hexsha
stringlengths 40
40
| size
int64 6
14.9M
| ext
stringclasses 1
value | lang
stringclasses 1
value | max_stars_repo_path
stringlengths 6
260
| max_stars_repo_name
stringlengths 6
119
| max_stars_repo_head_hexsha
stringlengths 40
41
| max_stars_repo_licenses
list | max_stars_count
int64 1
191k
⌀ | max_stars_repo_stars_event_min_datetime
stringlengths 24
24
⌀ | max_stars_repo_stars_event_max_datetime
stringlengths 24
24
⌀ | max_issues_repo_path
stringlengths 6
260
| max_issues_repo_name
stringlengths 6
119
| max_issues_repo_head_hexsha
stringlengths 40
41
| max_issues_repo_licenses
list | max_issues_count
int64 1
67k
⌀ | max_issues_repo_issues_event_min_datetime
stringlengths 24
24
⌀ | max_issues_repo_issues_event_max_datetime
stringlengths 24
24
⌀ | max_forks_repo_path
stringlengths 6
260
| max_forks_repo_name
stringlengths 6
119
| max_forks_repo_head_hexsha
stringlengths 40
41
| max_forks_repo_licenses
list | max_forks_count
int64 1
105k
⌀ | max_forks_repo_forks_event_min_datetime
stringlengths 24
24
⌀ | max_forks_repo_forks_event_max_datetime
stringlengths 24
24
⌀ | avg_line_length
float64 2
1.04M
| max_line_length
int64 2
11.2M
| alphanum_fraction
float64 0
1
| cells
list | cell_types
list | cell_type_groups
list |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
cb869bbad379b4d6359eca3c20b4c8a5220d23ff
| 141,710 |
ipynb
|
Jupyter Notebook
|
Task_3/Task_3.ipynb
|
scbayrak/inm707
|
4c573470271221741f5660b51ad59ad7ce2b527f
|
[
"MIT"
] | null | null | null |
Task_3/Task_3.ipynb
|
scbayrak/inm707
|
4c573470271221741f5660b51ad59ad7ce2b527f
|
[
"MIT"
] | null | null | null |
Task_3/Task_3.ipynb
|
scbayrak/inm707
|
4c573470271221741f5660b51ad59ad7ce2b527f
|
[
"MIT"
] | null | null | null | 434.693252 | 43,638 | 0.90904 |
[
[
[
"# INM707 Coursework - Task 3\nSelim Bayrak / 200056225 & Kristina Semenyuk / 200036364",
"_____no_output_____"
],
[
"### Model Training & Evaluation using Experiment Files\n\nTests are carried out automatically by experiment.py file which uses the test cases and hyper-parameters entered in tests.yaml file. There are 42 pre-defined tests cases. First 32 tests trial different hyper-parameters. Last 10 test cases are repeated with the selected optimum hyper-parameters. \n\n\n\n",
"_____no_output_____"
]
],
[
[
"from experiment import run_tests",
"_____no_output_____"
],
[
"df = run_tests()",
"Test No: 20 \t | \tEpisode: 10 \t | \taverage score: 18.5 \t | \tlearning_iterations: 9\nTest No: 20 \t | \tEpisode: 20 \t | \taverage score: 24.5 \t | \tlearning_iterations: 21\nTest No: 20 \t | \tEpisode: 30 \t | \taverage score: 19.6 \t | \tlearning_iterations: 31\nTest No: 20 \t | \tEpisode: 40 \t | \taverage score: 21.6 \t | \tlearning_iterations: 42\nTest No: 20 \t | \tEpisode: 50 \t | \taverage score: 20.1 \t | \tlearning_iterations: 52\nTest No: 20 \t | \tEpisode: 60 \t | \taverage score: 35.8 \t | \tlearning_iterations: 70\nTest No: 20 \t | \tEpisode: 70 \t | \taverage score: 29.4 \t | \tlearning_iterations: 84\nTest No: 20 \t | \tEpisode: 80 \t | \taverage score: 51.7 \t | \tlearning_iterations: 110\nTest No: 20 \t | \tEpisode: 90 \t | \taverage score: 102.6 \t | \tlearning_iterations: 161\nTest No: 20 \t | \tEpisode: 100 \t | \taverage score: 143.5 \t | \tlearning_iterations: 233\nTest No: 20 \t | \tEpisode: 110 \t | \taverage score: 169.2 \t | \tlearning_iterations: 318\nTest No: 20 \t | \tEpisode: 120 \t | \taverage score: 174.3 \t | \tlearning_iterations: 405\nTest No: 20 \t | \tEpisode: 130 \t | \taverage score: 129.5 \t | \tlearning_iterations: 470\nTest No: 20 \t | \tEpisode: 140 \t | \taverage score: 175.4 \t | \tlearning_iterations: 557\nTest No: 20 \t | \tEpisode: 150 \t | \taverage score: 136.0 \t | \tlearning_iterations: 625\nTest No: 20 \t | \tEpisode: 160 \t | \taverage score: 134.2 \t | \tlearning_iterations: 692\nTest No: 20 \t | \tEpisode: 170 \t | \taverage score: 76.8 \t | \tlearning_iterations: 731\nTest No: 20 \t | \tEpisode: 180 \t | \taverage score: 58.9 \t | \tlearning_iterations: 760\nTest No: 20 \t | \tEpisode: 190 \t | \taverage score: 78.2 \t | \tlearning_iterations: 799\nTest No: 20 \t | \tEpisode: 200 \t | \taverage score: 94.3 \t | \tlearning_iterations: 847\nTest No: 20 \t | \tEpisode: 210 \t | \taverage score: 165.0 \t | \tlearning_iterations: 929\nTest No: 20 \t | \tEpisode: 220 \t | \taverage score: 119.6 \t | \tlearning_iterations: 989\nTest No: 20 \t | \tEpisode: 230 \t | \taverage score: 126.4 \t | \tlearning_iterations: 1052\nTest No: 20 \t | \tEpisode: 240 \t | \taverage score: 187.9 \t | \tlearning_iterations: 1146\nTest No: 20 \t | \tEpisode: 250 \t | \taverage score: 180.2 \t | \tlearning_iterations: 1236\nTest No: 20 \t | \tEpisode: 260 \t | \taverage score: 68.5 \t | \tlearning_iterations: 1270\nTest No: 20 \t | \tEpisode: 270 \t | \taverage score: 143.5 \t | \tlearning_iterations: 1342\nTest No: 20 \t | \tEpisode: 280 \t | \taverage score: 198.5 \t | \tlearning_iterations: 1441\nTest No: 20 \t | \tEpisode: 290 \t | \taverage score: 198.3 \t | \tlearning_iterations: 1541\nTest No: 20 \t | \tEpisode: 300 \t | \taverage score: 200.0 \t | \tlearning_iterations: 1641\nMaximum score of 200 reached\nTest No: 27 \t | \tEpisode: 10 \t | \taverage score: 26.1 \t | \tlearning_iterations: 5\nTest No: 27 \t | \tEpisode: 20 \t | \taverage score: 19.1 \t | \tlearning_iterations: 9\nTest No: 27 \t | \tEpisode: 30 \t | \taverage score: 17.9 \t | \tlearning_iterations: 12\nTest No: 27 \t | \tEpisode: 40 \t | \taverage score: 25.2 \t | \tlearning_iterations: 17\nTest No: 27 \t | \tEpisode: 50 \t | \taverage score: 44.1 \t | \tlearning_iterations: 26\nTest No: 27 \t | \tEpisode: 60 \t | \taverage score: 82.6 \t | \tlearning_iterations: 43\nTest No: 27 \t | \tEpisode: 70 \t | \taverage score: 84.3 \t | \tlearning_iterations: 59\nTest No: 27 \t | \tEpisode: 80 \t | \taverage score: 163.6 \t | \tlearning_iterations: 92\nTest No: 27 \t | \tEpisode: 90 \t | \taverage score: 147.4 \t | \tlearning_iterations: 122\nTest No: 27 \t | \tEpisode: 100 \t | \taverage score: 148.1 \t | \tlearning_iterations: 151\nTest No: 27 \t | \tEpisode: 110 \t | \taverage score: 132.5 \t | \tlearning_iterations: 178\nTest No: 27 \t | \tEpisode: 120 \t | \taverage score: 134.6 \t | \tlearning_iterations: 205\nTest No: 27 \t | \tEpisode: 130 \t | \taverage score: 147.7 \t | \tlearning_iterations: 234\nTest No: 27 \t | \tEpisode: 140 \t | \taverage score: 175.9 \t | \tlearning_iterations: 269\nTest No: 27 \t | \tEpisode: 150 \t | \taverage score: 130.7 \t | \tlearning_iterations: 295\nTest No: 27 \t | \tEpisode: 160 \t | \taverage score: 165.2 \t | \tlearning_iterations: 329\nTest No: 27 \t | \tEpisode: 170 \t | \taverage score: 200.0 \t | \tlearning_iterations: 369\nMaximum score of 200 reached\nTest No: 29 \t | \tEpisode: 10 \t | \taverage score: 25.1 \t | \tlearning_iterations: 12\nTest No: 29 \t | \tEpisode: 20 \t | \taverage score: 33.1 \t | \tlearning_iterations: 29\nTest No: 29 \t | \tEpisode: 30 \t | \taverage score: 70.1 \t | \tlearning_iterations: 64\nTest No: 29 \t | \tEpisode: 40 \t | \taverage score: 134.3 \t | \tlearning_iterations: 131\nTest No: 29 \t | \tEpisode: 50 \t | \taverage score: 78.9 \t | \tlearning_iterations: 170\nTest No: 29 \t | \tEpisode: 60 \t | \taverage score: 132.6 \t | \tlearning_iterations: 237\nTest No: 29 \t | \tEpisode: 70 \t | \taverage score: 100.2 \t | \tlearning_iterations: 287\nTest No: 29 \t | \tEpisode: 80 \t | \taverage score: 151.1 \t | \tlearning_iterations: 362\nTest No: 29 \t | \tEpisode: 90 \t | \taverage score: 137.9 \t | \tlearning_iterations: 431\nTest No: 29 \t | \tEpisode: 100 \t | \taverage score: 133.0 \t | \tlearning_iterations: 498\nTest No: 29 \t | \tEpisode: 110 \t | \taverage score: 174.9 \t | \tlearning_iterations: 585\nTest No: 29 \t | \tEpisode: 120 \t | \taverage score: 134.4 \t | \tlearning_iterations: 652\nTest No: 29 \t | \tEpisode: 130 \t | \taverage score: 149.8 \t | \tlearning_iterations: 727\nTest No: 29 \t | \tEpisode: 140 \t | \taverage score: 149.0 \t | \tlearning_iterations: 802\nTest No: 29 \t | \tEpisode: 150 \t | \taverage score: 122.0 \t | \tlearning_iterations: 863\nTest No: 29 \t | \tEpisode: 160 \t | \taverage score: 110.2 \t | \tlearning_iterations: 918\nTest No: 29 \t | \tEpisode: 170 \t | \taverage score: 126.2 \t | \tlearning_iterations: 981\nTest No: 29 \t | \tEpisode: 180 \t | \taverage score: 185.4 \t | \tlearning_iterations: 1074\nTest No: 29 \t | \tEpisode: 190 \t | \taverage score: 147.7 \t | \tlearning_iterations: 1147\nTest No: 29 \t | \tEpisode: 200 \t | \taverage score: 182.4 \t | \tlearning_iterations: 1239\nTest No: 29 \t | \tEpisode: 210 \t | \taverage score: 200.0 \t | \tlearning_iterations: 1339\nMaximum score of 200 reached\n"
],
[
"df.head()",
"_____no_output_____"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
]
] |
cb86a031c91c7dfe4ae191b70e7bc293ccbef026
| 146,043 |
ipynb
|
Jupyter Notebook
|
_notebooks/2021-07-27-helloworld.ipynb
|
rvibek/fastpages
|
986d372b569dfecb5d3f6a7a29804ec29f3ef239
|
[
"Apache-2.0"
] | null | null | null |
_notebooks/2021-07-27-helloworld.ipynb
|
rvibek/fastpages
|
986d372b569dfecb5d3f6a7a29804ec29f3ef239
|
[
"Apache-2.0"
] | null | null | null |
_notebooks/2021-07-27-helloworld.ipynb
|
rvibek/fastpages
|
986d372b569dfecb5d3f6a7a29804ec29f3ef239
|
[
"Apache-2.0"
] | null | null | null | 63.194721 | 95,665 | 0.497545 |
[
[
[
"# hello world, Fastpages - setup done!\n\n> Say goodbye broken links on your static sites. Platform independent, fast, and built in python.\n- author: \"<a href='https://twitter.com/rvibek'>Vibek Raj Maurya</a>\"\n- toc: false\n- image: images/copied_from_nb/fastlinkcheck_images/fastlinkcheck.png\n- comments: true\n- categories: [fastpages, hello world]\n- permalink: /helloworld/\n- badges: true\n",
"_____no_output_____"
],
[
"# Curabitur porta senectus \nNibh proin eleifend odio integer venenatis vestibulum amet consequat mollis pharetra tempus, adipiscing massa accumsan eget quis imperdiet malesuada nunc lacus cubilia vitae tincidunt mauris. Molestie vel tellus in rhoncus elit torquent lobortis, dictum senectus tempor fringilla curabitur malesuada, nullam vehicula himenaeos convallis sociosqu gravida. Placerat fermentum class feugiat massa nulla iaculis habitasse congue, praesent vehicula varius velit etiam ultricies malesuada sit curae, in cum mauris eros risus adipiscing duis.\n",
"_____no_output_____"
]
],
[
[
"import altair as alt\nfrom vega_datasets import data\nimport plotly.express as px\n\nsource = data.cars()",
"_____no_output_____"
],
[
"alt.Chart(source).mark_circle(size=60).encode(\n x='Horsepower',\n y='Miles_per_Gallon',\n color='Origin',\n tooltip=['Name', 'Origin', 'Horsepower', 'Miles_per_Gallon']\n).interactive()",
"_____no_output_____"
],
[
"px.scatter(source, \n x='Horsepower',\n y='Miles_per_Gallon',\n color='Origin',\n hover_name='Name', \n template='plotly_white')\n",
"_____no_output_____"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
]
] |
cb86c4e629d0b08141ee6a00c856d5a23a06a3b3
| 84,878 |
ipynb
|
Jupyter Notebook
|
pipeline3.ipynb
|
chrisart10/DeepLearning.ai-Summary
|
edbfb84165f9031c561f56233d2da2924bc91dfd
|
[
"MIT"
] | null | null | null |
pipeline3.ipynb
|
chrisart10/DeepLearning.ai-Summary
|
edbfb84165f9031c561f56233d2da2924bc91dfd
|
[
"MIT"
] | null | null | null |
pipeline3.ipynb
|
chrisart10/DeepLearning.ai-Summary
|
edbfb84165f9031c561f56233d2da2924bc91dfd
|
[
"MIT"
] | null | null | null | 97.114416 | 19,054 | 0.7708 |
[
[
[
"<a href=\"https://colab.research.google.com/github/chrisart10/DeepLearning.ai-Summary/blob/master/pipeline3.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# Definir dimension de imagen",
"_____no_output_____"
]
],
[
[
"input_shape =300",
"_____no_output_____"
]
],
[
[
"# Importar modelos mediante tranfer learning",
"_____no_output_____"
]
],
[
[
"import os\r\n\r\nfrom tensorflow.keras import layers\r\nfrom tensorflow.keras import Model\r\n!wget --no-check-certificate \\\r\n https://storage.googleapis.com/mledu-datasets/inception_v3_weights_tf_dim_ordering_tf_kernels_notop.h5 \\\r\n -O /tmp/inception_v3_weights_tf_dim_ordering_tf_kernels_notop.h5\r\n \r\nfrom tensorflow.keras.applications.inception_v3 import InceptionV3\r\n\r\nlocal_weights_file = '/tmp/inception_v3_weights_tf_dim_ordering_tf_kernels_notop.h5'\r\n\r\npre_trained_model = InceptionV3(input_shape = (input_shape, input_shape, 3), \r\n include_top = False, \r\n weights = None)\r\n\r\npre_trained_model.load_weights(local_weights_file)\r\n\r\nfor layer in pre_trained_model.layers:\r\n layer.trainable = False\r\n \r\n# pre_trained_model.summary()\r\n\r\nlast_layer = pre_trained_model.get_layer('mixed7')\r\nprint('last layer output shape: ', last_layer.output_shape)\r\nlast_output = last_layer.output",
"--2021-02-12 19:19:21-- https://storage.googleapis.com/mledu-datasets/inception_v3_weights_tf_dim_ordering_tf_kernels_notop.h5\nResolving storage.googleapis.com (storage.googleapis.com)... 172.253.62.128, 172.253.115.128, 172.253.122.128, ...\nConnecting to storage.googleapis.com (storage.googleapis.com)|172.253.62.128|:443... connected.\nHTTP request sent, awaiting response... 200 OK\nLength: 87910968 (84M) [application/x-hdf]\nSaving to: ‘/tmp/inception_v3_weights_tf_dim_ordering_tf_kernels_notop.h5’\n\n/tmp/inception_v3_w 100%[===================>] 83.84M 290MB/s in 0.3s \n\n2021-02-12 19:19:22 (290 MB/s) - ‘/tmp/inception_v3_weights_tf_dim_ordering_tf_kernels_notop.h5’ saved [87910968/87910968]\n\nlast layer output shape: (None, 17, 17, 768)\n"
]
],
[
[
"# Ultima capa de aprendizaje",
"_____no_output_____"
]
],
[
[
"#from tensorflow.keras.optimizers import RMSprop\r\nfrom tensorflow.keras.optimizers import Adam\r\n# Flatten the output layer to 1 dimension\r\nx = layers.Flatten()(last_output)\r\n# Add a fully connected layer with 1,024 hidden units and ReLU activation\r\nx = layers.Dense(1024, activation='relu')(x)\r\n# Add a dropout rate of 0.2\r\nx = layers.Dropout(0.2)(x) \r\n# Add a final sigmoid layer for classification\r\nx = layers.Dense (6, activation='softmax')(x) \r\n\r\nmodel = Model( pre_trained_model.input, x) \r\n\r\nmodel.compile(optimizer = Adam(lr=0.0001), \r\n loss = 'categorical_crossentropy', \r\n metrics = ['accuracy'])\r\n\r\n## Rocket science\r\n",
"_____no_output_____"
],
[
"#model.summary()",
"_____no_output_____"
]
],
[
[
"# Importar Dataset desde kaggle",
"_____no_output_____"
]
],
[
[
"! pip install -q kaggle",
"_____no_output_____"
],
[
"from google.colab import files\r\nfiles.upload()",
"_____no_output_____"
],
[
"! mkdir ~/.kaggle\r\n! cp kaggle.json ~/.kaggle/\r\n! chmod 600 ~/.kaggle/kaggle.json",
"_____no_output_____"
]
],
[
[
"# Pegar Api del dataset",
"_____no_output_____"
]
],
[
[
"!kaggle datasets download -d sriramr/fruits-fresh-and-rotten-for-classification",
"Downloading fruits-fresh-and-rotten-for-classification.zip to /content\n100% 3.57G/3.58G [01:05<00:00, 45.5MB/s]\n100% 3.58G/3.58G [01:05<00:00, 58.6MB/s]\n"
],
[
"#!kaggle datasets download -d kmader/skin-cancer-mnist-ham10000",
"Downloading skin-cancer-mnist-ham10000.zip to /content\n100% 5.19G/5.20G [01:49<00:00, 36.5MB/s]\n100% 5.20G/5.20G [01:49<00:00, 50.8MB/s]\n"
],
[
"",
"_____no_output_____"
]
],
[
[
"# Extraer zip",
"_____no_output_____"
]
],
[
[
"import os\r\nimport zipfile\r\n\r\n#local_zip = '/content/skin-cancer-mnist-ham10000.zip'\r\nlocal_zip = \"/content/fruits-fresh-and-rotten-for-classification.zip\"\r\nzip_ref = zipfile.ZipFile(local_zip, 'r')\r\nzip_ref.extractall('/tmp')\r\nzip_ref.close()",
"_____no_output_____"
]
],
[
[
"# Preparar Dataset y asignar data augmentation",
"_____no_output_____"
]
],
[
[
"from tensorflow.keras.preprocessing.image import ImageDataGenerator\r\n# Define our example directories and files\r\nbase_dir = '/tmp/dataset/'\r\n\r\ntrain_dir = os.path.join( base_dir, 'train')\r\nvalidation_dir = os.path.join( base_dir, 'test')\r\n\r\n# Add our data-augmentation parameters to ImageDataGenerator\r\n\r\ntrain_datagen = ImageDataGenerator(rescale = 1./255.,\r\n rotation_range = 40,\r\n width_shift_range = 0.2,\r\n height_shift_range = 0.2,\r\n shear_range = 0.2,\r\n zoom_range = 0.2,\r\n horizontal_flip = True)\r\n\r\n# Note that the validation data should not be augmented!\r\ntest_datagen = ImageDataGenerator( rescale = 1.0/255. )\r\n\r\n# Flow training images in batches of 20 using train_datagen generator\r\ntrain_generator = train_datagen.flow_from_directory(train_dir,\r\n batch_size = 20,\r\n class_mode = 'categorical', \r\n target_size = (input_shape, input_shape)) \r\n\r\n# Flow validation images in batches of 20 using test_datagen generator\r\nvalidation_generator = test_datagen.flow_from_directory( validation_dir,\r\n batch_size = 20,\r\n class_mode = 'categorical', \r\n target_size = (input_shape, input_shape))",
"Found 10901 images belonging to 6 classes.\nFound 2698 images belonging to 6 classes.\n"
]
],
[
[
"# callback con early stopping\r\n",
"_____no_output_____"
]
],
[
[
"import tensorflow as tf\r\nclass myCallback(tf.keras.callbacks.Callback):\r\n def on_epoch_end(self, epoch, logs={}):\r\n if(logs.get('accuracy') >= 0.98):\r\n print(\"\\nReached 98% accuracy so cancelling training!\")\r\n self.model.stop_training = True\r\ncallbacks = myCallback()",
"_____no_output_____"
]
],
[
[
"# Entrenar el modelo",
"_____no_output_____"
]
],
[
[
"history = model.fit(\r\n train_generator,\r\n validation_data = validation_generator,\r\n steps_per_epoch = 32,\r\n epochs = 100,\r\n callbacks=[callbacks],\r\n validation_steps = 32,\r\n verbose = 1)",
"Epoch 1/100\n32/32 [==============================] - 28s 580ms/step - loss: 2.5920 - accuracy: 0.4076 - val_loss: 0.4284 - val_accuracy: 0.8469\nEpoch 2/100\n32/32 [==============================] - 18s 561ms/step - loss: 0.4661 - accuracy: 0.8251 - val_loss: 0.2546 - val_accuracy: 0.9047\nEpoch 3/100\n32/32 [==============================] - 18s 558ms/step - loss: 0.4498 - accuracy: 0.8402 - val_loss: 0.2065 - val_accuracy: 0.9375\nEpoch 4/100\n32/32 [==============================] - 18s 563ms/step - loss: 0.2827 - accuracy: 0.9074 - val_loss: 0.1710 - val_accuracy: 0.9500\nEpoch 5/100\n32/32 [==============================] - 18s 563ms/step - loss: 0.2748 - accuracy: 0.9185 - val_loss: 0.0917 - val_accuracy: 0.9672\nEpoch 6/100\n32/32 [==============================] - 18s 575ms/step - loss: 0.1707 - accuracy: 0.9463 - val_loss: 0.0741 - val_accuracy: 0.9766\nEpoch 7/100\n32/32 [==============================] - 18s 567ms/step - loss: 0.2256 - accuracy: 0.9358 - val_loss: 0.1114 - val_accuracy: 0.9578\nEpoch 8/100\n32/32 [==============================] - 18s 561ms/step - loss: 0.1793 - accuracy: 0.9477 - val_loss: 0.0608 - val_accuracy: 0.9859\nEpoch 9/100\n32/32 [==============================] - 18s 562ms/step - loss: 0.1342 - accuracy: 0.9527 - val_loss: 0.0770 - val_accuracy: 0.9812\nEpoch 10/100\n32/32 [==============================] - 18s 559ms/step - loss: 0.1484 - accuracy: 0.9475 - val_loss: 0.0741 - val_accuracy: 0.9734\nEpoch 11/100\n32/32 [==============================] - 18s 570ms/step - loss: 0.1727 - accuracy: 0.9394 - val_loss: 0.0783 - val_accuracy: 0.9719\nEpoch 12/100\n32/32 [==============================] - 18s 560ms/step - loss: 0.1492 - accuracy: 0.9358 - val_loss: 0.0622 - val_accuracy: 0.9781\nEpoch 13/100\n32/32 [==============================] - 18s 561ms/step - loss: 0.1133 - accuracy: 0.9582 - val_loss: 0.0365 - val_accuracy: 0.9875\nEpoch 14/100\n32/32 [==============================] - 18s 561ms/step - loss: 0.1555 - accuracy: 0.9484 - val_loss: 0.0604 - val_accuracy: 0.9766\nEpoch 15/100\n32/32 [==============================] - 18s 564ms/step - loss: 0.2132 - accuracy: 0.9282 - val_loss: 0.0403 - val_accuracy: 0.9875\nEpoch 16/100\n32/32 [==============================] - 18s 565ms/step - loss: 0.1421 - accuracy: 0.9457 - val_loss: 0.0685 - val_accuracy: 0.9719\nEpoch 17/100\n32/32 [==============================] - 18s 562ms/step - loss: 0.1388 - accuracy: 0.9437 - val_loss: 0.0925 - val_accuracy: 0.9578\nEpoch 18/100\n32/32 [==============================] - 18s 562ms/step - loss: 0.1721 - accuracy: 0.9367 - val_loss: 0.1248 - val_accuracy: 0.9563\nEpoch 19/100\n32/32 [==============================] - 18s 558ms/step - loss: 0.1981 - accuracy: 0.9272 - val_loss: 0.1051 - val_accuracy: 0.9578\nEpoch 20/100\n32/32 [==============================] - 18s 568ms/step - loss: 0.1336 - accuracy: 0.9520 - val_loss: 0.0839 - val_accuracy: 0.9656\nEpoch 21/100\n32/32 [==============================] - 18s 559ms/step - loss: 0.1797 - accuracy: 0.9336 - val_loss: 0.0613 - val_accuracy: 0.9812\nEpoch 22/100\n32/32 [==============================] - 18s 556ms/step - loss: 0.1418 - accuracy: 0.9586 - val_loss: 0.0214 - val_accuracy: 0.9937\nEpoch 23/100\n32/32 [==============================] - 18s 565ms/step - loss: 0.1252 - accuracy: 0.9433 - val_loss: 0.0531 - val_accuracy: 0.9859\nEpoch 24/100\n32/32 [==============================] - 18s 559ms/step - loss: 0.1316 - accuracy: 0.9486 - val_loss: 0.0452 - val_accuracy: 0.9891\nEpoch 25/100\n32/32 [==============================] - 18s 562ms/step - loss: 0.0736 - accuracy: 0.9739 - val_loss: 0.0322 - val_accuracy: 0.9891\nEpoch 26/100\n32/32 [==============================] - 18s 550ms/step - loss: 0.0983 - accuracy: 0.9773 - val_loss: 0.0579 - val_accuracy: 0.9766\nEpoch 27/100\n32/32 [==============================] - 18s 559ms/step - loss: 0.1114 - accuracy: 0.9575 - val_loss: 0.0542 - val_accuracy: 0.9734\nEpoch 28/100\n32/32 [==============================] - 18s 562ms/step - loss: 0.1157 - accuracy: 0.9547 - val_loss: 0.0376 - val_accuracy: 0.9844\nEpoch 29/100\n32/32 [==============================] - 18s 558ms/step - loss: 0.1232 - accuracy: 0.9579 - val_loss: 0.0622 - val_accuracy: 0.9797\nEpoch 30/100\n32/32 [==============================] - 18s 549ms/step - loss: 0.1045 - accuracy: 0.9678 - val_loss: 0.0416 - val_accuracy: 0.9906\nEpoch 31/100\n32/32 [==============================] - 18s 562ms/step - loss: 0.0965 - accuracy: 0.9684 - val_loss: 0.0502 - val_accuracy: 0.9844\nEpoch 32/100\n32/32 [==============================] - 18s 561ms/step - loss: 0.0930 - accuracy: 0.9623 - val_loss: 0.0996 - val_accuracy: 0.9688\nEpoch 33/100\n32/32 [==============================] - 18s 561ms/step - loss: 0.0871 - accuracy: 0.9635 - val_loss: 0.0355 - val_accuracy: 0.9906\nEpoch 34/100\n32/32 [==============================] - 18s 564ms/step - loss: 0.1064 - accuracy: 0.9686 - val_loss: 0.0345 - val_accuracy: 0.9875\nEpoch 35/100\n32/32 [==============================] - 18s 562ms/step - loss: 0.1110 - accuracy: 0.9607 - val_loss: 0.0471 - val_accuracy: 0.9859\nEpoch 36/100\n32/32 [==============================] - 18s 561ms/step - loss: 0.1171 - accuracy: 0.9563 - val_loss: 0.0181 - val_accuracy: 0.9953\nEpoch 37/100\n32/32 [==============================] - 18s 557ms/step - loss: 0.0964 - accuracy: 0.9595 - val_loss: 0.0287 - val_accuracy: 0.9828\nEpoch 38/100\n32/32 [==============================] - 18s 561ms/step - loss: 0.1364 - accuracy: 0.9684 - val_loss: 0.0308 - val_accuracy: 0.9875\nEpoch 39/100\n32/32 [==============================] - 18s 559ms/step - loss: 0.0600 - accuracy: 0.9784 - val_loss: 0.0394 - val_accuracy: 0.9844\n\nReached 98% accuracy so cancelling training!\n"
]
],
[
[
"# visualizacion del aprendizaje",
"_____no_output_____"
]
],
[
[
"#-----------------------------------------------------------\r\n# Retrieve a list of list results on training and test data\r\n# sets for each training epoch\r\n#-----------------------------------------------------------\r\nimport matplotlib.pyplot as plt\r\nacc = history.history[ 'accuracy' ]\r\nval_acc = history.history[ 'val_accuracy' ]\r\nloss = history.history[ 'loss' ]\r\nval_loss = history.history['val_loss' ]\r\n\r\nepochs = range(len(acc)) # Get number of epochs\r\n\r\n#------------------------------------------------\r\n# Plot training and validation accuracy per epoch\r\n#------------------------------------------------\r\n\r\nplt.plot(epochs, acc, 'r', label='Training accuracy')\r\nplt.plot(epochs, val_acc, 'b', label='Validation accuracy')\r\nplt.title ('Training and validation accuracy')\r\nplt.legend(loc=0)\r\nplt.figure()\r\n\r\n#------------------------------------------------\r\n# Plot training and validation loss per epoch\r\n#------------------------------------------------\r\nplt.plot ( epochs, loss, 'r', label='Training loss')\r\nplt.plot ( epochs, val_loss, 'b', label='Validation loss')\r\nplt.title ('Training and validation loss' )\r\nplt.legend(loc=0)\r\nplt.figure()",
"_____no_output_____"
]
],
[
[
"# Test del modelo ",
"_____no_output_____"
]
],
[
[
"import numpy as np\r\nfrom google.colab import files\r\nfrom keras.preprocessing import image\r\n\r\nuploaded = files.upload()\r\n\r\nfor fn in uploaded.keys():\r\n \r\n # predicting images\r\n path = '/content/' + fn\r\n img = image.load_img(path, target_size=(input_shape, input_shape))\r\n x = image.img_to_array(img)\r\n x = np.expand_dims(x, axis=0)\r\n\r\n images = np.vstack([x])\r\n classes = model.predict(images, batch_size=32)\r\n print(classes[0])\r\n # if classes[0]>0.5:\r\n # print(fn + \" is a dog\")\r\n # else:\r\n # print(fn + \" is a cat\")",
"_____no_output_____"
]
],
[
[
"# forma de guardar Opcion 1",
"_____no_output_____"
]
],
[
[
"import time\r\npath = '/tmp/simple_keras_model'\r\nmodel.save(saved_model_path)",
"_____no_output_____"
],
[
"new_model = tf.keras.models.load_model('/tmp/saved_models/1612553978/')\r\n\r\n# Check its architecture\r\n#new_model.summary()\r\n",
"_____no_output_____"
]
],
[
[
"# Forma de guardar opcion 2",
"_____no_output_____"
]
],
[
[
"# Save the entire model to a HDF5 file.\r\n# The '.h5' extension indicates that the model should be saved to HDF5.\r\nmodel.save('/tmp/saved_models/versions/my_model1.h5')",
"_____no_output_____"
],
[
"# Recreate the exact same model, including its weights and the optimizer\r\nnew_model = tf.keras.models.load_model('/tmp/saved_models/versions/my_model1.h5')\r\n\r\n# Show the model architecture\r\n#new_model.summary()",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
cb8710900caf23abf8a0dc241d035dee562cadc5
| 21,349 |
ipynb
|
Jupyter Notebook
|
Python for Data Science/Python for Data Science - Week 2 - Class 3 - Dictionaries.ipynb
|
ipiyushsonar/coursera-labs
|
39a6a528980d3298ceb52bf9e729d046f3b5cbfa
|
[
"MIT"
] | null | null | null |
Python for Data Science/Python for Data Science - Week 2 - Class 3 - Dictionaries.ipynb
|
ipiyushsonar/coursera-labs
|
39a6a528980d3298ceb52bf9e729d046f3b5cbfa
|
[
"MIT"
] | null | null | null |
Python for Data Science/Python for Data Science - Week 2 - Class 3 - Dictionaries.ipynb
|
ipiyushsonar/coursera-labs
|
39a6a528980d3298ceb52bf9e729d046f3b5cbfa
|
[
"MIT"
] | null | null | null | 24.150452 | 716 | 0.529158 |
[
[
[
"<div class=\"alert alert-block alert-info\" style=\"margin-top: 20px\">\n <a href=\"http://cocl.us/topNotebooksPython101Coursera\">\n <img src=\"https://s3-api.us-geo.objectstorage.softlayer.net/cf-courses-data/CognitiveClass/PY0101EN/Ad/TopAd.png\" width=\"750\" align=\"center\">\n </a>\n</div>",
"_____no_output_____"
],
[
"<a href=\"https://cognitiveclass.ai/\">\n <img src=\"https://s3-api.us-geo.objectstorage.softlayer.net/cf-courses-data/CognitiveClass/PY0101EN/Ad/CCLog.png\" width=\"200\" align=\"center\">\n</a>",
"_____no_output_____"
],
[
"<h1>Dictionaries in Python</h1>",
"_____no_output_____"
],
[
"<p><strong>Welcome!</strong> This notebook will teach you about the dictionaries in the Python Programming Language. By the end of this lab, you'll know the basics dictionary operations in Python, including what it is, and the operations on it.</p>",
"_____no_output_____"
],
[
"<h2>Table of Contents</h2>\n<div class=\"alert alert-block alert-info\" style=\"margin-top: 20px\">\n <ul>\n <li>\n <a href=\"#dic\">Dictionaries</a>\n <ul>\n <li><a href=\"content\">What are Dictionaries?</a></li>\n <li><a href=\"key\">Keys</a></li>\n </ul>\n </li>\n <li>\n <a href=\"#quiz\">Quiz on Dictionaries</a>\n </li>\n </ul>\n <p>\n Estimated time needed: <strong>20 min</strong>\n </p>\n</div>\n\n<hr>",
"_____no_output_____"
],
[
"<h2 id=\"Dic\">Dictionaries</h2>",
"_____no_output_____"
],
[
"<h3 id=\"content\">What are Dictionaries?</h3>",
"_____no_output_____"
],
[
"A dictionary consists of keys and values. It is helpful to compare a dictionary to a list. Instead of the numerical indexes such as a list, dictionaries have keys. These keys are the keys that are used to access values within a dictionary.",
"_____no_output_____"
],
[
"<img src=\"https://s3-api.us-geo.objectstorage.softlayer.net/cf-courses-data/CognitiveClass/PY0101EN/Chapter%202/Images/DictsList.png\" width=\"650\" />",
"_____no_output_____"
],
[
"An example of a Dictionary <code>Dict</code>:",
"_____no_output_____"
]
],
[
[
"# Create the dictionary\n\nDict = {\"key1\": 1, \"key2\": \"2\", \"key3\": [3, 3, 3], \"key4\": (4, 4, 4), ('key5'): 5, (0, 1): 6}\nDict",
"_____no_output_____"
]
],
[
[
"The keys can be strings:",
"_____no_output_____"
]
],
[
[
"# Access to the value by the key\n\nDict[\"key1\"]",
"_____no_output_____"
]
],
[
[
"Keys can also be any immutable object such as a tuple: ",
"_____no_output_____"
]
],
[
[
"# Access to the value by the key\n\nDict[(0, 1)]",
"_____no_output_____"
]
],
[
[
" Each key is separated from its value by a colon \"<code>:</code>\". Commas separate the items, and the whole dictionary is enclosed in curly braces. An empty dictionary without any items is written with just two curly braces, like this \"<code>{}</code>\".",
"_____no_output_____"
]
],
[
[
"# Create a sample dictionary\n\nrelease_year_dict = {\"Thriller\": \"1982\", \"Back in Black\": \"1980\", \\\n \"The Dark Side of the Moon\": \"1973\", \"The Bodyguard\": \"1992\", \\\n \"Bat Out of Hell\": \"1977\", \"Their Greatest Hits (1971-1975)\": \"1976\", \\\n \"Saturday Night Fever\": \"1977\", \"Rumours\": \"1977\"}\nrelease_year_dict",
"_____no_output_____"
]
],
[
[
"In summary, like a list, a dictionary holds a sequence of elements. Each element is represented by a key and its corresponding value. Dictionaries are created with two curly braces containing keys and values separated by a colon. For every key, there can only be one single value, however, multiple keys can hold the same value. Keys can only be strings, numbers, or tuples, but values can be any data type.",
"_____no_output_____"
],
[
"It is helpful to visualize the dictionary as a table, as in the following image. The first column represents the keys, the second column represents the values.",
"_____no_output_____"
],
[
"<img src=\"https://s3-api.us-geo.objectstorage.softlayer.net/cf-courses-data/CognitiveClass/PY0101EN/Chapter%202/Images/DictsStructure.png\" width=\"650\" />",
"_____no_output_____"
],
[
"<h3 id=\"key\">Keys</h3>",
"_____no_output_____"
],
[
"You can retrieve the values based on the names:",
"_____no_output_____"
]
],
[
[
"# Get value by keys\n\nrelease_year_dict['Thriller'] ",
"_____no_output_____"
]
],
[
[
"This corresponds to: \n",
"_____no_output_____"
],
[
"<img src=\"https://s3-api.us-geo.objectstorage.softlayer.net/cf-courses-data/CognitiveClass/PY0101EN/Chapter%202/Images/DictsKeyOne.png\" width=\"500\" />",
"_____no_output_____"
],
[
"Similarly for <b>The Bodyguard</b>",
"_____no_output_____"
]
],
[
[
"# Get value by key\n\nrelease_year_dict['The Bodyguard'] ",
"_____no_output_____"
]
],
[
[
"<img src=\"https://s3-api.us-geo.objectstorage.softlayer.net/cf-courses-data/CognitiveClass/PY0101EN/Chapter%202/Images/DictsKeyTwo.png\" width=\"500\" />",
"_____no_output_____"
],
[
"Now let you retrieve the keys of the dictionary using the method <code>release_year_dict()</code>:",
"_____no_output_____"
]
],
[
[
"# Get all the keys in dictionary\n\nrelease_year_dict.keys() ",
"_____no_output_____"
]
],
[
[
"You can retrieve the values using the method <code>values()</code>:",
"_____no_output_____"
]
],
[
[
"# Get all the values in dictionary\n\nrelease_year_dict.values() ",
"_____no_output_____"
]
],
[
[
"We can add an entry:",
"_____no_output_____"
]
],
[
[
"# Append value with key into dictionary\n\nrelease_year_dict['Graduation'] = '2007'\nrelease_year_dict",
"_____no_output_____"
]
],
[
[
"We can delete an entry: ",
"_____no_output_____"
]
],
[
[
"# Delete entries by key\n\ndel(release_year_dict['Thriller'])\ndel(release_year_dict['Graduation'])\nrelease_year_dict",
"_____no_output_____"
]
],
[
[
" We can verify if an element is in the dictionary: ",
"_____no_output_____"
]
],
[
[
"# Verify the key is in the dictionary\n\n'The Bodyguard' in release_year_dict",
"_____no_output_____"
]
],
[
[
"<hr>",
"_____no_output_____"
],
[
"<h2 id=\"quiz\">Quiz on Dictionaries</h2>",
"_____no_output_____"
],
[
"<b>You will need this dictionary for the next two questions:</b>",
"_____no_output_____"
]
],
[
[
"# Question sample dictionary\n\nsoundtrack_dic = {\"The Bodyguard\":\"1992\", \"Saturday Night Fever\":\"1977\"}\nsoundtrack_dic ",
"_____no_output_____"
]
],
[
[
"a) In the dictionary <code>soundtrack_dict</code> what are the keys ?",
"_____no_output_____"
]
],
[
[
"# Write your code below and press Shift+Enter to execute\nsoundtrack_dic.keys()",
"_____no_output_____"
]
],
[
[
"Double-click __here__ for the solution.\n\n<!-- Your answer is below:\nsoundtrack_dic.keys() # The Keys \"The Bodyguard\" and \"Saturday Night Fever\" \n-->",
"_____no_output_____"
],
[
"b) In the dictionary <code>soundtrack_dict</code> what are the values ?",
"_____no_output_____"
]
],
[
[
"# Write your code below and press Shift+Enter to execute\nsoundtrack_dic.values()",
"_____no_output_____"
]
],
[
[
"Double-click __here__ for the solution.\n\n<!-- Your answer is below:\nsoundtrack_dic.values() # The values are \"1992\" and \"1977\"\n-->",
"_____no_output_____"
],
[
"<hr>",
"_____no_output_____"
],
[
"<b>You will need this dictionary for the following questions:</b>",
"_____no_output_____"
],
[
"The Albums <b>Back in Black</b>, <b>The Bodyguard</b> and <b>Thriller</b> have the following music recording sales in millions 50, 50 and 65 respectively:",
"_____no_output_____"
],
[
"a) Create a dictionary <code>album_sales_dict</code> where the keys are the album name and the sales in millions are the values. ",
"_____no_output_____"
]
],
[
[
"# Write your code below and press Shift+Enter to execute\nalbum_sales_dict = {\"Back in Black\":50,\"The Bodyguard\":50,\"Thriller\":65}\nalbum_sales_dict",
"_____no_output_____"
]
],
[
[
"Double-click __here__ for the solution.\n\n<!-- Your answer is below:\nalbum_sales_dict = {\"The Bodyguard\":50, \"Back in Black\":50, \"Thriller\":65}\n-->",
"_____no_output_____"
],
[
"b) Use the dictionary to find the total sales of <b>Thriller</b>:",
"_____no_output_____"
]
],
[
[
"# Write your code below and press Shift+Enter to execute\nalbum_sales_dict[\"Thriller\"]",
"_____no_output_____"
]
],
[
[
"Double-click __here__ for the solution.\n\n<!-- Your answer is below:\nalbum_sales_dict[\"Thriller\"]\n-->",
"_____no_output_____"
],
[
"c) Find the names of the albums from the dictionary using the method <code>keys</code>:",
"_____no_output_____"
]
],
[
[
"# Write your code below and press Shift+Enter to execute\nalbum_sales_dict.keys()",
"_____no_output_____"
]
],
[
[
"Double-click __here__ for the solution.\n\n<!-- Your answer is below:\nalbum_sales_dict.keys()\n-->",
"_____no_output_____"
],
[
"d) Find the names of the recording sales from the dictionary using the method <code>values</code>:",
"_____no_output_____"
]
],
[
[
"# Write your code below and press Shift+Enter to execute\nalbum_sales_dict.value",
"_____no_output_____"
]
],
[
[
"Double-click __here__ for the solution.\n\n<!-- Your answer is below:\nalbum_sales_dict.values()\n-->",
"_____no_output_____"
],
[
"<hr>\n<h2>The last exercise!</h2>\n<p>Congratulations, you have completed your first lesson and hands-on lab in Python. However, there is one more thing you need to do. The Data Science community encourages sharing work. The best way to share and showcase your work is to share it on GitHub. By sharing your notebook on GitHub you are not only building your reputation with fellow data scientists, but you can also show it off when applying for a job. Even though this was your first piece of work, it is never too early to start building good habits. So, please read and follow <a href=\"https://cognitiveclass.ai/blog/data-scientists-stand-out-by-sharing-your-notebooks/\" target=\"_blank\">this article</a> to learn how to share your work.\n<hr>",
"_____no_output_____"
],
[
"<div class=\"alert alert-block alert-info\" style=\"margin-top: 20px\">\n<h2>Get IBM Watson Studio free of charge!</h2>\n <p><a href=\"https://cocl.us/bottemNotebooksPython101Coursera\"><img src=\"https://s3-api.us-geo.objectstorage.softlayer.net/cf-courses-data/CognitiveClass/PY0101EN/Ad/BottomAd.png\" width=\"750\" align=\"center\"></a></p>\n</div>",
"_____no_output_____"
],
[
"<h3>About the Authors:</h3> \n<p><a href=\"https://www.linkedin.com/in/joseph-s-50398b136/\" target=\"_blank\">Joseph Santarcangelo</a> is a Data Scientist at IBM, and holds a PhD in Electrical Engineering. His research focused on using Machine Learning, Signal Processing, and Computer Vision to determine how videos impact human cognition. Joseph has been working for IBM since he completed his PhD.</p>",
"_____no_output_____"
],
[
"Other contributors: <a href=\"www.linkedin.com/in/jiahui-mavis-zhou-a4537814a\">Mavis Zhou</a>",
"_____no_output_____"
],
[
"<hr>",
"_____no_output_____"
],
[
"<p>Copyright © 2018 IBM Developer Skills Network. This notebook and its source code are released under the terms of the <a href=\"https://cognitiveclass.ai/mit-license/\">MIT License</a>.</p>",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
cb871393e71393eb95f1baab339eab59cfbd1cd7
| 97,579 |
ipynb
|
Jupyter Notebook
|
programming_for_biology/arrays/smoothing.ipynb
|
janhohenheim/programming-for-biology
|
d895a20a9591888616af2bae2e2d87feebf6d60a
|
[
"MIT"
] | null | null | null |
programming_for_biology/arrays/smoothing.ipynb
|
janhohenheim/programming-for-biology
|
d895a20a9591888616af2bae2e2d87feebf6d60a
|
[
"MIT"
] | null | null | null |
programming_for_biology/arrays/smoothing.ipynb
|
janhohenheim/programming-for-biology
|
d895a20a9591888616af2bae2e2d87feebf6d60a
|
[
"MIT"
] | null | null | null | 750.607692 | 93,942 | 0.948903 |
[
[
[
"# Exercise\n\nNoise in images can be reduced in several ways. One way is to give a pixel the mean value of 9 pixels, of itself and of its 8 nearest neighbor pixels. Program a function containing this algorithm (the code from the smoothing video does something else!) and use it to smoothen this stink bug image. Leave the boundary pixels identical to the original image. Again, download the image by right-clicking on the mouse.\n\nKeep in mind that each pixel value must be a float between 0.0 and 1.0 or a uint8 in the range (0,255).\n",
"_____no_output_____"
]
],
[
[
"from os.path import join, abspath\nimport matplotlib.pyplot as plt\nimport matplotlib.image as img\nfrom matplotlib import rcParams\nimport numpy as np\n\n%matplotlib inline\n\ndef read_jpg(filename):\n path = join(abspath(\"\"), \"data\", filename)\n return img.imread(path)\n\n\ndef smooth_image(image):\n # fmt: off\n return np.array([[[\n _get_average_of_neighbors(image, row, col, channel)\n for channel in range(0, 3)]\n for col in range(0, image.shape[1])]\n for row in range(0, image.shape[0])]\n )\n # fmt: on\n\n\ndef _get_average_of_neighbors(image, row, col, channel):\n max_row_index = image.shape[0] - 1\n max_col_index = image.shape[1] - 1\n return (\n np.mean(image[row - 1 : row + 2, col - 1 : col + 2, channel])\n if 0 < row < max_row_index and 0 < col < max_col_index\n else image[row, col, channel]\n )\n",
"_____no_output_____"
],
[
"image = read_jpg(\"stinkbug.png\")\nsmoothed = smooth_image(image)\n\nprint(f\"Red value at position [5,499]: {smoothed[5,499,0]:.3}\")\nprint(f\"Green value at position [181, 260]: {smoothed[181, 260, 1]:.2}\")\nprint(f\"Grand mean pixel value: {np.mean(smoothed):.6}\")",
"Red value at position [5,499]: 0.439\nGreen value at position [181, 260]: 0.33\nGrand mean pixel value: 0.551117\n"
],
[
"rcParams['figure.figsize'] = 11 ,8\n\nfig, ax = plt.subplots(1,2)\nax[0].set_title(\"Original image\")\nax[0].imshow(image)\n\nax[1].set_title(\"Smoothed image\")\nax[1].imshow(smoothed)\nplt.show()",
"_____no_output_____"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
cb871dbb1d725f8cf9eed703a40dfce9f6f4df36
| 1,765 |
ipynb
|
Jupyter Notebook
|
Array/1227/1343. Number of Sub-arrays of Size K and Average Greater than or Equal to Threshold.ipynb
|
YuHe0108/Leetcode
|
90d904dde125dd35ee256a7f383961786f1ada5d
|
[
"Apache-2.0"
] | 1 |
2020-08-05T11:47:47.000Z
|
2020-08-05T11:47:47.000Z
|
Array/1227/1343. Number of Sub-arrays of Size K and Average Greater than or Equal to Threshold.ipynb
|
YuHe0108/LeetCode
|
b9e5de69b4e4d794aff89497624f558343e362ad
|
[
"Apache-2.0"
] | null | null | null |
Array/1227/1343. Number of Sub-arrays of Size K and Average Greater than or Equal to Threshold.ipynb
|
YuHe0108/LeetCode
|
b9e5de69b4e4d794aff89497624f558343e362ad
|
[
"Apache-2.0"
] | null | null | null | 19.831461 | 74 | 0.453824 |
[
[
[
"class Solution:\n def numOfSubarrays(self, arr, k: int, threshold: int) -> int:\n val = k * threshold\n presum = [0]\n for n in arr:\n presum.append(presum[-1] + n)\n \n cnt = 0\n for start in range(len(arr) - k + 1):\n end = start + k\n if presum[end] - presum[start] >= val:\n cnt += 1\n return cnt",
"_____no_output_____"
],
[
"solution = Solution()\nsolution.numOfSubarrays([1,1,1,1,1], 1, 0)",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code"
]
] |
cb8721f46a52a29c744bdb75bd1d3d97dfffcb94
| 5,547 |
ipynb
|
Jupyter Notebook
|
NoteBooks/KNN.ipynb
|
guptachetan1997/HeartDiseaseMLProject
|
d5c5e8f31a244eb444f70943393c2e4bd29ef3d2
|
[
"MIT"
] | 8 |
2016-09-07T08:19:33.000Z
|
2021-06-21T06:00:10.000Z
|
NoteBooks/KNN.ipynb
|
guptachetan1997/HeartDiseaseMLProject
|
d5c5e8f31a244eb444f70943393c2e4bd29ef3d2
|
[
"MIT"
] | null | null | null |
NoteBooks/KNN.ipynb
|
guptachetan1997/HeartDiseaseMLProject
|
d5c5e8f31a244eb444f70943393c2e4bd29ef3d2
|
[
"MIT"
] | 6 |
2016-08-13T17:53:45.000Z
|
2019-10-30T09:53:51.000Z
| 23.909483 | 284 | 0.561024 |
[
[
[
"# KNN",
"_____no_output_____"
],
[
"Importing required python modules\n---------------------------------",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nfrom sklearn.neighbors import KNeighborsClassifier \nfrom sklearn.cross_validation import train_test_split\nfrom sklearn import metrics\nfrom sklearn.preprocessing import normalize,scale\nfrom sklearn.cross_validation import cross_val_score\nimport numpy as np\nimport pandas as pd ",
"_____no_output_____"
]
],
[
[
"The following libraries have been used :\n* **Pandas** : pandas is an open source, BSD-licensed library providing high-performance, easy-to-use data structures and data analysis tools for the Python programming language.\n* **Numpy** : NumPy is the fundamental package for scientific computing with Python.\n* **Matplotlib** : matplotlib is a python 2D plotting library which produces publication quality figures in a variety of hardcopy formats and interactive environments .\n* **Sklearn** : It features various classification, regression and clustering algorithms including support vector machines, random forests, gradient boosting, k-means and DBSCAN, and is designed to interoperate with the Python numerical and scientific libraries NumPy and SciPy.",
"_____no_output_____"
],
[
"Retrieving the dataset\n----------------------",
"_____no_output_____"
]
],
[
[
"data = pd.read_csv('heart.csv', header=None)\n\ndf = pd.DataFrame(data)\n\n\nx = df.iloc[:, 0:5]\nx = x.drop(x.columns[1:3], axis=1)\nx = pd.DataFrame(scale(x))\n\ny = df.iloc[:, 13]\ny = y-1",
"_____no_output_____"
]
],
[
[
"1. Dataset is imported.\n2. The imported dataset is converted into a pandas DataFrame.\n3. Attributes(x) and labels(y) are extracted.",
"_____no_output_____"
]
],
[
[
"x_train, x_test, y_train, y_test = train_test_split(x, y, test_size=0.4)",
"_____no_output_____"
]
],
[
[
"Train/Test split is 0.4",
"_____no_output_____"
],
[
"Plotting the dataset\n--------------------",
"_____no_output_____"
]
],
[
[
"fig = plt.figure()\nax1 = fig.add_subplot(1,2,1)\nax1.scatter(x[1],x[2], c=y)\nax1.set_title(\"Original Data\")",
"_____no_output_____"
]
],
[
[
"Matplotlib is used to plot the loaded pandas DataFrame.",
"_____no_output_____"
],
[
"Learning from the data\n----------------------",
"_____no_output_____"
]
],
[
[
"model = KNeighborsClassifier(n_neighbors=5)\n\n\nscores = cross_val_score(model, x, y, scoring='accuracy', cv=10)\nprint (\"10-Fold Accuracy : \", scores.mean()*100)\n\nmodel.fit(x_train,y_train)\nprint (\"Testing Accuracy : \",model.score(x_test, y_test)*100)\npredicted = model.predict(x)\n",
"_____no_output_____"
]
],
[
[
"Here **model** is an instance of KNeighborsClassifier method from sklearn.neighbors. 10 Fold Cross Validation is used to verify the results.",
"_____no_output_____"
]
],
[
[
"ax2 = fig.add_subplot(1,2,2)\nax2.scatter(x[1],x[2], c=predicted)\nax2.set_title(\"KNearestNeighbours\")",
"_____no_output_____"
]
],
[
[
"The learned data is plotted.",
"_____no_output_____"
]
],
[
[
"cm = metrics.confusion_matrix(y, predicted)\nprint (cm/len(y))\nprint (metrics.classification_report(y, predicted))\n\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"Compute confusion matrix to evaluate the accuracy of a classification and build a text report showing the main classification metrics.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
cb87274b133a27bd44232f77ee1e42742edcd1e4
| 22,819 |
ipynb
|
Jupyter Notebook
|
_notebooks/2021-06-26-Jupyter-SQL-Notebook.ipynb
|
noklam/blog
|
0565d908ca10a043bd9dfc25d8a572b191be3950
|
[
"Apache-2.0"
] | 1 |
2021-03-31T12:51:36.000Z
|
2021-03-31T12:51:36.000Z
|
_notebooks/2021-06-26-Jupyter-SQL-Notebook.ipynb
|
noklam/blog
|
0565d908ca10a043bd9dfc25d8a572b191be3950
|
[
"Apache-2.0"
] | 9 |
2021-03-05T15:08:34.000Z
|
2021-09-28T01:15:51.000Z
|
_notebooks/2021-06-26-Jupyter-SQL-Notebook.ipynb
|
noklam/blog
|
0565d908ca10a043bd9dfc25d8a572b191be3950
|
[
"Apache-2.0"
] | null | null | null | 52.457471 | 7,880 | 0.72054 |
[
[
[
"# Jupyter Superpower - Extend SQL analysis with Python\n> Making collboration with Notebook possible and share perfect SQL analysis with Notebook.\n\n\n- toc: true \n- badges: true\n- comments: true\n- author: noklam\n- categories: [\"python\", \"reviewnb\", \"sql\"]\n- hide: false\n- canonical_url: https://blog.reviewnb.com/jupyter-sql-notebook/",
"_____no_output_____"
],
[
"If you have ever written SQL queries to extract data from a database, chances are you are familiar with an IDE like the screenshot below. The IDE offers features like auto-completion, visualize the query output, display the table schema and the ER diagram. Whenever you need to write a query, this is your go-to tool. However, you may want to add `Jupyter Notebook` into your toolkit. It improves my productivity by complementing some missing features in IDE.",
"_____no_output_____"
],
[
"\")",
"_____no_output_____"
]
],
[
[
"#collapse-hide\n# !pip install ipython_sql\n%load_ext sql\n%config SqlMagic.displaycon = False\n%config SqlMagic.feedback = False\n# Download the file from https://github.com/cwoodruff/ChinookDatabase/blob/master/Scripts/Chinook_Sqlite.sqlite\n%sql sqlite:///sales.sqlite.db\n \nfrom pathlib import Path\nDATA_DIR = Path('../_demo/sql_notebook')\n",
"_____no_output_____"
],
[
"%%sql\nselect ProductId, Sum(Unit) from Sales group by ProductId;",
"_____no_output_____"
]
],
[
[
"## Notebook as a self-contained report",
"_____no_output_____"
],
[
"As a data scientist/data analyst, you write SQL queries for ad-hoc analyses all the time. After getting the right data, you make nice-looking charts and put them in a PowerPoint and you are ready to present your findings. Unlike a well-defined ETL job, you are exploring the data and testing your hypotheses all the time. You make assumptions, which is often wrong but you only realized it after a few weeks. But all you got is a CSV that you cannot recall how it was generated in the first place.\n\nData is not stationary, why should your analysis be? I have seen many screenshots, fragmented scripts flying around in organizations. As a data scientist, I learned that you need to be cautious about what you heard. Don't trust peoples' words easily, verify the result! To achieve that, we need to know exactly how the data was extracted, what kind of assumptions have been made? Unfortunately, this information usually is not available. As a result, people are redoing the same analysis over and over. You will be surprised that this is very common in organizations. In fact, numbers often do not align because every department has its own definition for a given metric. It is not shared among the organization, and verbal communication is inaccurate and error-prone. It would be really nice if anyone in the organization can reproduce the same result with just a single click. Jupyter Notebook can achieve that reproducibility and keep your entire analysis (documentation, data, and code) in the same place.",
"_____no_output_____"
],
[
"## Notebook as an extension of IDE",
"_____no_output_____"
],
[
"Writing SQL queries in a notebook gives you extra flexibility of a full programming language alongside SQL.\nFor example:\n\n* Write complex processing logic that is not easy in pure SQL\n* Create visualizations directly from SQL results without exporting to an intermediate CSV\n\nFor instance, you can pipe your `SQL` query with `pandas` and then make a plot. It allows you to generate analysis with richer content. If you find bugs in your code, you can modify the code and re-run the analysis. This reduces the hustles to reproduce an analysis greatly. In contrast, if your analysis is reading data from an anonymous exported CSV, it is almost guaranteed that the definition of the data will be lost. No one will be able to reproduce the dataset.",
"_____no_output_____"
],
[
"You can make use of the `ipython_sql` library to make queries in a notebook. To do this, you need to use the **magic** function with the inline magic `%` or cell magic `%%`.",
"_____no_output_____"
]
],
[
[
"sales = %sql SELECT * from sales LIMIT 3\nsales",
"_____no_output_____"
]
],
[
[
"To make it fancier, you can even parameterize your query with variables. Tools like [papermill](https://www.bing.com/search?q=github+paramter+notebook&cvid=5b17218ec803438fb1ca41212d53d90a&FORM=ANAB01&PC=U531) allows you to parameterize your notebook. If you execute the notebook regularly with a scheduler, you can get a updated dashboard. To reference the python variable, the `$` sign is used.",
"_____no_output_____"
]
],
[
[
"table = \"sales\"\nquery = f\"SELECT * from {table} LIMIT 3\"\nsales = %sql $query\nsales",
"_____no_output_____"
]
],
[
[
"With a little bit of python code, you can make a nice plot to summarize your finding. You can even make an interactive plot if you want. This is a very powerful way to extend your analysis.",
"_____no_output_____"
]
],
[
[
"import seaborn as sns\nsales = %sql SELECT * FROM SALES\nsales_df = sales.DataFrame()\nsales_df = sales_df.groupby('ProductId', as_index=False).sum()\nax = sns.barplot(x='ProductId', y='Unit', data=sales_df)\nax.set_title('Sales by ProductId');",
"_____no_output_____"
]
],
[
[
"## Notebook as a collaboration tool\nJupyter Notebook is flexible and it fits extremely well with exploratory data analysis. To share to a non-coder, you can share the notebook or export it as an HTML file. They can read the report or any cached executed result. If they need to verify the data or add some extra plots, they can do it easily themselves. \n \nIt is true that Jupyter Notebook has an infamous reputation. It is not friendly to version control, it's hard to collaborate with notebooks. Luckily, there are efforts that make collaboration in notebook a lot easier now.",
"_____no_output_____"
],
[
"Here what I did not show you is that the table has an `isDeleted` column. Some of the records are invalid and we should exclude them. In reality, this happens frequently when you are dealing with hundreds of tables that you are not familiar with. These tables are made for applications, transactions, and they do not have analytic in mind. Data Analytic is usually an afterthought. Therefore, you need to consult the SME or the maintainer of that tables. It takes many iterations to get the correct data that can be used to produce useful insight.",
"_____no_output_____"
],
[
"With [ReviewNB](https://www.reviewnb.com/), you can publish your result and invite some domain expert to review your analysis. This is where notebook shine, this kind of workflow is not possible with just the SQL script or a screenshot of your finding. The notebook itself is a useful documentation and collaboration tool.",
"_____no_output_____"
],
[
"### Step 1 - Review PR online",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"You can view your notebook and add comments on a particular cell on [ReviewNB](https://www.reviewnb.com/). This lowers the technical barrier as your analysts do not have to understand Git. He can review changes and make comments on the web without the need to pull code at all. As soon as your analyst makes a suggestion, you can make changes.",
"_____no_output_____"
],
[
"### Step 2 - Review Changes",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"Once you have made changes to the notebook, you can review it side by side. This is very trivial to do it in your local machine. Without ReviewNB, you have to pull both notebooks separately. As Git tracks line-level changes, you can't really read the changes as it consists of a lot of confusing noise. It would also be impossible to view changes about the chart with git.",
"_____no_output_____"
],
[
"### Step 3 - Resolve Discussion",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"Once the changes are reviewed, you can resolve the discussion and share your insight with the team. You can publish the notebook to internal sharing platform like [knowledge-repo](https://github.com/airbnb/knowledge-repo) to organize the analysis.",
"_____no_output_____"
],
[
"I hope this convince you that Notebook is a good choice for adhoc analytics. It is possible to collaborate with notebook with proper software in place. Regarless if you use notebook or not, you should try your best to document the process. Let's make more reproducible analyses!",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
cb8736760f8329bd5d0a22b82ab3371f9ae76a11
| 14,634 |
ipynb
|
Jupyter Notebook
|
math/0308-6.5-6.6_pgmpy.ipynb
|
pearl-lee/TIL
|
531469156bc92b971ddcb582fc4f79e5ffc22245
|
[
"MIT"
] | null | null | null |
math/0308-6.5-6.6_pgmpy.ipynb
|
pearl-lee/TIL
|
531469156bc92b971ddcb582fc4f79e5ffc22245
|
[
"MIT"
] | null | null | null |
math/0308-6.5-6.6_pgmpy.ipynb
|
pearl-lee/TIL
|
531469156bc92b971ddcb582fc4f79e5ffc22245
|
[
"MIT"
] | null | null | null | 20.905714 | 115 | 0.333128 |
[
[
[
"### pgmpy패키지",
"_____no_output_____"
],
[
"#### 이산확률모형 구현",
"_____no_output_____"
]
],
[
[
"from pgmpy.factors.discrete import JointProbabilityDistribution as JPD\n# 확률변수 X의 확률\npx = JPD(['X'], [2], np.array([12, 8]) / 20)\nprint(px)",
"+-----+--------+\n| X | P(X) |\n+=====+========+\n| X_0 | 0.6000 |\n+-----+--------+\n| X_1 | 0.4000 |\n+-----+--------+\n"
],
[
"# 확률변수 Y의 확률\npy = JPD(['Y'], [2], np.array([10, 10]) / 20)\nprint(py)",
"+-----+--------+\n| Y | P(Y) |\n+=====+========+\n| Y_0 | 0.5000 |\n+-----+--------+\n| Y_1 | 0.5000 |\n+-----+--------+\n"
],
[
"# X, Y의 결합확률\npxy = JPD(['X', 'Y'], [2, 2], np.array([3, 9, 7, 1]) / 20)\nprint(pxy)",
"+-----+-----+----------+\n| X | Y | P(X,Y) |\n+=====+=====+==========+\n| X_0 | Y_0 | 0.1500 |\n+-----+-----+----------+\n| X_0 | Y_1 | 0.4500 |\n+-----+-----+----------+\n| X_1 | Y_0 | 0.3500 |\n+-----+-----+----------+\n| X_1 | Y_1 | 0.0500 |\n+-----+-----+----------+\n"
],
[
"# 주변확률 계산 1\npx = pxy.marginal_distribution(['X'], inplace=False)\nprint(px)",
"+-----+--------+\n| X | P(X) |\n+=====+========+\n| X_0 | 0.6000 |\n+-----+--------+\n| X_1 | 0.4000 |\n+-----+--------+\n"
],
[
"# 주변확률 계산 2\npy = pxy.marginalize(['X'], inplace=False)\nprint(py)",
"+-----+--------+\n| Y | P(Y) |\n+=====+========+\n| Y_0 | 0.5000 |\n+-----+--------+\n| Y_1 | 0.5000 |\n+-----+--------+\n"
],
[
"# 조건부확률 게산\npy_on_x0 = pxy.conditional_distribution([('X', 0)], inplace=False)\nprint((py_on_x0))",
"+-----+--------+\n| Y | P(Y) |\n+=====+========+\n| Y_0 | 0.2500 |\n+-----+--------+\n| Y_1 | 0.7500 |\n+-----+--------+\n"
],
[
"py_on_x1 = pxy.conditional_distribution([('X', 1)], inplace=False)\nprint((py_on_x1))",
"+-----+--------+\n| Y | P(Y) |\n+=====+========+\n| Y_0 | 0.8750 |\n+-----+--------+\n| Y_1 | 0.1250 |\n+-----+--------+\n"
],
[
"px_on_y0 = pxy.conditional_distribution([('Y', 0)], inplace=False)\nprint(px_on_y0)",
"+-----+--------+\n| X | P(X) |\n+=====+========+\n| X_0 | 0.3000 |\n+-----+--------+\n| X_1 | 0.7000 |\n+-----+--------+\n"
],
[
"px_on_y1 = pxy.conditional_distribution([('Y', 1)], inplace=False )\nprint(px_on_y1)",
"+-----+--------+\n| X | P(X) |\n+=====+========+\n| X_0 | 0.9000 |\n+-----+--------+\n| X_1 | 0.1000 |\n+-----+--------+\n"
],
[
"# 두 변수간 독립 확인\npxy.check_independence(['X'], ['Y'])\n# False : px * py != pxy",
"_____no_output_____"
],
[
"print(px * py)",
"+-----+-----+----------+\n| X | Y | P(X,Y) |\n+=====+=====+==========+\n| X_0 | Y_0 | 0.3000 |\n+-----+-----+----------+\n| X_0 | Y_1 | 0.3000 |\n+-----+-----+----------+\n| X_1 | Y_0 | 0.2000 |\n+-----+-----+----------+\n| X_1 | Y_1 | 0.2000 |\n+-----+-----+----------+\n"
],
[
"print(pxy)",
"+-----+-----+----------+\n| X | Y | P(X,Y) |\n+=====+=====+==========+\n| X_0 | Y_0 | 0.1500 |\n+-----+-----+----------+\n| X_0 | Y_1 | 0.4500 |\n+-----+-----+----------+\n| X_1 | Y_0 | 0.3500 |\n+-----+-----+----------+\n| X_1 | Y_1 | 0.0500 |\n+-----+-----+----------+\n"
],
[
"pxy2 = JPD(['X', 'Y'], [2, 2], np.array([6, 6, 4, 4]) / 20)\nprint(pxy2)",
"+-----+-----+----------+\n| X | Y | P(X,Y) |\n+=====+=====+==========+\n| X_0 | Y_0 | 0.3000 |\n+-----+-----+----------+\n| X_0 | Y_1 | 0.3000 |\n+-----+-----+----------+\n| X_1 | Y_0 | 0.2000 |\n+-----+-----+----------+\n| X_1 | Y_1 | 0.2000 |\n+-----+-----+----------+\n"
],
[
"px2 = pxy2.marginal_distribution(['X'], inplace=False)\nprint(px2)",
"+-----+--------+\n| X | P(X) |\n+=====+========+\n| X_0 | 0.6000 |\n+-----+--------+\n| X_1 | 0.4000 |\n+-----+--------+\n"
],
[
"py2 = pxy2.marginal_distribution(['Y'], inplace=False)\nprint(py2)",
"+-----+--------+\n| Y | P(Y) |\n+=====+========+\n| Y_0 | 0.5000 |\n+-----+--------+\n| Y_1 | 0.5000 |\n+-----+--------+\n"
],
[
"px2_on_y0 = pxy2.conditional_distribution([('Y', 0)], inplace=False)\nprint(px2_on_y0)",
"+-----+--------+\n| X | P(X) |\n+=====+========+\n| X_0 | 0.6000 |\n+-----+--------+\n| X_1 | 0.4000 |\n+-----+--------+\n"
],
[
"px2_on_y1 = pxy2.conditional_distribution([('Y', 1)], inplace=False)\nprint(px2_on_y1)",
"+-----+--------+\n| X | P(X) |\n+=====+========+\n| X_0 | 0.6000 |\n+-----+--------+\n| X_1 | 0.4000 |\n+-----+--------+\n"
],
[
"py2_on_x0 = pxy2.conditional_distribution([('X', 0)], inplace=False)\nprint(py2_on_x0)",
"+-----+--------+\n| Y | P(Y) |\n+=====+========+\n| Y_0 | 0.5000 |\n+-----+--------+\n| Y_1 | 0.5000 |\n+-----+--------+\n"
],
[
"py2_on_x1 = pxy2.conditional_distribution([('X', 1)], inplace=False)\nprint(py2_on_x1)",
"+-----+--------+\n| Y | P(Y) |\n+=====+========+\n| Y_0 | 0.5000 |\n+-----+--------+\n| Y_1 | 0.5000 |\n+-----+--------+\n"
],
[
"pxy2.check_independence(['X'], ['Y'])",
"_____no_output_____"
],
[
"print(px2 * py2)",
"+-----+-----+----------+\n| X | Y | P(X,Y) |\n+=====+=====+==========+\n| X_0 | Y_0 | 0.3000 |\n+-----+-----+----------+\n| X_0 | Y_1 | 0.3000 |\n+-----+-----+----------+\n| X_1 | Y_0 | 0.2000 |\n+-----+-----+----------+\n| X_1 | Y_1 | 0.2000 |\n+-----+-----+----------+\n"
],
[
"print(pxy2)",
"+-----+-----+----------+\n| X | Y | P(X,Y) |\n+=====+=====+==========+\n| X_0 | Y_0 | 0.3000 |\n+-----+-----+----------+\n| X_0 | Y_1 | 0.3000 |\n+-----+-----+----------+\n| X_1 | Y_0 | 0.2000 |\n+-----+-----+----------+\n| X_1 | Y_1 | 0.2000 |\n+-----+-----+----------+\n"
]
],
[
[
"#### 베이즈정리",
"_____no_output_____"
]
],
[
[
"from pgmpy.factors.discrete import TabularCPD\n\n# 사전확률\ncpd_x = TabularCPD('X', 2, [[1 - 0.002, 0.002]])\nprint(cpd_x)",
"+-----+-------+\n| X_0 | 0.998 |\n+-----+-------+\n| X_1 | 0.002 |\n+-----+-------+\n"
],
[
"# 조건부확률\ncpd_y_on_x = TabularCPD('Y', 2, np.array([[0.95, 0.01], [0.05, 0.99]]), evidence=['X'], evidence_card=[2])\nprint(cpd_y_on_x)",
"+-----+------+------+\n| X | X_0 | X_1 |\n+-----+------+------+\n| Y_0 | 0.95 | 0.01 |\n+-----+------+------+\n| Y_1 | 0.05 | 0.99 |\n+-----+------+------+\n"
],
[
"# 베이지안 모델\nfrom pgmpy.models import BayesianModel\n\nmodel = BayesianModel([('X', 'Y')]) \nmodel.add_cpds(cpd_x, cpd_y_on_x) # 조건부 확률 추가\nmodel.check_model() # 정상적인 모델인지 확인",
"_____no_output_____"
],
[
"from pgmpy.inference import VariableElimination\n\ninference = VariableElimination(model)\nposterior = inference.query(['X'], evidence={'Y': 1})\nprint(posterior['X'])",
"+-----+----------+\n| X | phi(X) |\n+=====+==========+\n| X_0 | 0.9618 |\n+-----+----------+\n| X_1 | 0.0382 |\n+-----+----------+\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
cb8742aea38acfd3b537cdf3a85e350d364bcadc
| 5,443 |
ipynb
|
Jupyter Notebook
|
Elements/Tuples.ipynb
|
jtentor/PyB2I
|
d3e9e5a6ce67ea877331149888a5a2e172c34774
|
[
"CC0-1.0"
] | null | null | null |
Elements/Tuples.ipynb
|
jtentor/PyB2I
|
d3e9e5a6ce67ea877331149888a5a2e172c34774
|
[
"CC0-1.0"
] | null | null | null |
Elements/Tuples.ipynb
|
jtentor/PyB2I
|
d3e9e5a6ce67ea877331149888a5a2e172c34774
|
[
"CC0-1.0"
] | 3 |
2022-03-22T02:54:57.000Z
|
2022-03-27T06:57:39.000Z
| 29.421622 | 238 | 0.561271 |
[
[
[
"<img src=\"../Images/Level1Beginner.png\" alt=\"Beginner\" width=\"128\" height=\"128\" align=\"right\">\n\n## Tuplas en Python\n\nUna tupla es una secuencia **inmutable** de elementos de cualquier tipo.\n\nSe comporta como una lista en la que no se puede modificar los elementos individuales.\n\nLa discusión sobre listas y tuplas tiene que ver con los mecanismos de gestión de memoria en un entorno de ejecución y sobre la necesidad de contar con valores inmutables para controlar la identidad de claves en conjuntos y mapas.\n",
"_____no_output_____"
],
[
"Una tupla se crea mediante paréntesis encerrando los elementos separados por comas.\n\nEs posible indicar los elementos sin paréntesis asegurándose de incluir una coma al final.\n\nLa función **len()** aplicada a una tupla devuelve la cantidad de elementos en ella.\n",
"_____no_output_____"
]
],
[
[
"\nsample_tuple = (1, 2, 3, 7, 8,5)\n\nprint(\"Ejemplo de tupla:\", sample_tuple)\nprint(\"Tipo:\", type(sample_tuple), \"\\n\")\n\nempty_tuple = ()\n\nprint(\"Tupla vacía:\", empty_tuple, \"tiene\", len(empty_tuple), \"elementos\")\nprint(\"Tipo:\", type(empty_tuple), \"\\n\")\n\none_element_tuple = (\"único elemento\", )\n\nprint(\"Tupla con único elemento:\", one_element_tuple)\nprint(\"Tipo:\", type(one_element_tuple), \"\\n\")\n\nanother_element_tuple = 1,\n\nprint(\"Otra tupla con único elemento:\", another_element_tuple)\nprint(\"Tipo:\", type(another_element_tuple), \"\\n\")\n",
"_____no_output_____"
]
],
[
[
"### Concatenar y Multiplicar\n\nLos operadores “**+**” para concatenar y “**\\***” para multiplicar se pueden utilizar con tuplas.\n",
"_____no_output_____"
]
],
[
[
"\none_element_tuple = (\"único elemento\", )\nanother_element_tuple = 1,\n\nconcatenated_tuple = one_element_tuple + sample_tuple\n\nprint(\"Tupla concatenada:\", concatenated_tuple,\n \"tiene\", len(concatenated_tuple), \"elementos\")\nprint(\"Tipo:\", type(concatenated_tuple), \"\\n\")\n\nmultiplied_tuple = one_element_tuple * 3\n\nprint(\"Tupla multiplicada:\", multiplied_tuple,\n \"tiene\", len(multiplied_tuple), \"elementos\")\nprint(\"Tipo:\", type(multiplied_tuple), \"\\n\")\n",
"_____no_output_____"
]
],
[
[
"### Convertir tuplas a y desde listas\n\nLos **constructores** **list(...)** y **tuple(...)** permiten crear una lista o tupla a partir de una secuencia como argumento. \n",
"_____no_output_____"
]
],
[
[
"sample_tuple = (1, 2, 3, 7, 8,5)\n\nprint(\"Ejemplo de tupla:\", sample_tuple)\nprint(\"Tipo:\", type(sample_tuple), \"\\n\")\n\nsample_list = list(sample_tuple)\n\nprint(\"Ejemplo de lista:\", sample_list)\nprint(\"Tipo:\", type(sample_list), \"\\n\")\n\nanother_tuple = tuple(sample_list)\nprint(\"Otra tupla:\", another_tuple)\nprint(\"Tipo:\", type(another_tuple), \"\\n\")\n",
"_____no_output_____"
]
],
[
[
"<img src=\"../Images/Level2Intermediate.png\" alt=\"Intermediate\" width=\"128\" height=\"128\" align=\"right\">",
"_____no_output_____"
],
[
"### Empaquetar y desempaquetar\n\nLas asignaciones conocidadas como **packing** y **unpacking** permiten asignar multiples valores a una tupla y asignar una tupla a multiples variables.\n",
"_____no_output_____"
]
],
[
[
"print(\"packing ...\")\none_tuple = True, 145, \"Python\"\nprint(\"one_tuple\", one_tuple, \"tipo\", type(one_tuple), \"tiene\", len(one_tuple), \"elementos\")\n\nprint(\"\\nunpacking ...\")\nboolean_var, integer_var, string_var = one_tuple\nprint(\"boolean_var\", boolean_var, \"\\ninteger_var\", integer_var, \"\\nstring_var\", string_var)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
]
] |
cb87518b65c376a4bb18a4d96fc533e318da67f9
| 86,993 |
ipynb
|
Jupyter Notebook
|
PyCitySchools_original_info.ipynb
|
Jimmy-1310/School_District_Analysis
|
705b2986f52f30f1e57798e7db026322da0a2a76
|
[
"MIT"
] | null | null | null |
PyCitySchools_original_info.ipynb
|
Jimmy-1310/School_District_Analysis
|
705b2986f52f30f1e57798e7db026322da0a2a76
|
[
"MIT"
] | null | null | null |
PyCitySchools_original_info.ipynb
|
Jimmy-1310/School_District_Analysis
|
705b2986f52f30f1e57798e7db026322da0a2a76
|
[
"MIT"
] | null | null | null | 35.377389 | 212 | 0.416884 |
[
[
[
"#import pandas\nimport pandas as pd",
"_____no_output_____"
],
[
"import os\n#Load files\nSchool_info_path=os.path.join(\"Resources\",\"schools_complete.csv\")\nStudent_info_path=os.path.join(\"Resources\",\"students_complete.csv\")",
"_____no_output_____"
],
[
"#Read school files\nschool_data_df=pd.read_csv(School_info_path)",
"_____no_output_____"
],
[
"#Read the student info\nstudent_data_df=pd.read_csv(Student_info_path)",
"_____no_output_____"
],
[
"#Determine if there are any missing values in the school data\nschool_data_df.count()\n#Determine data types for the school dataframe\nschool_data_df.dtypes",
"_____no_output_____"
],
[
"#Determine if there are any missing values in student data\nstudent_data_df.count()\n#Determine student data types\nstudent_data_df.dtypes",
"_____no_output_____"
],
[
"#Clean student names\nprefixes_suffixes=[\"Dr. \", \"Mr. \",\"Ms. \", \"Mrs. \", \"Miss \", \" MD\", \" DDS\", \" DVM\", \" PhD\"]\nfor word in prefixes_suffixes:\n student_data_df[\"student_name\"]=student_data_df[\"student_name\"].str.replace(word,\"\")\nstudent_data_df.head(10)",
"C:\\Users\\jaime_1s7\\anaconda3\\envs\\PythonData\\lib\\site-packages\\ipykernel_launcher.py:4: FutureWarning: The default value of regex will change from True to False in a future version.\n after removing the cwd from sys.path.\n"
],
[
"#Merge the school and student dfs\nschool_data_complete_df=pd.merge(student_data_df,school_data_df, on=[\"school_name\",\"school_name\"])",
"_____no_output_____"
],
[
"#Find Student Count\nstudent_count=school_data_complete_df[\"Student ID\"].count()",
"_____no_output_____"
],
[
"#Find school count\nschool_count=len(school_data_complete_df[\"school_name\"].unique())\n",
"_____no_output_____"
],
[
"#Find total budget of district\ntotal_budget=school_data_df[\"budget\"].sum()",
"_____no_output_____"
],
[
"#Calculate reading average\naverage_reading_score=school_data_complete_df[\"reading_score\"].mean()",
"_____no_output_____"
],
[
"#Claculate math average\naverage_math_score=school_data_complete_df[\"math_score\"].mean()",
"_____no_output_____"
],
[
"#Calculate passing math students\npassing_math=school_data_complete_df[school_data_complete_df[\"math_score\"]>=70]\npassing_math_count=passing_math[\"student_name\"].count()",
"_____no_output_____"
],
[
"#Calculate passing reading student\npassing_reading=school_data_complete_df[school_data_complete_df[\"reading_score\"]>=70]\npassing_reading_count=passing_reading[\"student_name\"].count()",
"_____no_output_____"
],
[
"#calculate % of passing math\npassing_math_percentage=passing_math_count/float(student_count) * 100\n#calculate % of passing reading\npassing_reading_percentage=passing_reading_count/float(student_count) * 100",
"_____no_output_____"
],
[
"#get the overral passing student count\npassing_math_reading=school_data_complete_df[(school_data_complete_df[\"math_score\"]>=70) & (school_data_complete_df[\"reading_score\"]>=70)]",
"_____no_output_____"
],
[
"#Calculate overall %\npassing_math_reading_count=passing_math_reading[\"student_name\"].count()\noverall_passing_percentage=passing_math_reading_count/float(student_count) * 100",
"_____no_output_____"
],
[
"#Add metrics to a df\ndistrict_summary_df=pd.DataFrame([{\n \"Total Schools\":school_count,\n \"Total Students\":student_count,\n \"Total Budget\" :total_budget,\n \"Average Math Score\":average_math_score,\n \"Average Reading Score\":average_reading_score,\n \"% Passing Math\":passing_math_percentage,\n \"% Passing Reading\":passing_reading_percentage,\n \"% Overall Passing\":overall_passing_percentage}])",
"_____no_output_____"
],
[
"#Formatting collumns\ndistrict_summary_df[\"Total Students\"]=district_summary_df[\"Total Students\"].map(\"{:,}\".format)\ndistrict_summary_df[\"Total Budget\"]=district_summary_df[\"Total Budget\"].map(\"${:,.2f}\".format)\ndistrict_summary_df[\"Average Reading Score\"]=district_summary_df[\"Average Reading Score\"].map(\"{:.1f}\".format)\ndistrict_summary_df[\"Average Math Score\"]=district_summary_df[\"Average Math Score\"].map(\"{:.1f}\".format)\ndistrict_summary_df[\"% Passing Math\"]=district_summary_df[\"% Passing Math\"].map(\"{:.1f}\".format)\ndistrict_summary_df[\"% Passing Reading\"]=district_summary_df[\"% Passing Reading\"].map(\"{:.1f}\".format)\ndistrict_summary_df[\"% Overall Passing\"]=district_summary_df[\"% Overall Passing\"].map(\"{:.1f}\".format)\n\ndistrict_summary_df",
"_____no_output_____"
],
[
"per_school_types=school_data_df.set_index([\"school_name\"])[\"type\"]",
"_____no_output_____"
],
[
"per_school_df=pd.DataFrame(per_school_types)",
"_____no_output_____"
],
[
"#Get the total students in each hs\nper_school_counts=school_data_df.set_index([\"school_name\"])[\"size\"]",
"_____no_output_____"
],
[
"#Calculate the budget per student in each hs\nper_school_budget=school_data_df.set_index([\"school_name\"])[\"budget\"]\nper_school_capita=per_school_budget/per_school_counts",
"_____no_output_____"
],
[
"#Calculate math scores.\nper_school_math=school_data_complete_df.groupby([\"school_name\"]).mean()[\"math_score\"]\n#Calculate reading scores\nper_school_reading=school_data_complete_df.groupby([\"school_name\"]).mean()[\"reading_score\"]",
"_____no_output_____"
],
[
"#Count the ammount of students passing math\nschool_passing_math=school_data_complete_df[(school_data_complete_df[\"math_score\"]>=70)].groupby([\"school_name\"]).count()[\"student_name\"]\n#Count the ammount of students passing reading\nschool_passing_reading=school_data_complete_df[(school_data_complete_df[\"reading_score\"]>=70)].groupby([\"school_name\"]).count()[\"student_name\"]",
"_____no_output_____"
],
[
"#calculate the % of students passing math\nper_school_passing_math_percentage=school_passing_math/per_school_counts * 100\n#Calculate the % of students passing reding\nper_school_passing_reading_percentage=school_passing_reading/per_school_counts * 100",
"_____no_output_____"
],
[
"#Calculate the students who passed math and reading \nper_passing_math_reading=school_data_complete_df[(school_data_complete_df[\"math_score\"]>=70) & (school_data_complete_df[\"reading_score\"]>=70)].groupby(\"school_name\").count()[\"student_name\"]\nper_school_overall_passing=per_passing_math_reading/per_school_counts * 100",
"_____no_output_____"
],
[
"#Add the per-school metrics to a data frame\nper_school_summary_df=pd.DataFrame({\n \"School Type\":per_school_types,\n \"Total Students\":per_school_counts,\n \"Total School Budget\":per_school_budget,\n \"Per Student Budget\":per_school_capita,\n \"Average Math Score\":per_school_math,\n \"Average Reading Score\":per_school_reading,\n \"% Passing Math\":per_school_passing_math_percentage,\n \"% Passing Reading\":per_school_passing_reading_percentage,\n \"% Overall Passing\":per_school_overall_passing})\n\n",
"_____no_output_____"
],
[
"#Formatting the df\nper_school_summary_df[\"Total School Budget\"] = per_school_summary_df[\"Total School Budget\"].map(\"${:,.2f}\".format)\n\nper_school_summary_df[\"Per Student Budget\"] = per_school_summary_df[\"Per Student Budget\"].map(\"${:,.2f}\".format)\n\n\n# Display the data frame\nper_school_summary_df",
"_____no_output_____"
],
[
"top_schools=per_school_summary_df.sort_values([\"% Overall Passing\"],ascending=False)\ntop_schools.head()",
"_____no_output_____"
],
[
"#Sort and show bottom five schools\nbottom_schools=per_school_summary_df.sort_values([\"% Overall Passing\"],ascending=True)\nbottom_schools.head()",
"_____no_output_____"
],
[
"#Get the students of each grade\nninth_graders=school_data_complete_df[(school_data_complete_df[\"grade\"]==\"9th\")]\ntenth_graders=school_data_complete_df[(school_data_complete_df[\"grade\"]==\"10th\")]\neleventh_graders=school_data_complete_df[(school_data_complete_df[\"grade\"]==\"11th\")]\ntwelfth_graders=school_data_complete_df[(school_data_complete_df[\"grade\"]==\"12th\")] ",
"_____no_output_____"
],
[
"#Get the average math scores for each grade\nninth_graders_math_scores=ninth_graders.groupby([\"school_name\"]).mean()[\"math_score\"]\ntenth_graders_math_scores=tenth_graders.groupby([\"school_name\"]).mean()[\"math_score\"]\neleventh_graders_math_scores=eleventh_graders.groupby([\"school_name\"]).mean()[\"math_score\"]\ntwelfth_graders_math_scores=twelfth_graders.groupby([\"school_name\"]).mean()[\"math_score\"]\n",
"_____no_output_____"
],
[
"#Get the average reading scores for each grade\nninth_graders_reading_scores=ninth_graders.groupby([\"school_name\"]).mean()[\"reading_score\"]\ntenth_graders_reading_scores=tenth_graders.groupby([\"school_name\"]).mean()[\"reading_score\"]\neleventh_graders_reading_scores=eleventh_graders.groupby([\"school_name\"]).mean()[\"reading_score\"]\ntwelfth_graders_reading_scores=twelfth_graders.groupby([\"school_name\"]).mean()[\"reading_score\"]",
"_____no_output_____"
],
[
"#Create a DataFrame with the average math score for each grade\naverage_math_grade={\"9th Grade\":ninth_graders_math_scores,\"10th Grade\":tenth_graders_math_scores,\"11th Grade\":eleventh_graders_math_scores,\"12th Grade\":twelfth_graders_math_scores}\nmath_scores_by_grade=pd.DataFrame(average_math_grade)\n",
"_____no_output_____"
],
[
" # Format each grade column.\nmath_scores_by_grade[\"9th Grade\"] = math_scores_by_grade[\"9th Grade\"].map(\"{:.1f}\".format)\n\nmath_scores_by_grade[\"10th Grade\"] = math_scores_by_grade[\"10th Grade\"].map(\"{:.1f}\".format)\n\nmath_scores_by_grade[\"11th Grade\"] = math_scores_by_grade[\"11th Grade\"].map(\"{:.1f}\".format)\n\nmath_scores_by_grade[\"12th Grade\"] = math_scores_by_grade[\"12th Grade\"].map(\"{:.1f}\".format)\n\n# Remove the index name.\nmath_scores_by_grade.index.name = None\n# Display the DataFrame.\nmath_scores_by_grade",
"_____no_output_____"
],
[
"#Create a DataFrame with the average reading score for each grade\naverage_reading_grade={\"9th Grade\":ninth_graders_reading_scores,\"10th Grade\":tenth_graders_reading_scores,\"11th Grade\":eleventh_graders_reading_scores,\"12th Grade\":twelfth_graders_reading_scores}\nreading_scores_by_grade=pd.DataFrame(average_reading_grade)\nreading_scores_by_grade.head()",
"_____no_output_____"
],
[
"reading_scores_by_grade[\"9th Grade\"] = reading_scores_by_grade[\"9th Grade\"].map(\"{:.1f}\".format)\n\nreading_scores_by_grade[\"10th Grade\"] = reading_scores_by_grade[\"10th Grade\"].map(\"{:.1f}\".format)\n\nreading_scores_by_grade[\"11th Grade\"] = reading_scores_by_grade[\"11th Grade\"].map(\"{:.1f}\".format)\n\nreading_scores_by_grade[\"12th Grade\"] = reading_scores_by_grade[\"12th Grade\"].map(\"{:.1f}\".format)\n\n# Remove the index name.\nreading_scores_by_grade.index.name = None\n# Display the DataFrame.\nreading_scores_by_grade",
"_____no_output_____"
],
[
"#Get the info of the spending bins\nper_school_capita.describe()\n#write the bins\nspending_bins=[0,585,630,645,675]\ngroup_names=[\"<$584\",\"$585-629\",\"$630-644\",\"$645-675\"]",
"_____no_output_____"
],
[
"#Cut the budget per student into the bins\nper_school_capita.groupby(pd.cut(per_school_capita,spending_bins,labels=group_names)).count()",
"_____no_output_____"
],
[
"#Categorize spending base on the bins\nper_school_summary_df[\"Spending Ranges (Per Student)\"]=pd.cut(per_school_capita,spending_bins,labels=group_names)\nper_school_summary_df.index.name=None\nper_school_summary_df.head()",
"_____no_output_____"
],
[
"#Calculate averages for the desire columns\nspending_math_score=per_school_summary_df.groupby([\"Spending Ranges (Per Student)\"]).mean()[\"Average Math Score\"]\nspending_reading_score=per_school_summary_df.groupby([\"Spending Ranges (Per Student)\"]).mean()[\"Average Reading Score\"]\nspending_passing_math=per_school_summary_df.groupby([\"Spending Ranges (Per Student)\"]).mean()[\"% Passing Math\"]\nspending_passing_reading=per_school_summary_df.groupby([\"Spending Ranges (Per Student)\"]).mean()[\"% Passing Reading\"]\nspending_passing_overall=per_school_summary_df.groupby([\"Spending Ranges (Per Student)\"]).mean()[\"% Overall Passing\"]",
"_____no_output_____"
],
[
"#Create the new DataFrame\nspending_summary_df=pd.DataFrame({\n \"Average Math Score\":spending_math_score,\n \"Average Reading Score\":spending_reading_score,\n \"% Passing Math\":spending_passing_math,\n \"% Passing Reading\":spending_passing_reading,\n \"% Overall Passing\":spending_passing_overall})",
"_____no_output_____"
],
[
"# Formatting\nspending_summary_df[\"Average Math Score\"] = spending_summary_df[\"Average Math Score\"].map(\"{:.1f}\".format)\n\nspending_summary_df[\"Average Reading Score\"] = spending_summary_df[\"Average Reading Score\"].map(\"{:.1f}\".format)\n\nspending_summary_df[\"% Passing Math\"] = spending_summary_df[\"% Passing Math\"].map(\"{:.1f}\".format)\n\nspending_summary_df[\"% Passing Reading\"] = spending_summary_df[\"% Passing Reading\"].map(\"{:.1f}\".format)\n\nspending_summary_df[\"% Overall Passing\"] = spending_summary_df[\"% Overall Passing\"].map(\"{:.1f}\".format)\n\nspending_summary_df",
"_____no_output_____"
],
[
"#Devide the schools intro population bins\npopulation_bins=[0,1000,2000,5000]\npopulation_names=[\"Small (<1000)\",\"Medium (1000-2000)\",\"Large (2000-5000)\"]",
"_____no_output_____"
],
[
"#Cut the data into the population bins and add it to the DataFrame\nper_school_summary_df[\"School Size\"]=pd.cut(per_school_summary_df[\"Total Students\"],population_bins,labels=population_names)",
"_____no_output_____"
],
[
"#Get the scores and average for each population size\nsize_math_score=per_school_summary_df.groupby([\"School Size\"]).mean()[\"Average Math Score\"]\nsize_reading_score=per_school_summary_df.groupby([\"School Size\"]).mean()[\"Average Reading Score\"]\nsize_passing_math=per_school_summary_df.groupby([\"School Size\"]).mean()[\"% Passing Math\"]\nsize_passing_reading=per_school_summary_df.groupby([\"School Size\"]).mean()[\"% Passing Reading\"]\nsize_overall_passing=per_school_summary_df.groupby([\"School Size\"]).mean()[\"% Overall Passing\"]",
"_____no_output_____"
],
[
"#Add the info to a data Frame\nsize_summary_df=pd.DataFrame({\n \"Average Math Score\":size_math_score,\n \"Average Reading Score\":size_reading_score,\n \"% Passing Math\":size_passing_math,\n \"% Passing Reading\":size_passing_reading,\n \"% Overall Passing\":size_overall_passing})",
"_____no_output_____"
],
[
"#Format the data frame\nsize_summary_df[\"Average Math Score\"]=size_summary_df[\"Average Math Score\"].map(\"{:.1f}\".format)\nsize_summary_df[\"Average Reading Score\"]=size_summary_df[\"Average Reading Score\"].map(\"{:.1f}\".format)\nsize_summary_df[\"% Passing Math\"]=size_summary_df[\"% Passing Math\"].map(\"{:.1f}\".format)\nsize_summary_df[\"% Passing Reading\"]=size_summary_df[\"% Passing Reading\"].map(\"{:.1f}\".format)\nsize_summary_df[\"% Overall Passing\"]=size_summary_df[\"% Overall Passing\"].map(\"{:.1f}\".format)\nsize_summary_df",
"_____no_output_____"
],
[
"#Get the average and scores by school types\ntype_math_score=per_school_summary_df.groupby([\"School Type\"]).mean()[\"Average Math Score\"]\ntype_reading_score=per_school_summary_df.groupby([\"School Type\"]).mean()[\"Average Reading Score\"]\ntype_passing_math=per_school_summary_df.groupby([\"School Type\"]).mean()[\"% Passing Math\"]\ntype_passing_reading=per_school_summary_df.groupby([\"School Type\"]).mean()[\"% Passing Reading\"]\ntype_overall_passing=per_school_summary_df.groupby([\"School Type\"]).mean()[\"% Overall Passing\"]",
"_____no_output_____"
],
[
"#Add the metrics into a Data Frame\ntype_summary_df=pd.DataFrame({\n \"Average Math Score\":type_math_score,\n \"Average Reading Score\":type_reading_score,\n \"% Passing Math\":type_passing_math,\n \"% Passing Reading\":type_passing_reading,\n \"% Overall Passing\":type_overall_passing})",
"_____no_output_____"
],
[
"#Formatting\n#Format the data frame\ntype_summary_df[\"Average Math Score\"]=type_summary_df[\"Average Math Score\"].map(\"{:.1f}\".format)\ntype_summary_df[\"Average Reading Score\"]=type_summary_df[\"Average Reading Score\"].map(\"{:.1f}\".format)\ntype_summary_df[\"% Passing Math\"]=type_summary_df[\"% Passing Math\"].map(\"{:.1f}\".format)\ntype_summary_df[\"% Passing Reading\"]=type_summary_df[\"% Passing Reading\"].map(\"{:.1f}\".format)\ntype_summary_df[\"% Overall Passing\"]=type_summary_df[\"% Overall Passing\"].map(\"{:.1f}\".format)\ntype_summary_df",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb875c6aaba7683e13390e4b5b27c112e6bd091e
| 2,560 |
ipynb
|
Jupyter Notebook
|
ass1day8.ipynb
|
varunchenna1234/ass1day8-b7
|
257d6f93101a7be84682b5df95f4cc5ff8e4b950
|
[
"Apache-2.0"
] | null | null | null |
ass1day8.ipynb
|
varunchenna1234/ass1day8-b7
|
257d6f93101a7be84682b5df95f4cc5ff8e4b950
|
[
"Apache-2.0"
] | null | null | null |
ass1day8.ipynb
|
varunchenna1234/ass1day8-b7
|
257d6f93101a7be84682b5df95f4cc5ff8e4b950
|
[
"Apache-2.0"
] | null | null | null | 27.234043 | 234 | 0.405859 |
[
[
[
"<a href=\"https://colab.research.google.com/github/varunchenna1234/ass1day8-b7/blob/master/ass1day8.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"ass1 day8-b7------finding fibonacci series using decorators",
"_____no_output_____"
]
],
[
[
"def getInput(calc_arg_fun):\n def wrapfun():\n nterms = int(input(\"Enter number of terms \"))\n calc_arg_fun(nterms)\n return wrapfun()\n@getInput\ndef fibonacci(num):\n n1, n2 = 0, 1\n count = 0\n if nterms <= 0:\n print(\"Please enter a positive integer\")\n elif nterms == 1:\n print(\"Fibonacci sequence upto\",nterms,\":\")\n print(n1)\n else:\n while count < nterms:\n print(n1)\n nth = n1 + n2\n # update values\n n1 = n2\n n2 = nth\n count += 1",
"Enter number of terms 9\n0\n1\n1\n2\n3\n5\n8\n13\n21\n34\n"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
]
] |
cb876d69e66aa740738de1f14c0ad789d287b811
| 400,840 |
ipynb
|
Jupyter Notebook
|
notebook.ipynb
|
timstoenner/going_deep_with_deepnote
|
594734ba0bb1347980a7c488def8473dd6918c42
|
[
"MIT"
] | null | null | null |
notebook.ipynb
|
timstoenner/going_deep_with_deepnote
|
594734ba0bb1347980a7c488def8473dd6918c42
|
[
"MIT"
] | null | null | null |
notebook.ipynb
|
timstoenner/going_deep_with_deepnote
|
594734ba0bb1347980a7c488def8473dd6918c42
|
[
"MIT"
] | null | null | null | 124.368601 | 275,205 | 0.782669 |
[
[
[
"empty"
]
]
] |
[
"empty"
] |
[
[
"empty"
]
] |
cb87716bcbecc8d7a60027922f7091403fad196e
| 23,921 |
ipynb
|
Jupyter Notebook
|
7.20.ipynb
|
wireboyzz/python_zz1
|
09b34fa10d97a7c10d11b2d0507459836ece64ca
|
[
"Apache-2.0"
] | null | null | null |
7.20.ipynb
|
wireboyzz/python_zz1
|
09b34fa10d97a7c10d11b2d0507459836ece64ca
|
[
"Apache-2.0"
] | null | null | null |
7.20.ipynb
|
wireboyzz/python_zz1
|
09b34fa10d97a7c10d11b2d0507459836ece64ca
|
[
"Apache-2.0"
] | null | null | null | 22.482143 | 1,394 | 0.462146 |
[
[
[
"# 函数\n\n- 函数可以用来定义可重复代码,组织和简化\n- 一般来说一个函数在实际开发中为一个小功能\n- 一个类为一个大功能\n- 同样函数的长度不要超过一屏",
"_____no_output_____"
],
[
"Python中的所有函数实际上都是有返回值(return None),\n\n如果你没有设置return,那么Python将不显示None.\n\n如果你设置return,那么将返回出return这个值.",
"_____no_output_____"
]
],
[
[
"def HJN():\n print('Hello')\n return 1000",
"_____no_output_____"
],
[
"b=HJN()\nprint(b)",
"Hello\n1000\n"
],
[
"HJN",
"_____no_output_____"
],
[
"def panduan(number):\n if number % 2 == 0:\n print('O')\n else:\n print('J')",
"_____no_output_____"
],
[
"panduan(number=1)",
"J\n"
],
[
"panduan(2)",
"O\n"
]
],
[
[
"## 定义一个函数\n\ndef function_name(list of parameters):\n \n do something\n\n- 以前使用的random 或者range 或者print.. 其实都是函数或者类",
"_____no_output_____"
],
[
"函数的参数如果有默认值的情况,当你调用该函数的时候:\n可以不给予参数值,那么就会走该参数的默认值\n否则的话,就走你给予的参数值.",
"_____no_output_____"
]
],
[
[
"import random",
"_____no_output_____"
],
[
"def hahah():\n n = random.randint(0,5)\n while 1:\n N = eval(input('>>'))\n if n == N:\n print('smart')\n break\n elif n < N:\n print('太小了')\n elif n > N:\n print('太大了')\n",
"_____no_output_____"
]
],
[
[
"## 调用一个函数\n- functionName()\n- \"()\" 就代表调用",
"_____no_output_____"
]
],
[
[
"def H():\n print('hahaha')",
"_____no_output_____"
],
[
"def B():\n H()",
"_____no_output_____"
],
[
"B()",
"hahaha\n"
],
[
"def A(f):\n f()",
"_____no_output_____"
],
[
"A(B)",
"hahaha\n"
]
],
[
[
"",
"_____no_output_____"
],
[
"## 带返回值和不带返回值的函数\n- return 返回的内容\n- return 返回多个值\n- 一般情况下,在多个函数协同完成一个功能的时候,那么将会有返回值",
"_____no_output_____"
],
[
"\n\n- 当然也可以自定义返回None",
"_____no_output_____"
],
[
"## EP:\n",
"_____no_output_____"
]
],
[
[
"def main():\n print(min(min(5,6),(51,6)))\ndef min(n1,n2):\n a = n1\n if n2 < a:\n a = n2",
"_____no_output_____"
],
[
"main()",
"_____no_output_____"
]
],
[
[
"## 类型和关键字参数\n- 普通参数\n- 多个参数\n- 默认值参数\n- 不定长参数",
"_____no_output_____"
],
[
"## 普通参数",
"_____no_output_____"
],
[
"## 多个参数",
"_____no_output_____"
],
[
"## 默认值参数",
"_____no_output_____"
],
[
"## 强制命名",
"_____no_output_____"
]
],
[
[
"def U(str_):\n xiaoxie = 0\n for i in str_:\n ASCII = ord(i)\n if 97<=ASCII<=122:\n xiaoxie +=1\n elif xxxx:\n daxie += 1\n elif xxxx:\n shuzi += 1\n return xiaoxie,daxie,shuzi",
"_____no_output_____"
],
[
"U('HJi12')",
"H\nJ\ni\n1\n2\n"
]
],
[
[
"## 不定长参数\n- \\*args\n> - 不定长,来多少装多少,不装也是可以的\n - 返回的数据类型是元组\n - args 名字是可以修改的,只是我们约定俗成的是args\n- \\**kwargs \n> - 返回的字典\n - 输入的一定要是表达式(键值对)\n- name,\\*args,name2,\\**kwargs 使用参数名",
"_____no_output_____"
]
],
[
[
"def TT(a,b)",
"_____no_output_____"
],
[
"def TT(*args,**kwargs):\n print(kwargs)\n print(args)\nTT(1,2,3,4,6,a=100,b=1000)",
"{'a': 100, 'b': 1000}\n(1, 2, 3, 4, 6)\n"
],
[
"{'key':'value'}",
"()\n"
],
[
"TT(1,2,4,5,7,8,9,)",
"(1, 2, 4, 5, 7, 8, 9)\n"
],
[
"def B(name1,nam3):\n pass",
"_____no_output_____"
],
[
"B(name1=100,2)",
"_____no_output_____"
],
[
"def sum_(*args,A='sum'):\n \n res = 0\n count = 0\n for i in args:\n res +=i\n count += 1\n if A == \"sum\":\n return res\n elif A == \"mean\":\n mean = res / count\n return res,mean\n else:\n print(A,'还未开放')\n \n ",
"_____no_output_____"
],
[
"sum_(-1,0,1,4,A='var')",
"var 还未开放\n"
],
[
"'aHbK134'.__iter__",
"_____no_output_____"
],
[
"b = 'asdkjfh'\nfor i in b :\n print(i)",
"a\ns\nd\nk\nj\nf\nh\n"
],
[
"2,5\n2 + 22 + 222 + 2222 + 22222",
"_____no_output_____"
]
],
[
[
"## 变量的作用域\n- 局部变量 local\n- 全局变量 global\n- globals 函数返回一个全局变量的字典,包括所有导入的变量\n- locals() 函数会以字典类型返回当前位置的全部局部变量。",
"_____no_output_____"
]
],
[
[
"a = 1000\nb = 10\ndef Y():\n global a,b\n a += 100\n print(a)\nY()",
"1100\n"
],
[
"def YY(a1):\n a1 += 100\n print(a1)\nYY(a)\nprint(a)",
"1200\n1100\n"
]
],
[
[
"## 注意:\n- global :在进行赋值操作的时候需要声明\n- 官方解释:This is because when you make an assignment to a variable in a scope, that variable becomes local to that scope and shadows any similarly named variable in the outer scope.\n- ",
"_____no_output_____"
],
[
"# Homework\n- 1\n",
"_____no_output_____"
]
],
[
[
"def getPentagonalNumber(n):\n \"\"\"\n 功能:计算五角数\n 参数:n:循环的数值\n \"\"\"\n return n*(3*n-1)/2\ncount =0\nfor n in range(1,101):\n if count <9:\n print( \"%.0f \"%getPentagonalNumber(n),end=\"\")\n count += 1\n else:\n print( \"%.0f\"%getPentagonalNumber(n))\n count = 0\n",
"1 5 12 22 35 51 70 92 117 145\n176 210 247 287 330 376 425 477 532 590\n651 715 782 852 925 1001 1080 1162 1247 1335\n1426 1520 1617 1717 1820 1926 2035 2147 2262 2380\n2501 2625 2752 2882 3015 3151 3290 3432 3577 3725\n3876 4030 4187 4347 4510 4676 4845 5017 5192 5370\n5551 5735 5922 6112 6305 6501 6700 6902 7107 7315\n7526 7740 7957 8177 8400 8626 8855 9087 9322 9560\n9801 10045 10292 10542 10795 11051 11310 11572 11837 12105\n12376 12650 12927 13207 13490 13776 14065 14357 14652 14950\n"
]
],
[
[
"- 2 \n",
"_____no_output_____"
]
],
[
[
"n = float(input())\ndef sumDigits(n):\n bai = n // 100\n shi = n // 10 % 10\n ge = n % 100 % 10\n sum = bai + shi + ge\n return sum\nprint(sumDigits(n))",
"234\n9.0\n"
]
],
[
[
"- 3\n",
"_____no_output_____"
]
],
[
[
"n = float(input())\ndef sumDigits(n):\n bai = n // 100\n shi = n // 10 % 10\n ge = n % 100 % 10\n sum = bai + shi + ge\n return sum\nprint(sumDigits(n))",
"_____no_output_____"
]
],
[
[
"- 4\n",
"_____no_output_____"
]
],
[
[
"touzie = input('The amount invested : ')\nnialilv = input('Annual interest rate : ')\ndef FutureInvestmentValue(investmentAmount,monthlyinterestRate,years):\n \n ",
"_____no_output_____"
]
],
[
[
"- 5\n",
"_____no_output_____"
]
],
[
[
"def printChars(ch1,ch2,numberPerLine):\n '''\n 参数没用\n 功能遍历从1到Z所有字符\n '''\n print(ord(\"1\"),ord(\"Z\"))\nfor i in range(49,91):\n if i%10 == 0:\n print(' ')\n print(chr(i),end = ' ')",
"1 \n2 3 4 5 6 7 8 9 : ; \n< = > ? @ A B C D E \nF G H I J K L M N O \nP Q R S T U V W X Y \nZ "
]
],
[
[
"- 6\n",
"_____no_output_____"
]
],
[
[
"def numberOfDaysInAYear(year):\n \"\"\"\n 功能:计算闰年\n 参数:\n year:年份\n \"\"\"\n days = 0\n if (year % 400 == 0) or (year % 4 == 0) and (year % 100 != 0):\n days = 366\n else:\n days = 365\n return days\nday1 = int(input(\">>\"))\nday2 = int(input(\">>\"))\nsum = 0\nfor i in range(day1,day2+1):\n sum = numberOfDaysInAYear(i)\n print(\"%d 年的天数为:%d\"%(i,sum))",
"_____no_output_____"
]
],
[
[
"- 7\n",
"_____no_output_____"
]
],
[
[
"重写个锤子!!!",
"_____no_output_____"
]
],
[
[
"- 8\n",
"_____no_output_____"
],
[
"- 9\n\n",
"_____no_output_____"
]
],
[
[
"def haomiaoshu():\n import time\n localtime = time.asctime(time.localtime(time.time()))\n print(\"本地时间为 :\", localtime)\n",
"本地时间为 : Sat May 11 08:37:21 2019\n"
]
],
[
[
"- 10\n",
"_____no_output_____"
]
],
[
[
"import random\ndef dice(x,y):\n \"\"\" \n 功能:计算骰子数的和,进行判断胜负\n 参数: x,y:随机生成的骰子数\n \"\"\"\n ying = [7,11]\n shu = [2,3,12]\n other = [4,5,6,8,9,10]\n count = 0\n if(x+y in shu):\n print(\"You shu\")\n elif(x+y in ying):\n print(\"You ying\")\n elif (x+y in other):\n count += 1 \n print(\"point is %d\"%(x+y))\n num1 = random.randint(1,6)\n num2 = random.randint(1,6)\n print(\"You rolled %d + %d = %d\"%(num1,num2,num1+num2))\n while num1+num2 != x+y or num1+num2 != 7:\n if num1+num2 == 7:\n print(\"ni shu le\")\n break\n if num1+num2 == x+y:\n print(\"ni ying le\")\n break\n num1 = random.randint(1,6)\n num2 = random.randint(1,6)\n print(\"You rolled %d + %d = %d\"%(num1,num2,num1+num2))\n\nx = random.randint(1,6)\ny = random.randint(1,6) \nprint(\"You rolled %d + %d = %d\"%(x,y,x+y))\ndice(x,y)",
"_____no_output_____"
]
],
[
[
"- 11 \n### 去网上寻找如何用Python代码发送邮件",
"_____no_output_____"
]
],
[
[
"import smtplib\nimport email\nfrom email.mime.text import MIMEText\nfrom email.mime.image import MIMEImage\nfrom email.mime.multipart import MIMEMultipart\n# 设置邮箱的域名\nHOST = 'smtp.qq.com'\n# 设置邮件标题\nSUBJECT = 'csdn博客代码'\n# 设置发件人邮箱\nFROM = '发件人邮箱@qq.com'\n# 设置收件人邮箱\nTO = '邮箱[email protected],邮箱[email protected]'\nmessage = MIMEMultipart('related')\n \n#--------------------------------------发送文本-----------------\n# 发送邮件主体到对方的邮箱中\nmessage_html = MIMEText('<h2 style=\"color:red;font-size:100px\">CSDN博客超级好</h2><img src=\"cid:big\">','html','utf-8')\nmessage.attach(message_html)\n \n#-------------------------------------发送图片--------------------\n# rb 读取二进制文件\n# 要确定当前目录有1.jpg这个文件\nimage_data = open('1.jpg','rb')\n# 设置读取获取的二进制数据\nmessage_image = MIMEImage(image_data.read())\n# 关闭刚才打开的文件\nimage_data.close()\nmessage_image.add_header('Content-ID','big')\n# 添加图片文件到邮件信息当中去\n# message.attach(message_image)\n \n#-------------------------------------添加文件---------------------\n# 要确定当前目录有table.xls这个文件\nmessage_xlsx = MIMEText(open('table.xls','rb').read(),'base64','utf-8')\n# 设置文件在附件当中的名字\nmessage_xlsx['Content-Disposition'] = 'attachment;filename=\"test1111.xlsx\"'\nmessage.attach(message_xlsx)\n \n# 设置邮件发件人\nmessage['From'] = FROM\n# 设置邮件收件人\nmessage['To'] = TO\n# 设置邮件标题\nmessage['Subject'] = SUBJECT\n \n# 获取简单邮件传输协议的证书\nemail_client = smtplib.SMTP_SSL()\n# 设置发件人邮箱的域名和端口,端口为465\nemail_client.connect(HOST,'465')\n# ---------------------------邮箱授权码------------------------------\nresult = email_client.login(FROM,'邮箱授权码')\nprint('登录结果',result)\nemail_client.sendmail(from_addr=FROM,to_addrs=TO.split(','),msg=message.as_string())\n# 关闭邮件发送客户端\nemail_client.close()",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb8779252a0db1a63711b94d3a599e7a50a9c37b
| 221,105 |
ipynb
|
Jupyter Notebook
|
Clustering neighbourhoods in Toronto.ipynb
|
ZRQ-rikkie/coursera-python
|
3be75d9da2ed86ca9bff8d231d26667540c7cc52
|
[
"Apache-2.0"
] | null | null | null |
Clustering neighbourhoods in Toronto.ipynb
|
ZRQ-rikkie/coursera-python
|
3be75d9da2ed86ca9bff8d231d26667540c7cc52
|
[
"Apache-2.0"
] | null | null | null |
Clustering neighbourhoods in Toronto.ipynb
|
ZRQ-rikkie/coursera-python
|
3be75d9da2ed86ca9bff8d231d26667540c7cc52
|
[
"Apache-2.0"
] | null | null | null | 717.873377 | 59,044 | 0.787685 |
[
[
[
"<h2>Segmenting and Clustering Neighbourhoods in Toronto</h2>\n\nThe project includes scraping the Wikipedia page for the postal codes of Canada and then process and clean the data for the clustering. The clustering is carried out by K Means and the clusters are plotted using the Folium Library. The Boroughs containing the name 'Toronto' in it are first plotted and then clustered and plotted again.\n\n\n\n<h3>All the 3 tasks of <i>web scraping</i>, <i>cleaning</i> and <i>clustering</i> are implemented in the same notebook for the ease of evaluation.</h3>",
"_____no_output_____"
],
[
"<h3>Installing and Importing the required Libraries</h3>",
"_____no_output_____"
]
],
[
[
"!pip install beautifulsoup4\n!pip install lxml\nimport requests # library to handle requests\nimport pandas as pd # library for data analsysis\nimport numpy as np # library to handle data in a vectorized manner\nimport random # library for random number generation\n\n#!conda install -c conda-forge geopy --yes \nfrom geopy.geocoders import Nominatim # module to convert an address into latitude and longitude values\n\n# libraries for displaying images\nfrom IPython.display import Image \nfrom IPython.core.display import HTML \n\n\nfrom IPython.display import display_html\nimport pandas as pd\nimport numpy as np\n \n# tranforming json file into a pandas dataframe library\nfrom pandas.io.json import json_normalize\n\n!conda install -c conda-forge folium=0.5.0 --yes\nimport folium # plotting library\nfrom bs4 import BeautifulSoup\nfrom sklearn.cluster import KMeans\nimport matplotlib.cm as cm\nimport matplotlib.colors as colors\n\nprint('Folium installed')\nprint('Libraries imported.')",
"Requirement already satisfied: beautifulsoup4 in /opt/conda/envs/Python36/lib/python3.6/site-packages (4.7.1)\nRequirement already satisfied: soupsieve>=1.2 in /opt/conda/envs/Python36/lib/python3.6/site-packages (from beautifulsoup4) (1.7.1)\nRequirement already satisfied: lxml in /opt/conda/envs/Python36/lib/python3.6/site-packages (4.3.1)\nSolving environment: done\n\n## Package Plan ##\n\n environment location: /opt/conda/envs/Python36\n\n added / updated specs: \n - folium=0.5.0\n\n\nThe following packages will be downloaded:\n\n package | build\n ---------------------------|-----------------\n altair-3.1.0 | py36_0 724 KB conda-forge\n vincent-0.4.4 | py_1 28 KB conda-forge\n certifi-2019.6.16 | py36_1 149 KB conda-forge\n openssl-1.1.1c | h516909a_0 2.1 MB conda-forge\n branca-0.3.1 | py_0 25 KB conda-forge\n folium-0.5.0 | py_0 45 KB conda-forge\n ca-certificates-2019.6.16 | hecc5488_0 145 KB conda-forge\n ------------------------------------------------------------\n Total: 3.2 MB\n\nThe following NEW packages will be INSTALLED:\n\n altair: 3.1.0-py36_0 conda-forge\n branca: 0.3.1-py_0 conda-forge\n folium: 0.5.0-py_0 conda-forge\n vincent: 0.4.4-py_1 conda-forge\n\nThe following packages will be UPDATED:\n\n ca-certificates: 2019.5.15-0 --> 2019.6.16-hecc5488_0 conda-forge\n certifi: 2019.6.16-py36_0 --> 2019.6.16-py36_1 conda-forge\n\nThe following packages will be DOWNGRADED:\n\n openssl: 1.1.1c-h7b6447c_1 --> 1.1.1c-h516909a_0 conda-forge\n\n\nDownloading and Extracting Packages\naltair-3.1.0 | 724 KB | ##################################### | 100% \nvincent-0.4.4 | 28 KB | ##################################### | 100% \ncertifi-2019.6.16 | 149 KB | ##################################### | 100% \nopenssl-1.1.1c | 2.1 MB | ##################################### | 100% \nbranca-0.3.1 | 25 KB | ##################################### | 100% \nfolium-0.5.0 | 45 KB | ##################################### | 100% \nca-certificates-2019 | 145 KB | ##################################### | 100% \nPreparing transaction: done\nVerifying transaction: done\nExecuting transaction: done\nFolium installed\nLibraries imported.\n"
]
],
[
[
"<h3>Scraping the Wikipedia page for the table of postal codes of Canada</h3>\n\nBeautifulSoup Library of Python is used for web scraping of table from the Wikipedia. The title of the webpage is printed to check if the page has been scraped successfully or not. Then the table of postal codes of Canada is printed.",
"_____no_output_____"
]
],
[
[
"source = requests.get('https://en.wikipedia.org/wiki/List_of_postal_codes_of_Canada:_M').text\nsoup=BeautifulSoup(source,'lxml')\nprint(soup.title)\nfrom IPython.display import display_html\ntab = str(soup.table)\ndisplay_html(tab,raw=True)",
"<title>List of postal codes of Canada: M - Wikipedia</title>\n"
]
],
[
[
"<h3>The html table is converted to Pandas DataFrame for cleaning and preprocessing.</h3>",
"_____no_output_____"
]
],
[
[
"dfs = pd.read_html(tab)\ndf=dfs[0]\ndf.head()",
"_____no_output_____"
]
],
[
[
"<h3>Data preprocessing and cleaning</h3>",
"_____no_output_____"
]
],
[
[
"# Dropping the rows where Borough is 'Not assigned'\ndf1 = df[df.Borough != 'Not assigned']\n\n# Combining the neighbourhoods with same Postalcode\ndf2 = df1.groupby(['Postcode','Borough'], sort=False).agg(', '.join)\ndf2.reset_index(inplace=True)\n\n# Replacing the name of the neighbourhoods which are 'Not assigned' with names of Borough\ndf2['Neighbourhood'] = np.where(df2['Neighbourhood'] == 'Not assigned',df2['Borough'], df2['Neighbourhood'])\n\ndf2",
"_____no_output_____"
],
[
"# Shape of data frame\ndf2.shape",
"_____no_output_____"
]
],
[
[
"<h3>Importing the csv file conatining the latitudes and longitudes for various neighbourhoods in Canada</h3>",
"_____no_output_____"
]
],
[
[
"lat_lon = pd.read_csv('https://cocl.us/Geospatial_data')\nlat_lon.head()",
"_____no_output_____"
]
],
[
[
"<h3>Merging the two tables for getting the Latitudes and Longitudes for various neighbourhoods in Canada</h3>",
"_____no_output_____"
]
],
[
[
"lat_lon.rename(columns={'Postal Code':'Postcode'},inplace=True)\ndf3 = pd.merge(df2,lat_lon,on='Postcode')\ndf3.head()",
"_____no_output_____"
]
],
[
[
"<h2>The notebook from here includes the Clustering and the plotting of the neighbourhoods of Canada which contain Toronto in their Borough</h2>",
"_____no_output_____"
],
[
"<h3>Getting all the rows from the data frame which contains Toronto in their Borough.</h3>",
"_____no_output_____"
]
],
[
[
"df4 = df3[df3['Borough'].str.contains('Toronto',regex=False)]\ndf4",
"_____no_output_____"
]
],
[
[
"<h3>Visualizing all the Neighbourhoods of the above data frame using Folium</h3>",
"_____no_output_____"
]
],
[
[
"map_toronto = folium.Map(location=[43.651070,-79.347015],zoom_start=10)\n\nfor lat,lng,borough,neighbourhood in zip(df4['Latitude'],df4['Longitude'],df4['Borough'],df4['Neighbourhood']):\n label = '{}, {}'.format(neighbourhood, borough)\n label = folium.Popup(label, parse_html=True)\n folium.CircleMarker(\n [lat,lng],\n radius=5,\n popup=label,\n color='blue',\n fill=True,\n fill_color='#3186cc',\n fill_opacity=0.7,\n parse_html=False).add_to(map_toronto)\nmap_toronto",
"_____no_output_____"
]
],
[
[
"<h3>The map might not be visible on Github. Check out the README for the map.</h3>",
"_____no_output_____"
],
[
"<h3>Using KMeans clustering for the clsutering of the neighbourhoods</h3>",
"_____no_output_____"
]
],
[
[
"k=5\ntoronto_clustering = df4.drop(['Postcode','Borough','Neighbourhood'],1)\nkmeans = KMeans(n_clusters = k,random_state=0).fit(toronto_clustering)\nkmeans.labels_\ndf4.insert(0, 'Cluster Labels', kmeans.labels_)\n",
"_____no_output_____"
],
[
"df4",
"_____no_output_____"
],
[
"# create map\nmap_clusters = folium.Map(location=[43.651070,-79.347015],zoom_start=10)\n\n# set color scheme for the clusters\nx = np.arange(k)\nys = [i + x + (i*x)**2 for i in range(k)]\ncolors_array = cm.rainbow(np.linspace(0, 1, len(ys)))\nrainbow = [colors.rgb2hex(i) for i in colors_array]\n\n# add markers to the map\nmarkers_colors = []\nfor lat, lon, neighbourhood, cluster in zip(df4['Latitude'], df4['Longitude'], df4['Neighbourhood'], df4['Cluster Labels']):\n label = folium.Popup(' Cluster ' + str(cluster), parse_html=True)\n folium.CircleMarker(\n [lat, lon],\n radius=5,\n popup=label,\n color=rainbow[cluster-1],\n fill=True,\n fill_color=rainbow[cluster-1],\n fill_opacity=0.7).add_to(map_clusters)\n \nmap_clusters",
"_____no_output_____"
]
],
[
[
"<h3>The map might not be visible on Github. Check out the README for the map.</h3>",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
]
] |
cb877aa4558752d447a896bc380c72783c832317
| 47,415 |
ipynb
|
Jupyter Notebook
|
ANN_model/create_credible_file_structures_and_convert_articles_requests_to_json.ipynb
|
Femme-js/Hoaxify-
|
1e4cde470274a9d0a2f4e0b4499ac7d0491956ab
|
[
"MIT"
] | 11 |
2020-06-26T06:11:20.000Z
|
2021-12-17T11:26:31.000Z
|
ANN_model/create_credible_file_structures_and_convert_articles_requests_to_json.ipynb
|
Femme-js/Hoaxify-
|
1e4cde470274a9d0a2f4e0b4499ac7d0491956ab
|
[
"MIT"
] | 35 |
2020-12-29T14:18:27.000Z
|
2021-02-23T04:18:13.000Z
|
ANN_model/create_credible_file_structures_and_convert_articles_requests_to_json.ipynb
|
Femme-js/Hoaxify-
|
1e4cde470274a9d0a2f4e0b4499ac7d0491956ab
|
[
"MIT"
] | 19 |
2020-07-14T07:40:26.000Z
|
2021-02-21T07:30:07.000Z
| 67.542735 | 216 | 0.690583 |
[
[
[
"#Import Required Packages\nimport requests\nimport time\nimport schedule\nimport os\nimport json\nimport newspaper\nfrom bs4 import BeautifulSoup\nfrom datetime import datetime\nfrom newspaper import fulltext\nimport newspaper\nimport pandas as pd\nimport numpy as np\nimport pickle",
"_____no_output_____"
],
[
"#Set Today's Date\n#dates = [datetime.today().strftime('%m-%d-%y')]\ndates = [datetime.today().strftime('%m-%d')]",
"_____no_output_____"
]
],
[
[
"### Define Urls for Newsapi",
"_____no_output_____"
]
],
[
[
"#Define urls for newsapi\nurls=[\n 'https://newsapi.org/v2/top-headlines?sources=associated-press&apiKey=3fb3c0c1e622430b8df3f9693c7a55b4',\n 'https://newsapi.org/v2/top-headlines?sources=independent&apiKey=3fb3c0c1e622430b8df3f9693c7a55b4',\n 'https://newsapi.org/v2/top-headlines?sources=bbc-news&apiKey=3fb3c0c1e622430b8df3f9693c7a55b4',\n 'https://newsapi.org/v2/top-headlines?sources=reuters&apiKey=3fb3c0c1e622430b8df3f9693c7a55b4',\n 'https://newsapi.org/v2/top-headlines?sources=the-wall-street-journal&apiKey=3fb3c0c1e622430b8df3f9693c7a55b4',\n 'https://newsapi.org/v2/top-headlines?sources=the-washington-post&apiKey=3fb3c0c1e622430b8df3f9693c7a55b4',\n 'https://newsapi.org/v2/top-headlines?sources=national-geographic&apiKey=3fb3c0c1e622430b8df3f9693c7a55b4',\n 'https://newsapi.org/v2/top-headlines?sources=usa-today&apiKey=3fb3c0c1e622430b8df3f9693c7a55b4',\n 'https://newsapi.org/v2/top-headlines?sources=cnn&apiKey=3fb3c0c1e622430b8df3f9693c7a55b4',\n 'https://newsapi.org/v2/top-headlines?sources=fox-news&apiKey=3fb3c0c1e622430b8df3f9693c7a55b4',\n 'https://newsapi.org/v2/top-headlines?sources=al-jazeera-english&apiKey=3fb3c0c1e622430b8df3f9693c7a55b4',\n 'https://newsapi.org/v2/top-headlines?sources=bloomberg&apiKey=3fb3c0c1e622430b8df3f9693c7a55b4',\n 'https://newsapi.org/v2/top-headlines?sources=business-insider&apiKey=3fb3c0c1e622430b8df3f9693c7a55b4',\n 'https://newsapi.org/v2/top-headlines?sources=cnbc&apiKey=3fb3c0c1e622430b8df3f9693c7a55b4',\n 'https://newsapi.org/v2/top-headlines?sources=the-new-york-times&apiKey=3fb3c0c1e622430b8df3f9693c7a55b4',\n 'https://newsapi.org/v2/top-headlines?sources=new-scientist&apiKey=3fb3c0c1e622430b8df3f9693c7a55b4',\n 'https://newsapi.org/v2/top-headlines?sources=news-com-au&apiKey=3fb3c0c1e622430b8df3f9693c7a55b4',\n 'https://newsapi.org/v2/top-headlines?sources=newsweek&apiKey=3fb3c0c1e622430b8df3f9693c7a55b4',\n 'https://newsapi.org/v2/top-headlines?sources=the-economist&apiKey=3fb3c0c1e622430b8df3f9693c7a55b4',\n 'https://newsapi.org/v2/top-headlines?sources=the-hill&apiKey=3fb3c0c1e622430b8df3f9693c7a55b4',\n 'https://newsapi.org/v2/top-headlines?sources=the-huffington-post&apiKey=3fb3c0c1e622430b8df3f9693c7a55b4',\n 'https://newsapi.org/v2/top-headlines?sources=the-next-web&apiKey=3fb3c0c1e622430b8df3f9693c7a55b4',\n 'https://newsapi.org/v2/top-headlines?sources=the-telegraph&apiKey=3fb3c0c1e622430b8df3f9693c7a55b4',\n 'https://newsapi.org/v2/top-headlines?sources=the-washington-times&apiKey=3fb3c0c1e622430b8df3f9693c7a55b4',\n 'https://newsapi.org/v2/top-headlines?sources=time&apiKey=3fb3c0c1e622430b8df3f9693c7a55b4',\n 'https://newsapi.org/v2/top-headlines?sources=the-jerusalem-post&apiKey=3fb3c0c1e622430b8df3f9693c7a55b4',\n 'https://newsapi.org/v2/top-headlines?sources=the-irish-times&apiKey=3fb3c0c1e622430b8df3f9693c7a55b4',\n 'https://newsapi.org/v2/top-headlines?sources=the-globe-and-mail&apiKey=3fb3c0c1e622430b8df3f9693c7a55b4',\n 'https://newsapi.org/v2/top-headlines?sources=the-american-conservative&apiKey=3fb3c0c1e622430b8df3f9693c7a55b4',\n 'https://newsapi.org/v2/top-headlines?sources=techcrunch-cn&apiKey=3fb3c0c1e622430b8df3f9693c7a55b4',\n 'https://newsapi.org/v2/top-headlines?sources=recode&apiKey=3fb3c0c1e622430b8df3f9693c7a55b4'\n]",
"_____no_output_____"
]
],
[
[
"### Develop news site folder structure and write top 10 headline urls to API file",
"_____no_output_____"
]
],
[
[
"for date in dates: \n print('saving {} ...'.format(date))\n for url in urls:\n r = requests.get(url)\n source = url.replace('https://newsapi.org/v2/top-headlines?sources=','').replace('&apiKey=3fb3c0c1e622430b8df3f9693c7a55b4','')\n print(source)\n filename = './data/Credible/{0}/articles/{1}/api.txt'.format(source, date)\n os.makedirs(os.path.dirname(filename), exist_ok=True)\n \n with open(filename, 'w') as f:\n json.dump(json.loads(r.text), f)\n \n print('Finished')",
"saving 05-24 ...\nassociated-press\nindependent\nbbc-news\nreuters\nthe-wall-street-journal\nthe-washington-post\nnational-geographic\nusa-today\ncnn\nfox-news\nal-jazeera-english\nbloomberg\nbusiness-insider\ncnbc\nthe-new-york-times\nnew-scientist\nnews-com-au\nnewsweek\nthe-economist\nthe-hill\nthe-huffington-post\nthe-next-web\nthe-telegraph\nthe-washington-times\ntime\nthe-jerusalem-post\nthe-irish-times\nthe-globe-and-mail\nthe-american-conservative\ntechcrunch-cn\nrecode\nFinished\n"
]
],
[
[
"### From individual API files, download news source link and extract text using newspaper python package",
"_____no_output_____"
]
],
[
[
"def saving_json():\n \n print('saving ...')\n for url in urls:\n url = url.strip()\n for date in dates: \n source = url.replace('https://newsapi.org/v2/top-headlines?sources=','').replace('&apiKey=3fb3c0c1e622430b8df3f9693c7a55b4','')\n print(source)\n sourcename = './data/Credible/{0}/articles/{1}/api.txt'.format(source, date)\n os.makedirs(os.path.dirname(sourcename), exist_ok=True)\n with open (sourcename) as f:\n jdata = json.load(f)\n jdata2=jdata['articles']\n \n for i in range(0,len(jdata2)):\n r=jdata2[i]['url']\n print(r)\n link = newspaper.Article(r)\n link.download()\n html = link.html\n \n if 'video' in r:\n pass\n \n elif link: \n try:\n link.parse()\n text = fulltext(html)\n date_longform = dates[0]\n\n article = {}\n article[\"html\"] = html\n article[\"title\"] = link.title\n article[\"url\"] = link.url\n article[\"date\"] = date_longform\n article[\"source\"] = source\n article[\"text\"] = link.text\n article[\"images\"] = list(link.images)\n article[\"videos\"] = link.movies\n count=i+1\n filename = './data/Credible/{0}/articles/{1}/article_{2}.txt'.format(source, date, count)\n os.makedirs(os.path.dirname(filename), exist_ok=True)\n with open(filename, 'w',encoding=\"utf8\",newline='') as file:\n json.dump(article,file)\n except:\n pass\n \n else:\n pass\n \n print('Finished')\n return None",
"_____no_output_____"
],
[
"saving_json()",
"saving ...\nassociated-press\nhttps://apnews.com/97b63e02c7454d7da03fb5df459062e6\nhttps://apnews.com/1a68c31f2cb54b15af536cbe168442bf\nhttps://apnews.com/a9889fc3693848008259be9f1615a05d\nhttps://apnews.com/055c34f044284f21b0fa5cb3e0ada520\nhttps://apnews.com/c5eb55bf3b074f8cb18471343136cd11\nhttps://apnews.com/3052974992744dad9858a6b246166086\nhttps://apnews.com/44494a468abe4e009b0388798c16a197\nhttps://apnews.com/67e83f62aa7340249637c83c77a7597b\nhttps://apnews.com/eb4c8e7b1a8c4a5d9d618464249a8be8\nhttps://apnews.com/84a47d2e26d4419c8b616db5c5f0fa6d\nindependent\nhttp://www.independent.co.uk/news/world/asia/india-election-result-narendra-modi-win-bjp-party-bharatiya-janata-party-a8928931.html\nhttps://www.indy100.com/article/larry-the-cat-theresa-may-resigns-speech-10-downing-street-video-8928691\nhttp://www.independent.co.uk/news/uk/politics/theresa-may-resigns-new-prime-minister-brexit-eu-election-results-a8928111.html\nhttp://www.independent.co.uk/news/uk/politics/theresa-may-resigns-latest-brexit-deal-conservative-leadership-race-european-elections-tory-party-a8928321.html\nhttp://www.independent.co.uk/arts-entertainment/music/features/taylor-swift-me-interview-new-music-video-latest-a8928306.html\nhttp://www.independent.co.uk/sport/football/live/transfer-news-live-rumours-gossip-manchester-united-liverpool-arsenal-transfer-news-today-now-a8928116.html\nhttp://www.independent.co.uk/voices/zelensky-ukraine-west-russia-poroshenko-nato-imf-a8927526.html\nhttp://www.independent.co.uk/independentminds/politics-explained/brexit-deal-next-prime-minister-theresa-may-withdrawal-agreement-a8926736.html\nhttp://www.independent.co.uk/news/long_reads/brexit-britain-economy-crisis-vince-cable-prosperity-future-liberal-a8887471.html\nhttp://www.independent.co.uk/news/long_reads/whale-intelligence-science-ocean-a8892636.html\nbbc-news\nhttp://www.bbc.co.uk/news/world-europe-48398074\nhttp://www.bbc.co.uk/news/science-environment-48387033\nhttp://www.bbc.co.uk/news/world-asia-48395241\nhttp://www.bbc.co.uk/news/uk-politics-48379730\nhttp://www.bbc.co.uk/news/uk-politics-48395905\nhttp://www.bbc.co.uk/news/world-us-canada-48393512\nhttp://www.bbc.co.uk/news/world-us-canada-48393665\nhttp://www.bbc.co.uk/news/world-us-canada-48393721\nhttp://www.bbc.co.uk/news/science-environment-48289204\nhttp://www.bbc.co.uk/news/world-asia-48378390\nreuters\nhttp://feeds.reuters.com/~r/reuters/topNews/~3/iAChlkHCchs/uk-foreign-secretary-hunt-to-stand-for-conservative-leadership-local-paper-idUSKCN1SU1G7\nhttp://feeds.reuters.com/~r/reuters/topNews/~3/i8UqdZKH2Ho/boris-johnson-says-next-british-pm-must-deliver-proper-brexit-idUSKCN1SU1FX\nhttp://feeds.reuters.com/~r/reuters/topNews/~3/gAxwjfI-tOE/palace-banquet-meeting-with-may-on-trumps-uk-trip-idUSKCN1SU1S0\nhttp://feeds.reuters.com/~r/reuters/topNews/~3/vLRkRqgnABQ/harvey-weinstein-and-accusers-reach-tentative-compensation-deal-wsj-idUSKCN1SU015\nhttp://feeds.reuters.com/~r/reuters/topNews/~3/CKgQjhHwQKA/ex-u-s-marine-held-by-russia-on-spying-charge-complains-of-threats-idUSKCN1SU0VV\nhttp://feeds.reuters.com/~r/reuters/topNews/~3/PyeQNPaIwh0/in-western-france-a-village-remembers-d-days-secret-massacre-idUSKCN1ST0KW\nhttp://feeds.reuters.com/~r/reuters/topNews/~3/0ZVhNEtYfeE/u-s-charges-wikileaks-founder-julian-assange-with-espionage-idUSKCN1ST2L4\nhttp://feeds.reuters.com/~r/reuters/topNews/~3/Dt6M0xx46Kg/north-korea-blames-u-s-for-failed-summit-urges-new-calculation-idUSKCN1SU0Z6\nhttp://feeds.reuters.com/~r/reuters/topNews/~3/VsNvtiUFY3Y/wall-street-gains-as-trade-tensions-ease-idUSKCN1SU1CL\nhttp://feeds.reuters.com/~r/reuters/topNews/~3/wmFfOjOcxuU/brexit-brings-down-may-johnson-stakes-leadership-claim-idUSKCN1SU0FI\nthe-wall-street-journal\nhttps://www.wsj.com/articles/think-before-you-fish-for-bargains-in-chinese-stocks-11558710015?mod=hp_lead_pos9\nhttps://www.wsj.com/articles/trump-administration-moves-to-roll-back-protections-for-transgender-patients-11558705347?mod=article_inline&mod=hp_lead_pos3\nhttps://www.wsj.com/articles/airlines-and-fliers-brace-for-summer-snarls-11558696527?mod=hp_lead_pos8\nhttps://www.wsj.com/articles/what-happens-in-a-new-mexico-town-when-u-s-border-patrol-drops-off-thousands-of-migrants-11558696442?mod=hp_lead_pos10\nhttps://www.wsj.com/articles/its-a-lifelong-burden-the-mixed-blessing-of-the-medal-of-honor-11558695600?mod=hp_lead_pos5\nhttps://www.wsj.com/articles/political-risks-loom-over-european-markets-11558692659?mod=hp_lead_pos2\nhttps://www.wsj.com/articles/trump-administration-preparing-executive-order-on-health-cost-disclosure-11558690320?mod=hp_lead_pos7\nhttps://www.wsj.com/articles/unicorns-unicorns-qualified-cfos-11558690202?mod=hp_lead_pos6\nhttps://www.wsj.com/articles/british-prime-minister-theresa-may-says-she-will-quit-11558688912?mod=hp_lead_pos1\nhttps://www.wsj.com/graphics/bankruptcy-power-players/?mod=article_inline&mod=hp_lead_pos4\nthe-washington-post\nhttps://www.washingtonpost.com/politics/schumer-calls-trump-a-get-nothing-done-president-as-blame-game-continues-after-aborted-white-house-meeting/2019/05/23/304db368-7d4e-11e9-a5b3-34f3edf1351e_story.html\nhttps://www.washingtonpost.com/lifestyle/travel/10-common-travel-scams--and-how-to-avoid-them/2019/05/23/a2f4565e-3b92-11e9-aaae-69364b2ed137_story.html\nhttps://www.washingtonpost.com/sports/2019/05/23/velocity-is-number-one-thing-this-high-tech-biomechanics-lab-is-changing-baseball/\nhttps://www.washingtonpost.com/local/legal-issues/wikileaks-founder-julian-assange-charged-with-violating-espionage-act/2019/05/23/42a2c6cc-7d6a-11e9-a5b3-34f3edf1351e_story.html\nhttps://www.washingtonpost.com/local/legal-issues/us-judge-hears-house-lawsuit-to-stop-trump-border-wall-construction/2019/05/23/234cfa4c-7caa-11e9-8bb7-0fc796cf2ec0_story.html\nhttps://www.washingtonpost.com/business/economy/house-lawmakers-to-leave-washington-with-billions-in-emergency-aid-stuck-in-gridlock/2019/05/23/b40652a8-7cd8-11e9-8ede-f4abf521ef17_story.html\nhttps://www.washingtonpost.com/sports/2019/05/23/pete-buttigieg-i-had-put-my-life-line-defend-nfl-players-right-protest/\nhttps://www.washingtonpost.com/politics/buttigieg-its-disgusting-that-trump-is-reportedly-considering-pardoning-soldiers-convicted-of-war-crimes/2019/05/23/5ccae858-7d61-11e9-a5b3-34f3edf1351e_story.html\nhttps://www.washingtonpost.com/world/national-security/putin-out-prepared-trump-in-key-meeting-rex-tillerson-told-house-panel/2019/05/22/9523c60f-865d-4917-898b-cb32b922e93c_story.html\nhttps://www.washingtonpost.com/sports/2019/05/21/velocity-is-strangling-baseball-its-grip-keeps-tightening/\nnational-geographic\nhttps://www.nationalgeographic.com/photography/your-shot/photos-of-the-week/2019/05/best-pictures-week-of-may-17-from-our-community.html\nhttps://www.nationalgeographic.com/science/2019/05/bermuda-volcano-hybrid-unlike-others-on-earth-new-way-to-make-volcanoes.html\nhttps://www.nationalgeographic.com/travel/destinations/north-america/united-states/lgbtq-pride-june-celebrations-where-to-go-what-to-do.html\nhttps://www.nationalgeographic.com/travel/destinations/asia/japan/kyoto/explore-beautiful-photos-of-things-to-do-and-see.html\nhttps://www.nationalgeographic.com/travel/features/photography/colorful-travel-around-world.html\nhttps://www.nationalgeographic.com/science/2019/05/mouse-lizard-dinosaur-crawled-then-walked-baby-mussaurus-patagonicus.html\nhttps://www.nationalgeographic.com/environment/2019/05/before-plastic-rubber-filled-american-homes.html\nhttps://www.nationalgeographic.com/science/2018/11/kilogram-forever-changed-why-mass-matters.html\nhttps://www.nationalgeographic.com/animals/reference/safety-animals-wildlife-attacks-national-parks.html\nhttps://www.nationalgeographic.com/science/2019/05/dear-hubble-space-telescope-transformed-astronomy-and-us-all.html\nusa-today\nhttps://www.usatoday.com/story/life/tv/2019/05/24/the-voice-gwen-stefani-replace-adam-levine-upcoming-season/1219646001/?utm_source=google&utm_medium=amp&utm_campaign=speakable\nhttps://www.usatoday.com/story/life/people/2019/05/24/emilia-clarke-didnt-want-nude-fifty-shades-grey/1219449001/?utm_source=google&utm_medium=amp&utm_campaign=speakable\nhttps://www.usatoday.com/story/news/world/2019/05/24/iran-threat-pentagon-increase-military-deployments-middle-east/1210096001/?utm_source=google&utm_medium=amp&utm_campaign=speakable\n"
],
[
"# #Create initial modeling DataFrame - Only Ran Once then commented out\n# modeling = pd.DataFrame(columns=('label', 'text', 'title'))\n\n# #Save initial DataFrame - Only Ran Once - Only Ran Once then commented out\n# with open('./data/credible_news_df.pickle', 'wb') as file:\n# pickle.dump(modeling, file)",
"_____no_output_____"
],
[
"#Open Corpus of News Article Text\nwith open('./data/credible_news_df.pickle', 'rb') as file:\n credible_news_df = pickle.load(file)",
"_____no_output_____"
],
[
"i = credible_news_df.shape[0] #Will start adding at the last row of the dataframe\nfor source in os.listdir(\"./data/Credible/\"):\n for file in os.listdir('./data/Credible/'+source+'/articles/'+dates[0]):\n if file.endswith(\".txt\") and 'api' not in file:\n curr_file = os.path.join('./data/Credible/'+source+'/articles/'+dates[0], file)\n #print curr_file\n with open(curr_file) as json_file:\n try:\n data = json.load(json_file)\n credible_news_df.loc[i] = [0,data[\"text\"],data[\"title\"]]\n i = i + 1\n except ValueError:\n continue",
"_____no_output_____"
],
[
"#Will Increase Daily\ncredible_news_df.shape",
"_____no_output_____"
],
[
"#Save Updated Data Frame\nwith open('./data/credible_news_df.pickle', 'wb') as file:\n pickle.dump(credible_news_df, file)",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb8780f6b048c1511205488853f612fd17e03935
| 27,005 |
ipynb
|
Jupyter Notebook
|
read_HX711.ipynb
|
AlvaritoMoscoso/PEC00144
|
a96662869d3822823455281474733ce7f69881c9
|
[
"MIT"
] | 29 |
2018-09-24T12:58:16.000Z
|
2021-09-05T09:56:35.000Z
|
read_HX711.ipynb
|
AlvaritoMoscoso/PEC00144
|
a96662869d3822823455281474733ce7f69881c9
|
[
"MIT"
] | null | null | null |
read_HX711.ipynb
|
AlvaritoMoscoso/PEC00144
|
a96662869d3822823455281474733ce7f69881c9
|
[
"MIT"
] | 7 |
2018-10-03T16:29:41.000Z
|
2021-09-15T19:21:59.000Z
| 115.405983 | 21,396 | 0.87169 |
[
[
[
"Universidade Federal do Rio Grande do Sul (UFRGS) \nPrograma de Pós-Graduação em Engenharia Civil (PPGEC) \n\n# PEC00144: Experimental Methods in Civil Engineering\n\n### Reading the serial port of an Arduino device \n\n---\n_Prof. Marcelo M. Rocha, Dr.techn._ [(ORCID)](https://orcid.org/0000-0001-5640-1020) \n_Porto Alegre, RS, Brazil_ \n",
"_____no_output_____"
]
],
[
[
"# Importing Python modules required for this notebook\n# (this cell must be executed with \"shift+enter\" before any other Python cell)\n\nimport sys\nimport time\nimport serial\nimport numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\n\nfrom MRPy import MRPy\n",
"_____no_output_____"
]
],
[
[
"### 1. Setup serial communication\n\nIn order to run this notebook, the Python module ``pyserial`` must be installed.\nTo ensure the module availability, open a conda terminal and issue the command:\n\n conda install -c anaconda pyserial\n\nBefore openning the serial port, verify with Arduino IDE which USB identifier the \nboard has be assigned (in Windows it has the form \"COMxx\", while in Linux it\nit is something like \"/dev/ttyXXXX\").\n",
"_____no_output_____"
]
],
[
[
"#port = '/dev/ttyUSB0' \n#baud = 9600\n\nport = 'COM5' # change this address according to your computer\nbaud = 9600 # match this number with the Arduino's output baud rate\n\nArdn = serial.Serial(port, baud, timeout=1)\n\ntime.sleep(3) # this is important to give time for serial settling\n",
"_____no_output_____"
]
],
[
[
"### 2. Define function for reading one incoming line",
"_____no_output_____"
]
],
[
[
"def ReadSerial(nchar, nvar, nlines=1):\n\n Ardn.write(str(nlines).encode())\n data = np.zeros((nlines,nvar))\n\n for k in range(nlines):\n\n wait = True\n while(wait):\n \n if (Ardn.inWaiting() >= nchar):\n wait = False\n \n bdat = Ardn.readline() \n sdat = bdat.decode()\n sdat = sdat.replace('\\n',' ').split()\n \n data[k, :] = np.array(sdat[0:nvar], dtype='int')\n \n return data\n",
"_____no_output_____"
]
],
[
[
"### 3. Acquire data lines from serial port",
"_____no_output_____"
]
],
[
[
"try:\n data = ReadSerial(16, 2, nlines=64)\n t = data[:,0]\n LC = data[:,1]\n\n Ardn.close()\n print('Acquisition ok!')\n \nexcept:\n Ardn.close()\n sys.exit('Acquisition failure!')\n",
"Acquisition ok!\n"
]
],
[
[
"### 4. Create ``MRPy`` instance and save to file ",
"_____no_output_____"
]
],
[
[
"ti = (t - t[0])/1000\nLC = (LC + 1270)/2**23\n \ndata = MRPy.resampling(ti, LC)\ndata.to_file('read_HX711', form='excel')\n\nprint('Average sampling rate is {0:5.1f}Hz.'.format(data.fs))\nprint('Total record duration is {0:5.1f}Hz.'.format(data.Td))\n\nprint((2**23)*data.mean())",
"Average sampling rate is 1.1Hz.\nTotal record duration is 57.3Hz.\n13324.396215560728\n"
]
],
[
[
"### 5. Data visualization",
"_____no_output_____"
]
],
[
[
"fig1 = data.plot_time(fig=1, figsize=(12,8), axis_t=[0, data.Td, -0.01, 0.01])\n",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb87a26accb32e7d720cbbdfc98579769eec87ad
| 60,666 |
ipynb
|
Jupyter Notebook
|
Language model-Copy1.ipynb
|
abdullah-taha/Sentiment-Classification-Using-ULMFI-and-Tranfer-learning
|
fa2aaa825cc6f9e2f1b8e2bd3ec03d839d8b02ba
|
[
"MIT"
] | 1 |
2020-05-04T20:12:22.000Z
|
2020-05-04T20:12:22.000Z
|
Language model-Copy1.ipynb
|
abdullah-taha/Sentiment-Classification-Using-ULMFI-and-Tranfer-learning
|
fa2aaa825cc6f9e2f1b8e2bd3ec03d839d8b02ba
|
[
"MIT"
] | null | null | null |
Language model-Copy1.ipynb
|
abdullah-taha/Sentiment-Classification-Using-ULMFI-and-Tranfer-learning
|
fa2aaa825cc6f9e2f1b8e2bd3ec03d839d8b02ba
|
[
"MIT"
] | null | null | null | 50.17866 | 11,528 | 0.603979 |
[
[
[
"%reload_ext autoreload\n%autoreload 2\n%matplotlib inline",
"_____no_output_____"
],
[
"from fastai.text import *",
"_____no_output_____"
],
[
"path = Path('./WikiTextTR')",
"_____no_output_____"
],
[
"path.ls()",
"_____no_output_____"
],
[
"LANG_FILENAMES = [str(f) for f in path.rglob(\"*/*\")]\nprint(len(LANG_FILENAMES))\nprint(LANG_FILENAMES[:5])",
"510\n['WikiTextTR\\\\AA\\\\wiki_00', 'WikiTextTR\\\\AA\\\\wiki_01', 'WikiTextTR\\\\AA\\\\wiki_02', 'WikiTextTR\\\\AA\\\\wiki_03', 'WikiTextTR\\\\AA\\\\wiki_04']\n"
],
[
"LANG_TEXT = []\nfor i in LANG_FILENAMES:\n try:\n for line in open(i, encoding=\"utf-8\"):\n LANG_TEXT.append(json.loads(line))\n except:\n break\nLANG_TEXT = pd.DataFrame(LANG_TEXT)",
"_____no_output_____"
],
[
"LANG_TEXT.head()",
"_____no_output_____"
],
[
"LANG_TEXT.to_csv(f\"{path}/Wiki_Turkish_Corpus.csv\", index=False)",
"_____no_output_____"
],
[
"LANG_TEXT = pd.read_csv(f\"{path}/Wiki_Turkish_Corpus.csv\")",
"_____no_output_____"
],
[
"LANG_TEXT.head()",
"_____no_output_____"
],
[
"LANG_TEXT.drop([\"id\",\"url\",\"title\"],axis=1,inplace=True)",
"_____no_output_____"
],
[
"LANG_TEXT = (LANG_TEXT.assign(labels = 0)\n .pipe(lambda x: x[['labels', 'text']])\n .to_csv(f\"{path}/Wiki_Turkish_Corpus2.csv\", index=False))\nLANG_TEXT.head()",
"_____no_output_____"
],
[
"LANG_TEXT = pd.read_csv(f\"{path}/Wiki_Turkish_Corpus2.csv\")\nLANG_TEXT.head()",
"_____no_output_____"
],
[
"def split_title_from_text(text):\n words = text.split(\"\\n\\n\")\n if len(words) >= 2:\n return ''.join(words[1:])\n else:\n return ''.join(words)\n \nLANG_TEXT['text'] = LANG_TEXT['text'].apply(lambda x: split_title_from_text(x))",
"_____no_output_____"
],
[
"\nLANG_TEXT.isna().any()",
"_____no_output_____"
],
[
"LANG_TEXT.shape",
"_____no_output_____"
],
[
"LANG_TEXT['text'].apply(lambda x: len(x.split(\" \"))).sum()",
"_____no_output_____"
],
[
"re1 = re.compile(r' +')\ndef fixup(x):\n x = x.replace('ü', \"u\").replace('Ü', 'U').replace('ı', \"i\").replace(\n 'ğ', 'g').replace('İ', 'I').replace('Ğ', \"G\").replace('ö', \"o\").replace(\n 'Ö', \"o\").replace('\\n\\n', ' ').replace(\"\\'\",' ').replace('\\n\\nSection::::',' ').replace(\n '\\n',' ').replace('\\\\', ' \\\\ ').replace('ç', 'c').replace('Ç', 'C').replace('ş', 's').replace('Ş', 'S')\n return re1.sub(' ', html.unescape(x))\n",
"_____no_output_____"
],
[
"LANG_TEXT.to_csv(f\"{path}/Wiki_Turkish_Corpus3.csv\", index=False)",
"_____no_output_____"
],
[
"LANG_TEXT = pd.read_csv(f\"{path}/Wiki_Turkish_Corpus3.csv\")#, chunksize=5000)\nLANG_TEXT.head()",
"_____no_output_____"
],
[
"import torch\ntorch.cuda.device(0)\ntorch.cuda.get_device_name(0)",
"_____no_output_____"
],
[
"LANG_TEXT.dropna(axis=0, inplace=True)",
"_____no_output_____"
],
[
"df = LANG_TEXT.iloc[np.random.permutation(len(LANG_TEXT))]",
"_____no_output_____"
],
[
"cut1 = int(0.8 * len(df)) + 1\ncut1",
"_____no_output_____"
],
[
"df_train, df_valid = df[:cut1], df[cut1:]",
"_____no_output_____"
],
[
"df = LANG_TEXT.iloc[np.random.permutation(len(LANG_TEXT))]\ncut1 = int(0.8 * len(df)) + 1\ndf_train, df_valid = df[:cut1], df[cut1:]",
"_____no_output_____"
],
[
"df_train.shape, df_valid.shape",
"_____no_output_____"
],
[
"df_train.head()",
"_____no_output_____"
],
[
"data_lm = TextLMDataBunch.from_df(path, train_df=df_train, valid_df= df_valid, label_cols=\"labels\", text_cols=\"text\")",
"_____no_output_____"
],
[
"data_lm.save('data_lm.pkl')",
"_____no_output_____"
],
[
"bs=16\ndata_lm = load_data(path, 'data_lm.pkl', bs=bs)",
"_____no_output_____"
],
[
"data_lm.show_batch()",
"_____no_output_____"
],
[
"learner = language_model_learner(data_lm, AWD_LSTM, pretrained=False, drop_mult=0.5)",
"_____no_output_____"
],
[
"learner.lr_find()",
"_____no_output_____"
],
[
"learner.recorder.plot()",
"_____no_output_____"
],
[
"learner.fit_one_cycle(1,1e-2)",
"_____no_output_____"
],
[
"learner.save(\"model1\")",
"_____no_output_____"
],
[
"learner.load(\"model1\")",
"_____no_output_____"
],
[
"TEXT = \"Birinci dünya savaşında\"\nN_WORDS = 40\nN_SENTENCES = 2\nprint(\"\\n\".join(learner.predict(TEXT, N_WORDS, temperature=0.75) for _ in range(N_SENTENCES)))",
"Birinci dünya savaşında Ahmet Derlediği adında bir kısmı San Hasan Rüstem tarafından evlendi ve bu sırada Facebook de Harekatın olarak verilmiştir . Önemli bir yasal Google Beyin Kaya Seferi ve\nBirinci dünya savaşında en büyük ve en fazla kitap , en iyi iki Amerikan bölgesinde . Devlet , Türkiye'nin , Avrupa'nın kısa ve Kadın Fakültesi ( İngilizce : Memlük ) , İsa Sonradan\n"
],
[
"file = open(\"itos.pkl\",\"wb\")\npickle.dump(data_lm.vocab.itos, file)",
"_____no_output_____"
],
[
"file.close()",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb87a40eaebabf8aaf577fc0600c7f38ce0015b1
| 33,697 |
ipynb
|
Jupyter Notebook
|
comp_assignment-1.ipynb
|
sju-chem264-2019/list-tutorials-Winboo1
|
13e68fc34476ed3270c737426b4fb326e9229a1d
|
[
"MIT"
] | null | null | null |
comp_assignment-1.ipynb
|
sju-chem264-2019/list-tutorials-Winboo1
|
13e68fc34476ed3270c737426b4fb326e9229a1d
|
[
"MIT"
] | null | null | null |
comp_assignment-1.ipynb
|
sju-chem264-2019/list-tutorials-Winboo1
|
13e68fc34476ed3270c737426b4fb326e9229a1d
|
[
"MIT"
] | null | null | null | 25.374247 | 1,033 | 0.50595 |
[
[
[
"# Computational Assignment 1",
"_____no_output_____"
],
[
" **Assigned Monday, 9-9-19.**, **Due Thursday, 9-12-19.**",
"_____no_output_____"
],
[
"Most of the problems we encounter in computational chemistry are multidimensional. This means that we need to be able to work with vectors and matrices in our code. Even when we consider a 1-dimensional function, we still need to code all of the data points into a list. \n\nAdditionally, when we need to analyze data, we don't want to reinvent the wheel. It can be useful to code your own math operation once to learn how it work, but most of the time you should be using existing libraries in your work. We will cover how to use math and science libraries to run calcualtions.\n\nThis notebook will cover the following concepts:\n\n1. Lists and arrays\n1. Defining lists\n1. Accessing list values\n1. Changing lists\n1. Counting, sorting, and looping methods",
