex, NumberOfDocs):\n for Term in invertedIndex: # For each term\n \n # How many times that term occured in the corpus\n TotalOccurance = sum(doc[1] for doc in invertedIndex[Term])\n \n # How many documents are containig that term \n NumberOfTDocs = len(invertedIndex[Term])\n \n # Computing the idf of a term \n idf = log(NumberOfDocs / NumberOfTDocs)\n \n for docs in invertedIndex[Term]: # For each document that is contaning the term \n \n # Put the tfidf (tf*idf) instead of the number of occurence of that term in the document\n docs[1] = (docs[1] / TotalOccurance) * idf\n \n return invertedIndex",
"_____no_output_____"
]
],
[
[
"### 2.2.1.2 Create a suitable inverted index",
"_____no_output_____"
],
[
"Now we should write a code that build another type of inverted index, which each term corresponds to the documents that are contaning that term and also the number of occurence of the term in each of those documents. This inverted index will be saved in 'InvertedIndex_2.json' file. ",
"_____no_output_____"
]
],
[
[
"# To get the term_id of each term\nVocabularies = json.load(open(\"Vocabulary.json\", \"r\"))\n\n# To store the inverted index\n# We will store our inverted index in this dictionary\n# Keys: term\n# Value: a list of (document_id (which is a pair of page and anime index), #occurance of the term in this document)\nInvertedIndexes = defaultdict(set)\n\n# For each page\nfor i in range(1, 384):\n # For each anime \n for j in range((50 if i != 383 else 28)):\n \n # Read the '.csv' file for a specific anime\n Path = '/home/mehrdad/ADM-HW3/HTMLS/page'\n File = open(Path + str(i) + '/anime_' + str(j) +'_synopsisPrep.csv', mode = 'r')\n \n # Extract the words in the synopsis of that anime\n DescriptionWord = File.read().split(',')\n \n # Check the number of the occurances of each word in the synopsis of a specific anime\n NumberOfOccurenceInDoc = Counter(DescriptionWord)\n \n # For each term we will add this anime and its total number of occurance in this anime\n for term in NumberOfOccurenceInDoc:\n InvertedIndexes[Vocabularies[term]].add(((i,j), NumberOfOccurenceInDoc[term]))\n \n \n \n# Sorting the document ids\nfor term in InvertedIndexes:\n InvertedIndexes[term] = sorted(InvertedIndexes[term])\n \n# Store this inverted index in '.json' file to get the information pretty easily\nwith open(\"InvertedIndex_2.json\", \"w\") as write_file:\n json.dump(InvertedIndexes, write_file)\n",
"_____no_output_____"
]
],
[
[
"Now we are able to compute the tf, idf and ea the end the tfidf of each term and docment. ",
"_____no_output_____"
],
[
"### 2.2.1.3 Main Creating the new Inverted Index",
"_____no_output_____"
],
[
"Now here we will create a new inverted index such that each term has the information about each document that is containig that specific term and also the tfidf of that term and that specific document. This new inverted index will be saved in a file named **'InvertedIndex_3.json'**.",
"_____no_output_____"
]
],
[
[
"# Loading the second inverted index term -> documents, # Occurence\nInvertedIndex = json.load(open(\"InvertedIndex_2.json\", \"r\"))\n\n# Number of the docuements in the corpus \nNumOfDocs = len(AllURLs) # Number of documents are the number of the anime\n\n# Computing the tfidf for each term and document\nNewInvertedIndex = GetTfIdf(InvertedIndex, NumOfDocs)\n\n# Saving the new inverted index into a new file named 'InvertedIndex_3.json'\nwith open(\"InvertedIndex_3.json\", \"w\") as write_file:\n json.dump(NewInvertedIndex, write_file)",
"_____no_output_____"
]
],
[
[
"### 2.2.1.4 Function query to tfidf",
"_____no_output_____"
],
[
"This function given the query, the second inverted index which each term corresonds to the docuemnts that are containig the number of occurence of that term in those documents, the vocabulary, and the number of all the docuemnts in the corpus will compute the tfidf of the term in the query. ",
"_____no_output_____"
]
],
[
[
"def QueryToTfidf(Query, invertedIndex,Vocabulary, NumberOfDocs):\n \n \n # Mapping the words in query to their term_id\n PreProcessedQuery = StringPreProcess(Query)\n Query = [str(Vocabulary.get(Term)) for Term in PreProcessedQuery]\n \n # Check the number of occurence of each term in the query \n QueryDict = Counter(Query)\n \n for Term in QueryDict:# for each term in the query\n \n # If that term is in the inverted index\n if invertedIndex.get(Term):\n\n # How many times that term occured in the corpus\n TotalOccurance = sum(doc[1] for doc in invertedIndex[Term])\n\n # How many documents are containig that term \n NumberOfTDocs = len(invertedIndex[Term])\n\n # Computing the idf of a term \n idf = log(NumberOfDocs / NumberOfTDocs)\n\n # Put the tfidf (tf*idf) instead of the number of occurence of that term in the query\n QueryDict[Term] = (QueryDict[Term]/ TotalOccurance) * idf\n else:\n QueryDict[Term] = 0\n \n return QueryDict",
"_____no_output_____"
]
],
[
[
"### 2.2.1.5 Function Dot Product ",
"_____no_output_____"
],
[
"In this function given two vectors, we will compute the dot product of the two and return back the result. ",
"_____no_output_____"
]
],
[
[
"def DotProduct(V1, V2): # The two given vectors should have the same length\n return sum(V1[i] * V2[i] for i in range(len(V1)))",
"_____no_output_____"
]
],
[
[
"### 2.2.1.6 Function Euclidean norm ",
"_____no_output_____"
],
[
"This function given a vector, will compute the Euclidean norm of that vector and return the result. ",
"_____no_output_____"
]
],
[
[
"# Computes the Eucledean norm the given vector\ndef Euclidean(Vector): \n return (sum(Vector[i]**2 for i in range(len(Vector)))) ** .5",
"_____no_output_____"
]
],
[
[
"### 2.2.1.7 Function Query to Documents",
"_____no_output_____"
],
[
"In this function, given the query and the 3rd inverted index, we will extract the documents that are contaning all the terms in the query. The result will be a dictinoary which the keys are the documents and the keys are a list which is containing all the tfirdf of the terms in the query. ",
"_____no_output_____"
]
],
[
[
"def QueryToDocuemnts(Query, invertedIndex):\n \n AllTermsDict = []\n for term in Query: # For each term in the query\n TermDict = {} # The dictionary which contains the documents that are containing that term \n \n # For each document which is contaning the term \n if invertedIndex.get(term):\n for doc in invertedIndex[term]:\n\n # Add the document and the corresponded tfidf for that term in that document\n TermDict[tuple(doc[0])] = doc[1]\n # Add the dictionary for a specific term in the query which is containig all \n # the document that are containig that term \n\n AllTermsDict.append(TermDict.copy())\n \n # Extract the documents that are containing all the terms in the query\n \n # All the documents in each dictionary \n AllDocs = [set(termDict.keys()) for termDict in AllTermsDict]\n \n AllIncludedDocs = AllDocs[0].intersection(*AllDocs) # Docs that are containing all the terms in the query\n \n # Here we are creating a dictionary which keys are the documents that are containig all the terms in the query\n # and the keys are the tfidf of all the term in the query in sequence. \n DocsToTfidf = defaultdict(list)\n for termDict in AllTermsDict:\n for doc in AllIncludedDocs:\n DocsToTfidf[doc].append(termDict[doc])\n \n \n return DocsToTfidf",
"_____no_output_____"
]
],
[
[
"### 2.2.1.8 Function Similarity ",
"_____no_output_____"
],
[
"In this function given the query tfidf values and all the documents that are containig all the terms in the query, we want to compute the similarity of the query with each docuement.",
"_____no_output_____"
],
[
"* The cosine similarity between two vectores will be the **dot product of the values** in these two vectors, **divided** by **the multiplication of the euclidean norm** of each of the vectors. ",
"_____no_output_____"
]
],
[
[
"def Similarity(QueryTfidf, Documents):\n \n for doc in Documents:\n \n # Compute the dot product of tfidf of the terms in the query and a specific document \n DotProductRes = DotProduct(QueryTfidf, Documents[doc])\n \n # Computer the similarity of query and a specific document \n Similarity = DotProductRes/(Euclidean(QueryTfidf)*Euclidean(Documents[doc]))\n \n # Append the value of similarity at the end of the least of each document. \n Documents[doc].append(Similarity)\n \n return Documents",
"_____no_output_____"
]
],
[
[
"* We are asked to sort the similar documents using heap sort. In order to use heap sort, we should have a class for the object that we are passing to the heap. ",
"_____no_output_____"
],
[
"### 2.2.1.9 Function Docs Class",
"_____no_output_____"
]
],
[
[
"class Docs: \n \n # Storing the passed information for each object\n def __init__(self,DocInfo): \n self.docInfo = DocInfo\n \n # Overriding the operator '<'. Here as we need the descending sort, we override in reverse. \n def __lt__(self, other): \n return self.docInfo[-1] > other.docInfo[-1]\n \n # Overriding the operator '>'\n def __gt__(self, other): \n return other.__lt__(self) ",
"_____no_output_____"
]
],
[
[
"### 2.2.1.10 Function HeapSort",
"_____no_output_____"
],
[
"This function given a list of object will sort them by using heap tree and return the result. ",
"_____no_output_____"
]
],
[
[
"def heapsort(objects): \n \n # Heap Tree\n heap = [] \n \n # Add all the given object to the tree\n for element in objects: \n heappush(heap, element) \n \n # Will store the ordered version of objects\n ordered = [] \n \n # Until we have still object in tree\n while heap: \n #Take the root of the tree -> Which is the highest value. \n ordered.append(heappop(heap)) \n \n return ordered ",
"_____no_output_____"
]
],
[
[
"### 2.2.1.10 Main function Ranking",
"_____no_output_____"
]
],
[
[
"# Get the query from the user\nQuery = input('Please enter your query here: ')\n\n# First K most similar documents\nK = int(input('Please enter the value of k here: '))\n\n# Loading the second inverted index term -> documents, # Occurence\nSecondInvertedIndex = json.load(open(\"InvertedIndex_2.json\", \"r\"))\n\n# Loading the third inverted index term -> documents, # tfidf\nThirdInvertedIndex = json.load(open(\"InvertedIndex_3.json\", \"r\"))\n\n# Loading the vocabulary file which maps each term to a specific ID\nVocabularies = json.load(open(\"Vocabulary.json\", \"r\"))\n\n# Compute the TFIDF of the query\nQueryTFIDF = QueryToTfidf(Query, SecondInvertedIndex,Vocabularies, len(AllURLs))\n\n# Take the documents that are containing all the words\nRelatedDocuments = QueryToDocuemnts(QueryTFIDF.keys(), ThirdInvertedIndex)\n\n# Compute the similarity of each document with query\nDocumentsSimilarity = Similarity(list(QueryTFIDF.values()), deepcopy(RelatedDocuments))\n\n# Here we will store the details of each similar documents \nData = []\nfor Res in DocumentsSimilarity:\n Path = '/home/mehrdad/ADM-HW3/HTMLS/page'\n FileURL = open('URLs.txt', mode = 'r')\n FileInfo = open(Path + str(Res[0]) + '/anime_' + str(Res[1]) +'.tsv', mode = 'r')\n\n Information = FileInfo.read().split('\\n')[1].split('\\t')\n Title = Information[0]\n Description = Information[10]\n URL = FileURL.read().split('\\n')[(Res[0]-1)*50 + Res[1]]\n Data.append([Title, Description, URL, DocumentsSimilarity[Res][-1]])\n \n# Here we will sort the documents based on the similarity using heap tree\nData = [Doc.docInfo for Doc in heapsort([Docs(i) for i in Data])]\n\n# Setting the similarity to 2 float places \nfor doc in Data:\n doc[-1] = \"{:.2f}\".format(doc[-1])\n\n# Here we will pick the first K documents that are more similar with respect to the given query\nDataToDataFrame(['animeTitle','animeDescriptio', 'Url', 'Similarity'], Data[:K])\n",
"Please enter your query here: saiyan race\nPlease enter the value of k here: 10\n"
]
],
[
[
"# 3. Define a new score",
"_____no_output_____"
],
[
"In this question we let the user to include more information in his query. Most of the provided information will be working as a filter except the which user gives us for the synopsis. What we let the user include in his query and in which format: ",
"_____no_output_____"
],
[
"### Filters",
"_____no_output_____"
],
[
"1- **The type of anime:** If a user wants to include the type of the anime in the query, he should use this notation -> 1- {The type of anime}, ex. 1- tv",
"_____no_output_____"
],
[
"2- **The date of releasing:** If a user want to include a starting point which our search engine consider the anime which their release that is after that starting poin, he should use this notation -> 2- {released year}, ex. 2- 2009",
"_____no_output_____"
],
[
"3- **The date of ending:** If the user wants to include an end point in which our search engine not consider the anime that have been ended after this time then he should use this notation: 3- {end year}, ex. 3- 2020",
"_____no_output_____"
],
[
"4- **Some words in the synopsis:** If the user wants to include some words that those words has been occured in the resulted anime's synopsis, he should use this notation: 4- {words in synopsis}, ex. 4- saiyan race",
"_____no_output_____"
],
[
"* **Note:** We won't consider the documents that are missing some of the information provided in the query. ",
"_____no_output_____"
],
[
"### Scoring",
"_____no_output_____"
],
[
"To score the related documents and show them in order, we will consider these variables: ",
"_____no_output_____"
],
[
"**The normalized value for value x in a sequence of values X will be defined as (x - min(X))/(max(X) - min(X))**",
"_____no_output_____"
],
[
"1- **The cosine similarity:** The docuemnts that have a higher cosine similarity with respect to the words in synopsis given in the query, will have a higher priority.**To mention this value is between 1 and 0**",
"_____no_output_____"
],
[
"2- **Anime score and anime user:** We will consider a combination of anime score and its number of user. We will use a multiplication of the number of the users and the score of the anime. Then when we had this value for all the related animes, we will normalize the values according to its formula.",
"_____no_output_____"
],
[
"3- **Number of members:** As we know here the higher the member, the more popular the anime. So here we will normalize these values using the mentioned formula for normalization. ",
"_____no_output_____"
],
[
"**Final score: The final score of a document in our approach will be the sum of all of these values. The the ones that having higher score, will have a higher priority.**",
"_____no_output_____"
],
[
"* **Note:** If in any case we had some missing values, we will consider those values as 0. ",
"_____no_output_____"
],
[
"### *The main function for this part is at the end of this section*.",
"_____no_output_____"
],
[
"Now we go throguh the steps and the functions needed to build this search engine. ",
"_____no_output_____"
],
[
"## 1. Filtering",
"_____no_output_____"
],
[
"**Here we first filter the the documents with respect to the given query**",
"_____no_output_____"
],
[
"### 3.1.1 Function Query Process",
"_____no_output_____"
]
],
[
[
"# With the help of this function we will be able to extract each parameter in the query\ndef QueryProcess(Query):\n Strings = re.split('[1|2|3|4]-', Query.lower())[1:] \n Numbers = re.findall('[1|2|3|4]+-', Query.lower())\n GivenConditions = [\"\".join(i) for i in zip(Numbers, Strings)]\n return GivenConditions",
"_____no_output_____"
]
],
[
[
"After getting the query, now we should filter the documents with respect to the given conditions. Here we will filter the by using 'Anime Type', 'Anime Released Data' and 'Anime End Data'. ",
"_____no_output_____"
],
[
"### 3.1.2 Function Filter Documents",
"_____no_output_____"
]
],
[
[
"def FilterDocs(Conditions):\n \n # This dictionary will contain the filtered documents and their necessary information\n FilteredDocs = {}\n \n # To have the URL of all the anime\n FileURL = open('URLs.txt', mode = 'r').read().split('\\n')\n \n # For each page\n for page in range(1, 384):\n \n # For each anime\n for anime in range((50 if page != 383 else 28)):\n \n # Read the '.tsv' file of each specific anime\n Path = '/home/mehrdad/ADM-HW3/HTMLS/page'\n File = open(Path + str(page) + '/anime_' + str(anime) +'.tsv', mode = 'r')\n \n # Take the URL of a specific anime\n URL = FileURL[(page-1)*50 + anime]\n \n # The extracted information for that anime\n AnimeInfo = FilterInfo(File)\n \n # A flag which specifies if we should consider this docuemnt or it should be filtered\n Pick = True\n \n # For each condition that we have in the query\n for Con in Conditions:\n \n # Here will check each condition given in the query\n # Anime type\n # Releasing date\n # End date\n \n Con = list(map(str.strip, Con.split('-')))\n if Con[0] == '1' and AnimeInfo['animeType'] != Con[1]:\n Pick = False\n elif (Con[0] == '2' and AnimeInfo['releaseDate'] < Con[1]) or not AnimeInfo['releaseDate'].isdigit():\n Pick = False\n elif (Con[0] == '3' and (AnimeInfo['endDate'] > Con[1]) or not AnimeInfo['endDate'].isdigit()):\n Pick = False \n if Pick:\n AnimeInfo['URL'] = URL\n FilteredDocs[(page,anime)] = deepcopy(AnimeInfo)\n return FilteredDocs",
"_____no_output_____"
]
],
[
[
"### 3.1.3 Function Filter Necessary Information",
"_____no_output_____"
],
[
"In this function given a document we will extract the necessary information which will be 'Anime Title', 'Anime Score', 'Anime Users' and 'Anime members'. ",
"_____no_output_____"
]
],
[
[
"def FilterInfo(Document):\n \n # We will filter some useful information the given document\n ReadDocument = Document.read().split('\\n')[1].split('\\t')\n InfoDict = {'animeTitle':ReadDocument[0], 'animeScore': ReadDocument[6], 'animeUser': ReadDocument[7],\n 'animeMembers': ReadDocument[5], 'releaseDate': ReadDocument[3].split(',')[-1].strip(), \n 'endDate':ReadDocument[4].split(',')[-1].strip(),'animeType': ReadDocument[1]}\n \n # We will lower case everything except Anime Title\n for i in list(InfoDict.keys())[1:]:\n InfoDict[i] = InfoDict[i].lower()\n \n return deepcopy(InfoDict)",
"_____no_output_____"
]
],
[
[
"## 3.1.4 Main Function Filter",
"_____no_output_____"
],
[
"Here we will get the query from the user in the format stated above.",
"_____no_output_____"
]
],
[
[
"def Filtering(Query):\n FilteredDocuments = FilterDocs(Query)\n\n # Then we should compute check the similarity between the query and the given filtered documents \n # So we will add another property to each filtered data name synopsisSimilarity.\n # This value will be by default 0. \n\n for Doc in FilteredDocuments:\n FilteredDocuments[Doc]['synopsisSimilarity'] = 0 \n\n # Then we will check the cosine similarity between the query and the synosis of the anime if this information\n # was provided in the query \n \n SynopsisQuery = ''\n \n for q in Query:\n if '4-' in q:\n SynopsisQuery = q.split('- ')[-1]\n\n if SynopsisQuery:\n # Loading the second inverted index term -> documents, # Occurence\n SecondInvertedIndex = json.load(open(\"InvertedIndex_2.json\", \"r\")) \n \n # Loading the third inverted index term -> documents, # tfidf\n ThirdInvertedIndex = json.load(open(\"InvertedIndex_3.json\", \"r\"))\n \n # Loading the vocabulary file which maps each term to a specific ID\n Vocabularies = json.load(open(\"Vocabulary.json\", \"r\"))\n\n # Compute the TFIDF of the query\n QueryTFIDF = QueryToTfidf(SynopsisQuery, SecondInvertedIndex,Vocabularies, len(AllURLs))\n\n # Take the documents that are containing all the words\n RelatedDocuments = QueryToDocuemnts(QueryTFIDF.keys(), ThirdInvertedIndex)\n\n # Compute the similarity of each document with query\n DocumentsSimilarity = Similarity(list(QueryTFIDF.values()), deepcopy(RelatedDocuments))\n # Here we will pick the similarity between our filtered data and given query\n for doc in FilteredDocuments:\n SimilarityOfDoc = DocumentsSimilarity.get(doc)\n if SimilarityOfDoc:\n FilteredDocuments[doc]['synopsisSimilarity'] = SimilarityOfDoc[-1]\n return FilteredDocuments",
"_____no_output_____"
]
],
[
[
"After having all the filtered documents and checking their similarity with respect to the given query, we will now add new scores to each documents. ",
"_____no_output_____"
],
[
"## Scoring",
"_____no_output_____"
],
[
"In the scoring part as we mentioned above, we will add new scores to each of the filtered documents. ",
"_____no_output_____"
],
[
"### 3.2.1 Function Score and User",
"_____no_output_____"
],
[
"This function, given all the filtered documents will consider a new score name 'animeSUScore' which is the multiplication of each 'animeScore' and 'animeUser'. At the end we will normalize the values. ",
"_____no_output_____"
]
],
[
[
"def RankUser(Docs):\n \n # Pick the default values for max and min \n Min, Max = float(\"inf\"), 0\n \n # For each document\n for doc in Docs: \n \n # Try to multiply animeScore and animeUser\n try:\n Docs[doc]['animeSUScore'] = float(Docs[doc]['animeScore']) * float(Docs[doc]['animeUser'])\n \n # In case of an error we find out the values are missed or corrupted\n except: \n \n # Consider the result of the multiplication 0\n Docs[doc]['animeSUScore'] = 0 \n \n # Update the min and max values\n Min, Max = min(Min, Docs[doc]['animeSUScore']), max(Max, Docs[doc]['animeSUScore'])\n \n # Normalize the values of animeSUScore\n for doc in Docs: \n Docs[doc]['animeSUScore'] = ( Docs[doc]['animeSUScore'] - Min)/(Max - Min)",
"_____no_output_____"
]
],
[
[
"### 3.2.2 Function Member Score",
"_____no_output_____"
],
[
"This function given all the filtered documents will normalize the number of members of each document and consider that as a new score. ",
"_____no_output_____"
]
],
[
[
"def MemberScore(Docs):\n \n # Setting the default values for min and max\n Min, Max = float('inf'), 0\n \n # For each document\n for doc in Docs:\n \n # Try to conver the value of animeMembers to float\n try:\n Docs[doc]['animeMembers'] = float(Docs[doc]['animeMembers'])\n \n # If corrupted or missed \n except: \n \n # Set it to 0\n Docs[doc]['animeMembers'] = 0 \n \n # Update the min and max\n Min, Max = min(Min, Docs[doc]['animeMembers']), max(Max, Docs[doc]['animeMembers'])\n \n # Normalize the value of memberScore\n for doc in Docs:\n Docs[doc]['memberScore'] = (Docs[doc]['animeMembers'] - Min)/(Max - Min)",
"_____no_output_____"
]
],
[
[
"### 3.2.3 Function Final Score",
"_____no_output_____"
],
[
"This function given all the documents will compute the final score of each document which will be the sum of all the scores we just defined and also the cosine similarity. ",
"_____no_output_____"
],
[
"* **None:** As the cosine similarity between the query and the synopsis of the anime gives us more detail about the anime, we will give a coefficient of '3' to this value so that this value dominate the others. ",
"_____no_output_____"
]
],
[
[
"def FinalScore(Docs):\n # For each document we will compute the final score\n for doc in Docs:\n Docs[doc]['finalScore'] = Docs[doc]['memberScore'] + Docs[doc]['animeSUScore'] + 3 * Docs[doc]['synopsisSimilarity']",
"_____no_output_____"
]
],
[
[
"## 3.2.4 Main Score ",
"_____no_output_____"
],
[
"This function will go over all the filtered documents and compute the socre of each of the documents and show the documents in order of their final score. ",
"_____no_output_____"
]
],
[
[
"# We will give all the filtered documents to be scored\ndef Scoring(FilteredDocuments):\n RankUser(FilteredDocuments)\n MemberScore(FilteredDocuments)\n FinalScore(FilteredDocuments)\n return FilteredDocuments",
"_____no_output_____"
]
],
[
[
"## 3.3 Main Function Query to Documents",
"_____no_output_____"
],
[
"Here we take a query from the user and give back him the resulted documents with respect to the given query. ",
"_____no_output_____"
],
[
