hexsha
stringlengths 40
40
| size
int64 6
14.9M
| ext
stringclasses 1
value | lang
stringclasses 1
value | max_stars_repo_path
stringlengths 6
260
| max_stars_repo_name
stringlengths 6
119
| max_stars_repo_head_hexsha
stringlengths 40
41
| max_stars_repo_licenses
list | max_stars_count
int64 1
191k
⌀ | max_stars_repo_stars_event_min_datetime
stringlengths 24
24
⌀ | max_stars_repo_stars_event_max_datetime
stringlengths 24
24
⌀ | max_issues_repo_path
stringlengths 6
260
| max_issues_repo_name
stringlengths 6
119
| max_issues_repo_head_hexsha
stringlengths 40
41
| max_issues_repo_licenses
list | max_issues_count
int64 1
67k
⌀ | max_issues_repo_issues_event_min_datetime
stringlengths 24
24
⌀ | max_issues_repo_issues_event_max_datetime
stringlengths 24
24
⌀ | max_forks_repo_path
stringlengths 6
260
| max_forks_repo_name
stringlengths 6
119
| max_forks_repo_head_hexsha
stringlengths 40
41
| max_forks_repo_licenses
list | max_forks_count
int64 1
105k
⌀ | max_forks_repo_forks_event_min_datetime
stringlengths 24
24
⌀ | max_forks_repo_forks_event_max_datetime
stringlengths 24
24
⌀ | avg_line_length
float64 2
1.04M
| max_line_length
int64 2
11.2M
| alphanum_fraction
float64 0
1
| cells
list | cell_types
list | cell_type_groups
list |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
cb8b6a528fd3f13bfccac00fadcf27afc18aad56
| 146,146 |
ipynb
|
Jupyter Notebook
|
malaria5_1_mobileNet.ipynb
|
RitwizKamal/Transfer-Learning
|
6aab6b94a3a7fcfe903f43f7b17896014a3f699a
|
[
"MIT"
] | 1 |
2019-12-23T07:21:53.000Z
|
2019-12-23T07:21:53.000Z
|
malaria5_1_mobileNet.ipynb
|
RitwizKamal/Transfer-Learning
|
6aab6b94a3a7fcfe903f43f7b17896014a3f699a
|
[
"MIT"
] | null | null | null |
malaria5_1_mobileNet.ipynb
|
RitwizKamal/Transfer-Learning
|
6aab6b94a3a7fcfe903f43f7b17896014a3f699a
|
[
"MIT"
] | null | null | null | 143.140059 | 60,132 | 0.830471 |
[
[
[
"import os\nimport glob\nbase_dir = os.path.join('F:/0Sem 7/B.TECH PROJECT/0Image data/cell_images')\ninfected_dir = os.path.join(base_dir,'Parasitized')\nhealthy_dir = os.path.join(base_dir,'Uninfected')\ninfected_files = glob.glob(infected_dir+'/*.png')\nhealthy_files = glob.glob(healthy_dir+'/*.png')\nprint(\"Infected samples:\",len(infected_files))\nprint(\"Uninfected samples:\",len(healthy_files))",
"Infected samples: 13779\nUninfected samples: 13779\n"
],
[
"import numpy as np\nimport pandas as pd\n\nnp.random.seed(42)\n\nfiles_df = pd.DataFrame({\n 'filename': infected_files + healthy_files,\n 'label': ['malaria'] * len(infected_files) + ['healthy'] * len(healthy_files)\n}).sample(frac=1, random_state=42).reset_index(drop=True)\n\nfiles_df.head()",
"_____no_output_____"
],
[
"from sklearn.model_selection import train_test_split\nfrom collections import Counter\n\ntrain_files, test_files, train_labels, test_labels = train_test_split(files_df['filename'].values,\n files_df['label'].values, \n test_size=0.3, random_state=42)\ntrain_files, val_files, train_labels, val_labels = train_test_split(train_files,\n train_labels, \n test_size=0.1, random_state=42)\n\nprint(train_files.shape, val_files.shape, test_files.shape)\nprint('Train:', Counter(train_labels), '\\nVal:', Counter(val_labels), '\\nTest:', Counter(test_labels))",
"(17361,) (1929,) (8268,)\nTrain: Counter({'healthy': 8734, 'malaria': 8627}) \nVal: Counter({'healthy': 970, 'malaria': 959}) \nTest: Counter({'malaria': 4193, 'healthy': 4075})\n"
],
[
"import cv2\nfrom concurrent import futures\nimport threading\n\ndef get_img_shape_parallel(idx, img, total_imgs):\n if idx % 5000 == 0 or idx == (total_imgs - 1):\n print('{}: working on img num: {}'.format(threading.current_thread().name,\n idx))\n return cv2.imread(img).shape\n \nex = futures.ThreadPoolExecutor(max_workers=None)\ndata_inp = [(idx, img, len(train_files)) for idx, img in enumerate(train_files)]\nprint('Starting Img shape computation:')\ntrain_img_dims_map = ex.map(get_img_shape_parallel, \n [record[0] for record in data_inp],\n [record[1] for record in data_inp],\n [record[2] for record in data_inp])\ntrain_img_dims = list(train_img_dims_map)\nprint('Min Dimensions:', np.min(train_img_dims, axis=0)) \nprint('Avg Dimensions:', np.mean(train_img_dims, axis=0))\nprint('Median Dimensions:', np.median(train_img_dims, axis=0))\nprint('Max Dimensions:', np.max(train_img_dims, axis=0))",
"Starting Img shape computation:\nThreadPoolExecutor-0_0: working on img num: 0\nThreadPoolExecutor-0_12: working on img num: 5000\nThreadPoolExecutor-0_3: working on img num: 10000\nThreadPoolExecutor-0_18: working on img num: 15000\nThreadPoolExecutor-0_8: working on img num: 17360\nMin Dimensions: [46 49 3]\nAvg Dimensions: [132.89856575 132.50751685 3. ]\nMedian Dimensions: [130. 130. 3.]\nMax Dimensions: [382 394 3]\n"
],
[
"IMG_DIMS = (32, 32)\n\ndef get_img_data_parallel(idx, img, total_imgs):\n if idx % 5000 == 0 or idx == (total_imgs - 1):\n print('{}: working on img num: {}'.format(threading.current_thread().name,\n idx))\n img = cv2.imread(img)\n img = cv2.resize(img, dsize=IMG_DIMS, \n interpolation=cv2.INTER_CUBIC)\n img = np.array(img, dtype=np.float32)\n return img\n\nex = futures.ThreadPoolExecutor(max_workers=None)\ntrain_data_inp = [(idx, img, len(train_files)) for idx, img in enumerate(train_files)]\nval_data_inp = [(idx, img, len(val_files)) for idx, img in enumerate(val_files)]\ntest_data_inp = [(idx, img, len(test_files)) for idx, img in enumerate(test_files)]\n\nprint('Loading Train Images:')\ntrain_data_map = ex.map(get_img_data_parallel, \n [record[0] for record in train_data_inp],\n [record[1] for record in train_data_inp],\n [record[2] for record in train_data_inp])\ntrain_data = np.array(list(train_data_map))\n\nprint('\\nLoading Validation Images:')\nval_data_map = ex.map(get_img_data_parallel, \n [record[0] for record in val_data_inp],\n [record[1] for record in val_data_inp],\n [record[2] for record in val_data_inp])\nval_data = np.array(list(val_data_map))\n\nprint('\\nLoading Test Images:')\ntest_data_map = ex.map(get_img_data_parallel, \n [record[0] for record in test_data_inp],\n [record[1] for record in test_data_inp],\n [record[2] for record in test_data_inp])\ntest_data = np.array(list(test_data_map))\n\ntrain_data.shape, val_data.shape, test_data.shape ",
"Loading Train Images:\nThreadPoolExecutor-1_0: working on img num: 0\nThreadPoolExecutor-1_12: working on img num: 5000\nThreadPoolExecutor-1_9: working on img num: 10000\nThreadPoolExecutor-1_15: working on img num: 15000\nThreadPoolExecutor-1_14: working on img num: 17360\n\nLoading Validation Images:\nThreadPoolExecutor-1_6: working on img num: 0\nThreadPoolExecutor-1_0: working on img num: 1928\n\nLoading Test Images:\nThreadPoolExecutor-1_18: working on img num: 0\nThreadPoolExecutor-1_11: working on img num: 5000\nThreadPoolExecutor-1_12: working on img num: 8267\n"
],
[
"import matplotlib.pyplot as plt\n%matplotlib inline\n\nplt.figure(1 , figsize = (8 , 8))\nn = 0 \nfor i in range(16):\n n += 1 \n r = np.random.randint(0 , train_data.shape[0] , 1)\n plt.subplot(4 , 4 , n)\n plt.subplots_adjust(hspace = 0.5 , wspace = 0.5)\n plt.imshow(train_data[r[0]]/255.)\n plt.title('{}'.format(train_labels[r[0]]))\n plt.xticks([]) , plt.yticks([])",
"_____no_output_____"
],
[
"BATCH_SIZE = 32\nNUM_CLASSES = 2\nEPOCHS = 25\nINPUT_SHAPE = (32, 32, 3)\n\ntrain_imgs_scaled = train_data / 255.\nval_imgs_scaled = val_data / 255.\n\n# encode text category labels\nfrom sklearn.preprocessing import LabelEncoder\n\nle = LabelEncoder()\nle.fit(train_labels)\ntrain_labels_enc = le.transform(train_labels)\nval_labels_enc = le.transform(val_labels)\n\nprint(train_labels[:6], train_labels_enc[:6])",
"['malaria' 'malaria' 'malaria' 'healthy' 'healthy' 'malaria'] [1 1 1 0 0 1]\n"
],
[
"import tensorflow as tf",
"_____no_output_____"
],
[
"vgg = tf.keras.applications.mobilenet.MobileNet(include_top=False, alpha=1.0, weights='imagenet', \n input_shape=INPUT_SHAPE)\n# Freeze the layers\nvgg.trainable = True\n\nset_trainable = False\nfor layer in vgg.layers:\n layer.trainable = False\n \nbase_vgg = vgg\nbase_out = base_vgg.output\npool_out = tf.keras.layers.Flatten()(base_out)\nhidden1 = tf.keras.layers.Dense(512, activation='relu')(pool_out)\ndrop1 = tf.keras.layers.Dropout(rate=0.3)(hidden1)\nhidden2 = tf.keras.layers.Dense(512, activation='relu')(drop1)\ndrop2 = tf.keras.layers.Dropout(rate=0.3)(hidden2)\n\nout = tf.keras.layers.Dense(1, activation='sigmoid')(drop2)\n\nmodel = tf.keras.Model(inputs=base_vgg.input, outputs=out)\n\nfrom tensorflow.keras.optimizers import Adam\nadam = Adam(lr=0.0001)\n\nmodel.compile(optimizer=adam,\n loss='binary_crossentropy',\n metrics=['accuracy'])\n\nprint(\"Total Layers:\", len(model.layers))\nprint(\"Total trainable layers:\", sum([1 for l in model.layers if l.trainable]))\n",
"C:\\Users\\Ritwiz\\Anaconda3\\envs\\newtfgpu\\lib\\site-packages\\keras_applications\\mobilenet.py:207: UserWarning: `input_shape` is undefined or non-square, or `rows` is not in [128, 160, 192, 224]. Weights for input shape (224, 224) will be loaded as the default.\n warnings.warn('`input_shape` is undefined or non-square, '\n"
],
[
"print(model.summary())",
"Model: \"model\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\ninput_1 (InputLayer) [(None, 32, 32, 3)] 0 \n_________________________________________________________________\nconv1_pad (ZeroPadding2D) (None, 33, 33, 3) 0 \n_________________________________________________________________\nconv1 (Conv2D) (None, 16, 16, 32) 864 \n_________________________________________________________________\nconv1_bn (BatchNormalization (None, 16, 16, 32) 128 \n_________________________________________________________________\nconv1_relu (ReLU) (None, 16, 16, 32) 0 \n_________________________________________________________________\nconv_dw_1 (DepthwiseConv2D) (None, 16, 16, 32) 288 \n_________________________________________________________________\nconv_dw_1_bn (BatchNormaliza (None, 16, 16, 32) 128 \n_________________________________________________________________\nconv_dw_1_relu (ReLU) (None, 16, 16, 32) 0 \n_________________________________________________________________\nconv_pw_1 (Conv2D) (None, 16, 16, 64) 2048 \n_________________________________________________________________\nconv_pw_1_bn (BatchNormaliza (None, 16, 16, 64) 256 \n_________________________________________________________________\nconv_pw_1_relu (ReLU) (None, 16, 16, 64) 0 \n_________________________________________________________________\nconv_pad_2 (ZeroPadding2D) (None, 17, 17, 64) 0 \n_________________________________________________________________\nconv_dw_2 (DepthwiseConv2D) (None, 8, 8, 64) 576 \n_________________________________________________________________\nconv_dw_2_bn (BatchNormaliza (None, 8, 8, 64) 256 \n_________________________________________________________________\nconv_dw_2_relu (ReLU) (None, 8, 8, 64) 0 \n_________________________________________________________________\nconv_pw_2 (Conv2D) (None, 8, 8, 128) 8192 \n_________________________________________________________________\nconv_pw_2_bn (BatchNormaliza (None, 8, 8, 128) 512 \n_________________________________________________________________\nconv_pw_2_relu (ReLU) (None, 8, 8, 128) 0 \n_________________________________________________________________\nconv_dw_3 (DepthwiseConv2D) (None, 8, 8, 128) 1152 \n_________________________________________________________________\nconv_dw_3_bn (BatchNormaliza (None, 8, 8, 128) 512 \n_________________________________________________________________\nconv_dw_3_relu (ReLU) (None, 8, 8, 128) 0 \n_________________________________________________________________\nconv_pw_3 (Conv2D) (None, 8, 8, 128) 16384 \n_________________________________________________________________\nconv_pw_3_bn (BatchNormaliza (None, 8, 8, 128) 512 \n_________________________________________________________________\nconv_pw_3_relu (ReLU) (None, 8, 8, 128) 0 \n_________________________________________________________________\nconv_pad_4 (ZeroPadding2D) (None, 9, 9, 128) 0 \n_________________________________________________________________\nconv_dw_4 (DepthwiseConv2D) (None, 4, 4, 128) 1152 \n_________________________________________________________________\nconv_dw_4_bn (BatchNormaliza (None, 4, 4, 128) 512 \n_________________________________________________________________\nconv_dw_4_relu (ReLU) (None, 4, 4, 128) 0 \n_________________________________________________________________\nconv_pw_4 (Conv2D) (None, 4, 4, 256) 32768 \n_________________________________________________________________\nconv_pw_4_bn (BatchNormaliza (None, 4, 4, 256) 1024 \n_________________________________________________________________\nconv_pw_4_relu (ReLU) (None, 4, 4, 256) 0 \n_________________________________________________________________\nconv_dw_5 (DepthwiseConv2D) (None, 4, 4, 256) 2304 \n_________________________________________________________________\nconv_dw_5_bn (BatchNormaliza (None, 4, 4, 256) 1024 \n_________________________________________________________________\nconv_dw_5_relu (ReLU) (None, 4, 4, 256) 0 \n_________________________________________________________________\nconv_pw_5 (Conv2D) (None, 4, 4, 256) 65536 \n_________________________________________________________________\nconv_pw_5_bn (BatchNormaliza (None, 4, 4, 256) 1024 \n_________________________________________________________________\nconv_pw_5_relu (ReLU) (None, 4, 4, 256) 0 \n_________________________________________________________________\nconv_pad_6 (ZeroPadding2D) (None, 5, 5, 256) 0 \n_________________________________________________________________\nconv_dw_6 (DepthwiseConv2D) (None, 2, 2, 256) 2304 \n_________________________________________________________________\nconv_dw_6_bn (BatchNormaliza (None, 2, 2, 256) 1024 \n_________________________________________________________________\nconv_dw_6_relu (ReLU) (None, 2, 2, 256) 0 \n_________________________________________________________________\nconv_pw_6 (Conv2D) (None, 2, 2, 512) 131072 \n_________________________________________________________________\nconv_pw_6_bn (BatchNormaliza (None, 2, 2, 512) 2048 \n_________________________________________________________________\nconv_pw_6_relu (ReLU) (None, 2, 2, 512) 0 \n_________________________________________________________________\nconv_dw_7 (DepthwiseConv2D) (None, 2, 2, 512) 4608 \n_________________________________________________________________\nconv_dw_7_bn (BatchNormaliza (None, 2, 2, 512) 2048 \n_________________________________________________________________\nconv_dw_7_relu (ReLU) (None, 2, 2, 512) 0 \n_________________________________________________________________\nconv_pw_7 (Conv2D) (None, 2, 2, 512) 262144 \n_________________________________________________________________\nconv_pw_7_bn (BatchNormaliza (None, 2, 2, 512) 2048 \n_________________________________________________________________\nconv_pw_7_relu (ReLU) (None, 2, 2, 512) 0 \n_________________________________________________________________\nconv_dw_8 (DepthwiseConv2D) (None, 2, 2, 512) 4608 \n_________________________________________________________________\nconv_dw_8_bn (BatchNormaliza (None, 2, 2, 512) 2048 \n_________________________________________________________________\nconv_dw_8_relu (ReLU) (None, 2, 2, 512) 0 \n_________________________________________________________________\nconv_pw_8 (Conv2D) (None, 2, 2, 512) 262144 \n_________________________________________________________________\nconv_pw_8_bn (BatchNormaliza (None, 2, 2, 512) 2048 \n_________________________________________________________________\nconv_pw_8_relu (ReLU) (None, 2, 2, 512) 0 \n_________________________________________________________________\nconv_dw_9 (DepthwiseConv2D) (None, 2, 2, 512) 4608 \n_________________________________________________________________\nconv_dw_9_bn (BatchNormaliza (None, 2, 2, 512) 2048 \n_________________________________________________________________\nconv_dw_9_relu (ReLU) (None, 2, 2, 512) 0 \n_________________________________________________________________\nconv_pw_9 (Conv2D) (None, 2, 2, 512) 262144 \n_________________________________________________________________\nconv_pw_9_bn (BatchNormaliza (None, 2, 2, 512) 2048 \n_________________________________________________________________\nconv_pw_9_relu (ReLU) (None, 2, 2, 512) 0 \n_________________________________________________________________\nconv_dw_10 (DepthwiseConv2D) (None, 2, 2, 512) 4608 \n_________________________________________________________________\nconv_dw_10_bn (BatchNormaliz (None, 2, 2, 512) 2048 \n_________________________________________________________________\nconv_dw_10_relu (ReLU) (None, 2, 2, 512) 0 \n_________________________________________________________________\nconv_pw_10 (Conv2D) (None, 2, 2, 512) 262144 \n_________________________________________________________________\nconv_pw_10_bn (BatchNormaliz (None, 2, 2, 512) 2048 \n_________________________________________________________________\nconv_pw_10_relu (ReLU) (None, 2, 2, 512) 0 \n_________________________________________________________________\nconv_dw_11 (DepthwiseConv2D) (None, 2, 2, 512) 4608 \n_________________________________________________________________\nconv_dw_11_bn (BatchNormaliz (None, 2, 2, 512) 2048 \n_________________________________________________________________\nconv_dw_11_relu (ReLU) (None, 2, 2, 512) 0 \n_________________________________________________________________\nconv_pw_11 (Conv2D) (None, 2, 2, 512) 262144 \n_________________________________________________________________\nconv_pw_11_bn (BatchNormaliz (None, 2, 2, 512) 2048 \n_________________________________________________________________\nconv_pw_11_relu (ReLU) (None, 2, 2, 512) 0 \n_________________________________________________________________\nconv_pad_12 (ZeroPadding2D) (None, 3, 3, 512) 0 \n_________________________________________________________________\nconv_dw_12 (DepthwiseConv2D) (None, 1, 1, 512) 4608 \n_________________________________________________________________\nconv_dw_12_bn (BatchNormaliz (None, 1, 1, 512) 2048 \n_________________________________________________________________\nconv_dw_12_relu (ReLU) (None, 1, 1, 512) 0 \n_________________________________________________________________\nconv_pw_12 (Conv2D) (None, 1, 1, 1024) 524288 \n_________________________________________________________________\nconv_pw_12_bn (BatchNormaliz (None, 1, 1, 1024) 4096 \n_________________________________________________________________\nconv_pw_12_relu (ReLU) (None, 1, 1, 1024) 0 \n_________________________________________________________________\nconv_dw_13 (DepthwiseConv2D) (None, 1, 1, 1024) 9216 \n_________________________________________________________________\nconv_dw_13_bn (BatchNormaliz (None, 1, 1, 1024) 4096 \n_________________________________________________________________\nconv_dw_13_relu (ReLU) (None, 1, 1, 1024) 0 \n_________________________________________________________________\nconv_pw_13 (Conv2D) (None, 1, 1, 1024) 1048576 \n_________________________________________________________________\nconv_pw_13_bn (BatchNormaliz (None, 1, 1, 1024) 4096 \n_________________________________________________________________\nconv_pw_13_relu (ReLU) (None, 1, 1, 1024) 0 \n_________________________________________________________________\nflatten (Flatten) (None, 1024) 0 \n_________________________________________________________________\ndense (Dense) (None, 512) 524800 \n_________________________________________________________________\ndropout (Dropout) (None, 512) 0 \n_________________________________________________________________\ndense_1 (Dense) (None, 512) 262656 \n_________________________________________________________________\ndropout_1 (Dropout) (None, 512) 0 \n_________________________________________________________________\ndense_2 (Dense) (None, 1) 513 \n=================================================================\nTotal params: 4,016,833\nTrainable params: 787,969\nNon-trainable params: 3,228,864\n_________________________________________________________________\nNone\n"
],
[
"history = model.fit(x=train_imgs_scaled, y=train_labels_enc, \n batch_size=BATCH_SIZE,\n epochs=EPOCHS, \n validation_data=(val_imgs_scaled, val_labels_enc),\n verbose=1)",
"Train on 17361 samples, validate on 1929 samples\nEpoch 1/25\n17361/17361 [==============================] - 15s 867us/sample - loss: 0.6412 - accuracy: 0.6746 - val_loss: 0.6768 - val_accuracy: 0.5023\nEpoch 2/25\n17361/17361 [==============================] - 7s 412us/sample - loss: 0.5482 - accuracy: 0.7255 - val_loss: 0.6768 - val_accuracy: 0.5023\nEpoch 3/25\n17361/17361 [==============================] - 6s 366us/sample - loss: 0.5197 - accuracy: 0.7446 - val_loss: 0.6715 - val_accuracy: 0.5029\nEpoch 4/25\n17361/17361 [==============================] - 6s 367us/sample - loss: 0.5048 - accuracy: 0.7531 - val_loss: 0.6737 - val_accuracy: 0.5039\nEpoch 5/25\n17361/17361 [==============================] - 6s 366us/sample - loss: 0.4909 - accuracy: 0.7650 - val_loss: 0.6781 - val_accuracy: 0.5034\nEpoch 6/25\n17361/17361 [==============================] - 6s 367us/sample - loss: 0.4897 - accuracy: 0.7634 - val_loss: 0.6796 - val_accuracy: 0.5034\nEpoch 7/25\n17361/17361 [==============================] - 6s 367us/sample - loss: 0.4832 - accuracy: 0.7641 - val_loss: 0.6821 - val_accuracy: 0.5029\nEpoch 8/25\n17361/17361 [==============================] - 6s 369us/sample - loss: 0.4787 - accuracy: 0.7691 - val_loss: 0.6878 - val_accuracy: 0.5034\nEpoch 9/25\n17361/17361 [==============================] - 6s 368us/sample - loss: 0.4680 - accuracy: 0.7769 - val_loss: 0.6910 - val_accuracy: 0.5039\nEpoch 10/25\n17361/17361 [==============================] - 7s 386us/sample - loss: 0.4689 - accuracy: 0.7756 - val_loss: 0.6890 - val_accuracy: 0.5034\nEpoch 11/25\n17361/17361 [==============================] - 7s 379us/sample - loss: 0.4655 - accuracy: 0.7756 - val_loss: 0.6936 - val_accuracy: 0.5029\nEpoch 12/25\n17361/17361 [==============================] - 7s 403us/sample - loss: 0.4592 - accuracy: 0.7788 - val_loss: 0.6865 - val_accuracy: 0.5034\nEpoch 13/25\n17361/17361 [==============================] - 7s 397us/sample - loss: 0.4631 - accuracy: 0.7775 - val_loss: 0.6941 - val_accuracy: 0.5029\nEpoch 14/25\n17361/17361 [==============================] - 7s 392us/sample - loss: 0.4606 - accuracy: 0.7799 - val_loss: 0.7021 - val_accuracy: 0.5034\nEpoch 15/25\n17361/17361 [==============================] - 7s 376us/sample - loss: 0.4545 - accuracy: 0.7811 - val_loss: 0.6920 - val_accuracy: 0.5039\nEpoch 16/25\n17361/17361 [==============================] - 7s 381us/sample - loss: 0.4491 - accuracy: 0.7860 - val_loss: 0.6935 - val_accuracy: 0.5039\nEpoch 17/25\n17361/17361 [==============================] - 7s 386us/sample - loss: 0.4466 - accuracy: 0.7865 - val_loss: 0.7009 - val_accuracy: 0.5034\nEpoch 18/25\n17361/17361 [==============================] - 7s 385us/sample - loss: 0.4439 - accuracy: 0.7902 - val_loss: 0.7030 - val_accuracy: 0.5054\nEpoch 19/25\n17361/17361 [==============================] - 7s 381us/sample - loss: 0.4420 - accuracy: 0.7871 - val_loss: 0.7014 - val_accuracy: 0.5049\nEpoch 20/25\n17361/17361 [==============================] - 7s 388us/sample - loss: 0.4439 - accuracy: 0.7883 - val_loss: 0.6996 - val_accuracy: 0.5034\nEpoch 21/25\n17361/17361 [==============================] - 7s 405us/sample - loss: 0.4385 - accuracy: 0.7911 - val_loss: 0.7024 - val_accuracy: 0.5034\nEpoch 22/25\n17361/17361 [==============================] - 7s 406us/sample - loss: 0.4384 - accuracy: 0.7897 - val_loss: 0.7063 - val_accuracy: 0.5044\nEpoch 23/25\n17361/17361 [==============================] - 7s 408us/sample - loss: 0.4314 - accuracy: 0.7951 - val_loss: 0.7077 - val_accuracy: 0.5044\nEpoch 24/25\n17361/17361 [==============================] - 7s 382us/sample - loss: 0.4315 - accuracy: 0.7963 - val_loss: 0.7177 - val_accuracy: 0.5034\nEpoch 25/25\n17361/17361 [==============================] - 7s 380us/sample - loss: 0.4287 - accuracy: 0.7950 - val_loss: 0.7209 - val_accuracy: 0.5034\n"
],
[
"f, (ax1, ax2) = plt.subplots(1, 2, figsize=(12, 4))\nt = f.suptitle('Basic CNN Performance', fontsize=12)\nf.subplots_adjust(top=0.85, wspace=0.3)\n\nmax_epoch = len(history.history['accuracy'])+1\nepoch_list = list(range(1,max_epoch))\nax1.plot(epoch_list, history.history['accuracy'], label='Train Accuracy')\nax1.plot(epoch_list, history.history['val_accuracy'], label='Validation Accuracy')\nax1.set_xticks(np.arange(1, max_epoch, 5))\nax1.set_ylabel('Accuracy Value')\nax1.set_xlabel('Epoch')\nax1.set_title('Accuracy')\nl1 = ax1.legend(loc=\"best\")\n\nax2.plot(epoch_list, history.history['loss'], label='Train Loss')\nax2.plot(epoch_list, history.history['val_loss'], label='Validation Loss')\nax2.set_xticks(np.arange(1, max_epoch, 5))\nax2.set_ylabel('Loss Value')\nax2.set_xlabel('Epoch')\nax2.set_title('Loss')\nl2 = ax2.legend(loc=\"best\")",
"_____no_output_____"
],
[
"test_imgs_scaled = test_data/255.\ntest_labels_enc = le.transform(test_labels)\n\n# evaluate the model\n_, train_acc = model.evaluate(train_imgs_scaled, train_labels_enc, verbose=0)\n_, test_acc = model.evaluate(test_imgs_scaled, test_labels_enc, verbose=0)\nprint('Train: %.3f, Test: %.3f' % (train_acc, test_acc))",
"Train: 0.503, Test: 0.493\n"
],
[
"print(model.summary())",
"Model: \"model\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\ninput_1 (InputLayer) [(None, 32, 32, 3)] 0 \n_________________________________________________________________\nconv1_pad (ZeroPadding2D) (None, 33, 33, 3) 0 \n_________________________________________________________________\nconv1 (Conv2D) (None, 16, 16, 32) 864 \n_________________________________________________________________\nconv1_bn (BatchNormalization (None, 16, 16, 32) 128 \n_________________________________________________________________\nconv1_relu (ReLU) (None, 16, 16, 32) 0 \n_________________________________________________________________\nconv_dw_1 (DepthwiseConv2D) (None, 16, 16, 32) 288 \n_________________________________________________________________\nconv_dw_1_bn (BatchNormaliza (None, 16, 16, 32) 128 \n_________________________________________________________________\nconv_dw_1_relu (ReLU) (None, 16, 16, 32) 0 \n_________________________________________________________________\nconv_pw_1 (Conv2D) (None, 16, 16, 64) 2048 \n_________________________________________________________________\nconv_pw_1_bn (BatchNormaliza (None, 16, 16, 64) 256 \n_________________________________________________________________\nconv_pw_1_relu (ReLU) (None, 16, 16, 64) 0 \n_________________________________________________________________\nconv_pad_2 (ZeroPadding2D) (None, 17, 17, 64) 0 \n_________________________________________________________________\nconv_dw_2 (DepthwiseConv2D) (None, 8, 8, 64) 576 \n_________________________________________________________________\nconv_dw_2_bn (BatchNormaliza (None, 8, 8, 64) 256 \n_________________________________________________________________\nconv_dw_2_relu (ReLU) (None, 8, 8, 64) 0 \n_________________________________________________________________\nconv_pw_2 (Conv2D) (None, 8, 8, 128) 8192 \n_________________________________________________________________\nconv_pw_2_bn (BatchNormaliza (None, 8, 8, 128) 512 \n_________________________________________________________________\nconv_pw_2_relu (ReLU) (None, 8, 8, 128) 0 \n_________________________________________________________________\nconv_dw_3 (DepthwiseConv2D) (None, 8, 8, 128) 1152 \n_________________________________________________________________\nconv_dw_3_bn (BatchNormaliza (None, 8, 8, 128) 512 \n_________________________________________________________________\nconv_dw_3_relu (ReLU) (None, 8, 8, 128) 0 \n_________________________________________________________________\nconv_pw_3 (Conv2D) (None, 8, 8, 128) 16384 \n_________________________________________________________________\nconv_pw_3_bn (BatchNormaliza (None, 8, 8, 128) 512 \n_________________________________________________________________\nconv_pw_3_relu (ReLU) (None, 8, 8, 128) 0 \n_________________________________________________________________\nconv_pad_4 (ZeroPadding2D) (None, 9, 9, 128) 0 \n_________________________________________________________________\nconv_dw_4 (DepthwiseConv2D) (None, 4, 4, 128) 1152 \n_________________________________________________________________\nconv_dw_4_bn (BatchNormaliza (None, 4, 4, 128) 512 \n_________________________________________________________________\nconv_dw_4_relu (ReLU) (None, 4, 4, 128) 0 \n_________________________________________________________________\nconv_pw_4 (Conv2D) (None, 4, 4, 256) 32768 \n_________________________________________________________________\nconv_pw_4_bn (BatchNormaliza (None, 4, 4, 256) 1024 \n_________________________________________________________________\nconv_pw_4_relu (ReLU) (None, 4, 4, 256) 0 \n_________________________________________________________________\nconv_dw_5 (DepthwiseConv2D) (None, 4, 4, 256) 2304 \n_________________________________________________________________\nconv_dw_5_bn (BatchNormaliza (None, 4, 4, 256) 1024 \n_________________________________________________________________\nconv_dw_5_relu (ReLU) (None, 4, 4, 256) 0 \n_________________________________________________________________\nconv_pw_5 (Conv2D) (None, 4, 4, 256) 65536 \n_________________________________________________________________\nconv_pw_5_bn (BatchNormaliza (None, 4, 4, 256) 1024 \n_________________________________________________________________\nconv_pw_5_relu (ReLU) (None, 4, 4, 256) 0 \n_________________________________________________________________\nconv_pad_6 (ZeroPadding2D) (None, 5, 5, 256) 0 \n_________________________________________________________________\nconv_dw_6 (DepthwiseConv2D) (None, 2, 2, 256) 2304 \n_________________________________________________________________\nconv_dw_6_bn (BatchNormaliza (None, 2, 2, 256) 1024 \n_________________________________________________________________\nconv_dw_6_relu (ReLU) (None, 2, 2, 256) 0 \n_________________________________________________________________\nconv_pw_6 (Conv2D) (None, 2, 2, 512) 131072 \n_________________________________________________________________\nconv_pw_6_bn (BatchNormaliza (None, 2, 2, 512) 2048 \n_________________________________________________________________\nconv_pw_6_relu (ReLU) (None, 2, 2, 512) 0 \n_________________________________________________________________\nconv_dw_7 (DepthwiseConv2D) (None, 2, 2, 512) 4608 \n_________________________________________________________________\nconv_dw_7_bn (BatchNormaliza (None, 2, 2, 512) 2048 \n_________________________________________________________________\nconv_dw_7_relu (ReLU) (None, 2, 2, 512) 0 \n_________________________________________________________________\nconv_pw_7 (Conv2D) (None, 2, 2, 512) 262144 \n_________________________________________________________________\nconv_pw_7_bn (BatchNormaliza (None, 2, 2, 512) 2048 \n_________________________________________________________________\nconv_pw_7_relu (ReLU) (None, 2, 2, 512) 0 \n_________________________________________________________________\nconv_dw_8 (DepthwiseConv2D) (None, 2, 2, 512) 4608 \n_________________________________________________________________\nconv_dw_8_bn (BatchNormaliza (None, 2, 2, 512) 2048 \n_________________________________________________________________\nconv_dw_8_relu (ReLU) (None, 2, 2, 512) 0 \n_________________________________________________________________\nconv_pw_8 (Conv2D) (None, 2, 2, 512) 262144 \n_________________________________________________________________\nconv_pw_8_bn (BatchNormaliza (None, 2, 2, 512) 2048 \n_________________________________________________________________\nconv_pw_8_relu (ReLU) (None, 2, 2, 512) 0 \n_________________________________________________________________\nconv_dw_9 (DepthwiseConv2D) (None, 2, 2, 512) 4608 \n_________________________________________________________________\nconv_dw_9_bn (BatchNormaliza (None, 2, 2, 512) 2048 \n_________________________________________________________________\nconv_dw_9_relu (ReLU) (None, 2, 2, 512) 0 \n_________________________________________________________________\nconv_pw_9 (Conv2D) (None, 2, 2, 512) 262144 \n_________________________________________________________________\nconv_pw_9_bn (BatchNormaliza (None, 2, 2, 512) 2048 \n_________________________________________________________________\nconv_pw_9_relu (ReLU) (None, 2, 2, 512) 0 \n_________________________________________________________________\nconv_dw_10 (DepthwiseConv2D) (None, 2, 2, 512) 4608 \n_________________________________________________________________\nconv_dw_10_bn (BatchNormaliz (None, 2, 2, 512) 2048 \n_________________________________________________________________\nconv_dw_10_relu (ReLU) (None, 2, 2, 512) 0 \n_________________________________________________________________\nconv_pw_10 (Conv2D) (None, 2, 2, 512) 262144 \n_________________________________________________________________\nconv_pw_10_bn (BatchNormaliz (None, 2, 2, 512) 2048 \n_________________________________________________________________\nconv_pw_10_relu (ReLU) (None, 2, 2, 512) 0 \n_________________________________________________________________\nconv_dw_11 (DepthwiseConv2D) (None, 2, 2, 512) 4608 \n_________________________________________________________________\nconv_dw_11_bn (BatchNormaliz (None, 2, 2, 512) 2048 \n_________________________________________________________________\nconv_dw_11_relu (ReLU) (None, 2, 2, 512) 0 \n_________________________________________________________________\nconv_pw_11 (Conv2D) (None, 2, 2, 512) 262144 \n_________________________________________________________________\nconv_pw_11_bn (BatchNormaliz (None, 2, 2, 512) 2048 \n_________________________________________________________________\nconv_pw_11_relu (ReLU) (None, 2, 2, 512) 0 \n_________________________________________________________________\nconv_pad_12 (ZeroPadding2D) (None, 3, 3, 512) 0 \n_________________________________________________________________\nconv_dw_12 (DepthwiseConv2D) (None, 1, 1, 512) 4608 \n_________________________________________________________________\nconv_dw_12_bn (BatchNormaliz (None, 1, 1, 512) 2048 \n_________________________________________________________________\nconv_dw_12_relu (ReLU) (None, 1, 1, 512) 0 \n_________________________________________________________________\nconv_pw_12 (Conv2D) (None, 1, 1, 1024) 524288 \n_________________________________________________________________\nconv_pw_12_bn (BatchNormaliz (None, 1, 1, 1024) 4096 \n_________________________________________________________________\nconv_pw_12_relu (ReLU) (None, 1, 1, 1024) 0 \n_________________________________________________________________\nconv_dw_13 (DepthwiseConv2D) (None, 1, 1, 1024) 9216 \n_________________________________________________________________\nconv_dw_13_bn (BatchNormaliz (None, 1, 1, 1024) 4096 \n_________________________________________________________________\nconv_dw_13_relu (ReLU) (None, 1, 1, 1024) 0 \n_________________________________________________________________\nconv_pw_13 (Conv2D) (None, 1, 1, 1024) 1048576 \n_________________________________________________________________\nconv_pw_13_bn (BatchNormaliz (None, 1, 1, 1024) 4096 \n_________________________________________________________________\nconv_pw_13_relu (ReLU) (None, 1, 1, 1024) 0 \n_________________________________________________________________\nflatten (Flatten) (None, 1024) 0 \n_________________________________________________________________\ndense (Dense) (None, 512) 524800 \n_________________________________________________________________\ndropout (Dropout) (None, 512) 0 \n_________________________________________________________________\ndense_1 (Dense) (None, 512) 262656 \n_________________________________________________________________\ndropout_1 (Dropout) (None, 512) 0 \n_________________________________________________________________\ndense_2 (Dense) (None, 1) 513 \n=================================================================\nTotal params: 4,016,833\nTrainable params: 787,969\nNon-trainable params: 3,228,864\n_________________________________________________________________\nNone\n"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb8b86d3be08a64ef6691b316b50fae9e2fabf44
| 51,646 |
ipynb
|
Jupyter Notebook
|
.ipynb_checkpoints/imdb-checkpoint.ipynb
|
mdhoffschmidt/deeplearning
|
4c0242baad394abee8d08d8899c2c27e66da011f
|
[
"MIT"
] | null | null | null |
.ipynb_checkpoints/imdb-checkpoint.ipynb
|
mdhoffschmidt/deeplearning
|
4c0242baad394abee8d08d8899c2c27e66da011f
|
[
"MIT"
] | null | null | null |
.ipynb_checkpoints/imdb-checkpoint.ipynb
|
mdhoffschmidt/deeplearning
|
4c0242baad394abee8d08d8899c2c27e66da011f
|
[
"MIT"
] | null | null | null | 112.273913 | 35,256 | 0.833985 |
[
[
[
"# Imdb sentiment classification.\n\nDataset of 25,000 movies reviews from IMDB, labeled by sentiment (positive/negative). Reviews have been preprocessed, and each review is encoded as a sequence of word indexes (integers). For convenience, words are indexed by overall frequency in the dataset, so that for instance the integer \"3\" encodes the 3rd most frequent word in the data. This allows for quick filtering operations such as: \"only consider the top 10,000 most common words, but eliminate the top 20 most common words\".",
"_____no_output_____"
]
],
[
[
"# Basic packages.\nimport os\nimport sys\nimport numpy as np\nimport matplotlib.pyplot as plt\nfrom sklearn.metrics import classification_report\n\n# Keras specific packages.\nfrom keras import Input\nfrom keras import Model\nfrom keras import regularizers\nfrom keras import optimizers\nfrom keras.layers import Dense, Activation, Flatten, GRU\nfrom keras.layers import Dropout\nfrom keras.layers import Conv1D, MaxPooling1D\nfrom keras.layers import Embedding\nfrom keras.preprocessing.text import Tokenizer\nfrom keras.preprocessing.sequence import pad_sequences\nfrom keras.utils import to_categorical\nfrom keras.datasets import imdb",
"_____no_output_____"
],
[
"MAX_NUM_WORDS = 10000\nMAX_SEQUENCE_LENGTH = 1000\nEMBEDDING_DIM = 100\nVALIDATION_SPLIT = 0.25\nTEXT_DATA_DIR = \"dataset/20_newsgroup\"\nGLOVE_DIR = \"dataset/glove\"\nEPOCHS = 10\nBATCH_SIZE = 129",
"_____no_output_____"
]
],
[
[
"## 1. Load the dataset.",
"_____no_output_____"
]
],
[
[
"# Load the data.\n(x_train, y_train), (x_test, y_test) = imdb.load_data(num_words=MAX_NUM_WORDS)",
"_____no_output_____"
],
[
"# Get the word to index dict.\nword_to_index = imdb.get_word_index()\n\n# Get the index to word dict.\nindex_to_word = dict(\n[(value, key) for (key, value) in word_to_index.items()])",
"_____no_output_____"
],
[
"# Display\nprint(\"Length dictionnary = {}\".format(len(word_to_index)))\n\nmax_row = []\nfor i in range(x_train.shape[0]):\n max_row.append(len(x_train[i]))\nprint(max(max_row))",
"Length dictionnary = 88584\n2494\n"
]
],
[
[
"## 2. Preparing the pretrained embedding layer.",
"_____no_output_____"
]
],
[
[
"embeddings_index = {}\nf = open(os.path.join(GLOVE_DIR, \"glove.6B.{}d.txt\".format(EMBEDDING_DIM)))\nfor line in f:\n values = line.split()\n word = values[0]\n coefs = np.asarray(values[1:], dtype=\"float32\")\n embeddings_index[word] = coefs\nf.close()\n\nprint(\"Found %s word vectors.\" % len(embeddings_index))",
"Found 400000 word vectors.\n"
],
[
"embedding_matrix = np.zeros((len(word_to_index) + 1, EMBEDDING_DIM))\nfor word, i in word_to_index.items():\n embedding_vector = embeddings_index.get(word)\n if embedding_vector is not None:\n # words not found in embedding index will be all-zeros.\n embedding_matrix[i] = embedding_vector",
"_____no_output_____"
],
[
"embedding_layer = Embedding(len(word_to_index) + 1,\n EMBEDDING_DIM,\n weights=[embedding_matrix],\n input_length=MAX_SEQUENCE_LENGTH,\n trainable=False)",
"_____no_output_____"
]
],
[
[
"## 3. Handle the dataset.\n\nHere we gather the features of words $X \\in \\mathbb{R}^{m \\times n}$ where $m$ is the total number of samples and $n$ is the features length. For the current example $n$ is equal to $10000$.\n",
"_____no_output_____"
]
],
[
[
"# Pad the training and test features.\nx_tr = pad_sequences(x_train, maxlen=MAX_SEQUENCE_LENGTH)\nx_te = pad_sequences(x_test, maxlen=MAX_SEQUENCE_LENGTH)\n\n# Display the size.\nprint(\"Size x_tr = {}\".format(x_tr.shape))\nprint(\"Size x_te = {}\".format(x_te.shape))",
"Size x_tr = (25000, 1000)\nSize x_te = (25000, 1000)\n"
],
[
"# Handle the training and test labels.\ny_tr = y_train.reshape(-1, 1)\ny_te = y_test.reshape(-1, 1)\n\n# Display the shapes.\nprint(\"y_train \", y_tr.shape)\nprint(\"y_test \", y_te.shape)",
"y_train (25000, 1)\ny_test (25000, 1)\n"
]
],
[
[
"## 3. Build the model.",
"_____no_output_____"
]
],
[
[
"# Set the input.\nsequence_input = Input(shape=(MAX_SEQUENCE_LENGTH,), dtype=\"int32\")\n\n# Set the embedding layer.\nembedded_sequences = embedding_layer(sequence_input)\n\n# Conv layer 1.\n\"\"\"\nx = Conv1D(64, 5, kernel_regularizer=regularizers.l2(0.001))(embedded_sequences)\nx = Activation(\"relu\")(x)\nx = MaxPooling1D(5)(x)\nX = Dropout(0.5)(x)\n\n# Conv Layer 2.\nx = Conv1D(64, 5, kernel_regularizer=regularizers.l2(0.001))(x)\nx = Activation(\"relu\")(x)\nx = MaxPooling1D(5)(x)\nX = Dropout(0.5)(x)\n\n# Conv Layer 3.\nx = Conv1D(64, 5, kernel_regularizer=regularizers.l2(0.001))(x)\nx = Activation(\"relu\")(x)\nx = MaxPooling1D(35)(x)\nX = Dropout(0.5)(x)\n\n# Output layer.\nx = Flatten()(x)\nx = Dense(128)(x)\nx = Activation(\"relu\")(x)\nX = Dropout(0.5)(x)\n\n\"\"\"\n\n#x = Flatten()(x)\n#x = Dense(128)(x)\n#x = Activation(\"relu\")(x)\n#X = Dropout(0.5)(x)\n\nx = GRU(128, return_sequences=False)(embedded_sequences)\n\n# Softmax layer.\npreds = Dense(1, activation=\"sigmoid\")(x)\n\n# Build the model.\nmodel = Model(sequence_input, preds)\n\n# Set the optimizer.\noptim = optimizers.Adam(lr=0.001)\n\n# Compile the model.\nmodel.compile(loss=\"binary_crossentropy\", optimizer=optim, metrics=[\"acc\"])\n\n# Set the fitting parameters.\nfit_params = {\n \"epochs\": EPOCHS,\n \"batch_size\": BATCH_SIZE,\n \"validation_split\": VALIDATION_SPLIT,\n \"shuffle\": True\n}\n\n# Print the model.\nmodel.summary()",
"_____no_output_____"
],
[
"# Fit the model.\nhistory = model.fit(x_tr, y_tr, **fit_params)",
"Train on 18750 samples, validate on 6250 samples\nEpoch 1/10\n18750/18750 [==============================] - 79s 4ms/step - loss: 0.8134 - acc: 0.5122 - val_loss: 0.7612 - val_acc: 0.5168\nEpoch 2/10\n18750/18750 [==============================] - 79s 4ms/step - loss: 0.7261 - acc: 0.5756 - val_loss: 0.6950 - val_acc: 0.6310\nEpoch 3/10\n18750/18750 [==============================] - 80s 4ms/step - loss: 0.6694 - acc: 0.6398 - val_loss: 0.6301 - val_acc: 0.6781\nEpoch 4/10\n18750/18750 [==============================] - 79s 4ms/step - loss: 0.6122 - acc: 0.6942 - val_loss: 0.6031 - val_acc: 0.7018\nEpoch 5/10\n18750/18750 [==============================] - 83s 4ms/step - loss: 0.5898 - acc: 0.7152 - val_loss: 0.5790 - val_acc: 0.7277\nEpoch 6/10\n18750/18750 [==============================] - 80s 4ms/step - loss: 0.5273 - acc: 0.7627 - val_loss: 0.5727 - val_acc: 0.7310\nEpoch 7/10\n18750/18750 [==============================] - 78s 4ms/step - loss: 0.4907 - acc: 0.7914 - val_loss: 0.5617 - val_acc: 0.7480\nEpoch 8/10\n18750/18750 [==============================] - 78s 4ms/step - loss: 0.4446 - acc: 0.8226 - val_loss: 0.5502 - val_acc: 0.7566\nEpoch 9/10\n18750/18750 [==============================] - 80s 4ms/step - loss: 0.4153 - acc: 0.8401 - val_loss: 0.5626 - val_acc: 0.7560\nEpoch 10/10\n18750/18750 [==============================] - 79s 4ms/step - loss: 0.3831 - acc: 0.8599 - val_loss: 0.5941 - val_acc: 0.7534\n"
],
[
"# Visualise the training resuls.\nplt.figure(figsize=(15,5))\nplt.subplot(121)\nplt.plot(history.history[\"loss\"], color=\"b\", label=\"tr\")\nplt.plot(history.history[\"val_loss\"], color=\"r\", label=\"te\")\nplt.ylabel(\"loss\")\nplt.xlabel(\"epochs\")\nplt.grid()\nplt.legend()\nplt.subplot(122)\nplt.plot(history.history[\"acc\"], color=\"b\", label=\"tr\")\nplt.plot(history.history[\"val_acc\"], color=\"r\", label=\"te\")\nplt.ylabel(\"acc\")\nplt.xlabel(\"epochs\")\nplt.grid()\nplt.legend()\nplt.show()",
"_____no_output_____"
]
],
[
[
"## 4. Evaluation",
"_____no_output_____"
]
],
[
[
"# Get the predictions for the test dataset.\ny_pred = model.predict(x_te)\n\n# Update the predictions.\ny_pred = 1.0 * (y_pred > 0.5 )\n\n# Display the classification report.\nprint(classification_report(y_te, y_pred))",
" precision recall f1-score support\n\n 0 0.79 0.69 0.74 12500\n 1 0.72 0.82 0.77 12500\n\navg / total 0.76 0.75 0.75 25000\n\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb8b986324b63e8c03f200969316f6eed605ba58
| 19,463 |
ipynb
|
Jupyter Notebook
|
docs/tutorials/barren_plateaus.ipynb
|
Pranay144/quantum
|
76d58fd774e003d02a8261f4b9f17991cab7e9a7
|
[
"Apache-2.0"
] | 1 |
2020-03-10T20:24:36.000Z
|
2020-03-10T20:24:36.000Z
|
docs/tutorials/barren_plateaus.ipynb
|
rish-16/quantum
|
f781f1bb65df746dc76ce93f54c6d599fc54a1cb
|
[
"Apache-2.0"
] | null | null | null |
docs/tutorials/barren_plateaus.ipynb
|
rish-16/quantum
|
f781f1bb65df746dc76ce93f54c6d599fc54a1cb
|
[
"Apache-2.0"
] | null | null | null | 37.646035 | 611 | 0.533114 |
[
[
[
"##### Copyright 2020 The TensorFlow Authors.",
"_____no_output_____"
]
],
[
[
"#@title Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# https://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.",
"_____no_output_____"
]
],
[
[
"# Barren plateaus",
"_____no_output_____"
],
[
"<table class=\"tfo-notebook-buttons\" align=\"left\">\n <td>\n <a target=\"_blank\" href=\"https://www.tensorflow.org/quantum/tutorials/barren_plateaus\"><img src=\"https://www.tensorflow.org/images/tf_logo_32px.png\" />View on TensorFlow.org</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://colab.research.google.com/github/tensorflow/quantum/blob/master/docs/tutorials/barren_plateaus.ipynb\"><img src=\"https://www.tensorflow.org/images/colab_logo_32px.png\" />Run in Google Colab</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://github.com/tensorflow/quantum/blob/master/docs/tutorials/barren_plateaus.ipynb\"><img src=\"https://www.tensorflow.org/images/GitHub-Mark-32px.png\" />View source on GitHub</a>\n </td>\n <td>\n <a href=\"https://storage.googleapis.com/tensorflow_docs/quantum/docs/tutorials/barren_plateaus.ipynb\"><img src=\"https://www.tensorflow.org/images/download_logo_32px.png\" />Download notebook</a>\n </td>\n</table>",
"_____no_output_____"
],
[
"In this example you will explore the result of <a href=\"https://www.nature.com/articles/s41467-018-07090-4\" class=\"external\">McClean, 2019</a> that says not just any quantum neural network structure will do well when it comes to learning. In particular you will see that a certain large family of random quantum circuits do not serve as good quantum neural networks, because they have gradients that vanish almost everywhere. In this example you won't be training any models for a specific learning problem, but instead focusing on the simpler problem of understanding the behaviors of gradients.",
"_____no_output_____"
],
[
"## Setup",
"_____no_output_____"
]
],
[
[
"try:\n %tensorflow_version 2.x\nexcept Exception:\n pass",
"_____no_output_____"
]
],
[
[
"Install TensorFlow Quantum:",
"_____no_output_____"
]
],
[
[
"!pip install tensorflow-quantum",
"_____no_output_____"
]
],
[
[
"Now import TensorFlow and the module dependencies:",
"_____no_output_____"
]
],
[
[
"import tensorflow as tf\nimport tensorflow_quantum as tfq\n\nimport cirq\nimport sympy\nimport numpy as np\n\n# visualization tools\n%matplotlib inline\nimport matplotlib.pyplot as plt\nfrom cirq.contrib.svg import SVGCircuit\n\nnp.random.seed(1234)",
"_____no_output_____"
]
],
[
[
"## 1. Summary\n\nRandom quantum circuits with many blocks that look like this ($R_{P}(\\theta)$ is a random Pauli rotation):<br/>\n<img src=\"./images/barren_2.png\" width=700>\n\nWhere if $f(x)$ is defined as the expectation value w.r.t. $Z_{a}Z_{b}$ for any qubits $a$ and $b$, then there is a problem that $f'(x)$ has a mean very close to 0 and does not vary much. You will see this below:",
"_____no_output_____"
],
[
"## 2. Generating random circuits\n\nThe construction from the paper is straightforward to follow. The following implements a simple function that generates a random quantum circuit—sometimes referred to as a *quantum neural network* (QNN)—with the given depth on a set of qubits:",
"_____no_output_____"
]
],
[
[
"def generate_random_qnn(qubits, symbol, depth):\n \"\"\"Generate random QNN's with the same structure from McClean et al.\"\"\"\n circuit = cirq.Circuit()\n for qubit in qubits:\n circuit += cirq.Ry(np.pi / 4.0)(qubit)\n\n for d in range(depth):\n # Add a series of single qubit rotations.\n for i, qubit in enumerate(qubits):\n random_n = np.random.uniform()\n random_rot = np.random.uniform(\n ) * 2.0 * np.pi if i != 0 or d != 0 else symbol\n if random_n > 2. / 3.:\n # Add a Z.\n circuit += cirq.Rz(random_rot)(qubit)\n elif random_n > 1. / 3.:\n # Add a Y.\n circuit += cirq.Ry(random_rot)(qubit)\n else:\n # Add a X.\n circuit += cirq.Rx(random_rot)(qubit)\n\n # Add CZ ladder.\n for src, dest in zip(qubits, qubits[1:]):\n circuit += cirq.CZ(src, dest)\n\n return circuit\n\n\ngenerate_random_qnn(cirq.GridQubit.rect(1, 3), sympy.Symbol('theta'), 2)",
"_____no_output_____"
]
],
[
[
"The authors investigate the gradient of a single parameter $\\theta_{1,1}$. Let's follow along by placing a `sympy.Symbol` in the circuit where $\\theta_{1,1}$ would be. Since the authors do not analyze the statistics for any other symbols in the circuit, let's replace them with random values now instead of later.",
"_____no_output_____"
],
[
"## 3. Running the circuits\n\nGenerate a few of these circuits along with an observable to test the claim that the gradients don't vary much. First, generate a batch of random circuits. Choose a random *ZZ* observable and batch calculate the gradients and variance using TensorFlow Quantum.",
"_____no_output_____"
],
[
"### 3.1 Batch variance computation\n\nLet's write a helper function that computes the variance of the gradient of a given observable over a batch of circuits:",
"_____no_output_____"
]
],
[
[
"def process_batch(circuits, symbol, op):\n \"\"\"Compute the variance of a batch of expectations w.r.t. op on each circuit that \n contains `symbol`. Note that this method sets up a new compute graph every time it is\n called so it isn't as performant as possible.\"\"\"\n\n # Setup a simple layer to batch compute the expectation gradients.\n expectation = tfq.layers.Expectation()\n\n # Prep the inputs as tensors\n circuit_tensor = tfq.convert_to_tensor(circuits)\n values_tensor = tf.convert_to_tensor(\n np.random.uniform(0, 2 * np.pi, (n_circuits, 1)).astype(np.float32))\n\n # Use TensorFlow GradientTape to track gradients.\n with tf.GradientTape() as g:\n g.watch(values_tensor)\n forward = expectation(circuit_tensor,\n operators=op,\n symbol_names=[symbol],\n symbol_values=values_tensor)\n\n # Return variance of gradients across all circuits.\n grads = g.gradient(forward, values_tensor)\n grad_var = tf.math.reduce_std(grads, axis=0)\n return grad_var.numpy()[0]",
"_____no_output_____"
]
],
[
[
"### 3.1 Set up and run\n\nChoose the number of random circuits to generate along with their depth and the amount of qubits they should act on. Then plot the results.",
"_____no_output_____"
]
],
[
[
"n_qubits = [2 * i for i in range(2, 7)\n ] # Ranges studied in paper are between 2 and 24.\ndepth = 50 # Ranges studied in paper are between 50 and 500.\nn_circuits = 200\ntheta_var = []\n\nfor n in n_qubits:\n # Generate the random circuits and observable for the given n.\n qubits = cirq.GridQubit.rect(1, n)\n symbol = sympy.Symbol('theta')\n circuits = [\n generate_random_qnn(qubits, symbol, depth) for _ in range(n_circuits)\n ]\n op = cirq.Z(qubits[0]) * cirq.Z(qubits[1])\n theta_var.append(process_batch(circuits, symbol, op))\n\nplt.semilogy(n_qubits, theta_var)\nplt.title('Gradient Variance in QNNs')\nplt.xlabel('n_qubits')\nplt.ylabel('$\\\\partial \\\\theta$ variance')\nplt.show()",
"_____no_output_____"
]
],
[
[
"This plot shows that for quantum machine learning problems, you can't simply guess a random QNN ansatz and hope for the best. Some structure must be present in the model circuit in order for gradients to vary to the point where learning can happen.",
"_____no_output_____"
],
[
"## 4. Heuristics\n\nAn interesting heuristic by <a href=\"https://arxiv.org/pdf/1903.05076.pdf\" class=\"external\">Grant, 2019</a> allows one to start very close to random, but not quite. Using the same circuits as McClean et al., the authors propose a different initialization technique for the classical control parameters to avoid barren plateaus. The initialization technique starts some layers with totally random control parameters—but, in the layers immediately following, choose parameters such that the initial transformation made by the first few layers is undone. The authors call this an *identity block*.\n\nThe advantage of this heuristic is that by changing just a single parameter, all other blocks outside of the current block will remain the identity—and the gradient signal comes through much stronger than before. This allows the user to pick and choose which variables and blocks to modify to get a strong gradient signal. This heuristic does not prevent the user from falling in to a barren plateau during the training phase (and restricts a fully simultaneous update), it just guarantees that you can start outside of a plateau.",
"_____no_output_____"
],
[
"### 4.1 New QNN construction\n\nNow construct a function to generate identity block QNNs. This implementation is slightly different than the one from the paper. For now, look at the behavior of the gradient of a single parameter so it is consistent with McClean et al, so some simplifications can be made.\n\nTo generate an identity block and train the model, generally you need $U1(\\theta_{1a}) U1(\\theta_{1b})^{\\dagger}$ and not $U1(\\theta_1) U1(\\theta_1)^{\\dagger}$. Initially $\\theta_{1a}$ and $\\theta_{1b}$ are the same angles but they are learned independently. Otherwise, you will always get the identity even after training. The choice for the number of identity blocks is empirical. The deeper the block, the smaller the variance in the middle of the block. But at the start and end of the block, the variance of the parameter gradients should be large. ",
"_____no_output_____"
]
],
[
[
"def generate_identity_qnn(qubits, symbol, block_depth, total_depth):\n \"\"\"Generate random QNN's with the same structure from Grant et al.\"\"\"\n circuit = cirq.Circuit()\n\n # Generate initial block with symbol.\n prep_and_U = generate_random_qnn(qubits, symbol, block_depth)\n circuit += prep_and_U\n\n # Generate dagger of initial block without symbol.\n U_dagger = (prep_and_U[1:])**-1\n circuit += cirq.resolve_parameters(\n U_dagger, param_resolver={symbol: np.random.uniform() * 2 * np.pi})\n\n for d in range(total_depth - 1):\n # Get a random QNN.\n prep_and_U_circuit = generate_random_qnn(\n qubits,\n np.random.uniform() * 2 * np.pi, block_depth)\n\n # Remove the state-prep component\n U_circuit = prep_and_U_circuit[1:]\n\n # Add U\n circuit += U_circuit\n\n # Add U^dagger\n circuit += U_circuit**-1\n\n return circuit\n\n\ngenerate_identity_qnn(cirq.GridQubit.rect(1, 3), sympy.Symbol('theta'), 2, 2)",
"_____no_output_____"
]
],
[
[
"### 4.2 Comparison\n\nHere you can see that the heuristic does help to keep the variance of the gradient from vanishing as quickly:",
"_____no_output_____"
]
],
[
[
"block_depth = 10\ntotal_depth = 5\n\nheuristic_theta_var = []\n\nfor n in n_qubits:\n # Generate the identity block circuits and observable for the given n.\n qubits = cirq.GridQubit.rect(1, n)\n symbol = sympy.Symbol('theta')\n circuits = [\n generate_identity_qnn(qubits, symbol, block_depth, total_depth)\n for _ in range(n_circuits)\n ]\n op = cirq.Z(qubits[0]) * cirq.Z(qubits[1])\n heuristic_theta_var.append(process_batch(circuits, symbol, op))\n\nplt.semilogy(n_qubits, theta_var)\nplt.semilogy(n_qubits, heuristic_theta_var)\nplt.title('Heuristic vs. Random')\nplt.xlabel('n_qubits')\nplt.ylabel('$\\\\partial \\\\theta$ variance')\nplt.show()",
"_____no_output_____"
]
],
[
[
"This is a great improvement in getting stronger gradient signals from (near) random QNNs.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
cb8b9c468d3958d98d18b08262554b6495d5f659
| 76,668 |
ipynb
|
Jupyter Notebook
|
wholesalecumter_kmeans.ipynb
|
SJ-16/test_machinelearning
|
8ccddcd9503e2ec65a1d319382fcbdf217d85768
|
[
"Apache-2.0"
] | null | null | null |
wholesalecumter_kmeans.ipynb
|
SJ-16/test_machinelearning
|
8ccddcd9503e2ec65a1d319382fcbdf217d85768
|
[
"Apache-2.0"
] | null | null | null |
wholesalecumter_kmeans.ipynb
|
SJ-16/test_machinelearning
|
8ccddcd9503e2ec65a1d319382fcbdf217d85768
|
[
"Apache-2.0"
] | null | null | null | 124.663415 | 36,358 | 0.822416 |
[
[
[
"<a href=\"https://colab.research.google.com/github/SJ-16/test_machinelearning/blob/master/wholesalecumter_kmeans.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"!ls",
" sample_data 'Wholesale customers data.csv'\n"
],
[
"!ls -l",
"total 20\ndrwxr-xr-x 1 root root 4096 Jun 15 13:37 sample_data\n-rw-r--r-- 1 root root 15021 Jul 2 04:46 'Wholesale customers data.csv'\n"
],
[
"!pwd",
"/content\n"
],
[
"!ls -l ./sample_data",
"total 55504\n-rwxr-xr-x 1 root root 1697 Jan 1 2000 anscombe.json\n-rw-r--r-- 1 root root 301141 Jun 15 13:37 california_housing_test.csv\n-rw-r--r-- 1 root root 1706430 Jun 15 13:37 california_housing_train.csv\n-rw-r--r-- 1 root root 18289443 Jun 15 13:37 mnist_test.csv\n-rw-r--r-- 1 root root 36523880 Jun 15 13:37 mnist_train_small.csv\n-rwxr-xr-x 1 root root 930 Jan 1 2000 README.md\n"
],
[
"!ls -l ./Wholesale\\ customers\\ data.csv",
"-rw-r--r-- 1 root root 15021 Jul 2 04:46 './Wholesale customers data.csv'\n"
],
[
"import pandas as pd\ndf = pd.read_csv('./Wholesale customers data.csv')\ndf.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 440 entries, 0 to 439\nData columns (total 8 columns):\n # Column Non-Null Count Dtype\n--- ------ -------------- -----\n 0 Channel 440 non-null int64\n 1 Region 440 non-null int64\n 2 Fresh 440 non-null int64\n 3 Milk 440 non-null int64\n 4 Grocery 440 non-null int64\n 5 Frozen 440 non-null int64\n 6 Detergents_Paper 440 non-null int64\n 7 Delicassen 440 non-null int64\ndtypes: int64(8)\nmemory usage: 27.6 KB\n"
],
[
"X = df.iloc[:,:]",
"_____no_output_____"
],
[
"X.shape",
"_____no_output_____"
],
[
"from sklearn.preprocessing import StandardScaler\nscaler = StandardScaler()\nscaler.fit(X)\nX = scaler.transform(X)",
"_____no_output_____"
],
[
"from sklearn import cluster\nkmeans = cluster.KMeans(n_clusters=5)",
"_____no_output_____"
],
[
"kmeans.fit(X)",
"_____no_output_____"
],
[
"kmeans.labels_",
"_____no_output_____"
],
[
"df['label'] = kmeans.labels_",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
],
[
"df.plot(kind='scatter', x= 'Grocery', y='Frozen', c='label', cmap='Set1',figsize=(10,10) )",
"_____no_output_____"
],
[
"# for ...:\n# if ~((df['label'] == 0) | (df['label'] ==4 )):\ndfx = df[~((df['label'] == 0) | (df['label'] ==4 ))]\ndf.shape, dfx.shape",
"_____no_output_____"
],
[
"dfx.plot(kind='scatter', x= 'Grocery', y='Frozen', c='label', cmap='Set1')",
"_____no_output_____"
],
[
"df.to_excel('./wholesale.xls')",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb8b9c908194c85a1dce8f4990c50ecc0cd5199e
| 24,207 |
ipynb
|
Jupyter Notebook
|
01-Linear_Algebra/Week_05/notebooks/PageRank-SOLVED.ipynb
|
goosen78/Coursera-Specialization___Mathematics-for-ML
|
1b9a74ca10b7a1a68f85ff4515ca075cfe7647e1
|
[
"MIT"
] | null | null | null |
01-Linear_Algebra/Week_05/notebooks/PageRank-SOLVED.ipynb
|
goosen78/Coursera-Specialization___Mathematics-for-ML
|
1b9a74ca10b7a1a68f85ff4515ca075cfe7647e1
|
[
"MIT"
] | null | null | null |
01-Linear_Algebra/Week_05/notebooks/PageRank-SOLVED.ipynb
|
goosen78/Coursera-Specialization___Mathematics-for-ML
|
1b9a74ca10b7a1a68f85ff4515ca075cfe7647e1
|
[
"MIT"
] | null | null | null | 35.862222 | 285 | 0.574173 |
[
[
[
"# PageRank\nIn this notebook, you'll build on your knowledge of eigenvectors and eigenvalues by exploring the PageRank algorithm.\nThe notebook is in two parts, the first is a worksheet to get you up to speed with how the algorithm works - here we will look at a micro-internet with fewer than 10 websites and see what it does and what can go wrong.\nThe second is an assessment which will test your application of eigentheory to this problem by writing code and calculating the page rank of a large network representing a sub-section of the internet.",
"_____no_output_____"
],
[
"## Part 1 - Worksheet\n### Introduction\n\nPageRank (developed by Larry Page and Sergey Brin) revolutionized web search by generating a\nranked list of web pages based on the underlying connectivity of the web. The PageRank algorithm is\nbased on an ideal random web surfer who, when reaching a page, goes to the next page by clicking on a\nlink. The surfer has equal probability of clicking any link on the page and, when reaching a page with no\nlinks, has equal probability of moving to any other page by typing in its URL. In addition, the surfer may\noccasionally choose to type in a random URL instead of following the links on a page. The PageRank is\nthe ranked order of the pages from the most to the least probable page the surfer will be viewing.\n",
"_____no_output_____"
]
],
[
[
"# Before we begin, let's load the libraries.\n%matplotlib widget\n\nimport matplotlib.pyplot as plt\nimport numpy as np\nimport numpy.linalg as la\nfrom readonly.PageRankFunctions import *\nnp.set_printoptions(suppress=True)",
"_____no_output_____"
]
],
[
[
"### PageRank as a linear algebra problem\nLet's imagine a micro-internet, with just 6 websites (**A**vocado, **B**ullseye, **C**atBabel, **D**romeda, **e**Tings, and **F**aceSpace).\nEach website links to some of the others, and this forms a network as shown,\n\n\n\nThe design principle of PageRank is that important websites will be linked to by important websites.\nThis somewhat recursive principle will form the basis of our thinking.\n\nImagine we have 100 *Procrastinating Pat*s on our micro-internet, each viewing a single website at a time.\nEach minute the Pats follow a link on their website to another site on the micro-internet.\nAfter a while, the websites that are most linked to will have more Pats visiting them, and in the long run, each minute for every Pat that leaves a website, another will enter keeping the total numbers of Pats on each website constant.\nThe PageRank is simply the ranking of websites by how many Pats they have on them at the end of this process.\n\nWe represent the number of Pats on each website with the vector,\n$$\\mathbf{r} = \\begin{bmatrix} r_A \\\\ r_B \\\\ r_C \\\\ r_D \\\\ r_E \\\\ r_F \\end{bmatrix}$$\nAnd say that the number of Pats on each website in minute $i+1$ is related to those at minute $i$ by the matrix transformation\n\n$$ \\mathbf{r}^{(i+1)} = L \\,\\mathbf{r}^{(i)}$$\nwith the matrix $L$ taking the form,\n$$ L = \\begin{bmatrix}\nL_{A→A} & L_{B→A} & L_{C→A} & L_{D→A} & L_{E→A} & L_{F→A} \\\\\nL_{A→B} & L_{B→B} & L_{C→B} & L_{D→B} & L_{E→B} & L_{F→B} \\\\\nL_{A→C} & L_{B→C} & L_{C→C} & L_{D→C} & L_{E→C} & L_{F→C} \\\\\nL_{A→D} & L_{B→D} & L_{C→D} & L_{D→D} & L_{E→D} & L_{F→D} \\\\\nL_{A→E} & L_{B→E} & L_{C→E} & L_{D→E} & L_{E→E} & L_{F→E} \\\\\nL_{A→F} & L_{B→F} & L_{C→F} & L_{D→F} & L_{E→F} & L_{F→F} \\\\\n\\end{bmatrix}\n$$\nwhere the columns represent the probability of leaving a website for any other website, and sum to one.\nThe rows determine how likely you are to enter a website from any other, though these need not add to one.\nThe long time behaviour of this system is when $ \\mathbf{r}^{(i+1)} = \\mathbf{r}^{(i)}$, so we'll drop the superscripts here, and that allows us to write,\n$$ L \\,\\mathbf{r} = \\mathbf{r}$$\n\nwhich is an eigenvalue equation for the matrix $L$, with eigenvalue 1 (this is guaranteed by the probabalistic structure of the matrix $L$).\n\nComplete the matrix $L$ below, we've left out the column for which websites the *FaceSpace* website (F) links to.\nRemember, this is the probability to click on another website from this one, so each column should add to one (by scaling by the number of links).",
"_____no_output_____"
]
],
[
[
"# Replace the ??? here with the probability of clicking a link to each website when leaving Website F (FaceSpace).\nL = np.array([[0, 1/2, 1/3, 0, 0, 1/3 ],\n [1/3, 0, 0, 0, 1/2, 0 ],\n [1/3, 1/2, 0, 1, 0, 1/2 ],\n [1/3, 0, 1/3, 0, 1/2, 0 ],\n [0, 0, 0, 0, 0, 1/6 ],\n [0, 0, 1/3, 0, 0, 0 ]])",
"_____no_output_____"
]
],
[
[
"In principle, we could use a linear algebra library, as below, to calculate the eigenvalues and vectors.\nAnd this would work for a small system. But this gets unmanagable for large systems.\nAnd since we only care about the principal eigenvector (the one with the largest eigenvalue, which will be 1 in this case), we can use the *power iteration method* which will scale better, and is faster for large systems.\n\nUse the code below to peek at the PageRank for this micro-internet.",
"_____no_output_____"
]
],
[
[
"eVals, eVecs = la.eig(L) # Gets the eigenvalues and vectors\norder = np.absolute(eVals).argsort()[::-1] # Orders them by their eigenvalues\neVals = eVals[order]\neVecs = eVecs[:,order]\n\nr = eVecs[:, 0] # Sets r to be the principal eigenvector\n100 * np.real(r / np.sum(r)) # Make this eigenvector sum to one, then multiply by 100 Procrastinating Pats",
"_____no_output_____"
]
],
[
[
"We can see from this list, the number of Procrastinating Pats that we expect to find on each website after long times.\nPutting them in order of *popularity* (based on this metric), the PageRank of this micro-internet is:\n\n**C**atBabel, **D**romeda, **A**vocado, **F**aceSpace, **B**ullseye, **e**Tings\n\nReferring back to the micro-internet diagram, is this what you would have expected?\nConvince yourself that based on which pages seem important given which others link to them, that this is a sensible ranking.\n\nLet's now try to get the same result using the Power-Iteration method that was covered in the video.\nThis method will be much better at dealing with large systems.\n\nFirst let's set up our initial vector, $\\mathbf{r}^{(0)}$, so that we have our 100 Procrastinating Pats equally distributed on each of our 6 websites.",
"_____no_output_____"
]
],
[
[
"r = 100 * np.ones(6) / 6 # Sets up this vector (6 entries of 1/6 × 100 each)\nr # Shows it's value",
"_____no_output_____"
]
],
[
[
"Next, let's update the vector to the next minute, with the matrix $L$.\nRun the following cell multiple times, until the answer stabilises.",
"_____no_output_____"
]
],
[
[
"r = L @ r # Apply matrix L to r\nr # Show it's value\n# Re-run this cell multiple times to converge to the correct answer.",
"_____no_output_____"
]
],
[
[
"We can automate applying this matrix multiple times as follows,",
"_____no_output_____"
]
],
[
[
"r = 100 * np.ones(6) / 6 # Sets up this vector (6 entries of 1/6 × 100 each)\nfor i in np.arange(100) : # Repeat 100 times\n r = L @ r\nr",
"_____no_output_____"
]
],
[
[
"Or even better, we can keep running until we get to the required tolerance.",
"_____no_output_____"
]
],
[
[
"r = 100 * np.ones(6) / 6 # Sets up this vector (6 entries of 1/6 × 100 each)\nlastR = r\nr = L @ r\ni = 0\nwhile la.norm(lastR - r) > 0.01 :\n lastR = r\n r = L @ r\n i += 1\nprint(str(i) + \" iterations to convergence.\")\nr",
"20 iterations to convergence.\n"
]
],
[
[
"See how the PageRank order is established fairly quickly, and the vector converges on the value we calculated earlier after a few tens of repeats.\n\nCongratulations! You've just calculated your first PageRank!",
"_____no_output_____"
],
[
"### Damping Parameter\nThe system we just studied converged fairly quickly to the correct answer.\nLet's consider an extension to our micro-internet where things start to go wrong.\n\nSay a new website is added to the micro-internet: *Geoff's* Website.\nThis website is linked to by *FaceSpace* and only links to itself.\n\n\nIntuitively, only *FaceSpace*, which is in the bottom half of the page rank, links to this website amongst the two others it links to,\nso we might expect *Geoff's* site to have a correspondingly low PageRank score.\n\nBuild the new $L$ matrix for the expanded micro-internet, and use Power-Iteration on the Procrastinating Pat vector.\nSee what happens…",
"_____no_output_____"
]
],
[
[
" # We'll call this one L2, to distinguish it from the previous L.\nL2 = np.array([[0, 1/2, 1/3, 0, 0, 1/3, 0 ],\n [1/3, 0, 0, 0, 1/2, 1/3, 0 ],\n [1/3, 1/2, 0, 1, 0, 0, 0 ],\n [1/3, 0, 1/3, 0, 1/2, 0, 0 ],\n [0, 0, 0, 0, 0, 1/3, 0 ],\n [0, 0, 1/3, 0, 0, 0, 0 ],\n [0, 0, 0, 0, 0, 0, 1 ]])",
"_____no_output_____"
],
[
"r = 100 * np.ones(7) / 7 # Sets up this vector (6 entries of 1/6 × 100 each)\nlastR = r\nr = L2 @ r\ni = 0\nwhile la.norm(lastR - r) > 0.01 :\n lastR = r\n r = L2 @ r\n i += 1\nprint(str(i) + \" iterations to convergence.\")\nr",
"12 iterations to convergence.\n"
]
],
[
[
"That's no good! *Geoff* seems to be taking all the traffic on the micro-internet, and somehow coming at the top of the PageRank.\nThis behaviour can be understood, because once a Pat get's to *Geoff's* Website, they can't leave, as all links head back to Geoff.\n\nTo combat this, we can add a small probability that the Procrastinating Pats don't follow any link on a webpage, but instead visit a website on the micro-internet at random.\nWe'll say the probability of them following a link is $d$ and the probability of choosing a random website is therefore $1-d$.\nWe can use a new matrix to work out where the Pat's visit each minute.\n$$ M = d \\, L + \\frac{1-d}{n} \\, J $$\nwhere $J$ is an $n\\times n$ matrix where every element is one.\n\nIf $d$ is one, we have the case we had previously, whereas if $d$ is zero, we will always visit a random webpage and therefore all webpages will be equally likely and equally ranked.\nFor this extension to work best, $1-d$ should be somewhat small - though we won't go into a discussion about exactly how small.\n\nLet's retry this PageRank with this extension.",
"_____no_output_____"
]
],
[
[
"d = 0.5 # Feel free to play with this parameter after running the code once.\nM = d * L2 + (1-d)/7 * np.ones([7, 7]) # np.ones() is the J matrix, with ones for each entry.",
"_____no_output_____"
],
[
"r = 100 * np.ones(7) / 7 # Sets up this vector (6 entries of 1/6 × 100 each)\nlastR = r\nr = M @ r\ni = 0\nwhile la.norm(lastR - r) > 0.01 :\n lastR = r\n r = M @ r\n i += 1\nprint(str(i) + \" iterations to convergence.\")\nr",
"5 iterations to convergence.\n"
]
],
[
[
"This is certainly better, the PageRank gives sensible numbers for the Procrastinating Pats that end up on each webpage.\nThis method still predicts Geoff has a high ranking webpage however.\nThis could be seen as a consequence of using a small network. We could also get around the problem by not counting self-links when producing the L matrix (an if a website has no outgoing links, make it link to all websites equally).\nWe won't look further down this route, as this is in the realm of improvements to PageRank, rather than eigenproblems.\n\nYou are now in a good position, having gained an understanding of PageRank, to produce your own code to calculate the PageRank of a website with thousands of entries.\n\nGood Luck!",
"_____no_output_____"
],
[
"## Part 2 - Assessment\nIn this assessment, you will be asked to produce a function that can calculate the PageRank for an arbitrarily large probability matrix.\nThis, the final assignment of the course, will give less guidance than previous assessments.\nYou will be expected to utilise code from earlier in the worksheet and re-purpose it to your needs.\n\n### How to submit\nEdit the code in the cell below to complete the assignment.\nOnce you are finished and happy with it, press the *Submit Assignment* button at the top of this notebook.\n\nPlease don't change any of the function names, as these will be checked by the grading script.\n\nIf you have further questions about submissions or programming assignments, here is a [list](https://www.coursera.org/learn/linear-algebra-machine-learning/discussions/weeks/1/threads/jB4klkn5EeibtBIQyzFmQg) of Q&A. You can also raise an issue on the discussion forum. Good luck!",
"_____no_output_____"
]
],
[
[
"# PACKAGE\n# Here are the imports again, just in case you need them.\n# There is no need to edit or submit this cell.\nimport numpy as np\nimport numpy.linalg as la\nfrom readonly.PageRankFunctions import *\nnp.set_printoptions(suppress=True)",
"_____no_output_____"
],
[
"# GRADED FUNCTION\n# Complete this function to provide the PageRank for an arbitrarily sized internet.\n# I.e. the principal eigenvector of the damped system, using the power iteration method.\n# (Normalisation doesn't matter here)\n# The functions inputs are the linkMatrix, and d the damping parameter - as defined in this worksheet.\ndef pageRank(linkMatrix, d) :\n n = linkMatrix.shape[0]\n M = d * linkMatrix + (1-d)/n * np.ones([n, n]) # np.ones() is the J matrix, with ones for each entry.\n r = 100 * np.ones(n) / n\n lastR = r\n r = M @ r\n while la.norm (lastR - r) > 0.01 :\n lastR = r\n r = M @ r\n \n return r\n",
"_____no_output_____"
]
],
[
[
"## Test your code before submission\nTo test the code you've written above, run the cell (select the cell above, then press the play button [ ▶| ] or press shift-enter).\nYou can then use the code below to test out your function.\nYou don't need to submit this cell; you can edit and run it as much as you like.",
"_____no_output_____"
]
],
[
[
"# Use the following function to generate internets of different sizes.\ngenerate_internet(5)",
"_____no_output_____"
],
[
"# Test your PageRank method against the built in \"eig\" method.\n# You should see yours is a lot faster for large internets\nL = generate_internet(10)",
"_____no_output_____"
],
[
"pageRank(L, 1)",
"_____no_output_____"
],
[
"# Do note, this is calculating the eigenvalues of the link matrix, L,\n# without any damping. It may give different results that your pageRank function.\n# If you wish, you could modify this cell to include damping.\n# (There is no credit for this though)\neVals, eVecs = la.eig(L) # Gets the eigenvalues and vectors\norder = np.absolute(eVals).argsort()[::-1] # Orders them by their eigenvalues\neVals = eVals[order]\neVecs = eVecs[:,order]\n\nr = eVecs[:, 0]\n100 * np.real(r / np.sum(r))",
"_____no_output_____"
],
[
"\n# You may wish to view the PageRank graphically.\n# This code will draw a bar chart, for each (numbered) website on the generated internet,\n# The height of each bar will be the score in the PageRank.\n# Run this code to see the PageRank for each internet you generate.\n# Hopefully you should see what you might expect\n# - there are a few clusters of important websites, but most on the internet are rubbish!\n%matplotlib widget\n\nimport matplotlib.pyplot as plt\nimport numpy as np\nr = pageRank(generate_internet(100), 0.9)\nplt.bar(np.arange(r.shape[0]), r);",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
cb8baef749939524d553a736c64d3707fcc98e46
| 1,968 |
ipynb
|
Jupyter Notebook
|
project/Transparency/preprocess/Tweets/Tweet_ADR.ipynb
|
hancia/ToxicSpansDetection
|
4a10600292af90a936767aee09559b39380e3d5e
|
[
"Apache-2.0"
] | 3 |
2021-03-23T08:07:54.000Z
|
2021-11-13T07:13:32.000Z
|
project/Transparency/preprocess/Tweets/Tweet_ADR.ipynb
|
hancia/ToxicSpansDetection
|
4a10600292af90a936767aee09559b39380e3d5e
|
[
"Apache-2.0"
] | null | null | null |
project/Transparency/preprocess/Tweets/Tweet_ADR.ipynb
|
hancia/ToxicSpansDetection
|
4a10600292af90a936767aee09559b39380e3d5e
|
[
"Apache-2.0"
] | null | null | null | 21.626374 | 151 | 0.553354 |
[
[
[
"## Obtaining the datasets\n\nFor obtaining ADR tweets data, please contact the authors of 'Attention is Not Explanation' paper (https://arxiv.org/abs/1902.10186). \n",
"_____no_output_____"
]
],
[
[
"import pandas as pd",
"_____no_output_____"
],
[
"df = pd.read_csv('adr_dataset.csv')\n\nfrom sklearn.model_selection import train_test_split\ntrain_idx, dev_idx = train_test_split(df.index[df.exp_split == 'train'], test_size=0.15, random_state=16377)",
"_____no_output_____"
],
[
"df.loc[dev_idx, 'exp_split'] = 'dev'\ndf.to_csv('adr_dataset_split.csv', index=False)",
"_____no_output_____"
],
[
"%run \"../preprocess_data_BC.py\" --data_file adr_dataset_split.csv --output_file ./vec_adr.p --word_vectors_type fasttext.simple.300d --min_df 2",
"Vocabulary size : 6845\nFound 5645 words in model out of 6845\n"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
cb8bb984eb9f48e1561e05157a1d2f4f6e9e9ece
| 7,894 |
ipynb
|
Jupyter Notebook
|
AM/Saved_Model_Checking.ipynb
|
Abdulk084/Hybrid_Model_Tox21
|
df2e14a6a621dafbfb08b2bb43d9790f4d96c4d1
|
[
"MIT"
] | 7 |
2018-11-19T20:06:48.000Z
|
2021-01-05T10:27:31.000Z
|
AM/Saved_Model_Checking.ipynb
|
Abdulk084/Hybrid_Model_Tox21
|
df2e14a6a621dafbfb08b2bb43d9790f4d96c4d1
|
[
"MIT"
] | 1 |
2020-09-23T14:51:38.000Z
|
2021-03-08T02:22:41.000Z
|
AM/Saved_Model_Checking.ipynb
|
Abdulk084/Hybrid_Model_Tox21
|
df2e14a6a621dafbfb08b2bb43d9790f4d96c4d1
|
[
"MIT"
] | 5 |
2020-02-23T14:56:27.000Z
|
2021-10-29T22:35:17.000Z
| 29.129151 | 259 | 0.554472 |
[
[
[
"import pandas as pd\nimport numpy as np\nimport h5py\nimport matplotlib.pyplot as plt\nimport scipy\nfrom PIL import Image\nfrom scipy import ndimage\n#from dnn_app_utils_v2 import *\nimport pandas as pd\n%matplotlib inline\nfrom pandas import ExcelWriter\nfrom pandas import ExcelFile\n%load_ext autoreload\n%autoreload 2\nfrom sklearn.utils import resample\nimport tensorflow as tf\nfrom tensorflow.python.framework import ops\nimport openpyxl\nimport keras\nimport xlsxwriter\n\n\n\n\n\nfrom keras.layers import Dense, Dropout\nfrom keras import optimizers\n\n\n\n\n\nimport pandas as pd\nimport numpy as np\nimport h5py\nimport matplotlib.pyplot as plt\nimport scipy\nfrom PIL import Image\nfrom scipy import ndimage\n#from dnn_app_utils_v2 import *\nimport pandas as pd\n%matplotlib inline\nfrom pandas import ExcelWriter\nfrom pandas import ExcelFile\n%load_ext autoreload\n%autoreload 2\nfrom sklearn.utils import resample\nimport tensorflow as tf\nfrom tensorflow.python.framework import ops\nimport openpyxl\nimport keras\nimport xlsxwriter\n\n\n\n\nfrom keras.models import Sequential\nfrom keras.layers import Dense\nfrom keras.wrappers.scikit_learn import KerasClassifier\nfrom sklearn.model_selection import cross_val_score\nfrom sklearn.preprocessing import LabelEncoder\nfrom sklearn.model_selection import StratifiedKFold\nfrom sklearn.preprocessing import StandardScaler\nfrom sklearn.pipeline import Pipeline\n\nprint(\" All the necessary Libraries have been loaded\")\nprint(\" \")\n\n\nprint(\" \")\nprint(\" The code after this is for loading your data into train and test. Make sure you load the correct features\")\n\n\nxls = pd.ExcelFile(\"test_selected.xlsx\")\n\ntest_selected_x = pd.read_excel(xls, 'test_selected_x')\n\ntest_selected_y = pd.read_excel(xls, 'test_selected_y')\n\n\nprint(\" The selected important features data for spesific model is loaded into train, and test\")\nprint(\" \")\n\n\n\n\n\ntest_selected_x=test_selected_x.values\ntest_selected_y=test_selected_y.values\n\n\n\n\n\nprint(\"##################################################################################################\")\n\n\n\n\nprint(\"Now you load the model but with correct model name\")\n\n\nprint(\" loading the trained model \")\nprint(\" \")\n\nfrom keras.models import model_from_json\n# load json and create model\njson_file = open('1_model.json', 'r')\nloaded_model_json = json_file.read()\njson_file.close()\nloaded_model_1 = model_from_json(loaded_model_json)\n# load weights into new model\nloaded_model_1.load_weights(\"1_model.h5\")\nprint(\"Loaded model from disk\")\nprint(\" \")\n\n\n\njson_file = open('2_model.json', 'r')\nloaded_model_json = json_file.read()\njson_file.close()\nloaded_model_2 = model_from_json(loaded_model_json)\n# load weights into new model\nloaded_model_2.load_weights(\"2_model.h5\")\nprint(\"Loaded model from disk\")\nprint(\" \")\n\n\njson_file = open('3_model.json', 'r')\nloaded_model_json = json_file.read()\njson_file.close()\nloaded_model_3 = model_from_json(loaded_model_json)\n# load weights into new model\nloaded_model_3.load_weights(\"3_model.h5\")\nprint(\"Loaded model from disk\")\nprint(\" \")\n\njson_file = open('4_model.json', 'r')\nloaded_model_json = json_file.read()\njson_file.close()\nloaded_model_4 = model_from_json(loaded_model_json)\n# load weights into new model\nloaded_model_4.load_weights(\"4_model.h5\")\nprint(\"Loaded model from disk\")\nprint(\" \")",
"C:\\ProgramData\\Anaconda3\\lib\\site-packages\\h5py\\__init__.py:34: FutureWarning: Conversion of the second argument of issubdtype from `float` to `np.floating` is deprecated. In future, it will be treated as `np.float64 == np.dtype(float).type`.\n from ._conv import register_converters as _register_converters\nUsing TensorFlow backend.\n"
],
[
"print(\" Computing the AUCROC using the loded model for checking \")\nprint(\" \")\n\n\n\n\n\n\n\n\n\n\n\n\nfrom sklearn.metrics import roc_auc_score, roc_curve\n\n \npred_test_1 = loaded_model_1.predict(test_selected_x)\npred_test_2 = loaded_model_2.predict(test_selected_x)\npred_test_3 = loaded_model_3.predict(test_selected_x)\npred_test_4 = loaded_model_4.predict(test_selected_x)\npred_test=(pred_test_1+pred_test_2+pred_test_3+pred_test_4)/4\n\nauc_test = roc_auc_score(test_selected_y, pred_test)\n\n\nprint (\"AUROC_test: \" + str(auc_test))\n \n\n",
" Computing the AUCROC using the loded model for checking \n \nAUROC_test: 0.8781239660175341\n"
]
]
] |
[
"code"
] |
[
[
"code",
"code"
]
] |
cb8bbf65d11c028a638dfe8baaccf34f30d4203c
| 450,118 |
ipynb
|
Jupyter Notebook
|
sigMF GPU STFT SVD voice reconstruction v6 old.ipynb
|
rdbadger/RF_SVD
|
d9d2da1c6f6b08df8968944ff463d738420984d7
|
[
"MIT"
] | null | null | null |
sigMF GPU STFT SVD voice reconstruction v6 old.ipynb
|
rdbadger/RF_SVD
|
d9d2da1c6f6b08df8968944ff463d738420984d7
|
[
"MIT"
] | null | null | null |
sigMF GPU STFT SVD voice reconstruction v6 old.ipynb
|
rdbadger/RF_SVD
|
d9d2da1c6f6b08df8968944ff463d738420984d7
|
[
"MIT"
] | null | null | null | 694.626543 | 228,180 | 0.948325 |
[
[
[
"## sigMF STFT on GPU and CPU",
"_____no_output_____"
]
],
[
[
"import os\nimport itertools\nfrom sklearn.utils import shuffle\nimport torch, torchvision\nimport torch.nn as nn\nimport torch.nn.functional as d\nimport torch.optim as optim\nimport torch.nn.functional as F\nimport torch.nn.modules as mod\nimport torch.utils.data\nimport torch.utils.data as data\nfrom torch.nn.utils.rnn import pack_padded_sequence\nfrom torch.nn.utils.rnn import pad_packed_sequence\nfrom torch.autograd import Variable\nimport numpy as np\nimport sys\nimport importlib\nimport time\nimport matplotlib.pyplot as plt\nimport torchvision.datasets as datasets\nimport torchvision.transforms as transforms\nfrom torchvision.utils import save_image\nimport librosa\nfrom scipy import signal\nfrom scipy import stats\nfrom scipy.special import comb\nimport matplotlib.pyplot as plt\nimport glob\nimport json\nimport pickle\nfrom random import randint, choice\nimport random\nfrom timeit import default_timer as timer\n# from torchaudio.functional import istft\nfrom torch import istft\nfrom sklearn.decomposition import NMF\nplt.style.use('default')\ndevice = torch.device('cuda:0')\nprint('Torch version =', torch.__version__, 'CUDA version =', torch.version.cuda)\nprint('CUDA Device:', device)\nprint('Is cuda available? =',torch.cuda.is_available())",
"Torch version = 1.6.0 CUDA version = 10.2\nCUDA Device: cuda:0\nIs cuda available? = True\n"
],
[
"# %matplotlib notebook\n# %matplotlib inline",
"_____no_output_____"
]
],
[
[
"#### Machine paths",
"_____no_output_____"
]
],
[
[
"path_save = \"/home/david/sigMF_ML/SVD/saved/\" \npath = \"/home/david/sigMF_ML/SVD/\" \nprint(path)",
"/home/david/sigMF_ML/SVD/\n"
]
],
[
[
"#### reading sigmf meta data and encoder function",
"_____no_output_____"
]
],
[
[
"# START OF FUNCTIONS ****************************************************\ndef meta_encoder(meta_list, num_classes): \n a = np.asarray(meta_list, dtype=int)\n# print('a = ', a)\n return a \n\ndef read_meta(meta_files):\n meta_list = []\n for meta in meta_files:\n all_meta_data = json.load(open(meta))\n meta_list.append(all_meta_data['global'][\"core:class\"])\n meta_list = list(map(int, meta_list))\n return meta_list\n\ndef read_num_val(x):\n x = len(meta_list_val)\n return x",
"_____no_output_____"
],
[
"print(path)\nos.chdir(path)\ndata_files = sorted(glob.glob('*.sigmf-data'))\nmeta_files = sorted(glob.glob('*.sigmf-meta'))",
"/home/david/sigMF_ML/SVD/\n"
],
[
"for meta in meta_files:\n all_meta_data = json.load(open(meta))\n print(\"file name = \", meta)",
"file name = UHF_UV5R_V2_plus_voice__snr_hi.sigmf-meta\n"
]
],
[
[
"#### torch GPU Cuda stft",
"_____no_output_____"
]
],
[
[
"def gpu(db, n_fft):\n I = db[0::2]\n Q = db[1::2]\n start = timer()\n w = n_fft\n win = torch.hann_window(w, periodic=True, dtype=None, layout=torch.strided, requires_grad=False).cuda()\n I_stft = torch.stft(torch.tensor(I).cuda(), n_fft=n_fft, hop_length=n_fft//2, win_length=w, window=win, center=True, normalized=True, onesided=False)\n Q_stft = torch.stft(torch.tensor(Q).cuda(), n_fft=n_fft, hop_length=n_fft//2, win_length=w, window=win, center=True, normalized=True, onesided=False)\n X_stft = I_stft[...,0] + Q_stft[...,0] + I_stft[...,1] + -1*Q_stft[...,1]\n X_stft = torch.cat((X_stft[n_fft//2:],X_stft[:n_fft//2]))\n end = timer() # mag spec of sum of I and Q complex channels; sqrt((a+bj)^2+(c+dj)^2)\n print(end - start)\n torch.cuda.empty_cache()\n return X_stft, I_stft, Q_stft",
"_____no_output_____"
]
],
[
[
"#### scipy CPU stft function",
"_____no_output_____"
]
],
[
[
"def cpu(db, n_fft):\n t = len(db)\n db2 = db[0::]\n start = timer()\n db = db.astype(np.float32).view(np.complex64)\n Fs = 1e6\n I_t, I_f, Z = signal.stft(db, fs=Fs, nperseg=n_fft, return_onesided=False)\n Z = np.vstack([Z[n_fft//2:], Z[:n_fft//2]])\n end = timer()\n print(end - start)\n return Z",
"_____no_output_____"
]
],
[
[
"### GPU Timing: Slow the first time running",
"_____no_output_____"
]
],
[
[
"n_fft = 1024\nfor file in data_files:\n db = np.fromfile(file, dtype=\"float32\")\n X_stft, I_stft, Q_stft = gpu(db, n_fft)",
"0.041077464004047215\n"
],
[
"plt.imshow(20*np.log10(np.abs(X_stft.cpu()+1e-8)), aspect='auto', origin='lower')\nplt.show()",
"_____no_output_____"
]
],
[
[
"### CPU Timing",
"_____no_output_____"
]
],
[
[
"for file in data_files:\n db = np.fromfile(file, dtype=\"float32\")\n stft_cpu = cpu(db, 1000)",
"0.20826889807358384\n"
]
],
[
[
"### CPU load stft to Cuda Time",
"_____no_output_____"
]
],
[
[
"start = timer()\nIQ_tensor = torch.tensor(np.abs(stft_cpu)).cuda()\nend = timer()\nprint(end - start)\ntorch.cuda.empty_cache()",
"0.06956048600841314\n"
],
[
"plt.imshow(20*np.log10(np.abs(stft_cpu)+1e-8), aspect='auto', origin='lower')\nplt.show()",
"_____no_output_____"
]
],
[
[
"#### GPU SVD",
"_____no_output_____"
]
],
[
[
"def udv_stft(I_stft,Q_stft):\n start = timer()\n U_I0, D_I0, V_I0 = torch.svd(I_stft[...,0]) \n U_I1, D_I1, V_I1 = torch.svd(I_stft[...,1]) \n U_Q0, D_Q0, V_Q0 = torch.svd(Q_stft[...,0]) \n U_Q1, D_Q1, V_Q1 = torch.svd(Q_stft[...,1]) \n end = timer()\n print('SVD time: ',end - start)\n return U_I0, D_I0, V_I0, U_I1, D_I1, V_I1, U_Q0, D_Q0, V_Q0, U_Q1, D_Q1, V_Q1",
"_____no_output_____"
]
],
[
[
"#### Inverse stft ",
"_____no_output_____"
]
],
[
[
"# def ISTFT(db, n_fft):# We are matching scipy.signal behavior (setting noverlap=frame_length - hop) \n# w = 512\n# win = torch.hann_window(w, periodic=True, dtype=None, layout=torch.strided, requires_grad=False).cuda()\n# start = timer()\n# Z = istft(db, n_fft=n_fft, hop_length=n_fft//2, win_length=w, window=win, center=True, normalized=True, onesided=False)\n# end = timer()\n# print('ISTFT time = ',end - start)\n# torch.cuda.empty_cache()\n# return Z",
"_____no_output_____"
],
[
"def ISTFT(db, n_fft):# We are matching scipy.signal behavior (setting noverlap=frame_length - hop) \n w = n_fft\n win = torch.hann_window(w, periodic=True, dtype=None, layout=torch.strided, requires_grad=False).cuda()\n start = timer()\n Z = istft(db, n_fft=n_fft, hop_length=n_fft//2, win_length=n_fft, window=win, center=True, normalized=True, onesided=False)\n end = timer()\n print('ISTFT time = ',end - start)\n torch.cuda.empty_cache()\n return Z",
"_____no_output_____"
]
],
[
[
"#### Re-combine UDV to approximate original signal",
"_____no_output_____"
]
],
[
[
"def udv(u, d, v, k): # like ----> np.matrix(U[:, :k]) * np.diag(D[:k]) * V[:k, :]\n start = timer()\n UD = torch.mul(u[:, :k], d[:k])\n v = torch.transpose(v,1,0)\n UDV = torch.mm(UD, v[:k, :])\n end = timer()\n print('UDV time: ',end - start)\n return UDV",
"_____no_output_____"
]
],
[
[
"### Main function to run all sub function calls",
"_____no_output_____"
]
],
[
[
"def complete(I_stft,Q_stft, num, n_fft):\n U_I0, D_I0, V_I0, U_I1, D_I1, V_I1, U_Q0, D_Q0, V_Q0, U_Q1, D_Q1, V_Q1 = udv_stft(I_stft,Q_stft)\n torch.cuda.empty_cache()\n print('UDV I0 shapes = ',U_I0.shape, D_I0.shape, V_I0.shape)\n print('UDV I1 shapes = ',U_I1.shape, D_I1.shape, V_I1.shape)\n print('UDV Q0 shapes = ', U_Q0.shape, D_Q0.shape, V_Q0.shape)\n print('UDV Q1 shapes = ', U_Q1.shape, D_Q1.shape, V_Q1.shape)\n udv_I0 = udv(U_I0, D_I0, V_I0,num)\n udv_I1 = udv(U_I1, D_I1, V_I1,num)\n udv_Q0 = udv(U_Q0, D_Q0, V_Q0,num)\n udv_Q1 = udv(U_Q1, D_Q1, V_Q1,num)\n torch.cuda.empty_cache()\n print('udv I shapes = ',udv_I0.shape,udv_I1.shape)\n print('udv Q shapes = ',udv_Q0.shape,udv_Q1.shape)\n # -------------stack and transpose----------------------------------------\n UDV_I = torch.stack([udv_I0,udv_I1])\n UDV_I = torch.transpose(UDV_I,2,0)\n UDV_I = torch.transpose(UDV_I,1,0)\n UDV_Q = torch.stack([udv_Q0,udv_Q1])\n UDV_Q = torch.transpose(UDV_Q,2,0)\n UDV_Q = torch.transpose(UDV_Q,1,0)\n torch.cuda.empty_cache()\n #--------------------------------------------------------------------------\n I = ISTFT(UDV_I, n_fft)\n Q = ISTFT(UDV_Q, n_fft)\n torch.cuda.empty_cache()\n I = I.detach().cpu().numpy()\n Q = Q.detach().cpu().numpy()\n end = len(I)*2\n IQ_SVD = np.zeros(len(I)*2)\n IQ_SVD[0:end:2] = I\n IQ_SVD[1:end:2] = Q \n IQ_SVD = IQ_SVD.astype(np.float32).view(np.complex64)\n return IQ_SVD",
"_____no_output_____"
]
],
[
[
"### Perform SVD on IQ stft data",
"_____no_output_____"
]
],
[
[
"num = 3 # number to reconstruct SVD matrix from\nIQ_SVD = complete(I_stft,Q_stft, num, n_fft)",
"SVD time: 2.275431339046918\nUDV I0 shapes = torch.Size([1024, 1024]) torch.Size([1024]) torch.Size([9766, 1024])\nUDV I1 shapes = torch.Size([1024, 1024]) torch.Size([1024]) torch.Size([9766, 1024])\nUDV Q0 shapes = torch.Size([1024, 1024]) torch.Size([1024]) torch.Size([9766, 1024])\nUDV Q1 shapes = torch.Size([1024, 1024]) torch.Size([1024]) torch.Size([9766, 1024])\nUDV time: 0.0010549119906499982\nUDV time: 0.00031954399310052395\nUDV time: 0.0002644420601427555\nUDV time: 0.00025576597545295954\nudv I shapes = torch.Size([1024, 9766]) torch.Size([1024, 9766])\nudv Q shapes = torch.Size([1024, 9766]) torch.Size([1024, 9766])\nISTFT time = 0.008287633070722222\nISTFT time = 0.009978348040021956\n"
],
[
"torch.cuda.empty_cache()",
"_____no_output_____"
]
],
[
[
"### Write reconstructed IQ file to file",
"_____no_output_____"
]
],
[
[
"from array import array\nIQ_file = open(\"UV5R_voice2\", 'wb')\nIQ_SVD.tofile(IQ_file)\nIQ_file.close()",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb8bc578b60a77a76ec94f6be46f04acb6371423
| 11,552 |
ipynb
|
Jupyter Notebook
|
Neural Network Numpy.ipynb
|
heytanay/neural-net-scratch
|
a5535b36819f5b359a673ee37db01aa510992ca9
|
[
"MIT"
] | null | null | null |
Neural Network Numpy.ipynb
|
heytanay/neural-net-scratch
|
a5535b36819f5b359a673ee37db01aa510992ca9
|
[
"MIT"
] | null | null | null |
Neural Network Numpy.ipynb
|
heytanay/neural-net-scratch
|
a5535b36819f5b359a673ee37db01aa510992ca9
|
[
"MIT"
] | null | null | null | 31.391304 | 147 | 0.441309 |
[
[
[
"import numpy as np\nfrom matplotlib import pyplot as plt",
"_____no_output_____"
],
[
"def relu(z):\n return max(0, z)\n\ndef sigmoid(z):\n return (1/(1+np.exp(-z)))",
"_____no_output_____"
],
[
"def layer_sizes(X, Y):\n \"\"\"\n Arguments:\n X -- input dataset of shape (input size, number of examples)\n Y -- labels of shape (output size, number of examples)\n \"\"\"\n \n n_x = X.shape[0] # size of input layer\n n_h = 4\n n_y = Y.shape[0] # size of output layer\n \n return (n_x, n_h, n_y)",
"_____no_output_____"
],
[
"def init_params(n_x, n_h, n_y):\n \"\"\"\n Argument:\n n_x -- size of the input layer\n n_h -- size of the hidden layer\n n_y -- size of the output layer\n \n Function:\n Generates Weights (random) and Biases (zeros) for the 2 layer neural network\n \n \"\"\"\n \n W1 = np.random.randn(n_h, n_x) * 0.01\n b1 = np.zeros((n_h, 1))\n W2 = np.random.randn(n_y, n_h) * 0.01\n b2 = np.zeros((n_y, 1))\n \n paramters = {\n 'W1': W1,\n 'b1': b1,\n 'W2': W2,\n 'b2': b2,\n }\n \n return paramters",
"_____no_output_____"
],
[
"def forward_propagation(X, parameters):\n \"\"\"\n Argument:\n X -- input data of size (n_x, m)\n parameters -- python dictionary containing your parameters (output of initialization function)\n \n \"\"\"\n # Extract the Weights and Bias from parameters dictionary\n W1 = parameters['W1']\n b1 = parameters['b1']\n W2 = parameters['W2']\n b2 = parameters['b2']\n \n # Calculate multiple steps of forward propagation and at the end calculate A2 Probabilities\n Z1 = np.matmul(W1, X) + b1\n A1 = np.tanh(Z1)\n Z2 = np.matmul(W2, A1) + b2\n A2 = sigmoid(Z2)\n \n cache = {\n 'Z1': Z1,\n 'A1': A1,\n 'Z2': Z2,\n 'A2': A2\n }\n \n return cache",
"_____no_output_____"
],
[
"def calc_cost(A2, Y, parameters):\n \"\"\"\n Computes the cross-entropy cost\n \n Arguments:\n A2 -- The sigmoid output of the second activation, of shape (1, number of examples)\n Y -- \"true\" labels vector of shape (1, number of examples)\n parameters -- python dictionary containing your parameters W1, b1, W2 and b2\n \n \"\"\"\n # Get the total number of examples\n m = Y.shape[1]\n \n # Compute the Cross-entropy cost\n logprob = (np.multiply(np.log(A2), Y) + np.multiply(np.log(1-A2), (1-Y)))\n \n # Squeeze the Numpy array (removes the extra dimensions)\n cost = np.squeeze(-(1/m) * np.sum(logprob))\n \n return cost",
"_____no_output_____"
],
[
"def backward_prop(parameters, cache, X, Y):\n \"\"\"\n Arguments:\n parameters -- python dictionary containing our parameters \n cache -- a dictionary containing \"Z1\", \"A1\", \"Z2\" and \"A2\".\n X -- input data of shape (2, number of examples)\n Y -- \"true\" labels vector of shape (1, number of examples)\n \"\"\"\n \n m = X.shape[1]\n \n # Get the Weights from 'paramters' dictionary\n W1 = parameters['W1']\n W2 = parameters['W2']\n \n # Retrieve the respective activations from 'cache' dictionary\n A1 = cache['A1']\n A2 = cache['A2']\n \n # Calculate respective derivatives\n dZ2 = A2 - Y # Derivative of Final layer output is final Activation - Target value (Predicted - Original)\n dW2 = (1/m) * np.matmul(dZ2, A1.T) # Derivative of Second layer weights is multiplication of dZ2 and A1.T, averaged over all samples\n db2 = (1/m) * np.sum(dZ2, axis=1, keepdims=True)\n dZ1 = np.matmul(W2.T, dZ2) * (1 - np.power(A1, 2))\n dW1 = (1/m) * np.matmul(dZ1, X.T)\n db1 = (1/m) * np.sum(dZ1, axis=1, keepdims=True)\n \n gradients = {\n 'dW1': dW1,\n 'db1': db1,\n 'dW2': dW2,\n 'db2': db2,\n }",
"_____no_output_____"
],
[
"def update_paramters(parameters, gradients, learning_rate = 1):\n \"\"\"\n Updates parameters using the gradient descent update rule given above\n \n Arguments:\n parameters -- python dictionary containing your parameters \n grads -- python dictionary containing your gradients \n \n \"\"\"\n \n # Get the Paramters from the Dictionary\n W1 = paramters['W1']\n b1 = parameters['b1']\n W2 = parameters['W2']\n b2 = parameters['b2']\n \n # Get the gradients from the dictionary\n dW1 = gradients['dW1']\n db1 = gradients['db1']\n dW2 = gradients['dW2']\n db2 = gradients['db2']\n \n # Run Gradient Descent for all weights and biases\n W1 = W1 - learning_rate * dW1\n b1 = b1 - learning_rate * db1\n W2 = W2 - learning_rate * dW2\n b2 = b2 - learning_rate * db2\n \n # Pack the parameters into a dictionary\n paramters = {\n 'W1': W1,\n 'b1': b1,\n 'W2': W2,\n 'b2': b2,\n }\n \n return paramters",
"_____no_output_____"
],
[
"def fit_model(X, Y, n_h, epochs=10000, print_cost = False):\n \"\"\"\n Arguments:\n X -- dataset of shape (2, number of examples)\n Y -- labels of shape (1, number of examples)\n n_h -- size of the hidden layer\n num_iterations -- Number of iterations in gradient descent loop\n print_cost -- if True, print the cost every 1000 iterations\n \n \"\"\"\n \n # Make new n_x and n_y\n n_x = layer_sizes(X, Y)[0]\n n_y = layer_sizes(X, Y)[2]\n \n # Initialize paramters\n parameters = initialize_parameters(n_x, n_h, n_y)\n W1 = parameters['W1']\n b1 = parameters['b1']\n W2 = parameters['W2']\n b2 = parameters['b2']\n \n \n # Run the loop for training\n for epoch in range(epochs):\n cache = forward_propagation(X, parameters=parameters)\n \n A2 = cache['A2']\n \n cost = calc_cost(A2, Y, parameters)\n \n grads = backward_prop(parameters, cache, X, Y)\n \n parameters = update_paramters(parameters, grads)\n \n if print_cost and epoch % 1000 == 0:\n print(\"Cost after iteration %i: %f\" %(i, cost))\n \n return parameters",
"_____no_output_____"
],
[
"def predict(parameters, X):\n \"\"\"\n Using the learned parameters, predicts a class for each example in X\n \n Arguments:\n parameters -- python dictionary containing your parameters \n X -- input data of size (n_x, m)\n \"\"\"\n \n cache = forward_propagation(X, parameters)\n A2 = cache['A2']\n \n predictions = (A2 > 0.5)\n \n return predictions",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb8bc650060df1b96edff658a89921fe60c114ed
| 14,225 |
ipynb
|
Jupyter Notebook
|
assets/nb-lessons/.ipynb_checkpoints/06_training_and_datasets-Copy3-checkpoint.ipynb
|
ravinpoudel/ml-training-site
|
1734d8769a51c4444c33bb21d0a1abd1e6f18e51
|
[
"MIT"
] | null | null | null |
assets/nb-lessons/.ipynb_checkpoints/06_training_and_datasets-Copy3-checkpoint.ipynb
|
ravinpoudel/ml-training-site
|
1734d8769a51c4444c33bb21d0a1abd1e6f18e51
|
[
"MIT"
] | null | null | null |
assets/nb-lessons/.ipynb_checkpoints/06_training_and_datasets-Copy3-checkpoint.ipynb
|
ravinpoudel/ml-training-site
|
1734d8769a51c4444c33bb21d0a1abd1e6f18e51
|
[
"MIT"
] | null | null | null | 23.708333 | 680 | 0.525132 |
[
[
[
"# Training and Data Sets\nAuthor: Ravin Poudel",
"_____no_output_____"
],
[
"Main goal in the statistical or machine learning model is to biuld a generalized predictive-model. Often we start with a set of data to build a model and describe the model fit and other properties. However, it is equally important to test the model with new data (the data that has not been used in fitting a model), and check the model performace. From agricultural perspective, basically we need to run an additional experiment to generate a data for purpose of model validation. Instead what we can do is to __randomly__ divide the a single dataset into two parts, and use one part for the purpose of learnign whereas the other part for testing the model performacne.\n\n<img src=\"../nb-images/Train_test.png\">\n\n> Train data set: A data set used to __construct/train__ a model. \n\n> Test data set: A data set used to __evaluate__ the model.\n\n\n\n#### How do we spilit a single dataset into two?\n\nThere is not a single or one best solution. Its convention to use more data for training the model than to test/evaluate the moddel. Often convention such as `75%/ 25% train/ test` or `90%/10% train/test` scheme are used. Larger the training dataset allows to learn better model, while the larger testing dataset, the better condifence in the model evaluation. \n> Can we apply similar data-splitting scheme when we have a small dataset? Often the case in agriculure or lifescience - \"as of now\".\n\n> Does a single random split make our predictive model random? Do we want a stable model or a random model?\n",
"_____no_output_____"
],
[
"We will be using an `iris dataset` to explore the concept of data-spiliting. The data set contains:\n\n- 50 samples of 3 different species of iris folower (150 samples total)\n- Iris folower: Setosa, Versicolour, and Virginica\n- Measurements: sepal length, sepal width, petal length, petal width\n",
"_____no_output_____"
]
],
[
[
"# Import data and modules\nimport numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nfrom sklearn import datasets\nfrom sklearn.model_selection import train_test_split\n",
"_____no_output_____"
],
[
"# import iris data from scikit and data preparato\niris = datasets.load_iris()\n",
"_____no_output_____"
],
[
"# check data shape\niris_X.data.shape\n",
"_____no_output_____"
],
[
"print(iris_y)",
"[0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1\n 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 2 2 2 2 2 2 2 2 2 2 2\n 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2\n 2 2]\n"
],
[
"print(names)",
"[]\n"
],
[
"print(feature_names)",
"['sepal length (cm)', 'sepal width (cm)', 'petal length (cm)', 'petal width (cm)']\n"
],
[
"# splitting into train and test data\n# test dataset = 20% of the original dataset\n\nX_train, X_test, y_train, y_test = train_test_split(iris_X, iris_y, test_size=0.2, random_state=0)",
"_____no_output_____"
],
[
"# shape of train dataset\nX_train.shape, y_train.shape",
"_____no_output_____"
],
[
"# shape of test dataset\nX_test.shape, y_test.shape\n",
"_____no_output_____"
],
[
"# instantiate a K-Nearest Neighbors(KNN) model, and fit with X and y\nmodel = KNeighborsClassifier()\nmodel_tt = model.fit(X_train, y_train)",
"_____no_output_____"
],
[
"# check the accuracy on the training set\nmodel_tt.score(X_test, y_test)\n",
"_____no_output_____"
],
[
"# predict class labels for the test set\npredicted = model_tt.predict(X_test)\nprint (predicted)",
"[2 1 0 2 0 2 0 1 1 1 2 1 1 1 2 0 1 1 0 0 2 1 0 0 2 0 0 1 1 0]\n"
],
[
"print(y_test)",
"[2 1 0 2 0 2 0 1 1 1 2 1 1 1 1 0 1 1 0 0 2 1 0 0 2 0 0 1 1 0]\n"
],
[
"# generate evaluation metrics\nfrom sklearn import metrics\nprint (metrics.accuracy_score(y_test, predicted))",
"0.9666666666666667\n"
],
[
"print (metrics.confusion_matrix(y_test, predicted))\n",
"[[11 0 0]\n [ 0 12 1]\n [ 0 0 6]]\n"
],
[
"print (metrics.classification_report(y_test, predicted))",
" precision recall f1-score support\n\n 0 1.00 1.00 1.00 11\n 1 1.00 0.92 0.96 13\n 2 0.86 1.00 0.92 6\n\n accuracy 0.97 30\n macro avg 0.95 0.97 0.96 30\nweighted avg 0.97 0.97 0.97 30\n\n"
]
],
[
[
"### Model Evaluation Using Cross-Validation",
"_____no_output_____"
]
],
[
[
"# evaluate the model using 10-fold cross-validation\nfrom sklearn.model_selection import cross_val_score\nscores = cross_val_score(KNeighborsClassifier(), iris_X, iris_y, cv=5)\nprint (scores)\n",
"[0.96666667 1. 0.93333333 0.96666667 1. ]\n"
],
[
"print (scores.mean())",
"0.9733333333333334\n"
],
[
"# The mean score and the 95% confidence interval of the score estimate are hence given by:\nprint(\"Accuracy: %.3f%% (%.3f%%)\" % (scores.mean()*100.0, scores.std()*100.0))",
"Accuracy: 97.333% (2.494%)\n"
]
],
[
[
"### K fold ",
"_____no_output_____"
]
],
[
[
"model = KNeighborsClassifier()\nkfold = model_selection.KFold(n_splits=5, random_state=12323, shuffle=False)",
"_____no_output_____"
],
[
"results = model_selection.cross_val_score(model, iris_X, iris_y, cv=kfold)\nresults",
"_____no_output_____"
],
[
"print(\"Accuracy: %.3f%% (%.3f%%)\" % (results.mean()*100.0, results.std()*100.0))",
"Accuracy: 91.333% (8.327%)\n"
]
],
[
[
"### K fold with randomization in split? -- might help us to understand my accurracy of model in cv is different from k fold?",
"_____no_output_____"
]
],
[
[
"model = KNeighborsClassifier()\nkfold = model_selection.KFold(n_splits=5, random_state=12323, shuffle=True)",
"_____no_output_____"
],
[
"results = model_selection.cross_val_score(model, iris_X, iris_y, cv=kfold)\nresults",
"_____no_output_____"
],
[
"print(\"Accuracy: %.3f%% (%.3f%%)\" % (results.mean()*100.0, results.std()*100.0))",
"Accuracy: 96.667% (2.108%)\n"
]
],
[
[
"### LOOCV",
"_____no_output_____"
]
],
[
[
"model = KNeighborsClassifier()\nloocv = model_selection.LeaveOneOut()\nresults = model_selection.cross_val_score(model, iris_X, iris_y, cv=loocv)\nprint(\"Accuracy: %.3f%% (%.3f%%)\" % (results.mean()*100.0, results.std()*100.0))",
"Accuracy: 96.667% (17.951%)\n"
]
],
[
[
"Resources: \n- https://towardsdatascience.com/train-test-split-and-cross-validation-in-python-80b61beca4b6\n- https://scikit-learn.org/stable/modules/cross_validation.html\n- https://blog.goodaudience.com/classifying-flowers-using-logistic-regression-in-sci-kit-learn-38262416e4c6\n- https://machinelearningmastery.com/machine-learning-in-python-step-by-step/\n- https://nbviewer.jupyter.org/gist/justmarkham/6d5c061ca5aee67c4316471f8c2ae976\n- https://machinelearningmastery.com/evaluate-performance-machine-learning-algorithms-python-using-resampling/\n- https://machinelearningmastery.com/k-fold-cross-validation/\n\n\n\n\n",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
cb8bcee5900d5bf02d416ab47fbad832adc0c9c0
| 714,676 |
ipynb
|
Jupyter Notebook
|
docs/source/notebooks/LKJ.ipynb
|
andbmme/pymc3
|
2aca43e2744b215449eb49c81e2df85139b0b333
|
[
"Apache-2.0"
] | 1 |
2020-05-08T00:38:00.000Z
|
2020-05-08T00:38:00.000Z
|
docs/source/notebooks/LKJ.ipynb
|
andbmme/pymc3
|
2aca43e2744b215449eb49c81e2df85139b0b333
|
[
"Apache-2.0"
] | null | null | null |
docs/source/notebooks/LKJ.ipynb
|
andbmme/pymc3
|
2aca43e2744b215449eb49c81e2df85139b0b333
|
[
"Apache-2.0"
] | null | null | null | 950.367021 | 268,720 | 0.948121 |
[
[
[
"# LKJ Cholesky Covariance Priors for Multivariate Normal Models",
"_____no_output_____"
],
[
"While the [inverse-Wishart distribution](https://en.wikipedia.org/wiki/Inverse-Wishart_distribution) is the conjugate prior for the covariance matrix of a multivariate normal distribution, it is [not very well-suited](https://github.com/pymc-devs/pymc3/issues/538#issuecomment-94153586) to modern Bayesian computational methods. For this reason, the [LKJ prior](http://www.sciencedirect.com/science/article/pii/S0047259X09000876) is recommended when modeling the covariance matrix of a multivariate normal distribution.\n\nTo illustrate modelling covariance with the LKJ distribution, we first generate a two-dimensional normally-distributed sample data set.",
"_____no_output_____"
]
],
[
[
"import arviz as az\nimport numpy as np\nimport pymc3 as pm\nimport seaborn as sns\nimport warnings\n\nfrom matplotlib.patches import Ellipse\nfrom matplotlib import pyplot as plt",
"_____no_output_____"
],
[
"az.style.use(\"arviz-darkgrid\")\nwarnings.simplefilter(action=\"ignore\", category=FutureWarning)\nRANDOM_SEED = 8924\nnp.random.seed(3264602) # from random.org",
"_____no_output_____"
],
[
"N = 10000\n\nμ_actual = np.array([1.0, -2.0])\nsigmas_actual = np.array([0.7, 1.5])\nRho_actual = np.matrix([[1.0, -0.4], [-0.4, 1.0]])\n\nΣ_actual = np.diag(sigmas_actual) * Rho_actual * np.diag(sigmas_actual)\n\nx = np.random.multivariate_normal(μ_actual, Σ_actual, size=N)\nΣ_actual",
"_____no_output_____"
],
[
"var, U = np.linalg.eig(Σ_actual)\nangle = 180.0 / np.pi * np.arccos(np.abs(U[0, 0]))\n\nfig, ax = plt.subplots(figsize=(8, 6))\n\nblue, _, red, *_ = sns.color_palette()\n\ne = Ellipse(\n μ_actual, 2 * np.sqrt(5.991 * var[0]), 2 * np.sqrt(5.991 * var[1]), angle=angle\n)\ne.set_alpha(0.5)\ne.set_facecolor(blue)\ne.set_zorder(10)\nax.add_artist(e)\n\nax.scatter(x[:, 0], x[:, 1], c=\"k\", alpha=0.05, zorder=11)\n\nrect = plt.Rectangle((0, 0), 1, 1, fc=blue, alpha=0.5)\nax.legend([rect], [\"95% density region\"], loc=2);",
"_____no_output_____"
]
],
[
[
"The sampling distribution for the multivariate normal model is $\\mathbf{x} \\sim N(\\mu, \\Sigma)$, where $\\Sigma$ is the covariance matrix of the sampling distribution, with $\\Sigma_{ij} = \\textrm{Cov}(x_i, x_j)$. The density of this distribution is\n\n$$f(\\mathbf{x}\\ |\\ \\mu, \\Sigma^{-1}) = (2 \\pi)^{-\\frac{k}{2}} |\\Sigma|^{-\\frac{1}{2}} \\exp\\left(-\\frac{1}{2} (\\mathbf{x} - \\mu)^{\\top} \\Sigma^{-1} (\\mathbf{x} - \\mu)\\right).$$\n\nThe LKJ distribution provides a prior on the correlation matrix, $\\mathbf{C} = \\textrm{Corr}(x_i, x_j)$, which, combined with priors on the standard deviations of each component, [induces](http://www3.stat.sinica.edu.tw/statistica/oldpdf/A10n416.pdf) a prior on the covariance matrix, $\\Sigma$. Since inverting $\\Sigma$ is numerically unstable and inefficient, it is computationally advantageous to use the [Cholesky decompositon](https://en.wikipedia.org/wiki/Cholesky_decomposition) of $\\Sigma$, $\\Sigma = \\mathbf{L} \\mathbf{L}^{\\top}$, where $\\mathbf{L}$ is a lower-triangular matrix. This decompositon allows computation of the term $(\\mathbf{x} - \\mu)^{\\top} \\Sigma^{-1} (\\mathbf{x} - \\mu)$ using back-substitution, which is more numerically stable and efficient than direct matrix inversion.\n\nPyMC3 supports LKJ priors for the Cholesky decomposition of the covariance matrix via the [LKJCholeskyCov](../api/distributions/multivariate.rst) distribution. This distribution has parameters `n` and `sd_dist`, which are the dimension of the observations, $\\mathbf{x}$, and the PyMC3 distribution of the component standard deviations, respectively. It also has a hyperparamter `eta`, which controls the amount of correlation between components of $\\mathbf{x}$. The LKJ distribution has the density $f(\\mathbf{C}\\ |\\ \\eta) \\propto |\\mathbf{C}|^{\\eta - 1}$, so $\\eta = 1$ leads to a uniform distribution on correlation matrices, while the magnitude of correlations between components decreases as $\\eta \\to \\infty$.\n\nIn this example, we model the standard deviations with $\\textrm{Exponential}(1.0)$ priors, and the correlation matrix as $\\mathbf{C} \\sim \\textrm{LKJ}(\\eta = 2)$.",
"_____no_output_____"
]
],
[
[
"with pm.Model() as m:\n packed_L = pm.LKJCholeskyCov(\n \"packed_L\", n=2, eta=2.0, sd_dist=pm.Exponential.dist(1.0)\n )",
"_____no_output_____"
]
],
[
[
"Since the Cholesky decompositon of $\\Sigma$ is lower triangular, `LKJCholeskyCov` only stores the diagonal and sub-diagonal entries, for efficiency:",
"_____no_output_____"
]
],
[
[
"packed_L.tag.test_value.shape",
"_____no_output_____"
]
],
[
[
"We use [expand_packed_triangular](../api/math.rst) to transform this vector into the lower triangular matrix $\\mathbf{L}$, which appears in the Cholesky decomposition $\\Sigma = \\mathbf{L} \\mathbf{L}^{\\top}$.",
"_____no_output_____"
]
],
[
[
"with m:\n L = pm.expand_packed_triangular(2, packed_L)\n Σ = L.dot(L.T)\n\nL.tag.test_value.shape",
"_____no_output_____"
]
],
[
[
"Often however, you'll be interested in the posterior distribution of the correlations matrix and of the standard deviations, not in the posterior Cholesky covariance matrix *per se*. Why? Because the correlations and standard deviations are easier to interpret and often have a scientific meaning in the model. As of PyMC 3.9, there is a way to tell PyMC to automatically do these computations and store the posteriors in the trace. You just have to specify `compute_corr=True` in `pm.LKJCholeskyCov`:",
"_____no_output_____"
]
],
[
[
"with pm.Model() as model:\n chol, corr, stds = pm.LKJCholeskyCov(\n \"chol\", n=2, eta=2.0, sd_dist=pm.Exponential.dist(1.0), compute_corr=True\n )\n cov = pm.Deterministic(\"cov\", chol.dot(chol.T))",
"_____no_output_____"
]
],
[
[
"To complete our model, we place independent, weakly regularizing priors, $N(0, 1.5),$ on $\\mu$:",
"_____no_output_____"
]
],
[
[
"with model:\n μ = pm.Normal(\"μ\", 0.0, 1.5, shape=2, testval=x.mean(axis=0))\n obs = pm.MvNormal(\"obs\", μ, chol=chol, observed=x)",
"_____no_output_____"
]
],
[
[
"We sample from this model using NUTS and give the trace to [ArviZ](https://arviz-devs.github.io/arviz/):",
"_____no_output_____"
]
],
[
[
"with model:\n trace = pm.sample(random_seed=RANDOM_SEED, init=\"adapt_diag\")\nidata = az.from_pymc3(trace)\naz.summary(idata, var_names=[\"~chol\"], round_to=2)",
"Auto-assigning NUTS sampler...\nInitializing NUTS using adapt_diag...\nMultiprocess sampling (4 chains in 4 jobs)\nNUTS: [μ, chol]\n"
]
],
[
[
"Sampling went smoothly: no divergences and good r-hats. You can also see that the sampler recovered the true means, correlations and standard deviations. As often, that will be clearer in a graph:",
"_____no_output_____"
]
],
[
[
"az.plot_trace(\n idata,\n var_names=[\"~chol\"],\n compact=True,\n lines=[\n (\"μ\", {}, μ_actual),\n (\"cov\", {}, Σ_actual),\n (\"chol_stds\", {}, sigmas_actual),\n (\"chol_corr\", {}, Rho_actual),\n ],\n);",
"_____no_output_____"
]
],
[
[
"The posterior expected values are very close to the true value of each component! How close exactly? Let's compute the percentage of closeness for $\\mu$ and $\\Sigma$:",
"_____no_output_____"
]
],
[
[
"μ_post = trace[\"μ\"].mean(axis=0)\n(1 - μ_post / μ_actual).round(2)",
"_____no_output_____"
],
[
"Σ_post = trace[\"cov\"].mean(axis=0)\n(1 - Σ_post / Σ_actual).round(2)",
"_____no_output_____"
]
],
[
[
"So the posterior means are within 3% of the true values of $\\mu$ and $\\Sigma$.\n\nNow let's replicate the plot we did at the beginning, but let's overlay the posterior distribution on top of the true distribution -- you'll see there is excellent visual agreement between both:",
"_____no_output_____"
]
],
[
[
"var_post, U_post = np.linalg.eig(Σ_post)\nangle_post = 180.0 / np.pi * np.arccos(np.abs(U_post[0, 0]))\n\nfig, ax = plt.subplots(figsize=(8, 6))\n\ne = Ellipse(\n μ_actual, 2 * np.sqrt(5.991 * var[0]), 2 * np.sqrt(5.991 * var[1]), angle=angle\n)\ne.set_alpha(0.5)\ne.set_facecolor(blue)\ne.set_zorder(10)\nax.add_artist(e)\n\ne_post = Ellipse(\n μ_post,\n 2 * np.sqrt(5.991 * var_post[0]),\n 2 * np.sqrt(5.991 * var_post[1]),\n angle=angle_post,\n)\ne_post.set_alpha(0.5)\ne_post.set_facecolor(red)\ne_post.set_zorder(10)\nax.add_artist(e_post)\n\nax.scatter(x[:, 0], x[:, 1], c=\"k\", alpha=0.05, zorder=11)\n\nrect = plt.Rectangle((0, 0), 1, 1, fc=blue, alpha=0.5)\nrect_post = plt.Rectangle((0, 0), 1, 1, fc=red, alpha=0.5)\nax.legend(\n [rect, rect_post],\n [\"True 95% density region\", \"Estimated 95% density region\"],\n loc=2,\n);",
"_____no_output_____"
],
[
"%load_ext watermark\n%watermark -n -u -v -iv -w",
"pymc3 3.8\narviz 0.7.0\nseaborn 0.9.0\nnumpy 1.17.5\nlast updated: Wed Apr 22 2020 \n\nCPython 3.8.0\nIPython 7.11.0\nwatermark 2.0.2\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
cb8bcf12166d0c83295192e37773473351961ae4
| 505,620 |
ipynb
|
Jupyter Notebook
|
1_code/Master_notebook_submit.ipynb
|
herbsh/USAID_Forecast_submit
|
758963829e3766c4f9466b2238e676ea401838af
|
[
"MIT"
] | 1 |
2021-01-11T18:55:35.000Z
|
2021-01-11T18:55:35.000Z
|
1_code/Master_notebook_submit.ipynb
|
herbsh/USAID_Forecast_submit
|
758963829e3766c4f9466b2238e676ea401838af
|
[
"MIT"
] | null | null | null |
1_code/Master_notebook_submit.ipynb
|
herbsh/USAID_Forecast_submit
|
758963829e3766c4f9466b2238e676ea401838af
|
[
"MIT"
] | null | null | null | 164.322392 | 49,896 | 0.859501 |
[
[
[
"# Forecasting on Contraceptive Use - A Multi-step Ensemble Approach¶\n\nUpdate: 09/07/2020\n\nGithub Repository: https://github.com/herbsh/USAID_Forecast_submit \n\n\n## key idea\n\n- The goal is to forecast on site_code & product_code level demand.\n- The site_code & product_code level demand fluctuates too much and doesn't have any obvious pattern. \n- The aggregate level is easier to forecast. The noise cancels out.\n- We don't know what is the best level to aggregate. It's possible that it varies regarding each site too.\n- we aggregate on various levels, and use \"Ensemble Learning\" to to determine the final result\n\n## Aggregate \nHow to aggregate? Here is the structure of the supply chain and products. We can use this guide our aggregation: \n- site_code -> district -> region\n- product_code -> product_type\nAfter we aggregate on some level, we get a time series of stock_distributed, {Y}.\nWe supplement some external data and match/aggregate to the same level, {X}.\nwe use a time-series model to forecast {Y} with {Y} and {X}.\n\n## Forecast the Aggregate : Time Series Modeling, Auto_SARIMAX\n- We use a SARIMAX stucture (ARIMA (AR, MA with a trend) with Seasonality and external data )\n- The specific order of SARIMAX is determined within each time series with an Auto_ARIMA function.\n- Use BIC criteria to pick the optimal order for ARIMA (p, d, q) and seasonality (P,D,Q) (BIC : best for forecast power) (range of p,d,q and P,D,Q - small, less than 4)\n- use the time series model to make aggregate level forecast. Store the results for later use. \n\n## De-aggregate (Distribute) : Machine Learning Modeling\n\n- We use machine learning to learn the variable \"share\", the share of the specific stock_distributed as a fraction of the aggregate sum.\n- training data:\n - all the data, excluding the last 3 month. Encode the year, month, region, district, product type, code, plus all available external data, matched to site_code+product level.\n- target: actual shares \n- model: RandomForest Decision Regression Tree\n - use the fitted model to make prediction on shares. \"Distribute\" aggregate stocks to individual forecasts.\n \n## Emsemble\n \n- From Aggregate and Distribute, we arrive at X different forecast numbers for each site+product_code. (We also have a lot of intermediary forecasts that could be of interests to various parties). \n- We introduce another model to perform the ensemble\n- For each training observation, we have multiple fourcasts and one actual realization, denote them as F1, F2, F3, F4 and Y (ommitting site, product, t time subscripts). We also have all the features X ( temperature, roads, etc).\n- The emsemble part estimate another model to take inputs (F1..F4, and features X) to arrive at an estimated Y_hat that minimizes its MSE to Y ( actual stock_distributed).\n- We used XGBoost to perform the ensemble learning part. \n\n## Key Takeaway of this approach \n- Combines traditional forecast methods (SARIMAX) and Machine Learning (XGBoost) \n- It's very transferable to other scenarios \n- It uses external data and it's easy to plug in more external data to improve the forecasts.\n- The ensemble piece makes adding model possible and easy",
"_____no_output_____"
]
],
[
[
"# suppress warning to make cleaner output \nimport warnings\nwarnings.filterwarnings(\"ignore\")",
"_____no_output_____"
]
],
[
[
"# Outline ",
"_____no_output_____"
],
[
"## Step 1: Data Cleaning \n### Upsample (fill in gaps in time series)\n- notebook: datacleaning_upsample.ipynb ",
"_____no_output_____"
]
],
[
[
"%run datacleaning_upsample.ipynb ",
"_____no_output_____"
]
],
[
[
"- inputs: \"..\\0_data\\contraceptive_logistics_data.csv\"\n- output: \"..\\2_pipeline\\df_upsample.csv\"\n- steps: \n - upsample - make sure all individual product-site series has no gaps in time even though they may differ in length \n - fill in 0 for NA in stock_distributed",
"_____no_output_____"
],
[
"### Supplement \n(very import thing about supplement data - if we are to use any supplement data, the value should exist for the 3 months that are to be forecasted) ( 10, 11, 12) Must be careful when constructing supplement dataset\n- notebook: datacleaning_prep_supplement.ipynb",
"_____no_output_____"
],
[
"- input:\n - \"..\\0_data\\supplement_data_raw.csv\"\n- steps:\n - time invariant supplement data:\n - identifiers: site_code product_code region district\n - information: (currently) road condition, product type\n - output: \"../0_data/time_invariant_supplement.dta\"\n\n - time variant supplement data: (include rows for time that need to be forecasted)\n - identifiers: temp_timeindex year month site_code product_code region district\n - information: maxtemp temp pressure relative rain visibility windspeed maxsus* storm fog\n - output: \"../0_data/time_variant_supplement.dta\"",
"_____no_output_____"
],
[
"### Combine ",
"_____no_output_____"
],
[
"- notebook: datacleaning_combine.ipynb ",
"_____no_output_____"
]
],
[
[
"%run datacleaning_combine.ipynb",
"import df_upsample from datacleaning_upsample.ipynb step\n year month stock_distributed\ncount 41348.00000 41348.000000 41348.000000\nmean 2017.52225 6.305964 14.033399\nstd 1.06620 3.328858 39.523746\nmin 2016.00000 1.000000 0.000000\n25% 2017.00000 3.000000 0.000000\n50% 2018.00000 6.000000 0.000000\n75% 2018.00000 9.000000 12.000000\nmax 2019.00000 12.000000 1805.000000\n\n prepare submission data site_code, product_code and time identifiers \n\n year month site_code product_code predicted_value\n0 2019 10 C4001 AS27134 0\n1 2019 10 C4001 AS27132 0\n2 2019 10 C4001 AS27000 0\n3 2019 10 C4001 AS27137 0\n4 2019 10 C4001 AS27138 0\n\n import time_invariant_supplement.dta from datacleaning_prep_supplement.ipynb step \n\n\n add site_type information from service_delivery_site_data.csv \n\n\n add product_type information from product.csv \n\n\n import time_variant_supplement.dta from datacleaning_prep_supplement.ipynb step \n\n\n Ready with df_combined.csv data. Variables include: \nyear\nmonth\nstock_distributed\nregion\ndistrict\nregionroads\nregionasphaltroads\nregionearthroads\nregionsurfacetreatmentroads\nregionpoorroads\npoorroads\nearthroads\nasphaltroads\nsite_type\nproduct_type\ntemp\nmaxtemp\npressure\nrelativehumidity\nrainfallsnowmelt\nvisibility\nwindspeed\nmaxsustainedwindspeed\nrainordrizzle\nstorm\nfog\n\n Export training data df_training for model development. Exclude the last three month \n\n\n Export full data for final forecast.\n"
]
],
[
[
"- input: \n - \"../2_pipeline/df_upsample.csv\"\n - '../0_data/submission_format.csv' \n - \"../0_data/time_invariant_supplement.dta\" \n - \"../0_data/time_variant_supplement.dta\"\n - '../0_data/service_delivery_site_data.csv' \n \n- output: a site_code & product_code & date level logistics data with time variant and time invariant exogenous features :\n - for development: '../0_data/df_training.csv'\n - for final prediction(contained 3 last month exog vars and space holder) '../0_data/df_combined_fullsample.csv'",
"_____no_output_____"
],
[
"## Step 2: Multiple Agg-Forecast-Distribute Models in parallel ",
"_____no_output_____"
],
[
"### Region Level ",
"_____no_output_____"
],
[
"- notebook: model_SARIMAX_Distribute_region.ipynb ",
"_____no_output_____"
],
[
"%run model_SARIMAX_Distribute_region.ipynb ",
"_____no_output_____"
],
[
" - input: \n - '../0_data/df_combined_fullsample.csv' \n - output:\n 1. ../2_pipeline/final_pred_region_lev.csv\n 2. ./2_pipeline/final_distribute_regionlev.csv'\n ",
"_____no_output_____"
],
[
"### District Level",
"_____no_output_____"
],
[
"- notebook: model_SARIMAX_Distribute_District.ipynb",
"_____no_output_____"
]
],
[
[
"%run model_SARIMAX_Distribute_District.ipynb",
"AGBOVILLE\nAS27138\n(0, 0, 0)\n(0, 0, 0, 12)\n"
]
],
[
[
" - input: \n - '../0_data/df_combined_fullsample.csv' \n - output::\n 1. '../2_pipeline/final_pred_district_lev.csv'\n 2. '../2_pipeline/final_distribute_districtlev.csv'\n",
"_____no_output_____"
],
[
"### Region-Product_type level",
"_____no_output_____"
],
[
" \n- notebook: model_SARIMAX_Distribute_regionproducttype.ipynb",
"_____no_output_____"
]
],
[
[
"%run model_SARIMAX_Distribute_regionproducttype.ipynb",
"GBEKE\nOral Contraceptive (Pill)\n(1, 0, 0)\n(0, 0, 1, 12)\n"
]
],
[
[
" - input: \n - '../0_data/df_combined_fullsample.csv'\n - output: \n 1. ../2_pipeline/final_pred_region_producttype_lev.csv\n 2. '../2_pipeline/final_distribute_regionproducttypelev.csv'",
"_____no_output_____"
],
[
"### Individual Level, with raw data, winsorized data, and rolling smoothed data ",
"_____no_output_____"
],
[
" \n- notebook: model_SARIMAX_individual.ipyn",
"_____no_output_____"
]
],
[
[
"%run model_SARIMAX_individual.ipynb ",
"C1054\nAS27138\n(2, 0, 0)\n(0, 0, 0, 12)\n"
]
],
[
[
" - input: \n - '../0_data/df_combined_fullsample.csv'\n - output: \n - '../2_pipeline/final_pred_ind_lev.csv'",
"_____no_output_____"
],
[
"- notebook: model_SARIMAX_individual_winsorized.ipynb",
"_____no_output_____"
]
],
[
[
"%run model_SARIMAX_individual_winsorized.ipynb",
"C1054\nAS27138\n(1, 0, 0)\n(0, 0, 0, 12)\n"
]
],
[
[
" - input: \n - '../0_data/df_combined_fullsample.csv'\n - output: \n\n - '../2_pipeline/final_pred_ind_winsorized_lev.csv'",
"_____no_output_____"
],
[
"- notebook: model_SARIMAX_individual_rollingsmoothed.ipynb ",
"_____no_output_____"
]
],
[
[
"%run model_SARIMAX_individual_rollingsmoothed.ipynb",
"C1054\nAS27138\n(3, 0, 0)\n(0, 0, 0, 12)\n"
]
],
[
[
" - input: \n - '../0_data/df_combined_fullsample.csv'\n - output: \n - '../2_pipeline/final_pred_ind_rollingsmoothed_lev.csv' ",
"_____no_output_____"
],
[
" \n \n## Step 3: Ensemble, learn the ensemble model, make final prediction \n- notebook: ensemble.ipynb ",
"_____no_output_____"
]
],
[
[
"%run ensemble.ipynb ",
"\n importing results from the distribution stage of various aggregation levels \n\n['../2_pipeline\\\\final_distribute_districtlev.csv', '../2_pipeline\\\\final_distribute_regionlev.csv', '../2_pipeline\\\\final_distribute_regionproducttypelev.csv']\nImport SARIMAX_agg model results and merge with predicted distribute values\n\n Import three individual level sarimax results \n\n['../2_pipeline\\\\final_pred_ind_lev.csv', '../2_pipeline\\\\final_pred_ind_rollingsmoothed_lev.csv', '../2_pipeline\\\\final_pred_ind_winsorized_lev.csv']\n"
]
],
[
[
" - input: \n - Distribution model results: glob.glob('../2_pipeline/final_distribute_*.csv') \n - SARIMAX results: glob.glob('../2_pipeline/final_pred_ind*.csv') \n - output: \n ",
"_____no_output_____"
],
[
"# Ensemble Model Details",
"_____no_output_____"
],
[
"## Import : data with actual stock distributed and exogenous variables ",
"_____no_output_____"
]
],
[
[
"import pandas as pd\n\ndf_combined=pd.read_csv('../0_data/df_combined_fullsample.csv')",
"_____no_output_____"
]
],
[
[
"## Import results from distribution(shares) models ",
"_____no_output_____"
]
],
[
[
"import glob\ntemp=glob.glob('../2_pipeline/final_distribute_*.csv')\nprint('\\n importing results from the distribution stage of various aggregation levels \\n')\nprint(temp)",
"\n importing results from the distribution stage of various aggregation levels \n\n['../2_pipeline\\\\final_distribute_districtlev.csv', '../2_pipeline\\\\final_distribute_regionlev.csv', '../2_pipeline\\\\final_distribute_regionproducttypelev.csv']\n"
],
[
"distribute_districtlev=pd.read_csv('../2_pipeline\\\\final_distribute_districtlev.csv').drop(columns=['Unnamed: 0'])\ndistribute_regionlev=pd.read_csv('../2_pipeline\\\\final_distribute_regionlev.csv').drop(columns=['Unnamed: 0'])\ndistribute_regionproducttypelev=pd.read_csv('../2_pipeline\\\\final_distribute_regionproducttypelev.csv').drop(columns=['Unnamed: 0']) ",
"_____no_output_____"
]
],
[
[
"## Import SARIMAX_agg model results and merge with distribute ",
"_____no_output_____"
]
],
[
[
"print('Import SARIMAX_agg model results and merge with predicted distribute values')",
"Import SARIMAX_agg model results and merge with predicted distribute values\n"
],
[
"sarimax_pred_region=pd.read_csv('../2_pipeline/final_pred_region_lev.csv').rename(columns={'Unnamed: 0':'date',}).rename(columns={'stock_distributed_forecasted':'stock_forecast_agg_region'})\nsarimax_pred_regionproducttype=pd.read_csv('../2_pipeline/final_pred_region_producttype_lev.csv').rename(columns={'Unnamed: 0':'date'}).rename(columns={'stock_distributed_forecasted':'stock_forecast_agg_regionproducttype'})\nsarimax_pred_district=pd.read_csv('../2_pipeline/final_pred_district_lev.csv').rename(columns={'Unnamed: 0':'date'}).rename(columns={'stock_distributed_forecasted':'stock_forecast_agg_district'})",
"_____no_output_____"
]
],
[
[
"- merge sarimax_pred_region with distribute_region",
"_____no_output_____"
]
],
[
[
"pred_agg_region=pd.merge(left=sarimax_pred_region,right=distribute_regionlev,on=['date','region','product_code'],how='right')\npred_agg_region.describe()",
"_____no_output_____"
]
],
[
[
"- merge sarimax_pred_regionproducttype with distribute_regionproducttype",
"_____no_output_____"
]
],
[
[
"pred_agg_regionproducttype=pd.merge(left=sarimax_pred_regionproducttype,right=distribute_regionproducttypelev,on=['date','region','product_type'],how='right')\npred_agg_regionproducttype.describe()",
"_____no_output_____"
]
],
[
[
"- merge sarimax_pred_district with distribute_districtlev",
"_____no_output_____"
]
],
[
[
"pred_agg_district=pd.merge(left=sarimax_pred_district,right=distribute_districtlev,on=['date','district','product_code'],how='right')\npred_agg_district.describe()",
"_____no_output_____"
]
],
[
[
"## Import three individual level sarimax results ",
"_____no_output_____"
]
],
[
[
"import glob\ntemp=glob.glob('../2_pipeline/final_pred_ind*.csv')\nprint('\\n Import three individual level sarimax results \\n')\nprint(temp)",
"\n Import three individual level sarimax results \n\n['../2_pipeline\\\\final_pred_ind_lev.csv', '../2_pipeline\\\\final_pred_ind_rollingsmoothed_lev.csv', '../2_pipeline\\\\final_pred_ind_winsorized_lev.csv']\n"
],
[
"sarimax_ind=pd.read_csv('../2_pipeline/final_pred_ind_lev.csv').rename(columns={'Unnamed: 0':'date','stock_distributed_forecasted':'stock_forecast_agg_ind'})\nsarimax_ind.head(2)",
"_____no_output_____"
],
[
"sarimax_ind_smooth=pd.read_csv('../2_pipeline/final_pred_ind_rollingsmoothed_lev.csv').rename(columns={'Unnamed: 0':'date','stock_distributed_forecasted':'stock_forecast_agg_ind_smooth'})\nsarimax_ind_smooth.head(2)",
"_____no_output_____"
],
[
"sarimax_ind_winsorized=pd.read_csv('../2_pipeline/final_pred_ind_winsorized_lev.csv').rename(columns={'Unnamed: 0':'date','stock_distributed_forecasted':'stock_forecast_agg_ind_winsorized'})\n\nsarimax_ind_winsorized.head(2)",
"_____no_output_____"
],
[
"df_ensemble=pd.merge(left=df_combined,right=pred_agg_region.drop(columns=['agg_level']),on=['date','region','product_code','site_code'],how='left')",
"_____no_output_____"
],
[
"len(df_ensemble)",
"_____no_output_____"
],
[
"df_ensemble=pd.merge(left=df_ensemble,right=pred_agg_regionproducttype.drop(columns=['agg_level']),on=['date','region','product_type','site_code','product_code'],how='left')\nlen(df_ensemble)",
"_____no_output_____"
],
[
"df_ensemble=pd.merge(left=df_ensemble,right=pred_agg_district.drop(columns=['agg_level']),on=['date','district','site_code','product_code'],how='left')\n\n \ndf_ensemble=pd.merge(left=df_ensemble,right=sarimax_ind,on=['date','site_code','product_code'],how='left')\n\ndf_ensemble=pd.merge(left=df_ensemble,right=sarimax_ind_smooth,on=['date','site_code','product_code'],how='left')\n\n\ndf_ensemble=pd.merge(left=df_ensemble,right=sarimax_ind_winsorized,on=['date','site_code','product_code'],how='left')",
"_____no_output_____"
],
[
"df_ensemble['date']=pd.to_datetime(df_ensemble['date'])",
"_____no_output_____"
],
[
"df_ensemble.set_index('date')['2019-10':].describe()",
"_____no_output_____"
],
[
"df_ensemble=df_ensemble.fillna(0)",
"_____no_output_____"
],
[
"df_ensemble.head()",
"_____no_output_____"
]
],
[
[
"## Sort df_ensemble dataframe by date to ensure the train-test data are set up correctly ",
"_____no_output_____"
]
],
[
[
"df_ensemble.sort_values(by='date',inplace=True)",
"_____no_output_____"
]
],
[
[
"## Feature Engineering",
"_____no_output_____"
],
[
"### Add a few interactions ",
"_____no_output_____"
]
],
[
[
"df_ensemble['interaction_1']=df_ensemble['pred_share_regionlev']*df_ensemble['stock_forecast_agg_regionproducttype']\ndf_ensemble['interaction_2']=df_ensemble['pred_share_districtlev']*df_ensemble['stock_forecast_agg_regionproducttype']\ndf_ensemble['weather_interaction']=df_ensemble['maxtemp']*df_ensemble['rainfallsnowmelt']",
"_____no_output_____"
],
[
"columns_to_encode=['site_code', 'product_code', 'year', 'month',\n 'region', 'district', 'product_type','site_type'] \ncolumns_continuous_exog=['regionroads',\n 'regionasphaltroads', 'regionearthroads', 'regionsurfacetreatmentroads',\n 'regionpoorroads', 'poorroads', 'earthroads', 'asphaltroads', 'temp',\n 'maxtemp', 'pressure', 'relativehumidity', 'rainfallsnowmelt',\n 'visibility', 'windspeed', 'maxsustainedwindspeed', 'rainordrizzle',\n 'storm', 'fog','weather_interaction']\n\ncolumns_continuous_frommodel=['stock_forecast_agg_region', 'pred_share_regionlev',\n 'stock_forecast_agg_regionproducttype',\n 'pred_share_regionproducttype_tlev', 'stock_forecast_agg_district',\n 'pred_share_districtlev', 'stock_forecast_agg_ind',\n 'stock_forecast_agg_ind_smooth', 'stock_forecast_agg_ind_winsorized','interaction_1','interaction_2']",
"_____no_output_____"
]
],
[
[
"## Setting up target ",
"_____no_output_____"
],
[
"\n- the y vector should have all 0s for the last 3 months worth of data ",
"_____no_output_____"
]
],
[
[
"y=df_ensemble.stock_distributed",
"_____no_output_____"
]
],
[
[
"## Setting up features ",
"_____no_output_____"
]
],
[
[
"# Import libraries and download example data\nfrom sklearn.preprocessing import OneHotEncoder\nohe = OneHotEncoder(sparse=False,categories='auto')\nencoded_columns = ohe.fit_transform(df_ensemble[columns_to_encode])\nimport numpy as np\nnp.shape(encoded_columns)",
"_____no_output_____"
]
],
[
[
"- produce one-hot encoding for categorical values",
"_____no_output_____"
]
],
[
[
"features=pd.DataFrame(data=encoded_columns,columns=ohe.get_feature_names(columns_to_encode))\nfeatures.describe()",
"_____no_output_____"
]
],
[
[
"- add continuous values. Put everything to a X matrix",
"_____no_output_____"
]
],
[
[
"X=features\nX[columns_continuous_exog]=df_ensemble[columns_continuous_exog]\nX[columns_continuous_frommodel]=df_ensemble[columns_continuous_frommodel]",
"_____no_output_____"
],
[
"X.to_csv('x.csv')",
"_____no_output_____"
]
],
[
[
"## Scale X ",
"_____no_output_____"
]
],
[
[
"from sklearn.preprocessing import scale\nXs = scale(X) ",
"_____no_output_____"
]
],
[
[
"# Use XGboost to make final prediction",
"_____no_output_____"
],
[
"## XGBoost\n ",
"_____no_output_____"
]
],
[
[
"import xgboost as xgb\nfrom sklearn.metrics import mean_squared_error\nimport pandas as pd\nimport numpy as np",
"_____no_output_____"
],
[
"df_train=df_ensemble.set_index('date')[:'2019-9'] \ndf_pred=df_ensemble.set_index('date')['2019-10':'2019-12'] \nXs_train=Xs[:df_train.shape[0]]\ny_train=y[:df_train.shape[0]]\nXs_pred=Xs[-df_pred.shape[0]:]\n\n\ndata_dmatrix = xgb.DMatrix(data=Xs,label=y)\n\nxg_reg = xgb.XGBRegressor(objective ='reg:squarederror', colsample_bytree = 0.3, learning_rate = 0.1,\n max_depth = 30, alpha = 10, n_estimators = 20)\nxg_reg.fit(Xs_train,y_train)\n\npreds = xg_reg.predict(Xs_pred)\n",
"_____no_output_____"
],
[
"len(preds)",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt\n%matplotlib inline\nxgb.plot_importance(xg_reg)\nplt.rcParams['figure.figsize'] = [8,10]\nplt.savefig('../2_pipeline/xgboost_plot_importance.jpg')\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Collect Results ",
"_____no_output_____"
]
],
[
[
"temp=df_pred[['year','month','site_code','product_code']].copy()\n",
"_____no_output_____"
],
[
"temp['predicted_value']=preds",
"_____no_output_____"
],
[
"temp=temp.reset_index() ",
"_____no_output_____"
],
[
"temp=temp.drop(columns='date')",
"_____no_output_____"
],
[
"submission_format=pd.read_csv('../0_data/submission_format.csv')",
"_____no_output_____"
],
[
"submission=pd.merge(left=submission_format.drop(columns='predicted_value'),right=temp,on=['year','month','site_code','product_code'],how='left')\nsubmission.describe()",
"_____no_output_____"
],
[
"submission['predicted_value']=submission['predicted_value'].apply(lambda x: max(x,0))",
"_____no_output_____"
],
[
"submission.describe()",
"_____no_output_____"
],
[
"submission.head()",
"_____no_output_____"
],
[
"submission[['year','month','site_code','product_code','predicted_value']].to_csv('../submission.csv')",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb8bcfdc52fa1d96769142a1a7319979738049f0
| 8,140 |
ipynb
|
Jupyter Notebook
|
_notebooks/2021-08-19-thirty-days-of-kaggle-day-10-model-validation.ipynb
|
duffymo/fatigue-failure
|
7567ed34af9779354d692c3ef8af82d9bb9868e8
|
[
"Apache-2.0"
] | null | null | null |
_notebooks/2021-08-19-thirty-days-of-kaggle-day-10-model-validation.ipynb
|
duffymo/fatigue-failure
|
7567ed34af9779354d692c3ef8af82d9bb9868e8
|
[
"Apache-2.0"
] | 2 |
2021-05-22T10:58:56.000Z
|
2021-05-22T11:01:19.000Z
|
_notebooks/2021-08-19-thirty-days-of-kaggle-day-10-model-validation.ipynb
|
duffymo/fatigue-failure
|
7567ed34af9779354d692c3ef8af82d9bb9868e8
|
[
"Apache-2.0"
] | null | null | null | 28.16609 | 324 | 0.579115 |
[
[
[
"% 30 Days of Kaggle - Day 10: (https://www.kaggle.com/dansbecker/underfitting-and-overfitting)[Over-Fitting and Under-Fitting].\n\nNow that I can create models I need to be able to evaluate their accuracy.\n\nI calculated mean absolute error in the last notebook using sklearn.\n\nMAE = \\frac{\\sum_0^N | predicted - actual |}{N}\n\nThe lesson notes give a great explanation of under- and over-fitting:\n\n\n\nThey use the example of housing data. Decision tree depth is the variable to watch. A binary tree of depth n will have 2^n leaf nodes. If n is too small we may be under-fitting. If n is too large we eventually end up with one case in each leaf node. There's a sweet spot that we have to find for training data.\n\nUse this utility method to compare MAE for different max leaf nodes:",
"_____no_output_____"
]
],
[
[
"from sklearn.metrics import mean_absolute_error\nfrom sklearn.tree import DecisionTreeRegressor\n\ndef get_mae(max_leaf_nodes, train_X, val_X, train_y, val_y):\n model = DecisionTreeRegressor(max_leaf_nodes = max_leaf_nodes, random_state = 0)\n model.fit(train_X, train_y)\n predictions_val = model.predict(val_X)\n mae = mean_absolute_error(val_y, predictions_val)\n return mae",
"_____no_output_____"
]
],
[
[
"These cells repeat the earlier calculations for the Melbourne housing data:",
"_____no_output_____"
]
],
[
[
"# Data Loading Code Runs At This Point\nimport pandas as pd\n\n# Load data\nmelbourne_file_path = '../datasets/kaggle/melbourne-house-prices/melb_data.csv'\nmelbourne_data = pd.read_csv(melbourne_file_path)\n# Filter rows with missing values\nfiltered_melbourne_data = melbourne_data.dropna(axis=0)\n# Choose target and features\ny = filtered_melbourne_data.Price\nmelbourne_features = ['Rooms', 'Bathroom', 'Landsize', 'BuildingArea',\n 'YearBuilt', 'Lattitude', 'Longtitude']\nX = filtered_melbourne_data[melbourne_features]\n",
"_____no_output_____"
]
],
[
[
"Split data into train and test sets:",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import train_test_split\n\n# split data into training and validation data, for both features and target\ntrain_X, val_X, train_y, val_y = train_test_split(X, y,random_state = 0)",
"_____no_output_____"
]
],
[
[
"Now let's calculate MEA with differing values of max_leaf_nodes:",
"_____no_output_____"
]
],
[
[
"for max_leaf_nodes in [5, 50, 500, 5000]:\n my_mae = get_mae(max_leaf_nodes, train_X, val_X, train_y, val_y)\n print(\"Max leaf nodes: %4d \\t\\t Mean Absolute Error: %d\" %(max_leaf_nodes, my_mae))",
"Max leaf nodes: 5 \t\t Mean Absolute Error: 347380\nMax leaf nodes: 50 \t\t Mean Absolute Error: 258171\nMax leaf nodes: 500 \t\t Mean Absolute Error: 243495\nMax leaf nodes: 5000 \t\t Mean Absolute Error: 254983\n"
]
],
[
[
"Exercises: do the same thing with the Iowa housing model.",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nfrom sklearn.metrics import mean_absolute_error\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.tree import DecisionTreeRegressor\n\niowa_file_path = '../datasets/kaggle/iowa-house-prices/train.csv'\nhome_data = pd.read_csv(iowa_file_path)\ny = home_data.SalePrice\nfeature_columns = ['Lot Area', 'Year Built', '1st Flr SF', '2nd Flr SF', 'Bedroom AbvGr', 'TotRms AbvGrd']\nX = home_data[feature_columns]\niowa_model = DecisionTreeRegressor(random_state=1)\niowa_model.fit(train_X, train_y)\nval_predictions = iowa_model.predict(val_X)\nval_mae = mean_absolute_error(val_predictions, val_y)\nprint(\"Validation MAE: {:,.0f}\".format(val_mae))",
"Validation MAE: 262,494\n"
],
[
"candidate_max_leaf_nodes = [5, 25, 50, 100, 250, 500, 600, 700, 800, 900, 1000]\nfor max_leaf_nodes in candidate_max_leaf_nodes:\n mae = get_mae(max_leaf_nodes, train_X, val_X, train_y, val_y)\n print(\"Max leaf nodes: %4d \\t\\t Mean Absolute Error: %d\" %(max_leaf_nodes, mae))",
"Max leaf nodes: 5 \t\t Mean Absolute Error: 347380\nMax leaf nodes: 25 \t\t Mean Absolute Error: 271044\nMax leaf nodes: 50 \t\t Mean Absolute Error: 258171\nMax leaf nodes: 100 \t\t Mean Absolute Error: 248734\nMax leaf nodes: 250 \t\t Mean Absolute Error: 247206\nMax leaf nodes: 500 \t\t Mean Absolute Error: 243495\nMax leaf nodes: 600 \t\t Mean Absolute Error: 243951\nMax leaf nodes: 700 \t\t Mean Absolute Error: 242954\nMax leaf nodes: 800 \t\t Mean Absolute Error: 244042\nMax leaf nodes: 900 \t\t Mean Absolute Error: 246292\nMax leaf nodes: 1000 \t\t Mean Absolute Error: 247345\n"
]
],
[
[
"Now that we know that we want 500 leaf nodes we can use all the data to create the final model.",
"_____no_output_____"
]
],
[
[
"final_model = DecisionTreeRegressor(max_leaf_nodes=500)\nfinal_model.fit(X, y)\nprint(final_model)",
"DecisionTreeRegressor(max_leaf_nodes=500)\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb8be5b9ded5e04ba6111d1e4b2abca3754a46fe
| 1,451 |
ipynb
|
Jupyter Notebook
|
test_chemspi_web_db.ipynb
|
Andreymcz/jmapy
|
b1d3238910c9b126a80ed4edd1ad4a7df9d3e4d4
|
[
"MIT"
] | null | null | null |
test_chemspi_web_db.ipynb
|
Andreymcz/jmapy
|
b1d3238910c9b126a80ed4edd1ad4a7df9d3e4d4
|
[
"MIT"
] | null | null | null |
test_chemspi_web_db.ipynb
|
Andreymcz/jmapy
|
b1d3238910c9b126a80ed4edd1ad4a7df9d3e4d4
|
[
"MIT"
] | null | null | null | 20.43662 | 89 | 0.523777 |
[
[
[
"import chemspi_web_db as chemsearch\nimport cutils as utils\n\nchempider_web_db = chemsearch.ChemspiWebDB(\"9GS3pzBwGsrdu0agqqP7buFcYwaaX2GH\")\n\ncompound_csids = \"388469\" \n\n\nfor csid in utils.parse_generated_CSID(compound_csids):\n compound = chempider_web_db.find_compound_by_id(csid)\n \n print(compound.iupac_name)\n for k, v in compound.details.items():\n print(k, v)\n \n \n",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code"
]
] |
cb8be9c15ea48f81e8a8202a80c26b242b24fdc1
| 174,409 |
ipynb
|
Jupyter Notebook
|
notebooks/basic_prediction_weather.ipynb
|
erezinman/FlightDelay
|
4d74dcb5d1775ec4386cc073ac65ee968d9d8dff
|
[
"Apache-2.0"
] | null | null | null |
notebooks/basic_prediction_weather.ipynb
|
erezinman/FlightDelay
|
4d74dcb5d1775ec4386cc073ac65ee968d9d8dff
|
[
"Apache-2.0"
] | null | null | null |
notebooks/basic_prediction_weather.ipynb
|
erezinman/FlightDelay
|
4d74dcb5d1775ec4386cc073ac65ee968d9d8dff
|
[
"Apache-2.0"
] | null | null | null | 217.467581 | 78,972 | 0.902442 |
[
[
[
"import numpy as np\nimport pandas as pd\n# from sklearn.preprocessing import\nfrom sklearn.model_selection import train_test_split\nfrom random import randint\nimport sklearn.metrics as skm\nfrom xgboost import XGBClassifier\nimport xgboost as xgb\nfrom sklearn.metrics import roc_curve\nfrom matplotlib import pyplot as plt",
"_____no_output_____"
],
[
"def replace_rare_entries(df, columns, threshold_frac):\n tot_instances = df.shape[0]\n threshold = tot_instances * threshold_frac\n df = df.apply(lambda x: x.mask(x.map(x.value_counts()) < threshold, 'RARE') if x.name in columns else x)\n return df\n ",
"_____no_output_____"
],
[
"categoricals = ['OP_UNIQUE_CARRIER', 'DEST', 'DEP_TIME_BLK', 'DAY_OF_MONTH', 'DAY_OF_WEEK', 'MONTH','weather_label']\nnumericals = ['precipitation_intensity','precipitation_probability','visibility','cloud_cover','humidity','wind_bearing','wind_speed','uv_index','temperature','moon_phase','dew_point','pressure','sunrise_time','sunset_time']",
"_____no_output_____"
],
[
"df = pd.read_csv('../Data/new_york/year_lga_dep_weather.csv')",
"_____no_output_____"
],
[
"data = df.drop([col for col in df.columns if (col not in categoricals and col not in numericals)], axis=1)\ndata = replace_rare_entries(data, ['DEST'], 0.005)\ndata = replace_rare_entries(data, ['UNIQUE_CARRIER'], 0.005)\ndata = pd.get_dummies(data, columns=categoricals)\nlabel = df['DEP_DEL15']",
"_____no_output_____"
],
[
"data.columns",
"_____no_output_____"
],
[
"data = data[~label.isna()]\nlabel = label[~label.isna()]",
"_____no_output_____"
],
[
"print('Rows: {}\\nFeatures: {}\\nLabel-1 Fraction: {}'\n .format(data.shape[0], data.shape[1], label.sum() / label.shape[0]))",
"Rows: 181088\nFeatures: 158\nLabel-1 Fraction: 0.21473537727513695\n"
],
[
"thres = np.linspace(0, 1, 500)",
"_____no_output_____"
],
[
"x, x_test, y, y_test = train_test_split(data, label, test_size=0.2, \n random_state=randint(1, 500),\n stratify=label)",
"_____no_output_____"
],
[
"x_train, x_val, y_train, y_val = train_test_split(x, y, test_size=0.2, \n random_state=randint(1, 500),\n stratify=y)\nDtrain = xgb.DMatrix(x_train, label=y_train)\nDval = xgb.DMatrix(x_val, label=y_val)",
"/home/erez/shared_venv_3.7/lib/python3.7/site-packages/xgboost/core.py:587: FutureWarning: Series.base is deprecated and will be removed in a future version\n if getattr(data, 'base', None) is not None and \\\n"
],
[
"y_train.shape[0]",
"_____no_output_____"
],
[
"param = { # General guidelines for initial paramaters:\n 'min_child_weight': 1, # 1 (choose small for high class imbalance)\n 'gamma': 0.3, # 0.1-0.2\n 'lambda': 0, #1 # L2 Regulariztion - default = 1\n 'scale_pos_weight': 4, # 1 (choose small for high class imbalance)\n 'subsample': 0.6, # 0.5-0.9\n 'colsample_bytree': 0.8, # 0.5-0.9\n 'colsample_bylevel': 0.7, # 0.5-0.9\n 'max_depth': 6, #5 # 3-10 \n 'eta': 0.1, # 0.05-0.3\n 'silent': 0, # 0 - prints progress 1 - quiet\n 'objective': 'binary:logistic', \n 'num_class': 1, \n 'eval_metric': 'auc'} \nnum_round = 10000 # the number of training iterations if not stopped early\nevallist = [(Dtrain, 'train'), (Dval, 'eval')] # Specify validation set to watch performance",
"_____no_output_____"
],
[
"# Train the model on the training set to get an initial impression on the performance\nmodel = xgb.train(param, Dtrain, num_round, evallist, early_stopping_rounds=10)\nprint(\"Best error: {:.2f} with {} rounds\".format(\n model.best_score,\n model.best_iteration+1))",