"_____no_output_____"
],
[
"## 1. Lists and Arrays",
"_____no_output_____"
],
[
"Computers regularly deal with multidimensional data. Even if you have one-dimensional data from some sort of function, $F(t)$, you would write the data points into a list of numbers. When talking about code, we call these lists or arrays. ",
"_____no_output_____"
]
],
[
[
"my_list=[1,2,3,4,5,6,7]",
"_____no_output_____"
]
],
[
[
"Python specifically has a few of ways to handle arrays. The most basic data object for handeling data is the **list**. The other two data objects are **tuples** and **dictionaries**. \n\nLists are defined with brackets `[]`\nTuples are defined with parenthessis `()`\nDictionaries are defined with curly brackets `{}`\n\nWe won't spend much time with tuples. They behave a lot like lists, but once make one, you can't change any of the values in the tuple. This can be useful if you want to make sure data values are not changed through a calculation. They also tend be faster to process, but you won't notice the speedup unless you are trying to process lots of data (Gb of data). ",
"_____no_output_____"
],
[
"## 1.1 Defining Lists",
"_____no_output_____"
],
[
"In python, we create **lists** by defining a set of numbers in a bracket ",
"_____no_output_____"
]
],
[
[
"[1,2,3]",
"_____no_output_____"
]
],
[
[
"You should also make sure to define your list as a variable so that you can use it later. Otherwise there is no way to call it. If you don't want to overwrite your variable, use a different name for the new varaiable",
"_____no_output_____"
]
],
[
[
"my_list2=[1,2,3]\nprint(my_list2)",
"[1, 2, 3]\n"
]
],
[
[
"You can also define an empty list. This is sometimes usefull if you plan to add alements to the list (more on the later ",
"_____no_output_____"
]
],
[
[
"empty=[]",
"_____no_output_____"
]
],
[
[
"Lists also don't have to be numbers. They can be a collection of strings (text)",
"_____no_output_____"
]
],
[
[
"sonoran_plants=[\"saguaro\",\"ocotillo\",\"pitaya\",\"creosote\"]",
"_____no_output_____"
]
],
[
[
"Your list can be a list of lists! Notice the line break. You can do that when there is a comma. This helps make your code readable, which is one of the underlying philosophies of python",
"_____no_output_____"
]
],
[
[
"sonoran_plants_animals=[[\"saguaro\",\"ocotillo\",\"pitaya\",\"creosote\"],\n [\"coyote\",\"kangaroo mouse\",\"javalina\",\"gila monster\"]]",
"_____no_output_____"
]
],
[
[
"Notice that the two nested lists are different sizes. Mathematically, these lists are not going to behave like vectors or matrices. Python will not add or multiply a list according to the rules of matrix algebra. This is fine. When we need matrices and vectors, it will make more sense to use arrays associated with the math library Numpy. And the Numpy library can usually understand most python lists. Most of the time, if you are using a list, you are mostly trying to organize data, not run a heavy calculation.",
"_____no_output_____"
],
[
"### Exercise 1.1: \nMake your own lists. \n\n1. Make a one-dimensional list of numbers \n1. A three dimensional list of numbers (a list of three lists)\n1. Make a lists of strings\n1. Make a two-dimensional list of strings\n1. Print all of your lists\n\nMake sure to define your lists with variables. We will use your lists later on",
"_____no_output_____"
]
],
[
[
"trio=[1,2,3]\nsingle_digit_trios=[[1,2,3],\n [4,5,6],\n [7,8,9]]\ndunkin_order=[\"large\",\"iced\",\"black\"]\ncoffee_pref=[[\"dunkin\",\"good\"],\n [\"starbucks\",\"bad\"]]\n\nprint(trio)\nprint(single_digit_trios)\nprint(dunkin_order)\nprint(coffee_pref)",
"[1, 2, 3]\n[[1, 2, 3], [4, 5, 6], [7, 8, 9]]\n['large', 'iced', 'black']\n[['dunkin', 'good'], ['starbucks', 'bad']]\n"
]
],
[
[
"## 1.2 List indexing ",
"_____no_output_____"
],
[
"Once you've made a list, you need to know how to get values from the list. To get a value from list use square brackets next to the variable name. `my_list[index]`. The first thing to note is that list indeces start at 0",
"_____no_output_____"
]
],
[
[
"# Printing the list to remind us of the elements\nprint(my_list)",
"[1, 2, 3, 4, 5, 6, 7]\n"
],
[
"my_list[0]",
"_____no_output_____"
]
],
[
[
"Your indexing can also be negative. The list indexing is cyclic, so if the first element is 0, the last element is -1, the second to last element is -2, etc. ",
"_____no_output_____"
]
],
[
[
"my_list[-1]",
"_____no_output_____"
],
[
"my_list[-2]",
"_____no_output_____"
]
],
[
[
"If your list is a list of lists, calling an index will give you the nested list. You need two indices to get individual items",
"_____no_output_____"
]
],
[
[
"# Printing the list to remind us of the elements\nprint(sonoran_plants_animals)",
"[['saguaro', 'ocotillo', 'pitaya', 'creosote'], ['coyote', 'kangaroo mouse', 'javalina', 'gila monster']]\n"
],
[
"sonoran_plants_animals[0]",
"_____no_output_____"
],
[
"sonoran_plants_animals[0][-1]",
"_____no_output_____"
]
],
[
[
"You can also make a new sublist by calling a range of indices",
"_____no_output_____"
]
],
[
[
"# Printing the list to remind us of the elements\nprint(my_list)",
"[1, 2, 3, 4, 5, 6, 7]\n"
],
[
"my_list[2:5]",
"_____no_output_____"
]
],
[
[
"You can also make a range with negatice indices. Order matters here, more negative number must be first",
"_____no_output_____"
]
],
[
[
"my_list[-4:-1]",
"_____no_output_____"
]
],
[
[
"### Exercise 1.2\n\nUsing the lists you made from up above do the following:\n\n1. For each list you made before, print the first and last values\n1. For each multi-dimensional list print the first and last entry of each nested list\n1. For each one-dimensional list, use a range of indices to make a new sublist",
"_____no_output_____"
]
],
[
[
"trio=[1,2,3]\nsingle_digit_trios=[[1,2,3],\n [4,5,6],\n [7,8,9]]\ndunkin_order=[\"large\",\"iced\",\"black\"]\ncoffee_pref=[[\"dunkin\",\"good\"],\n [\"starbucks\",\"bad\"]]\n\nprint(trio[0],\n trio[2])\nprint(single_digit_trios[0][0],\n single_digit_trios[0][2],\n single_digit_trios[1][0],\n single_digit_trios[1][2],\n single_digit_trios[2][0],\n single_digit_trios[2][2])\nprint(dunkin_order[0],\n dunkin_order[2])\nprint(coffee_pref[0][0],\n coffee_pref[0][1],\n coffee_pref[1][0],\n coffee_pref[1][1],)\nprint(trio[0:2])\nprint(dunkin_order[0:2])",
"1 3\n1 3 4 6 7 9\nlarge black\ndunkin good starbucks bad\n[1, 2]\n['large', 'iced']\n"
]
],
[
[
"## 1.3 Changing lists",
"_____no_output_____"
],
[
"First, we can get the length of a list. Many times, our list is very long or read in from a file. We may need to knwo how long the list actuall is",
"_____no_output_____"
]
],
[
[
"# Printing the list to remind us of the elements\nprint(sonoran_plants)",
"['saguaro', 'ocotillo', 'pitaya', 'creosote']\n"
],
[
"len(sonoran_plants)",
"_____no_output_____"
]
],
[
[
"We can change our lists after we make them. We can change individual values or we can add or remove values from a list. Note that tuples cannot be changed (they are called immutable) ",
"_____no_output_____"
]
],
[
[
"# Printing the list to remind us of the elements\nprint(my_list)",
"[1, 2, 3, 4, 5, 6, 7]\n"
]
],
[
[
"Individual values in a list can be changed",
"_____no_output_____"
]
],
[
[
"my_list[2] = -3\nprint(my_list)",
"[1, 2, -3, 4, 5, 6, 7, 8, 8, 8, 8]\n"
]
],
[
[
"Values can be added to a list",
"_____no_output_____"
]
],
[
[
"my_list.append(8)\nprint(my_list)",
"[1, 2, 4, 5, 6, 7, 8, 8, 8, 8, 8, 8, 8]\n"
]
],
[
[
"Values can be removed from a list",
"_____no_output_____"
]
],
[
[
"my_list.remove(-3)\nprint(my_list)",
"_____no_output_____"
]
],
[
[
"A quick note about objects. Python is an object oriented language. It's underlying philosophy is that everything is an object. An object has atributes and methods. You can get information about the attributes and you can use the methods to change the properties of the object. \n\nIn python you call the object attributes or methods using this format: `object_variable.attribute` For a list, you add values by changing the attribute `list.append(x)`, `list.remove(x)`.",
"_____no_output_____"
],
[
"We can add the elements of a list to our list",
"_____no_output_____"
]
],
[
[
"my_list.extend([1,2,3,4])",
"_____no_output_____"
],
[
"print(my_list)",
"[1, 2, 4, 5, 6, 7, 8, 1, 2, 3, 4]\n"
]
],
[
[
"We can insert values at a given index. When using insert, the first value is the index, the second value is the new list element",
"_____no_output_____"
]
],
[
[
"my_list.insert(0,15)\nprint(my_list)",
"[15, 1, 2, 4, 5, 6, 7, 8, 1, 2, 3, 4]\n"
]
],
[
[
"We can remove elements at a given index",
"_____no_output_____"
]
],
[
[
"my_list.pop(3)\nprint(my_list)",
"[15, 1, 2, 5, 6, 7, 8, 1, 2, 3, 4]\n"
]
],
[
[
"### Exercise 1.3 \n\nFor each one-dimensional list from above \n\n1. Append a new element\n1. Remove a previous element\n1. Extend the lists with new lists of elements\n1. Insert a value at the fourth index\n1. Pop the last value ",
"_____no_output_____"
]
],
[
[
"trio=[1,2,3]\ndunkin_order=[\"large\",\"iced\",\"black\"]\n\nprint(trio)\ntrio.append(4)\nprint(trio)\ntrio.remove(1)\nprint(trio)\ntrio.extend([5,6,7])\nprint(trio)\ntrio.insert(3,4)\nprint(trio)\ntrio.pop(6)\nprint(trio)\n\nprint(dunkin_order)\ndunkin_order.append(\"less ice\")\nprint(dunkin_order)\ndunkin_order.remove(\"black\")\nprint(dunkin_order)\ndunkin_order.extend([\"no milk\",\"no sugar\"])\nprint(dunkin_order)\ndunkin_order.insert(3,\"cold brew\")\nprint(dunkin_order)\ndunkin_order.pop(5)\nprint(dunkin_order)",
"[1, 2, 3]\n[1, 2, 3, 4]\n[2, 3, 4]\n[2, 3, 4, 5, 6, 7]\n[2, 3, 4, 4, 5, 6, 7]\n[2, 3, 4, 4, 5, 6]\n['large', 'iced', 'black']\n['large', 'iced', 'black', 'less ice']\n['large', 'iced', 'less ice']\n['large', 'iced', 'less ice', 'no milk', 'no sugar']\n['large', 'iced', 'less ice', 'cold brew', 'no milk', 'no sugar']\n['large', 'iced', 'less ice', 'cold brew', 'no milk']\n"
]
],
[
[
"## 1.4 Counting, sorting, and looping methods",
"_____no_output_____"
],
[
"There are a number of other list methods you can use to change your list. To demonstrate these methods, we will make a list of random integers using the append method",
"_____no_output_____"
]
],
[
[
"# Build a list of random integers\nimport random # import random number generator library\n\nrand_list=[] # Note this starts as an empty list\n\nfor i in range(0,100):\n rand_num=random.randrange(0,10)\n rand_list.append(rand_num)\n \nprint(rand_list)",
"[5, 2, 7, 8, 0, 1, 1, 6, 0, 6, 6, 3, 4, 0, 1, 6, 5, 8, 5, 9, 3, 1, 8, 3, 6, 4, 7, 5, 0, 4, 0, 1, 0, 7, 7, 3, 5, 2, 5, 7, 9, 5, 1, 0, 5, 5, 3, 0, 0, 7, 6, 1, 8, 3, 9, 2, 4, 6, 9, 4, 6, 9, 8, 9, 2, 3, 2, 0, 9, 2, 9, 6, 7, 1, 9, 8, 1, 4, 5, 4, 9, 6, 8, 9, 1, 0, 0, 5, 9, 8, 1, 2, 8, 1, 4, 0, 3, 3, 3, 4]\n"
]
],
[
[
"We can count the number of times a value is found in our list. This can be really useful for analysis",
"_____no_output_____"
]
],
[
[
"rand_list.count(3)",
"_____no_output_____"
]
],
[
[
"We can determine the index of the first instance of a value",
"_____no_output_____"
]
],
[
[
"rand_list.index(3)",
"_____no_output_____"
]
],
[
[
"The first time that a 3 is found is at list index 0. If you want to keep finding values of 3, you can use a range index to get the other values",
"_____no_output_____"
]
],
[
[
"rand_list[1:-1].index(3)",
"_____no_output_____"
],
[
"rand_list[19:-1].index(3)",
"_____no_output_____"
]
],
[
[
"The reason this gives you 1 is that the next instance of 3 is the $20^{\\text{th}}$ element $(19+1)$",
"_____no_output_____"
],
[
"We can also sort the list",
"_____no_output_____"
]
],
[
[
"rand_list.sort()\nprint(rand_list)",
"[0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 1, 1, 1, 1, 1, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 7, 7, 7, 7, 7, 8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 9, 9, 9, 9, 9, 9]\n"
]
],
[
[
"We can reverse the list",
"_____no_output_____"
]
],
[
[
"rand_list.reverse()\nprint(rand_list)",
"[9, 9, 9, 9, 9, 9, 8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 7, 7, 7, 7, 7, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 1, 1, 1, 1, 1, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0]\n"
]
],
[
[
"Lastly, any list can be looped over in python",
"_____no_output_____"
]
],
[
[
"sonoran_plants=[\"saguaro\",\"ocotillo\",\"pitaya\",\"creosote\"]\n\nfor plants in sonoran_plants:\n print(plants)",
"saguaro\nocotillo\npitaya\ncreosote\n"
]
],
[
[
"Multidimensional lists can also be looped over. The first loop counts over the nested loops, the second loop counts over the list elements",
"_____no_output_____"
]
],
[
[
"sonoran_plants_animals=[[\"saguaro\",\"ocotillo\",\"pitaya\",\"creosote\"],\n [\"coyote\",\"kangaroo mouse\",\"javalina\",\"gila monster\"]]\n\nfor collections in sonoran_plants_animals:\n for name in collections:\n print(name)",
"saguaro\nocotillo\npitaya\ncreosote\ncoyote\nkangaroo mouse\njavalina\ngila monster\n"
]
],
[
[
"### Excercise 1.4",
"_____no_output_____"
],
[
"For this exercise, we will creat a list of random number",
"_____no_output_____"
]
],
[
[
"rand1=[]\n\nfor i in range(0,100):\n rand_num1=random.randrange(0,10)\n rand1.append(rand_num1)",
"_____no_output_____"
],
[
"print(rand1)",
"[7, 8, 3, 0, 1, 1, 4, 3, 4, 1, 5, 0, 8, 4, 2, 2, 5, 4, 9, 1, 5, 3, 2, 5, 7, 8, 7, 5, 8, 7, 7, 6, 7, 4, 2, 4, 0, 0, 9, 7, 1, 7, 6, 4, 8, 4, 2, 3, 5, 5, 4, 1, 4, 4, 0, 8, 8, 4, 5, 3, 7, 7, 9, 4, 0, 1, 8, 3, 3, 6, 2, 6, 0, 6, 4, 4, 5, 3, 3, 6, 2, 9, 5, 8, 7, 0, 9, 3, 2, 3, 2, 7, 2, 2, 1, 0, 9, 0, 0, 7]\n"
]
],
[
[
"Using the list above, \n\n1. Count and print the nunber of instances of each integer (**Hint:** you can use a loop ranging from 0-9 to do this)\n2. Loop over the elements of rand1 and make a new list that labels each element as even or odd. For example given the list `[1,6,9]`, you would have a list that looked like `[\"odd\",\"even\",\"odd\"]`. \n\nHint: You can use the modulo operator `a%b`. This operator give you the remainder when you divide a by b. If `a/b` has no remainder, then `a%b=0` See the examples below.\n",
"_____no_output_____"
]
],
[
[
"# Modulo example\nprint(4%2)\nprint(4%3)",
"0\n1\n"
],
[
"# Module conditional\ni=4\nif (i%2==0):\n print(\"even\")\nelse:\n print(\"odd\")",
"even\n"
]
],
[
[
"Remember that you can always make new cells. Use them to test parts of you code along the way and to seperate your code to make it readable",
"_____no_output_____"
]
],
[
[
"import random\n\nrand=[]\n\nfor i in range(0,100):\n randnum=random.randrange(0,10)\n rand.append(randnum)\n \nprint(rand)\n\nprint()\n\nfor number in range(10):\n print(number,rand.count(number))",
"[1, 8, 3, 3, 3, 6, 2, 3, 6, 0, 8, 7, 2, 0, 2, 0, 9, 0, 0, 5, 6, 3, 3, 0, 0, 7, 3, 6, 7, 8, 4, 6, 5, 7, 2, 4, 9, 1, 5, 3, 3, 0, 6, 4, 6, 1, 3, 5, 4, 5, 8, 2, 8, 5, 6, 3, 6, 7, 7, 1, 5, 6, 5, 8, 2, 2, 8, 9, 1, 0, 6, 3, 8, 9, 4, 6, 3, 1, 9, 8, 4, 3, 6, 7, 7, 2, 8, 4, 5, 5, 9, 8, 8, 3, 3, 3, 1, 3, 3, 2]\n\n0 9\n1 7\n2 9\n3 19\n4 7\n5 10\n6 13\n7 8\n8 12\n9 6\n"
],
[
"import random\n\nrand=[]\n\nfor i in range(0,100):\n randnum=random.randrange(0,10)\n rand.append(randnum)\n \nprint(rand)\n\nprint()\n\nrand_parity=[]\n\nfor number in rand:\n if(number%2==0):\n rand_parity.append(\"even\")\n else:\n rand_parity.append(\"odd\")\n\nprint(rand_parity)",
"[2, 6, 3, 6, 5, 2, 0, 6, 4, 3, 1, 5, 2, 2, 0, 0, 0, 9, 0, 4, 8, 2, 8, 3, 2, 4, 2, 0, 4, 2, 6, 7, 1, 8, 3, 7, 5, 8, 2, 3, 4, 8, 1, 3, 0, 6, 8, 3, 9, 1, 1, 8, 1, 1, 2, 8, 5, 7, 2, 8, 6, 0, 6, 1, 0, 9, 3, 0, 0, 1, 5, 6, 0, 7, 3, 8, 0, 0, 7, 3, 9, 2, 4, 1, 0, 7, 5, 8, 6, 4, 9, 1, 0, 8, 1, 0, 6, 8, 2, 0]\n\n['even', 'even', 'odd', 'even', 'odd', 'even', 'even', 'even', 'even', 'odd', 'odd', 'odd', 'even', 'even', 'even', 'even', 'even', 'odd', 'even', 'even', 'even', 'even', 'even', 'odd', 'even', 'even', 'even', 'even', 'even', 'even', 'even', 'odd', 'odd', 'even', 'odd', 'odd', 'odd', 'even', 'even', 'odd', 'even', 'even', 'odd', 'odd', 'even', 'even', 'even', 'odd', 'odd', 'odd', 'odd', 'even', 'odd', 'odd', 'even', 'even', 'odd', 'odd', 'even', 'even', 'even', 'even', 'even', 'odd', 'even', 'odd', 'odd', 'even', 'even', 'odd', 'odd', 'even', 'even', 'odd', 'odd', 'even', 'even', 'even', 'odd', 'odd', 'odd', 'even', 'even', 'odd', 'even', 'odd', 'odd', 'even', 'even', 'even', 'odd', 'odd', 'even', 'even', 'odd', 'even', 'even', 'even', 'even', 'even']\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
cb87b1580bd58f6eafb542371764bd02bf14390a
| 806,031 |
ipynb
|
Jupyter Notebook
|
02/Thuc-CS480_Assignment_2.ipynb
|
ThucNguyen007/cs480student
|
0150a1590dd0fe6c697785e3d299905a563bbe14
|
[
"MIT"
] | null | null | null |
02/Thuc-CS480_Assignment_2.ipynb
|
ThucNguyen007/cs480student
|
0150a1590dd0fe6c697785e3d299905a563bbe14
|
[
"MIT"
] | null | null | null |
02/Thuc-CS480_Assignment_2.ipynb
|
ThucNguyen007/cs480student
|
0150a1590dd0fe6c697785e3d299905a563bbe14
|
[
"MIT"
] | null | null | null | 922.232265 | 132,688 | 0.950606 |
[
[
[
"\n#Assignment 2",
"_____no_output_____"
]
],
[
[
"%pylab inline",
"Populating the interactive namespace from numpy and matplotlib\n"
],
[
"# we will be using the EEG/MEG analysis library MNE\n# documentation is available here: https://mne.tools/stable/index.html\n!pip install -U mne",
"Requirement already satisfied: mne in c:\\users\\thucd\\appdata\\local\\programs\\python\\python38\\lib\\site-packages (0.23.0)\nRequirement already satisfied: scipy>=1.1.0 in c:\\users\\thucd\\appdata\\local\\programs\\python\\python38\\lib\\site-packages (from mne) (1.4.1)\nRequirement already satisfied: numpy>=1.15.4 in c:\\users\\thucd\\appdata\\local\\programs\\python\\python38\\lib\\site-packages (from mne) (1.20.2)\n"
],
[
"import mne # let's import MNE\n\n# .. and the sample dataset\nfrom mne.datasets import sample\nfrom mne.channels import combine_channels\nfrom mne.evoked import combine_evoked\n\n# These data were acquired with the Neuromag Vectorview system at \n# MGH/HMS/MIT Athinoula A. Martinos Center Biomedical Imaging. \n# EEG data from an electrode cap was acquired simultaneously with the MEG. \n\n### EXPERIMENT DESCRIPTION ###\n# In this experiment, checkerboard patterns were presented to the subject into\n# the left and right visual field, interspersed by tones to the left or right ear.\n# The interval between the stimuli was 750 ms. Occasionally a smiley face was \n# presented at the center of the visual field. The subject was asked to press a \n# key with the right index finger as soon as possible after the appearance of the face.",
"_____no_output_____"
],
[
"# and let's load it!\ndata_path = sample.data_path()\nraw = mne.io.read_raw_fif(data_path + '/MEG/sample/sample_audvis_raw.fif')",
"Opening raw data file C:\\Users\\thucd\\mne_data\\MNE-sample-data/MEG/sample/sample_audvis_raw.fif...\n Read a total of 3 projection items:\n PCA-v1 (1 x 102) idle\n PCA-v2 (1 x 102) idle\n PCA-v3 (1 x 102) idle\n Range : 25800 ... 192599 = 42.956 ... 320.670 secs\nReady.\n"
]
],
[
[
"**Task 1:** How many EEG channels were used when acquiring the data? [15 Points]\n",
"_____no_output_____"
]
],
[
[
"# Hint: You can use raw.info or raw.ch_names to figure this out.",
"_____no_output_____"
],
[
"EEG_count = 0\nfor ch_name in raw.ch_names:\n if 'EEG' in ch_name:\n EEG_count += 1",
"_____no_output_____"
],
[
"n_channel = len(raw.ch_names)\nprint(n_channel)",
"376\n"
],
[
"print(f\"There were {EEG_count} EEG channels among {len(raw.ch_names)} channels used when acquiring the data.\")",
"There were 60 EEG channels among 376 channels used when acquiring the data.\n"
]
],
[
[
"* There are in total of 60 collection of channels",
"_____no_output_____"
],
[
"**Task 2:** Let's look at some channels! [20 Points]",
"_____no_output_____"
]
],
[
[
"# the code below plots EEG channels 1-8 for 3 seconds after 2 minutes\nchs = ['EEG 001', 'EEG 002', 'EEG 003', 'EEG 004', 'EEG 005', 'EEG 006', 'EEG 007', 'EEG 008']\nchan_idxs = [raw.ch_names.index(ch) for ch in chs]\necg1to8 = raw.plot(order=chan_idxs, start=120, duration=3)",
"_____no_output_____"
],
[
"# plot EEG channels 50-60 for 1 second after 200 seconds.",
"_____no_output_____"
],
[
"# List comprehension and string concatenation to make the list of channel names\nchs = ['EEG 0' + str(i) for i in range(50,61)]\n# Get the channel indices\nchan_idxs = [raw.ch_names.index(ch) for ch in chs]\n# Plot\necg50to60 = raw.plot(order=chan_idxs, start=200, duration=1)",
"_____no_output_____"
]
],
[
[
"**Task 3:** How long between event and brain activity? [30 Points]",
"_____no_output_____"
]
],
[
[
"# the following code plots the stimulus channel for the same time\nchannel_indices = [raw.ch_names.index('STI 014')]\nstim = raw.plot(order=channel_indices, start=200, duration=1)",
"_____no_output_____"
],
[
"# combine the last two plots into one.\r\n# the plot should show EEG channels 50-60 and the stimulus channel together.",
"_____no_output_____"
],
[
"# filter by EEG name and then filter again by number 50-60\nfiltered = filter(lambda x: x[:3] == 'EEG',raw.ch_names)\nfiltered = filter(lambda x: 50 <= int(x[-3:]) <= 60,filtered)\nchs = list(filtered)\n# Added this check here to avoid re-appending each time I re-run this cell\nif 'STI 014' not in chs:\n chs.append('STI 014')\nchan_idxs = [raw.ch_names.index(ch) for ch in chs]\necg1to8 = raw.plot(order=chan_idxs, start=200, duration=1)\n",
"_____no_output_____"
],
[
"# Estimate the time between stimulus and brain activity.",
"_____no_output_____"
],
[
"# filter by EEG name and then filter again by number 50-60\nfiltered = filter(lambda x: x[:3] == 'EEG',raw.ch_names)\nfiltered = filter(lambda x: 50 <= int(x[-3:]) <= 60,filtered)\nchs = list(filtered)\n\n# let's copy to be data safe and cause no weird errors\n# we must load first all the data to allow filtering by frequency\n# I bandpassed the data with a lowcut to get rid of the baseline shift and a \n# high cut to get rid of the noise\n# now I can trust that finding the max corresponds to the first spike\nbandpassed_raw = raw.copy()\nbandpassed_raw.load_data()\nbandpassed_raw = bandpassed_raw.filter(l_freq=2,h_freq=20)\n\n# I took the data and put into into numpy form\nbandpassed_data = bandpassed_raw.get_data(chs)\n# The data is given back flattened so I arranged it into rows for each channel\nbandpassed_data = np.reshape(bandpassed_data, (len(chs), len(raw.times)) )\n# I took the mean across the channels axis to get the mean filtered signal\nmean_bandpassed_data = np.mean(bandpassed_data, axis=0)\n\n# I got the first sample index of the 200 to 201 duration sample range \n# and I also got its last sample index\nindex_range = np.argwhere((raw.times >= 200) & (raw.times <= 201 ))\nfirstRangeIndex = index_range[0][0]\nlastRangeIndex = index_range[-1][0]\n\nprint(\"ANSWERS:\")\n# The index of the peak is the starting sample index of the duration range plus\n # the index of the max value within the duration range\npeak_index = firstRangeIndex + mean_bandpassed_data[firstRangeIndex:lastRangeIndex+1].argmax()\nraw.times[peak_index]\nprint(\"The time in seconds of the first spike after the stimulus is: {}\".format(raw.times[peak_index]))\n\n# Now all we have to do is take the argmax value of the stimulus channel to find its starting index in a similar way to the mean filtered EEG data\nstimulus_data = raw.get_data('STI 014')\nstim_data_inRange = stimulus_data[0,firstRangeIndex:lastRangeIndex+1]\nstimulus_start_index = firstRangeIndex + stim_data_inRange.argmax()\nprint(\"The time in seconds of the beginning of the stimulus is: {}\".format(raw.times[stimulus_start_index]))\nprint()\n\n# let's subtract the first stimulus presentation time from the first spike in brain activity time to get the duration between stimulus to response\nprint(\"Hence the estimated time in seconds between stimulus and brain activity is: {}\".format(raw.times[peak_index]-raw.times[stimulus_start_index]))\nprint(\"Also the time in milliseconds is {}\".format((raw.times[peak_index]-raw.times[stimulus_start_index])*1000))",
"Reading 0 ... 166799 = 0.000 ... 277.714 secs...\nFiltering raw data in 1 contiguous segment\nSetting up band-pass filter from 2 - 20 Hz\n\nFIR filter parameters\n---------------------\nDesigning a one-pass, zero-phase, non-causal bandpass filter:\n- Windowed time-domain design (firwin) method\n- Hamming window with 0.0194 passband ripple and 53 dB stopband attenuation\n- Lower passband edge: 2.00\n- Lower transition bandwidth: 2.00 Hz (-6 dB cutoff frequency: 1.00 Hz)\n- Upper passband edge: 20.00 Hz\n- Upper transition bandwidth: 5.00 Hz (-6 dB cutoff frequency: 22.50 Hz)\n- Filter length: 993 samples (1.653 sec)\n\nANSWERS:\nThe time in seconds of the first spike after the stimulus is: 200.36296455577963\nThe time in seconds of the beginning of the stimulus is: 200.26972678963904\n\nHence the estimated time in seconds between stimulus and brain activity is: 0.09323776614058943\nAlso the time in milliseconds is 93.23776614058943\n"
]
],
[
[
"** Estimation **\n* As displayed on the graph of EEG 50-60, the highest peak is shown in the EEG-054, the number is roughly 200.36 secs after 200 seconds.\n* About 200.25 secs after 200 secs, the number of seconds is represented as peaking for stimilus 014 channel.\n* Therefore, the estimation between stimulus and brain activity are 200.36 - 200.25 = 0.11 secs\n",
"_____no_output_____"
],
[
"**Task 4:** Localize different brain waves for different stimuli! [35 Points]",
"_____no_output_____"
]
],
[
[
"# the following code groups all stimuli together\n# and allows the visualization of average brain activity per stimuli.\nevents = mne.find_events(raw, stim_channel='STI 014')\nevent_dict = {'auditory/left': 1, \n 'auditory/right': 2, \n 'visual/left': 3,\n 'visual/right': 4,\n 'face': 5, \n 'button': 32}\n\npicks = mne.pick_types(raw.info, eeg=True)\n\nepochs = mne.Epochs(raw, events, event_id=event_dict, picks=picks,\n preload=True)",
"320 events found\nEvent IDs: [ 1 2 3 4 5 32]\nNot setting metadata\nNot setting metadata\n320 matching events found\nSetting baseline interval to [-0.19979521315838786, 0.0] sec\nApplying baseline correction (mode: mean)\n3 projection items activated\nLoading data for 320 events and 421 original time points ...\n0 bad epochs dropped\n"
],
[
"# here we see the average localized brain activity for the right visual stimuli\nvisual_activity = epochs['visual/right'].plot_psd_topomap()",
" Using multitaper spectrum estimation with 7 DPSS windows\n"
],
[
"# here we see the average localized brain activity for the shown 'face'\nface_activity = epochs['face'].plot_psd_topomap()",
" Using multitaper spectrum estimation with 7 DPSS windows\n"
],
[
"# TODO Please visualize the average brain activity when the subject pushes the button",
"_____no_output_____"
],
[
"button_activity = epochs['button'].plot_psd_topomap()",
" Using multitaper spectrum estimation with 7 DPSS windows\n"
],
[
"# Which difference do you see between the visual/right, the face, and the button event?\r\n# Which brain region seems active during the button event?\r\n# visual/right and face seem more similar to the button event.",
"_____no_output_____"
]
],
[
[
"* The most common and equivalent brain scanning to the three activities, including visual/right, face, and pressing button, is having Beta and Gamma stimuli pretty much the same by looking at visualization and the approximation of numbers\n* The differences are displayed clearly in the Delta, Theta and Alpha of each individuals.\n* For instance, The Delta and Gamma stimuli of the Face activity is covered with red area on the left side of the brain. While, pressing button activity is widely covered on the front side.\n* Furthermore, the Alpha stimuli is also displayed the brain analysis differently when comparing between visual/right and button activity. \n* However, the visualizations have shown a bit similarities between Delta and Theta stimuli of visual/right and button activities.",
"_____no_output_____"
],
[
"**Bonus Task:** What type of event happened in Task 3? [33 Points]",
"_____no_output_____"
]
],
[
[
"# which event type happened?",
"_____no_output_____"
],
[
"# the following code groups all stimuli together\n# and allows the visualization of average brain activity per stimuli.\nevents = mne.find_events(raw, stim_channel='STI 014')\nevent_dict = {'auditory/left': 1, \n 'auditory/right': 2, \n 'visual/left': 3,\n 'visual/right': 4, \n 'face': 5,\n 'button': 32}\n\npicks = mne.pick_types(raw.info, eeg=True)\n\nepochs = mne.Epochs(raw, events, event_id=event_dict, picks=picks,\n preload=True)",
"320 events found\nEvent IDs: [ 1 2 3 4 5 32]\nNot setting metadata\nNot setting metadata\n320 matching events found\nSetting baseline interval to [-0.19979521315838786, 0.0] sec\nApplying baseline correction (mode: mean)\n3 projection items activated\nLoading data for 320 events and 421 original time points ...\n0 bad epochs dropped\n"
],
[
"# Display the remaining of activities:\n\n# here we see the average localized brain activity for the left visual stimuli\nleft_visual_activity = epochs['visual/left'].plot_psd_topomap()\n# here we see the average localized brain activity for the left auditory stimuli\nleft_audio_activity = epochs['auditory/left'].plot_psd_topomap()\n# here we see the average localized brain activity for the right auditory stimuli\nright_audio_activity = epochs['auditory/right'].plot_psd_topomap()",
" Using multitaper spectrum estimation with 7 DPSS windows\n"
],
[
"ecg_stimuli = raw.plot(order=chan_idxs, start=200, \n duration=1, events=events,\n event_color= {\n 1: 'chocolate', \n 2: 'darksalmon',\n 3: 'navy', \n 4: 'hotpink', \n 5: 'saddlebrown', \n 32: 'gold' }\n )",
"_____no_output_____"
]
],
[
[
"** Therefore, the brain activity as shown on the line pointed out the number 4, is visual/right brain stimuli **",
"_____no_output_____"
]
],
[
[
"# You did it!!\n#\n# ┈┈┈┈┈┈▕▔╲\n# ┈┈┈┈┈┈┈▏▕\n# ┈┈┈┈┈┈┈▏▕▂▂▂\n# ▂▂▂▂▂▂╱┈▕▂▂▂▏\n# ▉▉▉▉▉┈┈┈▕▂▂▂▏\n# ▉▉▉▉▉┈┈┈▕▂▂▂▏\n# ▔▔▔▔▔▔╲▂▕▂▂|\n#",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb87b541c6200028d0d6ac035c95308760e62fab
| 684,247 |
ipynb
|
Jupyter Notebook
|
benchmarking/one_bend_transient/caseB_integral_term.ipynb
|
ChristopherMayes/PyCSR2D
|
0b15cec4a8a5580fb0e3fa67fa0c13e4a70a369e
|
[
"Apache-2.0"
] | 4 |
2020-09-02T00:35:59.000Z
|
2021-06-02T04:22:16.000Z
|
benchmarking/one_bend_transient/caseB_integral_term.ipynb
|
ChristopherMayes/PyCSR2D
|
0b15cec4a8a5580fb0e3fa67fa0c13e4a70a369e
|
[
"Apache-2.0"
] | 3 |
2021-03-04T00:09:01.000Z
|
2021-03-31T00:48:39.000Z
|
benchmarking/one_bend_transient/caseB_integral_term.ipynb
|
ChristopherMayes/PyCSR2D
|
0b15cec4a8a5580fb0e3fa67fa0c13e4a70a369e
|
[
"Apache-2.0"
] | 1 |
2021-03-03T23:54:05.000Z
|
2021-03-03T23:54:05.000Z
| 877.239744 | 427,088 | 0.950841 |
[
[
[
"import numpy as np",
"_____no_output_____"
],
[
"import numpy as np\n\nimport matplotlib.pyplot as plt\nimport matplotlib\n%matplotlib notebook\n%matplotlib inline\n%config InlineBackend.figure_format = 'retina'\nfont = {'weight' : 'medium',\n 'size' : 13}\nmatplotlib.rc('font', **font)\n\n\nimport time\n\nimport concurrent.futures as cf\n\nimport warnings\nwarnings.filterwarnings(\"ignore\")\n\nimport scipy.constants\nmec2 = scipy.constants.value('electron mass energy equivalent in MeV')*1e6\nc_light = scipy.constants.c\ne_charge = scipy.constants.e\nr_e = scipy.constants.value('classical electron radius')",
"_____no_output_____"
]
],
[
[
"### Parameters",
"_____no_output_____"
]
],
[
[
"gamma = 5000\nrho = 1.5 # Bend radius in m\n\nbeta = (1-1/gamma**2)**(1/2)\n\nsigma_x = 50e-6\nsigma_z = 50e-6\n\n# Entrance angle\nphi = 0.1/rho",
"_____no_output_____"
]
],
[
[
"## code",
"_____no_output_____"
]
],
[
[
"from csr2d.core2 import psi_s, psi_x0_hat\nimport numpy as np",
"_____no_output_____"
],
[
"gamma = 5000\nrho = 1.5 # Bend radius in m\n\nbeta = (1-1/gamma**2)**(1/2)\n\nsigma_x = 50e-6\nsigma_z = 50e-6\nnz = 100\nnx = 100\n\ndz = (10*sigma_z) / (nz - 1)\ndx = (10*sigma_x) / (nx - 1)\n\nzvec = np.linspace(-5*sigma_z, 5*sigma_z, nz)\nxvec = np.linspace(-5*sigma_x, 5*sigma_x, nx)\nzm, xm = np.meshgrid(zvec, xvec, indexing='ij')\npsi_s_grid = psi_s(zm, xm, beta)",
"_____no_output_____"
],
[
"psi_x_grid = psi_x0_hat(zm, xm, beta, dx)",
"_____no_output_____"
],
[
"from csr2d.core2 import psi_s, psi_x_hat, psi_x0_hat\nfrom scipy.interpolate import RectBivariateSpline\nfrom numba import njit, vectorize, float64\nfrom csr2d.kick2 import green_meshes_hat, green_meshes",
"_____no_output_____"
],
[
"# Bypassing the beam, use smooth Gaussian distribution for testing\n\ndef lamb_2d(z,x):\n return 1/(2*np.pi*sigma_x*sigma_z)* np.exp(-z**2 / 2 / sigma_z**2 - x**2 / 2 / sigma_x**2)\ndef lamb_2d_prime(z,x):\n return 1/(2*np.pi*sigma_x*sigma_z)* np.exp(-z**2 / 2 / sigma_z**2 - x**2 / 2 / sigma_x**2) * (-z / sigma_z**2)",
"_____no_output_____"
],
[
"nz = 100\nnx = 100\n\nzvec = np.linspace(-5*sigma_z, 5*sigma_z, nz)\nxvec = np.linspace(-5*sigma_x, 5*sigma_x, nx)\nzm, xm = np.meshgrid(zvec, xvec, indexing='ij')\n\nlambda_grid_filtered = lamb_2d(zm,xm)\nlambda_grid_filtered_prime = lamb_2d_prime(zm,xm)\n\ndz = (10*sigma_z) / (nz - 1)\ndx = (10*sigma_x) / (nx - 1)",
"_____no_output_____"
],
[
"psi_s_grid = psi_s(zm, xm, beta)",
"_____no_output_____"
],
[
"\npsi_s_grid, psi_x_grid, zvec2, xvec2 = green_meshes_hat(nz, nx, dz, dx, rho=rho, beta=beta) \n",
"_____no_output_____"
]
],
[
[
"# Integral term code development",
"_____no_output_____"
]
],
[
[
"# Convolution for a specific observatino point only\n@njit\ndef my_2d_convolve2(g1, g2, ix1, ix2):\n d1, d2 = g1.shape\n g2_flip = np.flip(g2)\n g2_cut = g2_flip[d1-ix1:2*d1-ix1, d2-ix2:2*d2-ix2]\n \n sums = 0\n for i in range(d1):\n for j in range(d2):\n sums+= g1[i,j]*g2_cut[i,j]\n return sums",
"_____no_output_____"
],
[
"#@njit\n# njit doesn't like the condition grid and interpolation....\n\ndef transient_calc_lambda(phi, z_observe, x_observe, zvec, xvec, dz, dx, lambda_grid_filtered_prime, psi_s_grid, psi_x_grid):\n\n x_observe_index = np.argmin(np.abs(xvec - x_observe))\n #print('x_observe_index :', x_observe_index )\n z_observe_index = np.argmin(np.abs(zvec - z_observe))\n #print('z_observe_index :', z_observe_index )\n\n # Boundary condition \n temp = (x_observe - xvec)/rho\n zi_vec = rho*( phi - beta*np.sqrt(temp**2 + 4*(1 + temp)*np.sin(phi/2)**2))\n zo_vec = -beta*np.abs(x_observe - xvec)\n\n condition_grid = np.array([(zvec > z_observe - zo_vec[i]) | (zvec < z_observe - zi_vec[i]) for i in range(len(xvec))])\n\n lambda_grid_filtered_prime_bounded = np.where(condition_grid.T, 0, lambda_grid_filtered_prime)\n \n conv_s = my_2d_convolve2(lambda_grid_filtered_prime_bounded, psi_s_grid, z_observe_index, x_observe_index) \n conv_x = my_2d_convolve2(lambda_grid_filtered_prime_bounded, psi_x_grid, z_observe_index, x_observe_index) \n ##conv_s, conv_x = fftconvolve2(lambda_grid_filtered_prime_bounded, psi_s_grid, psi_x_grid)\n \n #Ws_grid = (beta**2 / abs(rho)) * (conv_s) * (dz * dx)\n #Wx_grid = (beta**2 / abs(rho)) * (conv_x) * (dz * dx)\n\n #lambda_interp = RectBivariateSpline(zvec, xvec, lambda_grid_filtered) # lambda lives in the observation grid\n #lambda_zi_vec = lambda_interp.ev( z_observe - zi_vec, xvec ) \n #psi_x_zi_vec = psi_x0(zi_vec/2/rho, temp, beta, dx)\n #Wx_zi = (beta**2 / rho) * np.dot(psi_x_zi_vec, lambda_zi_vec)*dx\n\n #lambda_zo_vec = lambda_interp.ev( z_observe - zo_vec, xvec ) \n #psi_x_zo_vec = psi_x0(zo_vec/2/rho, temp, beta, dx)\n #Wx_zo = (beta**2 / rho) * np.dot(psi_x_zo_vec, lambda_zo_vec)*dx\n \n #return Wx_grid[ z_observe_index ][ x_observe_index ], Wx_zi, Wx_zo\n #return conv_x, Wx_zi, Wx_zo\n return conv_x\n #return condition_grid",
"_____no_output_____"
],
[
"@njit\ndef transient_calc_lambda_2(phi, z_observe, x_observe, zvec, xvec, dz, dx, lambda_grid_filtered_prime, psi_s_grid, psi_x_grid):\n\n x_observe_index = np.argmin(np.abs(xvec - x_observe))\n #print('x_observe_index :', x_observe_index )\n z_observe_index = np.argmin(np.abs(zvec - z_observe))\n #print('z_observe_index :', z_observe_index )\n\n # Boundary condition \n temp = (x_observe - xvec)/rho\n zi_vec = rho*( phi - beta*np.sqrt(temp**2 + 4*(1 + temp)*np.sin(phi/2)**2))\n zo_vec = -beta*np.abs(x_observe - xvec)\n \n nz = len(zvec)\n nx = len(xvec)\n \n # Allocate array for histogrammed data\n cond = np.zeros( (nz,nx) )\n \n for i in range(nx):\n cond[:,i] = (zvec > z_observe - zo_vec[i]) | (zvec < z_observe - zi_vec[i])\n \n #condition_grid = np.array([(zvec < z_observe - zi_vec[i]) for i in range(len(xvec))])\n #condition_grid = np.array([(zvec > z_observe - zo_vec[i]) | (zvec < z_observe - zi_vec[i]) for i in range(len(xvec))])\n\n lambda_grid_filtered_prime_bounded = np.where(cond, 0, lambda_grid_filtered_prime)\n \n conv_s = my_2d_convolve2(lambda_grid_filtered_prime_bounded, psi_s_grid, z_observe_index, x_observe_index) \n conv_x = my_2d_convolve2(lambda_grid_filtered_prime_bounded, psi_x_grid, z_observe_index, x_observe_index) \n \n \n return conv_x",
"_____no_output_____"
]
],
[
[
"# Applying the codes",
"_____no_output_____"
],
[
"### Note that numba-jitted code are slower the FIRST time",
"_____no_output_____"
]
],
[
[
"t1 = time.time()\nr1 = transient_calc_lambda(phi, 2*sigma_z, sigma_x, zvec, xvec, dz, dx,lambda_grid_filtered_prime, psi_s_grid, psi_x_grid) \nprint(r1)\nt2 = time.time()\nprint('Mapping takes:', t2-t1, 'sec')",
"-816013178.6486729\nMapping takes: 0.0027959346771240234 sec\n"
],
[
"t1 = time.time()\nr1 = transient_calc_lambda_2(phi, 2*sigma_z, sigma_x, zvec, xvec, dz, dx,lambda_grid_filtered_prime, psi_s_grid, psi_x_grid) \nprint(r1)\nt2 = time.time()\nprint('Mapping takes:', t2-t1, 'sec')",
"_____no_output_____"
]
],
[
[
"## super version for parallelism",
"_____no_output_____"
]
],
[
[
"def transient_calc_lambda_super(z_observe, x_observe):\n return transient_calc_lambda(phi, z_observe, x_observe, zvec, xvec, dz, dx,lambda_grid_filtered_prime, psi_s_grid, psi_x_grid)",
"_____no_output_____"
],
[
"#@njit\n@vectorize([float64(float64,float64)], target='parallel')\ndef transient_calc_lambda_2_super(z_observe, x_observe):\n return transient_calc_lambda_2(phi, z_observe, x_observe, zvec, xvec, dz, dx,lambda_grid_filtered_prime, psi_s_grid, psi_x_grid)",
"_____no_output_____"
],
[
"t1 = time.time()\nwith cf.ProcessPoolExecutor(max_workers=20) as executor:\n result = executor.map(transient_calc_lambda_super, zm.flatten(), xm.flatten())\n g1 = np.array(list(result)).reshape(zm.shape)\n \nt2 = time.time()\nprint('Mapping takes:', t2-t1, 'sec')",
"_____no_output_____"
],
[
"t1 = time.time()\ng4 = transient_calc_lambda_boundary_super_new(zm,xm)\nt2 = time.time()\nprint('Mapping takes:', t2-t1, 'sec')",
"_____no_output_____"
],
[
"fig, ax = plt.subplots(figsize=(8,8))\nax = plt.axes(projection='3d')\nax.plot_surface(zm*1e5, xm*1e5, yaya , cmap='inferno', zorder=1)\n\nax.set_xlabel(r'z $(10^{-5}m)$')\nax.set_ylabel(r'x $(10^{-5}m)$')\nax.set_zlabel(r'$W_x$ $(\\times 10^3/m^2)$ ')\nax.zaxis.labelpad = 10\nax.set_title(r'$W_x$ benchmarking')",
"_____no_output_____"
],
[
"# To be fixed\nfrom scipy.integrate import quad\n\ndef transient_calc_lambda_boundary_quad(phi, z_observe, x_observe, dx):\n \n def integrand_zi(xp):\n\n temp = (x_observe - xp)/rho\n zi = rho*( phi - beta*np.sqrt(temp**2 + 4*(1 + temp)*np.sin(phi/2)**2))\n\n #return psi_x_hat(zi/2/rho, temp, beta)*lamb_2d(z_observe - zi, xp)\n return psi_x0_hat(zi/2/rho, temp, beta, dx)*lamb_2d(z_observe - zi, xp)\n \n def integrand_zo(xp):\n\n zo = -beta*np.abs(x_observe - xp)\n \n #return psi_x_hat(zo/2/rho, temp, beta)*lamb_2d(z_observe - zo, xp)\n return psi_x0_hat(zo/2/rho, temp, beta, dx)*lamb_2d(z_observe - zo, xp) \n \n return quad(integrand_zi, -5*sigma_x, 5*sigma_x)[0]/dx\n \n ",
"_____no_output_____"
],
[
"factor = (beta**2 / rho)*dx",
"_____no_output_____"
],
[
"diff = np.abs((g4.reshape(zm.shape) - g3.reshape(zm.shape))/g3.reshape(zm.shape) )* 100",
"_____no_output_____"
],
[
"diff = np.abs((g0 - g3.reshape(zm.shape))/g3.reshape(zm.shape)) * 100",
"_____no_output_____"
],
[
"g3.shape",
"_____no_output_____"
],
[
"fig, ax = plt.subplots(figsize=(8,8))\nax = plt.axes(projection='3d')\nax.plot_surface(zm*1e5, xm*1e5, factor*g3, cmap='inferno', zorder=1)\n\nax.set_xlabel(r'z $(10^{-5}m)$')\nax.set_ylabel(r'x $(10^{-5}m)$')\nax.set_zlabel(r'$W_x$ $(m^{-2}$) ')\nax.zaxis.labelpad = 10\nax.set_title(r'$W_x$ benchmarking')",
"_____no_output_____"
],
[
"fig, ax = plt.subplots(figsize=(8,8))\nax = plt.axes(projection='3d')\nax.plot_surface(zm*1e5, xm*1e5, diff, cmap='inferno', zorder=1)\n\nax.set_xlabel(r'z $(10^{-5}m)$')\nax.set_ylabel(r'x $(10^{-5}m)$')\nax.set_zlabel(r'$W_x$ $(\\times 10^3/m^2)$ ')\nax.zaxis.labelpad = 10\nax.set_title(r'$W_x$ benchmarking')",
"_____no_output_____"
],
[
"ax.zaxis.set_scale('log')",
"_____no_output_____"
],
[
"plt.plot(diff[30:100,100])",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb87c17acd7056ffe04330240f2afa58e8020a9f
| 164,814 |
ipynb
|
Jupyter Notebook
|
Jupyter/BasicGraphAssignment.ipynb
|
denis83/denis83.github.io
|
fa778bc1f8d8c31fb4a0a03183907a13372651ab
|
[
"CC0-1.0"
] | null | null | null |
Jupyter/BasicGraphAssignment.ipynb
|
denis83/denis83.github.io
|
fa778bc1f8d8c31fb4a0a03183907a13372651ab
|
[
"CC0-1.0"
] | null | null | null |
Jupyter/BasicGraphAssignment.ipynb
|
denis83/denis83.github.io
|
fa778bc1f8d8c31fb4a0a03183907a13372651ab
|
[
"CC0-1.0"
] | null | null | null | 661.903614 | 76,512 | 0.948342 |
[
[
[
"# M2: Basic Graphing Assignment - Denis Pelevin",
"_____no_output_____"
]
],
[
[
"# Import matplotlib and Pandas\nimport matplotlib.pyplot as plt\nimport pandas as pd\n\n# Enable in-cell graphs\n%matplotlib inline\n\n# Read-in the input files and tore in Data frames\ndf_opiods = pd.read_csv('OpiodsVA.csv')\ndf_pres = pd.read_csv('presidents.csv')\ndf_cars = pd.read_csv('TOTALNSA.csv')",
"_____no_output_____"
]
],
[
[
"### Problem 1\n**Question 1:** Do opioid overdoes tend to be associated with less affluent areas? That is, areas where\nfamilies have lower incomes?\n\n**Answer:** I chose scatter plots for this problem because they are the best at showing relationship between two numerical values.",
"_____no_output_____"
]
],
[
[
"# Plotting the data and adjusting the apperance of the plots\nfig,ax = plt.subplots()\n\n# Setting properties of graph - Opioids ODs vs Median Income\nax.scatter(df_opiods['MedianHouseholdIncome'], df_opiods['FPOO-Rate'],c = 'blue', alpha = 0.5,s = 50)\nax.xaxis.set_label_text('Median Household Income', fontsize = 14)\nax.yaxis.set_label_text('Opioid Overdoses', fontsize = 14)\n\n# Formating the graph using Figure methods\nfig.suptitle('Opioid ODs vs Median Income', fontsize = 18)\nfig.set_size_inches(8,5)\n\n# Display graph\nplt.show()",
"_____no_output_____"
]
],
[
[
"**Question 2:** What is the relationship in Virginia counties between opioid overdoses and heroin overdoses?\n\n**Answer:** I chose scatter plot for this problem because they are the best at showing relationship between two numerical values.",
"_____no_output_____"
]
],
[
[
"# Plotting the data and adjusting the apperance of the plots\nfig,ax = plt.subplots()\n\n# Setting properties of graph - Opioid ODs vs Heroin ODs\nax.scatter(df_opiods['FPOO-Rate'], df_opiods['FFHO-Rate'], c = 'green', alpha = 0.5,s = 50)\nax.xaxis.set_label_text('Opioid Overdoses', fontsize = 14)\nax.yaxis.set_label_text('Heroin Overdoses', fontsize = 14)\n\n# Formating the graph using Figure methods\nfig.suptitle('Opioid ODs vs Heroin ODs', fontsize = 18)\nfig.set_size_inches(8,5)\n\n# Display graph\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Problem 2\n**Question:** Which states are associated with the greatest number of United States presidents in terms of the presidents’ birthplaces?\n\n**Answer:** I chose bar graph (or Histogram) since it's the best when comparing frequences of events.",
"_____no_output_____"
]
],
[
[
"# Counting Values and storing them in a dictionary\nstateCounts = dict(df_pres['State'].value_counts())\n\n# Sorting the dictionary by frequency values and storing key/value pairs as sub-tuples\nsortedList = sorted(stateCounts.items(), key = lambda x: x[1], reverse = True)\n\n# storing sorted values in x and y\nx = [state for [state,freq] in sortedList]\ny = [freq for [state,freq] in sortedList]\n\n# Plotting the data and adjusting the apperance of the plots\nfig, ax = plt.subplots()\n\n# Setting properties of axes\nax.bar(x = x,height = y, color = 'blue', alpha = 0.75)\nax.yaxis.set_label_text('Number of Presidents', fontsize = 14)\nax.xaxis.set_label_text('States', fontsize = 14)\nplt.xticks(rotation = 90)\n\n# Setting properties of the graph\nfig.suptitle('Presidents Birthplace State Frequency Histogram', fontsize = 18)\nfig.set_size_inches(10,5)\n\n# Display graph\nplt.show()",
"_____no_output_____"
]
],
[
[
"# Problem 3\n**Question:** How have vehicle sales in the United States varied over time?\n\n**Answer:** I chose a line graph since they are the best at showing changes over periods of time (timelines).",
"_____no_output_____"
]
],
[
[
"# Plotting the data and adjusting the apperance of the plots\nfig,ax = plt.subplots()\n\n# Setting properties of the graph - Car Sales Overtime\nax.plot(df_cars['DATE'], df_cars['TOTALNSA'],c = 'blue')\nax.xaxis.set_label_text('Date', fontsize = 14)\nax.yaxis.set_label_text('Car Sales', fontsize = 14)\nax.xaxis.set_major_locator(plt.MaxNLocator(50)) # displaying 1 tick per year\nplt.xticks(rotation = 90)\n\n# Formating the graph using Figure methods\nfig.suptitle('US Car Sales', fontsize = 18)\nfig.set_size_inches(16,5)\n\n# Display graph\nplt.show()",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb87c57f4bcb6177364b8b8e39648ca06592e340
| 4,057 |
ipynb
|
Jupyter Notebook
|
pycaret_classification/seldom_server/Install_docker.ipynb
|
InfuseAI/showcase
|
54a177168c6e173580ed5c58fe97fe6018ecd06e
|
[
"Apache-2.0"
] | 1 |
2022-03-22T03:54:41.000Z
|
2022-03-22T03:54:41.000Z
|
pycaret_classification/seldom_server/Install_docker.ipynb
|
InfuseAI/showcase
|
54a177168c6e173580ed5c58fe97fe6018ecd06e
|
[
"Apache-2.0"
] | 4 |
2021-11-01T05:24:11.000Z
|
2022-03-29T05:21:02.000Z
|
pycaret_classification/seldom_server/Install_docker.ipynb
|
InfuseAI/showcase
|
54a177168c6e173580ed5c58fe97fe6018ecd06e
|
[
"Apache-2.0"
] | null | null | null | 30.051852 | 194 | 0.552872 |
[
[
[
"# The process to install docker service.",
"_____no_output_____"
]
],
[
[
"## Check docker service is not found.\n!docker",
"/bin/bash: docker: command not found\n"
],
[
"# Curl docker shell file.\n!curl -fsSL https://get.docker.com -o get-docker.sh",
"_____no_output_____"
],
[
"# Install docker service.\n!sudo sh get-docker.sh",
"# Executing docker install script, commit: 93d2499759296ac1f9c510605fef85052a2c32be\n+ sh -c apt-get update -qq >/dev/null\n+ sh -c DEBIAN_FRONTEND=noninteractive apt-get install -y -qq apt-transport-https ca-certificates curl gnupg >/dev/null\ndebconf: delaying package configuration, since apt-utils is not installed\n+ sh -c curl -fsSL \"https://download.docker.com/linux/ubuntu/gpg\" | gpg --dearmor --yes -o /usr/share/keyrings/docker-archive-keyring.gpg\n+ sh -c echo \"deb [arch=amd64 signed-by=/usr/share/keyrings/docker-archive-keyring.gpg] https://download.docker.com/linux/ubuntu bionic stable\" > /etc/apt/sources.list.d/docker.list\n+ sh -c apt-get update -qq >/dev/null\n+ sh -c DEBIAN_FRONTEND=noninteractive apt-get install -y -qq --no-install-recommends docker-ce-cli docker-scan-plugin docker-ce >/dev/null\ndebconf: delaying package configuration, since apt-utils is not installed\n+ version_gte 20.10\n+ [ -z ]\n+ return 0\n+ sh -c DEBIAN_FRONTEND=noninteractive apt-get install -y -qq docker-ce-rootless-extras >/dev/null\ndebconf: delaying package configuration, since apt-utils is not installed\n\n================================================================================\n\nTo run Docker as a non-privileged user, consider setting up the\nDocker daemon in rootless mode for your user:\n\n dockerd-rootless-setuptool.sh install\n\nVisit https://docs.docker.com/go/rootless/ to learn about rootless mode.\n\n\nTo run the Docker daemon as a fully privileged service, but granting non-root\nusers access, refer to https://docs.docker.com/go/daemon-access/\n\nWARNING: Access to the remote API on a privileged Docker daemon is equivalent\n to root access on the host. Refer to the 'Docker daemon attack surface'\n documentation for details: https://docs.docker.com/go/attack-surface/\n\n================================================================================\n\n"
],
[
"## Check docker service is successfully installed.\n!docker --version",
"Docker version 20.10.8, build 3967b7d\n"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
cb87e327d344ab34356b0cf0744eb136423389c5
| 75,741 |
ipynb
|
Jupyter Notebook
|
homework/Homework_5.ipynb
|
standingtree/finm-analytics-2021
|
6d056655f8f6bb21d67fb53351971804521925ac
|
[
"MIT"
] | 1 |
2021-08-05T21:37:27.000Z
|
2021-08-05T21:37:27.000Z
|
homework/Homework_5.ipynb
|
standingtree/finm-analytics-2021
|
6d056655f8f6bb21d67fb53351971804521925ac
|
[
"MIT"
] | null | null | null |
homework/Homework_5.ipynb
|
standingtree/finm-analytics-2021
|
6d056655f8f6bb21d67fb53351971804521925ac
|
[
"MIT"
] | 3 |
2021-08-05T21:37:35.000Z
|
2021-08-08T14:37:41.000Z
| 33.118059 | 365 | 0.344991 |
[
[
[
"# Data Analysis\n# FINM September Launch\n# Homework Solution 5",
"_____no_output_____"
],
[
"## Imports",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nimport statsmodels.api as sm\nfrom sklearn.linear_model import LinearRegression\nfrom sklearn.decomposition import PCA\nfrom sklearn.cross_decomposition import PLSRegression\nfrom numpy.linalg import svd\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nsns.set()\nimport warnings\nwarnings.filterwarnings(\"ignore\")",
"_____no_output_____"
]
],
[
[
"## Data",
"_____no_output_____"
]
],
[
[
"data = pd.read_excel(\"../data/single_name_return_data.xlsx\", sheet_name=\"total returns\").set_index(\"Date\")\ndata.head()",
"_____no_output_____"
],
[
"equities = data.drop(columns=['SPY', 'SHV'])\nequities.head()",
"_____no_output_____"
]
],
[
[
"## 1 Principal Components",
"_____no_output_____"
],
[
"#### 1.1 \n**Calculate the principal components of the return series.**",
"_____no_output_____"
],
[
"Using linear algebra:",
"_____no_output_____"
]
],
[
[
"clean = equities - equities.mean()\nu, s, vh = svd(clean)\nfactors = clean @ vh.T\nfactors.columns = np.arange(1,23)\nfactors",
"_____no_output_____"
]
],
[
[
"Using a package:",
"_____no_output_____"
]
],
[
[
"pca = PCA(svd_solver='full')\npca.fit(equities.values)\n\npca_factors = pd.DataFrame(pca.transform(equities.values), \n columns=['Factor {}'.format(i+1) for i in range(pca.n_components_)], \n index=equities.index)\npca_factors",
"_____no_output_____"
]
],
[
[
"#### 1.2\n**Report the eigenvalues associated with these principal components. Report each eigenvalue as a percentage of the sum of all the eigenvalues. This is the total variation each PCA explains.**",
"_____no_output_____"
],
[
"Using linear algebra:",
"_____no_output_____"
]
],
[
[
"PCA_eigenvals = pd.DataFrame(index=factors.columns, columns=['Eigen Value', 'Percentage Explained'])\nPCA_eigenvals['Eigen Value'] = s**2\nPCA_eigenvals['Percentage Explained'] = s**2 / (s**2).sum()\nPCA_eigenvals",
"_____no_output_____"
]
],
[
[
"Using package (no eignvalues method):",
"_____no_output_____"
]
],
[
[
"pkg_explained_var = pd.DataFrame(data = pca.explained_variance_ratio_, \n index = factors.columns, \n columns = ['Explained Variance'])\n\npkg_explained_var",
"_____no_output_____"
]
],
[
[
"#### 1.3\n**How many PCs are needed to explain 75% of the variation?**",
"_____no_output_____"
]
],
[
[
"pkg_explained_var.cumsum()",
"_____no_output_____"
]
],
[
[
"We need the first 5 PCs to explain at least 75% of the variation",
"_____no_output_____"
],
[
"#### 1.4\n**Calculate the correlation between the first (largest eigenvalue) principal component with each of the 22 single-name equities. Which correlation is highest?**",
"_____no_output_____"
]
],
[
[
"corr_4 = equities.copy()\ncorr_4['factor 1'] = factors[1]\n\ncorr_equities = corr_4.corr()['factor 1'].to_frame('Correlation to factor 1')\ncorr_equities.iloc[:len(equities.columns)]",
"_____no_output_____"
]
],
[
[
"#### 1.5\n**Calculate the correlation between the SPY and the first, second, third principal components.**",
"_____no_output_____"
]
],
[
[
"fac_corr = factors[[1,2,3]]\nfac_corr['SPY'] = data['SPY']\n\nSPY_corr = fac_corr.corr()['SPY'].to_frame('Correlation to SPY').iloc[:3]\n\nSPY_corr",
"_____no_output_____"
]
],
[
[
"## 2 PCR and PLS",
"_____no_output_____"
],
[
"#### 2.1\n**Principal Component Regression (PCR) refers to using PCA for dimension reduction, and then\nutilizing the principal components in a regression. Try this by regressing SPY on the first 3 PCs\ncalculated in the previous section. Report the r-squared.**",
"_____no_output_____"
]
],
[
[
"y_PCR = data['SPY']\nX_PCR = factors[[1,2,3]]\n\nmodel_PCR = LinearRegression().fit(X_PCR,y_PCR)\nprint('PCR R-squared: ' + str(round(model_PCR.score(X_PCR, y_PCR),3)))",
"PCR R-squared: 0.922\n"
]
],
[
[
"#### 2.2\n**Calculate the Partial Least Squares estimation of SPY on the 22 single-name equities. Model it for 3 factors. Report the r-squared.**",
"_____no_output_____"
]
],
[
[
"X_PLS = equities\ny_PLS = data['SPY']\n\nmodel_PLS = PLSRegression(n_components=3).fit(X_PLS, y_PLS)\n\nprint('PLS R-squared: ' + str(round(model_PLS.score(X_PLS, y_PLS),3)))",
"PLS R-squared: 0.961\n"
]
],
[
[
"#### 2.3\n**Compare the results between these two approaches and against penalized regression seen in the past homework.**",
"_____no_output_____"
],
[
"PCR and PLS both seek to maximize the ability to explain the variation in y variable, and therefore they will have high $R^{2}$ in-sample. When using LASSO or Ridge as our model, we are conservatively forming factors, and penalizing for additional factors. This makes in-sample $R^{2}$ lower as we saw in Homework #4, but may make more robust OOS predictions.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
cb87f3c0e999b193202c128bdd916d9552a512ec
| 42,473 |
ipynb
|
Jupyter Notebook
|
Advanced Computer Vision with TensorFlow/Code/8609_course-working-files/Section 4/Lesson 4.4/Training-ACGAN.ipynb
|
MachineLearningWithHuman/ComputerVision
|
9929a3115241067da2dd4bcbdd628d4c78fa8072
|
[
"Apache-2.0"
] | 3 |
2019-07-10T15:29:59.000Z
|
2020-06-15T17:10:15.000Z
|
Advanced Computer Vision with TensorFlow/Code/8609_course-working-files/Section 4/Lesson 4.4/Training-ACGAN.ipynb
|
MachineLearningWithHuman/ComputerVision
|
9929a3115241067da2dd4bcbdd628d4c78fa8072
|
[
"Apache-2.0"
] | null | null | null |
Advanced Computer Vision with TensorFlow/Code/8609_course-working-files/Section 4/Lesson 4.4/Training-ACGAN.ipynb
|
MachineLearningWithHuman/ComputerVision
|
9929a3115241067da2dd4bcbdd628d4c78fa8072
|
[
"Apache-2.0"
] | 1 |
2020-06-15T16:27:44.000Z
|
2020-06-15T16:27:44.000Z
| 43.076065 | 109 | 0.428013 |
[
[
[
"# Training and Evaluating ACGAN Model\n*by Marvin Bertin*\n<img src=\"../../images/keras-tensorflow-logo.jpg\" width=\"400\">",
"_____no_output_____"
],
[
"# Imports",
"_____no_output_____"
]
],
[
[
"from __future__ import absolute_import\nfrom __future__ import division\nfrom __future__ import print_function\n\nimport tensorflow as tf\nimport numpy as np\nfrom collections import defaultdict\nimport cPickle as pickle\nimport matplotlib.pyplot as plt\nimport pandas as pd\nfrom PIL import Image\nfrom six.moves import range\nfrom glob import glob",
"_____no_output_____"
],
[
"models = tf.contrib.keras.models\nlayers = tf.contrib.keras.layers\nutils = tf.contrib.keras.utils\nlosses = tf.contrib.keras.losses\noptimizers = tf.contrib.keras.optimizers \nmetrics = tf.contrib.keras.metrics\npreprocessing_image = tf.contrib.keras.preprocessing.image\ndatasets = tf.contrib.keras.datasets",
"_____no_output_____"
]
],
[
[
"# Construct Generator",
"_____no_output_____"
]
],
[
[
"def generator(latent_size, classes=10):\n \n def up_sampling_block(x, filter_size):\n x = layers.UpSampling2D(size=(2, 2))(x)\n x = layers.Conv2D(filter_size, (5,5), padding='same', activation='relu')(x)\n return x\n \n # Input 1\n # image class label\n image_class = layers.Input(shape=(1,), dtype='int32', name='image_class')\n \n # class embeddings\n emb = layers.Embedding(classes, latent_size,\n embeddings_initializer='glorot_normal')(image_class)\n \n # 10 classes in MNIST\n cls = layers.Flatten()(emb)\n \n # Input 2\n # latent noise vector\n latent_input = layers.Input(shape=(latent_size,), name='latent_noise')\n \n # hadamard product between latent embedding and a class conditional embedding\n h = layers.multiply([latent_input, cls])\n \n # Conv generator\n x = layers.Dense(1024, activation='relu')(h)\n x = layers.Dense(128 * 7 * 7, activation='relu')(x)\n x = layers.Reshape((7, 7, 128))(x)\n \n # upsample to (14, 14, 128)\n x = up_sampling_block(x, 256)\n \n # upsample to (28, 28, 256)\n x = up_sampling_block(x, 128)\n \n # reduce channel into binary image (28, 28, 1)\n generated_img = layers.Conv2D(1, (2,2), padding='same', activation='tanh')(x)\n \n return models.Model(inputs=[latent_input, image_class],\n outputs=generated_img,\n name='generator') ",
"_____no_output_____"
]
],
[
[
"# Construct Discriminator",
"_____no_output_____"
]
],
[
[
"def discriminator(input_shape=(28, 28, 1)):\n \n def conv_block(x, filter_size, stride):\n x = layers.Conv2D(filter_size, (3,3), padding='same', strides=stride)(x)\n x = layers.LeakyReLU()(x)\n x = layers.Dropout(0.3)(x)\n return x\n \n input_img = layers.Input(shape=input_shape)\n \n x = conv_block(input_img, 32, (2,2))\n x = conv_block(x, 64, (1,1))\n x = conv_block(x, 128, (2,2))\n x = conv_block(x, 256, (1,1))\n \n features = layers.Flatten()(x)\n \n # binary classifier, image fake or real\n fake = layers.Dense(1, activation='sigmoid', name='generation')(features)\n \n # multi-class classifier, image digit class\n aux = layers.Dense(10, activation='softmax', name='auxiliary')(features)\n \n \n return models.Model(inputs=input_img, outputs=[fake, aux], name='discriminator')",
"_____no_output_____"
]
],
[
[
"# Combine Generator with Discriminator",
"_____no_output_____"
]
],
[
[
"# Adam parameters suggested in paper\nadam_lr = 0.0002\nadam_beta_1 = 0.5\n\ndef ACGAN(latent_size = 100):\n # build the discriminator\n dis = discriminator()\n dis.compile(\n optimizer=optimizers.Adam(lr=adam_lr, beta_1=adam_beta_1),\n loss=['binary_crossentropy', 'sparse_categorical_crossentropy']\n )\n\n # build the generator\n gen = generator(latent_size)\n gen.compile(optimizer=optimizers.Adam(lr=adam_lr, beta_1=adam_beta_1),\n loss='binary_crossentropy')\n\n # Inputs\n latent = layers.Input(shape=(latent_size, ), name='latent_noise')\n image_class = layers.Input(shape=(1,), dtype='int32', name='image_class')\n\n # Get a fake image\n fake_img = gen([latent, image_class])\n\n # Only train generator in combined model\n dis.trainable = False\n fake, aux = dis(fake_img)\n combined = models.Model(inputs=[latent, image_class],\n outputs=[fake, aux],\n name='ACGAN')\n\n combined.compile(\n optimizer=optimizers.Adam(lr=adam_lr, beta_1=adam_beta_1),\n loss=['binary_crossentropy', 'sparse_categorical_crossentropy']\n )\n \n return combined, dis, gen",
"_____no_output_____"
]
],
[
[
"# Load and Normalize MNIST Dataset",
"_____no_output_____"
]
],
[
[
"# reshape to (..., 28, 28, 1)\n# normalize dataset with range [-1, 1]\n(X_train, y_train), (X_test, y_test) = datasets.mnist.load_data()\n\n# normalize and reshape train set\nX_train = (X_train.astype(np.float32) - 127.5) / 127.5\nX_train = np.expand_dims(X_train, axis=-1)\n\n# normalize and reshape test set\nX_test = (X_test.astype(np.float32) - 127.5) / 127.5\nX_test = np.expand_dims(X_test, axis=-1)\n\nnb_train, nb_test = X_train.shape[0], X_test.shape[0]",
"_____no_output_____"
]
],
[
[
"# Training Helper Functions",
"_____no_output_____"
]
],
[
[
"def print_logs(metrics_names, train_history, test_history):\n\n print('{0:<22s} | {1:4s} | {2:15s} | {3:5s}'.format(\n 'component', *metrics_names))\n print('-' * 65)\n\n ROW_FMT = '{0:<22s} | {1:<4.2f} | {2:<15.2f} | {3:<5.2f}'\n print(ROW_FMT.format('generator (train)',\n *train_history['generator'][-1]))\n print(ROW_FMT.format('generator (test)',\n *test_history['generator'][-1]))\n print(ROW_FMT.format('discriminator (train)',\n *train_history['discriminator'][-1]))\n print(ROW_FMT.format('discriminator (test)',\n *test_history['discriminator'][-1]))\n \ndef generate_batch_noise_and_labels(batch_size, latent_size):\n\n # generate a new batch of noise\n noise = np.random.uniform(-1, 1, (batch_size, latent_size))\n\n # sample some labels\n sampled_labels = np.random.randint(0, 10, batch_size)\n\n return noise, sampled_labels",
"_____no_output_____"
]
],
[
[
"# Train and Evaluate ACGAN on MNIST",
"_____no_output_____"
]
],
[
[
"nb_epochs = 50\nbatch_size = 100\n\ntrain_history = defaultdict(list)\ntest_history = defaultdict(list)\n\ncombined, dis, gen = ACGAN(latent_size = 100)\n\nfor epoch in range(nb_epochs):\n print('Epoch {} of {}'.format(epoch + 1, nb_epochs))\n\n nb_batches = int(X_train.shape[0] / batch_size)\n progress_bar = utils.Progbar(target=nb_batches)\n\n epoch_gen_loss = []\n epoch_disc_loss = []\n\n for index in range(nb_batches):\n progress_bar.update(index)\n \n ### Train Discriminator ###\n \n # generate noise and labels\n noise, sampled_labels = generate_batch_noise_and_labels(batch_size, latent_size)\n \n # generate a batch of fake images, using the generated labels as a conditioner\n generated_images = gen.predict([noise, sampled_labels.reshape((-1, 1))], verbose=0)\n\n # get a batch of real images\n image_batch = X_train[index * batch_size:(index + 1) * batch_size]\n label_batch = y_train[index * batch_size:(index + 1) * batch_size]\n\n # construct discriminator dataset\n X = np.concatenate((image_batch, generated_images))\n y = np.array([1] * batch_size + [0] * batch_size)\n aux_y = np.concatenate((label_batch, sampled_labels), axis=0)\n\n # train discriminator\n epoch_disc_loss.append(dis.train_on_batch(X, [y, aux_y]))\n\n ### Train Generator ###\n \n # generate 2 * batch size here such that we have\n # the generator optimize over an identical number of images as the\n # discriminator \n noise, sampled_labels = generate_batch_noise_and_labels(2 * batch_size, latent_size)\n\n # we want to train the generator to trick the discriminator\n # so all the labels should be not-fake (1)\n trick = np.ones(2 * batch_size)\n\n epoch_gen_loss.append(combined.train_on_batch(\n [noise, sampled_labels.reshape((-1, 1))], [trick, sampled_labels]))\n\n print('\\nTesting for epoch {}:'.format(epoch + 1))\n\n ### Evaluate Discriminator ###\n\n # generate a new batch of noise\n noise, sampled_labels = generate_batch_noise_and_labels(nb_test, latent_size)\n\n # generate images\n generated_images = gen.predict(\n [noise, sampled_labels.reshape((-1, 1))], verbose=False)\n\n # construct discriminator evaluation dataset\n X = np.concatenate((X_test, generated_images))\n y = np.array([1] * nb_test + [0] * nb_test)\n aux_y = np.concatenate((y_test, sampled_labels), axis=0)\n\n # evaluate discriminator\n # test loss\n discriminator_test_loss = dis.evaluate(X, [y, aux_y], verbose=False)\n # train loss\n discriminator_train_loss = np.mean(np.array(epoch_disc_loss), axis=0)\n \n ### Evaluate Generator ###\n\n # make new noise\n noise, sampled_labels = generate_batch_noise_and_labels(2 * nb_test, latent_size)\n\n # create labels\n trick = np.ones(2 * nb_test)\n\n # evaluate generator\n # test loss\n generator_test_loss = combined.evaluate(\n [noise, sampled_labels.reshape((-1, 1))],\n [trick, sampled_labels], verbose=False)\n\n # train loss\n generator_train_loss = np.mean(np.array(epoch_gen_loss), axis=0)\n\n ### Save Losses per Epoch ###\n \n # append train losses\n train_history['generator'].append(generator_train_loss)\n train_history['discriminator'].append(discriminator_train_loss)\n\n # append test losses\n test_history['generator'].append(generator_test_loss)\n test_history['discriminator'].append(discriminator_test_loss)\n \n # print training and test losses\n print_logs(dis.metrics_names, train_history, test_history)\n \n # save weights every epoch\n gen.save_weights(\n '../logs/params_generator_epoch_{0:03d}.hdf5'.format(epoch), True)\n dis.save_weights(\n '../logs/params_discriminator_epoch_{0:03d}.hdf5'.format(epoch), True)\n\n# Save train test loss history\npickle.dump({'train': train_history, 'test': test_history},\n open('../logs/acgan-history.pkl', 'wb'))",
"Epoch 1 of 50\n599/600 [============================>.] - ETA: 0s\nTesting for epoch 1:\ncomponent | loss | generation_loss | auxiliary_loss\n-----------------------------------------------------------------\ngenerator (train) | 8.10 | 5.75 | 2.35 \ngenerator (test) | 8.77 | 6.44 | 2.32 \ndiscriminator (train) | 1.46 | 0.04 | 1.43 \ndiscriminator (test) | 1.27 | 0.00 | 1.27 \nEpoch 2 of 50\n599/600 [============================>.] - ETA: 0s\nTesting for epoch 2:\ncomponent | loss | generation_loss | auxiliary_loss\n-----------------------------------------------------------------\ngenerator (train) | 8.74 | 6.42 | 2.32 \ngenerator (test) | 8.47 | 6.16 | 2.31 \ndiscriminator (train) | 1.26 | 0.00 | 1.26 \ndiscriminator (test) | 1.21 | 0.00 | 1.21 \nEpoch 3 of 50\n599/600 [============================>.] - ETA: 0s\nTesting for epoch 3:\ncomponent | loss | generation_loss | auxiliary_loss\n-----------------------------------------------------------------\ngenerator (train) | 8.90 | 6.58 | 2.32 \ngenerator (test) | 8.32 | 6.01 | 2.31 \ndiscriminator (train) | 1.23 | 0.00 | 1.22 \ndiscriminator (test) | 1.20 | 0.00 | 1.19 \nEpoch 4 of 50\n599/600 [============================>.] - ETA: 0s\nTesting for epoch 4:\ncomponent | loss | generation_loss | auxiliary_loss\n-----------------------------------------------------------------\ngenerator (train) | 9.08 | 6.76 | 2.31 \ngenerator (test) | 8.81 | 6.50 | 2.31 \ndiscriminator (train) | 1.21 | 0.00 | 1.21 \ndiscriminator (test) | 1.19 | 0.00 | 1.19 \nEpoch 5 of 50\n599/600 [============================>.] - ETA: 0s\nTesting for epoch 5:\ncomponent | loss | generation_loss | auxiliary_loss\n-----------------------------------------------------------------\ngenerator (train) | 9.28 | 6.97 | 2.31 \ngenerator (test) | 8.75 | 6.45 | 2.31 \ndiscriminator (train) | 1.20 | 0.00 | 1.20 \ndiscriminator (test) | 1.18 | 0.00 | 1.18 \nEpoch 6 of 50\n599/600 [============================>.] - ETA: 0s\nTesting for epoch 6:\ncomponent | loss | generation_loss | auxiliary_loss\n-----------------------------------------------------------------\ngenerator (train) | 9.41 | 7.10 | 2.31 \ngenerator (test) | 8.70 | 6.39 | 2.31 \ndiscriminator (train) | 1.20 | 0.00 | 1.20 \ndiscriminator (test) | 1.18 | 0.00 | 1.18 \nEpoch 7 of 50\n599/600 [============================>.] - ETA: 0s\nTesting for epoch 7:\ncomponent | loss | generation_loss | auxiliary_loss\n-----------------------------------------------------------------\ngenerator (train) | 9.58 | 7.27 | 2.31 \ngenerator (test) | 9.61 | 7.30 | 2.30 \ndiscriminator (train) | 1.19 | 0.00 | 1.19 \ndiscriminator (test) | 1.18 | 0.00 | 1.17 \nEpoch 8 of 50\n599/600 [============================>.] - ETA: 0s\nTesting for epoch 8:\ncomponent | loss | generation_loss | auxiliary_loss\n-----------------------------------------------------------------\ngenerator (train) | 9.68 | 7.37 | 2.31 \ngenerator (test) | 9.65 | 7.34 | 2.30 \ndiscriminator (train) | 1.19 | 0.00 | 1.19 \ndiscriminator (test) | 1.17 | 0.00 | 1.17 \nEpoch 9 of 50\n599/600 [============================>.] - ETA: 0s\nTesting for epoch 9:\ncomponent | loss | generation_loss | auxiliary_loss\n-----------------------------------------------------------------\ngenerator (train) | 9.90 | 7.60 | 2.31 \ngenerator (test) | 10.32 | 8.02 | 2.30 \ndiscriminator (train) | 1.18 | 0.00 | 1.18 \ndiscriminator (test) | 1.17 | 0.00 | 1.17 \nEpoch 10 of 50\n599/600 [============================>.] - ETA: 0s\nTesting for epoch 10:\ncomponent | loss | generation_loss | auxiliary_loss\n-----------------------------------------------------------------\ngenerator (train) | 10.07 | 7.77 | 2.31 \ngenerator (test) | 9.83 | 7.53 | 2.30 \ndiscriminator (train) | 1.18 | 0.00 | 1.18 \ndiscriminator (test) | 1.17 | 0.00 | 1.17 \nEpoch 11 of 50\n599/600 [============================>.] - ETA: 0s\nTesting for epoch 11:\ncomponent | loss | generation_loss | auxiliary_loss\n-----------------------------------------------------------------\ngenerator (train) | 10.25 | 7.94 | 2.31 \ngenerator (test) | 10.46 | 8.15 | 2.30 \ndiscriminator (train) | 1.18 | 0.00 | 1.18 \ndiscriminator (test) | 1.17 | 0.00 | 1.17 \nEpoch 12 of 50\n599/600 [============================>.] - ETA: 0s\nTesting for epoch 12:\ncomponent | loss | generation_loss | auxiliary_loss\n-----------------------------------------------------------------\ngenerator (train) | 10.47 | 8.17 | 2.31 \ngenerator (test) | 10.20 | 7.89 | 2.30 \ndiscriminator (train) | 1.18 | 0.00 | 1.18 \ndiscriminator (test) | 1.17 | 0.00 | 1.17 \nEpoch 13 of 50\n599/600 [============================>.] - ETA: 0s\nTesting for epoch 13:\ncomponent | loss | generation_loss | auxiliary_loss\n-----------------------------------------------------------------\ngenerator (train) | 10.54 | 8.24 | 2.31 \ngenerator (test) | 10.85 | 8.55 | 2.30 \ndiscriminator (train) | 1.18 | 0.00 | 1.18 \ndiscriminator (test) | 1.17 | 0.00 | 1.17 \nEpoch 14 of 50\n599/600 [============================>.] - ETA: 0s\nTesting for epoch 14:\ncomponent | loss | generation_loss | auxiliary_loss\n-----------------------------------------------------------------\ngenerator (train) | 10.76 | 8.46 | 2.31 \ngenerator (test) | 10.97 | 8.66 | 2.31 \ndiscriminator (train) | 1.18 | 0.00 | 1.18 \ndiscriminator (test) | 1.17 | 0.00 | 1.17 \nEpoch 15 of 50\n599/600 [============================>.] - ETA: 0s\nTesting for epoch 15:\ncomponent | loss | generation_loss | auxiliary_loss\n-----------------------------------------------------------------\ngenerator (train) | 10.94 | 8.63 | 2.31 \ngenerator (test) | 11.28 | 8.98 | 2.30 \ndiscriminator (train) | 1.18 | 0.00 | 1.18 \ndiscriminator (test) | 1.17 | 0.00 | 1.17 \nEpoch 16 of 50\n599/600 [============================>.] - ETA: 0s\nTesting for epoch 16:\ncomponent | loss | generation_loss | auxiliary_loss\n-----------------------------------------------------------------\ngenerator (train) | 11.08 | 8.77 | 2.30 \ngenerator (test) | 11.82 | 9.52 | 2.30 \ndiscriminator (train) | 1.17 | 0.00 | 1.17 \ndiscriminator (test) | 1.17 | 0.00 | 1.17 \nEpoch 17 of 50\n599/600 [============================>.] - ETA: 0s\nTesting for epoch 17:\ncomponent | loss | generation_loss | auxiliary_loss\n-----------------------------------------------------------------\ngenerator (train) | 11.35 | 9.04 | 2.31 \ngenerator (test) | 11.01 | 8.71 | 2.30 \ndiscriminator (train) | 1.17 | 0.00 | 1.17 \ndiscriminator (test) | 1.17 | 0.00 | 1.17 \nEpoch 18 of 50\n599/600 [============================>.] - ETA: 0s\nTesting for epoch 18:\ncomponent | loss | generation_loss | auxiliary_loss\n-----------------------------------------------------------------\ngenerator (train) | 11.43 | 9.13 | 2.30 \ngenerator (test) | 11.49 | 9.19 | 2.30 \ndiscriminator (train) | 1.17 | 0.00 | 1.17 \ndiscriminator (test) | 1.17 | 0.00 | 1.17 \nEpoch 19 of 50\n599/600 [============================>.] - ETA: 0s\nTesting for epoch 19:\ncomponent | loss | generation_loss | auxiliary_loss\n-----------------------------------------------------------------\ngenerator (train) | 11.66 | 9.36 | 2.30 \ngenerator (test) | 11.59 | 9.28 | 2.30 \ndiscriminator (train) | 1.17 | 0.00 | 1.17 \ndiscriminator (test) | 1.17 | 0.00 | 1.17 \nEpoch 20 of 50\n"
]
],
[
[
"# Generator Loss History",
"_____no_output_____"
]
],
[
[
"hist = pickle.load(open('../logs/acgan-history.pkl'))\n\nfor p in ['train', 'test']:\n for g in ['discriminator', 'generator']:\n hist[p][g] = pd.DataFrame(hist[p][g], columns=['loss', 'generation_loss', 'auxiliary_loss'])\n plt.plot(hist[p][g]['generation_loss'], label='{} ({})'.format(g, p))\n\n# get the NE and show as an equilibrium point\nplt.hlines(-np.log(0.5), 0, hist[p][g]['generation_loss'].shape[0], label='Nash Equilibrium')\nplt.legend()\nplt.title(r'$L_s$ (generation loss) per Epoch')\nplt.xlabel('Epoch')\nplt.ylabel(r'$L_s$')\nplt.show()",
"_____no_output_____"
]
],
[
[
"<img src=\"../../images/gen-loss.png\" width=\"500\">\n\n** Generator Loss: **\n- loss associated with tricking the discriminator\n- training losses converges to the Nash Equilibrium point\n- shakiness comes from the generator and the discriminator competing at the equilibrium.\n",
"_____no_output_____"
],
[
"# Label Classification Loss History",
"_____no_output_____"
]
],
[
[
"for g in ['discriminator', 'generator']:\n for p in ['train', 'test']:\n plt.plot(hist[p][g]['auxiliary_loss'], label='{} ({})'.format(g, p))\n\nplt.legend()\nplt.title(r'$L_c$ (classification loss) per Epoch')\nplt.xlabel('Epoch')\nplt.ylabel(r'$L_c$')\nplt.semilogy()\nplt.show()",
"_____no_output_____"
]
],
[
[
"<img src=\"../../images/class-loss.png\" width=\"500\">\n\n** Label classification loss: **\n- loss associated with the discriminator getting the correct label\n- discriminator and generator loss reach stable congerence point\n",
"_____no_output_____"
],
[
"# Generate Digits Conditioned on Class Label",
"_____no_output_____"
]
],
[
[
"# load the weights from the last epoch\ngen.load_weights(sorted(glob('../logs/params_generator*'))[-1])\n\n# construct batch of noise and labels\nnoise = np.tile(np.random.uniform(-1, 1, (10, latent_size)), (10, 1))\nsampled_labels = np.array([[i] * 10 for i in range(10)]).reshape(-1, 1)\n\n# generate digits\ngenerated_images = gen.predict([noise, sampled_labels], verbose=0)\n\n# arrange them into a grid and un-normalize the pixels\nimg = (np.concatenate([r.reshape(-1, 28)\n for r in np.split(generated_images, 10)\n ], axis=-1) * 127.5 + 127.5).astype(np.uint8)\n\n# plot images\nplt.imshow(img, cmap='gray')\n_ = plt.axis('off')",
"_____no_output_____"
]
],
[
[
"<img src=\"../../images/generated-digits.png\" width=\"500\">",
"_____no_output_____"
],
[
"## End of Section\n\n<img src=\"../../images/divider.png\" width=\"100\">",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
cb8802eaf782032aa8562b365ff389f4e21f667d
| 9,990 |
ipynb
|
Jupyter Notebook
|
d2l/tensorflow/chapter_preliminaries/autograd.ipynb
|
541979210/xdf
|
ab99d242fbabe56a7b4d7723605cc17aab7888b2
|
[
"Apache-2.0"
] | 2 |
2021-12-11T07:19:34.000Z
|
2022-03-11T09:29:49.000Z
|
d2l/tensorflow/chapter_preliminaries/autograd.ipynb
|
541979210/xdf
|
ab99d242fbabe56a7b4d7723605cc17aab7888b2
|
[
"Apache-2.0"
] | null | null | null |
d2l/tensorflow/chapter_preliminaries/autograd.ipynb
|
541979210/xdf
|
ab99d242fbabe56a7b4d7723605cc17aab7888b2
|
[
"Apache-2.0"
] | null | null | null | 19.782178 | 97 | 0.482683 |
[
[
[
"# 自动微分\n:label:`sec_autograd`\n\n正如我们在 :numref:`sec_calculus`中所说的那样,求导是几乎所有深度学习优化算法的关键步骤。\n虽然求导的计算很简单,只需要一些基本的微积分。\n但对于复杂的模型,手工进行更新是一件很痛苦的事情(而且经常容易出错)。\n\n深度学习框架通过自动计算导数,即*自动微分*(automatic differentiation)来加快求导。\n实际中,根据我们设计的模型,系统会构建一个*计算图*(computational graph),\n来跟踪计算是哪些数据通过哪些操作组合起来产生输出。\n自动微分使系统能够随后反向传播梯度。\n这里,*反向传播*(backpropagate)意味着跟踪整个计算图,填充关于每个参数的偏导数。\n\n## 一个简单的例子\n\n作为一个演示例子,(**假设我们想对函数$y=2\\mathbf{x}^{\\top}\\mathbf{x}$关于列向量$\\mathbf{x}$求导**)。\n首先,我们创建变量`x`并为其分配一个初始值。\n",
"_____no_output_____"
]
],
[
[
"import tensorflow as tf\n\nx = tf.range(4, dtype=tf.float32)\nx",
"_____no_output_____"
]
],
[
[
"[**在我们计算$y$关于$\\mathbf{x}$的梯度之前,我们需要一个地方来存储梯度。**]\n重要的是,我们不会在每次对一个参数求导时都分配新的内存。\n因为我们经常会成千上万次地更新相同的参数,每次都分配新的内存可能很快就会将内存耗尽。\n注意,一个标量函数关于向量$\\mathbf{x}$的梯度是向量,并且与$\\mathbf{x}$具有相同的形状。\n",
"_____no_output_____"
]
],
[
[
"x = tf.Variable(x)",
"_____no_output_____"
]
],
[
[
"(**现在让我们计算$y$。**)\n",
"_____no_output_____"
]
],
[
[
"# 把所有计算记录在磁带上\nwith tf.GradientTape() as t:\n y = 2 * tf.tensordot(x, x, axes=1)\ny",
"_____no_output_____"
]
],
[
[
"`x`是一个长度为4的向量,计算`x`和`x`的点积,得到了我们赋值给`y`的标量输出。\n接下来,我们[**通过调用反向传播函数来自动计算`y`关于`x`每个分量的梯度**],并打印这些梯度。\n",
"_____no_output_____"
]
],
[
[
"x_grad = t.gradient(y, x)\nx_grad",
"_____no_output_____"
]
],
[
[
"函数$y=2\\mathbf{x}^{\\top}\\mathbf{x}$关于$\\mathbf{x}$的梯度应为$4\\mathbf{x}$。\n让我们快速验证这个梯度是否计算正确。\n",
"_____no_output_____"
]
],
[
[
"x_grad == 4 * x",
"_____no_output_____"
]
],
[
[
"[**现在让我们计算`x`的另一个函数。**]\n",
"_____no_output_____"
]
],
[
[
"with tf.GradientTape() as t:\n y = tf.reduce_sum(x)\nt.gradient(y, x) # 被新计算的梯度覆盖",
"_____no_output_____"
]
],
[
[
"## 非标量变量的反向传播\n\n当`y`不是标量时,向量`y`关于向量`x`的导数的最自然解释是一个矩阵。\n对于高阶和高维的`y`和`x`,求导的结果可以是一个高阶张量。\n\n然而,虽然这些更奇特的对象确实出现在高级机器学习中(包括[**深度学习中**]),\n但当我们调用向量的反向计算时,我们通常会试图计算一批训练样本中每个组成部分的损失函数的导数。\n这里(**,我们的目的不是计算微分矩阵,而是单独计算批量中每个样本的偏导数之和。**)\n",
"_____no_output_____"
]
],
[
[
"with tf.GradientTape() as t:\n y = x * x\nt.gradient(y, x) # 等价于y=tf.reduce_sum(x*x)",
"_____no_output_____"
]
],
[
[
"## 分离计算\n\n有时,我们希望[**将某些计算移动到记录的计算图之外**]。\n例如,假设`y`是作为`x`的函数计算的,而`z`则是作为`y`和`x`的函数计算的。\n想象一下,我们想计算`z`关于`x`的梯度,但由于某种原因,我们希望将`y`视为一个常数,\n并且只考虑到`x`在`y`被计算后发挥的作用。\n\n在这里,我们可以分离`y`来返回一个新变量`u`,该变量与`y`具有相同的值,\n但丢弃计算图中如何计算`y`的任何信息。\n换句话说,梯度不会向后流经`u`到`x`。\n因此,下面的反向传播函数计算`z=u*x`关于`x`的偏导数,同时将`u`作为常数处理,\n而不是`z=x*x*x`关于`x`的偏导数。\n",
"_____no_output_____"
]
],
[
[
"# 设置persistent=True来运行t.gradient多次\nwith tf.GradientTape(persistent=True) as t:\n y = x * x\n u = tf.stop_gradient(y)\n z = u * x\n\nx_grad = t.gradient(z, x)\nx_grad == u",
"_____no_output_____"
]
],
[
[
"由于记录了`y`的计算结果,我们可以随后在`y`上调用反向传播,\n得到`y=x*x`关于的`x`的导数,即`2*x`。\n",
"_____no_output_____"
]
],
[
[
"t.gradient(y, x) == 2 * x",
"_____no_output_____"
]
],
[
[
"## Python控制流的梯度计算\n\n使用自动微分的一个好处是:\n[**即使构建函数的计算图需要通过Python控制流(例如,条件、循环或任意函数调用),我们仍然可以计算得到的变量的梯度**]。\n在下面的代码中,`while`循环的迭代次数和`if`语句的结果都取决于输入`a`的值。\n",
"_____no_output_____"
]
],
[
[
"def f(a):\n b = a * 2\n while tf.norm(b) < 1000:\n b = b * 2\n if tf.reduce_sum(b) > 0:\n c = b\n else:\n c = 100 * b\n return c",
"_____no_output_____"
]
],
[
[
"让我们计算梯度。\n",
"_____no_output_____"
]
],
[
[
"a = tf.Variable(tf.random.normal(shape=()))\nwith tf.GradientTape() as t:\n d = f(a)\nd_grad = t.gradient(d, a)\nd_grad",
"_____no_output_____"
]
],
[
[
"我们现在可以分析上面定义的`f`函数。\n请注意,它在其输入`a`中是分段线性的。\n换言之,对于任何`a`,存在某个常量标量`k`,使得`f(a)=k*a`,其中`k`的值取决于输入`a`。\n因此,我们可以用`d/a`验证梯度是否正确。\n",
"_____no_output_____"
]
],
[
[
"d_grad == d / a",
"_____no_output_____"
]
],
[
[
"## 小结\n\n* 深度学习框架可以自动计算导数:我们首先将梯度附加到想要对其计算偏导数的变量上。然后我们记录目标值的计算,执行它的反向传播函数,并访问得到的梯度。\n\n## 练习\n\n1. 为什么计算二阶导数比一阶导数的开销要更大?\n1. 在运行反向传播函数之后,立即再次运行它,看看会发生什么。\n1. 在控制流的例子中,我们计算`d`关于`a`的导数,如果我们将变量`a`更改为随机向量或矩阵,会发生什么?\n1. 重新设计一个求控制流梯度的例子,运行并分析结果。\n1. 使$f(x)=\\sin(x)$,绘制$f(x)$和$\\frac{df(x)}{dx}$的图像,其中后者不使用$f'(x)=\\cos(x)$。\n",
"_____no_output_____"
],
[
"[Discussions](https://discuss.d2l.ai/t/1757)\n",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
cb88073c0b6fbde81f6efd2a616a607ecf3b3eac
| 73,808 |
ipynb
|
Jupyter Notebook
|
dev_nbs/course/rossman_data_clean.ipynb
|
EtienneT/fastai
|
d7ea7c628b5bdb955804aeba237dd93591c1c0cd
|
[
"Apache-2.0"
] | 5 |
2020-08-27T00:52:27.000Z
|
2022-03-31T02:46:05.000Z
|
dev_nbs/course/rossman_data_clean.ipynb
|
EtienneT/fastai
|
d7ea7c628b5bdb955804aeba237dd93591c1c0cd
|
[
"Apache-2.0"
] | 22 |
2021-01-07T23:35:00.000Z
|
2022-03-20T00:16:40.000Z
|
dev_nbs/course/rossman_data_clean.ipynb
|
EtienneT/fastai
|
d7ea7c628b5bdb955804aeba237dd93591c1c0cd
|
[
"Apache-2.0"
] | 2 |
2021-04-17T03:33:21.000Z
|
2022-02-25T19:32:34.000Z
| 33.112607 | 673 | 0.403479 |
[
[
[
"# Rossman data preparation",
"_____no_output_____"
],
[
"To illustrate the techniques we need to apply before feeding all the data to a Deep Learning model, we are going to take the example of the [Rossmann sales Kaggle competition](https://www.kaggle.com/c/rossmann-store-sales). Given a wide range of information about a store, we are going to try predict their sale number on a given day. This is very useful to be able to manage stock properly and be able to properly satisfy the demand without wasting anything. The official training set was giving a lot of informations about various stores in Germany, but it was also allowed to use additional data, as long as it was made public and available to all participants.\n\nWe are going to reproduce most of the steps of one of the winning teams that they highlighted in [Entity Embeddings of Categorical Variables](https://arxiv.org/pdf/1604.06737.pdf). In addition to the official data, teams in the top of the leaderboard also used information about the weather, the states of the stores or the Google trends of those days. We have assembled all that additional data in one file available for download [here](http://files.fast.ai/part2/lesson14/rossmann.tgz) if you want to replicate those steps.",
"_____no_output_____"
],
[
"### A first look at the data",
"_____no_output_____"
],
[
"First things first, let's import everything we will need.",
"_____no_output_____"
]
],
[
[
"from fastai.tabular.all import *",
"_____no_output_____"
]
],
[
[
"If you have download the previous file and decompressed it in a folder named rossmann in the fastai data folder, you should see the following list of files with this instruction:",
"_____no_output_____"
]
],
[
[
"path = Config().data/'rossmann'\npath.ls()",
"_____no_output_____"
]
],
[
[
"The data that comes from Kaggle is in 'train.csv', 'test.csv', 'store.csv' and 'sample_submission.csv'. The other files are the additional data we were talking about. Let's start by loading everything using pandas.",
"_____no_output_____"
]
],
[
[
"table_names = ['train', 'store', 'store_states', 'state_names', 'googletrend', 'weather', 'test']\ntables = [pd.read_csv(path/f'{fname}.csv', low_memory=False) for fname in table_names]\ntrain, store, store_states, state_names, googletrend, weather, test = tables",
"_____no_output_____"
]
],
[
[
"To get an idea of the amount of data available, let's just look at the length of the training and test tables.",
"_____no_output_____"
]
],
[
[
"len(train), len(test)",
"_____no_output_____"
]
],
[
[
"So we have more than one million records available. Let's have a look at what's inside:",
"_____no_output_____"
]
],
[
[
"train.head()",
"_____no_output_____"
]
],
[
[
"The `Store` column contains the id of the stores, then we are given the id of the day of the week, the exact date, if the store was open on that day, if there were any promotion in that store during that day, and if it was a state or school holiday. The `Customers` column is given as an indication, and the `Sales` column is what we will try to predict.\n\nIf we look at the test table, we have the same columns, minus `Sales` and `Customers`, and it looks like we will have to predict on dates that are after the ones of the train table.",
"_____no_output_____"
]
],
[
[
"test.head()",
"_____no_output_____"
]
],
[
[
"The other table given by Kaggle contains some information specific to the stores: their type, what the competition looks like, if they are engaged in a permanent promotion program, and if so since then.",
"_____no_output_____"
]
],
[
[
"store.head().T",
"_____no_output_____"
]
],
[
[
"Now let's have a quick look at our four additional dataframes. `store_states` just gives us the abbreviated name of the sate of each store.",
"_____no_output_____"
]
],
[
[
"store_states.head()",
"_____no_output_____"
]
],
[
[
"We can match them to their real names with `state_names`.",
"_____no_output_____"
]
],
[
[
"state_names.head()",
"_____no_output_____"
]
],
[
[
"Which is going to be necessary if we want to use the `weather` table:",
"_____no_output_____"
]
],
[
[
"weather.head().T",
"_____no_output_____"
]
],
[
[
"Lastly the googletrend table gives us the trend of the brand in each state and in the whole of Germany.",
"_____no_output_____"
]
],
[
[
"googletrend.head()",
"_____no_output_____"
]
],
[
[
"Before we apply the fastai preprocessing, we will need to join the store table and the additional ones with our training and test table. Then, as we saw in our first example in chapter 1, we will need to split our variables between categorical and continuous. Before we do that, though, there is one type of variable that is a bit different from the others: dates.\n\nWe could turn each particular day in a category but there are cyclical information in dates we would miss if we did that. We already have the day of the week in our tables, but maybe the day of the month also bears some significance. People might be more inclined to go shopping at the beggining or the end of the month. The number of the week/month is also important to detect seasonal influences.\n\nThen we will try to exctract meaningful information from those dates. For instance promotions on their own are important inputs, but maybe the number of running weeks with promotion is another useful information as it will influence customers. A state holiday in itself is important, but it's more significant to know if we are the day before or after such a holiday as it will impact sales. All of those might seem very specific to this dataset, but you can actually apply them in any tabular data containing time information.\n\nThis first step is called feature-engineering and is extremely important: your model will try to extract useful information from your data but any extra help you can give it in advance is going to make training easier, and the final result better. In Kaggle Competitions using tabular data, it's often the way people prepared their data that makes the difference in the final leaderboard, not the exact model used.",
"_____no_output_____"
],
[
"### Feature Engineering",
"_____no_output_____"
],
[
"#### Merging tables",
"_____no_output_____"
],
[
"To merge tables together, we will use this little helper function that relies on the pandas library. It will merge the tables `left` and `right` by looking at the column(s) which names are in `left_on` and `right_on`: the information in `right` will be added to the rows of the tables in `left` when the data in `left_on` inside `left` is the same as the data in `right_on` inside `right`. If `left_on` and `right_on` are the same, we don't have to pass `right_on`. We keep the fields in `right` that have the same names as fields in `left` and add a `_y` suffix (by default) to those field names.",
"_____no_output_____"
]
],
[
[
"def join_df(left, right, left_on, right_on=None, suffix='_y'):\n if right_on is None: right_on = left_on\n return left.merge(right, how='left', left_on=left_on, right_on=right_on, \n suffixes=(\"\", suffix))",
"_____no_output_____"
]
],
[
[
"First, let's replace the state names in the weather table by the abbreviations, since that's what is used in the other tables.",
"_____no_output_____"
]
],
[
[
"weather = join_df(weather, state_names, \"file\", \"StateName\")\nweather[['file', 'Date', 'State', 'StateName']].head()",
"_____no_output_____"
]
],
[
[
"To double-check the merge happened without incident, we can check that every row has a `State` with this line:",
"_____no_output_____"
]
],
[
[
"len(weather[weather.State.isnull()])",
"_____no_output_____"
]
],
[
[
"We can now safely remove the columns with the state names (`file` and `StateName`) since they we'll use the short codes.",
"_____no_output_____"
]
],
[
[
"weather.drop(columns=['file', 'StateName'], inplace=True)",
"_____no_output_____"
]
],
[
[
"To add the weather informations to our `store` table, we first use the table `store_states` to match a store code with the corresponding state, then we merge with our weather table.",
"_____no_output_____"
]
],
[
[
"store = join_df(store, store_states, 'Store')\nstore = join_df(store, weather, 'State')",
"_____no_output_____"
]
],
[
[
"And again, we can check if the merge went well by looking if new NaNs where introduced.",
"_____no_output_____"
]
],
[
[
"len(store[store.Mean_TemperatureC.isnull()])",
"_____no_output_____"
]
],
[
[
"Next, we want to join the `googletrend` table to this `store` table. If you remember from our previous look at it, it's not exactly in the same format:",
"_____no_output_____"
]
],
[
[
"googletrend.head()",
"_____no_output_____"
]
],
[
[
"We will need to change the column with the states and the columns with the dates:\n- in the column `fil`, the state names contain `Rossmann_DE_XX` with `XX` being the code of the state, so we want to remove `Rossmann_DE`. We will do this by creating a new column containing the last part of a split of the string by '\\_'.\n- in the column `week`, we will extract the date corresponding to the beginning of the week in a new column by taking the last part of a split on ' - '.\n\nIn pandas, creating a new column is very easy: you just have to define them.",
"_____no_output_____"
]
],
[
[
"googletrend['Date'] = googletrend.week.str.split(' - ', expand=True)[0]\ngoogletrend['State'] = googletrend.file.str.split('_', expand=True)[2]\ngoogletrend.head()",
"_____no_output_____"
]
],
[
[
"Let's check everything went well by looking at the values in the new `State` column of our `googletrend` table.",
"_____no_output_____"
]
],
[
[
"store['State'].unique(),googletrend['State'].unique()",
"_____no_output_____"
]
],
[
[
"We have two additional values in the second (`None` and 'SL') but this isn't a problem since they'll be ignored when we join. One problem however is that 'HB,NI' in the first table is named 'NI' in the second one, so we need to change that.",
"_____no_output_____"
]
],
[
[
"googletrend.loc[googletrend.State=='NI', \"State\"] = 'HB,NI'",
"_____no_output_____"
]
],
[
[
"Why do we have a `None` in state? As we said before, there is a global trend for Germany that corresponds to `Rosmann_DE` in the field `file`. For those, the previous split failed which gave the `None` value. We will keep this global trend and put it in a new column.",
"_____no_output_____"
]
],
[
[
"trend_de = googletrend[googletrend.file == 'Rossmann_DE'][['Date', 'trend']]",
"_____no_output_____"
]
],
[
[
"Then we can merge it with the rest of our trends, by adding the suffix '\\_DE' to know it's the general trend.",
"_____no_output_____"
]
],
[
[
"googletrend = join_df(googletrend, trend_de, 'Date', suffix='_DE')",
"_____no_output_____"
]
],
[
[
"Then at this stage, we can remove the columns `file` and `week`since they won't be useful anymore, as well as the rows where `State` is `None` (since they correspond to the global trend that we saved in another column).",
"_____no_output_____"
]
],
[
[
"googletrend.drop(columns=['file', 'week'], axis=1, inplace=True)\ngoogletrend = googletrend[~googletrend['State'].isnull()]",
"_____no_output_____"
]
],
[
[
"The last thing missing to be able to join this with or store table is to extract the week from the date in this table and in the store table: we need to join them on week values since each trend is given for the full week that starts on the indicated date. This is linked to the next topic in feature engineering: extracting dateparts.",
"_____no_output_____"
],
[
"#### Adding dateparts",
"_____no_output_____"
],
[
"If your table contains dates, you will need to split the information there in several column for your Deep Learning model to be able to train properly. There is the basic stuff, such as the day number, week number, month number or year number, but anything that can be relevant to your problem is also useful. Is it the beginning or the end of the month? Is it a holiday?\n\nTo help with this, the fastai library as a convenience function called `add_datepart`. It will take a dataframe and a column you indicate, try to read it as a date, then add all those new columns. If we go back to our `googletrend` table, we now have gour columns.",
"_____no_output_____"
]
],
[
[
"googletrend.head()",
"_____no_output_____"
]
],
[
[
"If we add the dateparts, we will gain a lot more",
"_____no_output_____"
]
],
[
[
"googletrend = add_datepart(googletrend, 'Date', drop=False)",
"_____no_output_____"
],
[
"googletrend.head().T",
"_____no_output_____"
]
],
[
[
"We chose the option `drop=False` as we want to keep the `Date` column for now. Another option is to add the `time` part of the date, but it's not relevant to our problem here. \n\nNow we can join our Google trends with the information in the `store` table, it's just a join on \\['Week', 'Year'\\] once we apply `add_datepart` to that table. Note that we only keep the initial columns of `googletrend` with `Week` and `Year` to avoid all the duplicates.",
"_____no_output_____"
]
],
[
[
"googletrend = googletrend[['trend', 'State', 'trend_DE', 'Week', 'Year']]\nstore = add_datepart(store, 'Date', drop=False)\nstore = join_df(store, googletrend, ['Week', 'Year', 'State'])",
"_____no_output_____"
]
],
[
[
"At this stage, `store` contains all the information about the stores, the weather on that day and the Google trends applicable. We only have to join it with our training and test table. We have to use `make_date` before being able to execute that merge, to convert the `Date` column of `train` and `test` to proper date format.",
"_____no_output_____"
]
],
[
[
"make_date(train, 'Date')\nmake_date(test, 'Date')\ntrain_fe = join_df(train, store, ['Store', 'Date'])\ntest_fe = join_df(test, store, ['Store', 'Date'])",
"_____no_output_____"
]
],
[
[
"#### Elapsed times",
"_____no_output_____"
],
[
"Another feature that can be useful is the elapsed time before/after a certain event occurs. For instance the number of days since the last promotion or before the next school holiday. Like for the date parts, there is a fastai convenience function that will automatically add them.\n\nOne thing to take into account here is that you will need to use that function on the whole time series you have, even the test data: there might be a school holiday that takes place during the training data and it's going to impact those new features in the test data.",
"_____no_output_____"
]
],
[
[
"all_ftrs = train_fe.append(test_fe, sort=False)",
"_____no_output_____"
]
],
[
[
"We will consider the elapsed times for three events: 'Promo', 'StateHoliday' and 'SchoolHoliday'. Note that those must correspondon to booleans in your dataframe. 'Promo' and 'SchoolHoliday' already are (only 0s and 1s) but 'StateHoliday' has multiple values.",
"_____no_output_____"
]
],
[
[
"all_ftrs['StateHoliday'].unique()",
"_____no_output_____"
]
],
[
[
"If we refer to the explanation on Kaggle, 'b' is for Easter, 'c' for Christmas and 'a' for the other holidays. We will just converts this into a boolean that flags any holiday.",
"_____no_output_____"
]
],
[
[
"all_ftrs.StateHoliday = all_ftrs.StateHoliday!='0'",
"_____no_output_____"
]
],
[
[
"Now we can add, for each store, the number of days since or until the next promotion, state or school holiday. This will take a little while since the whole table is big.",
"_____no_output_____"
]
],
[
[
"all_ftrs = add_elapsed_times(all_ftrs, ['Promo', 'StateHoliday', 'SchoolHoliday'], \n date_field='Date', base_field='Store')",
"_____no_output_____"
]
],
[
[
"It added a four new features. If we look at 'StateHoliday' for instance:",
"_____no_output_____"
]
],
[
[
"[c for c in all_ftrs.columns if 'StateHoliday' in c]",
"_____no_output_____"
]
],
[
[
"The column 'AfterStateHoliday' contains the number of days since the last state holiday, 'BeforeStateHoliday' the number of days until the next one. As for 'StateHoliday_bw' and 'StateHoliday_fw', they contain the number of state holidays in the past or future seven days respectively. The same four columns have been added for 'Promo' and 'SchoolHoliday'.\n\nNow that we have added those features, we can split again our tables between the training and the test one.",
"_____no_output_____"
]
],
[
[
"train_df = all_ftrs.iloc[:len(train_fe)]\ntest_df = all_ftrs.iloc[len(train_fe):]",
"_____no_output_____"
]
],
[
[
"One last thing the authors of this winning solution did was to remove the rows with no sales, which correspond to exceptional closures of the stores. This might not have been a good idea since even if we don't have access to the same features in the test data, it can explain why we have some spikes in the training data.",
"_____no_output_____"
]
],
[
[
"train_df = train_df[train_df.Sales != 0.]",
"_____no_output_____"
]
],
[
[
"We will use those for training but since all those steps took a bit of time, it's a good idea to save our progress until now. We will just pickle those tables on the hard drive.",
"_____no_output_____"
]
],
[
[
"train_df.to_pickle(path/'train_clean')\ntest_df.to_pickle(path/'test_clean')",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb880a9a120b297fbf2535435dcf875bb0f19e8c
| 106,854 |
ipynb
|
Jupyter Notebook
|
tfmodule/ipynb_files/Stock_Predict_keras_1.ipynb
|
jwkanggist/tf-keras-stock-pred
|
c10a7ea9934443511bcf4b16096c0f574c4f5b03
|
[
"Apache-2.0"
] | 4 |
2018-10-06T11:46:26.000Z
|
2018-10-16T07:06:03.000Z
|
tfmodule/ipynb_files/Stock_Predict_keras_1.ipynb
|
jwkanggist/tf-keras-rnn-time-pred
|
c10a7ea9934443511bcf4b16096c0f574c4f5b03
|
[
"Apache-2.0"
] | null | null | null |
tfmodule/ipynb_files/Stock_Predict_keras_1.ipynb
|
jwkanggist/tf-keras-rnn-time-pred
|
c10a7ea9934443511bcf4b16096c0f574c4f5b03
|
[
"Apache-2.0"
] | 11 |
2018-10-09T10:49:21.000Z
|
2018-10-16T06:10:29.000Z
| 102.154876 | 21,816 | 0.718279 |
[
[
[
"empty"
]
]
] |
[
"empty"
] |
[
[
"empty"
]
] |
cb880b7bdfff3ab8b50a6a40d7134a53b2d37ffd
| 455,155 |
ipynb
|
Jupyter Notebook
|
Using Python and a Rigol DS1054Z Oscilloscope for Spectral Analysis.ipynb
|
RobotSquirrelProd/Shorts
|
dc8cea72ae2248a70b5d6aa3709644a1649eef57
|
[
"MIT"
] | null | null | null |
Using Python and a Rigol DS1054Z Oscilloscope for Spectral Analysis.ipynb
|
RobotSquirrelProd/Shorts
|
dc8cea72ae2248a70b5d6aa3709644a1649eef57
|
[
"MIT"
] | null | null | null |
Using Python and a Rigol DS1054Z Oscilloscope for Spectral Analysis.ipynb
|
RobotSquirrelProd/Shorts
|
dc8cea72ae2248a70b5d6aa3709644a1649eef57
|
[
"MIT"
] | null | null | null | 434.308206 | 158,700 | 0.94434 |
[
[
[
"### Introduction",
"_____no_output_____"
],
[
"I am testing the idea of using the juyter notebook as my script so the comments are verbose. Hopefully this helps synchronize the notebook content with the video. Comments on this approach are welcome.",
"_____no_output_____"
],
[
"More content like this can be found at [robotsquirrelproductions.com](https://robotsquirrelproductions.com/)",
"_____no_output_____"
],
[
"Today, I will take you through the steps to control and download data from your Rigol DS1054Z oscilloscope using Python in a Jupyter notebook. This tutorial uses the [DS1054Z library](https://ds1054z.readthedocs.io/en/stable/) written by Philipp Klaus. This library is required to replicate these examples in your environment.\n\nI want to mention some sites I found helpful in learning about this. [Ken Shirrif's blog](http://www.righto.com/2013/07/rigol-oscilloscope-hacks-with-python.html) includes spectrographic analysis and how-to export to .wav file. Of course, the [programming manual](https://beyondmeasure.rigoltech.com/acton/attachment/1579/f-0386/1/-/-/-/-/DS1000Z_Programming%20Guide_EN.pdf) itself also proved helpful.",
"_____no_output_____"
],
[
"### Set up the notebook and import the libraries",
"_____no_output_____"
],
[
"I want to document the Python version used in this example. Import the `sys` library and print the version information.",
"_____no_output_____"
]
],
[
[
"import sys\nprint(sys.version)",
"3.8.12 (default, Oct 12 2021, 03:01:40) [MSC v.1916 64 bit (AMD64)]\n"
]
],
[
[
"Begin by importing libraries to connect to the oscilloscope and to display the data. The [Matplotlib package](https://matplotlib.org/) provides data plotting functionality.",
"_____no_output_____"
]
],
[
[
"from ds1054z import DS1054Z\nimport matplotlib.pyplot as plt\nfrom matplotlib.ticker import FormatStrFormatter\nimport numpy as np",
"_____no_output_____"
]
],
[
[
"The [IPython package](https://ipython.org/) provides libraries needed to display the oscilloscope bitmap images.",
"_____no_output_____"
]
],
[
[
"from IPython.display import Image, display",
"_____no_output_____"
]
],
[
[
"Finally, include the libraries from the [SciPy package](https://scipy.org/). These libraries enable single-sided spectral analysis.",
"_____no_output_____"
]
],
[
[
"from scipy.fft import rfft, rfftfreq",
"_____no_output_____"
]
],
[
[
"### Connect to oscilloscope",
"_____no_output_____"
],
[
"Next, I need to connect to the oscilloscope to verify it responds to basic commands.",
"_____no_output_____"
]
],
[
[
"scope = DS1054Z('192.168.1.206')\nprint(scope.idn)",
"RIGOL TECHNOLOGIES,DS1054Z,DS1ZA200902668,00.04.04.SP3\n"
]
],
[
[
"The scope should respond with the make, model, and serial number. It looks like it has, so we have a good connection. \n\nEnsure the scope is in the run mode to collect some data.",
"_____no_output_____"
]
],
[
[
"scope.run()",
"_____no_output_____"
]
],
[
[
"### Download a waveform (source screen)",
"_____no_output_____"
],
[
"#### Configure the oscilloscope",
"_____no_output_____"
],
[
"I have channel 1 connected to a magnetic pickup. For details on which magnetic pickup I used and how I wired it to oscilloscope, check out my blog post on [selecting magnetic pickups](https://robotsquirrelproductions.com/selecting-a-magnetic-pickup/).\n\nThe pickup views a shaft rotating about 600 RPM. I would like to see about 5-6 revolutions on the screen. For this reason, I will set the horizontal time scaling to 50 ms/division. I will also set the probe ratio to unity and the vertical scale to 125 mV/division. These commands use the DS1054Z library, but VISA commands could also be used.",
"_____no_output_____"
]
],
[
[
"scope.set_probe_ratio(1, 1)\nscope.set_channel_scale(1, 0.125)\nscope.timebase_scale = 0.050",
"_____no_output_____"
]
],
[
[
"#### Configure the trigger",
"_____no_output_____"
],
[
"I want to set the trigger to capture a similar waveform each time I read data from the oscilloscope. Setting the trigger fixes the signal with respect to the grid. The magnetic pickup signal rises to approximately 200 mV. For this reason I put the trigger level at 100 mV, about half of the peak.\n\nI switched to VISA commands instead of using the DS1054Z functions. The `write` function sends the VISA commands. ",
"_____no_output_____"
]
],
[
[
"d_trigger_level = 0.1\nscope.write(':trigger:edge:source CHAN1')\nscope.write(':trigger:edge:level ' + format(d_trigger_level))",
"_____no_output_____"
]
],
[
[
"#### Download Rigol screen bitmap",
"_____no_output_____"
],
[
"With the signals captured, I place the oscilloscope in **STOP** mode. This ensures the buffer contents do not change as I pull configuration information.",
"_____no_output_____"
]
],
[
[
"scope.stop()",
"_____no_output_____"
]
],
[
[
"Take a screenshot of the data from the scope. A bitmap showing the oscilloscope configuration can be a helpful reference to check some of the calculations below.",
"_____no_output_____"
]
],
[
[
"bmap_scope = scope.display_data\ndisplay(Image(bmap_scope, width=800))",
"_____no_output_____"
]
],
[
[
"The screen capture confirms the configuration parameters. For example, the top right shows the trigger configured for a rising edge with a threshold value of 100 mV. In the top left, beside the Rigol logo, the image shows the scope is in stop mode and that the horizontal (\"H\") axis has 50 ms/division. The bottom left corner indicates that channel 1 has been configured for 125 mV/division.\n\nThe screen also has information about the memory layout. The top middle of the screenshot shows a wavy line. The wavy line may have greyed-out areas; reference the image below. The wavy line in the transparent region represents the screen buffer. The origin of the screen buffer begins at the left of this transparent area. In contrast, the RAW memory starts at the left of the wavy line regardless of shading. ",
"_____no_output_____"
]
],
[
[
"Image(filename=\"RigolBufferLayout.png\", width=800)",
"_____no_output_____"
]
],
[
[
"#### Download oscilloscope configuration",
"_____no_output_____"
],
[
"I now begin preparing for data collection. First,save off the vertical scale characteristics. Next, store the timescale value.",
"_____no_output_____"
]
],
[
[
"d_voltage_scale = scope.get_channel_scale(1)\nprint('Vertical scale: %0.3f volts' % d_voltage_scale)\nd_timebase_scale_actual = float(scope.query(':TIMebase:SCAle?'))\nprint('Horizontal time scale: %0.3f seconds' % d_timebase_scale_actual)",
"Vertical scale: 0.125 volts\nHorizontal time scale: 0.050 seconds\n"
]
],
[
[
"I like to store the instrument identification to describe the device that acquired the signal. I include this descriptive identifier on the plots along with the data. This can be helpful for troubleshooting. For example, if you later find a problem instrument, this helps identify projects or work that might be impacted.",
"_____no_output_____"
]
],
[
[
"str_idn = scope.idn\nprint(str_idn)",
"RIGOL TECHNOLOGIES,DS1054Z,DS1ZA200902668,00.04.04.SP3\n"
]
],
[
[
"#### Download the signal and plot it",
"_____no_output_____"
],
[
"Next, I use the `get_waveform_samples` function to download the waveform. I set the mode to **NORM** to capture the screen buffer.",
"_____no_output_____"
]
],
[
[
"d_ch1 = scope.get_waveform_samples(1, mode='NORM')",
"_____no_output_____"
]
],
[
[
"The DS1054Z scope should always return 1200 samples. I use the `scope.memory_depth_curr_waveform` command to pull the number of samples.",
"_____no_output_____"
]
],
[
[
"i_ns = scope.memory_depth_curr_waveform\nprint('Number of samples: %0.f' % i_ns)",
"Number of samples: 1200\n"
]
],
[
[
"The scope has twelve horizontal divisions, so the total time for the sample is 50 ms * 12 = 600 ms. Knowing the number of samples and the total length of time, I estimate the sampling frequency as 1200/600 ms = 2000 hertz.",
"_____no_output_____"
]
],
[
[
"d_fs = i_ns/(12.0 * d_timebase_scale_actual)\nprint('Sampling frequency: %0.3f hertz' % d_fs)",
"Sampling frequency: 2000.000 hertz\n"
]
],
[
[
"Next, I create a time series vector for the independent axis in the plot.",
"_____no_output_____"
]
],
[
[
"np_d_time = np.linspace(0,(i_ns-1),i_ns)/d_fs",
"_____no_output_____"
]
],
[
[
"Restore the oscilloscope to **RUN** mode.",
"_____no_output_____"
]
],
[
[
"scope.run()",
"_____no_output_____"
]
],
[
[
"Calculation of the time series completes the plotting preparation. Next, I write up the lines needed to create the [timebase plot](https://robotsquirrelproductions.com/vibration-data-visualization/#timebase-plot). To zoom in and see more detail, change the x-axis limits to `plt.xlim([0, 0.1])` and comment out both `plt.xticks(np.linspace(0, xmax, 13))` and `ax.xaxis.set_major_formatter(FormatStrFormatter('%.2f'))` lines. With these changes, the plot will show the first 100 ms.",
"_____no_output_____"
]
],
[
[
"plt.rcParams['figure.figsize'] = [16, 4]\nplt.figure\nplt.plot(np_d_time, d_ch1)\nplt.grid()\n\nplt.xlabel('Time, seconds')\nxmax = 12.0*d_timebase_scale_actual\nplt.xlim([0, xmax])\nplt.xticks(np.linspace(0, xmax, 13))\nax = plt.gca()\nax.xaxis.set_major_formatter(FormatStrFormatter('%.2f'))\n\nplt.ylabel('Amplitude, volts')\nplt.ylim([-4.0*d_voltage_scale, 4.0 *d_voltage_scale])\nplt.title(str_idn + '\\nChannel 1 | No. Samples %0.f | Sampling freq.: ' % i_ns)\n\nfigure = plt.gcf()\nfigure.set_size_inches(4*1.6, 4)\nplt.savefig('Timebase_Screen.pdf')\n\nplt.show",
"_____no_output_____"
]
],
[
[
"One note of caution: I do not know if the oscilloscope anti-aliased the signal before downsampling for the screen. The programming manual does not provide much detail. It only says it samples at equal time intervals to rebuild the waveform. Not knowing more details, I believe the RAW waveform should be collected to avoid aliasing problems.\n\nThe plot matches the signal presented in the screen capture. Next, we will take a spectrum of this data to examine the frequency content.",
"_____no_output_____"
],
[
"#### Spectral analysis (source: screen)",
"_____no_output_____"
],
[
"A separate [video](https://youtu.be/8KWPlno6VP0) and [blog post](https://robotsquirrelproductions.com/spectral-analysis-in-python/) covers the basics of single-sided spectral analysis in Python. For this reason, I only present the code here. These commands calculate the single-sided spectrum and labels for the frequency axis.",
"_____no_output_____"
]
],
[
[
"cx_y = rfft(d_ch1)/float(i_ns/2.)\nd_ws = rfftfreq(i_ns,1./d_fs)",
"_____no_output_____"
]
],
[
[
"Create the plot and display the single-sided spectrum. I used the plot function to create linear scales in the code below. In the video I also use a logarithmic vertical axis scale (\"log scale\"). Log scales show details that linear scales may miss. For example, to see the noise floor change the second line below to `plt.semilogy(d_ws, abs(cx_y))`. Also, change the fifth line to `plt.xlim([0, 1000])` so that the x-axis limits to 0 to 1000 hertz. The flat section of the spectrum from 200 to 1000 hertz shows the noise floor.",
"_____no_output_____"
]
],
[
[
"plt.figure()\nplt.plot(d_ws, abs(cx_y))\nplt.grid()\nplt.xlabel('Frequency, hertz')\nplt.xlim([0, 200])\nplt.ylabel('Amplitude, volts')\nplt.title('Signal spectrum (Screen)')\n\nfigure = plt.gcf()\nfigure.set_size_inches(4*1.6, 4)\nplt.savefig('Spectrum_Screen.pdf')",
"_____no_output_____"
]
],
[
[
"### Download a waveform (RAW)",
"_____no_output_____"
],
[
"The previous example downloaded the samples from the screen buffer. This example takes it further and downloads the data stored in memory. The overall workflow will be similar, but some details are different.",
"_____no_output_____"
],
[
"Begin by setting the scope mode to **STOP**.",
"_____no_output_____"
]
],
[
[
"scope.stop()",
"_____no_output_____"
]
],
[
[
"Next, get the information needed to make sense of the signals. I start with the vertical scale.",
"_____no_output_____"
]
],
[
[
"d_voltage_scale_raw = scope.get_channel_scale(1)\nprint('Vertical scale: %0.3f volts' % d_voltage_scale_raw)",
"Vertical scale: 0.125 volts\n"
]
],
[
[
"The sampling frequency can be downloaded directly from the oscilloscope for this example. The value returned by `:ACQuire:SRATe?` should match the value in the screenshot, highlighted by a red rectangle in the image below.",
"_____no_output_____"
]
],
[
[
"d_fs_raw = float(scope.query(\":ACQuire:SRATe?\"))\nprint(\"Sampling rate: %0.1f Msp/s \" % (d_fs_raw/1e6))\nImage(filename=\"RigolSampling.png\", width=800)",
"Sampling rate: 10.0 Msp/s \n"
]
],
[
[
"The `memory_depth_internal_currently_shown` returns the number of samples in raw (or deep) memory. This describes the number of samples in the raw memory for the screen's current signal.",
"_____no_output_____"
]
],
[
[
"i_ns_raw = scope.memory_depth_internal_currently_shown\nprint('Number of samples: %0.f' % i_ns_raw)",
"Number of samples: 6000000\n"
]
],
[
[
"In keeping with good practices, I pull the instrument identification again.",
"_____no_output_____"
]
],
[
[
"str_idn_raw = scope.idn\nprint(str_idn_raw)",
"RIGOL TECHNOLOGIES,DS1054Z,DS1ZA200902668,00.04.04.SP3\n"
]
],
[
[
"Next, I enter the Python command to download data from the oscilloscope. Downloading the signal takes a lot of time, on the order of 5-8 minutes for my arrangement. I have found that the cell must be run manually, using `Ctrl-Enter`. Alternatively, the `time.sleep()` function could pause the notebook execution and allow the upload to complete.",
"_____no_output_____"
]
],
[
[
"d_ch1_raw = scope.get_waveform_samples(1, mode='RAW')",
"_____no_output_____"
]
],
[
[
"Lastly, I set up the time series for this raw waveform.",
"_____no_output_____"
]
],
[
[
"np_d_time_raw = np.linspace(0,(i_ns_raw-1), i_ns_raw)/d_fs_raw",
"_____no_output_____"
]
],
[
[
"Place the oscilloscope in **RUN** mode.",
"_____no_output_____"
]
],
[
[
"scope.run()",
"_____no_output_____"
]
],
[
[
"Now I can plot this channel signal data. To zoom in and see more detail, change the x-axis limits to `plt.xlim([0, 0.1])` and comment out both `plt.xticks(np.linspace(0, xmax, 13))` and `ax.xaxis.set_major_formatter(FormatStrFormatter('%.2f'))` lines. With these changes, the plot will show the first 100 ms.",
"_____no_output_____"
]
],
[
[
"plt.rcParams['figure.figsize'] = [16, 4]\nplt.figure\nplt.plot(np_d_time_raw, d_ch1_raw)\nplt.grid()\n\nplt.xlabel('Time, seconds')\nxmax = float(i_ns_raw)/d_fs_raw\nplt.xlim([0, xmax])\nplt.xticks(np.linspace(0, xmax, 13))\nax = plt.gca()\nax.xaxis.set_major_formatter(FormatStrFormatter('%0.2f'))\n\nplt.ylabel('Amplitude, volts')\nplt.ylim([-4.0*d_voltage_scale_raw, 4.0 *d_voltage_scale_raw])\nplt.title(str_idn_raw + '\\n' + 'Raw Channel 1')\n\nfigure = plt.gcf()\nfigure.set_size_inches(4*1.6, 4)\nplt.savefig('Timebase_Raw.pdf')\n\nplt.show",
"_____no_output_____"
]
],
[
[
"#### Spectral analysis (source: RAW)",
"_____no_output_____"
],
[
"These commands calculate the single-sided spectrum and labels for the frequency axis for the raw data.",
"_____no_output_____"
]
],
[
[
"cx_y_raw = rfft(d_ch1_raw)/float(i_ns_raw/2.)\nd_ws_raw = rfftfreq(i_ns_raw,1./d_fs_raw)",
"_____no_output_____"
]
],
[
[
"Create the plot and display the single-sided spectrum. To replicate the results in the video, floor change the second line below to `plt.semilogy(d_ws, abs(cx_y))`. Also, change the fifth line to `plt.xlim([0, 1000])` so that the x-axis limits to 0 to 1000 hertz. The flat section of the spectrum from 200 to 1000 hertz shows the noise floor.",
"_____no_output_____"
]
],
[
[
"plt.figure()\nplt.plot(d_ws_raw, abs(cx_y_raw))\nplt.grid()\nplt.xlabel('Frequency, hertz')\nplt.xlim([0, 200])\nplt.ylabel('Amplitude, -')\nplt.title('Signal spectrum (RAW)')\n\nfigure = plt.gcf()\nfigure.set_size_inches(4*1.6, 4)\nplt.savefig('Spectrum_Raw.pdf')",
"_____no_output_____"
]
],
[
[
"### Conclusion",
"_____no_output_____"
],
[
"I used Python to pull data from a Rigol DS1054Z and plot both timebase and spectrum domain data in this posting. The notebook belongs to a collection of [short examples](https://github.com/RobotSquirrelProd/Shorts) on Github. Here is the [link](https://github.com/RobotSquirrelProd/Shorts/blob/main/Using%20Python%20and%20a%20Rigol%20DS1054Z%20Oscilloscope%20for%20Spectral%20Analysis.ipynb) to the Jupyter notebook. I hope you find the notes helpful, and I look forward to reading your comments.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
cb8823ac7147a7d34ecb8fa88a2f2709823866db
| 6,330 |
ipynb
|
Jupyter Notebook
|
examples/tutorial/scipy20.ipynb
|
meverson86/holoviz
|
3da05f204c22dd53b7fe017138eca83013d29be4
|
[
"BSD-3-Clause"
] | null | null | null |
examples/tutorial/scipy20.ipynb
|
meverson86/holoviz
|
3da05f204c22dd53b7fe017138eca83013d29be4
|
[
"BSD-3-Clause"
] | null | null | null |
examples/tutorial/scipy20.ipynb
|
meverson86/holoviz
|
3da05f204c22dd53b7fe017138eca83013d29be4
|
[
"BSD-3-Clause"
] | null | null | null | 51.463415 | 421 | 0.64534 |
[
[
[
"<style>div.container { width: 100% }</style>\n<img style=\"float:left; vertical-align:text-bottom;\" height=\"65\" width=\"172\" src=\"../assets/holoviz-logo-unstacked.svg\" />\n<div style=\"float:right; vertical-align:text-bottom;\"><h2>SciPy 2020 Tutorial Index</h2></div>",
"_____no_output_____"
],
[
"<div class=\"alert alert-warning\" role=\"alert\"> <strong>NOTE:</strong> This material is subject to change before the tutorial begins. Check out the <a href=\"https://github.com/pyviz/holoviz/tree/scipy20\">scipy20 tag</a> once the tutorial date approaches to access the materials included in the tutorial. For the latest version of the tutorial, visit <a href=\"https://holoviz.org/tutorial\">holoviz.org</a>.\n</div>",
"_____no_output_____"
],
[
"This tutorial will take you through all of the steps involved in exploring data of many different types and sizes, building simple and complex figures, working with billions of data points, adding interactive behavior, widgets and controls, and deploying full dashboards and applications.\n\nWe'll be using a wide range of open-source Python libraries, but focusing on the tools we help maintain as part of the HoloViz project: \n[Panel](https://panel.pyviz.org), \n[hvPlot](https://hvplot.pyviz.org),\n[HoloViews](http://holoviews.org),\n[GeoViews](http://geoviews.org),\n[Datashader](http://datashader.org),\n[Param](http://param.pyviz.org), and \n[Colorcet](http://colorcet.pyviz.org).\n\n<img width=\"800\" src=\"../assets/pn_hp_hv_gv_ds_pa_cs.png\"/>\n\nThese tools were previously part of [PyViz.org](http://pyviz.org), but have been pulled out into [HoloViz.org](http://holoviz.org) to allow PyViz to be fully neutral and general.\n\nThe HoloViz tools have been carefully designed to work together with each other and with the SciPy ecosystem to address a very wide range of data-analysis and visualization tasks, making it simple to discover, understand, and communicate the important properties of your data.\n\n<img align=\"center\" src=\"../assets/earthquakes.png\"></img>\n\nThis notebook serves as the homepage of the tutorial, including a table of contents letting you launch each tutorial section.",
"_____no_output_____"
],
[
"## Index and Schedule\n\n- **Introduction and setup**\n * **5 min** [Setup](./00_Setup.ipynb): Setting up the environment and data files.\n * **20 min** [Overview](./01_Overview.ipynb): Overview of the HoloViz tools, philosophy, and approach.\n\n- **Building dashboards using Panel**\n * **15 min** [Building_Panels](./02_Building_Panels.ipynb): How to make apps and dashboards from Python objects.\n * **5 min** [*Exercise 1*](./exercises/Building_a_Dashboard.ipynb#Exercise-1): Using a mix of visualizable types, create a panel and serve it.\n * **10 min** [Interlinked Panels](./03_Interlinked_Panels.ipynb): Customizing linkages between widgets and displayable objects.\n * **5 min** [*Exercise 2*](./exercises/Building_a_Dashboard.ipynb#Exercise-2): Add widgets to control your dashboard.\n * **10 min** *Break*\n \n \n- **The `.plot` API: a data-centric approach to visualization** \n * **30 min** [Basic Plotting](./04_Basic_Plotting.ipynb): Quick introduction to the `.plot` interface.\n * **10 min** [Composing Plots](./05_Composing_Plots.ipynb): Overlaying and laying out `.hvplot` outputs to show relationships.\n * **10 min** [*Exercise 3*](./exercises/Plotting.ipynb#Exercise-3): Add some `.plot` or `.hvplot` visualizations to your dashboard.\n * **10 min** *Break*\n\n \n- **Custom interactivity**\n * **25 min** [Interlinked Plots](./06_Interlinked_Plots.ipynb): Connecting HoloViews \"streams\" to customize behavior.\n * **10 min** [*Exercise 4*](./exercises/Plotting.ipynb#Exercise-4): Add a linked visualization with HoloViews.\n\n\n- **Working with large datasets**\n * **20 min** [Large Data](./07_Large_Data.ipynb): Using Datashader to pre-render data in Python\n * **10 min** *Break*\n\n\n- **Building advanced dashboards**\n * **15 min** [Advanced Dashboards](./08_Advanced_Dashboards.ipynb): Using Panel to create an advanced dashboard with linked plots and streams.\n * **30 min** [*Exercise 5*](./exercises/Advanced_Dashboarding.ipynb): Build a new dashboard using everything you've learned so far.",
"_____no_output_____"
],
[
"## Related links\n\nYou will find extensive support material on the websites for each package. You may find these links particularly useful during the tutorial:\n\n* [hvPlot user guide](https://hvplot.pyviz.org/user_guide): Guide to the plots available via `.hvplot()`\n* [HoloViews reference gallery](http://holoviews.org/reference/index.html): Visual reference of all HoloViews elements and containers, along with some other components\n* [Panel reference gallery](http://panel.pyviz.org/reference/index.html): Visual reference of all panes, layouts and widgets.\n* [PyViz Examples](http://examples.pyviz.org): Example projects using HoloViz and other PyViz tools",
"_____no_output_____"
]
]
] |
[
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
cb882d8194230d3440aabae6b8e60c31fa532bf1
| 12,079 |
ipynb
|
Jupyter Notebook
|
_book/3_Materialization/Tutorial7/Materialisation_Test.ipynb
|
BlockResearchGroup/CSD2_2022
|
6ecd461937d855397b62ac3ad896b4cbe708ca93
|
[
"MIT"
] | null | null | null |
_book/3_Materialization/Tutorial7/Materialisation_Test.ipynb
|
BlockResearchGroup/CSD2_2022
|
6ecd461937d855397b62ac3ad896b4cbe708ca93
|
[
"MIT"
] | 1 |
2022-02-21T09:09:01.000Z
|
2022-02-21T09:09:01.000Z
|
_book/3_Materialization/Tutorial7/Materialisation_Test.ipynb
|
BlockResearchGroup/CSD2_2022
|
6ecd461937d855397b62ac3ad896b4cbe708ca93
|
[
"MIT"
] | null | null | null | 44.737037 | 5,520 | 0.656097 |
[
[
[
"print('Materialisation Data Test')",
"Materialisation Data Test\n"
],
[
"import os\nimport compas\nfrom compas.datastructures import Mesh, mesh_bounding_box_xy\nfrom compas.geometry import Vector, Frame, Scale\n\nHERE = os.getcwd()\n\nFILE_I = os.path.join(HERE, 'blocks and ribs_RHINO', 'sessions', 'bm_vertical_equilibrium', 'simple_tripod.rv2')\nFILE_O1 = os.path.join(HERE, 'blocks and ribs_RHINO', 'data', 'form.json')\nFILE_O2 = os.path.join(HERE, 'blocks and ribs_RHINO', 'data', 'scaled_form.json')\n\nsession = compas.json_load(FILE_I)\n\nmesh = Mesh.from_data(session['data']['form'])",
"_____no_output_____"
]
],
[
[
"### to delete extra faces(more than 4 edges) if subdivided with catmulclark or other weird subdivision that connects the mesh with the ground",
"_____no_output_____"
]
],
[
[
"delete_faces =[]\n\nfor fkey in mesh.faces():\n if len(mesh.face_vertices(fkey)) > 4:\n delete_faces.append(fkey)\n\nfor fkey in delete_faces: \n mesh.delete_face(fkey)\n mesh.remove_unused_vertices()",
"_____no_output_____"
]
],
[
[
"### scale up the form if needed",
"_____no_output_____"
]
],
[
[
"scaled_mesh = mesh.copy()\n\nbox_points = mesh_bounding_box_xy(scaled_mesh)\nbase_mesh = scaled_mesh.from_points(box_points)\ncentroid = base_mesh.centroid()\n#print (centroid)\nframe = Frame(centroid,Vector(1,0,0),Vector(0,1,0))\n\nS = Scale.from_factors([100, 100, 100], frame)\nscaled_mesh.transform(S)",
"_____no_output_____"
]
],
[
[
"### Visualise and export Initial Mesh",
"_____no_output_____"
]
],
[
[
"mesh.to_json(FILE_O1)\nscaled_mesh.to_json(FILE_O2)\n\nprint(mesh)",
"<Mesh with 37 vertices, 24 faces, 60 edges>\n"
],
[
"from pythreejs import *\nimport numpy as np\nfrom IPython.display import display\n\nvertices = []\nfor face in mesh.faces():\n for v in mesh.face_vertices(face):\n xyz = mesh.vertex_attributes(v, \"xyz\")\n vertices.append(xyz)\n\nprint(vertices)",
"[[-4.223588094538546, -2.049343512703559, 11.75487914807182], [3.3168890776086184, -0.29503424492501495, 14.727819542584887], [-4.305358833080639, 8.309585822394906, 10.17640364037847], [-7.479312274451397, 6.859611040439988, 8.334298209825157], [-4.305358833080639, 8.309585822394906, 10.17640364037847], [-10.705645161290324, 15.92741935483871, 0.0], [-11.9007168252293, 15.10626581888064, 0.0], [-7.479312274451397, 6.859611040439988, 8.334298209825157], [-11.9007168252293, 15.10626581888064, 0.0], [-13.095788489168275, 14.285112282922572, 0.0], [-9.487585344957099, 6.071936033187963, 5.364442791079558], [-7.479312274451397, 6.859611040439988, 8.334298209825157], [-9.487585344957099, 6.071936033187963, 5.364442791079558], [-7.348177668464897, -2.7173239661514548, 7.5029938028193035], [-4.223588094538546, -2.049343512703559, 11.75487914807182], [-7.479312274451397, 6.859611040439988, 8.334298209825157], [5.99353911452859, 6.19208075151476, 11.503994506875527], [3.3168890776086184, -0.29503424492501495, 14.727819542584887], [16.207620220466968, 3.4433146390599068, 10.547679138694978], [16.325436936214732, 6.403473868581464, 8.344654638991253], [16.207620220466968, 3.4433146390599068, 10.547679138694978], [27.31854838709678, 7.056451612903227, 0.0], [26.73106786848563, 8.382104679062955, 0.0], [16.325436936214732, 6.403473868581464, 8.344654638991253], [26.73106786848563, 8.382104679062955, 0.0], [26.143587349874483, 9.707757745222683, 0.0], [16.369520541377753, 7.596395404688231, 6.665962599718423], [16.325436936214732, 6.403473868581464, 8.344654638991253], [16.369520541377753, 7.596395404688231, 6.665962599718423], [6.685197418480475, 7.913536243176651, 9.37090383117994], [5.99353911452859, 6.19208075151476, 11.503994506875527], [16.325436936214732, 6.403473868581464, 8.344654638991253], [8.158548979430396, -5.420329682885874, 13.257094068175222], [3.3168890776086184, -0.29503424492501495, 14.727819542584887], [-1.9294269211550636, -12.245513486930228, 10.50843944668955], [0.9418966861015068, -14.102070241972992, 9.432166081597474], [-1.9294269211550636, -12.245513486930228, 10.50843944668955], [-6.59274193548387, -22.540322580645164, 0.0], [-5.297501359829306, -23.192121063877785, 0.0], [0.9418966861015068, -14.102070241972992, 9.432166081597474], [-5.297501359829306, -23.192121063877785, 0.0], [-4.002260784174742, -23.843919547110403, 0.0], [2.8359660459600002, -15.550316734198093, 5.254973820898563], [0.9418966861015068, -14.102070241972992, 9.432166081597474], [2.8359660459600002, -15.550316734198093, 5.254973820898563], [10.505803673063719, -7.892581564325436, 7.361776481406822], [8.158548979430396, -5.420329682885874, 13.257094068175222], [0.9418966861015068, -14.102070241972992, 9.432166081597474], [-9.510573497351348, 16.74857289079678, 0.0], [-10.705645161290324, 15.92741935483871, 0.0], [-4.305358833080639, 8.309585822394906, 10.17640364037847], [-2.35336697418946, 10.74634713922594, 8.215366394202327], [-4.305358833080639, 8.309585822394906, 10.17640364037847], [3.3168890776086184, -0.29503424492501495, 14.727819542584887], [5.99353911452859, 6.19208075151476, 11.503994506875527], [-2.35336697418946, 10.74634713922594, 8.215366394202327], [5.99353911452859, 6.19208075151476, 11.503994506875527], [6.685197418480475, 7.913536243176651, 9.37090383117994], [-1.5910746808544416, 11.735149092197313, 6.6341145665105286], [-2.35336697418946, 10.74634713922594, 8.215366394202327], [-1.5910746808544416, 11.735149092197313, 6.6341145665105286], [-8.315501833412373, 17.569726426754848, 0.0], [-9.510573497351348, 16.74857289079678, 0.0], [-2.35336697418946, 10.74634713922594, 8.215366394202327], [27.906028905707927, 5.730798546743499, 0.0], [27.31854838709678, 7.056451612903227, 0.0], [16.207620220466968, 3.4433146390599068, 10.547679138694978], [17.869606480947738, 0.6083673196799507, 9.367713345164582], [16.207620220466968, 3.4433146390599068, 10.547679138694978], [3.3168890776086184, -0.29503424492501495, 14.727819542584887], [8.158548979430396, -5.420329682885874, 13.257094068175222], [17.869606480947738, 0.6083673196799507, 9.367713345164582], [8.158548979430396, -5.420329682885874, 13.257094068175222], [10.505803673063719, -7.892581564325436, 7.361776481406822], [19.206227818185663, -1.320314224197729, 5.270032040832946], [17.869606480947738, 0.6083673196799507, 9.367713345164582], [19.206227818185663, -1.320314224197729, 5.270032040832946], [28.493509424319075, 4.405145480583772, 0.0], [27.906028905707927, 5.730798546743499, 0.0], [17.869606480947738, 0.6083673196799507, 9.367713345164582], [-7.887982511138435, -21.888524097412542, 0.0], [-6.59274193548387, -22.540322580645164, 0.0], [-1.9294269211550636, -12.245513486930228, 10.50843944668955], [-5.38375151284651, -12.044626880177754, 8.469455382504629], [-1.9294269211550636, -12.245513486930228, 10.50843944668955], [3.3168890776086184, -0.29503424492501495, 14.727819542584887], [-4.223588094538546, -2.049343512703559, 11.75487914807182], [-5.38375151284651, -12.044626880177754, 8.469455382504629], [-4.223588094538546, -2.049343512703559, 11.75487914807182], [-7.348177668464897, -2.7173239661514548, 7.5029938028193035], [-7.494008524553994, -11.995126423664688, 5.330874543648329], [-5.38375151284651, -12.044626880177754, 8.469455382504629], [-7.494008524553994, -11.995126423664688, 5.330874543648329], [-9.183223086793, -21.236725614179925, 0.0], [-7.887982511138435, -21.888524097412542, 0.0], [-5.38375151284651, -12.044626880177754, 8.469455382504629]]\n"
],
[
"vertices = BufferAttribute(\n array = np.array(vertices,dtype=np.float32),\n normalized = False)\n\ngeometry = BufferGeometry(\n attribute={'position': vertices})\n\ngeometry.exec_three_obj_method('computeVertexNormals')\n\nmesh_3j = Mesh(geometry=geometry,\n material=MeshPhongMaterial(color='#0092D2'),\n position=[0,0,0])",
"_____no_output_____"
],
[
"c = PerspectiveCamera(position = [0, 5, 5], up = [0, 1, 0],\n children=[DirectionalLight(color='white', position=[3,5,1], intensity=0.5)])\n\nscene=Scene(children=[mesh_3j,c, AmbientLight(color='#777777')])\n\nrenderer = Renderer(camera=c, scene=scene, controls=[OrbitControls(controlling=c)],\n width=800, height=600)\ndisplay(renderer)\n\nprint(geometry)",
"_____no_output_____"
],
[
"from pythreejs._example_helper import use_example_model_ids\nuse_example_model_ids()\nBoxGeometry(\n width=5,\n height=10,\n depth=15,\n widthSegments=5,\n heightSegments=10,\n depthSegments=15)",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
cb883d99e489dae64f2635852edf1e6e1fa84607
| 1,002,002 |
ipynb
|
Jupyter Notebook
|
data processing and analysis/Investigating the Gender Wage Gap.ipynb
|
sengkchu/gender-gap-visualization
|
a57f7b86baacb2f7eb2dc891d32943104459d334
|
[
"MIT"
] | 4 |
2018-07-27T16:09:56.000Z
|
2019-05-01T15:30:05.000Z
|
data processing and analysis/Investigating the Gender Wage Gap.ipynb
|
sengkchu/gendergapvisualization
|
a57f7b86baacb2f7eb2dc891d32943104459d334
|
[
"MIT"
] | 2 |
2020-01-11T03:02:48.000Z
|
2021-06-01T21:43:00.000Z
|
data processing and analysis/Investigating the Gender Wage Gap.ipynb
|
sengkchu/gendergapvisualization
|
a57f7b86baacb2f7eb2dc891d32943104459d334
|
[
"MIT"
] | 2 |
2020-01-27T07:25:23.000Z
|
2021-03-22T03:07:55.000Z
| 351.579649 | 120,908 | 0.917044 |
[
[
[