"**Important: Don't forget to use the format of the query when you want to issue a query\"**",
"_____no_output_____"
],
[
"1- {Anime Type} 2- {Anime released date} 3- {Anime end date} 4- {Words in synopsis}<br/><br/>\n**FYI:** You can issue a subset of the parameters in the query. ",
"_____no_output_____"
]
],
[
[
"# Processing the query and filter the related documents \nQuery = QueryProcess(input(\"Please enter your query here: \"))\n\n# Filtering documents based on the given parameters in the query\nFilteredDocuments = Filtering(Query)\n\n# Scoring the filtered documents\nScoredDocuments = Scoring(FilteredDocuments)\n\n# Here we will store the details of each related documents \nData = []\nfor Res in ScoredDocuments:\n Path = '/home/mehrdad/ADM-HW3/HTMLS/page'\n FileInfo = open(Path + str(Res[0]) + '/anime_' + str(Res[1]) +'.tsv', mode = 'r')\n Description = FileInfo.read().split('\\n')[1].split('\\t')[10]\n\n Data.append([ScoredDocuments[Res]['animeTitle'], Description, ScoredDocuments[Res]['URL'],\n ScoredDocuments[Res]['finalScore']])\n\nData = [Doc.docInfo for Doc in heapsort([Docs(i) for i in Data])]\n\n# Setting the similarity to 2 float places \nfor doc in Data:\n doc[-1] = \"{:.2f}\".format(doc[-1])\n \n# Here we will pick the first K documents that are more similar with respect to the given query\nDataToDataFrame(['animeTitle','animeDescription', 'Url', 'Final score'], Data[:10])",
"Please enter your query here: 1- tv 4- saiyan race\n"
]
],
[
[
"## 3.4 Comparing",
"_____no_output_____"
],
[
"In this section we will compare the results that we got from this search engine the previous search engine that we have been built in the section 2.2.",
"_____no_output_____"
],
[
"The search engine in part 3.3 is more powerful than the search engine in 2.2 due to these: ",
"_____no_output_____"
],
[
"- Search engine in 3.3 gives the user the oppurtunity to use a kind of filtering based on the type, released data and the end date of the anime, while we didn't have this feature in search engine 2.2.",
"_____no_output_____"
],
[
"- Search engine in 3.3 for the resulted anime that have the same cosine similarity will with the help of the other information of the anime can sort the documents with respect to their popularity and rank. The ones that are better will be higher in the result. ",
"_____no_output_____"
],
[
"- Search engine in 3.3 even if can not find any document that contains all the word given for the synopsis will give us the result based on the other parameters of filtering. ",
"_____no_output_____"
],
[
"- If for a query in synopsis we have less than K related docuements, the search engine in 3.3 can give the result based on the other parameters of the filtering. ",
"_____no_output_____"
],
[
"**In the end we discovered that our search engine in 3.3 works much better than the search engine in 2.2.**",
"_____no_output_____"
],
[
"# 5. Algorithmic question",
"_____no_output_____"
],
[
"## 5.1 Algorithm",
"_____no_output_____"
],
[
"Basically in this case we are trying to find the maximum of the elements of an array while these elements are not adjacent. Here we will explain how does our algorithm works. ",
"_____no_output_____"
],
[
"We can solve this quetion with a greedy algorithm which solves this question with time complexity of O(n) as we can compute this value by iterating over the values just once. Here you can find the algorithm:",
"_____no_output_____"
],
[
"At each step, we will consider two sums. First the maximum sum up to this iteration with the previous element included and another sum which is the maximum sum up to this iteration without the previous element. We call these two sums as 1-SumWithPrevious, 2- SumWithoutPreviuos",
"_____no_output_____"
],
[
"1- We go through the elements. ",
"_____no_output_____"
],
[
"2- As we assume that the previous element has been picked so we add the current element to the SumWithoutPrevious sum. ",
"_____no_output_____"
],
[
"3- We check if this value (SumWithoutPrevious) is greater than the value of SumWithPrevious or not. If this was the case then we will swap these two values as for the next item we have picked its adjacent item.",
"_____no_output_____"
],
[
"4- If at some point the value of SumWithoutPrevious + the current element was not greater than SumWithPrevious, it means that with or without the current element we had the maximum sum so far. As we didn't include the element in the sums so we won't have a problem for the next item to be considered. Here we can assign the SumWithoutPrevios to SumWithPrevious. ",
"_____no_output_____"
],
[
"5- At the end we will return the maximum of these two values, which will be store in SumWithPrevious. ",
"_____no_output_____"
],
[
"**You can check a detailed example in 5.3 section.**",
"_____no_output_____"
],
[
"## 5.2 Implementation",
"_____no_output_____"
]
],
[
[
"def MaximumSumWithoutAdjacent(List):\n # define two sums and necessary datastructures\n SumWithoutPrevious, SumWithPrevious = 0, 0\n \n # Showing the elements in the list\n print('The elements in the list: ', List)\n \n # Iterating over the elements\n for i in List:\n \n # Add the current value to the sum without previous\n SumWithoutPrevious += i\n \n # Check the condition \n if SumWithoutPrevious > SumWithPrevious:\n \n # As we gain more score by picking the current element so we swap the values. \n SumWithoutPrevious, SumWithPrevious = SumWithPrevious, SumWithoutPrevious\n \n # If by considering the current element, we don't gain more score \n else:\n\n # Up to this point we have the maximum score by not picking the previous element\n SumWithoutPrevious = SumWithPrevious\n \n # Showing the last result. \n print('\\nResult:')\n print('\\nMaximum sum without considering adjacent elements: \\'', SumWithPrevious, '\\'', sep = '')",
"_____no_output_____"
]
],
[
[
"### Give me an example here: ",
"_____no_output_____"
]
],
[
[
"MyList = list(map(int, input(\"Please give me the elements of your list: \").split()))\nMaximumSumWithoutAdjacent(MyList)",
"Please give me the elements of your list: 30 40 25 50 30 20\nThe elements in the list: [30, 40, 25, 50, 30, 20]\n\nResult:\n\nMaximum sum without considering adjacent elements: '110'\n"
]
],
[
[
"## 5.3 Steps of the algorithm",
"_____no_output_____"
]
],
[
[
"def MaximumSumWithoutAdjacentSteps(List):\n # define two sums and necessary datastructures\n SumWithoutPrevious, SumWithPrevious, NotPickLast = 0, 0, False\n \n # To keep track the picked elements in each sum \n WithPrevious, WithoutPrevious = [], []\n \n # Showing the elements in the list\n print('The elements in the list: ', List)\n \n # Iterating over the elements\n for Index, i in enumerate(List):\n \n # To distinguish between the steps\n print('-' * 50)\n print('* Step '+str(Index+1)+' *\\n')\n \n # Explaining of the procedure\n print('We are considering number \\'', i, '\\':', sep = '')\n print('\\nCurrent value of SumWithPrevious:', SumWithPrevious)\n print('Current value of SumWithoutPrevious:', SumWithoutPrevious, end = '\\n\\n')\n \n # Add the current value to the sum without previous\n SumWithoutPrevious += i\n print('Adding value \\'', i, '\\' to SumWithoutPrevious -> SumWithoutPrevious = ', SumWithoutPrevious, sep = '', end = '\\n\\n')\n \n # Check the condition \n if SumWithoutPrevious > SumWithPrevious:\n \n # As we gain more score by picking the current element so we swap the values. \n SumWithoutPrevious, SumWithPrevious = SumWithPrevious, SumWithoutPrevious\n \n # If we didn't pick the previous element\n if NotPickLast:\n print('We didn\\'t pick the previous element!!!')\n \n # As we didn't pick the previous element, we add current value to with previous elements \n WithPrevious.append(i)\n \n # If we have considered the previous element\n else:\n \n # If this is not the first iteration \n if Index:\n print('Swap two lists!!!')\n \n # Now we add current element to sumwithout previous set and swap two lists\n WithoutPrevious.append(i)\n WithPrevious, WithoutPrevious = WithoutPrevious[:], WithPrevious[:]\n \n # Showing the values at the current iteration \n print('--->> Pick the current element!!!')\n print('\\nSumWithoutPrevious is greater than SumWithPrevious -> Swap two maximum!')\n print('SumWithPrevious = ', SumWithPrevious, ' SumWithoutPrevious = ', SumWithoutPrevious)\n \n # We removed the previous element -> we didn't pick the previous element \n NotPickLast = False\n \n # If by considering the current element, we don't gain more score \n else:\n\n # Up to this point we have the maximum score by not picking the previous element\n SumWithoutPrevious = SumWithPrevious\n \n WithoutPrevious = WithPrevious[:]\n \n # We are not picking the current element, so for next iteration we didn't pick the previous one.\n NotPickLast = True\n \n # Showing the details of what we did exactly\n print('\\n--->>Don\\'t pick the element!!!')\n print('\\nSumWithoutPrevious is not greater than SumWithPrevious -> Both take maximum!')\n print('SumWithPrevious = ', SumWithPrevious, ' SumWithoutPrevious = ', SumWithoutPrevious)\n \n # Showing the picked element up to this point. \n print('\\nMaximum sum with current element: ', WithPrevious)\n print('\\nMaximum sum without current element: ', WithoutPrevious)\n \n \n # Showing the last result. \n print('\\n***************')\n print('Result:\\n')\n print('All elements:', List)\n print('\\nMaximum sum without considering adjacent elements: \\'', SumWithPrevious, '\\'', sep = '')\n print('By picking these values: ', WithPrevious)",
"_____no_output_____"
]
],
[
[
"### Run the detailed function here.",
"_____no_output_____"
]
],
[
[
"MaximumSumWithoutAdjacentSteps(MyList)",
"The elements in the list: [30, 40, 25, 50, 30, 20]\n--------------------------------------------------\n* Step 1 *\n\nWe are considering number '30':\n\nCurrent value of SumWithPrevious: 0\nCurrent value of SumWithoutPrevious: 0\n\nAdding value '30' to SumWithoutPrevious -> SumWithoutPrevious = 30\n\n--->> Pick the current element!!!\n\nSumWithoutPrevious is greater than SumWithPrevious -> Swap two maximum!\nSumWithPrevious = 30 SumWithoutPrevious = 0\n\nMaximum sum with current element: [30]\n\nMaximum sum without current element: []\n--------------------------------------------------\n* Step 2 *\n\nWe are considering number '40':\n\nCurrent value of SumWithPrevious: 30\nCurrent value of SumWithoutPrevious: 0\n\nAdding value '40' to SumWithoutPrevious -> SumWithoutPrevious = 40\n\nSwap two lists!!!\n--->> Pick the current element!!!\n\nSumWithoutPrevious is greater than SumWithPrevious -> Swap two maximum!\nSumWithPrevious = 40 SumWithoutPrevious = 30\n\nMaximum sum with current element: [40]\n\nMaximum sum without current element: [30]\n--------------------------------------------------\n* Step 3 *\n\nWe are considering number '25':\n\nCurrent value of SumWithPrevious: 40\nCurrent value of SumWithoutPrevious: 30\n\nAdding value '25' to SumWithoutPrevious -> SumWithoutPrevious = 55\n\nSwap two lists!!!\n--->> Pick the current element!!!\n\nSumWithoutPrevious is greater than SumWithPrevious -> Swap two maximum!\nSumWithPrevious = 55 SumWithoutPrevious = 40\n\nMaximum sum with current element: [30, 25]\n\nMaximum sum without current element: [40]\n--------------------------------------------------\n* Step 4 *\n\nWe are considering number '50':\n\nCurrent value of SumWithPrevious: 55\nCurrent value of SumWithoutPrevious: 40\n\nAdding value '50' to SumWithoutPrevious -> SumWithoutPrevious = 90\n\nSwap two lists!!!\n--->> Pick the current element!!!\n\nSumWithoutPrevious is greater than SumWithPrevious -> Swap two maximum!\nSumWithPrevious = 90 SumWithoutPrevious = 55\n\nMaximum sum with current element: [40, 50]\n\nMaximum sum without current element: [30, 25]\n--------------------------------------------------\n* Step 5 *\n\nWe are considering number '30':\n\nCurrent value of SumWithPrevious: 90\nCurrent value of SumWithoutPrevious: 55\n\nAdding value '30' to SumWithoutPrevious -> SumWithoutPrevious = 85\n\n\n--->>Don't pick the element!!!\n\nSumWithoutPrevious is not greater than SumWithPrevious -> Both take maximum!\nSumWithPrevious = 90 SumWithoutPrevious = 90\n\nMaximum sum with current element: [40, 50]\n\nMaximum sum without current element: [40, 50]\n--------------------------------------------------\n* Step 6 *\n\nWe are considering number '20':\n\nCurrent value of SumWithPrevious: 90\nCurrent value of SumWithoutPrevious: 90\n\nAdding value '20' to SumWithoutPrevious -> SumWithoutPrevious = 110\n\nWe didn't pick the previous element!!!\n--->> Pick the current element!!!\n\nSumWithoutPrevious is greater than SumWithPrevious -> Swap two maximum!\nSumWithPrevious = 110 SumWithoutPrevious = 90\n\nMaximum sum with current element: [40, 50, 20]\n\nMaximum sum without current element: [40, 50]\n\n***************\nResult:\n\nAll elements: [30, 40, 25, 50, 30, 20]\n\nMaximum sum without considering adjacent elements: '110'\nBy picking these values: [40, 50, 20]\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
c501699e021be77824a0d317cf030beb4b17d7d1
| 11,500 |
ipynb
|
Jupyter Notebook
|
assignments/assignment1/softmax.ipynb
|
KimuraTian/stanford-cs231
|
8408c84dc3080408fca401f62f86035083e1d2f8
|
[
"MIT"
] | 282 |
2016-09-06T04:05:54.000Z
|
2022-01-03T05:39:28.000Z
|
assignments/assignment1/softmax.ipynb
|
kedar94/stanford-cs231
|
8408c84dc3080408fca401f62f86035083e1d2f8
|
[
"MIT"
] | 2 |
2017-01-28T17:13:25.000Z
|
2018-12-16T16:48:31.000Z
|
assignments/assignment1/softmax.ipynb
|
kedar94/stanford-cs231
|
8408c84dc3080408fca401f62f86035083e1d2f8
|
[
"MIT"
] | 138 |
2016-09-10T13:41:56.000Z
|
2021-12-19T11:23:06.000Z
| 37.337662 | 290 | 0.573652 |
[
[
[
"# Softmax exercise\n\n*Complete and hand in this completed worksheet (including its outputs and any supporting code outside of the worksheet) with your assignment submission. For more details see the [assignments page](http://vision.stanford.edu/teaching/cs231n/assignments.html) on the course website.*\n\nThis exercise is analogous to the SVM exercise. You will:\n\n- implement a fully-vectorized **loss function** for the Softmax classifier\n- implement the fully-vectorized expression for its **analytic gradient**\n- **check your implementation** with numerical gradient\n- use a validation set to **tune the learning rate and regularization** strength\n- **optimize** the loss function with **SGD**\n- **visualize** the final learned weights\n",
"_____no_output_____"
]
],
[
[
"import random\nimport numpy as np\nfrom cs231n.data_utils import load_CIFAR10\nimport matplotlib.pyplot as plt\n%matplotlib inline\nplt.rcParams['figure.figsize'] = (10.0, 8.0) # set default size of plots\nplt.rcParams['image.interpolation'] = 'nearest'\nplt.rcParams['image.cmap'] = 'gray'\n\n# for auto-reloading extenrnal modules\n# see http://stackoverflow.com/questions/1907993/autoreload-of-modules-in-ipython\n%load_ext autoreload\n%autoreload 2",
"_____no_output_____"
],
[
"def get_CIFAR10_data(num_training=49000, num_validation=1000, num_test=1000, num_dev=500):\n \"\"\"\n Load the CIFAR-10 dataset from disk and perform preprocessing to prepare\n it for the linear classifier. These are the same steps as we used for the\n SVM, but condensed to a single function. \n \"\"\"\n # Load the raw CIFAR-10 data\n cifar10_dir = 'cs231n/datasets/cifar-10-batches-py'\n X_train, y_train, X_test, y_test = load_CIFAR10(cifar10_dir)\n \n # subsample the data\n mask = range(num_training, num_training + num_validation)\n X_val = X_train[mask]\n y_val = y_train[mask]\n mask = range(num_training)\n X_train = X_train[mask]\n y_train = y_train[mask]\n mask = range(num_test)\n X_test = X_test[mask]\n y_test = y_test[mask]\n mask = np.random.choice(num_training, num_dev, replace=False)\n X_dev = X_train[mask]\n y_dev = y_train[mask]\n \n # Preprocessing: reshape the image data into rows\n X_train = np.reshape(X_train, (X_train.shape[0], -1))\n X_val = np.reshape(X_val, (X_val.shape[0], -1))\n X_test = np.reshape(X_test, (X_test.shape[0], -1))\n X_dev = np.reshape(X_dev, (X_dev.shape[0], -1))\n \n # Normalize the data: subtract the mean image\n mean_image = np.mean(X_train, axis = 0)\n X_train -= mean_image\n X_val -= mean_image\n X_test -= mean_image\n X_dev -= mean_image\n \n # add bias dimension and transform into columns\n X_train = np.hstack([X_train, np.ones((X_train.shape[0], 1))])\n X_val = np.hstack([X_val, np.ones((X_val.shape[0], 1))])\n X_test = np.hstack([X_test, np.ones((X_test.shape[0], 1))])\n X_dev = np.hstack([X_dev, np.ones((X_dev.shape[0], 1))])\n \n return X_train, y_train, X_val, y_val, X_test, y_test, X_dev, y_dev\n\n\n# Invoke the above function to get our data.\nX_train, y_train, X_val, y_val, X_test, y_test, X_dev, y_dev = get_CIFAR10_data()\nprint 'Train data shape: ', X_train.shape\nprint 'Train labels shape: ', y_train.shape\nprint 'Validation data shape: ', X_val.shape\nprint 'Validation labels shape: ', y_val.shape\nprint 'Test data shape: ', X_test.shape\nprint 'Test labels shape: ', y_test.shape\nprint 'dev data shape: ', X_dev.shape\nprint 'dev labels shape: ', y_dev.shape",
"_____no_output_____"
]
],
[
[
"## Softmax Classifier\n\nYour code for this section will all be written inside **cs231n/classifiers/softmax.py**. \n",
"_____no_output_____"
]
],
[
[
"# First implement the naive softmax loss function with nested loops.\n# Open the file cs231n/classifiers/softmax.py and implement the\n# softmax_loss_naive function.\n\nfrom cs231n.classifiers.softmax import softmax_loss_naive\nimport time\n\n# Generate a random softmax weight matrix and use it to compute the loss.\nW = np.random.randn(3073, 10) * 0.0001\nloss, grad = softmax_loss_naive(W, X_dev, y_dev, 0.0)\n\n# As a rough sanity check, our loss should be something close to -log(0.1).\nprint 'loss: %f' % loss\nprint 'sanity check: %f' % (-np.log(0.1))",
"_____no_output_____"
]
],
[
[
"## Inline Question 1:\nWhy do we expect our loss to be close to -log(0.1)? Explain briefly.**\n\n**Your answer:** *Fill this in*\n",
"_____no_output_____"
]
],
[
[
"# Complete the implementation of softmax_loss_naive and implement a (naive)\n# version of the gradient that uses nested loops.\nloss, grad = softmax_loss_naive(W, X_dev, y_dev, 0.0)\n\n# As we did for the SVM, use numeric gradient checking as a debugging tool.\n# The numeric gradient should be close to the analytic gradient.\nfrom cs231n.gradient_check import grad_check_sparse\nf = lambda w: softmax_loss_naive(w, X_dev, y_dev, 0.0)[0]\ngrad_numerical = grad_check_sparse(f, W, grad, 10)\n\n# similar to SVM case, do another gradient check with regularization\nloss, grad = softmax_loss_naive(W, X_dev, y_dev, 1e2)\nf = lambda w: softmax_loss_naive(w, X_dev, y_dev, 1e2)[0]\ngrad_numerical = grad_check_sparse(f, W, grad, 10)",
"_____no_output_____"
],
[
"# Now that we have a naive implementation of the softmax loss function and its gradient,\n# implement a vectorized version in softmax_loss_vectorized.\n# The two versions should compute the same results, but the vectorized version should be\n# much faster.\ntic = time.time()\nloss_naive, grad_naive = softmax_loss_naive(W, X_dev, y_dev, 0.00001)\ntoc = time.time()\nprint 'naive loss: %e computed in %fs' % (loss_naive, toc - tic)\n\nfrom cs231n.classifiers.softmax import softmax_loss_vectorized\ntic = time.time()\nloss_vectorized, grad_vectorized = softmax_loss_vectorized(W, X_dev, y_dev, 0.00001)\ntoc = time.time()\nprint 'vectorized loss: %e computed in %fs' % (loss_vectorized, toc - tic)\n\n# As we did for the SVM, we use the Frobenius norm to compare the two versions\n# of the gradient.\ngrad_difference = np.linalg.norm(grad_naive - grad_vectorized, ord='fro')\nprint 'Loss difference: %f' % np.abs(loss_naive - loss_vectorized)\nprint 'Gradient difference: %f' % grad_difference",
"_____no_output_____"
],
[
"# Use the validation set to tune hyperparameters (regularization strength and\n# learning rate). You should experiment with different ranges for the learning\n# rates and regularization strengths; if you are careful you should be able to\n# get a classification accuracy of over 0.35 on the validation set.\nfrom cs231n.classifiers import Softmax\nresults = {}\nbest_val = -1\nbest_softmax = None\nlearning_rates = [1e-7, 5e-7]\nregularization_strengths = [5e4, 1e8]\n\n################################################################################\n# TODO: #\n# Use the validation set to set the learning rate and regularization strength. #\n# This should be identical to the validation that you did for the SVM; save #\n# the best trained softmax classifer in best_softmax. #\n################################################################################\npass\n################################################################################\n# END OF YOUR CODE #\n################################################################################\n \n# Print out results.\nfor lr, reg in sorted(results):\n train_accuracy, val_accuracy = results[(lr, reg)]\n print 'lr %e reg %e train accuracy: %f val accuracy: %f' % (\n lr, reg, train_accuracy, val_accuracy)\n \nprint 'best validation accuracy achieved during cross-validation: %f' % best_val",
"_____no_output_____"
],
[
"# evaluate on test set\n# Evaluate the best softmax on test set\ny_test_pred = best_softmax.predict(X_test)\ntest_accuracy = np.mean(y_test == y_test_pred)\nprint 'softmax on raw pixels final test set accuracy: %f' % (test_accuracy, )",
"_____no_output_____"
],
[
"# Visualize the learned weights for each class\nw = best_softmax.W[:-1,:] # strip out the bias\nw = w.reshape(32, 32, 3, 10)\n\nw_min, w_max = np.min(w), np.max(w)\n\nclasses = ['plane', 'car', 'bird', 'cat', 'deer', 'dog', 'frog', 'horse', 'ship', 'truck']\nfor i in xrange(10):\n plt.subplot(2, 5, i + 1)\n \n # Rescale the weights to be between 0 and 255\n wimg = 255.0 * (w[:, :, :, i].squeeze() - w_min) / (w_max - w_min)\n plt.imshow(wimg.astype('uint8'))\n plt.axis('off')\n plt.title(classes[i])",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
c5017b0dcae6c92d0626d50a33dfea088d3bab86
| 9,760 |
ipynb
|
Jupyter Notebook
|
15-Music-the-magic-of-12/15.3a-Constructing-the-musical-scale.ipynb
|
misterhay/RabbitMath
|
8089e6cdf5ee0d70827b65c8be64619892b22b63
|
[
"MIT"
] | null | null | null |
15-Music-the-magic-of-12/15.3a-Constructing-the-musical-scale.ipynb
|
misterhay/RabbitMath
|
8089e6cdf5ee0d70827b65c8be64619892b22b63
|
[
"MIT"
] | null | null | null |
15-Music-the-magic-of-12/15.3a-Constructing-the-musical-scale.ipynb
|
misterhay/RabbitMath
|
8089e6cdf5ee0d70827b65c8be64619892b22b63
|
[
"MIT"
] | null | null | null | 41.355932 | 417 | 0.620594 |
[
[
[
"# Constructing the musical scale\n\nSo far we have discovered two important facts about how we hear and interpret musical notes. We will call them axioms because they are fundamental to our task of constructing the music scale.\n\n**Axiom 1** The ear compares different frequencies *multiplicatively* rather than *additively*\n\n**Axiom 2** Two notes of different frequency sound “good” together when the ratio of their frequencies can be expressed as a quotient of two small integers.\n\nOkay––let’s construct a musical scale. Now what do I mean by that? Recall that a note is a certain frequency of oscillation. There are an infinite number of possible frequencies available, so we need to pick out a certain finite subset and agree that these will be the notes to “use” (tune our instruments to) when we play music. But which ones shall we pick and what properties do we want the scale to have?\n\nThere are lots of ways to make a scale, in fact different solutions have been adopted by cultures througout history. The system we use now has dominated Western culture for 500 years, and that suggests that it might just have something going for it.\n\nIn this system we’ve grouped notes into octaves each with 12 notes, and the frequencies in each octave are twice those in the previous octave. In the table of frequencies generated by the code below, each row is an octave.",