"[0]\ttrain-auc:0.707297\teval-auc:0.699337\nMultiple eval metrics have been passed: 'eval-auc' will be used for early stopping.\n\nWill train until eval-auc hasn't improved in 10 rounds.\n[1]\ttrain-auc:0.726162\teval-auc:0.717123\n[2]\ttrain-auc:0.735941\teval-auc:0.72378\n[3]\ttrain-auc:0.738586\teval-auc:0.727576\n[4]\ttrain-auc:0.742002\teval-auc:0.729966\n[5]\ttrain-auc:0.74413\teval-auc:0.732707\n[6]\ttrain-auc:0.74832\teval-auc:0.736977\n[7]\ttrain-auc:0.751503\teval-auc:0.740192\n[8]\ttrain-auc:0.75306\teval-auc:0.741764\n[9]\ttrain-auc:0.75478\teval-auc:0.743582\n[10]\ttrain-auc:0.756732\teval-auc:0.745311\n[11]\ttrain-auc:0.75847\teval-auc:0.746524\n[12]\ttrain-auc:0.75978\teval-auc:0.747948\n[13]\ttrain-auc:0.761955\teval-auc:0.750082\n[14]\ttrain-auc:0.762793\teval-auc:0.750688\n[15]\ttrain-auc:0.763681\teval-auc:0.751193\n[16]\ttrain-auc:0.76595\teval-auc:0.753217\n[17]\ttrain-auc:0.767739\teval-auc:0.754294\n[18]\ttrain-auc:0.768983\teval-auc:0.755108\n[19]\ttrain-auc:0.770205\teval-auc:0.756079\n[20]\ttrain-auc:0.771801\teval-auc:0.75764\n[21]\ttrain-auc:0.772715\teval-auc:0.758054\n[22]\ttrain-auc:0.774657\teval-auc:0.758999\n[23]\ttrain-auc:0.775508\teval-auc:0.759609\n[24]\ttrain-auc:0.776531\teval-auc:0.760477\n[25]\ttrain-auc:0.77759\teval-auc:0.761029\n[26]\ttrain-auc:0.778824\teval-auc:0.761777\n[27]\ttrain-auc:0.779725\teval-auc:0.762529\n[28]\ttrain-auc:0.780671\teval-auc:0.763253\n[29]\ttrain-auc:0.781633\teval-auc:0.763846\n[30]\ttrain-auc:0.782886\teval-auc:0.76489\n[31]\ttrain-auc:0.783984\teval-auc:0.765565\n[32]\ttrain-auc:0.785452\teval-auc:0.766802\n[33]\ttrain-auc:0.786463\teval-auc:0.767637\n[34]\ttrain-auc:0.788146\teval-auc:0.76898\n[35]\ttrain-auc:0.789027\teval-auc:0.769698\n[36]\ttrain-auc:0.789734\teval-auc:0.770398\n[37]\ttrain-auc:0.790379\teval-auc:0.770925\n[38]\ttrain-auc:0.791034\teval-auc:0.771148\n[39]\ttrain-auc:0.79214\teval-auc:0.772081\n[40]\ttrain-auc:0.792817\teval-auc:0.772538\n[41]\ttrain-auc:0.793326\teval-auc:0.772874\n[42]\ttrain-auc:0.793875\teval-auc:0.773069\n[43]\ttrain-auc:0.79492\teval-auc:0.773737\n[44]\ttrain-auc:0.795458\teval-auc:0.77394\n[45]\ttrain-auc:0.796215\teval-auc:0.774279\n[46]\ttrain-auc:0.796577\teval-auc:0.774466\n[47]\ttrain-auc:0.797219\teval-auc:0.77481\n[48]\ttrain-auc:0.79775\teval-auc:0.774958\n[49]\ttrain-auc:0.798681\teval-auc:0.775533\n[50]\ttrain-auc:0.799314\teval-auc:0.776089\n[51]\ttrain-auc:0.799818\teval-auc:0.776432\n[52]\ttrain-auc:0.800295\teval-auc:0.776735\n[53]\ttrain-auc:0.801346\teval-auc:0.777711\n[54]\ttrain-auc:0.801916\teval-auc:0.778315\n[55]\ttrain-auc:0.802783\teval-auc:0.778701\n[56]\ttrain-auc:0.80339\teval-auc:0.779018\n[57]\ttrain-auc:0.803799\teval-auc:0.779182\n[58]\ttrain-auc:0.804614\teval-auc:0.779868\n[59]\ttrain-auc:0.805204\teval-auc:0.780428\n[60]\ttrain-auc:0.805583\teval-auc:0.780582\n[61]\ttrain-auc:0.805956\teval-auc:0.780642\n[62]\ttrain-auc:0.806676\teval-auc:0.78132\n[63]\ttrain-auc:0.806936\teval-auc:0.781512\n[64]\ttrain-auc:0.807296\teval-auc:0.781751\n[65]\ttrain-auc:0.807553\teval-auc:0.781815\n[66]\ttrain-auc:0.808228\teval-auc:0.782179\n[67]\ttrain-auc:0.808493\teval-auc:0.78233\n[68]\ttrain-auc:0.808772\teval-auc:0.782424\n[69]\ttrain-auc:0.809166\teval-auc:0.782597\n[70]\ttrain-auc:0.809738\teval-auc:0.782957\n[71]\ttrain-auc:0.810177\teval-auc:0.783246\n[72]\ttrain-auc:0.810724\teval-auc:0.783611\n[73]\ttrain-auc:0.811079\teval-auc:0.783698\n[74]\ttrain-auc:0.811419\teval-auc:0.783833\n[75]\ttrain-auc:0.811858\teval-auc:0.783724\n[76]\ttrain-auc:0.812304\teval-auc:0.783941\n[77]\ttrain-auc:0.81279\teval-auc:0.784353\n[78]\ttrain-auc:0.813292\teval-auc:0.784627\n[79]\ttrain-auc:0.81394\teval-auc:0.784956\n[80]\ttrain-auc:0.814233\teval-auc:0.784957\n[81]\ttrain-auc:0.814571\teval-auc:0.785098\n[82]\ttrain-auc:0.815088\teval-auc:0.785283\n[83]\ttrain-auc:0.815555\teval-auc:0.78537\n[84]\ttrain-auc:0.815797\teval-auc:0.785524\n[85]\ttrain-auc:0.816151\teval-auc:0.785701\n[86]\ttrain-auc:0.816565\teval-auc:0.78605\n[87]\ttrain-auc:0.81692\teval-auc:0.785972\n[88]\ttrain-auc:0.817342\teval-auc:0.786268\n[89]\ttrain-auc:0.817608\teval-auc:0.786452\n[90]\ttrain-auc:0.817941\teval-auc:0.786446\n[91]\ttrain-auc:0.818382\teval-auc:0.786745\n[92]\ttrain-auc:0.818744\teval-auc:0.78685\n[93]\ttrain-auc:0.819163\teval-auc:0.787042\n[94]\ttrain-auc:0.819363\teval-auc:0.787085\n[95]\ttrain-auc:0.81967\teval-auc:0.787291\n[96]\ttrain-auc:0.819855\teval-auc:0.787236\n[97]\ttrain-auc:0.820313\teval-auc:0.787481\n[98]\ttrain-auc:0.820808\teval-auc:0.787878\n[99]\ttrain-auc:0.821015\teval-auc:0.788154\n[100]\ttrain-auc:0.821374\teval-auc:0.788396\n[101]\ttrain-auc:0.821761\teval-auc:0.78854\n[102]\ttrain-auc:0.822358\teval-auc:0.78873\n[103]\ttrain-auc:0.822872\teval-auc:0.789068\n[104]\ttrain-auc:0.823166\teval-auc:0.789247\n[105]\ttrain-auc:0.823536\teval-auc:0.789391\n[106]\ttrain-auc:0.824018\teval-auc:0.789495\n[107]\ttrain-auc:0.824486\teval-auc:0.789753\n[108]\ttrain-auc:0.824811\teval-auc:0.789912\n[109]\ttrain-auc:0.825128\teval-auc:0.790003\n[110]\ttrain-auc:0.82546\teval-auc:0.790173\n[111]\ttrain-auc:0.825707\teval-auc:0.790378\n[112]\ttrain-auc:0.826141\teval-auc:0.790631\n[113]\ttrain-auc:0.826326\teval-auc:0.790667\n[114]\ttrain-auc:0.826648\teval-auc:0.79077\n[115]\ttrain-auc:0.826932\teval-auc:0.790923\n[116]\ttrain-auc:0.82724\teval-auc:0.790858\n[117]\ttrain-auc:0.827466\teval-auc:0.790947\n[118]\ttrain-auc:0.827846\teval-auc:0.791101\n[119]\ttrain-auc:0.828161\teval-auc:0.791351\n[120]\ttrain-auc:0.828351\teval-auc:0.791508\n[121]\ttrain-auc:0.828684\teval-auc:0.791623\n[122]\ttrain-auc:0.829053\teval-auc:0.79171\n[123]\ttrain-auc:0.829483\teval-auc:0.791926\n[124]\ttrain-auc:0.829942\teval-auc:0.79219\n[125]\ttrain-auc:0.830347\teval-auc:0.792183\n[126]\ttrain-auc:0.830538\teval-auc:0.792099\n[127]\ttrain-auc:0.830851\teval-auc:0.792256\n[128]\ttrain-auc:0.831326\teval-auc:0.792365\n[129]\ttrain-auc:0.831512\teval-auc:0.792409\n[130]\ttrain-auc:0.831825\teval-auc:0.79261\n[131]\ttrain-auc:0.832248\teval-auc:0.792765\n[132]\ttrain-auc:0.832402\teval-auc:0.792799\n[133]\ttrain-auc:0.832559\teval-auc:0.792869\n[134]\ttrain-auc:0.832872\teval-auc:0.793001\n[135]\ttrain-auc:0.83317\teval-auc:0.793211\n[136]\ttrain-auc:0.833536\teval-auc:0.793198\n[137]\ttrain-auc:0.833757\teval-auc:0.793322\n[138]\ttrain-auc:0.834058\teval-auc:0.793244\n[139]\ttrain-auc:0.834338\teval-auc:0.793313\n[140]\ttrain-auc:0.834657\teval-auc:0.793486\n[141]\ttrain-auc:0.834956\teval-auc:0.793674\n[142]\ttrain-auc:0.835172\teval-auc:0.793835\n[143]\ttrain-auc:0.835335\teval-auc:0.793853\n[144]\ttrain-auc:0.835627\teval-auc:0.793976\n[145]\ttrain-auc:0.835878\teval-auc:0.793939\n[146]\ttrain-auc:0.836289\teval-auc:0.794222\n[147]\ttrain-auc:0.836609\teval-auc:0.794306\n[148]\ttrain-auc:0.836966\teval-auc:0.794476\n[149]\ttrain-auc:0.837348\teval-auc:0.794648\n[150]\ttrain-auc:0.837599\teval-auc:0.794785\n[151]\ttrain-auc:0.838042\teval-auc:0.794759\n[152]\ttrain-auc:0.838407\teval-auc:0.794825\n[153]\ttrain-auc:0.838526\teval-auc:0.794828\n[154]\ttrain-auc:0.838849\teval-auc:0.794922\n[155]\ttrain-auc:0.839125\teval-auc:0.795043\n[156]\ttrain-auc:0.839372\teval-auc:0.795096\n[157]\ttrain-auc:0.839657\teval-auc:0.795166\n[158]\ttrain-auc:0.839911\teval-auc:0.795236\n[159]\ttrain-auc:0.840213\teval-auc:0.795348\n[160]\ttrain-auc:0.840354\teval-auc:0.795432\n[161]\ttrain-auc:0.840508\teval-auc:0.795435\n[162]\ttrain-auc:0.840687\teval-auc:0.795534\n[163]\ttrain-auc:0.840962\teval-auc:0.795655\n[164]\ttrain-auc:0.841157\teval-auc:0.795739\n[165]\ttrain-auc:0.841298\teval-auc:0.79576\n[166]\ttrain-auc:0.841609\teval-auc:0.795776\n[167]\ttrain-auc:0.841853\teval-auc:0.795676\n[168]\ttrain-auc:0.842016\teval-auc:0.795779\n[169]\ttrain-auc:0.84231\teval-auc:0.795922\n[170]\ttrain-auc:0.84255\teval-auc:0.795926\n[171]\ttrain-auc:0.842772\teval-auc:0.795895\n[172]\ttrain-auc:0.84299\teval-auc:0.795883\n[173]\ttrain-auc:0.843162\teval-auc:0.79588\n[174]\ttrain-auc:0.843348\teval-auc:0.795945\n[175]\ttrain-auc:0.84364\teval-auc:0.795947\n[176]\ttrain-auc:0.843802\teval-auc:0.795993\n[177]\ttrain-auc:0.844022\teval-auc:0.795958\n[178]\ttrain-auc:0.844231\teval-auc:0.795927\n[179]\ttrain-auc:0.844482\teval-auc:0.79605\n[180]\ttrain-auc:0.844712\teval-auc:0.796092\n[181]\ttrain-auc:0.844839\teval-auc:0.796082\n[182]\ttrain-auc:0.845097\teval-auc:0.796036\n[183]\ttrain-auc:0.845256\teval-auc:0.796062\n[184]\ttrain-auc:0.845536\teval-auc:0.796268\n[185]\ttrain-auc:0.845677\teval-auc:0.796232\n[186]\ttrain-auc:0.846002\teval-auc:0.796177\n[187]\ttrain-auc:0.846119\teval-auc:0.796141\n[188]\ttrain-auc:0.846228\teval-auc:0.796116\n[189]\ttrain-auc:0.846388\teval-auc:0.796124\n[190]\ttrain-auc:0.84663\teval-auc:0.796077\n"
],
[
"Dtest = xgb.DMatrix(x_test, label=y_test)\nprobas = model.predict(Dtest)\ny_test = Dtest.get_label()",
"_____no_output_____"
],
[
"accs, recalls, precs, f1s = [], [], [], []\nfor thr in thres:\n y_pred = (probas > thr).astype(int)\n accs.append(skm.accuracy_score(y_test, y_pred))\n recalls.append(skm.recall_score(y_test, y_pred))\n precs.append(skm.precision_score(y_test, y_pred))\n f1s.append(skm.f1_score(y_test, y_pred))\n \nfig = plt.figure(figsize=(20, 10))\nfig.subplots_adjust(hspace=0.4, wspace=0.4)\nfor i, (metric, name) in enumerate(zip([accs, recalls, precs, f1s], ['acc', 'rcl', 'prc', 'f1']), start=1):\n fig.add_subplot(2, 2, i)\n plt.plot(thres, metric)\n plt.title(name)\n",
"/home/erez/shared_venv_3.7/lib/python3.7/site-packages/sklearn/metrics/classification.py:1437: UndefinedMetricWarning: Precision is ill-defined and being set to 0.0 due to no predicted samples.\n 'precision', 'predicted', average, warn_for)\n/home/erez/shared_venv_3.7/lib/python3.7/site-packages/sklearn/metrics/classification.py:1437: UndefinedMetricWarning: F-score is ill-defined and being set to 0.0 due to no predicted samples.\n 'precision', 'predicted', average, warn_for)\n"
],
[
"tpr, fpr, _ = roc_curve(y_test, probas)\nplt.plot(tpr, fpr);",
"_____no_output_____"
],
[
"best_thres = thres[np.argmax(f1s)]\ny_pred = (probas > best_thres).astype(int)\nprint('Acc for max f1 threshold: ', skm.accuracy_score(y_test, y_pred))\nprint('Max acc : ', max(accs))\nprint('Precision : ', skm.precision_score(y_test, y_pred))\nprint('Recall : ', skm.recall_score(y_test, y_pred))\nprint('AUC : ', skm.auc(tpr, fpr))",
"Acc for max f1 threshold: 0.8089900049699045\nMax acc : 0.8369871334695456\nPrecision : 0.5528745537363043\nRecall : 0.5774720329175774\nAUC : 0.8021782895262507\n"
],
[
"xgb.plot_importance(model, max_num_features=20, importance_type='gain');",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb8bf1211be467aeece41cfce42273104e61a4c9
| 46,262 |
ipynb
|
Jupyter Notebook
|
nlp/bag-of-words/my_natural_language_processing_svm.ipynb
|
mirokuru/ml_toolkit
|
df488cc2ee833d16be9d8faaa0d2400347dcdf31
|
[
"CC0-1.0"
] | 3 |
2021-08-21T17:51:59.000Z
|
2021-08-23T16:03:33.000Z
|
nlp/bag-of-words/my_natural_language_processing_svm.ipynb
|
mirokuru/ml_toolkit
|
df488cc2ee833d16be9d8faaa0d2400347dcdf31
|
[
"CC0-1.0"
] | null | null | null |
nlp/bag-of-words/my_natural_language_processing_svm.ipynb
|
mirokuru/ml_toolkit
|
df488cc2ee833d16be9d8faaa0d2400347dcdf31
|
[
"CC0-1.0"
] | 2 |
2021-08-22T03:37:43.000Z
|
2021-08-23T04:49:04.000Z
| 92.155378 | 36,271 | 0.683066 |
[
[
[
"# Natural Language Processing",
"_____no_output_____"
],
[
"## Importing the libraries",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\nimport pandas as pd",
"_____no_output_____"
]
],
[
[
"## Importing the dataset",
"_____no_output_____"
]
],
[
[
"dataset = pd.read_csv('Restaurant_Reviews.tsv', delimiter = '\\t', quoting = 3)",
"_____no_output_____"
]
],
[
[
"## Cleaning the texts",
"_____no_output_____"
]
],
[
[
"import regex\nimport nltk\nnltk.download('stopwords')\nfrom nltk.corpus import stopwords\nfrom nltk.stem.porter import PorterStemmer\ncorpus = []\nfor i in range(0, len(dataset)):\n review = regex.sub('[^\\p{L}]', ' ', dataset['Review'][i])\n review = review.lower()\n review = review.split()\n ps = PorterStemmer()\n all_stopwords = stopwords.words('english')\n all_stopwords.remove('not')\n review = [ps.stem(word) for word in review if not word in set(all_stopwords)]\n review = ' '.join(review)\n corpus.append(review)",
"[nltk_data] Downloading package stopwords to\n[nltk_data] C:\\Users\\Admin\\AppData\\Roaming\\nltk_data...\n[nltk_data] Package stopwords is already up-to-date!\n"
],
[
"print(corpus)",
"['wow love place', 'crust not good', 'not tasti textur nasti', 'stop late may bank holiday rick steve recommend love', 'select menu great price', 'get angri want damn pho', 'honeslti tast fresh', 'potato like rubber could tell made ahead time kept warmer', 'fri great', 'great touch', 'servic prompt', 'would not go back', 'cashier care ever say still end wayyy overpr', 'tri cape cod ravoli chicken cranberri mmmm', 'disgust pretti sure human hair', 'shock sign indic cash', 'highli recommend', 'waitress littl slow servic', 'place not worth time let alon vega', 'not like', 'burritto blah', 'food amaz', 'servic also cute', 'could care less interior beauti', 'perform', 'right red velvet cake ohhh stuff good', 'never brought salad ask', 'hole wall great mexican street taco friendli staff', 'took hour get food tabl restaur food luke warm sever run around like total overwhelm', 'worst salmon sashimi', 'also combo like burger fri beer decent deal', 'like final blow', 'found place accid could not happier', 'seem like good quick place grab bite familiar pub food favor look elsewher', 'overal like place lot', 'redeem qualiti restaur inexpens', 'ampl portion good price', 'poor servic waiter made feel like stupid everi time came tabl', 'first visit hiro delight', 'servic suck', 'shrimp tender moist', 'not deal good enough would drag establish', 'hard judg whether side good gross melt styrofoam want eat fear get sick', 'posit note server attent provid great servic', 'frozen puck disgust worst peopl behind regist', 'thing like prime rib dessert section', 'bad food damn gener', 'burger good beef cook right', 'want sandwich go firehous', 'side greek salad greek dress tasti pita hummu refresh', 'order duck rare pink tender insid nice char outsid', 'came run us realiz husband left sunglass tabl', 'chow mein good', 'horribl attitud toward custom talk one custom enjoy food', 'portion huge', 'love friendli server great food wonder imagin menu', 'heart attack grill downtown vega absolut flat line excus restaur', 'not much seafood like string pasta bottom', 'salad right amount sauc not power scallop perfectli cook', 'rip banana not rip petrifi tasteless', 'least think refil water struggl wave minut', 'place receiv star appet', 'cocktail handmad delici', 'definit go back', 'glad found place', 'great food servic huge portion give militari discount', 'alway great time do gringo', 'updat went back second time still amaz', 'got food appar never heard salt batter fish chewi', 'great way finish great', 'deal includ tast drink jeff went beyond expect', 'realli realli good rice time', 'servic meh', 'took min get milkshak noth chocol milk', 'guess known place would suck insid excalibur use common sens', 'scallop dish quit appal valu well', 'time bad custom servic', 'sweet potato fri good season well', 'today second time lunch buffet pretti good', 'much good food vega feel cheat wast eat opportun go rice compani', 'come like experienc underwhelm relationship parti wait person ask break', 'walk place smell like old greas trap other eat', 'turkey roast beef bland', 'place', 'pan cake everyon rave tast like sugari disast tailor palat six year old', 'love pho spring roll oh yummi tri', 'poor batter meat ratio made chicken tender unsatisfi', 'say food amaz', 'omelet die', 'everyth fresh delici', 'summari larg disappoint dine experi', 'like realli sexi parti mouth outrag flirt hottest person parti', 'never hard rock casino never ever step forward', 'best breakfast buffet', 'say bye bye tip ladi', 'never go', 'back', 'food arriv quickli', 'not good', 'side cafe serv realli good food', 'server fantast found wife love roast garlic bone marrow ad extra meal anoth marrow go', 'good thing waiter help kept bloddi mari come', 'best buffet town price cannot beat', 'love mussel cook wine reduct duck tender potato dish delici', 'one better buffet', 'went tigerlilli fantast afternoon', 'food delici bartend attent person got great deal', 'ambienc wonder music play', 'go back next trip', 'sooooo good', 'real sushi lover let honest yama not good', 'least min pass us order food arriv busi', 'realli fantast thai restaur definit worth visit', 'nice spici tender', 'good price', 'check', 'pretti gross', 'better atmospher', 'kind hard mess steak', 'although much like look sound place actual experi bit disappoint', 'know place manag serv blandest food ever eaten prepar indian cuisin', 'worst servic boot least worri', 'servic fine waitress friendli', 'guy steak steak love son steak best worst place said best steak ever eaten', 'thought ventur away get good sushi place realli hit spot night', 'host staff lack better word bitch', 'bland not like place number reason want wast time bad review leav', 'phenomen food servic ambianc', 'return', 'definit worth ventur strip pork belli return next time vega', 'place way overpr mediocr food', 'penn vodka excel', 'good select food includ massiv meatloaf sandwich crispi chicken wrap delish tuna melt tasti burger', 'manag rude', 'delici nyc bagel good select cream chees real lox caper even', 'great subway fact good come everi subway not meet expect', 'serious solid breakfast', 'one best bar food vega', 'extrem rude realli mani restaur would love dine weekend vega', 'drink never empti made realli great menu suggest', '', 'waiter help friendli rare check us', 'husband ate lunch disappoint food servic', 'red curri much bamboo shoot tasti', 'nice blanket moz top feel like done cover subpar food', 'bathroom clean place well decor', 'menu alway chang food qualiti go servic extrem slow', 'servic littl slow consid serv peopl server food come slow pace', 'give thumb', 'watch waiter pay lot attent tabl ignor us', 'fiancé came middl day greet seat right away', 'great restaur mandalay bay', 'wait forti five minut vain', 'crostini came salad stale', 'highlight great qualiti nigiri', 'staff friendli joint alway clean', 'differ cut piec day still wonder tender well well flavor', 'order voodoo pasta first time realli excel pasta sinc go gluten free sever year ago', 'place good', 'unfortun must hit bakeri leftov day everyth order stale', 'came back today sinc reloc still not impress', 'seat immedi', 'menu divers reason price', 'avoid cost', 'restaur alway full never wait', 'delici', 'place hand one best place eat phoenix metro area', 'go look good food', 'never treat bad', 'bacon hella salti', 'also order spinach avocado salad ingredi sad dress liter zero tast', 'realli vega fine dine use right menu hand ladi price list', 'waitress friendli', 'lordi khao soi dish not miss curri lover', 'everyth menu terrif also thrill made amaz accommod vegetarian daughter', 'perhap caught night judg review not inspir go back', 'servic leav lot desir', 'atmospher modern hip maintain touch cozi', 'not weekli haunt definit place come back everi', 'liter sat minut one ask take order', 'burger absolut flavor meat total bland burger overcook charcoal flavor', 'also decid not send back waitress look like verg heart attack', 'dress treat rude', 'probabl dirt', 'love place hit spot want someth healthi not lack quantiti flavor', 'order lemon raspberri ice cocktail also incred', 'food suck expect suck could imagin', 'interest decor', 'realli like crepe station', 'also serv hot bread butter home made potato chip bacon bit top origin good', 'watch prepar delici food', 'egg roll fantast', 'order arriv one gyro miss', 'salad wing ice cream dessert left feel quit satisfi', 'not realli sure joey vote best hot dog valley reader phoenix magazin', 'best place go tasti bowl pho', 'live music friday total blow', 'never insult felt disrespect', 'friendli staff', 'worth drive', 'heard good thing place exceed everi hope could dream', 'food great serivc', 'warm beer help', 'great brunch spot', 'servic friendli invit', 'good lunch spot', 'live sinc first last time step foot place', 'worst experi ever', 'must night place', 'side delish mix mushroom yukon gold pure white corn beateou', 'bug never show would given sure side wall bug climb kitchen', 'minut wait salad realiz come time soon', 'friend love salmon tartar', 'go back', 'extrem tasti', 'waitress good though', 'soggi not good', 'jamaican mojito delici', 'small not worth price', 'food rich order accordingli', 'shower area outsid rins not take full shower unless mind nude everyon see', 'servic bit lack', 'lobster bisqu bussel sprout risotto filet need salt pepper cours none tabl', 'hope bode go busi someon cook come', 'either cold not enough flavor bad', 'love bacon wrap date', 'unbeliev bargain', 'folk otto alway make us feel welcom special', 'main also uninspir', 'place first pho amaz', 'wonder experi made place must stop whenev town', 'food bad enough enjoy deal world worst annoy drunk peopl', 'fun chef', 'order doubl cheeseburg got singl patti fall apart pictur upload yeah still suck', 'great place coupl drink watch sport event wall cover tv', 'possibl give zero star', 'descript said yum yum sauc anoth said eel sauc yet anoth said spici mayo well none roll sauc', 'say would hardest decis honestli dish tast suppos tast amaz', 'not roll eye may stay not sure go back tri', 'everyon attent provid excel custom servic', 'horribl wast time money', 'dish quit flavour', 'time side restaur almost empti excus', 'busi either also build freez cold', 'like review said pay eat place', 'drink took close minut come one point', 'serious flavor delight folk', 'much better ayc sushi place went vega', 'light dark enough set mood', 'base sub par servic receiv effort show gratitud busi go back', 'owner realli great peopl', 'noth privileg work eat', 'greek dress creami flavor', 'overal think would take parent place made similar complaint silent felt', 'pizza good peanut sauc tasti', 'tabl servic pretti fast', 'fantast servic', 'well would given godfath zero star possibl', 'know make', 'tough short flavor', 'hope place stick around', 'bar vega not ever recal charg tap water', 'restaur atmospher exquisit', 'good servic clean inexpens boot', 'seafood fresh gener portion', 'plu buck', 'servic not par either', 'thu far visit twice food absolut delici time', 'good year ago', 'self proclaim coffe cafe wildli disappoint', 'veggitarian platter world', 'cant go wrong food', 'beat', 'stop place madison ironman friendli kind staff', 'chef friendli good job', 'better not dedic boba tea spot even jenni pho', 'like patio servic outstand', 'goat taco skimp meat wow flavor', 'think not', 'mac salad pretti bland not get', 'went bachi burger friend recommend not disappoint', 'servic stink', 'wait wait', 'place not qualiti sushi not qualiti restaur', 'would definit recommend wing well pizza', 'great pizza salad', 'thing went wrong burn saganaki', 'wait hour breakfast could done time better home', 'place amaz', 'hate disagre fellow yelper husband disappoint place', 'wait hour never got either pizza mani around us came later', 'know slow', 'staff great food delish incred beer select', 'live neighborhood disappoint back conveni locat', 'know pull pork could soooo delici', 'get incred fresh fish prepar care', 'go gave star rate pleas know third time eat bachi burger write review', 'love fact everyth menu worth', 'never dine place', 'food excel servic good', 'good beer drink select good food select', 'pleas stay away shrimp stir fri noodl', 'potato chip order sad could probabl count mani chip box probabl around', 'food realli bore', 'good servic check', 'greedi corpor never see anoth dime', 'never ever go back', 'much like go back get pass atroci servic never return', 'summer dine charm outdoor patio delight', 'not expect good', 'fantast food', 'order toast english muffin came untoast', 'food good', 'never go back', 'great food price high qualiti hous made', 'bu boy hand rude', 'point friend basic figur place joke mind make publicli loudli known', 'back good bbq lighter fare reason price tell public back old way', 'consid two us left full happi go wrong', 'bread made hous', 'downsid servic', 'also fri without doubt worst fri ever', 'servic except food good review', 'coupl month later return amaz meal', 'favorit place town shawarrrrrrma', 'black eye pea sweet potato unreal', 'disappoint', 'could serv vinaigrett may make better overal dish still good', 'go far mani place never seen restaur serv egg breakfast especi', 'mom got home immedi got sick bite salad', 'server not pleasant deal alway honor pizza hut coupon', 'truli unbeliev good glad went back', 'fantast servic pleas atmospher', 'everyth gross', 'love place', 'great servic food', 'first bathroom locat dirti seat cover not replenish plain yucki', 'burger got gold standard burger kind disappoint', 'omg food delicioso', 'noth authent place', 'spaghetti noth special whatsoev', 'dish salmon best great', 'veget fresh sauc feel like authent thai', 'worth drive tucson', 'select probabl worst seen vega none', 'pretti good beer select', 'place like chipotl better', 'classi warm atmospher fun fresh appet succul steak basebal steak', 'star brick oven bread app', 'eaten multipl time time food delici', 'sat anoth ten minut final gave left', 'terribl', 'everyon treat equal special', 'take min pancak egg', 'delici', 'good side staff genuin pleasant enthusiast real treat', 'sadli gordon ramsey steak place shall sharpli avoid next trip vega', 'alway even wonder food delici', 'best fish ever life', 'bathroom next door nice', 'buffet small food offer bland', 'outstand littl restaur best food ever tast', 'pretti cool would say', 'definit turn doubt back unless someon els buy', 'server great job handl larg rowdi tabl', 'find wast food despic food', 'wife lobster bisqu soup lukewarm', 'would come back sushi crave vega', 'staff great ambianc great', 'deserv star', 'left stomach ach felt sick rest day', 'drop ball', 'dine space tini elegantli decor comfort', 'custom order way like usual eggplant green bean stir fri love', 'bean rice mediocr best', 'best taco town far', 'took back money got outta', 'interest part town place amaz', 'rude inconsider manag', 'staff not friendli wait time serv horribl one even say hi first minut', 'back', 'great dinner', 'servic outshin definit recommend halibut', 'food terribl', 'never ever go back told mani peopl happen', 'recommend unless car break front starv', 'come back everi time vega', 'place deserv one star food', 'disgrac', 'def come back bowl next time', 'want healthi authent ethic food tri place', 'continu come ladi night andddd date night highli recommend place anyon area', 'sever time past experi alway great', 'walk away stuf happi first vega buffet experi', 'servic excel price pretti reason consid vega locat insid crystal shop mall aria', 'summar food incred nay transcend noth bring joy quit like memori pneumat condiment dispens', 'probabl one peopl ever go ian not like', 'kid pizza alway hit lot great side dish option kiddo', 'servic perfect famili atmospher nice see', 'cook perfect servic impecc', 'one simpli disappoint', 'overal disappoint qualiti food bouchon', 'account know get screw', 'great place eat remind littl mom pop shop san francisco bay area', 'today first tast buldogi gourmet hot dog tell ever thought possibl', 'left frustrat', 'definit soon', 'food realli good got full petti fast', 'servic fantast', 'total wast time', 'know kind best ice tea', 'come hungri leav happi stuf', 'servic give star', 'assur disappoint', 'take littl bad servic food suck', 'gave tri eat crust teeth still sore', 'complet gross', 'realli enjoy eat', 'first time go think quickli becom regular', 'server nice even though look littl overwhelm need stay profession friendli end', 'dinner companion told everyth fresh nice textur tast', 'ground right next tabl larg smear step track everywher pile green bird poop', 'furthermor even find hour oper websit', 'tri like place time think done', 'mistak', 'complaint', 'serious good pizza expert connisseur topic', 'waiter jerk', 'strike want rush', 'nicest restaur owner ever come across', 'never come', 'love biscuit', 'servic quick friendli', 'order appet took minut pizza anoth minut', 'absolutley fantast', 'huge awkward lb piec cow th gristl fat', 'definit come back', 'like steiner dark feel like bar', 'wow spici delici', 'not familiar check', 'take busi dinner dollar elsewher', 'love go back', 'anyway fs restaur wonder breakfast lunch', 'noth special', 'day week differ deal delici', 'not mention combin pear almond bacon big winner', 'not back', 'sauc tasteless', 'food delici spici enough sure ask spicier prefer way', 'ribey steak cook perfectli great mesquit flavor', 'think go back anytim soon', 'food gooodd', 'far sushi connoisseur definit tell differ good food bad food certainli bad food', 'insult', 'last time lunch bad', 'chicken wing contain driest chicken meat ever eaten', 'food good enjoy everi mouth enjoy relax venu coupl small famili group etc', 'nargil think great', 'best tater tot southwest', 'love place', 'definit not worth paid', 'vanilla ice cream creami smooth profiterol choux pastri fresh enough', 'im az time new spot', 'manag worst', 'insid realli quit nice clean', 'food outstand price reason', 'think run back carli anytim soon food', 'due fact took minut acknowledg anoth minut get food kept forget thing', 'love margarita', 'first vega buffet not disappoint', 'good though', 'one note ventil could use upgrad', 'great pork sandwich', 'wast time', 'total letdown would much rather go camelback flower shop cartel coffe', 'third chees friend burger cold', 'enjoy pizza brunch', 'steak well trim also perfectli cook', 'group claim would handl us beauti', 'love', 'ask bill leav without eat bring either', 'place jewel la vega exactli hope find nearli ten year live', 'seafood limit boil shrimp crab leg crab leg definit not tast fresh', 'select food not best', 'delici absolut back', 'small famili restaur fine dine establish', 'toro tartar cavier extraordinari like thinli slice wagyu white truffl', 'dont think back long time', 'attach ga station rare good sign', 'awesom', 'back mani time soon', 'menu much good stuff could not decid', 'wors humili worker right front bunch horribl name call', 'conclus fill meal', 'daili special alway hit group', 'tragedi struck', 'pancak also realli good pretti larg', 'first crawfish experi delici', 'monster chicken fri steak egg time favorit', 'waitress sweet funni', 'also tast mom multi grain pumpkin pancak pecan butter amaz fluffi delici', 'rather eat airlin food serious', 'cant say enough good thing place', 'ambianc incred', 'waitress manag friendli', 'would not recommend place', 'overal impress noca', 'gyro basic lettuc', 'terribl servic', 'thoroughli disappoint', 'much pasta love homemad hand made pasta thin pizza', 'give tri happi', 'far best cheesecurd ever', 'reason price also', 'everyth perfect night', 'food good typic bar food', 'drive get', 'first glanc love bakeri cafe nice ambianc clean friendli staff', 'anyway not think go back', 'point finger item menu order disappoint', 'oh thing beauti restaur', 'gone go', 'greasi unhealthi meal', 'first time might last', 'burger amaz', 'similarli deliveri man not say word apolog food minut late', 'way expens', 'sure order dessert even need pack go tiramisu cannoli die', 'first time wait next', 'bartend also nice', 'everyth good tasti', 'place two thumb way', 'best place vega breakfast check sat sun', 'love authent mexican food want whole bunch interest yet delici meat choos need tri place', 'terribl manag', 'excel new restaur experienc frenchman', 'zero star would give zero star', 'great steak great side great wine amaz dessert', 'worst martini ever', 'steak shrimp opinion best entre gc', 'opportun today sampl amaz pizza', 'wait thirti minut seat although vacant tabl folk wait', 'yellowtail carpaccio melt mouth fresh', 'tri go back even empti', 'go eat potato found stranger hair', 'spici enough perfect actual', 'last night second time dine happi decid go back', 'not even hello right', 'dessert bit strang', 'boyfriend came first time recent trip vega could not pleas qualiti food servic', 'realli recommend place go wrong donut place', 'nice ambianc', 'would recommend save room', 'guess mayb went night disgrac', 'howev recent experi particular locat not good', 'know not like restaur someth', 'avoid establish', 'think restaur suffer not tri hard enough', 'tapa dish delici', 'heart place', 'salad bland vinegrett babi green heart palm', 'two felt disgust', 'good time', 'believ place great stop huge belli hanker sushi', 'gener portion great tast', 'never go back place never ever recommend place anyon', 'server went back forth sever time not even much help', 'food delici', 'hour serious', 'consid theft', 'eew locat need complet overhaul', 'recent wit poor qualiti manag toward guest well', 'wait wait wait', 'also came back check us regularli excel servic', 'server super nice check us mani time', 'pizza tast old super chewi not good way', 'swung give tri deepli disappoint', 'servic good compani better', 'staff also friendli effici', 'servic fan quick serv nice folk', 'boy sucker dri', 'rate', 'look authent thai food go els', 'steak recommend', 'pull car wait anoth minut acknowledg', 'great food great servic clean friendli set', 'assur back', 'hate thing much cheap qualiti black oliv', 'breakfast perpar great beauti present giant slice toast lightli dust powder sugar', 'kid play area nasti', 'great place fo take eat', 'waitress friendli happi accomod vegan veggi option', 'omg felt like never eaten thai food dish', 'extrem crumbi pretti tasteless', 'pale color instead nice char flavor', 'crouton also tast homemad extra plu', 'got home see driest damn wing ever', 'regular stop trip phoenix', 'realli enjoy crema café expand even told friend best breakfast', 'not good money', 'miss wish one philadelphia', 'got sit fairli fast end wait minut place order anoth minut food arriv', 'also best chees crisp town', 'good valu great food great servic', 'ask satisfi meal', 'food good', 'awesom', 'want leav', 'made drive way north scottsdal not one bit disappoint', 'not eat', 'owner realli realli need quit soooooo cheap let wrap freak sandwich two paper not one', 'check place coupl year ago not impress', 'chicken got definit reheat ok wedg cold soggi', 'sorri not get food anytim soon', 'absolut must visit', 'cow tongu cheek taco amaz', 'friend not like bloodi mari', 'despit hard rate busi actual rare give star', 'realli want make experi good one', 'not return', 'chicken pho tast bland', 'disappoint', 'grill chicken tender yellow saffron season', 'drive thru mean not want wait around half hour food somehow end go make us wait wait', 'pretti awesom place', 'ambienc perfect', 'best luck rude non custom servic focus new manag', 'grandmoth make roast chicken better one', 'ask multipl time wine list time ignor went hostess got one', 'staff alway super friendli help especi cool bring two small boy babi', 'four star food guy blue shirt great vibe still let us eat', 'roast beef sandwich tast realli good', 'even drastic sick', 'high qualiti chicken chicken caesar salad', 'order burger rare came done', 'promptli greet seat', 'tri go lunch madhous', 'proven dead wrong sushi bar not qualiti great servic fast food impecc', 'wait hour seat not greatest mood', 'good joint', 'macaron insan good', 'not eat', 'waiter attent friendli inform', 'mayb cold would somewhat edibl', 'place lot promis fail deliv', 'bad experi', 'mistak', 'food averag best', 'great food', 'go back anytim soon', 'disappoint order big bay plater', 'great place relax awesom burger beer', 'perfect sit famili meal get togeth friend', 'not much flavor poorli construct', 'patio seat comfort', 'fri rice dri well', 'hand favorit italian restaur', 'scream legit book somethat also pretti rare vega', 'not fun experi', 'atmospher great love duo violinist play song request', 'person love hummu pita baklava falafel baba ganoush amaz eggplant', 'conveni sinc stay mgm', 'owner super friendli staff courteou', 'great', 'eclect select', 'sweet potato tot good onion ring perfect close', 'staff attent', 'chef gener time even came around twice take pictur', 'owner use work nobu place realli similar half price', 'googl mediocr imagin smashburg pop', 'dont go', 'promis disappoint', 'sushi lover avoid place mean', 'great doubl cheeseburg', 'awesom servic food', 'fantast neighborhood gem', 'wait go back', 'plantain worst ever tast', 'great place highli recommend', 'servic slow not attent', 'gave star give star', 'staff spend time talk', 'dessert panna cotta amaz', 'good food great atmospher', 'damn good steak', 'total brunch fail', 'price reason flavor spot sauc home made slaw not drench mayo', 'decor nice piano music soundtrack pleasant', 'steak amaz rge fillet relleno best seafood plate ever', 'good food good servic', 'absolut amaz', 'probabl back honest', 'definit back', 'sergeant pepper beef sandwich auju sauc excel sandwich well', 'hawaiian breez mango magic pineappl delight smoothi tri far good', 'went lunch servic slow', 'much say place walk expect amaz quickli disappoint', 'mortifi', 'needless say never back', 'anyway food definit not fill price pay expect', 'chip came drip greas mostli not edibl', 'realli impress strip steak', 'go sinc everi meal awesom', 'server nice attent serv staff', 'cashier friendli even brought food', 'work hospit industri paradis valley refrain recommend cibo longer', 'atmospher fun', 'would not recommend other', 'servic quick even go order like like', 'mean realli get famou fish chip terribl', 'said mouth belli still quit pleas', 'not thing', 'thumb', 'read pleas go', 'love grill pizza remind legit italian pizza', 'pro larg seat area nice bar area great simpl drink menu best brick oven pizza homemad dough', 'realli nice atmospher', 'tonight elk filet special suck', 'one bite hook', 'order old classic new dish go time sore disappoint everyth', 'cute quaint simpl honest', 'chicken delici season perfect fri outsid moist chicken insid', 'food great alway compliment chef', 'special thank dylan recommend order yummi tummi', 'awesom select beer', 'great food awesom servic', 'one nice thing ad gratuiti bill sinc parti larger expect tip', 'fli appl juic fli', 'han nan chicken also tasti', 'servic thought good', 'food bare lukewarm must sit wait server bring us', 'ryan bar definit one edinburgh establish revisit', 'nicest chines restaur', 'overal like food servic', 'also serv indian naan bread hummu spici pine nut sauc world', 'probabl never come back recommend', 'friend pasta also bad bare touch', 'tri airport experi tasti food speedi friendli servic', 'love decor chines calligraphi wall paper', 'never anyth complain', 'restaur clean famili restaur feel', 'way fri', 'not sure long stood long enough begin feel awkwardli place', 'open sandwich impress not good way', 'not back', 'warm feel servic felt like guest special treat', 'extens menu provid lot option breakfast', 'alway order vegetarian menu dinner wide array option choos', 'watch price inflat portion get smaller manag attitud grow rapidli', 'wonder lil tapa ambienc made feel warm fuzzi insid', 'got enjoy seafood salad fabul vinegrett', 'wonton thin not thick chewi almost melt mouth', 'level spici perfect spice whelm soup', 'sat right time server get go fantast', 'main thing enjoy crowd older crowd around mid', 'side town definit spot hit', 'wait minut get drink longer get arepa', 'great place eat', 'jalapeno bacon soooo good', 'servic poor that nice', 'food good servic good price good', 'place not clean food oh stale', 'chicken dish ok beef like shoe leather', 'servic beyond bad', 'happi', 'tast like dirt', 'one place phoenix would defin go back', 'block amaz', 'close hous low key non fanci afford price good food', 'hot sour egg flower soup absolut star', 'sashimi poor qualiti soggi tasteless', 'great time famili dinner sunday night', 'food not tasti not say real tradit hunan style', 'bother slow servic', 'flair bartend absolut amaz', 'frozen margarita way sugari tast', 'good order twice', 'nutshel restaraunt smell like combin dirti fish market sewer', 'girlfriend veal bad', 'unfortun not good', 'pretti satifi experi', 'join club get awesom offer via email', 'perfect someon like beer ice cold case even colder', 'bland flavorless good way describ bare tepid meat', 'chain fan beat place easili', 'nacho must', 'not come back', 'mani word say place everyth pretti well', 'staff super nice quick even crazi crowd downtown juri lawyer court staff', 'great atmospher friendli fast servic', 'receiv pita huge lot meat thumb', 'food arriv meh', 'pay hot dog fri look like came kid meal wienerschnitzel not idea good meal', 'classic main lobster roll fantast', 'brother law work mall ate day guess sick night', 'good go review place twice herea tribut place tribut event held last night', 'chip salsa realli good salsa fresh', 'place great', 'mediocr food', 'get insid impress place', 'super pissd', 'servic super friendli', 'sad littl veget overcook', 'place nice surpris', 'golden crispi delici', 'high hope place sinc burger cook charcoal grill unfortun tast fell flat way flat', 'could eat bruschetta day devin', 'not singl employe came see ok even need water refil final serv us food', 'lastli mozzarella stick best thing order', 'first time ever came amaz experi still tell peopl awesom duck', 'server neglig need made us feel unwelcom would not suggest place', 'servic terribl though', 'place overpr not consist boba realli overpr', 'pack', 'love place', 'say dessert yummi', 'food terribl', 'season fruit fresh white peach pure', 'kept get wors wors offici done', 'place honestli blown', 'definit would not eat', 'not wast money', 'love put food nice plastic contain oppos cram littl paper takeout box', 'crêpe delic thin moist', 'aw servic', 'ever go', 'food qualiti horribl', 'price think place would much rather gone', 'servic fair best', 'love sushi found kabuki price hip servic', 'favor stay away dish', 'poor servic', 'one tabl thought food averag worth wait', 'best servic food ever maria server good friendli made day', 'excel', 'paid bill not tip felt server terribl job', 'lunch great experi', 'never bland food surpris consid articl read focus much spice flavor', 'food way overpr portion fuck small', 'recent tri caballero back everi week sinc', 'buck head realli expect better food', 'food came good pace', 'ate twice last visit especi enjoy salmon salad', 'back', 'could not believ dirti oyster', 'place deserv star', 'would not recommend place', 'fact go round star awesom', 'disbelief dish qualifi worst version food ever tast', 'bad day not low toler rude custom servic peopl job nice polit wash dish otherwis', 'potato great biscuit', 'probabl would not go', 'flavor perfect amount heat', 'price reason servic great', 'wife hate meal coconut shrimp friend realli not enjoy meal either', 'fella got huevo ranchero look appeal', 'went happi hour great list wine', 'may say buffet pricey think get pay place get quit lot', 'probabl come back', 'worst food servic', 'place pretti good nice littl vibe restaur', 'talk great custom servic cours back', 'hot dish not hot cold dish close room temp watch staff prepar food bare hand glove everyth deep fri oil', 'love fri bean', 'alway pleasur deal', 'plethora salad sandwich everyth tri get seal approv', 'place awesom want someth light healthi summer', 'sushi strip place go', 'servic great even manag came help tabl', 'feel dine room colleg cook cours high class dine servic slow best', 'start review two star edit give one', 'worst sushi ever eat besid costco', 'excel restaur highlight great servic uniqu menu beauti set', 'boyfriend sat bar complet delight experi', 'weird vibe owner', 'hardli meat', 'better bagel groceri store', 'go place gyro', 'love owner chef one authent japanes cool dude', 'burger good pizza use amaz doughi flavorless', 'found six inch long piec wire salsa', 'servic terribl food mediocr', 'defin enjoy', 'order albondiga soup warm tast like tomato soup frozen meatbal', 'three differ occas ask well done medium well three time got bloodiest piec meat plate', 'two bite refus eat anymor', 'servic extrem slow', 'minut wait got tabl', 'serious killer hot chai latt', 'allergi warn menu waitress absolut clue meal not contain peanut', 'boyfriend tri mediterranean chicken salad fell love', 'rotat beer tap also highlight place', 'price bit concern mellow mushroom', 'worst thai ever', 'stay vega must get breakfast least', 'want first say server great perfect servic', 'pizza select good', 'strawberri tea good', 'highli unprofession rude loyal patron', 'overal great experi', 'spend money elsewher', 'regular toast bread equal satisfi occasion pat butter mmmm', 'buffet bellagio far anticip', 'drink weak peopl', 'order not correct', 'also feel like chip bought not made hous', 'disappoint dinner went elsewher dessert', 'chip sal amaz', 'return', 'new fav vega buffet spot', 'serious cannot believ owner mani unexperienc employe run around like chicken head cut', 'sad', 'felt insult disrespect could talk judg anoth human like', 'call steakhous properli cook steak understand', 'not impress concept food', 'thing crazi guacamol like puré', 'realli noth postino hope experi better', 'got food poison buffet', 'brought fresh batch fri think yay someth warm', 'hilari yummi christma eve dinner rememb biggest fail entir trip us', 'needless say go back anytim soon', 'place disgust', 'everi time eat see care teamwork profession degre', 'ri style calamari joke', 'howev much garlic fondu bare edibl', 'could bare stomach meal complain busi lunch', 'bad lost heart finish', 'also took forev bring us check ask', 'one make scene restaur get definit lost love one', 'disappoint experi', 'food par denni say not good', 'want wait mediocr food downright terribl servic place', 'waaaaaayyyyyyyyyi rate say', 'go back', 'place fairli clean food simpli worth', 'place lack style', 'sangria half glass wine full ridicul', 'bother come', 'meat pretti dri slice brisket pull pork', 'build seem pretti neat bathroom pretti trippi eat', 'equal aw', 'probabl not hurri go back', 'slow seat even reserv', 'not good stretch imagin', 'cashew cream sauc bland veget undercook', 'chipolt ranch dip saus tasteless seem thin water heat', 'bit sweet not realli spici enough lack flavor', 'disappoint', 'place horribl way overpr', 'mayb vegetarian fare twice thought averag best', 'busi know', 'tabl outsid also dirti lot time worker not alway friendli help menu', 'ambianc not feel like buffet set douchey indoor garden tea biscuit', 'con spotti servic', 'fri not hot neither burger', 'came back cold', 'food came disappoint ensu', 'real disappoint waiter', 'husband said rude not even apolog bad food anyth', 'reason eat would fill night bing drink get carb stomach', 'insult profound deuchebaggeri go outsid smoke break serv solidifi', 'someon order two taco think may part custom servic ask combo ala cart', 'quit disappoint although blame need place door', 'rave review wait eat disappoint', 'del taco pretti nasti avoid possibl', 'not hard make decent hamburg', 'like', 'hell go back', 'gotten much better servic pizza place next door servic receiv restaur', 'know big deal place back ya', 'immedi said want talk manag not want talk guy shot firebal behind bar', 'ambianc much better', 'unfortun set us disapppoint entre', 'food good', 'server suck wait correct server heimer suck', 'happen next pretti put', 'bad caus know famili own realli want like place', 'overpr get', 'vomit bathroom mid lunch', 'kept look time soon becom minut yet still food', 'place eat circumst would ever return top list', 'start tuna sashimi brownish color obvious fresh', 'food averag', 'sure beat nacho movi would expect littl bit come restaur', 'ha long bay bit flop', 'problem charg sandwich bigger subway sub offer better amount veget', 'shrimp unwrap live mile brushfir liter ice cold', 'lack flavor seem undercook dri', 'realli impress place close', 'would avoid place stay mirag', 'refri bean came meal dri crusti food bland', 'spend money time place els', 'ladi tabl next us found live green caterpillar salad', 'present food aw', 'tell disappoint', 'think food flavor textur lack', 'appetit instantli gone', 'overal not impress would not go back', 'whole experi underwhelm think go ninja sushi next time', 'wast enough life pour salt wound draw time took bring check']\n"
]
],
[
[
"## Creating the Bag of Words model",
"_____no_output_____"
]
],
[
[
"from sklearn.feature_extraction.text import CountVectorizer\ncv = CountVectorizer(max_features = 1500) # set after getting number of all words\nX = cv.fit_transform(corpus).toarray()\ny = dataset.iloc[:, -1].values",
"_____no_output_____"
],
[
"len(X[0])",
"_____no_output_____"
]
],
[
[
"## Splitting the dataset into the Training set and Test set",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import train_test_split\nX_train, X_test, y_train, y_test = train_test_split(X, y, test_size = 0.2, random_state = 0)",
"_____no_output_____"
]
],
[
[
"## Training the Linear Support Vector Machine model on the Training set",
"_____no_output_____"
]
],
[
[
"from sklearn.svm import SVC\nclassifier = SVC(kernel = 'linear', random_state = 0)\nclassifier.fit(X_train, y_train)",
"_____no_output_____"
]
],
[
[
"## Predicting the Test set results",
"_____no_output_____"
]
],
[
[
"y_pred = classifier.predict(X_test)\n# evaluate performance by comparing the predicted review and the ground truth\nprint(np.concatenate(\n (\n y_pred.reshape(len(y_pred), 1),\n y_test.reshape(len(y_test), 1)\n ),\n axis=1))",
"[[0 0]\n [0 0]\n [0 0]\n [0 0]\n [0 0]\n [0 0]\n [1 1]\n [0 0]\n [0 0]\n [1 1]\n [1 1]\n [1 1]\n [1 0]\n [1 1]\n [1 1]\n [1 1]\n [0 0]\n [0 0]\n [0 0]\n [1 1]\n [1 0]\n [1 1]\n [1 1]\n [1 0]\n [1 0]\n [1 1]\n [0 1]\n [1 1]\n [1 1]\n [0 0]\n [1 1]\n [0 1]\n [0 1]\n [0 1]\n [1 1]\n [0 0]\n [0 0]\n [0 0]\n [0 0]\n [1 1]\n [1 1]\n [1 0]\n [1 1]\n [0 0]\n [0 0]\n [0 0]\n [1 0]\n [1 0]\n [1 0]\n [0 0]\n [1 1]\n [1 1]\n [1 1]\n [1 1]\n [0 0]\n [0 0]\n [0 1]\n [0 1]\n [0 0]\n [1 1]\n [0 0]\n [0 0]\n [0 0]\n [1 0]\n [1 1]\n [0 0]\n [1 1]\n [0 1]\n [0 1]\n [0 0]\n [1 1]\n [1 1]\n [0 1]\n [1 1]\n [0 0]\n [1 0]\n [1 1]\n [1 1]\n [0 0]\n [1 1]\n [0 0]\n [1 1]\n [1 1]\n [0 0]\n [1 1]\n [1 1]\n [1 0]\n [0 0]\n [1 1]\n [1 0]\n [0 0]\n [1 1]\n [0 0]\n [0 0]\n [0 0]\n [0 1]\n [0 0]\n [0 1]\n [0 1]\n [1 0]\n [0 1]\n [1 1]\n [1 1]\n [1 0]\n [1 1]\n [0 0]\n [1 1]\n [1 1]\n [0 0]\n [0 1]\n [0 1]\n [1 1]\n [0 0]\n [1 0]\n [0 1]\n [0 0]\n [1 1]\n [1 1]\n [1 1]\n [1 1]\n [1 1]\n [0 0]\n [1 1]\n [0 0]\n [0 0]\n [0 0]\n [1 1]\n [0 0]\n [0 0]\n [0 1]\n [0 0]\n [1 1]\n [0 0]\n [0 0]\n [1 1]\n [1 1]\n [1 1]\n [1 1]\n [1 1]\n [0 0]\n [0 1]\n [1 1]\n [0 1]\n [0 0]\n [0 0]\n [0 0]\n [0 0]\n [0 1]\n [0 1]\n [1 1]\n [0 1]\n [1 1]\n [1 1]\n [1 1]\n [1 0]\n [0 0]\n [1 1]\n [1 1]\n [1 1]\n [0 0]\n [0 0]\n [0 0]\n [1 1]\n [1 1]\n [1 0]\n [0 0]\n [0 0]\n [0 0]\n [0 0]\n [0 1]\n [0 0]\n [1 1]\n [1 1]\n [0 0]\n [0 0]\n [0 1]\n [0 0]\n [1 1]\n [0 0]\n [0 1]\n [1 1]\n [1 0]\n [0 0]\n [0 0]\n [0 0]\n [1 1]\n [0 0]\n [1 1]\n [0 0]\n [1 1]\n [1 1]\n [0 0]\n [0 0]\n [0 0]\n [1 1]\n [0 0]\n [1 1]\n [1 1]\n [0 0]\n [1 1]]\n"
]
],
[
[
"## Making the Confusion Matrix",
"_____no_output_____"
]
],
[
[
"from sklearn.metrics import confusion_matrix, accuracy_score, precision_score, recall_score, f1_score\ncm = confusion_matrix(y_test, y_pred)\nprint(cm)\nprint('Accuracy: {0:.2g}'.format(accuracy_score(y_test, y_pred)))\nprint('Precision: {0:.2g}'.format(precision_score(y_test, y_pred)))\nprint('Recall: {0:.2g}'.format(recall_score(y_test, y_pred)))\nprint('F1 Score: {0:.2g}'.format(f1_score(y_test, y_pred)))",
"[[79 18]\n [25 78]]\nAccuracy: 0.79\nPrecision: 0.81\nRecall: 0.76\nF1 Score: 0.78\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb8c2d3d0995ce1879b9dc733015fe84fee60678
| 132,539 |
ipynb
|
Jupyter Notebook
|
Code/analyze_large_shock.ipynb
|
MichiKal/healthcare-resilience
|
ad67db3f9ff9af52e8d80a52f15bd2a9147ed155
|
[
"MIT"
] | null | null | null |
Code/analyze_large_shock.ipynb
|
MichiKal/healthcare-resilience
|
ad67db3f9ff9af52e8d80a52f15bd2a9147ed155
|
[
"MIT"
] | null | null | null |
Code/analyze_large_shock.ipynb
|
MichiKal/healthcare-resilience
|
ad67db3f9ff9af52e8d80a52f15bd2a9147ed155
|
[
"MIT"
] | null | null | null | 232.117338 | 29,068 | 0.880133 |
[
[
[
"import pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nfrom os.path import join\nimport seaborn as sns",
"_____no_output_____"
]
],
[
[
"# Build results tables",
"_____no_output_____"
]
],
[
[
"N_patients = {\n \"AM\":14646484, \n \"AU\":973941,\n \"CH\":521211,\n \"DER\":771281,\n \"GGH\":748889,\n \"HNO\":564501,\n \"IM\":1693363,\n \"KI\":743235,\n \"NEU\":212302,\n \"ORTR\":1518719,\n \"PSY\":170016,\n \"RAD\":1593710,\n \"URO\":394209\n}\n\nspecialty_map = {\n \"AM\":\"GP\", \n \"AU\":\"OPH\",\n \"CH\":\"SRG\",\n \"DER\":\"DER\",\n \"GGH\":\"OBGYN\",\n \"HNO\":\"ENT\",\n \"IM\":\"IM\",\n \"KI\":\"PED\",\n \"NEU\":\"NEU\",\n \"ORTR\":\"ORTH\",\n \"PSY\":\"PSY\",\n \"RAD\":\"RAD\",\n \"URO\":\"OPT\"\n}",
"_____no_output_____"
],
[
"fname = \"states_doc_info_total_hour-based_quarterly.csv\"\nN_patients = pd.read_csv(join(src, fname))",
"_____no_output_____"
],
[
"N_patients",
"_____no_output_____"
],
[
"src = \"results\"\nfname = \"states_doc_info_total_hour-based_quarterly.csv\"\nN_patients = pd.read_csv(join(src, fname))\nfname = \"searching_pats_{}_iter10_shocksize{}.csv\"\nspecialties = [\"AM\", \"AU\", \"CH\", \"DER\", \"GGH\", \"HNO\", \"IM\", \"KI\",\n \"NEU\", \"ORTR\", \"PSY\", \"RAD\", \"URO\"]\nshocksizes = [7, 10, 15, 20]\n\nresults = pd.DataFrame()\nfor shocksize in shocksizes:\n for spec in specialties:\n df = pd.read_csv(join(src, fname.format(spec, shocksize)),\n names=[f\"step_{i}\" for i in range(1, 11)], header=None)\n for col in df.columns:\n df[col] = df[col] / N_patients[f\"{spec}_total\"].sum() * 100\n df[\"run\"] = range(1, 11)\n df[\"specialty\"] = specialty_map[spec]\n df[\"shocksize\"] = shocksize\n \n cols = [\"step_1\", \"step_10\", \"run\", \"specialty\", \"shocksize\"]\n results = pd.concat([results, df[cols]])\nresults = results.reset_index(drop=True)",
"_____no_output_____"
],
[
"ylims = {7:[5, 0.5], 10:[8, 1], 15:[10, 2], 20:[14, 3]}\nyticks_t1 = {\n 7:[1, 2, 3, 4, 5], \n 10:[0, 2, 4, 6, 8], \n 15:[0, 2, 4, 6, 8, 10, 12], \n 20:[0, 2, 4, 6, 8, 10, 12, 14, 16]}\nyticks_t10 = {\n 7:[0, 0.1, 0.2, 0.3, 0.4, 0.5],\n 10:[0, 0.2, 0.4, 0.6, 0.8, 1.0, 1.2], \n 15:[0., 0.5, 1.0, 1.5, 2],\n 20:[0, 1, 2, 3, 4]\n}\nylabels = {1: \"% searching patients\",\n 10: \"% lost patients\"}\n\nfor shock_size in [7, 10, 15, 20]:\n df = results[results[\"shocksize\"] == shock_size]\n fig, axes = plt.subplots(1, 2, figsize=(14, 4))\n cmap = plt.get_cmap(\"RdYlGn_r\")\n \n for i, step in enumerate([1, 10]):\n ax = axes[i]\n agg = df[[\"specialty\", f\"step_{step}\"]]\\\n .groupby(\"specialty\")\\\n .agg(\"median\")\\\n .sort_values(by=f\"step_{step}\")\n order = agg.index\n max_val = agg[f\"step_{step}\"][-1]\n palette = [cmap(i) for i in agg[f\"step_{step}\"] / max_val]\n \n sns.boxplot(\n ax=ax,\n x=\"specialty\",\n y=f\"step_{step}\",\n data=df,\n order=order,\n palette=palette\n )\n ax.spines['right'].set_visible(False)\n ax.spines['top'].set_visible(False)\n ax.set_ylabel(ylabels[step], fontsize=16)\n ax.set_xlabel(\"\")\n ax.set_title(f\"displacement step {step}\", fontsize=16)\n ax.set_xticks(range(len(labels)))\n ax.set_xticklabels(labels, fontsize=11)\n\n axes[0].set_ylim(0, ylims[shock_size][0])\n axes[0].set_yticks(yticks_t1[shock_size])\n axes[0].set_yticklabels(yticks_t1[shock_size], fontsize=12)\n axes[1].set_ylim(0, ylims[shock_size][1])\n axes[1].set_yticks(yticks_t10[shock_size])\n axes[1].set_yticklabels(yticks_t10[shock_size], fontsize=12)\n fig.tight_layout()\n plt.savefig(f\"figures/shock_results_{shock_size}.svg\")",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code"
] |
[
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
cb8c3e702b2c24565646fda1c591978e37393c3a
| 60,760 |
ipynb
|
Jupyter Notebook
|
mission_to_mars.ipynb
|
markrotil/web-scraping-challenge
|
6fc026275bf2d91913de7fe07ab85402cad5e59a
|
[
"ADSL"
] | null | null | null |
mission_to_mars.ipynb
|
markrotil/web-scraping-challenge
|
6fc026275bf2d91913de7fe07ab85402cad5e59a
|
[
"ADSL"
] | null | null | null |
mission_to_mars.ipynb
|
markrotil/web-scraping-challenge
|
6fc026275bf2d91913de7fe07ab85402cad5e59a
|
[
"ADSL"
] | null | null | null | 52.469775 | 9,588 | 0.502156 |
[
[
[
"# Import Dependencies\n\nimport pandas as pd\nfrom bs4 import BeautifulSoup as bs\nimport requests\nfrom splinter import Browser\nfrom splinter.exceptions import ElementDoesNotExist\nfrom IPython.display import HTML\n#browser = Browser()",
"_____no_output_____"
],
[
"# Create a path to use for splinter\nexecutable_path = {'executable_path' : 'chromedriver.exe'}\nbrowser = Browser('chrome', **executable_path, headless=False)",
"_____no_output_____"
],
[
"# Create shortcut for URL of main page\nurl = 'https://mars.nasa.gov/news/'",
"_____no_output_____"
],
[
"# Get response of web page\nresponse = requests.get(url)",
"_____no_output_____"
],
[
"# html parser with beautiful soup\nsoup = bs(response.text, 'html.parser')\n\n# check to see if it parses\nprint(soup.prettify())\n\n",
"<!DOCTYPE html>\n<html lang=\"en\" xml:lang=\"en\" xmlns=\"http://www.w3.org/1999/xhtml\">\n <head>\n <meta content=\"text/html; charset=utf-8\" http-equiv=\"Content-Type\"/>\n <!-- Always force latest IE rendering engine or request Chrome Frame -->\n <meta content=\"IE=edge,chrome=1\" http-equiv=\"X-UA-Compatible\"/>\n <!-- Responsiveness -->\n <meta content=\"width=device-width, initial-scale=1.0\" name=\"viewport\"/>\n <!-- Favicon -->\n <link href=\"/apple-touch-icon.png\" rel=\"apple-touch-icon\" sizes=\"180x180\"/>\n <link href=\"/favicon-32x32.png\" rel=\"icon\" sizes=\"32x32\" type=\"image/png\"/>\n <link href=\"/favicon-16x16.png\" rel=\"icon\" sizes=\"16x16\" type=\"image/png\"/>\n <link href=\"/manifest.json\" rel=\"manifest\"/>\n <link color=\"#e48b55\" href=\"/safari-pinned-tab.svg\" rel=\"mask-icon\"/>\n <meta content=\"#000000\" name=\"theme-color\"/>\n <meta content=\"authenticity_token\" name=\"csrf-param\">\n <meta content=\"S2BvoaJX2aI00Kq/ixYR3movbv2n2V0TO7fGe8cZDTtemhTQuluj5IQ+5VhpqvdflS/sRekX7gTZvu6o19o4iA==\" name=\"csrf-token\">\n <title>\n News – NASA’s Mars Exploration Program\n </title>\n <meta content=\"NASA’s Mars Exploration Program \" property=\"og:site_name\"/>\n <meta content=\"mars.nasa.gov\" name=\"author\"/>\n <meta content=\"Mars, missions, NASA, rover, Curiosity, Opportunity, InSight, Mars Reconnaissance Orbiter, facts\" name=\"keywords\"/>\n <meta content=\"NASA’s real-time portal for Mars exploration, featuring the latest news, images, and discoveries from the Red Planet.\" name=\"description\"/>\n <meta content=\"NASA’s real-time portal for Mars exploration, featuring the latest news, images, and discoveries from the Red Planet.\" property=\"og:description\"/>\n <meta content=\"News – NASA’s Mars Exploration Program \" property=\"og:title\"/>\n <meta content=\"https://mars.nasa.gov/news\" property=\"og:url\"/>\n <meta content=\"article\" property=\"og:type\"/>\n <meta content=\"2017-09-22 19:53:22 UTC\" property=\"og:updated_time\"/>\n <meta content=\"https://mars.nasa.gov/system/site_config_values/meta_share_images/1_mars-nasa-gov.jpg\" property=\"og:image\"/>\n <meta content=\"https://mars.nasa.gov/system/site_config_values/meta_share_images/1_mars-nasa-gov.jpg\" name=\"twitter:image\"/>\n <link href=\"https://mars.nasa.gov/system/site_config_values/meta_share_images/1_mars-nasa-gov.jpg\" rel=\"image_src\"/>\n <meta content=\"195570401081308\" property=\"fb:app_id\"/>\n <link href=\"https://fonts.googleapis.com/css?family=Montserrat:200,300,400,500,600,700|Raleway:300,400\" rel=\"stylesheet\"/>\n <link href=\"/assets/public_manifest-552f2e86bf99497d3f90f135feca40db5511b30599898647e0506f50582dc94c.css\" media=\"all\" rel=\"stylesheet\">\n <link href=\"/assets/mbcms/vendor/jquery.fancybox3-d5d81bdfc05a59e4ea72bca1d8b7fcc399bd3b61f7c06af95a8a48795df69d7a.css\" media=\"screen\" rel=\"stylesheet\">\n <link href=\"/assets/gulp/print-240f8bfaa7f6402dfd6c49ee3c1ffea57a89ddd4c8c90e2f2a5c7d63c5753e32.css\" media=\"print\" rel=\"stylesheet\">\n <script src=\"/assets/public_manifest-63a5b4071bf0b92dd1f67e47c49ab61a6fc0053457345d4c132ae36b10bde324.js\">\n </script>\n <script src=\"/assets/mbcms/vendor/jquery.fancybox3-bd48876205805faa43a79e74b656191a4ad37809923b4f3247b571ba82d4458c.js\">\n </script>\n <script src=\"/assets/mb_manifest-a0ae601bc18c852649e350709ab440161da58529f782ae84172c21f8ea27b714.js\">\n </script>\n <!--[if gt IE 8]><!-->\n <script src=\"/assets/not_ie8_manifest.js\">\n </script>\n <!--[if !IE]>-->\n <script src=\"/assets/not_ie8_manifest.js\">\n </script>\n <!--<![endif]-->\n <!-- /twitter cards -->\n <meta content=\"summary_large_image\" name=\"twitter:card\"/>\n <meta content=\"News \" name=\"twitter:title\"/>\n <meta content=\"NASA’s real-time portal for Mars exploration, featuring the latest news, images, and discoveries from the Red Planet.\" name=\"twitter:description\"/>\n <meta content=\"https://mars.nasa.gov/system/site_config_values/meta_share_images/1_mars-nasa-gov.jpg\" name=\"twitter:image\"/>\n </link>\n </link>\n </link>\n </meta>\n </meta>\n </head>\n <body id=\"news\">\n <svg display=\"none\" height=\"0\" width=\"0\">\n <symbol height=\"30\" id=\"circle_plus\" viewbox=\"0 0 30 30\" width=\"30\">\n <g fill-rule=\"evenodd\" transform=\"translate(1 1)\">\n <circle cx=\"14\" cy=\"14\" fill=\"#fff\" fill-opacity=\".1\" fill-rule=\"nonzero\" r=\"14\" stroke=\"inherit\" stroke-width=\"1\">\n </circle>\n <path class=\"the_plus\" d=\"m18.856 12.96v1.738h-4.004v3.938h-1.848v-3.938h-4.004v-1.738h4.004v-3.96h1.848v3.96z\" fill=\"inherit\" stroke-width=\"0\">\n </path>\n </g>\n </symbol>\n <symbol height=\"30\" id=\"circle_arrow\" viewbox=\"0 0 30 30\" width=\"30\" xmlns=\"http://www.w3.org/2000/svg\">\n <g transform=\"translate(1 1)\">\n <circle cx=\"14\" cy=\"14\" fill=\"#fff\" fill-opacity=\".1\" r=\"14\" stroke=\"inherit\" stroke-width=\"1\">\n </circle>\n <path class=\"the_arrow\" d=\"m8.5 15.00025h7.984l-2.342 2.42c-.189.197-.189.518 0 .715l.684.717c.188.197.494.197.684 0l4.35-4.506c.188-.199.188-.52 0-.717l-4.322-4.48c-.189-.199-.496-.199-.684 0l-.684.716c-.189.197-.189.519 0 .716l2.341 2.419h-8.011c-.276 0-.5.223-.5.5v1c0 .275.224.5.5.5z\" fill=\"inherit\" stroke-width=\"0\">\n </path>\n </g>\n </symbol>\n <symbol height=\"30\" id=\"circle_close\" viewbox=\"0 0 30 30\" width=\"30\">\n <g fill-rule=\"evenodd\" transform=\"translate(1 1)\">\n <circle cx=\"14\" cy=\"14\" fill=\"blue\" fill-opacity=\"1\" fill-rule=\"nonzero\" r=\"14\" stroke=\"inherit\" stroke-width=\"1\">\n </circle>\n <path class=\"the_plus\" d=\"m18.856 12.96v1.738h-4.004v3.938h-1.848v-3.938h-4.004v-1.738h4.004v-3.96h1.848v3.96z\" fill=\"inherit\" stroke-width=\"0\">\n </path>\n </g>\n </symbol>\n <symbol height=\"30\" id=\"circle_close_hover\" viewbox=\"0 0 30 30\" width=\"30\">\n <g fill-rule=\"evenodd\" transform=\"translate(1 1)\">\n <circle cx=\"14\" cy=\"14\" fill=\"white\" fill-opacity=\"1\" fill-rule=\"nonzero\" r=\"14\" stroke=\"inherit\" stroke-width=\"1\">\n </circle>\n <path class=\"the_plus\" d=\"m18.856 12.96v1.738h-4.004v3.938h-1.848v-3.938h-4.004v-1.738h4.004v-3.96h1.848v3.96z\" fill=\"inherit\" stroke-width=\"0\">\n </path>\n </g>\n </symbol>\n <symbol height=\"6\" id=\"chevron_down\" viewbox=\"0 0 10 6\" width=\"10\" xmlns=\"http://www.w3.org/2000/svg\">\n <path d=\"m59 7v2.72727273l5 3.27272727 5-3.27272727v-2.72727273l-5 3.2727273z\" transform=\"translate(-59 -7)\">\n </path>\n </symbol>\n <symbol height=\"16\" id=\"gear\" viewbox=\"0 0 16 16\" width=\"16\" xmlns=\"http://www.w3.org/2000/svg\">\n <path d=\"m68 9h-1.09c-.15-.91-.5-1.75-1.021-2.471l.761-.77c.39-.39.39-1.029 0-1.42-.391-.39-1.021-.39-1.41 0l-.771.77c-.719-.519-1.469-.869-2.469-1.019v-1.09c0-.55-.45-1-1-1s-1 .45-1 1v1.09c-1 .15-1.75.5-2.47 1.02l-.77-.77c-.389-.39-1.029-.39-1.42 0-.39.391-.39 1.03 0 1.42l.771.77c-.521.72-.871 1.56-1.021 2.47h-1.09c-.55 0-1 .48-1 1.029 0 .551.45.971 1.12.971h.97c.15.91.5 1.75 1.021 2.471l-.771.769c-.39.39-.39 1.029 0 1.42.391.39 1.021.39 1.41 0l.78-.77c.72.52 1.47.87 2.47 1.02v1.09c0 .55.45 1 1 1s1-.45 1-1v-1.09c1-.15 1.75-.5 2.47-1.02l.771.77c.391.39 1.02.39 1.41 0 .39-.391.39-1.03 0-1.42l-.761-.76c.51-.72.87-1.56 1.02-2.48h1.09c.55 0 1-.45 1-1s-.45-1-1-1zm-7 4c-1.66 0-3-1.35-3-3s1.34-3 3-3c1.65 0 3 1.35 3 3s-1.35 3-3 3z\" fill=\"#a79693\" transform=\"translate(-53 -2)\">\n </path>\n </symbol>\n </svg>\n <div data-react-cache-id=\"BrowseHappier-0\" data-react-class=\"BrowseHappier\" data-react-props='{\"gt\":1,\"lt\":11}'>\n </div>\n <div data-react-cache-id=\"HiPO-0\" data-react-class=\"HiPO\" data-react-props=\"{}\">\n </div>\n <div id=\"main_container\">\n <div id=\"site_body\">\n <div class=\"site_header_area\">\n <header class=\"site_header\">\n <div class=\"brand_area\">\n <div class=\"brand1\">\n <a class=\"nasa_logo\" href=\"http://www.nasa.gov\" target=\"_blank\" title=\"visit nasa.gov\">\n NASA\n </a>\n </div>\n <div class=\"brand2\">\n <a class=\"top_logo\" href=\"https://science.nasa.gov/\" target=\"_blank\" title=\"Explore NASA Science\">\n NASA Science\n </a>\n <a class=\"sub_logo\" href=\"/mars-exploration/#\" title=\"Mars\">\n Mars Exploration Program\n </a>\n </div>\n <img alt=\"\" class=\"print_only print_logo\" src=\"/assets/[email protected]\"/>\n </div>\n <a class=\"visuallyhidden focusable\" href=\"#page\">\n Skip Navigation\n </a>\n <div class=\"right_header_container\">\n <a class=\"menu_button\" href=\"javascript:void(0);\" id=\"menu_button\">\n <span class=\"menu_icon\">\n menu\n </span>\n </a>\n <a class=\"modal_close\" id=\"modal_close\">\n <span class=\"modal_close_icon\">\n </span>\n </a>\n <div class=\"nav_area\">\n <div id=\"site_nav_container\">\n <nav class=\"site_nav\" data-react-cache-id=\"Meganav-0\" data-react-class=\"Meganav\" data-react-props=\"{"nav_items":[{"name":"Mars Now","style":"icon","li_class":"nav_icon mars_now","target":"_self","link":"/explore/mars-now","svg_icon_id":"nav_icon","id":261,"features":[{"title":"Mars Now","body":"View a 3D visualization of all the missions exploring the Red Planet","image_src":"/system/basic_html_elements/225_mars_now_nav.jpg","link":"/explore/mars-now/","target":"_self","categories":[]}],"title":"","short_description":"View the current location and spacecraft communications activity of operating landers, rovers and orbiters using the NASA’s Mars Relay Network."},{"name":"The Red Planet","link":"/#red_planet","target":"_self","sections":[{"items":[{"name":"Dashboard","link":"/#red_planet/0","target":"_self","id":9},{"name":"Science Goals","link":"/#red_planet/1","target":"_self","id":13},{"name":"The Planet","link":"/#red_planet/2","target":"_self","id":14},{"name":"Atmosphere","link":"/#red_planet/3","target":"_self","id":16},{"name":"Astrobiology","link":"/#red_planet/4","target":"_self","id":17},{"name":"Past, Present, Future, Timeline","link":"/#red_planet/5","target":"_self","id":18}]}],"id":3,"meganav_style":"","features":[],"short_description":null},{"name":"The Program","link":"/#mars_exploration_program","target":"_self","sections":[{"items":[{"name":"Mission Statement","link":"/#mars_exploration_program/0","target":"_self","id":8},{"name":"About the Program","link":"/#mars_exploration_program/1","target":"_self","id":42},{"name":"Organization","link":"/#mars_exploration_program/2","target":"_self","id":43},{"name":"Why Mars?","link":"/#mars_exploration_program/3","target":"_self","id":51},{"name":"Research Programs","link":"/#mars_exploration_program/4","target":"_self","id":44},{"name":"Planetary Resources","link":"/#mars_exploration_program/5","target":"_self","id":52},{"name":"Technologies","link":"/#mars_exploration_program/6","target":"_self","id":56}]}],"id":2,"meganav_style":"","features":[],"short_description":null},{"name":"News \\u0026 Events","link":"/#news_and_events","target":"_self","sections":[{"items":[{"name":"News","link":"/news","target":"_self","id":92},{"name":"Events","link":"/events","target":"_self","id":93}]}],"id":4,"meganav_style":"","features":[],"short_description":null},{"name":"Multimedia","link":"/#multimedia","target":"_self","sections":[{"items":[{"name":"Images","link":"/multimedia/images/","target":"_self","id":90},{"name":"Videos","link":"/multimedia/videos/","target":"_self","id":91},{"name":"More Resources","link":"/multimedia/more-resources/","target":"_self","id":413}]}],"id":5,"meganav_style":"","features":[],"short_description":null},{"name":"Missions","link":"/#missions_gallery_subnav","target":"_self","sections":[{"items":[{"name":"Past","link":"/mars-exploration/missions/?category=167","target":"_self","id":38},{"name":"Present","link":"/mars-exploration/missions/?category=170","target":"_self","id":59},{"name":"Future","link":"/mars-exploration/missions/?category=171","target":"_self","id":60},{"name":"International Partners","link":"/mars-exploration/partners","target":"_self","id":40}]}],"id":6,"meganav_style":"","features":[],"short_description":null},{"name":"More","link":"/#more","target":"_self","sections":[],"id":7,"meganav_style":"","features":[],"short_description":null}],"gallery_subnav_items":[{"thumb":"/system/missions/list_view_images/23_PIA23764-RoverNamePlateonMars-320x240.jpg","id":23,"title":"Mars 2020 Perseverance Rover","description":"A mission to investigate key questions about potential life on Mars. ","date":"July 17, 2020","url":"/mars-exploration/missions/mars2020/","link_text":"","target":"_blank","mi_traveled":null,"gallery_subnav_link":"https://mars.nasa.gov/mars2020/"},{"thumb":"/system/missions/list_view_images/2_PIA14175-thmfeat.jpg","id":2,"title":"Curiosity Rover","description":"The largest and most capable rover ever sent to Mars.","date":"November 26, 2011","url":"/mars-exploration/missions/mars-science-laboratory","link_text":"","target":"_blank","mi_traveled":14.79,"gallery_subnav_link":"https://mars.nasa.gov/msl/home/"},{"thumb":"/system/missions/list_view_images/21_PIA22743-320x240.jpg","id":21,"title":"InSight Lander","description":"A mission to study the deep interior of Mars. ","date":"November 26, 2018","url":"/mars-exploration/missions/insight/","link_text":"","target":"_blank","mi_traveled":null,"gallery_subnav_link":"https://mars.nasa.gov/insight/"},{"thumb":"/system/missions/list_view_images/6_maven_320x240.jpg","id":6,"title":"MAVEN","description":"Measures Mars' atmosphere to understand its climate change.","date":"November 18, 2013","url":"/mars-exploration/missions/maven","link_text":"","target":"_blank","mi_traveled":null,"gallery_subnav_link":"https://mars.nasa.gov/maven/"},{"thumb":"/system/missions/list_view_images/8_MRO_320x240.jpg","id":8,"title":"Mars Reconnaissance Orbiter","description":"Takes high-resolution imagery of Martian terrain with extraordinary clarity. ","date":"August 12, 2012","url":"/mars-exploration/missions/mars-reconnaissance-orbiter","link_text":"","target":"_blank","mi_traveled":null,"gallery_subnav_link":"https://mars.nasa.gov/mro/"},{"thumb":"/system/missions/list_view_images/5_mars_odyssey320x240.jpg","id":5,"title":"2001 Mars Odyssey","description":"NASA's longest-lasting spacecraft at Mars. ","date":"April 7, 2001","url":"/mars-exploration/missions/odyssey","link_text":"","target":"_blank","mi_traveled":null,"gallery_subnav_link":"https://mars.nasa.gov/odyssey/"}],"search":true,"search_placeholder":{"placeholder":""},"highlight_current":{"highlight":true,"current_id":83,"parent_ids":[]},"search_submit":"/search/"}\">\n </nav>\n </div>\n </div>\n </div>\n </header>\n </div>\n <div id=\"sticky_nav_spacer\">\n </div>\n <div id=\"page\">\n <div class=\"page_cover\">\n </div>\n <!-- title to go in the page_header -->\n <div class=\"header_mask\">\n <section class=\"content_page module\">\n </section>\n </div>\n <div class=\"grid_list_page module content_page\">\n <div class=\"grid_layout\">\n <article>\n <header id=\"page_header\">\n </header>\n <div class=\"react_grid_list grid_list_container\" data-react-cache-id=\"GridListPage-0\" data-react-class=\"GridListPage\" data-react-props='{\"left_column\":false,\"class_name\":\"\",\"default_view\":\"list_view\",\"model\":\"news_items\",\"view_toggle\":false,\"search\":\"true\",\"list_item\":\"News\",\"title\":\"News\",\"categories\":[\"19,165,184,204\"],\"order\":\"publish_date desc,created_at desc\",\"no_items_text\":\"There are no items matching these criteria.\",\"site_title\":\"NASA’s Mars Exploration Program \",\"short_title\":\"Mars\",\"site_share_image\":\"/system/site_config_values/meta_share_images/1_mars-nasa-gov.jpg\",\"per_page\":null,\"filters\":\"[ [ \\\"date\\\", [ [ \\\"2020\\\", \\\"2020\\\" ], [ \\\"2019\\\", \\\"2019\\\" ], [ \\\"2018\\\", \\\"2018\\\" ], [ \\\"2017\\\", \\\"2017\\\" ], [ \\\"2016\\\", \\\"2016\\\" ], [ \\\"2015\\\", \\\"2015\\\" ], [ \\\"2014\\\", \\\"2014\\\" ], [ \\\"2013\\\", \\\"2013\\\" ], [ \\\"2012\\\", \\\"2012\\\" ], [ \\\"2011\\\", \\\"2011\\\" ], [ \\\"2010\\\", \\\"2010\\\" ], [ \\\"2009\\\", \\\"2009\\\" ], [ \\\"2008\\\", \\\"2008\\\" ], [ \\\"2007\\\", \\\"2007\\\" ], [ \\\"2006\\\", \\\"2006\\\" ], [ \\\"2005\\\", \\\"2005\\\" ], [ \\\"2004\\\", \\\"2004\\\" ], [ \\\"2003\\\", \\\"2003\\\" ], [ \\\"2002\\\", \\\"2002\\\" ], [ \\\"2001\\\", \\\"2001\\\" ], [ \\\"2000\\\", \\\"2000\\\" ] ], [ \\\"Latest\\\", \\\"\\\" ], false, false ], [ \\\"categories\\\", [ [ \\\"Feature Stories\\\", 165 ], [ \\\"Press Releases\\\", 19 ], [ \\\"Spotlights\\\", 184 ], [ \\\"Status Reports\\\", 204 ] ], [ \\\"All Categories\\\", \\\"\\\" ], false, false ] ]\",\"conditions\":null,\"scope_in_title\":true,\"options\":{\"blank_scope\":\"Latest\"},\"results_in_title\":false}'>\n </div>\n </article>\n </div>\n </div>\n <section class=\"module suggested_features\">\n <div class=\"grid_layout\">\n <header>\n <h2 class=\"module_title\">\n You Might Also Like\n </h2>\n </header>\n <section>\n <script>\n $(document).ready(function(){\n $(\".features\").slick({\n dots: false,\n infinite: true,\n speed: 300,\n slide: '.features .slide',\n slidesToShow: 3,\n slidesToScroll: 3,\n lazyLoad: 'ondemand',\n centerMode: false,\n arrows: true,\n appendArrows: '.features .slick-nav',\n appendDots: \".features .slick-nav\",\n responsive: [{\"breakpoint\":953,\"settings\":{\"slidesToShow\":2,\"slidesToScroll\":2,\"centerMode\":false}},{\"breakpoint\":480,\"settings\":{\"slidesToShow\":1,\"slidesToScroll\":1,\"centerMode\":true,\"arrows\":false,\"centerPadding\":\"25px\"}}]\n });\n });\n </script>\n <div class=\"features\">\n <div class=\"slide\">\n <div class=\"image_and_description_container\">\n <a href=\"/news/8716/nasa-to-broadcast-mars-2020-perseverance-launch-prelaunch-activities/\">\n <div class=\"rollover_description\">\n <div class=\"rollover_description_inner\">\n Starting July 27, news activities will cover everything from mission engineering and science to returning samples from Mars to, of course, the launch itself.\n </div>\n <div class=\"overlay_arrow\">\n <img alt=\"More\" src=\"/assets/overlay-arrow.png\"/>\n </div>\n </div>\n <img alt=\"NASA to Broadcast Mars 2020 Perseverance Launch, Prelaunch Activities\" class=\"img-lazy\" data-lazy=\"/system/news_items/list_view_images/8716_PIA23499-320x240.jpg\" src=\"/assets/loading_320x240.png\"/>\n </a>\n </div>\n <div class=\"content_title\">\n <a href=\"/news/8716/nasa-to-broadcast-mars-2020-perseverance-launch-prelaunch-activities/\">\n NASA to Broadcast Mars 2020 Perseverance Launch, Prelaunch Activities\n </a>\n </div>\n </div>\n <div class=\"slide\">\n <div class=\"image_and_description_container\">\n <a href=\"/news/8695/the-launch-is-approaching-for-nasas-next-mars-rover-perseverance/\">\n <div class=\"rollover_description\">\n <div class=\"rollover_description_inner\">\n The Red Planet's surface has been visited by eight NASA spacecraft. The ninth will be the first that includes a roundtrip ticket in its flight plan.\n </div>\n <div class=\"overlay_arrow\">\n <img alt=\"More\" src=\"/assets/overlay-arrow.png\"/>\n </div>\n </div>\n <img alt=\"The Launch Is Approaching for NASA's Next Mars Rover, Perseverance\" class=\"img-lazy\" data-lazy=\"/system/news_items/list_view_images/8695_24732_PIA23499-226.jpg\" src=\"/assets/loading_320x240.png\"/>\n </a>\n </div>\n <div class=\"content_title\">\n <a href=\"/news/8695/the-launch-is-approaching-for-nasas-next-mars-rover-perseverance/\">\n The Launch Is Approaching for NASA's Next Mars Rover, Perseverance\n </a>\n </div>\n </div>\n <div class=\"slide\">\n <div class=\"image_and_description_container\">\n <a href=\"/news/8692/nasa-to-hold-mars-2020-perseverance-rover-launch-briefing/\">\n <div class=\"rollover_description\">\n <div class=\"rollover_description_inner\">\n Learn more about the agency's next Red Planet mission during a live event on June 17.\n </div>\n <div class=\"overlay_arrow\">\n <img alt=\"More\" src=\"/assets/overlay-arrow.png\"/>\n </div>\n </div>\n <img alt=\"NASA to Hold Mars 2020 Perseverance Rover Launch Briefing\" class=\"img-lazy\" data-lazy=\"/system/news_items/list_view_images/8692_PIA23920-320x240.jpg\" src=\"/assets/loading_320x240.png\"/>\n </a>\n </div>\n <div class=\"content_title\">\n <a href=\"/news/8692/nasa-to-hold-mars-2020-perseverance-rover-launch-briefing/\">\n NASA to Hold Mars 2020 Perseverance Rover Launch Briefing\n </a>\n </div>\n </div>\n <div class=\"slide\">\n <div class=\"image_and_description_container\">\n <a href=\"/news/8659/alabama-high-school-student-names-nasas-mars-helicopter/\">\n <div class=\"rollover_description\">\n <div class=\"rollover_description_inner\">\n Vaneeza Rupani's essay was chosen as the name for the small spacecraft, which will mark NASA's first attempt at powered flight on another planet.\n </div>\n <div class=\"overlay_arrow\">\n <img alt=\"More\" src=\"/assets/overlay-arrow.png\"/>\n </div>\n </div>\n <img alt=\"Alabama High School Student Names NASA's Mars Helicopter\" class=\"img-lazy\" data-lazy=\"/system/news_items/list_view_images/8659_1-PIA23883-MAIN-320x240.jpg\" src=\"/assets/loading_320x240.png\"/>\n </a>\n </div>\n <div class=\"content_title\">\n <a href=\"/news/8659/alabama-high-school-student-names-nasas-mars-helicopter/\">\n Alabama High School Student Names NASA's Mars Helicopter\n </a>\n </div>\n </div>\n <div class=\"slide\">\n <div class=\"image_and_description_container\">\n <a href=\"/news/8645/mars-helicopter-attached-to-nasas-perseverance-rover/\">\n <div class=\"rollover_description\">\n <div class=\"rollover_description_inner\">\n The team also fueled the rover's sky crane to get ready for this summer's history-making launch.\n </div>\n <div class=\"overlay_arrow\">\n <img alt=\"More\" src=\"/assets/overlay-arrow.png\"/>\n </div>\n </div>\n <img alt=\"Mars Helicopter Attached to NASA's Perseverance Rover\" class=\"img-lazy\" data-lazy=\"/system/news_items/list_view_images/8645_PIA23824-RoverWithHelicopter-32x24.jpg\" src=\"/assets/loading_320x240.png\"/>\n </a>\n </div>\n <div class=\"content_title\">\n <a href=\"/news/8645/mars-helicopter-attached-to-nasas-perseverance-rover/\">\n Mars Helicopter Attached to NASA's Perseverance Rover\n </a>\n </div>\n </div>\n <div class=\"slide\">\n <div class=\"image_and_description_container\">\n <a href=\"/news/8641/nasas-perseverance-mars-rover-gets-its-wheels-and-air-brakes/\">\n <div class=\"rollover_description\">\n <div class=\"rollover_description_inner\">\n After the rover was shipped from JPL to Kennedy Space Center, the team is getting closer to finalizing the spacecraft for launch later this summer.\n </div>\n <div class=\"overlay_arrow\">\n <img alt=\"More\" src=\"/assets/overlay-arrow.png\"/>\n </div>\n </div>\n <img alt=\"NASA's Perseverance Mars Rover Gets Its Wheels and Air Brakes\" class=\"img-lazy\" data-lazy=\"/system/news_items/list_view_images/8641_PIA-23821-320x240.jpg\" src=\"/assets/loading_320x240.png\"/>\n </a>\n </div>\n <div class=\"content_title\">\n <a href=\"/news/8641/nasas-perseverance-mars-rover-gets-its-wheels-and-air-brakes/\">\n NASA's Perseverance Mars Rover Gets Its Wheels and Air Brakes\n </a>\n </div>\n </div>\n <div class=\"grid_layout\">\n <div class=\"slick-nav_container\">\n <div class=\"slick-nav\">\n </div>\n </div>\n </div>\n </div>\n </section>\n </div>\n </section>\n </div>\n <footer id=\"site_footer\">\n <div class=\"grid_layout\">\n <section class=\"upper_footer\">\n <div class=\"share_newsletter_container\">\n <div class=\"newsletter\">\n <h2>\n Get the Mars Newsletter\n </h2>\n <form action=\"/newsletter-subscribe\">\n <input id=\"email\" name=\"email\" placeholder=\"enter email address\" type=\"email\" value=\"\"/>\n <input data-disable-with=\"\" name=\"commit\" type=\"submit\" value=\"\"/>\n </form>\n </div>\n <div class=\"share\">\n <h2>\n Follow the Journey\n </h2>\n <div class=\"social_icons\">\n <!-- AddThis Button BEGIN -->\n <div class=\"addthis_toolbox addthis_default_style addthis_32x32_style\">\n <a addthis:userid=\"MarsCuriosity\" class=\"addthis_button_twitter_follow icon\">\n <img alt=\"twitter\" src=\"/assets/[email protected]\"/>\n </a>\n <a addthis:userid=\"MarsCuriosity\" class=\"addthis_button_facebook_follow icon\">\n <img alt=\"facebook\" src=\"/assets/[email protected]\"/>\n </a>\n <a addthis:userid=\"nasa\" class=\"addthis_button_instagram_follow icon\">\n <img alt=\"instagram\" src=\"/assets/[email protected]\"/>\n </a>\n <a addthis:url=\"https://mars.nasa.gov/rss/api/?feed=news&category=all&feedtype=rss\" class=\"addthis_button_rss_follow icon\">\n <img alt=\"rss\" src=\"/assets/[email protected]\"/>\n </a>\n </div>\n </div>\n <script src=\"//s7.addthis.com/js/300/addthis_widget.js#pubid=ra-5a690e4c1320e328\">\n </script>\n </div>\n </div>\n <div class=\"gradient_line\">\n </div>\n </section>\n <section class=\"sitemap\">\n <div class=\"sitemap_directory\" id=\"sitemap_directory\">\n <div class=\"sitemap_block\">\n <div class=\"footer_sitemap_item\">\n <h3 class=\"sitemap_title\">\n <a href=\"/#red_planet\">\n The Red Planet\n </a>\n </h3>\n <ul>\n <li>\n <div class=\"global_subnav_container\">\n <ul class=\"subnav\">\n <li>\n <a href=\"/#red_planet/0\" target=\"_self\">\n Dashboard\n </a>\n </li>\n <li>\n <a href=\"/#red_planet/1\" target=\"_self\">\n Science Goals\n </a>\n </li>\n <li>\n <a href=\"/#red_planet/2\" target=\"_self\">\n The Planet\n </a>\n </li>\n <li>\n <a href=\"/#red_planet/3\" target=\"_self\">\n Atmosphere\n </a>\n </li>\n <li>\n <a href=\"/#red_planet/4\" target=\"_self\">\n Astrobiology\n </a>\n </li>\n <li>\n <a href=\"/#red_planet/5\" target=\"_self\">\n Past, Present, Future, Timeline\n </a>\n </li>\n </ul>\n </div>\n </li>\n </ul>\n </div>\n </div>\n <div class=\"sitemap_block\">\n <div class=\"footer_sitemap_item\">\n <h3 class=\"sitemap_title\">\n <a href=\"/#mars_exploration_program\">\n The Program\n </a>\n </h3>\n <ul>\n <li>\n <div class=\"global_subnav_container\">\n <ul class=\"subnav\">\n <li>\n <a href=\"/#mars_exploration_program/0\" target=\"_self\">\n Mission Statement\n </a>\n </li>\n <li>\n <a href=\"/#mars_exploration_program/1\" target=\"_self\">\n About the Program\n </a>\n </li>\n <li>\n <a href=\"/#mars_exploration_program/2\" target=\"_self\">\n Organization\n </a>\n </li>\n <li>\n <a href=\"/#mars_exploration_program/3\" target=\"_self\">\n Why Mars?\n </a>\n </li>\n <li>\n <a href=\"/#mars_exploration_program/4\" target=\"_self\">\n Research Programs\n </a>\n </li>\n <li>\n <a href=\"/#mars_exploration_program/5\" target=\"_self\">\n Planetary Resources\n </a>\n </li>\n <li>\n <a href=\"/#mars_exploration_program/6\" target=\"_self\">\n Technologies\n </a>\n </li>\n </ul>\n </div>\n </li>\n </ul>\n </div>\n </div>\n <div class=\"sitemap_block\">\n <div class=\"footer_sitemap_item\">\n <h3 class=\"sitemap_title\">\n <a href=\"/#news_and_events\">\n News & Events\n </a>\n </h3>\n <ul>\n <li>\n <div class=\"global_subnav_container\">\n <ul class=\"subnav\">\n <li class=\"current\">\n <a href=\"/news\" target=\"_self\">\n News\n </a>\n </li>\n <li>\n <a href=\"/events\" target=\"_self\">\n Events\n </a>\n </li>\n </ul>\n </div>\n </li>\n </ul>\n </div>\n </div>\n <div class=\"sitemap_block\">\n <div class=\"footer_sitemap_item\">\n <h3 class=\"sitemap_title\">\n <a href=\"/#multimedia\">\n Multimedia\n </a>\n </h3>\n <ul>\n <li>\n <div class=\"global_subnav_container\">\n <ul class=\"subnav\">\n <li>\n <a href=\"/multimedia/images/\" target=\"_self\">\n Images\n </a>\n </li>\n <li>\n <a href=\"/multimedia/videos/\" target=\"_self\">\n Videos\n </a>\n </li>\n <li>\n <a href=\"/multimedia/more-resources/\" target=\"_self\">\n More Resources\n </a>\n </li>\n </ul>\n </div>\n </li>\n </ul>\n </div>\n </div>\n <div class=\"sitemap_block\">\n <div class=\"footer_sitemap_item\">\n <h3 class=\"sitemap_title\">\n <a href=\"/#missions_gallery_subnav\">\n Missions\n </a>\n </h3>\n <ul>\n <li>\n <div class=\"global_subnav_container\">\n <ul class=\"subnav\">\n <li>\n <a href=\"/mars-exploration/missions/?category=167\" target=\"_self\">\n Past\n </a>\n </li>\n <li>\n <a href=\"/mars-exploration/missions/?category=170\" target=\"_self\">\n Present\n </a>\n </li>\n <li>\n <a href=\"/mars-exploration/missions/?category=171\" target=\"_self\">\n Future\n </a>\n </li>\n <li>\n <a href=\"/mars-exploration/partners\" target=\"_self\">\n International Partners\n </a>\n </li>\n </ul>\n </div>\n </li>\n </ul>\n </div>\n </div>\n <div class=\"sitemap_block\">\n <div class=\"footer_sitemap_item\">\n <h3 class=\"sitemap_title\">\n <a href=\"/#more\">\n More\n </a>\n </h3>\n <ul>\n <li>\n <div class=\"global_subnav_container\">\n <ul class=\"subnav\">\n </ul>\n </div>\n </li>\n </ul>\n </div>\n </div>\n </div>\n <div class=\"gradient_line\">\n </div>\n </section>\n <section class=\"lower_footer\">\n <div class=\"nav_container\">\n <nav>\n <ul>\n <li>\n <a href=\"http://science.nasa.gov/\" target=\"_blank\">\n NASA Science Mission Directorate\n </a>\n </li>\n <li>\n <a href=\"https://www.jpl.nasa.gov/copyrights.php\" target=\"_blank\">\n Privacy\n </a>\n </li>\n <li>\n <a href=\"http://www.jpl.nasa.gov/imagepolicy/\" target=\"_blank\">\n Image Policy\n </a>\n </li>\n <li>\n <a href=\"https://mars.nasa.gov/feedback/\" target=\"_self\">\n Feedback\n </a>\n </li>\n </ul>\n </nav>\n </div>\n <div class=\"credits\">\n <div class=\"footer_brands_top\">\n <p>\n Managed by the Mars Exploration Program and the Jet Propulsion Laboratory for NASA’s Science Mission Directorate\n </p>\n </div>\n <!-- .footer_brands -->\n <!-- %a.jpl{href: \"\", target: \"_blank\"}Institution -->\n <!-- -->\n <!-- %a.caltech{href: \"\", target: \"_blank\"}Institution -->\n <!-- .staff -->\n <!-- %p -->\n <!-- - get_staff_for_category(get_field_from_admin_config(:web_staff_category_id)) -->\n <!-- - @staff.each_with_index do |staff, idx| -->\n <!-- - unless staff.is_in_footer == 0 -->\n <!-- = staff.title + \": \" -->\n <!-- - if staff.contact_link =~ /@/ -->\n <!-- = mail_to staff.contact_link, staff.name, :subject => \"[#{@site_title}]\" -->\n <!-- - elsif staff.contact_link.present? -->\n <!-- = link_to staff.name, staff.contact_link -->\n <!-- - else -->\n <!-- = staff.name -->\n <!-- - unless (idx + 1 == @staff.size) -->\n <!-- %br -->\n </div>\n </section>\n </div>\n </footer>\n </div>\n </div>\n <script id=\"_fed_an_ua_tag\" src=\"https://dap.digitalgov.gov/Universal-Federated-Analytics-Min.js?agency=NASA&subagency=JPL-Mars-MEPJPL&pua=UA-9453474-9,UA-118212757-11&dclink=true&sp=searchbox&exts=tif,tiff,wav\" type=\"text/javascript\">\n </script>\n </body>\n</html>\n\n"
],
[
"#locates most recent articel title\n\ntitle = soup.find(\"div\", class_= \"content_title\").text\nprint(title)",
"\n\nNASA to Broadcast Mars 2020 Perseverance Launch, Prelaunch Activities\n\n\n"
],
[
"# Locates the paragraph within the most recent story\nparagraph= soup.find(\"div\", class_= \"rollover_description_inner\").text\nprint(paragraph)",
"\nStarting July 27, news activities will cover everything from mission engineering and science to returning samples from Mars to, of course, the launch itself.\n\n"
],
[
"# Visit's the website below in the new browser\nbrowser.visit('https://www.jpl.nasa.gov/spaceimages/?search=&category=Mars')",
"_____no_output_____"
],
[
"# Manually clicks the link in the browser\nbrowser.click_link_by_id('full_image')",
"_____no_output_____"
],
[
"# manually clicks the \"more info\" link\nbrowser.click_link_by_partial_text('more info')",
"C:\\Users\\markr\\anaconda3\\lib\\site-packages\\splinter\\driver\\webdriver\\__init__.py:493: FutureWarning: browser.find_link_by_partial_text is deprecated. Use browser.links.find_by_partial_text instead.\n FutureWarning,\n"
],
[
"# html parser\nhtml=browser.html\nsoup = bs(html, \"html.parser\")",
"_____no_output_____"
],
[
"image= soup.select_one('figure.lede a img').get('src')\nimage",
"_____no_output_____"
],
[
"main_url= \"https://www.jpl.nasa.gov\"\nfeatured_image_url = main_url+image\nfeatured_image_url",
"_____no_output_____"
],
[
"# Use Pandas to scrape data\ntables = pd.read_html('https://space-facts.com/mars/')\ntables\n\n# Creates a dataframe from the list that is \"tables\"\nmars_df = pd.DataFrame(tables[0])\n# Changes the name of the columns\nmars_df.rename(columns={0:\"Information\", 1:\"Values\"})",
"_____no_output_____"
],
[
"# Transforms dataframe so it is readible in html\nmars_html_table = [mars_df.to_html(classes='data_table', index=False, header=False, border=0)]\nmars_html_table ",
"_____no_output_____"
],
[
"mars_df.rename = mars_df.rename.to_html()",
"_____no_output_____"
],
[
"# Visits the website below \nbrowser.visit('https://astrogeology.usgs.gov/search/results?q=hemisphere+enhanced&k1=target&v1=Mars')",
"_____no_output_____"
],
[
"# html and parser\nhtml = browser.html\nsoup = bs(html, 'html.parser')\n\n# Creates an empty list that will contian the names of the hemispheres \nhemi_names = []\n\n# Search for the names of all four hemispheres\nresults = soup.find_all('div', class_=\"collapsible results\")\nhemispheres = results[0].find_all('h3')\n\n# Get text and store in list\nfor name in hemispheres:\n hemi_names.append(name.text)\n\nhemi_names",
"_____no_output_____"
],
[
"\n# Search for thumbnail links\nthumbnail_results = results[0].find_all('a')\nthumbnail_links = []\n\nfor thumbnail in thumbnail_results:\n \n # If the thumbnail element has an image...\n if (thumbnail.img):\n \n # then grab the attached link\n thumbnail_url = 'https://astrogeology.usgs.gov/' + thumbnail['href']\n \n # Append list with links\n thumbnail_links.append(thumbnail_url)\n\nthumbnail_links",
"_____no_output_____"
],
[
"\nfull_imgs = []\n\nfor url in thumbnail_links:\n \n # Click through each thumbanil link\n browser.visit(url)\n \n html = browser.html\n soup = bs(html, 'html.parser')\n \n # Scrape each page for the relative image path\n results = soup.find_all('img', class_='wide-image')\n relative_img_path = results[0]['src']\n \n # Combine the reltaive image path to get the full url\n img_link = 'https://astrogeology.usgs.gov/' + relative_img_path\n \n # Add full image links to a list\n full_imgs.append(img_link)\n\nfull_imgs",
"_____no_output_____"
],
[
"# Zip together the list of hemisphere names and hemisphere image links\nmars_hemi_zip = zip(hemi_names, full_imgs)\n\nhemisphere_image_urls = []\n\n# Iterate through the zipped object\nfor title, img in mars_hemi_zip:\n \n mars_hemi_dict = {}\n \n # Add hemisphere title to dictionary\n mars_hemi_dict['title'] = title\n \n # Add image url to dictionary\n mars_hemi_dict['img_url'] = img\n \n # Append the list with dictionaries\n hemisphere_image_urls.append(mars_hemi_dict)\n\nhemisphere_image_urls",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb8c408c60b3ad576853f17fbf95b8828cdf7b55
| 48,911 |
ipynb
|
Jupyter Notebook
|
tensorflow-compress.ipynb
|
vasiliyeskin/tensorflow-compress
|
078d3c813d8a92e404c2bfb9ec4ed66dc609bda6
|
[
"Unlicense"
] | null | null | null |
tensorflow-compress.ipynb
|
vasiliyeskin/tensorflow-compress
|
078d3c813d8a92e404c2bfb9ec4ed66dc609bda6
|
[
"Unlicense"
] | null | null | null |
tensorflow-compress.ipynb
|
vasiliyeskin/tensorflow-compress
|
078d3c813d8a92e404c2bfb9ec4ed66dc609bda6
|
[
"Unlicense"
] | null | null | null | 45.329935 | 621 | 0.598393 |
[
[
[
"# tensorflow-compress",
"_____no_output_____"
],
[
"[](https://colab.research.google.com/github/byronknoll/tensorflow-compress/blob/master/tensorflow-compress.ipynb)\n\nMade by Byron Knoll. GitHub repository: https://github.com/byronknoll/tensorflow-compress\n\n### Description\n\ntensorflow-compress performs lossless data compression using neural networks in TensorFlow. It can run on GPUs with a large batch size to get a substantial speed improvement. It is made using Colab, which should make it easy to run through a web browser. You can choose a file, perform compression (or decompression), and download the result.\n\ntensorflow-compress is open source and the code should hopefully be easy to understand and modify. Feel free to experiment with the code and create pull requests with improvements.\n\nThe neural network is trained from scratch during compression and decompression, so the model weights do not need to be stored. Arithmetic coding is used to encode the model predictions to a file.\n\nFeel free to contact me at [email protected] if you have any questions.\n\n### Instructions:\n\nBasic usage: configure all the fields in the \"Parameters\" section and select Runtime->Run All.\n\nAdvanced usage: save a copy of this notebook and modify the code.\n\n### Related Projects\n* [NNCP](https://bellard.org/nncp/) - this uses a similar LSTM architecture to tensorflow-compress. It is limited to running only on CPUs.\n* [lstm-compress](https://github.com/byronknoll/lstm-compress) - similar to NNCP, but has a batch size limit of one (so it is significantly slower).\n* [cmix](http://www.byronknoll.com/cmix.html) - shares the same LSTM code as lstm-compress, but contains a bunch of other components to get better compression rate.\n* [DeepZip](https://github.com/mohit1997/DeepZip) - this also performs compression using TensorFlow. However, it has some substantial architecture differences to tensorflow-compress: it uses pretraining (using multiple passes over the training data) and stores the model weights in the compressed file.\n\n### Benchmarks\nThese benchmarks were performed using tensorflow-compress v3 with the default parameter settings. Some parameters differ between enwik8 and enwik9 as noted in the parameter comments. Colab Pro was used with Tesla V100 GPU. Compression time and decompression time are approximately the same.\n* enwik8: compressed to 16,128,954 bytes in 32,113.38 seconds. NNCP preprocessing time: 206.38 seconds. Dictionary size: 65,987 bytes.\n* enwik9: compressed to 118,938,744 bytes in 297,505.98 seconds. NNCP preprocessing time: 2,598.77 seconds. Dictionary size: 79,876 bytes. Since Colab has a 24 hour time limit, the preprocessed enwik9 file was split into four parts using [this notebook](https://colab.sandbox.google.com/github/byronknoll/tensorflow-compress/blob/master/nncp-splitter.ipynb). The \"checkpoint\" option was used to save/load model weights between processing each part. For the first part, start_learning_rate=0.0007 and end_learning_rate=0.0005 was used. For the remaining three parts, a constant 0.00035 learning rate was used.\n\nSee the [Large Text Compression Benchmark](http://mattmahoney.net/dc/text.html) for more information about the test files and a comparison with other programs.\n\n### Versions\n* v3 - released November 28, 2020. Changes from v2:\n * Parameter tuning\n * [New notebook](https://colab.sandbox.google.com/github/byronknoll/tensorflow-compress/blob/master/nncp-splitter.ipynb) for file splitting\n * Support for learning rate decay\n* v2 - released September 6, 2020. Changes from v1:\n * 16 bit floats for improved speed\n * Weight updates occur at every timestep (instead of at spaced intervals)\n * Support for saving/loading model weights\n* v1 - released July 20, 2020.",
"_____no_output_____"
],
[
"## Parameters",
"_____no_output_____"
]
],
[
[
"batch_size = 96 #@param {type:\"integer\"}\n#@markdown >_This will split the file into N batches, and process them in parallel. Increasing this will improve speed but can make compression rate worse. Make this a multiple of 8 to improve speed on certain GPUs._\nseq_length = 11 #@param {type:\"integer\"}\n#@markdown >_This determines the horizon for back propagation through time. Reducing this will improve speed, but can make compression rate worse._\nrnn_units = 1400 #@param {type:\"integer\"}\n#@markdown >_This is the number of units to use within each LSTM layer. Reducing this will improve speed, but can make compression rate worse. Make this a multiple of 8 to improve speed on certain GPUs._\nnum_layers = 6 #@param {type:\"integer\"}\n#@markdown >_This is the number of LSTM layers to use. Reducing this will improve speed, but can make compression rate worse._\nstart_learning_rate = 0.00075 #@param {type:\"number\"}\n#@markdown >_Learning rate for Adam optimizer. Recommended value for enwik8: 0.00075. For enwik9, see the notes in the \"Benchmarks\" section for the recommended learning rate._\nend_learning_rate = 0.00075 #@param {type:\"number\"}\n#@markdown >_Typically this should be set to the same value as the \"start_learning_rate\" parameter above. If this is set to a different value, the learning rate will start at \"start_learning_rate\" and linearly change to \"end_learning_rate\" by the end of the file. For large files this could be useful for learning rate decay._\nmode = 'compress' #@param [\"compress\", \"decompress\", \"both\", \"preprocess_only\"]\n#@markdown >_Whether to run compression only, decompression only, or both. \"preprocess_only\" will only run preprocessing and skip compression._\npreprocess = 'nncp' #@param [\"cmix\", \"nncp\", \"nncp-done\", \"none\"]\n#@markdown >_The choice of preprocessor. NNCP works better on enwik8/enwik9. NNCP preprocessing is slower since it constructs a custom dictionary, while cmix uses a pretrained dictionary. \"nncp_done\" is used for files which have already been preprocessed by NNCP (the dictionary must also be included)._\nn_words = 8192 #@param {type:\"integer\"}\n#@markdown >_Only used for NNCP preprocessor: this is the approximative maximum number of words of the dictionary. Recommended value for enwik8/enwik9: 8192._\nmin_freq = 64 #@param {type:\"integer\"}\n#@markdown >_Only used for NNCP preprocessor: this is the minimum frequency of the selected words. Recommended value for enwik8: 64, enwik9: 512._\npath_to_file = \"enwik8\" #@param [\"enwik4\", \"enwik6\", \"enwik8\", \"enwik9\", \"custom\"]\n#@markdown >_Name of the file to compress or decompress. If \"custom\" is selected, use the next parameter to set a custom path._\ncustom_path = '' #@param {type:\"string\"}\n#@markdown >_Use this if the previous parameter was set to \"custom\". Set this to the name of the file you want to compress/decompress. You can transfer files using the \"http_path\" or \"local_upload\" options below._\nhttp_path = '' #@param {type:\"string\"}\n#@markdown >_The file from this URL will be downloaded. It is recommended to use Google Drive URLs to get fast transfer speed. Use this format for Google Drive files: https://drive.google.com/uc?id= and paste the file ID at the end of the URL. You can find the file ID from the \"Get Link\" URL in Google Drive. You can enter multiple URLs here, space separated._\nlocal_upload = False #@param {type:\"boolean\"}\n#@markdown >_If enabled, you will be prompted in the \"Setup Files\" section to select files to upload from your local computer. You can upload multiple files. Note: the upload speed can be quite slow (use \"http_path\" for better transfer speeds)._\ndownload_option = \"no_download\" #@param [\"no_download\", \"local\", \"google_drive\"]\n#@markdown >_If this is set to \"local\", the output files will be downloaded to your computer after compression/decompression. If set to \"google_drive\", they will be copied to your Google Drive account (which is significantly faster than downloading locally)._\ncheckpoint = False #@param {type:\"boolean\"}\n#@markdown >_If this is enabled, a checkpoint of the model weights will be downloaded (using the \"download_option\" parameter). This can be useful for getting around session time limits for Colab, by splitting files into multiple segments and saving/loading the model weights between each segment. Checkpoints (if present) will automatically be loaded when starting compression._\n",
"_____no_output_____"
]
],
[
[
"## Setup",
"_____no_output_____"
]
],
[
[
"#@title Imports\n\nimport tensorflow as tf\nimport numpy as np\nimport random\nfrom google.colab import files\nimport time\nimport math\nimport sys\nimport subprocess\nimport contextlib\nimport os\nfrom tensorflow.keras.mixed_precision import experimental as mixed_precision\nfrom google.colab import drive\nos.environ['TF_DETERMINISTIC_OPS'] = '1'",
"_____no_output_____"
],
[
"#@title System Info\n\ndef system_info():\n \"\"\"Prints out system information.\"\"\"\n gpu_info = !nvidia-smi\n gpu_info = '\\n'.join(gpu_info)\n if gpu_info.find('failed') >= 0:\n print('Select the Runtime → \"Change runtime type\" menu to enable a GPU accelerator, ')\n print('and then re-execute this cell.')\n else:\n print(gpu_info)\n print(\"TensorFlow version: \", tf.__version__)\n !lscpu |grep 'Model name'\n !cat /proc/meminfo | head -n 3\n\nsystem_info()",
"_____no_output_____"
],
[
"#@title Mount Google Drive\nif download_option == \"google_drive\":\n drive.mount('/content/gdrive')",
"_____no_output_____"
],
[
"#@title Setup Files\n\n!mkdir -p \"data\"\n\nif local_upload:\n %cd data\n files.upload()\n %cd ..\n\nif path_to_file == 'enwik8' or path_to_file == 'enwik6' or path_to_file == 'enwik4':\n %cd data\n !gdown --id 1BUbuEUhPOBaVZDdOh0KG8hxvIDgsyiZp\n !unzip enwik8.zip\n !head -c 1000000 enwik8 > enwik6\n !head -c 10000 enwik8 > enwik4\n path_to_file = 'data/' + path_to_file\n %cd ..\n\nif path_to_file == 'enwik9':\n %cd data\n !gdown --id 1D2gCmf9AlXIBP62ARhy0XcIuIolOTRAE\n !unzip enwik9.zip\n path_to_file = 'data/' + path_to_file\n %cd ..\n\nif path_to_file == 'custom':\n path_to_file = 'data/' + custom_path\n\nif http_path:\n %cd data\n paths = http_path.split()\n for path in paths:\n !gdown $path\n %cd ..\n\nif preprocess == 'cmix':\n !gdown --id 1qa7K28tlUDs9GGYbaL_iE9M4m0L1bYm9\n !unzip cmix-v18.zip\n %cd cmix\n !make\n %cd ..\n\nif preprocess == 'nncp' or preprocess == 'nncp-done':\n !gdown --id 1EzVPbRkBIIbgOzvEMeM0YpibDi2R4SHD\n !tar -xf nncp-2019-11-16.tar.gz\n %cd nncp-2019-11-16/\n !make preprocess\n %cd ..",
"_____no_output_____"
],
[
"#@title Model Architecture\n\ndef build_model(vocab_size):\n \"\"\"Builds the model architecture.\n\n Args:\n vocab_size: Int, size of the vocabulary.\n \"\"\"\n policy = mixed_precision.Policy('mixed_float16')\n mixed_precision.set_policy(policy)\n inputs = [\n tf.keras.Input(batch_input_shape=[batch_size, seq_length, vocab_size])]\n # In addition to the primary input, there are also two \"state\" inputs for each\n # layer of the network.\n for i in range(num_layers):\n inputs.append(tf.keras.Input(shape=(None,)))\n inputs.append(tf.keras.Input(shape=(None,)))\n # Skip connections will be used to connect each LSTM layer output to the final\n # output layer. Each LSTM layer will get as input both the original input and\n # the output of the previous layer.\n skip_connections = []\n # In addition to the softmax output, there are also two \"state\" outputs for\n # each layer of the network.\n outputs = []\n predictions, state_h, state_c = tf.keras.layers.LSTM(rnn_units,\n return_sequences=True,\n return_state=True,\n recurrent_initializer='glorot_uniform',\n )(inputs[0], initial_state=[\n tf.cast(inputs[1], tf.float16),\n tf.cast(inputs[2], tf.float16)])\n skip_connections.append(predictions)\n outputs.append(state_h)\n outputs.append(state_c)\n for i in range(num_layers - 1):\n layer_input = tf.keras.layers.concatenate(\n [inputs[0], skip_connections[-1]])\n predictions, state_h, state_c = tf.keras.layers.LSTM(rnn_units,\n return_sequences=True,\n return_state=True,\n recurrent_initializer='glorot_uniform')(\n layer_input, initial_state=[tf.cast(inputs[i*2+3], tf.float16),\n tf.cast(inputs[i*2+4], tf.float16)])\n skip_connections.append(predictions)\n outputs.append(state_h)\n outputs.append(state_c)\n # The dense output layer only needs to be computed for the last timestep, so\n # we can discard the earlier outputs.\n last_timestep = []\n for i in range(num_layers):\n last_timestep.append(tf.slice(skip_connections[i], [0, seq_length - 1, 0],\n [batch_size, 1, rnn_units]))\n if num_layers == 1:\n layer_input = last_timestep[0]\n else:\n layer_input = tf.keras.layers.concatenate(last_timestep)\n dense = tf.keras.layers.Dense(vocab_size, name='dense_logits')(layer_input)\n output = tf.keras.layers.Activation('softmax', dtype='float32',\n name='predictions')(dense)\n outputs.insert(0, output)\n model = tf.keras.Model(inputs=inputs, outputs=outputs)\n return model",
"_____no_output_____"
],
[
"#@title Compression Library\n\ndef get_symbol(index, length, freq, coder, compress, data):\n \"\"\"Runs arithmetic coding and returns the next symbol.\n\n Args:\n index: Int, position of the symbol in the file.\n length: Int, size limit of the file.\n freq: ndarray, predicted symbol probabilities.\n coder: this is the arithmetic coder.\n compress: Boolean, True if compressing, False if decompressing.\n data: List containing each symbol in the file.\n \n Returns:\n The next symbol, or 0 if \"index\" is over the file size limit.\n \"\"\"\n symbol = 0\n if index < length:\n if compress:\n symbol = data[index]\n coder.write(freq, symbol)\n else:\n symbol = coder.read(freq)\n data[index] = symbol\n return symbol\n\ndef train(pos, seq_input, length, vocab_size, coder, model, optimizer, compress,\n data, states):\n \"\"\"Runs one training step.\n\n Args:\n pos: Int, position in the file for the current symbol for the *first* batch.\n seq_input: Tensor, containing the last seq_length inputs for the model.\n length: Int, size limit of the file.\n vocab_size: Int, size of the vocabulary.\n coder: this is the arithmetic coder.\n model: the model to generate predictions.\n optimizer: optimizer used to train the model.\n compress: Boolean, True if compressing, False if decompressing.\n data: List containing each symbol in the file.\n states: List containing state information for the layers of the model.\n \n Returns:\n seq_input: Tensor, containing the last seq_length inputs for the model.\n cross_entropy: cross entropy numerator.\n denom: cross entropy denominator.\n \"\"\"\n loss = cross_entropy = denom = 0\n split = math.ceil(length / batch_size)\n # Keep track of operations while running the forward pass for automatic\n # differentiation.\n with tf.GradientTape() as tape:\n # The model inputs contain both seq_input and the states for each layer.\n inputs = states.pop(0)\n inputs.insert(0, seq_input)\n # Run the model (for all batches in parallel) to get predictions for the\n # next characters.\n outputs = model(inputs)\n predictions = outputs.pop(0)\n states.append(outputs)\n p = predictions.numpy()\n symbols = []\n # When the last batch reaches the end of the file, we start giving it \"0\"\n # as input. We use a mask to prevent this from influencing the gradients.\n mask = []\n # Go over each batch to run the arithmetic coding and prepare the next\n # input.\n for i in range(batch_size):\n # The \"10000000\" is used to convert floats into large integers (since\n # the arithmetic coder works on integers).\n freq = np.cumsum(p[i][0] * 10000000 + 1)\n index = pos + 1 + i * split\n symbol = get_symbol(index, length, freq, coder, compress, data)\n symbols.append(symbol)\n if index < length:\n prob = p[i][0][symbol]\n if prob <= 0:\n # Set a small value to avoid error with log2.\n prob = 0.000001\n cross_entropy += math.log2(prob)\n denom += 1\n mask.append(1.0)\n else:\n mask.append(0.0)\n # \"input_one_hot\" will be used both for the loss function and for the next\n # input.\n input_one_hot = tf.expand_dims(tf.one_hot(symbols, vocab_size), 1)\n loss = tf.keras.losses.categorical_crossentropy(\n input_one_hot, predictions, from_logits=False) * tf.expand_dims(\n tf.convert_to_tensor(mask), 1)\n # Remove the oldest input and append the new one.\n seq_input = tf.slice(seq_input, [0, 1, 0],\n [batch_size, seq_length - 1, vocab_size])\n seq_input = tf.concat([seq_input, input_one_hot], 1)\n # Run the backwards pass to update model weights.\n grads = tape.gradient(loss, model.trainable_variables)\n # Gradient clipping to make training more robust.\n capped_grads = [tf.clip_by_value(grad, -5., 5.) for grad in grads]\n optimizer.apply_gradients(zip(capped_grads, model.trainable_variables))\n return (seq_input, cross_entropy, denom)\n\ndef reset_seed():\n \"\"\"Initializes various random seeds to help with determinism.\"\"\"\n SEED = 1234\n os.environ['PYTHONHASHSEED']=str(SEED)\n random.seed(SEED)\n np.random.seed(SEED)\n tf.random.set_seed(SEED)\n\ndef download(path):\n \"\"\"Downloads the file at the specified path.\"\"\"\n if download_option == 'local':\n files.download(path)\n elif download_option == 'google_drive':\n !cp -f $path /content/gdrive/My\\ Drive\n\ndef process(compress, length, vocab_size, coder, data):\n \"\"\"This runs compression/decompression.\n\n Args:\n compress: Boolean, True if compressing, False if decompressing.\n length: Int, size limit of the file.\n vocab_size: Int, size of the vocabulary.\n coder: this is the arithmetic coder.\n data: List containing each symbol in the file.\n \"\"\"\n start = time.time()\n reset_seed()\n model = build_model(vocab_size = vocab_size)\n checkpoint_path = tf.train.latest_checkpoint('./data')\n if checkpoint_path:\n model.load_weights(checkpoint_path)\n model.summary()\n\n # Try to split the file into equal size pieces for the different batches. The\n # last batch may have fewer characters if the file can't be split equally.\n split = math.ceil(length / batch_size)\n\n learning_rate_fn = tf.keras.optimizers.schedules.PolynomialDecay(\n start_learning_rate,\n split,\n end_learning_rate,\n power=1.0)\n optimizer = tf.keras.optimizers.Adam(\n learning_rate=learning_rate_fn, beta_1=0, beta_2=0.9999, epsilon=1e-5)\n\n hidden = model.reset_states()\n # Use a uniform distribution for predicting the first batch of symbols. The\n # \"10000000\" is used to convert floats into large integers (since the\n # arithmetic coder works on integers).\n freq = np.cumsum(np.full(vocab_size, (1.0 / vocab_size)) * 10000000 + 1)\n # Construct the first set of input characters for training.\n symbols = []\n for i in range(batch_size):\n symbols.append(get_symbol(i*split, length, freq, coder, compress, data))\n input_one_hot = tf.expand_dims(tf.one_hot(symbols, vocab_size), 1)\n # Replicate the input tensor seq_length times, to match the input format.\n seq_input = tf.tile(input_one_hot, [1, seq_length, 1])\n pos = cross_entropy = denom = last_output = 0\n template = '{:0.2f}%\\tcross entropy: {:0.2f}\\ttime: {:0.2f}'\n # This will keep track of layer states. Initialize them to zeros.\n states = []\n for i in range(seq_length):\n states.append([tf.zeros([batch_size, rnn_units])] * (num_layers * 2))\n # Keep repeating the training step until we get to the end of the file.\n while pos < split:\n seq_input, ce, d = train(pos, seq_input, length, vocab_size, coder, model,\n optimizer, compress, data, states)\n cross_entropy += ce\n denom += d\n pos += 1\n time_diff = time.time() - start\n # If it has been over 20 seconds since the last status message, display a\n # new one.\n if time_diff - last_output > 20:\n last_output = time_diff\n percentage = 100 * pos / split\n if percentage >= 100: continue\n print(template.format(percentage, -cross_entropy / denom, time_diff))\n if compress:\n coder.finish()\n print(template.format(100, -cross_entropy / length, time.time() - start))\n system_info()\n if mode != \"both\" or not compress:\n model.save_weights('./data/model')\n",
"_____no_output_____"
],
[
"#@title Arithmetic Coding Library\n\n# \n# Reference arithmetic coding\n# Copyright (c) Project Nayuki\n# \n# https://www.nayuki.io/page/reference-arithmetic-coding\n# https://github.com/nayuki/Reference-arithmetic-coding\n# \n\nimport sys\npython3 = sys.version_info.major >= 3\n\n\n# ---- Arithmetic coding core classes ----\n\n# Provides the state and behaviors that arithmetic coding encoders and decoders share.\nclass ArithmeticCoderBase(object):\n\t\n\t# Constructs an arithmetic coder, which initializes the code range.\n\tdef __init__(self, numbits):\n\t\tif numbits < 1:\n\t\t\traise ValueError(\"State size out of range\")\n\t\t\n\t\t# -- Configuration fields --\n\t\t# Number of bits for the 'low' and 'high' state variables. Must be at least 1.\n\t\t# - Larger values are generally better - they allow a larger maximum frequency total (maximum_total),\n\t\t# and they reduce the approximation error inherent in adapting fractions to integers;\n\t\t# both effects reduce the data encoding loss and asymptotically approach the efficiency\n\t\t# of arithmetic coding using exact fractions.\n\t\t# - But larger state sizes increase the computation time for integer arithmetic,\n\t\t# and compression gains beyond ~30 bits essentially zero in real-world applications.\n\t\t# - Python has native bigint arithmetic, so there is no upper limit to the state size.\n\t\t# For Java and C++ where using native machine-sized integers makes the most sense,\n\t\t# they have a recommended value of num_state_bits=32 as the most versatile setting.\n\t\tself.num_state_bits = numbits\n\t\t# Maximum range (high+1-low) during coding (trivial), which is 2^num_state_bits = 1000...000.\n\t\tself.full_range = 1 << self.num_state_bits\n\t\t# The top bit at width num_state_bits, which is 0100...000.\n\t\tself.half_range = self.full_range >> 1 # Non-zero\n\t\t# The second highest bit at width num_state_bits, which is 0010...000. This is zero when num_state_bits=1.\n\t\tself.quarter_range = self.half_range >> 1 # Can be zero\n\t\t# Minimum range (high+1-low) during coding (non-trivial), which is 0010...010.\n\t\tself.minimum_range = self.quarter_range + 2 # At least 2\n\t\t# Maximum allowed total from a frequency table at all times during coding. This differs from Java\n\t\t# and C++ because Python's native bigint avoids constraining the size of intermediate computations.\n\t\tself.maximum_total = self.minimum_range\n\t\t# Bit mask of num_state_bits ones, which is 0111...111.\n\t\tself.state_mask = self.full_range - 1\n\t\t\n\t\t# -- State fields --\n\t\t# Low end of this arithmetic coder's current range. Conceptually has an infinite number of trailing 0s.\n\t\tself.low = 0\n\t\t# High end of this arithmetic coder's current range. Conceptually has an infinite number of trailing 1s.\n\t\tself.high = self.state_mask\n\t\n\t\n\t# Updates the code range (low and high) of this arithmetic coder as a result\n\t# of processing the given symbol with the given frequency table.\n\t# Invariants that are true before and after encoding/decoding each symbol\n\t# (letting full_range = 2^num_state_bits):\n\t# - 0 <= low <= code <= high < full_range. ('code' exists only in the decoder.)\n\t# Therefore these variables are unsigned integers of num_state_bits bits.\n\t# - low < 1/2 * full_range <= high.\n\t# In other words, they are in different halves of the full range.\n\t# - (low < 1/4 * full_range) || (high >= 3/4 * full_range).\n\t# In other words, they are not both in the middle two quarters.\n\t# - Let range = high - low + 1, then full_range/4 < minimum_range\n\t# <= range <= full_range. These invariants for 'range' essentially\n\t# dictate the maximum total that the incoming frequency table can have.\n\tdef update(self, freqs, symbol):\n\t\t# State check\n\t\tlow = self.low\n\t\thigh = self.high\n\t\t# if low >= high or (low & self.state_mask) != low or (high & self.state_mask) != high:\n\t\t# \traise AssertionError(\"Low or high out of range\")\n\t\trange = high - low + 1\n\t\t# if not (self.minimum_range <= range <= self.full_range):\n\t\t# \traise AssertionError(\"Range out of range\")\n\t\t\n\t\t# Frequency table values check\n\t\ttotal = int(freqs[-1])\n\t\tsymlow = int(freqs[symbol-1]) if symbol > 0 else 0\n\t\tsymhigh = int(freqs[symbol])\n\t\t#total = freqs.get_total()\n\t\t#symlow = freqs.get_low(symbol)\n\t\t#symhigh = freqs.get_high(symbol)\n\t\t# if symlow == symhigh:\n\t\t# \traise ValueError(\"Symbol has zero frequency\")\n\t\t# if total > self.maximum_total:\n\t\t# \traise ValueError(\"Cannot code symbol because total is too large\")\n\t\t\n\t\t# Update range\n\t\tnewlow = low + symlow * range // total\n\t\tnewhigh = low + symhigh * range // total - 1\n\t\tself.low = newlow\n\t\tself.high = newhigh\n\t\t\n\t\t# While low and high have the same top bit value, shift them out\n\t\twhile ((self.low ^ self.high) & self.half_range) == 0:\n\t\t\tself.shift()\n\t\t\tself.low = ((self.low << 1) & self.state_mask)\n\t\t\tself.high = ((self.high << 1) & self.state_mask) | 1\n\t\t# Now low's top bit must be 0 and high's top bit must be 1\n\t\t\n\t\t# While low's top two bits are 01 and high's are 10, delete the second highest bit of both\n\t\twhile (self.low & ~self.high & self.quarter_range) != 0:\n\t\t\tself.underflow()\n\t\t\tself.low = (self.low << 1) ^ self.half_range\n\t\t\tself.high = ((self.high ^ self.half_range) << 1) | self.half_range | 1\n\t\n\t\n\t# Called to handle the situation when the top bit of 'low' and 'high' are equal.\n\tdef shift(self):\n\t\traise NotImplementedError()\n\t\n\t\n\t# Called to handle the situation when low=01(...) and high=10(...).\n\tdef underflow(self):\n\t\traise NotImplementedError()\n\n\n# Encodes symbols and writes to an arithmetic-coded bit stream.\nclass ArithmeticEncoder(ArithmeticCoderBase):\n\t\n\t# Constructs an arithmetic coding encoder based on the given bit output stream.\n\tdef __init__(self, numbits, bitout):\n\t\tsuper(ArithmeticEncoder, self).__init__(numbits)\n\t\t# The underlying bit output stream.\n\t\tself.output = bitout\n\t\t# Number of saved underflow bits. This value can grow without bound.\n\t\tself.num_underflow = 0\n\t\n\t\n\t# Encodes the given symbol based on the given frequency table.\n\t# This updates this arithmetic coder's state and may write out some bits.\n\tdef write(self, freqs, symbol):\n\t\tself.update(freqs, symbol)\n\t\n\t\n\t# Terminates the arithmetic coding by flushing any buffered bits, so that the output can be decoded properly.\n\t# It is important that this method must be called at the end of the each encoding process.\n\t# Note that this method merely writes data to the underlying output stream but does not close it.\n\tdef finish(self):\n\t\tself.output.write(1)\n\t\n\t\n\tdef shift(self):\n\t\tbit = self.low >> (self.num_state_bits - 1)\n\t\tself.output.write(bit)\n\t\t\n\t\t# Write out the saved underflow bits\n\t\tfor _ in range(self.num_underflow):\n\t\t\tself.output.write(bit ^ 1)\n\t\tself.num_underflow = 0\n\t\n\t\n\tdef underflow(self):\n\t\tself.num_underflow += 1\n\n\n# Reads from an arithmetic-coded bit stream and decodes symbols.\nclass ArithmeticDecoder(ArithmeticCoderBase):\n\t\n\t# Constructs an arithmetic coding decoder based on the\n\t# given bit input stream, and fills the code bits.\n\tdef __init__(self, numbits, bitin):\n\t\tsuper(ArithmeticDecoder, self).__init__(numbits)\n\t\t# The underlying bit input stream.\n\t\tself.input = bitin\n\t\t# The current raw code bits being buffered, which is always in the range [low, high].\n\t\tself.code = 0\n\t\tfor _ in range(self.num_state_bits):\n\t\t\tself.code = self.code << 1 | self.read_code_bit()\n\t\n\t\n\t# Decodes the next symbol based on the given frequency table and returns it.\n\t# Also updates this arithmetic coder's state and may read in some bits.\n\tdef read(self, freqs):\n\t\t#if not isinstance(freqs, CheckedFrequencyTable):\n\t\t#\tfreqs = CheckedFrequencyTable(freqs)\n\t\t\n\t\t# Translate from coding range scale to frequency table scale\n\t\ttotal = int(freqs[-1])\n\t\t#total = freqs.get_total()\n\t\t#if total > self.maximum_total:\n\t\t#\traise ValueError(\"Cannot decode symbol because total is too large\")\n\t\trange = self.high - self.low + 1\n\t\toffset = self.code - self.low\n\t\tvalue = ((offset + 1) * total - 1) // range\n\t\t#assert value * range // total <= offset\n\t\t#assert 0 <= value < total\n\t\t\n\t\t# A kind of binary search. Find highest symbol such that freqs.get_low(symbol) <= value.\n\t\tstart = 0\n\t\tend = len(freqs)\n\t\t#end = freqs.get_symbol_limit()\n\t\twhile end - start > 1:\n\t\t\tmiddle = (start + end) >> 1\n\t\t\tlow = int(freqs[middle-1]) if middle > 0 else 0\n\t\t\t#if freqs.get_low(middle) > value:\n\t\t\tif low > value:\n\t\t\t\tend = middle\n\t\t\telse:\n\t\t\t\tstart = middle\n\t\t#assert start + 1 == end\n\t\t\n\t\tsymbol = start\n\t\t#assert freqs.get_low(symbol) * range // total <= offset < freqs.get_high(symbol) * range // total\n\t\tself.update(freqs, symbol)\n\t\t#if not (self.low <= self.code <= self.high):\n\t\t#\traise AssertionError(\"Code out of range\")\n\t\treturn symbol\n\t\n\t\n\tdef shift(self):\n\t\tself.code = ((self.code << 1) & self.state_mask) | self.read_code_bit()\n\t\n\t\n\tdef underflow(self):\n\t\tself.code = (self.code & self.half_range) | ((self.code << 1) & (self.state_mask >> 1)) | self.read_code_bit()\n\t\n\t\n\t# Returns the next bit (0 or 1) from the input stream. The end\n\t# of stream is treated as an infinite number of trailing zeros.\n\tdef read_code_bit(self):\n\t\ttemp = self.input.read()\n\t\tif temp == -1:\n\t\t\ttemp = 0\n\t\treturn temp\n\n\n# ---- Bit-oriented I/O streams ----\n\n# A stream of bits that can be read. Because they come from an underlying byte stream,\n# the total number of bits is always a multiple of 8. The bits are read in big endian.\nclass BitInputStream(object):\n\t\n\t# Constructs a bit input stream based on the given byte input stream.\n\tdef __init__(self, inp):\n\t\t# The underlying byte stream to read from\n\t\tself.input = inp\n\t\t# Either in the range [0x00, 0xFF] if bits are available, or -1 if end of stream is reached\n\t\tself.currentbyte = 0\n\t\t# Number of remaining bits in the current byte, always between 0 and 7 (inclusive)\n\t\tself.numbitsremaining = 0\n\t\n\t\n\t# Reads a bit from this stream. Returns 0 or 1 if a bit is available, or -1 if\n\t# the end of stream is reached. The end of stream always occurs on a byte boundary.\n\tdef read(self):\n\t\tif self.currentbyte == -1:\n\t\t\treturn -1\n\t\tif self.numbitsremaining == 0:\n\t\t\ttemp = self.input.read(1)\n\t\t\tif len(temp) == 0:\n\t\t\t\tself.currentbyte = -1\n\t\t\t\treturn -1\n\t\t\tself.currentbyte = temp[0] if python3 else ord(temp)\n\t\t\tself.numbitsremaining = 8\n\t\tassert self.numbitsremaining > 0\n\t\tself.numbitsremaining -= 1\n\t\treturn (self.currentbyte >> self.numbitsremaining) & 1\n\t\n\t\n\t# Reads a bit from this stream. Returns 0 or 1 if a bit is available, or raises an EOFError\n\t# if the end of stream is reached. The end of stream always occurs on a byte boundary.\n\tdef read_no_eof(self):\n\t\tresult = self.read()\n\t\tif result != -1:\n\t\t\treturn result\n\t\telse:\n\t\t\traise EOFError()\n\t\n\t\n\t# Closes this stream and the underlying input stream.\n\tdef close(self):\n\t\tself.input.close()\n\t\tself.currentbyte = -1\n\t\tself.numbitsremaining = 0\n\n\n# A stream where bits can be written to. Because they are written to an underlying\n# byte stream, the end of the stream is padded with 0's up to a multiple of 8 bits.\n# The bits are written in big endian.\nclass BitOutputStream(object):\n\t\n\t# Constructs a bit output stream based on the given byte output stream.\n\tdef __init__(self, out):\n\t\tself.output = out # The underlying byte stream to write to\n\t\tself.currentbyte = 0 # The accumulated bits for the current byte, always in the range [0x00, 0xFF]\n\t\tself.numbitsfilled = 0 # Number of accumulated bits in the current byte, always between 0 and 7 (inclusive)\n\t\n\t\n\t# Writes a bit to the stream. The given bit must be 0 or 1.\n\tdef write(self, b):\n\t\tif b not in (0, 1):\n\t\t\traise ValueError(\"Argument must be 0 or 1\")\n\t\tself.currentbyte = (self.currentbyte << 1) | b\n\t\tself.numbitsfilled += 1\n\t\tif self.numbitsfilled == 8:\n\t\t\ttowrite = bytes((self.currentbyte,)) if python3 else chr(self.currentbyte)\n\t\t\tself.output.write(towrite)\n\t\t\tself.currentbyte = 0\n\t\t\tself.numbitsfilled = 0\n\t\n\t\n\t# Closes this stream and the underlying output stream. If called when this\n\t# bit stream is not at a byte boundary, then the minimum number of \"0\" bits\n\t# (between 0 and 7 of them) are written as padding to reach the next byte boundary.\n\tdef close(self):\n\t\twhile self.numbitsfilled != 0:\n\t\t\tself.write(0)\n\t\tself.output.close()",
"_____no_output_____"
]
],
[
[
"## Compress",
"_____no_output_____"
]
],
[
[
"#@title Preprocess\n\nif mode != 'decompress':\n input_path = path_to_file\n\n if preprocess == 'cmix':\n !./cmix/cmix -s ./cmix/dictionary/english.dic $path_to_file ./data/preprocessed.dat\n input_path = \"./data/preprocessed.dat\"\n\n # int_list will contain the characters of the file.\n int_list = []\n if preprocess == 'nncp' or preprocess == 'nncp-done':\n if preprocess == 'nncp':\n !time ./nncp-2019-11-16/preprocess c data/dictionary.words $path_to_file data/preprocessed.dat $n_words $min_freq\n else:\n !cp $path_to_file data/preprocessed.dat\n input_path = \"./data/preprocessed.dat\"\n orig = open(input_path, 'rb').read()\n for i in range(0, len(orig), 2):\n int_list.append(orig[i] * 256 + orig[i+1])\n vocab_size = int(subprocess.check_output(\n ['wc', '-l', 'data/dictionary.words']).split()[0])\n else:\n text = open(input_path, 'rb').read()\n vocab = sorted(set(text))\n vocab_size = len(vocab)\n # Creating a mapping from unique characters to indexes.\n char2idx = {u:i for i, u in enumerate(vocab)}\n for idx, c in enumerate(text):\n int_list.append(char2idx[c])\n\n # Round up to a multiple of 8 to improve performance.\n vocab_size = math.ceil(vocab_size/8) * 8\n file_len = len(int_list)\n print ('Length of file: {} symbols'.format(file_len))\n print ('Vocabulary size: {}'.format(vocab_size))",
"_____no_output_____"
],
[
"#@title Compression\n\nif mode == 'compress' or mode == 'both':\n original_file = path_to_file\n path_to_file = \"data/compressed.dat\"\n with open(path_to_file, \"wb\") as out, contextlib.closing(BitOutputStream(out)) as bitout:\n length = len(int_list)\n # Write the original file length to the compressed file header.\n out.write(length.to_bytes(5, byteorder='big', signed=False))\n if preprocess != 'nncp' and preprocess != 'nncp-done':\n # If NNCP was not used for preprocessing, write 256 bits to the compressed\n # file header to keep track of the vocabulary.\n for i in range(256):\n if i in char2idx:\n bitout.write(1)\n else:\n bitout.write(0)\n enc = ArithmeticEncoder(32, bitout)\n process(True, length, vocab_size, enc, int_list)\n print(\"Compressed size:\", os.path.getsize(path_to_file))",
"_____no_output_____"
],
[
"#@title Download Result\n\nif mode == 'preprocess_only':\n if preprocess == 'nncp':\n download('data/dictionary.words')\n download(input_path)\nelif mode != 'decompress':\n download('data/compressed.dat')\n if preprocess == 'nncp':\n download('data/dictionary.words')\n if checkpoint and mode != \"both\":\n download('data/model.index')\n download('data/model.data-00000-of-00001')\n download('data/checkpoint')",
"_____no_output_____"
]
],
[
[
"## Decompress",
"_____no_output_____"
]
],
[
[
"#@title Decompression\n\nif mode == 'decompress' or mode == 'both':\n output_path = \"data/decompressed.dat\"\n with open(path_to_file, \"rb\") as inp, open(output_path, \"wb\") as out:\n # Read the original file size from the header.\n length = int.from_bytes(inp.read()[:5], byteorder='big')\n inp.seek(5)\n # Create a list to store the file characters.\n output = [0] * length\n bitin = BitInputStream(inp)\n if preprocess == 'nncp' or preprocess == 'nncp-done':\n # If the preprocessor is NNCP, we can get the vocab_size from the\n # dictionary.\n vocab_size = int(subprocess.check_output(\n ['wc', '-l', 'data/dictionary.words']).split()[0])\n else:\n # If the preprocessor is not NNCP, we can get the vocabulary from the file\n # header.\n vocab = []\n for i in range(256):\n if bitin.read():\n vocab.append(i)\n vocab_size = len(vocab)\n # Round up to a multiple of 8 to improve performance.\n vocab_size = math.ceil(vocab_size/8) * 8\n dec = ArithmeticDecoder(32, bitin)\n process(False, length, vocab_size, dec, output)\n # The decompressed data is stored in the \"output\" list. We can now write the\n # data to file (based on the type of preprocessing used).\n if preprocess == 'nncp' or preprocess == 'nncp-done':\n for i in range(length):\n out.write(bytes(((output[i] // 256),)))\n out.write(bytes(((output[i] % 256),)))\n else:\n # Convert indexes back to the original characters.\n idx2char = np.array(vocab)\n for i in range(length):\n out.write(bytes((idx2char[output[i]],)))\n\n if preprocess == 'cmix':\n !./cmix/cmix -d ./cmix/dictionary/english.dic $output_path ./data/final.dat\n output_path = \"data/final.dat\"\n if preprocess == 'nncp' or preprocess == 'nncp-done':\n !./nncp-2019-11-16/preprocess d data/dictionary.words $output_path ./data/final.dat\n output_path = \"data/final.dat\"",
"_____no_output_____"
],
[
"#@title Download Result\n\nif mode == 'decompress':\n if preprocess == 'nncp-done':\n download('data/decompressed.dat')\n else:\n download(output_path)\n if checkpoint:\n download('data/model.index')\n download('data/model.data-00000-of-00001')\n download('data/checkpoint')",
"_____no_output_____"
],
[
"#@title Validation\n\nif mode == 'decompress' or mode == 'both':\n if preprocess == 'nncp-done':\n !md5sum data/decompressed.dat\n !md5sum $output_path\nif mode == 'both':\n !md5sum $original_file",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
cb8c610d68fb91da844c3160ccfe727b6b7741b7
| 16,475 |
ipynb
|
Jupyter Notebook
|
examples/brainstorm/notebooks/pandas dataframes.ipynb
|
rdelosreyes/myctapipe
|
dad0784b60de986d5ee871e7b61a951e948998d6
|
[
"BSD-3-Clause"
] | null | null | null |
examples/brainstorm/notebooks/pandas dataframes.ipynb
|
rdelosreyes/myctapipe
|
dad0784b60de986d5ee871e7b61a951e948998d6
|
[
"BSD-3-Clause"
] | null | null | null |
examples/brainstorm/notebooks/pandas dataframes.ipynb
|
rdelosreyes/myctapipe
|
dad0784b60de986d5ee871e7b61a951e948998d6
|
[
"BSD-3-Clause"
] | null | null | null | 27.458333 | 77 | 0.376692 |
[
[
[
"#test of using Pandas DataFrame as internal structure\n\nAdvantages\n* well supported\n* has connection to lots of I/O layers (even PyTables)\n* can add/remove columns easily\n\nProblems:\n* can't seem to store vector data in a column...",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np",
"_____no_output_____"
],
[
"evid = np.arange(10) + 1024\nX = np.linspace(-10,10,10, dtype=np.float32)\nY = np.linspace(-10,10,10, dtype=np.float32)\nV = np.random.uniform(size=10)",
"_____no_output_____"
],
[
"data = pd.DataFrame({ 'DETX':X,'DETY':Y,'VALUE':V}, index=evid)\ndata",
"_____no_output_____"
],
[
"data.describe()",
"_____no_output_____"
],
[
"print data.loc[1029]",
"DETX 1.111112\nDETY 1.111112\nVALUE 0.887062\nName: 1029, dtype: float64\n"
],
[
"for col in data:\n print col",
"DETX\nDETY\nVALUE\n"
]
],
[
[
"Adding a column:",
"_____no_output_____"
]
],
[
[
"energy = np.random.lognormal(2,1,size=10)\ndata['ENERGY'] = energy\ndata",
"_____no_output_____"
]
],
[
[
"adding array data:",
"_____no_output_____"
]
],
[
[
"pixval = np.arange(5*10).reshape(10,5)\nprint pixval\ntry:\n data['PIXVAL']=pixval\nexcept Exception,e:\n print \"FAILED:\",e\ndata",
"[[ 0 1 2 3 4]\n [ 5 6 7 8 9]\n [10 11 12 13 14]\n [15 16 17 18 19]\n [20 21 22 23 24]\n [25 26 27 28 29]\n [30 31 32 33 34]\n [35 36 37 38 39]\n [40 41 42 43 44]\n [45 46 47 48 49]]\nFAILED: Wrong number of items passed 5, placement implies 1\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb8c7c213e60c854605c355c41e11dc2f2472abd
| 36,805 |
ipynb
|
Jupyter Notebook
|
notebooks/QCB_chems_small/evaluation.ipynb
|
joewandy/vimms-gym
|
95cb6fa84ee6e3a64618b7a2a54c3835ad0d7867
|
[
"MIT"
] | null | null | null |
notebooks/QCB_chems_small/evaluation.ipynb
|
joewandy/vimms-gym
|
95cb6fa84ee6e3a64618b7a2a54c3835ad0d7867
|
[
"MIT"
] | null | null | null |
notebooks/QCB_chems_small/evaluation.ipynb
|
joewandy/vimms-gym
|
95cb6fa84ee6e3a64618b7a2a54c3835ad0d7867
|
[
"MIT"
] | null | null | null | 45.159509 | 1,880 | 0.608015 |
[
[
[
"%matplotlib inline\n\n%load_ext autoreload\n%autoreload 2",
"_____no_output_____"
],
[
"import os\nimport sys\nfrom os.path import exists\n\nsys.path.append('../..')",
"_____no_output_____"
],
[
"import pylab as plt\nimport pandas as pd\nimport numpy as np\nfrom loguru import logger\nimport seaborn as sns\n\nfrom stable_baselines3 import PPO, DQN",
"_____no_output_____"
],
[
"from vimms.Common import POSITIVE, set_log_level_warning, load_obj, save_obj\nfrom vimms.ChemicalSamplers import UniformRTAndIntensitySampler, GaussianChromatogramSampler, UniformMZFormulaSampler, \\\n MZMLFormulaSampler, MZMLRTandIntensitySampler, MZMLChromatogramSampler\nfrom vimms.Noise import UniformSpikeNoise\nfrom vimms.Evaluation import evaluate_real\nfrom vimms.Chemicals import ChemicalMixtureFromMZML\nfrom vimms.Roi import RoiBuilderParams, SmartRoiParams\n\nfrom mass_spec_utils.data_import.mzmine import load_picked_boxes\n\nfrom vimms_gym.env import DDAEnv\nfrom vimms_gym.chemicals import generate_chemicals\nfrom vimms_gym.evaluation import evaluate, run_method\nfrom vimms_gym.common import METHOD_RANDOM, METHOD_FULLSCAN, METHOD_TOPN, METHOD_PPO, METHOD_DQN",
"_____no_output_____"
]
],
[
[
"# 1. Parameters",
"_____no_output_____"
]
],
[
[
"n_chemicals = (20, 50)\nmz_range = (100, 110)\nrt_range = (0, 1440)\nintensity_range = (1E4, 1E20)",
"_____no_output_____"
],
[
"min_mz = mz_range[0]\nmax_mz = mz_range[1]\nmin_rt = rt_range[0]\nmax_rt = rt_range[1]\nmin_log_intensity = np.log(intensity_range[0])\nmax_log_intensity = np.log(intensity_range[1])",
"_____no_output_____"
],
[
"isolation_window = 0.7\nN = 10\nrt_tol = 120\nexclusion_t_0 = 15\nmz_tol = 10\nmin_ms1_intensity = 5000\nionisation_mode = POSITIVE\n\nenable_spike_noise = True\nnoise_density = 0.1\nnoise_max_val = 1E3",
"_____no_output_____"
],
[
"mzml_filename = '../fullscan_QCB.mzML'\nsamplers = None\nsamplers_pickle = 'samplers_fullscan_QCB_small.mzML.p'\nif exists(samplers_pickle):\n logger.info('Loaded %s' % samplers_pickle)\n samplers = load_obj(samplers_pickle)\n mz_sampler = samplers['mz']\n ri_sampler = samplers['rt_intensity']\n cr_sampler = samplers['chromatogram']\nelse:\n logger.info('Creating samplers from %s' % mzml_filename)\n mz_sampler = MZMLFormulaSampler(mzml_filename, min_mz=min_mz, max_mz=max_mz)\n ri_sampler = MZMLRTandIntensitySampler(mzml_filename, min_rt=min_rt, max_rt=max_rt,\n min_log_intensity=min_log_intensity,\n max_log_intensity=max_log_intensity)\n roi_params = RoiBuilderParams(min_roi_length=3, at_least_one_point_above=1000)\n cr_sampler = MZMLChromatogramSampler(mzml_filename, roi_params=roi_params)\n samplers = {\n 'mz': mz_sampler,\n 'rt_intensity': ri_sampler,\n 'chromatogram': cr_sampler\n }\n save_obj(samplers, samplers_pickle)",
"2022-06-13 12:29:13.904 | INFO | __main__:<module>:11 - Creating samplers from ../fullscan_QCB.mzML\n2022-06-13 12:29:15.091 | DEBUG | mass_spec_utils.data_import.mzml:_load_file:166 - Loaded 2471 scans\n2022-06-13 12:29:17.763 | DEBUG | mass_spec_utils.data_import.mzml:_load_file:166 - Loaded 2471 scans\n2022-06-13 12:32:19.012 | DEBUG | vimms.ChemicalSamplers:_extract_rois:491 - Extracted 43107 good ROIs from ../fullscan_QCB.mzML\n2022-06-13 12:32:19.013 | INFO | vimms.Common:save_obj:410 - Saving <class 'dict'> to ../samplers_fullscan_QCB_small.mzML.p\n"
],
[
"params = {\n 'chemical_creator': {\n 'mz_range': mz_range,\n 'rt_range': rt_range,\n 'intensity_range': intensity_range,\n 'n_chemicals': n_chemicals,\n 'mz_sampler': mz_sampler,\n 'ri_sampler': ri_sampler,\n 'cr_sampler': GaussianChromatogramSampler(),\n },\n 'noise': {\n 'enable_spike_noise': enable_spike_noise,\n 'noise_density': noise_density,\n 'noise_max_val': noise_max_val,\n 'mz_range': mz_range\n },\n 'env': {\n 'ionisation_mode': ionisation_mode,\n 'rt_range': rt_range,\n 'isolation_window': isolation_window,\n 'mz_tol': mz_tol,\n 'rt_tol': rt_tol,\n }\n}",
"_____no_output_____"
],
[
"max_peaks = 200\nin_dir = 'results'",
"_____no_output_____"
],
[
"n_eval_episodes = 1\ndeterministic = True",
"_____no_output_____"
]
],
[
[
"# 2. Evaluation",
"_____no_output_____"
],
[
"#### Generate some chemical sets",
"_____no_output_____"
]
],
[
[
"set_log_level_warning()",
"_____no_output_____"
],
[
"eval_dir = 'evaluation'\nmethods = [\n METHOD_TOPN,\n METHOD_RANDOM, \n METHOD_PPO,\n]",
"_____no_output_____"
],
[
"chemical_creator_params = params['chemical_creator']\n\nchem_list = []\nfor i in range(n_eval_episodes):\n print(i)\n chems = generate_chemicals(chemical_creator_params)\n chem_list.append(chems)",
"0\n"
]
],
[
[
"#### Run different methods",
"_____no_output_____"
]
],
[
[
"for chems in chem_list:\n print(len(chems))",
"21\n"
],
[
"max_peaks",
"_____no_output_____"
],
[
"out_dir = eval_dir\nin_dir, out_dir",
"_____no_output_____"
]
],
[
[
"#### Compare to Top-10",
"_____no_output_____"
]
],
[
[
"env_name = 'DDAEnv'\nmodel_name = 'PPO'\nintensity_threshold = 0.5",
"_____no_output_____"
],
[
"topN_N = 20\ntopN_rt_tol = 30",
"_____no_output_____"
],
[
"method_eval_results = {}\nfor method in methods:\n\n effective_rt_tol = rt_tol\n copy_params = dict(params) \n copy_params['env']['rt_tol'] = effective_rt_tol\n \n if method == METHOD_PPO:\n fname = os.path.join(in_dir, '%s_%s.zip' % (env_name, model_name))\n model = PPO.load(fname)\n elif method == METHOD_DQN:\n fname = os.path.join(in_dir, '%s_%s.zip' % (env_name, model_name))\n model = DQN.load(fname)\n else:\n model = None\n if method == METHOD_TOPN:\n N = topN_N\n effective_rt_tol = topN_rt_tol\n copy_params = dict(params) \n copy_params['env']['rt_tol'] = effective_rt_tol \n\n banner = 'method = %s max_peaks = %d N = %d rt_tol = %d' % (method, max_peaks, N, effective_rt_tol)\n print(banner)\n print() \n \n episodic_results = run_method(env_name, copy_params, max_peaks, chem_list, method, out_dir, \n N=N, min_ms1_intensity=min_ms1_intensity, model=model,\n print_eval=True, print_reward=False, intensity_threshold=intensity_threshold)\n eval_results = [er.eval_res for er in episodic_results]\n method_eval_results[method] = eval_results\n print()",
"method = topN max_peaks = 200 N = 20 rt_tol = 30\n\n{'coverage_prop': '1.000', 'intensity_prop': '0.481', 'ms1/ms2 ratio': '123.655', 'efficiency': '0.724', 'TP': '4', 'FP': '15', 'FN': '2', 'precision': '0.211', 'recall': '0.667', 'f1': '0.320'}\n\nmethod = random max_peaks = 200 N = 20 rt_tol = 120\n\n{'coverage_prop': '1.000', 'intensity_prop': '0.999', 'ms1/ms2 ratio': '0.654', 'efficiency': '0.007', 'TP': '19', 'FP': '1', 'FN': '1', 'precision': '0.950', 'recall': '0.950', 'f1': '0.950'}\n\n"
]
],
[
[
"#### Test classic controllers in ViMMS",
"_____no_output_____"
]
],
[
[
"from vimms.MassSpec import IndependentMassSpectrometer\nfrom vimms.Controller import TopNController, TopN_SmartRoiController, WeightedDEWController\nfrom vimms.Environment import Environment",
"_____no_output_____"
]
],
[
[
"Run Top-N Controller",
"_____no_output_____"
]
],
[
[
"method = 'TopN_Controller'\nprint('method = %s' % method)\nprint()\n\neffective_rt_tol = topN_rt_tol\neffective_N = topN_N\neval_results = []\nfor i in range(len(chem_list)):\n \n spike_noise = None\n if enable_spike_noise:\n noise_params = params['noise']\n noise_density = noise_params['noise_density']\n noise_max_val = noise_params['noise_max_val']\n noise_min_mz = noise_params['mz_range'][0]\n noise_max_mz = noise_params['mz_range'][1]\n spike_noise = UniformSpikeNoise(noise_density, noise_max_val, min_mz=noise_min_mz,\n max_mz=noise_max_mz)\n\n chems = chem_list[i]\n mass_spec = IndependentMassSpectrometer(ionisation_mode, chems, spike_noise=spike_noise)\n controller = TopNController(ionisation_mode, effective_N, isolation_window, mz_tol, effective_rt_tol,\n min_ms1_intensity)\n env = Environment(mass_spec, controller, min_rt, max_rt, progress_bar=False, out_dir=out_dir,\n out_file='%s_%d.mzML' % (method, i), save_eval=True)\n env.run()\n \n eval_res = evaluate(env, intensity_threshold)\n eval_results.append(eval_res)\n print('Episode %d finished' % i)\n print(eval_res)\n \nmethod_eval_results[method] = eval_results",
"_____no_output_____"
]
],
[
[
"Run SmartROI Controller",
"_____no_output_____"
]
],
[
[
"alpha = 2\nbeta = 0.1\nsmartroi_N = 20\nsmartroi_dew = 15",
"_____no_output_____"
],
[
"method = 'SmartROI_Controller'\nprint('method = %s' % method)\nprint()\n\neffective_rt_tol = exclusion_t_0\neval_results = []\nfor i in range(len(chem_list)):\n \n spike_noise = None\n if enable_spike_noise:\n noise_params = params['noise']\n noise_density = noise_params['noise_density']\n noise_max_val = noise_params['noise_max_val']\n noise_min_mz = noise_params['mz_range'][0]\n noise_max_mz = noise_params['mz_range'][1]\n spike_noise = UniformSpikeNoise(noise_density, noise_max_val, min_mz=noise_min_mz,\n max_mz=noise_max_mz)\n\n chems = chem_list[i]\n mass_spec = IndependentMassSpectrometer(ionisation_mode, chems, spike_noise=spike_noise)\n \n roi_params = RoiBuilderParams(min_roi_intensity=500, min_roi_length=0) \n smartroi_params = SmartRoiParams(intensity_increase_factor=alpha, drop_perc=beta/100.0)\n controller = TopN_SmartRoiController(ionisation_mode, isolation_window, smartroi_N, mz_tol, smartroi_dew,\n min_ms1_intensity, roi_params, smartroi_params)\n env = Environment(mass_spec, controller, min_rt, max_rt, progress_bar=False, out_dir=out_dir,\n out_file='%s_%d.mzML' % (method, i), save_eval=True)\n env.run()\n \n eval_res = evaluate(env, intensity_threshold)\n eval_results.append(eval_res)\n print('Episode %d finished' % i)\n print(eval_res)\n \nmethod_eval_results[method] = eval_results",
"_____no_output_____"
]
],
[
[
"Run WeightedDEW Controller",
"_____no_output_____"
]
],
[
[
"t0 = 15\nt1 = 60\nweighteddew_N = 20",
"_____no_output_____"
],
[
"method = 'WeightedDEW_Controller'\nprint('method = %s' % method)\nprint()\n\neval_results = []\nfor i in range(len(chem_list)):\n \n spike_noise = None\n if enable_spike_noise:\n noise_params = params['noise']\n noise_density = noise_params['noise_density']\n noise_max_val = noise_params['noise_max_val']\n noise_min_mz = noise_params['mz_range'][0]\n noise_max_mz = noise_params['mz_range'][1]\n spike_noise = UniformSpikeNoise(noise_density, noise_max_val, min_mz=noise_min_mz,\n max_mz=noise_max_mz)\n\n chems = chem_list[i]\n mass_spec = IndependentMassSpectrometer(ionisation_mode, chems, spike_noise=spike_noise)\n \n controller = WeightedDEWController(ionisation_mode, weighteddew_N, isolation_window, mz_tol, t1,\n min_ms1_intensity, exclusion_t_0=t0)\n env = Environment(mass_spec, controller, min_rt, max_rt, progress_bar=False, out_dir=out_dir,\n out_file='%s_%d.mzML' % (method, i), save_eval=True)\n env.run()\n \n eval_res = evaluate(env, intensity_threshold)\n eval_results.append(eval_res)\n print('Episode %d finished' % i)\n print(eval_res)\n \nmethod_eval_results[method] = eval_results",
"_____no_output_____"
]
],
[
[
"#### Plotting",
"_____no_output_____"
],
[
"Flatten data into dataframe",
"_____no_output_____"
]
],
[
[
"data = []\nfor method in method_eval_results:\n eval_results = method_eval_results[method]\n for eval_res in eval_results:\n row = (\n method, \n float(eval_res['coverage_prop']), \n float(eval_res['intensity_prop']), \n float(eval_res['ms1/ms2 ratio']), \n float(eval_res['efficiency']),\n float(eval_res['precision']),\n float(eval_res['recall']),\n float(eval_res['f1']), \n )\n data.append(row)\n \ndf = pd.DataFrame(data, columns=['method', 'coverage_prop', 'intensity_prop', 'ms1/ms2_ratio', 'efficiency', 'precision', 'recall', 'f1'])\n# df.set_index('method', inplace=True)\ndf.head()",
"_____no_output_____"
],
[
"sns.set_context(\"poster\")",
"_____no_output_____"
],
[
"plt.figure(figsize=(10, 5))\nsns.boxplot(data=df, x='method', y='coverage_prop')\nplt.xticks(rotation=90)\nplt.title('Coverage Proportion')",
"_____no_output_____"
],
[
"plt.figure(figsize=(10, 5))\nsns.boxplot(data=df, x='method', y='intensity_prop')\nplt.xticks(rotation=90)\nplt.title('Intensity Proportion')",
"_____no_output_____"
],
[
"plt.figure(figsize=(10, 5))\nsns.boxplot(data=df, x='method', y='ms1/ms2_ratio')\nplt.xticks(rotation=90)\nplt.title('MS1/MS2 Ratio')",
"_____no_output_____"
],
[
"plt.figure(figsize=(10, 5))\nsns.boxplot(data=df, x='method', y='efficiency')\nplt.xticks(rotation=90)\nplt.title('Efficiency')",
"_____no_output_____"
],
[
"plt.figure(figsize=(10, 5))\nsns.boxplot(data=df, x='method', y='precision')\nplt.xticks(rotation=90)\nplt.title('Precision')",
"_____no_output_____"
],
[
"plt.figure(figsize=(10, 5))\nsns.boxplot(data=df, x='method', y='recall')\nplt.xticks(rotation=90)\nplt.title('Recall')",
"_____no_output_____"
],
[
"plt.figure(figsize=(10, 5))\nsns.boxplot(data=df, x='method', y='f1')\nplt.xticks(rotation=90)\nplt.title('F1')",
"_____no_output_____"
],
[
"df.to_pickle('evaluation.p')",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb8c7e19ef1982a921d66328f70285775ecf0575
| 4,202 |
ipynb
|
Jupyter Notebook
|
rosette/Converting ADM TO LightTag Suggestions On Sixgill.ipynb
|
LightTag/lighttag-to-adm
|
00d44fc89ca92d2a77bae628d050d5aae94645c6
|
[
"MIT"
] | 1 |
2021-05-20T11:51:19.000Z
|
2021-05-20T11:51:19.000Z
|
rosette/Converting ADM TO LightTag Suggestions On Sixgill.ipynb
|
LightTag/lighttag-to-adm
|
00d44fc89ca92d2a77bae628d050d5aae94645c6
|
[
"MIT"
] | null | null | null |
rosette/Converting ADM TO LightTag Suggestions On Sixgill.ipynb
|
LightTag/lighttag-to-adm
|
00d44fc89ca92d2a77bae628d050d5aae94645c6
|
[
"MIT"
] | null | null | null | 26.764331 | 261 | 0.600428 |
[
[
[
"from lighttag_to_adm import convert_lighttag_example_to_adm\nfrom adm_to_lighttag import adm_doc_to_lighttag_suggetions,adm_document_list_to_lighttag_suggestions\nimport json\nimport os\nimport pandas as pd",
"_____no_output_____"
]
],
[
[
"## Get The ADMS\nIn this case we'll just make adms from LightTag data. You probably have ADMs in a file or in memory",
"_____no_output_____"
],
[
"## Notes on the adm_document_list_to_lighttag_suggestions function\nThis function takes two optional callback arguments\n\n* **example_id_fn** extracts the lighttag example id from the ADM Doc\n\n* **lighttag_tag_name_extractor** resolves the LightTag tag name of the entity\n\nBoth of these have defaults defined in adm_to_lighttag\n",
"_____no_output_____"
]
],
[
[
"adms = []\nfor root,d,files in os.walk('/tmp/sixgill/'):\n for f in files:\n if f.endswith('adm.json'):\n adms.append(json.load(open(os.path.join(root,f))))\n \nseen_examples = adm_document_list_to_lighttag_suggestions(adms)",
"_____no_output_____"
],
[
"suggestions = seen_examples['suggestions']\n\nseen_example_ids = seen_examples['seen_example_ids']",
"_____no_output_____"
]
],
[
[
"# TODO For Sixgill/ Nadav\n * Notice that the example_id is a list, please modify the example_id_fn to match your branch\n * Implement the lighttag_tag_name_extractor to get the lighttag tag name\n * Is Basis sending more tags than what is in your schema ? We'll reject those ",
"_____no_output_____"
]
],
[
[
"pd.DataFrame(suggestions)",
"_____no_output_____"
]
],
[
[
"# Sanity Check conflicts which LightTag might reject\n\nWe'll run the conflict resolveer, if you get any warnings here than you need to check why you have overlapping suggestions. ",
"_____no_output_____"
]
],
[
[
"import copy\nfrom itertools import groupby\npseudo_annotations = []\nfor sug in suggestions:\n x = copy.copy(sug)\n x['annotated_by'] = [\"my_model\"]\n pseudo_annotations.append(x)\n\npseudos_by_example = groupby(pseudo_annotations,key=lambda x:x['example_id'])\n",
"_____no_output_____"
],
[
"from conflict_resolver import resolve_annotation_conflicts\nfor example_id, example_suggestions in pseudos_by_example:\n resolve_annotation_conflicts(list(example_suggestions))\n",
"_____no_output_____"
]
],
[
[
"## Now Upload The Suggestions as Usual",
"_____no_output_____"
],
[
"From this point, you need to [register the model in LightTag](https://guide.lighttag.io/suggestions/suggestions/#2.-Registering-a-SuggestionModel) and [upload the suggestions ](https://guide.lighttag.io/suggestions/suggestions/#4.-Upload-Your-Suggestions)",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
]
] |