"* [1.0 - Introduction](#1.0---Introduction)\n - [1.1 - Library imports and loading the data from SQL to pandas](#1.1---Library-imports-and-loading-the-data-from-SQL-to-pandas)\n \n \n* [2.0 - Data Cleaning](#2.0---Data-Cleaning)\n - [2.1 - Pre-cleaning, investigating data types](#2.1---Pre-cleaning,-investigating-data-types)\n - [2.2 - Dealing with non-numerical values](#2.2---Dealing-with-non-numerical-values)\n \n \n* [3.0 - Creating New Features](#)\n - [3.1 - Creating the 'gender' column](#3.1---Creating-the-'gender'-column)\n - [3.2 - Categorizing job titles](#3.2---Categorizing-job-titles) \n \n\n* [4.0 - Data Analysis and Visualizations](#4.0---Data-Analysis-and-Visualizations)\n - [4.1 - Overview of the gender gap](#4.1---Overview-of-the-gender-gap) \n - [4.2 - Exploring the year column](#4.2---Exploring-the-year-column)\n - [4.3 - Full time vs. part time employees](#4.3---Full-time-vs.-part-time-employees)\n - [4.4 - Breaking down the total pay](#4.4---Breaking-down-the-total-pay)\n - [4.5 - Breaking down the base pay by job category](#4.5---Breaking-down-the-base-pay-by-job-category) \n - [4.6 - Gender representation by job category](#4.6---Gender-representation-by-job-category)\n - [4.7 - Significance testing by exact job title](#4.7---Significance-testing-by-exact-job-title)\n \n\n* [5.0 - San Francisco vs. Newport Beach](#5.0---San Francisco-vs.-Newport-Beach)\n - [5.1 - Part time vs. full time workers](#5.1---Part-time-vs.-full-time-workers) \n - [5.2 - Comparisons by job cateogry](#5.2---Comparisons-by-job-cateogry) \n - [5.3 - Gender representation by job category](#5.3---Gender-representation-by-job-category) \n \n \n* [6.0 - Conclusion](#6.0---Conclusion)\n\n \n ",
"_____no_output_____"
],
[
"### 1.0 - Introduction\n\nIn this notebook, I will focus on data analysis and preprocessing for the gender wage gap. Specifically, I am going to focus on public jobs in the city of San Francisco and Newport Beach. This data set is publically available on [Kaggle](https://www.kaggle.com/kaggle/sf-salaries) and [Transparent California](https://transparentcalifornia.com/). \n\nI also created a web application based on this dataset. You can play arround with it [here](https://gendergapvisualization.herokuapp.com/). For a complete list of requirements and files used for my web app, check out my GitHub repository [here](https://github.com/sengkchu/gendergapvisualization).\n\nIn this notebook following questions will be explored:\n\n+ Is there an overall gender wage gap for public jobs in San Francisco?\n+ Is the gender gap really 78 cents on the dollar?\n+ Is there a gender wage gap for full time employees?\n+ Is there a gender wage gap for part time employees?\n+ Is there a gender wage gap if the employees were grouped by job categories?\n+ Is there a gender wage gap if the employees were grouped by exact job title?\n+ If the gender wage gap exists, is the data statistically significant?\n+ If the gender wage gap exists, how does the gender wage gap in San Francisco compare with more conservative cities in California?\n\nLastly, I want to mention that I am not affiliated with any political group, everything I write in this project is based on my perspective of the data alone.",
"_____no_output_____"
],
[
"#### 1.1 - Library imports and loading the data from SQL to pandas\n\nThe SQL database is about 18 megabytes, which is small enough for my computer to handle. So I've decided to just load the entire database into memory using pandas. However, I created a function that takes in a SQL query and returns the result as a pandas dataframe just in case I need to use SQL queries.",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nimport sqlite3\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nimport gender_guesser.detector as gender\nimport time\nimport collections\n%matplotlib inline\nsns.set(font_scale=1.5)\n\ndef run_query(query):\n with sqlite3.connect('database.sqlite') as conn:\n return pd.read_sql(query, conn)\n\n#Read the data from SQL->Pandas\nq1 = '''\nSELECT * FROM Salaries\n'''\n\ndata = run_query(q1)\ndata.head()",
"_____no_output_____"
]
],
[
[
"### 2.0 - Data Cleaning\n\nFortunately, this data set is already very clean. However, we should still look into every column. Specifically, we are interested in the data types of each column, and check for null values within the rows.",
"_____no_output_____"
],
[
"#### 2.1 - Pre-cleaning, investigating data types\n\nBefore we do anything to the dataframe, we are going to simply explore the data a little bit.",
"_____no_output_____"
]
],
[
[
"data.dtypes",
"_____no_output_____"
],
[
"data['JobTitle'].nunique()",
"_____no_output_____"
]
],
[
[
"There is no gender column, so we'll have to create one. In addition, we'll need to reduce the number of unique values in the `'JobTitle'` column. `'BasePay'`, `'OvertimePay'`, `'OtherPay'`, and `'Benefits'` are all object columns. We'll need to find a way to covert these into numeric values.\n\nLet's take a look at the rest of the columns using the `.value_counts()` method.",
"_____no_output_____"
]
],
[
[
"data['Year'].value_counts()",
"_____no_output_____"
],
[
"data['Notes'].value_counts()",
"_____no_output_____"
],
[
"data['Agency'].value_counts()",
"_____no_output_____"
],
[
"data['Status'].value_counts()",
"_____no_output_____"
]
],
[
[
"It looks like the data is split into 4 years. The `'Notes'` column is empty for 148654 rows, so we should just remove it. The `'Agency'` column is also not useful, because we already know the data is for San Francisco.\n\nThe `'Status'` column shows a separation for full time employees and part time employees. We should leave that alone for now.",
"_____no_output_____"
],
[
"#### 2.2 - Dealing with non-numerical values\n\nLet's tackle the object columns first, we are going to convert everything into integers using the `pandas.to_numeric()` function. If we run into any errors, the returned value will be NaN.",
"_____no_output_____"
]
],
[
[
"def process_pay(df):\n cols = ['BasePay','OvertimePay', 'OtherPay', 'Benefits']\n \n print('Checking for nulls:')\n for col in cols:\n df[col] = pd.to_numeric(df[col], errors ='coerce')\n print(len(col)*'-')\n print(col)\n print(len(col)*'-')\n print(df[col].isnull().value_counts())\n \n return df\n\ndata = process_pay(data.copy())",
"Checking for nulls:\n-------\nBasePay\n-------\nFalse 148045\nTrue 609\nName: BasePay, dtype: int64\n-----------\nOvertimePay\n-----------\nFalse 148650\nTrue 4\nName: OvertimePay, dtype: int64\n--------\nOtherPay\n--------\nFalse 148650\nTrue 4\nName: OtherPay, dtype: int64\n--------\nBenefits\n--------\nFalse 112491\nTrue 36163\nName: Benefits, dtype: int64\n"
]
],
[
[
"Looking at our results above, we found 609 null values in `BasePay` and 36163 null values in `Benefits`. We are going to drop the rows with null values in `BasePay`. Not everyone will recieve benefits for their job, so it makes more sense to fill in the null values for `Benefits` with zeroes.",
"_____no_output_____"
]
],
[
[
"def process_pay2(df):\n df['Benefits'] = df['Benefits'].fillna(0)\n \n df = df.dropna()\n print(df['BasePay'].isnull().value_counts())\n return df\n\ndata = process_pay2(data)",
"False 148045\nName: BasePay, dtype: int64\n"
]
],
[
[
"Lastly, let's drop the `Agency` and `Notes` columns as they do not provide any information.",
"_____no_output_____"
]
],
[
[
"data = data.drop(columns=['Agency', 'Notes'])",
"_____no_output_____"
]
],
[
[
"### 3.0 - Creating New Features\n\nUnfortunately, this data set does not include demographic information. Since this project is focused on investigating the gender wage gap, we need a way to classify a person's gender. Furthermore, the `JobTitle` column has 2159 unique values. We'll need to simplify this column. ",
"_____no_output_____"
],
[
"#### 3.1 - Creating the 'gender' column\n\nDue to the limitations of this data set. We'll have to assume the gender of the employee by using their first name. The `gender_guesser` library is very useful for this. ",
"_____no_output_____"
]
],
[
[
"#Create the 'Gender' column based on employee's first name.\nd = gender.Detector(case_sensitive=False)\ndata['FirstName'] = data['EmployeeName'].str.split().apply(lambda x: x[0])\ndata['Gender'] = data['FirstName'].apply(lambda x: d.get_gender(x))\ndata['Gender'].value_counts()",
"_____no_output_____"
]
],
[
[
"We are just going to remove employees with ambiguous or gender neutral first names from our analysis.",
"_____no_output_____"
]
],
[
[
"#Retain data with 'male' and 'female' names.\nmale_female_only = data[(data['Gender'] == 'male') | (data['Gender'] == 'female')].copy()\nmale_female_only['Gender'].value_counts()",
"_____no_output_____"
]
],
[
[
"#### 3.2 - Categorizing job titles\n\nNext, we'll have to simplify the `JobTitles` column. To do this, we'll use the brute force method. I created an ordered dictionary with keywords and their associated job category. The generic titles are at the bottom of the dictionary, and the more specific titles are at the top of the dictionary. Then we are going to use a for loop in conjunction with the `.map()` method on the column.\n\nI used the same labels as this [kernel](https://www.kaggle.com/mevanoff24/data-exploration-predicting-salaries) on Kaggle, but I heavily modified the code for readability.",
"_____no_output_____"
]
],
[
[
"def find_job_title2(row):\n \n #Prioritize specific titles on top \n titles = collections.OrderedDict([\n ('Police',['police', 'sherif', 'probation', 'sergeant', 'officer', 'lieutenant']),\n ('Fire', ['fire']),\n ('Transit',['mta', 'transit']),\n ('Medical',['anesth', 'medical', 'nurs', 'health', 'physician', 'orthopedic', 'pharm', 'care']),\n ('Architect', ['architect']),\n ('Court',['court', 'legal']),\n ('Mayor Office', ['mayoral']),\n ('Library', ['librar']),\n ('Public Works', ['public']),\n ('Attorney', ['attorney']),\n ('Custodian', ['custodian']),\n ('Gardener', ['garden']),\n ('Recreation Leader', ['recreation']),\n ('Automotive',['automotive', 'mechanic', 'truck']),\n ('Engineer',['engineer', 'engr', 'eng', 'program']),\n ('General Laborer',['general laborer', 'painter', 'inspector', 'carpenter', 'electrician', 'plumber', 'maintenance']),\n ('Food Services', ['food serv']),\n ('Clerk', ['clerk']),\n ('Porter', ['porter']),\n ('Airport Staff', ['airport']),\n ('Social Worker',['worker']), \n ('Guard', ['guard']),\n ('Assistant',['aide', 'assistant', 'secretary', 'attendant']), \n ('Analyst', ['analy']),\n ('Manager', ['manager']) \n ]) \n \n #Loops through the dictionaries\n for group, keywords in titles.items():\n for keyword in keywords:\n if keyword in row.lower():\n return group\n return 'Other'\n\nstart_time = time.time() \nmale_female_only[\"Job_Group\"] = male_female_only[\"JobTitle\"].map(find_job_title2)\nprint(\"--- Run Time: %s seconds ---\" % (time.time() - start_time))\n\nmale_female_only['Job_Group'].value_counts()",
"--- Run Time: 2.2815001010894775 seconds ---\n"
]
],
[
[
"### 4.0 - Data Analysis and Visualizations\n\nIn this section, we are going to use the data to answer the questions stated in the [introduction section](#1.0---Introduction).",
"_____no_output_____"
],
[
"#### 4.1 - Overview of the gender gap\n\nLet's begin by splitting the data set in half, one for females and one for males. Then we'll plot the overall income distribution using kernel density estimation based on the gausian function.",
"_____no_output_____"
]
],
[
[
"fig = plt.figure(figsize=(10, 5))\nmale_only = male_female_only[male_female_only['Gender'] == 'male']\nfemale_only = male_female_only[male_female_only['Gender'] == 'female']\n\n\nax = sns.kdeplot(male_only['TotalPayBenefits'], color ='Blue', label='Male', shade=True)\nax = sns.kdeplot(female_only['TotalPayBenefits'], color='Red', label='Female', shade=True)\n\nplt.yticks([])\nplt.title('Overall Income Distribution')\nplt.ylabel('Density of Employees')\nplt.xlabel('Total Pay + Benefits ($)')\nplt.xlim(0, 350000)\nplt.show()",
"_____no_output_____"
]
],
[
[
"The income distribution plot is bimodal. In addition, we see a gender wage gap in favor of males in between the ~110000 and the ~275000 region. But, this plot doesn't capture the whole story. We need to break down the data some more. But first, let's explore the percentage of employees based on gender.",
"_____no_output_____"
]
],
[
[
"fig = plt.figure(figsize=(5, 5))\n\ncolors = ['#AFAFF5', '#EFAFB5']\nlabels = ['Male', 'Female']\nsizes = [len(male_only), len(female_only)]\nexplode = (0.05, 0)\nsns.set(font_scale=1.5)\nax = plt.pie(sizes, labels=labels, explode=explode, colors=colors, shadow=True, startangle=90, autopct='%1.f%%')\n\nplt.title('Estimated Percentages of Employees: Overall')\nplt.show()",
"_____no_output_____"
]
],
[
[
"Another key factor we have to consider is the number of employees. How do we know if there are simply more men working at higher paying jobs? How can we determine if social injustice has occured?\n\nThe chart above only tells us the total percentage of employees across all job categories, but it does give us an overview of the data.",
"_____no_output_____"
],
[
"#### 4.2 - Exploring the year column\n\nThe data set contain information on employees between 2011-2014. Let's take a look at an overview of the income based on the `Year` column regardless of gender.",
"_____no_output_____"
]
],
[
[
"data_2011 = male_female_only[male_female_only['Year'] == 2011]\ndata_2012 = male_female_only[male_female_only['Year'] == 2012]\ndata_2013 = male_female_only[male_female_only['Year'] == 2013]\ndata_2014 = male_female_only[male_female_only['Year'] == 2014]\n\n\nplt.figure(figsize=(10,7.5))\nax = plt.boxplot([data_2011['TotalPayBenefits'].values, data_2012['TotalPayBenefits'].values, \\\n data_2013['TotalPayBenefits'].values, data_2014['TotalPayBenefits'].values])\nplt.ylim(0, 350000)\nplt.xticks([1, 2, 3, 4], ['2011', '2012', '2013', '2014'])\nplt.xlabel('Year')\nplt.ylabel('Total Pay + Benefits ($)')\nplt.tight_layout()",
"_____no_output_____"
]
],
[
[
"From the boxplots, we see that the total pay is increasing for every year. We'll have to consider inflation in our analysis. In addition, it is very possible for an employee to stay at their job for multiple years. We don't want to double sample on these employees. \n\nTo simplify the data for the purpose of investigating the gender gap. It makes more sense to only choose only one year for our analysis. From our data exploration, we noticed that the majority of the `status` column was blank. Let's break the data down by year using the `.value_counts()` method.",
"_____no_output_____"
]
],
[
[
"years = ['2011', '2012', '2013', '2014']\nall_data = [data_2011, data_2012, data_2013, data_2014]\n\nfor i in range(4):\n print(len(years[i])*'-')\n print(years[i])\n print(len(years[i])*'-')\n print(all_data[i]['Status'].value_counts())",
"----\n2011\n----\n 29473\nName: Status, dtype: int64\n----\n2012\n----\n 29798\nName: Status, dtype: int64\n----\n2013\n----\n 29838\nName: Status, dtype: int64\n----\n2014\n----\nFT 18295\nPT 12299\nName: Status, dtype: int64\n"
]
],
[
[
"The status of the employee is critical to our analysis, only year 2014 has this information. So it makes sense to focus on analysis on 2014. ",
"_____no_output_____"
]
],
[
[
"data_2014_FT = data_2014[data_2014['Status'] == 'FT']\ndata_2014_PT = data_2014[data_2014['Status'] == 'PT']",
"_____no_output_____"
]
],
[
[
"#### 4.3 - Full time vs. part time employees\n\nLet's take a look at the kernal density estimation plot for part time and full time employees.",
"_____no_output_____"
]
],
[
[
"fig = plt.figure(figsize=(10, 5))\nax = sns.kdeplot(data_2014_PT['TotalPayBenefits'], color = 'Orange', label='Part Time Workers', shade=True)\nax = sns.kdeplot(data_2014_FT['TotalPayBenefits'], color = 'Green', label='Full Time Workers', shade=True)\nplt.yticks([])\n\nplt.title('Part Time Workers vs. Full Time Workers')\nplt.ylabel('Density of Employees')\nplt.xlabel('Total Pay + Benefits ($)')\nplt.xlim(0, 350000)\nplt.show()",
"_____no_output_____"
]
],
[
[
"If we split the data by employment status, we can see that the kernal distribution plot is no longer bimodal. Next, let's see how these two plots look if we seperate the data by gender.",
"_____no_output_____"
]
],
[
[
"fig = plt.figure(figsize=(10, 10))\nfig.subplots_adjust(hspace=.5) \n\n#Generate the top plot\nmale_only = data_2014_FT[data_2014_FT['Gender'] == 'male']\nfemale_only = data_2014_FT[data_2014_FT['Gender'] == 'female']\nax = fig.add_subplot(2, 1, 1)\nax = sns.kdeplot(male_only['TotalPayBenefits'], color ='Blue', label='Male', shade=True)\nax = sns.kdeplot(female_only['TotalPayBenefits'], color='Red', label='Female', shade=True)\nplt.title('Full Time Workers')\nplt.ylabel('Density of Employees')\nplt.xlabel('Total Pay & Benefits ($)')\nplt.xlim(0, 350000)\nplt.yticks([])\n\n#Generate the bottom plot\nmale_only = data_2014_PT[data_2014_PT['Gender'] == 'male']\nfemale_only = data_2014_PT[data_2014_PT['Gender'] == 'female']\nax2 = fig.add_subplot(2, 1, 2)\nax2 = sns.kdeplot(male_only['TotalPayBenefits'], color ='Blue', label='Male', shade=True)\nax2 = sns.kdeplot(female_only['TotalPayBenefits'], color='Red', label='Female', shade=True)\nplt.title('Part Time Workers')\nplt.ylabel('Density of Employees')\nplt.xlabel('Total Pay & Benefits ($)')\nplt.xlim(0, 350000)\nplt.yticks([])\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"For part time workers, the KDE plot is nearly identical for both males and females.\n\nFor full time workers, we still see a gender gap. We'll need to break down the data some more.",
"_____no_output_____"
],
[
"#### 4.4 - Breaking down the total pay\n\nWe used total pay including benefits for the x-axis for the KDE plot in the previous section. Is this a fair way to analyze the data? What if men work more overtime hours than women? Can we break down the data some more?",
"_____no_output_____"
]
],
[
[
"male_only = data_2014_FT[data_2014_FT['Gender'] == 'male']\nfemale_only = data_2014_FT[data_2014_FT['Gender'] == 'female']\n\nfig = plt.figure(figsize=(10, 15))\nfig.subplots_adjust(hspace=.5) \n\n#Generate the top plot \nax = fig.add_subplot(3, 1, 1)\nax = sns.kdeplot(male_only['OvertimePay'], color ='Blue', label='Male', shade=True)\nax = sns.kdeplot(female_only['OvertimePay'], color='Red', label='Female', shade=True)\nplt.title('Full Time Workers')\nplt.ylabel('Density of Employees')\nplt.xlabel('Overtime Pay ($)')\nplt.xlim(0, 60000)\nplt.yticks([])\n\n#Generate the middle plot\nax2 = fig.add_subplot(3, 1, 2)\nax2 = sns.kdeplot(male_only['Benefits'], color ='Blue', label='Male', shade=True)\nax2 = sns.kdeplot(female_only['Benefits'], color='Red', label='Female', shade=True)\nplt.ylabel('Density of Employees')\nplt.xlabel('Benefits Only ($)')\nplt.xlim(0, 75000)\nplt.yticks([])\n\n#Generate the bottom plot\nax3 = fig.add_subplot(3, 1, 3)\nax3 = sns.kdeplot(male_only['BasePay'], color ='Blue', label='Male', shade=True)\nax3 = sns.kdeplot(female_only['BasePay'], color='Red', label='Female', shade=True)\nplt.ylabel('Density of Employees')\nplt.xlabel('Base Pay Only ($)')\nplt.xlim(0, 300000)\nplt.yticks([])\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"We see a gender gap for all three plots above. Looks like we'll have to dig even deeper and analyze the data by job cateogries.\n\nBut first, let's take a look at the overall correlation for the data set.",
"_____no_output_____"
]
],
[
[
"data_2014_FT.corr()",
"_____no_output_____"
]
],
[
[
"The correlation table above uses Pearson's R to determine the values. The `BasePay` and `Benefits` column are very closely related. We can visualize this relationship using a scatter plot.",
"_____no_output_____"
]
],
[
[
"fig = plt.figure(figsize=(10, 5))\n\nax = plt.scatter(data_2014_FT['BasePay'], data_2014_FT['Benefits'])\n\nplt.ylabel('Benefits ($)')\nplt.xlabel('Base Pay ($)')\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"This makes a lot of sense because an employee's benefits is based on a percentage of their base pay. The San Francisco Human Resources department includes this information on their website [here](http://sfdhr.org/benefits-overview).\n\nAs we move further into our analysis of the data, it makes the most sense to focus on the `BasePay` column. Both `Benefits` and `OvertimePay` are dependent of the `BasePay`. ",
"_____no_output_____"
],
[
"#### 4.5 - Breaking down the base pay by job category",
"_____no_output_____"
],
[
"Next we'll analyze the base pay of full time workers by job category.",
"_____no_output_____"
]
],
[
[
"pal = sns.diverging_palette(0, 255, n=2)\nax = sns.factorplot(x='BasePay', y='Job_Group', hue='Gender', data=data_2014_FT,\n size=10, kind=\"bar\", palette=pal, ci=None)\n\n\nplt.title('Full Time Workers')\nplt.xlabel('Base Pay ($)')\nplt.ylabel('Job Group')\nplt.show()",
"_____no_output_____"
]
],
[
[
"At a glance, we can't really draw any conclusive statements about the gender wage gap. Some job categories favor females, some favor males. It really depends on what job group the employee is actually in. Maybe it makes more sense to calculate the the difference between these two bars.",
"_____no_output_____"
]
],
[
[
"salaries_by_group = pd.pivot_table(data = data_2014_FT, \n values = 'BasePay',\n columns = 'Job_Group', index='Gender',\n aggfunc = np.mean)\n\ncount_by_group = pd.pivot_table(data = data_2014_FT, \n values = 'Id',\n columns = 'Job_Group', index='Gender',\n aggfunc = len)\n\nsalaries_by_group",
"_____no_output_____"
],
[
"fig = plt.figure(figsize=(10, 15))\nsns.set(font_scale=1.5)\n\ndifferences = (salaries_by_group.loc['female'] - salaries_by_group.loc['male'])*100/salaries_by_group.loc['male']\n\nlabels = differences.sort_values().index\n\nx = differences.sort_values()\ny = [i for i in range(len(differences))]\npalette = sns.diverging_palette(240, 10, n=28, center ='dark')\nax = sns.barplot(x, y, orient = 'h', palette = palette)\n\n#Draws the two arrows\nbbox_props = dict(boxstyle=\"rarrow,pad=0.3\", fc=\"white\", ec=\"black\", lw=1)\nt = plt.text(5.5, 12, \"Higher pay for females\", ha=\"center\", va=\"center\", rotation=0,\n size=15,\n bbox=bbox_props)\nbbox_props2 = dict(boxstyle=\"larrow,pad=0.3\", fc=\"white\", ec=\"black\", lw=1)\nt = plt.text(-5.5, 12, \"Higher pay for males\", ha=\"center\", va=\"center\", rotation=0,\n size=15,\n bbox=bbox_props2)\n\n#Labels each bar with the percentage of females\npercent_labels = count_by_group[labels].iloc[0]*100 \\\n /(count_by_group[labels].iloc[0] + count_by_group[labels].iloc[1])\nfor i in range(len(ax.patches)):\n p = ax.patches[i]\n width = p.get_width()*1+1\n ax.text(15,\n p.get_y()+p.get_height()/2+0.3,\n '{:1.0f}'.format(percent_labels[i])+' %',\n ha=\"center\") \n ax.text(15, -1+0.3, 'Female Representation',\n ha=\"center\", fontname='Arial', rotation = 0) \n\n \nplt.yticks(range(len(differences)), labels)\nplt.title('Full Time Workers (Base Pay)')\nplt.xlabel('Mean Percent Difference in Pay (Females - Males)')\nplt.xlim(-11, 11)\nplt.show()",
"_____no_output_____"
]
],
[
[
"I believe this is a better way to represent the gender wage gap. I calculated the mean difference between female and male pay based on job categories. Then I converted the values into a percentage by using this formula:\n\n$$ \\text{Mean Percent Difference} = \\frac{\\text{(Female Mean Pay - Male Mean Pay)*100}} {\\text{Male Mean Pay}} $$\n\nThe theory stating that women makes 78 cents for every dollar men makes implies a 22% pay difference. None of these percentages were more than 10%, and not all of these percentage values showed favoritism towards males. However, we should keep in mind that this data set only applies to San Francisco public jobs. We should also keep in mind that we do not have access to job experience data which would directly correlate with base pay.\n\nIn addition, I included a short table of female representation for each job group on the right side of the graph. We'll dig further into this on the next section.",
"_____no_output_____"
],
[
"#### 4.6 - Gender representation by job category",
"_____no_output_____"
]
],
[
[
"contingency_table = pd.crosstab(\n data_2014_FT['Gender'],\n data_2014_FT['Job_Group'],\n margins = True\n)\ncontingency_table",
"_____no_output_____"
],
[
"#Assigns the frequency values\nfemalecount = contingency_table.iloc[0][0:-1].values\nmalecount = contingency_table.iloc[1][0:-1].values\n\ntotals = contingency_table.iloc[2][0:-1]\nfemalepercentages = femalecount*100/totals\nmalepercentages = malecount*100/totals\n\n\nmalepercentages=malepercentages.sort_values(ascending=True)\nfemalepercentages=femalepercentages.sort_values(ascending=False)\nlength = range(len(femalepercentages))\n\n#Plots the bar chart\nfig = plt.figure(figsize=(10, 12))\nsns.set(font_scale=1.5)\np1 = plt.barh(length, malepercentages.values, 0.55, label='Male', color='#AFAFF5')\np2 = plt.barh(length, femalepercentages, 0.55, left=malepercentages, color='#EFAFB5', label='Female')\n\n\n\nlabels = malepercentages.index\nplt.yticks(range(len(malepercentages)), labels)\nplt.xticks([0, 25, 50, 75, 100], ['0 %', '25 %', '50 %', '75 %', '100 %'])\nplt.xlabel('Percentage of Males')\nplt.title('Gender Representation by Job Group')\nplt.legend(bbox_to_anchor=(0, 1, 1, 0), loc=3,\n ncol=2, mode=\"expand\", borderaxespad=0)\nplt.show()",
"_____no_output_____"
]
],
[
[
"The chart above does not include any information based on pay. I wanted to show an overview of gender representation based on job category. It is safe to say, women don't like working with automotives with <1% female representation. Where as female representation is highest for medical jobs at 73%.",
"_____no_output_____"
],
[
"#### 4.7 - Significance testing by exact job title\n\nSo what if breaking down the wage gap by job category is not good enough? Should we break down the gender gap by exact job title? Afterall, the argument is for equal pay for equal work. We can assume equal work if the job titles are exactly the same.\n\nWe can use hypothesis testing using the Welch's t-test to determine if there is a statistically significant result between male and female wages. The Welch's t-test is very robust as it doesn't assume equal variance and equal sample size. It does however, assume a normal distrbution which is well represented by the KDE plots. I talk about this in detail in my blog post [here](https://codingdisciple.com/hypothesis-testing-welch-python.html).\n\nLet's state our null and alternative hypothesis:\n\n$ H_0 : \\text{There is no statistically significant relationship between gender and pay.} $\n\n$ H_a : \\text{There is a statistically significant relationship between gender and pay.} $\n\nWe are going to use only job titles with more than 100 employees, and job titles with more than 30 females and 30 males for this t-test. Using a for loop, we'll perform the Welch's t-test on every job title tat matches our criteria.",
"_____no_output_____"
]
],
[
[
"from scipy import stats\n\n#Significance testing by job title\njob_titles = data_2014['JobTitle'].value_counts(dropna=True)\njob_titles_over_100 = job_titles[job_titles > 100 ]\n\nt_scores = {}\n\nfor title,count in job_titles_over_100.iteritems():\n male_pay = pd.to_numeric(male_only[male_only['JobTitle'] == title]['BasePay'])\n female_pay = pd.to_numeric(female_only[female_only['JobTitle'] == title]['BasePay'])\n \n if female_pay.shape[0] < 30:\n continue\n if male_pay.shape[0] < 30:\n continue\n\n t_scores[title] = stats.ttest_ind_from_stats( \n mean1=male_pay.mean(), std1=(male_pay.std()), nobs1= male_pay.shape[0], \\\n mean2=female_pay.mean(), std2=(female_pay.std()), nobs2=female_pay.shape[0], \\\n equal_var=False)\n \nfor key, value in t_scores.items():\n if value[1] < 0.05:\n print(len(key)*'-') \n print(key)\n print(len(key)*'-')\n print(t_scores[key])\n print(' ')\n print('Male: {}'.format((male_only[male_only['JobTitle'] == key]['BasePay']).mean()))\n print('sample size: {}'.format(male_only[male_only['JobTitle'] == key].shape[0]))\n print(' ')\n print('Female: {}'.format((female_only[female_only['JobTitle'] == key]['BasePay']).mean()))\n print('sample size: {}'.format(female_only[female_only['JobTitle'] == key].shape[0]))\n",
"------------------------------\nCommunity Police Services Aide\n------------------------------\nTtest_indResult(statistic=2.1634922455155117, pvalue=0.03368488556993355)\n \nMale: 64833.20256410256\nsample size: 78\n \nFemale: 63633.422115384616\nsample size: 52\n----------\nManager II\n----------\nTtest_indResult(statistic=2.4258239418558945, pvalue=0.017341194165396163)\n \nMale: 121976.36624999999\nsample size: 56\n \nFemale: 117550.0123529412\nsample size: 51\n----------------\nTransit Operator\n----------------\nTtest_indResult(statistic=5.20560164116152, pvalue=3.9803552807992223e-07)\n \nMale: 64068.532049433576\nsample size: 971\n \nFemale: 62491.15360215054\nsample size: 186\n------------\nSenior Clerk\n------------\nTtest_indResult(statistic=-2.5391065634795362, pvalue=0.013575306848745547)\n \nMale: 51291.90378378379\nsample size: 37\n \nFemale: 53830.58425\nsample size: 120\n----------------\nRegistered Nurse\n----------------\nTtest_indResult(statistic=-3.7087643141584667, pvalue=0.00033539561477398915)\n \nMale: 128033.80955882354\nsample size: 68\n \nFemale: 132992.0894059406\nsample size: 202\n"
],
[
"len(t_scores)",
"_____no_output_____"
]
],
[
[
"Out of the 25 jobs that were tested using the Welch's t-test, 5 jobs resulted in a p-value of less than 0.05. However, not all jobs showed favoritism towards males. 'Registered Nurse' and 'Senior Clerk' both showed an average pay in favor of females. However, we should take the Welch's t-test results with a grain of salt. We do not have data on the work experience of the employees. Maybe female nurses have more work experience over males. Maybe male transit operators have more work experience over females. We don't actually know. Since `BasePay` is a function of work experience, without this critical piece of information, we can not make any conclusions based on the t-test alone. All we know is that a statistically significant difference exists.",
"_____no_output_____"
],
[
"### 5.0 - San Francisco vs. Newport Beach",
"_____no_output_____"
],
[
"Let's take a look at more a more conservative city such as Newport Beach. This data can be downloaded at Transparent California [here](https://transparentcalifornia.com/salaries/2016/newport-beach/).\n\nWe can process the data similar to the San Francisco data set. The following code performs the following:\n\n+ Read the data using pandas\n+ Create the `Job_Group` column\n+ Create the `Gender` column\n+ Create two new dataframes: one for part time workers and one for full time workers",
"_____no_output_____"
]
],
[
[
"#Reads in the data\nnb_data = pd.read_csv('newport-beach-2016.csv')\n\n#Creates job groups\ndef find_job_title_nb(row):\n titles = collections.OrderedDict([\n ('Police',['police', 'sherif', 'probation', 'sergeant', 'officer', 'lieutenant']),\n ('Fire', ['fire']),\n ('Transit',['mta', 'transit']),\n ('Medical',['anesth', 'medical', 'nurs', 'health', 'physician', 'orthopedic', 'pharm', 'care']),\n ('Architect', ['architect']),\n ('Court',['court', 'legal']),\n ('Mayor Office', ['mayoral']),\n ('Library', ['librar']),\n ('Public Works', ['public']),\n ('Attorney', ['attorney']),\n ('Custodian', ['custodian']),\n ('Gardener', ['garden']),\n ('Recreation Leader', ['recreation']),\n ('Automotive',['automotive', 'mechanic', 'truck']),\n ('Engineer',['engineer', 'engr', 'eng', 'program']),\n ('General Laborer',['general laborer', 'painter', 'inspector', 'carpenter', 'electrician', 'plumber', 'maintenance']),\n ('Food Services', ['food serv']),\n ('Clerk', ['clerk']),\n ('Porter', ['porter']),\n ('Airport Staff', ['airport']),\n ('Social Worker',['worker']), \n ('Guard', ['guard']),\n ('Assistant',['aide', 'assistant', 'secretary', 'attendant']), \n ('Analyst', ['analy']),\n ('Manager', ['manager']) \n ]) \n \n #Loops through the dictionaries\n for group, keywords in titles.items():\n for keyword in keywords:\n if keyword in row.lower():\n return group\n return 'Other'\n\nstart_time = time.time() \nnb_data[\"Job_Group\"]=data[\"JobTitle\"].map(find_job_title_nb)\n\n#Create the 'Gender' column based on employee's first name.\nd = gender.Detector(case_sensitive=False)\nnb_data['FirstName'] = nb_data['Employee Name'].str.split().apply(lambda x: x[0])\nnb_data['Gender'] = nb_data['FirstName'].apply(lambda x: d.get_gender(x))\nnb_data['Gender'].value_counts()\n\n#Retain data with 'male' and 'female' names.\nnb_male_female_only = nb_data[(nb_data['Gender'] == 'male') | (nb_data['Gender'] == 'female')]\nnb_male_female_only['Gender'].value_counts()\n\n#Seperates full time/part time data\nnb_data_FT = nb_male_female_only[nb_male_female_only['Status'] == 'FT']\nnb_data_PT = nb_male_female_only[nb_male_female_only['Status'] == 'PT']\n\nnb_data_FT.head()",
"_____no_output_____"
]
],
[
[
"#### 5.1 - Part time vs. full time workers",
"_____no_output_____"
]
],
[
[
"fig = plt.figure(figsize=(10, 5))\n\nnb_male_only = nb_data_PT[nb_data_PT['Gender'] == 'male']\nnb_female_only = nb_data_PT[nb_data_PT['Gender'] == 'female']\nax = fig.add_subplot(1, 1, 1)\nax = sns.kdeplot(nb_male_only['Total Pay & Benefits'], color ='Blue', label='Male', shade=True)\nax = sns.kdeplot(nb_female_only['Total Pay & Benefits'], color='Red', label='Female', shade=True)\nplt.title('Newport Beach: Part Time Workers')\nplt.ylabel('Density of Employees')\nplt.xlabel('Total Pay + Benefits ($)')\nplt.xlim(0, 400000)\nplt.yticks([])\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"Similar to the KDE plot for San Francisco, the KDE plot is nearly identical for both males and females for part time workers.\n\nLet's take a look at the full time workers.",
"_____no_output_____"
]
],
[
[
"fig = plt.figure(figsize=(10, 10))\nfig.subplots_adjust(hspace=.5) \n\n#Generate the top chart\nnb_male_only = nb_data_FT[nb_data_FT['Gender'] == 'male']\nnb_female_only = nb_data_FT[nb_data_FT['Gender'] == 'female']\nax = fig.add_subplot(2, 1, 1)\nax = sns.kdeplot(nb_male_only['Total Pay & Benefits'], color ='Blue', label='Male', shade=True)\nax = sns.kdeplot(nb_female_only['Total Pay & Benefits'], color='Red', label='Female', shade=True)\nplt.title('Newport Beach: Full Time Workers')\nplt.ylabel('Density of Employees')\nplt.xlabel('Total Pay + Benefits ($)')\nplt.xlim(0, 400000)\nplt.yticks([])\n\n#Generate the bottom chart\nmale_only = data_2014_FT[data_2014_FT['Gender'] == 'male']\nfemale_only = data_2014_FT[data_2014_FT['Gender'] == 'female']\nax2 = fig.add_subplot(2, 1, 2)\nax2 = sns.kdeplot(male_only['TotalPayBenefits'], color ='Blue', label='Male', shade=True)\nax2 = sns.kdeplot(female_only['TotalPayBenefits'], color='Red', label='Female', shade=True)\nplt.title('San Francisco: Full Time Workers')\nplt.ylabel('Density of Employees')\nplt.xlabel('Total Pay + Benefits ($)')\nplt.xlim(0, 400000)\nplt.yticks([])\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"The kurtosis of the KDE plot for Newport Beach full time workers is lower than KDE plot for San Francisco full time workers. We can see a higher gender wage gap for Newport beach workers than San Francisco workers. However, these two plots do not tell us the full story. We need to break down the data by job category.",
"_____no_output_____"
],
[
"#### 5.2 - Comparisons by job cateogry",
"_____no_output_____"
]
],
[
[
"nb_salaries_by_group = pd.pivot_table(data = nb_data_FT, \n values = 'Base Pay',\n columns = 'Job_Group', index='Gender',\n aggfunc = np.mean,)\n\nnb_salaries_by_group",
"_____no_output_____"
],
[
"fig = plt.figure(figsize=(10, 7.5))\nsns.set(font_scale=1.5)\n\ndifferences = (nb_salaries_by_group.loc['female'] - nb_salaries_by_group.loc['male'])*100/nb_salaries_by_group.loc['male']\nnb_labels = differences.sort_values().index\nx = differences.sort_values()\ny = [i for i in range(len(differences))]\nnb_palette = sns.diverging_palette(240, 10, n=9, center ='dark')\nax = sns.barplot(x, y, orient = 'h', palette = nb_palette)\n\n\nplt.yticks(range(len(differences)), nb_labels)\nplt.title('Newport Beach: Full Time Workers (Base Pay)')\nplt.xlabel('Mean Percent Difference in Pay (Females - Males)')\nplt.xlim(-25, 25)\nplt.show()",
"_____no_output_____"
]
],
[
[
"Most of these jobs shows a higher average pay for males. The only job category where females were paid higher on average was 'Manager'. Some of these job categories do not even have a single female within the category, so the difference cannot be calculated. We should create a contingency table to check the sample size of our data.",
"_____no_output_____"
],
[
"#### 5.3 - Gender representation by job category",
"_____no_output_____"
]
],
[
[
"nb_contingency_table = pd.crosstab(\n nb_data_FT['Gender'],\n nb_data_FT['Job_Group'],\n margins = True\n)\nnb_contingency_table",
"_____no_output_____"
]
],
[
[
"The number of public jobs is much lower in Newport Beach compared to San Francisco. With only 3 female managers working full time in Newport Beach, we can't really say female managers make more money on average than male managers.",
"_____no_output_____"
]
],
[
[
"#Assigns the frequency values\nnb_femalecount = nb_contingency_table.iloc[0][0:-1].values\nnb_malecount = nb_contingency_table.iloc[1][0:-1].values\n\nnb_totals = nb_contingency_table.iloc[2][0:-1]\nnb_femalepercentages = nb_femalecount*100/nb_totals\nnb_malepercentages = nb_malecount*100/nb_totals\n\n\nnb_malepercentages=nb_malepercentages.sort_values(ascending=True)\nnb_femalepercentages=nb_femalepercentages.sort_values(ascending=False)\nnb_length = range(len(nb_malepercentages))\n\n#Plots the bar chart\nfig = plt.figure(figsize=(10, 10))\nsns.set(font_scale=1.5)\np1 = plt.barh(nb_length, nb_malepercentages.values, 0.55, label='Male', color='#AFAFF5')\np2 = plt.barh(nb_length, nb_femalepercentages, 0.55, left=nb_malepercentages, color='#EFAFB5', label='Female')\nlabels = nb_malepercentages.index\nplt.yticks(range(len(nb_malepercentages)), labels)\nplt.xticks([0, 25, 50, 75, 100], ['0 %', '25 %', '50 %', '75 %', '100 %'])\nplt.xlabel('Percentage of Males')\nplt.title('Gender Representation by Job Group')\nplt.legend(bbox_to_anchor=(0, 1, 1, 0), loc=3,\n ncol=2, mode=\"expand\", borderaxespad=0)\nplt.show()",
"_____no_output_____"
],
[
"fig = plt.figure(figsize=(10, 5))\n\ncolors = ['#AFAFF5', '#EFAFB5']\nlabels = ['Male', 'Female']\nsizes = [len(nb_male_only), len(nb_female_only)]\nexplode = (0.05, 0)\nsns.set(font_scale=1.5)\nax = fig.add_subplot(1, 2, 1)\nax = plt.pie(sizes, labels=labels, explode=explode, colors=colors, shadow=True, startangle=90, autopct='%1.f%%')\nplt.title('Newport Beach: Full Time')\n\n\nsizes = [len(male_only), len(female_only)]\nexplode = (0.05, 0)\nsns.set(font_scale=1.5)\nax2 = fig.add_subplot(1, 2, 2)\nax2 = plt.pie(sizes, labels=labels, explode=explode, colors=colors, shadow=True, startangle=90, autopct='%1.f%%')\nplt.title('San Francisco: Full Time')\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"Looking at the plots above. There are fewer females working full time public jobs in Newport Beach compared to San Francisco. ",
"_____no_output_____"
],
[
"### 6.0 - Conclusion",
"_____no_output_____"
],
[
"It is very easy for people to say there is a gender wage gap and make general statements about it. But the real concern is whether if there is social injustice and discrimination involved. Yes, there is an overall gender wage gap for both San Francisco and Newport Beach. In both cases, the income distribution for part time employees were nearly identical for both males and females.\n\nFor full time public positions in San Francisco, an overall gender wage gap can be observed. When the full time positions were broken down to job categories, the gender wage gap went both ways. Some jobs favored men, some favored women. For full time public positions in Newport Beach, the majority of the jobs favored men.\n\nHowever, we were missing a critical piece of information in this entire analysis. We don't have any information on the job experience of the employees. Maybe the men just had more job experience in Newport Beach, we don't actually know. For San Francisco, we assumed equal experience by comparing employees with the same exact job titles. Only job titles with a size greater than 100 were chosen. Out of the 25 job titles that were selected, 5 of them showed a statistically significant result with the Welch's t-test. Two of those jobs showed an average base pay in favor of females.\n\nOverall, I do not believe the '78 cents to a dollar' is a fair statement. It generalizes the data and oversimplifies the problem. There are many hidden factors that is not shown by the data. Maybe women are less likely to ask for a promotion. Maybe women perform really well in the medical world. Maybe the men's body is more suitable for the police officer role. Maybe women are more organized than men and make better libarians. The list goes on and on, the point is, we should always be skeptical of what the numbers tell us. The truth is, men and women are different on a fundamental level. Social injustices and gender discrimination should be analyzed on a case by case basis. ",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
cb88434b9461f469c6ffec7e6ffd2491a88dd61d
| 887 |
ipynb
|
Jupyter Notebook
|
Think21_Lab_MLOps.ipynb
|
romeokienzler/covid-trusted-ai-pipeline
|
65e38c5077e9998a30918696e79dfc699ff7eb2c
|
[
"Apache-2.0"
] | null | null | null |
Think21_Lab_MLOps.ipynb
|
romeokienzler/covid-trusted-ai-pipeline
|
65e38c5077e9998a30918696e79dfc699ff7eb2c
|
[
"Apache-2.0"
] | null | null | null |
Think21_Lab_MLOps.ipynb
|
romeokienzler/covid-trusted-ai-pipeline
|
65e38c5077e9998a30918696e79dfc699ff7eb2c
|
[
"Apache-2.0"
] | null | null | null | 19.282609 | 87 | 0.553551 |
[
[
[
"!git clone --recursive https://github.com/romeokienzler/covid-trusted-ai-pipeline",
"_____no_output_____"
],
[
"!cd covid-trusted-ai-pipeline/component-library && git checkout elyra1407",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code"
]
] |
cb8843a4031bcbf3e03558b025741877725246c3
| 252,917 |
ipynb
|
Jupyter Notebook
|
lung-damage/Lung damage - Linear regression.ipynb
|
CesarRamosA/lung-damage-linearreg-MLmodel
|
519e29a8eb59b5847c4de1533a5f2bccde34479c
|
[
"MIT"
] | null | null | null |
lung-damage/Lung damage - Linear regression.ipynb
|
CesarRamosA/lung-damage-linearreg-MLmodel
|
519e29a8eb59b5847c4de1533a5f2bccde34479c
|
[
"MIT"
] | null | null | null |
lung-damage/Lung damage - Linear regression.ipynb
|
CesarRamosA/lung-damage-linearreg-MLmodel
|
519e29a8eb59b5847c4de1533a5f2bccde34479c
|
[
"MIT"
] | null | null | null | 118.351427 | 87,968 | 0.786978 |
[
[
[
"\n# Lung damage - linear regression model\n",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nfrom sklearn.metrics import mean_squared_error\nfrom sklearn import linear_model\nfrom sklearn.model_selection import train_test_split\nimport seaborn as sns\nimport matplotlib.pyplot as plt\nfrom urls import lung_damage_url",
"_____no_output_____"
],
[
"#CSV are read into a dataframe\n#Data is read from AWS\nlung_damage_df = pd.read_csv(lung_damage_url)\nlung_damage_df",
"_____no_output_____"
],
[
"#We drop individual_id as this doesn't provide any relevant information to the model\nlung_damage_df = lung_damage_df.drop(['individual_id'], axis = 'columns')\nlung_damage_df",
"_____no_output_____"
]
],
[
[
"\n### Testing if a data present a normal distribution\n",
"_____no_output_____"
]
],
[
[
"fig = plt.figure(figsize = (15,20))\nax = fig.gca()\nlung_damage_df.hist(ax = ax)\n#lung_damage_df.hist()",
"C:\\Users\\cesar\\AppData\\Local\\Temp/ipykernel_33872/2778530183.py:3: UserWarning: To output multiple subplots, the figure containing the passed axes is being cleared\n lung_damage_df.hist(ax = ax)\n"
]
],
[
[
"\n### Certain values are normally distributed, which suggest linear regression\n",
"_____no_output_____"
],
[
"\n## Correlation analysis\n",
"_____no_output_____"
]
],
[
[
"fig, axes = plt.subplots(ncols=1, nrows=1,figsize=(15,15))\ncorr_matrix = lung_damage_df.select_dtypes(include=['int64', 'float64']).corr(method = 'pearson')\nsns.heatmap(corr_matrix, annot = True)\naxes.set_xticklabels(labels=axes.get_xticklabels(),rotation=45)",
"_____no_output_____"
]
],
[
[
"\n### Lung damage stronly correlates to: 1. Weight, 2. height and 3. cigarettes a week\n",
"_____no_output_____"
],
[
"## Linear regresion model 1 - using all features",
"_____no_output_____"
]
],
[
[
"#One hot encoding by dummy variables\nsex_dummy = pd.get_dummies(lung_damage_df.sex)\ncancer_dummy = pd.get_dummies(lung_damage_df.ancestry_cancer_flag,prefix='cancer') \ndiabetes_dummy = pd.get_dummies(lung_damage_df.ancestry_diabetes_flag,prefix='diabetes')\noverweight_dummy = pd.get_dummies(lung_damage_df.ancestry_overweight_flag,prefix='overweight')",
"_____no_output_____"
],
[
"dummies = pd.concat([sex_dummy, cancer_dummy, diabetes_dummy, overweight_dummy], axis = 'columns')\nmerged_lung_dummies = pd.concat([lung_damage_df, dummies], axis = 'columns')",
"_____no_output_____"
],
[
"X = merged_lung_dummies.drop(['sex', 'ancestry_cancer_flag', 'ancestry_diabetes_flag',\n 'ancestry_overweight_flag', 'F', 'cancer_False', 'diabetes_False',\n 'overweight_False', 'lung_damage'], axis = 'columns') \n\n\ny = lung_damage_df['lung_damage'].to_frame()\n\nprint(\"X\")\nprint(X)\nprint(X.shape)\nprint(\"y\")\nprint(y)\nprint(y.shape)",
"X\n age weight_in_kg height_in_meters cigarettes_a_week \\\n0 42 70.3 1.65 0 \n1 35 60.0 1.66 0 \n2 46 70.6 1.58 0 \n3 45 87.0 1.75 0 \n4 29 68.8 1.66 0 \n... ... ... ... ... \n9995 45 74.3 1.75 0 \n9996 50 61.6 1.69 0 \n9997 38 67.3 1.63 0 \n9998 37 85.5 1.63 0 \n9999 34 82.6 1.72 18 \n\n alcohol_beverages_a_week workout_sessions_a_week oxigen_measure \\\n0 1 6 57.2 \n1 5 2 48.5 \n2 4 0 64.9 \n3 0 5 46.8 \n4 3 0 36.1 \n... ... ... ... \n9995 5 0 57.0 \n9996 4 0 45.4 \n9997 4 0 49.0 \n9998 1 2 69.5 \n9999 3 4 49.4 \n\n co2_measure M cancer_True diabetes_True overweight_True \n0 60.0 1 1 0 0 \n1 71.8 1 0 0 0 \n2 36.3 0 1 0 0 \n3 51.2 1 0 0 0 \n4 68.6 1 0 0 1 \n... ... .. ... ... ... \n9995 72.8 1 0 0 0 \n9996 83.0 1 0 0 0 \n9997 76.1 0 0 0 0 \n9998 65.4 1 1 1 0 \n9999 61.1 0 0 0 1 \n\n[10000 rows x 12 columns]\n(10000, 12)\ny\n lung_damage\n0 0.4157\n1 0.2391\n2 0.5526\n3 0.5636\n4 0.3898\n... ...\n9995 0.3442\n9996 0.2302\n9997 0.4002\n9998 0.8532\n9999 0.7094\n\n[10000 rows x 1 columns]\n(10000, 1)\n"
],
[
"#Algorithm\nl_reg = linear_model.LinearRegression()",
"_____no_output_____"
],
[
"#Looking if relation is appropiate for linear regression\n#We can only make a relationship for one feature at the time\nplt.scatter(X['weight_in_kg'], y) \nplt.show()\n\n#We want to know is if the data is appropiate for linear regresion, which is",
"_____no_output_____"
],
[
"l_reg = linear_model.LinearRegression()\n\nplt.scatter(X['height_in_meters'], y)\nplt.show()",
"_____no_output_____"
]
],
[
[
"\n### This suggest data is linear, so linear regression will be fine\n",
"_____no_output_____"
]
],
[
[
"#separating between the testing and training data\nX_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state = 1234, shuffle = True)",
"_____no_output_____"
],
[
"#train\nmodel = l_reg.fit(X_train, y_train)\n\npredictions = model.predict(X_test)\nmse = mean_squared_error(y_test, predictions)\n\nprint(\"predictions: \", predictions)\nprint(\"R^2: \", l_reg.score(X, y))\nprint(\"mse: \", mse)\nprint(\"coeff: \", l_reg.coef_)\nprint(\"intercept: \", l_reg.intercept_)",
"predictions: [[0.93242556]\n [0.70960976]\n [0.45091427]\n ...\n [0.7983847 ]\n [0.47001117]\n [0.59622372]]\nR^2: 0.9792732521816737\nmse: 0.0009237937195200156\ncoeff: [[ 2.43615191e-05 1.60672827e-02 -1.35944722e+00 8.62040093e-03\n 2.69780971e-05 2.78971986e-05 4.77077269e-05 -3.24909803e-05\n 2.16604935e-04 7.21697769e-04 1.57238029e-01 3.90796809e-04]]\nintercept: [1.54883834]\n"
],
[
"#lung_damage_df[lung_damage_df.lung_damage > 0.8]\n#lung_damage_df[lung_damage_df.lung_damage < 0.3]",
"_____no_output_____"
],
[
"test_de_prueba = [66, 91.9, 1.63, 20, 5, 0, 50, 70, 1, 1, 1, 0]\ntest_de_prueba = [test_de_prueba]\n\ntest_de_prueba = np.array(test_de_prueba).reshape(1,-1)\nprediction = model.predict(test_de_prueba)\nprint(prediction)",
"[[1.14196077]]\n"
]
],
[
[
"\n***Linear regression model predicted a value greater than 1. An arbitrary fix may be suitable***\n",
"_____no_output_____"
]
],
[
[
"test_de_prueba = [35, 70.9, 1.75, 2, 5, 0, 50, 60, 1, 0, 0, 0]\ntest_de_prueba = [test_de_prueba]\n\ntest_de_prueba = np.array(test_de_prueba).reshape(1,-1)\nprediction = model.predict(test_de_prueba)\nprint(prediction)",
"[[0.32785693]]\n"
]
],
[
[
"\n***Last two predictions are reasonable***\n",
"_____no_output_____"
],
[
"\n## Linear regresion model 2\n\n### 3 variables: Weight, height and cigarettes per week\n",
"_____no_output_____"
]
],
[
[
"X= pd.concat([lung_damage_df.weight_in_kg, lung_damage_df.height_in_meters, \n lung_damage_df.cigarettes_a_week], axis = 'columns')\ny = lung_damage_df['lung_damage'].to_frame()\n\nprint(\"X\")\nprint(X)\nprint(X.shape)\nprint(\"y\")\nprint(y)\nprint(y.shape)",
"X\n weight_in_kg height_in_meters cigarettes_a_week\n0 70.3 1.65 0\n1 60.0 1.66 0\n2 70.6 1.58 0\n3 87.0 1.75 0\n4 68.8 1.66 0\n... ... ... ...\n9995 74.3 1.75 0\n9996 61.6 1.69 0\n9997 67.3 1.63 0\n9998 85.5 1.63 0\n9999 82.6 1.72 18\n\n[10000 rows x 3 columns]\n(10000, 3)\ny\n lung_damage\n0 0.4157\n1 0.2391\n2 0.5526\n3 0.5636\n4 0.3898\n... ...\n9995 0.3442\n9996 0.2302\n9997 0.4002\n9998 0.8532\n9999 0.7094\n\n[10000 rows x 1 columns]\n(10000, 1)\n"
],
[
"l_reg = linear_model.LinearRegression()",
"_____no_output_____"
],
[
"#separating between the testing and training data\nX_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state = 1234, shuffle = True)",
"_____no_output_____"
],
[
"#train\nmodel = l_reg.fit(X_train, y_train)\n\npredictions = model.predict(X_test)\nmse = mean_squared_error(y_test, predictions)\n\nprint(\"predictions: \", predictions)\nprint(\"R^2: \", l_reg.score(X, y))\nprint(\"mse: \", mse)\nprint(\"coeff: \", l_reg.coef_)\nprint(\"intercept: \", l_reg.intercept_)",
"predictions: [[0.9642608 ]\n [0.73978894]\n [0.48201241]\n ...\n [0.8293483 ]\n [0.50140466]\n [0.62834863]]\nR^2: 0.8878097514398119\nmse: 0.00487656608813481\ncoeff: [[ 0.0160809 -1.3578138 0.00860991]]\nintercept: [1.5781583]\n"
],
[
"test_de_prueba = [91.9, 1.63, 20]\ntest_de_prueba = [test_de_prueba]\n\ntest_de_prueba = np.array(test_de_prueba).reshape(1,-1)\nprediction = model.predict(test_de_prueba)\nprint(prediction)",
"[[1.01495492]]\n"
],
[
"test_de_prueba = [70.9, 1.75, 2]\ntest_de_prueba = [test_de_prueba]\n\ntest_de_prueba = np.array(test_de_prueba).reshape(1,-1)\nprediction = model.predict(test_de_prueba)\nprint(prediction)",
"[[0.35933999]]\n"
]
],
[
[
"\n### *Linear regresion model 1 gave better results*\n",
"_____no_output_____"
],
[
"\n## Linear regresion model 1* \n\n#### Certain part of the data will not be considered for the split, as it will be used to test the predictions",
"_____no_output_____"
]
],
[
[
"#One hot encoding by dummy variables\nsex_dummy = pd.get_dummies(lung_damage_df.sex)\ncancer_dummy = pd.get_dummies(lung_damage_df.ancestry_cancer_flag,prefix='cancer') \ndiabetes_dummy = pd.get_dummies(lung_damage_df.ancestry_diabetes_flag,prefix='diabetes')\noverweight_dummy = pd.get_dummies(lung_damage_df.ancestry_overweight_flag,prefix='overweight')",
"_____no_output_____"
],
[
"dummies = pd.concat([sex_dummy, cancer_dummy, diabetes_dummy, overweight_dummy], axis = 'columns')\nmerged_lung_dummies = pd.concat([lung_damage_df, dummies], axis = 'columns')",
"_____no_output_____"
],
[
"X = merged_lung_dummies.drop(['sex', 'ancestry_cancer_flag', 'ancestry_diabetes_flag',\n 'ancestry_overweight_flag', 'F', 'cancer_False', 'diabetes_False',\n 'overweight_False', 'lung_damage'], axis = 'columns') \n\nX = X.iloc[:9000] #se dejan fuera los ultimos 1000 datos\nX_left = lung_damage_df.iloc[9000:10000] #los datos que quedan se guardan aqui\ny = lung_damage_df['lung_damage'].to_frame()\ny = y.iloc[:9000]\nprint(\"X\")\nprint(X)\nprint(X.shape)\nprint(\"y\")\nprint(y)\nprint(y.shape)",
"X\n age weight_in_kg height_in_meters cigarettes_a_week \\\n0 42 70.3 1.65 0 \n1 35 60.0 1.66 0 \n2 46 70.6 1.58 0 \n3 45 87.0 1.75 0 \n4 29 68.8 1.66 0 \n... ... ... ... ... \n8995 33 62.3 1.70 4 \n8996 37 70.4 1.60 0 \n8997 42 69.2 1.79 0 \n8998 50 51.3 1.65 0 \n8999 26 73.3 1.60 0 \n\n alcohol_beverages_a_week workout_sessions_a_week oxigen_measure \\\n0 1 6 57.2 \n1 5 2 48.5 \n2 4 0 64.9 \n3 0 5 46.8 \n4 3 0 36.1 \n... ... ... ... \n8995 4 0 42.3 \n8996 3 6 53.6 \n8997 5 7 68.0 \n8998 1 7 64.1 \n8999 3 0 41.2 \n\n co2_measure M cancer_True diabetes_True overweight_True \n0 60.0 1 1 0 0 \n1 71.8 1 0 0 0 \n2 36.3 0 1 0 0 \n3 51.2 1 0 0 0 \n4 68.6 1 0 0 1 \n... ... .. ... ... ... \n8995 54.9 1 0 1 0 \n8996 73.6 0 1 1 0 \n8997 105.3 0 0 1 0 \n8998 72.6 0 0 0 0 \n8999 88.5 1 0 0 0 \n\n[9000 rows x 12 columns]\n(9000, 12)\ny\n lung_damage\n0 0.4157\n1 0.2391\n2 0.5526\n3 0.5636\n4 0.3898\n... ...\n8995 0.4262\n8996 0.6907\n8997 0.3864\n8998 0.1455\n8999 0.5669\n\n[9000 rows x 1 columns]\n(9000, 1)\n"
],
[
"#Algorithm\nl_reg = linear_model.LinearRegression()",
"_____no_output_____"
],
[
"#separating between the testing and training data\nX_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state = 1785, shuffle = True)",
"_____no_output_____"
],
[
"#train\nmodel = l_reg.fit(X_train, y_train)\n\npredictions = model.predict(X_test)\nmse = mean_squared_error(y_test, predictions)\n\nprint(\"predictions: \", predictions)\nprint(\"R^2: \", l_reg.score(X, y))\nprint(\"mse: \", mse)\nprint(\"coeff: \", l_reg.coef_)\nprint(\"intercept: \", l_reg.intercept_)",
"predictions: [[0.46646301]\n [0.24434368]\n [0.71188913]\n ...\n [0.46155287]\n [0.25404845]\n [0.58611086]]\nR^2: 0.9792889268569628\nmse: 0.0009363014934790182\ncoeff: [[ 4.41936249e-05 1.60546795e-02 -1.36475466e+00 8.58038632e-03\n 3.25177643e-04 7.08036905e-06 8.34519765e-05 -2.52856766e-05\n 3.56334141e-04 -8.21012938e-05 1.57126546e-01 1.00722448e-03]]\nintercept: [1.55504396]\n"
]
],
[
[
"\n### R-squared is 97%, which is a great value. The data is ideal in this model.\n",
"_____no_output_____"
],
[
"\n## Testing predictions vs real\n",
"_____no_output_____"
]
],
[
[
"X",
"_____no_output_____"
],
[
"#lung_damage_df[lung_damage_df.lung_damage > 0.8]\n#lung_damage_df[lung_damage_df.lung_damage < 0.3]\nX_left",
"_____no_output_____"
],
[
"test_de_prueba = [46, 81.5, 1.64, 0, 2, 0, 53.5, 63.9, 1, 0, 0, 0]\ntest_de_prueba = [test_de_prueba]\ntest_de_prueba = np.array(test_de_prueba).reshape(1,-1)\n\nprediction = model.predict(test_de_prueba)\nprint(prediction)",
"[[0.63119121]]\n"
]
],
[
[
"\n***The prediction says 63% of lung damage, in comparison with 65% real data***\n",
"_____no_output_____"
]
],
[
[
"test_de_prueba = [32, 53.6, 1.60, 12, 5, 4, 28.1, 41.7, 0, 0, 1, 0]\ntest_de_prueba = [test_de_prueba]\n\ntest_de_prueba = np.array(test_de_prueba).reshape(1,-1)\nprediction = model.predict(test_de_prueba)\nprint(prediction)",
"[[0.49641749]]\n"
]
],
[
[
"\n***49% predicted vs 47% real***\n",
"_____no_output_____"
],
[
"## Conclusions:\n\n- Model seems great using all features\n\n- An arbitrary fix that maps all variables >1 to 1 will be suitable \n\n- This data is surely generated by a program and it's not real data, which could explain the accuracy of the model\n\n- Even though, this model is good which can be optimized using a correction to the linear regression model",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
cb88474e95f88ff55df01a943437f3bebaba9f08
| 96,112 |
ipynb
|
Jupyter Notebook
|
msda/msda_res_fig_12_id.ipynb
|
TuffDev/motion-sense
|
5429f8c7db5d4097494f823257f5dcecd3bd52da
|
[
"MIT"
] | 226 |
2018-03-01T17:24:36.000Z
|
2022-03-21T13:45:47.000Z
|
msda/msda_res_fig_12_id.ipynb
|
TuffDev/motion-sense
|
5429f8c7db5d4097494f823257f5dcecd3bd52da
|
[
"MIT"
] | 7 |
2018-05-20T12:33:09.000Z
|
2021-03-22T18:17:01.000Z
|
msda/msda_res_fig_12_id.ipynb
|
TuffDev/motion-sense
|
5429f8c7db5d4097494f823257f5dcecd3bd52da
|
[
"MIT"
] | 89 |
2018-04-16T17:25:27.000Z
|
2022-03-22T11:14:04.000Z
| 51.673118 | 258 | 0.548901 |
[
[
[
"import tensorflow as tf \nimport keras \nimport keras.backend as K\n\nfrom sklearn.utils import shuffle\nfrom sklearn.metrics import classification_report, confusion_matrix, accuracy_score, f1_score\nfrom collections import Counter\n\nfrom keras import regularizers\nfrom keras.models import Sequential, Model, load_model, model_from_json \nfrom keras.utils import to_categorical\nfrom keras.layers import Input, Dense, Flatten, Reshape, Concatenate, Dropout \nfrom keras.layers import Conv2D, MaxPooling2D, UpSampling2D, Conv2DTranspose\nfrom keras.layers.normalization import BatchNormalization\nfrom keras.callbacks import ModelCheckpoint\nfrom keras.utils import np_utils\nfrom keras.layers.advanced_activations import LeakyReLU\n\ndef get_class_weights(y):\n counter = Counter(y)\n majority = max(counter.values())\n return {cls: float(majority/count) for cls, count in counter.items()}\n\n\n\nclass Estimator:\n l2p = 0.001\n @staticmethod\n def early_layers(inp, fm = (1,3), hid_act_func=\"relu\"):\n # Start\n x = Conv2D(64, fm, padding=\"same\", kernel_regularizer=regularizers.l2(Estimator.l2p), activation=hid_act_func)(inp)\n x = BatchNormalization()(x)\n x = MaxPooling2D(pool_size=(1, 2))(x)\n x = Dropout(0.25)(x)\n \n # 1\n x = Conv2D(64, fm, padding=\"same\", kernel_regularizer=regularizers.l2(Estimator.l2p), activation=hid_act_func)(x)\n x = BatchNormalization()(x)\n x = MaxPooling2D(pool_size=(1, 2))(x)\n x = Dropout(0.25)(x)\n\n return x\n \n @staticmethod\n def late_layers(inp, num_classes, fm = (1,3), act_func=\"softmax\", hid_act_func=\"relu\", b_name=\"Identifier\"):\n # 2\n x = Conv2D(32, fm, padding=\"same\", kernel_regularizer=regularizers.l2(Estimator.l2p), activation=hid_act_func)(inp)\n x = BatchNormalization()(x)\n x = MaxPooling2D(pool_size=(1, 2))(x)\n x = Dropout(0.25)(x)\n \n # 3\n x = Conv2D(32, fm, padding=\"same\", kernel_regularizer=regularizers.l2(Estimator.l2p), activation=hid_act_func)(x)\n x = BatchNormalization()(x)\n x = MaxPooling2D(pool_size=(1, 2))(x)\n x = Dropout(0.25)(x)\n \n # End\n x = Flatten()(x)\n x = Dense(256, kernel_regularizer=regularizers.l2(Estimator.l2p), activation=hid_act_func)(x)\n x = BatchNormalization()(x)\n x = Dropout(0.5)(x)\n x = Dense(64, kernel_regularizer=regularizers.l2(Estimator.l2p), activation=hid_act_func)(x)\n x = BatchNormalization()(x)\n x = Dropout(0.5)(x)\n x = Dense(num_classes, activation=act_func, name = b_name)(x)\n\n return x\n \n @staticmethod\n def build(height, width, num_classes, name, fm = (1,3), act_func=\"softmax\",hid_act_func=\"relu\"):\n inp = Input(shape=(height, width, 1))\n early = Estimator.early_layers(inp, fm, hid_act_func=hid_act_func)\n late = Estimator.late_layers(early, num_classes, fm, act_func=act_func, hid_act_func=hid_act_func)\n model = Model(inputs=inp, outputs=late ,name=name)\n return model",
"/anaconda/envs/py35/lib/python3.5/site-packages/h5py/__init__.py:36: FutureWarning: Conversion of the second argument of issubdtype from `float` to `np.floating` is deprecated. In future, it will be treated as `np.float64 == np.dtype(float).type`.\n from ._conv import register_converters as _register_converters\nUsing TensorFlow backend.\n"
],
[
"import numpy as np\nimport pandas as pd\nfrom pandas.plotting import autocorrelation_plot\nimport matplotlib.pyplot as plt\n\n\ndef get_ds_infos():\n \"\"\"\n Read the file includes data subject information.\n \n Data Columns:\n 0: code [1-24]\n 1: weight [kg]\n 2: height [cm]\n 3: age [years]\n 4: gender [0:Female, 1:Male]\n \n Returns:\n A pandas DataFrame that contains inforamtion about data subjects' attributes \n \"\"\" \n\n dss = pd.read_csv(\"data_subjects_info.csv\")\n print(\"[INFO] -- Data subjects' information is imported.\")\n \n return dss\n\ndef set_data_types(data_types=[\"userAcceleration\"]):\n \"\"\"\n Select the sensors and the mode to shape the final dataset.\n \n Args:\n data_types: A list of sensor data type from this list: [attitude, gravity, rotationRate, userAcceleration] \n\n Returns:\n It returns a list of columns to use for creating time-series from files.\n \"\"\"\n dt_list = []\n for t in data_types:\n if t != \"attitude\":\n dt_list.append([t+\".x\",t+\".y\",t+\".z\"])\n else:\n dt_list.append([t+\".roll\", t+\".pitch\", t+\".yaw\"])\n\n return dt_list\n\n\ndef creat_time_series(dt_list, act_labels, trial_codes, mode=\"mag\", labeled=True, combine_grav_acc=False):\n \"\"\"\n Args:\n dt_list: A list of columns that shows the type of data we want.\n act_labels: list of activites\n trial_codes: list of trials\n mode: It can be \"raw\" which means you want raw data\n for every dimention of each data type,\n [attitude(roll, pitch, yaw); gravity(x, y, z); rotationRate(x, y, z); userAcceleration(x,y,z)].\n or it can be \"mag\" which means you only want the magnitude for each data type: (x^2+y^2+z^2)^(1/2)\n labeled: True, if we want a labeld dataset. False, if we only want sensor values.\n combine_grav_acc: True, means adding each axis of gravity to corresponding axis of userAcceleration.\n Returns: \n It returns a time-series of sensor data.\n \n \"\"\"\n num_data_cols = len(dt_list) if mode == \"mag\" else len(dt_list*3)\n\n if labeled:\n dataset = np.zeros((0,num_data_cols+7)) # \"7\" --> [act, code, weight, height, age, gender, trial] \n else:\n dataset = np.zeros((0,num_data_cols))\n \n ds_list = get_ds_infos()\n \n print(\"[INFO] -- Creating Time-Series\")\n for sub_id in ds_list[\"code\"]:\n for act_id, act in enumerate(act_labels):\n for trial in trial_codes[act_id]:\n fname = 'A_DeviceMotion_data/'+act+'_'+str(trial)+'/sub_'+str(int(sub_id))+'.csv'\n raw_data = pd.read_csv(fname)\n raw_data = raw_data.drop(['Unnamed: 0'], axis=1)\n vals = np.zeros((len(raw_data), num_data_cols))\n \n if combine_grav_acc:\n raw_data[\"userAcceleration.x\"] = raw_data[\"userAcceleration.x\"].add(raw_data[\"gravity.x\"])\n raw_data[\"userAcceleration.y\"] = raw_data[\"userAcceleration.y\"].add(raw_data[\"gravity.y\"])\n raw_data[\"userAcceleration.z\"] = raw_data[\"userAcceleration.z\"].add(raw_data[\"gravity.z\"])\n \n for x_id, axes in enumerate(dt_list):\n if mode == \"mag\":\n vals[:,x_id] = (raw_data[axes]**2).sum(axis=1)**0.5 \n else:\n vals[:,x_id*3:(x_id+1)*3] = raw_data[axes].values\n vals = vals[:,:num_data_cols]\n if labeled:\n lbls = np.array([[act_id,\n sub_id-1,\n ds_list[\"weight\"][sub_id-1],\n ds_list[\"height\"][sub_id-1],\n ds_list[\"age\"][sub_id-1],\n ds_list[\"gender\"][sub_id-1],\n trial \n ]]*len(raw_data))\n vals = np.concatenate((vals, lbls), axis=1)\n dataset = np.append(dataset,vals, axis=0)\n cols = []\n for axes in dt_list:\n if mode == \"raw\":\n cols += axes\n else:\n cols += [str(axes[0][:-2])]\n \n if labeled:\n cols += [\"act\", \"id\", \"weight\", \"height\", \"age\", \"gender\", \"trial\"]\n \n dataset = pd.DataFrame(data=dataset, columns=cols)\n return dataset\n#________________________________\n#________________________________\n\ndef ts_to_secs(dataset, w, s, standardize = False, **options):\n \n data = dataset[dataset.columns[:-7]].values \n act_labels = dataset[\"act\"].values\n id_labels = dataset[\"id\"].values\n trial_labels = dataset[\"trial\"].values\n\n mean = 0\n std = 1\n if standardize:\n ## Standardize each sensor’s data to have a zero mean and unity standard deviation.\n ## As usual, we normalize test dataset by training dataset's parameters \n if options:\n mean = options.get(\"mean\")\n std = options.get(\"std\")\n print(\"[INFO] -- Test/Val Data has been standardized\")\n else:\n mean = data.mean(axis=0)\n std = data.std(axis=0)\n print(\"[INFO] -- Training Data has been standardized: the mean is = \"+str(mean)+\" ; and the std is = \"+str(std)) \n\n data -= mean\n data /= std\n else:\n print(\"[INFO] -- Without Standardization.....\")\n\n ## We want the Rows of matrices show each Feature and the Columns show time points.\n data = data.T\n\n m = data.shape[0] # Data Dimension \n ttp = data.shape[1] # Total Time Points\n number_of_secs = int(round(((ttp - w)/s)))\n\n ## Create a 3D matrix for Storing Sections \n secs_data = np.zeros((number_of_secs , m , w ))\n act_secs_labels = np.zeros(number_of_secs)\n id_secs_labels = np.zeros(number_of_secs)\n\n k=0\n for i in range(0 , ttp-w, s):\n j = i // s\n if j >= number_of_secs:\n break\n if id_labels[i] != id_labels[i+w-1]: \n continue\n if act_labels[i] != act_labels[i+w-1]: \n continue\n if trial_labels[i] != trial_labels[i+w-1]:\n continue\n \n secs_data[k] = data[:, i:i+w]\n act_secs_labels[k] = act_labels[i].astype(int)\n id_secs_labels[k] = id_labels[i].astype(int)\n k = k+1\n \n secs_data = secs_data[0:k]\n act_secs_labels = act_secs_labels[0:k]\n id_secs_labels = id_secs_labels[0:k]\n return secs_data, act_secs_labels, id_secs_labels, mean, std\n##________________________________________________________________\n\n\nACT_LABELS = [\"dws\",\"ups\", \"wlk\", \"jog\", \"std\", \"sit\"]\nTRIAL_CODES = {\n ACT_LABELS[0]:[1,2,11],\n ACT_LABELS[1]:[3,4,12],\n ACT_LABELS[2]:[7,8,15],\n ACT_LABELS[3]:[9,16],\n ACT_LABELS[4]:[6,14],\n ACT_LABELS[5]:[5,13],\n}",
"_____no_output_____"
],
[
"#https://stackoverflow.com/a/45305384/5210098\ndef f1_metric(y_true, y_pred):\n def recall(y_true, y_pred):\n \"\"\"Recall metric.\n\n Only computes a batch-wise average of recall.\n\n Computes the recall, a metric for multi-label classification of\n how many relevant items are selected.\n \"\"\"\n true_positives = K.sum(K.round(K.clip(y_true * y_pred, 0, 1)))\n possible_positives = K.sum(K.round(K.clip(y_true, 0, 1)))\n recall = true_positives / (possible_positives + K.epsilon())\n return recall\n\n def precision(y_true, y_pred):\n \"\"\"Precision metric.\n\n Only computes a batch-wise average of precision.\n\n Computes the precision, a metric for multi-label classification of\n how many selected items are relevant.\n \"\"\"\n true_positives = K.sum(K.round(K.clip(y_true * y_pred, 0, 1)))\n predicted_positives = K.sum(K.round(K.clip(y_pred, 0, 1)))\n precision = true_positives / (predicted_positives + K.epsilon())\n return precision\n precision = precision(y_true, y_pred)\n recall = recall(y_true, y_pred)\n return 2*((precision*recall)/(precision+recall+K.epsilon()))\n\n\ndef eval_id(sdt, mode, ep, cga):\n\n print(\"[INFO] -- Selected sensor data types: \"+str(sdt)+\" -- Mode: \"+str(mode)+\" -- Grav+Acc: \"+str(cga)) \n act_labels = ACT_LABELS [0:4]\n\n print(\"[INFO] -- Selected activites: \"+str(act_labels)) \n trial_codes = [TRIAL_CODES[act] for act in act_labels]\n dt_list = set_data_types(sdt)\n dataset = creat_time_series(dt_list, act_labels, trial_codes, mode=mode, labeled=True, combine_grav_acc = cga)\n print(\"[INFO] -- Shape of time-Series dataset:\"+str(dataset.shape)) \n\n\n #*****************\n TRAIN_TEST_TYPE = \"trial\" # \"subject\" or \"trial\"\n #*****************\n\n if TRAIN_TEST_TYPE == \"subject\":\n test_ids = [4,9,11,21]\n print(\"[INFO] -- Test IDs: \"+str(test_ids))\n test_ts = dataset.loc[(dataset['id'].isin(test_ids))]\n train_ts = dataset.loc[~(dataset['id'].isin(test_ids))]\n else:\n test_trail = [11,12,13,14,15,16] \n print(\"[INFO] -- Test Trials: \"+str(test_trail))\n test_ts = dataset.loc[(dataset['trial'].isin(test_trail))]\n train_ts = dataset.loc[~(dataset['trial'].isin(test_trail))]\n\n print(\"[INFO] -- Shape of Train Time-Series :\"+str(train_ts.shape))\n print(\"[INFO] -- Shape of Test Time-Series :\"+str(test_ts.shape))\n \n print(\"___________________________________________________\")\n\n ## This Variable Defines the Size of Sliding Window\n ## ( e.g. 100 means in each snapshot we just consider 100 consecutive observations of each sensor) \n w = 128 # 50 Equals to 1 second for MotionSense Dataset (it is on 50Hz samplig rate)\n ## Here We Choose Step Size for Building Diffrent Snapshots from Time-Series Data\n ## ( smaller step size will increase the amount of the instances and higher computational cost may be incurred )\n s = 10\n train_data, act_train, id_train, train_mean, train_std = ts_to_secs(train_ts.copy(),\n w,\n s,\n standardize = True)\n \n \n s = 10\n test_data, act_test, id_test, test_mean, test_std = ts_to_secs(test_ts.copy(),\n w,\n s,\n standardize = True,\n mean = train_mean, \n std = train_std)\n \n print(\"[INFO] -- Training Sections: \"+str(train_data.shape))\n print(\"[INFO] -- Test Sections: \"+str(test_data.shape))\n\n\n id_train_labels = to_categorical(id_train)\n id_test_labels = to_categorical(id_test)\n \n act_train_labels = to_categorical(act_train)\n act_test_labels = to_categorical(act_test)\n \n ## Here we add an extra dimension to the datasets just to be ready for using with Convolution2D\n train_data = np.expand_dims(train_data,axis=3)\n print(\"[INFO] -- Training Sections:\"+str(train_data.shape))\n\n test_data = np.expand_dims(test_data,axis=3)\n print(\"[INFO] -- Test Sections:\"+str(test_data.shape))\n\n height = train_data.shape[1]\n width = train_data.shape[2]\n\n id_class_numbers = 24\n act_class_numbers = 4\n fm = (1,5)\n\n print(\"___________________________________________________\")\n ## Callbacks\n #eval_metric= \"val_acc\"\n eval_metric= \"val_f1_metric\" \n early_stop = keras.callbacks.EarlyStopping(monitor=eval_metric, mode='max', patience = 7)\n filepath=\"MID.best.hdf5\"\n checkpoint = ModelCheckpoint(filepath, monitor=eval_metric, verbose=0, save_best_only=True, mode='max')\n callbacks_list = [early_stop,\n checkpoint\n ]\n ## Callbacks\n eval_id = Estimator.build(height, width, id_class_numbers, name =\"EVAL_ID\", fm=fm, act_func=\"softmax\",hid_act_func=\"relu\")\n eval_id.compile( loss=\"categorical_crossentropy\", optimizer='adam', metrics=['acc', f1_metric])\n print(\"Model Size = \"+str(eval_id.count_params()))\n\n eval_id.fit(train_data, id_train_labels,\n validation_data = (test_data, id_test_labels),\n epochs = ep,\n batch_size = 128,\n verbose = 0,\n class_weight = get_class_weights(np.argmax(id_train_labels,axis=1)),\n callbacks = callbacks_list\n )\n\n eval_id.load_weights(\"MID.best.hdf5\")\n eval_id.compile( loss=\"categorical_crossentropy\", optimizer='adam', metrics=['acc',f1_metric])\n\n result1 = eval_id.evaluate(test_data, id_test_labels, verbose = 2)\n id_acc = result1[1]\n print(\"***[RESULT]*** ID Accuracy: \"+str(id_acc))\n rf1 = result1[2].round(4)*100\n print(\"***[RESULT]*** ID F1: \"+str(rf1))\n \n preds = eval_id.predict(test_data)\n preds = np.argmax(preds, axis=1)\n conf_mat = confusion_matrix(np.argmax(id_test_labels, axis=1), preds)\n conf_mat = conf_mat.astype('float') / conf_mat.sum(axis=1)[:, np.newaxis]\n print(\"***[RESULT]*** ID Confusion Matrix\")\n print((np.array(conf_mat).diagonal()).round(3)*100) \n \n d_test_ids = [4,9,11,21]\n to_avg = 0\n for i in range(len(d_test_ids)):\n true_positive = conf_mat[d_test_ids[i],d_test_ids[i]]\n print(\"True Positive Rate for \"+str(d_test_ids[i])+\" : \"+str(true_positive*100))\n to_avg+=true_positive\n atp = to_avg/len(d_test_ids) \n print(\"Average TP:\"+str(atp*100)) \n \n f1id = f1_score(np.argmax(id_test_labels, axis=1), preds, average=None).mean()\n print(\"***[RESULT]*** ID Averaged F-1 Score : \"+str(f1id))\n \n return [round(id_acc,4), round(f1id,4), round(atp,4)]",
"_____no_output_____"
],
[
"results ={}",
"_____no_output_____"
],
[
"## Here we set parameter to build labeld time-series from dataset of \"(A)DeviceMotion_data\"\n## attitude(roll, pitch, yaw); gravity(x, y, z); rotationRate(x, y, z); userAcceleration(x,y,z)\nsdt = [\"rotationRate\"]\nmode = \"mag\"\nep = 40\ncga = False # Add gravity to acceleration or not\nfor i in range(5):\n results[str(sdt)+\"--\"+str(mode)+\"--\"+str(cga)+\"--\"+str(i)] = eval_id(sdt, mode, ep, cga)",
"[INFO] -- Selected sensor data types: ['rotationRate'] -- Mode: mag -- Grav+Acc: False\n[INFO] -- Selected activites: ['dws', 'ups', 'wlk', 'jog']\n[INFO] -- Data subjects' information is imported.\n[INFO] -- Creating Time-Series\n[INFO] -- Shape of time-Series dataset:(767660, 8)\n[INFO] -- Test Trials: [11, 12, 13, 14, 15, 16]\n[INFO] -- Shape of Train Time-Series :(621973, 8)\n[INFO] -- Shape of Test Time-Series :(145687, 8)\n___________________________________________________\n[INFO] -- Training Data has been standardized: the mean is = [2.20896278] ; and the std is = [1.42146386]\n[INFO] -- Test/Val Data has been standardized\n[INFO] -- Training Sections: (60059, 1, 128)\n[INFO] -- Test Sections: (13344, 1, 128)\n[INFO] -- Training Sections:(60059, 1, 128, 1)\n[INFO] -- Test Sections:(13344, 1, 128, 1)\n___________________________________________________\nModel Size = 122200\n***[RESULT]*** ID Accuracy: 0.8687799760191847\n***[RESULT]*** ID F1: 86.37\n***[RESULT]*** ID Confusion Matrix\n[71.3 99.4 91.7 95. 88.1 85.1 87.8 76.5 77.8 88.1 81. 86.5 95. 75.5\n 81.6 95.9 88.3 66.2 81.5 84.1 93.1 96. 94.3 88.9]\nTrue Positive Rate for 4 : 88.13953488372093\nTrue Positive Rate for 9 : 88.09946714031972\nTrue Positive Rate for 11 : 86.45276292335116\nTrue Positive Rate for 21 : 96.03174603174604\nAverage TP:89.68087774478445\n***[RESULT]*** ID Averaged F-1 Score : 0.8661190556894892\n[INFO] -- Selected sensor data types: ['rotationRate'] -- Mode: mag -- Grav+Acc: False\n[INFO] -- Selected activites: ['dws', 'ups', 'wlk', 'jog']\n[INFO] -- Data subjects' information is imported.\n[INFO] -- Creating Time-Series\n[INFO] -- Shape of time-Series dataset:(767660, 8)\n[INFO] -- Test Trials: [11, 12, 13, 14, 15, 16]\n[INFO] -- Shape of Train Time-Series :(621973, 8)\n[INFO] -- Shape of Test Time-Series :(145687, 8)\n___________________________________________________\n[INFO] -- Training Data has been standardized: the mean is = [2.20896278] ; and the std is = [1.42146386]\n[INFO] -- Test/Val Data has been standardized\n[INFO] -- Training Sections: (60059, 1, 128)\n[INFO] -- Test Sections: (13344, 1, 128)\n[INFO] -- Training Sections:(60059, 1, 128, 1)\n[INFO] -- Test Sections:(13344, 1, 128, 1)\n___________________________________________________\nModel Size = 122200\n***[RESULT]*** ID Accuracy: 0.8428507194244604\n***[RESULT]*** ID F1: 83.57\n***[RESULT]*** ID Confusion Matrix\n[78.4 98.4 96.4 91.8 86.7 83.5 80. 63.3 71. 91.1 75.6 86.3 97.2 61.\n 78. 96.2 75. 68.7 85.6 83. 93.2 95. 83.2 87.7]\nTrue Positive Rate for 4 : 86.74418604651163\nTrue Positive Rate for 9 : 91.1190053285968\nTrue Positive Rate for 11 : 86.27450980392157\nTrue Positive Rate for 21 : 95.03968253968253\nAverage TP:89.79434592967814\n***[RESULT]*** ID Averaged F-1 Score : 0.8376911152398089\n[INFO] -- Selected sensor data types: ['rotationRate'] -- Mode: mag -- Grav+Acc: False\n[INFO] -- Selected activites: ['dws', 'ups', 'wlk', 'jog']\n[INFO] -- Data subjects' information is imported.\n[INFO] -- Creating Time-Series\n[INFO] -- Shape of time-Series dataset:(767660, 8)\n[INFO] -- Test Trials: [11, 12, 13, 14, 15, 16]\n[INFO] -- Shape of Train Time-Series :(621973, 8)\n[INFO] -- Shape of Test Time-Series :(145687, 8)\n___________________________________________________\n[INFO] -- Training Data has been standardized: the mean is = [2.20896278] ; and the std is = [1.42146386]\n[INFO] -- Test/Val Data has been standardized\n[INFO] -- Training Sections: (60059, 1, 128)\n[INFO] -- Test Sections: (13344, 1, 128)\n[INFO] -- Training Sections:(60059, 1, 128, 1)\n[INFO] -- Test Sections:(13344, 1, 128, 1)\n___________________________________________________\nModel Size = 122200\n***[RESULT]*** ID Accuracy: 0.856339928057554\n***[RESULT]*** ID F1: 85.21\n***[RESULT]*** ID Confusion Matrix\n[70.6 97.3 83.3 86.1 83. 79.9 93.1 92.1 80.8 85.6 86.5 80.6 94.4 73.8\n 84.1 92.5 87.9 71.8 67.4 86.8 94.2 94.4 90.3 83.2]\nTrue Positive Rate for 4 : 83.02325581395348\nTrue Positive Rate for 9 : 85.61278863232683\nTrue Positive Rate for 11 : 80.57040998217468\nTrue Positive Rate for 21 : 94.44444444444444\nAverage TP:85.91272471822487\n***[RESULT]*** ID Averaged F-1 Score : 0.8520730708032556\n[INFO] -- Selected sensor data types: ['rotationRate'] -- Mode: mag -- Grav+Acc: False\n[INFO] -- Selected activites: ['dws', 'ups', 'wlk', 'jog']\n[INFO] -- Data subjects' information is imported.\n[INFO] -- Creating Time-Series\n[INFO] -- Shape of time-Series dataset:(767660, 8)\n[INFO] -- Test Trials: [11, 12, 13, 14, 15, 16]\n[INFO] -- Shape of Train Time-Series :(621973, 8)\n[INFO] -- Shape of Test Time-Series :(145687, 8)\n___________________________________________________\n[INFO] -- Training Data has been standardized: the mean is = [2.20896278] ; and the std is = [1.42146386]\n[INFO] -- Test/Val Data has been standardized\n[INFO] -- Training Sections: (60059, 1, 128)\n[INFO] -- Test Sections: (13344, 1, 128)\n[INFO] -- Training Sections:(60059, 1, 128, 1)\n[INFO] -- Test Sections:(13344, 1, 128, 1)\n___________________________________________________\nModel Size = 122200\n***[RESULT]*** ID Accuracy: 0.8696043165467626\n***[RESULT]*** ID F1: 86.21\n***[RESULT]*** ID Confusion Matrix\n[80.2 98.9 93.7 87.7 90.5 89.2 87.7 82.1 83.6 88.1 85.5 91.1 93.5 51.2\n 83.5 97.4 81.7 67.8 81.5 88.9 92.4 98. 88.5 87.7]\nTrue Positive Rate for 4 : 90.46511627906978\nTrue Positive Rate for 9 : 88.09946714031972\nTrue Positive Rate for 11 : 91.0873440285205\nTrue Positive Rate for 21 : 98.01587301587301\nAverage TP:91.91695011594577\n***[RESULT]*** ID Averaged F-1 Score : 0.8617920403510498\n[INFO] -- Selected sensor data types: ['rotationRate'] -- Mode: mag -- Grav+Acc: False\n[INFO] -- Selected activites: ['dws', 'ups', 'wlk', 'jog']\n[INFO] -- Data subjects' information is imported.\n[INFO] -- Creating Time-Series\n[INFO] -- Shape of time-Series dataset:(767660, 8)\n[INFO] -- Test Trials: [11, 12, 13, 14, 15, 16]\n[INFO] -- Shape of Train Time-Series :(621973, 8)\n[INFO] -- Shape of Test Time-Series :(145687, 8)\n___________________________________________________\n[INFO] -- Training Data has been standardized: the mean is = [2.20896278] ; and the std is = [1.42146386]\n[INFO] -- Test/Val Data has been standardized\n[INFO] -- Training Sections: (60059, 1, 128)\n[INFO] -- Test Sections: (13344, 1, 128)\n[INFO] -- Training Sections:(60059, 1, 128, 1)\n[INFO] -- Test Sections:(13344, 1, 128, 1)\n___________________________________________________\nModel Size = 122200\n***[RESULT]*** ID Accuracy: 0.8554406474820144\n***[RESULT]*** ID F1: 84.74000000000001\n***[RESULT]*** ID Confusion Matrix\n[74.9 97.5 93.2 92.7 88.8 76.5 91.6 78.9 81.6 87.4 79.1 89.8 93.2 59.2\n 87.3 98. 73.2 65.8 66.7 87.9 93.8 95.4 93.4 86.9]\nTrue Positive Rate for 4 : 88.83720930232558\nTrue Positive Rate for 9 : 87.38898756660745\nTrue Positive Rate for 11 : 89.83957219251337\nTrue Positive Rate for 21 : 95.43650793650794\nAverage TP:90.37556924948859\n***[RESULT]*** ID Averaged F-1 Score : 0.8522158533718187\n"
],
[
"## Here we set parameter to build labeld time-series from dataset of \"(A)DeviceMotion_data\"\n## attitude(roll, pitch, yaw); gravity(x, y, z); rotationRate(x, y, z); userAcceleration(x,y,z)\nsdt = [\"rotationRate\"]\nmode = \"raw\"\nep = 40\ncga = False # Add gravity to acceleration or not\nfor i in range(5):\n results[str(sdt)+\"--\"+str(mode)+\"--\"+str(cga)+\"--\"+str(i)] = eval_id(sdt, mode, ep, cga)",
"[INFO] -- Selected sensor data types: ['rotationRate'] -- Mode: raw -- Grav+Acc: False\n[INFO] -- Selected activites: ['dws', 'ups', 'wlk', 'jog']\n[INFO] -- Data subjects' information is imported.\n[INFO] -- Creating Time-Series\n[INFO] -- Shape of time-Series dataset:(767660, 10)\n[INFO] -- Test Trials: [11, 12, 13, 14, 15, 16]\n[INFO] -- Shape of Train Time-Series :(621973, 10)\n[INFO] -- Shape of Test Time-Series :(145687, 10)\n___________________________________________________\n[INFO] -- Training Data has been standardized: the mean is = [0.00676741 0.02878308 0.02359966] ; and the std is = [1.74135109 1.64053436 1.08396877]\n[INFO] -- Test/Val Data has been standardized\n[INFO] -- Training Sections: (60059, 3, 128)\n[INFO] -- Test Sections: (13344, 3, 128)\n[INFO] -- Training Sections:(60059, 3, 128, 1)\n[INFO] -- Test Sections:(13344, 3, 128, 1)\n___________________________________________________\nModel Size = 253272\n***[RESULT]*** ID Accuracy: 0.9426708633093526\n***[RESULT]*** ID F1: 94.23\n***[RESULT]*** ID Confusion Matrix\n[ 95.6 100. 99.7 90.7 100. 97.6 89. 63.9 96.8 98.9 97.9 94.8\n 100. 65.3 89.6 99.5 98. 99.5 100. 98.6 98.7 97.4 92.2 99.4]\nTrue Positive Rate for 4 : 100.0\nTrue Positive Rate for 9 : 98.93428063943162\nTrue Positive Rate for 11 : 94.8306595365419\nTrue Positive Rate for 21 : 97.42063492063492\nAverage TP:97.79639377415211\n***[RESULT]*** ID Averaged F-1 Score : 0.9424289110626735\n[INFO] -- Selected sensor data types: ['rotationRate'] -- Mode: raw -- Grav+Acc: False\n[INFO] -- Selected activites: ['dws', 'ups', 'wlk', 'jog']\n[INFO] -- Data subjects' information is imported.\n[INFO] -- Creating Time-Series\n[INFO] -- Shape of time-Series dataset:(767660, 10)\n[INFO] -- Test Trials: [11, 12, 13, 14, 15, 16]\n[INFO] -- Shape of Train Time-Series :(621973, 10)\n[INFO] -- Shape of Test Time-Series :(145687, 10)\n___________________________________________________\n[INFO] -- Training Data has been standardized: the mean is = [0.00676741 0.02878308 0.02359966] ; and the std is = [1.74135109 1.64053436 1.08396877]\n[INFO] -- Test/Val Data has been standardized\n[INFO] -- Training Sections: (60059, 3, 128)\n[INFO] -- Test Sections: (13344, 3, 128)\n[INFO] -- Training Sections:(60059, 3, 128, 1)\n[INFO] -- Test Sections:(13344, 3, 128, 1)\n___________________________________________________\nModel Size = 253272\n***[RESULT]*** ID Accuracy: 0.9459682254196643\n***[RESULT]*** ID F1: 94.38\n***[RESULT]*** ID Confusion Matrix\n[ 97.2 100. 100. 98.6 100. 97. 71.4 70.4 97.2 100. 100. 95.4\n 100. 64.2 94.1 98.3 99.4 99.8 99.8 98.8 98. 99.8 94.2 97.7]\nTrue Positive Rate for 4 : 100.0\nTrue Positive Rate for 9 : 100.0\nTrue Positive Rate for 11 : 95.36541889483065\nTrue Positive Rate for 21 : 99.8015873015873\nAverage TP:98.79175154910449\n***[RESULT]*** ID Averaged F-1 Score : 0.9444730432088009\n[INFO] -- Selected sensor data types: ['rotationRate'] -- Mode: raw -- Grav+Acc: False\n[INFO] -- Selected activites: ['dws', 'ups', 'wlk', 'jog']\n[INFO] -- Data subjects' information is imported.\n[INFO] -- Creating Time-Series\n[INFO] -- Shape of time-Series dataset:(767660, 10)\n[INFO] -- Test Trials: [11, 12, 13, 14, 15, 16]\n[INFO] -- Shape of Train Time-Series :(621973, 10)\n[INFO] -- Shape of Test Time-Series :(145687, 10)\n___________________________________________________\n[INFO] -- Training Data has been standardized: the mean is = [0.00676741 0.02878308 0.02359966] ; and the std is = [1.74135109 1.64053436 1.08396877]\n[INFO] -- Test/Val Data has been standardized\n[INFO] -- Training Sections: (60059, 3, 128)\n[INFO] -- Test Sections: (13344, 3, 128)\n[INFO] -- Training Sections:(60059, 3, 128, 1)\n[INFO] -- Test Sections:(13344, 3, 128, 1)\n___________________________________________________\nModel Size = 253272\n***[RESULT]*** ID Accuracy: 0.9250599520383693\n***[RESULT]*** ID F1: 92.4\n***[RESULT]*** ID Confusion Matrix\n[ 88.5 100. 93.4 98.9 99.8 98.2 54.7 72.4 98.2 99.8 98.1 90.9\n 100. 60.5 85.5 99.8 98.8 99.6 100. 95.7 99.2 98. 91.8 98.3]\nTrue Positive Rate for 4 : 99.76744186046511\nTrue Positive Rate for 9 : 99.82238010657194\nTrue Positive Rate for 11 : 90.9090909090909\nTrue Positive Rate for 21 : 98.01587301587301\nAverage TP:97.12869647300025\n***[RESULT]*** ID Averaged F-1 Score : 0.9245642140496729\n[INFO] -- Selected sensor data types: ['rotationRate'] -- Mode: raw -- Grav+Acc: False\n[INFO] -- Selected activites: ['dws', 'ups', 'wlk', 'jog']\n[INFO] -- Data subjects' information is imported.\n[INFO] -- Creating Time-Series\n[INFO] -- Shape of time-Series dataset:(767660, 10)\n[INFO] -- Test Trials: [11, 12, 13, 14, 15, 16]\n[INFO] -- Shape of Train Time-Series :(621973, 10)\n[INFO] -- Shape of Test Time-Series :(145687, 10)\n___________________________________________________\n[INFO] -- Training Data has been standardized: the mean is = [0.00676741 0.02878308 0.02359966] ; and the std is = [1.74135109 1.64053436 1.08396877]\n[INFO] -- Test/Val Data has been standardized\n[INFO] -- Training Sections: (60059, 3, 128)\n[INFO] -- Test Sections: (13344, 3, 128)\n[INFO] -- Training Sections:(60059, 3, 128, 1)\n[INFO] -- Test Sections:(13344, 3, 128, 1)\n___________________________________________________\nModel Size = 253272\n***[RESULT]*** ID Accuracy: 0.9343525179856115\n***[RESULT]*** ID F1: 93.13\n***[RESULT]*** ID Confusion Matrix\n[ 90.8 100. 99.8 98.1 100. 98.8 75. 48.9 96.4 97.7 99.8 95.\n 100. 61.2 90.4 99.2 99.6 100. 100. 99.5 97.7 100. 97.7 98.6]\nTrue Positive Rate for 4 : 100.0\nTrue Positive Rate for 9 : 97.69094138543517\nTrue Positive Rate for 11 : 95.00891265597147\nTrue Positive Rate for 21 : 100.0\nAverage TP:98.17496351035166\n***[RESULT]*** ID Averaged F-1 Score : 0.9324872919486719\n[INFO] -- Selected sensor data types: ['rotationRate'] -- Mode: raw -- Grav+Acc: False\n[INFO] -- Selected activites: ['dws', 'ups', 'wlk', 'jog']\n[INFO] -- Data subjects' information is imported.\n[INFO] -- Creating Time-Series\n[INFO] -- Shape of time-Series dataset:(767660, 10)\n[INFO] -- Test Trials: [11, 12, 13, 14, 15, 16]\n[INFO] -- Shape of Train Time-Series :(621973, 10)\n[INFO] -- Shape of Test Time-Series :(145687, 10)\n___________________________________________________\n[INFO] -- Training Data has been standardized: the mean is = [0.00676741 0.02878308 0.02359966] ; and the std is = [1.74135109 1.64053436 1.08396877]\n[INFO] -- Test/Val Data has been standardized\n[INFO] -- Training Sections: (60059, 3, 128)\n[INFO] -- Test Sections: (13344, 3, 128)\n[INFO] -- Training Sections:(60059, 3, 128, 1)\n[INFO] -- Test Sections:(13344, 3, 128, 1)\n___________________________________________________\nModel Size = 253272\n***[RESULT]*** ID Accuracy: 0.9327038369304557\n***[RESULT]*** ID F1: 93.24\n***[RESULT]*** ID Confusion Matrix\n[ 98.6 100. 100. 97. 100. 97.6 76.2 41.5 96. 99.3 100. 94.7\n 99.7 65.9 92.2 99.1 100. 99.8 99.8 98.3 95.5 100. 94.8 99.7]\nTrue Positive Rate for 4 : 100.0\nTrue Positive Rate for 9 : 99.28952042628775\nTrue Positive Rate for 11 : 94.6524064171123\nTrue Positive Rate for 21 : 100.0\nAverage TP:98.48548171085001\n***[RESULT]*** ID Averaged F-1 Score : 0.9324549486716581\n"
],
[
"results",
"_____no_output_____"
],
[
"## Here we set parameter to build labeld time-series from dataset of \"(A)DeviceMotion_data\"\n## attitude(roll, pitch, yaw); gravity(x, y, z); rotationRate(x, y, z); userAcceleration(x,y,z)\nsdt = [\"userAcceleration\"]\nmode = \"mag\"\nep = 40\ncga = True # Add gravity to acceleration or not\nfor i in range(5):\n results[str(sdt)+\"--\"+str(mode)+\"--\"+str(cga)+\"--\"+str(i)] = eval_id(sdt, mode, ep, cga)",
"[INFO] -- Selected sensor data types: ['userAcceleration'] -- Mode: mag -- Grav+Acc: True\n[INFO] -- Selected activites: ['dws', 'ups', 'wlk', 'jog']\n[INFO] -- Data subjects' information is imported.\n[INFO] -- Creating Time-Series\n[INFO] -- Shape of time-Series dataset:(767660, 8)\n[INFO] -- Test Trials: [11, 12, 13, 14, 15, 16]\n[INFO] -- Shape of Train Time-Series :(621973, 8)\n[INFO] -- Shape of Test Time-Series :(145687, 8)\n___________________________________________________\n[INFO] -- Training Data has been standardized: the mean is = [1.19815844] ; and the std is = [0.70139403]\n[INFO] -- Test/Val Data has been standardized\n[INFO] -- Training Sections: (60059, 1, 128)\n[INFO] -- Test Sections: (13344, 1, 128)\n[INFO] -- Training Sections:(60059, 1, 128, 1)\n[INFO] -- Test Sections:(13344, 1, 128, 1)\n___________________________________________________\nModel Size = 122200\n***[RESULT]*** ID Accuracy: 0.8779976019184652\n***[RESULT]*** ID F1: 87.22\n***[RESULT]*** ID Confusion Matrix\n[76.3 93.5 94.3 86.3 83.7 74.5 89.8 96.9 92.2 92.2 71.1 87.3 90.1 32.3\n 82. 95.1 90.1 92. 97.2 92.2 97.9 97.8 81.5 96.6]\nTrue Positive Rate for 4 : 83.72093023255815\nTrue Positive Rate for 9 : 92.1847246891652\nTrue Positive Rate for 11 : 87.34402852049911\nTrue Positive Rate for 21 : 97.81746031746032\nAverage TP:90.2667859399207\n***[RESULT]*** ID Averaged F-1 Score : 0.8632631976169179\n[INFO] -- Selected sensor data types: ['userAcceleration'] -- Mode: mag -- Grav+Acc: True\n[INFO] -- Selected activites: ['dws', 'ups', 'wlk', 'jog']\n[INFO] -- Data subjects' information is imported.\n[INFO] -- Creating Time-Series\n[INFO] -- Shape of time-Series dataset:(767660, 8)\n[INFO] -- Test Trials: [11, 12, 13, 14, 15, 16]\n[INFO] -- Shape of Train Time-Series :(621973, 8)\n[INFO] -- Shape of Test Time-Series :(145687, 8)\n___________________________________________________\n[INFO] -- Training Data has been standardized: the mean is = [1.19815844] ; and the std is = [0.70139403]\n[INFO] -- Test/Val Data has been standardized\n[INFO] -- Training Sections: (60059, 1, 128)\n[INFO] -- Test Sections: (13344, 1, 128)\n[INFO] -- Training Sections:(60059, 1, 128, 1)\n[INFO] -- Test Sections:(13344, 1, 128, 1)\n___________________________________________________\nModel Size = 122200\n***[RESULT]*** ID Accuracy: 0.8872152278177458\n***[RESULT]*** ID F1: 87.91\n***[RESULT]*** ID Confusion Matrix\n[81.1 97.2 92.5 86.3 89.1 76.9 85.6 96.9 93.2 87.7 81.5 84.8 95. 44.5\n 71.6 94.4 92.3 90.6 96.9 94.3 99.3 94. 91.4 90.3]\nTrue Positive Rate for 4 : 89.06976744186046\nTrue Positive Rate for 9 : 87.74422735346359\nTrue Positive Rate for 11 : 84.84848484848484\nTrue Positive Rate for 21 : 94.04761904761905\nAverage TP:88.92752467285699\n***[RESULT]*** ID Averaged F-1 Score : 0.8761227862027668\n[INFO] -- Selected sensor data types: ['userAcceleration'] -- Mode: mag -- Grav+Acc: True\n[INFO] -- Selected activites: ['dws', 'ups', 'wlk', 'jog']\n[INFO] -- Data subjects' information is imported.\n[INFO] -- Creating Time-Series\n[INFO] -- Shape of time-Series dataset:(767660, 8)\n[INFO] -- Test Trials: [11, 12, 13, 14, 15, 16]\n[INFO] -- Shape of Train Time-Series :(621973, 8)\n[INFO] -- Shape of Test Time-Series :(145687, 8)\n___________________________________________________\n[INFO] -- Training Data has been standardized: the mean is = [1.19815844] ; and the std is = [0.70139403]\n[INFO] -- Test/Val Data has been standardized\n[INFO] -- Training Sections: (60059, 1, 128)\n[INFO] -- Test Sections: (13344, 1, 128)\n[INFO] -- Training Sections:(60059, 1, 128, 1)\n[INFO] -- Test Sections:(13344, 1, 128, 1)\n___________________________________________________\nModel Size = 122200\n***[RESULT]*** ID Accuracy: 0.8863908872901679\n***[RESULT]*** ID F1: 88.05\n***[RESULT]*** ID Confusion Matrix\n[77.9 96.6 95.3 78.1 85.6 84.1 83.9 92.4 95.2 92.2 65.4 90.9 95. 62.3\n 73.3 97.4 79.2 91.7 98.4 91.3 95.5 89.5 98.9 95.2]\nTrue Positive Rate for 4 : 85.5813953488372\nTrue Positive Rate for 9 : 92.1847246891652\nTrue Positive Rate for 11 : 90.9090909090909\nTrue Positive Rate for 21 : 89.48412698412699\nAverage TP:89.53983448280508\n***[RESULT]*** ID Averaged F-1 Score : 0.8776687976998061\n[INFO] -- Selected sensor data types: ['userAcceleration'] -- Mode: mag -- Grav+Acc: True\n[INFO] -- Selected activites: ['dws', 'ups', 'wlk', 'jog']\n[INFO] -- Data subjects' information is imported.\n[INFO] -- Creating Time-Series\n[INFO] -- Shape of time-Series dataset:(767660, 8)\n[INFO] -- Test Trials: [11, 12, 13, 14, 15, 16]\n[INFO] -- Shape of Train Time-Series :(621973, 8)\n[INFO] -- Shape of Test Time-Series :(145687, 8)\n___________________________________________________\n[INFO] -- Training Data has been standardized: the mean is = [1.19815844] ; and the std is = [0.70139403]\n[INFO] -- Test/Val Data has been standardized\n[INFO] -- Training Sections: (60059, 1, 128)\n[INFO] -- Test Sections: (13344, 1, 128)\n[INFO] -- Training Sections:(60059, 1, 128, 1)\n[INFO] -- Test Sections:(13344, 1, 128, 1)\n___________________________________________________\nModel Size = 122200\n***[RESULT]*** ID Accuracy: 0.8785971223021583\n***[RESULT]*** ID F1: 87.21\n***[RESULT]*** ID Confusion Matrix\n[ 78.6 94.4 90.7 81.7 75.8 81.5 88. 93.7 92. 95.2 70.6 85.4\n 90.4 60.1 73.5 93.6 87.5 91.5 100. 93.9 94.2 96. 85.4 94.9]\nTrue Positive Rate for 4 : 75.81395348837209\nTrue Positive Rate for 9 : 95.20426287744228\nTrue Positive Rate for 11 : 85.38324420677363\nTrue Positive Rate for 21 : 96.03174603174604\nAverage TP:88.10830165108351\n***[RESULT]*** ID Averaged F-1 Score : 0.8714816123809531\n[INFO] -- Selected sensor data types: ['userAcceleration'] -- Mode: mag -- Grav+Acc: True\n[INFO] -- Selected activites: ['dws', 'ups', 'wlk', 'jog']\n[INFO] -- Data subjects' information is imported.\n[INFO] -- Creating Time-Series\n[INFO] -- Shape of time-Series dataset:(767660, 8)\n[INFO] -- Test Trials: [11, 12, 13, 14, 15, 16]\n[INFO] -- Shape of Train Time-Series :(621973, 8)\n[INFO] -- Shape of Test Time-Series :(145687, 8)\n___________________________________________________\n[INFO] -- Training Data has been standardized: the mean is = [1.19815844] ; and the std is = [0.70139403]\n[INFO] -- Test/Val Data has been standardized\n[INFO] -- Training Sections: (60059, 1, 128)\n[INFO] -- Test Sections: (13344, 1, 128)\n[INFO] -- Training Sections:(60059, 1, 128, 1)\n[INFO] -- Test Sections:(13344, 1, 128, 1)\n___________________________________________________\nModel Size = 122200\n***[RESULT]*** ID Accuracy: 0.8828687050359713\n***[RESULT]*** ID F1: 87.72\n***[RESULT]*** ID Confusion Matrix\n[87.4 96.9 95.2 89.3 85.6 80.1 86.7 93.5 96.6 96.1 73.7 82.4 94.7 29.5\n 79.6 95.1 93.7 87.5 94.6 93.4 98.9 96.2 80.9 87.7]\nTrue Positive Rate for 4 : 85.5813953488372\nTrue Positive Rate for 9 : 96.0923623445826\nTrue Positive Rate for 11 : 82.35294117647058\nTrue Positive Rate for 21 : 96.23015873015873\nAverage TP:90.06421440001228\n***[RESULT]*** ID Averaged F-1 Score : 0.8675925381639025\n"
],
[
"results",
"_____no_output_____"
],
[
"## Here we set parameter to build labeld time-series from dataset of \"(A)DeviceMotion_data\"\n## attitude(roll, pitch, yaw); gravity(x, y, z); rotationRate(x, y, z); userAcceleration(x,y,z)\nsdt = [\"userAcceleration\"]\nmode = \"raw\"\nep = 40\ncga = True # Add gravity to acceleration or not\nfor i in range(5):\n results[str(sdt)+\"--\"+str(mode)+\"--\"+str(cga)+\"--\"+str(i)] = eval_id(sdt, mode, ep, cga)",
"[INFO] -- Selected sensor data types: ['userAcceleration'] -- Mode: raw -- Grav+Acc: True\n[INFO] -- Selected activites: ['dws', 'ups', 'wlk', 'jog']\n[INFO] -- Data subjects' information is imported.\n[INFO] -- Creating Time-Series\n[INFO] -- Shape of time-Series dataset:(767660, 10)\n[INFO] -- Test Trials: [11, 12, 13, 14, 15, 16]\n[INFO] -- Shape of Train Time-Series :(621973, 10)\n[INFO] -- Shape of Test Time-Series :(145687, 10)\n___________________________________________________\n[INFO] -- Training Data has been standardized: the mean is = [ 0.02367904 0.95806826 -0.05104623] ; and the std is = [0.48401853 0.74077811 0.47270486]\n[INFO] -- Test/Val Data has been standardized\n[INFO] -- Training Sections: (60059, 3, 128)\n[INFO] -- Test Sections: (13344, 3, 128)\n[INFO] -- Training Sections:(60059, 3, 128, 1)\n[INFO] -- Test Sections:(13344, 3, 128, 1)\n___________________________________________________\nModel Size = 253272\n***[RESULT]*** ID Accuracy: 0.9224370503597122\n***[RESULT]*** ID F1: 92.25\n***[RESULT]*** ID Confusion Matrix\n[ 63.4 99.9 97.9 93.1 100. 96.8 53.5 73.6 97.4 98.9 97.6 95.7\n 100. 75.5 93.3 98.9 94.6 100. 86.7 100. 98.7 98.2 98.3 99.4]\nTrue Positive Rate for 4 : 100.0\nTrue Positive Rate for 9 : 98.93428063943162\nTrue Positive Rate for 11 : 95.72192513368985\nTrue Positive Rate for 21 : 98.21428571428571\nAverage TP:98.2176228718518\n***[RESULT]*** ID Averaged F-1 Score : 0.9197941176289994\n[INFO] -- Selected sensor data types: ['userAcceleration'] -- Mode: raw -- Grav+Acc: True\n[INFO] -- Selected activites: ['dws', 'ups', 'wlk', 'jog']\n[INFO] -- Data subjects' information is imported.\n[INFO] -- Creating Time-Series\n[INFO] -- Shape of time-Series dataset:(767660, 10)\n[INFO] -- Test Trials: [11, 12, 13, 14, 15, 16]\n[INFO] -- Shape of Train Time-Series :(621973, 10)\n[INFO] -- Shape of Test Time-Series :(145687, 10)\n___________________________________________________\n[INFO] -- Training Data has been standardized: the mean is = [ 0.02367904 0.95806826 -0.05104623] ; and the std is = [0.48401853 0.74077811 0.47270486]\n[INFO] -- Test/Val Data has been standardized\n[INFO] -- Training Sections: (60059, 3, 128)\n[INFO] -- Test Sections: (13344, 3, 128)\n[INFO] -- Training Sections:(60059, 3, 128, 1)\n[INFO] -- Test Sections:(13344, 3, 128, 1)\n___________________________________________________\nModel Size = 253272\n***[RESULT]*** ID Accuracy: 0.9288069544364509\n***[RESULT]*** ID F1: 92.9\n***[RESULT]*** ID Confusion Matrix\n[ 57.9 100. 97.9 95.9 100. 99. 53. 71.9 85.4 100. 94.8 95.7\n 100. 96.1 97.1 99.4 97.2 99.8 92.9 99.7 97.3 99.8 97.7 99.4]\nTrue Positive Rate for 4 : 100.0\nTrue Positive Rate for 9 : 100.0\nTrue Positive Rate for 11 : 95.72192513368985\nTrue Positive Rate for 21 : 99.8015873015873\nAverage TP:98.88087810881927\n***[RESULT]*** ID Averaged F-1 Score : 0.929605296860835\n[INFO] -- Selected sensor data types: ['userAcceleration'] -- Mode: raw -- Grav+Acc: True\n[INFO] -- Selected activites: ['dws', 'ups', 'wlk', 'jog']\n[INFO] -- Data subjects' information is imported.\n[INFO] -- Creating Time-Series\n[INFO] -- Shape of time-Series dataset:(767660, 10)\n[INFO] -- Test Trials: [11, 12, 13, 14, 15, 16]\n[INFO] -- Shape of Train Time-Series :(621973, 10)\n[INFO] -- Shape of Test Time-Series :(145687, 10)\n___________________________________________________\n[INFO] -- Training Data has been standardized: the mean is = [ 0.02367904 0.95806826 -0.05104623] ; and the std is = [0.48401853 0.74077811 0.47270486]\n[INFO] -- Test/Val Data has been standardized\n[INFO] -- Training Sections: (60059, 3, 128)\n[INFO] -- Test Sections: (13344, 3, 128)\n[INFO] -- Training Sections:(60059, 3, 128, 1)\n[INFO] -- Test Sections:(13344, 3, 128, 1)\n___________________________________________________\nModel Size = 253272\n***[RESULT]*** ID Accuracy: 0.9276828537170264\n***[RESULT]*** ID F1: 92.73\n***[RESULT]*** ID Confusion Matrix\n[ 60.5 100. 97.6 88. 100. 98. 68.5 71.6 96.2 100. 98.1 93.2\n 100. 73.1 94.7 100. 96.6 99.5 96.1 99.1 94.9 98.6 99.1 99.7]\nTrue Positive Rate for 4 : 100.0\nTrue Positive Rate for 9 : 100.0\nTrue Positive Rate for 11 : 93.22638146167557\nTrue Positive Rate for 21 : 98.61111111111111\nAverage TP:97.95937314319667\n***[RESULT]*** ID Averaged F-1 Score : 0.9244424382727164\n[INFO] -- Selected sensor data types: ['userAcceleration'] -- Mode: raw -- Grav+Acc: True\n[INFO] -- Selected activites: ['dws', 'ups', 'wlk', 'jog']\n[INFO] -- Data subjects' information is imported.\n[INFO] -- Creating Time-Series\n[INFO] -- Shape of time-Series dataset:(767660, 10)\n[INFO] -- Test Trials: [11, 12, 13, 14, 15, 16]\n[INFO] -- Shape of Train Time-Series :(621973, 10)\n[INFO] -- Shape of Test Time-Series :(145687, 10)\n___________________________________________________\n[INFO] -- Training Data has been standardized: the mean is = [ 0.02367904 0.95806826 -0.05104623] ; and the std is = [0.48401853 0.74077811 0.47270486]\n[INFO] -- Test/Val Data has been standardized\n[INFO] -- Training Sections: (60059, 3, 128)\n[INFO] -- Test Sections: (13344, 3, 128)\n[INFO] -- Training Sections:(60059, 3, 128, 1)\n[INFO] -- Test Sections:(13344, 3, 128, 1)\n___________________________________________________\nModel Size = 253272\n***[RESULT]*** ID Accuracy: 0.9258093525179856\n***[RESULT]*** ID F1: 92.56\n***[RESULT]*** ID Confusion Matrix\n[ 62.8 100. 99.4 96.1 99.5 98.2 52.2 69.9 96.8 99.8 92.4 96.3\n 100. 85.2 92. 99.2 95.6 100. 92. 99.3 96.2 97.6 99.5 99.7]\nTrue Positive Rate for 4 : 99.53488372093024\nTrue Positive Rate for 9 : 99.82238010657194\nTrue Positive Rate for 11 : 96.2566844919786\nTrue Positive Rate for 21 : 97.61904761904762\nAverage TP:98.3082489846321\n***[RESULT]*** ID Averaged F-1 Score : 0.9240084516904764\n[INFO] -- Selected sensor data types: ['userAcceleration'] -- Mode: raw -- Grav+Acc: True\n[INFO] -- Selected activites: ['dws', 'ups', 'wlk', 'jog']\n[INFO] -- Data subjects' information is imported.\n[INFO] -- Creating Time-Series\n[INFO] -- Shape of time-Series dataset:(767660, 10)\n[INFO] -- Test Trials: [11, 12, 13, 14, 15, 16]\n[INFO] -- Shape of Train Time-Series :(621973, 10)\n[INFO] -- Shape of Test Time-Series :(145687, 10)\n___________________________________________________\n[INFO] -- Training Data has been standardized: the mean is = [ 0.02367904 0.95806826 -0.05104623] ; and the std is = [0.48401853 0.74077811 0.47270486]\n[INFO] -- Test/Val Data has been standardized\n[INFO] -- Training Sections: (60059, 3, 128)\n[INFO] -- Test Sections: (13344, 3, 128)\n[INFO] -- Training Sections:(60059, 3, 128, 1)\n[INFO] -- Test Sections:(13344, 3, 128, 1)\n___________________________________________________\nModel Size = 253272\n***[RESULT]*** ID Accuracy: 0.9274580335731415\n***[RESULT]*** ID F1: 92.73\n***[RESULT]*** ID Confusion Matrix\n[ 56.3 100. 88.9 90.1 100. 98. 81.4 64.4 94. 100. 88.6 96.8\n 99.7 78.5 97.1 99.8 98.8 100. 99.4 96.5 95.6 100. 98.3 99.1]\nTrue Positive Rate for 4 : 100.0\nTrue Positive Rate for 9 : 100.0\nTrue Positive Rate for 11 : 96.79144385026738\nTrue Positive Rate for 21 : 100.0\nAverage TP:99.19786096256684\n***[RESULT]*** ID Averaged F-1 Score : 0.9254639638126179\n"
],
[
"results ",
"_____no_output_____"
],
[
"## Here we set parameter to build labeld time-series from dataset of \"(A)DeviceMotion_data\"\n## attitude(roll, pitch, yaw); gravity(x, y, z); rotationRate(x, y, z); userAcceleration(x,y,z)\nsdt = [\"rotationRate\",\"userAcceleration\"]\nmode = \"mag\"\nep = 40\ncga = True # Add gravity to acceleration or not\nfor i in range(5):\n results[str(sdt)+\"--\"+str(mode)+\"--\"+str(cga)+\"--\"+str(i)] = eval_id(sdt, mode, ep, cga)",
"[INFO] -- Selected sensor data types: ['rotationRate', 'userAcceleration'] -- Mode: mag -- Grav+Acc: True\n[INFO] -- Selected activites: ['dws', 'ups', 'wlk', 'jog']\n[INFO] -- Data subjects' information is imported.\n[INFO] -- Creating Time-Series\n[INFO] -- Shape of time-Series dataset:(767660, 9)\n[INFO] -- Test Trials: [11, 12, 13, 14, 15, 16]\n[INFO] -- Shape of Train Time-Series :(621973, 9)\n[INFO] -- Shape of Test Time-Series :(145687, 9)\n___________________________________________________\n[INFO] -- Training Data has been standardized: the mean is = [2.20896278 1.19815844] ; and the std is = [1.42146386 0.70139403]\n[INFO] -- Test/Val Data has been standardized\n[INFO] -- Training Sections: (60059, 2, 128)\n[INFO] -- Test Sections: (13344, 2, 128)\n[INFO] -- Training Sections:(60059, 2, 128, 1)\n[INFO] -- Test Sections:(13344, 2, 128, 1)\n___________________________________________________\nModel Size = 187736\n***[RESULT]*** ID Accuracy: 0.9230365707434053\n***[RESULT]*** ID F1: 92.17999999999999\n***[RESULT]*** ID Confusion Matrix\n[74.7 98.8 95. 91.8 93.7 84.7 96.1 97.4 95.6 98.4 74.2 93.9 97.2 49.\n 90.4 99.4 95.8 91.9 98.5 94.3 99. 98.6 93.4 88.9]\nTrue Positive Rate for 4 : 93.72093023255815\nTrue Positive Rate for 9 : 98.40142095914743\nTrue Positive Rate for 11 : 93.93939393939394\nTrue Positive Rate for 21 : 98.61111111111111\nAverage TP:96.16821406055266\n***[RESULT]*** ID Averaged F-1 Score : 0.9114187439044757\n[INFO] -- Selected sensor data types: ['rotationRate', 'userAcceleration'] -- Mode: mag -- Grav+Acc: True\n[INFO] -- Selected activites: ['dws', 'ups', 'wlk', 'jog']\n[INFO] -- Data subjects' information is imported.\n[INFO] -- Creating Time-Series\n[INFO] -- Shape of time-Series dataset:(767660, 9)\n[INFO] -- Test Trials: [11, 12, 13, 14, 15, 16]\n[INFO] -- Shape of Train Time-Series :(621973, 9)\n[INFO] -- Shape of Test Time-Series :(145687, 9)\n___________________________________________________\n[INFO] -- Training Data has been standardized: the mean is = [2.20896278 1.19815844] ; and the std is = [1.42146386 0.70139403]\n[INFO] -- Test/Val Data has been standardized\n[INFO] -- Training Sections: (60059, 2, 128)\n[INFO] -- Test Sections: (13344, 2, 128)\n[INFO] -- Training Sections:(60059, 2, 128, 1)\n[INFO] -- Test Sections:(13344, 2, 128, 1)\n___________________________________________________\nModel Size = 187736\n***[RESULT]*** ID Accuracy: 0.9313549160671463\n***[RESULT]*** ID F1: 92.9\n***[RESULT]*** ID Confusion Matrix\n[87.8 99.5 97.7 90.9 97.7 85.7 91.6 97.1 91.6 97.2 82.2 91.8 98.8 66.4\n 84.3 98.5 95.2 88.1 99.8 92.5 99.7 98. 97.1 88.6]\nTrue Positive Rate for 4 : 97.67441860465115\nTrue Positive Rate for 9 : 97.15808170515098\nTrue Positive Rate for 11 : 91.80035650623886\nTrue Positive Rate for 21 : 98.01587301587301\nAverage TP:96.1621824579785\n***[RESULT]*** ID Averaged F-1 Score : 0.924153792706694\n[INFO] -- Selected sensor data types: ['rotationRate', 'userAcceleration'] -- Mode: mag -- Grav+Acc: True\n[INFO] -- Selected activites: ['dws', 'ups', 'wlk', 'jog']\n[INFO] -- Data subjects' information is imported.\n[INFO] -- Creating Time-Series\n[INFO] -- Shape of time-Series dataset:(767660, 9)\n[INFO] -- Test Trials: [11, 12, 13, 14, 15, 16]\n[INFO] -- Shape of Train Time-Series :(621973, 9)\n[INFO] -- Shape of Test Time-Series :(145687, 9)\n___________________________________________________\n[INFO] -- Training Data has been standardized: the mean is = [2.20896278 1.19815844] ; and the std is = [1.42146386 0.70139403]\n[INFO] -- Test/Val Data has been standardized\n[INFO] -- Training Sections: (60059, 2, 128)\n[INFO] -- Test Sections: (13344, 2, 128)\n[INFO] -- Training Sections:(60059, 2, 128, 1)\n[INFO] -- Test Sections:(13344, 2, 128, 1)\n___________________________________________________\nModel Size = 187736\n***[RESULT]*** ID Accuracy: 0.9392236211031175\n***[RESULT]*** ID F1: 93.78999999999999\n***[RESULT]*** ID Confusion Matrix\n[ 85.7 100. 97.9 90.4 89.3 88.4 87.2 95.4 97. 98.4 83.4 92.7\n 92.9 74.8 91.2 99.5 95. 93.9 99.8 97.6 99. 99.2 94.6 94.3]\nTrue Positive Rate for 4 : 89.30232558139535\nTrue Positive Rate for 9 : 98.40142095914743\nTrue Positive Rate for 11 : 92.6916221033868\nTrue Positive Rate for 21 : 99.20634920634922\nAverage TP:94.90042946256969\n***[RESULT]*** ID Averaged F-1 Score : 0.9338696272527999\n[INFO] -- Selected sensor data types: ['rotationRate', 'userAcceleration'] -- Mode: mag -- Grav+Acc: True\n[INFO] -- Selected activites: ['dws', 'ups', 'wlk', 'jog']\n[INFO] -- Data subjects' information is imported.\n[INFO] -- Creating Time-Series\n[INFO] -- Shape of time-Series dataset:(767660, 9)\n[INFO] -- Test Trials: [11, 12, 13, 14, 15, 16]\n[INFO] -- Shape of Train Time-Series :(621973, 9)\n[INFO] -- Shape of Test Time-Series :(145687, 9)\n___________________________________________________\n[INFO] -- Training Data has been standardized: the mean is = [2.20896278 1.19815844] ; and the std is = [1.42146386 0.70139403]\n[INFO] -- Test/Val Data has been standardized\n[INFO] -- Training Sections: (60059, 2, 128)\n[INFO] -- Test Sections: (13344, 2, 128)\n[INFO] -- Training Sections:(60059, 2, 128, 1)\n[INFO] -- Test Sections:(13344, 2, 128, 1)\n___________________________________________________\nModel Size = 187736\n***[RESULT]*** ID Accuracy: 0.938923860911271\n***[RESULT]*** ID F1: 93.7\n***[RESULT]*** ID Confusion Matrix\n[82.3 99.8 99.5 89.4 96.5 86.7 95.4 98.6 89.4 96.8 82.5 93.2 97.2 76.6\n 95.9 99.5 95. 81.9 99.7 97.2 99.4 96.8 97.7 87.2]\nTrue Positive Rate for 4 : 96.51162790697676\nTrue Positive Rate for 9 : 96.80284191829485\nTrue Positive Rate for 11 : 93.22638146167557\nTrue Positive Rate for 21 : 96.82539682539682\nAverage TP:95.841562028086\n***[RESULT]*** ID Averaged F-1 Score : 0.9332315997361625\n[INFO] -- Selected sensor data types: ['rotationRate', 'userAcceleration'] -- Mode: mag -- Grav+Acc: True\n[INFO] -- Selected activites: ['dws', 'ups', 'wlk', 'jog']\n[INFO] -- Data subjects' information is imported.\n[INFO] -- Creating Time-Series\n[INFO] -- Shape of time-Series dataset:(767660, 9)\n[INFO] -- Test Trials: [11, 12, 13, 14, 15, 16]\n[INFO] -- Shape of Train Time-Series :(621973, 9)\n[INFO] -- Shape of Test Time-Series :(145687, 9)\n___________________________________________________\n[INFO] -- Training Data has been standardized: the mean is = [2.20896278 1.19815844] ; and the std is = [1.42146386 0.70139403]\n[INFO] -- Test/Val Data has been standardized\n[INFO] -- Training Sections: (60059, 2, 128)\n[INFO] -- Test Sections: (13344, 2, 128)\n[INFO] -- Training Sections:(60059, 2, 128, 1)\n[INFO] -- Test Sections:(13344, 2, 128, 1)\n___________________________________________________\nModel Size = 187736\n***[RESULT]*** ID Accuracy: 0.9327038369304557\n***[RESULT]*** ID F1: 93.16\n***[RESULT]*** ID Confusion Matrix\n[ 74.9 99.7 93.4 89.1 96.5 83.1 95.9 98.9 94. 98.4 70.9 90.9\n 98.8 74.8 89.6 100. 95.6 93.3 100. 93.2 99.9 99. 95.1 94.6]\nTrue Positive Rate for 4 : 96.51162790697676\nTrue Positive Rate for 9 : 98.40142095914743\nTrue Positive Rate for 11 : 90.9090909090909\nTrue Positive Rate for 21 : 99.0079365079365\nAverage TP:96.2075190707879\n***[RESULT]*** ID Averaged F-1 Score : 0.9261680441674489\n"
],
[
"results",
"_____no_output_____"
],
[
"## Here we set parameter to build labeld time-series from dataset of \"(A)DeviceMotion_data\"\n## attitude(roll, pitch, yaw); gravity(x, y, z); rotationRate(x, y, z); userAcceleration(x,y,z)\nsdt = [\"rotationRate\",\"userAcceleration\"]\nmode = \"raw\"\nep = 40\ncga = True # Add gravity to acceleration or not\nfor i in range(5):\n results[str(sdt)+\"--\"+str(mode)+\"--\"+str(cga)+\"--\"+str(i)] = eval_id(sdt, mode, ep, cga)",
"[INFO] -- Selected sensor data types: ['rotationRate', 'userAcceleration'] -- Mode: raw -- Grav+Acc: True\n[INFO] -- Selected activites: ['dws', 'ups', 'wlk', 'jog']\n[INFO] -- Data subjects' information is imported.\n[INFO] -- Creating Time-Series\n[INFO] -- Shape of time-Series dataset:(767660, 13)\n[INFO] -- Test Trials: [11, 12, 13, 14, 15, 16]\n[INFO] -- Shape of Train Time-Series :(621973, 13)\n[INFO] -- Shape of Test Time-Series :(145687, 13)\n___________________________________________________\n[INFO] -- Training Data has been standardized: the mean is = [ 0.00676741 0.02878308 0.02359966 0.02367904 0.95806826 -0.05104623] ; and the std is = [1.74135109 1.64053436 1.08396877 0.48401853 0.74077811 0.47270486]\n[INFO] -- Test/Val Data has been standardized\n[INFO] -- Training Sections: (60059, 6, 128)\n[INFO] -- Test Sections: (13344, 6, 128)\n[INFO] -- Training Sections:(60059, 6, 128, 1)\n[INFO] -- Test Sections:(13344, 6, 128, 1)\n___________________________________________________\nModel Size = 449880\n***[RESULT]*** ID Accuracy: 0.9350269784172662\n***[RESULT]*** ID F1: 93.34\n***[RESULT]*** ID Confusion Matrix\n[ 92.9 100. 89.9 84.9 100. 99.2 70.3 69.4 97.4 98.8 98.8 96.8\n 100. 69.4 91.8 100. 97. 100. 100. 95. 97.9 100. 99.2 100. ]\nTrue Positive Rate for 4 : 100.0\nTrue Positive Rate for 9 : 98.75666074600356\nTrue Positive Rate for 11 : 96.79144385026738\nTrue Positive Rate for 21 : 100.0\nAverage TP:98.88702614906774\n***[RESULT]*** ID Averaged F-1 Score : 0.9325265689243271\n[INFO] -- Selected sensor data types: ['rotationRate', 'userAcceleration'] -- Mode: raw -- Grav+Acc: True\n[INFO] -- Selected activites: ['dws', 'ups', 'wlk', 'jog']\n[INFO] -- Data subjects' information is imported.\n[INFO] -- Creating Time-Series\n[INFO] -- Shape of time-Series dataset:(767660, 13)\n[INFO] -- Test Trials: [11, 12, 13, 14, 15, 16]\n[INFO] -- Shape of Train Time-Series :(621973, 13)\n[INFO] -- Shape of Test Time-Series :(145687, 13)\n___________________________________________________\n[INFO] -- Training Data has been standardized: the mean is = [ 0.00676741 0.02878308 0.02359966 0.02367904 0.95806826 -0.05104623] ; and the std is = [1.74135109 1.64053436 1.08396877 0.48401853 0.74077811 0.47270486]\n[INFO] -- Test/Val Data has been standardized\n[INFO] -- Training Sections: (60059, 6, 128)\n[INFO] -- Test Sections: (13344, 6, 128)\n[INFO] -- Training Sections:(60059, 6, 128, 1)\n[INFO] -- Test Sections:(13344, 6, 128, 1)\n___________________________________________________\nModel Size = 449880\n***[RESULT]*** ID Accuracy: 0.9302308153477218\n***[RESULT]*** ID F1: 92.86999999999999\n***[RESULT]*** ID Confusion Matrix\n[ 77.2 100. 100. 47.8 100. 98.8 93.4 60.8 98.2 100. 100. 97.\n 100. 72.5 99.8 99.1 97.6 100. 98.2 99.8 98.2 99.6 99.7 100. ]\nTrue Positive Rate for 4 : 100.0\nTrue Positive Rate for 9 : 100.0\nTrue Positive Rate for 11 : 96.96969696969697\nTrue Positive Rate for 21 : 99.60317460317461\nAverage TP:99.1432178932179\n***[RESULT]*** ID Averaged F-1 Score : 0.9290414241426904\n[INFO] -- Selected sensor data types: ['rotationRate', 'userAcceleration'] -- Mode: raw -- Grav+Acc: True\n[INFO] -- Selected activites: ['dws', 'ups', 'wlk', 'jog']\n[INFO] -- Data subjects' information is imported.\n[INFO] -- Creating Time-Series\n[INFO] -- Shape of time-Series dataset:(767660, 13)\n[INFO] -- Test Trials: [11, 12, 13, 14, 15, 16]\n[INFO] -- Shape of Train Time-Series :(621973, 13)\n[INFO] -- Shape of Test Time-Series :(145687, 13)\n___________________________________________________\n[INFO] -- Training Data has been standardized: the mean is = [ 0.00676741 0.02878308 0.02359966 0.02367904 0.95806826 -0.05104623] ; and the std is = [1.74135109 1.64053436 1.08396877 0.48401853 0.74077811 0.47270486]\n[INFO] -- Test/Val Data has been standardized\n[INFO] -- Training Sections: (60059, 6, 128)\n[INFO] -- Test Sections: (13344, 6, 128)\n[INFO] -- Training Sections:(60059, 6, 128, 1)\n[INFO] -- Test Sections:(13344, 6, 128, 1)\n___________________________________________________\nModel Size = 449880\n***[RESULT]*** ID Accuracy: 0.9288818944844125\n***[RESULT]*** ID F1: 92.69\n***[RESULT]*** ID Confusion Matrix\n[ 87.1 100. 96.2 38.3 100. 100. 97.2 60.8 96.8 99.1 99.1 98.8\n 100. 73.3 93.9 100. 100. 100. 100. 100. 99.7 98. 99.8 98. ]\nTrue Positive Rate for 4 : 100.0\nTrue Positive Rate for 9 : 99.11190053285968\nTrue Positive Rate for 11 : 98.75222816399287\nTrue Positive Rate for 21 : 98.01587301587301\nAverage TP:98.9700004281814\n***[RESULT]*** ID Averaged F-1 Score : 0.9291945175218213\n[INFO] -- Selected sensor data types: ['rotationRate', 'userAcceleration'] -- Mode: raw -- Grav+Acc: True\n[INFO] -- Selected activites: ['dws', 'ups', 'wlk', 'jog']\n[INFO] -- Data subjects' information is imported.\n[INFO] -- Creating Time-Series\n[INFO] -- Shape of time-Series dataset:(767660, 13)\n[INFO] -- Test Trials: [11, 12, 13, 14, 15, 16]\n[INFO] -- Shape of Train Time-Series :(621973, 13)\n[INFO] -- Shape of Test Time-Series :(145687, 13)\n___________________________________________________\n[INFO] -- Training Data has been standardized: the mean is = [ 0.00676741 0.02878308 0.02359966 0.02367904 0.95806826 -0.05104623] ; and the std is = [1.74135109 1.64053436 1.08396877 0.48401853 0.74077811 0.47270486]\n[INFO] -- Test/Val Data has been standardized\n[INFO] -- Training Sections: (60059, 6, 128)\n[INFO] -- Test Sections: (13344, 6, 128)\n[INFO] -- Training Sections:(60059, 6, 128, 1)\n[INFO] -- Test Sections:(13344, 6, 128, 1)\n___________________________________________________\nModel Size = 449880\n***[RESULT]*** ID Accuracy: 0.9248351318944844\n***[RESULT]*** ID F1: 92.36\n***[RESULT]*** ID Confusion Matrix\n[ 84.4 100. 98.2 47. 100. 99.4 53.5 90.4 99.8 100. 99.5 91.1\n 100. 73.3 94.9 100. 98.4 100. 100. 99.8 99. 99. 99.7 96. ]\nTrue Positive Rate for 4 : 100.0\nTrue Positive Rate for 9 : 100.0\nTrue Positive Rate for 11 : 91.0873440285205\nTrue Positive Rate for 21 : 99.0079365079365\nAverage TP:97.52382013411426\n***[RESULT]*** ID Averaged F-1 Score : 0.9188192865713979\n[INFO] -- Selected sensor data types: ['rotationRate', 'userAcceleration'] -- Mode: raw -- Grav+Acc: True\n[INFO] -- Selected activites: ['dws', 'ups', 'wlk', 'jog']\n[INFO] -- Data subjects' information is imported.\n[INFO] -- Creating Time-Series\n[INFO] -- Shape of time-Series dataset:(767660, 13)\n[INFO] -- Test Trials: [11, 12, 13, 14, 15, 16]\n[INFO] -- Shape of Train Time-Series :(621973, 13)\n[INFO] -- Shape of Test Time-Series :(145687, 13)\n___________________________________________________\n[INFO] -- Training Data has been standardized: the mean is = [ 0.00676741 0.02878308 0.02359966 0.02367904 0.95806826 -0.05104623] ; and the std is = [1.74135109 1.64053436 1.08396877 0.48401853 0.74077811 0.47270486]\n[INFO] -- Test/Val Data has been standardized\n[INFO] -- Training Sections: (60059, 6, 128)\n[INFO] -- Test Sections: (13344, 6, 128)\n[INFO] -- Training Sections:(60059, 6, 128, 1)\n[INFO] -- Test Sections:(13344, 6, 128, 1)\n___________________________________________________\nModel Size = 449880\n***[RESULT]*** ID Accuracy: 0.9425959232613909\n***[RESULT]*** ID F1: 93.67\n***[RESULT]*** ID Confusion Matrix\n[ 95.4 100. 96.5 64.8 100. 97.8 85.1 91. 80. 99.8 99.1 95.7\n 100. 72.2 92.2 97.7 99.2 99.5 100. 100. 99.9 99. 98.2 99.1]\nTrue Positive Rate for 4 : 100.0\nTrue Positive Rate for 9 : 99.82238010657194\nTrue Positive Rate for 11 : 95.72192513368985\nTrue Positive Rate for 21 : 99.0079365079365\nAverage TP:98.63806043704957\n***[RESULT]*** ID Averaged F-1 Score : 0.9417669448883043\n"
],
[
"results",
"_____no_output_____"
],
[
"#https://stackoverflow.com/a/45305384/5210098\ndef f1_metric(y_true, y_pred):\n def recall(y_true, y_pred):\n \"\"\"Recall metric.\n\n Only computes a batch-wise average of recall.\n\n Computes the recall, a metric for multi-label classification of\n how many relevant items are selected.\n \"\"\"\n true_positives = K.sum(K.round(K.clip(y_true * y_pred, 0, 1)))\n possible_positives = K.sum(K.round(K.clip(y_true, 0, 1)))\n recall = true_positives / (possible_positives + K.epsilon())\n return recall\n\n def precision(y_true, y_pred):\n \"\"\"Precision metric.\n\n Only computes a batch-wise average of precision.\n\n Computes the precision, a metric for multi-label classification of\n how many selected items are relevant.\n \"\"\"\n true_positives = K.sum(K.round(K.clip(y_true * y_pred, 0, 1)))\n predicted_positives = K.sum(K.round(K.clip(y_pred, 0, 1)))\n precision = true_positives / (predicted_positives + K.epsilon())\n return precision\n precision = precision(y_true, y_pred)\n recall = recall(y_true, y_pred)\n return 2*((precision*recall)/(precision+recall+K.epsilon()))\n\n\ndef eval_id(sdt, mode, ep, cga):\n\n print(\"[INFO] -- Selected sensor data types: \"+str(sdt)+\" -- Mode: \"+str(mode)+\" -- Grav+Acc: \"+str(cga)) \n act_labels = ACT_LABELS [0:4]\n\n print(\"[INFO] -- Selected activites: \"+str(act_labels)) \n trial_codes = [TRIAL_CODES[act] for act in act_labels]\n dt_list = set_data_types(sdt)\n dataset = creat_time_series(dt_list, act_labels, trial_codes, mode=mode, labeled=True, combine_grav_acc = cga)\n print(\"[INFO] -- Shape of time-Series dataset:\"+str(dataset.shape)) \n\n\n #*****************\n TRAIN_TEST_TYPE = \"trial\" # \"subject\" or \"trial\"\n #*****************\n\n if TRAIN_TEST_TYPE == \"subject\":\n test_ids = [4,9,11,21]\n print(\"[INFO] -- Test IDs: \"+str(test_ids))\n test_ts = dataset.loc[(dataset['id'].isin(test_ids))]\n train_ts = dataset.loc[~(dataset['id'].isin(test_ids))]\n else:\n test_trail = [11,12,13,14,15,16] \n print(\"[INFO] -- Test Trials: \"+str(test_trail))\n test_ts = dataset.loc[(dataset['trial'].isin(test_trail))]\n train_ts = dataset.loc[~(dataset['trial'].isin(test_trail))]\n\n print(\"[INFO] -- Shape of Train Time-Series :\"+str(train_ts.shape))\n print(\"[INFO] -- Shape of Test Time-Series :\"+str(test_ts.shape))\n \n# print(\"___________Train_VAL____________\")\n# val_trail = [11,12,13,14,15,16]\n# val_ts = train_ts.loc[(train_ts['trial'].isin(val_trail))]\n# train_ts = train_ts.loc[~(train_ts['trial'].isin(val_trail))]\n# print(\"[INFO] -- Training Time-Series :\"+str(train_ts.shape))\n# print(\"[INFO] -- Validation Time-Series :\"+str(val_ts.shape)) \n print(\"___________________________________________________\")\n\n ## This Variable Defines the Size of Sliding Window\n ## ( e.g. 100 means in each snapshot we just consider 100 consecutive observations of each sensor) \n w = 128 # 50 Equals to 1 second for MotionSense Dataset (it is on 50Hz samplig rate)\n ## Here We Choose Step Size for Building Diffrent Snapshots from Time-Series Data\n ## ( smaller step size will increase the amount of the instances and higher computational cost may be incurred )\n s = 10\n train_data, act_train, id_train, train_mean, train_std = ts_to_secs(train_ts.copy(),\n w,\n s,\n standardize = True)\n \n \n s = 10\n test_data, act_test, id_test, test_mean, test_std = ts_to_secs(test_ts.copy(),\n w,\n s,\n standardize = True,\n mean = train_mean, \n std = train_std)\n \n print(\"[INFO] -- Training Sections: \"+str(train_data.shape))\n print(\"[INFO] -- Test Sections: \"+str(test_data.shape))\n\n\n id_train_labels = to_categorical(id_train)\n id_test_labels = to_categorical(id_test)\n \n act_train_labels = to_categorical(act_train)\n act_test_labels = to_categorical(act_test)\n \n ## Here we add an extra dimension to the datasets just to be ready for using with Convolution2D\n train_data = np.expand_dims(train_data,axis=3)\n print(\"[INFO] -- Training Sections:\"+str(train_data.shape))\n\n test_data = np.expand_dims(test_data,axis=3)\n print(\"[INFO] -- Test Sections:\"+str(test_data.shape))\n\n height = train_data.shape[1]\n width = train_data.shape[2]\n\n id_class_numbers = 24\n act_class_numbers = 4\n fm = (2,5)\n\n print(\"___________________________________________________\")\n ## Callbacks\n #eval_metric= \"val_acc\"\n eval_metric= \"val_f1_metric\" \n early_stop = keras.callbacks.EarlyStopping(monitor=eval_metric, mode='max', patience = 7)\n filepath=\"MID.best.hdf5\"\n checkpoint = ModelCheckpoint(filepath, monitor=eval_metric, verbose=0, save_best_only=True, mode='max')\n callbacks_list = [early_stop,\n checkpoint\n ]\n ## Callbacks\n eval_id = Estimator.build(height, width, id_class_numbers, name =\"EVAL_ID\", fm=fm, act_func=\"softmax\",hid_act_func=\"relu\")\n eval_id.compile( loss=\"categorical_crossentropy\", optimizer='adam', metrics=['acc', f1_metric])\n print(\"Model Size = \"+str(eval_id.count_params()))\n\n eval_id.fit(train_data, id_train_labels,\n validation_data = (test_data, id_test_labels),\n epochs = ep,\n batch_size = 128,\n verbose = 0,\n class_weight = get_class_weights(np.argmax(id_train_labels,axis=1)),\n callbacks = callbacks_list\n )\n\n eval_id.load_weights(\"MID.best.hdf5\")\n eval_id.compile( loss=\"categorical_crossentropy\", optimizer='adam', metrics=['acc',f1_metric])\n\n result1 = eval_id.evaluate(test_data, id_test_labels, verbose = 2)\n id_acc = result1[1]\n print(\"***[RESULT]*** ID Accuracy: \"+str(id_acc))\n rf1 = result1[2].round(4)*100\n print(\"***[RESULT]*** ID F1: \"+str(rf1))\n \n preds = eval_id.predict(test_data)\n preds = np.argmax(preds, axis=1)\n conf_mat = confusion_matrix(np.argmax(id_test_labels, axis=1), preds)\n conf_mat = conf_mat.astype('float') / conf_mat.sum(axis=1)[:, np.newaxis]\n print(\"***[RESULT]*** ID Confusion Matrix\")\n print((np.array(conf_mat).diagonal()).round(3)*100) \n \n d_test_ids = [4,9,11,21]\n to_avg = 0\n for i in range(len(d_test_ids)):\n true_positive = conf_mat[d_test_ids[i],d_test_ids[i]]\n print(\"True Positive Rate for \"+str(d_test_ids[i])+\" : \"+str(true_positive*100))\n to_avg+=true_positive\n atp = to_avg/len(d_test_ids) \n print(\"Average TP:\"+str(atp*100)) \n \n f1id = f1_score(np.argmax(id_test_labels, axis=1), preds, average=None).mean()\n print(\"***[RESULT]*** ID Averaged F-1 Score : \"+str(f1id))\n \n return [round(id_acc,4), round(f1id,4), round(atp,4)]",
"_____no_output_____"
],
[
"## Here we set parameter to build labeld time-series from dataset of \"(A)DeviceMotion_data\"\n## attitude(roll, pitch, yaw); gravity(x, y, z); rotationRate(x, y, z); userAcceleration(x,y,z)\nsdt = [\"rotationRate\",\"userAcceleration\"]\nmode = \"mag\"\nep = 40\ncga = True # Add gravity to acceleration or not\nfor i in range(5):\n results[str(sdt)+\"-2D-\"+str(mode)+\"--\"+str(cga)+\"--\"+str(i)] = eval_id(sdt, mode, ep, cga)",
"[INFO] -- Selected sensor data types: ['rotationRate', 'userAcceleration'] -- Mode: mag -- Grav+Acc: True\n[INFO] -- Selected activites: ['dws', 'ups', 'wlk', 'jog']\n[INFO] -- Data subjects' information is imported.\n[INFO] -- Creating Time-Series\n[INFO] -- Shape of time-Series dataset:(767660, 9)\n[INFO] -- Test Trials: [11, 12, 13, 14, 15, 16]\n[INFO] -- Shape of Train Time-Series :(621973, 9)\n[INFO] -- Shape of Test Time-Series :(145687, 9)\n___________________________________________________\n[INFO] -- Training Data has been standardized: the mean is = [2.20896278 1.19815844] ; and the std is = [1.42146386 0.70139403]\n[INFO] -- Test/Val Data has been standardized\n[INFO] -- Training Sections: (60059, 2, 128)\n[INFO] -- Test Sections: (13344, 2, 128)\n[INFO] -- Training Sections:(60059, 2, 128, 1)\n[INFO] -- Test Sections:(13344, 2, 128, 1)\n___________________________________________________\nModel Size = 223896\n"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb8859289072e8544f277601889fa4e3e64efbe4
| 3,366 |
ipynb
|
Jupyter Notebook
|
cleaning/data_cleaning_10k.ipynb
|
SouthernMethodistUniversity/Python_Finance_TextMining
|
db40d4cba0e771339ace0c080b2c5800aad82013
|
[
"MIT"
] | null | null | null |
cleaning/data_cleaning_10k.ipynb
|
SouthernMethodistUniversity/Python_Finance_TextMining
|
db40d4cba0e771339ace0c080b2c5800aad82013
|
[
"MIT"
] | null | null | null |
cleaning/data_cleaning_10k.ipynb
|
SouthernMethodistUniversity/Python_Finance_TextMining
|
db40d4cba0e771339ace0c080b2c5800aad82013
|
[
"MIT"
] | null | null | null | 21.43949 | 102 | 0.483363 |
[
[
[
"# Table of Contents\n <p>",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nfrom bs4 import BeautifulSoup\nimport numpy as np\nimport os",
"_____no_output_____"
],
[
"def read_function(filename):\n ll = []\n with open(filename) as f:\n contents = f.read()\n contents = contents.split('<FILENAME>')[1]\n s = BeautifulSoup(contents, \"html.parser\")\n f = s.find_all('p')\n if len(f)<5:\n f = s.find_all('div')\n\n for i in range(len(f)):\n tt = f[i].text\n if tt is not None:\n ll = np.append(ll,tt)\n result = \" \".join(ll).replace(u'\\xa0', u' ').replace(u'\\n',u' ').replace(u'\\t',u' ')\n result = \" \".join(result.split())\n \n name = filename.split('_')[0]\n date = filename.split('_')[1].split('.')[0]\n \n print('Done: '+filename)\n \n #return s\n return (name, date, result)",
"_____no_output_____"
],
[
"filelist= os.listdir('data10k')\nfilelist",
"_____no_output_____"
],
[
"df =pd.DataFrame(columns=['Name','Date','Story'])\nfor f in filelist:\n info = read_function('data10k/'+f)\n df = df.append({'Name':info[0],'Date':info[1],\"Story\":info[2]}, ignore_index=True)",
"_____no_output_____"
],
[
"df[\"Date\"]=pd.to_datetime(df[\"Date\"])",
"_____no_output_____"
],
[
"df['Name'] = df['Name'].apply(lambda x: x.split('/')[-1])",
"_____no_output_____"
],
[
"df.to_csv('finance_data_10k.csv', index=False, encoding='utf8')",
"_____no_output_____"
],
[
"df",
"_____no_output_____"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb8875224d2c1a6184fc6b8ad6e991c27daae444
| 11,473 |
ipynb
|
Jupyter Notebook
|
LabFiles/Module 2/Ex2.3 UCB.ipynb
|
csastry12/DAT257x
|
5f786fb93eca660338e375f523e0022695668c79
|
[
"Unlicense"
] | null | null | null |
LabFiles/Module 2/Ex2.3 UCB.ipynb
|
csastry12/DAT257x
|
5f786fb93eca660338e375f523e0022695668c79
|
[
"Unlicense"
] | null | null | null |
LabFiles/Module 2/Ex2.3 UCB.ipynb
|
csastry12/DAT257x
|
5f786fb93eca660338e375f523e0022695668c79
|
[
"Unlicense"
] | null | null | null | 53.115741 | 1,330 | 0.545106 |
[
[
[
"# DAT257x: Reinforcement Learning Explained\n\n## Lab 2: Bandits\n\n### Exercise 2.3: UCB",
"_____no_output_____"
]
],
[
[
"# import numpy as np\n# import sys\n\n# if \"../\" not in sys.path:\n# sys.path.append(\"../\") \n\n# from lib.envs.bandit import BanditEnv\n# from lib.simulation import Experiment",
"_____no_output_____"
],
[
"# #Policy interface\n# class Policy:\n# #num_actions: (int) Number of arms [indexed by 0 ... num_actions-1]\n# def __init__(self, num_actions):\n# self.num_actions = num_actions\n \n# def act(self):\n# pass\n \n# def feedback(self, action, reward):\n# pass",
"_____no_output_____"
],
[
"# #Greedy policy\n# class Greedy(Policy):\n# def __init__(self, num_actions):\n# Policy.__init__(self, num_actions)\n# self.name = \"Greedy\"\n# self.total_rewards = np.zeros(num_actions, dtype = np.longdouble)\n# self.total_counts = np.zeros(num_actions, dtype = np.longdouble)\n \n# def act(self):\n# current_averages = np.divide(self.total_rewards, self.total_counts, where = self.total_counts > 0)\n# current_averages[self.total_counts <= 0] = 0.5 #Correctly handles Bernoulli rewards; over-estimates otherwise\n# current_action = np.argmax(current_averages)\n# return current_action\n \n# def feedback(self, action, reward):\n# self.total_rewards[action] += reward\n# self.total_counts[action] += 1",
"_____no_output_____"
],
[
"# #Epsilon Greedy policy\n# class EpsilonGreedy(Greedy):\n# def __init__(self, num_actions, epsilon):\n# Greedy.__init__(self, num_actions)\n# if (epsilon is None or epsilon < 0 or epsilon > 1):\n# print(\"EpsilonGreedy: Invalid value of epsilon\", flush = True)\n# sys.exit(0)\n \n# self.epsilon = epsilon\n# self.name = \"Epsilon Greedy\"\n \n# def act(self):\n# choice = None\n# if self.epsilon == 0:\n# choice = 0\n# elif self.epsilon == 1:\n# choice = 1\n# else:\n# choice = np.random.binomial(1, self.epsilon)\n \n# if choice == 1:\n# return np.random.choice(self.num_actions)\n# else:\n# current_averages = np.divide(self.total_rewards, self.total_counts, where = self.total_counts > 0)\n# current_averages[self.total_counts <= 0] = 0.5 #Correctly handles Bernoulli rewards; over-estimates otherwise\n# current_action = np.argmax(current_averages)\n# return current_action\n ",
"_____no_output_____"
]
],
[
[
"Now let's implement a UCB algorithm. \n\n",
"_____no_output_____"
]
],
[
[
"# xx = np.ones(10)\n# xy = np.divide(2, xx)\n# np.sqrt(xy)",
"_____no_output_____"
],
[