"_____no_output_____"
]
],
[
[
"import pandas as pd\n\nnoteList = ['C', 'C#', 'D', 'D#', 'E', 'F', 'F#', 'G', 'G#', 'A', 'A#', 'B']\n\ndef generateOctave(note, startingFrequency):\n frequencyList = [startingFrequency]\n for x in range(1,9):\n frequencyList.append(startingFrequency * 2**x)\n frequencyDf = pd.DataFrame({note:frequencyList})\n return frequencyDf\n \ndf = pd.DataFrame() # create an empty dataframe\nfor n, note in enumerate(noteList, start=1):\n startingFrequency = 2**(n/12) * 15.434 # calculate the new note's frequency\n frequencyDf = generateOctave(note, startingFrequency)\n df = pd.concat([df, frequencyDf], axis=1) # join the new column to the dataframe\n\ndf.style.set_precision(4) # display the dataframe with 4 significant figures",
"_____no_output_____"
]
],
[
[
"What patterns do you see in the frequency table?\n\nOut of all the possible alternatives for designing a scale, why did we choose to base it on 12? As a hint, it has something to do with factors and simple ratios.",
"_____no_output_____"
],
[
"We can reword the axioms above as:\n\n1. *Homogeneity* - No matter where we are in the scale, adjacent notes should have the same frequency ratio.\n\n2. *Nice combinations* - The scale should have lots of pairs of notes with frequencies in the ratio of small integers.\n\nLooking at the second axiom, the simplest ratio of integers is 1:2. If we play a note and then a note with double the frequency, they sound like different versions of the same note. So we give them the same letter name, for example $A_4$ is 440 Hz and $A_5$ is 880 Hz. We say these two notes are an [octave](https://en.wikipedia.org/wiki/Octave) apart.",
"_____no_output_____"
]
],
[
[
"from numpy import linspace, pi, sin\nfrom IPython.display import Audio, display\nimport time\n\nf1 = 440\nf2 = 880\n\nsampleRate = 32000\nduration = 0.5\nt = linspace(0, duration, int(sampleRate * duration))\n\ndisplay(Audio(sin(f1 * 2 * pi * t), rate = sampleRate, autoplay = True))\ntime.sleep(1)\ndisplay(Audio(sin(f2 * 2 * pi * t), rate = sampleRate, autoplay = True))",
"_____no_output_____"
]
],
[
[
"The question is, how many notes (frequencies) should we include in an octave? We know from axiom 1 that the frequencies of successive notes should be equally spaced multiplicatively. We also know from axiom 2 that we should get a note that is twice the frequency of the first.\n\nThis means that we must choose some ratio of frequencies, $r$, to an integer exponent, $n$, that results in 2. We can write that as $r^n=2$ where $n$ will be the number of notes in an octave.",
"_____no_output_____"
],
[
"The equation $r^n=2$ contains two unknowns though. Rather than trying to find a value for $r$, it will be easier to find a value for $n$ because we know it is an integer.",
"_____no_output_____"
],
[
"## The search for $n$\n\nSo let’s look for $n$, the number of notes in an octave. This is not so hard to do as we know that it has to be an integer and it can’t be too small or too large.\n\nIf it were small, like around 5, we wouldn’t have enough notes in an octave to give us interesting music, and if it were too large, like 20, we might well have more notes than we could cope with, both conceptually and mechanically.\n\nWe will judge $n$ to be \"good\" if we get lots of small integer ratios.",
"_____no_output_____"
],
[
"For example, if we take $n=10$ then the 10 notes in an octave will be: $r^0=1$, $r^1$, $r^2$... $r^{10}=2$.\n\nActually there are 11 notes in that list, but we usually take the last one, $r^n=2$, to be the start of the next octave.\n\nSince we know that $r^{10}=2$, then $r=2^{1/10} \\approx 1.0718$ means that the first ratio of notes is about 1:1.0718.",
"_____no_output_____"
],
[
"Each successive note will be equally spaced multiplicatively, so:\n\n$r^0 = (2^{1/10})^0 = 1$\n\n$r^1 = (2^{1/10})^1 \\approx 1.0718$\n\n...\n\n$r^10 = (2^{1/10})^{10} = 2$\n\nWhich means in general $r^i = (2^{1/n})^i = 2^{i/n}$\n\nNow that we know $r^i = 2^{i/n}$, let's try some different values for $n$:",
"_____no_output_____"
]
],
[
[
"valuesToCheck = range(4, 17) # we'll check values from 4 to 16\n\ndef generateFrequencyList(n):\n frequencyList = [1.0] # start a new list with 1.0\n for i in range(1,n+1): # iterate integers from 1 to n\n f = 2**(i/n) # calculate the frequency value\n frequencyList.append(f)\n return frequencyList\n\ndf = pd.DataFrame() # create an empty dataframe\nfor n in valuesToCheck:\n frequencyList = generateFrequencyList(n)\n frequencyDf = pd.DataFrame({'n = '+str(n): frequencyList}) # make a dataframe from the list\n df = pd.concat([df, frequencyDf], axis=1) # join the new dataframe to our existing one\n\ndf.fillna(value='', inplace=True) # replace all of the NaN values with blanks\npd.options.display.float_format = '{:,.3f}'.format # display values to 3 decimals\ndf # display the dataframe",
"_____no_output_____"
]
],
[
[
"That table shows us the decimal representations of the frequencies in an octave with $n$ equal to the number of notes.\n\nSo what makes on $n$ better than another? Axiom 2 tells us that we want lots of small integer ratios in our pairs of notes. That means that whatever colun we select shoul have a good supply of fractions like $\\frac{3}{2}$, $\\frac{4}{3}$, $\\frac{5}{3}$, $\\frac{5}{4}$, $\\frac{7}{4}$, etc.",
"_____no_output_____"
],
[
"First the bad news, the numbers $2^{i/n}$ will always be irrational and will *never* contain integer ratios (small or otherwise). This is a fascinating set of ideas in itself, and is pursued in problem 6.",
"_____no_output_____"
],
[
"But now for the good news, the ear can’t actually distinguish tiny variations in frequency so it’s good enough to be very close to lots of small integer ratios. And from the JND table of the last section, “very close” in the case of frequency discrimination means around half a percent. So let’s re-reformulate axiom 2:\n2. *Nice combinations* - The scale should have lots of pairs of notes with frequencies within half a percent of small-integer ratios.",
"_____no_output_____"
],
[
"Go on to [15.3b-Looking-for-a-good-ratio.ipynb](./15.3b-Looking-for-a-good-ratio.ipynb)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
c5017c3a10b24103a631743df436e3cddc0a1072
| 4,676 |
ipynb
|
Jupyter Notebook
|
examples/rsqsim_api/specfem_inputs/find_specfem_input_info.ipynb
|
uc-eqgeo/rsqsim-python-tools
|
35d65629809b7edc10053a464c212ea03616c8df
|
[
"MIT"
] | 1 |
2022-03-20T12:02:02.000Z
|
2022-03-20T12:02:02.000Z
|
examples/rsqsim_api/specfem_inputs/find_specfem_input_info.ipynb
|
uc-eqgeo/rsqsim-python-tools
|
35d65629809b7edc10053a464c212ea03616c8df
|
[
"MIT"
] | null | null | null |
examples/rsqsim_api/specfem_inputs/find_specfem_input_info.ipynb
|
uc-eqgeo/rsqsim-python-tools
|
35d65629809b7edc10053a464c212ea03616c8df
|
[
"MIT"
] | 1 |
2021-12-12T19:15:49.000Z
|
2021-12-12T19:15:49.000Z
| 23.979487 | 131 | 0.545979 |
[
[
[
"Find the info needed to run specfem for each event in catalogue:\n* average strike\n* average dip\n* average rake\n* fault coordinates\n* mo",
"_____no_output_____"
]
],
[
[
"# import modules\nimport math\nimport numpy as np\nimport pandas as pd\n\nfrom rsqsim_api.catalogue.catalogue import RsqSimCatalogue\nfrom rsqsim_api.fault.multifault import RsqSimMultiFault\nimport os\nfrom shapely.geometry.polygon import Polygon\nimport geopandas as gpd",
"_____no_output_____"
]
],
[
[
"Read in catalogue and fault model",
"_____no_output_____"
]
],
[
[
"script_dir = os.path.abspath('')\nfault_dir = \"../../../data/shaw2021/rundir5091\"\ncatalogue_dir = os.path.join(fault_dir,\"specfem\")\n\nfault_model = RsqSimMultiFault.read_fault_file_bruce(os.path.join(script_dir, fault_dir, \"zfault_Deepen.in\"),\n os.path.join(script_dir, fault_dir, \"znames_Deepen.in\"),\n transform_from_utm=True)\nwhole_catalogue = RsqSimCatalogue.from_csv_and_arrays(os.path.join(catalogue_dir, \"single_fault_200yr\"))",
"_____no_output_____"
],
[
"#get specific event of interest: 448545\nev_number=448545\nev_cat=whole_catalogue.filter_by_events(event_number=ev_number)\nEOI=ev_cat.first_event(fault_model)\nEOI_fault=EOI.faults[0]",
"_____no_output_____"
],
[
"#write out parameters of interest\nout_name=f'event_{ev_number}.txt'\nwith open(os.path.join(script_dir,out_name),'w') as f_out:\n f_out.write(f'#Evid mean_strike mean_dip mean_rake mean_slip(m) Mo\\n')\n f_out.write(f'{ev_number} {EOI.mean_strike:.1f} {EOI.mean_dip:.1f} {EOI.mean_rake:.1f} {EOI.mean_slip:.1f} {EOI.m0:.2e}')",
"_____no_output_____"
],
[
"#write out trace\nEOItrace=gpd.GeoSeries(EOI_fault.trace,crs=\"EPSG:2193\")\n#convert to lat/ lon\nEOItrace_ll=EOItrace.to_crs(\"EPSG:4326\")\n",
"_____no_output_____"
],
[
"EOI_outline=gpd.GeoSeries(EOI_fault.fault_outline,crs=\"EPSG:2193\")\nEOI_outline_ll=EOI_outline.to_crs(\"EPSG:4326\").iloc[0]\nas_xyz=np.reshape(EOI_outline_ll.exterior.coords[:],(len(EOI_outline_ll.exterior.coords[:]),3))\nEOI_pandas=pd.DataFrame(as_xyz)\nEOI_pandas.to_csv(f'event_{ev_number}_fault_outline.csv',header=False,index=False)",
"_____no_output_____"
],
[
"EOI.slip_dist_to_txt(os.path.join(script_dir,f'event_{ev_number}_slip.txt'),nztm_to_lonlat=True)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
c5017cd45a6db238d16ae2a3b2841c7917beae61
| 259,088 |
ipynb
|
Jupyter Notebook
|
sphinx/datascience/source/autograd-poisson-regression-gradient-descent.ipynb
|
oneoffcoder/books
|
84619477294a3e37e0d7538adf819113c9e8dcb8
|
[
"CC-BY-4.0"
] | 26 |
2020-05-05T08:07:43.000Z
|
2022-02-12T03:28:15.000Z
|
sphinx/datascience/source/autograd-poisson-regression-gradient-descent.ipynb
|
oneoffcoder/books
|
84619477294a3e37e0d7538adf819113c9e8dcb8
|
[
"CC-BY-4.0"
] | 19 |
2021-03-10T00:33:51.000Z
|
2022-03-02T13:04:32.000Z
|
sphinx/datascience/source/autograd-poisson-regression-gradient-descent.ipynb
|
oneoffcoder/books
|
84619477294a3e37e0d7538adf819113c9e8dcb8
|
[
"CC-BY-4.0"
] | 2 |
2022-01-09T16:48:21.000Z
|
2022-02-19T17:06:50.000Z
| 627.331719 | 106,188 | 0.947149 |
[
[
[
"# Poisson Regression, Gradient Descent\n\nIn this notebook, we will show how to use gradient descent to solve a [Poisson regression model](https://en.wikipedia.org/wiki/Poisson_regression). A Poisson regression model takes on the following form.\n\n$\\operatorname{E}(Y\\mid\\mathbf{x})=e^{\\boldsymbol{\\theta}' \\mathbf{x}}$\n\nwhere\n\n* $x$ is a vector of input values\n* $\\theta$ is a vector weights (the coefficients)\n* $y$ is the expected value of the parameter for a [Poisson distribution](https://en.wikipedia.org/wiki/Poisson_distribution), typically, denoted as $\\lambda$\n\nNote that [Scikit-Learn](http://scikit-learn.org/stable/modules/classes.html#module-sklearn.linear_model) does not provide a solver a Poisson regression model, but [statsmodels](http://www.statsmodels.org/dev/generated/statsmodels.discrete.discrete_model.Poisson.html) does, though examples for the latter is [thin](https://datascience.stackexchange.com/questions/23143/poisson-regression-options-in-python).",
"_____no_output_____"
],
[
"## Simulate data\n\nNow, let's simulate the data. Note that the coefficients are $[1, 0.5, 0.2]$ and that there is error $\\epsilon \\sim \\mathcal{N}(0, 1)$ added to the simulated data.\n\n$y=e^{1 + 0.5x_1 + 0.2x_2 + \\epsilon}$\n\nIn this notebook, the score is denoted as $z$ and $z = 1 + 0.5x_1 + 0.2x_2 + \\epsilon$. Additionally, $y$ is the mean for a Poisson distribution. The variables $X_1$ and $X_2$ are independently sampled from their own normal distribution $\\mathcal{N}(0, 1)$.\n\nAfter we simulate the data, we will plot the distribution of the scores and means. Note that the expected value of the output $y$ is 5.2.",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nfrom numpy.random import normal\nfrom scipy.stats import poisson\n\nnp.random.seed(37)\nsns.set(color_codes=True)\n\nn = 10000\nX = np.hstack([\n np.array([1 for _ in range(n)]).reshape(n, 1), \n normal(0.0, 1.0, n).reshape(n, 1), \n normal(0.0, 1.0, n).reshape(n, 1)\n])\nz = np.dot(X, np.array([1.0, 0.5, 0.2])) + normal(0.0, 1.0, n)\ny = np.exp(z)",
"_____no_output_____"
]
],
[
[
"## Visualize data",
"_____no_output_____"
]
],
[
[
"fig, ax = plt.subplots(1, 2, figsize=(20, 5))\n\nsns.kdeplot(z, ax=ax[0])\nax[0].set_title(r'Distribution of Scores')\nax[0].set_xlabel('score')\nax[0].set_ylabel('probability')\n\nsns.kdeplot(y, ax=ax[1])\nax[1].set_title(r'Distribution of Means')\nax[1].set_xlabel('mean')\nax[1].set_ylabel('probability')",
"_____no_output_____"
]
],
[
[
"## Solve for the Poisson regression model weights\n\nNow we learn the weights of the Poisson regression model using gradient descent. Notice that the loss function of a Poisson regression model is identical to an Ordinary Least Square (OLS) regression model?\n\n$L(\\theta) = \\frac{1}{n} (\\hat{y} - y)^2$\n\nWe do not have to worry about writing out the gradient of the loss function since we are using [Autograd](https://github.com/HIPS/autograd).",
"_____no_output_____"
]
],
[
[
"import autograd.numpy as np\nfrom autograd import grad\nfrom autograd.numpy import exp, log, sqrt\n\n# define the loss function\ndef loss(w, X, y):\n y_pred = np.exp(np.dot(X, w))\n loss = ((y_pred - y) ** 2.0)\n return loss.mean(axis=None)\n\n#the magic line that gives you the gradient of the loss function\nloss_grad = grad(loss)\n\ndef learn_weights(X, y, alpha=0.05, max_iter=30000, debug=False):\n w = np.array([0.0 for _ in range(X.shape[1])])\n \n if debug is True:\n print('initial weights = {}'.format(w))\n\n loss_trace = []\n weight_trace = []\n\n for i in range(max_iter):\n loss = loss_grad(w, X, y)\n w = w - (loss * alpha)\n if i % 2000 == 0 and debug is True:\n print('{}: loss = {}, weights = {}'.format(i, loss, w))\n\n loss_trace.append(loss)\n weight_trace.append(w)\n\n if debug is True:\n print('intercept + weights: {}'.format(w))\n \n loss_trace = np.array(loss_trace)\n weight_trace = np.array(weight_trace)\n \n return w, loss_trace, weight_trace\n\ndef plot_traces(w, loss_trace, weight_trace, alpha):\n fig, ax = plt.subplots(1, 2, figsize=(20, 5))\n\n ax[0].set_title(r'Log-loss of the weights over iterations, $\\alpha=${}'.format(alpha))\n ax[0].set_xlabel('iteration')\n ax[0].set_ylabel('log-loss')\n ax[0].plot(loss_trace[:, 0], label=r'$\\beta$')\n ax[0].plot(loss_trace[:, 1], label=r'$x_0$')\n ax[0].plot(loss_trace[:, 2], label=r'$x_1$')\n ax[0].legend()\n\n ax[1].set_title(r'Weight learning over iterations, $\\alpha=${}'.format(alpha))\n ax[1].set_xlabel('iteration')\n ax[1].set_ylabel('weight')\n ax[1].plot(weight_trace[:, 0], label=r'$\\beta={:.2f}$'.format(w[0]))\n ax[1].plot(weight_trace[:, 1], label=r'$x_0={:.2f}$'.format(w[1]))\n ax[1].plot(weight_trace[:, 2], label=r'$x_1={:.2f}$'.format(w[2]))\n ax[1].legend()",
"_____no_output_____"
]
],
[
[
"We try learning the coefficients with different learning weights $\\alpha$. Note the behavior of the traces of the loss and weights for different $\\alpha$? The loss function was the same one used for OLS regression, but the loss function for Poisson regression is defined differently. Nevertheless, we still get acceptable results.",
"_____no_output_____"
],
[
"### Use gradient descent with $\\alpha=0.001$",
"_____no_output_____"
]
],
[
[
"alpha = 0.001\nw, loss_trace, weight_trace = learn_weights(X, y, alpha=alpha, max_iter=1000)\nplot_traces(w, loss_trace, weight_trace, alpha=alpha)\nprint(w)",
"[1.50066529 0.49134304 0.20836951]\n"
]
],
[
[
"### Use gradient descent with $\\alpha=0.005$",
"_____no_output_____"
]
],
[
[
"alpha = 0.005\nw, loss_trace, weight_trace = learn_weights(X, y, alpha=alpha, max_iter=200)\nplot_traces(w, loss_trace, weight_trace, alpha=alpha)\nprint(w)",
"[1.50066529 0.49134304 0.20836951]\n"
]
],
[
[
"### Use gradient descent with $\\alpha=0.01$",
"_____no_output_____"
]
],
[
[
"alpha = 0.01\nw, loss_trace, weight_trace = learn_weights(X, y, alpha=alpha, max_iter=200)\nplot_traces(w, loss_trace, weight_trace, alpha=alpha)\nprint(w)",
"[1.50393889 0.49616052 0.21077159]\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
c5017daf3b98549ba9f4b11ef38975b99b91ae18
| 13,281 |
ipynb
|
Jupyter Notebook
|
jupyter_notebooks/Simulator.ipynb
|
wdr-data/unwetter2
|
72240b0b29e65e660dbf8f47fb79c31c90e990fa
|
[
"MIT"
] | 8 |
2018-10-09T12:36:30.000Z
|
2021-08-18T12:06:48.000Z
|
jupyter_notebooks/Simulator.ipynb
|
wdr-data/unwetter2
|
72240b0b29e65e660dbf8f47fb79c31c90e990fa
|
[
"MIT"
] | 30 |
2018-09-22T14:35:06.000Z
|
2022-02-28T04:20:11.000Z
|
jupyter_notebooks/Simulator.ipynb
|
wdr-data/unwetter2
|
72240b0b29e65e660dbf8f47fb79c31c90e990fa
|
[
"MIT"
] | 1 |
2020-06-09T19:53:59.000Z
|
2020-06-09T19:53:59.000Z
| 31.471564 | 174 | 0.495896 |
[
[
[
"# Unwetter Simulator",
"_____no_output_____"
]
],
[
[
"import os\nos.chdir('..')\n\n\nf'Working directory: {os.getcwd()}'",
"_____no_output_____"
],
[
"from unwetter import db, map\nfrom datetime import datetime",
"_____no_output_____"
],
[
"from unwetter import config\n\nconfig.SEVERITY_FILTER = ['Severe', 'Extreme']\nconfig.STATES_FILTER = ['NW']\nconfig.URGENCY_FILTER = ['Immediate']\n\nseverities = {\n 'Minor': 'Wetterwarnung',\n 'Moderate': 'Markante Wetterwarnung',\n 'Severe': '🔴 Amtliche Unwetterwarnung',\n 'Extreme': '🔴 Amtliche Extreme Unwetterwarnung',\n}",
"_____no_output_____"
],
[
"search_start = datetime(2019, 6, 19, 11, 0)\nsearch_end = datetime(2019, 6, 19, 22, 0)\n\nsearch_filter = {\n '$and': [\n {\n 'sent': {\n '$gt': search_start,\n },\n },\n {\n 'sent': {\n '$lt': search_end,\n },\n },\n ]\n}\n\nevents = list(db.collection.find(search_filter).sort([('sent', 1)]))",
"_____no_output_____"
],
[
"len(events)",
"_____no_output_____"
],
[
"len([e for e in events if e['published']])",
"_____no_output_____"
],
[
"for e in events:\n e['published'] = False",
"_____no_output_____"
],
[
"def mock_by_ids(ids):\n return [event for event in events if event['id'] in ids]\n\ndef mock_publish(ids):\n for event in events:\n if event['id'] in ids:\n event['published'] = True",
"_____no_output_____"
],
[
"from unwetter.dwd import special_type\n\ndef mock_has_changes(event, old_events):\n\n if not any(t['published'] for t in old_events):\n extended_references = set()\n extended_references.update(event.get('extended_references', event['references']))\n \n for old_event in old_events:\n if 'extended_references' in old_event:\n extended_references.update(old_event['extended_references'])\n elif 'references' in old_event:\n extended_references.update(old_event['references'])\n \n event['extended_references'] = sorted(extended_references, reverse=True)\n \n old_events = mock_by_ids(extended_references)\n \n event['has_changes'] = [\n {\n 'id': old_event['id'],\n 'changed': mock_changes(event, old_event),\n 'published': old_event['published'],\n }\n for old_event in old_events\n ]\n\n event['special_type'] = special_type(event, old_events)\n return event",
"_____no_output_____"
],
[
"from datetime import datetime, timedelta\nfrom unwetter.generate.blocks import expires, district_list, state_for_cell_id, region_list, dates\nfrom unwetter.generate.helpers import upper_first, local_time\n\nSTATES_FILTER = config.STATES_FILTER\n\n\ndef mock_changes_old(event, old_event):\n \"\"\"\n Generate a list of changes between two events\n :param event:\n :param old_event:\n :return: str\n \"\"\"\n text = ''\n\n simple_fields = {\n 'severity': 'Warnstufe',\n 'event': 'Wetterphänomen',\n 'certainty': 'Wahrscheinlichkeit',\n }\n\n for field in simple_fields:\n if old_event.get(field) != event.get(field):\n if field == 'severity' and event[field] in ['Minor', 'Moderate']:\n text += f'{simple_fields[field]}: Herabstufung auf {severities[event[field]]}\\n\\n'\n elif field == 'severity':\n text += f'{simple_fields[field]}: {severities[event[field]]} ' \\\n f'(zuvor \"{severities[old_event[field]]}\")\\n\\n'\n else:\n text += f'{simple_fields[field]}: {event[field]} ' \\\n f'(zuvor \"{old_event.get(field, \"Nicht angegeben\")}\")\\n\\n'\n\n # Editorial request to check only, if expires time changed, since every update has new onset-time\n if abs(event['onset'] - event['sent']) > timedelta(minutes=2) and dates(old_event) != dates(event):\n text += f'Gültigkeit: {dates(event)} (zuvor \"{dates(old_event)}\")\\n\\n'\n elif expires(old_event) != expires(event):\n text += f'Ende der Gültigkeit: {expires(event)} (zuvor \"{expires(old_event)}\")\\n\\n'\n\n if district_list(old_event) != district_list(event):\n districts_now = {\n district['name'] for district in event['districts']\n if state_for_cell_id(district['warn_cell_id']) in STATES_FILTER\n }\n districts_before = {\n district['name'] for district in old_event['districts']\n if state_for_cell_id(district['warn_cell_id']) in STATES_FILTER\n }\n\n added = districts_now - districts_before\n removed = districts_before - districts_now\n\n if added:\n text += f'Neue Kreise/Städte: {\", \".join(sorted(added))}\\n'\n\n if removed:\n text += f'Nicht mehr betroffene Kreise/Städte: {\", \".join(sorted(removed))}\\n'\n\n if region_list(old_event) != region_list(event):\n text += f'Regionale Zuordnung: {upper_first(region_list(event))} ' \\\n f'(zuvor: \"{upper_first(region_list(old_event))}\")\\n\\n'\n else:\n text += f'Regionale Zuordnung unverändert: {upper_first(region_list(event))}\\n\\n'\n\n '''\n # Editorial choice --> No relevant information due to relatively small area --> Thus, no update\n\n elif commune_list(old_event) != commune_list(event):\n text += 'Regionale Zuordnung: Änderung der betroffenen Gemeinden\\n\\n'\n '''\n \n return text",
"_____no_output_____"
],
[
"from datetime import datetime, timedelta\nimport re\n\nSTATES_FILTER = config.STATES_FILTER\n\n\ndef mock_changes(event, old_event):\n \"\"\"\n Generate a list of changes between two events\n :param event:\n :param old_event:\n :return: bool\n \"\"\"\n \n if any(event.get(field) != old_event.get(field) for field in ['severity', 'certainty']):\n return True\n \n # Notify about big hail sizes\n if 'Hagel' not in event['parameters']:\n if event['event'] != old_event['event'].replace(' und HAGEL', ''):\n return True\n else:\n hail_re = r'^.*?(\\d+).*?cm'\n hail_size_now = int(re.match(hail_re, event['parameters']['Hagel']).group(1))\n hail_size_before = int(re.match(hail_re, old_event['parameters'].get('Hagel', '0 cm')).group(1))\n \n if hail_size_now >= 3 and hail_size_before < 3:\n return True\n else:\n if event['event'].replace(' und HAGEL', '') != old_event['event'].replace(' und HAGEL', ''):\n return True\n \n if abs(event['onset'] - event['sent']) > timedelta(minutes=2) and event['sent'] - event['onset'] < timedelta(minutes=2) and old_event['onset'] != event['onset']:\n return True\n elif old_event['expires'] != event['expires']:\n return True\n \n if len(set(r[0] for r in event['regions']) - set(r[0] for r in old_event['regions'])) > 0:\n return True\n\n districts_now = {\n district['name'] for district in event['districts']\n if state_for_cell_id(district['warn_cell_id']) in STATES_FILTER\n }\n districts_before = {\n district['name'] for district in old_event['districts']\n if state_for_cell_id(district['warn_cell_id']) in STATES_FILTER\n }\n added = districts_now - districts_before\n \n if len(districts_before) <= 3 and added:\n return True\n \n return False",
"_____no_output_____"
],
[