cb8c7ff162a5265ecf52565f7c935e18cc8d2c9d
| 123,400 |
ipynb
|
Jupyter Notebook
|
MRC_Notebook/SBM_rewiring_example.ipynb
|
jeffalstott/network_clustering_growth
|
e3b2552f692236bb5a579965cf8aae387d96ac67
|
[
"MIT"
] | null | null | null |
MRC_Notebook/SBM_rewiring_example.ipynb
|
jeffalstott/network_clustering_growth
|
e3b2552f692236bb5a579965cf8aae387d96ac67
|
[
"MIT"
] | null | null | null |
MRC_Notebook/SBM_rewiring_example.ipynb
|
jeffalstott/network_clustering_growth
|
e3b2552f692236bb5a579965cf8aae387d96ac67
|
[
"MIT"
] | null | null | null | 642.708333 | 64,955 | 0.933947 |
[
[
[
"empty"
]
]
] |
[
"empty"
] |
[
[
"empty"
]
] |
cb8c8816ab7cedae523907242c44e63d510bbb69
| 695,214 |
ipynb
|
Jupyter Notebook
|
book/docs/lectures/stochastic_processes/random_walks_and_markov_processes.ipynb
|
finsberg/IN1910_H21
|
4bd8f49c0f6839884bfaf8a0b1e3a717c3041092
|
[
"CC-BY-4.0"
] | null | null | null |
book/docs/lectures/stochastic_processes/random_walks_and_markov_processes.ipynb
|
finsberg/IN1910_H21
|
4bd8f49c0f6839884bfaf8a0b1e3a717c3041092
|
[
"CC-BY-4.0"
] | null | null | null |
book/docs/lectures/stochastic_processes/random_walks_and_markov_processes.ipynb
|
finsberg/IN1910_H21
|
4bd8f49c0f6839884bfaf8a0b1e3a717c3041092
|
[
"CC-BY-4.0"
] | 2 |
2021-08-30T12:38:40.000Z
|
2021-11-05T14:14:59.000Z
| 590.6661 | 149,816 | 0.94122 |
[
[
[
"# Random Walks\n\nThis week we will discuss a new topic, *random walks*. Random walks are an example of a markov process, and we will also learn what this means, and how we can analyze the behavior of the random walker using a markov chain.\n\nThe exercises this week are slightly more extensive then other weeks, and is more of a project based work than earlier exercise sets as well. This is because the plan is to cover some of these exercises in L20, i.e., the lecture on Friday November 8th. It it therefore recommended that you work on the exercises before Thursday. If you cannot attend the lecture on Friday, it is strongly recommended to take a good look at the example solutions, which I will upload during Friday's lecture.",
"_____no_output_____"
],
[
"## Random Walks\n\nA random walk is a process where we follow some object taking *random steps*. The path the object walks then defines a random path, or trajectory. Random walks are powerful mathematical objects with a large number of use cases, but we will return to this point later, for now let us look at some actual random walks.",
"_____no_output_____"
],
[
"### The 1D Random Walker\n\nA random walk can refer to many different processes, but let us start of with perhaps the simplest of them all, a 1D random walk on a regular grid. Assume some walker starts of at $x=0$. Now it takes steps to the left or right at random, with equal probability.\n<img src=\"fig/1D_walk.png\" width=600>\n\nWe denote the position of the walker after $N$ steps by $X_N$. Because the walker is taking random steps, $X_N$ is what we call a *random* or *stochastic variable*, it won't have a specific value in general, but be different for each specific random walk, depending on what steps are actually taken.\n\nFor each step the walker takes, we move either 1 step to the left, or 1 step to the right. Thus\n\n$$X_{N+1} = X_{N} + K_N,$$\n\nwhere $K_N$ is the $N$'th step taken. We assume that all steps are independent of all others, and that each step has an equal chance of being to the left or to the right, so \n$$K_N = \\begin{cases}\n1 & \\mbox{with 50} \\% \\mbox{ chance} \\\\\n-1 & \\mbox{with 50}\\% \\mbox{ chance}\n\\end{cases}$$\n\nLet us look at how a random walk looks. To draw the step $K_N$ using numpy, we use `np.random.randint(2)`, but this gives us 0 or 1, so we instead use `2*np.random.randint(2) - 1`, which will then give us -1 or 1 with equal probability.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\nnp.random.seed(12345)\n\nnr_steps = 10\nX = np.zeros(nr_steps+1)\n\nX[0] = 0\nfor N in range(nr_steps):\n X[N+1] = X[N] + 2*np.random.randint(2) - 1\n \nplt.plot(range(nr_steps+1), X)\nplt.xlabel('Nr of steps taken')\nplt.ylabel(r'$X_N$')\nplt.show()\n",
"_____no_output_____"
]
],
[
[
"Simply using `plt.plot` here can be a bit misleading, so we can alternatively change the plotstyle, or we can change to use the `plt.step` function instead:",
"_____no_output_____"
]
],
[
[
"plt.step(range(nr_steps+1), X, where='mid')\nplt.xlabel('Nr of steps taken')\nplt.ylabel(r'$X_N$')\nplt.show()",
"_____no_output_____"
],
[
"nr_steps = 1000\nX = np.zeros(nr_steps+1)\n\nX[0] = 0\nfor N in range(nr_steps):\n X[N+1] = X[N] + 2*np.random.randint(2) - 1\n\nplt.step(range(nr_steps+1), X, where='mid')\nplt.xlabel('Nr of steps taken')\nplt.ylabel('Displacement')\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Vectorized Random Walk\n\nAs we saw last week, if we want to repeatedly draw and use random numbers, `np.random` can be used in a vectorized way to be more efficient. Let us see how we can do this for a random walk.\n\nDrawing the steps themselves is straight forward:",
"_____no_output_____"
]
],
[
[
"nr_steps = 1000\nsteps = 2*np.random.randint(2, size=nr_steps) - 1",
"_____no_output_____"
]
],
[
[
"But now we need to combine these steps into the variable $X_N$. Now, if we only want to know the final displacement after all the steps, then we could simply do the sum\n$$X_{1000} = \\sum_{i=1}^{1000} K_i.$$\nHowever, if we want to plot out the full trajectory of the walk, then we need to compute all the partial sums as well, i.e., find $X_N$ for $N=1, 2, 3, \\ldots 1000.$\n\nWe can do this with the function `np.cumsum`, which stands for *cumulative sum*. Taking the cumulative sum of a sequence gives a new sequence where element $n$ of the new sequence is the sum of the first $n$ elements of the input. Thus, the cumulative sum of $K_N$ will give $X_N$.",
"_____no_output_____"
]
],
[
[
"X = np.zeros(nr_steps + 1)\nX[0] = 0\nX[1:] = X[0] + np.cumsum(steps)",
"_____no_output_____"
]
],
[
[
"Note that we could have simply said `X = np.cumsum(steps)`, but in that case, $X_0$ wouldn't be 0, it would be -1 or 1. That's not a big deal, but we take the extra step of defining $X_0 = 0$, and then finding the rest of $X_N$ for $N > 100$.",
"_____no_output_____"
]
],
[
[
"plt.step(range(nr_steps+1), X, where='mid')\nplt.xlabel('Nr of steps taken')\nplt.ylabel('Displacement')\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Many Walkers\n\nBecause the walker is completely random, understanding how it behaves from looking at a single walker isn't that useful. Instead, we can look at a large *ensemble* of walkers, and then perhaps we can gleam some insight into how they behave.\n\nWe can also use the vectorization of `np.random` to draw the walks of many different walkers in a vectorized manner:",
"_____no_output_____"
]
],
[
[
"nr_steps = 100\nwalkers = 5\n\nX = np.zeros((nr_steps+1, walkers))\nX[0, :] = 0\n\nsteps = 2*np.random.randint(2, size=(nr_steps, walkers)) - 1\nX[1:, :] = np.cumsum(steps, axis=0)\n\nplt.plot(X)\nplt.xlabel('Nr of Steps')\nplt.ylabel('Displacement')\nplt.show()",
"_____no_output_____"
]
],
[
[
"Or with many more steps:",
"_____no_output_____"
]
],
[
[
"nr_steps = 10000\nwalkers = 5\n\nX = np.zeros((nr_steps+1, walkers))\nX[0, :] = 0\n\nsteps = 2*np.random.randint(2, size=(nr_steps, walkers)) - 1\nX[1:, :] = np.cumsum(steps, axis=0)\n\nplt.plot(X, linewidth=0.5)\nplt.xlabel('Nr of Steps')\nplt.ylabel('Displacement')\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Very Many Walkers\n\nWe have now seen how we can plot 5 walkers. But if we really want to understand the average behavior, we might want to plot a lot more walkers. With our code, this works just fine, but the output won't tell us to much, because it will become too chaotic:",
"_____no_output_____"
]
],
[
[
"nr_steps = 1000\nwalkers = 1000\n\nX = np.zeros((nr_steps+1, walkers))\nX[0, :] = 0\n\nsteps = 2*np.random.randint(2, size=(nr_steps, walkers)) - 1\nX[1:, :] = np.cumsum(steps, axis=0)\n\nplt.plot(X)\nplt.xlabel('Nr of Steps')\nplt.ylabel('Displacement')\nplt.show()",
"_____no_output_____"
]
],
[
[
"This plot shows a thousand random walks overlaying each other, but we cannot really see what is going on, because the different lines simply overlap and hide each other.\n\nTo fix this, instead of plotting all the walks over each other, we plot the *density* of walkers. We can accomplish this by using the `alpha` keyword to `plt.plot`. This keyword is used to make a line semi-transparent . Here, `alpha=1` is the default, non-transparent line, `alpha=0` is a completely transparent, and thus invisible, line. If we then set for example `alpha=0.1`, we get 10% transparent lines. \n\nWith semi-transparent lines, anywhere many lines overlap will give a strong color, if there are fewer lines, we get a weaker color. To emphasise this, let us also only plot black lines, and ignore colors.",
"_____no_output_____"
]
],
[
[
"nr_steps = 1000\nwalkers = 1000\n\nX = np.zeros((nr_steps+1, walkers))\nX[0, :] = 0\n\nsteps = 2*np.random.randint(2, size=(nr_steps, walkers)) - 1\nX[1:, :] = np.cumsum(steps, axis=0)\n\nplt.plot(X, alpha=0.01, color='k')\nplt.xlabel('Nr of Steps')\nplt.ylabel('Displacement')\nplt.axis((0, 1000, -100, 100))\nplt.show()",
"_____no_output_____"
]
],
[
[
"At the beginning, all the walkers are close to the origin, as they simply have not had time to get further away. As time progresses towards the right, the walkers spread out. The highest density of walkers is still found in the middle however, as the net sum of steps will tend towards a mean of 0.",
"_____no_output_____"
],
[
"### Analyzing the average behavior of a walker\n\nBecause the random walk is a random process, prediciting how a single walker will move is impossible. However, if we instead look at a lot of walkers, we can analyze their *average* activity. Because of the law of large numbers, we know that the average behavior for a large number of walkers will converge to a specific behavior.\n\nCompare for example the first plot of a single walker. If you rerun this code, you will get a dramatically different behavior, because one specific walk looks very different from one specific different walk. For the last Figure we made however, rerunning the code won't change much, because the average behavior of 1000 walkers will tend to be the same.\n\nOne way to explore the average behavior is of course to do simulation, and then simply taking the sample average. For more complex random walk behaviors, this is the only option. Our random walk however, is quite simple, and so we can also analyze it mathematically. Let us do this.\n\n#### Average Displacement\n\nFirst we want to know the average displacement of a large number of walkers? For a single walker, the position of the walker after $N$ steps was given by\n$$X_N = X_0 + \\sum_{i=1}^N K_i.$$\nor alternatively:\n$$X_{N+1} = X_{N} + K_N.$$\n\nNow, we want to compute the *average* of this variable, which we will denote $\\langle X_N \\rangle$, another word for this value is the *expected value* or *expectation*. If you have not heard these terms before, simply think of the value as the average of a large number of walkers.\n\nTaking the average of the $X_N$ gives:\n\n$$\\langle X_{N+1} \\rangle = \\langle X_N + K_N \\rangle.$$\n\nHowever, taking an average is a linear operation, and so we can split the right hand side into\n\n$$\\langle X_{N+1} \\rangle = \\langle X_N \\rangle + \\langle K_N \\rangle.$$\n\nNow, we don't know $\\langle X_{N} \\rangle,$ because this is what we are actually trying to find. However, $\\langle K_N \\rangle$, we know, because it is simply the average of the two outcomes:\n$$\\langle K_N \\rangle = \\frac{1}{2}\\cdot1 + \\frac{1}{2}\\cdot (-1) = \\frac{1}{2} - \\frac{1}{2} = 0.$$\n\nBecause there is an equal chance of taking a step to the left and the right, the *average* displacement for a single step will be 0. Inserting this gives\n\n$$\\langle X_{N+1} \\rangle = \\langle X_{N} \\rangle.$$\n\nIf the walkers start in $X_0 = 0$, then $\\langle X_0 \\rangle =0$, which in turn implies $\\langle X_1 = 0$, and then $\\langle X_2 \\rangle = 0$ and so on. Giving \n\n$$\\langle X_{N}\\rangle = 0.$$\n\nThis expression tells us that the average displacement of a large number of walkers will be 0, no matter how many steps they take. Is this not surprising? We have seen that the more steps the walkers take, the longer away from the origin they will tend to move, so why is the average 0?\n\nThe average is 0 because we are looking at a completely *uniform* and symmetric walker. The walkers have an equal chance of moving left, or right, from the origin, and the average will therefore tend to be 0, even if the walkers move away from the origin.",
"_____no_output_____"
],
[
"#### Averaged Square Displacement\n\nThe average displacement became 0 because the problem is completely symmetric. However, if we now instead look at the squared displacement $X_N^2$, we get a better feel for how far away from the origin things move, because the square is positive regardless of wether the walker moves away in the positive or negative direction.\n\nWe can write out an expression for $X_{N+1}^2$ as \n\n$$X_{N+1}^2 = (X_{N} + K_N)^2 = X_{N}^2 + 2X_N \\cdot K_N + K_N^2.$$\n\nAgain we care about the average, so we take the average of this expression:\n\n$$\\langle X_{N+1}^2 \\rangle = \\langle X_{N}^2 \\rangle + 2\\langle X_N \\cdot K_N \\rangle + \\langle K_N^2 \\rangle.$$\n\nNow, the term $\\langle X_N \\cdot K_N \\rangle$ will again be zero, because $K_N$ is independent of $X_N$ and has an equal chance of being positive and negative. So we get\n$$\\langle X_{N+1}^2 \\rangle = \\langle X_N^2 \\rangle + \\langle K_N^2 \\rangle.$$\nLet us compute $\\langle K_N^2 \\rangle$:\n$$\\langle K_N^2 \\rangle = \\frac{1}{2}(1)^2 + \\frac{1}{2}(-1)^2 = \\frac{1}{2} + \\frac{1}{2} = 1.$$\nThus we get\n\n$$\\langle X_{N+1}^2 \\rangle = \\langle X_N^2 \\rangle + 1.$$\n\nIf we say that $X_0 = 0$, we then get that $\\langle X_1 \\rangle = 1$, $\\langle X_2 \\rangle = 2$, and so on:\n\n$$\\langle X_N^2 \\rangle = N.$$\n\nSo we see that while the average displacement does not change over time: $\\langle X_N \\rangle = 0$, the average squared displacement does! In fact, the squared displacement grows linearily with the number of steps $N$. The longer a random walk carries on for, the further away from the origin the walker will tend to move.\n\nThis expression also tells us that the *variance* of the walkers, because the variance of random variable can always be written as \n\n$$\\text{Var}(X_N) = \\langle X_N^2 \\rangle - \\langle X_N \\rangle^2,$$\nand so in this case\n$$\\text{Var}(X_N) = N - 0^2 = N.$$\nSo thar variance of $X_N$ is also $N$.",
"_____no_output_____"
],
[
"#### Root Mean Square Displacement\n\nWhile it is clear from the expression \n$$\\langle X_N^2 \\rangle = N,$$\n\nthat the walkers will tend to move further away from the origin, this is the *squared* displacement. A more intuitive quantity would perhaps be the average absolute *displacement*, i.e., $\\langle |X_N| \\rangle$. This would be a useful quantity, but it turns out to be a bit tricky to compute.\n\nAs an easier solution, we just take the root of the mean squared displacement:\n\n$$\\text{RMS} = \\sqrt{\\langle X_N^2 \\rangle} = \\sqrt{N}.$$\n\nThis quantity is known as the *root mean square* displacement (RMS). It won't be exactly the same as $\\langle |X_N| \\rangle$, but it will be close to it.\n\nBecause the root mean square displacement grows as $\\sqrt{N}$, we see that a 1D random walker will tend to be about $\\sqrt{N}$ away from the origin after taking $N$ steps.\n",
"_____no_output_____"
],
[
"### Plotting the RMS\n\nLet us verify our statement. We repeat our density plot with 1000 walkers, but now we also plot in our expression for the RMS: $\\sqrt{N}$:",
"_____no_output_____"
]
],
[
[
"N = 1000\nwalkers = 1000\n\nk = 2*np.random.randint(2, size=(N, walkers)) - 1\nX = np.cumsum(k, axis=0)\n\nplt.plot(X, alpha=0.01, color='k')\nplt.plot(range(N), np.sqrt(2*np.arange(N)), color='C1')\nplt.plot(range(N), -np.sqrt(2*np.arange(N)), color='C1')\nplt.xlabel('Nr of Steps')\nplt.ylabel('Displacement')\nplt.axis((0, 1000, -100, 100))\nplt.show()",
"_____no_output_____"
]
],
[
[
"We see that the density of walkers inside the RMS curves is higher than outside it. This makes sense, because the root-mean-square will tend to give outliers more weight. The RMS curves still seem very reasonable, as they clearly indicate the rough region where most walkers will be found. We also see that the scaling seems reasonable.\n\nInstead of plotting it, we can also compute the actual root-mean-square of our 1000 walkers, which is then a *sample mean*, and compare it to our analytical expresison.",
"_____no_output_____"
]
],
[
[
"N = 1000\nwalkers = 1000\n\nk = 2*np.random.randint(2, size=(N, walkers)) - 1\nX = np.cumsum(k, axis=0)\n\nRMS = np.sqrt(np.mean(X**2, axis=1))\n\nplt.plot(np.arange(N), np.sqrt(np.arange(N)), '--', label=\"Analytic Mean\")\nplt.plot(np.arange(N), RMS, label=\"Sample mean\")\nplt.legend()\nplt.xlabel('Number of steps')\nplt.ylabel('Root Mean Square Displacement')\nplt.show()",
"_____no_output_____"
]
],
[
[
"So we see that our analytic expression looks very reasonable.",
"_____no_output_____"
],
[
"### Flipping Coins and the Law of Large Numbers\n\nSo far we have only look at the random walk as a completely theoretical exercise. As an example, let us now couple it to some more concrete situation.\n\nOur 1D random walk is the sum of a discrete random variable that has 2, equally likely outcomes. An example of this is flipping a coin. Thus, our random walk models the process of flipping a coin many times and keeping track of the total number of heads and tails we get.\n\nWe looked at this example last week as well:",
"_____no_output_____"
]
],
[
[
"def flip_coins(N):\n flips = np.random.randint(2, size=N)\n heads = np.sum(flips == 0)\n tails = N - heads\n return heads, tails\n \n \nprint(\"Flipping 1000 coins:\")\nheads, tails = flip_coins(1000)\nprint(\"Heads:\", heads)\nprint(\"Tail:\", tails)",
"Flipping 1000 coins:\nHeads: 477\nTail: 523\n"
]
],
[
[
"When we flip $N$ coins, we expect close an equal number of heads and tails, i.e., about $N/2$ of each. But should we expect exactly $N/2$ heads? The answer is *no*. The probability of getting a perfectly even distribution goes *down* with the number of throws $N$. Let us look at some numbers:",
"_____no_output_____"
]
],
[
[
"print(f\"{'N':>10} {'Heads':>10}|{'Tails':<6} {'Deviation':>12} {'Ratio':>10}\")\nprint(\"=\"*60)\n\nfor N in 10, 1000, 10**4, 10**5, 10**6:\n for i in range(3):\n heads, tails = flip_coins(N)\n print(f\"{N:>10} {heads:>10}|{tails:<6} {abs(N/2-heads):10} {heads/N:>10.1%}|{tails/N:<6.1%}\")\n print()\nprint(\"=\"*60)",
" N Heads|Tails Deviation Ratio\n============================================================\n 10 7|3 2.0 70.0%|30.0% \n 10 4|6 1.0 40.0%|60.0% \n 10 6|4 1.0 60.0%|40.0% \n\n 1000 500|500 0.0 50.0%|50.0% \n 1000 500|500 0.0 50.0%|50.0% \n 1000 489|511 11.0 48.9%|51.1% \n\n 10000 5020|4980 20.0 50.2%|49.8% \n 10000 4988|5012 12.0 49.9%|50.1% \n 10000 5017|4983 17.0 50.2%|49.8% \n\n 100000 49994|50006 6.0 50.0%|50.0% \n 100000 49856|50144 144.0 49.9%|50.1% \n 100000 50159|49841 159.0 50.2%|49.8% \n\n 1000000 499345|500655 655.0 49.9%|50.1% \n 1000000 499588|500412 412.0 50.0%|50.0% \n 1000000 499322|500678 678.0 49.9%|50.1% \n\n============================================================\n"
]
],
[
[
"Here, we explore how the *deviation*, which is the number of flips we are away from a perfectly even split, grows with $N$. What we call the deviation here is equivalent to the displacement of one of our random walkers, and as we have seen, the root mean square displacement grows as $\\sqrt{N}$. The more coins we flip $N$, the bigger deviation from the baseline we expect.\n\nNow, isn't this counterdicting the law of large numbers? No, it isn't, but it actually highlights an important point about the law of large numbers. The law of large numbers only guarantees that the *average* of many trials will approach the expected value for large numbers. Thus the law of large numbers states that the *ratio* of heads and tails will become 50%/50% in the long run, it gives not guarantee that we will have the same number of outcomes.\n\nIn fact ,we see that this is indeed the case for our results too, while the deviation grows with $N$, we can see for the exact same random sample that the *ratio* of heads and tails approaches 50/50! This is because the ratio is computed from\n\n$$P(\\text{heads}) \\approx \\frac{\\text{number of heads}}{N},$$\n\nbut we know that the deviation in the number of heads grows as $\\sqrt{N}$, but that means the deviation in the *ratio* grows as\n\n$$\\frac{\\sqrt{N}}{N} = \\frac{1}{\\sqrt{N}}.$$\nAnd so our results don't contradict the law of large numbers, it proves it. \n\nThe law of large numbers only talkes about averages, never about single events. However, it is a very common fallacy to think that the number of heads and tails have to *even out* in the long run. This is known as the *Gambler's Fallacy*.",
"_____no_output_____"
],
[
"### 2D Random Walk\n\nSo far we have only looked at a random walk in one dimension. Let us add another dimension, so we are looking at a random walker moving around in a 2D plane. We will still be looking at a random walk on a regular grid or lattice.\n\nFor every step, there are then 4 choices for our walker. If we envison our grid as the streets of a city seen from above, these directions would be *north*, *south*, *west*, and *east*. We now denote the displacement of the walker as \n\n$$\\vec{R}_N = (X_N, Y_N).$$\n\nLet us jump right into simulating a random walk.",
"_____no_output_____"
]
],
[
[
"possible_steps = [(1, 0), (-1, 0), (0, 1), (0, -1)]\n\nN = 100\nR = np.zeros((N+1, 2))\nR[0] = (0, 0)\n\nfor i in range(N):\n step = possible_steps[np.random.randint(4)]\n R[i+1] = R[i] + step\n \nX = R[:, 0]\nY = R[:, 1]\nplt.plot(X, Y)\n\nplt.axis('equal')\nplt.show()",
"_____no_output_____"
]
],
[
[
"Here we specify the possible steps, and then draw one of these at random for every step. Performing this vectorized is slightly tricky. to make things a lot simpler, we simply change the possible steps to saying that the walker takes a step in both dimensions for each step, so instead of\n\n$$(1, 0) \\quad (-1, 0) \\quad (0, 1) \\quad (0, -1),$$\nas our possibilities, we have\n$$(1, 1) \\quad (1, -1) \\quad (-1, 1) \\quad (-1, 1).$$\n\nThis makes things a lot easier, cause the steps in the $X$ and $Y$ direction are now decoupled.",
"_____no_output_____"
]
],
[
[
"N = 1000\nsteps = 2*np.random.randint(2, size=(N, 2)) - 1 \nR = np.cumsum(steps, axis=0)\n\nX = R[:, 0]\nY = R[:, 1]\n\nplt.plot(X, Y)\nplt.axis('equal')\nplt.show()",
"_____no_output_____"
]
],
[
[
"The only difference with our change to the steps is that our walker now walks a distance $\\sqrt{2}$ every step, instead of 1. The plot also looks like the diagonal version of the previous plot.",
"_____no_output_____"
],
[
"Let us try to plot many more steps:",
"_____no_output_____"
]
],
[
[
"N = 25000\nsteps = 2*np.random.randint(2, size=(N, 2)) - 1 \nR = np.cumsum(steps, axis=0)\n\nX = R[:, 0]\nY = R[:, 1]\n\nplt.plot(X, Y)\nplt.axis('equal')\nplt.show()",
"_____no_output_____"
]
],
[
[
"Because our walker is now using 2 spatial dimensions, we cannot plot this walk over time, we can only plot out the total trajectory over time. This has some drawbacks, as it is hard to understand how the walk builds up over time, and how much the walk doubles back over itself.",
"_____no_output_____"
],
[
"A fix to this is to create an animation of the walk over time. We won't take the time to do this here. But you can click the links under to see such animations:\n1. [Animated random walk in 2D with 2500 steps](https://upload.wikimedia.org/wikipedia/commons/f/f3/Random_walk_2500_animated.svg)\n2. [Animated random walk in 2D with 25000 steps](https://upload.wikimedia.org/wikipedia/commons/c/cb/Random_walk_25000.svg)",
"_____no_output_____"
],
[
"\n",
"_____no_output_____"
],
[
"### Plotting several walkers\n\nAgain we can plot several walks over each other",
"_____no_output_____"
]
],
[
[
"nr_steps = 500\nnr_walkers = 5\n\nfor walker in range(nr_walkers):\n steps = 2*np.random.randint(2, size=(nr_steps, 2)) - 1 \n R = np.cumsum(steps, axis=0)\n\n X = R[:, 0]\n Y = R[:, 1]\n\n plt.plot(X, Y, alpha=0.5)\n plt.axis('equal')\n\nplt.scatter(0, 0, marker='o', color='black', s=100)\nplt.show()",
"_____no_output_____"
]
],
[
[
"Here, we plot 5 random walks over each other, and mark the origin with a black circle.",
"_____no_output_____"
],
[
"### Analyzing the Mean Displacement\n\nWe can now return to analyze the average behavior of the 2D random walker, just like we did for the 1D case. However, it turns out we don't need to reinvent the wheel. We know that \n$$\\vec{R}_N = (X_N, Y_N).$$\nSo to find the mean displacement, we find\n$$\\langle \\vec{R}_N \\rangle = (\\langle X_N \\rangle, \\langle Y_N \\rangle).$$\n\nHowever, both $X_N$ and $Y_N$ behave exactly like a 1D-walker in their dimension, as they increase by -1 or 1 every step. So we have\n\n$$\\langle \\vec{R}_N \\rangle = (0, 0).$$\n\nWe could almost have guessed this, because the 2D problem is, just like the 1D problem, completely symmetric. The average will therefore tend to be the exact origin.\n\nBut what about the mean square displacement? In this case, taking the square of the vector means taking the dot product with itself, it is thus the square of the distance to the origin we are computing:\n$$\\langle |\\vec{R_N}|^2 \\rangle = \\langle X_N^2 \\rangle + \\langle Y_N^2 \\rangle.$$\n\nSo again we can simply insert the values we found earlier for the 1D walker:\n\n$$\\langle |\\vec{R_N}|^2 \\rangle = 2N.$$\n\nThus, the root mean square distance of a 2D random walker to the origin is given by\n\n$$\\text{RMS} = \\sqrt{\\langle |\\vec{R_N}|^2 \\rangle} = \\sqrt{2N}.$$\n\nWe can draw this into our 2D plot, to see if this seems reasonable.",
"_____no_output_____"
]
],
[
[
"nr_steps = 500\nnr_walkers = 5\n\n# Plot random walks\nfor walker in range(nr_walkers):\n steps = 2*np.random.randint(2, size=(nr_steps, 2)) - 1 \n R = np.cumsum(steps, axis=0)\n X = R[:, 0]\n Y = R[:, 1]\n plt.plot(X, Y, alpha=0.5)\n plt.axis('equal')\n\n# Plot origin\nplt.scatter(0, 0, marker='o', color='black', s=100)\n\n# Plot analytic RMS\nrms = np.sqrt(2*nr_steps)\ntheta = np.linspace(0, 2*np.pi, 1001)\nplt.plot(rms*np.cos(theta), rms*np.sin(theta), 'k--')\n\n# Plot \nplt.show()",
"_____no_output_____"
]
],
[
[
"## Why Random Walkers are so interesting\n\nThe random walker is an example of a process that is built up of simple, random steps, but whose net behavior can be complex. These kind of processes are found throughout the natural sciences and in mathematics. The list of applications of random walks is therefore very long and varied.\n\nSome examples of processess that can be modelled with random walks are:\n* The price of stocks in economics\n* Modeling of population dynamics in biology\n* The modeling of genetic drift\n* The study of polymers in material science use a special type of self-avoiding random walks\n* In image processing, images can be segmented by using an algorithm that randomly walks over the image\n* Twitter uses a random walk approach to make suggestions of who to follow\n\nThese are just *some* examples, and the list goes on and on. If you want more example, there is a more extensive list [here](https://en.wikipedia.org/wiki/Random_walk#Applications).",
"_____no_output_____"
],
[
"## Moving from a discrete to a continious model\n\nAs a final example, let us show how we can move from a discrete random walk model to a continious one. As we already have seen some examples of, when we move towards a large number of steps $N$, the movement of the random walker doesn't necessarily look so jagged and force anymore, but *seems* more like a continious process. And this is the whole trick to moving to a continious model, letting $N\\to\\infty$. We obviously cannot do this on a computer, but we can analyze the problem mathematically.",
"_____no_output_____"
],
[
"To keep things as simple as possible, we can consider the uniform 1D random walker. Instead of talking about the displacement $X_N$, we now define a function $P(x, t)$ that denotes the probability of finding the walker at position $x$ at time $t$.\n\nBecause we have a discrete model, we say that the walker moves a length $\\Delta x$ each step, so that the walker will be at a position\n\n$$x_i = i\\cdot \\Delta x,$$\n\nIn addition, we assume the walker takes one step every $\\Delta t$ timestep, so we can denote a given time as\n\n$$t_j = j\\cdot \\Delta t.$$\n\nThus, we are talking about the probability of finding the walker at position $x_i$ at time $t_j$, which is described by the function $P(x_i, t_j)$, or simply $P_{i, j}$ for short.\n\n",
"_____no_output_____"
],
[
"Now, our goal isn't necessarily to find an expression for $P$, to find an expression for how it develops over time. Or put more formally, we are trying to find an expression for the time-derivative\n$$\\frac{\\partial P(x, t)}{\\partial t},$$\n\ni.e., we are trying to find a differential equation. To find a time derivative, we want to find an expression on the form:\n\n$$\\frac{P(x_i, t_{j+1}) - P(x_i, t_j)}{\\Delta t}.$$\nBecause then we can take the limit $\\Delta t \\to 0$ to get a derivative.",
"_____no_output_____"
],
[
"As we are trying to find the time-derivative of $P(x, t)$, let us write out what we know about stepping forward in time with our model. The probability of finding the walker in position $x_i$ at the *next* time step, must be given by the chance of finding it at the two neighboring grid points at the current time step, so:\n$$P(x_i, t_{j+1}) = \\frac{1}{2}P(x_{i-1}, t_j) + \\frac{1}{2}P(x_{i+1}, t_j).$$\n\nThe reasons the two terms have a factor 1/2, is because there is only a 50% chance of a walker in those grid points moving the right direction.",
"_____no_output_____"
],
[
"Now, to find an expression for the time derivative, we need to subtract $P(x_i, t_j)$ from both sides.\n\n$$P(x_i, t_{j+1}) - P(x_i, t_j) = \\frac{1}{2}\\big(P(x_{i-1}, t_j) - 2P(x_i, t_j) + P(x_{i+1}, t_j)\\big).$$\n\nThe next step is then to divide by $\\Delta t$ on both sides\n\n$$\\frac{P(x_i, t_{j+1}) - P(x_i, t_j)}{\\Delta t}=\\frac{1}{2\\Delta t}\\big(P(x_{i-1}, t_j) - 2P(x_i, t_j) + P(x_{i+1}, t_j)\\big).$$\n\nNow we are getting very close! However, we cannot take the limit $\\Delta t \\to 0$ just yet, because then the expression on the right will blow up. However, we can fix this by expanding the fraction by a factor of $\\Delta x^2$\n\n$$\\frac{P(x_i, t_{j+1}) - P(x_i, t_j)}{\\Delta t}=\\frac{\\Delta x^2}{2\\Delta t}\\frac{P(x_{i-1}, t_j) - 2P(x_i, t_j) + P(x_{i+1}, t_j)}{\\Delta x^2}.$$",
"_____no_output_____"
],
[
"This helps, because we can now take the limit of $\\Delta t \\to 0$ and $\\Delta x^2 \\to 0$ at the *same* time. This way, we can enforce the constraint that we do it in such a manner that \n$$\\frac{\\Delta x^2}{2\\Delta t} = \\text{constant}.$$\n\nBecause this expression will be a constant, we name it $D$. We then have\n\n$$\\lim_{\\substack{\\Delta t \\to 0 \\\\ \\Delta x \\to 0 \\\\ D={\\rm const.}}} \\bigg[\\frac{P(x_i, t_{j+1}) - P(x_i, t_j)}{\\Delta t}= D \\frac{P(x_{i-1}, t_j) - 2P(x_i, t_j) + P(x_{i+1}, t_j)}{\\Delta x^2}\\bigg].$$",
"_____no_output_____"
],
[
"Now, the term on the left was equal to the time derivative of $P$. But the expression on the right is also a derivative, it is the second-order derivative with respect to $x$! So we get\n$$\\frac{\\partial P}{\\partial t} = D\\frac{\\partial^2 P}{\\partial x^2}.$$",
"_____no_output_____"
],
[
"Let us summarize what we have done, we have said our random walker takes steps of $\\Delta x$ in time $\\Delta t$, and then taken the limit where both of these go to 0. Effectively, we are saying the walker takes infinitesimally small steps, infinitely fast. This is effectively the same as letting the number of steps taken go to infinity ($N \\to \\infty$). But at the same time, we do this in the manner in which the total displacement of the walker stays bounded.",
"_____no_output_____"
],
[
"Taking the limit of a simple 1D walker has given us a partial differential equation known as the *Diffusion Equation*, or alternatively the *Heat Equation*. This is one of the most fundamental and important equations in the natural sciences, so it is quite astonishing that it can be derived from a simple random walker!\n\nFor more information and more detailed derivations, see for example:\n- [Mark Kac's classical paper from 1947](http://www.math.hawaii.edu/~xander/Fa06/Kac--Brownian_Motion.pdf)\n\nIn practice, one does not use a 1D diffusion equation, but a 3D one:\n$$\\frac{\\partial u}{\\partial t} = \\nabla^2 u.$$\n\nBut this pde can be found by taking the limit of a 3D random walker in exactly the same manner.",
"_____no_output_____"
],
[
"### Solving the Diffusion Equation\n\nWhat is very interesting about what we have just done, is that we have gone from a discrete, numerically solvable problem, into a continiouse partial differential equation. This is the opposite process of what we are used to dealing with when we are looking at numerics!\n\nIf we want to solve the diffusion equation numerically, we have to discretize the equation again, and move back to the effective 1D walker. If you want to read how that can be done, take a look at this supplemental notebook: [*Solving the 1D Diffusion Equation*](S19_solving_the_1D_diffusion_equation.ipynb).",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
cb8c8846de3c64fd4e480cd17e3d8bc516114be6
| 525,885 |
ipynb
|
Jupyter Notebook
|
2. Define the Network Architecture.ipynb
|
mandy1057/Facial_keypoints
|
d48c74f273b721a1ae10173097bc026d795fb184
|
[
"MIT"
] | null | null | null |
2. Define the Network Architecture.ipynb
|
mandy1057/Facial_keypoints
|
d48c74f273b721a1ae10173097bc026d795fb184
|
[
"MIT"
] | 3 |
2021-03-19T01:38:10.000Z
|
2022-01-13T01:21:49.000Z
|
2. Define the Network Architecture.ipynb
|
mrtechnoo/facial_keypoints
|
d48c74f273b721a1ae10173097bc026d795fb184
|
[
"MIT"
] | null | null | null | 364.944483 | 96,460 | 0.924006 |
[
[
[
"## Define the Convolutional Neural Network\n\nAfter you've looked at the data you're working with and, in this case, know the shapes of the images and of the keypoints, you are ready to define a convolutional neural network that can *learn* from this data.\n\nIn this notebook and in `models.py`, you will:\n1. Define a CNN with images as input and keypoints as output\n2. Construct the transformed FaceKeypointsDataset, just as before\n3. Train the CNN on the training data, tracking loss\n4. See how the trained model performs on test data\n5. If necessary, modify the CNN structure and model hyperparameters, so that it performs *well* **\\***\n\n**\\*** What does *well* mean?\n\n\"Well\" means that the model's loss decreases during training **and**, when applied to test image data, the model produces keypoints that closely match the true keypoints of each face. And you'll see examples of this later in the notebook.\n\n---\n",
"_____no_output_____"
],
[
"## CNN Architecture\n\nRecall that CNN's are defined by a few types of layers:\n* Convolutional layers\n* Maxpooling layers\n* Fully-connected layers\n\nYou are required to use the above layers and encouraged to add multiple convolutional layers and things like dropout layers that may prevent overfitting. You are also encouraged to look at literature on keypoint detection, such as [this paper](https://arxiv.org/pdf/1710.00977.pdf), to help you determine the structure of your network.\n\n\n### TODO: Define your model in the provided file `models.py` file\n\nThis file is mostly empty but contains the expected name and some TODO's for creating your model.\n\n---",
"_____no_output_____"
],
[
"## PyTorch Neural Nets\n\nTo define a neural network in PyTorch, you define the layers of a model in the function `__init__` and define the feedforward behavior of a network that employs those initialized layers in the function `forward`, which takes in an input image tensor, `x`. The structure of this Net class is shown below and left for you to fill in.\n\nNote: During training, PyTorch will be able to perform backpropagation by keeping track of the network's feedforward behavior and using autograd to calculate the update to the weights in the network.\n\n#### Define the Layers in ` __init__`\nAs a reminder, a conv/pool layer may be defined like this (in `__init__`):\n```\n# 1 input image channel (for grayscale images), 32 output channels/feature maps, 3x3 square convolution kernel\nself.conv1 = nn.Conv2d(1, 32, 3)\n\n# maxpool that uses a square window of kernel_size=2, stride=2\nself.pool = nn.MaxPool2d(2, 2) \n```\n\n#### Refer to Layers in `forward`\nThen referred to in the `forward` function like this, in which the conv1 layer has a ReLu activation applied to it before maxpooling is applied:\n```\nx = self.pool(F.relu(self.conv1(x)))\n```\n\nBest practice is to place any layers whose weights will change during the training process in `__init__` and refer to them in the `forward` function; any layers or functions that always behave in the same way, such as a pre-defined activation function, should appear *only* in the `forward` function.",
"_____no_output_____"
],
[
"#### Why models.py\n\nYou are tasked with defining the network in the `models.py` file so that any models you define can be saved and loaded by name in different notebooks in this project directory. For example, by defining a CNN class called `Net` in `models.py`, you can then create that same architecture in this and other notebooks by simply importing the class and instantiating a model:\n```\n from models import Net\n net = Net()\n```",
"_____no_output_____"
]
],
[
[
"# import the usual resources\nimport matplotlib.pyplot as plt\nimport numpy as np\n\n# watch for any changes in model.py, if it changes, re-load it automatically\n%load_ext autoreload\n%autoreload 2",
"_____no_output_____"
],
[
"## TODO: Define the Net in models.py\n\nimport torch\nimport torch.nn as nn\nimport torch.nn.functional as F\n\n## TODO: Once you've define the network, you can instantiate it\n# one example conv layer has been provided for you\nfrom models import Net\n\nnet = Net()\nprint(net)",
"Net(\n (pool): MaxPool2d(kernel_size=2, stride=2, padding=0, dilation=1, ceil_mode=False)\n (conv1): Conv2d(1, 32, kernel_size=(5, 5), stride=(1, 1))\n (conv1_drop): Dropout2d(p=0.1)\n (conv2): Conv2d(32, 48, kernel_size=(5, 5), stride=(1, 1))\n (conv2_drop): Dropout2d(p=0.1)\n (conv3): Conv2d(48, 72, kernel_size=(5, 5), stride=(1, 1))\n (conv3_drop): Dropout2d(p=0.1)\n (conv4): Conv2d(72, 96, kernel_size=(5, 5), stride=(1, 1))\n (conv4_drop): Dropout2d(p=0.1)\n (fc1): Linear(in_features=9600, out_features=2400, bias=True)\n (fc1_drop): Dropout(p=0.2)\n (fc2): Linear(in_features=2400, out_features=272, bias=True)\n (fc2_drop): Dropout(p=0.4)\n (fc3): Linear(in_features=272, out_features=136, bias=True)\n)\n"
]
],
[
[
"## Transform the dataset \n\nTo prepare for training, create a transformed dataset of images and keypoints.\n\n### TODO: Define a data transform\n\nIn PyTorch, a convolutional neural network expects a torch image of a consistent size as input. For efficient training, and so your model's loss does not blow up during training, it is also suggested that you normalize the input images and keypoints. The necessary transforms have been defined in `data_load.py` and you **do not** need to modify these; take a look at this file (you'll see the same transforms that were defined and applied in Notebook 1).\n\nTo define the data transform below, use a [composition](http://pytorch.org/tutorials/beginner/data_loading_tutorial.html#compose-transforms) of:\n1. Rescaling and/or cropping the data, such that you are left with a square image (the suggested size is 224x224px)\n2. Normalizing the images and keypoints; turning each RGB image into a grayscale image with a color range of [0, 1] and transforming the given keypoints into a range of [-1, 1]\n3. Turning these images and keypoints into Tensors\n\nThese transformations have been defined in `data_load.py`, but it's up to you to call them and create a `data_transform` below. **This transform will be applied to the training data and, later, the test data**. It will change how you go about displaying these images and keypoints, but these steps are essential for efficient training.\n\nAs a note, should you want to perform data augmentation (which is optional in this project), and randomly rotate or shift these images, a square image size will be useful; rotating a 224x224 image by 90 degrees will result in the same shape of output.",
"_____no_output_____"
]
],
[
[
"from torch.utils.data import Dataset, DataLoader\nfrom torchvision import transforms, utils\n\n# the dataset we created in Notebook 1 is copied in the helper file `data_load.py`\nfrom data_load import FacialKeypointsDataset\n# the transforms we defined in Notebook 1 are in the helper file `data_load.py`\nfrom data_load import Rescale, RandomCrop, Normalize, ToTensor\n\n\n## TODO: define the data_transform using transforms.Compose([all tx's, . , .])\n# order matters! i.e. rescaling should come before a smaller crop\ndata_transform = None\n\n#coding\ndata_transform =transforms.Compose([Rescale(250),\n RandomCrop(224),\n Normalize(),\n ToTensor()])\n\n# testing that you've defined a transform\nassert(data_transform is not None), 'Define a data_transform'",
"_____no_output_____"
],
[
"# create the transformed dataset\ntransformed_dataset = FacialKeypointsDataset(csv_file='data/training_frames_keypoints.csv',\n root_dir='data/training/',\n transform=data_transform)\n\n\nprint('Number of images: ', len(transformed_dataset))\n\n# iterate through the transformed dataset and print some stats about the first few samples\nfor i in range(4):\n sample = transformed_dataset[i]\n print(i, sample['image'].size(), sample['keypoints'].size())",
"Number of images: 3462\n0 torch.Size([1, 224, 224]) torch.Size([68, 2])\n1 torch.Size([1, 224, 224]) torch.Size([68, 2])\n2 torch.Size([1, 224, 224]) torch.Size([68, 2])\n3 torch.Size([1, 224, 224]) torch.Size([68, 2])\n"
]
],
[
[
"## Batching and loading data\n\nNext, having defined the transformed dataset, we can use PyTorch's DataLoader class to load the training data in batches of whatever size as well as to shuffle the data for training the model. You can read more about the parameters of the DataLoader, in [this documentation](http://pytorch.org/docs/master/data.html).\n\n#### Batch size\nDecide on a good batch size for training your model. Try both small and large batch sizes and note how the loss decreases as the model trains.\n\n**Note for Windows users**: Please change the `num_workers` to 0 or you may face some issues with your DataLoader failing.",
"_____no_output_____"
]
],
[
[
"# load training data in batches\nbatch_size = 10\n\ntrain_loader = DataLoader(transformed_dataset, \n batch_size=batch_size,\n shuffle=True, \n num_workers=4)\n",
"_____no_output_____"
]
],
[
[
"## Before training\n\nTake a look at how this model performs before it trains. You should see that the keypoints it predicts start off in one spot and don't match the keypoints on a face at all! It's interesting to visualize this behavior so that you can compare it to the model after training and see how the model has improved.\n\n#### Load in the test dataset\n\nThe test dataset is one that this model has *not* seen before, meaning it has not trained with these images. We'll load in this test data and before and after training, see how your model performs on this set!\n\nTo visualize this test data, we have to go through some un-transformation steps to turn our images into python images from tensors and to turn our keypoints back into a recognizable range. ",
"_____no_output_____"
]
],
[
[
"# load in the test data, using the dataset class\n# AND apply the data_transform you defined above\n\n# create the test dataset\ntest_dataset = FacialKeypointsDataset(csv_file='data/test_frames_keypoints.csv',\n root_dir='data/test/',\n transform=data_transform)\n\n",
"_____no_output_____"
],
[
"# load test data in batches\nbatch_size = 10\n\ntest_loader = DataLoader(test_dataset, \n batch_size=batch_size,\n shuffle=True, \n num_workers=4)",
"_____no_output_____"
]
],
[
[
"## Apply the model on a test sample\n\nTo test the model on a test sample of data, you have to follow these steps:\n1. Extract the image and ground truth keypoints from a sample\n2. Make sure the image is a FloatTensor, which the model expects.\n3. Forward pass the image through the net to get the predicted, output keypoints.\n\nThis function test how the network performs on the first batch of test data. It returns the images, the transformed images, the predicted keypoints (produced by the model), and the ground truth keypoints.",
"_____no_output_____"
]
],
[
[
"# test the model on a batch of test images\n\ndef net_sample_output():\n \n # iterate through the test dataset\n for i, sample in enumerate(test_loader):\n \n # get sample data: images and ground truth keypoints\n images = sample['image']\n key_pts = sample['keypoints']\n\n # convert images to FloatTensors\n images = images.type(torch.FloatTensor)\n\n # forward pass to get net output\n output_pts = net(images)\n \n # reshape to batch_size x 68 x 2 pts\n output_pts = output_pts.view(output_pts.size()[0], 68, -1)\n \n # break after first image is tested\n if i == 0:\n return images, output_pts, key_pts\n ",
"_____no_output_____"
]
],
[
[
"#### Debugging tips\n\nIf you get a size or dimension error here, make sure that your network outputs the expected number of keypoints! Or if you get a Tensor type error, look into changing the above code that casts the data into float types: `images = images.type(torch.FloatTensor)`.",
"_____no_output_____"
]
],
[
[
"# call the above function\n# returns: test images, test predicted keypoints, test ground truth keypoints\ntest_images, test_outputs, gt_pts = net_sample_output()\n\n# print out the dimensions of the data to see if they make sense\nprint(test_images.data.size())\nprint(test_outputs.data.size())\nprint(gt_pts.size())",
"torch.Size([10, 1, 224, 224])\ntorch.Size([10, 68, 2])\ntorch.Size([10, 68, 2])\n"
]
],
[
[
"## Visualize the predicted keypoints\n\nOnce we've had the model produce some predicted output keypoints, we can visualize these points in a way that's similar to how we've displayed this data before, only this time, we have to \"un-transform\" the image/keypoint data to display it.\n\nNote that I've defined a *new* function, `show_all_keypoints` that displays a grayscale image, its predicted keypoints and its ground truth keypoints (if provided).",
"_____no_output_____"
]
],
[
[
"def show_all_keypoints(image, predicted_key_pts, gt_pts=None):\n \"\"\"Show image with predicted keypoints\"\"\"\n # image is grayscale\n plt.imshow(image, cmap='gray')\n plt.scatter(predicted_key_pts[:, 0], predicted_key_pts[:, 1], s=20, marker='.', c='m')\n # plot ground truth points as green pts\n if gt_pts is not None:\n plt.scatter(gt_pts[:, 0], gt_pts[:, 1], s=20, marker='.', c='g')\n",
"_____no_output_____"
]
],
[
[
"#### Un-transformation\n\nNext, you'll see a helper function. `visualize_output` that takes in a batch of images, predicted keypoints, and ground truth keypoints and displays a set of those images and their true/predicted keypoints.\n\nThis function's main role is to take batches of image and keypoint data (the input and output of your CNN), and transform them into numpy images and un-normalized keypoints (x, y) for normal display. The un-transformation process turns keypoints and images into numpy arrays from Tensors *and* it undoes the keypoint normalization done in the Normalize() transform; it's assumed that you applied these transformations when you loaded your test data.",
"_____no_output_____"
]
],
[
[
"# visualize the output\n# by default this shows a batch of 10 images\ndef visualize_output(test_images, test_outputs, gt_pts=None, batch_size=10):\n\n for i in range(batch_size):\n plt.figure(figsize=(20,10))\n ax = plt.subplot(1, batch_size, i+1)\n\n # un-transform the image data\n image = test_images[i].data # get the image from it's wrapper\n image = image.numpy() # convert to numpy array from a Tensor\n image = np.transpose(image, (1, 2, 0)) # transpose to go from torch to numpy image\n\n # un-transform the predicted key_pts data\n predicted_key_pts = test_outputs[i].data\n predicted_key_pts = predicted_key_pts.numpy()\n # undo normalization of keypoints \n predicted_key_pts = predicted_key_pts*50.0+100\n \n # plot ground truth points for comparison, if they exist\n ground_truth_pts = None\n if gt_pts is not None:\n ground_truth_pts = gt_pts[i] \n ground_truth_pts = ground_truth_pts*50.0+100\n \n # call show_all_keypoints\n show_all_keypoints(np.squeeze(image), predicted_key_pts, ground_truth_pts)\n \n plt.axis('off')\n\n plt.show()\n \n# call it\nvisualize_output(test_images, test_outputs, gt_pts)",
"_____no_output_____"
]
],
[
[
"## Training\n\n#### Loss function\nTraining a network to predict keypoints is different than training a network to predict a class; instead of outputting a distribution of classes and using cross entropy loss, you may want to choose a loss function that is suited for regression, which directly compares a predicted value and target value. Read about the various kinds of loss functions (like MSE or L1/SmoothL1 loss) in [this documentation](http://pytorch.org/docs/master/_modules/torch/nn/modules/loss.html).\n\n### TODO: Define the loss and optimization\n\nNext, you'll define how the model will train by deciding on the loss function and optimizer.\n\n---",
"_____no_output_____"
]
],
[
[
"## TODO: Define the loss and optimization\nimport torch.optim as optim\n\n#criterion = None\ncriterion = nn.MSELoss()\n\n#optimizer = None\noptimizer = optim.Adam(net.parameters(),lr = 0.01)\n",
"_____no_output_____"
]
],
[
[
"## Training and Initial Observation\n\nNow, you'll train on your batched training data from `train_loader` for a number of epochs. \n\nTo quickly observe how your model is training and decide on whether or not you should modify it's structure or hyperparameters, you're encouraged to start off with just one or two epochs at first. As you train, note how your the model's loss behaves over time: does it decrease quickly at first and then slow down? Does it take a while to decrease in the first place? What happens if you change the batch size of your training data or modify your loss function? etc. \n\nUse these initial observations to make changes to your model and decide on the best architecture before you train for many epochs and create a final model.",
"_____no_output_____"
]
],
[
[
"def train_net(n_epochs):\n\n # prepare the net for training\n net.train()\n\n for epoch in range(n_epochs): # loop over the dataset multiple times\n \n running_loss = 0.0\n\n # train on batches of data, assumes you already have train_loader\n for batch_i, data in enumerate(train_loader):\n # get the input images and their corresponding labels\n images = data['image']\n key_pts = data['keypoints']\n\n # flatten pts\n key_pts = key_pts.view(key_pts.size(0), -1)\n\n # convert variables to floats for regression loss\n key_pts = key_pts.type(torch.FloatTensor)\n images = images.type(torch.FloatTensor)\n\n # forward pass to get outputs\n output_pts = net(images)\n\n # calculate the loss between predicted and target keypoints\n loss = criterion(output_pts, key_pts)\n\n # zero the parameter (weight) gradients\n optimizer.zero_grad()\n \n # backward pass to calculate the weight gradients\n loss.backward()\n\n # update the weights\n optimizer.step()\n\n # print loss statistics\n # to convert loss into a scalar and add it to the running_loss, use .item()\n running_loss += loss.item()\n if batch_i % 10 == 9: # print every 10 batches\n print('Epoch: {}, Batch: {}, Avg. Loss: {}'.format(epoch + 1, batch_i+1, running_loss/1000))\n running_loss = 0.0\n\n print('Finished Training')\n",
"_____no_output_____"
],
[
"# train your network\nn_epochs = 10 # start small, and increase when you've decided on your model structure and hyperparams\n\ntrain_net(n_epochs)",
"Epoch: 1, Batch: 10, Avg. Loss: 507.3042490871251\nEpoch: 1, Batch: 20, Avg. Loss: 10.675888020068408\nEpoch: 1, Batch: 30, Avg. Loss: 0.07985420599579811\nEpoch: 1, Batch: 40, Avg. Loss: 33.92743387827277\nEpoch: 1, Batch: 50, Avg. Loss: 0.013151312232017516\nEpoch: 1, Batch: 60, Avg. Loss: 0.0937241096496582\nEpoch: 1, Batch: 70, Avg. Loss: 0.00666309580206871\nEpoch: 1, Batch: 80, Avg. Loss: 0.009181032180786133\nEpoch: 1, Batch: 90, Avg. Loss: 0.005870131015777588\nEpoch: 1, Batch: 100, Avg. Loss: 0.007821534872055053\nEpoch: 1, Batch: 110, Avg. Loss: 0.0065793790221214295\nEpoch: 1, Batch: 120, Avg. Loss: 0.006224306970834732\nEpoch: 1, Batch: 130, Avg. Loss: 0.006179891020059586\nEpoch: 1, Batch: 140, Avg. Loss: 0.007807506382465363\nEpoch: 1, Batch: 150, Avg. Loss: 0.00737674868106842\nEpoch: 1, Batch: 160, Avg. Loss: 0.0064330708384513855\nEpoch: 1, Batch: 170, Avg. Loss: 0.006822253942489624\nEpoch: 1, Batch: 180, Avg. Loss: 0.0068513801097869875\nEpoch: 1, Batch: 190, Avg. Loss: 0.006971220672130585\nEpoch: 1, Batch: 200, Avg. Loss: 0.006951295554637909\nEpoch: 1, Batch: 210, Avg. Loss: 0.007007784605026245\nEpoch: 1, Batch: 220, Avg. Loss: 0.006982840359210968\nEpoch: 1, Batch: 230, Avg. Loss: 0.006287442952394485\nEpoch: 1, Batch: 240, Avg. Loss: 0.006352033466100693\nEpoch: 1, Batch: 250, Avg. Loss: 0.006270585834980011\nEpoch: 1, Batch: 260, Avg. Loss: 0.0061953898072242735\nEpoch: 1, Batch: 270, Avg. Loss: 0.006784923136234283\nEpoch: 1, Batch: 280, Avg. Loss: 0.006715250134468079\nEpoch: 1, Batch: 290, Avg. Loss: 0.006440463066101074\nEpoch: 1, Batch: 300, Avg. Loss: 0.005825052380561828\nEpoch: 1, Batch: 310, Avg. Loss: 0.005678276479244232\nEpoch: 1, Batch: 320, Avg. Loss: 0.006369813024997712\nEpoch: 1, Batch: 330, Avg. Loss: 0.005867576718330383\nEpoch: 1, Batch: 340, Avg. Loss: 0.005923893183469773\nEpoch: 2, Batch: 10, Avg. Loss: 0.0061584247648715975\nEpoch: 2, Batch: 20, Avg. Loss: 0.006436422675848008\nEpoch: 2, Batch: 30, Avg. Loss: 0.005458706676959991\nEpoch: 2, Batch: 40, Avg. Loss: 0.005443674892187118\nEpoch: 2, Batch: 50, Avg. Loss: 0.0062083094716072085\nEpoch: 2, Batch: 60, Avg. Loss: 0.0059262548685073855\nEpoch: 2, Batch: 70, Avg. Loss: 0.005132598280906678\nEpoch: 2, Batch: 80, Avg. Loss: 0.005338089764118195\nEpoch: 2, Batch: 90, Avg. Loss: 0.0051441526114940645\nEpoch: 2, Batch: 100, Avg. Loss: 0.006271188288927078\nEpoch: 2, Batch: 110, Avg. Loss: 0.006265846788883209\nEpoch: 2, Batch: 120, Avg. Loss: 0.005456428915262222\nEpoch: 2, Batch: 130, Avg. Loss: 0.005399909108877182\nEpoch: 2, Batch: 140, Avg. Loss: 0.006226599872112274\nEpoch: 2, Batch: 150, Avg. Loss: 0.005605380952358246\nEpoch: 2, Batch: 160, Avg. Loss: 0.004948697090148926\nEpoch: 2, Batch: 170, Avg. Loss: 0.0067890679836273195\nEpoch: 2, Batch: 180, Avg. Loss: 0.006465445786714554\nEpoch: 2, Batch: 190, Avg. Loss: 0.005668889164924622\nEpoch: 2, Batch: 200, Avg. Loss: 0.0070425220727920535\nEpoch: 2, Batch: 210, Avg. Loss: 0.005862577527761459\nEpoch: 2, Batch: 220, Avg. Loss: 0.006298950493335724\nEpoch: 2, Batch: 230, Avg. Loss: 0.006384554445743561\nEpoch: 2, Batch: 240, Avg. Loss: 0.0052345280647277835\nEpoch: 2, Batch: 250, Avg. Loss: 0.005523312032222748\nEpoch: 2, Batch: 260, Avg. Loss: 0.005630752265453338\nEpoch: 2, Batch: 270, Avg. Loss: 0.004935734122991562\nEpoch: 2, Batch: 280, Avg. Loss: 0.005242919564247131\nEpoch: 2, Batch: 290, Avg. Loss: 0.004648560017347336\nEpoch: 2, Batch: 300, Avg. Loss: 0.004900180578231812\nEpoch: 2, Batch: 310, Avg. Loss: 0.004549592822790146\nEpoch: 2, Batch: 320, Avg. Loss: 0.0043331669867038725\nEpoch: 2, Batch: 330, Avg. Loss: 0.0045249505043029785\nEpoch: 2, Batch: 340, Avg. Loss: 0.005463088899850845\nEpoch: 3, Batch: 10, Avg. Loss: 0.005006200671195984\nEpoch: 3, Batch: 20, Avg. Loss: 0.00550458100438118\nEpoch: 3, Batch: 30, Avg. Loss: 0.0050128822922706606\nEpoch: 3, Batch: 40, Avg. Loss: 0.004417843639850616\nEpoch: 3, Batch: 50, Avg. Loss: 0.004687974244356155\nEpoch: 3, Batch: 60, Avg. Loss: 0.004538481593132019\nEpoch: 3, Batch: 70, Avg. Loss: 0.004627327561378479\nEpoch: 3, Batch: 80, Avg. Loss: 0.004325562924146652\nEpoch: 3, Batch: 90, Avg. Loss: 0.0042263175547122955\nEpoch: 3, Batch: 100, Avg. Loss: 0.004323940992355346\nEpoch: 3, Batch: 110, Avg. Loss: 0.004600764155387878\nEpoch: 3, Batch: 120, Avg. Loss: 0.004572845697402954\nEpoch: 3, Batch: 130, Avg. Loss: 0.004371796518564224\nEpoch: 3, Batch: 140, Avg. Loss: 0.004456951469182968\nEpoch: 3, Batch: 150, Avg. Loss: 0.004546834677457809\nEpoch: 3, Batch: 160, Avg. Loss: 0.004450147211551667\nEpoch: 3, Batch: 170, Avg. Loss: 0.004451075613498688\nEpoch: 3, Batch: 180, Avg. Loss: 0.002990755349397659\nEpoch: 3, Batch: 190, Avg. Loss: 0.0031955537796020507\nEpoch: 3, Batch: 200, Avg. Loss: 0.0028393281549215317\nEpoch: 3, Batch: 210, Avg. Loss: 0.0033632365614175794\nEpoch: 3, Batch: 220, Avg. Loss: 0.003132236450910568\nEpoch: 3, Batch: 230, Avg. Loss: 0.004533502653241157\nEpoch: 3, Batch: 240, Avg. Loss: 0.0032863923758268358\nEpoch: 3, Batch: 250, Avg. Loss: 0.002887480765581131\nEpoch: 3, Batch: 260, Avg. Loss: 0.003464510038495064\nEpoch: 3, Batch: 270, Avg. Loss: 0.0032939517199993135\nEpoch: 3, Batch: 280, Avg. Loss: 0.0023032625168561936\nEpoch: 3, Batch: 290, Avg. Loss: 0.0025704858750104902\nEpoch: 3, Batch: 300, Avg. Loss: 0.002277796685695648\nEpoch: 3, Batch: 310, Avg. Loss: 0.002295256435871124\nEpoch: 3, Batch: 320, Avg. Loss: 0.0027350700348615646\nEpoch: 3, Batch: 330, Avg. Loss: 0.002554924041032791\nEpoch: 3, Batch: 340, Avg. Loss: 0.003393212988972664\nEpoch: 4, Batch: 10, Avg. Loss: 0.002868907153606415\nEpoch: 4, Batch: 20, Avg. Loss: 0.0034082791805267335\nEpoch: 4, Batch: 30, Avg. Loss: 0.0027797412127256393\nEpoch: 4, Batch: 40, Avg. Loss: 0.0024840635508298872\nEpoch: 4, Batch: 50, Avg. Loss: 0.003008595943450928\nEpoch: 4, Batch: 60, Avg. Loss: 0.0027287011221051216\nEpoch: 4, Batch: 70, Avg. Loss: 0.0025225503444671633\nEpoch: 4, Batch: 80, Avg. Loss: 0.0024126685336232187\nEpoch: 4, Batch: 90, Avg. Loss: 0.002343493953347206\nEpoch: 4, Batch: 100, Avg. Loss: 0.002594391778111458\nEpoch: 4, Batch: 110, Avg. Loss: 0.0025090036243200302\nEpoch: 4, Batch: 120, Avg. Loss: 0.002209158018231392\nEpoch: 4, Batch: 130, Avg. Loss: 0.0024918284118175507\nEpoch: 4, Batch: 140, Avg. Loss: 0.0023977086097002028\nEpoch: 4, Batch: 150, Avg. Loss: 0.0026046514958143236\nEpoch: 4, Batch: 160, Avg. Loss: 0.0023334139734506607\nEpoch: 4, Batch: 170, Avg. Loss: 0.002749546319246292\nEpoch: 4, Batch: 180, Avg. Loss: 0.0026977894008159637\nEpoch: 4, Batch: 190, Avg. Loss: 0.002754357025027275\nEpoch: 4, Batch: 200, Avg. Loss: 0.0026075479090213777\nEpoch: 4, Batch: 210, Avg. Loss: 0.002515153482556343\nEpoch: 4, Batch: 220, Avg. Loss: 0.002437335103750229\nEpoch: 4, Batch: 230, Avg. Loss: 0.002396150469779968\nEpoch: 4, Batch: 240, Avg. Loss: 0.0024830902367830275\nEpoch: 4, Batch: 250, Avg. Loss: 0.002703489542007446\nEpoch: 4, Batch: 260, Avg. Loss: 0.0035097808092832564\nEpoch: 4, Batch: 270, Avg. Loss: 0.00313472044467926\nEpoch: 4, Batch: 280, Avg. Loss: 0.002187839150428772\nEpoch: 4, Batch: 290, Avg. Loss: 0.001880323849618435\nEpoch: 4, Batch: 300, Avg. Loss: 0.003062489002943039\nEpoch: 4, Batch: 310, Avg. Loss: 0.0023360235542058943\nEpoch: 4, Batch: 320, Avg. Loss: 0.001972401425242424\nEpoch: 4, Batch: 330, Avg. Loss: 0.0023226851522922514\nEpoch: 4, Batch: 340, Avg. Loss: 0.0024514556378126143\nEpoch: 5, Batch: 10, Avg. Loss: 0.0026924779266119005\nEpoch: 5, Batch: 20, Avg. Loss: 0.003636874780058861\nEpoch: 5, Batch: 30, Avg. Loss: 0.003536754310131073\nEpoch: 5, Batch: 40, Avg. Loss: 0.002854443699121475\nEpoch: 5, Batch: 50, Avg. Loss: 0.002477498412132263\nEpoch: 5, Batch: 60, Avg. Loss: 0.002178221873939037\nEpoch: 5, Batch: 70, Avg. Loss: 0.0021282797306776046\nEpoch: 5, Batch: 80, Avg. Loss: 0.0022899614423513414\nEpoch: 5, Batch: 90, Avg. Loss: 0.002152713105082512\nEpoch: 5, Batch: 100, Avg. Loss: 0.0026276725083589554\nEpoch: 5, Batch: 110, Avg. Loss: 0.0019681509882211686\nEpoch: 5, Batch: 120, Avg. Loss: 0.003086076736450195\nEpoch: 5, Batch: 130, Avg. Loss: 0.0026583701074123383\nEpoch: 5, Batch: 140, Avg. Loss: 0.0025378719717264174\nEpoch: 5, Batch: 150, Avg. Loss: 0.002687408670783043\nEpoch: 5, Batch: 160, Avg. Loss: 0.0025377071350812914\n"
]
],
[
[
"## Test data\n\nSee how your model performs on previously unseen, test data. We've already loaded and transformed this data, similar to the training data. Next, run your trained model on these images to see what kind of keypoints are produced. You should be able to see if your model is fitting each new face it sees, if the points are distributed randomly, or if the points have actually overfitted the training data and do not generalize.",
"_____no_output_____"
]
],
[
[
"# get a sample of test data again\ntest_images, test_outputs, gt_pts = net_sample_output()\n\nprint(test_images.data.size())\nprint(test_outputs.data.size())\nprint(gt_pts.size())",
"torch.Size([10, 1, 224, 224])\ntorch.Size([10, 68, 2])\ntorch.Size([10, 68, 2])\n"
],
[
"## TODO: visualize your test output\n# you can use the same function as before, by un-commenting the line below:\n\n# visualize_output(test_images, test_outputs, gt_pts)\nvisualize_output(test_images, test_outputs, gt_pts)",
"_____no_output_____"
]
],
[
[
"Once you've found a good model (or two), save your model so you can load it and use it later!",
"_____no_output_____"
]
],
[
[
"## TODO: change the name to something uniqe for each new model\nmodel_dir = 'saved_models/'\nmodel_name = 'keypoints_model_1.pt'\n\n# after training, save your model parameters in the dir 'saved_models'\ntorch.save(net.state_dict(), model_dir+model_name)",
"_____no_output_____"
]
],
[
[
"After you've trained a well-performing model, answer the following questions so that we have some insight into your training and architecture selection process. Answering all questions is required to pass this project.",
"_____no_output_____"
],
[
"### Question 1: What optimization and loss functions did you choose and why?\n",
"_____no_output_____"
],
[
"**Answer**: write your answer here (double click to edit this cell)",
"_____no_output_____"
],
[
"### Question 2: What kind of network architecture did you start with and how did it change as you tried different architectures? Did you decide to add more convolutional layers or any layers to avoid overfitting the data?",
"_____no_output_____"
],
[
"**Answer**: write your answer here",
"_____no_output_____"
],
[
"### Question 3: How did you decide on the number of epochs and batch_size to train your model?",
"_____no_output_____"
],
[
"**Answer**: write your answer here",
"_____no_output_____"
],
[
"## Feature Visualization\n\nSometimes, neural networks are thought of as a black box, given some input, they learn to produce some output. CNN's are actually learning to recognize a variety of spatial patterns and you can visualize what each convolutional layer has been trained to recognize by looking at the weights that make up each convolutional kernel and applying those one at a time to a sample image. This technique is called feature visualization and it's useful for understanding the inner workings of a CNN.",
"_____no_output_____"
],
[
"In the cell below, you can see how to extract a single filter (by index) from your first convolutional layer. The filter should appear as a grayscale grid.",
"_____no_output_____"
]
],
[
[
"# Get the weights in the first conv layer, \"conv1\"\n# if necessary, change this to reflect the name of your first conv layer\nweights1 = net.conv1.weight.data\n\nw = weights1.numpy()\n\nfilter_index = 0\n\nprint(w[filter_index][0])\nprint(w[filter_index][0].shape)\n\n# display the filter weights\nplt.imshow(w[filter_index][0], cmap='gray')\n",
"[[ 0.04770301 -0.05072723 -0.17620866 -0.13470505 0.09507251]\n [-0.16137457 -0.01026666 -0.09733022 -0.13840328 -0.02536753]\n [-0.10547946 -0.05649187 0.14990291 0.17614274 -0.14610295]\n [-0.12937796 -0.19437549 0.16836514 -0.06317821 -0.16176204]\n [-0.11310177 -0.13403887 0.07403331 -0.15014146 -0.13802744]]\n(5, 5)\n"
]
],
[
[
"## Feature maps\n\nEach CNN has at least one convolutional layer that is composed of stacked filters (also known as convolutional kernels). As a CNN trains, it learns what weights to include in it's convolutional kernels and when these kernels are applied to some input image, they produce a set of **feature maps**. So, feature maps are just sets of filtered images; they are the images produced by applying a convolutional kernel to an input image. These maps show us the features that the different layers of the neural network learn to extract. For example, you might imagine a convolutional kernel that detects the vertical edges of a face or another one that detects the corners of eyes. You can see what kind of features each of these kernels detects by applying them to an image. One such example is shown below; from the way it brings out the lines in an the image, you might characterize this as an edge detection filter.\n\n<img src='images/feature_map_ex.png' width=50% height=50%/>\n\n\nNext, choose a test image and filter it with one of the convolutional kernels in your trained CNN; look at the filtered output to get an idea what that particular kernel detects.\n\n### TODO: Filter an image to see the effect of a convolutional kernel\n---",
"_____no_output_____"
]
],
[
[
"import cv2\n##TODO: load in and display any image from the transformed test dataset\nimage = test_images[i].data # get the image from it's wrapper\nimage = image.numpy() # convert to numpy array from a Tensor\nimage = np.transpose(image, (1, 2, 0)) # transpose to go from torch to numpy image\n\n\n## TODO: Using cv's filter2D function,\n## apply a specific set of filter weights (like the one displayed above) to the test image\n\n\n\n## TODO: Using cv's filter2D function,\n## apply a specific set of filter weights (like the one displayed above) to the test image\nfiltered_image = cv2.filter2D(image, -1, w[0][0])\n\nf, (ax1, ax2) = plt.subplots(1, 2, figsize=(20,10))\n\nax1.set_title('filter 0 - image in conv1')\nax1.imshow(filtered_image, cmap='gray')\n\nax2.set_title('filter 0 - image in conv2')\nax2.imshow( cv2.filter2D(image, -1, w[0][0]), cmap='gray')",
"_____no_output_____"
]
],
[
[
"### Question 4: Choose one filter from your trained CNN and apply it to a test image; what purpose do you think it plays? What kind of feature do you think it detects?\n",
"_____no_output_____"
],
[
"**Answer**: (does it detect vertical lines or does it blur out noise, etc.) write your answer here",
"_____no_output_____"
],
[
"---\n## Moving on!\n\nNow that you've defined and trained your model (and saved the best model), you are ready to move on to the last notebook, which combines a face detector with your saved model to create a facial keypoint detection system that can predict the keypoints on *any* face in an image!",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
cb8c8e3ec936dc94dea962c8fdb3c37ba850d45e
| 308,804 |
ipynb
|
Jupyter Notebook
|
numpy_class/phanguyen_numpy_class.ipynb
|
tuanthi/Machine-Learning-Course
|
591166a75860d1499fc8f9538e854a7b6f97f61a
|
[
"MIT"
] | null | null | null |
numpy_class/phanguyen_numpy_class.ipynb
|
tuanthi/Machine-Learning-Course
|
591166a75860d1499fc8f9538e854a7b6f97f61a
|
[
"MIT"
] | null | null | null |
numpy_class/phanguyen_numpy_class.ipynb
|
tuanthi/Machine-Learning-Course
|
591166a75860d1499fc8f9538e854a7b6f97f61a
|
[
"MIT"
] | null | null | null | 657.029787 | 153,928 | 0.946801 |
[
[
[
"# Ex1",
"_____no_output_____"
]
],
[
[
"import numpy as np\nfrom matplotlib import pyplot as plt\nimport pandas as pd",
"_____no_output_____"
],
[
"v = np.array([1/3, 1/3, 1/3])\nA = np.array([[0.3, 0.6, 0.1],\n [0.5, 0.2, 0.3],\n [0.4, 0.1, 0.5]])",
"_____no_output_____"
],
[
"dist = []\nfor i in range(25):\n vf = v.dot(A)\n dist.append(np.linalg.norm(v-vf))\n v = vf\nplt.plot(range(25), dist)\nplt.show()",
"_____no_output_____"
]
],
[
[
"# Ex2",
"_____no_output_____"
]
],
[
[
"y = []\nfor i in range(1000):\n x = np.random.uniform(0, 1, 1000)\n y.append(np.sum(x))\nplt.hist(y, bins = 30)\nplt.show()\nprint('Expected mean: ', np.mean(y))\nprint('Expected variance: ', np.var(y))",
"_____no_output_____"
]
],
[
[
"# Ex3",
"_____no_output_____"
]
],
[
[
"df = pd.read_csv('../../machine_learning_examples/large_files/train.csv')",
"_____no_output_____"
],
[
"for i in range(10):\n number = df[df.label == i].as_matrix()[:,1:].mean(axis = 0).reshape(28,28)\n plt.imshow(number)\n plt.show()",
"_____no_output_____"
]
],
[
[
"# Ex4",
"_____no_output_____"
]
],
[
[
"plt.imshow(np.rot90(number, 3))\nplt.show()",
"_____no_output_____"
],
[
"def rotate_img(img):\n _img = np.zeros(img.shape)\n for i in range(img.shape[0]):\n for j in range(img.shape[1]):\n _img[i, j] = img[img.shape[0] - j - 1, i]\n return _img\nplt.imshow(rotate_img(number))\nplt.show()",
"_____no_output_____"
]
],
[
[
"# Ex5",
"_____no_output_____"
]
],
[
[
"def np_is_symmetric(matrix):\n return np.allclose(matrix, matrix.T)",
"_____no_output_____"
],
[
"def manual_is_symmetric(matrix):\n for i in range(matrix.shape[0]):\n for j in range(matrix.shape[1]):\n if matrix[i, j] != matrix[j, i]: return False\n return True",
"_____no_output_____"
]
],
[
[
"# Ex6",
"_____no_output_____"
]
],
[
[
"def color(x, y):\n if (x > 0) ^ (y >0): return 'red'\n else: return 'blue'\n\na = np.random.uniform(-1, 1, 2000)\nb = np.random.uniform(-1, 1, 2000)\n\nfor i, j in zip(a, b):\n plt.scatter(i, j, c = color(i, j), edgecolor='black', alpha=0.3)\nplt.show()\n",
"_____no_output_____"
]
],
[
[
"# Ex7",
"_____no_output_____"
]
],
[
[
"import math\n\na = np.random.uniform(-30, 30, 3000)\nb = np.random.uniform(-30, 30, 3000)\n\ndef dist(x, y):\n return math.sqrt((x[0] - y[0])**2 + (x[1] - y[1])**2)\n\ndef draw(a, b, radius, size = 2):\n global count\n if (dist((a, b), (0, 0)) > radius - size) and (dist((a, b), (0, 0)) < radius + size): return True\n else: return False\n \nfor i, j in zip(a, b):\n if draw(i, j, 10): plt.scatter(i, j, c='blue', edgecolor='black', alpha=0.3)\n if draw(i, j, 20): plt.scatter(i, j, c='red', edgecolor='black', alpha=0.3)\n\nplt.axis('equal')\nplt.show()",
"_____no_output_____"
]
],
[
[
"# Ex8",
"_____no_output_____"
],
[
"# Ex9",
"_____no_output_____"
]
],
[
[
"x = []\nfor i, j in zip(a, b):\n if draw(i, j, 10): x.append([i, j, 0])\n if draw(i, j, 20): x.append([i, j, 1])\nx = np.array(x)\ndf = pd.DataFrame(x)\ndf.columns = ['x1', 'x2', 'y']\ndf.to_csv('donuts.csv')\nprint(df.head())",
" x1 x2 y\n0 -2.874956 -21.566285 1.0\n1 -19.992435 -1.937197 1.0\n2 -0.798693 -8.528103 0.0\n3 -4.270119 17.569628 1.0\n4 8.982702 0.762297 0.0\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
]
] |
cb8ca1aa382bf791efe1f391b3a77528e57e147a
| 40,230 |
ipynb
|
Jupyter Notebook
|
notebooks/data_vis_with_safas.ipynb
|
cskv/safas
|
3f62a90f05bc2c9d54bd744cb5dd373614def373
|
[
"MIT"
] | 2 |
2020-03-25T12:53:51.000Z
|
2021-12-27T07:15:29.000Z
|
notebooks/data_vis_with_safas.ipynb
|
cskv/safas
|
3f62a90f05bc2c9d54bd744cb5dd373614def373
|
[
"MIT"
] | 8 |
2021-08-05T02:15:57.000Z
|
2021-09-16T11:52:22.000Z
|
notebooks/data_vis_with_safas.ipynb
|
cskv/safas
|
3f62a90f05bc2c9d54bd744cb5dd373614def373
|
[
"MIT"
] | 1 |
2020-04-19T13:48:04.000Z
|
2020-04-19T13:48:04.000Z
| 245.304878 | 35,372 | 0.919115 |
[
[
[
"# Data Visualization With Safas\n\nThis notebook demonstrates plotting the results from Safas video analysis.",
"_____no_output_____"
],
[
"## Import modules and data\n\nImport safas and other components for display and analysis. safas has several example images in the safas/data directory. These images are accessible as attributes of the data module because the __init__ function of safas/data also acts as a loader. ",
"_____no_output_____"
]
],
[
[
"import sys\nfrom matplotlib import pyplot as plt\nfrom matplotlib.ticker import ScalarFormatter \n%matplotlib inline\n\nimport pandas as pd\nimport cv2\n\nfrom safas import filters\nfrom safas import data\nfrom safas.filters.sobel_focus import imfilter as sobel_filter\nfrom safas.filters.imfilters_module import add_contours",
"_____no_output_____"
]
],
[
[
"## Object properties\nUsers may interactively select and link objects in safas. When the data is saved, the data is written in tabular form where the properties of each oject are stored. \n\nThe object properties are calculated with the Scikit-Image function [regionprops.](https://scikit-image.org/docs/dev/api/skimage.measure.html#skimage.measure.regionprops). A complete description of these properties may be found in the [regionprops.](https://scikit-image.org/docs/dev/api/skimage.measure.html#skimage.measure.regionprops) documentation. At this time, the following properties are stored in a .xlsx file: \n\nproperty | unit |\n--- | --- |\narea | $\\mu m^2$ | \nequivalent_diameter | $\\mu m$ | \nperimeter | $\\mu m$ | \neuler_number | -- |\nminor_axis_length | $\\mu m$ | \nmajor_axis_length\t| $\\mu m$ | \nextent\t| -- | \n\nIf a selected object is linked to an object in the next frame, the instantaneous velocity will be calculated based on the displacement of the object centroid and the frame rate of the video. \n\nproperty | unit | description\n--- | --- | ---|\nvel_mean | [mm/s]\t| velocity |\nvel_N\t| [--] | number of objects linked|\nvel_std | [mm/s] | standard deviation of velocity |\n",
"_____no_output_____"
],
[
"## Plot a settling velocity versus floc size\n",
"_____no_output_____"
]
],
[
[
"# load the excel file as a Pandas DataFrame\ndf = pd.read_excel('data/floc_props.xlsx')\n# see the keys\nprint(df.keys())\n",
"Index(['Unnamed: 0', 'area', 'equivalent_diameter', 'perimeter',\n 'euler_number', 'minor_axis_length', 'major_axis_length', 'extent',\n 'vel_mean', 'vel_N', 'vel_std'],\n dtype='object')\n"
],
[
"# plot velocity vs major_axis_length\nf, ax = plt.subplots(1,1, figsize=(3.5, 2.2), dpi=250)\n\n# note: remove *10 factor if floc_props.xlsx file is updated: previous version was output in [cm/s]\nax.plot(df.major_axis_length, df.vel_mean*10, marker='o', linestyle='None')\nax.set_xlabel('Floc size [$\\mu m$]')\nax.set_ylabel('Settling velocity [mm/s]')\n\n# convert to log-log\nax.loglog()\nax.axis([100, 5000, 0.1, 100])\n\nfor axis in [ax.xaxis, ax.yaxis]:\n axis.set_major_formatter(ScalarFormatter())\n\nplt.tight_layout()\n\nsave = True\n\nif save: \n plt.savefig('png/vel_size.png', dpi=900)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
]
] |
cb8cb0df19390ee11704cdfc2f6db17b2248686a
| 13,646 |
ipynb
|
Jupyter Notebook
|
content/lessons/05/Class-Coding-Lab/CCL-Iterations.ipynb
|
auramnar/Summer2018Learn2Code
|
068b89215ad2b90f57a73511b0b1b6b126589c5f
|
[
"MIT"
] | null | null | null |
content/lessons/05/Class-Coding-Lab/CCL-Iterations.ipynb
|
auramnar/Summer2018Learn2Code
|
068b89215ad2b90f57a73511b0b1b6b126589c5f
|
[
"MIT"
] | null | null | null |
content/lessons/05/Class-Coding-Lab/CCL-Iterations.ipynb
|
auramnar/Summer2018Learn2Code
|
068b89215ad2b90f57a73511b0b1b6b126589c5f
|
[
"MIT"
] | null | null | null | 32.724221 | 394 | 0.524476 |
[
[
[
"# In-Class Coding Lab: Iterations\n\nThe goals of this lab are to help you to understand:\n\n- How loops work.\n- The difference between definite and indefinite loops, and when to use each.\n- How to build an indefinite loop with complex exit conditions.\n- How to create a program from a complex idea.\n",
"_____no_output_____"
],
[
"# Understanding Iterations\n\nIterations permit us to repeat code until a Boolean expression is `False`. Iterations or **loops** allow us to write succint, compact code. Here's an example, which counts to 3 before [Blitzing the Quarterback in backyard American Football](https://www.quora.com/What-is-the-significance-of-counting-one-Mississippi-two-Mississippi-and-so-on):",
"_____no_output_____"
]
],
[
[
"i = 1\nwhile i <= 4:\n print(i,\"Mississippi...\")\n i=i+1\nprint(\"Blitz!\")",
"1 Mississippi...\n2 Mississippi...\n3 Mississippi...\n4 Mississippi...\nBlitz!\n"
]
],
[
[
"## Breaking it down...\n\nThe `while` statement on line 2 starts the loop. The code indented beneath it (lines 3-4) will repeat, in a linear fashion until the Boolean expression on line 2 `i <= 3` is `False`, at which time the program continues with line 5.\n\n### Some Terminology\n\nWe call `i <=3` the loop's **exit condition**. The variable `i` inside the exit condition is the only thing that we can change to make the exit condition `False`, therefore it is the **loop control variable**. On line 4 we change the loop control variable by adding one to it, this is called an **increment**.\n\nFurthermore, we know how many times this loop will execute before it actually runs: 3. Even if we allowed the user to enter a number, and looped that many times, we would still know. We call this a **definite loop**. Whenever we iterate over a fixed number of values, regardless of whether those values are determined at run-time or not, we're using a definite loop.\n\nIf the loop control variable never forces the exit condition to be `False`, we have an **infinite loop**. As the name implies, an Infinite loop never ends and typically causes our computer to crash or lock up. ",
"_____no_output_____"
]
],
[
[
"## WARNING!!! INFINITE LOOP AHEAD\n## IF YOU RUN THIS CODE YOU WILL NEED TO KILL YOUR BROWSER AND SHUT DOWN JUPYTER NOTEBOOK\n\ni = 1\nwhile i <= 3:\n print(i,\"Mississippi...\")\n# i=i+1\nprint(\"Blitz!\")",
"_____no_output_____"
]
],
[
[
"### For loops\n\nTo prevent an infinite loop when the loop is definite, we use the `for` statement. Here's the same program using `for`:",
"_____no_output_____"
]
],
[
[
"for i in range(1,4):\n print(i,\"Mississippi...\")\nprint(\"Blitz!\")",
"1 Mississippi...\n2 Mississippi...\n3 Mississippi...\nBlitz!\n"
]
],
[
[
"One confusing aspect of this loop is `range(1,4)` why does this loop from 1 to 3? Why not 1 to 4? Well it has to do with the fact that computers start counting at zero. The easier way to understand it is if you subtract the two numbers you get the number of times it will loop. So for example, 4-1 == 3.\n\n### Now Try It\n\nIn the space below, Re-Write the above program to count from 10 to 15. Note: How many times will that loop?",
"_____no_output_____"
]
],
[
[
"# TODO Write code here\nfor i in range(10,16):\n print(i, \"Mississippi...\")\nprint(\"Blitz!\")\n",
"10 Mississippi...\n11 Mississippi...\n12 Mississippi...\n13 Mississippi...\n14 Mississippi...\n15 Mississippi...\nBlitz!\n"
]
],
[
[
"## Indefinite loops\n\nWith **indefinite loops** we do not know how many times the program will execute. This is typically based on user action, and therefore our loop is subject to the whims of whoever interacts with it. Most applications like spreadsheets, photo editors, and games use indefinite loops. They'll run on your computer, seemingly forever, until you choose to quit the application. \n\nThe classic indefinite loop pattern involves getting input from the user inside the loop. We then inspect the input and based on that input we might exit the loop. Here's an example:",
"_____no_output_____"
]
],
[
[
"name = \"\"\nwhile name != 'mike':\n name = input(\"Say my name! : \")\n print(\"Nope, my name is not %s! \" %(name))",
"Say my name! : bill\nNope, my name is not bill! \nSay my name! : mike\nNope, my name is not mike! \n"
]
],
[
[
"The classic problem with indefinite loops is that its really difficult to get the application's logic to line up with the exit condition. For example we need to set `name = \"\"` in line 1 so that line 2 start out as `True`. Also we have this wonky logic where when we say `'mike'` it still prints `Nope, my name is not mike!` before exiting.\n\n### Break statement\n\nThe solution to this problem is to use the break statement. **break** tells Python to exit the loop immediately. We then re-structure all of our indefinite loops to look like this:\n\n```\nwhile True:\n if exit-condition:\n break\n```\n\nHere's our program we-written with the break statement. This is the recommended way to write indefinite loops in this course.",
"_____no_output_____"
]
],
[
[
"while True:\n name = input(\"Say my name!: \")\n if name == 'mike':\n break\n print(\"Nope, my name is not %s!\" %(name))",
"Say my name!: bill\nNope, my name is not bill!\nSay my name!: dave\nNope, my name is not dave!\nSay my name!: mike\n"
]
],
[
[
"### Multiple exit conditions\n\nThis indefinite loop pattern makes it easy to add additional exit conditions. For example, here's the program again, but it now stops when you say my name or type in 3 wrong names. Make sure to run this program a couple of times. First enter mike to exit the program, next enter the wrong name 3 times.",
"_____no_output_____"
]
],
[
[
"times = 0\nwhile True:\n name = input(\"Say my name!: \")\n times = times + 1\n if name == 'mike':\n print(\"You got it!\")\n break\n if times == 3:\n print(\"Game over. Too many tries!\")\n break\n print(\"Nope, my name is not %s!\" %(name))",
"Say my name!: dsjfh\nNope, my name is not dsjfh!\nSay my name!: sdaf\nNope, my name is not sdaf!\nSay my name!: sdf\nGame over. Too many tries!\n"
]
],
[
[
"# Number sums\n\nLet's conclude the lab with you writing your own program which\n\nuses an indefinite loop. We'll provide the to-do list, you write the code. This program should ask for floating point numbers as input and stops looping when **the total of the numbers entered is over 100**, or **more than 5 numbers have been entered**. Those are your two exit conditions. After the loop stops print out the total of the numbers entered and the count of numbers entered. ",
"_____no_output_____"
]
],
[
[
"## TO-DO List\n\n#1 count = 0\n#2 total = 0\n#3 loop Indefinitely\n#4. input a number\n#5 increment count\n#6 add number to total\n#7 if count equals 5 stop looping\n#8 if total greater than 100 stop looping\n#9 print total and count",
"_____no_output_____"
],
[
"# Write Code here:\n\n# Title and Header\nprint(\"Race to 100 Plus!!!\")\nprint(\"*\" * 40)\nprint(\"This program adds 5 numbers the goal being to exceed 100. If you exceed 100 or enter 5 numbers program ends.\")\nprint(\"*\" * 40)\n\n#Program\n \ncount = 0\ntotal = 0\ntry:\n while True:\n number = float(input(\"Input a number less than or equal to 100: \"))\n count += 1\n total = total + number\n if count == 5:\n print(\"*\" * 40)\n print(\"You enter the max amout of numberers. Your total is.\")\n print(\"*\" * 40)\n break\n if total > 100:\n print(\"*\" * 40)\n print(\"You've exceeded 100 in less than 5 numbers! Your total is.\")\n print(\"*\" * 40)\n break\n print(\"*\" * 40)\n print(\"Please enter another number.\")\n print(\"*\" * 40)\n if total > 100:\n print(\"Your total is %.2f after %.0f entries. You exceeded 100!\" % (total, count))\n print(\"*\" * 40)\n else:\n print(\"Your total is %.2f after %.0f entries. You did not exceed 100!\" % (total, count))\n print(\"*\" * 40)\nexcept ValueError:\n print(\"*\" * 40)\n print(\"Error 10001: Numerical Value Error. You did not enter a number! \")\n print(\"*\" * 40)",
"Race to 100 Plus!!!\n****************************************\nThis program adds 5 numbers the goal being to exceed 100. If you exceed 100 or enter 5 numbers program ends.\n****************************************\nInput a number less than or equal to 100: 3\n****************************************\nPlease enter another number.\n****************************************\nInput a number less than or equal to 100: 3\n****************************************\nPlease enter another number.\n****************************************\nInput a number less than or equal to 100: 23\n****************************************\nPlease enter another number.\n****************************************\nInput a number less than or equal to 100: 32\n****************************************\nPlease enter another number.\n****************************************\nInput a number less than or equal to 100: 23\n****************************************\nYou enter the max amout of numberers. Your total is.\n****************************************\nYour total is 84.00 after 5 entries. You did not exceed 100!\n****************************************\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
cb8ccb1be4c7c5708310df96617e8d3a8dc2e006
| 9,070 |
ipynb
|
Jupyter Notebook
|
exercise_2/code/stat_modeling_primer.ipynb
|
euberdeveloper/statistische_methoden_der_systemgenetic
|
9b308b867e24ccb3801fc641b9604df77634156c
|
[
"MIT"
] | null | null | null |
exercise_2/code/stat_modeling_primer.ipynb
|
euberdeveloper/statistische_methoden_der_systemgenetic
|
9b308b867e24ccb3801fc641b9604df77634156c
|
[
"MIT"
] | null | null | null |
exercise_2/code/stat_modeling_primer.ipynb
|
euberdeveloper/statistische_methoden_der_systemgenetic
|
9b308b867e24ccb3801fc641b9604df77634156c
|
[
"MIT"
] | null | null | null | 27.319277 | 141 | 0.561191 |
[
[
[
"# 3. Example: Univariate Gaussian",
"_____no_output_____"
]
],
[
[
"# Install and load deps\n\nif (!require(\"stringr\")) {\n install.packages(\"stringr\")\n}\n\nlibrary(purrr)",
"_____no_output_____"
]
],
[
[
"As an example, we consider the heights in cm of 20 individuals:\n\nWe will model the heights using the univariate Gaussian. The univariate Gaussian has two\nparameters, its mean and variance, which we aim to estimate.",
"_____no_output_____"
]
],
[
[
".heights_str <- \"178 162 178 178 169 150 156 162 165 189 173 157 154 162 162 161 168 156 169 153\"\nheights <- unlist(\n map(\n str_split(.heights_str, \" \"),\n function(el) {\n as.numeric(el)\n }\n )\n)\n\nheights",
"_____no_output_____"
]
],
[
[
"### Step 1. Data generative model\n\nWe assume the data i.i.d. and choose the univariate Gaussian as model. The `p` will become `N` and the parameters (`mu`, `sigma^2`).\n\nFor the i.i.d., `p(X, params) = mult(p(x_i, params))\n",
"_____no_output_____"
],
[
"### Step 2. Simulate data\n\nWe can simulate data according to the model using the R function `rnorm()` which draws random samples of the normal distribution.",
"_____no_output_____"
]
],
[
[
"simulate <- function(n, theta) {\n x <- rnorm(n, mean = theta[[\"mu\"]], sd = theta[[\"sigma\"]])\n return(x)\n}\n\nn <- 20\ntheta <- c(mu = 175, sigma = 5)\n\nx <- simulate(n, theta)\nx",
"_____no_output_____"
]
],
[
[
"### Step 3. Parameter estimation procedure",
"_____no_output_____"
],
[
"We consider maximum likelihood estimation of the parameters, so the parameters that maximize `L(theta, X)`.\n\nTriks:\n* Minimize the negative log-likelihood `NLL(theta)` which is equal to `-log(L(theta, X))`\n* Reparametrize Gaussian using precision, which is `1/(sigma^2)`\n\nThe parameters estimates are gotten by setting the NLL to the null vector **0**.\n\n*Strange math calcs.......*\n\n`mu` is equal to the sample **mean** while `sigma` is equal to the **biased sample variance**.\n",
"_____no_output_____"
],
[
"### Step 4. Implementation and empirical verification",
"_____no_output_____"
],
[
"We code our estimation procedure into a R function `estimate()`",
"_____no_output_____"
]
],
[
[
"estimate <- function(x) {\n mean_x <- mean(x)\n\n theta_hat <- c(\n mu = mean_x,\n sigma = sqrt(mean((x - mean_x)^2))\n )\n return(theta_hat)\n}",
"_____no_output_____"
],
[
"n <- 20\nx <- simulate(n, theta = c(mu = 175, sigma = 5))\n\ntheta_hat <- estimate(x)\ntheta_hat",
"_____no_output_____"
]
],
[
[
"These estimates are not too far from the ground truth values. Our simple check is good enough\nfor this didactic toy example. The code would allow you to investigate more systematically\nthe relationships between estimates and ground truth with various values of the parameters\nand sample size n.",
"_____no_output_____"
],
[
"### Step 5. Application to real data",
"_____no_output_____"
],
[
"We finally apply our estimation to the original dataset:",
"_____no_output_____"
]
],
[
[
"theta_hat <- estimate(heights)\ntheta_hat",
"_____no_output_____"
]
],
[
[
"# 4. Assessing whether a distribution fits data with Q-Q plots",
"_____no_output_____"
],
[
"The strategy described in section 1 allows assessing whether an estimation procedure returns\nreasonable estimates on simulated data. It does not assess however ether the simulation\nassumptions, i.e. the data generative model, is a reasonable model for the data at hand.\nOne key modeling assumption of a data generative model is the choice of the distribution.\nThe quantile-quantile plot is a graphical tool to assess whether a distribution fits the data\nreasonably.\n\nAs a concrete example, let’s consider 50 data points coming from the uniform distribution\nin the [2,3] interval. If you assume your data comes from the uniform distribution in the\n[2,3] interval, you expect the first 10% of your data to fall in [2,2.1], the second 10% in\n[2.1,2.2] and so forth. A histogram could be used to visually assess this agreement. However,\nhistograms are shaky because of possible low counts in every bin:",
"_____no_output_____"
]
],
[
[
"par(cex = 0.7)\nu <- runif(50, min = 2, max = 3)\nhist(u, main = \"\")",
"_____no_output_____"
]
],
[
[
"Instead of the histogram, one could plot the deciles of the sample distribution against those\nof a theoretical distribution. Here are the deciles:",
"_____no_output_____"
]
],
[
[
"dec <- quantile(u, seq(0, 1, 0.1))\ndec",
"_____no_output_____"
],
[
"par(cex = 0.7)\nplot(seq(2, 3, 0.1),\n dec,\n xlim = c(2, 3), ylim = c(2, 3),\n xlab = \"Deciles of the uniform distribution over [2,3]\",\n ylab = \"Deciles of the dataset\"\n)\nabline(0, 1) ## diagonal y=x",
"_____no_output_____"
]
],
[
[
"Now we see a clear agreement between the expected values of the deciles of the theoretical\ndistribution (x-axis) and those empirically observed (y-axis). The advantage of this strategy is\nthat it also generalizes to other distributions (e.g. Normal), where the shape of the density\ncan be difficult to assess with a histogram.",
"_____no_output_____"
],
[
"For a finite sample we can estimate the quantile for every data point, not just the deciles.\nThe Q-Q plot scatter plots the quantiles of two distributions against each other. One way is\nto use as expected quantile (r − 0.5)/N (Hazen, 1914), where r is the rank of the data point.\nThe R function ppoints gives more accurate values:\n",
"_____no_output_____"
]
],
[
[
"par(cex = 0.7)\nplot(qunif(ppoints(length(u)), min = 2, max = 3), sort(u),\n xlim = c(2, 3), ylim = c(2, 3),\n xlab = \"Quantiles of the uniform distribution over [2,3]\",\n ylab = \"Quantiles of the dataset\"\n)\nabline(0, 1)",
"_____no_output_____"
]
],
[
[
"In R, Q-Q plots between two datasets can be generated using the function qqplot() . In the\nspecial case of a normal distribution use the function qqnorm() and the function qqline() ,\nwhich adds a line to the “theoretical” quantile-quantile plot passing through the first and\nthird quartiles.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
cb8cdfe5ca638e6d97277d2974e13f159b1a4458
| 39,359 |
ipynb
|
Jupyter Notebook
|
notebooks/decision tree-Copy1.ipynb
|
sborquez/HER2_Fuzzy_Logic
|
f685510f017850c2dd36fab9bce5297491a0dd2b
|
[
"MIT"
] | null | null | null |
notebooks/decision tree-Copy1.ipynb
|
sborquez/HER2_Fuzzy_Logic
|
f685510f017850c2dd36fab9bce5297491a0dd2b
|
[
"MIT"
] | null | null | null |
notebooks/decision tree-Copy1.ipynb
|
sborquez/HER2_Fuzzy_Logic
|
f685510f017850c2dd36fab9bce5297491a0dd2b
|
[
"MIT"
] | null | null | null | 81.657676 | 27,136 | 0.832287 |
[
[
[
"<h2 align=\"center\">INF575 - Fuzzy Logic</h2>\n<h1 align=\"center\">Segmentation of HER2 Overexpression in Histopathology Images with Fuzzy Decision Tree<h1>\n \n<center>\n <img src=\"https://rochepacientes.es/content/dam/roche-pacientes-2/es/assets/images/que-es-her2.jpg\" width=\"60%\"/>\n</center>\n\n<h2 align=\"center\">Classic Decision Tree</h2>\n\n<center>\n<i> Sebastián Bórquez G. - <a href=\"mailto://[email protected]\">[email protected]</a> - DI UTFSM - August 2020.</i>\n</center>",
"_____no_output_____"
]
],
[
[
"%cd ..",
"/mnt/d/sebas/Desktop/her2_FL\n"
],
[
"import cv2\nimport matplotlib.pyplot as plt\nimport pandas as pd\nimport numpy as np\nimport seaborn as sns; sns.set(palette=\"muted\")\nfrom IPython.display import display, HTML\n\nfrom load_features import *\n\nfrom sklearn.datasets import load_iris\nfrom sklearn.tree import DecisionTreeClassifier, plot_tree\nfrom sklearn.ensemble import RandomForestClassifier",
"_____no_output_____"
],
[
"selected_features = [\n 'mean_rawblue', \n 'mean_dab',\n 'mean_intentsity',\n 'mean_rawgreen',\n 'mean_eosin',\n 'mean_vertical',\n 'mean_rawbred',\n 'homogeneity_vertical',\n 'mean_hematoxylin',\n 'sobel_magnitud'\n]",
"_____no_output_____"
],
[
"train_csv_files = [\n \"./data/1+_2.csv\",\n \"./data/1+_20.csv\",\n \"./data/2+_1.csv\",\n \"./data/2+_8.csv\",\n \"./data/3+_16.csv\",\n \"./data/3+_15.csv\",\n]\ntrain_features = merge_features([load_features(csv_file, selected_features=selected_features) for csv_file in train_csv_files])\n(feature_names, target_col), (train_X, train_y) = split_features_target(train_features)\n\ntest_csv_files = [\n \"./data/1+_25.csv\",\n \"./data/2+_9.csv\",\n \"./data/3+_15.csv\",\n]\ntest_features = merge_features([load_features(csv_file, selected_features=selected_features) for csv_file in test_csv_files])\ntest_X, test_y = split_features_target(test_features)",
"_____no_output_____"
],
[
"# Parameters\nclass_weight = {0: 1., 1: 20.}\nmin_samples_leaf = 15\nmax_depth=5",
"_____no_output_____"
]
],
[
[
"## Train",
"_____no_output_____"
]
],
[
[
"# Train\nclf = DecisionTreeClassifier(class_weight=class_weight, min_samples_leaf=min_samples_leaf).fit(train_X, train_y)",
"_____no_output_____"
],
[
"# Train\nrf = RandomForestClassifier(n_estimators=30, class_weight=class_weight, min_samples_leaf=min_samples_leaf).fit(train_X, train_y)",
"_____no_output_____"
],
[
"train_images = train_features.image.unique()\nfor train_image in train_images:\n image_features = train_features[train_features.image == train_image]\n X_i, y_i = split_features_target(image_features, True)\n predicted = clf.predict_proba(X_i)[:,1]\n show_images_and_masks(train_image, image_features, predicted)",
"_____no_output_____"
],
[
"index=np.argsort(clf.feature_importances_,)\nplt.figure(figsize=(6,8))\nplt.title('DT - Feature Importance')\nplt.barh(np.arange(len(clf.feature_importances_)), clf.feature_importances_[index], tick_label=np.array(feature_names)[index])",
"_____no_output_____"
],
[
"train_images = train_features.image.unique()\nfor train_image in train_images:\n image_features = train_features[train_features.image == train_image]\n X_i, y_i = split_features_target(image_features, True)\n predicted = rf.predict_proba(X_i)[:,1]\n show_images_and_masks(train_image, image_features, predicted)",
"_____no_output_____"
],
[
"index=np.argsort(rf.feature_importances_)\nplt.figure(figsize=(6,8))\nplt.title('RF - Feature Importance')\nplt.barh(np.arange(len(rf.feature_importances_)), rf.feature_importances_[index], tick_label=np.array(feature_names)[index])",
"_____no_output_____"
]
],
[
[
"## Test",
"_____no_output_____"
]
],
[
[
"from time import time",
"_____no_output_____"
],
[
"test_images = test_features.image.unique()\ntrue_targets = []\nfor test_image in test_images:\n image_features = test_features[test_features.image == test_image]\n _, test_y_i = split_features_target(image_features, True)\n true_targets.append(test_y_i)\ntrue_targets = np.hstack(true_targets)",
"_____no_output_____"
],
[
"test_images = test_features.image.unique()\ndt_predicted = []\nstart = time()\nfor test_image in test_images:\n image_features = test_features[test_features.image == test_image]\n test_X_i, test_y_i = split_features_target(image_features, True)\n #dt_predicted_i = clf.predict_proba(test_X_i)[:,1]\n #dt_predicted.append(dt_predicted_i)\n #show_images_and_masks(test_image, image_features, dt_predicted_i)\nend = time()\n#dt_predicted = np.hstack(dt_predicted)",
"_____no_output_____"
],
[
"print(end - start)",
"1.215993881225586\n"
],
[
"test_images = test_features.image.unique()\nrf_predicted = []\nstart = time()\nfor test_image in test_images:\n image_features = test_features[test_features.image == test_image]\n test_X_i, test_y_i = split_features_target(image_features, True)\n #predicted = rf.predict_proba(test_X_i)[:,1]\n rf_predicted_i = rf.predict(test_X_i)\n #rf_predicted.append(rf_predicted_i)\n #show_images_and_masks(test_image, image_features, rf_predicted_i)\nend=time()\n#rf_predicted = np.hstack(rf_predicted)",
"_____no_output_____"
],
[
"print(end - start)",
"21.681201219558716\n"
],
[
"3*14.635354*60/3e6",
"_____no_output_____"
],
[
"1.215993881225586/3e6",
"_____no_output_____"
],
[
"21.681201219558716/3e6\n",
"_____no_output_____"
],
[
"results = pd.DataFrame({\n \"target\": true_targets,\n \"decision tree\": dt_predicted.astype(int),\n \"random forest\": rf_predicted.astype(int)\n})\nresults.to_csv(\"crisp_results.csv\", index=False)",
"_____no_output_____"
],
[
"from sklearn.metrics import classification_report\n\nprint(\"Decision Tree\")\nprint(classification_report(results[\"target\"], results[\"decision tree\"], target_names=[\"non-overexpression\", \"overexpression\"]))\n\n\nprint(\"Random Forest\")\nprint(classification_report(results[\"target\"], results[\"random forest\"], target_names=[\"non-overexpression\", \"overexpression\"]))",
"Decision Tree\n precision recall f1-score support\n\nnon-overexpression 0.93 1.00 0.96 2781471\n overexpression 0.97 0.04 0.08 211306\n\n accuracy 0.93 2992777\n macro avg 0.95 0.52 0.52 2992777\n weighted avg 0.93 0.93 0.90 2992777\n\nRandom Forest\n precision recall f1-score support\n\nnon-overexpression 0.99 0.92 0.95 2781471\n overexpression 0.46 0.87 0.60 211306\n\n accuracy 0.92 2992777\n macro avg 0.72 0.89 0.78 2992777\n weighted avg 0.95 0.92 0.93 2992777\n\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb8cea007cccbee4fee90968c3d1ee1963fb5fec
| 9,397 |
ipynb
|
Jupyter Notebook
|
examples/pyterrier_kwiksort_learned.ipynb
|
heinrichreimer/ir_axioms
|
f7349c4adde96cfa19c7247824a70a4662c07582
|
[
"MIT"
] | 5 |
2022-03-11T15:28:04.000Z
|
2022-03-11T15:28:58.000Z
|
examples/pyterrier_kwiksort_learned.ipynb
|
heinrichreimer/ir_axioms
|
f7349c4adde96cfa19c7247824a70a4662c07582
|
[
"MIT"
] | null | null | null |
examples/pyterrier_kwiksort_learned.ipynb
|
heinrichreimer/ir_axioms
|
f7349c4adde96cfa19c7247824a70a4662c07582
|
[
"MIT"
] | null | null | null | 28.56231 | 812 | 0.520166 |
[
[
[
"from sys import modules\n\nIN_COLAB = 'google.colab' in modules\nif IN_COLAB:\n !pip install -q ir_axioms[examples] python-terrier",
"_____no_output_____"
],
[
"# Start/initialize PyTerrier.\nfrom pyterrier import started, init\n\nif not started():\n init(tqdm=\"auto\", no_download=True)",
"PyTerrier 0.8.0 has loaded Terrier 5.6 (built by craigmacdonald on 2021-09-17 13:27)\n\nNo etc/terrier.properties, using terrier.default.properties for bootstrap configuration.\n"
],
[
"from pyterrier.datasets import get_dataset, Dataset\n\n# Load dataset.\ndataset_name = \"msmarco-passage\"\ndataset: Dataset = get_dataset(f\"irds:{dataset_name}\")\ndataset_train: Dataset = get_dataset(f\"irds:{dataset_name}/trec-dl-2019/judged\")\ndataset_test: Dataset = get_dataset(f\"irds:{dataset_name}/trec-dl-2020/judged\")",
"_____no_output_____"
],
[
"from pathlib import Path\n\ncache_dir = Path(\"cache/\")\nindex_dir = cache_dir / \"indices\" / dataset_name.split(\"/\")[0]",
"_____no_output_____"
],
[
"from pyterrier.index import IterDictIndexer\n\nif not index_dir.exists():\n indexer = IterDictIndexer(str(index_dir.absolute()))\n indexer.index(\n dataset.get_corpus_iter(),\n fields=[\"text\"]\n )",
"_____no_output_____"
],
[
"from pyterrier.batchretrieve import BatchRetrieve\n\n# BM25 baseline retrieval.\nbm25 = BatchRetrieve(str(index_dir.absolute()), wmodel=\"BM25\")",
"_____no_output_____"
],
[
"from ir_axioms.axiom import (\n ArgUC, QTArg, QTPArg, aSL, PROX1, PROX2, PROX3, PROX4, PROX5, TFC1, TFC3, RS_TF, RS_TF_IDF, RS_BM25, RS_PL2, RS_QL,\n AND, LEN_AND, M_AND, LEN_M_AND, DIV, LEN_DIV, M_TDC, LEN_M_TDC, STMC1, STMC1_f, STMC2, STMC2_f, LNC1, TF_LNC, LB1,\n REG, ANTI_REG, REG_f, ANTI_REG_f, ASPECT_REG, ASPECT_REG_f, ORIG\n)\n\naxioms = [\n ~ArgUC(), ~QTArg(), ~QTPArg(), ~aSL(),\n ~LNC1(), ~TF_LNC(), ~LB1(),\n ~PROX1(), ~PROX2(), ~PROX3(), ~PROX4(), ~PROX5(),\n ~REG(), ~REG_f(), ~ANTI_REG(), ~ANTI_REG_f(), ~ASPECT_REG(), ~ASPECT_REG_f(),\n ~AND(), ~LEN_AND(), ~M_AND(), ~LEN_M_AND(), ~DIV(), ~LEN_DIV(),\n ~RS_TF(), ~RS_TF_IDF(), ~RS_BM25(), ~RS_PL2(), ~RS_QL(),\n ~TFC1(), ~TFC3(), ~M_TDC(), ~LEN_M_TDC(),\n ~STMC1(), ~STMC1_f(), ~STMC2(), ~STMC2_f(),\n ORIG()\n]",
"_____no_output_____"
],
[
"from sklearn.ensemble import RandomForestClassifier\nfrom ir_axioms.modules.pivot import MiddlePivotSelection\nfrom ir_axioms.backend.pyterrier.estimator import EstimatorKwikSortReranker\n\nrandom_forest = RandomForestClassifier(\n max_depth=3,\n)\nkwiksort_random_forest = bm25 % 20 >> EstimatorKwikSortReranker(\n axioms=axioms,\n estimator=random_forest,\n index=index_dir,\n dataset=dataset_name,\n pivot_selection=MiddlePivotSelection(),\n cache_dir=cache_dir,\n verbose=True,\n)",
"_____no_output_____"
],
[
"kwiksort_random_forest.fit(dataset_train.get_topics(), dataset_train.get_qrels())",
"_____no_output_____"
],
[
"from pyterrier.pipelines import Experiment\nfrom ir_measures import nDCG, MAP, RR\n\nexperiment = Experiment(\n [bm25, kwiksort_random_forest ^ bm25],\n dataset_test.get_topics(),\n dataset_test.get_qrels(),\n [nDCG @ 10, RR, MAP],\n [\"BM25\", \"KwikSort Random Forest\"],\n verbose=True,\n)\nexperiment.sort_values(by=\"nDCG@10\", ascending=False, inplace=True)",
"_____no_output_____"
],
[
"experiment",
"_____no_output_____"
],
[
"random_forest.feature_importances_",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb8cf2baa7e88aab7dac093fb5ff9910fc366b1a
| 33,531 |
ipynb
|
Jupyter Notebook
|
Module5/Module5 - Lab5.ipynb
|
marselag/DAT210x
|
68e1743485a52a9c4623e3e7b478786dd434c452
|
[
"MIT"
] | null | null | null |
Module5/Module5 - Lab5.ipynb
|
marselag/DAT210x
|
68e1743485a52a9c4623e3e7b478786dd434c452
|
[
"MIT"
] | null | null | null |
Module5/Module5 - Lab5.ipynb
|
marselag/DAT210x
|
68e1743485a52a9c4623e3e7b478786dd434c452
|
[
"MIT"
] | null | null | null | 71.191083 | 20,860 | 0.802839 |
[
[
[
"# DAT210x - Programming with Python for DS",
"_____no_output_____"
],
[
"## Module5- Lab5",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport matplotlib\n\nmatplotlib.style.use('ggplot') # Look Pretty",
"_____no_output_____"
]
],
[
[
"### A Convenience Function",
"_____no_output_____"
]
],
[
[
"def plotDecisionBoundary(model, X, y):\n fig = plt.figure()\n ax = fig.add_subplot(111)\n\n padding = 0.6\n resolution = 0.0025\n colors = ['royalblue','forestgreen','ghostwhite']\n\n # Calculate the boundaris\n x_min, x_max = X[:, 0].min(), X[:, 0].max()\n y_min, y_max = X[:, 1].min(), X[:, 1].max()\n x_range = x_max - x_min\n y_range = y_max - y_min\n x_min -= x_range * padding\n y_min -= y_range * padding\n x_max += x_range * padding\n y_max += y_range * padding\n\n # Create a 2D Grid Matrix. The values stored in the matrix\n # are the predictions of the class at at said location\n xx, yy = np.meshgrid(np.arange(x_min, x_max, resolution),\n np.arange(y_min, y_max, resolution))\n\n # What class does the classifier say?\n Z = model.predict(np.c_[xx.ravel(), yy.ravel()])\n Z = Z.reshape(xx.shape)\n\n # Plot the contour map\n cs = plt.contourf(xx, yy, Z, cmap=plt.cm.terrain)\n\n # Plot the test original points as well...\n for label in range(len(np.unique(y))):\n indices = np.where(y == label)\n plt.scatter(X[indices, 0], X[indices, 1], c=colors[label], label=str(label), alpha=0.8)\n\n p = model.get_params()\n plt.axis('tight')\n plt.title('K = ' + str(p['n_neighbors']))",
"_____no_output_____"
]
],
[
[
"### The Assignment",
"_____no_output_____"
],
[
"Load up the dataset into a variable called `X`. Check `.head` and `dtypes` to make sure you're loading your data properly--don't fail on the 1st step!",
"_____no_output_____"
]
],
[
[
"# .. your code here ..\nX = pd.read_csv('C:/Users/mgavrilova/Desktop/DAT210x/Module5/Datasets/wheat.data')\nprint(X.head(5))\nX.dtypes",
" id area perimeter compactness length width asymmetry groove \\\n0 0 15.26 14.84 0.8710 5.763 3.312 2.221 5.220 \n1 1 14.88 14.57 0.8811 5.554 3.333 1.018 4.956 \n2 2 14.29 14.09 0.9050 5.291 3.337 2.699 4.825 \n3 3 13.84 13.94 0.8955 5.324 3.379 2.259 4.805 \n4 4 16.14 14.99 0.9034 5.658 3.562 1.355 5.175 \n\n wheat_type \n0 kama \n1 kama \n2 kama \n3 kama \n4 kama \n"
]
],
[
[
"Copy the `wheat_type` series slice out of `X`, and into a series called `y`. Then drop the original `wheat_type` column from the `X`:",
"_____no_output_____"
]
],
[
[
"# .. your code here ..\ny = X.wheat_type\nX = X.drop(columns=['id', 'wheat_type'], axis = 1)\nprint(X.head(3))",
" area perimeter compactness length width asymmetry groove\n0 15.26 14.84 0.8710 5.763 3.312 2.221 5.220\n1 14.88 14.57 0.8811 5.554 3.333 1.018 4.956\n2 14.29 14.09 0.9050 5.291 3.337 2.699 4.825\n"
]
],
[
[
"Do a quick, \"ordinal\" conversion of `y`. In actuality our classification isn't ordinal, but just as an experiment...",
"_____no_output_____"
]
],
[
[
"# .. your code here ..\ny = y.astype('category').cat.codes",
"_____no_output_____"
]
],
[
[
"Do some basic nan munging. Fill each row's nans with the mean of the feature:",
"_____no_output_____"
]
],
[
[
"# .. your code here ..\nX = X.fillna(X.mean())",
"_____no_output_____"
]
],
[
[
"Split `X` into training and testing data sets using `train_test_split()`. Use `0.33` test size, and use `random_state=1`. This is important so that your answers are verifiable. In the real world, you wouldn't specify a random_state:",
"_____no_output_____"
]
],
[
[
"# .. your code here ..\nfrom sklearn.cross_validation import train_test_split\nX_train, X_test, y_train, y_test = train_test_split(X, y, test_size = 0.33, random_state = 1)",
"_____no_output_____"
]
],
[
[
"Create an instance of SKLearn's Normalizer class and then train it using its .fit() method against your _training_ data. The reason you only fit against your training data is because in a real-world situation, you'll only have your training data to train with! In this lab setting, you have both train+test data; but in the wild, you'll only have your training data, and then unlabeled data you want to apply your models to.",
"_____no_output_____"
]
],
[
[
"# .. your code here ..\nfrom sklearn.preprocessing import Normalizer\n\nnorm = Normalizer()\nnorm.fit(X_train)",
"_____no_output_____"
]
],
[
[
"With your trained pre-processor, transform both your training AND testing data. Any testing data has to be transformed with your preprocessor that has ben fit against your training data, so that it exist in the same feature-space as the original data used to train your models.",
"_____no_output_____"
]
],
[
[
"# .. your code here ..\nX_train_norm = norm.transform(X_train)\n\nX_test_norm = norm.transform(X_test)",
"_____no_output_____"
]
],
[
[
"Just like your preprocessing transformation, create a PCA transformation as well. Fit it against your training data, and then project your training and testing features into PCA space using the PCA model's `.transform()` method. This has to be done because the only way to visualize the decision boundary in 2D would be if your KNN algo ran in 2D as well:",
"_____no_output_____"
]
],
[
[
"# .. your code here ..\nfrom sklearn.decomposition import PCA\n\npca = PCA(n_components=2, svd_solver='randomized')\npca.fit(X_train_norm)\npca_train = pca.transform(X_train_norm)\npca_test = pca.transform(X_test_norm)",
"_____no_output_____"
]
],
[
[
"Create and train a KNeighborsClassifier. Start with `K=9` neighbors. Be sure train your classifier against the pre-processed, PCA- transformed training data above! You do not, of course, need to transform your labels.",
"_____no_output_____"
]
],
[
[
"# .. your code here ..\nfrom sklearn.neighbors import KNeighborsClassifier\n\nknn = KNeighborsClassifier(n_neighbors=1)\nknn.fit(pca_train, y_train)",
"_____no_output_____"
],
[
"# I hope your KNeighbors classifier model from earlier was named 'knn'\n# If not, adjust the following line:\nplotDecisionBoundary(knn, pca_train, y_train)",
"_____no_output_____"
]
],
[
[
"Display the accuracy score of your test data/labels, computed by your KNeighbors model. You do NOT have to run `.predict` before calling `.score`, since `.score` will take care of running your predictions for you automatically.",
"_____no_output_____"
]
],
[
[
"# .. your code here ..\naccuracy_score = knn.score(pca_test, y_test)\naccuracy_score",
"_____no_output_____"
]
],
[
[
"### Bonus",
"_____no_output_____"
],
[
"Instead of the ordinal conversion, try and get this assignment working with a proper Pandas get_dummies for feature encoding. You might have to update some of the `plotDecisionBoundary()` code.",
"_____no_output_____"
]
],
[
[
"plt.show()",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
]
] |
cb8cf97420e6fcfd71891b854bf28bc4db63d8e1
| 7,267 |
ipynb
|
Jupyter Notebook
|
DS_HW1/DS_HW1.ipynb
|
AliAkber-79/data-science-coursework
|
03c5726dc932689db47a585cc3c7b704d5afb79f
|
[
"MIT"
] | 1 |
2022-03-01T02:57:26.000Z
|
2022-03-01T02:57:26.000Z
|
DS_HW1/DS_HW1.ipynb
|
AliAkber-79/data-science-coursework
|
03c5726dc932689db47a585cc3c7b704d5afb79f
|
[
"MIT"
] | null | null | null |
DS_HW1/DS_HW1.ipynb
|
AliAkber-79/data-science-coursework
|
03c5726dc932689db47a585cc3c7b704d5afb79f
|
[
"MIT"
] | null | null | null | 19.534946 | 151 | 0.400991 |
[
[
[
"#Q1\ninput_str = \"That pie looks scrumptious and sizzling and super and SWEET.\"\nx = input_str.split()\nfor a in x :\n if a[0] == 'S' or a[0] == 's' :\n print(a)\n ",
"scrumptious\nsizzling\nsuper\nSWEET.\n"
],
[
"#Q2\never_nos = list(range(0,11,2))\nprint(ever_nos)",
"[0, 2, 4, 6, 8, 10]\n"
],
[
"#Q3\ndivisible_by_three = [i for i in range(1,50) if i%3 == 0 ]\nprint(divisible_by_three)",
"[3, 6, 9, 12, 15, 18, 21, 24, 27, 30, 33, 36, 39, 42, 45, 48]\n"
],
[
"#Q4\ndef foo(str1):\n upper_case = 0\n lower_case = 0\n for x in str1:\n if(x.isupper()):\n upper_case= upper_case + 1\n elif(x.islower()):\n lower_case= lower_case + 1\n print(\"No. of Uppercase characters: \"+ str(upper_case))\n print(\"No. of Lowercase characters: \"+ str(lower_case))\n\nfoo(\"Hello Mr. Rogers, how are you this fine Tuesday?\")\n",
"No. of Uppercase characters: 4\nNo. of Lowercase characters: 33\n"
],
[
"#Q5\n\nfor x in range(1,101): \n if(x%3 == 0 and x%5 == 0):\n print(\"FizzBuzz\")\n elif(x%3 == 0):\n print(\"Fizz\")\n elif(x%5 == 0):\n print(\"Buzz\")\n else:\n print(x)\n \n ",
"1\n2\nFizz\n4\nBuzz\nFizz\n7\n8\nFizz\nBuzz\n11\nFizz\n13\n14\nFizzBuzz\n16\n17\nFizz\n19\nBuzz\nFizz\n22\n23\nFizz\nBuzz\n26\nFizz\n28\n29\nFizzBuzz\n31\n32\nFizz\n34\nBuzz\nFizz\n37\n38\nFizz\nBuzz\n41\nFizz\n43\n44\nFizzBuzz\n46\n47\nFizz\n49\nBuzz\nFizz\n52\n53\nFizz\nBuzz\n56\nFizz\n58\n59\nFizzBuzz\n61\n62\nFizz\n64\nBuzz\nFizz\n67\n68\nFizz\nBuzz\n71\nFizz\n73\n74\nFizzBuzz\n76\n77\nFizz\n79\nBuzz\nFizz\n82\n83\nFizz\nBuzz\n86\nFizz\n88\n89\nFizzBuzz\n91\n92\nFizz\n94\nBuzz\nFizz\n97\n98\nFizz\nBuzz\n"
],
[
"#Q6\nstr2 = 'Create a list of the first letters of every word in this string'\nstr3 = str2.split()\nstr4 = [x[0] for x in str3 ]\nprint(str4)",
"['C', 'a', 'l', 'o', 't', 'f', 'l', 'o', 'e', 'w', 'i', 't', 's']\n"
],
[
"#Q7\nstr5 = \"hello\"\nreverse_str5 = str5[-1:-6:-1]\nprint(reverse_str5)",
"olleh\n"
],
[
"#Q8\nd1 = {'simple_key':'hello'}\nd2 = {'k1':{'k2':'hello'}}\nd3 = {'k1':[{'nest_key':['this is deep',['hello']]}]}\n\nh1 = d1[\"simple_key\"]\nh2 = d2[\"k1\"][\"k2\"]\nh3 = d3[\"k1\"][0][\"nest_key\"][1][0]\n\nprint(h1)\nprint(h2)\nprint(h3)",
"hello\nhello\nhello\n"
],
[
"#Q9\nimport Calculator as clctr\n\nx = clctr.Calculator()\na = input(\"Enter first number: \")\nb = input(\"Enter second number: \")\n\ntup1=(int(a),int(b))\nend= False\nwhile(not end):\n Input = input(\"\\nEnter A for Addition,\\nS for subtraction,\\nM for multiplication,\\nP for Power,\\nor D for Division\\nor Q to Quit\")\n if(Input == 'A'):\n print(x.Addition(tup1))\n if(Input == 'S'):\n print(x.Subtraction(tup1))\n if(Input == 'M'):\n print(x.Multiplication(tup1))\n if(Input == 'D'):\n print(x.Division(tup1))\n if(Input == 'P'):\n print(x.Power(tup1))\n if(Input == 'Q'):\n end=True",
"_____no_output_____"
],
[
"#Q10\ndef gen_squares(N):\n squares = [i*i for i in range(N)]\n return squares\n\nfor x in gen_squares(10):\n print (x)\n",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb8d0bcc50f9d7e1fef7ceed000b602682bb561e
| 2,857 |
ipynb
|
Jupyter Notebook
|
aquitania/brains/notebooks/Performance Charts.ipynb
|
marcus-var/aquitania
|
36cd097f15bb49428e88620330c60c10775fc345
|
[
"MIT"
] | null | null | null |
aquitania/brains/notebooks/Performance Charts.ipynb
|
marcus-var/aquitania
|
36cd097f15bb49428e88620330c60c10775fc345
|
[
"MIT"
] | null | null | null |
aquitania/brains/notebooks/Performance Charts.ipynb
|
marcus-var/aquitania
|
36cd097f15bb49428e88620330c60c10775fc345
|
[
"MIT"
] | null | null | null | 21.162963 | 108 | 0.532727 |
[
[
[
"# Performance Charts",
"_____no_output_____"
]
],
[
[
"import pandas as pd",
"_____no_output_____"
],
[
"df = pd.read_csv('../../data/ai/all_trades.csv', index_col=[0], parse_dates=[0])",
"_____no_output_____"
]
],
[
[
"# 1. Financial Performance Chart",
"_____no_output_____"
]
],
[
[
" # Calculates 'start_balance'\nstart_balance_d = df.resample('D')['start_balance'].head(1)\n\n# Calculates 'end_balance'\nend_balance_d = df.resample('D')['end_balance'].tail(1)\n\n# Plots Financial Performance\nend_balance_d.plot()",
"_____no_output_____"
]
],
[
[
"# 2. Yearly Performance Graph",
"_____no_output_____"
]
],
[
[
" # Calculates 'start_balance'\nstart_balance = df.resample('Y')['start_balance'].head(1)\n\n# Calculates 'end_balance'\nend_balance = df.resample('Y')['end_balance'].tail(1)\n\n# Calculates yearly returns\nser = pd.Series(end_balance.values / start_balance.values, index=end_balance.index).subtract(1)\n\n# Gets right labels\nser.index = pd.to_datetime(ser.index).year\n\n# Plots Graph\nser.plot.bar()",
"_____no_output_____"
]
],
[
[
"# 3. Distribution of Trades by Year",
"_____no_output_____"
]
],
[
[
" # Calculates 'total_trades'\ntotal_trades = df[(df['kelly_coh'] > 0) | (df['kelly_coh_i'] > 0)]['results'].resample('Y').count()\n\n# Gets right labels\ntotal_trades.index = pd.to_datetime(total_trades.index).year\n\n# Plots Graph\ntotal_trades.plot.bar()",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb8d0f1ad78285907141164f7a8741e9dc40509a
| 3,385 |
ipynb
|
Jupyter Notebook
|
01B - Intro to the Azure ML SDK.ipynb
|
MicrosoftLearning/DP-100JA
|
b826d89262ea0d06075e22cb554c2b304b72a2b8
|
[
"MIT"
] | 9 |
2020-04-05T12:17:41.000Z
|
2021-05-27T11:56:26.000Z
|
01B - Intro to the Azure ML SDK.ipynb
|
MicrosoftLearning/DP-100JA
|
b826d89262ea0d06075e22cb554c2b304b72a2b8
|
[
"MIT"
] | null | null | null |
01B - Intro to the Azure ML SDK.ipynb
|
MicrosoftLearning/DP-100JA
|
b826d89262ea0d06075e22cb554c2b304b72a2b8
|
[
"MIT"
] | 12 |
2020-01-15T03:46:31.000Z
|
2021-07-17T05:39:10.000Z
| 29.692982 | 247 | 0.628951 |
[
[
[
"# Azure ML SDK の概要\n\nAzure Machine Learning (*Azure ML*) は、機械学習ソリューションを作成および管理するためのクラウドベースのサービスです。データ サイエンティストが既存のデータ処理とモデル開発のスキルとフレームワークを活用し、ワークロードをクラウドに拡大するのに役立つように設計されています。Azure ML SDK for Python には、Azure サブスクリプションで Azure ML を操作するために使用できるクラスが用意されています。\n\n## Azure ML SDK のバージョンを確認する\n\nまず、**azureml-core** パッケージをインポートし、インストールされている SDK のバージョンを確認します。",
"_____no_output_____"
]
],
[
[
"import azureml.core\nprint(\"Ready to use Azure ML\", azureml.core.VERSION)",
"_____no_output_____"
]
],
[
[
"## ワークスペースに接続する\n\nすべての実験と関連リソースは、Azure ML ワークスペース内で管理されます。既存のワークスペースに接続するか、Azure ML SDK を使用して新しいワークスペースを作成できます。\n\nほとんどの場合、ワークスペースの構成は JSON 構成ファイルに格納されます。これにより、Azure サブスクリプション ID などの詳細を覚えておく必要なく、簡単に再接続できます。Azure portal のワークスペースのブレードから JSON 構成ファイルをダウンロードできますが、ワークスペースでコンピューティング インスタンスを使用している場合、構成ファイルは既にルート フォルダーにダウンロードされています。\n\n次のコードでは、構成ファイルを使用してワークスペースに接続します。Notebook セッションで初めて実行するときは、Azure にサインインするように求められますので、https://microsoft.com/devicelogin リンクをクリックし、自動的に生成されたコードを入力して Azure にサインインしてください。正常にサインインした後、開いたブラウザー タブを閉じて、この Notebook に戻ることができます。",
"_____no_output_____"
]
],
[
[
"from azureml.core import Workspace\n\nws = Workspace.from_config()\nprint(ws.name, \"loaded\")",
"_____no_output_____"
]
],
[
[
"## Azure ML リソースの表示\n\nワークスペースへの接続ができたので、ワークスペースに含まれるリソースを表示できます。",
"_____no_output_____"
]
],
[
[
"from azureml.core import ComputeTarget, Datastore, Dataset\n\nprint(\"Compute Targets:\")\nfor compute_name in ws.compute_targets:\n compute = ws.compute_targets[compute_name]\n print(\"\\t\", compute.name, ':', compute.type)\n \nprint(\"Datastores:\")\nfor datastore_name in ws.datastores:\n datastore = Datastore.get(ws, datastore_name)\n print(\"\\t\", datastore.name, ':', datastore.datastore_type)\n \nprint(\"Datasets:\")\nfor dataset_name in list(ws.datasets.keys()):\n dataset = Dataset.get_by_name(ws, dataset_name)\n print(\"\\t\", dataset.name)",
"_____no_output_____"
]
],
[
[
"Azure ML SDK を使用してワークスペース内のリソースを表示する方法について説明しました。SDK は、Azure ML を使用して機械学習ワークロードを操作するために必要なリソースの作成と構成をスクリプト化する優れた方法を提供します。詳細については、[Azure ML SDK のドキュメント](https://docs.microsoft.com/python/api/overview/azure/ml/intro?view=azure-ml-py)を参照してください。",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
cb8d0fdafe8448bca5914af47c37d5d502027775
| 68,534 |
ipynb
|
Jupyter Notebook
|
old-exp/adapt/adapt16/make-evgmm0.1.1-EJM04.ipynb
|
Akio-m/evgmm
|
b7f89f4da34a1d4ca4b233dd2fab4d146512e8f7
|
[
"MIT"
] | 2 |
2018-10-29T06:50:24.000Z
|
2019-09-29T01:44:11.000Z
|
old-exp/adapt/adapt16/make-evgmm0.1.1-EJM04.ipynb
|
Akio-m/evgmm
|
b7f89f4da34a1d4ca4b233dd2fab4d146512e8f7
|
[
"MIT"
] | 2 |
2018-01-29T13:33:05.000Z
|
2018-02-03T15:06:30.000Z
|
old-exp/adapt/adapt16/make-evgmm0.1.1-EJM04.ipynb
|
Akio-m/evgmm
|
b7f89f4da34a1d4ca4b233dd2fab4d146512e8f7
|
[
"MIT"
] | null | null | null | 28.603506 | 307 | 0.442321 |
[
[
[
"# -*- coding: utf-8 -*-\n\n\"\"\"\nEVCのためのEV-GMMを構築します. そして, 適応学習する.\n詳細 : https://pdfs.semanticscholar.org/cbfe/71798ded05fb8bf8674580aabf534c4dbb8bc.pdf\n\nThis program make EV-GMM for EVC. Then, it make adaptation learning.\nCheck detail : https://pdfs.semanticscholar.org/cbfe/71798ded05fb8bf8674580abf534c4dbb8bc.pdf\n\"\"\"",
"_____no_output_____"
],
[
"from __future__ import division, print_function\n\nimport os\nfrom shutil import rmtree\nimport argparse\nimport glob\nimport pickle\nimport time\n\nimport numpy as np\nfrom numpy.linalg import norm \nfrom sklearn.decomposition import PCA\nfrom sklearn.mixture import GMM # sklearn 0.20.0から使えない\nfrom sklearn.preprocessing import StandardScaler\nimport scipy.signal\nimport scipy.sparse\n\n%matplotlib inline\nimport matplotlib.pyplot as plt\nimport IPython \nfrom IPython.display import Audio \n\nimport soundfile as sf\nimport wave \nimport pyworld as pw\nimport librosa.display\n\nfrom dtw import dtw\nimport warnings\nwarnings.filterwarnings('ignore')",
"_____no_output_____"
],
[
"\"\"\"\nParameters\n\n__Mixtured : GMM混合数\n__versions : 実験セット\n__convert_source : 変換元話者のパス\n__convert_target : 変換先話者のパス\n\"\"\"\n# parameters \n__Mixtured = 40\n__versions = 'pre-stored0.1.1'\n__convert_source = 'input/EJM10/V01/T01/TIMIT/000/*.wav' \n__convert_target = 'adaptation/EJM04/V01/T01/ATR503/A/*.wav'\n\n# settings\n__same_path = './utterance/' + __versions + '/'\n__output_path = __same_path + 'output/EJM04/' # EJF01, EJF07, EJM04, EJM05\n\nMixtured = __Mixtured\npre_stored_pickle = __same_path + __versions + '.pickle'\npre_stored_source_list = __same_path + 'pre-source/**/V01/T01/**/*.wav'\npre_stored_list = __same_path + \"pre/**/V01/T01/**/*.wav\"\n#pre_stored_target_list = \"\" (not yet)\npre_stored_gmm_init_pickle = __same_path + __versions + '_init-gmm.pickle'\npre_stored_sv_npy = __same_path + __versions + '_sv.npy'\n\nsave_for_evgmm_covarXX = __output_path + __versions + '_covarXX.npy'\nsave_for_evgmm_covarYX = __output_path + __versions + '_covarYX.npy'\nsave_for_evgmm_fitted_source = __output_path + __versions + '_fitted_source.npy'\nsave_for_evgmm_fitted_target = __output_path + __versions + '_fitted_target.npy'\nsave_for_evgmm_weights = __output_path + __versions + '_weights.npy'\nsave_for_evgmm_source_means = __output_path + __versions + '_source_means.npy'\n\nfor_convert_source = __same_path + __convert_source\nfor_convert_target = __same_path + __convert_target\nconverted_voice_npy = __output_path + 'sp_converted_' + __versions \nconverted_voice_wav = __output_path + 'sp_converted_' + __versions \nmfcc_save_fig_png = __output_path + 'mfcc3dim_' + __versions \nf0_save_fig_png = __output_path + 'f0_converted' + __versions\nconverted_voice_with_f0_wav = __output_path + 'sp_f0_converted' + __versions",
"_____no_output_____"
],
[
"EPSILON = 1e-8\n\nclass MFCC:\n \"\"\"\n MFCC() : メル周波数ケプストラム係数(MFCC)を求めたり、MFCCからスペクトルに変換したりするクラス.\n 動的特徴量(delta)が実装途中.\n ref : http://aidiary.hatenablog.com/entry/20120225/1330179868\n \"\"\"\n \n \n def __init__(self, frequency, nfft=1026, dimension=24, channels=24):\n \"\"\"\n 各種パラメータのセット\n nfft : FFTのサンプル点数\n frequency : サンプリング周波数\n dimension : MFCC次元数\n channles : メルフィルタバンクのチャンネル数(dimensionに依存)\n fscale : 周波数スケール軸\n filterbankl, fcenters : フィルタバンク行列, フィルタバンクの頂点(?)\n \"\"\"\n self.nfft = nfft\n self.frequency = frequency\n self.dimension = dimension\n self.channels = channels\n self.fscale = np.fft.fftfreq(self.nfft, d = 1.0 / self.frequency)[: int(self.nfft / 2)]\n self.filterbank, self.fcenters = self.melFilterBank()\n \n def hz2mel(self, f):\n \"\"\"\n 周波数からメル周波数に変換\n \"\"\"\n return 1127.01048 * np.log(f / 700.0 + 1.0)\n \n def mel2hz(self, m):\n \"\"\"\n メル周波数から周波数に変換\n \"\"\" \n return 700.0 * (np.exp(m / 1127.01048) - 1.0)\n\n def melFilterBank(self):\n \"\"\"\n メルフィルタバンクを生成する\n \"\"\" \n fmax = self.frequency / 2\n melmax = self.hz2mel(fmax)\n nmax = int(self.nfft / 2)\n df = self.frequency / self.nfft\n dmel = melmax / (self.channels + 1)\n melcenters = np.arange(1, self.channels + 1) * dmel\n fcenters = self.mel2hz(melcenters)\n indexcenter = np.round(fcenters / df)\n indexstart = np.hstack(([0], indexcenter[0:self.channels - 1]))\n indexstop = np.hstack((indexcenter[1:self.channels], [nmax]))\n\n filterbank = np.zeros((self.channels, nmax))\n for c in np.arange(0, self.channels):\n increment = 1.0 / (indexcenter[c] - indexstart[c])\n # np,int_ は np.arangeが[0. 1. 2. ..]となるのをintにする\n for i in np.int_(np.arange(indexstart[c], indexcenter[c])):\n filterbank[c, i] = (i - indexstart[c]) * increment\n decrement = 1.0 / (indexstop[c] - indexcenter[c])\n # np,int_ は np.arangeが[0. 1. 2. ..]となるのをintにする\n for i in np.int_(np.arange(indexcenter[c], indexstop[c])):\n filterbank[c, i] = 1.0 - ((i - indexcenter[c]) * decrement)\n\n return filterbank, fcenters\n \n def mfcc(self, spectrum):\n \"\"\"\n スペクトルからMFCCを求める.\n \"\"\"\n mspec = []\n mspec = np.log10(np.dot(spectrum, self.filterbank.T))\n mspec = np.array(mspec)\n \n return scipy.fftpack.realtransforms.dct(mspec, type=2, norm=\"ortho\", axis=-1)\n \n def delta(self, mfcc):\n \"\"\"\n MFCCから動的特徴量を求める.\n 現在は,求める特徴量フレームtをt-1とt+1の平均としている.\n \"\"\"\n mfcc = np.concatenate([\n [mfcc[0]], \n mfcc, \n [mfcc[-1]]\n ]) # 最初のフレームを最初に、最後のフレームを最後に付け足す\n delta = None\n for i in range(1, mfcc.shape[0] - 1):\n slope = (mfcc[i+1] - mfcc[i-1]) / 2\n if delta is None:\n delta = slope\n else:\n delta = np.vstack([delta, slope])\n return delta\n \n def imfcc(self, mfcc, spectrogram):\n \"\"\"\n MFCCからスペクトルを求める.\n \"\"\"\n im_sp = np.array([])\n for i in range(mfcc.shape[0]):\n mfcc_s = np.hstack([mfcc[i], [0] * (self.channels - self.dimension)])\n mspectrum = scipy.fftpack.idct(mfcc_s, norm='ortho')\n # splrep はスプライン補間のための補間関数を求める\n tck = scipy.interpolate.splrep(self.fcenters, np.power(10, mspectrum))\n # splev は指定座標での補間値を求める\n im_spectrogram = scipy.interpolate.splev(self.fscale, tck)\n im_sp = np.concatenate((im_sp, im_spectrogram), axis=0)\n \n return im_sp.reshape(spectrogram.shape)\n \n def trim_zeros_frames(x, eps=1e-7):\n \"\"\"\n 無音区間を取り除く.\n \"\"\"\n T, D = x.shape\n s = np.sum(np.abs(x), axis=1)\n s[s < 1e-7] = 0.\n return x[s > eps]",
"_____no_output_____"
],
[
"def analyse_by_world_with_harverst(x, fs):\n \"\"\"\n WORLD音声分析合成器で基本周波数F0,スペクトル包絡,非周期成分を求める.\n 基本周波数F0についてはharvest法により,より精度良く求める.\n \"\"\"\n # 4 Harvest with F0 refinement (using Stonemask)\n frame_period = 5\n _f0_h, t_h = pw.harvest(x, fs, frame_period=frame_period)\n f0_h = pw.stonemask(x, _f0_h, t_h, fs)\n sp_h = pw.cheaptrick(x, f0_h, t_h, fs)\n ap_h = pw.d4c(x, f0_h, t_h, fs)\n \n return f0_h, sp_h, ap_h\n\ndef wavread(file):\n \"\"\"\n wavファイルから音声トラックとサンプリング周波数を抽出する.\n \"\"\"\n wf = wave.open(file, \"r\")\n fs = wf.getframerate()\n x = wf.readframes(wf.getnframes())\n x = np.frombuffer(x, dtype= \"int16\") / 32768.0\n wf.close()\n return x, float(fs)\n\ndef preEmphasis(signal, p=0.97):\n \"\"\"\n MFCC抽出のための高域強調フィルタ.\n 波形を通すことで,高域成分が強調される.\n \"\"\"\n return scipy.signal.lfilter([1.0, -p], 1, signal)\n\ndef alignment(source, target, path):\n \"\"\"\n タイムアライメントを取る.\n target音声をsource音声の長さに合うように調整する.\n \"\"\"\n # ここでは814に合わせよう(targetに合わせる)\n # p_p = 0 if source.shape[0] > target.shape[0] else 1\n\n #shapes = source.shape if source.shape[0] > target.shape[0] else target.shape \n shapes = source.shape\n align = np.array([])\n for (i, p) in enumerate(path[0]):\n if i != 0:\n if j != p:\n temp = np.array(target[path[1][i]])\n align = np.concatenate((align, temp), axis=0)\n else:\n temp = np.array(target[path[1][i]])\n align = np.concatenate((align, temp), axis=0) \n \n j = p\n \n return align.reshape(shapes)",
"_____no_output_____"
],
[
"\"\"\"\npre-stored学習のためのパラレル学習データを作る。\n時間がかかるため、利用できるlearn-data.pickleがある場合はそれを利用する。\nそれがない場合は一から作り直す。\n\"\"\"\ntimer_start = time.time()\nif os.path.exists(pre_stored_pickle):\n print(\"exist, \", pre_stored_pickle)\n with open(pre_stored_pickle, mode='rb') as f:\n total_data = pickle.load(f)\n print(\"open, \", pre_stored_pickle)\n print(\"Load pre-stored time = \", time.time() - timer_start , \"[sec]\")\nelse:\n source_mfcc = []\n #source_data_sets = []\n for name in sorted(glob.iglob(pre_stored_source_list, recursive=True)):\n print(name)\n x, fs = sf.read(name)\n f0, sp, ap = analyse_by_world_with_harverst(x, fs)\n \n mfcc = MFCC(fs)\n source_mfcc_temp = mfcc.mfcc(sp)\n #source_data = np.hstack([source_mfcc_temp, mfcc.delta(source_mfcc_temp)]) # static & dynamic featuers\n source_mfcc.append(source_mfcc_temp)\n #source_data_sets.append(source_data)\n\n total_data = []\n \n i = 0\n _s_len = len(source_mfcc)\n for name in sorted(glob.iglob(pre_stored_list, recursive=True)):\n print(name, len(total_data))\n x, fs = sf.read(name)\n f0, sp, ap = analyse_by_world_with_harverst(x, fs)\n\n mfcc = MFCC(fs)\n target_mfcc = mfcc.mfcc(sp)\n\n dist, cost, acc, path = dtw(source_mfcc[i%_s_len], target_mfcc, dist=lambda x, y: norm(x - y, ord=1))\n #print('Normalized distance between the two sounds:' + str(dist))\n #print(\"target_mfcc = {0}\".format(target_mfcc.shape))\n\n aligned = alignment(source_mfcc[i%_s_len], target_mfcc, path)\n #target_data_sets = np.hstack([aligned, mfcc.delta(aligned)]) # static & dynamic features\n #learn_data = np.hstack((source_data_sets[i], target_data_sets))\n learn_data = np.hstack([source_mfcc[i%_s_len], aligned])\n\n total_data.append(learn_data)\n i += 1\n \n with open(pre_stored_pickle, 'wb') as output:\n pickle.dump(total_data, output)\n print(\"Make, \", pre_stored_pickle)\n print(\"Make pre-stored time = \", time.time() - timer_start , \"[sec]\")",
"exist, ./utterance/pre-stored0.1.1/pre-stored0.1.1.pickle\nopen, ./utterance/pre-stored0.1.1/pre-stored0.1.1.pickle\nLoad pre-stored time = 1.7890803813934326 [sec]\n"
],
[
"\"\"\"\n全事前学習出力話者からラムダを推定する.\nラムダは適応学習で変容する.\n\"\"\"\n\nS = len(total_data)\nD = int(total_data[0].shape[1] / 2)\nprint(\"total_data[0].shape = \", total_data[0].shape)\nprint(\"S = \", S)\nprint(\"D = \", D)\n\ntimer_start = time.time()\nif os.path.exists(pre_stored_gmm_init_pickle):\n print(\"exist, \", pre_stored_gmm_init_pickle)\n with open(pre_stored_gmm_init_pickle, mode='rb') as f:\n initial_gmm = pickle.load(f)\n print(\"open, \", pre_stored_gmm_init_pickle)\n print(\"Load initial_gmm time = \", time.time() - timer_start , \"[sec]\") \nelse:\n initial_gmm = GMM(n_components = Mixtured, covariance_type = 'full')\n initial_gmm.fit(np.vstack(total_data))\n with open(pre_stored_gmm_init_pickle, 'wb') as output:\n pickle.dump(initial_gmm, output)\n print(\"Make, \", initial_gmm)\n print(\"Make initial_gmm time = \", time.time() - timer_start , \"[sec]\") \n \nweights = initial_gmm.weights_\nsource_means = initial_gmm.means_[:, :D]\ntarget_means = initial_gmm.means_[:, D:]\ncovarXX = initial_gmm.covars_[:, :D, :D]\ncovarXY = initial_gmm.covars_[:, :D, D:]\ncovarYX = initial_gmm.covars_[:, D:, :D]\ncovarYY = initial_gmm.covars_[:, D:, D:]\n\nfitted_source = source_means\nfitted_target = target_means",
"total_data[0].shape = (959, 48)\nS = 1012\nD = 24\nexist, ./utterance/pre-stored0.1.1/pre-stored0.1.1_init-gmm.pickle\nopen, ./utterance/pre-stored0.1.1/pre-stored0.1.1_init-gmm.pickle\nLoad initial_gmm time = 0.0013790130615234375 [sec]\n"
],
[
"\"\"\"\nSVはGMMスーパーベクトルで、各pre-stored学習における出力話者について平均ベクトルを推定する。\nGMMの学習を見てみる必要があるか?\n\"\"\"\n\ntimer_start = time.time()\nif os.path.exists(pre_stored_sv_npy):\n print(\"exist, \", pre_stored_sv_npy)\n sv = np.load(pre_stored_sv_npy)\n print(\"open, \", pre_stored_sv_npy)\n print(\"Load pre_stored_sv time = \", time.time() - timer_start , \"[sec]\") \n \nelse:\n sv = []\n for i in range(S):\n gmm = GMM(n_components = Mixtured, params = 'm', init_params = '', covariance_type = 'full')\n gmm.weights_ = initial_gmm.weights_\n gmm.means_ = initial_gmm.means_\n gmm.covars_ = initial_gmm.covars_\n gmm.fit(total_data[i])\n sv.append(gmm.means_)\n sv = np.array(sv)\n np.save(pre_stored_sv_npy, sv)\n print(\"Make pre_stored_sv time = \", time.time() - timer_start , \"[sec]\") ",
"exist, ./utterance/pre-stored0.1.1/pre-stored0.1.1_sv.npy\nopen, ./utterance/pre-stored0.1.1/pre-stored0.1.1_sv.npy\nLoad pre_stored_sv time = 0.0048675537109375 [sec]\n"
],
[
"\"\"\"\n各事前学習出力話者のGMM平均ベクトルに対して主成分分析(PCA)を行う.\nPCAで求めた固有値と固有ベクトルからeigenvectorsとbiasvectorsを作る.\n\"\"\"\ntimer_start = time.time()\n#source_pca\nsource_n_component, source_n_features = sv[:, :, :D].reshape(S, Mixtured*D).shape\n# 標準化(分散を1、平均を0にする)\nsource_stdsc = StandardScaler()\n# 共分散行列を求める\nsource_X_std = source_stdsc.fit_transform(sv[:, :, :D].reshape(S, Mixtured*D)) \n\n# PCAを行う\nsource_cov = source_X_std.T @ source_X_std / (source_n_component - 1)\nsource_W, source_V_pca = np.linalg.eig(source_cov)\n\nprint(source_W.shape)\nprint(source_V_pca.shape)\n\n# データを主成分の空間に変換する\nsource_X_pca = source_X_std @ source_V_pca\nprint(source_X_pca.shape)\n\n#target_pca\ntarget_n_component, target_n_features = sv[:, :, D:].reshape(S, Mixtured*D).shape\n# 標準化(分散を1、平均を0にする)\ntarget_stdsc = StandardScaler()\n#共分散行列を求める\ntarget_X_std = target_stdsc.fit_transform(sv[:, :, D:].reshape(S, Mixtured*D)) \n\n#PCAを行う\ntarget_cov = target_X_std.T @ target_X_std / (target_n_component - 1)\ntarget_W, target_V_pca = np.linalg.eig(target_cov)\n\nprint(target_W.shape)\nprint(target_V_pca.shape)\n\n# データを主成分の空間に変換する\ntarget_X_pca = target_X_std @ target_V_pca\nprint(target_X_pca.shape)\n\neigenvectors = source_X_pca.reshape((Mixtured, D, S)), target_X_pca.reshape((Mixtured, D, S))\nsource_bias = np.mean(sv[:, :, :D], axis=0)\ntarget_bias = np.mean(sv[:, :, D:], axis=0)\nbiasvectors = source_bias.reshape((Mixtured, D)), target_bias.reshape((Mixtured, D))\n\nprint(\"Do PCA time = \", time.time() - timer_start , \"[sec]\") ",
"(960,)\n(960, 960)\n(1012, 960)\n(960,)\n(960, 960)\n(1012, 960)\nDo PCA time = 26.145132303237915 [sec]\n"
],
[
"\"\"\"\n声質変換に用いる変換元音声と目標音声を読み込む.\n\"\"\"\n\ntimer_start = time.time()\nsource_mfcc_for_convert = []\nsource_sp_for_convert = []\nsource_f0_for_convert = []\nsource_ap_for_convert = []\nfs_source = None\nfor name in sorted(glob.iglob(for_convert_source, recursive=True)):\n print(\"source = \", name)\n x_source, fs_source = sf.read(name)\n f0_source, sp_source, ap_source = analyse_by_world_with_harverst(x_source, fs_source)\n mfcc_source = MFCC(fs_source)\n #mfcc_s_tmp = mfcc_s.mfcc(sp)\n #source_mfcc_for_convert = np.hstack([mfcc_s_tmp, mfcc_s.delta(mfcc_s_tmp)])\n source_mfcc_for_convert.append(mfcc_source.mfcc(sp_source))\n source_sp_for_convert.append(sp_source)\n source_f0_for_convert.append(f0_source)\n source_ap_for_convert.append(ap_source)\n\ntarget_mfcc_for_fit = []\ntarget_f0_for_fit = []\ntarget_ap_for_fit = []\nfor name in sorted(glob.iglob(for_convert_target, recursive=True)):\n print(\"target = \", name)\n x_target, fs_target = sf.read(name)\n f0_target, sp_target, ap_target = analyse_by_world_with_harverst(x_target, fs_target)\n mfcc_target = MFCC(fs_target)\n #mfcc_target_tmp = mfcc_target.mfcc(sp_target)\n #target_mfcc_for_fit = np.hstack([mfcc_t_tmp, mfcc_t.delta(mfcc_t_tmp)])\n target_mfcc_for_fit.append(mfcc_target.mfcc(sp_target))\n target_f0_for_fit.append(f0_target)\n target_ap_for_fit.append(ap_target)\n\n# 全部numpy.arrrayにしておく\nsource_data_mfcc = np.array(source_mfcc_for_convert)\nsource_data_sp = np.array(source_sp_for_convert)\nsource_data_f0 = np.array(source_f0_for_convert)\nsource_data_ap = np.array(source_ap_for_convert)\n\ntarget_mfcc = np.array(target_mfcc_for_fit)\ntarget_f0 = np.array(target_f0_for_fit)\ntarget_ap = np.array(target_ap_for_fit)\n\nprint(\"Load Input and Target Voice time = \", time.time() - timer_start , \"[sec]\") ",
"source = ./utterance/pre-stored0.1.1/input/EJM10/V01/T01/TIMIT/000/A11.wav\nsource = ./utterance/pre-stored0.1.1/input/EJM10/V01/T01/TIMIT/000/A14.wav\nsource = ./utterance/pre-stored0.1.1/input/EJM10/V01/T01/TIMIT/000/A17.wav\nsource = ./utterance/pre-stored0.1.1/input/EJM10/V01/T01/TIMIT/000/A18.wav\nsource = ./utterance/pre-stored0.1.1/input/EJM10/V01/T01/TIMIT/000/A19.wav\nsource = ./utterance/pre-stored0.1.1/input/EJM10/V01/T01/TIMIT/000/A20.wav\nsource = ./utterance/pre-stored0.1.1/input/EJM10/V01/T01/TIMIT/000/A21.wav\nsource = ./utterance/pre-stored0.1.1/input/EJM10/V01/T01/TIMIT/000/A22.wav\nsource = ./utterance/pre-stored0.1.1/input/EJM10/V01/T01/TIMIT/000/A23.wav\nsource = ./utterance/pre-stored0.1.1/input/EJM10/V01/T01/TIMIT/000/A24.wav\nsource = ./utterance/pre-stored0.1.1/input/EJM10/V01/T01/TIMIT/000/A25.wav\nsource = ./utterance/pre-stored0.1.1/input/EJM10/V01/T01/TIMIT/000/A26.wav\nsource = ./utterance/pre-stored0.1.1/input/EJM10/V01/T01/TIMIT/000/A27.wav\nsource = ./utterance/pre-stored0.1.1/input/EJM10/V01/T01/TIMIT/000/A28.wav\nsource = ./utterance/pre-stored0.1.1/input/EJM10/V01/T01/TIMIT/000/A29.wav\nsource = ./utterance/pre-stored0.1.1/input/EJM10/V01/T01/TIMIT/000/A30.wav\nsource = ./utterance/pre-stored0.1.1/input/EJM10/V01/T01/TIMIT/000/A31.wav\nsource = ./utterance/pre-stored0.1.1/input/EJM10/V01/T01/TIMIT/000/A32.wav\nsource = ./utterance/pre-stored0.1.1/input/EJM10/V01/T01/TIMIT/000/A33.wav\nsource = ./utterance/pre-stored0.1.1/input/EJM10/V01/T01/TIMIT/000/A34.wav\nsource = ./utterance/pre-stored0.1.1/input/EJM10/V01/T01/TIMIT/000/A35.wav\nsource = ./utterance/pre-stored0.1.1/input/EJM10/V01/T01/TIMIT/000/A36.wav\nsource = ./utterance/pre-stored0.1.1/input/EJM10/V01/T01/TIMIT/000/A37.wav\nsource = ./utterance/pre-stored0.1.1/input/EJM10/V01/T01/TIMIT/000/A38.wav\nsource = ./utterance/pre-stored0.1.1/input/EJM10/V01/T01/TIMIT/000/A39.wav\nsource = ./utterance/pre-stored0.1.1/input/EJM10/V01/T01/TIMIT/000/A40.wav\nsource = ./utterance/pre-stored0.1.1/input/EJM10/V01/T01/TIMIT/000/A41.wav\nsource = ./utterance/pre-stored0.1.1/input/EJM10/V01/T01/TIMIT/000/A42.wav\nsource = ./utterance/pre-stored0.1.1/input/EJM10/V01/T01/TIMIT/000/A43.wav\nsource = ./utterance/pre-stored0.1.1/input/EJM10/V01/T01/TIMIT/000/A44.wav\nsource = ./utterance/pre-stored0.1.1/input/EJM10/V01/T01/TIMIT/000/A45.wav\nsource = ./utterance/pre-stored0.1.1/input/EJM10/V01/T01/TIMIT/000/A46.wav\nsource = ./utterance/pre-stored0.1.1/input/EJM10/V01/T01/TIMIT/000/A47.wav\nsource = ./utterance/pre-stored0.1.1/input/EJM10/V01/T01/TIMIT/000/A48.wav\nsource = ./utterance/pre-stored0.1.1/input/EJM10/V01/T01/TIMIT/000/A49.wav\nsource = ./utterance/pre-stored0.1.1/input/EJM10/V01/T01/TIMIT/000/A50.wav\nsource = ./utterance/pre-stored0.1.1/input/EJM10/V01/T01/TIMIT/000/A51.wav\nsource = ./utterance/pre-stored0.1.1/input/EJM10/V01/T01/TIMIT/000/A52.wav\nsource = ./utterance/pre-stored0.1.1/input/EJM10/V01/T01/TIMIT/000/A53.wav\nsource = ./utterance/pre-stored0.1.1/input/EJM10/V01/T01/TIMIT/000/A54.wav\nsource = ./utterance/pre-stored0.1.1/input/EJM10/V01/T01/TIMIT/000/A55.wav\nsource = ./utterance/pre-stored0.1.1/input/EJM10/V01/T01/TIMIT/000/A56.wav\nsource = ./utterance/pre-stored0.1.1/input/EJM10/V01/T01/TIMIT/000/A57.wav\nsource = ./utterance/pre-stored0.1.1/input/EJM10/V01/T01/TIMIT/000/A58.wav\nsource = ./utterance/pre-stored0.1.1/input/EJM10/V01/T01/TIMIT/000/A59.wav\ntarget = ./utterance/pre-stored0.1.1/adaptation/EJM04/V01/T01/ATR503/A/A01.wav\ntarget = ./utterance/pre-stored0.1.1/adaptation/EJM04/V01/T01/ATR503/A/A02.wav\ntarget = ./utterance/pre-stored0.1.1/adaptation/EJM04/V01/T01/ATR503/A/A03.wav\ntarget = ./utterance/pre-stored0.1.1/adaptation/EJM04/V01/T01/ATR503/A/A05.wav\ntarget = ./utterance/pre-stored0.1.1/adaptation/EJM04/V01/T01/ATR503/A/A06.wav\ntarget = ./utterance/pre-stored0.1.1/adaptation/EJM04/V01/T01/ATR503/A/A07.wav\ntarget = ./utterance/pre-stored0.1.1/adaptation/EJM04/V01/T01/ATR503/A/A08.wav\ntarget = ./utterance/pre-stored0.1.1/adaptation/EJM04/V01/T01/ATR503/A/A09.wav\ntarget = ./utterance/pre-stored0.1.1/adaptation/EJM04/V01/T01/ATR503/A/A10.wav\ntarget = ./utterance/pre-stored0.1.1/adaptation/EJM04/V01/T01/ATR503/A/A11.wav\ntarget = ./utterance/pre-stored0.1.1/adaptation/EJM04/V01/T01/ATR503/A/A13.wav\ntarget = ./utterance/pre-stored0.1.1/adaptation/EJM04/V01/T01/ATR503/A/A14.wav\ntarget = ./utterance/pre-stored0.1.1/adaptation/EJM04/V01/T01/ATR503/A/A15.wav\ntarget = ./utterance/pre-stored0.1.1/adaptation/EJM04/V01/T01/ATR503/A/A16.wav\ntarget = ./utterance/pre-stored0.1.1/adaptation/EJM04/V01/T01/ATR503/A/A17.wav\ntarget = ./utterance/pre-stored0.1.1/adaptation/EJM04/V01/T01/ATR503/A/A18.wav\nLoad Input and Target Voice time = 159.0263273715973 [sec]\n"
],
[
"\"\"\"\n適応話者学習を行う.\nつまり,事前学習出力話者から目標話者の空間を作りだす.\n\n適応話者文数ごとにfitted_targetを集めるのは未実装.\n\"\"\"\n\ntimer_start = time.time()\nepoch=100\n\npy = GMM(n_components = Mixtured, covariance_type = 'full')\npy.weights_ = weights\npy.means_ = target_means\npy.covars_ = covarYY\n\nfitted_target = None\n\nfor i in range(len(target_mfcc)):\n print(\"adaptation = \", i+1, \"/\", len(target_mfcc))\n target = target_mfcc[i]\n\n for x in range(epoch):\n print(\"epoch = \", x)\n predict = py.predict_proba(np.atleast_2d(target))\n y = np.sum([predict[:, i: i + 1] * (target - biasvectors[1][i])\n for i in range(Mixtured)], axis = 1)\n gamma = np.sum(predict, axis = 0)\n\n left = np.sum([gamma[i] * np.dot(eigenvectors[1][i].T,\n np.linalg.solve(py.covars_, eigenvectors[1])[i])\n for i in range(Mixtured)], axis=0)\n right = np.sum([np.dot(eigenvectors[1][i].T, \n np.linalg.solve(py.covars_, y)[i]) \n for i in range(Mixtured)], axis = 0)\n weight = np.linalg.solve(left, right)\n\n fitted_target = np.dot(eigenvectors[1], weight) + biasvectors[1]\n py.means_ = fitted_target\n \nprint(\"Load Input and Target Voice time = \", time.time() - timer_start , \"[sec]\") ",
"adaptation = 1 / 16\nepoch = 0\nepoch = 1\nepoch = 2\nepoch = 3\nepoch = 4\nepoch = 5\nepoch = 6\nepoch = 7\nepoch = 8\nepoch = 9\nepoch = 10\nepoch = 11\nepoch = 12\nepoch = 13\nepoch = 14\nepoch = 15\nepoch = 16\nepoch = 17\nepoch = 18\nepoch = 19\nepoch = 20\nepoch = 21\nepoch = 22\nepoch = 23\nepoch = 24\nepoch = 25\nepoch = 26\nepoch = 27\nepoch = 28\nepoch = 29\nepoch = 30\nepoch = 31\nepoch = 32\nepoch = 33\nepoch = 34\nepoch = 35\nepoch = 36\nepoch = 37\nepoch = 38\nepoch = 39\nepoch = 40\nepoch = 41\nepoch = 42\nepoch = 43\nepoch = 44\nepoch = 45\nepoch = 46\nepoch = 47\nepoch = 48\nepoch = 49\nepoch = 50\nepoch = 51\nepoch = 52\nepoch = 53\nepoch = 54\nepoch = 55\nepoch = 56\nepoch = 57\nepoch = 58\nepoch = 59\nepoch = 60\nepoch = 61\nepoch = 62\nepoch = 63\nepoch = 64\nepoch = 65\nepoch = 66\nepoch = 67\nepoch = 68\nepoch = 69\nepoch = 70\nepoch = 71\nepoch = 72\nepoch = 73\nepoch = 74\nepoch = 75\nepoch = 76\nepoch = 77\nepoch = 78\nepoch = 79\nepoch = 80\nepoch = 81\nepoch = 82\nepoch = 83\nepoch = 84\nepoch = 85\nepoch = 86\nepoch = 87\nepoch = 88\nepoch = 89\nepoch = 90\nepoch = 91\nepoch = 92\nepoch = 93\nepoch = 94\nepoch = 95\nepoch = 96\nepoch = 97\nepoch = 98\nepoch = 99\nadaptation = 2 / 16\nepoch = 0\nepoch = 1\nepoch = 2\nepoch = 3\nepoch = 4\nepoch = 5\nepoch = 6\nepoch = 7\nepoch = 8\nepoch = 9\nepoch = 10\nepoch = 11\nepoch = 12\nepoch = 13\nepoch = 14\nepoch = 15\nepoch = 16\nepoch = 17\nepoch = 18\nepoch = 19\nepoch = 20\nepoch = 21\nepoch = 22\nepoch = 23\nepoch = 24\nepoch = 25\nepoch = 26\nepoch = 27\nepoch = 28\nepoch = 29\nepoch = 30\nepoch = 31\nepoch = 32\nepoch = 33\nepoch = 34\nepoch = 35\nepoch = 36\nepoch = 37\nepoch = 38\nepoch = 39\nepoch = 40\nepoch = 41\nepoch = 42\nepoch = 43\nepoch = 44\nepoch = 45\nepoch = 46\nepoch = 47\nepoch = 48\nepoch = 49\nepoch = 50\nepoch = 51\nepoch = 52\nepoch = 53\nepoch = 54\nepoch = 55\nepoch = 56\nepoch = 57\nepoch = 58\nepoch = 59\nepoch = 60\nepoch = 61\nepoch = 62\nepoch = 63\nepoch = 64\nepoch = 65\nepoch = 66\nepoch = 67\nepoch = 68\nepoch = 69\nepoch = 70\nepoch = 71\nepoch = 72\nepoch = 73\nepoch = 74\nepoch = 75\nepoch = 76\nepoch = 77\nepoch = 78\nepoch = 79\nepoch = 80\nepoch = 81\nepoch = 82\nepoch = 83\nepoch = 84\nepoch = 85\nepoch = 86\nepoch = 87\nepoch = 88\nepoch = 89\nepoch = 90\nepoch = 91\nepoch = 92\nepoch = 93\nepoch = 94\nepoch = 95\nepoch = 96\nepoch = 97\nepoch = 98\nepoch = 99\nadaptation = 3 / 16\nepoch = 0\nepoch = 1\nepoch = 2\nepoch = 3\nepoch = 4\nepoch = 5\nepoch = 6\nepoch = 7\nepoch = 8\nepoch = 9\nepoch = 10\nepoch = 11\nepoch = 12\nepoch = 13\nepoch = 14\nepoch = 15\nepoch = 16\nepoch = 17\nepoch = 18\nepoch = 19\nepoch = 20\nepoch = 21\nepoch = 22\nepoch = 23\nepoch = 24\nepoch = 25\nepoch = 26\nepoch = 27\nepoch = 28\nepoch = 29\nepoch = 30\nepoch = 31\nepoch = 32\nepoch = 33\nepoch = 34\nepoch = 35\nepoch = 36\nepoch = 37\nepoch = 38\nepoch = 39\nepoch = 40\nepoch = 41\nepoch = 42\nepoch = 43\nepoch = 44\nepoch = 45\nepoch = 46\nepoch = 47\nepoch = 48\nepoch = 49\nepoch = 50\nepoch = 51\nepoch = 52\nepoch = 53\nepoch = 54\nepoch = 55\nepoch = 56\nepoch = 57\nepoch = 58\nepoch = 59\nepoch = 60\nepoch = 61\nepoch = 62\nepoch = 63\nepoch = 64\nepoch = 65\nepoch = 66\nepoch = 67\nepoch = 68\nepoch = 69\nepoch = 70\nepoch = 71\nepoch = 72\nepoch = 73\nepoch = 74\nepoch = 75\nepoch = 76\nepoch = 77\nepoch = 78\nepoch = 79\nepoch = 80\nepoch = 81\nepoch = 82\nepoch = 83\nepoch = 84\nepoch = 85\nepoch = 86\nepoch = 87\nepoch = 88\nepoch = 89\nepoch = 90\nepoch = 91\nepoch = 92\nepoch = 93\nepoch = 94\nepoch = 95\nepoch = 96\nepoch = 97\nepoch = 98\nepoch = 99\nadaptation = 4 / 16\nepoch = 0\nepoch = 1\nepoch = 2\nepoch = 3\nepoch = 4\nepoch = 5\nepoch = 6\nepoch = 7\nepoch = 8\nepoch = 9\nepoch = 10\nepoch = 11\nepoch = 12\nepoch = 13\nepoch = 14\nepoch = 15\nepoch = 16\nepoch = 17\nepoch = 18\nepoch = 19\nepoch = 20\nepoch = 21\nepoch = 22\nepoch = 23\nepoch = 24\nepoch = 25\nepoch = 26\nepoch = 27\nepoch = 28\nepoch = 29\nepoch = 30\nepoch = 31\nepoch = 32\nepoch = 33\nepoch = 34\nepoch = 35\nepoch = 36\nepoch = 37\nepoch = 38\nepoch = 39\nepoch = 40\nepoch = 41\nepoch = 42\nepoch = 43\nepoch = 44\nepoch = 45\nepoch = 46\nepoch = 47\nepoch = 48\nepoch = 49\nepoch = 50\nepoch = 51\nepoch = 52\nepoch = 53\nepoch = 54\nepoch = 55\nepoch = 56\nepoch = 57\nepoch = 58\nepoch = 59\nepoch = 60\nepoch = 61\nepoch = 62\nepoch = 63\nepoch = 64\nepoch = 65\nepoch = 66\nepoch = 67\nepoch = 68\nepoch = 69\nepoch = 70\nepoch = 71\nepoch = 72\nepoch = 73\nepoch = 74\nepoch = 75\nepoch = 76\nepoch = 77\nepoch = 78\nepoch = 79\nepoch = 80\nepoch = 81\nepoch = 82\nepoch = 83\nepoch = 84\nepoch = 85\nepoch = 86\nepoch = 87\nepoch = 88\nepoch = 89\nepoch = 90\nepoch = 91\nepoch = 92\nepoch = 93\nepoch = 94\nepoch = 95\nepoch = 96\nepoch = 97\nepoch = 98\nepoch = 99\nadaptation = 5 / 16\nepoch = 0\nepoch = 1\nepoch = 2\nepoch = 3\nepoch = 4\nepoch = 5\nepoch = 6\nepoch = 7\nepoch = 8\nepoch = 9\nepoch = 10\nepoch = 11\nepoch = 12\nepoch = 13\nepoch = 14\nepoch = 15\nepoch = 16\nepoch = 17\nepoch = 18\nepoch = 19\nepoch = 20\nepoch = 21\nepoch = 22\nepoch = 23\nepoch = 24\nepoch = 25\nepoch = 26\nepoch = 27\nepoch = 28\nepoch = 29\nepoch = 30\nepoch = 31\nepoch = 32\nepoch = 33\nepoch = 34\nepoch = 35\nepoch = 36\nepoch = 37\nepoch = 38\nepoch = 39\nepoch = 40\nepoch = 41\nepoch = 42\nepoch = 43\nepoch = 44\nepoch = 45\nepoch = 46\nepoch = 47\nepoch = 48\nepoch = 49\nepoch = 50\nepoch = 51\nepoch = 52\nepoch = 53\nepoch = 54\nepoch = 55\nepoch = 56\nepoch = 57\nepoch = 58\nepoch = 59\nepoch = 60\nepoch = 61\nepoch = 62\nepoch = 63\nepoch = 64\nepoch = 65\nepoch = 66\nepoch = 67\nepoch = 68\nepoch = 69\nepoch = 70\nepoch = 71\nepoch = 72\nepoch = 73\nepoch = 74\nepoch = 75\nepoch = 76\nepoch = 77\nepoch = 78\nepoch = 79\nepoch = 80\nepoch = 81\nepoch = 82\nepoch = 83\nepoch = 84\nepoch = 85\nepoch = 86\nepoch = 87\nepoch = 88\nepoch = 89\nepoch = 90\nepoch = 91\nepoch = 92\nepoch = 93\nepoch = 94\nepoch = 95\nepoch = 96\nepoch = 97\nepoch = 98\nepoch = 99\nadaptation = 6 / 16\nepoch = 0\nepoch = 1\nepoch = 2\nepoch = 3\nepoch = 4\nepoch = 5\nepoch = 6\nepoch = 7\nepoch = 8\nepoch = 9\nepoch = 10\nepoch = 11\nepoch = 12\nepoch = 13\nepoch = 14\nepoch = 15\nepoch = 16\nepoch = 17\nepoch = 18\nepoch = 19\nepoch = 20\nepoch = 21\nepoch = 22\nepoch = 23\nepoch = 24\nepoch = 25\nepoch = 26\nepoch = 27\nepoch = 28\nepoch = 29\nepoch = 30\nepoch = 31\nepoch = 32\nepoch = 33\nepoch = 34\nepoch = 35\nepoch = 36\nepoch = 37\nepoch = 38\nepoch = 39\nepoch = 40\nepoch = 41\nepoch = 42\nepoch = 43\nepoch = 44\nepoch = 45\nepoch = 46\nepoch = 47\nepoch = 48\nepoch = 49\nepoch = 50\nepoch = 51\nepoch = 52\nepoch = 53\nepoch = 54\nepoch = 55\nepoch = 56\nepoch = 57\nepoch = 58\nepoch = 59\nepoch = 60\nepoch = 61\nepoch = 62\nepoch = 63\nepoch = 64\nepoch = 65\nepoch = 66\nepoch = 67\nepoch = 68\nepoch = 69\nepoch = 70\nepoch = 71\nepoch = 72\nepoch = 73\nepoch = 74\nepoch = 75\nepoch = 76\nepoch = 77\nepoch = 78\nepoch = 79\nepoch = 80\nepoch = 81\nepoch = 82\nepoch = 83\nepoch = 84\nepoch = 85\nepoch = 86\nepoch = 87\nepoch = 88\nepoch = 89\nepoch = 90\nepoch = 91\nepoch = 92\nepoch = 93\nepoch = 94\nepoch = 95\nepoch = 96\nepoch = 97\nepoch = 98\nepoch = 99\nadaptation = 7 / 16\nepoch = 0\nepoch = 1\nepoch = 2\nepoch = 3\nepoch = 4\nepoch = 5\nepoch = 6\nepoch = 7\nepoch = 8\nepoch = 9\nepoch = 10\nepoch = 11\nepoch = 12\nepoch = 13\nepoch = 14\nepoch = 15\nepoch = 16\nepoch = 17\nepoch = 18\nepoch = 19\nepoch = 20\nepoch = 21\nepoch = 22\nepoch = 23\nepoch = 24\nepoch = 25\nepoch = 26\nepoch = 27\nepoch = 28\nepoch = 29\nepoch = 30\nepoch = 31\nepoch = 32\nepoch = 33\nepoch = 34\nepoch = 35\nepoch = 36\nepoch = 37\nepoch = 38\nepoch = 39\nepoch = 40\nepoch = 41\nepoch = 42\nepoch = 43\nepoch = 44\nepoch = 45\nepoch = 46\nepoch = 47\nepoch = 48\nepoch = 49\nepoch = 50\nepoch = 51\nepoch = 52\nepoch = 53\nepoch = 54\nepoch = 55\nepoch = 56\nepoch = 57\nepoch = 58\nepoch = 59\nepoch = 60\nepoch = 61\nepoch = 62\nepoch = 63\nepoch = 64\nepoch = 65\nepoch = 66\nepoch = 67\nepoch = 68\nepoch = 69\nepoch = 70\nepoch = 71\nepoch = 72\nepoch = 73\nepoch = 74\nepoch = 75\nepoch = 76\n"
],
[