"# #UCB policy\n# class UCB(Greedy):\n# def __init__(self, num_actions):\n# Greedy.__init__(self, num_actions)\n# self.name = \"UCB\"\n# self.round = 0\n \n# def act(self):\n# current_action = None\n# # self.round += 1\n# if self.round < self.num_actions:\n# \"\"\"The first k rounds, where k is the number of arms/actions, play each arm/action once\"\"\"\n# current_action = self.round\n# else:\n# \"\"\"At round t, play the arms with maximum average and exploration bonus\"\"\"\n \n# current_averages = np.divide(self.total_rewards, self.total_counts, where = self.total_counts > 0)\n# current_averages[self.total_counts <= 0] = 0.5 #Correctly handles Bernoulli rewards; over-estimates otherwise\n# exp_bonus = np.sqrt(np.divide(2.0 * np.log(self.round), self.total_counts, where = self.total_counts > 0))\n# current_averages = current_averages + exp_bonus\n# current_action = np.argmax(current_averages)\n \n# self.round += 1\n \n# return current_action",
"_____no_output_____"
],
[
"# class UCB(Greedy):\n# def __init__(self, num_actions, k):\n# Greedy.__init__(self, num_actions)\n# self.name = \"UCB\"\n# self.round = 0\n# self.k = k\n# self.previous_action = num_actions - 1\n# self.num_actions = num_actions\n# self.qvalues = np.zeros(num_actions)\n# self.counts = np.zeros(num_actions)\n# self.t = 0\n \n# def act(self):\n# if (self.round < self.k):\n# \"\"\"The first k rounds, play each arm/action once\"\"\"\n# current_action = self.previous_action + 1\n# if current_action >= self.num_actions:\n# self.round += 1\n# current_action = 0\n# self.previous_action = current_action\n \n# if (self.round >= self.k):\n# \"\"\"play the arms with maximum average and exploration bonus\"\"\"\n# r_hats = self.total_rewards/self.total_counts\n# scores = r_hats + np.sqrt(np.log(self.t)/self.total_counts)\n# current_action = np.argmax(scores)\n \n# return current_action\n \n# def feedback(self, action, reward):\n# self.total_rewards[action] += reward\n# self.total_counts[action] += 1\n# self.t += 1",
"_____no_output_____"
]
],
[
[
"Now let's prepare the simulation. ",
"_____no_output_____"
]
],
[
[
"# evaluation_seed = 1239\n# num_actions = 10\n# trials = 10000\n# distribution = \"bernoulli\"\n# # distribution = \"normal\"",
"_____no_output_____"
]
],
[
[
"What do you think the regret graph would look like?",
"_____no_output_____"
]
],
[
[
"# env = BanditEnv(num_actions, distribution, evaluation_seed)\n# agent = UCB(num_actions)\n# # K = 1\n# # agent = UCB(num_actions, K)\n# experiment = Experiment(env, agent)\n# experiment.run_bandit(trials)",
"_____no_output_____"
]
],
[
[
"# DAT257x: Reinforcement Learning Explained\n\n## Lab 2: Bandits\n\n### Exercise 2.4 Thompson Beta",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport sys\n\nif \"../\" not in sys.path:\n sys.path.append(\"../\") \n\nfrom lib.envs.bandit import BanditEnv\nfrom lib.simulation import Experiment",
"_____no_output_____"
],
[
"#Policy interface\nclass Policy:\n #num_actions: (int) Number of arms [indexed by 0 ... num_actions-1]\n def __init__(self, num_actions):\n self.num_actions = num_actions\n \n def act(self):\n pass\n \n def feedback(self, action, reward):\n pass",
"_____no_output_____"
]
],
[
[
"Now let's implement a Thompson Beta algorithm. ",
"_____no_output_____"
]
],
[
[
"#Tompson Beta policy\nclass ThompsonBeta(Policy):\n def __init__(self, num_actions):\n Policy.__init__(self, num_actions)\n #PRIOR Hyper-params: successes = 1; failures = 1\n self.total_counts = np.zeros(num_actions, dtype = np.longdouble)\n self.name = \"Thompson Beta\"\n \n #For each arm, maintain success and failures\n self.successes = np.ones(num_actions, dtype = np.int)\n self.failures = np.ones(num_actions, dtype = np.int)\n \n def act(self):\n \"\"\"Sample beta distribution from success and failures\"\"\"\n \n \"\"\"Play the max of the sampled values\"\"\"\n \n current_action = 0\n return current_action\n \n def feedback(self, action, reward):\n if reward > 0:\n self.successes[action] += 1\n else:\n self.failures[action] += 1\n self.total_counts[action] += 1",
"_____no_output_____"
]
],
[
[
"Now let's prepare the simulation. ",
"_____no_output_____"
]
],
[
[
"evaluation_seed = 1239\nnum_actions = 10\ntrials = 10000\ndistribution = \"bernoulli\"",
"_____no_output_____"
]
],
[
[
"What do you think the regret graph would look like?",
"_____no_output_____"
]
],
[
[
"env = BanditEnv(num_actions, distribution, evaluation_seed)\nagent = ThompsonBeta(num_actions)\nexperiment = Experiment(env, agent)\nexperiment.run_bandit(trials)",
"_____no_output_____"
]
],
[
[
"Now let's prepare another simulation by setting a different distribution, that is set distribion = \"normal\"",
"_____no_output_____"
],
[
"Run the simulation and observe the results.",
"_____no_output_____"
],
[
"What do you think the regret graph would look like?",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
cb887deafaf85f32c6ab403c98abe7e0991c25fc
| 69,083 |
ipynb
|
Jupyter Notebook
|
raw/exploratory_computing_with_python/notebook9_discrete_random_variables/py_exploratory_comp_9_sol.ipynb
|
MAMBA-python/raw_material
|
55029cfb4c64ffb5deebf04015fd85a94a0093be
|
[
"MIT"
] | 1 |
2018-11-30T21:16:40.000Z
|
2018-11-30T21:16:40.000Z
|
raw/exploratory_computing_with_python/notebook9_discrete_random_variables/py_exploratory_comp_9_sol.ipynb
|
MAMBA-python/raw_material
|
55029cfb4c64ffb5deebf04015fd85a94a0093be
|
[
"MIT"
] | null | null | null |
raw/exploratory_computing_with_python/notebook9_discrete_random_variables/py_exploratory_comp_9_sol.ipynb
|
MAMBA-python/raw_material
|
55029cfb4c64ffb5deebf04015fd85a94a0093be
|
[
"MIT"
] | null | null | null | 69.710394 | 8,160 | 0.78536 |
[
[
[
"<figure>\n <IMG SRC=\"https://raw.githubusercontent.com/mbakker7/exploratory_computing_with_python/master/tudelft_logo.png\" WIDTH=250 ALIGN=\"right\">\n</figure>\n\n# Exploratory Computing with Python\n*Developed by Mark Bakker*",
"_____no_output_____"
],
[
"## Notebook 9: Discrete random variables\nIn this Notebook you learn how to deal with discrete random variables. Many of the functions we will use are included in the `random` subpackage of `numpy`. We will import this package and call it `rnd` so that we don't have to type `np.random.` all the time.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\nimport numpy.random as rnd\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"### Random numbers\nA random number generator lets you draw, at random, a number from a specified distribution. Several random number generators are included in the `random` subpackage of `numpy`. For example, the `ranint(low, high, size)` function returns an integer array of shape `size` at random from `low` up to (but not including) `high`. For example, let's flip a coin 10 times and assign a 0 to heads and a 1 to tails. Note that the `high` is specified as `1 + 1`, which means it is `1` higher than the value we want.",
"_____no_output_____"
]
],
[
[
"rnd.randint(0, 1 + 1, 10)",
"_____no_output_____"
]
],
[
[
"If we call the `ran_int` function again, we get a different sequence of heads (zeros) and tails (ones):",
"_____no_output_____"
]
],
[
[
"rnd.randint(0, 1 + 1, 10)",
"_____no_output_____"
]
],
[
[
"Internally, the random number generator starts with what is called a *seed*. The seed is a number and is generated automatically (and supposedly at random) when you call the random number generator. The value of the seed exactly defines the sequence of random numbers that you get (so some people may argue that the generated sequence is at best pseudo-random, and you may not want to use the sequence for any serious cryptographic use, but for our purposes they are random enough). For example, let's set `seed` equal to 10",
"_____no_output_____"
]
],
[
[
"rnd.seed(10)\nrnd.randint(0, 1 + 1, 10)",
"_____no_output_____"
]
],
[
[
"If we now specify the seed again as 10, we can generate the exact same sequence",
"_____no_output_____"
]
],
[
[
"rnd.seed(10)\nrnd.randint(0, 1 + 1, 10)",
"_____no_output_____"
]
],
[
[
"The ability to generate the exact same sequence is useful during code development. For example, by seeding the random number generator, you can compare your output to output of others trying to solve the same problem.",
"_____no_output_____"
],
[
"### Flipping a coin\nEnough for now about random number generators. Let's flip a coin 100 times and count the number of heads (0-s) and the number of tails (1-s):",
"_____no_output_____"
]
],
[
[
"flip = rnd.randint(0, 1 + 1, 100)\nheadcount = 0\ntailcount = 0\nfor i in range(100):\n if flip[i] == 0:\n headcount += 1\n else:\n tailcount += 1\nprint('number of heads:', headcount)\nprint('number of tails:', tailcount)",
"number of heads: 59\nnumber of tails: 41\n"
]
],
[
[
"First of all, note that the number of heads and the number of tails add up to 100. Also, note how we counted the heads and tails. We created counters `headcount` and `tailcount`, looped through all flips, and added 1 to the appropriate counter. Instead of a loop, we could have used a condition for the indices combined with a summation as follows",
"_____no_output_____"
]
],
[
[
"headcount = np.count_nonzero(flip == 0)\ntailcount = np.count_nonzero(flip == 1)\nprint('headcount', headcount)\nprint('tailcount', tailcount)",
"headcount 59\ntailcount 41\n"
]
],
[
[
"How does that work? You may recall that the `flip == 0` statement returns an array with length 100 (equal to the lenght of `flip`) with the value `True` when the condition is met, and `False` when the condition is not met. The boolean `True` has the value 1, and the boolean `False` has the value 0. So we simply need to count the nonzero values using the `np.count_nonzero` function to find out how many items are `True`. \n\nThe code above is easy, but if we do an experiment with more than two outcomes, it may be cumbersome to count the non-zero items for every possible outcome. So let's try to rewrite this part of the code using a loop. For this specific case the number of lines of code doesn't decrease, but when we have an experiment with many different outcomes this will be much more efficient. Note that `dtype='int'` sets the array to integers.",
"_____no_output_____"
]
],
[
[
"outcomes = np.zeros(2, dtype='int') # Two outcomes. heads are stored in outcome[0], tails in outcome[1]\nfor i in range (2):\n outcomes[i] = np.count_nonzero(flip == i)\n print('outcome ', i, ' is ', outcomes[i])",
"outcome 0 is 59\noutcome 1 is 41\n"
]
],
[
[
"### Exercise 1. <a name=\"back1\"></a>Throwing a dice\nThrow a dice 100 times and report how many times you throw 1, 2, 3, 4, 5, and 6. Use a seed of 33. Make sure that the reported values add up to 100. Make sure you use a loop in your code as we did in the previous code cell.",
"_____no_output_____"
],
[
"<a href=\"#ex1answer\">Answers to Exercise 1</a>",
"_____no_output_____"
],
[
"### Flipping a coin twice\nNext we are going to flip a coin twice for 100 times and count the number of tails. We generate a random array of 0-s (heads) and 1-s (tails) with two rows (representing two coin flips) and 100 colums. The sum of the two rows represents the number of tails. The `np.sum` function takes an array and by default sums all the values in the array and returns one number. In this case we want to sum the rows. For that, the `sum` function has a keyword argument called `axis`, where `axis=0` sums over index 0 of the array (the rows), `axis=1` sums over the index 1 of the array (the columns), etc.",
"_____no_output_____"
]
],
[
[
"rnd.seed(55)\nflips = rnd.randint(low=0, high=1 + 1, size=(2, 100))\ntails = np.sum(flips, axis=0)\nnumber_of_tails = np.zeros(3, dtype='int')\nfor i in range(3):\n number_of_tails[i] = np.count_nonzero(tails == i)\nprint('number of 0, 1, 2 tails:', number_of_tails)",
"number of 0, 1, 2 tails: [27 47 26]\n"
]
],
[
[
"Another way to simulate flipping a coin twice, is to draw a number at random from a set of 2 numbers (0 and 1). You need to replace the number after every draw, of course. The `numpy` function to draw a random number from a given array is called `choice`. The `choice` function has a keyword to specify whether values are replaced or not. Hence the following two ways to generate 5 flips are identical.",
"_____no_output_____"
]
],
[
[
"rnd.seed(55)\nflips1 = rnd.randint(low=0, high=1 + 1, size=5)\nrnd.seed(55)\nflips2 = rnd.choice(range(2), size=5, replace=True)\nnp.alltrue(flips1 == flips2) # Check whether all values in the two arrays are equal",
"_____no_output_____"
]
],
[
[
"### Bar graph\nThe outcome of the experiment may also be plotted with a bar graph",
"_____no_output_____"
]
],
[
[
"plt.bar(range(0, 3), number_of_tails)\nplt.xticks(range(0, 3))\nplt.xlabel('number of tails')\nplt.ylabel('occurence in 100 trials');",
"_____no_output_____"
]
],
[
[
"### Cumulative Probability\nNext we compute the experimental probability of 0 tails, 1 tail, and 2 tails through division by the total number of trials (one trial is two coin flips). The three probabilities add up to 1. The cumulative probability distribution is obtained by cumulatively summing the probabilities using the `cumsum` function of `numpy`. The first value is the probability of throwing 0 tails. The second value is the probability of 1 or fewer tails, and the third value it the probability of 2 or fewer tails. The probability is computed as the number of tails divided by the total number of trials.",
"_____no_output_____"
]
],
[
[
"prob = number_of_tails / 100 # number_of_tails was computed two code cells back\ncum_prob = np.cumsum(prob) # So cum_prob[0] = prob[0], cum_prob[1] = prob[0] + prob[1], etc.\nprint('cum_prob ', cum_prob)",
"cum_prob [ 0.27 0.74 1. ]\n"
]
],
[
[
"The cumulative probability distribution is plotted with a bar graph, making sure that all the bars touch each other (by setting the width to 1, in the case below)",
"_____no_output_____"
]
],
[
[
"plt.bar(range(0, 3), cum_prob, width=1)\nplt.xticks(range(0, 3))\nplt.xlabel('number of tails in two flips')\nplt.ylabel('cumulative probability');",
"_____no_output_____"
]
],
[
[
"### Exercise 2. <a name=\"back2\"></a>Flip a coin five times\nFlip a coin five times in a row and record how many times you obtain tails (varying from 0-5). Perform the exeriment 1000 times. Make a bar graph with the total number of tails on the horizontal axis and the emperically computed probability to get that many tails, on the vertical axis. Execute your code several times (hit [shift]-[enter]) and see that the graph changes a bit every time, as the sequence of random numbers changes every time. ",
"_____no_output_____"
],
[
"Compute the cumulative probability. Print the values to the screen and make a plot of the cumulative probability function using a bar graph.",
"_____no_output_____"
],
[
"<a href=\"#ex2answer\">Answers to Exercise 2</a>",
"_____no_output_____"
],
[
"### Probability of a Bernouilli variable\nIn the previous exercise, we computed the probability of a certain number of heads in five flips experimentally. But we can, of course, compute the value exactly by using a few simple formulas. Consider the random variable $Y$, which is the outcome of an experiment with two possible values 0 and 1. Let $p$ be the probability of success, $p=P(Y=1)$. \nThen $Y$ is said to be a Bernoulli variable. The experiment is repeated $n$ times and we define $X$ as the number of successes in the experiment. The variable $X$ has a Binomial Distribution with parameters $n$ and $p$. The probability that $X$ takes value $k$ can be computed as (see for example [here](http://en.wikipedia.org/wiki/Binomial_distribution))\n\n$$P(X=k) = \\binom{n}{k}p^k(1-p)^{n-k}$$\n\nThe term $\\binom{n}{k}$ may be computed with the `comb` function, which needs to be imported from the `scipy.misc` package.",
"_____no_output_____"
],
[
"### Exercise 3. <a name=\"back3\"></a>Flip a coin 5 times revisited\nGo back to the experiment where we flip a coin five times in a row and record how many times we obtain tails.\nCompute the theoretical probability for 0, 1, 2, 3, 4, and 5 tails and compare your answer to the probability computed from 1000 trials, 10000 trials, and 100000 trials (use a loop for these three sets of trials). Do you approach the theoretical value with more trials?",
"_____no_output_____"
],
[
"<a href=\"#ex3answer\">Answers to Exercise 3</a>",
"_____no_output_____"
],
[
"### Exercise 4. <a name=\"back4\"></a>Maximum value of two dice throws\nThrow a dice two times and record the maximum value of the two throws. Use the `np.max` function to compute the maximum value. Like the `np.sum` function, the `np.max` function takes an array as input argument and an optional keyword argument named `axis`. Perform the experiment 1000 times and compute the probability that the highest value is 1, 2, 3, 4, 5, or 6. Make a graph of the cumulative probability distribution function using a step graph.",
"_____no_output_____"
],
[
"<a href=\"#ex4answer\">Answers to Exercise 4</a>",
"_____no_output_____"
],
[
"### Exercise 5. <a name=\"back5\"></a>Maximum value of two dice throws revisited\nRefer back to Exercise 4.\nCompute the theoretical value of the probability of the highest dice when throwing the dice twice (the throws are labeled T1 and T2, respectively). There are 36 possible outcomes for this experiment. Let $M$ denote the random variable corresponding to this experiment (this means for instance that $M=3$ when your first throw is a 2, and the second throw is a 3). All outcomes of $M$ can easily be written down, as shown in the following Table: \n\n| T1$\\downarrow$ T2$\\to$ | 1 | 2 | 3 | 4 | 5 | 6 |\n|-----------:|------------:|:------------:|\n| 1 | 1 | 2 | 3 | 4 | 5 | 6 |\n| 2 | 2 | 2 | 3 | 4 | 5 | 6 |\n| 3 | 3 | 3 | 3 | 4 | 5 | 6 |\n| 4 | 4 | 4 | 4 | 4 | 5 | 6 |\n| 5 | 5 | 5 | 5 | 5 | 5 | 6 |\n| 6 | 6 | 6 | 6 | 6 | 6 | 6 |\n\n\nUse the 36 possible outcomes shown in the Table to compute the theoretical probability of $M$ being 1, 2, 3, 4, 5, or 6. Compare the theoretical outcome with the experimental outcome for 100, 1000, and 10000 dice throws.",
"_____no_output_____"
],
[
"<a href=\"#ex5answer\">Answers to Exercise 5</a>",
"_____no_output_____"
],
[
"### Generate random integers with non-equal probabilities\nSo far, we have generated random numbers of which the probability of each outcome was the same (heads or tails, or the numbers on a dice, considering the throwing device was \"fair\"). What now if we want to generate outcomes that don't have the same probability? For example, consider the case that we have a bucket with 4 blue balls and 6 red balls. When you draw a ball at random, the probability of a blue ball is 0.4 and the probability of a red ball is 0.6. A sequence of drawing ten balls, with replacement, may be generated as follows",
"_____no_output_____"
]
],
[
[
"balls = np.zeros(10, dtype='int') # zero is blue\nballs[4:] = 1 # one is red\nprint('balls:', balls)\ndrawing = rnd.choice(balls, 10, replace=True)\nprint('drawing:', drawing)\nprint('blue balls:', np.count_nonzero(drawing == 0))\nprint('red balls:', np.count_nonzero(drawing == 1))",
"balls: [0 0 0 0 1 1 1 1 1 1]\ndrawing: [1 1 1 0 1 0 0 1 1 0]\nblue balls: 4\nred balls: 6\n"
]
],
[
[
"### Exercise 6. <a name=\"back6\"></a>Election poll\nConsider an election where one million people will vote. 490,000 people will vote for candidate $A$ and 510,000 people will vote for candidate $B$. One day before the election, the company of 'Maurice the Dog' conducts a pole among 1000 randomly chosen voters. Compute whether the Dog will predict the winner correctly using the approach explained above and a seed of 2.",
"_____no_output_____"
],
[
"Perform the pole 1000 times. Count how many times the outcome of the pole is that candidate $A$ wins and how many times the outcome of the pole is that candidate $B$ wins. What is the probability that the Dog will predict the correct winner based on these 1000 poles of 1000 people? ",
"_____no_output_____"
],
[
"Compute the probability that the Dog will predict the correct winner based on 1000 poles of 5000 people? Does the probability that The Dog predicts the correct winner increase significantly when he poles 5000 people?",
"_____no_output_____"
],
[
"<a href=\"#ex6answer\">Answers to Exercise 6</a>",
"_____no_output_____"
],
[
"### Answers to the exercises",
"_____no_output_____"
],
[
"<a name=\"ex1answer\">Answers to Exercise 1</a>",
"_____no_output_____"
]
],
[
[
"rnd.seed(33)\ndicethrow = rnd.randint(1, 6 + 1, 100)\nside = np.zeros(6, dtype='int')\nfor i in range(6):\n side[i] = np.count_nonzero(dicethrow == i + 1)\n print('number of times', i + 1, 'is', side[i])\nprint('total number of throws ', sum(side))",
"number of times 1 is 17\nnumber of times 2 is 17\nnumber of times 3 is 15\nnumber of times 4 is 24\nnumber of times 5 is 19\nnumber of times 6 is 8\ntotal number of throws 100\n"
]
],
[
[
"<a href=\"#back1\">Back to Exercise 1</a>\n\n<a name=\"ex2answer\">Answers to Exercise 2</a>",
"_____no_output_____"
]
],
[
[
"N = 1000\ntails = np.sum(rnd.randint(0, 1 + 1, (5, 1000)), axis=0)\ncounttails = np.zeros(6, dtype='int')\nfor i in range(6):\n counttails[i] = np.count_nonzero(tails == i)\nplt.bar(range(0, 6), counttails / N)\nplt.xlabel('number of tails in five flips')\nplt.ylabel('probability');",
"_____no_output_____"
],
[
"cumprob = np.cumsum(counttails / N)\nprint('cumprob:', cumprob)\nplt.bar(range(0, 6), cumprob, width=1)\nplt.xlabel('number of tails in five flips')\nplt.ylabel('cumulative probability');",
"cumprob: [ 0.033 0.192 0.491 0.812 0.968 1. ]\n"
]
],
[
[
"<a href=\"#back2\">Back to Exercise 2</a>\n\n<a name=\"ex3answer\">Answers to Exercise 3</a>",
"_____no_output_____"
]
],
[
[
"from scipy.misc import comb\nprint('Theoretical probabilities:')\nfor k in range(6):\n print(k, ' tails ', comb(5, k) * 0.5 ** k * 0.5 ** (5 - k))\nfor N in (1000, 10000, 100000):\n tails = np.sum(rnd.randint(0, 1 + 1, (5, N)), axis=0)\n counttails = np.zeros(6)\n for i in range(6):\n counttails[i] = np.count_nonzero(tails==i)\n print('Probability with', N, 'trials: ', counttails / float(N))",
"Theoretical probabilities:\n0 tails 0.03125\n1 tails 0.15625\n2 tails 0.3125\n3 tails 0.3125\n4 tails 0.15625\n5 tails 0.03125\nProbability with 1000 trials: [ 0.031 0.16 0.307 0.334 0.139 0.029]\nProbability with 10000 trials: [ 0.0315 0.1579 0.3083 0.3098 0.1634 0.0291]\nProbability with 100000 trials: [ 0.03093 0.15591 0.31454 0.31266 0.15484 0.03112]\n"
]
],
[
[
"<a href=\"#back3\">Back to Exercise 3</a>\n\n<a name=\"ex4answer\">Answers to Exercise 4</a>",
"_____no_output_____"
]
],
[
[
"dice = rnd.randint(1, 6 + 1, (2, 1000))\nhighest_dice = np.max(dice, 0)\noutcome = np.zeros(6)\nfor i in range(6):\n outcome[i] = np.sum(highest_dice == i + 1) / 1000\nplt.bar(left=np.arange(1, 7), height=outcome, width=1)\nplt.xlabel('highest dice in two throws')\nplt.ylabel('probability');",
"_____no_output_____"
]
],
[
[
"<a href=\"#back4\">Back to Exercise 4</a>\n\n<a name=\"ex5answer\">Answers to Exercise 5</a>",
"_____no_output_____"
]
],
[
[
"for N in [100, 1000, 10000]:\n dice = rnd.randint(1, 6 + 1, (2, N))\n highest_dice = np.max(dice, axis=0)\n outcome = np.zeros(6)\n for i in range(6):\n outcome[i] = np.sum(highest_dice == i + 1) / N\n print('Outcome for', N, 'throws: ', outcome)\n# Exact values\nexact = np.zeros(6)\nfor i, j in enumerate(range(1, 12, 2)):\n exact[i] = j / 36\nprint('Exact probabilities: ',exact)",
"Outcome for 100 throws: [ 0.01 0.03 0.19 0.13 0.27 0.37]\nOutcome for 1000 throws: [ 0.035 0.076 0.142 0.189 0.262 0.296]\nOutcome for 10000 throws: [ 0.0258 0.0862 0.1323 0.1924 0.248 0.3153]\nExact probabilities: [ 0.02777778 0.08333333 0.13888889 0.19444444 0.25 0.30555556]\n"
]
],
[
[
"<a href=\"#back5\">Back to Exercise 5</a>\n\n<a name=\"ex6answer\">Answers to Exercise 6</a>",
"_____no_output_____"
]
],
[
[
"rnd.seed(2)\npeople = np.zeros(1000000, dtype='int') # candidate A is 0\npeople[490000:] = 1 # candidate B is 1\npole = rnd.choice(people, 1000)\npoled_for_A = np.count_nonzero(pole == 0)\nprint('poled for A:', poled_for_A)\nif poled_for_A > 500: \n print('The Dog will predict the wrong winner')\nelse:\n print('The Dog will predict the correct winner')",
"poled for A: 508\nThe Dog will predict the wrong winner\n"
],
[
"Awins = 0\nBwins = 0\nfor i in range(1000):\n people = np.zeros(1000000, dtype='int') # candidate A is 0\n people[490000:] = 1 # candidate B is 1\n pole = rnd.choice(people, 1000)\n poled_for_A = np.count_nonzero(pole == 0)\n if poled_for_A > 500: \n Awins += 1\n else:\n Bwins += 1\nprint('1000 poles of 1000 people')\nprint('Probability that The Dog predicts candidate A to win:', Awins / 1000)",
"1000 poles of 1000 people\nProbability that The Dog predicts candidate A to win: 0.267\n"
],
[
"Awins = 0\nBwins = 0\nfor i in range(1000):\n people = np.zeros(1000000, dtype='int') # candidate A is 0\n people[490000:] = 1 # candidate B is 1\n pole = rnd.choice(people, 5000)\n poled_for_A = np.count_nonzero(pole == 0)\n if poled_for_A > 2500: \n Awins += 1\n else:\n Bwins += 1\nprint('1000 poles of 5000 people')\nprint('Probability that The Dog predicts candidate A to win:', Awins / 5000)",
"1000 poles of 5000 people\nProbability that The Dog predicts candidate A to win: 0.014\n"
]
],
[
[
"<a href=\"#back6\">Back to Exercise 6</a>",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
]
] |
cb8880c10ef9f2a2607d027b664070374beaba94
| 24,273 |
ipynb
|
Jupyter Notebook
|
module3-permutation-boosting/LS_DS_233.ipynb
|
CurdtMillion/DS-Unit-2-Applied-Modeling
|
89c475634f0b08de91e85c5e741d8ae687f292b3
|
[
"MIT"
] | 1 |
2022-02-23T17:22:01.000Z
|
2022-02-23T17:22:01.000Z
|
module3-permutation-boosting/LS_DS_233.ipynb
|
CurdtMillion/DS-Unit-2-Applied-Modeling
|
89c475634f0b08de91e85c5e741d8ae687f292b3
|
[
"MIT"
] | 2 |
2022-01-13T04:03:19.000Z
|
2022-03-12T01:04:52.000Z
|
module3-permutation-boosting/LS_DS_233.ipynb
|
CurdtMillion/DS-Unit-2-Applied-Modeling
|
89c475634f0b08de91e85c5e741d8ae687f292b3
|
[
"MIT"
] | 2 |
2019-08-08T02:25:10.000Z
|
2020-06-04T00:27:58.000Z
| 38.345972 | 1,022 | 0.629959 |
[
[
[
"Lambda School Data Science\n\n*Unit 2, Sprint 3, Module 3*\n\n---",
"_____no_output_____"
],
[
"# Permutation & Boosting\n\n- Get **permutation importances** for model interpretation and feature selection\n- Use xgboost for **gradient boosting**",
"_____no_output_____"
],
[
"### Setup\n\nRun the code cell below. You can work locally (follow the [local setup instructions](https://lambdaschool.github.io/ds/unit2/local/)) or on Colab.\n\nLibraries:\n\n- category_encoders\n- [**eli5**](https://eli5.readthedocs.io/en/latest/)\n- matplotlib\n- numpy\n- pandas\n- scikit-learn\n- [**xgboost**](https://xgboost.readthedocs.io/en/latest/)",
"_____no_output_____"
]
],
[
[
"%%capture\nimport sys\n\n# If you're on Colab:\nif 'google.colab' in sys.modules:\n DATA_PATH = 'https://raw.githubusercontent.com/LambdaSchool/DS-Unit-2-Applied-Modeling/master/data/'\n !pip install category_encoders==2.*\n !pip install eli5\n\n# If you're working locally:\nelse:\n DATA_PATH = '../data/'",
"_____no_output_____"
]
],
[
[
"We'll go back to Tanzania Waterpumps for this lesson.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nfrom sklearn.model_selection import train_test_split\n\n# Merge train_features.csv & train_labels.csv\ntrain = pd.merge(pd.read_csv(DATA_PATH+'waterpumps/train_features.csv'), \n pd.read_csv(DATA_PATH+'waterpumps/train_labels.csv'))\n\n# Read test_features.csv & sample_submission.csv\ntest = pd.read_csv(DATA_PATH+'waterpumps/test_features.csv')\nsample_submission = pd.read_csv(DATA_PATH+'waterpumps/sample_submission.csv')\n\n\n# Split train into train & val\ntrain, val = train_test_split(train, train_size=0.80, test_size=0.20, \n stratify=train['status_group'], random_state=42)\n\n\ndef wrangle(X):\n \"\"\"Wrangle train, validate, and test sets in the same way\"\"\"\n \n # Prevent SettingWithCopyWarning\n X = X.copy()\n \n # About 3% of the time, latitude has small values near zero,\n # outside Tanzania, so we'll treat these values like zero.\n X['latitude'] = X['latitude'].replace(-2e-08, 0)\n \n # When columns have zeros and shouldn't, they are like null values.\n # So we will replace the zeros with nulls, and impute missing values later.\n # Also create a \"missing indicator\" column, because the fact that\n # values are missing may be a predictive signal.\n cols_with_zeros = ['longitude', 'latitude', 'construction_year', \n 'gps_height', 'population']\n for col in cols_with_zeros:\n X[col] = X[col].replace(0, np.nan)\n X[col+'_MISSING'] = X[col].isnull()\n \n # Drop duplicate columns\n duplicates = ['quantity_group', 'payment_type']\n X = X.drop(columns=duplicates)\n \n # Drop recorded_by (never varies) and id (always varies, random)\n unusable_variance = ['recorded_by', 'id']\n X = X.drop(columns=unusable_variance)\n \n # Convert date_recorded to datetime\n X['date_recorded'] = pd.to_datetime(X['date_recorded'], infer_datetime_format=True)\n \n # Extract components from date_recorded, then drop the original column\n X['year_recorded'] = X['date_recorded'].dt.year\n X['month_recorded'] = X['date_recorded'].dt.month\n X['day_recorded'] = X['date_recorded'].dt.day\n X = X.drop(columns='date_recorded')\n \n # Engineer feature: how many years from construction_year to date_recorded\n X['years'] = X['year_recorded'] - X['construction_year']\n X['years_MISSING'] = X['years'].isnull()\n \n # return the wrangled dataframe\n return X\n\ntrain = wrangle(train)\nval = wrangle(val)\ntest = wrangle(test)",
"_____no_output_____"
],
[
"# Arrange data into X features matrix and y target vector\ntarget = 'status_group'\nX_train = train.drop(columns=target)\ny_train = train[target]\nX_val = val.drop(columns=target)\ny_val = val[target]\nX_test = test",
"_____no_output_____"
],
[
"import category_encoders as ce\nfrom sklearn.impute import SimpleImputer\nfrom sklearn.ensemble import RandomForestClassifier\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.pipeline import make_pipeline\n\npipeline = make_pipeline(\n ce.OrdinalEncoder(), \n SimpleImputer(strategy='median'), \n RandomForestClassifier(n_estimators=100, random_state=42, n_jobs=-1)\n)\n\n# Fit on train, score on val\npipeline.fit(X_train, y_train)\nprint('Validation Accuracy', pipeline.score(X_val, y_val))",
"_____no_output_____"
]
],
[
[
"# Get permutation importances for model interpretation and feature selection",
"_____no_output_____"
],
[
"## Overview",
"_____no_output_____"
],
[
"Default Feature Importances are fast, but Permutation Importances may be more accurate.\n\nThese links go deeper with explanations and examples:\n\n- Permutation Importances\n - [Kaggle / Dan Becker: Machine Learning Explainability](https://www.kaggle.com/dansbecker/permutation-importance)\n - [Christoph Molnar: Interpretable Machine Learning](https://christophm.github.io/interpretable-ml-book/feature-importance.html)\n- (Default) Feature Importances\n - [Ando Saabas: Selecting good features, Part 3, Random Forests](https://blog.datadive.net/selecting-good-features-part-iii-random-forests/)\n - [Terence Parr, et al: Beware Default Random Forest Importances](https://explained.ai/rf-importance/index.html)",
"_____no_output_____"
],
[
"There are three types of feature importances:",
"_____no_output_____"
],
[
"### 1. (Default) Feature Importances\n\nFastest, good for first estimates, but be aware:\n\n\n\n>**When the dataset has two (or more) correlated features, then from the point of view of the model, any of these correlated features can be used as the predictor, with no concrete preference of one over the others.** But once one of them is used, the importance of others is significantly reduced since effectively the impurity they can remove is already removed by the first feature. As a consequence, they will have a lower reported importance. This is not an issue when we want to use feature selection to reduce overfitting, since it makes sense to remove features that are mostly duplicated by other features. But when interpreting the data, it can lead to the incorrect conclusion that one of the variables is a strong predictor while the others in the same group are unimportant, while actually they are very close in terms of their relationship with the response variable. — [Selecting good features – Part III: random forests](https://blog.datadive.net/selecting-good-features-part-iii-random-forests/) \n\n\n \n > **The scikit-learn Random Forest feature importance ... tends to inflate the importance of continuous or high-cardinality categorical variables.** ... Breiman and Cutler, the inventors of Random Forests, indicate that this method of “adding up the gini decreases for each individual variable over all trees in the forest gives a **fast** variable importance that is often very consistent with the permutation importance measure.” — [Beware Default Random Forest Importances](https://explained.ai/rf-importance/index.html)\n\n \n",
"_____no_output_____"
]
],
[
[
"# Get feature importances\nrf = pipeline.named_steps['randomforestclassifier']\nimportances = pd.Series(rf.feature_importances_, X_train.columns)\n\n# Plot feature importances\n%matplotlib inline\nimport matplotlib.pyplot as plt\n\nn = 20\nplt.figure(figsize=(10,n/2))\nplt.title(f'Top {n} features')\nimportances.sort_values()[-n:].plot.barh(color='grey');",
"_____no_output_____"
]
],
[
[
"### 2. Drop-Column Importance\n\nThe best in theory, but too slow in practice",
"_____no_output_____"
]
],
[
[
"column = 'quantity'\n\n# Fit without column\npipeline = make_pipeline(\n ce.OrdinalEncoder(), \n SimpleImputer(strategy='median'), \n RandomForestClassifier(n_estimators=100, random_state=42, n_jobs=-1)\n)\npipeline.fit(X_train.drop(columns=column), y_train)\nscore_without = pipeline.score(X_val.drop(columns=column), y_val)\nprint(f'Validation Accuracy without {column}: {score_without}')\n\n# Fit with column\npipeline = make_pipeline(\n ce.OrdinalEncoder(), \n SimpleImputer(strategy='median'), \n RandomForestClassifier(n_estimators=100, random_state=42, n_jobs=-1)\n)\npipeline.fit(X_train, y_train)\nscore_with = pipeline.score(X_val, y_val)\nprint(f'Validation Accuracy with {column}: {score_with}')\n\n# Compare the error with & without column\nprint(f'Drop-Column Importance for {column}: {score_with - score_without}')",
"_____no_output_____"
]
],
[
[
"### 3. Permutation Importance\n\nPermutation Importance is a good compromise between Feature Importance based on impurity reduction (which is the fastest) and Drop Column Importance (which is the \"best.\")\n\n[The ELI5 library documentation explains,](https://eli5.readthedocs.io/en/latest/blackbox/permutation_importance.html)\n\n> Importance can be measured by looking at how much the score (accuracy, F1, R^2, etc. - any score we’re interested in) decreases when a feature is not available.\n>\n> To do that one can remove feature from the dataset, re-train the estimator and check the score. But it requires re-training an estimator for each feature, which can be computationally intensive. ...\n>\n>To avoid re-training the estimator we can remove a feature only from the test part of the dataset, and compute score without using this feature. It doesn’t work as-is, because estimators expect feature to be present. So instead of removing a feature we can replace it with random noise - feature column is still there, but it no longer contains useful information. This method works if noise is drawn from the same distribution as original feature values (as otherwise estimator may fail). The simplest way to get such noise is to shuffle values for a feature, i.e. use other examples’ feature values - this is how permutation importance is computed.\n>\n>The method is most suitable for computing feature importances when a number of columns (features) is not huge; it can be resource-intensive otherwise.",
"_____no_output_____"
],
[
"### Do-It-Yourself way, for intuition",
"_____no_output_____"
],
[
"### With eli5 library\n\nFor more documentation on using this library, see:\n- [eli5.sklearn.PermutationImportance](https://eli5.readthedocs.io/en/latest/autodocs/sklearn.html#eli5.sklearn.permutation_importance.PermutationImportance)\n- [eli5.show_weights](https://eli5.readthedocs.io/en/latest/autodocs/eli5.html#eli5.show_weights)\n- [scikit-learn user guide, `scoring` parameter](https://scikit-learn.org/stable/modules/model_evaluation.html#the-scoring-parameter-defining-model-evaluation-rules)\n\neli5 doesn't work with pipelines.",
"_____no_output_____"
]
],
[
[
"# Ignore warnings\n",
"_____no_output_____"
]
],
[
[
"### We can use importances for feature selection\n\nFor example, we can remove features with zero importance. The model trains faster and the score does not decrease.",
"_____no_output_____"
],
[
"# Use xgboost for gradient boosting",
"_____no_output_____"
],
[
"## Overview",
"_____no_output_____"
],
[
"In the Random Forest lesson, you learned this advice:\n\n#### Try Tree Ensembles when you do machine learning with labeled, tabular data\n- \"Tree Ensembles\" means Random Forest or **Gradient Boosting** models. \n- [Tree Ensembles often have the best predictive accuracy](https://arxiv.org/abs/1708.05070) with labeled, tabular data.\n- Why? Because trees can fit non-linear, non-[monotonic](https://en.wikipedia.org/wiki/Monotonic_function) relationships, and [interactions](https://christophm.github.io/interpretable-ml-book/interaction.html) between features.\n- A single decision tree, grown to unlimited depth, will [overfit](http://www.r2d3.us/visual-intro-to-machine-learning-part-1/). We solve this problem by ensembling trees, with bagging (Random Forest) or **[boosting](https://www.youtube.com/watch?v=GM3CDQfQ4sw)** (Gradient Boosting).\n- Random Forest's advantage: may be less sensitive to hyperparameters. **Gradient Boosting's advantage:** may get better predictive accuracy.",
"_____no_output_____"
],
[
"Like Random Forest, Gradient Boosting uses ensembles of trees. But the details of the ensembling technique are different:\n\n### Understand the difference between boosting & bagging\n\nBoosting (used by Gradient Boosting) is different than Bagging (used by Random Forests). \n\nHere's an excerpt from [_An Introduction to Statistical Learning_](http://www-bcf.usc.edu/~gareth/ISL/ISLR%20Seventh%20Printing.pdf) Chapter 8.2.3, Boosting:\n\n>Recall that bagging involves creating multiple copies of the original training data set using the bootstrap, fitting a separate decision tree to each copy, and then combining all of the trees in order to create a single predictive model.\n>\n>**Boosting works in a similar way, except that the trees are grown _sequentially_: each tree is grown using information from previously grown trees.**\n>\n>Unlike fitting a single large decision tree to the data, which amounts to _fitting the data hard_ and potentially overfitting, the boosting approach instead _learns slowly._ Given the current model, we fit a decision tree to the residuals from the model.\n>\n>We then add this new decision tree into the fitted function in order to update the residuals. Each of these trees can be rather small, with just a few terminal nodes. **By fitting small trees to the residuals, we slowly improve fˆ in areas where it does not perform well.**\n>\n>Note that in boosting, unlike in bagging, the construction of each tree depends strongly on the trees that have already been grown.\n\nThis high-level overview is all you need to know for now. If you want to go deeper, we recommend you watch the StatQuest videos on gradient boosting!",
"_____no_output_____"
],
[
"Let's write some code. We have lots of options for which libraries to use:\n\n#### Python libraries for Gradient Boosting\n- [scikit-learn Gradient Tree Boosting](https://scikit-learn.org/stable/modules/ensemble.html#gradient-boosting) — slower than other libraries, but [the new version may be better](https://twitter.com/amuellerml/status/1129443826945396737)\n - Anaconda: already installed\n - Google Colab: already installed\n- [xgboost](https://xgboost.readthedocs.io/en/latest/) — can accept missing values and enforce [monotonic constraints](https://xiaoxiaowang87.github.io/monotonicity_constraint/)\n - Anaconda, Mac/Linux: `conda install -c conda-forge xgboost`\n - Windows: `conda install -c anaconda py-xgboost`\n - Google Colab: already installed\n- [LightGBM](https://lightgbm.readthedocs.io/en/latest/) — can accept missing values and enforce [monotonic constraints](https://blog.datadive.net/monotonicity-constraints-in-machine-learning/)\n - Anaconda: `conda install -c conda-forge lightgbm`\n - Google Colab: already installed\n- [CatBoost](https://catboost.ai/) — can accept missing values and use [categorical features](https://catboost.ai/docs/concepts/algorithm-main-stages_cat-to-numberic.html) without preprocessing\n - Anaconda: `conda install -c conda-forge catboost`\n - Google Colab: `pip install catboost`",
"_____no_output_____"
],
[
"In this lesson, you'll use a new library, xgboost — But it has an API that's almost the same as scikit-learn, so it won't be a hard adjustment!\n\n#### [XGBoost Python API Reference: Scikit-Learn API](https://xgboost.readthedocs.io/en/latest/python/python_api.html#module-xgboost.sklearn)",
"_____no_output_____"
],
[
"#### [Avoid Overfitting By Early Stopping With XGBoost In Python](https://machinelearningmastery.com/avoid-overfitting-by-early-stopping-with-xgboost-in-python/)\n\nWhy is early stopping better than a For loop, or GridSearchCV, to optimize `n_estimators`?\n\nWith early stopping, if `n_iterations` is our number of iterations, then we fit `n_iterations` decision trees.\n\nWith a for loop, or GridSearchCV, we'd fit `sum(range(1,n_rounds+1))` trees.\n\nBut it doesn't work well with pipelines. You may need to re-run multiple times with different values of other parameters such as `max_depth` and `learning_rate`.\n\n#### XGBoost parameters\n- [Notes on parameter tuning](https://xgboost.readthedocs.io/en/latest/tutorials/param_tuning.html)\n- [Parameters documentation](https://xgboost.readthedocs.io/en/latest/parameter.html)\n",
"_____no_output_____"
],
[
"### Try adjusting these hyperparameters\n\n#### Random Forest\n- class_weight (for imbalanced classes)\n- max_depth (usually high, can try decreasing)\n- n_estimators (too low underfits, too high wastes time)\n- min_samples_leaf (increase if overfitting)\n- max_features (decrease for more diverse trees)\n\n#### Xgboost\n- scale_pos_weight (for imbalanced classes)\n- max_depth (usually low, can try increasing)\n- n_estimators (too low underfits, too high wastes time/overfits) — Use Early Stopping!\n- learning_rate (too low underfits, too high overfits)\n\nFor more ideas, see [Notes on Parameter Tuning](https://xgboost.readthedocs.io/en/latest/tutorials/param_tuning.html) and [DART booster](https://xgboost.readthedocs.io/en/latest/tutorials/dart.html).",
"_____no_output_____"
],
[
"## Challenge\n\nYou will use your portfolio project dataset for all assignments this sprint. Complete these tasks for your project, and document your work.\n\n- Continue to clean and explore your data. Make exploratory visualizations.\n- Fit a model. Does it beat your baseline?\n- Try xgboost.\n- Get your model's permutation importances.\n\nYou should try to complete an initial model today, because the rest of the week, we're making model interpretation visualizations.\n\nBut, if you aren't ready to try xgboost and permutation importances with your dataset today, you can practice with another dataset instead. You may choose any dataset you've worked with previously.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
cb88909679477ac9214b41529a8e851143d3e529
| 27,556 |
ipynb
|
Jupyter Notebook
|
sphinx/datascience/source/partial-correlation.ipynb
|
oneoffcoder/books
|
84619477294a3e37e0d7538adf819113c9e8dcb8
|
[
"CC-BY-4.0"
] | 26 |
2020-05-05T08:07:43.000Z
|
2022-02-12T03:28:15.000Z
|
sphinx/datascience/source/partial-correlation.ipynb
|
oneoffcoder/books
|
84619477294a3e37e0d7538adf819113c9e8dcb8
|
[
"CC-BY-4.0"
] | 19 |
2021-03-10T00:33:51.000Z
|
2022-03-02T13:04:32.000Z
|
sphinx/datascience/source/partial-correlation.ipynb
|
oneoffcoder/books
|
84619477294a3e37e0d7538adf819113c9e8dcb8
|
[
"CC-BY-4.0"
] | 2 |
2022-01-09T16:48:21.000Z
|
2022-02-19T17:06:50.000Z
| 62.913242 | 13,652 | 0.756568 |
[
[
[
"# Partial Correlation\n\nThe purpose of this notebook is to understand how to compute the [partial correlation](https://en.wikipedia.org/wiki/Partial_correlation) between two variables, $X$ and $Y$, given a third $Z$. In particular, these variables are assumed to be guassians (or, in general, multivariate gaussians). \n\nWhy is it important to estimate partial correlations? The primary reason for estimating a partial correlation is to use it to detect for [confounding](https://en.wikipedia.org/wiki/Confounding_variable) variables during causal analysis. \n\n## Simulation\n\nLet's start out by simulating 3 data sets. Graphically, these data sets comes from graphs represented by the following.\n\n* $X \\rightarrow Z \\rightarrow Y$ (serial)\n* $X \\leftarrow Z \\rightarrow Y$ (diverging)\n* $X \\rightarrow Z \\leftarrow Y$ (converging)",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nimport matplotlib.pyplot as plt\nimport networkx as nx\nimport warnings\n\nwarnings.filterwarnings('ignore')\nplt.style.use('ggplot')\n\ndef get_serial_graph():\n g = nx.DiGraph()\n \n g.add_node('X')\n g.add_node('Y')\n g.add_node('Z')\n \n g.add_edge('X', 'Z')\n g.add_edge('Z', 'Y')\n \n return g\n\ndef get_diverging_graph():\n g = nx.DiGraph()\n \n g.add_node('X')\n g.add_node('Y')\n g.add_node('Z')\n \n g.add_edge('Z', 'X')\n g.add_edge('Z', 'Y')\n \n return g\n\ndef get_converging_graph():\n g = nx.DiGraph()\n \n g.add_node('X')\n g.add_node('Y')\n g.add_node('Z')\n \n g.add_edge('X', 'Z')\n g.add_edge('Y', 'Z')\n \n return g\n\ng_serial = get_serial_graph()\ng_diverging = get_diverging_graph()\ng_converging = get_converging_graph()\n\np_serial = nx.nx_agraph.graphviz_layout(g_serial, prog='dot', args='-Kcirco')\np_diverging = nx.nx_agraph.graphviz_layout(g_diverging, prog='dot', args='-Kcirco')\np_converging = nx.nx_agraph.graphviz_layout(g_converging, prog='dot', args='-Kcirco')\n\nfig, ax = plt.subplots(3, 1, figsize=(5, 5))\n\nnx.draw(g_serial, pos=p_serial, with_labels=True, node_color='#e0e0e0', node_size=800, arrowsize=20, ax=ax[0])\nnx.draw(g_diverging, pos=p_diverging, with_labels=True, node_color='#e0e0e0', node_size=800, arrowsize=20, ax=ax[1])\nnx.draw(g_converging, pos=p_converging, with_labels=True, node_color='#e0e0e0', node_size=800, arrowsize=20, ax=ax[2])\n\nax[0].set_title('Serial')\nax[1].set_title('Diverging')\nax[2].set_title('Converging')\n\nplt.tight_layout()",
"_____no_output_____"
]
],
[
[
"In the serial graph, `X` causes `Z` and `Z` causes `Y`. In the diverging graph, `Z` causes both `X` and `Y`. In the converging graph, `X` and `Y` cause `Z`. Below, the serial, diverging, and converging data sets are named S, D, and C, correspondingly.\n\nNote that in the serial graph, the data is sampled as follows.\n\n* $X \\sim \\mathcal{N}(0, 1)$\n* $Z \\sim 2 + 1.8 \\times X$\n* $Y \\sim 5 + 2.7 \\times Z$\n\nIn the diverging graph, the data is sampled as follows.\n\n* $Z \\sim \\mathcal{N}(0, 1)$\n* $X \\sim 4.3 + 3.3 \\times Z$\n* $Y \\sim 5.0 + 2.7 \\times Z$\n\nLastly, in the converging graph, the data is sampled as follows.\n\n* $X \\sim \\mathcal{N}(0, 1)$\n* $Y \\sim \\mathcal{N}(5.5, 1)$\n* $Z \\sim 2.0 + 0.8 \\times X + 1.2 \\times Y$\n\nNote the ordering of the sampling with the variables follows the structure of the corresponding graph.",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\nnp.random.seed(37)\n\ndef get_error(N=10000, mu=0.0, std=0.2):\n return np.random.normal(mu, std, N)\n\ndef to_matrix(X, Z, Y):\n return np.concatenate([\n X.reshape(-1, 1), \n Z.reshape(-1, 1), \n Y.reshape(-1, 1)], axis=1)\n\ndef get_serial(N=10000, e_mu=0.0, e_std=0.2):\n X = np.random.normal(0, 1, N) + get_error(N, e_mu, e_std)\n Z = 2 + 1.8 * X + get_error(N, e_mu, e_std)\n Y = 5 + 2.7 * Z + get_error(N, e_mu, e_std)\n\n return to_matrix(X, Z, Y)\n\ndef get_diverging(N=10000, e_mu=0.0, e_std=0.2):\n Z = np.random.normal(0, 1, N) + get_error(N, e_mu, e_std)\n X = 4.3 + 3.3 * Z + get_error(N, e_mu, e_std)\n Y = 5 + 2.7 * Z + get_error(N, e_mu, e_std)\n\n return to_matrix(X, Z, Y)\n\ndef get_converging(N=10000, e_mu=0.0, e_std=0.2):\n X = np.random.normal(0, 1, N) + get_error(N, e_mu, e_std)\n Y = np.random.normal(5.5, 1, N) + get_error(N, e_mu, e_std)\n Z = 2 + 0.8 * X + 1.2 * Y + get_error(N, e_mu, e_std)\n \n return to_matrix(X, Z, Y)",
"_____no_output_____"
],
[
"S = get_serial()\nD = get_diverging()\nC = get_converging()",
"_____no_output_____"
]
],
[
[
"## Computation\n\nFor the three datasets, `S`, `D`, and `C`, we want to compute the partial correlation between $X$ and $Y$ given $Z$. The way to do this is as follows.\n\n* Regress $X$ on $Z$ and also $Y$ on $Z$\n * $X = b_X + w_X * Z$\n * $Y = b_Y + w_Y * Z$\n* With the new weights $(b_X, w_X)$ and $(b_Y, w_Y)$, predict $X$ and $Y$.\n * $\\hat{X} = b_X + w_X * Z$\n * $\\hat{Y} = b_Y + w_Y * Z$\n* Now compute the residuals between the true and predicted values.\n * $R_X = X - \\hat{X}$\n * $R_Y = Y - \\hat{Y}$\n* Finally, compute the Pearson correlation between $R_X$ and $R_Y$.\n\nThe correlation between the residuals is the partial correlation and runs from -1 to +1. More interesting is the test of significance. If $p > \\alpha$, where $\\alpha \\in [0.1, 0.05, 0.01]$, then assume independence. For example, assume $\\alpha = 0.01$ and $p = 0.002$, then $X$ is conditionally independent of $Y$ given $Z$.",
"_____no_output_____"
]
],
[
[
"from sklearn.linear_model import LinearRegression\nfrom sklearn.metrics import r2_score\nfrom scipy.stats import pearsonr\nfrom scipy import stats\n\ndef get_cond_indep_test(c_xy_z, N=10000, alpha=0.01):\n point = stats.norm.ppf(1 - (alpha / 2.0))\n z_transform = np.sqrt(N - 3) * np.abs(0.5 * np.log((1 + c_xy_z) / (1 - c_xy_z)))\n return z_transform, point, z_transform > point\n\ndef get_partial_corr(M):\n X = M[:, 0]\n Z = M[:, 1].reshape(-1, 1)\n Y = M[:, 2]\n\n mXZ = LinearRegression()\n mXZ.fit(Z, X)\n pXZ = mXZ.predict(Z)\n rXZ = X - pXZ\n\n mYZ = LinearRegression()\n mYZ.fit(Z, Y)\n pYZ = mYZ.predict(Z)\n rYZ = Y - pYZ\n\n c_xy, p_xy = pearsonr(X, Y)\n c_xy_z, p_xy_z = pearsonr(rXZ, rYZ)\n\n return c_xy, p_xy, c_xy_z, p_xy_z",
"_____no_output_____"
]
],
[
[
"## Serial graph data\n\nFor $X \\rightarrow Z \\rightarrow Y$, note that the marginal correlation is high (0.99) and the correlation is significant (p < 0.01). However, the correlation between X and Y vanishes given Z to -0.01 (p > 0.01). Note the conditional independence test fails to reject the null hypothesis.",
"_____no_output_____"
]
],
[
[
"c_xy, p_xy, c_xy_z, p_xy_z = get_partial_corr(S)\nprint(f'corr_xy={c_xy:.5f}, p_xy={p_xy:.5f}')\nprint(f'corr_xy_z={c_xy_z:.5f}, p_xy_z={p_xy_z:.5f}')\nprint(get_cond_indep_test(c_xy_z))",
"corr_xy=0.99331, p_xy=0.00000\ncorr_xy_z=-0.01493, p_xy_z=0.13543\n(1.4930316470699307, 2.5758293035489004, False)\n"
]
],
[
[
"## Diverging graph data\n\nFor $X \\leftarrow Z \\rightarrow Y$, note that the marginal correlation is high (0.99) and the correlation is significant (p < 0.01). However, the correlation between X and Y vanishes given Z to 0.01 (p > 0.01). Note the conditional independence test fails to reject the null hypothesis.",
"_____no_output_____"
]
],
[
[
"c_xy, p_xy, c_xy_z, p_xy_z = get_partial_corr(D)\nprint(f'corr_xy={c_xy:.5f}, p_xy={p_xy:.5f}')\nprint(f'corr_xy_z={c_xy_z:.5f}, p_xy_z={p_xy_z:.5f}')\nprint(get_cond_indep_test(c_xy_z))",
"corr_xy=0.99575, p_xy=0.00000\ncorr_xy_z=0.01155, p_xy_z=0.24815\n(1.1548311182263977, 2.5758293035489004, False)\n"
]
],
[
[
"## Converging graph data\n\nFor $X \\rightarrow Z \\leftarrow Y$, note that the correlation is low (-0.00) and the correlation is insignficiant (p > 0.01). However, the correlation between X and Y increases to -0.96 and becomes significant (p < 0.01)! Note the conditional independence test rejects the null hypothesis.",
"_____no_output_____"
]
],
[
[
"c_xy, p_xy, c_xy_z, p_xy_z = get_partial_corr(C)\nprint(f'corr_xy={c_xy:.5f}, p_xy={p_xy:.5f}')\nprint(f'corr_xy_z={c_xy_z:.5f}, p_xy_z={p_xy_z:.5f}')\nprint(get_cond_indep_test(c_xy_z))",
"corr_xy=-0.00269, p_xy=0.78774\ncorr_xy_z=-0.95791, p_xy_z=0.00000\n(191.9601051372688, 2.5758293035489004, True)\n"
]
],
[
[
"## Statistically Distinguishable\n\nThe `serial` and `diverging` graphs are said to be `statistically indistingishable` since $X$ and $Y$ are both `conditionally independent` given $Z$. However, the `converging` graph is `statistically distinguishable` since it is the only graph where $X$ and $Y$ are `conditionally dependent` given $Z$.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
cb889197c694caf619c27a0197395e5382278e87
| 3,153 |
ipynb
|
Jupyter Notebook
|
inclass/inclass3-abpwrs/check_env.ipynb
|
abpwrs/ece-5490-sp19
|
d7b6a0867eda6cf3a93f78673a6fc264175caf06
|
[
"MIT"
] | 13 |
2019-03-17T04:34:21.000Z
|
2022-02-06T06:58:19.000Z
|
inclass/inclass3-abpwrs/check_env.ipynb
|
abpwrs/ece-5490-sp19
|
d7b6a0867eda6cf3a93f78673a6fc264175caf06
|
[
"MIT"
] | 27 |
2019-03-01T14:39:38.000Z
|
2019-07-30T04:44:48.000Z
|
inclass/inclass3-abpwrs/check_env.ipynb
|
abpwrs/ece-5490-sp19
|
d7b6a0867eda6cf3a93f78673a6fc264175caf06
|
[
"MIT"
] | 7 |
2019-03-01T19:14:05.000Z
|
2020-11-05T09:43:20.000Z
| 32.173469 | 88 | 0.47764 |
[
[
[
"from __future__ import print_function\nfrom distutils.version import LooseVersion as Version\nimport sys\n\n\ntry:\n import curses\n curses.setupterm()\n assert curses.tigetnum(\"colors\") > 2\n OK = \"\\x1b[1;%dm[ OK ]\\x1b[0m\" % (30 + curses.COLOR_GREEN)\n FAIL = \"\\x1b[1;%dm[FAIL]\\x1b[0m\" % (30 + curses.COLOR_RED)\nexcept:\n OK = '[ OK ]'\n FAIL = '[FAIL]'\n\ntry:\n import importlib\nexcept ImportError:\n print(FAIL, \"Python version 3.5 (or 2.7) is required,\"\n \" but %s is installed.\" % sys.version)\n\n \ndef import_version(pkg, min_ver, fail_msg=\"\"):\n mod = None\n try:\n mod = importlib.import_module(pkg)\n if pkg in {'itk'}:\n ver = mod.Version.GetITKVersion()\n else:\n ver = mod.__version__\n if Version(ver) < min_ver:\n print(FAIL, \"%s version %s or higher required, but %s installed.\"\n % (lib, min_ver, ver))\n else:\n print(OK, '%s version %s' % (pkg, ver))\n except ImportError:\n print(FAIL, '%s not installed. %s' % (pkg, fail_msg))\n return mod\n\n\n# first check the python version\nprint('Using python in', sys.prefix)\nprint(sys.version)\npyversion = Version(sys.version)\nif pyversion >= \"3\":\n if pyversion < \"3.5\":\n print(FAIL, \"Python version 3.5 (or 2.7) is required,\"\n \" but %s is installed.\" % sys.version)\nelif pyversion >= \"2\":\n if pyversion < \"2.7\":\n print(FAIL, \"Python version 2.7 is required,\"\n \" but %s is installed.\" % sys.version)\nelse:\n print(FAIL, \"Unknown Python version: %s\" % sys.version)\n\nprint()\nrequirements = {'numpy': \"1.6.1\", 'scipy': \"0.9\", 'matplotlib': \"2.0\",\n 'IPython': \"3.0\", 'sklearn': \"0.20\", 'itk': \"5.0\",\n 'itkwidgets': \"0.16\", 'ipywidgets': '7.4', 'notebook': '5.4'}\n\n# now the dependencies\nfor lib, required_version in list(requirements.items()):\n import_version(lib, required_version)",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code"
]
] |
cb889551a781a3eee79455bd2b38233efb881afd
| 594,739 |
ipynb
|
Jupyter Notebook
|
.ipynb_checkpoints/Nanodegree Project-checkpoint.ipynb
|
lokesh051/Udacity-Starbuck-Capstone-Project
|
85d8f69677eed03104c2e5f94ea62b8dd343cdee
|
[
"Apache-2.0"
] | null | null | null |
.ipynb_checkpoints/Nanodegree Project-checkpoint.ipynb
|
lokesh051/Udacity-Starbuck-Capstone-Project
|
85d8f69677eed03104c2e5f94ea62b8dd343cdee
|
[
"Apache-2.0"
] | null | null | null |
.ipynb_checkpoints/Nanodegree Project-checkpoint.ipynb
|
lokesh051/Udacity-Starbuck-Capstone-Project
|
85d8f69677eed03104c2e5f94ea62b8dd343cdee
|
[
"Apache-2.0"
] | null | null | null | 89.006136 | 183,072 | 0.738704 |
[
[
[
"# Starbucks Capstone Challenge\n\n### Introduction\n\nThis data set contains simulated data that mimics customer behavior on the Starbucks rewards mobile app. Once every few days, Starbucks sends out an offer to users of the mobile app. An offer can be merely an advertisement for a drink or an actual offer such as a discount or BOGO (buy one get one free). Some users might not receive any offer during certain weeks. \n\nNot all users receive the same offer, and that is the challenge to solve with this data set.\n\nYour task is to combine transaction, demographic and offer data to determine which demographic groups respond best to which offer type. This data set is a simplified version of the real Starbucks app because the underlying simulator only has one product whereas Starbucks actually sells dozens of products.\n\nEvery offer has a validity period before the offer expires. As an example, a BOGO offer might be valid for only 5 days. You'll see in the data set that informational offers have a validity period even though these ads are merely providing information about a product; for example, if an informational offer has 7 days of validity, you can assume the customer is feeling the influence of the offer for 7 days after receiving the advertisement.\n\nYou'll be given transactional data showing user purchases made on the app including the timestamp of purchase and the amount of money spent on a purchase. This transactional data also has a record for each offer that a user receives as well as a record for when a user actually views the offer. There are also records for when a user completes an offer. \n\nKeep in mind as well that someone using the app might make a purchase through the app without having received an offer or seen an offer.\n\n### Example\n\nTo give an example, a user could receive a discount offer buy 10 dollars get 2 off on Monday. The offer is valid for 10 days from receipt. If the customer accumulates at least 10 dollars in purchases during the validity period, the customer completes the offer.\n\nHowever, there are a few things to watch out for in this data set. Customers do not opt into the offers that they receive; in other words, a user can receive an offer, never actually view the offer, and still complete the offer. For example, a user might receive the \"buy 10 dollars get 2 dollars off offer\", but the user never opens the offer during the 10 day validity period. The customer spends 15 dollars during those ten days. There will be an offer completion record in the data set; however, the customer was not influenced by the offer because the customer never viewed the offer.\n\n### Cleaning\n\nThis makes data cleaning especially important and tricky.\n\nYou'll also want to take into account that some demographic groups will make purchases even if they don't receive an offer. From a business perspective, if a customer is going to make a 10 dollar purchase without an offer anyway, you wouldn't want to send a buy 10 dollars get 2 dollars off offer. You'll want to try to assess what a certain demographic group will buy when not receiving any offers.\n\n### Final Advice\n\nBecause this is a capstone project, you are free to analyze the data any way you see fit. For example, you could build a machine learning model that predicts how much someone will spend based on demographics and offer type. Or you could build a model that predicts whether or not someone will respond to an offer. Or, you don't need to build a machine learning model at all. You could develop a set of heuristics that determine what offer you should send to each customer (i.e., 75 percent of women customers who were 35 years old responded to offer A vs 40 percent from the same demographic to offer B, so send offer A).",
"_____no_output_____"
],
[
"# Data Sets\n\nThe data is contained in three files:\n\n* portfolio.json - containing offer ids and meta data about each offer (duration, type, etc.)\n* profile.json - demographic data for each customer\n* transcript.json - records for transactions, offers received, offers viewed, and offers completed\n\nHere is the schema and explanation of each variable in the files:\n\n**portfolio.json**\n* id (string) - offer id\n* offer_type (string) - type of offer ie BOGO, discount, informational\n* difficulty (int) - minimum required spend to complete an offer\n* reward (int) - reward given for completing an offer\n* duration (int) - time for offer to be open, in days\n* channels (list of strings)\n\n**profile.json**\n* age (int) - age of the customer \n* became_member_on (int) - date when customer created an app account\n* gender (str) - gender of the customer (note some entries contain 'O' for other rather than M or F)\n* id (str) - customer id\n* income (float) - customer's income\n\n**transcript.json**\n* event (str) - record description (ie transaction, offer received, offer viewed, etc.)\n* person (str) - customer id\n* time (int) - time in hours since start of test. The data begins at time t=0\n* value - (dict of strings) - either an offer id or transaction amount depending on the record\n\n**Note:** If you are using the workspace, you will need to go to the terminal and run the command `conda update pandas` before reading in the files. This is because the version of pandas in the workspace cannot read in the transcript.json file correctly, but the newest version of pandas can. You can access the termnal from the orange icon in the top left of this notebook. \n\nYou can see how to access the terminal and how the install works using the two images below. First you need to access the terminal:\n\n<img src=\"pic1.png\"/>\n\nThen you will want to run the above command:\n\n<img src=\"pic2.png\"/>\n\nFinally, when you enter back into the notebook (use the jupyter icon again), you should be able to run the below cell without any errors.",
"_____no_output_____"
],
[
"## Problem Statement",
"_____no_output_____"
],
[
"In this project I will determine how likely is a customer to complete an offer. The end goal is:\n1. To determine, does sending more offers lead to a higher completion rate.\n2. Customers with lower completion rate should be sent offers or not",
"_____no_output_____"
],
[
"## Exploratory Data Analysis",
"_____no_output_____"
],
[
"### Read Data Files",
"_____no_output_____"
]
],
[
[
"#######Run This\nimport pandas as pd\nimport numpy as np\nimport math\nimport json\nimport os\n%matplotlib inline\n\nportfolio = pd.read_json('data/portfolio.json', orient='records', lines=True)\nprofile = pd.read_json('data/profile.json', orient='records', lines=True)\ntranscript = pd.read_json('data/transcript.json', orient='records', lines=True)",
"_____no_output_____"
]
],
[
[
"### Save The Data Files ",
"_____no_output_____"
]
],
[
[
"if not os.path.isdir('explore'):\n os.makedirs('explore')\n \ndef data_info(data, filename):\n path = os.path.join('explore', filename)\n if not os.path.isfile(path):\n pd.DataFrame(data).to_csv(path)\n print(data.shape)\n\ndata_info(portfolio, 'portfolio.csv')\ndata_info(profile, 'profile.csv')\ndata_info(transcript, 'transcript.csv')",
"(10, 6)\n(17000, 5)\n(306534, 4)\n"
]
],
[
[
"### Clean Portfolio\nBy looking at the portfolio file we can see that the channels column in grouped, so we'll use sklearn's MultiLabelBinarizer to unpack the channel column and then remove it from the DataFrame",
"_____no_output_____"
]
],
[
[
"from sklearn.preprocessing import MultiLabelBinarizer\n\ncleaned_portfolio = portfolio.copy()\ncleaned_portfolio.rename(columns={'id':'offer_id'}, inplace=True)\n\ns = cleaned_portfolio['channels']\nmlb = MultiLabelBinarizer()\nchannels = pd.DataFrame(mlb.fit_transform(s),columns=mlb.classes_, index=cleaned_portfolio.index)\n\ncleaned_portfolio = cleaned_portfolio.join(channels)\ncleaned_portfolio.drop(['channels'], axis=1, inplace=True)\ncleaned_portfolio\n",
"_____no_output_____"
]
],
[
[
"### Clean Profile\nBy looking at the profile data we can see that there are missing age values, we also observe that the people with missing age values have missing income values. Therefore for now we'll remove all the rows with NaN values(~2000), deduce inference and then later combine with missing values and compare results\n",
"_____no_output_____"
]
],
[
[
"#profile['became_member_on'] = pd.to_datetime(profile['became_member_on'], format='%Y%m%d')\nprofile.rename(columns={\"id\":\"person\"}, inplace=True)\nundefined_group = None\ncleaned_profile = None\n\n#cleaning profile and dividing it into cleaned_profile and undefined_group\nundefined_group = profile.copy()\nundefined_group['gender'] = undefined_group['gender'].fillna('U')\nundefined_group = undefined_group.loc[undefined_group['gender'] == 'U'].reset_index(drop=True)\ncleaned_profile = profile.dropna().reset_index(drop=True)\ncleaned_profile",
"_____no_output_____"
]
],
[
[
"### Clean Transcript\nFrom the transcript we can see that the column values has 2 values i.e it has an offer id or amount spent for that transaction, we'll split the value in 2 columns offer_id, amount and then drop the value column",
"_____no_output_____"
]
],
[
[
"cleaned_transcript = transcript.copy()\nvalue = cleaned_transcript['value']\ncleaned_transcript['amount'] = [int(i['amount']) if i.get('amount') else 0 for i in value]\n\ncleaned_transcript['offer_id'] = [i['offer_id'] if i.get('offer_id') else (i['offer id'] if i.get('offer id') else '0') for i in value]\n\ncleaned_transcript.drop(['value'], axis=1, inplace=True)\n\n#drop the profile which have no gender or income\ncleaned_transcript = cleaned_transcript[~cleaned_transcript.person.isin(undefined_group.person)]\n\nsort_df = cleaned_transcript.sort_values(by=['person', 'time'])\nsort_df",
"_____no_output_____"
]
],
[
[
"### Get Data\n\nmethod: get_valid_data\n\nparams: df {df is the set of all the events for a person, lets say offer received, viewed or completed}\n\n---- The idea is to parse a set of transaction entries for a person and then divide it into offer received, offer viewed and offer completed\n\n---- Then create a new column ['g'] which stores the cumalative count for every entry lets say offer id 'a' offered twice then the corresponding g column will store the count something like this:\n\n offer_id g\n a 0\n a 1\nThe idea behind g is that it will help us merge elements on [person, offer_id] and will prevent duplicates",
"_____no_output_____"
]
],
[
[
"def get_valid_data(df):\n \n offer_received = df.loc[df['event'] == 'offer received'].reset_index(drop=True)\n offer_viewed = df.loc[df['event'] == 'offer viewed'].reset_index(drop=True)\n offer_completed = df.loc[df['event'] == 'offer completed'].reset_index(drop=True)\n\n\n offer_received['g'] = offer_received.groupby('offer_id').cumcount()\n offer_viewed['g'] = offer_viewed.groupby('offer_id').cumcount()\n offer_completed['g'] = offer_completed.groupby('offer_id').cumcount()\n \n\n res = pd.merge(offer_received, offer_viewed, on=['person', 'offer_id', 'g'], how='outer')\n res = pd.merge(res, offer_completed, on=['person', 'offer_id', 'g'], how='outer')\n\n \n return res\n\noffers_completed = sort_df.groupby('person').apply(lambda x: get_valid_data(x))\noffers_completed = offers_completed.dropna()\noffers_completed = offers_completed.reset_index(drop=True)\noffers_completed",
"_____no_output_____"
]
],
[
[
"### Combine Portfolio with the offers completed for every entry \n\nmethod: valid_offer_completed\nparameter: df {offers completed}, cleaned_portfolio {cleaned_portfolio- information about every customer like age, income}\n\n##### Functions\n1. Drop columns like amount_x, amount_y since they only have value 0 and theh drop event like offer received etc\n2. Merge cleaned_portfolio[offer_type, duration] to df on offer_id \n3. Drop the columns where a user have completed and offer before and viewed it later i.e keep only those where time_y <= time ",
"_____no_output_____"
]
],
[
[
"def valid_offer_completed(df, cleaned_portfolio):\n df = df.rename(columns={\"offer_id_x\":\"offer_id\"})\n offers = cleaned_portfolio[['offer_id', 'offer_type', 'duration']]\n df = df.merge(offers,how='left', on='offer_id')\n \n \n df = df.drop(['amount_x', 'amount_y', 'amount', 'event_x', 'event_y', 'event', 'g'], axis=1).reset_index(drop=True)\n df = df[['person','offer_id','time_x','time_y', 'time', 'offer_type', 'duration']]\n \n df = df[(df.time_x <= df.time_y) & (df.time_y <= df.time)]\n return df\n\nvalid = valid_offer_completed(offers_completed, cleaned_portfolio)\nvalid = valid.reset_index(drop=True)\nvalid",
"_____no_output_____"
]
],
[
[
"### Find Informational Offers\n\nInformational offers do not have any offer completed record so we need to find the offer_completed time because we need to combine then with the valid dataframe later on\n\nso we'll caluate the offfer completed based on the duration of the information offer ",
"_____no_output_____"
]
],
[
[
"def info_offer(df):\n\n offer_received = df.loc[df['event'] == 'offer received'].reset_index(drop=True)\n offer_viewed = df.loc[df['event'] == 'offer viewed'].reset_index(drop=True)\n\n\n offer_received['g'] = offer_received.groupby('offer_id').cumcount()\n offer_viewed['g'] = offer_viewed.groupby('offer_id').cumcount()\n \n\n res = pd.merge(offer_received, offer_viewed, on=['person', 'offer_id', 'g'], how='outer')\n \n offers = cleaned_portfolio[['offer_id', 'offer_type', 'duration']]\n res = res.merge(offers,how='left', on='offer_id')\n \n res['time'] = res['time_x'] + res['duration'] * 24\n res = res.dropna()\n \n res = res[res.time_x <= res.time_y]\n res['response'] = np.where(res.time_y > res.time , 0, 1)\n res = res.loc[res.response == 1]\n \n \n res = res.drop(['response', 'amount_x', 'amount_y', 'event_x', 'event_y', 'g'], axis=1).reset_index(drop=True)\n res = res[['person','offer_id','time_x','time_y', 'time', 'offer_type', 'duration']]\n \n \n \n return res\n\ninfo_df = sort_df[sort_df['offer_id'].isin(['3f207df678b143eea3cee63160fa8bed', '5a8bc65990b245e5a138643cd4eb9837'])]\ninfo_data = info_df.groupby('person').apply(lambda x: info_offer(x))\ninfo_data =info_data.reset_index(drop=True)\ninfo_data",
"_____no_output_____"
]
],
[
[
"### Combine the valid and information dataframes",
"_____no_output_____"
]
],
[
[
"complete = pd.concat([valid, info_data], ignore_index=True, sort=False)\ncomplete",
"_____no_output_____"
]
],
[
[
"### Fill Profile \nmethod: fill_profile\nparams: gd {Grouped data is the grouped data which includes all the transction record per person}, df {df is the customer portfolio}\n\n1. Find the number of valid offers completed\n2. Append the total offers completed for every person in th customer portfolio",
"_____no_output_____"
]
],
[
[
"df = None\ndef fill_profile(gd, df):\n grouped_data = gd.groupby(['person'])\n \n invalid = []\n for index, row in df.iterrows():\n if row['person'] in grouped_data.groups.keys():\n offers = grouped_data.get_group(row['person'])['offer_type'].value_counts()\n df.at[index, 'offers completed'] = offers.sum()\n \n for offer, count in offers.items():\n df.at[index, offer] = count\n \n else:\n invalid.append(row['person'])\n print(len(invalid))\n df = df.fillna(0)\n return df",
"_____no_output_____"
],
[
"df = fill_profile(complete, cleaned_profile)\ndf",
"2133\n"
]
],
[
[
"### Find Data\n\nmethod: find_data\nparameters: gd {gd is the grouped data which includes all the transction record per person}, df {df is the customer portfolio}\n\n1. Find the total number of offers received for every customer from the original transcript not the updated one\n2. Calculate the completion rate\n3. Append the new details in the customer portfiolio dataframe for each user",
"_____no_output_____"
]
],
[
[
"def find_data(gd, df):\n gd = gd[(gd.event == 'offer received')].reset_index(drop=True)\n grouped_data = gd.groupby(['person'])\n\n \n for index, row in df.iterrows():\n if row['person'] in grouped_data.groups.keys():\n events = grouped_data.get_group(row['person'])['event'].count()\n df.at[index, 'offers received'] = events\n df.at[index, 'completion rate'] = row['offers completed'] * 100 / events\n \n return df",
"_____no_output_____"
],
[
"df = find_data(sort_df, df)\ndf",
"_____no_output_____"
]
],
[
[
"### Find amount\n1. Find the total amount spent by each user",
"_____no_output_____"
]
],
[
[
"def find_amount(df):\n amount = pd.DataFrame()\n values = df.groupby(['person']).sum()\n amount['person'] = values.index\n amount['total amount'] = values.amount.to_numpy()\n return amount\n \n\ntotal_amount = find_amount(cleaned_transcript)\ndf = df.merge(total_amount, on='person')\ndf = df.reset_index(drop=True)\n\n########### Convert gender to M-0/F-1/O-2\ndf['gender'] = df['gender'].map({'M': 0, 'F': 1, 'O': 2})\ndf = df.fillna(0)\ndf",
"_____no_output_____"
],
[
"data_info(df, 'complete_profile_with_missing_values.csv')\ndata_info(complete, 'transcript_with_missing_values.csv')",
"(14825, 12)\n(30956, 7)\n"
]
],
[
[
"# Data Visualization",
"_____no_output_____"
],
[
"### Visualising the Data in 1D Space",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nimport matplotlib\n\ncomplete.hist(bins=15, color='steelblue', edgecolor='black', linewidth=1.0,\n xlabelsize=8, ylabelsize=8, grid=False) \nplt.tight_layout(rect=(0, 0, 1.5, 1.5)) ",
"_____no_output_____"
],
[
"df.hist(bins=15, color='steelblue', edgecolor='black', linewidth=2.0,\n xlabelsize=20, ylabelsize=20, grid=False) \nplt.tight_layout(rect=(0, 0, 5.8, 10))",
"_____no_output_____"
],
[
"import seaborn as sns\n\nf, ax = plt.subplots(figsize=(12, 8))\ncorr = df.corr()\nhm = sns.heatmap(round(corr,2), annot=True, ax=ax, cmap=\"coolwarm\",fmt='.2f',\n linewidths=.05)\nf.subplots_adjust(top=0.93)\nt= f.suptitle('Profile Attributes Correlation Heatmap', fontsize=14)",
"_____no_output_____"
],
[
"plt.figure(figsize=(15,4))\nplt.plot(df['completion rate'].value_counts().sort_index())",
"_____no_output_____"
]
],
[
[
"# Unsupervised Learning\n\nWe will use 2 unsuoervised learning algorithms to check if our data is actually seprable in approximately 5 clusters\nThe reason being if we get good number of clusters(4 or 5) then we can go and label all the data points according to our logic for likeliness (will be discussed later)",
"_____no_output_____"
]
],
[
[
"df.index = df['person']\ndf = df.drop(['person'], axis = 1)",
"_____no_output_____"
]
],
[
[
"### Normalizing the data\n Using sklearn's Min Max Scaler we will normalize the data in the range of 0 to 1 so that it becomes easier to work with supervised or unsupervised algorithms ",
"_____no_output_____"
]
],
[
[
"from sklearn.preprocessing import MinMaxScaler\n\ndef normalize_data(df):\n scaler = MinMaxScaler()\n df_scaled = pd.DataFrame(scaler.fit_transform(df.astype(float)))\n df_scaled.columns = df.columns\n df_scaled.index = df.index\n \n return df_scaled\n\ndf_scaled = normalize_data(df)\ndf_scaled",
"_____no_output_____"
]
],
[
[
"### Agglomerative Clustering\n1. Plot the dendogram\n2. Based on the dendogram detemine the distance threshold\n3. Use the distance threshold to find the number of clusters\n4. Check the distribution of clusters",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nfrom hcluster import pdist, linkage, dendrogram\n\nX = df_scaled.T.values #Transpose values \nY = pdist(X)\nZ = linkage(Y)\nplt.subplots(figsize=(18,5))\ndendrogram(Z, labels = df_scaled.columns)",
"_____no_output_____"
]
],
[
[
"from the dendogram gram graph we cab determine the distance threshold i.e. the line from where we can cut the graph is about 40 on the y axis",
"_____no_output_____"
]
],
[
[
"from sklearn.cluster import AgglomerativeClustering\n\ncluster = AgglomerativeClustering(n_clusters=None, affinity='euclidean', linkage='ward', distance_threshold=40)\nagg_clusters = np.array(cluster.fit_predict(df_scaled))",
"_____no_output_____"
],
[
"unique, counts = np.unique(agg_clusters, return_counts=True)\ndict(zip(unique, counts))",
"_____no_output_____"
]
],
[
[
"### K-means Clustering\n 1. Apply kmeans and find out the optimal number of clusters using the elbow method\n 2. Analyse the no of clusters formed and select the one where the clusters are equally distributed",
"_____no_output_____"
]
],
[
[
"from sklearn.cluster import KMeans\nfrom sklearn import metrics\nfrom scipy.spatial.distance import cdist\nimport numpy as np\nimport matplotlib.pyplot as plt\n\n\n# create new plot and data\nplt.plot()\nX = df_scaled\n\n# k means determine k\ndistortions = []\nfor k in range(1,10):\n km = KMeans(n_clusters=k, n_init=30)\n km.fit(X)\n wcss = km.inertia_\n km_clusters = km.predict(X)\n unique, counts = np.unique(km_clusters, return_counts=True)\n print(\"Cluster \", k, dict(zip(unique, counts)))\n distortions.append(wcss)\n\n# Plot the elbow\nplt.plot(range(1,10), distortions, 'bx-')\nplt.xlabel('k')\nplt.ylabel('Distortion')\nplt.title('The Elbow Method showing the optimal k')\nplt.show()",
"Cluster 1 {0: 14825}\nCluster 2 {0: 7533, 1: 7292}\nCluster 3 {0: 3638, 1: 5816, 2: 5371}\nCluster 4 {0: 3948, 1: 3327, 2: 4534, 3: 3016}\nCluster 5 {0: 3125, 1: 3102, 2: 1708, 3: 2957, 4: 3933}\nCluster 6 {0: 2666, 1: 1921, 2: 2832, 3: 3749, 4: 1633, 5: 2024}\nCluster 7 {0: 1967, 1: 2151, 2: 2936, 3: 1338, 4: 1903, 5: 1855, 6: 2675}\nCluster 8 {0: 1661, 1: 2134, 2: 1250, 3: 1389, 4: 1871, 5: 2934, 6: 1731, 7: 1855}\nCluster 9 {0: 1569, 1: 1740, 2: 2703, 3: 1220, 4: 1171, 5: 1605, 6: 1388, 7: 1785, 8: 1644}\n"
]
],
[
[
"from the k-means clustering we can see that for cluster 4 and cluster 5, and also from the elbow graph we can see that around 5 clusters are suitable ",
"_____no_output_____"
]
],
[
[
"from sklearn.cluster import KMeans\n\nkmeans = KMeans(n_clusters=5)\nkmeans.fit(df_scaled)\n\nlabels = kmeans.predict(df_scaled)\ncentroids = kmeans.cluster_centers_\n\nunique, counts = np.unique(labels, return_counts=True)\nprint(\"Cluster 5\",dict(zip(unique, counts)))",
"Cluster 5 {0: 1711, 1: 3937, 2: 3105, 3: 3117, 4: 2955}\n"
]
],
[
[
"### Unsupervised Learning Algorithm Results\nAgglomerative clustering gives really bad results because of the variability in our dataset, whereas k-means gives an average result. We can also see that the elbow graph is not well formed but we do get an idea of the separablity in our dataset. Thereforw we will now use Supervised Learning Algorithms to properly label our data",
"_____no_output_____"
],
[
"## Supervised Learning for Multi-Label Classification",
"_____no_output_____"
],
[
"### Determining the likeliness\n\nlogic of determining the likeliness is column*weight,I am determining the likeliness using offer completion rate, and the the offer types i.e. bogo, informational and discount. We assign the completion rate a weightage of 3 and offer type a weightage of 1. We noramlize the dataframe so that all the values in the 3 offer type columns are in the range and therefore the logic begind weightage can be applied. \n\ntotal weights = 3(completion rate) + 1(offer types) = 4\n\nscore = { (bogo + informational + discount)/3 + completion_rate*3} / total_weight\n\nLabel 4 - Very Likely (score>= 80)\n\nLabel 3 - Likely (score>= 60)\n\nLabel 2 - Neutral(50% chance) (score>= 40)\n\nLabel 1 - Unlikely(score>= 20)\n\nLabel 0 - Very Unlikely (score < 20)",
"_____no_output_____"
]
],
[
[
"def calculate_likeliness(rate):\n if rate >= 80 and rate <= 100:\n return 4\n elif rate >= 60 and rate < 80:\n return 3\n elif rate >= 40 and rate < 60:\n return 2\n elif rate >= 20 and rate < 40:\n return 1\n else:\n return 0\n\n\ndef likelihood(row):\n completion_rate= row[9]\n discount = row[7]\n informational = row[6]\n bogo = row[5]\n \n \n \n rate = ((discount + informational + bogo)/3 + completion_rate*3)/4\n \n return calculate_likeliness(rate * 100)\n \n\ndf_scaled.apply(lambda x: likelihood(x), axis=1)\ndf_scaled['likeliness'] = df_scaled.apply(lambda x: likelihood(x), axis=1)\ndf_scaled\n",
"_____no_output_____"
],
[
"## we can see that the data is well distributed \ndf_scaled.likeliness.value_counts()",
"_____no_output_____"
],
[
"# drop the columns used for determing the likeliness, so that our supervised learning model is not able to cheat\ndf_scaled = df_scaled.drop(['bogo', 'informational', 'discount', 'completion rate'], axis=1)\ndf_scaled",
"_____no_output_____"
],
[
"from sklearn.model_selection import train_test_split\nfrom sklearn.metrics import accuracy_score\n\nmat = df_scaled.values\nmat = df_scaled.values\n\nX = mat[:,0:7]\nY = mat[:,7]\n\nseed = 7\ntest_size = 0.33\nX_train, X_test, y_train, y_test = train_test_split(X, Y, test_size=test_size, random_state=seed)",
"_____no_output_____"
],
[
"def find_accuracy(y_test, y_pred):\n accuracy = accuracy_score(y_test, y_pred)\n print(\"Accuracy: %.2f%%\" % (accuracy * 100.0))",
"_____no_output_____"
],
[
"from sklearn.svm import LinearSVC\nclf = LinearSVC(random_state=0, tol=1e-5)\nclf.fit(X_train, y_train)\n# print(clf.coef_)\n# print(clf.intercept_)\ny_pred = clf.predict(X_test)\nfind_accuracy(y_test, y_pred)",
"Accuracy: 74.62%\n"
],
[
"from sklearn.svm import SVC\nsvclassifier = SVC(kernel='rbf')\nsvclassifier.fit(X_train, y_train)\ny_pred = svclassifier.predict(X_test)\nfind_accuracy(y_test, y_pred)",
"/Users/lokesh/opt/anaconda3/lib/python3.7/site-packages/sklearn/svm/base.py:193: FutureWarning: The default value of gamma will change from 'auto' to 'scale' in version 0.22 to account better for unscaled features. Set gamma explicitly to 'auto' or 'scale' to avoid this warning.\n \"avoid this warning.\", FutureWarning)\n"
],
[
"from sklearn.svm import SVC\nsvclassifier = SVC(kernel='poly')\nsvclassifier.fit(X_train, y_train)\ny_pred = svclassifier.predict(X_test)\nfind_accuracy(y_test, y_pred)",
"/Users/lokesh/opt/anaconda3/lib/python3.7/site-packages/sklearn/svm/base.py:193: FutureWarning: The default value of gamma will change from 'auto' to 'scale' in version 0.22 to account better for unscaled features. Set gamma explicitly to 'auto' or 'scale' to avoid this warning.\n \"avoid this warning.\", FutureWarning)\n"
],
[
"from sklearn.svm import SVC\nsvclassifier = SVC(kernel='linear')\nsvclassifier.fit(X_train, y_train)\ny_pred = svclassifier.predict(X_test)\nfind_accuracy(y_test, y_pred)",
"Accuracy: 96.20%\n"
],
[
"from sklearn.ensemble import RandomForestClassifier\nrf_classifier = RandomForestClassifier()\nrf_classifier.fit(X_train, y_train)\nrf_pred = rf_classifier.predict(X_test)\nfind_accuracy(y_test, rf_pred)",
"Accuracy: 98.75%\n"
]
],
[
[
"## Benchmark Model\nXgboost Algorithm is our benchmark model because it performs tasks like multilabel classifcation with ease and with very High accuracy",
"_____no_output_____"
]
],
[
[
"from xgboost import XGBClassifier\nmodel = XGBClassifier()\nmodel.fit(X_train, y_train)\ny_pred = model.predict(X_test)\npredictions = [round(value) for value in y_pred]\nfind_accuracy(y_test, y_pred)",
"Accuracy: 100.00%\n"
]
],
[
[
"## Evaluating the Model",
"_____no_output_____"
],
[
"#### Now we will evaluate our random forest model",
"_____no_output_____"
]
],
[
[
"from sklearn import model_selection\nfrom sklearn.metrics import mean_absolute_error, mean_squared_error\nfrom math import sqrt\n\nfrom sklearn.metrics import classification_report, confusion_matrix\nprint(confusion_matrix(y_test, rf_pred))\nprint(classification_report(y_test, rf_pred))\n\nkfold = model_selection.KFold(n_splits=10, random_state=seed)\nscoring = 'accuracy'\nresults = model_selection.cross_val_score(model, X, Y, cv=kfold, scoring=scoring)\nprint('Accuracy -val set: %.2f%% (%.2f)' % (results.mean()*100, results.std()))\n\nprint(\"MAE test score:\", mean_absolute_error(y_test, rf_pred))\nprint(\"RMSE test score:\", sqrt(mean_squared_error(y_test, rf_pred)))",
"[[1164 0 0 0 0]\n [ 0 1213 1 0 0]\n [ 0 0 1304 1 0]\n [ 0 4 25 713 2]\n [ 0 0 3 25 438]]\n precision recall f1-score support\n\n 0.0 1.00 1.00 1.00 1164\n 1.0 1.00 1.00 1.00 1214\n 2.0 0.98 1.00 0.99 1305\n 3.0 0.96 0.96 0.96 744\n 4.0 1.00 0.94 0.97 466\n\n accuracy 0.99 4893\n macro avg 0.99 0.98 0.98 4893\nweighted avg 0.99 0.99 0.99 4893\n\nAccuracy -val set: 100.00% (0.00)\nMAE test score: 0.01389740445534437\nRMSE test score: 0.12945514583973036\n"
]
],
[
[
"#### We can observe that our model performs really well and it has accuracy nearly as our benchmark model with really low MAE and RMSE Scores",
"_____no_output_____"
],
[
"## Now solving the same problem by filling the missing values",
"_____no_output_____"
]
],
[
[
"undefined = None\nincome_max = cleaned_profile.income.describe().max()\nundefined_group = undefined_group.fillna(income_max)\n# undefined = pd.concat([undefined_group, cleaned_profile], ignore_index=True)\n\n# undefined\ncomplete_transcript = transcript.copy()\nvalue = complete_transcript['value']\ncomplete_transcript['amount'] = [int(i['amount']) if i.get('amount') else 0 for i in value]\n\ncomplete_transcript['offer_id'] = [i['offer_id'] if i.get('offer_id') else (i['offer id'] if i.get('offer id') else '0') for i in value]\n\ncomplete_transcript.drop(['value'], axis=1, inplace=True)\n\nsort_df = complete_transcript.sort_values(by=['person', 'time'])\nsort_df",
"_____no_output_____"
],
[
"users = sort_df.groupby('person').apply(lambda x: get_valid_data(x))\nusers = users.dropna() \nusers = users.reset_index(drop=True)\nusers",
"_____no_output_____"
],
[
"valid_df = valid_offer_completed(users, cleaned_portfolio)\nvalid_df",
"_____no_output_____"
],
[
"complete_info = sort_df[sort_df['offer_id'].isin(['3f207df678b143eea3cee63160fa8bed', '5a8bc65990b245e5a138643cd4eb9837'])]\ncomplete_info_data = complete_info.groupby('person').apply(lambda x: info_offer(x))\ncomplete_info_data =complete_info_data.reset_index(drop=True)\ncomplete_info_data",
"_____no_output_____"
],
[
"complete_df = pd.concat([valid_df, complete_info_data], ignore_index=True)\ncomplete_df",
"_____no_output_____"
],
[
"full_profile = profile.copy()\nfull_profile['gender'] = full_profile['gender'].fillna('U')\nfull_profile['income'] = full_profile['income'].fillna(income_max)\nfull_profile",
"_____no_output_____"
],
[
"df2 = fill_profile(complete_df, full_profile)\ndf2 = find_data(sort_df, df2)\ndf2 = df2.fillna(0)\ndf2",
"2887\n"
],
[
"total_amount = find_amount(complete_transcript)\ndf2 = df2.merge(total_amount, on='person')\ndf2 = df2.reset_index(drop=True)\n\ndf2.index = df2['person']\ndf2 = df2.drop(['person'], axis = 1)\n########### Convert gender to 0/1/2\ndf2['gender'] = df2['gender'].map({'M': 0, 'F': 1, 'O': 2, 'U':3})\ndf2_scaled = normalize_data(df2)\ndf2_scaled",
"_____no_output_____"
],
[
"data_info(df2, 'complete_profile.csv')\ndata_info(complete_df, 'complete_transcript.csv')",
"(17000, 11)\n(33257, 7)\n"
],
[
"df2_scaled.apply(lambda x: likelihood(x), axis=1)\ndf2_scaled['likeliness'] = df2_scaled.apply(lambda x: likelihood(x), axis=1)\ndf2_scaled",
"_____no_output_____"
],
[
"df2_scaled = df2_scaled.drop(['bogo', 'informational', 'discount', 'completion rate'], axis=1)\ndf2_scaled",
"_____no_output_____"
],
[
"mat2 = df2_scaled.values\n\nX2 = mat2[:,0:7]\nY2 = mat2[:,7]\n\nseed = 7\ntest_size = 0.20\nX2_train, X2_test, y2_train, y2_test = train_test_split(X2, Y2, test_size=test_size, random_state=seed)",
"_____no_output_____"
],
[
"##Random Forest\nrf2_classifier = RandomForestClassifier()\nrf2_classifier.fit(X2_train, y2_train)\nrf2_pred = rf2_classifier.predict(X2_test)\nfind_accuracy(y2_test, rf2_pred)",
"Accuracy: 99.68%\n"
],
[
"## XGBoost Algorithm\nmodel2 = XGBClassifier()\nmodel2.fit(X2_train, y2_train)\n\ny2_pred = model2.predict(X2_test)\npredictions2 = [round(value) for value in y2_pred]\n\nfind_accuracy(y2_test, predictions2)",
"Accuracy: 100.00%\n"
]
],
[
[
"## Conculsion",
"_____no_output_____"
],
[
"In the project I have tried to determine how likely is a user complete an offer. I have used some data visualition to explain some realtion in the data. Then I have used unsupervised learning techniques to determine how seprable the data is and if we are actually able to divide the data in like 5 clusters. Then I determined the likeliness of every data point and removed the columns used to calculate the likeliness. This means that our supervised leaning model will not be able to deduce any inference in determining the likeliness. Then I split the data into training and test set and passed it to several SVM models with different kernel. One thing, I observed that tree models like Random Forest or XGBoost Algorithm perform really well for multi labeling tasks like this. Hence we will choose Gradient Boost as the algorithm of our choice and XGBoost as a benchmark model. Hence we will choose Gradient Boost as the algorithm of our choice and XGBoost as a benchmark model. \nFor evaluating our model we look at the confusion matrix from where we can see High precision and High Recall which means that our results have been labeled correctly.\n\nAlthough we are having very good accuracy for our models but that does not mean that our model is perfect it simply means that our model has very less data for now and therefore classifying our data is an easy task, also because we do not have very high dimensional data. Multi-label Classification is easy for low dimensional data.\n\n#### Missing Values vs Non-Missing Values\n\nWhen we removed missing values from our data we found a good balance between various classes but when we added those missing values and performed some inference on that data we notice that our class imbalance increased. Otherwise, the performance is the same \n",
"_____no_output_____"
]
],
[
[
"df2['offers received'].describe()",
"_____no_output_____"
],
[
"df2['total amount'].describe()",
"_____no_output_____"
],
[
"df['likeliness'] = df_scaled['likeliness']\ndef find_info(df2, class_label):\n label = df2[df2['likeliness'] == class_label]\n print(\"Likeliness ==\", class_label)\n print(\"bogo\", label.bogo.sum())\n print(\"discount\", label.discount.sum())\n print(\"informational\", label.informational.sum())\n print(\"offers received\", label['offers received'].sum())\n\n print()\n\n \nfind_info(df, 4)\nfind_info(df, 3)\nfind_info(df, 2)\nfind_info(df, 1)\nfind_info(df, 0)",
"Likeliness == 4\nbogo 2244.0\ndiscount 2331.0\ninformational 1433.0\noffers received 6008.0\n\nLikeliness == 3\nbogo 2855.0\ndiscount 2871.0\ninformational 1931.0\noffers received 9628.0\n\nLikeliness == 2\nbogo 3674.0\ndiscount 3998.0\ninformational 2625.0\noffers received 17959.0\n\nLikeliness == 1\nbogo 1675.0\ndiscount 2101.0\ninformational 1749.0\noffers received 16269.0\n\nLikeliness == 0\nbogo 344.0\ndiscount 511.0\ninformational 614.0\noffers received 16637.0\n\n"
]
],
[
[
"## Final Verdict\n\nWe saw that the Random Forest model performs better than other models except for the XGBoost Algorithm but we could see an improvement in the performance of the random forest model when we added more data i.e. when we imputed the missing values and added to the training and test sets. Since our model is already so accurate we didn't perform any hyperparameter tuning. \n\n1. from the above distribution we can see that users who have received more informational offers have a lower completion rate than users who have received more bogo/discount offers\n2. We can also see that the users who have received the most offers have low completion rate\n3. Lastly, age, income, became_memeber_on, total_amount have no influence over the completion rate",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
]
] |
cb88b5b8226fccab3f80f0b1f1de6f6310839b58
| 47,808 |
ipynb
|
Jupyter Notebook
|
NLP/02_01_Recommender_System_Practice.ipynb
|
xinrui112/Recommendation-System
|
fd363059f1c8859ef08a1ad7b2de9a2c6bcff7f9
|
[
"MIT"
] | null | null | null |
NLP/02_01_Recommender_System_Practice.ipynb
|
xinrui112/Recommendation-System
|
fd363059f1c8859ef08a1ad7b2de9a2c6bcff7f9
|
[
"MIT"
] | null | null | null |
NLP/02_01_Recommender_System_Practice.ipynb
|
xinrui112/Recommendation-System
|
fd363059f1c8859ef08a1ad7b2de9a2c6bcff7f9
|
[
"MIT"
] | null | null | null | 31.619048 | 256 | 0.334798 |
[
[
[
"## Recommender System Algorithm\n\n\n### Objective\nWe want to help consumers find attorneys. To surface attorneys to consumers, sales consultants often have to help attorneys describe their areas of practice (areas like Criminal Defense, Business or Personal Injury). \nTo expand their practices, attorneys can branch into related areas of practice. This can allow attorneys to help different customers while remaining within the bounds of their experience.\n\nAttached is an anonymized dataset of attorneys and their specialties. The columns are anonymized attorney IDs and specialty IDs. Please design a process that returns the top 5 recommended practice areas for a given attorney with a set of specialties.",
"_____no_output_____"
],
[
"## Data",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nfrom sklearn.preprocessing import normalize\nimport numpy as np",
"_____no_output_____"
],
[
"# Import data\ndata = pd.read_excel('data.xlsx', 'data')",
"_____no_output_____"
],
[
"# View first few rows of the dataset\ndata.head()",
"_____no_output_____"
]
],
[
[
"## 3. Data Exploration",
"_____no_output_____"
]
],
[
[
"# Information of the dataset\ndata.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 200000 entries, 0 to 199999\nData columns (total 2 columns):\nattorney_id 200000 non-null int64\nspecialty_id 200000 non-null int64\ndtypes: int64(2)\nmemory usage: 3.1 MB\n"
],
[
"# Check missing values\ndata.isnull().sum()",
"_____no_output_____"
],
[
"# Check duplicates\ndata.duplicated().sum()",
"_____no_output_____"
],
[
"# Check unique value count for the two ID's\ndata['attorney_id'].nunique(), data['specialty_id'].nunique()",
"_____no_output_____"
],
[
"# Check number of specialties per attorney\ndata.groupby('attorney_id')['specialty_id'].nunique().sort_values()",
"_____no_output_____"
]
],
[
[
"The number of specialties of an attorney ranges from 1 to 28.",
"_____no_output_____"
]
],
[
[
"# View a sample: an attorney with 28 specialties\ndata[data['attorney_id']==157715]",
"_____no_output_____"
]
],
[
[
"## Recommendation System",
"_____no_output_____"
],
[
"### Recommendation for Top K Practice Areas based on Similarity for Specialties",
"_____no_output_____"
],
[
"#### Step 1: Build the specialty-attorney matrix",
"_____no_output_____"
]
],
[
[
"# Build the specialty-attorney matrix\nspecialty_attorney = data.groupby(['specialty_id','attorney_id'])['attorney_id'].count().unstack(fill_value=0)\nspecialty_attorney = (specialty_attorney > 0).astype(int)\nspecialty_attorney",
"_____no_output_____"
]
],
[
[
"#### Step 2: Build specialty-specialty similarity matrix",
"_____no_output_____"
]
],
[
[
"# Build specialty-specialty similarity matrix\nspecialty_attorney_norm = normalize(specialty_attorney, axis=1) \nsimilarity = np.dot(specialty_attorney_norm, specialty_attorney_norm.T)\ndf_similarity = pd.DataFrame(similarity, index=specialty_attorney.index, columns=specialty_attorney.index)\n\ndf_similarity",
"_____no_output_____"
]
],
[
[
"#### Step 3: Find the Top K most similar specialties",
"_____no_output_____"
]
],
[
[
"# Find the top k most similar specialties\ndef topk_specialty(specialty, similarity, k):\n result = similarity.loc[specialty].sort_values(ascending=False)[1:k + 1].reset_index()\n result = result.rename(columns={'specialty_id': 'Specialty_Recommend', specialty: 'Similarity'})\n return result",
"_____no_output_____"
]
],
[
[
"### Testing Recommender System based on Similarity\n#### Process:\n1. Ask user to input the ID of his/her obtained specialties\n2. The system will recommend top 5 practice areas for the user's specialties based on similarity",
"_____no_output_____"
]
],
[
[
"# Test on a specialty sample 1\nuser_input1 = int(input('Please input your specialty ID: '))\nrecommend_user1 = topk_specialty(specialty=user_input1, similarity=df_similarity, k=5)\nprint('Top 5 recommended practice areas for user 1:')\nprint('--------------------------------------------')\nprint(recommend_user1)",
"Please input your specialty ID: 909\nTop 5 recommended practice areas for user 1:\n--------------------------------------------\n Specialty_Recommend Similarity\n0 205 0.117307\n1 439 0.115609\n2 712 0.101321\n3 208 0.098208\n4 252 0.097700\n"
],
[
"# Test on a specialty sample 2\nuser_input2 = int(input('Please input your specialty ID: '))\nrecommend_user2 = topk_specialty(specialty=user_input2, similarity=df_similarity, k=5)\nprint('Top 5 recommended practice areas for user 2:')\nprint('--------------------------------------------')\nprint(recommend_user2)",
"Please input your specialty ID: 196\nTop 5 recommended practice areas for user 2:\n--------------------------------------------\n Specialty_Recommend Similarity\n0 436 0.103643\n1 263 0.080463\n2 218 0.066211\n3 429 0.063597\n4 667 0.057946\n"
]
],
[
[
"### Popularity-based Recommendation - If user requests recommedation based on popularity",
"_____no_output_____"
]
],
[
[
"# Get ranked specialties based on popularity\ndf_specialty_popular = data_recommend.groupby('specialty_id')['attorney_id'].nunique().sort_values(ascending=False)\ndf_specialty_popular",
"_____no_output_____"
],
[
"# Top 5 specialties based on popularity among attorneys\ndf_specialty_popular.columns = ['specialty_id', 'count_popular']\nprint('The 5 most popular specialties:')\nprint('--------------------------------')\nprint(df_specialty_popular.nlargest(5, keep='all'))",
"The 5 most popular specialties:\n--------------------------------\nspecialty_id\n218 14780\n258 9856\n429 9608\n257 8615\n373 8245\nName: attorney_id, dtype: int64\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
cb88be3b56770c9e8806b582760dd99fcd4cf217
| 67,045 |
ipynb
|
Jupyter Notebook
|
DataMasterClass.ipynb
|
toskpl/DataMasterClass
|
8d634c8c312b02ad250ebeea03e0e3a06dd904f7
|
[
"MIT"
] | null | null | null |
DataMasterClass.ipynb
|
toskpl/DataMasterClass
|
8d634c8c312b02ad250ebeea03e0e3a06dd904f7
|
[
"MIT"
] | null | null | null |
DataMasterClass.ipynb
|
toskpl/DataMasterClass
|
8d634c8c312b02ad250ebeea03e0e3a06dd904f7
|
[
"MIT"
] | null | null | null | 40.58414 | 135 | 0.358655 |
[
[
[
"# Import bibilotek",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\n\nimport xgboost as xgb\nfrom sklearn.tree import DecisionTreeRegressor\n\nfrom sklearn.model_selection import cross_val_score, KFold\nfrom sklearn.metrics import mean_absolute_error\n",
"_____no_output_____"
],
[
"pip install --upgrade tables",
"Requirement already satisfied: tables in c:\\users\\tskrzypczak\\anaconda3\\lib\\site-packages (3.6.1)\nRequirement already satisfied: numpy>=1.9.3 in c:\\users\\tskrzypczak\\anaconda3\\lib\\site-packages (from tables) (1.20.1)\nRequirement already satisfied: numexpr>=2.6.2 in c:\\users\\tskrzypczak\\anaconda3\\lib\\site-packages (from tables) (2.7.3)\nNote: you may need to restart the kernel to use updated packages.\n"
]
],
[
[
"# Odczyt danych z pliku h5",
"_____no_output_____"
]
],
[
[
"df_train = pd.read_hdf(\"train_data.h5\")\ndf_train['price'] = df_train['price'].map(parse_price)\n\ndf_test = pd.read_hdf(\"test_data.h5\")\n\ndf = pd.concat([df_train, df_test])\nprint(df_train.shape, df_test.shape)\n\ndf",
"(22732, 8) (11448, 7)\n"
]
],
[
[
"# Funkcje pomocniczne",
"_____no_output_____"
]
],
[
[
"def parse_price(val):\n if isinstance(val, str): \n if \"₽\" in val:\n val = val.split('₽')[0]\n \n val = val.replace(' ', '')\n return int(val) / 1000000\n \n return float(val)\n\ndef parse_area(val):\n if isinstance(val, int): return val\n if isinstance(val, float): return val\n \n return float(val.split(\"м\")[0].replace(\" \", \"\"))\n\ndef parse_floor(val):\n if isinstance(val, int): return val\n if isinstance(val, str):\n return val.split('/')[0]\n return val\n\ndef get_metro_station(row):\n for i in row:\n if 'МЦК' in i:\n return i\n \ndef check_log_model(df, feats, model, cv=5, scoring=\"neg_mean_absolute_error\"):\n df_train = df[ ~df[\"price\"].isnull() ].copy()\n\n X = df_train[feats]\n y = df_train[\"price\"]\n y_log = np.log(y)\n \n cv = KFold(n_splits=5, shuffle=True, random_state=0)\n scores = []\n for train_idx, test_idx in cv.split(X):\n X_train, X_test = X.iloc[train_idx], X.iloc[test_idx]\n y_log_train, y_test = y_log.iloc[train_idx], y.iloc[test_idx]\n\n model = xgb.XGBRegressor(max_depth=md, n_estimators=ne, learning_rate=lr, random_state=0)\n model.fit(X_train, y_log_train)\n y_log_pred = model.predict(X_test)\n y_pred = np.exp(y_log_pred)\n\n score = mean_absolute_error(y_test, y_pred)\n scores.append(score)\n\n return np.mean(scores), np.std(scores)\n",
"_____no_output_____"
]
],
[
[
"# Future ennginiring",
"_____no_output_____"
]
],
[
[
"params = df[\"params\"].apply(pd.Series)\n\nparams = params.fillna(-1)\n\nif \"Охрана:\" not in df:\n df = pd.concat([df, params], axis=1)\n \n obj_feats = params.select_dtypes(object).columns\n\n for feat in obj_feats:\n df[\"{}_cat\".format(feat)] = df[feat].factorize()[0]\n\ncat_feats = [x for x in df.columns if \"_cat\" in x]\n\ncat_feats",
"_____no_output_____"
],
[
"# powierzchnia mieszkania\ndf[\"area\"] = df[\"Общая площадь:\"].map(parse_area)",
"_____no_output_____"
],
[
"# powierzchnia kuchni\ndf[\"kitchen_area\"] = df[\"Площадь кухни:\"].map(parse_area)",
"_____no_output_____"
],
[
"geo_block = (\ndf[\"geo_block\"]\n .map(lambda x: x[:int(len(x)/2) ])\n .map(lambda x: {\"geo_block_{}\".format(idx):val for idx,val in enumerate(x) })\n .apply(pd.Series)\n)",
"_____no_output_____"
],
[
"for feat in geo_block.columns:\n df[\"{}_cat\".format(feat)] = geo_block[feat].factorize()[0]",
"_____no_output_____"
],
[
"geo_cat_feats = [x for x in df.columns if \"geo_block\" in x and \"_cat\" in x]",
"_____no_output_____"
],
[
"breadcrumbs = (\n df[\"breadcrumbs\"]\n .map(lambda x: {\"breadcrumbs_{}\".format(idx):val for idx,val in enumerate(x) })\n .apply(pd.Series)\n)",
"_____no_output_____"
],
[
"for feat in breadcrumbs.columns:\n df[\"{}_cat\".format(feat)] = breadcrumbs[feat].factorize()[0]\ndf",
"_____no_output_____"
],
[
"breadcrumbs_cat_feats = [x for x in df.columns if \"breadcrumbs\" in x and \"_cat\" in x]\nbreadcrumbs_cat_feats",
"_____no_output_____"
],
[
"metro_station = (\n df[\"breadcrumbs\"]\n .map(lambda x: get_metro_station(x))\n .apply(pd.Series)\n)\nmetro_station.columns = ['metro_station_name']",
"_____no_output_____"
],
[
"df[\"metro_station_cat\"] = metro_station.apply(lambda x : pd.factorize(x)[0])\n\ndf",
"_____no_output_____"
]
],
[
[
"# Model DecisionTreeRegressor",
"_____no_output_____"
]
],
[
[
"feats = [\"area\", \"kitchen_area\", \"metro_station_cat\"] + geo_cat_feats + cat_feats + breadcrumbs_cat_feats \n\nmodel = DecisionTreeRegressor(max_depth=20)\ncheck_log_model(df, feats, DecisionTreeRegressor(max_depth=20))",
"_____no_output_____"
]
],
[
[
"# Model XGBRegressor",
"_____no_output_____"
]
],
[
[
"\nmd = 20\nne = 700\nlr = 0.15",
"_____no_output_____"
],
[
"feats = [\"area\", \"kitchen_area\", \"metro_station_cat\"] + geo_cat_feats + cat_feats + breadcrumbs_cat_feats \n \ncheck_log_model(df, feats, xgb.XGBRegressor(max_depth=md, n_estimators=ne, learning_rate=lr, random_state=0))",
"_____no_output_____"
]
],
[
[
"# Kaggle submit",
"_____no_output_____"
]
],
[
[
"feats = [\"area\", \"kitchen_area\", \"metro_station_cat\"] + geo_cat_feats + cat_feats + breadcrumbs_cat_feats\n\ndf_train = df[ ~df[\"price\"].isnull() ].copy()\ndf_test = df[ df[\"price\"].isnull() ].copy()\n\nX_train = df_train[feats]\ny_train = df_train[\"price\"]\ny_log_train = np.log(y_train)\n\nX_test = df_test[feats]\n\nmodel = xgb.XGBRegressor(max_depth=8, n_estimators=700, learning_rate=0.1, random_state=0)\nmodel.fit(X_train, y_log_train)\ny_log_pred = model.predict(X_test)\ny_pred = np.exp(y_log_pred)\n\n\ndf_test[\"price\"] = y_pred\ndf_test[ [\"id\", \"price\"] ].to_csv(\"./xgb_location_log_area_v2.csv\", index=False)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb88bf07267bd2fa6750e93e5685acb1cc600c7f
| 78,264 |
ipynb
|
Jupyter Notebook
|
lec4.ipynb
|
epicha/ia340
|
55c2ffbfcd005d17adbb03d64e2008c342714c4b
|
[
"MIT"
] | null | null | null |
lec4.ipynb
|
epicha/ia340
|
55c2ffbfcd005d17adbb03d64e2008c342714c4b
|
[
"MIT"
] | null | null | null |
lec4.ipynb
|
epicha/ia340
|
55c2ffbfcd005d17adbb03d64e2008c342714c4b
|
[
"MIT"
] | null | null | null | 78.736419 | 22,836 | 0.796637 |
[
[
[
"print('hello')",
"hello\n"
],
[
"for number in [1,2,3]:\n print(number)",
"1\n2\n3\n"
],
[
"print('1+3 is {}'.format(1+3))",
"1+3 is 4\n"
],
[
"!pip install psycopg2",
"Requirement already satisfied: psycopg2 in /home/ec2-user/anaconda3/envs/python3/lib/python3.6/site-packages (2.7.5)\r\n"
],
[
"import pandas\nimport psycopg2\nimport configparser",
"/home/ec2-user/anaconda3/envs/python3/lib/python3.6/site-packages/psycopg2/__init__.py:144: UserWarning: The psycopg2 wheel package will be renamed from release 2.8; in order to keep installing from binary please use \"pip install psycopg2-binary\" instead. For details see: <http://initd.org/psycopg/docs/install.html#binary-install-from-pypi>.\n \"\"\")\n"
],
[
"config = configparser.ConfigParser()\nconfig.read('config.ini')\n\nhost=config['myaws']['host']\ndb=config['myaws']['db']\nuser=config['myaws']['user']\npwd = config['myaws']['pwd']\n\nconn = psycopg2.connect( host=host,\n user=user,\n password = pwd,\n dbname=db)",
"_____no_output_____"
],
[
"cur=conn.cursor()",
"_____no_output_____"
]
],
[
[
"Define SQL statement",
"_____no_output_____"
]
],
[
[
"sql_statement = \"\"\" select bathroom,bedroom\n from public.house_price_full\n where bathroom>2\"\"\"",
"_____no_output_____"
]
],
[
[
"Cursor Executes the SQL statement",
"_____no_output_____"
]
],
[
[
"cur.execute(sql_statement)",
"_____no_output_____"
],
[
"cur.fetchone()",
"_____no_output_____"
],
[
"for bathroom,bedroom in cur.fetchall()[:10]:\n print(bathroom,bedroom)",
"3 4\n3 5\n3 4\n3 4\n3 3\n3 5\n4 5\n3 4\n3 4\n3 4\n"
],
[
"df = pandas.read_sql_query(sql_statement,conn)\ndf[:]",
"_____no_output_____"
],
[
"sql_statement= \"\"\"\n select built_in,\n avg(price) as avg_price\n from public.house_price_full\n group by built_in\n order by built_in\n \"\"\"",
"_____no_output_____"
],
[
"df = pandas.read_sql_query(sql_statement,conn)\ndf[:10]",
"_____no_output_____"
],
[
"df_price=pandas.read_sql_query(sql_statement,conn)\n\ndf_price.plot(y='avg_price',x='built_in')",
"_____no_output_____"
],
[
"sql_statement= \"\"\"\n select price,area\n from public.house_price_full\n \"\"\"",
"_____no_output_____"
],
[
"df_price=pandas.read_sql_query(sql_statement,conn)\n\ndf_price[:10]",
"_____no_output_____"
],
[
"df_price=pandas.read_sql_query(sql_statement,conn)\n\ndf_price['area'].hist()",
"_____no_output_____"
],
[
"df_price=pandas.read_sql_query(sql_statement,conn)\n\ndf_price.plot.scatter(x='area',y='price')",
"_____no_output_____"
],
[
"sql_statement= \"\"\"\n select house_type,\n avg(price) as avg_price\n from public.house_price_full\n group by house_type\n order by avg_price desc\n \"\"\"",
"_____no_output_____"
],
[
"df_price=pandas.read_sql_query(sql_statement,conn)\n\ndf_price.plot.bar(x='house_type',y='avg_price')",
"_____no_output_____"
],
[
"sql_statement = \"\"\"\n insert into gp1.student(s_email,s_name,s_major)\n values('{}','{}','{}')\n \n \"\"\".format('[email protected]','s5','ia')\n\nprint(sql_statement)",
"\n insert into gp1.student(s_email,s_name,s_major)\n values('[email protected]','s5','ia')\n \n \n"
],
[
"conn.rollback()",
"_____no_output_____"
],
[
"sql_statement = \"\"\"\n insert into gp1.student(s_email,s_name,s_major)\n values('{}','{}','{}')\n \n \"\"\".format('[email protected]','s6','ia')",
"_____no_output_____"
],
[
"cur.execute(sql_statement)",
"_____no_output_____"
],
[
"conn.commit()",
"_____no_output_____"
],
[
"df_student=pandas.read_sql_query('select * from gp1.student',conn)\ndf_student[:]",
"_____no_output_____"
],
[
"sql_statement = \"\"\"\n delete from gp1.student\n where s_email = '{}'\n \"\"\".format('[email protected]')\nprint(sql_statement)",
"\n delete from gp1.student\n where s_email = '[email protected]'\n \n"
],
[
"cur.execute(sql_statement)",
"_____no_output_____"
],
[
"conn.commit()",
"_____no_output_____"
],
[
"cur.close()\nconn.close()",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb88cc5e7af1db1c13a6a6ee26f19c5f47ba1751
| 211,442 |
ipynb
|
Jupyter Notebook
|
13. Industry Applications/2. CNN for Chest X-Ray Images/.ipynb_checkpoints/Working_With_Chest_XRay_Images--checkpoint.ipynb
|
vshubh24/machineLearning
|
eedd4432ce7c58e20503611c3d801a53b74e3c2a
|
[
"Apache-2.0"
] | null | null | null |
13. Industry Applications/2. CNN for Chest X-Ray Images/.ipynb_checkpoints/Working_With_Chest_XRay_Images--checkpoint.ipynb
|
vshubh24/machineLearning
|
eedd4432ce7c58e20503611c3d801a53b74e3c2a
|
[
"Apache-2.0"
] | null | null | null |
13. Industry Applications/2. CNN for Chest X-Ray Images/.ipynb_checkpoints/Working_With_Chest_XRay_Images--checkpoint.ipynb
|
vshubh24/machineLearning
|
eedd4432ce7c58e20503611c3d801a53b74e3c2a
|
[
"Apache-2.0"
] | null | null | null | 262.66087 | 123,752 | 0.908528 |
[
[
[
"# Analysis of Chest X-Ray images",
"_____no_output_____"
],
[
"Neural networks have revolutionised image processing in several different domains. Among these is the field of medical imaging. In the following notebook, we will get some hands-on experience in working with Chest X-Ray (CXR) images.\n\nThe objective of this exercise is to identify images where an \"effusion\" is present. This is a classification problem, where we will be dealing with two classes - 'effusion' and 'nofinding'. Here, the latter represents a \"normal\" X-ray image.\n\nThis same methodology can be used to spot various other illnesses that can be detected via a chest x-ray. For the scope of this demonstration, we will specifically deal with \"effusion\".",
"_____no_output_____"
],
[
"## 1. Data Pre-processing",
"_____no_output_____"
],
[
"Our data is in the form of grayscale (black and white) images of chest x-rays. To perform our classification task effectively, we need to perform some pre-processing of the data.\n\nFirst, we load all the relevant libraries.",
"_____no_output_____"
]
],
[
[
"from skimage import io\nimport os\nimport glob\nimport numpy as np\nimport matplotlib.pyplot as plt\n\nimport warnings\nwarnings.simplefilter('ignore')",
"_____no_output_____"
]
],
[
[
"Point a variable to the path where the data resides. Note that to use the code below you will need to move the folders effusion/ and nofinding/ into one common folder. You can do something like this:\n\n```\nmkdir CXR_Data\nmove effusion CXR_Data\nmove nofinding CXR_Data\n```",
"_____no_output_____"
]
],
[
[
"DATASET_PATH = './CXR_data/'\n\n# There are two classes of images that we will deal with\ndisease_cls = ['effusion', 'nofinding']",
"_____no_output_____"
]
],
[
[
"Next, we read the \"effusion\" and \"nofinding\" images.",
"_____no_output_____"
]
],
[
[
"effusion_path = os.path.join(DATASET_PATH, disease_cls[0], '*')\neffusion = glob.glob(effusion_path)\neffusion = io.imread(effusion[0])\n\nnormal_path = os.path.join(DATASET_PATH, disease_cls[1], '*')\nnormal = glob.glob(normal_path)\nnormal = io.imread(normal[0])\n\nf, axes = plt.subplots(1, 2, sharey=True)\nf.set_figwidth(10)\n \naxes[0].imshow(effusion, cmap='gray')\naxes[1].imshow(normal, cmap='gray')",
"_____no_output_____"
],
[
"effusion.shape",
"_____no_output_____"
],
[
"normal.shape",
"_____no_output_____"
]
],
[
[
"### Data Augmentation ###\n\nNow that we have read the images, the next step is data augmentation. We use the concept of a \"data generator\" that you learnt in the last section.",
"_____no_output_____"
]
],
[
[
"from skimage.transform import rescale\nfrom tensorflow.keras.preprocessing.image import ImageDataGenerator\n\ndatagen = ImageDataGenerator(\n featurewise_center=True,\n featurewise_std_normalization=True,\n rotation_range=10,\n width_shift_range=0,\n height_shift_range=0,\n vertical_flip=False,)\n\ndef preprocess_img(img, mode):\n img = (img - img.min())/(img.max() - img.min())\n img = rescale(img, 0.25, multichannel=True, mode='constant')\n \n if mode == 'train':\n if np.random.randn() > 0:\n img = datagen.random_transform(img)\n return img",
"_____no_output_____"
]
],
[
[
"## 2. Model building",
"_____no_output_____"
],
[
"We will be using a Resnet in this (you learnt about Resnets previously). \n\nFor this to work, the script that defines the resnet model (resnet.py) should reside in the same folder as this notebook",
"_____no_output_____"
]
],
[
[
"import resnet\n\nimg_channels = 1\nimg_rows = 256\nimg_cols = 256\n\nnb_classes = 2",
"_____no_output_____"
],
[
"import numpy as np\nimport tensorflow as tf\n\nclass AugmentedDataGenerator(tf.keras.utils.Sequence):\n 'Generates data for Keras'\n def __init__(self, mode='train', ablation=None, disease_cls = ['nofinding', 'effusion'], \n batch_size=32, dim=(256, 256), n_channels=1, shuffle=True):\n 'Initialization'\n self.dim = dim\n self.batch_size = batch_size\n self.labels = {}\n self.list_IDs = []\n self.mode = mode\n \n for i, cls in enumerate(disease_cls):\n paths = glob.glob(os.path.join(DATASET_PATH, cls, '*'))\n brk_point = int(len(paths)*0.8)\n if self.mode == 'train':\n paths = paths[:brk_point]\n else:\n paths = paths[brk_point:]\n if ablation is not None:\n paths = paths[:int(len(paths)*ablation/100)]\n self.list_IDs += paths\n self.labels.update({p:i for p in paths})\n \n \n self.n_channels = n_channels\n self.n_classes = len(disease_cls)\n self.shuffle = shuffle\n self.on_epoch_end()\n\n def __len__(self):\n 'Denotes the number of batches per epoch'\n return int(np.floor(len(self.list_IDs) / self.batch_size))\n\n def __getitem__(self, index):\n 'Generate one batch of data'\n\n indexes = self.indexes[index*self.batch_size:(index+1)*self.batch_size]\n list_IDs_temp = [self.list_IDs[k] for k in indexes]\n\n X, y = self.__data_generation(list_IDs_temp)\n\n return X, y\n\n def on_epoch_end(self):\n 'Updates indexes after each epoch'\n self.indexes = np.arange(len(self.list_IDs))\n if self.shuffle == True:\n np.random.shuffle(self.indexes)\n\n def __data_generation(self, list_IDs_temp):\n 'Generates data containing batch_size samples' # X : (n_samples, *dim, n_channels)\n # Initialization\n X = np.empty((self.batch_size, *self.dim, self.n_channels))\n y = np.empty((self.batch_size), dtype=int)\n \n delete_rows = []\n\n # Generate data\n for i, ID in enumerate(list_IDs_temp):\n img = io.imread(ID)\n img = img[:, :, np.newaxis]\n if img.shape == (1024, 1024,1):\n img = preprocess_img(img, self.mode)\n X[i,] = img\n y[i] = self.labels[ID]\n else:\n delete_rows.append(i)\n continue\n \n X = np.delete(X, delete_rows, axis=0)\n y = np.delete(y, delete_rows, axis=0)\n \n return X, tf.keras.utils.to_categorical(y, num_classes=self.n_classes)",
"_____no_output_____"
]
],
[
[
"## 3. Ablation Run",
"_____no_output_____"
],
[
"In the previous notebook, you learnt about Ablation. Briefly, an ablation run is when you systematically modify certain parts of the input, in order to observe the equivalent change in the input.\n\nFor the following section, we'll be using the Data Generator concept that you previously worked on.",
"_____no_output_____"
]
],
[
[
"model = resnet.ResnetBuilder.build_resnet_18((img_channels, img_rows, img_cols), nb_classes)\nmodel.compile(loss='categorical_crossentropy',optimizer='SGD',\n metrics=['accuracy'])\ntraining_generator = AugmentedDataGenerator('train', ablation=5)\nvalidation_generator = AugmentedDataGenerator('val', ablation=5)\n\nmodel.fit(training_generator, epochs=1, validation_data=validation_generator)",
"1/1 [==============================] - 0s 3ms/step - loss: 1.3989 - accuracy: 0.9667\n"
],
[
"model = resnet.ResnetBuilder.build_resnet_18((img_channels, img_rows, img_cols), nb_classes)\nmodel.compile(loss='categorical_crossentropy',optimizer='SGD',\n metrics=['accuracy'])\n\ntraining_generator = AugmentedDataGenerator('train', ablation=5)\nvalidation_generator = AugmentedDataGenerator('val', ablation=5)\n\nmodel.fit(training_generator, epochs=5, validation_data=None)",
"Epoch 1/5\n1/1 [==============================] - 0s 1ms/step - loss: 1.7565 - accuracy: 0.1000\nEpoch 2/5\n1/1 [==============================] - 0s 2ms/step - loss: 1.4482 - accuracy: 0.9333\nEpoch 3/5\n1/1 [==============================] - 0s 1ms/step - loss: 1.3399 - accuracy: 0.9355\nEpoch 4/5\n1/1 [==============================] - 0s 2ms/step - loss: 1.2365 - accuracy: 0.9667\nEpoch 5/5\n1/1 [==============================] - 0s 2ms/step - loss: 1.1930 - accuracy: 0.9667\n"
],
[
"from sklearn.metrics import roc_auc_score\nfrom tensorflow.keras import optimizers\nfrom tensorflow.keras.callbacks import *\n\nclass roc_callback(Callback):\n \n def on_train_begin(self, logs={}):\n logs['val_auc'] = 0\n\n def on_epoch_end(self, epoch, logs={}):\n y_p = []\n y_v = []\n for i in range(len(validation_generator)):\n x_val, y_val = validation_generator[i]\n y_pred = self.model.predict(x_val)\n y_p.append(y_pred)\n y_v.append(y_val)\n y_p = np.concatenate(y_p)\n y_v = np.concatenate(y_v)\n roc_auc = roc_auc_score(y_v, y_p)\n print ('\\nVal AUC for epoch{}: {}'.format(epoch, roc_auc))\n logs['val_auc'] = roc_auc",
"_____no_output_____"
],
[
"model = resnet.ResnetBuilder.build_resnet_18((img_channels, img_rows, img_cols), nb_classes)\nmodel.compile(loss='categorical_crossentropy',optimizer='SGD',\n metrics=['accuracy'])\n\ntraining_generator = AugmentedDataGenerator('train', ablation=20)\nvalidation_generator = AugmentedDataGenerator('val', ablation=20)\n\nauc_logger = roc_callback()\n\nmodel.fit(training_generator, epochs=5, validation_data=validation_generator, callbacks=[auc_logger])",
"Epoch 1/5\n5/5 [==============================] - ETA: 0s - loss: 1.5154 - accuracy: 0.7089\nVal AUC for epoch0: 0.4655172413793104\n5/5 [==============================] - 11s 2s/step - loss: 1.5154 - accuracy: 0.7089 - val_loss: 2.9160 - val_accuracy: 0.9000 - val_auc: 0.4655\nEpoch 2/5\n5/5 [==============================] - ETA: 0s - loss: 1.2867 - accuracy: 0.9177\nVal AUC for epoch1: 0.41379310344827586\n5/5 [==============================] - 10s 2s/step - loss: 1.2867 - accuracy: 0.9177 - val_loss: 1.8222 - val_accuracy: 0.9355 - val_auc: 0.4138\nEpoch 3/5\n5/5 [==============================] - ETA: 0s - loss: 1.2691 - accuracy: 0.9114\nVal AUC for epoch2: 0.5714285714285714\n5/5 [==============================] - 10s 2s/step - loss: 1.2691 - accuracy: 0.9114 - val_loss: 1.5441 - val_accuracy: 0.9355 - val_auc: 0.5714\nEpoch 4/5\n5/5 [==============================] - ETA: 0s - loss: 1.2725 - accuracy: 0.9057\nVal AUC for epoch3: 0.48275862068965514\n5/5 [==============================] - 10s 2s/step - loss: 1.2725 - accuracy: 0.9057 - val_loss: 1.5360 - val_accuracy: 0.9032 - val_auc: 0.4828\nEpoch 5/5\n5/5 [==============================] - ETA: 0s - loss: 1.2732 - accuracy: 0.9051\nVal AUC for epoch4: 0.033333333333333326\n5/5 [==============================] - 10s 2s/step - loss: 1.2732 - accuracy: 0.9051 - val_loss: 1.1250 - val_accuracy: 0.9667 - val_auc: 0.0333\n"
],
[
"from functools import partial\nimport tensorflow.keras.backend as K\nfrom itertools import product\n\ndef w_categorical_crossentropy(y_true, y_pred, weights):\n nb_cl = len(weights)\n final_mask = K.zeros_like(y_pred[:, 0])\n y_pred_max = K.max(y_pred, axis=1)\n y_pred_max = K.reshape(y_pred_max, (K.shape(y_pred)[0], 1))\n y_pred_max_mat = K.cast(K.equal(y_pred, y_pred_max), K.floatx())\n for c_p, c_t in product(range(nb_cl), range(nb_cl)):\n final_mask += (weights[c_t, c_p] * y_pred_max_mat[:, c_p] * y_true[:, c_t])\n cross_ent = K.categorical_crossentropy(y_true, y_pred, from_logits=False)\n return cross_ent * final_mask\n\nbin_weights = np.ones((2,2))\nbin_weights[0, 1] = 5\nbin_weights[1, 0] = 5\nncce = partial(w_categorical_crossentropy, weights=bin_weights)\nncce.__name__ ='w_categorical_crossentropy'",
"_____no_output_____"
],
[
"model = resnet.ResnetBuilder.build_resnet_18((img_channels, img_rows, img_cols), nb_classes)\nmodel.compile(loss=ncce, optimizer='SGD',\n metrics=['accuracy'])\n\ntraining_generator = AugmentedDataGenerator('train', ablation=5)\nvalidation_generator = AugmentedDataGenerator('val', ablation=5)\n\nmodel.fit(training_generator, epochs=1, validation_data=None)",
"1/1 [==============================] - 0s 1ms/step - loss: 5.5409 - accuracy: 0.0645\n"
]
],
[
[
"## 4. Final Run",
"_____no_output_____"
],
[
"After deeply examining our data and building some preliminary models, we are finally ready to build a model that will perform our prediction task.",
"_____no_output_____"
]
],
[
[
"class DecayLR(tf.keras.callbacks.Callback):\n def __init__(self, base_lr=0.01, decay_epoch=1):\n super(DecayLR, self).__init__()\n self.base_lr = base_lr\n self.decay_epoch = decay_epoch \n self.lr_history = []\n \n def on_train_begin(self, logs={}):\n K.set_value(self.model.optimizer.lr, self.base_lr)\n\n def on_epoch_end(self, epoch, logs={}):\n new_lr = self.base_lr * (0.5 ** (epoch // self.decay_epoch))\n self.lr_history.append(K.get_value(self.model.optimizer.lr))\n K.set_value(self.model.optimizer.lr, new_lr)",
"_____no_output_____"
],
[
"model = resnet.ResnetBuilder.build_resnet_18((img_channels, img_rows, img_cols), nb_classes)\nsgd = optimizers.SGD(lr=0.005)\n\nbin_weights = np.ones((2,2))\nbin_weights[1, 1] = 10\nbin_weights[1, 0] = 10\nncce = partial(w_categorical_crossentropy, weights=bin_weights)\nncce.__name__ ='w_categorical_crossentropy'\n\nmodel.compile(loss=ncce,optimizer= sgd,\n metrics=['accuracy'])\ntraining_generator = AugmentedDataGenerator('train', ablation=50)\nvalidation_generator = AugmentedDataGenerator('val', ablation=50)\n\nauc_logger = roc_callback()\nfilepath = 'models/best_model.hdf5'\ncheckpoint = ModelCheckpoint(filepath, monitor='val_auc', verbose=1, save_best_only=True, mode='max')\n\ndecay = DecayLR()\n\nmodel.fit(training_generator, epochs=10, validation_data=validation_generator, callbacks=[auc_logger, decay, checkpoint])",
"Epoch 1/10\n13/13 [==============================] - ETA: 0s - loss: 2.2712 - accuracy: 0.5725\nVal AUC for epoch0: 0.5701058201058202\n\nEpoch 00001: val_auc improved from -inf to 0.57011, saving model to models/best_model.hdf5\n13/13 [==============================] - 39s 3s/step - loss: 2.2712 - accuracy: 0.5725 - val_loss: 5.3443 - val_accuracy: 0.1075 - val_auc: 0.5701\nEpoch 2/10\n13/13 [==============================] - ETA: 0s - loss: 2.1631 - accuracy: 0.5591\nVal AUC for epoch1: 0.5852941176470587\n\nEpoch 00002: val_auc improved from 0.57011 to 0.58529, saving model to models/best_model.hdf5\n13/13 [==============================] - 25s 2s/step - loss: 2.1631 - accuracy: 0.5591 - val_loss: 3.5104 - val_accuracy: 0.0968 - val_auc: 0.5853\nEpoch 3/10\n13/13 [==============================] - ETA: 0s - loss: 2.0383 - accuracy: 0.6355\nVal AUC for epoch2: 0.6411764705882352\n\nEpoch 00003: val_auc improved from 0.58529 to 0.64118, saving model to models/best_model.hdf5\n13/13 [==============================] - 25s 2s/step - loss: 2.0383 - accuracy: 0.6355 - val_loss: 3.3596 - val_accuracy: 0.0860 - val_auc: 0.6412\nEpoch 4/10\n13/13 [==============================] - ETA: 0s - loss: 2.0308 - accuracy: 0.7118\nVal AUC for epoch3: 0.5180722891566265\n\nEpoch 00004: val_auc did not improve from 0.64118\n13/13 [==============================] - 23s 2s/step - loss: 2.0308 - accuracy: 0.7118 - val_loss: 3.2708 - val_accuracy: 0.0860 - val_auc: 0.5181\nEpoch 5/10\n13/13 [==============================] - ETA: 0s - loss: 2.0061 - accuracy: 0.7346\nVal AUC for epoch4: 0.5797619047619048\n\nEpoch 00005: val_auc did not improve from 0.64118\n13/13 [==============================] - 28s 2s/step - loss: 2.0061 - accuracy: 0.7346 - val_loss: 2.9860 - val_accuracy: 0.1075 - val_auc: 0.5798\nEpoch 6/10\n13/13 [==============================] - ETA: 0s - loss: 1.9883 - accuracy: 0.7365\nVal AUC for epoch5: 0.524547803617571\n\nEpoch 00006: val_auc did not improve from 0.64118\n13/13 [==============================] - 26s 2s/step - loss: 1.9883 - accuracy: 0.7365 - val_loss: 2.7612 - val_accuracy: 0.1170 - val_auc: 0.5245\nEpoch 7/10\n13/13 [==============================] - ETA: 0s - loss: 2.0141 - accuracy: 0.7414\nVal AUC for epoch6: 0.6718954248366014\n\nEpoch 00007: val_auc improved from 0.64118 to 0.67190, saving model to models/best_model.hdf5\n13/13 [==============================] - 20s 2s/step - loss: 2.0141 - accuracy: 0.7414 - val_loss: 2.6299 - val_accuracy: 0.1158 - val_auc: 0.6719\nEpoch 8/10\n13/13 [==============================] - ETA: 0s - loss: 2.0081 - accuracy: 0.7537\nVal AUC for epoch7: 0.6482683982683983\n\nEpoch 00008: val_auc did not improve from 0.67190\n13/13 [==============================] - 21s 2s/step - loss: 2.0081 - accuracy: 0.7537 - val_loss: 2.4338 - val_accuracy: 0.1277 - val_auc: 0.6483\nEpoch 9/10\n13/13 [==============================] - ETA: 0s - loss: 2.0309 - accuracy: 0.7475\nVal AUC for epoch8: 0.7307189542483661\n\nEpoch 00009: val_auc improved from 0.67190 to 0.73072, saving model to models/best_model.hdf5\n13/13 [==============================] - 21s 2s/step - loss: 2.0309 - accuracy: 0.7475 - val_loss: 2.3805 - val_accuracy: 0.1684 - val_auc: 0.7307\nEpoch 10/10\n13/13 [==============================] - ETA: 0s - loss: 2.0132 - accuracy: 0.7463\nVal AUC for epoch9: 0.7191176470588236\n\nEpoch 00010: val_auc did not improve from 0.73072\n13/13 [==============================] - 27s 2s/step - loss: 2.0132 - accuracy: 0.7463 - val_loss: 2.2621 - val_accuracy: 0.1915 - val_auc: 0.7191\n"
]
],
[
[
"## 5. Making a Prediction",
"_____no_output_____"
]
],
[
[
"val_model = resnet.ResnetBuilder.build_resnet_18((img_channels, img_rows, img_cols), nb_classes)\nval_model.load_weights('models/best_model.hdf5')",
"_____no_output_____"
],
[
"effusion_path = os.path.join(DATASET_PATH, disease_cls[0], '*')\neffusion = glob.glob(effusion_path)\neffusion = io.imread(effusion[-8])\nplt.imshow(effusion,cmap='gray')",
"_____no_output_____"
],
[
"img = preprocess_img(effusion[:, :, np.newaxis], 'validation')\nval_model.predict(img[np.newaxis,:])",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
cb88d323c736462d43ab501b58a3ca383c2bd612
| 5,774 |
ipynb
|
Jupyter Notebook
|
index.ipynb
|
gagandeepreehal/einsteinpy
|
d72d780f27a1a35938847e791872a8f9f27c71ca
|
[
"MIT"
] | null | null | null |
index.ipynb
|
gagandeepreehal/einsteinpy
|
d72d780f27a1a35938847e791872a8f9f27c71ca
|
[
"MIT"
] | 2 |
2019-04-08T17:39:50.000Z
|
2019-04-11T03:10:09.000Z
|
index.ipynb
|
gagandeepreehal/einsteinpy
|
d72d780f27a1a35938847e791872a8f9f27c71ca
|
[
"MIT"
] | null | null | null | 52.018018 | 240 | 0.712331 |
[
[
[
"# Gallery of examples\n\n\n\nHere you can browse a gallery of examples using EinsteinPy in the form of Jupyter notebooks.",
"_____no_output_____"
],
[
"## [Analyzing Earth using EinsteinPy!](docs/source/examples/Analyzing%20Earth%20using%20EinsteinPy!.ipynb)\n\n[](docs/source/examples/Analyzing%20Earth%20using%20EinsteinPy!.ipynb)\n\n\n## [Animations in EinsteinPy!](docs/source/examples/Animations%20in%20EinsteinPy.ipynb)\n\n[](docs/source/examples/Animations%20in%20EinsteinPy.ipynb)\n\n\n## [Einstein Tensor calculations using Symbolic module](docs/source/examples/Einstein%20Tensor%20symbolic%20calculation.ipynb)\n\n[](docs/source/examples/Einstein%20Tensor%20symbolic%20calculation.ipynb)\n\n\n## [Lambdify in Symbolic module](docs/source/examples/Lambdify%20symbolic%20calculation.ipynb)\n\n[](docs/source/examples/Lambdify%20symbolic%20calculation.ipynb)\n\n\n## [Playing with Contravariant and Covariant Indices in Tensors(Symbolic)](docs/source/examples/Playing%20with%20Contravariant%20and%20Covariant%20Indices%20in%20Tensors(Symbolic).ipynb)\n\n[](docs/source/examples/Playing%20with%20Contravariant%20and%20Covariant%20Indices%20in%20Tensors(Symbolic).ipynb)\n\n\n## [Predefined Metrics in Symbolic Module](docs/source/examples/Predefined%20Metrics%20in%20Symbolic%20Module.ipynb)\n\n[](docs/source/examples/Predefined%20Metrics%20in%20Symbolic%20Module.ipynb)\n\n\n## [Ricci Tensor and Scalar Curvature calculations using Symbolic module](docs/source/examples/Ricci%20Tensor%20and%20Scalar%20Curvature%20symbolic%20calculation.ipynb)\n\n[](docs/source/examples/Ricci%20Tensor%20and%20Scalar%20Curvature%20symbolic%20calculation.ipynb)\n<center><em>Gregorio Ricci-Curbastro</em></center>\n\n## [Shadow cast by an thin emission disk around a black hole](docs/source/examples/Shadow%20cast%20by%20an%20thin%20emission%20disk%20around%20a%20black%20hole.ipynb)\n\n[](docs/source/examples/Shadow%20cast%20by%20an%20thin%20emission%20disk%20around%20a%20black%20hole.ipynb)\n\n\n## [Spatial Hypersurface Embedding for Schwarzschild Space-Time!](docs/source/examples/Plotting%20spacial%20hypersurface%20embedding%20for%20schwarzschild%20spacetime.ipynb)\n\n[](docs/source/examples/Plotting%20spacial%20hypersurface%20embedding%20for%20schwarzschild%20spacetime.ipynb)\n\n\n## [Symbolically Understanding Christoffel Symbol and Riemann Metric Tensor using EinsteinPy](docs/source/examples/Symbolically%20Understanding%20Christoffel%20Symbol%20and%20Riemann%20Curvature%20Tensor%20using%20EinsteinPy.ipynb)\n\n[](docs/source/examples/Symbolically%20Understanding%20Christoffel%20Symbol%20and%20Riemann%20Curvature%20Tensor%20using%20EinsteinPy.ipynb)\n\n\n## [Visualizing Event Horizon and Ergosphere (Singularities) of Kerr Metric or Black Hole](docs/source/examples/Visualizing%20Event%20Horizon%20and%20Ergosphere%20(Singularities)%20of%20Kerr%20Metric%20or%20Black%20Hole.ipynb)\n\n[](docs/source/examples/Visualizing%20Event%20Horizon%20and%20Ergosphere%20(Singularities)%20of%20Kerr%20Metric%20or%20Black%20Hole.ipynb)\n\n\n## [Visualizing Frame Dragging in Kerr Spacetime](docs/source/examples/Visualizing%20Frame%20Dragging%20in%20Kerr%20Spacetime.ipynb)\n\n[](docs/source/examples/Visualizing%20Frame%20Dragging%20in%20Kerr%20Spacetime.ipynb)\n\n\n## [Visualizing Precession in Schwarzschild Spacetime](docs/source/examples/Visualizing%20Precession%20in%20Schwarzschild%20Spacetime.ipynb)\n\n[](docs/source/examples/Visualizing%20Precession%20in%20Schwarzschild%20Spacetime.ipynb)\n\n\n## [Weyl Tensor calculations using Symbolic module](docs/source/examples/Weyl%20Tensor%20symbolic%20calculation.ipynb)\n\n[](docs/source/examples/Weyl%20Tensor%20symbolic%20calculation.ipynb)\n<center><em>Hermann Weyl</em></center>",