"from unwetter.config import filter_event\n\ndef mock_update(new_events):\n\n filtered = []\n for event in new_events:\n if filter_event(event):\n if event['msg_type'] in ['Alert', 'Cancel']:\n filtered.append(event)\n\n elif any(t['changed'] and t['published'] for t in event['has_changes']):\n filtered.append(event)\n \n elif event['special_type'] == 'UpdateAlert':\n filtered.append(event)\n\n elif not any(t['changed'] and t['published'] for t in event['has_changes']):\n continue\n\n else:\n print(f'Event was not filtered 1: {event[\"id\"]}')\n\n mock_publish([event['id'] for event in filtered])\n return filtered",
"_____no_output_____"
],
[
"from unwetter.generate.blocks import changes\n\ncurrent_sent = events[0]['sent']\n\nbins = []\ncurrent_bin = []\n\nfor event in events:\n if event['sent'] != current_sent:\n current_sent = event['sent']\n bins.append(current_bin)\n current_bin = []\n \n current_bin.append(event)\n\nbins.append(current_bin)",
"_____no_output_____"
],
[
"processed = []\n\nfor bin in bins:\n for event in bin:\n if 'references' in event:\n old_events = mock_by_ids(event.get('extended_references', event['references']))\n mock_has_changes(event, old_events)\n \n processed.append(mock_update(bin))",
"_____no_output_____"
],
[
"sum(len(bin) for bin in processed)",
"_____no_output_____"
],
[
"[print(event['event'], event['sent'] + timedelta(hours=2), event.get('special_type'), event['id']) for bin in processed for event in bin]",
"_____no_output_____"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
c5018474a6274d6ea9b222f3e02cc4f3b757f2e1
| 130,806 |
ipynb
|
Jupyter Notebook
|
explainers/notebooks/mli-byor.ipynb
|
dolcos/driverlessai-recipes
|
e1eb2787daeaae387d80ce24d4d3398c4fddb585
|
[
"Apache-2.0"
] | 194 |
2019-04-23T10:25:13.000Z
|
2022-03-29T04:19:28.000Z
|
explainers/notebooks/mli-byor.ipynb
|
dolcos/driverlessai-recipes
|
e1eb2787daeaae387d80ce24d4d3398c4fddb585
|
[
"Apache-2.0"
] | 50 |
2019-06-24T20:17:51.000Z
|
2022-03-16T20:05:37.000Z
|
explainers/notebooks/mli-byor.ipynb
|
dolcos/driverlessai-recipes
|
e1eb2787daeaae387d80ce24d4d3398c4fddb585
|
[
"Apache-2.0"
] | 85 |
2019-03-27T12:26:43.000Z
|
2022-01-27T13:15:37.000Z
| 48.339246 | 2,457 | 0.492913 |
[
[
[
"# MLI BYOR: Custom Explainers\nThis notebook is a demo of MLI **bring your own explainer recipe** (BYOR) Python API.\n\n**Ad-hoc OOTB and/or custom explainer run** scenario:\n* **Upload** interpretation recipe.\n * Determine recipe upload job **status**.\n* **Run** ad-hoc recipe run job.\n * Determine ad-hoc recipe job **status**.\n* **Get** explainer type for given job.\n* **List** explanation types for given explainer run.\n* **List** explanation representations (formats) URLs for given explanation type.\n* **Download** explanation representation from ^ URL. \n* **Download** interpretation recipe job result **data**.\n\n**Interpretation explainers run** scenario:\n* **List** available/compatible explainers.\n* **Choose** subset or all ^ compatible explainers.\n* **Run** interpretation job.\n * Determine **status** of interpretation job.\n * Determine **status** per explainer job (running within the scope of interpretation).\n* **List** explainer types which were ran within interpretation.\n* **List** explainer runs for given explainer type within the interpretation.\n* **List** explanation types for given explainer run.\n* **List** explanation representations (formats) URLs for given explanation type.\n* **Download** explanation representation from ^ URL.",
"_____no_output_____"
],
[
"### Virtual Environment and Dependencies\nPrepare and activate virtual environment for this notebook by running:\n```\n. .env/bin/activate\npip install ipykernel\nipython kernel install --user --name=dai\n```",
"_____no_output_____"
]
],
[
[
"import os\nimport pprint\nimport time\nfrom random import randint\nfrom h2oaicore.mli.oss.commons import MimeType, MliJobStatus\nfrom h2oaicore.messages import (\n CommonDaiExplainerParameters,\n CommonExplainerParameters,\n DatasetReference,\n Explainer,\n ExplainerDescriptor,\n ExplainerJobStatus,\n ExplainerRunJob,\n ExplainersRunJob,\n InterpretationJob,\n ModelReference,\n)\n\nimport h2o\nimport h2oai_client\nfrom h2oai_client import Client",
"_____no_output_____"
]
],
[
[
"**Connect** to DAI server:",
"_____no_output_____"
]
],
[
[
"# connect to Driverless AI server - make sure to use the same \n# user name and password as when signing in through the GUI\nhostname = '127.0.0.1'\naddress = 'http://' + hostname + ':12345'\nusername = 'h2oai'\npassword = 'h2oai'\n\n# h2oai = Client(\"http://localhost:12345\", \"h2oai\", \"h2oai\")\nh2oai = Client(address = address, username = username, password = password)",
"_____no_output_____"
]
],
[
[
"# Upload \nUpload BYOR interpretation [recipe](http://192.168.59.141/mli-byor/mli_byor_foo.py) to Driverless AI server.",
"_____no_output_____"
]
],
[
[
"# URL of the recipe to upload\n\n# Custom Morris sensitivity analysis\nURL_BYOR_EXPLAINER = \"https://h2o-public-test-data.s3.amazonaws.com/recipes/explainers/morris_sensitivity_explainer.py\"\nBYOR_EXPLAINER_NAME = \"Morris Sensitivity Analysis\"",
"_____no_output_____"
]
],
[
[
"**Upload recipe** to DAI server:",
"_____no_output_____"
]
],
[
[
"recipe_job_key = h2oai.create_custom_recipe_from_url(URL_BYOR_EXPLAINER)\nrecipe_job_key",
"_____no_output_____"
],
[
"recipe_job = h2oai._wait_for_recipe_load(recipe_job_key)\npprint.pprint(recipe_job.dump())",
"{'created': 1601291428.8856423,\n 'entity': {'data_file': '',\n 'data_files': [],\n 'datas': [],\n 'explainers': [],\n 'fpath': None,\n 'key': '3782c148-017b-11eb-a66d-207918bc8e4b',\n 'models': [],\n 'name': 'morris_sensitivity_explainer',\n 'pretransformers': [],\n 'scorers': [],\n 'transformers': [],\n 'type': '',\n 'url': 'https://h2o-public-test-data.s3.amazonaws.com/recipes/explainers/morris_sensitivity_explainer.py'},\n 'error': '',\n 'message': 'MorrisSensitivityLeExplainer:\\nInstalling Class Packages',\n 'progress': 1,\n 'status': 0}\n"
],
[
"if recipe_job.entity.explainers:\n uploaded_explainer_id:str = recipe_job.entity.explainers[0].id \nelse:\n # explainer already deployed (look it up)\n explainers = h2oai.list_explainers(\n experiment_types=None, \n explanation_scopes=None,\n dai_model_key=None,\n keywords=None,\n explainer_filter=[]\n )\n uploaded_explainer_id = [explainer.id for explainer in explainers if BYOR_EXPLAINER_NAME == explainer.name][0]\n\nprint(f\"Uploaded recipe ID: {uploaded_explainer_id}'\")",
"Uploaded recipe ID: False_morris_sensitivity_explainer_d935205d_contentexplainer.MorrisSensitivityLeExplainer'\n"
]
],
[
[
"Driverless AI **model** and **dataset**:",
"_____no_output_____"
]
],
[
[
"# *) hardcoded DAI keys\n# DATASET_KEY = \"f12f69b4-475b-11ea-bf67-9cb6d06b189b\"\n# MODEL_KEY = \"f268e364-475b-11ea-bf67-9cb6d06b189b\"\n\n# *) lookup compatible DAI keys\ncompatible_models = h2oai.list_explainable_models(\n explainer_id=uploaded_explainer_id, offset=0, size=30\n)\nif compatible_models.models:\n MODEL_KEY = compatible_models.models[0].key\n DATASET_KEY = compatible_models.models[0].parameters.dataset.key\nelse:\n raise RuntimeError(\"No compatible models found: please train an IID regression/binomial experiment\")\n\ntarget_col = h2oai.get_model_summary(key=MODEL_KEY).parameters.target_col\n \nprint(f\"Model : {MODEL_KEY}\\nDataset: {DATASET_KEY}\\nTarget : {target_col}\")",
"Model : 34edc42e-0152-11eb-a66d-207918bc8e4b\nDataset: dataset_33ab9348-0152-11eb-a66d-207918bc8e4b\nTarget : default payment next month\n"
]
],
[
[
"# List Explainers",
"_____no_output_____"
],
[
"List custom and OOTB recipes.",
"_____no_output_____"
]
],
[
[
"# list available server recipes\nexplainers = h2oai.list_explainers(\n experiment_types=None, \n explanation_scopes=None,\n dai_model_key=None,\n keywords=None,\n explainer_filter=[]\n)\n\nfor e in explainers:\n pprint.pprint(e.dump())",
"{'can_explain': ['regression', 'binomial'],\n 'explanation_scopes': ['global_scope'],\n 'explanations': [{'category': None,\n 'explanation_type': 'global-sensitivity-analysis',\n 'formats': [],\n 'has_local': None,\n 'name': 'SaExplanation',\n 'scope': 'global'},\n {'category': None,\n 'explanation_type': 'global-work-dir-archive',\n 'formats': [],\n 'has_local': None,\n 'name': 'WorkDirArchiveExplanation',\n 'scope': 'global'}],\n 'id': 'h2oaicore.mli.byor.recipes.sa_explainer.SaExplainer',\n 'keywords': ['run-by-default'],\n 'model_types': ['iid'],\n 'name': 'SA explainer'}\n{'can_explain': ['regression', 'binomial'],\n 'explanation_scopes': ['global_scope'],\n 'explanations': [{'category': None,\n 'explanation_type': 'global-disparate-impact-analysis',\n 'formats': [],\n 'has_local': None,\n 'name': 'DiaExplanation',\n 'scope': 'global'},\n {'category': None,\n 'explanation_type': 'global-work-dir-archive',\n 'formats': [],\n 'has_local': None,\n 'name': 'WorkDirArchiveExplanation',\n 'scope': 'global'}],\n 'id': 'h2oaicore.mli.byor.recipes.dia_explainer.DiaExplainer',\n 'keywords': ['run-by-default'],\n 'model_types': ['iid'],\n 'name': 'DIA explainer'}\n{'can_explain': ['regression', 'binomial'],\n 'explanation_scopes': ['global_scope'],\n 'explanations': [{'category': None,\n 'explanation_type': 'global-feature-importance',\n 'formats': [],\n 'has_local': None,\n 'name': 'GlobalFeatImpExplanation',\n 'scope': 'global'}],\n 'id': 'h2oaicore.mli.byor.recipes.permutation_feat_imp_absolute_explainer.AbsolutePermutationFeatureImportanceExplainer',\n 'keywords': [],\n 'model_types': ['iid'],\n 'name': 'Absolute permutation-based feature importance explainer'}\n{'can_explain': ['regression', 'binomial', 'multinomial'],\n 'explanation_scopes': ['global_scope', 'local_scope'],\n 'explanations': [{'category': None,\n 'explanation_type': 'global-partial-dependence',\n 'formats': [],\n 'has_local': None,\n 'name': 'PartialDependenceExplanation',\n 'scope': 'global'},\n {'category': None,\n 'explanation_type': 'local-individual-conditional-explanation',\n 'formats': [],\n 'has_local': None,\n 'name': 'IndividualConditionalExplanation',\n 'scope': 'local'}],\n 'id': 'h2oaicore.mli.byor.recipes.dai_pd_ice_explainer.DaiPdIceExplainer',\n 'keywords': ['run-by-default'],\n 'model_types': ['iid'],\n 'name': 'DAI PD/ICE explainer'}\n{'can_explain': ['regression', 'binomial'],\n 'explanation_scopes': ['global_scope'],\n 'explanations': [{'category': None,\n 'explanation_type': 'global-report',\n 'formats': [],\n 'has_local': None,\n 'name': 'ReportExplanation',\n 'scope': 'global'}],\n 'id': 'h2oaicore.mli.byor.recipes.mock.mock_markdown_explainer.MockMarkdownVegaExplainer',\n 'keywords': ['mock'],\n 'model_types': ['iid'],\n 'name': 'Mock Markdown with Vega explainer'}\n{'can_explain': ['regression', 'binomial', 'multinomial'],\n 'explanation_scopes': ['global_scope', 'local_scope'],\n 'explanations': [{'category': None,\n 'explanation_type': 'global-partial-dependence',\n 'formats': [],\n 'has_local': None,\n 'name': 'PartialDependenceExplanation',\n 'scope': 'global'},\n {'category': None,\n 'explanation_type': 'local-individual-conditional-explanation',\n 'formats': [],\n 'has_local': None,\n 'name': 'IndividualConditionalExplanation',\n 'scope': 'local'},\n {'category': None,\n 'explanation_type': 'global-work-dir-archive',\n 'formats': [],\n 'has_local': None,\n 'name': 'WorkDirArchiveExplanation',\n 'scope': 'global'}],\n 'id': 'h2oaicore.mli.byor.recipes.mock.mock_pd_explainer.MockPartialDependenceExplainer',\n 'keywords': ['mock'],\n 'model_types': ['iid'],\n 'name': 'Mock PartialDependence explainer'}\n{'can_explain': ['regression', 'binomial'],\n 'explanation_scopes': ['global_scope'],\n 'explanations': [{'category': None,\n 'explanation_type': 'global-decision-tree',\n 'formats': [],\n 'has_local': None,\n 'name': 'GlobalDtExplanation',\n 'scope': 'global'}],\n 'id': 'h2oaicore.mli.byor.recipes.mock.mock_dt_explainer.MockDecisionTreeExplainer',\n 'keywords': ['mock'],\n 'model_types': ['iid'],\n 'name': 'Mock DecisionTree explainer'}\n{'can_explain': ['regression', 'binomial'],\n 'explanation_scopes': ['global_scope'],\n 'explanations': [{'category': None,\n 'explanation_type': 'global-report',\n 'formats': [],\n 'has_local': None,\n 'name': 'ReportExplanation',\n 'scope': 'global'}],\n 'id': 'h2oaicore.mli.byor.recipes.mock.mock_markdown_pd_explainer.MockMarkdownPdExplainer',\n 'keywords': ['mock'],\n 'model_types': ['iid'],\n 'name': 'Mock Markdown explainer'}\n{'can_explain': ['regression', 'binomial'],\n 'explanation_scopes': ['global_scope'],\n 'explanations': [{'category': None,\n 'explanation_type': 'global-feature-importance',\n 'formats': [],\n 'has_local': None,\n 'name': 'GlobalFeatImpExplanation',\n 'scope': 'global'}],\n 'id': 'h2oaicore.mli.byor.recipes.mock.mock_featimp_explainer.MockFeatureImportanceExplainer',\n 'keywords': ['mock'],\n 'model_types': ['iid'],\n 'name': 'Mock Feature Importance explainer'}\n{'can_explain': ['regression', 'binomial'],\n 'explanation_scopes': ['global_scope'],\n 'explanations': [{'category': None,\n 'explanation_type': 'global-scatter-plot',\n 'formats': [],\n 'has_local': None,\n 'name': 'GlobalScatterPlotExplanation',\n 'scope': 'global'}],\n 'id': 'h2oaicore.mli.byor.recipes.mock.mock_scatter_plot_explainer.MockScatterPlotExplainer',\n 'keywords': ['mock'],\n 'model_types': ['iid'],\n 'name': 'Mock ScatterPlot explainer'}\n{'can_explain': ['regression', 'binomial', 'multinomial'],\n 'explanation_scopes': ['global_scope', 'local_scope'],\n 'explanations': [{'category': None,\n 'explanation_type': 'global-work-dir-archive',\n 'formats': [],\n 'has_local': None,\n 'name': 'WorkDirArchiveExplanation',\n 'scope': 'global'}],\n 'id': 'h2oaicore.mli.byor.recipes.surrogates_n_shapleys_explainer.MainSurrogatesAndShapleysExplainer',\n 'keywords': ['run-by-default'],\n 'model_types': ['iid'],\n 'name': 'Surrogates and Shapleys explainer'}\n{'can_explain': ['regression', 'binomial', 'multinomial'],\n 'explanation_scopes': ['global_scope', 'local_scope'],\n 'explanations': [{'category': None,\n 'explanation_type': 'local-proxy_explanation',\n 'formats': [],\n 'has_local': None,\n 'name': 'ProxyExplanation',\n 'scope': 'local'},\n {'category': None,\n 'explanation_type': 'global-feature-importance',\n 'formats': [],\n 'has_local': None,\n 'name': 'GlobalFeatImpExplanation',\n 'scope': 'global'}],\n 'id': 'h2oaicore.mli.byor.recipes.permutation_feat_imp_relative_explainer.RelativePermutationFeatureImportanceExplainer',\n 'keywords': [],\n 'model_types': ['iid'],\n"
],
[
"# list server recipes for given experiment type\nexplainers = h2oai.list_explainers(\n experiment_types=[\"binomial\"], \n explanation_scopes=None,\n dai_model_key=None,\n keywords=None,\n explainer_filter=[]\n)\n\nfor e in explainers:\n pprint.pprint(e.dump())",
"{'can_explain': ['regression', 'binomial'],\n 'explanation_scopes': ['global_scope'],\n 'explanations': [{'category': None,\n 'explanation_type': 'global-sensitivity-analysis',\n 'formats': [],\n 'has_local': None,\n 'name': 'SaExplanation',\n 'scope': 'global'},\n {'category': None,\n 'explanation_type': 'global-work-dir-archive',\n 'formats': [],\n 'has_local': None,\n 'name': 'WorkDirArchiveExplanation',\n 'scope': 'global'}],\n 'id': 'h2oaicore.mli.byor.recipes.sa_explainer.SaExplainer',\n 'keywords': ['run-by-default'],\n 'model_types': ['iid'],\n 'name': 'SA explainer'}\n{'can_explain': ['regression', 'binomial'],\n 'explanation_scopes': ['global_scope'],\n 'explanations': [{'category': None,\n 'explanation_type': 'global-disparate-impact-analysis',\n 'formats': [],\n 'has_local': None,\n 'name': 'DiaExplanation',\n 'scope': 'global'},\n {'category': None,\n 'explanation_type': 'global-work-dir-archive',\n 'formats': [],\n 'has_local': None,\n 'name': 'WorkDirArchiveExplanation',\n 'scope': 'global'}],\n 'id': 'h2oaicore.mli.byor.recipes.dia_explainer.DiaExplainer',\n 'keywords': ['run-by-default'],\n 'model_types': ['iid'],\n 'name': 'DIA explainer'}\n{'can_explain': ['regression', 'binomial'],\n 'explanation_scopes': ['global_scope'],\n 'explanations': [{'category': None,\n 'explanation_type': 'global-feature-importance',\n 'formats': [],\n 'has_local': None,\n 'name': 'GlobalFeatImpExplanation',\n 'scope': 'global'}],\n 'id': 'h2oaicore.mli.byor.recipes.permutation_feat_imp_absolute_explainer.AbsolutePermutationFeatureImportanceExplainer',\n 'keywords': [],\n 'model_types': ['iid'],\n 'name': 'Absolute permutation-based feature importance explainer'}\n{'can_explain': ['regression', 'binomial', 'multinomial'],\n 'explanation_scopes': ['global_scope', 'local_scope'],\n 'explanations': [{'category': None,\n 'explanation_type': 'global-partial-dependence',\n 'formats': [],\n 'has_local': None,\n 'name': 'PartialDependenceExplanation',\n 'scope': 'global'},\n {'category': None,\n 'explanation_type': 'local-individual-conditional-explanation',\n 'formats': [],\n 'has_local': None,\n 'name': 'IndividualConditionalExplanation',\n 'scope': 'local'}],\n 'id': 'h2oaicore.mli.byor.recipes.dai_pd_ice_explainer.DaiPdIceExplainer',\n 'keywords': ['run-by-default'],\n 'model_types': ['iid'],\n 'name': 'DAI PD/ICE explainer'}\n{'can_explain': ['regression', 'binomial'],\n 'explanation_scopes': ['global_scope'],\n 'explanations': [{'category': None,\n 'explanation_type': 'global-report',\n 'formats': [],\n 'has_local': None,\n 'name': 'ReportExplanation',\n 'scope': 'global'}],\n 'id': 'h2oaicore.mli.byor.recipes.mock.mock_markdown_explainer.MockMarkdownVegaExplainer',\n 'keywords': ['mock'],\n 'model_types': ['iid'],\n 'name': 'Mock Markdown with Vega explainer'}\n{'can_explain': ['regression', 'binomial', 'multinomial'],\n 'explanation_scopes': ['global_scope', 'local_scope'],\n 'explanations': [{'category': None,\n 'explanation_type': 'global-partial-dependence',\n 'formats': [],\n 'has_local': None,\n 'name': 'PartialDependenceExplanation',\n 'scope': 'global'},\n {'category': None,\n 'explanation_type': 'local-individual-conditional-explanation',\n 'formats': [],\n 'has_local': None,\n 'name': 'IndividualConditionalExplanation',\n 'scope': 'local'},\n {'category': None,\n 'explanation_type': 'global-work-dir-archive',\n 'formats': [],\n 'has_local': None,\n 'name': 'WorkDirArchiveExplanation',\n 'scope': 'global'}],\n 'id': 'h2oaicore.mli.byor.recipes.mock.mock_pd_explainer.MockPartialDependenceExplainer',\n 'keywords': ['mock'],\n 'model_types': ['iid'],\n 'name': 'Mock PartialDependence explainer'}\n{'can_explain': ['regression', 'binomial'],\n 'explanation_scopes': ['global_scope'],\n 'explanations': [{'category': None,\n 'explanation_type': 'global-decision-tree',\n 'formats': [],\n 'has_local': None,\n 'name': 'GlobalDtExplanation',\n 'scope': 'global'}],\n 'id': 'h2oaicore.mli.byor.recipes.mock.mock_dt_explainer.MockDecisionTreeExplainer',\n 'keywords': ['mock'],\n 'model_types': ['iid'],\n 'name': 'Mock DecisionTree explainer'}\n{'can_explain': ['regression', 'binomial'],\n 'explanation_scopes': ['global_scope'],\n 'explanations': [{'category': None,\n 'explanation_type': 'global-report',\n 'formats': [],\n 'has_local': None,\n 'name': 'ReportExplanation',\n 'scope': 'global'}],\n 'id': 'h2oaicore.mli.byor.recipes.mock.mock_markdown_pd_explainer.MockMarkdownPdExplainer',\n 'keywords': ['mock'],\n 'model_types': ['iid'],\n 'name': 'Mock Markdown explainer'}\n{'can_explain': ['regression', 'binomial'],\n 'explanation_scopes': ['global_scope'],\n 'explanations': [{'category': None,\n 'explanation_type': 'global-feature-importance',\n 'formats': [],\n 'has_local': None,\n 'name': 'GlobalFeatImpExplanation',\n 'scope': 'global'}],\n 'id': 'h2oaicore.mli.byor.recipes.mock.mock_featimp_explainer.MockFeatureImportanceExplainer',\n 'keywords': ['mock'],\n 'model_types': ['iid'],\n 'name': 'Mock Feature Importance explainer'}\n{'can_explain': ['regression', 'binomial'],\n 'explanation_scopes': ['global_scope'],\n 'explanations': [{'category': None,\n 'explanation_type': 'global-scatter-plot',\n 'formats': [],\n 'has_local': None,\n 'name': 'GlobalScatterPlotExplanation',\n 'scope': 'global'}],\n 'id': 'h2oaicore.mli.byor.recipes.mock.mock_scatter_plot_explainer.MockScatterPlotExplainer',\n 'keywords': ['mock'],\n 'model_types': ['iid'],\n 'name': 'Mock ScatterPlot explainer'}\n{'can_explain': ['regression', 'binomial', 'multinomial'],\n 'explanation_scopes': ['global_scope', 'local_scope'],\n 'explanations': [{'category': None,\n 'explanation_type': 'global-work-dir-archive',\n 'formats': [],\n 'has_local': None,\n 'name': 'WorkDirArchiveExplanation',\n 'scope': 'global'}],\n 'id': 'h2oaicore.mli.byor.recipes.surrogates_n_shapleys_explainer.MainSurrogatesAndShapleysExplainer',\n 'keywords': ['run-by-default'],\n 'model_types': ['iid'],\n 'name': 'Surrogates and Shapleys explainer'}\n{'can_explain': ['regression', 'binomial', 'multinomial'],\n 'explanation_scopes': ['global_scope', 'local_scope'],\n 'explanations': [{'category': None,\n 'explanation_type': 'local-proxy_explanation',\n 'formats': [],\n 'has_local': None,\n 'name': 'ProxyExplanation',\n 'scope': 'local'},\n {'category': None,\n 'explanation_type': 'global-feature-importance',\n 'formats': [],\n 'has_local': None,\n 'name': 'GlobalFeatImpExplanation',\n 'scope': 'global'}],\n 'id': 'h2oaicore.mli.byor.recipes.permutation_feat_imp_relative_explainer.RelativePermutationFeatureImportanceExplainer',\n 'keywords': [],\n 'model_types': ['iid'],\n"
],
[
"# list server recipes compatible with given DAI model\nexplainers = h2oai.list_explainers(\n dai_model_key=MODEL_KEY, \n experiment_types=None,\n explanation_scopes=None,\n keywords=None,\n explainer_filter=[]\n)\n\nfor e in explainers:\n pprint.pprint(e.dump())",