"\"\"\"\n変換に必要なものを残しておく.\n\"\"\"\nnp.save(save_for_evgmm_covarXX, covarXX)\nnp.save(save_for_evgmm_covarYX, covarYX)\nnp.save(save_for_evgmm_fitted_source, fitted_source)\nnp.save(save_for_evgmm_fitted_target, fitted_target)\nnp.save(save_for_evgmm_weights, weights)\nnp.save(save_for_evgmm_source_means, source_means)",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb8d139b10a5dca855bfdc5a83176ad4423f033c
| 41,561 |
ipynb
|
Jupyter Notebook
|
python/notebook/1.POP/2.str.ipynb
|
dunitian/BaseCode
|
4855ef4c6dd7c95d7239d2048832d8acfe26e084
|
[
"Apache-2.0"
] | 25 |
2018-06-13T08:13:44.000Z
|
2020-11-19T14:02:11.000Z
|
python/notebook/1.POP/2.str.ipynb
|
dunitian/BaseCode
|
4855ef4c6dd7c95d7239d2048832d8acfe26e084
|
[
"Apache-2.0"
] | null | null | null |
python/notebook/1.POP/2.str.ipynb
|
dunitian/BaseCode
|
4855ef4c6dd7c95d7239d2048832d8acfe26e084
|
[
"Apache-2.0"
] | 13 |
2018-06-13T08:13:38.000Z
|
2022-01-06T06:45:07.000Z
| 24.290473 | 539 | 0.45663 |
[
[
[
"Code:<a href=\"https://github.com/lotapp/BaseCode\" target=\"_blank\">https://github.com/lotapp/BaseCode</a>\n\n多图旧排版:<a href=\"https://www.cnblogs.com/dunitian/p/9119986.html\" target=\"_blank\">https://www.cnblogs.com/dunitian/p/9119986.html</a>\n\n在线编程:<a href=\"https://mybinder.org/v2/gh/lotapp/BaseCode/master\" target=\"_blank\">https://mybinder.org/v2/gh/lotapp/BaseCode/master</a>\n\nPython设计的目的就是 ==> **让程序员解放出来,不要过于关注代码本身**\n\n步入正题:**欢迎提出更简单或者效率更高的方法**\n\n**基础系列**:(这边重点说说`Python`,上次讲过的东西我就一笔带过了)\n\n## 1.基础回顾\n\n### 1.1.输出+类型转换",
"_____no_output_____"
]
],
[
[
"user_num1=input(\"输入第一个数:\")\nuser_num2=input(\"输入第二个数:\")\n\nprint(\"两数之和:%d\"%(int(user_num1)+int(user_num2)))",
"输入第一个数:1\n输入第二个数:2\n两数之和:3\n"
]
],
[
[
"### 1.2.字符串拼接+拼接输出方式",
"_____no_output_____"
]
],
[
[
"user_name=input(\"输入昵称:\")\nuser_pass=input(\"输入密码:\")\nuser_url=\"192.168.1.121\"\n\n#拼接输出方式一:\nprint(\"ftp://\"+user_name+\":\"+user_pass+\"@\"+user_url)\n\n#拼接输出方式二:\nprint(\"ftp://%s:%s@%s\"%(user_name,user_pass,user_url))",
"输入昵称:tom\n输入密码:1379\nftp://tom:[email protected]\nftp://tom:[email protected]\n"
]
],
[
[
"## 2.字符串遍历、下标、切片\n\n### 2.1.Python\n\n重点说下`python`的 **下标**,有点意思,最后一个元素,我们一般都是`len(str)-1`,他可以直接用`-1`,倒2自然就是`-2`了\n\n**最后一个元素:`user_str[-1]`**\n\nuser_str[-1]\n\nuser_str[len(user_str)-1] #其他编程语言写法\n\n**倒数第二个元素:`user_str[-2]`**\n\nuser_str[-1]\n\nuser_str[len(user_str)-2] #其他编程语言写法",
"_____no_output_____"
]
],
[
[
"user_str=\"七大姑曰:工作了吗?八大姨问:买房了吗?异性说:结婚了吗?\"",
"_____no_output_____"
],
[
"#遍历\nfor item in user_str:\n print(item,end=\" \") # 不换行,以“ ”方式拼接",
"七 大 姑 曰 : 工 作 了 吗 ? 八 大 姨 问 : 买 房 了 吗 ? 异 性 说 : 结 婚 了 吗 ? "
],
[
"#长度:len(user_str)\nlen(user_str)",
"29\n"
],
[
"# #第一个元素:user_str[0]\nuser_str[0]",
"_____no_output_____"
],
[
"# 最后一个元素:user_str[-1]\nprint(user_str[-1])\nprint(user_str[len(user_str)-1])#其他编程语言写法",
"?\n?\n"
],
[
"#倒数第二个元素:user_str[-2]\nprint(user_str[-2])\nprint(user_str[len(user_str)-2])#其他编程语言写法",
"吗\n吗\n"
]
],
[
[
"**python切片语法**:`[start_index:end_index:step]` (**end_index取不到**)\n\neg:`str[1:4]` 取str[1]、str[2]、str[3]\n\neg:`str[2:]` 取下标为2开始到最后的元素\n\neg:`str[2:-1]` 取下标为2~到倒数第二个元素(end_index取不到)\n\neg:`str[1:6:2]` 隔着取~str[1]、str[3]、str[5](案例会详细说)\n\neg:`str[::-1]` 逆向输出(案例会详细说)",
"_____no_output_____"
]
],
[
[
"it_str=\"我爱编程,编程爱它,它是程序,程序是谁?\"",
"_____no_output_____"
],
[
"# eg:取“编程爱它” it_str[5:9]\nprint(it_str[5:9])\nprint(it_str[5:-11]) # end_index用-xx也一样\nprint(it_str[-15:-11])# start_index用-xx也可以",
"编程爱它\n编程爱它\n编程爱它\n"
],
[
"# eg:取“编程爱它,它是程序,程序是谁?” it_str[5:]\nprint(it_str[5:])# 不写默认取到最后一个",
"编程爱它,它是程序,程序是谁?\n"
],
[
"# eg:一个隔一个跳着取(\"我编,程它它程,序谁\") it_str[0::2]\nprint(it_str[0::2])# step=△index(eg:0,1,2,3。这里的step=> 2-0 => 间隔1)",
"我编,程它它程,序谁\n"
],
[
"# eg:倒序输出 it_str[::-1]\n# end_index不写默认是取到最后一个,是正取(从左往右)还是逆取(从右往左),就看step是正是负\nprint(it_str[::-1])\nprint(it_str[-1::-1])# 等价于上一个",
"?谁是序程,序程是它,它爱程编,程编爱我\n?谁是序程,序程是它,它爱程编,程编爱我\n"
]
],
[
[
"### 2.2.CSharp\n\n这次为了更加形象对比,一句一句翻译成C#\n\n有没有发现规律,`user_str[user_str.Length-1]`==> -1是最后一个\n\n`user_str[user_str.Length-2]`==> -2是最后第二个\n\npython的切片其实就是在这方面简化了",
"_____no_output_____"
]
],
[
[
"%%script csharp\n//# # 字符串遍历、下标、切片\n//# user_str=\"七大姑曰:工作了吗?八大姨问:买房了吗?异性说:结婚了吗?\"\nvar user_str = \"七大姑曰:工作了吗?八大姨问:买房了吗?异性说:结婚了吗?\";\n\n//# #遍历\n//# for item in user_str:\n//# print(item,end=\" \")\nforeach (var item in user_str)\n{\n Console.Write(item);\n}\n\n//# #长度:len(user_str)\n//# print(len(user_str))\nConsole.WriteLine(user_str.Length);\n\n//# #第一个元素:user_str[0]\n//# print(user_str[0])\nConsole.WriteLine(user_str[0]);\n\n//# #最后一个元素:user_str[-1]\n//# print(user_str[-1])\n//# print(user_str[len(user_str)-1])#其他编程语言写法\nConsole.WriteLine(user_str[user_str.Length - 1]);\n//\n//# #倒数第二个元素:user_str[-2]\n//# print(user_str[-2])\nConsole.WriteLine(user_str[user_str.Length - 2]);",
"七大姑曰:工作了吗?八大姨问:买房了吗?异性说:结婚了吗?29\n七\n?\n吗\n"
]
],
[
[
"其实你用`Pytho`n跟其他语言对比反差更大,`net`真的很强大了。\n\n补充(对比看就清楚`Python`的`step`为什么是2了,i+=2==>2)",
"_____no_output_____"
]
],
[
[
"%%script csharp\n//# 切片:[start_index:end_index:step] (end_index取不到)\n//# eg:str[1:4] 取str[1]、str[2]、str[3]\n//# eg:str[2:] 取下标为2开始到最后的元素\n//# eg:str[2:-1] 取下标为2~到倒数第二个元素(end_index取不到)\n//# eg:str[1:6:2] 隔着取~str[1]、str[3]、str[5](案例会详细说)\n//# eg:str[::-1] 逆向输出(案例会详细说,)\n//\nvar it_str = \"我爱编程,编程爱它,它是程序,程序是谁?\";\n//\n//#eg:取“编程爱它” it_str[5:9]\n// print(it_str[5:9])\n// print(it_str[5:-11]) #end_index用-xx也一样\n// print(it_str[-15:-11])#start_index用-xx也可以\n\n//Substring(int startIndex, int length)\nConsole.WriteLine(it_str.Substring(5, 4));//第二个参数是长度\n\n//\n//#eg:取“编程爱它,它是程序,程序是谁?” it_str[5:]\n// print(it_str[5:])#不写默认取到最后一个\nConsole.WriteLine(it_str.Substring(5));//不写默认取到最后一个\n\n//#eg:一个隔一个跳着取(\"我编,程它它程,序谁\") it_str[0::2]\n// print(it_str[0::2])#step=△index(eg:0,1,2,3。这里的step=> 2-0 => 间隔1)\n\n//这个我第一反应是用linq ^_^\nfor (int i = 0; i < it_str.Length; i += 2)//对比看就清除Python的step为什么是2了,i+=2==》2\n{\n Console.Write(it_str[i]);\n}\n\nConsole.WriteLine(\"\\n倒序:\");\n//#eg:倒序输出 it_str[::-1]\n//# end_index不写默认是取到最后一个,是正取(从左往右)还是逆取(从右往左),就看step是正是负\n// print(it_str[::-1])\n// print(it_str[-1::-1])#等价于上一个\nfor (int i = it_str.Length - 1; i >= 0; i--)\n{\n Console.Write(it_str[i]);\n}\n//其实可以用Linq:Console.WriteLine(new string(it_str.ToCharArray().Reverse().ToArray()));\n",
"编程爱它\n编程爱它,它是程序,程序是谁?\n我编,程它它程,序谁\n倒序:\n?谁是序程,序程是它,它爱程编,程编爱我"
]
],
[
[
"## 3.Python字符串方法系列\n\n### 3.1.Python查找\n\n`find`,`rfind`,`index`,`rindex`\n\nPython查找 **推荐**你用`find`和`rfind`",
"_____no_output_____"
]
],
[
[
"test_str = \"ABCDabcdefacddbdf\"\n# 查找:find,rfind,index,rindex\n# xxx.find(str, start, end)\nprint(test_str.find(\"cd\"))#从左往右\nprint(test_str.rfind(\"cd\"))#从右往左\nprint(test_str.find(\"dnt\"))#find和rfind找不到就返回-1",
"6\n11\n-1\n"
],
[
"# index和rindex用法和find一样,只是找不到会报错(以后用find系即可)\nprint(test_str.index(\"dnt\"))",
"_____no_output_____"
]
],
[
[
"### 3.2.Python计数\n\npython:`xxx.count(str, start, end)`",
"_____no_output_____"
]
],
[
[
"# 计数:count\n# xxx.count(str, start, end)\nprint(test_str.count(\"d\"))#4\nprint(test_str.count(\"cd\"))#2",
"4\n2\n"
]
],
[
[
"### 3.3.Python替换\n\nPython:`xxx.replace(str1, str2, 替换次数)`",
"_____no_output_____"
]
],
[
[
"# 替换:replace\n# xxx.replace(str1, str2, 替换次数)\n\nprint(test_str)\nprint(test_str.replace(\"b\",\"B\"))#并没有改变原字符串,只是生成了一个新的字符串\nprint(test_str)",
"ABCDabcdefacddbdf\nABCDaBcdefacddBdf\nABCDabcdefacddbdf\n"
],
[
"# replace可以指定替换几次\nprint(test_str.replace(\"b\",\"B\",1))#ABCDaBcdefacddbdf",
"ABCDaBcdefacddbdf\n"
]
],
[
[
"### 3.4.Python分割\n\n`split`(按指定字符分割),`splitlines`(按行分割)\n\n`partition`(以str分割成三部分,str前,str和str后),`rpartition`(从右边开始)\n\n说下 **split的切片用法**:`print(test_input.split(\" \",3))` 在第三个空格处切片,后面的不切了\n",
"_____no_output_____"
]
],
[
[
"# 分割:split(按指定字符分割),splitlines(按行分割),partition(以str分割成三部分,str前,str和str后),rpartition\ntest_list=test_str.split(\"a\")#a有两个,按照a分割,那么会分成三段,返回类型是列表(List),并且返回结果中没有a\nprint(test_list)",
"['ABCD', 'bcdef', 'cddbdf']\n"
],
[
"test_input=\"hi my name is dnt\"\nprint(test_input.split(\" \")) #返回列表格式(后面会说)['hi', 'my', 'name', 'is', 'dnt']\nprint(test_input.split(\" \",3))#在第三个空格处切片,后面的不管了",
"['hi', 'my', 'name', 'is', 'dnt']\n['hi', 'my', 'name', 'is dnt']\n"
]
],
[
[
"继续说说`splitlines`(按行分割),和`split(\"\\n\")`的区别:",
"_____no_output_____"
]
],
[
[
"# splitlines()按行分割,返回类型为List\ntest_line_str=\"abc\\nbca\\ncab\\n\"\nprint(test_line_str.splitlines())#['abc', 'bca', 'cab']\nprint(test_line_str.split(\"\\n\"))#看出区别了吧:['abc', 'bca', 'cab', '']",
"['abc', 'bca', 'cab']\n['abc', 'bca', 'cab', '']\n"
],
[
"# splitlines(按行分割),和split(\"\\n\")的区别没看出来就再来个案例\ntest_line_str2=\"abc\\nbca\\ncab\\nLLL\"\nprint(test_line_str2.splitlines())#['abc', 'bca', 'cab', 'LLL']\nprint(test_line_str2.split(\"\\n\"))#再提示一下,最后不是\\n就和上面一样效果",
"['abc', 'bca', 'cab', 'LLL']\n['abc', 'bca', 'cab', 'LLL']\n"
]
],
[
[
"扩展:`split()`,默认按 **空字符**切割(`空格、\\t、\\n`等等,不用担心返回`''`)",
"_____no_output_____"
]
],
[
[
"# 扩展:split(),默认按空字符切割(空格、\\t、\\n等等,不用担心返回'')\nprint(\"hi my name is dnt\\t\\n m\\n\\t\\n\".split())",
"['hi', 'my', 'name', 'is', 'dnt', 'm']\n"
]
],
[
[
"最后说一下`partition`和`rpartition`: 返回是元祖类型(后面会说的)\n\n方式和find一样,找到第一个匹配的就罢工了【**注意一下没找到的情况**】",
"_____no_output_____"
]
],
[
[
"# partition(以str分割成三部分,str前,str和str后)\n# 返回是元祖类型(后面会说的),方式和find一样,找到第一个匹配的就罢工了【注意一下没找到的情况】\n\nprint(test_str.partition(\"cd\"))#('ABCDab', 'cd', 'efacddbdf')\nprint(test_str.rpartition(\"cd\"))#('ABCDabcdefa', 'cd', 'dbdf')\nprint(test_str.partition(\"感觉自己萌萌哒\"))#没找到:('ABCDabcdefacddbdf', '', '')",
"('ABCDab', 'cd', 'efacddbdf')\n('ABCDabcdefa', 'cd', 'dbdf')\n('ABCDabcdefacddbdf', '', '')\n"
]
],
[
[
"### 3.5.Python字符串连接\n\n**join** :`\"-\".join(test_list)`",
"_____no_output_____"
]
],
[
[
"# 连接:join\n# separat.join(xxx)\n# 错误用法:xxx.join(\"-\")\nprint(\"-\".join(test_list))",
"ABCD-bcdef-cddbdf\n"
]
],
[
[
"### 3.6.Python头尾判断\n\n`startswith`(以。。。开头),`endswith`(以。。。结尾)",
"_____no_output_____"
]
],
[
[
"# 头尾判断:startswith(以。。。开头),endswith(以。。。结尾)\n# test_str.startswith(以。。。开头)\nstart_end_str=\"http://www.baidu.net\"\nprint(start_end_str.startswith(\"https://\") or start_end_str.startswith(\"http://\"))\nprint(start_end_str.endswith(\".com\"))",
"True\nFalse\n"
]
],
[
[
"### 3.7.Python大小写系\n\n`lower`(字符串转换为小写),`upper`(字符串转换为大写)\n\n`title`(单词首字母大写),`capitalize`(第一个字符大写,其他变小写)",
"_____no_output_____"
]
],
[
[
"# 大小写系:lower(字符串转换为小写),upper(字符串转换为大写)\n# title(单词首字母大写),capitalize(第一个字符大写,其他变小写)\n\nprint(test_str)\nprint(test_str.upper())#ABCDABCDEFACDDBDF\nprint(test_str.lower())#abcdabcdefacddbdf\nprint(test_str.capitalize())#第一个字符大写,其他变小写",
"ABCDabcdefacddbdf\nABCDABCDEFACDDBDF\nabcdabcdefacddbdf\nAbcdabcdefacddbdf\n"
]
],
[
[
"### 3.8.Python格式系列\n\n`lstrip`(去除左边空格),`rstrip`(去除右边空格)\n\n**`strip`** (去除两边空格)美化输出系列:`ljust`,`rjust`,`center`\n\n`ljust,rjust,center`这些就不说了,python经常在linux终端中输出,所以这几个用的比较多",
"_____no_output_____"
]
],
[
[
"# 格式系列:lstrip(去除左边空格),rstrip(去除右边空格),strip(去除两边空格)美化输出系列:ljust,rjust,center\nstrip_str=\" I Have a Dream \"\nprint(strip_str.strip()+\"|\")#我加 | 是为了看清后面空格,没有别的用处\nprint(strip_str.lstrip()+\"|\")\nprint(strip_str.rstrip()+\"|\")\n\n#这个就是格式化输出,就不讲了\nprint(test_str.ljust(50))\nprint(test_str.rjust(50))\nprint(test_str.center(50))",
"I Have a Dream|\nI Have a Dream |\n I Have a Dream|\nABCDabcdefacddbdf \n ABCDabcdefacddbdf\n ABCDabcdefacddbdf \n"
]
],
[
[
"### 3.9.Python验证系列\n\n`isalpha`(是否是纯字母),`isalnum`(是否是数字|字母)\n\n`isdigit`(是否是纯数字),`isspace`(是否是纯空格)\n\n注意~ `test_str5=\" \\t \\n \"` # **isspace() ==>true**",
"_____no_output_____"
]
],
[
[
"# 验证系列:isalpha(是否是纯字母),isalnum(是否是数字|字母),isdigit(是否是纯数字),isspace(是否是纯空格)\n# 注意哦~ test_str5=\" \\t \\n \" #isspace() ==>true\n\ntest_str2=\"Abcd123\"\ntest_str3=\"123456\"\ntest_str4=\" \\t\" #isspace() ==>true\ntest_str5=\" \\t \\n \" #isspace() ==>true",
"_____no_output_____"
],
[
"test_str.isalpha() #是否是纯字母",
"_____no_output_____"
],
[
"test_str.isalnum() #是否是数字|字母",
"_____no_output_____"
],
[
"test_str.isdigit() #是否是纯数字",
"_____no_output_____"
],
[
"test_str.isspace() #是否是纯空格",
"_____no_output_____"
],
[
"test_str2.isalnum() #是否是数字和字母组成",
"_____no_output_____"
],
[
"test_str2.isdigit() #是否是纯数字",
"_____no_output_____"
],
[
"test_str3.isdigit() #是否是纯数字",
"_____no_output_____"
],
[
"test_str5.isspace() #是否是纯空格",
"_____no_output_____"
],
[
"test_str4.isspace() #是否是纯空格",
"_____no_output_____"
]
],
[
[
"### Python补充\n\n像这些方法练习用`ipython3`就好了(`sudo apt-get install ipython3`)\n\ncode的话需要一个个的print,比较麻烦(我这边因为需要写文章,所以只能一个个code)\n\n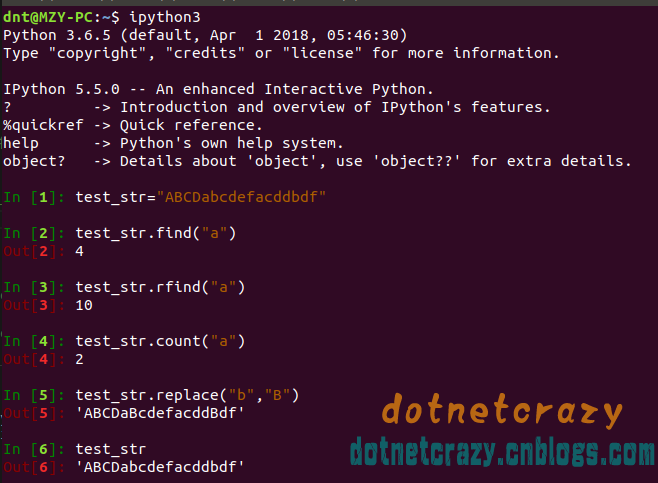",
"_____no_output_____"
],
[
"## 4.CSharp字符串方法系列\n\n### 4.1.查找\n\n`index0f`就相当于python里面的`find`\n\n`LastIndexOf` ==> `rfind`",
"_____no_output_____"
]
],
[
[
"%%script csharp\nvar test_str = \"ABCDabcdefacddbdf\";\n\n//# # 查找:find,rfind,index,rindex\n//# # xxx.find(str, start, end)\n//# print(test_str.find(\"cd\"))#从左往右\nConsole.WriteLine(test_str.IndexOf('a'));//4\nConsole.WriteLine(test_str.IndexOf(\"cd\"));//6\n\n//# print(test_str.rfind(\"cd\"))#从右往左\nConsole.WriteLine(test_str.LastIndexOf(\"cd\"));//11\n\n//# print(test_str.find(\"dnt\"))#find和rfind找不到就返回-1\nConsole.WriteLine(test_str.IndexOf(\"dnt\"));//-1",
"4\n6\n11\n-1\n"
]
],
[
[
"### 4.2.计数\n\n这个真用基础来解决的话,两种方法:\n\n第一种自己变形一下:(原字符串长度 - 替换后的长度) / 字符串长度\n\n```csharp\n//# # 计数:count\n//# # xxx.count(str, start, end)\n// print(test_str.count(\"d\"))#4\n// print(test_str.count(\"cd\"))#2\n// 第一反应,字典、正则、linq,后来想怎么用基础知识解决,于是有了这个~(原字符串长度-替换后的长度)/字符串长度\n\nConsole.WriteLine(test_str.Length - test_str.Replace(\"d\", \"\").Length);//统计单个字符就简单了\nConsole.WriteLine((test_str.Length - test_str.Replace(\"cd\", \"\").Length) / \"cd\".Length);\nConsole.WriteLine(test_str);//不用担心原字符串改变(python和C#都是有字符串不可变性的)\n```\n\n字符串统计另一种方法(<a href=\"https://github.com/dunitian/LoTCodeBase/tree/master/NetCode/2.面向对象/4.字符串\" target=\"_blank\">就用index</a>)\n\n```csharp\nint count = 0;\nint index = input.IndexOf(\"abc\");\n\nwhile (index != -1)\n{\n count++;\n index = input.IndexOf(\"abc\", index + 3);//index指向abc的后一位\n}\n```",
"_____no_output_____"
],
[
"### 4.3.替换\n\n替换指定次数的功能有点业余,就不说了,你可以自行思考哦~",
"_____no_output_____"
]
],
[
[
"%%script csharp\nvar test_str = \"ABCDabcdefacddbdf\";\nConsole.WriteLine(test_str.Replace(\"b\", \"B\"));",
"ABCDaBcdefacddBdf\n"
]
],
[
[
"### 4.4.分割\n\n`split`里面很多重载方法,可以自己去查看下\n\neg:`Split(\"\\n\",StringSplitOptions.RemoveEmptyEntries)`\n\n再说一下这个:`test_str.Split('a');` //返回数组\n\n如果要和Python一样返回列表==》`test_str.Split('a').ToList();` 【需要引用linq的命名空间哦】\n```csharp\nvar test_array = test_str.Split('a');//返回数组(如果要返回列表==》test_str.Split('a').ToList();)\nvar test_input = \"hi my name is dnt\";\n//# print(test_input.split(\" \")) #返回列表格式(后面会说)['hi', 'my', 'name', 'is', 'dnt']\ntest_input.Split(\" \");\n//# 按行分割,返回类型为List\nvar test_line_str = \"abc\\nbca\\ncab\\n\";\n//# print(test_line_str.splitlines())#['abc', 'bca', 'cab']\ntest_line_str.Split(\"\\n\", StringSplitOptions.RemoveEmptyEntries);\n```",
"_____no_output_____"
],
[
"### 4.5.连接\n\n**`string.Join(分隔符,数组)`**\n```csharp\nConsole.WriteLine(string.Join(\"-\", test_array));//test_array是数组 ABCD-bcdef-cddbdf\n\n```\n\n### 4.6.头尾判断\n\n`StartsWith`(以。。。开头),`EndsWith`(以。。。结尾)",
"_____no_output_____"
]
],
[
[
"%%script csharp\nvar start_end_str = \"http://www.baidu.net\";\n//# print(start_end_str.startswith(\"https://\") or start_end_str.startswith(\"http://\"))\nSystem.Console.WriteLine(start_end_str.StartsWith(\"https://\") || start_end_str.StartsWith(\"http://\"));\n//# print(start_end_str.endswith(\".com\"))\nSystem.Console.WriteLine(start_end_str.EndsWith(\".com\"));",
"True\nFalse\n"
]
],
[
[
"### 4.7.大小写系\n\n```csharp\n//# print(test_str.upper())#ABCDABCDEFACDDBDF\nConsole.WriteLine(test_str.ToUpper());\n//# print(test_str.lower())#abcdabcdefacddbdf\nConsole.WriteLine(test_str.ToLower());\n```\n\n### 4.8.格式化系\n\n`Tirm`很强大,除了去空格还可以去除你想去除的任意字符\n\nnet里面`string.Format`各种格式化输出,可以参考,这边就不讲了",
"_____no_output_____"
]
],
[
[
"%%script csharp\nvar strip_str = \" I Have a Dream \";\n//# print(strip_str.strip()+\"|\")#我加 | 是为了看清后面空格,没有别的用处\nConsole.WriteLine(strip_str.Trim() + \"|\");\n//# print(strip_str.lstrip()+\"|\")\nConsole.WriteLine(strip_str.TrimStart() + \"|\");\n//# print(strip_str.rstrip()+\"|\")\nConsole.WriteLine(strip_str.TrimEnd() + \"|\");",
"I Have a Dream|\nI Have a Dream |\n I Have a Dream|\n"
]
],
[
[
"### 4.9.验证系列\n`string.IsNullOrEmpty` 和 `string.IsNullOrWhiteSpace` 是系统自带的",
"_____no_output_____"
]
],
[
[
"%%script csharp\nvar test_str4 = \" \\t\";\nvar test_str5 = \" \\t \\n \"; //#isspace() ==>true\n// string.IsNullOrEmpty 和 string.IsNullOrWhiteSpace 是系统自带的,其他的你需要自己封装一个扩展类\nConsole.WriteLine(string.IsNullOrEmpty(test_str4)); //false\nConsole.WriteLine(string.IsNullOrWhiteSpace(test_str4));//true\nConsole.WriteLine(string.IsNullOrEmpty(test_str5));//false\nConsole.WriteLine(string.IsNullOrWhiteSpace(test_str5));//true",
"False\nTrue\nFalse\nTrue\n"
]
],
[
[
"其他的你需要自己封装一个扩展类(eg:<a href=\"https://github.com/dunitian/LoTCodeBase/blob/master/NetCode/5.逆天类库/LoTLibrary/Validation/ValidationHelper.cs\" target=\"_blank\">简单封装</a>)\n\n```csharp\nusing System;\nusing System.Collections.Generic;\nusing System.Linq;\nusing System.Text.RegularExpressions;\n\npublic static partial class ValidationHelper\n{\n #region 常用验证\n\n #region 集合系列\n /// <summary>\n /// 判断集合是否有数据\n /// </summary>\n /// <typeparam name=\"T\"></typeparam>\n /// <param name=\"list\"></param>\n /// <returns></returns>\n public static bool ExistsData<T>(this IEnumerable<T> list)\n {\n bool b = false;\n if (list != null && list.Count() > 0)\n {\n b = true;\n }\n return b;\n } \n #endregion\n\n #region Null判断系列\n /// <summary>\n /// 判断是否为空或Null\n /// </summary>\n /// <param name=\"objStr\"></param>\n /// <returns></returns>\n public static bool IsNullOrWhiteSpace(this string objStr)\n {\n if (string.IsNullOrWhiteSpace(objStr))\n {\n return true;\n }\n else\n {\n return false;\n }\n }\n\n /// <summary>\n /// 判断类型是否为可空类型\n /// </summary>\n /// <param name=\"theType\"></param>\n /// <returns></returns>\n public static bool IsNullableType(Type theType)\n {\n return (theType.IsGenericType && theType.GetGenericTypeDefinition().Equals(typeof(Nullable<>)));\n }\n #endregion\n\n #region 数字字符串检查\n /// <summary>\n /// 是否数字字符串(包括小数)\n /// </summary>\n /// <param name=\"objStr\">输入字符串</param>\n /// <returns></returns>\n public static bool IsNumber(this string objStr)\n {\n try\n {\n return Regex.IsMatch(objStr, @\"^\\d+(\\.\\d+)?$\");\n }\n catch\n {\n return false;\n }\n }\n\n /// <summary>\n /// 是否是浮点数\n /// </summary>\n /// <param name=\"objStr\">输入字符串</param>\n /// <returns></returns>\n public static bool IsDecimal(this string objStr)\n {\n try\n {\n return Regex.IsMatch(objStr, @\"^(-?\\d+)(\\.\\d+)?$\");\n }\n catch\n {\n return false;\n }\n }\n #endregion\n\n #endregion\n\n #region 业务常用\n\n #region 中文检测\n /// <summary>\n /// 检测是否有中文字符\n /// </summary>\n /// <param name=\"objStr\"></param>\n /// <returns></returns>\n public static bool IsZhCN(this string objStr)\n {\n try\n {\n return Regex.IsMatch(objStr, \"[\\u4e00-\\u9fa5]\");\n }\n catch\n {\n return false;\n }\n }\n #endregion\n\n #region 邮箱验证\n /// <summary>\n /// 判断邮箱地址是否正确\n /// </summary>\n /// <param name=\"objStr\"></param>\n /// <returns></returns>\n public static bool IsEmail(this string objStr)\n {\n try\n {\n return Regex.IsMatch(objStr, @\"^([\\w-\\.]+)@((\\[[0-9]{1,3}\\.[0-9]{1,3}\\.[0-9]{1,3}\\.)|(([\\w-]+\\.)+))([a-zA-Z]{2,4}|[0-9]{1,3})(\\]?)$\");\n }\n catch\n {\n return false;\n }\n }\n #endregion\n\n #region IP系列验证\n /// <summary>\n /// 是否为ip\n /// </summary>\n /// <param name=\"objStr\"></param>\n /// <returns></returns>\n public static bool IsIP(this string objStr)\n {\n return Regex.IsMatch(objStr, @\"^((2[0-4]\\d|25[0-5]|[01]?\\d\\d?)\\.){3}(2[0-4]\\d|25[0-5]|[01]?\\d\\d?)$\");\n }\n\n /// <summary> \n /// 判断输入的字符串是否是表示一个IP地址 \n /// </summary> \n /// <param name=\"objStr\">被比较的字符串</param> \n /// <returns>是IP地址则为True</returns> \n public static bool IsIPv4(this string objStr)\n {\n string[] IPs = objStr.Split('.');\n for (int i = 0; i < IPs.Length; i++)\n {\n if (!Regex.IsMatch(IPs[i], @\"^\\d+$\"))\n {\n return false;\n }\n if (Convert.ToUInt16(IPs[i]) > 255)\n {\n return false;\n }\n }\n return true;\n }\n\n /// <summary>\n /// 判断输入的字符串是否是合法的IPV6 地址 \n /// </summary>\n /// <param name=\"input\"></param>\n /// <returns></returns>\n public static bool IsIPV6(string input)\n {\n string temp = input;\n string[] strs = temp.Split(':');\n if (strs.Length > 8)\n {\n return false;\n }\n int count = input.GetStrCount(\"::\");\n if (count > 1)\n {\n return false;\n }\n else if (count == 0)\n {\n return Regex.IsMatch(input, @\"^([\\da-f]{1,4}:){7}[\\da-f]{1,4}$\");\n }\n else\n {\n return Regex.IsMatch(input, @\"^([\\da-f]{1,4}:){0,5}::([\\da-f]{1,4}:){0,5}[\\da-f]{1,4}$\");\n }\n }\n #endregion\n\n #region 网址系列验证\n /// <summary>\n /// 验证网址是否正确(http:或者https:)【后期添加 // 的情况】\n /// </summary>\n /// <param name=\"objStr\">地址</param>\n /// <returns></returns>\n public static bool IsWebUrl(this string objStr)\n {\n try\n {\n return Regex.IsMatch(objStr, @\"http://([\\w-]+\\.)+[\\w-]+(/[\\w- ./?%&=]*)?|https://([\\w-]+\\.)+[\\w-]+(/[\\w- ./?%&=]*)?\");\n }\n catch\n {\n return false;\n }\n }\n\n /// <summary>\n /// 判断输入的字符串是否是一个超链接 \n /// </summary>\n /// <param name=\"objStr\"></param>\n /// <returns></returns>\n public static bool IsURL(this string objStr)\n {\n string pattern = @\"^[a-zA-Z]+://(\\w+(-\\w+)*)(\\.(\\w+(-\\w+)*))*(\\?\\S*)?$\";\n return Regex.IsMatch(objStr, pattern);\n }\n #endregion\n\n #region 邮政编码验证\n /// <summary>\n /// 验证邮政编码是否正确\n /// </summary>\n /// <param name=\"objStr\">输入字符串</param>\n /// <returns></returns>\n public static bool IsZipCode(this string objStr)\n {\n try\n {\n return Regex.IsMatch(objStr, @\"\\d{6}\");\n }\n catch\n {\n return false;\n }\n }\n #endregion\n\n #region 电话+手机验证\n /// <summary>\n /// 验证手机号是否正确\n /// </summary>\n /// <param name=\"objStr\">手机号</param>\n /// <returns></returns>\n public static bool IsMobile(this string objStr)\n {\n try\n {\n return Regex.IsMatch(objStr, @\"^13[0-9]{9}|15[012356789][0-9]{8}|18[0123456789][0-9]{8}|147[0-9]{8}$\");\n }\n catch\n {\n return false;\n }\n }\n\n /// <summary>\n /// 匹配3位或4位区号的电话号码,其中区号可以用小括号括起来,也可以不用,区号与本地号间可以用连字号或空格间隔,也可以没有间隔 \n /// </summary>\n /// <param name=\"objStr\"></param>\n /// <returns></returns>\n public static bool IsPhone(this string objStr)\n {\n try\n {\n return Regex.IsMatch(objStr, \"^\\\\(0\\\\d{2}\\\\)[- ]?\\\\d{8}$|^0\\\\d{2}[- ]?\\\\d{8}$|^\\\\(0\\\\d{3}\\\\)[- ]?\\\\d{7}$|^0\\\\d{3}[- ]?\\\\d{7}$\");\n }\n catch\n {\n return false;\n }\n }\n #endregion\n\n #region 字母或数字验证\n /// <summary>\n /// 是否只是字母或数字\n /// </summary>\n /// <param name=\"objStr\"></param>\n /// <returns></returns>\n public static bool IsAbcOr123(this string objStr)\n {\n try\n {\n return Regex.IsMatch(objStr, @\"^[0-9a-zA-Z\\$]+$\");\n }\n catch\n {\n return false;\n }\n }\n #endregion\n\n #endregion\n}\n```",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
cb8d1528b101a5aa649a4ebcd6c1d6a9b9f903b4
| 10,102 |
ipynb
|
Jupyter Notebook
|
lessons/03_Lesson03_Assignment.ipynb
|
goodsang1023/aeropython
|
dbb780745fc93f3d6fd173b7b55ba23cda3c1d8d
|
[
"CC-BY-4.0"
] | null | null | null |
lessons/03_Lesson03_Assignment.ipynb
|
goodsang1023/aeropython
|
dbb780745fc93f3d6fd173b7b55ba23cda3c1d8d
|
[
"CC-BY-4.0"
] | null | null | null |
lessons/03_Lesson03_Assignment.ipynb
|
goodsang1023/aeropython
|
dbb780745fc93f3d6fd173b7b55ba23cda3c1d8d
|
[
"CC-BY-4.0"
] | 1 |
2021-01-31T22:54:57.000Z
|
2021-01-31T22:54:57.000Z
| 34.59589 | 345 | 0.514354 |
[
[
[
"###### Text provided under a Creative Commons Attribution license, CC-BY. Code under MIT license. (c)2014 Lorena A. Barba, Pi-Yueh Chuang. Thanks: NSF for support via CAREER award #1149784.",
"_____no_output_____"
],
[
"# Source Distribution on an Airfoil",
"_____no_output_____"
],
[
"In [Lesson 3](03_Lesson03_doublet.ipynb) of *AeroPython*, you learned that it is possible to represent potential flow around a circular cylinder using the superposition of a doublet singularity and a free stream. But potential flow is even more powerful: you can represent the flow around *any* shape. How is it possible, you might ask?\n\nFor non-lifting bodies, you can use a source distribution on the body surface, superposed with a free stream. In this assignment, you will build the flow around a NACA0012 airfoil, using a set of sources.\n\nBefore you start, take a moment to think: in flow around a symmetric airfoil at $0^{\\circ}$ angle of attack,\n\n* Where is the point of maximum pressure?\n* What do we call that point?\n* Will the airfoil generate any lift?\n\nAt the end of this assignment, come back to these questions, and see if it all makes sense.",
"_____no_output_____"
],
[
"## Problem Setup",
"_____no_output_____"
],
[
"You will read data files containing information about the location and the strength of a set of sources located on the surface of a NACA0012 airfoil. \n\nThere are three data files: NACA0012_x.txt, NACA0012_y.txt, and NACA0012_sigma.txt. To load each file into a NumPy array, you need the function [`numpy.loadtxt`](http://docs.scipy.org/doc/numpy/reference/generated/numpy.loadtxt.html). The files should be found in the `resources` folder of the `lessons`. ",
"_____no_output_____"
],
[
"Using 51 mesh points in each direction, and a domain $[-1, 2]\\times[-0.5, 0.5]$, compute the velocity due to the set of sources plus a free stream in the $x$-direction with $U_{\\infty}=1$. Also compute the coefficient of pressure on your grid points.",
"_____no_output_____"
],
[
"## Questions:",
"_____no_output_____"
],
[
"1. What is the value of maximum pressure coefficient, $C_p$?\n2. What are the array indices for the maximum value of $C_p$?\n\nMake the following plots to visualize and inspect the resulting flow pattern:\n\n* Stream lines in the domain and the profile of our NACA0012 airfoil, in one plot\n* Distribution of the pressure coefficient and a single marker on the location of the maximum pressure",
"_____no_output_____"
],
[
"**Hint**: You might use the following NumPy functions: [`numpy.unravel_index`](http://docs.scipy.org/doc/numpy/reference/generated/numpy.unravel_index.html) and [`numpy.argmax`](http://docs.scipy.org/doc/numpy/reference/generated/numpy.argmax.html)",
"_____no_output_____"
],
[
"##### Think",
"_____no_output_____"
],
[
"1. Do the stream lines look like you expected?\n2. What does the distribution of pressure tell you about lift generated by the airfoil?\n3. Does the location of the point of maximum pressure seem right to you?",
"_____no_output_____"
]
],
[
[
"from IPython.core.display import HTML\ndef css_styling(filepath):\n styles = open(filepath, 'r').read()\n return HTML(styles)\ncss_styling('../styles/custom.css')",
"_____no_output_____"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
]
] |
cb8d1c779229bc43f9e42b73e13ee30f5792a03b
| 44,098 |
ipynb
|
Jupyter Notebook
|
Pandas-and-Requests-Example.ipynb
|
libcce/lc-lesson-materials
|
c100a0f1e4a3bbba062605f19e9f0fc8d21e7358
|
[
"Apache-2.0"
] | 1 |
2018-09-21T13:42:56.000Z
|
2018-09-21T13:42:56.000Z
|
Pandas-and-Requests-Example.ipynb
|
libcce/lc-lesson-materials
|
c100a0f1e4a3bbba062605f19e9f0fc8d21e7358
|
[
"Apache-2.0"
] | null | null | null |
Pandas-and-Requests-Example.ipynb
|
libcce/lc-lesson-materials
|
c100a0f1e4a3bbba062605f19e9f0fc8d21e7358
|
[
"Apache-2.0"
] | null | null | null | 68.368992 | 16,660 | 0.695111 |
[
[
[
"import pandas as pd",
"_____no_output_____"
],
[
"# Read csv into pandas - CrossRef example from OpenRefine lesson\ndata = pandas.read_csv(\"https://raw.githubusercontent.com/LibraryCarpentry/lc-open-refine/gh-pages/data/doaj-article-sample.csv\")",
"_____no_output_____"
],
[
"# Get the first 10 rows of data\ndata.head()",
"_____no_output_____"
],
[
"# Get the last 10 rows of data from CrossRef csv\ndata.tail()",
"_____no_output_____"
],
[
"# Get the count of licenses by type\ndata.groupby(['Licence']).size()",
"_____no_output_____"
],
[
"# Get the shape of the data, number of rows and columns\ndata.shape",
"_____no_output_____"
],
[
"# Plot number of licenses by type\ndata.groupby(['Licence']).size().plot(kind='bar')",
"_____no_output_____"
]
],
[
[
"For a nice tutorial on Pandas and Bokeh, see:\nhttps://programminghistorian.org/en/lessons/visualizing-with-bokeh",
"_____no_output_____"
]
],
[
[
"# Example of installing Python module\n!pip install plotly",
"Collecting plotly\n Downloading https://files.pythonhosted.org/packages/25/e6/716d30f51604daedaf12b1064e1d1e48d1c0a224ac41a83496b39862124d/plotly-3.2.1-py2.py3-none-any.whl (37.0MB)\n\u001b[K 100% |████████████████████████████████| 37.0MB 34kB/s eta 0:00:011\n\u001b[?25hCollecting retrying>=1.3.3 (from plotly)\n Downloading https://files.pythonhosted.org/packages/44/ef/beae4b4ef80902f22e3af073397f079c96969c69b2c7d52a57ea9ae61c9d/retrying-1.3.3.tar.gz\nRequirement already satisfied: pytz in /anaconda3/lib/python3.6/site-packages (from plotly)\nRequirement already satisfied: nbformat>=4.2 in /anaconda3/lib/python3.6/site-packages (from plotly)\nRequirement already satisfied: decorator>=4.0.6 in /anaconda3/lib/python3.6/site-packages (from plotly)\nRequirement already satisfied: requests in /anaconda3/lib/python3.6/site-packages (from plotly)\nRequirement already satisfied: six in /anaconda3/lib/python3.6/site-packages (from plotly)\nRequirement already satisfied: ipython_genutils in /anaconda3/lib/python3.6/site-packages (from nbformat>=4.2->plotly)\nRequirement already satisfied: traitlets>=4.1 in /anaconda3/lib/python3.6/site-packages (from nbformat>=4.2->plotly)\nRequirement already satisfied: jsonschema!=2.5.0,>=2.4 in /anaconda3/lib/python3.6/site-packages (from nbformat>=4.2->plotly)\nRequirement already satisfied: jupyter_core in /anaconda3/lib/python3.6/site-packages (from nbformat>=4.2->plotly)\nRequirement already satisfied: chardet<3.1.0,>=3.0.2 in /anaconda3/lib/python3.6/site-packages (from requests->plotly)\nRequirement already satisfied: idna<2.7,>=2.5 in /anaconda3/lib/python3.6/site-packages (from requests->plotly)\nRequirement already satisfied: urllib3<1.23,>=1.21.1 in /anaconda3/lib/python3.6/site-packages (from requests->plotly)\nRequirement already satisfied: certifi>=2017.4.17 in /anaconda3/lib/python3.6/site-packages (from requests->plotly)\nBuilding wheels for collected packages: retrying\n Running setup.py bdist_wheel for retrying ... \u001b[?25ldone\n\u001b[?25h Stored in directory: /Users/cerdmann/Library/Caches/pip/wheels/d7/a9/33/acc7b709e2a35caa7d4cae442f6fe6fbf2c43f80823d46460c\nSuccessfully built retrying\nInstalling collected packages: retrying, plotly\nSuccessfully installed plotly-3.2.1 retrying-1.3.3\n\u001b[33mYou are using pip version 9.0.1, however version 18.0 is available.\nYou should consider upgrading via the 'pip install --upgrade pip' command.\u001b[0m\n"
],
[
"import requests\nimport json",
"_____no_output_____"
],
[
"crossref_url = 'http://api.crossref.org/journals/1099-4300'\nresponse = requests.get(crossref_url)\nprint(response.text)",
"{\"status\":\"ok\",\"message-type\":\"journal\",\"message-version\":\"1.0.0\",\"message\":{\"last-status-check-time\":1537311111267,\"counts\":{\"total-dois\":3583,\"current-dois\":1850,\"backfile-dois\":1733},\"breakdowns\":{\"dois-by-issued-year\":[[2018,697],[2017,694],[2016,459],[2015,453],[2014,328],[2013,284],[2012,137],[2010,119],[2011,101],[2009,69],[2008,56],[2003,39],[2004,33],[2001,21],[2005,20],[2006,18],[2007,17],[1999,16],[2000,12],[2002,10]]},\"publisher\":\"MDPI AG\",\"coverage\":{\"affiliations-current\":0.0,\"similarity-checking-current\":0.999459445476532,\"funders-backfile\":0.06231967732310295,\"licenses-backfile\":0.9976918697357178,\"funders-current\":0.26972973346710205,\"affiliations-backfile\":0.0,\"resource-links-backfile\":0.0,\"orcids-backfile\":0.00865551084280014,\"update-policies-current\":0.0,\"open-references-backfile\":1.0,\"orcids-current\":0.36918920278549194,\"similarity-checking-backfile\":0.9976918697357178,\"references-backfile\":0.9699942469596863,\"award-numbers-backfile\":0.06116560846567154,\"update-policies-backfile\":0.0,\"licenses-current\":0.999459445476532,\"award-numbers-current\":0.2572973072528839,\"abstracts-backfile\":0.0,\"resource-links-current\":0.0,\"abstracts-current\":0.10378378629684448,\"open-references-current\":1.0,\"references-current\":0.9897297024726868},\"title\":\"Entropy\",\"subjects\":[{\"name\":\"General Physics and Astronomy\",\"ASJC\":3100}],\"coverage-type\":{\"all\":{\"last-status-check-time\":1537311109329,\"affiliations\":0.0,\"abstracts\":0.0535714291036129,\"orcids\":0.1947544664144516,\"licenses\":0.9983258843421936,\"references\":0.9799107313156128,\"funders\":0.1693638414144516,\"similarity-checking\":0.9983258843421936,\"award-numbers\":0.1623883992433548,\"update-policies\":0.0,\"resource-links\":0.0,\"open-references\":1.0},\"backfile\":{\"last-status-check-time\":1537311108830,\"affiliations\":0.0,\"abstracts\":0.0,\"orcids\":0.00865551084280014,\"licenses\":0.9976918697357178,\"references\":0.9699942469596863,\"funders\":0.06231967732310295,\"similarity-checking\":0.9976918697357178,\"award-numbers\":0.06116560846567154,\"update-policies\":0.0,\"resource-links\":0.0,\"open-references\":1.0},\"current\":{\"last-status-check-time\":1537311108125,\"affiliations\":0.0,\"abstracts\":0.10378378629684448,\"orcids\":0.36918920278549194,\"licenses\":0.999459445476532,\"references\":0.9897297024726868,\"funders\":0.26972973346710205,\"similarity-checking\":0.999459445476532,\"award-numbers\":0.2572973072528839,\"update-policies\":0.0,\"resource-links\":0.0,\"open-references\":1.0}},\"flags\":{\"deposits-abstracts-current\":true,\"deposits-orcids-current\":true,\"deposits\":true,\"deposits-affiliations-backfile\":false,\"deposits-update-policies-backfile\":false,\"deposits-similarity-checking-backfile\":true,\"deposits-award-numbers-current\":true,\"deposits-resource-links-current\":false,\"deposits-articles\":true,\"deposits-affiliations-current\":false,\"deposits-funders-current\":true,\"deposits-references-backfile\":true,\"deposits-abstracts-backfile\":false,\"deposits-licenses-backfile\":true,\"deposits-award-numbers-backfile\":true,\"deposits-open-references-backfile\":true,\"deposits-open-references-current\":true,\"deposits-references-current\":true,\"deposits-resource-links-backfile\":false,\"deposits-orcids-backfile\":true,\"deposits-funders-backfile\":true,\"deposits-update-policies-current\":false,\"deposits-similarity-checking-current\":true,\"deposits-licenses-current\":true},\"ISSN\":[\"1099-4300\"],\"issn-type\":[{\"value\":\"1099-4300\",\"type\":\"electronic\"}]}}\n"
],
[
"json_data = json.loads(response.text)",
"_____no_output_____"
],
[
"pprint(json_data)",
"Pretty printing has been turned ON\n"
],
[
"print(json_data['message']['publisher'])",
"MDPI AG\n"
],
[
"# Command line example, print working directory\n!pwd",
"/Users/cerdmann\r\n"
],
[
"# Command line example, list files\n!ls",
"\u001b[34mAnacondaProjects\u001b[m\u001b[m \u001b[34mDownloads\u001b[m\u001b[m \u001b[34mPictures\u001b[m\u001b[m\r\n\u001b[34mApplications\u001b[m\u001b[m \u001b[34mLibrary\u001b[m\u001b[m \u001b[34mPublic\u001b[m\u001b[m\r\n\u001b[34mBox Sync\u001b[m\u001b[m \u001b[34mMovies\u001b[m\u001b[m \u001b[34mZotero\u001b[m\u001b[m\r\n\u001b[34mDesktop\u001b[m\u001b[m \u001b[34mMusic\u001b[m\u001b[m \u001b[34mstudio-link\u001b[m\u001b[m\r\n\u001b[34mDocuments\u001b[m\u001b[m Pandas Example.ipynb\r\n"
],
[
"# Command line example, create a file\n!touch abc.txt",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb8d2a0fdd0f8d5fcf75ce5f3b0785e1f74d6972
| 244,951 |
ipynb
|
Jupyter Notebook
|
socraticmodels/SocraticModels_ImageCaptioning.ipynb
|
nhutnamhcmus/google-research
|
5b645f0005fb5a8d96c58d8d84017d97662d564d
|
[
"Apache-2.0"
] | null | null | null |
socraticmodels/SocraticModels_ImageCaptioning.ipynb
|
nhutnamhcmus/google-research
|
5b645f0005fb5a8d96c58d8d84017d97662d564d
|
[
"Apache-2.0"
] | null | null | null |
socraticmodels/SocraticModels_ImageCaptioning.ipynb
|
nhutnamhcmus/google-research
|
5b645f0005fb5a8d96c58d8d84017d97662d564d
|
[
"Apache-2.0"
] | null | null | null | 293.003589 | 196,221 | 0.88692 |
[
[
[
"Copyright 2021 Google LLC.\nSPDX-License-Identifier: Apache-2.0\n\nLicensed under the Apache License, Version 2.0 (the \"License\");\nyou may not use this file except in compliance with the License.\nYou may obtain a copy of the License at\n\nhttps://www.apache.org/licenses/LICENSE-2.0\n\nUnless required by applicable law or agreed to in writing, software\ndistributed under the License is distributed on an \"AS IS\" BASIS,\nWITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\nSee the License for the specific language governing permissions and\nlimitations under the License.\n\n# **Socratic Models: Image Captioning**\n\nSocratic Models (SMs) is a framework that composes multiple pre-existing foundation models (e.g., large language models, visual language models, audio-language models) to provide results for new multimodal tasks, without any model finetuning.\n\nThis colab runs an example of SMs for image captioning.\n\nThis is a reference implementation of one task demonstrated in the work: [Socratic Models: Composing Zero-Shot Multimodal Reasoning with Language](https://socraticmodels.github.io/)\n\n**Disclaimer:** this colab uses CLIP and GPT-3 as foundation models, and may be subject to unwanted biases. This code should be used with caution (and checked for correctness) in downstream applications.\n\n### **Quick Start:**\n\n**Step 1.** Register for an [OpenAI API key](https://openai.com/blog/openai-api/) to use GPT-3 (there's a free trial) and enter it below\n\n**Step 2.** Menu > Change runtime type > Hardware accelerator > \"GPU\"\n\n**Step 3.** Menu > Runtime > Run all\n\n",
"_____no_output_____"
]
],
[
[
"openai_api_key = \"your-api-key\"",
"_____no_output_____"
]
],
[
[
"## **Setup**\nThis installs a few dependencies: PyTorch, CLIP, GPT-3.",
"_____no_output_____"
]
],
[
[
"!pip install ftfy regex tqdm fvcore imageio imageio-ffmpeg openai pattern\n!pip install git+https://github.com/openai/CLIP.git\n!pip install -U --no-cache-dir gdown --pre\n!pip install profanity-filter\n!nvidia-smi # Show GPU info.",
"Collecting ftfy\n Downloading ftfy-6.1.1-py3-none-any.whl (53 kB)\n\u001b[?25l\r\u001b[K |██████▏ | 10 kB 17.6 MB/s eta 0:00:01\r\u001b[K |████████████▍ | 20 kB 12.3 MB/s eta 0:00:01\r\u001b[K |██████████████████▌ | 30 kB 7.2 MB/s eta 0:00:01\r\u001b[K |████████████████████████▊ | 40 kB 3.6 MB/s eta 0:00:01\r\u001b[K |██████████████████████████████▉ | 51 kB 4.1 MB/s eta 0:00:01\r\u001b[K |████████████████████████████████| 53 kB 833 kB/s \n\u001b[?25hRequirement already satisfied: regex in /usr/local/lib/python3.7/dist-packages (2019.12.20)\nRequirement already satisfied: tqdm in /usr/local/lib/python3.7/dist-packages (4.63.0)\nCollecting fvcore\n Downloading fvcore-0.1.5.post20220305.tar.gz (50 kB)\n\u001b[K |████████████████████████████████| 50 kB 3.5 MB/s \n\u001b[?25hRequirement already satisfied: imageio in /usr/local/lib/python3.7/dist-packages (2.4.1)\nCollecting imageio-ffmpeg\n Downloading imageio_ffmpeg-0.4.5-py3-none-manylinux2010_x86_64.whl (26.9 MB)\n\u001b[K |████████████████████████████████| 26.9 MB 7.5 MB/s \n\u001b[?25hCollecting openai\n Downloading openai-0.16.0.tar.gz (41 kB)\n\u001b[K |████████████████████████████████| 41 kB 203 kB/s \n\u001b[?25h Installing build dependencies ... \u001b[?25l\u001b[?25hdone\n Getting requirements to build wheel ... \u001b[?25l\u001b[?25hdone\n Preparing wheel metadata ... \u001b[?25l\u001b[?25hdone\nCollecting pattern\n Downloading Pattern-3.6.0.tar.gz (22.2 MB)\n\u001b[K |████████████████████████████████| 22.2 MB 1.7 MB/s \n\u001b[?25hRequirement already satisfied: wcwidth>=0.2.5 in /usr/local/lib/python3.7/dist-packages (from ftfy) (0.2.5)\nRequirement already satisfied: numpy in /usr/local/lib/python3.7/dist-packages (from fvcore) (1.21.5)\nCollecting yacs>=0.1.6\n Downloading yacs-0.1.8-py3-none-any.whl (14 kB)\nCollecting pyyaml>=5.1\n Downloading PyYAML-6.0-cp37-cp37m-manylinux_2_5_x86_64.manylinux1_x86_64.manylinux_2_12_x86_64.manylinux2010_x86_64.whl (596 kB)\n\u001b[K |████████████████████████████████| 596 kB 28.6 MB/s \n\u001b[?25hRequirement already satisfied: termcolor>=1.1 in /usr/local/lib/python3.7/dist-packages (from fvcore) (1.1.0)\nRequirement already satisfied: Pillow in /usr/local/lib/python3.7/dist-packages (from fvcore) (7.1.2)\nRequirement already satisfied: tabulate in /usr/local/lib/python3.7/dist-packages (from fvcore) (0.8.9)\nCollecting iopath>=0.1.7\n Downloading iopath-0.1.9-py3-none-any.whl (27 kB)\nCollecting portalocker\n Downloading portalocker-2.4.0-py2.py3-none-any.whl (16 kB)\nRequirement already satisfied: pandas>=1.2.3 in /usr/local/lib/python3.7/dist-packages (from openai) (1.3.5)\nRequirement already satisfied: openpyxl>=3.0.7 in /usr/local/lib/python3.7/dist-packages (from openai) (3.0.9)\nRequirement already satisfied: requests>=2.20 in /usr/local/lib/python3.7/dist-packages (from openai) (2.23.0)\nCollecting pandas-stubs>=1.1.0.11\n Downloading pandas_stubs-1.2.0.56-py3-none-any.whl (162 kB)\n\u001b[K |████████████████████████████████| 162 kB 48.9 MB/s \n\u001b[?25hRequirement already satisfied: et-xmlfile in /usr/local/lib/python3.7/dist-packages (from openpyxl>=3.0.7->openai) (1.1.0)\nRequirement already satisfied: python-dateutil>=2.7.3 in /usr/local/lib/python3.7/dist-packages (from pandas>=1.2.3->openai) (2.8.2)\nRequirement already satisfied: pytz>=2017.3 in /usr/local/lib/python3.7/dist-packages (from pandas>=1.2.3->openai) (2018.9)\nRequirement already satisfied: typing-extensions>=3.7.4.3 in /usr/local/lib/python3.7/dist-packages (from pandas-stubs>=1.1.0.11->openai) (3.10.0.2)\nRequirement already satisfied: six>=1.5 in /usr/local/lib/python3.7/dist-packages (from python-dateutil>=2.7.3->pandas>=1.2.3->openai) (1.15.0)\nRequirement already satisfied: chardet<4,>=3.0.2 in /usr/local/lib/python3.7/dist-packages (from requests>=2.20->openai) (3.0.4)\nRequirement already satisfied: urllib3!=1.25.0,!=1.25.1,<1.26,>=1.21.1 in /usr/local/lib/python3.7/dist-packages (from requests>=2.20->openai) (1.24.3)\nRequirement already satisfied: idna<3,>=2.5 in /usr/local/lib/python3.7/dist-packages (from requests>=2.20->openai) (2.10)\nRequirement already satisfied: certifi>=2017.4.17 in /usr/local/lib/python3.7/dist-packages (from requests>=2.20->openai) (2021.10.8)\nRequirement already satisfied: future in /usr/local/lib/python3.7/dist-packages (from pattern) (0.16.0)\nCollecting backports.csv\n Downloading backports.csv-1.0.7-py2.py3-none-any.whl (12 kB)\nCollecting mysqlclient\n Downloading mysqlclient-2.1.0.tar.gz (87 kB)\n\u001b[K |████████████████████████████████| 87 kB 6.7 MB/s \n\u001b[?25hRequirement already satisfied: beautifulsoup4 in /usr/local/lib/python3.7/dist-packages (from pattern) (4.6.3)\nRequirement already satisfied: lxml in /usr/local/lib/python3.7/dist-packages (from pattern) (4.2.6)\nCollecting feedparser\n Downloading feedparser-6.0.8-py3-none-any.whl (81 kB)\n\u001b[K |████████████████████████████████| 81 kB 9.8 MB/s \n\u001b[?25hCollecting pdfminer.six\n Downloading pdfminer.six-20220319-py3-none-any.whl (5.6 MB)\n\u001b[K |████████████████████████████████| 5.6 MB 35.2 MB/s \n\u001b[?25hRequirement already satisfied: scipy in /usr/local/lib/python3.7/dist-packages (from pattern) (1.4.1)\nRequirement already satisfied: nltk in /usr/local/lib/python3.7/dist-packages (from pattern) (3.2.5)\nCollecting python-docx\n Downloading python-docx-0.8.11.tar.gz (5.6 MB)\n\u001b[K |████████████████████████████████| 5.6 MB 38.0 MB/s \n\u001b[?25hCollecting cherrypy\n Downloading CherryPy-18.6.1-py2.py3-none-any.whl (419 kB)\n\u001b[K |████████████████████████████████| 419 kB 47.6 MB/s \n\u001b[?25hCollecting jaraco.collections\n Downloading jaraco.collections-3.5.1-py3-none-any.whl (10 kB)\nCollecting cheroot>=8.2.1\n Downloading cheroot-8.6.0-py2.py3-none-any.whl (104 kB)\n\u001b[K |████████████████████████████████| 104 kB 50.6 MB/s \n\u001b[?25hCollecting zc.lockfile\n Downloading zc.lockfile-2.0-py2.py3-none-any.whl (9.7 kB)\nCollecting portend>=2.1.1\n Downloading portend-3.1.0-py3-none-any.whl (5.3 kB)\nRequirement already satisfied: more-itertools in /usr/local/lib/python3.7/dist-packages (from cherrypy->pattern) (8.12.0)\nCollecting jaraco.functools\n Downloading jaraco.functools-3.5.0-py3-none-any.whl (7.0 kB)\nCollecting tempora>=1.8\n Downloading tempora-5.0.1-py3-none-any.whl (15 kB)\nCollecting sgmllib3k\n Downloading sgmllib3k-1.0.0.tar.gz (5.8 kB)\nCollecting jaraco.classes\n Downloading jaraco.classes-3.2.1-py3-none-any.whl (5.6 kB)\nCollecting jaraco.text\n Downloading jaraco.text-3.7.0-py3-none-any.whl (8.6 kB)\nCollecting jaraco.context>=4.1\n Downloading jaraco.context-4.1.1-py3-none-any.whl (4.4 kB)\nRequirement already satisfied: importlib-resources in /usr/local/lib/python3.7/dist-packages (from jaraco.text->jaraco.collections->cherrypy->pattern) (5.4.0)\nRequirement already satisfied: zipp>=3.1.0 in /usr/local/lib/python3.7/dist-packages (from importlib-resources->jaraco.text->jaraco.collections->cherrypy->pattern) (3.7.0)\nCollecting cryptography\n Downloading cryptography-36.0.2-cp36-abi3-manylinux_2_24_x86_64.whl (3.6 MB)\n\u001b[K |████████████████████████████████| 3.6 MB 38.9 MB/s \n\u001b[?25hRequirement already satisfied: cffi>=1.12 in /usr/local/lib/python3.7/dist-packages (from cryptography->pdfminer.six->pattern) (1.15.0)\nRequirement already satisfied: pycparser in /usr/local/lib/python3.7/dist-packages (from cffi>=1.12->cryptography->pdfminer.six->pattern) (2.21)\nRequirement already satisfied: setuptools in /usr/local/lib/python3.7/dist-packages (from zc.lockfile->cherrypy->pattern) (57.4.0)\nBuilding wheels for collected packages: fvcore, openai, pattern, mysqlclient, python-docx, sgmllib3k\n Building wheel for fvcore (setup.py) ... \u001b[?25l\u001b[?25hdone\n Created wheel for fvcore: filename=fvcore-0.1.5.post20220305-py3-none-any.whl size=61214 sha256=983578915a9fcc79fcd9cc600b21557c2c3c2a6678e4e64c13380ca5bcb08f5d\n Stored in directory: /root/.cache/pip/wheels/b5/b7/6e/43b1693d06fac3633af48db68557513b0a37ab38b0a8b798f9\n Building wheel for openai (PEP 517) ... \u001b[?25l\u001b[?25hdone\n Created wheel for openai: filename=openai-0.16.0-py3-none-any.whl size=50784 sha256=49563d8fd3ed946ac624ac5af4bafab0ac2e0a63e2ec55848ebba713e116daa6\n Stored in directory: /root/.cache/pip/wheels/c1/f3/50/adfd6d5b5a417fef651921a8c5f77c0e644265ae000f3fb69b\n Building wheel for pattern (setup.py) ... \u001b[?25l\u001b[?25hdone\n Created wheel for pattern: filename=Pattern-3.6-py3-none-any.whl size=22332721 sha256=4e0291fdb369102a3ff22a8417d3dd7af3395d9ca564b5dcc1776cbc6e79c713\n Stored in directory: /root/.cache/pip/wheels/8d/1f/4e/9b67afd2430d55dee90bd57618dd7d899f1323e5852c465682\n Building wheel for mysqlclient (setup.py) ... \u001b[?25l\u001b[?25hdone\n Created wheel for mysqlclient: filename=mysqlclient-2.1.0-cp37-cp37m-linux_x86_64.whl size=99974 sha256=34f2d11e5991ff51a7b3ae035f0734463ca0bba6f79aae239229d19939c25285\n Stored in directory: /root/.cache/pip/wheels/97/d4/df/08cd6e1fa4a8691b268ab254bd0fa589827ab5b65638c010b4\n Building wheel for python-docx (setup.py) ... \u001b[?25l\u001b[?25hdone\n Created wheel for python-docx: filename=python_docx-0.8.11-py3-none-any.whl size=184507 sha256=d384403bbaff25c5c3cf0b777bd429e6ef9a4dea53d26f6fde239ad26ddf8536\n Stored in directory: /root/.cache/pip/wheels/f6/6f/b9/d798122a8b55b74ad30b5f52b01482169b445fbb84a11797a6\n Building wheel for sgmllib3k (setup.py) ... \u001b[?25l\u001b[?25hdone\n Created wheel for sgmllib3k: filename=sgmllib3k-1.0.0-py3-none-any.whl size=6066 sha256=307845fda19d662854fa7c869f6fb5c610f5d94bcc682c16999161a26cd4e728\n Stored in directory: /root/.cache/pip/wheels/73/ad/a4/0dff4a6ef231fc0dfa12ffbac2a36cebfdddfe059f50e019aa\nSuccessfully built fvcore openai pattern mysqlclient python-docx sgmllib3k\nInstalling collected packages: jaraco.functools, jaraco.context, tempora, jaraco.text, jaraco.classes, zc.lockfile, sgmllib3k, pyyaml, portend, portalocker, jaraco.collections, cryptography, cheroot, yacs, python-docx, pdfminer.six, pandas-stubs, mysqlclient, iopath, feedparser, cherrypy, backports.csv, pattern, openai, imageio-ffmpeg, fvcore, ftfy\n Attempting uninstall: pyyaml\n Found existing installation: PyYAML 3.13\n Uninstalling PyYAML-3.13:\n Successfully uninstalled PyYAML-3.13\nSuccessfully installed backports.csv-1.0.7 cheroot-8.6.0 cherrypy-18.6.1 cryptography-36.0.2 feedparser-6.0.8 ftfy-6.1.1 fvcore-0.1.5.post20220305 imageio-ffmpeg-0.4.5 iopath-0.1.9 jaraco.classes-3.2.1 jaraco.collections-3.5.1 jaraco.context-4.1.1 jaraco.functools-3.5.0 jaraco.text-3.7.0 mysqlclient-2.1.0 openai-0.16.0 pandas-stubs-1.2.0.56 pattern-3.6 pdfminer.six-20220319 portalocker-2.4.0 portend-3.1.0 python-docx-0.8.11 pyyaml-6.0 sgmllib3k-1.0.0 tempora-5.0.1 yacs-0.1.8 zc.lockfile-2.0\nCollecting git+https://github.com/openai/CLIP.git\n Cloning https://github.com/openai/CLIP.git to /tmp/pip-req-build-jjedlelt\n Running command git clone -q https://github.com/openai/CLIP.git /tmp/pip-req-build-jjedlelt\nRequirement already satisfied: ftfy in /usr/local/lib/python3.7/dist-packages (from clip==1.0) (6.1.1)\nRequirement already satisfied: regex in /usr/local/lib/python3.7/dist-packages (from clip==1.0) (2019.12.20)\nRequirement already satisfied: tqdm in /usr/local/lib/python3.7/dist-packages (from clip==1.0) (4.63.0)\nRequirement already satisfied: torch in /usr/local/lib/python3.7/dist-packages (from clip==1.0) (1.10.0+cu111)\nRequirement already satisfied: torchvision in /usr/local/lib/python3.7/dist-packages (from clip==1.0) (0.11.1+cu111)\nRequirement already satisfied: wcwidth>=0.2.5 in /usr/local/lib/python3.7/dist-packages (from ftfy->clip==1.0) (0.2.5)\nRequirement already satisfied: typing-extensions in /usr/local/lib/python3.7/dist-packages (from torch->clip==1.0) (3.10.0.2)\nRequirement already satisfied: pillow!=8.3.0,>=5.3.0 in /usr/local/lib/python3.7/dist-packages (from torchvision->clip==1.0) (7.1.2)\nRequirement already satisfied: numpy in /usr/local/lib/python3.7/dist-packages (from torchvision->clip==1.0) (1.21.5)\nBuilding wheels for collected packages: clip\n Building wheel for clip (setup.py) ... \u001b[?25l\u001b[?25hdone\n Created wheel for clip: filename=clip-1.0-py3-none-any.whl size=1369221 sha256=874d6c99f592bae9ca231a2e100f92cf2e5e09bdb1d54e46b3c2c5d0a90c066d\n Stored in directory: /tmp/pip-ephem-wheel-cache-i_djooze/wheels/fd/b9/c3/5b4470e35ed76e174bff77c92f91da82098d5e35fd5bc8cdac\nSuccessfully built clip\nInstalling collected packages: clip\nSuccessfully installed clip-1.0\nRequirement already satisfied: gdown in /usr/local/lib/python3.7/dist-packages (4.2.2)\nCollecting gdown\n Downloading gdown-4.4.0.tar.gz (14 kB)\n Installing build dependencies ... \u001b[?25l\u001b[?25hdone\n Getting requirements to build wheel ... \u001b[?25l\u001b[?25hdone\n Preparing wheel metadata ... \u001b[?25l\u001b[?25hdone\nRequirement already satisfied: filelock in /usr/local/lib/python3.7/dist-packages (from gdown) (3.6.0)\nRequirement already satisfied: requests[socks] in /usr/local/lib/python3.7/dist-packages (from gdown) (2.23.0)\nRequirement already satisfied: six in /usr/local/lib/python3.7/dist-packages (from gdown) (1.15.0)\nRequirement already satisfied: tqdm in /usr/local/lib/python3.7/dist-packages (from gdown) (4.63.0)\nRequirement already satisfied: beautifulsoup4 in /usr/local/lib/python3.7/dist-packages (from gdown) (4.6.3)\nRequirement already satisfied: urllib3!=1.25.0,!=1.25.1,<1.26,>=1.21.1 in /usr/local/lib/python3.7/dist-packages (from requests[socks]->gdown) (1.24.3)\nRequirement already satisfied: chardet<4,>=3.0.2 in /usr/local/lib/python3.7/dist-packages (from requests[socks]->gdown) (3.0.4)\nRequirement already satisfied: idna<3,>=2.5 in /usr/local/lib/python3.7/dist-packages (from requests[socks]->gdown) (2.10)\nRequirement already satisfied: certifi>=2017.4.17 in /usr/local/lib/python3.7/dist-packages (from requests[socks]->gdown) (2021.10.8)\nRequirement already satisfied: PySocks!=1.5.7,>=1.5.6 in /usr/local/lib/python3.7/dist-packages (from requests[socks]->gdown) (1.7.1)\nBuilding wheels for collected packages: gdown\n Building wheel for gdown (PEP 517) ... \u001b[?25l\u001b[?25hdone\n Created wheel for gdown: filename=gdown-4.4.0-py3-none-any.whl size=14774 sha256=285fefc33c01fab68afc821d709aa5beb61b7694fb278ac30ca71b2f457141fe\n Stored in directory: /tmp/pip-ephem-wheel-cache-9e1t22tz/wheels/fb/c3/0e/c4d8ff8bfcb0461afff199471449f642179b74968c15b7a69c\nSuccessfully built gdown\nInstalling collected packages: gdown\n Attempting uninstall: gdown\n Found existing installation: gdown 4.2.2\n Uninstalling gdown-4.2.2:\n Successfully uninstalled gdown-4.2.2\nSuccessfully installed gdown-4.4.0\nCollecting profanity-filter\n Downloading profanity_filter-1.3.3-py3-none-any.whl (45 kB)\n\u001b[K |████████████████████████████████| 45 kB 2.0 MB/s \n\u001b[?25hCollecting poetry-version<0.2.0,>=0.1.3\n Downloading poetry_version-0.1.5-py2.py3-none-any.whl (13 kB)\nCollecting pydantic<2.0,>=1.3\n Downloading pydantic-1.9.0-cp37-cp37m-manylinux_2_17_x86_64.manylinux2014_x86_64.whl (10.9 MB)\n\u001b[K |████████████████████████████████| 10.9 MB 8.4 MB/s \n\u001b[?25hRequirement already satisfied: spacy<3.0,>=2.0 in /usr/local/lib/python3.7/dist-packages (from profanity-filter) (2.2.4)\nCollecting ruamel.yaml<0.16.0,>=0.15.89\n Downloading ruamel.yaml-0.15.100-cp37-cp37m-manylinux1_x86_64.whl (654 kB)\n\u001b[K |████████████████████████████████| 654 kB 49.1 MB/s \n\u001b[?25hRequirement already satisfied: cached-property<2.0,>=1.5 in /usr/local/lib/python3.7/dist-packages (from profanity-filter) (1.5.2)\nCollecting redis<4.0,>=3.2\n Downloading redis-3.5.3-py2.py3-none-any.whl (72 kB)\n\u001b[K |████████████████████████████████| 72 kB 571 kB/s \n\u001b[?25hCollecting ordered-set<4.0,>=3.0\n Downloading ordered-set-3.1.1.tar.gz (10 kB)\nRequirement already satisfied: more-itertools<9.0,>=8.0 in /usr/local/lib/python3.7/dist-packages (from profanity-filter) (8.12.0)\nCollecting ordered-set-stubs<0.2.0,>=0.1.3\n Downloading ordered_set_stubs-0.1.3-py2.py3-none-any.whl (4.8 kB)\nCollecting tomlkit<0.6.0,>=0.4.6\n Downloading tomlkit-0.5.11-py2.py3-none-any.whl (31 kB)\nRequirement already satisfied: typing-extensions>=3.7.4.3 in /usr/local/lib/python3.7/dist-packages (from pydantic<2.0,>=1.3->profanity-filter) (3.10.0.2)\nRequirement already satisfied: cymem<2.1.0,>=2.0.2 in /usr/local/lib/python3.7/dist-packages (from spacy<3.0,>=2.0->profanity-filter) (2.0.6)\nRequirement already satisfied: blis<0.5.0,>=0.4.0 in /usr/local/lib/python3.7/dist-packages (from spacy<3.0,>=2.0->profanity-filter) (0.4.1)\nRequirement already satisfied: srsly<1.1.0,>=1.0.2 in /usr/local/lib/python3.7/dist-packages (from spacy<3.0,>=2.0->profanity-filter) (1.0.5)\nRequirement already satisfied: tqdm<5.0.0,>=4.38.0 in /usr/local/lib/python3.7/dist-packages (from spacy<3.0,>=2.0->profanity-filter) (4.63.0)\nRequirement already satisfied: preshed<3.1.0,>=3.0.2 in /usr/local/lib/python3.7/dist-packages (from spacy<3.0,>=2.0->profanity-filter) (3.0.6)\nRequirement already satisfied: thinc==7.4.0 in /usr/local/lib/python3.7/dist-packages (from spacy<3.0,>=2.0->profanity-filter) (7.4.0)\nRequirement already satisfied: murmurhash<1.1.0,>=0.28.0 in /usr/local/lib/python3.7/dist-packages (from spacy<3.0,>=2.0->profanity-filter) (1.0.6)\nRequirement already satisfied: numpy>=1.15.0 in /usr/local/lib/python3.7/dist-packages (from spacy<3.0,>=2.0->profanity-filter) (1.21.5)\nRequirement already satisfied: requests<3.0.0,>=2.13.0 in /usr/local/lib/python3.7/dist-packages (from spacy<3.0,>=2.0->profanity-filter) (2.23.0)\nRequirement already satisfied: catalogue<1.1.0,>=0.0.7 in /usr/local/lib/python3.7/dist-packages (from spacy<3.0,>=2.0->profanity-filter) (1.0.0)\nRequirement already satisfied: setuptools in /usr/local/lib/python3.7/dist-packages (from spacy<3.0,>=2.0->profanity-filter) (57.4.0)\nRequirement already satisfied: wasabi<1.1.0,>=0.4.0 in /usr/local/lib/python3.7/dist-packages (from spacy<3.0,>=2.0->profanity-filter) (0.9.0)\nRequirement already satisfied: plac<1.2.0,>=0.9.6 in /usr/local/lib/python3.7/dist-packages (from spacy<3.0,>=2.0->profanity-filter) (1.1.3)\nRequirement already satisfied: importlib-metadata>=0.20 in /usr/local/lib/python3.7/dist-packages (from catalogue<1.1.0,>=0.0.7->spacy<3.0,>=2.0->profanity-filter) (4.11.3)\nRequirement already satisfied: zipp>=0.5 in /usr/local/lib/python3.7/dist-packages (from importlib-metadata>=0.20->catalogue<1.1.0,>=0.0.7->spacy<3.0,>=2.0->profanity-filter) (3.7.0)\nRequirement already satisfied: certifi>=2017.4.17 in /usr/local/lib/python3.7/dist-packages (from requests<3.0.0,>=2.13.0->spacy<3.0,>=2.0->profanity-filter) (2021.10.8)\nRequirement already satisfied: urllib3!=1.25.0,!=1.25.1,<1.26,>=1.21.1 in /usr/local/lib/python3.7/dist-packages (from requests<3.0.0,>=2.13.0->spacy<3.0,>=2.0->profanity-filter) (1.24.3)\nRequirement already satisfied: chardet<4,>=3.0.2 in /usr/local/lib/python3.7/dist-packages (from requests<3.0.0,>=2.13.0->spacy<3.0,>=2.0->profanity-filter) (3.0.4)\nRequirement already satisfied: idna<3,>=2.5 in /usr/local/lib/python3.7/dist-packages (from requests<3.0.0,>=2.13.0->spacy<3.0,>=2.0->profanity-filter) (2.10)\nBuilding wheels for collected packages: ordered-set\n Building wheel for ordered-set (setup.py) ... \u001b[?25l\u001b[?25hdone\n Created wheel for ordered-set: filename=ordered_set-3.1.1-py2.py3-none-any.whl size=7822 sha256=ac5ba57c4f7434187f6893782b8c75654d7945f11ecd7603739b21ddbaee9ab3\n Stored in directory: /root/.cache/pip/wheels/10/91/db/f8476865ccd2187467d2926d9f965673b4886175b6babca6b9\nSuccessfully built ordered-set\nInstalling collected packages: tomlkit, ruamel.yaml, redis, pydantic, poetry-version, ordered-set-stubs, ordered-set, profanity-filter\nSuccessfully installed ordered-set-3.1.1 ordered-set-stubs-0.1.3 poetry-version-0.1.5 profanity-filter-1.3.3 pydantic-1.9.0 redis-3.5.3 ruamel.yaml-0.15.100 tomlkit-0.5.11\nFri Apr 1 21:40:51 2022 \n+-----------------------------------------------------------------------------+\n| NVIDIA-SMI 460.32.03 Driver Version: 460.32.03 CUDA Version: 11.2 |\n|-------------------------------+----------------------+----------------------+\n| GPU Name Persistence-M| Bus-Id Disp.A | Volatile Uncorr. ECC |\n| Fan Temp Perf Pwr:Usage/Cap| Memory-Usage | GPU-Util Compute M. |\n| | | MIG M. |\n|===============================+======================+======================|\n| 0 Tesla K80 Off | 00000000:00:04.0 Off | 0 |\n| N/A 73C P8 32W / 149W | 0MiB / 11441MiB | 0% Default |\n| | | N/A |\n+-------------------------------+----------------------+----------------------+\n \n+-----------------------------------------------------------------------------+\n| Processes: |\n| GPU GI CI PID Type Process name GPU Memory |\n| ID ID Usage |\n|=============================================================================|\n| No running processes found |\n+-----------------------------------------------------------------------------+\n"
],
[
"import datetime\nimport json\nimport os\nimport re\nimport time\n\nimport requests\nimport clip\nimport cv2\nimport matplotlib.pyplot as plt\nimport numpy as np\nimport openai\nfrom PIL import Image\nfrom profanity_filter import ProfanityFilter\nimport torch\n\nopenai.api_key = openai_api_key",
"_____no_output_____"
]
],
[
[
"## **Foundation Models**\nSelect which foundation models to use.\n\n**Defaults:** [CLIP](https://arxiv.org/abs/2103.00020) VIT-L/14 as the VLM, and [GPT-3](https://arxiv.org/abs/2005.14165) \"Davinci\" as the LM.",
"_____no_output_____"
]
],
[
[
"clip_version = \"ViT-L/14\" #@param [\"RN50\", \"RN101\", \"RN50x4\", \"RN50x16\", \"RN50x64\", \"ViT-B/32\", \"ViT-B/16\", \"ViT-L/14\"] {type:\"string\"}\ngpt_version = \"text-davinci-002\" #@param [\"text-davinci-001\", \"text-davinci-002\", \"text-curie-001\", \"text-babbage-001\", \"text-ada-001\"] {type:\"string\"}\n\nclip_feat_dim = {'RN50': 1024, 'RN101': 512, 'RN50x4': 640, 'RN50x16': 768, 'RN50x64': 1024, 'ViT-B/32': 512, 'ViT-B/16': 512, 'ViT-L/14': 768}[clip_version]",
"_____no_output_____"
]
],
[
[
"## **Getting Started**\nDownload CLIP model weights, and define helper functions. This might take a few minutes.",
"_____no_output_____"
],
[
"##### Download [CLIP](https://arxiv.org/abs/2103.00020) model weights.",
"_____no_output_____"
]
],
[
[
"# torch.cuda.set_per_process_memory_fraction(0.9, None) # Only needed if session crashes.\nmodel, preprocess = clip.load(clip_version) # clip.available_models()\nmodel.cuda().eval()\n\ndef num_params(model):\n return np.sum([int(np.prod(p.shape)) for p in model.parameters()])\nprint(\"Model parameters (total):\", num_params(model))\nprint(\"Model parameters (image encoder):\", num_params(model.visual))\nprint(\"Model parameters (text encoder):\", num_params(model.token_embedding) + num_params(model.transformer))\nprint(\"Input image resolution:\", model.visual.input_resolution)\nprint(\"Context length:\", model.context_length)\nprint(\"Vocab size:\", model.vocab_size)\nimg_size = model.visual.input_resolution",
"100%|███████████████████████████████████████| 890M/890M [00:14<00:00, 65.3MiB/s]\n"
]
],
[
[
"##### Define CLIP helper functions (e.g., nearest neighbor search).",
"_____no_output_____"
]
],
[
[
"def get_text_feats(in_text, batch_size=64):\n text_tokens = clip.tokenize(in_text).cuda()\n text_id = 0\n text_feats = np.zeros((len(in_text), clip_feat_dim), dtype=np.float32)\n while text_id < len(text_tokens): # Batched inference.\n batch_size = min(len(in_text) - text_id, batch_size)\n text_batch = text_tokens[text_id:text_id+batch_size]\n with torch.no_grad():\n batch_feats = model.encode_text(text_batch).float()\n batch_feats /= batch_feats.norm(dim=-1, keepdim=True)\n batch_feats = np.float32(batch_feats.cpu())\n text_feats[text_id:text_id+batch_size, :] = batch_feats\n text_id += batch_size\n return text_feats\n\ndef get_img_feats(img):\n img_pil = Image.fromarray(np.uint8(img))\n img_in = preprocess(img_pil)[None, ...]\n with torch.no_grad():\n img_feats = model.encode_image(img_in.cuda()).float()\n img_feats /= img_feats.norm(dim=-1, keepdim=True)\n img_feats = np.float32(img_feats.cpu())\n return img_feats\n\ndef get_nn_text(raw_texts, text_feats, img_feats):\n scores = text_feats @ img_feats.T\n scores = scores.squeeze()\n high_to_low_ids = np.argsort(scores).squeeze()[::-1]\n high_to_low_texts = [raw_texts[i] for i in high_to_low_ids]\n high_to_low_scores = np.sort(scores).squeeze()[::-1]\n return high_to_low_texts, high_to_low_scores",
"_____no_output_____"
]
],
[
[
"##### Define [GPT-3](https://arxiv.org/abs/2005.14165) helper functions.",
"_____no_output_____"
]
],
[
[
"def prompt_llm(prompt, max_tokens=64, temperature=0, stop=None):\n response = openai.Completion.create(engine=gpt_version, prompt=prompt, max_tokens=max_tokens, temperature=temperature, stop=stop)\n return response[\"choices\"][0][\"text\"].strip()",
"_____no_output_____"
]
],
[
[
"##### Load scene categories from [Places365](http://places2.csail.mit.edu/download.html) and compute their CLIP features.",
"_____no_output_____"
]
],
[
[
"# Load scene categories from Places365.\nif not os.path.exists('categories_places365.txt'):\n ! wget https://raw.githubusercontent.com/zhoubolei/places_devkit/master/categories_places365.txt\nplace_categories = np.loadtxt('categories_places365.txt', dtype=str)\nplace_texts = []\nfor place in place_categories[:, 0]:\n place = place.split('/')[2:]\n if len(place) > 1:\n place = place[1] + ' ' + place[0]\n else:\n place = place[0]\n place = place.replace('_', ' ')\n place_texts.append(place)\nplace_feats = get_text_feats([f'Photo of a {p}.' for p in place_texts])",
"--2022-04-01 21:42:39-- https://raw.githubusercontent.com/zhoubolei/places_devkit/master/categories_places365.txt\nResolving raw.githubusercontent.com (raw.githubusercontent.com)... 185.199.108.133, 185.199.109.133, 185.199.110.133, ...\nConnecting to raw.githubusercontent.com (raw.githubusercontent.com)|185.199.108.133|:443... connected.\nHTTP request sent, awaiting response... 200 OK\nLength: 6833 (6.7K) [text/plain]\nSaving to: ‘categories_places365.txt’\n\ncategories_places36 100%[===================>] 6.67K --.-KB/s in 0s \n\n2022-04-01 21:42:39 (43.9 MB/s) - ‘categories_places365.txt’ saved [6833/6833]\n\n"
]
],
[
[
"##### Load object categories from [Tencent ML Images](https://arxiv.org/pdf/1901.01703.pdf) and compute their CLIP features. This might take a few minutes.",
"_____no_output_____"
]
],
[
[
"# Load object categories from Tencent ML Images.\nif not os.path.exists('dictionary_and_semantic_hierarchy.txt'):\n ! wget https://raw.githubusercontent.com/Tencent/tencent-ml-images/master/data/dictionary_and_semantic_hierarchy.txt\nwith open('dictionary_and_semantic_hierarchy.txt') as fid:\n object_categories = fid.readlines()\nobject_texts = []\npf = ProfanityFilter()\nfor object_text in object_categories[1:]:\n object_text = object_text.strip()\n object_text = object_text.split('\\t')[3]\n safe_list = ''\n for variant in object_text.split(','):\n text = variant.strip()\n if pf.is_clean(text):\n safe_list += f'{text}, '\n safe_list = safe_list[:-2]\n if len(safe_list) > 0:\n object_texts.append(safe_list)\nobject_texts = [o for o in list(set(object_texts)) if o not in place_texts] # Remove redundant categories.\nobject_feats = get_text_feats([f'Photo of a {o}.' for o in object_texts])",
"--2022-04-01 21:42:47-- https://raw.githubusercontent.com/Tencent/tencent-ml-images/master/data/dictionary_and_semantic_hierarchy.txt\nResolving raw.githubusercontent.com (raw.githubusercontent.com)... 185.199.108.133, 185.199.109.133, 185.199.110.133, ...\nConnecting to raw.githubusercontent.com (raw.githubusercontent.com)|185.199.108.133|:443... connected.\nHTTP request sent, awaiting response... 200 OK\nLength: 488167 (477K) [text/plain]\nSaving to: ‘dictionary_and_semantic_hierarchy.txt’\n\ndictionary_and_sema 100%[===================>] 476.73K --.-KB/s in 0.04s \n\n2022-04-01 21:42:48 (10.9 MB/s) - ‘dictionary_and_semantic_hierarchy.txt’ saved [488167/488167]\n\n"
]
],
[
[
"## **Demo:** Image Captioning\nRun image captioning on an Internet image (linked via URL).\n\n**Note:** due to the non-zero temperature used for sampling from the generative language model, results from this approach are stochastic, but comparable results are producible.\n\n",
"_____no_output_____"
]
],
[
[
"# Download image.\nimg_url = \"https://github.com/rmokady/CLIP_prefix_caption/raw/main/Images/COCO_val2014_000000165547.jpg\" #@param {type:\"string\"}\nfname = 'demo_img.png'\nwith open(fname, 'wb') as f:\n f.write(requests.get(img_url).content)\n\nverbose = True #@param {type:\"boolean\"}\n\n# Load image.\nimg = cv2.cvtColor(cv2.imread(fname), cv2.COLOR_BGR2RGB)\nimg_feats = get_img_feats(img)\nplt.imshow(img); plt.show()\n\n# Zero-shot VLM: classify image type.\nimg_types = ['photo', 'cartoon', 'sketch', 'painting']\nimg_types_feats = get_text_feats([f'This is a {t}.' for t in img_types])\nsorted_img_types, img_type_scores = get_nn_text(img_types, img_types_feats, img_feats)\nimg_type = sorted_img_types[0]\n\n# Zero-shot VLM: classify number of people.\nppl_texts = ['no people', 'people']\nppl_feats = get_text_feats([f'There are {p} in this photo.' for p in ppl_texts])\nsorted_ppl_texts, ppl_scores = get_nn_text(ppl_texts, ppl_feats, img_feats)\nppl_result = sorted_ppl_texts[0]\nif ppl_result == 'people':\n ppl_texts = ['is one person', 'are two people', 'are three people', 'are several people', 'are many people']\n ppl_feats = get_text_feats([f'There {p} in this photo.' for p in ppl_texts])\n sorted_ppl_texts, ppl_scores = get_nn_text(ppl_texts, ppl_feats, img_feats)\n ppl_result = sorted_ppl_texts[0]\nelse:\n ppl_result = f'are {ppl_result}'\n\n# Zero-shot VLM: classify places.\nplace_topk = 3\nplace_feats = get_text_feats([f'Photo of a {p}.' for p in place_texts ])\nsorted_places, places_scores = get_nn_text(place_texts, place_feats, img_feats)\n\n# Zero-shot VLM: classify objects.\nobj_topk = 10\nsorted_obj_texts, obj_scores = get_nn_text(object_texts, object_feats, img_feats)\nobject_list = ''\nfor i in range(obj_topk):\n object_list += f'{sorted_obj_texts[i]}, '\nobject_list = object_list[:-2]\n\n# Zero-shot LM: generate captions.\nnum_captions = 10\nprompt = f'''I am an intelligent image captioning bot.\nThis image is a {img_type}. There {ppl_result}.\nI think this photo was taken at a {sorted_places[0]}, {sorted_places[1]}, or {sorted_places[2]}.\nI think there might be a {object_list} in this {img_type}.\nA creative short caption I can generate to describe this image is:'''\ncaption_texts = [prompt_llm(prompt, temperature=0.9) for _ in range(num_captions)]\n\n# Zero-shot VLM: rank captions.\ncaption_feats = get_text_feats(caption_texts)\nsorted_captions, caption_scores = get_nn_text(caption_texts, caption_feats, img_feats)\nprint(f'{sorted_captions[0]}\\n')\n\nif verbose:\n print(f'VLM: This image is a:')\n for img_type, score in zip(sorted_img_types, img_type_scores):\n print(f'{score:.4f} {img_type}')\n\n print(f'\\nVLM: There:')\n for ppl_text, score in zip(sorted_ppl_texts, ppl_scores):\n print(f'{score:.4f} {ppl_text}')\n\n print(f'\\nVLM: I think this photo was taken at a:')\n for place, score in zip(sorted_places[:place_topk], places_scores[:place_topk]):\n print(f'{score:.4f} {place}')\n\n print(f'\\nVLM: I think there might be a:')\n for obj_text, score in zip(sorted_obj_texts[:obj_topk], obj_scores[:obj_topk]):\n print(f'{score:.4f} {obj_text}')\n\n print(f'\\nLM generated captions ranked by VLM scores:')\n for caption, score in zip(sorted_captions, caption_scores):\n print(f'{score:.4f} {caption}')",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb8d2ab9a4af6e5dfdf5e35f1e9bdd2d5d12e6f2
| 57,596 |
ipynb
|
Jupyter Notebook
|
CB multi regression.ipynb
|
Jdijedi/CB
|
90c838730495ac45a32800bceee947f67934036b
|
[
"MIT"
] | null | null | null |
CB multi regression.ipynb
|
Jdijedi/CB
|
90c838730495ac45a32800bceee947f67934036b
|
[
"MIT"
] | null | null | null |
CB multi regression.ipynb
|
Jdijedi/CB
|
90c838730495ac45a32800bceee947f67934036b
|
[
"MIT"
] | null | null | null | 29.581921 | 1,377 | 0.399941 |
[
[
[
"import pandas as pd",
"_____no_output_____"
],
[
"import numpy as np\nfrom sklearn import linear_model",
"_____no_output_____"
],
[
"df = pd.read_csv('C:\\Codebasics\\exercise.csv')\n",
"_____no_output_____"
],
[
"df",
"_____no_output_____"
],
[
"import math",
"_____no_output_____"
],
[
"from word2number import w2n",
"_____no_output_____"
],
[
"import word2number",
"_____no_output_____"
],
[
"from word2number import w2n",
"_____no_output_____"
],
[
"print(w2n.word_to_num(df.experience.values)",
"_____no_output_____"
],
[
"df",
"_____no_output_____"
],
[
"df=df.rename(columns = {'test_score(out of 10)':'test_s'})",
"_____no_output_____"
],
[
"df=df.rename(columns = {'interview_score(out of 10)':'interview_s'})",
"_____no_output_____"
],
[
"df=df.rename(columns = {'salary($)':'salary'})",
"_____no_output_____"
],
[
"df",
"_____no_output_____"
],
[
"median_test = math.floor(df.test_s.median())",
"_____no_output_____"
]
],
[
[
"print(median_test)",
"_____no_output_____"
]
],
[
[
"print(median_test)",
"8\n"
],
[
"df.test_s = df.test_s.fillna(median_test)",
"_____no_output_____"
],
[
"df",
"_____no_output_____"
],
[
"df",
"_____no_output_____"
],
[
"df.experience = df.experience.fillna(0)",
"_____no_output_____"
],
[
"df",
"_____no_output_____"
],
[
"df",
"_____no_output_____"
],
[
"for index, row in df.iterrows():\n print (index)",
"0\n1\n2\n3\n4\n5\n6\n7\n"
],
[
"df['experience'] ",
"_____no_output_____"
],
[
"df.experience",
"_____no_output_____"
],
[
"df.experience = w2n.word_to_num(df.experience)",
"_____no_output_____"
]
],
[
[
"df",
"_____no_output_____"
]
],
[
[
"df",
"_____no_output_____"
],
[
"for index, row in df.iterrows():\n row[index] = w2n.word_to_num(row['experience'])",
"_____no_output_____"
],
[
"w2n.word_to_num(df.experience[1])",
"_____no_output_____"
],
[
"for index, row in df.iterrows():\n df.experience[index] = w2n.word_to_num(df.experience[index])",
"_____no_output_____"
],
[
"for x in range(2, 8):\n df.experience[x] = w2n.word_to_num(df.experience[x])",
"C:\\Users\\sharma3n\\AppData\\Local\\Continuum\\Anaconda3\\lib\\site-packages\\ipykernel_launcher.py:2: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n \n"
],
[
"df",
"_____no_output_____"
],
[
"reg = linear_model.LinearRegression()",
"_____no_output_____"
],
[
"reg.fit(df[['experience','test_s','interview_s']],df.salary)",
"_____no_output_____"
],
[
"reg.coef_",
"_____no_output_____"
],
[
"reg.intercept_",
"_____no_output_____"
],
[
"reg.predict([[2, 9, 6]])",
"_____no_output_____"
],
[
"reg.predict([[12, 10, 10]])",
"_____no_output_____"
],
[
"df.test_s[6] = 7\ndf",
"C:\\Users\\sharma3n\\AppData\\Local\\Continuum\\Anaconda3\\lib\\site-packages\\ipykernel_launcher.py:1: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n \"\"\"Entry point for launching an IPython kernel.\n"
],
[
"df.salary",
"_____no_output_____"
],
[
"df.salary.iloc[x:x+2]",
"_____no_output_____"
],
[
"x=2",
"_____no_output_____"
],
[
"x",
"_____no_output_____"
],
[
"df[['experience','test_s','interview_s']].iloc[x:x+4]",
"_____no_output_____"
],
[
"reg.fit(df[['experience','test_s','interview_s']].iloc[x:x+5],df.salary.iloc[x:x+5])",
"_____no_output_____"
],
[
"reg.coef_, reg.intercept_",
"_____no_output_____"
],
[
"reg.get_params",
"_____no_output_____"
],
[
"df",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb8d3695f449ea5b64b99fefb91075f1055b4e5e
| 55,967 |
ipynb
|
Jupyter Notebook
|
C5_Sequence Models/Week 3/Trigger Word Detection/Trigger word detection - v1.ipynb
|
aurimas13/Deep-Learning-Specialization-solutions---notes
|
f2ca6b19b7a568de1e0e12c6341c0605e007e186
|
[
"MIT"
] | null | null | null |
C5_Sequence Models/Week 3/Trigger Word Detection/Trigger word detection - v1.ipynb
|
aurimas13/Deep-Learning-Specialization-solutions---notes
|
f2ca6b19b7a568de1e0e12c6341c0605e007e186
|
[
"MIT"
] | null | null | null |
C5_Sequence Models/Week 3/Trigger Word Detection/Trigger word detection - v1.ipynb
|
aurimas13/Deep-Learning-Specialization-solutions---notes
|
f2ca6b19b7a568de1e0e12c6341c0605e007e186
|
[
"MIT"
] | null | null | null | 42.080451 | 882 | 0.609984 |
[
[
[
"## Trigger Word Detection\n\nWelcome to the final programming assignment of this specialization! \n\nIn this week's videos, you learned about applying deep learning to speech recognition. In this assignment, you will construct a speech dataset and implement an algorithm for trigger word detection (sometimes also called keyword detection, or wakeword detection). Trigger word detection is the technology that allows devices like Amazon Alexa, Google Home, Apple Siri, and Baidu DuerOS to wake up upon hearing a certain word. \n\nFor this exercise, our trigger word will be \"Activate.\" Every time it hears you say \"activate,\" it will make a \"chiming\" sound. By the end of this assignment, you will be able to record a clip of yourself talking, and have the algorithm trigger a chime when it detects you saying \"activate.\" \n\nAfter completing this assignment, perhaps you can also extend it to run on your laptop so that every time you say \"activate\" it starts up your favorite app, or turns on a network connected lamp in your house, or triggers some other event? \n\n<img src=\"images/sound.png\" style=\"width:1000px;height:150px;\">\n\nIn this assignment you will learn to: \n- Structure a speech recognition project\n- Synthesize and process audio recordings to create train/dev datasets\n- Train a trigger word detection model and make predictions\n\nLets get started! Run the following cell to load the package you are going to use. \n",
"_____no_output_____"
]
],
[
[
"import numpy as np\nfrom pydub import AudioSegment\nimport random\nimport sys\nimport io\nimport os\nimport glob\nimport IPython\nfrom td_utils import *\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"# 1 - Data synthesis: Creating a speech dataset \n\nLet's start by building a dataset for your trigger word detection algorithm. A speech dataset should ideally be as close as possible to the application you will want to run it on. In this case, you'd like to detect the word \"activate\" in working environments (library, home, offices, open-spaces ...). You thus need to create recordings with a mix of positive words (\"activate\") and negative words (random words other than activate) on different background sounds. Let's see how you can create such a dataset. \n\n## 1.1 - Listening to the data \n\nOne of your friends is helping you out on this project, and they've gone to libraries, cafes, restaurants, homes and offices all around the region to record background noises, as well as snippets of audio of people saying positive/negative words. This dataset includes people speaking in a variety of accents. \n\nIn the raw_data directory, you can find a subset of the raw audio files of the positive words, negative words, and background noise. You will use these audio files to synthesize a dataset to train the model. The \"activate\" directory contains positive examples of people saying the word \"activate\". The \"negatives\" directory contains negative examples of people saying random words other than \"activate\". There is one word per audio recording. The \"backgrounds\" directory contains 10 second clips of background noise in different environments.\n\nRun the cells below to listen to some examples.",
"_____no_output_____"
]
],
[
[
"IPython.display.Audio(\"./raw_data/activates/1.wav\")",
"_____no_output_____"
],
[
"IPython.display.Audio(\"./raw_data/negatives/4.wav\")",
"_____no_output_____"
],
[
"IPython.display.Audio(\"./raw_data/backgrounds/1.wav\")",
"_____no_output_____"
]
],
[
[
"You will use these three type of recordings (positives/negatives/backgrounds) to create a labelled dataset.",
"_____no_output_____"
],
[
"## 1.2 - From audio recordings to spectrograms\n\nWhat really is an audio recording? A microphone records little variations in air pressure over time, and it is these little variations in air pressure that your ear also perceives as sound. You can think of an audio recording is a long list of numbers measuring the little air pressure changes detected by the microphone. We will use audio sampled at 44100 Hz (or 44100 Hertz). This means the microphone gives us 44100 numbers per second. Thus, a 10 second audio clip is represented by 441000 numbers (= $10 \\times 44100$). \n\nIt is quite difficult to figure out from this \"raw\" representation of audio whether the word \"activate\" was said. In order to help your sequence model more easily learn to detect triggerwords, we will compute a *spectrogram* of the audio. The spectrogram tells us how much different frequencies are present in an audio clip at a moment in time. \n\n(If you've ever taken an advanced class on signal processing or on Fourier transforms, a spectrogram is computed by sliding a window over the raw audio signal, and calculates the most active frequencies in each window using a Fourier transform. If you don't understand the previous sentence, don't worry about it.) \n\nLets see an example. ",
"_____no_output_____"
]
],
[
[
"IPython.display.Audio(\"audio_examples/example_train.wav\")",
"_____no_output_____"
],
[
"x = graph_spectrogram(\"audio_examples/example_train.wav\")",
"_____no_output_____"
]
],
[
[
"The graph above represents how active each frequency is (y axis) over a number of time-steps (x axis). \n\n<img src=\"images/spectrogram.png\" style=\"width:500px;height:200px;\">\n<center> **Figure 1**: Spectrogram of an audio recording, where the color shows the degree to which different frequencies are present (loud) in the audio at different points in time. Green squares means a certain frequency is more active or more present in the audio clip (louder); blue squares denote less active frequencies. </center>\n\nThe dimension of the output spectrogram depends upon the hyperparameters of the spectrogram software and the length of the input. In this notebook, we will be working with 10 second audio clips as the \"standard length\" for our training examples. The number of timesteps of the spectrogram will be 5511. You'll see later that the spectrogram will be the input $x$ into the network, and so $T_x = 5511$.\n",
"_____no_output_____"
]
],
[
[
"_, data = wavfile.read(\"audio_examples/example_train.wav\")\nprint(\"Time steps in audio recording before spectrogram\", data[:,0].shape)\nprint(\"Time steps in input after spectrogram\", x.shape)",
"_____no_output_____"
]
],
[
[
"Now, you can define:",
"_____no_output_____"
]
],
[
[
"Tx = 5511 # The number of time steps input to the model from the spectrogram\nn_freq = 101 # Number of frequencies input to the model at each time step of the spectrogram",
"_____no_output_____"
]
],
[
[
"Note that even with 10 seconds being our default training example length, 10 seconds of time can be discretized to different numbers of value. You've seen 441000 (raw audio) and 5511 (spectrogram). In the former case, each step represents $10/441000 \\approx 0.000023$ seconds. In the second case, each step represents $10/5511 \\approx 0.0018$ seconds. \n\nFor the 10sec of audio, the key values you will see in this assignment are:\n\n- $441000$ (raw audio)\n- $5511 = T_x$ (spectrogram output, and dimension of input to the neural network). \n- $10000$ (used by the `pydub` module to synthesize audio) \n- $1375 = T_y$ (the number of steps in the output of the GRU you'll build). \n\nNote that each of these representations correspond to exactly 10 seconds of time. It's just that they are discretizing them to different degrees. All of these are hyperparameters and can be changed (except the 441000, which is a function of the microphone). We have chosen values that are within the standard ranges uses for speech systems. \n\nConsider the $T_y = 1375$ number above. This means that for the output of the model, we discretize the 10s into 1375 time-intervals (each one of length $10/1375 \\approx 0.0072$s) and try to predict for each of these intervals whether someone recently finished saying \"activate.\" \n\nConsider also the 10000 number above. This corresponds to discretizing the 10sec clip into 10/10000 = 0.001 second itervals. 0.001 seconds is also called 1 millisecond, or 1ms. So when we say we are discretizing according to 1ms intervals, it means we are using 10,000 steps. \n",
"_____no_output_____"
]
],
[
[
"Ty = 1375 # The number of time steps in the output of our model",
"_____no_output_____"
]
],
[
[
"## 1.3 - Generating a single training example\n\nBecause speech data is hard to acquire and label, you will synthesize your training data using the audio clips of activates, negatives, and backgrounds. It is quite slow to record lots of 10 second audio clips with random \"activates\" in it. Instead, it is easier to record lots of positives and negative words, and record background noise separately (or download background noise from free online sources). \n\nTo synthesize a single training example, you will:\n\n- Pick a random 10 second background audio clip\n- Randomly insert 0-4 audio clips of \"activate\" into this 10sec clip\n- Randomly insert 0-2 audio clips of negative words into this 10sec clip\n\nBecause you had synthesized the word \"activate\" into the background clip, you know exactly when in the 10sec clip the \"activate\" makes its appearance. You'll see later that this makes it easier to generate the labels $y^{\\langle t \\rangle}$ as well. \n\nYou will use the pydub package to manipulate audio. Pydub converts raw audio files into lists of Pydub data structures (it is not important to know the details here). Pydub uses 1ms as the discretization interval (1ms is 1 millisecond = 1/1000 seconds) which is why a 10sec clip is always represented using 10,000 steps. ",
"_____no_output_____"
]
],
[
[
"# Load audio segments using pydub \nactivates, negatives, backgrounds = load_raw_audio()\n\nprint(\"background len: \" + str(len(backgrounds[0]))) # Should be 10,000, since it is a 10 sec clip\nprint(\"activate[0] len: \" + str(len(activates[0]))) # Maybe around 1000, since an \"activate\" audio clip is usually around 1 sec (but varies a lot)\nprint(\"activate[1] len: \" + str(len(activates[1]))) # Different \"activate\" clips can have different lengths ",
"_____no_output_____"
]
],
[
[
"**Overlaying positive/negative words on the background**:\n\nGiven a 10sec background clip and a short audio clip (positive or negative word), you need to be able to \"add\" or \"insert\" the word's short audio clip onto the background. To ensure audio segments inserted onto the background do not overlap, you will keep track of the times of previously inserted audio clips. You will be inserting multiple clips of positive/negative words onto the background, and you don't want to insert an \"activate\" or a random word somewhere that overlaps with another clip you had previously added. \n\nFor clarity, when you insert a 1sec \"activate\" onto a 10sec clip of cafe noise, you end up with a 10sec clip that sounds like someone saying \"activate\" in a cafe, with \"activate\" superimposed on the background cafe noise. You do *not* end up with an 11 sec clip. You'll see later how pydub allows you to do this. \n\n**Creating the labels at the same time you overlay**:\n\nRecall also that the labels $y^{\\langle t \\rangle}$ represent whether or not someone has just finished saying \"activate.\" Given a background clip, we can initialize $y^{\\langle t \\rangle}=0$ for all $t$, since the clip doesn't contain any \"activates.\" \n\nWhen you insert or overlay an \"activate\" clip, you will also update labels for $y^{\\langle t \\rangle}$, so that 50 steps of the output now have target label 1. You will train a GRU to detect when someone has *finished* saying \"activate\". For example, suppose the synthesized \"activate\" clip ends at the 5sec mark in the 10sec audio---exactly halfway into the clip. Recall that $T_y = 1375$, so timestep $687 = $ `int(1375*0.5)` corresponds to the moment at 5sec into the audio. So, you will set $y^{\\langle 688 \\rangle} = 1$. Further, you would quite satisfied if the GRU detects \"activate\" anywhere within a short time-internal after this moment, so we actually set 50 consecutive values of the label $y^{\\langle t \\rangle}$ to 1. Specifically, we have $y^{\\langle 688 \\rangle} = y^{\\langle 689 \\rangle} = \\cdots = y^{\\langle 737 \\rangle} = 1$. \n\nThis is another reason for synthesizing the training data: It's relatively straightforward to generate these labels $y^{\\langle t \\rangle}$ as described above. In contrast, if you have 10sec of audio recorded on a microphone, it's quite time consuming for a person to listen to it and mark manually exactly when \"activate\" finished. \n\nHere's a figure illustrating the labels $y^{\\langle t \\rangle}$, for a clip which we have inserted \"activate\", \"innocent\", activate\", \"baby.\" Note that the positive labels \"1\" are associated only with the positive words. \n\n<img src=\"images/label_diagram.png\" style=\"width:500px;height:200px;\">\n<center> **Figure 2** </center>\n\nTo implement the training set synthesis process, you will use the following helper functions. All of these function will use a 1ms discretization interval, so the 10sec of audio is always discretized into 10,000 steps. \n\n1. `get_random_time_segment(segment_ms)` gets a random time segment in our background audio\n2. `is_overlapping(segment_time, existing_segments)` checks if a time segment overlaps with existing segments\n3. `insert_audio_clip(background, audio_clip, existing_times)` inserts an audio segment at a random time in our background audio using `get_random_time_segment` and `is_overlapping`\n4. `insert_ones(y, segment_end_ms)` inserts 1's into our label vector y after the word \"activate\"",
"_____no_output_____"
],
[
"The function `get_random_time_segment(segment_ms)` returns a random time segment onto which we can insert an audio clip of duration `segment_ms`. Read through the code to make sure you understand what it is doing. \n",
"_____no_output_____"
]
],
[
[
"def get_random_time_segment(segment_ms):\n \"\"\"\n Gets a random time segment of duration segment_ms in a 10,000 ms audio clip.\n \n Arguments:\n segment_ms -- the duration of the audio clip in ms (\"ms\" stands for \"milliseconds\")\n \n Returns:\n segment_time -- a tuple of (segment_start, segment_end) in ms\n \"\"\"\n \n segment_start = np.random.randint(low=0, high=10000-segment_ms) # Make sure segment doesn't run past the 10sec background \n segment_end = segment_start + segment_ms - 1\n \n return (segment_start, segment_end)",
"_____no_output_____"
]
],
[
[
"Next, suppose you have inserted audio clips at segments (1000,1800) and (3400,4500). I.e., the first segment starts at step 1000, and ends at step 1800. Now, if we are considering inserting a new audio clip at (3000,3600) does this overlap with one of the previously inserted segments? In this case, (3000,3600) and (3400,4500) overlap, so we should decide against inserting a clip here. \n\nFor the purpose of this function, define (100,200) and (200,250) to be overlapping, since they overlap at timestep 200. However, (100,199) and (200,250) are non-overlapping. \n\n**Exercise**: Implement `is_overlapping(segment_time, existing_segments)` to check if a new time segment overlaps with any of the previous segments. You will need to carry out 2 steps:\n\n1. Create a \"False\" flag, that you will later set to \"True\" if you find that there is an overlap.\n2. Loop over the previous_segments' start and end times. Compare these times to the segment's start and end times. If there is an overlap, set the flag defined in (1) as True. You can use:\n```python\nfor ....:\n if ... <= ... and ... >= ...:\n ...\n```\nHint: There is overlap if the segment starts before the previous segment ends, and the segment ends after the previous segment starts.",
"_____no_output_____"
]
],
[
[
"# GRADED FUNCTION: is_overlapping\n\ndef is_overlapping(segment_time, previous_segments):\n \"\"\"\n Checks if the time of a segment overlaps with the times of existing segments.\n \n Arguments:\n segment_time -- a tuple of (segment_start, segment_end) for the new segment\n previous_segments -- a list of tuples of (segment_start, segment_end) for the existing segments\n \n Returns:\n True if the time segment overlaps with any of the existing segments, False otherwise\n \"\"\"\n \n segment_start, segment_end = segment_time\n \n ### START CODE HERE ### (≈ 4 line)\n # Step 1: Initialize overlap as a \"False\" flag. (≈ 1 line)\n overlap = False\n \n # Step 2: loop over the previous_segments start and end times.\n # Compare start/end times and set the flag to True if there is an overlap (≈ 3 lines)\n for previous_start, previous_end in previous_segments:\n if segment_start <= previous_end and segment_end >= previous_start:\n overlap = True\n ### END CODE HERE ###\n\n return overlap",
"_____no_output_____"
],
[
"overlap1 = is_overlapping((950, 1430), [(2000, 2550), (260, 949)])\noverlap2 = is_overlapping((2305, 2950), [(824, 1532), (1900, 2305), (3424, 3656)])\nprint(\"Overlap 1 = \", overlap1)\nprint(\"Overlap 2 = \", overlap2)",
"_____no_output_____"
]
],
[
[
"**Expected Output**:\n\n<table>\n <tr>\n <td>\n **Overlap 1**\n </td>\n <td>\n False\n </td>\n </tr>\n <tr>\n <td>\n **Overlap 2**\n </td>\n <td>\n True\n </td>\n </tr>\n</table>",
"_____no_output_____"
],
[
"Now, lets use the previous helper functions to insert a new audio clip onto the 10sec background at a random time, but making sure that any newly inserted segment doesn't overlap with the previous segments. \n\n**Exercise**: Implement `insert_audio_clip()` to overlay an audio clip onto the background 10sec clip. You will need to carry out 4 steps:\n\n1. Get a random time segment of the right duration in ms.\n2. Make sure that the time segment does not overlap with any of the previous time segments. If it is overlapping, then go back to step 1 and pick a new time segment.\n3. Add the new time segment to the list of existing time segments, so as to keep track of all the segments you've inserted. \n4. Overlay the audio clip over the background using pydub. We have implemented this for you.",
"_____no_output_____"
]
],
[
[
"# GRADED FUNCTION: insert_audio_clip\n\ndef insert_audio_clip(background, audio_clip, previous_segments):\n \"\"\"\n Insert a new audio segment over the background noise at a random time step, ensuring that the \n audio segment does not overlap with existing segments.\n \n Arguments:\n background -- a 10 second background audio recording. \n audio_clip -- the audio clip to be inserted/overlaid. \n previous_segments -- times where audio segments have already been placed\n \n Returns:\n new_background -- the updated background audio\n \"\"\"\n \n # Get the duration of the audio clip in ms\n segment_ms = len(audio_clip)\n \n ### START CODE HERE ### \n # Step 1: Use one of the helper functions to pick a random time segment onto which to insert \n # the new audio clip. (≈ 1 line)\n segment_time = get_random_time_segment(segment_ms)\n \n # Step 2: Check if the new segment_time overlaps with one of the previous_segments. If so, keep \n # picking new segment_time at random until it doesn't overlap. (≈ 2 lines)\n while is_overlapping(segment_time, previous_segments):\n segment_time = get_random_time_segment(segment_ms)\n\n # Step 3: Add the new segment_time to the list of previous_segments (≈ 1 line)\n previous_segments.append(segment_time)\n ### END CODE HERE ###\n \n # Step 4: Superpose audio segment and background\n new_background = background.overlay(audio_clip, position = segment_time[0])\n \n return new_background, segment_time",
"_____no_output_____"
],
[
"np.random.seed(5)\naudio_clip, segment_time = insert_audio_clip(backgrounds[0], activates[0], [(3790, 4400)])\naudio_clip.export(\"insert_test.wav\", format=\"wav\")\nprint(\"Segment Time: \", segment_time)\nIPython.display.Audio(\"insert_test.wav\")",
"_____no_output_____"
]
],
[
[
"**Expected Output**\n\n<table>\n <tr>\n <td>\n **Segment Time**\n </td>\n <td>\n (2254, 3169)\n </td>\n </tr>\n</table>",
"_____no_output_____"
]
],
[
[
"# Expected audio\nIPython.display.Audio(\"audio_examples/insert_reference.wav\")",
"_____no_output_____"
]
],
[
[
"Finally, implement code to update the labels $y^{\\langle t \\rangle}$, assuming you just inserted an \"activate.\" In the code below, `y` is a `(1,1375)` dimensional vector, since $T_y = 1375$. \n\nIf the \"activate\" ended at time step $t$, then set $y^{\\langle t+1 \\rangle} = 1$ as well as for up to 49 additional consecutive values. However, make sure you don't run off the end of the array and try to update `y[0][1375]`, since the valid indices are `y[0][0]` through `y[0][1374]` because $T_y = 1375$. So if \"activate\" ends at step 1370, you would get only `y[0][1371] = y[0][1372] = y[0][1373] = y[0][1374] = 1`\n\n**Exercise**: Implement `insert_ones()`. You can use a for loop. (If you are an expert in python's slice operations, feel free also to use slicing to vectorize this.) If a segment ends at `segment_end_ms` (using a 10000 step discretization), to convert it to the indexing for the outputs $y$ (using a $1375$ step discretization), we will use this formula: \n```\n segment_end_y = int(segment_end_ms * Ty / 10000.0)\n```",
"_____no_output_____"
]
],
[
[
"# GRADED FUNCTION: insert_ones\n\ndef insert_ones(y, segment_end_ms):\n \"\"\"\n Update the label vector y. The labels of the 50 output steps strictly after the end of the segment \n should be set to 1. By strictly we mean that the label of segment_end_y should be 0 while, the\n 50 following labels should be ones.\n \n \n Arguments:\n y -- numpy array of shape (1, Ty), the labels of the training example\n segment_end_ms -- the end time of the segment in ms\n \n Returns:\n y -- updated labels\n \"\"\"\n \n # duration of the background (in terms of spectrogram time-steps)\n segment_end_y = int(segment_end_ms * Ty / 10000.0)\n \n # Add 1 to the correct index in the background label (y)\n ### START CODE HERE ### (≈ 3 lines)\n for i in range(segment_end_y + 1, segment_end_y + 51):\n if i < Ty:\n y[0, i] = 1\n ### END CODE HERE ###\n \n return y",
"_____no_output_____"
],
[
"arr1 = insert_ones(np.zeros((1, Ty)), 9700)\nplt.plot(insert_ones(arr1, 4251)[0,:])\nprint(\"sanity checks:\", arr1[0][1333], arr1[0][634], arr1[0][635])",
"_____no_output_____"
]
],
[
[
"**Expected Output**\n<table>\n <tr>\n <td>\n **sanity checks**:\n </td>\n <td>\n 0.0 1.0 0.0\n </td>\n </tr>\n</table>\n<img src=\"images/ones_reference.png\" style=\"width:320;height:240px;\">",
"_____no_output_____"
],
[
"Finally, you can use `insert_audio_clip` and `insert_ones` to create a new training example.\n\n**Exercise**: Implement `create_training_example()`. You will need to carry out the following steps:\n\n1. Initialize the label vector $y$ as a numpy array of zeros and shape $(1, T_y)$.\n2. Initialize the set of existing segments to an empty list.\n3. Randomly select 0 to 4 \"activate\" audio clips, and insert them onto the 10sec clip. Also insert labels at the correct position in the label vector $y$.\n4. Randomly select 0 to 2 negative audio clips, and insert them into the 10sec clip. \n",
"_____no_output_____"
]
],
[
[
"# GRADED FUNCTION: create_training_example\n\ndef create_training_example(background, activates, negatives):\n \"\"\"\n Creates a training example with a given background, activates, and negatives.\n \n Arguments:\n background -- a 10 second background audio recording\n activates -- a list of audio segments of the word \"activate\"\n negatives -- a list of audio segments of random words that are not \"activate\"\n \n Returns:\n x -- the spectrogram of the training example\n y -- the label at each time step of the spectrogram\n \"\"\"\n \n # Set the random seed\n np.random.seed(18)\n \n # Make background quieter\n background = background - 20\n\n ### START CODE HERE ###\n # Step 1: Initialize y (label vector) of zeros (≈ 1 line)\n y = np.zeros((1, Ty))\n\n # Step 2: Initialize segment times as empty list (≈ 1 line)\n previous_segments = []\n ### END CODE HERE ###\n \n # Select 0-4 random \"activate\" audio clips from the entire list of \"activates\" recordings\n number_of_activates = np.random.randint(0, 5)\n random_indices = np.random.randint(len(activates), size=number_of_activates)\n random_activates = [activates[i] for i in random_indices]\n \n ### START CODE HERE ### (≈ 3 lines)\n # Step 3: Loop over randomly selected \"activate\" clips and insert in background\n for random_activate in random_activates:\n # Insert the audio clip on the background\n background, segment_time = insert_audio_clip(background, random_activate, previous_segments)\n # Retrieve segment_start and segment_end from segment_time\n segment_start, segment_end = segment_time\n # Insert labels in \"y\"\n y = insert_ones(y, segment_end_ms=segment_end)\n ### END CODE HERE ###\n\n # Select 0-2 random negatives audio recordings from the entire list of \"negatives\" recordings\n number_of_negatives = np.random.randint(0, 3)\n random_indices = np.random.randint(len(negatives), size=number_of_negatives)\n random_negatives = [negatives[i] for i in random_indices]\n\n ### START CODE HERE ### (≈ 2 lines)\n # Step 4: Loop over randomly selected negative clips and insert in background\n for random_negative in random_negatives:\n # Insert the audio clip on the background \n background, _ = insert_audio_clip(background, random_negative, previous_segments)\n ### END CODE HERE ###\n \n # Standardize the volume of the audio clip \n background = match_target_amplitude(background, -20.0)\n\n # Export new training example \n file_handle = background.export(\"train\" + \".wav\", format=\"wav\")\n print(\"File (train.wav) was saved in your directory.\")\n \n # Get and plot spectrogram of the new recording (background with superposition of positive and negatives)\n x = graph_spectrogram(\"train.wav\")\n \n return x, y",
"_____no_output_____"
],
[
"x, y = create_training_example(backgrounds[0], activates, negatives)",
"_____no_output_____"
]
],
[
[
"**Expected Output**\n<img src=\"images/train_reference.png\" style=\"width:320;height:240px;\">",
"_____no_output_____"
],
[
"Now you can listen to the training example you created and compare it to the spectrogram generated above.",
"_____no_output_____"
]
],
[
[
"IPython.display.Audio(\"train.wav\")",
"_____no_output_____"
]
],
[
[
"**Expected Output**",
"_____no_output_____"
]
],
[
[
"IPython.display.Audio(\"audio_examples/train_reference.wav\")",
"_____no_output_____"
]
],
[
[
"Finally, you can plot the associated labels for the generated training example.",
"_____no_output_____"
]
],
[
[
"plt.plot(y[0])",
"_____no_output_____"
]
],
[
[
"**Expected Output**\n<img src=\"images/train_label.png\" style=\"width:320;height:240px;\">",
"_____no_output_____"
],
[
"## 1.4 - Full training set\n\nYou've now implemented the code needed to generate a single training example. We used this process to generate a large training set. To save time, we've already generated a set of training examples. ",
"_____no_output_____"
]
],
[
[
"# Load preprocessed training examples\nX = np.load(\"./XY_train/X.npy\")\nY = np.load(\"./XY_train/Y.npy\")",
"_____no_output_____"
]
],
[
[
"## 1.5 - Development set\n\nTo test our model, we recorded a development set of 25 examples. While our training data is synthesized, we want to create a development set using the same distribution as the real inputs. Thus, we recorded 25 10-second audio clips of people saying \"activate\" and other random words, and labeled them by hand. This follows the principle described in Course 3 that we should create the dev set to be as similar as possible to the test set distribution; that's why our dev set uses real rather than synthesized audio. \n",
"_____no_output_____"
]
],
[
[
"# Load preprocessed dev set examples\nX_dev = np.load(\"./XY_dev/X_dev.npy\")\nY_dev = np.load(\"./XY_dev/Y_dev.npy\")",
"_____no_output_____"
]
],
[
[
"# 2 - Model\n\nNow that you've built a dataset, lets write and train a trigger word detection model! \n\nThe model will use 1-D convolutional layers, GRU layers, and dense layers. Let's load the packages that will allow you to use these layers in Keras. This might take a minute to load. ",
"_____no_output_____"
]
],
[
[
"from keras.callbacks import ModelCheckpoint\nfrom keras.models import Model, load_model, Sequential\nfrom keras.layers import Dense, Activation, Dropout, Input, Masking, TimeDistributed, LSTM, Conv1D\nfrom keras.layers import GRU, Bidirectional, BatchNormalization, Reshape\nfrom keras.optimizers import Adam",
"_____no_output_____"
]
],
[
[
"## 2.1 - Build the model\n\nHere is the architecture we will use. Take some time to look over the model and see if it makes sense. \n\n<img src=\"images/model.png\" style=\"width:600px;height:600px;\">\n<center> **Figure 3** </center>\n\nOne key step of this model is the 1D convolutional step (near the bottom of Figure 3). It inputs the 5511 step spectrogram, and outputs a 1375 step output, which is then further processed by multiple layers to get the final $T_y = 1375$ step output. This layer plays a role similar to the 2D convolutions you saw in Course 4, of extracting low-level features and then possibly generating an output of a smaller dimension. \n\nComputationally, the 1-D conv layer also helps speed up the model because now the GRU has to process only 1375 timesteps rather than 5511 timesteps. The two GRU layers read the sequence of inputs from left to right, then ultimately uses a dense+sigmoid layer to make a prediction for $y^{\\langle t \\rangle}$. Because $y$ is binary valued (0 or 1), we use a sigmoid output at the last layer to estimate the chance of the output being 1, corresponding to the user having just said \"activate.\"\n\nNote that we use a uni-directional RNN rather than a bi-directional RNN. This is really important for trigger word detection, since we want to be able to detect the trigger word almost immediately after it is said. If we used a bi-directional RNN, we would have to wait for the whole 10sec of audio to be recorded before we could tell if \"activate\" was said in the first second of the audio clip. \n",
"_____no_output_____"
],
[
"Implementing the model can be done in four steps:\n \n**Step 1**: CONV layer. Use `Conv1D()` to implement this, with 196 filters, \na filter size of 15 (`kernel_size=15`), and stride of 4. [[See documentation.](https://keras.io/layers/convolutional/#conv1d)]\n\n**Step 2**: First GRU layer. To generate the GRU layer, use:\n```\nX = GRU(units = 128, return_sequences = True)(X)\n```\nSetting `return_sequences=True` ensures that all the GRU's hidden states are fed to the next layer. Remember to follow this with Dropout and BatchNorm layers. \n\n**Step 3**: Second GRU layer. This is similar to the previous GRU layer (remember to use `return_sequences=True`), but has an extra dropout layer. \n\n**Step 4**: Create a time-distributed dense layer as follows: \n```\nX = TimeDistributed(Dense(1, activation = \"sigmoid\"))(X)\n```\nThis creates a dense layer followed by a sigmoid, so that the parameters used for the dense layer are the same for every time step. [[See documentation](https://keras.io/layers/wrappers/).]\n\n**Exercise**: Implement `model()`, the architecture is presented in Figure 3.",
"_____no_output_____"
]
],
[
[
"# GRADED FUNCTION: model\n\ndef model(input_shape):\n \"\"\"\n Function creating the model's graph in Keras.\n \n Argument:\n input_shape -- shape of the model's input data (using Keras conventions)\n\n Returns:\n model -- Keras model instance\n \"\"\"\n \n X_input = Input(shape = input_shape)\n \n ### START CODE HERE ###\n \n # Step 1: CONV layer (≈4 lines)\n X = Conv1D(196, 15, strides=4)(X_input) # CONV1D\n X = BatchNormalization()(X) # Batch normalization\n X = Activation('relu')(X) # ReLu activation\n X = Dropout(0.8)(X) # dropout (use 0.8)\n\n # Step 2: First GRU Layer (≈4 lines)\n X = GRU(units = 128, return_sequences=True)(X) # GRU (use 128 units and return the sequences)\n X = Dropout(0.8)(X) # dropout (use 0.8)\n X = BatchNormalization()(X) # Batch normalization\n \n # Step 3: Second GRU Layer (≈4 lines)\n X = GRU(units = 128, return_sequences=True)(X) # GRU (use 128 units and return the sequences)\n X = Dropout(0.8)(X) # dropout (use 0.8)\n X = BatchNormalization()(X) # Batch normalization\n X = Dropout(0.8)(X) # dropout (use 0.8)\n \n # Step 4: Time-distributed dense layer (≈1 line)\n X = TimeDistributed(Dense(1, activation = \"sigmoid\"))(X) # time distributed (sigmoid)\n\n ### END CODE HERE ###\n\n model = Model(inputs = X_input, outputs = X)\n \n return model ",
"_____no_output_____"
],
[
"model = model(input_shape = (Tx, n_freq))",
"_____no_output_____"
]
],
[
[
"Let's print the model summary to keep track of the shapes.",
"_____no_output_____"
]
],
[
[
"model.summary()",
"_____no_output_____"
]
],
[
[
"**Expected Output**:\n\n<table>\n <tr>\n <td>\n **Total params**\n </td>\n <td>\n 522,561\n </td>\n </tr>\n <tr>\n <td>\n **Trainable params**\n </td>\n <td>\n 521,657\n </td>\n </tr>\n <tr>\n <td>\n **Non-trainable params**\n </td>\n <td>\n 904\n </td>\n </tr>\n</table>",
"_____no_output_____"
],
[
"The output of the network is of shape (None, 1375, 1) while the input is (None, 5511, 101). The Conv1D has reduced the number of steps from 5511 at spectrogram to 1375. ",
"_____no_output_____"
],
[
"## 2.2 - Fit the model",
"_____no_output_____"
],
[
"Trigger word detection takes a long time to train. To save time, we've already trained a model for about 3 hours on a GPU using the architecture you built above, and a large training set of about 4000 examples. Let's load the model. ",
"_____no_output_____"
]
],
[
[
"model = load_model('./models/tr_model.h5')",
"_____no_output_____"
]
],
[
[
"You can train the model further, using the Adam optimizer and binary cross entropy loss, as follows. This will run quickly because we are training just for one epoch and with a small training set of 26 examples. ",
"_____no_output_____"
]
],
[
[
"opt = Adam(lr=0.0001, beta_1=0.9, beta_2=0.999, decay=0.01)\nmodel.compile(loss='binary_crossentropy', optimizer=opt, metrics=[\"accuracy\"])",
"_____no_output_____"
],
[
"model.fit(X, Y, batch_size = 5, epochs=1)",
"_____no_output_____"
]
],
[
[
"## 2.3 - Test the model\n\nFinally, let's see how your model performs on the dev set.",
"_____no_output_____"
]
],
[
[
"loss, acc = model.evaluate(X_dev, Y_dev)\nprint(\"Dev set accuracy = \", acc)",
"_____no_output_____"
]
],
[
[
"This looks pretty good! However, accuracy isn't a great metric for this task, since the labels are heavily skewed to 0's, so a neural network that just outputs 0's would get slightly over 90% accuracy. We could define more useful metrics such as F1 score or Precision/Recall. But let's not bother with that here, and instead just empirically see how the model does. ",
"_____no_output_____"
],
[
"# 3 - Making Predictions\n\nNow that you have built a working model for trigger word detection, let's use it to make predictions. This code snippet runs audio (saved in a wav file) through the network. \n\n<!--\ncan use your model to make predictions on new audio clips.\n\nYou will first need to compute the predictions for an input audio clip.\n\n**Exercise**: Implement predict_activates(). You will need to do the following:\n\n1. Compute the spectrogram for the audio file\n2. Use `np.swap` and `np.expand_dims` to reshape your input to size (1, Tx, n_freqs)\n5. Use forward propagation on your model to compute the prediction at each output step\n!-->",
"_____no_output_____"
]
],
[
[
"def detect_triggerword(filename):\n plt.subplot(2, 1, 1)\n\n x = graph_spectrogram(filename)\n # the spectogram outputs (freqs, Tx) and we want (Tx, freqs) to input into the model\n x = x.swapaxes(0,1)\n x = np.expand_dims(x, axis=0)\n predictions = model.predict(x)\n \n plt.subplot(2, 1, 2)\n plt.plot(predictions[0,:,0])\n plt.ylabel('probability')\n plt.show()\n return predictions",
"_____no_output_____"
]
],
[
[
"Once you've estimated the probability of having detected the word \"activate\" at each output step, you can trigger a \"chiming\" sound to play when the probability is above a certain threshold. Further, $y^{\\langle t \\rangle}$ might be near 1 for many values in a row after \"activate\" is said, yet we want to chime only once. So we will insert a chime sound at most once every 75 output steps. This will help prevent us from inserting two chimes for a single instance of \"activate\". (This plays a role similar to non-max suppression from computer vision.) \n\n<!-- \n**Exercise**: Implement chime_on_activate(). You will need to do the following:\n\n1. Loop over the predicted probabilities at each output step\n2. When the prediction is larger than the threshold and more than 75 consecutive time steps have passed, insert a \"chime\" sound onto the original audio clip\n\nUse this code to convert from the 1,375 step discretization to the 10,000 step discretization and insert a \"chime\" using pydub:\n\n` audio_clip = audio_clip.overlay(chime, position = ((i / Ty) * audio.duration_seconds)*1000)\n`\n!--> ",
"_____no_output_____"
]
],
[
[
"chime_file = \"audio_examples/chime.wav\"\ndef chime_on_activate(filename, predictions, threshold):\n audio_clip = AudioSegment.from_wav(filename)\n chime = AudioSegment.from_wav(chime_file)\n Ty = predictions.shape[1]\n # Step 1: Initialize the number of consecutive output steps to 0\n consecutive_timesteps = 0\n # Step 2: Loop over the output steps in the y\n for i in range(Ty):\n # Step 3: Increment consecutive output steps\n consecutive_timesteps += 1\n # Step 4: If prediction is higher than the threshold and more than 75 consecutive output steps have passed\n if predictions[0,i,0] > threshold and consecutive_timesteps > 75:\n # Step 5: Superpose audio and background using pydub\n audio_clip = audio_clip.overlay(chime, position = ((i / Ty) * audio_clip.duration_seconds)*1000)\n # Step 6: Reset consecutive output steps to 0\n consecutive_timesteps = 0\n \n audio_clip.export(\"chime_output.wav\", format='wav')",
"_____no_output_____"
]
],
[
[
"## 3.3 - Test on dev examples",
"_____no_output_____"
],
[
"Let's explore how our model performs on two unseen audio clips from the development set. Lets first listen to the two dev set clips. ",
"_____no_output_____"
]
],
[
[
"IPython.display.Audio(\"./raw_data/dev/1.wav\")",
"_____no_output_____"
],
[
"IPython.display.Audio(\"./raw_data/dev/2.wav\")",
"_____no_output_____"
]
],
[
[
"Now lets run the model on these audio clips and see if it adds a chime after \"activate\"!",
"_____no_output_____"
]
],
[
[
"filename = \"./raw_data/dev/1.wav\"\nprediction = detect_triggerword(filename)\nchime_on_activate(filename, prediction, 0.5)\nIPython.display.Audio(\"./chime_output.wav\")",
"_____no_output_____"
],
[
"filename = \"./raw_data/dev/2.wav\"\nprediction = detect_triggerword(filename)\nchime_on_activate(filename, prediction, 0.5)\nIPython.display.Audio(\"./chime_output.wav\")",
"_____no_output_____"
]
],
[
[
"# Congratulations \n\nYou've come to the end of this assignment! \n\nHere's what you should remember:\n- Data synthesis is an effective way to create a large training set for speech problems, specifically trigger word detection. \n- Using a spectrogram and optionally a 1D conv layer is a common pre-processing step prior to passing audio data to an RNN, GRU or LSTM.\n- An end-to-end deep learning approach can be used to built a very effective trigger word detection system. \n\n*Congratulations* on finishing the final assignment! \n\nThank you for sticking with us through the end and for all the hard work you've put into learning deep learning. We hope you have enjoyed the course! \n",
"_____no_output_____"
],
[
"# 4 - Try your own example! (OPTIONAL/UNGRADED)\n\nIn this optional and ungraded portion of this notebook, you can try your model on your own audio clips! \n\nRecord a 10 second audio clip of you saying the word \"activate\" and other random words, and upload it to the Coursera hub as `myaudio.wav`. Be sure to upload the audio as a wav file. If your audio is recorded in a different format (such as mp3) there is free software that you can find online for converting it to wav. If your audio recording is not 10 seconds, the code below will either trim or pad it as needed to make it 10 seconds. \n",
"_____no_output_____"
]
],
[
[
"# Preprocess the audio to the correct format\ndef preprocess_audio(filename):\n # Trim or pad audio segment to 10000ms\n padding = AudioSegment.silent(duration=10000)\n segment = AudioSegment.from_wav(filename)[:10000]\n segment = padding.overlay(segment)\n # Set frame rate to 44100\n segment = segment.set_frame_rate(44100)\n # Export as wav\n segment.export(filename, format='wav')",
"_____no_output_____"
]
],
[
[
"Once you've uploaded your audio file to Coursera, put the path to your file in the variable below.",
"_____no_output_____"
]
],
[
[
"your_filename = \"audio_examples/my_audio.wav\"",
"_____no_output_____"
],
[
"preprocess_audio(your_filename)\nIPython.display.Audio(your_filename) # listen to the audio you uploaded ",
"_____no_output_____"
]
],
[
[
"Finally, use the model to predict when you say activate in the 10 second audio clip, and trigger a chime. If beeps are not being added appropriately, try to adjust the chime_threshold.",
"_____no_output_____"
]
],
[
[
"chime_threshold = 0.5\nprediction = detect_triggerword(your_filename)\nchime_on_activate(your_filename, prediction, chime_threshold)\nIPython.display.Audio(\"./chime_output.wav\")",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb8d51d66319c679052f647580d4bb98a1f34570
| 1,015,763 |
ipynb
|
Jupyter Notebook
|
Module-2.ipynb
|
Rakesh148/CareerLauncher-ML-Intern
|
4f58c196a1416c17ccdeae58711d139c7508016b
|
[
"Apache-2.0"
] | null | null | null |
Module-2.ipynb
|
Rakesh148/CareerLauncher-ML-Intern
|
4f58c196a1416c17ccdeae58711d139c7508016b
|
[
"Apache-2.0"
] | 1 |
2020-07-30T15:05:53.000Z
|
2020-07-30T15:05:53.000Z
|
Module-2.ipynb
|
Rakesh148/CareerLauncher-ML-Intern
|
4f58c196a1416c17ccdeae58711d139c7508016b
|
[
"Apache-2.0"
] | null | null | null | 1,220.86899 | 243,964 | 0.956106 |
[
[
[
"# Module - 2: Data visualization and Technical Analysis",
"_____no_output_____"
],
[
"###### Loading required libraries",
"_____no_output_____"
]
],
[
[
"import pandas as pd # data loading tool\nimport matplotlib.pyplot as plt #ploting tool\nimport seaborn as sns\nimport numpy as np",
"_____no_output_____"
]
],
[
[
"## 2.1 Loading dataset and changing the Date format",
"_____no_output_____"
]
],
[
[
"mod2_data = pd.read_csv('week2.csv')\ndel mod2_data['Unnamed: 0'] #deleting Unnammed column\nmod2_data.Date = pd.to_datetime(mod2_data['Date'])\nmod2_data= mod2_data.set_index('Date')\nprint(mod2_data.index.dtype == \"datetime64[ns]\")\nmod2_data.head()",
"True\n"
],
[
"fig, ax = plt.subplots(figsize=(10, 6))\nsns.lineplot(data=mod2_data, x=mod2_data.index, y='Close Price', ax=ax)\nplt.tight_layout()\nplt.show()",
"_____no_output_____"
]
],
[
[
"###### News:\n1. Between 2017-10 and 2018-02. \n***Tata Power Company (TPCL) has posted an improved performance in 1QFY18 with its consolidated net profit rising by 126% YoY to Rs1.64bn (vs. Rs0.72bn in 1QFY17) due to strong performance by the coal subsidiaries, renewable business and better operational performance. Notably, renewable business generated Rs1.09bn PAT in 1QFY18 compared to Rs0.26bn in 1QFY17. Consolidated revenue rose by 2% YoY to Rs67.2bn mainly due to improved revenue from Welspun Renewable Energy (WREPL).***\nSource:https://www.moneycontrol.com/news/business/stocks/buy-tata-power-target-of-rs-88-reliance-securities-2370965.html\n2. Between 2018-02 and 2018-11.\n***Tata Power Company's third quarter consolidated profit is expected to fall 22 percent to Rs 466 crore compared to Rs 599 crore in year-ago quarter.Revenue from operations may grow 11 percent to Rs 7,445 crore compared to Rs 6,684 crore in same quarter last fiscal, according to average of estimates of analysts polled by CNBC-TV18. Operating profit is likely to increase 15 percent year-on-year to Rs 1,611 crore and margin may expand 70 basis points to 21.6 percent in Q3.Year-on-year profit comparison may not be valid due to (1) higher interest cost in Q3FY18 to fund renewable asset acquisition and (2) tax reversal in Q3FY17, despite stable operations in the core distribution business.Analysts expect generation volumes to remain sluggish and realisations to remain flattish. They further expect coal business and renewable business to maintain strong momentum.More than numbers, the Street will watch out for restructuring news. Tata Power had guided for simplification of group structure in FY18 at the beginning of the year.***\nSource:https://www.moneycontrol.com/news/business/earnings/tata-power-q3-profit-seen-down-22-generation-volumes-may-remain-sluggish-2507829.html\n3. Between 2018-10 and 2019-01.\n***Tata Power, HPCL join hands to set up EV charging stations***\nsource:https://www.moneycontrol.com/news/india/tata-power-hpcl-join-hands-to-set-up-ev-charging-stations-2991981.html\n4. Between 2019-01 and 2019-03.\n***Fuel cost of the company rose to Rs 3,189.87 crore from Rs 2,491.24 crore in the year-ago period. Similarly, the finance cost rose to Rs 1,013.96 crore from Rs 855.28 crore a year ago.***\nSource:https://www.moneycontrol.com/news/business/tata-power-q3-profit-plunges-67-to-rs-205-cr-in-q3-3445841.html\n5. After 2019-04.\n***Tata Power Q4 net drops 92% to Rs 107.32 cr; declares dividend of Rs 1.30/share***\nSource:https://www.moneycontrol.com/news/business/tata-power-q4-net-drops-92-to-rs-107-32-cr-declares-dividend-of-rs-1-30share-3924591.html",
"_____no_output_____"
],
[
"## 2.2 Stem plot",
"_____no_output_____"
]
],
[
[
"fig, ax = plt.subplots(figsize=(10, 6))\nax.stem(mod2_data.index, mod2_data.Day_Perc_Change, 'g', label='Percente Change')\nplt.tight_layout()\nplt.legend()\nplt.show()\n",
"C:\\Users\\user\\anaconda3\\lib\\site-packages\\ipykernel_launcher.py:2: UserWarning: In Matplotlib 3.3 individual lines on a stem plot will be added as a LineCollection instead of individual lines. This significantly improves the performance of a stem plot. To remove this warning and switch to the new behaviour, set the \"use_line_collection\" keyword argument to True.\n \n"
]
],
[
[
"## 2.3 Daily volume and comparison with %stem plot",
"_____no_output_____"
]
],
[
[
"volume_scaled = mod2_data['No. of Trades'] - mod2_data['No. of Trades'].min()\nvolume_scaled = volume_scaled/volume_scaled.max()*mod2_data.Day_Perc_Change.max()\nfig, ax = plt.subplots(figsize=(10, 6))\nax.plot(mod2_data.index, volume_scaled, label='Volume')\nax.set_xlabel('Date')\nplt.legend(loc=2)\n\nplt.tight_layout()\nplt.show()",
"_____no_output_____"
],
[
"fig, ax = plt.subplots(figsize=(10, 6))\n\nax.stem(mod2_data.index, mod2_data.Day_Perc_Change , 'g', label='Percente Change')\nax.plot(mod2_data.index, volume_scaled, 'k', label='Volume')\n\nax.set_xlabel('Date')\nplt.legend(loc=2)\n\nplt.tight_layout()\nplt.show()",
"C:\\Users\\user\\anaconda3\\lib\\site-packages\\ipykernel_launcher.py:3: UserWarning: In Matplotlib 3.3 individual lines on a stem plot will be added as a LineCollection instead of individual lines. This significantly improves the performance of a stem plot. To remove this warning and switch to the new behaviour, set the \"use_line_collection\" keyword argument to True.\n This is separate from the ipykernel package so we can avoid doing imports until\n"
]
],
[
[
"###### Relationship between volume and daily percentage change\nAs the volume increases the percentage change becomes positive and vice versa.",
"_____no_output_____"
],
[
"## 2.4 Pie chart and Bar plot ",
"_____no_output_____"
]
],
[
[
"gridsize = (2, 6)\nfig = plt.figure(figsize=(14, 10))\nax1 = plt.subplot2grid(gridsize, (0, 0), colspan=2, rowspan=1)\nax2 = plt.subplot2grid(gridsize, (0, 3), colspan=3)\nax3 = plt.subplot2grid(gridsize, (1, 0), colspan=6)\n\nmod2_data['ones'] = np.ones((mod2_data.shape[0]))\nsums = mod2_data.ones.groupby(mod2_data.Trend).sum()\nexplod = [0.2, 0.2, 0.5, 0, 0, 0, 0 ,0,0]\nax1.pie(sums, labels=sums.index, autopct='%1.1f%%', explode=explod)\nax2.title.set_text('Trend')\nmod2_data = mod2_data.drop(['ones'], axis=1)\n\nbard1 = mod2_data[['Trend', 'Total Traded Quantity']].groupby(['Trend'], as_index=False).mean()\nbar1 = sns.barplot(\"Trend\", 'Total Traded Quantity', data=bard1, ci=None, ax=ax2)\nfor item in bar1.get_xticklabels():\n item.set_rotation(45)\nax2.set_ylabel('') \nax2.title.set_text('Trend to mean of Total Traded Quantity')\n\nbard2 = mod2_data[['Trend', 'Total Traded Quantity']].groupby(['Trend'], as_index=False).median()\nbar2 = sns.barplot(\"Trend\", 'Total Traded Quantity', data=bard2, ci=None, ax=ax3)\nfor item in bar2.get_xticklabels():\n item.set_rotation(45)\nax3.set_ylabel('') \nax3.title.set_text('Trend to meadian of Total Traded Quantity')\n\nplt.tight_layout()\nplt.show()",
"_____no_output_____"
]
],
[
[
"## 2.5 Daily returns",
"_____no_output_____"
]
],
[
[
"fig, ax = plt.subplots(figsize=(10, 6))\nax.hist(mod2_data.Day_Perc_Change, bins=50)\nax.set_ylabel('Percent Change')\nplt.show()",
"_____no_output_____"
]
],
[
[
"## 2.6 Correlation",
"_____no_output_____"
]
],
[
[
"five_stocks = ['AMARAJABAT.csv', 'CUMMINSIND.csv', 'JINDALSTEL.csv', 'MRPL.csv', 'VOLTAS.csv']\ndfs = {}\nfor i in five_stocks:\n stock = i.split('.')[0]\n temp_df = pd.read_csv(i)\n temp_df = temp_df[temp_df[\"Series\"] == \"EQ\"]\n temp_df['Day_Perc_Change'] = temp_df['Close Price'].pct_change()*100\n temp_df = temp_df['Day_Perc_Change']\n temp_df = temp_df.drop(temp_df.index[0])\n dfs[stock] = temp_df\n\ndfs = pd.DataFrame(dfs)\nsns.pairplot(dfs)\nplt.show()",
"_____no_output_____"
]
],
[
[
"There is no correlation among almost all the stocks, which is good thing. To get the profit from the stock market, the company's stocks trend should be independent from the other stock's trend",
"_____no_output_____"
],
[
"## 2.7 Volatility",
"_____no_output_____"
]
],
[
[
"rolling1 = dfs.rolling(7).std()\nrolling1.dropna()\nfig, ax = plt.subplots(figsize=(15, 5))\nax.plot(np.arange(len(rolling1.VOLTAS)), rolling1.VOLTAS, 'k')\nplt.title('VOLTAS Volatility')\nplt.show()",
"_____no_output_____"
]
],
[
[
"## 2.8 Comparing with Nifty50",
"_____no_output_____"
]
],
[
[
"nifty = pd.read_csv('Nifty50.csv')\nnifty['Day_Perc_Change'] = nifty['Close'].pct_change()*100\nrolling2 = nifty['Day_Perc_Change'].rolling(7).std()\nrolling2 = rolling2.dropna()",
"_____no_output_____"
],
[
"fig, ax = plt.subplots(figsize=(15, 5))\nax.plot(np.arange(len(rolling1.VOLTAS)), rolling1.VOLTAS, label = 'VOLTAS')\nax.plot(np.arange(len(rolling2)), rolling2, 'k', label = 'Nifty')\nplt.legend()\nplt.tight_layout()\nplt.show()",
"_____no_output_____"
]
],
[
[
"## 2.9 Trade calls",
"_____no_output_____"
]
],
[
[
"nifty['roll21'] = nifty['Close'].rolling(21).mean()\nnifty['roll34'] = nifty['Close'].rolling(34).mean()\nnifty = nifty.dropna()",
"_____no_output_____"
],
[
"nifty.Date = pd.to_datetime(nifty['Date'])\nfig, ax = plt.subplots(figsize=(15, 7))\n\ndef cross(values):\n l=[]\n were = values[0]\n flag = True\n for i, ele in enumerate(values):\n if were==ele:\n l.append(0)\n else:\n l.append(1)\n were = ele\n return l\n\nnifty['buy'] = nifty['roll21'] > nifty['roll34']\nnifty['sell'] = nifty['roll21'] < nifty['roll34']\n\nnifty['buy_change'] = np.array(cross(nifty.buy.values.reshape(1, len(nifty.buy)).flatten())) #reshaping from (461, )\nnifty['sell_change'] = np.array(cross(nifty.sell.values.reshape(1, len(nifty.sell)).flatten())) #reshaping from(461, )\n\nnifty['buy'] = nifty['buy_change'].where(nifty['buy']==True)\nnifty['buy'] = nifty['roll21'].where(nifty['buy']==1)\n\nnifty['sell'] = nifty['sell_change'].where(nifty['sell']==True)\nnifty['sell'] = nifty['roll21'].where(nifty['sell']==1)\n\nax.plot(nifty.Date, nifty.Close, 'r')\nax.plot(nifty.Date, nifty.roll34, 'b', label='34_SMA')\nax.plot(nifty.Date, nifty.roll21, 'g', label='21_SMA')\nax.plot(nifty.Date, nifty.buy, \"g^\")\nax.plot(nifty.Date, nifty.sell, \"kv\")\n\nax.set_xlabel('Date')\nplt.legend(loc=2)\nplt.tight_layout()\nplt.show()\n ",
"_____no_output_____"
]
],
[
[
"## 2.10 Trade call - using Bollinger band",
"_____no_output_____"
]
],
[
[
"nifty['roll14'] = nifty['Close'].rolling(14).mean()\nstd = nifty['Close'].rolling(14).std()\nnifty['upper_band'] = nifty['roll14']+2*std\nnifty['lower_band'] = nifty['roll14']-2*std\n\nfig, ax = plt.subplots(figsize=(15, 7))\nax.plot(nifty.Date, nifty['Close'],'k' ,label = 'avg price')\nax.plot(nifty.Date, nifty.roll14, label= 'roll14')\nax.plot(nifty.Date, nifty.upper_band, 'r', label = 'upper_band')\nax.plot(nifty.Date, nifty.lower_band,'g', label = 'lower_band')\n\nax.set_xlabel('Date')\nplt.legend(loc=2)\nplt.tight_layout()\nplt.show()",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb8d597bcda19ecf9236da50a30b8b39760c87f7
| 1,011,048 |
ipynb
|
Jupyter Notebook
|
examples/xarray_and_cartopy.ipynb
|
dlnash/pyclivac
|
8ba30e5ab883f6f13105494615a433c6a04673c8
|
[
"MIT"
] | 5 |
2019-11-21T15:24:09.000Z
|
2020-06-18T18:50:16.000Z
|
examples/xarray_and_cartopy.ipynb
|
CLIVAC/pyclivac
|
8ba30e5ab883f6f13105494615a433c6a04673c8
|
[
"MIT"
] | null | null | null |
examples/xarray_and_cartopy.ipynb
|
CLIVAC/pyclivac
|
8ba30e5ab883f6f13105494615a433c6a04673c8
|
[
"MIT"
] | 1 |
2021-12-06T23:01:03.000Z
|
2021-12-06T23:01:03.000Z
| 325.305019 | 224,180 | 0.893839 |
[
[
[
"## Importing and mapping netCDF data with xarray and cartopy\n\n- Read data from a netCDF file with xarray\n- Select (index) and modify variables using xarray\n- Create user-defined functions\n- Set up map features with cartopy (lat/lon tickmarks, continents, country/state borders); create a function to automate these steps\n- Overlay various plot types: contour lines, filled contours, vectors, and barbs\n- Customize plot elements such as the colorbar and titles\n- Save figure\n ",
"_____no_output_____"
]
],
[
[
"## Imports\n\nimport os, sys\nimport numpy as np\nimport xarray as xr\nimport matplotlib.pyplot as plt\nimport cartopy.crs as ccrs\nimport cartopy.feature as cfeature\nfrom cartopy.mpl.geoaxes import GeoAxes\nfrom cartopy.mpl.ticker import LongitudeFormatter, LatitudeFormatter\n",
"_____no_output_____"
]
],
[
[
"### Load netcdf data with Xarray\n\nThis example demonstrates importing and mapping ERA5 reanalysis data for an AR-Thunderstorm event that occurred in Santa Barbara County on 6 March 2019. The data file created for this example can be found in the `sample-data` folder that 6-hourly ERA5 Reanalysis on a 0.5 x 0.5 deg lat-lon grid 4-8 March. ERA5 data was retrieved from the Climate Data Store and subset to a regional domain over the Western US/N. Pacific. \n\nThe xarray package provides an easy interface for importing and analyzing multidimensional data. Because xarray was designed around the netCDF data model, it is an exceptionally powerful tool for working with weather and climate data.\n\n\nXarray has two fundamental **data structures**: \n\n**1)** a **`DataArray`**, which holds a single n-dimensional variable. Elements of a DataArray include:\n - `values`: numpy array of data values\n - `dims`: list of named dimensions (for example, `['time','lat','lon']`)\n - `coords`: coordinate arrays (e.g., vectors of lat/lon values or datetime data)\n - `atts`: variable attributes such as `units` and `standard_name` \n \n**2)** a **`Dataset`**, which holds multiple n-dimensional variables (shared coordinates). Elements of a Dataset: data variables, dimensions, coordinates, and attributes.\n\n\nIn the cell below, we will load the ERA5 data (netcdf file) into an xarray dataset.\n",
"_____no_output_____"
]
],
[
[
"# Path to ERA5 data\nfilepath = \"../sample-data/era5.6hr.AR-thunderstorm.20190304_08.nc\"\n\n# Read nc file into xarray dataset\nds = xr.open_dataset(filepath)\n\n# Print dataset contents \nprint(ds)\n",
"<xarray.Dataset>\nDimensions: (latitude: 61, level: 3, longitude: 91, time: 20)\nCoordinates:\n * longitude (longitude) float32 -145.0 -144.5 -144.0 ... -101.0 -100.5 -100.0\n * latitude (latitude) float32 52.0 51.5 51.0 50.5 ... 23.5 23.0 22.5 22.0\n * level (level) float64 250.0 500.0 850.0\n * time (time) datetime64[ns] 2019-03-04 ... 2019-03-08T18:00:00\nData variables:\n zg (time, level, latitude, longitude) float64 ...\n u (time, level, latitude, longitude) float64 ...\n v (time, level, latitude, longitude) float64 ...\n msl (time, latitude, longitude) float64 ...\n prec (time, latitude, longitude) float64 ...\nAttributes:\n CDI: Climate Data Interface version 1.8.2 (http://m...\n Conventions: CF-1.6\n NCO: 4.6.7\n nco_openmp_thread_number: 1\n CDO: Climate Data Operators version 1.8.2 (http://m...\n title: ERA5 Reanalysis\n institution: European Centre for Medium-Range Weather Forec...\n grid_resolution: 0.5 x 0.5 deg lat-lon\n frequency: 6 hours\n"
]
],
[
[
"### Selecting/Indexing data with xarray\n\nWe can always use regular numpy indexing and slicing on DataArrays and Datasets; however, \nit is often more powerful and easier to use xarray’s `.sel()` method of label-based indexing.\n",
"_____no_output_____"
]
],
[
[
"# Select a single time\n\nds.sel(time='2019-03-05T18:00:00') # 5 March 2019 at 18 UTC\n",
"_____no_output_____"
],
[
"# Select all times within a single day \n\nds.sel(time='2019-03-06')\n",
"_____no_output_____"
],
[
"# Select times at 06 UTC\n\nidx = (ds['time.hour'] == 6) # selection uses boolean indexing\nhr06 = ds.sel(time=idx) # statements could be combined into a single line\n\nprint(hr06)\nprint(hr06.time.values) # check time coordinates in new dataset",
"['2019-03-04T06:00:00.000000000' '2019-03-05T06:00:00.000000000'\n '2019-03-06T06:00:00.000000000' '2019-03-07T06:00:00.000000000'\n '2019-03-08T06:00:00.000000000']\n"
]
],
[
[
"In the previous block, we used the `ds['time.hour']` to access the 'hour' component of a datetime object. Other datetime components include 'year', 'month', 'day', 'dayofyear', and 'season'. The 'season' component is unqiue to xarray; valid seasons include 'DJF', 'MAM', 'JJA', and 'SON'.",
"_____no_output_____"
]
],
[
[
"# Select a single grid point\n\nds.sel(latitude=40, longitude=-120)\n",
"_____no_output_____"
],
[
"# Select the grid point nearest to 34.4208° N, 119.6982°W;\n\nds.sel(latitude=34.4208, longitude=-119.6982, method='nearest')\n",
"_____no_output_____"
],
[
"# Select range of lats (30-40 N)\n# because ERA5 data latitudes are listed from 90N to 90S\n# you have to slice from latmax to latmin\n\nlatmin=30\nlatmax=40\nds.sel(latitude=slice(latmax,latmin))\n",
"_____no_output_____"
]
],
[
[
"Select data at the peak of the AR-Thunderstorm event (06-Mar-2019, 18UTC).",
"_____no_output_____"
]
],
[
[
"# Select the date/time of the AR event (~06 March 2019 at 06 UTC); \n# assign subset selection to new dataset `dsAR`\n\ndsAR = ds.sel(time='2019-03-06T06:00:00')\nprint(dsAR)\n",
"<xarray.Dataset>\nDimensions: (latitude: 61, level: 3, longitude: 91)\nCoordinates:\n * longitude (longitude) float32 -145.0 -144.5 -144.0 ... -101.0 -100.5 -100.0\n * latitude (latitude) float32 52.0 51.5 51.0 50.5 ... 23.5 23.0 22.5 22.0\n * level (level) float64 250.0 500.0 850.0\n time datetime64[ns] 2019-03-06T06:00:00\nData variables:\n zg (level, latitude, longitude) float64 ...\n u (level, latitude, longitude) float64 ...\n v (level, latitude, longitude) float64 ...\n msl (latitude, longitude) float64 ...\n prec (latitude, longitude) float64 ...\nAttributes:\n CDI: Climate Data Interface version 1.8.2 (http://m...\n Conventions: CF-1.6\n NCO: 4.6.7\n nco_openmp_thread_number: 1\n CDO: Climate Data Operators version 1.8.2 (http://m...\n title: ERA5 Reanalysis\n institution: European Centre for Medium-Range Weather Forec...\n grid_resolution: 0.5 x 0.5 deg lat-lon\n frequency: 6 hours\n"
],
[
"# Select data on a single pressure level `plev`\n\nplev = '250'\ndsAR = dsAR.sel(level=plev)\n",
"_____no_output_____"
]
],
[
[
"In the following code block, we select the data and coordinate variables needed to create a map of 250-hPa heights and winds at the time of the AR-Thunderstorm event.",
"_____no_output_____"
]
],
[
[
"# coordinate arrays\nlats = dsAR['latitude'].values # .values extracts var as numpy array\nlons = dsAR['longitude'].values \n#print(lats.shape, lons.shape)\n#print(lats)\n\n# data variables\nuwnd = dsAR['u'].values\nvwnd = dsAR['v'].values\nhgts = dsAR['zg'].values\n\n# check the shape and values of \nprint(hgts.shape)\nprint(hgts)\n",
"(61, 91)\n[[10208.39410476 10203.69141899 10197.37218498 ... 9871.41727741\n 9858.92576832 9846.87513603]\n [10210.01065299 10205.89580294 10199.8704868 ... 9885.81925258\n 9873.18078457 9860.2483987 ]\n [10211.92111909 10207.80626904 10202.51574754 ... 9902.57257065\n 9889.34626691 9876.11996318]\n ...\n [10755.22828469 10754.93436683 10754.7874079 ... 10892.78184352\n 10892.78184352 10891.75313101]\n [10771.09984917 10770.80593131 10770.51201345 ... 10899.39499539\n 10898.36628287 10897.19061143]\n [10787.11837258 10786.97141365 10786.67749579 ... 10904.39159902\n 10903.65680437 10902.48113292]]\n"
]
],
[
[
"### Simple arithmetic \n\nCalculate the magnitude of horizontal wind (wind speed) from its u and v components.\nConvert wspd data from m/s to knots.",
"_____no_output_____"
]
],
[
[
"# Define a function to calculate wind speed from u and v wind components\n\ndef calc_wspd(u, v):\n \"\"\"Computes wind speed from u and v components\"\"\" \n \n wspd = np.sqrt(u**2 + v**2) \n \n return wspd\n",
"_____no_output_____"
],
[
"# Use calc_wspd() function on uwnd & vwnd\n\nwspd = calc_wspd(uwnd, vwnd)\n",
"_____no_output_____"
],
[
"# Define a function to convert m/s to knots\n# Hint: 1 m/s = 1.9438445 knots\n\ndef to_knots(x):\n \n x_kt = x * 1.9438445\n \n return x_kt\n",
"_____no_output_____"
],
[
"# Convert wspd data to knots, save as separate array\n\nwspd_kt = to_knots(wspd)\n\nprint(wspd_kt)\n",
"[[ 19.01482381 25.49648009 32.78745376 ... 61.74856793 60.4400341\n 59.21274167]\n [ 17.96029277 24.06775504 31.07673251 ... 68.3190908 66.87967492\n 65.3504397 ]\n [ 16.78149734 22.54974541 29.22114385 ... 74.79678391 73.36355303\n 71.75952882]\n ...\n [101.25875455 101.15119681 100.91540436 ... 40.10411396 38.28553726\n 37.20120173]\n [101.73871083 101.62514342 101.42265795 ... 35.4757521 35.16105775\n 35.53265168]\n [104.33690552 104.27150023 103.96300562 ... 30.7666449 32.75753808\n 34.22663849]]\n"
]
],
[
[
"### Plotting with Cartopy\n\nMap 250-hPa height lines, isotachs (in knots), and wind vectors or barbs. ",
"_____no_output_____"
]
],
[
[
"# Set up map properties\n\n# Projection/Coordinate systems\ndatacrs = ccrs.PlateCarree() # data/source\nmapcrs = ccrs.PlateCarree() # map/destination\n\n# Map extent \nlonmin = lons.min() \nlonmax = lons.max() \nlatmin = lats.min() \nlatmax = lats.max()\n\n# Tickmark Locations\ndx = 10; dy = 10 \nxticks = np.arange(lonmin, lonmax+1, dx) # np.arange(start, stop, interval) returns 1d array \nyticks = np.arange(latmin, latmax+1, dy) # that ranges from `start` to `stop-1` by `interval`\nprint('xticks:', xticks)\nprint('yticks:', yticks)\n",
"xticks: [-145. -135. -125. -115. -105.]\nyticks: [22. 32. 42. 52.]\n"
]
],
[
[
"First, we need to create a basemap to plot our data on. In creating the basemap, we will set the map extent, draw lat/lon tickmarks, and add/customize map features such as coastlines and country borders. Next, use the `contour()` function to draw lines of 250-hPa geopotential heights. ",
"_____no_output_____"
]
],
[
[
"# Create figure\nfig = plt.figure(figsize=(11,8)) \n\n# Add plot axes \nax = fig.add_subplot(111, projection=mapcrs)\nax.set_extent([lonmin,lonmax,latmin,latmax], crs=mapcrs)\n\n# xticks (longitude tickmarks)\nax.set_xticks(xticks, crs=mapcrs) \nlon_formatter = LongitudeFormatter()\nax.xaxis.set_major_formatter(lon_formatter)\n# yticks (latitude tickmarks)\nax.set_yticks(yticks, crs=mapcrs)\nlat_formatter = LatitudeFormatter()\nax.yaxis.set_major_formatter(lat_formatter)\n# format tickmarks\nax.tick_params(direction='out', # draws ticks outside of plot (`out`,`in`,`inout)\n labelsize=8.5, # font size of ticklabel,\n length=5, # lenght of tickmark in points\n pad=2, # points between tickmark anmd label\n color='black')\n\n# Add map features\nax.add_feature(cfeature.LAND, facecolor='0.9') # color fill land gray\nax.add_feature(cfeature.COASTLINE, edgecolor='k', linewidth=1.0) # coastlines\nax.add_feature(cfeature.BORDERS, edgecolor='0.1', linewidth=0.7) # country borders\nax.add_feature(cfeature.STATES, edgecolor='0.1', linewidth=0.7) # state borders\n\n# Create arr of contour levels using np.arange(start,stop,interval)\nclevs_hgts = np.arange(8400,12800,120)\n#print(clevs_hgts)\n\n# Draw contour lines for geop heights \ncs = ax.contour(lons, lats, hgts, transform=datacrs, # first line= required\n levels=clevs_hgts, # contour levels\n colors='blue', # line color\n linewidths=1.2) # line thickness (default=1.0)\n \n# Add labels to contour lines\nplt.clabel(cs, fmt='%d', fontsize=9, inline_spacing=5) \n\n# # Show\nplt.show() \n",
"_____no_output_____"
]
],
[
[
"Create a function that will create and return a figure with a background map. This saves us from having to copy/paste lines 1-27 in the previous block each time we create a new map. ",
"_____no_output_____"
]
],
[
[
"def draw_basemap():\n\n # Create figure\n fig = plt.figure(figsize=(11,9)) \n\n # Add plot axes and draw basemap\n ax = fig.add_subplot(111, projection=mapcrs)\n ax.set_extent([lonmin,lonmax,latmin,latmax], crs=mapcrs)\n\n # xticks\n ax.set_xticks(xticks, crs=mapcrs) \n lon_formatter = LongitudeFormatter()\n ax.xaxis.set_major_formatter(lon_formatter)\n # yticks\n ax.set_yticks(yticks, crs=mapcrs)\n lat_formatter = LatitudeFormatter()\n ax.yaxis.set_major_formatter(lat_formatter)\n # tick params\n ax.tick_params(direction='out', labelsize=8.5, length=5, pad=2, color='black') \n\n # Map features\n ax.add_feature(cfeature.LAND, facecolor='0.9') \n ax.add_feature(cfeature.COASTLINE, edgecolor='k', linewidth=1.0)\n ax.add_feature(cfeature.BORDERS, edgecolor='0.1', linewidth=0.7)\n ax.add_feature(cfeature.STATES, edgecolor='0.1', linewidth=0.7)\n \n return fig, ax\n",
"_____no_output_____"
]
],
[
[
"Use your `draw_basemap` function to create a new figure and background map. Plot height contours, then use `contourf()` to plot filled contours for wind speed (knots). \n",
"_____no_output_____"
]
],
[
[
"# Draw basemap\nfig, ax = draw_basemap()\n\n# Geopotential Heights (contour lines)\nclevs_hgts = np.arange(8400,12800,120)\ncs = ax.contour(lons, lats, hgts, transform=datacrs,\n levels=clevs_hgts, # contour levels\n colors='b', # line color\n linewidths=1.2) # line thickness\n \n# Add labels to contour lines\nplt.clabel(cs, fmt='%d',fontsize=8.5, inline_spacing=5)\n\n# Wind speed - contour fill\nclevs_wspd = np.arange(70,121,10)\ncf = ax.contourf(lons, lats, wspd_kt, transform=datacrs,\n levels=clevs_wspd, \n cmap='BuPu', # colormap \n extend='max', \n alpha=0.8) # transparency (0=transparent, 1=opaque)\n\n\n# show\nplt.show()\n",
"_____no_output_____"
]
],
[
[
"Add wind vectors using `quiver()`",
"_____no_output_____"
]
],
[
[
"# Draw basemap\nfig, ax = draw_basemap()\n\n# Geopotenital height lines\nclevs_hgts = np.arange(840,1280,12)\ncs = ax.contour(lons, lats, hgts/10., transform=datacrs,\n levels=clevs_hgts,\n colors='b', # line color\n linewidths=1.2) # line thickness\n \n# Add labels to contour lines\nplt.clabel(cs, fmt='%d',fontsize=9, inline_spacing=5)\n\n# Wind speed - contour fill\nclevs_wspd = np.arange(70,131,10)\ncf = ax.contourf(lons, lats, wspd_kt, transform=datacrs,\n levels=clevs_wspd, \n cmap='BuPu', \n extend='max', # use if max data value \n alpha=0.8) # transparency (0=transparent, 1=opaque)\n\n# Wind vectors \nax.quiver(lons, lats, uwnd, vwnd, transform=datacrs, \n color='k', \n pivot='middle',\n regrid_shape=12) # increasing regrid_shape increases the number/density of vectors \n\n# show\nplt.show()\n",
"_____no_output_____"
]
],
[
[
"Plot barbs instead of vectors using `barbs()`",
"_____no_output_____"
]
],
[
[
"# Draw basemap\nfig, ax = draw_basemap()\n\n# Geopotenital height lines\nclevs_hgts = np.arange(840,1280,12)\ncs = ax.contour(lons, lats, hgts/10., transform=datacrs,\n levels=clevs_hgts,\n colors='b', # line color\n linewidths=1.25) # line thickness\n \n# Add labels to contour lines\nplt.clabel(cs, fmt='%d',fontsize=9, inline_spacing=5)\n\n# Wind speed - contour fill\nclevs_wspd = np.arange(70,131,10)\ncf = ax.contourf(lons, lats, wspd_kt, transform=datacrs,\n levels=clevs_wspd, \n cmap='BuPu', \n extend='max',\n alpha=0.8) # transparency (0=transparent, 1=opaque)\n\n# Wind barbs\nax.barbs(lons, lats, uwnd, vwnd, transform=datacrs, # uses the same args as quiver\n color='k', regrid_shape=12, pivot='middle') \n\n# show\nplt.show()\n",
"_____no_output_____"
]
],
[
[
"Add plot elements such as a colorbar and title. Option to save figure.",
"_____no_output_____"
]
],
[
[
"# Draw basemap\nfig, ax = draw_basemap()\n\n# Geopotenital height lines\nclevs_hgts = np.arange(8400,12800,120)\ncs = ax.contour(lons, lats, hgts, transform=datacrs,\n levels=clevs_hgts,\n colors='b', # line color\n linewidths=1.2) # line thickness \n# Add labels to contour lines\nplt.clabel(cs, fmt='%d',fontsize=8.5, inline_spacing=5)\n\n# Wind speed - contour fill\nclevs_wspd = np.arange(70,131,10)\ncf = ax.contourf(lons, lats, wspd_kt, transform=datacrs,\n levels=clevs_wspd, \n cmap='BuPu', \n extend='max',\n alpha=0.8) # transparency (0=transparent, 1=opaque)\n\n# Wind barbs\nax.barbs(lons, lats, uwnd, vwnd, transform=datacrs, # uses the same args as quiver\n color='k', regrid_shape=12, pivot='middle') \n\n# Add colorbar\ncb = plt.colorbar(cf, orientation='vertical', # 'horizontal' or 'vertical'\n shrink=0.7, pad=0.03) # fraction to shrink cb by; pad= space between cb and plot\ncb.set_label('knots')\n\n# Plot title\ntitlestring = f\"{plev}-hPa Hgts/Wind\" # uses new f-string formatting\nax.set_title(titlestring, loc='center',fontsize=13) # loc: {'center','right','left'}\n\n# Save figure\noutfile = 'map-250hPa.png'\nplt.savefig(outfile, \n bbox_inches='tight', # trims excess whitespace from around figure\n dpi=300) # resolution in dots per inch\n\n# show\nplt.show()\n",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb8d7fb2ac10fd513b5ffaa7eff07e70c3bca706
| 13,889 |
ipynb
|
Jupyter Notebook
|
notebooks/insert_allWISE.ipynb
|
vbrinnel/extcats
|
fd37ad19c8b613cca6410c303416d26210392a02
|
[
"MIT"
] | 2 |
2019-05-07T12:57:49.000Z
|
2020-04-07T14:38:54.000Z
|
notebooks/insert_allWISE.ipynb
|
vbrinnel/extcats
|
fd37ad19c8b613cca6410c303416d26210392a02
|
[
"MIT"
] | 5 |
2018-03-06T20:14:54.000Z
|
2018-11-01T13:59:03.000Z
|
notebooks/insert_allWISE.ipynb
|
vbrinnel/extcats
|
fd37ad19c8b613cca6410c303416d26210392a02
|
[
"MIT"
] | 5 |
2018-02-28T15:25:58.000Z
|
2021-02-01T10:33:37.000Z
| 41.708709 | 1,254 | 0.573115 |
[
[
[
"# Add external catalog for source matching: allWISE catalog\n\nThis notebook will create a dabase containing the allWISE all-sky mid-infrared catalog. As the catalogs grows (the allWISE catalog we are inserting contains of the order of hundreds of millions sources), using an index on the geoJSON corrdinate type to support the queries becomes unpractical, as such an index does not compress well. In this case, and healpix based indexing offers a good compromise. We will use an healpix grid of order 16, which has a resolution of ~ 3 arcseconds, simlar to the FWHM of ZTF images. \n\nReferences, data access, and documentation on the catalog can be found at:\n\nhttp://wise2.ipac.caltech.edu/docs/release/allwise/\n\nhttp://irsa.ipac.caltech.edu/data/download/wise-allwise/\n\nThis notebook is straight to the point, more like an actual piece of code than a demo. For an explanation of the various steps needed in the see the 'insert_example' notebook in this same folder.",
"_____no_output_____"
],
[
"## 1) Inserting:",
"_____no_output_____"
]
],
[
[
"import numpy as np\nfrom healpy import ang2pix\nfrom extcats import CatalogPusher\n\n# build the pusher object and point it to the raw files.\nwisep = CatalogPusher.CatalogPusher(\n catalog_name = 'wise',\n data_source = '../testdata/AllWISE/',\n file_type = \".bz2\")\n\n\n# read column names and types from schema file\nschema_file = \"../testdata/AllWISE/wise-allwise-cat-schema.txt\"\nnames, types = [], {}\nwith open(schema_file) as schema:\n for l in schema:\n if \"#\" in l or (not l.strip()):\n continue\n name, dtype = zip(\n [p.strip() for p in l.strip().split(\" \") if not p in [\"\"]])\n name, dtype = name[0], dtype[0]\n #print (name, dtype)\n names.append(name)\n # convert the data type\n if \"char\" in dtype:\n types[name] = str\n elif \"decimal\" in dtype:\n types[name] = np.float64\n elif \"serial\" in dtype or \"integer\" in dtype:\n types[name] = int\n elif \"smallfloat\" in dtype:\n types[name] = np.float16\n elif \"smallint\" in dtype:\n types[name] = np.int16\n elif dtype == \"int8\":\n types[name] = np.int8\n else:\n print(\"unknown data type: %s\"%dtype)\n\n# select the columns you want to use.\nuse_cols = []\nselect = [\"Basic Position and Identification Information\", \n \"Primary Photometric Information\", \n \"Measurement Quality and Source Reliability Information\",\n \"2MASS PSC Association Information\"]\nwith open(schema_file) as schema:\n blocks = schema.read().split(\"#\")\n for block in blocks:\n if any([k in block for k in select]):\n for l in block.split(\"\\n\")[1:]:\n if \"#\" in l or (not l.strip()):\n continue\n name, dtype = zip(\n [p.strip() for p in l.strip().split(\" \") if not p in [\"\"]])\n use_cols.append(name[0])\nprint(\"we will be using %d columns out of %d\"%(len(use_cols), len(names)))\n\n# now assign the reader to the catalog pusher object\nimport pandas as pd\nwisep.assign_file_reader(\n reader_func = pd.read_csv, \n read_chunks = True,\n names = names,\n usecols = lambda x : x in use_cols,\n #dtype = types, #this mess up with NaN values\n chunksize=5000,\n header=None,\n engine='c',\n sep='|',\n na_values = 'nnnn')\n\n\n# define the dictionary modifier that will act on the single entries\ndef modifier(srcdict):\n srcdict['hpxid_16'] = int(\n ang2pix(2**16, srcdict['ra'], srcdict['dec'], lonlat = True, nest = True))\n #srcdict['_id'] = srcdict.pop('source_id') doesn't work, seems it is not unique\n return srcdict\nwisep.assign_dict_modifier(modifier)\n\n\n# finally push it in the databse\nwisep.push_to_db(\n coll_name = 'srcs', \n index_on = \"hpxid_16\",\n overwrite_coll = True, \n append_to_coll = False)\n\n\n# if needed print extensive info on database\n#wisep.info()",