"_____no_output_____"
]
]
] |
[
"markdown"
] |
[
[
"markdown",
"markdown"
]
] |
cb88d3cd4412fa62c1e137606a58944dbf2790b7
| 6,221 |
ipynb
|
Jupyter Notebook
|
digital-image-processing/notebooks/edges/plot_line_hough_transform.ipynb
|
sinamedialab/courses
|
720a78ebf4b4fb77f57a73870480233646f9a51d
|
[
"MIT"
] | null | null | null |
digital-image-processing/notebooks/edges/plot_line_hough_transform.ipynb
|
sinamedialab/courses
|
720a78ebf4b4fb77f57a73870480233646f9a51d
|
[
"MIT"
] | null | null | null |
digital-image-processing/notebooks/edges/plot_line_hough_transform.ipynb
|
sinamedialab/courses
|
720a78ebf4b4fb77f57a73870480233646f9a51d
|
[
"MIT"
] | null | null | null | 78.746835 | 2,556 | 0.656486 |
[
[
[
"%matplotlib inline",
"_____no_output_____"
]
],
[
[
"\n# Straight line Hough transform\n\nThe Hough transform in its simplest form is a method to detect straight lines\n[1]_.\n\nIn the following example, we construct an image with a line intersection. We\nthen use the `Hough transform <https://en.wikipedia.org/wiki/Hough_transform>`__.\nto explore a parameter space for straight lines that may run through the image.\n\n## Algorithm overview\n\nUsually, lines are parameterised as $y = mx + c$, with a gradient\n$m$ and y-intercept `c`. However, this would mean that $m$ goes to\ninfinity for vertical lines. Instead, we therefore construct a segment\nperpendicular to the line, leading to the origin. The line is represented by\nthe length of that segment, $r$, and the angle it makes with the x-axis,\n$\\theta$.\n\nThe Hough transform constructs a histogram array representing the parameter\nspace (i.e., an $M \\times N$ matrix, for $M$ different values of\nthe radius and $N$ different values of $\\theta$). For each\nparameter combination, $r$ and $\\theta$, we then find the number\nof non-zero pixels in the input image that would fall close to the\ncorresponding line, and increment the array at position $(r, \\theta)$\nappropriately.\n\nWe can think of each non-zero pixel \"voting\" for potential line candidates. The\nlocal maxima in the resulting histogram indicates the parameters of the most\nprobably lines. In our example, the maxima occur at 45 and 135 degrees,\ncorresponding to the normal vector angles of each line.\n\nAnother approach is the Progressive Probabilistic Hough Transform [2]_. It is\nbased on the assumption that using a random subset of voting points give a good\napproximation to the actual result, and that lines can be extracted during the\nvoting process by walking along connected components. This returns the\nbeginning and end of each line segment, which is useful.\n\nThe function `probabilistic_hough` has three parameters: a general threshold\nthat is applied to the Hough accumulator, a minimum line length and the line\ngap that influences line merging. In the example below, we find lines longer\nthan 10 with a gap less than 3 pixels.\n\n## References\n\n.. [1] Duda, R. O. and P. E. Hart, \"Use of the Hough Transformation to\n Detect Lines and Curves in Pictures,\" Comm. ACM, Vol. 15,\n pp. 11-15 (January, 1972)\n\n.. [2] C. Galamhos, J. Matas and J. Kittler,\"Progressive probabilistic\n Hough transform for line detection\", in IEEE Computer Society\n Conference on Computer Vision and Pattern Recognition, 1999.\n",
"_____no_output_____"
],
[
"### Line Hough Transform\n\n",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\nfrom skimage.transform import hough_line, hough_line_peaks\nfrom skimage.feature import canny\nfrom skimage import data\n\nimport matplotlib.pyplot as plt\nfrom matplotlib import cm\n\n\n# Constructing test image\nimage = np.zeros((200, 200))\nidx = np.arange(25, 175)\nimage[idx[::-1], idx] = 255\nimage[idx, idx] = 255\n\n# Classic straight-line Hough transform\n# Set a precision of 0.5 degree.\ntested_angles = np.linspace(-np.pi / 2, np.pi / 2, 360)\nh, theta, d = hough_line(image, theta=tested_angles)\n\n# Generating figure 1\nfig, axes = plt.subplots(1, 3, figsize=(15, 6))\nax = axes.ravel()\n\nax[0].imshow(image, cmap=cm.gray)\nax[0].set_title('Input image')\nax[0].set_axis_off()\n\nax[1].imshow(np.log(1 + h),\n extent=[np.rad2deg(theta[-1]), np.rad2deg(theta[0]), d[-1], d[0]],\n cmap=cm.gray, aspect=1/1.5)\nax[1].set_title('Hough transform')\nax[1].set_xlabel('Angles (degrees)')\nax[1].set_ylabel('Distance (pixels)')\nax[1].axis('image')\n\nax[2].imshow(image, cmap=cm.gray)\norigin = np.array((0, image.shape[1]))\nfor _, angle, dist in zip(*hough_line_peaks(h, theta, d)):\n y0, y1 = (dist - origin * np.cos(angle)) / np.sin(angle)\n ax[2].plot(origin, (y0, y1), '-r')\nax[2].set_xlim(origin)\nax[2].set_ylim((image.shape[0], 0))\nax[2].set_axis_off()\nax[2].set_title('Detected lines')\n\nplt.tight_layout()\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Probabilistic Hough Transform\n\n",
"_____no_output_____"
]
],
[
[
"from skimage.transform import probabilistic_hough_line\n\n# Line finding using the Probabilistic Hough Transform\nimage = data.camera()\nedges = canny(image, 2, 1, 25)\nlines = probabilistic_hough_line(edges, threshold=10, line_length=5,\n line_gap=3)\n\n# Generating figure 2\nfig, axes = plt.subplots(1, 3, figsize=(15, 5), sharex=True, sharey=True)\nax = axes.ravel()\n\nax[0].imshow(image, cmap=cm.gray)\nax[0].set_title('Input image')\n\nax[1].imshow(edges, cmap=cm.gray)\nax[1].set_title('Canny edges')\n\nax[2].imshow(edges * 0)\nfor line in lines:\n p0, p1 = line\n ax[2].plot((p0[0], p1[0]), (p0[1], p1[1]))\nax[2].set_xlim((0, image.shape[1]))\nax[2].set_ylim((image.shape[0], 0))\nax[2].set_title('Probabilistic Hough')\n\nfor a in ax:\n a.set_axis_off()\n\nplt.tight_layout()\nplt.show()",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb88dfd60c0c0b0feba617de9237f9e7c897fdcc
| 2,811 |
ipynb
|
Jupyter Notebook
|
Download_Prep_L8.ipynb
|
allenpope/Landsat8_Velocity_LarsenC
|
6a26d690b7fee0a7306dd234df40d8a32c15372a
|
[
"MIT"
] | 2 |
2017-07-10T12:03:07.000Z
|
2021-01-15T13:24:55.000Z
|
Download_Prep_L8.ipynb
|
allenpope/Landsat8_Velocity_LarsenC
|
6a26d690b7fee0a7306dd234df40d8a32c15372a
|
[
"MIT"
] | null | null | null |
Download_Prep_L8.ipynb
|
allenpope/Landsat8_Velocity_LarsenC
|
6a26d690b7fee0a7306dd234df40d8a32c15372a
|
[
"MIT"
] | 2 |
2018-07-24T14:13:42.000Z
|
2019-03-08T08:00:29.000Z
| 25.554545 | 101 | 0.499822 |
[
[
[
"#http://docs.python-guide.org/en/latest/dev/virtualenvs/\n#move to virtual environment and load it\n#os.system(\"cd /Users/apope/Documents/Landsat_downloads/\")\n#os.system(\"source venv/bin/activate\")",
"_____no_output_____"
],
[
"#Download Landsat Scenes\n\n#Import list from file\nin_file = \"/Users/apope/Dropbox/Transfer/To NSIDC/LANDSAT_LarsenC.csv\"\noutpath = \"/Users/apope/Desktop/pycorr/In/LarsenC/\"\n\nimport csv\nimport os\n\nwith open(in_file, 'rU') as inputfile:\n l = list(csv.reader(inputfile))\n\nscenes = list([item for sublist in l for item in sublist]) #list of lists to just a list \nprint \"Scenes:\"\nprint scenes\n\nfor x in scenes:\n os.system(\"landsat download -b 8 \" + x)\n \n \n#Unzip and Move Scenes as appropriate\nimport shutil\n\nos.chdir(\"/Users/apope/landsat/downloads/\")\n\nfor x in scenes:\n if os.path.isfile(x + \".tar.bz\") is True:\n print \"Processing \" + x\n os.system(\"tar -xjvf \" + x + \".tar.bz \" + x + \"_B8.TIF\")\n shutil.move(x + \"_B8.TIF\", outpath + x + \"_B8.TIF\")\n os.remove(x + \".tar.bz\")\n if os.path.exists(x) is True:\n shutil.rmtree(x)\n elif os.path.isfile(x + \"/\" + x + \"_B8.TIF\") is True:\n print \"Processing \" + x\n shutil.move(x + \"/\" + x + \"_B8.TIF\", outpath + x + \"_B8.TIF\")\n shutil.rmtree(x)\n else:\n print \"No File Named \" + x",
"Scenes:\n['LC82181062016326LGN00']\nProcessing LC82181062016326LGN00\n"
],
[
"#deactivate",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code"
]
] |
cb88e16f4520167eec9cef3d070ab197111f3e91
| 35,782 |
ipynb
|
Jupyter Notebook
|
code/.ipynb_checkpoints/ResultsAnalysis-checkpoint.ipynb
|
drigobon/FFC_Pentlandians_Code
|
ad20d22643688b991d17eade84407fbc24bf8750
|
[
"MIT"
] | 1 |
2019-03-11T21:39:58.000Z
|
2019-03-11T21:39:58.000Z
|
code/.ipynb_checkpoints/ResultsAnalysis-checkpoint.ipynb
|
drigobon/FFC_Pentlandians_Code
|
ad20d22643688b991d17eade84407fbc24bf8750
|
[
"MIT"
] | null | null | null |
code/.ipynb_checkpoints/ResultsAnalysis-checkpoint.ipynb
|
drigobon/FFC_Pentlandians_Code
|
ad20d22643688b991d17eade84407fbc24bf8750
|
[
"MIT"
] | 2 |
2017-11-08T16:33:22.000Z
|
2021-03-19T21:54:37.000Z
| 43.267231 | 594 | 0.597926 |
[
[
[
"# Purpose: Analyze results from Predictions Files created by Models\n# Inputs: Prediction files from Random Forest, Elastic Net, XGBoost, and Team Ensembles\n# Outputs: Figures (some included in the paper, some in SI)",
"_____no_output_____"
],
[
"import pandas as pd\nimport numpy as np\nimport seaborn as sns\nimport matplotlib.pyplot as plt\nimport astropy.stats as AS\nfrom scipy.stats.stats import pearsonr \nfrom os import listdir\nfrom sklearn.decomposition import PCA\n\n%matplotlib inline\n",
"_____no_output_____"
]
],
[
[
"## Reading Data, generating In-Sample Scores",
"_____no_output_____"
]
],
[
[
"name_dict = {'lassoRF_prediction': 'Lasso RF','elastic_prediction': 'Elastic Net','RF_prediction': 'Ensemble RF',\n 'LR_prediction': 'Ensemble LR','weighted_multiRF_prediction': 'Nested RF',\n 'weighted_avrg_prediction': 'Weighted Team Avg', 'avrg_prediction': 'Team Avg',\n 'xgboost_prediction': 'Gradient Boosted Tree'}",
"_____no_output_____"
],
[
"training=pd.read_csv('../data/train.csv',index_col = 'challengeID')\nbaseline=np.mean(training, axis=0)",
"_____no_output_____"
],
[
"BL_CV_scores = pd.DataFrame(columns = ['outcome','type','model','score_avg'])\n\nfor outcome in training.columns.values:\n y = training[outcome].dropna()\n y_hat = baseline[outcome]\n partition_scores = list()\n\n for i in range(10,110,10):\n bools = y.index<np.percentile(y.index,i)\n y_curr=y[bools]\n partition_scores.append(np.linalg.norm(y_curr-y_hat)/len(y_curr))\n\n bootstrapped_means = AS.bootstrap(np.array(partition_scores),samples = 10, bootnum = 100, bootfunc = np.mean)\n to_add = pd.DataFrame({'outcome':list(len(bootstrapped_means)*[outcome]),'type':len(bootstrapped_means)*['In-Sample Error'],'model':len(bootstrapped_means)*['Baseline'],'score_avg':bootstrapped_means})\n\n BL_CV_scores = BL_CV_scores.append(to_add, ignore_index = True)",
"_____no_output_____"
],
[
"name_dict",
"_____no_output_____"
],
[
"bootstrapped_scores_all = {}",
"_____no_output_____"
],
[
"for name in list(name_dict.keys()):\n model_name = name_dict[name]\n \n data=pd.read_csv(str('../output/final_pred/'+name+'.csv'), index_col = 'challengeID')\n\n CV_scores = pd.DataFrame(columns = ['outcome','type','model','score_avg'])\n for outcome in training.columns.values:\n y = training[outcome].dropna()\n y_hat = data[outcome][np.in1d(data.index,y.index)] \n partition_scores = list()\n\n for i in range(10,110,10):\n bools = y.index<np.percentile(y.index,i)\n y_curr=y[bools]\n y_hat_curr = y_hat[bools]\n partition_scores.append(np.linalg.norm(y_curr-y_hat_curr)/len(y_curr))\n\n bootstrapped_means = AS.bootstrap(np.array(partition_scores),samples = 10, bootnum = 100, bootfunc = np.mean)\n \n bootstrapped_means = (1-np.divide(bootstrapped_means,BL_CV_scores.score_avg[BL_CV_scores.outcome==outcome]))*100\n to_add = pd.DataFrame({'outcome':list(len(bootstrapped_means)*[outcome]),'type':len(bootstrapped_means)*['In-Sample Error'],'model':len(bootstrapped_means)*[model_name],'score_avg':bootstrapped_means})\n\n CV_scores = CV_scores.append(to_add, ignore_index = True)\n bootstrapped_scores_all[name] = CV_scores\n",
"_____no_output_____"
]
],
[
[
"## Individual Model Scores",
"_____no_output_____"
]
],
[
[
"GBT_CV = bootstrapped_scores_all['xgboost_prediction']\nGBT_leaderboard = pd.DataFrame({'outcome':['gpa','grit','materialHardship','eviction','layoff','jobTraining'],'type':6*['Leaderboard'],'model':6*['Gradient Boosted Tree'],'score_avg':[0.37543,0.22008,0.02437,0.05453,0.17406,0.19676]})\nGBT_holdout = pd.DataFrame({'outcome':['gpa','grit','materialHardship','eviction','layoff','jobTraining'],'type':6*['Holdout'],'model':6*['Gradient Boosted Tree'],'score_avg':[0.34379983,0.238180899,0.019950074,0.056877623,0.167392429,0.177202581]})\nGBT_scores = GBT_CV.append(GBT_leaderboard.append(GBT_holdout,ignore_index = True),ignore_index = True)\n\navrg_CV = bootstrapped_scores_all['avrg_prediction']\navrg_leaderboard = pd.DataFrame({'outcome':['gpa','grit','materialHardship','eviction','layoff','jobTraining'],'type':6*['Leaderboard'],'model':6*['Team Avg'],'score_avg':[0.36587,0.21287,0.02313,0.05025,0.17467,0.20058]})\navrg_holdout = pd.DataFrame({'outcome':['gpa','grit','materialHardship','eviction','layoff','jobTraining'],'type':6*['Holdout'],'model':6*['Team Avg'],'score_avg':[0.352115776,0.241462042,0.019888218,0.053480264,0.169287396,0.181767792]})\navrg_scores = avrg_CV.append(avrg_leaderboard.append(avrg_holdout,ignore_index = True),ignore_index = True)\n\nweighted_avrg_CV = bootstrapped_scores_all['weighted_avrg_prediction']\nweighted_avrg_leaderboard = pd.DataFrame({'outcome':['gpa','grit','materialHardship','eviction','layoff','jobTraining'],'type':6*['Leaderboard'],'model':6*['Weighted Team Avg'],'score_avg':[0.36587,0.21287,0.02301,0.04917,0.1696,0.19782]})\nweighted_avrg_holdout = pd.DataFrame({'outcome':['gpa','grit','materialHardship','eviction','layoff','jobTraining'],'type':6*['Holdout'],'model':6*['Weighted Team Avg'],'score_avg':[0.352115776,0.241462042,0.020189616,0.053818827,0.162462938,0.178098036]})\nweighted_avrg_scores = weighted_avrg_CV.append(weighted_avrg_leaderboard.append(weighted_avrg_holdout,ignore_index = True),ignore_index = True)\n\nmulti_RF_CV = bootstrapped_scores_all['weighted_multiRF_prediction']\nmulti_RF_leaderboard = pd.DataFrame({'outcome':['gpa','grit','materialHardship','eviction','layoff','jobTraining'],'type':6*['Leaderboard'],'model':6*['Nested RF'],'score_avg':[0.38766,0.22353,0.02542,0.05446,0.20228,0.22092]})\nmulti_RF_holdout = pd.DataFrame({'outcome':['gpa','grit','materialHardship','eviction','layoff','jobTraining'],'type':6*['Holdout'],'model':6*['Nested RF'],'score_avg':[0.365114483,0.248124154,0.021174361,0.063930882,0.207400541,0.191352482]})\nmulti_RF_scores = multi_RF_CV.append(multi_RF_leaderboard.append(multi_RF_holdout,ignore_index = True),ignore_index = True)\n\nLR_CV = bootstrapped_scores_all['LR_prediction']\nLR_leaderboard = pd.DataFrame({'outcome':['gpa','grit','materialHardship','eviction','layoff','jobTraining'],'type':6*['Leaderboard'],'model':6*['Ensemble LR'],'score_avg':[0.37674,0.2244,0.02715,0.05092,0.18341,0.22311]})\nLR_holdout = pd.DataFrame({'outcome':['gpa','grit','materialHardship','eviction','layoff','jobTraining'],'type':6*['Holdout'],'model':6*['Ensemble LR'],'score_avg':[0.364780108,0.247382526,0.021359837,0.058200047,0.181441591,0.194502527]})\nLR_scores = LR_CV.append(LR_leaderboard.append(LR_holdout,ignore_index = True),ignore_index = True)\n\nRF_CV = bootstrapped_scores_all['RF_prediction']\nRF_leaderboard = pd.DataFrame({'outcome':['gpa','grit','materialHardship','eviction','layoff','jobTraining'],'type':6*['Leaderboard'],'model':6*['Ensemble RF'],'score_avg':[0.38615,0.22342,0.02547,0.05475,0.20346,0.22135]})\nRF_holdout = pd.DataFrame({'outcome':['gpa','grit','materialHardship','eviction','layoff','jobTraining'],'type':6*['Holdout'],'model':6*['Ensemble RF'],'score_avg':[0.364609923,0.247940405,0.021135379,0.064494339,0.208869867,0.191742726]})\nRF_scores = RF_CV.append(RF_leaderboard.append(RF_holdout,ignore_index = True),ignore_index = True)\n\nlasso_RF_CV = bootstrapped_scores_all['lassoRF_prediction']\nlasso_RF_leaderboard = pd.DataFrame({'outcome':['gpa','grit','materialHardship','eviction','layoff','jobTraining'],'type':6*['Leaderboard'],'model':6*['Lasso RF'],'score_avg':[0.37483,0.21686,0.02519,0.05226,0.17223,0.20028]})\nlasso_RF_holdout = pd.DataFrame({'outcome':['gpa','grit','materialHardship','eviction','layoff','jobTraining'],'type':6*['Holdout'],'model':6*['Lasso RF'],'score_avg':[0.361450643,0.243745261,0.020491841,0.054397319,0.165154165,0.180446409]})\nlasso_scores = lasso_RF_CV.append(lasso_RF_leaderboard.append(lasso_RF_holdout,ignore_index = True),ignore_index = True)\n\neNet_CV = bootstrapped_scores_all['elastic_prediction']\neNet_leaderboard = pd.DataFrame({'outcome':['gpa','grit','materialHardship','eviction','layoff','jobTraining'],'type':6*['Leaderboard'],'model':6*['Elastic Net'],'score_avg':[0.36477,0.21252,0.02353,0.05341,0.17435,0.20224]})\neNet_holdout = pd.DataFrame({'outcome':['gpa','grit','materialHardship','eviction','layoff','jobTraining'],'type':6*['Holdout'],'model':6*['Elastic Net'],'score_avg':[0.350083,0.239361,0.019791,0.055458,0.167224,0.185329]})\neNet_scores = eNet_CV.append(eNet_leaderboard.append(eNet_holdout,ignore_index = True),ignore_index = True)\n\n#bools = np.in1d(eNet_scores.outcome,['gpa','grit','materialHardship'])\n#eNet_scores = eNet_scores.loc[bools]",
"_____no_output_____"
]
],
[
[
"## Score Aggregation and Plotting",
"_____no_output_____"
]
],
[
[
"## Baseline Scores:\nBL_LB = pd.DataFrame({'outcome':['gpa','grit','materialHardship','eviction','layoff','jobTraining'],'type':6*['Leaderboard'],'model':6*['Baseline'],'score_avg':[0.39273,0.21997,0.02880,0.05341,0.17435,0.20224]})\nBL_HO = pd.DataFrame({'outcome':['gpa','grit','materialHardship','eviction','layoff','jobTraining'],'type':6*['Holdout'],'model':6*['Baseline'],'score_avg':[0.425148881,0.252983596,0.024905617,0.055457913,0.167223718,0.185329492]})",
"_____no_output_____"
],
[
"scores_all = eNet_scores.append(lasso_scores.append(RF_scores.append(LR_scores.append(multi_RF_scores.append(weighted_avrg_scores.append(avrg_scores.append(GBT_scores,ignore_index = True),ignore_index = True),ignore_index = True),ignore_index = True),ignore_index = True),ignore_index = True), ignore_index = True)\nscores_ADJ = scores_all\n",
"_____no_output_____"
],
[
"scores = scores_all.loc[scores_all.type != 'In-Sample Error']\nfor OUTCOME in training.columns.values:\n f, ax = plt.subplots(nrows=1, ncols=1, figsize=(12, 7), sharex=True)\n\n temp=scores.loc[scores.outcome==OUTCOME]\n temp.score_avg.loc[temp.type=='Leaderboard']=(1-np.divide(temp.score_avg.loc[temp.type=='Leaderboard'],BL_LB.score_avg.loc[BL_LB.outcome==OUTCOME]))*100\n temp.score_avg.loc[temp.type=='Holdout']=(1-np.divide(temp.score_avg.loc[temp.type=='Holdout'],BL_HO.score_avg.loc[BL_HO.outcome==OUTCOME]))*100\n \n \n scores_ADJ.score_avg.loc[(scores_ADJ.outcome==OUTCOME) & (scores_ADJ.type=='Leaderboard')] = (1-np.divide(scores_ADJ.score_avg.loc[(scores_ADJ.outcome==OUTCOME) & (scores_ADJ.type=='Leaderboard')],BL_LB.score_avg.loc[BL_LB.outcome==OUTCOME]))*100\n scores_ADJ.score_avg.loc[(scores_ADJ.outcome==OUTCOME) & (scores_ADJ.type=='Holdout')] = (1-np.divide(scores_ADJ.score_avg.loc[(scores_ADJ.outcome==OUTCOME) & (scores_ADJ.type=='Holdout')],BL_HO.score_avg.loc[BL_HO.outcome==OUTCOME]))*100\n\n \n \n sns.barplot('model','score_avg',hue = 'type', data = temp, ci = 'sd', ax=ax)\n\n ax.set_title(str(OUTCOME))\n ax.set_xlabel('Model')\n ax.set_ylabel('Accuracy Improvement over Baseline (%)')\n plt.setp( ax.xaxis.get_majorticklabels(), rotation=30)\n ax.tick_params(labelsize=18)\n plt.savefig(str('../output/fig/'+OUTCOME+'.pdf'))\n bools_L = (scores.type=='Leaderboard') & (scores.outcome==OUTCOME)\n bools_H = (scores.type=='Holdout') & (scores.outcome==OUTCOME)\n print(OUTCOME)\n print('Best Leaderboard Model: ',scores.loc[(bools_L)&(scores.loc[bools_L].score_avg==max(scores.loc[bools_L].score_avg))].model)\n print('Best Holdout Model: ',scores.loc[(bools_H)&(scores.loc[bools_H].score_avg==max(scores.loc[bools_H].score_avg))].model)\n print()\n\n",
"_____no_output_____"
],
[
"scores = scores_all.loc[scores_all.type=='In-Sample Error']\nf, ax = plt.subplots(nrows=1, ncols=1, figsize=(24, 7), sharex=True)\n \nsns.barplot('model','score_avg', hue = 'outcome', data = scores, ci = 'sd', ax=ax)\n\nax.set_title('In-Sample Model Performance Improvement')\nax.set_xlabel('Model')\nax.set_ylabel('Accuracy Improvement over Baseline (%)')\nplt.setp( ax.xaxis.get_majorticklabels(), rotation=30)\nplt.ylim([-20,100])\nax.tick_params(labelsize=18)\nplt.savefig(str('../output/fig/ALL_IS.pdf'))",
"_____no_output_____"
],
[
"scores = scores_all.loc[scores_all.type=='In-Sample Error']\nfor OUTCOME in training.columns.values:\n f, ax = plt.subplots(nrows=1, ncols=1, figsize=(12, 7), sharex=True)\n\n temp=scores.loc[scores.outcome==OUTCOME]\n \n sns.barplot('model','score_avg', data = temp, ci = 'sd', ax=ax, color = 'red')\n\n ax.set_title(str(OUTCOME))\n ax.set_xlabel('Model')\n ax.set_ylabel('Accuracy Improvement over Baseline (%)')\n plt.setp( ax.xaxis.get_majorticklabels(), rotation=30)\n ax.tick_params(labelsize=18)\n plt.savefig(str('../output/fig/'+OUTCOME+'_IS.pdf'))\n bools_L = (scores.type=='Leaderboard') & (scores.outcome==OUTCOME)\n bools_H = (scores.type=='Holdout') & (scores.outcome==OUTCOME)\n \n\n",
"_____no_output_____"
]
],
[
[
"# Data Partition Performance",
"_____no_output_____"
]
],
[
[
"scores_PLT = scores_ADJ\n\nscores_PLT = scores_PLT.loc[~((scores_ADJ.model=='Elastic Net') & np.in1d(scores_ADJ.outcome,['eviction','layoff','jobTraining']))]\nscores_PLT['color'] = [-1]*np.shape(scores_PLT)[0]\n\nfor i,OUTCOME in enumerate(['gpa', 'grit', 'materialHardship', 'eviction', 'layoff', 'jobTraining']):\n scores_PLT.color.loc[scores_PLT.outcome==OUTCOME] = i",
"_____no_output_____"
],
[
"# LEADERBOARD vs HOLDOUT\n\n\nscores_X = scores_PLT.loc[scores_PLT.type=='Leaderboard']\nscores_Y = scores_PLT.loc[scores_PLT.type=='Holdout'] \n\ntxt = [str(a) for a,b in zip(scores_X.model,scores_X.outcome)]\n\n\nf, ax = plt.subplots(nrows=1, ncols=1, figsize=(12, 12), sharex=True)\ncolors = ['red','blue','green','black','yellow','orange']\nfor i in range(6):\n corr_temp = np.round(pearsonr(scores_X.score_avg.loc[scores_X.color==i],\n scores_Y.score_avg.loc[scores_Y.color==i]),decimals = 3)\n plt.scatter(x = scores_X.score_avg.loc[scores_X.color==i], \n s=20, y = scores_Y.score_avg.loc[scores_Y.color==i],\n c = colors[i],label=str(scores_X.outcome.loc[scores_X.color==i].iloc[0])+': r^2='+str(corr_temp[0])+' p='+str(corr_temp[1])) \n print(i)\n print(len(scores_X.score_avg.loc[scores_X.color==i]),\n len(scores_Y.score_avg.loc[scores_Y.color==i]))\nax.set_xlabel('Leaderboard Improvement Over Baseline (%)')\nax.set_ylabel('Holdout Improvement Over Baseline (%)')\nax.tick_params(labelsize=18)\nplt.ylim([-26, 22])\nplt.xlim([-26, 22])\nax.plot([-26,22],[-26,22], 'k-')\nax.legend()\n\n\nfor i,n in enumerate(txt):\n ax.annotate(n,(scores_X.score_avg.iloc[i],scores_Y.score_avg.iloc[i]),\n size = 10,textcoords='data')\n\nplt.savefig(str('../output/fig/LB_vs_HO.pdf'))\n",
"_____no_output_____"
],
[
"# LEADERBOARD VS IN-SAMPLE\n\n\nscores_X = scores_PLT.loc[scores_PLT.type=='Leaderboard']\nscores_Y = scores_PLT.loc[scores_PLT.type=='In-Sample Error'] \nscores_Y = pd.DataFrame(scores_Y.groupby([scores_Y.model,scores_Y.outcome]).mean())\n\n\ntxt = [str(a) for a,b in zip(scores_X.model,scores_X.outcome)]\n\n\nf, ax = plt.subplots(nrows=1, ncols=1, figsize=(12, 12), sharex=True)\ncolors = ['red','blue','green','black','yellow','orange']\nfor i in range(6):\n corr_temp = np.round(pearsonr(scores_X.score_avg.loc[scores_X.color==i],\n scores_Y.score_avg.loc[scores_Y.color==i]),decimals = 3)\n plt.scatter(x = scores_X.score_avg.loc[scores_X.color==i], \n s=20, y = scores_Y.score_avg.loc[scores_Y.color==i],\n c = colors[i],label=str(scores_X.outcome.loc[scores_X.color==i].iloc[0])+': r^2='+str(corr_temp[0])+' p='+str(corr_temp[1])) \n print(i)\n print(len(scores_X.score_avg.loc[scores_X.color==i]),\n len(scores_Y.score_avg.loc[scores_Y.color==i]))\nax.set_xlabel('Leaderboard Improvement Over Baseline (%)')\nax.set_ylabel('In-Sample Error Improvement Over Baseline (%)')\nax.tick_params(labelsize=18)\n#plt.ylim([-26, 22])\n#plt.xlim([-26, 22])\n#ax.plot([-26,22],[-26,22], 'k-')\nax.legend()\n\n\nfor i,n in enumerate(txt):\n ax.annotate(n,(scores_X.score_avg.iloc[i],scores_Y.score_avg.iloc[i]),\n size = 10,textcoords='data')\n\nplt.savefig(str('../output/fig/LB_vs_IS.pdf'))",
"_____no_output_____"
],
[
"# HOLDOUT VS IN-SAMPLE\n\n\nscores_X = scores_PLT.loc[scores_PLT.type=='Holdout']\nscores_Y = scores_PLT.loc[scores_PLT.type=='In-Sample Error'] \nscores_Y = scores_Y.groupby([scores_Y.model,scores_Y.outcome]).mean().reset_index()\n# UNCOMMENT if STD\n#scores_Y.color = [0, 1, 2, 3, 0, 1, 5, 4, 2, 3, 0, 1, 5, 4, 2, 3, 0, 1, 5, 4, 2, 3, 0,\n# 1, 5, 4, 2, 3, 0, 1, 5, 4, 2, 3, 0, 1, 5, 4, 2, 3, 0, 1, 5, 4, 2]\n\ntxt = [str(a) for a,b in zip(scores_X.model,scores_X.outcome)]\n\n\nf, ax = plt.subplots(nrows=1, ncols=1, figsize=(12, 12), sharex=True)\ncolors = ['red','blue','green','black','yellow','orange']\nfor i in range(6):\n corr_temp = np.round(pearsonr(scores_X.score_avg.loc[scores_X.color==i],\n scores_Y.score_avg.loc[scores_Y.color==i]),decimals = 3)\n plt.scatter(x = scores_X.score_avg.loc[scores_X.color==i], \n s=20, y = scores_Y.score_avg.loc[scores_Y.color==i],\n c = colors[i],label=str(scores_X.outcome.loc[scores_X.color==i].iloc[0])+': r^2='+str(corr_temp[0])+' p='+str(corr_temp[1])) \n print(i)\n print(len(scores_X.score_avg.loc[scores_X.color==i]),\n len(scores_Y.score_avg.loc[scores_Y.color==i]))\nax.set_xlabel('Holdout Improvement Over Baseline (%)')\nax.set_ylabel('In-Sample Error Improvement Over Baseline (%)')\nax.tick_params(labelsize=18)\n#plt.ylim([-26, 22])\n#plt.xlim([-26, 22])\n#ax.plot([-26,22],[-26,22], 'k-')\nax.legend()\n\n\nfor i,n in enumerate(txt):\n ax.annotate(n,(scores_X.score_avg.iloc[i],scores_Y.score_avg.iloc[i]),\n size = 10,textcoords='data')\n\nplt.savefig(str('../output/fig/HO_vs_IS.pdf'))",
"_____no_output_____"
]
],
[
[
"### Bootstrapping Correlation Values",
"_____no_output_____"
]
],
[
[
"bootnum = 10000\n\nall_keys_boot = ['gpa']*bootnum\ntemp = ['grit']*bootnum\nall_keys_boot.extend(temp)\ntemp = ['materialHardship']*bootnum\nall_keys_boot.extend(temp)\ntemp = ['eviction']*bootnum\nall_keys_boot.extend(temp)\ntemp = ['layoff']*bootnum\nall_keys_boot.extend(temp)\ntemp = ['jobTraining']*bootnum\nall_keys_boot.extend(temp)\ntemp = ['overall']*bootnum\nall_keys_boot.extend(temp)\n\nscores_ADJ = scores_all\n\nkeys = ['gpa', 'grit', 'materialHardship', 'eviction', 'layoff', 'jobTraining','overall']\n\n\nt1 = ['In-Sample Error']*14\ntemp = ['Leaderboard']*7\nt1.extend(temp)\nt2 = ['Leaderboard']*7\ntemp = ['Holdout']*14\nt2.extend(temp)\n\nall_keys_boot = ['gpa']*bootnum\ntemp = ['grit']*bootnum\nall_keys_boot.extend(temp)\ntemp = ['materialHardship']*bootnum\nall_keys_boot.extend(temp)\ntemp = ['eviction']*bootnum\nall_keys_boot.extend(temp)\ntemp = ['layoff']*bootnum\nall_keys_boot.extend(temp)\ntemp = ['jobTraining']*bootnum\nall_keys_boot.extend(temp)\ntemp = ['overall']*bootnum\nall_keys_boot.extend(temp)\n\ndf_full = pd.DataFrame(columns = ['T1-T2', 'condition', 'avg_corr','sd_corr'])\n\n\nfor [T1,T2] in [['In-Sample Error','Leaderboard'],['In-Sample Error','Holdout'],['Leaderboard','Holdout']]:\n\n X_type = scores_ADJ.loc[scores_ADJ.type==T1]\n Y_type = scores_ADJ.loc[scores_ADJ.type==T2]\n\n avg_corr = list([])\n\n # For Ind. Outcomes\n for OUTCOME in ['gpa', 'grit', 'materialHardship', 'eviction', 'layoff', 'jobTraining']:\n corr = np.zeros(bootnum)\n\n X_OC = X_type.loc[X_type.outcome==OUTCOME]\n Y_OC = Y_type.loc[Y_type.outcome==OUTCOME]\n\n X_curr = X_OC.groupby(X_OC.model).score_avg.mean()\n Y_curr = Y_OC.groupby(Y_OC.model).score_avg.mean()\n\n\n for i in range(bootnum):\n index = np.random.choice(list(range(len(X_curr))),len(X_curr))\n avg_corr.append(pearsonr(X_curr[index].values,Y_curr[index].values)[0])\n\n # For Overall\n X_curr = X_type.groupby([X_type.model,X_type.outcome]).score_avg.mean()\n Y_curr = Y_type.groupby([Y_type.model,Y_type.outcome]).score_avg.mean()\n corr = np.zeros(bootnum)\n\n for i in range(bootnum):\n index = np.random.choice(list(range(len(X_curr))),len(X_curr))\n avg_corr.append(pearsonr(X_curr[index].values,Y_curr[index].values)[0])\n\n to_add = pd.DataFrame({'T1-T2':7*bootnum*[str(T1)+' w/ '+str(T2)], 'condition': all_keys_boot,\n 'avg_corr':avg_corr})\n \n df_full = df_full.append(to_add)\n",
"_____no_output_____"
],
[
"f, ax = plt.subplots(nrows=1, ncols=1, figsize=(12, 7), sharex=True)\n \nsns.barplot('T1-T2','avg_corr', hue = 'condition', data = df_full, ci = 'sd', ax=ax)\n\nax.set_title('Correlation Comparison')\nax.set_xlabel('Data Partitions Compared')\nax.set_ylabel('Avg. Correlation')\nplt.setp( ax.xaxis.get_majorticklabels(), rotation=30)\nplt.ylim([-1.3,1.2])\nax.tick_params(labelsize=18)\nplt.savefig(str('../output/fig/Correlation_Comparison.pdf'))\n",
"_____no_output_____"
],
[
"f, ax = plt.subplots(nrows=1, ncols=1, figsize=(12, 7), sharex=True)\n \nsns.barplot('T1-T2','avg_corr', hue = 'condition', data = df_full.loc[df_full.condition=='overall'], ci = 'sd', ax=ax)\n\nax.set_title('Correlation Comparison')\nax.set_xlabel('Data Partitions Compared')\nax.set_ylabel('Avg. Correlation')\nplt.setp( ax.xaxis.get_majorticklabels(), rotation=30)\nplt.ylim([0,1])\nax.tick_params(labelsize=18)\nplt.savefig(str('../output/fig/Correlations_Overall.pdf'))",
"_____no_output_____"
]
],
[
[
"## Feature Importance XGBoost",
"_____no_output_____"
]
],
[
[
"father = pd.DataFrame({'outcome': ['gpa','eviction','grit','materialHardship','jobTraining','layoff'],\n 'characteristic': 6*['Father'],'score': [0.199531305,0.140893472,0.221546773,0.1923971,0.130434782,0.27181208]})\n\nhomevisit = pd.DataFrame({'outcome': ['gpa','eviction','grit','materialHardship','jobTraining','layoff'],\n 'characteristic': 6*['Home Visit'],'score': [0.203213929,0.209621994,0.189125295,0.112949541,0.036789297,0.187919463]})\n\nchild = pd.DataFrame({'outcome': ['gpa','eviction','grit','materialHardship','jobTraining','layoff'],\n 'characteristic': 6*['Child'],'score': [0.044861065,0.003436426,0.082404594,0.01572542,0.006688963,0.023489933]})\n\nkinder = pd.DataFrame({'outcome': ['gpa','eviction','grit','materialHardship','jobTraining','layoff'],\n 'characteristic': 6*['Kindergarden'],'score': [0.003347841,0.003436426,0.00810537,0.008432472,0.003344482,0.006711409]})\n\nmother = pd.DataFrame({'outcome': ['gpa','eviction','grit','materialHardship','jobTraining','layoff'],\n 'characteristic': 6*['Mother'],'score': [0.349849352,0.515463913,0.360351229,0.569032313,0.66889632,0.395973155]})\n\nother = pd.DataFrame({'outcome': ['gpa','eviction','grit','materialHardship','jobTraining','layoff'],\n 'characteristic': 6*['Other'],'score': [0.016069635,0.01718213,0.003377237,0.0097999,0.006688963,0.016778523]})\n\ncare = pd.DataFrame({'outcome': ['gpa','eviction','grit','materialHardship','jobTraining','layoff'],\n 'characteristic': 6*['Caregiver'],'score': [0.085369937,0.048109966,0.10570753,0.060713797,0.140468227,0.080536912]})\n\nteacher = pd.DataFrame({'outcome': ['gpa','eviction','grit','materialHardship','jobTraining','layoff'],\n 'characteristic': 6*['Teacher'],'score': [0.087378641,0.058419244,0.023302938,0.02306395,0.006688963,0.016778524]})\n\n\nwav1 = pd.DataFrame({'outcome': ['gpa','eviction','grit','materialHardship','jobTraining','layoff'],\n 'characteristic': 6*['Wave 1'],'score': [0.109809175,0.048109966,0.101654846,0.317288843,0.046822742,0.104026846]})\n\nwav2 = pd.DataFrame({'outcome': ['gpa','eviction','grit','materialHardship','jobTraining','layoff'],\n 'characteristic': 6*['Wave 2'],'score': [0.126548378,0.085910654,0.125295507,0.122612698,0.117056855,0.073825504]})\n\nwav3 = pd.DataFrame({'outcome': ['gpa','eviction','grit','materialHardship','jobTraining','layoff'],\n 'characteristic': 6*['Wave 3'],'score': [0.189822567,0.206185568,0.173252278,0.162496011,0.143812707,0.271812079]})\n\nwav4 = pd.DataFrame({'outcome': ['gpa','eviction','grit','materialHardship','jobTraining','layoff'],\n 'characteristic': 6*['Wave 4'],'score': [0.172079012,0.230240552,0.205336034,0.166826199,0.217391305,0.241610739]})\n\nwav5 = pd.DataFrame({'outcome': ['gpa','eviction','grit','materialHardship','jobTraining','layoff'],\n 'characteristic': 6*['Wave 5'],'score': [0.388014734,0.422680407,0.380276931,0.214458269,0.471571907,0.302013422]})\n\n\n\n\n\nwho_df = pd.concat([mother,father,care,homevisit,child,teacher,kinder,other],ignore_index = True)\nwhen_df = pd.concat([wav1,wav2,wav3,wav4,wav5],ignore_index = True)",
"_____no_output_____"
],
[
"f, ax = plt.subplots(nrows=1, ncols=1, figsize=(12, 5), sharex=True)\n\nsns.barplot('characteristic','score', hue = 'outcome', data = who_df,\n ci = None,ax=ax)\n\nax.set_ylabel('Feature Importance (Sum)')\nax.tick_params(labelsize=13)\nax.set_ylim(0,0.7)\nplt.savefig('../output/fig/Who_Feature_Importance.pdf')\n\n\nf, ax = plt.subplots(nrows=1, ncols=1, figsize=(12, 5), sharex=True)\n\nsns.barplot('characteristic','score', hue = 'outcome', data = when_df,\n ci = None,ax=ax)\n\nax.set_ylabel('Feature Importance (Sum)')\nax.tick_params(labelsize=13)\nax.set_ylim(0,0.7)\nplt.savefig('../output/fig/When_Feature_Importance.pdf')",
"_____no_output_____"
]
],
[
[
"## Comparison of Feature Selection Methods",
"_____no_output_____"
]
],
[
[
"LASSO_files = listdir('../output/LASSO_ALL/')\nMI_files = ['data_univariate_feature_selection_5.csv','data_univariate_feature_selection_15.csv','data_univariate_feature_selection_50.csv','data_univariate_feature_selection_100.csv','data_univariate_feature_selection_200.csv','data_univariate_feature_selection_300.csv','data_univariate_feature_selection_500.csv','data_univariate_feature_selection_700.csv','data_univariate_feature_selection_1000.csv','data_univariate_feature_selection_1500.csv','data_univariate_feature_selection_2000.csv','data_univariate_feature_selection_3000.csv','data_univariate_feature_selection_4000.csv']\n\nmsk = [i!='.DS_Store' for i in LASSO_files]\nLASSO_files = [i for i,j in zip(LASSO_files,msk) if j]\nLASSO_files = np.sort(LASSO_files)\n\nMI_file = MI_files[0]\nL_file = LASSO_files[0]\n\nperc_similar = np.zeros((len(LASSO_files),len(MI_files)))\nPC1_corr = np.zeros((len(LASSO_files),len(MI_files)))\nL_names = []\nMI_names = []\n\nfor i,L_file in enumerate(LASSO_files):\n temp_L = pd.read_csv(('../output/LASSO_ALL/'+L_file))\n L_names.append(np.shape(temp_L.columns.values)[0])\n L_PC = PCA(n_components=2).fit_transform(temp_L)\n\n for j,MI_file in enumerate(MI_files):\n temp_M = pd.read_csv(('../output/MI/'+MI_file))\n MI_names.append(np.shape(temp_M.columns.values)[0])\n MI_PC = PCA(n_components=2).fit_transform(temp_M)\n\n PC1_corr[i,j] = pearsonr(L_PC[:,0],MI_PC[:,0])[0]\n perc_similar[i,j]= sum(np.in1d(temp_L.columns.values,temp_M.columns.values))\n \n \n\ndata_named = pd.DataFrame(perc_similar,index = L_names, columns = np.unique(MI_names))\ncolumns = data_named.columns.tolist()\ncolumns = columns[::-1]\ndata_named = data_named[columns]\n\nf, ax = plt.subplots(nrows=1, ncols=1, figsize=(15, 15), sharex=True)\nsns.heatmap(data_named, annot = True)\nplt.savefig('../output/fig/feature_heatmap.png')\n\n\n\n\nL_names = [str('r^2='+str(i)) for i in np.linspace(0.1,0.9,9)]\nMI_names = [str('K='+str(i)) for i in [5,15,50,100,200,300,500,700,1000,1500,2000,3000,4000]]\ndata_PC = pd.DataFrame(PC1_corr,index = L_names, columns = MI_names)\ncolumns = data_named.columns.tolist()\ncolumns = columns[::-1]\ndata_named = data_named[columns]\n\nf, ax = plt.subplots(nrows=1, ncols=1, figsize=(15, 15), sharex=True)\nsns.heatmap(data_PC, annot = True)\nplt.savefig('../output/fig/PC1_heatmap.png')\n\n",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb88e26f17db341ffce4843d0080491efd1fa910
| 939,894 |
ipynb
|
Jupyter Notebook
|
sentiment-analysis-network/Sentiment_Classification_Projects.ipynb
|
xxiMiaxx/deep-learning-v2-pytorch
|
0efbd54935c8d0bf214e01627da973f260f5bc90
|
[
"MIT"
] | null | null | null |
sentiment-analysis-network/Sentiment_Classification_Projects.ipynb
|
xxiMiaxx/deep-learning-v2-pytorch
|
0efbd54935c8d0bf214e01627da973f260f5bc90
|
[
"MIT"
] | null | null | null |
sentiment-analysis-network/Sentiment_Classification_Projects.ipynb
|
xxiMiaxx/deep-learning-v2-pytorch
|
0efbd54935c8d0bf214e01627da973f260f5bc90
|
[
"MIT"
] | null | null | null | 81.123252 | 46,924 | 0.714228 |
[
[
[
"# Sentiment Classification & How To \"Frame Problems\" for a Neural Network\n\nby Andrew Trask\n\n- **Twitter**: @iamtrask\n- **Blog**: http://iamtrask.github.io",
"_____no_output_____"
],
[
"### What You Should Already Know\n\n- neural networks, forward and back-propagation\n- stochastic gradient descent\n- mean squared error\n- and train/test splits\n\n### Where to Get Help if You Need it\n- Re-watch previous Udacity Lectures\n- Leverage the recommended Course Reading Material - [Grokking Deep Learning](https://www.manning.com/books/grokking-deep-learning) (Check inside your classroom for a discount code)\n- Shoot me a tweet @iamtrask\n\n\n### Tutorial Outline:\n\n- Intro: The Importance of \"Framing a Problem\" (this lesson)\n\n- [Curate a Dataset](#lesson_1)\n- [Developing a \"Predictive Theory\"](#lesson_2)\n- [**PROJECT 1**: Quick Theory Validation](#project_1)\n\n\n- [Transforming Text to Numbers](#lesson_3)\n- [**PROJECT 2**: Creating the Input/Output Data](#project_2)\n\n\n- Putting it all together in a Neural Network (video only - nothing in notebook)\n- [**PROJECT 3**: Building our Neural Network](#project_3)\n\n\n- [Understanding Neural Noise](#lesson_4)\n- [**PROJECT 4**: Making Learning Faster by Reducing Noise](#project_4)\n\n\n- [Analyzing Inefficiencies in our Network](#lesson_5)\n- [**PROJECT 5**: Making our Network Train and Run Faster](#project_5)\n\n\n- [Further Noise Reduction](#lesson_6)\n- [**PROJECT 6**: Reducing Noise by Strategically Reducing the Vocabulary](#project_6)\n\n\n- [Analysis: What's going on in the weights?](#lesson_7)",
"_____no_output_____"
],
[
"# Lesson: Curate a Dataset<a id='lesson_1'></a>\nThe cells from here until Project 1 include code Andrew shows in the videos leading up to mini project 1. We've included them so you can run the code along with the videos without having to type in everything.",
"_____no_output_____"
]
],
[
[
"def pretty_print_review_and_label(i):\n print(labels[i] + \"\\t:\\t\" + reviews[i][:80] + \"...\")\n\ng = open('reviews.txt','r') # What we know!\nreviews = list(map(lambda x:x[:-1],g.readlines()))\ng.close()\n\ng = open('labels.txt','r') # What we WANT to know!\nlabels = list(map(lambda x:x[:-1].upper(),g.readlines()))\ng.close()",
"_____no_output_____"
]
],
[
[
"**Note:** The data in `reviews.txt` we're using has already been preprocessed a bit and contains only lower case characters. If we were working from raw data, where we didn't know it was all lower case, we would want to add a step here to convert it. That's so we treat different variations of the same word, like `The`, `the`, and `THE`, all the same way.",
"_____no_output_____"
]
],
[
[
"len(reviews)",
"_____no_output_____"
],
[
"reviews[0]",
"_____no_output_____"
],
[
"labels[0]",
"_____no_output_____"
]
],
[
[
"# Lesson: Develop a Predictive Theory<a id='lesson_2'></a>",
"_____no_output_____"
]
],
[
[
"print(\"labels.txt \\t : \\t reviews.txt\\n\")\npretty_print_review_and_label(2137)\npretty_print_review_and_label(12816)\npretty_print_review_and_label(6267)\npretty_print_review_and_label(21934)\npretty_print_review_and_label(5297)\npretty_print_review_and_label(4998)",
"labels.txt \t : \t reviews.txt\n\nNEGATIVE\t:\tthis movie is terrible but it has some good effects . ...\nPOSITIVE\t:\tadrian pasdar is excellent is this film . he makes a fascinating woman . ...\nNEGATIVE\t:\tcomment this movie is impossible . is terrible very improbable bad interpretat...\nPOSITIVE\t:\texcellent episode movie ala pulp fiction . days suicides . it doesnt get more...\nNEGATIVE\t:\tif you haven t seen this it s terrible . it is pure trash . i saw this about ...\nPOSITIVE\t:\tthis schiffer guy is a real genius the movie is of excellent quality and both e...\n"
]
],
[
[
"# Project 1: Quick Theory Validation<a id='project_1'></a>\n\nThere are multiple ways to implement these projects, but in order to get your code closer to what Andrew shows in his solutions, we've provided some hints and starter code throughout this notebook.\n\nYou'll find the [Counter](https://docs.python.org/2/library/collections.html#collections.Counter) class to be useful in this exercise, as well as the [numpy](https://docs.scipy.org/doc/numpy/reference/) library.",
"_____no_output_____"
]
],
[
[
"from collections import Counter\nimport numpy as np",
"_____no_output_____"
]
],
[
[
"We'll create three `Counter` objects, one for words from postive reviews, one for words from negative reviews, and one for all the words.",
"_____no_output_____"
]
],
[
[
"# Create three Counter objects to store positive, negative and total counts\npositive_counts = Counter()\nnegative_counts = Counter()\ntotal_counts = Counter()",
"_____no_output_____"
]
],
[
[
"**TODO:** Examine all the reviews. For each word in a positive review, increase the count for that word in both your positive counter and the total words counter; likewise, for each word in a negative review, increase the count for that word in both your negative counter and the total words counter.\n\n**Note:** Throughout these projects, you should use `split(' ')` to divide a piece of text (such as a review) into individual words. If you use `split()` instead, you'll get slightly different results than what the videos and solutions show.",
"_____no_output_____"
]
],
[
[
"# TODO: Loop over all the words in all the reviews and increment the counts in the appropriate counter objects\nfor i in range(len(reviews)):\n if labels[i]=='POSITIVE':\n every_word_in_reviews = reviews[i].split(' ')\n for word in every_word_in_reviews:\n positive_counts[word] += 1\n total_counts[word] += 1\n else:\n every_word_in_reviews = reviews[i].split(' ')\n for word in every_word_in_reviews:\n negative_counts[word]+= 1\n total_counts[word] += 1\n ",
"_____no_output_____"
]
],
[
[
"Run the following two cells to list the words used in positive reviews and negative reviews, respectively, ordered from most to least commonly used. ",
"_____no_output_____"
]
],
[
[
"# Examine the counts of the most common words in positive reviews\npositive_counts.most_common()",
"_____no_output_____"
],
[
"# Examine the counts of the most common words in negative reviews\nnegative_counts.most_common()",
"_____no_output_____"
]
],
[
[
"As you can see, common words like \"the\" appear very often in both positive and negative reviews. Instead of finding the most common words in positive or negative reviews, what you really want are the words found in positive reviews more often than in negative reviews, and vice versa. To accomplish this, you'll need to calculate the **ratios** of word usage between positive and negative reviews.\n\n**TODO:** Check all the words you've seen and calculate the ratio of postive to negative uses and store that ratio in `pos_neg_ratios`. \n>Hint: the positive-to-negative ratio for a given word can be calculated with `positive_counts[word] / float(negative_counts[word]+1)`. Notice the `+1` in the denominator – that ensures we don't divide by zero for words that are only seen in positive reviews.",
"_____no_output_____"
]
],
[
[
"# Create Counter object to store positive/negative ratios\npos_neg_ratios = Counter()\n\n# TODO: Calculate the ratios of positive and negative uses of the most common words\n# Consider words to be \"common\" if they've been used at least 100 times\nfor term,cnt in list(total_counts.most_common()):\n if(cnt > 100):\n pos_neg_ratio = positive_counts[term] / float(negative_counts[term]+1)\n pos_neg_ratios[term] = pos_neg_ratio",
"_____no_output_____"
]
],
[
[
"Examine the ratios you've calculated for a few words:",
"_____no_output_____"
]
],
[
[
"print(\"Pos-to-neg ratio for 'the' = {}\".format(pos_neg_ratios[\"the\"]))\nprint(\"Pos-to-neg ratio for 'amazing' = {}\".format(pos_neg_ratios[\"amazing\"]))\nprint(\"Pos-to-neg ratio for 'terrible' = {}\".format(pos_neg_ratios[\"terrible\"]))",
"Pos-to-neg ratio for 'the' = 1.0607993145235326\nPos-to-neg ratio for 'amazing' = 4.022813688212928\nPos-to-neg ratio for 'terrible' = 0.17744252873563218\n"
]
],
[
[
"Looking closely at the values you just calculated, we see the following:\n\n* Words that you would expect to see more often in positive reviews – like \"amazing\" – have a ratio greater than 1. The more skewed a word is toward postive, the farther from 1 its positive-to-negative ratio will be.\n* Words that you would expect to see more often in negative reviews – like \"terrible\" – have positive values that are less than 1. The more skewed a word is toward negative, the closer to zero its positive-to-negative ratio will be.\n* Neutral words, which don't really convey any sentiment because you would expect to see them in all sorts of reviews – like \"the\" – have values very close to 1. A perfectly neutral word – one that was used in exactly the same number of positive reviews as negative reviews – would be almost exactly 1. The `+1` we suggested you add to the denominator slightly biases words toward negative, but it won't matter because it will be a tiny bias and later we'll be ignoring words that are too close to neutral anyway.\n\nOk, the ratios tell us which words are used more often in postive or negative reviews, but the specific values we've calculated are a bit difficult to work with. A very positive word like \"amazing\" has a value above 4, whereas a very negative word like \"terrible\" has a value around 0.18. Those values aren't easy to compare for a couple of reasons:\n\n* Right now, 1 is considered neutral, but the absolute value of the postive-to-negative rations of very postive words is larger than the absolute value of the ratios for the very negative words. So there is no way to directly compare two numbers and see if one word conveys the same magnitude of positive sentiment as another word conveys negative sentiment. So we should center all the values around netural so the absolute value fro neutral of the postive-to-negative ratio for a word would indicate how much sentiment (positive or negative) that word conveys.\n* When comparing absolute values it's easier to do that around zero than one. \n\nTo fix these issues, we'll convert all of our ratios to new values using logarithms.\n\n**TODO:** Go through all the ratios you calculated and convert them to logarithms. (i.e. use `np.log(ratio)`)\n\nIn the end, extremely positive and extremely negative words will have positive-to-negative ratios with similar magnitudes but opposite signs.",
"_____no_output_____"
]
],
[
[
"# TODO: Convert ratios to logs\nfor word,ratio in pos_neg_ratios.most_common():\n pos_neg_ratios[word] = np.log(ratio)",
"_____no_output_____"
]
],
[
[
"Examine the new ratios you've calculated for the same words from before:",
"_____no_output_____"
]
],
[
[
"print(\"Pos-to-neg ratio for 'the' = {}\".format(pos_neg_ratios[\"the\"]))\nprint(\"Pos-to-neg ratio for 'amazing' = {}\".format(pos_neg_ratios[\"amazing\"]))\nprint(\"Pos-to-neg ratio for 'terrible' = {}\".format(pos_neg_ratios[\"terrible\"]))",
"Pos-to-neg ratio for 'the' = 0.05902269426102881\nPos-to-neg ratio for 'amazing' = 1.3919815802404802\nPos-to-neg ratio for 'terrible' = -1.7291085042663878\n"
]
],
[
[
"If everything worked, now you should see neutral words with values close to zero. In this case, \"the\" is near zero but slightly positive, so it was probably used in more positive reviews than negative reviews. But look at \"amazing\"'s ratio - it's above `1`, showing it is clearly a word with positive sentiment. And \"terrible\" has a similar score, but in the opposite direction, so it's below `-1`. It's now clear that both of these words are associated with specific, opposing sentiments.\n\nNow run the following cells to see more ratios. \n\nThe first cell displays all the words, ordered by how associated they are with postive reviews. (Your notebook will most likely truncate the output so you won't actually see *all* the words in the list.)\n\nThe second cell displays the 30 words most associated with negative reviews by reversing the order of the first list and then looking at the first 30 words. (If you want the second cell to display all the words, ordered by how associated they are with negative reviews, you could just write `reversed(pos_neg_ratios.most_common())`.)\n\nYou should continue to see values similar to the earlier ones we checked – neutral words will be close to `0`, words will get more positive as their ratios approach and go above `1`, and words will get more negative as their ratios approach and go below `-1`. That's why we decided to use the logs instead of the raw ratios.",
"_____no_output_____"
]
],
[
[
"# words most frequently seen in a review with a \"POSITIVE\" label\npos_neg_ratios.most_common()",
"_____no_output_____"
],
[
"# words most frequently seen in a review with a \"NEGATIVE\" label\nlist(reversed(pos_neg_ratios.most_common()))[0:30]\n\n# Note: Above is the code Andrew uses in his solution video, \n# so we've included it here to avoid confusion.\n# If you explore the documentation for the Counter class, \n# you will see you could also find the 30 least common\n# words like this: pos_neg_ratios.most_common()[:-31:-1]",
"_____no_output_____"
]
],
[
[
"# End of Project 1. \n## Watch the next video to see Andrew's solution, then continue on to the next lesson.\n\n# Transforming Text into Numbers<a id='lesson_3'></a>\nThe cells here include code Andrew shows in the next video. We've included it so you can run the code along with the video without having to type in everything.",
"_____no_output_____"
]
],
[
[
"from IPython.display import Image\n\nreview = \"This was a horrible, terrible movie.\"\n\nImage(filename='sentiment_network.png')",
"_____no_output_____"
],
[
"review = \"The movie was excellent\"\n\nImage(filename='sentiment_network_pos.png')",
"_____no_output_____"
]
],
[
[
"# Project 2: Creating the Input/Output Data<a id='project_2'></a>\n\n**TODO:** Create a [set](https://docs.python.org/3/tutorial/datastructures.html#sets) named `vocab` that contains every word in the vocabulary.",
"_____no_output_____"
]
],
[
[
"# TODO: Create set named \"vocab\" containing all of the words from all of the reviews\nvocab = set(total_counts.keys())",
"_____no_output_____"
]
],
[
[
"Run the following cell to check your vocabulary size. If everything worked correctly, it should print **74074**",
"_____no_output_____"
]
],
[
[
"vocab_size = len(vocab)\nprint(vocab_size)",
"74074\n"
]
],
[
[
"Take a look at the following image. It represents the layers of the neural network you'll be building throughout this notebook. `layer_0` is the input layer, `layer_1` is a hidden layer, and `layer_2` is the output layer.",
"_____no_output_____"
]
],
[
[
"from IPython.display import Image\nImage(filename='sentiment_network_2.png')",
"_____no_output_____"
]
],
[
[
"**TODO:** Create a numpy array called `layer_0` and initialize it to all zeros. You will find the [zeros](https://docs.scipy.org/doc/numpy/reference/generated/numpy.zeros.html) function particularly helpful here. Be sure you create `layer_0` as a 2-dimensional matrix with 1 row and `vocab_size` columns. ",
"_____no_output_____"
]
],
[
[
"# TODO: Create layer_0 matrix with dimensions 1 by vocab_size, initially filled with zeros\nlayer_0 = np.zeros((1, vocab_size))",
"_____no_output_____"
]
],
[
[
"Run the following cell. It should display `(1, 74074)`",
"_____no_output_____"
]
],
[
[
"layer_0.shape",
"_____no_output_____"
],
[
"from IPython.display import Image\nImage(filename='sentiment_network.png')",
"_____no_output_____"
]
],
[
[
"`layer_0` contains one entry for every word in the vocabulary, as shown in the above image. We need to make sure we know the index of each word, so run the following cell to create a lookup table that stores the index of every word.",
"_____no_output_____"
]
],
[
[
"# Create a dictionary of words in the vocabulary mapped to index positions\n# (to be used in layer_0)\nword2index = {}\nfor i,word in enumerate(vocab):\n word2index[word] = i\n \n# display the map of words to indices\nword2index",
"_____no_output_____"
]
],
[
[
"**TODO:** Complete the implementation of `update_input_layer`. It should count \n how many times each word is used in the given review, and then store\n those counts at the appropriate indices inside `layer_0`.",
"_____no_output_____"
]
],
[
[
"def update_input_layer(review):\n \"\"\" Modify the global layer_0 to represent the vector form of review.\n The element at a given index of layer_0 should represent\n how many times the given word occurs in the review.\n Args:\n review(string) - the string of the review\n Returns:\n None\n \"\"\"\n global layer_0\n # clear out previous state by resetting the layer to be all 0s\n layer_0 *= 0\n \n # TODO: count how many times each word is used in the given review and store the results in layer_0\n for word in review.split(\" \"):\n layer_0[0][word2index[word]] += 1\n ",
"_____no_output_____"
]
],
[
[
"Run the following cell to test updating the input layer with the first review. The indices assigned may not be the same as in the solution, but hopefully you'll see some non-zero values in `layer_0`. ",
"_____no_output_____"
]
],
[
[
"update_input_layer(reviews[0])\nlayer_0",
"_____no_output_____"
]
],
[
[
"**TODO:** Complete the implementation of `get_target_for_labels`. It should return `0` or `1`, \n depending on whether the given label is `NEGATIVE` or `POSITIVE`, respectively.",
"_____no_output_____"
]
],
[
[
"def get_target_for_label(label):\n \"\"\"Convert a label to `0` or `1`.\n Args:\n label(string) - Either \"POSITIVE\" or \"NEGATIVE\".\n Returns:\n `0` or `1`.\n \"\"\"\n # TODO: Your code here\n if (label == \"POSITIVE\"):\n return 1\n else:\n return 0\n ",
"_____no_output_____"
]
],
[
[
"Run the following two cells. They should print out`'POSITIVE'` and `1`, respectively.",
"_____no_output_____"
]
],
[
[
"labels[0]",
"_____no_output_____"
],
[
"get_target_for_label(labels[0])",
"_____no_output_____"
]
],
[
[
"Run the following two cells. They should print out `'NEGATIVE'` and `0`, respectively.",
"_____no_output_____"
]
],
[
[
"labels[1]",
"_____no_output_____"
],
[
"get_target_for_label(labels[1])",
"_____no_output_____"
]
],
[
[
"# End of Project 2. \n## Watch the next video to see Andrew's solution, then continue on to the next lesson.",
"_____no_output_____"
],
[
"# Project 3: Building a Neural Network<a id='project_3'></a>",
"_____no_output_____"
],
[
"**TODO:** We've included the framework of a class called `SentimentNetork`. Implement all of the items marked `TODO` in the code. These include doing the following:\n- Create a basic neural network much like the networks you've seen in earlier lessons and in Project 1, with an input layer, a hidden layer, and an output layer. \n- Do **not** add a non-linearity in the hidden layer. That is, do not use an activation function when calculating the hidden layer outputs.\n- Re-use the code from earlier in this notebook to create the training data (see `TODO`s in the code)\n- Implement the `pre_process_data` function to create the vocabulary for our training data generating functions\n- Ensure `train` trains over the entire corpus",
"_____no_output_____"
],
[
"### Where to Get Help if You Need it\n- Re-watch earlier Udacity lectures\n- Chapters 3-5 - [Grokking Deep Learning](https://www.manning.com/books/grokking-deep-learning) - (Check inside your classroom for a discount code)",
"_____no_output_____"
]
],
[
[
"import time\nimport sys\nimport numpy as np\n\n# Encapsulate our neural network in a class\nclass SentimentNetwork:\n def __init__(self, reviews, labels, hidden_nodes = 10, learning_rate = 0.1):\n \"\"\"Create a SentimenNetwork with the given settings\n Args:\n reviews(list) - List of reviews used for training\n labels(list) - List of POSITIVE/NEGATIVE labels associated with the given reviews\n hidden_nodes(int) - Number of nodes to create in the hidden layer\n learning_rate(float) - Learning rate to use while training\n \n \"\"\"\n # Assign a seed to our random number generator to ensure we get\n # reproducable results during development \n np.random.seed(1)\n\n # process the reviews and their associated labels so that everything\n # is ready for training\n self.pre_process_data(reviews, labels)\n \n # Build the network to have the number of hidden nodes and the learning rate that\n # were passed into this initializer. Make the same number of input nodes as\n # there are vocabulary words and create a single output node.\n self.init_network(len(self.review_vocab),hidden_nodes, 1, learning_rate)\n\n def pre_process_data(self, reviews, labels):\n \n review_vocab = set()\n # TODO: populate review_vocab with all of the words in the given reviews\n # Remember to split reviews into individual words \n # using \"split(' ')\" instead of \"split()\".\n review_vocab = set()\n for review in reviews:\n for word in review.split(\" \"):\n review_vocab.add(word)\n \n # Convert the vocabulary set to a list so we can access words via indices\n self.review_vocab = list(review_vocab)\n \n label_vocab = set()\n # TODO: populate label_vocab with all of the words in the given labels.\n # There is no need to split the labels because each one is a single word.\n #label_vocab = set()\n for label in labels:\n label_vocab.add(label)\n \n # Convert the label vocabulary set to a list so we can access labels via indices\n self.label_vocab = list(label_vocab)\n \n # Store the sizes of the review and label vocabularies.\n self.review_vocab_size = len(self.review_vocab)\n self.label_vocab_size = len(self.label_vocab)\n \n # Create a dictionary of words in the vocabulary mapped to index positions\n self.word2index = {}\n # TODO: populate self.word2index with indices for all the words in self.review_vocab\n # like you saw earlier in the notebook\n for i, word in enumerate(self.review_vocab):\n self.word2index[word] = i\n \n # Create a dictionary of labels mapped to index positions\n self.label2index = {}\n # TODO: do the same thing you did for self.word2index and self.review_vocab, \n # but for self.label2index and self.label_vocab instead\n for i, label in enumerate(self.label_vocab):\n self.label2index[label] = i\n \n def init_network(self, input_nodes, hidden_nodes, output_nodes, learning_rate):\n # Store the number of nodes in input, hidden, and output layers.\n self.input_nodes = input_nodes\n self.hidden_nodes = hidden_nodes\n self.output_nodes = output_nodes\n\n # Store the learning rate\n self.learning_rate = learning_rate\n\n # Initialize weights\n \n # TODO: initialize self.weights_0_1 as a matrix of zeros. These are the weights between\n # the input layer and the hidden layer.\n self.weights_0_1 = np.zeros((self.input_nodes,self.hidden_nodes))\n \n # TODO: initialize self.weights_1_2 as a matrix of random values. \n # These are the weights between the hidden layer and the output layer.\n self.weights_1_2 = np.random.normal(0.0, self.output_nodes**-0.5, \n (self.hidden_nodes, self.output_nodes))\n \n # TODO: Create the input layer, a two-dimensional matrix with shape \n # 1 x input_nodes, with all values initialized to zero\n self.layer_0 = np.zeros((1,input_nodes))\n \n \n def update_input_layer(self,review):\n # TODO: You can copy most of the code you wrote for update_input_layer \n # earlier in this notebook. \n self.layer_0 *= 0\n for word in review.split(\" \"):\n if(word in self.word2index.keys()):\n self.layer_0[0][self.word2index[word]] += 1\n # However, MAKE SURE YOU CHANGE ALL VARIABLES TO REFERENCE\n # THE VERSIONS STORED IN THIS OBJECT, NOT THE GLOBAL OBJECTS.\n # For example, replace \"layer_0 *= 0\" with \"self.layer_0 *= 0\"\n \n \n def get_target_for_label(self,label):\n # TODO: Copy the code you wrote for get_target_for_label \n # earlier in this notebook. \n if(label == 'POSITIVE'):\n return 1\n else:\n return 0\n \n def sigmoid(self,x):\n # TODO: Return the result of calculating the sigmoid activation function\n # shown in the lectures\n return 1 / (1 + np.exp(-x))\n \n def sigmoid_output_2_derivative(self,output):\n # TODO: Return the derivative of the sigmoid activation function, \n # where \"output\" is the original output from the sigmoid function \n return output * (1 - output)\n\n def train(self, training_reviews, training_labels):\n \n # make sure out we have a matching number of reviews and labels\n assert(len(training_reviews) == len(training_labels))\n \n # Keep track of correct predictions to display accuracy during training \n correct_so_far = 0\n \n # Remember when we started for printing time statistics\n start = time.time()\n\n # loop through all the given reviews and run a forward and backward pass,\n # updating weights for every item\n for i in range(len(training_reviews)):\n \n # TODO: Get the next review and its correct label\n review = training_reviews[i]\n label = training_labels[i]\n # TODO: Implement the forward pass through the network. \n # That means use the given review to update the input layer, \n # then calculate values for the hidden layer,\n # and finally calculate the output layer.\n # \n # Do not use an activation function for the hidden layer,\n # but use the sigmoid activation function for the output layer.\n ### Forward pass ###\n\n # Input Layer\n self.update_input_layer(review)\n\n # Hidden layer\n layer_1 = self.layer_0.dot(self.weights_0_1)\n\n # Output layer\n layer_2 = self.sigmoid(layer_1.dot(self.weights_1_2))\n \n # TODO: Implement the back propagation pass here. \n # That means calculate the error for the forward pass's prediction\n # and update the weights in the network according to their\n # contributions toward the error, as calculated via the\n # gradient descent and back propagation algorithms you \n # learned in class.\n #### Implement the backward pass here ####\n ### Backward pass ###\n\n # Output error\n layer_2_error = layer_2 - self.get_target_for_label(label) # Output layer error is the difference between desired target and actual output.\n layer_2_delta = layer_2_error * self.sigmoid_output_2_derivative(layer_2)\n\n # Backpropagated error\n layer_1_error = layer_2_delta.dot(self.weights_1_2.T) # errors propagated to the hidden layer\n layer_1_delta = layer_1_error # hidden layer gradients - no nonlinearity so it's the same as the error\n\n # Update the weights\n self.weights_1_2 -= layer_1.T.dot(layer_2_delta) * self.learning_rate # update hidden-to-output weights with gradient descent step\n self.weights_0_1 -= self.layer_0.T.dot(layer_1_delta) * self.learning_rate\n \n # TODO: Keep track of correct predictions. To determine if the prediction was\n # correct, check that the absolute value of the output error \n # is less than 0.5. If so, add one to the correct_so_far count.\n \n if(layer_2 >= 0.5 and label == 'POSITIVE'):\n correct_so_far += 1\n elif(layer_2 < 0.5 and label == 'NEGATIVE'):\n correct_so_far += 1\n \n # For debug purposes, print out our prediction accuracy and speed \n # throughout the training process. \n\n elapsed_time = float(time.time() - start)\n reviews_per_second = i / elapsed_time if elapsed_time > 0 else 0\n \n sys.stdout.write(\"\\rProgress:\" + str(100 * i/float(len(training_reviews)))[:4] \\\n + \"% Speed(reviews/sec):\" + str(reviews_per_second)[0:5] \\\n + \" #Correct:\" + str(correct_so_far) + \" #Trained:\" + str(i+1) \\\n + \" Training Accuracy:\" + str(correct_so_far * 100 / float(i+1))[:4] + \"%\")\n if(i % 2500 == 0):\n print(\"\")\n \n def test(self, testing_reviews, testing_labels):\n \"\"\"\n Attempts to predict the labels for the given testing_reviews,\n and uses the test_labels to calculate the accuracy of those predictions.\n \"\"\"\n \n # keep track of how many correct predictions we make\n correct = 0\n\n # we'll time how many predictions per second we make\n start = time.time()\n\n # Loop through each of the given reviews and call run to predict\n # its label. \n for i in range(len(testing_reviews)):\n pred = self.run(testing_reviews[i])\n if(pred == testing_labels[i]):\n correct += 1\n \n # For debug purposes, print out our prediction accuracy and speed \n # throughout the prediction process. \n\n elapsed_time = float(time.time() - start)\n reviews_per_second = i / elapsed_time if elapsed_time > 0 else 0\n \n sys.stdout.write(\"\\rProgress:\" + str(100 * i/float(len(testing_reviews)))[:4] \\\n + \"% Speed(reviews/sec):\" + str(reviews_per_second)[0:5] \\\n + \" #Correct:\" + str(correct) + \" #Tested:\" + str(i+1) \\\n + \" Testing Accuracy:\" + str(correct * 100 / float(i+1))[:4] + \"%\")\n \n def run(self, review):\n \"\"\"\n Returns a POSITIVE or NEGATIVE prediction for the given review.\n \"\"\"\n # TODO: Run a forward pass through the network, like you did in the\n # \"train\" function. That means use the given review to \n # update the input layer, then calculate values for the hidden layer,\n # and finally calculate the output layer.\n #\n # Note: The review passed into this function for prediction \n # might come from anywhere, so you should convert it \n # to lower case prior to using it.\n # Input Layer\n self.update_input_layer(review.lower())\n\n # Hidden layer\n layer_1 = self.layer_0.dot(self.weights_0_1)\n\n # Output layer\n layer_2 = self.sigmoid(layer_1.dot(self.weights_1_2))\n # TODO: The output layer should now contain a prediction. \n # Return `POSITIVE` for predictions greater-than-or-equal-to `0.5`, \n # and `NEGATIVE` otherwise.\n if(layer_2[0] >= 0.5):\n return \"POSITIVE\"\n else:\n return \"NEGATIVE\"\n",
"_____no_output_____"
]
],
[
[
"Run the following cell to create a `SentimentNetwork` that will train on all but the last 1000 reviews (we're saving those for testing). Here we use a learning rate of `0.1`.",
"_____no_output_____"
]
],
[
[
"mlp = SentimentNetwork(reviews[:-1000],labels[:-1000], learning_rate=0.1)",
"_____no_output_____"
]
],
[
[
"Run the following cell to test the network's performance against the last 1000 reviews (the ones we held out from our training set). \n\n**We have not trained the model yet, so the results should be about 50% as it will just be guessing and there are only two possible values to choose from.**",
"_____no_output_____"
]
],
[
[
"mlp.test(reviews[-1000:],labels[-1000:])",
"\rProgress:0.0% Speed(reviews/sec):0.0 #Correct:1 #Tested:1 Testing Accuracy:100.%\rProgress:0.1% Speed(reviews/sec):57.81 #Correct:1 #Tested:2 Testing Accuracy:50.0%\rProgress:0.2% Speed(reviews/sec):108.9 #Correct:2 #Tested:3 Testing Accuracy:66.6%\rProgress:0.3% Speed(reviews/sec):148.2 #Correct:2 #Tested:4 Testing Accuracy:50.0%\rProgress:0.4% Speed(reviews/sec):187.9 #Correct:3 #Tested:5 Testing Accuracy:60.0%\rProgress:0.5% Speed(reviews/sec):221.3 #Correct:3 #Tested:6 Testing Accuracy:50.0%\rProgress:0.6% Speed(reviews/sec):246.6 #Correct:4 #Tested:7 Testing Accuracy:57.1%\rProgress:0.7% Speed(reviews/sec):267.4 #Correct:4 #Tested:8 Testing Accuracy:50.0%\rProgress:0.8% Speed(reviews/sec):289.6 #Correct:5 #Tested:9 Testing Accuracy:55.5%\rProgress:0.9% Speed(reviews/sec):290.2 #Correct:5 #Tested:10 Testing Accuracy:50.0%\rProgress:1.0% Speed(reviews/sec):301.1 #Correct:6 #Tested:11 Testing Accuracy:54.5%\rProgress:1.1% Speed(reviews/sec):320.3 #Correct:6 #Tested:12 Testing Accuracy:50.0%\rProgress:1.2% Speed(reviews/sec):337.8 #Correct:7 #Tested:13 Testing Accuracy:53.8%\rProgress:1.3% Speed(reviews/sec):331.2 #Correct:7 #Tested:14 Testing Accuracy:50.0%\rProgress:1.4% Speed(reviews/sec):342.7 #Correct:8 #Tested:15 Testing Accuracy:53.3%\rProgress:1.5% Speed(reviews/sec):356.7 #Correct:8 #Tested:16 Testing Accuracy:50.0%\rProgress:1.6% Speed(reviews/sec):364.0 #Correct:9 #Tested:17 Testing Accuracy:52.9%\rProgress:1.7% Speed(reviews/sec):374.6 #Correct:9 #Tested:18 Testing Accuracy:50.0%\rProgress:1.8% Speed(reviews/sec):387.9 #Correct:10 #Tested:19 Testing Accuracy:52.6%\rProgress:1.9% Speed(reviews/sec):397.9 #Correct:10 #Tested:20 Testing Accuracy:50.0%\rProgress:2.0% Speed(reviews/sec):405.3 #Correct:11 #Tested:21 Testing Accuracy:52.3%\rProgress:2.1% Speed(reviews/sec):416.0 #Correct:11 #Tested:22 Testing Accuracy:50.0%\rProgress:2.2% Speed(reviews/sec):425.1 #Correct:12 #Tested:23 Testing Accuracy:52.1%\rProgress:2.3% Speed(reviews/sec):431.9 #Correct:12 #Tested:24 Testing Accuracy:50.0%\rProgress:2.4% Speed(reviews/sec):435.7 #Correct:13 #Tested:25 Testing Accuracy:52.0%\rProgress:2.5% Speed(reviews/sec):444.3 #Correct:13 #Tested:26 Testing Accuracy:50.0%\rProgress:2.6% Speed(reviews/sec):453.1 #Correct:14 #Tested:27 Testing Accuracy:51.8%\rProgress:2.7% Speed(reviews/sec):451.0 #Correct:14 #Tested:28 Testing Accuracy:50.0%\rProgress:2.8% Speed(reviews/sec):457.5 #Correct:15 #Tested:29 Testing Accuracy:51.7%\rProgress:2.9% Speed(reviews/sec):462.8 #Correct:15 #Tested:30 Testing Accuracy:50.0%\rProgress:3.0% Speed(reviews/sec):467.5 #Correct:16 #Tested:31 Testing Accuracy:51.6%\rProgress:3.1% Speed(reviews/sec):474.5 #Correct:16 #Tested:32 Testing Accuracy:50.0%\rProgress:3.2% Speed(reviews/sec):480.2 #Correct:17 #Tested:33 Testing Accuracy:51.5%\rProgress:3.3% Speed(reviews/sec):486.1 #Correct:17 #Tested:34 Testing Accuracy:50.0%\rProgress:3.4% Speed(reviews/sec):490.1 #Correct:18 #Tested:35 Testing Accuracy:51.4%\rProgress:3.5% Speed(reviews/sec):493.7 #Correct:18 #Tested:36 Testing Accuracy:50.0%\rProgress:3.6% Speed(reviews/sec):496.9 #Correct:19 #Tested:37 Testing Accuracy:51.3%\rProgress:3.7% Speed(reviews/sec):503.4 #Correct:19 #Tested:38 Testing Accuracy:50.0%\rProgress:3.8% Speed(reviews/sec):506.4 #Correct:20 #Tested:39 Testing Accuracy:51.2%\rProgress:3.9% Speed(reviews/sec):506.6 #Correct:20 #Tested:40 Testing Accuracy:50.0%\rProgress:4.0% Speed(reviews/sec):510.6 #Correct:21 #Tested:41 Testing Accuracy:51.2%\rProgress:4.1% Speed(reviews/sec):515.2 #Correct:21 #Tested:42 Testing Accuracy:50.0%\rProgress:4.2% Speed(reviews/sec):513.4 #Correct:22 #Tested:43 Testing Accuracy:51.1%\rProgress:4.3% Speed(reviews/sec):515.2 #Correct:22 #Tested:44 Testing Accuracy:50.0%\rProgress:4.4% Speed(reviews/sec):514.5 #Correct:23 #Tested:45 Testing Accuracy:51.1%\rProgress:4.5% Speed(reviews/sec):510.7 #Correct:23 #Tested:46 Testing Accuracy:50.0%\rProgress:4.6% Speed(reviews/sec):511.6 #Correct:24 #Tested:47 Testing Accuracy:51.0%\rProgress:4.7% Speed(reviews/sec):516.7 #Correct:24 #Tested:48 Testing Accuracy:50.0%\rProgress:4.8% Speed(reviews/sec):517.7 #Correct:25 #Tested:49 Testing Accuracy:51.0%\rProgress:4.9% Speed(reviews/sec):514.4 #Correct:25 #Tested:50 Testing Accuracy:50.0%\rProgress:5.0% Speed(reviews/sec):516.6 #Correct:26 #Tested:51 Testing Accuracy:50.9%\rProgress:5.1% Speed(reviews/sec):520.9 #Correct:26 #Tested:52 Testing Accuracy:50.0%\rProgress:5.2% Speed(reviews/sec):521.7 #Correct:27 #Tested:53 Testing Accuracy:50.9%\rProgress:5.3% Speed(reviews/sec):525.1 #Correct:27 #Tested:54 Testing Accuracy:50.0%\rProgress:5.4% Speed(reviews/sec):529.0 #Correct:28 #Tested:55 Testing Accuracy:50.9%\rProgress:5.5% Speed(reviews/sec):531.1 #Correct:28 #Tested:56 Testing Accuracy:50.0%\rProgress:5.6% Speed(reviews/sec):521.1 #Correct:29 #Tested:57 Testing Accuracy:50.8%\rProgress:5.7% Speed(reviews/sec):525.3 #Correct:29 #Tested:58 Testing Accuracy:50.0%\rProgress:5.8% Speed(reviews/sec):529.0 #Correct:30 #Tested:59 Testing Accuracy:50.8%\rProgress:5.9% Speed(reviews/sec):532.5 #Correct:30 #Tested:60 Testing Accuracy:50.0%\rProgress:6.0% Speed(reviews/sec):533.3 #Correct:31 #Tested:61 Testing Accuracy:50.8%\rProgress:6.1% Speed(reviews/sec):535.3 #Correct:31 #Tested:62 Testing Accuracy:50.0%\rProgress:6.2% Speed(reviews/sec):539.2 #Correct:32 #Tested:63 Testing Accuracy:50.7%\rProgress:6.3% Speed(reviews/sec):542.7 #Correct:32 #Tested:64 Testing Accuracy:50.0%\rProgress:6.4% Speed(reviews/sec):543.1 #Correct:33 #Tested:65 Testing Accuracy:50.7%\rProgress:6.5% Speed(reviews/sec):546.1 #Correct:33 #Tested:66 Testing Accuracy:50.0%\rProgress:6.6% Speed(reviews/sec):548.5 #Correct:34 #Tested:67 Testing Accuracy:50.7%\rProgress:6.7% Speed(reviews/sec):551.1 #Correct:34 #Tested:68 Testing Accuracy:50.0%\rProgress:6.8% Speed(reviews/sec):553.8 #Correct:35 #Tested:69 Testing Accuracy:50.7%\rProgress:6.9% Speed(reviews/sec):555.5 #Correct:35 #Tested:70 Testing Accuracy:50.0%\rProgress:7.0% Speed(reviews/sec):556.6 #Correct:36 #Tested:71 Testing Accuracy:50.7%\rProgress:7.1% Speed(reviews/sec):559.7 #Correct:36 #Tested:72 Testing Accuracy:50.0%\rProgress:7.2% Speed(reviews/sec):562.1 #Correct:37 #Tested:73 Testing Accuracy:50.6%\rProgress:7.3% Speed(reviews/sec):564.8 #Correct:37 #Tested:74 Testing Accuracy:50.0%\rProgress:7.4% Speed(reviews/sec):567.0 #Correct:38 #Tested:75 Testing Accuracy:50.6%\rProgress:7.5% Speed(reviews/sec):569.6 #Correct:38 #Tested:76 Testing Accuracy:50.0%\rProgress:7.6% Speed(reviews/sec):572.4 #Correct:39 #Tested:77 Testing Accuracy:50.6%\rProgress:7.7% Speed(reviews/sec):575.1 #Correct:39 #Tested:78 Testing Accuracy:50.0%\rProgress:7.8% Speed(reviews/sec):577.6 #Correct:40 #Tested:79 Testing Accuracy:50.6%\rProgress:7.9% Speed(reviews/sec):579.5 #Correct:40 #Tested:80 Testing Accuracy:50.0%\rProgress:8.0% Speed(reviews/sec):580.2 #Correct:41 #Tested:81 Testing Accuracy:50.6%\rProgress:8.1% Speed(reviews/sec):572.5 #Correct:41 #Tested:82 Testing Accuracy:50.0%\rProgress:8.2% Speed(reviews/sec):573.1 #Correct:42 #Tested:83 Testing Accuracy:50.6%\rProgress:8.3% Speed(reviews/sec):574.3 #Correct:42 #Tested:84 Testing Accuracy:50.0%\rProgress:8.4% Speed(reviews/sec):576.6 #Correct:43 #Tested:85 Testing Accuracy:50.5%\rProgress:8.5% Speed(reviews/sec):578.2 #Correct:43 #Tested:86 Testing Accuracy:50.0%\rProgress:8.6% Speed(reviews/sec):578.6 #Correct:44 #Tested:87 Testing Accuracy:50.5%\rProgress:8.7% Speed(reviews/sec):580.9 #Correct:44 #Tested:88 Testing Accuracy:50.0%\rProgress:8.8% Speed(reviews/sec):583.2 #Correct:45 #Tested:89 Testing Accuracy:50.5%\rProgress:8.9% Speed(reviews/sec):581.1 #Correct:45 #Tested:90 Testing Accuracy:50.0%\rProgress:9.0% Speed(reviews/sec):582.2 #Correct:46 #Tested:91 Testing Accuracy:50.5%\rProgress:9.1% Speed(reviews/sec):584.2 #Correct:46 #Tested:92 Testing Accuracy:50.0%\rProgress:9.2% Speed(reviews/sec):586.1 #Correct:47 #Tested:93 Testing Accuracy:50.5%\rProgress:9.3% Speed(reviews/sec):587.7 #Correct:47 #Tested:94 Testing Accuracy:50.0%\rProgress:9.4% Speed(reviews/sec):589.4 #Correct:48 #Tested:95 Testing Accuracy:50.5%\rProgress:9.5% Speed(reviews/sec):590.2 #Correct:48 #Tested:96 Testing Accuracy:50.0%\rProgress:9.6% Speed(reviews/sec):590.8 #Correct:49 #Tested:97 Testing Accuracy:50.5%\rProgress:9.7% Speed(reviews/sec):591.4 #Correct:49 #Tested:98 Testing Accuracy:50.0%\rProgress:9.8% Speed(reviews/sec):593.5 #Correct:50 #Tested:99 Testing Accuracy:50.5%\rProgress:9.9% Speed(reviews/sec):595.6 #Correct:50 #Tested:100 Testing Accuracy:50.0%\rProgress:10.0% Speed(reviews/sec):597.4 #Correct:51 #Tested:101 Testing Accuracy:50.4%\rProgress:10.1% Speed(reviews/sec):599.0 #Correct:51 #Tested:102 Testing Accuracy:50.0%\rProgress:10.2% Speed(reviews/sec):600.6 #Correct:52 #Tested:103 Testing Accuracy:50.4%\rProgress:10.3% Speed(reviews/sec):596.8 #Correct:52 #Tested:104 Testing Accuracy:50.0%\rProgress:10.4% Speed(reviews/sec):597.6 #Correct:53 #Tested:105 Testing Accuracy:50.4%\rProgress:10.5% Speed(reviews/sec):599.7 #Correct:53 #Tested:106 Testing Accuracy:50.0%\rProgress:10.6% Speed(reviews/sec):599.4 #Correct:54 #Tested:107 Testing Accuracy:50.4%\rProgress:10.7% Speed(reviews/sec):599.7 #Correct:54 #Tested:108 Testing Accuracy:50.0%\rProgress:10.8% Speed(reviews/sec):600.1 #Correct:55 #Tested:109 Testing Accuracy:50.4%\rProgress:10.9% Speed(reviews/sec):601.1 #Correct:55 #Tested:110 Testing Accuracy:50.0%\rProgress:11.0% Speed(reviews/sec):602.5 #Correct:56 #Tested:111 Testing Accuracy:50.4%\rProgress:11.1% Speed(reviews/sec):603.8 #Correct:56 #Tested:112 Testing Accuracy:50.0%\rProgress:11.2% Speed(reviews/sec):604.8 #Correct:57 #Tested:113 Testing Accuracy:50.4%\rProgress:11.3% Speed(reviews/sec):602.8 #Correct:57 #Tested:114 Testing Accuracy:50.0%\rProgress:11.4% Speed(reviews/sec):603.9 #Correct:58 #Tested:115 Testing Accuracy:50.4%\rProgress:11.5% Speed(reviews/sec):605.7 #Correct:58 #Tested:116 Testing Accuracy:50.0%\rProgress:11.6% Speed(reviews/sec):607.4 #Correct:59 #Tested:117 Testing Accuracy:50.4%\rProgress:11.7% Speed(reviews/sec):608.3 #Correct:59 #Tested:118 Testing Accuracy:50.0%\rProgress:11.8% Speed(reviews/sec):609.5 #Correct:60 #Tested:119 Testing Accuracy:50.4%\rProgress:11.9% Speed(reviews/sec):610.4 #Correct:60 #Tested:120 Testing Accuracy:50.0%\rProgress:12.0% Speed(reviews/sec):610.7 #Correct:61 #Tested:121 Testing Accuracy:50.4%\rProgress:12.1% Speed(reviews/sec):611.5 #Correct:61 #Tested:122 Testing Accuracy:50.0%\rProgress:12.2% Speed(reviews/sec):612.4 #Correct:62 #Tested:123 Testing Accuracy:50.4%\rProgress:12.3% Speed(reviews/sec):610.5 #Correct:62 #Tested:124 Testing Accuracy:50.0%\rProgress:12.4% Speed(reviews/sec):608.5 #Correct:63 #Tested:125 Testing Accuracy:50.4%\rProgress:12.5% Speed(reviews/sec):606.7 #Correct:63 #Tested:126 Testing Accuracy:50.0%\rProgress:12.6% Speed(reviews/sec):607.8 #Correct:64 #Tested:127 Testing Accuracy:50.3%\rProgress:12.7% Speed(reviews/sec):606.5 #Correct:64 #Tested:128 Testing Accuracy:50.0%\rProgress:12.8% Speed(reviews/sec):607.5 #Correct:65 #Tested:129 Testing Accuracy:50.3%\rProgress:12.9% Speed(reviews/sec):608.3 #Correct:65 #Tested:130 Testing Accuracy:50.0%\rProgress:13.0% Speed(reviews/sec):609.3 #Correct:66 #Tested:131 Testing Accuracy:50.3%\rProgress:13.1% Speed(reviews/sec):610.3 #Correct:66 #Tested:132 Testing Accuracy:50.0%"
]
],
[
[
"Run the following cell to actually train the network. During training, it will display the model's accuracy repeatedly as it trains so you can see how well it's doing.",
"_____no_output_____"
]
],
[
[
"mlp.train(reviews[:-1000],labels[:-1000])",
"Progress:0.0% Speed(reviews/sec):0.0 #Correct:1 #Trained:1 Training Accuracy:100.%\nProgress:10.4% Speed(reviews/sec):166.3 #Correct:1251 #Trained:2501 Training Accuracy:50.0%\nProgress:20.8% Speed(reviews/sec):175.6 #Correct:2501 #Trained:5001 Training Accuracy:50.0%\nProgress:31.2% Speed(reviews/sec):165.5 #Correct:3751 #Trained:7501 Training Accuracy:50.0%\nProgress:41.6% Speed(reviews/sec):158.0 #Correct:5001 #Trained:10001 Training Accuracy:50.0%\nProgress:52.0% Speed(reviews/sec):163.4 #Correct:6251 #Trained:12501 Training Accuracy:50.0%\nProgress:62.5% Speed(reviews/sec):167.9 #Correct:7501 #Trained:15001 Training Accuracy:50.0%\nProgress:72.9% Speed(reviews/sec):170.7 #Correct:8751 #Trained:17501 Training Accuracy:50.0%\nProgress:83.3% Speed(reviews/sec):168.5 #Correct:10001 #Trained:20001 Training Accuracy:50.0%\nProgress:93.7% Speed(reviews/sec):167.3 #Correct:11251 #Trained:22501 Training Accuracy:50.0%\nProgress:99.9% Speed(reviews/sec):167.2 #Correct:12000 #Trained:24000 Training Accuracy:50.0%"
]
],
[
[
"That most likely didn't train very well. Part of the reason may be because the learning rate is too high. Run the following cell to recreate the network with a smaller learning rate, `0.01`, and then train the new network.",
"_____no_output_____"
]
],
[
[
"mlp = SentimentNetwork(reviews[:-1000],labels[:-1000], learning_rate=0.01)\nmlp.train(reviews[:-1000],labels[:-1000])",
"Progress:0.0% Speed(reviews/sec):0.0 #Correct:1 #Trained:1 Training Accuracy:100.%\nProgress:10.4% Speed(reviews/sec):162.4 #Correct:1248 #Trained:2501 Training Accuracy:49.9%\nProgress:20.8% Speed(reviews/sec):161.0 #Correct:2498 #Trained:5001 Training Accuracy:49.9%\nProgress:31.2% Speed(reviews/sec):148.8 #Correct:3748 #Trained:7501 Training Accuracy:49.9%\nProgress:41.6% Speed(reviews/sec):141.5 #Correct:4998 #Trained:10001 Training Accuracy:49.9%\nProgress:52.0% Speed(reviews/sec):142.3 #Correct:6248 #Trained:12501 Training Accuracy:49.9%\nProgress:62.5% Speed(reviews/sec):143.7 #Correct:7491 #Trained:15001 Training Accuracy:49.9%\nProgress:72.9% Speed(reviews/sec):145.9 #Correct:8765 #Trained:17501 Training Accuracy:50.0%\nProgress:76.7% Speed(reviews/sec):147.1 #Correct:9226 #Trained:18417 Training Accuracy:50.0%"
]
],
[
[
"That probably wasn't much different. Run the following cell to recreate the network one more time with an even smaller learning rate, `0.001`, and then train the new network.",
"_____no_output_____"
]
],
[
[
"mlp = SentimentNetwork(reviews[:-1000],labels[:-1000], learning_rate=0.001)\nmlp.train(reviews[:-1000],labels[:-1000])",
"Progress:0.0% Speed(reviews/sec):0.0 #Correct:1 #Trained:1 Training Accuracy:100.%\nProgress:10.4% Speed(reviews/sec):162.3 #Correct:1284 #Trained:2501 Training Accuracy:51.3%\nProgress:20.8% Speed(reviews/sec):149.3 #Correct:2622 #Trained:5001 Training Accuracy:52.4%\nProgress:31.2% Speed(reviews/sec):147.9 #Correct:4086 #Trained:7501 Training Accuracy:54.4%\nProgress:41.6% Speed(reviews/sec):151.0 #Correct:5666 #Trained:10001 Training Accuracy:56.6%\nProgress:52.0% Speed(reviews/sec):152.2 #Correct:7249 #Trained:12501 Training Accuracy:57.9%\nProgress:62.5% Speed(reviews/sec):158.0 #Correct:8829 #Trained:15001 Training Accuracy:58.8%\nProgress:72.9% Speed(reviews/sec):162.5 #Correct:10487 #Trained:17501 Training Accuracy:59.9%\nProgress:83.3% Speed(reviews/sec):166.1 #Correct:12129 #Trained:20001 Training Accuracy:60.6%\nProgress:93.7% Speed(reviews/sec):165.6 #Correct:13822 #Trained:22501 Training Accuracy:61.4%\nProgress:99.9% Speed(reviews/sec):163.3 #Correct:14872 #Trained:24000 Training Accuracy:61.9%"
]
],
[
[
"With a learning rate of `0.001`, the network should finally have started to improve during training. It's still not very good, but it shows that this solution has potential. We will improve it in the next lesson.",
"_____no_output_____"
],
[
"# End of Project 3. \n## Watch the next video to see Andrew's solution, then continue on to the next lesson.",
"_____no_output_____"
],
[
"# Understanding Neural Noise<a id='lesson_4'></a>\n\nThe following cells include includes the code Andrew shows in the next video. We've included it here so you can run the cells along with the video without having to type in everything.",
"_____no_output_____"
]
],
[
[
"from IPython.display import Image\nImage(filename='sentiment_network.png')",
"_____no_output_____"
],
[
"def update_input_layer(review):\n \n global layer_0\n \n # clear out previous state, reset the layer to be all 0s\n layer_0 *= 0\n for word in review.split(\" \"):\n layer_0[0][word2index[word]] += 1\n\nupdate_input_layer(reviews[0])",
"_____no_output_____"
],
[
"layer_0",
"_____no_output_____"
],
[
"review_counter = Counter()",
"_____no_output_____"
],
[
"for word in reviews[0].split(\" \"):\n review_counter[word] += 1",
"_____no_output_____"
],
[
"review_counter.most_common()",
"_____no_output_____"
]
],
[
[
"# Project 4: Reducing Noise in Our Input Data<a id='project_4'></a>\n\n**TODO:** Attempt to reduce the noise in the input data like Andrew did in the previous video. Specifically, do the following:\n* Copy the `SentimentNetwork` class you created earlier into the following cell.\n* Modify `update_input_layer` so it does not count how many times each word is used, but rather just stores whether or not a word was used. ",
"_____no_output_____"
]
],
[
[
"# TODO: -Copy the SentimentNetwork class from Projet 3 lesson\n# -Modify it to reduce noise, like in the video \nimport time\nimport sys\nimport numpy as np\n\n# Encapsulate our neural network in a class\nclass SentimentNetwork:\n def __init__(self, reviews,labels,hidden_nodes = 10, learning_rate = 0.1):\n \"\"\"Create a SentimenNetwork with the given settings\n Args:\n reviews(list) - List of reviews used for training\n labels(list) - List of POSITIVE/NEGATIVE labels associated with the given reviews\n hidden_nodes(int) - Number of nodes to create in the hidden layer\n learning_rate(float) - Learning rate to use while training\n \n \"\"\"\n # Assign a seed to our random number generator to ensure we get\n # reproducable results during development \n np.random.seed(1)\n\n # process the reviews and their associated labels so that everything\n # is ready for training\n self.pre_process_data(reviews, labels)\n \n # Build the network to have the number of hidden nodes and the learning rate that\n # were passed into this initializer. Make the same number of input nodes as\n # there are vocabulary words and create a single output node.\n self.init_network(len(self.review_vocab),hidden_nodes, 1, learning_rate)\n\n def pre_process_data(self, reviews, labels):\n \n # populate review_vocab with all of the words in the given reviews\n review_vocab = set()\n for review in reviews:\n for word in review.split(\" \"):\n review_vocab.add(word)\n\n # Convert the vocabulary set to a list so we can access words via indices\n self.review_vocab = list(review_vocab)\n \n # populate label_vocab with all of the words in the given labels.\n label_vocab = set()\n for label in labels:\n label_vocab.add(label)\n \n # Convert the label vocabulary set to a list so we can access labels via indices\n self.label_vocab = list(label_vocab)\n \n # Store the sizes of the review and label vocabularies.\n self.review_vocab_size = len(self.review_vocab)\n self.label_vocab_size = len(self.label_vocab)\n \n # Create a dictionary of words in the vocabulary mapped to index positions\n self.word2index = {}\n for i, word in enumerate(self.review_vocab):\n self.word2index[word] = i\n \n # Create a dictionary of labels mapped to index positions\n self.label2index = {}\n for i, label in enumerate(self.label_vocab):\n self.label2index[label] = i\n \n def init_network(self, input_nodes, hidden_nodes, output_nodes, learning_rate):\n # Set number of nodes in input, hidden and output layers.\n self.input_nodes = input_nodes\n self.hidden_nodes = hidden_nodes\n self.output_nodes = output_nodes\n\n # Store the learning rate\n self.learning_rate = learning_rate\n\n # Initialize weights\n\n # These are the weights between the input layer and the hidden layer.\n self.weights_0_1 = np.zeros((self.input_nodes,self.hidden_nodes))\n \n # These are the weights between the hidden layer and the output layer.\n self.weights_1_2 = np.random.normal(0.0, self.output_nodes**-0.5, \n (self.hidden_nodes, self.output_nodes))\n \n # The input layer, a two-dimensional matrix with shape 1 x input_nodes\n self.layer_0 = np.zeros((1,input_nodes))\n \n \n def update_input_layer(self,review):\n\n # clear out previous state, reset the layer to be all 0s\n self.layer_0 *= 0\n \n for word in review.split(\" \"):\n # NOTE: This if-check was not in the version of this method created in Project 2,\n # and it appears in Andrew's Project 3 solution without explanation. \n # It simply ensures the word is actually a key in word2index before\n # accessing it, which is important because accessing an invalid key\n # with raise an exception in Python. This allows us to ignore unknown\n # words encountered in new reviews.\n if(word in self.word2index.keys()):\n ## New for Project 4: changed to set to 1 instead of add 1\n self.layer_0[0][self.word2index[word]] = 1\n \n def get_target_for_label(self,label):\n if(label == 'POSITIVE'):\n return 1\n else:\n return 0\n \n def sigmoid(self,x):\n return 1 / (1 + np.exp(-x))\n \n def sigmoid_output_2_derivative(self,output):\n return output * (1 - output)\n \n def train(self, training_reviews, training_labels):\n \n # make sure out we have a matching number of reviews and labels\n assert(len(training_reviews) == len(training_labels))\n \n # Keep track of correct predictions to display accuracy during training \n correct_so_far = 0\n\n # Remember when we started for printing time statistics\n start = time.time()\n \n # loop through all the given reviews and run a forward and backward pass,\n # updating weights for every item\n for i in range(len(training_reviews)):\n \n # Get the next review and its correct label\n review = training_reviews[i]\n label = training_labels[i]\n \n #### Implement the forward pass here ####\n ### Forward pass ###\n\n # Input Layer\n self.update_input_layer(review)\n\n # Hidden layer\n layer_1 = self.layer_0.dot(self.weights_0_1)\n\n # Output layer\n layer_2 = self.sigmoid(layer_1.dot(self.weights_1_2))\n \n #### Implement the backward pass here ####\n ### Backward pass ###\n\n # Output error\n layer_2_error = layer_2 - self.get_target_for_label(label) # Output layer error is the difference between desired target and actual output.\n layer_2_delta = layer_2_error * self.sigmoid_output_2_derivative(layer_2)\n\n # Backpropagated error\n layer_1_error = layer_2_delta.dot(self.weights_1_2.T) # errors propagated to the hidden layer\n layer_1_delta = layer_1_error # hidden layer gradients - no nonlinearity so it's the same as the error\n\n # Update the weights\n self.weights_1_2 -= layer_1.T.dot(layer_2_delta) * self.learning_rate # update hidden-to-output weights with gradient descent step\n self.weights_0_1 -= self.layer_0.T.dot(layer_1_delta) * self.learning_rate # update input-to-hidden weights with gradient descent step\n\n # Keep track of correct predictions.\n if(layer_2 >= 0.5 and label == 'POSITIVE'):\n correct_so_far += 1\n elif(layer_2 < 0.5 and label == 'NEGATIVE'):\n correct_so_far += 1\n \n # For debug purposes, print out our prediction accuracy and speed \n # throughout the training process. \n elapsed_time = float(time.time() - start)\n reviews_per_second = i / elapsed_time if elapsed_time > 0 else 0\n \n sys.stdout.write(\"\\rProgress:\" + str(100 * i/float(len(training_reviews)))[:4] \\\n + \"% Speed(reviews/sec):\" + str(reviews_per_second)[0:5] \\\n + \" #Correct:\" + str(correct_so_far) + \" #Trained:\" + str(i+1) \\\n + \" Training Accuracy:\" + str(correct_so_far * 100 / float(i+1))[:4] + \"%\")\n if(i % 2500 == 0):\n print(\"\")\n \n def test(self, testing_reviews, testing_labels):\n \"\"\"\n Attempts to predict the labels for the given testing_reviews,\n and uses the test_labels to calculate the accuracy of those predictions.\n \"\"\"\n \n # keep track of how many correct predictions we make\n correct = 0\n\n # we'll time how many predictions per second we make\n start = time.time()\n\n # Loop through each of the given reviews and call run to predict\n # its label. \n for i in range(len(testing_reviews)):\n pred = self.run(testing_reviews[i])\n if(pred == testing_labels[i]):\n correct += 1\n \n # For debug purposes, print out our prediction accuracy and speed \n # throughout the prediction process. \n\n elapsed_time = float(time.time() - start)\n reviews_per_second = i / elapsed_time if elapsed_time > 0 else 0\n \n sys.stdout.write(\"\\rProgress:\" + str(100 * i/float(len(testing_reviews)))[:4] \\\n + \"% Speed(reviews/sec):\" + str(reviews_per_second)[0:5] \\\n + \" #Correct:\" + str(correct) + \" #Tested:\" + str(i+1) \\\n + \" Testing Accuracy:\" + str(correct * 100 / float(i+1))[:4] + \"%\")\n \n def run(self, review):\n \"\"\"\n Returns a POSITIVE or NEGATIVE prediction for the given review.\n \"\"\"\n # Run a forward pass through the network, like in the \"train\" function.\n \n # Input Layer\n self.update_input_layer(review.lower())\n\n # Hidden layer\n layer_1 = self.layer_0.dot(self.weights_0_1)\n\n # Output layer\n layer_2 = self.sigmoid(layer_1.dot(self.weights_1_2))\n \n # Return POSITIVE for values above greater-than-or-equal-to 0.5 in the output layer;\n # return NEGATIVE for other values\n if(layer_2[0] >= 0.5):\n return \"POSITIVE\"\n else:\n return \"NEGATIVE\"\n ",
"_____no_output_____"
]
],
[
[
"Run the following cell to recreate the network and train it. Notice we've gone back to the higher learning rate of `0.1`.",
"_____no_output_____"
]
],
[
[
"mlp = SentimentNetwork(reviews[:-1000],labels[:-1000], learning_rate=0.1)\nmlp.train(reviews[:-1000],labels[:-1000])",
"Progress:0.0% Speed(reviews/sec):0.0 #Correct:1 #Trained:1 Training Accuracy:100.%\nProgress:10.4% Speed(reviews/sec):178.9 #Correct:1824 #Trained:2501 Training Accuracy:72.9%\nProgress:20.8% Speed(reviews/sec):158.8 #Correct:3813 #Trained:5001 Training Accuracy:76.2%\nProgress:31.2% Speed(reviews/sec):162.5 #Correct:5889 #Trained:7501 Training Accuracy:78.5%\nProgress:41.6% Speed(reviews/sec):161.3 #Correct:8040 #Trained:10001 Training Accuracy:80.3%\nProgress:52.0% Speed(reviews/sec):162.5 #Correct:10185 #Trained:12501 Training Accuracy:81.4%\nProgress:62.5% Speed(reviews/sec):161.9 #Correct:12315 #Trained:15001 Training Accuracy:82.0%\nProgress:72.9% Speed(reviews/sec):162.5 #Correct:14436 #Trained:17501 Training Accuracy:82.4%\nProgress:83.3% Speed(reviews/sec):163.5 #Correct:16630 #Trained:20001 Training Accuracy:83.1%\nProgress:93.7% Speed(reviews/sec):164.3 #Correct:18816 #Trained:22501 Training Accuracy:83.6%\nProgress:99.9% Speed(reviews/sec):164.8 #Correct:20138 #Trained:24000 Training Accuracy:83.9%"
]
],
[
[
"That should have trained much better than the earlier attempts. It's still not wonderful, but it should have improved dramatically. Run the following cell to test your model with 1000 predictions.",
"_____no_output_____"
]
],
[
[
"mlp.test(reviews[-1000:],labels[-1000:])",
"Progress:99.9% Speed(reviews/sec):608.0 #Correct:853 #Tested:1000 Testing Accuracy:85.3%"
]
],
[
[
"# End of Project 4. \n## Andrew's solution was actually in the previous video, so rewatch that video if you had any problems with that project. Then continue on to the next lesson.\n# Analyzing Inefficiencies in our Network<a id='lesson_5'></a>\nThe following cells include the code Andrew shows in the next video. We've included it here so you can run the cells along with the video without having to type in everything.",
"_____no_output_____"
]
],
[
[
"Image(filename='sentiment_network_sparse.png')",
"_____no_output_____"
],
[
"layer_0 = np.zeros(10)",
"_____no_output_____"
],
[
"layer_0",
"_____no_output_____"
],
[
"layer_0[4] = 1\nlayer_0[9] = 1",
"_____no_output_____"
],
[
"layer_0",
"_____no_output_____"
],
[
"weights_0_1 = np.random.randn(10,5)",
"_____no_output_____"
],
[
"layer_0.dot(weights_0_1)",
"_____no_output_____"
],
[
"indices = [4,9]",
"_____no_output_____"
],
[
"layer_1 = np.zeros(5)",
"_____no_output_____"
],
[
"for index in indices:\n layer_1 += (1 * weights_0_1[index])",
"_____no_output_____"
],
[
"layer_1",
"_____no_output_____"
],
[
"Image(filename='sentiment_network_sparse_2.png')",
"_____no_output_____"
],
[
"layer_1 = np.zeros(5)",
"_____no_output_____"
],
[
"for index in indices:\n layer_1 += (weights_0_1[index])",
"_____no_output_____"
],
[
"layer_1",
"_____no_output_____"
]
],
[
[
"# Project 5: Making our Network More Efficient<a id='project_5'></a>\n**TODO:** Make the `SentimentNetwork` class more efficient by eliminating unnecessary multiplications and additions that occur during forward and backward propagation. To do that, you can do the following:\n* Copy the `SentimentNetwork` class from the previous project into the following cell.\n* Remove the `update_input_layer` function - you will not need it in this version.\n* Modify `init_network`:\n>* You no longer need a separate input layer, so remove any mention of `self.layer_0`\n>* You will be dealing with the old hidden layer more directly, so create `self.layer_1`, a two-dimensional matrix with shape 1 x hidden_nodes, with all values initialized to zero\n* Modify `train`:\n>* Change the name of the input parameter `training_reviews` to `training_reviews_raw`. This will help with the next step.\n>* At the beginning of the function, you'll want to preprocess your reviews to convert them to a list of indices (from `word2index`) that are actually used in the review. This is equivalent to what you saw in the video when Andrew set specific indices to 1. Your code should create a local `list` variable named `training_reviews` that should contain a `list` for each review in `training_reviews_raw`. Those lists should contain the indices for words found in the review.\n>* Remove call to `update_input_layer`\n>* Use `self`'s `layer_1` instead of a local `layer_1` object.\n>* In the forward pass, replace the code that updates `layer_1` with new logic that only adds the weights for the indices used in the review.\n>* When updating `weights_0_1`, only update the individual weights that were used in the forward pass.\n* Modify `run`:\n>* Remove call to `update_input_layer` \n>* Use `self`'s `layer_1` instead of a local `layer_1` object.\n>* Much like you did in `train`, you will need to pre-process the `review` so you can work with word indices, then update `layer_1` by adding weights for the indices used in the review.",
"_____no_output_____"
]
],
[
[
"# TODO: -Copy the SentimentNetwork class from Project 4 lesson\n# -Modify it according to the above instructions \nimport time\nimport sys\nimport numpy as np\n\n# Encapsulate our neural network in a class\nclass SentimentNetwork:\n def __init__(self, reviews,labels,hidden_nodes = 10, learning_rate = 0.1):\n \"\"\"Create a SentimenNetwork with the given settings\n Args:\n reviews(list) - List of reviews used for training\n labels(list) - List of POSITIVE/NEGATIVE labels associated with the given reviews\n hidden_nodes(int) - Number of nodes to create in the hidden layer\n learning_rate(float) - Learning rate to use while training\n \n \"\"\"\n # Assign a seed to our random number generator to ensure we get\n # reproducable results during development \n np.random.seed(1)\n\n # process the reviews and their associated labels so that everything\n # is ready for training\n self.pre_process_data(reviews, labels)\n \n # Build the network to have the number of hidden nodes and the learning rate that\n # were passed into this initializer. Make the same number of input nodes as\n # there are vocabulary words and create a single output node.\n self.init_network(len(self.review_vocab),hidden_nodes, 1, learning_rate)\n\n def pre_process_data(self, reviews, labels):\n \n # populate review_vocab with all of the words in the given reviews\n review_vocab = set()\n for review in reviews:\n for word in review.split(\" \"):\n review_vocab.add(word)\n\n # Convert the vocabulary set to a list so we can access words via indices\n self.review_vocab = list(review_vocab)\n \n # populate label_vocab with all of the words in the given labels.\n label_vocab = set()\n for label in labels:\n label_vocab.add(label)\n \n # Convert the label vocabulary set to a list so we can access labels via indices\n self.label_vocab = list(label_vocab)\n \n # Store the sizes of the review and label vocabularies.\n self.review_vocab_size = len(self.review_vocab)\n self.label_vocab_size = len(self.label_vocab)\n \n # Create a dictionary of words in the vocabulary mapped to index positions\n self.word2index = {}\n for i, word in enumerate(self.review_vocab):\n self.word2index[word] = i\n \n # Create a dictionary of labels mapped to index positions\n self.label2index = {}\n for i, label in enumerate(self.label_vocab):\n self.label2index[label] = i\n\n def init_network(self, input_nodes, hidden_nodes, output_nodes, learning_rate):\n # Set number of nodes in input, hidden and output layers.\n self.input_nodes = input_nodes\n self.hidden_nodes = hidden_nodes\n self.output_nodes = output_nodes\n\n # Store the learning rate\n self.learning_rate = learning_rate\n\n # Initialize weights\n\n # These are the weights between the input layer and the hidden layer.\n self.weights_0_1 = np.zeros((self.input_nodes,self.hidden_nodes))\n\n # These are the weights between the hidden layer and the output layer.\n self.weights_1_2 = np.random.normal(0.0, self.output_nodes**-0.5, \n (self.hidden_nodes, self.output_nodes))\n \n ## New for Project 5: Removed self.layer_0; added self.layer_1\n # The input layer, a two-dimensional matrix with shape 1 x hidden_nodes\n self.layer_1 = np.zeros((1,hidden_nodes))\n \n ## New for Project 5: Removed update_input_layer function\n \n def get_target_for_label(self,label):\n if(label == 'POSITIVE'):\n return 1\n else:\n return 0\n \n def sigmoid(self,x):\n return 1 / (1 + np.exp(-x))\n \n def sigmoid_output_2_derivative(self,output):\n return output * (1 - output)\n \n ## New for Project 5: changed name of first parameter form 'training_reviews' \n # to 'training_reviews_raw'\n def train(self, training_reviews_raw, training_labels):\n\n ## New for Project 5: pre-process training reviews so we can deal \n # directly with the indices of non-zero inputs\n training_reviews = list()\n for review in training_reviews_raw:\n indices = set()\n for word in review.split(\" \"):\n if(word in self.word2index.keys()):\n indices.add(self.word2index[word])\n training_reviews.append(list(indices))\n\n # make sure out we have a matching number of reviews and labels\n assert(len(training_reviews) == len(training_labels))\n \n # Keep track of correct predictions to display accuracy during training \n correct_so_far = 0\n\n # Remember when we started for printing time statistics\n start = time.time()\n \n # loop through all the given reviews and run a forward and backward pass,\n # updating weights for every item\n for i in range(len(training_reviews)):\n \n # Get the next review and its correct label\n review = training_reviews[i]\n label = training_labels[i]\n \n #### Implement the forward pass here ####\n ### Forward pass ###\n\n ## New for Project 5: Removed call to 'update_input_layer' function\n # because 'layer_0' is no longer used\n\n # Hidden layer\n ## New for Project 5: Add in only the weights for non-zero items\n self.layer_1 *= 0\n for index in review:\n self.layer_1 += self.weights_0_1[index]\n\n # Output layer\n ## New for Project 5: changed to use 'self.layer_1' instead of 'local layer_1'\n layer_2 = self.sigmoid(self.layer_1.dot(self.weights_1_2)) \n \n #### Implement the backward pass here ####\n ### Backward pass ###\n\n # Output error\n layer_2_error = layer_2 - self.get_target_for_label(label) # Output layer error is the difference between desired target and actual output.\n layer_2_delta = layer_2_error * self.sigmoid_output_2_derivative(layer_2)\n\n # Backpropagated error\n layer_1_error = layer_2_delta.dot(self.weights_1_2.T) # errors propagated to the hidden layer\n layer_1_delta = layer_1_error # hidden layer gradients - no nonlinearity so it's the same as the error\n\n # Update the weights\n ## New for Project 5: changed to use 'self.layer_1' instead of local 'layer_1'\n self.weights_1_2 -= self.layer_1.T.dot(layer_2_delta) * self.learning_rate # update hidden-to-output weights with gradient descent step\n \n ## New for Project 5: Only update the weights that were used in the forward pass\n for index in review:\n self.weights_0_1[index] -= layer_1_delta[0] * self.learning_rate # update input-to-hidden weights with gradient descent step\n\n # Keep track of correct predictions.\n if(layer_2 >= 0.5 and label == 'POSITIVE'):\n correct_so_far += 1\n elif(layer_2 < 0.5 and label == 'NEGATIVE'):\n correct_so_far += 1\n \n # For debug purposes, print out our prediction accuracy and speed \n # throughout the training process. \n elapsed_time = float(time.time() - start)\n reviews_per_second = i / elapsed_time if elapsed_time > 0 else 0\n \n sys.stdout.write(\"\\rProgress:\" + str(100 * i/float(len(training_reviews)))[:4] \\\n + \"% Speed(reviews/sec):\" + str(reviews_per_second)[0:5] \\\n + \" #Correct:\" + str(correct_so_far) + \" #Trained:\" + str(i+1) \\\n + \" Training Accuracy:\" + str(correct_so_far * 100 / float(i+1))[:4] + \"%\")\n if(i % 2500 == 0):\n print(\"\")\n \n def test(self, testing_reviews, testing_labels):\n \"\"\"\n Attempts to predict the labels for the given testing_reviews,\n and uses the test_labels to calculate the accuracy of those predictions.\n \"\"\"\n \n # keep track of how many correct predictions we make\n correct = 0\n\n # we'll time how many predictions per second we make\n start = time.time()\n\n # Loop through each of the given reviews and call run to predict\n # its label. \n for i in range(len(testing_reviews)):\n pred = self.run(testing_reviews[i])\n if(pred == testing_labels[i]):\n correct += 1\n \n # For debug purposes, print out our prediction accuracy and speed \n # throughout the prediction process. \n\n elapsed_time = float(time.time() - start)\n reviews_per_second = i / elapsed_time if elapsed_time > 0 else 0\n \n sys.stdout.write(\"\\rProgress:\" + str(100 * i/float(len(testing_reviews)))[:4] \\\n + \"% Speed(reviews/sec):\" + str(reviews_per_second)[0:5] \\\n + \" #Correct:\" + str(correct) + \" #Tested:\" + str(i+1) \\\n + \" Testing Accuracy:\" + str(correct * 100 / float(i+1))[:4] + \"%\")\n \n def run(self, review):\n \"\"\"\n Returns a POSITIVE or NEGATIVE prediction for the given review.\n \"\"\"\n # Run a forward pass through the network, like in the \"train\" function.\n \n ## New for Project 5: Removed call to update_input_layer function\n # because layer_0 is no longer used\n\n # Hidden layer\n ## New for Project 5: Identify the indices used in the review and then add\n # just those weights to layer_1 \n self.layer_1 *= 0\n unique_indices = set()\n for word in review.lower().split(\" \"):\n if word in self.word2index.keys():\n unique_indices.add(self.word2index[word])\n for index in unique_indices:\n self.layer_1 += self.weights_0_1[index]\n \n # Output layer\n ## New for Project 5: changed to use self.layer_1 instead of local layer_1\n layer_2 = self.sigmoid(self.layer_1.dot(self.weights_1_2))\n \n # Return POSITIVE for values above greater-than-or-equal-to 0.5 in the output layer;\n # return NEGATIVE for other values\n if(layer_2[0] >= 0.5):\n return \"POSITIVE\"\n else:\n return \"NEGATIVE\"\n\n",
"_____no_output_____"
]
],
[
[
"Run the following cell to recreate the network and train it once again.",
"_____no_output_____"
]
],
[
[
"mlp = SentimentNetwork(reviews[:-1000],labels[:-1000], learning_rate=0.1)\nmlp.train(reviews[:-1000],labels[:-1000])",
"\rProgress:0.0% Speed(reviews/sec):0.0 #Correct:1 #Trained:1 Training Accuracy:100.%\n"
]
],
[
[
"That should have trained much better than the earlier attempts. Run the following cell to test your model with 1000 predictions.",
"_____no_output_____"
]
],
[
[
"mlp.test(reviews[-1000:],labels[-1000:])",
"\rProgress:0.0% Speed(reviews/sec):0.0 #Correct:1 #Tested:1 Testing Accuracy:100.%\rProgress:0.1% Speed(reviews/sec):75.04 #Correct:1 #Tested:2 Testing Accuracy:50.0%\rProgress:0.2% Speed(reviews/sec):88.08 #Correct:2 #Tested:3 Testing Accuracy:66.6%\rProgress:0.3% Speed(reviews/sec):118.1 #Correct:3 #Tested:4 Testing Accuracy:75.0%\rProgress:0.4% Speed(reviews/sec):136.7 #Correct:4 #Tested:5 Testing Accuracy:80.0%\rProgress:0.5% Speed(reviews/sec):153.3 #Correct:5 #Tested:6 Testing Accuracy:83.3%\rProgress:0.6% Speed(reviews/sec):154.6 #Correct:6 #Tested:7 Testing Accuracy:85.7%\rProgress:0.7% Speed(reviews/sec):173.7 #Correct:7 #Tested:8 Testing Accuracy:87.5%\rProgress:0.8% Speed(reviews/sec):186.8 #Correct:8 #Tested:9 Testing Accuracy:88.8%\rProgress:0.9% Speed(reviews/sec):183.7 #Correct:9 #Tested:10 Testing Accuracy:90.0%\rProgress:1.0% Speed(reviews/sec):191.8 #Correct:10 #Tested:11 Testing Accuracy:90.9%\rProgress:1.1% Speed(reviews/sec):203.5 #Correct:11 #Tested:12 Testing Accuracy:91.6%\rProgress:1.2% Speed(reviews/sec):215.8 #Correct:12 #Tested:13 Testing Accuracy:92.3%\rProgress:1.3% Speed(reviews/sec):223.6 #Correct:13 #Tested:14 Testing Accuracy:92.8%\rProgress:1.4% Speed(reviews/sec):236.5 #Correct:14 #Tested:15 Testing Accuracy:93.3%\rProgress:1.5% Speed(reviews/sec):250.6 #Correct:15 #Tested:16 Testing Accuracy:93.7%\rProgress:1.6% Speed(reviews/sec):253.0 #Correct:16 #Tested:17 Testing Accuracy:94.1%\rProgress:1.7% Speed(reviews/sec):262.3 #Correct:17 #Tested:18 Testing Accuracy:94.4%\rProgress:1.8% Speed(reviews/sec):274.7 #Correct:18 #Tested:19 Testing Accuracy:94.7%\rProgress:1.9% Speed(reviews/sec):284.8 #Correct:18 #Tested:20 Testing Accuracy:90.0%\rProgress:2.0% Speed(reviews/sec):290.9 #Correct:19 #Tested:21 Testing Accuracy:90.4%\rProgress:2.1% Speed(reviews/sec):299.8 #Correct:20 #Tested:22 Testing Accuracy:90.9%\rProgress:2.2% Speed(reviews/sec):305.4 #Correct:21 #Tested:23 Testing Accuracy:91.3%\rProgress:2.3% Speed(reviews/sec):312.8 #Correct:22 #Tested:24 Testing Accuracy:91.6%\rProgress:2.4% Speed(reviews/sec):316.5 #Correct:23 #Tested:25 Testing Accuracy:92.0%\rProgress:2.5% Speed(reviews/sec):325.7 #Correct:24 #Tested:26 Testing Accuracy:92.3%\rProgress:2.6% Speed(reviews/sec):335.2 #Correct:25 #Tested:27 Testing Accuracy:92.5%\rProgress:2.7% Speed(reviews/sec):337.4 #Correct:26 #Tested:28 Testing Accuracy:92.8%\rProgress:2.8% Speed(reviews/sec):345.0 #Correct:27 #Tested:29 Testing Accuracy:93.1%\rProgress:2.9% Speed(reviews/sec):347.0 #Correct:28 #Tested:30 Testing Accuracy:93.3%\rProgress:3.0% Speed(reviews/sec):347.8 #Correct:29 #Tested:31 Testing Accuracy:93.5%\rProgress:3.1% Speed(reviews/sec):352.0 #Correct:30 #Tested:32 Testing Accuracy:93.7%\rProgress:3.2% Speed(reviews/sec):358.7 #Correct:31 #Tested:33 Testing Accuracy:93.9%\rProgress:3.3% Speed(reviews/sec):366.1 #Correct:32 #Tested:34 Testing Accuracy:94.1%\rProgress:3.4% Speed(reviews/sec):371.9 #Correct:33 #Tested:35 Testing Accuracy:94.2%\rProgress:3.5% Speed(reviews/sec):378.8 #Correct:34 #Tested:36 Testing Accuracy:94.4%\rProgress:3.6% Speed(reviews/sec):384.0 #Correct:35 #Tested:37 Testing Accuracy:94.5%\rProgress:3.7% Speed(reviews/sec):391.9 #Correct:35 #Tested:38 Testing Accuracy:92.1%\rProgress:3.8% Speed(reviews/sec):397.9 #Correct:36 #Tested:39 Testing Accuracy:92.3%\rProgress:3.9% Speed(reviews/sec):401.4 #Correct:36 #Tested:40 Testing Accuracy:90.0%\rProgress:4.0% Speed(reviews/sec):406.5 #Correct:37 #Tested:41 Testing Accuracy:90.2%\rProgress:4.1% Speed(reviews/sec):411.9 #Correct:38 #Tested:42 Testing Accuracy:90.4%\rProgress:4.2% Speed(reviews/sec):406.6 #Correct:39 #Tested:43 Testing Accuracy:90.6%\rProgress:4.3% Speed(reviews/sec):410.8 #Correct:40 #Tested:44 Testing Accuracy:90.9%\rProgress:4.4% Speed(reviews/sec):413.1 #Correct:41 #Tested:45 Testing Accuracy:91.1%\rProgress:4.5% Speed(reviews/sec):412.6 #Correct:41 #Tested:46 Testing Accuracy:89.1%\rProgress:4.6% Speed(reviews/sec):414.5 #Correct:42 #Tested:47 Testing Accuracy:89.3%\rProgress:4.7% Speed(reviews/sec):421.2 #Correct:43 #Tested:48 Testing Accuracy:89.5%\rProgress:4.8% Speed(reviews/sec):425.6 #Correct:44 #Tested:49 Testing Accuracy:89.7%\rProgress:4.9% Speed(reviews/sec):431.1 #Correct:45 #Tested:50 Testing Accuracy:90.0%\rProgress:5.0% Speed(reviews/sec):438.0 #Correct:46 #Tested:51 Testing Accuracy:90.1%\rProgress:5.1% Speed(reviews/sec):444.3 #Correct:47 #Tested:52 Testing Accuracy:90.3%\rProgress:5.2% Speed(reviews/sec):446.9 #Correct:48 #Tested:53 Testing Accuracy:90.5%\rProgress:5.3% Speed(reviews/sec):450.5 #Correct:49 #Tested:54 Testing Accuracy:90.7%\rProgress:5.4% Speed(reviews/sec):455.2 #Correct:50 #Tested:55 Testing Accuracy:90.9%\rProgress:5.5% Speed(reviews/sec):459.8 #Correct:51 #Tested:56 Testing Accuracy:91.0%\rProgress:5.6% Speed(reviews/sec):455.5 #Correct:52 #Tested:57 Testing Accuracy:91.2%\rProgress:5.7% Speed(reviews/sec):459.8 #Correct:53 #Tested:58 Testing Accuracy:91.3%\rProgress:5.8% Speed(reviews/sec):465.9 #Correct:54 #Tested:59 Testing Accuracy:91.5%\rProgress:5.9% Speed(reviews/sec):470.8 #Correct:55 #Tested:60 Testing Accuracy:91.6%\rProgress:6.0% Speed(reviews/sec):474.9 #Correct:56 #Tested:61 Testing Accuracy:91.8%\rProgress:6.1% Speed(reviews/sec):479.4 #Correct:57 #Tested:62 Testing Accuracy:91.9%\rProgress:6.2% Speed(reviews/sec):486.1 #Correct:58 #Tested:63 Testing Accuracy:92.0%\rProgress:6.3% Speed(reviews/sec):491.8 #Correct:59 #Tested:64 Testing Accuracy:92.1%\rProgress:6.4% Speed(reviews/sec):495.7 #Correct:59 #Tested:65 Testing Accuracy:90.7%\rProgress:6.5% Speed(reviews/sec):501.1 #Correct:59 #Tested:66 Testing Accuracy:89.3%\rProgress:6.6% Speed(reviews/sec):507.5 #Correct:60 #Tested:67 Testing Accuracy:89.5%\rProgress:6.7% Speed(reviews/sec):513.3 #Correct:61 #Tested:68 Testing Accuracy:89.7%\rProgress:6.8% Speed(reviews/sec):516.4 #Correct:62 #Tested:69 Testing Accuracy:89.8%\rProgress:6.9% Speed(reviews/sec):517.1 #Correct:63 #Tested:70 Testing Accuracy:90.0%\rProgress:7.0% Speed(reviews/sec):520.1 #Correct:63 #Tested:71 Testing Accuracy:88.7%\rProgress:7.1% Speed(reviews/sec):525.2 #Correct:64 #Tested:72 Testing Accuracy:88.8%\rProgress:7.2% Speed(reviews/sec):531.1 #Correct:65 #Tested:73 Testing Accuracy:89.0%\rProgress:7.3% Speed(reviews/sec):537.6 #Correct:66 #Tested:74 Testing Accuracy:89.1%\rProgress:7.4% Speed(reviews/sec):541.7 #Correct:66 #Tested:75 Testing Accuracy:88.0%\rProgress:7.5% Speed(reviews/sec):547.1 #Correct:67 #Tested:76 Testing Accuracy:88.1%\rProgress:7.6% Speed(reviews/sec):552.4 #Correct:68 #Tested:77 Testing Accuracy:88.3%\rProgress:7.7% Speed(reviews/sec):558.3 #Correct:69 #Tested:78 Testing Accuracy:88.4%\rProgress:7.8% Speed(reviews/sec):563.8 #Correct:70 #Tested:79 Testing Accuracy:88.6%\rProgress:7.9% Speed(reviews/sec):568.2 #Correct:71 #Tested:80 Testing Accuracy:88.7%\rProgress:8.0% Speed(reviews/sec):573.7 #Correct:72 #Tested:81 Testing Accuracy:88.8%\rProgress:8.1% Speed(reviews/sec):574.0 #Correct:72 #Tested:82 Testing Accuracy:87.8%\rProgress:8.2% Speed(reviews/sec):577.9 #Correct:73 #Tested:83 Testing Accuracy:87.9%\rProgress:8.3% Speed(reviews/sec):582.7 #Correct:74 #Tested:84 Testing Accuracy:88.0%\rProgress:8.4% Speed(reviews/sec):587.8 #Correct:75 #Tested:85 Testing Accuracy:88.2%\rProgress:8.5% Speed(reviews/sec):592.8 #Correct:76 #Tested:86 Testing Accuracy:88.3%\rProgress:8.6% Speed(reviews/sec):596.3 #Correct:77 #Tested:87 Testing Accuracy:88.5%\rProgress:8.7% Speed(reviews/sec):600.8 #Correct:77 #Tested:88 Testing Accuracy:87.5%\rProgress:8.8% Speed(reviews/sec):605.1 #Correct:78 #Tested:89 Testing Accuracy:87.6%\rProgress:8.9% Speed(reviews/sec):603.8 #Correct:79 #Tested:90 Testing Accuracy:87.7%\rProgress:9.0% Speed(reviews/sec):607.4 #Correct:80 #Tested:91 Testing Accuracy:87.9%\rProgress:9.1% Speed(reviews/sec):612.6 #Correct:81 #Tested:92 Testing Accuracy:88.0%\rProgress:9.2% Speed(reviews/sec):616.7 #Correct:82 #Tested:93 Testing Accuracy:88.1%\rProgress:9.3% Speed(reviews/sec):621.4 #Correct:83 #Tested:94 Testing Accuracy:88.2%\rProgress:9.4% Speed(reviews/sec):626.5 #Correct:84 #Tested:95 Testing Accuracy:88.4%\rProgress:9.5% Speed(reviews/sec):629.7 #Correct:85 #Tested:96 Testing Accuracy:88.5%\rProgress:9.6% Speed(reviews/sec):634.2 #Correct:86 #Tested:97 Testing Accuracy:88.6%\rProgress:9.7% Speed(reviews/sec):637.3 #Correct:87 #Tested:98 Testing Accuracy:88.7%\rProgress:9.8% Speed(reviews/sec):642.6 #Correct:88 #Tested:99 Testing Accuracy:88.8%\rProgress:9.9% Speed(reviews/sec):646.3 #Correct:89 #Tested:100 Testing Accuracy:89.0%\rProgress:10.0% Speed(reviews/sec):651.0 #Correct:90 #Tested:101 Testing Accuracy:89.1%\rProgress:10.1% Speed(reviews/sec):655.3 #Correct:91 #Tested:102 Testing Accuracy:89.2%\rProgress:10.2% Speed(reviews/sec):659.2 #Correct:92 #Tested:103 Testing Accuracy:89.3%\rProgress:10.3% Speed(reviews/sec):661.3 #Correct:92 #Tested:104 Testing Accuracy:88.4%\rProgress:10.4% Speed(reviews/sec):665.6 #Correct:93 #Tested:105 Testing Accuracy:88.5%\rProgress:10.5% Speed(reviews/sec):671.4 #Correct:94 #Tested:106 Testing Accuracy:88.6%\rProgress:10.6% Speed(reviews/sec):675.4 #Correct:95 #Tested:107 Testing Accuracy:88.7%\rProgress:10.7% Speed(reviews/sec):678.8 #Correct:96 #Tested:108 Testing Accuracy:88.8%\rProgress:10.8% Speed(reviews/sec):683.6 #Correct:97 #Tested:109 Testing Accuracy:88.9%\rProgress:10.9% Speed(reviews/sec):688.1 #Correct:97 #Tested:110 Testing Accuracy:88.1%\rProgress:11.0% Speed(reviews/sec):690.5 #Correct:98 #Tested:111 Testing Accuracy:88.2%\rProgress:11.1% Speed(reviews/sec):694.7 #Correct:99 #Tested:112 Testing Accuracy:88.3%\rProgress:11.2% Speed(reviews/sec):698.5 #Correct:100 #Tested:113 Testing Accuracy:88.4%\rProgress:11.3% Speed(reviews/sec):701.7 #Correct:101 #Tested:114 Testing Accuracy:88.5%\rProgress:11.4% Speed(reviews/sec):704.3 #Correct:102 #Tested:115 Testing Accuracy:88.6%\rProgress:11.5% Speed(reviews/sec):708.8 #Correct:102 #Tested:116 Testing Accuracy:87.9%\rProgress:11.6% Speed(reviews/sec):712.4 #Correct:103 #Tested:117 Testing Accuracy:88.0%\rProgress:11.7% Speed(reviews/sec):715.7 #Correct:104 #Tested:118 Testing Accuracy:88.1%\rProgress:11.8% Speed(reviews/sec):718.0 #Correct:105 #Tested:119 Testing Accuracy:88.2%\rProgress:11.9% Speed(reviews/sec):718.3 #Correct:106 #Tested:120 Testing Accuracy:88.3%\rProgress:12.0% Speed(reviews/sec):719.3 #Correct:107 #Tested:121 Testing Accuracy:88.4%\rProgress:12.1% Speed(reviews/sec):722.1 #Correct:108 #Tested:122 Testing Accuracy:88.5%\rProgress:12.2% Speed(reviews/sec):726.6 #Correct:109 #Tested:123 Testing Accuracy:88.6%\rProgress:12.3% Speed(reviews/sec):728.2 #Correct:110 #Tested:124 Testing Accuracy:88.7%\rProgress:12.4% Speed(reviews/sec):730.0 #Correct:111 #Tested:125 Testing Accuracy:88.8%\rProgress:12.5% Speed(reviews/sec):733.2 #Correct:112 #Tested:126 Testing Accuracy:88.8%\rProgress:12.6% Speed(reviews/sec):737.0 #Correct:113 #Tested:127 Testing Accuracy:88.9%\rProgress:12.7% Speed(reviews/sec):739.5 #Correct:114 #Tested:128 Testing Accuracy:89.0%\rProgress:12.8% Speed(reviews/sec):742.7 #Correct:115 #Tested:129 Testing Accuracy:89.1%\rProgress:12.9% Speed(reviews/sec):746.9 #Correct:116 #Tested:130 Testing Accuracy:89.2%\rProgress:13.0% Speed(reviews/sec):750.9 #Correct:116 #Tested:131 Testing Accuracy:88.5%\rProgress:13.1% Speed(reviews/sec):754.0 #Correct:117 #Tested:132 Testing Accuracy:88.6%\rProgress:13.2% Speed(reviews/sec):758.1 #Correct:118 #Tested:133 Testing Accuracy:88.7%\rProgress:13.3% Speed(reviews/sec):762.3 #Correct:119 #Tested:134 Testing Accuracy:88.8%\rProgress:13.4% Speed(reviews/sec):764.1 #Correct:120 #Tested:135 Testing Accuracy:88.8%\rProgress:13.5% Speed(reviews/sec):766.9 #Correct:120 #Tested:136 Testing Accuracy:88.2%\rProgress:13.6% Speed(reviews/sec):770.2 #Correct:121 #Tested:137 Testing Accuracy:88.3%\rProgress:13.7% Speed(reviews/sec):774.3 #Correct:122 #Tested:138 Testing Accuracy:88.4%\rProgress:13.8% Speed(reviews/sec):777.4 #Correct:123 #Tested:139 Testing Accuracy:88.4%\rProgress:13.9% Speed(reviews/sec):778.2 #Correct:124 #Tested:140 Testing Accuracy:88.5%\rProgress:14.0% Speed(reviews/sec):782.1 #Correct:125 #Tested:141 Testing Accuracy:88.6%\rProgress:14.1% Speed(reviews/sec):785.2 #Correct:126 #Tested:142 Testing Accuracy:88.7%\rProgress:14.2% Speed(reviews/sec):788.0 #Correct:127 #Tested:143 Testing Accuracy:88.8%\rProgress:14.3% Speed(reviews/sec):790.7 #Correct:128 #Tested:144 Testing Accuracy:88.8%\rProgress:14.4% Speed(reviews/sec):793.1 #Correct:129 #Tested:145 Testing Accuracy:88.9%\rProgress:14.5% Speed(reviews/sec):796.0 #Correct:130 #Tested:146 Testing Accuracy:89.0%\rProgress:14.6% Speed(reviews/sec):797.9 #Correct:131 #Tested:147 Testing Accuracy:89.1%\rProgress:14.7% Speed(reviews/sec):800.1 #Correct:132 #Tested:148 Testing Accuracy:89.1%\rProgress:14.8% Speed(reviews/sec):803.6 #Correct:133 #Tested:149 Testing Accuracy:89.2%\rProgress:14.9% Speed(reviews/sec):806.1 #Correct:134 #Tested:150 Testing Accuracy:89.3%\rProgress:15.0% Speed(reviews/sec):809.9 #Correct:135 #Tested:151 Testing Accuracy:89.4%\rProgress:15.1% Speed(reviews/sec):813.9 #Correct:136 #Tested:152 Testing Accuracy:89.4%\rProgress:15.2% Speed(reviews/sec):814.5 #Correct:137 #Tested:153 Testing Accuracy:89.5%\rProgress:15.3% Speed(reviews/sec):816.2 #Correct:138 #Tested:154 Testing Accuracy:89.6%\rProgress:15.4% Speed(reviews/sec):818.6 #Correct:139 #Tested:155 Testing Accuracy:89.6%\rProgress:15.5% Speed(reviews/sec):822.1 #Correct:140 #Tested:156 Testing Accuracy:89.7%\rProgress:15.6% Speed(reviews/sec):823.6 #Correct:141 #Tested:157 Testing Accuracy:89.8%\rProgress:15.7% Speed(reviews/sec):825.5 #Correct:141 #Tested:158 Testing Accuracy:89.2%\rProgress:15.8% Speed(reviews/sec):828.5 #Correct:142 #Tested:159 Testing Accuracy:89.3%\rProgress:15.9% Speed(reviews/sec):831.0 #Correct:143 #Tested:160 Testing Accuracy:89.3%\rProgress:16.0% Speed(reviews/sec):833.7 #Correct:143 #Tested:161 Testing Accuracy:88.8%\rProgress:16.1% Speed(reviews/sec):834.8 #Correct:143 #Tested:162 Testing Accuracy:88.2%\rProgress:16.2% Speed(reviews/sec):836.7 #Correct:144 #Tested:163 Testing Accuracy:88.3%\rProgress:16.3% Speed(reviews/sec):839.0 #Correct:145 #Tested:164 Testing Accuracy:88.4%\rProgress:16.4% Speed(reviews/sec):841.9 #Correct:146 #Tested:165 Testing Accuracy:88.4%\rProgress:16.5% Speed(reviews/sec):842.2 #Correct:147 #Tested:166 Testing Accuracy:88.5%\rProgress:16.6% Speed(reviews/sec):843.2 #Correct:148 #Tested:167 Testing Accuracy:88.6%\rProgress:16.7% Speed(reviews/sec):841.1 #Correct:149 #Tested:168 Testing Accuracy:88.6%\rProgress:16.8% Speed(reviews/sec):843.3 #Correct:150 #Tested:169 Testing Accuracy:88.7%\rProgress:16.9% Speed(reviews/sec):843.6 #Correct:151 #Tested:170 Testing Accuracy:88.8%\rProgress:17.0% Speed(reviews/sec):845.4 #Correct:151 #Tested:171 Testing Accuracy:88.3%\rProgress:17.1% Speed(reviews/sec):848.2 #Correct:152 #Tested:172 Testing Accuracy:88.3%\rProgress:17.2% Speed(reviews/sec):847.6 #Correct:153 #Tested:173 Testing Accuracy:88.4%\rProgress:17.3% Speed(reviews/sec):848.3 #Correct:153 #Tested:174 Testing Accuracy:87.9%\rProgress:17.4% Speed(reviews/sec):850.7 #Correct:154 #Tested:175 Testing Accuracy:88.0%\rProgress:17.5% Speed(reviews/sec):853.4 #Correct:155 #Tested:176 Testing Accuracy:88.0%\rProgress:17.6% Speed(reviews/sec):856.9 #Correct:156 #Tested:177 Testing Accuracy:88.1%\rProgress:17.7% Speed(reviews/sec):859.5 #Correct:157 #Tested:178 Testing Accuracy:88.2%\rProgress:17.8% Speed(reviews/sec):861.8 #Correct:158 #Tested:179 Testing Accuracy:88.2%\rProgress:17.9% Speed(reviews/sec):864.9 #Correct:159 #Tested:180 Testing Accuracy:88.3%\rProgress:18.0% Speed(reviews/sec):867.1 #Correct:160 #Tested:181 Testing Accuracy:88.3%\rProgress:18.1% Speed(reviews/sec):869.4 #Correct:161 #Tested:182 Testing Accuracy:88.4%\rProgress:18.2% Speed(reviews/sec):872.3 #Correct:162 #Tested:183 Testing Accuracy:88.5%\rProgress:18.3% Speed(reviews/sec):875.0 #Correct:163 #Tested:184 Testing Accuracy:88.5%\rProgress:18.4% Speed(reviews/sec):874.7 #Correct:164 #Tested:185 Testing Accuracy:88.6%\rProgress:18.5% Speed(reviews/sec):875.6 #Correct:165 #Tested:186 Testing Accuracy:88.7%\rProgress:18.6% Speed(reviews/sec):879.2 #Correct:165 #Tested:187 Testing Accuracy:88.2%\rProgress:18.7% Speed(reviews/sec):880.9 #Correct:166 #Tested:188 Testing Accuracy:88.2%\rProgress:18.8% Speed(reviews/sec):882.6 #Correct:167 #Tested:189 Testing Accuracy:88.3%\rProgress:18.9% Speed(reviews/sec):884.3 #Correct:168 #Tested:190 Testing Accuracy:88.4%\rProgress:19.0% Speed(reviews/sec):887.7 #Correct:169 #Tested:191 Testing Accuracy:88.4%\rProgress:19.1% Speed(reviews/sec):891.1 #Correct:170 #Tested:192 Testing Accuracy:88.5%\rProgress:19.2% Speed(reviews/sec):893.0 #Correct:170 #Tested:193 Testing Accuracy:88.0%\rProgress:19.3% Speed(reviews/sec):893.9 #Correct:170 #Tested:194 Testing Accuracy:87.6%\rProgress:19.4% Speed(reviews/sec):896.5 #Correct:171 #Tested:195 Testing Accuracy:87.6%\rProgress:19.5% Speed(reviews/sec):898.5 #Correct:171 #Tested:196 Testing Accuracy:87.2%\rProgress:19.6% Speed(reviews/sec):900.1 #Correct:172 #Tested:197 Testing Accuracy:87.3%\rProgress:19.7% Speed(reviews/sec):901.8 #Correct:173 #Tested:198 Testing Accuracy:87.3%\rProgress:19.8% Speed(reviews/sec):905.4 #Correct:174 #Tested:199 Testing Accuracy:87.4%\rProgress:19.9% Speed(reviews/sec):907.2 #Correct:175 #Tested:200 Testing Accuracy:87.5%\rProgress:20.0% Speed(reviews/sec):910.1 #Correct:176 #Tested:201 Testing Accuracy:87.5%\rProgress:20.1% Speed(reviews/sec):912.1 #Correct:177 #Tested:202 Testing Accuracy:87.6%\rProgress:20.2% Speed(reviews/sec):915.1 #Correct:178 #Tested:203 Testing Accuracy:87.6%\rProgress:20.3% Speed(reviews/sec):915.6 #Correct:179 #Tested:204 Testing Accuracy:87.7%\rProgress:20.4% Speed(reviews/sec):918.8 #Correct:180 #Tested:205 Testing Accuracy:87.8%\rProgress:20.5% Speed(reviews/sec):921.2 #Correct:181 #Tested:206 Testing Accuracy:87.8%\rProgress:20.6% Speed(reviews/sec):923.6 #Correct:182 #Tested:207 Testing Accuracy:87.9%\rProgress:20.7% Speed(reviews/sec):925.8 #Correct:183 #Tested:208 Testing Accuracy:87.9%\rProgress:20.8% Speed(reviews/sec):928.1 #Correct:184 #Tested:209 Testing Accuracy:88.0%\rProgress:20.9% Speed(reviews/sec):928.9 #Correct:184 #Tested:210 Testing Accuracy:87.6%\rProgress:21.0% Speed(reviews/sec):931.3 #Correct:184 #Tested:211 Testing Accuracy:87.2%\rProgress:21.1% Speed(reviews/sec):934.2 #Correct:185 #Tested:212 Testing Accuracy:87.2%\rProgress:21.2% Speed(reviews/sec):937.6 #Correct:186 #Tested:213 Testing Accuracy:87.3%\rProgress:21.3% Speed(reviews/sec):937.3 #Correct:187 #Tested:214 Testing Accuracy:87.3%\rProgress:21.4% Speed(reviews/sec):939.3 #Correct:188 #Tested:215 Testing Accuracy:87.4%\rProgress:21.5% Speed(reviews/sec):940.4 #Correct:189 #Tested:216 Testing Accuracy:87.5%\rProgress:21.6% Speed(reviews/sec):942.7 #Correct:190 #Tested:217 Testing Accuracy:87.5%\rProgress:21.7% Speed(reviews/sec):944.1 #Correct:191 #Tested:218 Testing Accuracy:87.6%\rProgress:21.8% Speed(reviews/sec):942.7 #Correct:192 #Tested:219 Testing Accuracy:87.6%\rProgress:21.9% Speed(reviews/sec):944.3 #Correct:193 #Tested:220 Testing Accuracy:87.7%\rProgress:22.0% Speed(reviews/sec):943.0 #Correct:194 #Tested:221 Testing Accuracy:87.7%\rProgress:22.1% Speed(reviews/sec):944.1 #Correct:195 #Tested:222 Testing Accuracy:87.8%\rProgress:22.2% Speed(reviews/sec):945.9 #Correct:196 #Tested:223 Testing Accuracy:87.8%\rProgress:22.3% Speed(reviews/sec):947.5 #Correct:197 #Tested:224 Testing Accuracy:87.9%\rProgress:22.4% Speed(reviews/sec):945.1 #Correct:198 #Tested:225 Testing Accuracy:88.0%\rProgress:22.5% Speed(reviews/sec):945.9 #Correct:199 #Tested:226 Testing Accuracy:88.0%\rProgress:22.6% Speed(reviews/sec):945.7 #Correct:200 #Tested:227 Testing Accuracy:88.1%\rProgress:22.7% Speed(reviews/sec):947.3 #Correct:201 #Tested:228 Testing Accuracy:88.1%\rProgress:22.8% Speed(reviews/sec):949.3 #Correct:202 #Tested:229 Testing Accuracy:88.2%\rProgress:22.9% Speed(reviews/sec):950.7 #Correct:203 #Tested:230 Testing Accuracy:88.2%\rProgress:23.0% Speed(reviews/sec):950.8 #Correct:204 #Tested:231 Testing Accuracy:88.3%\rProgress:23.1% Speed(reviews/sec):951.8 #Correct:205 #Tested:232 Testing Accuracy:88.3%\rProgress:23.2% Speed(reviews/sec):955.0 #Correct:206 #Tested:233 Testing Accuracy:88.4%\rProgress:23.3% Speed(reviews/sec):957.6 #Correct:207 #Tested:234 Testing Accuracy:88.4%\rProgress:23.4% Speed(reviews/sec):957.0 #Correct:208 #Tested:235 Testing Accuracy:88.5%"
]
],
[
[
"# End of Project 5. \n## Watch the next video to see Andrew's solution, then continue on to the next lesson.\n# Further Noise Reduction<a id='lesson_6'></a>",
"_____no_output_____"
]
],
[
[
"Image(filename='sentiment_network_sparse_2.png')",
"_____no_output_____"
],
[
"# words most frequently seen in a review with a \"POSITIVE\" label\npos_neg_ratios.most_common()",
"_____no_output_____"
],
[
"# words most frequently seen in a review with a \"NEGATIVE\" label\nlist(reversed(pos_neg_ratios.most_common()))[0:30]",
"_____no_output_____"
],
[
"from bokeh.models import ColumnDataSource, LabelSet\nfrom bokeh.plotting import figure, show, output_file\nfrom bokeh.io import output_notebook\noutput_notebook()",
"_____no_output_____"
],
[
"hist, edges = np.histogram(list(map(lambda x:x[1],pos_neg_ratios.most_common())), density=True, bins=100, normed=True)\n\np = figure(tools=\"pan,wheel_zoom,reset,save\",\n toolbar_location=\"above\",\n title=\"Word Positive/Negative Affinity Distribution\")\np.quad(top=hist, bottom=0, left=edges[:-1], right=edges[1:], line_color=\"#555555\")\nshow(p)",
"/Users/Mia/opt/anaconda3/envs/style-transfer/lib/python3.7/site-packages/ipykernel_launcher.py:1: DeprecationWarning: The normed argument is ignored when density is provided. In future passing both will result in an error.\n \"\"\"Entry point for launching an IPython kernel.\n"
],
[
"frequency_frequency = Counter()\n\nfor word, cnt in total_counts.most_common():\n frequency_frequency[cnt] += 1",
"_____no_output_____"
],
[
"hist, edges = np.histogram(list(map(lambda x:x[1],frequency_frequency.most_common())), density=True, bins=100, normed=True)\n\np = figure(tools=\"pan,wheel_zoom,reset,save\",\n toolbar_location=\"above\",\n title=\"The frequency distribution of the words in our corpus\")\np.quad(top=hist, bottom=0, left=edges[:-1], right=edges[1:], line_color=\"#555555\")\nshow(p)",
"_____no_output_____"
]
],
[
[
"# Project 6: Reducing Noise by Strategically Reducing the Vocabulary<a id='project_6'></a>\n\n**TODO:** Improve `SentimentNetwork`'s performance by reducing more noise in the vocabulary. Specifically, do the following:\n* Copy the `SentimentNetwork` class from the previous project into the following cell.\n* Modify `pre_process_data`:\n>* Add two additional parameters: `min_count` and `polarity_cutoff`\n>* Calculate the positive-to-negative ratios of words used in the reviews. (You can use code you've written elsewhere in the notebook, but we are moving it into the class like we did with other helper code earlier.)\n>* Andrew's solution only calculates a postive-to-negative ratio for words that occur at least 50 times. This keeps the network from attributing too much sentiment to rarer words. You can choose to add this to your solution if you would like. \n>* Change so words are only added to the vocabulary if they occur in the vocabulary more than `min_count` times.\n>* Change so words are only added to the vocabulary if the absolute value of their postive-to-negative ratio is at least `polarity_cutoff`\n* Modify `__init__`:\n>* Add the same two parameters (`min_count` and `polarity_cutoff`) and use them when you call `pre_process_data`",