"{'can_explain': ['regression', 'binomial'],\n 'explanation_scopes': ['global_scope'],\n 'explanations': [{'category': None,\n 'explanation_type': 'global-sensitivity-analysis',\n 'formats': [],\n 'has_local': None,\n 'name': 'SaExplanation',\n 'scope': 'global'},\n {'category': None,\n 'explanation_type': 'global-work-dir-archive',\n 'formats': [],\n 'has_local': None,\n 'name': 'WorkDirArchiveExplanation',\n 'scope': 'global'}],\n 'id': 'h2oaicore.mli.byor.recipes.sa_explainer.SaExplainer',\n 'keywords': ['run-by-default'],\n 'model_types': ['iid'],\n 'name': 'SA explainer'}\n{'can_explain': ['regression', 'binomial'],\n 'explanation_scopes': ['global_scope'],\n 'explanations': [{'category': None,\n 'explanation_type': 'global-disparate-impact-analysis',\n 'formats': [],\n 'has_local': None,\n 'name': 'DiaExplanation',\n 'scope': 'global'},\n {'category': None,\n 'explanation_type': 'global-work-dir-archive',\n 'formats': [],\n 'has_local': None,\n 'name': 'WorkDirArchiveExplanation',\n 'scope': 'global'}],\n 'id': 'h2oaicore.mli.byor.recipes.dia_explainer.DiaExplainer',\n 'keywords': ['run-by-default'],\n 'model_types': ['iid'],\n 'name': 'DIA explainer'}\n{'can_explain': ['regression', 'binomial'],\n 'explanation_scopes': ['global_scope'],\n 'explanations': [{'category': None,\n 'explanation_type': 'global-feature-importance',\n 'formats': [],\n 'has_local': None,\n 'name': 'GlobalFeatImpExplanation',\n 'scope': 'global'}],\n 'id': 'h2oaicore.mli.byor.recipes.permutation_feat_imp_absolute_explainer.AbsolutePermutationFeatureImportanceExplainer',\n 'keywords': [],\n 'model_types': ['iid'],\n 'name': 'Absolute permutation-based feature importance explainer'}\n{'can_explain': ['regression', 'binomial', 'multinomial'],\n 'explanation_scopes': ['global_scope', 'local_scope'],\n 'explanations': [{'category': None,\n 'explanation_type': 'global-partial-dependence',\n 'formats': [],\n 'has_local': None,\n 'name': 'PartialDependenceExplanation',\n 'scope': 'global'},\n {'category': None,\n 'explanation_type': 'local-individual-conditional-explanation',\n 'formats': [],\n 'has_local': None,\n 'name': 'IndividualConditionalExplanation',\n 'scope': 'local'}],\n 'id': 'h2oaicore.mli.byor.recipes.dai_pd_ice_explainer.DaiPdIceExplainer',\n 'keywords': ['run-by-default'],\n 'model_types': ['iid'],\n 'name': 'DAI PD/ICE explainer'}\n{'can_explain': ['regression', 'binomial'],\n 'explanation_scopes': ['global_scope'],\n 'explanations': [{'category': None,\n 'explanation_type': 'global-report',\n 'formats': [],\n 'has_local': None,\n 'name': 'ReportExplanation',\n 'scope': 'global'}],\n 'id': 'h2oaicore.mli.byor.recipes.mock.mock_markdown_explainer.MockMarkdownVegaExplainer',\n 'keywords': ['mock'],\n 'model_types': ['iid'],\n 'name': 'Mock Markdown with Vega explainer'}\n{'can_explain': ['regression', 'binomial', 'multinomial'],\n 'explanation_scopes': ['global_scope', 'local_scope'],\n 'explanations': [{'category': None,\n 'explanation_type': 'global-partial-dependence',\n 'formats': [],\n 'has_local': None,\n 'name': 'PartialDependenceExplanation',\n 'scope': 'global'},\n {'category': None,\n 'explanation_type': 'local-individual-conditional-explanation',\n 'formats': [],\n 'has_local': None,\n 'name': 'IndividualConditionalExplanation',\n 'scope': 'local'},\n {'category': None,\n 'explanation_type': 'global-work-dir-archive',\n 'formats': [],\n 'has_local': None,\n 'name': 'WorkDirArchiveExplanation',\n 'scope': 'global'}],\n 'id': 'h2oaicore.mli.byor.recipes.mock.mock_pd_explainer.MockPartialDependenceExplainer',\n 'keywords': ['mock'],\n 'model_types': ['iid'],\n 'name': 'Mock PartialDependence explainer'}\n{'can_explain': ['regression', 'binomial'],\n 'explanation_scopes': ['global_scope'],\n 'explanations': [{'category': None,\n 'explanation_type': 'global-decision-tree',\n 'formats': [],\n 'has_local': None,\n 'name': 'GlobalDtExplanation',\n 'scope': 'global'}],\n 'id': 'h2oaicore.mli.byor.recipes.mock.mock_dt_explainer.MockDecisionTreeExplainer',\n 'keywords': ['mock'],\n 'model_types': ['iid'],\n 'name': 'Mock DecisionTree explainer'}\n{'can_explain': ['regression', 'binomial'],\n 'explanation_scopes': ['global_scope'],\n 'explanations': [{'category': None,\n 'explanation_type': 'global-report',\n 'formats': [],\n 'has_local': None,\n 'name': 'ReportExplanation',\n 'scope': 'global'}],\n 'id': 'h2oaicore.mli.byor.recipes.mock.mock_markdown_pd_explainer.MockMarkdownPdExplainer',\n 'keywords': ['mock'],\n 'model_types': ['iid'],\n 'name': 'Mock Markdown explainer'}\n{'can_explain': ['regression', 'binomial'],\n 'explanation_scopes': ['global_scope'],\n 'explanations': [{'category': None,\n 'explanation_type': 'global-feature-importance',\n 'formats': [],\n 'has_local': None,\n 'name': 'GlobalFeatImpExplanation',\n 'scope': 'global'}],\n 'id': 'h2oaicore.mli.byor.recipes.mock.mock_featimp_explainer.MockFeatureImportanceExplainer',\n 'keywords': ['mock'],\n 'model_types': ['iid'],\n 'name': 'Mock Feature Importance explainer'}\n{'can_explain': ['regression', 'binomial'],\n 'explanation_scopes': ['global_scope'],\n 'explanations': [{'category': None,\n 'explanation_type': 'global-scatter-plot',\n 'formats': [],\n 'has_local': None,\n 'name': 'GlobalScatterPlotExplanation',\n 'scope': 'global'}],\n 'id': 'h2oaicore.mli.byor.recipes.mock.mock_scatter_plot_explainer.MockScatterPlotExplainer',\n 'keywords': ['mock'],\n 'model_types': ['iid'],\n 'name': 'Mock ScatterPlot explainer'}\n{'can_explain': ['regression', 'binomial', 'multinomial'],\n 'explanation_scopes': ['global_scope', 'local_scope'],\n 'explanations': [{'category': None,\n 'explanation_type': 'global-work-dir-archive',\n 'formats': [],\n 'has_local': None,\n 'name': 'WorkDirArchiveExplanation',\n 'scope': 'global'}],\n 'id': 'h2oaicore.mli.byor.recipes.surrogates_n_shapleys_explainer.MainSurrogatesAndShapleysExplainer',\n 'keywords': ['run-by-default'],\n 'model_types': ['iid'],\n 'name': 'Surrogates and Shapleys explainer'}\n{'can_explain': ['regression', 'binomial', 'multinomial'],\n 'explanation_scopes': ['global_scope', 'local_scope'],\n 'explanations': [{'category': None,\n 'explanation_type': 'local-proxy_explanation',\n 'formats': [],\n 'has_local': None,\n 'name': 'ProxyExplanation',\n 'scope': 'local'},\n {'category': None,\n 'explanation_type': 'global-feature-importance',\n 'formats': [],\n 'has_local': None,\n 'name': 'GlobalFeatImpExplanation',\n 'scope': 'global'}],\n 'id': 'h2oaicore.mli.byor.recipes.permutation_feat_imp_relative_explainer.RelativePermutationFeatureImportanceExplainer',\n 'keywords': [],\n 'model_types': ['iid'],\n"
]
],
[
[
"# Ad-hoc Run of Built-in Explainer Recipe",
"_____no_output_____"
],
[
"Run OOTB explainer recipe shipped w/ DAI server:",
"_____no_output_____"
]
],
[
[
"sa_explainer_id = [explainer.id for explainer in explainers if \"SA explainer\" == explainer.name][0]\nsa_explainer_id",
"_____no_output_____"
],
[
"# prepare explaination parameters\nexplanation_params=Client.build_common_dai_explainer_params(\n target_col=target_col,\n model_key=MODEL_KEY,\n dataset_key=DATASET_KEY,\n)\nexplanation_params.dump()",
"_____no_output_____"
],
[
"# run explainer\nexplainer_id = sa_explainer_id\n\nprint(f\"Running OOTB explainer: {explainer_id}\")\n\nrun_job = h2oai.run_explainers(\n explainers=[Explainer(\n explainer_id=explainer_id,\n explainer_params=\"\",\n )],\n params=explanation_params,\n)\nrun_job.dump()",
"Running OOTB explainer: h2oaicore.mli.byor.recipes.sa_explainer.SaExplainer\n"
],
[
"# wait for explainer to finish\nexplainer_job_statuses = h2oai.wait_for_explainers(run_job.mli_key)\nfor job_status in explainer_job_statuses:\n pprint.pprint(job_status.dump())\n \nmli_key = job_status.mli_key\nexplainer_job_key = job_status.explainer_job_key\nexplainer_job = job_status.explainer_job",
"{'explainer_job': {'child_explainers_job_keys': [],\n 'created': 1601291446.8943293,\n 'duration': 4.786961078643799,\n 'entity': {'can_explain': ['regression', 'binomial'],\n 'explanation_scopes': ['global_scope'],\n 'explanations': [{'category': 'DAI MODEL',\n 'explanation_type': 'global-work-dir-archive',\n 'formats': ['application/zip'],\n 'has_local': None,\n 'name': 'SA Data ZIP Archive',\n 'scope': 'global'},\n {'category': 'DAI MODEL',\n 'explanation_type': 'global-sensitivity-analysis',\n 'formats': ['text/plain'],\n 'has_local': None,\n 'name': 'Sensitivity Analysis '\n '(SA)',\n 'scope': 'global'}],\n 'id': 'h2oaicore.mli.byor.recipes.sa_explainer.SaExplainer',\n 'keywords': ['run-by-default'],\n 'model_types': ['iid'],\n 'name': 'SA explainer'},\n 'error': '',\n 'message': 'Explainer 423581b8-017b-11eb-a66d-207918bc8e4b '\n 'run successfully finished',\n 'progress': 1.0,\n 'status': 0},\n 'explainer_job_key': '423581b8-017b-11eb-a66d-207918bc8e4b',\n 'mli_key': '423581b6-017b-11eb-a66d-207918bc8e4b'}\n"
]
],
[
[
"## Get Recipe Result",
"_____no_output_____"
]
],
[
[
"# get recipe result FORMATS/TYPES (representations of recipe output)\nexplainer_job.entity.can_explain",
"_____no_output_____"
],
[
"explainer_job.entity.explanation_scopes",
"_____no_output_____"
],
[
"for explanation in explainer_job.entity.explanations: pprint.pprint(explanation.dump())",
"{'category': 'DAI MODEL',\n 'explanation_type': 'global-work-dir-archive',\n 'formats': ['application/zip'],\n 'has_local': None,\n 'name': 'SA Data ZIP Archive',\n 'scope': 'global'}\n{'category': 'DAI MODEL',\n 'explanation_type': 'global-sensitivity-analysis',\n 'formats': ['text/plain'],\n 'has_local': None,\n 'name': 'Sensitivity Analysis (SA)',\n 'scope': 'global'}\n"
],
[
"# choose the most suitable format (if more than one) and get the result\nBASE_URL = f\"{address}/files/\"\n\nfor explanation in explainer_job.entity.explanations:\n for e_format in explanation.formats:\n server_path: str = h2oai.get_explainer_result_url_path(\n mli_key=mli_key,\n explainer_job_key=explainer_job_key,\n explanation_type=explanation.explanation_type,\n explanation_format=e_format\n )\n print(f\"Explanation {explanation.explanation_type}:\\n {e_format}:\\n {BASE_URL}{server_path}\")\n \n download_dir = \"/tmp\"\n h2oai.download(server_path, download_dir)",
"Explanation global-work-dir-archive:\n application/zip:\n http://127.0.0.1:12345/files/h2oai/mli_experiment_423581b6-017b-11eb-a66d-207918bc8e4b/explainer_h2oaicore_mli_byor_recipes_sa_explainer_SaExplainer_423581b8-017b-11eb-a66d-207918bc8e4b/global_work_dir_archive/application_zip/explanation.zip\nExplanation global-sensitivity-analysis:\n text/plain:\n http://127.0.0.1:12345/files/h2oai/mli_experiment_423581b6-017b-11eb-a66d-207918bc8e4b/explainer_h2oaicore_mli_byor_recipes_sa_explainer_SaExplainer_423581b8-017b-11eb-a66d-207918bc8e4b/global_sensitivity_analysis/text_plain/explanation.txt\n"
],
[
"!ls -l {download_dir}/explanation.zip",
"-rw-r--r-- 1 dvorka dvorka 1842252 Sep 28 13:10 /tmp/explanation.zip\r\n"
]
],
[
[
"URL from above can be used to **download** choosen recipe result representation. Explainer log can be downloaded from:",
"_____no_output_____"
]
],
[
[
"server_path = h2oai.get_explainer_run_log_url_path(\n mli_key=mli_key,\n explainer_job_key=explainer_job_key,\n)\nprint(f\"{BASE_URL}{server_path}\")",
"http://127.0.0.1:12345/files/h2oai/mli_experiment_423581b6-017b-11eb-a66d-207918bc8e4b/explainer_h2oaicore_mli_byor_recipes_sa_explainer_SaExplainer_423581b8-017b-11eb-a66d-207918bc8e4b/log/explainer_run_423581b8-017b-11eb-a66d-207918bc8e4b.log\n"
]
],
[
[
"# Ad-hoc Run of Custom Explainer Recipe",
"_____no_output_____"
],
[
"Running previously uploaded custom explainer.",
"_____no_output_____"
]
],
[
[
"# run custom explainer - use previously uploaded recipe ID\nif uploaded_explainer_id:\n explainer_id = uploaded_explainer_id\nelse:\n # explainer has been uploaded before\n explainers = h2oai.list_explainers(\n time_series=False, \n dai_model_key=MODEL_KEY, \n experiment_types=None,\n explanation_scopes=None,\n keywords=None,\n )\n for e in explainers:\n if e.name == BYOR_EXPLAINER_NAME:\n explainer_id = e.id\n\nprint(f\"Running CUSTOM explainer: {explainer_id}\")\n\nrun_job = h2oai.run_explainers(\n explainers=[\n Explainer(explainer_id=explainer_id, explainer_params=None)\n ],\n params=Client.build_common_dai_explainer_params(\n target_col=target_col,\n model_key=MODEL_KEY,\n dataset_key=DATASET_KEY,\n ),\n)\nrun_job.dump()",
"Running CUSTOM explainer: False_morris_sensitivity_explainer_d935205d_contentexplainer.MorrisSensitivityLeExplainer\n"
],
[
"# wait for explainer to finish\nexplainer_job_statuses = h2oai.wait_for_explainers(run_job.mli_key)\nfor job_status in explainer_job_statuses:\n pprint.pprint(job_status.dump())",
"{'explainer_job': {'child_explainers_job_keys': [],\n 'created': 1601291453.4908412,\n 'duration': 3.8387792110443115,\n 'entity': {'can_explain': ['regression', 'binomial'],\n 'explanation_scopes': ['global_scope'],\n 'explanations': [{'category': 'CUSTOM',\n 'explanation_type': 'global-feature-importance',\n 'formats': ['application/vnd.h2oai.json+datatable.jay',\n 'application/json'],\n 'has_local': None,\n 'name': 'Morris Sensitivity '\n 'Analysis',\n 'scope': 'global'}],\n 'id': 'False_morris_sensitivity_explainer_d935205d_contentexplainer.MorrisSensitivityLeExplainer',\n 'keywords': [],\n 'model_types': ['iid'],\n 'name': 'Morris Sensitivity Analysis'},\n 'error': '',\n 'message': 'Explainer 46260112-017b-11eb-a66d-207918bc8e4b '\n 'run successfully finished',\n 'progress': 1.0,\n 'status': 0},\n 'explainer_job_key': '46260112-017b-11eb-a66d-207918bc8e4b',\n 'mli_key': '46260110-017b-11eb-a66d-207918bc8e4b'}\n"
],
[
"server_path = h2oai.get_explainer_result_url_path(\n mli_key=job_status.mli_key,\n explainer_job_key=job_status.explainer_job_key,\n explanation_type='global-feature-importance',\n explanation_format='application/json'\n)\nprint(f\"{BASE_URL}{server_path}\")",
"http://127.0.0.1:12345/files/h2oai/mli_experiment_46260110-017b-11eb-a66d-207918bc8e4b/explainer_False_morris_sensitivity_explainer_d935205d_contentexplainer_MorrisSensitivityLeExplainer_46260112-017b-11eb-a66d-207918bc8e4b/global_feature_importance/application_json/explanation.json\n"
]
],
[
[
"URL from above can be used to **download** choosen **custom** recipe result representation.",
"_____no_output_____"
],
[
"# Explain (Model) with All Compatible or Selected Explainers",
"_____no_output_____"
]
],
[
[
"# get IDs of previously listed recipes\nexplainer_ids = [explainer.id for explainer in explainers]\nexplainer_ids",
"_____no_output_____"
],
[
"# run explainers: list of IDs OR empty list\n# - empty explainer IDs list means \"run all model COMPATIBLE explainers with default parameters\")\n\nprint(f\"All explainers:\\n{explainer_ids}\")\nrun_job: ExplainersRunJob = h2oai.run_explainers(\n explainers=[],\n params=Client.build_common_dai_explainer_params(\n target_col=target_col,\n model_key=MODEL_KEY,\n dataset_key=DATASET_KEY,\n ),\n)\nrun_job.dump()",
"All explainers:\n['h2oaicore.mli.byor.recipes.sa_explainer.SaExplainer', 'h2oaicore.mli.byor.recipes.dia_explainer.DiaExplainer', 'h2oaicore.mli.byor.recipes.permutation_feat_imp_absolute_explainer.AbsolutePermutationFeatureImportanceExplainer', 'h2oaicore.mli.byor.recipes.dai_pd_ice_explainer.DaiPdIceExplainer', 'h2oaicore.mli.byor.recipes.mock.mock_markdown_explainer.MockMarkdownVegaExplainer', 'h2oaicore.mli.byor.recipes.mock.mock_pd_explainer.MockPartialDependenceExplainer', 'h2oaicore.mli.byor.recipes.mock.mock_dt_explainer.MockDecisionTreeExplainer', 'h2oaicore.mli.byor.recipes.mock.mock_markdown_pd_explainer.MockMarkdownPdExplainer', 'h2oaicore.mli.byor.recipes.mock.mock_featimp_explainer.MockFeatureImportanceExplainer', 'h2oaicore.mli.byor.recipes.mock.mock_scatter_plot_explainer.MockScatterPlotExplainer', 'h2oaicore.mli.byor.recipes.surrogates_n_shapleys_explainer.MainSurrogatesAndShapleysExplainer', 'h2oaicore.mli.byor.recipes.permutation_feat_imp_relative_explainer.RelativePermutationFeatureImportanceExplainer', 'h2oaicore.mli.byor.recipes.orig_kernel_shap_explainer.OriginalKernelShapExplainer', 'h2oaicore.mli.byor.recipes.surrogates.rf_pd_explainer.RandomForestPartialDependenceExplainer', 'h2oaicore.mli.byor.recipes.surrogates.rf_feat_imp_explainer.RandomForestFeatureImportanceExplainer', 'h2oaicore.mli.byor.recipes.surrogates.original_feat_imp_explainer.OriginalFeatureImportanceExplainer', 'h2oaicore.mli.byor.recipes.surrogates.dt_surrogate_explainer.DecisionTreeSurrogateExplainer', 'h2oaicore.mli.byor.recipes.surrogates.klime_explainer.KLimeExplainer', 'h2oaicore.mli.byor.recipes.surrogates.rf_loco_explainer.RandomForestLocoExplainer', 'h2oaicore.mli.byor.recipes.text.nlp_tokenizer_explainer.NlpTokenizerExplainer', 'h2oaicore.mli.byor.recipes.text.nlp_loco_explainer.NlpLocoExplainer', 'h2oaicore.mli.byor.recipes.autoreport_explainer.AutoReportExplainer', 'h2oaicore.mli.byor.recipes.transformed_feat_imp_explainer.TransformedFeatureImportanceExplainer', 'h2oaicore.mli.byor.recipes.transformed_shapley_explainer.TransformedShapleyFeatureImportanceExplainer', 'False_morris_sensitivity_explainer_d935205d_contentexplainer.MorrisSensitivityLeExplainer', 'False_test_mock_ex_setup_explainer_28f31b80_contentexplainer.TestMockExSetupExplainer', 'False_test_mock_ex_fit_explainer_1b629753_contentexplainer.TestMockExFitExplainer', 'False_test_mock_ex_explain_explainer_4cbf4487_contentexplainer.TestMockExExplainExplainer']\n"
],
[
"# check interpretation job status (legacy RPC API)\ni_job: InterpretationJob = h2oai.get_interpretation_job(run_job.mli_key)\n# note per-explainer (subtask) ID and display name\ni_job.dump()",
"_____no_output_____"
],
[
"# check particular sub-job status (existing RPC API reused)\nh2oai.get_explainer_job_status(run_job.mli_key, run_job.explainer_job_keys[0]).dump()",
"_____no_output_____"
],
[
"# check sub-jobs statuses (existing RPC API reused)\njob_statuses = h2oai.get_explainer_job_statuses(run_job.mli_key, run_job.explainer_job_keys)\nfor js in job_statuses:\n pprint.pprint(js.dump())",
"{'explainer_job': {'child_explainers_job_keys': [],\n 'created': 1601291459.8144472,\n 'duration': 0.6814770698547363,\n 'entity': {'can_explain': ['regression', 'binomial'],\n 'explanation_scopes': ['global_scope'],\n 'explanations': [],\n 'id': 'h2oaicore.mli.byor.recipes.sa_explainer.SaExplainer',\n 'keywords': ['run-by-default'],\n 'model_types': ['iid'],\n 'name': 'SA explainer'},\n 'error': '',\n 'message': 'Entering sequential execution gate...',\n 'progress': 0,\n 'status': -1},\n 'explainer_job_key': '49ec5ea4-017b-11eb-a66d-207918bc8e4b',\n 'mli_key': '49ec5ea2-017b-11eb-a66d-207918bc8e4b'}\n{'explainer_job': {'child_explainers_job_keys': [],\n 'created': 1601291459.8862343,\n 'duration': 0.6159448623657227,\n 'entity': {'can_explain': ['regression', 'binomial'],\n 'explanation_scopes': ['global_scope'],\n 'explanations': [],\n 'id': 'h2oaicore.mli.byor.recipes.dia_explainer.DiaExplainer',\n 'keywords': ['run-by-default'],\n 'model_types': ['iid'],\n 'name': 'DIA explainer'},\n 'error': '',\n 'message': 'Entering sequential execution gate...',\n 'progress': 0,\n 'status': -1},\n 'explainer_job_key': '49ec5ea6-017b-11eb-a66d-207918bc8e4b',\n 'mli_key': '49ec5ea2-017b-11eb-a66d-207918bc8e4b'}\n{'explainer_job': {'child_explainers_job_keys': ['49ec5eaa-017b-11eb-a66d-207918bc8e4b',\n '49ec5eac-017b-11eb-a66d-207918bc8e4b',\n '49ec5eae-017b-11eb-a66d-207918bc8e4b',\n '49ec5eb0-017b-11eb-a66d-207918bc8e4b',\n '49ec5eb2-017b-11eb-a66d-207918bc8e4b',\n '49ec5eb4-017b-11eb-a66d-207918bc8e4b',\n '49ec5eb6-017b-11eb-a66d-207918bc8e4b',\n '49ec5eb8-017b-11eb-a66d-207918bc8e4b'],\n 'created': 1601291459.927952,\n 'duration': 0.5900819301605225,\n 'entity': {'can_explain': ['regression',\n 'binomial',\n 'multinomial'],\n 'explanation_scopes': ['global_scope',\n 'local_scope'],\n 'explanations': [],\n 'id': 'h2oaicore.mli.byor.recipes.surrogates_n_shapleys_explainer.MainSurrogatesAndShapleysExplainer',\n 'keywords': ['run-by-default'],\n 'model_types': ['iid'],\n 'name': 'Surrogates and Shapleys explainer'},\n 'error': '',\n 'message': 'Entering sequential execution gate...',\n 'progress': 0,\n 'status': -1},\n 'explainer_job_key': '49ec5ea8-017b-11eb-a66d-207918bc8e4b',\n 'mli_key': '49ec5ea2-017b-11eb-a66d-207918bc8e4b'}\n{'explainer_job': {'child_explainers_job_keys': [],\n 'created': 1601291459.9655921,\n 'duration': 0.5583939552307129,\n 'entity': {'can_explain': ['regression', 'binomial'],\n 'explanation_scopes': ['global_scope',\n 'local_scope'],\n 'explanations': [],\n 'id': 'h2oaicore.mli.byor.recipes.surrogates.rf_pd_explainer.RandomForestPartialDependenceExplainer',\n 'keywords': ['run-by-default', 'proxy-explainer'],\n 'model_types': ['iid'],\n 'name': 'Surrogate RF PD/ICE explainer'},\n 'error': '',\n 'message': 'Entering sequential execution gate...',\n 'progress': 0,\n 'status': -1},\n 'explainer_job_key': '49ec5eaa-017b-11eb-a66d-207918bc8e4b',\n 'mli_key': '49ec5ea2-017b-11eb-a66d-207918bc8e4b'}\n{'explainer_job': {'child_explainers_job_keys': [],\n 'created': 1601291460.0036705,\n 'duration': 0.5254826545715332,\n 'entity': {'can_explain': ['regression',\n 'binomial',\n 'multinomial'],\n 'explanation_scopes': ['global_scope',\n 'local_scope'],\n 'explanations': [],\n 'id': 'h2oaicore.mli.byor.recipes.surrogates.rf_feat_imp_explainer.RandomForestFeatureImportanceExplainer',\n 'keywords': ['run-by-default', 'proxy-explainer'],\n 'model_types': ['iid'],\n 'name': 'Surrogate RF feature importance '\n 'explainer'},\n 'error': '',\n 'message': 'Entering sequential execution gate...',\n 'progress': 0,\n 'status': -1},\n 'explainer_job_key': '49ec5eac-017b-11eb-a66d-207918bc8e4b',\n 'mli_key': '49ec5ea2-017b-11eb-a66d-207918bc8e4b'}\n{'explainer_job': {'child_explainers_job_keys': [],\n 'created': 1601291460.0425496,\n 'duration': 0.4986147880554199,\n 'entity': {'can_explain': ['regression',\n 'binomial',\n 'multinomial'],\n 'explanation_scopes': ['global_scope',\n 'local_scope'],\n 'explanations': [],\n 'id': 'h2oaicore.mli.byor.recipes.surrogates.original_feat_imp_explainer.OriginalFeatureImportanceExplainer',\n 'keywords': ['run-by-default', 'proxy-explainer'],\n 'model_types': ['iid'],\n 'name': 'Original feature importance explainer'},\n 'error': '',\n 'message': 'Entering sequential execution gate...',\n 'progress': 0,\n 'status': -1},\n 'explainer_job_key': '49ec5eae-017b-11eb-a66d-207918bc8e4b',\n 'mli_key': '49ec5ea2-017b-11eb-a66d-207918bc8e4b'}\n{'explainer_job': {'child_explainers_job_keys': [],\n 'created': 1601291460.0819962,\n 'duration': 0.46430420875549316,\n 'entity': {'can_explain': ['regression',\n 'binomial',\n 'multinomial'],\n 'explanation_scopes': ['global_scope',\n 'local_scope'],\n 'explanations': [],\n 'id': 'h2oaicore.mli.byor.recipes.surrogates.dt_surrogate_explainer.DecisionTreeSurrogateExplainer',\n 'keywords': ['run-by-default'],\n 'model_types': ['iid'],\n 'name': 'DT Surrogate explainer'},\n 'error': '',\n 'message': 'Entering sequential execution gate...',\n 'progress': 0,\n 'status': -1},\n 'explainer_job_key': '49ec5eb0-017b-11eb-a66d-207918bc8e4b',\n 'mli_key': '49ec5ea2-017b-11eb-a66d-207918bc8e4b'}\n{'explainer_job': {'child_explainers_job_keys': [],\n"
],
[
"# wait for ALL explainers to finish\nexplainer_statuses=h2oai.wait_for_explainers(run_job.mli_key)\nexplainer_statuses",
"_____no_output_____"
],
[
"# download explanation type in desired format\nDOWNLOAD_DIR = f\"/tmp/interpretation_run_{randint(0,1_000_000)}\"\n\nexplainer_job_key = explainer_statuses[0].explainer_job_key\nexplanations = h2oai.list_explainer_results(explainer_job_key=explainer_job_key).explanations\n# explanations\nfor explanation in explanations:\n # explantion's formats\n for explanation_format in explanation.formats:\n # format's URL\n result_path: str = h2oai.get_explainer_result_url_path(\n mli_key=run_job.mli_key,\n explainer_job_key=explainer_job_key,\n explanation_type=explanation.explanation_type,\n explanation_format=explanation_format,\n )\n \n # where to download\n EXPLANATION_DIR = f\"{DOWNLOAD_DIR}/explanation_{randint(0,1_000_000)}\"\n os.makedirs(EXPLANATION_DIR, exist_ok=True)\n \n # download\n h2oai.download(result_path, EXPLANATION_DIR)\n\n print(\n f\"Explanation {explanation.explanation_type}:\\n\"\n f\" {explanation_format}:\\n\"\n f\" {BASE_URL}{result_path}\"\n )\n \nprint(f\"\\nDownloaded explanations in {DOWNLOAD_DIR}:\")\n!ls -l {DOWNLOAD_DIR}",
"Explanation global-work-dir-archive:\n application/zip:\n http://127.0.0.1:12345/files/h2oai/mli_experiment_49ec5ea2-017b-11eb-a66d-207918bc8e4b/explainer_h2oaicore_mli_byor_recipes_sa_explainer_SaExplainer_49ec5ea4-017b-11eb-a66d-207918bc8e4b/global_work_dir_archive/application_zip/explanation.zip\nExplanation global-sensitivity-analysis:\n text/plain:\n http://127.0.0.1:12345/files/h2oai/mli_experiment_49ec5ea2-017b-11eb-a66d-207918bc8e4b/explainer_h2oaicore_mli_byor_recipes_sa_explainer_SaExplainer_49ec5ea4-017b-11eb-a66d-207918bc8e4b/global_sensitivity_analysis/text_plain/explanation.txt\n\nDownloaded explanations in /tmp/interpretation_run_388854:\ntotal 8\ndrwxr-xr-x 2 dvorka dvorka 4096 Sep 28 13:16 explanation_753563\ndrwxr-xr-x 2 dvorka dvorka 4096 Sep 28 13:16 explanation_809124\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
c50186d80edd2b745e84c522eaa80e047a21d142
| 25,800 |
ipynb
|
Jupyter Notebook
|
.ipynb_checkpoints/Part C - Stacked Ensemble-checkpoint.ipynb
|
prashantk016/Predictive-Analysis-for-E-Commerce-Website-and-Product-Recommendation
|
393a446d8667ffff6ae5437a5f33cb6f32b8837b
|
[
"MIT"
] | null | null | null |
.ipynb_checkpoints/Part C - Stacked Ensemble-checkpoint.ipynb
|
prashantk016/Predictive-Analysis-for-E-Commerce-Website-and-Product-Recommendation
|
393a446d8667ffff6ae5437a5f33cb6f32b8837b
|
[
"MIT"
] | null | null | null |
.ipynb_checkpoints/Part C - Stacked Ensemble-checkpoint.ipynb
|
prashantk016/Predictive-Analysis-for-E-Commerce-Website-and-Product-Recommendation
|
393a446d8667ffff6ae5437a5f33cb6f32b8837b
|
[
"MIT"
] | null | null | null | 42.715232 | 8,152 | 0.699574 |
[
[
[