"INFO:extcats.CatalogPusher:found 1 files for catalog wise in data source: ['../testdata/AllWISE/']\nINFO:extcats.CatalogPusher:checking raw files for existence and consistency..\nINFO:extcats.CatalogPusher:all files exists and have consistent type.\nINFO:extcats.CatalogPusher:file reader read_csv assigned to pusher.\nINFO:extcats.CatalogPusher:source document modifer modifier assigned to pusher.\nINFO:extcats.CatalogPusher:using mongo client at localhost:27017\nINFO:extcats.CatalogPusher:connecting to database wise. Here some stats:\nINFO:extcats.CatalogPusher:{\n \"db\": \"wise\",\n \"collections\": 1,\n \"views\": 0,\n \"objects\": 15575416,\n \"avgObjSize\": 1284.6681279652498,\n \"dataSize\": 20009240515.0,\n \"storageSize\": 8750858240.0,\n \"numExtents\": 0,\n \"indexes\": 2,\n \"indexSize\": 469798912.0,\n \"fsUsedSize\": 220678574080.0,\n \"fsTotalSize\": 231446335488.0,\n \"ok\": 1.0\n}\nWARNING:extcats.CatalogPusher:overwrite_coll asserted: collection srcs will be dropped.\nINFO:extcats.CatalogPusher:collection has the following indexes: _id_, hpxid_16_1\nINFO:extcats.CatalogPusher:inserting ../testdata/AllWISE/wise-allwise-cat-part33.bz2 in collection wise.srcs.\n"
]
],
[
[
"## 2) Testing the catalog\n\nAt this stage, a simple test is run on the database, consisting in crossmatching with a set of randomly distributed points.",
"_____no_output_____"
]
],
[
[
"# now test the database for query performances. We use \n# a sample of randomly distributed points on a sphere\n# as targets. \n\n# define the funtion to test coordinate based queries:\nfrom healpy import ang2pix, get_all_neighbours\nfrom astropy.table import Table\nfrom astropy.coordinates import SkyCoord\n\nreturn_fields = ['designation', 'ra', 'dec']\nproject = {}\nfor field in return_fields: project[field] = 1\nprint (project)\n\n\nhp_order, rs_arcsec = 16, 30.\ndef test_query(ra, dec, coll):\n \"\"\"query collection for points within rs of target ra, dec.\n The results as returned as an astropy Table.\"\"\"\n \n # find the index of the target pixel and its neighbours \n target_pix = int( ang2pix(2**hp_order, ra, dec, nest = True, lonlat = True) )\n neighbs = get_all_neighbours(2**hp_order, ra, dec, nest = True, lonlat = True)\n\n # remove non-existing neigbours (in case of E/W/N/S) and add center pixel\n pix_group = [int(pix_id) for pix_id in neighbs if pix_id != -1] + [target_pix]\n \n # query the database for sources in these pixels\n qfilter = { 'hpxid_%d'%hp_order: { '$in': pix_group } }\n qresults = [o for o in coll.find(qfilter)]\n if len(qresults)==0:\n return None\n \n # then use astropy to find the closest match\n tab = Table(qresults)\n target = SkyCoord(ra, dec, unit = 'deg')\n matches_pos = SkyCoord(tab['ra'], tab['dec'], unit = 'deg')\n d2t = target.separation(matches_pos).arcsecond\n match_id = np.argmin(d2t)\n\n # if it's too far away don't use it\n if d2t[match_id]>rs_arcsec:\n return None\n return tab[match_id]\n\n# run the test\nwisep.run_test(test_query, npoints = 10000)\n",
"INFO:extcats.CatalogPusher:running test queries using 10000 random points\n 1%| | 54/10000 [00:00<00:18, 536.21it/s]"
]
],
[
[
"# 3) Adding metadata\n\nOnce the database is set up and the query performance are satisfactory, metadata describing the catalog content, contact person, and query strategies have to be added to the catalog database. If presents, the keys and parameters for the healpix partitioning of the sources are also to be given, as well as the name of the compound geoJSON/legacy pair entry in the documents.\n\nThis information will be added into the 'metadata' collection of the database which will be accessed by the CatalogQuery. The metadata will be stored in a dedicated collection so that the database containig a given catalog will have two collections:\n - db['srcs'] : contains the sources.\n - db['meta'] : describes the catalog.",
"_____no_output_____"
]
],
[
[
"mqp.healpix_meta(healpix_id_key = 'hpxid_16', order = 16, is_indexed = True, nest = True)\nmqp.science_meta(\n contact = 'C. Norris', \n email = '[email protected]', \n description = 'allWISE infrared catalog',\n reference = 'http://wise2.ipac.caltech.edu/docs/release/allwise/')",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb8d82beb8429c86c03b52bb1005be09b8c3f8c1
| 32,327 |
ipynb
|
Jupyter Notebook
|
.ipynb_checkpoints/model_1-checkpoint.ipynb
|
CrisDimacali/machine-learning-challenge
|
1d47aa1114d7345d97ef78912a6e6e41cf53d388
|
[
"ADSL"
] | null | null | null |
.ipynb_checkpoints/model_1-checkpoint.ipynb
|
CrisDimacali/machine-learning-challenge
|
1d47aa1114d7345d97ef78912a6e6e41cf53d388
|
[
"ADSL"
] | null | null | null |
.ipynb_checkpoints/model_1-checkpoint.ipynb
|
CrisDimacali/machine-learning-challenge
|
1d47aa1114d7345d97ef78912a6e6e41cf53d388
|
[
"ADSL"
] | null | null | null | 42.368283 | 1,445 | 0.52965 |
[
[
[
"# Update sklearn to prevent version mismatches\n!pip install sklearn --upgrade",
"Requirement already up-to-date: sklearn in c:\\users\\criselda\\anaconda3\\envs\\myenv\\lib\\site-packages (0.0)\nRequirement already satisfied, skipping upgrade: scikit-learn in c:\\users\\criselda\\anaconda3\\envs\\myenv\\lib\\site-packages (from sklearn) (0.23.1)\nRequirement already satisfied, skipping upgrade: numpy>=1.13.3 in c:\\users\\criselda\\anaconda3\\envs\\myenv\\lib\\site-packages (from scikit-learn->sklearn) (1.19.1)\nRequirement already satisfied, skipping upgrade: joblib>=0.11 in c:\\users\\criselda\\anaconda3\\envs\\myenv\\lib\\site-packages (from scikit-learn->sklearn) (0.16.0)\nRequirement already satisfied, skipping upgrade: threadpoolctl>=2.0.0 in c:\\users\\criselda\\anaconda3\\envs\\myenv\\lib\\site-packages (from scikit-learn->sklearn) (2.1.0)\nRequirement already satisfied, skipping upgrade: scipy>=0.19.1 in c:\\users\\criselda\\anaconda3\\envs\\myenv\\lib\\site-packages (from scikit-learn->sklearn) (1.5.2)\n"
],
[
"# install joblib. This will be used to save your model. \n# Restart your kernel after installing \n!pip install joblib",
"Requirement already satisfied: joblib in c:\\users\\criselda\\anaconda3\\envs\\myenv\\lib\\site-packages (0.16.0)\n"
],
[
"import pandas as pd",
"_____no_output_____"
]
],
[
[
"# Read the CSV and Perform Basic Data Cleaning",
"_____no_output_____"
]
],
[
[
"df = pd.read_csv(\"exoplanet_data.csv\")\n# Drop the null columns where all values are null\ndf = df.dropna(axis='columns', how='all')\n# Drop the null rows\ndf = df.dropna()\ndf.head()",
"_____no_output_____"
]
],
[
[
"# Select your features (columns)",
"_____no_output_____"
]
],
[
[
"# Set features. This will also be used as your x values.\nselected_features = df[['koi_period', 'koi_period_err1', 'koi_time0bk', 'koi_slogg', 'koi_srad']]",
"_____no_output_____"
]
],
[
[
"# Create a Train Test Split\n\nUse `koi_disposition` for the y values",
"_____no_output_____"
]
],
[
[
"#import dependencies\nfrom sklearn.model_selection import train_test_split\n\n#assign x and y values\nX = selected_features\ny = df[\"koi_disposition\"]\n\n#split training and testing data\nX_train, X_test, y_train, y_test = train_test_split(X, y, random_state=1, stratify=y)",
"_____no_output_____"
],
[
"X_train.head()",
"_____no_output_____"
],
[
"#assign x and y values\nX = selected_features\ny = df[\"koi_disposition\"]\n\n#split training and testing data\nX_train, X_test, y_train, y_test = train_test_split(X, y, random_state=1, stratify=y)",
"_____no_output_____"
]
],
[
[
"# Pre-processing\n\nScale the data using the MinMaxScaler and perform some feature selection",
"_____no_output_____"
]
],
[
[
"# !pip install --upgrade tensorflow",
"Collecting tensorflow\n Downloading tensorflow-2.3.0-cp36-cp36m-win_amd64.whl (342.5 MB)\nCollecting astunparse==1.6.3\n Downloading astunparse-1.6.3-py2.py3-none-any.whl (12 kB)\nRequirement already satisfied, skipping upgrade: six>=1.12.0 in c:\\users\\criselda\\anaconda3\\envs\\myenv\\lib\\site-packages (from tensorflow) (1.15.0)\nRequirement already satisfied, skipping upgrade: wheel>=0.26 in c:\\users\\criselda\\anaconda3\\envs\\myenv\\lib\\site-packages (from tensorflow) (0.34.2)\nCollecting tensorflow-estimator<2.4.0,>=2.3.0\n Downloading tensorflow_estimator-2.3.0-py2.py3-none-any.whl (459 kB)\nRequirement already satisfied, skipping upgrade: wrapt>=1.11.1 in c:\\users\\criselda\\anaconda3\\envs\\myenv\\lib\\site-packages (from tensorflow) (1.12.1)\nRequirement already satisfied, skipping upgrade: scipy==1.4.1 in c:\\users\\criselda\\anaconda3\\envs\\myenv\\lib\\site-packages (from tensorflow) (1.4.1)\nCollecting gast==0.3.3\n Downloading gast-0.3.3-py2.py3-none-any.whl (9.7 kB)\nCollecting tensorboard<3,>=2.3.0\n Downloading tensorboard-2.3.0-py3-none-any.whl (6.8 MB)\nRequirement already satisfied, skipping upgrade: termcolor>=1.1.0 in c:\\users\\criselda\\anaconda3\\envs\\myenv\\lib\\site-packages (from tensorflow) (1.1.0)\nRequirement already satisfied, skipping upgrade: protobuf>=3.9.2 in c:\\users\\criselda\\anaconda3\\envs\\myenv\\lib\\site-packages (from tensorflow) (3.12.3)\nRequirement already satisfied, skipping upgrade: google-pasta>=0.1.8 in c:\\users\\criselda\\anaconda3\\envs\\myenv\\lib\\site-packages (from tensorflow) (0.2.0)\nRequirement already satisfied, skipping upgrade: opt-einsum>=2.3.2 in c:\\users\\criselda\\anaconda3\\envs\\myenv\\lib\\site-packages (from tensorflow) (3.3.0)\nCollecting numpy<1.19.0,>=1.16.0\n Downloading numpy-1.18.5-cp36-cp36m-win_amd64.whl (12.7 MB)\nRequirement already satisfied, skipping upgrade: grpcio>=1.8.6 in c:\\users\\criselda\\anaconda3\\envs\\myenv\\lib\\site-packages (from tensorflow) (1.30.0)\nRequirement already satisfied, skipping upgrade: h5py<2.11.0,>=2.10.0 in c:\\users\\criselda\\anaconda3\\envs\\myenv\\lib\\site-packages (from tensorflow) (2.10.0)\nRequirement already satisfied, skipping upgrade: keras-preprocessing<1.2,>=1.1.1 in c:\\users\\criselda\\anaconda3\\envs\\myenv\\lib\\site-packages (from tensorflow) (1.1.2)\nRequirement already satisfied, skipping upgrade: absl-py>=0.7.0 in c:\\users\\criselda\\anaconda3\\envs\\myenv\\lib\\site-packages (from tensorflow) (0.9.0)\nRequirement already satisfied, skipping upgrade: google-auth-oauthlib<0.5,>=0.4.1 in c:\\users\\criselda\\anaconda3\\envs\\myenv\\lib\\site-packages (from tensorboard<3,>=2.3.0->tensorflow) (0.4.1)\nRequirement already satisfied, skipping upgrade: google-auth<2,>=1.6.3 in c:\\users\\criselda\\anaconda3\\envs\\myenv\\lib\\site-packages (from tensorboard<3,>=2.3.0->tensorflow) (1.17.2)\nRequirement already satisfied, skipping upgrade: markdown>=2.6.8 in c:\\users\\criselda\\anaconda3\\envs\\myenv\\lib\\site-packages (from tensorboard<3,>=2.3.0->tensorflow) (3.2.2)\nRequirement already satisfied, skipping upgrade: setuptools>=41.0.0 in c:\\users\\criselda\\anaconda3\\envs\\myenv\\lib\\site-packages (from tensorboard<3,>=2.3.0->tensorflow) (49.2.0.post20200714)\nRequirement already satisfied, skipping upgrade: werkzeug>=0.11.15 in c:\\users\\criselda\\anaconda3\\envs\\myenv\\lib\\site-packages (from tensorboard<3,>=2.3.0->tensorflow) (1.0.1)\nRequirement already satisfied, skipping upgrade: tensorboard-plugin-wit>=1.6.0 in c:\\users\\criselda\\anaconda3\\envs\\myenv\\lib\\site-packages (from tensorboard<3,>=2.3.0->tensorflow) (1.6.0)\nRequirement already satisfied, skipping upgrade: requests<3,>=2.21.0 in c:\\users\\criselda\\anaconda3\\envs\\myenv\\lib\\site-packages (from tensorboard<3,>=2.3.0->tensorflow) (2.24.0)\nRequirement already satisfied, skipping upgrade: requests-oauthlib>=0.7.0 in c:\\users\\criselda\\anaconda3\\envs\\myenv\\lib\\site-packages (from google-auth-oauthlib<0.5,>=0.4.1->tensorboard<3,>=2.3.0->tensorflow) (1.3.0)\nRequirement already satisfied, skipping upgrade: cachetools<5.0,>=2.0.0 in c:\\users\\criselda\\anaconda3\\envs\\myenv\\lib\\site-packages (from google-auth<2,>=1.6.3->tensorboard<3,>=2.3.0->tensorflow) (4.1.0)\nRequirement already satisfied, skipping upgrade: rsa<5,>=3.1.4; python_version >= \"3\" in c:\\users\\criselda\\anaconda3\\envs\\myenv\\lib\\site-packages (from google-auth<2,>=1.6.3->tensorboard<3,>=2.3.0->tensorflow) (4.0)\nRequirement already satisfied, skipping upgrade: pyasn1-modules>=0.2.1 in c:\\users\\criselda\\anaconda3\\envs\\myenv\\lib\\site-packages (from google-auth<2,>=1.6.3->tensorboard<3,>=2.3.0->tensorflow) (0.2.7)\nRequirement already satisfied, skipping upgrade: importlib-metadata; python_version < \"3.8\" in c:\\users\\criselda\\anaconda3\\envs\\myenv\\lib\\site-packages (from markdown>=2.6.8->tensorboard<3,>=2.3.0->tensorflow) (1.7.0)\nRequirement already satisfied, skipping upgrade: chardet<4,>=3.0.2 in c:\\users\\criselda\\anaconda3\\envs\\myenv\\lib\\site-packages (from requests<3,>=2.21.0->tensorboard<3,>=2.3.0->tensorflow) (3.0.4)\nRequirement already satisfied, skipping upgrade: urllib3!=1.25.0,!=1.25.1,<1.26,>=1.21.1 in c:\\users\\criselda\\anaconda3\\envs\\myenv\\lib\\site-packages (from requests<3,>=2.21.0->tensorboard<3,>=2.3.0->tensorflow) (1.25.9)\nRequirement already satisfied, skipping upgrade: certifi>=2017.4.17 in c:\\users\\criselda\\anaconda3\\envs\\myenv\\lib\\site-packages (from requests<3,>=2.21.0->tensorboard<3,>=2.3.0->tensorflow) (2020.6.20)\nRequirement already satisfied, skipping upgrade: idna<3,>=2.5 in c:\\users\\criselda\\anaconda3\\envs\\myenv\\lib\\site-packages (from requests<3,>=2.21.0->tensorboard<3,>=2.3.0->tensorflow) (2.10)\nRequirement already satisfied, skipping upgrade: oauthlib>=3.0.0 in c:\\users\\criselda\\anaconda3\\envs\\myenv\\lib\\site-packages (from requests-oauthlib>=0.7.0->google-auth-oauthlib<0.5,>=0.4.1->tensorboard<3,>=2.3.0->tensorflow) (3.1.0)\nRequirement already satisfied, skipping upgrade: pyasn1>=0.1.3 in c:\\users\\criselda\\anaconda3\\envs\\myenv\\lib\\site-packages (from rsa<5,>=3.1.4; python_version >= \"3\"->google-auth<2,>=1.6.3->tensorboard<3,>=2.3.0->tensorflow) (0.4.8)\nRequirement already satisfied, skipping upgrade: zipp>=0.5 in c:\\users\\criselda\\anaconda3\\envs\\myenv\\lib\\site-packages (from importlib-metadata; python_version < \"3.8\"->markdown>=2.6.8->tensorboard<3,>=2.3.0->tensorflow) (3.1.0)\nInstalling collected packages: astunparse, tensorflow-estimator, gast, numpy, tensorboard, tensorflow\n Attempting uninstall: tensorflow-estimator\n Found existing installation: tensorflow-estimator 2.1.0\n Uninstalling tensorflow-estimator-2.1.0:\n Successfully uninstalled tensorflow-estimator-2.1.0\n Attempting uninstall: gast\n Found existing installation: gast 0.2.2\n Uninstalling gast-0.2.2:\n Successfully uninstalled gast-0.2.2\n Attempting uninstall: numpy\n Found existing installation: numpy 1.19.1\n Uninstalling numpy-1.19.1:\n Successfully uninstalled numpy-1.19.1\n Attempting uninstall: tensorboard\n Found existing installation: tensorboard 2.1.1\n Uninstalling tensorboard-2.1.1:\n Successfully uninstalled tensorboard-2.1.1\n Attempting uninstall: tensorflow\n Found existing installation: tensorflow 2.1.0\n Uninstalling tensorflow-2.1.0:\n Successfully uninstalled tensorflow-2.1.0\nSuccessfully installed astunparse-1.6.3 gast-0.3.3 numpy-1.18.5 tensorboard-2.3.0 tensorflow-2.3.0 tensorflow-estimator-2.3.0\n"
],
[
"!conda install tensorflow",
"Collecting package metadata (current_repodata.json): ...working... done\nSolving environment: ...working... done\n\n# All requested packages already installed.\n\n"
],
[
"import tensorflow",
"_____no_output_____"
],
[
"# Scale your data\nfrom sklearn.preprocessing import LabelEncoder, MinMaxScaler\nfrom tensorflow.keras.utils import to_categorical\nX_train, X_test, y_train, y_test = train_test_split(X, y, random_state=1, stratify=y)",
"_____no_output_____"
],
[
"X_scaler = MinMaxScaler().fit(X_train)\nX_train_scaled = X_scaler.transform(X_train)\nX_test_scaled = X_scaler.transform(X_test)",
"_____no_output_____"
],
[
"# Step 1: Label-encode data set\nlabel_encoder = LabelEncoder()\nlabel_encoder.fit(y_train)\nencoded_y_train = label_encoder.transform(y_train)\nencoded_y_test = label_encoder.transform(y_test)",
"_____no_output_____"
],
[
"# Step 2: Convert encoded labels to one-hot-encoding\ny_train_categorical = to_categorical(encoded_y_train)\ny_test_categorical = to_categorical(encoded_y_test)",
"_____no_output_____"
]
],
[
[
"# Train the Model\n\n",
"_____no_output_____"
]
],
[
[
"print(f\"Training Data Score: {model2.score(X_train_scaled, y_train)}\")\nprint(f\"Testing Data Score: {model2.score(X_test_scaled, y_test)}\")",
"_____no_output_____"
]
],
[
[
"# Hyperparameter Tuning\n\nUse `GridSearchCV` to tune the model's parameters",
"_____no_output_____"
]
],
[
[
"# Create the GridSearchCV model",
"_____no_output_____"
],
[
"# Train the model with GridSearch",
"_____no_output_____"
],
[
"print(grid2.best_params_)\nprint(grid2.best_score_)",
"_____no_output_____"
]
],
[
[
"# Save the Model",
"_____no_output_____"
]
],
[
[
"# save your model by updating \"your_name\" with your name\n# and \"your_model\" with your model variable\n# be sure to turn this in to BCS\n# if joblib fails to import, try running the command to install in terminal/git-bash\nimport joblib\nfilename = 'your_name.sav'\njoblib.dump(your_model, filename)",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb8da4fb93f40b863f9512d1f037ace2ce9ce41d
| 23,224 |
ipynb
|
Jupyter Notebook
|
notebooks/6_AB_check_raw_processed.ipynb
|
AlessandroVol23/kdd-cup-2019
|
7f140d1d6213dc0d05d07a2c8bff9fe949b72ed8
|
[
"MIT"
] | 10 |
2019-07-24T19:14:59.000Z
|
2022-01-04T06:35:27.000Z
|
notebooks/6_AB_check_raw_processed.ipynb
|
AlessandroVol23/kdd-cup-2019
|
7f140d1d6213dc0d05d07a2c8bff9fe949b72ed8
|
[
"MIT"
] | 2 |
2020-07-02T15:53:45.000Z
|
2020-07-29T04:40:33.000Z
|
notebooks/6_AB_check_raw_processed.ipynb
|
AlessandroVol23/kdd-cup-2019
|
7f140d1d6213dc0d05d07a2c8bff9fe949b72ed8
|
[
"MIT"
] | 8 |
2019-07-21T20:05:09.000Z
|
2021-12-28T03:03:40.000Z
| 23.434914 | 100 | 0.354935 |
[
[
[
"import pandas as pd",
"_____no_output_____"
],
[
"feattype = 'all'\ntraindf = pd.read_pickle('../data/processed_'+feattype+'/train_'+feattype+'_last.pickle')\ntraindf2 = pd.read_pickle('../data/processed_'+feattype+'/train_'+feattype+'_first.pickle')\ntraindf1 = pd.read_pickle('../data/processed_'+feattype+'/train_'+feattype+'_row.pickle')",
"_____no_output_____"
],
[
"traindf.shape, traindf1.shape",
"_____no_output_____"
],
[
"traindf.tail(5)",
"_____no_output_____"
],
[
"list(traindf.columns)",
"_____no_output_____"
],
[
"traindf = traindf.drop('click_time', axis=1)\ntraindf1 = traindf1.drop('click_time', axis=1)\ntraindf2 = traindf2.drop('click_time', axis=1)\n\ntraindf = traindf.drop('weather', axis=1)\ntraindf1 = traindf1.drop('weather', axis=1)\ntraindf2 = traindf2.drop('weather', axis=1)",
"_____no_output_____"
],
[
"traindf.weather.sample(20)",
"_____no_output_____"
],
[
"traincols = traindf.columns.tolist()\ntestcols = testdf.columns.tolist()\ntraincols1 = traindf1.columns.tolist()\ntestcols1 = testdf1.columns.tolist()",
"_____no_output_____"
],
[
"set(traincols) - set(testcols)",
"_____no_output_____"
],
[
"set(traincols1) - set(traincols)",
"_____no_output_____"
],
[
"traincols",
"_____no_output_____"
],
[
"type = 'row'\ntraindf = pd.read_pickle('../data/processed_all/train_all_'+type+'.pickle')\ntestdf = pd.read_pickle('../data/processed_all/test_all_'+type+'.pickle')",
"_____no_output_____"
],
[
"traindf.shape, testdf.shape",
"_____no_output_____"
],
[
"traindf.columns.to_list()",
"_____no_output_____"
],
[
"df_profiles = pd.read_csv('../data/raw/data_set_phase1/profiles.csv')\nprint(traindf1.shape)\ntraindf1 = pd.merge(traindf1, df_profiles, how='outer')",
"(2101060, 15)\n"
],
[
"traindf1 = traindf1[pd.notnull(traindf1['o_long'])]\nprint(traindf1.shape)",
"(2101060, 81)\n"
],
[
"testdf1 = pd.merge(testdf1, df_profiles, how='outer')",
"_____no_output_____"
],
[
"testdf1 = testdf1[pd.notnull(testdf1['o_long'])]\nprint(testdf1.shape)",
"(429803, 79)\n"
],
[
"traindf.to_pickle('../data/processed_all/train_all_last.pickle')",
"_____no_output_____"
],
[
"traindf1.to_pickle('../data/processed_all/train_all_first.pickle')",
"_____no_output_____"
],
[
"traindf2.to_pickle('../data/processed_all/train_all_row.pickle')",
"_____no_output_____"
],
[
"testdf1.to_pickle('../data/processed_all/test_all_row.pickle')",
"_____no_output_____"
],
[
"traincols = traindf.columns.tolist()\ntestcols = testdf.columns.tolist()\ntraincols1 = traindf1.columns.tolist()\ntestcols1 = testdf1.columns.tolist()",
"_____no_output_____"
],
[
"len(set(traincols) - set(traincols1))",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb8db96d7fcb10131af22c618c0b293fd39320bb
| 167,952 |
ipynb
|
Jupyter Notebook
|
PA3/PA3.ipynb
|
Magicicesea/Python-Machine-Learning-algorithm-implement
|
8a766932d0e65cbf65ad695f9d2c6a92857b41a3
|
[
"MIT"
] | null | null | null |
PA3/PA3.ipynb
|
Magicicesea/Python-Machine-Learning-algorithm-implement
|
8a766932d0e65cbf65ad695f9d2c6a92857b41a3
|
[
"MIT"
] | null | null | null |
PA3/PA3.ipynb
|
Magicicesea/Python-Machine-Learning-algorithm-implement
|
8a766932d0e65cbf65ad695f9d2c6a92857b41a3
|
[
"MIT"
] | null | null | null | 358.10661 | 150,804 | 0.922341 |
[
[
[
"# Homework #3 Programming Assignment\nCSCI567, Spring 2019<br>Victor Adamchik<br>**Due: 11:59 pm, March 3rd 2019**\n\n\n### Before you start: \nOn Vocareum, when you submit your homework, it takes around 5-6 minutes to run the grading scripts and evaluate your code. So, please be patient regarding the same.<br>\n\n\n## Office Hour for Project Assignment 3\nOffice hours for Anirudh: <br>\nFebruary 15th, 2pm - 3pm<br>\nFebruary 22nd, 2pm - 3pm<br>\nMarch 1st, 2pm - 4pm<br>\n<br>\nOffice hours for Piyush:<br>\nFebruary 13th, 2pm - 3pm<br>\nFebruary 21nd, 2pm - 3pm<br>\nMarch 4th, 2pm - 4pm<br>\n<br>\nAlso, you can post your question on Piazza under pa-3 folder. We will try our best to answer all questions as soon as possible. Please make sure you read previous posts before creating a new post in case your question has been answered before. However, if you have any urgent issue, please feel free to send an email to both of us. <br>\nAnirudh, Kashi: [email protected]<br>\nPiyush, Umate: [email protected]<br>\n\n\n\n## Problem 1 Neural Networks (40 points)\n\n<br><br>\nFor this Assignment, you are asked to implement neural networks. We will be using this neural network to classify MNIST database of handwritten digits (0-9). The architecture of the multi-layer perceptron (MLP, just another term for fully connected feedforward networks we discussed in the lecture) you will be implementing is shown in figure 1. Following MLP is designed for a K-class classification problem. \n\nLet $(x\\in\\mathbb{R}^D, y\\in\\{1,2,\\cdots,K\\})$ be a labeled instance, such an MLP performs the following computations.\n<br><br><br><br>\n$$\n\\begin{align}\n \\textbf{input features}: \\hspace{15pt} & x \\in \\mathbb{R}^D \\\\\n \\textbf{linear}^{(1)}: \\hspace{15pt} & u = W^{(1)}x + b^{(1)} \\hspace{2em}, W^{(1)} \\in \\mathbb{R}^{M\\times D} \\text{ and } b^{(1)} \\in \\mathbb{R}^{M} \\label{linear_forward}\\\\\n \\textbf{tanh}:\\hspace{15pt} & h =\\cfrac{2}{1+e^{-2u}}-1 \\label{tanh_forward}\\\\\n \\textbf{relu}: \\hspace{15pt} & h = max\\{0, u\\} =\n\\begin{bmatrix}\n\\max\\{0, u_1\\}\\\\\n\\vdots \\\\\n\\max\\{0, u_M\\}\\\\\n\\end{bmatrix} \\label{relu_forward}\\\\\n \\textbf{linear}^{(2)}: \\hspace{15pt} & a = W^{(2)}h + b^{(2)} \\hspace{2em}, W^{(2)} \\in \\mathbb{R}^{K\\times M} \\text{ and } b^{(2)} \\in \\mathbb{R}^{K} \\label{linear2_forward}\\\\\n \\textbf{softmax}: \\hspace{15pt} & z = \\begin{bmatrix}\n\\cfrac{e^{a_1}}{\\sum_{k} e^{a_{k}}}\\\\\n\\vdots \\\\\n\\cfrac{e^{a_K}}{\\sum_{k} e^{a_{k}}} \\\\\n\\end{bmatrix}\\\\\n \\textbf{predicted label}: \\hspace{15pt} & \\hat{y} = argmax_k z_k.\n%& l = -\\sum_{k} y_{k}\\log{\\hat{y_{k}}} \\hspace{2em}, \\vy \\in \\mathbb{R}^{k} \\text{ and } y_k=1 \\text{ if } \\vx \\text{ belongs to the } k' \\text{-th class}.\n\\end{align}\n$$\n\n\nFor a $K$-class classification problem, one popular loss function for training (i.e., to learn $W^{(1)}$, $W^{(2)}$, $b^{(1)}$, $b^{(2)}$) is the cross-entropy loss.\nSpecifically we denote the cross-entropy loss with respect to the training example $(x, y)$ by $l$:\n<br><br>\n$$\n\\begin{align}\n l = -\\log (z_y) = \\log \\left( 1 + \\sum_{k\\neq y} e^{a_k - a_y} \\right)\n\\end{align}\n$$\n<br><br>\nNote that one should look at $l$ as a function of the parameters of the network, that is, $W^{(1)}, b^{(1)}, W^{(2)}$ and $b^{(2)}$.\nFor ease of notation, let us define the one-hot (i.e., 1-of-$K$) encoding of a class $y$ as\n\n\\begin{align}\ny \\in \\mathbb{R}^K \\text{ and }\ny_k =\n\\begin{cases}\n1, \\text{ if }y = k,\\\\\n0, \\text{ otherwise}.\n\\end{cases} \n\\end{align}\nso that\n\\begin{align} \nl = -\\sum_{k} y_{k}\\log{z_k} = \n-y^T\n\\begin{bmatrix}\n\\log z_1\\\\\n\\vdots \\\\\n\\log z_K\\\\\n\\end{bmatrix}\n= -y^T\\log{z}.\n\\end{align}\n\nWe can then perform error-backpropagation, a way to compute partial derivatives (or gradients) w.r.t the parameters of a neural network, and use gradient-based optimization to learn the parameters. \n\n\nSubmission: All you need to submit is neural_networks.py\n\n### Q1.1 Mini batch Gradient Descent \nFirst, You need to implement mini-batch gradient descent which is a gradient-based optimization to learn the parameters of the neural network. \n<br>\n$$\n\\begin{align}\n\\upsilon = \\alpha \\upsilon - \\eta \\delta_t\\\\\nw_t = w_{t-1} + \\upsilon\n\\end{align}\n$$\n<br>\nYou can use the formula above to update the weights using momentum. <br>\nHere,\n$\\alpha$ is the discount factor such that $\\alpha \\in (0, 1)$ <br>\n$\\upsilon$ is the velocity update<br>\n$\\eta$ is the learning rate<br>\n$\\delta_t$ is the gradient<br>\n\nYou need to handle with as well without momentum scenario in the ```miniBatchGradientDescent``` function.\n\n* ```TODO 1```\nYou need to complete ```def miniBatchGradientDescent(model, momentum, _lambda, _alpha, _learning_rate)``` in ```neural_networks.py```\n\n### Q1.2 Linear Layer (10 points)\nSecond, You need to implement the linear layer of MLP. In this part, you need to implement 3 python functions in ```class linear_layer```. In ```def __init__(self, input_D, output_D)``` function, you need to initialize W with random values using np.random.normal such that the mean is 0 and standard deviation is 0.1. You also need to initialize gradients to zeroes in the same function. \n\n$$\n\\begin{align}\n\\text{forward pass:}\\hspace{2em} &\nu = \\text{linear}^{(1)}\\text{.forward}(x) = W^{(1)}x + b^{(1)},\\\\\n&\\text{where } W^{(1)} \\text{ and } b^{(1)} \\text{ are its parameters.}\\nonumber\\\\ \n\\nonumber\\\\\n\\text{backward pass:}\\hspace{2em} &[\\frac{\\partial l}{\\partial x}, \\frac{\\partial l}{\\partial W^{(1)}}, \\frac{\\partial l}{\\partial b^{(1)}}] = \\text{linear}^{(1)}\\text{.backward}(x, \\frac{\\partial l}{\\partial u}).\n\\end{align}\n$$\n\nYou can use the above formula as a reference to implement the ```def forward(self, X)``` forward pass and ```def backward(self, X, grad)``` backward pass in class linear_layer. In backward pass, you only need to return the backward_output. You also need to compute gradients of W and b in backward pass. \n\n* ```TODO 2```\nYou need to complete ```def __init__(self, input_D, output_D)``` in ```class linear_layer``` of ```neural_networks.py```\n* ```TODO 3```\nYou need to complete ```def forward(self, X)``` in ```class linear_layer``` of ```neural_networks.py```\n* ```TODO 4```\nYou need to complete ```def backward(self, X, grad)``` in ```class linear_layer``` of ```neural_networks.py```\n\n### Q1.3 Activation function - tanh (10 points)\nNow, you need to implement the activation function tanh. In this part, you need to implement 2 python functions in ```class tanh```. In ```def forward(self, X)```, you need to implement the forward pass and you need to compute the derivative and accordingly implement ```def backward(self, X, grad)```, i.e. the backward pass.\n$$\n\\begin{align}\n\\textbf{tanh}:\\hspace{15pt} & h =\\cfrac{2}{1+e^{-2u}}-1\\\\\n\\end{align}\n$$\nYou can use the above formula for tanh as a reference.\n* ```TODO 5```\nYou need to complete ```def forward(self, X)``` in ```class tanh``` of ```neural_networks.py```\n* ```TODO 6```\nYou need to complete ```def backward(self, X, grad)``` in ```class tanh``` of ```neural_networks.py```\n\n\n\n\n### Q1.4 Activation function - relu (10 points)\nYou need to implement another activation function called relu. In this part, you need to implement 2 python functions in ```class relu```. In ```def forward(self, X)```, you need to implement the forward pass and you need to compute the derivative and accordingly implement ```def backward(self, X, grad)```, i.e. the backward pass.\n$$\n\\begin{align}\n\\textbf{relu}: \\hspace{15pt} & h = max\\{0, u\\} =\n\\begin{bmatrix}\n\\max\\{0, u_1\\}\\\\\n\\vdots \\\\\n\\max\\{0, u_M\\}\\\\\n\\end{bmatrix}\n\\end{align}\n$$\nYou can use the above formula for relu as a reference.\n* ```TODO 7```\nYou need to complete ```def forward(self, X)``` in ```class relu``` of ```neural_networks.py```\n* ```TODO 8```\nYou need to complete ```def backward(self, X, grad)``` in ```class relu``` of ```neural_networks.py```\n\n\n\n### Q1.5 Dropout (10 points)\nTo prevent overfitting, we usually add regularization. Dropout is another way of handling overfitting. In this part, you will initially read and understand ```def forward(self, X, is_train)``` i.e. the forward pass of ```class dropout``` and derive partial derivatives accordingly to implement ```def backward(self, X, grad)``` i.e. the backward pass of ```class dropout```. We define the forward and the backward passes as follows.\n\n\\begin{align}\n\\text{forward pass:}\\hspace{2em} &\n{s} = \\text{dropout}\\text{.forward}({q}\\in\\mathbb{R}^J) = \\frac{1}{1-r}\\times\n\\begin{bmatrix}\n\\textbf{1}[p_1 >= r] \\times q_1\\\\\n\\vdots \\\\\n\\textbf{1}[p_J >= r] \\times q_J\\\\\n\\end{bmatrix},\n\\\\\n\\nonumber\\\\\n&\\text{where } p_j \\text{ is sampled uniformly from }[0, 1), \\forall j\\in\\{1,\\cdots,J\\}, \\nonumber\\\\\n&\\text{and } r\\in [0, 1) \\text{ is a pre-defined scalar named dropout rate}.\n\\end{align}\n\\begin{align}\n\\text{backward pass:}\\hspace{2em} &\\frac{\\partial l}{\\partial {q}} = \\text{dropout}\\text{.backward}({q}, \\frac{\\partial l}{\\partial {s}})=\n\\frac{1}{1-r}\\times\n\\begin{bmatrix}\n\\textbf{1}[p_1 >= r] \\times \\cfrac{\\partial l}{\\partial s_1}\\\\\n\\vdots \\\\\n\\textbf{1}[p_J >= r] \\times \\cfrac{\\partial l}{\\partial s_J}\\\\\n\\end{bmatrix}.\n\\end{align}\n\nNote that $p_j, j\\in\\{1,\\cdots,J\\}$ and $r$ are not be learned so we do not need to compute the derivatives w.r.t. to them. Moreover, $p_j, j\\in\\{1,\\cdots,J\\}$ are re-sampled every forward pass, and are kept for the following backward pass. The dropout rate $r$ is set to 0 during testing.\n\n* ```TODO 9```\nYou need to complete ```def backward(self, X, grad)``` in ```class dropout``` of ```neural_networks.py```\n\n### Q1.6 Connecting the dots\nIn this part, you will combine the modules written from question Q1.1 to Q1.5 by implementing TODO snippets in the ```def main(main_params, optimization_type=\"minibatch_sgd\")``` i.e. main function. After implementing forward and backward passes of MLP layers in Q1.1 to Q1.5,now in the main function you will call the forward methods and backward methods of every layer in the model in an appropriate order based on the architecture.\n\n* ```TODO 10```\nYou need to complete ```main(main_params, optimization_type=\"minibatch_sgd\")``` in ```neural_networks.py```\n\n\n\n### Grading\nYour code will be graded on Vocareum with autograding script. For your reference, the solution code takes around 5 minutes to execute. As long as your code can finish grading on Vocareum, you should be good. When you finish all ```TODO``` parts, please click submit button on Vocareum. Sometimes you may need to come back to check grading report later.\n\nYour code will be tested on the correctness of modules you have implemented as well as certain custom testcases. 40 points are assigned for Question 1 while 60 points are assigned to custom testcases.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nprint(np.version)",
"<module 'numpy.version' from '/anaconda3/lib/python3.6/site-packages/numpy/version.py'>\n"
],
[
"a = np.array([[1,-2],[3,-2],[2,-2]])",
"_____no_output_____"
],
[
"a.shape[0]",
"_____no_output_____"
],
[
"a * a",
"_____no_output_____"
],
[
"np.tanh(a)",
"_____no_output_____"
],
[
"np.tanh(a)**2",
"_____no_output_____"
],
[
"for row in a:\n for element in row:\n print(element)\n if element < 0:\n element = 0\nprint(a)",
"1\n-2\n1\n-2\n1\n-2\n[[ 1 -2]\n [ 1 -2]\n [ 1 -2]]\n"
],
[
"print(np.maximum(a,0))",
"[[1 0]\n [1 0]\n [1 0]]\n"
],
[
"a",
"_____no_output_____"
],
[
"x = a\nx[x<=0] = 0\nx[x>0] = 1",
"_____no_output_____"
],
[
"x",
"_____no_output_____"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb8dbaafe2140f828628b5d0de9a59c9c71f3fa9
| 17,226 |
ipynb
|
Jupyter Notebook
|
user-story-10-grant-outputs/py-grant-outputs.ipynb
|
ArtemisLav/pidgraph-notebooks-python
|
7224adf16d1c5c47f6accb43c6b1814f2f2d0ac6
|
[
"MIT"
] | null | null | null |
user-story-10-grant-outputs/py-grant-outputs.ipynb
|
ArtemisLav/pidgraph-notebooks-python
|
7224adf16d1c5c47f6accb43c6b1814f2f2d0ac6
|
[
"MIT"
] | null | null | null |
user-story-10-grant-outputs/py-grant-outputs.ipynb
|
ArtemisLav/pidgraph-notebooks-python
|
7224adf16d1c5c47f6accb43c6b1814f2f2d0ac6
|
[
"MIT"
] | null | null | null | 39.15 | 422 | 0.569778 |
[
[
[
"  | [FREYA](https://www.project-freya.eu/en) WP2 [User Story 10](https://github.com/datacite/freya/issues/45) | As a funder, we want to be able to find all the outputs related to our awarded grants, including block grants such as doctoral training grants, for management info and looking at impact. \n :------------- | :------------- | :-------------\n\nFunders are interested in monitoring the output of grants they award - while the grant is active as well as retrospectively. The quality, quantity and types of the grant's outputs are useful proxies for the value obtained as a result of the funder's investment.<p />\nThis notebook uses the [DataCite GraphQL API](https://api.datacite.org/graphql) to retrieve all outputs of [FREYA grant award](https://cordis.europa.eu/project/id/777523) from [European Union](https://doi.org/10.13039/501100000780) to date. \n\n**Goal**: By the end of this notebook you should be able to:\n- Retrieve all outputs of a grant award from a specific funder; \n- Plot number of outputs per year-quarter of the grant award duration;\n- Display de-duplicated outputs in tabular format, including the number of their citations, views and downloads;\n- Plot a pie chart of the number of outputs per resource type;\n- Display an interactive chord plot of co-authorship relationships across all outputs, e.g. <br> <img src=\"example_plot.png\" width=\"318\" height=\"309\" />",
"_____no_output_____"
],
[
"## Install libraries and prepare GraphQL client",
"_____no_output_____"
]
],
[
[
"%%capture\n# Install required Python packages\n!pip install gql requests chord numpy",
"_____no_output_____"
],
[
"# Prepare the GraphQL client\nimport requests\nfrom IPython.display import display, Markdown\nfrom gql import gql, Client\nfrom gql.transport.requests import RequestsHTTPTransport\n\n_transport = RequestsHTTPTransport(\n url='https://api.datacite.org/graphql',\n use_json=True,\n)\n\nclient = Client(\n transport=_transport,\n fetch_schema_from_transport=True,\n)",
"_____no_output_____"
]
],
[
[
"## Define and run GraphQL query\nDefine the GraphQL query to find all outputs of [FREYA grant award](https://cordis.europa.eu/project/id/777523) from [European Union](https://doi.org/10.13039/501100000780) to date.",
"_____no_output_____"
]
],
[
[
"# Generate the GraphQL query: find all outputs of FREYA grant award (https://cordis.europa.eu/project/id/777523) from funder (EU) to date\nquery_params = {\n \"funderId\" : \"https://doi.org/10.13039/501100000780\",\n \"funderAwardQuery\" : \"fundingReferences.awardNumber:777523\",\n \"maxWorks\" : 75\n}\n\nquery = gql(\"\"\"query getGrantOutputsForFunderAndAward($funderId: ID!, $funderAwardQuery: String!, $maxWorks: Int!)\n{\nfunder(id: $funderId) {\n name\n works(query: $funderAwardQuery, first: $maxWorks) {\n totalCount\n nodes {\n id\n formattedCitation(style: \"vancouver\")\n titles {\n title\n }\n descriptions {\n description\n } \n types {\n resourceType\n }\n dates {\n date\n dateType\n }\n versionOfCount\n creators {\n id\n name\n }\n fundingReferences {\n funderIdentifier\n funderName\n awardNumber\n awardTitle\n }\n citationCount\n viewCount\n downloadCount\n }\n }\n }\n}\n\"\"\")",
"_____no_output_____"
]
],
[
[
"Run the above query via the GraphQL client",
"_____no_output_____"
]
],
[
[
"import json\ndata = client.execute(query, variable_values=json.dumps(query_params))",
"_____no_output_____"
]
],
[
[
"## Display total number of works \nDisplay the total number of [FREYA grant award](https://cordis.europa.eu/project/id/777523) outputs to date.",
"_____no_output_____"
]
],
[
[
"# Get the total number of outputs to date\nfunder = data['funder']['works']\ndisplay(Markdown(str(funder['totalCount'])))",
"_____no_output_____"
]
],
[
[
"## Plot number of works per quarter\nDisplay a bar plot of number of [FREYA grant award](https://cordis.europa.eu/project/id/777523) outputs to date, per each quarter of project's duration.",
"_____no_output_____"
]
],
[
[
"# Plot the number of FREYA outputs to date, by year\nimport matplotlib.pyplot as plt\nfrom matplotlib.ticker import FormatStrFormatter\nimport numpy as np\n\n# Return quarter (number) given month (number)\ndef get_quarter(month):\n return (month - 1) // 3 + 1\n\n# Return list of consecutive years between min_year_quarter and max_year_quarter inclusive\ndef get_consecutive_year_quarters(min_year_quarter, max_year_quarter):\n year_quarters = [\"%d Q%d\" % (min_year_quarter[0],min_year_quarter[1])]\n yq = min_year_quarter\n while yq != max_year_quarter:\n year = yq[0]\n quarter = yq[1]\n if quarter == 4:\n year += 1\n quarter = 1\n else:\n quarter += 1\n yq = (year, quarter)\n year_quarters.append(\"%d Q%d\" % (year,quarter))\n year_quarters.append(\"%d Q%d\" % (max_year_quarter[0],max_year_quarter[1])) \n return year_quarters\n\nplt.rcdefaults()\n\n# Retrieve works counts by year-quarter from nodes\n# Pick out date of type: 'Issued'; failing that use 'Created' date.\nnum_outputs_dict = {}\nfunder = data['funder']['works']\nfor r in funder['nodes']:\n node_date = None\n for date_dict in r['dates']:\n ym = date_dict['date'].split('-')[0:2]\n if len(ym) < 2:\n continue\n yq = ym[0] + \" Q\" + str(get_quarter(int(ym[1])))\n if node_date is None:\n if date_dict['dateType'] in ['Issued', 'Created']:\n node_date = yq\n else:\n if date_dict['dateType'] in ['Issued']:\n node_date = yq\n if node_date:\n if node_date not in num_outputs_dict:\n num_outputs_dict[node_date] = 0\n num_outputs_dict[node_date] += 1;\n \n# Sort works counts by year-quarter in chronological order\nsorted_year_quarters = sorted(list(num_outputs_dict.keys()))\n# Get all consecutive year-quarters FREYA-specific start-end year-quarter\nyear_quarters = get_consecutive_year_quarters((2017,4), (2020,4)) \n# Populate non-zero counts for year_quarters \nnum_outputs = []\nfor yq in year_quarters:\n if yq in sorted_year_quarters:\n num_outputs.append(num_outputs_dict[yq])\n else:\n num_outputs.append(0)\n \n# Generate a plot of number of grant outputs by year - quarter\nfig, ax = plt.subplots(1, 1, figsize = (10, 5))\nx_pos = np.arange(len(year_quarters))\nax.bar(x_pos, num_outputs, align='center', color='blue', edgecolor='black', linewidth = 0.1, alpha=0.5)\nax.set_xticks(x_pos)\nax.set_xticklabels(year_quarters, rotation='vertical')\nax.set_ylabel('Number of outputs')\nax.set_xlabel('Year Quarter')\nax.set_title('Number of Grant Award Outputs per Year-Quarter')\nplt.show()\n",
"_____no_output_____"
]
],
[
[
"## Display de-duplicated works in tabular format\nDisplay the outputs of [FREYA grant award](https://cordis.europa.eu/project/id/777523) in a html table, including the number of their citations, views and downloads. Note that the outputs are de-duplicated, i.e. outputs that are versions of another output are excluded.",
"_____no_output_____"
]
],
[
[
"from IPython.core.display import display, HTML\nimport textwrap\nxstr = lambda s: 'General' if s is None else str(s)\n\n# Get details for each output\noutputs = [['ID','Type','Publication Date','Formatted Citation','Descriptions', 'Number of Citations', 'Number of Views', 'Number of Downloads']]\n# Since there is scope for duplicates in Zenodo, versions of previously seen nodes are considered duplicates and stored in duplicate_versions so that \n# they can be excluded if seen later\nfor r in funder['nodes']:\n id = '<a href=\"%s\">%s</a></html>' % (r['id'], '/'.join(r['id'].split(\"/\")[3:]))\n if r['versionOfCount'] > 0:\n # If the current output is a version of another one, exclude it\n continue\n # As Publication Date, pick out date of type: 'Issued'; failing that use 'Created' date.\n pub_date = None\n for date_dict in r['dates']:\n if pub_date is None:\n if date_dict['dateType'] in ['Issued', 'Created']:\n pub_date = date_dict['date'];\n else:\n if date_dict['dateType'] in ['Issued']:\n pub_date = date_dict['date'];\n \n titles = '; '.join([s['title'] for s in r['titles']])\n creators = '; '.join(['<a href=\"%s\">%s</a>' % (s['id'],s['name']) for s in r['creators']])\n formatted_citation = \"%s. %s. %s; Available from: %s\" % (creators, titles, pub_date, id) \n resource_type = xstr(r['types']['resourceType'])\n descriptions = textwrap.shorten('; '.join([s['description'] for s in r['descriptions']]), width=200, placeholder=\"...\")\n output = [id, resource_type, pub_date, formatted_citation, descriptions, str(r['citationCount']), str(r['viewCount']), str(r['downloadCount'])]\n outputs += [output]\n \n# Display outputs as html table \nhtml_table = '<html><table>' \nhtml_table += '<tr><th style=\"text-align:center;\">' + '</th><th style=\"text-align:center;\">'.join(outputs[0]) + '</th></tr>'\nfor row in outputs[1:]:\n html_table += '<tr><td style=\"text-align:left;\">' + '</td><td style=\"text-align:left;\">'.join(row) + '</td></tr>'\nhtml_table += '</table></html>'\ndisplay(HTML(html_table))",
"_____no_output_____"
]
],
[
[
"## Plot number of outputs per resource type\nPlot as a pie chart the number of [FREYA grant award](https://cordis.europa.eu/project/id/777523) outputs per resource type.",
"_____no_output_____"
]
],
[
[
"# Plot as a pie chart the number of outputs per resource type\nimport matplotlib.pyplot as plt\nfrom matplotlib.ticker import FormatStrFormatter\nimport numpy as np\nimport operator\n\nxstr = lambda s: 'General' if s is None else str(s)\n\nplt.rcdefaults()\n\n# Retrieve works counts by resource type from nodes\n# Pick out date of type: 'Issued'; failing that use 'Created' date.\nfunder = data['funder']['works']\nnum_outputs_dict = {}\nfor r in funder['nodes']:\n resource_type = xstr(r['types']['resourceType'])\n if resource_type not in num_outputs_dict:\n num_outputs_dict[resource_type] = 0\n num_outputs_dict[resource_type] += 1;\n \n# Sort resource types by count of work desc\nsorted_num_outputs = sorted(num_outputs_dict.items(),key=operator.itemgetter(1),reverse=True)\n# Populate lists needed for pie chart\nresource_types = [s[0] for s in sorted_num_outputs] \nnum_outputs = [s[1] for s in sorted_num_outputs] \n\n# Generate a pie chart of number of grant outputs by resource type\nfig = plt.figure()\nax = fig.add_axes([0,0,1,1])\nax.set_title('Number of Grant Outputs per Resource Type')\nax.axis('equal')\nax.pie(num_outputs, labels = resource_types,autopct='%1.2f%%')\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Display an interactive plot of co-authorship relationships across all outputs\nDisplay an interactive chord plot representing co-authorship relationships across all [FREYA grant award](https://cordis.europa.eu/project/id/777523) outputs.",
"_____no_output_____"
]
],
[
[
"# Generate a Chord plot representing co-authorship relationships across all grant award outputs\nfrom chord import Chord \nfrom IPython.display import IFrame\n\nall_creator_names_by_node = []\nall_creator_names_set = set([])\nfunder = data['funder']['works']\nfor r in funder['nodes']:\n if r['versionOfCount'] > 0:\n # If the current output is a version of another one, exclude it\n continue\n # To minimise cropping of names in the below, retain just the first letter of the first name\n # if the author name is well formatted \n creator_names = []\n for name in [s['name'] for s in r['creators'] if s['name']]:\n if name.find(\",\") > 0:\n creator_names.append(name[0:name.index(\",\") + 3])\n elif name.find(\",\") == 0:\n creator_names.append(name[1:].strip())\n else:\n creator_names.append(name)\n all_creator_names_by_node.append(creator_names)\n all_creator_names_set.update(creator_names)\n \n# Assemble data structures for the co-authorship chord diagram\nall_creator_names = sorted(list(all_creator_names_set))\n\n# Initialise chord data matrix\nlength = len(all_creator_names)\ncoauthorship_matrix = []\nfor i in range(length):\n r = []\n for j in range(length):\n r.append(0)\n coauthorship_matrix.append(r)\n \n# Populate chord data matrix\nfor node_creators in all_creator_names_by_node:\n for creator in node_creators:\n c_pos = all_creator_names.index(creator)\n for co_creator in node_creators:\n co_pos = all_creator_names.index(co_creator)\n if c_pos != co_pos:\n coauthorship_matrix[c_pos][co_pos] += 1\n\n# display co-authorship cord diagram\nplot = Chord(coauthorship_matrix, all_creator_names, padding=0.04, wrap_labels=False, margin=130, width=1000).to_html()\nIFrame(src=\"./out.html\", width=1000, height=1000)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb8dc407d353d61ec5362c8d782272cfe7466864
| 20,230 |
ipynb
|
Jupyter Notebook
|
jwst_validation_notebooks/extract_2d/jwst_extract_2d_nirspec_test/jwst_extract_2d_nirspec_test.ipynb
|
york-stsci/jwst_validation_notebooks
|
1a244c927a027f89dcb01d146964fef71b5ea029
|
[
"BSD-3-Clause"
] | 4 |
2019-02-28T21:21:20.000Z
|
2022-01-31T04:24:12.000Z
|
jwst_validation_notebooks/extract_2d/jwst_extract_2d_nirspec_test/jwst_extract_2d_nirspec_test.ipynb
|
york-stsci/jwst_validation_notebooks
|
1a244c927a027f89dcb01d146964fef71b5ea029
|
[
"BSD-3-Clause"
] | 58 |
2020-02-17T14:54:30.000Z
|
2022-03-10T14:53:00.000Z
|
jwst_validation_notebooks/extract_2d/jwst_extract_2d_nirspec_test/jwst_extract_2d_nirspec_test.ipynb
|
york-stsci/jwst_validation_notebooks
|
1a244c927a027f89dcb01d146964fef71b5ea029
|
[
"BSD-3-Clause"
] | 20 |
2019-03-11T17:24:03.000Z
|
2022-01-07T20:57:13.000Z
| 43.042553 | 432 | 0.544736 |
[
[
[
"<a id=\"title_ID\"></a>\n# JWST Pipeline Validation Testing Notebook: spec2, extract_2d step\n\n<span style=\"color:red\"> **Instruments Affected**</span>: NIRSpec \n\nTested on CV3 data\n\n### Table of Contents\n<div style=\"text-align: left\"> \n\n<br> [Imports](#imports_ID) <br> [Introduction](#intro_ID) <br> [Testing Data Set](#data_ID) <br> [Run the JWST pipeline and assign_wcs validation tests](#pipeline_ID): [FS Full-Frame test](#FULLFRAME), [FS ALLSLITS test](#ALLSLITS), [MOS test](#MOS) <br> [About This Notebook](#about_ID)<br> [Results](#results) <br>\n\n</div>",
"_____no_output_____"
],
[
"<a id=\"imports_ID\"></a>\n# Imports\nThe library imports relevant to this notebook are aready taken care of by importing PTT.\n\n* astropy.io for opening fits files\n* jwst.module.PipelineStep is the pipeline step being tested\n* matplotlib.pyplot.plt to generate plot\n\nNOTE: This notebook assumes that the pipeline version to be tested is already installed and its environment is activated.\n\nTo be able to run this notebook you need to install nptt. \n\nIf all goes well you will be able to import PTT.\n\n[Top of Page](#title_ID)",
"_____no_output_____"
]
],
[
[
"# Create a temporary directory to hold notebook output, and change the working directory to that directory.\nfrom tempfile import TemporaryDirectory\nimport os\nimport shutil\ndata_dir = TemporaryDirectory()\n\nos.chdir(data_dir.name)",
"_____no_output_____"
],
[
"# Choose CRDS cache location\nuse_local_crds_cache = False\ncrds_cache_tempdir = False\ncrds_cache_notebook_dir = True\ncrds_cache_home = False\ncrds_cache_custom_dir = False\ncrds_cache_dir_name = \"\"\n\nif use_local_crds_cache:\n if crds_cache_tempdir:\n os.environ['CRDS_PATH'] = os.path.join(os.getcwd(), \"crds\")\n elif crds_cache_notebook_dir:\n try:\n os.environ['CRDS_PATH'] = os.path.join(orig_dir, \"crds\")\n except Exception as e:\n os.environ['CRDS_PATH'] = os.path.join(os.getcwd(), \"crds\")\n elif crds_cache_home:\n os.environ['CRDS_PATH'] = os.path.join(os.environ['HOME'], 'crds', 'cache')\n elif crds_cache_custom_dir:\n os.environ['CRDS_PATH'] = crds_cache_dir_name",
"_____no_output_____"
],
[
"import warnings\nimport psutil\nfrom astropy.io import fits\n\n# Only print a DeprecationWarning the first time it shows up, not every time.\nwith warnings.catch_warnings():\n warnings.simplefilter(\"once\", category=DeprecationWarning)\n import jwst\n from jwst.pipeline.calwebb_detector1 import Detector1Pipeline\n from jwst.assign_wcs.assign_wcs_step import AssignWcsStep\n from jwst.msaflagopen.msaflagopen_step import MSAFlagOpenStep\n from jwst.extract_2d.extract_2d_step import Extract2dStep\n\n# The latest version of NPTT is installed in the requirements text file at:\n# /jwst_validation_notebooks/environment.yml\n\n# import NPTT\nimport nirspec_pipe_testing_tool as nptt\n\n# To get data from Artifactory\nfrom ci_watson.artifactory_helpers import get_bigdata\n",
"_____no_output_____"
],
[
"# Print the versions used for the pipeline and NPTT\n\npipeline_version = jwst.__version__\nnptt_version = nptt.__version__\n\nprint(\"Using jwst pipeline version: \", pipeline_version)\nprint(\"Using NPTT version: \", nptt_version)",
"_____no_output_____"
]
],
[
[
"<a id=\"intro_ID\"></a>\n# Test Description\n\nWe compared Institute's pipeline product of the assign_wcs step with our benchmark files, or with the intermediary products from the ESA pipeline, which is completely independent from the Institute's. The comparison file is referred to as 'truth'. We calculated the relative difference and expected it to be equal to or less than computer precision: relative_difference = absolute_value( (Truth - ST)/Truth ) <= 1x10^-7. \n\nFor the test to be considered PASSED, every single slit (for FS data), slitlet (for MOS data) or slice (for IFU data) in the input file has to pass. If there is any failure, the whole test will be considered as FAILED. \n\nThe code for this test can be obtained at: https://github.com/spacetelescope/nirspec_pipe_testing_tool/blob/master/nirspec_pipe_testing_tool/calwebb_spec2_pytests/auxiliary_code/check_corners_extract2d.py. Multi Object Spectroscopy (MOS), the code is in the same repository but is named ```compare_wcs_mos.py```, and for Integral Field Unit (IFU) data, the test is named ```compare_wcs_ifu.py```.\nThe input file is defined in the variable ```input_file``` (see section [Testing Data Set and Variable Setup](#data_ID)).\n\nStep description: https://jwst-pipeline.readthedocs.io/en/latest/jwst/extract_2d/main.html\n\nPipeline code: https://github.com/spacetelescope/jwst/tree/master/jwst/extract_2d\n\n\n### Results\n\nIf the test **PASSED** this means that all slits, slitlets, or slices individually passed the test. However, if ony one individual slit (for FS data), slitlet (for MOS data) or slice (for IFU data) test failed, the whole test will be reported as **FAILED**.\n\n### Calibration WG Requested Algorithm: \n\nA short description and link to the page: \nhttps://outerspace.stsci.edu/display/JWSTCC/Vanilla+Path-Loss+Correction\n\n\n### Defining Term\nAcronymns used un this notebook:\n\npipeline: calibration pipeline\n\nspec2: spectroscopic calibration pipeline level 2b\n\nPTT: NIRSpec pipeline testing tool (https://github.com/spacetelescope/nirspec_pipe_testing_tool)\n\n\n[Top of Page](#title_ID)",
"_____no_output_____"
],
[
"<a id=\"pipeline_ID\"></a>\n# Run the JWST pipeline and extract_2d validation tests\n\nThe pipeline can be run from the command line in two variants: full or per step.\n\nTu run the spec2 pipeline in full use the command: \n\n$ strun jwst.pipeline.Spec2Pipeline jwtest_rate.fits\n\nTu only run the extract_2d step, use the command:\n\n$ strun jwst.extract_2d.Extract2dStep jwtest_previous_step_output.fits\n\n\nThese options are also callable from a script with the testing environment active. The Python call for running the pipeline in full or by step are:\n\n$\\gt$ from jwst.pipeline.calwebb_spec2 import Spec2Pipeline\n\n$\\gt$ Spec2Pipeline.call(jwtest_rate.fits)\n \nor\n \n$\\gt$ from jwst.extract_2d import Extract2dStep\n \n$\\gt$ Extract2dStep.call(jwtest_previous_step_output.fits)\n\n\nPTT can run the spec2 pipeline either in full or per step, as well as the imaging pipeline in full. In this notebook we will use PTT to run the pipeline and the validation tests. To run PTT, follow the directions in the corresponding repo page.\n\n[Top of Page](#title_ID)",
"_____no_output_____"
],
[
"<a id=\"data_ID\"></a>\n# Testing Data Set\n\nAll testing data is from the CV3 campaign. We chose these files because this is our most complete data set, i.e. all modes and filter-grating combinations.\n\nData used was for testing was only FS and MOS, since extract_2d is skipped for IFU. Data sets are:\n- FS_PRISM_CLEAR\n- FS_FULLFRAME_G395H_F290LP\n- FS_ALLSLITS_G140H_F100LP \n- MOS_G140M_LINE1 \n- MOS_PRISM_CLEAR\n\n\n[Top of Page](#title_ID)",
"_____no_output_____"
]
],
[
[
"testing_data = {'fs_prism_clear':{\n 'uncal_file_nrs1': 'fs_prism_nrs1_uncal.fits',\n 'uncal_file_nrs2': 'fs_prism_nrs2_uncal.fits',\n 'truth_file_nrs1': 'fs_prism_nrs1_extract_2d_truth.fits',\n 'truth_file_nrs2': 'fs_prism_nrs2_extract_2d_truth.fits',\n 'msa_shutter_config': None },\n \n 'fs_fullframe_g395h_f290lp':{\n 'uncal_file_nrs1': 'fs_fullframe_g35h_f290lp_nrs1_uncal.fits',\n 'uncal_file_nrs2': 'fs_fullframe_g35h_f290lp_nrs2_uncal.fits',\n 'truth_file_nrs1': 'fs_fullframe_g35h_f290lp_nrs1_extract_2d_truth.fits',\n 'truth_file_nrs2': 'fs_fullframe_g35h_f290lp_nrs2_extract_2d_truth.fits', \n 'msa_shutter_config': None },\n \n 'fs_allslits_g140h_f100lp':{\n 'uncal_file_nrs1': 'fs_allslits_g140h_f100lp_nrs1_uncal.fits',\n 'uncal_file_nrs2': 'fs_allslits_g140h_f100lp_nrs2_uncal.fits',\n 'truth_file_nrs1': 'fs_allslits_g140h_f100lp_nrs1_extract_2d_truth.fits',\n 'truth_file_nrs2': 'fs_allslits_g140h_f100lp_nrs2_extract_2d_truth.fits',\n 'msa_shutter_config': None },\n \n # Commented out because the pipeline is failing with this file\n #'bots_g235h_f170lp':{\n # 'uncal_file_nrs1': 'bots_g235h_f170lp_nrs1_uncal.fits',\n # 'uncal_file_nrs2': 'bots_g235h_f170lp_nrs2_uncal.fits',\n # 'truth_file_nrs1': 'bots_g235h_f170lp_nrs1_extract_2d_truth.fits',\n # 'truth_file_nrs2': 'bots_g235h_f170lp_nrs2_extract_2d_truth.fits',\n # 'msa_shutter_config': None },\n \n 'mos_prism_clear':{\n 'uncal_file_nrs1': 'mos_prism_nrs1_uncal.fits',\n 'uncal_file_nrs2': 'mos_prism_nrs2_uncal.fits',\n 'truth_file_nrs1': 'mos_prism_nrs1_extract_2d_truth.fits',\n 'truth_file_nrs2': None,\n 'msa_shutter_config': 'V0030006000104_msa.fits' },\n \n 'mos_g140m_f100lp':{\n 'uncal_file_nrs1': 'mos_g140m_line1_NRS1_uncal.fits',\n 'uncal_file_nrs2': 'mos_g140m_line1_NRS2_uncal.fits', \n 'truth_file_nrs1': 'mos_g140m_line1_nrs1_extract_2d_truth.fits',\n 'truth_file_nrs2': 'mos_g140m_line1_nrs2_extract_2d_truth.fits',\n 'msa_shutter_config': 'V8460001000101_msa.fits' },\n \n\n }\n",
"_____no_output_____"
],
[
"# define function to pull data from Artifactory\ndef get_artifactory_file(data_set_dict, detector):\n \"\"\"This function creates a list with all the files needed per detector to run the test.\n Args:\n data_set_dict: dictionary, contains inputs for a specific mode and configuration\n detector: string, either nrs1 or nrs2\n Returns:\n data: list, contains all files needed to run test\n \"\"\"\n files2obtain = ['uncal_file_nrs1', 'truth_file_nrs1', 'msa_shutter_config']\n data = []\n for file in files2obtain:\n data_file = None\n try: \n if '_nrs' in file and '2' in detector:\n file = file.replace('_nrs1', '_nrs2')\n\n data_file = get_bigdata('jwst_validation_notebooks',\n 'validation_data',\n 'nirspec_data', \n data_set_dict[file])\n except TypeError:\n data.append(None)\n continue\n\n data.append(data_file)\n\n return data",
"_____no_output_____"
],
[
"# Set common NPTT switches for NPTT and run the test for both detectors in each data set\n\n# define benchmark (or 'truth') file\ncompare_assign_wcs_and_extract_2d_with_esa = False\n\n# accepted threshold difference with respect to benchmark files\nextract_2d_threshold_diff = 4\n\n# define benchmark (or 'truth') file\nesa_files_path, raw_data_root_file = None, None\ncompare_assign_wcs_and_extract_2d_with_esa = False\n\n# Get the data\nresults_dict = {}\ndetectors = ['nrs1', 'nrs2']\nfor mode_config, data_set_dict in testing_data.items():\n for det in detectors:\n print('Testing files for detector: ', det)\n data = get_artifactory_file(data_set_dict, det)\n uncal_file, truth_file, msa_shutter_config = data\n print('Working with uncal_file: ', uncal_file)\n uncal_basename = os.path.basename(uncal_file)\n \n # Make sure that there is an assign_wcs truth product to compare to, else skip this data set\n if truth_file is None:\n print('No truth file to compare to for this detector, skipping this file. \\n')\n skip_file = True\n else:\n skip_file = False\n\n if not skip_file: \n # Run the stage 1 pipeline \n rate_object = Detector1Pipeline.call(uncal_file)\n # Make sure the MSA shutter configuration file is set up correctly\n if msa_shutter_config is not None:\n msa_metadata = rate_object.meta.instrument.msa_metadata_file\n print(msa_metadata)\n if msa_metadata is None or msa_metadata == 'N/A':\n rate_object.meta.instrument.msa_metadata_file = msa_shutter_config\n\n # Run the stage 2 pipeline steps\n pipe_object = AssignWcsStep.call(rate_object)\n if 'mos' in uncal_basename.lower():\n pipe_object = MSAFlagOpenStep.call(pipe_object)\n extract_2d_object = Extract2dStep.call(pipe_object)\n\n # Run the validation test\n %matplotlib inline\n\n if 'fs' in uncal_file.lower():\n print('Running test for FS...')\n result, _ = nptt.calwebb_spec2_pytests.auxiliary_code.check_corners_extract2d.find_FSwindowcorners(\n extract_2d_object, \n truth_file=truth_file,\n esa_files_path=esa_files_path,\n extract_2d_threshold_diff=extract_2d_threshold_diff)\n if 'mos' in uncal_file.lower():\n print('Running test for MOS...')\n result, _ = nptt.calwebb_spec2_pytests.auxiliary_code.check_corners_extract2d.find_MOSwindowcorners(\n extract_2d_object,\n msa_shutter_config,\n truth_file=truth_file,\n esa_files_path=esa_files_path,\n extract_2d_threshold_diff= extract_2d_threshold_diff)\n\n\n else:\n result = 'skipped'\n\n # Did the test passed \n print(\"Did assign_wcs validation test passed? \", result, \"\\n\\n\")\n rd = {uncal_basename: result}\n results_dict.update(rd)\n \n # close all open files\n psutil.Process().open_files()\n closing_files = []\n for fd in psutil.Process().open_files():\n if data_dir.name in fd.path:\n closing_files.append(fd)\n for fd in closing_files:\n try:\n print('Closing file: ', fd)\n open(fd.fd).close()\n except:\n print('File already closed: ', fd)\n\n",
"_____no_output_____"
],
[
"# Quickly see if the test passed \n\nprint('These are the final results of the tests: ')\nfor key, val in results_dict.items():\n print(key, val)\n",
"_____no_output_____"
]
],
[
[
"<a id=\"about_ID\"></a>\n## About this Notebook\n**Author:** Maria A. Pena-Guerrero, Staff Scientist II - Systems Science Support, NIRSpec\n<br>**Updated On:** Mar/24/2021",
"_____no_output_____"
],
[
"[Top of Page](#title_ID)\n<img style=\"float: right;\" src=\"./stsci_pri_combo_mark_horizonal_white_bkgd.png\" alt=\"stsci_pri_combo_mark_horizonal_white_bkgd\" width=\"200px\"/> ",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
]
] |
cb8dc55fa900cc102173ac5c766f8596b10cce15
| 22,420 |
ipynb
|
Jupyter Notebook
|
Guides/Pump/Pump+Calculations.ipynb
|
rocketproplab/Guides
|
165f0ffc6ed2ea746de08941077e2c2e0c2af554
|
[
"MIT"
] | 9 |
2017-04-11T01:10:37.000Z
|
2020-11-14T23:08:28.000Z
|
Guides/Pump/Pump+Calculations.ipynb
|
rocketproplab/Guides
|
165f0ffc6ed2ea746de08941077e2c2e0c2af554
|
[
"MIT"
] | null | null | null |
Guides/Pump/Pump+Calculations.ipynb
|
rocketproplab/Guides
|
165f0ffc6ed2ea746de08941077e2c2e0c2af554
|
[
"MIT"
] | 6 |
2017-04-15T22:36:07.000Z
|
2020-11-14T23:08:29.000Z
| 55.771144 | 3,714 | 0.642016 |
[
[
[
"# Pump Calculations",
"_____no_output_____"
]
],
[
[
"import numpy as np",
"_____no_output_____"
]
],
[
[
"## Power Input",
"_____no_output_____"
]
],
[
[
"#Constants and inputs\ng = 32.174; #gravitational acceleration, ft/s^2\nrho_LOx = 71.27; #Density of Liquid Oxygen- lbm/ft^3\nrho_LCH4 = 26.3; #Density of Liquid Methane- lbm/ft^3\n\nDifferential = #Desired pressure differential (psi)\n\nmLOx = #Mass flow of Liquid Oxygen (lb/s)\nmLCH4 = #Mass Flow of Liquid Methane (lb/s)",
"_____no_output_____"
],
[
"#Head Calculations\nHLOx = (((Differential)*144)/(rho_LOx * g))*32.174 #Head of Liquid Oxygen - ft\nHLCH4 = (((Differential)*144)/(rho_LCH4 * g))*32.174 #Head of Liquid Methane - ft",
"_____no_output_____"
],
[
"#Power Calculations - Assume a 75% effiency (Minimum value that we can reach)\nPower_LOx = (((mLOx * g * HLOx)/0.75)/32.174) * 1.36; #Output is in Watts\nPower_LCH4 = (((mLCH4 * g * HLCH4)/0.75)/32.174) * 1.36; #Output is in Watts",
"_____no_output_____"
]
],
[
[
"## Impeller Calculations",
"_____no_output_____"
],
[
"### Constants",
"_____no_output_____"
]
],
[
[
"QLOx = mLOx/rho_LOx #Volumetric flow rate of Liquid Oxygen in ft^3/s\nQLCH4 = mLCH4/rho_LCH4 #Volumetric flow rate of Liquid Methane in ft^3/s\n\nEff_Vol = #Volumetric effiency is a measure of how much fluid is lost due to leakages, estimate the value\n\nQImp_LOx = QLox/Eff_Vol #Impeller flow rate of Liquid oxygen in ft^3/s\nQImp_LCH4 = QLCH4/Eff_Vol #Impeller flow rate of Liquid methane in ft^3/s\n\nn = #RPM of impeller, pick such that nq_LOx is low but not too low\n\nnq_LOx = n * (QImp_LOx ** 0.5)/(HLOx ** 0.75) #Specific speed of Liquid Oxygen\nnq_LCH4 = n * (QImp_LCH4 ** 0.5)/(HLCH4 ** 0.75) #Specific speed of Liquid Methane\n\nomegas_LOx = nq_LOx/52.9 #Universal specific speed\nomegas_LCH4 = nq_LCH4/52.9 #Universal specific speed\n\ntau = #Shear stress of desired metal (Pa)\n\nfq = 1 #Number of impeller inlets, either 1 or 2\nf_t = 1.1 #Given earlier in the text\n\nPC_LOx = 1.21*f_t*(np.exp(-0.408*omegas_LOx))* nq #Pressure coefficient of static pressure rise in impeller of Liquid Oxygen, the equation given uses nq_ref, but I just use nq because I didn't define an nq_ref\nPC_LCH4 = 1.21*f_t*(np.exp(-0.408*omegas_LCH4))* nq #Pressure coefficienct of static pressure rise in impeller of Liquid Methane\n",
"_____no_output_____"
]
],
[
[
"#### Shaft diameter",
"_____no_output_____"
]
],
[
[
"dw_LOx = 3.65(Power_LOx)/(rpm*tau) #Shaft diameter of Liquid Oxygen Impeller\ndw_LCH4 = 3.65(Power_LCH4)/(rpm*tau) #Shaft diameter of Liquid Methane Impeller",
"_____no_output_____"
]
],
[
[
"#### Specific Speed",
"_____no_output_____"
]
],
[
[
"q_LOx = QLOx * 3600 * (.3048 ** 3) #converts ft^3/s to m^3/h\nq_LCH4 = QLCH4 * 3600 * (.3048 ** 3) #converts ft^3/s to m^3/h\n\n\nps = 200 #static pressure in fluid close to impeller in psi\npv_LOX = \npv_LCH4 = \nA_LOx = #see two lines below to see what to do \nA_LCH4 = #see two lines below to see what to do\nv_LOx = (mLOx / rho_LOx) / A_LOx #Define A above as the area of the inlet pipe in ft^2\nv_LCH4 = (mLCH4 / rho_LCH4) / A_LCH4 #Define A above as the area of the inlet pipe in ft^2\n\n\nNPSH_LOx = ps/rho_LOx * (v_LOx ** 2)/(2*9.81) - pv/rho_LOx #substitue pv as Vapor Pressure of Oxygen at temperature in psi above \nNPSH_LCH4 = ps/rho_LOx * (v_LOx ** 2)/(2*9.81) - pv/rho_LOx #substitue pv as Vapor Pressure of Methane at temperature in psi above \n\n\nnss_LOx = n*(q_LOx ** 0.5)/(NPSH_LOx ** 0.75)\nnss_LCH4 = n*(q_LCH4 ** 0.5)/(NPSH_LCH4 ** 0.75)",
"_____no_output_____"
]
],
[
[
"#### Inlet diameter",
"_____no_output_____"
]
],
[
[
"#Note: The equation given in the book uses a (1+tan(Beta1)/tan(alpha1)) term, but since the impeller is radial, alpha1 is 90 so the term goes to infinity and therefore results in a multiplication by 1\n#Beta1 is determined by finding the specific suction speed** and reading off of the graph, or using:\n#kn = 1 - (dn ** 2)/(d1 ** 2); Just choose a value (I assumed inlet diameter ~ 1.15x the size of dn, the hub diameter) since d1 depends on the value of kn and vice versa\n tan_Beta1_LOx = (kn) ** 1.1 * (125/nss_LOx) ** 2.2 * (nq_LOx/27) ** 0.418 #Calculates Beta with a 40% std deviation, so a large amount of values is determined with this formula\n tan_Beta1_LCH4 = (kn) ** 1.1 * (125/nss_LCH4) ** 2.2 * (nq_LCH4/27) ** 0.418 #Calculates Beta with a 40% std deviation, so a large amount of values is determined with this formula\n\nd1_LOx = 2.9 * (QImp_LOx/(fq*n*kn*tan_Beta1_LOx))^(1/3)\nd1_LCH4 = 2.9 * (QImp_LCH4/(fq*n*kn*tan_Beta1_LCH4))^(1/3)",
"_____no_output_____"
]
],
[
[
"#### Exit Diameter",
"_____no_output_____"
]
],
[
[
"d2_LOx = 60/(np.pi * n) * (2 * 9.81 * (HLOx * 0.3048)/(PC_LOX)) ** 0.5\nd2_LCH4 = 60/(np.pi * n) * (2 * 9.81 * (HLCH4 * 0.3048)/(PC_LCH4)) ** 0.5",
"_____no_output_____"
]
],
[
[
"#### Blade Thickness",
"_____no_output_____"
]
],
[
[
"e_LOx = 0.022 * d2_LOx #Blade thickness for LOx, this number may have to go up for manufacturing purposes\ne_LCH4 = 0.022 * d2_LCH4 #Blade thickness for LCH4, this number may have to go up for manufacturing purposes",
"_____no_output_____"
]
],
[
[
"#### Leading and Trailing Edge Profiles",
"_____no_output_____"
]
],
[
[
"cp_min_sf = 0.155\nLp1_LOx = (2 + (4 + 4 * ((cp_min_sf/0.373)/e_LOx)*(0.373 * e_LOx)) ** 0.5)/ (2 * (cp_min_sf/0.373)/e_LOx) #Leading Edge profile, simplification of formula in Centrifugal Pumps in terms of Quadratic formula\nLp2_LOx = (2 - (4 + 4 * ((cp_min_sf/0.373)/e_LOx)*(0.373 * e_LOx)) ** 0.5)/ (2 * (cp_min_sf/0.373)/e_LOx) #Leading Edge profile, simplification of formula in Centrifugal Pumps in terms of Quadratic formula\n\nLp1_LCH4 = (2 + (4 + 4 * ((cp_min_sf/0.373)/e_LCH4)*(0.373 * e_LCH4)) ** 0.5)/ (2 * (cp_min_sf/0.373)/e_LCH4) #Leading Edge profile, simplification of formula in Centrifugal Pumps in terms of Quadratic formula\nLp2_LCH4 = (2 - (4 + 4 * ((cp_min_sf/0.373)/e_LCH4)*(0.373 * e_LCH4)) ** 0.5)/ (2 * (cp_min_sf/0.373)/e_LCH4) #Leading Edge profile, simplification of formula in Centrifugal Pumps in terms of Quadratic formula\n#Take whichever value above comes out positive, assumed an elliptical profile where cp,min,sf was given as 0.155. Formula changes if cp_min_sf changes\n\nTE_LOx = e_LOx/2 #Trailing edge for Liquid Oxygen using the most simple formula given\nTE_LCH4 = e_LCH4/2 #Trailing edge for Liquid Methane using the most simple formula given",
"_____no_output_____"
]
],
[
[
"# Impeller Calcuations",
"_____no_output_____"
]
],
[
[
"#Reference values given on page 667 of Centrifugal Pumps and then converted to imperial from metric\nnq_ref = 40 #unitless\nHref = 3280.84 #meters to feet\nrho_ref = 62.428 #lb/ft^3 \n\n\ntau3 = 1 #given\nepsilon_sp = np.pi #Radians. Guessed from the fact that doube volutes are generally at 180\n\nQLe_LOx = QImp_LOx/0.95 * 0.0283168 #m^3/s. Assume that the leakages due to the volute are really low\nQLe_LCH4 = QImp_LCH4/0.95 * 0.0283168 #m^3/s\n\nb3_LOx = 1 #Guess; Width of the diffuser inlet (cm)\nb3_LCH4 = 1 #Guess; Width of the diffuser inlet (cm)\n\nu2_LOX = (np.pi*d2_LOx*n)/60 #Circumferential speed at the outer diameter of the impeller for Liquid Oxygen\nu2_LCH4 = (np.pi*d2_LCH4*n)/60 #Circumferential speed at the outer diameter of the impeller for Liquid Methane\n\nu1m_LOx = (np.pi*d1_LOx*n)/60 #Circumferential speed at the inner diameter of the impeller for Liquid Oxygen\nu1m_LCH4 = (np.pi*d1_LOx*n)/60 #Circumferential speed at the inner diameter of the impeller for Liquid Methane\n\nc1u = 1 #Formula is c1m/tan(alpha1) but alpha1 is 90 degrees, so it simplifies to 1\nQref = 1 #Since Volumetric Flow was calculated absolutely, the \"reference\" value is 1\n\na = 1 #Taken from book for Q less than or equal to 1 m^3/s\nm_LOx = 0.08 * a * (Qref/QImp_LOx) ** 0.15 * (45/nq_LOx) ** 0.06 #Exponential to find hydraulic efficiency\nm_LCH4 = 0.08 * a * (Qref/QImp_LCH4) ** 0.15 * (45/nq_LCH4) ** 0.06 #Expoential to find hydraulic efficiency\n\nEff_Hyd_LOx = 1 - 0.055 * (Qref/QImp_LOx) ** m_LOx - 0.2 * (0.26 - np.log10(nq_LOx/25)) ** 2 #Hydraulic Efficiency of LOx Pump\nEff_Hyd_LCH4 = 1 - 0.055 * (Qref/QImp_LCH4) ** m_LCH4 - 0.2 * (0.26 - np.log10(nq_LCH4/25)) ** 2 #Hydraulic Efficiency of LCH4 Pump\n\nc2u_LOx = (g*HLOx)/(Eff_Hyd_LOx*u2_LOx)+(u1m_LOx*c1u)/u2_LOx #Circumferential component of absolute velocity at impeller outlet for Liquid Oxygen \nc2u_LCH4 = (g*HLCH4)/(Eff_Hyd_LCH4*u2_LCH4)+(u1m_LCH4*c1u)/u2_LCH4 #Circumferential component of absolute velocity at impeller outlet for Liquid Methane\n\nd3_LOx = d2_LOx * (1.03 + 0.1*(nq_LOx/nq_ref)*0.07(rho_LOx * HLOX)/(rho_ref*Href)) #distance of the gap bewteen the impeller and volute for Liquid Oxygen\nd3_LCH4 = d2_LCH4 * (1.03 + 0.1*(nq_LCH4/nq_ref)*0.07(rho_LCH4 * HLCH4)/(rho_ref*Href)) #distance of the gap bewteen the impeller and volute for Liquid Methane\n\nc3u_LOx = d2_LOx * c2u_LOx / d3_LOx #Circumferential component of absolute velocity at diffuser inlet for Liquid Oxygen\nc3u_LCH4 = d2_LCH4 * c2u_LCH4 / d3_LCH4 #Circumferential component of absolute velocity at diffuser inlet for Liquid Methane\n\nc3m_LOx = QLe_LOx*tau3/(np.pi*d3_LOx*b3_LOx) #Meridional component of absolute velocity at diffuser inlet for Liquid Oxygen\nc3m_LCH4 = QLe_LCH4 * tau3/(np.pi*d3_LCH4*b3_LCH4) #Meridional component of absolute velocity at diffuser inlet for Liquid Methane\n\ntan_alpha3_LOx = c3m_LOx/c3u_LOx #Flow angle at diffuser inlet with blockage for Liquid Oxygen\ntan_alpha3_LCH4 = c3m_LCH4/c3u_LCH4 #Flow angle at diffuser inlet with blockage for Liquid Methane\n\nalpha3b_LOx = np.degrees(np.arctan(tan_alpha3_LOx)) + 3 #Degrees. Diffuser vane inlet, can change the scalar 3 anywhere in the realm of real numbers of [-3,3] for Liquid Oxygen\nalpha3b_LCH4 = np.degrees(np.arctan(tan_alpha3_LCH4)) + 3 #Degrees. Diffuser vane inlet, can change the scalar 3 anywhere in the realm of real numbers of [-3,3] for Liquid Methane\n\nr2_LOx = d2_LOx/2 #Radius of the impeller outlet for Liquid Oxygen\nr2_LCH4 = d2_LCH4/2 #Radius of the impeller outlet for Liquid Methane\n\n#Throat area calculations, many variables are used that aren't entirely explained\nXsp_LOx = (QLe_LOx * epsilon_sp)/(np.pi*c2u_LOx*r2_LOx * 2 * np.pi) \nXsp_LCH4 = (QLe_LCH4 * epsilon_sp)/(np.pi*c2u_LCH4*r2_LCH4 * 2 * np.pi)\n\nd3q_LOx = Xsp_LOx + (2*d3_LOx*Xsp_LOx) ** 0.5\nd3q_LCH4 = Xsp_LCH4 + (2*d3_LCH4*Xsp_LCH4) ** 0.5\n\nA3q_LOx = np.pi*((d3q_LOx) ** 2)/4 \nA3q_LCH4 = np.pi*((d3q_LCH4) ** 2)/4",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb8dcc7ba7d6205fa29e88d62c1e84a2a65cb6bc
| 532,720 |
ipynb
|
Jupyter Notebook
|
mnist_dnn/mnist_dnn.ipynb
|
mett29/Deep-Learning
|
96e8bc1fdd191beaa2af9795c7620bada42c1a0a
|
[
"MIT"
] | 8 |
2018-05-17T13:09:09.000Z
|
2020-11-10T18:48:00.000Z
|
mnist_dnn/mnist_dnn.ipynb
|
mett29/Deep-Learning
|
96e8bc1fdd191beaa2af9795c7620bada42c1a0a
|
[
"MIT"
] | null | null | null |
mnist_dnn/mnist_dnn.ipynb
|
mett29/Deep-Learning
|
96e8bc1fdd191beaa2af9795c7620bada42c1a0a
|
[
"MIT"
] | 2 |
2020-01-29T05:57:22.000Z
|
2020-05-21T15:21:18.000Z
| 71.458082 | 423 | 0.666795 |
[
[
[
"import numpy as np\nimport tensorflow as tf\nfrom tensorflow.examples.tutorials.mnist import input_data\nfrom functools import partial",
"C:\\Miniconda3\\lib\\site-packages\\h5py\\__init__.py:34: FutureWarning: Conversion of the second argument of issubdtype from `float` to `np.floating` is deprecated. In future, it will be treated as `np.float64 == np.dtype(float).type`.\n from ._conv import register_converters as _register_converters\n"
],
[
"n_inputs = 28*28\nn_hidden1 = 100\nn_hidden2 = 100\nn_hidden3 = 100\nn_hidden4 = 100\nn_hidden5 = 100\nn_outputs = 5\n\n# Let's define the placeholders for the inputs and the targets\nX = tf.placeholder(tf.float32, shape=(None, n_inputs), name=\"X\")\ny = tf.placeholder(tf.int64, shape=(None), name=\"y\")",
"_____no_output_____"
],
[
"# Let's create the DNN\nhe_init = tf.contrib.layers.variance_scaling_initializer()\nmy_dense_layer = partial(\n tf.layers.dense, activation=tf.nn.elu, \n kernel_initializer=he_init)\n\nwith tf.name_scope(\"dnn\"):\n hidden1 = my_dense_layer(X, n_hidden1, name=\"hidden1\")\n hidden2 = my_dense_layer(hidden1, n_hidden2, name=\"hidden2\")\n hidden3 = my_dense_layer(hidden2, n_hidden3, name=\"hidden3\")\n hidden4 = my_dense_layer(hidden3, n_hidden4, name=\"hidden4\")\n hidden5 = my_dense_layer(hidden4, n_hidden5, name=\"hidden5\")\n logits = my_dense_layer(hidden5, n_outputs, activation=None, name=\"outputs\")\n Y_proba = tf.nn.softmax(logits, name=\"Y_proba\")\n \nlearning_rate = 0.01\n\nwith tf.name_scope(\"loss\"):\n xentropy = tf.nn.sparse_softmax_cross_entropy_with_logits(labels=y, logits=logits)\n loss = tf.reduce_mean(xentropy, name=\"loss\")\n\nwith tf.name_scope(\"train\"):\n optimizer = tf.train.AdamOptimizer(learning_rate)\n training_op = optimizer.minimize(loss, name=\"training_op\")\n\nwith tf.name_scope(\"eval\"):\n correct = tf.nn.in_top_k(logits, y , 1)\n accuracy = tf.reduce_mean(tf.cast(correct, tf.float32), name=\"accuracy\")\n\ninit = tf.global_variables_initializer()\nsaver = tf.train.Saver()",
"_____no_output_____"
],
[
"mnist = input_data.read_data_sets(\"/tmp/data/\")",
"Extracting /tmp/data/train-images-idx3-ubyte.gz\nExtracting /tmp/data/train-labels-idx1-ubyte.gz\nExtracting /tmp/data/t10k-images-idx3-ubyte.gz\nExtracting /tmp/data/t10k-labels-idx1-ubyte.gz\n"
],
[
"X_train1 = mnist.train.images[mnist.train.labels < 5]\ny_train1 = mnist.train.labels[mnist.train.labels < 5]\nX_valid1 = mnist.validation.images[mnist.validation.labels < 5]\ny_valid1 = mnist.validation.labels[mnist.validation.labels < 5]\nX_test1 = mnist.test.images[mnist.test.labels < 5]\ny_test1 = mnist.test.labels[mnist.test.labels < 5]",
"_____no_output_____"
],
[
"n_epochs = 1000\nbatch_size = 20\n\nmax_checks_without_progress = 20\nchecks_without_progress = 0\nbest_loss = np.infty\n\nwith tf.Session() as sess:\n init.run()\n\n for epoch in range(n_epochs):\n rnd_idx = np.random.permutation(len(X_train1))\n for rnd_indices in np.array_split(rnd_idx, len(X_train1) // batch_size):\n X_batch, y_batch = X_train1[rnd_indices], y_train1[rnd_indices]\n sess.run(training_op, feed_dict={X: X_batch, y: y_batch})\n # Calculate loss and acc on the validation set to do early stopping\n loss_val, acc_val = sess.run([loss, accuracy], feed_dict={X: X_valid1, y: y_valid1})\n if loss_val < best_loss:\n save_path = saver.save(sess, \"./my_mnist_model_0_to_4.ckpt\")\n best_loss = loss_val\n checks_without_progress = 0\n else:\n checks_without_progress += 1\n if checks_without_progress > max_checks_without_progress:\n print(\"Early stopping!\")\n break\n print(\"{}\\tValidation loss: {:.6f}\\tBest loss: {:.6f}\\tAccuracy: {:.2f}%\".format(\n epoch, loss_val, best_loss, acc_val * 100))\n\nwith tf.Session() as sess:\n saver.restore(sess, \"./my_mnist_model_0_to_4.ckpt\")\n acc_test = accuracy.eval(feed_dict={X: X_test1, y: y_test1})\n print(\"Final test accuracy: {:.2f}%\".format(acc_test * 100))",
"0\tValidation loss: 0.181033\tBest loss: 0.181033\tAccuracy: 95.27%\n1\tValidation loss: 0.125367\tBest loss: 0.125367\tAccuracy: 97.19%\n2\tValidation loss: 0.258047\tBest loss: 0.125367\tAccuracy: 97.30%\n3\tValidation loss: 0.137535\tBest loss: 0.125367\tAccuracy: 97.07%\n4\tValidation loss: 0.120659\tBest loss: 0.120659\tAccuracy: 97.11%\n5\tValidation loss: 0.467715\tBest loss: 0.120659\tAccuracy: 75.76%\n6\tValidation loss: 0.246886\tBest loss: 0.120659\tAccuracy: 95.97%\n7\tValidation loss: 0.234324\tBest loss: 0.120659\tAccuracy: 97.15%\n8\tValidation loss: 0.136194\tBest loss: 0.120659\tAccuracy: 97.58%\n9\tValidation loss: 0.360426\tBest loss: 0.120659\tAccuracy: 96.87%\n10\tValidation loss: 0.769926\tBest loss: 0.120659\tAccuracy: 74.39%\n11\tValidation loss: 0.742954\tBest loss: 0.120659\tAccuracy: 78.81%\n12\tValidation loss: 0.542269\tBest loss: 0.120659\tAccuracy: 77.37%\n13\tValidation loss: 0.487362\tBest loss: 0.120659\tAccuracy: 79.20%\n14\tValidation loss: 0.510532\tBest loss: 0.120659\tAccuracy: 79.24%\n15\tValidation loss: 0.466710\tBest loss: 0.120659\tAccuracy: 79.79%\n16\tValidation loss: 0.795642\tBest loss: 0.120659\tAccuracy: 60.52%\n17\tValidation loss: 1.274218\tBest loss: 0.120659\tAccuracy: 40.03%\n18\tValidation loss: 1.294121\tBest loss: 0.120659\tAccuracy: 41.71%\n19\tValidation loss: 1.312004\tBest loss: 0.120659\tAccuracy: 38.39%\n20\tValidation loss: 1.325414\tBest loss: 0.120659\tAccuracy: 38.47%\n21\tValidation loss: 1.069673\tBest loss: 0.120659\tAccuracy: 67.63%\n22\tValidation loss: 0.762168\tBest loss: 0.120659\tAccuracy: 91.95%\n23\tValidation loss: 0.281776\tBest loss: 0.120659\tAccuracy: 94.96%\n24\tValidation loss: 0.311277\tBest loss: 0.120659\tAccuracy: 97.42%\nEarly stopping!\nINFO:tensorflow:Restoring parameters from ./my_mnist_model_0_to_4.ckpt\nFinal test accuracy: 97.92%\n"
]
],
[
[
"<h1>DNNClassifier</h1>",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport tensorflow as tf\nfrom sklearn.base import BaseEstimator, ClassifierMixin\nfrom sklearn.exceptions import NotFittedError\n\nclass DNNClassifier(BaseEstimator, ClassifierMixin):\n \n def __init__(self, n_hidden_layers=5, n_neurons=100, optimizer_class=tf.train.AdamOptimizer,\n learning_rate=0.01, batch_size=20, activation=tf.nn.elu, initializer=he_init,\n batch_norm_momentum=None, dropout_rate=None, random_state=None):\n \"\"\"Initialize the DNNClassifier by simply storing all the hyperparameters.\"\"\"\n self.n_hidden_layers = n_hidden_layers\n self.n_neurons = n_neurons\n self.optimizer_class = optimizer_class\n self.learning_rate = learning_rate\n self.batch_size = batch_size\n self.activation = activation\n self.initializer = initializer\n self.batch_norm_momentum = batch_norm_momentum\n self.dropout_rate = dropout_rate\n self.random_state = random_state\n self._session = None\n\n def _dnn(self, inputs):\n \"\"\"Build the hidden layers, with support for batch normalization and dropout.\"\"\"\n for layer in range(self.n_hidden_layers):\n if self.dropout_rate:\n inputs = tf.layers.dropout(inputs, self.dropout_rate, training=self._training)\n inputs = tf.layers.dense(inputs, self.n_neurons,\n kernel_initializer=self.initializer,\n name=\"hidden%d\" % (layer + 1))\n if self.batch_norm_momentum:\n inputs = tf.layers.batch_normalization(inputs, momentum=self.batch_norm_momentum,\n training=self._training)\n inputs = self.activation(inputs, name=\"hidden%d_out\" % (layer + 1))\n return inputs\n\n def _build_graph(self, n_inputs, n_outputs):\n \"\"\"Build the same model as earlier\"\"\"\n if self.random_state is not None:\n tf.set_random_seed(self.random_state)\n np.random.seed(self.random_state)\n\n X = tf.placeholder(tf.float32, shape=(None, n_inputs), name=\"X\")\n y = tf.placeholder(tf.int32, shape=(None), name=\"y\")\n\n if self.batch_norm_momentum or self.dropout_rate:\n self._training = tf.placeholder_with_default(False, shape=(), name='training')\n else:\n self._training = None\n\n dnn_outputs = self._dnn(X)\n\n logits = tf.layers.dense(dnn_outputs, n_outputs, kernel_initializer=he_init, name=\"logits\")\n Y_proba = tf.nn.softmax(logits, name=\"Y_proba\")\n\n xentropy = tf.nn.sparse_softmax_cross_entropy_with_logits(labels=y,\n logits=logits)\n loss = tf.reduce_mean(xentropy, name=\"loss\")\n\n optimizer = self.optimizer_class(learning_rate=self.learning_rate)\n training_op = optimizer.minimize(loss)\n\n correct = tf.nn.in_top_k(logits, y, 1)\n accuracy = tf.reduce_mean(tf.cast(correct, tf.float32), name=\"accuracy\")\n\n init = tf.global_variables_initializer()\n saver = tf.train.Saver()\n\n # Make the important operations available easily through instance variables\n self._X, self._y = X, y\n self._Y_proba, self._loss = Y_proba, loss\n self._training_op, self._accuracy = training_op, accuracy\n self._init, self._saver = init, saver\n\n def close_session(self):\n if self._session:\n self._session.close()\n\n def _get_model_params(self):\n \"\"\"Get all variable values (used for early stopping, faster than saving to disk)\"\"\"\n with self._graph.as_default():\n gvars = tf.get_collection(tf.GraphKeys.GLOBAL_VARIABLES)\n return {gvar.op.name: value for gvar, value in zip(gvars, self._session.run(gvars))}\n\n def _restore_model_params(self, model_params):\n \"\"\"Set all variables to the given values (for early stopping, faster than loading from disk)\"\"\"\n gvar_names = list(model_params.keys())\n assign_ops = {gvar_name: self._graph.get_operation_by_name(gvar_name + \"/Assign\")\n for gvar_name in gvar_names}\n init_values = {gvar_name: assign_op.inputs[1] for gvar_name, assign_op in assign_ops.items()}\n feed_dict = {init_values[gvar_name]: model_params[gvar_name] for gvar_name in gvar_names}\n self._session.run(assign_ops, feed_dict=feed_dict)\n\n def fit(self, X, y, n_epochs=100, X_valid=None, y_valid=None):\n \"\"\"Fit the model to the training set. If X_valid and y_valid are provided, use early stopping.\"\"\"\n self.close_session()\n\n # infer n_inputs and n_outputs from the training set.\n n_inputs = X.shape[1]\n self.classes_ = np.unique(y)\n n_outputs = len(self.classes_)\n \n # Translate the labels vector to a vector of sorted class indices, containing\n # integers from 0 to n_outputs - 1.\n # For example, if y is equal to [8, 8, 9, 5, 7, 6, 6, 6], then the sorted class\n # labels (self.classes_) will be equal to [5, 6, 7, 8, 9], and the labels vector\n # will be translated to [3, 3, 4, 0, 2, 1, 1, 1]\n self.class_to_index_ = {label: index\n for index, label in enumerate(self.classes_)}\n y = np.array([self.class_to_index_[label]\n for label in y], dtype=np.int32)\n \n self._graph = tf.Graph()\n with self._graph.as_default():\n self._build_graph(n_inputs, n_outputs)\n # extra ops for batch normalization\n extra_update_ops = tf.get_collection(tf.GraphKeys.UPDATE_OPS)\n\n # needed in case of early stopping\n max_checks_without_progress = 20\n checks_without_progress = 0\n best_loss = np.infty\n best_params = None\n \n # Now train the model!\n self._session = tf.Session(graph=self._graph)\n with self._session.as_default() as sess:\n self._init.run()\n for epoch in range(n_epochs):\n rnd_idx = np.random.permutation(len(X))\n for rnd_indices in np.array_split(rnd_idx, len(X) // self.batch_size):\n X_batch, y_batch = X[rnd_indices], y[rnd_indices]\n feed_dict = {self._X: X_batch, self._y: y_batch}\n if self._training is not None:\n feed_dict[self._training] = True\n sess.run(self._training_op, feed_dict=feed_dict)\n if extra_update_ops:\n sess.run(extra_update_ops, feed_dict=feed_dict)\n if X_valid is not None and y_valid is not None:\n loss_val, acc_val = sess.run([self._loss, self._accuracy],\n feed_dict={self._X: X_valid,\n self._y: y_valid})\n if loss_val < best_loss:\n best_params = self._get_model_params()\n best_loss = loss_val\n checks_without_progress = 0\n else:\n checks_without_progress += 1\n print(\"{}\\tValidation loss: {:.6f}\\tBest loss: {:.6f}\\tAccuracy: {:.2f}%\".format(\n epoch, loss_val, best_loss, acc_val * 100))\n if checks_without_progress > max_checks_without_progress:\n print(\"Early stopping!\")\n break\n else:\n loss_train, acc_train = sess.run([self._loss, self._accuracy],\n feed_dict={self._X: X_batch,\n self._y: y_batch})\n print(\"{}\\tLast training batch loss: {:.6f}\\tAccuracy: {:.2f}%\".format(\n epoch, loss_train, acc_train * 100))\n # If we used early stopping then rollback to the best model found\n if best_params:\n self._restore_model_params(best_params)\n return self\n\n def predict_proba(self, X):\n if not self._session:\n raise NotFittedError(\"This %s instance is not fitted yet\" % self.__class__.__name__)\n with self._session.as_default() as sess:\n return self._Y_proba.eval(feed_dict={self._X: X})\n\n def predict(self, X):\n class_indices = np.argmax(self.predict_proba(X), axis=1)\n return np.array([[self.classes_[class_index]]\n for class_index in class_indices], np.int32)\n\n def save(self, path):\n self._saver.save(self._session, path)",
"_____no_output_____"
],
[
"dnn_clf = DNNClassifier(random_state=42)\ndnn_clf.fit(X_train1, y_train1, n_epochs=1000, X_valid=X_valid1, y_valid=y_valid1)",
"0\tValidation loss: 0.190826\tBest loss: 0.190826\tAccuracy: 96.64%\n1\tValidation loss: 1.689649\tBest loss: 0.190826\tAccuracy: 18.73%\n2\tValidation loss: 1.660114\tBest loss: 0.190826\tAccuracy: 20.91%\n3\tValidation loss: 1.778077\tBest loss: 0.190826\tAccuracy: 22.01%\n4\tValidation loss: 1.667106\tBest loss: 0.190826\tAccuracy: 22.01%\n5\tValidation loss: 1.654532\tBest loss: 0.190826\tAccuracy: 22.01%\n6\tValidation loss: 1.680933\tBest loss: 0.190826\tAccuracy: 18.73%\n7\tValidation loss: 1.779077\tBest loss: 0.190826\tAccuracy: 22.01%\n8\tValidation loss: 1.699482\tBest loss: 0.190826\tAccuracy: 19.27%\n9\tValidation loss: 1.767771\tBest loss: 0.190826\tAccuracy: 20.91%\n10\tValidation loss: 1.629350\tBest loss: 0.190826\tAccuracy: 22.01%\n11\tValidation loss: 1.812643\tBest loss: 0.190826\tAccuracy: 22.01%\n12\tValidation loss: 1.675939\tBest loss: 0.190826\tAccuracy: 18.73%\n13\tValidation loss: 1.633259\tBest loss: 0.190826\tAccuracy: 20.91%\n14\tValidation loss: 1.652904\tBest loss: 0.190826\tAccuracy: 20.91%\n15\tValidation loss: 1.635943\tBest loss: 0.190826\tAccuracy: 20.91%\n16\tValidation loss: 1.718915\tBest loss: 0.190826\tAccuracy: 19.08%\n17\tValidation loss: 1.682456\tBest loss: 0.190826\tAccuracy: 19.27%\n18\tValidation loss: 1.675366\tBest loss: 0.190826\tAccuracy: 18.73%\n19\tValidation loss: 1.645805\tBest loss: 0.190826\tAccuracy: 19.08%\n20\tValidation loss: 1.722336\tBest loss: 0.190826\tAccuracy: 22.01%\n21\tValidation loss: 1.656422\tBest loss: 0.190826\tAccuracy: 22.01%\nEarly stopping!\n"
],
[
"from sklearn.metrics import accuracy_score\n\ny_pred = dnn_clf.predict(X_test1)\naccuracy_score(y_test1, y_pred)",
"_____no_output_____"
],
[
"from sklearn.model_selection import RandomizedSearchCV\n\ndef leaky_relu(alpha=0.01):\n def parametrized_leaky_relu(z, name=None):\n return tf.maximum(alpha * z, z, name=name)\n return parametrized_leaky_relu\n\nparam_distribs = {\n \"n_neurons\": [10, 30, 50, 70, 90, 100, 120, 140, 160],\n \"batch_size\": [10, 50, 100, 500],\n \"learning_rate\": [0.01, 0.02, 0.05, 0.1],\n \"activation\": [tf.nn.relu, tf.nn.elu, leaky_relu(alpha=0.01), leaky_relu(alpha=0.1)],\n # you could also try exploring different numbers of hidden layers, different optimizers, etc.\n #\"n_hidden_layers\": [0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10],\n #\"optimizer_class\": [tf.train.AdamOptimizer, partial(tf.train.MomentumOptimizer, momentum=0.95)],\n}\n\nrnd_search = RandomizedSearchCV(DNNClassifier(random_state=42), param_distribs, n_iter=50,\n fit_params={\"X_valid\": X_valid1, \"y_valid\": y_valid1, \"n_epochs\": 1000},\n random_state=42, verbose=2)\nrnd_search.fit(X_train1, y_train1)",
"Fitting 3 folds for each of 50 candidates, totalling 150 fits\n[CV] n_neurons=10, batch_size=100, activation=<function elu at 0x000001A7BE2E2268>, learning_rate=0.05 \n0\tValidation loss: 0.131135\tBest loss: 0.131135\tAccuracy: 96.36%\n1\tValidation loss: 0.130872\tBest loss: 0.130872\tAccuracy: 96.17%\n2\tValidation loss: 0.153402\tBest loss: 0.130872\tAccuracy: 96.36%\n3\tValidation loss: 0.145776\tBest loss: 0.130872\tAccuracy: 96.76%\n4\tValidation loss: 0.132235\tBest loss: 0.130872\tAccuracy: 96.87%\n5\tValidation loss: 0.140970\tBest loss: 0.130872\tAccuracy: 96.99%\n6\tValidation loss: 0.150057\tBest loss: 0.130872\tAccuracy: 96.25%\n7\tValidation loss: 0.138547\tBest loss: 0.130872\tAccuracy: 96.64%\n8\tValidation loss: 0.129539\tBest loss: 0.129539\tAccuracy: 96.05%\n9\tValidation loss: 0.131695\tBest loss: 0.129539\tAccuracy: 96.44%\n10\tValidation loss: 0.135688\tBest loss: 0.129539\tAccuracy: 96.68%\n11\tValidation loss: 1.787319\tBest loss: 0.129539\tAccuracy: 19.12%\n12\tValidation loss: 1.630504\tBest loss: 0.129539\tAccuracy: 20.91%\n13\tValidation loss: 1.644414\tBest loss: 0.129539\tAccuracy: 18.73%\n14\tValidation loss: 1.617375\tBest loss: 0.129539\tAccuracy: 22.01%\n15\tValidation loss: 1.611893\tBest loss: 0.129539\tAccuracy: 22.01%\n16\tValidation loss: 1.612366\tBest loss: 0.129539\tAccuracy: 20.91%\n17\tValidation loss: 1.616471\tBest loss: 0.129539\tAccuracy: 19.27%\n18\tValidation loss: 1.616871\tBest loss: 0.129539\tAccuracy: 19.27%\n19\tValidation loss: 1.624991\tBest loss: 0.129539\tAccuracy: 22.01%\n20\tValidation loss: 1.626455\tBest loss: 0.129539\tAccuracy: 19.08%\n21\tValidation loss: 1.632699\tBest loss: 0.129539\tAccuracy: 19.27%\n22\tValidation loss: 1.620706\tBest loss: 0.129539\tAccuracy: 18.73%\n23\tValidation loss: 1.613954\tBest loss: 0.129539\tAccuracy: 20.91%\n24\tValidation loss: 1.626613\tBest loss: 0.129539\tAccuracy: 22.01%\n25\tValidation loss: 1.620113\tBest loss: 0.129539\tAccuracy: 22.01%\n26\tValidation loss: 1.643669\tBest loss: 0.129539\tAccuracy: 18.73%\n27\tValidation loss: 1.622781\tBest loss: 0.129539\tAccuracy: 19.27%\n28\tValidation loss: 1.624650\tBest loss: 0.129539\tAccuracy: 19.27%\n29\tValidation loss: 1.615069\tBest loss: 0.129539\tAccuracy: 22.01%\nEarly stopping!\n[CV] n_neurons=10, batch_size=100, activation=<function elu at 0x000001A7BE2E2268>, learning_rate=0.05, total= 12.1s\n[CV] n_neurons=10, batch_size=100, activation=<function elu at 0x000001A7BE2E2268>, learning_rate=0.05 \n"
],
[
"rnd_search.best_params_",
"_____no_output_____"
],
[
"y_pred = rnd_search.predict(X_test1)\naccuracy_score(y_test1, y_pred)",
"_____no_output_____"
],
[
"rnd_search.best_estimator_.save(\"./my_best_mnist_model_0_to_4\")",
"_____no_output_____"
],
[
"# Let's train the best model found, once again, to see how fast it converges\ndnn_clf = DNNClassifier(activation=leaky_relu(alpha=0.1), batch_size=500, learning_rate=0.01,\n n_neurons=50, random_state=42)\ndnn_clf.fit(X_train1, y_train1, n_epochs=1000, X_valid=X_valid1, y_valid=y_valid1)",
"0\tValidation loss: 0.072400\tBest loss: 0.072400\tAccuracy: 97.62%\n1\tValidation loss: 0.067381\tBest loss: 0.067381\tAccuracy: 97.93%\n2\tValidation loss: 0.050353\tBest loss: 0.050353\tAccuracy: 98.40%\n3\tValidation loss: 0.057750\tBest loss: 0.050353\tAccuracy: 98.28%\n4\tValidation loss: 0.052084\tBest loss: 0.050353\tAccuracy: 98.51%\n5\tValidation loss: 0.048122\tBest loss: 0.048122\tAccuracy: 98.59%\n6\tValidation loss: 0.053389\tBest loss: 0.048122\tAccuracy: 98.67%\n7\tValidation loss: 0.046709\tBest loss: 0.046709\tAccuracy: 98.91%\n8\tValidation loss: 0.051118\tBest loss: 0.046709\tAccuracy: 98.71%\n9\tValidation loss: 0.049088\tBest loss: 0.046709\tAccuracy: 98.87%\n10\tValidation loss: 0.064235\tBest loss: 0.046709\tAccuracy: 98.36%\n11\tValidation loss: 0.045773\tBest loss: 0.045773\tAccuracy: 98.79%\n12\tValidation loss: 0.052007\tBest loss: 0.045773\tAccuracy: 98.79%\n13\tValidation loss: 0.054095\tBest loss: 0.045773\tAccuracy: 98.87%\n14\tValidation loss: 0.051559\tBest loss: 0.045773\tAccuracy: 98.87%\n15\tValidation loss: 0.063819\tBest loss: 0.045773\tAccuracy: 98.79%\n16\tValidation loss: 0.062628\tBest loss: 0.045773\tAccuracy: 98.67%\n17\tValidation loss: 0.085544\tBest loss: 0.045773\tAccuracy: 98.32%\n18\tValidation loss: 0.057520\tBest loss: 0.045773\tAccuracy: 98.83%\n19\tValidation loss: 0.043413\tBest loss: 0.043413\tAccuracy: 99.06%\n20\tValidation loss: 0.044119\tBest loss: 0.043413\tAccuracy: 99.06%\n21\tValidation loss: 0.041425\tBest loss: 0.041425\tAccuracy: 99.18%\n22\tValidation loss: 0.054479\tBest loss: 0.041425\tAccuracy: 99.06%\n23\tValidation loss: 0.077733\tBest loss: 0.041425\tAccuracy: 98.94%\n24\tValidation loss: 0.098854\tBest loss: 0.041425\tAccuracy: 98.59%\n25\tValidation loss: 0.056819\tBest loss: 0.041425\tAccuracy: 99.06%\n26\tValidation loss: 0.063282\tBest loss: 0.041425\tAccuracy: 98.44%\n27\tValidation loss: 0.062965\tBest loss: 0.041425\tAccuracy: 98.71%\n28\tValidation loss: 0.056530\tBest loss: 0.041425\tAccuracy: 98.79%\n29\tValidation loss: 0.054551\tBest loss: 0.041425\tAccuracy: 98.98%\n30\tValidation loss: 0.071203\tBest loss: 0.041425\tAccuracy: 98.87%\n31\tValidation loss: 0.054987\tBest loss: 0.041425\tAccuracy: 98.67%\n32\tValidation loss: 0.074646\tBest loss: 0.041425\tAccuracy: 98.71%\n33\tValidation loss: 0.067929\tBest loss: 0.041425\tAccuracy: 98.83%\n34\tValidation loss: 0.066096\tBest loss: 0.041425\tAccuracy: 98.83%\n35\tValidation loss: 0.072558\tBest loss: 0.041425\tAccuracy: 98.87%\n36\tValidation loss: 0.077099\tBest loss: 0.041425\tAccuracy: 98.79%\n37\tValidation loss: 0.095041\tBest loss: 0.041425\tAccuracy: 98.67%\n38\tValidation loss: 0.047264\tBest loss: 0.041425\tAccuracy: 98.91%\n39\tValidation loss: 0.076432\tBest loss: 0.041425\tAccuracy: 98.67%\n40\tValidation loss: 0.068125\tBest loss: 0.041425\tAccuracy: 98.83%\n41\tValidation loss: 0.056462\tBest loss: 0.041425\tAccuracy: 98.87%\n42\tValidation loss: 0.075779\tBest loss: 0.041425\tAccuracy: 98.75%\nEarly stopping!\n"
],
[
"y_pred = dnn_clf.predict(X_test1)\naccuracy_score(y_test1, y_pred)\n\n# Here the accuracy is different because I put leaky_relu in the training instead of relu as the rnd_search_best_params says.\n# However, the accuracy is better...Wtf?!",
"_____no_output_____"
],
[
"# Let's try to add Batch Normalization\ndnn_clf_bn = DNNClassifier(activation=leaky_relu(alpha=0.1), batch_size=500, learning_rate=0.01,\n n_neurons=50, random_state=42,\n batch_norm_momentum=0.95)\ndnn_clf_bn.fit(X_train1, y_train1, n_epochs=1000, X_valid=X_valid1, y_valid=y_valid1)",
"0\tValidation loss: 0.057204\tBest loss: 0.057204\tAccuracy: 98.24%\n1\tValidation loss: 0.046775\tBest loss: 0.046775\tAccuracy: 98.20%\n2\tValidation loss: 0.038639\tBest loss: 0.038639\tAccuracy: 98.75%\n3\tValidation loss: 0.039907\tBest loss: 0.038639\tAccuracy: 98.75%\n4\tValidation loss: 0.045205\tBest loss: 0.038639\tAccuracy: 98.59%\n5\tValidation loss: 0.034912\tBest loss: 0.034912\tAccuracy: 98.94%\n6\tValidation loss: 0.032480\tBest loss: 0.032480\tAccuracy: 99.02%\n7\tValidation loss: 0.045129\tBest loss: 0.032480\tAccuracy: 98.98%\n8\tValidation loss: 0.044904\tBest loss: 0.032480\tAccuracy: 98.87%\n9\tValidation loss: 0.031486\tBest loss: 0.031486\tAccuracy: 99.10%\n10\tValidation loss: 0.049647\tBest loss: 0.031486\tAccuracy: 98.79%\n11\tValidation loss: 0.045901\tBest loss: 0.031486\tAccuracy: 98.91%\n12\tValidation loss: 0.043318\tBest loss: 0.031486\tAccuracy: 98.79%\n13\tValidation loss: 0.040476\tBest loss: 0.031486\tAccuracy: 99.02%\n14\tValidation loss: 0.043031\tBest loss: 0.031486\tAccuracy: 99.30%\n15\tValidation loss: 0.044041\tBest loss: 0.031486\tAccuracy: 98.87%\n16\tValidation loss: 0.036324\tBest loss: 0.031486\tAccuracy: 99.22%\n17\tValidation loss: 0.055593\tBest loss: 0.031486\tAccuracy: 98.87%\n18\tValidation loss: 0.049460\tBest loss: 0.031486\tAccuracy: 98.98%\n19\tValidation loss: 0.035340\tBest loss: 0.031486\tAccuracy: 99.26%\n20\tValidation loss: 0.034851\tBest loss: 0.031486\tAccuracy: 99.26%\n21\tValidation loss: 0.036083\tBest loss: 0.031486\tAccuracy: 99.06%\n22\tValidation loss: 0.043626\tBest loss: 0.031486\tAccuracy: 99.22%\n23\tValidation loss: 0.040572\tBest loss: 0.031486\tAccuracy: 99.06%\n24\tValidation loss: 0.049133\tBest loss: 0.031486\tAccuracy: 98.98%\n25\tValidation loss: 0.043373\tBest loss: 0.031486\tAccuracy: 98.87%\n26\tValidation loss: 0.040943\tBest loss: 0.031486\tAccuracy: 99.06%\n27\tValidation loss: 0.027661\tBest loss: 0.027661\tAccuracy: 99.22%\n28\tValidation loss: 0.035219\tBest loss: 0.027661\tAccuracy: 99.22%\n29\tValidation loss: 0.029462\tBest loss: 0.027661\tAccuracy: 99.37%\n30\tValidation loss: 0.033848\tBest loss: 0.027661\tAccuracy: 99.22%\n31\tValidation loss: 0.026993\tBest loss: 0.026993\tAccuracy: 99.37%\n32\tValidation loss: 0.032726\tBest loss: 0.026993\tAccuracy: 99.45%\n33\tValidation loss: 0.028211\tBest loss: 0.026993\tAccuracy: 99.45%\n34\tValidation loss: 0.049205\tBest loss: 0.026993\tAccuracy: 99.14%\n35\tValidation loss: 0.051756\tBest loss: 0.026993\tAccuracy: 98.94%\n36\tValidation loss: 0.027438\tBest loss: 0.026993\tAccuracy: 99.18%\n37\tValidation loss: 0.042923\tBest loss: 0.026993\tAccuracy: 99.10%\n38\tValidation loss: 0.041161\tBest loss: 0.026993\tAccuracy: 98.79%\n39\tValidation loss: 0.037147\tBest loss: 0.026993\tAccuracy: 99.18%\n40\tValidation loss: 0.037225\tBest loss: 0.026993\tAccuracy: 99.18%\n41\tValidation loss: 0.046190\tBest loss: 0.026993\tAccuracy: 99.14%\n42\tValidation loss: 0.034444\tBest loss: 0.026993\tAccuracy: 99.22%\n43\tValidation loss: 0.034811\tBest loss: 0.026993\tAccuracy: 99.14%\n44\tValidation loss: 0.040818\tBest loss: 0.026993\tAccuracy: 99.37%\n45\tValidation loss: 0.041241\tBest loss: 0.026993\tAccuracy: 99.06%\n46\tValidation loss: 0.045522\tBest loss: 0.026993\tAccuracy: 99.18%\n47\tValidation loss: 0.041817\tBest loss: 0.026993\tAccuracy: 99.34%\n48\tValidation loss: 0.032834\tBest loss: 0.026993\tAccuracy: 99.34%\n49\tValidation loss: 0.045425\tBest loss: 0.026993\tAccuracy: 98.94%\n50\tValidation loss: 0.038260\tBest loss: 0.026993\tAccuracy: 99.22%\n51\tValidation loss: 0.035299\tBest loss: 0.026993\tAccuracy: 99.26%\n52\tValidation loss: 0.031154\tBest loss: 0.026993\tAccuracy: 99.30%\nEarly stopping!\n"
],
[
"y_pred = dnn_clf_bn.predict(X_test1)\naccuracy_score(y_test1, y_pred)",
"_____no_output_____"
],
[
"# Mmm, Batch Normalization did not improve the accuracy. We should try to do another tuning for hyperparameters with BN\n# and try again.\n# ...",
"_____no_output_____"
],
[
"# Now let's go back to our previous model and see how well perform on the training set\ny_pred = dnn_clf.predict(X_train1)\naccuracy_score(y_train1, y_pred)",
"_____no_output_____"
],
[
"# Much better than the test set, so probably it is overfitting the training set. Let's try using dropout\ndnn_clf_dropout = DNNClassifier(activation=leaky_relu(alpha=0.1), batch_size=500, learning_rate=0.01,\n n_neurons=50, random_state=42,\n dropout_rate=0.5)\ndnn_clf_dropout.fit(X_train1, y_train1, n_epochs=1000, X_valid=X_valid1, y_valid=y_valid1)",
"0\tValidation loss: 0.502725\tBest loss: 0.502725\tAccuracy: 84.83%\n1\tValidation loss: 0.174035\tBest loss: 0.174035\tAccuracy: 94.72%\n2\tValidation loss: 0.132589\tBest loss: 0.132589\tAccuracy: 96.05%\n3\tValidation loss: 0.122869\tBest loss: 0.122869\tAccuracy: 96.64%\n4\tValidation loss: 0.114765\tBest loss: 0.114765\tAccuracy: 96.87%\n5\tValidation loss: 0.105026\tBest loss: 0.105026\tAccuracy: 97.46%\n6\tValidation loss: 0.096043\tBest loss: 0.096043\tAccuracy: 97.19%\n7\tValidation loss: 0.103087\tBest loss: 0.096043\tAccuracy: 97.62%\n8\tValidation loss: 0.093458\tBest loss: 0.093458\tAccuracy: 97.30%\n9\tValidation loss: 0.096742\tBest loss: 0.093458\tAccuracy: 97.46%\n10\tValidation loss: 0.098123\tBest loss: 0.093458\tAccuracy: 97.58%\n11\tValidation loss: 0.097413\tBest loss: 0.093458\tAccuracy: 97.38%\n12\tValidation loss: 0.089873\tBest loss: 0.089873\tAccuracy: 97.69%\n13\tValidation loss: 0.089845\tBest loss: 0.089845\tAccuracy: 97.62%\n14\tValidation loss: 0.082723\tBest loss: 0.082723\tAccuracy: 97.73%\n15\tValidation loss: 0.082565\tBest loss: 0.082565\tAccuracy: 97.65%\n16\tValidation loss: 0.077797\tBest loss: 0.077797\tAccuracy: 97.93%\n17\tValidation loss: 0.083845\tBest loss: 0.077797\tAccuracy: 98.01%\n18\tValidation loss: 0.072469\tBest loss: 0.072469\tAccuracy: 98.20%\n19\tValidation loss: 0.085881\tBest loss: 0.072469\tAccuracy: 98.01%\n20\tValidation loss: 0.084050\tBest loss: 0.072469\tAccuracy: 97.97%\n21\tValidation loss: 0.079772\tBest loss: 0.072469\tAccuracy: 98.01%\n22\tValidation loss: 0.081636\tBest loss: 0.072469\tAccuracy: 97.97%\n23\tValidation loss: 0.077294\tBest loss: 0.072469\tAccuracy: 97.97%\n24\tValidation loss: 0.078276\tBest loss: 0.072469\tAccuracy: 97.93%\n25\tValidation loss: 0.089046\tBest loss: 0.072469\tAccuracy: 98.01%\n26\tValidation loss: 0.085872\tBest loss: 0.072469\tAccuracy: 97.89%\n27\tValidation loss: 0.075593\tBest loss: 0.072469\tAccuracy: 98.20%\n28\tValidation loss: 0.078363\tBest loss: 0.072469\tAccuracy: 98.05%\n29\tValidation loss: 0.079937\tBest loss: 0.072469\tAccuracy: 98.01%\n30\tValidation loss: 0.075742\tBest loss: 0.072469\tAccuracy: 98.16%\n31\tValidation loss: 0.084797\tBest loss: 0.072469\tAccuracy: 97.81%\n32\tValidation loss: 0.077950\tBest loss: 0.072469\tAccuracy: 97.97%\n33\tValidation loss: 0.078195\tBest loss: 0.072469\tAccuracy: 98.08%\n34\tValidation loss: 0.081641\tBest loss: 0.072469\tAccuracy: 97.85%\n35\tValidation loss: 0.077142\tBest loss: 0.072469\tAccuracy: 97.89%\n36\tValidation loss: 0.074055\tBest loss: 0.072469\tAccuracy: 98.08%\n37\tValidation loss: 0.072604\tBest loss: 0.072469\tAccuracy: 98.12%\n38\tValidation loss: 0.080937\tBest loss: 0.072469\tAccuracy: 98.16%\n39\tValidation loss: 0.071516\tBest loss: 0.071516\tAccuracy: 98.24%\n40\tValidation loss: 0.072892\tBest loss: 0.071516\tAccuracy: 98.12%\n41\tValidation loss: 0.073297\tBest loss: 0.071516\tAccuracy: 98.16%\n42\tValidation loss: 0.071817\tBest loss: 0.071516\tAccuracy: 97.97%\n43\tValidation loss: 0.074031\tBest loss: 0.071516\tAccuracy: 98.05%\n44\tValidation loss: 0.076460\tBest loss: 0.071516\tAccuracy: 97.89%\n45\tValidation loss: 0.075179\tBest loss: 0.071516\tAccuracy: 97.97%\n46\tValidation loss: 0.078572\tBest loss: 0.071516\tAccuracy: 98.12%\n47\tValidation loss: 0.073359\tBest loss: 0.071516\tAccuracy: 98.40%\n48\tValidation loss: 0.067496\tBest loss: 0.067496\tAccuracy: 98.24%\n49\tValidation loss: 0.071818\tBest loss: 0.067496\tAccuracy: 97.93%\n50\tValidation loss: 0.069599\tBest loss: 0.067496\tAccuracy: 98.08%\n51\tValidation loss: 0.076246\tBest loss: 0.067496\tAccuracy: 98.20%\n52\tValidation loss: 0.067484\tBest loss: 0.067484\tAccuracy: 98.28%\n53\tValidation loss: 0.078478\tBest loss: 0.067484\tAccuracy: 97.93%\n54\tValidation loss: 0.074413\tBest loss: 0.067484\tAccuracy: 98.28%\n55\tValidation loss: 0.071111\tBest loss: 0.067484\tAccuracy: 98.12%\n56\tValidation loss: 0.074798\tBest loss: 0.067484\tAccuracy: 98.16%\n57\tValidation loss: 0.069056\tBest loss: 0.067484\tAccuracy: 98.12%\n58\tValidation loss: 0.070102\tBest loss: 0.067484\tAccuracy: 98.20%\n59\tValidation loss: 0.065691\tBest loss: 0.065691\tAccuracy: 98.24%\n60\tValidation loss: 0.067241\tBest loss: 0.065691\tAccuracy: 98.28%\n61\tValidation loss: 0.079002\tBest loss: 0.065691\tAccuracy: 98.20%\n62\tValidation loss: 0.073623\tBest loss: 0.065691\tAccuracy: 98.16%\n63\tValidation loss: 0.068377\tBest loss: 0.065691\tAccuracy: 98.32%\n64\tValidation loss: 0.067814\tBest loss: 0.065691\tAccuracy: 98.20%\n65\tValidation loss: 0.068876\tBest loss: 0.065691\tAccuracy: 98.24%\n66\tValidation loss: 0.071318\tBest loss: 0.065691\tAccuracy: 98.28%\n67\tValidation loss: 0.069749\tBest loss: 0.065691\tAccuracy: 98.32%\n68\tValidation loss: 0.068016\tBest loss: 0.065691\tAccuracy: 98.20%\n69\tValidation loss: 0.070979\tBest loss: 0.065691\tAccuracy: 98.16%\n70\tValidation loss: 0.072989\tBest loss: 0.065691\tAccuracy: 98.20%\n71\tValidation loss: 0.067382\tBest loss: 0.065691\tAccuracy: 98.20%\n72\tValidation loss: 0.076456\tBest loss: 0.065691\tAccuracy: 98.28%\n73\tValidation loss: 0.079694\tBest loss: 0.065691\tAccuracy: 98.36%\n74\tValidation loss: 0.073880\tBest loss: 0.065691\tAccuracy: 97.89%\n75\tValidation loss: 0.066215\tBest loss: 0.065691\tAccuracy: 98.28%\n76\tValidation loss: 0.069724\tBest loss: 0.065691\tAccuracy: 98.20%\n77\tValidation loss: 0.067828\tBest loss: 0.065691\tAccuracy: 98.40%\n78\tValidation loss: 0.070395\tBest loss: 0.065691\tAccuracy: 98.24%\n79\tValidation loss: 0.066952\tBest loss: 0.065691\tAccuracy: 98.40%\n80\tValidation loss: 0.072089\tBest loss: 0.065691\tAccuracy: 97.97%\nEarly stopping!\n"
],
[
"y_pred = dnn_clf_dropout.predict(X_test1)\naccuracy_score(y_test1, y_pred)",
"_____no_output_____"
],
[
"# Dropout doesn't seem to help. As said before, we could try to tune the network with dropout and see what we got.\n# ...",
"_____no_output_____"
]
],
[
[
"<h1>Transfer Learning</h1>\n\n<p>Let's try to reuse the previous model on digits from 5 to 9, using only 100 images per digit!</p>",
"_____no_output_____"
]
],
[
[
"restore_saver = tf.train.import_meta_graph(\"./my_best_mnist_model_0_to_4.meta\")\n\nX = tf.get_default_graph().get_tensor_by_name(\"X:0\")\ny = tf.get_default_graph().get_tensor_by_name(\"y:0\")\nloss = tf.get_default_graph().get_tensor_by_name(\"loss:0\")\nY_proba = tf.get_default_graph().get_tensor_by_name(\"Y_proba:0\")\nlogits = Y_proba.op.inputs[0]\naccuracy = tf.get_default_graph().get_tensor_by_name(\"accuracy:0\")",
"_____no_output_____"
],
[
"learning_rate = 0.01\n\noutput_layer_vars = tf.get_collection(tf.GraphKeys.TRAINABLE_VARIABLES, scope=\"logits\")\noptimizer = tf.train.AdamOptimizer(learning_rate, name=\"Adam2\")\n# Freeze all the hidden layers\ntraining_op = optimizer.minimize(loss, var_list=output_layer_vars)",
"_____no_output_____"
],
[
"correct = tf.nn.in_top_k(logits, y, 1)\naccuracy = tf.reduce_mean(tf.cast(correct, tf.float32), name=\"accuracy\")\n\ninit = tf.global_variables_initializer()\nfive_frozen_saver = tf.train.Saver()",
"_____no_output_____"
],
[
"X_train2_full = mnist.train.images[mnist.train.labels >= 5]\ny_train2_full = mnist.train.labels[mnist.train.labels >= 5] - 5\nX_valid2_full = mnist.validation.images[mnist.validation.labels >= 5]\ny_valid2_full = mnist.validation.labels[mnist.validation.labels >= 5] - 5\nX_test2 = mnist.test.images[mnist.test.labels >= 5]\ny_test2 = mnist.test.labels[mnist.test.labels >= 5] - 5",
"_____no_output_____"
],
[
"def sample_n_instances_per_class(X, y, n=100):\n Xs, ys = [], []\n for label in np.unique(y):\n idx = (y == label)\n Xc = X[idx][:n]\n yc = y[idx][:n]\n Xs.append(Xc)\n ys.append(yc)\n return np.concatenate(Xs), np.concatenate(ys)",
"_____no_output_____"
],
[
"X_train2, y_train2 = sample_n_instances_per_class(X_train2_full, y_train2_full, n=100)\nX_valid2, y_valid2 = sample_n_instances_per_class(X_valid2_full, y_valid2_full, n=30)",
"_____no_output_____"
],
[
"import time\n\nn_epochs = 1000\nbatch_size = 20\n\nmax_checks_without_progress = 20\nchecks_without_progress = 0\nbest_loss = np.infty\n\nwith tf.Session() as sess:\n init.run()\n restore_saver.restore(sess, \"./my_best_mnist_model_0_to_4\")\n for var in output_layer_vars:\n var.initializer.run()\n\n t0 = time.time()\n \n for epoch in range(n_epochs):\n rnd_idx = np.random.permutation(len(X_train2))\n for rnd_indices in np.array_split(rnd_idx, len(X_train2) // batch_size):\n X_batch, y_batch = X_train2[rnd_indices], y_train2[rnd_indices]\n sess.run(training_op, feed_dict={X: X_batch, y: y_batch})\n loss_val, acc_val = sess.run([loss, accuracy], feed_dict={X: X_valid2, y: y_valid2})\n if loss_val < best_loss:\n save_path = five_frozen_saver.save(sess, \"./my_mnist_model_5_to_9_five_frozen\")\n best_loss = loss_val\n checks_without_progress = 0\n else:\n checks_without_progress += 1\n if checks_without_progress > max_checks_without_progress:\n print(\"Early stopping!\")\n break\n print(\"{}\\tValidation loss: {:.6f}\\tBest loss: {:.6f}\\tAccuracy: {:.2f}%\".format(\n epoch, loss_val, best_loss, acc_val * 100))\n\n t1 = time.time()\n print(\"Total training time: {:.1f}s\".format(t1 - t0))\n\nwith tf.Session() as sess:\n five_frozen_saver.restore(sess, \"./my_mnist_model_5_to_9_five_frozen\")\n acc_test = accuracy.eval(feed_dict={X: X_test2, y: y_test2})\n print(\"Final test accuracy: {:.2f}%\".format(acc_test * 100))",
"INFO:tensorflow:Restoring parameters from ./my_best_mnist_model_0_to_4\n0\tValidation loss: 1.076381\tBest loss: 1.076381\tAccuracy: 58.00%\n1\tValidation loss: 0.995478\tBest loss: 0.995478\tAccuracy: 55.33%\n2\tValidation loss: 0.954706\tBest loss: 0.954706\tAccuracy: 64.67%\n3\tValidation loss: 0.941471\tBest loss: 0.941471\tAccuracy: 66.67%\n4\tValidation loss: 0.930525\tBest loss: 0.930525\tAccuracy: 66.67%\n5\tValidation loss: 0.912695\tBest loss: 0.912695\tAccuracy: 71.33%\n6\tValidation loss: 0.920844\tBest loss: 0.912695\tAccuracy: 65.33%\n7\tValidation loss: 0.890324\tBest loss: 0.890324\tAccuracy: 69.33%\n8\tValidation loss: 0.916895\tBest loss: 0.890324\tAccuracy: 64.67%\n9\tValidation loss: 0.965269\tBest loss: 0.890324\tAccuracy: 62.67%\n10\tValidation loss: 0.875946\tBest loss: 0.875946\tAccuracy: 67.33%\n11\tValidation loss: 0.953211\tBest loss: 0.875946\tAccuracy: 65.33%\n12\tValidation loss: 0.894069\tBest loss: 0.875946\tAccuracy: 66.67%\n13\tValidation loss: 0.890609\tBest loss: 0.875946\tAccuracy: 64.67%\n14\tValidation loss: 0.871395\tBest loss: 0.871395\tAccuracy: 72.00%\n15\tValidation loss: 0.860191\tBest loss: 0.860191\tAccuracy: 70.00%\n16\tValidation loss: 0.887385\tBest loss: 0.860191\tAccuracy: 65.33%\n17\tValidation loss: 0.888017\tBest loss: 0.860191\tAccuracy: 64.67%\n18\tValidation loss: 0.883475\tBest loss: 0.860191\tAccuracy: 67.33%\n19\tValidation loss: 0.965452\tBest loss: 0.860191\tAccuracy: 61.33%\n20\tValidation loss: 0.874323\tBest loss: 0.860191\tAccuracy: 67.33%\n21\tValidation loss: 0.872406\tBest loss: 0.860191\tAccuracy: 66.00%\n22\tValidation loss: 0.866727\tBest loss: 0.860191\tAccuracy: 71.33%\n23\tValidation loss: 0.857422\tBest loss: 0.857422\tAccuracy: 66.67%\n24\tValidation loss: 0.858661\tBest loss: 0.857422\tAccuracy: 72.00%\n25\tValidation loss: 0.906020\tBest loss: 0.857422\tAccuracy: 68.00%\n26\tValidation loss: 0.857865\tBest loss: 0.857422\tAccuracy: 71.33%\n27\tValidation loss: 0.844817\tBest loss: 0.844817\tAccuracy: 69.33%\n28\tValidation loss: 0.860182\tBest loss: 0.844817\tAccuracy: 69.33%\n29\tValidation loss: 0.864159\tBest loss: 0.844817\tAccuracy: 69.33%\n30\tValidation loss: 0.899200\tBest loss: 0.844817\tAccuracy: 64.00%\n31\tValidation loss: 0.876889\tBest loss: 0.844817\tAccuracy: 72.67%\n32\tValidation loss: 0.850177\tBest loss: 0.844817\tAccuracy: 67.33%\n33\tValidation loss: 0.841878\tBest loss: 0.841878\tAccuracy: 70.00%\n34\tValidation loss: 0.835707\tBest loss: 0.835707\tAccuracy: 69.33%\n35\tValidation loss: 0.850293\tBest loss: 0.835707\tAccuracy: 70.00%\n36\tValidation loss: 0.859399\tBest loss: 0.835707\tAccuracy: 69.33%\n37\tValidation loss: 0.883369\tBest loss: 0.835707\tAccuracy: 68.00%\n38\tValidation loss: 0.895023\tBest loss: 0.835707\tAccuracy: 67.33%\n39\tValidation loss: 0.857958\tBest loss: 0.835707\tAccuracy: 70.00%\n40\tValidation loss: 0.863078\tBest loss: 0.835707\tAccuracy: 69.33%\n41\tValidation loss: 0.860008\tBest loss: 0.835707\tAccuracy: 70.00%\n42\tValidation loss: 0.882908\tBest loss: 0.835707\tAccuracy: 67.33%\n43\tValidation loss: 0.867568\tBest loss: 0.835707\tAccuracy: 72.00%\n44\tValidation loss: 0.840496\tBest loss: 0.835707\tAccuracy: 69.33%\n45\tValidation loss: 0.883272\tBest loss: 0.835707\tAccuracy: 68.67%\n46\tValidation loss: 0.865739\tBest loss: 0.835707\tAccuracy: 72.00%\n47\tValidation loss: 0.983563\tBest loss: 0.835707\tAccuracy: 62.00%\n48\tValidation loss: 0.877031\tBest loss: 0.835707\tAccuracy: 68.00%\n49\tValidation loss: 0.891799\tBest loss: 0.835707\tAccuracy: 70.67%\n50\tValidation loss: 0.882461\tBest loss: 0.835707\tAccuracy: 72.00%\n51\tValidation loss: 0.879556\tBest loss: 0.835707\tAccuracy: 68.00%\n52\tValidation loss: 0.878090\tBest loss: 0.835707\tAccuracy: 70.67%\n53\tValidation loss: 0.919329\tBest loss: 0.835707\tAccuracy: 64.67%\n54\tValidation loss: 0.896891\tBest loss: 0.835707\tAccuracy: 66.67%\nEarly stopping!\nTotal training time: 11.7s\nINFO:tensorflow:Restoring parameters from ./my_mnist_model_5_to_9_five_frozen\nFinal test accuracy: 63.14%\n"
]
],
[
[
"<p>As we can see, not so good...But of course, we're using 100 images per digit and we only changed the output layer.</p>",
"_____no_output_____"
]
],
[
[
"# Let's try to reuse only 4 hidden layers instead of 5\nn_outputs = 5\n\nrestore_saver = tf.train.import_meta_graph(\"./my_best_mnist_model_0_to_4.meta\")\n\nX = tf.get_default_graph().get_tensor_by_name(\"X:0\")\ny = tf.get_default_graph().get_tensor_by_name(\"y:0\")\n\nhidden4_out = tf.get_default_graph().get_tensor_by_name(\"hidden4_out:0\")\nlogits = tf.layers.dense(hidden4_out, n_outputs, kernel_initializer=he_init, name=\"new_logits\")\nY_proba = tf.nn.softmax(logits)\nxentropy = tf.nn.sparse_softmax_cross_entropy_with_logits(labels=y, logits=logits)\nloss = tf.reduce_mean(xentropy)\ncorrect = tf.nn.in_top_k(logits, y, 1)\naccuracy = tf.reduce_mean(tf.cast(correct, tf.float32), name=\"accuracy\")",
"_____no_output_____"
],
[
"learning_rate = 0.01\n\noutput_layer_vars = tf.get_collection(tf.GraphKeys.TRAINABLE_VARIABLES, scope=\"new_logits\")\noptimizer = tf.train.AdamOptimizer(learning_rate, name=\"Adam2\")\ntraining_op = optimizer.minimize(loss, var_list=output_layer_vars)\n\ninit = tf.global_variables_initializer()\nfour_frozen_saver = tf.train.Saver()",
"_____no_output_____"
],
[
"n_epochs = 1000\nbatch_size = 20\n\nmax_checks_without_progress = 20\nchecks_without_progress = 0\nbest_loss = np.infty\n\nwith tf.Session() as sess:\n init.run()\n restore_saver.restore(sess, \"./my_best_mnist_model_0_to_4\")\n \n for epoch in range(n_epochs):\n rnd_idx = np.random.permutation(len(X_train2))\n for rnd_indices in np.array_split(rnd_idx, len(X_train2) // batch_size):\n X_batch, y_batch = X_train2[rnd_indices], y_train2[rnd_indices]\n sess.run(training_op, feed_dict={X: X_batch, y: y_batch})\n loss_val, acc_val = sess.run([loss, accuracy], feed_dict={X: X_valid2, y: y_valid2})\n if loss_val < best_loss:\n save_path = four_frozen_saver.save(sess, \"./my_mnist_model_5_to_9_four_frozen\")\n best_loss = loss_val\n checks_without_progress = 0\n else:\n checks_without_progress += 1\n if checks_without_progress > max_checks_without_progress:\n print(\"Early stopping!\")\n break\n print(\"{}\\tValidation loss: {:.6f}\\tBest loss: {:.6f}\\tAccuracy: {:.2f}%\".format(\n epoch, loss_val, best_loss, acc_val * 100))\n\nwith tf.Session() as sess:\n four_frozen_saver.restore(sess, \"./my_mnist_model_5_to_9_four_frozen\")\n acc_test = accuracy.eval(feed_dict={X: X_test2, y: y_test2})\n print(\"Final test accuracy: {:.2f}%\".format(acc_test * 100))",
"INFO:tensorflow:Restoring parameters from ./my_best_mnist_model_0_to_4\n0\tValidation loss: 1.303004\tBest loss: 1.303004\tAccuracy: 52.00%\n1\tValidation loss: 1.046397\tBest loss: 1.046397\tAccuracy: 54.00%\n2\tValidation loss: 0.990561\tBest loss: 0.990561\tAccuracy: 57.33%\n3\tValidation loss: 0.943077\tBest loss: 0.943077\tAccuracy: 61.33%\n4\tValidation loss: 0.924572\tBest loss: 0.924572\tAccuracy: 61.33%\n5\tValidation loss: 0.889116\tBest loss: 0.889116\tAccuracy: 63.33%\n6\tValidation loss: 0.896398\tBest loss: 0.889116\tAccuracy: 64.00%\n7\tValidation loss: 0.858603\tBest loss: 0.858603\tAccuracy: 66.67%\n8\tValidation loss: 0.873803\tBest loss: 0.858603\tAccuracy: 69.33%\n9\tValidation loss: 0.841726\tBest loss: 0.841726\tAccuracy: 68.00%\n10\tValidation loss: 0.862488\tBest loss: 0.841726\tAccuracy: 67.33%\n11\tValidation loss: 0.821854\tBest loss: 0.821854\tAccuracy: 70.67%\n12\tValidation loss: 0.809741\tBest loss: 0.809741\tAccuracy: 71.33%\n13\tValidation loss: 0.810555\tBest loss: 0.809741\tAccuracy: 70.00%\n14\tValidation loss: 0.820386\tBest loss: 0.809741\tAccuracy: 68.67%\n15\tValidation loss: 0.812963\tBest loss: 0.809741\tAccuracy: 71.33%\n16\tValidation loss: 0.795716\tBest loss: 0.795716\tAccuracy: 70.00%\n17\tValidation loss: 0.799856\tBest loss: 0.795716\tAccuracy: 69.33%\n18\tValidation loss: 0.800295\tBest loss: 0.795716\tAccuracy: 72.00%\n19\tValidation loss: 0.795355\tBest loss: 0.795355\tAccuracy: 70.67%\n20\tValidation loss: 0.800698\tBest loss: 0.795355\tAccuracy: 72.00%\n21\tValidation loss: 0.791533\tBest loss: 0.791533\tAccuracy: 71.33%\n22\tValidation loss: 0.789892\tBest loss: 0.789892\tAccuracy: 71.33%\n23\tValidation loss: 0.790024\tBest loss: 0.789892\tAccuracy: 70.67%\n24\tValidation loss: 0.785441\tBest loss: 0.785441\tAccuracy: 72.00%\n25\tValidation loss: 0.792833\tBest loss: 0.785441\tAccuracy: 69.33%\n26\tValidation loss: 0.771823\tBest loss: 0.771823\tAccuracy: 72.67%\n27\tValidation loss: 0.778180\tBest loss: 0.771823\tAccuracy: 70.67%\n28\tValidation loss: 0.779182\tBest loss: 0.771823\tAccuracy: 73.33%\n29\tValidation loss: 0.785683\tBest loss: 0.771823\tAccuracy: 74.00%\n30\tValidation loss: 0.776967\tBest loss: 0.771823\tAccuracy: 74.00%\n31\tValidation loss: 0.804916\tBest loss: 0.771823\tAccuracy: 71.33%\n32\tValidation loss: 0.789834\tBest loss: 0.771823\tAccuracy: 72.00%\n33\tValidation loss: 0.770294\tBest loss: 0.770294\tAccuracy: 72.67%\n34\tValidation loss: 0.766526\tBest loss: 0.766526\tAccuracy: 74.00%\n35\tValidation loss: 0.848782\tBest loss: 0.766526\tAccuracy: 68.67%\n36\tValidation loss: 0.814959\tBest loss: 0.766526\tAccuracy: 70.67%\n37\tValidation loss: 0.803090\tBest loss: 0.766526\tAccuracy: 70.67%\n38\tValidation loss: 0.779465\tBest loss: 0.766526\tAccuracy: 73.33%\n39\tValidation loss: 0.804749\tBest loss: 0.766526\tAccuracy: 71.33%\n40\tValidation loss: 0.812036\tBest loss: 0.766526\tAccuracy: 73.33%\n41\tValidation loss: 0.777164\tBest loss: 0.766526\tAccuracy: 74.67%\n42\tValidation loss: 0.788647\tBest loss: 0.766526\tAccuracy: 72.00%\n43\tValidation loss: 0.756715\tBest loss: 0.756715\tAccuracy: 74.00%\n44\tValidation loss: 0.828021\tBest loss: 0.756715\tAccuracy: 67.33%\n45\tValidation loss: 0.846436\tBest loss: 0.756715\tAccuracy: 68.00%\n46\tValidation loss: 0.807225\tBest loss: 0.756715\tAccuracy: 70.67%\n47\tValidation loss: 0.812321\tBest loss: 0.756715\tAccuracy: 68.67%\n48\tValidation loss: 0.790139\tBest loss: 0.756715\tAccuracy: 73.33%\n49\tValidation loss: 0.805462\tBest loss: 0.756715\tAccuracy: 70.67%\n50\tValidation loss: 0.782307\tBest loss: 0.756715\tAccuracy: 73.33%\n51\tValidation loss: 0.822047\tBest loss: 0.756715\tAccuracy: 70.67%\n52\tValidation loss: 0.791988\tBest loss: 0.756715\tAccuracy: 72.00%\n53\tValidation loss: 0.832699\tBest loss: 0.756715\tAccuracy: 68.67%\n54\tValidation loss: 0.791335\tBest loss: 0.756715\tAccuracy: 73.33%\n55\tValidation loss: 0.806211\tBest loss: 0.756715\tAccuracy: 72.67%\n56\tValidation loss: 0.810055\tBest loss: 0.756715\tAccuracy: 71.33%\n57\tValidation loss: 0.809657\tBest loss: 0.756715\tAccuracy: 70.00%\n58\tValidation loss: 0.804617\tBest loss: 0.756715\tAccuracy: 72.67%\n59\tValidation loss: 0.789282\tBest loss: 0.756715\tAccuracy: 70.00%\n60\tValidation loss: 0.775632\tBest loss: 0.756715\tAccuracy: 74.00%\n61\tValidation loss: 0.775557\tBest loss: 0.756715\tAccuracy: 73.33%\n62\tValidation loss: 0.803277\tBest loss: 0.756715\tAccuracy: 70.67%\n63\tValidation loss: 0.819929\tBest loss: 0.756715\tAccuracy: 71.33%\nEarly stopping!\nINFO:tensorflow:Restoring parameters from ./my_mnist_model_5_to_9_four_frozen\nFinal test accuracy: 66.28%\n"
]
],
[
[
"<p>Well, a bit better...</p>",
"_____no_output_____"
]
],
[
[
"# Let's try now to unfreeze the last two layers\nlearning_rate = 0.01\n\nunfrozen_vars = tf.get_collection(tf.GraphKeys.TRAINABLE_VARIABLES, scope=\"hidden[34]|new_logits\")\noptimizer = tf.train.AdamOptimizer(learning_rate, name=\"Adam3\")\ntraining_op = optimizer.minimize(loss, var_list=unfrozen_vars)\n\ninit = tf.global_variables_initializer()\ntwo_frozen_saver = tf.train.Saver()",
"_____no_output_____"
],
[
"n_epochs = 1000\nbatch_size = 20\n\nmax_checks_without_progress = 20\nchecks_without_progress = 0\nbest_loss = np.infty\n\nwith tf.Session() as sess:\n init.run()\n four_frozen_saver.restore(sess, \"./my_mnist_model_5_to_9_four_frozen\")\n \n for epoch in range(n_epochs):\n rnd_idx = np.random.permutation(len(X_train2))\n for rnd_indices in np.array_split(rnd_idx, len(X_train2) // batch_size):\n X_batch, y_batch = X_train2[rnd_indices], y_train2[rnd_indices]\n sess.run(training_op, feed_dict={X: X_batch, y: y_batch})\n loss_val, acc_val = sess.run([loss, accuracy], feed_dict={X: X_valid2, y: y_valid2})\n if loss_val < best_loss:\n save_path = two_frozen_saver.save(sess, \"./my_mnist_model_5_to_9_two_frozen\")\n best_loss = loss_val\n checks_without_progress = 0\n else:\n checks_without_progress += 1\n if checks_without_progress > max_checks_without_progress:\n print(\"Early stopping!\")\n break\n print(\"{}\\tValidation loss: {:.6f}\\tBest loss: {:.6f}\\tAccuracy: {:.2f}%\".format(\n epoch, loss_val, best_loss, acc_val * 100))\n\nwith tf.Session() as sess:\n two_frozen_saver.restore(sess, \"./my_mnist_model_5_to_9_two_frozen\")\n acc_test = accuracy.eval(feed_dict={X: X_test2, y: y_test2})\n print(\"Final test accuracy: {:.2f}%\".format(acc_test * 100))",
"INFO:tensorflow:Restoring parameters from ./my_mnist_model_5_to_9_four_frozen\n0\tValidation loss: 0.853785\tBest loss: 0.853785\tAccuracy: 72.67%\n1\tValidation loss: 0.870564\tBest loss: 0.853785\tAccuracy: 67.33%\n2\tValidation loss: 0.652311\tBest loss: 0.652311\tAccuracy: 76.00%\n3\tValidation loss: 0.698115\tBest loss: 0.652311\tAccuracy: 78.00%\n4\tValidation loss: 0.690634\tBest loss: 0.652311\tAccuracy: 75.33%\n5\tValidation loss: 0.768487\tBest loss: 0.652311\tAccuracy: 77.33%\n6\tValidation loss: 0.848543\tBest loss: 0.652311\tAccuracy: 80.00%\n7\tValidation loss: 0.697138\tBest loss: 0.652311\tAccuracy: 83.33%\n8\tValidation loss: 0.712297\tBest loss: 0.652311\tAccuracy: 84.67%\n9\tValidation loss: 0.779837\tBest loss: 0.652311\tAccuracy: 80.00%\n10\tValidation loss: 0.833671\tBest loss: 0.652311\tAccuracy: 81.33%\n11\tValidation loss: 0.803134\tBest loss: 0.652311\tAccuracy: 82.00%\n12\tValidation loss: 0.954865\tBest loss: 0.652311\tAccuracy: 78.67%\n13\tValidation loss: 0.921988\tBest loss: 0.652311\tAccuracy: 80.00%\n14\tValidation loss: 1.050293\tBest loss: 0.652311\tAccuracy: 79.33%\n15\tValidation loss: 1.037723\tBest loss: 0.652311\tAccuracy: 80.00%\n16\tValidation loss: 1.128598\tBest loss: 0.652311\tAccuracy: 82.00%\n17\tValidation loss: 1.228396\tBest loss: 0.652311\tAccuracy: 80.00%\n18\tValidation loss: 1.048694\tBest loss: 0.652311\tAccuracy: 80.67%\n19\tValidation loss: 1.131650\tBest loss: 0.652311\tAccuracy: 80.67%\n20\tValidation loss: 1.170808\tBest loss: 0.652311\tAccuracy: 80.67%\n21\tValidation loss: 1.108722\tBest loss: 0.652311\tAccuracy: 82.67%\n22\tValidation loss: 1.137113\tBest loss: 0.652311\tAccuracy: 82.67%\nEarly stopping!\nINFO:tensorflow:Restoring parameters from ./my_mnist_model_5_to_9_two_frozen\nFinal test accuracy: 73.42%\n"
]
],
[
[
"<p>Not bad...And what if we unfreeze all the layers?</p>",
"_____no_output_____"
]
],
[
[
"learning_rate = 0.01\n\noptimizer = tf.train.AdamOptimizer(learning_rate, name=\"Adam4\")\ntraining_op = optimizer.minimize(loss)\n\ninit = tf.global_variables_initializer()\nno_frozen_saver = tf.train.Saver()",
"_____no_output_____"
],
[
"n_epochs = 1000\nbatch_size = 20\n\nmax_checks_without_progress = 20\nchecks_without_progress = 0\nbest_loss = np.infty\n\nwith tf.Session() as sess:\n init.run()\n two_frozen_saver.restore(sess, \"./my_mnist_model_5_to_9_two_frozen\")\n \n for epoch in range(n_epochs):\n rnd_idx = np.random.permutation(len(X_train2))\n for rnd_indices in np.array_split(rnd_idx, len(X_train2) // batch_size):\n X_batch, y_batch = X_train2[rnd_indices], y_train2[rnd_indices]\n sess.run(training_op, feed_dict={X: X_batch, y: y_batch})\n loss_val, acc_val = sess.run([loss, accuracy], feed_dict={X: X_valid2, y: y_valid2})\n if loss_val < best_loss:\n save_path = no_frozen_saver.save(sess, \"./my_mnist_model_5_to_9_no_frozen\")\n best_loss = loss_val\n checks_without_progress = 0\n else:\n checks_without_progress += 1\n if checks_without_progress > max_checks_without_progress:\n print(\"Early stopping!\")\n break\n print(\"{}\\tValidation loss: {:.6f}\\tBest loss: {:.6f}\\tAccuracy: {:.2f}%\".format(\n epoch, loss_val, best_loss, acc_val * 100))\n\nwith tf.Session() as sess:\n no_frozen_saver.restore(sess, \"./my_mnist_model_5_to_9_no_frozen\")\n acc_test = accuracy.eval(feed_dict={X: X_test2, y: y_test2})\n print(\"Final test accuracy: {:.2f}%\".format(acc_test * 100))",
"INFO:tensorflow:Restoring parameters from ./my_mnist_model_5_to_9_two_frozen\n0\tValidation loss: 0.753302\tBest loss: 0.753302\tAccuracy: 79.33%\n1\tValidation loss: 0.518203\tBest loss: 0.518203\tAccuracy: 87.33%\n2\tValidation loss: 0.449906\tBest loss: 0.449906\tAccuracy: 89.33%\n3\tValidation loss: 0.387903\tBest loss: 0.387903\tAccuracy: 92.00%\n4\tValidation loss: 0.649829\tBest loss: 0.387903\tAccuracy: 88.00%\n5\tValidation loss: 0.645186\tBest loss: 0.387903\tAccuracy: 91.33%\n6\tValidation loss: 1.225373\tBest loss: 0.387903\tAccuracy: 88.67%\n7\tValidation loss: 1.132690\tBest loss: 0.387903\tAccuracy: 88.67%\n8\tValidation loss: 0.816616\tBest loss: 0.387903\tAccuracy: 90.00%\n9\tValidation loss: 0.769069\tBest loss: 0.387903\tAccuracy: 90.67%\n10\tValidation loss: 0.990865\tBest loss: 0.387903\tAccuracy: 89.33%\n11\tValidation loss: 1.153069\tBest loss: 0.387903\tAccuracy: 89.33%\n12\tValidation loss: 1.028986\tBest loss: 0.387903\tAccuracy: 90.00%\n13\tValidation loss: 1.057120\tBest loss: 0.387903\tAccuracy: 92.00%\n14\tValidation loss: 1.062341\tBest loss: 0.387903\tAccuracy: 92.00%\n15\tValidation loss: 1.062881\tBest loss: 0.387903\tAccuracy: 92.00%\n16\tValidation loss: 1.062505\tBest loss: 0.387903\tAccuracy: 92.00%\n17\tValidation loss: 1.062810\tBest loss: 0.387903\tAccuracy: 92.00%\n18\tValidation loss: 1.063364\tBest loss: 0.387903\tAccuracy: 92.00%\n19\tValidation loss: 1.063347\tBest loss: 0.387903\tAccuracy: 92.00%\n20\tValidation loss: 1.064176\tBest loss: 0.387903\tAccuracy: 92.00%\n21\tValidation loss: 1.064999\tBest loss: 0.387903\tAccuracy: 92.00%\n22\tValidation loss: 1.065630\tBest loss: 0.387903\tAccuracy: 92.00%\n23\tValidation loss: 1.067343\tBest loss: 0.387903\tAccuracy: 92.00%\nEarly stopping!\nINFO:tensorflow:Restoring parameters from ./my_mnist_model_5_to_9_no_frozen\nFinal test accuracy: 89.34%\n"
],
[
"# Let's compare this result with a DNN trained from scratch\ndnn_clf_5_to_9 = DNNClassifier(n_hidden_layers=4, random_state=42)\ndnn_clf_5_to_9.fit(X_train2, y_train2, n_epochs=1000, X_valid=X_valid2, y_valid=y_valid2)",
"0\tValidation loss: 0.803556\tBest loss: 0.803556\tAccuracy: 71.33%\n1\tValidation loss: 0.966740\tBest loss: 0.803556\tAccuracy: 85.33%\n2\tValidation loss: 1.158966\tBest loss: 0.803556\tAccuracy: 78.00%\n3\tValidation loss: 0.615953\tBest loss: 0.615953\tAccuracy: 88.00%\n4\tValidation loss: 0.612615\tBest loss: 0.612615\tAccuracy: 92.00%\n5\tValidation loss: 0.686583\tBest loss: 0.612615\tAccuracy: 89.33%\n6\tValidation loss: 0.804757\tBest loss: 0.612615\tAccuracy: 89.33%\n7\tValidation loss: 0.748284\tBest loss: 0.612615\tAccuracy: 88.00%\n8\tValidation loss: 0.947906\tBest loss: 0.612615\tAccuracy: 84.00%\n9\tValidation loss: 1.652254\tBest loss: 0.612615\tAccuracy: 89.33%\n10\tValidation loss: 0.982898\tBest loss: 0.612615\tAccuracy: 92.67%\n11\tValidation loss: 1.143990\tBest loss: 0.612615\tAccuracy: 90.00%\n12\tValidation loss: 1.167457\tBest loss: 0.612615\tAccuracy: 92.00%\n13\tValidation loss: 1.120672\tBest loss: 0.612615\tAccuracy: 92.00%\n14\tValidation loss: 1.773683\tBest loss: 0.612615\tAccuracy: 82.00%\n15\tValidation loss: 0.980001\tBest loss: 0.612615\tAccuracy: 91.33%\n16\tValidation loss: 1.402388\tBest loss: 0.612615\tAccuracy: 86.00%\n17\tValidation loss: 0.935867\tBest loss: 0.612615\tAccuracy: 94.00%\n18\tValidation loss: 1.124392\tBest loss: 0.612615\tAccuracy: 93.33%\n19\tValidation loss: 1.193039\tBest loss: 0.612615\tAccuracy: 93.33%\n20\tValidation loss: 1.209065\tBest loss: 0.612615\tAccuracy: 93.33%\n21\tValidation loss: 1.217737\tBest loss: 0.612615\tAccuracy: 93.33%\n22\tValidation loss: 1.221018\tBest loss: 0.612615\tAccuracy: 93.33%\n23\tValidation loss: 1.224113\tBest loss: 0.612615\tAccuracy: 93.33%\n24\tValidation loss: 1.228211\tBest loss: 0.612615\tAccuracy: 93.33%\n25\tValidation loss: 1.231094\tBest loss: 0.612615\tAccuracy: 93.33%\nEarly stopping!\n"
],
[
"from sklearn.metrics import accuracy_score\n\ny_pred = dnn_clf_5_to_9.predict(X_test2)\naccuracy_score(y_test2, y_pred)",
"_____no_output_____"
]
],
[
[
"<p>Unfortunately in this case transfer learning did not help too much.</p>",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
cb8df2500523c076b9caeebeac563cca21843be8
| 76,228 |
ipynb
|
Jupyter Notebook
|
examples/Notebooks/LB_Colloids_ColloidModel.ipynb
|
jdlarsen-UA/LB-colloids
|
92ad3cc8a08e8bdf4f468e55a3f5cf7bcc319a67
|
[
"BSD-3-Clause"
] | 1 |
2020-09-17T02:45:12.000Z
|
2020-09-17T02:45:12.000Z
|
examples/Notebooks/LB_Colloids_ColloidModel.ipynb
|
Gweiqi/LB-colloids
|
92ad3cc8a08e8bdf4f468e55a3f5cf7bcc319a67
|
[
"BSD-3-Clause"
] | null | null | null |
examples/Notebooks/LB_Colloids_ColloidModel.ipynb
|
Gweiqi/LB-colloids
|
92ad3cc8a08e8bdf4f468e55a3f5cf7bcc319a67
|
[
"BSD-3-Clause"
] | 3 |
2019-12-28T21:06:42.000Z
|
2020-09-17T02:45:10.000Z
| 164.284483 | 60,608 | 0.872881 |
[
[
[
"# LB-Colloids Colloid particle tracking\n\nLB-Colloids allows the user to perform colloid and nanoparticle tracking simulations on Computational Fluid Dynamics domains. As the user, you supply the chemical and physical properties, and the code performs the mathematics and particle tracking!\n\nLet's set up our workspace to begin. And we will use the Synthetic5 example problem to parameterize run LB-Colloids",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nimport matplotlib.pyplot as plt\nimport matplotlib.image as mpimg\nimport os\nfrom lb_colloids import LBImage, LB2DModel\nfrom lb_colloids import ColloidModel, cIO\n\nworkspace = os.path.join(\"..\", \"data\")\ndomain = \"Synth100_5.png\"\nlb_name = \"s5.hdf5\"\nendpoint = \"s5.endpoint\"",
"_____no_output_____"
]
],
[
[
"First thing, let's run a lattice Boltzmann model to get our fluid domain. For more details see the LB2D Notebook.",
"_____no_output_____"
]
],
[
[
"lbi = LBImage.Images(os.path.join(workspace, domain))\nbc = LBImage.BoundaryCondition(lbi.arr, fluidvx=[253], solidvx=[0], nlayers=5)\nlbm = LB2DModel(bc.binarized)\nlbm.niters = 1000\nlbm.run(output=os.path.join(workspace, lb_name), verbose=1000)",
"('Porosity: ', 0.5658)\n1000\nIter: 01000\n[Writing to: ../data/s5.hdf5]\n"
]
],
[
[
"## Setting up a Colloids particle tracking model\n\nWe can begin setting up a Colloids model by using the `ColloidsConfig()` class. This class ensures that valid values are supplied to particle tracking variables and allows the user to write an external particle tracking configuration file for documentation and later use if wanted.\n\nLet's generate an empty `ColloidsConfig` instance\n\n",
"_____no_output_____"
]
],
[
[
"io = cIO.ColloidsConfig()",
"_____no_output_____"
]
],
[
[
"`ColloidsConfig()` uses dictionary keys to be parameterized. Common parameterization variables include\n\n`lbmodel`: required parameter that points to the CFD fluid domain\n\n`ncols`: required parameter that describes the number of colloids released\n\n`iters`: number of time steps to simulate transport\n\n`lbres`: the lattice Boltzmann simulation resolution in meters\n\n`gridref`: optional grid refinement parameter, uses bi-linear interpolation\n\n`ac`: colloid radius in meters\n\n`timestep`: the timestep length in seconds. Recommend very small timesteps!\n\n`continuous`: flag for continuous release. If 0 one release of colloids occurs, if > 0 a release of colloids occurs at continuous number of timesteps\n\n`i`: fluid ionic strength in M\n\n`print_time`: how often iteration progress prints to the screen\n\n`endpoint`: endpoint file name to store breakthrough information\n\n`store_time`: internal function that can be used to reduce memory requirements, a higher store_time equals less memory devoted to storing colloid positions (old positions are striped every store_time timesteps).\n\n`zeta_colloid`: zeta potential of the colloid in V\n\n`zeta_solid`: zeta potential of the solid in V\n\n`plot`: boolean flag that generates a plot at the end of the model run\n\n`showfig`: boolean flag that determines weather to show the figure or save it to disk\n\nA complete listing of these are available in the user guide.",
"_____no_output_____"
]
],
[
[
"# model parameters\nio[\"lbmodel\"] = os.path.join(workspace, lb_name)\nio['ncols'] = 2000\nio['iters'] = 50000\nio['lbres'] = 1e-6\nio['gridref'] = 10\nio['ac'] = 1e-06\nio['timestep'] = 1e-06 # should be less than or equal to colloid radius!\nio['continuous'] = 0\n\n# chemical parameters\nio['i'] = 1e-03 # Molar ionic strength of solution\nio['zeta_colloid'] = -49.11e-3 # zeta potential of Na-Kaolinite at 1e-03 M NaCl\nio['zeta_solid'] = -61.76e-3 # zeta potential of Glass Beads at 1e-03 M NaCl\n\n# output control\nio['print_time'] = 10000\nio['endpoint'] = os.path.join(workspace, endpoint)\nio['store_time'] = 100\nio['plot'] = True\nio['showfig'] = True",
"_____no_output_____"
]
],
[
[
"We can now look at the parameter dictionaries `ColloidConfig` creates!",
"_____no_output_____"
]
],
[
[
"io.model_parameters, io.chemical_parameters, io.physical_parameters, io.output_control_parameters",
"_____no_output_____"
]
],
[
[
"we can also write a config file for documentation and later runs! And see the information that will be written to the config file by using the `io.config` call",
"_____no_output_____"
]
],
[
[
"io.write(os.path.join(workspace, \"s2.config\"))\nio.config",
"_____no_output_____"
]
],
[
[
"the `ColloidsConfig` can be directly be used with the `Config` reader to instanstiate a LB-Colloids model",
"_____no_output_____"
]
],
[
[
"config = cIO.Config(io.config)",
"_____no_output_____"
]
],
[
[
"and we can run the model using the `ColloidModel.run()` call",
"_____no_output_____"
]
],
[
[
"ColloidModel.run(config)",
"2000\n(110, 102)\n(110, 102)\n(10000, '0.010')\n(20000, '0.020')\n(30000, '0.030')\n(40000, '0.040')\n(50000, '0.050')\n"
]
],
[
[
"The output image shows the path of colloids which haven't yet broke through the model domain!\n\n### For ColloidModel outputs please see the LB_Colloids_output_contol notebook",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
cb8df704a9c8b6f8406d988b07b53b50282b3950
| 24,023 |
ipynb
|
Jupyter Notebook
|
Grid Search/Example 0/Grid_Search_example0.ipynb
|
alphoenixbiz/Machine-Learning-Python
|
74266fcb54fcdc8b214441090b4cff611342f612
|
[
"Apache-2.0"
] | 3 |
2019-05-04T11:26:25.000Z
|
2019-05-26T16:44:19.000Z
|
Grid Search/Example 0/Grid_Search_example0.ipynb
|
alphoenixbiz/Machine-Learning-Using-Python
|
74266fcb54fcdc8b214441090b4cff611342f612
|
[
"Apache-2.0"
] | null | null | null |
Grid Search/Example 0/Grid_Search_example0.ipynb
|
alphoenixbiz/Machine-Learning-Using-Python
|
74266fcb54fcdc8b214441090b4cff611342f612
|
[
"Apache-2.0"
] | null | null | null | 42.821747 | 8,990 | 0.599051 |
[
[
[
"<a href=\"https://colab.research.google.com/github/alphoenixbiz/Machine-Learning-Using-Python/blob/master/Grid%20Search/Example%200/Grid_Search_example0.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nfrom matplotlib import pyplot as plt\n%matplotlib inline",
"_____no_output_____"
],
[
"from google.colab import drive \ndrive.mount('/content/gdrive')",
"_____no_output_____"
],
[
"df = pd.read_csv(\"gdrive/My Drive/Colab Notebooks/DATA/Social_Network_Ads.csv\")\ndf.head()",
"_____no_output_____"
],
[
"X = df.iloc[:, [2, 3]].values\ny = df.iloc[:, 4].values\n",
"_____no_output_____"
],
[
"from sklearn.model_selection import train_test_split\nX_train, X_test, y_train, y_test = train_test_split(X, y, test_size = 0.25, random_state = 0)",
"_____no_output_____"
]
],
[
[
"**Feature Scaling**",
"_____no_output_____"
]
],
[
[
"# Feature Scaling\nfrom sklearn.preprocessing import StandardScaler\nsc = StandardScaler()\nX_train = sc.fit_transform(X_train)\nX_test = sc.transform(X_test)",
"_____no_output_____"
],
[
"from sklearn.svm import SVC\nclassifier = SVC(kernel = 'rbf', random_state = 0, gamma ='auto')\nclassifier.fit(X_train, y_train)",
"_____no_output_____"
],
[
"y_pred = classifier.predict(X_test)\ny_pred",
"_____no_output_____"
],
[
"classifier.score(X_test, y_test)",
"_____no_output_____"
]
],
[
[
"**Making the Confusion Matrix**",
"_____no_output_____"
]
],
[
[
"from sklearn.metrics import confusion_matrix\ncm = confusion_matrix(y_test, y_pred)\ncm",
"_____no_output_____"
],
[
"import seaborn as sn\nplt.figure(figsize = (10,7))\nsn.heatmap(cm, annot=True)\nplt.xlabel('Predicted')\nplt.ylabel('Actual')",
"_____no_output_____"
]
],
[
[
"**Applying k-Fold Cross Validation**",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import cross_val_score\naccuracies = cross_val_score(estimator = classifier, X = X_train, y = y_train, cv = 10)\naccuracies.mean()\naccuracies.std()",
"_____no_output_____"
]
],
[
[
"**Applying Grid Search to find the best model and the best parameters**",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import GridSearchCV\nparameters = [{'C': [1, 10, 100, 1000], 'kernel': ['linear']},\n {'C': [1, 10, 100, 1000], 'kernel': ['rbf'], 'gamma': [0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 0.9]}]",
"_____no_output_____"
],
[
"grid_search = GridSearchCV(estimator = classifier,\n param_grid = parameters,\n scoring = 'accuracy',\n cv = 10,\n n_jobs = -1,\n iid = False)",
"_____no_output_____"
],
[
"grid_search = grid_search.fit(X_train, y_train)",
"_____no_output_____"
],
[
"best_accuracy = grid_search.best_score_\nbest_accuracy",
"_____no_output_____"
]
],
[
[
"Best parameters",
"_____no_output_____"
]
],
[
[
"best_parameters = grid_search.best_params_\nbest_parameters",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb8df9680ee824bbdc41f7c9de17cd48be1ee8e4
| 483,574 |
ipynb
|
Jupyter Notebook
|
hw10/HW10_AYeganov.ipynb
|
ayeganov/RBE500
|
a7a3c091aaec47ed5414454db06775e84c700a28
|
[
"MIT"
] | null | null | null |
hw10/HW10_AYeganov.ipynb
|
ayeganov/RBE500
|
a7a3c091aaec47ed5414454db06775e84c700a28
|
[
"MIT"
] | null | null | null |
hw10/HW10_AYeganov.ipynb
|
ayeganov/RBE500
|
a7a3c091aaec47ed5414454db06775e84c700a28
|
[
"MIT"
] | null | null | null | 898.836431 | 47,906 | 0.942412 |
[
[
[
"%%javascript\nIPython.OutputArea.prototype._should_scroll = function(lines) {\n return false;\n}",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt\nimport numpy as np\n\ndt = 0.1\n\ndef draw_plot(measurements, mlabel=None, estimates=None, estlabel=None, title=None, xlabel=None, ylabel=None):\n xvals = np.linspace(0, dt * len(measurements), len(measurements))\n plt.title(title, fontsize=12)\n \n xlabel and plt.xlabel(\"Time in seconds\")\n ylabel and plt.ylabel(\"Distance to Wall in cm\")\n \n ax = plt.subplot(111)\n \n ax.plot(measurements, label=mlabel)\n np.any(estimates) and estlabel and ax.plot(estimates, label=estlabel)\n \n box = ax.get_position()\n ax.set_position([box.x0, box.y0 + box.height * 0.1,\n box.width, box.height * 1.1])\n\n ax.legend(loc='upper center', bbox_to_anchor=(.5, -0.05), ncol=3)\n plt.show()\n \ndef add_noise(data, size=20):\n noise = np.random.uniform(-1, 1, len(data)) * size\n return data + noise",
"_____no_output_____"
]
],
[
[
"### Part A",
"_____no_output_____"
]
],
[
[
"from kalman import predict, update, dot3\n\nplt.rcParams[\"figure.figsize\"] = [12, 9]\n\nwall_file = \"RBE500-F17-100ms-Constant-Vel.csv\"\n\ndata = add_noise(np.loadtxt(wall_file, delimiter=\",\"), 0)\n\n# Setup initial variables and matrices\ninitial_pos = 2530\nvelocity = -10.0\nvariance = 10.0\ndt = 0.1\nF = np.array([[1., dt],\n [0., 1.]])\nP = np.array([[100., 0],\n [0, 100.0]])\nH = np.array([[1., 0.]])\nR = np.array([[variance]])\nQ = np.array([[0.1, 1], [1, 10.]])\nx = np.array([initial_pos, velocity]).T\n\npredicted_xs = []\n\ndef run(x, P, R, Q, dt, zs):\n # run the kalman filter and store the results\n xs, cov = [], []\n for z in zs:\n x, P = predict(x, P, F, Q)\n x, P = update(x, P, z, R, H)\n xs.append(x)\n cov.append(P)\n\n xs, cov = np.array(xs), np.array(cov)\n return xs, cov\n\ntime_values = np.linspace(0, dt * len(data), len(data))\n\nest_x, est_P = run(x, P, R, Q, dt, data)\nest_pos = [v[0] for v in est_x]\nest_vel = [v[1] for v in est_x]\ndraw_plot(data, \n estimates=est_pos, \n title=\"Raw Data VS Kalman Estimation\", \n mlabel=\"Measurements\",\n estlabel=\"Kalman Estimates\",\n xlabel=\"Time in Seconds\",\n ylabel=\"Distance to Wall in CM\")\n\nplt.plot(time_values, est_vel)\nplt.xlabel(\"Time in seconds\")\nplt.ylabel(\"Velocity in cm/sec\")\nplt.title(\"Velocity Over Time\")\nplt.show()\n\npos_std = [p[0][0]**0.5 for p in est_P]\nvel_var = [p[1][1]**0.5 for p in est_P]\npos_vel_corr = [p[0][1] for p in est_P]\nplt.plot(time_values, pos_std)\nplt.ylabel(\"Position StdDev\")\nplt.xlabel(\"Time in Seconds\")\nplt.title(\"Position StdDev Over Time\")\nplt.show()\n\nplt.plot(time_values, vel_var)\nplt.ylabel(\"Velocity StdDev\")\nplt.xlabel(\"Time in Seconds\")\nplt.title(\"Velocity StdDev Over Time\")\nplt.show()\n\nplt.plot(time_values, pos_vel_corr)\nplt.ylabel(\"Velocity-Position Correlation\")\nplt.xlabel(\"Time in Seconds\")\nplt.title(\"Velocity-Position Correlation Over Time\")\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Part B",
"_____no_output_____"
]
],
[
[
"data = add_noise(np.loadtxt(wall_file, delimiter=\",\"), 0)\n\n# Setup initial variables and matrices\ninitial_pos = 2530\nvelocity = -10.0\nvariance = 10.0\ndt = 0.1\nF = np.array([[1., dt],\n [0., 1.]])\nP = np.array([[100., 0],\n [0, 100.0]])\nH = np.array([[1., 0.]])\nR = np.array([[variance]])\nQ = np.array([[0.1, 1], [1, 10.]])\nx = np.array([initial_pos, velocity]).T\n\npredicted_xs = []\n\ndef run(x, P, R=0, Q=0, dt=0.1, zs=None):\n # run the kalman filter and store the results\n xs, cov = [], []\n for z in zs:\n x, P = predict(x, P, F, Q)\n S = dot3(H, P, H.T) + R\n n = z - np.dot(H, x)\n d = n*n / S\n \n if d < 9.0:\n x, P = update(x, P, z, R, H)\n\n xs.append(x)\n cov.append(P)\n\n xs, cov = np.array(xs), np.array(cov)\n return xs, cov\n\ntime_values = np.linspace(0, dt * len(data), len(data))\n\nest_x, est_P = run(x, P, R, Q, dt, data)\nest_pos = [v[0] for v in est_x]\ndraw_plot(data, \n estimates=est_pos, \n title=\"Raw Data VS Kalman Estimation\", \n mlabel=\"Measurements\",\n estlabel=\"Kalman Estimates\",\n xlabel=\"Time in Seconds\",\n ylabel=\"Distance to Wall in CM\")\n\nest_vel = [v[1] for v in est_x]\nplt.plot(time_values, est_vel)\nplt.xlabel(\"Time in seconds\")\nplt.ylabel(\"Speed in cm/sec\")\nplt.title(\"Velocity Over Time\")\nplt.show()\n\npos_std = [p[0][0]**0.5 for p in est_P]\nvel_var = [p[1][1]**0.5 for p in est_P]\npos_vel_corr = [p[0][1] for p in est_P]\nplt.plot(time_values, pos_std)\nplt.ylabel(\"Position StdDev\")\nplt.xlabel(\"Time in Seconds\")\nplt.title(\"Position StdDev Over Time\")\nplt.show()\n\nplt.plot(time_values, vel_var)\nplt.ylabel(\"Velocity StdDev\")\nplt.xlabel(\"Time in Seconds\")\nplt.title(\"Velocity StdDev Over Time\")\nplt.show()\n\nplt.plot(time_values, pos_vel_corr)\nplt.ylabel(\"Velocity-Position Correlation\")\nplt.xlabel(\"Time in Seconds\")\nplt.title(\"Velocity-Position Correlation Over Time\")\nplt.show()\n",
"_____no_output_____"
]
],
[
[
"### Part C",
"_____no_output_____"
]
],
[
[
"data = add_noise(np.loadtxt(wall_file, delimiter=\",\"), 0)\n\ndef gaussian(x, mu, sig):\n return (1/(np.sqrt(2 * np.pi * np.power(sig, 2.)))) * np.exp(-np.power(x - mu, 2.) / (2 * np.power(sig, 2.)))\n\n# Setup initial variables and matrices\ninitial_pos = 2530\nvelocity = -10.0\nvariance = 10.0\ndt = 0.1\nF = np.array([[1., dt],\n [0., 1.]])\nP = np.array([[100., 0],\n [0, 100.0]])\nH = np.array([[1., 0.]])\nR = np.array([[variance]])\nQ = np.array([[0.1, 1], [1, 10.]])\nx = np.array([initial_pos, velocity]).T\nxs = []\n\nobject_c = 0.2\nwall_c = 0.8\nlamb = 0.0005\n\ndef object_pdf(z):\n return object_c * lamb * np.exp(-lamb * z)\n\ndef wall_pdf(z, wall_mean):\n return wall_c * gaussian(z, wall_mean, variance)\n\ndef run(x, P, R, Q, dt, zs):\n # run the kalman filter and store the results\n xs, cov = [], []\n for z in zs:\n x, P = predict(x, P, F, Q)\n prob_wall = wall_pdf(x[0], z)\n prob_obj = object_pdf(z)\n\n if prob_obj < prob_wall:\n x, P = update(x, P, z, R, H)\n\n xs.append(x)\n cov.append(P)\n\n xs, cov = np.array(xs), np.array(cov)\n return xs, cov\n\nold_est_x = est_x\nest_x, est_P = run(x, P, R, Q, dt, data)\n\nest_pos = [v[0] for v in est_x]\ndraw_plot(data, \n estimates=est_pos, \n title=\"Raw Data VS Kalman Estimation\", \n mlabel=\"Measurements\",\n estlabel=\"Kalman Estimates\",\n xlabel=\"Time in Seconds\",\n ylabel=\"Distance to Wall in CM\")\n\nest_vel = [v[1] for v in est_x]\nplt.plot(time_values, est_vel)\nplt.xlabel(\"Time in seconds\")\nplt.ylabel(\"Speed in cm/sec\")\nplt.title(\"Velocity Over Time\")\nplt.show()\n\npos_std = [p[0][0]**0.5 for p in est_P]\nvel_var = [p[1][1]**0.5 for p in est_P]\npos_vel_corr = [p[0][1] for p in est_P]\nplt.plot(time_values, pos_std)\nplt.ylabel(\"Position StdDev\")\nplt.xlabel(\"Time in Seconds\")\nplt.title(\"Position StdDev Over Time\")\nplt.show()\n\nplt.plot(time_values, vel_var)\nplt.ylabel(\"Velocity StdDev\")\nplt.xlabel(\"Time in Seconds\")\nplt.title(\"Velocity StdDev Over Time\")\nplt.show()\n\nplt.plot(time_values, pos_vel_corr)\nplt.ylabel(\"Velocity-Position Correlation\")\nplt.xlabel(\"Time in Seconds\")\nplt.title(\"Velocity-Position Correlation Over Time\")\nplt.show()\n",
"_____no_output_____"
]
],
[
[
"### Part D\n\n1. Were any anomolous points processed by mistake?\n - Kalman filter without outlier detection processed object readings. Both methods of outlier detection succesfully removed object readings.\n2. Were any valid points rejected?\n - Neither outlier detection method rejected valid points.\n3. How can each of these methods fail?\n - As long as the data supplied is linear and linearly separable these methods should work robustly. If any of these constraints are broken all 3 methods would likely fail.",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
cb8dfb8039b3e3719cf46908deee776ad0adf4d7
| 6,338 |
ipynb
|
Jupyter Notebook
|
NLP/3_nltk/basic_nltk.ipynb
|
ybdesire/machinelearning
|
0224746332e1085336e0b02e0ca3b11d74bd9a91
|
[
"MIT"
] | 30 |
2017-02-28T13:52:58.000Z
|
2022-03-24T10:28:43.000Z
|
NLP/3_nltk/basic_nltk.ipynb
|
ybdesire/machinelearning
|
0224746332e1085336e0b02e0ca3b11d74bd9a91
|
[
"MIT"
] | null | null | null |
NLP/3_nltk/basic_nltk.ipynb
|
ybdesire/machinelearning
|
0224746332e1085336e0b02e0ca3b11d74bd9a91
|
[
"MIT"
] | 17 |
2017-03-03T12:38:04.000Z
|
2022-03-11T01:53:20.000Z
| 25.250996 | 176 | 0.527138 |
[
[
[
"# nltk basic",
"_____no_output_____"
],
[
"## 1. install",
"_____no_output_____"
]
],
[
[
"! pip install -U nltk",