"_____no_output_____"
]
],
[
[
"# TODO: -Copy the SentimentNetwork class from Project 5 lesson\n# -Modify it according to the above instructions ",
"_____no_output_____"
]
],
[
[
"Run the following cell to train your network with a small polarity cutoff.",
"_____no_output_____"
]
],
[
[
"mlp = SentimentNetwork(reviews[:-1000],labels[:-1000],min_count=20,polarity_cutoff=0.05,learning_rate=0.01)\nmlp.train(reviews[:-1000],labels[:-1000])",
"_____no_output_____"
]
],
[
[
"And run the following cell to test it's performance. It should be ",
"_____no_output_____"
]
],
[
[
"mlp.test(reviews[-1000:],labels[-1000:])",
"_____no_output_____"
]
],
[
[
"Run the following cell to train your network with a much larger polarity cutoff.",
"_____no_output_____"
]
],
[
[
"mlp = SentimentNetwork(reviews[:-1000],labels[:-1000],min_count=20,polarity_cutoff=0.8,learning_rate=0.01)\nmlp.train(reviews[:-1000],labels[:-1000])",
"_____no_output_____"
]
],
[
[
"And run the following cell to test it's performance.",
"_____no_output_____"
]
],
[
[
"mlp.test(reviews[-1000:],labels[-1000:])",
"_____no_output_____"
]
],
[
[
"# End of Project 6. \n## Watch the next video to see Andrew's solution, then continue on to the next lesson.",
"_____no_output_____"
],
[
"# Analysis: What's Going on in the Weights?<a id='lesson_7'></a>",
"_____no_output_____"
]
],
[
[
"mlp_full = SentimentNetwork(reviews[:-1000],labels[:-1000],min_count=0,polarity_cutoff=0,learning_rate=0.01)",
"_____no_output_____"
],
[
"mlp_full.train(reviews[:-1000],labels[:-1000])",
"_____no_output_____"
],
[
"Image(filename='sentiment_network_sparse.png')",
"_____no_output_____"
],
[
"def get_most_similar_words(focus = \"horrible\"):\n most_similar = Counter()\n\n for word in mlp_full.word2index.keys():\n most_similar[word] = np.dot(mlp_full.weights_0_1[mlp_full.word2index[word]],mlp_full.weights_0_1[mlp_full.word2index[focus]])\n \n return most_similar.most_common()",
"_____no_output_____"
],
[
"get_most_similar_words(\"excellent\")",
"_____no_output_____"
],
[
"get_most_similar_words(\"terrible\")",
"_____no_output_____"
],
[
"import matplotlib.colors as colors\n\nwords_to_visualize = list()\nfor word, ratio in pos_neg_ratios.most_common(500):\n if(word in mlp_full.word2index.keys()):\n words_to_visualize.append(word)\n \nfor word, ratio in list(reversed(pos_neg_ratios.most_common()))[0:500]:\n if(word in mlp_full.word2index.keys()):\n words_to_visualize.append(word)",
"_____no_output_____"
],
[
"pos = 0\nneg = 0\n\ncolors_list = list()\nvectors_list = list()\nfor word in words_to_visualize:\n if word in pos_neg_ratios.keys():\n vectors_list.append(mlp_full.weights_0_1[mlp_full.word2index[word]])\n if(pos_neg_ratios[word] > 0):\n pos+=1\n colors_list.append(\"#00ff00\")\n else:\n neg+=1\n colors_list.append(\"#000000\")",
"_____no_output_____"
],
[
"from sklearn.manifold import TSNE\ntsne = TSNE(n_components=2, random_state=0)\nwords_top_ted_tsne = tsne.fit_transform(vectors_list)",
"_____no_output_____"
],
[
"p = figure(tools=\"pan,wheel_zoom,reset,save\",\n toolbar_location=\"above\",\n title=\"vector T-SNE for most polarized words\")\n\nsource = ColumnDataSource(data=dict(x1=words_top_ted_tsne[:,0],\n x2=words_top_ted_tsne[:,1],\n names=words_to_visualize,\n color=colors_list))\n\np.scatter(x=\"x1\", y=\"x2\", size=8, source=source, fill_color=\"color\")\n\nword_labels = LabelSet(x=\"x1\", y=\"x2\", text=\"names\", y_offset=6,\n text_font_size=\"8pt\", text_color=\"#555555\",\n source=source, text_align='center')\np.add_layout(word_labels)\n\nshow(p)\n\n# green indicates positive words, black indicates negative words",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb88f30ada551021411ee29cbed2385f8a534cba
| 15,155 |
ipynb
|
Jupyter Notebook
|
Perceptron/Perceptron.ipynb
|
bhanu0925/DeepLearning
|
747445df8622e74eff8fae6aabb8cb156cbc16da
|
[
"MIT"
] | null | null | null |
Perceptron/Perceptron.ipynb
|
bhanu0925/DeepLearning
|
747445df8622e74eff8fae6aabb8cb156cbc16da
|
[
"MIT"
] | null | null | null |
Perceptron/Perceptron.ipynb
|
bhanu0925/DeepLearning
|
747445df8622e74eff8fae6aabb8cb156cbc16da
|
[
"MIT"
] | null | null | null | 25.686441 | 105 | 0.434048 |
[
[
[
"## Perceptron Implemetation",
"_____no_output_____"
],
[
"Data set or gate ",
"_____no_output_____"
]
],
[
[
"import os\nimport matplotlib.pyplot as plt\nimport joblib\nimport numpy as np\nimport pandas as pd\nplt.style.use(\"fivethirtyeight\")\n",
"_____no_output_____"
],
[
"class Perceptron:\n def __init__(self,eta,epochs):\n self.weights = np.random.randn(3) * 1e-4 # weights\n self.eta = eta # learning rate\n self.epochs = epochs # iterations\n \n def _z_outcome(self,inputs,weights):\n return np.dot(inputs,weights)\n \n def activation_function(self,z):\n return np.where(z>0,1,0)\n \n \n def fit(self,X,y):\n self.X = X\n self.y = y\n X_with_bias = np.c_[self.X,-np.ones((len(self.X),1))]\n print(f\"x_with_bias : \\n{X_with_bias}\")\n \n for epoc in range(self.epochs):\n z = self._z_outcome(X_with_bias,self.weights)\n y_hat = self.activation_function(z)\n \n print(f\"predicted value after forward pass: \\n{y_hat}\")\n \n self.error = self.y - y_hat\n print(f\"Error : \\n{self.error}\")\n \n # backword propagation \n self.weights = self.weights + self.eta * np.dot(X_with_bias.T, self.error)\n print(f\"updated epochs after epoch {epoc +1 } >> {self.epochs}:\\n{self.weights}\")\n \n def predict(self,X):\n X_with_bias = np.c_[X,-np.ones((len(X),1))]\n z = self._z_outcome(X_with_bias,self.weights)\n y_hat = self.activation_function(z)\n return y_hat\n \n",
"_____no_output_____"
],
[
"def prep_data(df,target_col = \"y\"):\n X = df.drop(target_col,axis = 1)\n y = df[target_col]\n return X,y",
"_____no_output_____"
]
],
[
[
"## OR",
"_____no_output_____"
]
],
[
[
"OR = {\n \"A\": [0,0,1,1],\n \"B\": [0,1,0,1],\n \"y\": [0,1,1,1]\n}\n\ndf_OR = pd.DataFrame(OR)\ndf_OR\n\nX,y = prep_data(df_OR)\nETA = 0.01\nEPOCHS = 10\nmodel_or = Perceptron(eta = ETA, epochs=EPOCHS)\n\nmodel_or.fit(X,y)\nmodel_or.predict(X)",
"x_with_bias : \n[[ 0. 0. -1.]\n [ 0. 1. -1.]\n [ 1. 0. -1.]\n [ 1. 1. -1.]]\npredicted value after forward pass: \n[1 1 1 1]\nError : \n0 -1\n1 0\n2 0\n3 0\nName: y, dtype: int64\nupdated epochs after epoch 1 >> 10:\n[7.35325501e-05 7.13490154e-05 9.99766552e-03]\npredicted value after forward pass: \n[0 0 0 0]\nError : \n0 0\n1 1\n2 1\n3 1\nName: y, dtype: int64\nupdated epochs after epoch 2 >> 10:\n[ 0.02007353 0.02007135 -0.02000233]\npredicted value after forward pass: \n[1 1 1 1]\nError : \n0 -1\n1 0\n2 0\n3 0\nName: y, dtype: int64\nupdated epochs after epoch 3 >> 10:\n[ 0.02007353 0.02007135 -0.01000233]\npredicted value after forward pass: \n[1 1 1 1]\nError : \n0 -1\n1 0\n2 0\n3 0\nName: y, dtype: int64\nupdated epochs after epoch 4 >> 10:\n[ 2.00735326e-02 2.00713490e-02 -2.33448496e-06]\npredicted value after forward pass: \n[1 1 1 1]\nError : \n0 -1\n1 0\n2 0\n3 0\nName: y, dtype: int64\nupdated epochs after epoch 5 >> 10:\n[0.02007353 0.02007135 0.00999767]\npredicted value after forward pass: \n[0 1 1 1]\nError : \n0 0\n1 0\n2 0\n3 0\nName: y, dtype: int64\nupdated epochs after epoch 6 >> 10:\n[0.02007353 0.02007135 0.00999767]\npredicted value after forward pass: \n[0 1 1 1]\nError : \n0 0\n1 0\n2 0\n3 0\nName: y, dtype: int64\nupdated epochs after epoch 7 >> 10:\n[0.02007353 0.02007135 0.00999767]\npredicted value after forward pass: \n[0 1 1 1]\nError : \n0 0\n1 0\n2 0\n3 0\nName: y, dtype: int64\nupdated epochs after epoch 8 >> 10:\n[0.02007353 0.02007135 0.00999767]\npredicted value after forward pass: \n[0 1 1 1]\nError : \n0 0\n1 0\n2 0\n3 0\nName: y, dtype: int64\nupdated epochs after epoch 9 >> 10:\n[0.02007353 0.02007135 0.00999767]\npredicted value after forward pass: \n[0 1 1 1]\nError : \n0 0\n1 0\n2 0\n3 0\nName: y, dtype: int64\nupdated epochs after epoch 10 >> 10:\n[0.02007353 0.02007135 0.00999767]\n"
]
],
[
[
"## AND",
"_____no_output_____"
]
],
[
[
"AND = {\n \"A\": [0,0,1,1],\n \"B\": [0,1,0,1],\n \"y\": [0,0,0,1]\n}\n\ndf_AND = pd.DataFrame(AND)\ndf_AND\n\nX,y = prep_data(df_AND)\nETA = 0.01\nEPOCHS = 10\nmodel_or = Perceptron(eta = ETA, epochs=EPOCHS)\n\nmodel_or.fit(X,y)\nmodel_or.predict(X)",
"x_with_bias : \n[[ 0. 0. -1.]\n [ 0. 1. -1.]\n [ 1. 0. -1.]\n [ 1. 1. -1.]]\npredicted value after forward pass: \n[0 1 0 1]\nError : \n0 0\n1 -1\n2 0\n3 0\nName: y, dtype: int64\nupdated epochs after epoch 1 >> 10:\n[ 7.11624898e-06 -9.95644238e-03 1.00131207e-02]\npredicted value after forward pass: \n[0 0 0 0]\nError : \n0 0\n1 0\n2 0\n3 1\nName: y, dtype: int64\nupdated epochs after epoch 2 >> 10:\n[1.00071162e-02 4.35576159e-05 1.31206511e-05]\npredicted value after forward pass: \n[0 1 1 1]\nError : \n0 0\n1 -1\n2 -1\n3 0\nName: y, dtype: int64\nupdated epochs after epoch 3 >> 10:\n[ 7.11624898e-06 -9.95644238e-03 2.00131207e-02]\npredicted value after forward pass: \n[0 0 0 0]\nError : \n0 0\n1 0\n2 0\n3 1\nName: y, dtype: int64\nupdated epochs after epoch 4 >> 10:\n[1.00071162e-02 4.35576159e-05 1.00131207e-02]\npredicted value after forward pass: \n[0 0 0 1]\nError : \n0 0\n1 0\n2 0\n3 0\nName: y, dtype: int64\nupdated epochs after epoch 5 >> 10:\n[1.00071162e-02 4.35576159e-05 1.00131207e-02]\npredicted value after forward pass: \n[0 0 0 1]\nError : \n0 0\n1 0\n2 0\n3 0\nName: y, dtype: int64\nupdated epochs after epoch 6 >> 10:\n[1.00071162e-02 4.35576159e-05 1.00131207e-02]\npredicted value after forward pass: \n[0 0 0 1]\nError : \n0 0\n1 0\n2 0\n3 0\nName: y, dtype: int64\nupdated epochs after epoch 7 >> 10:\n[1.00071162e-02 4.35576159e-05 1.00131207e-02]\npredicted value after forward pass: \n[0 0 0 1]\nError : \n0 0\n1 0\n2 0\n3 0\nName: y, dtype: int64\nupdated epochs after epoch 8 >> 10:\n[1.00071162e-02 4.35576159e-05 1.00131207e-02]\npredicted value after forward pass: \n[0 0 0 1]\nError : \n0 0\n1 0\n2 0\n3 0\nName: y, dtype: int64\nupdated epochs after epoch 9 >> 10:\n[1.00071162e-02 4.35576159e-05 1.00131207e-02]\npredicted value after forward pass: \n[0 0 0 1]\nError : \n0 0\n1 0\n2 0\n3 0\nName: y, dtype: int64\nupdated epochs after epoch 10 >> 10:\n[1.00071162e-02 4.35576159e-05 1.00131207e-02]\n"
]
],
[
[
"## XOR",
"_____no_output_____"
]
],
[
[
"XOR = {\n \"A\": [0,0,1,1],\n \"B\": [0,1,0,1],\n \"y\": [0,1,1,0]\n}\n\ndf_XOR = pd.DataFrame(XOR)\ndf_XOR\n\nX,y = prep_data(df_XOR)\nETA = 0.01\nEPOCHS = 10\nmodel_or = Perceptron(eta = ETA, epochs=EPOCHS)\n\nmodel_or.fit(X,y)\nmodel_or.predict(X)",
"x_with_bias : \n[[ 0. 0. -1.]\n [ 0. 1. -1.]\n [ 1. 0. -1.]\n [ 1. 1. -1.]]\npredicted value after forward pass: \n[0 0 1 1]\nError : \n0 0\n1 1\n2 0\n3 -1\nName: y, dtype: int64\nupdated epochs after epoch 1 >> 10:\n[-0.00986156 0.00011289 0.00013145]\npredicted value after forward pass: \n[0 0 0 0]\nError : \n0 0\n1 1\n2 1\n3 0\nName: y, dtype: int64\nupdated epochs after epoch 2 >> 10:\n[ 0.00013844 0.01011289 -0.01986855]\npredicted value after forward pass: \n[1 1 1 1]\nError : \n0 -1\n1 0\n2 0\n3 -1\nName: y, dtype: int64\nupdated epochs after epoch 3 >> 10:\n[-0.00986156 0.00011289 0.00013145]\npredicted value after forward pass: \n[0 0 0 0]\nError : \n0 0\n1 1\n2 1\n3 0\nName: y, dtype: int64\nupdated epochs after epoch 4 >> 10:\n[ 0.00013844 0.01011289 -0.01986855]\npredicted value after forward pass: \n[1 1 1 1]\nError : \n0 -1\n1 0\n2 0\n3 -1\nName: y, dtype: int64\nupdated epochs after epoch 5 >> 10:\n[-0.00986156 0.00011289 0.00013145]\npredicted value after forward pass: \n[0 0 0 0]\nError : \n0 0\n1 1\n2 1\n3 0\nName: y, dtype: int64\nupdated epochs after epoch 6 >> 10:\n[ 0.00013844 0.01011289 -0.01986855]\npredicted value after forward pass: \n[1 1 1 1]\nError : \n0 -1\n1 0\n2 0\n3 -1\nName: y, dtype: int64\nupdated epochs after epoch 7 >> 10:\n[-0.00986156 0.00011289 0.00013145]\npredicted value after forward pass: \n[0 0 0 0]\nError : \n0 0\n1 1\n2 1\n3 0\nName: y, dtype: int64\nupdated epochs after epoch 8 >> 10:\n[ 0.00013844 0.01011289 -0.01986855]\npredicted value after forward pass: \n[1 1 1 1]\nError : \n0 -1\n1 0\n2 0\n3 -1\nName: y, dtype: int64\nupdated epochs after epoch 9 >> 10:\n[-0.00986156 0.00011289 0.00013145]\npredicted value after forward pass: \n[0 0 0 0]\nError : \n0 0\n1 1\n2 1\n3 0\nName: y, dtype: int64\nupdated epochs after epoch 10 >> 10:\n[ 0.00013844 0.01011289 -0.01986855]\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb88f9bb8be00ee326032deeaa9cea9eaaf87085
| 61,588 |
ipynb
|
Jupyter Notebook
|
ch4/ch4_nb5_explore_imagenet_and_its_tiny_version.ipynb
|
Aldream/Hands-On-Computer-Vision-with-Tensorflow
|
3076dd279dbd87d4dda233823d6d558035aad523
|
[
"MIT"
] | null | null | null |
ch4/ch4_nb5_explore_imagenet_and_its_tiny_version.ipynb
|
Aldream/Hands-On-Computer-Vision-with-Tensorflow
|
3076dd279dbd87d4dda233823d6d558035aad523
|
[
"MIT"
] | null | null | null |
ch4/ch4_nb5_explore_imagenet_and_its_tiny_version.ipynb
|
Aldream/Hands-On-Computer-Vision-with-Tensorflow
|
3076dd279dbd87d4dda233823d6d558035aad523
|
[
"MIT"
] | 1 |
2020-10-28T06:56:56.000Z
|
2020-10-28T06:56:56.000Z
| 72.117096 | 25,384 | 0.74409 |
[
[
[
"<p style=\"border: 1px solid #e7692c; border-left: 15px solid #e7692c; padding: 10px; text-align:justify;\">\n <strong style=\"color: #e7692c\">Tip.</strong> <a style=\"color: #000000;\" href=\"https://nbviewer.jupyter.org/github/PacktPublishing/Hands-On-Computer-Vision-with-Tensorflow/blob/master/ch4/ch4_nb5_explore_imagenet_and_its_tiny_version.ipynb\" title=\"View with Jupyter Online\">Click here to view this notebook on <code>nbviewer.jupyter.org</code></a>. \n <br/>These notebooks are better read there, as Github default viewer ignores some of the formatting and interactive content.\n </p>",
"_____no_output_____"
],
[
"<table style=\"font-size: 1em; padding: 0; margin: 0;\">\n <tr style=\"vertical-align: top; padding: 0; margin: 0;\">\n <td style=\"vertical-align: top; padding: 0; margin: 0; padding-right: 15px;\">\n <p style=\"background: #363636; color:#ffffff; text-align:justify; padding: 10px 25px;\">\n <strong style=\"font-size: 1.0em;\"><span style=\"font-size: 1.2em;\"><span style=\"color: #e7692c;\">Hands-on</span> Computer Vision with TensorFlow 2</span><br/>by <em>Eliot Andres</em> & <em>Benjamin Planche</em> (Packt Pub.)</strong><br/><br/>\n <strong>> Chapter 4: Influential Classification Tools</strong><br/>\n </p>\n\n<h1 style=\"width: 100%; text-align: left; padding: 0px 25px;\"><small style=\"color: #e7692c;\">\n Notebook 5:</small><br/>Exploring ImageNet and Tiny-ImageNet</h1>\n<br/>\n<p style=\"border-left: 15px solid #363636; text-align:justify; padding: 0 10px;\">\n In this additional notebook, we demonstrate how those interested can acquire <em><strong>ImageNet</em></strong> and its smaller version <em><strong>Tiny-ImageNet</em></strong>, and can set up training pipelines using them. With this notebook, we will also briefly introduce the <code>tf.data</code> API.\n</p>\n<br/>\n<p style=\"border-left: 15px solid #e7692c; padding: 0 10px; text-align:justify;\">\n <strong style=\"color: #e7692c;\">Tip.</strong> The notebooks shared on this git repository illustrate some of notions from the book \"<em><strong>Hands-on Computer Vision with TensorFlow 2</strong></em>\" written by Eliot Andres and Benjamin Planche and published by Packt. If you enjoyed the insights shared here, <strong>please consider acquiring the book!</strong>\n<br/><br/>\nThe book provides further guidance for those eager to learn about computer vision and to harness the power of TensorFlow 2 and Keras to build performant recognition systems for object detection, segmentation, video processing, smartphone applications, and more.</p>\n </td>\n <td style=\"vertical-align: top; padding: 0; margin: 0; width: 250px;\">\n <a href=\"https://www.packtpub.com\" title=\"Buy on Packt!\">\n <img src=\"../banner_images/book_cover.png\" width=250>\n </a>\n <p style=\"background: #e7692c; color:#ffffff; padding: 10px; text-align:justify;\"><strong>Leverage deep learning to create powerful image processing apps with TensorFlow 2 and Keras. <br/></strong>Get the book for more insights!</p>\n <ul style=\"height: 32px; white-space: nowrap; text-align: center; margin: 0px; padding: 0px; padding-top: 10px;\">\n <li style=\"display: inline-block; height: 100%; vertical-align: middle; float: left; margin: 5px; padding: 0px;\">\n <a href=\"https://www.packtpub.com\" title=\"Get the book on Amazon!\">\n <img style=\"vertical-align: middle; max-width: 72px; max-height: 32px;\" src=\"../banner_images/logo_amazon.png\" width=\"75px\">\n </a>\n </li>\n <li style=\"display: inline-block; height: 100%; vertical-align: middle; float: left; margin: 5px; padding: 0px;\">\n <a href=\"https://www.packtpub.com\" title=\"Get your Packt book!\">\n <img style=\"vertical-align: middle; max-width: 72px; max-height: 32px;\" src=\"../banner_images/logo_packt.png\" width=\"75px\">\n </a>\n </li>\n <li style=\"display: inline-block; height: 100%; vertical-align: middle; float: left; margin: 5px; padding: 0px;\">\n <a href=\"https://www.packtpub.com\" title=\"Get the book on O'Reilly Safari!\">\n <img style=\"vertical-align: middle; max-width: 72px; max-height: 32px;\" src=\"../banner_images/logo_oreilly.png\" width=\"75px\">\n </a>\n </li>\n </ul>\n </td>\n </tr>\n </table>",
"_____no_output_____"
]
],
[
[
"import os\nimport glob\nimport tensorflow as tf\nfrom matplotlib import pyplot as plt",
"_____no_output_____"
]
],
[
[
"## Tiny-ImageNet",
"_____no_output_____"
],
[
"### Presentation",
"_____no_output_____"
],
[
"As presented in the chapter, the *ImageNet* dataset ([http://image-net.org](http://image-net.org)) and its yearly competition pushed forward the development of performant CNNs for image recognition[$^1$](#ref).\nWhile it could have been interesting to reuse this dataset to reproduce the results listed in the book, its huge size makes _ImageNet_ difficult to deploy on most machines (memory-wise). Training on such a dataset would also be a long, expensive task.\n\nAnother solution could have been to use only a portion of _ImageNet_. Indeed, the people at Standford University already compiled such a dataset for one of their famous classes (\"_CS231n: Convolutional Neural Networks for Visual Recognition_\" - http://cs231n.stanford.edu/). This dataset, _Tiny-ImageNet_ ([https://tiny-imagenet.herokuapp.com](https://tiny-imagenet.herokuapp.com)) contains 200 different classes (against the 1,000 of ImageNet). For each class, it offers 500 training images, 50 validation images, and 50 test ones.",
"_____no_output_____"
],
[
"### Setup",
"_____no_output_____"
],
[
"Tiny-ImageNet can be downloaded at [https://tiny-imagenet.herokuapp.com](https://tiny-imagenet.herokuapp.com) or [http://image-net.org/download-images](http://image-net.org/download-images) (users need the proper access).\n\n***Note:*** Makee sure to check the _ImageNet_ terms of use: [http://image-net.org/download-faq](http://image-net.org/download-faq).\n\nOnce downloaded, the archive can be unzipped (`unzip tiny-imagenet-200.zip`) at a proper location. Its path is stored into a variable:",
"_____no_output_____"
]
],
[
[
"ROOT_FOLDER = os.path.expanduser('~/datasets/tiny-imagenet-200/')",
"_____no_output_____"
]
],
[
[
"Let us have a look at the directory structure of the dataset:",
"_____no_output_____"
],
[
" - <ROOT_FOLDER>/tiny-imagenet-200/\n - wnids.txt <-- File with the list of class IDs in the dataset\n \n - words.txt <-- File with the mapping from class IDs to readable labels\n \n - train/ <-- Training folder\n - <class_i>/ <-- Folder containing training data of class <class_i> \n - images/ <-- Sub-folder with all the images for this class\n - ***.JPEG\n - n01443537_boxes.txt <-- Annotations for detection tasks (unused)\n \n - val/ <-- Validation folder\n - images/ <-- Folder with all the validation images\n - val_annotations.txt <-- File with the list of eval image filenames and\n the corresponding class IDs\n \n - test/ <-- Test folder\n - images/ <-- Folder containing all the test images",
"_____no_output_____"
],
[
"Finally, we define some additional dataset-related constants useful for later:",
"_____no_output_____"
]
],
[
[
"IMAGENET_IDS_FILE_BASENAME = 'wnids.txt' # File in ROOT_FOLDER containing the list of class IDs\nIMAGENET_WORDS_FILE_BASENAME = 'words.txt' # File in ROOT_FOLDER containing the mapping from class IDs to readable labels\nIMG_HEIGHT, IMG_WIDTH, IMG_CHANNELS = 64, 64, 3 # Image dimensions",
"_____no_output_____"
]
],
[
[
"## Input Pipeline",
"_____no_output_____"
],
[
"Datasets come in all forms and sizes. As training a CNN is a complex and heavy process, it is important to have an efficient data pipeline to provide the training batches on time to avoid performance bottlenecks.\n\nIn the following section, we will set up an input pipeline for a Tensorflow model, using Tiny-ImageNet as an example.",
"_____no_output_____"
],
[
"### Parsing the Labels",
"_____no_output_____"
],
[
"_Tiny-ImageNet_ is mainly organized by class. Therefore, let us start by listing and parsing those various classes.\n\nWe will use the two text files at the root of _Tiny-ImageNet_ to:\n - List the IDs corresponding to the 200 classes. This list will allow us to assign to each ID (IDs are 9-character-long strings) an integer from 0 to 199 (the ID position in the list);\n - Build a dictionary to map the IDs to human-readable labels (e.g., '_n01443537_' $ \\rightarrow$ '_goldfish, Carassius auratus_')\n\nThe first list is the most important, as it defines the categories (mapping the string IDs to numbers) which will be the target of our recognition models. The second structure, the dictionary, will simply allow us at the end to get understandable results.",
"_____no_output_____"
]
],
[
[
"def _get_class_information(ids_file, words_file):\n \"\"\"\n Extract the class IDs and corresponding human-readable labels from metadata files.\n :param ids_file: IDs filename (contains list of unique string class IDs)\n :param words_file: Words filename (contains list of tuples <ID, human-readable label>)\n :return: List of IDs, Dictionary of labels\n \"\"\"\n with open(ids_file, \"r\") as f:\n class_ids = [line[:-1] for line in f.readlines()] # removing the `\\n` for each line\n\n with open(words_file, \"r\") as f:\n words_lines = f.readlines()\n class_readable_labels = {}\n for line in words_lines:\n # We split the line between the ID (9-char long) and the human readable label:\n class_id = line[:9]\n class_label = line[10:-1]\n\n # If this class is in our dataset, we add it to our id-to-label dictionary:\n if class_id in class_ids:\n class_readable_labels[class_id] = class_label\n\n return class_ids, class_readable_labels\n",
"_____no_output_____"
]
],
[
[
"We can directly test this function:",
"_____no_output_____"
]
],
[
[
"ids_file = os.path.join(ROOT_FOLDER, IMAGENET_IDS_FILE_BASENAME)\nwords_file = os.path.join(ROOT_FOLDER, IMAGENET_WORDS_FILE_BASENAME)\nclass_ids, class_readable_labels = _get_class_information(ids_file, words_file)\n",
"_____no_output_____"
],
[
"# Let's for example print the 10 first IDs and their human-readable labels:\nfor i in range(10):\n id = class_ids[i]\n print('\"{}\" --> \"{}\"'.format(id, class_readable_labels[id]))\n",
"\"n02124075\" --> \"Egyptian cat\"\n\"n04067472\" --> \"reel\"\n\"n04540053\" --> \"volleyball\"\n\"n04099969\" --> \"rocking chair, rocker\"\n\"n07749582\" --> \"lemon\"\n\"n01641577\" --> \"bullfrog, Rana catesbeiana\"\n\"n02802426\" --> \"basketball\"\n\"n09246464\" --> \"cliff, drop, drop-off\"\n\"n07920052\" --> \"espresso\"\n\"n03970156\" --> \"plunger, plumber's helper\"\n"
]
],
[
[
"### Listing All Images and Labels",
"_____no_output_____"
],
[
"Now that we have the categories defined, we can list all the images along with their respective categorical labels.\n\nSince the dataset structure is different for training/validation/testing splits, we have to cover them separately. This happens often in practice, as defining a normalized structure for datasets is a complicated task (image format, annotation types, folder structure, etc. are heavily affected by the use-cases).\n\nIn this example, we will cover only the training and validation split:",
"_____no_output_____"
]
],
[
[
"def _get_train_image_files_and_labels(root_folder, class_ids):\n \"\"\"\n Fetch the lists of training images and numerical labels.\n We assume the images are stored as \"<root_folder>/train/<class_id>/images/*.JPEG\"\n :param root_folder: Dataset root folder\n :param class_ids: List of class IDs\n :return: List of image filenames and List of corresponding labels\n \"\"\"\n image_files, labels = [], []\n\n for i in range(len(class_ids)):\n class_id = class_ids[i]\n # Grabbing all the image files for this class:\n class_image_paths = os.path.join(root_folder, 'train', class_id, 'images', '*.JPEG')\n class_images = glob.glob(class_image_paths)\n # Creating as many numerical labels:\n class_labels = [i] * len(class_images)\n\n image_files += class_images\n labels += class_labels\n\n return image_files, labels",
"_____no_output_____"
],
[
"def _get_val_image_files_and_labels(root_folder, class_ids):\n \"\"\"\n Fetch the lists of validation images and numerical labels.\n We assume the images are stored as \"<root_folder>/train/<class_id>/images/*.JPEG\"\n :param root_folder: Dataset root folder\n :param class_ids: List of class IDs\n :return: List of image filenames and List of corresponding labels\n \"\"\"\n image_files, labels = [], []\n\n # The file 'val_annotations.txt' contains for each line the image filename and its annotations.\n # We parse it to build our dataset lists:\n val_annotation_file = os.path.join(root_folder, 'val', 'val_annotations.txt')\n with open(val_annotation_file, \"r\") as f:\n anno_lines = f.readlines()\n for line in anno_lines:\n split_line = line.split('\\t') # Splitting the line to extract the various pieces of info\n if len(split_line) > 1:\n image_file, image_class_id = split_line[0], split_line[1]\n class_num_id = class_ids.index(image_class_id)\n if class_num_id >= 0: # If the label belongs to our dataset, we add them:\n image_files.append(image_file)\n labels.append(class_num_id)\n\n return image_files, labels\n",
"_____no_output_____"
]
],
[
[
"If we call the method for the training data, we obtain our list of 500 * 200 = 100,000 images and their labels:",
"_____no_output_____"
]
],
[
[
"image_files, image_labels = _get_train_image_files_and_labels(ROOT_FOLDER, class_ids)\nprint(\"Number of training images: {}\".format(len(image_files)))",
"Number of training images: 100000\n"
]
],
[
[
"### Building an Iterable Dataset with Tensorflow",
"_____no_output_____"
],
[
"We need to convert this list of filenames into images, and generate a list of batches our model could iterate over during its training. There are however lots of elements to take into consideration. \n\nFor instance, pre-loading all the images may not be possible for modest machines (at least for bigger datasets); but loading images on the fly would cause continuous delays. Also, in several papers we presented in Chapter 4, data scientists are applying random transformations to the images at each iteration (cropping, scaling, etc.). Those operations are also consuming.\n\nAll in all, we would probably need some multi-thread pipeline for our inputs. Thankfully, Tensorflow provides us with an efficient solution. Its **`tf.data`** API contains several methods to build **`tf.data.Dataset()`** instances, a dataset structure which can be converted into batch iterators for TF models.\n\n***Note:*** The `tf.data` API is thoroughfully detailed later in Chapter [7](./ch7).",
"_____no_output_____"
],
[
"For instance, a `Dataset` can be created from tensors containing lists of elements. Therefore, we can easily wrap our `image_files` and `image_labels` into a `Dataset`, first converting them into tensors: ",
"_____no_output_____"
]
],
[
[
"image_files = tf.constant(image_files)\nimage_labels = tf.constant(image_labels)\ndataset = tf.data.Dataset.from_tensor_slices((image_files, image_labels))\ndataset\n",
"_____no_output_____"
]
],
[
[
"This object has multiple methods to transform its content, batch the elements, shuffle them, etc. Once defined, those operations will be applied only when necessary / called by the framework (like any other operation in TF graphs).\n\nOur goal is to have this dataset output batches of images and their labels. So first thing first, let us add an operation to obtain the images from the filenames:\n",
"_____no_output_____"
]
],
[
[
"def _parse_function(filename, label, size=[IMG_HEIGHT, IMG_WIDTH]):\n \"\"\"\n Parse the provided tensors, loading and resizing the corresponding image.\n Code snippet from https://www.tensorflow.org/guide/datasets#decoding_image_data_and_resizing_it (Apache 2.0 License).\n :param filename: Image filename (String Tensor)\n :param label: Image label\n :param size: Size to resize the images to\n :return: Image, Label\n \"\"\"\n # Reading the file and returning its content as bytes:\n image_string = tf.io.read_file(filename)\n # Decoding those into the image\n # (with `channels=3`, TF will duplicate the channels of grayscale images so they have 3 channels too):\n image_decoded = tf.io.decode_jpeg(image_string, channels=3)\n # Converting to float:\n image_float = tf.image.convert_image_dtype(image_decoded, tf.float32)\n # Resizing the image to the expected dimensions:\n image_resized = tf.image.resize(image_float, size)\n return image_resized, label\n\ndataset = dataset.map(_parse_function)\n",
"_____no_output_____"
]
],
[
[
"\n`dataset.map(fn)` tells the dataset to apply the function `fn` to each element requested at a given iteration. These functions can be chained. For example, we can add another function to randomly transform the training images, to artificially increase the number of different images our model can train on:",
"_____no_output_____"
]
],
[
[
"def _training_augmentation_fn(image, label):\n \"\"\"\n Apply random transformations to augment the training images.\n :param images: Images\n :param label: Labels\n :return: Augmented Images, Labels\n \"\"\"\n\n # Randomly applied horizontal flip:\n image = tf.image.random_flip_left_right(image)\n\n # Random B/S changes:\n image = tf.image.random_brightness(image, max_delta=0.1)\n image = tf.image.random_saturation(image, lower=0.5, upper=1.5)\n image = tf.clip_by_value(image, 0.0, 1.0) # keeping pixel values in check\n\n # Random resize and random crop back to expected size:\n original_shape = tf.shape(image)\n random_scale_factor = tf.random.uniform([1], minval=0.7, maxval=1.3, dtype=tf.float32)\n scaled_height = tf.cast(tf.cast(original_shape[0], tf.float32) * random_scale_factor, \n tf.int32)\n scaled_width = tf.cast(tf.cast(original_shape[1], tf.float32) * random_scale_factor, \n tf.int32)\n scaled_shape = tf.squeeze(tf.stack([scaled_height, scaled_width]))\n image = tf.image.resize(image, scaled_shape)\n image = tf.image.random_crop(image, original_shape)\n\n return image, label\n\ndataset.map(_training_augmentation_fn)\n",
"_____no_output_____"
]
],
[
[
"We can also specify if we want the dataset to be suffled, or sepcify how many elements we want at each iteration in a batch, how many times we want the dataset to be repeated (for multiple epochs), how many batches to pre-fetch, etc:\n",
"_____no_output_____"
]
],
[
[
"batch_size = 32\nnum_epochs = 30\n\ndataset = dataset.shuffle(buffer_size=10000)\ndataset = dataset.batch(batch_size)\ndataset = dataset.repeat(num_epochs)\ndataset = dataset.prefetch(1)\n",
"_____no_output_____"
]
],
[
[
"***Note:*** More detailed explanations on `Dataset` and its methods, as well as performance recommendations, will be provided in Chapter 7 and its [notebooks](../ch7).",
"_____no_output_____"
],
[
"Our dataset is ready, and we can now simply iterate over it to obtain our batches:",
"_____no_output_____"
]
],
[
[
"images, labels = next(dataset.__iter__())",
"_____no_output_____"
],
[
"# Displaying an example:\ni = 0\nclass_id = class_ids[labels[i]]\nreadable_label = class_readable_labels[class_id]\nprint(readable_label)\nplt.imshow(images[i])\n",
"organ, pipe organ\n"
]
],
[
[
"As we saw through the previous notebooks, this `tf.data.Dataset` instances can be simply passed to Keras models for their training.\n\n\n### Wrapping Up for Estimators",
"_____no_output_____"
],
[
"If we want to pass our dataset to an Estimator, we can wrap the iterable inputs (`images` here) into a dictionary in order to name the content.\n",
"_____no_output_____"
]
],
[
[
"batch = {'image': images, 'label': labels}",
"_____no_output_____"
]
],
[
[
"We know have our input pipeline ready. We will reuse these variables in the next notebooks. For clarity, we wrap their definition into easy-to-call functions:",
"_____no_output_____"
]
],
[
[
"def _input_fn(image_files, image_labels,\n shuffle=True, batch_size=32, num_epochs=None,\n augmentation_fn=None, wrap_for_estimator=True, resize_to=None):\n \"\"\"\n Prepares and returns the iterators for a dataset.\n :param image_files: List of image files\n :param image_labels: List of image labels\n :param shuffle: Flag to shuffle the dataset (if True)\n :param batch_size: Batch size\n :param num_epochs: Number of epochs (to repeat the iteration - infinite if None)\n :param augmentation_fn: opt. Augmentation function\n :param wrap_for_estimator: Flag to wrap the inputs to be passed for Estimators\n :param resize_to: (opt) Dimensions (h x w) to resize the images to\n :return: Iterable batched images and labels\n \"\"\"\n\n # Converting to TF dataset:\n image_files = tf.constant(image_files)\n image_labels = tf.constant(image_labels)\n dataset = tf.data.Dataset.from_tensor_slices((image_files, image_labels))\n if shuffle:\n dataset = dataset.shuffle(buffer_size=50000)\n # Adding parsing operation, to open and decode images:\n if resize_to is None:\n parse_fn = _parse_function\n else:\n # We specify to which dimensions to resize the images, if requested:\n parse_fn = partial(_parse_function, size=resize_to)\n dataset = dataset.map(parse_fn, num_parallel_calls=4)\n # Opt. adding some further transformations:\n if augmentation_fn is not None:\n dataset.map(augmentation_fn, num_parallel_calls=4)\n # Further preparing for iterating on:\n dataset = dataset.batch(batch_size)\n dataset = dataset.repeat(num_epochs)\n dataset = dataset.prefetch(1)\n if wrap_for_estimator:\n dataset = dataset.map(lambda img, label: {'image': img, 'label': label})\n return dataset\n\n\ndef tiny_imagenet(phase='train', shuffle=True, batch_size=32, num_epochs=None,\n augmentation_fn=_training_augmentation_fn, wrap_for_estimator=True,\n root_folder=ROOT_FOLDER, resize_to=None):\n \"\"\"\n Instantiate a Tiny-Image training or validation dataset, which can be passed to any model.\n :param phase: Phase ('train' or 'val')\n :param shuffle: Flag to shuffle the dataset (if True)\n :param batch_size: Batch size\n :param num_epochs: Number of epochs (to repeat the iteration - infinite if None)\n :param augmentation_fn: opt. Augmentation function\n :param wrap_for_estimator: Flag to wrap the inputs to be passed for Estimators\n :param root_folder: Dataset root folder\n :param resize_to: (opt) Dimensions (h x w) to resize the images to\n :return: Dataset pipeline, IDs List, Dictionary to read labels\n \"\"\"\n\n ids_file = os.path.join(root_folder, IMAGENET_IDS_FILE_BASENAME)\n words_file = os.path.join(root_folder, IMAGENET_WORDS_FILE_BASENAME)\n class_ids, class_readable_labels = _get_class_information(ids_file, words_file)\n if phase == 'train':\n image_files, image_labels = _get_train_image_files_and_labels(root_folder, class_ids)\n elif phase == 'val':\n image_files, image_labels = _get_val_image_files_and_labels(root_folder, class_ids)\n else:\n raise ValueError(\"Unknown phase ('train' or 'val' only)\")\n\n dataset = _input_fn(image_files, image_labels,\n shuffle, batch_size, num_epochs, augmentation_fn,\n wrap_for_estimator, resize_to)\n\n return dataset, class_ids, class_readable_labels",
"_____no_output_____"
]
],
[
[
"## ImageNet\n\nFor our more ambitious readers, the same process can be followed with the original *ImageNet* dataset, after acquiring it from its website ([http://image-net.org](http://image-net.org)).\n\nHowever, TensorFlow developers have made public the `tensorflow-datasets` package ([https://github.com/tensorflow/datasets](https://github.com/tensorflow/datasets)), which greatly simplifies the download and usage of many standard datasets (it is still up to the users to make sure they have the proper authorizations / they respect the terms of use for the datasets they download this way).\n\nWe will not extend further in this notebook, as `tensorflow-datasets` has been already properly introduced in a previous [notebook](./ch4_nb1_implement_resnet_from_scratch.ipynb). The explanations shared there can be directly applied to the _ImageNet_ version provided by these package ([details](https://github.com/tensorflow/datasets/blob/master/docs/datasets.md#imagenet2012)):",
"_____no_output_____"
]
],
[
[
"# !pip install tensorflow-datasets # Uncomment to install the module\nimport tensorflow_datasets as tfds\n\nimagenet_builder = tfds.builder(\"imagenet2012\")\nprint(imagenet_builder.info)\n\n# Uncommment to download and get started (check terms of use!):\n# imagenet_builder.download_and_prepare()",
"tfds.core.DatasetInfo(\n name='imagenet2012',\n version=2.0.1,\n description='ILSVRC 2012, aka ImageNet is an image dataset organized according to the\nWordNet hierarchy. Each meaningful concept in WordNet, possibly described by\nmultiple words or word phrases, is called a \"synonym set\" or \"synset\". There are\nmore than 100,000 synsets in WordNet, majority of them are nouns (80,000+). In\nImageNet, we aim to provide on average 1000 images to illustrate each synset.\nImages of each concept are quality-controlled and human-annotated. In its\ncompletion, we hope ImageNet will offer tens of millions of cleanly sorted\nimages for most of the concepts in the WordNet hierarchy.\n',\n urls=['http://image-net.org/'],\n features=FeaturesDict({\n 'file_name': Text(shape=(), dtype=tf.string, encoder=None),\n 'image': Image(shape=(None, None, 3), dtype=tf.uint8),\n 'label': ClassLabel(shape=(), dtype=tf.int64, num_classes=1000)\n },\n total_num_examples=1331167,\n splits={\n 'train': <tfds.core.SplitInfo num_examples=1281167>,\n 'validation': <tfds.core.SplitInfo num_examples=50000>\n },\n supervised_keys=('image', 'label'),\n citation='\"\"\"\n @article{ILSVRC15,\n Author = {Olga Russakovsky and Jia Deng and Hao Su and Jonathan Krause and Sanjeev Satheesh and Sean Ma and Zhiheng Huang and Andrej Karpathy and Aditya Khosla and Michael Bernstein and Alexander C. Berg and Li Fei-Fei},\n Title = {{ImageNet Large Scale Visual Recognition Challenge}},\n Year = {2015},\n journal = {International Journal of Computer Vision (IJCV)},\n doi = {10.1007/s11263-015-0816-y},\n volume={115},\n number={3},\n pages={211-252}\n }\n \n \"\"\"',\n redistribution_info=,\n)\n\n"
]
],
[
[
"<a id=\"ref\"></a>\n#### References\n\n1. Russakovsky, O., Deng, J., Su, H., Krause, J., Satheesh, S., Ma, S., Huang, Z., Karpathy, A., Khosla, A., Bernstein, M., Berg, A.C., Fei-Fei, L., 2014. ImageNet Large Scale Visual Recognition Challenge. arXiv:1409.0575 [cs].",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
cb8908ece9eebc2211016b9c26a5853c10849a8e
| 16,937 |
ipynb
|
Jupyter Notebook
|
prototypical-net-withinverse.ipynb
|
abgoswam/Prototypical-Networks
|
396b26bd1ba67c9cbfe0b90d3618b665aebd54f0
|
[
"MIT"
] | null | null | null |
prototypical-net-withinverse.ipynb
|
abgoswam/Prototypical-Networks
|
396b26bd1ba67c9cbfe0b90d3618b665aebd54f0
|
[
"MIT"
] | null | null | null |
prototypical-net-withinverse.ipynb
|
abgoswam/Prototypical-Networks
|
396b26bd1ba67c9cbfe0b90d3618b665aebd54f0
|
[
"MIT"
] | null | null | null | 33.340551 | 458 | 0.551337 |
[
[
[
"# Few-Shot Learning With Prototypical Networks",
"_____no_output_____"
]
],
[
[
"import torch\nimport numpy as np # linear algebra\nimport pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)\nfrom matplotlib import pyplot as plt\nimport cv2\nfrom tensorboardX import SummaryWriter\nfrom torch import optim\nfrom tqdm import tqdm\nimport multiprocessing as mp\nfrom preprocessing import read_images\nfrom prototypicalNet import PrototypicalNet, train_step, test_step, load_weights\ntqdm.pandas(desc=\"my bar!\")",
"C:\\ProgramData\\Anaconda3\\lib\\site-packages\\tqdm\\std.py:656: FutureWarning: The Panel class is removed from pandas. Accessing it from the top-level namespace will also be removed in the next version\n from pandas import Panel\nC:\\ProgramData\\Anaconda3\\lib\\site-packages\\tqdm\\std.py:656: FutureWarning: The Panel class is removed from pandas. Accessing it from the top-level namespace will also be removed in the next version\n from pandas import Panel\n"
]
],
[
[
"## Data Reading and Augmentation\nThe Omniglot data set is designed for developing more human-like learning algorithms. It contains 1623 different handwritten characters from 50 different alphabets. Then to increase the number of classes, all the images are rotated by 90, 180 and 270 degrees and each rotation resulted in one more class. Hence the total count of classes reached to 6492(1623 * 4) classes. We split images of 4200 classes to training data and the rest went to test set.",
"_____no_output_____"
]
],
[
[
"# Reading the data\nprint(\"Reading background images\")\ntrainx, trainy = read_images(r'D:\\_hackerreborn\\Prototypical-Networks\\input\\omniglot\\images_background')\nprint(trainx.shape)\nprint(trainy.shape)",
"Reading background images\n(77120, 28, 28, 3)\n(77120,)\n"
],
[
"# Checking if GPU is available\nuse_gpu = torch.cuda.is_available()\n\n# Converting input to pytorch Tensor\ntrainx = torch.from_numpy(trainx).float()\n\nif use_gpu:\n trainx = trainx.cuda()",
"_____no_output_____"
],
[
"trainx.shape, trainy.shape",
"_____no_output_____"
]
],
[
[
"## Model",
"_____no_output_____"
]
],
[
[
"import torch\nimport torchvision\nimport torch.nn as nn\nimport torch.nn.functional as F\nimport torch.optim as optim\nfrom torch.autograd import Variable\nfrom tqdm import trange\nfrom time import sleep\nimport numpy as np\nfrom sklearn.preprocessing import OneHotEncoder, LabelEncoder",
"_____no_output_____"
],
[
"class Net(nn.Module):\n \"\"\"\n Image2Vector CNN which takes image of dimension (28x28x3) and return column vector length 64\n \"\"\"\n def sub_block(self, in_channels, out_channels=64, kernel_size=3):\n block = torch.nn.Sequential(\n torch.nn.Conv2d(kernel_size=kernel_size, in_channels=in_channels, out_channels=out_channels, padding=1),\n torch.nn.BatchNorm2d(out_channels),\n torch.nn.ReLU(),\n torch.nn.MaxPool2d(kernel_size=2)\n )\n return block\n \n def __init__(self):\n super(Net, self).__init__()\n self.convnet1 = self.sub_block(3)\n self.convnet2 = self.sub_block(64)\n self.convnet3 = self.sub_block(64)\n self.convnet4 = self.sub_block(64)\n\n def forward(self, x):\n x = self.convnet1(x)\n x = self.convnet2(x)\n x = self.convnet3(x)\n x = self.convnet4(x)\n x = torch.flatten(x, start_dim=1)\n return x",
"_____no_output_____"
],
[
"class PrototypicalNet(nn.Module):\n def __init__(self, use_gpu=False):\n super(PrototypicalNet, self).__init__()\n self.f = Net()\n self.gpu = use_gpu\n if self.gpu:\n self.f = self.f.cuda()\n \n def forward(self, datax, datay, Ns,Nc, Nq, total_classes):\n \"\"\"\n Implementation of one episode in Prototypical Net\n datax: Training images\n datay: Corresponding labels of datax\n Nc: Number of classes per episode\n Ns: Number of support data per class\n Nq: Number of query data per class\n total_classes: Total classes in training set\n \"\"\"\n k = total_classes.shape[0]\n K = np.random.choice(total_classes, Nc, replace=False)\n Query_x = torch.Tensor()\n if(self.gpu):\n Query_x = Query_x.cuda()\n Query_y = []\n Query_y_count = []\n centroid_per_class = {}\n class_label = {}\n label_encoding = 0\n for cls in K:\n S_cls, Q_cls = self.random_sample_cls(datax, datay, Ns, Nq, cls)\n centroid_per_class[cls] = self.get_centroid(S_cls, Nc)\n class_label[cls] = label_encoding\n label_encoding += 1\n Query_x = torch.cat((Query_x, Q_cls), 0) # Joining all the query set together\n Query_y += [cls]\n Query_y_count += [Q_cls.shape[0]]\n Query_y, Query_y_labels = self.get_query_y(Query_y, Query_y_count, class_label)\n Query_x = self.get_query_x(Query_x, centroid_per_class, Query_y_labels)\n return Query_x, Query_y\n \n def random_sample_cls(self, datax, datay, Ns, Nq, cls):\n \"\"\"\n Randomly samples Ns examples as support set and Nq as Query set\n \"\"\"\n data = datax[(datay == cls).nonzero()]\n perm = torch.randperm(data.shape[0])\n idx = perm[:Ns]\n S_cls = data[idx]\n idx = perm[Ns : Ns+Nq]\n Q_cls = data[idx]\n if self.gpu:\n S_cls = S_cls.cuda()\n Q_cls = Q_cls.cuda()\n return S_cls, Q_cls\n \n def get_centroid(self, S_cls, Nc):\n \"\"\"\n Returns a centroid vector of support set for a class\n \"\"\"\n return torch.sum(self.f(S_cls), 0).unsqueeze(1).transpose(0,1) / Nc\n \n def get_query_y(self, Qy, Qyc, class_label):\n \"\"\"\n Returns labeled representation of classes of Query set and a list of labels.\n \"\"\"\n labels = []\n m = len(Qy)\n for i in range(m):\n labels += [Qy[i]] * Qyc[i]\n labels = np.array(labels).reshape(len(labels), 1)\n label_encoder = LabelEncoder()\n Query_y = torch.Tensor(label_encoder.fit_transform(labels).astype(int)).long()\n if self.gpu:\n Query_y = Query_y.cuda()\n Query_y_labels = np.unique(labels)\n return Query_y, Query_y_labels\n \n def get_centroid_matrix(self, centroid_per_class, Query_y_labels):\n \"\"\"\n Returns the centroid matrix where each column is a centroid of a class.\n \"\"\"\n centroid_matrix = torch.Tensor()\n if(self.gpu):\n centroid_matrix = centroid_matrix.cuda()\n for label in Query_y_labels:\n centroid_matrix = torch.cat((centroid_matrix, centroid_per_class[label]))\n if self.gpu:\n centroid_matrix = centroid_matrix.cuda()\n return centroid_matrix\n \n def get_query_x(self, Query_x, centroid_per_class, Query_y_labels):\n \"\"\"\n Returns distance matrix from each Query image to each centroid.\n \"\"\"\n centroid_matrix = self.get_centroid_matrix(centroid_per_class, Query_y_labels)\n Query_x = self.f(Query_x)\n m = Query_x.size(0)\n n = centroid_matrix.size(0)\n # The below expressions expand both the matrices such that they become compatible to each other in order to caclulate L2 distance.\n centroid_matrix = centroid_matrix.expand(m, centroid_matrix.size(0), centroid_matrix.size(1)) # Expanding centroid matrix to \"m\".\n Query_matrix = Query_x.expand(n, Query_x.size(0), Query_x.size(1)).transpose(0,1) # Expanding Query matrix \"n\" times\n Qx = torch.pairwise_distance(centroid_matrix.transpose(1,2), Query_matrix.transpose(1,2))\n return Qx",
"_____no_output_____"
],
[
"protonet = PrototypicalNet(use_gpu=use_gpu)\noptimizer = optim.SGD(protonet.parameters(), lr = 0.01, momentum=0.99)",
"_____no_output_____"
]
],
[
[
"## Training",
"_____no_output_____"
]
],
[
[
"def train_step(datax, datay, Ns,Nc, Nq):\n optimizer.zero_grad()\n Qx, Qy= protonet(datax, datay, Ns, Nc, Nq, np.unique(datay))\n Qx = Qx.max() - Qx\n pred = torch.log_softmax(Qx, dim=-1)\n loss = F.nll_loss(pred, Qy)\n loss.backward()\n optimizer.step()\n acc = torch.mean((torch.argmax(pred, 1) == Qy).float())\n return loss, acc",
"_____no_output_____"
],
[
"num_episode = 16000\nframe_size = 1000\ntrainx = trainx.permute(0, 3, 1, 2)",
"_____no_output_____"
],
[
"frame_loss = 0\nframe_acc = 0\nfor i in range(num_episode):\n loss, acc = train_step(trainx, trainy, 5, 60, 5)\n frame_loss += loss.data\n frame_acc += acc.data\n if( (i+1) % frame_size == 0):\n print(\"Frame Number:\", ((i+1) // frame_size), 'Frame Loss: ', frame_loss.data.cpu().numpy().tolist()/ frame_size, 'Frame Accuracy:', (frame_acc.data.cpu().numpy().tolist() * 100) / frame_size)\n frame_loss = 0\n frame_acc = 0",
"C:\\ProgramData\\Anaconda3\\lib\\site-packages\\sklearn\\preprocessing\\label.py:235: DataConversionWarning: A column-vector y was passed when a 1d array was expected. Please change the shape of y to (n_samples, ), for example using ravel().\n y = column_or_1d(y, warn=True)\n"
]
],
[
[
"## Testing",
"_____no_output_____"
]
],
[
[
"def test_step(datax, datay, Ns,Nc, Nq):\n Qx, Qy= protonet(datax, datay, Ns, Nc, Nq, np.unique(datay))\n pred = torch.log_softmax(Qx, dim=-1)\n loss = F.nll_loss(pred, Qy)\n acc = torch.mean((torch.argmax(pred, 1) == Qy).float())\n return loss, acc",
"_____no_output_____"
],
[
"num_test_episode = 2000",
"_____no_output_____"
],
[
"avg_loss = 0\navg_acc = 0\nfor _ in range(num_test_episode):\n loss, acc = test_step(testx, testy, 5, 60, 15)\n avg_loss += loss.data\n avg_acc += acc.data\nprint('Avg Loss: ', avg_loss.data.cpu().numpy().tolist() / num_test_episode , 'Avg Accuracy:', (avg_acc.data.cpu().numpy().tolist() * 100) / num_test_episode)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
cb890aace6bb06f40edc1e718625d1b98f4677e7
| 9,325 |
ipynb
|
Jupyter Notebook
|
jupyter_dev/Untitled.ipynb
|
ngocpham97/vnd-ai-chabot-nlp
|
c498b176295b92ae411ac1e42c2d5943eaf4043b
|
[
"Apache-2.0"
] | null | null | null |
jupyter_dev/Untitled.ipynb
|
ngocpham97/vnd-ai-chabot-nlp
|
c498b176295b92ae411ac1e42c2d5943eaf4043b
|
[
"Apache-2.0"
] | null | null | null |
jupyter_dev/Untitled.ipynb
|
ngocpham97/vnd-ai-chabot-nlp
|
c498b176295b92ae411ac1e42c2d5943eaf4043b
|
[
"Apache-2.0"
] | null | null | null | 44.404762 | 368 | 0.654048 |
[
[
[
"from transformers import RobertaTokenizer, TFRobertaModel",
"2021-08-03 06:55:56.101430: W tensorflow/stream_executor/platform/default/dso_loader.cc:59] Could not load dynamic library 'libcudart.so.10.1'; dlerror: libcudart.so.10.1: cannot open shared object file: No such file or directory\n2021-08-03 06:55:56.101483: I tensorflow/stream_executor/cuda/cudart_stub.cc:29] Ignore above cudart dlerror if you do not have a GPU set up on your machine.\n"
],
[
"import tensorflow as tf\n",
"_____no_output_____"
],
[
"tokenizer = RobertaTokenizer.from_pretrained('roberta-base')",
"_____no_output_____"
],
[
"model = TFRobertaModel.from_pretrained('roberta-base')",
"_____no_output_____"
],
[
"inputs = tokenizer(\"Hello, my dog is cute\", return_tensors=\"tf\")",
"_____no_output_____"
],
[
"inputs",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb8943f651946fd3d4178f8eb966bec2e6b9c4e6
| 38,810 |
ipynb
|
Jupyter Notebook
|
AGII_chap05_modelling_sphere.ipynb
|
georgkaufmann/lecture_agII
|
d042dae2a9ab3e47387744da7d6c47c382130acb
|
[
"MIT"
] | null | null | null |
AGII_chap05_modelling_sphere.ipynb
|
georgkaufmann/lecture_agII
|
d042dae2a9ab3e47387744da7d6c47c382130acb
|
[
"MIT"
] | null | null | null |
AGII_chap05_modelling_sphere.ipynb
|
georgkaufmann/lecture_agII
|
d042dae2a9ab3e47387744da7d6c47c382130acb
|
[
"MIT"
] | null | null | null | 176.409091 | 31,964 | 0.888457 |
[
[
[
"<table>\n<tr><td><img style=\"height: 150px;\" src=\"images/geo_hydro1.jpg\"></td>\n<td bgcolor=\"#FFFFFF\">\n <p style=\"font-size: xx-large; font-weight: 900; line-height: 100%\">AG Dynamics of the Earth</p>\n <p style=\"font-size: large; color: rgba(0,0,0,0.5);\">Jupyter notebooks</p>\n <p style=\"font-size: large; color: rgba(0,0,0,0.5);\">Georg Kaufmann</p>\n </td>\n</tr>\n</table>",
"_____no_output_____"
],
[
"# Angewandte Geophysik II: Kap 5: Gravimetrie\n# Schweremodellierung\n----\n*Georg Kaufmann,\nGeophysics Section,\nInstitute of Geological Sciences,\nFreie Universität Berlin,\nGermany*",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\nimport ipywidgets as widgets",
"_____no_output_____"
],
[
"# define profile\nxmin = -400.\nxmax = +400.\nxstep = 101\nx = np.linspace(xmin,xmax,xstep)",
"_____no_output_____"
]
],
[
[
"## 3D sphere\n\n<img src=figures/sketch_kugel.jpg style=width:10cm>\n\n$$\n g(x) = {{4}\\over{3}} \\pi G \\Delta\\rho R^3 {{D}\\over{(x^2 + D^2)^{3/2}}}\n$$",
"_____no_output_____"
]
],
[
[
"def boug_sphere(x,D=100.,R=50.,drho=500.):\n # Bouguer gravity of solid sphere\n G = 6.672e-11 # m^3/kg/s^2\n boug = 4./3.*np.pi*G*drho * R**3*D/(x**2+D**2)**(3/2)\n return boug",
"_____no_output_____"
],
[
"def plot_sphere(f1=False,f2=False,f3=False,f4=False,f5=False):\n fig,axs = plt.subplots(2,1,figsize=(12,8))\n\n axs[0].set_xlim([-400,400])\n axs[0].set_xticks([x for x in np.linspace(-300,300,7)])\n #axs[0].set_xlabel('Profile [m]')\n axs[0].set_ylim([0,0.4])\n axs[0].set_yticks([y for y in np.linspace(0,0.4,5)])\n axs[0].set_ylabel('Gravity [mGal]')\n axs[0].plot(x,1.e5*boug_sphere(x),linewidth=1.0,linestyle=':',color='black',label='sphere')\n if (f1):\n axs[0].plot(x,1.e5*boug_sphere(x),linewidth=2.0,linestyle='-',color='red',label='R=50m, D=100m')\n if (f2):\n axs[0].plot(x,1.e5*boug_sphere(x,D=80),linewidth=2.0,linestyle='--',color='red',label='R=50m, D=80m') \n if (f3):\n axs[0].plot(x,1.e5*boug_sphere(x,D=120),linewidth=2.0,linestyle=':',color='red',label='R=50m, D=120m')\n if (f4):\n axs[0].plot(x,1.e5*boug_sphere(x,R=40),linewidth=2.0,linestyle='-',color='green',label='R=40m, D=100m')\n if (f5):\n axs[0].plot(x,1.e5*boug_sphere(x,R=60),linewidth=2.0,linestyle='-',color='blue',label='R=60m, D=100m')\n axs[0].legend()\n\n axs[1].set_xlim([-400,400])\n axs[1].set_xticks([x for x in np.linspace(-300,300,7)])\n axs[1].set_xlabel('Profile [m]')\n axs[1].set_ylim([250,0])\n axs[1].set_yticks([y for y in np.linspace(0.,200.,5)])\n axs[1].set_ylabel('Depth [m]')\n angle = [theta for theta in np.linspace(0,2*np.pi,41)]\n R1=50.;D1=100.\n R2=50.;D2=80.\n R3=50.;D3=120.\n R4=40.;D4=100.\n R5=60.;D5=100.\n if (f1):\n axs[1].plot(R1*np.cos(angle),D1+R1*np.sin(angle),linewidth=2.0,linestyle='-',color='red',label='R=50m, D=100m')\n if (f2):\n axs[1].plot(R2*np.cos(angle),D2+R2*np.sin(angle),linewidth=2.0,linestyle='--',color='red',label='R=50m, D=80m')\n if (f3):\n axs[1].plot(R3*np.cos(angle),D3+R3*np.sin(angle),linewidth=2.0,linestyle=':',color='red',label='R=50m, D=120m')\n if (f4):\n axs[1].plot(R4*np.cos(angle),D4+R4*np.sin(angle),linewidth=2.0,linestyle='-',color='green',label='R=40m, D=100m')\n if (f5):\n axs[1].plot(R5*np.cos(angle),D5+R5*np.sin(angle),linewidth=2.0,linestyle='-',color='blue',label='R=60m, D=100m')",
"_____no_output_____"
],
[
"plot_sphere(f3=True)",
"_____no_output_____"
],
[
"# call interactive module\nw = dict(\nf1=widgets.Checkbox(value=True,description='eins',continuous_update=False,disabled=False),\n#a1=widgets.FloatSlider(min=0.,max=2.,step=0.1,value=1.0),\nf2=widgets.Checkbox(value=False,description='zwei',continuous_update=False,disabled=False),\nf3=widgets.Checkbox(value=False,description='drei',continuous_update=False,disabled=False),\nf4=widgets.Checkbox(value=False,description='vier',continuous_update=False,disabled=False),\nf5=widgets.Checkbox(value=False,description='fuenf',continuous_update=False,disabled=False))\noutput = widgets.interactive_output(plot_sphere, w)\nbox = widgets.HBox([widgets.VBox([*w.values()]), output])\ndisplay(box)",
"_____no_output_____"
]
],
[
[
"... done",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
cb894c1dab60c830105ec85a01f0c41037c40975
| 65,272 |
ipynb
|
Jupyter Notebook
|
Final_Project/scripts/.ipynb_checkpoints/fetch_game_reviews-checkpoint.ipynb
|
cbroker1/text-as-data
|
b28c7e48d10a155861445e9918f7ee29222e1cff
|
[
"MIT"
] | null | null | null |
Final_Project/scripts/.ipynb_checkpoints/fetch_game_reviews-checkpoint.ipynb
|
cbroker1/text-as-data
|
b28c7e48d10a155861445e9918f7ee29222e1cff
|
[
"MIT"
] | null | null | null |
Final_Project/scripts/.ipynb_checkpoints/fetch_game_reviews-checkpoint.ipynb
|
cbroker1/text-as-data
|
b28c7e48d10a155861445e9918f7ee29222e1cff
|
[
"MIT"
] | null | null | null | 36.628507 | 186 | 0.43858 |
[
[
[
"# 1. User Reviews via Steam API (https://partner.steamgames.com/doc/store/getreviews)",
"_____no_output_____"
]
],
[
[
"# import packages\nimport os\nimport sys\nimport time\nimport json\nimport numpy as np\nimport urllib.parse\nimport urllib.request\nfrom tqdm import tqdm\nimport plotly.express as px\nfrom datetime import datetime\nfrom googletrans import Translator\n\nimport pandas as pd\nfrom pandas import json_normalize\n\n# list package ver. etc.\nprint(\"Python version\")\nprint (sys.version)\nprint(\"Version info.\")\nprint (sys.version_info)\nprint('---------------')",
"Python version\n3.8.3 (default, May 19 2020, 18:47:26) \n[GCC 7.3.0]\nVersion info.\nsys.version_info(major=3, minor=8, micro=3, releaselevel='final', serial=0)\n---------------\n"
]
],
[
[
"---\n### Data Dictionary:\n\n- Response:\n - success - 1 if the query was successful\n - query_summary - Returned in the first request\n - recommendationid - The unique id of the recommendation\n - author\n - steamid - the user’s SteamID\n - um_games_owned - number of games owned by the user\n - num_reviews - number of reviews written by the user\n - playtime_forever - lifetime playtime tracked in this app\n - playtime_last_two_weeks - playtime tracked in the past two weeks for this app\n - playtime_at_review - playtime when the review was written\n - last_played - time for when the user last played\n - language - language the user indicated when authoring the review\n - review - text of written review\n - timestamp_created - date the review was created (unix timestamp)\n - timestamp_updated - date the review was last updated (unix timestamp)\n - voted_up - true means it was a positive recommendation\n - votes_up - the number of users that found this review helpful\n - votes_funny - the number of users that found this review funny\n - weighted_vote_score - helpfulness score\n - comment_count - number of comments posted on this review\n - steam_purchase - true if the user purchased the game on Steam\n - received_for_free - true if the user checked a box saying they got the app for free\n - written_during_early_access - true if the user posted this review while the game was in Early Access\n - developer_response - text of the developer response, if any\n - timestamp_dev_responded - Unix timestamp of when the developer responded, if applicable\n\n---\nSource: https://partner.steamgames.com/doc/store/getreviews",
"_____no_output_____"
],
[
"## 1.1 Import",
"_____no_output_____"
]
],
[
[
"# generate game review df\n\n#steam 'chunks' their json files (the game reviews) in sets of 100\n#ending with a signature, a 'cursor'. This cursor is then pasted\n#onto the the same url, to 'grab' the next chunk and so on. \n#This sequence block with an 'end cursor' of 'AoJ4tey90tECcbOXSw=='\n\n#set variables\nurl_base = 'https://store.steampowered.com/appreviews/393380?json=1&filter=updated&language=all&review_type=all&purchase_type=all&num_per_page=100&cursor='\n\n#first pass\nurl = urllib.request.urlopen(\"https://store.steampowered.com/appreviews/393380?json=1&filter=updated&language=all&review_type=all&purchase_type=all&num_per_page=100&cursor=*\")\ndata = json.loads(url.read().decode())\nnext_cursor = data['cursor']\nnext_cursor = next_cursor.replace('+', '%2B')\ndf1 = json_normalize(data['reviews'])\nprint(next_cursor)\n\n#add results till stopcursor met, then send all results to csv\nwhile True:\n time.sleep(0.5) # Sleep for one second\n url_temp = url_base + next_cursor\n url = urllib.request.urlopen(url_temp)\n data = json.loads(url.read().decode())\n next_cursor = data['cursor']\n next_cursor = next_cursor.replace('+', '%2B')\n df2 = json_normalize(data['reviews'])\n df1 = pd.concat([df1, df2])\n print(next_cursor)\n if next_cursor == 'AoJ44PCp0tECd4WXSw==' or next_cursor == '*':\n df_steam_reviews = df1\n df1 = None\n break\n \n#the hash below is each 'cursor' I loop through until the 'end cursor'.\n#this is just my way to monitor the download.",
"AoJwtMGZ6PICfJ%2BKjwI=\nAoJwrcGT4fICf5OSiAI=\nAoJ4v%2B6N2/ICcsuPjgI=\nAoJwheLr1PICf9HSjQI=\nAoJ416WgzvICc%2BaYjQI=\nAoJ4quzJyPICecbmjAI=\nAoJw0Mf4wvICdautjAI=\nAoJw1ovEvvICe/iEjAI=\nAoJ4qaXyufICfMXbiwI=\nAoJw65nYtfICe6e1iwI=\nAoJ4j53er/ICdsf6igI=\nAoJwkLeKqvICd63EigI=\nAoJwjdHhpvICeK2iigI=\nAoJ44M%2BYovICd%2BbwiQI=\nAoJ4t9nBnvICftXFiQI=\nAoJ40N6RmvICe8%2BPiQI=\nAoJwycW%2BlfICc9PeiAI=\nAoJwpezBkfICfsqyiAI=\nAoJw4fzBjfICeMaAiAI=\nAoJ47faaivICcqXchwI=\nAoJ43eSShvICcZCyhwI=\nAoJw4Ke2g/ICcemVhwI=\nAoJ4nPXrgPICevn7hgI=\nAoJ4ltHU/vECe6/0igE=\nAoJ4yIGA/PECeZ7IhgI=\nAoJwxLKL%2BvECd8rK%2BQE=\nAoJw%2BJiD%2BfECdpqphgI=\nAoJ4kbjh%2BPECcIOl0QE=\nAoJwk%2B%2BQ%2BPECcJKfhgI=\nAoJwmNz99vECcrjExAE=\nAoJ40Zfg9fECfoOFhgI=\nAoJ4mPjM8/ECfb3thQI=\nAoJwt%2Bmj8fECdvPRhQI=\nAoJ4sOfb7/ECdf%2B%2BhQI=\nAoJwo7bb7vECeey0hQI=\nAoJwwqX87fECf8yshQI=\nAoJwx7/X7fECeuqohQI=\nAoJ41djX7PECc62chQI=\nAoJ4m9fF6/ECeoyQhQI=\nAoJ4yp3k6vECd5GFhQI=\nAoJw0dP36fECeZv7hAI=\nAoJ4nsb96PECf9vwhAI=\nAoJ4zLCe6PECe/WR4QE=\nAoJ477395vECf47XhAI=\nAoJwmNeO5PECe/G1hAI=\nAoJ4rMSr3vECffHygwI=\nAoJ4z/Di2PECe4qxgwI=\nAoJ44pCH0vECcMTmggI=\nAoJ43/XOy/ECeu6WggI=\nAoJw8LTixPECd8XFgQI=\nAoJ4xLK4v/ECeb2EgQI=\nAoJ46aqOuvECd%2BC%2BgAI=\nAoJ4zJaZtfECf6%2BAgAI=\nAoJ4kKPXr/ECdNG3/wE=\nAoJ499TpqfECcuTr/gE=\nAoJwkvfQofECeMbw/QE=\nAoJw%2Bubqm/ECfvWZ/QE=\nAoJwuOOxlfECeNur/AE=\nAoJ46K2ZkfECfZzx%2BwE=\nAoJwo4rsjPECfKqx%2BwE=\nAoJ4/Y%2BVh/ECf67h%2BgE=\nAoJwspvcgfECcfiQ%2BgE=\nAoJwtLDL%2B/ACefK1%2BQE=\nAoJw7oCy9PACfZn74gE=\nAoJ4w9Xg7fACeeretAE=\nAoJ4hveo5/ACdpXO9wE=\nAoJ49af73vACfPL%2B9gE=\nAoJw/5/e2PACe5O99gE=\nAoJwlb6v0fACdIP69QE=\nAoJ48dK3y/ACcpDE9QE=\nAoJ4ko6JxfACfYmA9QE=\nAoJwsve2vvACfdjD9AE=\nAoJwieKcuPACe%2ByD9AE=\nAoJw8ZXQsPACfuXMlgE=\nAoJw8aaJqvACfLT18gE=\nAoJwi7CgpfACdZi/8gE=\nAoJ4r%2B3mn/ACeo%2BD8gE=\nAoJ41vzImPACcMO88QE=\nAoJw5Jvhk/ACe9mG8QE=\nAoJ42euYjvACfJPO8AE=\nAoJwwtrbhvACcNyJ8AE=\nAoJ4wOumgfACffvQ7wE=\nAoJwjerF%2B%2B8CcaeP7wE=\nAoJ4tpH39e8Cdq/U7gE=\nAoJwieL38O8CesCb7gE=\nAoJw3InS7O8CcKfp7QE=\nAoJw87yI5%2B8CeJ217QE=\nAoJ4pcLt4e8CfM2G7QE=\nAoJwkL2H3e8CdsTY7AE=\nAoJws4Sz2O8CeZir7AE=\nAoJw8pm00%2B8Cfp6A7AE=\nAoJ448//zu8Cc6TZ6wE=\nAoJ4rNuby%2B8CeOax6wE=\nAoJwsb3vx%2B8Cfq%2BK6wE=\nAoJw%2B%2Bb5we8CfojS6gE=\nAoJw4rOAve8Cc7if6gE=\nAoJ4mpiWue8CeOHw6QE=\nAoJ4r5edtu8CeuTH6QE=\nAoJ4sOv8su8Ce7%2BY6QE=\nAoJw9I7er%2B8Cc7Tq6AE=\nAoJwkPaKrO8Cc6G46AE=\nAoJ4k%2BOSqe8CdKWM6AE=\nAoJwmMa8pe8CcdvT5wE=\nAoJ4h%2B2lou8Cdc%2Bl5wE=\nAoJ45vWFn%2B8Cdd/VVA==\nAoJ48MiUmu8CeJ%2B55gE=\nAoJw59L4lu8CeIaM5gE=\nAoJwyv3ok%2B8Ceo7f5QE=\nAoJw28%2B5kO8CcIu45QE=\nAoJw/cqyje8CeI2b5QE=\nAoJwnqadie8Cfuzy5AE=\nAoJ4lPLuhO8CdJvB5AE=\nAoJ4gKqzgO8CdpyI5AE=\nAoJw0K/5%2Bu4CdcjP4wE=\nAoJ4rozI9u4CdOWk4wE=\nAoJ4iKaV8%2B4CdYGA4wE=\nAoJ43vna8O4CdbHd4gE=\nAoJ4ufKh7u4CdZC64gE=\nAoJw5LDe6%2B4CdKSa4gE=\nAoJwjIbm5%2B4CeJDp4QE=\nAoJwh42J5e4CdsW44QE=\nAoJ48bmz4%2B4Cd8eT4QE=\nAoJw25G04e4CeK3q4AE=\nAoJ4np7t3%2B4Cdd664AE=\nAoJwgeTV3u4CeunUsgE=\nAoJ4krvB3e4CfI3w3wE=\nAoJwrZup3O4CcbTO3wE=\nAoJw3uWA2%2B4CdMCj3wE=\nAoJ44c3a2e4CfsyC3wE=\nAoJw363h2O4CfvDa3gE=\nAoJwwNy81%2B4Ceay23gE=\nAoJ4j6%2BE1%2B4Cd6fIzgE=\nAoJwm%2BvM1u4Ccvv83QE=\nAoJw16ff1e4CdNnK3QE=\nAoJ47qqP1e4Ce%2B2p3QE=\nAoJw4bnN1O4Cc%2BX93AE=\nAoJ4gKWY0u4Cd%2BrS3AE=\nAoJ4grjSz%2B4CeLu73AE=\nAoJ4lPbrze4CfrWj3AE=\nAoJ4ubvJy%2B4CfNiE3AE=\nAoJ4nOm1ye4Ceonq2wE=\nAoJ4gey9x%2B4Cc/LY2wE=\nAoJ435b0xO4CdKPA2wE=\nAoJw%2Bb7Vwu4CfPCp2wE=\nAoJwsd/YwO4CerOU2wE=\nAoJ4z/7wv%2B4CeuaL2wE=\nAoJwv4%2BFv%2B4CdJOC2wE=\nAoJ4zfnqve4CdsX42gE=\nAoJwjY6Dve4Ccfns2gE=\nAoJw/fqbvO4Ccs7g2gE=\nAoJw6KKUu%2B4CfYzU2gE=\nAoJwmbanuu4CcevF2gE=\nAoJ4/If%2BuO4CdvKz2gE=\nAoJws8Hht%2B4Cf7Si2gE=\nAoJ4/JaCtu4CdJjR0wE=\nAoJw7PuAs%2B4CfN3u2QE=\nAoJwrcrQru4CcdLC2QE=\nAoJ46sTpq%2B4Cfayj2QE=\nAoJ4xb2wqe4CeP7%2B2AE=\nAoJw3eaDp%2B4CcpDY2AE=\nAoJ48ZKDpO4CdIOx2AE=\nAoJ4qvSPoe4CcOiM2AE=\nAoJ46pPNnu4Ce97s1wE=\nAoJwle%2BpnO4CfdPN1wE=\nAoJw6dqDmu4CeK%2Bs1wE=\nAoJ40O/fmO4Cd%2BiR1wE=\nAoJwoe63l%2B4CdfT31gE=\nAoJ44buvlu4Cc5je1gE=\nAoJ43ab0lO4CfaG71gE=\nAoJ4rdHuk%2B4Cffag1gE=\nAoJ485rEku4CdJH%2B1QE=\nAoJwo5TLke4Cfdzn1QE=\nAoJwy5LmkO4CcanJ1QE=\nAoJw%2Bdnoj%2B4Ce6Sw1QE=\nAoJw0LH4i%2B4CctiV1QE=\nAoJ4/73kg%2B4CftPt1AE=\nAoJwn4eC%2BO0Cd9DjlAE=\nAoJ4rvLH7u0Cceak1AE=\nAoJwsPeB4u0CcYSD1AE=\nAoJ4o%2BqU0u0CfObW0wE=\nAoJwxc6cwe0Cds6S0wE=\nAoJ4x9/Wru0Cc63e0gE=\nAoJwscODne0Cf6WaZQ==\nAoJwuajJje0CctH%2B0QE=\nAoJw0Y6I%2B%2BwCf8zM0QE=\nAoJw1pOU7ewCf96j0QE=\nAoJ4uMWG3ewCdvDt0AE=\nAoJw9vmPy%2BwCca%2B00AE=\nAoJ4gOWkv%2BwCfMWQ0AE=\nAoJwhenVtewCcLPyzwE=\nAoJ4yPzxruwCevTezwE=\nAoJ4wLeTp%2BwCd9bGzwE=\nAoJ45qzin%2BwCcpGwzwE=\nAoJwu9PLnewCe7KpzwE=\nAoJ4kO3RnOwCd4qmzwE=\nAoJw8fiVm%2BwCc6GhzwE=\nAoJw/vLhmewCd52czwE=\nAoJ4gMSnmOwCf%2BmXzwE=\nAoJ4gf7fluwCe42SzwE=\nAoJw4K3AkewCepaBzwE=\nAoJ4/tqbh%2BwCddHczgE=\nAoJ4q4ip/OsCd9C6zgE=\nAoJwqJLU7%2BsCcqSBzgE=\nAoJwqIiZ6OsCd9zpzQE=\nAoJ4yOuu5esCe/nBzQE=\nAoJ48ZjZ4%2BsCdM2YzQE=\nAoJw2ZTq4esCeIjvzAE=\nAoJwsaml4OsCd4rEzAE=\nAoJwqarx3usCf6GdzAE=\nAoJ4y6C13esCe6n1ywE=\nAoJ48smm3OsCebPPywE=\nAoJ49OeD2%2BsCfcikywE=\nAoJ4pbqC2usCd8P/ygE=\nAoJ49MOV2esCfd/TygE=\nAoJ4ocGU2OsCf8KsygE=\nAoJw8/LK1%2BsCdJ%2BOygE=\nAoJ49PmJ1%2BsCdaTjyQE=\nAoJ4q%2BzQ1usCdvW8yQE=\nAoJ4zpX61esCd76TyQE=\nAoJ4uKSy1esCeOHyyAE=\nAoJwtceE1esCdcbPyAE=\nAoJ4/f3j1OsCc5GryAE=\nAoJwlIeg1OsCc4P/xwE=\nAoJwvYHY0%2BsCfaDWxwE=\nAoJw4%2BLu0usCefumxwE=\nAoJwiPCu0usCcKv5xgE=\nAoJw9aOE0usCc%2BTKxgE=\nAoJwgNfW0esCfp2TxgE=\nAoJ4trqp0esCefnTxQE=\nAoJwkOzs0OsCf/COxQE=\nAoJwoZrA0OsCcMSHaw==\nAoJ4gq3Ty%2BsCdJS0xAE=\nAoJ4rpbev%2BsCecaAxAE=\nAoJwpeLcrOsCdN63wwE=\nAoJwhIbYnesCf7GHwwE=\nAoJwm8q%2BlOsCeJnnwgE=\nAoJwsLygg%2BsCeKytwgE=\nAoJ4tMbh8%2BoCduSbkwE=\nAoJ48t6T4%2BoCfou4rgE=\nAoJw7fix1OoCcZqCwQE=\nAoJ4zqzew%2BoCdM/NwAE=\nAoJ4vry8tuoCf6CgwAE=\nAoJ44O%2BIoOoCdOzBYw==\nAoJwv%2BL0geoCf/X3vgE=\nAoJw6LiG6OkCdvOYvgE=\nAoJ47quqzekCcOK3vQE=\nAoJ404vurekCf%2BnEvAE=\nAoJwkMr8j%2BkCe73rUA==\nAoJw2Pq78%2BgCf5rmugE=\nAoJ4wJXH3egCdbyCugE=\nAoJwru3OxugCf62ouQE=\nAoJ4742eqegCcK3XbQ==\nAoJ4i435k%2BgCftPetwE=\nAoJw76/ogegCeNaEtwE=\nAoJwiLCf%2BOcCfoXOtgE=\nAoJ44/no6ecCcI74tQE=\nAoJwwfOP2OcCctOrtQE=\nAoJ4ua6/yecCcoOlSw==\nAoJ4r8HdtOcCe7aLtAE=\nAoJ4z8PeqecCcarXswE=\nAoJw2MnrpucCcNOkswE=\nAoJ47vCgpecCdJH3sgE=\nAoJ4m6m5o%2BcCdurIsgE=\nAoJwl6WpoucCcejOjgE=\nAoJ4gO%2BBoecCcJn6sQE=\nAoJw39uVoOcCfaHOsQE=\nAoJ4yK7rnucCc4ilsQE=\nAoJ4uYulnucCcqSCsQE=\nAoJ4oZ34necCdZTesAE=\nAoJw2vSynecCe4WtsAE=\nAoJw7NjmnOcCcN/ahwE=\nAoJwtryinOcCdMLhrwE=\nAoJ4gLH7m%2BcCftu9rwE=\nAoJ46d6Zl%2BcCcfuOrwE=\nAoJwnvuZi%2BcCcu/lrgE=\nAoJwifmrhucCdYLYrgE=\nAoJ4sMyLhOcCdKzRrgE=\nAoJw4ZDvgecCfZnKrgE=\nAoJw7IiugOcCfq3ErgE=\nAoJw7fCq/%2BYCesXArgE=\nAoJ4z8S4/uYCebK8rgE=\nAoJ4kICy/OYCeLSLmAE=\nAoJ4nJf34uYCdOXSrQE=\nAoJ4/MD9yuYCdquKrQE=\nAoJ43t/NruYCfKCFnAE=\nAoJwrueQjuYCdYvGqwE=\nAoJwiejS8OUCf/nfqgE=\nAoJwxvCAzOUCe56RjwE=\nAoJw353HuOUCeq2gqQE=\nAoJw9p6OpeUCcujWqAE=\nAoJws8qnkeUCcZWwSw==\nAoJwvL7v%2BuQCf7TddQ==\nAoJ45tXo6OQCd/PrpgE=\nAoJ4v6Cr2uQCfPm4pgE=\nAoJw4smIy%2BQCdYz/pQE=\nAoJ465qQveQCfZHIpQE=\nAoJwrceisuQCdMKSpQE=\nAoJ4z/nnqeQCdbnnpAE=\nAoJ4m6KJoeQCcpC3pAE=\nAoJw%2BL%2BRmuQCcMmLpAE=\nAoJwlfWWluQCe7vyowE=\nAoJ46KC8kuQCeYLaowE=\nAoJ49J/lhOQCfImTowE=\nAoJwnven9OMCfL7LogE=\nAoJ42NH14%2BMCe6DkZA==\nAoJ4uOe91OMCcbDFoQE=\nAoJ4w4LIxOMCedP0oAE=\nAoJ4r4nwsuMCefa3oAE=\nAoJ4nqO7oeMCesCzggE=\nAoJ4z8eTluMCeavTnwE=\nAoJw9ZHFg%2BMCeviQnwE=\nAoJwzf%2B58%2BICc5jYngE=\nAoJwkva74%2BICdaKkngE=\nAoJ47v2kzeICe%2BbNnQE=\nAoJwrtK0uuICe7n5nAE=\nAoJw/rf%2BpuICeY%2BnnAE=\nAoJw5LzXk%2BICefrbmwE=\nAoJwocLHg%2BICffmOmwE=\nAoJwy//V9uECcsjUmgE=\nAoJ4tubc5%2BECf4yQmgE=\nAoJ41eDE3OECfcXTmQE=\nAoJ4yt390%2BECePmkmQE=\nAoJ4q5vmzOECcuv7mAE=\nAoJ4x5ebxOECcojUmAE=\nAoJwgs7Qv%2BECfbG%2BmAE=\nAoJw%2BvTFuuECe/WnmAE=\nAoJ4rsLut%2BECdPecmAE=\nAoJ4tfnTteECf52UmAE=\nAoJ41c3ys%2BECfYK7ag==\nAoJ4n7zyqeECfsjflwE=\nAoJwo5e/kuECdOrlSw==\nAoJwy%2BWl/uACdomclgE=\nAoJ4oLSD6eACceaxlQE=\nAoJwweDv3eACfcXmlAE=\nAoJw%2BP/F0%2BACerKclAE=\nAoJw152rzOACcLvlkwE=\nAoJw2avVweACeNWTkwE=\nAoJ4y6f5r%2BACdfOmkgE=\nAoJw7pXYmOACcIPWiwE=\nAoJ4vJCei%2BACdMn5kAE=\nAoJ4mtWNgeACede6kAE=\nAoJ4scGr/d8CcJj3jwE=\nAoJw3LCr%2B98CfYrBjwE=\nAoJ45%2BHQ%2Bd8Ce9uPjwE=\nAoJ4hPiD%2BN8CcOThjgE=\nAoJwjK/f9t8CdvGBYQ==\nAoJwj%2B6j9d8Cc%2BfxjQE=\nAoJ4rpmq9N8CfYOgjQE=\nAoJ4rLuU898CdZLVjAE=\nAoJ4tK7S8t8CffqgjAE=\nAoJ4oO7D7N8CcdSmZQ==\nAoJ4i8Lp5d8Ces2udg==\nAoJ4%2BNOL3t8CcJ6niwE=\nAoJw962M298CdtabiwE=\nAoJ4hfLU2d8Ce8qWiwE=\nAoJ4r%2B/92N8Ccf2TiwE=\nAoJ4yI2Q2N8CeYOPiwE=\nAoJ4zKT91t8Cd%2BiJiwE=\nAoJ43c7%2B1d8CdO%2BEiwE=\nAoJw6YP21N8Cfvv%2BigE=\nAoJ42eXh098Cfav6igE=\nAoJ4oLb60t8Cd%2Bz1igE=\nAoJw4IKt0d8CeZ7vigE=\nAoJ4/LHNx98CeYbDigE=\nAoJwiufSuN8Cd8bIiQE=\nAoJw5cHHqt8CeLqoiQE=\nAoJ46br2lN8CdpPEiAE=\nAoJwr86j/94CcPffhwE=\nAoJwodSt6d4Cfq/lhgE=\nAoJ4mJGj1t4CfaCHhgE=\nAoJ42r6rvt4CfsWdhQE=\nAoJ47a/cqt4Cep/RhAE=\nAoJ4%2BaTNlN4CfYLogwE=\nAoJ4lvj4gN4Cd4yVgwE=\nAoJwl7756N0CcvisggE=\nAoJ4r92Y2N0Cc%2BPngQE=\nAoJwkfPRxt0Cc72cgQE=\nAoJ4%2BLKjtt0CdM7WgAE=\nAoJwxbOPpd0CesujdA==\nAoJ4jZOvmd0Cf73Sfw==\nAoJ4m6qujd0Cfq2Sfw==\nAoJwoZv%2BhN0Cd8fIZQ==\nAoJwr9KN/twCetuRfg==\nAoJwopzt99wCd7DXfQ==\nAoJ4t7bq8twCfYWcfQ==\nAoJwwMbT7NwCdLjnfA==\nAoJ42Kq%2B5dwCeoW3fA==\nAoJwqsPl0NwCeMvGew==\nAoJ4ptr0vdwCcLfxeg==\nAoJ415ekr9wCc7eyeg==\nAoJw%2BKfPoNwCe/n0eQ==\nAoJ459mLj9wCdPq2eQ==\nAoJwt7es/tsCef/2eA==\nAoJ4w5Ld79sCc8TAeA==\nAoJ4jZiL4tsCeayOeA==\nAoJw39DF0dsCdszLdw==\nAoJ4%2BbzNxNsCe%2BGWdw==\nAoJ47sKJutsCecbydg==\nAoJ4gtSks9sCdN7Xdg==\nAoJwhtvTrtsCe6Wsdg==\nAoJw9s%2BZrNsCct%2B%2Bdg==\nAoJw7eSWq9sCdqu7dg==\nAoJw1oatqtsCeva4dg==\nAoJ49J/HqdsCdv20dg==\nAoJ4vPj4qNsCeoCydg==\nAoJw4rn3p9sCe9Oudg==\nAoJwp%2BeNp9sCccWqdg==\nAoJ4nuyyptsCfJSndg==\nAoJwofe4pdsCf5ikdg==\nAoJwrp/JpNsCf6mddg==\nAoJ4ps7So9sCdYyddg==\nAoJw8bC0otsCeMKZdg==\nAoJ4zP%2Bok9sCcpfedQ==\nAoJ4tYHo%2BtoCd8mGZg==\nAoJ4ufnW4NoCfZabdA==\nAoJ4jZjZy9oCcJbOcw==\nAoJ4hfj0s9oCc4/tcg==\nAoJ41%2BCzmtoCet2Acg==\nAoJ4hZyCgtoCc9GWcQ==\nAoJ4yZb27NkCe5DDcA==\nAoJw9Pve19kCevTwbw==\nAoJ4tL7dx9kCcd2tbw==\nAoJ47MOWt9kCeMLfbg==\nAoJ4r5XYrdkCdbulbg==\nAoJwsI/xo9kCfZHgbQ==\nAoJ4uee9mtkCda%2BYbQ==\nAoJ45bmpjdkCdMzSbA==\nAoJ42Yrt/NgCepmSbA==\nAoJ4i56l69gCd5nXaw==\nAoJ4qIGc2dgCeLn7UQ==\nAoJwytWu1NgCc5i7ag==\nAoJw16/e0dgCfdLyaQ==\nAoJww8vPz9gCdKymaQ==\nAoJ4lPXhzdgCfOnYaA==\nAoJwrqvoy9gCfqWFaA==\nAoJw18TsytgCe6SsZw==\nAoJ42e3oydgCfvvoZg==\nAoJwzN6wydgCf4SxZg==\nAoJ426SgxNgCdv3%2BZQ==\nAoJ4zvXZu9gCfee9ZQ==\nAoJ4v9rJttgCdLrKZQ==\nAoJ4mtyQs9gCfKzBZQ==\nAoJ46Zq/sdgCff27ZQ==\nAoJwuJmzsNgCdKq5ZQ==\nAoJ47ZLjr9gCcumnZQ==\nAoJ4tvOtr9gCfZK1ZQ==\nAoJwr/TvrtgCesuyZQ==\nAoJ46siBrtgCfravZQ==\nAoJwlPCjrdgCeOysZQ==\nAoJ4nKfurNgCetKqZQ==\nAoJ4zY%2B5rNgCc6%2BoZQ==\nAoJ42%2BKArNgCdZqmZQ==\nAoJ4uLGzq9gCcJ%2BkZQ==\nAoJ4v8jzqtgCec2iZQ==\nAoJ4sbDFqtgCd/mgZQ==\nAoJ4y/CRqtgCf/ieZQ==\nAoJwx53FqdgCcKKcZQ==\nAoJ4m7KxqNgCdJGZZQ==\nAoJ43rDYotgCdaSIZQ==\nAoJw8pPUkdgCcd7KZA==\nAoJ4553Z3tcCc5ScYw==\nAoJw9p%2B%2BsdcCcM2EYg==\nAoJ4pJLrjdcCecKVYQ==\nAoJw5fD16NYCd4SVYA==\nAoJw%2B4nVz9YCe%2BGuXw==\nAoJw27j6utYCc9zUXg==\nAoJ4%2B77SrNYCdK2mXg==\nAoJ4y4Deh9YCd4yiXQ==\nAoJ4577C6NUCdaevXA==\nAoJ4yqjO1dUCc9vMWw==\nAoJws6LDytUCefuMWw==\nAoJwkIyLwNUCeLfIWg==\nAoJ43rGuq9UCf4r%2BWQ==\nAoJ4k4fRi9UCd9iQWQ==\nAoJ4tq3e6tQCdIacWA==\nAoJ4/JjZy9QCcu%2BmVw==\nAoJ4/oHpsdQCdJTRVg==\nAoJwpqK/jtQCfarOVQ==\nAoJ47/zY8tMCd9nvVA==\nAoJ4i%2BrR1dMCe8KGVA==\nAoJwi%2BnltdMCe/2QUw==\nAoJ42vfknNMCdrGvUg==\nAoJwkfiTidMCeKLbUQ==\nAoJ4ntGc9dICcL2JUQ==\nAoJwrImh5tICeJnDUA==\nAoJ44Iqz19ICfIP6Tw==\nAoJw7db0xNICe/axTw==\nAoJwjP7ZtdICfqv1Tg==\nAoJw5sbcqdICcePKTg==\nAoJ4r6DhotICd5OtTg==\nAoJwgpGHnNICdPKXTg==\nAoJ4%2B%2B3YldICcP/%2BTQ==\nAoJwmOOsj9ICccniTQ==\nAoJwk439h9ICdcW%2BTQ==\nAoJwxMLngtICc%2BqbTQ==\nAoJ4nJ%2BQ/dECco/1TA==\nAoJw58uP%2BNECcIfVTA==\nAoJ4rfXE9NECdPC2TA==\nAoJ4iLHP8NECdf2cTA==\nAoJ43NrE7NECeOyFTA==\nAoJwkOu%2B6NECecfvSw==\nAoJwu9/i5dECep3dSw==\nAoJ4kOH24tECepWtSw==\nAoJ48Ino39ECftvFSw==\nAoJ4q7Lr3NECdbm5Sw==\nAoJwiZLB2tECdIqxSw==\nAoJwv4qa2NECcNiYSw==\nAoJ43sPI1tECe%2BmlSw==\nAoJ4xO2y1dECc4yhSw==\nAoJ4roGW1NECevidSw==\nAoJ4xZzF09ECdcCbSw==\nAoJ4/7T/0tECcfiYSw==\nAoJwv4Su0tECcIaXSw==\nAoJ44PCp0tECd4WXSw==\n"
],
[
"# inspect columns\nprint(df_steam_reviews.info(verbose=True))",
"<class 'pandas.core.frame.DataFrame'>\nInt64Index: 51321 entries, 0 to 20\nData columns (total 21 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 recommendationid 51321 non-null object \n 1 language 51321 non-null object \n 2 review 51321 non-null object \n 3 timestamp_created 51321 non-null int64 \n 4 timestamp_updated 51321 non-null int64 \n 5 voted_up 51321 non-null bool \n 6 votes_up 51321 non-null int64 \n 7 votes_funny 51321 non-null int64 \n 8 weighted_vote_score 51321 non-null object \n 9 comment_count 51321 non-null int64 \n 10 steam_purchase 51321 non-null bool \n 11 received_for_free 51321 non-null bool \n 12 written_during_early_access 51321 non-null bool \n 13 author.steamid 51321 non-null object \n 14 author.num_games_owned 51321 non-null int64 \n 15 author.num_reviews 51321 non-null int64 \n 16 author.playtime_forever 51321 non-null int64 \n 17 author.playtime_last_two_weeks 51321 non-null int64 \n 18 author.last_played 51321 non-null int64 \n 19 timestamp_dev_responded 1 non-null float64\n 20 developer_response 1 non-null object \ndtypes: bool(4), float64(1), int64(10), object(6)\nmemory usage: 7.2+ MB\nNone\n"
],
[
"# inspect shape\nprint(df_steam_reviews.shape)",
"(51321, 21)\n"
],
[
"# inspect df\ndf_steam_reviews",
"_____no_output_____"
],
[
"# save that sheet\ndf_steam_reviews.to_csv('squad_reviews.csv', index=False)",
"_____no_output_____"
]
],
[
[
"## 1.2 Clean",
"_____no_output_____"
]
],
[
[
"#search for presence of empty cells\ndf_steam_reviews.isnull().sum(axis = 0)",
"_____no_output_____"
],
[
"#drop empty cols 'timestamp_dev_responded' and 'developer_response'\ndf_steam_reviews = df_steam_reviews.drop(['timestamp_dev_responded', 'developer_response'], axis=1)",
"_____no_output_____"
],
[
"# convert unix timestamp columns to datetime format\ndef time_to_clean(x):\n return datetime.fromtimestamp(x)\n\ndf_steam_reviews['timestamp_created'] = df_steam_reviews['timestamp_created'].apply(time_to_clean)\ndf_steam_reviews['timestamp_updated'] = df_steam_reviews['timestamp_updated'].apply(time_to_clean)\ndf_steam_reviews['author.last_played'] = df_steam_reviews['author.last_played'].apply(time_to_clean)",
"_____no_output_____"
],
[
"# inspect\ndf_steam_reviews",
"_____no_output_____"
],
[
"# save that sheet\ndf_steam_reviews.to_csv('squad_reviews.csv', index=False)",
"_____no_output_____"
]
],
[
[
"# Misc",
"_____no_output_____"
]
],
[
[
"# list of free weekends:\nSquad Free Weekend - Nov 2016\nSquad Free Weekend - Apr 2017\nSquad Free Weekend - Nov 2017\nSquad Free Weekend - Jun 2018\nSquad Free Weekend - Nov 2018\nSquad Free Weekend - Jul 2019\nSquad Free Weekend - Nov 2019\n\n# list of major patch days:\nv1 - July 1 2015\nv2 - Oct 31 2015\nv3 - Dec 15 2015\nv4 - ?\nv5 - Mar 30 2016\nv6 - May 26 2016\nv7 - Aug 7 2016\nv8 - Nov 1 2016\nv9 - Mar 9 2017\nv10 Feb 5 2018\nv11 Jun 6 2018\nv12 Nov 29 2018\nv13 May ? 2019\nv14 Jun 28 2019\nv15 Jul 22 2019\nv16 Oct 10 2019\nv17 Nov 25 2019\nv18 ?\nv19 May 2 2020",
"_____no_output_____"
]
],
[
[
"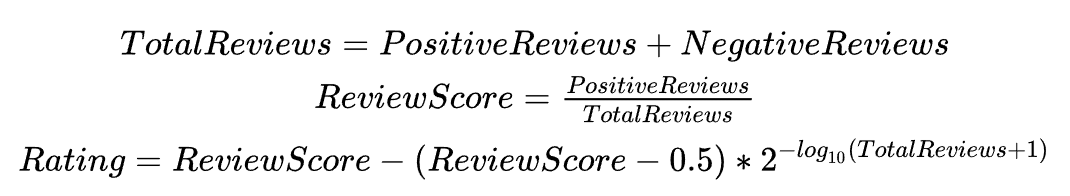",
"_____no_output_____"
]
],
[
[
"#v2 (fromhttps://cloud.google.com/translate/docs/simple-translate-call#translate_translate_text-python)\n# translate/spellcheck via googletranslate pkg\nfrom google.cloud import translate_v2 as translate\n\ndef time_to_translate(x):\n if x == None: # ignore the 'NaN' reviews\n return 'NaN'\n else:\n translate_client = translate.Client()\n if isinstance(x, six.binary_type):\n text = x.decode('utf-8')\n return text\n \n#print(time_to_translate('hola'))",
"_____no_output_____"
],
[
"# scratch\ndf_steam_reviews = pd.read_csv('squad_reviews.csv', low_memory=False)",
"_____no_output_____"
],
[
"df_steam_reviews",
"_____no_output_____"
],
[
"# display reviews \nfig = px.histogram(df_steam_reviews, x=\"timestamp_created\", color=\"voted_up\", width=1000, height=500, title='Positive(True)/Negative(False) Reviews')\nfig.show()",
"_____no_output_____"
],
[
"# translate/spellcheck t\nt['review.translated'] = t['review'].progress_apply(time_to_translate)\nt.to_csv('t.csv', index=False)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
cb895bd27e68957df90dca9381f88225f0637887
| 122,866 |
ipynb
|
Jupyter Notebook
|
docs/gallery/permanent_tutorial.ipynb
|
NunoEdgarGFlowHub/thewalrus
|
487957ec04a7d7da4a5007a0a9b9d209c4bee51f
|
[
"Apache-2.0"
] | 60 |
2019-08-13T18:28:00.000Z
|
2022-03-07T17:37:10.000Z
|
docs/gallery/permanent_tutorial.ipynb
|
NunoEdgarGFlowHub/thewalrus
|
487957ec04a7d7da4a5007a0a9b9d209c4bee51f
|
[
"Apache-2.0"
] | 280 |
2019-08-19T00:28:31.000Z
|
2022-03-28T19:25:12.000Z
|
docs/gallery/permanent_tutorial.ipynb
|
NunoEdgarGFlowHub/thewalrus
|
487957ec04a7d7da4a5007a0a9b9d209c4bee51f
|
[
"Apache-2.0"
] | 36 |
2019-09-18T18:23:28.000Z
|
2022-02-20T07:01:29.000Z
| 41.805376 | 319 | 0.478765 |
[
[
[
"# Benchmarking the Permanent\n\nThis tutorial shows how to use the permanent function using The Walrus, which calculates the permanent using Ryser's algorithm",
"_____no_output_____"
],
[
"### The Permanent\nThe permanent of an $n$-by-$n$ matrix A = $a_{i,j}$ is defined as\n\n$\\text{perm}(A)=\\sum_{\\sigma\\in S_n}\\prod_{i=1}^n a_{i,\\sigma(i)}.$\n\nThe sum here extends over all elements $\\sigma$ of the symmetric group $S_n$; i.e. over all permutations of the numbers $1, 2, \\ldots, n$. ([see Wikipedia](https://en.wikipedia.org/wiki/Permanent)).\n\nThe function `thewalrus.perm` implements [Ryser's algorithm](https://en.wikipedia.org/wiki/Computing_the_permanent#Ryser_formula) to calculate the permanent of an arbitrary matrix using [Gray code](https://en.wikipedia.org/wiki/Gray_code) ordering.",
"_____no_output_____"
],
[
"## Using the library",
"_____no_output_____"
],
[
"Once installed or compiled, one imports the library in the usual way:",
"_____no_output_____"
]
],
[
[
"from thewalrus import perm",
"_____no_output_____"
]
],
[
[
"To use it we need to pass square numpy arrays thus we also import NumPy:",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport time",
"_____no_output_____"
]
],
[
[
"The library provides functions to compute permanents of real and complex matrices. The functions take as arguments the matrix; the number of threads to be used to do the computation are determined using OpenMP.",
"_____no_output_____"
]
],
[
[
"size = 20\nmatrix = np.ones([size,size])\nperm(matrix)",
"_____no_output_____"
],
[
"size = 20\nmatrix = np.ones([size,size], dtype=np.complex128)\nperm(matrix)",
"_____no_output_____"
]
],
[
[
"Not surprisingly, the permanent of a matrix containing only ones equals the factorial of the dimension of the matrix, in our case $20!$.",
"_____no_output_____"
]
],
[
[
"from math import factorial\nfactorial(20)",
"_____no_output_____"
]
],
[
[
"### Benchmarking the performance of the code",
"_____no_output_____"
],
[
"For sizes $n=1,28$ we will generate random unitary matrices and measure the (average) amount of time it takes to calculate their permanent. The number of samples for each will be geometrically distirbuted with a 1000 samples for size $n=1$ and 10 samples for $n=28$. The unitaries will be random Haar distributed.",
"_____no_output_____"
]
],
[
[
"a0 = 1000.\nanm1 = 10.\nn = 28\nr = (anm1/a0)**(1./(n-1))\nnreps = [(int)(a0*(r**((i)))) for i in range(n)]",
"_____no_output_____"
],
[
"nreps",
"_____no_output_____"
]
],
[
[
"The following function generates random Haar unitaries of dimensions $n$",
"_____no_output_____"
]
],
[
[
"from scipy import diagonal, randn\nfrom scipy.linalg import qr\ndef haar_measure(n):\n '''A Random matrix distributed with Haar measure\n See https://arxiv.org/abs/math-ph/0609050\n How to generate random matrices from the classical compact groups\n by Francesco Mezzadri '''\n z = (randn(n,n) + 1j*randn(n,n))/np.sqrt(2.0)\n q,r = qr(z)\n d = diagonal(r)\n ph = d/np.abs(d)\n q = np.multiply(q,ph,q)\n return q",
"_____no_output_____"
]
],
[
[
"Now let's bench mark the scaling of the calculation with the matrix size:",
"_____no_output_____"
]
],
[
[
"times = np.empty(n)\nfor ind, reps in enumerate(nreps):\n #print(ind+1,reps)\n start = time.time()\n for i in range(reps):\n size = ind+1\n nth = 1\n matrix = haar_measure(size)\n res = perm(matrix)\n end = time.time()\n times[ind] = (end - start)/reps\n print(ind+1, times[ind])",
"1 0.00028934645652770995\n2 0.0001495122061081204\n3 0.00015489853603739133\n4 0.0004637452318194713\n5 0.00017665730844629873\n6 0.0006603159255265071\n7 0.0006937515768832151\n8 0.0008643358060629061\n9 0.0004252480525596469\n10 0.0008683936540470567\n11 0.0006751460923674357\n12 0.001460242115594203\n13 0.002330223719278971\n14 0.00457644021069562\n15 0.010608830294766268\n16 0.01871370959591556\n17 0.04012102713951698\n18 0.08530152927745473\n19 0.17479530106420102\n20 0.3602719612610646\n21 0.7553631681384463\n22 1.603381006805985\n23 3.432816049327021\n24 7.144709775322362\n25 14.937034338712692\n26 31.058412892477854\n27 64.35744528336959\n28 136.51278076171874\n"
]
],
[
[
"We can now plot the (average) time it takes to calculate the permanent vs. the size of the matrix:",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\n%matplotlib inline\n%config InlineBackend.figure_formats=['svg']",
"_____no_output_____"
],
[
"plt.semilogy(np.arange(1,n+1),times,\"+\")\nplt.xlabel(r\"Matrix size $n$\")\nplt.ylabel(r\"Time in seconds for 4 threads\")",
"_____no_output_____"
]
],
[
[
"We can also fit to the theoretical scaling of $ c n 2^n$ and use it to extrapolate for larger sizes:",
"_____no_output_____"
]
],
[
[
"def fit(n,c):\n return c*n*2**n",
"_____no_output_____"
],
[
"from scipy.optimize import curve_fit\npopt, pcov = curve_fit(fit, np.arange(1,n+1)[15:-1],times[15:-1])",
"_____no_output_____"
]
],
[
[
"The scaling prefactor is ",
"_____no_output_____"
]
],
[
[
"popt[0]",
"_____no_output_____"
]
],
[
[
"And we can use it to extrapolate the time it takes to calculate permanents of bigger dimensions",
"_____no_output_____"
]
],
[
[
"flags = [3600,3600*24*7, 3600*24*365, 3600*24*365*1000]\nlabels = [\"1 hour\", \"1 week\", \"1 year\", \"1000 years\"]\nplt.semilogy(np.arange(1,n+1), times, \"+\", np.arange(1,61), fit(np.arange(1,61),popt[0]))\nplt.xlabel(r\"Matrix size $n$\")\nplt.ylabel(r\"Time in seconds for single thread\")\nplt.hlines(flags,0,60,label=\"1 hr\",linestyles=u'dotted')\nfor i in range(len(flags)):\n plt.text(0,2*flags[i], labels[i])",
"_____no_output_____"
]
],
[
[
"The specs of the computer on which this benchmark was performed are:",
"_____no_output_____"
]
],
[
[
"!cat /proc/cpuinfo|head -19 ",
"processor\t: 0\nvendor_id\t: AuthenticAMD\ncpu family\t: 21\nmodel\t\t: 101\nmodel name\t: AMD A12-9800 RADEON R7, 12 COMPUTE CORES 4C+8G\nstepping\t: 1\nmicrocode\t: 0x6006118\ncpu MHz\t\t: 2709.605\ncache size\t: 1024 KB\nphysical id\t: 0\nsiblings\t: 4\ncore id\t\t: 0\ncpu cores\t: 2\napicid\t\t: 16\ninitial apicid\t: 0\nfpu\t\t: yes\nfpu_exception\t: yes\ncpuid level\t: 13\nwp\t\t: yes\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb897846d7a43fa02b1fa4d7849567ca7d99847e
| 165,737 |
ipynb
|
Jupyter Notebook
|
Convolutional Neural Networks/3 Transfer Learning/Transfer Learning.ipynb
|
ibadrather/Computer_Vision
|
30bcec1cfdbf38781d17f31a1279130dbdae3f5e
|
[
"MIT"
] | 1 |
2021-05-11T02:22:53.000Z
|
2021-05-11T02:22:53.000Z
|
Convolutional Neural Networks/3 Transfer Learning/Transfer Learning.ipynb
|
ibadrather/Computer_Vision
|
30bcec1cfdbf38781d17f31a1279130dbdae3f5e
|
[
"MIT"
] | null | null | null |
Convolutional Neural Networks/3 Transfer Learning/Transfer Learning.ipynb
|
ibadrather/Computer_Vision
|
30bcec1cfdbf38781d17f31a1279130dbdae3f5e
|
[
"MIT"
] | null | null | null | 98.652976 | 19,196 | 0.681857 |
[
[
[
"#@title Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# https://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.",
"_____no_output_____"
],
[
"# Import all the necessary files!\nimport os\nimport tensorflow as tf\nfrom tensorflow.keras import layers\nfrom tensorflow.keras import Model",
"_____no_output_____"
],
[
"# Download the inception v3 weights\n!wget --no-check-certificate \\\n https://storage.googleapis.com/mledu-datasets/inception_v3_weights_tf_dim_ordering_tf_kernels_notop.h5 \\\n -O /tmp/inception_v3_weights_tf_dim_ordering_tf_kernels_notop.h5\n\n# Import the inception model \nfrom tensorflow.keras.applications.inception_v3 import InceptionV3\n\n# Create an instance of the inception model from the local pre-trained weights\nlocal_weights_file = '/tmp/inception_v3_weights_tf_dim_ordering_tf_kernels_notop.h5'\n\n# Your Code Here\npre_trained_model = InceptionV3(input_shape=(150,150,3),\n include_top = False,\n weights = None)\n\n\npre_trained_model.load_weights(local_weights_file)\n\n# Make all the layers in the pre-trained model non-trainable\nfor layer in pre_trained_model.layers:\n # Your Code Here\n layer.trainable = False\n \n# Print the model summary\npre_trained_model.summary()\n\n# Expected Output is extremely large, but should end with:\n\n#batch_normalization_v1_281 (Bat (None, 3, 3, 192) 576 conv2d_281[0][0] \n#__________________________________________________________________________________________________\n#activation_273 (Activation) (None, 3, 3, 320) 0 batch_normalization_v1_273[0][0] \n#__________________________________________________________________________________________________\n#mixed9_1 (Concatenate) (None, 3, 3, 768) 0 activation_275[0][0] \n# activation_276[0][0] \n#__________________________________________________________________________________________________\n#concatenate_5 (Concatenate) (None, 3, 3, 768) 0 activation_279[0][0] \n# activation_280[0][0] \n#__________________________________________________________________________________________________\n#activation_281 (Activation) (None, 3, 3, 192) 0 batch_normalization_v1_281[0][0] \n#__________________________________________________________________________________________________\n#mixed10 (Concatenate) (None, 3, 3, 2048) 0 activation_273[0][0] \n# mixed9_1[0][0] \n# concatenate_5[0][0] \n# activation_281[0][0] \n#==================================================================================================\n#Total params: 21,802,784\n#Trainable params: 0\n#Non-trainable params: 21,802,784",
"--2021-04-08 03:29:14-- https://storage.googleapis.com/mledu-datasets/inception_v3_weights_tf_dim_ordering_tf_kernels_notop.h5\nResolving storage.googleapis.com (storage.googleapis.com)... 74.125.140.128, 108.177.15.128, 173.194.76.128, ...\nConnecting to storage.googleapis.com (storage.googleapis.com)|74.125.140.128|:443... connected.\nHTTP request sent, awaiting response... 200 OK\nLength: 87910968 (84M) [application/x-hdf]\nSaving to: ‘/tmp/inception_v3_weights_tf_dim_ordering_tf_kernels_notop.h5’\n\n/tmp/inception_v3_w 100%[===================>] 83.84M 16.0MB/s in 5.2s \n\n2021-04-08 03:29:20 (16.0 MB/s) - ‘/tmp/inception_v3_weights_tf_dim_ordering_tf_kernels_notop.h5’ saved [87910968/87910968]\n\nModel: \"inception_v3\"\n__________________________________________________________________________________________________\nLayer (type) Output Shape Param # Connected to \n==================================================================================================\ninput_2 (InputLayer) [(None, 150, 150, 3) 0 \n__________________________________________________________________________________________________\nconv2d_94 (Conv2D) (None, 74, 74, 32) 864 input_2[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_94 (BatchNo (None, 74, 74, 32) 96 conv2d_94[0][0] \n__________________________________________________________________________________________________\nactivation_94 (Activation) (None, 74, 74, 32) 0 batch_normalization_94[0][0] \n__________________________________________________________________________________________________\nconv2d_95 (Conv2D) (None, 72, 72, 32) 9216 activation_94[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_95 (BatchNo (None, 72, 72, 32) 96 conv2d_95[0][0] \n__________________________________________________________________________________________________\nactivation_95 (Activation) (None, 72, 72, 32) 0 batch_normalization_95[0][0] \n__________________________________________________________________________________________________\nconv2d_96 (Conv2D) (None, 72, 72, 64) 18432 activation_95[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_96 (BatchNo (None, 72, 72, 64) 192 conv2d_96[0][0] \n__________________________________________________________________________________________________\nactivation_96 (Activation) (None, 72, 72, 64) 0 batch_normalization_96[0][0] \n__________________________________________________________________________________________________\nmax_pooling2d_4 (MaxPooling2D) (None, 35, 35, 64) 0 activation_96[0][0] \n__________________________________________________________________________________________________\nconv2d_97 (Conv2D) (None, 35, 35, 80) 5120 max_pooling2d_4[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_97 (BatchNo (None, 35, 35, 80) 240 conv2d_97[0][0] \n__________________________________________________________________________________________________\nactivation_97 (Activation) (None, 35, 35, 80) 0 batch_normalization_97[0][0] \n__________________________________________________________________________________________________\nconv2d_98 (Conv2D) (None, 33, 33, 192) 138240 activation_97[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_98 (BatchNo (None, 33, 33, 192) 576 conv2d_98[0][0] \n__________________________________________________________________________________________________\nactivation_98 (Activation) (None, 33, 33, 192) 0 batch_normalization_98[0][0] \n__________________________________________________________________________________________________\nmax_pooling2d_5 (MaxPooling2D) (None, 16, 16, 192) 0 activation_98[0][0] \n__________________________________________________________________________________________________\nconv2d_102 (Conv2D) (None, 16, 16, 64) 12288 max_pooling2d_5[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_102 (BatchN (None, 16, 16, 64) 192 conv2d_102[0][0] \n__________________________________________________________________________________________________\nactivation_102 (Activation) (None, 16, 16, 64) 0 batch_normalization_102[0][0] \n__________________________________________________________________________________________________\nconv2d_100 (Conv2D) (None, 16, 16, 48) 9216 max_pooling2d_5[0][0] \n__________________________________________________________________________________________________\nconv2d_103 (Conv2D) (None, 16, 16, 96) 55296 activation_102[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_100 (BatchN (None, 16, 16, 48) 144 conv2d_100[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_103 (BatchN (None, 16, 16, 96) 288 conv2d_103[0][0] \n__________________________________________________________________________________________________\nactivation_100 (Activation) (None, 16, 16, 48) 0 batch_normalization_100[0][0] \n__________________________________________________________________________________________________\nactivation_103 (Activation) (None, 16, 16, 96) 0 batch_normalization_103[0][0] \n__________________________________________________________________________________________________\naverage_pooling2d_9 (AveragePoo (None, 16, 16, 192) 0 max_pooling2d_5[0][0] \n__________________________________________________________________________________________________\nconv2d_99 (Conv2D) (None, 16, 16, 64) 12288 max_pooling2d_5[0][0] \n__________________________________________________________________________________________________\nconv2d_101 (Conv2D) (None, 16, 16, 64) 76800 activation_100[0][0] \n__________________________________________________________________________________________________\nconv2d_104 (Conv2D) (None, 16, 16, 96) 82944 activation_103[0][0] \n__________________________________________________________________________________________________\nconv2d_105 (Conv2D) (None, 16, 16, 32) 6144 average_pooling2d_9[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_99 (BatchNo (None, 16, 16, 64) 192 conv2d_99[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_101 (BatchN (None, 16, 16, 64) 192 conv2d_101[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_104 (BatchN (None, 16, 16, 96) 288 conv2d_104[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_105 (BatchN (None, 16, 16, 32) 96 conv2d_105[0][0] \n__________________________________________________________________________________________________\nactivation_99 (Activation) (None, 16, 16, 64) 0 batch_normalization_99[0][0] \n__________________________________________________________________________________________________\nactivation_101 (Activation) (None, 16, 16, 64) 0 batch_normalization_101[0][0] \n__________________________________________________________________________________________________\nactivation_104 (Activation) (None, 16, 16, 96) 0 batch_normalization_104[0][0] \n__________________________________________________________________________________________________\nactivation_105 (Activation) (None, 16, 16, 32) 0 batch_normalization_105[0][0] \n__________________________________________________________________________________________________\nmixed0 (Concatenate) (None, 16, 16, 256) 0 activation_99[0][0] \n activation_101[0][0] \n activation_104[0][0] \n activation_105[0][0] \n__________________________________________________________________________________________________\nconv2d_109 (Conv2D) (None, 16, 16, 64) 16384 mixed0[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_109 (BatchN (None, 16, 16, 64) 192 conv2d_109[0][0] \n__________________________________________________________________________________________________\nactivation_109 (Activation) (None, 16, 16, 64) 0 batch_normalization_109[0][0] \n__________________________________________________________________________________________________\nconv2d_107 (Conv2D) (None, 16, 16, 48) 12288 mixed0[0][0] \n__________________________________________________________________________________________________\nconv2d_110 (Conv2D) (None, 16, 16, 96) 55296 activation_109[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_107 (BatchN (None, 16, 16, 48) 144 conv2d_107[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_110 (BatchN (None, 16, 16, 96) 288 conv2d_110[0][0] \n__________________________________________________________________________________________________\nactivation_107 (Activation) (None, 16, 16, 48) 0 batch_normalization_107[0][0] \n__________________________________________________________________________________________________\nactivation_110 (Activation) (None, 16, 16, 96) 0 batch_normalization_110[0][0] \n__________________________________________________________________________________________________\naverage_pooling2d_10 (AveragePo (None, 16, 16, 256) 0 mixed0[0][0] \n__________________________________________________________________________________________________\nconv2d_106 (Conv2D) (None, 16, 16, 64) 16384 mixed0[0][0] \n__________________________________________________________________________________________________\nconv2d_108 (Conv2D) (None, 16, 16, 64) 76800 activation_107[0][0] \n__________________________________________________________________________________________________\nconv2d_111 (Conv2D) (None, 16, 16, 96) 82944 activation_110[0][0] \n__________________________________________________________________________________________________\nconv2d_112 (Conv2D) (None, 16, 16, 64) 16384 average_pooling2d_10[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_106 (BatchN (None, 16, 16, 64) 192 conv2d_106[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_108 (BatchN (None, 16, 16, 64) 192 conv2d_108[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_111 (BatchN (None, 16, 16, 96) 288 conv2d_111[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_112 (BatchN (None, 16, 16, 64) 192 conv2d_112[0][0] \n__________________________________________________________________________________________________\nactivation_106 (Activation) (None, 16, 16, 64) 0 batch_normalization_106[0][0] \n__________________________________________________________________________________________________\nactivation_108 (Activation) (None, 16, 16, 64) 0 batch_normalization_108[0][0] \n__________________________________________________________________________________________________\nactivation_111 (Activation) (None, 16, 16, 96) 0 batch_normalization_111[0][0] \n__________________________________________________________________________________________________\nactivation_112 (Activation) (None, 16, 16, 64) 0 batch_normalization_112[0][0] \n__________________________________________________________________________________________________\nmixed1 (Concatenate) (None, 16, 16, 288) 0 activation_106[0][0] \n activation_108[0][0] \n activation_111[0][0] \n activation_112[0][0] \n__________________________________________________________________________________________________\nconv2d_116 (Conv2D) (None, 16, 16, 64) 18432 mixed1[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_116 (BatchN (None, 16, 16, 64) 192 conv2d_116[0][0] \n__________________________________________________________________________________________________\nactivation_116 (Activation) (None, 16, 16, 64) 0 batch_normalization_116[0][0] \n__________________________________________________________________________________________________\nconv2d_114 (Conv2D) (None, 16, 16, 48) 13824 mixed1[0][0] \n__________________________________________________________________________________________________\nconv2d_117 (Conv2D) (None, 16, 16, 96) 55296 activation_116[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_114 (BatchN (None, 16, 16, 48) 144 conv2d_114[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_117 (BatchN (None, 16, 16, 96) 288 conv2d_117[0][0] \n__________________________________________________________________________________________________\nactivation_114 (Activation) (None, 16, 16, 48) 0 batch_normalization_114[0][0] \n__________________________________________________________________________________________________\nactivation_117 (Activation) (None, 16, 16, 96) 0 batch_normalization_117[0][0] \n__________________________________________________________________________________________________\naverage_pooling2d_11 (AveragePo (None, 16, 16, 288) 0 mixed1[0][0] \n__________________________________________________________________________________________________\nconv2d_113 (Conv2D) (None, 16, 16, 64) 18432 mixed1[0][0] \n__________________________________________________________________________________________________\nconv2d_115 (Conv2D) (None, 16, 16, 64) 76800 activation_114[0][0] \n__________________________________________________________________________________________________\nconv2d_118 (Conv2D) (None, 16, 16, 96) 82944 activation_117[0][0] \n__________________________________________________________________________________________________\nconv2d_119 (Conv2D) (None, 16, 16, 64) 18432 average_pooling2d_11[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_113 (BatchN (None, 16, 16, 64) 192 conv2d_113[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_115 (BatchN (None, 16, 16, 64) 192 conv2d_115[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_118 (BatchN (None, 16, 16, 96) 288 conv2d_118[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_119 (BatchN (None, 16, 16, 64) 192 conv2d_119[0][0] \n__________________________________________________________________________________________________\nactivation_113 (Activation) (None, 16, 16, 64) 0 batch_normalization_113[0][0] \n__________________________________________________________________________________________________\nactivation_115 (Activation) (None, 16, 16, 64) 0 batch_normalization_115[0][0] \n__________________________________________________________________________________________________\nactivation_118 (Activation) (None, 16, 16, 96) 0 batch_normalization_118[0][0] \n__________________________________________________________________________________________________\nactivation_119 (Activation) (None, 16, 16, 64) 0 batch_normalization_119[0][0] \n__________________________________________________________________________________________________\nmixed2 (Concatenate) (None, 16, 16, 288) 0 activation_113[0][0] \n activation_115[0][0] \n activation_118[0][0] \n activation_119[0][0] \n__________________________________________________________________________________________________\nconv2d_121 (Conv2D) (None, 16, 16, 64) 18432 mixed2[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_121 (BatchN (None, 16, 16, 64) 192 conv2d_121[0][0] \n__________________________________________________________________________________________________\nactivation_121 (Activation) (None, 16, 16, 64) 0 batch_normalization_121[0][0] \n__________________________________________________________________________________________________\nconv2d_122 (Conv2D) (None, 16, 16, 96) 55296 activation_121[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_122 (BatchN (None, 16, 16, 96) 288 conv2d_122[0][0] \n__________________________________________________________________________________________________\nactivation_122 (Activation) (None, 16, 16, 96) 0 batch_normalization_122[0][0] \n__________________________________________________________________________________________________\nconv2d_120 (Conv2D) (None, 7, 7, 384) 995328 mixed2[0][0] \n__________________________________________________________________________________________________\nconv2d_123 (Conv2D) (None, 7, 7, 96) 82944 activation_122[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_120 (BatchN (None, 7, 7, 384) 1152 conv2d_120[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_123 (BatchN (None, 7, 7, 96) 288 conv2d_123[0][0] \n__________________________________________________________________________________________________\nactivation_120 (Activation) (None, 7, 7, 384) 0 batch_normalization_120[0][0] \n__________________________________________________________________________________________________\nactivation_123 (Activation) (None, 7, 7, 96) 0 batch_normalization_123[0][0] \n__________________________________________________________________________________________________\nmax_pooling2d_6 (MaxPooling2D) (None, 7, 7, 288) 0 mixed2[0][0] \n__________________________________________________________________________________________________\nmixed3 (Concatenate) (None, 7, 7, 768) 0 activation_120[0][0] \n activation_123[0][0] \n max_pooling2d_6[0][0] \n__________________________________________________________________________________________________\nconv2d_128 (Conv2D) (None, 7, 7, 128) 98304 mixed3[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_128 (BatchN (None, 7, 7, 128) 384 conv2d_128[0][0] \n__________________________________________________________________________________________________\nactivation_128 (Activation) (None, 7, 7, 128) 0 batch_normalization_128[0][0] \n__________________________________________________________________________________________________\nconv2d_129 (Conv2D) (None, 7, 7, 128) 114688 activation_128[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_129 (BatchN (None, 7, 7, 128) 384 conv2d_129[0][0] \n__________________________________________________________________________________________________\nactivation_129 (Activation) (None, 7, 7, 128) 0 batch_normalization_129[0][0] \n__________________________________________________________________________________________________\nconv2d_125 (Conv2D) (None, 7, 7, 128) 98304 mixed3[0][0] \n__________________________________________________________________________________________________\nconv2d_130 (Conv2D) (None, 7, 7, 128) 114688 activation_129[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_125 (BatchN (None, 7, 7, 128) 384 conv2d_125[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_130 (BatchN (None, 7, 7, 128) 384 conv2d_130[0][0] \n__________________________________________________________________________________________________\nactivation_125 (Activation) (None, 7, 7, 128) 0 batch_normalization_125[0][0] \n__________________________________________________________________________________________________\nactivation_130 (Activation) (None, 7, 7, 128) 0 batch_normalization_130[0][0] \n__________________________________________________________________________________________________\nconv2d_126 (Conv2D) (None, 7, 7, 128) 114688 activation_125[0][0] \n__________________________________________________________________________________________________\nconv2d_131 (Conv2D) (None, 7, 7, 128) 114688 activation_130[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_126 (BatchN (None, 7, 7, 128) 384 conv2d_126[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_131 (BatchN (None, 7, 7, 128) 384 conv2d_131[0][0] \n__________________________________________________________________________________________________\nactivation_126 (Activation) (None, 7, 7, 128) 0 batch_normalization_126[0][0] \n__________________________________________________________________________________________________\nactivation_131 (Activation) (None, 7, 7, 128) 0 batch_normalization_131[0][0] \n__________________________________________________________________________________________________\naverage_pooling2d_12 (AveragePo (None, 7, 7, 768) 0 mixed3[0][0] \n__________________________________________________________________________________________________\nconv2d_124 (Conv2D) (None, 7, 7, 192) 147456 mixed3[0][0] \n__________________________________________________________________________________________________\nconv2d_127 (Conv2D) (None, 7, 7, 192) 172032 activation_126[0][0] \n__________________________________________________________________________________________________\nconv2d_132 (Conv2D) (None, 7, 7, 192) 172032 activation_131[0][0] \n__________________________________________________________________________________________________\nconv2d_133 (Conv2D) (None, 7, 7, 192) 147456 average_pooling2d_12[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_124 (BatchN (None, 7, 7, 192) 576 conv2d_124[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_127 (BatchN (None, 7, 7, 192) 576 conv2d_127[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_132 (BatchN (None, 7, 7, 192) 576 conv2d_132[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_133 (BatchN (None, 7, 7, 192) 576 conv2d_133[0][0] \n__________________________________________________________________________________________________\nactivation_124 (Activation) (None, 7, 7, 192) 0 batch_normalization_124[0][0] \n__________________________________________________________________________________________________\nactivation_127 (Activation) (None, 7, 7, 192) 0 batch_normalization_127[0][0] \n__________________________________________________________________________________________________\nactivation_132 (Activation) (None, 7, 7, 192) 0 batch_normalization_132[0][0] \n__________________________________________________________________________________________________\nactivation_133 (Activation) (None, 7, 7, 192) 0 batch_normalization_133[0][0] \n__________________________________________________________________________________________________\nmixed4 (Concatenate) (None, 7, 7, 768) 0 activation_124[0][0] \n activation_127[0][0] \n activation_132[0][0] \n activation_133[0][0] \n__________________________________________________________________________________________________\nconv2d_138 (Conv2D) (None, 7, 7, 160) 122880 mixed4[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_138 (BatchN (None, 7, 7, 160) 480 conv2d_138[0][0] \n__________________________________________________________________________________________________\nactivation_138 (Activation) (None, 7, 7, 160) 0 batch_normalization_138[0][0] \n__________________________________________________________________________________________________\nconv2d_139 (Conv2D) (None, 7, 7, 160) 179200 activation_138[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_139 (BatchN (None, 7, 7, 160) 480 conv2d_139[0][0] \n__________________________________________________________________________________________________\nactivation_139 (Activation) (None, 7, 7, 160) 0 batch_normalization_139[0][0] \n__________________________________________________________________________________________________\nconv2d_135 (Conv2D) (None, 7, 7, 160) 122880 mixed4[0][0] \n__________________________________________________________________________________________________\nconv2d_140 (Conv2D) (None, 7, 7, 160) 179200 activation_139[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_135 (BatchN (None, 7, 7, 160) 480 conv2d_135[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_140 (BatchN (None, 7, 7, 160) 480 conv2d_140[0][0] \n__________________________________________________________________________________________________\nactivation_135 (Activation) (None, 7, 7, 160) 0 batch_normalization_135[0][0] \n__________________________________________________________________________________________________\nactivation_140 (Activation) (None, 7, 7, 160) 0 batch_normalization_140[0][0] \n__________________________________________________________________________________________________\nconv2d_136 (Conv2D) (None, 7, 7, 160) 179200 activation_135[0][0] \n__________________________________________________________________________________________________\nconv2d_141 (Conv2D) (None, 7, 7, 160) 179200 activation_140[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_136 (BatchN (None, 7, 7, 160) 480 conv2d_136[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_141 (BatchN (None, 7, 7, 160) 480 conv2d_141[0][0] \n__________________________________________________________________________________________________\nactivation_136 (Activation) (None, 7, 7, 160) 0 batch_normalization_136[0][0] \n__________________________________________________________________________________________________\nactivation_141 (Activation) (None, 7, 7, 160) 0 batch_normalization_141[0][0] \n__________________________________________________________________________________________________\naverage_pooling2d_13 (AveragePo (None, 7, 7, 768) 0 mixed4[0][0] \n__________________________________________________________________________________________________\nconv2d_134 (Conv2D) (None, 7, 7, 192) 147456 mixed4[0][0] \n__________________________________________________________________________________________________\nconv2d_137 (Conv2D) (None, 7, 7, 192) 215040 activation_136[0][0] \n__________________________________________________________________________________________________\nconv2d_142 (Conv2D) (None, 7, 7, 192) 215040 activation_141[0][0] \n__________________________________________________________________________________________________\nconv2d_143 (Conv2D) (None, 7, 7, 192) 147456 average_pooling2d_13[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_134 (BatchN (None, 7, 7, 192) 576 conv2d_134[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_137 (BatchN (None, 7, 7, 192) 576 conv2d_137[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_142 (BatchN (None, 7, 7, 192) 576 conv2d_142[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_143 (BatchN (None, 7, 7, 192) 576 conv2d_143[0][0] \n__________________________________________________________________________________________________\nactivation_134 (Activation) (None, 7, 7, 192) 0 batch_normalization_134[0][0] \n__________________________________________________________________________________________________\nactivation_137 (Activation) (None, 7, 7, 192) 0 batch_normalization_137[0][0] \n__________________________________________________________________________________________________\nactivation_142 (Activation) (None, 7, 7, 192) 0 batch_normalization_142[0][0] \n__________________________________________________________________________________________________\nactivation_143 (Activation) (None, 7, 7, 192) 0 batch_normalization_143[0][0] \n__________________________________________________________________________________________________\nmixed5 (Concatenate) (None, 7, 7, 768) 0 activation_134[0][0] \n activation_137[0][0] \n activation_142[0][0] \n activation_143[0][0] \n__________________________________________________________________________________________________\nconv2d_148 (Conv2D) (None, 7, 7, 160) 122880 mixed5[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_148 (BatchN (None, 7, 7, 160) 480 conv2d_148[0][0] \n__________________________________________________________________________________________________\nactivation_148 (Activation) (None, 7, 7, 160) 0 batch_normalization_148[0][0] \n__________________________________________________________________________________________________\nconv2d_149 (Conv2D) (None, 7, 7, 160) 179200 activation_148[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_149 (BatchN (None, 7, 7, 160) 480 conv2d_149[0][0] \n__________________________________________________________________________________________________\nactivation_149 (Activation) (None, 7, 7, 160) 0 batch_normalization_149[0][0] \n__________________________________________________________________________________________________\nconv2d_145 (Conv2D) (None, 7, 7, 160) 122880 mixed5[0][0] \n__________________________________________________________________________________________________\nconv2d_150 (Conv2D) (None, 7, 7, 160) 179200 activation_149[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_145 (BatchN (None, 7, 7, 160) 480 conv2d_145[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_150 (BatchN (None, 7, 7, 160) 480 conv2d_150[0][0] \n__________________________________________________________________________________________________\nactivation_145 (Activation) (None, 7, 7, 160) 0 batch_normalization_145[0][0] \n__________________________________________________________________________________________________\nactivation_150 (Activation) (None, 7, 7, 160) 0 batch_normalization_150[0][0] \n__________________________________________________________________________________________________\nconv2d_146 (Conv2D) (None, 7, 7, 160) 179200 activation_145[0][0] \n__________________________________________________________________________________________________\nconv2d_151 (Conv2D) (None, 7, 7, 160) 179200 activation_150[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_146 (BatchN (None, 7, 7, 160) 480 conv2d_146[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_151 (BatchN (None, 7, 7, 160) 480 conv2d_151[0][0] \n__________________________________________________________________________________________________\nactivation_146 (Activation) (None, 7, 7, 160) 0 batch_normalization_146[0][0] \n__________________________________________________________________________________________________\nactivation_151 (Activation) (None, 7, 7, 160) 0 batch_normalization_151[0][0] \n__________________________________________________________________________________________________\naverage_pooling2d_14 (AveragePo (None, 7, 7, 768) 0 mixed5[0][0] \n__________________________________________________________________________________________________\nconv2d_144 (Conv2D) (None, 7, 7, 192) 147456 mixed5[0][0] \n__________________________________________________________________________________________________\nconv2d_147 (Conv2D) (None, 7, 7, 192) 215040 activation_146[0][0] \n__________________________________________________________________________________________________\nconv2d_152 (Conv2D) (None, 7, 7, 192) 215040 activation_151[0][0] \n__________________________________________________________________________________________________\nconv2d_153 (Conv2D) (None, 7, 7, 192) 147456 average_pooling2d_14[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_144 (BatchN (None, 7, 7, 192) 576 conv2d_144[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_147 (BatchN (None, 7, 7, 192) 576 conv2d_147[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_152 (BatchN (None, 7, 7, 192) 576 conv2d_152[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_153 (BatchN (None, 7, 7, 192) 576 conv2d_153[0][0] \n__________________________________________________________________________________________________\nactivation_144 (Activation) (None, 7, 7, 192) 0 batch_normalization_144[0][0] \n__________________________________________________________________________________________________\nactivation_147 (Activation) (None, 7, 7, 192) 0 batch_normalization_147[0][0] \n__________________________________________________________________________________________________\nactivation_152 (Activation) (None, 7, 7, 192) 0 batch_normalization_152[0][0] \n__________________________________________________________________________________________________\nactivation_153 (Activation) (None, 7, 7, 192) 0 batch_normalization_153[0][0] \n__________________________________________________________________________________________________\nmixed6 (Concatenate) (None, 7, 7, 768) 0 activation_144[0][0] \n activation_147[0][0] \n activation_152[0][0] \n activation_153[0][0] \n__________________________________________________________________________________________________\nconv2d_158 (Conv2D) (None, 7, 7, 192) 147456 mixed6[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_158 (BatchN (None, 7, 7, 192) 576 conv2d_158[0][0] \n__________________________________________________________________________________________________\nactivation_158 (Activation) (None, 7, 7, 192) 0 batch_normalization_158[0][0] \n__________________________________________________________________________________________________\nconv2d_159 (Conv2D) (None, 7, 7, 192) 258048 activation_158[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_159 (BatchN (None, 7, 7, 192) 576 conv2d_159[0][0] \n__________________________________________________________________________________________________\nactivation_159 (Activation) (None, 7, 7, 192) 0 batch_normalization_159[0][0] \n__________________________________________________________________________________________________\nconv2d_155 (Conv2D) (None, 7, 7, 192) 147456 mixed6[0][0] \n__________________________________________________________________________________________________\nconv2d_160 (Conv2D) (None, 7, 7, 192) 258048 activation_159[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_155 (BatchN (None, 7, 7, 192) 576 conv2d_155[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_160 (BatchN (None, 7, 7, 192) 576 conv2d_160[0][0] \n__________________________________________________________________________________________________\nactivation_155 (Activation) (None, 7, 7, 192) 0 batch_normalization_155[0][0] \n__________________________________________________________________________________________________\nactivation_160 (Activation) (None, 7, 7, 192) 0 batch_normalization_160[0][0] \n__________________________________________________________________________________________________\nconv2d_156 (Conv2D) (None, 7, 7, 192) 258048 activation_155[0][0] \n__________________________________________________________________________________________________\nconv2d_161 (Conv2D) (None, 7, 7, 192) 258048 activation_160[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_156 (BatchN (None, 7, 7, 192) 576 conv2d_156[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_161 (BatchN (None, 7, 7, 192) 576 conv2d_161[0][0] \n__________________________________________________________________________________________________\nactivation_156 (Activation) (None, 7, 7, 192) 0 batch_normalization_156[0][0] \n__________________________________________________________________________________________________\nactivation_161 (Activation) (None, 7, 7, 192) 0 batch_normalization_161[0][0] \n__________________________________________________________________________________________________\naverage_pooling2d_15 (AveragePo (None, 7, 7, 768) 0 mixed6[0][0] \n__________________________________________________________________________________________________\nconv2d_154 (Conv2D) (None, 7, 7, 192) 147456 mixed6[0][0] \n__________________________________________________________________________________________________\nconv2d_157 (Conv2D) (None, 7, 7, 192) 258048 activation_156[0][0] \n__________________________________________________________________________________________________\nconv2d_162 (Conv2D) (None, 7, 7, 192) 258048 activation_161[0][0] \n__________________________________________________________________________________________________\nconv2d_163 (Conv2D) (None, 7, 7, 192) 147456 average_pooling2d_15[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_154 (BatchN (None, 7, 7, 192) 576 conv2d_154[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_157 (BatchN (None, 7, 7, 192) 576 conv2d_157[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_162 (BatchN (None, 7, 7, 192) 576 conv2d_162[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_163 (BatchN (None, 7, 7, 192) 576 conv2d_163[0][0] \n__________________________________________________________________________________________________\nactivation_154 (Activation) (None, 7, 7, 192) 0 batch_normalization_154[0][0] \n__________________________________________________________________________________________________\nactivation_157 (Activation) (None, 7, 7, 192) 0 batch_normalization_157[0][0] \n__________________________________________________________________________________________________\nactivation_162 (Activation) (None, 7, 7, 192) 0 batch_normalization_162[0][0] \n__________________________________________________________________________________________________\nactivation_163 (Activation) (None, 7, 7, 192) 0 batch_normalization_163[0][0] \n__________________________________________________________________________________________________\nmixed7 (Concatenate) (None, 7, 7, 768) 0 activation_154[0][0] \n activation_157[0][0] \n activation_162[0][0] \n activation_163[0][0] \n__________________________________________________________________________________________________\nconv2d_166 (Conv2D) (None, 7, 7, 192) 147456 mixed7[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_166 (BatchN (None, 7, 7, 192) 576 conv2d_166[0][0] \n__________________________________________________________________________________________________\nactivation_166 (Activation) (None, 7, 7, 192) 0 batch_normalization_166[0][0] \n__________________________________________________________________________________________________\nconv2d_167 (Conv2D) (None, 7, 7, 192) 258048 activation_166[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_167 (BatchN (None, 7, 7, 192) 576 conv2d_167[0][0] \n__________________________________________________________________________________________________\nactivation_167 (Activation) (None, 7, 7, 192) 0 batch_normalization_167[0][0] \n__________________________________________________________________________________________________\nconv2d_164 (Conv2D) (None, 7, 7, 192) 147456 mixed7[0][0] \n__________________________________________________________________________________________________\nconv2d_168 (Conv2D) (None, 7, 7, 192) 258048 activation_167[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_164 (BatchN (None, 7, 7, 192) 576 conv2d_164[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_168 (BatchN (None, 7, 7, 192) 576 conv2d_168[0][0] \n__________________________________________________________________________________________________\nactivation_164 (Activation) (None, 7, 7, 192) 0 batch_normalization_164[0][0] \n__________________________________________________________________________________________________\nactivation_168 (Activation) (None, 7, 7, 192) 0 batch_normalization_168[0][0] \n__________________________________________________________________________________________________\nconv2d_165 (Conv2D) (None, 3, 3, 320) 552960 activation_164[0][0] \n__________________________________________________________________________________________________\nconv2d_169 (Conv2D) (None, 3, 3, 192) 331776 activation_168[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_165 (BatchN (None, 3, 3, 320) 960 conv2d_165[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_169 (BatchN (None, 3, 3, 192) 576 conv2d_169[0][0] \n__________________________________________________________________________________________________\nactivation_165 (Activation) (None, 3, 3, 320) 0 batch_normalization_165[0][0] \n__________________________________________________________________________________________________\nactivation_169 (Activation) (None, 3, 3, 192) 0 batch_normalization_169[0][0] \n__________________________________________________________________________________________________\nmax_pooling2d_7 (MaxPooling2D) (None, 3, 3, 768) 0 mixed7[0][0] \n__________________________________________________________________________________________________\nmixed8 (Concatenate) (None, 3, 3, 1280) 0 activation_165[0][0] \n activation_169[0][0] \n max_pooling2d_7[0][0] \n__________________________________________________________________________________________________\nconv2d_174 (Conv2D) (None, 3, 3, 448) 573440 mixed8[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_174 (BatchN (None, 3, 3, 448) 1344 conv2d_174[0][0] \n__________________________________________________________________________________________________\nactivation_174 (Activation) (None, 3, 3, 448) 0 batch_normalization_174[0][0] \n__________________________________________________________________________________________________\nconv2d_171 (Conv2D) (None, 3, 3, 384) 491520 mixed8[0][0] \n__________________________________________________________________________________________________\nconv2d_175 (Conv2D) (None, 3, 3, 384) 1548288 activation_174[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_171 (BatchN (None, 3, 3, 384) 1152 conv2d_171[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_175 (BatchN (None, 3, 3, 384) 1152 conv2d_175[0][0] \n__________________________________________________________________________________________________\nactivation_171 (Activation) (None, 3, 3, 384) 0 batch_normalization_171[0][0] \n__________________________________________________________________________________________________\nactivation_175 (Activation) (None, 3, 3, 384) 0 batch_normalization_175[0][0] \n__________________________________________________________________________________________________\nconv2d_172 (Conv2D) (None, 3, 3, 384) 442368 activation_171[0][0] \n__________________________________________________________________________________________________\nconv2d_173 (Conv2D) (None, 3, 3, 384) 442368 activation_171[0][0] \n__________________________________________________________________________________________________\nconv2d_176 (Conv2D) (None, 3, 3, 384) 442368 activation_175[0][0] \n__________________________________________________________________________________________________\nconv2d_177 (Conv2D) (None, 3, 3, 384) 442368 activation_175[0][0] \n__________________________________________________________________________________________________\naverage_pooling2d_16 (AveragePo (None, 3, 3, 1280) 0 mixed8[0][0] \n__________________________________________________________________________________________________\nconv2d_170 (Conv2D) (None, 3, 3, 320) 409600 mixed8[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_172 (BatchN (None, 3, 3, 384) 1152 conv2d_172[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_173 (BatchN (None, 3, 3, 384) 1152 conv2d_173[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_176 (BatchN (None, 3, 3, 384) 1152 conv2d_176[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_177 (BatchN (None, 3, 3, 384) 1152 conv2d_177[0][0] \n__________________________________________________________________________________________________\nconv2d_178 (Conv2D) (None, 3, 3, 192) 245760 average_pooling2d_16[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_170 (BatchN (None, 3, 3, 320) 960 conv2d_170[0][0] \n__________________________________________________________________________________________________\nactivation_172 (Activation) (None, 3, 3, 384) 0 batch_normalization_172[0][0] \n__________________________________________________________________________________________________\nactivation_173 (Activation) (None, 3, 3, 384) 0 batch_normalization_173[0][0] \n__________________________________________________________________________________________________\nactivation_176 (Activation) (None, 3, 3, 384) 0 batch_normalization_176[0][0] \n__________________________________________________________________________________________________\nactivation_177 (Activation) (None, 3, 3, 384) 0 batch_normalization_177[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_178 (BatchN (None, 3, 3, 192) 576 conv2d_178[0][0] \n__________________________________________________________________________________________________\nactivation_170 (Activation) (None, 3, 3, 320) 0 batch_normalization_170[0][0] \n__________________________________________________________________________________________________\nmixed9_0 (Concatenate) (None, 3, 3, 768) 0 activation_172[0][0] \n activation_173[0][0] \n__________________________________________________________________________________________________\nconcatenate_2 (Concatenate) (None, 3, 3, 768) 0 activation_176[0][0] \n activation_177[0][0] \n__________________________________________________________________________________________________\nactivation_178 (Activation) (None, 3, 3, 192) 0 batch_normalization_178[0][0] \n__________________________________________________________________________________________________\nmixed9 (Concatenate) (None, 3, 3, 2048) 0 activation_170[0][0] \n mixed9_0[0][0] \n concatenate_2[0][0] \n activation_178[0][0] \n__________________________________________________________________________________________________\nconv2d_183 (Conv2D) (None, 3, 3, 448) 917504 mixed9[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_183 (BatchN (None, 3, 3, 448) 1344 conv2d_183[0][0] \n__________________________________________________________________________________________________\nactivation_183 (Activation) (None, 3, 3, 448) 0 batch_normalization_183[0][0] \n__________________________________________________________________________________________________\nconv2d_180 (Conv2D) (None, 3, 3, 384) 786432 mixed9[0][0] \n__________________________________________________________________________________________________\nconv2d_184 (Conv2D) (None, 3, 3, 384) 1548288 activation_183[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_180 (BatchN (None, 3, 3, 384) 1152 conv2d_180[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_184 (BatchN (None, 3, 3, 384) 1152 conv2d_184[0][0] \n__________________________________________________________________________________________________\nactivation_180 (Activation) (None, 3, 3, 384) 0 batch_normalization_180[0][0] \n__________________________________________________________________________________________________\nactivation_184 (Activation) (None, 3, 3, 384) 0 batch_normalization_184[0][0] \n__________________________________________________________________________________________________\nconv2d_181 (Conv2D) (None, 3, 3, 384) 442368 activation_180[0][0] \n__________________________________________________________________________________________________\nconv2d_182 (Conv2D) (None, 3, 3, 384) 442368 activation_180[0][0] \n__________________________________________________________________________________________________\nconv2d_185 (Conv2D) (None, 3, 3, 384) 442368 activation_184[0][0] \n__________________________________________________________________________________________________\nconv2d_186 (Conv2D) (None, 3, 3, 384) 442368 activation_184[0][0] \n__________________________________________________________________________________________________\naverage_pooling2d_17 (AveragePo (None, 3, 3, 2048) 0 mixed9[0][0] \n__________________________________________________________________________________________________\nconv2d_179 (Conv2D) (None, 3, 3, 320) 655360 mixed9[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_181 (BatchN (None, 3, 3, 384) 1152 conv2d_181[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_182 (BatchN (None, 3, 3, 384) 1152 conv2d_182[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_185 (BatchN (None, 3, 3, 384) 1152 conv2d_185[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_186 (BatchN (None, 3, 3, 384) 1152 conv2d_186[0][0] \n__________________________________________________________________________________________________\nconv2d_187 (Conv2D) (None, 3, 3, 192) 393216 average_pooling2d_17[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_179 (BatchN (None, 3, 3, 320) 960 conv2d_179[0][0] \n__________________________________________________________________________________________________\nactivation_181 (Activation) (None, 3, 3, 384) 0 batch_normalization_181[0][0] \n__________________________________________________________________________________________________\nactivation_182 (Activation) (None, 3, 3, 384) 0 batch_normalization_182[0][0] \n__________________________________________________________________________________________________\nactivation_185 (Activation) (None, 3, 3, 384) 0 batch_normalization_185[0][0] \n__________________________________________________________________________________________________\nactivation_186 (Activation) (None, 3, 3, 384) 0 batch_normalization_186[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_187 (BatchN (None, 3, 3, 192) 576 conv2d_187[0][0] \n__________________________________________________________________________________________________\nactivation_179 (Activation) (None, 3, 3, 320) 0 batch_normalization_179[0][0] \n__________________________________________________________________________________________________\nmixed9_1 (Concatenate) (None, 3, 3, 768) 0 activation_181[0][0] \n activation_182[0][0] \n__________________________________________________________________________________________________\nconcatenate_3 (Concatenate) (None, 3, 3, 768) 0 activation_185[0][0] \n activation_186[0][0] \n__________________________________________________________________________________________________\nactivation_187 (Activation) (None, 3, 3, 192) 0 batch_normalization_187[0][0] \n__________________________________________________________________________________________________\nmixed10 (Concatenate) (None, 3, 3, 2048) 0 activation_179[0][0] \n mixed9_1[0][0] \n concatenate_3[0][0] \n activation_187[0][0] \n==================================================================================================\nTotal params: 21,802,784\nTrainable params: 0\nNon-trainable params: 21,802,784\n__________________________________________________________________________________________________\n"