"from sklearn import datasets\nfrom sklearn.cross_validation import cross_val_score, train_test_split\nfrom sklearn.model_selection import cross_val_score\nfrom sklearn.linear_model import LinearRegression,LogisticRegression\nfrom sklearn.naive_bayes import GaussianNB\nfrom sklearn.ensemble import RandomForestClassifier\nfrom sklearn.svm import SVC\nfrom sklearn.neighbors import KNeighborsClassifier\nfrom sklearn.ensemble import VotingClassifier\nimport pandas as pd\nimport warnings\nimport numpy as np\nfrom sklearn.model_selection import RepeatedKFold\nfrom sklearn.model_selection import KFold\nfrom sklearn.metrics import accuracy_score\nwarnings.filterwarnings('ignore')",
"_____no_output_____"
],
[
"df = pd.read_csv(\"Replaced.csv\", encoding='ISO-8859-1')",
"_____no_output_____"
],
[
"df.didPurchase = (df.didPurchase)*1\ndf.doRecommend = (df.doRecommend)*1\n\ndf['doRecommend'] = df['doRecommend'].fillna(1)\ndf['didPurchase'] = df['didPurchase'].fillna(1)",
"_____no_output_____"
]
],
[
[
"## Voting Classifier Ensemble",
"_____no_output_____"
]
],
[
[
"X=df[['didPurchase','rating']]\ny=df['doRecommend']\n\nclf1 = LogisticRegression(random_state=1)\nclf2 = RandomForestClassifier(random_state=1)\nclf3 = GaussianNB()\nclf4 = KNeighborsClassifier(n_neighbors=10)\neclf = VotingClassifier(estimators=[('lr', clf1), ('rf', clf2), ('gnb', clf3),('knn',clf4)], voting='hard')\nX_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, random_state=None)",
"_____no_output_____"
],
[
"eclf.fit(X_train, y_train)",
"_____no_output_____"
],
[
"eclf.score( X_test, y_test)",
"_____no_output_____"
]
],
[
[
"Voting classifier gives an accuracy of 95%",
"_____no_output_____"
]
],
[
[
"for clf, label in zip([clf1, clf2, clf3,clf4, eclf], ['Logistic Regression', 'Random Forest', 'naive Bayes', 'KNN','Ensemble']):\n scores = cross_val_score(clf, X, y, cv=20, scoring='accuracy')\n print(\"Accuracy: %0.3f (+/- %0.3f) [%s]\" % (scores.mean(), scores.std(), label))",
"Accuracy: 0.944 (+/- 0.035) [Logistic Regression]\nAccuracy: 0.954 (+/- 0.029) [Random Forest]\nAccuracy: 0.948 (+/- 0.036) [naive Bayes]\nAccuracy: 0.947 (+/- 0.037) [KNN]\nAccuracy: 0.946 (+/- 0.037) [Ensemble]\n"
]
],
[
[
"***\n# Staked Ensemble",
"_____no_output_____"
]
],
[
[
"training, valid, ytraining, yvalid = train_test_split(X_train, y_train, test_size=0.3, random_state=0)",
"_____no_output_____"
],
[
"clf2.fit(training,ytraining)",
"_____no_output_____"
],
[
"clf4.fit(training,ytraining)",
"_____no_output_____"
],
[
"preds1=clf2.predict(valid)\npreds2=clf4.predict(valid)",
"_____no_output_____"
],
[
"test_preds1=clf2.predict(X_test)\ntest_preds2=clf4.predict(X_test)",
"_____no_output_____"
],
[
"stacked_predictions=np.column_stack((preds1,preds2))\nstacked_test_predictions=np.column_stack((test_preds1,test_preds2))",
"_____no_output_____"
],
[
"meta_model=LinearRegression()\nmeta_model.fit(stacked_predictions,yvalid)",
"_____no_output_____"
],
[
"final_predictions=meta_model.predict(stacked_test_predictions)\n",
"_____no_output_____"
],
[
"count=[];\ny_list=y_test.tolist()\nfor i in range(len(y_list)):\n if (y_list[i]==np.round(final_predictions[i])):\n count.append(\"Correct\")\n else:\n count.append(\"Incorrect\")",
"_____no_output_____"
],
[
"import seaborn as sns\nsns.countplot(x=count)",
"_____no_output_____"
],
[
"accuracy_score(y_test,np.round(final_predictions))",
"_____no_output_____"
]
],
[
[
"***\n## Stacked Ensemble with Cross validation ",
"_____no_output_____"
]
],
[
[
"def fit_stack(clf, training, valid, ytraining,X_test,stacked_predictions,stacked_test_predictions):\n clf.fit(training,ytraining)\n preds=clf.predict(valid)\n test_preds=clf.predict(X_test)\n stacked_predictions=np.append(stacked_predictions,preds)\n stacked_test_predictions=np.append(stacked_test_predictions,test_preds)\n return {'pred':stacked_predictions,'test':stacked_test_predictions}",
"_____no_output_____"
],
[
"def stacked():\n X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, random_state=None)\n training, valid, ytraining, yvalid = train_test_split(X_train, y_train, test_size=0.3, random_state=None) \n #kf = RepeatedKFold(n_splits=5, n_repeats=5, random_state=None) \n #for train_index, test_index in kf.split(X):\n # training, valid = X.iloc[train_index], X.iloc[test_index] \n # ytraining, yvalid = y.iloc[train_index], y.iloc[test_index]\n clf2.fit(training,ytraining)\n clf4.fit(training,ytraining)\n preds1=clf2.predict(valid)\n preds2=clf4.predict(valid)\n test_preds1=clf2.predict(X_test)\n test_preds2=clf4.predict(X_test)\n stacked_predictions=np.column_stack((preds1,preds2))\n stacked_test_predictions=np.column_stack((test_preds1,test_preds2))\n #print(stacked_predictions.shape)\n #print(stacked_test_predictions.shape)\n meta_model=KNeighborsClassifier(n_neighbors=10)\n meta_model.fit(stacked_predictions,yvalid)\n final_predictions=meta_model.predict(stacked_test_predictions)\n return accuracy_score(y_test,final_predictions)",
"_____no_output_____"
],
[
"accuracies=[]\nfor i in range(10):\n accuracies.append(stacked())\n print(\"%0.3f\" % accuracies[i])\nprint(accuracies)\nmean_acc=sum(accuracies) / float(len(accuracies))\nprint(\"Mean %0.3f\" % mean_acc)",
"0.955\n0.957\n0.957\n0.956\n0.957\n0.956\n0.959\n0.955\n0.959\n0.955\n[0.9551664241115441, 0.9565278625416647, 0.9568564856110042, 0.9564339702361392, 0.9566217548471903, 0.9559645087085114, 0.9589690624853293, 0.9552133702643069, 0.9591099009436177, 0.9552133702643069]\nMean 0.957\n"
]
],
[
[
"### Repeated KFold with X and y",
"_____no_output_____"
]
],
[
[
"accuracies=[]\ndef stacked2():\n from sklearn.metrics import accuracy_score\n #X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, random_state=None)\n #training, valid, ytraining, yvalid = train_test_split(X_train, y_train, test_size=0.3, random_state=None) \n kf = RepeatedKFold(n_splits=5, n_repeats=2, random_state=None) \n for train_index, test_index in kf.split(X):\n \n X_train, X_test = X.iloc[train_index], X.iloc[test_index] \n y_train, y_test = y.iloc[train_index], y.iloc[test_index]\n training, valid, ytraining, yvalid = train_test_split(X_train, y_train, test_size=0.3, random_state=None) \n stacked_predictions=[]\n stacked_test_predictions=[]\n result=fit_stack(clf2, training, valid, ytraining,X_test,stacked_predictions,stacked_test_predictions)\n stacked_predictions=result['pred']\n stacked_test_predictions=result['test']\n result=fit_stack(clf4, training, valid, ytraining,X_test,stacked_predictions,stacked_test_predictions)\n stacked_predictions=result['pred']\n stacked_test_predictions=result['test']\n result=fit_stack(clf3, training, valid, ytraining,X_test,stacked_predictions,stacked_test_predictions)\n stacked_predictions=result['pred']\n stacked_test_predictions=result['test']\n meta_model=KNeighborsClassifier(n_neighbors=10)\n meta_model.fit(stacked_predictions.reshape(-1,3),yvalid)\n final_predictions=meta_model.predict(stacked_test_predictions.reshape(-1,3))\n acc=accuracy_score(y_test,final_predictions)\n accuracies.append(acc)\n print(accuracies)\n mean_acc=sum(accuracies) / float(len(accuracies))\n print(\"Mean %0.3f\" % mean_acc)",
"_____no_output_____"
],
[
"stacked2()",
"[0.9308499401450602, 0.9345820716850926, 0.9332441377367791, 0.9311971830985916, 0.9290845070422535, 0.9292303358918386, 0.9323991268220548, 0.9326103795507359, 0.9348591549295775, 0.9298591549295775]\nMean 0.932\n"
]
],
[
[
"### Repeated KFold with X_train and y_train",
"_____no_output_____"
]
],
[
[
"accuracies=[]\ndef stacked1():\n X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, random_state=None)\n kf = RepeatedKFold(n_splits=5, n_repeats=1, random_state=None) \n for train_index, test_index in kf.split(X_train):\n \n training, valid = X_train.iloc[train_index], X_train.iloc[test_index]\n ytraining, yvalid = y_train.iloc[train_index], y_train.iloc[test_index]\n stacked_predictions=[]\n stacked_test_predictions=[]\n result=fit_stack(clf2, training, valid, ytraining,X_test,stacked_predictions,stacked_test_predictions)\n stacked_predictions=result['pred']\n stacked_test_predictions=result['test']\n result=fit_stack(clf4, training, valid, ytraining,X_test,stacked_predictions,stacked_test_predictions)\n stacked_predictions=result['pred']\n stacked_test_predictions=result['test']\n result=fit_stack(clf3, training, valid, ytraining,X_test,stacked_predictions,stacked_test_predictions)\n stacked_predictions=result['pred']\n stacked_test_predictions=result['test']\n meta_model=KNeighborsClassifier(n_neighbors=10)\n meta_model.fit(stacked_predictions.reshape(-1,3),yvalid)\n final_predictions=meta_model.predict(stacked_test_predictions.reshape(-1,3))\n acc=accuracy_score(y_test,final_predictions)\n accuracies.append(acc)\n print(accuracies)\n mean_acc=sum(accuracies) / float(len(accuracies))\n print(\"Mean %0.3f\" % mean_acc)",
"_____no_output_____"
],
[
"stacked1()",
"[0.9301910708417446, 0.9301910708417446, 0.9301910708417446, 0.9301910708417446, 0.9301910708417446]\nMean 0.930\n"
]
],
[
[
"### How did to combine the models? Cross-validate the model. How well did it do? ",
"_____no_output_____"
],
[
"Cross validation did not increase the acccuray. It decreased the accuracy by approx by 1.5%. \nBut this gives us a more clear picture of the actual accuracy of the model which around 94%",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
]
] |
c50188b0fdd327c9540d768512e66d735fec7796
| 12,186 |
ipynb
|
Jupyter Notebook
|
site/en/r2/tutorials/images/intro_to_cnns.ipynb
|
ecrows/docs
|
d169cb190d4907044f750d669303bd9af6b7650b
|
[
"Apache-2.0"
] | null | null | null |
site/en/r2/tutorials/images/intro_to_cnns.ipynb
|
ecrows/docs
|
d169cb190d4907044f750d669303bd9af6b7650b
|
[
"Apache-2.0"
] | null | null | null |
site/en/r2/tutorials/images/intro_to_cnns.ipynb
|
ecrows/docs
|
d169cb190d4907044f750d669303bd9af6b7650b
|
[
"Apache-2.0"
] | null | null | null | 32.935135 | 467 | 0.542015 |
[
[
[
"##### Copyright 2019 The TensorFlow Authors.",
"_____no_output_____"
]
],
[
[
"#@title Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# https://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.",
"_____no_output_____"
]
],
[
[
"# Convolutional Neural Networks",
"_____no_output_____"
],
[
"<table class=\"tfo-notebook-buttons\" align=\"left\">\n <td>\n <a target=\"_blank\" href=\"https://www.tensorflow.org/beta/tutorials/images/intro_to_cnns\">\n <img src=\"https://www.tensorflow.org/images/tf_logo_32px.png\" />\n View on TensorFlow.org</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://colab.research.google.com/github/tensorflow/docs/blob/master/site/en/r2/tutorials/images/intro_to_cnns.ipynb\">\n <img src=\"https://www.tensorflow.org/images/colab_logo_32px.png\" />\n Run in Google Colab</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://github.com/tensorflow/docs/blob/master/site/en/r2/tutorials/images/intro_to_cnns.ipynb\">\n <img src=\"https://www.tensorflow.org/images/GitHub-Mark-32px.png\" />\n View source on GitHub</a>\n </td>\n <td>\n <a href=\"https://storage.googleapis.com/tensorflow_docs/site/en/r2/tutorials/images/intro_to_cnns.ipynb\"><img src=\"https://www.tensorflow.org/images/download_logo_32px.png\" />Download notebook</a>\n </td>\n</table>",
"_____no_output_____"
],
[
"This tutorial demonstrates training a simple [Convolutional Neural Network](https://developers.google.com/machine-learning/glossary/#convolutional_neural_network) (CNN) to classify MNIST digits. This simple network will achieve over 99% accuracy on the MNIST test set. Because this tutorial uses the [Keras Sequential API](https://www.tensorflow.org/guide/keras), creating and training our model will take just a few lines of code.\n\nNote: CNNs train faster with a GPU. If you are running this notebook with Colab, you can enable the free GPU via * Edit -> Notebook settings -> Hardware accelerator -> GPU*.",
"_____no_output_____"
],
[
"### Import TensorFlow",
"_____no_output_____"
]
],
[
[
"from __future__ import absolute_import, division, print_function, unicode_literals\n\n!pip install tensorflow-gpu==2.0.0-beta0\nimport tensorflow as tf\n\nfrom tensorflow.keras import datasets, layers, models",
"_____no_output_____"
]
],
[
[
"### Download and prepare the MNIST dataset",
"_____no_output_____"
]
],
[
[
"(train_images, train_labels), (test_images, test_labels) = datasets.mnist.load_data()\n\ntrain_images = train_images.reshape((60000, 28, 28, 1))\ntest_images = test_images.reshape((10000, 28, 28, 1))\n\n# Normalize pixel values to be between 0 and 1\ntrain_images, test_images = train_images / 255.0, test_images / 255.0",
"_____no_output_____"
]
],
[
[
"### Create the convolutional base",
"_____no_output_____"
],
[
"The 6 lines of code below define the convolutional base using a common pattern: a stack of [Conv2D](https://www.tensorflow.org/api_docs/python/tf/keras/layers/Conv2D) and [MaxPooling2D](https://www.tensorflow.org/api_docs/python/tf/keras/layers/MaxPool2D) layers.\n\nAs input, a CNN takes tensors of shape (image_height, image_width, color_channels), ignoring the batch size. If you are new to color channels, MNIST has one (because the images are grayscale), whereas a color image has three (R,G,B). In this example, we will configure our CNN to process inputs of shape (28, 28, 1), which is the format of MNIST images. We do this by passing the argument `input_shape` to our first layer.\n\n",
"_____no_output_____"
]
],
[
[
"model = models.Sequential()\nmodel.add(layers.Conv2D(32, (3, 3), activation='relu', input_shape=(28, 28, 1)))\nmodel.add(layers.MaxPooling2D((2, 2)))\nmodel.add(layers.Conv2D(64, (3, 3), activation='relu'))\nmodel.add(layers.MaxPooling2D((2, 2)))\nmodel.add(layers.Conv2D(64, (3, 3), activation='relu'))",
"_____no_output_____"
]
],
[
[
"Let display the architecture of our model so far.",
"_____no_output_____"
]
],
[
[
"model.summary()",
"_____no_output_____"
]
],
[
[
"Above, you can see that the output of every Conv2D and MaxPooling2D layer is a 3D tensor of shape (height, width, channels). The width and height dimensions tend to shrink as we go deeper in the network. The number of output channels for each Conv2D layer is controlled by the first argument (e.g., 32 or 64). Typically, as the width and height shrink, we can afford (computationally) to add more output channels in each Conv2D layer.",
"_____no_output_____"
],
[
"### Add Dense layers on top\nTo complete our model, we will feed the last output tensor from the convolutional base (of shape (3, 3, 64)) into one or more Dense layers to perform classification. Dense layers take vectors as input (which are 1D), while the current output is a 3D tensor. First, we will flatten (or unroll) the 3D output to 1D, then add one or more Dense layers on top. MNIST has 10 output classes, so we use a final Dense layer with 10 outputs and a softmax activation.",
"_____no_output_____"
]
],
[
[
"model.add(layers.Flatten())\nmodel.add(layers.Dense(64, activation='relu'))\nmodel.add(layers.Dense(10, activation='softmax'))",
"_____no_output_____"
]
],
[
[
" Here's the complete architecture of our model.",
"_____no_output_____"
]
],
[
[
"model.summary()",
"_____no_output_____"
]
],
[
[
"As you can see, our (3, 3, 64) outputs were flattened into vectors of shape (576) before going through two Dense layers.",
"_____no_output_____"
],
[
"### Compile and train the model",
"_____no_output_____"
]
],
[
[
"model.compile(optimizer='adam',\n loss='sparse_categorical_crossentropy',\n metrics=['accuracy'])\n\nmodel.fit(train_images, train_labels, epochs=5)",
"_____no_output_____"
]
],
[
[
"### Evaluate the model",
"_____no_output_____"
]
],
[
[
"test_loss, test_acc = model.evaluate(test_images, test_labels)",
"_____no_output_____"
],
[
"print(test_acc)",
"_____no_output_____"
]
],
[
[
"As you can see, our simple CNN has achieved a test accuracy of over 99%. Not bad for a few lines of code! For another style of writing a CNN (using the Keras Subclassing API and a GradientTape) head [here](https://github.com/tensorflow/docs/blob/master/site/en/r2/tutorials/quickstart/advanced.ipynb).",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
c5019bbbd717e510c8bca8bb7bee4348e407a599
| 183,353 |
ipynb
|
Jupyter Notebook
|
notebooks/04_Sensitivity_Analysis.ipynb
|
thinkingmachines/geoai-immap
|
99634812602db6ceec9f55bfbacbd917eec2c407
|
[
"MIT"
] | 23 |
2020-08-03T06:27:12.000Z
|
2022-03-07T17:44:37.000Z
|
notebooks/04_Sensitivity_Analysis.ipynb
|
thinkingmachines/geoai-immap
|
99634812602db6ceec9f55bfbacbd917eec2c407
|
[
"MIT"
] | null | null | null |
notebooks/04_Sensitivity_Analysis.ipynb
|
thinkingmachines/geoai-immap
|
99634812602db6ceec9f55bfbacbd917eec2c407
|
[
"MIT"
] | 5 |
2020-08-07T03:52:24.000Z
|
2022-01-19T15:44:09.000Z
| 216.21816 | 42,800 | 0.893375 |
[
[
[
"# Sensitivity Analysis",
"_____no_output_____"
]
],
[
[
"import os\nimport itertools\nimport random\n\nimport pandas as pd\nimport numpy as np\nimport scipy\n\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nsns.set(style=\"whitegrid\")\n\nimport sys\nsys.path.insert(0, '../utils')\nimport model_utils\nimport geoutils\n\nimport logging\nimport warnings\nlogging.getLogger().setLevel(logging.ERROR)\nwarnings.filterwarnings(\"ignore\")\n\nSEED = 42\n\n%load_ext autoreload\n%autoreload 2",
"_____no_output_____"
]
],
[
[
"## File Locations",
"_____no_output_____"
]
],
[
[
"lr_index, rf_index, svc_index = 7, 3, 3\noutput_dir = \"../outputs/\"\nneg_samples_strs = ['10k', '30k', '50k']\nneg_samples_dirs = [\n output_dir + '10k_results/',\n output_dir + '30k_results/',\n output_dir + '50k_results/'\n]\nmodel_types = [\n 'logistic_regression', \n 'random_forest', \n 'linear_svc'\n]",
"_____no_output_____"
]
],
[
[
"## Load Results",
"_____no_output_____"
]
],
[
[
"results_dict = model_utils.load_neg_sample_results(model_types, neg_samples_strs, neg_samples_dirs)\nresults_dict['logistic_regression']['10k_per_area'][0]['pixel_preds'][lr_index].head(3)",
"_____no_output_____"
]
],
[
[
"## Generate Sensitivity Analysis Matrix",
"_____no_output_____"
]
],
[
[
"lr_area_dict = {\n '10k LR' : results_dict['logistic_regression']['10k_per_area'],\n '30k LR' : results_dict['logistic_regression']['30k_per_area'],\n '50k LR' : results_dict['logistic_regression']['50k_per_area'],\n}\nmodel_utils.generate_iou_matrix_per_area(\n lr_area_dict, lr_index, model_utils.AREA_CODES, percent=0.20\n)",
"_____no_output_____"
],
[
"rf_area_dict = {\n '10k RF' : results_dict['random_forest']['10k_per_area'],\n '30k RF' : results_dict['random_forest']['30k_per_area'],\n '50k RF' : results_dict['random_forest']['50k_per_area'],\n}\nmodel_utils.generate_iou_matrix_per_area(\n rf_area_dict, rf_index, model_utils.AREA_CODES, percent=0.20\n)",
"_____no_output_____"
],
[
"svc_area_dict = {\n '10k SVC' : results_dict['linear_svc']['10k_per_area'],\n '30k SVC' : results_dict['linear_svc']['30k_per_area'],\n '50k SVC' : results_dict['linear_svc']['50k_per_area'],\n}\nmodel_utils.generate_iou_matrix_per_area(\n svc_area_dict, svc_index, model_utils.AREA_CODES, percent=0.20\n)",
"_____no_output_____"
]
],
[
[
"## Sensitivity Analysis on Unseen Test Set",
"_____no_output_____"
]
],
[
[
"from tqdm import tqdm\nfrom sklearn.linear_model import LogisticRegression\nfrom sklearn.ensemble import RandomForestClassifier\nfrom sklearn.pipeline import Pipeline\nfrom sklearn.preprocessing import (\n MinMaxScaler,\n StandardScaler\n)\nSEED = 42",
"_____no_output_____"
]
],
[
[
"### File Locations",
"_____no_output_____"
]
],
[
[
"version = '20200509'\ndata_dir = \"../data/\"\n\ninput_file = data_dir + '{}_dataset.csv'.format(version)\noutput_dir = \"../outputs/sensitivity/\"\ntmp_dir = data_dir + 'tmp/'\n\nimages_dir = data_dir + 'images/'\nindices_dir = data_dir + 'indices/'\n\nif not os.path.exists(output_dir):\n os.makedirs(output_dir)\nif not os.path.exists(tmp_dir):\n os.makedirs(tmp_dir)",
"_____no_output_____"
],
[
"#!gsutil -q -m cp gs://immap-images/20200525/medellin_*.tif {images_dir}\n#!gsutil -q -m cp gs://immap-indices/20200525/indices_medellin_*.tif {indices_dir}\n#!gsutil -q -m cp gs://immap-images/20200518/cali_*.tif {images_dir}\n#!gsutil -q -m cp gs://immap-indices/20200518/indices_cali_*.tif {indices_dir}\n#!gsutil -q -m cp gs://immap-images/20200508/malambo_*.tif {images_dir}\n#!gsutil -q -m cp gs://immap-indices/20200508/indices_malambo_*.tif {indices_dir}",
"_____no_output_____"
]
],
[
[
"### Load Data",
"_____no_output_____"
]
],
[
[
"raw_data = pd.read_csv(input_file).reset_index(drop=True)\nprint('Data dimensions: {}'.format(raw_data.shape))\nraw_data.head(3)",
"Data dimensions: (1029869, 69)\n"
]
],
[
[
"### Check Hyperparameters of Best Model",
"_____no_output_____"
]
],
[
[
"print('Logistic Regression Parameters: {}'.format(\n results_dict['logistic_regression']['30k']['labels'][lr_index]\n))\nprint('Random Forest Parameters: {}'.format(\n results_dict['random_forest']['30k']['labels'][rf_index]\n))",
"Logistic Regression Parameters: penalty=l1, C=1.000\nRandom Forest Parameters: n_estimators=800, max_depth=12, min_samples_split=15, min_samples_leaf=2\n"
]
],
[
[
"### Instantiate Models",
"_____no_output_____"
]
],
[
[
"lr = LogisticRegression(penalty='l1', C=1.0)\nrf = RandomForestClassifier(\n n_estimators=800, \n max_depth=12, \n min_samples_split=15,\n min_samples_leaf=2,\n random_state=42\n)\n\nneg_samples_list = [10000, 30000, 50000]\nmodels, model_strs = [lr, rf], ['LR', 'RF']\nareas = ['medellin', 'cali', 'malambo']\narea_dict = geoutils.get_filepaths(areas, images_dir, indices_dir)",
"_____no_output_____"
]
],
[
[
"### Run Model for 10k, 30k, and 50k Negative Samples",
"_____no_output_____"
]
],
[
[
"for num_neg_samples, neg_samples_str in zip(neg_samples_list, neg_samples_strs):\n for model, model_str in zip(models, model_strs):\n model, features = model_utils.train_model(model, raw_data, num_neg_samples, SEED)\n for area in areas:\n output = output_dir + '{}_{}_{}_{}.tif'.format(version, area, model_str, neg_samples_str)\n geoutils.get_preds_windowing(\n area=area, \n area_dict=area_dict,\n model=model, \n tmp_dir=tmp_dir,\n best_features=features, \n output=output, \n grid_blocks=9,\n threshold=0\n )",
"100%|██████████| 81/81 [01:42<00:00, 1.27s/it]\n100%|██████████| 81/81 [02:20<00:00, 1.74s/it]\n100%|██████████| 81/81 [00:27<00:00, 2.97it/s]\n100%|██████████| 81/81 [06:45<00:00, 5.00s/it]\n100%|██████████| 81/81 [09:34<00:00, 7.09s/it]\n100%|██████████| 81/81 [01:49<00:00, 1.35s/it]\n100%|██████████| 81/81 [01:42<00:00, 1.27s/it]\n100%|██████████| 81/81 [02:20<00:00, 1.74s/it]\n100%|██████████| 81/81 [00:27<00:00, 2.98it/s]\n100%|██████████| 81/81 [06:48<00:00, 5.05s/it]\n100%|██████████| 81/81 [09:41<00:00, 7.18s/it]\n100%|██████████| 81/81 [01:49<00:00, 1.35s/it]\n100%|██████████| 81/81 [01:46<00:00, 1.31s/it]\n100%|██████████| 81/81 [02:28<00:00, 1.84s/it]\n100%|██████████| 81/81 [00:29<00:00, 2.72it/s]\n100%|██████████| 81/81 [06:48<00:00, 5.05s/it]\n100%|██████████| 81/81 [09:42<00:00, 7.19s/it]\n100%|██████████| 81/81 [01:49<00:00, 1.35s/it]\n"
],
[
"for file in os.listdir(output_dir):\n if '.ipynb' not in file:\n out_file = output_dir + file\n !gsutil -q cp {out_file} gs://immap-results/probmaps/",
"_____no_output_____"
]
],
[
[
"## Test on Unseen Data",
"_____no_output_____"
]
],
[
[
"import geopandas as gpd",
"_____no_output_____"
],
[
"areas = ['medellin', 'malambo', 'cali']\n\ndata_dir = \"../data/\"\ngrid_dirs = [data_dir + 'grids/grid-' + area + '.gpkg' for area in areas]\ngrid_gpkgs = {area: gpd.read_file(file) for area, file in zip(areas, grid_dirs)}\n\ngrid_gpkgs['medellin'].head(3)",
"_____no_output_____"
],
[