"Collecting nltk\nRequirement not upgraded as not directly required: six in /root/anaconda3/envs/envtf/lib/python3.5/site-packages (from nltk) (1.11.0)\nCollecting singledispatch (from nltk)\n Using cached https://files.pythonhosted.org/packages/c5/10/369f50bcd4621b263927b0a1519987a04383d4a98fb10438042ad410cf88/singledispatch-3.4.0.3-py2.py3-none-any.whl\n\u001b[31mtwisted 18.7.0 requires PyHamcrest>=1.9.0, which is not installed.\u001b[0m\n\u001b[31mmkl-fft 1.0.4 requires cython, which is not installed.\u001b[0m\n\u001b[31mmkl-random 1.0.1 requires cython, which is not installed.\u001b[0m\n\u001b[31mtensorflow 1.10.0 has requirement numpy<=1.14.5,>=1.13.3, but you'll have numpy 1.15.0 which is incompatible.\u001b[0m\n\u001b[31mtensorflow 1.10.0 has requirement setuptools<=39.1.0, but you'll have setuptools 40.0.0 which is incompatible.\u001b[0m\nInstalling collected packages: singledispatch, nltk\nSuccessfully installed nltk-3.4 singledispatch-3.4.0.3\n\u001b[33mYou are using pip version 10.0.1, however version 18.1 is available.\nYou should consider upgrading via the 'pip install --upgrade pip' command.\u001b[0m\n"
]
],
[
[
"## 2. Tokenize (英文分词)",
"_____no_output_____"
]
],
[
[
"import nltk\nsentence = \"\"\"At eight o'clock on Thursday morning Arthur didn't feel very good.\"\"\"\ntokens = nltk.word_tokenize(sentence)",
"_____no_output_____"
],
[
"tokens",
"_____no_output_____"
]
],
[
[
"## 3. tag (词性标注)",
"_____no_output_____"
]
],
[
[
"nltk.download('averaged_perceptron_tagger')",
"[nltk_data] Downloading package averaged_perceptron_tagger to\n[nltk_data] /root/nltk_data...\n[nltk_data] Unzipping taggers/averaged_perceptron_tagger.zip.\n"
],
[
"tagged = nltk.pos_tag(tokens)",
"_____no_output_____"
],
[
"tagged",
"_____no_output_____"
]
],
[
[
"## 4. tag meanings",
"_____no_output_____"
],
[
"* CC coordinating conjunction\n* CD cardinal digit\n* DT determiner\n* EX existential there (like: “there is” … think of it like “there exists”)\n* FW foreign word\n* IN preposition/subordinating conjunction\n* JJ adjective ‘big’\n* JJR adjective, comparative ‘bigger’\n* JJS adjective, superlative ‘biggest’\n* LS list marker 1)\n* MD modal could, will\n* NN noun, singular ‘desk’\n* NNS noun plural ‘desks’\n* NNP proper noun, singular ‘Harrison’\n* NNPS proper noun, plural ‘Americans’\n* PDT predeterminer ‘all the kids’\n* POS possessive ending parent’s\n* PRP personal pronoun I, he, she\n* PRP+dollar possessive pronoun my, his, hers\n* RB adverb very, silently,\n* RBR adverb, comparative better\n* RBS adverb, superlative best\n* RP particle give up\n* TO, to go ‘to’ the store.\n* UH interjection, errrrrrrrm\n* VB verb, base form take\n* VBD verb, past tense took\n* VBG verb, gerund/present participle taking\n* VBN verb, past participle taken\n* VBP verb, sing. present, non-3d take\n* VBZ verb, 3rd person sing. present takes\n* WDT wh-determiner which\n* WP wh-pronoun who, what\n* WP+dollar possessive wh-pronoun whose\n* WRB wh-abverb where, when\n",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
]
] |
cb8dfe0b4b40dd9030e1bd2337df47a3e909eb64
| 49,935 |
ipynb
|
Jupyter Notebook
|
Traffic_Sign_Classifier.ipynb
|
manheima/CarND-Traffic-Sign-Classifier-Project
|
1ef77f37b622d44598d4a734373610de901a6561
|
[
"MIT"
] | null | null | null |
Traffic_Sign_Classifier.ipynb
|
manheima/CarND-Traffic-Sign-Classifier-Project
|
1ef77f37b622d44598d4a734373610de901a6561
|
[
"MIT"
] | null | null | null |
Traffic_Sign_Classifier.ipynb
|
manheima/CarND-Traffic-Sign-Classifier-Project
|
1ef77f37b622d44598d4a734373610de901a6561
|
[
"MIT"
] | null | null | null | 62.653701 | 10,644 | 0.728968 |
[
[
[
"# Self-Driving Car Engineer Nanodegree\n\n## Deep Learning\n\n## Project: Build a Traffic Sign Recognition Classifier\n\nIn this notebook, a template is provided for you to implement your functionality in stages, which is required to successfully complete this project. If additional code is required that cannot be included in the notebook, be sure that the Python code is successfully imported and included in your submission if necessary. \n\n> **Note**: Once you have completed all of the code implementations, you need to finalize your work by exporting the iPython Notebook as an HTML document. Before exporting the notebook to html, all of the code cells need to have been run so that reviewers can see the final implementation and output. You can then export the notebook by using the menu above and navigating to \\n\",\n \"**File -> Download as -> HTML (.html)**. Include the finished document along with this notebook as your submission. \n\nIn addition to implementing code, there is a writeup to complete. The writeup should be completed in a separate file, which can be either a markdown file or a pdf document. There is a [write up template](https://github.com/udacity/CarND-Traffic-Sign-Classifier-Project/blob/master/writeup_template.md) that can be used to guide the writing process. Completing the code template and writeup template will cover all of the [rubric points](https://review.udacity.com/#!/rubrics/481/view) for this project.\n\nThe [rubric](https://review.udacity.com/#!/rubrics/481/view) contains \"Stand Out Suggestions\" for enhancing the project beyond the minimum requirements. The stand out suggestions are optional. If you decide to pursue the \"stand out suggestions\", you can include the code in this Ipython notebook and also discuss the results in the writeup file.\n\n\n>**Note:** Code and Markdown cells can be executed using the **Shift + Enter** keyboard shortcut. In addition, Markdown cells can be edited by typically double-clicking the cell to enter edit mode.",
"_____no_output_____"
],
[
"---\n## Step 0: Load The Data",
"_____no_output_____"
]
],
[
[
"# Load pickled data\nimport pickle\n\n# TODO: Fill this in based on where you saved the training and testing data\n\ntraining_file = \"traffic-signs-data/train.p\"\nvalidation_file= \"traffic-signs-data/valid.p\"\ntesting_file = \"traffic-signs-data/test.p\"\n\nwith open(training_file, mode='rb') as f:\n train = pickle.load(f)\nwith open(validation_file, mode='rb') as f:\n valid = pickle.load(f)\nwith open(testing_file, mode='rb') as f:\n test = pickle.load(f)\n \nX_train, y_train = train['features'], train['labels']\nX_valid, y_validation = valid['features'], valid['labels']\nX_test, y_test = test['features'], test['labels']",
"_____no_output_____"
]
],
[
[
"---\n\n## Step 1: Dataset Summary & Exploration\n\nThe pickled data is a dictionary with 4 key/value pairs:\n\n- `'features'` is a 4D array containing raw pixel data of the traffic sign images, (num examples, width, height, channels).\n- `'labels'` is a 1D array containing the label/class id of the traffic sign. The file `signnames.csv` contains id -> name mappings for each id.\n- `'sizes'` is a list containing tuples, (width, height) representing the original width and height the image.\n- `'coords'` is a list containing tuples, (x1, y1, x2, y2) representing coordinates of a bounding box around the sign in the image. **THESE COORDINATES ASSUME THE ORIGINAL IMAGE. THE PICKLED DATA CONTAINS RESIZED VERSIONS (32 by 32) OF THESE IMAGES**\n\nComplete the basic data summary below. Use python, numpy and/or pandas methods to calculate the data summary rather than hard coding the results. For example, the [pandas shape method](http://pandas.pydata.org/pandas-docs/stable/generated/pandas.DataFrame.shape.html) might be useful for calculating some of the summary results. ",
"_____no_output_____"
],
[
"### Provide a Basic Summary of the Data Set Using Python, Numpy and/or Pandas",
"_____no_output_____"
]
],
[
[
"### Replace each question mark with the appropriate value. \n### Use python, pandas or numpy methods rather than hard coding the results\n\n# TODO: Number of training examples\nn_train = X_train.shape[0]\n\n# TODO: Number of validation examples\nn_validation = X_valid.shape[0]\n\n# TODO: Number of testing examples.\nn_test = X_test.shape[0]\n\n# TODO: What's the shape of an traffic sign image?\nimage_shape = X_train[0].shape\n\n# TODO: How many unique classes/labels there are in the dataset.\nn_classes = 43\n\nprint(\"Number of training examples =\", n_train)\nprint(\"Number of testing examples =\", n_test)\nprint(\"Image data shape =\", image_shape)\nprint(\"Number of classes =\", n_classes)",
"Number of training examples = 34799\nNumber of testing examples = 12630\nImage data shape = (32, 32, 3)\nNumber of classes = 43\n"
]
],
[
[
"### Include an exploratory visualization of the dataset",
"_____no_output_____"
],
[
"Visualize the German Traffic Signs Dataset using the pickled file(s). This is open ended, suggestions include: plotting traffic sign images, plotting the count of each sign, etc. \n\nThe [Matplotlib](http://matplotlib.org/) [examples](http://matplotlib.org/examples/index.html) and [gallery](http://matplotlib.org/gallery.html) pages are a great resource for doing visualizations in Python.\n\n**NOTE:** It's recommended you start with something simple first. If you wish to do more, come back to it after you've completed the rest of the sections. It can be interesting to look at the distribution of classes in the training, validation and test set. Is the distribution the same? Are there more examples of some classes than others?",
"_____no_output_____"
]
],
[
[
"import random\nimport numpy as np\nimport matplotlib.pyplot as plt\n%matplotlib inline\n\nindex = random.randint(0, len(X_train))\nimage = X_train[index].squeeze()\n\nplt.figure(figsize=(1,1))\nplt.imshow(image)\nprint(y_train[index])",
"32\n"
]
],
[
[
"----\n\n## Step 2: Design and Test a Model Architecture\n\nDesign and implement a deep learning model that learns to recognize traffic signs. Train and test your model on the [German Traffic Sign Dataset](http://benchmark.ini.rub.de/?section=gtsrb&subsection=dataset).\n\nThe LeNet-5 implementation shown in the [classroom](https://classroom.udacity.com/nanodegrees/nd013/parts/fbf77062-5703-404e-b60c-95b78b2f3f9e/modules/6df7ae49-c61c-4bb2-a23e-6527e69209ec/lessons/601ae704-1035-4287-8b11-e2c2716217ad/concepts/d4aca031-508f-4e0b-b493-e7b706120f81) at the end of the CNN lesson is a solid starting point. You'll have to change the number of classes and possibly the preprocessing, but aside from that it's plug and play! \n\nWith the LeNet-5 solution from the lecture, you should expect a validation set accuracy of about 0.89. To meet specifications, the validation set accuracy will need to be at least 0.93. It is possible to get an even higher accuracy, but 0.93 is the minimum for a successful project submission. \n\nThere are various aspects to consider when thinking about this problem:\n\n- Neural network architecture (is the network over or underfitting?)\n- Play around preprocessing techniques (normalization, rgb to grayscale, etc)\n- Number of examples per label (some have more than others).\n- Generate fake data.\n\nHere is an example of a [published baseline model on this problem](http://yann.lecun.com/exdb/publis/pdf/sermanet-ijcnn-11.pdf). It's not required to be familiar with the approach used in the paper but, it's good practice to try to read papers like these.",
"_____no_output_____"
],
[
"### Pre-process the Data Set (normalization, grayscale, etc.)",
"_____no_output_____"
],
[
"Minimally, the image data should be normalized so that the data has mean zero and equal variance. For image data, `(pixel - 128)/ 128` is a quick way to approximately normalize the data and can be used in this project. \n\nOther pre-processing steps are optional. You can try different techniques to see if it improves performance. \n\nUse the code cell (or multiple code cells, if necessary) to implement the first step of your project.",
"_____no_output_____"
]
],
[
[
"### Preprocess the data here. It is required to normalize the data. Other preprocessing steps could include \n### converting to grayscale, etc.\n### Feel free to use as many code cells as needed.\n\n#First shuffle the data\nfrom sklearn.utils import shuffle\n\nX_train, y_train = shuffle(X_train, y_train)\n\n#Now normalize the data. Shift pixel rgb. \nX_train = (X_train.astype(float)-128)/128\nX_valid = (X_valid.astype(float)-128)/128\nX_test = (X_test.astype(float)-128)/128",
"_____no_output_____"
]
],
[
[
"### Model Architecture",
"_____no_output_____"
]
],
[
[
"### Setup Tesnorflow\nimport tensorflow as tf\n\nEPOCHS = 100\nBATCH_SIZE = 64 #128\n\nfrom tensorflow.contrib.layers import flatten\n\n######AAron idea: try dropout insted of max pooling?\ndef LeNet(x): \n # Arguments used for tf.truncated_normal, randomly defines variables for the weights and biases for each layer\n mu = 0\n sigma = 0.1\n \n # SOLUTION: Layer 1: Convolutional. Input = 32x32x3. Output = 28x28x6.\n conv1_W = tf.Variable(tf.truncated_normal(shape=(5, 5, 3, 6), mean = mu, stddev = sigma))\n conv1_b = tf.Variable(tf.zeros(6))\n conv1 = tf.nn.conv2d(x, conv1_W, strides=[1, 1, 1, 1], padding='VALID') + conv1_b\n\n # SOLUTION: Activation.\n conv1 = tf.nn.relu(conv1)\n\n # SOLUTION: Pooling. Input = 28x28x6. Output = 14x14x6.\n conv1 = tf.nn.max_pool(conv1, ksize=[1, 2, 2, 1], strides=[1, 2, 2, 1], padding='VALID')\n\n # SOLUTION: Layer 2: Convolutional. Output = 10x10x16.\n conv2_W = tf.Variable(tf.truncated_normal(shape=(5, 5, 6, 16), mean = mu, stddev = sigma))\n conv2_b = tf.Variable(tf.zeros(16))\n conv2 = tf.nn.conv2d(conv1, conv2_W, strides=[1, 1, 1, 1], padding='VALID') + conv2_b\n \n # SOLUTION: Activation.\n conv2 = tf.nn.relu(conv2)\n\n # SOLUTION: Pooling. Input = 10x10x16. Output = 5x5x16.\n conv2 = tf.nn.max_pool(conv2, ksize=[1, 2, 2, 1], strides=[1, 2, 2, 1], padding='VALID')\n\n # SOLUTION: Flatten. Input = 5x5x16. Output = 400.\n fc0 = flatten(conv2)\n \n # SOLUTION: Layer 3: Fully Connected. Input = 400. Output = 120.\n fc1_W = tf.Variable(tf.truncated_normal(shape=(400, 120), mean = mu, stddev = sigma))\n fc1_b = tf.Variable(tf.zeros(120))\n fc1 = tf.matmul(fc0, fc1_W) + fc1_b\n \n # SOLUTION: Activation.\n fc1 = tf.nn.relu(fc1)\n\n # SOLUTION: Layer 4: Fully Connected. Input = 120. Output = 84.\n fc2_W = tf.Variable(tf.truncated_normal(shape=(120, 84), mean = mu, stddev = sigma))\n fc2_b = tf.Variable(tf.zeros(84))\n fc2 = tf.matmul(fc1, fc2_W) + fc2_b\n \n # SOLUTION: Activation.\n fc2 = tf.nn.relu(fc2)\n\n # SOLUTION: Layer 5: Fully Connected. Input = 84. Output = 43.\n fc3_W = tf.Variable(tf.truncated_normal(shape=(84, 43), mean = mu, stddev = sigma))\n fc3_b = tf.Variable(tf.zeros(43))\n logits = tf.matmul(fc2, fc3_W) + fc3_b\n \n return logits",
"_____no_output_____"
]
],
[
[
"### Train, Validate and Test the Model",
"_____no_output_____"
],
[
"A validation set can be used to assess how well the model is performing. A low accuracy on the training and validation\nsets imply underfitting. A high accuracy on the training set but low accuracy on the validation set implies overfitting.",
"_____no_output_____"
]
],
[
[
"### Calculate and report the accuracy on the training and validation set.\n### Once a final model architecture is selected, \n### the accuracy on the test set should be calculated and reported as well.\n### Feel free to use as many code cells as needed.\n\n## Features and Lables. \n# x is a placeholder for a batch of input images. y is a placeholder for a batch of output labels\nx = tf.placeholder(tf.float32, (None, 32, 32, 3))\ny = tf.placeholder(tf.int32, (None))\none_hot_y = tf.one_hot(y, 43)\n\n### Training Pipeline\nrate = .0004 #0.001\n\nlogits = LeNet(x)\ncross_entropy = tf.nn.softmax_cross_entropy_with_logits(labels=one_hot_y, logits=logits)\nloss_operation = tf.reduce_mean(cross_entropy)\noptimizer = tf.train.AdamOptimizer(learning_rate = rate)\ntraining_operation = optimizer.minimize(loss_operation)\n\n\n### Model Evaluation\n# Evaluate the loss and accuracy of the model for a given dataset\ncorrect_prediction = tf.equal(tf.argmax(logits, 1), tf.argmax(one_hot_y, 1))\naccuracy_operation = tf.reduce_mean(tf.cast(correct_prediction, tf.float32))\nsaver = tf.train.Saver()\nprediction = tf.argmax(logits, 1)\n\ndef evaluate(X_data, y_data):\n num_examples = len(X_data)\n total_accuracy = 0\n sess = tf.get_default_session()\n for offset in range(0, num_examples, BATCH_SIZE):\n batch_x, batch_y = X_data[offset:offset+BATCH_SIZE], y_data[offset:offset+BATCH_SIZE]\n accuracy = sess.run(accuracy_operation, feed_dict={x: batch_x, y: batch_y})\n total_accuracy += (accuracy * len(batch_x))\n return total_accuracy / num_examples\n\ndef predictClass(X_data):\n sess = tf.get_default_session()\n predictions = sess.run(prediction, feed_dict={x: X_data})\n return predictions\n\ndef getLogits(X_data):\n sess = tf.get_default_session()\n return sess.run(logits, feed_dict={x: X_data})",
"_____no_output_____"
]
],
[
[
"## Run the training algorithm",
"_____no_output_____"
]
],
[
[
"### Train model\n#Run the training data through the training pipeline to train the model.\n#Before each epoch, shuffle the training set.\n#After each epoch, measure the loss and accuracy of the validation set.\n#Save the model after training.\nwith tf.Session() as sess:\n sess.run(tf.global_variables_initializer())\n num_examples = len(X_train)\n print(\"Training...\")\n print()\n for i in range(EPOCHS):\n X_train, y_train = shuffle(X_train, y_train)\n for offset in range(0, num_examples, BATCH_SIZE):\n end = offset + BATCH_SIZE\n batch_x, batch_y = X_train[offset:end], y_train[offset:end]\n sess.run(training_operation, feed_dict={x: batch_x, y: batch_y})\n \n validation_accuracy = evaluate(X_valid, y_validation)\n print(\"EPOCH {} ...\".format(i+1))\n print(\"Validation Accuracy = {:.3f}\".format(validation_accuracy))\n print()\n \n saver.save(sess, './lenet')\n print(\"Model saved\")",
"_____no_output_____"
]
],
[
[
"## Check model on Test Images - only do this once if possible",
"_____no_output_____"
]
],
[
[
"with tf.Session() as sess:\n saver = tf.train.import_meta_graph('lenet.meta')\n saver.restore(sess, tf.train.latest_checkpoint('.'))\n\n test_accuracy = evaluate(X_test, y_test)\n print(\"Test Accuracy = {:.3f}\".format(test_accuracy))",
"INFO:tensorflow:Restoring parameters from ./lenet\nTest Accuracy = 0.905\n"
]
],
[
[
"---\n\n## Step 3: Test a Model on New Images\n\nTo give yourself more insight into how your model is working, download at least five pictures of German traffic signs from the web and use your model to predict the traffic sign type.\n\nYou may find `signnames.csv` useful as it contains mappings from the class id (integer) to the actual sign name.",
"_____no_output_____"
],
[
"### Load and Output the Images",
"_____no_output_____"
]
],
[
[
"### Load the images and plot them here.\n### Feel free to use as many code cells as needed.\nimport cv2\nimport matplotlib.pyplot as plt\nimport matplotlib.image as mpimg\nimport numpy as np\nimage1 = mpimg.imread('traffic-signs-web-images/BumpyRoad_22.jpg')\nimage2 = mpimg.imread('traffic-signs-web-images/SlipperyRoad_23.jpg')\nimage3 = mpimg.imread('traffic-signs-web-images/NoEntry_17.jpg')\nimage4 = mpimg.imread('traffic-signs-web-images/WildAnimalsCrossing_31.jpg')\nimage5 = mpimg.imread('traffic-signs-web-images/Yield_13.jpg')\nprint('This image is:', type(image), 'with dimensions:', image.shape)\n\n\n#Put all the images in a lsit\nX_web = np.zeros([5,32,32,3],dtype=np.uint8)\nX_web[0] = image1 #Failed to classify\nX_web[1] = image2 #Failed to classify\nX_web[2] = image3 #Failed to classify\nX_web[3] = image4 #Classified!\nX_web[4] = image5 #Classified!\n\n#Put the labels in a list\ny_web = np.array([22,23,17,31,13])\n\n\nplt.imshow(X_web[2])",
"This image is: <class 'numpy.ndarray'> with dimensions: (32, 32, 3)\n"
]
],
[
[
"### Predict the Sign Type for Each Image",
"_____no_output_____"
]
],
[
[
"### Run the predictions here and use the model to output the prediction for each image.\n### Make sure to pre-process the images with the same pre-processing pipeline used earlier.\n### Feel free to use as many code cells as needed.\n\n#Normalize the data\nX_web = (X_web.astype(float)-128)/128\n\n#Now make the predictions for each image\nwith tf.Session() as sess:\n sess.run(tf.global_variables_initializer())\n saver = tf.train.import_meta_graph('lenet-Copy1.meta')\n saver.restore(sess, tf.train.latest_checkpoint('.'))\n y_hat = predictClass(X_web)\n \n \nprint(\"Predictions: \"+str(y_hat))\nprint(\"Actual: \"+str(y_web))",
"INFO:tensorflow:Restoring parameters from ./lenet\nPredictions: [29 11 12 25 13]\nActual: [22 23 17 31 13]\n"
]
],
[
[
"### Analyze Performance",
"_____no_output_____"
]
],
[
[
"### Calculate the accuracy for these 5 new images. \n### For example, if the model predicted 1 out of 5 signs correctly, it's 20% accurate on these new images.\nwith tf.Session() as sess:\n saver = tf.train.import_meta_graph('lenet.meta')\n saver.restore(sess, tf.train.latest_checkpoint('.'))\n\n test_accuracy = evaluate(X_web, y_web)\n print(\"Test Accuracy = {:.3f}\".format(test_accuracy))",
"INFO:tensorflow:Restoring parameters from ./lenet\nTest Accuracy = 0.200\n"
]
],
[
[
"### Output Top 5 Softmax Probabilities For Each Image Found on the Web",
"_____no_output_____"
],
[
"For each of the new images, print out the model's softmax probabilities to show the **certainty** of the model's predictions (limit the output to the top 5 probabilities for each image). [`tf.nn.top_k`](https://www.tensorflow.org/versions/r0.12/api_docs/python/nn.html#top_k) could prove helpful here. \n\nThe example below demonstrates how tf.nn.top_k can be used to find the top k predictions for each image.\n\n`tf.nn.top_k` will return the values and indices (class ids) of the top k predictions. So if k=3, for each sign, it'll return the 3 largest probabilities (out of a possible 43) and the correspoding class ids.\n\nTake this numpy array as an example. The values in the array represent predictions. The array contains softmax probabilities for five candidate images with six possible classes. `tf.nn.top_k` is used to choose the three classes with the highest probability:\n\n```\n# (5, 6) array\na = np.array([[ 0.24879643, 0.07032244, 0.12641572, 0.34763842, 0.07893497,\n 0.12789202],\n [ 0.28086119, 0.27569815, 0.08594638, 0.0178669 , 0.18063401,\n 0.15899337],\n [ 0.26076848, 0.23664738, 0.08020603, 0.07001922, 0.1134371 ,\n 0.23892179],\n [ 0.11943333, 0.29198961, 0.02605103, 0.26234032, 0.1351348 ,\n 0.16505091],\n [ 0.09561176, 0.34396535, 0.0643941 , 0.16240774, 0.24206137,\n 0.09155967]])\n```\n\nRunning it through `sess.run(tf.nn.top_k(tf.constant(a), k=3))` produces:\n\n```\nTopKV2(values=array([[ 0.34763842, 0.24879643, 0.12789202],\n [ 0.28086119, 0.27569815, 0.18063401],\n [ 0.26076848, 0.23892179, 0.23664738],\n [ 0.29198961, 0.26234032, 0.16505091],\n [ 0.34396535, 0.24206137, 0.16240774]]), indices=array([[3, 0, 5],\n [0, 1, 4],\n [0, 5, 1],\n [1, 3, 5],\n [1, 4, 3]], dtype=int32))\n```\n\nLooking just at the first row we get `[ 0.34763842, 0.24879643, 0.12789202]`, you can confirm these are the 3 largest probabilities in `a`. You'll also notice `[3, 0, 5]` are the corresponding indices.",
"_____no_output_____"
]
],
[
[
"### Print out the top five softmax probabilities for the predictions on the German traffic sign images found on the web. \n### Feel free to use as many code cells as needed.\nwith tf.Session() as sess:\n sess.run(tf.global_variables_initializer())\n saver = tf.train.import_meta_graph('lenet-Copy1.meta')\n saver.restore(sess, tf.train.latest_checkpoint('.'))\n theLogits = getLogits(X_web)\n probs = sess.run(tf.nn.softmax(theLogits))\n print(sess.run(tf.nn.top_k(tf.constant(probs), k=5)))\n \n",
"INFO:tensorflow:Restoring parameters from ./lenet\n"
]
],
[
[
"### Project Writeup\n\nOnce you have completed the code implementation, document your results in a project writeup using this [template](https://github.com/udacity/CarND-Traffic-Sign-Classifier-Project/blob/master/writeup_template.md) as a guide. The writeup can be in a markdown or pdf file. ",
"_____no_output_____"
],
[
"> **Note**: Once you have completed all of the code implementations and successfully answered each question above, you may finalize your work by exporting the iPython Notebook as an HTML document. You can do this by using the menu above and navigating to \\n\",\n \"**File -> Download as -> HTML (.html)**. Include the finished document along with this notebook as your submission.",
"_____no_output_____"
],
[
"---\n\n## Step 4 (Optional): Visualize the Neural Network's State with Test Images\n\n This Section is not required to complete but acts as an additional excersise for understaning the output of a neural network's weights. While neural networks can be a great learning device they are often referred to as a black box. We can understand what the weights of a neural network look like better by plotting their feature maps. After successfully training your neural network you can see what it's feature maps look like by plotting the output of the network's weight layers in response to a test stimuli image. From these plotted feature maps, it's possible to see what characteristics of an image the network finds interesting. For a sign, maybe the inner network feature maps react with high activation to the sign's boundary outline or to the contrast in the sign's painted symbol.\n\n Provided for you below is the function code that allows you to get the visualization output of any tensorflow weight layer you want. The inputs to the function should be a stimuli image, one used during training or a new one you provided, and then the tensorflow variable name that represents the layer's state during the training process, for instance if you wanted to see what the [LeNet lab's](https://classroom.udacity.com/nanodegrees/nd013/parts/fbf77062-5703-404e-b60c-95b78b2f3f9e/modules/6df7ae49-c61c-4bb2-a23e-6527e69209ec/lessons/601ae704-1035-4287-8b11-e2c2716217ad/concepts/d4aca031-508f-4e0b-b493-e7b706120f81) feature maps looked like for it's second convolutional layer you could enter conv2 as the tf_activation variable.\n\nFor an example of what feature map outputs look like, check out NVIDIA's results in their paper [End-to-End Deep Learning for Self-Driving Cars](https://devblogs.nvidia.com/parallelforall/deep-learning-self-driving-cars/) in the section Visualization of internal CNN State. NVIDIA was able to show that their network's inner weights had high activations to road boundary lines by comparing feature maps from an image with a clear path to one without. Try experimenting with a similar test to show that your trained network's weights are looking for interesting features, whether it's looking at differences in feature maps from images with or without a sign, or even what feature maps look like in a trained network vs a completely untrained one on the same sign image.\n\n<figure>\n <img src=\"visualize_cnn.png\" width=\"380\" alt=\"Combined Image\" />\n <figcaption>\n <p></p> \n <p style=\"text-align: center;\"> Your output should look something like this (above)</p> \n </figcaption>\n</figure>\n <p></p> \n",
"_____no_output_____"
]
],
[
[
"### Visualize your network's feature maps here.\n### Feel free to use as many code cells as needed.\n\n# image_input: the test image being fed into the network to produce the feature maps\n# tf_activation: should be a tf variable name used during your training procedure that represents the calculated state of a specific weight layer\n# activation_min/max: can be used to view the activation contrast in more detail, by default matplot sets min and max to the actual min and max values of the output\n# plt_num: used to plot out multiple different weight feature map sets on the same block, just extend the plt number for each new feature map entry\n\ndef outputFeatureMap(image_input, tf_activation, activation_min=-1, activation_max=-1 ,plt_num=1):\n # Here make sure to preprocess your image_input in a way your network expects\n # with size, normalization, ect if needed\n # image_input =\n # Note: x should be the same name as your network's tensorflow data placeholder variable\n # If you get an error tf_activation is not defined it may be having trouble accessing the variable from inside a function\n activation = tf_activation.eval(session=sess,feed_dict={x : image_input})\n featuremaps = activation.shape[3]\n plt.figure(plt_num, figsize=(15,15))\n for featuremap in range(featuremaps):\n plt.subplot(6,8, featuremap+1) # sets the number of feature maps to show on each row and column\n plt.title('FeatureMap ' + str(featuremap)) # displays the feature map number\n if activation_min != -1 & activation_max != -1:\n plt.imshow(activation[0,:,:, featuremap], interpolation=\"nearest\", vmin =activation_min, vmax=activation_max, cmap=\"gray\")\n elif activation_max != -1:\n plt.imshow(activation[0,:,:, featuremap], interpolation=\"nearest\", vmax=activation_max, cmap=\"gray\")\n elif activation_min !=-1:\n plt.imshow(activation[0,:,:, featuremap], interpolation=\"nearest\", vmin=activation_min, cmap=\"gray\")\n else:\n plt.imshow(activation[0,:,:, featuremap], interpolation=\"nearest\", cmap=\"gray\")",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
]
] |
cb8dfeddea1dadb73cc1f9d23b7cffd179fe02b3
| 185,970 |
ipynb
|
Jupyter Notebook
|
MNIST/MNIST_CNN/.ipynb_checkpoints/ex_lass_signum_no_fixed-step-length-MNIST-Exp1-15-checkpoint.ipynb
|
yashkant/Decision-Flip-Experiments
|
02a65218c58ba8996584a5e2c36bb5dc03c5b45d
|
[
"Apache-2.0"
] | 7 |
2018-11-08T18:38:22.000Z
|
2018-11-28T14:30:39.000Z
|
MNIST/MNIST_CNN/.ipynb_checkpoints/ex_lass_signum_no_fixed-step-length-MNIST-Exp1-15-checkpoint.ipynb
|
yashkant/Decision-Flip-Experiments
|
02a65218c58ba8996584a5e2c36bb5dc03c5b45d
|
[
"Apache-2.0"
] | null | null | null |
MNIST/MNIST_CNN/.ipynb_checkpoints/ex_lass_signum_no_fixed-step-length-MNIST-Exp1-15-checkpoint.ipynb
|
yashkant/Decision-Flip-Experiments
|
02a65218c58ba8996584a5e2c36bb5dc03c5b45d
|
[
"Apache-2.0"
] | 1 |
2018-03-16T03:00:55.000Z
|
2018-03-16T03:00:55.000Z
| 169.680657 | 78,548 | 0.887251 |
[
[
[
"import os\npath = '/home/yash/Desktop/tensorflow-adversarial/tf_example'\nos.chdir(path)\n# supress tensorflow logging other than errors\nos.environ['TF_CPP_MIN_LOG_LEVEL'] = '3'\nimport numpy as np\nimport tensorflow as tf\nfrom tensorflow.contrib.learn import ModeKeys, Estimator\n\nimport matplotlib\nmatplotlib.use('Agg')\nimport matplotlib.pyplot as plt\nimport matplotlib.gridspec as gridspec\nfrom fgsm4 import fgsm\nimport mnist\n\nimg_rows = 28\nimg_cols = 28\nimg_chas = 1\ninput_shape = (img_rows, img_cols, img_chas)\nn_classes = 10\n\nprint('\\nLoading mnist')\n(X_train, y_train), (X_test, y_test) = mnist.load_data()\n\nX_train = X_train.astype('float32') / 255.\nX_test = X_test.astype('float32') / 255.\n\nX_train = X_train.reshape(-1, img_rows, img_cols, img_chas)\nX_test = X_test.reshape(-1, img_rows, img_cols, img_chas)\n\n# one hot encoding, basically creates hte si\ndef _to_categorical(x, n_classes):\n x = np.array(x, dtype=int).ravel()\n n = x.shape[0]\n ret = np.zeros((n, n_classes))\n ret[np.arange(n), x] = 1\n return ret\n\ndef find_l2(X_test, X_adv):\n a=X_test.reshape(-1,28*28)\n b=X_adv.reshape(-1,28*28)\n l2_unsquared = np.sum(np.square(a-b),axis=1)\n return l2_unsquared\n\ny_train = _to_categorical(y_train, n_classes)\ny_test = _to_categorical(y_test, n_classes)\nprint('\\nShuffling training data')\nind = np.random.permutation(X_train.shape[0])\nX_train, y_train = X_train[ind], y_train[ind]\n\n# X_train = X_train[:1000]\n# y_train = y_train[:1000]\n\n\n# split training/validation dataset\nvalidation_split = 0.1\nn_train = int(X_train.shape[0]*(1-validation_split))\nX_valid = X_train[n_train:]\nX_train = X_train[:n_train]\ny_valid = y_train[n_train:]\ny_train = y_train[:n_train]\n\nclass Dummy:\n pass\nenv = Dummy()\n\n",
"\nLoading mnist\n\nShuffling training data\n"
],
[
"def model(x, logits=False, training=False):\n conv0 = tf.layers.conv2d(x, filters=32, kernel_size=[3, 3],\n padding='same', name='conv0',\n activation=tf.nn.relu)\n \n pool0 = tf.layers.max_pooling2d(conv0, pool_size=[2, 2],\n strides=2, name='pool0')\n \n conv1 = tf.layers.conv2d(pool0, filters=64,\n kernel_size=[3, 3], padding='same',\n name='conv1', activation=tf.nn.relu)\n \n pool1 = tf.layers.max_pooling2d(conv1, pool_size=[2, 2],\n strides=2, name='pool1')\n \n flat = tf.reshape(pool1, [-1, 7*7*64], name='flatten')\n dense1 = tf.layers.dense(flat, units=1024, activation=tf.nn.relu,\n name='dense1')\n dense2 = tf.layers.dense(dense1, units=128, activation=tf.nn.relu,\n name='dense2')\n logits_ = tf.layers.dense(dense2, units=10, name='logits') #removed dropout\n y = tf.nn.softmax(logits_, name='ybar')\n if logits:\n return y, logits_\n return y",
"_____no_output_____"
],
[
"# We need a scope since the inference graph will be reused later\nwith tf.variable_scope('model'):\n env.x = tf.placeholder(tf.float32, (None, img_rows, img_cols,\n img_chas), name='x')\n env.y = tf.placeholder(tf.float32, (None, n_classes), name='y')\n env.training = tf.placeholder(bool, (), name='mode')\n\n env.ybar, logits = model(env.x, logits=True,\n training=env.training)\n\n z = tf.argmax(env.y, axis=1)\n zbar = tf.argmax(env.ybar, axis=1)\n env.count = tf.cast(tf.equal(z, zbar), tf.float32)\n env.acc = tf.reduce_mean(env.count, name='acc')\n\n xent = tf.nn.softmax_cross_entropy_with_logits(labels=env.y,\n logits=logits)\n env.loss = tf.reduce_mean(xent, name='loss')\n\nextra_update_ops = tf.get_collection(tf.GraphKeys.UPDATE_OPS)\n\nwith tf.control_dependencies(extra_update_ops):\n env.optim = tf.train.AdamOptimizer(beta1=0.9, beta2=0.999, epsilon=1e-08,).minimize(env.loss) \n",
"_____no_output_____"
],
[
"with tf.variable_scope('model', reuse=True):\n env.x_adv, env.all_flipped = fgsm(model, env.x, step_size=.05, bbox_semi_side=10) #epochs is redundant now!",
"_____no_output_____"
],
[
"sess = tf.InteractiveSession()\nsess.run(tf.global_variables_initializer())\nsess.run(tf.local_variables_initializer())",
"_____no_output_____"
],
[
"def save_model(label):\n saver = tf.train.Saver()\n saver.save(sess, './models/mnist/' + label)\n \ndef restore_model(label):\n saver = tf.train.Saver()\n saver.restore(sess, './models/mnist/' + label)",
"_____no_output_____"
],
[
"def _evaluate(X_data, y_data, env):\n print('\\nEvaluating')\n n_sample = X_data.shape[0]\n batch_size = 128\n n_batch = int(np.ceil(n_sample/batch_size))\n loss, acc = 0, 0\n ns = 0\n for ind in range(n_batch):\n print(' batch {0}/{1}'.format(ind+1, n_batch), end='\\r')\n start = ind*batch_size\n end = min(n_sample, start+batch_size)\n batch_loss, batch_count, batch_acc = sess.run(\n [env.loss, env.count, env.acc],\n feed_dict={env.x: X_data[start:end],\n env.y: y_data[start:end],\n env.training: False})\n loss += batch_loss*batch_size\n# print('batch count: {0}'.format(np.sum(batch_count)))\n ns+=batch_size\n acc += batch_acc*batch_size\n loss /= ns\n acc /= ns\n# print (ns)\n# print (n_sample)\n print(' loss: {0:.4f} acc: {1:.4f}'.format(loss, acc))\n return loss, acc",
"_____no_output_____"
],
[
"def _predict(X_data, env):\n print('\\nPredicting')\n n_sample = X_data.shape[0]\n batch_size = 128\n n_batch = int(np.ceil(n_sample/batch_size))\n yval = np.empty((X_data.shape[0], n_classes))\n for ind in range(n_batch):\n print(' batch {0}/{1}'.format(ind+1, n_batch), end='\\r')\n start = ind*batch_size\n end = min(n_sample, start+batch_size)\n batch_y = sess.run(env.ybar, feed_dict={\n env.x: X_data[start:end], env.training: False})\n yval[start:end] = batch_y\n return yval\n\ndef train(label):\n print('\\nTraining')\n n_sample = X_train.shape[0]\n batch_size = 128\n n_batch = int(np.ceil(n_sample/batch_size))\n n_epoch = 50\n for epoch in range(n_epoch):\n print('Epoch {0}/{1}'.format(epoch+1, n_epoch))\n for ind in range(n_batch):\n print(' batch {0}/{1}'.format(ind+1, n_batch), end='\\r')\n start = ind*batch_size\n end = min(n_sample, start+batch_size)\n sess.run(env.optim, feed_dict={env.x: X_train[start:end],\n env.y: y_train[start:end],\n env.training: True})\n if(epoch%5 == 0):\n model_label = label+ '{0}'.format(epoch)\n print(\"saving model \" + model_label)\n save_model(model_label)\n \n save_model(label)\n ",
"_____no_output_____"
],
[
"def create_adv(X, Y, label):\n print('\\nCrafting adversarial')\n n_sample = X.shape[0]\n batch_size = 1\n n_batch = int(np.ceil(n_sample/batch_size))\n n_epoch = 20\n X_adv = np.empty_like(X)\n for ind in range(n_batch):\n print(' batch {0}/{1}'.format(ind+1, n_batch), end='\\r')\n start = ind*batch_size\n end = min(n_sample, start+batch_size)\n tmp, all_flipped = sess.run([env.x_adv, env.all_flipped], feed_dict={env.x: X[start:end],\n env.y: Y[start:end],\n env.training: False})\n# _evaluate(tmp, Y[start:end],env)\n X_adv[start:end] = tmp\n# print(all_flipped)\n print('\\nSaving adversarial')\n os.makedirs('data', exist_ok=True)\n np.save('data/mnist/' + label + '.npy', X_adv)\n return X_adv",
"_____no_output_____"
],
[
"label = \"mnist_with_cnn\"\n# train(label) # else \n#Assuming that you've started a session already else do that first!\nrestore_model(label + '5')\n# restore_model(label + '10')\n# restore_model(label + '50')\n# restore_model(label + '100')\n_evaluate(X_train, y_train, env)",
"INFO:tensorflow:Restoring parameters from ./models/mnist/mnist_with_cnn0\n\nEvaluating\nbatch count: 126.0\nbatch count: 127.0\nbatch count: 124.0\nbatch count: 128.0\nbatch count: 128.0\nbatch count: 128.0\nbatch count: 126.0\nbatch count: 125.0\nbatch count: 125.0\nbatch count: 127.0\nbatch count: 124.0\nbatch count: 126.0\nbatch count: 128.0\nbatch count: 126.0\nbatch count: 126.0\nbatch count: 125.0\nbatch count: 124.0\nbatch count: 125.0\nbatch count: 127.0\nbatch count: 125.0\nbatch count: 127.0\nbatch count: 126.0\nbatch count: 125.0\nbatch count: 125.0\nbatch count: 128.0\nbatch count: 126.0\nbatch count: 128.0\nbatch count: 125.0\nbatch count: 123.0\nbatch count: 128.0\nbatch count: 125.0\nbatch count: 125.0\nbatch count: 128.0\nbatch count: 125.0\nbatch count: 125.0\nbatch count: 125.0\nbatch count: 126.0\nbatch count: 126.0\nbatch count: 127.0\nbatch count: 122.0\nbatch count: 127.0\nbatch count: 124.0\nbatch count: 125.0\nbatch count: 127.0\nbatch count: 126.0\nbatch count: 126.0\nbatch count: 126.0\nbatch count: 126.0\nbatch count: 126.0\nbatch count: 127.0\nbatch count: 125.0\nbatch count: 126.0\nbatch count: 125.0\nbatch count: 126.0\nbatch count: 125.0\nbatch count: 126.0\nbatch count: 127.0\nbatch count: 125.0\nbatch count: 126.0\nbatch count: 126.0\nbatch count: 127.0\nbatch count: 126.0\nbatch count: 125.0\nbatch count: 127.0\nbatch count: 126.0\nbatch count: 123.0\nbatch count: 125.0\nbatch count: 125.0\nbatch count: 122.0\nbatch count: 127.0\nbatch count: 127.0\nbatch count: 127.0\nbatch count: 125.0\nbatch count: 127.0\nbatch count: 127.0\nbatch count: 126.0\nbatch count: 127.0\nbatch count: 128.0\nbatch count: 128.0\nbatch count: 124.0\nbatch count: 128.0\nbatch count: 125.0\nbatch count: 127.0\nbatch count: 128.0\nbatch count: 125.0\nbatch count: 126.0\nbatch count: 126.0\nbatch count: 125.0\nbatch count: 127.0\nbatch count: 126.0\nbatch count: 128.0\nbatch count: 126.0\nbatch count: 126.0\nbatch count: 127.0\nbatch count: 126.0\nbatch count: 123.0\nbatch count: 126.0\nbatch count: 126.0\nbatch count: 128.0\nbatch count: 125.0\nbatch count: 124.0\nbatch count: 123.0\nbatch count: 124.0\nbatch count: 128.0\nbatch count: 127.0\nbatch count: 126.0\nbatch count: 126.0\nbatch count: 128.0\nbatch count: 126.0\nbatch count: 124.0\nbatch count: 124.0\nbatch count: 126.0\nbatch count: 125.0\nbatch count: 126.0\nbatch count: 126.0\nbatch count: 125.0\nbatch count: 122.0\nbatch count: 125.0\nbatch count: 124.0\nbatch count: 123.0\nbatch count: 126.0\nbatch count: 122.0\nbatch count: 123.0\nbatch count: 124.0\nbatch count: 125.0\nbatch count: 124.0\nbatch count: 124.0\nbatch count: 127.0\nbatch count: 126.0\nbatch count: 128.0\nbatch count: 127.0\nbatch count: 125.0\nbatch count: 125.0\nbatch count: 126.0\nbatch count: 124.0\nbatch count: 122.0\nbatch count: 124.0\nbatch count: 127.0\nbatch count: 127.0\nbatch count: 125.0\nbatch count: 127.0\nbatch count: 125.0\nbatch count: 124.0\nbatch count: 125.0\nbatch count: 127.0\nbatch count: 127.0\nbatch count: 126.0\nbatch count: 127.0\nbatch count: 124.0\nbatch count: 127.0\nbatch count: 123.0\nbatch count: 127.0\nbatch count: 126.0\nbatch count: 128.0\nbatch count: 128.0\nbatch count: 124.0\nbatch count: 125.0\nbatch count: 127.0\nbatch count: 127.0\nbatch count: 125.0\nbatch count: 124.0\nbatch count: 127.0\nbatch count: 125.0\nbatch count: 127.0\nbatch count: 126.0\nbatch count: 125.0\nbatch count: 125.0\nbatch count: 124.0\nbatch count: 126.0\nbatch count: 127.0\nbatch count: 123.0\nbatch count: 127.0\nbatch count: 128.0\nbatch count: 125.0\nbatch count: 126.0\nbatch count: 123.0\nbatch count: 125.0\nbatch count: 125.0\nbatch count: 127.0\nbatch count: 127.0\nbatch count: 127.0\nbatch count: 127.0\nbatch count: 125.0\nbatch count: 126.0\nbatch count: 125.0\nbatch count: 124.0\nbatch count: 123.0\nbatch count: 123.0\nbatch count: 125.0\nbatch count: 126.0\nbatch count: 126.0\nbatch count: 127.0\nbatch count: 125.0\nbatch count: 128.0\nbatch count: 127.0\nbatch count: 125.0\nbatch count: 124.0\nbatch count: 125.0\nbatch count: 125.0\nbatch count: 126.0\nbatch count: 126.0\nbatch count: 127.0\nbatch count: 126.0\nbatch count: 125.0\nbatch count: 127.0\nbatch count: 127.0\nbatch count: 126.0\nbatch count: 126.0\nbatch count: 127.0\nbatch count: 124.0\nbatch count: 124.0\nbatch count: 125.0\nbatch count: 126.0\nbatch count: 125.0\nbatch count: 126.0\nbatch count: 126.0\nbatch count: 127.0\nbatch count: 126.0\nbatch count: 128.0\nbatch count: 124.0\nbatch count: 126.0\nbatch count: 126.0\nbatch count: 127.0\nbatch count: 124.0\nbatch count: 126.0\nbatch count: 126.0\nbatch count: 123.0\nbatch count: 122.0\nbatch count: 126.0\nbatch count: 123.0\nbatch count: 127.0\nbatch count: 127.0\nbatch count: 127.0\nbatch count: 126.0\nbatch count: 128.0\nbatch count: 126.0\nbatch count: 127.0\nbatch count: 124.0\nbatch count: 127.0\nbatch count: 126.0\nbatch count: 123.0\nbatch count: 127.0\nbatch count: 125.0\nbatch count: 125.0\nbatch count: 125.0\nbatch count: 125.0\nbatch count: 126.0\nbatch count: 127.0\nbatch count: 126.0\nbatch count: 126.0\nbatch count: 127.0\nbatch count: 126.0\nbatch count: 126.0\nbatch count: 127.0\nbatch count: 127.0\nbatch count: 126.0\nbatch count: 126.0\nbatch count: 126.0\nbatch count: 125.0\nbatch count: 126.0\nbatch count: 126.0\nbatch count: 124.0\nbatch count: 126.0\nbatch count: 124.0\nbatch count: 126.0\nbatch count: 127.0\nbatch count: 126.0\nbatch count: 127.0\nbatch count: 126.0\nbatch count: 126.0\nbatch count: 128.0\nbatch count: 127.0\nbatch count: 124.0\nbatch count: 127.0\nbatch count: 125.0\nbatch count: 128.0\nbatch count: 125.0\nbatch count: 127.0\nbatch count: 124.0\nbatch count: 127.0\nbatch count: 123.0\nbatch count: 124.0\nbatch count: 124.0\nbatch count: 123.0\nbatch count: 126.0\nbatch count: 123.0\nbatch count: 124.0\nbatch count: 124.0\nbatch count: 126.0\nbatch count: 127.0\nbatch count: 128.0\nbatch count: 124.0\nbatch count: 123.0\nbatch count: 125.0\nbatch count: 126.0\nbatch count: 125.0\nbatch count: 126.0\nbatch count: 125.0\nbatch count: 125.0\nbatch count: 127.0\nbatch count: 125.0\nbatch count: 123.0\nbatch count: 126.0\nbatch count: 121.0\nbatch count: 127.0\nbatch count: 126.0\nbatch count: 125.0\nbatch count: 126.0\nbatch count: 127.0\nbatch count: 125.0\nbatch count: 124.0\nbatch count: 128.0\nbatch count: 124.0\nbatch count: 127.0\nbatch count: 126.0\nbatch count: 126.0\nbatch count: 127.0\nbatch count: 128.0\nbatch count: 124.0\nbatch count: 127.0\nbatch count: 123.0\nbatch count: 126.0\nbatch count: 126.0\nbatch count: 126.0\nbatch count: 125.0\nbatch count: 125.0\nbatch count: 127.0\nbatch count: 126.0\nbatch count: 126.0\nbatch count: 123.0\nbatch count: 125.0\nbatch count: 124.0\nbatch count: 125.0\nbatch count: 125.0\nbatch count: 125.0\nbatch count: 126.0\nbatch count: 125.0\nbatch count: 126.0\nbatch count: 126.0\nbatch count: 128.0\nbatch count: 126.0\nbatch count: 125.0\nbatch count: 126.0\nbatch count: 125.0\nbatch count: 125.0\nbatch count: 123.0\nbatch count: 125.0\nbatch count: 128.0\nbatch count: 126.0\nbatch count: 128.0\nbatch count: 128.0\nbatch count: 127.0\nbatch count: 127.0\nbatch count: 126.0\nbatch count: 127.0\nbatch count: 128.0\nbatch count: 126.0\nbatch count: 126.0\nbatch count: 126.0\nbatch count: 123.0\nbatch count: 128.0\nbatch count: 125.0\nbatch count: 127.0\nbatch count: 126.0\nbatch count: 126.0\nbatch count: 124.0\nbatch count: 127.0\nbatch count: 127.0\nbatch count: 123.0\nbatch count: 126.0\nbatch count: 128.0\nbatch count: 125.0\nbatch count: 125.0\nbatch count: 128.0\nbatch count: 126.0\nbatch count: 127.0\nbatch count: 125.0\nbatch count: 128.0\nbatch count: 126.0\nbatch count: 128.0\nbatch count: 125.0\nbatch count: 126.0\nbatch count: 126.0\nbatch count: 126.0\nbatch count: 127.0\nbatch count: 127.0\nbatch count: 125.0\nbatch count: 126.0\nbatch count: 126.0\nbatch count: 125.0\nbatch count: 125.0\nbatch count: 125.0\nbatch count: 126.0\nbatch count: 124.0\nbatch count: 127.0\nbatch count: 127.0\nbatch count: 123.0\nbatch count: 127.0\nbatch count: 126.0\nbatch count: 125.0\nbatch count: 126.0\nbatch count: 126.0\nbatch count: 125.0\nbatch count: 124.0\nbatch count: 126.0\nbatch count: 125.0\nbatch count: 125.0\nbatch count: 126.0\nbatch count: 124.0\nbatch count: 126.0\nbatch count: 128.0\nbatch count: 123.0\nbatch count: 127.0\nbatch count: 127.0\nbatch count: 124.0\nbatch count: 126.0\nbatch count: 124.0\nbatch count: 126.0\nbatch count: 125.0\nbatch count: 128.0\nbatch count: 125.0\nbatch count: 108.0\n loss: 0.0558 acc: 0.9820\n"
],
[
"def random_normal_func(X):\n X=X.reshape(-1,28*28)\n print(X.shape)\n mean, std = np.mean(X, axis=0), np.std(X,axis=0)\n randomX = np.zeros([10000,X[0].size])\n print(randomX.shape)\n for i in range(X[0].size):\n randomX[:,i] = np.random.normal(mean[i],std[i],10000)\n randomX = randomX.reshape(-1,28,28,1)\n ans = sess.run(env.ybar, feed_dict={env.x: randomX,env.training: False})\n labels = _to_categorical(np.argmax(ans,axis=1), n_classes)\n return randomX,labels\n ",
"_____no_output_____"
],
[
"test = \"test_fs_exp1_0\"\ntrain = \"train_fs_exp1_0\"\nrandom = \"random_fs_exp1_0\"\nrandom_normal= \"random_normal_fs_exp1_0\"\n\n\nX_train_sub = X_train[:10000]\ny_train_sub = sess.run(env.ybar, feed_dict={env.x: X_train_sub,env.training: False})\ny_train_sub = _to_categorical(np.argmax(y_train_sub, axis=1), n_classes)\n\ny_test_sub = sess.run(env.ybar, feed_dict={env.x: X_test,env.training: False})\ny_test_sub = _to_categorical(np.argmax(y_test_sub, axis=1), n_classes)\n\nX_random = np.random.rand(10000,28,28,1)\nX_random = X_random[:10000]\ny_random = sess.run(env.ybar, feed_dict={env.x: X_random,env.training: False})\ny_random = _to_categorical(np.argmax(y_random, axis=1), n_classes)\nX_random_normal, y_random_normal = random_normal_func(X_train)\n\nX_adv_test = create_adv(X_test, y_test_sub, test)\nX_adv_train = create_adv(X_train_sub, y_train_sub, train)\nX_adv_random = create_adv(X_random,y_random, random)\nX_adv_random_normal = create_adv(X_random_normal, y_random_normal, random_normal)\n\n# X_adv_test = np.load('data/mnist/' + test + '.npy')\n# X_adv_train = np.load('data/mnist/' + train + '.npy')\n# X_adv_random = np.load('data/mnist/' + random + '.npy')\n# X_adv_random_normal = np.load('data/mnist/' + random_normal + '.npy')",
"(54000, 784)\n(10000, 784)\n\nCrafting adversarial\n batch 10000/10000\nSaving adversarial\n\nCrafting adversarial\n batch 10000/10000\nSaving adversarial\n\nCrafting adversarial\n batch 10000/10000\nSaving adversarial\n\nCrafting adversarial\n batch 10000/10000\nSaving adversarial\n"
],
[
"l2_test = find_l2(X_adv_test,X_test)\nl2_train = find_l2(X_adv_train, X_train_sub)\nl2_random = find_l2(X_adv_random,X_random)\nl2_random_normal = find_l2(X_adv_random_normal,X_random_normal)",
"_____no_output_____"
],
[
"print(l2_train)",
"[ 0.82968032 0.88142085 4.80068111 ..., 5.88712502 4.3569665\n 2.70526552]\n"
],
[
"print(X_adv_random_normal[0][3])",
"[[ 0.00000000e+00]\n [ 0.00000000e+00]\n [ 0.00000000e+00]\n [ 2.45368428e-04]\n [ 3.59936239e-04]\n [ 9.65054613e-03]\n [ 1.36153949e-02]\n [ 2.59003509e-02]\n [ 1.33717284e-02]\n [ 0.00000000e+00]\n [ 1.27349779e-01]\n [ 1.80915192e-01]\n [ 1.29013415e-02]\n [ 5.22749424e-02]\n [ 7.28076976e-03]\n [ 2.12018564e-02]\n [ 2.54217625e-01]\n [ 1.93438619e-01]\n [ 3.24693695e-02]\n [ 1.60969615e-01]\n [ 1.64130822e-01]\n [ 1.82762295e-02]\n [ 2.11828023e-01]\n [ 6.01061471e-02]\n [ 2.00864542e-02]\n [ 8.41681287e-03]\n [ 7.61829130e-03]\n [ 1.70961842e-02]]\n"
],
[
"%matplotlib inline\n# evenly sampled time at 200ms intervals\nt = np.arange(1,10001, 1)\n\n# red dashes, blue squares and green triangles\nplt.plot(t, l2_test, 'r--', t, l2_train, 'b--', t, l2_random, 'y--', l2_random_normal, 'g--')\nplt.show()",
"_____no_output_____"
],
[
"import matplotlib.patches as mpatches\n\n%matplotlib inline\n# evenly sampled time at 200ms intervals\nt = np.arange(1,101, 1)\n\n# red dashes, blue squares and green triangles\nplt.plot(t, l2_test[:100], 'r--', t, l2_train[:100], 'b--',t, l2_random[:100], 'y--',l2_random_normal[:100], 'g--')\nblue_patch = mpatches.Patch(color='blue', label='Train Data')\nplt.legend(handles=[blue_patch])\n\nplt.show()",
"_____no_output_____"
],
[
"%matplotlib inline\nplt.hist(l2_test,100)\nplt.title(\"L2 distance of test data\")\nplt.xlabel(\"Distance\")\nplt.ylabel(\"Frequency\")\nplt.show()",
"_____no_output_____"
],
[
"%matplotlib inline\nplt.hist(l2_train,100)\nplt.title(\"L2 distance of train data\")\nplt.xlabel(\"Distance\")\nplt.ylabel(\"Frequency\")\nplt.show()",
"_____no_output_____"
],
[
"%matplotlib inline\nplt.hist(l2_random,100)\nplt.title(\"L2 distance of random data\")\nplt.xlabel(\"Distance\")\nplt.ylabel(\"Frequency\")\nplt.show()",
"_____no_output_____"
],
[
"%matplotlib inline\nplt.hist(l2_random_normal,100)\nplt.title(\"L2 distance of random normal data\")\nplt.xlabel(\"Distance\")\nplt.ylabel(\"Frequency\")\nplt.show()",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb8dff368bf75a46846d215927645adec088f500
| 52,486 |
ipynb
|
Jupyter Notebook
|
Lab 1/ATOC5860_applicationlab1_bootstrapping.ipynb
|
wk2500/ATOC5860_Spring2022
|
b108fcaaedddc9cce50ce4c00f133af32a5a0324
|
[
"MIT"
] | null | null | null |
Lab 1/ATOC5860_applicationlab1_bootstrapping.ipynb
|
wk2500/ATOC5860_Spring2022
|
b108fcaaedddc9cce50ce4c00f133af32a5a0324
|
[
"MIT"
] | null | null | null |
Lab 1/ATOC5860_applicationlab1_bootstrapping.ipynb
|
wk2500/ATOC5860_Spring2022
|
b108fcaaedddc9cce50ce4c00f133af32a5a0324
|
[
"MIT"
] | null | null | null | 68.252276 | 14,252 | 0.769024 |
[
[
[
"###### Applications Lab #1-- ATOC7500 Objective Analysis - bootstrapping\n##### Originally coded by Prof. Kay (CU) with input from Vineel Yettella (CU ATOC Ph.D. 2018)\n##### last updated September 2, 2020\n\n###LEARNING GOALS:\n###1) Working in an ipython notebook: read in csv file, make histogram plot\n###2) Assessing statistical significance using bootstrapping (and t-test)\n\n### GENERAL SETUP\n%matplotlib inline \n# this enables plotting within notebook\n\nimport matplotlib # library for plotting\nimport matplotlib.pyplot as plt # later you will type plt.$COMMAND\nimport numpy as np # basic math library you will type np.$STUFF e.g., np.cos(1)\nimport pandas as pd # library for data analysis for text files (everything but netcdf files)\nimport scipy.stats as stats # imports stats functions https://docs.scipy.org/doc/scipy/reference/stats.html ",
"_____no_output_____"
],
[
"### Read in the data\nfilename='snow_enso_data.csv'\ndata=pd.read_csv(filename,sep=',')\ndata.head()",
"_____no_output_____"
],
[
"### Print the data column names\nprint(data.columns[0])\nprint(data.columns[1])\nprint(data.columns[2])",
"Year\nLovelandPass_April1SWE_inches\nNino34_anomaly_prevDec\n"
],
[
"### Print the data values - LOOK AT YOUR DATA. If new to Python - check out what happens when you remove .values.\nprint(data['Year'].values)\nprint(data['LovelandPass_April1SWE_inches'].values)\nprint(data['Nino34_anomaly_prevDec'].values)",
"[1936 1937 1938 1939 1940 1941 1942 1943 1944 1945 1946 1947 1948 1949\n 1950 1951 1952 1953 1954 1955 1956 1957 1958 1959 1960 1961 1962 1963\n 1964 1965 1966 1967 1968 1969 1970 1971 1972 1973 1974 1975 1976 1977\n 1978 1979 1980 1981 1982 1983 1984 1985 1986 1987 1988 1989 1990 1991\n 1992 1993 1994 1995 1996 1997 1998 1999 2000 2001 2002 2003 2004 2005\n 2006 2007 2008 2009 2010 2011 2012 2013 2014 2015 2016]\n[15.8 10.1 12.1 18.2 10. 9.9 13.9 17.9 11.4 13.3 11.4 17.4 14. 14.\n 14. 19. 24. 17.3 11.2 13.5 21.2 17.5 14.9 18.7 16.4 15.3 19.2 12.\n 12.7 19.9 6.3 14.6 16.1 14.2 20.7 19. 16.2 12. 18.8 19.6 15.1 11.\n 22.1 16.7 20. 9. 19.8 15.5 18. 12.3 18.2 11.3 15.9 12.1 13.8 13.2\n 14.2 21.8 17.8 17.2 28.7 22.6 17.2 16.5 19.6 17.2 10. 19.9 13.1 15.6\n 22.4 19.7 21.2 21.6 14.3 27.3 12.8 14.9 24.4 16.4 19. ]\n[-0.04 0.63 0.22 -0.79 0.05 1.39 1.27 -1.24 -0.42 -0.26 -0.54 0.03\n 0.18 0.38 -1.2 -1.04 0.71 -0.55 0.15 -0.79 -1.52 -0.58 1.18 0.3\n -0.05 0.02 -0.29 -0.49 1.05 -1.07 1.45 -0.26 -0.35 0.76 0.68 -1.11\n -0.89 2.19 -2.18 -0.86 -1.64 0.64 1.09 0.07 0.69 0.36 0.07 2.33\n -0.95 -1.53 -0.4 0.98 1.05 -1.98 -0.13 0.32 1.62 0.04 0.19 1.21\n -0.72 -0.43 2.3 -1.51 -1.54 -0.92 -0.46 1.41 0.32 0.71 -0.75 1.1\n -1.61 -0.9 1.81 -1.63 -1.05 -0.13 -0.09 0.77 2.56]\n"
],
[
"### Calculate the average snowfall on April 1 at Loveland Pass, Colorado\nSWE_avg=data['LovelandPass_April1SWE_inches'].mean()\nSWE_std=data['LovelandPass_April1SWE_inches'].std()\nN_SWE=len(data.LovelandPass_April1SWE_inches)\nprint('Average SWE (inches):',np.str(np.round(SWE_avg,2)))\nprint('Standard Deviation SWE (inches):',np.str(np.round(SWE_std,2)))\nprint('N:',np.str(N_SWE))",
"Average SWE (inches): 16.33\nStandard Deviation SWE (inches): 4.22\nN: 81\n"
],
[
"### Print to figure out how to condition and make sure it is working. Check out if new to Python.\n#print(data.Nino34_anomaly_prevDec>1) ## this gives True/False\n#print(data[data.Nino34_anomaly_prevDec>1]) ## where it is True, values will print\n\n### Calculate the average SWE when it was an el nino year\nSWE_avg_nino=data[data.Nino34_anomaly_prevDec>1.0]['LovelandPass_April1SWE_inches'].mean()\nSWE_std_nino=data[data.Nino34_anomaly_prevDec>1.0]['LovelandPass_April1SWE_inches'].std()\nN_SWE_nino=len(data[data.Nino34_anomaly_prevDec>1.0].LovelandPass_April1SWE_inches)\nprint('Average SWE El Nino (inches):',np.str(np.round(SWE_avg_nino,2)))\nprint('Standard Deviation SWE El Nino (inches):',np.str(np.round(SWE_std_nino,2)))\nprint('N El Nino:',np.str(N_SWE_nino))",
"Average SWE El Nino (inches): 15.29\nStandard Deviation SWE El Nino (inches): 4.0\nN El Nino: 16\n"
],
[
"### Calculate the average SWE when it was an la nina year\nSWE_avg_nina=data[data.Nino34_anomaly_prevDec<-1.0]['LovelandPass_April1SWE_inches'].mean()\nSWE_std_nina=data[data.Nino34_anomaly_prevDec<-1.0]['LovelandPass_April1SWE_inches'].std()\nN_SWE_nina=len(data[data.Nino34_anomaly_prevDec<-1.0].LovelandPass_April1SWE_inches)\nprint('Average SWE La Nina (inches):',np.str(np.round(SWE_avg_nina,2)))\nprint('Standard Deviation SWE La Nina (inches):',np.str(np.round(SWE_std_nina,2)))\nprint('N La Nina:',np.str(N_SWE_nina))",
"Average SWE La Nina (inches): 17.78\nStandard Deviation SWE La Nina (inches): 4.11\nN La Nina: 15\n"
],
[
"### Bootstrap!! Generate random samples of size N_SWE_nino and N_SWE_nina. Do it once to see if it works.\nP_random=np.random.choice(data.LovelandPass_April1SWE_inches,N_SWE_nino)\nprint(P_random) ## LOOK AT YOUR DATA\nprint(len(P_random))",
"[20.7 15.6 22.6 17.3 14. 12. 14.2 13.2 13.3 27.3 19. 11.2 16.1 14.\n 24. 13.2]\n16\n"
],
[
"### Now Bootstrap Nbs times to generate a distribution of randomly selected mean SWE.\nNbs=1000\n## initialize array\nP_Bootstrap=np.empty((Nbs,N_SWE_nino))\n\nprint(P_Bootstrap.shape)",
"(1000000, 16)\n"
],
[
"### Now Bootstrap Nbs times to generate a distribution of randomly selected mean SWE.\nNbs=100000\n## initialize array\nP_Bootstrap=np.empty((Nbs,N_SWE_nino))\n## loop over to fill in array with randomly selected values\nfor ii in range(Nbs):\n P_Bootstrap[ii,:]=np.random.choice(data.LovelandPass_April1SWE_inches,N_SWE_nino)\n\n## Calculate the means of your randomly selected SWE values.\nP_Bootstrap_mean=np.mean(P_Bootstrap,axis=1)\nprint(len(P_Bootstrap_mean)) ## check length to see if you averaged across the correct axis\nprint(np.shape(P_Bootstrap_mean)) ## another option to look at the dimensions of a variable\n#print(P_Bootstrap_mean)\n\nP_Bootstrap_mean_avg=np.mean(P_Bootstrap_mean)\nprint(P_Bootstrap_mean_avg)\nP_Bootstrap_mean_std=np.std(P_Bootstrap_mean)\nprint(P_Bootstrap_mean_std)\nP_Bootstrap_mean_min=np.min(P_Bootstrap_mean)\nprint(P_Bootstrap_mean_min)\nP_Bootstrap_mean_max=np.max(P_Bootstrap_mean)\nprint(P_Bootstrap_mean_max)",
"100000\n(100000,)\n16.338942624999998\n1.0451323563408652\n11.6\n21.35\n"
],
[
"### Use matplotlib to plot a histogram of the bootstrapped means to compare to the conditioned SWE mean\nbinsize=0.1\nmin4hist=np.round(np.min(P_Bootstrap_mean),1)-binsize\nmax4hist=np.round(np.max(P_Bootstrap_mean),1)+binsize\nnbins=int((max4hist-min4hist)/binsize)\n\nplt.hist(P_Bootstrap_mean,nbins,edgecolor='black')\nplt.xlabel('Mean SWE (inches)');\nplt.ylabel('Count');\nplt.title('Bootstrapped Randomly Selected Mean SWE Values');",
"_____no_output_____"
],
[
"## What is the probability that the snowfall was lower during El Nino by chance?\n## Using Barnes equation (83) on page 15 to calculate probability using z-statistic\nsample_mean=SWE_avg_nino\nsample_N=1\npopulation_mean=np.mean(P_Bootstrap_mean)\npopulation_std=np.std(P_Bootstrap_mean)\nxstd=population_std/np.sqrt(sample_N)\nz_nino=(sample_mean-population_mean)/xstd\nprint(\"sample_mean - El Nino: \",np.str(np.round(sample_mean,2))) ############\nprint(\"population_mean: \",np.str(np.round(population_mean,2)))\nprint(\"population_std: \",np.str(np.round(population_std,2)))\nprint(\"Z-statistic (number of standard errors that the sample mean deviates from the population mean:\")\nprint(np.round(z_nino,2))\nprob=(1-stats.norm.cdf(np.abs(z_nino)))*100 ##this is a one-sided test\nprint(\"Probability one-tailed test (percent):\")\nprint(np.round(prob,2)) ",
"sample_mean - El Nino: 15.29\npopulation_mean: 16.34\npopulation_std: 1.05\nZ-statistic (number of standard errors that the sample mean deviates from the population mean:\n-1.0\nProbability one-tailed test (percent):\n15.86\n"
],
[
"## What is the probability that the snowfall that the El Nino mean differs from the mean by chance?\n## Using Barnes equation (83) on page 15 to calculate probability using z-statistic\nsample_mean=SWE_avg_nino\nsample_N=1\npopulation_mean=np.mean(P_Bootstrap_mean)\npopulation_std=np.std(P_Bootstrap_mean)\nxstd=population_std/np.sqrt(sample_N)\nz_nino=(sample_mean-population_mean)/xstd\nprint(\"sample_mean - El Nino: \",np.str(np.round(sample_mean,2)))\nprint(\"population_mean: \",np.str(np.round(population_mean,2)))\nprint(\"population_std: \",np.str(np.round(population_std,2)))\nprint(\"Z-statistic (number of standard errors that the sample mean deviates from the population mean):\")\nprint(np.round(z_nino,2))\nprob=(1-stats.norm.cdf(np.abs(z_nino)))*2*100 ##this is a two-sided test\nprint(\"Probability - two-tailed test (percent):\")\nprint(np.round(prob,2)) ",
"sample_mean - El Nino: 15.29\npopulation_mean: 16.34\npopulation_std: 1.05\nZ-statistic (number of standard errors that the sample mean deviates from the population mean):\n-1.0\nProbability - two-tailed test (percent):\n31.73\n"
],
[
"## What is the probability that the snowfall was higher during La Nina just due to chance?\n## Using Barnes equation (83) on page 15 to calculate probability using z-statistic\nsample_mean=SWE_avg_nina\nsample_N=1\npopulation_mean=np.mean(P_Bootstrap_mean)\npopulation_std=np.std(P_Bootstrap_mean)\nxstd=population_std/np.sqrt(sample_N)\nz_nina=(sample_mean-population_mean)/xstd\n\nprint(\"sample_mean - La Nina: \",np.str(np.round(sample_mean,2)))\nprint(\"population_mean: \",np.str(np.round(population_mean,2)))\nprint(\"population_std: \",np.str(np.round(population_std,2)))\nprint(\"Z-statistic (number of standard errors that the sample mean deviates from the population mean:\")\nprint(np.round(z_nina,2))\nprob=(1-stats.norm.cdf(np.abs(z_nina)))*100 ##this is a one-sided test\nprint(\"Probability one-tailed test (percent):\")\nprint(np.round(prob,2)) ",
"sample_mean - La Nina: 17.78\npopulation_mean: 16.34\npopulation_std: 1.05\nZ-statistic (number of standard errors that the sample mean deviates from the population mean:\n1.38\nProbability one-tailed test (percent):\n8.4\n"
],
[
"## What is the probability that the snowfall during La Nina differed just due to chance?\n## Using Barnes equation (83) on page 15 to calculate probability using z-statistic\nsample_mean=SWE_avg_nina\nsample_N=1\npopulation_mean=np.mean(P_Bootstrap_mean)\npopulation_std=np.std(P_Bootstrap_mean)\nxstd=population_std/np.sqrt(sample_N)\nz_nina=(sample_mean-population_mean)/xstd\n\nprint(\"sample_mean - La Nina: \",np.str(np.round(sample_mean,2)))\nprint(\"population_mean: \",np.str(np.round(population_mean,2)))\nprint(\"population_std: \",np.str(np.round(population_std,2)))\nprint(\"Z-statistic (number of standard errors that the sample mean deviates from the population mean):\")\nprint(np.round(z_nina,2))\nprob=(1-stats.norm.cdf(np.abs(z_nina)))*2*100 ##this is a two-sided test\nprint(\"Probability - two-tailed test (percent):\")\nprint(np.round(prob,2)) ",
"sample_mean - La Nina: 17.78\npopulation_mean: 16.34\npopulation_std: 1.05\nZ-statistic (number of standard errors that the sample mean deviates from the population mean):\n1.38\nProbability - two-tailed test (percent):\n16.79\n"
],
[
"### Strategy #2: Forget bootstrapping, let's use a t-test...\n## Apply a t-test to test the null hypothesis that the means of the two samples \n## are the same at the 95% confidence level (alpha=0.025, two-sided test)\n## If pvalue < alpha - reject null hypothesis.\nprint('Null Hypothesis: ENSO snow years have the same mean as the full record.')\nt=stats.ttest_ind(data[data.Nino34_anomaly_prevDec>1.0]['LovelandPass_April1SWE_inches'],data['LovelandPass_April1SWE_inches'],equal_var=False)\nprint(t)\nprint('Cannot reject the null hypthesis.')\n\n#### Wait a second - What is that function doing??? Let's check it with the Barnes notes.",
"Null Hypothesis: ENSO snow years have the same mean as the full record.\nTtest_indResult(statistic=-0.9419860439508277, pvalue=0.35637575995310133)\nCannot reject the null hypthesis.\n"
],
[
"### Always code it yourself and understand what the function is doing. \n### Word to the wise - do not use python functions without checking them!!\n### Let's find out what stats.ttest_ind is doing - It doesn't look like it is calculating the t-statistic\n### as the difference between the sample mean and the population mean. That calculation is below...\n\n## Calculate the t-statistic using the Barnes Notes - Compare a sample mean and a population mean.\n## Barnes Eq. (96)\nN=len(data[data.Nino34_anomaly_prevDec>1.0]['LovelandPass_April1SWE_inches'])\nprint(N)\nsample_mean=np.mean(data[data.Nino34_anomaly_prevDec>1.0]['LovelandPass_April1SWE_inches'])\nprint(sample_mean)\nsample_std=np.std(data[data.Nino34_anomaly_prevDec>1.0]['LovelandPass_April1SWE_inches'])\nprint(sample_std)\npopulation_mean=np.mean(data['LovelandPass_April1SWE_inches'])\n\n## Using Barnes equation (96) to calculate probability using the t-statistic\nprint(\"T-statistic:\")\nt=(sample_mean-population_mean)/(sample_std/(np.sqrt(N-1)))\nprint(np.round(t,2))\nprint(\"Probability (percent):\")\nprob=(1-stats.t.cdf(t,N-1))*100\nprint(np.round(prob,2))",
"16\n15.29375\n3.8746723651813446\nT-statistic:\n-1.04\nProbability (percent):\n84.27\n"
],
[
"## Calculate the t-statistic using the Barnes Notes - Compare two sample means. Equation (110)\n## This is also called Welch's t-test\n## It doesn't look like the function is calculating the t-statistic using Welch's t-test!\n## as the difference between the sample mean and the population mean. That calculation is below...\n## Guess using the two sample means test (i.e., Eq. 100) vs sample/population means test (i.e., Barnes Eq. )\n\nsampledata1=data['LovelandPass_April1SWE_inches']\nsampledata2=data[data.Nino34_anomaly_prevDec>1.0]['LovelandPass_April1SWE_inches']\n\nN1=len(sampledata1)\nN2=len(sampledata2)\nprint(N1)\nprint(N2)\nsample_mean1=np.mean(sampledata1)\nsample_mean2=np.mean(sampledata2)\nprint(sample_mean1)\nprint(sample_mean2)\nsample_std1=np.std(sampledata1)\nsample_std2=np.std(sampledata2)\nprint(sample_std1)\nprint(sample_std2)\n\n## Using Barnes equation (96) to calculate probability using the t-statistic\nprint(\"T-statistic using Welch's t-test:\")\ns=np.sqrt((N1*sample_std1**2+N2*sample_std2**2)/(N1+N2-2))\nprint(s)\n#t=(sample_mean1-sample_mean2-0)/(s*np.sqrt(1/N1+1/N2))\nprint(np.round(t,2))\nprint(\"Probability (percent):\")\nprob=(1-stats.t.cdf(t,N-1))*100\nprint(np.round(prob,2))",
"81\n16\n16.33456790123457\n15.29375\n4.194975256885701\n3.8746723651813446\nT-statistic using Welch's t-test:\n4.1872394937055875\n-1.04\nProbability (percent):\n84.27\n"
],
[
"### Strategy #3 (provided by Vineel Yettella)\nSWE = data['LovelandPass_April1SWE_inches']\nSWE_nino = data[data.Nino34_anomaly_prevDec>1.0]['LovelandPass_April1SWE_inches']\n\n#We start by setting up a null hypothesis H0. \n#Our H0 will be that the difference in means of the two populations that the samples came from is equal to zero.\n#We will use the bootstrap to test this null hypothesis.\n\n#We next choose a significance level for the hypothesis test\nalpha = 0.05\n\n#All hypothesis tests need a test statistic.\n#Here, we'll use the difference in sample means as the test statistic.\n#create array to hold bootstrapped test statistic values\nbootstrap_statistic = np.empty(10000)\n\n#bootstrap 10000 times\nfor i in range(1,10000):\n \n #create a resample of SWE by sampling with replacement (same length as SWE)\n resample_original = np.random.choice(SWE, len(SWE), replace=True)\n \n #create a resample of SWE_nino by sampling with replacement (same length as SWE_nino)\n resample_nino = np.random.choice(SWE_nino, len(SWE_nino), replace=True)\n \n #Compute the test statistic from the resampled data, i.e., the difference in means\n bootstrap_statistic[i] = np.mean(resample_original) - np.mean(resample_nino)\n\n#Let's plot the distribution of the test statistic\nplt.hist(bootstrap_statistic,[-5,-4,-3,-2,-1,0,1,2,3,4,5],edgecolor='black')\nplt.xlabel('Difference in sample means')\nplt.ylabel('Count')\nplt.title('Bootstrap distribution of difference in sample means')\n\n#Create 95% CI from the bootstrapped distribution. The upper limit of the CI is defined as the 97.5% percentile\n#and the lower limit as the 2.5% percentile of the boostrap distribution, so that 95% of the \n#distribution lies within the two limits\n\nCI_up = np.percentile(bootstrap_statistic, 100*(1 - alpha/2.0))\nCI_lo = np.percentile(bootstrap_statistic, 100*(alpha/2.0))\n\nprint(CI_up)\nprint(CI_lo)\n\n#We see that the confidence interval contains zero, so we fail to reject the null hypothesis that the difference\n#in means is equal to zero",
"3.1893614969135817\n-0.9720814043209867\n"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb8e1cc47e1deef8e918173209768c02e9ab4638
| 27,083 |
ipynb
|
Jupyter Notebook
|
section2/feature_selection.ipynb
|
Jeffresh/deployment-of-machine-learning-models
|
bc4a5ff5713601a1a4b2c73292b1f276f22bf258
|
[
"MIT"
] | null | null | null |
section2/feature_selection.ipynb
|
Jeffresh/deployment-of-machine-learning-models
|
bc4a5ff5713601a1a4b2c73292b1f276f22bf258
|
[
"MIT"
] | null | null | null |
section2/feature_selection.ipynb
|
Jeffresh/deployment-of-machine-learning-models
|
bc4a5ff5713601a1a4b2c73292b1f276f22bf258
|
[
"MIT"
] | 1 |
2021-02-10T13:03:04.000Z
|
2021-02-10T13:03:04.000Z
| 78.501449 | 12,218 | 0.457889 |
[
[
[
"# Feature selection\n\nWe will select a group of variables, the most predictive ones, to build our machine learning model\n\n## Why do we select variables?\n- For production: Fewer variables mean smaller client input requeriments(e.q customers filling out a form on a webiste or mobile app), and hence less code for error handling. This reduces the chances of introducing bugs.\n\n- For model performance: Fewer variables mean simpler, more interpretable, better generalizing models.\n\nWe will select variables using the Lasso regression: Lasso has the property of setting the coefficient of non-informative varbiales to zero. This way we can identify those variables and remove then from our final model.",
"_____no_output_____"
],
[
"## Imports",
"_____no_output_____"
]
],
[
[
"import pandas as pd \nimport numpy as np\n\n# for plotting\nimport matplotlib.pyplot as plt\n\n# to build the models\nfrom sklearn.linear_model import Lasso \nfrom sklearn.feature_selection import SelectFromModel\n\n# to visualise all the columns in the dataframe\npd.pandas.set_option('display.max_columns', None)\n",
"_____no_output_____"
],
[
"# load the train and test set with the engineered variables\n\nX_train = pd.read_csv('xtrain.csv')\nX_test = pd.read_csv('xtest.csv')\n\nX_train.head()",
"_____no_output_____"
],
[
"# Capture the target (remember that the target is log transformed)\ny_train = X_train['SalePrice']\ny_test = X_test['SalePrice']\n\n# drop unnecessary variables from our training and testing sets\n\nX_train.drop(['Id', 'SalePrice'], axis=1, inplace=True)\nX_test.drop(['Id', 'SalePrice'], axis=1, inplace=True)",
"_____no_output_____"
]
],
[
[
"## Feature Selection\n\nSelect a subsset of the most predictive features. There is an element of randomness in the Lasso regression, ro remembmer to set the seed.",
"_____no_output_____"
]
],
[
[
"# We will do the model fititng and feature selection altogether in a few lines of code\n\n# first, we specigy the Lasso Regression mode, and we slect a suitable alpha (equivalent penalty).\n# The bigger the alpha the less features that will be selected.\n\n# Then ewe use the selectFromModel object from sklearn, which will select automatically the features which coefficients are non-zero\n\n# remember to set the seed, the random state in this function\n\nsel_ = SelectFromModel(Lasso(alpha=0.005, random_state=0))\n\nsel_.fit(X_train, y_train)",
"_____no_output_____"
],
[
"# let's visualize those features that were selected.\n# (selected features marked with True)\n\nsel_.get_support()",
"_____no_output_____"
],
[
"# let's print the number of total and selected features\n\n# this is how we ca nmake a list of the selected features\n\nselected_feats = X_train.columns[sel_.get_support()]\n\n# let's print some stats\nprint('total features: {}'.format(X_train.shape[1]))\nprint('selected features: {}'.format(len(selected_feats)))\nprint('features with coefficients sharnk to zero: {}'.format(np.sum(sel_.estimator_.coef_ == 0)))",
"total features: 82\nselected features: 22\nfeatures with coefficients sharnk to zero: 60\n"
],
[
"## Identify the selected variables\n\n# this is an alternative way of identifying the selected features\n# based on the non-zero regularisation coefficients:\n\nselected_feats = X_train.columns[(sel_.estimator_.coef_ != 0).ravel().tolist()]\nselected_feats",
"_____no_output_____"
],
[
"pd.Series(selected_feats).to_csv('selected_features.csv', index=False)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
cb8e27f80aa9f0695ed592a6f1e78e4d4fa2ac3c
| 13,976 |
ipynb
|
Jupyter Notebook
|
notebook/Twitter-NER.ipynb
|
bobflagg/deepER
|
2a574fe3d0689f41e82d1e891de92fa2aabaa049
|
[
"Apache-2.0"
] | null | null | null |
notebook/Twitter-NER.ipynb
|
bobflagg/deepER
|
2a574fe3d0689f41e82d1e891de92fa2aabaa049
|
[
"Apache-2.0"
] | null | null | null |
notebook/Twitter-NER.ipynb
|
bobflagg/deepER
|
2a574fe3d0689f41e82d1e891de92fa2aabaa049
|
[
"Apache-2.0"
] | null | null | null | 35.382278 | 716 | 0.515813 |
[
[
[
"# A Baseline Named Entity Recognizer for Twitter\n\nIn this notebook I'll follow the example presented in [Named entities and random fields](http://www.orbifold.net/default/2017/06/29/dutch-ner/) to train a conditional random field to recognize named entities in Twitter data. The data and some of the code below are taken from a programming assignment in the amazing class [Natural Language Processing](https://www.coursera.org/learn/language-processing) offered by [Coursera](https://www.coursera.org/). In the assignment we were shown how to build a named entity recognizer using deep learning with a bidirectional LSTM, which is a pretty complicated approach and I wanted to have a baseline model to see what sort of accuracy should be expected on this data.",
"_____no_output_____"
],
[
"### 1. Preparing the Data\n\nFirst load the text and tags for training, validation and test data:",
"_____no_output_____"
]
],
[
[
"def read_data(file_path):\n tokens = []\n tags = []\n \n tweet_tokens = []\n tweet_tags = []\n for line in open(file_path, encoding='utf-8'):\n line = line.strip()\n if not line:\n if tweet_tokens:\n tokens.append(tweet_tokens)\n tags.append(tweet_tags)\n tweet_tokens = []\n tweet_tags = []\n else:\n token, tag = line.split()\n # Replace all urls with <URL> token\n # Replace all users with <USR> token\n if token.startswith(\"http://\") or token.startswith(\"https://\"): token = \"<URL>\"\n elif token.startswith(\"@\"): token = \"<USR>\"\n tweet_tokens.append(token)\n tweet_tags.append(tag) \n return tokens, tags\ntrain_tokens, train_tags = read_data('data/train.txt')\nvalidation_tokens, validation_tags = read_data('data/validation.txt')\ntest_tokens, test_tags = read_data('data/test.txt')",
"_____no_output_____"
]
],
[
[
"The CRF model uses part of speech tags as features so we'll need to add those to the datasets.",
"_____no_output_____"
]
],
[
[
"%%time\nimport nltk\n\ndef build_sentence(tokens, tags):\n pos_tags = [item[-1] for item in nltk.pos_tag(tokens)]\n return list(zip(tokens, pos_tags, tags))\n\ndef build_sentences(tokens_set, tags_set):\n return [build_sentence(tokens, tags) for tokens, tags in zip(tokens_set, tags_set)]\n\ntrain_sents = build_sentences(train_tokens, train_tags)\nvalidation_sents = build_sentences(validation_tokens, validation_tags)\ntest_sents = build_sentences(test_tokens, test_tags)",
"CPU times: user 7.06 s, sys: 192 ms, total: 7.26 s\nWall time: 7.26 s\n"
]
],
[
[
"### 2. Computing Features",
"_____no_output_____"
]
],
[
[
"\ndef word2features(sent, i):\n word = sent[i][0]\n postag = sent[i][1]\n\n features = {\n 'bias': 1.0,\n 'word.lower()': word.lower(),\n 'word[-3:]': word[-3:],\n 'word[-2:]': word[-2:],\n 'word.isupper()': word.isupper(),\n 'word.istitle()': word.istitle(),\n 'word.isdigit()': word.isdigit(),\n 'postag': postag,\n 'postag[:2]': postag[:2],\n }\n if i > 0:\n word1 = sent[i - 1][0]\n postag1 = sent[i - 1][1]\n features.update({\n '-1:word.lower()': word1.lower(),\n '-1:word.istitle()': word1.istitle(),\n '-1:word.isupper()': word1.isupper(),\n '-1:postag': postag1,\n '-1:postag[:2]': postag1[:2],\n })\n else:\n features['BOS'] = True\n\n if i < len(sent) - 1:\n word1 = sent[i + 1][0]\n postag1 = sent[i + 1][1]\n features.update({\n '+1:word.lower()': word1.lower(),\n '+1:word.istitle()': word1.istitle(),\n '+1:word.isupper()': word1.isupper(),\n '+1:postag': postag1,\n '+1:postag[:2]': postag1[:2],\n })\n else:\n features['EOS'] = True\n\n return features\n\ndef sent2features(sent):\n return [word2features(sent, i) for i in range(len(sent))]\n\n\ndef sent2labels(sent):\n return [label for token, postag, label in sent]\n\n\ndef sent2tokens(sent):\n return [token for token, postag, label in sent]\n\n\nX_train = [sent2features(s) for s in train_sents]\ny_train = [sent2labels(s) for s in train_sents]\n\nX_validation = [sent2features(s) for s in validation_sents]\ny_validation = [sent2labels(s) for s in validation_sents]\n\nX_test = [sent2features(s) for s in test_sents]\ny_test = [sent2labels(s) for s in test_sents]",
"_____no_output_____"
]
],
[
[
"### 3. Train the Model",
"_____no_output_____"
]
],
[
[
"import sklearn_crfsuite",
"_____no_output_____"
],
[
"crf = sklearn_crfsuite.CRF(\n algorithm='lbfgs',\n c1=0.12,\n c2=0.01,\n max_iterations=100,\n all_possible_transitions=True\n)\ncrf.fit(X_train, y_train)",
"_____no_output_____"
]
],
[
[
"### 4. Evaluate the Model\n\nWe evaluate the model using the CoNLL shared task evaluation script.",
"_____no_output_____"
]
],
[
[
"from evaluation import precision_recall_f1\n\ndef eval_conll(model, tokens, tags, short_report=True):\n \"\"\"Computes NER quality measures using CONLL shared task script.\"\"\"\n tags_pred = model.predict(tokens)\n y_true = [y for s in tags for y in s] \n y_pred = [y for s in tags_pred for y in s] \n results = precision_recall_f1(y_true, y_pred, print_results=True, short_report=short_report)\n return results",
"_____no_output_____"
],
[
"print('-' * 20 + ' Train set quality: ' + '-' * 20)\ntrain_results = eval_conll(crf, X_train, y_train, short_report=False)\n\nprint('-' * 20 + ' Validation set quality: ' + '-' * 20)\nvalidation_results = eval_conll(crf, X_validation, y_validation, short_report=False)\n\nprint('-' * 20 + ' Test set quality: ' + '-' * 20)\ntest_results = eval_conll(crf, X_test, y_test, short_report=False)",
"-------------------- Train set quality: --------------------\nprocessed 99983 tokens with 4489 phrases; found: 4476 phrases; correct: 4433.\n\nprecision: 99.04%; recall: 98.75%; F1: 98.90\n\n\t company: precision: 98.75%; recall: 98.13%; F1: 98.44; predicted: 639\n\n\t facility: precision: 97.76%; recall: 97.13%; F1: 97.44; predicted: 312\n\n\t geo-loc: precision: 99.20%; recall: 99.40%; F1: 99.30; predicted: 998\n\n\t movie: precision: 100.00%; recall: 100.00%; F1: 100.00; predicted: 68\n\n\t musicartist: precision: 97.85%; recall: 98.28%; F1: 98.06; predicted: 233\n\n\t other: precision: 98.94%; recall: 98.68%; F1: 98.81; predicted: 755\n\n\t person: precision: 99.32%; recall: 98.98%; F1: 99.15; predicted: 883\n\n\t product: precision: 99.68%; recall: 99.06%; F1: 99.37; predicted: 316\n\n\t sportsteam: precision: 100.00%; recall: 99.54%; F1: 99.77; predicted: 216\n\n\t tvshow: precision: 100.00%; recall: 96.55%; F1: 98.25; predicted: 56\n\n-------------------- Validation set quality: --------------------\nprocessed 12112 tokens with 537 phrases; found: 317 phrases; correct: 213.\n\nprecision: 67.19%; recall: 39.66%; F1: 49.88\n\n\t company: precision: 78.67%; recall: 56.73%; F1: 65.92; predicted: 75\n\n\t facility: precision: 76.92%; recall: 29.41%; F1: 42.55; predicted: 13\n\n\t geo-loc: precision: 76.25%; recall: 53.98%; F1: 63.21; predicted: 80\n\n\t movie: precision: 100.00%; recall: 14.29%; F1: 25.00; predicted: 1\n\n\t musicartist: precision: 55.56%; recall: 17.86%; F1: 27.03; predicted: 9\n\n\t other: precision: 52.17%; recall: 29.63%; F1: 37.80; predicted: 46\n\n\t person: precision: 67.12%; recall: 43.75%; F1: 52.97; predicted: 73\n\n\t product: precision: 14.29%; recall: 5.88%; F1: 8.33; predicted: 14\n\n\t sportsteam: precision: 33.33%; recall: 10.00%; F1: 15.38; predicted: 6\n\n\t tvshow: precision: 0.00%; recall: 0.00%; F1: 0.00; predicted: 0\n\n-------------------- Test set quality: --------------------\nprocessed 12534 tokens with 604 phrases; found: 383 phrases; correct: 273.\n\nprecision: 71.28%; recall: 45.20%; F1: 55.32\n\n\t company: precision: 85.71%; recall: 57.14%; F1: 68.57; predicted: 56\n\n\t facility: precision: 72.73%; recall: 51.06%; F1: 60.00; predicted: 33\n\n\t geo-loc: precision: 82.91%; recall: 58.79%; F1: 68.79; predicted: 117\n\n\t movie: precision: 0.00%; recall: 0.00%; F1: 0.00; predicted: 2\n\n\t musicartist: precision: 33.33%; recall: 7.41%; F1: 12.12; predicted: 6\n\n\t other: precision: 55.07%; recall: 36.89%; F1: 44.19; predicted: 69\n\n\t person: precision: 64.20%; recall: 50.00%; F1: 56.22; predicted: 81\n\n\t product: precision: 37.50%; recall: 10.71%; F1: 16.67; predicted: 8\n\n\t sportsteam: precision: 81.82%; recall: 29.03%; F1: 42.86; predicted: 11\n\n\t tvshow: precision: 0.00%; recall: 0.00%; F1: 0.00; predicted: 0\n\n"
]
],
[
[
"### 5. Tuning Parameters\n\nI tried tuning the parameters c1 and c2 of the model using [randomized grid search](http://scikit-learn.org/stable/modules/generated/sklearn.model_selection.RandomizedSearchCV.html) but was not able to improve the results that way. I plan to try [GPyOpt](https://github.com/SheffieldML/GPyOpt) to see if that will do better but don't have time to do that here.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
cb8e2ec3368402e228e7d2a880b693fcfaf7db47
| 7,773 |
ipynb
|
Jupyter Notebook
|
CryptoLab5.ipynb
|
raghavverma651/CryptoLabs
|
fe0ec7e9adb1d77966a7d42f8c540724fc6359b5
|
[
"BSD-3-Clause"
] | 2 |
2020-09-01T15:24:30.000Z
|
2020-09-10T04:36:41.000Z
|
CryptoLab5.ipynb
|
raghavverma651/SecurityLabs
|
fe0ec7e9adb1d77966a7d42f8c540724fc6359b5
|
[
"BSD-3-Clause"
] | null | null | null |
CryptoLab5.ipynb
|
raghavverma651/SecurityLabs
|
fe0ec7e9adb1d77966a7d42f8c540724fc6359b5
|
[
"BSD-3-Clause"
] | null | null | null | 22.272206 | 97 | 0.42995 |
[
[
[
"# Function for Mod Inverse",
"_____no_output_____"
]
],
[
[
"def findModInverse(a, m):\n if gcd(a, m) != 1:\n return None\n u1, u2, u3 = 1, 0, a\n v1, v2, v3 = 0, 1, m\n while v3 != 0:\n q = u3 // v3\n v1, v2, v3, u1, u2, u3 = (u1 - q * v1), (u2 - q * v2), (u3 - q * v3), v1, v2, v3\n return u1 % m",
"_____no_output_____"
]
],
[
[
"# RSA Algorithm",
"_____no_output_____"
]
],
[
[
"from random import choice \nlower=100\nupper=1000\nprimes=[]\niterr=0\nfor x in range(lower, upper + 1):\n \n if x > 1:\n for i in range(2, x):\n if (x % i) == 0:\n break\n else:\n primes.append(x)\np=choice(primes)\nq=choice(primes)\nprint(\"p taken -\",p)\nprint(\"q taken -\",q)\nn=p*q\nphi=(p-1)*(q-1)\nfrom math import gcd\ne=0\nfor i in range(2,phi):\n if(gcd(i,phi)==1):\n e=i\n break\nd=0\nfor i in range(1,10): \n x = 1 + i*phi \n if x % e == 0: \n d = int(x/e) \n break\nprint(\"Public key -\",(e,n))\nprint(\"Private key -\",(d,n))\nwhile(iterr!=2):\n cipher={chr(i+97):i+1 for i in range(26)}\n revcipher={i+1:chr(i+97) for i in range(26)}\n ch=int(input(\"\\n1 - Encrypt\\n2 - Decrypt\\nEnter choice : \"))\n if(ch==1):\n plain=input(\"\\nEnter text to encrypt - \")\n plain=[cipher[i] for i in plain]\n num=\"\"\n for i in plain:\n if(i>9):\n num+=str(i)\n else:\n num+=\"0\"+str(i)\n encrypted=[]\n for i in range(0,len(num),2):\n smt=((int(str(num[i]+num[i+1])))**e)%n\n encrypted.append(str(smt))\n print(' '.join(encrypted))\n elif(ch==2):\n enc=list(map(int,input(\"Enter text to decrypt (numeric and spaced)\").split()))\n decrypted=[]\n for i in enc:\n decrypted.append((i**d)%n)\n print(\"Decrypted message -\",''.join([revcipher[i] for i in decrypted]))\n iterr+=1",
"p taken - 101\nq taken - 359\nPublic key - (3, 36259)\nPrivate key - (23867, 36259)\n\n1 - Encrypt\n2 - Decrypt\nEnter choice : 1\n\nEnter text to encrypt - plain\n4096 1728 1 729 2744\n\n1 - Encrypt\n2 - Decrypt\nEnter choice : 2\nEnter text to decrypt (numeric and spaced)4096 1728 1 729 2744\nDecrypted message - plain\n"
],
[
"a=[1,2,3,4,5,6,7]\na=[str(i) for i in a]",
"_____no_output_____"
],
[
"' '.join(a)",
"_____no_output_____"
],
[
"a=\"1 2 3 4 5 6 7\"",
"_____no_output_____"
],
[
"a.split()",
"_____no_output_____"
],
[
"a=[int(i) for i in a.split()]\na",
"_____no_output_____"
],
[
"revcipher={i+1:chr(i+97) for i in range(26)}\nrevcipher",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb8e4c790358cb30e212a52a8319a52a7aab6c70
| 14,525 |
ipynb
|
Jupyter Notebook
|
.ipynb_checkpoints/CLIQUE-checkpoint.ipynb
|
rdedo099/HonoursProject2021
|
94c61218371587fd4dd9dacaa5e8f0ce7f44875d
|
[
"MIT"
] | null | null | null |
.ipynb_checkpoints/CLIQUE-checkpoint.ipynb
|
rdedo099/HonoursProject2021
|
94c61218371587fd4dd9dacaa5e8f0ce7f44875d
|
[
"MIT"
] | null | null | null |
.ipynb_checkpoints/CLIQUE-checkpoint.ipynb
|
rdedo099/HonoursProject2021
|
94c61218371587fd4dd9dacaa5e8f0ce7f44875d
|
[
"MIT"
] | null | null | null | 29.764344 | 371 | 0.555456 |
[
[
[
"!pip install pyclustering",
"Collecting pyclustering\n Downloading pyclustering-0.10.1.2.tar.gz (2.6 MB)\nRequirement already satisfied: scipy>=1.1.0 in c:\\users\\riley\\appdata\\local\\packages\\pythonsoftwarefoundation.python.3.9_qbz5n2kfra8p0\\localcache\\local-packages\\python39\\site-packages (from pyclustering) (1.7.0)\nRequirement already satisfied: matplotlib>=3.0.0 in c:\\users\\riley\\appdata\\local\\packages\\pythonsoftwarefoundation.python.3.9_qbz5n2kfra8p0\\localcache\\local-packages\\python39\\site-packages (from pyclustering) (3.4.2)\nRequirement already satisfied: numpy>=1.15.2 in c:\\users\\riley\\appdata\\local\\packages\\pythonsoftwarefoundation.python.3.9_qbz5n2kfra8p0\\localcache\\local-packages\\python39\\site-packages (from pyclustering) (1.21.0)\nRequirement already satisfied: Pillow>=5.2.0 in c:\\users\\riley\\appdata\\local\\packages\\pythonsoftwarefoundation.python.3.9_qbz5n2kfra8p0\\localcache\\local-packages\\python39\\site-packages (from pyclustering) (8.2.0)\nRequirement already satisfied: python-dateutil>=2.7 in c:\\users\\riley\\appdata\\local\\packages\\pythonsoftwarefoundation.python.3.9_qbz5n2kfra8p0\\localcache\\local-packages\\python39\\site-packages (from matplotlib>=3.0.0->pyclustering) (2.8.1)\nRequirement already satisfied: cycler>=0.10 in c:\\users\\riley\\appdata\\local\\packages\\pythonsoftwarefoundation.python.3.9_qbz5n2kfra8p0\\localcache\\local-packages\\python39\\site-packages (from matplotlib>=3.0.0->pyclustering) (0.10.0)\nRequirement already satisfied: kiwisolver>=1.0.1 in c:\\users\\riley\\appdata\\local\\packages\\pythonsoftwarefoundation.python.3.9_qbz5n2kfra8p0\\localcache\\local-packages\\python39\\site-packages (from matplotlib>=3.0.0->pyclustering) (1.3.1)\nRequirement already satisfied: pyparsing>=2.2.1 in c:\\users\\riley\\appdata\\local\\packages\\pythonsoftwarefoundation.python.3.9_qbz5n2kfra8p0\\localcache\\local-packages\\python39\\site-packages (from matplotlib>=3.0.0->pyclustering) (2.4.7)\nRequirement already satisfied: six in c:\\users\\riley\\appdata\\local\\packages\\pythonsoftwarefoundation.python.3.9_qbz5n2kfra8p0\\localcache\\local-packages\\python39\\site-packages (from cycler>=0.10->matplotlib>=3.0.0->pyclustering) (1.16.0)\nUsing legacy 'setup.py install' for pyclustering, since package 'wheel' is not installed.\nInstalling collected packages: pyclustering\n Running setup.py install for pyclustering: started\n Running setup.py install for pyclustering: finished with status 'done'\nSuccessfully installed pyclustering-0.10.1.2\n"
],
[
"%load_ext autoreload\n%autoreload 2",
"_____no_output_____"
],
[
"from Clique.Clique import *",
"_____no_output_____"
],
[
"from load_logs import *\nfrom evaluation import *\nfrom features import *\nfrom visualize import *",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt\nimport numpy as np",
"_____no_output_____"
],
[
"logs, log_labels = read_logs_and_labels(\"./Saved/logs.txt\", \"./Saved/labels.txt\")",
"_____no_output_____"
],
[
"np.set_printoptions(threshold=np.inf)",
"_____no_output_____"
],
[
"X = get_features(logs, 2, 70)\nX = X.toarray()",
"_____no_output_____"
],
[
"np.count_nonzero(X[13456,:])",
"_____no_output_____"
],
[
"intervals = [3, 5, 10, 20, 30, 40, 50]\nthresholds = [0.1, 0.2, 0.3, 0.4, 0.5]",
"_____no_output_____"
],
[
"def grid_search(logs, labels_, gram, min_df):\n X = get_features(logs, gram, min_df)\n X = X.toarray()\n \n idxs = np.where(np.all(X == 0, axis=1))\n \n X = np.delete(X, idxs, axis=0)\n labels_ = np.delete(labels_, idxs)\n \n result_header = [\"Interval\", \"Threshold\", \"VMeasure\", \"Fowlkes-Mallows\"]\n results = []\n \n for interval in intervals:\n for threshold in thresholds:\n \n clusters = run_clique(data=X,\n xsi=interval,\n tau=threshold)\n \n print(\"Clique eval for interval %2d, and threshold %2d\" % (interval, threshold))\n evaluate_clustering_performance(clusters, labels_)\n \n# vm = evaluate_vmeasure(labels_, labels)\n# fm = evaluate_fm(labels_, labels)\n\n# result = [interval, threshold, vm, fm]\n# results.append(result)\n\n# tab_results(result_header, results) \n# tab_results(result_header, results) ",
"_____no_output_____"
],
[
"# def grid_search(logs, labels_, gram, min_df):\n# X = get_features(logs, gram, min_df)\n# X = X.toarray()\n \n# idxs = np.where(np.all(X == 0, axis=1))\n \n# X = np.delete(X, idxs, axis=0)\n# labels_ = np.delete(labels_, idxs)\n \n# result_header = [\"Interval\", \"Threshold\", \"VMeasure\", \"Fowlkes-Mallows\"]\n# results = []\n \n# for interval in intervals:\n# for threshold in thresholds:\n \n# clique_instance = clique(X, 40, 0)\n# clique_instance.process()\n \n# labels = clique_instance.get_clusters()\n# #labels = labels.reshape\n# print(labels)\n# #print(labels.shape)\n \n# cells = clique_instance.get_cells()\n# #print(cells)\n \n# vm = evaluate_vmeasure(labels_, labels)\n# fm = evaluate_fm(labels_, labels)\n\n# result = [metric, linkage, vm, fm]\n# results.append(result)\n\n# tab_results(result_header, results) \n# tab_results(result_header, results) ",
"_____no_output_____"
],
[
"grid_search(logs, log_labels, 2, 70)",
"_____no_output_____"
],
[
"grid_search(logs, log_labels, 3, 90)",
"_____no_output_____"
],
[
"X = get_features(logs, 2, 100)\nX = X.toarray()\n\nidxs = np.where(np.all(X == 0, axis=1))\n\nX = np.delete(X, idxs, axis=0)",
"_____no_output_____"
],
[
"from pyclustering.cluster.clique import clique, clique_visualizer\nfrom pyclustering.utils import read_sample\nfrom pyclustering.samples.definitions import FCPS_SAMPLES\n# read two-dimensional input data 'Target'\ndata = X\n# create CLIQUE algorithm for processing\nintervals = 1 # defines amount of cells in grid in each dimension\nthreshold = 1 # lets consider each point as non-outlier\nclique_instance = clique(data, intervals, threshold)\n# start clustering process and obtain results\nclique_instance.process()\nclusters = clique_instance.get_clusters() # allocated clusters\nnoise = clique_instance.get_noise() # points that are considered as outliers (in this example should be empty)\ncells = clique_instance.get_cells() # CLIQUE blocks that forms grid\nprint(\"Amount of clusters:\", len(clusters))\n# visualize clustering results\nclique_visualizer.show_grid(cells, data[:,:2]) # show grid that has been formed by the algorithm\nclique_visualizer.show_clusters(data[:,:2], clusters, noise) # show clustering results",
"_____no_output_____"
],
[
"data[:, :2]",
"_____no_output_____"
]
],
[
[
"## Bigram Feature Vectorizer",
"_____no_output_____"
]
],
[
[
"eval_results = []",
"_____no_output_____"
],
[
"labels_ = log_labels\n \nX = get_features(logs, 2, 70)\nX = X.toarray()\n\nidxs = np.where(np.all(X == 0, axis=1))\n\nX = np.delete(X, idxs, axis=0)\nlabels_ = np.delete(labels_, idxs)",
"_____no_output_____"
],
[
"clique_instance = clique(X, interval, threshold)\nclique_instance.process()\n\nlabels = clique_instance.get_clusters()",
"_____no_output_____"
],
[
"plot_clusters(\"CLIQUE Bigram Clustering using UMAP\", X, labels)",
"_____no_output_____"
],
[
"results = evaluate_clustering('CLIQUE Bigram Clustering', X, labels_, labels)\nprint(results)",
"_____no_output_____"
],
[
"eval_results.append(results)",
"_____no_output_____"
]
],
[
[
"## Trigram Feature Vectorizer",
"_____no_output_____"
]
],
[
[
"labels_ = log_labels\n \nX = get_features(logs, 3, 90)\nX = X.toarray()\n\nidxs = np.where(np.all(X == 0, axis=1))\n\nX = np.delete(X, idxs, axis=0)\nlabels_ = np.delete(labels_, idxs)",
"_____no_output_____"
],
[
"clique_instance = clique(X, interval, threshold)\nclique_instance.process()\n\nlabels = clique_instance.get_clusters()",
"_____no_output_____"
],
[
"plot_clusters(\"CLIQUE Bigram Clustering using UMAP\", X, labels)",
"_____no_output_____"
],
[
"results = evaluate_clustering('CLIQUE Bigram Clustering', X, labels_, labels)\nprint(results)",
"_____no_output_____"
],
[
"eval_results.append(results)",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
cb8e5a419fa02e2c538a721e2f2f26903b2b95de
| 9,139 |
ipynb
|
Jupyter Notebook
|
dev_notebooks/test TL.ipynb
|
agrogeophy/geometadp
|
aa69abf97546e59307abdeb6d365c6619c3db737
|
[
"MIT"
] | 1 |
2021-07-05T13:46:16.000Z
|
2021-07-05T13:46:16.000Z
|
dev_notebooks/test TL.ipynb
|
agrogeophy/geometadp
|
aa69abf97546e59307abdeb6d365c6619c3db737
|
[
"MIT"
] | null | null | null |
dev_notebooks/test TL.ipynb
|
agrogeophy/geometadp
|
aa69abf97546e59307abdeb6d365c6619c3db737
|
[
"MIT"
] | 1 |
2021-03-17T10:36:31.000Z
|
2021-03-17T10:36:31.000Z
| 29.576052 | 157 | 0.53299 |
[
[
[
"import ipywidgets\n\ntabs = ipywidgets.Tab()\ntabs.children = [ipywidgets.Label(value='tab1'), ipywidgets.Label(value='tab2'), ipywidgets.Label(value='tab3'), ipywidgets.Label(value='tab4')]\ntabs.observe(lambda change: print(f\"selected index: {change['new']}\") , names='selected_index')\n\ndef change_children(_):\n id = tabs.selected_index\n tabs.selected_index = None # Warning : this will emit a change event\n tabs.children = [ipywidgets.Label(value='tab1'), ipywidgets.Label(value='tab2'), ipywidgets.Label(value='tab3'), ipywidgets.Label(value='tab4')]\n tabs.selected_index = id\n\nbtn = ipywidgets.Button(description='change_children')\nbtn.on_click(change_children)\n\nipywidgets.VBox([tabs, btn])",
"_____no_output_____"
],
[
"import ipywidgets as widgets\n\ntab_contents = ['P0', 'P1']\nchildren = [widgets.Text(description=name) for name in tab_contents]\ntab = widgets.Tab()\ntab.children = children\nfor i in range(len(children)):\n tab.set_title(i, str(i))\n\ndef tab_toggle_var(*args):\n global vartest\n if tab.selected_index ==0:\n vartest = 0\n else:\n vartest = 1\ntab.observe(tab_toggle_var)\ntab_toggle_var()\nprint(children)\n\nmetadata={}\n\ndef _observe_test(change):\n print(change)\n \ndef _observe_config(change):\n print('_observe_config')\n metadata[ widget_elec_config.description] = widget_elec_config.value\n metadata_json_raw = json.dumps(metadata, indent=4)\n export.value = \"<pre>{}</pre>\".format(\n html.escape(metadata_json_raw))\n \nexport = widgets.HTML()\nvbox_metadata = widgets.VBox(\n [\n widgets.HTML('''\n <h4>Preview of metadata export:</h4>\n <hr style=\"height:1px;border-width:0;color:black;background-color:gray\">\n '''),\n export\n ]\n)\n\nfor child in tab.children:\n print(child)\n child.observe(_observe_test)\n\ndisplay(tab)\n",
"[Text(value='', description='P0'), Text(value='', description='P1')]\nText(value='', description='P0')\nText(value='', description='P1')\n"
],
[
"w = widgets.Dropdown(\n options=['Addition', 'Multiplication', 'Subtraction', 'Division'],\n value='Addition',\n description='Task:',\n)\n\ndef on_change(change):\n if change['type'] == 'change' and change['name'] == 'value':\n print(\"changed to %s\" % change['new'])\n\nw.observe(on_change)\n\ndisplay(w)",
"_____no_output_____"
],
[
"from IPython.display import display\nimport ipywidgets as widgets\n\nint_range0_slider = widgets.IntSlider()\nint_range1_slider = widgets.IntSlider()\noutput = widgets.Output()\n\ndef interactive_function(inp0,inp1):\n with output:\n print('ie changed. int_range0_slider: '+str(inp0)+' int_range1_slider: '+str(inp1))\n return\n\ndef report_int_range0_change(change):\n with output:\n print('int_range0 change observed'+str(change))\n return\n\ndef report_ie_change(change):\n with output:\n print('ie change observed'+str(change))\n return\n\nie = widgets.interactive(interactive_function, inp0=int_range0_slider,inp1=int_range1_slider)\n\n# print(int_range0_slider.observe)\n# print(ie.observe)\n# int_range0_slider.observe(report_int_range0_change, names='value')\nfor child in ie.children:\n child.observe(report_ie_change)\n\ndisplay(int_range0_slider,int_range1_slider,output)",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code"
]
] |
cb8e5bcbafdba625324585afded32861c3d6083b
| 39,059 |
ipynb
|
Jupyter Notebook
|
06_Kaggle_HomeCredit/code/03_XGBoost.ipynb
|
KartikKannapur/Kaggle
|
bec2f3f4444956a1d0ce2ee0215be049c543c637
|
[
"MIT"
] | 2 |
2020-12-15T06:25:07.000Z
|
2020-12-17T16:18:29.000Z
|
06_Kaggle_HomeCredit/code/03_XGBoost.ipynb
|
KartikKannapur/Kaggle
|
bec2f3f4444956a1d0ce2ee0215be049c543c637
|
[
"MIT"
] | null | null | null |
06_Kaggle_HomeCredit/code/03_XGBoost.ipynb
|
KartikKannapur/Kaggle
|
bec2f3f4444956a1d0ce2ee0215be049c543c637
|
[
"MIT"
] | null | null | null | 31.222222 | 399 | 0.448603 |
[
[
[
"# Home Credit Default Risk\n\nCan you predict how capable each applicant is of repaying a loan?",
"_____no_output_____"
],
[
"Many people struggle to get loans due to **insufficient or non-existent credit histories**. And, unfortunately, this population is often taken advantage of by untrustworthy lenders.\n\nHome Credit strives to broaden financial inclusion for the **unbanked population by providing a positive and safe borrowing experience**. In order to make sure this underserved population has a positive loan experience, Home Credit makes use of a variety of alternative data--including telco and transactional information--to predict their clients' repayment abilities.\n\nWhile Home Credit is currently using various statistical and machine learning methods to make these predictions, they're challenging Kagglers to help them unlock the full potential of their data. Doing so will ensure that clients capable of repayment are not rejected and that loans are given with a principal, maturity, and repayment calendar that will empower their clients to be successful.",
"_____no_output_____"
],
[
"**Submissions are evaluated on area under the ROC curve between the predicted probability and the observed target.**",
"_____no_output_____"
],
[
"# Dataset",
"_____no_output_____"
]
],
[
[
"# #Python Libraries\nimport numpy as np\nimport scipy as sp\nimport pandas as pd\nimport statsmodels\nimport pandas_profiling\n\n%matplotlib inline\nimport matplotlib.pyplot as plt\nimport seaborn as sns\n\nimport os\nimport sys\nimport time\nimport requests\nimport datetime\n\nimport missingno as msno\nimport math\nimport sys\nimport gc\nimport os\n\n# #sklearn\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.model_selection import cross_val_score\nfrom sklearn.model_selection import RandomizedSearchCV\nfrom sklearn.model_selection import GridSearchCV\nfrom sklearn.ensemble import RandomForestRegressor\nfrom sklearn import preprocessing\n\n# #sklearn - metrics\nfrom sklearn.metrics import mean_squared_error\nfrom sklearn.metrics import mean_absolute_error\nfrom sklearn.metrics import r2_score\n\n# #XGBoost & LightGBM\nimport xgboost as xgb\nimport lightgbm as lgb\n\n# #Missing value imputation\nfrom fancyimpute import KNN, MICE",
"_____no_output_____"
]
],
[
[
"## Data Dictionary",
"_____no_output_____"
]
],
[
[
"!ls -l ../data/",
"total 2621364\n-rw-r--r-- 1 karti 197609 26567651 May 17 18:06 application_test.csv\n-rw-r--r-- 1 karti 197609 166133370 May 17 18:06 application_train.csv\n-rw-r--r-- 1 karti 197609 170016717 May 17 18:08 bureau.csv\n-rw-r--r-- 1 karti 197609 375592889 May 17 18:08 bureau_balance.csv\n-rw-r--r-- 1 karti 197609 424582605 May 17 18:10 credit_card_balance.csv\n-rw-r--r-- 1 karti 197609 37383 May 20 19:55 HomeCredit_columns_description.csv\n-rw-r--r-- 1 karti 197609 723118349 May 17 18:13 installments_payments.csv\n-rw-r--r-- 1 karti 197609 392703158 May 17 18:14 POS_CASH_balance.csv\n-rw-r--r-- 1 karti 197609 404973293 May 17 18:15 previous_application.csv\n-rw-r--r-- 1 karti 197609 536202 May 17 18:06 sample_submission.csv\n"
]
],
[
[
"- application_{train|test}.csv\n\nThis is the main table, broken into two files for Train (**with TARGET**) and Test (without TARGET).\nStatic data for all applications. **One row represents one loan in our data sample.**\n\n- bureau.csv\n\nAll client's previous credits provided by other financial institutions that were reported to Credit Bureau (for clients who have a loan in our sample).\nFor every loan in our sample, there are as many rows as number of credits the client had in Credit Bureau before the application date.\n\n- bureau_balance.csv\n\nMonthly balances of previous credits in Credit Bureau.\nThis table has one row for each month of history of every previous credit reported to Credit Bureau – i.e the table has (#loans in sample * # of relative previous credits * # of months where we have some history observable for the previous credits) rows.\n\n- POS_CASH_balance.csv\n\nMonthly balance snapshots of previous POS (point of sales) and cash loans that the applicant had with Home Credit.\nThis table has one row for each month of history of every previous credit in Home Credit (consumer credit and cash loans) related to loans in our sample – i.e. the table has (#loans in sample * # of relative previous credits * # of months in which we have some history observable for the previous credits) rows.\n\n- credit_card_balance.csv\n\nMonthly balance snapshots of previous credit cards that the applicant has with Home Credit.\nThis table has one row for each month of history of every previous credit in Home Credit (consumer credit and cash loans) related to loans in our sample – i.e. the table has (#loans in sample * # of relative previous credit cards * # of months where we have some history observable for the previous credit card) rows.\n\n- previous_application.csv\n\nAll previous applications for Home Credit loans of clients who have loans in our sample.\nThere is one row for each previous application related to loans in our data sample.\n\n- installments_payments.csv\n\nRepayment history for the previously disbursed credits in Home Credit related to the loans in our sample.\nThere is a) one row for every payment that was made plus b) one row each for missed payment.\nOne row is equivalent to one payment of one installment OR one installment corresponding to one payment of one previous Home Credit credit related to loans in our sample.\n\n- HomeCredit_columns_description.csv\n\nThis file contains descriptions for the columns in the various data files.",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"# Data Pre-processing",
"_____no_output_____"
]
],
[
[
"df_application_train = pd.read_csv(\"../data/application_train.csv\")",
"_____no_output_____"
],
[
"df_application_train.head()",
"_____no_output_____"
],
[
"df_application_test = pd.read_csv(\"../data/application_test.csv\")",
"_____no_output_____"
],
[
"df_application_test.head()",
"_____no_output_____"
]
],
[
[
"## Missing Value Imputation",
"_____no_output_____"
]
],
[
[
"df_application_train_imputed = pd.read_csv(\"../transformed_data/application_train_imputed.csv\")",
"_____no_output_____"
],
[
"df_application_test_imputed = pd.read_csv(\"../transformed_data/application_test_imputed.csv\")",
"_____no_output_____"
],
[
"df_application_train.shape, df_application_test.shape",
"_____no_output_____"
],
[
"df_application_train_imputed.shape, df_application_test_imputed.shape",
"_____no_output_____"
],
[
"df_application_train.isnull().sum(axis = 0).sum(), df_application_test.isnull().sum(axis = 0).sum()",
"_____no_output_____"
],
[
"df_application_train_imputed.isnull().sum(axis = 0).sum(), df_application_test_imputed.isnull().sum(axis = 0).sum()",
"_____no_output_____"
]
],
[
[
"# Model Building",
"_____no_output_____"
],
[
"## Encode categorical columns",
"_____no_output_____"
]
],
[
[
"# arr_categorical_columns = df_application_train.select_dtypes(['object']).columns\n# for var_col in arr_categorical_columns:\n# df_application_train[var_col] = df_application_train[var_col].astype('category').cat.codes\n\n# arr_categorical_columns = df_application_test.select_dtypes(['object']).columns\n# for var_col in arr_categorical_columns:\n# df_application_test[var_col] = df_application_test[var_col].astype('category').cat.codes",
"_____no_output_____"
]
],
[
[
"## Train-Validation Split",
"_____no_output_____"
]
],
[
[
"input_columns = df_application_train_imputed.columns\ninput_columns = input_columns[input_columns != 'TARGET']\ntarget_column = 'TARGET'\n\nX = df_application_train_imputed[input_columns]\ny = df_application_train_imputed[target_column]\nX_train, X_test, y_train, y_test = train_test_split(X, y, random_state=42)",
"_____no_output_____"
],
[
"xgb_params = {\n 'seed': 0,\n 'colsample_bytree': 0.8,\n 'silent': 1,\n 'subsample': 0.6,\n 'learning_rate': 0.01,\n 'objective': 'binary:logistic',\n 'eval_metric': 'auc', \n 'max_depth': 6,\n 'num_parallel_tree': 1,\n 'min_child_weight': 5,\n}\n\n",
"_____no_output_____"
],
[
"watchlist = [(xgb.DMatrix(X_train, y_train), 'train'), (xgb.DMatrix(X_test, y_test), 'valid')]\nmodel = xgb.train(xgb_params, xgb.DMatrix(X_train, y_train), 270, watchlist, maximize=True, verbose_eval=100)",
"[0]\ttrain-auc:0.719021\tvalid-auc:0.712287\n[100]\ttrain-auc:0.748328\tvalid-auc:0.737499\n[200]\ttrain-auc:0.754814\tvalid-auc:0.740613\n[269]\ttrain-auc:0.760755\tvalid-auc:0.743433\n"
],
[
"df_predict = model.predict(xgb.DMatrix(df_application_test_imputed), ntree_limit=model.best_ntree_limit)",
"_____no_output_____"
],
[
"submission = pd.DataFrame()\nsubmission[\"SK_ID_CURR\"] = df_application_test[\"SK_ID_CURR\"]\nsubmission[\"TARGET\"] = df_predict\n\nsubmission.to_csv(\"../submissions/model_1_xgbstarter_missingdata_MICE_imputed.csv\", index=False)",
"_____no_output_____"
],
[
"submission.shape",
"_____no_output_____"
],
[
"input_columns = df_application_train.columns\ninput_columns = input_columns[input_columns != 'TARGET']\ntarget_column = 'TARGET'\n\nX = df_application_train[input_columns]\ny = df_application_train[target_column]\nX_train, X_test, y_train, y_test = train_test_split(X, y, random_state=42)",
"_____no_output_____"
],
[
"watchlist = [(xgb.DMatrix(X_train, y_train), 'train'), (xgb.DMatrix(X_test, y_test), 'valid')]\nmodel = xgb.train(xgb_params, xgb.DMatrix(X_train, y_train), 270, watchlist, maximize=True, verbose_eval=100)",
"[0]\ttrain-auc:0.716955\tvalid-auc:0.708265\n[100]\ttrain-auc:0.749595\tvalid-auc:0.738329\n[200]\ttrain-auc:0.755462\tvalid-auc:0.741356\n[269]\ttrain-auc:0.760462\tvalid-auc:0.744093\n"
],
[
"df_predict = model.predict(xgb.DMatrix(df_application_test), ntree_limit=model.best_ntree_limit)",
"_____no_output_____"
],
[
"submission = pd.DataFrame()\nsubmission[\"SK_ID_CURR\"] = df_application_test[\"SK_ID_CURR\"]\nsubmission[\"TARGET\"] = df_predict\n\nsubmission.to_csv(\"../submissions/model_1_xgbstarter_missingdata_MICE_nonimputed_hypothesis.csv\", index=False)",
"_____no_output_____"
],
[
"submission.shape",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb8e82ef685db9b92ff6ebca8fdbd53286c7bcf4
| 280,387 |
ipynb
|
Jupyter Notebook
|
content/notebooks/6-Bootstrap_rTPC_tutorial_Nesbit.ipynb
|
nesbitm/Vector_BiTE_21
|
9d396610fad20ce5caf8865eba9a95988238682c
|
[
"MIT"
] | 1 |
2021-05-07T09:28:51.000Z
|
2021-05-07T09:28:51.000Z
|
content/notebooks/6-Bootstrap_rTPC_tutorial_Nesbit.ipynb
|
nesbitm/Vector_BiTE_21
|
9d396610fad20ce5caf8865eba9a95988238682c
|
[
"MIT"
] | null | null | null |
content/notebooks/6-Bootstrap_rTPC_tutorial_Nesbit.ipynb
|
nesbitm/Vector_BiTE_21
|
9d396610fad20ce5caf8865eba9a95988238682c
|
[
"MIT"
] | null | null | null | 402.276901 | 97,854 | 0.919005 |
[
[
[
"library(repr) ; options(repr.plot.width = 5, repr.plot.height = 6) # Change plot sizes (in cm)",
"_____no_output_____"
]
],
[
[
"# Bootstrapping using rTPC package",
"_____no_output_____"
],
[
"## Introduction\nIn this Chapter we will work through an example of model fitting using the rTPC package in R. This references the previous chapters' work, especially [Model Fitting the Bayesian way](https://www.youtube.com/watch?v=dQw4w9WgXcQ).\n\nLets start with the requirements!",
"_____no_output_____"
]
],
[
[
"require('ggplot2')\nrequire('nls.multstart')\nrequire('broom')\nrequire('tidyverse')\nrequire('rTPC')\nrequire('dplyr')\nrequire('data.table')\nrequire('car')\nrequire('boot')\nrequire('patchwork')\nrequire('minpack.lm')\nrequire(\"tidyr\")\nrequire('purrr')\n# update.packages(ask = FALSE)\n\nrm(list=ls())\ngraphics.off()\nsetwd(\"/home/primuser/Documents/VByte/VecMismatchPaper1/code/\")",
"Loading required package: ggplot2\n\nLoading required package: nls.multstart\n\nLoading required package: broom\n\nLoading required package: tidyverse\n\n── \u001b[1mAttaching packages\u001b[22m ─────────────────────────────────────── tidyverse 1.3.1 ──\n\n\u001b[32m✔\u001b[39m \u001b[34mtibble \u001b[39m 3.1.2 \u001b[32m✔\u001b[39m \u001b[34mdplyr \u001b[39m 1.0.6\n\u001b[32m✔\u001b[39m \u001b[34mtidyr \u001b[39m 1.1.3 \u001b[32m✔\u001b[39m \u001b[34mstringr\u001b[39m 1.4.0\n\u001b[32m✔\u001b[39m \u001b[34mreadr \u001b[39m 1.4.0 \u001b[32m✔\u001b[39m \u001b[34mforcats\u001b[39m 0.5.1\n\u001b[32m✔\u001b[39m \u001b[34mpurrr \u001b[39m 0.3.4 \n\n── \u001b[1mConflicts\u001b[22m ────────────────────────────────────────── tidyverse_conflicts() ──\n\u001b[31m✖\u001b[39m \u001b[34mdplyr\u001b[39m::\u001b[32mfilter()\u001b[39m masks \u001b[34mstats\u001b[39m::filter()\n\u001b[31m✖\u001b[39m \u001b[34mdplyr\u001b[39m::\u001b[32mlag()\u001b[39m masks \u001b[34mstats\u001b[39m::lag()\n\nLoading required package: rTPC\n\nLoading required package: data.table\n\n\nAttaching package: ‘data.table’\n\n\nThe following objects are masked from ‘package:dplyr’:\n\n between, first, last\n\n\nThe following object is masked from ‘package:purrr’:\n\n transpose\n\n\nLoading required package: car\n\nLoading required package: carData\n\n\nAttaching package: ‘car’\n\n\nThe following object is masked from ‘package:dplyr’:\n\n recode\n\n\nThe following object is masked from ‘package:purrr’:\n\n some\n\n\nLoading required package: boot\n\n\nAttaching package: ‘boot’\n\n\nThe following object is masked from ‘package:car’:\n\n logit\n\n\nLoading required package: patchwork\n\nLoading required package: minpack.lm\n\n"
]
],
[
[
"Now that we have the background requirements going, we can start using the rTPC package. Lets look through the different models available!",
"_____no_output_____"
]
],
[
[
"#take a look at the different models available\nget_model_names()",
"_____no_output_____"
]
],
[
[
"There are 24 models to choose from. For our purposes in this chapter we will be using the sharpesschoolhigh_1981 model. More information on the model can be found [here](https://padpadpadpad.github.io/rTPC/reference/sharpeschoolhigh_1981.html).\n\nFrom here lets load in our data from the overall repository. This will be called '../data/Final_Traitofinterest.csv'.",
"_____no_output_____"
]
],
[
[
"#read in the trait data\nfinal_trait_data <- read.csv('../data/Final_Traitofinterest.csv')",
"_____no_output_____"
]
],
[
[
"Lets reduce this to a single trait. This data comes from the [VectorBiTE database](https://legacy.vectorbyte.org/) and so has unique IDs. We will use this to get our species and trait of interest isolated from the larger dataset. In this example we will be looking at Development Rate across temperatures for Aedes albopictus, which we can find an example of in csm7I.",
"_____no_output_____"
]
],
[
[
"df1 <- final_trait_data %>%\n dplyr::select('originalid', 'originaltraitname', 'originaltraitunit', 'originaltraitvalue', 'interactor1', 'ambienttemp', 'citation')\n#filter to single species and trait\ndf2 <- dplyr::filter(df1, originalid == 'csm7I')",
"_____no_output_____"
]
],
[
[
"Now lets visualize our data in ggplot.",
"_____no_output_____"
]
],
[
[
"#visualize\nggplot(df2, aes(ambienttemp, originaltraitvalue))+\n geom_point()+\n theme_bw(base_size = 12) +\n labs(x = 'Temperature (ºC)',\n y = 'Development Rate',\n title = 'Development Rate across temperatures for Aedes albopictus')",
"_____no_output_____"
]
],
[
[
"We will need to write which model we are using (sharpschoolhigh_1981). From here we can actually build our fit. We will use ''nls_multstart'' to automatically find our starting values. This lets us skip the [starting value problem](https://mhasoba.github.io/TheMulQuaBio/notebooks/20-ModelFitting-NLLS.html#the-starting-values-problem). From here we build our predicted line.",
"_____no_output_____"
]
],
[
[
"# choose model\nmod = 'sharpschoolhigh_1981'\nd<- df2 %>%\n rename(temp = ambienttemp,\n rate = originaltraitvalue)",
"_____no_output_____"
],
[
"# fit Sharpe-Schoolfield model\nd_fit <- nest(d, data = c(temp, rate)) %>%\n mutate(sharpeschoolhigh = map(data, ~nls_multstart(rate~sharpeschoolhigh_1981(temp = temp, r_tref,e,eh,th, tref = 15),\n data = .x,\n iter = c(3,3,3,3),\n start_lower = get_start_vals(.x$temp, .x$rate, model_name = 'sharpeschoolhigh_1981') - 10,\n start_upper = get_start_vals(.x$temp, .x$rate, model_name = 'sharpeschoolhigh_1981') + 10,\n lower = get_lower_lims(.x$temp, .x$rate, model_name = 'sharpeschoolhigh_1981'),\n upper = get_upper_lims(.x$temp, .x$rate, model_name = 'sharpeschoolhigh_1981'),\n supp_errors = 'Y',\n convergence_count = FALSE)),\n \n # create new temperature data\n new_data = map(data, ~tibble(temp = seq(min(.x$temp), max(.x$temp), length.out = 100))),\n # predict over that data,\n preds = map2(sharpeschoolhigh, new_data, ~augment(.x, newdata = .y)))",
"_____no_output_____"
],
[
"# unnest predictions\nd_preds <- select(d_fit, preds) %>%\n unnest(preds)",
"_____no_output_____"
]
],
[
[
"Lets visualize the line:",
"_____no_output_____"
]
],
[
[
"# plot data and predictions\nggplot() +\n geom_line(aes(temp, .fitted), d_preds, col = 'blue') +\n geom_point(aes(temp, rate), d, size = 2, alpha = 0.5) +\n theme_bw(base_size = 12) +\n labs(x = 'Temperature (ºC)',\n y = 'Growth rate',\n title = 'Growth rate across temperatures')",
"_____no_output_____"
]
],
[
[
"This looks like a good fit! We can start exploring using bootstrapping. Lets start with refitting the model using nlsLM.",
"_____no_output_____"
]
],
[
[
"# refit model using nlsLM\nfit_nlsLM <- minpack.lm::nlsLM(rate~sharpeschoolhigh_1981(temp = temp, r_tref,e,eh,th, tref = 15),\n data = d,\n start = coef(d_fit$sharpeschoolhigh[[1]]),\n lower = get_lower_lims(d$temp, d$rate, model_name = 'sharpeschoolhigh_1981'),\n upper = get_upper_lims(d$temp, d$rate, model_name = 'sharpeschoolhigh_1981'),\n weights = rep(1, times = nrow(d)))",
"_____no_output_____"
]
],
[
[
"Now we can actually bootstrap.",
"_____no_output_____"
]
],
[
[
"# bootstrap using case resampling\nboot1 <- Boot(fit_nlsLM, method = 'case')",
"Warning message in nls.lm(par = start, fn = FCT, jac = jac, control = control, lower = lower, :\n“lmdif: info = -1. Number of iterations has reached `maxiter' == 50.\n”\nWarning message in nls.lm(par = start, fn = FCT, jac = jac, control = control, lower = lower, :\n“lmdif: info = -1. Number of iterations has reached `maxiter' == 50.\n”\n"
]
],
[
[
"It is a good idea to explore the data again now.",
"_____no_output_____"
]
],
[
[
"# look at the data\nhead(boot1$t)",
"_____no_output_____"
],
[
"hist(boot1, layout = c(2,2))",
"Warning message in norm.inter(t, adj.alpha):\n“extreme order statistics used as endpoints”\nWarning message in norm.inter(t, adj.alpha):\n“extreme order statistics used as endpoints”\nWarning message in norm.inter(t, adj.alpha):\n“extreme order statistics used as endpoints”\n"
]
],
[
[
"Now we use the bootstrapped model to build predictions which we can explore visually.",
"_____no_output_____"
]
],
[
[
"# create predictions of each bootstrapped model\nboot1_preds <- boot1$t %>%\n as.data.frame() %>%\n drop_na() %>%\n mutate(iter = 1:n()) %>%\n group_by_all() %>%\n do(data.frame(temp = seq(min(d$temp), max(d$temp), length.out = 100))) %>%\n ungroup() %>%\n mutate(pred = sharpeschoolhigh_1981(temp, r_tref, e, eh, th, tref = 15))",
"_____no_output_____"
],
[
"# calculate bootstrapped confidence intervals\nboot1_conf_preds <- group_by(boot1_preds, temp) %>%\n summarise(conf_lower = quantile(pred, 0.025),\n conf_upper = quantile(pred, 0.975)) %>%\n ungroup()",
"_____no_output_____"
],
[
"# plot bootstrapped CIs\np1 <- ggplot() +\n geom_line(aes(temp, .fitted), d_preds, col = 'blue') +\n geom_ribbon(aes(temp, ymin = conf_lower, ymax = conf_upper), boot1_conf_preds, fill = 'blue', alpha = 0.3) +\n geom_point(aes(temp, rate), d, size = 2, alpha = 0.5) +\n theme_bw(base_size = 12) +\n labs(x = 'Temperature (ºC)',\n y = 'Growth rate',\n title = 'Growth rate across temperatures')\n\n# plot bootstrapped predictions\np2 <- ggplot() +\n geom_line(aes(temp, .fitted), d_preds, col = 'blue') +\n geom_line(aes(temp, pred, group = iter), boot1_preds, col = 'blue', alpha = 0.007) +\n geom_point(aes(temp, rate), d, size = 2, alpha = 0.5) +\n theme_bw(base_size = 12) +\n labs(x = 'Temperature (ºC)',\n y = 'Growth rate',\n title = 'Growth rate across temperatures')",
"_____no_output_____"
],
[
"p1 + p2",
"_____no_output_____"
]
],
[
[
"We can see here that when we bootstrap this data, the fit is not as good as we would expect from the initial exploration. We do not necessarily get a good thermal optima from this data. However, this does show how to use this function in the future. Please see Daniel Padfields [git](https://padpadpadpad.github.io/rTPC/articles/rTPC.html) for more information on using the rTPC package.",
"_____no_output_____"
],
[
"# Please go to the [landing page](https://www.youtube.com/watch?v=YddwkMJG1Jo) and proceed on to the next stage of the training!",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
]
] |
cb8e8c7ddfcfc0d48214cde0679be7f045fb817c
| 34,146 |
ipynb
|
Jupyter Notebook
|
JupyterNotebooks/ReadMe.ipynb
|
fawadkarimi/JupyterNotebooks2
|
323b679c0ae1fbf241e2a728c0568edc7d969617
|
[
"MIT"
] | null | null | null |
JupyterNotebooks/ReadMe.ipynb
|
fawadkarimi/JupyterNotebooks2
|
323b679c0ae1fbf241e2a728c0568edc7d969617
|
[
"MIT"
] | 1 |
2018-09-06T05:01:31.000Z
|
2018-09-06T05:01:31.000Z
|
JupyterNotebooks/ReadMe.ipynb
|
daroox/python-scripts
|
323b679c0ae1fbf241e2a728c0568edc7d969617
|
[
"MIT"
] | null | null | null | 96.185915 | 13,656 | 0.855561 |
[
[
[
"# Read Me\nThe idea behind this *project* or Notebook is to create a script which loops over everything: folders and files and plots everything.\n\nShould loop over folders plot `.dat` or `.txt` files and then enter folders one by one and plot everything there",
"_____no_output_____"
]
],
[
[
"# to create folders in a loop\nimport os \nmonths = ['april', 'may', 'june', 'july']\nfor i in months:\n os.makedirs(i)",
"_____no_output_____"
]
],
[
[
"import os\nyear = os.listdir()\nyear",
"_____no_output_____"
],
[
"datFiles = []; txtFiles = []; matFiles = []; folders = []\n\nfor m in year:\n if m.endswith('.dat'):\n datFiles.append(m)\n if m.endswith('.mat'):\n matFiles.append(m)\n if m.endswith('.txt'):\n txtFiles.append(m)\n if os.path.isdir(m) and not (m.startswith('.')): # we dont want hidden folders strating with '.' for example: .folder\n folders.append(m)",
"_____no_output_____"
],
[
"folders",
"_____no_output_____"
],
[
"matFiles",
"_____no_output_____"
],
[
"len(matFiles)",
"_____no_output_____"
],
[
"import pandas as pd\n%matplotlib inline",
"_____no_output_____"
],
[
"if len(matFiles)> 0:\n for file in matFiles:\n data = pd.read_csv(file,sep=' ')\nelse:\n print('no matFiles found')",
"no matFiles found\n"
],
[
"if len(datFiles)> 0:\n for file in datFiles:\n data = pd.read_csv(file,sep=' ',names = ['a','b'])\n data.plot('a','b')\nelse:\n print('no datFiles found')",
"_____no_output_____"
],
[
"!head -7 data1.txt",
"a b\n1 2\n2 4\n5 6\n"
],
[
"if len(txtFiles)> 0:\n for file in txtFiles:\n data = pd.read_csv(file,sep=' ')\n data.plot('a','b')\nelse:\n print('no txtFiles found')",
"_____no_output_____"
],
[
"data.head()",
"_____no_output_____"
]
],
[
[
"I have created a `.txt` file in the folder `april`. Now I want to write a script which would access that `.txt` file but the script would execute from this folder!",
"_____no_output_____"
]
],
[
[
"ls .\\\\april",
"_____no_output_____"
]
],
[
[
"import os\npath ='.\\\\april'\nos.chdir(path)",
"_____no_output_____"
]
],
[
[
"pwd",
"_____no_output_____"
]
]
] |
[
"markdown",
"raw",
"code",
"markdown",
"raw",
"code",
"raw"
] |
[
[
"markdown"
],
[
"raw"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"raw"
],
[
"code"
],
[
"raw"
]
] |
cb8e9bb522a1ba035971a9a467ba51346bc3be2b
| 107,574 |
ipynb
|
Jupyter Notebook
|
SIRmodels.ipynb
|
rtwhite1546/COVID-19-FSRP
|
b4b76d0bc9793c29a4a2af9c980cc47fa03f158a
|
[
"MIT"
] | null | null | null |
SIRmodels.ipynb
|
rtwhite1546/COVID-19-FSRP
|
b4b76d0bc9793c29a4a2af9c980cc47fa03f158a
|
[
"MIT"
] | null | null | null |
SIRmodels.ipynb
|
rtwhite1546/COVID-19-FSRP
|
b4b76d0bc9793c29a4a2af9c980cc47fa03f158a
|
[
"MIT"
] | null | null | null | 106.298419 | 35,040 | 0.807937 |
[
[
[
"# import libraries\nimport numpy as np\nimport pandas as pd\nfrom numpy import genfromtxt\nimport math\nfrom scipy import optimize\nimport matplotlib.pyplot as plt\nimport csv\nimport sqlite3\nimport os\nimport urllib.request",
"_____no_output_____"
],
[
"# Function for the SIR model with two levels of alpha and beta -- O(n) speed\n#\n# INPUTS\n# \n# S0 - initial number of susceptible people\n# I0 - initial number of infected people\n# R0 - initial number of recovered people (including those who died)\n# alpha1 - initial recovery rate\n# alpha2 - later recovery rate\n# beta1 - initial contact rate\n# beta2 - later recovery rate\n# n1 - time when alpha transitions\n# n2 - time when beta transitions\n# n - amount of days to simulate\n#\n# OUTPUTS\n#\n# SIR - a 3-by-(n+1) matrix storing the simulated paths for S(t), I(t), and R(t) for t = 0, 1, 2, ..., n\n\ndef sir22(S0,I0,R0,alpha1,alpha2,beta1,beta2,n1,n2,n):\n SIR = np.zeros((3,n+1))\n \n # Fill in initial data\n SIR[:,0] = np.array([S0, I0, R0])\n \n alpha = alpha1\n beta = beta1\n \n for i in range(n):\n SIR[:,i+1] = SIR[:,i] + np.array([-beta*SIR[0,i]*SIR[1,i], beta*SIR[0,i]*SIR[1,i] - alpha*SIR[1,i], alpha*SIR[1,i]])\n \n if i is n1:\n alpha = alpha2\n \n if i is n2:\n beta = beta2\n \n return SIR",
"_____no_output_____"
],
[
"# Function for the standard SIR model -- O(n) speed\n#\n# INPUTS\n# \n# S0 - initial number of susceptible people\n# I0 - initial number of infected people\n# R0 - initial number of recovered people (including those who died)\n# alpha - recovery rate\n# beta - contact rate\n# n - amount of days to simulate\n#\n# OUTPUTS\n#\n# SIR - a 3-by-(n+1) matrix storing the simulated paths for S(t), I(t), and R(t) for t = 0, 1, 2, ..., n\n\ndef sir11(S0,I0,R0,alpha,beta,n):\n SIR = np.zeros((3,n+1))\n \n # Fill in initial data\n SIR[:,0] = np.array([S0, I0, R0])\n \n for i in range(n):\n SIR[:,i+1] = SIR[:,i] + np.array([-beta*SIR[0,i]*SIR[1,i], beta*SIR[0,i]*SIR[1,i] - alpha*SIR[1,i], alpha*SIR[1,i]])\n \n return SIR",
"_____no_output_____"
],
[
"# Function to compute the error between predicted data and real data\n#\n# INPUTS\n#\n# data - 3-by-(n+1) matrix of real data for S(t), I(t), R(t) at times t = 0, 1, ..., l\n# precition - 3-by-(n+1) matrix of simulated data for S(t), I(t), and R(t) for t = 0, 1, ..., l\n#\n# OUTPUTS\n#\n# error - the sum of squared differences between real and simulated data (using root of sum of squared L2 norms)\n\ndef findError(data,prediction):\n return math.sqrt(np.sum((data - prediction)**2))",
"_____no_output_____"
],
[
"# Read CSV file into dataframe and clean it\ndef cleanCSV(filename):\n # Read the csv file into a Pandas dataframe\n df = pd.read_csv(filename)\n\n # Replace slashes in header with _\n df.columns =[column.replace(\"/\", \"_\") for column in df.columns]\n \n return df",
"_____no_output_____"
],
[
"def downloadDataIntoCleanRows():\n # Download and clean confirmed cases by country/date\n url = 'https://raw.githubusercontent.com/CSSEGISandData/COVID-19/master/csse_covid_19_data/csse_covid_19_time_series/time_series_covid19_confirmed_global.csv'\n filename = 'time_series_covid19_confirmed_global.csv'\n urllib.request.urlretrieve(url, filename)\n confirmed = cleanCSV(filename)\n\n # Download and clean recovered cases by country/date\n url = 'https://raw.githubusercontent.com/CSSEGISandData/COVID-19/master/csse_covid_19_data/csse_covid_19_time_series/time_series_covid19_recovered_global.csv'\n filename = 'time_series_covid19_recovered_global.csv'\n urllib.request.urlretrieve(url, filename)\n recovered = cleanCSV(filename)\n\n # Download and clean deaths by country/date\n url = 'https://raw.githubusercontent.com/CSSEGISandData/COVID-19/master/csse_covid_19_data/csse_covid_19_time_series/time_series_covid19_deaths_global.csv'\n filename = 'time_series_covid19_deaths_global.csv'\n urllib.request.urlretrieve(url, filename)\n deaths = cleanCSV(filename)\n \n return confirmed, recovered, deaths",
"_____no_output_____"
],
[
"# Get data for queried country\ndef queryForCountry(confirmed,recovered,deaths,country):\n popData = pd.read_csv('WorldDevelopmentIndicatorsApr92020.csv')\n popData.columns =[column.replace(\" \", \"_\") for column in popData.columns]\n population = popData.query(\"Country_Name == @country\").to_numpy()[0,1]\n \n # Query for country data\n confirmed = confirmed.query(\"Country_Region == @country\")\n recovered = recovered.query(\"Country_Region == @country\")\n dead = deaths.query(\"Country_Region == @country\")\n \n # Prep data and drop columns when there are no cases\n confirmed = confirmed.drop(['Province_State', 'Country_Region', 'Lat', 'Long'], axis=1)\n confirmed = confirmed.to_numpy()[0]\n \n # Index of first case plus 9 days\n firstCase = np.nonzero(confirmed)[0][0] + 9\n print(firstCase)\n \n # Cut dates before the first infected case\n confirmed = confirmed[firstCase:]\n\n recovered = recovered.drop(['Province_State', 'Country_Region', 'Lat', 'Long'], axis=1)\n recovered = recovered.to_numpy()[0]\n recovered = recovered[firstCase:]\n\n dead = dead.drop(['Province_State', 'Country_Region', 'Lat', 'Long'], axis=1)\n dead = dead.to_numpy()[0]\n dead = dead[firstCase:]\n\n # Turn the data into S I R data\n R = recovered + dead\n I = confirmed - R\n S = population - I - R\n\n # Create data array\n data = np.vstack((S, I, R))\n\n # Convert data array to float\n data = np.array(list(data[:, :]), dtype=np.float64)\n\n # Find number of days of data\n lastData = np.size(data,1) - 1\n \n return data, lastData, firstCase",
"_____no_output_____"
],
[
"# Find values for the parameters minimizing the error\n\nconfirmed, recovered, deaths = downloadDataIntoCleanRows()\n\ndata, lastData, firstCase = queryForCountry(confirmed,recovered,deaths,'Jordan')\n\n# Read the initial data from the first column of data\nS0 = data[0,0]\nI0 = data[1,0]\nR0 = data[2,0]\n\n# Create a function of the parameters that runs the simulator and measures the error from the real data\nf = lambda x: findError(sir22(S0,I0,R0,x[0],x[1],x[2]/S0,x[3]/S0,18,10,lastData),data)",
"50\n"
],
[
"# Optimize the parameters (via gradient descent)\nresult = optimize.minimize(f,[0.1, 0.1, 0.2, 0.2],bounds = ((0,1),(0,1),(0,1),(0,1)))\n\n# Let x be the parameters found above\nx = result.x\n\n# Return details about the optimization\nresult",
"_____no_output_____"
],
[
"# Simulate SIR again with optimal parameters and plot it with the real data\nSIR = sir22(S0,I0,R0,x[0],x[1],x[2]/S0,x[3]/S0,18,10,200)\n\nfig, ax1 = plt.subplots()\n\nax1.set_xlabel('Time')\nax1.set_ylabel('Susceptible')\nax1.plot(SIR[0,:],color='tab:blue')\nax1.tick_params(axis='y')\n\nax2 = ax1.twinx() # instantiate a second axes that shares the same x-axis\n\nax2.set_ylabel('Infectious / Recovered') # we already handled the x-label with ax1\nax2.plot(SIR[1,:],color='tab:orange')\nax2.plot(SIR[2,:],color='tab:green')\nax2.tick_params(axis='y')\n\nax1.plot(data[0,:],'.',color='tab:blue')\nax2.plot(data[1,:],'.',color='tab:orange')\nax2.plot(data[2,:],'.',color='tab:green')\n\nax1.ticklabel_format(useOffset=False)\n\nfig.tight_layout() # otherwise the right y-label is slightly clipped\nplt.show()",
"_____no_output_____"
],
[
"plt.plot(SIR[0,:])\nplt.plot(SIR[1,:])\nplt.plot(SIR[2,:])\n\nplt.gca().set_prop_cycle(None)\n\nplt.plot(data[0,:],'.')\nplt.plot(data[1,:],'.')\nplt.plot(data[2,:],'.')",
"_____no_output_____"
],
[
"bestError = [0,0,0,0,0,0,1000000]\n\n# This code optimizes the parameters but also times when alpha and beta change\n# Note that it is brute force in the times, so this runs SLOWLY\n\nfor n1 in range(int(lastData/3),lastData):\n \n mError = bestError[-1]\n \n print(n1)\n for n2 in range(int(lastData/5),lastData):\n # optimize the alpha and beta parameters for given n1 and n2\n f = lambda x: findError(sir22(S0,I0,R0,x[0],x[1],x[2]/S0,x[3]/S0,n1,n2,lastData),data)\n result = optimize.minimize(f,[0.1, 0.1, 0.2, 0.2], bounds = ((0,1),(0,1),(0,1),(0,1)))\n \n # If we find a lower error than we found previously, record the parameters and error\n if result.fun < bestError[-1]:\n x = result.x\n bestError = [x[0], x[1], x[2], x[3], n1, n2, result.fun]\n \n if bestError[-1] == mError:\n break\n \n # Display the best parameters and error we found so far\n print(bestError)",
"12\n[0.0, 0.055440692479481406, 0.5627167455394336, 0.04435742399958146, 12, 11, 252.8106338745577]\n13\n[0.0, 0.05869163949938361, 0.5604921626134632, 0.04483960083109135, 13, 11, 234.61793279662672]\n14\n[0.0, 0.06222341857779198, 0.5584069535192409, 0.04517744453643131, 14, 11, 217.7068142931486]\n15\n[0.0, 0.06610434446365042, 0.5564864533640101, 0.04536670027532379, 15, 11, 202.18593504415728]\n16\n[0.0, 0.07167614546875158, 0.6055818492631427, 0.04843963043508369, 16, 10, 189.79811991833478]\n17\n[0.00023039803847530046, 0.07746963019478668, 0.6672172453343894, 0.0508557233767315, 17, 9, 188.11772956283]\n18\n[0.007093195936275441, 0.07774867076730349, 0.6117010657109857, 0.04764959634242392, 18, 10, 176.88260915505188]\n19\n[0.013580298811566607, 0.07784351532388996, 0.5672124201508237, 0.04419987472828944, 19, 11, 173.93406814647855]\n20\n[0.017243897256569815, 0.08040351840049202, 0.5710721885272736, 0.04376283570061301, 20, 11, 172.7672953609359]\n21\n"
],
[
"# plot with the optimal parameters\nSIR = sir22(S0,I0,R0,bestError[0],bestError[1],bestError[2]/S0,bestError[3]/S0,bestError[4],bestError[5],100)\n\n#fig = plt.figure(figsize=(10, 8), dpi= 80, facecolor='w', edgecolor='k')\n\nfig, ax1 = plt.subplots()\n\nax1.set_xlabel('Time')\nax1.set_ylabel('Susceptible (blue)')\nax1.plot(SIR[0,:],color='tab:blue')\nax1.tick_params(axis='y')\n\nax2 = ax1.twinx() # instantiate a second axes that shares the same x-axis\n\nax2.set_ylabel('Infectious (orange) / Recovered (green)') # we already handled the x-label with ax1\nax2.plot(SIR[1,:],color='tab:orange')\nax2.plot(SIR[2,:],color='tab:green')\nax2.tick_params(axis='y')\n\nax1.plot(data[0,:],'.',color='tab:blue')\nax2.plot(data[1,:],'.',color='tab:orange')\nax2.plot(data[2,:],'.',color='tab:green')\n\n#ax1.set_ylim([9719000,9720000])\nax1.ticklabel_format(useOffset=False)\n\nfig.tight_layout() # otherwise the right y-label is slightly clipped\nplt.show()\n\n#plt.plot(SIR[0,:])\n#plt.plot(SIR[1,:])\n#plt.plot(SIR[2,:])\n\n#plt.gca().set_prop_cycle(None)\n\n#plt.plot(data[0,:],'.')\n#plt.plot(data[1,:],'.')\n#plt.plot(data[2,:],'.')",
"_____no_output_____"
],
[
"## FIND BEST PARAMETERS FOR COUNTRIES IN THE LIST\n\n# Download data and put into rows\nconfirmed, recovered, deaths = downloadDataIntoCleanRows()\n\ncountries = ['Algeria', 'Bahrain', 'Cyprus', 'Egypt', 'Iran', 'Iraq', 'Israel', 'Jordan', 'Lebanon', 'Morocco', 'Oman', 'Qatar', 'Saudi Arabia', 'Tunisia', 'Turkey', 'United Arab Emirates', 'West Bank and Gaza']\n\n#countries = ['Algeria', 'Bahrain', 'Cyprus']\n\nerrorStore = []\n\nfor country in countries:\n print(country)\n \n data, lastData, firstCase = queryForCountry(confirmed,recovered,deaths,country)\n\n # Read the initial data from the first column of data\n S0 = data[0,0]\n I0 = data[1,0]\n R0 = data[2,0]\n \n # bestError = [country,firstCase, alpha1, alpha2, beta1, beta2, n1, n2, error]\n bestError = ['a',0,0,0,0,0,0,0,1000000]\n\n # This code optimizes the parameters but also times when alpha and beta change\n # Note that it is brute force in the times, so this runs SLOWLY\n\n for n1 in range(int(lastData/4),lastData):\n\n mError = bestError[-1]\n\n print(n1)\n for n2 in range(int(lastData/5),lastData):\n # optimize the alpha and beta parameters for given n1 and n2\n f = lambda x: findError(sir22(S0,I0,R0,x[0],x[1],x[2]/S0,x[3]/S0,n1,n2,lastData),data)\n result = optimize.minimize(f,[0.1, 0.1, 0.2, 0.2], bounds = ((0,1),(0,1),(0,1),(0,1)))\n\n # If we find a lower error than we found previously, record the parameters and error\n if result.fun < bestError[-1]:\n x = result.x\n bestError = [firstCase, x[0], x[1], x[2], x[3], n1, n2, result.fun]\n\n if bestError[-1] == mError:\n break\n \n errorStore.append(bestError)",
"Algeria\n10\n11\nBahrain\n11\n12\nCyprus\n7\n8\n9\n10\n11\n12\n13\n14\n15\n16\n17\nEgypt\n13\n14\nIran\n12\n13\nIraq\n11\n12\n13\n14\n15\n16\n17\n18\n19\n20\n21\n22\n23\n24\n25\n26\n27\n28\nIsrael\n11\n12\n13\n14\n15\n16\n17\n18\n19\n20\n21\n22\n23\n24\n25\n26\n27\nJordan\n9\n10\n11\n12\n13\n14\n15\n16\n17\n18\n19\n20\n21\nLebanon\n11\n12\nMorocco\n9\n10\nOman\n11\n12\nQatar\n9\n10\n11\n12\n13\n14\nSaudi Arabia\n9\n10\n11\n12\n13\n14\n15\n16\n17\n18\n19\n20\n21\n22\n23\n24\n25\n26\n27\n28\nTunisia\n8\n9\n10\n11\nTurkey\n7\n8\n9\nUnited Arab Emirates\n17\n18\nWest Bank and Gaza\n8\n9\n"
],
[
"df = pd.DataFrame(data=errorStore,index=countries,columns=['day of first case+10', 'initial alpha', 'final alpha', 'initial beta', 'final beta', 'alpha switchover', 'beta switchover', 'error'])",
"_____no_output_____"
],
[
"df",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb8eae567d156f4ec2f42b5c1b166fc79896504a
| 565 |
ipynb
|
Jupyter Notebook
|
1_introduction/1_HelloWorld.ipynb
|
AlbeBertin/evolution
|
06b8971c1df0fb08b18d011f67634a3bdcbc9ed4
|
[
"Apache-2.0"
] | null | null | null |
1_introduction/1_HelloWorld.ipynb
|
AlbeBertin/evolution
|
06b8971c1df0fb08b18d011f67634a3bdcbc9ed4
|
[
"Apache-2.0"
] | null | null | null |
1_introduction/1_HelloWorld.ipynb
|
AlbeBertin/evolution
|
06b8971c1df0fb08b18d011f67634a3bdcbc9ed4
|
[
"Apache-2.0"
] | null | null | null | 15.27027 | 35 | 0.476106 |
[
[
[
"empty"
]
]
] |
[
"empty"
] |
[
[
"empty"
]
] |
cb8eb197d4c263a375047ea40d517c2992eab97c
| 2,445 |
ipynb
|
Jupyter Notebook
|
nbs/dl1/telegram_notify_test.ipynb
|
hwasiti/fastai-course-v3
|
9401277f830035caf2b6c753e063d868f30408c1
|
[
"Apache-2.0"
] | 1 |
2019-01-13T14:06:17.000Z
|
2019-01-13T14:06:17.000Z
|
nbs/dl1/telegram_notify_test.ipynb
|
hwasiti/fastai-course-v3
|
9401277f830035caf2b6c753e063d868f30408c1
|
[
"Apache-2.0"
] | null | null | null |
nbs/dl1/telegram_notify_test.ipynb
|
hwasiti/fastai-course-v3
|
9401277f830035caf2b6c753e063d868f30408c1
|
[
"Apache-2.0"
] | 2 |
2019-01-16T02:15:20.000Z
|
2019-12-07T16:37:56.000Z
| 20.546218 | 91 | 0.478119 |
[
[
[
"import sys\nsys.executable",
"_____no_output_____"
],
[
"%connect_info",
"{\n \"shell_port\": 3435,\n \"iopub_port\": 3436,\n \"stdin_port\": 3437,\n \"control_port\": 3438,\n \"hb_port\": 3439,\n \"ip\": \"127.0.0.1\",\n \"key\": \"634acea5-84371de725ee3f1e211f462c\",\n \"transport\": \"tcp\",\n \"signature_scheme\": \"hmac-sha256\",\n \"kernel_name\": \"\"\n}\n\nPaste the above JSON into a file, and connect with:\n $> jupyter <app> --existing <file>\nor, if you are local, you can connect with just:\n $> jupyter <app> --existing kernel-5f307a2f-9b4f-4231-9636-b19f029affe5.json\nor even just:\n $> jupyter <app> --existing\nif this is the most recent Jupyter kernel you have started.\n"
],
[
"from my_python_tricks import *",
"_____no_output_____"
],
[
"a= [4,5,6,7]",
"_____no_output_____"
],
[
"brkpt()",
"_____no_output_____"
],
[
"a= [10, 11]",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb8ec580cd1cb5953c976f1dceec15ecbc84b588
| 28,371 |
ipynb
|
Jupyter Notebook
|
processing_xgboost.ipynb
|
melanie531/amazon-sagemaker-immersion-day-custom
|
e0bbf60b92419579f527ba10d92b9a40a731c6d8
|
[
"MIT-0"
] | 2 |
2021-09-13T08:13:56.000Z
|
2021-11-19T00:42:34.000Z
|
processing_xgboost.ipynb
|
dgallitelli/amazon-sagemaker-immersion-day
|
e4ad58c179dba3eb00ec501026cd5d75127b7cb5
|
[
"MIT-0"
] | null | null | null |
processing_xgboost.ipynb
|
dgallitelli/amazon-sagemaker-immersion-day
|
e4ad58c179dba3eb00ec501026cd5d75127b7cb5
|
[
"MIT-0"
] | 1 |
2021-11-17T08:28:11.000Z
|
2021-11-17T08:28:11.000Z
| 40.704448 | 793 | 0.621374 |
[
[
[
"# Targeting Direct Marketing with Amazon SageMaker XGBoost\n_**Supervised Learning with Gradient Boosted Trees: A Binary Prediction Problem With Unbalanced Classes**_\n\n---\n\n## Background\nDirect marketing, either through mail, email, phone, etc., is a common tactic to acquire customers. Because resources and a customer's attention is limited, the goal is to only target the subset of prospects who are likely to engage with a specific offer. Predicting those potential customers based on readily available information like demographics, past interactions, and environmental factors is a common machine learning problem.\n\nThis notebook presents an example problem to predict if a customer will enroll for a term deposit at a bank, after one or more phone calls. The steps include:\n\n* Preparing your Amazon SageMaker notebook\n* Downloading data from the internet into Amazon SageMaker\n* Investigating and transforming the data so that it can be fed to Amazon SageMaker algorithms\n* Estimating a model using the Gradient Boosting algorithm\n* Evaluating the effectiveness of the model\n* Setting the model up to make on-going predictions\n\n---\n\n## Preparation\n\n_This notebook was created and tested on an ml.m4.xlarge notebook instance._\n\nLet's start by specifying:\n\n- The S3 bucket and prefix that you want to use for training and model data. This should be within the same region as the Notebook Instance, training, and hosting.\n- The IAM role arn used to give training and hosting access to your data. See the documentation for how to create these. Note, if more than one role is required for notebook instances, training, and/or hosting, please replace the boto regexp with a the appropriate full IAM role arn string(s).",
"_____no_output_____"
]
],
[
[
"# cell 01\nimport sagemaker\nbucket=sagemaker.Session().default_bucket()\nprefix = 'sagemaker/DEMO-xgboost-dm'\n \n# Define IAM role\nimport boto3\nimport re\nfrom sagemaker import get_execution_role\n\nrole = get_execution_role()",
"_____no_output_____"
]
],
[
[
"Now let's bring in the Python libraries that we'll use throughout the analysis",
"_____no_output_____"
]
],
[
[
"# cell 02\nimport numpy as np # For matrix operations and numerical processing\nimport pandas as pd # For munging tabular data\nimport matplotlib.pyplot as plt # For charts and visualizations\nfrom IPython.display import Image # For displaying images in the notebook\nfrom IPython.display import display # For displaying outputs in the notebook\nfrom time import gmtime, strftime # For labeling SageMaker models, endpoints, etc.\nimport sys # For writing outputs to notebook\nimport math # For ceiling function\nimport json # For parsing hosting outputs\nimport os # For manipulating filepath names\nimport sagemaker \nimport zipfile # Amazon SageMaker's Python SDK provides many helper functions",
"_____no_output_____"
]
],
[
[
"---\n\n## Data\nLet's start by downloading the [direct marketing dataset](https://sagemaker-sample-data-us-west-2.s3-us-west-2.amazonaws.com/autopilot/direct_marketing/bank-additional.zip) from the sample data s3 bucket. \n\n\\[Moro et al., 2014\\] S. Moro, P. Cortez and P. Rita. A Data-Driven Approach to Predict the Success of Bank Telemarketing. Decision Support Systems, Elsevier, 62:22-31, June 2014\n",
"_____no_output_____"
]
],
[
[
"# cell 03\n!wget https://sagemaker-sample-data-us-west-2.s3-us-west-2.amazonaws.com/autopilot/direct_marketing/bank-additional.zip\n\nwith zipfile.ZipFile('bank-additional.zip', 'r') as zip_ref:\n zip_ref.extractall('.')",
"_____no_output_____"
]
],
[
[
"Now lets read this into a Pandas data frame and take a look.",
"_____no_output_____"
]
],
[
[
"# cell 04\ndata = pd.read_csv('./bank-additional/bank-additional-full.csv')\npd.set_option('display.max_columns', 500) # Make sure we can see all of the columns\npd.set_option('display.max_rows', 20) # Keep the output on one page\ndata",
"_____no_output_____"
]
],
[
[
"We will store this natively in S3 to then process it with SageMaker Processing.",
"_____no_output_____"
]
],
[
[
"# cell 05\nfrom sagemaker import Session\n\nsess = Session()\ninput_source = sess.upload_data('./bank-additional/bank-additional-full.csv', bucket=bucket, key_prefix=f'{prefix}/input_data')\ninput_source",
"_____no_output_____"
]
],
[
[
"# Feature Engineering with Amazon SageMaker Processing\n\nAmazon SageMaker Processing allows you to run steps for data pre- or post-processing, feature engineering, data validation, or model evaluation workloads on Amazon SageMaker. Processing jobs accept data from Amazon S3 as input and store data into Amazon S3 as output.\n\n\n\nHere, we'll import the dataset and transform it with SageMaker Processing, which can be used to process terabytes of data in a SageMaker-managed cluster separate from the instance running your notebook server. In a typical SageMaker workflow, notebooks are only used for prototyping and can be run on relatively inexpensive and less powerful instances, while processing, training and model hosting tasks are run on separate, more powerful SageMaker-managed instances. SageMaker Processing includes off-the-shelf support for Scikit-learn, as well as a Bring Your Own Container option, so it can be used with many different data transformation technologies and tasks. \n\nTo use SageMaker Processing, simply supply a Python data preprocessing script as shown below. For this example, we're using a SageMaker prebuilt Scikit-learn container, which includes many common functions for processing data. There are few limitations on what kinds of code and operations you can run, and only a minimal contract: input and output data must be placed in specified directories. If this is done, SageMaker Processing automatically loads the input data from S3 and uploads transformed data back to S3 when the job is complete.",
"_____no_output_____"
]
],
[
[
"# cell 06\n%%writefile preprocessing.py\n\nimport pandas as pd\nimport numpy as np\nimport argparse\nimport os\nfrom sklearn.preprocessing import OrdinalEncoder\n\ndef _parse_args():\n\n parser = argparse.ArgumentParser()\n\n # Data, model, and output directories\n # model_dir is always passed in from SageMaker. By default this is a S3 path under the default bucket.\n parser.add_argument('--filepath', type=str, default='/opt/ml/processing/input/')\n parser.add_argument('--filename', type=str, default='bank-additional-full.csv')\n parser.add_argument('--outputpath', type=str, default='/opt/ml/processing/output/')\n parser.add_argument('--categorical_features', type=str, default='y, job, marital, education, default, housing, loan, contact, month, day_of_week, poutcome')\n\n return parser.parse_known_args()\n\nif __name__==\"__main__\":\n # Process arguments\n args, _ = _parse_args()\n # Load data\n df = pd.read_csv(os.path.join(args.filepath, args.filename))\n # Change the value . into _\n df = df.replace(regex=r'\\.', value='_')\n df = df.replace(regex=r'\\_$', value='')\n # Add two new indicators\n df[\"no_previous_contact\"] = (df[\"pdays\"] == 999).astype(int)\n df[\"not_working\"] = df[\"job\"].isin([\"student\", \"retired\", \"unemployed\"]).astype(int)\n df = df.drop(['duration', 'emp.var.rate', 'cons.price.idx', 'cons.conf.idx', 'euribor3m', 'nr.employed'], axis=1)\n # Encode the categorical features\n df = pd.get_dummies(df)\n # Train, test, validation split\n train_data, validation_data, test_data = np.split(df.sample(frac=1, random_state=42), [int(0.7 * len(df)), int(0.9 * len(df))]) # Randomly sort the data then split out first 70%, second 20%, and last 10%\n # Local store\n pd.concat([train_data['y_yes'], train_data.drop(['y_yes','y_no'], axis=1)], axis=1).to_csv(os.path.join(args.outputpath, 'train/train.csv'), index=False, header=False)\n pd.concat([validation_data['y_yes'], validation_data.drop(['y_yes','y_no'], axis=1)], axis=1).to_csv(os.path.join(args.outputpath, 'validation/validation.csv'), index=False, header=False)\n test_data['y_yes'].to_csv(os.path.join(args.outputpath, 'test/test_y.csv'), index=False, header=False)\n test_data.drop(['y_yes','y_no'], axis=1).to_csv(os.path.join(args.outputpath, 'test/test_x.csv'), index=False, header=False)\n print(\"## Processing complete. Exiting.\")",
"_____no_output_____"
]
],
[
[
"Before starting the SageMaker Processing job, we instantiate a `SKLearnProcessor` object. This object allows you to specify the instance type to use in the job, as well as how many instances.",
"_____no_output_____"
]
],
[
[
"# cell 07\ntrain_path = f\"s3://{bucket}/{prefix}/train\"\nvalidation_path = f\"s3://{bucket}/{prefix}/validation\"\ntest_path = f\"s3://{bucket}/{prefix}/test\"",
"_____no_output_____"
],
[
"# cell 08\nfrom sagemaker.sklearn.processing import SKLearnProcessor\nfrom sagemaker.processing import ProcessingInput, ProcessingOutput\nfrom sagemaker import get_execution_role\n\n\nsklearn_processor = SKLearnProcessor(\n framework_version=\"0.23-1\",\n role=get_execution_role(),\n instance_type=\"ml.m5.large\",\n instance_count=1, \n base_job_name='sm-immday-skprocessing'\n)\n\nsklearn_processor.run(\n code='preprocessing.py',\n # arguments = ['arg1', 'arg2'],\n inputs=[\n ProcessingInput(\n source=input_source, \n destination=\"/opt/ml/processing/input\",\n s3_input_mode=\"File\",\n s3_data_distribution_type=\"ShardedByS3Key\"\n )\n ],\n outputs=[\n ProcessingOutput(\n output_name=\"train_data\", \n source=\"/opt/ml/processing/output/train\",\n destination=train_path,\n ),\n ProcessingOutput(output_name=\"validation_data\", source=\"/opt/ml/processing/output/validation\", destination=validation_path),\n ProcessingOutput(output_name=\"test_data\", source=\"/opt/ml/processing/output/test\", destination=test_path),\n ]\n)",
"_____no_output_____"
],
[
"# cell 09\n!aws s3 ls $train_path/",
"2021-06-10 17:39:22 3545009 train.csv\n"
]
],
[
[
"---\n\n## End of Lab 1\n",
"_____no_output_____"
],
[
"---\n\n## Training\nNow we know that most of our features have skewed distributions, some are highly correlated with one another, and some appear to have non-linear relationships with our target variable. Also, for targeting future prospects, good predictive accuracy is preferred to being able to explain why that prospect was targeted. Taken together, these aspects make gradient boosted trees a good candidate algorithm.\n\nThere are several intricacies to understanding the algorithm, but at a high level, gradient boosted trees works by combining predictions from many simple models, each of which tries to address the weaknesses of the previous models. By doing this the collection of simple models can actually outperform large, complex models. Other Amazon SageMaker notebooks elaborate on gradient boosting trees further and how they differ from similar algorithms.\n\n`xgboost` is an extremely popular, open-source package for gradient boosted trees. It is computationally powerful, fully featured, and has been successfully used in many machine learning competitions. Let's start with a simple `xgboost` model, trained using Amazon SageMaker's managed, distributed training framework.\n\nFirst we'll need to specify the ECR container location for Amazon SageMaker's implementation of XGBoost.",
"_____no_output_____"
]
],
[
[
"# cell 10\ncontainer = sagemaker.image_uris.retrieve(region=boto3.Session().region_name, framework='xgboost', version='latest')",
"_____no_output_____"
]
],
[
[
"Then, because we're training with the CSV file format, we'll create `s3_input`s that our training function can use as a pointer to the files in S3, which also specify that the content type is CSV.",
"_____no_output_____"
]
],
[
[
"# cell 11\ns3_input_train = sagemaker.inputs.TrainingInput(s3_data=train_path.format(bucket, prefix), content_type='csv')\ns3_input_validation = sagemaker.inputs.TrainingInput(s3_data=validation_path.format(bucket, prefix), content_type='csv')",
"_____no_output_____"
]
],
[
[
"First we'll need to specify training parameters to the estimator. This includes:\n1. The `xgboost` algorithm container\n1. The IAM role to use\n1. Training instance type and count\n1. S3 location for output data\n1. Algorithm hyperparameters\n\nAnd then a `.fit()` function which specifies:\n1. S3 location for output data. In this case we have both a training and validation set which are passed in.",
"_____no_output_____"
]
],
[
[