],
[
"last_layer = pre_trained_model.get_layer('mixed7')\nprint('last layer output shape: ', last_layer.output_shape)\nlast_output = last_layer.output# Your Code Here\n\n# Expected Output:\n# ('last layer output shape: ', (None, 7, 7, 768))",
"last layer output shape: (None, 7, 7, 768)\n"
],
[
"# Define a Callback class that stops training once accuracy reaches 99.9%\nclass myCallback(tf.keras.callbacks.Callback):\n def on_epoch_end(self, epoch, logs={}):\n if logs.get('accuracy'):\n if(logs.get('accuracy')>0.999):\n print(\"\\nReached 99.9% accuracy so cancelling training!\")\n self.model.stop_training = True\n\n ",
"_____no_output_____"
],
[
"from tensorflow.keras.optimizers import RMSprop\n\n# Flatten the output layer to 1 dimension\nx = layers.Flatten()(last_output)\n# Add a fully connected layer with 1,024 hidden units and ReLU activation\nx = layers.Dense(1024, activation='relu')(x)\n# Add a dropout rate of 0.2\nx = layers.Dropout(0.2)(x) \n# Add a final sigmoid layer for classification\nx = layers.Dense (1, activation='sigmoid')(x) \n\nmodel = Model(pre_trained_model.input, x) \n\nmodel.compile(optimizer = RMSprop(lr=0.0001), \n loss = 'binary_crossentropy', \n metrics = ['acc'])\n\nmodel.summary()\n\n# Expected output will be large. Last few lines should be:\n\n# mixed7 (Concatenate) (None, 7, 7, 768) 0 activation_248[0][0] \n# activation_251[0][0] \n# activation_256[0][0] \n# activation_257[0][0] \n# __________________________________________________________________________________________________\n# flatten_4 (Flatten) (None, 37632) 0 mixed7[0][0] \n# __________________________________________________________________________________________________\n# dense_8 (Dense) (None, 1024) 38536192 flatten_4[0][0] \n# __________________________________________________________________________________________________\n# dropout_4 (Dropout) (None, 1024) 0 dense_8[0][0] \n# __________________________________________________________________________________________________\n# dense_9 (Dense) (None, 1) 1025 dropout_4[0][0] \n# ==================================================================================================\n# Total params: 47,512,481\n# Trainable params: 38,537,217\n# Non-trainable params: 8,975,264\n",
"Model: \"model_1\"\n__________________________________________________________________________________________________\nLayer (type) Output Shape Param # Connected to \n==================================================================================================\ninput_2 (InputLayer) [(None, 150, 150, 3) 0 \n__________________________________________________________________________________________________\nconv2d_94 (Conv2D) (None, 74, 74, 32) 864 input_2[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_94 (BatchNo (None, 74, 74, 32) 96 conv2d_94[0][0] \n__________________________________________________________________________________________________\nactivation_94 (Activation) (None, 74, 74, 32) 0 batch_normalization_94[0][0] \n__________________________________________________________________________________________________\nconv2d_95 (Conv2D) (None, 72, 72, 32) 9216 activation_94[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_95 (BatchNo (None, 72, 72, 32) 96 conv2d_95[0][0] \n__________________________________________________________________________________________________\nactivation_95 (Activation) (None, 72, 72, 32) 0 batch_normalization_95[0][0] \n__________________________________________________________________________________________________\nconv2d_96 (Conv2D) (None, 72, 72, 64) 18432 activation_95[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_96 (BatchNo (None, 72, 72, 64) 192 conv2d_96[0][0] \n__________________________________________________________________________________________________\nactivation_96 (Activation) (None, 72, 72, 64) 0 batch_normalization_96[0][0] \n__________________________________________________________________________________________________\nmax_pooling2d_4 (MaxPooling2D) (None, 35, 35, 64) 0 activation_96[0][0] \n__________________________________________________________________________________________________\nconv2d_97 (Conv2D) (None, 35, 35, 80) 5120 max_pooling2d_4[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_97 (BatchNo (None, 35, 35, 80) 240 conv2d_97[0][0] \n__________________________________________________________________________________________________\nactivation_97 (Activation) (None, 35, 35, 80) 0 batch_normalization_97[0][0] \n__________________________________________________________________________________________________\nconv2d_98 (Conv2D) (None, 33, 33, 192) 138240 activation_97[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_98 (BatchNo (None, 33, 33, 192) 576 conv2d_98[0][0] \n__________________________________________________________________________________________________\nactivation_98 (Activation) (None, 33, 33, 192) 0 batch_normalization_98[0][0] \n__________________________________________________________________________________________________\nmax_pooling2d_5 (MaxPooling2D) (None, 16, 16, 192) 0 activation_98[0][0] \n__________________________________________________________________________________________________\nconv2d_102 (Conv2D) (None, 16, 16, 64) 12288 max_pooling2d_5[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_102 (BatchN (None, 16, 16, 64) 192 conv2d_102[0][0] \n__________________________________________________________________________________________________\nactivation_102 (Activation) (None, 16, 16, 64) 0 batch_normalization_102[0][0] \n__________________________________________________________________________________________________\nconv2d_100 (Conv2D) (None, 16, 16, 48) 9216 max_pooling2d_5[0][0] \n__________________________________________________________________________________________________\nconv2d_103 (Conv2D) (None, 16, 16, 96) 55296 activation_102[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_100 (BatchN (None, 16, 16, 48) 144 conv2d_100[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_103 (BatchN (None, 16, 16, 96) 288 conv2d_103[0][0] \n__________________________________________________________________________________________________\nactivation_100 (Activation) (None, 16, 16, 48) 0 batch_normalization_100[0][0] \n__________________________________________________________________________________________________\nactivation_103 (Activation) (None, 16, 16, 96) 0 batch_normalization_103[0][0] \n__________________________________________________________________________________________________\naverage_pooling2d_9 (AveragePoo (None, 16, 16, 192) 0 max_pooling2d_5[0][0] \n__________________________________________________________________________________________________\nconv2d_99 (Conv2D) (None, 16, 16, 64) 12288 max_pooling2d_5[0][0] \n__________________________________________________________________________________________________\nconv2d_101 (Conv2D) (None, 16, 16, 64) 76800 activation_100[0][0] \n__________________________________________________________________________________________________\nconv2d_104 (Conv2D) (None, 16, 16, 96) 82944 activation_103[0][0] \n__________________________________________________________________________________________________\nconv2d_105 (Conv2D) (None, 16, 16, 32) 6144 average_pooling2d_9[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_99 (BatchNo (None, 16, 16, 64) 192 conv2d_99[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_101 (BatchN (None, 16, 16, 64) 192 conv2d_101[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_104 (BatchN (None, 16, 16, 96) 288 conv2d_104[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_105 (BatchN (None, 16, 16, 32) 96 conv2d_105[0][0] \n__________________________________________________________________________________________________\nactivation_99 (Activation) (None, 16, 16, 64) 0 batch_normalization_99[0][0] \n__________________________________________________________________________________________________\nactivation_101 (Activation) (None, 16, 16, 64) 0 batch_normalization_101[0][0] \n__________________________________________________________________________________________________\nactivation_104 (Activation) (None, 16, 16, 96) 0 batch_normalization_104[0][0] \n__________________________________________________________________________________________________\nactivation_105 (Activation) (None, 16, 16, 32) 0 batch_normalization_105[0][0] \n__________________________________________________________________________________________________\nmixed0 (Concatenate) (None, 16, 16, 256) 0 activation_99[0][0] \n activation_101[0][0] \n activation_104[0][0] \n activation_105[0][0] \n__________________________________________________________________________________________________\nconv2d_109 (Conv2D) (None, 16, 16, 64) 16384 mixed0[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_109 (BatchN (None, 16, 16, 64) 192 conv2d_109[0][0] \n__________________________________________________________________________________________________\nactivation_109 (Activation) (None, 16, 16, 64) 0 batch_normalization_109[0][0] \n__________________________________________________________________________________________________\nconv2d_107 (Conv2D) (None, 16, 16, 48) 12288 mixed0[0][0] \n__________________________________________________________________________________________________\nconv2d_110 (Conv2D) (None, 16, 16, 96) 55296 activation_109[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_107 (BatchN (None, 16, 16, 48) 144 conv2d_107[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_110 (BatchN (None, 16, 16, 96) 288 conv2d_110[0][0] \n__________________________________________________________________________________________________\nactivation_107 (Activation) (None, 16, 16, 48) 0 batch_normalization_107[0][0] \n__________________________________________________________________________________________________\nactivation_110 (Activation) (None, 16, 16, 96) 0 batch_normalization_110[0][0] \n__________________________________________________________________________________________________\naverage_pooling2d_10 (AveragePo (None, 16, 16, 256) 0 mixed0[0][0] \n__________________________________________________________________________________________________\nconv2d_106 (Conv2D) (None, 16, 16, 64) 16384 mixed0[0][0] \n__________________________________________________________________________________________________\nconv2d_108 (Conv2D) (None, 16, 16, 64) 76800 activation_107[0][0] \n__________________________________________________________________________________________________\nconv2d_111 (Conv2D) (None, 16, 16, 96) 82944 activation_110[0][0] \n__________________________________________________________________________________________________\nconv2d_112 (Conv2D) (None, 16, 16, 64) 16384 average_pooling2d_10[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_106 (BatchN (None, 16, 16, 64) 192 conv2d_106[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_108 (BatchN (None, 16, 16, 64) 192 conv2d_108[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_111 (BatchN (None, 16, 16, 96) 288 conv2d_111[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_112 (BatchN (None, 16, 16, 64) 192 conv2d_112[0][0] \n__________________________________________________________________________________________________\nactivation_106 (Activation) (None, 16, 16, 64) 0 batch_normalization_106[0][0] \n__________________________________________________________________________________________________\nactivation_108 (Activation) (None, 16, 16, 64) 0 batch_normalization_108[0][0] \n__________________________________________________________________________________________________\nactivation_111 (Activation) (None, 16, 16, 96) 0 batch_normalization_111[0][0] \n__________________________________________________________________________________________________\nactivation_112 (Activation) (None, 16, 16, 64) 0 batch_normalization_112[0][0] \n__________________________________________________________________________________________________\nmixed1 (Concatenate) (None, 16, 16, 288) 0 activation_106[0][0] \n activation_108[0][0] \n activation_111[0][0] \n activation_112[0][0] \n__________________________________________________________________________________________________\nconv2d_116 (Conv2D) (None, 16, 16, 64) 18432 mixed1[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_116 (BatchN (None, 16, 16, 64) 192 conv2d_116[0][0] \n__________________________________________________________________________________________________\nactivation_116 (Activation) (None, 16, 16, 64) 0 batch_normalization_116[0][0] \n__________________________________________________________________________________________________\nconv2d_114 (Conv2D) (None, 16, 16, 48) 13824 mixed1[0][0] \n__________________________________________________________________________________________________\nconv2d_117 (Conv2D) (None, 16, 16, 96) 55296 activation_116[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_114 (BatchN (None, 16, 16, 48) 144 conv2d_114[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_117 (BatchN (None, 16, 16, 96) 288 conv2d_117[0][0] \n__________________________________________________________________________________________________\nactivation_114 (Activation) (None, 16, 16, 48) 0 batch_normalization_114[0][0] \n__________________________________________________________________________________________________\nactivation_117 (Activation) (None, 16, 16, 96) 0 batch_normalization_117[0][0] \n__________________________________________________________________________________________________\naverage_pooling2d_11 (AveragePo (None, 16, 16, 288) 0 mixed1[0][0] \n__________________________________________________________________________________________________\nconv2d_113 (Conv2D) (None, 16, 16, 64) 18432 mixed1[0][0] \n__________________________________________________________________________________________________\nconv2d_115 (Conv2D) (None, 16, 16, 64) 76800 activation_114[0][0] \n__________________________________________________________________________________________________\nconv2d_118 (Conv2D) (None, 16, 16, 96) 82944 activation_117[0][0] \n__________________________________________________________________________________________________\nconv2d_119 (Conv2D) (None, 16, 16, 64) 18432 average_pooling2d_11[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_113 (BatchN (None, 16, 16, 64) 192 conv2d_113[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_115 (BatchN (None, 16, 16, 64) 192 conv2d_115[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_118 (BatchN (None, 16, 16, 96) 288 conv2d_118[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_119 (BatchN (None, 16, 16, 64) 192 conv2d_119[0][0] \n__________________________________________________________________________________________________\nactivation_113 (Activation) (None, 16, 16, 64) 0 batch_normalization_113[0][0] \n__________________________________________________________________________________________________\nactivation_115 (Activation) (None, 16, 16, 64) 0 batch_normalization_115[0][0] \n__________________________________________________________________________________________________\nactivation_118 (Activation) (None, 16, 16, 96) 0 batch_normalization_118[0][0] \n__________________________________________________________________________________________________\nactivation_119 (Activation) (None, 16, 16, 64) 0 batch_normalization_119[0][0] \n__________________________________________________________________________________________________\nmixed2 (Concatenate) (None, 16, 16, 288) 0 activation_113[0][0] \n activation_115[0][0] \n activation_118[0][0] \n activation_119[0][0] \n__________________________________________________________________________________________________\nconv2d_121 (Conv2D) (None, 16, 16, 64) 18432 mixed2[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_121 (BatchN (None, 16, 16, 64) 192 conv2d_121[0][0] \n__________________________________________________________________________________________________\nactivation_121 (Activation) (None, 16, 16, 64) 0 batch_normalization_121[0][0] \n__________________________________________________________________________________________________\nconv2d_122 (Conv2D) (None, 16, 16, 96) 55296 activation_121[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_122 (BatchN (None, 16, 16, 96) 288 conv2d_122[0][0] \n__________________________________________________________________________________________________\nactivation_122 (Activation) (None, 16, 16, 96) 0 batch_normalization_122[0][0] \n__________________________________________________________________________________________________\nconv2d_120 (Conv2D) (None, 7, 7, 384) 995328 mixed2[0][0] \n__________________________________________________________________________________________________\nconv2d_123 (Conv2D) (None, 7, 7, 96) 82944 activation_122[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_120 (BatchN (None, 7, 7, 384) 1152 conv2d_120[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_123 (BatchN (None, 7, 7, 96) 288 conv2d_123[0][0] \n__________________________________________________________________________________________________\nactivation_120 (Activation) (None, 7, 7, 384) 0 batch_normalization_120[0][0] \n__________________________________________________________________________________________________\nactivation_123 (Activation) (None, 7, 7, 96) 0 batch_normalization_123[0][0] \n__________________________________________________________________________________________________\nmax_pooling2d_6 (MaxPooling2D) (None, 7, 7, 288) 0 mixed2[0][0] \n__________________________________________________________________________________________________\nmixed3 (Concatenate) (None, 7, 7, 768) 0 activation_120[0][0] \n activation_123[0][0] \n max_pooling2d_6[0][0] \n__________________________________________________________________________________________________\nconv2d_128 (Conv2D) (None, 7, 7, 128) 98304 mixed3[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_128 (BatchN (None, 7, 7, 128) 384 conv2d_128[0][0] \n__________________________________________________________________________________________________\nactivation_128 (Activation) (None, 7, 7, 128) 0 batch_normalization_128[0][0] \n__________________________________________________________________________________________________\nconv2d_129 (Conv2D) (None, 7, 7, 128) 114688 activation_128[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_129 (BatchN (None, 7, 7, 128) 384 conv2d_129[0][0] \n__________________________________________________________________________________________________\nactivation_129 (Activation) (None, 7, 7, 128) 0 batch_normalization_129[0][0] \n__________________________________________________________________________________________________\nconv2d_125 (Conv2D) (None, 7, 7, 128) 98304 mixed3[0][0] \n__________________________________________________________________________________________________\nconv2d_130 (Conv2D) (None, 7, 7, 128) 114688 activation_129[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_125 (BatchN (None, 7, 7, 128) 384 conv2d_125[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_130 (BatchN (None, 7, 7, 128) 384 conv2d_130[0][0] \n__________________________________________________________________________________________________\nactivation_125 (Activation) (None, 7, 7, 128) 0 batch_normalization_125[0][0] \n__________________________________________________________________________________________________\nactivation_130 (Activation) (None, 7, 7, 128) 0 batch_normalization_130[0][0] \n__________________________________________________________________________________________________\nconv2d_126 (Conv2D) (None, 7, 7, 128) 114688 activation_125[0][0] \n__________________________________________________________________________________________________\nconv2d_131 (Conv2D) (None, 7, 7, 128) 114688 activation_130[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_126 (BatchN (None, 7, 7, 128) 384 conv2d_126[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_131 (BatchN (None, 7, 7, 128) 384 conv2d_131[0][0] \n__________________________________________________________________________________________________\nactivation_126 (Activation) (None, 7, 7, 128) 0 batch_normalization_126[0][0] \n__________________________________________________________________________________________________\nactivation_131 (Activation) (None, 7, 7, 128) 0 batch_normalization_131[0][0] \n__________________________________________________________________________________________________\naverage_pooling2d_12 (AveragePo (None, 7, 7, 768) 0 mixed3[0][0] \n__________________________________________________________________________________________________\nconv2d_124 (Conv2D) (None, 7, 7, 192) 147456 mixed3[0][0] \n__________________________________________________________________________________________________\nconv2d_127 (Conv2D) (None, 7, 7, 192) 172032 activation_126[0][0] \n__________________________________________________________________________________________________\nconv2d_132 (Conv2D) (None, 7, 7, 192) 172032 activation_131[0][0] \n__________________________________________________________________________________________________\nconv2d_133 (Conv2D) (None, 7, 7, 192) 147456 average_pooling2d_12[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_124 (BatchN (None, 7, 7, 192) 576 conv2d_124[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_127 (BatchN (None, 7, 7, 192) 576 conv2d_127[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_132 (BatchN (None, 7, 7, 192) 576 conv2d_132[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_133 (BatchN (None, 7, 7, 192) 576 conv2d_133[0][0] \n__________________________________________________________________________________________________\nactivation_124 (Activation) (None, 7, 7, 192) 0 batch_normalization_124[0][0] \n__________________________________________________________________________________________________\nactivation_127 (Activation) (None, 7, 7, 192) 0 batch_normalization_127[0][0] \n__________________________________________________________________________________________________\nactivation_132 (Activation) (None, 7, 7, 192) 0 batch_normalization_132[0][0] \n__________________________________________________________________________________________________\nactivation_133 (Activation) (None, 7, 7, 192) 0 batch_normalization_133[0][0] \n__________________________________________________________________________________________________\nmixed4 (Concatenate) (None, 7, 7, 768) 0 activation_124[0][0] \n activation_127[0][0] \n activation_132[0][0] \n activation_133[0][0] \n__________________________________________________________________________________________________\nconv2d_138 (Conv2D) (None, 7, 7, 160) 122880 mixed4[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_138 (BatchN (None, 7, 7, 160) 480 conv2d_138[0][0] \n__________________________________________________________________________________________________\nactivation_138 (Activation) (None, 7, 7, 160) 0 batch_normalization_138[0][0] \n__________________________________________________________________________________________________\nconv2d_139 (Conv2D) (None, 7, 7, 160) 179200 activation_138[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_139 (BatchN (None, 7, 7, 160) 480 conv2d_139[0][0] \n__________________________________________________________________________________________________\nactivation_139 (Activation) (None, 7, 7, 160) 0 batch_normalization_139[0][0] \n__________________________________________________________________________________________________\nconv2d_135 (Conv2D) (None, 7, 7, 160) 122880 mixed4[0][0] \n__________________________________________________________________________________________________\nconv2d_140 (Conv2D) (None, 7, 7, 160) 179200 activation_139[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_135 (BatchN (None, 7, 7, 160) 480 conv2d_135[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_140 (BatchN (None, 7, 7, 160) 480 conv2d_140[0][0] \n__________________________________________________________________________________________________\nactivation_135 (Activation) (None, 7, 7, 160) 0 batch_normalization_135[0][0] \n__________________________________________________________________________________________________\nactivation_140 (Activation) (None, 7, 7, 160) 0 batch_normalization_140[0][0] \n__________________________________________________________________________________________________\nconv2d_136 (Conv2D) (None, 7, 7, 160) 179200 activation_135[0][0] \n__________________________________________________________________________________________________\nconv2d_141 (Conv2D) (None, 7, 7, 160) 179200 activation_140[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_136 (BatchN (None, 7, 7, 160) 480 conv2d_136[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_141 (BatchN (None, 7, 7, 160) 480 conv2d_141[0][0] \n__________________________________________________________________________________________________\nactivation_136 (Activation) (None, 7, 7, 160) 0 batch_normalization_136[0][0] \n__________________________________________________________________________________________________\nactivation_141 (Activation) (None, 7, 7, 160) 0 batch_normalization_141[0][0] \n__________________________________________________________________________________________________\naverage_pooling2d_13 (AveragePo (None, 7, 7, 768) 0 mixed4[0][0] \n__________________________________________________________________________________________________\nconv2d_134 (Conv2D) (None, 7, 7, 192) 147456 mixed4[0][0] \n__________________________________________________________________________________________________\nconv2d_137 (Conv2D) (None, 7, 7, 192) 215040 activation_136[0][0] \n__________________________________________________________________________________________________\nconv2d_142 (Conv2D) (None, 7, 7, 192) 215040 activation_141[0][0] \n__________________________________________________________________________________________________\nconv2d_143 (Conv2D) (None, 7, 7, 192) 147456 average_pooling2d_13[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_134 (BatchN (None, 7, 7, 192) 576 conv2d_134[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_137 (BatchN (None, 7, 7, 192) 576 conv2d_137[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_142 (BatchN (None, 7, 7, 192) 576 conv2d_142[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_143 (BatchN (None, 7, 7, 192) 576 conv2d_143[0][0] \n__________________________________________________________________________________________________\nactivation_134 (Activation) (None, 7, 7, 192) 0 batch_normalization_134[0][0] \n__________________________________________________________________________________________________\nactivation_137 (Activation) (None, 7, 7, 192) 0 batch_normalization_137[0][0] \n__________________________________________________________________________________________________\nactivation_142 (Activation) (None, 7, 7, 192) 0 batch_normalization_142[0][0] \n__________________________________________________________________________________________________\nactivation_143 (Activation) (None, 7, 7, 192) 0 batch_normalization_143[0][0] \n__________________________________________________________________________________________________\nmixed5 (Concatenate) (None, 7, 7, 768) 0 activation_134[0][0] \n activation_137[0][0] \n activation_142[0][0] \n activation_143[0][0] \n__________________________________________________________________________________________________\nconv2d_148 (Conv2D) (None, 7, 7, 160) 122880 mixed5[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_148 (BatchN (None, 7, 7, 160) 480 conv2d_148[0][0] \n__________________________________________________________________________________________________\nactivation_148 (Activation) (None, 7, 7, 160) 0 batch_normalization_148[0][0] \n__________________________________________________________________________________________________\nconv2d_149 (Conv2D) (None, 7, 7, 160) 179200 activation_148[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_149 (BatchN (None, 7, 7, 160) 480 conv2d_149[0][0] \n__________________________________________________________________________________________________\nactivation_149 (Activation) (None, 7, 7, 160) 0 batch_normalization_149[0][0] \n__________________________________________________________________________________________________\nconv2d_145 (Conv2D) (None, 7, 7, 160) 122880 mixed5[0][0] \n__________________________________________________________________________________________________\nconv2d_150 (Conv2D) (None, 7, 7, 160) 179200 activation_149[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_145 (BatchN (None, 7, 7, 160) 480 conv2d_145[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_150 (BatchN (None, 7, 7, 160) 480 conv2d_150[0][0] \n__________________________________________________________________________________________________\nactivation_145 (Activation) (None, 7, 7, 160) 0 batch_normalization_145[0][0] \n__________________________________________________________________________________________________\nactivation_150 (Activation) (None, 7, 7, 160) 0 batch_normalization_150[0][0] \n__________________________________________________________________________________________________\nconv2d_146 (Conv2D) (None, 7, 7, 160) 179200 activation_145[0][0] \n__________________________________________________________________________________________________\nconv2d_151 (Conv2D) (None, 7, 7, 160) 179200 activation_150[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_146 (BatchN (None, 7, 7, 160) 480 conv2d_146[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_151 (BatchN (None, 7, 7, 160) 480 conv2d_151[0][0] \n__________________________________________________________________________________________________\nactivation_146 (Activation) (None, 7, 7, 160) 0 batch_normalization_146[0][0] \n__________________________________________________________________________________________________\nactivation_151 (Activation) (None, 7, 7, 160) 0 batch_normalization_151[0][0] \n__________________________________________________________________________________________________\naverage_pooling2d_14 (AveragePo (None, 7, 7, 768) 0 mixed5[0][0] \n__________________________________________________________________________________________________\nconv2d_144 (Conv2D) (None, 7, 7, 192) 147456 mixed5[0][0] \n__________________________________________________________________________________________________\nconv2d_147 (Conv2D) (None, 7, 7, 192) 215040 activation_146[0][0] \n__________________________________________________________________________________________________\nconv2d_152 (Conv2D) (None, 7, 7, 192) 215040 activation_151[0][0] \n__________________________________________________________________________________________________\nconv2d_153 (Conv2D) (None, 7, 7, 192) 147456 average_pooling2d_14[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_144 (BatchN (None, 7, 7, 192) 576 conv2d_144[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_147 (BatchN (None, 7, 7, 192) 576 conv2d_147[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_152 (BatchN (None, 7, 7, 192) 576 conv2d_152[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_153 (BatchN (None, 7, 7, 192) 576 conv2d_153[0][0] \n__________________________________________________________________________________________________\nactivation_144 (Activation) (None, 7, 7, 192) 0 batch_normalization_144[0][0] \n__________________________________________________________________________________________________\nactivation_147 (Activation) (None, 7, 7, 192) 0 batch_normalization_147[0][0] \n__________________________________________________________________________________________________\nactivation_152 (Activation) (None, 7, 7, 192) 0 batch_normalization_152[0][0] \n__________________________________________________________________________________________________\nactivation_153 (Activation) (None, 7, 7, 192) 0 batch_normalization_153[0][0] \n__________________________________________________________________________________________________\nmixed6 (Concatenate) (None, 7, 7, 768) 0 activation_144[0][0] \n activation_147[0][0] \n activation_152[0][0] \n activation_153[0][0] \n__________________________________________________________________________________________________\nconv2d_158 (Conv2D) (None, 7, 7, 192) 147456 mixed6[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_158 (BatchN (None, 7, 7, 192) 576 conv2d_158[0][0] \n__________________________________________________________________________________________________\nactivation_158 (Activation) (None, 7, 7, 192) 0 batch_normalization_158[0][0] \n__________________________________________________________________________________________________\nconv2d_159 (Conv2D) (None, 7, 7, 192) 258048 activation_158[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_159 (BatchN (None, 7, 7, 192) 576 conv2d_159[0][0] \n__________________________________________________________________________________________________\nactivation_159 (Activation) (None, 7, 7, 192) 0 batch_normalization_159[0][0] \n__________________________________________________________________________________________________\nconv2d_155 (Conv2D) (None, 7, 7, 192) 147456 mixed6[0][0] \n__________________________________________________________________________________________________\nconv2d_160 (Conv2D) (None, 7, 7, 192) 258048 activation_159[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_155 (BatchN (None, 7, 7, 192) 576 conv2d_155[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_160 (BatchN (None, 7, 7, 192) 576 conv2d_160[0][0] \n__________________________________________________________________________________________________\nactivation_155 (Activation) (None, 7, 7, 192) 0 batch_normalization_155[0][0] \n__________________________________________________________________________________________________\nactivation_160 (Activation) (None, 7, 7, 192) 0 batch_normalization_160[0][0] \n__________________________________________________________________________________________________\nconv2d_156 (Conv2D) (None, 7, 7, 192) 258048 activation_155[0][0] \n__________________________________________________________________________________________________\nconv2d_161 (Conv2D) (None, 7, 7, 192) 258048 activation_160[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_156 (BatchN (None, 7, 7, 192) 576 conv2d_156[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_161 (BatchN (None, 7, 7, 192) 576 conv2d_161[0][0] \n__________________________________________________________________________________________________\nactivation_156 (Activation) (None, 7, 7, 192) 0 batch_normalization_156[0][0] \n__________________________________________________________________________________________________\nactivation_161 (Activation) (None, 7, 7, 192) 0 batch_normalization_161[0][0] \n__________________________________________________________________________________________________\naverage_pooling2d_15 (AveragePo (None, 7, 7, 768) 0 mixed6[0][0] \n__________________________________________________________________________________________________\nconv2d_154 (Conv2D) (None, 7, 7, 192) 147456 mixed6[0][0] \n__________________________________________________________________________________________________\nconv2d_157 (Conv2D) (None, 7, 7, 192) 258048 activation_156[0][0] \n__________________________________________________________________________________________________\nconv2d_162 (Conv2D) (None, 7, 7, 192) 258048 activation_161[0][0] \n__________________________________________________________________________________________________\nconv2d_163 (Conv2D) (None, 7, 7, 192) 147456 average_pooling2d_15[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_154 (BatchN (None, 7, 7, 192) 576 conv2d_154[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_157 (BatchN (None, 7, 7, 192) 576 conv2d_157[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_162 (BatchN (None, 7, 7, 192) 576 conv2d_162[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_163 (BatchN (None, 7, 7, 192) 576 conv2d_163[0][0] \n__________________________________________________________________________________________________\nactivation_154 (Activation) (None, 7, 7, 192) 0 batch_normalization_154[0][0] \n__________________________________________________________________________________________________\nactivation_157 (Activation) (None, 7, 7, 192) 0 batch_normalization_157[0][0] \n__________________________________________________________________________________________________\nactivation_162 (Activation) (None, 7, 7, 192) 0 batch_normalization_162[0][0] \n__________________________________________________________________________________________________\nactivation_163 (Activation) (None, 7, 7, 192) 0 batch_normalization_163[0][0] \n__________________________________________________________________________________________________\nmixed7 (Concatenate) (None, 7, 7, 768) 0 activation_154[0][0] \n activation_157[0][0] \n activation_162[0][0] \n activation_163[0][0] \n__________________________________________________________________________________________________\nflatten_1 (Flatten) (None, 37632) 0 mixed7[0][0] \n__________________________________________________________________________________________________\ndense_2 (Dense) (None, 1024) 38536192 flatten_1[0][0] \n__________________________________________________________________________________________________\ndropout_1 (Dropout) (None, 1024) 0 dense_2[0][0] \n__________________________________________________________________________________________________\ndense_3 (Dense) (None, 1) 1025 dropout_1[0][0] \n==================================================================================================\nTotal params: 47,512,481\nTrainable params: 38,537,217\nNon-trainable params: 8,975,264\n__________________________________________________________________________________________________\n"
],
[
"# Get the Horse or Human dataset\n!wget --no-check-certificate https://storage.googleapis.com/laurencemoroney-blog.appspot.com/horse-or-human.zip -O /tmp/horse-or-human.zip\n\n# Get the Horse or Human Validation dataset\n!wget --no-check-certificate https://storage.googleapis.com/laurencemoroney-blog.appspot.com/validation-horse-or-human.zip -O /tmp/validation-horse-or-human.zip \n \nfrom tensorflow.keras.preprocessing.image import ImageDataGenerator\n\nimport os\nimport zipfile\n\nlocal_zip = '//tmp/horse-or-human.zip'\nzip_ref = zipfile.ZipFile(local_zip, 'r')\nzip_ref.extractall('/tmp/training')\nzip_ref.close()\n\nlocal_zip = '//tmp/validation-horse-or-human.zip'\nzip_ref = zipfile.ZipFile(local_zip, 'r')\nzip_ref.extractall('/tmp/validation')\nzip_ref.close()",
"--2021-04-08 03:29:23-- https://storage.googleapis.com/laurencemoroney-blog.appspot.com/horse-or-human.zip\nResolving storage.googleapis.com (storage.googleapis.com)... 64.233.166.128, 64.233.184.128, 74.125.133.128, ...\nConnecting to storage.googleapis.com (storage.googleapis.com)|64.233.166.128|:443... connected.\nHTTP request sent, awaiting response... 200 OK\nLength: 149574867 (143M) [application/zip]\nSaving to: ‘/tmp/horse-or-human.zip’\n\n/tmp/horse-or-human 100%[===================>] 142.65M 38.6MB/s in 3.7s \n\n2021-04-08 03:29:28 (38.6 MB/s) - ‘/tmp/horse-or-human.zip’ saved [149574867/149574867]\n\n--2021-04-08 03:29:28-- https://storage.googleapis.com/laurencemoroney-blog.appspot.com/validation-horse-or-human.zip\nResolving storage.googleapis.com (storage.googleapis.com)... 74.125.140.128, 108.177.15.128, 173.194.76.128, ...\nConnecting to storage.googleapis.com (storage.googleapis.com)|74.125.140.128|:443... connected.\nHTTP request sent, awaiting response... 200 OK\nLength: 11480187 (11M) [application/zip]\nSaving to: ‘/tmp/validation-horse-or-human.zip’\n\n/tmp/validation-hor 100%[===================>] 10.95M 62.2MB/s in 0.2s \n\n2021-04-08 03:29:28 (62.2 MB/s) - ‘/tmp/validation-horse-or-human.zip’ saved [11480187/11480187]\n\n"
],
[
"train_horses_dir = \"/tmp/training/horses/\" # Your Code Here\ntrain_humans_dir = \"/tmp/training/humans\" # Your Code Here\nvalidation_horses_dir = \"/tmp/validation/horses\" # Your Code Here\nvalidation_humans_dir = \"/tmp/validation/humans\" # Your Code Here\n\ntrain_horses_fnames = len(os.listdir(train_horses_dir))\ntrain_humans_fnames = len(os.listdir(train_humans_dir)) # Your Code Here\nvalidation_horses_fnames = len(os.listdir(validation_horses_dir)) # Your Code Here\nvalidation_humans_fnames = len(os.listdir(validation_humans_dir)) # Your Code Here\n\nprint(train_horses_fnames)\nprint(train_humans_fnames)\nprint(validation_horses_fnames)\nprint(validation_humans_fnames)\n# Expected Output:\n# 500\n# 527\n# 128\n# 128",
"500\n527\n128\n128\n"
],
[
"# Define our example directories and files\ntrain_dir = '/tmp/training'\nvalidation_dir = '/tmp/validation'\n\ntrain_datagen = ImageDataGenerator(rescale=1./255,\n rotation_range=40,\n width_shift_range=0.2,\n height_shift_range=0.2,\n shear_range=0.2,\n zoom_range=0.2,\n horizontal_flip=True,\n fill_mode='nearest')\n\n# Note that the validation data should not be augmented!\ntest_datagen = ImageDataGenerator( rescale = 1.0/255.)\n\n# Flow training images in batches of 20 using train_datagen generator\ntrain_generator = train_datagen.flow_from_directory(train_dir,\n batch_size=20,\n class_mode='binary',\n target_size=(150, 150)) \n\n# Flow validation images in batches of 20 using test_datagen generator\nvalidation_generator = test_datagen.flow_from_directory(validation_dir,\n batch_size=20,\n class_mode = 'binary',\n target_size = (150, 150))\n\n# Expected Output:\n# Found 1027 images belonging to 2 classes.\n# Found 256 images belonging to 2 classes.",
"Found 1027 images belonging to 2 classes.\nFound 256 images belonging to 2 classes.\n"
],
[
"# Run this and see how many epochs it should take before the callback\n# fires, and stops training at 99.9% accuracy\n# (It should take less than 100 epochs)\n\ncallbacks = myCallback()# Your Code Here\nhistory = model.fit_generator(train_generator,\n epochs=3,\n verbose=1,\n validation_data=validation_generator,\n callbacks=[callbacks]) ",
"/usr/local/lib/python3.7/dist-packages/tensorflow/python/keras/engine/training.py:1844: UserWarning: `Model.fit_generator` is deprecated and will be removed in a future version. Please use `Model.fit`, which supports generators.\n warnings.warn('`Model.fit_generator` is deprecated and '\n"
],
[
"\nimport matplotlib.pyplot as plt\nacc = history.history['acc']\nval_acc = history.history['val_acc']\nloss = history.history['loss']\nval_loss = history.history['val_loss']\n\nepochs = range(len(acc))\n\nplt.plot(epochs, acc, 'r', label='Training accuracy')\nplt.plot(epochs, val_acc, 'b', label='Validation accuracy')\nplt.title('Training and validation accuracy')\nplt.legend(loc=0)\nplt.figure()\n\n\nplt.show()",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb8982906e9f044246f0990bd11014179e3185e7
| 911,215 |
ipynb
|
Jupyter Notebook
|
archive/genres.ipynb
|
Patrickbfuller/classify_genre
|
5ac2720bb557f603148e8a0aab21875710e51e00
|
[
"MIT"
] | null | null | null |
archive/genres.ipynb
|
Patrickbfuller/classify_genre
|
5ac2720bb557f603148e8a0aab21875710e51e00
|
[
"MIT"
] | null | null | null |
archive/genres.ipynb
|
Patrickbfuller/classify_genre
|
5ac2720bb557f603148e8a0aab21875710e51e00
|
[
"MIT"
] | null | null | null | 309.305838 | 59,792 | 0.918645 |
[
[
[
"# BUSINESS UNDERSTANDING",
"_____no_output_____"
],
[
"# DATA UNDERSTANDING\n### Collecting The Sonic Features\nCollecting implicitly labeled songs from playlists such as 'top 100 country songs'. Experiment can be rerun with different genres.",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nimport itertools\nplt.style.use('seaborn')\n\nfrom genres import collect_genre_features\n%load_ext autoreload\n%autoreload 2",
"/Applications/anaconda3/envs/learn-env/lib/python3.6/importlib/_bootstrap.py:219: RuntimeWarning: numpy.ufunc size changed, may indicate binary incompatibility. Expected 192 from C header, got 216 from PyObject\n return f(*args, **kwds)\n"
],
[
"genres = {\n 'country': 'https://www.youtube.com/playlist?list=PL3oW2tjiIxvQW6c-4Iry8Bpp3QId40S5S', \n 'jazz': 'https://www.youtube.com/playlist?list=PL8F6B0753B2CCA128', \n 'hip_hop': 'https://www.youtube.com/playlist?list=PLAPo1R_GVX4IZGbDvUH60bOwIOnZplZzM', \n 'classical': 'https://www.youtube.com/playlist?list=PLRb-5mC4V_Lop8KLXqSqMv4_mqw5M9jjW', \n 'metal': 'https://www.youtube.com/playlist?list=PLfY-m4YMsF-OM1zG80pMguej_Ufm8t0VC', \n 'electronic': 'https://www.youtube.com/playlist?list=PLDDAxmBan0BKeIxuYWjMPBWGXDqNRaW5S'\n}",
"_____no_output_____"
],
[
"# collect_genre_features(genres) # Started 8:55 done around 11:28",
"_____no_output_____"
],
[
"df = pd.read_json('data/genre_features.json', lines=True)",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
]
],
[
[
"Each row contains the sonic features for a unique 10 second audio portion of a song's video. \nIf a song is longer than 4 minutes, we only have the first 4 minutes of it. \nThere may be statistical noise in the form of cinematic intros and dialogue. ",
"_____no_output_____"
]
],
[
[
"df.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 14221 entries, 0 to 14220\nData columns (total 22 columns):\nsong 14221 non-null object\ngenre 14221 non-null object\ntempo 14221 non-null float64\nbeats 14221 non-null int64\nchroma_stft 14221 non-null float64\nrms 14221 non-null float64\nspec_cent 14221 non-null float64\nspec_bw 14221 non-null float64\nrolloff 14221 non-null float64\nzcr 14221 non-null float64\nmfcc1 14221 non-null float64\nmfcc2 14221 non-null float64\nmfcc3 14221 non-null float64\nmfcc4 14221 non-null float64\nmfcc5 14221 non-null float64\nmfcc6 14221 non-null float64\nmfcc7 14221 non-null float64\nmfcc8 14221 non-null float64\nmfcc9 14221 non-null float64\nmfcc10 14221 non-null float64\nmfcc11 14221 non-null float64\nmfcc12 14221 non-null float64\ndtypes: float64(19), int64(1), object(2)\nmemory usage: 2.4+ MB\n"
],
[
"df.describe()",
"_____no_output_____"
]
],
[
[
"Tempo and Beats columns both have a minimum of 0.",
"_____no_output_____"
]
],
[
[
"df[df.tempo == 0]",
"_____no_output_____"
],
[
"display(df[df.song.str.contains('Her World')].head(2))\ndf[df.song.str.contains('Her World')].tail(2)",
"_____no_output_____"
],
[
"df[df.song.str.contains('Marry Me')].tail(3)",
"_____no_output_____"
]
],
[
[
"These are intros and outros. \nA case could be made to: \n1. drop them as statistical noise in the tempo and beats columns. \n2. keep them as relevant audio in the spectral features. \n3. replace the 0s with the average tempo of the rest of the song.\n\nAs this is a first iteration we will include the rows with 0s unaltered",
"_____no_output_____"
],
[
"### Examine the distribution of data points among genres.",
"_____no_output_____"
]
],
[
[
"df.groupby('genre').song.count()",
"_____no_output_____"
],
[
"df.groupby('genre').song.nunique()",
"_____no_output_____"
]
],
[
[
"The dataset is roughly balanced among genres",
"_____no_output_____"
],
[
"### Examine the distribution of features among genres",
"_____no_output_____"
]
],
[
[
"c_options = ['darkorange', 'green', 'deepskyblue', 'mediumblue', 'black', 'deeppink']\ncolors = {k:v for k, v in zip(genres.keys(), c_options)}\ncolors",
"_____no_output_____"
],
[
"cols = ['chroma_stft', 'spec_cent', 'mfcc1', 'mfcc3']\nfor g1, g2 in itertools.combinations(df.genre.unique(), 2):\n plt.figure(figsize=(18,2))\n for i, col in enumerate(cols, start=1):\n plt.subplot(1, len(cols), i)\n s1 = df.loc[df.genre == g1, col]\n s2 = df.loc[df.genre == g2, col]\n sns.distplot(s1, label=g1, color=colors[g1])\n sns.distplot(s2, label=g2, color=colors[g2])\n plt.title(col)\n plt.legend()\n plt.show()",
"_____no_output_____"
]
],
[
[
"# DATA PREPARATION",
"_____no_output_____"
],
[
"### Train Test Split",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import train_test_split",
"_____no_output_____"
],
[
"y = df['genre']\nX = df.drop(['genre', 'song'], axis=1)\nX_work, X_holdout, y_work, y_holdout = train_test_split(X, y, test_size=0.2, random_state=111)\nX_train, X_test, y_train, y_test = train_test_split(X_work, y_work, test_size=0.2, random_state=111)",
"_____no_output_____"
],
[
"y_train.value_counts()",
"_____no_output_____"
]
],
[
[
"Training set is well balanced. Should be no need for class weighting.",
"_____no_output_____"
],
[
"### Data Transformation\n- For Non-Tree models, numerical scaling is requried.\n - Also power_transforming will be tested\n- PCA will be tested for predictive improvements \n\nBoth of these will be added into pipelines",
"_____no_output_____"
],
[
"# __MODELING__",
"_____no_output_____"
]
],
[
[
"from sklearn.preprocessing import StandardScaler, PowerTransformer\nfrom sklearn.decomposition import PCA\nfrom sklearn.pipeline import Pipeline\nfrom sklearn.metrics import log_loss, jaccard_score",
"_____no_output_____"
]
],
[
[
"#### Out of the box (Non Grid Search)\n- Logistic Regression\n- SVM\n- Random Forest \n-\n- GBTrees",
"_____no_output_____"
]
],
[
[
"labels = list(genres.keys())",
"_____no_output_____"
]
],
[
[
"## Logistic Regression",
"_____no_output_____"
]
],
[
[
"from sklearn.linear_model import LogisticRegression",
"_____no_output_____"
],
[
"logit_model = LogisticRegression(solver='lbfgs', multi_class='ovr', n_jobs=-1)\nlogreg = Pipeline([\n ('scaler', StandardScaler()),\n ('model', logit_model)\n ])\nlogreg.fit(X_train, y_train)\npred_probas = logreg.predict_proba(X_test)\npreds = logreg.predict(X_test)",
"_____no_output_____"
]
],
[
[
"#### Log Loss - Logistic Regression",
"_____no_output_____"
]
],
[
[
"log_loss(y_test, pred_probas, labels=labels)",
"_____no_output_____"
]
],
[
[
"#### Jaccard Score - Logistic Regression",
"_____no_output_____"
]
],
[
[
"jaccard_score(y_test, preds, average='macro').round(5)",
"_____no_output_____"
],
[
"print(list(zip(labels, jaccard_score(y_test, preds, average=None, labels=labels).round(5))))",
"[('country', 0.35196), ('jazz', 0.53704), ('hip_hop', 0.47182), ('classical', 0.67467), ('metal', 0.51941), ('electronic', 0.41532)]\n"
]
],
[
[
"#### Power Transformation - Logistic Regression",
"_____no_output_____"
]
],
[
[
"def eval_model(y_test, preds, pred_probas):\n print(f\"\"\"Log Loss:\n {log_loss(y_test, pred_probas, labels=labels)}\n Jaccard:\n {jaccard_score(y_test, preds, average='macro').round(5)}\n {list(zip(labels, jaccard_score(y_test, preds, average=None, labels=labels).round(5)))}\"\"\")",
"_____no_output_____"
],
[
"from genres import eval_model",
"_____no_output_____"
],
[
"logreg = Pipeline([\n ('scaler', PowerTransformer()),\n ('model', logit_model)\n ])\nlogreg.fit(X_train, y_train)\npred_probas = logreg.predict_proba(X_test)\npreds = logreg.predict(X_test)\neval_model(y_test, preds, pred_probas, labels=labels)",
"\n Log Loss:\n 0.9867394808313655\n Jaccard:\n 0.50895\n\t-Country: 0.36174)\n\t-Jazz: 0.55037)\n\t-Hip_Hop: 0.48434)\n\t-Classical: 0.71076)\n\t-Metal: 0.53096)\n\t-Electronic: 0.41551)\n"
]
],
[
[
"PowerTransformer provides a narrow improvement over StandardScaler.",
"_____no_output_____"
],
[
"#### PCA - Logistic Regression",
"_____no_output_____"
]
],
[
[
"logreg = Pipeline([\n ('scaler', PowerTransformer()),\n ('pca', PCA()),\n ('model', logit_model)\n ])\nlogreg.fit(X_train, y_train)\npred_probas = logreg.predict_proba(X_test)\npreds = logreg.predict(X_test)\neval_model(y_test, preds, pred_probas, labels=labels)",
"Log Loss:\n 0.9867394810831467\n Jaccard:\n 0.50895\n [('country', 0.36174), ('jazz', 0.55037), ('hip_hop', 0.48434), ('classical', 0.71076), ('metal', 0.53096), ('electronic', 0.41551)]\n"
]
],
[
[
"PCA does not seem to improve performance.",
"_____no_output_____"
],
[
"---\n---\n### Support Vector Machine",
"_____no_output_____"
]
],
[
[
"from sklearn.svm import SVC",
"_____no_output_____"
],
[
"svc_model = SVC(gamma='scale', probability=True)\nsvc = Pipeline([\n ('scaler', StandardScaler()),\n ('model', svc_model)\n])\nsvc.fit(X_train, y_train)\npred_probas = svc.predict_proba(X_test)\npreds = svc.predict(X_test)\neval_model(y_test, preds, pred_probas, labels=labels)",
"Log Loss:\n 0.5885192667060156\n Jaccard:\n 0.66298\n [('country', 0.56167), ('jazz', 0.78556), ('hip_hop', 0.59556), ('classical', 0.8101), ('metal', 0.68444), ('electronic', 0.54054)]\n"
]
],
[
[
"Support Vectors seem to improve predictions. Will PowerTransformer and PCA have any effect?",
"_____no_output_____"
],
[
"### Support Vector Machine - Power Transform",
"_____no_output_____"
]
],
[
[
"svc = Pipeline([\n ('scaler', PowerTransformer()),\n ('model', svc_model)\n])\nsvc.fit(X_train, y_train)\npred_probas = svc.predict_proba(X_test)\npreds = svc.predict(X_test)\neval_model(y_test, preds, pred_probas, labels=labels)",
"Log Loss:\n 0.581016254959359\n Jaccard:\n 0.67544\n [('country', 0.58077), ('jazz', 0.80132), ('hip_hop', 0.61224), ('classical', 0.81535), ('metal', 0.68202), ('electronic', 0.56092)]\n"
]
],
[
[
"Again, PowerTransformer provides a modest increase in performance.",
"_____no_output_____"
],
[
"### Support Vector Machine - PCA",
"_____no_output_____"
]
],
[
[
"svc = Pipeline([\n ('pca', PCA()),\n ('model', svc_model)\n])\nsvc.fit(X_train, y_train)\npred_probas = svc.predict_proba(X_test)\npreds = svc.predict(X_test)\neval_model(y_test, preds, pred_probas, labels=labels)",
"Log Loss:\n 1.349898292688635\n Jaccard:\n 0.30463\n [('country', 0.26933), ('jazz', 0.33206), ('hip_hop', 0.12424), ('classical', 0.56827), ('metal', 0.29103), ('electronic', 0.24283)]\n"
]
],
[
[
"Again, PCA does not improve performance.",
"_____no_output_____"
],
[
"### Random Forest",
"_____no_output_____"
]
],
[
[
"from sklearn.ensemble import RandomForestClassifier",
"_____no_output_____"
],
[
"rfc_model = RandomForestClassifier(n_estimators=200, n_jobs=-1)\nrfc_model.fit(X_train, y_train)\npred_probas = rfc_model.predict_proba(X_test)\npreds = rfc_model.predict(X_test)\neval_model(y_test, preds, pred_probas, labels=labels)",
"Log Loss:\n 0.6642586281615095\n Jaccard:\n 0.68764\n [('country', 0.61614), ('jazz', 0.79476), ('hip_hop', 0.65668), ('classical', 0.78009), ('metal', 0.72055), ('electronic', 0.5576)]\n"
]
],
[
[
"### Random Forest - PCA",
"_____no_output_____"
]
],
[
[
"rfc = Pipeline([\n ('pca', PCA()),\n ('model', rfc_model)\n])\nrfc.fit(X_train, y_train)\npred_probas = rfc.predict_proba(X_test)\npreds = rfc.predict(X_test)\neval_model(y_test, preds, pred_probas, labels=labels)",
"Log Loss:\n 0.6793650321890452\n Jaccard:\n 0.70219\n [('country', 0.61866), ('jazz', 0.7987), ('hip_hop', 0.69606), ('classical', 0.80048), ('metal', 0.71689), ('electronic', 0.58237)]\n"
]
],
[
[
"Random Forest seems to perform comparably to Support Vectors and PCA *is* helpful for the Forest model.",
"_____no_output_____"
],
[
"### Gradient Learning - Trees",
"_____no_output_____"
]
],
[
[
"from sklearn.ensemble import GradientBoostingClassifier # 57 and 70",
"/Applications/anaconda3/envs/learn-env/lib/python3.6/importlib/_bootstrap.py:219: RuntimeWarning: numpy.ufunc size changed, may indicate binary incompatibility. Expected 192 from C header, got 216 from PyObject\n return f(*args, **kwds)\n"
],
[
"gbt_model = GradientBoostingClassifier(max_depth=10)\ngbt = Pipeline([\n ('pca', PCA()),\n ('model', gbt_model)\n])\ngbt.fit(X_train, y_train)\npred_probas = gbt.predict_proba(X_test)\npreds = gbt.predict(X_test)\neval_model(y_test, preds, pred_probas, labels=labels)",
"\n Log Loss:\n 0.5798826061517595\n Jaccard:\n 0.71282\n\t-Country: 0.64609)\n\t-Jazz: 0.79914)\n\t-Hip_Hop: 0.69816)\n\t-Classical: 0.81509)\n\t-Metal: 0.71136)\n\t-Electronic: 0.60706)\n"
]
],
[
[
"The Gradient Boosted Trees Jaccard score is comparable to the Random Forest models but the log loss is an improvement.",
"_____no_output_____"
],
[
"### __Grid Searching__\n1. Support Vector\n3. Gradient Boosted Trees",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import GridSearchCV\nfrom sklearn.metrics import make_scorer\n\nscorer = make_scorer(log_loss, greater_is_better=False, needs_proba=True)",
"_____no_output_____"
]
],
[
[
"#### Support Vector - Grid Search",
"_____no_output_____"
]
],
[
[
"# svc = Pipeline([\n# ('scaler', PowerTransformer()),\n# ('model', svc_model)\n# ])\n# params = {\n# 'model__C': [0.5, 1, 10],\n# 'model__kernel': ['rbf', 'poly', 'sigmoid'],\n# 'model__gamma': [0.1, 0.25, 0.5] \n# }\n# svc_grid = GridSearchCV(\n# estimator=svc,\n# param_grid=params,\n# cv=5,\n# scoring=scorer,\n# n_jobs=-1\n# )\n# svc_grid.fit(X_train, y_train) # Takes some time to run",
"_____no_output_____"
],
[
"svc_grid.best_params_",
"_____no_output_____"
],
[
"eval_model(\n y_test,\n svc_grid.predict(X_test),\n svc_grid.predict_proba(X_test),\n labels=labels\n)",
"_____no_output_____"
]
],
[
[
"#### Support Vector Best Performer\n```python\n{'model__C': 10, 'model__gamma': 0.25, 'model__kernel': 'rbf'}\nLog Loss: \n 0.38742063263445364 \n Jaccard: \n 0.77164 \n [('country', 0.70789), ('jazz', 0.89671), ('hip_hop', 0.71136), ('classical', 0.84577), ('metal', 0.811), ('electronic', 0.65707)]\n ```",
"_____no_output_____"
],
[
"#### Gradient Boosting Classifier - Grid Search",
"_____no_output_____"
]
],
[
[
"# params = {\n# 'model__n_estimators': [100,150,200],\n# 'model__max_depth': [6,8,10,12,15],\n# 'model__subsample': [1, 0.9, 0.8] \n# }\n# gbt_grid = GridSearchCV(\n# estimator=gbt,\n# param_grid=params,\n# cv=5,\n# scoring=scorer,\n# n_jobs=-1\n# )\n# gbt_grid.fit(X_train, y_train)",
"_____no_output_____"
],
[
"gbt_grid.best_params_",
"_____no_output_____"
],
[
"eval_model(\n y_test=y_test,\n preds=gbt_grid.predict(X_test),\n pred_probas=gbt_grid.predict_proba(X_test),\n labels=labels\n)",
"_____no_output_____"
]
],
[
[
"#### Gradient Boosting Best Performer\n```python\n{'model__max_depth': 6, 'model__n_estimators': 200, 'model__subsample': 0.8}\nLog Loss: \n 0.4983750092724258 \n Jaccard: \n 0.72957 \n [('country', 0.65496), ('jazz', 0.82589), ('hip_hop', 0.71101), ('classical', 0.83127), ('metal', 0.73708), ('electronic', 0.61722)]\n ```",
"_____no_output_____"
],
[
"The Gradient Boosting Trees model does not improve upon the Support Vector Machine. \nIt will, however, scale better and therefor can be a viable option. ",
"_____no_output_____"
],
[
"### Deep Learning",
"_____no_output_____"
]
],
[
[
"from keras.layers import Dense, Dropout\nfrom keras.models import Sequential\nfrom keras.optimizers import SGD\nfrom keras.utils.np_utils import to_categorical\nfrom keras.wrappers.scikit_learn import KerasClassifier\nfrom keras.regularizers import l2",
"_____no_output_____"
],
[
"genre_map = {genre:i for i, genre in enumerate(genres.keys())}",
"_____no_output_____"
],
[
"labels = y.map(genre_map)\nY = to_categorical(labels)\nY_work, Y_holdout = train_test_split(Y, test_size=0.2, random_state=111)\nY_train, Y_test = train_test_split(Y_work, test_size=0.2, random_state=111)",
"_____no_output_____"
],
[
"def build_model(dropout=0.3, optimizer='adam'):\n model = Sequential()\n model.add(Dense(\n 40,\n activation='relu',\n input_shape=(20,),\n kernel_regularizer=l2(),\n bias_regularizer=l2()\n ))\n model.add(Dropout(rate=dropout))\n model.add(Dense(\n 20,\n activation='relu',\n kernel_regularizer=l2(),\n bias_regularizer=l2()\n ))\n model.add(Dropout(rate=dropout))\n model.add(Dense(\n 6,\n activation='softmax',\n kernel_regularizer=l2(),\n bias_regularizer=l2()\n ))\n model.compile(\n optimizer=optimizer,\n loss='categorical_crossentropy',\n metrics=['accuracy']\n )\n return model\n\nclf = KerasClassifier(build_fn=build_model)\n\npipe = Pipeline([\n ('pca', PCA()),\n# ('scaling', PowerTransformer()),\n ('model', clf)\n])\nhistory = pipe.fit(\n X_train,\n Y_train,\n model__epochs=200,\n model__batch_size=50,\n model__validation_data=(X_test,Y_test)\n)",
"_____no_output_____"
]
],
[
[
"### Log Loss",
"_____no_output_____"
]
],
[
[
"target = y_test.map(genre_map)\npreds = history.predict(X_test)\npred_probas = history.predict_proba(X_test)",
"_____no_output_____"
],
[
"log_loss(target, pred_probas)",
"_____no_output_____"
]
],
[
[
"### Jaccard",
"_____no_output_____"
]
],
[
[
"jaccard_score(target, preds, average='macro')",
"_____no_output_____"
],
[
"js = jaccard_score(target, preds, average=None, labels=[0,1,2,3,4,5])\nlist(zip(genres.keys(), js))",
"_____no_output_____"
]
],
[
[
"Deep Learning has not yet produce a robust model.",
"_____no_output_____"
],
[
"# EVALUATION",
"_____no_output_____"
],
[
"Holdout Time!",
"_____no_output_____"
]
],
[
[
"from sklearn.metrics import confusion_matrix",
"_____no_output_____"
],
[
"svc_model = SVC(\n C=10,\n gamma=0.25,\n probability=True,\n)\nsvc = Pipeline([\n ('scaling', PowerTransformer()),\n ('model', svc_model)\n])\nsvc.fit(X_work, y_work)\npred_probas = svc.predict_proba(X_holdout)\npreds = svc.predict(X_holdout)\neval_model(y_holdout, preds, pred_probas, labels)",
"Log Loss:\n 0.38242710444074923\n Jaccard:\n 0.7861\n [('country', 0.71743), ('jazz', 0.88025), ('hip_hop', 0.71553), ('classical', 0.86729), ('metal', 0.83093), ('electronic', 0.70517)]\n"
]
],
[
[
"#### Confusion Matrix",
"_____no_output_____"
]
],
[
[
"cm = confusion_matrix(y_holdout, preds, labels=labels)\nax = sns.heatmap(cm, cmap='gnuplot_r')\nax.set_ylim(6,0)\nax.set_ylabel('True Label', labelpad=20)\nax.set_xlabel('Predicted Label', labelpad=30)\nax.set_xticklabels(labels, rotation=20)\nax.set_yticklabels(labels, rotation=20)\nthresh = cm.max()/2\nfor i,j in itertools.product(range(6),range(6)):\n val = cm[i,j]\n plt.text(\n x=j+0.35,\n y=i+0.5,\n s=val,\n color='white' if val>thresh else 'black',\n size=15\n )\nplt.title(\n 'Genre Confusion Matrix',\n pad=20,\n fontdict={'fontweight': 'bold', 'fontsize': 28}\n);",
"_____no_output_____"
]
],
[
[
"### How did each category do?\nPercent of true labels depicted in Confusion Matrix",
"_____no_output_____"
]
],
[
[
"from genres import percentify_cm",
"_____no_output_____"
],
[
"pcm = percentify_cm(cm)\nax = sns.heatmap(pcm, cmap='gnuplot_r')\nax.set_ylim(6,0)\nax.set_ylabel('True Label', labelpad=20)\nax.set_xlabel('Predicted Label', labelpad=30)\nax.set_xticklabels(labels, rotation=20)\nax.set_yticklabels(labels, rotation=20)\nthresh = pcm.max()/2\nfor i,j in itertools.product(range(6),range(6)):\n val = pcm[i,j]\n plt.text(\n x=j+0.35,\n y=i+0.5,\n s=val,\n color='white' if val>thresh else 'black',\n size=15\n )\nplt.title(\n 'Confusion Matrix: Percent of True Label',\n pad=20,\n fontdict={'fontweight': 'bold', 'fontsize': 22}\n);",
"_____no_output_____"
]
],
[
[
"# DEPLOYMENT",
"_____no_output_____"
]
],
[
[
"import pickle",
"_____no_output_____"
],
[
"with open('genre_clf.pkl', 'wb') as f:\n pickle.dump(svc, f)",
"_____no_output_____"
],
[
"svc.classes_",
"_____no_output_____"
],
[
"pred_probas.sum(axis=0)/ pred_probas.sum()",
"_____no_output_____"
],
[
"y_holdout",
"_____no_output_____"
],
[
"from genres import classify_rows",
"_____no_output_____"
],
[
"classify_rows(df[:10])",
"_____no_output_____"
],
[
"from genres import classify",
"_____no_output_____"
],
[
"original_fp = '/Users/patrickfuller/Music/iTunes/iTunes Media/Music/Unknown Artist/Unknown Album/Waiting Dare.mp3'\nclassify(m4a_fp=original_fp)",
"_____no_output_____"
],
[
"moonlight_sonata_url = 'https://www.youtube.com/watch?v=4591dCHe_sE'\nclassify(url=moonlight_sonata_url)",
"_____no_output_____"
],
[
"luigi_url = 'https://www.youtube.com/watch?v=EHQ43ObPMHQ'\nclassify(url=luigi_url)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb8988ce8ef478dabbaefa19d1c71c6884e9ba4b
| 600 |
ipynb
|
Jupyter Notebook
|
home-data-for-ml-course/Untitled.ipynb
|
solomonxie/ml-practice
|
0f85aa771ce29603a92a51836c0b2060f1817ae0
|
[
"MIT"
] | 1 |
2019-03-19T06:42:06.000Z
|
2019-03-19T06:42:06.000Z
|
home-data-for-ml-course/Untitled.ipynb
|
solomonxie/ml-practice
|
0f85aa771ce29603a92a51836c0b2060f1817ae0
|
[
"MIT"
] | null | null | null |
home-data-for-ml-course/Untitled.ipynb
|
solomonxie/ml-practice
|
0f85aa771ce29603a92a51836c0b2060f1817ae0
|
[
"MIT"
] | null | null | null | 17.142857 | 40 | 0.533333 |
[
[
[
"# ISLR chapter 3 Linear Regression",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code"
]
] |
cb898f546779b432c3c47b8d2c89cc86e4294035
| 7,227 |
ipynb
|
Jupyter Notebook
|
gatesbezos/.ipynb_checkpoints/Border Collie or Yellow Lab-checkpoint.ipynb
|
jrjames83/Keras-Gates-vs-Bezos-Image-Classifier
|
399c4325ee081a0c3fec3cff8b6773265bfe925e
|
[
"MIT"
] | 7 |
2018-04-10T12:39:01.000Z
|
2021-08-23T06:15:35.000Z
|
gatesbezos/.ipynb_checkpoints/Border Collie or Yellow Lab-checkpoint.ipynb
|
jrjames83/Keras-Gates-vs-Bezos-Image-Classifier
|
399c4325ee081a0c3fec3cff8b6773265bfe925e
|
[
"MIT"
] | null | null | null |
gatesbezos/.ipynb_checkpoints/Border Collie or Yellow Lab-checkpoint.ipynb
|
jrjames83/Keras-Gates-vs-Bezos-Image-Classifier
|
399c4325ee081a0c3fec3cff8b6773265bfe925e
|
[
"MIT"
] | 8 |
2018-06-06T14:55:08.000Z
|
2020-05-04T06:01:31.000Z
| 29.141129 | 429 | 0.541719 |
[
[
[
"from keras import applications\n# python image_scraper.py \"yellow labrador retriever\" --count 500 --label labrador\nfrom keras.preprocessing.image import ImageDataGenerator\nfrom keras_tqdm import TQDMNotebookCallback\n\n\nfrom keras import optimizers\nfrom keras.models import Sequential, Model\nfrom keras.layers import (Dropout, Flatten, Dense, Conv2D, \n Activation, MaxPooling2D)\n\nfrom keras.applications.inception_v3 import InceptionV3\nfrom keras.layers import GlobalAveragePooling2D\nfrom keras import backend as K\n\nfrom sklearn.cross_validation import train_test_split\n\nimport os, glob\nfrom tqdm import tqdm\nfrom collections import Counter\nimport pandas as pd\nfrom sklearn.utils import shuffle\nimport numpy as np\nimport shutil",
"Using TensorFlow backend.\nC:\\Users\\jeffr\\AppData\\Local\\Continuum\\Anaconda3\\envs\\python3\\lib\\site-packages\\sklearn\\cross_validation.py:44: DeprecationWarning: This module was deprecated in version 0.18 in favor of the model_selection module into which all the refactored classes and functions are moved. Also note that the interface of the new CV iterators are different from that of this module. This module will be removed in 0.20.\n \"This module will be removed in 0.20.\", DeprecationWarning)\n"
],
[
"more_im = glob.glob(\"collie_lab/*/*.jpg\")\nmore_im = shuffle(more_im)\ncollie = [x for x in more_im if \"coll\" in x.split(\"\\\\\")[-2]]\nlab = [x for x in shuffle(more_im) if \"lab\" in x.split(\"\\\\\")[-2]]\n\nprint(len(collie))\nprint(len(lab))\n\nfor_labeling = collie + lab\nfor_labeling = shuffle(for_labeling)\n\nCounter([x.split(\"\\\\\")[-2] for x in more_im]).most_common()",
"600\n596\n"
],
[
"import shutil\nfrom tqdm import tqdm\n\n%mkdir collie_lab_train\n%mkdir collie_lab_valid\n\n%mkdir collie_lab_train\\\\collie\n%mkdir collie_lab_train\\\\lab\n\n%mkdir collie_lab_valid\\\\collie\n%mkdir collie_lab_valid\\\\lab\n\n\nfor index, image in tqdm(enumerate(for_labeling)):\n \n if index < 1000:\n label = image.split(\"\\\\\")[-2]\n image_name = image.split(\"\\\\\")[-1]\n if \"coll\" in label:\n shutil.copy(image, 'collie_lab_train\\\\collie\\\\{}'.format(image_name))\n if \"lab\" in label:\n shutil.copy(image, 'collie_lab_train\\\\lab\\\\{}'.format(image_name))\n \n if index > 1000:\n label = image.split(\"\\\\\")[-2]\n image_name = image.split(\"\\\\\")[-1]\n if \"coll\" in label:\n shutil.copy(image, 'collie_lab_valid\\\\collie\\\\{}'.format(image_name))\n if \"lab\" in label:\n shutil.copy(image, 'collie_lab_valid\\\\lab\\\\{}'.format(image_name)) ",
"A subdirectory or file collie_lab_train already exists.\nA subdirectory or file collie_lab_valid already exists.\n1196it [00:04, 244.83it/s]\n"
],
[
"train_datagen = ImageDataGenerator(\n rescale=1. / 255,\n shear_range=0.2,\n zoom_range=0.2,\n horizontal_flip=False)\n\n\ntest_datagen = ImageDataGenerator(rescale=1. / 255)\n\ntrain_generator = train_datagen.flow_from_directory(\n 'collie_lab_train/',\n target_size=(150, 150),\n batch_size=32,\n shuffle=True,\n class_mode='binary')\n\nvalidation_generator = test_datagen.flow_from_directory(\n 'collie_lab_valid/',\n target_size=(150, 150),\n batch_size=32,\n class_mode='binary')",
"Found 1000 images belonging to 2 classes.\nFound 195 images belonging to 2 classes.\n"
],
[
"model = Sequential()\n\nmodel.add(Conv2D(32, (3, 3), input_shape=(150, 150, 3)))\nmodel.add(Activation('relu')) #tanh\nmodel.add(MaxPooling2D(pool_size=(2, 2)))\nmodel.add(Dropout(0.5))\n\nmodel.add(Conv2D(32, (3, 3)))\nmodel.add(Activation('relu')) #tanh\nmodel.add(MaxPooling2D(pool_size=(2, 2)))\nmodel.add(Dropout(0.5))\n\nmodel.add(Conv2D(64, (3, 3)))\nmodel.add(Activation('relu'))\nmodel.add(MaxPooling2D(pool_size=(2, 2)))\nmodel.add(Dropout(0.5))\n\nmodel.add(Flatten())\nmodel.add(Dense(96))\nmodel.add(Activation('relu'))\nmodel.add(Dropout(0.5))\nmodel.add(Dense(1)) # binary\nmodel.add(Activation('sigmoid'))\n\nmodel.compile(loss='binary_crossentropy',\n optimizer='adam',\n metrics=['accuracy'])",
"_____no_output_____"
],
[
"model.fit_generator(\n train_generator,\n steps_per_epoch= 3000 // 32, # give me more data\n epochs=30,\n callbacks=[TQDMNotebookCallback()],\n verbose=0,\n validation_data=validation_generator,\n validation_steps= 300 // 32)",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb89a067d7a20a6234aae819c2c07763fcd830c5
| 15,302 |
ipynb
|
Jupyter Notebook
|
data/data-pipeline/data_pipeline/ipython/ACS Validate.ipynb
|
widal001/justice40-tool
|
356e16950f5f36909681c4d7d5ed78d1ced55baf
|
[
"CC0-1.0"
] | 59 |
2021-05-10T21:43:36.000Z
|
2022-03-30T17:57:17.000Z
|
data/data-pipeline/data_pipeline/ipython/ACS Validate.ipynb
|
widal001/justice40-tool
|
356e16950f5f36909681c4d7d5ed78d1ced55baf
|
[
"CC0-1.0"
] | 1,259 |
2021-05-10T18:21:26.000Z
|
2022-03-31T21:35:49.000Z
|
data/data-pipeline/data_pipeline/ipython/ACS Validate.ipynb
|
widal001/justice40-tool
|
356e16950f5f36909681c4d7d5ed78d1ced55baf
|
[
"CC0-1.0"
] | 24 |
2021-05-15T00:58:39.000Z
|
2022-03-24T23:18:17.000Z
| 27.227758 | 75 | 0.352895 |
[
[
[
"import pandas as pd\nimport csv\nfrom pathlib import Path\nimport os\nimport sys",
"_____no_output_____"
],
[
"module_path = os.path.abspath(os.path.join(\"..\"))\nif module_path not in sys.path:\n sys.path.append(module_path)",
"_____no_output_____"
],
[
"DATA_PATH = Path.cwd().parent / \"data\"\nTMP_PATH: Path = DATA_PATH / \"tmp\"\nACS_YEAR = \"2019\"\nOUTPUT_PATH = DATA_PATH / \"dataset\" / f\"census_acs_{ACS_YEAR}\"\nCENSUS_USA_CSV = DATA_PATH / \"census\" / \"csv\" / \"us.csv\"",
"_____no_output_____"
],
[
"cbg_usa_df = pd.read_csv(\n CENSUS_USA_CSV,\n names=[\"GEOID10\"],\n dtype={\"GEOID10\": \"string\"},\n low_memory=False,\n header=None,\n)",
"_____no_output_____"
],
[
"cbg_usa_df.head()",
"_____no_output_____"
],
[
"cbg_usa_df.dtypes",
"_____no_output_____"
],
[
"acs_df = pd.read_csv(\n OUTPUT_PATH / \"usa.csv\",\n dtype={\"GEOID10\": \"string\"},\n low_memory=False,\n)",
"_____no_output_____"
],
[
"acs_df.head()",
"_____no_output_____"
],
[
"acs_df.dtypes",
"_____no_output_____"
],
[
"merged_df = cbg_usa_df.merge(acs_df, on=\"GEOID10\", how=\"left\")",
"_____no_output_____"
],
[
"merged_df.head()",
"_____no_output_____"
],
[
"merged_df[merged_df[\"Unemployed civilians (percent)\"].isnull()]",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb89ab608661babf11de1f242d6467233134971b
| 23,262 |
ipynb
|
Jupyter Notebook
|
notebooks/TAi_development.ipynb
|
Global-Green-Growth-Institute/GraphModels
|
6a1c1a545df24daf2f7571fc6be8d47bece55c55
|
[
"MIT"
] | null | null | null |
notebooks/TAi_development.ipynb
|
Global-Green-Growth-Institute/GraphModels
|
6a1c1a545df24daf2f7571fc6be8d47bece55c55
|
[
"MIT"
] | null | null | null |
notebooks/TAi_development.ipynb
|
Global-Green-Growth-Institute/GraphModels
|
6a1c1a545df24daf2f7571fc6be8d47bece55c55
|
[
"MIT"
] | null | null | null | 31.392713 | 178 | 0.396741 |
[
[
[
"import os\npath_parent = os.path.dirname(os.getcwd())\nos.chdir(path_parent)",
"_____no_output_____"
],
[
"from data_utils.utils import get_X_y_from_data, data_dict_from_df_tables\nfrom ggmodel_dev.models.landuse.BE2 import model_dictionnary\n\nimport pandas as pd\nimport numpy as np",
"_____no_output_____"
],
[
"from ggmodel_dev.graphmodel import GraphModel, concatenate_graph_specs\nfrom ggmodel_dev.validation import score_model, plot_diagnostic, score_model_dictionnary",
"_____no_output_____"
],
[
"import os\nos.environ[\"GGGI_db_username\"] = 'postgres'\nos.environ[\"GGGI_db_password\"] = 'lHeJnnRINyWCzfkDOzKl'\nos.environ['GGGI_db_endpoint'] ='database-gggi-1.cg4tog4qy0py.ap-northeast-2.rds.amazonaws.com'\nos.environ['GGGI_db_port'] = '5432'\n\nfrom ggmodel_dev.database import get_variables_df",
"_____no_output_____"
],
[
"def prepare_TAI_data():\n data_dict = get_variables_df(['ANPi', 'AYi', 'FPi', 'PTTAi', 'TAi'], exclude_tables=['variabledata', 'foodbalancesheet'])\n data_dict['livestock'] = data_dict['livestock'].drop(columns=['Description_y', 'Unit_y']).rename(columns={'Description_x': 'Description', 'Unit_x': 'Unit'})\n data_dict['foodbalancesheet_new'] = data_dict['foodbalancesheet_new'].query(\"group == 'animal'\")\n \n return data_dict",
"_____no_output_____"
],
[
"data_dict = prepare_TAI_data()",
"emissions TAi: Done\nfoodbalancesheet FPi: Excluded\nfoodbalancesheet_new FPi: Done\nlivestock FPi, ANPi, AYi, PTTAi: Done\nvariabledata TAi: Excluded\n"
],
[
"data_dict['emissions']",
"_____no_output_____"
],
[
"test = data_dict_from_df_tables([data_dict['livestock']])",
"_____no_output_____"
],
[
"data_dict['livestock'].query(\"Variable == 'FPi'\")",
"_____no_output_____"
],
[
"test = data_dict_from_df_tables([data_dict['livestock']])\n\ntest['ANPi'] = test['ANPi'].droplevel(['FBS_item', 'table']).dropna()\ntest['AYi'] = test['AYi'].droplevel(['FBS_item', 'table']).dropna()\ntest['FPi'] = test['FPi'].droplevel(['FBS_item', 'table']).dropna()\ntest['PTTAi'] = test['PTTAi'].droplevel(['FBS_item', 'table', 'Item']).rename_axis(index={\"emi_item\": 'Item'})\n\ntest['TAi'] = data_dict['emissions'].set_index(['ISO', 'Year', 'Item'])['Value']",
"_____no_output_____"
],
[
"TAi_nodes = {'FPi': {'type': 'input',\n 'unit': '1000 tonnes',\n 'name': 'Food production per food group'},\n 'AYi': {'type': 'input',\n 'unit': 'tonnes/head',\n 'name': 'Vector of animal yields'},\n 'ANPi': {'type': 'variable',\n 'unit': 'head',\n 'name': 'Animals needed for production per animal type',\n 'computation': lambda FPi, AYi, **kwargs: 1e3 * FPi / AYi\n },\n 'PTTAi': {'type': 'parameter',\n 'unit': '1',\n 'name': 'Production to animal population ratio',\n },\n 'TAi': {'type': 'output',\n 'unit': 'head',\n 'name': 'Animal population',\n 'computation': lambda ANPi, PTTAi, **kwargs: PTTAi * ANPi.groupby(level=['ISO', 'Year', 'emi_item']).sum().rename_axis(index={\"emi_item\": 'Item'})\n },\n }\n\nmodel = GraphModel(TAi_nodes)",
"_____no_output_____"
],
[
"res = model.run(test)",
"_____no_output_____"
],
[
"scores = score_model(model, test)",
"_____no_output_____"
],
[
"scores['score_by_Variable']",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb89c0ac43307d406485e93aeba73ef9a0fc5bb9
| 1,763 |
ipynb
|
Jupyter Notebook
|
Problemas 5.2/09.ipynb
|
mateuschaves/CALCULO-DIFERENCIAL-E-INTEGRAL-II
|
a643b8c86cacb3b3d4eb61c258e013fe2cdadba0
|
[
"MIT"
] | 2 |
2018-09-15T18:05:00.000Z
|
2018-10-13T01:55:55.000Z
|
Problemas 5.2/09.ipynb
|
mateuschaves/CALCULO-DIFERENCIAL-E-INTEGRAL-II
|
a643b8c86cacb3b3d4eb61c258e013fe2cdadba0
|
[
"MIT"
] | null | null | null |
Problemas 5.2/09.ipynb
|
mateuschaves/CALCULO-DIFERENCIAL-E-INTEGRAL-II
|
a643b8c86cacb3b3d4eb61c258e013fe2cdadba0
|
[
"MIT"
] | null | null | null | 17.116505 | 76 | 0.456041 |
[
[
[
"<b>Calcule a integral dada</b>",
"_____no_output_____"
],
[
"<b>$9. \\int xe^{x^2}$</b>",
"_____no_output_____"
],
[
"$u = x^2$",
"_____no_output_____"
],
[
"$du = 2x dx$",
"_____no_output_____"
],
[
"$\\frac{du}{2} = xdx$",
"_____no_output_____"
],
[
"<b>Aplicando a substituição</b>",
"_____no_output_____"
],
[
"$\\int xe^{x^2} \\rightarrow \\frac{1}{2} \\cdot \\int e^u du$",
"_____no_output_____"
],
[
"<b>Integrando $\\frac{1}{2} \\cdot \\int e^u du$</b>",
"_____no_output_____"
],
[
"$\\frac{1}{2} \\cdot \\int e^u du = \\frac{1}{2} \\cdot e^u + C$",
"_____no_output_____"
],
[
"<b>Desfazendo a substituição</b>",
"_____no_output_____"
],
[
"$\\frac{1}{2} \\cdot \\int e^{u} du = \\frac{1}{2} \\cdot e^{x^2} + C$",
"_____no_output_____"
]
]
] |
[
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
cb89c456ad0b22b73e9292b124a62c4f5fae9d82
| 131,158 |
ipynb
|
Jupyter Notebook
|
Recommendations_with_IBM.ipynb
|
speQtrum/Udacity-Recommendation-Engine-Project
|
a70314ad1a4914118e9cbc097cefaeebdb9d93a8
|
[
"MIT"
] | null | null | null |
Recommendations_with_IBM.ipynb
|
speQtrum/Udacity-Recommendation-Engine-Project
|
a70314ad1a4914118e9cbc097cefaeebdb9d93a8
|
[
"MIT"
] | null | null | null |
Recommendations_with_IBM.ipynb
|
speQtrum/Udacity-Recommendation-Engine-Project
|
a70314ad1a4914118e9cbc097cefaeebdb9d93a8
|
[
"MIT"
] | null | null | null | 62.725012 | 24,300 | 0.713155 |
[
[
[
"# Recommendations with IBM\n\nIn this notebook, you will be putting your recommendation skills to use on real data from the IBM Watson Studio platform. \n\n\nYou may either submit your notebook through the workspace here, or you may work from your local machine and submit through the next page. Either way assure that your code passes the project [RUBRIC](https://review.udacity.com/#!/rubrics/2322/view). **Please save regularly.**\n\nBy following the table of contents, you will build out a number of different methods for making recommendations that can be used for different situations. \n\n\n## Table of Contents\n\nI. [Exploratory Data Analysis](#Exploratory-Data-Analysis)<br>\nII. [Rank Based Recommendations](#Rank)<br>\nIII. [User-User Based Collaborative Filtering](#User-User)<br>\nIV. [Content Based Recommendations (EXTRA - NOT REQUIRED)](#Content-Recs)<br>\nV. [Matrix Factorization](#Matrix-Fact)<br>\nVI. [Extras & Concluding](#conclusions)\n\nAt the end of the notebook, you will find directions for how to submit your work. Let's get started by importing the necessary libraries and reading in the data.",
"_____no_output_____"
]
],
[
[
"!pip install progressbar",
"Collecting progressbar\n Downloading https://files.pythonhosted.org/packages/a3/a6/b8e451f6cff1c99b4747a2f7235aa904d2d49e8e1464e0b798272aa84358/progressbar-2.5.tar.gz\nBuilding wheels for collected packages: progressbar\n Running setup.py bdist_wheel for progressbar ... \u001b[?25ldone\n\u001b[?25h Stored in directory: /root/.cache/pip/wheels/c0/e9/6b/ea01090205e285175842339aa3b491adeb4015206cda272ff0\nSuccessfully built progressbar\nInstalling collected packages: progressbar\nSuccessfully installed progressbar-2.5\n"
],
[
"# Import necessary packages\nimport pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nimport re\nimport progressbar\nfrom sklearn.feature_extraction.text import TfidfVectorizer\nfrom sklearn.metrics import accuracy_score\nimport project_tests as t\nimport pickle\n\n\n# nltk\nimport nltk\nfrom nltk.corpus import stopwords\nfrom nltk.stem.wordnet import WordNetLemmatizer\nfrom nltk.tokenize import word_tokenize\nnltk.download(['punkt', 'wordnet', 'stopwords',\n 'averaged_perceptron_tagger'])\n\n# Pretty display for notebooks\n%matplotlib inline\n%config InlineBachend.figure_format = 'retina'\n\n",
"[nltk_data] Downloading package punkt to /root/nltk_data...\n[nltk_data] Package punkt is already up-to-date!\n[nltk_data] Downloading package wordnet to /root/nltk_data...\n[nltk_data] Package wordnet is already up-to-date!\n[nltk_data] Downloading package stopwords to /root/nltk_data...\n[nltk_data] Package stopwords is already up-to-date!\n[nltk_data] Downloading package averaged_perceptron_tagger to\n[nltk_data] /root/nltk_data...\n[nltk_data] Package averaged_perceptron_tagger is already up-to-\n[nltk_data] date!\n"
],
[
"df = pd.read_csv('data/user-item-interactions.csv')\ndf_content = pd.read_csv('data/articles_community.csv')\ndel df['Unnamed: 0']\ndel df_content['Unnamed: 0']\n\n# Show df to get an idea of the data\ndf.head()",
"_____no_output_____"
],
[
"# Show df_content to get an idea of the data\ndf_content.head()",
"_____no_output_____"
]
],
[
[
"### <a class=\"anchor\" id=\"Exploratory-Data-Analysis\">Part I : Exploratory Data Analysis</a>\n\nUse the dictionary and cells below to provide some insight into the descriptive statistics of the data.\n\n`1.` What is the distribution of how many articles a user interacts with in the dataset? Provide a visual and descriptive statistics to assist with giving a look at the number of times each user interacts with an article. ",
"_____no_output_____"
]
],
[
[
"# Check null values in df\ndf.isnull().sum()",
"_____no_output_____"
],
[
"# Check null values in df_content\ndf_content.isnull().sum()",
"_____no_output_____"
],
[
"# Count user interaction\nuser_interaction = df.email.value_counts(dropna=False)",
"_____no_output_____"
],
[
"# Distribution of how many articles a user interacts with in the dataset\nplt.figure()\nplt.hist(user_interaction.values, bins=100)\nplt.title('Distribution of how many articles a user \\\n interacts with in the dataset')\nplt.xlabel('interactions')\nplt.ylabel('count')\nplt.show()",
"_____no_output_____"
],
[
"\n\nmost_articles = df.article_id.value_counts(dropna=False)\n\ncum_user = np.cumsum(most_articles.values)\n\n# 50% of individuals interact with ____ number of articles or fewer.\nmedian_val = len(cum_user[cum_user <= len(user_interaction)/2])\n\n# The maximum number of user-article interactions by any 1 user is ______.\nmax_views_by_user = user_interaction.iloc[0]\n",
"_____no_output_____"
]
],
[
[
"`2.` Explore and remove duplicate articles from the **df_content** dataframe. ",
"_____no_output_____"
]
],
[
[
"# Find and explore duplicate articles\narticle_count = df_content.article_id.value_counts(dropna=False)\ndup_articles = article_count[article_count > 1]\n\nprint('number of duplicate articles is: ', len(dup_articles))",
"number of duplicate articles is: 5\n"
],
[
"# Remove any rows that have the same article_id - only keep the first\ndf_content.drop_duplicates(subset=['article_id'], inplace=True)",
"_____no_output_____"
]
],
[
[
"`3.` Use the cells below to find:\n\n**a.** The number of unique articles that have an interaction with a user. \n**b.** The number of unique articles in the dataset (whether they have any interactions or not).<br>\n**c.** The number of unique users in the dataset. (excluding null values) <br>\n**d.** The number of user-article interactions in the dataset.",
"_____no_output_____"
]
],
[
[
"# The number of unique articles that have at least one interaction\nunique_articles = len(most_articles)\n\n# The number of unique articles on the IBM platform\ntotal_articles = df_content.shape[0]\n\n# The number of unique users\nunique_users = len(user_interaction)-1\n\n# The number of user-article interactions\nuser_article_interactions = len(df)",
"_____no_output_____"
]
],
[
[
"`4.` Use the cells below to find the most viewed **article_id**, as well as how often it was viewed. After talking to the company leaders, the `email_mapper` function was deemed a reasonable way to map users to ids. There were a small number of null values, and it was found that all of these null values likely belonged to a single user (which is how they are stored using the function below).",
"_____no_output_____"
]
],
[
[
"# The most viewed article in the dataset\n# as a string with one value following the decimal\nmost_viewed_article_id = str(most_articles.index[0])\n# The most viewed article in the dataset was viewed how many times?\nmax_views = most_articles.iloc[0]",
"_____no_output_____"
],
[
"## No need to change the code here - this will be helpful for later parts of the notebook\n# Run this cell to map the user email to a user_id column and remove the email column\n\ndef email_mapper():\n coded_dict = dict()\n cter = 1\n email_encoded = []\n \n for val in df['email']:\n if val not in coded_dict:\n coded_dict[val] = cter\n cter+=1\n \n email_encoded.append(coded_dict[val])\n return email_encoded\n\nemail_encoded = email_mapper()\ndel df['email']\ndf['user_id'] = email_encoded\n\n# show header\ndf.head()",
"_____no_output_____"
],
[
"## If you stored all your results in the variable names above, \n## you shouldn't need to change anything in this cell\n\nsol_1_dict = {\n '`50% of individuals have _____ or fewer interactions.`': median_val,\n '`The total number of user-article interactions in the dataset is ______.`': user_article_interactions,\n '`The maximum number of user-article interactions by any 1 user is ______.`': max_views_by_user,\n '`The most viewed article in the dataset was viewed _____ times.`': max_views,\n '`The article_id of the most viewed article is ______.`': most_viewed_article_id,\n '`The number of unique articles that have at least 1 rating ______.`': unique_articles,\n '`The number of unique users in the dataset is ______`': unique_users,\n '`The number of unique articles on the IBM platform`': total_articles\n}\n\n# Test your dictionary against the solution\nt.sol_1_test(sol_1_dict)",
"It looks like you have everything right here! Nice job!\n"
]
],
[
[
"### <a class=\"anchor\" id=\"Rank\">Part II: Rank-Based Recommendations</a>\n\nUnlike in the earlier lessons, we don't actually have ratings for whether a user liked an article or not. We only know that a user has interacted with an article. In these cases, the popularity of an article can really only be based on how often an article was interacted with.\n\n`1.` Fill in the function below to return the **n** top articles ordered with most interactions as the top. Test your function using the tests below.",
"_____no_output_____"
]
],
[
[
"def get_top_articles(n, df=df):\n '''\n INPUT:\n n - (int) the number of top articles to return\n df - (pandas dataframe) df as defined at the top of the notebook\n\n OUTPUT:\n top_articles - (list) A list of the top 'n' article titles\n\n '''\n\n # Get articles ids\n user_by_article = df.groupby(['user_id',\n 'article_id'])['title'].count().unstack()\n articles_interaction = user_by_article.sum().sort_values(ascending=False)\n articles_index = articles_interaction.iloc[:n].index\n\n # Get articles titles\n df_art_title = df.drop_duplicates(subset=['article_id'])[['article_id',\n 'title']]\n df_art_title.index = df_art_title.article_id\n\n # get list of the top n article titles\n top_articles = list(df_art_title.loc[articles_index].title)\n\n return top_articles\n\n\ndef get_top_article_ids(n, df=df):\n '''\n INPUT:\n n - (int) the number of top articles to return\n df - (pandas dataframe) df as defined at the top of the notebook\n\n OUTPUT:\n top_articles - (list) A list of the top 'n' article titles\n\n '''\n\n user_by_article = df.groupby(['user_id',\n 'article_id'])['title'].count().unstack()\n articles_interaction = user_by_article.sum().sort_values(ascending=False)\n top_articles = list(articles_interaction.iloc[:n].index)\n\n return top_articles # Return the top article ids",
"_____no_output_____"
],
[
"print(get_top_articles(10))\nprint(get_top_article_ids(10))",
"['use deep learning for image classification', 'insights from new york car accident reports', 'visualize car data with brunel', 'use xgboost, scikit-learn & ibm watson machine learning apis', 'predicting churn with the spss random tree algorithm', 'healthcare python streaming application demo', 'finding optimal locations of new store using decision optimization', 'apache spark lab, part 1: basic concepts', 'analyze energy consumption in buildings', 'gosales transactions for logistic regression model']\n[1429.0, 1330.0, 1431.0, 1427.0, 1364.0, 1314.0, 1293.0, 1170.0, 1162.0, 1304.0]\n"
],
[
"# Test your function by returning the top 5, 10, and 20 articles\ntop_5 = get_top_articles(5)\ntop_10 = get_top_articles(10)\ntop_20 = get_top_articles(20)\n\n# Test each of your three lists from above\nt.sol_2_test(get_top_articles)",
"Your top_5 looks like the solution list! Nice job.\nYour top_10 looks like the solution list! Nice job.\nYour top_20 looks like the solution list! Nice job.\n"
]
],
[
[
"### <a class=\"anchor\" id=\"User-User\">Part III: User-User Based Collaborative Filtering</a>\n\n\n`1.` Use the function below to reformat the **df** dataframe to be shaped with users as the rows and articles as the columns. \n\n* Each **user** should only appear in each **row** once.\n\n\n* Each **article** should only show up in one **column**. \n\n\n* **If a user has interacted with an article, then place a 1 where the user-row meets for that article-column**. It does not matter how many times a user has interacted with the article, all entries where a user has interacted with an article should be a 1. \n\n\n* **If a user has not interacted with an item, then place a zero where the user-row meets for that article-column**. \n\nUse the tests to make sure the basic structure of your matrix matches what is expected by the solution.",
"_____no_output_____"
]
],
[
[
"# create the user-article matrix with 1's and 0's\n\ndef create_user_item_matrix(df):\n '''\n INPUT:\n df - pandas dataframe with article_id, title, user_id columns\n\n OUTPUT:\n user_item - user item matrix\n\n Description:\n Return a matrix with user ids as rows and article ids on the columns\n with 1 values where a user interacted with an article and a 0 otherwise\n '''\n\n # Fill in the function here\n user_item = df.groupby(['user_id',\n 'article_id'])['title'].agg(lambda x: 1).unstack()\n user_item.fillna(0, inplace=True)\n\n return user_item # return the user_item matrix\n\n\nuser_item = create_user_item_matrix(df)\n\n# save the matrix in a pickle file\nuser_item.to_pickle('user_item_matrix.p')",
"_____no_output_____"
],
[
"## Tests: You should just need to run this cell. Don't change the code.\nassert user_item.shape[0] == 5149, \"Oops! The number of users in the user-article matrix doesn't look right.\"\nassert user_item.shape[1] == 714, \"Oops! The number of articles in the user-article matrix doesn't look right.\"\nassert user_item.sum(axis=1)[1] == 36, \"Oops! The number of articles seen by user 1 doesn't look right.\"\nprint(\"You have passed our quick tests! Please proceed!\")",
"You have passed our quick tests! Please proceed!\n"
]
],
[
[
"`2.` Complete the function below which should take a user_id and provide an ordered list of the most similar users to that user (from most similar to least similar). The returned result should not contain the provided user_id, as we know that each user is similar to him/herself. Because the results for each user here are binary, it (perhaps) makes sense to compute similarity as the dot product of two users. \n\nUse the tests to test your function.",
"_____no_output_____"
]
],
[
[
"def find_similar_users(user_id, user_item=user_item):\n '''\n INPUT:\n user_id - (int) a user_id\n user_item - (pandas dataframe) matrix of users by articles:\n 1's when a user has interacted with an article, 0 otherwise\n\n OUTPUT:\n similar_users - (list) an ordered list where the closest users\n (largest dot product users) are listed first\n\n Description:\n Computes the similarity of every pair of users based on the dot product\n Returns an ordered\n\n '''\n\n # Compute similarity of each user to the provided user\n user_vector = np.array(user_item.loc[user_id]).reshape(-1, 1)\n Matrix_item = user_item.drop(user_id)\n similarity = np.dot(Matrix_item.values, user_vector)\n # sort by similarity\n df_smly = pd.DataFrame({'user_id': Matrix_item.index,\n 'similarity': similarity.flatten()})\n df_smly.sort_values(by=['similarity'], inplace=True, ascending=False)\n\n # Create list of just the ids\n most_similar_users = list(df_smly.user_id)\n\n return most_similar_users",
"_____no_output_____"
],
[
"# Do a spot check of your function\nprint(\"The 10 most similar users to user 1 are: {}\".format(find_similar_users(1)[:10]))\nprint(\"The 5 most similar users to user 3933 are: {}\".format(find_similar_users(3933)[:5]))\nprint(\"The 3 most similar users to user 46 are: {}\".format(find_similar_users(46)[:3]))",
"The 10 most similar users to user 1 are: [3933, 23, 3782, 203, 4459, 3870, 131, 46, 4201, 395]\nThe 5 most similar users to user 3933 are: [1, 23, 3782, 4459, 203]\nThe 3 most similar users to user 46 are: [4201, 23, 3782]\n"
]
],
[
[
"`3.` Now that you have a function that provides the most similar users to each user, you will want to use these users to find articles you can recommend. Complete the functions below to return the articles you would recommend to each user. ",
"_____no_output_____"
]
],
[
[
"def get_article_names(article_ids, df=df):\n '''\n INPUT:\n article_ids - (list) a list of article ids (str)\n df - (pandas dataframe) df as defined at the top of the notebook\n\n OUTPUT:\n article_names - (list) a list of article names associated with the list\n of article ids (this is identified by the title column)\n '''\n\n article_ids = [float(x) for x in article_ids]\n df_2 = df.drop_duplicates(subset=['article_id'])\n df_2.set_index('article_id', inplace=True)\n article_names = list(df_2.loc[article_ids]['title'])\n\n return article_names\n\n\ndef get_user_articles(user_id, user_item=user_item):\n '''\n INPUT:\n user_id - (int) a user id\n user_item - (pandas dataframe) matrix of users by articles:\n 1's when a user has interacted with an article, 0 otherwise\n\n OUTPUT:\n article_ids - (list) a list of the article ids seen by the user\n article_names - (list) a list of article names associated with\n the list of article ids\n\n Description:\n Provides a list of the article_ids and article titles that have\n been seen by a user\n '''\n row_user = user_item.loc[user_id]\n article_ids = list(row_user[row_user > 0].index)\n article_ids = [str(x) for x in article_ids]\n article_names = get_article_names(article_ids)\n return article_ids, article_names\n\n\ndef user_user_recs(user_id, m=10):\n '''\n INPUT:\n user_id - (int) a user id\n m - (int) the number of recommendations you want for the user\n\n OUTPUT:\n recs - (list) a list of recommendations for the user\n\n Description:\n Loops through the users based on closeness to the input user_id\n For each user - finds articles the user hasn't seen before and\n provides them as recs\n Does this until m recommendations are found\n\n Notes:\n Users who are the same closeness are chosen arbitrarily as the 'next' user\n\n For the user where the number of recommended articles starts below m\n and ends exceeding m, the last items are chosen arbitrarily\n\n '''\n\n # Get user articles\n article_ids, _ = get_user_articles(user_id)\n # Find similar users\n most_similar_users = find_similar_users(user_id)\n # How many users for progress bar\n n_users = len(most_similar_users)\n\n recs = []\n # Create the progressbar\n cnter = 0\n bar = progressbar.ProgressBar(maxval=n_users+1,\n widgets=[progressbar.Bar('=', '[', ']'),\n ' ', progressbar.Percentage()])\n bar.start()\n\n for user in most_similar_users:\n\n # Update the progress bar\n cnter += 1\n bar.update(cnter)\n\n # Get user articles\n ids, _ = get_user_articles(user)\n article_not_seen = np.setdiff1d(np.array(ids), np.array(article_ids))\n article_not_recs = np.setdiff1d(article_not_seen, np.array(recs))\n recs.extend(list(article_not_recs))\n\n # If there are more than\n if len(recs) > m:\n break\n\n bar.finish()\n\n recs = recs[:10]\n\n return recs\n",
"_____no_output_____"
],
[
"# Check Results\nget_article_names(user_user_recs(1, 10)) # Return 10 recommendations for user 1",
"[========================================================================] 100%\n"
],
[
"# Test your functions here - No need to change this code - just run this cell\nassert set(get_article_names(['1024.0', '1176.0', '1305.0', '1314.0', '1422.0', '1427.0'])) == set(['using deep learning to reconstruct high-resolution audio', 'build a python app on the streaming analytics service', 'gosales transactions for naive bayes model', 'healthcare python streaming application demo', 'use r dataframes & ibm watson natural language understanding', 'use xgboost, scikit-learn & ibm watson machine learning apis']), \"Oops! Your the get_article_names function doesn't work quite how we expect.\"\nassert set(get_article_names(['1320.0', '232.0', '844.0'])) == set(['housing (2015): united states demographic measures','self-service data preparation with ibm data refinery','use the cloudant-spark connector in python notebook']), \"Oops! Your the get_article_names function doesn't work quite how we expect.\"\nassert set(get_user_articles(20)[0]) == set(['1320.0', '232.0', '844.0'])\nassert set(get_user_articles(20)[1]) == set(['housing (2015): united states demographic measures', 'self-service data preparation with ibm data refinery','use the cloudant-spark connector in python notebook'])\nassert set(get_user_articles(2)[0]) == set(['1024.0', '1176.0', '1305.0', '1314.0', '1422.0', '1427.0'])\nassert set(get_user_articles(2)[1]) == set(['using deep learning to reconstruct high-resolution audio', 'build a python app on the streaming analytics service', 'gosales transactions for naive bayes model', 'healthcare python streaming application demo', 'use r dataframes & ibm watson natural language understanding', 'use xgboost, scikit-learn & ibm watson machine learning apis'])\nprint(\"If this is all you see, you passed all of our tests! Nice job!\")",
"If this is all you see, you passed all of our tests! Nice job!\n"
]
],
[
[
"`4.` Now we are going to improve the consistency of the **user_user_recs** function from above. \n\n* Instead of arbitrarily choosing when we obtain users who are all the same closeness to a given user - choose the users that have the most total article interactions before choosing those with fewer article interactions.\n\n\n* Instead of arbitrarily choosing articles from the user where the number of recommended articles starts below m and ends exceeding m, choose articles with the articles with the most total interactions before choosing those with fewer total interactions. This ranking should be what would be obtained from the **top_articles** function you wrote earlier.",
"_____no_output_____"
]
],
[
[
"def get_top_sorted_users(user_id, df=df, user_item=user_item):\n '''\n INPUT:\n user_id - (int)\n df - (pandas dataframe) df as defined at the top of the notebook\n user_item - (pandas dataframe) matrix of users by articles:\n 1's when a user has interacted with an article, 0 otherwise\n\n OUTPUT:\n neighbors_df - (pandas dataframe) a dataframe with:\n neighbor_id - is a neighbor user_id\n similarity - measure of the similarity of each\n user to the provided user_id\n num_interactions - the number of articles viewed\n by the user - if a u\n\n Other Details - sort the neighbors_df by the similarity and then by number\n of interactions where highest of each is higher in the dataframe\n\n '''\n\n # similarity\n user_vector = np.array(user_item.loc[user_id]).reshape(-1, 1)\n Matrix_item = user_item.drop(user_id)\n similarity = np.dot(Matrix_item.values, user_vector)\n # sort by similarity\n df_smly = pd.DataFrame({'neighbor_id': Matrix_item.index,\n 'similarity': similarity.flatten()})\n\n # Number of interaction\n count_inter = df.groupby('user_id')['article_id'].count()\n df_inter = pd.DataFrame({'neighbor_id': count_inter.index,\n 'num_interactions': count_inter.values})\n\n # Merging the two dataframes\n neighbors_df = df_smly.merge(df_inter)\n\n # sort the neighbors_df\n neighbors_df.sort_values(by=['similarity', 'num_interactions'],\n inplace=True, ascending=False)\n\n return neighbors_df\n\n\ndef user_user_recs_part2(user_id, m=10):\n '''\n INPUT:\n user_id - (int) a user id\n m - (int) the number of recommendations you want for the user\n\n OUTPUT:\n recs - (list) a list of recommendations for the user by article id\n rec_names - (list) a list of recommendations for the user by article title\n\n Description:\n Loops through the users based on closeness to the input user_id\n For each user - finds articles the user hasn't seen before and\n provides them as recs\n Does this until m recommendations are found\n\n Notes:\n * Choose the users that have the most total article interactions\n before choosing those with fewer article interactions.\n\n * Choose articles with the articles with the most total interactions\n before choosing those with fewer total interactions.\n\n '''\n # get user articles\n article_ids, _ = get_user_articles(user_id)\n # find similar users\n most_similar_users = list(get_top_sorted_users(user_id).neighbor_id)\n # How many users for progress bar\n n_users = len(most_similar_users)\n\n recs = []\n # Create the progressbar\n cnter = 0\n bar = progressbar.ProgressBar(maxval=n_users+1,\n widgets=[progressbar.Bar('=', '[', ']'), ' ',\n progressbar.Percentage()])\n bar.start()\n\n for user in most_similar_users:\n\n # Update the progress bar\n cnter += 1\n bar.update(cnter)\n\n # get user articles\n ids, _ = get_user_articles(user)\n article_not_seen = np.setdiff1d(np.array(ids), np.array(article_ids))\n article_not_recs = np.setdiff1d(article_not_seen, np.array(recs))\n recs.extend(list(article_not_recs))\n\n # If there are more than\n if len(recs) > m:\n break\n\n bar.finish()\n\n recs = recs[:10]\n rec_names = get_article_names(recs)\n\n return recs, rec_names\n",
"_____no_output_____"
],
[
"# Quick spot check - don't change this code - just use it to test your functions\nrec_ids, rec_names = user_user_recs_part2(20, 10)\nprint(\"The top 10 recommendations for user 20 are the following article ids:\")\nprint(rec_ids)\nprint()\nprint(\"The top 10 recommendations for user 20 are the following article names:\")\nprint(rec_names)",
"[========================================================================] 100%\r"
]
],
[
[
"`5.` Use your functions from above to correctly fill in the solutions to the dictionary below. Then test your dictionary against the solution. Provide the code you need to answer each following the comments below.",
"_____no_output_____"
]
],
[
[
"# Tests with a dictionary of results\nuser1_most_sim = get_top_sorted_users(1).neighbor_id.values[0] # Find the user that is most similar to user 1 \nuser131_10th_sim = get_top_sorted_users(131).neighbor_id.values[9] # Find the 10th most similar user to user 131",
"_____no_output_____"
],
[
"## Dictionary Test Here\nsol_5_dict = {\n 'The user that is most similar to user 1.': user1_most_sim, \n 'The user that is the 10th most similar to user 131': user131_10th_sim,\n}\n\nt.sol_5_test(sol_5_dict)",
"This all looks good! Nice job!\n"
]
],
[
[
"`6.` If we were given a new user, which of the above functions would you be able to use to make recommendations? Explain. Can you think of a better way we might make recommendations? Use the cell below to explain a better method for new users.",
"_____no_output_____"
],
[
"**Provide your response here.**\n\n\nFor a new user, we can use get_top_articles function to suggest top articles.\n\nWe can improve our recommendations for a new user by using a Knowledge-Based Recommendations where we will ask the user to provide pieces of information about the types of articles they are interested in and look throughout our data for articles that meet the user specifications.\n",
"_____no_output_____"
],
[
"`7.` Using your existing functions, provide the top 10 recommended articles you would provide for the a new user below. You can test your function against our thoughts to make sure we are all on the same page with how we might make a recommendation.",
"_____no_output_____"
]
],
[
[
"\n\nnew_user = '0.0'\n\n# List of the top 10 article ids you would give to\nnew_user_recs = [str(x) for x in get_top_article_ids(10)]",
"_____no_output_____"
],
[
"assert set(new_user_recs) == set(['1314.0','1429.0','1293.0','1427.0','1162.0','1364.0','1304.0','1170.0','1431.0','1330.0']), \"Oops! It makes sense that in this case we would want to recommend the most popular articles, because we don't know anything about these users.\"\n\nprint(\"That's right! Nice job!\")",
"That's right! Nice job!\n"
]
],
[
[
"### <a class=\"anchor\" id=\"Content-Recs\">Part IV: Content Based Recommendations (EXTRA - NOT REQUIRED)</a>\n\nAnother method we might use to make recommendations is to perform a ranking of the highest ranked articles associated with some term. You might consider content to be the **doc_body**, **doc_description**, or **doc_full_name**. There isn't one way to create a content based recommendation, especially considering that each of these columns hold content related information. \n\n`1.` Use the function body below to create a content based recommender. Since there isn't one right answer for this recommendation tactic, no test functions are provided. Feel free to change the function inputs if you decide you want to try a method that requires more input values. The input values are currently set with one idea in mind that you may use to make content based recommendations. One additional idea is that you might want to choose the most popular recommendations that meet your 'content criteria', but again, there is a lot of flexibility in how you might make these recommendations.\n\n### This part is NOT REQUIRED to pass this project. However, you may choose to take this on as an extra way to show off your skills.",
"_____no_output_____"
]
],
[
[
"def make_content_recs():\n '''\n INPUT:\n \n OUTPUT:\n \n '''",
"_____no_output_____"
]
],
[
[
"`2.` Now that you have put together your content-based recommendation system, use the cell below to write a summary explaining how your content based recommender works. Do you see any possible improvements that could be made to your function? Is there anything novel about your content based recommender?\n\n### This part is NOT REQUIRED to pass this project. However, you may choose to take this on as an extra way to show off your skills.",
"_____no_output_____"
],
[
"**Write an explanation of your content based recommendation system here.**",
"_____no_output_____"
],
[
"`3.` Use your content-recommendation system to make recommendations for the below scenarios based on the comments. Again no tests are provided here, because there isn't one right answer that could be used to find these content based recommendations.\n\n### This part is NOT REQUIRED to pass this project. However, you may choose to take this on as an extra way to show off your skills.",
"_____no_output_____"
]
],
[
[
"# make recommendations for a brand new user\n\n\n# make a recommendations for a user who only has interacted with article id '1427.0'\n\n",
"_____no_output_____"
]
],
[
[
"### <a class=\"anchor\" id=\"Matrix-Fact\">Part V: Matrix Factorization</a>\n\nIn this part of the notebook, you will build use matrix factorization to make article recommendations to the users on the IBM Watson Studio platform.\n\n`1.` You should have already created a **user_item** matrix above in **question 1** of **Part III** above. This first question here will just require that you run the cells to get things set up for the rest of **Part V** of the notebook. ",
"_____no_output_____"
]
],
[
[
"# Load the matrix here\nuser_item_matrix = pd.read_pickle('user_item_matrix.p')",
"_____no_output_____"
],
[
"# quick look at the matrix\nuser_item_matrix.head()",
"_____no_output_____"
]
],
[
[
"`2.` In this situation, you can use Singular Value Decomposition from [numpy](https://docs.scipy.org/doc/numpy-1.14.0/reference/generated/numpy.linalg.svd.html) on the user-item matrix. Use the cell to perform SVD, and explain why this is different than in the lesson.",
"_____no_output_____"
]
],
[
[
"\n\n# Perform SVD on the User-Item Matrix Here\n\n# use the built in to get the three matrices\nu, s, vt = np.linalg.svd(user_item_matrix)\ns.shape, u.shape, vt.shape\n\n",
"_____no_output_____"
]
],
[
[
"**Provide your response here.**\n\n\nThe lesson provides a data structure with numeric values representing a rating and nulls representing non-interaction. This is not a matrix in the linear algebra sense and cannot be operated on (eg by SVD). Funk SVD would have to be used to provide a numeric approximation.\n\nThe matrix in this exercise contains binary values with a zero representing non-interaction and a one representing interaction. Although not invertible, this matrix can be factored by SVD.\n",
"_____no_output_____"
],
[
"`3.` Now for the tricky part, how do we choose the number of latent features to use? Running the below cell, you can see that as the number of latent features increases, we obtain a lower error rate on making predictions for the 1 and 0 values in the user-item matrix. Run the cell below to get an idea of how the accuracy improves as we increase the number of latent features.",
"_____no_output_____"
]
],
[
[
"num_latent_feats = np.arange(10,700+10,20)\nsum_errs = []\n\nfor k in num_latent_feats:\n # restructure with k latent features\n s_new, u_new, vt_new = np.diag(s[:k]), u[:, :k], vt[:k, :]\n \n # take dot product\n user_item_est = np.around(np.dot(np.dot(u_new, s_new), vt_new))\n \n # compute error for each prediction to actual value\n diffs = np.subtract(user_item_matrix, user_item_est)\n \n # total errors and keep track of them\n err = np.sum(np.sum(np.abs(diffs)))\n sum_errs.append(err)\n \n \nplt.plot(num_latent_feats, 1 - np.array(sum_errs)/df.shape[0]);\nplt.xlabel('Number of Latent Features');\nplt.ylabel('Accuracy');\nplt.title('Accuracy vs. Number of Latent Features');",
"_____no_output_____"
]
],
[
[
"`4.` From the above, we can't really be sure how many features to use, because simply having a better way to predict the 1's and 0's of the matrix doesn't exactly give us an indication of if we are able to make good recommendations. Instead, we might split our dataset into a training and test set of data, as shown in the cell below. \n\nUse the code from question 3 to understand the impact on accuracy of the training and test sets of data with different numbers of latent features. Using the split below: \n\n* How many users can we make predictions for in the test set? \n* How many users are we not able to make predictions for because of the cold start problem?\n* How many articles can we make predictions for in the test set? \n* How many articles are we not able to make predictions for because of the cold start problem?",
"_____no_output_____"
]
],
[
[
"df_train = df.head(40000)\ndf_test = df.tail(5993)\n\n\ndef create_test_and_train_user_item(df_train, df_test):\n '''\n INPUT:\n df_train - training dataframe\n df_test - test dataframe\n\n OUTPUT:\n user_item_train - a user-item matrix of the training dataframe\n (unique users for each row and unique articles\n for each column)\n user_item_test - a user-item matrix of the testing dataframe\n (unique users for each row and unique articles for\n each column)\n test_idx - all of the test user ids\n test_arts - all of the test article ids\n\n '''\n\n # user-item matrix of the training dataframe\n user_item_train = create_user_item_matrix(df_train)\n\n # user-item matrix of the testing dataframe\n user_item_test = create_user_item_matrix(df_test)\n\n test_idx = list(user_item_train.index) # test user ids\n test_arts = list(user_item_train.columns) # test article ids\n\n return user_item_train, user_item_test, test_idx, test_arts\n\n\nuser_item_train, user_item_test, test_idx, \\\n test_arts = create_test_and_train_user_item(df_train, df_test)",
"_____no_output_____"
],
[
"# Replace the values in the dictionary below\na = 662 \nb = 574 \nc = 20 \nd = 0 \n\n\nsol_4_dict = {\n 'How many users can we make predictions for in the test set?': c, \n 'How many users in the test set are we not able to make predictions for because of the cold start problem?': a, \n 'How many movies can we make predictions for in the test set?': b,\n 'How many movies in the test set are we not able to make predictions for because of the cold start problem?':d\n}\n\nt.sol_4_test(sol_4_dict)",
"Awesome job! That's right! All of the test movies are in the training data, but there are only 20 test users that were also in the training set. All of the other users that are in the test set we have no data on. Therefore, we cannot make predictions for these users using SVD.\n"
]
],
[
[
"`5.` Now use the **user_item_train** dataset from above to find U, S, and V transpose using SVD. Then find the subset of rows in the **user_item_test** dataset that you can predict using this matrix decomposition with different numbers of latent features to see how many features makes sense to keep based on the accuracy on the test data. This will require combining what was done in questions `2` - `4`.\n\nUse the cells below to explore how well SVD works towards making predictions for recommendations on the test data. ",
"_____no_output_____"
]
],
[
[
"# Fit SVD on the user_item_train matrix\nu_train, s_train, vt_train = np.linalg.svd(user_item_train)",
"_____no_output_____"
],
[
"# Use these cells to see how well you can use the training \n# decomposition to predict on test data",
"_____no_output_____"
],
[
"# Subset of rows in the user_item_test dataset that you can predict\n\n# Rows that match the test set\ntest_idx = user_item_test.index\nrow_idxs = user_item_train.index.isin(test_idx)\nu_test = u_train[row_idxs, :]\n\n# Columns that match the test set\ntest_col = user_item_test.columns\ncol_idxs = user_item_train.columns.isin(test_col)\nvt_test = vt_train[:, col_idxs]",
"_____no_output_____"
],
[
"# Test data\ntrain_idx = user_item_train.index\nrow_idxs_2 = user_item_test.index.isin(train_idx)\nsub_user_item_test = user_item_test.loc[row_idxs_2]",
"_____no_output_____"
],
[
"latent_feats = np.arange(10, 700+10, 20)\n\nall_errs, train_errs, test_errs = [], [], []\n\n\nfor k in latent_feats:\n # restructure with k latent features\n s_train_lat, u_train_lat, vt_train_lat = np.diag(s_train[:k]), u_train[:, :k], vt_train[:k, :]\n \n u_test_lat, vt_test_lat = u_test[:, :k], vt_test[:k, :]\n \n # take dot product\n user_item_train_preds = np.around(np.dot(np.dot(u_train_lat, s_train_lat), vt_train_lat))\n user_item_test_preds = np.around(np.dot(np.dot(u_test_lat, s_train_lat), vt_test_lat))\n \n all_errs.append(1 - ((np.sum(user_item_test_preds)+np.sum(np.sum(sub_user_item_test)))/(sub_user_item_test.shape[0]*sub_user_item_test.shape[1])))\n\n # compute prediction accuracy\n train_errs.append(accuracy_score(user_item_train.values.flatten(), user_item_train_preds.flatten()))\n test_errs.append(accuracy_score(sub_user_item_test.values.flatten(), user_item_test_preds.flatten()))\n\n\nplt.figure()\nplt.plot(latent_feats, all_errs, label='All Errors')\nplt.plot(latent_feats, train_errs, label='Train')\nplt.plot(latent_feats, test_errs, label='Test')\nplt.xlabel('Number of Latent Features')\nplt.ylabel('Accuracy')\nplt.title('Accuracy vs. Number of Latent Features')\nplt.legend()\nplt.show()\n\n",
"_____no_output_____"
]
],
[
[
"`6.` Use the cell below to comment on the results you found in the previous question. Given the circumstances of your results, discuss what you might do to determine if the recommendations you make with any of the above recommendation systems are an improvement to how users currently find articles? ",
"_____no_output_____"
],
[
"**Your response here.**\nThe figure above shows that in overall the accuracy of the model is very high. But it is a misleading result because we have a class imbalance in your data. In fact, the data contain much more zeros than ones.\n\nThe training accuracy increase to near 100% as the number of latent features increases. While the testing accuracy decrease as the number of latent features increases. This could be due to a limited variety in the datasets. A solution to this problem could be to perform Cross Validation to determine the number of latent features which allow the model to see different subsets of the datasets.\n",
"_____no_output_____"
],
[
"<a id='conclusions'></a>\n### Extras\nUsing your workbook, you could now save your recommendations for each user, develop a class to make new predictions and update your results, and make a flask app to deploy your results. These tasks are beyond what is required for this project. However, from what you learned in the lessons, you certainly capable of taking these tasks on to improve upon your work here!\n\n\n## Conclusion\n\n> Congratulations! You have reached the end of the Recommendations with IBM project! \n\n> **Tip**: Once you are satisfied with your work here, check over your report to make sure that it is satisfies all the areas of the [rubric](https://review.udacity.com/#!/rubrics/2322/view). You should also probably remove all of the \"Tips\" like this one so that the presentation is as polished as possible.\n\n\n## Directions to Submit\n\n> Before you submit your project, you need to create a .html or .pdf version of this notebook in the workspace here. To do that, run the code cell below. If it worked correctly, you should get a return code of 0, and you should see the generated .html file in the workspace directory (click on the orange Jupyter icon in the upper left).\n\n> Alternatively, you can download this report as .html via the **File** > **Download as** submenu, and then manually upload it into the workspace directory by clicking on the orange Jupyter icon in the upper left, then using the Upload button.\n\n> Once you've done this, you can submit your project by clicking on the \"Submit Project\" button in the lower right here. This will create and submit a zip file with this .ipynb doc and the .html or .pdf version you created. Congratulations! ",
"_____no_output_____"
]
],
[
[
"from subprocess import call\ncall(['python', '-m', 'nbconvert', 'Recommendations_with_IBM.ipynb'])",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
]
] |
cb89c612a3afdaae0695e6105c52972f8d82550e
| 261,096 |
ipynb
|
Jupyter Notebook
|
notebooks/McDavid.ipynb
|
ManchesterBioinference/GrandPrix
|
2790cb01d6cc8a1bd5a62de642c09be3a56324c2
|
[
"Apache-2.0"
] | 14 |
2017-12-04T18:39:32.000Z
|
2022-03-26T06:46:07.000Z
|
notebooks/McDavid.ipynb
|
ManchesterBioinference/GrandPrix
|
2790cb01d6cc8a1bd5a62de642c09be3a56324c2
|
[
"Apache-2.0"
] | 1 |
2018-10-31T09:41:08.000Z
|
2019-05-22T12:03:34.000Z
|
notebooks/McDavid.ipynb
|
ManchesterBioinference/GrandPrix
|
2790cb01d6cc8a1bd5a62de642c09be3a56324c2
|
[
"Apache-2.0"
] | 7 |
2017-12-04T14:55:20.000Z
|
2019-11-06T10:54:40.000Z
| 405.428571 | 239,578 | 0.913675 |
[
[
[
"# Applying GrandPrix on the cell cycle single cell nCounter data of PC3 human prostate cancer\n_Sumon Ahmed_, 2017, 2018\n\nThis notebooks describes how GrandPrix with informative prior over the latent space can be used to infer the cell cycle stages from the single cell nCounter data of the PC3 human prostate cancer cell line.",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nfrom GrandPrix import GrandPrix",
"_____no_output_____"
]
],
[
[
"# Data decription\n<a href=\"https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4102402/\" terget=\"_blank\">McDavid et al. (2014)</a> assayed the expression profiles of the PC3 human prostate cancer cell line. They identified the cells in G0/G1, S and G2/M cell cycle stages. The cells identified as G0/G1, S and G2/M have been mapped to the capture times of 1, 2 and 3, respectively. Due to the additional challenge of optimizing pseudotime parameters for periodic data, random pseudotimes having the largest log likelihood to estimate cell cycle peak time points have been used to initilize the prior.\n\n\nThe __McDavidtrainingData.csv__ file contains the expression profiles of the top __56__ differentially expressed genes in __361__ cells from the PC3 human prostate cancer cell line which have been used in the inference.\n\nThe __McDavidCellMeta.csv__ file contains the additional information of the data such as capture time of each cells, different initializations of pseudotimes, etc.",
"_____no_output_____"
]
],
[
[
"Y = pd.read_csv('../data/McDavid/McDavidtrainingData.csv', index_col=[0]).T\nmData = pd.read_csv('../data/McDavid/McDavidCellMeta.csv', index_col=[0])",
"_____no_output_____"
],
[
"N, D = Y.shape\nprint('Time Points: %s, Genes: %s'%(N, D))",
"Time Points: 361, Genes: 56\n"
],
[
"mData.head()",
"_____no_output_____"
]
],
[
[
"## Model with Informative prior\n\nCapture time points have been used as the informative prior information over pseudotime. Following arguments have been passed to initialize the model.\n\n<!--\n- __data__: _array-like, shape N x D_. Observed data, where N is the number of time points and D is the number of genes.\n- __latent_prior_mean__: _array-like, shape N_ x 1, _optional (default:_ __0__). > Mean of the prior distribution over pseudotime.\n- __latent_prior_var__: _array-like, shape N_ x 1, _optional (default:_ __1.__). Variance of the prior distribution over pseudotime.\n- __latent_mean__: _array-like, shape N_ x 1, _optional (default:_ __1.__). Initial mean values of the approximate posterior distribution over pseudotime.\n- __latent_var__: _array-like, shape N_ x 1, _optional (default:_ __1.__). Initial variance of the approximate posterior distribution over pseudotime.\n- __kernel:__ _optional (default: RBF kernel with lengthscale and variance set to 1.0)_. Covariance function to define the mapping from the latent space to the data space in Gaussian process prior. \n\n-->\n\n- __data__: _array-like, shape N x D_. Observed data, where N is the number of time points and D is the number of genes.\n\n\n- __latent_prior_mean__: _array-like, shape N_ x 1. Mean of the prior distribution over pseudotime.\n\n\n- __latent_prior_mean__: _array-like, shape N_ x 1. Mean of the prior distribution over pseudotime.\n\n\n- __latent_prior_var__: _array-like, shape N_ x 1. Variance of the prior distribution over pseudotime.\n\n\n- __latent_mean__: _array-like, shape N_ x 1. Initial mean values of the approximate posterior distribution over pseudotime.\n\n<!--\n- __latent_var__: _array-like, shape N_ x 1. Initial variance of the approximate posterior distribution over pseudotime.\n-->\n\n- __kernel__: Covariance function to define the mapping from the latent space to the data space in Gaussian process prior. Here we have used the standard periodic covariance function <a href=\"http://www.ics.uci.edu/~welling/teaching/KernelsICS273B/gpB.pdf\" terget=\"_blank\">(MacKay, 1998)</a>, to restrict the Gaussian Process (GP) prior to periodic functions only.\n\n\n- __predict__: _int_. The number of new points. The mean of the expression level and associated variance of these new data points will be predicted. ",
"_____no_output_____"
]
],
[
[
"np.random.seed(10)\nsigma_t = .5\nprior_mean = mData['prior'].values[:, None]\n\ninit_mean = mData['capture.orig'].values[:, None]\nX_mean = [init_mean[i, 0] + sigma_t * np.random.randn(1) for i in range(0, N)] # initialisation of latent_mean ",
"_____no_output_____"
],
[
"mp = GrandPrix.fit_model(data=Y.values, n_inducing_points = 20, latent_prior_mean=prior_mean, latent_prior_var=np.square(sigma_t),\n latent_mean=np.asarray(X_mean), kernel={'name':'Periodic', 'ls':5.0, 'var':1.0}, predict=100)",
"/Users/mqbpwsae/newInstall/GPflow_1_1_0/gpflow/expectations_quadrature.py:65: UserWarning: Quadrature is used to calculate the expectation. This means that an analytical implementations is not available for the given combination.\n warnings.warn(\"Quadrature is used to calculate the expectation. This means that \"\n"
],
[
"pseudotimes = mp[0]\nposterior_var = mp[1]\nmean = mp[2] # mean of predictive distribution\nvar = mp[3] # variance of predictive distribution",
"_____no_output_____"
],
[
"Xnew = np.linspace(min(pseudotimes), max(pseudotimes), 100)[:, None]",
"_____no_output_____"
]
],
[
[
"# Visualize the results\nThe expression profile of some interesting genes have been plotted against the estimated pseudotime. Each point corresponds to a particular gene expression in a cell. \n\nThe points are coloured based on cell cycle stages according to <a href=\"https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4102402/\" terget=\"_blank\" style=\"text-decoration:none;\">McDavid et al. (2014)</a>. The circular horizontal axis (where both first and last labels are G2/M) represents the periodicity realized by the method in pseudotime inference. \n\nThe solid black line is the posterior predicted mean of expression profiles while the grey ribbon depicts the 95% confidence interval. \n\nThe vertical dotted lines are the CycleBase peak times for the selected genes.\n\nTo see the expression profiles of a different set of genes a list containing gene names shound be passed to the function `plot_genes`.",
"_____no_output_____"
]
],
[
[
"selectedGenes = ['CDC6', 'MKI67', 'NUF2', 'PRR11', 'PTTG1', 'TPX2']",
"_____no_output_____"
],
[
"geneProfiles = pd.DataFrame({selectedGenes[i]: Y[selectedGenes[i]] for i in range(len(selectedGenes))})",
"_____no_output_____"
]
],
[
[
"## Binding gene names with predictive mean and variations",
"_____no_output_____"
]
],
[
[
"geneNames = Y.columns.values\nname = [_ for _ in geneNames]\nposterior_mean = pd.DataFrame(mean, columns=name)\nposterior_var = pd.DataFrame(var, columns=name)",
"_____no_output_____"
]
],
[
[
"## geneData description\nThe __\"McDavidgene.csv\"__ file contains gene specific information such as peak time, etc. for the top 56 differentially expressed genes. ",
"_____no_output_____"
]
],
[
[