"lr_area_dict = {\n '10k LR' : {area: {'grid_preds' : [grid_gpkgs[area][['id', 'LR_10k_mean']]]} for area in areas},\n '30k LR' : {area: {'grid_preds' : [grid_gpkgs[area][['id', 'LR_30k_mean']]]} for area in areas},\n '50k LR' : {area: {'grid_preds' : [grid_gpkgs[area][['id', 'LR_50k_mean']]]} for area in areas},\n}\nmodel_utils.generate_iou_matrix_per_area(\n lr_area_dict, 0, areas, percent=0.20, nrows=1, ncols=3, figsize=(8,2.5)\n)",
"_____no_output_____"
],
[
"rf_area_dict = {\n '10k RF' : {area: {'grid_preds' : [grid_gpkgs[area][['id', 'RF_10K_mean']]]} for area in areas},\n '30k RF' : {area: {'grid_preds' : [grid_gpkgs[area][['id', 'RF_30K_mean']]]} for area in areas},\n '50k RF' : {area: {'grid_preds' : [grid_gpkgs[area][['id', 'RF_50K_mean']]]} for area in areas},\n}\nmodel_utils.generate_iou_matrix_per_area(\n rf_area_dict, 0, areas, percent=0.20, nrows=1, ncols=3, figsize=(8,2.5)\n)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
c5019ed758f501cee5cbcf645216db3712517b30
| 178,621 |
ipynb
|
Jupyter Notebook
|
TrainingScript/ssd_keras/weight_sampling_tutorial.ipynb
|
chaliburton/CarND-Capstone
|
4fcf4f69a7a4892946bb2a97ada0e3c4afcd485f
|
[
"MIT"
] | null | null | null |
TrainingScript/ssd_keras/weight_sampling_tutorial.ipynb
|
chaliburton/CarND-Capstone
|
4fcf4f69a7a4892946bb2a97ada0e3c4afcd485f
|
[
"MIT"
] | 22 |
2020-01-28T23:14:49.000Z
|
2022-03-12T00:12:09.000Z
|
TrainingScript/ssd_keras/weight_sampling_tutorial.ipynb
|
chaliburton/CarND-Capstone
|
4fcf4f69a7a4892946bb2a97ada0e3c4afcd485f
|
[
"MIT"
] | null | null | null | 203.905251 | 139,484 | 0.890892 |
[
[
[
"# Weight Sampling Tutorial\n\nIf you want to fine-tune one of the trained original SSD models on your own dataset, chances are that your dataset doesn't have the same number of classes as the trained model you're trying to fine-tune.\n\nThis notebook explains a few options for how to deal with this situation. In particular, one solution is to sub-sample (or up-sample) the weight tensors of all the classification layers so that their shapes correspond to the number of classes in your dataset.\n\nThis notebook explains how this is done.",
"_____no_output_____"
],
[
"## 0. Our example\n\nI'll use a concrete example to make the process clear, but of course the process explained here is the same for any dataset.\n\nConsider the following example. You have a dataset on road traffic objects. Let this dataset contain annotations for the following object classes of interest:\n\n`['car', 'truck', 'pedestrian', 'bicyclist', 'traffic_light', 'motorcycle', 'bus', 'stop_sign']`\n\nThat is, your dataset contains annotations for 8 object classes.\n\nYou would now like to train an SSD300 on this dataset. However, instead of going through all the trouble of training a new model from scratch, you would instead like to use the fully trained original SSD300 model that was trained on MS COCO and fine-tune it on your dataset.\n\nThe problem is: The SSD300 that was trained on MS COCO predicts 80 different classes, but your dataset has only 8 classes. The weight tensors of the classification layers of the MS COCO model don't have the right shape for your model that is supposed to learn only 8 classes. Bummer.\n\nSo what options do we have?\n\n### Option 1: Just ignore the fact that we need only 8 classes\n\nThe maybe not so obvious but totally obvious option is: We could just ignore the fact that the trained MS COCO model predicts 80 different classes, but we only want to fine-tune it on 8 classes. We could simply map the 8 classes in our annotated dataset to any 8 indices out of the 80 that the MS COCO model predicts. The class IDs in our dataset could be indices 1-8, they could be the indices `[0, 3, 8, 1, 2, 10, 4, 6, 12]`, or any other 8 out of the 80. Whatever we would choose them to be. The point is that we would be training only 8 out of every 80 neurons that predict the class for a given box and the other 72 would simply not be trained. Nothing would happen to them, because the gradient for them would always be zero, because these indices don't appear in our dataset.\n\nThis would work, and it wouldn't even be a terrible option. Since only 8 out of the 80 classes would get trained, the model might get gradually worse at predicting the other 72 clases, but we don't care about them anyway, at least not right now. And if we ever realize that we now want to predict more than 8 different classes, our model would be expandable in that sense. Any new class we want to add could just get any one of the remaining free indices as its ID. We wouldn't need to change anything about the model, it would just be a matter of having the dataset annotated accordingly.\n\nStill, in this example we don't want to take this route. We don't want to carry around the computational overhead of having overly complex classifier layers, 90 percent of which we don't use anyway, but still their whole output needs to be computed in every forward pass.\n\nSo what else could we do instead?\n\n### Option 2: Just ignore those weights that are causing problems\n\nWe could build a new SSD300 with 8 classes and load into it the weights of the MS COCO SSD300 for all layers except the classification layers. Would that work? Yes, that would work. The only conflict is with the weights of the classification layers, and we can avoid this conflict by simply ignoring them. While this solution would be easy, it has a significant downside: If we're not loading trained weights for the classification layers of our new SSD300 model, then they will be initialized randomly. We'd still benefit from the trained weights for all the other layers, but the classifier layers would need to be trained from scratch.\n\nNot the end of the world, but we like pre-trained stuff, because it saves us a lot of training time. So what else could we do?\n\n### Option 3: Sub-sample the weights that are causing problems\n\nInstead of throwing the problematic weights away like in option 2, we could also sub-sample them. If the weight tensors of the classification layers of the MS COCO model don't have the right shape for our new model, we'll just **make** them have the right shape. This way we can still benefit from the pre-trained weights in those classification layers. Seems much better than option 2.\n\nThe great thing in this example is: MS COCO happens to contain all of the eight classes that we care about. So when we sub-sample the weight tensors of the classification layers, we won't just do so randomly. Instead, we'll pick exactly those elements from the tensor that are responsible for the classification of the 8 classes that we care about.\n\nHowever, even if the classes in your dataset were entirely different from the classes in any of the fully trained models, it would still make a lot of sense to use the weights of the fully trained model. Any trained weights are always a better starting point for the training than random initialization, even if your model will be trained on entirely different object classes.\n\nAnd of course, in case you happen to have the opposite problem, where your dataset has **more** classes than the trained model you would like to fine-tune, then you can simply do the same thing in the opposite direction: Instead of sub-sampling the classification layer weights, you would then **up-sample** them. Works just the same way as what we'll be doing below.\n\nLet's get to it.",
"_____no_output_____"
]
],
[
[
"import h5py\nimport numpy as np\nimport shutil\n\nfrom misc_utils.tensor_sampling_utils import sample_tensors",
"_____no_output_____"
]
],
[
[
"## 1. Load the trained weights file and make a copy\n\nFirst, we'll load the HDF5 file that contains the trained weights that we need (the source file). In our case this is \"`VGG_coco_SSD_300x300_iter_400000.h5`\" (download link available in the README of this repo), which are the weights of the original SSD300 model that was trained on MS COCO.\n\nThen, we'll make a copy of that weights file. That copy will be our output file (the destination file).",
"_____no_output_____"
]
],
[
[
"# TODO: Set the path for the source weights file you want to load.\ndataFolder = '../Models/'\n\nweights_source_path = dataFolder + 'VGG_coco_SSD_300x300_iter_400000.h5'\n\n# TODO: Set the path and name for the destination weights file\n# that you want to create.\n\nweights_destination_path = dataFolder + 'VGG_coco_SSD_300x300_iter_400000_subsampled_3_classes3.h5'\n\n# Make a copy of the weights file.\nshutil.copy(weights_source_path, weights_destination_path)",
"_____no_output_____"
],
[
"# Load both the source weights file and the copy we made.\n# We will load the original weights file in read-only mode so that we can't mess up anything.\nweights_source_file = h5py.File(weights_source_path, 'r')\nweights_destination_file = h5py.File(weights_destination_path)",
"/home/anaconda3/envs/deep-learning/lib/python3.6/site-packages/ipykernel_launcher.py:4: H5pyDeprecationWarning: The default file mode will change to 'r' (read-only) in h5py 3.0. To suppress this warning, pass the mode you need to h5py.File(), or set the global default h5.get_config().default_file_mode, or set the environment variable H5PY_DEFAULT_READONLY=1. Available modes are: 'r', 'r+', 'w', 'w-'/'x', 'a'. See the docs for details.\n after removing the cwd from sys.path.\n"
]
],
[
[
"## 2. Figure out which weight tensors we need to sub-sample\n\nNext, we need to figure out exactly which weight tensors we need to sub-sample. As mentioned above, the weights for all layers except the classification layers are fine, we don't need to change anything about those.\n\nSo which are the classification layers in SSD300? Their names are:",
"_____no_output_____"
]
],
[
[
"classifier_names = ['conv4_3_norm_mbox_conf',\n 'fc7_mbox_conf',\n 'conv6_2_mbox_conf',\n 'conv7_2_mbox_conf',\n 'conv8_2_mbox_conf',\n 'conv9_2_mbox_conf']",
"_____no_output_____"
]
],
[
[
"## 3. Figure out which slices to pick\n\nThe following section is optional. I'll look at one classification layer and explain what we want to do, just for your understanding. If you don't care about that, just skip ahead to the next section.\n\nWe know which weight tensors we want to sub-sample, but we still need to decide which (or at least how many) elements of those tensors we want to keep. Let's take a look at the first of the classifier layers, \"`conv4_3_norm_mbox_conf`\". Its two weight tensors, the kernel and the bias, have the following shapes:",
"_____no_output_____"
]
],
[
[
"conv4_3_norm_mbox_conf_kernel = weights_source_file[classifier_names[0]][classifier_names[0]]['kernel:0']\nconv4_3_norm_mbox_conf_bias = weights_source_file[classifier_names[0]][classifier_names[0]]['bias:0']\n\nprint(\"Shape of the '{}' weights:\".format(classifier_names[0]))\nprint()\nprint(\"kernel:\\t\", conv4_3_norm_mbox_conf_kernel.shape)\nprint(\"bias:\\t\", conv4_3_norm_mbox_conf_bias.shape)",
"Shape of the 'conv4_3_norm_mbox_conf' weights:\n\nkernel:\t (3, 3, 512, 324)\nbias:\t (324,)\n"
]
],
[
[
"So the last axis has 324 elements. Why is that?\n\n- MS COCO has 80 classes, but the model also has one 'backgroud' class, so that makes 81 classes effectively.\n- The 'conv4_3_norm_mbox_loc' layer predicts 4 boxes for each spatial position, so the 'conv4_3_norm_mbox_conf' layer has to predict one of the 81 classes for each of those 4 boxes.\n\nThat's why the last axis has 4 * 81 = 324 elements.\n\nSo how many elements do we want in the last axis for this layer?\n\nLet's do the same calculation as above:\n\n- Our dataset has 8 classes, but our model will also have a 'background' class, so that makes 9 classes effectively.\n- We need to predict one of those 9 classes for each of the four boxes at each spatial position.\n\nThat makes 4 * 9 = 36 elements.\n\nNow we know that we want to keep 36 elements in the last axis and leave all other axes unchanged. But which 36 elements out of the original 324 elements do we want?\n\nShould we just pick them randomly? If the object classes in our dataset had absolutely nothing to do with the classes in MS COCO, then choosing those 36 elements randomly would be fine (and the next section covers this case, too). But in our particular example case, choosing these elements randomly would be a waste. Since MS COCO happens to contain exactly the 8 classes that we need, instead of sub-sampling randomly, we'll just take exactly those elements that were trained to predict our 8 classes.\n\nHere are the indices of the 9 classes in MS COCO that we are interested in:\n\n`[0, 1, 2, 3, 4, 6, 8, 10, 12]`\n\nThe indices above represent the following classes in the MS COCO datasets:\n\n`['background', 'person', 'bicycle', 'car', 'motorcycle', 'bus', 'truck', 'traffic_light', 'stop_sign']`\n\nHow did I find out those indices? I just looked them up in the annotations of the MS COCO dataset.\n\nWhile these are the classes we want, we don't want them in this order. In our dataset, the classes happen to be in the following order as stated at the top of this notebook:\n\n`['background', 'car', 'truck', 'pedestrian', 'bicyclist', 'traffic_light', 'motorcycle', 'bus', 'stop_sign']`\n\nFor example, '`traffic_light`' is class ID 5 in our dataset but class ID 10 in the SSD300 MS COCO model. So the order in which I actually want to pick the 9 indices above is this:\n\n`[0, 3, 8, 1, 2, 10, 4, 6, 12]`\n\nSo out of every 81 in the 324 elements, I want to pick the 9 elements above. This gives us the following 36 indices:",
"_____no_output_____"
]
],
[
[
"n_classes_source = 81\nclasses_of_interest = [0, 3, 8, 1, 2, 10, 4, 6, 12]\nclasses_of_interest = [0, 10, 10, 10]\n \nsubsampling_indices = []\nfor i in range(int(324/n_classes_source)):\n indices = np.array(classes_of_interest) + i * n_classes_source\n subsampling_indices.append(indices)\nsubsampling_indices = list(np.concatenate(subsampling_indices))\n\nprint(subsampling_indices)",
"[0, 10, 10, 10, 81, 91, 91, 91, 162, 172, 172, 172, 243, 253, 253, 253]\n"
]
],
[
[
"These are the indices of the 36 elements that we want to pick from both the bias vector and from the last axis of the kernel tensor.\n\nThis was the detailed example for the '`conv4_3_norm_mbox_conf`' layer. And of course we haven't actually sub-sampled the weights for this layer yet, we have only figured out which elements we want to keep. The piece of code in the next section will perform the sub-sampling for all the classifier layers.",
"_____no_output_____"
],
[
"## 4. Sub-sample the classifier weights\n\nThe code in this section iterates over all the classifier layers of the source weights file and performs the following steps for each classifier layer:\n\n1. Get the kernel and bias tensors from the source weights file.\n2. Compute the sub-sampling indices for the last axis. The first three axes of the kernel remain unchanged.\n3. Overwrite the corresponding kernel and bias tensors in the destination weights file with our newly created sub-sampled kernel and bias tensors.\n\nThe second step does what was explained in the previous section.\n\nIn case you want to **up-sample** the last axis rather than sub-sample it, simply set the `classes_of_interest` variable below to the length you want it to have. The added elements will be initialized either randomly or optionally with zeros. Check out the documentation of `sample_tensors()` for details.",
"_____no_output_____"
]
],
[
[
"# TODO: Set the number of classes in the source weights file. Note that this number must include\n# the background class, so for MS COCO's 80 classes, this must be 80 + 1 = 81.\nn_classes_source = 81\n# TODO: Set the indices of the classes that you want to pick for the sub-sampled weight tensors.\n# In case you would like to just randomly sample a certain number of classes, you can just set\n# `classes_of_interest` to an integer instead of the list below. Either way, don't forget to\n# include the background class. That is, if you set an integer, and you want `n` positive classes,\n# then you must set `classes_of_interest = n + 1`.\nclasses_of_interest = [0, 10, 13, 14]\n#classes_of_interest = [0, 3, 8, 1, 2, 10, 4, 6, 12]\n# classes_of_interest = 9 # Uncomment this in case you want to just randomly sub-sample the last axis instead of providing a list of indices.\n\nfor name in classifier_names:\n # Get the trained weights for this layer from the source HDF5 weights file.\n kernel = weights_source_file[name][name]['kernel:0'].value\n bias = weights_source_file[name][name]['bias:0'].value\n\n # Get the shape of the kernel. We're interested in sub-sampling\n # the last dimension, 'o'.\n height, width, in_channels, out_channels = kernel.shape\n \n # Compute the indices of the elements we want to sub-sample.\n # Keep in mind that each classification predictor layer predicts multiple\n # bounding boxes for every spatial location, so we want to sub-sample\n # the relevant classes for each of these boxes.\n if isinstance(classes_of_interest, (list, tuple)):\n subsampling_indices = []\n for i in range(int(out_channels/n_classes_source)):\n indices = np.array(classes_of_interest) + i * n_classes_source\n subsampling_indices.append(indices)\n subsampling_indices = list(np.concatenate(subsampling_indices))\n elif isinstance(classes_of_interest, int):\n subsampling_indices = int(classes_of_interest * (out_channels/n_classes_source))\n else:\n raise ValueError(\"`classes_of_interest` must be either an integer or a list/tuple.\")\n \n # Sub-sample the kernel and bias.\n # The `sample_tensors()` function used below provides extensive\n # documentation, so don't hesitate to read it if you want to know\n # what exactly is going on here.\n new_kernel, new_bias = sample_tensors(weights_list=[kernel, bias],\n sampling_instructions=[height, width, in_channels, subsampling_indices],\n axes=[[3]], # The one bias dimension corresponds to the last kernel dimension.\n init=['gaussian', 'zeros'],\n mean=0.0,\n stddev=0.005)\n \n # Delete the old weights from the destination file.\n del weights_destination_file[name][name]['kernel:0']\n del weights_destination_file[name][name]['bias:0']\n # Create new datasets for the sub-sampled weights.\n weights_destination_file[name][name].create_dataset(name='kernel:0', data=new_kernel)\n weights_destination_file[name][name].create_dataset(name='bias:0', data=new_bias)\n\n# Make sure all data is written to our output file before this sub-routine exits.\nweights_destination_file.flush()",
"/home/anaconda3/envs/deep-learning/lib/python3.6/site-packages/ipykernel_launcher.py:15: H5pyDeprecationWarning: dataset.value has been deprecated. Use dataset[()] instead.\n from ipykernel import kernelapp as app\n/home/anaconda3/envs/deep-learning/lib/python3.6/site-packages/ipykernel_launcher.py:16: H5pyDeprecationWarning: dataset.value has been deprecated. Use dataset[()] instead.\n app.launch_new_instance()\n"
]
],
[
[
"That's it, we're done.\n\nLet's just quickly inspect the shapes of the weights of the '`conv4_3_norm_mbox_conf`' layer in the destination weights file:",
"_____no_output_____"
]
],
[
[
"conv4_3_norm_mbox_conf_kernel = weights_destination_file[classifier_names[0]][classifier_names[0]]['kernel:0']\nconv4_3_norm_mbox_conf_bias = weights_destination_file[classifier_names[0]][classifier_names[0]]['bias:0']\n\nprint(\"Shape of the '{}' weights:\".format(classifier_names[0]))\nprint()\nprint(\"kernel:\\t\", conv4_3_norm_mbox_conf_kernel.shape)\nprint(\"bias:\\t\", conv4_3_norm_mbox_conf_bias.shape)",
"Shape of the 'conv4_3_norm_mbox_conf' weights:\n\nkernel:\t (3, 3, 512, 16)\nbias:\t (16,)\n"
]
],
[
[
"Nice! Exactly what we wanted, 36 elements in the last axis. Now the weights are compatible with our new SSD300 model that predicts 8 positive classes.\n\nThis is the end of the relevant part of this tutorial, but we can do one more thing and verify that the sub-sampled weights actually work. Let's do that in the next section.",
"_____no_output_____"
],
[
"## 5. Verify that our sub-sampled weights actually work\n\nIn our example case above we sub-sampled the fully trained weights of the SSD300 model trained on MS COCO from 80 classes to just the 8 classes that we needed.\n\nWe can now create a new SSD300 with 8 classes, load our sub-sampled weights into it, and see how the model performs on a few test images that contain objects for some of those 8 classes. Let's do it.",
"_____no_output_____"
]
],
[
[
"from keras.optimizers import Adam\nfrom keras import backend as K\nfrom keras.models import load_model\n\nfrom models.keras_ssd300 import ssd_300\nfrom keras_loss_function.keras_ssd_loss import SSDLoss\nfrom keras_layers.keras_layer_AnchorBoxes import AnchorBoxes\nfrom keras_layers.keras_layer_DecodeDetections import DecodeDetections\nfrom keras_layers.keras_layer_DecodeDetectionsFast import DecodeDetectionsFast\nfrom keras_layers.keras_layer_L2Normalization import L2Normalization\n\nfrom data_generator.object_detection_2d_data_generator import DataGenerator\nfrom data_generator.object_detection_2d_photometric_ops import ConvertTo3Channels\nfrom data_generator.object_detection_2d_patch_sampling_ops import RandomMaxCropFixedAR\nfrom data_generator.object_detection_2d_geometric_ops import Resize",
"_____no_output_____"
]
],
[
[
"### 5.1. Set the parameters for the model.\n\nAs always, set the parameters for the model. We're going to set the configuration for the SSD300 MS COCO model.",
"_____no_output_____"
]
],
[
[
"img_height = 300 # Height of the input images\nimg_width = 300 # Width of the input images\nimg_channels = 3 # Number of color channels of the input images\nsubtract_mean = [123, 117, 104] # The per-channel mean of the images in the dataset\nswap_channels = [2, 1, 0] # The color channel order in the original SSD is BGR, so we should set this to `True`, but weirdly the results are better without swapping.\n# TODO: Set the number of classes.\nn_classes = 3 # Number of positive classes, e.g. 20 for Pascal VOC, 80 for MS COCO\nscales = [0.07, 0.15, 0.33, 0.51, 0.69, 0.87, 1.05] # The anchor box scaling factors used in the original SSD300 for the MS COCO datasets.\n# scales = [0.1, 0.2, 0.37, 0.54, 0.71, 0.88, 1.05] # The anchor box scaling factors used in the original SSD300 for the Pascal VOC datasets.\naspect_ratios = [[1.0, 2.0, 0.5],\n [1.0, 2.0, 0.5, 3.0, 1.0/3.0],\n [1.0, 2.0, 0.5, 3.0, 1.0/3.0],\n [1.0, 2.0, 0.5, 3.0, 1.0/3.0],\n [1.0, 2.0, 0.5],\n [1.0, 2.0, 0.5]] # The anchor box aspect ratios used in the original SSD300; the order matters\ntwo_boxes_for_ar1 = True\nsteps = [8, 16, 32, 64, 100, 300] # The space between two adjacent anchor box center points for each predictor layer.\noffsets = [0.5, 0.5, 0.5, 0.5, 0.5, 0.5] # The offsets of the first anchor box center points from the top and left borders of the image as a fraction of the step size for each predictor layer.\nclip_boxes = False # Whether or not you want to limit the anchor boxes to lie entirely within the image boundaries\nvariances = [0.1, 0.1, 0.2, 0.2] # The variances by which the encoded target coordinates are scaled as in the original implementation\nnormalize_coords = True",
"_____no_output_____"
]
],
[
[
"### 5.2. Build the model\n\nBuild the model and load our newly created, sub-sampled weights into it.",
"_____no_output_____"
]
],
[
[
"# 1: Build the Keras model\n\nK.clear_session() # Clear previous models from memory.\n\nmodel = ssd_300(image_size=(img_height, img_width, img_channels),\n n_classes=n_classes,\n mode='inference',\n l2_regularization=0.0005,\n scales=scales,\n aspect_ratios_per_layer=aspect_ratios,\n two_boxes_for_ar1=two_boxes_for_ar1,\n steps=steps,\n offsets=offsets,\n clip_boxes=clip_boxes,\n variances=variances,\n normalize_coords=normalize_coords,\n subtract_mean=subtract_mean,\n divide_by_stddev=None,\n swap_channels=swap_channels,\n confidence_thresh=0.5,\n iou_threshold=0.45,\n top_k=200,\n nms_max_output_size=400,\n return_predictor_sizes=False)\n\nprint(\"Model built.\")\n\n# 2: Load the sub-sampled weights into the model.\n\n# Load the weights that we've just created via sub-sampling.\nweights_path = weights_destination_path\n\nmodel.load_weights(weights_path, by_name=True)\n\nprint(\"Weights file loaded:\", weights_path)\n\n# 3: Instantiate an Adam optimizer and the SSD loss function and compile the model.\n\nadam = Adam(lr=0.001, beta_1=0.9, beta_2=0.999, epsilon=1e-08, decay=0.0)\n\nssd_loss = SSDLoss(neg_pos_ratio=3, alpha=1.0)\n\nmodel.compile(optimizer=adam, loss=ssd_loss.compute_loss)",