"# cell 12\nsess = sagemaker.Session()\n\nxgb = sagemaker.estimator.Estimator(container,\n role, \n instance_count=1, \n instance_type='ml.m4.xlarge',\n output_path='s3://{}/{}/output'.format(bucket, prefix),\n sagemaker_session=sess)\nxgb.set_hyperparameters(max_depth=5,\n eta=0.2,\n gamma=4,\n min_child_weight=6,\n subsample=0.8,\n silent=0,\n objective='binary:logistic',\n num_round=100)\n\nxgb.fit({'train': s3_input_train, 'validation': s3_input_validation}) ",
"_____no_output_____"
]
],
[
[
"---\n\n## Hosting\nNow that we've trained the `xgboost` algorithm on our data, let's deploy a model that's hosted behind a real-time endpoint.",
"_____no_output_____"
]
],
[
[
"# cell 13\nxgb_predictor = xgb.deploy(initial_instance_count=1,\n instance_type='ml.m4.xlarge')",
"_____no_output_____"
]
],
[
[
"---\n\n## Evaluation\nThere are many ways to compare the performance of a machine learning model, but let's start by simply comparing actual to predicted values. In this case, we're simply predicting whether the customer subscribed to a term deposit (`1`) or not (`0`), which produces a simple confusion matrix.\n\nFirst we'll need to determine how we pass data into and receive data from our endpoint. Our data is currently stored as NumPy arrays in memory of our notebook instance. To send it in an HTTP POST request, we'll serialize it as a CSV string and then decode the resulting CSV.\n\n*Note: For inference with CSV format, SageMaker XGBoost requires that the data does NOT include the target variable.*",
"_____no_output_____"
]
],
[
[
"# cell 14\nxgb_predictor.serializer = sagemaker.serializers.CSVSerializer()",
"_____no_output_____"
]
],
[
[
"Now, we'll use a simple function to:\n1. Loop over our test dataset\n1. Split it into mini-batches of rows \n1. Convert those mini-batches to CSV string payloads (notice, we drop the target variable from our dataset first)\n1. Retrieve mini-batch predictions by invoking the XGBoost endpoint\n1. Collect predictions and convert from the CSV output our model provides into a NumPy array",
"_____no_output_____"
]
],
[
[
"# cell 15\n!aws s3 cp $test_path/test_x.csv /tmp/test_x.csv\n!aws s3 cp $test_path/test_y.csv /tmp/test_y.csv",
"_____no_output_____"
],
[
"# cell 16\ndef predict(data, predictor, rows=500 ):\n split_array = np.array_split(data, int(data.shape[0] / float(rows) + 1))\n predictions = ''\n for array in split_array:\n predictions = ','.join([predictions, predictor.predict(array).decode('utf-8')])\n\n return np.fromstring(predictions[1:], sep=',')\n\ntest_x = pd.read_csv('/tmp/test_x.csv', names=[f'{i}' for i in range(59)])\ntest_y = pd.read_csv('/tmp/test_y.csv', names=['y'])\npredictions = predict(test_x.drop(test_x.columns[0], axis=1).to_numpy(), xgb_predictor)",
"_____no_output_____"
]
],
[
[
"Now we'll check our confusion matrix to see how well we predicted versus actuals.",
"_____no_output_____"
]
],
[
[
"# cell 17\npd.crosstab(index=test_y['y'].values, columns=np.round(predictions), rownames=['actuals'], colnames=['predictions'])",
"_____no_output_____"
]
],
[
[
"So, of the ~4000 potential customers, we predicted 136 would subscribe and 94 of them actually did. We also had 389 subscribers who subscribed that we did not predict would. This is less than desirable, but the model can (and should) be tuned to improve this. Most importantly, note that with minimal effort, our model produced accuracies similar to those published [here](http://media.salford-systems.com/video/tutorial/2015/targeted_marketing.pdf).\n\n_Note that because there is some element of randomness in the algorithm's subsample, your results may differ slightly from the text written above._",
"_____no_output_____"
],
[
"## Automatic model Tuning (optional)\nAmazon SageMaker automatic model tuning, also known as hyperparameter tuning, finds the best version of a model by running many training jobs on your dataset using the algorithm and ranges of hyperparameters that you specify. It then chooses the hyperparameter values that result in a model that performs the best, as measured by a metric that you choose.\nFor example, suppose that you want to solve a binary classification problem on this marketing dataset. Your goal is to maximize the area under the curve (auc) metric of the algorithm by training an XGBoost Algorithm model. You don't know which values of the eta, alpha, min_child_weight, and max_depth hyperparameters to use to train the best model. To find the best values for these hyperparameters, you can specify ranges of values that Amazon SageMaker hyperparameter tuning searches to find the combination of values that results in the training job that performs the best as measured by the objective metric that you chose. Hyperparameter tuning launches training jobs that use hyperparameter values in the ranges that you specified, and returns the training job with highest auc.\n",
"_____no_output_____"
]
],
[
[
"# cell 18\nfrom sagemaker.tuner import IntegerParameter, CategoricalParameter, ContinuousParameter, HyperparameterTuner\nhyperparameter_ranges = {'eta': ContinuousParameter(0, 1),\n 'min_child_weight': ContinuousParameter(1, 10),\n 'alpha': ContinuousParameter(0, 2),\n 'max_depth': IntegerParameter(1, 10)}\n",
"_____no_output_____"
],
[
"# cell 19\nobjective_metric_name = 'validation:auc'",
"_____no_output_____"
],
[
"# cell 20\ntuner = HyperparameterTuner(xgb,\n objective_metric_name,\n hyperparameter_ranges,\n max_jobs=20,\n max_parallel_jobs=3)\n",
"_____no_output_____"
],
[
"# cell 21\ntuner.fit({'train': s3_input_train, 'validation': s3_input_validation})",
"_____no_output_____"
],
[
"# cell 22\nboto3.client('sagemaker').describe_hyper_parameter_tuning_job(\nHyperParameterTuningJobName=tuner.latest_tuning_job.job_name)['HyperParameterTuningJobStatus']",
"_____no_output_____"
],
[
"# cell 23\n# return the best training job name\ntuner.best_training_job()",
"_____no_output_____"
],
[
"# cell 24\n# Deploy the best trained or user specified model to an Amazon SageMaker endpoint\ntuner_predictor = tuner.deploy(initial_instance_count=1,\n instance_type='ml.m4.xlarge')",
"_____no_output_____"
],
[
"# cell 25\n# Create a serializer\ntuner_predictor.serializer = sagemaker.serializers.CSVSerializer()",
"_____no_output_____"
],
[
"# cell 26\n# Predict\npredictions = predict(test_x.to_numpy(),tuner_predictor)",
"_____no_output_____"
],
[
"# cell 27\n# Collect predictions and convert from the CSV output our model provides into a NumPy array\npd.crosstab(index=test_y['y'].values, columns=np.round(predictions), rownames=['actuals'], colnames=['predictions'])",
"_____no_output_____"
]
],
[
[
"---\n\n## Extensions\n\nThis example analyzed a relatively small dataset, but utilized Amazon SageMaker features such as distributed, managed training and real-time model hosting, which could easily be applied to much larger problems. In order to improve predictive accuracy further, we could tweak value we threshold our predictions at to alter the mix of false-positives and false-negatives, or we could explore techniques like hyperparameter tuning. In a real-world scenario, we would also spend more time engineering features by hand and would likely look for additional datasets to include which contain customer information not available in our initial dataset.",
"_____no_output_____"
],
[
"### (Optional) Clean-up\n\nIf you are done with this notebook, please run the cell below. This will remove the hosted endpoint you created and avoid any charges from a stray instance being left on.",
"_____no_output_____"
]
],
[
[
"# cell 28\nxgb_predictor.delete_endpoint(delete_endpoint_config=True)",
"_____no_output_____"
],
[
"# cell 29\ntuner_predictor.delete_endpoint(delete_endpoint_config=True)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
]
] |
cb8ec7b3dbb5432d69dc145f87e109c9e2b95892
| 103,877 |
ipynb
|
Jupyter Notebook
|
notebooks/1-RedCard-EDA/1-Redcard-Dataset.ipynb
|
stiero/legendary-tribble
|
8574b3b41f546fd3b2634248fe490cee9b0b1bea
|
[
"MIT"
] | null | null | null |
notebooks/1-RedCard-EDA/1-Redcard-Dataset.ipynb
|
stiero/legendary-tribble
|
8574b3b41f546fd3b2634248fe490cee9b0b1bea
|
[
"MIT"
] | null | null | null |
notebooks/1-RedCard-EDA/1-Redcard-Dataset.ipynb
|
stiero/legendary-tribble
|
8574b3b41f546fd3b2634248fe490cee9b0b1bea
|
[
"MIT"
] | null | null | null | 31.563962 | 596 | 0.390414 |
[
[
[
"# Redcard Exploratory Data Analysis\n\nThis dataset is taken from a fantastic paper that looks to see how analytical choices made by different data science teams on the same dataset in an attempt to answer the same research question affect the final outcome.\n\n[Many analysts, one dataset: Making transparent how variations in analytical choices affect results](https://osf.io/gvm2z/)\n\nThe data can be found [here](https://osf.io/47tnc/).\n\n",
"_____no_output_____"
],
[
"## The Task\n\nDo an Exploratory Data Analysis on the redcard dataset. Keeping in mind the question is the following: **Are soccer referees more likely to give red cards to dark-skin-toned players than light-skin-toned players?**\n",
"_____no_output_____"
]
],
[
[
"!conda install -c conda-forge pandas-profiling -y \n\n!pip install missingno",
"Fetching package metadata ...........\nSolving package specifications: .\n\nPackage plan for installation in environment /home/nbcommon/anaconda3_410:\n\nThe following NEW packages will be INSTALLED:\n\n pandas-profiling: 1.3.0-py35_0 conda-forge\n\nThe following packages will be SUPERSEDED by a higher-priority channel:\n\n conda: 4.3.17-py35_0 --> 4.2.13-py35_0 conda-forge\n conda-env: 2.6.0-0 --> 2.6.0-0 conda-forge\n\nconda-env-2.6. 100% |################################| Time: 0:00:00 1.17 MB/s\nconda-4.2.13-p 100% |################################| Time: 0:00:00 1.44 MB/s\npandas-profili 100% |################################| Time: 0:00:00 663.79 kB/s\nCollecting missingno\n Downloading missingno-0.3.5.tar.gz\nRequirement already satisfied (use --upgrade to upgrade): numpy in /home/nbcommon/anaconda3_410/lib/python3.5/site-packages (from missingno)\nRequirement already satisfied (use --upgrade to upgrade): matplotlib in /home/nbcommon/anaconda3_410/lib/python3.5/site-packages (from missingno)\nRequirement already satisfied (use --upgrade to upgrade): scipy in /home/nbcommon/anaconda3_410/lib/python3.5/site-packages (from missingno)\nRequirement already satisfied (use --upgrade to upgrade): seaborn in /home/nbcommon/anaconda3_410/lib/python3.5/site-packages (from missingno)\nRequirement already satisfied (use --upgrade to upgrade): python-dateutil in /home/nbcommon/anaconda3_410/lib/python3.5/site-packages (from matplotlib->missingno)\nRequirement already satisfied (use --upgrade to upgrade): pytz in /home/nbcommon/anaconda3_410/lib/python3.5/site-packages (from matplotlib->missingno)\nRequirement already satisfied (use --upgrade to upgrade): cycler in /home/nbcommon/anaconda3_410/lib/python3.5/site-packages (from matplotlib->missingno)\nRequirement already satisfied (use --upgrade to upgrade): pyparsing!=2.0.4,>=1.5.6 in /home/nbcommon/anaconda3_410/lib/python3.5/site-packages (from matplotlib->missingno)\nRequirement already satisfied (use --upgrade to upgrade): six>=1.5 in /home/nbcommon/anaconda3_410/lib/python3.5/site-packages (from python-dateutil->matplotlib->missingno)\nBuilding wheels for collected packages: missingno\n Running setup.py bdist_wheel for missingno ... \u001b[?25l-\b \b\\\b \bdone\n\u001b[?25h Stored in directory: /home/nbuser/.cache/pip/wheels/f3/9f/31/38d2fad2bd1ac3ac70a2d159c61515de5825296429a4f13056\nSuccessfully built missingno\nInstalling collected packages: missingno\nSuccessfully installed missingno-0.3.5\n\u001b[33mYou are using pip version 8.1.2, however version 9.0.1 is available.\nYou should consider upgrading via the 'pip install --upgrade pip' command.\u001b[0m\n"
],
[
"from __future__ import absolute_import, division, print_function\n\n\n%matplotlib inline\n%config InlineBackend.figure_format='retina'\n\nimport matplotlib as mpl\nfrom matplotlib import pyplot as plt\nfrom matplotlib.pyplot import GridSpec\nimport seaborn as sns\nimport numpy as np\nimport pandas as pd\nimport os, sys\nfrom tqdm import tqdm\nimport warnings\nwarnings.filterwarnings('ignore')\nsns.set_context(\"poster\", font_scale=1.3)\n\nimport missingno as msno\nimport pandas_profiling\n\nfrom sklearn.datasets import make_blobs\nimport time",
"_____no_output_____"
]
],
[
[
"## About the Data\n\n> The dataset is available as a list with 146,028 dyads of players and referees and includes details from players, details from referees and details regarding the interactions of player-referees. A summary of the variables of interest can be seen below. A detailed description of all variables included can be seen in the README file on the project website. \n\n> From a company for sports statistics, we obtained data and profile photos from all soccer players (N = 2,053) playing in the first male divisions of England, Germany, France and Spain in the 2012-2013 season and all referees (N = 3,147) that these players played under in their professional career (see Figure 1). We created a dataset of playerâreferee dyads including the number of matches players and referees encountered each other and our dependent variable, the number of red cards given to a player by a particular referee throughout all matches the two encountered each other.\n\n> -- https://docs.google.com/document/d/1uCF5wmbcL90qvrk_J27fWAvDcDNrO9o_APkicwRkOKc/edit\n\n\n| Variable Name: | Variable Description: | \n| -- | -- | \n| playerShort | short player ID | \n| player | player name | \n| club | player club | \n| leagueCountry | country of player club (England, Germany, France, and Spain) | \n| height | player height (in cm) | \n| weight | player weight (in kg) | \n| position | player position | \n| games | number of games in the player-referee dyad | \n| goals | number of goals in the player-referee dyad | \n| yellowCards | number of yellow cards player received from the referee | \n| yellowReds | number of yellow-red cards player received from the referee | \n| redCards | number of red cards player received from the referee | \n| photoID | ID of player photo (if available) | \n| rater1 | skin rating of photo by rater 1 | \n| rater2 | skin rating of photo by rater 2 | \n| refNum | unique referee ID number (referee name removed for anonymizing purposes) | \n| refCountry | unique referee country ID number | \n| meanIAT | mean implicit bias score (using the race IAT) for referee country | \n| nIAT | sample size for race IAT in that particular country | \n| seIAT | standard error for mean estimate of race IAT | \n| meanExp | mean explicit bias score (using a racial thermometer task) for referee country | \n| nExp | sample size for explicit bias in that particular country | \n| seExp | standard error for mean estimate of explicit bias measure | \n\n",
"_____no_output_____"
],
[
"## What the teams found\n\n\n### Choices in model features\n\nThe following is the covariates chosen for the respective models: \n\n<img src=\"figures/covariates.png\" width=80%;>\n\n\n### Choices in modeling\n\nOf the many choices made by the team, here is a small selection of the models used to answer this question:\n\n\n<img src=\"figures/models.png\" width=80%;>\n\n\n## Final Results\n\n - 0 teams: negative effect\n - 9 teams: no significant relationship\n - 20 teams: finding a positive effect\n\n<img src=\"figures/results.png\" width=80%;>\n\nAbove image from: http://fivethirtyeight.com/features/science-isnt-broken/#part2\n\n\n> â¦selecting randomly from the present teams, there would have been a 69% probability of reporting a positive result and a 31% probability of reporting a null effect. This raises the possibility that many research projects contain hidden uncertainty due to the wide range of analytic choices available to the researchers. -- Silberzahn, R., Uhlmann, E. L., Martin, D. P., Pasquale, Aust, F., Awtrey, E. C., ⦠Nosek, B. A. (2015, August 20). Many analysts, one dataset: Making transparent how variations in analytical choices affect results. Retrieved from osf.io/gvm2z\n\n\nImages and data from: Silberzahn, R., Uhlmann, E. L., Martin, D. P., Pasquale, Aust, F., Awtrey, E. C., ⦠Nosek, B. A. (2015, August 20). Many analysts, one dataset: Making transparent how variations in analytical choices affect results. Retrieved from osf.io/gvm2z",
"_____no_output_____"
],
[
"## General tips\n\n- Before plotting/joining/doing something, have a question or hypothesis that you want to investigate\n- Draw a plot of what you want to see on paper to sketch the idea\n- Write it down, then make the plan on how to get there\n- How do you know you aren't fooling yourself\n- What else can I check if this is actually true?\n- What evidence could there be that it's wrong?\n",
"_____no_output_____"
]
],
[
[
"# Uncomment one of the following lines and run the cell:\n\ndf = pd.read_csv(\"/home/msr/git/pycon-2017-eda-tutorial/data/redcard/redcard.csv.gz\", compression='gzip')\n# df = pd.read_csv(\"https://github.com/cmawer/pycon-2017-eda-tutorial/raw/master/data/redcard/redcard.csv.gz\",\n# compression='gzip')",
"_____no_output_____"
],
[
"df.shape",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
],
[
"df.describe().T",
"_____no_output_____"
],
[
"df.dtypes",
"_____no_output_____"
],
[
"all_columns = df.columns.tolist()\nall_columns",
"_____no_output_____"
]
],
[
[
"# Challenge\n\nBefore looking below, try to answer some high level questions about the dataset. \n\n\nHow do we operationalize the question of referees giving more red cards to dark skinned players?\n* Counterfactual: if the player were lighter, a ref is more likely to have given a yellow or no card **for the same offense under the same conditions**\n* Regression: accounting for confounding, darker players have positive coefficient on regression against proportion red/total card\n\nPotential issues\n* How to combine rater1 and rater2? Average them? What if they disagree? Throw it out?\n* Is data imbalanced, i.e. red cards are very rare?\n* Is data biased, i.e. players have different amounts of play time? Is this a summary of their whole career?\n* How do I know I've accounted for all forms of confounding?\n\n**First, is there systematic discrimination across all refs?**\n\nExploration/hypotheses:\n* Distribution of games played\n* red cards vs games played\n* Reds per game played vs total cards per game played by skin color\n* Distribution of # red, # yellow, total cards, and fraction red per game played for all players by avg skin color\n* How many refs did players encounter?\n* Do some clubs play more aggresively and get carded more? Or are more reserved and get less?\n* Does carding vary by leagueCountry?\n* Do high scorers get more slack (fewer cards) for the same position?\n* Are there some referees that give more red/yellow cards than others?\n* how consistent are raters? Check with Cohen's kappa.\n* how do red cards vary by position? e.g. defenders get more?\n* Do players with more games get more cards, and is there difference across skin color?\n* indication of bias depending on refCountry?",
"_____no_output_____"
],
[
"## Understand how the data's organized\n\nThe dataset is a single csv where it aggregated every interaction between referee and player into a single row. In other words: Referee A refereed Player B in, say, 10 games, and gave 2 redcards during those 10 games. Then there would be a unique row in the dataset that said: \n\n Referee A, Player B, 2 redcards, ... \n\nThis has several implications that make this first step to understanding and dealing with this data a bit tricky. First, is that the information about Player B is repeated each time -- meaning if we did a simple average of some metric of we would likely get a misleading result. \n\nFor example, asking \"what is the average `weight` of the players?\"",
"_____no_output_____"
]
],
[
[
"df.height.mean()",
"_____no_output_____"
],
[
"df['height'].mean()",
"_____no_output_____"
],
[
"np.mean(df.groupby('playerShort').height.mean())",
"_____no_output_____"
]
],
[
[
"Doing a simple average over the rows will risk double-counting the same player multiple times, for a skewed average. The simple (incorrect) average is ~76.075 kg, but the average weight of the players is ~75.639 kg. There are multiple ways of doing this, but doing a groupby on player makes it so that so each player gets counted exactly once.\n\nNot a huge difference in this case but already an illustration of some difficulty.",
"_____no_output_____"
],
[
"## Tidy Data\n\nHadley Wickham's concept of a **tidy dataset** summarized as:\n\n> - Each variable forms a column\n> - Each observation forms a row\n> - Each type of observational unit forms a table\n\nA longer paper describing this can be found in this [pdf](https://www.jstatsoft.org/article/view/v059i10/v59i10.pdf).\n\nHaving datasets in this form allows for much simpler analyses. So the first step is to try and clean up the dataset into a tidy dataset. \n\nThe first step that I am going to take is to break up the dataset into the different observational units. By that I'm going to have separate tables (or dataframes) for: \n\n - players\n - clubs\n - referees\n - countries\n - dyads",
"_____no_output_____"
],
[
"## Create Tidy Players Table",
"_____no_output_____"
]
],
[
[
"player_index = 'playerShort'\nplayer_cols = [#'player', # drop player name, we have unique identifier\n 'birthday',\n 'height',\n 'weight',\n 'position',\n 'photoID',\n 'rater1',\n 'rater2',\n ]",
"_____no_output_____"
],
[
"# Count the unique variables (if we got different weight values, \n# for example, then we should get more than one unique value in this groupby)\nall_cols_unique_players = df.groupby('playerShort').agg({col:'nunique' for col in player_cols})",
"_____no_output_____"
],
[
"all_cols_unique_players.head()",
"_____no_output_____"
],
[
"# If all values are the same per player then this should be empty (and it is!)\nall_cols_unique_players[all_cols_unique_players > 1].dropna().head()",
"_____no_output_____"
],
[
"# A slightly more elegant way to test the uniqueness\nall_cols_unique_players[all_cols_unique_players > 1].dropna().shape[0] == 0",
"_____no_output_____"
]
],
[
[
"Hooray, our data passed our sanity check. Let's create a function to create a table and run this check for each table that we create.",
"_____no_output_____"
]
],
[
[
"def get_subgroup(dataframe, g_index, g_columns):\n \"\"\"Helper function that creates a sub-table from the columns and runs a quick uniqueness test.\"\"\"\n g = dataframe.groupby(g_index).agg({col:'nunique' for col in g_columns})\n if g[g > 1].dropna().shape[0] != 0:\n print(\"Warning: you probably assumed this had all unique values but it doesn't.\")\n return dataframe.groupby(g_index).agg({col:'max' for col in g_columns})",
"_____no_output_____"
],
[
"players = get_subgroup(df, player_index, player_cols)\nplayers.head()",
"_____no_output_____"
],
[
"def save_subgroup(dataframe, g_index, subgroup_name, prefix='raw_'):\n save_subgroup_filename = \"\".join([prefix, subgroup_name, \".csv.gz\"])\n dataframe.to_csv(save_subgroup_filename, compression='gzip', encoding='UTF-8')\n test_df = pd.read_csv(save_subgroup_filename, compression='gzip', index_col=g_index, encoding='UTF-8')\n # Test that we recover what we send in\n if dataframe.equals(test_df):\n print(\"Test-passed: we recover the equivalent subgroup dataframe.\")\n else:\n print(\"Warning -- equivalence test!!! Double-check.\")",
"_____no_output_____"
],
[
"save_subgroup(players, player_index, \"players\")",
"Test-passed: we recover the equivalent subgroup dataframe.\n"
]
],
[
[
"## Create Tidy Clubs Table\n\nCreate the clubs table.",
"_____no_output_____"
]
],
[
[
"club_index = 'club'\nclub_cols = ['leagueCountry']\nclubs = get_subgroup(df, club_index, club_cols)\nclubs.head()",
"_____no_output_____"
],
[
"clubs['leagueCountry'].value_counts()",
"_____no_output_____"
],
[
"save_subgroup(clubs, club_index, \"clubs\", )",
"Test-passed: we recover the equivalent subgroup dataframe.\n"
]
],
[
[
"## Create Tidy Referees Table",
"_____no_output_____"
]
],
[
[
"referee_index = 'refNum'\nreferee_cols = ['refCountry']\nreferees = get_subgroup(df, referee_index, referee_cols)\nreferees.head()",
"_____no_output_____"
],
[
"referees.refCountry.nunique()",
"_____no_output_____"
],
[
"referees.tail()",
"_____no_output_____"
],
[
"referees.shape",
"_____no_output_____"
],
[
"save_subgroup(referees, referee_index, \"referees\")",
"Test-passed: we recover the equivalent subgroup dataframe.\n"
]
],
[
[
"## Create Tidy Countries Table",
"_____no_output_____"
]
],
[
[
"country_index = 'refCountry'\ncountry_cols = ['Alpha_3', # rename this name of country\n 'meanIAT',\n 'nIAT',\n 'seIAT',\n 'meanExp',\n 'nExp',\n 'seExp',\n ]\ncountries = get_subgroup(df, country_index, country_cols)\ncountries.head()",
"_____no_output_____"
],
[
"rename_columns = {'Alpha_3':'countryName', }\ncountries = countries.rename(columns=rename_columns)\ncountries.head()",
"_____no_output_____"
],
[
"countries.shape",
"_____no_output_____"
],
[
"save_subgroup(countries, country_index, \"countries\")",
"Warning -- equivalence test!!! Double-check.\n"
],
[
"# Ok testing this out: \ntest_df = pd.read_csv(\"raw_countries.csv.gz\", compression='gzip', index_col=country_index)",
"_____no_output_____"
],
[
"for (_, row1), (_, row2) in zip(test_df.iterrows(), countries.iterrows()):\n if not row1.equals(row2):\n print(row1)\n print()\n print(row2)\n print()\n break",
"countryName LUX\nmeanIAT 0.325185\nnIAT 127\nseIAT 0.00329681\nmeanExp 0.538462\nnExp 130\nseExp 0.0137522\nName: 4, dtype: object\n\ncountryName LUX\nmeanIAT 0.325185\nnIAT 127\nseIAT 0.00329681\nmeanExp 0.538462\nnExp 130\nseExp 0.0137522\nName: 4, dtype: object\n\n"
],
[
"row1.eq(row2)",
"_____no_output_____"
],
[
"row1.seIAT - row2.seIAT",
"_____no_output_____"
],
[
"countries.dtypes",
"_____no_output_____"
],
[
"test_df.dtypes",
"_____no_output_____"
],
[
"countries.head()",
"_____no_output_____"
],
[
"test_df.head()",
"_____no_output_____"
]
],
[
[
"Looks like precision error, so I'm not concerned. All other sanity checks pass.",
"_____no_output_____"
]
],
[
[
"countries.tail()",
"_____no_output_____"
],
[
"test_df.tail()",
"_____no_output_____"
]
],
[
[
"## Create separate (not yet Tidy) Dyads Table\n\nThis is one of the more complex tables to reason about -- so we'll save it for a bit later. ",
"_____no_output_____"
]
],
[
[
"dyad_index = ['refNum', 'playerShort']\ndyad_cols = ['games',\n 'victories',\n 'ties',\n 'defeats',\n 'goals',\n 'yellowCards',\n 'yellowReds',\n 'redCards',\n ]",
"_____no_output_____"
],
[
"dyads = get_subgroup(df, g_index=dyad_index, g_columns=dyad_cols)",
"_____no_output_____"
],
[
"dyads.head(10)",
"_____no_output_____"
],
[
"dyads.shape",
"_____no_output_____"
],
[
"dyads[dyads.redCards > 1].head(10)",
"_____no_output_____"
],
[
"save_subgroup(dyads, dyad_index, \"dyads\")",
"Test-passed: we recover the equivalent subgroup dataframe.\n"
],
[
"dyads.redCards.max()",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb8eddd556f86a27035f66351cb52ef4c3d8edcd
| 2,761 |
ipynb
|
Jupyter Notebook
|
intro-neural-networks/student-admissions/StudentAdmissionsSolutions.ipynb
|
DhruvBajaj01/deep-learning-v2-pytorch
|
a03b9f809d21520b73c6da87381bfc2df506052d
|
[
"MIT"
] | 1 |
2022-03-01T06:38:34.000Z
|
2022-03-01T06:38:34.000Z
|
intro-neural-networks/student-admissions/StudentAdmissionsSolutions.ipynb
|
DhruvBajaj01/deep-learning-v2-pytorch
|
a03b9f809d21520b73c6da87381bfc2df506052d
|
[
"MIT"
] | null | null | null |
intro-neural-networks/student-admissions/StudentAdmissionsSolutions.ipynb
|
DhruvBajaj01/deep-learning-v2-pytorch
|
a03b9f809d21520b73c6da87381bfc2df506052d
|
[
"MIT"
] | null | null | null | 20.451852 | 94 | 0.523723 |
[
[
[
"# Solutions",
"_____no_output_____"
],
[
"### One-hot encoding the rank",
"_____no_output_____"
]
],
[
[
"## One solution\n# Make dummy variables for rank\none_hot_data = pd.concat([data, pd.get_dummies(data['rank'], prefix='rank')], axis=1)\n\n# Drop the previous rank column\none_hot_data = one_hot_data.drop('rank', axis=1)\n\n# Print the first 10 rows of our data\none_hot_data[:10]",
"_____no_output_____"
],
[
"## Alternative solution ##\n# if you're using an up-to-date version of pandas, \n# you can also use selection by columns\n\n# an equally valid solution\none_hot_data = pd.get_dummies(data, columns=['rank'])",
"_____no_output_____"
]
],
[
[
"### Scaling the data",
"_____no_output_____"
]
],
[
[
"# Copying our data\nprocessed_data = one_hot_data[:]\n\n# Scaling the columns\nprocessed_data['gre'] = processed_data['gre']/800\nprocessed_data['gpa'] = processed_data['gpa']/4.0\nprocessed_data[:10]",
"_____no_output_____"
]
],
[
[
"### Backpropagating the data",
"_____no_output_____"
]
],
[
[
"def error_term_formula(x, y, output):\n# for binary cross entropy loss\n return (y - output)*x\n# for mean square error\n# return (y - output)*sigmoid_prime(x)*x",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb8efae05112f4ed07f0345c7c426d3f974cf513
| 6,618 |
ipynb
|
Jupyter Notebook
|
src/00_Practical_Introduction.ipynb
|
Kaushal1011/machine-learning
|
07d08ee39d5ff2237b5174beb127ea1cc6224e1b
|
[
"BSD-3-Clause"
] | 6 |
2020-03-08T12:56:34.000Z
|
2022-02-21T14:02:49.000Z
|
src/00_Practical_Introduction.ipynb
|
Kaushal1011/machine-learning
|
07d08ee39d5ff2237b5174beb127ea1cc6224e1b
|
[
"BSD-3-Clause"
] | 2 |
2020-03-08T13:10:36.000Z
|
2020-10-05T15:13:15.000Z
|
src/00_Practical_Introduction.ipynb
|
Kaushal1011/machine-learning
|
07d08ee39d5ff2237b5174beb127ea1cc6224e1b
|
[
"BSD-3-Clause"
] | null | null | null | 54.694215 | 473 | 0.676488 |
[
[
[
"# Machine Learning\n\n## Types of learning\n- Whether or not they are trained with human supervision (supervised, unsupervised, semisupervised, and Reinforcement Learning)\n- Whether or not they can learn incrementally on the fly (online versus batch learning)\n- Whether they work by simply comparing new data points to known data points, or instead detect patterns in the training data and build a predictive model, much like scientists do (instance-based versus model-based learning)\n",
"_____no_output_____"
],
[
"# Types Of Machine Learning Systems\n\n## Supervised VS Unsupervised learning\n\n### Supervised Learning\n- In supervised learning, the training data you feed to the algorithm includes the desired solutions, called labels \n - k-Nearest Neighbors\n - Linear Regression\n - Logistic Regression\n - Support Vector Machines (SVMs)\n - Decision Trees and Random Forests\n \n - Neural networks\n\n### Unsupervised Learning\n- In unsupervised learning, as you might guess, the training data is unlabeled. The system tries to learn without a teacher\n - Clustering\n - k-Means\n - Hierarchical Cluster Analysis (HCA)\n - Expectation Maximization\n - DBSCAN\n - Visualization and dimensionality reduction\n - Principal Component Analysis (PCA)\n - Kernel PCA\n - Locally-Linear Embedding (LLE)\n - t-distributed Stochastic Neighbor Embedding (t-SNE)\n - Association rule learning\n - Apriori\n - Eclat\n - Anamoly detection and novelty detection\n - One-Class SVM\n - Isolation Forest\n\n## Semisupervised Learning\n- Some algorithms can deal with partially labeled training data, usually a lot of unla beled data and a little bit of labeled data. This is called semisupervised learning\n- Most semisupervised learning algorithms are combinations of unsupervised and supervised algorithms. For example, deep belief networks (DBNs) are based on unsu‐ pervised components called restricted Boltzmann machines (RBMs) stacked on top of one another. RBMs are trained sequentially in an unsupervised manner, and then the whole system is fine-tuned using supervised learning techniques.\n\n## Reinforcement Learning\n- Reinforcement Learning is a very different beast. The learning system, called an agent in this context, can observe the environment, select and perform actions, and get rewards in return (or penalties in the form of negative rewards. It must then learn by itself what is the best strategy, called a policy, to get the most reward over time. A policy defines what action the agent should choose when it is in a given situation.\n\n## Batch And Online Learning\n\n### Batch Learning\n- In batch learning, the system is incapable of learning incrementally: it must be trained using all the available data. This will generally take a lot of time and computing resources, so it is typically done offline. First the system is trained, and then it is launched into production and runs without learning anymore; it just applies what it has learned. This is called offline learning.\n- If you want a batch learning system to know about new data (such as a new type of spam), you need to train a new version of the system from scratch on the full dataset (not just the new data, but also the old data), then stop the old system and replace it with the new one.\n\n### Online Learning\n- In online learning, you train the system incrementally by feeding it data instances sequentially, either individually or by small groups called mini-batches. Each learning step is fast and cheap, so the system can learn about new data on the fly, as it arrives\n- Online learning is great for systems that receive data as a continuous flow (e.g., stock prices) and need to adapt to change rapidly or autonomously. It is also a good option if you have limited computing resources: once an online learning system has learned about new data instances, it does not need them anymore, so you can discard them (unless you want to be able to roll back to a previous state and “replay” the data). This can save a huge amount of space.\n- Online learning algorithms can also be used to train systems on huge datasets that cannot fit in one machine’s main memory (this is called out-of-core learning). The algorithm loads part of the data, runs a training step on that data, and repeats the process until it has run on all of the data\n - This whole process is usually done offline (i.e., not on the live system), so online learning can be a confusing name. Think of it as incremental learning\n- Importance of Learning Rate in Online Learning\n\n## Instance-Based Vs Model-Based Learning\n\n### Instance-Based Learning\n- the system learns the examples by heart, then generalizes to new cases using a similarity measure \n\n### Model-Based Learning\n- Another way to generalize from a set of examples is to build a model of these examples, then use that model to make predictions. This is called model-based learning\n\n",
"_____no_output_____"
],
[
"# Challenges of Machine Learning\n- Insuffcient Quantity of Training Data\n- Nonrepresentative Training Data\n- Poor-Quality Data\n- Irrelevant Features\n- Overfitting the Training Data\n- Underfitting the Training Data\n\n\nMost Common Supervised Learning Tasks are Classification(predicting classes) And Regression(predicting Values)",
"_____no_output_____"
]
]
] |
[
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown"
]
] |
cb8f059b3b0e36346ef6e6c70ddeef8652db9b92
| 81,145 |
ipynb
|
Jupyter Notebook
|
terra/qis_intro/superposition.ipynb
|
lukasszz/qiskit-tutorials-community
|
1cf11ad977aec8e0680e32c38110aa8cf3677e60
|
[
"Apache-2.0"
] | 293 |
2020-05-29T17:03:04.000Z
|
2022-03-31T07:09:50.000Z
|
terra/qis_intro/superposition.ipynb
|
SalahuddinNur/qiskit-community-tutorials
|
f589e0aa4b31de30aed01f8219abdd3e56771653
|
[
"Apache-2.0"
] | 30 |
2020-06-23T19:11:32.000Z
|
2021-12-20T22:25:54.000Z
|
terra/qis_intro/superposition.ipynb
|
SalahuddinNur/qiskit-community-tutorials
|
f589e0aa4b31de30aed01f8219abdd3e56771653
|
[
"Apache-2.0"
] | 204 |
2020-06-08T12:55:52.000Z
|
2022-03-31T08:37:14.000Z
| 122.760968 | 11,204 | 0.871033 |
[
[
[
"<img src=\"https://raw.githubusercontent.com/Qiskit/qiskit-tutorials/master/images/qiskit-heading.png\" alt=\"Note: In order for images to show up in this jupyter notebook you need to select File => Trusted Notebook\" width=\"500 px\" align=\"left\">",
"_____no_output_____"
],
[
"## _*Superposition*_ \n\n\nThe latest version of this notebook is available on https://github.com/qiskit/qiskit-tutorial.\n\n***\n### Contributors\nJay Gambetta, Antonio Córcoles, Andrew Cross, Anna Phan\n\n### Qiskit Package Versions",
"_____no_output_____"
]
],
[
[
"import qiskit\nqiskit.__qiskit_version__",
"_____no_output_____"
]
],
[
[
"## Introduction\nMany people tend to think quantum physics is hard math, but this is not actually true. Quantum concepts are very similar to those seen in the linear algebra classes you may have taken as a freshman in college, or even in high school. The challenge of quantum physics is the necessity to accept counter-intuitive ideas, and its lack of a simple underlying theory. We believe that if you can grasp the following two Principles, you will have a good start: \n1. A physical system in a definite state can still behave randomly.\n2. Two systems that are too far apart to influence each other can nevertheless behave in ways that, though individually random, are somehow strongly correlated.\n\nIn this tutorial, we will be discussing the first of these Principles, the second is discussed in [this other tutorial](entanglement_introduction.ipynb).",
"_____no_output_____"
]
],
[
[
"# useful additional packages \nimport matplotlib.pyplot as plt\n%matplotlib inline\nimport numpy as np\n\n# importing Qiskit\nfrom qiskit import QuantumCircuit, QuantumRegister, ClassicalRegister, execute\nfrom qiskit import BasicAer, IBMQ\n\n# import basic plot tools\nfrom qiskit.tools.visualization import plot_histogram",
"_____no_output_____"
],
[
"backend = BasicAer.get_backend('qasm_simulator') # run on local simulator by default\n\n# Uncomment the following lines to run on a real device\n#IBMQ.load_accounts()\n#from qiskit.providers.ibmq import least_busy\n#backend = least_busy(IBMQ.backends(operational=True, simulator=False))\n#print(\"the best backend is \" + backend.name())",
"_____no_output_____"
]
],
[
[
"## Quantum States - Basis States and Superpositions<a id='section1'></a>\n\nThe first Principle above tells us that the results of measuring a quantum state may be random or deterministic, depending on what basis is used. To demonstrate, we will first introduce the computational (or standard) basis for a qubit. \n\nThe computational basis is the set containing the ground and excited state $\\{|0\\rangle,|1\\rangle\\}$, which also corresponds to the following vectors: \n\n$$|0\\rangle =\\begin{pmatrix} 1 \\\\ 0 \\end{pmatrix}$$\n$$|1\\rangle =\\begin{pmatrix} 0 \\\\ 1 \\end{pmatrix}$$\n\nIn Python these are represented by",
"_____no_output_____"
]
],
[
[
"zero = np.array([[1],[0]])\none = np.array([[0],[1]])",
"_____no_output_____"
]
],
[
[
"In our quantum processor system (and many other physical quantum processors) it is natural for all qubits to start in the $|0\\rangle$ state, known as the ground state. To make the $|1\\rangle$ (or excited) state, we use the operator \n\n$$ X =\\begin{pmatrix} 0 & 1 \\\\ 1 & 0 \\end{pmatrix}.$$\n\nThis $X$ operator is often called a bit-flip because it exactly implements the following:\n\n$$X: |0\\rangle \\rightarrow |1\\rangle$$\n$$X: |1\\rangle \\rightarrow |0\\rangle.$$\n\nIn Python this can be represented by the following:",
"_____no_output_____"
]
],
[
[
"X = np.array([[0,1],[1,0]])\nprint(np.dot(X,zero))\nprint(np.dot(X,one))",
"[[0]\n [1]]\n[[1]\n [0]]\n"
]
],
[
[
"Next, we give the two quantum circuits for preparing and measuring a single qubit in the ground and excited states using Qiskit.",
"_____no_output_____"
]
],
[
[
"# Creating registers\nqr = QuantumRegister(1)\ncr = ClassicalRegister(1)\n\n# Quantum circuit ground \nqc_ground = QuantumCircuit(qr, cr)\nqc_ground.measure(qr[0], cr[0])\n\n# Quantum circuit excited \nqc_excited = QuantumCircuit(qr, cr)\nqc_excited.x(qr)\nqc_excited.measure(qr[0], cr[0])",
"_____no_output_____"
],
[
"qc_ground.draw(output='mpl')",
"_____no_output_____"
],
[
"qc_excited.draw(output='mpl')",
"_____no_output_____"
]
],
[
[
"Here we have created two jobs with different quantum circuits; the first to prepare the ground state, and the second to prepare the excited state. Now we can run the prepared jobs.",
"_____no_output_____"
]
],
[
[
"circuits = [qc_ground, qc_excited]\njob = execute(circuits, backend)\nresult = job.result()",
"_____no_output_____"
]
],
[
[
"After the run has been completed, the data can be extracted from the API output and plotted.",
"_____no_output_____"
]
],
[
[
"plot_histogram(result.get_counts(qc_ground))",
"_____no_output_____"
],
[
"plot_histogram(result.get_counts(qc_excited))",
"_____no_output_____"
]
],
[
[
"Here we see that the qubit is in the $|0\\rangle$ state with 100% probability for the first circuit and in the $|1\\rangle$ state with 100% probability for the second circuit. If we had run on a quantum processor rather than the simulator, there would be a difference from the ideal perfect answer due to a combination of measurement error, preparation error, and gate error (for the $|1\\rangle$ state). \n\nUp to this point, nothing is different from a classical system of a bit. To go beyond, we must explore what it means to make a superposition. The operation in the quantum circuit language for generating a superposition is the Hadamard gate, $H$. Let's assume for now that this gate is like flipping a fair coin. The result of a flip has two possible outcomes, heads or tails, each occurring with equal probability. If we repeat this simple thought experiment many times, we would expect that on average we will measure as many heads as we do tails. Let heads be $|0\\rangle$ and tails be $|1\\rangle$. \n\nLet's run the quantum version of this experiment. First we prepare the qubit in the ground state $|0\\rangle$. We then apply the Hadamard gate (coin flip). Finally, we measure the state of the qubit. Repeat the experiment 1024 times (shots). As you likely predicted, half the outcomes will be in the $|0\\rangle$ state and half will be in the $|1\\rangle$ state.\n\nTry the program below.",
"_____no_output_____"
]
],
[
[
"# Quantum circuit superposition \nqc_superposition = QuantumCircuit(qr, cr)\nqc_superposition.h(qr)\nqc_superposition.measure(qr[0], cr[0])\n\nqc_superposition.draw()",
"_____no_output_____"
],
[
"job = execute(qc_superposition, backend, shots = 1024)\nresult = job.result()\n\nplot_histogram(result.get_counts(qc_superposition))",
"_____no_output_____"
]
],
[
[
"Indeed, much like a coin flip, the results are close to 50/50 with some non-ideality due to errors (again due to state preparation, measurement, and gate errors). So far, this is still not unexpected. Let's run the experiment again, but this time with two $H$ gates in succession. If we consider the $H$ gate to be analog to a coin flip, here we would be flipping it twice, and still expecting a 50/50 distribution. ",
"_____no_output_____"
]
],
[
[
"# Quantum circuit two Hadamards \nqc_twohadamard = QuantumCircuit(qr, cr)\nqc_twohadamard.h(qr)\nqc_twohadamard.barrier()\nqc_twohadamard.h(qr)\nqc_twohadamard.measure(qr[0], cr[0])\n\nqc_twohadamard.draw(output='mpl')",
"_____no_output_____"
],
[
"job = execute(qc_twohadamard, backend)\nresult = job.result()\n\nplot_histogram(result.get_counts(qc_twohadamard))",
"_____no_output_____"
]
],
[
[
"This time, the results are surprising. Unlike the classical case, with high probability the outcome is not random, but in the $|0\\rangle$ state. *Quantum randomness* is not simply like a classical random coin flip. In both of the above experiments, the system (without noise) is in a definite state, but only in the first case does it behave randomly. This is because, in the first case, via the $H$ gate, we make a uniform superposition of the ground and excited state, $(|0\\rangle+|1\\rangle)/\\sqrt{2}$, but then follow it with a measurement in the computational basis. The act of measurement in the computational basis forces the system to be in either the $|0\\rangle$ state or the $|1\\rangle$ state with an equal probability (due to the uniformity of the superposition). In the second case, we can think of the second $H$ gate as being a part of the final measurement operation; it changes the measurement basis from the computational basis to a *superposition* basis. The following equations illustrate the action of the $H$ gate on the computational basis states:\n$$H: |0\\rangle \\rightarrow |+\\rangle=\\frac{|0\\rangle+|1\\rangle}{\\sqrt{2}}$$\n$$H: |1\\rangle \\rightarrow |-\\rangle=\\frac{|0\\rangle-|1\\rangle}{\\sqrt{2}}.$$\nWe can redefine this new transformed basis, the superposition basis, as the set {$|+\\rangle$, $|-\\rangle$}. We now have a different way of looking at the second experiment above. The first $H$ gate prepares the system into a superposition state, namely the $|+\\rangle$ state. The second $H$ gate followed by the standard measurement changes it into a measurement in the superposition basis. If the measurement gives 0, we can conclude that the system was in the $|+\\rangle$ state before the second $H$ gate, and if we obtain 1, it means the system was in the $|-\\rangle$ state. In the above experiment we see that the outcome is mainly 0, suggesting that our system was in the $|+\\rangle$ superposition state before the second $H$ gate. \n\n\nThe math is best understood if we represent the quantum superposition state $|+\\rangle$ and $|-\\rangle$ by: \n\n$$|+\\rangle =\\frac{1}{\\sqrt{2}}\\begin{pmatrix} 1 \\\\ 1 \\end{pmatrix}$$\n$$|-\\rangle =\\frac{1}{\\sqrt{2}}\\begin{pmatrix} 1 \\\\ -1 \\end{pmatrix}$$\n\nA standard measurement, known in quantum mechanics as a projective or von Neumann measurement, takes any superposition state of the qubit and projects it to either the state $|0\\rangle$ or the state $|1\\rangle$ with a probability determined by:\n\n$$P(i|\\psi) = |\\langle i|\\psi\\rangle|^2$$ \n\nwhere $P(i|\\psi)$ is the probability of measuring the system in state $i$ given preparation $\\psi$.\n\nWe have written the Python function ```state_overlap``` to return this: ",
"_____no_output_____"
]
],
[
[
"state_overlap = lambda state1, state2: np.absolute(np.dot(state1.conj().T,state2))**2",
"_____no_output_____"
]
],
[
[
"Now that we have a simple way of going from a state to the probability distribution of a standard measurement, we can go back to the case of a superposition made from the Hadamard gate. The Hadamard gate is defined by the matrix:\n\n$$ H =\\frac{1}{\\sqrt{2}}\\begin{pmatrix} 1 & 1 \\\\ 1 & -1 \\end{pmatrix}$$\n\nThe $H$ gate acting on the state $|0\\rangle$ gives:",
"_____no_output_____"
]
],
[
[
"Hadamard = np.array([[1,1],[1,-1]],dtype=complex)/np.sqrt(2)\npsi1 = np.dot(Hadamard,zero)\nP0 = state_overlap(zero,psi1)\nP1 = state_overlap(one,psi1)\nplot_histogram({'0' : P0.item(0), '1' : P1.item(0)})",
"_____no_output_____"
]
],
[
[
"which is the ideal version of the first superposition experiment. \n\nThe second experiment involves applying the Hadamard gate twice. While matrix multiplication shows that the product of two Hadamards is the identity operator (meaning that the state $|0\\rangle$ remains unchanged), here (as previously mentioned) we prefer to interpret this as doing a measurement in the superposition basis. Using the above definitions, you can show that $H$ transforms the computational basis to the superposition basis.",
"_____no_output_____"
]
],
[
[
"print(np.dot(Hadamard,zero))\nprint(np.dot(Hadamard,one))",
"[[0.70710678+0.j]\n [0.70710678+0.j]]\n[[ 0.70710678+0.j]\n [-0.70710678+0.j]]\n"
]
],
[
[
"This is just the beginning of how a quantum state differs from a classical state. Please continue to [Amplitude and Phase](amplitude_and_phase.ipynb) to explore further!",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
cb8f1dfcddbd339587f6f57d9cbd708027ef6793
| 767,211 |
ipynb
|
Jupyter Notebook
|
Notebooks/need_to_org/old_Trials_Structures_Zones.ipynb
|
alexgonzl/TreeMazeAnalyses2
|
9bd20328368a915a0d9b81c02ae7af37c5c0c839
|
[
"MIT"
] | null | null | null |
Notebooks/need_to_org/old_Trials_Structures_Zones.ipynb
|
alexgonzl/TreeMazeAnalyses2
|
9bd20328368a915a0d9b81c02ae7af37c5c0c839
|
[
"MIT"
] | null | null | null |
Notebooks/need_to_org/old_Trials_Structures_Zones.ipynb
|
alexgonzl/TreeMazeAnalyses2
|
9bd20328368a915a0d9b81c02ae7af37c5c0c839
|
[
"MIT"
] | null | null | null | 106.690446 | 193,448 | 0.76455 |
[
[
[
"%matplotlib inline\n\nimport numpy as np\nimport pandas as pd\nfrom scipy import signal, ndimage, interpolate, stats\nfrom scipy.interpolate import CubicSpline\nfrom itertools import combinations\n\nimport matplotlib.pyplot as plt\nimport matplotlib.ticker as ticker\nfrom matplotlib.ticker import FormatStrFormatter\nfrom matplotlib.offsetbox import AnchoredText\nimport statsmodels.api as sm\nfrom sklearn.model_selection import train_test_split\nimport statsmodels.formula.api as smf\n\nimport seaborn as sns\nfont = {'family' : 'sans-serif',\n 'size' : 20}\n\nplt.rc('font', **font)\nplt.rc('text',usetex=False)\n\nfrom pathlib import Path\nimport os,sys\nimport h5py, json\nimport sys \nimport pickle as pkl\nimport time\n\nimport nept\nsys.path.append('../PreProcessing/') \nsys.path.append('../TrackingAnalyses/') \nsys.path.append('../Lib/')\nsys.path.append('../Analyses/')\nfrom filters_ag import *\n\nfrom importlib import reload # Python 3.4+ only.\nimport pre_process_neuralynx as PPN\nimport TreeMazeFunctions as TMF\nimport spike_functions as SF\nimport spatial_tuning as ST\nimport stats_functions as StatsF\nimport plot_functions as PF\nimport zone_analyses_session as ZA\n",
"_____no_output_____"
],
[
"animal = 'Li'\ntask = 'T3g'\ndate = '062718'\nsession = animal+'_'+task+'_'+date",
"_____no_output_____"
],
[
"sessionPaths = ZA.getSessionPaths(oakPaths,session)\nPosDat = TMF.getBehTrackData(sessionPaths,0)\ncell_FR, mua_FR = SF.getSessionFR(sessionPaths)\n",
"Loading Beh Tracking Data\nComplete.\nLoading Spikes...\nBinned Spike Files Loaded.\nLoading FRs ...\nFR Loaded.\n"
],
[
"import numpy as np\nfrom scipy.sparse.linalg import svds\nfrom functools import partial\n\n\ndef emsvd(Y, k=None, tol=1E-3, maxiter=None):\n \"\"\"\n Approximate SVD on data with missing values via expectation-maximization\n\n Inputs:\n -----------\n Y: (nobs, ndim) data matrix, missing values denoted by NaN/Inf\n k: number of singular values/vectors to find (default: k=ndim)\n tol: convergence tolerance on change in trace norm\n maxiter: maximum number of EM steps to perform (default: no limit)\n\n Returns:\n -----------\n Y_hat: (nobs, ndim) reconstructed data matrix\n mu_hat: (ndim,) estimated column means for reconstructed data\n U, s, Vt: singular values and vectors (see np.linalg.svd and \n scipy.sparse.linalg.svds for details)\n \"\"\"\n\n if k is None:\n svdmethod = partial(np.linalg.svd, full_matrices=False)\n else:\n svdmethod = partial(svds, k=k)\n if maxiter is None:\n maxiter = np.inf\n\n # initialize the missing values to their respective column means\n mu_hat = np.nanmean(Y, axis=0, keepdims=1)\n valid = np.isfinite(Y)\n Y_hat = np.where(valid, Y, mu_hat)\n\n halt = False\n ii = 1\n v_prev = 0\n\n while not halt:\n\n # SVD on filled-in data\n U, s, Vt = svdmethod(Y_hat - mu_hat)\n\n # impute missing values\n Y_hat[~valid] = (U.dot(np.diag(s)).dot(Vt) + mu_hat)[~valid]\n\n # update bias parameter\n mu_hat = Y_hat.mean(axis=0, keepdims=1)\n\n # test convergence using relative change in trace norm\n v = s.sum()\n if ii >= maxiter or ((v - v_prev) / v_prev) < tol:\n halt = True\n ii += 1\n v_prev = v\n\n return Y_hat, mu_hat, U, s, Vt",
"_____no_output_____"
],
[
"def getPosSequence(PosZones,startID,endID):\n nSamps = len(PosZones)\n pos = []\n samp = []\n \n pos.append(PosZones[0])\n samp.append(0)\n for p in np.arange(nSamps-1):\n p0 = PosZones[p]\n p1 = PosZones[p+1]\n if p0!=p1:\n pos.append(p1)\n samp.append(p+1)\n pos = np.array(pos)\n samp = np.array(samp) + startID\n nPos = len(pos)\n dur = np.zeros(nPos,dtype=int)\n for p in np.arange(nPos-1):\n dur[p] = samp[p+1]-samp[p]\n dur[-1] = endID-samp[-1]\n \n return pos, samp, dur\n\ndef cmp(a,b):\n return (a>b)-(a<b)\n\ndef getTrials(dat,**kwargs):\n nTr = dat.shape[0]\n trials = set(np.arange(1,nTr+1))\n try:\n for k,v in kwargs.items():\n trials = trials & set(np.where(dat[k]==v)[0]+1)\n except:\n print('Invalid Selection {} {}'.format(k,v))\n pass\n return np.sort(np.array(list(trials)))\n\ndef zscore(x,mu,sig):\n return (x-mu)/sig\n\ndef getFR_TrZone(TrialInfo, FRMat):\n nCells = FRMat.shape[0]\n TrZnFR = {} # FR for every zone visited in that trial\n OTrZnFR = {} # FR for every zone visited in that trial\n \n for tr in TrialInfo['All']['Trials']:\n nPos = len(TrialInfo['TrSeq']['Pos'][tr])\n trSpPos = np.zeros((nCells,nPos))\n for p in np.arange(nPos):\n s=TrialInfo['TrSeq']['Samp'][tr][p]\n d=TrialInfo['TrSeq']['Dur'][tr][p] \n samps = np.arange(s,s+d)\n for cell in np.arange(nCells):\n trSpPos[cell, p]=np.mean(FRMat[cell][samps])\n \n nPos = len(TrialInfo['OffTrSeq']['Pos'][tr])\n otrSpPos = np.zeros((nCells,nPos))\n for p in np.arange(nPos):\n s=TrialInfo['OffTrSeq']['Samp'][tr][p]\n d=TrialInfo['OffTrSeq']['Dur'][tr][p] \n samps = np.arange(s,s+d)\n for cell in np.arange(nCells):\n otrSpPos[cell, p]=np.mean(FRMat[cell][samps])\n \n TrZnFR[tr] = trSpPos\n OTrZnFR[tr] = otrSpPos\n return TrZnFR, OTrZnFR\ndef AICc(model):\n n = model.nobs\n llf = model.llf\n k = len(model.params)\n AIC = 2*(k-llf)\n c = 2*k*(k+1)/(n-k-1)\n return AIC+c\n\ndef R2(x,y):\n return (np.corrcoef(x,y)**2)[0,1]\n\ndef aR2(model,y,fit=[]):\n if fit==[]:\n fit = model.fittedvalues\n r2 = R2(fit,y)\n n = model.nobs\n p = len(model.params)-1\n aR2 = 1-(1-r2)*(n-1)/(n-p-1)\n return aR2\n\ndef getParamSet():\n '''\n Returns a dictionary of parameter sets for modeling.\n '''\n params = ['Loc','IO','Cue','Desc','Co']\n combs = []\n\n for i in np.arange(1, len(params)+1):\n combs+= [list(x) for x in combinations(params, i)]\n param_set = {}\n cnt=0\n for c in combs:\n param_set[cnt] = c\n cnt+=1\n\n for c in combs:\n if ('IO' in c) and ('Loc' in c):\n param_set[cnt] = ['Loc:IO']+c\n cnt+=1\n return param_set\n\ndef getModel_testR2(dat,formula='',params=[],mixedlm=True, verbose=False):\n '''\n Obtains the test R2 based on even/odd splits of the data\n '''\n \n if len(params)>0 and len(formula)==0:\n formula = getFormula(params)\n else:\n print('No Method of selecting parameters provided.')\n return np.nan,np.nan,[]\n print('\\nComputing mixedlm with formula: {}'.format(formula))\n \n dat_even = dat[dat['EvenTrial']==True]\n dat_odd = dat[dat['EvenTrial']==False]\n \n if mixedlm:\n md_even = smf.mixedlm(formula, data=dat_even,groups=dat_even[\"trID\"])\n else:\n md_even = smf.ols(formula + 'trID', data=dat_even)\n mdf_even = md_even.fit()\n pred_odd = mdf_even.predict(dat_odd)\n \n\n if mixedlm:\n md_odd = smf.mixedlm(formula, data=dat_odd,groups=dat_odd[\"trID\"])\n else:\n md_odd = smf.ols(formula + 'trID', data=md_odd)\n \n mdf_odd = md_odd.fit()\n pred_even = mdf_odd.predict(dat_even)\n \n if verbose:\n print('\\nPerformance Train-Even:Test-Odd')\n print(\"Train_aR2 = {0:.3f}\".format(aR2(mdf_even,dat_even['zFR'])))\n print(\"Model_AICc = {0:.3f}\".format(AICc(mdf_even)))\n print(\"Test_R2 = {0:.3f}\".format(R2(pred_odd,dat_odd['zFR'])))\n print('\\nPerformance Train-Odd:Test-Even')\n print(\"Train_aR2 = {0:.3f}\".format(aR2(mdf_odd,dat_odd['zFR'])))\n print(\"Model_AICc = {0:.3f}\".format(AICc(mdf_odd)))\n print(\"Test_R2 = {0:.3f}\".format(R2(pred_even,dat_even['zFR'])))\n \n dat['Pred']=np.zeros(dat.shape[0])\n dat.loc[dat['EvenTrial']==True,'Pred']=pred_even\n dat.loc[dat['EvenTrial']==False,'Pred']=pred_odd\n \n r2 = R2(dat['zFR'],dat['Pred'])\n print('\\nOverall test R2: {0:.3f}'.format(r2))\n return r2\n\ndef getFormula(params):\n formula = 'zFR ~ '\n nP = len(params)\n cnt=1\n for i in params:\n formula += i\n if cnt<nP:\n formula +='+'\n cnt+=1\n return formula\n\ndef getModelPerf(dat,formula='',params=[],mixedlm=True):\n '''\n Obtains the train adjusted R2, and AIC for data.\n returns aR2, AIC, and the fitted model. \n '''\n print('\\nComputing mixedlm with formula: {}'.format(formula))\n\n if len(params)>0 and len(formula)==0:\n formula = getFormula(params)\n else:\n print('No Method of selecting parameters provided.')\n return np.nan,np.nan,[]\n \n if mixedlm:\n md = smf.mixedlm(formula, data=dat, groups=dat[\"trID\"])\n else:\n md = smf.ols(formula + '+trID', data=dat)\n \n mdf = md.fit()\n print('\\n Model Performance:')\n train_aR2 = aR2(mdf,dat['zFR'])\n print(\"Train_aR2 = {0:.3f}\".format(train_aR2))\n aic = AICc(mdf)\n print(\"Model_AICc = {0:.3f}\".format(aic))\n \n return train_aR2, aic, mdf",
"_____no_output_____"
],
[
"ValidTraj = {'R_S1':['Home','SegA','Center','SegB','I1','SegC','G1'],\n 'R_S2':['Home','SegA','Center','SegB','I1','SegD','G2'],\n 'R_L1':['Home','SegA','Center','SegB','I1','SegD','G2','SegD','I1','SegC','G1'],\n 'R_L2':['Home','SegA','Center','SegB','I1','SegC','G1','SegC','I1','SegD','G2'],\n 'L_S3':['Home','SegA','Center','SegE','I2','SegF','G3'],\n 'L_S4':['Home','SegA','Center','SegE','I2','SegG','G4'],\n 'L_L3':['Home','SegA','Center','SegE','I2','SegG','G4','SegG','I2','SegF','G3'],\n 'L_L4':['Home','SegA','Center','SegE','I2','SegF','G3','SegF','I2','SegG','G4'],\n }\nValidOffTraj = {}\nfor k,v in ValidTraj.items():\n ValidOffTraj[k] = v[::-1]\n\n# get trial durations and samples\nTrialVec = PosDat['EventDat']['TrID']\nnTr=TrialVec.max()\n\nstartIDs = np.zeros(nTr,dtype=int)\nendIDs = np.zeros(nTr,dtype=int)\nfor tr in np.arange(nTr):\n trIDs = np.where(TrialVec==(tr+1))[0]\n startIDs[tr]=trIDs[0]\n endIDs[tr] = trIDs[-1]\n\nTrialDurs = endIDs-startIDs\n\nOffTrialDurs=np.concatenate((startIDs[1:],[len(PosDat['t'])]))-endIDs\nOffTrialVec = np.full_like(TrialVec,0)\n\nfor tr in np.arange(nTr):\n idx = np.arange(endIDs[tr],endIDs[tr]+OffTrialDurs[tr])\n OffTrialVec[idx]=tr+1\n \n# Pre allocated Trial Info structure.\nTrialInfo = {'All':{'Trials':[],'Co':[],'InCo':[]},'L':{'Trials':[],'Co':[],'InCo':[]},\n 'R':{'Trials':[],'Co':[],'InCo':[]},'BadTr':[],'Cues':np.full(nTr,''),'Desc':np.full(nTr,''),\n 'DurThr':45,'TrDurs':TrialDurs,\n 'TrialVec':TrialVec,'TrStSamp':startIDs,'TrEnSamp':endIDs,'TrSeq':{'Pos':{},'Samp':{},'Dur':{}},\n 'OffTrStSamp':endIDs,'OffTrEnSamp':endIDs+OffTrialDurs,'OffTrDurs':OffTrialDurs,\n 'OffTrialVec':OffTrialVec, 'OffTrSeq':{'Pos':{},'Samp':{},'Dur':{}},\n 'ValidSeqTrials':[],'ValidSeqOffTrials':[],'ValidSeqTrID':[],'ValidSeqOffTrID':[],\n 'ValidSeqNames':ValidTraj,'ValidSeqOffNames':ValidOffTraj}\n\nTrialInfo['All']['Trials']=np.arange(nTr)+1\n#get separate trials and allocate by correct/incorrect\nfor tr in TrialInfo['All']['Trials']:\n idx= TrialVec==tr\n for s in ['L','R']:\n c = PosDat['EventDat']['C'+s][idx] \n d = PosDat['EventDat'][s+'Ds'][idx]\n if np.mean(d)>0.5: # descicion\n TrialInfo['Desc'][tr-1]=s\n if np.mean(c)>0.5: # cue\n TrialInfo[s]['Trials'].append(tr)\n TrialInfo['Cues'][tr-1]=s\n if np.mean(d&c)>0.5: # correct descicion\n TrialInfo[s]['Co'].append(tr)\n else:\n TrialInfo[s]['InCo'].append(tr)\nassert set(TrialInfo['R']['Trials']) & set(TrialInfo['L']['Trials']) == set(), 'Trial classified as both left and right.'\nassert len(TrialInfo['Cues']) ==len(TrialInfo['Desc']), 'Number of trials mismatch'\nassert len(TrialInfo['Cues']) ==nTr, 'Number of trials mismatch'\n\nfor trC in ['Co', 'InCo']:\n TrialInfo['All'][trC] = np.sort(TrialInfo['L'][trC]+TrialInfo['R'][trC])\n\nfor i in ['All','L','R']:\n for j in ['Trials','Co','InCo']:\n TrialInfo[i]['n'+j]=len(TrialInfo[i][j])\n TrialInfo[i][j]=np.array(TrialInfo[i][j])\n\n# determine if the trials are too long to be included.\nTrialInfo['BadTr'] = np.where(TrialInfo['TrDurs']*PosDat['step']>TrialInfo['DurThr'])[0]\n\n# get positions for each trial\nfor tr in TrialInfo['All']['Trials']:\n \n idx = TrialInfo['TrialVec']==tr\n sID = TrialInfo['TrStSamp'][tr-1]\n eID = TrialInfo['TrEnSamp'][tr-1]\n \n p,s,d=getPosSequence(PosDat['PosZones'][idx],sID,eID)\n #p,s,d=getPosSequence(p4[idx],sID,eID)\n \n TrialInfo['TrSeq']['Pos'][tr]=p\n TrialInfo['TrSeq']['Samp'][tr]=s\n TrialInfo['TrSeq']['Dur'][tr]=d\n \n idx = TrialInfo['OffTrialVec']==tr\n sID = TrialInfo['OffTrStSamp'][tr-1]\n eID = TrialInfo['OffTrEnSamp'][tr-1]\n \n p,s,d=getPosSequence(PosDat['PosZones'][idx],sID,eID) \n\n TrialInfo['OffTrSeq']['Pos'][tr]=p\n TrialInfo['OffTrSeq']['Samp'][tr]=s\n TrialInfo['OffTrSeq']['Dur'][tr]=d\n\n# determine if the sequence of positions are valid for each trial\nTrSeqs = {}\nvTr = []\nOffTrSeqs = {}\nvOTr = []\nfor tr in TrialInfo['All']['Trials']:\n seq = [TMF.Zones[a] for a in TrialInfo['TrSeq']['Pos'][tr]]\n match = 0\n for vSeqN, vSeq in ValidTraj.items():\n if cmp(seq,vSeq)==0:\n match = 1\n vTr.append(tr)\n TrSeqs[tr]=vSeqN\n break\n if match==0:\n TrSeqs[tr]=[]\n \n seq = [TMF.Zones[a] for a in TrialInfo['OffTrSeq']['Pos'][tr]]\n match = 0\n for vSeqN, vSeq in ValidOffTraj.items():\n if cmp(seq,vSeq)==0:\n match = 1\n vOTr.append(tr)\n OffTrSeqs[tr]=vSeqN\n break\n if match==0:\n OffTrSeqs[tr]=[]\n \nTrialInfo['ValidSeqTrials'] = vTr\nTrialInfo['ValidSeqOffTrials'] = vOTr\nTrialInfo['ValidSeqTrID'] = TrSeqs\nTrialInfo['ValidSeqOffTrID'] = OffTrSeqs\n",
"_____no_output_____"
],
[
"conds = ['Cues','Desc','Co','Traj','OTraj','Dur','Good','Length','OLength']\nTrCondMat = pd.DataFrame(np.full((nTr,len(conds)),np.nan),index=TrialInfo['All']['Trials'],columns=conds)\n\nTrCondMat['Cues'] = TrialInfo['Cues']\nTrCondMat['Desc'] = TrialInfo['Desc']\nTrCondMat['Dur'] = TrialDurs\n\nTrCondMat['Co'].loc[TrialInfo['All']['Co']]='Co'\nTrCondMat['Co'].loc[TrialInfo['All']['InCo']]='InCo'\n\nvseq=TrialInfo['ValidSeqTrials']\nTrCondMat['Traj'].loc[vseq]=[TrialInfo['ValidSeqTrID'][s] for s in vseq]\n\nvseq=TrialInfo['ValidSeqOffTrials']\nTrCondMat['OTraj'].loc[vseq]=[TrialInfo['ValidSeqOffTrID'][s] for s in vseq]\n\nTrCondMat['Good'] = (~TrCondMat['Traj'].isnull()) & (TrialDurs*PosDat['step']<TrialInfo['DurThr'])\n\nx=np.full(nTr,'')\nfor k,v in TrialInfo['ValidSeqTrID'].items():\n if len(v)>0: \n x[k-1]=v[2]\nTrCondMat['Length']= x\n\nx=np.full(nTr,'')\nfor k,v in TrialInfo['ValidSeqOffTrID'].items():\n if len(v)>0: \n x[k-1]=v[2]\nTrCondMat['OLength']= x\n",
"_____no_output_____"
],
[
"# working version of long DF trialxPos matrix. this takes either short or long trajectories in the outbound but only short trajectories inbound\nnMaxPos = 11\nnMinPos = 7\nnTr =len(TrialInfo['All']['Trials'])\n\nnCells = cell_FR.shape[0]\nnMua = mua_FR.shape[0]\nnTotalUnits = nCells+nMua\nnUnits = {'cell':nCells,'mua':nMua}\n\nTrZn={'cell':[],'mua':[]}\nOTrZn={'cell':[],'mua':[]}\n\nTrZn['cell'],OTrZn['cell'] = getFR_TrZone(TrialInfo,cell_FR)\nTrZn['mua'],OTrZn['mua'] = getFR_TrZone(TrialInfo,mua_FR)\n\ncellCols = ['cell_'+str(i) for i in np.arange(nCells)]\nmuaCols = ['mua_'+str(i) for i in np.arange(nMua)]\nunitCols = {'cell':cellCols,'mua':muaCols}\nallUnits = cellCols+muaCols\n\nmu = {'cell':np.mean(cell_FR,1),'mua':np.mean(mua_FR,1)}\nsig = {'cell':np.std(cell_FR,1),'mua':np.std(mua_FR,1)}\n\nOut=pd.DataFrame(np.full((nTr*nMaxPos,nTotalUnits),np.nan),columns=allUnits)\nIn=pd.DataFrame(np.full((nTr*nMaxPos,nTotalUnits),np.nan),columns=allUnits)\nO_I=pd.DataFrame(np.full((nTr*nMaxPos,nTotalUnits),np.nan),columns=allUnits)\n\nOut = Out.assign(trID = np.tile(TrialInfo['All']['Trials'],nMaxPos))\nIn = In.assign(trID = np.tile(TrialInfo['All']['Trials'],nMaxPos))\nO_I = O_I.assign(trID = np.tile(TrialInfo['All']['Trials'],nMaxPos))\n\nOut = Out.assign(Pos = np.repeat(np.arange(nMaxPos),nTr))\nIn = In.assign(Pos = np.repeat(np.arange(nMaxPos),nTr))\nO_I = O_I.assign(Pos = np.repeat(np.arange(nMaxPos),nTr))\n\nOut = Out.assign(IO = ['Out']*(nTr*nMaxPos))\nIn = In.assign(IO = ['In']*(nTr*nMaxPos))\nO_I = O_I.assign(IO = ['O_I']*(nTr*nMaxPos))\n\nfor ut in ['cell','mua']:\n for cell in np.arange(nUnits[ut]):\n X=pd.DataFrame(np.full((nTr,nMaxPos),np.nan),index=TrialInfo['All']['Trials'],columns=np.arange(nMaxPos))\n Y=pd.DataFrame(np.full((nTr,nMaxPos),np.nan),index=TrialInfo['All']['Trials'],columns=np.arange(nMaxPos))\n Z=pd.DataFrame(np.full((nTr,nMaxPos),np.nan),index=TrialInfo['All']['Trials'],columns=np.arange(nMaxPos))\n\n m = mu[ut][cell]\n s = sig[ut][cell]\n\n for tr in TrialInfo['All']['Trials']:\n traj = TrialInfo['ValidSeqTrID'][tr]\n if traj in ValidTrajNames:\n if traj[2]=='S':\n X.loc[tr][0:nMinPos] = zscore(TrZn[ut][tr][cell],m,s)\n else:\n X.loc[tr] = zscore(TrZn[ut][tr][cell],m,s)\n\n otraj = TrialInfo['ValidSeqOffTrID'][tr]\n if otraj in ValidTrajNames:\n if otraj[2]=='S':\n Y.loc[tr][4:] = zscore(OTrZn[ut][tr][cell],m,s)\n else:\n Y.loc[tr] = zscore(OTrZn[ut][tr][cell],m,s)\n if (traj in ValidTrajNames) and (otraj in ValidTrajNames):\n if traj==otraj:\n Z.loc[tr] = X.loc[tr].values-Y.loc[tr][::-1].values\n elif traj[2]=='L' and otraj[2]=='S': # ambigous interserction position, skipping that computation\n Z.loc[tr][[0,1,2,3]] = X.loc[tr][[0,1,2,3]].values-Y.loc[tr][[10,9,8,7]].values\n Z.loc[tr][[5,6]] = X.loc[tr][[9,10]].values-Y.loc[tr][[5,4]].values\n elif traj[2]=='S' and otraj[2]=='L':\n Z.loc[tr][[0,1,2,3]] = X.loc[tr][[0,1,2,3]].values-Y.loc[tr][[10,9,8,7]].values\n Z.loc[tr][[5,6]] = X.loc[tr][[5,6]].values-Y.loc[tr][[1,0]].values\n\n Out[unitCols[ut][cell]]=X.melt(value_name='zFR')['zFR']\n In[unitCols[ut][cell]]=Y.melt(value_name='zFR')['zFR']\n O_I[unitCols[ut][cell]]=Z.melt(value_name='zFR')['zFR']\n\nData = pd.DataFrame()\nData = pd.concat([Data,Out])\nData = pd.concat([Data,In])\nData = pd.concat([Data,O_I])\nData = Data.reset_index()",
"_____no_output_____"
],
[
"Data",
"_____no_output_____"
],
[
"nMaxPos = 11\nnTr =len(TrialInfo['All']['Trials'])\n\nCols = ['trID','Pos','IO','Cue','Desc','Traj','Loc','OTraj', 'Goal','ioMatch','Co','Valid']\nnCols = len(Cols) \nnDatStack = 3 # Out, In, O-I\nTrialMatInfo = pd.DataFrame(np.full((nTr*nMaxPos*nDatStack,nCols),np.nan),columns=Cols)\n\nTrialMatInfo['trID'] = np.tile(np.tile(TrialInfo['All']['Trials'],nMaxPos),nDatStack)\nTrialMatInfo['Pos'] = np.tile(np.repeat(np.arange(nMaxPos),nTr),nDatStack)\nTrialMatInfo['IO'] = np.repeat(['Out','In','O_I'],nTr*nMaxPos)\n\nTrialMatInfo['Traj'] = np.tile(np.tile(TrCondMat['Traj'],nMaxPos),nDatStack)\nTrialMatInfo['OTraj'] = np.tile(np.tile(TrCondMat['OTraj'],nMaxPos),nDatStack)\nTrialMatInfo['Co'] = np.tile(np.tile(TrCondMat['Co'],nMaxPos),nDatStack)\nTrialMatInfo['Cue'] = np.tile(np.tile(TrCondMat['Cues'],nMaxPos),nDatStack)\nTrialMatInfo['Desc'] = np.tile(np.tile(TrCondMat['Desc'],nMaxPos),nDatStack)\nTrialMatInfo['ioMatch'] = [traj==otraj for traj,otraj in zip(TrialMatInfo['Traj'],TrialMatInfo['OTraj'])]\nTrialMatInfo['Goal'] = [traj[3] if traj==traj else '' for traj in TrialMatInfo['Traj']]\nTrialMatInfo['Len'] = [traj[2] if traj==traj else '' for traj in TrialMatInfo['Traj']]\nTrialMatInfo['OLen'] = [traj[2] if traj==traj else '' for traj in TrialMatInfo['OTraj']]\n\n# get true location in each trials sequence 'Loc'\n# note that 'Pos' is a numerical indicator of the order in a sequence\noutTrSeq = pd.DataFrame(np.full((nTr,nMaxPos),np.nan),index=TrialInfo['All']['Trials'])\ninTrSeq = pd.DataFrame(np.full((nTr,nMaxPos),np.nan),index=TrialInfo['All']['Trials'])\noiTrSeq = pd.DataFrame(np.full((nTr,nMaxPos),np.nan),index=TrialInfo['All']['Trials'])\n#allTrValid = []\nfor tr in TrialInfo['All']['Trials']:\n traj = TrialInfo['ValidSeqTrID'][tr]\n if traj in ValidTrajNames:\n seq = TrialInfo['ValidSeqNames'][traj]\n if len(seq)==nMaxPos: \n outTrSeq.loc[tr]=seq\n else:\n outTrSeq.loc[tr]= seq + [np.nan]*4\n else:\n outTrSeq.loc[tr] = [np.nan]*nMaxPos\n \n otraj = TrialInfo['ValidSeqOffTrID'][tr]\n if otraj in ValidTrajNames:\n oseq = TrialInfo['ValidSeqOffNames'][otraj]\n if len(oseq)==nMaxPos:\n inTrSeq.loc[tr]=oseq\n else:\n inTrSeq.loc[tr]=[np.nan]*4+oseq\n else:\n inTrSeq.loc[tr]=[np.nan]*nMaxPos\n \n if (traj in ValidTrajNames) and (otraj in ValidTrajNames):\n if traj==otraj:\n if len(seq)==nMaxPos:\n oiTrSeq.loc[tr]=seq\n else:\n oiTrSeq.loc[tr] = seq + [np.nan]*4\n elif traj[2]=='L' and otraj[2]=='S':\n oiTrSeq.loc[tr] = seq[:4]+[np.nan]+seq[9:]+[np.nan]*4\n elif traj[2]=='S' and otraj[2]=='L':\n oiTrSeq.loc[tr] = seq[:4]+[np.nan]+seq[5:]+[np.nan]*4\n else:\n oiTrSeq.loc[tr] = [np.nan]*nMaxPos\n\nTrialMatInfo['Loc'] = pd.concat([pd.concat([outTrSeq.melt(value_name='Loc')['Loc'],\n inTrSeq.melt(value_name='Loc')['Loc']]),\n oiTrSeq.melt(value_name='Loc')['Loc']]).values\nTrialMatInfo['Valid'] = ~TrialMatInfo['Loc'].isnull()\nTrialMatInfo['EvenTrial'] = TrialMatInfo['trID']%2==0",
"_____no_output_____"
],
[
"TrialMatInfo",
"_____no_output_____"
],
[
"traj = 'R_L1'\ngoal = traj[3]\nnPos = 11\nif traj[2]=='S':\n nOutPos = 7\nelif traj[2]=='L':\n nOutPos = 11\n\noutTrials = getTrials(TrCondMat,Good=True,Traj=traj)\nnOutTr = len(outTrials)\nif nOutTr > 1:\n # Valid outbound trials\n outLocs = ValidTraj[traj]\n x = pd.DataFrame(np.full((nOutTr,nPos),np.nan),columns=np.arange(nPos))\n cnt = 0\n for tr in outTrials:\n x.iloc[cnt][0:nOutPos]=X.loc[tr][outLocs].values\n cnt+=1\n\n cTr = TrCondMat.loc[outTrials]['Co']\n cues = TrCondMat.loc[outTrials]['Cues']\n desc = TrCondMat.loc[outTrials]['Desc']\n x = x.melt(var_name='Pos',value_name='zFR')\n x = x.assign(Traj = [traj]*(nOutTr*nPos) )\n x = x.assign(IO = ['Out']*(nOutTr*nPos) )\n x = x.assign(Goal = [goal]*(nOutTr*nPos) )\n x = x.assign(Cue = np.tile(cues,nPos) )\n x = x.assign(Desc = np.tile(desc,nPos) )\n x = x.assign(trID = np.tile(outTrials, nPos).astype(int))\n x = x.assign(Co = np.tile(cTr,nPos))\n \n# y: subset of outbound trials that are directly 'short' inbound\n# z: difference in the overlapping positions between outbount/inbound\nbadTrials = getTrials(TrCondMat,Good=True,Traj=traj,OLength='')\ninTrials = np.setdiff1d(outTrials,badTrials)\nnInTr = len(inTrials)\n\ny = pd.DataFrame(np.full((nInTr,nPos),np.nan),columns=np.arange(nPos))\nz = pd.DataFrame(np.full((nInTr,nPos),np.nan),columns=np.arange(nPos))\ncTr = TrCondMat.loc[inTrials]['Co']\ncues = TrCondMat.loc[inTrials]['Cues']\ndesc = TrCondMat.loc[inTrials]['Desc']\ncnt = 0\nfor tr in inTrials:\n inLocs = ValidOffTraj[TrCondMat.loc[tr,'OTraj']]\n if len(inLocs)==nPos:\n y.iloc[cnt]=Y.loc[tr][inLocs].values\n z.iloc[cnt]=Z.loc[tr][inLocs].values\n else:\n y.iloc[cnt][(nPos-len(inLocs)):]=Y.loc[tr][inLocs].values\n z.iloc[cnt][(nPos-len(inLocs)):]=Z.loc[tr][inLocs].values\n cnt+=1\n\ny = y.melt(var_name='Pos',value_name='zFR')\ny = y.assign(Traj = [traj]*(nInTr*nPos) )\ny = y.assign(IO = ['In']*(nInTr*nPos) )\ny = y.assign(Goal = [goal]*(nInTr*nPos) )\ny = y.assign(Cue = np.tile(cues, nPos))\ny = y.assign(Desc = np.tile(desc,nPos) )\ny = y.assign(trID = np.tile(inTrials, nPos).astype(int))\ny = y.assign(Co = np.tile(cTr,nPos))\ncellDat = pd.concat([cellDat,y])\n\nz = z.melt(var_name='Pos',value_name='zFR')\nz = z.assign(Traj = [traj]*(nInTr*nPos) )\nz = z.assign(IO = ['O-I']*(nInTr*nPos) )\nz = z.assign(Goal = [goal]*(nInTr*nPos) )\nz = z.assign(Cue = np.tile(cues, nPos))\nz = z.assign(Desc = np.tile(desc,nPos) )\nz = z.assign(trID = np.tile(inTrials, nPos).astype(int))\nz = z.assign(Co = np.tile(cTr,nPos))\ncellDat = pd.concat([cellDat,z])",
"_____no_output_____"
],
[
"cell = 10\n\ncellDat = TrialMatInfo.copy()\ncellDat['zFR'] = Data[cellCols[cell]]\n\nsns.set()\nsns.set(style=\"whitegrid\",context='notebook',font_scale=1.5,rc={ \n 'axes.spines.bottom': False,\n 'axes.spines.left': False,\n 'axes.spines.right': False,\n 'axes.spines.top': False,\n 'axes.edgecolor':'0.5'})\n\npal = sns.xkcd_palette(['green','purple'])\n\nf,ax = plt.subplots(2,3, figsize=(15,6))\nw = 0.25\nh = 0.43\nratio = 6.5/10.5 \nhsp = 0.05\nvsp = 0.05\nW = [w,w*ratio,w*ratio]\nyPos = [vsp,2*vsp+h]\nxPos = [hsp,1.5*hsp+W[0],2.5*hsp+W[1]+W[0]]\nxlims = [[-0.25,10.25],[3.75,10.25],[-0.25,6.25]]\nfor i in [0,1]:\n for j in np.arange(3):\n ax[i][j].set_position([xPos[j],yPos[i],W[j],h])\n ax[i][j].set_xlim(xlims[j])\n\nxPosLabels = {}\nxPosLabels[0] = ['Home','SegA','Center','SegBE','Int','CDFG','Goals','CDFG','Int','CDFG','Goals'] \nxPosLabels[2] = ['Home','SegA','Center','SegBE','Int','CDFG','Goals']\nxPosLabels[1] = xPosLabels[2][::-1]\n\nplotAll = False\nalpha=0.15\nmlw = 1\nwith sns.color_palette(pal):\n coSets = ['InCo','Co']\n for i in [0,1]:\n if i==0:\n leg=False\n else:\n leg='brief'\n \n if plotAll:\n subset = (cellDat['IO']=='Out') & (cellDat['Co']==coSets[i]) & (cellDat['Valid'])\n ax[i][0] = sns.lineplot(x='Pos',y='zFR',hue='Cue',style='Goal',ci=None,data=cellDat[subset],\n ax=ax[i][0],legend=False,lw=3,hue_order=['L','R'],style_order=['1','2','3','4'])\n ax[i][0] = sns.lineplot(x='Pos',y='zFR',hue='Desc',estimator=None,units='trID',data=cellDat[subset],\n ax=ax[i][0],legend=False,lw=mlw,alpha=alpha,hue_order=['L','R'])\n \n subset = (cellDat['IO']=='In') & (cellDat['Co']==coSets[i]) & (cellDat['Pos']>=4) & (cellDat['Valid'])\n ax[i][1] = sns.lineplot(x='Pos',y='zFR',hue='Cue',style='Goal',ci=None,data=cellDat[subset],\n ax=ax[i][1],legend=False,lw=3,hue_order=['L','R'],style_order=['1','2','3','4'])\n ax[i][1] = sns.lineplot(x='Pos',y='zFR',hue='Cue',estimator=None,units='trID',data=cellDat[subset],\n ax=ax[i][1],legend=False,lw=mlw,alpha=alpha,hue_order=['L','R'])\n \n subset = (cellDat['IO']=='O_I') & (cellDat['Co']==coSets[i])& (cellDat['Valid']) \n ax[i][2] = sns.lineplot(x='Pos',y='zFR',hue='Cue',style='Goal',ci=None,data=cellDat[subset],\n ax=ax[i][2],legend=leg,lw=3,hue_order=['L','R'],style_order=['1','2','3','4'])\n ax[i][2] = sns.lineplot(x='Pos',y='zFR',hue='Cue',estimator=None,units='trID',data=cellDat[subset],\n ax=ax[i][2],legend=False,lw=mlw,alpha=alpha,hue_order=['L','R'])\n\n else:\n subset = (cellDat['IO']=='Out') & (cellDat['Co']==coSets[i]) & (cellDat['Valid'])\n ax[i][0] = sns.lineplot(x='Pos',y='zFR',hue='Cue',style='Goal',data=cellDat[subset],\n ax=ax[i][0],lw=2,legend=False,hue_order=['L','R'],style_order=['1','2','3','4'])\n subset = (cellDat['IO']=='In') & (cellDat['Co']==coSets[i]) & (cellDat['Pos']>=4) & (cellDat['Valid'])\n ax[i][1] = sns.lineplot(x='Pos',y='zFR',hue='Cue',style='Goal',data=cellDat[subset],\n ax=ax[i][1],lw=2,legend=False,hue_order=['L','R'],style_order=['1','2','3','4'])\n subset = (cellDat['IO']=='O_I') & (cellDat['Co']==coSets[i])& (cellDat['Valid'])\n ax[i][2] = sns.lineplot(x='Pos',y='zFR',hue='Cue',style='Goal',data=cellDat[subset],\n ax=ax[i][2],legend=leg,lw=2,hue_order=['L','R'],style_order=['1','2','3','4'])\n \n ax[i][1].set_xticks(np.arange(4,nMaxPos))\n ax[i][0].set_xticks(np.arange(nMaxPos)) \n ax[i][2].set_xticks(np.arange(nMinPos))\n \n for j in np.arange(3):\n ax[i][j].set_xlabel('')\n ax[i][j].set_ylabel('')\n ax[i][j].tick_params(axis='x', rotation=60)\n \n ax[i][0].set_ylabel('{} zFR'.format(coSets[i]))\n ax[i][1].set_yticklabels('') \n \n if i==0:\n for j in np.arange(3):\n ax[i][j].set_xticklabels(xPosLabels[j]) \n else:\n ax[i][0].set_title('Out')\n ax[i][1].set_title('In')\n ax[i][2].set_title('O-I')\n for j in np.arange(3):\n ax[i][j].set_xticklabels('')\n l =ax[1][2].get_legend()\n plt.legend(bbox_to_anchor=(1.05, 0), loc=6, borderaxespad=0.,frameon=False)\n l.set_frame_on(False)\n\n # out/in limits\n lims = np.zeros((4,2))\n cnt =0\n for i in [0,1]:\n for j in [0,1]:\n lims[cnt]=np.array(ax[i][j].get_ylim())\n cnt+=1\n minY = np.floor(np.min(lims[:,0])*20)/20\n maxY = np.ceil(np.max(lims[:,1]*20))/20\n for i in [0,1]:\n for j in [0,1]:\n ax[i][j].set_ylim([minY,maxY])\n \n # o-i limits\n lims = np.zeros((2,2))\n cnt =0\n for i in [0,1]:\n lims[cnt]=np.array(ax[i][2].get_ylim())\n cnt+=1\n minY = np.floor(np.min(lims[:,0])*20)/20\n maxY = np.ceil(np.max(lims[:,1]*20))/20\n for i in [0,1]:\n ax[i][2].set_ylim([minY,maxY])\n \n ",
"_____no_output_____"
],
[
"f,ax = plt.subplots(1,2, figsize=(10,4))\n\nsns.set(style=\"whitegrid\",font_scale=1.5,rc={ \n 'axes.spines.bottom': False,\n 'axes.spines.left': False,\n 'axes.spines.right': False,\n 'axes.spines.top': False,\n 'axes.edgecolor':'0.5'})\n\npal = sns.xkcd_palette(['spring green','light purple'])\n\nsubset = cellDat['Co']=='Co'\ndat =[]\ndat = cellDat[subset].groupby(['trID','IO','Cue','Desc']).mean()\ndat = dat.reset_index()\nwith sns.color_palette(pal):\n ax[0]=sns.violinplot(y='zFR',x='IO',hue='Desc',data=dat,split=True, ax=ax[0],\n scale='count',inner='quartile',hue_order=['L','R'],saturation=0.5,order=['Out','In','O_I'])\npal = sns.xkcd_palette(['emerald green','medium purple'])\nwith sns.color_palette(pal):\n ax[0]=sns.swarmplot(y='zFR',x='IO',hue='Desc',data=dat,dodge=True,hue_order=['L','R'],alpha=0.7,ax=ax[0],\n edgecolor='gray',order=['Out','In','O_I'])\nl=ax[0].get_legend()\nl.set_visible(False)\nax[0].set_xlabel('Direction')\n\npal = sns.xkcd_palette(['spring green','light purple'])\nsubset= cellDat['IO']=='Out'\ndat = cellDat[subset].groupby(['trID','Cue','Co','Desc']).mean()\ndat = dat.reset_index()\n\nwith sns.color_palette(pal):\n ax[1]=sns.violinplot(y='zFR',x='Desc',hue='Cue',data=dat,split=True,scale='width',ax=ax[1],\n inner='quartile',order=['L','R'],hue_order=['L','R'],saturation=0.5)\npal = sns.xkcd_palette(['emerald green','medium purple'])\nwith sns.color_palette(pal):\n ax[1]=sns.swarmplot(y='zFR',x='Desc',hue='Cue',data=dat,dodge=True,order=['L','R'],ax=ax[1],\n hue_order=['L','R'],alpha=0.7,edgecolor='gray')\nax[1].set_xlabel('Decision')\nax[1].set_ylabel('')\nl=ax[1].get_legend()\nhandles, labels = ax[1].get_legend_handles_labels()\nl.set_visible(False)\nplt.legend(handles[2:],labels[2:],bbox_to_anchor=(1.05, 0), loc=3, borderaxespad=0.,frameon=False,title='Cue')\n",
"_____no_output_____"
],
[
"cell = 10\n\ncellDat = TrialMatInfo.copy()\ncellDat['zFR'] = Data[cellCols[cell]]\n\nsubset = (cellDat['Valid']) & ~(cellDat['IO']=='O_I')\ndat = cellDat[subset] \ndat = dat.reset_index()\n#dat['Pos'] = dat['Pos'].astype('category')\nmd = smf.mixedlm(\"zFR ~ Loc*IO+Co+Cue+Desc\", data=dat,groups=dat[\"trID\"])\n#md = smf.mixedlm(\"zFR ~ Loc*IO\", data=dat,groups=dat[\"trID\"])\n#md = smf.mixedlm(\"zFR ~ IO\", data=dat,groups=dat[\"trID\"])\n\nmdf = md.fit()\ndat['Fit'] = mdf.fittedvalues\ndat['Res'] = mdf.resid\nprint(mdf.summary())\nprint(\"R2 = {0:.3f}\".format((np.corrcoef(dat['Fit'],dat['zFR'])**2)[0,1]))\nprint(mdf.wald_test_terms())\n\n\nmd = smf.mixedlm(\"Res ~ Cue+Desc+Co\", data=dat,groups=dat[\"trID\"])\nmdf = md.fit()\nprint(mdf.summary())\nprint(\"R2 = {0:.3f}\".format((np.corrcoef(mdf.fittedvalues,dat['Res'])**2)[0,1]))\nprint(mdf.wald_test_terms())\n",
"/home/alexg8/anaconda3/lib/python3.7/site-packages/statsmodels/base/model.py:508: ConvergenceWarning: Maximum Likelihood optimization failed to converge. Check mle_retvals\n \"Check mle_retvals\", ConvergenceWarning)\n"
],
[
"f,ax = plt.subplots(1,2, figsize=(10,4))\n\nsns.set(style=\"whitegrid\",font_scale=1.5,rc={ \n 'axes.spines.bottom': False,\n 'axes.spines.left': False,\n 'axes.spines.right': False,\n 'axes.spines.top': False,\n 'axes.edgecolor':'0.5'})\n\npal = sns.xkcd_palette(['spring green','light purple'])\ny = 'Fit'\nsubset = dat['Co']=='Co'\ndat2 =[]\ndat2 = dat[subset].groupby(['trID','IO','Cue','Desc']).mean()\ndat2 = dat2.reset_index()\nwith sns.color_palette(pal):\n ax[0]=sns.violinplot(y=y,x='IO',hue='Desc',data=dat2,split=True, ax=ax[0],\n scale='count',inner='quartile',hue_order=['L','R'],saturation=0.5,order=['Out','In'])\npal = sns.xkcd_palette(['emerald green','medium purple'])\nwith sns.color_palette(pal):\n ax[0]=sns.swarmplot(y=y,x='IO',hue='Desc',data=dat2,dodge=True,hue_order=['L','R'],alpha=0.7,ax=ax[0],\n edgecolor='gray',order=['Out','In'])\nl=ax[0].get_legend()\nl.set_visible(False)\nax[0].set_xlabel('Direction')\n\npal = sns.xkcd_palette(['spring green','light purple'])\nsubset= dat['IO']=='Out'\ndat2 =[]\ndat2 = dat[subset].groupby(['trID','Cue','Co','Desc']).mean()\ndat2 = dat2.reset_index()\n\nwith sns.color_palette(pal):\n ax[1]=sns.violinplot(y=y,x='Desc',hue='Cue',data=dat2,split=True,scale='width',ax=ax[1],\n inner='quartile',order=['L','R'],hue_order=['L','R'],saturation=0.5)\npal = sns.xkcd_palette(['emerald green','medium purple'])\nwith sns.color_palette(pal):\n ax[1]=sns.swarmplot(y=y,x='Desc',hue='Cue',data=dat2,dodge=True,order=['L','R'],ax=ax[1],\n hue_order=['L','R'],alpha=0.7,edgecolor='gray')\nax[1].set_xlabel('Decision')\nax[1].set_ylabel('')\nl=ax[1].get_legend()\nhandles, labels = ax[1].get_legend_handles_labels()\nl.set_visible(False)\nplt.legend(handles[2:],labels[2:],bbox_to_anchor=(1.05, 0), loc=3, borderaxespad=0.,frameon=False,title='Cue')\n",
"_____no_output_____"
],
[
"TrialInfo['TrDurs']",
"_____no_output_____"
],
[
"cell = 10\ncellDat = TrialMatInfo.copy()\ncellDat['zFR'] = Data[cellCols[cell]] \n\ndat = []\ndat = cellDat[~(cellDat['IO']=='O_I') & (cellDat['Valid'])].copy()\ndat['trID'] = dat['trID'].astype('category')\ndat = dat.reset_index()\n\ndat_even = dat[dat['EvenTrial']==True]\ndat_odd = dat[dat['EvenTrial']==False]\n\n#md1 = smf.mixedlm(\"zFR ~ Loc*IO+Cue+Desc+Co\", data=dat1,groups=dat1[\"trID\"])\n#md_even = sm.OLS.from_formula(\"zFR ~ Loc:IO+Loc+IO+Cue+Desc+Co+trID\", data=dat_even)\nmd_even = smf.mixedlm(\"zFR ~ Loc:IO+Loc+IO+Cue+Desc+Co\", data=dat_even,groups=dat_even[\"trID\"])\nmdf_even = md_even.fit()\n\nprint('\\nPerformance Train-Even:Test-Odd')\nprint(\"Train_aR2 = {0:.3f}\".format(aR2(mdf_even,dat_even['zFR'])))\nprint(\"Model_AICc = {0:.3f}\".format(AICc(mdf_even)))\n#print(mdf.wald_test_terms())\npred_odd = mdf_even.predict(dat_odd)\nprint(\"Test_R2 = {0:.3f}\".format(R2(pred_odd,dat_odd['zFR'])))\n\nprint('\\nPerformance Train-Odd:Test-Even')\n#md2 = smf.mixedlm(\"zFR ~ Loc*IO+Cue+Desc+Co\", data=dat2,groups=dat2[\"trID\"])\n#md_odd = sm.OLS.from_formula(\"zFR ~ Loc:IO+Loc+IO+Cue+Desc+Co+trID\", data=dat_odd)\nmd_odd = smf.mixedlm(\"zFR ~ Loc:IO+Loc+IO+Cue+Desc+Co\", data=dat_odd,groups=dat_odd[\"trID\"])\nmdf_odd = md_odd.fit()\n\nprint(\"Train_aR2 = {0:.3f}\".format(aR2(mdf_odd,dat_odd['zFR'])))\nprint(\"Model_AICc = {0:.3f}\".format(AICc(mdf_odd)))\n#print(mdf.wald_test_terms())\npred_even = mdf_odd.predict(dat_even)\nprint(\"Test_R2 = {0:.3f}\".format(R2(pred_even,dat_even['zFR'])))\ndat['Pred']=np.zeros(dat.shape[0])\ndat.loc[dat['EvenTrial']==True,'Pred']=pred_even\ndat.loc[dat['EvenTrial']==False,'Pred']=pred_odd",
"/home/alexg8/anaconda3/lib/python3.7/site-packages/statsmodels/base/model.py:508: ConvergenceWarning: Maximum Likelihood optimization failed to converge. Check mle_retvals\n \"Check mle_retvals\", ConvergenceWarning)\n/home/alexg8/anaconda3/lib/python3.7/site-packages/statsmodels/base/model.py:508: ConvergenceWarning: Maximum Likelihood optimization failed to converge. Check mle_retvals\n \"Check mle_retvals\", ConvergenceWarning)\n/home/alexg8/anaconda3/lib/python3.7/site-packages/statsmodels/base/model.py:508: ConvergenceWarning: Maximum Likelihood optimization failed to converge. Check mle_retvals\n \"Check mle_retvals\", ConvergenceWarning)\n/home/alexg8/anaconda3/lib/python3.7/site-packages/statsmodels/base/model.py:508: ConvergenceWarning: Maximum Likelihood optimization failed to converge. Check mle_retvals\n \"Check mle_retvals\", ConvergenceWarning)\n/home/alexg8/anaconda3/lib/python3.7/site-packages/statsmodels/base/model.py:508: ConvergenceWarning: Maximum Likelihood optimization failed to converge. Check mle_retvals\n \"Check mle_retvals\", ConvergenceWarning)\n/home/alexg8/anaconda3/lib/python3.7/site-packages/statsmodels/regression/mixed_linear_model.py:2026: ConvergenceWarning: Gradient optimization failed.\n warnings.warn(msg, ConvergenceWarning)\n"
],
[
"\n\ncell = 10\nR2thr = 0.2\n\nparam_set = getParamSet()\nnModels = len(param_set)\n\ncellDat = TrialMatInfo.copy()\ncellDat['zFR'] = Data[cellCols[cell]] \n\ndat = []\ndat = cellDat[~(cellDat['IO']=='O_I') & (cellDat['Valid'])].copy()\ndat['trID'] = dat['trID'].astype('category')\ndat = dat.reset_index()\n\nparams = ['Loc:IO','Loc','IO','Cue','Desc','Co']\nform = getFormula(params)\nt1 = time.time()\ntR2 = getModel_testR2(form,dat)\n\nif tR2>=R2thr:\n for k,params in param_set.items():\n form = getFormula(params)\n t2 = time.time()\n trainR2[k],trainAIC[k],_ = getModelPerf(form,dat)\n t3 = time.time()\n print('Time to fit model {0} : {1} = {2:0.3f}s'.format(k,form,t3-t2))\n \nprint('Fitting Completed for cell {0}, total time = {1:0.3f}s'.format(cell,t3-t1))\n\n ",
"\nComputing mixedlm with formula: zFR ~ Loc:IO+Loc+IO+Cue+Desc+Co\n"
],
[
"from joblib import Parallel, delayed\nif not sys.warnoptions:\n import warnings\n warnings.simplefilter(\"ignore\")\n\n\nall_params = ['Loc:IO','Loc','IO','Cue','Desc','Co']\nparam_set = getParamSet()\nnModels = len(param_set)\nR2thr = 0.2\n \nnTr =len(TrialInfo['All']['Trials'])\ncellColIDs = [i for i,item in enumerate(Data.columns.values) if 'cell' in item]\nnCells = len(cellColIDs)\nmuaColIDs = [i for i,item in enumerate(Data.columns.values) if 'mua' in item]\nnMua = len(musColIDs)\nnTotalUnits = nCells+nMua\nnUnits = {'cell':nCells,'mua':nMua}\n\ncellCols = Data.columns[cellColIDs]\nmuaCols = Data.columns[muaColIDs]\nunitCols = {'cell':cellCols,'mua':muaCols}\n\nperfCols = ['FullMod_tR2','modelNum','trainR2','AICc','testR2'] \nCols = ['ut']+perfCols+ all_params\nnCols = len(Cols)\nLM_Dat = pd.DataFrame(np.full((nTotalUnits,nCols),np.nan),columns=Cols)\nLM_Dat.loc[:,'ut'] = ['cell']*nCells+['mua']*nMua\n\ndatSubset = ~(TrialMatInfo['IO']=='O_I') & (TrialMatInfo['Valid'])\ndat = []\ndat = TrialMatInfo[datSubset].copy()\ndat['trID'] = dat['trID'].astype('category')\ndat = dat.reset_index()\nN = dat.shape[0]\ndat['zFR'] = np.zeros(N)\n\nt0 = time.time()\nwith Parallel(n_jobs=16) as parallel:\n cnt=0\n for ut in ['cell','mua']:\n for cell in np.arange(nUnits[ut]): \n print('\\n\\nAnalyzing {} {}'.format(ut,cell))\n\n dat.loc[:,'zFR'] = Data.loc[datSubset,unitCols[ut][cell]].values \n\n t1 = time.time()\n tR2 = getModel_testR2(dat,params=all_params)\n t2 = time.time()\n LM_Dat.loc[cnt,'FullMod_tR2'] = tR2\n\n print('Full Model Test Set Fit completed. Time = {}'.format(t2-t1))\n if tR2>=R2thr:\n print('Full Model passed the threshold, looking for optimal submodel.')\n r = parallel(delayed(getModelPerf)(dat,params=params) for params in param_set.values())\n trainR2,trainAICc,_ = zip(*r)\n\n t3 = time.time()\n print('\\nFitting Completed for {0} {1}, total time = {2:0.3f}s'.format(ut,cell,t3-t1))\n selMod = np.argmin(trainAICc)\n\n selMod_tR2 = getModel_testR2(dat,params=param_set[selMod])\n print('Selected Model = {}, AICc = {}, testR2 = {} '.format(selMod,trainAICc[selMod],selMod_tR2))\n\n LM_Dat.loc[cnt,'modelNum']=selMod\n LM_Dat.loc[cnt,'trainR2']=trainR2[selMod]\n LM_Dat.loc[cnt,'AICc'] = trainAICc[selMod]\n LM_Dat.loc[cnt,'testR2'] = selMod_tR2\n\n temp = r[selMod][2].wald_test_terms()\n LM_Dat.loc[cnt,param_set[selMod]] = np.sqrt(temp.summary_frame()['chi2'][param_set[selMod]])\n cnt+=1\nprint('Model Fitting completed. Time = {}s'.format(time.time()-t0))",
"\nAnalyzing cell 0\n\nComputing mixedlm with formula: zFR ~ Loc:IO+Loc+IO+Cue+Desc+Co\n\nOverall test R2: 0.010\nFull Model Test Set Fit completed. Time = 1.9928991794586182\n\nAnalyzing cell 1\n\nComputing mixedlm with formula: zFR ~ Loc:IO+Loc+IO+Cue+Desc+Co\n\nOverall test R2: 0.084\nFull Model Test Set Fit completed. Time = 0.7516374588012695\n\nAnalyzing cell 2\n\nComputing mixedlm with formula: zFR ~ Loc:IO+Loc+IO+Cue+Desc+Co\n\nOverall test R2: 0.208\nFull Model Test Set Fit completed. Time = 0.7196385860443115\nFull Model passed the threshold, looking for optimal submodel.\n\n\nFitting Completed for cell 2, total time = 19.928s\n\nComputing mixedlm with formula: zFR ~ Loc:IO+Loc+IO+Co\n\nOverall test R2: 0.209\nSelected Model = 34, AICc = 6689.746441247439, testR2 = 0.20896430342682012 \n\nAnalyzing cell 3\n\nComputing mixedlm with formula: zFR ~ Loc:IO+Loc+IO+Cue+Desc+Co\n\nOverall test R2: 0.578\nFull Model Test Set Fit completed. Time = 0.6880843639373779\nFull Model passed the threshold, looking for optimal submodel.\n\n\nFitting Completed for cell 3, total time = 30.939s\n\nComputing mixedlm with formula: zFR ~ Loc:IO+Loc+IO+Cue\n\nOverall test R2: 0.577\nSelected Model = 32, AICc = 5927.329701055561, testR2 = 0.5774549131497138 \n\nAnalyzing cell 4\n\nComputing mixedlm with formula: zFR ~ Loc:IO+Loc+IO+Cue+Desc+Co\n\nOverall test R2: 0.006\nFull Model Test Set Fit completed. Time = 12.057710647583008\n\nAnalyzing cell 5\n\nComputing mixedlm with formula: zFR ~ Loc:IO+Loc+IO+Cue+Desc+Co\n\nOverall test R2: 0.448\nFull Model Test Set Fit completed. Time = 10.569126844406128\nFull Model passed the threshold, looking for optimal submodel.\n\n\nFitting Completed for cell 5, total time = 35.403s\n\nComputing mixedlm with formula: zFR ~ Loc:IO+Loc+IO\n\nOverall test R2: 0.448\nSelected Model = 31, AICc = 5765.774490190265, testR2 = 0.44822254858184435 \n\nAnalyzing cell 6\n\nComputing mixedlm with formula: zFR ~ Loc:IO+Loc+IO+Cue+Desc+Co\n\nOverall test R2: 0.498\nFull Model Test Set Fit completed. Time = 12.613003730773926\nFull Model passed the threshold, looking for optimal submodel.\n\n\nFitting Completed for cell 6, total time = 34.868s\n\nComputing mixedlm with formula: zFR ~ Loc:IO+Loc+IO\n\nOverall test R2: 0.497\nSelected Model = 31, AICc = 5618.716019879345, testR2 = 0.49687025481845226 \n\nAnalyzing cell 7\n\nComputing mixedlm with formula: zFR ~ Loc:IO+Loc+IO+Cue+Desc+Co\n\nOverall test R2: 0.137\nFull Model Test Set Fit completed. Time = 0.7056155204772949\n\nAnalyzing cell 8\n\nComputing mixedlm with formula: zFR ~ Loc:IO+Loc+IO+Cue+Desc+Co\n\nOverall test R2: 0.412\nFull Model Test Set Fit completed. Time = 1.6463806629180908\nFull Model passed the threshold, looking for optimal submodel.\n\n\nFitting Completed for cell 8, total time = 47.433s\n\nComputing mixedlm with formula: zFR ~ Loc:IO+Loc+IO+Co\n\nOverall test R2: 0.411\nSelected Model = 34, AICc = 5944.003239274817, testR2 = 0.4111197902344131 \n\nAnalyzing cell 9\n\nComputing mixedlm with formula: zFR ~ Loc:IO+Loc+IO+Cue+Desc+Co\n\nOverall test R2: 0.171\nFull Model Test Set Fit completed. Time = 0.7182857990264893\n\nAnalyzing cell 10\n\nComputing mixedlm with formula: zFR ~ Loc:IO+Loc+IO+Cue+Desc+Co\n\nOverall test R2: 0.645\nFull Model Test Set Fit completed. Time = 13.802710771560669\nFull Model passed the threshold, looking for optimal submodel.\n\n\nFitting Completed for cell 10, total time = 65.870s\n\nComputing mixedlm with formula: zFR ~ Loc:IO+Loc+IO\n\nOverall test R2: 0.644\nSelected Model = 31, AICc = 4664.870817096485, testR2 = 0.6443346385531358 \n\nAnalyzing cell 11\n\nComputing mixedlm with formula: zFR ~ Loc:IO+Loc+IO+Cue+Desc+Co\n\nOverall test R2: 0.563\nFull Model Test Set Fit completed. Time = 0.6449635028839111\nFull Model passed the threshold, looking for optimal submodel.\n\n\nFitting Completed for cell 11, total time = 4.171s\n\nComputing mixedlm with formula: zFR ~ Loc:IO+Loc+IO+Desc+Co\n\nOverall test R2: 0.563\nSelected Model = 37, AICc = 4986.247546517854, testR2 = 0.5634069061513584 \n\nAnalyzing cell 12\n\nComputing mixedlm with formula: zFR ~ Loc:IO+Loc+IO+Cue+Desc+Co\n\nOverall test R2: 0.069\nFull Model Test Set Fit completed. Time = 0.6185505390167236\n\nAnalyzing cell 13\n\nComputing mixedlm with formula: zFR ~ Loc:IO+Loc+IO+Cue+Desc+Co\n\nOverall test R2: 0.450\nFull Model Test Set Fit completed. Time = 0.708378791809082\nFull Model passed the threshold, looking for optimal submodel.\n\n\nFitting Completed for cell 13, total time = 24.273s\n\nComputing mixedlm with formula: zFR ~ Loc:IO+Loc+IO\n\nOverall test R2: 0.452\nSelected Model = 31, AICc = 5789.485214371848, testR2 = 0.4524037158939023 \n\nAnalyzing cell 14\n\nComputing mixedlm with formula: zFR ~ Loc:IO+Loc+IO+Cue+Desc+Co\n\nOverall test R2: 0.646\nFull Model Test Set Fit completed. Time = 0.5949862003326416\nFull Model passed the threshold, looking for optimal submodel.\n\n\nFitting Completed for cell 14, total time = 26.002s\n\nComputing mixedlm with formula: zFR ~ Loc:IO+Loc+IO+Co\n\nOverall test R2: 0.645\nSelected Model = 34, AICc = 3925.8227058922334, testR2 = 0.6452431495960822 \n\nAnalyzing cell 15\n\nComputing mixedlm with formula: zFR ~ Loc:IO+Loc+IO+Cue+Desc+Co\n\nOverall test R2: 0.717\nFull Model Test Set Fit completed. Time = 0.614811897277832\nFull Model passed the threshold, looking for optimal submodel.\n\n\nFitting Completed for cell 15, total time = 31.046s\n\nComputing mixedlm with formula: zFR ~ Loc:IO+Loc+IO+Desc\n\nOverall test R2: 0.722\nSelected Model = 33, AICc = 4164.820285970919, testR2 = 0.7215479136992394 \n\nAnalyzing mua 0\n\nComputing mixedlm with formula: zFR ~ Loc:IO+Loc+IO+Cue+Desc+Co\n\nOverall test R2: 0.000\nFull Model Test Set Fit completed. Time = 0.7227976322174072\n\nAnalyzing mua 1\n\nComputing mixedlm with formula: zFR ~ Loc:IO+Loc+IO+Cue+Desc+Co\n\nOverall test R2: 0.503\nFull Model Test Set Fit completed. Time = 0.6638846397399902\nFull Model passed the threshold, looking for optimal submodel.\n\n\nFitting Completed for mua 1, total time = 4.084s\n\nComputing mixedlm with formula: zFR ~ Loc:IO+Loc+IO+Desc+Co\n\nOverall test R2: 0.511\nSelected Model = 37, AICc = 4833.269252797552, testR2 = 0.5113005520140127 \n\nAnalyzing mua 2\n\nComputing mixedlm with formula: zFR ~ Loc:IO+Loc+IO+Cue+Desc+Co\n\nOverall test R2: 0.483\nFull Model Test Set Fit completed. Time = 0.6899373531341553\nFull Model passed the threshold, looking for optimal submodel.\n\n\nFitting Completed for mua 2, total time = 19.451s\n\nComputing mixedlm with formula: zFR ~ Loc:IO+Loc+IO+Desc+Co\n\nOverall test R2: 0.496\nSelected Model = 37, AICc = 5008.825982701441, testR2 = 0.4955666250185201 \n\nAnalyzing mua 3\n\nComputing mixedlm with formula: zFR ~ Loc:IO+Loc+IO+Cue+Desc+Co\n\nOverall test R2: 0.002\nFull Model Test Set Fit completed. Time = 0.6976866722106934\n\nAnalyzing mua 4\n\nComputing mixedlm with formula: zFR ~ Loc:IO+Loc+IO+Cue+Desc+Co\n\nOverall test R2: 0.301\nFull Model Test Set Fit completed. Time = 0.8428401947021484\nFull Model passed the threshold, looking for optimal submodel.\n\n\nFitting Completed for mua 4, total time = 4.264s\n\nComputing mixedlm with formula: zFR ~ Loc:IO+Loc+IO+Co\n\nOverall test R2: 0.303\nSelected Model = 34, AICc = 5760.10879100552, testR2 = 0.3030687037637767 \n\nAnalyzing mua 5\n\nComputing mixedlm with formula: zFR ~ Loc:IO+Loc+IO+Cue+Desc+Co\n\nOverall test R2: 0.314\nFull Model Test Set Fit completed. Time = 1.3789558410644531\nFull Model passed the threshold, looking for optimal submodel.\n\n\nFitting Completed for mua 5, total time = 27.293s\n\nComputing mixedlm with formula: zFR ~ Loc:IO+Loc+IO\n\nOverall test R2: 0.320\nSelected Model = 31, AICc = 6252.23501403625, testR2 = 0.3198025984073697 \n\nAnalyzing mua 6\n\nComputing mixedlm with formula: zFR ~ Loc:IO+Loc+IO+Cue+Desc+Co\n\nOverall test R2: 0.277\nFull Model Test Set Fit completed. Time = 0.6061422824859619\nFull Model passed the threshold, looking for optimal submodel.\n\n\nFitting Completed for mua 6, total time = 3.987s\n\nComputing mixedlm with formula: zFR ~ Loc:IO+Loc+IO+Cue\n\nOverall test R2: 0.277\nSelected Model = 32, AICc = 5715.728683399003, testR2 = 0.2774639920891122 \n\nAnalyzing mua 7\n\nComputing mixedlm with formula: zFR ~ Loc:IO+Loc+IO+Cue+Desc+Co\n\nOverall test R2: 0.711\nFull Model Test Set Fit completed. Time = 0.6644766330718994\nFull Model passed the threshold, looking for optimal submodel.\n\n\nFitting Completed for mua 7, total time = 24.661s\n\nComputing mixedlm with formula: zFR ~ Loc:IO+Loc+IO+Desc+Co\n\nOverall test R2: 0.722\nSelected Model = 37, AICc = 3625.7856429581802, testR2 = 0.7223539080607021 \n\nAnalyzing mua 8\n\nComputing mixedlm with formula: zFR ~ Loc:IO+Loc+IO+Cue+Desc+Co\n\nOverall test R2: 0.704\nFull Model Test Set Fit completed. Time = 0.5107736587524414\nFull Model passed the threshold, looking for optimal submodel.\n\n\nFitting Completed for mua 8, total time = 4.228s\n\nComputing mixedlm with formula: zFR ~ Loc:IO+Loc+IO+Desc\n\nOverall test R2: 0.705\nSelected Model = 33, AICc = 4076.7573485964735, testR2 = 0.7046681176524413 \n\nAnalyzing mua 9\n\nComputing mixedlm with formula: zFR ~ Loc:IO+Loc+IO+Cue+Desc+Co\n\nOverall test R2: 0.505\nFull Model Test Set Fit completed. Time = 0.7258498668670654\nFull Model passed the threshold, looking for optimal submodel.\n\n\nFitting Completed for mua 9, total time = 28.991s\n\nComputing mixedlm with formula: zFR ~ Loc:IO+Loc+IO+Desc\n\nOverall test R2: 0.515\nSelected Model = 33, AICc = 4634.873328449968, testR2 = 0.5145496375795725 \n\nAnalyzing mua 10\n\nComputing mixedlm with formula: zFR ~ Loc:IO+Loc+IO+Cue+Desc+Co\n\nOverall test R2: 0.549\nFull Model Test Set Fit completed. Time = 0.8372843265533447\nFull Model passed the threshold, looking for optimal submodel.\n\n\nFitting Completed for mua 10, total time = 30.210s\n\nComputing mixedlm with formula: zFR ~ Loc:IO+Loc+IO\n\nOverall test R2: 0.550\nSelected Model = 31, AICc = 4989.564295878295, testR2 = 0.5503158713252031 \n\nAnalyzing mua 11\n\nComputing mixedlm with formula: zFR ~ Loc:IO+Loc+IO+Cue+Desc+Co\n\nOverall test R2: 0.024\nFull Model Test Set Fit completed. Time = 0.5470905303955078\n\nAnalyzing mua 12\n\nComputing mixedlm with formula: zFR ~ Loc:IO+Loc+IO+Cue+Desc+Co\n\nOverall test R2: 0.001\nFull Model Test Set Fit completed. Time = 1.2361078262329102\nModel Fitting completed. Time = 500.83789443969727s\n"
],
[
"LM_Dat",
"_____no_output_____"
],
[
"param_set",
"_____no_output_____"
],
[
"sns.lmplot(x='zFR',y='fit',hue='Cue',col='Desc',data=dat2,col_wrap=3,sharex=False, sharey=False, robust=True)",
"_____no_output_____"
],
[
"sns.set_style(\"whitegrid\")\nsns.jointplot(dat2['zFR'],dat2['fit'],kind='reg')",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb8f1f1f0edd7170d268bd542e2401e3a2aa77be
| 79,330 |
ipynb
|
Jupyter Notebook
|
Option/Options.ipynb
|
youtside/notebook
|
9a5e4eeffc05bca8e626b681ed06b1bab59483cc
|
[
"Apache-2.0"
] | null | null | null |
Option/Options.ipynb
|
youtside/notebook
|
9a5e4eeffc05bca8e626b681ed06b1bab59483cc
|
[
"Apache-2.0"
] | null | null | null |
Option/Options.ipynb
|
youtside/notebook
|
9a5e4eeffc05bca8e626b681ed06b1bab59483cc
|
[
"Apache-2.0"
] | null | null | null | 65.453795 | 33,380 | 0.739493 |
[
[
[
"# BSM\n## Assumptions:\n- Price of underlying asset follows a lognormal dist; return ~ normal\n- $r_f^c$ is known and constant\n- volatility $\\sigma$ of underlying asset is known and constant\n- Frictionless market\n- No cash flow* (dividend)\n- European options\n\n## Formula\n### European Call\n\n$$c_0= S_0e^{-qT}*N(d1)- Xe^{-R_f^cxT} * N(d2)$$\n\n$$d1 = \\frac{ln(S_0/X) + (r-q+\\sigma^2/2)T} {\\sigma \\sqrt{T}}$$\n\n$$d2 = d1 - \\sigma \\sqrt{T}$$\n\n### European Put\n\n$$p_0= Xe^{-R_f^cxT}*N(-d2) - S_0e^{-qT}*N(-d1)$$\n\n## Notes\n- Call: `S0xN(d1)`: buy #delta stock & `-X` term: borrow money\n- Put: `X` term: buy in a bond & `-S0xN(-d1)`: Short Stock\n- Roughly: N(D1) = Prob. ITM before T\n- N(D2) = Prlb(S_T > X) = Prob. Exercise at T",
"_____no_output_____"
],
[
"rfd quotes\n\nMy personal setup with options is to do non-directional short term trades to benefit of the rising implied volatility or simply sell expensive premium without betting on a single market direction. \n\nTo make the best of options, one should implement complex strategies, which improves risk reward ratio. Butterflies, Iron Butterflies, double calendar, hedged sttaddle or strangle amd put/call ratio are better vehicles for the short term, which can greatly benefit from the increasing volatility of a nervous market without making bets on a single direction. \n\nIf the goal is simplicity, I prefer to sell puts (get paid to buy the stock at the price you want) and once assigned, sell calls (to increase income and sell for a guaranteed profit). Once in cash again, rinse and repeat. If the market tanks when holding the stock, sell calls further out and have the temperament for market gyrations - it's temporary. \n\nThis works very well with Canadian bank stocks or companies like Costco. Sell a put to buy with a 10% discount from current price. When it tanks enough and you are assigned, sell a call to have a 5% or 10% profit in a month or 2 out. Keep receiving the dividends and call premium meanwhile, until it's sold for your profit price. Repeat again, sell puts (collecting premium) until the price drops again, and so forth. This works forever on quality companies - main Canadian banks and Costco are just some examples. Also works on BCE and AT&T. Again, simple income strategy with fairly low risk. \n\nhttps://forums.redflagdeals.com/day-trading-option-2259692/\nI personally find swing trading with options (larger timeframe) a lot easier than day trading. Technical analysis and multiple indicators help to validate trend / support / resistance, where you can place directional or non-directional trades. \n",
"_____no_output_____"
]
],
[
[
"def newton(f, Df, x0, epsilon, max_iter):\n '''Approximate solution of f(x)=0 by Newton's method.\n https://www.math.ubc.ca/~pwalls/math-python/roots-optimization/newton/\n \n Parameters\n ----------\n f : function\n Function for which we are searching for a solution f(x)=0.\n Df : function\n Derivative of f(x).\n x0 : number\n Initial guess for a solution f(x)=0.\n epsilon : number\n Stopping criteria is abs(f(x)) < epsilon.\n max_iter : integer\n Maximum number of iterations of Newton's method.\n\n Returns\n -------\n xn : number\n Implement Newton's method: compute the linear approximation\n of f(x) at xn and find x intercept by the formula\n x = xn - f(xn)/Df(xn)\n Continue until abs(f(xn)) < epsilon and return xn.\n If Df(xn) == 0, return None. If the number of iterations\n exceeds max_iter, then return None.\n\n Examples\n --------\n >>> f = lambda x: x**2 - x - 1\n >>> Df = lambda x: 2*x - 1\n >>> newton(f,Df,1,1e-8,10)\n Found solution after 5 iterations.\n 1.618033988749989\n '''\n xn = x0\n for n in range(0,max_iter):\n fxn = f(xn)\n if abs(fxn) < epsilon:\n print('Found solution after',n,'iterations.')\n return xn\n Dfxn = Df(xn)\n if Dfxn == 0:\n print('Zero derivative. No solution found.')\n return None\n xn = xn - fxn/Dfxn\n print('Exceeded maximum iterations. No solution found.')\n return None",
"_____no_output_____"
],
[
"import numpy as np\nimport matplotlib.pyplot as plt\nimport pandas as pd\nfrom math import log, sqrt, exp\nfrom scipy import stats\nfrom functools import partial\n\nclass Bsm(object):\n def __init__(self, s0, k, t, r, sigma=0.5, q=0):\n self.s0 = s0 # current stock price\n self.k = k # strike price\n self.t = t # time to expiration in years\n self.r = r # continuous risk free rate\n self.q = q # dividend rate\n self.sigma = sigma # sd\n \n def d1(self, sigma_est=None):\n sigma = sigma_est or self.sigma\n return (log(self.s0/self.k) + (self.r - self.q + 0.5*sigma**2)*self.t)\\\n / (sigma * sqrt(self.t))\n \n def d2(self, sigma_est=None):\n sigma = sigma_est or self.sigma\n return self.d1(sigma_est) - (sigma * sqrt(self.t))\n \n def call_value(self, sigma_est=None):\n return self.s0 * stats.norm.cdf(self.d1(sigma_est),0.,1.) - \\\n self.k * exp(-self.r*self.t) * stats.norm.cdf(self.d2(sigma_est),0.,1.)\n \n def put_value(self, sigma_est=None):\n return self.k * exp(-self.r*self.t) * stats.norm.cdf(-self.d2(sigma_est),0.,1.) - \\\n self.s0 * stats.norm.cdf(-self.d1(sigma_est),0.,1.)\n \n def vega(self, sigma_est=None):\n return self.s0 * stats.norm.pdf(self.d1(sigma_est),0.,1.) * sqrt(self.t)\n \n def call_iv_newton(self, c0, sigma_est=1., n=100):\n self.sigma_est = sigma_est\n f = lambda x: self.call_value(x) - c0\n df = lambda x: self.vega(x)\n return newton(f, df, sigma_est, 0.001, 100)\n\n def call_iv_dichotomy(self, c0):\n c_est = 0\n high = 3\n low = 0\n sigma = (high + low) / 2\n \n while abs(c0 - c_est) > 1e-8:\n c_est = self.call_value(sigma)\n # print(f'c_est = {c_est}, sigma = {sigma}')\n if c0 - c_est > 0:\n low = sigma\n sigma = (sigma + high) / 2\n else:\n high = sigma\n sigma = (low + sigma) / 2\n return sigma\n \n def __repr__(self):\n return f'Base(s0={self.s0}, k={self.k}, t={self.t}, r={self.r})'",
"_____no_output_____"
],
[
"bsm1 = Bsm(s0=100, k=100, t=1.0, r=0.05, q=0, sigma=0.3)\nn_d1 = stats.norm.cdf(bsm1.d1(), 0., 1.)\nbsm1.call_value()",
"_____no_output_____"
],
[
"bsm2 = Bsm(s0=100, k=100, t=1.0, r=0.05, q=0,)\n#bsm2.call_iv_newton(c0=14.2313, n=10)\nbsm2.call_iv_dichotomy(c0=14.2313)",
"_____no_output_____"
],
[
"def iv(df, current_date, strike_date, s0, rf):\n k = df['Strike']\n call = (df['Bid'] + df['Ask']) / 2\n t = (pd.Timestamp(strike_date) - pd.Timestamp(current_date)).days / 365\n \n sigma_init = 1\n sigma_newton = []\n sigma_dichotomy = []\n\n for i in range(df.shape[0]):\n model = Bsm(s0, k[i], t, rf)\n try:\n sigma_newton.append(model.call_iv_newton(c0=call[i], sigma_est=sigma_init))\n #sigma_dichotomy.append(model.call_iv_dichotomy(c0=call[i]))\n except ZeroDivisionError as zde:\n print(f'{zde!r}: {model!r}')\n sigma_dichotomy.append(None)\n return sigma_newton, sigma_dichotomy\n",
"_____no_output_____"
],
[
"s0 = 1290.69\nrf = 0.0248 # libor\n\npd_read_excel = partial(pd.read_excel, 'OEX1290.69.xlsx', skiprows=3)\ndf_05 = pd_read_excel(sheet_name='20190517')\ndf_06 = pd_read_excel(sheet_name='20190621')\ndf_07 = pd_read_excel(sheet_name='20190719')\ndf_09 = pd_read_excel(sheet_name='20190930')\n\n\niv_05 = iv(df_05, '20190422', '20190517', s0, rf)\niv_06 = iv(df_06, '20190422', '20190621', s0, rf)\niv_07 = iv(df_07, '20190422', '20190719', s0, rf)\niv_09 = iv(df_09, '20190422', '20190930', s0, rf)",
"Found solution after 6 iterations.\nZero derivative. No solution found.\nFound solution after 5 iterations.\nZero derivative. No solution found.\nFound solution after 4 iterations.\nZero derivative. No solution found.\nFound solution after 3 iterations.\nZero derivative. No solution found.\nFound solution after 2 iterations.\nZero derivative. No solution found.\nFound solution after 3 iterations.\nZero derivative. No solution found.\nFound solution after 4 iterations.\nZero derivative. No solution found.\nFound solution after 4 iterations.\nFound solution after 4 iterations.\nZero derivative. No solution found.\nFound solution after 4 iterations.\nZero derivative. No solution found.\nFound solution after 4 iterations.\nZero derivative. No solution found.\nFound solution after 5 iterations.\nZero derivative. No solution found.\nFound solution after 5 iterations.\nFound solution after 5 iterations.\nZero derivative. No solution found.\nFound solution after 5 iterations.\nZero derivative. No solution found.\nFound solution after 5 iterations.\nZero derivative. No solution found.\nFound solution after 5 iterations.\nZero derivative. No solution found.\nFound solution after 5 iterations.\nExceeded maximum iterations. No solution found.\nFound solution after 5 iterations.\nZero derivative. No solution found.\nFound solution after 5 iterations.\nZero derivative. No solution found.\nFound solution after 5 iterations.\nZero derivative. No solution found.\nFound solution after 5 iterations.\nZero derivative. No solution found.\nFound solution after 5 iterations.\nZero derivative. No solution found.\nFound solution after 5 iterations.\nZero derivative. No solution found.\nFound solution after 5 iterations.\nZero derivative. No solution found.\nFound solution after 5 iterations.\nZero derivative. No solution found.\nFound solution after 5 iterations.\nZero derivative. No solution found.\nFound solution after 5 iterations.\nZero derivative. No solution found.\nFound solution after 5 iterations.\nZero derivative. No solution found.\nFound solution after 5 iterations.\nZero derivative. No solution found.\nFound solution after 5 iterations.\nZero derivative. No solution found.\nFound solution after 5 iterations.\nZero derivative. No solution found.\nFound solution after 4 iterations.\nZero derivative. No solution found.\nFound solution after 4 iterations.\nZero derivative. No solution found.\nFound solution after 4 iterations.\nFound solution after 5 iterations.\nFound solution after 4 iterations.\nFound solution after 5 iterations.\nFound solution after 4 iterations.\nFound solution after 4 iterations.\nFound solution after 4 iterations.\nFound solution after 4 iterations.\nFound solution after 4 iterations.\nFound solution after 4 iterations.\nFound solution after 3 iterations.\nFound solution after 3 iterations.\nFound solution after 3 iterations.\nFound solution after 3 iterations.\nFound solution after 2 iterations.\nFound solution after 2 iterations.\nFound solution after 2 iterations.\nFound solution after 2 iterations.\nFound solution after 2 iterations.\nFound solution after 2 iterations.\nFound solution after 3 iterations.\nFound solution after 3 iterations.\nFound solution after 4 iterations.\nFound solution after 4 iterations.\nFound solution after 4 iterations.\n"
],
[
"dfs = [df_05, df_06, df_07, df_09]\nivs = [iv_05, iv_06, iv_07, iv_09]\ncolor = (c for c in ['r', 'g', 'k', 'c'])\n\nfor result in zip(dfs, ivs):\n k = result[0]['Strike']\n iv_newton = result[1][0]\n print(len(k), len(iv_newton))\n plt.plot(k, iv_newton, lw=1.5)\n plt.plot(k, iv_newton, next(color))\n\nplt.grid(True)\nplt.xlabel('Strike')\nplt.ylabel('Implied volatility')\nplt.legend()\nplt.xlim(1100, 1800)\nplt.ylim(0, .4)\nplt.show()",
"No handles with labels found to put in legend.\n"
]
],
[
[
"# Option Greeks\nBSM 5 inputs:\n1. Underlying asset price -> $\\frac{\\Delta C}{\\Delta S}$ Delta ---> Gamma\n1. Volatility -> $\\frac{\\Delta C}{\\Delta \\sigma}$ Vega\n1. risk-free rate -> $\\frac{\\Delta C}{\\Delta r_f}$ Rho (rate)\n1. time to expiration -> $\\frac{\\Delta C}{\\Delta t}$ Theta\n1. strike price -> $\\frac{\\Delta C}{\\Delta X}$ X/K\n\n\n- `S+` --> `C+` (0<=Delta<=1)\n- `S-` --> `P+` (-1<=Delta<=0)\n- $\\sigma$+ --> `C+` & `P+` (Vega>0)\n- $r_f$+ --> `C+` (Rho>0) (C=P+S-K/(1+__r__)^T)\n- $r_f$- --> `P+` (Rho<0)\n- Theta < 0; Time decay*\n- `X-` --> `C+`\n- `X+` --> `P+`\n- Long: `Gamma>0`; Short: `Gamma<0`\n- `Gamma` max at ATM, approaches 0 when deep ITM or OTM",
"_____no_output_____"
],
[
"## Delta\n- Sensitivity of the option price to a change in the price of the underlying asset\n- One option = delta stock\n- $delta_{call} = \\Delta C / \\Delta S = N(d1)$\n- $delta_{put} = \\Delta P / \\Delta S = delta_{call} - 1 = N(d1) - 1$\n\n- 0 when deep OTM\n - option price not censitive to $\\delta S$\n- $\\pm1$ when deep ITM\n - very sensitive. 1-1 ratio on stock price\n- $\\pm0.5$ around `X`\n- When t->T\n - ITM delta_c --> 1\n - OTM delta_c --> 0\n \n### delta_p = delta_c - 1\n- Forward on S = Call on S - Put on S\n- FP = C - P --> $dC/dS - dP/dS = d{FP}/dS$\n - FP and S relation: 1:1 \n - delta_p = delta_c - 1\n\n### N(d1) = delta_c\n- $\\frac{dBSM}{dS} = N(d1) + 0 = delta_c$\n\n### Dynamic hedging\n- TL;DR: adjusting # of call options to make delta-neutral portfolio\n- delta-neutral protfolio. value of the portfolio remains unchanged\n - `+ S - h*C`, h=1/delta\n - when S increased by n dollar, call decrease by $delta \\times n$; delta calls decrease n dollar\n- hedge ratio depends on which hedges which\n - Long Stock &\n - short call / long put\n- portfolio value unchanged: \n - need: $\\Delta_{portfolio} / \\Delta S = 0$\n - $nS \\times \\Delta S - nC \\times \\Delta C = 0$\n - $\\frac{\\Delta C}{\\Delta S} = \\frac{nS}{nC} = delta_{call}$\n- Dynamic\n - as t -> T, delta is changing (to OTM 0 or ITM 1)\n - $nS$ remains unchanged, need to change $nC$\n- Maximum Cost\n - change of delta is fast (Gamma max)\n - max at ATM\n - rebalance portfolio more frequently -> higher transaction cost",
"_____no_output_____"
],
[
"## Gamma\n- Sensitivity of the option delta to a change in the price of the underlying asset\n- $gamma = \\Delta delta / \\Delta S$\n- Call and put options on the same stock with same T and X have equal gammas\n",
"_____no_output_____"
],
[
"## Vega\n- sensitivity of the option value to a change in the volatility of the underlying asset",
"_____no_output_____"
],
[
"## Theta\n- sensitivity of the option value to a change in the calendar time\n- Time decay",
"_____no_output_____"
],
[
"## Rho\n- sensitivity of the option value to a change in the $r_f$\n- leverage effect. rho>0 for call\n- small impact compared to vega",
"_____no_output_____"
],
[
"## Volatility\n1. Historical Volatility\n - using historical data to calculate the variance and s.d. of the continuously compounded returns\n - $S_{R_i^c}^2 = \\frac{\\Sigma_{i=1}^N(R_i^c-\\bar{R_i^c})^2}{N-1}$\n - $\\sigma = \\sqrt{S_{R_i^c}^2}$\n1. Implied Volatility\n - Calculate $\\sigma$ backwards\n - IV=20%, Historical=10% -> Option Overvalued",
"_____no_output_____"
],
[
"# Strategies\n## Synthetic\n- Synthetic long/short asset\n - C - P = +S = Forward\n - P - C = -S\n- Synthetic call/put\n - C = S + P\n - P = -S + C\n- Synthetic Stock\n - Equity = rf + forward/futures\n - Synthetic Equity = risk-free asset + stock futures\n- Synthetic Cash\n - rf = Stock - Forward\n - Synthetic risk-free asset = Stock - stock futures\n - S0=10, 1yr FP, rf=10% no div\n - Long stock - forward; \n - $FP=S_0*(1+r_f)^T$\n - if S1=12: stock+=2, (-forward)-=1 => portfolio+=1\n - if S1=8: stock-=2, (-forward)+=3 => portfolio+=1",
"_____no_output_____"
],
[
"## Fiduciary call: C + bond\n- C(X,T)\n- Pure-discount bond pays X in T years\n- Payoff:\n - $S_T \\le X$: $X$\n - $S_T > X$: $S_T$",
"_____no_output_____"
],
[
"## Covered call: $S - C$\n- Neutral\n- Call covered by a stock\n- 股价温和上涨, 达不到X; CG + call premium\n- Same as: Short Put\n- profit: $(S_T-S_0) - max\\{0, (S_T-X)\\} + C$\n - Cost / break even: $S_T = S_0 - C$\n - Max loss at $S_T=0$\n - All the cost: $-(S_0 - C)$\n - Max profit at $S_T \\ge X$\n - Constant: $X - (S_0 - C)$ ",
"_____no_output_____"
],
[
"## Protective put: $S + P_{atm}$\n- Bullish\n- Married Put\n- Same as: long call\n- Pros: \n - unlimited profit\n- Cons:\n - Pay put premium\n - Lower total return\n - Put will expire\n- Deductible: $S_0 - X$\n - If lower the cost, higher deductible\n - i.e., more OTM\n- profit: $(S_T-S_0) + max\\{0,(X-S_T)\\} - P$\n - Cost: $P + S_0$\n - Max loss at $S_T<X$\n - $ X - (P + S_0)$: Strike price - cost\n - Gain at $S_T > P_0 + S_0$ ",
"_____no_output_____"
],
[
"## Bull Call Spread\n- (bullish) benefit from a stock's limited increase in price.\n- Long 1 call & \n- short 1 call, the same expiry, __higher__ X\n- Trade-off:\n - Reduce promium\n - Limit upside profit potential\n- Max loss: \n - Net_premium_spent x 100\n- Max upside profit potential:\n - (call spread width - premium spent) x 100\n- Breakeven:\n - Lower X + net premium spent",
"_____no_output_____"
],
[
"## Bear Put Spread\n- bearish. moderate decline\n- Long 1 put & Short 1 put at lower strike",
"_____no_output_____"
],
[
"## Collar\n- protect against large losses & limits large gains\n- Currently long 100 shares with a gain\n- Sell 1 OTM call & \n- buy 1 OTM put, same expiry\n- Max Loss: Limited\n - Net debit: Put X - Stock purchase price - net premium paid\n - Net credit: Put X - Stock purchase price + net premium collected\n- Max Profit: Limited\n - Net debit: Call X - Stock purchase price - net premium paid\n - Net credit: Call X - Stock purchase price + net premium collected\n- Breakeven:\n - Net debit: Stock purchase price + net premium paid\n - Net credit: Stock purchase price - net premium collected",
"_____no_output_____"
],
[
"## Straddle\n- Profit from a very strong move in either direction\n- Move from low volatility to high volatility\n- `\\/`\n- Long 1 call ATM &\n- Long 1 put ATM on same S, same T and same X\n - X is very close to ATM\n- Max Loss: Limited\n - at X: (Call + Put premium) x 100\n- Max Profit: Unlimited\n- Breakeven:\n - Up: Call strike + call premium + put premium\n - Down: Put strike - call premium - put premium",
"_____no_output_____"
],
[
"## Strangle\n- Long 1 OTM Call & \n- Long 1 OTM Put, Put X < Call X\n- Net debit\n- `\\_/`\n- Breakeven:\n - Up: Call strike + put premium + call premium\n - Down: Put strike - put premium - call premium\n- Max loss: limited\n - put premium + call premium + commission\n - between put X and call X",
"_____no_output_____"
],
[
"## Butterfly\n- Neutral\n- net debit\n- a bull spread + a bear spread\n - Long 1 ITM Call\n - Short 2 ATM Call\n - Long 1 OTM Call\n- `_/\\_`\n- Net premium = C_ITM - 2xC_ATM + C_OTM\n- Breakeven:\n - Lower: Long Call Lower X + Net premium\n - Upper: Long Call Higher X - Net premium\n- Max loss: limited\n - Net premium + commission\n - when underlying <= Long Call Lower X OR underlying >= Long Call Upper X\n- Max profit: limited\n - Short Call X - Long Call Lower X - Net premium - commission\n - when underlying = Short Call X\n ",
"_____no_output_____"
],
[
"## Condor\n- Limited profit: Low IV\n- Long outside X, Short middle X\n - Long 1 ITM Call (Lower X)\n - Short 1 ITM Call\n - Short 1 OTM Call\n - Long 1 OTM Call (Higher X)\n- Same T, \n- Long Condor: `_/T\\_`. Cut off butterfly\n\n- Breakeven:\n - Lower: Long Call Lower X + Net premium\n - Upper: Long Call Higher X - Net premium\n- Max loss: limited\n - Net premium + commission\n - when underlying <= Long Call Lower X OR underlying >= Long Call Upper X\n- Max profit: limited\n - Short Call X - Long Call Lower X - Net premium - commission\n - when underlying between 2 short calls",
"_____no_output_____"
],
[
"## Iron Condor:\n- Call Bull spread + Put Bear spread\n - Long 1 ITM Call\n - Short 1 ITM Call\n - Short 1 ITM Put\n - Long 1 ITM Put\n\n- Breakeven:\n - Lower: Short Call X + Net premium\n - Upper: Short Put X - Net premium\n- Max profit: limited\n - Net premium + commission\n - when underlying between 2 short calls\n- Max loss: limited\n - Long Call X - Short Call X - Net premium - commission\n - when underlying <= Long Call X OR underlying >= Long Put X",
"_____no_output_____"
],
[
"## Iron Butterfly\n- When IV is Ext. high\n- Long 1 OTM Put\n- Short 1 ATM Put\n- Short 1 ATM Call\n- Long 1 OTM Call",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
cb8f2757a60cc9c7e0592522b7cc791d10c286b9
| 672,648 |
ipynb
|
Jupyter Notebook
|
ejemplos/Caso de uso - Agrupar textos por temas.ipynb
|
sergiomora03/ConTexto
|
6e67f13222c9b6310b4c3d112bd5c7fead7fe100
|
[
"X11"
] | null | null | null |
ejemplos/Caso de uso - Agrupar textos por temas.ipynb
|
sergiomora03/ConTexto
|
6e67f13222c9b6310b4c3d112bd5c7fead7fe100
|
[
"X11"
] | null | null | null |
ejemplos/Caso de uso - Agrupar textos por temas.ipynb
|
sergiomora03/ConTexto
|
6e67f13222c9b6310b4c3d112bd5c7fead7fe100
|
[
"X11"
] | null | null | null | 1,774.796834 | 356,264 | 0.958088 |
[
[
[
"# Caso de uso: Agrupación de textos por temáticas similares\n**Autor:** Unidad de Científicos de Datos (UCD)\n\n---\nEste es un caso de uso que utiliza varias funcionalidades de la libtería **ConTexto** para procesar y vectorizar textos de noticias sobre diferentes temas. Luego, sobre estos vectores se aplica t-SNE, una técnica no lineal de reducción de dimensionalidad, para transformar los vectores a un espacio bidimensional.\n\nUna vez se tengan los vectores en 2 dimensiones, se graficarán con un color correspondiente al tema de cada texto. Esto permite observar si los puntos de cada tema quedan juntos entre sí y separados de los otros temas.",
"_____no_output_____"
],
[
"---\n\n## 1. Cargar librerías necesarias y definir parámetros importantes para el caso de uso\n\nEl primer paso es cargar los módulos y las librerías necesarias para correr el caso de estudio. De parte de **ConTexto** se necesitan funciones del módulo de `limpieza`, para hacer un procesamiento básico de los textos y remover *stopwords* y el módulo de `vectorización`, para generar las representaciones vectoriales de los textos.\n\nAdicionalmente, se importan los siguientes paquetes externos:\n\n- `pyplot`: para generar y mostrar las gráficas\n- `manifold`, de `sklearn`: para hacer la reducción de dimensionalidad por medio de t-SNE\n- De `sklearn.datasets` se importa la función `fetch_20newsgroups`, que permite descargar noticias en inglés sobre 20 temas distintos. Para más información sobre este conjunto de datos, se puede consultar <a href=\"https://scikit-learn.org/0.19/datasets/twenty_newsgroups.html\" target=\"_blank\">su documentación</a>.\n",
"_____no_output_____"
]
],
[
[
"# Componentes de ConTexto necesarios\nfrom contexto.limpieza import limpieza_texto, lista_stopwords\nfrom contexto.vectorizacion import *\nfrom contexto.utils.auxiliares import verificar_crear_dir\n\n# Librerías adicionales\nimport matplotlib.pyplot as plt\nfrom sklearn import manifold\nfrom sklearn.datasets import fetch_20newsgroups\n",
"_____no_output_____"
]
],
[
[
"De los 20 temas disponibles en el conjunto de datos, se arman dos grupos, de 4 temas cada uno. La siguiente tabla muestra los temas que contiene cada grupo:\n\n| Grupo 1 | Grupo 2 |\n|----------------------|------------------------|\n|Computación - Gráficos|Religión - Cristiana |\n|Deportes - Béisbol |Alternativo - Ateísmo |\n|Ciencia - Medicina |Religión - Miscelánea |\n|Política - Armas |Política - Medio Oriente|\n\nComo se puede ver, el grupo 1 tiene temas que son más diferentes entre sí, y por lo tanto los vectores resultantes deberían poderse ver más separados entre sí al graficarlos en dos dimensiones. Por el otro lado, el grupo 2 tiene temas con más cosas en común, por lo que en primera instancia puede que no sea tan fácil verlos separados.\n\nFinalmente, se define una lista de vectorizadores a utilizar. Se utilizan los siguientes:\n\n- BOW\n- TF-IDF\n- Hashing\n- Doc2Vec\n- Word2Vec\n- Word2Vec - ignorando palabras desconocidas (Para mayor información sobre esto, consultar el ejemplo de vectorización)",
"_____no_output_____"
]
],
[
[
"# Temas que en principio son bastante diferentes entre sí, por lo que deberían\n# ser más fáciles de agrupar\ngrupo_1 = ['comp.graphics', 'rec.sport.baseball', 'sci.med', 'talk.politics.guns']\n\n# Temas mucho más similares entre sí\ngrupo_2 = ['soc.religion.christian', 'alt.atheism', 'talk.religion.misc', 'talk.politics.mideast']\n\n# Vectorizadores a considerar\nvectorizadores = ['bow', 'tfidf', 'hash', 'doc2vec', 'word2vec', 'word2vec_conocidas']",
"_____no_output_____"
]
],
[
[
"---\n\n## 2. Funciones de apoyo\n\nA continuación se definen dos funciones, que se encargan de llevar a cabo lo siguiente:\n\n- `graficar_textos`: Recibe un arreglo de vectores (de dos dimensiones) y sus respectivas etiquetas (el tema de cada texto). Esta función se encarga de pintar los vectores bidimensionales y asignar un color a cada punto, dependiendo de su respectivo tema. El parámetro *dir_salida* permite definir la carpeta en donde se guardarán los resultados. El nombre de cada gráfica depende del grupo de temas utilizado, de si se realizó normalización sobre los vectores de los textos y de un título que se asigne a la gráfica (parámetro *titulo*).\n- `comparacion_vectorizadores`: Recibe el número que indica qué grupo de temas se va a tratar, una lista de vectorizadores a utilizar y un parámetro *normalizar*, que indica si se quiere hacer normalización min-max sobre los vectores generados para el grupo de textos. Esta normalización puede mejorar los resultados para algunos vectorizadores, en particular los basados en frecuencias (menos el TF-IDF, que ya hace una especie de normalización al tener en cuenta la frecuencia inversa en documentos de los términos). Ests función realiza todo el proceso, que consiste en:\n - Extraer las noticias de los temas del grupo determinado, utilizando la función `fetch_20newsgroups`\n - Pre-procesar los textos, removiendo signos de puntuación y *stopwords*, además de pasar todo el texto a minúsculas\n - Inicializar y ajustar (si aplica) los vectorizadores sobre el corpus de noticias\n - Aplicar los vectorizadores para obtener las representaciones vectoriales de los textos\n - Normalizar los vectores utilizando min-max, solo si se indicó en el parámetro *normalizar* de la función\n - Aplicar la reducción de dimensionalidad, para llevar los vectores a 2 dimensiones. Este paso puede ser un poco demorado\n - Utilizar la función `graficar_textos` para producir las gráficas\n ",
"_____no_output_____"
]
],
[
[
"# Función para graficar los puntos\ndef graficar_textos(X, y, titulo, num_grupo, norm, nombres, dir_salida='salida/caso_uso_vectores/'): \n num_cats = len(np.unique(y))\n # Hasta 8 diferentes categorías\n colores = ['black', 'blue', 'yellow', 'red', 'green', 'orange', 'brown', 'purple']\n color_dict = {i:colores[i] for i in range(num_cats)}\n label_dict = {i:nombres[i] for i in range(num_cats)}\n fig, ax = plt.subplots(figsize=(10,10))\n for g in range(num_cats):\n ix = np.where(y == g)\n ax.scatter(X[ix,0], X[ix,1], c=color_dict[g], label=label_dict[g])\n # Convenciones\n plt.legend(loc=\"lower right\", title=\"Clases\")\n plt.xticks([]), plt.yticks([])\n plt.title(titulo)\n # Guardar la imagen resultante\n verificar_crear_dir(dir_salida)\n norm_str = '_norm' if norm else ''\n nombre_archivo = f'grupo_{num_grupo}_{titulo}{norm_str}.jpg'\n plt.savefig(dir_salida + nombre_archivo)\n plt.close()",
"_____no_output_____"
],
[
"def comparacion_vectorizadores(num_grupo, normalizar, vectorizadores=vectorizadores, dir_salida='salida/caso_uso_vectores/'):\n grupo = grupo_1 if num_grupo == 1 else grupo_2 \n # Obtener dataset de las categorías seleccionadas\n dataset = fetch_20newsgroups(subset='all', categories=grupo, shuffle=True, random_state=42)\n clases = dataset.target\n nombres_clases = dataset.target_names\n # Limpieza básica a los textos para quitar ruido\n # Tener en cuenta que los textos están en inglés\n textos_limpios = [limpieza_texto(i, lista_stopwords('en')) for i in dataset.data]\n # Inicializar los 5 vectorizadores. Todos se configuran para tener 300 elementos,\n # de modo que estén en igualdad de condiciones\n v_bow = VectorizadorFrecuencias(tipo='bow', max_elementos=500)\n v_tfidf = VectorizadorFrecuencias(tipo='tfidf', max_elementos=500)\n v_hash = VectorizadorHash(n_elementos=500)\n v_word2vec = VectorizadorWord2Vec('en')\n v_doc2vec = VectorizadorDoc2Vec(n_elementos=300)\n # Ajustar los modelos que deben ser ajustados sobre el corpus\n v_bow.ajustar(textos_limpios)\n v_tfidf.ajustar(textos_limpios)\n v_doc2vec.entrenar_modelo(textos_limpios)\n # Obtener los vectores para cada vectorizador\n dict_vectores = {}\n for v in vectorizadores:\n print(f'Vectorizando con técnica {v}...')\n if 'conocidas' in v:\n v_mod = v.split('_')[0]\n dict_vectores[v] = eval(f'v_{v_mod}.vectorizar(textos_limpios, quitar_desconocidas=True)')\n else:\n dict_vectores[v] = eval(f'v_{v}.vectorizar(textos_limpios)')\n # Normalizar los vectores\n if normalizar:\n for v in vectorizadores:\n min_v = dict_vectores[v].min(axis=0)\n max_v = dict_vectores[v].max(axis=0)\n dict_vectores[v] = (dict_vectores[v] - min_v) / (max_v - min_v)\n # Aplicar t-sne para dejar vectores en 2 dimensiones\n dict_tsne = {}\n for v in vectorizadores:\n print(f'Reducción de dimensionalidad a vector {v}...')\n dict_tsne[v] = manifold.TSNE(n_components=2, init=\"pca\").fit_transform(dict_vectores[v])\n # Graficar los puntos para cada técnica\n for v in vectorizadores:\n graficar_textos(dict_tsne[v], clases, v, num_grupo, normalizar, nombres_clases, dir_salida=dir_salida)\n",
"_____no_output_____"
]
],
[
[
"---\n\n## 3. Realizar el barrido, para generar las gráficas y comparar\n\nA continuación se hace un barrido para ambos grupos de noticias (1 y 2) y ambas opciones de normalizar (hacerlo o no hacerlo), para generar todas las gráficas, y así poder determinar qué vectorizadores generar vectores más \"separables\" en cada caso.",
"_____no_output_____"
]
],
[
[
"# Barrido para realizar las pruebas\nfor num_grupo in [1, 2]:\n for normalizar in [True, False]:\n print(f'\\n -------------- Grupo: {num_grupo}, normalizar: {normalizar}')\n comparacion_vectorizadores(num_grupo, normalizar, vectorizadores=vectorizadores)",
"\n -------------- Grupo: 1, normalizar: True\nVectorizando con técnica bow...\nVectorizando con técnica tfidf...\nVectorizando con técnica hash...\nVectorizando con técnica doc2vec...\nVectorizando con técnica word2vec...\nVectorizando con técnica word2vec_conocidas...\nReducción de dimensionalidad a vector bow...\nReducción de dimensionalidad a vector tfidf...\nReducción de dimensionalidad a vector hash...\nReducción de dimensionalidad a vector doc2vec...\nReducción de dimensionalidad a vector word2vec...\nReducción de dimensionalidad a vector word2vec_conocidas...\n\n -------------- Grupo: 1, normalizar: False\nVectorizando con técnica bow...\nVectorizando con técnica tfidf...\nVectorizando con técnica hash...\nVectorizando con técnica doc2vec...\nVectorizando con técnica word2vec...\nVectorizando con técnica word2vec_conocidas...\nReducción de dimensionalidad a vector bow...\nReducción de dimensionalidad a vector tfidf...\nReducción de dimensionalidad a vector hash...\nReducción de dimensionalidad a vector doc2vec...\nReducción de dimensionalidad a vector word2vec...\nReducción de dimensionalidad a vector word2vec_conocidas...\n\n -------------- Grupo: 2, normalizar: True\nVectorizando con técnica bow...\nVectorizando con técnica tfidf...\nVectorizando con técnica hash...\nVectorizando con técnica doc2vec...\nVectorizando con técnica word2vec...\nVectorizando con técnica word2vec_conocidas...\nReducción de dimensionalidad a vector bow...\nReducción de dimensionalidad a vector tfidf...\nReducción de dimensionalidad a vector hash...\nReducción de dimensionalidad a vector doc2vec...\nReducción de dimensionalidad a vector word2vec...\nReducción de dimensionalidad a vector word2vec_conocidas...\n\n -------------- Grupo: 2, normalizar: False\nVectorizando con técnica bow...\nVectorizando con técnica tfidf...\nVectorizando con técnica hash...\nVectorizando con técnica doc2vec...\nVectorizando con técnica word2vec...\nVectorizando con técnica word2vec_conocidas...\nReducción de dimensionalidad a vector bow...\nReducción de dimensionalidad a vector tfidf...\nReducción de dimensionalidad a vector hash...\nReducción de dimensionalidad a vector doc2vec...\nReducción de dimensionalidad a vector word2vec...\nReducción de dimensionalidad a vector word2vec_conocidas...\n"
]
],
[
[
"---\n\n## 4. Resultados\nLas imágenes de los resultados quedarán guardadas en la carpeta especificada en el parámetro *dir_salida*. Todas las combinaciones de vectorizadores, grupo y normalización producen 24 imágenes.\n\nA continuación se muestra la imagen obtenida para el grupo uno con el vectorizador Word2Vec, sin tener en cuenta palabras desconocidas y sin normalizar. Se puede ver que en este caso los textos aparecen agrupados por temas, con algunas pocas excepciones.",
"_____no_output_____"
]
],
[
[
"import matplotlib.image as mpimg\n\nimg = mpimg.imread('salida/caso_uso_vectores/grupo_1_word2vec_conocidas.jpg')\nplt.figure(figsize=(10,10))\nimgplot = plt.imshow(img)\nplt.axis('off')\nplt.show()",
"_____no_output_____"
]
],
[
[
"Por otro lado, en el grupo 2 los resultados no son tan buenos. Esto era de esperarse, dado que los temas en este grupo son más cercanos entre sí. A continuación se muestra la gráfica obtenida con el vectoriador Doc2Vec sin normalizar. Se puede ver que, aunque los textos salen mucho más mezclados entre sí, el tema de *Política - Medio oriente*, que en el papel era el más distinto de los 4, sale más separado del resto.\n\nEs importante recordar que en este caso de uso solo se hizo una vectorización, seguida de reducción de dimensionalidad. El entrenamiento de otros modelos no supervisados (*clustering*) o supervisados (modelos de clasificación multiclase) pueden llevar a mejores resultados.",
"_____no_output_____"
]
],
[
[
"img = mpimg.imread('salida/caso_uso_vectores/grupo_2_doc2vec.jpg')\nplt.figure(figsize=(10,10))\nimgplot = plt.imshow(img)\nplt.axis('off')\nplt.show()",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb8f3c5f7f3e4ec92e4566b26c247d57b3ffc317
| 10,590 |
ipynb
|
Jupyter Notebook
|
demo.ipynb
|
joaopalmeiro/pycoas
|
6fd16d720a111ac8e366e80e16ae4b4802a86742
|
[
"MIT"
] | null | null | null |
demo.ipynb
|
joaopalmeiro/pycoas
|
6fd16d720a111ac8e366e80e16ae4b4802a86742
|
[
"MIT"
] | 5 |
2019-08-08T20:00:30.000Z
|
2021-12-03T10:39:20.000Z
|
demo.ipynb
|
joaopalmeiro/pycoas
|
6fd16d720a111ac8e366e80e16ae4b4802a86742
|
[
"MIT"
] | null | null | null | 34.271845 | 248 | 0.395845 |
[
[
[
"%load_ext autoreload\n%autoreload 2",
"_____no_output_____"
],
[
"import pandas as pd\n\nimport pycoas\nfrom pycoas import pd_utils",
"_____no_output_____"
],
[
"pycoas.__version__",
"_____no_output_____"
],
[
"df = pd.read_json(\n \"https://raw.githubusercontent.com/vega/vega-datasets/master/data/penguins.json\"\n)\ndf.head()",
"_____no_output_____"
],
[
"display(*pd_utils.show_unique_values(df, verbose=True))",
"Number of columns: 7\nNumber of object columns: 3\n"
]
],
[
[
"---",
"_____no_output_____"
]
]
] |
[
"code",
"markdown"
] |
[
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
cb8f42b5f76337b70540f5e2fc21bae2ece4f497
| 10,358 |
ipynb
|
Jupyter Notebook
|
JuliaImplementation.ipynb
|
aadimator/label-graph-partitioning
|
2720b0687765b7964d5954a3752e0d617d085817
|
[
"MIT"
] | 2 |
2022-02-18T09:26:24.000Z
|
2022-02-24T05:09:14.000Z
|
JuliaImplementation.ipynb
|
aadimator/label-graph-partitioning
|
2720b0687765b7964d5954a3752e0d617d085817
|
[
"MIT"
] | null | null | null |
JuliaImplementation.ipynb
|
aadimator/label-graph-partitioning
|
2720b0687765b7964d5954a3752e0d617d085817
|
[
"MIT"
] | 1 |
2022-02-18T09:26:25.000Z
|
2022-02-18T09:26:25.000Z
| 30.464706 | 761 | 0.439177 |
[
[
[
"empty"
]
]
] |
[
"empty"
] |
[
[
"empty"
]
] |
cb8f6b485e595d3b8f84323a52d2c650dbbc6893
| 35,014 |
ipynb
|
Jupyter Notebook
|
data/NearestNeighbors.ipynb
|
Build-Week-AirBnb-TT3/airbnb-predictor
|
ffe9647f6a669c459434ebe96a8e20dbc8129c1f
|
[
"MIT"
] | null | null | null |
data/NearestNeighbors.ipynb
|
Build-Week-AirBnb-TT3/airbnb-predictor
|
ffe9647f6a669c459434ebe96a8e20dbc8129c1f
|
[
"MIT"
] | null | null | null |
data/NearestNeighbors.ipynb
|
Build-Week-AirBnb-TT3/airbnb-predictor
|
ffe9647f6a669c459434ebe96a8e20dbc8129c1f
|
[
"MIT"
] | null | null | null | 37.649462 | 147 | 0.450477 |
[
[
[
"### imports\n#\nimport pandas as pd\nimport numpy as np\n#\nimport gzip\nimport csv\nimport json\nimport string\nimport warnings\nwarnings.filterwarnings('ignore')\n#\nfrom distutils.util import strtobool\n# pickle\nimport pickle\n#\nfrom sklearn.neighbors import NearestNeighbors",
"_____no_output_____"
]
],
[
[
"EDA",
"_____no_output_____"
]
],
[
[
"# convert files \n\nasheville = pd.read_csv('ashevillelisting.csv.gz', compression='gzip', header=0, sep=',', quotechar='\"', error_bad_lines=False)\naustin = pd.read_csv('austinlisting.csv.gz', compression='gzip', header=0, sep=',', quotechar='\"', error_bad_lines=False)\nboston = pd.read_csv('bostonlisting.csv.gz', compression='gzip', header=0, sep=',', quotechar='\"', error_bad_lines=False)\nbroward = pd.read_csv('browardcountylisting.csv.gz', compression='gzip', header=0, sep=',', quotechar='\"', error_bad_lines=False)\ncambridge = pd.read_csv('cambridgelisting.csv.gz', compression='gzip', header=0, sep=',', quotechar='\"', error_bad_lines=False)\n\nchicago = pd.read_csv('chicagolisting.csv.gz', compression='gzip', header=0, sep=',', quotechar='\"', error_bad_lines=False)\nclarkcounty = pd.read_csv('clarkcountylisting.csv.gz', compression='gzip', header=0, sep=',', quotechar='\"', error_bad_lines=False)\ncolumbus = pd.read_csv('columbuslisting.csv.gz', compression='gzip', header=0, sep=',', quotechar='\"', error_bad_lines=False)\ndc = pd.read_csv('dclisting.csv.gz', compression='gzip', header=0, sep=',', quotechar='\"', error_bad_lines=False)\ndenver = pd.read_csv('denverlisting.csv.gz', compression='gzip', header=0, sep=',', quotechar='\"', error_bad_lines=False)\n\nhawaii = pd.read_csv('hawaiilisting.csv.gz', compression='gzip', header=0, sep=',', quotechar='\"', error_bad_lines=False)\njerseycity = pd.read_csv('jerseycitylisting.csv.gz', compression='gzip', header=0, sep=',', quotechar='\"', error_bad_lines=False)\n\nlosangeles = pd.read_csv('losangeleslisting.csv.gz', compression='gzip', header=0, sep=',', quotechar='\"', error_bad_lines=False)\nnashville = pd.read_csv('nashvillelisting.csv.gz', compression='gzip', header=0, sep=',', quotechar='\"', error_bad_lines=False)\nneworleans = pd.read_csv('neworleanslisting.csv.gz', compression='gzip', header=0, sep=',', quotechar='\"', error_bad_lines=False)\n\nnyc = pd.read_csv('nyclisting.csv.gz', compression='gzip', header=0, sep=',', quotechar='\"', error_bad_lines=False)\noakland = pd.read_csv('oaklandlisting.csv.gz', compression='gzip', header=0, sep=',', quotechar='\"', error_bad_lines=False)\npacificgrove = pd.read_csv('pacificfgrovelisting.csv.gz', compression='gzip', header=0, sep=',', quotechar='\"', error_bad_lines=False)\nportland = pd.read_csv('portlandlisting.csv.gz', compression='gzip', header=0, sep=',', quotechar='\"', error_bad_lines=False)\nri = pd.read_csv('rhodeislandlisting.csv.gz', compression='gzip', header=0, sep=',', quotechar='\"', error_bad_lines=False)\n\nsalem = pd.read_csv('salemlisting.csv.gz', compression='gzip', header=0, sep=',', quotechar='\"', error_bad_lines=False)\nsandiego = pd.read_csv('sandiegolisting.csv.gz', compression='gzip', header=0, sep=',', quotechar='\"', error_bad_lines=False)\nsanfran = pd.read_csv('sanfranciscolisting.csv.gz', compression='gzip', header=0, sep=',', quotechar='\"', error_bad_lines=False)\nsanmateo = pd.read_csv('sanmateocountylisting.csv.gz', compression='gzip', header=0, sep=',', quotechar='\"', error_bad_lines=False)\n\n\nsantacruz = pd.read_csv('santacruzlisting.csv.gz', compression='gzip', header=0, sep=',', quotechar='\"', error_bad_lines=False)\nseattle = pd.read_csv('seattlelisting.csv.gz', compression='gzip', header=0, sep=',', quotechar='\"', error_bad_lines=False)\ntwincities = pd.read_csv('twincitieslisting.csv.gz', compression='gzip', header=0, sep=',', quotechar='\"', error_bad_lines=False)",
"_____no_output_____"
],
[
"# get city\nasheville['location'] = 'Asheville'\naustin['location'] = 'Austin'\nboston['location'] = 'Boston'\nbroward['location'] = 'Broward County'\ncambridge['location'] = 'Cambridge'\nchicago['location'] = 'Chicago'\nclarkcounty['location'] = 'Clark County'\ncolumbus['location'] = 'Columbus'\ndc['location'] = 'Washington, D.C.'\ndenver['location'] = 'Denver'\nhawaii['location'] = 'Anywhere in Hawaii'\njerseycity['location'] = 'Jersey City'\nlosangeles['location'] = 'Los Angeles'\nnashville['location'] = 'Nashville'\nneworleans['location'] = 'New Orleans'\nnyc['location'] = 'New York City'\noakland['location'] = 'Oakland'\npacificgrove['location'] = 'Pacific Grove'\nportland['location'] = 'Portland'\nri['location'] = 'Any City in Rhode Island'\nsalem['location'] = 'Salem'\nsandiego['location'] = 'San Diego'\nsanfran['location'] = 'San Francisco'\nsanmateo['location'] = 'San Mateo County'\nsantacruz['location'] = 'Santa Cruz County'\nseattle['location'] = 'Seattle'\ntwincities['location'] = 'Twin Cities'\n# get states \nasheville['state'] = 'NC'\naustin['state'] = 'TX'\nboston['state'] = 'MA'\nbroward['state'] = 'FL'\ncambridge['state'] = 'MA'\nchicago['state'] = 'IL'\nclarkcounty['state'] = 'NV'\ncolumbus['state'] = 'OH'\ndc['state'] = 'DC'\ndenver['state'] = 'CO'\nhawaii['state'] = 'HI'\njerseycity['state'] = 'NJ'\nlosangeles['state'] = 'CA'\nnashville['state'] = 'TN'\nneworleans['state'] = 'LA'\nnyc['state'] = 'NY'\noakland['state'] = 'CA'\npacificgrove['state'] = 'CA'\nportland['state'] = 'OR'\nri['state'] = 'RI'\nsalem['state'] = 'OR'\nsandiego['state'] = 'CA'\nsanfran['state'] = 'CA'\nsanmateo['state'] = 'CA'\nsantacruz['state'] = 'CA'\nseattle['state'] = 'WA'\ntwincities['state'] = 'MN'",
"_____no_output_____"
],
[
"# concat all the dfs \n\ndf_first = pd.concat([asheville,austin,boston,broward,cambridge,\n chicago,clarkcounty,columbus,dc,denver,\n hawaii,jerseycity,losangeles,nashville,neworleans,\n nyc,oakland,pacificgrove,portland,ri,\n salem,sandiego,sanfran,sanmateo,\n santacruz,seattle,twincities\n ])",
"_____no_output_____"
],
[
"# dropping cols\n\ndf_first = df_first.drop(['calculated_host_listings_count',\n 'calculated_host_listings_count_entire_homes',\n 'calculated_host_listings_count_private_rooms',\n 'calculated_host_listings_count_shared_rooms',\n 'first_review',\n 'minimum_minimum_nights',\n 'maximum_minimum_nights',\n 'minimum_maximum_nights',\n 'maximum_maximum_nights',\n 'has_availability',\n 'availability_30',\n 'availability_60',\n 'availability_90',\n 'availability_365',\n 'number_of_reviews_ltm',\n 'number_of_reviews_l30d',\n 'host_response_time',\n 'host_response_rate',\n 'host_acceptance_rate',\n 'host_listings_count',\n 'host_total_listings_count',\n 'calendar_updated', \n 'reviews_per_month',\n 'neighbourhood',\n 'neighbourhood_cleansed', \n 'neighbourhood_group_cleansed',\n 'latitude',\n 'longitude',\n 'license', \n 'name',\n 'neighborhood_overview',\n 'host_about',\n 'host_id',\n 'host_since',\n 'host_verifications',\n 'review_scores_rating',\n 'review_scores_accuracy',\n 'review_scores_cleanliness',\n 'review_scores_checkin',\n 'review_scores_communication',\n 'review_scores_location',\n 'review_scores_value',\n 'host_location',\n 'last_scraped',\n 'last_review',\n 'minimum_nights',\n 'maximum_nights',\n 'minimum_nights_avg_ntm',\n 'maximum_nights_avg_ntm',\n 'calendar_last_scraped',\n 'scrape_id', \n 'description',\n 'picture_url',\n 'host_url',\n 'host_name',\n 'host_thumbnail_url',\n 'host_picture_url',\n 'host_neighbourhood',\n 'bathrooms',\n 'host_has_profile_pic',\n 'host_identity_verified',\n 'number_of_reviews'\n \n \n \n ],\n axis=1)",
"_____no_output_____"
],
[
"df1 = df_first",
"_____no_output_____"
],
[
"# replace nulls\ndf1 = df1.replace(np.nan, 0)",
"_____no_output_____"
],
[
"# cleaning on bathrooms_text\ndf1['bathrooms_text'] = df1['bathrooms_text'].str.rstrip(string.ascii_letters)\ndf1['bathrooms_text'] = df1['bathrooms_text'].str.strip()\ndf1['bathrooms_text'] = df1['bathrooms_text'].str.lower()\n# create and apply subset\ndf1 = df1[df1['bathrooms_text'].notna()]\nsubset = df1['bathrooms_text'].str.contains('half')\ndf1['bathrooms_text'] = df1['bathrooms_text'].where(~subset, other=0.5)\n\n# cleaning on bathrooms_text\ndf1['bathrooms_text'] = df1['bathrooms_text'].str.rstrip(string.ascii_letters)\ndf1['bathrooms_text'] = df1['bathrooms_text'].str.strip()\ndf1['bathrooms_text'] = df1['bathrooms_text'].str.lower()\n# create and apply subset\ndf1 = df1[df1['bathrooms_text'].notna()]\nsubset = df1['bathrooms_text'].str.contains('half')\ndf1['bathrooms_text'] = df1['bathrooms_text'].where(~subset, other=0.5)\n\n\n# for some reason you get an error trying to convert it to float if I don't run the above line twice\n# change bathroom_text to float\ndf1['bathrooms_text'] = df1['bathrooms_text'].astype(float)",
"_____no_output_____"
],
[
"df1.info()",
"<class 'pandas.core.frame.DataFrame'>\nInt64Index: 184339 entries, 0 to 3909\nData columns (total 14 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 id 184339 non-null int64 \n 1 listing_url 184339 non-null object \n 2 host_is_superhost 184339 non-null object \n 3 property_type 184339 non-null object \n 4 room_type 184339 non-null object \n 5 accommodates 184339 non-null int64 \n 6 bathrooms_text 184339 non-null float64\n 7 bedrooms 184339 non-null float64\n 8 beds 184339 non-null float64\n 9 amenities 184339 non-null object \n 10 price 184339 non-null object \n 11 instant_bookable 184339 non-null object \n 12 location 184339 non-null object \n 13 state 184339 non-null object \ndtypes: float64(3), int64(2), object(9)\nmemory usage: 21.1+ MB\n"
],
[
"# lowcase other cols that besides state and city/location\ndf1['amenities'] = df1['amenities'].str.lower()\ndf1['property_type'] = df1['property_type'].str.lower()\ndf1['room_type'] = df1['room_type'].str.lower()\n",
"_____no_output_____"
],
[
"# feat engineer amenities if in coloumns to help model\ndf1['hot_water'] = df1['amenities'].str.contains('hot water')\ndf1['air_conditioning'] = df1['amenities'].str.contains('air conditioning')\ndf1['parking'] = df1['amenities'].str.contains('parking')\ndf1['refrigerator'] = df1['amenities'].str.contains('refrigerator')\ndf1['patio_balcony'] = df1['amenities'].str.contains('patio')\ndf1['wifi'] = df1['amenities'].str.contains('wifi')\ndf1['breakfast'] = df1['amenities'].str.contains('breakfast')\ndf1['hair_dryer'] = df1['amenities'].str.contains('hair dryer')\ndf1['waterfront'] = df1['amenities'].str.contains('waterfront')\ndf1['workspace'] = df1['amenities'].str.contains('workspace')\ndf1['kitchen'] = df1['amenities'].str.contains('kitchen')\ndf1['fireplace'] = df1['amenities'].str.contains('fireplace')\ndf1['tv'] = df1['amenities'].str.contains('tv')\ndf1['clothes_dryer'] = df1['amenities'].str.contains('dryer')",
"_____no_output_____"
],
[
"df1.head()\n",
"_____no_output_____"
]
],
[
[
"### load in models and apply knn",
"_____no_output_____"
]
],
[
[
"tfidf = pickle.load(open('tfidf.pkl', 'rb'))",
"_____no_output_____"
],
[
"dtm = tfidf.fit_transform(df1['amenities'])\ndtm = pd.DataFrame(dtm.todense(), columns=tfidf.get_feature_names())",
"_____no_output_____"
],
[
"# knn \nknn = NearestNeighbors(n_neighbors=10, metric='cosine')\nknn.fit(dtm)",
"_____no_output_____"
],
[
"def recommender(text):\n x = pd.DataFrame(columns=df1.columns)\n input_features = tfidf.transform(text)\n for i in knn.kneighbors(input_features, n_neighbors=5, return_distance=False)[0]:\n x = x.append(df1.iloc[[i]])\n return x",
"_____no_output_____"
]
],
[
[
"Takes fourteen popular AirBnB amenities to help with accuracy along with other listings \n\nto assist in determining house listing price using nearest neighbor\n1. hot water\n2. air_conditioning\n3. parking\n4. refrigerator\n5. patio_balcony\n6. wifi\n7. breakfast\n8. hair_dryer\n9. waterfront\n10. workspace\n11. kitchen\n12. fireplace\n13. tv\n14. clothes_dryer",
"_____no_output_____"
]
],
[
[
"recommender(['studio', 'tv', 'kitchen'])",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb8f70fd0c9e5f83dc38f7e664bea78b45885f94
| 5,764 |
ipynb
|
Jupyter Notebook
|
cosmos-gremlin.ipynb
|
cjoakim/azure-jupyter
|
498c91efa8ed7c7e366d7de056e214be537b8a19
|
[
"MIT"
] | 1 |
2022-01-15T21:33:49.000Z
|
2022-01-15T21:33:49.000Z
|
cosmos-gremlin.ipynb
|
cjoakim/azure-jupyter
|
498c91efa8ed7c7e366d7de056e214be537b8a19
|
[
"MIT"
] | null | null | null |
cosmos-gremlin.ipynb
|
cjoakim/azure-jupyter
|
498c91efa8ed7c7e366d7de056e214be537b8a19
|
[
"MIT"
] | 1 |
2022-01-15T21:34:00.000Z
|
2022-01-15T21:34:00.000Z
| 27.061033 | 107 | 0.5144 |
[
[
[
"## CosmosDB/Gremlin Client with gremlinpython library\n\n- https://docs.microsoft.com/en-us/azure/cosmos-db/graph/create-graph-python\n- https://pypi.org/project/gremlinpython/",
"_____no_output_____"
],
[
"### Enable just a single Python Async Event Loop with nest_asyncio",
"_____no_output_____"
]
],
[
[
"import nest_asyncio\nnest_asyncio.apply()",
"_____no_output_____"
]
],
[
[
"### Create Gremlin Client object from environment variable configuration values",
"_____no_output_____"
]
],
[
[
"import json\nimport os\nimport sys\nimport traceback\n\nfrom gremlin_python.driver import client, serializer, protocol\nfrom gremlin_python.driver.protocol import GremlinServerError\n\nacct = os.environ['AZURE_COSMOSDB_GRAPHDB_ACCT']\nkey = os.environ['AZURE_COSMOSDB_GRAPHDB_KEY']\ndbname = os.environ['AZURE_COSMOSDB_GRAPHDB_DBNAME']\ngraph = os.environ['AZURE_COSMOSDB_GRAPHDB_GRAPH']\n\ndbname = 'december'\ngraph = 'rdf1'\nurl = 'wss://{}.gremlin.cosmosdb.azure.com:443/'.format(acct)\nusername = '/dbs/{}/colls/{}'.format(dbname, graph)\n\nprint('account: {}'.format(acct))\nprint('key length: {}'.format(len(key)))\nprint('url: {}'.format(url))\nprint('username: {}'.format(username))\n\nclient = client.Client(url, 'g', username=username, password=key, ssl=True,\n message_serializer=serializer.GraphSONSerializersV2d0())\n",
"account: cjoakimcosmosgremlinbom\nkey length: 88\nurl: wss://cjoakimcosmosgremlinbom.gremlin.cosmosdb.azure.com:443/\nusername: /dbs/december/colls/rdf1\n"
]
],
[
[
"### Count the Vertices and Edges in the CosmosDB/Gremlin Graph",
"_____no_output_____"
]
],
[
[
"query = 'g.V().count()'\ncallback = client.submitAsync(query)\nif callback.result() is not None:\n print(\"vertex count: {0}\".format(callback.result().all().result()))\nelse:\n print(\"unable to execute query: {0}\".format(query))\n\nquery = 'g.E().count()'\ncallback = client.submitAsync(query)\nif callback.result() is not None:\n print(\"edge count: {0}\".format(callback.result().all().result()))\nelse:\n print(\"unable to execute query: {0}\".format(query))\n",
"vertex count: [1570]\nedge count: [3413]\n"
]
],
[
[
"### Query a given Vertex",
"_____no_output_____"
]
],
[
[
"pk = \"z584e6bb0-2497-d97a-a0af-41f37462c9a9\"\nid = \"z584e6bb0-2497-d97a-a0af-41f37462c9a9\"\nquery = 'g.V(\"{}\", \"{}\")'.format(pk, id)\nprint('query: {}'.format(query))\n\ncallback = client.submitAsync(query)\nif callback.result() is not None:\n obj = callback.result().all().result()\n pretty_json = json.dumps(obj, sort_keys=False, indent=2)\n print(pretty_json)\nelse:\n print(\"unable to execute query: {0}\".format(query))",
"query: g.V(\"z584e6bb0-2497-d97a-a0af-41f37462c9a9\", \"z584e6bb0-2497-d97a-a0af-41f37462c9a9\")\n[\n {\n \"id\": \"z584e6bb0-2497-d97a-a0af-41f37462c9a9\",\n \"label\": \"resources\",\n \"type\": \"vertex\",\n \"properties\": {\n \"pk\": [\n {\n \"id\": \"z584e6bb0-2497-d97a-a0af-41f37462c9a9|pk\",\n \"value\": \"z584e6bb0-2497-d97a-a0af-41f37462c9a9\"\n }\n ],\n \"core#prefLabel\": [\n {\n \"id\": \"e7321288-e3ae-4c54-9dc3-cce8b6a65aea\",\n \"value\": \"Tony Stark\"\n }\n ],\n \"core#prefLabel_lang\": [\n {\n \"id\": \"dd2a31d2-d0cb-4295-9e25-b54c006b6dcf\",\n \"value\": \"en\"\n }\n ]\n }\n }\n]\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb8f7218a3295ddbbd5fac1e2bf687879e3eb636
| 21,507 |
ipynb
|
Jupyter Notebook
|
.ipynb_checkpoints/Untitled1-checkpoint.ipynb
|
Jeevi10/AICR
|
53f16cf05a4f0ac315de81c5c94c5717356e8ac9
|
[
"MIT"
] | null | null | null |
.ipynb_checkpoints/Untitled1-checkpoint.ipynb
|
Jeevi10/AICR
|
53f16cf05a4f0ac315de81c5c94c5717356e8ac9
|
[
"MIT"
] | null | null | null |
.ipynb_checkpoints/Untitled1-checkpoint.ipynb
|
Jeevi10/AICR
|
53f16cf05a4f0ac315de81c5c94c5717356e8ac9
|
[
"MIT"
] | null | null | null | 35.785358 | 1,050 | 0.536291 |
[
[
[
"import torch\nimport torch.nn as nn\nimport torch.nn.functional as F\nfrom torch.autograd import Variable\n\nimport torchvision\nfrom torchvision import datasets\nfrom torchvision import transforms\nfrom torchvision.utils import save_image\nfrom torchsummary import summary\nfrom matplotlib import pyplot as plt\n\n#from pushover import notify\nfrom random import randint\n\nfrom IPython.display import Image\nfrom IPython.core.display import Image, display\nimport dataloader as dl\nimport model as m\nimport networks\nfrom networks import LeNet, ClassificationNet\nfrom testers import attack_test\nfrom resnet import ResNet\nimport gmm as gmm\nimport parameters as p\nimport helper\nimport misc\n\nfrom mpl_toolkits.mplot3d import Axes3D\nimport matplotlib.pyplot as plt\n\n\n%load_ext autoreload\n%autoreload 2",
"_____no_output_____"
],
[
"torch.cuda.set_device(1)",
"_____no_output_____"
],
[
"bs = 256\ntrain_loader,test_loader,loader_list = misc.get_dataloaders(\"Lenet\")",
"_____no_output_____"
],
[
"fixed_x, _ = next(iter(loader_list[0]))\nsave_image(fixed_x, 'real_image.png')\n\nImage('real_image.png')\n\n\ndef in_top_k(targets, preds, k):\n topk = preds.topk(k,largest=False)[1]\n return (targets.unsqueeze(1) == topk).any(dim=1)\n\n\ndef cross_corr(centers):\n c = centers.view(-1,10*centers.size(1))\n corr =torch.matmul(c.T,c)\n loss = torch.norm(torch.triu(corr, diagonal=1, out=None))\n return 2*loss/corr.size(0)",
"_____no_output_____"
],
[
"class Proximity(nn.Module):\n\n def __init__(self, num_classes=100, feat_dim=1024, use_gpu=True, margin = 0.0 ):\n super(Proximity, self).__init__()\n self.num_classes = num_classes\n self.feat_dim = feat_dim\n self.use_gpu = use_gpu\n self.device = torch.device(\"cuda:1\")\n self.margin = margin\n\n if self.use_gpu:\n self.centers = nn.Parameter(torch.randn(self.num_classes, self.feat_dim).cuda())\n else:\n self.centers = nn.Parameter(torch.randn(self.num_classes, self.feat_dim))\n\n def forward(self, x , labels):\n batch_size = x.size(0)\n distmat = torch.pow(x, 2).sum(dim=1, keepdim=True).expand(batch_size, self.num_classes) + \\\n torch.pow(self.centers, 2).sum(dim=1, keepdim=True).expand(self.num_classes, batch_size).t()\n distmat.addmm_(1, -2, x, self.centers.t())\n\n classes = torch.arange(self.num_classes).long()\n if self.use_gpu: classes = classes.to(self.device)\n labels = labels.unsqueeze(1).expand(batch_size, self.num_classes)\n mask = labels.eq(classes.expand(batch_size, self.num_classes))\n d_y = distmat[mask.clone()]\n \n \n values, indices = torch.topk(distmat,2, dim=1, largest=False, sorted=True, out=None)\n d_1 = values[:,0]\n d_2 = values[:,1]\n \n indicators = in_top_k(labels,distmat,1)[:,0]\n con_indicators = ~ indicators.clone()\n \n d_c = d_2*indicators + d_1*con_indicators\n \n loss = F.relu((d_y-d_c)/(d_y+d_c) + self.margin)\n mean_loss = loss.mean()\n return mean_loss, torch.argmin(distmat,dim=1)\n ",
"_____no_output_____"
],
[
"image_channels = fixed_x.size(1)",
"_____no_output_____"
],
[
"embedding_net = LeNet()\nmodel = ClassificationNet(embedding_net, n_classes=p.n_classes).cuda()\ngmm = gmm.GaussianMixturePrior(p.num_classes, network_weights=list(model.embedding_net.layers.parameters()), pi_zero=0.99).cuda()\n \ncriterion_prox_256 = Proximity(num_classes=10, feat_dim=256, use_gpu=True,margin=0.75)\ncriterion_prox_1024 = Proximity(num_classes=10, feat_dim=1024, use_gpu=True, margin=0.75)",
"_____no_output_____"
],
[
"optimizer_pre = torch.optim.Adam([{'params':model.parameters()}], lr=1e-3, weight_decay=1e-7)\n#optimizer_post = torch.optim.Adam([{'params':model.parameters()},\n# {'params': gmm.means, 'lr': p.lr_mu},\n# {'params': gmm.gammas, 'lr': p.lr_gamma},\n# {'params': gmm.rhos, 'lr': p.lr_rho}], lr=p.lr_post)\noptimizer_post = torch.optim.Adam([{'params':model.parameters()}], lr=5e-3, weight_decay=1e-7)\n#optimizer_prox_1024 = torch.optim.SGD(criterion_prox_1024.parameters(), lr=0.1)\n#optimizer_conprox_1024 = torch.optim.SGD(criterion_conprox_1024.parameters(), lr=0.0001)\n \n \noptimizer_prox_256 = torch.optim.SGD(criterion_prox_256.parameters(), lr=0.01)\noptimizer_prox_1024 = torch.optim.SGD(criterion_prox_1024.parameters(), lr=0.01)\n\n\ncriterion = nn.CrossEntropyLoss()",
"_____no_output_____"
],
[
"!rm -rfr reconstructed\n!rm -rfr softmaxreconstructed\n!rm -rfr figs\n!mkdir reconstructed\n!mkdir softmaxreconstructed\n!mkdir figs",
"_____no_output_____"
],
[
"epochs_0 = 50\nepochs_1 = 60",
"_____no_output_____"
],
[
"import time\nimport pandas as pd\nimport matplotlib.patheffects as PathEffects\n%matplotlib inline\nimport seaborn as sns\nimport numpy as np\nimport matplotlib\nimport matplotlib.pyplot as plt\n\nsns.set_style('darkgrid')\nsns.set_palette('muted')\nsns.set_context(\"notebook\", font_scale=1.5,\n rc={\"lines.linewidth\": 2.5})\nRS = 123\n\nfrom sklearn.manifold import TSNE\nfrom sklearn.decomposition import PCA\n\n\ncolors = ['#1f77b4', '#ff7f0e', '#2ca02c', '#d62728',\n '#9467bd', '#8c564b', '#e377c2', '#7f7f7f',\n '#bcbd22', '#17becf']\n\nmnist_classes = ['0', '1', '2', '3', '4', '5', '6', '7', '8', '9']\n\ndef t_sne_gen(data):\n fashion_tsne = TSNE(random_state=RS).fit_transform(data.numpy())\n #fashion_pca = PCA(n_components=2, svd_solver='full').fit(data.numpy())\n #x = fashion_pca.transform(data.numpy())\n return fashion_tsne\n\n\ndef fashion_scatter(x, colors,name,folder):\n # choose a color palette with seaborn.\n num_classes = len(np.unique(colors))\n palette = np.array(sns.color_palette(\"hls\", num_classes))\n\n # create a scatter plot.\n f = plt.figure(figsize=(8, 8))\n ax = plt.subplot(aspect='equal')\n sc = ax.scatter(x[:,0], x[:,1], lw=0, s=40, c=palette[colors.astype(np.int)])\n plt.title(name)\n plt.xlim(-25, 25)\n plt.ylim(-25, 25)\n ax.axis('off')\n ax.axis('tight')\n\n # add the labels for each digit corresponding to the label\n txts = []\n\n for i in range(num_classes):\n\n # Position of each label at median of data points.\n\n xtext, ytext = np.median(x[colors == i, :], axis=0)\n txt = ax.text(xtext, ytext, str(i), fontsize=24)\n txt.set_path_effects([\n PathEffects.Stroke(linewidth=5, foreground=\"w\"),\n PathEffects.Normal()])\n txts.append(txt)\n \n plt.savefig(folder+name+'.png')\n\n return f, ax, sc, txts\n\n\ndef plot_embeddings(embeddings, targets, xlim=None, ylim=None):\n fig = plt.figure(figsize=(10,10))\n ax = fig.add_subplot(111, projection='3d')\n for i in range(10):\n #ax = fig.add_subplot(111, projection='3d')\n inds = np.where(targets==i)[0]\n ax.scatter(embeddings[inds,0], embeddings[inds,1], embeddings[inds,2], alpha=0.5, color=colors[i])\n if xlim:\n plt.xlim(xlim[0], xlim[1])\n if ylim:\n plt.ylim(ylim[0], ylim[1])\n plt.legend(mnist_classes)\n\ndef extract_embeddings(dataloader, model, pretrain):\n with torch.no_grad():\n model.eval()\n embeddings_1 = np.zeros((len(dataloader.dataset), networks.vis_size))\n embeddings_2 = np.zeros((len(dataloader.dataset), networks.vis_size))\n labels = np.zeros(len(dataloader.dataset))\n k = 0\n for images, target in dataloader:\n \n images = images.cuda()\n emb_1, emb_2= model.get_embedding(images, pretrain)\n emb_1, emb_2 = emb_1.cpu(), emb_2.cpu()\n embeddings_1[k:k+len(images)] = emb_1\n embeddings_2[k:k+len(images)] = emb_2\n labels[k:k+len(images)] = target.numpy()\n k += len(images)\n return embeddings_1, embeddings_2, labels\n\n\n\n",
"_____no_output_____"
],
[
"import copy\n\ncorrect =0\nnum_example =0\ntest_loss_bce=0\ntest_correct=0\ntest_num_example =0\nfor epoch in range(epochs_0):\n model.train()\n for idx, (images, target) in enumerate(train_loader):\n images, target= images.cuda(), target.cuda()\n out, rep_1, rep_2 = model(images, test= False)\n loss_bce = criterion(out,target)\n #loss_prox_1024 = criterion_prox_1024(rep_1, target) \n #loss_conprox_1024 = criterion_conprox_1024(rep_1, target) \n #loss_prox_256 = criterion_prox_256(rep_2, target) \n #loss_conprox_256= criterion_conprox_256(rep_2, target) \n loss = loss_bce #+ loss_prox_1024 + loss_prox_256 - loss_conprox_1024*0.0001 - loss_conprox_256*0.0001\n preds = out.data.max(1, keepdim=True)[1]\n correct += preds.eq(target.data.view_as(preds)).sum()\n num_example += len(target)\n optimizer_pre.zero_grad()\n loss.backward()\n optimizer_pre.step()\n \n to_print = \"Epoch[{}/{}] Loss: {:.3f} Accuracy: {}\".format(epoch+1,epochs_0, loss.item()/bs, correct.item()/num_example)\n \n \n if idx % 500 == 0:\n print(to_print)\n \n \n \n model.eval()\n \n with torch.no_grad():\n for images, target in test_loader:\n images, target = images.cuda(), target.cuda()\n out, rep_1, rep_2= model(images, test=False)\n loss_bce = criterion(out,target)\n preds = out.data.max(1, keepdim=True)[1]\n test_correct += preds.eq(target.data.view_as(preds)).sum()\n test_num_example += len(target)\n test_loss_bce+=loss_bce.item()\n \n \n \n test_loss_bce /= len(test_loader.dataset)\n print( \"test_Loss: {:.3f} Test accuracy: {}\".format( test_loss_bce, test_correct.item()/test_num_example))\n if epoch %10==0:\n val_embeddings_1, val_embeddings_2, val_labels_baseline = extract_embeddings(test_loader, model,False)\n plot_embeddings(val_embeddings_1, val_labels_baseline) \n plot_embeddings(val_embeddings_2, val_labels_baseline) \n #fashion_scatter(t_sne_gen(rep_2.cpu()), target.cpu().numpy(),\"Clean_data: \"+\"VAE_\"+str(epoch)+\"softmax_rep2\",\"./softmaxreconstructed/\") \n #fashion_scatter(t_sne_gen(rep_1.cpu()), target.cpu().numpy(),\"Clean_data: \"+\"VAE_\"+str(epoch)+\"softmax_rep1\",\"./softmaxreconstructed/\")\n attack_test(model, test_loader, nn.CrossEntropyLoss() )",
"_____no_output_____"
],
[
"import copy\ncorrect =0\nnum_example =0\ntest_loss_bce=0\ntest_correct=0\ntest_num_example =0\npre_wts= copy.deepcopy(list(model.embedding_net.layers.parameters()))\nfor epoch in range(epochs_1):\n model.train()\n for idx, (images, target) in enumerate(train_loader):\n images, target= images.cuda(), target.cuda()\n out, rep_1, rep_2 = model(images,test=False)\n #loss_bce = criterion(out,target)\n loss_prox_1024, _ = criterion_prox_1024(rep_1, target) \n loss_prox_256, preds = criterion_prox_256(rep_2, target) \n loss = loss_prox_256 + loss_prox_1024 + 0.1 * cross_corr(criterion_prox_256.centers)\n #preds = out.data.max(1, keepdim=True)[1]\n correct += preds.eq(target.data.view_as(preds)).sum()\n num_example += len(target)\n optimizer_post.zero_grad()\n optimizer_prox_1024.zero_grad() \n optimizer_prox_256.zero_grad() \n loss.backward()\n optimizer_post.step()\n \n for param in criterion_prox_256.parameters():\n param.grad.data *= (1. /1)\n optimizer_prox_256.step()\n \n \n for param in criterion_prox_1024.parameters():\n param.grad.data *= (1. /1)\n optimizer_prox_256.step()\n \n \n to_print = \"Epoch[{}/{}] Loss: {:.3f} Accuracy: {}\".format(epoch+1,epochs_1, loss.item()/bs, correct.item()/num_example)\n \n \n if idx % 500 == 0:\n print(to_print)\n \n #helper.plot_histogram(epoch,idx, pre_wts, list(model.embedding_net.layers.parameters()), list(gmm.parameters()), correct.item()/num_example,\"./figs/\") \n \n model.eval()\n \n with torch.no_grad():\n for images, target in test_loader:\n images, target = images.cuda(), target.cuda()\n out, rep_1, rep_2= model(images, test=False)\n loss_bce = criterion(out,target)\n loss_prox_256, preds = criterion_prox_256(rep_2, target) \n #preds = out.data.max(1, keepdim=True)[1]\n test_correct += preds.eq(target.data.view_as(preds)).sum()\n test_num_example += len(target)\n test_loss_bce+=loss_bce.item()\n \n \n \n test_loss_bce /= len(test_loader.dataset)\n print( \"test_Loss: {:.3f} Test accuracy: {}\".format( test_loss_bce, test_correct.item()/test_num_example))\n \n \n if epoch %10==0:\n val_embeddings_1, val_embeddings_2, val_labels_baseline = extract_embeddings(test_loader, model,True)\n plot_embeddings(val_embeddings_1, val_labels_baseline) \n plot_embeddings(val_embeddings_2, val_labels_baseline) \n #fashion_scatter(t_sne_gen(rep_2.cpu()), target.cpu().numpy(),\"Clean_data: \"+\"VAE_\"+str(epoch)+\"softmax_rep2\",\"./softmaxreconstructed/\") \n #fashion_scatter(t_sne_gen(rep_1.cpu()), target.cpu().numpy(),\"Clean_data: \"+\"VAE_\"+str(epoch)+\"softmax_rep1\",\"./softmaxreconstructed/\")\n attack_test(model, test_loader, nn.CrossEntropyLoss() )",
"_____no_output_____"
],
[
"\n",
"_____no_output_____"
],
[
"\n",
"_____no_output_____"
],
[
"\n",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb8f74bb750a6c4326108e4f2fd7b16b0f8fe965
| 12,373 |
ipynb
|
Jupyter Notebook
|
proof_of_work/multiagent/turn_based/v6/original.ipynb
|
michaelneuder/parkes_lab_fa19
|
18d9f564e0df9c17ac5d54619ed869d778d4f6a4
|
[
"MIT"
] | null | null | null |
proof_of_work/multiagent/turn_based/v6/original.ipynb
|
michaelneuder/parkes_lab_fa19
|
18d9f564e0df9c17ac5d54619ed869d778d4f6a4
|
[
"MIT"
] | null | null | null |
proof_of_work/multiagent/turn_based/v6/original.ipynb
|
michaelneuder/parkes_lab_fa19
|
18d9f564e0df9c17ac5d54619ed869d778d4f6a4
|
[
"MIT"
] | null | null | null | 66.521505 | 6,912 | 0.775479 |
[
[
[
"import mdptoolbox\nimport matplotlib.pyplot as plt\nimport numpy as np\nimport scipy.sparse as ss\nimport seaborn as sns\nimport warnings\nwarnings.filterwarnings('ignore', category=ss.SparseEfficiencyWarning)",
"_____no_output_____"
],
[
"# params\nalpha = 0.9\nT = 8\nstate_count = (T+1) * (T+1)\nepsilon = 10e-5\n\n# game\naction_count = 3\nadopt = 0; override = 1; wait = 2\n\n# mapping utils\nstate_mapping = {}\nstates = []\ncount = 0\nfor a in range(T+1):\n for h in range(T+1):\n state_mapping[(a, h)] = count\n states.append((a, h))\n count += 1\n\n# initialize matrices\ntransitions = []; reward_selfish = []; reward_honest = []\nfor _ in range(action_count):\n transitions.append(ss.csr_matrix(np.zeros(shape=(state_count, state_count))))\n reward_selfish.append(ss.csr_matrix(np.zeros(shape=(state_count, state_count))))\n reward_honest.append(ss.csr_matrix(np.zeros(shape=(state_count, state_count))))",
"_____no_output_____"
],
[
"# populate matrices\nfor state_index in range(state_count):\n a, h = states[state_index]\n\n # adopt transitions\n transitions[adopt][state_index, state_mapping[1, 0]] = alpha\n transitions[adopt][state_index, state_mapping[0, 1]] = 1 - alpha\n # adopt rewards\n reward_honest[adopt][state_index, state_mapping[1, 0]] = h\n reward_honest[adopt][state_index, state_mapping[0, 1]] = h\n\n # override\n if a > h:\n transitions[override][state_index, state_mapping[a-h, 0]] = alpha\n reward_selfish[override][state_index, state_mapping[a-h, 0]] = h+1\n transitions[override][state_index, state_mapping[a-h-1, 1]] = 1 - alpha\n reward_selfish[override][state_index, state_mapping[a-h-1, 1]] = h+1\n else:\n transitions[override][state_index, 0] = 1\n reward_honest[override][state_index, 0] = 10000\n\n # wait transitions\n if (a < T) and (h < T):\n transitions[wait][state_index, state_mapping[a+1, h]] = alpha\n transitions[wait][state_index, state_mapping[a, h+1]] = 1 - alpha\n else:\n transitions[wait][state_index, 0] = 1\n reward_honest[wait][state_index, 0] = 10000",
"_____no_output_____"
],
[
"low = 0; high = 1\nwhile (high - low) > epsilon / 8:\n rho = (low + high) / 2\n print(low, high, rho)\n total_reward = []\n for i in range(action_count):\n total_reward.append((1-rho)*reward_selfish[i] - rho*reward_honest[i])\n rvi = mdptoolbox.mdp.RelativeValueIteration(transitions, total_reward, epsilon/8)\n rvi.run()\n if rvi.average_reward > 0:\n low = rho\n else:\n high = rho\npolicy = rvi.policy\nprint('alpha: ', alpha, 'lower bound reward:', rho)",
"0 1 0.5\n0.5 1 0.75\n0.75 1 0.875\n0.875 1 0.9375\n0.9375 1 0.96875\n0.96875 1 0.984375\n0.984375 1 0.9921875\n0.9921875 1 0.99609375\n0.99609375 1 0.998046875\n0.998046875 1 0.9990234375\n0.9990234375 1 0.99951171875\n0.99951171875 1 0.999755859375\n0.999755859375 1 0.9998779296875\n0.9998779296875 1 0.99993896484375\n0.99993896484375 1 0.999969482421875\n0.999969482421875 1 0.9999847412109375\n0.9999847412109375 1 0.9999923706054688\nalpha: 0.9 lower bound reward: 0.9999923706054688\n"
],
[
"f, ax = plt.subplots(figsize=(6,6))\nax.imshow(np.reshape(policy, (9,9)))\nax = sns.heatmap(np.reshape(policy, (9,9)), annot=True, cmap='viridis')\ncb = ax.collections[-1].colorbar \ncb.remove()\nplt.xticks([])\nplt.yticks([])\nplt.show()",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code"
]
] |
cb8f79d65c1199bb617427731e1d298bd386180f
| 23,022 |
ipynb
|
Jupyter Notebook
|
code/.ipynb_checkpoints/Classification_svm_nb_baseline-augmentation-checkpoint.ipynb
|
InscribeDeeper/Text-Classification
|
9cd3def58b5bd4b722a5b8fdff60a07d977234aa
|
[
"MIT"
] | null | null | null |
code/.ipynb_checkpoints/Classification_svm_nb_baseline-augmentation-checkpoint.ipynb
|
InscribeDeeper/Text-Classification
|
9cd3def58b5bd4b722a5b8fdff60a07d977234aa
|
[
"MIT"
] | null | null | null |
code/.ipynb_checkpoints/Classification_svm_nb_baseline-augmentation-checkpoint.ipynb
|
InscribeDeeper/Text-Classification
|
9cd3def58b5bd4b722a5b8fdff60a07d977234aa
|
[
"MIT"
] | null | null | null | 27.871671 | 380 | 0.539527 |
[
[
[
"<h1>Table of Contents<span class=\"tocSkip\"></span></h1>\n<div class=\"toc\"><ul class=\"toc-item\"><li><span><a href=\"#Grid-Search\" data-toc-modified-id=\"Grid-Search-1\"><span class=\"toc-item-num\">1 </span>Grid Search</a></span></li><li><span><a href=\"#Best-params-result\" data-toc-modified-id=\"Best-params-result-2\"><span class=\"toc-item-num\">2 </span>Best params result</a></span></li></ul></div>",
"_____no_output_____"
]
],
[
[
"nb_name = 'Classification_svm_nb_baseline-augmentation'",
"_____no_output_____"
],
[
"from classification_utils import *\nfrom clustering_utils import *\nfrom eda_utils import *\nfrom nn_utils_keras import *\nfrom feature_engineering_utils import *\nfrom data_utils import *\nimport warnings \nwarnings.filterwarnings(\"ignore\")",
"[nltk_data] Downloading package punkt to\n[nltk_data] C:\\Users\\Administrator\\AppData\\Roaming\\nltk_data...\n[nltk_data] Package punkt is already up-to-date!\n[nltk_data] Downloading package stopwords to\n[nltk_data] C:\\Users\\Administrator\\AppData\\Roaming\\nltk_data...\n[nltk_data] Package stopwords is already up-to-date!\n[nltk_data] Downloading package words to\n[nltk_data] C:\\Users\\Administrator\\AppData\\Roaming\\nltk_data...\n[nltk_data] Package words is already up-to-date!\n"
],
[
"train, test = load_data()\n# train, upsampling_info = upsampling_train(train)\n\ntrain_text, train_label = train_augmentation(train, select_comb=[['text'], ['reply', 'reference_one']])\ntest_text, test_label = test['text'], test['label']\n\n# test_text = test_text.apply(lambda x: normal_string(x))\n# train_text = train_text.apply(lambda x: normal_string(x))",
"\nmay use cols: \n ['global_index', 'doc_path', 'label', 'reply', 'reference_one', 'reference_two', 'tag_reply', 'tag_reference_one', 'tag_reference_two', 'Subject', 'From', 'Lines', 'Organization', 'contained_emails', 'long_string', 'text', 'error_message']\ncombination 1 train: ['text']\ncombination 2 train: ['reply', 'reference_one']\n"
],
[
"####################################\n### label mapper\n####################################\nlabels = sorted(train_label.unique())\nlabel_mapper = dict(zip(labels, range(len(labels))))\ntrain_label = train_label.map(label_mapper)\ntest_label = test_label.map(label_mapper)\ny_train = train_label\ny_test = test_label\n\nprint(train_text.shape)\nprint(test_text.shape)\nprint(train_label.shape)\nprint(test_label.shape)",
"(22166,)\n(7761,)\n(22166,)\n(7761,)\n"
],
[
"set(train_label.tolist())",
"_____no_output_____"
]
],
[
[
"# Grid Search",
"_____no_output_____"
]
],
[
[
"metric = \"f1_macro\"\n\ntext_clf = Pipeline([('tfidf', TfidfVectorizer()), ('clf', MultinomialNB())])\nparameters = {'tfidf__min_df': [1, 3, 5], 'tfidf__stop_words': [None, 'english'], 'tfidf__use_idf': [True, False], 'tfidf__binary': [True, False],\n 'clf__alpha': [0.2, 0.4, 0.6, 0.8, 1]}\ngs_clf = GridSearchCV(text_clf, scoring=metric, param_grid=parameters, cv=4)\ngs_clf = gs_clf.fit(train_text, y_train)\n\nfor param_name in gs_clf.best_params_:\n print(\"{0}:\\t{1}\".format(param_name, gs_clf.best_params_[param_name]))\n\nprint(\"best f1 score: {:.3f}\".format(gs_clf.best_score_))\ncv_results = pd.DataFrame(gs_clf.cv_results_)\ncv_results.to_excel(f\"NB_cv_result_{nb_name}.xlsx\")",
"clf__alpha:\t0.2\ntfidf__binary:\tTrue\ntfidf__min_df:\t3\ntfidf__stop_words:\tenglish\ntfidf__use_idf:\tTrue\nbest f1 score: 0.939\n"
],
[
"metric = \"f1_macro\"\n\ntext_clf = Pipeline([('tfidf', TfidfVectorizer()), ('clf', LinearSVC())])\nparameters = {'tfidf__min_df': [1, 3, 5], 'tfidf__stop_words': [None, 'english'], 'tfidf__use_idf': [True, False], 'tfidf__binary': [True, False],\n 'clf__penalty':['l2'], 'clf__C':[1,2,3]}\ngs_clf = GridSearchCV(text_clf, scoring=metric, param_grid=parameters, cv=4)\ngs_clf = gs_clf.fit(train_text, y_train)\n\nfor param_name in gs_clf.best_params_:\n print(\"{0}:\\t{1}\".format(param_name, gs_clf.best_params_[param_name]))\n\nprint(\"best f1 score: {:.3f}\".format(gs_clf.best_score_))\ncv_results = pd.DataFrame(gs_clf.cv_results_)\ncv_results.to_excel(f\"SVC_cv_result_{nb_name}.xlsx\")",
"clf__C:\t3\nclf__penalty:\tl2\ntfidf__binary:\tTrue\ntfidf__min_df:\t1\ntfidf__stop_words:\tNone\ntfidf__use_idf:\tTrue\nbest f1 score: 0.992\n"
]
],
[
[
"# Best params result",
"_____no_output_____"
]
],
[
[
"X_train, X_test, word_to_idx, tfidf_vect = tfidf_vectorizer(train_text, test_text, stop_words=None, binary=True, min_df=1)\n# tfidf_vectorizer(train_text, test_text, min_df=2, max_df=100)\n# X_train, transform_mapper = dimension_reduction(X_train, out_dim=500)\n# X_test = transform_mapper.transform(X_test)\n\nprint('X_train.shape', X_train.shape)\nprint('X_test.shape', X_test.shape)",
"num of words: 130526\nX_train.shape (22166, 130526)\nX_test.shape (7761, 130526)\n"
],
[
"# clf = LinearSVC(penalty=\"l2\", multi_class='ovr', C=3.0, dual=True,)\n# clf.fit(X_train, y_train)\npred = clf.predict(X_test)\nclassification_report = evaluation_report(y_test, pred, labels=labels)\n# roc_auc(y_test, pred)\n\n\n##########################################\n## CV shows the stable result\n##########################################\n# cv_metrics = [\"precision_macro\",\"accuracy\", \"f1_macro\"]\n# cv = cross_validate(clf, X_train, y_train,scoring=cv_metrics, cv=4, return_train_score=True)\n# cv = pd.DataFrame(cv)\n# f1 = cv['test_f1_macro'].mean()\n# print(\"cv average f1 macro: \", f1)\n\n# cv",
"classification_report:\nf1: {'micro avg': 0.847313490529571, 'macro avg': 0.8349258727936671} \n\n precision recall f1-score support\nalt.atheism 0.812 0.774 0.793 319.0\ncomp.graphics 0.757 0.810 0.783 389.0\ncomp.os.ms-windows.misc 0.770 0.731 0.750 394.0\ncomp.sys.ibm.pc.hardware 0.743 0.753 0.748 392.0\ncomp.sys.mac.hardware 0.829 0.857 0.843 385.0\ncomp.windows.x 0.880 0.815 0.846 395.0\nmisc.forsale 0.857 0.908 0.882 390.0\nrec.autos 0.907 0.916 0.912 395.0\nrec.motorcycles 0.952 0.957 0.955 398.0\nrec.sport.baseball 0.822 0.965 0.888 397.0\nrec.sport.hockey 0.992 0.889 0.938 827.0\nsci.crypt 0.934 0.922 0.928 396.0\nsci.electronics 0.797 0.738 0.766 393.0\nsci.med 0.724 0.939 0.818 198.0\nsci.space 0.892 0.919 0.905 394.0\nsoc.religion.christian 0.835 0.915 0.873 398.0\ntalk.politics.guns 0.716 0.915 0.803 364.0\ntalk.politics.mideast 0.974 0.886 0.928 376.0\ntalk.politics.misc 0.813 0.561 0.664 310.0\ntalk.religion.misc 0.741 0.625 0.678 251.0\naccuracy 0.847 0.847 0.847 7761.0\nmacro avg 0.837 0.840 0.835 7761.0\nweighted avg 0.851 0.847 0.846 7761.0\nmicro avg 0.847 0.847 0.847 7761.0\n"
],
[
"clf = MultinomialNB()\nclf.fit(X_train, y_train)\npred = clf.predict(X_test)\nclassification_report = evaluation_report(y_test, pred, labels=labels)\n\n##########################################\n## CV shows the stable result\n##########################################\n# cv_metrics = [\"precision_macro\",\"accuracy\", \"f1_macro\"]\n# cv = cross_validate(clf, X_train, y_train,scoring=cv_metrics, cv=4, return_train_score=True)\n# cv = pd.DataFrame(cv)\n# f1 = cv['test_f1_macro'].mean()\n# print(\"cv average f1 macro: \", f1)\n\n# cv",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
cb8f838cf533f7577e211c1d185e8f18dc844e37
| 496,515 |
ipynb
|
Jupyter Notebook
|
04_Projects/01_Ants_vrs_Bees/.ipynb_checkpoints/04_Conv_NN-Image Classification-checkpoint.ipynb
|
CrispenGari/pytorch-python
|
e5e8f8d48aa17ed4eb2236e53cbea78d6beeacf6
|
[
"MIT"
] | 1 |
2021-11-08T07:37:16.000Z
|
2021-11-08T07:37:16.000Z
|
04_Projects/01_Ants_vrs_Bees/.ipynb_checkpoints/04_Conv_NN-Image Classification-checkpoint.ipynb
|
CrispenGari/pytorch-python
|
e5e8f8d48aa17ed4eb2236e53cbea78d6beeacf6
|
[
"MIT"
] | null | null | null |
04_Projects/01_Ants_vrs_Bees/.ipynb_checkpoints/04_Conv_NN-Image Classification-checkpoint.ipynb
|
CrispenGari/pytorch-python
|
e5e8f8d48aa17ed4eb2236e53cbea78d6beeacf6
|
[
"MIT"
] | 1 |
2021-11-22T17:52:50.000Z
|
2021-11-22T17:52:50.000Z
| 822.044702 | 200,656 | 0.954664 |
[
[
[
"### Image Classification - Conv Nets -Pytorch",
"_____no_output_____"
],
[
"> Classifying if an image is a `bee` of an `ant` using `ConvNets` in pytorch",
"_____no_output_____"
],
[
"### Imports",
"_____no_output_____"
]
],
[
[
"import cv2\nimport matplotlib.pyplot as plt\nimport numpy as np\nfrom sklearn.model_selection import train_test_split\nimport torch\nfrom torch import nn\nimport torch.nn.functional as F\nimport os",
"_____no_output_____"
]
],
[
[
"### Data Preparation",
"_____no_output_____"
]
],
[
[
"class Insect:\n BEE = 'BEE'\n ANT = \"ANT\"\n BEES_IMAGES_PATH = 'data/colored/rgb/bees'\n ANTS_IMAGES_PATH = 'data/colored/rgb/ants'\n \nclasses = {'bee': 0, 'ant' : 1}\n\nclasses =dict([(i, j) for (j, i) in classes.items()])\nclasses",
"_____no_output_____"
],
[
"os.path.exists(Insect.BEES_IMAGES_PATH)",
"_____no_output_____"
],
[
"insects = []\nfor path in os.listdir(Insect.BEES_IMAGES_PATH):\n img_path = os.path.join(Insect.BEES_IMAGES_PATH, path)\n image = np.array(cv2.imread(img_path, cv2.IMREAD_UNCHANGED), dtype='float32')\n image = image / 255\n insects.append([image, 0])\nfor path in os.listdir(Insect.ANTS_IMAGES_PATH):\n img_path = os.path.join(Insect.ANTS_IMAGES_PATH, path)\n image = np.array(cv2.imread(img_path, cv2.IMREAD_UNCHANGED), dtype='float32')\n image = image / 255\n insects.append([image, 1])\ninsects = np.array(insects)",
"<ipython-input-4-6b8198280b07>:12: VisibleDeprecationWarning: Creating an ndarray from ragged nested sequences (which is a list-or-tuple of lists-or-tuples-or ndarrays with different lengths or shapes) is deprecated. If you meant to do this, you must specify 'dtype=object' when creating the ndarray\n insects = np.array(insects)\n"
],
[
"np.random.shuffle(insects)",
"_____no_output_____"
]
],
[
[
"### Visualization",
"_____no_output_____"
]
],
[
[
"plt.imshow(insects[7][0], cmap=\"gray\"), insects[10][0].shape",
"_____no_output_____"
]
],
[
[
"> Seperating Labels and features",
"_____no_output_____"
]
],
[
[
"X = np.array([insect[0] for insect in insects])\ny = np.array([insect[1] for insect in insects])",
"_____no_output_____"
],
[
"X[0].shape",
"_____no_output_____"
]
],
[
[
"> Splitting the data into training and test.",
"_____no_output_____"
]
],
[
[
"X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=33, test_size=.2)",
"_____no_output_____"
],
[
"X_train.shape, y_train.shape, y_test.shape, X_test.shape",
"_____no_output_____"
]
],
[
[
"> Converting the data into `torch` tensor.",
"_____no_output_____"
]
],
[
[
"X_train = torch.from_numpy(X_train.astype('float32'))\nX_test = torch.from_numpy(X_test.astype('float32'))\n\ny_train = torch.Tensor(y_train)\ny_test = torch.Tensor(y_test)",
"_____no_output_____"
]
],
[
[
"### Model Creation",
"_____no_output_____"
]
],
[
[
"class Net(nn.Module):\n def __init__(self):\n super().__init__()\n self.conv1 = nn.Conv2d(in_channels=3, out_channels= 32, kernel_size=(3, 3))\n self.conv2 = nn.Conv2d(32, 64, (3, 3))\n self.conv3 = nn.Conv2d(64, 64, (3, 3))\n \n self._to_linear = None # protected variable\n self.x = torch.randn(3, 200, 200).view(-1, 3, 200, 200)\n self.conv(self.x)\n \n self.fc1 = nn.Linear(self._to_linear, 64)\n self.fc2 = nn.Linear(64, 2)\n \n def conv(self, x):\n x = F.max_pool2d(F.relu(self.conv1(x)), (2, 2))\n x = F.max_pool2d(F.relu(self.conv2(x)), (2, 2))\n x = F.max_pool2d(F.relu(self.conv3(x)), (2, 2))\n \n if self._to_linear is None:\n self._to_linear = x.shape[1] * x.shape[2] * x.shape[3]\n \n return x\n \n def forward(self, x):\n x = self.conv(x)\n x = x.view(-1, self._to_linear)\n x = F.relu(self.fc1(x))\n return x\n \nnet = Net()\nnet",
"_____no_output_____"
],
[
"optimizer = torch.optim.SGD(net.parameters(), lr=1e-3)\nloss_function = nn.CrossEntropyLoss()",
"_____no_output_____"
],
[
"EPOCHS = 10\nBATCH_SIZE = 5\nfor epoch in range(EPOCHS):\n print(f'Epochs: {epoch+1}/{EPOCHS}')\n for i in range(0, len(y_train), BATCH_SIZE):\n X_batch = X_train[i: i+BATCH_SIZE].view(-1, 3, 200, 200)\n y_batch = y_train[i: i+BATCH_SIZE].long()\n \n net.zero_grad() ## or you can say optimizer.zero_grad()\n \n outputs = net(X_batch)\n loss = loss_function(outputs, y_batch)\n loss.backward()\n optimizer.step()\n print(\"Loss\", loss)",
"Epochs: 1/10\nLoss tensor(0.6264, grad_fn=<NllLossBackward>)\nEpochs: 2/10\nLoss tensor(0.4958, grad_fn=<NllLossBackward>)\nEpochs: 3/10\nLoss tensor(0.3589, grad_fn=<NllLossBackward>)\nEpochs: 4/10\nLoss tensor(0.2593, grad_fn=<NllLossBackward>)\nEpochs: 5/10\nLoss tensor(0.2091, grad_fn=<NllLossBackward>)\nEpochs: 6/10\nLoss tensor(0.1895, grad_fn=<NllLossBackward>)\nEpochs: 7/10\nLoss tensor(0.1824, grad_fn=<NllLossBackward>)\nEpochs: 8/10\nLoss tensor(0.1800, grad_fn=<NllLossBackward>)\nEpochs: 9/10\nLoss tensor(0.1793, grad_fn=<NllLossBackward>)\nEpochs: 10/10\nLoss tensor(0.1795, grad_fn=<NllLossBackward>)\n"
]
],
[
[
"### Evaluating the model",
"_____no_output_____"
],
[
"### Test set",
"_____no_output_____"
]
],
[
[
"total, correct = 0, 0\nwith torch.no_grad():\n for i in range(len(X_test)):\n correct_label = torch.argmax(y_test[i])\n prediction = torch.argmax(net(X_test[i].view(-1, 3, 200, 200))[0])\n if prediction == correct_label:\n correct+=1\n total +=1\n \n print(f\"Accuracy: {correct/total}\")",
"Accuracy: 0.6354166666666666\n"
],
[
"torch.argmax(net(X_test[1].view(-1, 3, 200, 200))), y_test[0]",
"_____no_output_____"
]
],
[
[
"### Train set",
"_____no_output_____"
]
],
[
[
"total, correct = 0, 0\nwith torch.no_grad():\n for i in range(len(X_train)):\n correct_label = torch.argmax(y_train[i])\n prediction = torch.argmax(net(X_train[i].view(-1, 3, 200, 200))[0])\n if prediction == correct_label:\n correct+=1\n total +=1\n \n print(f\"Accuracy: {correct/total}\")",
"Accuracy: 0.6510416666666666\n"
]
],
[
[
"### Making Predictions",
"_____no_output_____"
]
],
[
[
"plt.imshow(X_test[12])\nplt.title(classes[torch.argmax(net(X_test[12].view(-1, 3, 200, 200))).item()].title(), fontsize=16)\nplt.show()",
"_____no_output_____"
],
[
"\nfig, ax = plt.subplots(nrows=3, ncols=3, figsize=(10, 10))\nfor row in ax:\n for col in row:\n col.imshow(X_test[2])\nplt.show()",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
cb8f9975b61ce53a4728a42b76ea8890a3f3237e
| 21,361 |
ipynb
|
Jupyter Notebook
|
Untitled.ipynb
|
adrianpearl/OpenNMT-py
|
a62e37eb0f092d36e8467031972c979dc6aea86a
|
[
"MIT"
] | null | null | null |
Untitled.ipynb
|
adrianpearl/OpenNMT-py
|
a62e37eb0f092d36e8467031972c979dc6aea86a
|
[
"MIT"
] | null | null | null |
Untitled.ipynb
|
adrianpearl/OpenNMT-py
|
a62e37eb0f092d36e8467031972c979dc6aea86a
|
[
"MIT"
] | null | null | null | 33.015456 | 105 | 0.450634 |
[
[
[
"import torch",
"_____no_output_____"
],
[
"vocab = torch.load('../Data_Collection_old/data_small/CNNDM.vocab.pt')",
"_____no_output_____"
],
[
"vocab",
"_____no_output_____"
],
[
"vocab['src_map']",
"_____no_output_____"
],
[
"train = torch.load('../data_full/split/BERT/BERT.train.0.pt')",
"_____no_output_____"
],
[
"t1, t2 = train[0], train[1]",
"_____no_output_____"
],
[
"t1.__dict__.keys()",
"_____no_output_____"
],
[
"t2.src[0]",
"_____no_output_____"
],
[
"from pytorch_pretrained_bert import BertTokenizer, BertModel",
"_____no_output_____"
],
[
"tokenizer = BertTokenizer.from_pretrained('bert-base-uncased')",
"_____no_output_____"
],
[
"tok1, tok2 = tokenizer.convert_tokens_to_ids(t1.src[0]), tokenizer.convert_tokens_to_ids(t2.src[0])",
"_____no_output_____"
],
[
"tok1 += [0]*(40 - len(tok1))\ntok2 += [0]*(40 - len(tok2))",
"_____no_output_____"
],
[
"tensor = torch.tensor([tok1, tok2])",
"_____no_output_____"
],
[
"segment = torch.zeros_like(tensor)",
"_____no_output_____"
],
[
"bm = BertModel.from_pretrained('bert-base-uncased')\nbm.eval()",
"_____no_output_____"
],
[
"with torch.no_grad():\n encoded_layers, _ = bm(tensor, segment)",
"_____no_output_____"
],
[
"len(encoded_layers)",
"_____no_output_____"
],
[
"fl = encoded_layers[-1]",
"_____no_output_____"
],
[
"fl.shape",
"_____no_output_____"
],
[
"fl[:,0,:].shape",
"_____no_output_____"
],
[
"fl[0,-3:,:]",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb8fb585c81104fb9f56e569e368d4ae1a325b3c
| 4,892 |
ipynb
|
Jupyter Notebook
|
src/1. Data preprocessing.ipynb
|
aaronfinlay123/FaceMaskDetector
|
4ad434b972957cd99673aa2b992d68d114e6a094
|
[
"CC0-1.0"
] | null | null | null |
src/1. Data preprocessing.ipynb
|
aaronfinlay123/FaceMaskDetector
|
4ad434b972957cd99673aa2b992d68d114e6a094
|
[
"CC0-1.0"
] | null | null | null |
src/1. Data preprocessing.ipynb
|
aaronfinlay123/FaceMaskDetector
|
4ad434b972957cd99673aa2b992d68d114e6a094
|
[
"CC0-1.0"
] | null | null | null | 31.159236 | 489 | 0.59955 |
[
[
[
"# Data Pre-Processing",
"_____no_output_____"
],
[
"When training any sort of model using a machine learning algorithm, a large dataset is first needed to train that model off of. Data can be anything that would help benefit with the training of the model. In this case, images of people facing the camera head on wearing/not wearing a face mask is the type of data that is being used.\n\n# Data preparation\n\nRaw data is first collected. These are just images of people wearing/not wearing face masks. This is not enough by itself as the data must the be divided into two groups, i.e into 'with_mask' and 'without_mask'.\n\n# Categorisation and labeling\n\nNext the data must be categorised and labeled as such.",
"_____no_output_____"
]
],
[
[
"import cv2,os\n\ndata_path='dataset'\ncategories=os.listdir(data_path)\nlabels=[i for i in range(len(categories))]\n\nlabel_dict=dict(zip(categories,labels)) #empty dictionary\n\nprint(label_dict)\nprint(categories)\nprint(labels)",
"{'without_mask': 0, 'with_mask': 1}\n['without_mask', 'with_mask']\n[0, 1]\n"
]
],
[
[
"# Resizing and reshaping the data\nWhen feeding any sort of data into an algorithm, it is important that we normalise the data. In this case of working with many images in the dataset, the images must be resized and shaped so that they are all fixed and common in size. For the purpose of this project, each image was resized to be 50 pixels by 50 pixels and were converted to greyscale. It is also important to note that each image was added to the data array, and its corresponding label is added to the target array",
"_____no_output_____"
]
],
[
[
"img_size=100\ndata=[]\ntarget=[]\n\n\nfor category in categories:\n folder_path=os.path.join(data_path,category)\n img_names=os.listdir(folder_path)\n \n for img_name in img_names:\n img_path=os.path.join(folder_path,img_name)\n img=cv2.imread(img_path)\n\n try:\n gray=cv2.cvtColor(img,cv2.COLOR_BGR2GRAY) \n resized=cv2.resize(gray,(img_size,img_size))\n data.append(resized)\n target.append(label_dict[category])\n\n except Exception as e:\n print('Exception:',e)",
"_____no_output_____"
]
],
[
[
"# Serialising the resulting pre processed data\nNow that the dataset has been preprocessed and sorted into arrays, it must now be serialised so it can be used in the training process. To serialise the data, numpy is used as it is capable of serialising arrays and deserialising them later on for use (also known as flattening and unflattening)",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\ndata=np.array(data)/255.0\ndata=np.reshape(data,(data.shape[0],img_size,img_size,1))\ntarget=np.array(target)\n\nfrom keras.utils import np_utils\n\nnew_target=np_utils.to_categorical(target)\n\nnp.save('data',data)\nnp.save('target',new_target)",
"_____no_output_____"
]
],
[
[
"# Finishing up\nNow that the data has been preprocessed and serialised, it is now ready to be used in the training process. It is important to note that this must be done any time data needs to be added or removed from the dataset so it is not uncommon to have to use this multiple times throughout the development of the project",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
cb8fb8bc2c958ff150249910c2636c801838b605
| 14,544 |
ipynb
|
Jupyter Notebook
|
QiskitEducation/withoutQE.ipynb
|
veenaiyuri/qiskit-education
|
0b58c8985b9377b4bc3404d205ea6235877a9778
|
[
"Apache-2.0"
] | 6 |
2019-02-03T10:03:12.000Z
|
2020-11-05T18:13:08.000Z
|
QiskitEducation/withoutQE.ipynb
|
veenaiyuri/qiskit-education
|
0b58c8985b9377b4bc3404d205ea6235877a9778
|
[
"Apache-2.0"
] | null | null | null |
QiskitEducation/withoutQE.ipynb
|
veenaiyuri/qiskit-education
|
0b58c8985b9377b4bc3404d205ea6235877a9778
|
[
"Apache-2.0"
] | 2 |
2019-01-31T16:48:17.000Z
|
2019-01-31T17:00:59.000Z
| 158.086957 | 12,624 | 0.906697 |
[
[
[
"from qiskit import *\nfrom qiskit.tools.visualization import plot_histogram, circuit_drawer\n\nq = QuantumRegister(3)\nc = ClassicalRegister(3)\nqc = QuantumCircuit(q, c)",
"_____no_output_____"
],
[
"qc.h(q[0])\nqc.h(q[1])\nqc.cx(q[1], q[2])\nqc.cx(q[0], q[1])\nqc.h(q[0])\nqc.measure(q[0], c[0])\nqc.measure(q[1], c[1])\nqc.cx(q[1], q[2])\nqc.cz(q[0], q[2])\nqc.h(q[2])\nqc.measure(q[2], c[2])\n\nbackend = Aer.get_backend('qasm_simulator')\njob_sim = execute(qc, backend, shots=1024)\nsim_result = job_sim.result()\nmeasurement_result = sim_result.get_counts(qc)\nprint(measurement_result)\nplot_histogram(measurement_result)",
"{'011': 267, '001': 254, '010': 259, '000': 244}\n"
]
]
] |
[
"code"
] |
[
[
"code",
"code"
]
] |
cb8fe43e7693863efcc4cb3380c06198ed7909b8
| 6,637 |
ipynb
|
Jupyter Notebook
|
NetworksAsLayers.ipynb
|
Beyond-ML-Labs/mann-notebooks
|
d1b3780990e34f40d06cca7f2652672601ac767d
|
[
"MIT"
] | 1 |
2022-01-27T16:05:08.000Z
|
2022-01-27T16:05:08.000Z
|
NetworksAsLayers.ipynb
|
AISquaredInc/mann-notebooks
|
d1b3780990e34f40d06cca7f2652672601ac767d
|
[
"MIT"
] | null | null | null |
NetworksAsLayers.ipynb
|
AISquaredInc/mann-notebooks
|
d1b3780990e34f40d06cca7f2652672601ac767d
|
[
"MIT"
] | 1 |
2022-02-19T04:20:57.000Z
|
2022-02-19T04:20:57.000Z
| 33.690355 | 448 | 0.625584 |
[
[
[
"# Using Models as Layers in Another Model\n\nIn this notebook, we show how you can use Keras models as Layers within a larger model and still perform pruning on that model.",
"_____no_output_____"
]
],
[
[
"# Import required packages\n\nimport tensorflow as tf\nimport mann\n\nfrom sklearn.metrics import confusion_matrix, classification_report",
"_____no_output_____"
],
[
"# Load the data\n(x_train, y_train), (x_test, y_test) = tf.keras.datasets.fashion_mnist.load_data()\n\n# Convert images from grayscale to RGB\nx_train = tf.image.grayscale_to_rgb(tf.Variable(x_train.reshape(-1, 28, 28, 1)))\nx_test = tf.image.grayscale_to_rgb(tf.Variable(x_test.reshape(-1, 28, 28, 1)))",
"_____no_output_____"
]
],
[
[
"## Model Creation\n\nIn the following cells, we create two models and put them together to create a larger model. The first model, called the `preprocess_model`, takes in images, divides the pixel values by 255 to ensure all values are between 0 and 1, resized the image to a height and width of 40 pixels. It then performs training data augmentation by randomly flips some images across the y-axis, randomly rotates images, and randomly translates the images.\n\nThe second model, called the `true_model`, contains the logic for performing prediction on images. It contains blocks of convolutional layers followed by max pooling and dropout layers. The output of these blocks is flattened and passed through fully-connected layers to output predicted class probabilities.\n\nThese two models are combined in the `training_model` to be trained.",
"_____no_output_____"
]
],
[
[
"preprocess_model = tf.keras.models.Sequential()\npreprocess_model.add(tf.keras.layers.Rescaling(1./255))\npreprocess_model.add(tf.keras.layers.Resizing(40, 40, input_shape = (None, None, 3)))\npreprocess_model.add(tf.keras.layers.RandomFlip('horizontal'))\npreprocess_model.add(tf.keras.layers.RandomRotation(0.1))\npreprocess_model.add(tf.keras.layers.RandomTranslation(0.1, 0.1))\n\ntrue_model = tf.keras.models.Sequential()\ntrue_model.add(mann.layers.MaskedConv2D(16, padding = 'same', input_shape = (40, 40, 3)))\ntrue_model.add(mann.layers.MaskedConv2D(16, padding = 'same'))\ntrue_model.add(tf.keras.layers.MaxPool2D())\ntrue_model.add(tf.keras.layers.Dropout(0.2))\ntrue_model.add(mann.layers.MaskedConv2D(32, padding = 'same', activation = 'relu'))\ntrue_model.add(mann.layers.MaskedConv2D(32, padding = 'same', activation = 'relu'))\ntrue_model.add(tf.keras.layers.MaxPool2D())\ntrue_model.add(tf.keras.layers.Dropout(0.2))\ntrue_model.add(mann.layers.MaskedConv2D(64, padding = 'same', activation = 'relu'))\ntrue_model.add(mann.layers.MaskedConv2D(64, padding = 'same', activation = 'relu'))\ntrue_model.add(tf.keras.layers.MaxPool2D())\ntrue_model.add(tf.keras.layers.Dropout(0.2))\ntrue_model.add(tf.keras.layers.Flatten())\ntrue_model.add(mann.layers.MaskedDense(256, activation = 'relu'))\ntrue_model.add(mann.layers.MaskedDense(256, activation = 'relu'))\ntrue_model.add(mann.layers.MaskedDense(10, activation = 'softmax'))\n\ntraining_input = tf.keras.layers.Input((None, None, 3))\ntraining_x = preprocess_model(training_input)\ntraining_output = true_model(training_x)\ntraining_model = tf.keras.models.Model(\n training_input,\n training_output\n)\n\ntraining_model.compile(\n loss = 'sparse_categorical_crossentropy',\n metrics = ['accuracy'],\n optimizer = 'adam'\n)\n\ntraining_model.summary()",
"_____no_output_____"
]
],
[
[
"## Model Training\n\nIn this cell, we create the `ActiveSparsification` object to continually sparsify the model as it trains, and train the model.",
"_____no_output_____"
]
],
[
[
"callback = mann.utils.ActiveSparsification(\n 0.80,\n sparsification_rate = 5\n)\n\ntraining_model.fit(\n x_train,\n y_train,\n epochs = 200,\n batch_size = 512,\n validation_split = 0.2,\n callbacks = [callback]\n)",
"_____no_output_____"
]
],
[
[
"## Convert the model to not have masking layers\n\nIn the following cell, we configure the model to remove masking layers and replace them with non-masking native TensorFlow layers. We then perform prediction on the resulting model and present the results.",
"_____no_output_____"
]
],
[
[
"model = mann.utils.remove_layer_masks(training_model)\npreds = model.predict(x_test).argmax(axis = 1)\nprint(confusion_matrix(y_test, preds))\nprint(classification_report(y_test, preds))",
"_____no_output_____"
]
],
[
[
"## Save only the model that performs prediction\n\nLastly, save only the part of the model that performs prediction",
"_____no_output_____"
]
],
[
[
"model.layers[2].save('ModelLayer.h5')",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb8ffdbdbf3889e358dc068a8f3d31b512460551
| 355,536 |
ipynb
|
Jupyter Notebook
|
1_Introduction_to_plotly/notebook/Plotly_data_visualization.ipynb
|
hirenhk15/ga-code-alongs
|
fc8ba845eb60668e297fd53061c607f6986a8c25
|
[
"Apache-2.0"
] | null | null | null |
1_Introduction_to_plotly/notebook/Plotly_data_visualization.ipynb
|
hirenhk15/ga-code-alongs
|
fc8ba845eb60668e297fd53061c607f6986a8c25
|
[
"Apache-2.0"
] | null | null | null |
1_Introduction_to_plotly/notebook/Plotly_data_visualization.ipynb
|
hirenhk15/ga-code-alongs
|
fc8ba845eb60668e297fd53061c607f6986a8c25
|
[
"Apache-2.0"
] | null | null | null | 33.159485 | 23,751 | 0.376527 |
[
[
[
"# Data Visualization Using Plotly",
"_____no_output_____"
],
[
"##### To visualize plots in this notebook please click [here](https://nbviewer.jupyter.org/github/hirenhk15/ga-code-alongs/blob/main/1_Introduction_to_plotly/notebook/Plotly_data_visualization.ipynb)",
"_____no_output_____"
]
],
[
[
"# Import packages\nimport cufflinks\nimport plotly\nimport plotly.graph_objects as go\nfrom plotly.offline import iplot\nfrom plotly.offline import init_notebook_mode\nimport pandas as pd\nimport matplotlib.pyplot as plt\n\nimport warnings\nwarnings.filterwarnings('ignore')",
"_____no_output_____"
],
[
"# Sample graph using plotly\nfig = go.Figure(data=go.Bar(y=[2, 3, 1]))\nfig.write_html('first_figure.html', auto_open=True)",
"_____no_output_____"
]
],
[
[
"# Loading Data",
"_____no_output_____"
]
],
[
[
"timesData = pd.read_csv('../data/timesData.csv')\ntimesData.head()",
"_____no_output_____"
]
],
[
[
"# Bar plot using dataframe",
"_____no_output_____"
],
[
"## Citations of top 4 universities in 2015",
"_____no_output_____"
]
],
[
[
"# Settings to enable dataframe plotting\ncufflinks.go_offline(connected=True)\ninit_notebook_mode(connected=True)\n\n# Creating the dataframe\ndf = timesData[timesData.year == 2015].iloc[:4,]\ndf2 = df[['citations','university_name']].set_index('university_name')\n\n# Plotting the bar plot\ndf2.iplot(kind='bar', xTitle='University', yTitle='Citation Score', title='University Citations')",
"_____no_output_____"
]
],
[
[
"# Bar plot using plotly graph objects",
"_____no_output_____"
],
[
"### Citations of top 4 universities in 2015",
"_____no_output_____"
]
],
[
[
"df_2015 = timesData[timesData.year == 2015].iloc[:4,:]\n\ntrace = go.Bar(\n x=df_2015.university_name,\n y=df_2015.citations,\n name='Citations',\n text=df_2015.country\n )\n\nfig = go.Figure(data=[trace])\nfig.show()\n#iplot(fig)\n#plotly.offline.plot(fig, filename='bar.html')",
"_____no_output_____"
]
],
[
[
"### Citations and Research score of top 4 universities in 2015 - Vertical",
"_____no_output_____"
]
],
[
[
"trace1 = go.Bar(\n x=df_2015.university_name,\n y=df_2015.citations,\n name='Citations',\n marker=dict(color='rgba(26, 118, 255, 0.5)',\n line=dict(color='rgb(0, 0, 0)', width=1.5)),\n text=df_2015.country\n )\n\ntrace2 = go.Bar(\n x=df_2015.university_name,\n y=df_2015.research,\n name='Research',\n marker=dict(color='rgba(249, 6, 6, 0.5)',\n line=dict(color='rgb(0, 0, 0)', width=1.5)),\n text=df_2015.country\n )\n\ndata = [trace1, trace2]\nfig = go.Figure(data=data)\nfig.show()",
"_____no_output_____"
]
],
[
[
"### Citations and Research score of top 4 universities in 2015 - Horizontal",
"_____no_output_____"
]
],
[
[
"trace1 = go.Bar(\n y=df_2015.university_name,\n x=df_2015.citations,\n name='Citations',\n orientation='h',\n marker=dict(color='rgba(26, 118, 255, 0.5)',\n line=dict(color='rgb(0, 0, 0)', width=1.5)),\n text=df_2015.country\n )\n\ntrace2 = go.Bar(\n y=df_2015.university_name,\n x=df_2015.research,\n name='Research',\n orientation='h',\n marker=dict(color='rgba(249, 6, 6, 0.5)',\n line=dict(color='rgb(0, 0, 0)', width=1.5)),\n text=df_2015.country\n )\n\ndata = [trace1, trace2]\nfig = go.Figure(data=data)\nfig.show()",
"_____no_output_____"
]
],
[
[
"# Scatter Plot",
"_____no_output_____"
],
[
"### Citation vs world rank of top 100 universities with 2014, 2015 and 2016 years",
"_____no_output_____"
]
],
[
[
"df_2014 = timesData[timesData.year==2014].iloc[:100,]\ndf_2015 = timesData[timesData.year==2015].iloc[:100,]\ndf_2016 = timesData[timesData.year==2016].iloc[:100,]\n\ntrace1 = go.Scatter(\n x=df_2014.world_rank,\n y=df_2014.citations,\n name='2014',\n mode='markers',\n marker=dict(color='rgba(255, 128, 255, 0.8)'),\n text=df_2014.university_name\n )\n\ntrace2 = go.Scatter(\n x=df_2015.world_rank,\n y=df_2015.citations,\n name='2015',\n mode='markers',\n marker=dict(color='rgba(255, 128, 2, 0.8)'),\n text=df_2015.university_name\n )\n\ntrace3 = go.Scatter(\n x=df_2016.world_rank,\n y=df_2016.citations,\n name='2016',\n mode='markers',\n marker=dict(color='rgba(0, 255, 200, 0.8)'),\n text=df_2016.university_name\n )\n\ndata = [trace1, trace2, trace3]\nlayout = dict(title='Citation vs world rank of top 100 universities with 2014, 2015 and 2016 years',\n xaxis=dict(title='World Rank'),\n yaxis=dict(title='Citation'),\n template='plotly_dark'\n )\n\nfig = go.Figure(data=data, layout=layout)\nfig.show()",
"_____no_output_____"
]
],
[
[
"# Lineplot",
"_____no_output_____"
],
[
"### Citation vs world rank of top 100 universities with 2014, 2015 and 2016 years",
"_____no_output_____"
]
],
[
[
"df_2014 = timesData[timesData.year==2014].iloc[:100,]\ndf_2015 = timesData[timesData.year==2015].iloc[:100,]\ndf_2016 = timesData[timesData.year==2016].iloc[:100,]\n\ntrace1 = go.Scatter(\n x=df_2014.world_rank,\n y=df_2014.citations,\n name='2014',\n mode='lines',\n marker=dict(color='rgba(255, 128, 255, 0.8)'),\n text=df_2014.university_name\n )\n\ntrace2 = go.Scatter(\n x=df_2015.world_rank,\n y=df_2015.citations,\n name='2015',\n mode='lines',\n marker=dict(color='rgba(255, 128, 2, 0.8)'),\n text=df_2015.university_name\n )\n\ntrace3 = go.Scatter(\n x=df_2016.world_rank,\n y=df_2016.citations,\n name='2016',\n mode='lines',\n marker=dict(color='rgba(0, 255, 200, 0.8)'),\n text=df_2016.university_name\n )\n\ndata = [trace1, trace2, trace3]\nlayout = dict(title='Citation vs world rank of top 100 universities with 2014, 2015 and 2016 years',\n xaxis=dict(title='World Rank'),\n yaxis=dict(title='Citation'),\n template='plotly_dark'\n )\n\nfig = go.Figure(data=data, layout=layout)\nfig.show()",
"_____no_output_____"
]
],
[
[
"# Bubble Plot",
"_____no_output_____"
],
[
"### University world rank (first 20) vs teaching score with number of students(size) and international score (color) in 2015",
"_____no_output_____"
]
],
[
[
"df_2015 = timesData[timesData.year == 2015].iloc[:20,]\n\n# Converting following columns to float\nnum_students_size = df_2015.num_students.apply(lambda x: float(x.replace(',', '.'))).tolist()\ninternational_color = df_2015.international.astype('float').tolist()\n\n# Plotting bubble plot\ntrace = go.Scatter(\n x=df_2015.world_rank,\n y=df_2015.teaching,\n mode='markers',\n marker=dict(color=international_color, size=num_students_size, showscale=True),\n text= df_2015.university_name\n )\n\nfig = go.Figure(data=[trace])\nfig.show()",
"_____no_output_____"
]
],
[
[
"# Histogram",
"_____no_output_____"
],
[
"### Histogram of students-staff ratio in 2011 and 2016 year",
"_____no_output_____"
]
],
[
[
"x2011 = timesData.student_staff_ratio[timesData.year == 2011]\nx2016 = timesData.student_staff_ratio[timesData.year == 2016]\n\n# Creating histogram trace\ntrace1 = go.Histogram(\n x=x2011,\n opacity=0.75,\n name='2011',\n marker=dict(color='rgba(171, 50, 96, 0.6)')\n )\n\ntrace2 = go.Histogram(\n x=x2016,\n opacity=0.75,\n name='2016',\n marker=dict(color='rgba(12, 50, 196, 0.6)')\n )\n\ndata = [trace1, trace2]\nlayout = go.Layout(\n barmode='overlay',\n title='Student-Staff Ratio in 2011 and 2016',\n xaxis=dict(title='Student-Staff Ratio'),\n yaxis=dict(title='Count')\n )\n\nfig = go.Figure(data=data, layout=layout)\nfig.show()",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
]
] |
cb90077a0fdeed8a7633c601e0b376393dbd2ed0
| 123,602 |
ipynb
|
Jupyter Notebook
|
examples/imbalanced_datasets.ipynb
|
jaswinder9051998/ATOM
|
486396c9be8b5d7d6927f6cb0480cbce985a1563
|
[
"MIT"
] | null | null | null |
examples/imbalanced_datasets.ipynb
|
jaswinder9051998/ATOM
|
486396c9be8b5d7d6927f6cb0480cbce985a1563
|
[
"MIT"
] | null | null | null |
examples/imbalanced_datasets.ipynb
|
jaswinder9051998/ATOM
|
486396c9be8b5d7d6927f6cb0480cbce985a1563
|
[
"MIT"
] | null | null | null | 234.094697 | 51,087 | 0.88196 |
[
[
[
"# Imbalanced datasets\n------------------------------------\n\nThis example shows how ATOM can help you handle imbalanced datasets. We will evaluate the performance of three different Random Forest models: one trained directly on the imbalanced dataset, one trained on an oversampled dataset and the last one trained on an undersampled dataset.",
"_____no_output_____"
],
[
"## Load the data",
"_____no_output_____"
]
],
[
[
"# Import packages\nfrom atom import ATOMClassifier\nfrom sklearn.datasets import make_classification",
"_____no_output_____"
],
[
"# Create a mock imbalanced dataset\nX, y = make_classification(\n n_samples=5000,\n n_features=30,\n n_informative=20,\n weights=(0.95,),\n random_state=1,\n)",
"_____no_output_____"
]
],
[
[
"## Run the pipeline",
"_____no_output_____"
]
],
[
[
"# Initialize atom\natom = ATOMClassifier(X, y, test_size=0.2, verbose=2, random_state=1)",
"<< ================== ATOM ================== >>\nAlgorithm task: binary classification.\n\nDataset stats ==================== >>\nShape: (5000, 31)\nMemory: 1.22 MB\nScaled: False\nOutlier values: 565 (0.5%)\n-------------------------------------\nTrain set size: 4000\nTest set size: 1000\n-------------------------------------\n| | dataset | train | test |\n| - | ------------ | ------------ | ------------ |\n| 0 | 4731 (17.6) | 3785 (17.6) | 946 (17.5) |\n| 1 | 269 (1.0) | 215 (1.0) | 54 (1.0) |\n\n"
],
[
"# Let's have a look at the data. Note that, since the input wasn't\n# a dataframe, atom has given default names to the columns.\natom.head()",
"_____no_output_____"
],
[
"# Let's start reducing the number of features\natom.feature_selection(\"RFE\", solver=\"RF\", n_features=12)",
"Fitting FeatureSelector...\nPerforming feature selection ...\n --> RFE selected 12 features from the dataset.\n >>> Dropping feature feature_2 (rank 12).\n >>> Dropping feature feature_3 (rank 8).\n >>> Dropping feature feature_5 (rank 2).\n >>> Dropping feature feature_7 (rank 17).\n >>> Dropping feature feature_8 (rank 14).\n >>> Dropping feature feature_11 (rank 19).\n >>> Dropping feature feature_12 (rank 3).\n >>> Dropping feature feature_13 (rank 11).\n >>> Dropping feature feature_14 (rank 9).\n >>> Dropping feature feature_15 (rank 13).\n >>> Dropping feature feature_17 (rank 5).\n >>> Dropping feature feature_19 (rank 16).\n >>> Dropping feature feature_20 (rank 4).\n >>> Dropping feature feature_23 (rank 7).\n >>> Dropping feature feature_24 (rank 10).\n >>> Dropping feature feature_25 (rank 18).\n >>> Dropping feature feature_26 (rank 6).\n >>> Dropping feature feature_27 (rank 15).\n"
],
[
"# Fit a model directly on the imbalanced data\natom.run(\"RF\", metric=\"ba\")",
"\nTraining ========================= >>\nModels: RF\nMetric: balanced_accuracy\n\n\nResults for Random Forest:\nFit ---------------------------------------------\nTrain evaluation --> balanced_accuracy: 1.0\nTest evaluation --> balanced_accuracy: 0.6111\nTime elapsed: 0.814s\n-------------------------------------------------\nTotal time: 0.815s\n\n\nFinal results ==================== >>\nDuration: 0.816s\n-------------------------------------\nRandom Forest --> balanced_accuracy: 0.6111 ~\n"
],
[
"# The transformer and the models have been added to the branch\natom.branch",
"_____no_output_____"
]
],
[
[
"## Oversampling",
"_____no_output_____"
]
],
[
[
"# Create a new branch for oversampling\natom.branch = \"oversample\"",
"New branch oversample successfully created.\n"
],
[
"# Perform oversampling of the minority class\natom.balance(strategy=\"smote\")",
"Oversampling with SMOTE...\n --> Adding 3570 samples to class 1.\n"
],
[
"atom.classes # Check the balanced training set!",
"_____no_output_____"
],
[
"# Train another model on the new branch. Add a tag after \n# the model's acronym to distinguish it from the first model\natom.run(\"rf_os\") # os for oversample",
"\nTraining ========================= >>\nModels: RF_os\nMetric: balanced_accuracy\n\n\nResults for Random Forest:\nFit ---------------------------------------------\nTrain evaluation --> balanced_accuracy: 1.0\nTest evaluation --> balanced_accuracy: 0.7214\nTime elapsed: 1.485s\n-------------------------------------------------\nTotal time: 1.485s\n\n\nFinal results ==================== >>\nDuration: 1.485s\n-------------------------------------\nRandom Forest --> balanced_accuracy: 0.7214 ~\n"
]
],
[
[
"## Undersampling",
"_____no_output_____"
]
],
[
[
"# Create the undersampling branch\n# Split from master to not adopt the oversmapling transformer\natom.branch = \"undersample_from_master\"",
"New branch undersample successfully created.\n"
],
[
"atom.classes # In this branch, the data is still imbalanced",
"_____no_output_____"
],
[
"# Perform undersampling of the majority class\natom.balance(strategy=\"NearMiss\")",
"Undersampling with NearMiss...\n --> Removing 3570 samples from class 0.\n"
],
[
"atom.run(\"rf_us\")",
"\nTraining ========================= >>\nModels: RF_us\nMetric: balanced_accuracy\n\n\nResults for Random Forest:\nFit ---------------------------------------------\nTrain evaluation --> balanced_accuracy: 1.0\nTest evaluation --> balanced_accuracy: 0.7225\nTime elapsed: 0.156s\n-------------------------------------------------\nTotal time: 0.156s\n\n\nFinal results ==================== >>\nDuration: 0.156s\n-------------------------------------\nRandom Forest --> balanced_accuracy: 0.7225 ~\n"
],
[
"# Check that the branch only contains the desired transformers \natom.branch",
"_____no_output_____"
]
],
[
[
"## Analyze results",
"_____no_output_____"
]
],
[
[
"atom.evaluate()",
"The frame.append method is deprecated and will be removed from pandas in a future version. Use pandas.concat instead.\nThe frame.append method is deprecated and will be removed from pandas in a future version. Use pandas.concat instead.\nThe frame.append method is deprecated and will be removed from pandas in a future version. Use pandas.concat instead.\n"
],
[
"atom.plot_prc()",
"_____no_output_____"
],
[
"atom.plot_roc()",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
cb900904a9722f6edc28f6918429c0248889ac16
| 9,148 |
ipynb
|
Jupyter Notebook
|
examples/protyping/icom/007-extract-patient-from-archive.ipynb
|
lipteck/pymedphys
|
6e8e2b5db8173eafa6006481ceeca4f4341789e0
|
[
"Apache-2.0"
] | 2 |
2020-02-04T03:21:20.000Z
|
2020-04-11T14:17:53.000Z
|
prototyping/icom/007-extract-patient-from-archive.ipynb
|
SimonBiggs/pymedphys
|
83f02eac6549ac155c6963e0a8d1f9284359b652
|
[
"Apache-2.0"
] | 6 |
2020-10-06T15:36:46.000Z
|
2022-02-27T05:15:17.000Z
|
prototyping/icom/007-extract-patient-from-archive.ipynb
|
SimonBiggs/pymedphys
|
83f02eac6549ac155c6963e0a8d1f9284359b652
|
[
"Apache-2.0"
] | 1 |
2020-12-20T14:14:00.000Z
|
2020-12-20T14:14:00.000Z
| 22.927318 | 103 | 0.53498 |
[
[
[
"import pathlib\nimport lzma\nimport re\nimport os\nimport datetime\nimport copy\nimport functools\n\nimport numpy as np\nimport pandas as pd",
"_____no_output_____"
],
[
"# Makes it so any changes in pymedphys is automatically\n# propagated into the notebook without needing a kernel reset.\nfrom IPython.lib.deepreload import reload\n%load_ext autoreload\n%autoreload 2",
"_____no_output_____"
],
[
"import pymedphys._icom.extract\nimport pymedphys.mudensity\nimport pymedphys",
"_____no_output_____"
],
[
"patient_id = '008566'",
"_____no_output_____"
],
[
"patients_dir = pathlib.Path(r'\\\\physics-server\\iComLogFiles\\patients')\npatient_data_paths = list(patients_dir.glob(f'{patient_id}_*/*.xz'))\npatient_data_paths",
"_____no_output_____"
],
[
"patient_data_path = patient_data_paths[0]\nwith lzma.open(patient_data_path, 'r') as f:\n patient_data = f.read()",
"_____no_output_____"
],
[
"DATE_PATTERN = re.compile(rb\"\\d\\d\\d\\d-\\d\\d-\\d\\d\\d\\d:\\d\\d:\\d\\d.\")\n\ndef get_data_points(data):\n date_index = [m.span() for m in DATE_PATTERN.finditer(data)]\n start_points = [span[0] - 8 for span in date_index]\n\n end_points = start_points[1::] + [None]\n\n data_points = [data[start:end] for start, end in zip(start_points, end_points)]\n return data_points",
"_____no_output_____"
],
[
"patient_data_list = get_data_points(patient_data)\nlen(patient_data_list)",
"_____no_output_____"
],
[
"@functools.lru_cache()\ndef get_coll_regex(label, number):\n header = rb\"0\\xb8\\x00DS\\x00R.\\x00\\x00\\x00\" + label + b\"\\n\"\n item = rb\"0\\x1c\\x01DS\\x00R.\\x00\\x00\\x00(-?\\d+\\.\\d+)\"\n\n regex = re.compile(header + b\"\\n\".join([item] * number))\n return regex\n\n\ndef extract_coll(data, label, number):\n regex = get_coll_regex(label, number)\n\n match = regex.search(data)\n span = match.span()\n\n data = data[0 : span[0]] + data[span[1] + 1 : :]\n items = np.array([float(item) for item in match.groups()])\n\n return data, items",
"_____no_output_____"
],
[
"def get_delivery_data_items(single_icom_stream):\n shrunk_stream, mu = pymedphys._icom.extract.extract(single_icom_stream, \"Delivery MU\")\n shrunk_stream, gantry = pymedphys._icom.extract.extract(shrunk_stream, \"Gantry\")\n shrunk_stream, collimator = pymedphys._icom.extract.extract(shrunk_stream, \"Collimator\")\n \n shrunk_stream, mlc = extract_coll(shrunk_stream, b\"MLCX\", 160)\n mlc = mlc.reshape((80,2))\n mlc = np.fliplr(np.flipud(mlc * 10))\n mlc[:,1] = -mlc[:,1]\n mlc = np.round(mlc,10)\n# shrunk_stream, result[\"ASYMX\"] = extract_coll(shrunk_stream, b\"ASYMX\", 2)\n shrunk_stream, jaw = extract_coll(shrunk_stream, b\"ASYMY\", 2)\n jaw = np.round(np.array(jaw) * 10, 10)\n jaw = np.flipud(jaw)\n \n return mu, gantry, collimator, mlc, jaw",
"_____no_output_____"
],
[
"mu, gantry, collimator, mlc, jaw = get_delivery_data_items(patient_data_list[250])",
"_____no_output_____"
],
[
"gantry",
"_____no_output_____"
],
[
"collimator",
"_____no_output_____"
],
[
"mu",
"_____no_output_____"
],
[
"jaw",
"_____no_output_____"
],
[
"len(patient_data_list)",
"_____no_output_____"
],
[
"delivery_raw = [\n get_delivery_data_items(single_icom_stream)\n for single_icom_stream in patient_data_list\n]",
"_____no_output_____"
],
[
"mu = np.array([item[0] for item in delivery_raw])\ndiff_mu = np.concatenate([[0], np.diff(mu)])\ndiff_mu[diff_mu<0] = 0\nmu = np.cumsum(diff_mu)",
"_____no_output_____"
],
[
"gantry = np.array([item[1] for item in delivery_raw])\ncollimator = np.array([item[2] for item in delivery_raw])\nmlc = np.array([item[3] for item in delivery_raw])\njaw = np.array([item[4] for item in delivery_raw])",
"_____no_output_____"
],
[
"icom_delivery = pymedphys.Delivery(mu, gantry, collimator, mlc, jaw)\nicom_delivery = icom_delivery._filter_cps()",
"_____no_output_____"
],
[
"monaco_directory = pathlib.Path(r'\\\\monacoda\\FocalData\\RCCC\\1~Clinical')",
"_____no_output_____"
],
[
"tel_path = list(monaco_directory.glob(f'*~{patient_id}/plan/*/tel.1'))[-1]\ntel_path",
"_____no_output_____"
],
[
"GRID = pymedphys.mudensity.grid()\n\ndelivery_tel = pymedphys.Delivery.from_monaco(tel_path)\nmudensity_tel = delivery_tel.mudensity()\npymedphys.mudensity.display(GRID, mudensity_tel)",
"_____no_output_____"
],
[
"mudensity_icom = icom_delivery.mudensity()\npymedphys.mudensity.display(GRID, mudensity_icom)",
"_____no_output_____"
],
[
"icom_delivery.mu[16]",
"_____no_output_____"
],
[
"delivery_tel.mu[1]",
"_____no_output_____"
],
[
"delivery_tel.mlc[1]",
"_____no_output_____"
],
[
"icom_delivery.mlc[16]",
"_____no_output_____"
],
[
"delivery_tel.jaw[1]",
"_____no_output_____"
],
[
"icom_delivery.jaw[16]",
"_____no_output_____"
],
[
"# new_mlc = np.fliplr(np.flipud(np.array(icom_delivery.mlc[16]) * 10))\n# new_mlc[:,1] = -new_mlc[:,1]\n# new_mlc",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb9013baf394bb75ee2af3c1b4ab5213043bc015
| 39,306 |
ipynb
|
Jupyter Notebook
|
nbs/70_callback.wandb.ipynb
|
EmbraceLife/fastai
|
85258502eff144708d657aa4b4d2ab4c2a2b3a0b
|
[
"Apache-2.0"
] | null | null | null |
nbs/70_callback.wandb.ipynb
|
EmbraceLife/fastai
|
85258502eff144708d657aa4b4d2ab4c2a2b3a0b
|
[
"Apache-2.0"
] | null | null | null |
nbs/70_callback.wandb.ipynb
|
EmbraceLife/fastai
|
85258502eff144708d657aa4b4d2ab4c2a2b3a0b
|
[
"Apache-2.0"
] | null | null | null | 38.573111 | 2,291 | 0.526103 |
[
[
[
"#|hide\n#|skip\n! [ -e /content ] && pip install -Uqq fastai # upgrade fastai on colab",
"_____no_output_____"
],
[
"#|all_slow",
"_____no_output_____"
],
[
"#|export\nfrom __future__ import annotations\nfrom fastai.basics import *\nfrom fastai.callback.progress import *\nfrom fastai.text.data import TensorText\nfrom fastai.tabular.all import TabularDataLoaders, Tabular\nfrom fastai.callback.hook import total_params",
"_____no_output_____"
],
[
"#|hide\nfrom nbdev.showdoc import *",
"_____no_output_____"
],
[
"#|default_exp callback.wandb",
"_____no_output_____"
]
],
[
[
"# Wandb\n\n> Integration with [Weights & Biases](https://docs.wandb.com/library/integrations/fastai) ",
"_____no_output_____"
],
[
"First thing first, you need to install wandb with\n```\npip install wandb\n```\nCreate a free account then run \n``` \nwandb login\n```\nin your terminal. Follow the link to get an API token that you will need to paste, then you're all set!",
"_____no_output_____"
]
],
[
[
"#|export\nimport wandb",
"_____no_output_____"
],
[
"#|export\nclass WandbCallback(Callback):\n \"Saves model topology, losses & metrics\"\n remove_on_fetch,order = True,Recorder.order+1\n # Record if watch has been called previously (even in another instance)\n _wandb_watch_called = False\n\n def __init__(self, \n log:str=None, # What to log (can be `gradients`, `parameters`, `all` or None) \n log_preds:bool=True, # Whether to log model predictions on a `wandb.Table`\n log_preds_every_epoch:bool=False, # Whether to log predictions every epoch or at the end\n log_model:bool=False, # Whether to save the model checkpoint to a `wandb.Artifact` \n model_name:str=None, # The name of the `model_name` to save, overrides `SaveModelCallback`\n log_dataset:bool=False, # Whether to log the dataset to a `wandb.Artifact`\n dataset_name:str=None, # A name to log the dataset with\n valid_dl:TfmdDL=None, # If `log_preds=True`, then the samples will be drawn from `valid_dl`\n n_preds:int=36, # How many samples to log predictions \n seed:int=12345, # The seed of the samples drawn\n reorder=True):\n store_attr()\n \n def after_create(self):\n # log model\n if self.log_model:\n if not hasattr(self, 'save_model'):\n # does not have the SaveModelCallback\n self.learn.add_cb(SaveModelCallback(fname=ifnone(self.model_name, 'model')))\n else:\n # override SaveModelCallback\n if self.model_name is not None:\n self.save_model.fname = self.model_name\n \n def before_fit(self):\n \"Call watch method to log model topology, gradients & weights\"\n # Check if wandb.init has been called\n if wandb.run is None:\n raise ValueError('You must call wandb.init() before WandbCallback()')\n # W&B log step\n self._wandb_step = wandb.run.step - 1 # -1 except if the run has previously logged data (incremented at each batch)\n self._wandb_epoch = 0 if not(wandb.run.step) else math.ceil(wandb.run.summary['epoch']) # continue to next epoch\n \n self.run = not hasattr(self.learn, 'lr_finder') and not hasattr(self, \"gather_preds\") and rank_distrib()==0\n if not self.run: return\n\n # Log config parameters\n log_config = self.learn.gather_args()\n _format_config(log_config)\n try:\n wandb.config.update(log_config, allow_val_change=True)\n except Exception as e:\n print(f'WandbCallback could not log config parameters -> {e}')\n\n if not WandbCallback._wandb_watch_called:\n WandbCallback._wandb_watch_called = True\n # Logs model topology and optionally gradients and weights\n if self.log is not None:\n wandb.watch(self.learn.model, log=self.log)\n\n # log dataset\n assert isinstance(self.log_dataset, (str, Path, bool)), 'log_dataset must be a path or a boolean'\n if self.log_dataset is True:\n if Path(self.dls.path) == Path('.'):\n print('WandbCallback could not retrieve the dataset path, please provide it explicitly to \"log_dataset\"')\n self.log_dataset = False\n else:\n self.log_dataset = self.dls.path\n if self.log_dataset:\n self.log_dataset = Path(self.log_dataset)\n assert self.log_dataset.is_dir(), f'log_dataset must be a valid directory: {self.log_dataset}'\n metadata = {'path relative to learner': os.path.relpath(self.log_dataset, self.learn.path)}\n log_dataset(path=self.log_dataset, name=self.dataset_name, metadata=metadata)\n\n if self.log_preds:\n try:\n if not self.valid_dl:\n #Initializes the batch watched\n wandbRandom = random.Random(self.seed) # For repeatability\n self.n_preds = min(self.n_preds, len(self.dls.valid_ds))\n idxs = wandbRandom.sample(range(len(self.dls.valid_ds)), self.n_preds)\n if isinstance(self.dls, TabularDataLoaders):\n test_items = getattr(self.dls.valid_ds.items, 'iloc', self.dls.valid_ds.items)[idxs]\n self.valid_dl = self.dls.test_dl(test_items, with_labels=True, process=False)\n else:\n test_items = [getattr(self.dls.valid_ds.items, 'iloc', self.dls.valid_ds.items)[i] for i in idxs]\n self.valid_dl = self.dls.test_dl(test_items, with_labels=True)\n self.learn.add_cb(FetchPredsCallback(dl=self.valid_dl, with_input=True, with_decoded=True, reorder=self.reorder))\n except Exception as e:\n self.log_preds = False\n print(f'WandbCallback was not able to prepare a DataLoader for logging prediction samples -> {e}')\n \n def before_batch(self): \n self.ti_batch = time.perf_counter()\n \n def after_batch(self):\n \"Log hyper-parameters and training loss\"\n if self.training:\n batch_time = time.perf_counter() - self.ti_batch\n self._wandb_step += 1\n self._wandb_epoch += 1/self.n_iter\n hypers = {f'{k}_{i}':v for i,h in enumerate(self.opt.hypers) for k,v in h.items()}\n wandb.log({'epoch': self._wandb_epoch, 'train_loss': self.smooth_loss, 'raw_loss': self.loss, **hypers}, step=self._wandb_step)\n wandb.log({'train_samples_per_sec': len(self.xb[0]) / batch_time}, step=self._wandb_step)\n \n def log_predictions(self):\n try:\n inp,preds,targs,out = self.learn.fetch_preds.preds\n b = tuplify(inp) + tuplify(targs)\n x,y,its,outs = self.valid_dl.show_results(b, out, show=False, max_n=self.n_preds)\n wandb.log(wandb_process(x, y, its, outs, preds), step=self._wandb_step)\n except Exception as e:\n self.log_preds = False\n self.remove_cb(FetchPredsCallback)\n print(f'WandbCallback was not able to get prediction samples -> {e}')\n \n\n def after_epoch(self):\n \"Log validation loss and custom metrics & log prediction samples\"\n # Correct any epoch rounding error and overwrite value\n self._wandb_epoch = round(self._wandb_epoch)\n if self.log_preds and self.log_preds_every_epoch:\n self.log_predictions()\n wandb.log({'epoch': self._wandb_epoch}, step=self._wandb_step)\n wandb.log({n:s for n,s in zip(self.recorder.metric_names, self.recorder.log) if n not in ['train_loss', 'epoch', 'time']}, step=self._wandb_step)\n \n \n \n def after_fit(self):\n if self.log_preds and not self.log_preds_every_epoch:\n self.log_predictions()\n if self.log_model:\n if self.save_model.last_saved_path is None:\n print('WandbCallback could not retrieve a model to upload')\n else:\n metadata = {n:s for n,s in zip(self.recorder.metric_names, self.recorder.log) if n not in ['train_loss', 'epoch', 'time']}\n log_model(self.save_model.last_saved_path, name=self.save_model.fname, metadata=metadata) \n self.run = True\n if self.log_preds: self.remove_cb(FetchPredsCallback)\n \n wandb.log({}) # ensure sync of last step\n self._wandb_step += 1\n ",
"_____no_output_____"
]
],
[
[
"Optionally logs weights and or gradients depending on `log` (can be \"gradients\", \"parameters\", \"all\" or None), sample predictions if ` log_preds=True` that will come from `valid_dl` or a random sample of the validation set (determined by `seed`). `n_preds` are logged in this case.\n\nIf used in combination with `SaveModelCallback`, the best model is saved as well (can be deactivated with `log_model=False`).\n\nDatasets can also be tracked:\n* if `log_dataset` is `True`, tracked folder is retrieved from `learn.dls.path`\n* `log_dataset` can explicitly be set to the folder to track\n* the name of the dataset can explicitly be given through `dataset_name`, otherwise it is set to the folder name\n* *Note: the subfolder \"models\" is always ignored*\n\nFor custom scenarios, you can also manually use functions `log_dataset` and `log_model` to respectively log your own datasets and models.",
"_____no_output_____"
]
],
[
[
"#|export\n@patch\ndef gather_args(self:Learner):\n \"Gather config parameters accessible to the learner\"\n # args stored by `store_attr`\n cb_args = {f'{cb}':getattr(cb,'__stored_args__',True) for cb in self.cbs}\n args = {'Learner':self, **cb_args}\n # input dimensions\n try:\n n_inp = self.dls.train.n_inp\n args['n_inp'] = n_inp\n xb = self.dls.valid.one_batch()[:n_inp]\n args.update({f'input {n+1} dim {i+1}':d for n in range(n_inp) for i,d in enumerate(list(detuplify(xb[n]).shape))})\n except: print(f'Could not gather input dimensions')\n # other useful information\n with ignore_exceptions():\n args['batch size'] = self.dls.bs\n args['batch per epoch'] = len(self.dls.train)\n args['model parameters'] = total_params(self.model)[0]\n args['device'] = self.dls.device.type\n args['frozen'] = bool(self.opt.frozen_idx)\n args['frozen idx'] = self.opt.frozen_idx\n args['dataset.tfms'] = f'{self.dls.dataset.tfms}'\n args['dls.after_item'] = f'{self.dls.after_item}'\n args['dls.before_batch'] = f'{self.dls.before_batch}'\n args['dls.after_batch'] = f'{self.dls.after_batch}'\n return args",
"_____no_output_____"
],
[
"#|export\ndef _make_plt(img):\n \"Make plot to image resolution\"\n # from https://stackoverflow.com/a/13714915\n my_dpi = 100\n fig = plt.figure(frameon=False, dpi=my_dpi)\n h, w = img.shape[:2]\n fig.set_size_inches(w / my_dpi, h / my_dpi)\n ax = plt.Axes(fig, [0., 0., 1., 1.])\n ax.set_axis_off()\n fig.add_axes(ax)\n return fig, ax",
"_____no_output_____"
],
[
"#|export\ndef _format_config_value(v):\n if isinstance(v, list):\n return [_format_config_value(item) for item in v]\n elif hasattr(v, '__stored_args__'):\n return {**_format_config(v.__stored_args__), '_name': v}\n return v",
"_____no_output_____"
],
[
"#|export\ndef _format_config(config):\n \"Format config parameters before logging them\"\n for k,v in config.items():\n if isinstance(v, dict):\n config[k] = _format_config(v)\n else:\n config[k] = _format_config_value(v)\n return config",
"_____no_output_____"
],
[
"#|export\ndef _format_metadata(metadata):\n \"Format metadata associated to artifacts\"\n for k,v in metadata.items(): metadata[k] = str(v)",
"_____no_output_____"
],
[
"#|export\ndef log_dataset(path, name=None, metadata={}, description='raw dataset'):\n \"Log dataset folder\"\n # Check if wandb.init has been called in case datasets are logged manually\n if wandb.run is None:\n raise ValueError('You must call wandb.init() before log_dataset()')\n path = Path(path)\n if not path.is_dir():\n raise f'path must be a valid directory: {path}'\n name = ifnone(name, path.name)\n _format_metadata(metadata)\n artifact_dataset = wandb.Artifact(name=name, type='dataset', metadata=metadata, description=description)\n # log everything except \"models\" folder\n for p in path.ls():\n if p.is_dir():\n if p.name != 'models': artifact_dataset.add_dir(str(p.resolve()), name=p.name)\n else: artifact_dataset.add_file(str(p.resolve()))\n wandb.run.use_artifact(artifact_dataset)",
"_____no_output_____"
],
[
"#|export\ndef log_model(path, name=None, metadata={}, description='trained model'):\n \"Log model file\"\n if wandb.run is None:\n raise ValueError('You must call wandb.init() before log_model()')\n path = Path(path)\n if not path.is_file():\n raise f'path must be a valid file: {path}'\n name = ifnone(name, f'run-{wandb.run.id}-model')\n _format_metadata(metadata) \n artifact_model = wandb.Artifact(name=name, type='model', metadata=metadata, description=description)\n with artifact_model.new_file(name, mode='wb') as fa:\n fa.write(path.read_bytes())\n wandb.run.log_artifact(artifact_model)",
"_____no_output_____"
],
[
"#|export\n@typedispatch\ndef wandb_process(x:TensorImage, y, samples, outs, preds):\n \"Process `sample` and `out` depending on the type of `x/y`\"\n res_input, res_pred, res_label = [],[],[]\n for s,o in zip(samples, outs):\n img = s[0].permute(1,2,0)\n res_input.append(wandb.Image(img, caption='Input_data'))\n for t, capt, res in ((o[0], \"Prediction\", res_pred), (s[1], \"Ground_Truth\", res_label)):\n fig, ax = _make_plt(img)\n # Superimpose label or prediction to input image\n ax = img.show(ctx=ax)\n ax = t.show(ctx=ax)\n res.append(wandb.Image(fig, caption=capt))\n plt.close(fig)\n return {\"Inputs\":res_input, \"Predictions\":res_pred, \"Ground_Truth\":res_label}",
"_____no_output_____"
],
[
"#export\ndef _unlist(l):\n \"get element of lists of lenght 1\"\n if isinstance(l, (list, tuple)):\n if len(l) == 1: return l[0]\n else: return l",
"_____no_output_____"
],
[
"#|export\n@typedispatch\ndef wandb_process(x:TensorImage, y:(TensorCategory,TensorMultiCategory), samples, outs, preds):\n table = wandb.Table(columns=[\"Input image\", \"Ground_Truth\", \"Predictions\"])\n for (image, label), pred_label in zip(samples,outs):\n table.add_data(wandb.Image(image.permute(1,2,0)), label, _unlist(pred_label))\n return {\"Prediction_Samples\": table}",
"_____no_output_____"
],
[
"#|export\n@typedispatch\ndef wandb_process(x:TensorImage, y:TensorMask, samples, outs, preds):\n res = []\n codes = getattr(outs[0][0], 'codes', None)\n if codes is not None:\n class_labels = [{'name': name, 'id': id} for id, name in enumerate(codes)] \n else:\n class_labels = [{'name': i, 'id': i} for i in range(preds.shape[1])]\n table = wandb.Table(columns=[\"Input Image\", \"Ground_Truth\", \"Predictions\"])\n for (image, label), pred_label in zip(samples, outs):\n img = image.permute(1,2,0)\n table.add_data(wandb.Image(img),\n wandb.Image(img, masks={\"Ground_Truth\": {'mask_data': label.numpy().astype(np.uint8)}}, classes=class_labels), \n wandb.Image(img, masks={\"Prediction\": {'mask_data': pred_label[0].numpy().astype(np.uint8)}}, classes=class_labels) \n )\n return {\"Prediction_Samples\": table}",
"_____no_output_____"
],
[
"#|export\n@typedispatch\ndef wandb_process(x:TensorText, y:(TensorCategory,TensorMultiCategory), samples, outs, preds):\n data = [[s[0], s[1], o[0]] for s,o in zip(samples,outs)]\n return {\"Prediction_Samples\": wandb.Table(data=data, columns=[\"Text\", \"Target\", \"Prediction\"])}",
"_____no_output_____"
],
[
"#|export\n@typedispatch\ndef wandb_process(x:Tabular, y:Tabular, samples, outs, preds):\n df = x.all_cols\n for n in x.y_names: df[n+'_pred'] = y[n].values\n return {\"Prediction_Samples\": wandb.Table(dataframe=df)}",
"_____no_output_____"
]
],
[
[
"## Example of use:\n\nOnce your have defined your `Learner`, before you call to `fit` or `fit_one_cycle`, you need to initialize wandb:\n```\nimport wandb\nwandb.init()\n```\nTo use Weights & Biases without an account, you can call `wandb.init(anonymous='allow')`.\n\nThen you add the callback to your `learner` or call to `fit` methods, potentially with `SaveModelCallback` if you want to save the best model:\n```\nfrom fastai.callback.wandb import *\n\n# To log only during one training phase\nlearn.fit(..., cbs=WandbCallback())\n\n# To log continuously for all training phases\nlearn = learner(..., cbs=WandbCallback())\n```\nDatasets and models can be tracked through the callback or directly through `log_model` and `log_dataset` functions.\n\nFor more details, refer to [W&B documentation](https://docs.wandb.com/library/integrations/fastai).",
"_____no_output_____"
]
],
[
[
"#|hide\n#|slow\nfrom fastai.vision.all import *\nimport tempfile\n\npath = untar_data(URLs.MNIST_TINY)\nitems = get_image_files(path)\ntds = Datasets(items, [PILImageBW.create, [parent_label, Categorize()]], splits=GrandparentSplitter()(items))\ndls = tds.dataloaders(after_item=[ToTensor(), IntToFloatTensor()])\n\nos.environ['WANDB_MODE'] = 'dryrun' # run offline\nwith tempfile.TemporaryDirectory() as wandb_local_dir:\n wandb.init(anonymous='allow', dir=wandb_local_dir)\n learn = vision_learner(dls, resnet18, loss_func=CrossEntropyLossFlat(), cbs=WandbCallback(log_model=False))\n learn.fit(1)\n\n # add more data from a new learner on same run\n learn = vision_learner(dls, resnet18, loss_func=CrossEntropyLossFlat(), cbs=WandbCallback(log_model=False))\n learn.fit(1, lr=slice(0.005))\n \n # save model\n learn = cnn_learner(dls, resnet18, loss_func=CrossEntropyLossFlat(), cbs=WandbCallback(log_model=True))\n learn.fit(1, lr=slice(0.005))\n \n # save model override name\n learn = cnn_learner(dls, resnet18, loss_func=CrossEntropyLossFlat(), cbs=[WandbCallback(log_model=True, model_name=\"good_name\"), SaveModelCallback(fname=\"bad_name\")])\n learn.fit(1, lr=slice(0.005))\n \n # finish writing files to temporary folder\n wandb.finish()",
"_____no_output_____"
],
[
"#|export\n_all_ = ['wandb_process']",
"_____no_output_____"
]
],
[
[
"## Export -",
"_____no_output_____"
]
],
[
[
"#|hide\nfrom nbdev.export import *\nnotebook2script()",
"Converted 00_torch_core.ipynb.\nConverted 01_layers.ipynb.\nConverted 01a_losses.ipynb.\nConverted 02_data.load.ipynb.\nConverted 03_data.core.ipynb.\nConverted 04_data.external.ipynb.\nConverted 05_data.transforms.ipynb.\nConverted 06_data.block.ipynb.\nConverted 07_vision.core.ipynb.\nConverted 08_vision.data.ipynb.\nConverted 09_vision.augment.ipynb.\nConverted 09b_vision.utils.ipynb.\nConverted 09c_vision.widgets.ipynb.\nConverted 10_tutorial.pets.ipynb.\nConverted 10b_tutorial.albumentations.ipynb.\nConverted 11_vision.models.xresnet.ipynb.\nConverted 12_optimizer.ipynb.\nConverted 13_callback.core.ipynb.\nConverted 13a_learner.ipynb.\nConverted 13b_metrics.ipynb.\nConverted 14_callback.schedule.ipynb.\nConverted 14a_callback.data.ipynb.\nConverted 15_callback.hook.ipynb.\nConverted 15a_vision.models.unet.ipynb.\nConverted 16_callback.progress.ipynb.\nConverted 17_callback.tracker.ipynb.\nConverted 18_callback.fp16.ipynb.\nConverted 18a_callback.training.ipynb.\nConverted 18b_callback.preds.ipynb.\nConverted 19_callback.mixup.ipynb.\nConverted 20_interpret.ipynb.\nConverted 20a_distributed.ipynb.\nConverted 21_vision.learner.ipynb.\nConverted 22_tutorial.imagenette.ipynb.\nConverted 23_tutorial.vision.ipynb.\nConverted 24_tutorial.image_sequence.ipynb.\nConverted 24_tutorial.siamese.ipynb.\nConverted 24_vision.gan.ipynb.\nConverted 30_text.core.ipynb.\nConverted 31_text.data.ipynb.\nConverted 32_text.models.awdlstm.ipynb.\nConverted 33_text.models.core.ipynb.\nConverted 34_callback.rnn.ipynb.\nConverted 35_tutorial.wikitext.ipynb.\nConverted 37_text.learner.ipynb.\nConverted 38_tutorial.text.ipynb.\nConverted 39_tutorial.transformers.ipynb.\nConverted 40_tabular.core.ipynb.\nConverted 41_tabular.data.ipynb.\nConverted 42_tabular.model.ipynb.\nConverted 43_tabular.learner.ipynb.\nConverted 44_tutorial.tabular.ipynb.\nConverted 45_collab.ipynb.\nConverted 46_tutorial.collab.ipynb.\nConverted 50_tutorial.datablock.ipynb.\nConverted 60_medical.imaging.ipynb.\nConverted 61_tutorial.medical_imaging.ipynb.\nConverted 65_medical.text.ipynb.\nConverted 70_callback.wandb.ipynb.\nConverted 71_callback.tensorboard.ipynb.\nConverted 72_callback.neptune.ipynb.\nConverted 73_callback.captum.ipynb.\nConverted 74_callback.azureml.ipynb.\nConverted 97_test_utils.ipynb.\nConverted 99_pytorch_doc.ipynb.\nConverted dev-setup.ipynb.\nConverted app_examples.ipynb.\nConverted camvid.ipynb.\nConverted migrating_catalyst.ipynb.\nConverted migrating_ignite.ipynb.\nConverted migrating_lightning.ipynb.\nConverted migrating_pytorch.ipynb.\nConverted migrating_pytorch_verbose.ipynb.\nConverted ulmfit.ipynb.\nConverted index.ipynb.\nConverted quick_start.ipynb.\nConverted tutorial.ipynb.\n"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb9018eba92cf066a9078e69e9d3880aaa70d872
| 15,661 |
ipynb
|
Jupyter Notebook
|
chapter2/cnn.ipynb
|
christian-deeplearner/programming-pytorch
|
630bc7b075e967c7ce313248a8dd4d1f8348654a
|
[
"MIT"
] | null | null | null |
chapter2/cnn.ipynb
|
christian-deeplearner/programming-pytorch
|
630bc7b075e967c7ce313248a8dd4d1f8348654a
|
[
"MIT"
] | null | null | null |
chapter2/cnn.ipynb
|
christian-deeplearner/programming-pytorch
|
630bc7b075e967c7ce313248a8dd4d1f8348654a
|
[
"MIT"
] | null | null | null | 31.073413 | 1,450 | 0.555265 |
[
[
[
"import torch\nimport torchvision\nimport torch.nn as nn\nimport torch.nn.functional as F\nfrom torchvision import transforms\nimport torch.utils.data as data",
"_____no_output_____"
],
[
"train_data_path = \"./train\"\ntransform = transforms.Compose([\n transforms.Resize((64, 64)),\n transforms.ToTensor(),\n transforms.Normalize(mean=[0.485, 0.456, 0.406], \n std=[0.229, 0.224, 0.225])\n])\n\ntrain_data = torchvision.datasets.ImageFolder(root=train_data_path, transform=transform)",
"_____no_output_____"
],
[
"transform",
"_____no_output_____"
],
[
"val_data_path = \"./val/\"\nval_data = torchvision.datasets.ImageFolder(root=val_data_path, transform=transform)\n\ntest_data_path = \"./test/\"\ntest_data = torchvision.datasets.ImageFolder(root=test_data_path, transform=transform)",
"_____no_output_____"
],
[
"batch_size = 64\ntrain_data_loader = data.DataLoader(train_data, batch_size=batch_size)\nval_data_loader = data.DataLoader(val_data, batch_size=batch_size)\ntest_data_loader = data.DataLoader(test_data, batch_size=batch_size)",
"_____no_output_____"
],
[
"image, label = next(iter(train_data_loader))",
"_____no_output_____"
],
[
"image.shape, label.shape",
"_____no_output_____"
],
[
"class SimpleNet(nn.Module):\n def __init__(self):\n super(SimpleNet, self).__init__()\n self.fc1 = nn.Linear(12288, 84)\n self.fc2 = nn.Linear(84, 50)\n self.fc3 = nn.Linear(50, 2)\n\n def forward(self, x):\n x = x.view(-1, 12288)\n x = F.relu(self.fc1(x))\n x = F.relu(self.fc2(x))\n x = self.fc3(x)\n return x\n \n ",
"_____no_output_____"
],
[
"simplenet = SimpleNet()",
"_____no_output_____"
],
[
"simplenet",
"_____no_output_____"
],
[
"import torch.optim as optim\noptimizer = optim.Adam(simplenet.parameters(), lr=0.001)",
"_____no_output_____"
],
[
"if torch.cuda.is_available():\n device = torch.device(\"cuda\")\nelse:\n device = torch.device(\"cpu\")\n\n# model.to(device)",
"_____no_output_____"
],
[
"device",
"_____no_output_____"
],
[
"def train(model, optimizer, loss_fn, train_loader, val_loader, epochs=20, device=\"cpu\"):\n print(device)\n for epoch in range(epochs):\n training_loss = 0.0\n valid_loss = 0.0\n model.train()\n\n for batch in train_loader:\n optimizer.zero_grad()\n input, target = batch\n input = input.to(device)\n target = target.to(device)\n\n output = model(input)\n\n loss = loss_fn(output, target)\n loss.backward()\n optimizer.step()\n\n training_loss += loss.data.item()\n training_loss /= len(train_loader)\n\n model.eval()\n num_correct = 0\n num_examples = 0\n\n for batch in val_loader:\n input, target = batch\n input = input.to(device)\n output = model(input)\n\n target = target.to(device)\n\n loss = loss_fn(output, target)\n valid_loss += loss.data.item()\n\n correct = torch.eq(torch.max(F.softmax(output), dim=1)[1],\n\t\t\t\t\t\t\t target).view(-1)\n\n num_correct += torch.sum(correct).item()\n num_examples += correct.shape[0]\n\n valid_loss /= len(val_loader)\n\n print(\"Epoch: {}, Training Loss: {:.2f}, Validation Loss: {:.2f}, accuracy = {:.2f}\".format(epoch, training_loss,\n valid_loss, num_correct / num_examples))\n\n",
"_____no_output_____"
],
[
"device",
"_____no_output_____"
],
[
"train(simplenet, optimizer, torch.nn.CrossEntropyLoss(),train_data_loader, val_data_loader, 20, device)",
"cpu\nEpoch: 0, Training Loss: 3.22, Validation Loss: 2.61, accuracy = 0.41\nEpoch: 1, Training Loss: 1.97, Validation Loss: 1.58, accuracy = 0.67\nEpoch: 2, Training Loss: 1.24, Validation Loss: 0.84, accuracy = 0.61\nEpoch: 3, Training Loss: 0.60, Validation Loss: 0.65, accuracy = 0.73\nEpoch: 4, Training Loss: 0.52, Validation Loss: 0.62, accuracy = 0.68\nEpoch: 5, Training Loss: 0.39, Validation Loss: 0.66, accuracy = 0.70\nEpoch: 6, Training Loss: 0.36, Validation Loss: 0.63, accuracy = 0.72\nEpoch: 7, Training Loss: 0.32, Validation Loss: 0.63, accuracy = 0.72\nEpoch: 8, Training Loss: 0.29, Validation Loss: 0.63, accuracy = 0.72\nEpoch: 9, Training Loss: 0.26, Validation Loss: 0.66, accuracy = 0.72\nEpoch: 10, Training Loss: 0.24, Validation Loss: 0.65, accuracy = 0.71\nEpoch: 11, Training Loss: 0.22, Validation Loss: 0.67, accuracy = 0.71\nEpoch: 12, Training Loss: 0.20, Validation Loss: 0.68, accuracy = 0.71\nEpoch: 13, Training Loss: 0.18, Validation Loss: 0.70, accuracy = 0.72\nEpoch: 14, Training Loss: 0.16, Validation Loss: 0.73, accuracy = 0.72\nEpoch: 15, Training Loss: 0.14, Validation Loss: 0.75, accuracy = 0.71\nEpoch: 16, Training Loss: 0.12, Validation Loss: 0.84, accuracy = 0.68\nEpoch: 17, Training Loss: 0.13, Validation Loss: 0.77, accuracy = 0.72\nEpoch: 18, Training Loss: 0.10, Validation Loss: 0.82, accuracy = 0.70\nEpoch: 19, Training Loss: 0.09, Validation Loss: 0.82, accuracy = 0.72\n"
],
[
"torch.save(simplenet, \"simplenet\")",
"_____no_output_____"
],
[
"img = test_data[0][0]",
"_____no_output_____"
],
[
"img.numpy().shape",
"_____no_output_____"
],
[
"img = Image.fromarray(img.numpy())",
"_____no_output_____"
],
[
"classes = train_data_loader.dataset.classes",
"_____no_output_____"
],
[
"classes",
"_____no_output_____"
],
[
"cat = \"/Users/christian_acuna/workspace/beginners-pytorch-deep-learning/chapter2/test/cat/3156111_a9dba42579.jpg\"",
"_____no_output_____"
],
[
"fish = \"/Users/christian_acuna/workspace/beginners-pytorch-deep-learning/chapter2/test/fish/35099721_fbc694fa1b.jpg\"",
"_____no_output_____"
],
[
"from PIL import Image\n\ndef inference(filename, classes, model):\n labels = ['cat', 'fish']\n img = Image.open(filename)\n img = transform(img)\n img = img.unsqueeze(0)\n\n prediction = model(img)\n prediction = prediction.argmax()\n print(classes[prediction])",
"_____no_output_____"
],
[
"inference(cat, classes, simplenet)",
"cat\n"
],
[
"inference(fish, classes, simplenet)",
"fish\n"
],
[
"net = torch.load(\"simplenet\")",
"_____no_output_____"
],
[
"net",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb901d146f29711ef45ef04b682f7a80e8d4fbe6
| 9,139 |
ipynb
|
Jupyter Notebook
|
p8/ex19_Covid.ipynb
|
fh-swf-hgi/skriptsprachen-ex
|
ddc878d9bcc52f12fd74f6f780291181690eca53
|
[
"MIT"
] | null | null | null |
p8/ex19_Covid.ipynb
|
fh-swf-hgi/skriptsprachen-ex
|
ddc878d9bcc52f12fd74f6f780291181690eca53
|
[
"MIT"
] | null | null | null |
p8/ex19_Covid.ipynb
|
fh-swf-hgi/skriptsprachen-ex
|
ddc878d9bcc52f12fd74f6f780291181690eca53
|
[
"MIT"
] | null | null | null | 31.405498 | 411 | 0.599847 |
[
[
[
"<figure>\n <IMG SRC=\"https://upload.wikimedia.org/wikipedia/commons/thumb/d/d5/Fachhochschule_Südwestfalen_20xx_logo.svg/320px-Fachhochschule_Südwestfalen_20xx_logo.svg.png\" WIDTH=250 ALIGN=\"right\">\n</figure>\n\n# Skriptsprachen\n### Sommersemester 2021\nProf. Dr. Heiner Giefers",
"_____no_output_____"
],
[
"# Covid-19 Daten visualisieren\n\nIn dieser kleinen Aufgabe geht es darum, die weltweiten Covid-19 Zahlen zu visualisieren.\nWir wollen die die Daten von einer URL herunterladen und als *Pandas DataFrame* anlegen.\nDamit können wir die Daten vorverarbeiten und schließlich auch plotten.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport csv\nimport requests\nimport io\nimport pandas as pd\nimport plotly.express as px\nimport geojson",
"_____no_output_____"
],
[
"url = \"https://covid.ourworldindata.org/data/owid-covid-data.csv\"\ns=requests.get(url).content\ndf=pd.read_csv(io.StringIO(s.decode('utf-8')))",
"_____no_output_____"
],
[
"print(f\"Die Anzahl aller Einträge ist {df.size}\") ",
"_____no_output_____"
]
],
[
[
"Um sich einen ersten Eindruck von der Tabelle zu machen, kann man eine Reihe von Pandas-Methoden aufrufen:\n- `df.head(k)` zeigt die ersten `k` Einträge der Tabelle. Sie werden sehen, dass die Daten nach Ländern sortiert sind\n- `df.info()` zeigt Informationen zu den Spalten der Tabelle\n- `df.describe()` Gibt einige statistische Kennzahlen zu den Daten aus",
"_____no_output_____"
]
],
[
[
"df.head(10)",
"_____no_output_____"
]
],
[
[
"Wenn Sie die Daten nach einer anderen Spalte sortieren wollen, geht das mit der `sort_by_values` Methode:",
"_____no_output_____"
]
],
[
[
"df_date = df.sort_values(by='date')\ndf_date",
"_____no_output_____"
]
],
[
[
"Wir wollen nun das in Deutschland häufig verwendete Maß der *7-Tage Inzidenz* darstellen. Dieses ist in der Tabelle aber nicht direkt enthalten.\nWir können es allerdings aus den neuen Fällen pro Tag berechnen. Um eine Normalisierung gemäß der Einwohnerzahlen zu erreichen, verwendenden wir die Spalte `new_cases_per_million`.\nDiese kann allerdings fehlende Werte enthalten, z.B. weil für einige Länder an bestimmten Tagen keine Daten vorlagen.\nUm diese fehlenden Werte zu *schätzen*, interpolieren wir. D.h. wir nehmen an, bei einer *Lücke* würden sie Werte linear fortlaufen. Also bei der folge `1, 2, 3, NaN, 7, 8, 9` würde das `NaN` durch `5` ersetzt.\n\nNun gibt es noch ein weiteres Problem: Wir haben die Tabelle nach Daten sortiert, alle Länder stehen also vermischt in der Tabelle.\nBeim Aufsummieren der *7-Tage-Inzidenz* sollen aber natürlich nur Daten innerhalb eines Landes betrachtet werden.\nUm dies zu erreichen, können wir die `groupby`-Methode verwenden.\nAls Parameter erfhält `groupby` eine Funktion, die auf die gruppierten Daten angewendet wird.\n\n**Aufgabe:** Implementieren Sie die Funktion `berechne_inzidenz(x)`, die dem DataFrame `x` eine Spalte `Inzidenz` hinzufügt. Dazu soll zuerst die Spalte `new_cases_per_million` mit der Funktion `interpolate()` interpoliert werden. Anschließend soll die 7-Tage Inzidenz ausgerechnet werden. Sie können dazu die Methode `rolling(k)` verwenden, die ein gleitendes *Fenster* über `k`-Werte der Spalte liefert.",
"_____no_output_____"
]
],
[
[
"# Falsche Werte aussortieren\nindexEntries = df_date[df_date['new_cases_per_million'] < 0 ].index\ndf_cleaned = df_date.drop(indexEntries)\n\ndef berechne_inzidenz(x):\n # YOUR CODE HERE\n raise NotImplementedError()\n return x\n \ndf_cleaned = df_cleaned.groupby('iso_code').apply(berechne_inzidenz)",
"_____no_output_____"
]
],
[
[
"Nun können wir die Inzidenz-Werte anzeigen.\nDafür eignet sich gut eine Darstellung als Weltkarte, die wir z.B. mit der *Plotly* Methode `choropleth` erzeugen können.",
"_____no_output_____"
]
],
[
[
"fig = px.choropleth(df_cleaned, locations=\"iso_code\",\n color=\"Inzidenz\",\n #scope='europe',\n range_color = [0,200],\n hover_name=\"location\",\n animation_frame=\"date\",\n title = \"Corvid: weltweite 7-Tages Inzidenz\",\n color_continuous_scale=px.colors.sequential.Jet)\n \n \nfig[\"layout\"].pop(\"updatemenus\")\nfig.show()",
"_____no_output_____"
]
],
[
[
"Um bestimmte Zeilen eines DataFrames herauszufiltern, kann man bei der Auswahl der Spalten Bedingungen angeben.\nSo können wir z.B. die Werte aus Deutschland aus der Tabelle herausfiltern:",
"_____no_output_____"
]
],
[
[
"df_de = df[df['iso_code']=='DEU']\n\nprint(f\"Die Anzahl aller Einträge aus Deutschland ist {df_de.size}\")\ndf_de.head()",
"_____no_output_____"
]
],
[
[
"**Aufgabe:** Plotten Sie die Inzidenz-Werte für Deutschland (`DEU`), Großbritannien (`GBR`) und USA (`USA`) in einen gemeinsamen Graphen.\nVerwenden sie dau die *Matplotlib*-Methode `plot()`.",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nfrom matplotlib.ticker import MaxNLocator\n%matplotlib inline\n\nfig, axes = plt.subplots(1,1,figsize=(16,8))\n\ndf_cleaned['dedate'] = pd.to_datetime(df_cleaned.date).dt.strftime('%d.%m.%Y')\n# YOUR CODE HERE\nraise NotImplementedError()\naxes.xaxis.set_major_locator(MaxNLocator(15))\nplt.xticks(rotation = 45) \nplt.legend()\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Datenquelle\n\nDie verwendeten Daten stammen von [_Our World in Data_](https://ourworldindata.org/) und wurden dem Git-Repository [https://github.com/owid/covid-19-data](https://github.com/owid/covid-19-data) entnommen\n\nDetails zum Datensatz findet man in der folgenden Publikation:\n\n> Hasell, J., Mathieu, E., Beltekian, D. _et al._ A cross-country database of COVID-19 testing. _Sci Data_ **7**, 345 (2020). [https://doi.org/10.1038/s41597-020-00688-8](https://doi.org/10.1038/s41597-020-00688-8)\n\n",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
cb902d09405bde9c6c36dbfbe52ccc89197437c1
| 345,805 |
ipynb
|
Jupyter Notebook
|
pymaceuticals_starter.ipynb
|
ElenaSezionova/matplotlib-challenge
|
831d3eb71cf473ecc08574aed30f9ecffa43fb3e
|
[
"ADSL"
] | null | null | null |
pymaceuticals_starter.ipynb
|
ElenaSezionova/matplotlib-challenge
|
831d3eb71cf473ecc08574aed30f9ecffa43fb3e
|
[
"ADSL"
] | null | null | null |
pymaceuticals_starter.ipynb
|
ElenaSezionova/matplotlib-challenge
|
831d3eb71cf473ecc08574aed30f9ecffa43fb3e
|
[
"ADSL"
] | null | null | null | 198.967204 | 103,572 | 0.879311 |
[
[
[
"# Load Necessary Libraries",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nimport matplotlib.pyplot as plt\nimport pandas as pd\nimport numpy as np",
"_____no_output_____"
]
],
[
[
"# Hide warning messages in notebook",
"_____no_output_____"
]
],
[
[
"import warnings\nwarnings.filterwarnings('ignore')",
"_____no_output_____"
]
],
[
[
"# Load in data",
"_____no_output_____"
]
],
[
[
"mouse_drug_data = pd.read_csv('data/mouse_drug_data.csv')\nmouse_drug_data.head()",
"_____no_output_____"
],
[
"clinical_trial_data = pd.read_csv('data/clinicaltrial_data.csv')\nclinical_trial_data.head()",
"_____no_output_____"
]
],
[
[
"# Combine the data into a single dataset",
"_____no_output_____"
]
],
[
[
"mouse_drug_data.sort_values(by=\"Mouse ID\", inplace=True)\nclinical_trial_data.sort_values(by=\"Mouse ID\", inplace=True)\n\nmouse_drug_data = mouse_drug_data.reset_index(drop=True)\nclinical_trial_data = clinical_trial_data.reset_index(drop=True)\n\ndf = pd.merge(mouse_drug_data, clinical_trial_data, on=\"Mouse ID\", how=\"left\")\ndf.head()",
"_____no_output_____"
]
],
[
[
"# Tumor Response to Treatment\nStore the Mean Tumor Volume Data grouped by Drug and Timepoint",
"_____no_output_____"
]
],
[
[
"df_groupby = df.groupby(['Drug', 'Timepoint'])\nmean_tumor_volume_df = df_groupby['Tumor Volume (mm3)'].mean()\nmean_tumor_volume_df\n\ntumor_response = pd.DataFrame(mean_tumor_volume_df).reset_index()\ntumor_response.head()",
"_____no_output_____"
]
],
[
[
"Store the Standart Error of Tumor Volumes grouped by Drug and Timepoint",
"_____no_output_____"
]
],
[
[
"tumor_error = df_groupby['Tumor Volume (mm3)'].sem()\ntumor_response_error = pd.DataFrame(tumor_error).reset_index()\ntumor_response_error.head()",
"_____no_output_____"
]
],
[
[
"Minor Data Munging to Re-Format the Data Frames",
"_____no_output_____"
]
],
[
[
"reformat_df = tumor_response.pivot(index = 'Timepoint', columns = 'Drug', values = 'Tumor Volume (mm3)')\nreformat_df.head()",
"_____no_output_____"
]
],
[
[
"# Generate the Plot (with Error Bars)",
"_____no_output_____"
]
],
[
[
"plt.figure(figsize=(12,8))\n\nplt.title('Tumor Response to Treatment', fontdict={'fontweight':'bold', 'fontsize':18})\n\nCapomulin_df = tumor_response_error.loc[tumor_response_error['Drug'] == 'Capomulin', 'Tumor Volume (mm3)']\nCeftamin_df = tumor_response_error.loc[tumor_response_error['Drug'] == 'Ceftamin', 'Tumor Volume (mm3)']\nInfubinol_df = tumor_response_error.loc[tumor_response_error['Drug'] == 'Infubinol', 'Tumor Volume (mm3)']\nKetapril_df = tumor_response_error.loc[tumor_response_error['Drug'] == 'Ketapril', 'Tumor Volume (mm3)']\nNaftisol_df = tumor_response_error.loc[tumor_response_error['Drug'] == 'Naftisol', 'Tumor Volume (mm3)']\nPlacebo_df = tumor_response_error.loc[tumor_response_error['Drug'] == 'Placebo', 'Tumor Volume (mm3)']\nPropriva_df = tumor_response_error.loc[tumor_response_error['Drug'] == 'Propriva', 'Tumor Volume (mm3)']\nRamicane_df = tumor_response_error.loc[tumor_response_error['Drug'] == 'Ramicane', 'Tumor Volume (mm3)']\nStelasyn_df = tumor_response_error.loc[tumor_response_error['Drug'] == 'Stelasyn', 'Tumor Volume (mm3)']\nZoniferol_df = tumor_response_error.loc[tumor_response_error['Drug'] == 'Zoniferol', 'Tumor Volume (mm3)']\n\nTime = [0, 5, 10, 15, 20, 25, 30, 35, 40, 45]\n\nplt.errorbar(Time, reformat_df['Capomulin'], yerr = Capomulin_df, color = 'b', linestyle = '--', marker = 'o', label='Capomulin')\nplt.errorbar(Time, reformat_df[\"Ceftamin\"], yerr = Ceftamin_df, color = 'r', linestyle = '--', marker = '*', label=\"Ceftamin\")\nplt.errorbar(Time, reformat_df[\"Infubinol\"], yerr = Infubinol_df, color = 'y', linestyle = '--', marker = '^', label=\"Infubinol\")\nplt.errorbar(Time, reformat_df[\"Ketapril\"], yerr = Ketapril_df, color = 'g', linestyle = '--', marker = 's', label=\"Ketapril\")\nplt.errorbar(Time, reformat_df[\"Naftisol\"], yerr = Naftisol_df, color = '#abcdef', linestyle = '--', marker = 'D', label=\"Naftisol\")\nplt.errorbar(Time, reformat_df[\"Placebo\"], yerr = Placebo_df, color = '#adefab', linestyle = '--', marker = 'v', label=\"Placebo\")\nplt.errorbar(Time, reformat_df[\"Propriva\"], yerr = Propriva_df, color = '#d0abef', linestyle = '--', marker = '>', label=\"Propriva\")\nplt.errorbar(Time, reformat_df[\"Ramicane\"], yerr = Ramicane_df, color = '#efabec', linestyle = '--', marker = '<', label=\"Ramicane\")\nplt.errorbar(Time, reformat_df[\"Stelasyn\"], yerr = Stelasyn_df, color = '#c2abef', linestyle = '--', marker = '3', label=\"Stelasyn\")\nplt.errorbar(Time, reformat_df[\"Zoniferol\"], yerr = Zoniferol_df, color = 'grey', linestyle = '--', marker = '4', label=\"Zoniferol\")\n\nfor Drug in reformat_df:\n if Drug !='Tumor Volume (mm3)':\n print(Drug)\n\nplt.xlabel('Time (Days)')\nplt.ylabel ('Tumor Volume (mm3)')\n\nplt.legend()\nplt.grid()\n\nplt.savefig('Tumor_Response_to_Treatment', dpi=300)\nplt.show()\n\n\n",
"Capomulin\nCeftamin\nInfubinol\nKetapril\nNaftisol\nPlacebo\nPropriva\nRamicane\nStelasyn\nZoniferol\n"
]
],
[
[
"# Metastatic Response to Treatment\nStore the Mean Met. Site Data Grouped by Drug and Timepoint ",
"_____no_output_____"
]
],
[
[
"df_groupby = df.groupby(['Drug', 'Timepoint'])\nmean_tumor_volume_df = df_groupby['Metastatic Sites'].mean()\nmean_tumor_volume_df\n\ntumor_response = pd.DataFrame(mean_tumor_volume_df).reset_index()\ntumor_response.head()",
"_____no_output_____"
]
],
[
[
"Store the Standard Error associated with Met. Sites Grouped by Drug and Timepoint ",
"_____no_output_____"
]
],
[
[
"tumor_error = df_groupby['Metastatic Sites'].sem()\ntumor_error = pd.DataFrame(tumor_error).reset_index()\ntumor_error.head()",
"_____no_output_____"
]
],
[
[
"Minor Data Munging to Re-Format the Data Frames",
"_____no_output_____"
]
],
[
[
"metastatic_df = tumor_response.pivot(index = 'Timepoint', columns = 'Drug', values = 'Metastatic Sites')\nmetastatic_df.head()",
"_____no_output_____"
]
],
[
[
"Generate the Plot (with Error Bars)",
"_____no_output_____"
]
],
[
[
"plt.figure(figsize=(12,8))\n\nplt.title('Metastatic Spread During Treatment', fontdict={'fontweight':'bold', 'fontsize':16})\n\nCapomulin_df = tumor_error.loc[tumor_response_error['Drug'] == 'Capomulin', 'Metastatic Sites']\nCeftamin_df = tumor_error.loc[tumor_response_error['Drug'] == 'Ceftamin', 'Metastatic Sites']\nInfubinol_df = tumor_error.loc[tumor_response_error['Drug'] == 'Infubinol', 'Metastatic Sites']\nKetapril_df = tumor_error.loc[tumor_response_error['Drug'] == 'Ketapril', 'Metastatic Sites']\nNaftisol_df = tumor_error.loc[tumor_response_error['Drug'] == 'Naftisol', 'Metastatic Sites']\nPlacebo_df = tumor_error.loc[tumor_response_error['Drug'] == 'Placebo', 'Metastatic Sites']\nPropriva_df = tumor_error.loc[tumor_response_error['Drug'] == 'Propriva', 'Metastatic Sites']\nRamicane_df = tumor_error.loc[tumor_response_error['Drug'] == 'Ramicane', 'Metastatic Sites']\nStelasyn_df = tumor_error.loc[tumor_response_error['Drug'] == 'Stelasyn', 'Metastatic Sites']\nZoniferol_df = tumor_error.loc[tumor_response_error['Drug'] == 'Zoniferol', 'Metastatic Sites']\n\nTime = [0, 5, 10, 15, 20, 25, 30, 35, 40, 45]\n\nplt.errorbar(Time, metastatic_df['Capomulin'], yerr = Capomulin_df, color = '#4287f5', linestyle = 'solid', marker = '*', label='Capomulin')\nplt.errorbar(Time, metastatic_df[\"Ceftamin\"], yerr = Ceftamin_df, color = '#48f542', linestyle = '-.', marker = 'p', label=\"Ceftamin\")\nplt.errorbar(Time, metastatic_df[\"Infubinol\"], yerr = Infubinol_df, color = '#42f5d4', linestyle = '-', marker = 'h', label=\"Infubinol\")\nplt.errorbar(Time, metastatic_df[\"Ketapril\"], yerr = Ketapril_df, color = '#f542ec', linestyle = '-', marker = 's', label=\"Ketapril\")\nplt.errorbar(Time, metastatic_df[\"Naftisol\"], yerr = Naftisol_df, color = '#f56042', linestyle = 'solid', marker = 'X', label=\"Naftisol\")\nplt.errorbar(Time, metastatic_df[\"Placebo\"], yerr = Placebo_df, color = '#adefab', linestyle = '-.', marker = 'd', label=\"Placebo\")\nplt.errorbar(Time, metastatic_df[\"Propriva\"], yerr = Propriva_df, color = '#d0abef', linestyle = 'solid', marker = '>', label=\"Propriva\")\nplt.errorbar(Time, metastatic_df[\"Ramicane\"], yerr = Ramicane_df, color = '#efabec', linestyle = '-', marker = '<', label=\"Ramicane\")\nplt.errorbar(Time, metastatic_df[\"Stelasyn\"], yerr = Stelasyn_df, color = '#c2abef', linestyle = 'solid', marker = '.', label=\"Stelasyn\")\nplt.errorbar(Time, metastatic_df[\"Zoniferol\"], yerr = Zoniferol_df, color = '#f5425a', linestyle = '-', marker = '8', label=\"Zoniferol\")\n\nfor Drug in reformat_df:\n if Drug !='Metastatic Sites)':\n print(Drug)\n\nplt.xlabel('Treatment Duration (Days)')\nplt.ylabel ('Met Sites')\n\nplt.legend()\nplt.grid()\n\nplt.savefig('Metastatic Spread During Treatment', dpi=300)\nplt.show()\n\n\n\n",
"Capomulin\nCeftamin\nInfubinol\nKetapril\nNaftisol\nPlacebo\nPropriva\nRamicane\nStelasyn\nZoniferol\n"
]
],
[
[
"# Survival Rates",
"_____no_output_____"
],
[
"Store the Count of Mice Grouped by Drug and Timepoint (W can pass any metric)",
"_____no_output_____"
]
],
[
[
"df_groupby = df.groupby(['Drug', 'Timepoint'])\nmice_count = df_groupby['Mouse ID'].count()\n\nmice_df = pd.DataFrame(mice_count).reset_index()\n\nrename_mice_df = mice_df.rename(columns={'Mouse ID': 'Mouse Count'})\nrename_mice_df.head()",
"_____no_output_____"
]
],
[
[
"Minor Data Munging to Re-Format the Data Frames",
"_____no_output_____"
]
],
[
[
"mouse_df = mice_df.pivot(index = 'Timepoint', columns = 'Drug', values = 'Mouse ID')\nmouse_df.head()",
"_____no_output_____"
]
],
[
[
"Generate the Plot (Accounting for percentages)",
"_____no_output_____"
]
],
[
[
"plt.figure(figsize=(8,5))\n\nplt.title('Survival During Treatment', fontdict={'fontweight':'bold', 'fontsize':14})\n\nTime = [0, 5, 10, 15, 20, 25, 30, 35, 40, 45]\n\nplt.plot(Time, (mouse_df['Capomulin']/25)*100, 'b.-', marker = '*', label='Capomulin')\nplt.plot(Time, (mouse_df['Ceftamin']/25)*100, 'r.-', marker = 'p', label='Ceftamin')\nplt.plot(Time, (mouse_df['Infubinol']/25)*100, 'g.-', marker = 'h', label='Infubinol')\nplt.plot(Time, (mouse_df['Ketapril']/25)*100, 'y.-', marker = 's', label='Ketapril')\nplt.plot(Time, (mouse_df['Naftisol']/25)*100, 'm.-', marker = 'X', label='Naftisol')\nplt.plot(Time, (mouse_df['Placebo']/25)*100, 'c.-', marker = 'D', label='Placebo')\nplt.plot(Time, (mouse_df['Propriva']/25)*100, color = '#adefab', linestyle = '-', marker = '>', label='Propriva')\nplt.plot(Time, (mouse_df['Ramicane']/25)*100, color = '#d0abef', linestyle = '-', marker = '<', label='Ramicane')\nplt.plot(Time, (mouse_df['Stelasyn']/25)*100, color = '#f542ec', linestyle = '-', marker = '.', label='Stelasyn')\nplt.plot(Time, (mouse_df['Zoniferol']/25)*100, color = '#f5425a', linestyle = '-', marker = '8', label='Zoniferoln')\n\n\nplt.xlabel('Time (Days)')\nplt.ylabel ('Survival Rate (%)')\n\nplt.legend()\nplt.grid()\n\nplt.savefig('Survival During Treatment', dpi=300)\nplt.show()",
"_____no_output_____"
]
],
[
[
"# Summary Bar Graph\nCalculate the percent changes for each drug",
"_____no_output_____"
]
],
[
[
"tumor_volume = 45\npercent_changes = ((reformat_df.loc[45, :] - tumor_volume)/tumor_volume)*100\n\npercent_changes",
"_____no_output_____"
]
],
[
[
"Store all Relevant Percent Changes into a Tuple",
"_____no_output_____"
]
],
[
[
"tuple_changes = tuple(zip(percent_changes.index, percent_changes))\ntuple_changes_list = list(tuple_changes)\ntuple_changes_list",
"_____no_output_____"
]
],
[
[
"Splice the data between passing and failing drugs",
"_____no_output_____"
]
],
[
[
"passing_drugs =[]\nfailing_drugs =[]\nindx_pass_drugs =[]\nindx_fail_drugs =[]\n\nfor j,elements in tuple_changes_list:\n if elements>0:\n pass_drugs = elements\n passing_drugs.append(elements)\n indx_pass_drugs.append(j)\n else:\n fail_drugs = elements\n failing_drugs.append(elements)\n indx_fail_drugs.append(j)\n \npassingDrugs = list(zip(indx_pass_drugs, passing_drugs))\nfailingDrugs = list(zip(indx_fail_drugs, failing_drugs))\nprint(passingDrugs)\nprint(failingDrugs)",
"[('Ceftamin', 42.51649185589744), ('Infubinol', 46.12347172785187), ('Ketapril', 57.028794686606076), ('Naftisol', 53.92334713476923), ('Placebo', 51.29796048315153), ('Propriva', 47.24117486320637), ('Stelasyn', 52.08513428789896), ('Zoniferol', 46.57975086509525)]\n[('Capomulin', -19.475302667894173), ('Ramicane', -22.32090046276664)]\n"
]
],
[
[
"Orient widths. Add labels, tick marks, etc.",
"_____no_output_____"
]
],
[
[
"fig, fig_df = plt.subplots(figsize=(8, 5))\nfig_df.set_title('Tumor Change Over 45 Day Treatment', fontdict={'fontweight':'bold', 'fontsize':12})\n\ny = [percent_changes['Ceftamin'], percent_changes['Infubinol'], \n percent_changes['Ketapril'], percent_changes['Naftisol'], \n percent_changes['Placebo'], percent_changes['Propriva'], \n percent_changes['Stelasyn'], percent_changes['Zoniferol']\n ]\nx_axis = [0]\nx_axis1 = [1]\nx_axis2 =[2, 3, 4, 5, 6, 7, 8, 9]\n\n\n\nbars = fig_df.bar(x_axis, percent_changes['Capomulin'], color = 'g', alpha=0.8, align='edge', width = -1)\nbars1 = fig_df.bar(x_axis1, percent_changes['Ramicane'], color = 'g', alpha=0.8, align='edge', width = -1)\nbars2 = fig_df.bar(x_axis2, y, color='m', alpha=0.8, align='edge', width = -1) \n\nx_labels=[\"Capomulin\", \"Ramicane\", \"Ceftamin\", \"Infubinol\", \"Ketapril\", \"Naftisol\", \"Placebo\", \n \"Propriva\", \"Stelasyn\", \"Zoniferol\"]\n\nplt.setp(fig_df, xticks=[0, 1, 2, 3, 4, 5, 6, 7, 8, 9], xticklabels=[\"Capomulin\", \"Ramicane\", \"Ceftamin\", \"Infubinol\", \"Ketapril\", \"Naftisol\", \"Placebo\", \n \"Propriva\", \"Stelasyn\", \"Zoniferol\"], \n yticks=[-20, 0, 20, 40, 60])\n\nplt.xticks(rotation='vertical')\nplt.subplots_adjust(bottom=0.10)\n\nfig_df.set_ylabel('% Tumor Volume Change')\n\n\nfig_df.grid()\n\ndef autolabel(rects):\n for rect in rects:\n height = rect.get_height()\n fig_df.text(rect.get_x() + rect.get_width()/2, .1*height, \"%d\" %int(height)+ \"%\",\n horizontalalignment='center', verticalalignment='top', color=\"black\")\nautolabel(bars)\nautolabel(bars1)\nautolabel(bars2)\nfig.tight_layout()\n\nfig.savefig('Tumor Change Over 45 Day Treatment', dpi=300)\nfig.show()\n",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb903efeefe0c0841b4fea4ffd0f65f019d0f500
| 329,049 |
ipynb
|
Jupyter Notebook
|
sandbox/data_science/skill_lda.ipynb
|
shiftorg/skills
|
ac64f60bf215ac69373ea956870961fd77cc83a3
|
[
"MIT"
] | 2 |
2018-08-28T15:32:29.000Z
|
2019-02-13T18:52:05.000Z
|
sandbox/data_science/skill_lda.ipynb
|
shiftorg/skills
|
ac64f60bf215ac69373ea956870961fd77cc83a3
|
[
"MIT"
] | 11 |
2018-01-31T05:17:10.000Z
|
2018-04-12T06:26:57.000Z
|
sandbox/data_science/skill_lda.ipynb
|
shiftorg/skills
|
ac64f60bf215ac69373ea956870961fd77cc83a3
|
[
"MIT"
] | 3 |
2018-03-20T17:51:05.000Z
|
2022-02-16T23:30:37.000Z
| 209.185633 | 275,318 | 0.676443 |
[
[
[
"# Transform JD text files into an LDA model and pyLDAvis visualization\n\n### Steps:\n1. Use spaCy phrase matching to identify skills\n2. Parse the job descriptions. A full, readable job description gets turned into a bunch of newline-delimited skills.\n3. Create a Gensim corpus and dictionary from the parsed skills\n4. Train an LDA model using the corpus and dictionary\n5. Visualize the LDA model\n6. Compare user input to the LDA model; get out a list of relevant skills",
"_____no_output_____"
]
],
[
[
"# Modeling and visualization\nimport gensim\nfrom gensim.corpora import Dictionary, MmCorpus\nfrom gensim.models.ldamodel import LdaModel\nimport pyLDAvis\nimport pyLDAvis.gensim\n\n# Utilities\nimport codecs\nimport pickle\nimport os\nimport warnings\n\n# Black magic\nimport spacy \nfrom spacy.matcher import Matcher \nfrom spacy.attrs import *\nnlp = spacy.load('en') ",
"_____no_output_____"
]
],
[
[
"### 1. Use spaCy phrase matching to ID skills in job descriptions\n\n**First, we read in a pickled dictionary that contains the word patterns we'll use to extract skills from JDs. Here's what the first few patterns look like:**\n\n``` Python\n{\n 0 : [{\"lower\": \"after\"}, {\"lower\": \"effects\"}],\n 1 : [{\"lower\": \"amazon\"}, {\"lower\": \"web\"}, {\"lower\": \"services\"}],\n 2 : [{\"lower\": \"angular\"}, {\"lower\": \"js\"}],\n 3 : [{\"lower\": \"ansible\"}],\n 4 : [{\"lower\": \"bash\"}, {\"lower\": \"shell\"}],\n 5 : [{\"lower\": \"business\"}, {\"lower\": \"intelligence\"}]\n}\n```\n\n**We generated the pickled dictionary through some (rather heavy) preprocessing steps:**\n\n1. Train a word2vec model on all of the job descriptions. Cluster the word embeddings, identify clusters associated with hard skills, and annotate all of the words in those clusters. Save those words as a \"skill repository\" (a text document that we'll use as the canonical list of hard tech skills).\n2. Clean the skill repository. Inevitably, terms that are not hard skills made it into the word2vec \"skill\" clusters. Remove them. In this case, we defined a \"skill\" as \"a tool, platform, or language that would make sense as a skill to learn or improve.\"\n3. Use the skill repository to train an Named Entity Recognition model (in our case, using Prodigy). Use the training process to identify hard skills that we previously did not have in our repository. Add the new skills to the repository.\n4. Create a Python dictionary of the skills. Format the dictionary so that the values can be ingested as spaCy language patterns.\n\nSee spaCy's [matcher documentation](https://spacy.io/api/matcher#init) for more details.\n",
"_____no_output_____"
]
],
[
[
"# read pickled dict() object\nwith open('skill_dict.pkl', 'rb') as f:\n skill_dict = pickle.load(f)",
"_____no_output_____"
],
[
"%%time\n# Read JDs into memory\nimport os\ndirectory = os.fsencode('../local_data/')\n\njds = []\n\nfor file in os.listdir(directory):\n filename = os.fsdecode(file)\n path = '../local_data/' + filename\n with open(path, 'r') as infile:\n jds.append(infile.read())\n\nprint(len(jds), \"JDs\")\nimport sys\nprint(sys.getsizeof(jds)/1000000, \"Megabytes\")",
"_____no_output_____"
]
],
[
[
"### 2. Parse job descriptions\nFrom each JD, generate a list of skills.",
"_____no_output_____"
]
],
[
[
"%%time\n\n# Write skill-parsed JDs to file. \n# This took about three hours for 106k jobs.\n\nfor idx, jd in enumerate(jds):\n out_path = '../skill_parsed/'+ str(idx+1) + '.txt'\n with open(out_path, 'w') as outfile:\n # Creating a matcher object\n doc = nlp(jd) \n matcher = Matcher(nlp.vocab) \n for label, pattern in skill_dict.items():\n matcher.add(label, None, pattern)\n matches = matcher(doc)\n for match in matches:\n # match object returns a tuple with (id, startpos, endpos)\n output = str(doc[match[1]:match[2]]).replace(' ', '_').lower()\n outfile.write(output)\n outfile.write('\\n')",
"_____no_output_____"
]
],
[
[
"### 3. Generate a Gensim corpus and dictionary from the parsed skill documents",
"_____no_output_____"
]
],
[
[
"%%time\n# Load parsed items back into memory\ndirectory = os.fsencode('skill_parsed//')\n\nparsed_jds = []\n\nfor file in os.listdir(directory):\n filename = os.fsdecode(file)\n path = 'skill_parsed/' + filename\n # Ran into an encoding issue; changing to latin-1 fixed it\n with codecs.open(path, 'r', encoding='latin-1') as infile:\n parsed_jds.append(infile.read())",
"CPU times: user 6.09 s, sys: 8.46 s, total: 14.5 s\nWall time: 42.3 s\n"
],
[
"%%time\n'''\nGensim needs documents to be formatted as a list-of-lists, where the inner\nlists are simply lists including the tokens (skills) from a given document.\nIt's important to note that any bigram or trigram skills are already tokenized\nwith underscores instead of spaces to preserve them as tokens.\n'''\nnested_dict_corpus = [text.split() for text in parsed_jds]\nprint(nested_dict_corpus[222:226])",
"[['artificial_intelligence', 'newly', 'artificial_intelligence', 'ai', 'computer_science'], ['excel', 'word'], ['sql', 'word', 'excel', 'power_point', 'statistics', 'computer_science'], ['aws', 'computer_science', 'java', 'amazon_web_services', 'web_services', 'aws', 'azure', 'unix', 'linux', 'agile']]\nCPU times: user 264 ms, sys: 286 ms, total: 550 ms\nWall time: 728 ms\n"
],
[
"from gensim.corpora import Dictionary, MmCorpus\n\ngensim_skills_dict = Dictionary(nested_dict_corpus)\n\n# save the dict\ngensim_skills_dict.save('gensim_skills.dict')",
"_____no_output_____"
],
[
"corpus = [gensim_skills_dict.doc2bow(text) for text in nested_dict_corpus]",
"_____no_output_____"
],
[
"# Save the corpus\ngensim.corpora.MmCorpus.serialize('skill_bow_corpus.mm', corpus, id2word=gensim_skills_dict)",
"_____no_output_____"
],
[
"# Load up the dictionary\ngensim_skills_dict = Dictionary.load('gensim_skills.dict')\n\n# Load the corpus\nbow_corpus = MmCorpus('skill_bow_corpus.mm')",
"_____no_output_____"
]
],
[
[
"### 4. Create the LDA model using Gensim",
"_____no_output_____"
]
],
[
[
"%%time\n\nwith warnings.catch_warnings():\n warnings.simplefilter('ignore')\n\n lda_alpha_auto = LdaModel(bow_corpus, \n id2word=gensim_skills_dict, \n num_topics=20)\n \n lda_alpha_auto.save('lda/skills_lda')",
"CPU times: user 19.4 s, sys: 366 ms, total: 19.8 s\nWall time: 20 s\n"
],
[
"# load the finished LDA model from disk\nlda = LdaModel.load('lda/skills_lda')",
"_____no_output_____"
]
],
[
[
"### 5. Visualize using pyLDAvis",
"_____no_output_____"
]
],
[
[
"LDAvis_data_filepath = 'lda/ldavis/ldavis'",
"_____no_output_____"
],
[
"%%time\n\nLDAvis_prepared = pyLDAvis.gensim.prepare(lda, bow_corpus,\n gensim_skills_dict)\n\nwith open(LDAvis_data_filepath, 'wb') as f:\n pickle.dump(LDAvis_prepared, f)",
"/usr/local/lib/python3.6/site-packages/pyLDAvis/_prepare.py:387: DeprecationWarning: \n.ix is deprecated. Please use\n.loc for label based indexing or\n.iloc for positional indexing\n\nSee the documentation here:\nhttp://pandas.pydata.org/pandas-docs/stable/indexing.html#ix-indexer-is-deprecated\n topic_term_dists = topic_term_dists.ix[topic_order]\n"
],
[
"# load the pre-prepared pyLDAvis data from disk\nwith open(LDAvis_data_filepath, 'rb') as f:\n LDAvis_prepared = pickle.load(f)",
"_____no_output_____"
],
[
"pyLDAvis.display(LDAvis_prepared)",
"_____no_output_____"
],
[
"# Save the file as HTML\npyLDAvis.save_html(LDAvis_prepared, 'lda/html/lda.html')",
"_____no_output_____"
]
],
[
[
"### 6. Compare user input to the LDA model\nOutput the skills a user has and does not have from various topics.",
"_____no_output_____"
]
],
[
[
"# Look at the topics\ndef explore_topic(topic_number, topn=20):\n \"\"\"\n accept a topic number and print out a \n formatted list of the top terms\n \"\"\"\n print(u'{:20} {}'.format(u'term', u'frequency') + u'')\n\n for term, frequency in lda.show_topic(topic_number, topn=40):\n print(u'{:20} {:.3f}'.format(term, round(frequency, 3)))\n \nfor i in range(20): # Same number as the types of jobs we scraped initially\n print(\"\\n\\nTopic %s\" % i)\n explore_topic(topic_number=i)",
"\n\nTopic 0\nterm frequency\naws 0.193\nbig_data 0.061\nweb_services 0.043\njava 0.041\nkafka 0.036\namazon_web_services 0.032\nnosql 0.031\npython 0.030\napache 0.028\ncassandra 0.027\ncomputer_science 0.026\nec2 0.024\ns3 0.024\nelasticsearch 0.017\nscala 0.017\nlinux 0.017\nmysql 0.016\nmongodb 0.014\nelastic 0.014\nrds 0.013\nhive 0.012\nlambda 0.011\npostgres 0.011\nmapreduce 0.011\nredis 0.010\netl 0.009\ndynamodb 0.009\ndata_pipeline 0.009\nagile 0.008\ntuning 0.006\npig 0.006\nnginx 0.006\nubuntu 0.006\nzookeeper 0.005\nruby 0.005\npostgresql 0.005\nelastic_search 0.005\nsqs 0.005\nelb 0.005\nprivate_cloud 0.005\n\n\nTopic 1\nterm frequency\nc 0.215\n.net 0.135\nsql 0.107\nasp.net 0.058\ncomputer_science 0.046\nsql_server 0.045\nmvc 0.035\nweb_services 0.019\ntfs 0.018\njavascript 0.018\nc++ 0.018\napi 0.016\ncustom 0.015\nunit_testing 0.014\nwcf 0.013\nwpf 0.013\niis 0.012\nasp 0.011\nobject_oriented 0.010\nagile 0.009\nvb 0.009\ntestng 0.008\njquery 0.007\ninformation_systems 0.007\norm 0.006\nsmb 0.006\nwireshark 0.006\ndbs 0.006\nuft 0.005\nprogramming_languages 0.005\nangular 0.005\nrestful 0.005\nrelational_databases 0.005\nsoa 0.004\nneuroscience 0.004\nhtml 0.004\nwinforms 0.004\nversion_control 0.004\nrelational_database 0.004\noo 0.004\n\n\nTopic 2\nterm frequency\nruby 0.087\npython 0.076\nmysql 0.051\nrails 0.044\njava 0.042\nnosql 0.040\nmongodb 0.039\njavascript 0.039\npostgresql 0.038\nscala 0.029\ncomputer_science 0.026\ndjango 0.026\nsql 0.024\nnode.js 0.024\nrelational_databases 0.023\nredis 0.021\napi 0.020\ncustomize 0.018\nphp 0.016\nd3 0.015\nneo4j 0.014\nrestful 0.013\nreact_native 0.013\nblockchain 0.013\ncassandra 0.012\nprogramming_languages 0.011\nreact.js 0.011\nelasticsearch 0.011\ncaching 0.010\nes6 0.010\nlooker 0.009\nsingle_page 0.009\nwms 0.008\ncustom 0.007\nrelational_database 0.007\nd3.js 0.007\nstats 0.007\nprogramming_language 0.006\nember 0.005\nrabbitmq 0.005\n\n\nTopic 3\nterm frequency\nlinux 0.190\nunix 0.091\njava 0.072\npython 0.064\ncomputer_science 0.061\nc++ 0.048\nperl 0.045\nshell 0.043\nc 0.032\noperating_system 0.030\nj2ee 0.028\nbash 0.019\nscripting_language 0.017\napache 0.014\nsql 0.013\nmysql 0.011\ncustom 0.011\njboss 0.010\ninformation_systems 0.009\nphp 0.009\nscm 0.009\nwebsphere 0.008\nversion_control 0.008\ntuning 0.008\ngit 0.008\nbaseline 0.007\nruby 0.007\ncustomization 0.007\ncommand_line 0.007\nweblogic 0.007\neclipse 0.007\nredhat 0.005\necs 0.005\ndbms 0.005\nsubversion 0.004\njdbc 0.004\njms 0.004\nlucene 0.004\nvbscript 0.004\nesb 0.004\n\n\nTopic 4\nterm frequency\nsql 0.383\nsql_server 0.137\nazure 0.097\ntuning 0.071\nvmware 0.041\nrelational_database 0.022\ndb2 0.022\ncouchbase 0.020\ncomputer_science 0.020\nmysql 0.017\npowershell 0.016\ninformation_systems 0.011\nrdbms 0.011\nrelational_databases 0.010\nmainframe 0.009\nolap 0.008\niis 0.006\nnetezza 0.005\npostgresql 0.005\nvsphere 0.004\nnosql 0.004\naurora 0.004\netl 0.004\ndatabase_servers 0.004\npentaho 0.004\nsitecore 0.004\nbackbone.js 0.004\ntestrail 0.003\nmemcache 0.003\nshell 0.003\npolymer 0.003\nairwatch 0.003\ncobol 0.003\ncustom 0.003\nvcenter 0.003\npostgres 0.002\nloadrunner 0.002\njcl 0.002\noperating_system 0.002\nprogramming_languages 0.002\n\n\nTopic 5\nterm frequency\nagile 0.487\nscrum 0.211\ncustom 0.060\njira 0.059\ncomputer_science 0.047\nproduct_owner 0.036\nconfluence 0.022\ncsm 0.016\natlassian 0.014\nrally 0.011\ncontinuous_integration 0.007\ncertified_scrum_master 0.004\ntrello 0.003\nversionone 0.003\nprism 0.002\nworkbench 0.002\nacp 0.002\nrazor 0.002\nglue 0.001\naurelia 0.001\ninformation_systems 0.001\nbitbucket 0.001\nvm 0.001\netcd 0.001\ngremlin 0.001\nsparql 0.001\ncertified_scrummaster 0.000\nproject_management 0.000\natdd 0.000\nregression 0.000\nbaseline 0.000\nvisio 0.000\ndotnetnuke 0.000\ndrupal 0.000\npivotal 0.000\nsharepoint 0.000\nproduct_management 0.000\nborland 0.000\njava 0.000\nc 0.000\n\n\nTopic 6\nterm frequency\nproject_management 0.882\ncitrix 0.022\nsugarcrm 0.012\ncomputer_science 0.012\ninformation_systems 0.011\nbaseline 0.010\nr2 0.006\nsharepoint 0.005\nvisio 0.004\noltp 0.004\ntrigger 0.003\ndatabase_schema 0.003\ne2e 0.003\nespresso 0.003\nr12 0.002\ndatastage 0.002\nsccm 0.002\ncustomization 0.002\nmailchimp 0.002\nrxjava 0.001\nxunit 0.001\nvisualisation 0.001\nelm 0.001\niseries 0.001\nopenldap 0.001\nredgate 0.000\npersistence_layer 0.000\nopenwrt 0.000\nsql 0.000\ndbms 0.000\noperating_system 0.000\ncustom 0.000\nfoxpro 0.000\nproper 0.000\nsql_server 0.000\ncustomized 0.000\nexcel 0.000\nagile 0.000\nvm 0.000\ncustomize 0.000\n\n\nTopic 7\nterm frequency\nproduct_management 0.498\nproper 0.157\ninformation_systems 0.124\ncomputer_science 0.080\nmetric 0.022\ngtm 0.020\nbusiness_knowledge 0.013\nnetapp 0.011\nchecklist 0.010\nspa 0.009\nvetting 0.008\nblueprint 0.006\npearl 0.006\ncanvas 0.005\nhaskell 0.004\nstandard_operating_procedure 0.004\nrpc 0.004\nfilemaker 0.004\nesx 0.003\nucs 0.003\ndatamart 0.002\nocr 0.002\nfreebsd 0.002\nntp 0.001\nnetty 0.001\nbig_data 0.001\nagile 0.000\nproduct_owner 0.000\nbde 0.000\nbdb 0.000\nweb_services 0.000\nsgi 0.000\nvirtual_machine 0.000\npivotal 0.000\ntuning 0.000\nmssql 0.000\nlinux 0.000\nrelational_database 0.000\nunix 0.000\nc 0.000\n\n\nTopic 8\nterm frequency\nexcel 0.393\ncrm 0.157\nmicrosoft_excel 0.090\nword 0.075\ngaap 0.074\nms_excel 0.055\nspreadsheet 0.038\npower_point 0.032\nproper 0.031\ncharles 0.030\nscp 0.008\npowerpoint 0.005\natf 0.002\ncontrol_group 0.002\nwebpage 0.002\nvisio 0.001\ntcpip 0.001\nbagging 0.001\nwix 0.001\ncmdb 0.001\nchecklist 0.000\nfreescale 0.000\nzoho 0.000\ngoogle_doc 0.000\nproject_management 0.000\ninformation_systems 0.000\ncustom 0.000\nworkflow 0.000\nblockchain 0.000\nstatistics 0.000\nr 0.000\ngoogle_sheet 0.000\npivotal 0.000\nc 0.000\ncustomization 0.000\nprogram_management 0.000\nnewly 0.000\nsql 0.000\nautocad 0.000\nproduct_management 0.000\n\n\nTopic 9\nterm frequency\nprogram_management 0.436\nworkflow 0.272\nnewly 0.078\npanda 0.031\nalm 0.027\nssa 0.022\nlamp 0.020\nangular_js 0.019\nsybase 0.018\nkarma 0.015\ntemplate 0.013\nomniture 0.010\ncomputer_science 0.006\nafter_effects 0.006\nplsql 0.006\nzend 0.005\nsh 0.005\nuser_experience_research 0.005\njama 0.002\nshard 0.001\nsmarty 0.001\nproject_management 0.000\ninformation_systems 0.000\nbdb 0.000\njasmine 0.000\nhaxe 0.000\ncustom 0.000\nmetric 0.000\nramda.js 0.000\nlodash.js 0.000\ntimeline.js 0.000\njs 0.000\noscommerce 0.000\nagile 0.000\npivotal 0.000\nsql 0.000\nc 0.000\nphp 0.000\nweb_services 0.000\njavascript 0.000\n\n\nTopic 10\nterm frequency\nexcel 0.281\nword 0.271\npowerpoint 0.214\nsharepoint 0.071\nvisio 0.045\ncustomized 0.044\nautocad 0.023\nwebdriver 0.013\njmeter 0.008\nsoapui 0.007\nbaseline 0.004\ngreenplum 0.003\nmainframe 0.003\nember.js 0.002\neis 0.002\nstata 0.002\nfoss 0.002\nudeploy 0.001\nnsx 0.001\ntestcomplete 0.001\nproject_management 0.000\nmessaging_protocol 0.000\nsphinx 0.000\ncustom 0.000\ncustomize 0.000\nbdb 0.000\nwireframing 0.000\nspss 0.000\nmetric 0.000\ncomputer_science 0.000\nsql 0.000\nxna 0.000\nms_excel 0.000\nvetting 0.000\nstatistics 0.000\ntkinter 0.000\nvb 0.000\nrpc 0.000\npython 0.000\nlinux 0.000\n\n\nTopic 11\nterm frequency\ndocker 0.154\naws 0.061\njenkins 0.056\nci 0.051\npython 0.049\npuppet 0.048\nchef 0.048\nansible 0.045\nlinux 0.041\ncontinuous_integration 0.038\nkubernetes 0.037\nazure 0.024\ngit 0.023\nruby 0.021\nbash 0.019\nagile 0.013\nshell 0.010\nmaven 0.010\nopenstack 0.010\ngithub 0.009\nvmware 0.008\nnagios 0.008\nnexus 0.008\npowershell 0.008\ngcp 0.008\njava 0.008\nxamarin 0.008\ncomputer_science 0.007\nvb.net 0.006\nflask 0.006\nperl 0.006\nmesos 0.006\ngolang 0.006\ngitlab 0.005\nsvn 0.005\nopenshift 0.004\nteamcity 0.004\ngradle 0.003\ncontainerized 0.003\ngroovy 0.003\n\n\nTopic 12\nterm frequency\njavascript 0.198\ncss 0.117\nhtml 0.115\nangular 0.048\njquery 0.047\nhtml5 0.043\nangularjs 0.033\njs 0.031\ncomputer_science 0.028\najax 0.023\nphp 0.022\nnode.js 0.019\ncss3 0.018\nbootstrap 0.018\nagile 0.018\nmvc 0.016\njava 0.015\nreactjs 0.013\ntypescript 0.013\nweb_services 0.012\nsass 0.010\nredux 0.009\nmysql 0.009\nsmoke 0.008\nnodejs 0.007\nsql 0.007\ncustom 0.006\nnpm 0.006\nwireframing 0.006\nobject_oriented 0.006\nbalsamiq 0.005\nnode 0.005\nangular.js 0.005\ngit 0.004\nunit_testing 0.004\nclearcase 0.004\ndrupal 0.004\nmocha 0.004\ncreative_cloud 0.003\noop 0.003\n\n\nTopic 13\nterm frequency\nvisualization 0.139\nsql 0.101\nexcel 0.068\ndashboard 0.068\nssis 0.059\nssrs 0.059\nhyperion 0.053\nvba 0.037\nspss 0.031\njmp 0.028\nsop 0.025\ntableau 0.025\nmacros 0.024\ncomputer_science 0.020\ncustom 0.019\nqlik 0.019\narcgis 0.017\nobiee 0.017\nvisual_basic 0.015\nwireframe 0.014\ninformation_systems 0.013\nstatistics 0.013\nstructured_query_language 0.012\npowerbi 0.012\nsql_server 0.011\nrelational_database 0.009\npython 0.008\nadhoc 0.008\nwebgl 0.008\nnatural_language_understanding 0.006\nbpo 0.006\nlua 0.006\nsqlite 0.006\nmathematica 0.005\nminitab 0.005\nelectron 0.005\nbigtable 0.004\nrelational_databases 0.004\nms_excel 0.004\nvideo_editing 0.003\n\n\nTopic 14\nterm frequency\nsql 0.109\nbi 0.108\nbusiness_intelligence 0.092\ntableau 0.079\netl 0.073\nsas 0.070\nstatistics 0.070\nr 0.063\ndata_warehouse 0.056\ncomputer_science 0.025\ninformatica 0.025\npython 0.022\nvisualization 0.021\nbig_data 0.020\nteradata 0.017\ncognos 0.014\nmicrostrategy 0.012\npower_bi 0.012\ninformation_systems 0.010\nqlikview 0.010\nrelational_databases 0.009\nhive 0.007\ntalend 0.007\nregression 0.006\nhana 0.006\nspotfire 0.005\nprogramming_languages 0.004\ncustom 0.004\npredictive_modeling 0.004\ndomo 0.003\ndata_science 0.003\nerwin 0.003\npaxata 0.002\nexcel 0.002\nsap_hana 0.002\nrdbms 0.002\nmssql 0.002\ntoad 0.002\nprogramming_language 0.002\nddl 0.001\n\n\nTopic 15\nterm frequency\ngit 0.104\napi 0.104\nversion_control 0.043\nagile 0.039\njenkins 0.038\njira 0.036\ncontinuous_integration 0.036\ngithub 0.033\nrestful 0.032\nnode 0.028\njavascript 0.027\nsvn 0.027\npivotal 0.021\nangular 0.019\nsubversion 0.019\nci 0.016\nmaven 0.016\ngradle 0.016\njava 0.015\njs 0.015\nnodejs 0.015\nconfluence 0.015\ngulp 0.012\nbitbucket 0.012\nunit_testing 0.012\nwebpack 0.011\ngrunt 0.011\nvue 0.011\ncomputer_science 0.010\natlassian 0.009\njasmine 0.009\nbranching 0.008\ncloud_foundry 0.008\ncordova 0.007\nstash 0.007\nember 0.007\noauth 0.006\nclojure 0.006\nintellij 0.005\npython 0.005\n\n\nTopic 16\nterm frequency\nmath 0.242\ncomputer_science 0.184\ngoogle_cloud 0.171\niaas 0.092\npaas 0.056\npublic_cloud 0.048\nlabview 0.025\nmacro 0.024\nmfc 0.014\ntuning 0.012\nscada 0.012\noracle_11 0.011\nsdl 0.010\noracle_12c 0.010\nrman 0.008\ndatabase_server 0.008\nsnowflake 0.007\nvdi 0.007\ntile 0.007\nc++ 0.006\ntoad 0.005\nprolog 0.005\ncolumnar 0.005\nsharding 0.004\nocp 0.004\npython 0.003\nwin32 0.003\ncloud_compute 0.002\npascal 0.002\nmodel_view_controller 0.002\nbig_data 0.002\nextract_transform_load 0.002\ndataguard 0.001\nc 0.001\nstatistics 0.001\npdb 0.001\nlinux 0.000\njava 0.000\nlvm 0.000\nconcurrency 0.000\n\n\nTopic 17\nterm frequency\njava 0.181\ncomputer_science 0.100\nc 0.085\nc++ 0.078\nagile 0.049\nweb_services 0.038\nselenium 0.035\npython 0.031\nobject_oriented 0.029\nprogramming_languages 0.024\nregression 0.024\nsql 0.020\nswift 0.018\njavascript 0.016\napi 0.015\nprogramming_language 0.015\njunit 0.013\nunit_testing 0.013\ncontinuous_integration 0.012\nrestful 0.012\nsoa 0.010\ncucumber 0.009\noo 0.008\nlinux 0.007\njenkins 0.007\neclipse 0.007\noop 0.006\nobjective_c 0.006\nnosql 0.006\ncode_review 0.005\nbdd 0.005\nxcode 0.005\nmaven 0.005\nconcurrency 0.004\nruby 0.004\ngit 0.004\nrelational_databases 0.004\nqtp 0.004\nrevision_control 0.003\nood 0.003\n\n\nTopic 18\nterm frequency\nmachine_learning 0.173\ndata_science 0.091\npython 0.081\nbig_data 0.075\ncomputer_science 0.059\nai 0.045\nstatistics 0.043\nr 0.036\nartificial_intelligence 0.030\ndeep_learning 0.026\njava 0.025\nc++ 0.022\nml 0.021\nsql 0.018\nvisualization 0.017\ncomputer_vision 0.016\nscala 0.016\nc 0.014\nregression 0.014\nnatural_language_processing 0.014\nhive 0.013\nnlp 0.012\nprogramming_languages 0.012\npredictive_modeling 0.009\npig 0.008\nnosql 0.008\nmapreduce 0.007\nnumpy 0.005\nlinux 0.005\nbayesian 0.005\nprogramming_language 0.004\nhpc 0.004\nanomaly_detection 0.004\ntoolchain 0.003\ncustom 0.003\nstata 0.003\napache 0.003\nhypothesis_testing 0.003\ncmake 0.003\ncrucible 0.002\n\n\nTopic 19\nterm frequency\nsketch 0.132\nphotoshop 0.132\nillustrator 0.110\ninvision 0.092\naxure 0.067\ndetailed_description 0.065\nindesign 0.047\ncreative_suite 0.042\ncom 0.039\nwordpress 0.039\nomnigraffle 0.036\nagile 0.026\nsvm 0.021\nvms 0.019\nhtml 0.015\nelixir 0.015\nxd 0.013\nuxpin 0.010\ndatameer 0.009\ndml 0.009\nmsbuild 0.008\njava8 0.007\ndreamweaver 0.007\nsupervised_learning 0.007\nflinto 0.005\nffmpeg 0.005\nrdf 0.004\nvb6 0.003\nrdb 0.002\nproto.io 0.002\ningres 0.002\nfsm 0.002\nopenroad 0.001\ncss 0.001\nopenvms 0.001\ncomputer_science 0.000\nweb_services 0.000\namazon_web_services 0.000\ncss3 0.000\nbootstrap 0.000\n"
],
[
"# A stab at naming the topics\ntopic_names = {1: u'Data Engineering (Big Data Focus)',\n 2: u'Microsoft OOP Engineering (C, C++, .NET)',\n 3: u'Web Application Development (Ruby, Rails, JS, Databases)',\n 4: u'Linux/Unix, Software Engineering, and Scripting',\n 5: u'Database Administration',\n 6: u'Project Management (Agile Focus)',\n 7: u'Project Management (General Software)',\n 8: u'Product Management',\n 9: u'General Management & Productivity (Microsoft Office Focus)',\n 10: u'Software Program Management',\n 11: u'Project and Program Management',\n 12: u'DevOps and Cloud Computing/Infrastructure',\n 13: u'Frontend Software Engineering and Design',\n 14: u'Business Intelligence',\n 15: u'Analytics',\n 16: u'Quality Engineering, Version Control, & Build',\n 17: u'Big Data Analytics; Hardware & Scientific Computing',\n 18: u'Software Engineering',\n 19: u'Data Science, Machine Learning, and AI',\n 20: u'Design'}",
"_____no_output_____"
]
],
[
[
"#### Ingest user input & transform into list of skills",
"_____no_output_____"
]
],
[
[
"matcher = Matcher(nlp.vocab) \nuser_input = '''\nMy skills are Postgresql, and Python.\n\nExperience with Chef Puppet and Docker required.\n\nI also happen to know Blastoise and Charzard. Also NeuRal neTwOrk.\n\nI use Git, Github, svn, Subversion, but not git, github or subversion.\n\nAdditionally, I can program using Perl, Java, and Haskell. But not perl, java, or haskell.'''\n\n# Construct matcher object\ndoc = nlp(user_input) \nfor label, pattern in skill_dict.items():\n matcher.add(label, None, pattern)\n\n# Compare input to pre-defined skill patterns\nuser_skills = []\nmatches = matcher(doc) \nfor match in matches:\n if match is not None:\n # match object returns a tuple with (id, startpos, endpos)\n output = str(doc[match[1]:match[2]]).lower()\n user_skills.append(output)\n\nprint(\"*** User skills: *** \")\nfor skill in user_skills:\n print(skill)",
"*** User skills: *** \npostgresql\npython\nchef\npuppet\ndocker\nneural network\ngit\ngithub\nsvn\nsubversion\ngit\ngithub\nsubversion\nperl\njava\nhaskell\nperl\njava\nhaskell\n"
]
],
[
[
"#### Compare user skills to the LDA model",
"_____no_output_____"
]
],
[
[
"def top_match_items(input_doc, lda_model, input_dictionary, num_terms=20):\n \"\"\"\n (1) parse input doc with spaCy, apply text pre-proccessing steps,\n (3) create a bag-of-words representation (4) create an LDA representation\n \"\"\"\n doc_bow = gensim_skills_dict.doc2bow(input_doc)\n\n # create an LDA representation\n document_lda = lda_model[doc_bow]\n \n # Sort in descending order\n sorted_doc_lda = sorted(document_lda, key=lambda review_lda: -review_lda[1])\n \n topic_number, freq = sorted_doc_lda[0][0], sorted_doc_lda[0][1]\n highest_probability_topic = topic_names[topic_number+1]\n \n top_topic_skills = []\n for term, term_freq in lda.show_topic(topic_number, topn=num_terms):\n top_topic_skills.append(term)\n return highest_probability_topic, round(freq, 3), top_topic_skills\n\nmatched_topic, matched_freq, top_topic_skills = top_match_items(user_skills, lda, gensim_skills_dict)",
"_____no_output_____"
],
[
"def common_skills(top_topic_skills, user_skills):\n return [item for item in top_topic_skills if item in user_skills]\n\ndef non_common_skills(top_topic_skills, user_skills):\n return [item for item in top_topic_skills if item not in user_skills]",
"_____no_output_____"
],
[
"print(\"**** User's matched topic and percent match:\")\nprint(matched_topic, matched_freq)\nprint(\"\\n**** Skills user has in common with topic:\")\nfor skill in common_skills(top_topic_skills, user_skills):\n print(skill)\nprint(\"\\n**** Skills user does NOT have in common with topic:\")\nfor skill in non_common_skills(top_topic_skills, user_skills):\n print(skill)",
"**** User's matched topic and percent match:\nQuality Engineering, Version Control, & Build 0.35\n\n**** Skills user has in common with topic:\ngit\ngithub\nsvn\nsubversion\njava\n\n**** Skills user does NOT have in common with topic:\napi\nversion_control\nagile\njenkins\njira\ncontinuous_integration\nrestful\nnode\njavascript\npivotal\nangular\nci\nmaven\ngradle\njs\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
cb9057913141d7e374563d12f112d2a036258492
| 18,121 |
ipynb
|
Jupyter Notebook
|
Demo.ipynb
|
johnsoong216/Texas-Holdem-Probability-Calculator
|
929471621aa545cbac67a21e26a43469a46afa4a
|
[
"MIT"
] | 3 |
2019-02-21T23:55:23.000Z
|
2019-12-01T19:50:33.000Z
|
Demo.ipynb
|
johnsoong216/PokerOddsCalc
|
929471621aa545cbac67a21e26a43469a46afa4a
|
[
"MIT"
] | null | null | null |
Demo.ipynb
|
johnsoong216/PokerOddsCalc
|
929471621aa545cbac67a21e26a43469a46afa4a
|
[
"MIT"
] | 1 |
2021-09-02T07:45:51.000Z
|
2021-09-02T07:45:51.000Z
| 22.622971 | 155 | 0.458694 |
[
[
[
"from table import HoldemTable, OmahaTable",
"_____no_output_____"
]
],
[
[
"# PokerOddsCalc\n\n---\n\n**PokerOddsCalc** is a simple poker hand probability calculation tool that supports Texas Holdem and Omaha.\n\n\n### Initialization\n\n--- \n\nInitialize with number of players and deck type(full or short deck(6 and above))",
"_____no_output_____"
]
],
[
[
"holdem_game = HoldemTable(num_players=5, deck_type='full')",
"_____no_output_____"
]
],
[
[
"### Deal Player Cards\n\n--- \n\n- Can Either Manually Add or Random Hand out Cards\n- Check the Current Table",
"_____no_output_____"
]
],
[
[
"holdem_game.add_to_hand(1, ['Td', 'Ts'])",
"_____no_output_____"
],
[
"holdem_game.next_round() # Will hand out remaining players starting hand",
"INFO:root:Giving Player 2 4h 8s\nINFO:root:Giving Player 3 Tc 7c\nINFO:root:Giving Player 4 6s 3c\nINFO:root:Giving Player 5 2d 5c\n"
],
[
"holdem_game.view_table()",
"_____no_output_____"
]
],
[
[
"### Simulation\n\n--- \n\n- Set Number of Scenarios, default is 150000 for Holdem and 25000 for Omaha\n- Return Final Hand Probability\n- Return Three Types of Odds Calculation (Default is tie_win)\n - win_any, any win counts as a win scenario for a player\n - tie_win, any exclusive win counts as win, any tied win or tie counts as a tie\n - precise, every possible outcome",
"_____no_output_____"
]
],
[
[
"holdem_game.simulate()",
"INFO:root:15750000 Simulations in 2.77s\n"
],
[
"holdem_game.simulate(num_scenarios=500000, odds_type='precise')",
"INFO:root:52500000 Simulations in 10.32s\n"
],
[
"holdem_game.simulate(odds_type='win_any')",
"INFO:root:15750000 Simulations in 2.69s\n"
],
[
"win_draw_loss, final_hand = holdem_game.simulate(final_hand=True)\nfinal_hand",
"INFO:root:15750000 Simulations in 2.97s\n"
]
],
[
[
"### Next Round\n\n--- \n\n- Run Next Round and Update Odds\n- Check Game Result\n- Check Current Best Hand for each player",
"_____no_output_____"
]
],
[
[
"holdem_game.next_round()",
"INFO:root:Flop card: 8c 4s Qh\n"
],
[
"holdem_game.view_result()",
"_____no_output_____"
],
[
"holdem_game.view_hand()",
"_____no_output_____"
],
[
"holdem_game.simulate()",
"INFO:root:77805 Simulations in 0.21s\n"
],
[
"holdem_game.next_round()",
"INFO:root:Turn card: 9s\n"
],
[
"holdem_game.view_result()",
"_____no_output_____"
],
[
"holdem_game.simulate(final_hand=True)",
"INFO:root:3990 Simulations in 0.11s\n"
],
[
"holdem_game.view_hand()",
"_____no_output_____"
],
[
"holdem_game.next_round()",
"_____no_output_____"
],
[
"holdem_game.view_result()",
"_____no_output_____"
]
],
[
[
"## Omaha\n---\n\nFunctions are exactly identical in Omaha. However calculations are slower in Omaha because 60 different combinations are possible with one simulation",
"_____no_output_____"
]
],
[
[
"omaha_game = OmahaTable(num_players=3, deck_type='short')",
"_____no_output_____"
],
[
"omaha_game.next_round()\nomaha_game.view_table()",
"INFO:root:Giving Player 1 7d Qs 8d Qd\nINFO:root:Giving Player 2 7h 9c Ac Ah\nINFO:root:Giving Player 3 Ad 8c Js Qc\n"
],
[
"win_tie_loss, final_hand = omaha_game.simulate(final_hand=True)",
"INFO:root:4500000 Simulations in 3.07s\n"
],
[
"win_tie_loss",
"_____no_output_____"
],
[
"final_hand",
"_____no_output_____"
],
[
"omaha_game.next_round()\nomaha_game.simulate()",
"INFO:root:Flop card: 7s Ts As\nINFO:root:37800 Simulations in 0.21s\n"
],
[
"omaha_game.view_result()",
"_____no_output_____"
],
[
"omaha_game.view_hand()",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb905decc53a9cb0c292832f9ca2a1b71d1319d0
| 36,578 |
ipynb
|
Jupyter Notebook
|
[Lab] Day1-00 Unconstraint opt. with NN modules.ipynb
|
Junyoungpark/2021-lg-AI-camp
|
3c0e5dd689e8e3dd61cc80243ad90cab951c06de
|
[
"MIT"
] | 4 |
2021-11-14T14:25:02.000Z
|
2021-11-23T06:23:51.000Z
|
[Lab] Day1-00 Unconstraint opt. with NN modules.ipynb
|
Junyoungpark/2021-lg-AI-camp
|
3c0e5dd689e8e3dd61cc80243ad90cab951c06de
|
[
"MIT"
] | null | null | null |
[Lab] Day1-00 Unconstraint opt. with NN modules.ipynb
|
Junyoungpark/2021-lg-AI-camp
|
3c0e5dd689e8e3dd61cc80243ad90cab951c06de
|
[
"MIT"
] | 2 |
2021-11-15T02:11:21.000Z
|
2021-11-15T23:57:47.000Z
| 141.775194 | 28,960 | 0.882826 |
[
[
[
"# Unconstrainted optimization with NN models\n\nIn this tutorial we will go over type 1 optimization problem which entails nn.Module rerpesented cost function and __no constarint__ at all. This type of problem is often written as follows:\n\n$$ \\min_{x} f_{\\theta}(x) $$\nwe can find Type1 problems quite easily. For instance assuming you are the manager of some manufactoring facilities, then your primary objective would be to maximize the yield of the manufactoring process. In industrial grade of manufactoring process the model of process is often __unknown__. hence we may need to learn the model through your favorite differentiable models such as neural networks and perform the graident based optimization to find the (local) optimums that minimize (or maximize) the yield.\n\n### General problem solving tricks; Cast your problem into QP, approximately.\n\nAs far as I know, Convex optimization is the most general class of optmization problems where we have algorithms that can solve the problem optimally. Qudartic progamming (QP) is a type of convex optimization problems which is well developed in the side of theory and computations. We will heavily utilize QPs to solve the optimziation problems that have dependency with `torch` models.\n\nOur general problem solving tricks are as follows:\n1. Construct the cost or constraint models from the data\n2. By utilizting `torch` automatic differentiation functionality, compute the jacobian or hessians of the moodels.\n3. solve (possibley many times) QP with the estimated jacobian and hessians.\n\n> It is noteworthy that even we locally cast the problem into QP, that doesn't mean our original problem is convex. Therefore, we cannot say that this approahces we will look over can find the global optimum.",
"_____no_output_____"
]
],
[
[
"import torch\nimport numpy as np\nimport matplotlib.pyplot as plt\nfrom torch.utils.data import TensorDataset, DataLoader\n\nfrom src.utils import generate_y\nfrom src.nn.MLP import MLP",
"_____no_output_____"
]
],
[
[
"## Generate training dataset\n",
"_____no_output_____"
]
],
[
[
"x_min, x_max = -4.0, 4.0\n\nxs_linspace = torch.linspace(-4, 4, 2000).view(-1, 1)\nys_linspace = generate_y(xs_linspace)\n\n# samples to construct training dataset\nx_dist = torch.distributions.uniform.Uniform(-4.0, 4.0)\nxs = x_dist.sample(sample_shape=(500, 1))\nys = generate_y(xs)",
"_____no_output_____"
],
[
"BS = 64 # Batch size\nds = TensorDataset(xs, ys)\nloader = DataLoader(ds, batch_size=BS, shuffle=True)",
"_____no_output_____"
],
[
"input_dim, output_dim = 1, 1\nm = MLP(input_dim, output_dim, num_neurons=[128, 128])\nmse_criteria = torch.nn.MSELoss()\nopt = torch.optim.Adam(m.parameters(), lr=1e-3)",
"_____no_output_____"
],
[
"n_update = 0\nprint_every = 500\nepochs = 200\nfor _ in range(epochs):\n for x, y in loader: \n y_pred = m(x)\n loss = mse_criteria(y_pred, y)\n opt.zero_grad()\n loss.backward()\n opt.step()\n\n n_update += 1\n if n_update % print_every == 0:\n print(n_update, loss.item())",
"500 0.02176450751721859\n1000 0.003090523649007082\n1500 0.0011741176713258028\n"
],
[
"# save model for the later usages\ntorch.save(m.state_dict(), './model.pt')",
"_____no_output_____"
]
],
[
[
"## Solve the unconstraint optimization problem\n\nLet's solve the unconstraint optimization problem with torch estmiated graidents and simple gradient descent method.\n",
"_____no_output_____"
]
],
[
[
"def minimize_y(x_init, model, num_steps=15, step_size=1e-1):\n def _grad(model, x):\n return torch.autograd.functional.jacobian(model, x).squeeze()\n \n x = x_init\n \n xs = [x]\n ys = [model(x)]\n gs = [_grad(model, x)]\n for _ in range(num_steps):\n grad = _grad(model, x)\n x = (x- step_size * grad).clone()\n y = model(x)\n \n xs.append(x)\n ys.append(y)\n gs.append(grad)\n \n \n xs = torch.stack(xs).detach().numpy()\n ys = torch.stack(ys).detach().numpy()\n gs = torch.stack(gs).detach().numpy() \n return xs, ys, gs",
"_____no_output_____"
],
[
"x_min, x_max = -4.0, 4.0\nn_steps = 40\n\nx_init = torch.tensor(np.random.uniform(x_min, x_max, 1)).float()\nopt_xs, opt_ys, grad = minimize_y(x_init, m, n_steps)",
"_____no_output_____"
],
[
"pred_ys = m(xs_linspace).detach()\n\nfig, axes = plt.subplots(1, 1, figsize=(10, 5))\naxes.grid()\naxes.plot(xs_linspace, ys_linspace, label='Ground truth')\naxes.plot(xs_linspace, pred_ys, label='Model prediction')\n\naxes.scatter(opt_xs[0], opt_ys[0], label='Opt start', \n c='green', marker='*', s=100.0)\naxes.scatter(opt_xs[1:], opt_ys[1:], label='NN opt', c='green')\n_ = axes.legend()",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.