"Model built.\nWeights file loaded: ../Models/VGG_coco_SSD_300x300_iter_400000_subsampled_3_classes3.h5\n"
]
],
[
[
"### 5.3. Load some images to test our model on\n\nWe sub-sampled some of the road traffic categories from the trained SSD300 MS COCO weights, so let's try out our model on a few road traffic images. The Udacity road traffic dataset linked to in the `ssd7_training.ipynb` notebook lends itself to this task. Let's instantiate a `DataGenerator` and load the Udacity dataset. Everything here is preset already, but if you'd like to learn more about the data generator and its capabilities, take a look at the detailed tutorial in [this](https://github.com/pierluigiferrari/data_generator_object_detection_2d) repository.",
"_____no_output_____"
]
],
[
[
"dataset = DataGenerator()\n\n# TODO: Set the paths to your dataset here.\nimages_path = '../TrainData/Simulator/'\nlabels_path = '../TrainData/Simulator/label_sim_training.csv'\n\ndataset.parse_csv(images_dir=images_path,\n labels_filename=labels_path,\n input_format=['image_name', 'xmin', 'xmax', 'ymin', 'ymax', 'class_id'], # This is the order of the first six columns in the CSV file that contains the labels for your dataset. If your labels are in XML format, maybe the XML parser will be helpful, check the documentation.\n include_classes='all',\n random_sample=False)\n\nprint(\"Number of images in the dataset:\", dataset.get_dataset_size())",
"Number of images in the dataset: 487\n"
]
],
[
[
"Make sure the batch generator generates images of size `(300, 300)`. We'll first randomly crop the largest possible patch with aspect ratio 1.0 and then resize to `(300, 300)`.",
"_____no_output_____"
]
],
[
[
"convert_to_3_channels = ConvertTo3Channels()\nrandom_max_crop = RandomMaxCropFixedAR(patch_aspect_ratio=img_width/img_height)\nresize = Resize(height=img_height, width=img_width)\n\ngenerator = dataset.generate(batch_size=1,\n shuffle=True,\n transformations=[convert_to_3_channels,\n random_max_crop,\n resize],\n returns={'processed_images',\n 'processed_labels',\n 'filenames'},\n keep_images_without_gt=False)",
"_____no_output_____"
]
],
[
[
"### 5.4. Make predictions and visualize them",
"_____no_output_____"
]
],
[
[
"# Make a prediction\ny_pred = model.predict(batch_images)\n\nprint(y_pred)",
"_____no_output_____"
],
[
"# Generate samples\n\nbatch_images, batch_labels, batch_filenames = next(generator)\n\ni = 0 # Which batch item to look at\n\nprint(\"Image:\", batch_filenames[i])\nprint()\nprint(\"Ground truth boxes:\\n\")\nprint(batch_labels[i])\n\n\n# Make a prediction\n\ny_pred = model.predict(batch_images)\n\n# Decode the raw prediction.\n\ni = 0\n\nconfidence_threshold = 0.10\n\ny_pred_thresh = [y_pred[k][y_pred[k,:,1] > confidence_threshold] for k in range(y_pred.shape[0])]\n\nnp.set_printoptions(precision=2, suppress=True, linewidth=90)\nprint(\"Predicted boxes:\\n\")\nprint(' class conf xmin ymin xmax ymax')\nprint(y_pred_thresh[0])",
"Image: ../TrainData/Simulator/1_33img.jpeg\n\nGround truth boxes:\n\n[[ 2 0 16 6 116]\n [ 2 160 17 208 120]]\nPredicted boxes:\n\n class conf xmin ymin xmax ymax\n[[ 1. 0.8 160.72 17.36 207.62 120.47]]\n"
],
[
"# Visualize the predictions.\n\nfrom matplotlib import pyplot as plt\n\n%matplotlib inline\n\nplt.figure(figsize=(20,12))\nplt.imshow(batch_images[i])\n\ncurrent_axis = plt.gca()\n\nclasses = ['background', 'car', 'truck', 'pedestrian', 'bicyclist',\n 'traffic_light', 'motorcycle', 'bus', 'stop_sign'] # Just so we can print class names onto the image instead of IDs\n\n# Draw the predicted boxes in blue\nfor box in y_pred_thresh[i]:\n class_id = box[0]\n confidence = box[1]\n xmin = box[2]\n ymin = box[3]\n xmax = box[4]\n ymax = box[5]\n label = '{}: {:.2f}'.format(classes[int(class_id)], confidence)\n current_axis.add_patch(plt.Rectangle((xmin, ymin), xmax-xmin, ymax-ymin, color='blue', fill=False, linewidth=2)) \n current_axis.text(xmin, ymin, label, size='x-large', color='white', bbox={'facecolor':'blue', 'alpha':1.0})\n\n# Draw the ground truth boxes in green (omit the label for more clarity)\nfor box in batch_labels[i]:\n class_id = box[0]\n xmin = box[1]\n ymin = box[2]\n xmax = box[3]\n ymax = box[4]\n label = '{}'.format(classes[int(class_id)])\n current_axis.add_patch(plt.Rectangle((xmin, ymin), xmax-xmin, ymax-ymin, color='green', fill=False, linewidth=2)) \n #current_axis.text(box[1], box[3], label, size='x-large', color='white', bbox={'facecolor':'green', 'alpha':1.0})",
"_____no_output_____"
]
],
[
[
"Seems as if our sub-sampled weights were doing a good job, sweet. Now we can fine-tune this model on our dataset with 8 classes.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
]
] |
c501b117de922f1869f4e935a11543367dedb021
| 150,575 |
ipynb
|
Jupyter Notebook
|
Localization/Kalmanfilter_basics.ipynb
|
SaintWarri0r/PythonRobotics
|
17920dcbf489dff0f804ac541973c1c7b8ed77f0
|
[
"MIT"
] | 14 |
2021-09-26T06:37:01.000Z
|
2022-03-31T12:30:58.000Z
|
Localization/Kalmanfilter_basics.ipynb
|
SaintWarri0r/PythonRobotics
|
17920dcbf489dff0f804ac541973c1c7b8ed77f0
|
[
"MIT"
] | 65 |
2020-07-28T09:41:44.000Z
|
2022-03-14T20:01:08.000Z
|
Localization/Kalmanfilter_basics.ipynb
|
lgkimjy/PythonRobotics
|
4fe851cc1d1f2e5d21684e24267d78c5e0299e36
|
[
"MIT"
] | 7 |
2020-11-18T02:15:30.000Z
|
2022-03-13T06:47:00.000Z
| 195.806242 | 51,568 | 0.903656 |
[
[
[
"## KF Basics - Part I\n",
"_____no_output_____"
],
[
"### Introduction\n#### What is the need to describe belief in terms of PDF's?\nThis is because robot environments are stochastic. A robot environment may have cows with Tesla by side. That is a robot and it's environment cannot be deterministically modelled(e.g as a function of something like time t). In the real world sensors are also error prone, and hence there'll be a set of values with a mean and variance that it can take. Hence, we always have to model around some mean and variances associated.",
"_____no_output_____"
],
[
"#### What is Expectation of a Random Variables?\n Expectation is nothing but an average of the probabilites\n \n$$\\mathbb E[X] = \\sum_{i=1}^n p_ix_i$$\n\nIn the continous form,\n\n$$\\mathbb E[X] = \\int_{-\\infty}^\\infty x\\, f(x) \\,dx$$\n",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport random\nx=[3,1,2]\np=[0.1,0.3,0.4]\nE_x=np.sum(np.multiply(x,p))\nprint(E_x)",
"1.4000000000000001\n"
]
],
[
[
"#### What is the advantage of representing the belief as a unimodal as opposed to multimodal?\nObviously, it makes sense because we can't multiple probabilities to a car moving for two locations. This would be too confusing and the information will not be useful.",
"_____no_output_____"
],
[
"### Variance, Covariance and Correlation\n\n#### Variance\nVariance is the spread of the data. The mean does'nt tell much **about** the data. Therefore the variance tells us about the **story** about the data meaning the spread of the data.\n\n$$\\mathit{VAR}(X) = \\frac{1}{n}\\sum_{i=1}^n (x_i - \\mu)^2$$",
"_____no_output_____"
]
],
[
[
"x=np.random.randn(10)\nnp.var(x)",
"_____no_output_____"
]
],
[
[
"#### Covariance\n\nThis is for a multivariate distribution. For example, a robot in 2-D space can take values in both x and y. To describe them, a normal distribution with mean in both x and y is needed.\n\nFor a multivariate distribution, mean $\\mu$ can be represented as a matrix, \n\n$$\n\\mu = \\begin{bmatrix}\\mu_1\\\\\\mu_2\\\\ \\vdots \\\\\\mu_n\\end{bmatrix}\n$$\n\n\nSimilarly, variance can also be represented.\n\nBut an important concept is that in the same way as every variable or dimension has a variation in its values, it is also possible that there will be values on how they **together vary**. This is also a measure of how two datasets are related to each other or **correlation**.\n\nFor example, as height increases weight also generally increases. These variables are correlated. They are positively correlated because as one variable gets larger so does the other.\n\nWe use a **covariance matrix** to denote covariances of a multivariate normal distribution:\n$$\n\\Sigma = \\begin{bmatrix}\n \\sigma_1^2 & \\sigma_{12} & \\cdots & \\sigma_{1n} \\\\\n \\sigma_{21} &\\sigma_2^2 & \\cdots & \\sigma_{2n} \\\\\n \\vdots & \\vdots & \\ddots & \\vdots \\\\\n \\sigma_{n1} & \\sigma_{n2} & \\cdots & \\sigma_n^2\n \\end{bmatrix}\n$$\n\n**Diagonal** - Variance of each variable associated. \n\n**Off-Diagonal** - covariance between ith and jth variables.\n\n$$\\begin{aligned}VAR(X) = \\sigma_x^2 &= \\frac{1}{n}\\sum_{i=1}^n(X - \\mu)^2\\\\\nCOV(X, Y) = \\sigma_{xy} &= \\frac{1}{n}\\sum_{i=1}^n[(X-\\mu_x)(Y-\\mu_y)\\big]\\end{aligned}$$",
"_____no_output_____"
]
],
[
[
"x=np.random.random((3,3))\nnp.cov(x)",
"_____no_output_____"
]
],
[
[
"Covariance taking the data as **sample** with $\\frac{1}{N-1}$",
"_____no_output_____"
]
],
[
[
"x_cor=np.random.rand(1,10)\ny_cor=np.random.rand(1,10)\nnp.cov(x_cor,y_cor)",
"_____no_output_____"
]
],
[
[
"Covariance taking the data as **population** with $\\frac{1}{N}$",
"_____no_output_____"
]
],
[
[
"np.cov(x_cor,y_cor,bias=1)",
"_____no_output_____"
]
],
[
[
"### Gaussians \n\n#### Central Limit Theorem\n\nAccording to this theorem, the average of n samples of random and independent variables tends to follow a normal distribution as we increase the sample size.(Generally, for n>=30)",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nimport random\na=np.zeros((100,))\nfor i in range(100):\n x=[random.uniform(1,10) for _ in range(1000)]\n a[i]=np.sum(x,axis=0)/1000\nplt.hist(a)",
"_____no_output_____"
]
],
[
[
"#### Gaussian Distribution\nA Gaussian is a *continuous probability distribution* that is completely described with two parameters, the mean ($\\mu$) and the variance ($\\sigma^2$). It is defined as:\n\n$$ \nf(x, \\mu, \\sigma) = \\frac{1}{\\sigma\\sqrt{2\\pi}} \\exp\\big [{-\\frac{(x-\\mu)^2}{2\\sigma^2} }\\big ]\n$$\nRange is $$[-\\inf,\\inf] $$\n\n\nThis is just a function of mean($\\mu$) and standard deviation ($\\sigma$) and what gives the normal distribution the charecteristic **bell curve**. ",
"_____no_output_____"
]
],
[
[
"import matplotlib.mlab as mlab\nimport math\nimport scipy.stats\n\nmu = 0\nvariance = 5\nsigma = math.sqrt(variance)\nx = np.linspace(mu - 5*sigma, mu + 5*sigma, 100)\nplt.plot(x,scipy.stats.norm.pdf(x, mu, sigma))\nplt.show()\n",
"_____no_output_____"
]
],
[
[
"#### Why do we need Gaussian distributions?\nSince it becomes really difficult in the real world to deal with multimodal distribution as we cannot put the belief in two seperate location of the robots. This becomes really confusing and in practice impossible to comprehend. \nGaussian probability distribution allows us to drive the robots using only one mode with peak at the mean with some variance.",
"_____no_output_____"
],
[
"### Gaussian Properties\n\n**Multiplication**\n\n\nFor the measurement update in a Bayes Filter, the algorithm tells us to multiply the Prior P(X_t) and measurement P(Z_t|X_t) to calculate the posterior:\n\n$$P(X \\mid Z) = \\frac{P(Z \\mid X)P(X)}{P(Z)}$$\n\nHere for the numerator, $P(Z \\mid X),P(X)$ both are gaussian.\n\n$N(\\mu_1, \\sigma_1^2)$ and $N(\\mu_2, \\sigma_2^2)$ are their mean and variances.\n\nNew mean is \n\n$$\\mu_\\mathtt{new} = \\frac{\\mu_1 \\sigma_2^2 + \\mu_2 \\sigma_1^2}{\\sigma_1^2+\\sigma_2^2}$$\nNew variance is\n$$\\sigma_\\mathtt{new} = \\frac{\\sigma_1^2\\sigma_2^2}{\\sigma_1^2+\\sigma_2^2}$$",
"_____no_output_____"
]
],
[
[
"import matplotlib.mlab as mlab\nimport math\nmu1 = 0\nvariance1 = 2\nsigma = math.sqrt(variance1)\nx1 = np.linspace(mu1 - 3*sigma, mu1 + 3*sigma, 100)\nplt.plot(x1,scipy.stats.norm.pdf(x1, mu1, sigma),label='prior')\n\nmu2 = 10\nvariance2 = 2\nsigma = math.sqrt(variance2)\nx2 = np.linspace(mu2 - 3*sigma, mu2 + 3*sigma, 100)\nplt.plot(x2,scipy.stats.norm.pdf(x2, mu2, sigma),\"g-\",label='measurement')\n\n\nmu_new=(mu1*variance2+mu2*variance1)/(variance1+variance2)\nprint(\"New mean is at: \",mu_new)\nvar_new=(variance1*variance2)/(variance1+variance2)\nprint(\"New variance is: \",var_new)\nsigma = math.sqrt(var_new)\nx3 = np.linspace(mu_new - 3*sigma, mu_new + 3*sigma, 100)\nplt.plot(x3,scipy.stats.norm.pdf(x3, mu_new, var_new),label=\"posterior\")\nplt.legend(loc='upper left')\nplt.xlim(-10,20)\nplt.show()\n",
"New mean is at: 5.0\nNew variance is: 1.0\n"
]
],
[
[
"**Addition**\n\nThe motion step involves a case of adding up probability (Since it has to abide the Law of Total Probability). This means their beliefs are to be added and hence two gaussians. They are simply arithmetic additions of the two.\n\n$$\\begin{gathered}\\mu_x = \\mu_p + \\mu_z \\\\\n\\sigma_x^2 = \\sigma_z^2+\\sigma_p^2\\, \\square\\end{gathered}$$",
"_____no_output_____"
]
],
[
[
"import matplotlib.mlab as mlab\nimport math\nmu1 = 5\nvariance1 = 1\nsigma = math.sqrt(variance1)\nx1 = np.linspace(mu1 - 3*sigma, mu1 + 3*sigma, 100)\nplt.plot(x1,scipy.stats.norm.pdf(x1, mu1, sigma),label='prior')\n\nmu2 = 10\nvariance2 = 1\nsigma = math.sqrt(variance2)\nx2 = np.linspace(mu2 - 3*sigma, mu2 + 3*sigma, 100)\nplt.plot(x2,scipy.stats.norm.pdf(x2, mu2, sigma),\"g-\",label='measurement')\n\n\nmu_new=mu1+mu2\nprint(\"New mean is at: \",mu_new)\nvar_new=(variance1+variance2)\nprint(\"New variance is: \",var_new)\nsigma = math.sqrt(var_new)\nx3 = np.linspace(mu_new - 3*sigma, mu_new + 3*sigma, 100)\nplt.plot(x3,scipy.stats.norm.pdf(x3, mu_new, var_new),label=\"posterior\")\nplt.legend(loc='upper left')\nplt.xlim(-10,20)\nplt.show()",
"New mean is at: 15\nNew variance is: 2\n"
],
[
"#Example from:\n#https://scipython.com/blog/visualizing-the-bivariate-gaussian-distribution/\nimport numpy as np\nimport matplotlib.pyplot as plt\nfrom matplotlib import cm\nfrom mpl_toolkits.mplot3d import Axes3D\n\n# Our 2-dimensional distribution will be over variables X and Y\nN = 60\nX = np.linspace(-3, 3, N)\nY = np.linspace(-3, 4, N)\nX, Y = np.meshgrid(X, Y)\n\n# Mean vector and covariance matrix\nmu = np.array([0., 1.])\nSigma = np.array([[ 1. , -0.5], [-0.5, 1.5]])\n\n# Pack X and Y into a single 3-dimensional array\npos = np.empty(X.shape + (2,))\npos[:, :, 0] = X\npos[:, :, 1] = Y\n\ndef multivariate_gaussian(pos, mu, Sigma):\n \"\"\"Return the multivariate Gaussian distribution on array pos.\n\n pos is an array constructed by packing the meshed arrays of variables\n x_1, x_2, x_3, ..., x_k into its _last_ dimension.\n\n \"\"\"\n\n n = mu.shape[0]\n Sigma_det = np.linalg.det(Sigma)\n Sigma_inv = np.linalg.inv(Sigma)\n N = np.sqrt((2*np.pi)**n * Sigma_det)\n # This einsum call calculates (x-mu)T.Sigma-1.(x-mu) in a vectorized\n # way across all the input variables.\n fac = np.einsum('...k,kl,...l->...', pos-mu, Sigma_inv, pos-mu)\n\n return np.exp(-fac / 2) / N\n\n# The distribution on the variables X, Y packed into pos.\nZ = multivariate_gaussian(pos, mu, Sigma)\n\n# Create a surface plot and projected filled contour plot under it.\nfig = plt.figure()\nax = fig.gca(projection='3d')\nax.plot_surface(X, Y, Z, rstride=3, cstride=3, linewidth=1, antialiased=True,\n cmap=cm.viridis)\n\ncset = ax.contourf(X, Y, Z, zdir='z', offset=-0.15, cmap=cm.viridis)\n\n# Adjust the limits, ticks and view angle\nax.set_zlim(-0.15,0.2)\nax.set_zticks(np.linspace(0,0.2,5))\nax.view_init(27, -21)\n\nplt.show()\n",
"_____no_output_____"
]
],
[
[
"This is a 3D projection of the gaussians involved with the lower surface showing the 2D projection of the 3D projection above. The innermost ellipse represents the highest peak, that is the maximum probability for a given (X,Y) value.\n\n\n\n\n** numpy einsum examples **",
"_____no_output_____"
]
],
[
[
"a = np.arange(25).reshape(5,5)\nb = np.arange(5)\nc = np.arange(6).reshape(2,3)\nprint(a)\nprint(b)\nprint(c)\n",
"[[ 0 1 2 3 4]\n [ 5 6 7 8 9]\n [10 11 12 13 14]\n [15 16 17 18 19]\n [20 21 22 23 24]]\n[0 1 2 3 4]\n[[0 1 2]\n [3 4 5]]\n"
],
[
"#this is the diagonal sum, i repeated means the diagonal\nnp.einsum('ij', a)\n#this takes the output ii which is the diagonal and outputs to a\nnp.einsum('ii->i',a)\n#this takes in the array A represented by their axes 'ij' and B by its only axes'j' \n#and multiples them element wise\nnp.einsum('ij,j',a, b)",
"_____no_output_____"
],
[
"A = np.arange(3).reshape(3,1)\nB = np.array([[ 0, 1, 2, 3],\n [ 4, 5, 6, 7],\n [ 8, 9, 10, 11]])\nC=np.multiply(A,B)\nnp.sum(C,axis=1)",
"_____no_output_____"
],
[
"D = np.array([0,1,2])\nE = np.array([[ 0, 1, 2, 3],\n [ 4, 5, 6, 7],\n [ 8, 9, 10, 11]])\n\nnp.einsum('i,ij->i',D,E)",
"_____no_output_____"
],
[
"from scipy.stats import multivariate_normal\nx, y = np.mgrid[-5:5:.1, -5:5:.1]\npos = np.empty(x.shape + (2,))\npos[:, :, 0] = x; pos[:, :, 1] = y\nrv = multivariate_normal([0.5, -0.2], [[2.0, 0.9], [0.9, 0.5]])\nplt.contourf(x, y, rv.pdf(pos))\n\n",
"_____no_output_____"
]
],
[
[
"### References:\n\n1. Roger Labbe's [repo](https://github.com/rlabbe/Kalman-and-Bayesian-Filters-in-Python) on Kalman Filters. (Majority of the examples in the notes are from this)\n\n\n\n2. Probabilistic Robotics by Sebastian Thrun, Wolfram Burgard and Dieter Fox, MIT Press.\n\n\n\n3. Scipy [Documentation](https://scipython.com/blog/visualizing-the-bivariate-gaussian-distribution/)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
c501b16e44c5093f44b08fe0eda1ac497f2bb5ca
| 35,343 |
ipynb
|
Jupyter Notebook
|
.ipynb_checkpoints/1main_npred500-checkpoint.ipynb
|
danhtaihoang/expectation-reflection
|
ae89c77da1e47ffc0ea09fb2e919d29308998b95
|
[
"MIT"
] | null | null | null |
.ipynb_checkpoints/1main_npred500-checkpoint.ipynb
|
danhtaihoang/expectation-reflection
|
ae89c77da1e47ffc0ea09fb2e919d29308998b95
|
[
"MIT"
] | null | null | null |
.ipynb_checkpoints/1main_npred500-checkpoint.ipynb
|
danhtaihoang/expectation-reflection
|
ae89c77da1e47ffc0ea09fb2e919d29308998b95
|
[
"MIT"
] | null | null | null | 92.279373 | 12,756 | 0.840676 |
[
[
[
"## Expectation Reflection for Classification",
"_____no_output_____"
]
],
[
[
"import numpy as np\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.model_selection import KFold\nfrom sklearn.metrics import accuracy_score\n\nimport expectation_reflection as ER\nfrom sklearn.linear_model import LogisticRegression\n\nimport matplotlib.pyplot as plt\n%matplotlib inline",
"_____no_output_____"
],
[
"np.random.seed(1)",
"_____no_output_____"
],
[
"l = 1000\nn = 20\ng = 3.",
"_____no_output_____"
]
],
[
[
"### Categorical variables",
"_____no_output_____"
]
],
[
[
"def synthesize_data(l,n,g,data_type='discrete'): \n if data_type == 'binary':\n X = np.sign(np.random.rand(l,n)-0.5)\n w = np.random.normal(0.,g/np.sqrt(n),size=n)\n \n if data_type == 'discrete':\n X = 2*np.random.rand(l,n)-1\n w = np.random.normal(0.,g/np.sqrt(n),size=n)\n \n if data_type == 'categorical': \n from sklearn.preprocessing import OneHotEncoder\n m = 5 # categorical number for each variables\n # initial s (categorical variables)\n s = np.random.randint(0,m,size=(l,n)) # integer values\n onehot_encoder = OneHotEncoder(sparse=False,categories='auto')\n X = onehot_encoder.fit_transform(s)\n w = np.random.normal(0.,g/np.sqrt(n),size=n*m)\n \n h = X.dot(w)\n p = 1/(1+np.exp(-2*h)) # kinetic\n #p = 1/(1+np.exp(-h)) # logistic regression\n y = np.sign(p - np.random.rand(l))\n return w,X,y",
"_____no_output_____"
],
[
"w0,X,y = synthesize_data(l,n,g,data_type='categorical')",
"_____no_output_____"
],
[
"from sklearn.preprocessing import MinMaxScaler\nX = MinMaxScaler().fit_transform(X)",
"_____no_output_____"
],
[
"h0,w = ER.fit(X,y,niter_max=100,regu=0.005)",
"_____no_output_____"
],
[
"plt.figure(figsize=(4,3))\n\nplt.plot([-2,2],[-2,2],'r--')\nplt.scatter(w0,w)\nplt.xlabel('actual interactions')\nplt.ylabel('inferred interactios')\n\nplt.show()",
"_____no_output_____"
],
[
"y_pred = ER.predict(X,h0,w)\naccuracy = accuracy_score(y,y_pred)\nmse = ((w0-w)**2).mean()\nprint(mse,accuracy)",
"(0.1531093835397234, 0.908)\n"
],
[
"kf = 5\ndef ER_inference(X,y,kf=5,regu=0.005): \n #x_train,x_test,y_train,y_test = train_test_split(X1,y,test_size=0.3,random_state = 100) \n kfold = KFold(n_splits=kf,shuffle=False,random_state=1)\n accuracy = np.zeros(kf)\n \n for i,(train_index,test_index) in enumerate(kfold.split(y)):\n X_train, X_test = X[train_index], X[test_index]\n y_train, y_test = y[train_index], y[test_index]\n\n # predict with ER\n h0,w = ER.fit(X_train,y_train,niter_max=100,regu=0.005)\n y_pred = ER.predict(X_test,h0,w)\n accuracy[i] = accuracy_score(y_test,y_pred)\n #print(accuracy[i])\n return accuracy.mean(),accuracy.std()",
"_____no_output_____"
],
[
"regu_list = [0.0,0.001,0.002,0.003,0.004,0.005,0.01,0.02,0.1,0.2,0.5,0.6,0.8,1.]\nfor regu in regu_list:\n accuracy_mean,accuracy_std = ER_inference(X,y,kf,regu)\n print('ER:',accuracy_mean,accuracy_std,regu)",
"('ER:', 0.845, 0.017888543819998288, 0.0)\n('ER:', 0.845, 0.017888543819998288, 0.001)\n('ER:', 0.845, 0.017888543819998288, 0.002)\n('ER:', 0.845, 0.017888543819998288, 0.003)\n('ER:', 0.845, 0.017888543819998288, 0.004)\n('ER:', 0.845, 0.017888543819998288, 0.005)\n('ER:', 0.845, 0.017888543819998288, 0.01)\n('ER:', 0.845, 0.017888543819998288, 0.02)\n('ER:', 0.845, 0.017888543819998288, 0.1)\n('ER:', 0.845, 0.017888543819998288, 0.2)\n('ER:', 0.845, 0.017888543819998288, 0.5)\n('ER:', 0.845, 0.017888543819998288, 0.6)\n('ER:', 0.845, 0.017888543819998288, 0.8)\n('ER:', 0.845, 0.017888543819998288, 1.0)\n"
]
],
[
[
"#### Logistic Regression",
"_____no_output_____"
]
],
[
[
"model = LogisticRegression(solver='liblinear')\nmodel.fit(X, y)\nw_lg = 0.5*model.coef_",
"_____no_output_____"
],
[
"plt.figure(figsize=(4,3))\n\nplt.plot([-2,2],[-2,2],'r--')\nplt.scatter(w0,w_lg)\nplt.xlabel('actual interactions')\nplt.ylabel('inferred interactios')\n\nplt.show()",
"_____no_output_____"
],
[
"y_pred = model.predict(X)\naccuracy = accuracy_score(y,y_pred)\nmse = ((w0-w_lg)**2).mean()\nprint(mse,accuracy)",
"(0.15543397153876845, 0.913)\n"
],
[
"def inference(X,y,kf=5,method='naive_bayes'): \n kfold = KFold(n_splits=kf,shuffle=False,random_state=1) \n accuracy = np.zeros(kf)\n \n if method == 'logistic_regression':\n model = LogisticRegression(solver='liblinear')\n\n if method == 'naive_bayes': \n model = GaussianNB()\n \n if method == 'random_forest':\n model = RandomForestClassifier(criterion = \"gini\", random_state = 1,\n max_depth=3, min_samples_leaf=5,n_estimators=100) \n if method == 'decision_tree':\n model = DecisionTreeClassifier()\n \n if method == 'knn': \n model = KNeighborsClassifier()\n \n if method == 'svm': \n model = SVC(gamma='scale') \n \n for i,(train_index,test_index) in enumerate(kfold.split(y)):\n X_train, X_test = X[train_index], X[test_index]\n y_train, y_test = y[train_index], y[test_index]\n\n # fit and predict\n model.fit(X_train, y_train)\n y_pred = model.predict(X_test)\n \n accuracy[i] = accuracy_score(y_test,y_pred)\n #print(accuracy[i])\n return accuracy.mean(),accuracy.std()",
"_____no_output_____"
],
[
"other_methods=['logistic_regression']\n\nfor i,method in enumerate(other_methods):\n accuracy_mean,accuracy_std = inference(X,y,kf,method)\n print('% 20s :'%method,accuracy_mean,accuracy_std)",
"(' logistic_regression :', 0.842, 0.013266499161421611)\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.