"geneData = pd.read_csv('../data/McDavid/McDavid_gene.csv', index_col=0).T\n",
"_____no_output_____"
],
[
"geneData.head()",
"_____no_output_____"
],
[
"%matplotlib inline\nfrom utils import plot_genes\ncpt = mData['capture.orig'].values\nplot_genes(pseudotimes, geneProfiles, geneData, cpt, prediction=(Xnew, posterior_mean, posterior_var))",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
cb89c616f6d1c1455099a6b06af18f4f1f6727e1
| 44,993 |
ipynb
|
Jupyter Notebook
|
Applied Data Science with Python Specialzation/Introduction to Data Science in Python/Week+1.ipynb
|
lynnxlmiao/Coursera
|
8dc4073e29429dac14998689814388ee84435824
|
[
"MIT"
] | null | null | null |
Applied Data Science with Python Specialzation/Introduction to Data Science in Python/Week+1.ipynb
|
lynnxlmiao/Coursera
|
8dc4073e29429dac14998689814388ee84435824
|
[
"MIT"
] | null | null | null |
Applied Data Science with Python Specialzation/Introduction to Data Science in Python/Week+1.ipynb
|
lynnxlmiao/Coursera
|
8dc4073e29429dac14998689814388ee84435824
|
[
"MIT"
] | null | null | null | 18.599835 | 296 | 0.491432 |
[
[
[
"---\n\n_You are currently looking at **version 1.1** of this notebook. To download notebooks and datafiles, as well as get help on Jupyter notebooks in the Coursera platform, visit the [Jupyter Notebook FAQ](https://www.coursera.org/learn/python-data-analysis/resources/0dhYG) course resource._\n\n---",
"_____no_output_____"
],
[
"# The Python Programming Language: Functions",
"_____no_output_____"
],
[
"<br>\n`add_numbers` is a function that takes two numbers and adds them together.",
"_____no_output_____"
]
],
[
[
"def add_numbers(x, y):\n return x + y\n\nadd_numbers(1, 2)",
"_____no_output_____"
]
],
[
[
"<br>\n`add_numbers` updated to take an optional 3rd parameter. Using `print` allows printing of multiple expressions within a single cell.",
"_____no_output_____"
]
],
[
[
"def add_numbers(x,y,z=None):\n if (z==None):\n return x+y\n else:\n return x+y+z\n\nprint(add_numbers(1, 2))\nprint(add_numbers(1, 2, 3))",
"_____no_output_____"
]
],
[
[
"<br>\n`add_numbers` updated to take an optional flag parameter.",
"_____no_output_____"
]
],
[
[
"def add_numbers(x, y, z=None, flag=False):\n if (flag):\n print('Flag is true!')\n if (z==None):\n return x + y\n else:\n return x + y + z\n \nprint(add_numbers(1, 2, flag=True))",
"_____no_output_____"
]
],
[
[
"<br>\nAssign function `add_numbers` to variable `a`.",
"_____no_output_____"
]
],
[
[
"def add_numbers(x,y):\n return x+y\n\na = add_numbers\na(1,2)",
"_____no_output_____"
]
],
[
[
"<br>\n# The Python Programming Language: Types and Sequences",
"_____no_output_____"
],
[
"<br>\nUse `type` to return the object's type.",
"_____no_output_____"
]
],
[
[
"type('This is a string')",
"_____no_output_____"
],
[
"type(None)",
"_____no_output_____"
],
[
"type(1)",
"_____no_output_____"
],
[
"type(1.0)",
"_____no_output_____"
],
[
"type(add_numbers)",
"_____no_output_____"
]
],
[
[
"<br>\nTuples are an immutable data structure (cannot be altered).",
"_____no_output_____"
]
],
[
[
"x = (1, 'a', 2, 'b')\ntype(x)",
"_____no_output_____"
]
],
[
[
"<br>\nLists are a mutable data structure.",
"_____no_output_____"
]
],
[
[
"x = [1, 'a', 2, 'b']\ntype(x)",
"_____no_output_____"
]
],
[
[
"<br>\nUse `append` to append an object to a list.",
"_____no_output_____"
]
],
[
[
"x.append(3.3)\nprint(x)",
"_____no_output_____"
]
],
[
[
"<br>\nThis is an example of how to loop through each item in the list.",
"_____no_output_____"
]
],
[
[
"for item in x:\n print(item)",
"_____no_output_____"
]
],
[
[
"<br>\nOr using the indexing operator:",
"_____no_output_____"
]
],
[
[
"i=0\nwhile( i != len(x) ):\n print(x[i])\n i = i + 1",
"_____no_output_____"
]
],
[
[
"<br>\nUse `+` to concatenate lists.",
"_____no_output_____"
]
],
[
[
"[1,2] + [3,4]",
"_____no_output_____"
]
],
[
[
"<br>\nUse `*` to repeat lists.",
"_____no_output_____"
]
],
[
[
"[1]*3",
"_____no_output_____"
]
],
[
[
"<br>\nUse the `in` operator to check if something is inside a list.",
"_____no_output_____"
]
],
[
[
"1 in [1, 2, 3]",
"_____no_output_____"
]
],
[
[
"<br>\nNow let's look at strings. Use bracket notation to slice a string.",
"_____no_output_____"
]
],
[
[
"x = 'This is a string'\nprint(x[0]) #first character\nprint(x[0:1]) #first character, but we have explicitly set the end character\nprint(x[0:2]) #first two characters\n",
"_____no_output_____"
]
],
[
[
"<br>\nThis will return the last element of the string.",
"_____no_output_____"
]
],
[
[
"x[-1]",
"_____no_output_____"
]
],
[
[
"<br>\nThis will return the slice starting from the 4th element from the end and stopping before the 2nd element from the end.",
"_____no_output_____"
]
],
[
[
"x[-4:-2]",
"_____no_output_____"
]
],
[
[
"<br>\nThis is a slice from the beginning of the string and stopping before the 3rd element.",
"_____no_output_____"
]
],
[
[
"x[:3]",
"_____no_output_____"
]
],
[
[
"<br>\nAnd this is a slice starting from the 4th element of the string and going all the way to the end.",
"_____no_output_____"
]
],
[
[
"x[3:]",
"_____no_output_____"
],
[
"firstname = 'Christopher'\nlastname = 'Brooks'\n\nprint(firstname + ' ' + lastname)\nprint(firstname*3)\nprint('Chris' in firstname)\n",
"_____no_output_____"
]
],
[
[
"<br>\n`split` returns a list of all the words in a string, or a list split on a specific character.",
"_____no_output_____"
]
],
[
[
"firstname = 'Christopher Arthur Hansen Brooks'.split(' ')[0] # [0] selects the first element of the list\nlastname = 'Christopher Arthur Hansen Brooks'.split(' ')[-1] # [-1] selects the last element of the list\nprint(firstname)\nprint(lastname)",
"_____no_output_____"
]
],
[
[
"<br>\nMake sure you convert objects to strings before concatenating.",
"_____no_output_____"
]
],
[
[
"'Chris' + 2",
"_____no_output_____"
],
[
"'Chris' + str(2)",
"_____no_output_____"
]
],
[
[
"<br>\nDictionaries associate keys with values.",
"_____no_output_____"
]
],
[
[
"x = {'Christopher Brooks': '[email protected]', 'Bill Gates': '[email protected]'}\nx['Christopher Brooks'] # Retrieve a value by using the indexing operator\n",
"_____no_output_____"
],
[
"x['Kevyn Collins-Thompson'] = None\nx['Kevyn Collins-Thompson']",
"_____no_output_____"
]
],
[
[
"<br>\nIterate over all of the keys:",
"_____no_output_____"
]
],
[
[
"for name in x:\n print(x[name])",
"_____no_output_____"
]
],
[
[
"<br>\nIterate over all of the values:",
"_____no_output_____"
]
],
[
[
"for email in x.values():\n print(email)",
"_____no_output_____"
]
],
[
[
"<br>\nIterate over all of the items in the list:",
"_____no_output_____"
]
],
[
[
"for name, email in x.items():\n print(name)\n print(email)",
"_____no_output_____"
]
],
[
[
"<br>\nYou can unpack a sequence into different variables:",
"_____no_output_____"
]
],
[
[
"x = ('Christopher', 'Brooks', '[email protected]')\nfname, lname, email = x",
"_____no_output_____"
],
[
"fname",
"_____no_output_____"
],
[
"lname",
"_____no_output_____"
]
],
[
[
"<br>\nMake sure the number of values you are unpacking matches the number of variables being assigned.",
"_____no_output_____"
]
],
[
[
"x = ('Christopher', 'Brooks', '[email protected]', 'Ann Arbor')\nfname, lname, email = x",
"_____no_output_____"
]
],
[
[
"<br>\n# The Python Programming Language: More on Strings",
"_____no_output_____"
]
],
[
[
"print('Chris' + 2)",
"_____no_output_____"
],
[
"print('Chris' + str(2))",
"_____no_output_____"
]
],
[
[
"<br>\nPython has a built in method for convenient string formatting.",
"_____no_output_____"
]
],
[
[
"sales_record = {\n'price': 3.24,\n'num_items': 4,\n'person': 'Chris'}\n\nsales_statement = '{} bought {} item(s) at a price of {} each for a total of {}'\n\nprint(sales_statement.format(sales_record['person'],\n sales_record['num_items'],\n sales_record['price'],\n sales_record['num_items']*sales_record['price']))\n",
"_____no_output_____"
]
],
[
[
"<br>\n# Reading and Writing CSV files",
"_____no_output_____"
],
[
"<br>\nLet's import our datafile mpg.csv, which contains fuel economy data for 234 cars.\n\n* mpg : miles per gallon\n* class : car classification\n* cty : city mpg\n* cyl : # of cylinders\n* displ : engine displacement in liters\n* drv : f = front-wheel drive, r = rear wheel drive, 4 = 4wd\n* fl : fuel (e = ethanol E85, d = diesel, r = regular, p = premium, c = CNG)\n* hwy : highway mpg\n* manufacturer : automobile manufacturer\n* model : model of car\n* trans : type of transmission\n* year : model year",
"_____no_output_____"
]
],
[
[
"import csv\n\n%precision 2\n\nwith open('mpg.csv') as csvfile:\n mpg = list(csv.DictReader(csvfile))\n \nmpg[:3] # The first three dictionaries in our list.",
"_____no_output_____"
]
],
[
[
"<br>\n`csv.Dictreader` has read in each row of our csv file as a dictionary. `len` shows that our list is comprised of 234 dictionaries.",
"_____no_output_____"
]
],
[
[
"len(mpg)",
"_____no_output_____"
]
],
[
[
"<br>\n`keys` gives us the column names of our csv.",
"_____no_output_____"
]
],
[
[
"mpg[0].keys()",
"_____no_output_____"
]
],
[
[
"<br>\nThis is how to find the average cty fuel economy across all cars. All values in the dictionaries are strings, so we need to convert to float.",
"_____no_output_____"
]
],
[
[
"sum(float(d['cty']) for d in mpg) / len(mpg)",
"_____no_output_____"
]
],
[
[
"<br>\nSimilarly this is how to find the average hwy fuel economy across all cars.",
"_____no_output_____"
]
],
[
[
"sum(float(d['hwy']) for d in mpg) / len(mpg)",
"_____no_output_____"
]
],
[
[
"<br>\nUse `set` to return the unique values for the number of cylinders the cars in our dataset have.",
"_____no_output_____"
]
],
[
[
"cylinders = set(d['cyl'] for d in mpg)\ncylinders",
"_____no_output_____"
]
],
[
[
"<br>\nHere's a more complex example where we are grouping the cars by number of cylinder, and finding the average cty mpg for each group.",
"_____no_output_____"
]
],
[
[
"CtyMpgByCyl = []\n\nfor c in cylinders: # iterate over all the cylinder levels\n summpg = 0\n cyltypecount = 0\n for d in mpg: # iterate over all dictionaries\n if d['cyl'] == c: # if the cylinder level type matches,\n summpg += float(d['cty']) # add the cty mpg\n cyltypecount += 1 # increment the count\n CtyMpgByCyl.append((c, summpg / cyltypecount)) # append the tuple ('cylinder', 'avg mpg')\n\nCtyMpgByCyl.sort(key=lambda x: x[0])\nCtyMpgByCyl",
"_____no_output_____"
]
],
[
[
"<br>\nUse `set` to return the unique values for the class types in our dataset.",
"_____no_output_____"
]
],
[
[
"vehicleclass = set(d['class'] for d in mpg) # what are the class types\nvehicleclass",
"_____no_output_____"
]
],
[
[
"<br>\nAnd here's an example of how to find the average hwy mpg for each class of vehicle in our dataset.",
"_____no_output_____"
]
],
[
[
"HwyMpgByClass = []\n\nfor t in vehicleclass: # iterate over all the vehicle classes\n summpg = 0\n vclasscount = 0\n for d in mpg: # iterate over all dictionaries\n if d['class'] == t: # if the cylinder amount type matches,\n summpg += float(d['hwy']) # add the hwy mpg\n vclasscount += 1 # increment the count\n HwyMpgByClass.append((t, summpg / vclasscount)) # append the tuple ('class', 'avg mpg')\n\nHwyMpgByClass.sort(key=lambda x: x[1])\nHwyMpgByClass",
"_____no_output_____"
]
],
[
[
"<br>\n# The Python Programming Language: Dates and Times",
"_____no_output_____"
]
],
[
[
"import datetime as dt\nimport time as tm",
"_____no_output_____"
]
],
[
[
"<br>\n`time` returns the current time in seconds since the Epoch. (January 1st, 1970)",
"_____no_output_____"
]
],
[
[
"tm.time()",
"_____no_output_____"
]
],
[
[
"<br>\nConvert the timestamp to datetime.",
"_____no_output_____"
]
],
[
[
"dtnow = dt.datetime.fromtimestamp(tm.time())\ndtnow",
"_____no_output_____"
]
],
[
[
"<br>\nHandy datetime attributes:",
"_____no_output_____"
]
],
[
[
"dtnow.year, dtnow.month, dtnow.day, dtnow.hour, dtnow.minute, dtnow.second # get year, month, day, etc.from a datetime",
"_____no_output_____"
]
],
[
[
"<br>\n`timedelta` is a duration expressing the difference between two dates.",
"_____no_output_____"
]
],
[
[
"delta = dt.timedelta(days = 100) # create a timedelta of 100 days\ndelta",
"_____no_output_____"
]
],
[
[
"<br>\n`date.today` returns the current local date.",
"_____no_output_____"
]
],
[
[
"today = dt.date.today()",
"_____no_output_____"
],
[
"today - delta # the date 100 days ago",
"_____no_output_____"
],
[
"today > today-delta # compare dates",
"_____no_output_____"
]
],
[
[
"<br>\n# The Python Programming Language: Objects and map()",
"_____no_output_____"
],
[
"<br>\nAn example of a class in python:",
"_____no_output_____"
]
],
[
[
"class Person:\n department = 'School of Information' #a class variable\n\n def set_name(self, new_name): #a method\n self.name = new_name\n def set_location(self, new_location):\n self.location = new_location",
"_____no_output_____"
],
[
"person = Person()\nperson.set_name('Christopher Brooks')\nperson.set_location('Ann Arbor, MI, USA')\nprint('{} live in {} and works in the department {}'.format(person.name, person.location, person.department))",
"_____no_output_____"
]
],
[
[
"<br>\nHere's an example of mapping the `min` function between two lists.",
"_____no_output_____"
]
],
[
[
"store1 = [10.00, 11.00, 12.34, 2.34]\nstore2 = [9.00, 11.10, 12.34, 2.01]\ncheapest = map(min, store1, store2)\ncheapest",
"_____no_output_____"
]
],
[
[
"<br>\nNow let's iterate through the map object to see the values.",
"_____no_output_____"
]
],
[
[
"for item in cheapest:\n print(item)",
"_____no_output_____"
]
],
[
[
"<br>\n# The Python Programming Language: Lambda and List Comprehensions",
"_____no_output_____"
],
[
"<br>\nHere's an example of lambda that takes in three parameters and adds the first two.",
"_____no_output_____"
]
],
[
[
"my_function = lambda a, b, c : a + b",
"_____no_output_____"
],
[
"my_function(1, 2, 3)",
"_____no_output_____"
]
],
[
[
"<br>\nLet's iterate from 0 to 999 and return the even numbers.",
"_____no_output_____"
]
],
[
[
"my_list = []\nfor number in range(0, 1000):\n if number % 2 == 0:\n my_list.append(number)\nmy_list",
"_____no_output_____"
]
],
[
[
"<br>\nNow the same thing but with list comprehension.",
"_____no_output_____"
]
],
[
[
"my_list = [number for number in range(0,1000) if number % 2 == 0]\nmy_list",
"_____no_output_____"
]
],
[
[
"<br>\n# The Python Programming Language: Numerical Python (NumPy)",
"_____no_output_____"
]
],
[
[
"import numpy as np",
"_____no_output_____"
]
],
[
[
"<br>\n## Creating Arrays",
"_____no_output_____"
],
[
"Create a list and convert it to a numpy array",
"_____no_output_____"
]
],
[
[
"mylist = [1, 2, 3]\nx = np.array(mylist)\nx",
"_____no_output_____"
]
],
[
[
"<br>\nOr just pass in a list directly",
"_____no_output_____"
]
],
[
[
"y = np.array([4, 5, 6])\ny",
"_____no_output_____"
]
],
[
[
"<br>\nPass in a list of lists to create a multidimensional array.",
"_____no_output_____"
]
],
[
[
"m = np.array([[7, 8, 9], [10, 11, 12]])\nm",
"_____no_output_____"
]
],
[
[
"<br>\nUse the shape method to find the dimensions of the array. (rows, columns)",
"_____no_output_____"
]
],
[
[
"m.shape",
"_____no_output_____"
]
],
[
[
"<br>\n`arange` returns evenly spaced values within a given interval.",
"_____no_output_____"
]
],
[
[
"n = np.arange(0, 30, 2) # start at 0 count up by 2, stop before 30\nn",
"_____no_output_____"
]
],
[
[
"<br>\n`reshape` returns an array with the same data with a new shape.",
"_____no_output_____"
]
],
[
[
"n = n.reshape(3, 5) # reshape array to be 3x5\nn",
"_____no_output_____"
]
],
[
[
"<br>\n`linspace` returns evenly spaced numbers over a specified interval.",
"_____no_output_____"
]
],
[
[
"o = np.linspace(0, 4, 9) # return 9 evenly spaced values from 0 to 4\no",
"_____no_output_____"
]
],
[
[
"<br>\n`resize` changes the shape and size of array in-place.",
"_____no_output_____"
]
],
[
[
"o.resize(3, 3)\no",
"_____no_output_____"
]
],
[
[
"<br>\n`ones` returns a new array of given shape and type, filled with ones.",
"_____no_output_____"
]
],
[
[
"np.ones((3, 2))",
"_____no_output_____"
]
],
[
[
"<br>\n`zeros` returns a new array of given shape and type, filled with zeros.",
"_____no_output_____"
]
],
[
[
"np.zeros((2, 3))",
"_____no_output_____"
]
],
[
[
"<br>\n`eye` returns a 2-D array with ones on the diagonal and zeros elsewhere.",
"_____no_output_____"
]
],
[
[
"np.eye(3)",
"_____no_output_____"
]
],
[
[
"<br>\n`diag` extracts a diagonal or constructs a diagonal array.",
"_____no_output_____"
]
],
[
[
"np.diag(y)",
"_____no_output_____"
]
],
[
[
"<br>\nCreate an array using repeating list (or see `np.tile`)",
"_____no_output_____"
]
],
[
[
"np.array([1, 2, 3] * 3)",
"_____no_output_____"
]
],
[
[
"<br>\nRepeat elements of an array using `repeat`.",
"_____no_output_____"
]
],
[
[
"np.repeat([1, 2, 3], 3)",
"_____no_output_____"
]
],
[
[
"<br>\n#### Combining Arrays",
"_____no_output_____"
]
],
[
[
"p = np.ones([2, 3], int)\np",
"_____no_output_____"
]
],
[
[
"<br>\nUse `vstack` to stack arrays in sequence vertically (row wise).",
"_____no_output_____"
]
],
[
[
"np.vstack([p, 2*p])",
"_____no_output_____"
]
],
[
[
"<br>\nUse `hstack` to stack arrays in sequence horizontally (column wise).",
"_____no_output_____"
]
],
[
[
"np.hstack([p, 2*p])",
"_____no_output_____"
]
],
[
[
"<br>\n## Operations",
"_____no_output_____"
],
[
"Use `+`, `-`, `*`, `/` and `**` to perform element wise addition, subtraction, multiplication, division and power.",
"_____no_output_____"
]
],
[
[
"print(x + y) # elementwise addition [1 2 3] + [4 5 6] = [5 7 9]\nprint(x - y) # elementwise subtraction [1 2 3] - [4 5 6] = [-3 -3 -3]",
"_____no_output_____"
],
[
"print(x * y) # elementwise multiplication [1 2 3] * [4 5 6] = [4 10 18]\nprint(x / y) # elementwise divison [1 2 3] / [4 5 6] = [0.25 0.4 0.5]",
"_____no_output_____"
],
[
"print(x**2) # elementwise power [1 2 3] ^2 = [1 4 9]",
"_____no_output_____"
]
],
[
[
"<br>\n**Dot Product:** \n\n$ \\begin{bmatrix}x_1 \\ x_2 \\ x_3\\end{bmatrix}\n\\cdot\n\\begin{bmatrix}y_1 \\\\ y_2 \\\\ y_3\\end{bmatrix}\n= x_1 y_1 + x_2 y_2 + x_3 y_3$",
"_____no_output_____"
]
],
[
[
"x.dot(y) # dot product 1*4 + 2*5 + 3*6",
"_____no_output_____"
],
[
"z = np.array([y, y**2])\nprint(len(z)) # number of rows of array",
"_____no_output_____"
]
],
[
[
"<br>\nLet's look at transposing arrays. Transposing permutes the dimensions of the array.",
"_____no_output_____"
]
],
[
[
"z = np.array([y, y**2])\nz",
"_____no_output_____"
]
],
[
[
"<br>\nThe shape of array `z` is `(2,3)` before transposing.",
"_____no_output_____"
]
],
[
[
"z.shape",
"_____no_output_____"
]
],
[
[
"<br>\nUse `.T` to get the transpose.",
"_____no_output_____"
]
],
[
[
"z.T",
"_____no_output_____"
]
],
[
[
"<br>\nThe number of rows has swapped with the number of columns.",
"_____no_output_____"
]
],
[
[
"z.T.shape",
"_____no_output_____"
]
],
[
[
"<br>\nUse `.dtype` to see the data type of the elements in the array.",
"_____no_output_____"
]
],
[
[
"z.dtype",
"_____no_output_____"
]
],
[
[
"<br>\nUse `.astype` to cast to a specific type.",
"_____no_output_____"
]
],
[
[
"z = z.astype('f')\nz.dtype",
"_____no_output_____"
]
],
[
[
"<br>\n## Math Functions",
"_____no_output_____"
],
[
"Numpy has many built in math functions that can be performed on arrays.",
"_____no_output_____"
]
],
[
[
"a = np.array([-4, -2, 1, 3, 5])",
"_____no_output_____"
],
[
"a.sum()",
"_____no_output_____"
],
[
"a.max()",
"_____no_output_____"
],
[
"a.min()",
"_____no_output_____"
],
[
"a.mean()",
"_____no_output_____"
],
[
"a.std()",
"_____no_output_____"
]
],
[
[
"<br>\n`argmax` and `argmin` return the index of the maximum and minimum values in the array.",
"_____no_output_____"
]
],
[
[
"a.argmax()",
"_____no_output_____"
],
[
"a.argmin()",
"_____no_output_____"
]
],
[
[
"<br>\n## Indexing / Slicing",
"_____no_output_____"
]
],
[
[
"s = np.arange(13)**2\ns",
"_____no_output_____"
]
],
[
[
"<br>\nUse bracket notation to get the value at a specific index. Remember that indexing starts at 0.",
"_____no_output_____"
]
],
[
[
"s[0], s[4], s[-1]",
"_____no_output_____"
]
],
[
[
"<br>\nUse `:` to indicate a range. `array[start:stop]`\n\n\nLeaving `start` or `stop` empty will default to the beginning/end of the array.",
"_____no_output_____"
]
],
[
[
"s[1:5]",
"_____no_output_____"
]
],
[
[
"<br>\nUse negatives to count from the back.",
"_____no_output_____"
]
],
[
[
"s[-4:]",
"_____no_output_____"
]
],
[
[
"<br>\nA second `:` can be used to indicate step-size. `array[start:stop:stepsize]`\n\nHere we are starting 5th element from the end, and counting backwards by 2 until the beginning of the array is reached.",
"_____no_output_____"
]
],
[
[
"s[-5::-2]",
"_____no_output_____"
]
],
[
[
"<br>\nLet's look at a multidimensional array.",
"_____no_output_____"
]
],
[
[
"r = np.arange(36)\nr.resize((6, 6))\nr",
"_____no_output_____"
]
],
[
[
"<br>\nUse bracket notation to slice: `array[row, column]`",
"_____no_output_____"
]
],
[
[
"r[2, 2]",
"_____no_output_____"
]
],
[
[
"<br>\nAnd use : to select a range of rows or columns",
"_____no_output_____"
]
],
[
[
"r[3, 3:6]",
"_____no_output_____"
]
],
[
[
"<br>\nHere we are selecting all the rows up to (and not including) row 2, and all the columns up to (and not including) the last column.",
"_____no_output_____"
]
],
[
[
"r[:2, :-1]",
"_____no_output_____"
]
],
[
[
"<br>\nThis is a slice of the last row, and only every other element.",
"_____no_output_____"
]
],
[
[
"r[-1, ::2]",
"_____no_output_____"
]
],
[
[
"<br>\nWe can also perform conditional indexing. Here we are selecting values from the array that are greater than 30. (Also see `np.where`)",
"_____no_output_____"
]
],
[
[
"r[r > 30]",
"_____no_output_____"
]
],
[
[
"<br>\nHere we are assigning all values in the array that are greater than 30 to the value of 30.",
"_____no_output_____"
]
],
[
[
"r[r > 30] = 30\nr",
"_____no_output_____"
]
],
[
[
"<br>\n## Copying Data",
"_____no_output_____"
],
[
"Be careful with copying and modifying arrays in NumPy!\n\n\n`r2` is a slice of `r`",
"_____no_output_____"
]
],
[
[
"r2 = r[:3,:3]\nr2",
"_____no_output_____"
]
],
[
[
"<br>\nSet this slice's values to zero ([:] selects the entire array)",
"_____no_output_____"
]
],
[
[
"r2[:] = 0\nr2",
"_____no_output_____"
]
],
[
[
"<br>\n`r` has also been changed!",
"_____no_output_____"
]
],
[
[
"r",
"_____no_output_____"
]
],
[
[
"<br>\nTo avoid this, use `r.copy` to create a copy that will not affect the original array",
"_____no_output_____"
]
],
[
[
"r_copy = r.copy()\nr_copy",
"_____no_output_____"
]
],
[
[
"<br>\nNow when r_copy is modified, r will not be changed.",
"_____no_output_____"
]
],
[
[
"r_copy[:] = 10\nprint(r_copy, '\\n')\nprint(r)",
"_____no_output_____"
]
],
[
[
"<br>\n### Iterating Over Arrays",
"_____no_output_____"
],
[
"Let's create a new 4 by 3 array of random numbers 0-9.",
"_____no_output_____"
]
],
[
[
"test = np.random.randint(0, 10, (4,3))\ntest",
"_____no_output_____"
]
],
[
[
"<br>\nIterate by row:",
"_____no_output_____"
]
],
[
[
"for row in test:\n print(row)",
"_____no_output_____"
]
],
[
[
"<br>\nIterate by index:",
"_____no_output_____"
]
],
[
[
"for i in range(len(test)):\n print(test[i])",
"_____no_output_____"
]
],
[
[
"<br>\nIterate by row and index:",
"_____no_output_____"
]
],
[
[
"for i, row in enumerate(test):\n print('row', i, 'is', row)",
"_____no_output_____"
]
],
[
[
"<br>\nUse `zip` to iterate over multiple iterables.",
"_____no_output_____"
]
],
[
[
"test2 = test**2\ntest2",
"_____no_output_____"
],
[
"for i, j in zip(test, test2):\n print(i,'+',j,'=',i+j)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
cb89cb8ad9667feb97d3dc2c12c4c9c2603cafd5
| 19,910 |
ipynb
|
Jupyter Notebook
|
remote-technical_challenge/test_bench.ipynb
|
marwin-ko/projects
|
97a857bf05d9ac95f7d73c9f70fdff20b4793c5a
|
[
"MIT"
] | null | null | null |
remote-technical_challenge/test_bench.ipynb
|
marwin-ko/projects
|
97a857bf05d9ac95f7d73c9f70fdff20b4793c5a
|
[
"MIT"
] | null | null | null |
remote-technical_challenge/test_bench.ipynb
|
marwin-ko/projects
|
97a857bf05d9ac95f7d73c9f70fdff20b4793c5a
|
[
"MIT"
] | null | null | null | 28.730159 | 199 | 0.453742 |
[
[
[
"from elasticsearch import Elasticsearch\nfrom random import randint",
"_____no_output_____"
],
[
"es = Elasticsearch([{'host': 'localhost', 'port': 9200}], http_auth=('xxxxxxx', 'xxxxxxxxx'))",
"_____no_output_____"
],
[
"# ~ 6,000,000 companies \n# ~ 4,000 colleges\nratio ==> 1500 companies per one college",
"_____no_output_____"
],
[
"6000000/4000",
"_____no_output_____"
],
[
"doc = {'email':'name_'+str(i)+'@email.com',\n 'number': randint(1,100),\n 'company': 'company_'+str(randint(1,100)),\n 'school': 'school_'+str(randint(1,10))}\nres = es.index(index=\"test-index\", doc_type='tweet', body=doc)",
"_____no_output_____"
],
[
"es.count(index='users')['count']",
"_____no_output_____"
]
],
[
[
"## POST (adding new user)",
"_____no_output_____"
]
],
[
[
"def add_user():\n # Can also implement request.json\n print('Please fill out the following information.')\n email = input('email: ')\n number = input('number: ')\n company = input('company: ')\n school = input('school: ')\n doc = {'email': email,\n 'number': int(number),\n 'company': company,\n 'school': school}\n#es.index(index='users',doc_type='people',id=es.count(index='users')['count']+1,body=doc)\nadd_user()",
"Please fill out the following information.\nemail: fds\nnumber: 129394\ncompany: dklfdj\nschool: dfsd\n{'number': 129394, 'company': 'dklfdj', 'school': 'dfsd', 'email': 'fds'}\n"
]
],
[
[
"## DELETE (delete existing user)",
"_____no_output_____"
]
],
[
[
"def delete_user():\n # Delete user based off ID in the users index\n print('###########################################')\n print('################# WARNING #################')\n print('###########################################')\n print('You are about to delete a user from Remote.')\n print(' ')\n answer = input('Do you wish to continue? Y/N ')\n if answer.upper() == 'Y':\n user_id = input('Enter user ID: ')\n# es.delete(index='users',doc_type='people',id=int(user_id))\n print('You have removed %s from Remote.com. :(' % user_id)\n else:\n pass\ndelete_user()",
"###########################################\n################# WARNING #################\n###########################################\nYou are about to delete a user from Remote.\n \nDo you wish to continue? Y/N y\nEnter user ID: 24039\nYou have removed 24039 from Remote.com. :(\n"
]
],
[
[
"## PUT (update user)",
"_____no_output_____"
]
],
[
[
"def update_user():\n print('You are about to update a user\\'s information.')\n print(' ')\n answer = input('Do you wish to continue? Y/N ')\n if answer.upper() == 'Y':\n user_id = input('Enter user id: ')\n print('Please update the following information.')\n email = input('email: ')\n number = input('number: ')\n company = input('company: ')\n school = input('school: ')\n doc = {'email': email,\n 'number': int(number),\n 'company': company,\n 'school': school}\n# es.index(index, doc_type, body, id=user_id)\n# return jsonify({'Update': True})\n else:\n pass\n# return jsonify({'Update': False})\nupdate_user()",
"You are about to update a user's information.\n \nDo you wish to continue? Y/N n\n"
]
],
[
[
"## GET (view user info)",
"_____no_output_____"
]
],
[
[
"def get_user_info():\n user_id = input('Enter user id: ')\n return jsonify(es.search(index='users',body={'query': {'match': {'_id':user_id}}})['hits']['hits'][0]['_source'])",
"_____no_output_____"
],
[
"es.search(index='users',body={'query': {'match': {'_id':'2600'}}})['hits']['hits'][0]['_source']",
"_____no_output_____"
],
[
"user = es.search(index='users',body={'query': {'match': {'_id':'2600'}}})['hits']['hits'][0]\nprint(user)",
"{'_score': 1.0, '_source': {'number': 986, 'email': '[email protected]', 'school': 'school_287', 'company': 'company_5836'}, '_id': '2600', '_type': 'people', '_index': 'users'}\n"
],
[
"es.search(index='users',body={'query': {'match': {'company':'company_100'}}})",
"_____no_output_____"
]
],
[
[
"## GET (user's 1st connection)",
"_____no_output_____"
]
],
[
[
"def user_1st_degree():\n user_id = input('Enter user id: ')\n user_info = es.search(index='users',body={'query': {'match': {'_id':user_id}}})['hits']['hits'][0]['_source']\n coworkers = es.search(index='users',body={'query': {'match': {'company':user_info['company']}}})['hits']['hits']\n coworker_ids = [coworker['_id'] for coworker in coworkers]\n classmates = es.search(index='users',body={'query': {'match': {'school':user_info['school']}}})['hits']['hits']\n classmate_ids = [classmate['_id'] for classmate in classmates]\n first_deg_conns = list(set(coworker_ids+classmate_ids))\n return first_deg_conns\nuser_1st_degree()",
"Enter user id: 100\n"
],
[
"coworkers = es.search(index='users',body={'query': {'match': {'company':'company_7092'}}})['hits']['hits']\ncoworker_ids = [coworker['_id'] for coworker in coworkers]\n# print(coworker_ids)\n\nclassmates = es.search(index='users',body={'query': {'match': {'school':'school_303'}}})['hits']['hits']\nclassmate_ids = [classmate['_id'] for classmate in classmates]\n# print(classmate_ids)\n\ntotal = classmate_ids+ coworker_ids\nprint(list(set(total)))",
"['46958', '54872', '61922', '920211', '37042', '46507', '70260', '354142', '649661', '663234', '834550', '10577', '40005', '332767', '475258', '1108020', '6257', '50640', '40318', '942818']\n"
],
[
"es.search(index='users',body={'query': {'match': {'company':'company_7092'}}})",
"_____no_output_____"
]
],
[
[
"## GET (user's 2nd degree connections)\n\nFrom your 1st degree connections, get their 1st degree connections...this will yield your 2nd degree connections",
"_____no_output_____"
]
],
[
[
"def user_1st_degreex(user_id):\n user_info = es.search(index='users',body={'query': {'match': {'_id':user_id}}})['hits']['hits'][0]['_source']\n coworkers = es.search(index='users',body={'query': {'match': {'company':user_info['company']}}})['hits']['hits']\n coworker_ids = [coworker['_id'] for coworker in coworkers]\n classmates = es.search(index='users',body={'query': {'match': {'school':user_info['school']}}})['hits']['hits']\n classmate_ids = [classmate['_id'] for classmate in classmates]\n first_conns = list(set(coworker_ids+classmate_ids))\n return first_conns\n\ndef user_2nd_degree():\n user_id = input('Enter user id: ')\n first_conns = user_1st_degree(user_id)\n second_conns = []\n for conn in first_conns:\n second_conns.extend(user_1st_degree(conn))\n unique_second_conns = list(set(second_conns))\n return unique_second_conns\n\nuser_2nd_degree()",
"Enter user id: 100\n"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb89cc1ccb67bb274b41f119514ea011d26929a1
| 435 |
ipynb
|
Jupyter Notebook
|
notebook.ipynb
|
twhyntie/image-heatmap
|
e953aced30f49371fd93bacb9951170b5424c408
|
[
"MIT"
] | null | null | null |
notebook.ipynb
|
twhyntie/image-heatmap
|
e953aced30f49371fd93bacb9951170b5424c408
|
[
"MIT"
] | null | null | null |
notebook.ipynb
|
twhyntie/image-heatmap
|
e953aced30f49371fd93bacb9951170b5424c408
|
[
"MIT"
] | null | null | null | 16.111111 | 67 | 0.471264 |
[
[
[
"empty"
]
]
] |
[
"empty"
] |
[
[
"empty"
]
] |
cb89dc93f763dbe92acbccecf4800da41cc44301
| 3,703 |
ipynb
|
Jupyter Notebook
|
cad/cradle/MomentofInertia.ipynb
|
psas/lv3.0-airframe
|
cabca568560ea2410fd4d29c69f59f0df3a2acd8
|
[
"BSD-2-Clause"
] | 17 |
2016-09-13T16:05:18.000Z
|
2022-03-25T19:13:46.000Z
|
cad/cradle/MomentofInertia.ipynb
|
psas/lv3.0-airframe
|
cabca568560ea2410fd4d29c69f59f0df3a2acd8
|
[
"BSD-2-Clause"
] | 30 |
2016-09-21T05:09:13.000Z
|
2018-07-17T23:07:25.000Z
|
cad/cradle/MomentofInertia.ipynb
|
psas/lv3.0-airframe
|
cabca568560ea2410fd4d29c69f59f0df3a2acd8
|
[
"BSD-2-Clause"
] | 21 |
2016-08-20T02:46:28.000Z
|
2022-03-25T19:14:22.000Z
| 29.388889 | 105 | 0.506346 |
[
[
[
"## Cradle Values and Analysis",
"_____no_output_____"
]
],
[
[
"import math as m\nimport numpy as np\n\n# Cradle Analysis, Quick\n# Geometry\nTdo = 0.5 # Tube, Outer Diameter [in.]\nTdi = 0.38 # Tube, Inner Diameter [in.]\nPw = 2 # Plate Width [in.]\nPt = 0.1 # Plate Thickness [in.]\nPdi = 5.4 # Plate, Inner Y Dist. from X=0 [in.]\nPdo = Pdi+Pt # Plate, Outer Y Dist. from X=0 [in.]\nTdc = 0.5*(4.58+0.25) # Tube, center to Y=0 Dist. [in.]\n\n# Material\nE = 400e3 # Elastic Modulus [psi]\n\n# Moment of Inertia (MoI)\n# Tube:\nTube_Ixo = (m.pi/4)*(Tdo/2)**4 # Outer Tube MoI (Relative to Tube Center)\nTube_Ixi = (m.pi/4)*(Tdi/2)**4 # Inner Tube MoI (Relative to Tube Center)\nTube_Ix = Tube_Ixo-Tube_Ixi # Tubing MoI [in.^4]\n# Plate:\nPlate_Ix = Pw*Pt*(Pdi+0.5*Pt)**2 + (Pw*Pt**3)/12 # Plate MoI, x [in.^4]\nPlate_Iy = (Pt*Pw**3)/12 # Plate MoI, y [in.^4]\n# Parallel Axis Theorem:\nTube_Ix_Corrected = Tube_Ix+(Tdc**2)*(m.pi*(0.5*Tdo)**2 - m.pi*(0.5*Tdi)**2) # Tube MoI [in.^4] \nTotal_Ix = 2*Tube_Ix_Corrected + 2*Plate_Ix # Total MoI [in.^4]\n\nprint('Moment of Inertia about x-axis')\nprint(' Plate: {0:.4f} [in.^4]'.format(Plate_Ix))\nprint(' Tube: {0:.4f} [in.^4]'.format(Tube_Ix_Corrected))\nprint(' Total: {0:.3f} [in.^4]'.format(Total_Ix))\n\n# SolidWorksI = np.array[125.99, 17.36, 123.99] # Solidworks MoI, [Ix, Iy, Iz]\n\n# Bolt Shear\n# Material Properties\nE = 28000e3 # Elastic Modulus, Stainless [psi]\nS = 12500e3 # Shear Modulus, Stainless [psi]\n\n# Geometry\nRbolt = 0.112/2 # Bolt Radius [in.]\nBoltA = m.pi*Rbolt**2 # Bolt x-section Area [in.^2]\nN = 12 # Quantity of Bolts\n\n# G-Forces\ng = 9.81 # Gravitational Acceleration [m/s^2]\na = 15*g # Acceleration Expected [m/s^2]\nmass = 1.25 # Mass [kg]\nFgNewton = mass*a # G-Force [N]\nFg = FgNewton*0.2248 # G-Force [lbf]\nShear = Fg/(N*BoltA) # Shear Stress, per bolt [psi]\n\nprint('')\nprint('Shear Stresses:')\nprint(' G-Forces: {0:.3f} [N]'.format(Fg))\nprint(' Total Bolt Area: {0:.4f} [in.^2]'.format(BoltA))\nprint(' Per Bolt: {0:.2f} [psi]'.format(Shear))",
"Moment of Inertia about x-axis\n Plate: 5.9407 [in.^4]\n Tube: 0.4858 [in.^4]\n Total: 12.853 [in.^4]\n\nShear Stresses:\n G-Forces: 41.349 [N]\n Total Bolt Area: 0.0099 [in.^2]\n Per Bolt: 349.75 [psi]\n"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
]
] |
cb89e1c8a5560a6e20d084094a1efc80f447e645
| 201,470 |
ipynb
|
Jupyter Notebook
|
ch02.ipynb
|
Chrisssq/machinelearning
|
7e6c95014e0548cfa7d3c0b2a2f6c1eea1a96e1f
|
[
"MIT"
] | null | null | null |
ch02.ipynb
|
Chrisssq/machinelearning
|
7e6c95014e0548cfa7d3c0b2a2f6c1eea1a96e1f
|
[
"MIT"
] | null | null | null |
ch02.ipynb
|
Chrisssq/machinelearning
|
7e6c95014e0548cfa7d3c0b2a2f6c1eea1a96e1f
|
[
"MIT"
] | null | null | null | 366.976321 | 40,940 | 0.918832 |
[
[
[
"import numpy as np\nclass Perception(object):\n '''\n Created on May 14th, 2017\n Perception: A very simple model for binary classification\n @author: Qi Gong\n '''\n def __init__(self, eta = 0.01, n_iter = 10):\n self.eta = eta\n self.n_iter = n_iter\n \n def fit(self, X, y):\n '''\n X : matrix, shape = [n_samples, n_features]. Traning data\n y : vector. label\n '''\n self.w_ = np.zeros(1 + X.shape[1])\n self.errors_ = []\n \n for _ in range(self.n_iter):\n errors = 0\n for xi, target in zip(X, y):\n update = self.eta * (target - self.predict(xi))\n self.w_[1:] += update * xi\n self.w_[0] += update\n errors += int(update != 0)\n self.errors_.append(errors)\n return self\n \n def net_input(self, X):\n '''\n Calculate net_input\n input: X. X is training data and a matrix.\n '''\n return np.dot(X, self.w_[1:]) + self.w_[0]\n \n def predict(self, X):\n '''\n Predict the label by input X\n input : X. X is a matrix.\n '''\n return np.where(self.net_input(X) > 0.0, 1, -1)",
"_____no_output_____"
],
[
"import pandas as pd\ndf = pd.read_csv('https://archive.ics.uci.edu/ml/machine-learning-databases/iris/iris.data', header=None)\ndf.tail()",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt\n\ny = df.iloc[0:100, 4].values\ny = np.where(y == 'Iris-setosa', 1, -1)\nX = df.iloc[0:100, [0, 2]].values\nplt.scatter(X[:50, 0], X[:50, 1], color = 'red', marker = 'o', label = 'setosa')\nplt.scatter(X[50:100, 0], X[50:100, 1], color = 'blue', marker = 'x', label = 'viginica')\nplt.xlabel('petal length')\nplt.ylabel('sepal length')\nplt.legend(loc = 'upper left')\nplt.show()",
"_____no_output_____"
],
[
"ppn = Perception(eta = 0.1, n_iter = 10)\nppn.fit(X, y)\nplt.plot(range(1, len(ppn.errors_) + 1), ppn.errors_, marker='o')\nplt.xlabel('Epoches')\nplt.ylabel('Number of misclassifications')\nplt.show()",
"_____no_output_____"
],
[
"from matplotlib.colors import ListedColormap\n\ndef plot_decision_regions(X, y, classifier, resolution = 0.02):\n '''\n visualize the decision boundaries for 2D datasheet\n '''\n markers = ('s', 'x', 'o', '^', 'v')\n colors = ('red', 'blue', 'lightgreen','gray','cyan')\n \n cmap = ListedColormap(colors[:len(np.unique(y))])\n \n #plot the decision surface\n x1_min, x1_max = X[:, 0].min() - 1, X[:, 0].max() + 1\n x2_min, x2_max = X[:, 1].min() - 1, X[:, 1].max() + 1\n xx1, xx2 = np.meshgrid(np.arange(x1_min, x1_max, resolution), \n np.arange(x2_min, x2_max, resolution))\n Z = classifier.predict(np.array([xx1.ravel(), xx2.ravel()]).T)\n Z = Z.reshape(xx1.shape)\n plt.contourf(xx1, xx2, Z, alpha = 0.4, cmap = cmap)\n plt.xlim(xx1.min(), xx1.max())\n plt.ylim(xx2.min(), xx2.max())\n \n #plot class samples\n for idx, cl in enumerate(np.unique(y)):\n plt.scatter(x = X[y == cl, 0], y = X[y == cl, 1], alpha = 0.8, c = cmap(idx),\n marker = markers[idx], label = cl)\n \nplot_decision_regions(X, y, classifier=ppn)\nplt.xlabel('sepal length [cm]')\nplt.ylabel('petal length [cm]')\nplt.legend(loc = 'upper left')\nplt.show()\n ",
"_____no_output_____"
]
],
[
[
"# Implementing an Adaptive Linear Neuron in Python",
"_____no_output_____"
]
],
[
[
"class AdalineGD(object):\n def __init__(self, eta = 0.01, n_iter = 50):\n self.eta = eta\n self.n_iter = n_iter\n \n def fit(self, X, y):\n self.w_ = np.zeros(1+X.shape[1])\n self.errors_ = []\n for i in range(self.n_iter):\n output = self.net_input(X)\n error = y - output\n self.w_[1:] += self.eta * X.T.dot(error)\n self.w_[0] += self.eta * error.sum()\n cost = (error**2).sum()/2.0\n self.errors_.append(cost)\n return self\n \n def net_input(self, X):\n return np.dot(X, self.w_[1:]) + self.w_[0]\n \n def activation(self, X):\n return self.net_input(X)\n \n def predict(self, X):\n return np.where(self.activation(X) > 0.0, 1, -1)\n ",
"_____no_output_____"
],
[
"fig, ax = plt.subplots(nrows = 1, ncols = 2, figsize=(8, 4))\nada1 = AdalineGD(n_iter = 10, eta = 0.01).fit(X, y)\nax[0].plot(range(1, len(ada1.errors_) + 1), np.log10(ada1.errors_), marker = 'o')\nax[0].set_xlabel('Epochs')\nax[0].set_ylabel('log(Sum-squared-error')\nax[0].set_title('AdalineGD Learning rate = 0.01')\nada2 = AdalineGD(n_iter = 10, eta = 0.0001).fit(X, y)\nax[1].plot(range(1, len(ada2.errors_) + 1), np.log10(ada2.errors_), marker = 'o')\nax[1].set_xlabel('Epochs')\nax[1].set_ylabel('log(Sum-squared-error)')\nax[1].set_title('AdalineGD learning rate = 0.0001')\nplt.show()",
"_____no_output_____"
]
],
[
[
"# Standardize the data",
"_____no_output_____"
]
],
[
[
"X_std = np.copy(X)\nX_std[:, 0] = (X[:, 0] - X[:, 0].mean()) / X[:, 0].std()\nX_std[:, 1] = (X[:, 1] - X[:, 1].mean()) / X[:, 1].std()\n\nada = AdalineGD(n_iter = 15, eta = 0.01)\nada.fit(X_std, y)\nplot_decision_regions(X_std, y, classifier=ada)\nplt.title('Adaline-Gradient Descent')\nplt.xlabel('sepal length [standardized]')\nplt.ylabel('petal length [standardized]')\nplt.show()\n\nplt.plot(range(1, len(ada.errors_) + 1), ada.errors_, marker = 'o')\nplt.xlabel('Epochs')\nplt.ylabel('Sum-squared-error')\nplt.show()\n",
"_____no_output_____"
]
],
[
[
"# Adaptive linear neuron Stochastic Gradient Descent",
"_____no_output_____"
]
],
[
[
"from numpy.random import seed\nclass AdalineSGD(object):\n def __init__(self, eta = 0.01, n_iter = 50, shuffle = True, random_state = None):\n self.eta = eta\n self.n_iter = n_iter\n self.w_initialized = False\n self.shuffle = shuffle\n if random_state:\n seed(random_state)\n \n def fit(self, X, y):\n self.__initialize_weights(X.shape[1])\n self.errors_ = []\n for i in range(self.n_iter):\n if self.shuffle:\n X, y = self.__shuffle(X, y)\n \n cost = []\n for xi, target in zip(X, y):\n cost.append(self.__update_weights(xi, target))\n avg_cost = sum(cost) / len(y)\n self.errors_.append(avg_cost)\n return self\n \n def partial_fit(self, X, y):\n if not self.w_initialized:\n self._initialize_weights(X.shape[1])\n if y.ravel().shape[0] > 1:\n for xi, target in zip(X, y):\n self.__update_weights(xi, target)\n else:\n self.__update_weights(X, y)\n return self\n \n def __initialize_weights(self, m):\n self.w_ = np.zeros(1+m)\n self.w_initialized = True\n \n def __shuffle(self, X, y):\n r = np.random.permutation(len(y))\n return X[r], y[r]\n \n def __update_weights(self, x, y):\n output = self.net_input(x)\n error = y - output\n self.w_[1:] += self.eta * x.dot(error)\n self.w_[0] += self.eta * error\n \n cost = 0.5 * error ** 2\n return cost\n \n def net_input(self, X):\n return np.dot(X, self.w_[1:]) + self.w_[0]\n \n def activation(self, X):\n return self.net_input(X)\n \n def predict(self, X):\n return np.where(self.activation(X) > 0.0, 1, -1)\n ",
"_____no_output_____"
]
],
[
[
"# Test",
"_____no_output_____"
]
],
[
[
"adasgd = AdalineSGD(n_iter = 15, eta = 0.01, random_state = 1)\nadasgd.fit(X_std, y)\nplot_decision_regions(X_std, y, classifier=adasgd)\nplt.title('Adaline-Gradient Stochastic Descent')\nplt.xlabel('sepal length [standardized]')\nplt.ylabel('petal length [standardized]')\nplt.legend(loc = 'upper left')\nplt.show()\n\nplt.plot(range(1, len(adasgd.errors_) + 1), adasgd.errors_, marker = 'o')\nplt.xlabel('Epochs')\nplt.ylabel('Sum-squared-error')\nplt.show()",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb89f7ef8d85c103e72e0e7c51e0f1c429b11261
| 15,368 |
ipynb
|
Jupyter Notebook
|
docs/tutorial/sequences.ipynb
|
klavinslab/coral
|
17f59591211562a59a051f474cd6cecba4829df9
|
[
"MIT"
] | 34 |
2015-12-26T22:13:51.000Z
|
2021-11-17T11:46:37.000Z
|
docs/tutorial/sequences.ipynb
|
klavinslab/coral
|
17f59591211562a59a051f474cd6cecba4829df9
|
[
"MIT"
] | 13 |
2015-09-11T23:27:51.000Z
|
2018-06-25T20:44:28.000Z
|
docs/tutorial/sequences.ipynb
|
klavinslab/coral
|
17f59591211562a59a051f474cd6cecba4829df9
|
[
"MIT"
] | 14 |
2015-10-08T17:08:48.000Z
|
2022-02-22T04:25:54.000Z
| 30.192534 | 1,031 | 0.615044 |
[
[
[
"# Sequences",
"_____no_output_____"
],
[
"## `sequence.DNA`\n\n`coral.DNA` is the core data structure of `coral`. If you are already familiar with core python data structures, it mostly acts like a container similar to lists or strings, but also provides further object-oriented methods for DNA-specific tasks, like reverse complementation. Most design functions in `coral` return a `coral.DNA` object or something that contains a `coral.DNA` object (like `coral.Primer`). In addition, there are related `coral.RNA` and `coral.Peptide` objects for representing RNA and peptide sequences and methods for converting between them.\n\nTo get started with `coral.DNA`, import `coral`:",
"_____no_output_____"
]
],
[
[
"import coral as cor",
"_____no_output_____"
]
],
[
[
"### Your first sequence\n\nLet's jump right into things. Let's make a sequence that's the first 30 bases of gfp from *A. victoria*. To initialize a sequence, you feed it a string of DNA characters.",
"_____no_output_____"
]
],
[
[
"example_dna = cor.DNA('atgagtaaaggagaagaacttttcactgga')\ndisplay(example_dna)",
"_____no_output_____"
]
],
[
[
"A few things just happened behind the scenes. First, the input was checked to make sure it's DNA (A, T, G, and C). For now, it supports only unambiguous letters - no N, Y, R, etc. Second, the internal representation is converted to an uppercase string - this way, DNA is displayed uniformly and functional elements (like annealing and overhang regions of primers) can be delineated using case. If you input a non-DNA sequence, a `ValueError` is raised.",
"_____no_output_____"
],
[
"For the most part, a `sequence.DNA` instance acts like a python container and many string-like operations work.",
"_____no_output_____"
]
],
[
[
"# Extract the first three bases\ndisplay(example_dna[0:3])",
"_____no_output_____"
],
[
"# Extract the last seven bases\ndisplay(example_dna[-7:])",
"_____no_output_____"
],
[
"# Reverse a sequence\ndisplay(example_dna[::-1])",
"_____no_output_____"
],
[
"# Grab every other base starting at index 0\ndisplay(example_dna[::2])",
"_____no_output_____"
],
[
"# Is the sequence 'AT' in our sequence? How about 'AC'?\nprint \"'AT' is in our sequence: {}.\".format(\"AT\" in example_dna)\nprint \"'ATT' is in our sequence: {}.\".format(\"ATT\" in example_dna)",
"'AT' is in our sequence: True.\n'ATT' is in our sequence: False.\n"
]
],
[
[
"Several other common special methods and operators are defined for sequences - you can concatenate DNA (so long as it isn't circular) using `+`, repeat linear sequences using `*` with an integer, check for equality with `==` and `!=` (note: features, not just sequences, must be identical), check the length with `len(dna_object)`, etc.",
"_____no_output_____"
],
[
"### Simple sequences - methods\n\nIn addition to slicing, `sequence.DNA` provides methods for common molecular manipulations. For example, reverse complementing a sequence is a single call:",
"_____no_output_____"
]
],
[
[
"example_dna.reverse_complement()",
"_____no_output_____"
]
],
[
[
"An extremely important method is the `.copy()` method. It may seem redundant to have an entire function for copying a sequence - why not just assign a `sequence.DNA` object to a new variable? As in most high-level languages, python does not actually copy entire objects in memory when assignment happens - it just adds another reference to the same data. The short of it is that the very common operation of generating a lot of new variants to a sequence, or copying a sequence, requires the use of a `.copy()` method. For example, if you want to generate a new list of variants where an 'a' is substituted one at a time at each part of the sequence, using `.copy()` returns the correct result (the first example) while directly accessing example_dna has horrible consequences (the edits build up, as they all modify the same piece of data sequentially):",
"_____no_output_____"
]
],
[
[
"example_dna.copy()",
"_____no_output_____"
],
[
"# Incorrect way (editing shared + mutable sequence):\nexample_dna = cor.DNA('atgagtaaaggagaagaacttttcactgga')\nvariant_list = []\nfor i, base in enumerate(example_dna):\n variant = example_dna\n variant.top[i] = 'A'\n variant.bottom[i] = 'T'\n variant_list.append(variant)\nprint [str(x) for x in variant_list]\n\nprint\n\n# Correct way (copy mutable sequence, then edit):\nexample_dna = cor.DNA('atgagtaaaggagaagaacttttcactgga')\nvariant_list = []\nfor i, base in enumerate(example_dna):\n variant = example_dna.copy()\n variant.top[i] = 'A'\n variant.bottom[i] = 'T'\n variant_list.append(variant)\nprint [str(x) for x in variant_list]",
"['AAAAAAAAAAAAAAAAAAAAAAAAAAAAAA', 'AAAAAAAAAAAAAAAAAAAAAAAAAAAAAA', 'AAAAAAAAAAAAAAAAAAAAAAAAAAAAAA', 'AAAAAAAAAAAAAAAAAAAAAAAAAAAAAA', 'AAAAAAAAAAAAAAAAAAAAAAAAAAAAAA', 'AAAAAAAAAAAAAAAAAAAAAAAAAAAAAA', 'AAAAAAAAAAAAAAAAAAAAAAAAAAAAAA', 'AAAAAAAAAAAAAAAAAAAAAAAAAAAAAA', 'AAAAAAAAAAAAAAAAAAAAAAAAAAAAAA', 'AAAAAAAAAAAAAAAAAAAAAAAAAAAAAA', 'AAAAAAAAAAAAAAAAAAAAAAAAAAAAAA', 'AAAAAAAAAAAAAAAAAAAAAAAAAAAAAA', 'AAAAAAAAAAAAAAAAAAAAAAAAAAAAAA', 'AAAAAAAAAAAAAAAAAAAAAAAAAAAAAA', 'AAAAAAAAAAAAAAAAAAAAAAAAAAAAAA', 'AAAAAAAAAAAAAAAAAAAAAAAAAAAAAA', 'AAAAAAAAAAAAAAAAAAAAAAAAAAAAAA', 'AAAAAAAAAAAAAAAAAAAAAAAAAAAAAA', 'AAAAAAAAAAAAAAAAAAAAAAAAAAAAAA', 'AAAAAAAAAAAAAAAAAAAAAAAAAAAAAA', 'AAAAAAAAAAAAAAAAAAAAAAAAAAAAAA', 'AAAAAAAAAAAAAAAAAAAAAAAAAAAAAA', 'AAAAAAAAAAAAAAAAAAAAAAAAAAAAAA', 'AAAAAAAAAAAAAAAAAAAAAAAAAAAAAA', 'AAAAAAAAAAAAAAAAAAAAAAAAAAAAAA', 'AAAAAAAAAAAAAAAAAAAAAAAAAAAAAA', 'AAAAAAAAAAAAAAAAAAAAAAAAAAAAAA', 'AAAAAAAAAAAAAAAAAAAAAAAAAAAAAA', 'AAAAAAAAAAAAAAAAAAAAAAAAAAAAAA', 'AAAAAAAAAAAAAAAAAAAAAAAAAAAAAA']\n\n['ATGAGTAAAGGAGAAGAACTTTTCACTGGA', 'AAGAGTAAAGGAGAAGAACTTTTCACTGGA', 'ATAAGTAAAGGAGAAGAACTTTTCACTGGA', 'ATGAGTAAAGGAGAAGAACTTTTCACTGGA', 'ATGAATAAAGGAGAAGAACTTTTCACTGGA', 'ATGAGAAAAGGAGAAGAACTTTTCACTGGA', 'ATGAGTAAAGGAGAAGAACTTTTCACTGGA', 'ATGAGTAAAGGAGAAGAACTTTTCACTGGA', 'ATGAGTAAAGGAGAAGAACTTTTCACTGGA', 'ATGAGTAAAAGAGAAGAACTTTTCACTGGA', 'ATGAGTAAAGAAGAAGAACTTTTCACTGGA', 'ATGAGTAAAGGAGAAGAACTTTTCACTGGA', 'ATGAGTAAAGGAAAAGAACTTTTCACTGGA', 'ATGAGTAAAGGAGAAGAACTTTTCACTGGA', 'ATGAGTAAAGGAGAAGAACTTTTCACTGGA', 'ATGAGTAAAGGAGAAAAACTTTTCACTGGA', 'ATGAGTAAAGGAGAAGAACTTTTCACTGGA', 'ATGAGTAAAGGAGAAGAACTTTTCACTGGA', 'ATGAGTAAAGGAGAAGAAATTTTCACTGGA', 'ATGAGTAAAGGAGAAGAACATTTCACTGGA', 'ATGAGTAAAGGAGAAGAACTATTCACTGGA', 'ATGAGTAAAGGAGAAGAACTTATCACTGGA', 'ATGAGTAAAGGAGAAGAACTTTACACTGGA', 'ATGAGTAAAGGAGAAGAACTTTTAACTGGA', 'ATGAGTAAAGGAGAAGAACTTTTCACTGGA', 'ATGAGTAAAGGAGAAGAACTTTTCAATGGA', 'ATGAGTAAAGGAGAAGAACTTTTCACAGGA', 'ATGAGTAAAGGAGAAGAACTTTTCACTAGA', 'ATGAGTAAAGGAGAAGAACTTTTCACTGAA', 'ATGAGTAAAGGAGAAGAACTTTTCACTGGA']\n"
]
],
[
[
"An important fact about `sequence.DNA` methods and slicing is that none of the operations modify the object directly (they don't mutate their parent) - if we look at example_dna, it has not been reverse-complemented itself. Running `example_dna.reverse_complement()` outputs a new sequence, so if you want to save your chance you need to assign a variable:",
"_____no_output_____"
]
],
[
[
"revcomp_dna = example_dna.reverse_complement()\ndisplay(example_dna)\ndisplay(revcomp_dna)",
"_____no_output_____"
]
],
[
[
"You also have direct access important attributes of a `sequence.DNA` object. The following are examples of how to get important sequences or information about a sequence.",
"_____no_output_____"
]
],
[
[
"# The top strand - a simple python string in the 5' -> 3' orientation.\nexample_dna.top",
"_____no_output_____"
],
[
"# The bottom strand - another python string, also in the 5' -> 3' orientation.\nexample_dna.bottom",
"_____no_output_____"
],
[
"# Sequences are double stranded, or 'ds' by default. \n# This is a directly accessible attribute, not a method, so () is not required.\nexample_dna.ds",
"_____no_output_____"
],
[
"# DNA can be linear or circular - check the boolean `circular` attribute.\nexample_dna.circular",
"_____no_output_____"
],
[
"# You can switch between topologies using the .circularize and .linearize methods.\n# Circular DNA has different properties:\n# 1) it can't be concatenated to\n# 2) sequence searches using .locate will search over the current origin (e.g. from -10 to +10 for a 20-base sequence).\ncircular_dna = example_dna.circularize()\ncircular_dna.circular",
"_____no_output_____"
],
[
"# Linearization is more complex - you can choose the index at which to linearize a circular sequence.\n# This simulates a precise double stranded break at the index of your choosing.\n# The following example shows the difference between linearizing at index 0 (default) versus index 2\n# (python 0-indexes, so index 2 = 3rd base, i.e. 'g' in 'atg')\nprint circular_dna.linearize()\nprint\nprint circular_dna.linearize(2)",
"ATGAGTAAAGGAGAAGAACTTTTCACTGGA\n\nGAGTAAAGGAGAAGAACTTTTCACTGGAAT\n"
],
[
"# Sometimes you just want to rotate the sequence around - i.e. switch the top and bottom strands. \n# For this, use the .flip() method\nexample_dna.flip()",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb89ff87a9ef1162d968b147f0910e4b79d73025
| 661,045 |
ipynb
|
Jupyter Notebook
|
HR Analytics classification Problem.ipynb
|
shailcasticofficial/Work-Profile-Data-Science
|
db56b4656b2abb41b966be4cedc4ce9c47ca3500
|
[
"MIT"
] | 1 |
2022-01-17T12:50:42.000Z
|
2022-01-17T12:50:42.000Z
|
HR Analytics classification Problem.ipynb
|
shailcasticofficial/Work-Profile-Data-Science
|
db56b4656b2abb41b966be4cedc4ce9c47ca3500
|
[
"MIT"
] | null | null | null |
HR Analytics classification Problem.ipynb
|
shailcasticofficial/Work-Profile-Data-Science
|
db56b4656b2abb41b966be4cedc4ce9c47ca3500
|
[
"MIT"
] | null | null | null | 187.850242 | 478,340 | 0.876191 |
[
[
[
"import os\nos.chdir('C:\\\\Users\\\\SHAILESH TIWARI\\\\Downloads\\\\Classification\\\\hr')",
"_____no_output_____"
],
[
"%matplotlib inline\nimport pandas as pd\nimport numpy as np\nimport seaborn as sns\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"train = pd.read_csv('train.csv')\n# getting their shapes\nprint(\"Shape of train :\", train.shape)\n#print(\"Shape of test :\", test.shape)",
"Shape of train : (54808, 14)\n"
],
[
"train.shape",
"_____no_output_____"
],
[
"train.head()",
"_____no_output_____"
],
[
"train.columns",
"_____no_output_____"
],
[
"train.isna().sum()",
"_____no_output_____"
],
[
"#calculation of percentage of missing data\ntotal = train.isnull().sum().sort_values(ascending=False)\npercent_1 = train.isnull().sum()/train.isnull().count()*100\npercent_2 = (round(percent_1, 1)).sort_values(ascending=False)\nmissing_data = pd.concat([total, percent_2], axis=1, keys=['Total', '%'])\nmissing_data.head(5)",
"_____no_output_____"
],
[
"train['is_promoted'].value_counts() #unbalanced",
"_____no_output_____"
],
[
"train.shape",
"_____no_output_____"
],
[
"# finding the %age of people promoted\n\npromoted = (4668/54808)*100\nprint(\"Percentage of Promoted Employees is {:.2f}%\".format(promoted))",
"Percentage of Promoted Employees is 8.52%\n"
],
[
"#plotting a scatter plot \n\nplt.hist(train['is_promoted'])\nplt.title('plot to show the gap in Promoted and Non-Promoted Employees', fontsize = 30)\nplt.xlabel('0 -No Promotion and 1- Promotion', fontsize = 20)\nplt.ylabel('count')\nplt.show()",
"_____no_output_____"
],
[
"s1=train.dtypes\ns1.groupby(s1).count()",
"_____no_output_____"
],
[
"train.dtypes",
"_____no_output_____"
],
[
"corr_matrix = train.corr(method='pearson')\ncorr_matrix['is_promoted'].sort_values(kind=\"quicksort\")",
"_____no_output_____"
],
[
"#dropping the column\ntrain.drop(['employee_id','region'], axis = 1, inplace = True)",
"_____no_output_____"
],
[
"train.head()",
"_____no_output_____"
],
[
"train.columns.values",
"_____no_output_____"
],
[
"#check for missing value, unique etc\nFileNameDesc = pd.DataFrame(columns = ['column_name','missing_count','percent_missing','unique_count'])\nfor col in list(train.columns.values):\n sum_missing = train[col].isnull().sum()\n percent_missing = sum_missing/len(train)*100\n uniq_count = (train.groupby([col])[col].count()).count()\n FileNameDesc = FileNameDesc.append({'column_name':col,'missing_count':sum_missing,\n 'percent_missing':percent_missing,'unique_count':uniq_count},\n ignore_index = True)",
"_____no_output_____"
],
[
"FileNameDesc",
"_____no_output_____"
],
[
"#Apply Mode strategy to populate the categorical data\ntrain.groupby('education').agg({'education': np.size})",
"_____no_output_____"
],
[
"train[\"education\"]=train[\"education\"].fillna('Attchd')\ntrain[\"education\"]=train[\"education\"].astype('category')\ntrain[\"education\"] = train[\"education\"].cat.codes",
"_____no_output_____"
],
[
"train.isnull().sum()",
"_____no_output_____"
],
[
"train['previous_year_rating'].unique()",
"_____no_output_____"
],
[
"train['previous_year_rating'].mode()",
"_____no_output_____"
],
[
"train['previous_year_rating'].fillna(1, inplace = True)",
"_____no_output_____"
],
[
"train.isnull().sum()",
"_____no_output_____"
],
[
"train.dtypes",
"_____no_output_____"
],
[
"data=pd.get_dummies(train,columns=['department','gender','recruitment_channel','previous_year_rating'],drop_first=True)",
"_____no_output_____"
],
[
"data",
"_____no_output_____"
],
[
"df1=data['is_promoted']",
"_____no_output_____"
],
[
"data.drop(['is_promoted'], axis = 1, inplace = True)",
"_____no_output_____"
],
[
"data=pd.concat([data,df1],axis=1)",
"_____no_output_____"
],
[
"#Key data analysis\nlen(data)\ndata.head()\ndata.isnull().any()\ndata.isnull().sum()",
"_____no_output_____"
],
[
"data.corr()\nsns.heatmap(data.corr(),annot=False)",
"_____no_output_____"
],
[
"data.columns",
"_____no_output_____"
],
[
"x = data.iloc[:,0:22].values\ny = data.iloc[:,-1:].values",
"_____no_output_____"
],
[
"x",
"_____no_output_____"
],
[
"y",
"_____no_output_____"
],
[
"from sklearn.preprocessing import StandardScaler # to make the data in standard format to read\nsc = StandardScaler() # feature scaling because salary and age are both in different scale\nx=sc.fit_transform(x)",
"_____no_output_____"
],
[
"pd.DataFrame(x)",
"_____no_output_____"
],
[
"from sklearn.model_selection import train_test_split\nx_train, x_test, y_train, y_test= train_test_split(x,y,test_size=0.20, random_state=0)",
"_____no_output_____"
],
[
"# applying logistic regression\nfrom sklearn.linear_model import LogisticRegression\nlogmodel = LogisticRegression()\nlogmodel.fit(x_train,y_train)",
"C:\\Users\\SHAILESH TIWARI\\anaconda3\\lib\\site-packages\\sklearn\\utils\\validation.py:760: DataConversionWarning: A column-vector y was passed when a 1d array was expected. Please change the shape of y to (n_samples, ), for example using ravel().\n y = column_or_1d(y, warn=True)\n"
],
[
"# prediction for x_test\ny_pred = logmodel.predict(x_test)",
"_____no_output_____"
],
[
"y_pred",
"_____no_output_____"
],
[
"y_test",
"_____no_output_____"
],
[
"# concept of confusion matrix\nfrom sklearn.metrics import confusion_matrix\nconfusion_matrix(y_test,y_pred)",
"_____no_output_____"
],
[
"from sklearn.metrics import accuracy_score\naccuracy_score(y_test,y_pred)",
"_____no_output_____"
],
[
"len(y_test)",
"_____no_output_____"
],
[
"sns.pairplot(train)",
"_____no_output_____"
],
[
"# applying cross validation on top of algo\nfrom sklearn.model_selection import cross_val_score\naccuracies = cross_val_score(estimator=logmodel, X=x_train,y=y_train,cv=10)",
"C:\\Users\\SHAILESH TIWARI\\anaconda3\\lib\\site-packages\\sklearn\\utils\\validation.py:760: DataConversionWarning: A column-vector y was passed when a 1d array was expected. Please change the shape of y to (n_samples, ), for example using ravel().\n y = column_or_1d(y, warn=True)\nC:\\Users\\SHAILESH TIWARI\\anaconda3\\lib\\site-packages\\sklearn\\utils\\validation.py:760: DataConversionWarning: A column-vector y was passed when a 1d array was expected. Please change the shape of y to (n_samples, ), for example using ravel().\n y = column_or_1d(y, warn=True)\nC:\\Users\\SHAILESH TIWARI\\anaconda3\\lib\\site-packages\\sklearn\\utils\\validation.py:760: DataConversionWarning: A column-vector y was passed when a 1d array was expected. Please change the shape of y to (n_samples, ), for example using ravel().\n y = column_or_1d(y, warn=True)\nC:\\Users\\SHAILESH TIWARI\\anaconda3\\lib\\site-packages\\sklearn\\utils\\validation.py:760: DataConversionWarning: A column-vector y was passed when a 1d array was expected. Please change the shape of y to (n_samples, ), for example using ravel().\n y = column_or_1d(y, warn=True)\nC:\\Users\\SHAILESH TIWARI\\anaconda3\\lib\\site-packages\\sklearn\\utils\\validation.py:760: DataConversionWarning: A column-vector y was passed when a 1d array was expected. Please change the shape of y to (n_samples, ), for example using ravel().\n y = column_or_1d(y, warn=True)\nC:\\Users\\SHAILESH TIWARI\\anaconda3\\lib\\site-packages\\sklearn\\utils\\validation.py:760: DataConversionWarning: A column-vector y was passed when a 1d array was expected. Please change the shape of y to (n_samples, ), for example using ravel().\n y = column_or_1d(y, warn=True)\nC:\\Users\\SHAILESH TIWARI\\anaconda3\\lib\\site-packages\\sklearn\\utils\\validation.py:760: DataConversionWarning: A column-vector y was passed when a 1d array was expected. Please change the shape of y to (n_samples, ), for example using ravel().\n y = column_or_1d(y, warn=True)\nC:\\Users\\SHAILESH TIWARI\\anaconda3\\lib\\site-packages\\sklearn\\utils\\validation.py:760: DataConversionWarning: A column-vector y was passed when a 1d array was expected. Please change the shape of y to (n_samples, ), for example using ravel().\n y = column_or_1d(y, warn=True)\nC:\\Users\\SHAILESH TIWARI\\anaconda3\\lib\\site-packages\\sklearn\\utils\\validation.py:760: DataConversionWarning: A column-vector y was passed when a 1d array was expected. Please change the shape of y to (n_samples, ), for example using ravel().\n y = column_or_1d(y, warn=True)\nC:\\Users\\SHAILESH TIWARI\\anaconda3\\lib\\site-packages\\sklearn\\utils\\validation.py:760: DataConversionWarning: A column-vector y was passed when a 1d array was expected. Please change the shape of y to (n_samples, ), for example using ravel().\n y = column_or_1d(y, warn=True)\n"
],
[
"accuracies",
"_____no_output_____"
],
[
"accuracies.mean()",
"_____no_output_____"
],
[
"# k nearest neighbour algo applying\nfrom sklearn.neighbors import KNeighborsClassifier\nclassifier_knn =KNeighborsClassifier(n_neighbors=11,metric='euclidean',p=2)\nclassifier_knn.fit(x_train,y_train)",
"C:\\Users\\SHAILESH TIWARI\\anaconda3\\lib\\site-packages\\ipykernel_launcher.py:4: DataConversionWarning: A column-vector y was passed when a 1d array was expected. Please change the shape of y to (n_samples, ), for example using ravel().\n after removing the cwd from sys.path.\n"
],
[
"y_pred_knn = classifier_knn.predict(x_test)",
"_____no_output_____"
],
[
"y_pred_knn",
"_____no_output_____"
],
[
"y_test",
"_____no_output_____"
],
[
"from sklearn.metrics import confusion_matrix\nconfusion_matrix(y_test,y_pred_knn)",
"_____no_output_____"
],
[
"from sklearn.metrics import accuracy_score\naccuracy_score(y_test,y_pred_knn)",
"_____no_output_____"
],
[
"from sklearn.model_selection import cross_val_score\naccuracies = cross_val_score(estimator=classifier_knn, X=x_train,y=y_train,cv=10)",
"C:\\Users\\SHAILESH TIWARI\\anaconda3\\lib\\site-packages\\sklearn\\model_selection\\_validation.py:515: DataConversionWarning: A column-vector y was passed when a 1d array was expected. Please change the shape of y to (n_samples, ), for example using ravel().\n estimator.fit(X_train, y_train, **fit_params)\nC:\\Users\\SHAILESH TIWARI\\anaconda3\\lib\\site-packages\\sklearn\\model_selection\\_validation.py:515: DataConversionWarning: A column-vector y was passed when a 1d array was expected. Please change the shape of y to (n_samples, ), for example using ravel().\n estimator.fit(X_train, y_train, **fit_params)\nC:\\Users\\SHAILESH TIWARI\\anaconda3\\lib\\site-packages\\sklearn\\model_selection\\_validation.py:515: DataConversionWarning: A column-vector y was passed when a 1d array was expected. Please change the shape of y to (n_samples, ), for example using ravel().\n estimator.fit(X_train, y_train, **fit_params)\nC:\\Users\\SHAILESH TIWARI\\anaconda3\\lib\\site-packages\\sklearn\\model_selection\\_validation.py:515: DataConversionWarning: A column-vector y was passed when a 1d array was expected. Please change the shape of y to (n_samples, ), for example using ravel().\n estimator.fit(X_train, y_train, **fit_params)\nC:\\Users\\SHAILESH TIWARI\\anaconda3\\lib\\site-packages\\sklearn\\model_selection\\_validation.py:515: DataConversionWarning: A column-vector y was passed when a 1d array was expected. Please change the shape of y to (n_samples, ), for example using ravel().\n estimator.fit(X_train, y_train, **fit_params)\nC:\\Users\\SHAILESH TIWARI\\anaconda3\\lib\\site-packages\\sklearn\\model_selection\\_validation.py:515: DataConversionWarning: A column-vector y was passed when a 1d array was expected. Please change the shape of y to (n_samples, ), for example using ravel().\n estimator.fit(X_train, y_train, **fit_params)\nC:\\Users\\SHAILESH TIWARI\\anaconda3\\lib\\site-packages\\sklearn\\model_selection\\_validation.py:515: DataConversionWarning: A column-vector y was passed when a 1d array was expected. Please change the shape of y to (n_samples, ), for example using ravel().\n estimator.fit(X_train, y_train, **fit_params)\nC:\\Users\\SHAILESH TIWARI\\anaconda3\\lib\\site-packages\\sklearn\\model_selection\\_validation.py:515: DataConversionWarning: A column-vector y was passed when a 1d array was expected. Please change the shape of y to (n_samples, ), for example using ravel().\n estimator.fit(X_train, y_train, **fit_params)\nC:\\Users\\SHAILESH TIWARI\\anaconda3\\lib\\site-packages\\sklearn\\model_selection\\_validation.py:515: DataConversionWarning: A column-vector y was passed when a 1d array was expected. Please change the shape of y to (n_samples, ), for example using ravel().\n estimator.fit(X_train, y_train, **fit_params)\nC:\\Users\\SHAILESH TIWARI\\anaconda3\\lib\\site-packages\\sklearn\\model_selection\\_validation.py:515: DataConversionWarning: A column-vector y was passed when a 1d array was expected. Please change the shape of y to (n_samples, ), for example using ravel().\n estimator.fit(X_train, y_train, **fit_params)\n"
],
[
"accuracies",
"_____no_output_____"
],
[
"accuracies.mean()",
"_____no_output_____"
],
[
"# naiye baise algo application\nfrom sklearn.naive_bayes import GaussianNB\nclassifier_nb =GaussianNB()\nclassifier_nb.fit(x_train,y_train)",
"C:\\Users\\SHAILESH TIWARI\\anaconda3\\lib\\site-packages\\sklearn\\naive_bayes.py:206: DataConversionWarning: A column-vector y was passed when a 1d array was expected. Please change the shape of y to (n_samples, ), for example using ravel().\n y = column_or_1d(y, warn=True)\n"
],
[
"y_pred_nb = classifier_nb.predict(x_test)",
"_____no_output_____"
],
[
"y_pred_nb",
"_____no_output_____"
],
[
"y_test",
"_____no_output_____"
],
[
"from sklearn.metrics import confusion_matrix\nconfusion_matrix(y_test,y_pred_nb)",
"_____no_output_____"
],
[
"from sklearn.metrics import accuracy_score\naccuracy_score(y_test,y_pred_nb)",
"_____no_output_____"
],
[
"from sklearn.model_selection import cross_val_score\naccuracies = cross_val_score(estimator=classifier_nb, X=x_train,y=y_train,cv=10)",
"C:\\Users\\SHAILESH TIWARI\\anaconda3\\lib\\site-packages\\sklearn\\naive_bayes.py:206: DataConversionWarning: A column-vector y was passed when a 1d array was expected. Please change the shape of y to (n_samples, ), for example using ravel().\n y = column_or_1d(y, warn=True)\nC:\\Users\\SHAILESH TIWARI\\anaconda3\\lib\\site-packages\\sklearn\\naive_bayes.py:206: DataConversionWarning: A column-vector y was passed when a 1d array was expected. Please change the shape of y to (n_samples, ), for example using ravel().\n y = column_or_1d(y, warn=True)\nC:\\Users\\SHAILESH TIWARI\\anaconda3\\lib\\site-packages\\sklearn\\naive_bayes.py:206: DataConversionWarning: A column-vector y was passed when a 1d array was expected. Please change the shape of y to (n_samples, ), for example using ravel().\n y = column_or_1d(y, warn=True)\nC:\\Users\\SHAILESH TIWARI\\anaconda3\\lib\\site-packages\\sklearn\\naive_bayes.py:206: DataConversionWarning: A column-vector y was passed when a 1d array was expected. Please change the shape of y to (n_samples, ), for example using ravel().\n y = column_or_1d(y, warn=True)\nC:\\Users\\SHAILESH TIWARI\\anaconda3\\lib\\site-packages\\sklearn\\naive_bayes.py:206: DataConversionWarning: A column-vector y was passed when a 1d array was expected. Please change the shape of y to (n_samples, ), for example using ravel().\n y = column_or_1d(y, warn=True)\nC:\\Users\\SHAILESH TIWARI\\anaconda3\\lib\\site-packages\\sklearn\\naive_bayes.py:206: DataConversionWarning: A column-vector y was passed when a 1d array was expected. Please change the shape of y to (n_samples, ), for example using ravel().\n y = column_or_1d(y, warn=True)\nC:\\Users\\SHAILESH TIWARI\\anaconda3\\lib\\site-packages\\sklearn\\naive_bayes.py:206: DataConversionWarning: A column-vector y was passed when a 1d array was expected. Please change the shape of y to (n_samples, ), for example using ravel().\n y = column_or_1d(y, warn=True)\nC:\\Users\\SHAILESH TIWARI\\anaconda3\\lib\\site-packages\\sklearn\\naive_bayes.py:206: DataConversionWarning: A column-vector y was passed when a 1d array was expected. Please change the shape of y to (n_samples, ), for example using ravel().\n y = column_or_1d(y, warn=True)\nC:\\Users\\SHAILESH TIWARI\\anaconda3\\lib\\site-packages\\sklearn\\naive_bayes.py:206: DataConversionWarning: A column-vector y was passed when a 1d array was expected. Please change the shape of y to (n_samples, ), for example using ravel().\n y = column_or_1d(y, warn=True)\nC:\\Users\\SHAILESH TIWARI\\anaconda3\\lib\\site-packages\\sklearn\\naive_bayes.py:206: DataConversionWarning: A column-vector y was passed when a 1d array was expected. Please change the shape of y to (n_samples, ), for example using ravel().\n y = column_or_1d(y, warn=True)\n"
],
[
"accuracies",
"_____no_output_____"
],
[
"accuracies.mean()",
"_____no_output_____"
],
[
"# support vector machine application through sigmoid kernel\nfrom sklearn.svm import SVC\nclassifier_svm_sig = SVC(kernel='sigmoid')\nclassifier_svm_sig.fit(x_train,y_train)",
"C:\\Users\\SHAILESH TIWARI\\anaconda3\\lib\\site-packages\\sklearn\\utils\\validation.py:760: DataConversionWarning: A column-vector y was passed when a 1d array was expected. Please change the shape of y to (n_samples, ), for example using ravel().\n y = column_or_1d(y, warn=True)\n"
],
[
"pred_svm_sig = classifier_svm_sig.predict(x_test)",
"_____no_output_____"
],
[
"pred_svm_sig",
"_____no_output_____"
],
[
"from sklearn.metrics import confusion_matrix\nconfusion_matrix(y_test,pred_svm_sig)",
"_____no_output_____"
],
[
"from sklearn.metrics import accuracy_score\naccuracy_score(y_test,pred_svm_sig)",
"_____no_output_____"
],
[
"# support vector machine application through linear kernel\nfrom sklearn.svm import SVC\nclassifier_svm_lin = SVC(kernel='linear')\nclassifier_svm_lin.fit(x_train,y_train)",
"C:\\Users\\SHAILESH TIWARI\\anaconda3\\lib\\site-packages\\sklearn\\utils\\validation.py:760: DataConversionWarning: A column-vector y was passed when a 1d array was expected. Please change the shape of y to (n_samples, ), for example using ravel().\n y = column_or_1d(y, warn=True)\n"
],
[
"y_pred_svm_lin = classifier_svm_lin.predict(x_test)",
"_____no_output_____"
],
[
"y_pred_svm_lin",
"_____no_output_____"
],
[
"from sklearn.metrics import confusion_matrix\nconfusion_matrix(y_test,y_pred_svm_lin)",
"_____no_output_____"
],
[
"from sklearn.metrics import accuracy_score\naccuracy_score(y_test,y_pred_svm_lin)",
"_____no_output_____"
],
[
"# support vector machine application through polynomial kernel\nfrom sklearn.svm import SVC\nclassifier_svm_poly = SVC(kernel='poly')\nclassifier_svm_poly.fit(x_train,y_train)",
"C:\\Users\\SHAILESH TIWARI\\anaconda3\\lib\\site-packages\\sklearn\\utils\\validation.py:760: DataConversionWarning: A column-vector y was passed when a 1d array was expected. Please change the shape of y to (n_samples, ), for example using ravel().\n y = column_or_1d(y, warn=True)\n"
],
[
"y_pred_svm_poly = classifier_svm_poly.predict(x_test)",
"_____no_output_____"
],
[
"y_pred_svm_poly",
"_____no_output_____"
],
[
"from sklearn.metrics import confusion_matrix\nconfusion_matrix(y_test,y_pred_svm_poly)",
"_____no_output_____"
],
[
"from sklearn.metrics import accuracy_score\naccuracy_score(y_test,y_pred_svm_poly)",
"_____no_output_____"
],
[
"# support vector machine application through rbf kernel\nfrom sklearn.svm import SVC\nclassifier_svm_rbf = SVC(kernel='rbf')\nclassifier_svm_rbf.fit(x_train,y_train)",
"C:\\Users\\SHAILESH TIWARI\\anaconda3\\lib\\site-packages\\sklearn\\utils\\validation.py:760: DataConversionWarning: A column-vector y was passed when a 1d array was expected. Please change the shape of y to (n_samples, ), for example using ravel().\n y = column_or_1d(y, warn=True)\n"
],
[
"y_pred_svm_rbf = classifier_svm_rbf.predict(x_test)",
"_____no_output_____"
],
[
"y_pred_svm_rbf",
"_____no_output_____"
],
[
"from sklearn.metrics import confusion_matrix\nconfusion_matrix(y_test,y_pred_svm_rbf)",
"_____no_output_____"
],
[
"#accuracy score\nfrom sklearn.metrics import accuracy_score\naccuracy_score(y_test,y_pred_svm_rbf)",
"_____no_output_____"
],
[
"#running decision tree algo\nfrom sklearn.tree import DecisionTreeClassifier\nclassifier_dt =DecisionTreeClassifier(criterion='entropy') # also can use gini\nclassifier_dt.fit(x_train,y_train)",
"_____no_output_____"
],
[
"y_pred_dt =classifier_dt.predict(x_test)",
"_____no_output_____"
],
[
"y_pred_dt",
"_____no_output_____"
],
[
"from sklearn.metrics import confusion_matrix\nconfusion_matrix(y_test,y_pred_dt)",
"_____no_output_____"
],
[
"# accuracy score calculation\nfrom sklearn.metrics import accuracy_score\naccuracy_score(y_test,y_pred_dt)",
"_____no_output_____"
],
[
"# running random forest algorithm\nfrom sklearn.ensemble import RandomForestClassifier\nclassifier_rf =RandomForestClassifier(n_estimators=3, criterion='entropy')\nclassifier_rf.fit(x_train,y_train)",
"C:\\Users\\SHAILESH TIWARI\\anaconda3\\lib\\site-packages\\ipykernel_launcher.py:4: DataConversionWarning: A column-vector y was passed when a 1d array was expected. Please change the shape of y to (n_samples,), for example using ravel().\n after removing the cwd from sys.path.\n"
],
[
"y_pred_rf =classifier_rf.predict(x_test)",
"_____no_output_____"
],
[
"y_pred_rf",
"_____no_output_____"
],
[
"from sklearn.metrics import confusion_matrix\nconfusion_matrix(y_test,y_pred_rf)",
"_____no_output_____"
],
[
"from sklearn.metrics import accuracy_score\naccuracy_score(y_test,y_pred_rf)",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb8a0f8d132ebcd63790feb93ddc99a9f9a721f8
| 1,476 |
ipynb
|
Jupyter Notebook
|
min_zero_transform_demo.ipynb
|
herwinvw/TabularPlayGroundMay2021
|
e92a519ba58db3eb509915dc6276d95c481eb29e
|
[
"MIT"
] | null | null | null |
min_zero_transform_demo.ipynb
|
herwinvw/TabularPlayGroundMay2021
|
e92a519ba58db3eb509915dc6276d95c481eb29e
|
[
"MIT"
] | null | null | null |
min_zero_transform_demo.ipynb
|
herwinvw/TabularPlayGroundMay2021
|
e92a519ba58db3eb509915dc6276d95c481eb29e
|
[
"MIT"
] | null | null | null | 23.0625 | 88 | 0.538618 |
[
[
[
"from sklearn.base import BaseEstimator, TransformerMixin # type: ignore\nfrom typing import List, Any\n\nimport pandas as pd\nclass MinZeroTransform(BaseEstimator, TransformerMixin):\n \"\"\"\n Transforms X to X-X.min(), so that the minimum is shifted to 0.\n X.min() is learned in the fit.\n \"\"\"\n def transform(self, df:pd.DataFrame, **transform_params:Any)->pd.DataFrame:\n return df - self.min()\n \n def fit(self, X:pd.DataFrame, y:Any=None, **fit_params:Any):\n self.min = X.min()",
"_____no_output_____"
],
[
"from min_zero_transform import MinZeroTransform",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code"
]
] |
cb8a254fad6a15f5db72c607f83f650d61291664
| 12,681 |
ipynb
|
Jupyter Notebook
|
CONS.ipynb
|
anilk991/rosalindsolutions
|
1548b57fa6fbf04ec9968bc8ab47668d37c765d7
|
[
"MIT"
] | null | null | null |
CONS.ipynb
|
anilk991/rosalindsolutions
|
1548b57fa6fbf04ec9968bc8ab47668d37c765d7
|
[
"MIT"
] | null | null | null |
CONS.ipynb
|
anilk991/rosalindsolutions
|
1548b57fa6fbf04ec9968bc8ab47668d37c765d7
|
[
"MIT"
] | null | null | null | 60.966346 | 1,909 | 0.522041 |
[
[
[
"### Consensus sequence matrix",
"_____no_output_____"
]
],
[
[
"import re",
"_____no_output_____"
],
[
"file = open(\"/Users/anilkumar/projectEEG/rosalind_cons (3).txt\")\nseqs = file.read()\nseqs = seqs.replace(\"\\n\", \"\")\nids = re.findall(r'>Rosalind_\\d{4}', seqs)\nseqs = re.split(r'>Rosalind_\\d{4}', seqs)[1:]",
"_____no_output_____"
],
[
"prof_mat = {'A': [], 'C': [], 'G':[], 'T': []}",
"_____no_output_____"
],
[
"num_seqs = len(seqs)\nlen_seqs = len(seqs[0])",
"_____no_output_____"
],
[
"for i in range(len_seqs):\n countA = 0\n countT = 0\n countG = 0\n countC = 0\n for j in range(num_seqs):\n if seqs[j][i] == 'A':\n countA += 1\n elif seqs[j][i] == 'T':\n countT += 1\n elif seqs[j][i] == 'G':\n countG += 1\n elif seqs[j][i] == 'C':\n countC += 1\n prof_mat['A'].append(countA)\n prof_mat['C'].append(countC)\n prof_mat['G'].append(countG)\n prof_mat['T'].append(countT)",
"_____no_output_____"
],
[
"def consensus(prof_mat):\n cons = []\n mat = np.array(list(prof_mat.values()))\n for i in range(len_seqs):\n max_res = np.argmax(np.array(list(prof_mat.values()))[:,i])\n cons.append(list(prof_mat.keys())[max_res])\n return \"\".join(cons)",
"_____no_output_____"
],
[
"print(consensus(prof_mat))\nfor k in ['A', 'C', 'G', 'T']:\n print(k + \": \" + \" \".join(str(x) for x in prof_mat[k]))",
"TAGAATTAAAACGCGCGATTCCAATAACGGTGGAAGGGGGATACGGTTAGGGAGTTTGACCATTGCGACTATATAACCCGGAGGCTCAGCATGAGACAGCGTACTCGACCATGAGCACGAAAAATGAACAGTACCCGGGCCCACCGAGACTAGCAACACGTCATATCAAGCCCCACGGGATCCTGAACATAACTACCTAACCGGCAAACGGGATTAGGAGCTCCAACAATCACACTTCAAGTGCCACGACAGAGCGGTAAAATTTCTTCCACATTCATAGGCCAGTATAAGTCCGGCAGAAATAGGAACACTCACACATAATCAAATAACAACACCCATCCGTATCAAATACTGAGCACTACGGGTATTGCCTGCGCCTAAAATTTCGGTTTTACATGACCCCAAGTCTCATGCAAATCCGGTGATCAGTGAACAGGGCGAACAATCAAATCATTAACAGAGGGACCCAGCGTTCTGTAACTTAAGCAGGTGAACTTTGACCGCACAGACGTAAACCTACGCAAGTAACGGTACAATAAATCTGATCCATTAGACCAATACCAGCATCGTAATAACCTCAGAACAGACTGACAAATACCATTGCTAAGACGAGAGCTCTTAATCGAGCATCAGGAGAACCGGTCAACGAAAACTCAGATGAGGCTTATATAAATACCGATCTTTGGCATTTACCCGGGCCAGAAGAAGTCTAGAGCCCGGGATAACCTACCTGGCAAACACCCAGCGCGCAGATCCAACGTCAATGTAAACTGCTTAATTACCTGAGCAACTTGGTCAAGAAGGCTGTTCCTAGGCATTCGAGTGAGGTACGTGAAAGCTCCGTACAATAAAAGCAAACAACAACTCCAGAAGAACGTGCCAGAGTAAAGCCCAAAGCTCATGGCTGTGCCATAGATCGGTTAGTACGGCATAAAAGGCCTCACAAAT\nA: 1 3 2 4 4 3 2 4 3 4 4 2 1 2 2 2 2 3 3 0 2 2 3 4 0 4 3 2 3 3 2 1 3 4 6 3 1 3 3 2 5 1 5 2 1 3 2 0 3 2 1 2 4 2 3 2 0 2 4 3 1 5 2 2 1 1 1 5 3 2 4 3 3 0 4 3 2 2 0 0 2 5 2 2 2 1 1 3 2 2 3 0 2 3 2 4 1 4 2 3 3 2 4 2 4 2 1 5 2 3 4 3 1 3 3 3 3 2 1 4 5 4 3 3 2 2 3 4 1 4 2 2 4 2 1 2 3 3 2 2 2 0 6 0 3 2 4 2 4 3 2 5 1 2 4 5 1 3 2 2 1 2 4 4 4 1 3 4 3 2 2 2 3 2 4 2 1 3 1 4 3 2 1 2 1 4 3 1 4 0 4 3 2 2 5 2 2 2 3 3 2 2 2 1 1 3 3 3 0 2 2 2 4 1 2 4 1 2 3 2 1 2 1 3 3 3 2 4 5 1 1 4 2 5 2 1 3 2 4 6 2 0 2 1 2 3 3 2 4 1 3 2 6 2 0 1 1 3 3 3 5 4 3 1 3 0 0 2 3 1 5 3 5 2 2 1 4 3 3 2 0 3 3 4 1 3 3 0 5 4 2 3 1 3 2 2 2 4 2 3 3 3 1 3 2 1 4 3 1 5 1 2 3 3 2 5 2 4 0 4 4 1 3 4 3 5 1 3 4 1 3 5 1 4 3 3 2 5 2 0 3 2 3 4 2 1 4 3 3 2 3 2 2 1 3 2 3 4 1 2 5 2 1 2 1 3 4 3 3 2 0 0 3 2 0 2 0 2 1 5 4 3 3 2 2 2 1 1 2 1 3 0 1 4 2 3 3 1 4 0 1 2 2 3 5 1 3 2 3 2 4 2 3 1 4 3 3 1 2 2 0 2 2 2 4 0 2 4 2 2 2 7 4 1 5 2 2 1 4 1 4 3 3 4 5 2 0 3 5 3 2 2 4 0 2 5 4 1 3 3 3 1 1 1 5 2 2 1 3 2 1 2 3 2 1 1 2 1 4 4 2 1 0 4 4 2 1 4 2 0 0 3 5 5 3 3 2 3 2 5 0 2 1 2 4 1 4 0 3 1 3 2 4 4 4 2 2 2 3 1 1 3 3 5 2 1 5 4 1 2 1 2 3 0 4 4 3 6 6 5 2 1 3 2 5 2 1 1 5 4 2 4 2 3 2 2 4 5 2 3 0 0 4 3 4 4 2 2 1 2 4 3 2 4 4 1 1 1 1 4 2 8 4 2 4 3 4 4 1 2 4 2 3 5 4 2 4 2 3 4 2 2 2 1 2 3 5 3 4 0 1 3 3 6 1 1 2 3 2 3 4 4 1 2 2 6 2 1 4 2 2 3 3 1 4 3 3 4 2 2 1 2 2 1 4 6 1 2 5 5 4 6 2 3 2 4 1 4 0 2 3 3 3 3 3 1 3 3 4 2 3 5 4 1 5 1 2 1 4 1 1 3 2 1 1 1 3 3 1 2 0 3 3 1 2 2 2 2 0 1 3 1 3 3 2 4 3 3 3 3 1 5 3 4 2 1 2 3 1 0 2 5 1 4 3 1 2 3 3 1 2 1 1 2 3 5 3 3 2 4 3 2 2 4 0 4 2 3 2 1 4 2 4 3 2 3 5 4 1 2 1 2 3 4 2 2 3 4 5 4 2 1 2 1 2 1 4 4 2 2 4 2 1 1 1 5 1 2 3 4 2 2 1 0 1 1 2 3 3 0 4 4 1 1 2 2 2 1 1 1 1 3 4 2 3 2 5 2 2 1 1 4 0 1 3 4 2 2 2 4 3 1 3 2 5 3 3 3 1 2 2 3 1 2 3 1 4 4 3 4 4 3 3 2 2 3 4 6 2 4 3 2 3 5 2 3 3 3 5 2 5 3 1 4 7 2 3 3 2 1 2 4 1 4 2 1 5 4 3 3 3 2 0 4 3 3 3 2 1 3 5 1 3 2 1 2 1 1 2 2 1 3 3 3 3 3 3 2 2 0 1 1 6 3 1 3 2 3 1 1 4 1 5 4 4 5 2 2 1 3 2 3 4 3 3 4 5 2\nC: 1 1 1 1 1 1 2 2 1 2 3 3 3 6 1 3 0 3 2 3 4 5 2 1 1 1 2 3 3 2 3 1 3 3 2 0 2 2 1 0 2 2 0 3 2 0 3 3 3 2 3 3 2 3 1 2 3 0 2 4 5 0 0 3 3 3 1 1 4 2 4 2 2 3 1 3 4 4 5 2 2 3 1 2 3 2 3 2 3 3 3 2 1 3 2 2 5 2 1 4 1 1 4 3 1 3 2 1 3 4 1 2 0 1 1 4 2 5 1 3 1 2 2 3 3 2 3 0 4 0 2 0 2 4 4 4 0 3 0 4 3 4 0 4 4 2 4 2 1 5 3 2 2 4 2 1 5 3 3 2 3 3 3 1 2 3 4 1 2 2 4 5 4 3 3 3 2 1 3 4 1 3 5 1 3 3 2 4 3 3 1 3 5 3 0 4 4 1 1 2 4 4 2 1 6 3 2 3 4 1 2 0 2 1 1 2 2 0 3 2 3 2 5 5 2 3 4 2 0 1 4 2 3 2 3 3 2 3 2 1 2 2 2 5 3 2 4 1 3 5 3 2 2 2 4 2 2 0 3 3 2 4 2 3 0 5 3 1 4 4 2 4 3 2 1 4 2 1 3 1 4 5 5 1 3 0 3 4 2 4 3 1 5 4 3 2 4 3 3 3 2 2 3 2 2 2 4 3 4 2 5 2 4 2 4 3 4 3 2 1 3 1 4 3 3 2 3 1 1 5 2 2 3 2 4 4 3 0 1 5 4 2 1 2 1 4 2 2 2 2 2 3 1 1 3 2 4 3 4 2 2 4 2 1 3 1 1 1 2 3 4 5 2 2 5 2 5 3 1 2 1 3 3 1 2 2 3 2 1 3 2 1 2 3 3 3 1 1 2 6 3 3 5 3 1 1 2 3 1 4 2 0 1 4 2 3 2 2 3 5 4 1 3 3 2 3 4 1 0 2 3 2 2 6 1 2 2 3 5 3 2 2 4 2 1 3 4 2 3 2 3 4 0 2 1 1 1 4 2 1 2 3 3 3 2 3 4 3 2 3 3 3 2 3 4 2 2 2 1 2 3 2 3 4 1 2 4 3 2 3 3 1 0 4 4 1 2 3 0 1 7 4 3 3 3 3 3 2 2 3 0 2 3 3 3 3 4 1 3 3 2 4 3 3 2 3 1 1 6 2 3 1 2 4 1 4 1 0 0 2 3 3 1 2 1 1 6 4 2 1 2 3 2 3 3 4 3 1 3 3 6 6 2 1 5 1 2 5 1 2 2 2 3 4 4 3 4 4 5 2 3 1 2 3 1 1 1 5 2 3 2 5 2 1 1 3 3 5 4 2 1 2 3 4 2 1 0 2 1 4 2 1 2 1 2 4 1 5 3 1 2 1 2 5 2 1 1 4 3 1 3 1 1 2 1 2 2 2 5 4 1 2 2 5 1 0 7 1 1 3 2 0 4 2 5 3 2 4 2 2 3 1 3 4 1 3 2 0 3 2 3 3 4 2 3 4 5 2 4 3 3 2 1 3 2 2 4 2 2 1 3 3 4 6 3 3 2 0 4 4 3 3 3 3 2 2 2 2 1 4 2 1 2 1 2 3 3 5 3 2 2 2 2 2 1 5 3 1 1 5 4 2 2 1 4 2 2 3 3 1 4 4 4 2 3 6 2 4 2 3 2 2 3 0 3 4 0 2 5 1 2 3 3 3 2 2 1 3 1 3 3 2 1 4 1 2 1 1 3 3 1 3 5 2 2 2 0 4 2 4 4 3 2 1 2 3 3 3 2 1 1 2 2 1 4 1 2 2 2 3 4 1 2 2 1 5 3 0 2 4 2 1 3 2 1 2 2 1 3 2 4 3 1 2 3 2 2 1 3 2 5 4 1 2 2 4 0 1 2 3 3 2 2 2 4 1 0 0 3 2 3 3 2 2 4 2 4 4 2 3 1 2 2 1 1 4 0 0 2 3 3 2 2 3 1 2 1 1 3 2 5 4 7 1 3 2 1 3 3 4 0 2 2 2 5 2 1 1 1 4 3 2 0 3 1 3 1 4 1 2 2 2 2 2 3 2 3 2 3 5 1 2 2 1 4 2 2 0 3 6 3 4 0 4 2 2 0 1\nG: 3 3 6 2 1 1 0 3 3 1 0 2 5 1 6 3 4 2 1 3 3 1 3 2 4 2 2 3 4 4 1 4 4 1 0 4 4 4 4 5 3 1 4 3 4 4 1 1 2 5 4 4 2 4 1 1 1 5 2 3 2 0 1 1 4 3 4 3 1 2 1 1 3 3 1 2 1 3 3 4 4 1 5 3 2 3 3 2 4 3 3 2 4 3 3 2 2 1 5 0 4 3 2 3 0 3 4 4 3 0 2 1 5 3 4 1 2 1 5 2 2 3 3 2 1 3 3 2 4 4 3 2 1 2 4 3 5 4 4 3 3 4 2 3 2 3 1 4 4 0 1 1 4 3 3 1 2 3 2 3 2 3 2 0 4 1 2 3 2 5 2 2 1 3 2 2 4 4 4 1 2 3 3 3 5 3 3 2 2 2 1 1 2 0 1 4 1 2 3 2 1 2 3 4 1 2 2 2 2 5 3 5 2 1 1 2 4 6 2 3 3 2 2 0 3 2 3 2 3 3 3 0 3 1 3 2 1 3 1 1 3 3 3 1 3 3 1 5 2 3 2 3 2 5 3 6 5 3 2 2 1 2 1 0 3 2 3 3 2 3 3 2 1 1 1 3 2 2 3 4 5 1 2 4 4 3 1 1 2 1 4 2 2 1 4 3 3 2 4 2 2 3 2 3 3 5 1 3 4 1 0 1 2 2 2 0 1 2 2 2 1 3 2 1 3 0 2 3 4 4 2 1 3 3 2 1 2 5 3 2 2 4 2 2 3 3 3 3 3 2 3 2 2 4 2 4 1 2 3 1 1 1 4 5 4 2 1 1 1 5 2 2 1 4 5 3 2 2 3 1 3 2 2 3 2 1 3 4 5 2 1 3 3 1 3 2 2 5 1 1 3 2 2 2 4 5 1 3 2 1 1 3 4 3 2 1 3 3 3 3 5 6 1 4 1 3 0 2 4 2 4 0 3 0 1 3 4 4 1 5 1 2 1 3 1 1 4 2 1 3 0 0 2 3 2 2 1 2 2 6 3 4 5 4 1 3 1 3 2 5 3 4 1 1 3 3 6 3 2 3 2 3 3 2 4 4 2 1 5 5 3 5 0 0 2 2 1 0 6 2 3 2 5 2 1 3 1 5 3 3 4 2 2 2 1 3 3 3 2 3 4 1 2 1 3 1 2 2 0 3 5 2 2 3 3 0 2 1 2 2 1 3 0 4 1 3 1 3 1 0 2 0 4 2 2 3 1 3 1 1 3 2 1 4 1 2 2 3 4 2 0 3 1 1 2 3 2 0 1 2 4 1 1 3 3 4 2 0 2 4 3 2 3 1 2 1 2 3 3 1 2 1 4 2 1 3 2 4 1 2 4 3 4 1 4 3 3 1 1 1 2 4 3 2 4 1 5 3 1 2 3 3 4 4 3 4 3 4 3 0 4 4 2 3 2 4 1 5 1 0 2 1 2 1 2 1 4 1 3 5 2 5 4 2 2 2 3 2 2 1 1 0 1 3 0 2 1 5 0 2 3 0 3 2 5 4 0 3 3 3 3 1 2 2 2 4 3 5 3 2 2 5 1 3 3 1 2 4 2 0 3 4 4 1 3 3 2 1 4 6 5 2 3 0 3 3 3 1 3 1 4 3 4 4 1 2 3 1 3 2 2 2 2 3 4 0 3 0 3 3 3 3 1 3 3 2 3 3 0 4 1 3 1 2 2 3 2 2 2 2 3 2 4 4 2 3 4 4 1 1 2 3 2 2 5 1 6 0 2 1 3 1 3 5 4 2 2 1 2 8 2 3 4 4 3 3 4 3 3 3 1 2 2 4 4 3 0 3 2 2 4 1 5 1 4 1 3 4 1 3 2 4 2 4 1 2 3 4 3 2 1 2 4 2 2 3 3 3 1 3 1 2 2 4 1 3 3 3 3 0 3 3 3 1 2 1 1 2 2 4 3 2 6 1 0 3 4 3 5 3 3 1 4 0 5 2 2 2 2 4 2 2 1 2 2 2 5 2 2 1 2 3 4 3 2 2 4 3 4 2 3 3 3 1 4 3 2 1 5 4 1 3 1 4 1 2 3 4 4 1 4 3 2 4 1 0 3 6 3 0 1 3 3 1 3 2 2 3\nT: 5 3 1 3 4 5 6 1 3 3 3 3 1 1 1 2 4 2 4 4 1 2 2 3 5 3 3 2 0 1 4 4 0 2 2 3 3 1 2 3 0 6 1 2 3 3 4 6 2 1 2 1 2 1 5 5 6 3 2 0 2 5 7 4 2 3 4 1 2 4 1 4 2 4 4 2 3 1 2 4 2 1 2 3 3 4 3 3 1 2 1 6 3 1 3 2 2 3 2 3 2 4 0 2 5 2 3 0 2 3 3 4 4 3 2 2 3 2 3 1 2 1 2 2 4 3 1 4 1 2 3 6 3 2 1 1 2 0 4 1 2 2 2 3 1 3 1 2 1 2 4 2 3 1 1 3 2 1 3 3 4 2 1 5 0 5 1 2 3 1 2 1 2 2 1 3 3 2 2 1 4 2 1 4 1 0 2 3 1 5 4 3 1 5 4 0 3 5 3 3 3 2 3 4 2 2 3 2 4 2 3 3 2 7 6 2 3 2 2 3 3 4 2 2 2 2 1 2 2 5 2 4 2 2 2 4 4 2 3 2 3 5 3 3 2 2 2 2 1 1 2 3 0 1 3 1 2 4 2 2 2 0 4 6 4 3 4 4 1 2 0 1 1 5 6 2 2 4 1 3 1 1 0 1 2 4 3 5 1 1 1 4 2 2 1 3 1 1 1 2 3 2 4 2 3 2 1 1 1 2 4 5 1 3 2 2 3 1 6 3 2 5 1 2 1 3 4 3 1 0 3 2 3 1 1 2 3 0 4 3 1 2 4 2 4 2 1 2 2 4 2 3 5 4 2 2 2 1 2 5 2 3 3 2 2 4 4 5 4 0 4 3 4 2 0 3 3 3 5 2 2 2 2 4 4 5 3 3 2 4 4 6 4 2 2 2 4 3 3 3 3 3 1 2 0 3 4 2 4 3 3 5 2 2 2 3 2 4 2 0 1 1 4 1 3 4 4 3 4 4 1 1 1 3 3 3 2 2 0 1 3 3 2 1 3 4 2 3 1 2 5 4 4 5 5 2 4 3 3 0 2 2 1 2 2 2 3 3 3 0 3 1 4 4 2 4 0 4 3 1 3 4 4 0 1 2 3 2 1 2 4 1 5 1 1 4 5 4 2 2 0 2 1 3 2 3 2 3 2 3 3 4 1 1 2 2 1 4 2 3 3 2 2 1 3 5 2 3 3 3 1 5 3 3 2 2 4 3 2 1 4 3 6 2 3 4 2 2 2 5 4 3 2 2 3 1 2 1 4 3 1 2 3 2 0 3 4 0 4 4 4 2 4 1 0 3 3 5 3 2 1 0 3 2 2 2 3 1 5 1 1 1 2 3 3 4 1 0 0 3 5 5 1 3 5 3 3 1 4 4 3 3 1 2 3 2 4 1 4 5 2 1 4 1 2 2 2 2 2 5 2 3 2 3 2 1 2 0 0 4 4 2 4 1 3 0 1 2 3 2 2 3 2 4 1 2 3 1 5 1 2 1 0 1 4 4 2 5 1 5 3 2 1 4 2 3 2 2 2 4 3 5 4 4 2 3 3 2 4 4 4 3 1 1 3 1 3 3 3 3 2 1 3 1 3 3 3 1 4 3 4 0 1 4 3 3 3 1 2 2 1 1 4 4 3 1 2 5 3 3 0 4 3 3 2 1 2 3 2 3 1 2 2 1 3 0 3 3 3 3 1 3 2 4 2 1 2 1 4 3 6 2 3 1 4 3 4 1 2 1 2 5 3 1 5 4 1 1 4 4 3 2 2 5 2 2 3 4 3 1 1 4 4 4 3 4 3 3 3 1 3 1 3 4 1 4 2 4 4 3 4 4 2 2 2 0 2 5 4 3 3 4 2 6 2 3 3 3 4 1 1 2 4 2 1 3 2 2 3 4 2 1 4 4 3 2 3 2 4 0 2 3 3 2 3 3 3 1 2 4 1 2 2 2 2 4 2 1 1 1 1 3 1 4 2 1 3 4 1 3 2 3 3 3 2 5 2 3 2 1 0 2 2 3 2 3 1 3 4 2 3 4 1 3 2 4 4 5 3 2 3 2 4 3 2 1 4 3 2 4 6 4 1 1 5 3 2 1 2 3 1 4 1 1 1 3 3 2 3 1 4 0 3 2 2 2 3 4\n"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb8a2b2c71c756e018f5a83d831468c15eb3900c
| 221,662 |
ipynb
|
Jupyter Notebook
|
DeepAnT.ipynb
|
dyadav4/2020-HackIllinois-Catepillar-Probabilistic-Anomaly-Detection
|
c81f54f3fd29b9484bfd4ed765b2dc6b1291ae66
|
[
"MIT"
] | null | null | null |
DeepAnT.ipynb
|
dyadav4/2020-HackIllinois-Catepillar-Probabilistic-Anomaly-Detection
|
c81f54f3fd29b9484bfd4ed765b2dc6b1291ae66
|
[
"MIT"
] | null | null | null |
DeepAnT.ipynb
|
dyadav4/2020-HackIllinois-Catepillar-Probabilistic-Anomaly-Detection
|
c81f54f3fd29b9484bfd4ed765b2dc6b1291ae66
|
[
"MIT"
] | null | null | null | 50.94507 | 15,144 | 0.397208 |
[
[
[
"import numpy as np\nfrom numpy import array\n\nimport random\nfrom random import randint\nimport os\n\nimport matplotlib.pyplot as plt\nimport pandas as pd\n\nimport keras\nfrom keras.models import Sequential\nfrom keras.layers import Dense, Conv1D, Flatten, Activation, MaxPooling1D, Dropout\nfrom keras.optimizers import SGD",
"_____no_output_____"
],
[
"os.environ[\"CUDA_DEVICE_ORDER\"]=\"PCI_BUS_ID\"\nos.environ[\"CUDA_VISIBLE_DEVICES\"]=\"1\" #model will be trained on GPU 1",
"_____no_output_____"
],
[
" \"\"\"Hyperparameters\"\"\"\nw = 17280-500 # History window (number of time stamps taken into account) \n # i.e., filter(kernel) size \np_w = 5000 # Prediction window (number of time stampes required to be \n # predicted)\nn_features = 1 # Univariate time series\n\nkernel_size = 2 # Size of filter in conv layers\nnum_filt_1 = 32 # Number of filters in first conv layer\nnum_filt_2 = 32 # Number of filters in second conv layer\nnum_nrn_dl = 40 # Number of neurons in dense layer\nnum_nrn_ol = p_w # Number of neurons in output layer\n\nconv_strides = 1\npool_size_1 = 2 # Length of window of pooling layer 1\npool_size_2 = 2 # Length of window of pooling layer 2\npool_strides_1 = 2 # Stride of window of pooling layer 1\npool_strides_2 = 2 # Stride of window of pooling layer 2\n\nepochs = 30\ndropout_rate = 0.5 # Dropout rate in the fully connected layer\nlearning_rate = 2e-5 \nanm_det_thr = 0.8 # Threshold for classifying anomaly (0.5~0.8)",
"_____no_output_____"
],
[
"from hdf_helper import *\nfrom stat_helper import *\nfrom data_cleaning import *\n\nimport h5py",
"_____no_output_____"
],
[
"#df = pd.read_csv('data/datch_3.csv').drop(['Unnamed: 0'], axis = 1)\n\npath = 'dat/cleaned_dat/ch_27.csv.csv'\ndf_test = pd.read_csv(path, nrows=100)\n\nfloat_cols = [c for i, c in enumerate(df_test.columns) if i != 0]\nfloat64_cols = {c: np.float64 for c in float_cols}\n\ndf = pd.read_csv(path, engine='c', dtype=float64_cols).drop(['Unnamed: 0'], axis = 1)\ndf = df.replace(np.NAN, 0.0)\n\nzero_outliers = df.loc[:, (df == 0.0).all(axis=0)]\nreg_data = df.loc[:,(df != 0.0).any(axis=0)]\n\n#df = reduce_dataset_size(df, cluster_size = 50)\ndf = smooth_values(df)\nscaler = RobustScaler()\ndf = pd.DataFrame(scaler.fit_transform(df))\n",
"_____no_output_____"
],
[
"w = len(df.index) - p_w",
"_____no_output_____"
],
[
"\"\"\"\nData preprocessing\n\n\n\"\"\"\n# split a univariate sequence into samples\ndef split_sequence(sequence):\n X, y = list(), list()\n for i in range(len(sequence)):\n # find the end of this pattern\n end_ix = i + w\n out_end_ix = end_ix + p_w\n # check if we are beyond the sequence\n if out_end_ix > len(sequence):\n break\n # gather input and output parts of the pattern\n seq_x, seq_y = sequence[i:end_ix], sequence[end_ix:out_end_ix]\n X.append(seq_x)\n y.append(seq_y)\n return array(X), array(y)",
"_____no_output_____"
],
[
"# # define input sequence\n# for col in reg_data.columns:\n# sampl, labl = split_sequence(list(reg_data[col]))\nsamples = []\nlabels = []\n\nbatch_sampl, batch_labl = split_sequence(list(reg_data.ix[:,0]))\nsamples.append(batch_sampl)\nlabels.append(batch_labl)\n\nprint()\n\nfor i in range(1, len(reg_data.columns)):\n batch_sampl, batch_labl = split_sequence(list(reg_data.ix[:,i]))\n samples.append(batch_sampl)\n labels.append(batch_labl)\n \nbatch_sample = np.array(samples)\nbatch_label = np.array(labels)\n \nprint(batch_sample.shape)\n\n# summarize the data\n# for i in range(5):\n# print(X[i], Y[i])\n \n# 2. reshape from [samples, timesteps] into [samples, timesteps, features]\n\n# need to convert batch into 3D tensor of the form [batch_size, input_seq_len, n_features]\nbatch_sample = batch_sample.reshape((batch_sample.shape[0], batch_sample.shape[2], n_features))",
"\n(57, 1, 12280)\n"
],
[
"batch_label = batch_label.reshape((batch_label.shape[0],batch_label.shape[1],batch_label.shape[2]))\nprint(batch_label.shape)\nprint(batch_sample.shape)",
"(57, 1, 5000)\n(57, 12280, 1)\n"
],
[
" \"\"\"Generate model for predictor\"\"\"\nmodel = Sequential()\n\n# Convolutional Layer #1\n# Computes 32 features using a 1D filter(kernel) of with w with ReLU activation. \n# Padding is added to preserve width.\n# Input Tensor Shape: [batch_size, w, 1] / batch_size = len(batch_sample)\n# Output Tensor Shape: [batch_size, w, num_filt_1] (num_filt_1 = 32 feature vectors)\nmodel.add(Conv1D(filters=num_filt_1,\n kernel_size=kernel_size,\n strides=conv_strides,\n padding='valid',\n activation='relu',\n input_shape=(w, n_features)))\n\n# Pooling Layer #1\n# First max pooling layer with a filter of length 2 and stride of 2\n# Input Tensor Shape: [batch_size, w, num_filt_1]\n# Output Tensor Shape: [batch_size, 0.5 * w, num_filt_1]\n\nmodel.add(MaxPooling1D(pool_size=pool_size_1)) \n # strides=pool_strides_1, \n # padding='valid'))\n\n# Convolutional Layer #2\n# Computes 64 features using a 5x5 filter.\n# Padding is added to preserve width and height.\n# Input Tensor Shape: [batch_size, 0.5 * w, 32]\n# Output Tensor Shape: [batch_size, 0.5 * w, num_filt_1 * num_filt_2]\nmodel.add(Conv1D(filters=num_filt_2,\n kernel_size=kernel_size,\n strides=conv_strides,\n padding='valid',\n activation='relu'))\n\n# Max Pooling Layer #2\n# Second max pooling layer with a 2x2 filter and stride of 2\n# Input Tensor Shape: [batch_size, 0.5 * w, num_filt_1 * num_filt_2]\n# Output Tensor Shape: [batch_size, 0.25 * w, num_filt_1 * num_filt_2]\nmodel.add(MaxPooling1D(pool_size=pool_size_2))\n # strides=pool_strides_2, \n # padding='valid'\n \n# Flatten tensor into a batch of vectors\n# Input Tensor Shape: [batch_size, 0.25 * w, num_filt_1 * num_filt_2]\n# Output Tensor Shape: [batch_size, 0.25 * w * num_filt_1 * num_filt_2]\nmodel.add(Flatten())\n\n# Dense Layer (Output layer)\n# Densely connected layer with 1024 neurons\n# Input Tensor Shape: [batch_size, 0.25 * w * num_filt_1 * num_filt_2]\n# Output Tensor Shape: [batch_size, 1024]\nmodel.add(Dense(units=num_nrn_dl, activation='relu')) \n\n# Dropout\n# Prevents overfitting in deep neural networks\nmodel.add(Dropout(dropout_rate))\n\n# Output layer\n# Input Tensor Shape: [batch_size, 1024]\n# Output Tensor Shape: [batch_size, p_w]\nmodel.add(Dense(units=num_nrn_ol))\n\n# Summarize model structure\nmodel.summary()",
"Model: \"sequential_35\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\nconv1d_68 (Conv1D) (None, 12279, 32) 96 \n_________________________________________________________________\nmax_pooling1d_68 (MaxPooling (None, 6139, 32) 0 \n_________________________________________________________________\nconv1d_69 (Conv1D) (None, 6138, 32) 2080 \n_________________________________________________________________\nmax_pooling1d_69 (MaxPooling (None, 3069, 32) 0 \n_________________________________________________________________\nflatten_36 (Flatten) (None, 98208) 0 \n_________________________________________________________________\ndense_68 (Dense) (None, 40) 3928360 \n_________________________________________________________________\ndropout_34 (Dropout) (None, 40) 0 \n_________________________________________________________________\ndense_69 (Dense) (None, 5000) 205000 \n=================================================================\nTotal params: 4,135,536\nTrainable params: 4,135,536\nNon-trainable params: 0\n_________________________________________________________________\n"
],
[
" '''configure model'''\nmodel.compile(optimizer='adam', \n loss='mean_absolute_error')\n\n# sgd = keras.optimizers.SGD(lr=learning_rate, \n# decay=1e-6, \n# momentum=0.9, \n# nesterov=True)\n# model.compile(optimizer='sgd', \n# loss='mean_absolute_error', \n# metrics=['accuracy'])",
"_____no_output_____"
],
[
"'''\n\nmodel_fit = model.fit(batch_sample,\n batch_label,\n epochs=epochs,\n verbose=1)\n'''",
"_____no_output_____"
],
[
"for i in range(len(reg_data.columns)):\n sampl = batch_sample[i].reshape((1,batch_sample.shape[1],batch_sample.shape[2]))\n print(sampl.shape)\n labl = batch_label[i].reshape((batch_label.shape[1],batch_label.shape[2]))\n model.fit(sampl,\n labl,\n epochs=epochs,\n verbose=1)",
"(1, 12280, 1)\nEpoch 1/30\n1/1 [==============================] - 1s 623ms/step - loss: 3.0526e-07\nEpoch 2/30\n1/1 [==============================] - 0s 42ms/step - loss: 6.0533e-05\nEpoch 3/30\n1/1 [==============================] - 0s 39ms/step - loss: 5.7316e-05\nEpoch 4/30\n1/1 [==============================] - 0s 45ms/step - loss: 3.2684e-05\nEpoch 5/30\n1/1 [==============================] - 0s 41ms/step - loss: 2.6355e-06\nEpoch 6/30\n1/1 [==============================] - 0s 40ms/step - loss: 1.4367e-05\nEpoch 7/30\n1/1 [==============================] - 0s 36ms/step - loss: 1.0496e-05\nEpoch 8/30\n1/1 [==============================] - 0s 89ms/step - loss: 4.4095e-06\nEpoch 9/30\n1/1 [==============================] - 0s 41ms/step - loss: 5.9122e-06\nEpoch 10/30\n1/1 [==============================] - 0s 38ms/step - loss: 2.8654e-06\nEpoch 11/30\n1/1 [==============================] - 0s 48ms/step - loss: 8.5628e-07\nEpoch 12/30\n1/1 [==============================] - 0s 39ms/step - loss: 9.8239e-06\nEpoch 13/30\n1/1 [==============================] - 0s 44ms/step - loss: 1.0444e-05\nEpoch 14/30\n1/1 [==============================] - 0s 49ms/step - loss: 2.7317e-06\nEpoch 15/30\n1/1 [==============================] - 0s 42ms/step - loss: 1.1961e-05\nEpoch 16/30\n1/1 [==============================] - 0s 41ms/step - loss: 1.7055e-05\nEpoch 17/30\n1/1 [==============================] - 0s 39ms/step - loss: 1.3969e-05\nEpoch 18/30\n1/1 [==============================] - 0s 47ms/step - loss: 3.8641e-06\nEpoch 19/30\n1/1 [==============================] - 0s 63ms/step - loss: 1.2297e-05\nEpoch 20/30\n1/1 [==============================] - 0s 36ms/step - loss: 1.9472e-05\nEpoch 21/30\n1/1 [==============================] - 0s 42ms/step - loss: 1.8820e-05\nEpoch 22/30\n1/1 [==============================] - 0s 41ms/step - loss: 1.1326e-05\nEpoch 23/30\n1/1 [==============================] - 0s 41ms/step - loss: 2.1696e-06\nEpoch 24/30\n1/1 [==============================] - 0s 46ms/step - loss: 7.4264e-06\nEpoch 25/30\n1/1 [==============================] - 0s 45ms/step - loss: 5.4183e-06\nEpoch 26/30\n1/1 [==============================] - 0s 39ms/step - loss: 3.0105e-06\nEpoch 27/30\n1/1 [==============================] - 0s 41ms/step - loss: 3.9522e-06\nEpoch 28/30\n1/1 [==============================] - 0s 44ms/step - loss: 1.7475e-06\nEpoch 29/30\n1/1 [==============================] - 0s 47ms/step - loss: 3.4425e-07\nEpoch 30/30\n1/1 [==============================] - 0s 46ms/step - loss: 7.3781e-06\n(1, 12280, 1)\nEpoch 1/30\n1/1 [==============================] - 0s 40ms/step - loss: 4.1362e-04\nEpoch 2/30\n1/1 [==============================] - 0s 44ms/step - loss: 5.2565e-04\nEpoch 3/30\n1/1 [==============================] - 0s 44ms/step - loss: 5.8413e-04\nEpoch 4/30\n1/1 [==============================] - 0s 40ms/step - loss: 3.0447e-04\nEpoch 5/30\n1/1 [==============================] - 0s 42ms/step - loss: 4.9769e-04\nEpoch 6/30\n1/1 [==============================] - 0s 44ms/step - loss: 0.0064\nEpoch 7/30\n1/1 [==============================] - 0s 38ms/step - loss: 6.8801e-04\nEpoch 8/30\n1/1 [==============================] - 0s 52ms/step - loss: 6.0893e-04\nEpoch 9/30\n1/1 [==============================] - 0s 43ms/step - loss: 4.5433e-04\nEpoch 10/30\n1/1 [==============================] - 0s 46ms/step - loss: 4.7742e-04\nEpoch 11/30\n1/1 [==============================] - 0s 39ms/step - loss: 3.6052e-04\nEpoch 12/30\n1/1 [==============================] - 0s 44ms/step - loss: 2.4124e-04\nEpoch 13/30\n1/1 [==============================] - 0s 45ms/step - loss: 3.7509e-04\nEpoch 14/30\n1/1 [==============================] - 0s 54ms/step - loss: 4.7417e-04\nEpoch 15/30\n1/1 [==============================] - 0s 41ms/step - loss: 4.5152e-04\nEpoch 16/30\n1/1 [==============================] - 0s 37ms/step - loss: 3.0472e-04\nEpoch 17/30\n1/1 [==============================] - 0s 45ms/step - loss: 2.3822e-04\nEpoch 18/30\n1/1 [==============================] - 0s 36ms/step - loss: 3.8082e-04\nEpoch 19/30\n1/1 [==============================] - 0s 39ms/step - loss: 3.8783e-04\nEpoch 20/30\n1/1 [==============================] - 0s 40ms/step - loss: 3.1896e-04\nEpoch 21/30\n1/1 [==============================] - 0s 45ms/step - loss: 3.3587e-04\nEpoch 22/30\n1/1 [==============================] - 0s 43ms/step - loss: 3.5273e-04\nEpoch 23/30\n1/1 [==============================] - 0s 43ms/step - loss: 2.5386e-04\nEpoch 24/30\n1/1 [==============================] - 0s 43ms/step - loss: 1.5816e-04\nEpoch 25/30\n1/1 [==============================] - 0s 49ms/step - loss: 3.8051e-04\nEpoch 26/30\n1/1 [==============================] - 0s 45ms/step - loss: 4.4162e-04\nEpoch 27/30\n1/1 [==============================] - 0s 51ms/step - loss: 4.6701e-04\nEpoch 28/30\n1/1 [==============================] - 0s 43ms/step - loss: 3.5862e-04\nEpoch 29/30\n1/1 [==============================] - 0s 35ms/step - loss: 2.8049e-04\nEpoch 30/30\n1/1 [==============================] - 0s 46ms/step - loss: 3.6990e-04\n(1, 12280, 1)\nEpoch 1/30\n1/1 [==============================] - 0s 42ms/step - loss: 0.1641\nEpoch 2/30\n1/1 [==============================] - 0s 43ms/step - loss: 0.1498\nEpoch 3/30\n1/1 [==============================] - 0s 47ms/step - loss: 0.0010\nEpoch 4/30\n1/1 [==============================] - 0s 43ms/step - loss: 0.1990\nEpoch 5/30\n1/1 [==============================] - 0s 45ms/step - loss: 0.0805\nEpoch 6/30\n1/1 [==============================] - 0s 44ms/step - loss: 8.7328e-04\nEpoch 7/30\n1/1 [==============================] - 0s 43ms/step - loss: 9.3038e-04\nEpoch 8/30\n1/1 [==============================] - 0s 37ms/step - loss: 9.1908e-04\nEpoch 9/30\n1/1 [==============================] - 0s 43ms/step - loss: 8.5664e-04\nEpoch 10/30\n1/1 [==============================] - 0s 42ms/step - loss: 7.4169e-04\nEpoch 11/30\n1/1 [==============================] - 0s 43ms/step - loss: 6.7453e-04\nEpoch 12/30\n1/1 [==============================] - 0s 49ms/step - loss: 5.3176e-04\nEpoch 13/30\n1/1 [==============================] - 0s 41ms/step - loss: 5.1982e-04\nEpoch 14/30\n1/1 [==============================] - 0s 52ms/step - loss: 5.1598e-04\nEpoch 15/30\n1/1 [==============================] - 0s 43ms/step - loss: 4.4826e-04\nEpoch 16/30\n1/1 [==============================] - 0s 44ms/step - loss: 4.3694e-04\nEpoch 17/30\n1/1 [==============================] - 0s 41ms/step - loss: 4.3430e-04\nEpoch 18/30\n1/1 [==============================] - 0s 36ms/step - loss: 4.0403e-04\nEpoch 19/30\n1/1 [==============================] - 0s 41ms/step - loss: 3.7721e-04\nEpoch 20/30\n1/1 [==============================] - 0s 42ms/step - loss: 3.5241e-04\nEpoch 21/30\n1/1 [==============================] - 0s 43ms/step - loss: 3.7252e-04\nEpoch 22/30\n1/1 [==============================] - 0s 38ms/step - loss: 3.4857e-04\nEpoch 23/30\n1/1 [==============================] - 0s 41ms/step - loss: 2.9579e-04\nEpoch 24/30\n1/1 [==============================] - 0s 41ms/step - loss: 2.4503e-04\nEpoch 25/30\n1/1 [==============================] - 0s 45ms/step - loss: 2.3804e-04\nEpoch 26/30\n1/1 [==============================] - 0s 49ms/step - loss: 2.7570e-04\nEpoch 27/30\n1/1 [==============================] - 0s 45ms/step - loss: 2.9022e-04\nEpoch 28/30\n1/1 [==============================] - 0s 44ms/step - loss: 2.7130e-04\nEpoch 29/30\n1/1 [==============================] - 0s 45ms/step - loss: 2.5925e-04\nEpoch 30/30\n1/1 [==============================] - 0s 37ms/step - loss: 2.5149e-04\n(1, 12280, 1)\nEpoch 1/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.0082\nEpoch 2/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.0056\nEpoch 3/30\n1/1 [==============================] - 0s 41ms/step - loss: 4.2101e-04\nEpoch 4/30\n1/1 [==============================] - 0s 41ms/step - loss: 4.5770e-04\nEpoch 5/30\n1/1 [==============================] - 0s 41ms/step - loss: 4.4413e-04\nEpoch 6/30\n1/1 [==============================] - 0s 43ms/step - loss: 0.0069\nEpoch 7/30\n1/1 [==============================] - 0s 55ms/step - loss: 3.8659e-04\nEpoch 8/30\n1/1 [==============================] - 0s 44ms/step - loss: 3.6342e-04\nEpoch 9/30\n1/1 [==============================] - 0s 41ms/step - loss: 3.4269e-04\nEpoch 10/30\n1/1 [==============================] - 0s 41ms/step - loss: 3.0469e-04\nEpoch 11/30\n1/1 [==============================] - 0s 41ms/step - loss: 2.5712e-04\nEpoch 12/30\n1/1 [==============================] - 0s 44ms/step - loss: 2.4372e-04\nEpoch 13/30\n1/1 [==============================] - 0s 55ms/step - loss: 2.3680e-04\nEpoch 14/30\n1/1 [==============================] - 0s 45ms/step - loss: 2.6274e-04\nEpoch 15/30\n1/1 [==============================] - 0s 45ms/step - loss: 2.7612e-04\nEpoch 16/30\n1/1 [==============================] - 0s 52ms/step - loss: 2.6318e-04\nEpoch 17/30\n1/1 [==============================] - 0s 50ms/step - loss: 2.3746e-04\nEpoch 18/30\n1/1 [==============================] - 0s 46ms/step - loss: 2.2091e-04\nEpoch 19/30\n1/1 [==============================] - 0s 42ms/step - loss: 2.0912e-04\nEpoch 20/30\n1/1 [==============================] - 0s 45ms/step - loss: 2.0833e-04\nEpoch 21/30\n1/1 [==============================] - 0s 40ms/step - loss: 2.2006e-04\nEpoch 22/30\n1/1 [==============================] - 0s 37ms/step - loss: 2.1138e-04\nEpoch 23/30\n1/1 [==============================] - 0s 41ms/step - loss: 1.9211e-04\nEpoch 24/30\n1/1 [==============================] - 0s 46ms/step - loss: 1.8912e-04\nEpoch 25/30\n1/1 [==============================] - 0s 40ms/step - loss: 1.8351e-04\nEpoch 26/30\n1/1 [==============================] - 0s 50ms/step - loss: 1.9590e-04\nEpoch 27/30\n1/1 [==============================] - 0s 47ms/step - loss: 2.0407e-04\nEpoch 28/30\n1/1 [==============================] - 0s 50ms/step - loss: 1.9372e-04\nEpoch 29/30\n1/1 [==============================] - 0s 47ms/step - loss: 1.8299e-04\nEpoch 30/30\n1/1 [==============================] - 0s 42ms/step - loss: 1.8193e-04\n(1, 12280, 1)\nEpoch 1/30\n1/1 [==============================] - 0s 42ms/step - loss: 0.0919\nEpoch 2/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.1224\nEpoch 3/30\n1/1 [==============================] - 0s 43ms/step - loss: 3.8441e-04\nEpoch 4/30\n1/1 [==============================] - 0s 41ms/step - loss: 4.1436e-04\nEpoch 5/30\n1/1 [==============================] - 0s 45ms/step - loss: 4.0197e-04\nEpoch 6/30\n1/1 [==============================] - 0s 64ms/step - loss: 3.5064e-04\nEpoch 7/30\n1/1 [==============================] - 0s 66ms/step - loss: 2.5101e-04\nEpoch 8/30\n1/1 [==============================] - 0s 46ms/step - loss: 2.4707e-04\nEpoch 9/30\n1/1 [==============================] - 0s 39ms/step - loss: 2.7529e-04\nEpoch 10/30\n1/1 [==============================] - 0s 64ms/step - loss: 2.4428e-04\nEpoch 11/30\n1/1 [==============================] - 0s 48ms/step - loss: 0.0044\nEpoch 12/30\n1/1 [==============================] - 0s 48ms/step - loss: 3.1527e-04\nEpoch 13/30\n1/1 [==============================] - 0s 36ms/step - loss: 3.0295e-04\nEpoch 14/30\n1/1 [==============================] - 0s 54ms/step - loss: 3.0318e-04\nEpoch 15/30\n1/1 [==============================] - 0s 39ms/step - loss: 3.0832e-04\nEpoch 16/30\n1/1 [==============================] - 0s 45ms/step - loss: 3.0638e-04\nEpoch 17/30\n1/1 [==============================] - 0s 42ms/step - loss: 2.5418e-04\nEpoch 18/30\n1/1 [==============================] - 0s 42ms/step - loss: 1.9289e-04\nEpoch 19/30\n1/1 [==============================] - 0s 40ms/step - loss: 1.9998e-04\nEpoch 20/30\n1/1 [==============================] - 0s 43ms/step - loss: 2.3737e-04\nEpoch 21/30\n1/1 [==============================] - 0s 39ms/step - loss: 2.3981e-04\nEpoch 22/30\n1/1 [==============================] - 0s 46ms/step - loss: 2.1439e-04\nEpoch 23/30\n1/1 [==============================] - 0s 43ms/step - loss: 1.9725e-04\nEpoch 24/30\n1/1 [==============================] - 0s 41ms/step - loss: 2.1465e-04\nEpoch 25/30\n1/1 [==============================] - 0s 37ms/step - loss: 1.9214e-04\nEpoch 26/30\n1/1 [==============================] - 0s 37ms/step - loss: 1.8072e-04\nEpoch 27/30\n1/1 [==============================] - 0s 52ms/step - loss: 2.0314e-04\nEpoch 28/30\n1/1 [==============================] - 0s 43ms/step - loss: 2.1294e-04\nEpoch 29/30\n1/1 [==============================] - 0s 39ms/step - loss: 1.9786e-04\nEpoch 30/30\n1/1 [==============================] - 0s 46ms/step - loss: 1.7323e-04\n(1, 12280, 1)\nEpoch 1/30\n1/1 [==============================] - 0s 37ms/step - loss: 1.6895e-04\nEpoch 2/30\n1/1 [==============================] - 0s 38ms/step - loss: 2.0164e-04\nEpoch 3/30\n1/1 [==============================] - 0s 40ms/step - loss: 2.0650e-04\nEpoch 4/30\n1/1 [==============================] - 0s 38ms/step - loss: 1.8578e-04\nEpoch 5/30\n1/1 [==============================] - 0s 37ms/step - loss: 1.7998e-04\nEpoch 6/30\n1/1 [==============================] - 0s 38ms/step - loss: 1.8315e-04\nEpoch 7/30\n1/1 [==============================] - 0s 43ms/step - loss: 1.7097e-04\nEpoch 8/30\n1/1 [==============================] - 0s 41ms/step - loss: 1.6704e-04\nEpoch 9/30\n1/1 [==============================] - 0s 42ms/step - loss: 1.8838e-04\nEpoch 10/30\n1/1 [==============================] - 0s 38ms/step - loss: 1.9824e-04\nEpoch 11/30\n1/1 [==============================] - 0s 49ms/step - loss: 1.7940e-04\nEpoch 12/30\n1/1 [==============================] - 0s 46ms/step - loss: 1.6098e-04\nEpoch 13/30\n1/1 [==============================] - 0s 39ms/step - loss: 1.5508e-04\nEpoch 14/30\n1/1 [==============================] - 0s 43ms/step - loss: 1.8299e-04\nEpoch 15/30\n1/1 [==============================] - 0s 45ms/step - loss: 1.9047e-04\nEpoch 16/30\n1/1 [==============================] - 0s 40ms/step - loss: 1.6811e-04\nEpoch 17/30\n1/1 [==============================] - 0s 40ms/step - loss: 1.7043e-04\nEpoch 18/30\n1/1 [==============================] - 0s 41ms/step - loss: 1.7793e-04\nEpoch 19/30\n1/1 [==============================] - 0s 42ms/step - loss: 1.6224e-04\nEpoch 20/30\n1/1 [==============================] - 0s 48ms/step - loss: 1.5460e-04\nEpoch 21/30\n1/1 [==============================] - 0s 40ms/step - loss: 1.6891e-04\nEpoch 22/30\n1/1 [==============================] - 0s 38ms/step - loss: 1.8489e-04\nEpoch 23/30\n1/1 [==============================] - 0s 47ms/step - loss: 1.7057e-04\nEpoch 24/30\n1/1 [==============================] - 0s 42ms/step - loss: 1.4622e-04\nEpoch 25/30\n1/1 [==============================] - 0s 41ms/step - loss: 1.5095e-04\nEpoch 26/30\n1/1 [==============================] - 0s 40ms/step - loss: 1.8091e-04\nEpoch 27/30\n1/1 [==============================] - 0s 43ms/step - loss: 1.9024e-04\nEpoch 28/30\n1/1 [==============================] - 0s 46ms/step - loss: 1.6938e-04\nEpoch 29/30\n1/1 [==============================] - 0s 39ms/step - loss: 1.6432e-04\nEpoch 30/30\n1/1 [==============================] - 0s 40ms/step - loss: 1.6883e-04\n(1, 12280, 1)\nEpoch 1/30\n1/1 [==============================] - 0s 48ms/step - loss: 0.0115\nEpoch 2/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.0623\nEpoch 3/30\n1/1 [==============================] - 0s 38ms/step - loss: 3.2203e-04\nEpoch 4/30\n1/1 [==============================] - 0s 45ms/step - loss: 3.4759e-04\nEpoch 5/30\n1/1 [==============================] - 0s 44ms/step - loss: 3.3675e-04\nEpoch 6/30\n1/1 [==============================] - 0s 46ms/step - loss: 2.8933e-04\nEpoch 7/30\n1/1 [==============================] - 0s 40ms/step - loss: 2.0271e-04\nEpoch 8/30\n1/1 [==============================] - 0s 40ms/step - loss: 2.1371e-04\nEpoch 9/30\n1/1 [==============================] - 0s 41ms/step - loss: 2.4048e-04\nEpoch 10/30\n1/1 [==============================] - 0s 40ms/step - loss: 2.1308e-04\nEpoch 11/30\n1/1 [==============================] - 0s 42ms/step - loss: 2.2087e-04\nEpoch 12/30\n1/1 [==============================] - 0s 44ms/step - loss: 2.1170e-04\nEpoch 13/30\n1/1 [==============================] - 0s 43ms/step - loss: 1.7696e-04\nEpoch 14/30\n1/1 [==============================] - 0s 35ms/step - loss: 2.0736e-04\nEpoch 15/30\n1/1 [==============================] - 0s 40ms/step - loss: 2.3431e-04\nEpoch 16/30\n1/1 [==============================] - 0s 43ms/step - loss: 2.2131e-04\nEpoch 17/30\n1/1 [==============================] - 0s 43ms/step - loss: 1.9850e-04\nEpoch 18/30\n1/1 [==============================] - 0s 40ms/step - loss: 1.6293e-04\nEpoch 19/30\n1/1 [==============================] - 0s 41ms/step - loss: 1.4182e-04\nEpoch 20/30\n1/1 [==============================] - 0s 41ms/step - loss: 1.6952e-04\nEpoch 21/30\n1/1 [==============================] - 0s 37ms/step - loss: 1.9831e-04\nEpoch 22/30\n1/1 [==============================] - 0s 37ms/step - loss: 1.9062e-04\nEpoch 23/30\n1/1 [==============================] - 0s 41ms/step - loss: 1.7962e-04\nEpoch 24/30\n1/1 [==============================] - 0s 37ms/step - loss: 1.6259e-04\nEpoch 25/30\n1/1 [==============================] - 0s 38ms/step - loss: 1.3248e-04\nEpoch 26/30\n1/1 [==============================] - 0s 36ms/step - loss: 1.4869e-04\nEpoch 27/30\n1/1 [==============================] - 0s 41ms/step - loss: 1.8134e-04\nEpoch 28/30\n1/1 [==============================] - 0s 37ms/step - loss: 1.8471e-04\nEpoch 29/30\n1/1 [==============================] - 0s 37ms/step - loss: 1.7581e-04\nEpoch 30/30\n1/1 [==============================] - 0s 40ms/step - loss: 1.4948e-04\n(1, 12280, 1)\nEpoch 1/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.1270\nEpoch 2/30\n1/1 [==============================] - 0s 43ms/step - loss: 0.3735\nEpoch 3/30\n1/1 [==============================] - 0s 40ms/step - loss: 3.4161e-04\nEpoch 4/30\n1/1 [==============================] - 0s 39ms/step - loss: 3.7920e-04\nEpoch 5/30\n1/1 [==============================] - 0s 40ms/step - loss: 3.7337e-04\nEpoch 6/30\n1/1 [==============================] - 0s 41ms/step - loss: 3.1766e-04\nEpoch 7/30\n1/1 [==============================] - 0s 43ms/step - loss: 2.1964e-04\nEpoch 8/30\n1/1 [==============================] - 0s 36ms/step - loss: 2.2751e-04\nEpoch 9/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.0143\nEpoch 10/30\n1/1 [==============================] - 0s 37ms/step - loss: 6.5357e-04\nEpoch 11/30\n1/1 [==============================] - 0s 38ms/step - loss: 3.7499e-04\nEpoch 12/30\n1/1 [==============================] - 0s 45ms/step - loss: 3.8571e-04\nEpoch 13/30\n1/1 [==============================] - 0s 40ms/step - loss: 3.5745e-04\nEpoch 14/30\n1/1 [==============================] - 0s 43ms/step - loss: 3.2577e-04\nEpoch 15/30\n1/1 [==============================] - 0s 38ms/step - loss: 2.4479e-04\nEpoch 16/30\n1/1 [==============================] - 0s 46ms/step - loss: 2.1102e-04\nEpoch 17/30\n1/1 [==============================] - 0s 41ms/step - loss: 2.4815e-04\nEpoch 18/30\n1/1 [==============================] - 0s 41ms/step - loss: 2.4619e-04\nEpoch 19/30\n1/1 [==============================] - 0s 41ms/step - loss: 2.3682e-04\nEpoch 20/30\n1/1 [==============================] - 0s 42ms/step - loss: 2.1905e-04\nEpoch 21/30\n1/1 [==============================] - 0s 38ms/step - loss: 0.0114\nEpoch 22/30\n1/1 [==============================] - 0s 41ms/step - loss: 2.8004e-04\nEpoch 23/30\n1/1 [==============================] - 0s 39ms/step - loss: 3.1124e-04\nEpoch 24/30\n1/1 [==============================] - 0s 42ms/step - loss: 2.8310e-04\nEpoch 25/30\n1/1 [==============================] - 0s 48ms/step - loss: 2.6030e-04\nEpoch 26/30\n1/1 [==============================] - 0s 47ms/step - loss: 2.3839e-04\nEpoch 27/30\n1/1 [==============================] - 0s 42ms/step - loss: 1.8367e-04\nEpoch 28/30\n1/1 [==============================] - 0s 38ms/step - loss: 1.6076e-04\nEpoch 29/30\n1/1 [==============================] - 0s 41ms/step - loss: 2.1109e-04\nEpoch 30/30\n1/1 [==============================] - 0s 47ms/step - loss: 2.3603e-04\n(1, 12280, 1)\nEpoch 1/30\n1/1 [==============================] - 0s 37ms/step - loss: 0.0851\nEpoch 2/30\n1/1 [==============================] - 0s 41ms/step - loss: 2.6031e-04\nEpoch 3/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.0371\nEpoch 4/30\n1/1 [==============================] - 0s 37ms/step - loss: 2.9267e-04\nEpoch 5/30\n1/1 [==============================] - 0s 42ms/step - loss: 2.8862e-04\nEpoch 6/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.0547\nEpoch 7/30\n1/1 [==============================] - 0s 38ms/step - loss: 2.9683e-04\nEpoch 8/30\n1/1 [==============================] - 0s 43ms/step - loss: 3.0024e-04\nEpoch 9/30\n1/1 [==============================] - 0s 40ms/step - loss: 2.8128e-04\nEpoch 10/30\n1/1 [==============================] - 0s 39ms/step - loss: 2.4109e-04\nEpoch 11/30\n1/1 [==============================] - 0s 40ms/step - loss: 1.9948e-04\nEpoch 12/30\n1/1 [==============================] - 0s 37ms/step - loss: 1.9997e-04\nEpoch 13/30\n1/1 [==============================] - 0s 41ms/step - loss: 2.1441e-04\nEpoch 14/30\n1/1 [==============================] - 0s 41ms/step - loss: 2.0813e-04\nEpoch 15/30\n1/1 [==============================] - 0s 38ms/step - loss: 2.1402e-04\nEpoch 16/30\n1/1 [==============================] - 0s 37ms/step - loss: 2.0776e-04\nEpoch 17/30\n1/1 [==============================] - 0s 41ms/step - loss: 1.8427e-04\nEpoch 18/30\n1/1 [==============================] - 0s 40ms/step - loss: 1.8657e-04\nEpoch 19/30\n1/1 [==============================] - 0s 40ms/step - loss: 1.9328e-04\nEpoch 20/30\n1/1 [==============================] - 0s 45ms/step - loss: 1.8659e-04\nEpoch 21/30\n1/1 [==============================] - 0s 36ms/step - loss: 1.8200e-04\nEpoch 22/30\n1/1 [==============================] - 0s 41ms/step - loss: 1.6553e-04\nEpoch 23/30\n1/1 [==============================] - 0s 43ms/step - loss: 1.5398e-04\nEpoch 24/30\n1/1 [==============================] - 0s 43ms/step - loss: 1.6337e-04\nEpoch 25/30\n1/1 [==============================] - 0s 42ms/step - loss: 1.6847e-04\nEpoch 26/30\n1/1 [==============================] - 0s 43ms/step - loss: 1.6469e-04\nEpoch 27/30\n1/1 [==============================] - 0s 41ms/step - loss: 1.6508e-04\nEpoch 28/30\n1/1 [==============================] - 0s 37ms/step - loss: 1.5605e-04\nEpoch 29/30\n1/1 [==============================] - 0s 50ms/step - loss: 1.4525e-04\nEpoch 30/30\n1/1 [==============================] - 0s 39ms/step - loss: 1.4819e-04\n(1, 12280, 1)\nEpoch 1/30\n1/1 [==============================] - 0s 47ms/step - loss: 0.0474\nEpoch 2/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.0802\nEpoch 3/30\n1/1 [==============================] - 0s 37ms/step - loss: 0.0677\nEpoch 4/30\n1/1 [==============================] - 0s 45ms/step - loss: 0.1398\nEpoch 5/30\n1/1 [==============================] - 0s 47ms/step - loss: 0.0251\nEpoch 6/30\n1/1 [==============================] - 0s 44ms/step - loss: 7.0391e-04\nEpoch 7/30\n1/1 [==============================] - 0s 40ms/step - loss: 7.3734e-04\nEpoch 8/30\n1/1 [==============================] - 0s 46ms/step - loss: 0.0020\nEpoch 9/30\n1/1 [==============================] - 0s 54ms/step - loss: 6.8841e-04\nEpoch 10/30\n1/1 [==============================] - 0s 45ms/step - loss: 6.3286e-04\nEpoch 11/30\n1/1 [==============================] - 0s 41ms/step - loss: 5.6043e-04\nEpoch 12/30\n1/1 [==============================] - 0s 42ms/step - loss: 4.6225e-04\nEpoch 13/30\n1/1 [==============================] - 0s 43ms/step - loss: 3.9959e-04\nEpoch 14/30\n1/1 [==============================] - 0s 43ms/step - loss: 3.5927e-04\nEpoch 15/30\n1/1 [==============================] - 0s 40ms/step - loss: 3.2299e-04\nEpoch 16/30\n1/1 [==============================] - 0s 47ms/step - loss: 3.4227e-04\nEpoch 17/30\n1/1 [==============================] - 0s 58ms/step - loss: 3.5030e-04\nEpoch 18/30\n1/1 [==============================] - 0s 42ms/step - loss: 3.4625e-04\nEpoch 19/30\n1/1 [==============================] - 0s 39ms/step - loss: 3.2948e-04\nEpoch 20/30\n1/1 [==============================] - 0s 86ms/step - loss: 3.0929e-04\nEpoch 21/30\n1/1 [==============================] - 0s 84ms/step - loss: 2.9777e-04\nEpoch 22/30\n1/1 [==============================] - 0s 40ms/step - loss: 2.7595e-04\nEpoch 23/30\n1/1 [==============================] - 0s 43ms/step - loss: 2.6226e-04\nEpoch 24/30\n1/1 [==============================] - 0s 40ms/step - loss: 2.3145e-04\nEpoch 25/30\n1/1 [==============================] - 0s 48ms/step - loss: 2.1957e-04\nEpoch 26/30\n1/1 [==============================] - 0s 41ms/step - loss: 2.3125e-04\nEpoch 27/30\n1/1 [==============================] - 0s 43ms/step - loss: 2.2795e-04\nEpoch 28/30\n1/1 [==============================] - 0s 45ms/step - loss: 2.3182e-04\nEpoch 29/30\n1/1 [==============================] - 0s 42ms/step - loss: 2.2497e-04\nEpoch 30/30\n1/1 [==============================] - 0s 41ms/step - loss: 2.1032e-04\n(1, 12280, 1)\nEpoch 1/30\n1/1 [==============================] - 0s 40ms/step - loss: 82.5153\nEpoch 2/30\n1/1 [==============================] - 0s 37ms/step - loss: 82.5152\nEpoch 3/30\n1/1 [==============================] - 0s 42ms/step - loss: 82.5152\nEpoch 4/30\n1/1 [==============================] - 0s 44ms/step - loss: 82.5152\nEpoch 5/30\n1/1 [==============================] - 0s 39ms/step - loss: 82.5151\nEpoch 6/30\n1/1 [==============================] - 0s 40ms/step - loss: 82.5150\nEpoch 7/30\n1/1 [==============================] - 0s 44ms/step - loss: 82.5149\nEpoch 8/30\n1/1 [==============================] - 0s 40ms/step - loss: 82.5148\nEpoch 9/30\n1/1 [==============================] - 0s 41ms/step - loss: 82.5148\nEpoch 10/30\n1/1 [==============================] - 0s 44ms/step - loss: 82.5147\nEpoch 11/30\n1/1 [==============================] - 0s 39ms/step - loss: 82.5145\nEpoch 12/30\n1/1 [==============================] - 0s 41ms/step - loss: 82.5144\nEpoch 13/30\n1/1 [==============================] - 0s 38ms/step - loss: 82.5143\nEpoch 14/30\n1/1 [==============================] - 0s 46ms/step - loss: 82.5142\nEpoch 15/30\n1/1 [==============================] - 0s 39ms/step - loss: 82.5140\nEpoch 16/30\n1/1 [==============================] - 0s 64ms/step - loss: 82.5139\nEpoch 17/30\n1/1 [==============================] - 0s 38ms/step - loss: 82.5138\nEpoch 18/30\n1/1 [==============================] - 0s 39ms/step - loss: 82.5136\nEpoch 19/30\n1/1 [==============================] - 0s 41ms/step - loss: 82.5135\nEpoch 20/30\n1/1 [==============================] - 0s 43ms/step - loss: 82.5133\nEpoch 21/30\n1/1 [==============================] - 0s 42ms/step - loss: 82.5132\nEpoch 22/30\n1/1 [==============================] - 0s 42ms/step - loss: 82.5130\nEpoch 23/30\n1/1 [==============================] - 0s 41ms/step - loss: 82.5129\nEpoch 24/30\n1/1 [==============================] - 0s 44ms/step - loss: 82.5127\nEpoch 25/30\n1/1 [==============================] - 0s 45ms/step - loss: 82.5126\nEpoch 26/30\n1/1 [==============================] - 0s 41ms/step - loss: 82.5124\nEpoch 27/30\n1/1 [==============================] - 0s 40ms/step - loss: 82.5122\nEpoch 28/30\n1/1 [==============================] - 0s 41ms/step - loss: 82.5121\nEpoch 29/30\n1/1 [==============================] - 0s 37ms/step - loss: 82.5119\nEpoch 30/30\n1/1 [==============================] - 0s 37ms/step - loss: 82.5117\n(1, 12280, 1)\nEpoch 1/30\n1/1 [==============================] - 0s 37ms/step - loss: 0.0038\nEpoch 2/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.0039\nEpoch 3/30\n1/1 [==============================] - 0s 44ms/step - loss: 0.0040\nEpoch 4/30\n1/1 [==============================] - 0s 42ms/step - loss: 0.0041\nEpoch 5/30\n1/1 [==============================] - 0s 43ms/step - loss: 0.0041\nEpoch 6/30\n1/1 [==============================] - 0s 43ms/step - loss: 0.0042\nEpoch 7/30\n1/1 [==============================] - 0s 37ms/step - loss: 0.0042\nEpoch 8/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.0042\nEpoch 9/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.0041\nEpoch 10/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.0041\nEpoch 11/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.0040\nEpoch 12/30\n1/1 [==============================] - 0s 37ms/step - loss: 0.0040\nEpoch 13/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.0039\nEpoch 14/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.0038\nEpoch 15/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.0037\nEpoch 16/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.0036\nEpoch 17/30\n1/1 [==============================] - 0s 38ms/step - loss: 0.0035\nEpoch 18/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.0034\nEpoch 19/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.0033\nEpoch 20/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.0031\nEpoch 21/30\n1/1 [==============================] - 0s 38ms/step - loss: 0.0030\nEpoch 22/30\n1/1 [==============================] - 0s 37ms/step - loss: 0.0029\nEpoch 23/30\n1/1 [==============================] - 0s 42ms/step - loss: 0.0027\nEpoch 24/30\n1/1 [==============================] - 0s 37ms/step - loss: 0.0026\nEpoch 25/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.0025\nEpoch 26/30\n1/1 [==============================] - 0s 46ms/step - loss: 0.0023\nEpoch 27/30\n1/1 [==============================] - 0s 43ms/step - loss: 0.0022\nEpoch 28/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.0020\nEpoch 29/30\n1/1 [==============================] - 0s 42ms/step - loss: 0.0019\nEpoch 30/30\n1/1 [==============================] - 0s 38ms/step - loss: 0.0017\n(1, 12280, 1)\nEpoch 1/30\n1/1 [==============================] - 0s 42ms/step - loss: 0.0015\nEpoch 2/30\n1/1 [==============================] - 0s 38ms/step - loss: 0.0014\nEpoch 3/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.0012\nEpoch 4/30\n1/1 [==============================] - 0s 46ms/step - loss: 0.0011\nEpoch 5/30\n1/1 [==============================] - 0s 44ms/step - loss: 9.1946e-04\nEpoch 6/30\n1/1 [==============================] - 0s 38ms/step - loss: 7.5297e-04\nEpoch 7/30\n1/1 [==============================] - 0s 38ms/step - loss: 5.9029e-04\nEpoch 8/30\n1/1 [==============================] - 0s 44ms/step - loss: 4.4553e-04\nEpoch 9/30\n1/1 [==============================] - 0s 43ms/step - loss: 3.5245e-04\nEpoch 10/30\n1/1 [==============================] - 0s 41ms/step - loss: 2.8021e-04\nEpoch 11/30\n1/1 [==============================] - 0s 41ms/step - loss: 3.2834e-04\nEpoch 12/30\n1/1 [==============================] - 0s 40ms/step - loss: 3.5718e-04\nEpoch 13/30\n1/1 [==============================] - 0s 39ms/step - loss: 4.3010e-04\nEpoch 14/30\n1/1 [==============================] - 0s 43ms/step - loss: 4.8415e-04\nEpoch 15/30\n1/1 [==============================] - 0s 39ms/step - loss: 5.1989e-04\nEpoch 16/30\n1/1 [==============================] - 0s 42ms/step - loss: 5.3502e-04\nEpoch 17/30\n1/1 [==============================] - 0s 40ms/step - loss: 5.3225e-04\nEpoch 18/30\n1/1 [==============================] - 0s 40ms/step - loss: 5.0509e-04\nEpoch 19/30\n1/1 [==============================] - 0s 39ms/step - loss: 4.6257e-04\nEpoch 20/30\n1/1 [==============================] - 0s 39ms/step - loss: 4.0940e-04\nEpoch 21/30\n1/1 [==============================] - 0s 38ms/step - loss: 3.4841e-04\nEpoch 22/30\n1/1 [==============================] - 0s 51ms/step - loss: 2.8580e-04\nEpoch 23/30\n1/1 [==============================] - 0s 37ms/step - loss: 2.4661e-04\nEpoch 24/30\n1/1 [==============================] - 0s 43ms/step - loss: 2.1325e-04\nEpoch 25/30\n1/1 [==============================] - 0s 41ms/step - loss: 2.2496e-04\nEpoch 26/30\n1/1 [==============================] - 0s 40ms/step - loss: 2.3916e-04\nEpoch 27/30\n1/1 [==============================] - 0s 42ms/step - loss: 2.6218e-04\nEpoch 28/30\n1/1 [==============================] - 0s 37ms/step - loss: 2.7097e-04\nEpoch 29/30\n1/1 [==============================] - 0s 47ms/step - loss: 2.6569e-04\nEpoch 30/30\n1/1 [==============================] - 0s 38ms/step - loss: 2.3671e-04\n(1, 12280, 1)\nEpoch 1/30\n1/1 [==============================] - 0s 38ms/step - loss: 2.1955e-04\nEpoch 2/30\n1/1 [==============================] - 0s 45ms/step - loss: 1.9214e-04\nEpoch 3/30\n1/1 [==============================] - 0s 41ms/step - loss: 1.9848e-04\nEpoch 4/30\n1/1 [==============================] - 0s 45ms/step - loss: 2.0403e-04\nEpoch 5/30\n1/1 [==============================] - 0s 41ms/step - loss: 2.1592e-04\nEpoch 6/30\n1/1 [==============================] - 0s 38ms/step - loss: 2.1725e-04\nEpoch 7/30\n1/1 [==============================] - 0s 50ms/step - loss: 2.0179e-04\nEpoch 8/30\n1/1 [==============================] - 0s 38ms/step - loss: 1.8657e-04\nEpoch 9/30\n1/1 [==============================] - 0s 42ms/step - loss: 1.7384e-04\nEpoch 10/30\n1/1 [==============================] - 0s 40ms/step - loss: 1.7313e-04\nEpoch 11/30\n1/1 [==============================] - 0s 45ms/step - loss: 1.8303e-04\nEpoch 12/30\n1/1 [==============================] - 0s 38ms/step - loss: 1.8070e-04\nEpoch 13/30\n1/1 [==============================] - 0s 38ms/step - loss: 1.7310e-04\nEpoch 14/30\n1/1 [==============================] - 0s 40ms/step - loss: 1.6717e-04\nEpoch 15/30\n1/1 [==============================] - 0s 39ms/step - loss: 1.5363e-04\nEpoch 16/30\n1/1 [==============================] - 0s 44ms/step - loss: 1.5924e-04\nEpoch 17/30\n1/1 [==============================] - 0s 40ms/step - loss: 1.6737e-04\nEpoch 18/30\n1/1 [==============================] - 0s 40ms/step - loss: 1.6503e-04\nEpoch 19/30\n1/1 [==============================] - 0s 37ms/step - loss: 1.6040e-04\nEpoch 20/30\n1/1 [==============================] - 0s 60ms/step - loss: 1.5707e-04\nEpoch 21/30\n1/1 [==============================] - 0s 35ms/step - loss: 1.4717e-04\nEpoch 22/30\n1/1 [==============================] - 0s 41ms/step - loss: 1.5573e-04\nEpoch 23/30\n1/1 [==============================] - 0s 43ms/step - loss: 1.5764e-04\nEpoch 24/30\n1/1 [==============================] - 0s 37ms/step - loss: 1.5369e-04\nEpoch 25/30\n1/1 [==============================] - 0s 39ms/step - loss: 1.5361e-04\nEpoch 26/30\n1/1 [==============================] - 0s 42ms/step - loss: 1.4866e-04\nEpoch 27/30\n1/1 [==============================] - 0s 40ms/step - loss: 1.4939e-04\nEpoch 28/30\n1/1 [==============================] - 0s 38ms/step - loss: 1.4805e-04\nEpoch 29/30\n1/1 [==============================] - 0s 37ms/step - loss: 1.5529e-04\nEpoch 30/30\n1/1 [==============================] - 0s 39ms/step - loss: 1.5101e-04\n(1, 12280, 1)\nEpoch 1/30\n1/1 [==============================] - 0s 40ms/step - loss: 1.4625e-04\nEpoch 2/30\n1/1 [==============================] - 0s 45ms/step - loss: 1.4398e-04\nEpoch 3/30\n1/1 [==============================] - 0s 39ms/step - loss: 1.4035e-04\nEpoch 4/30\n1/1 [==============================] - 0s 41ms/step - loss: 1.4604e-04\nEpoch 5/30\n1/1 [==============================] - 0s 38ms/step - loss: 1.4832e-04\nEpoch 6/30\n1/1 [==============================] - 0s 39ms/step - loss: 1.4087e-04\nEpoch 7/30\n1/1 [==============================] - 0s 37ms/step - loss: 1.3941e-04\nEpoch 8/30\n1/1 [==============================] - 0s 44ms/step - loss: 1.3690e-04\nEpoch 9/30\n1/1 [==============================] - 0s 36ms/step - loss: 1.3720e-04\nEpoch 10/30\n1/1 [==============================] - 0s 46ms/step - loss: 1.3731e-04\nEpoch 11/30\n1/1 [==============================] - 0s 41ms/step - loss: 1.4700e-04\nEpoch 12/30\n1/1 [==============================] - 0s 42ms/step - loss: 1.4312e-04\nEpoch 13/30\n1/1 [==============================] - 0s 40ms/step - loss: 1.4087e-04\nEpoch 14/30\n1/1 [==============================] - 0s 43ms/step - loss: 1.3825e-04\nEpoch 15/30\n1/1 [==============================] - 0s 37ms/step - loss: 1.3810e-04\nEpoch 16/30\n1/1 [==============================] - 0s 42ms/step - loss: 1.4201e-04\nEpoch 17/30\n1/1 [==============================] - 0s 43ms/step - loss: 1.4715e-04\nEpoch 18/30\n1/1 [==============================] - 0s 39ms/step - loss: 1.3986e-04\nEpoch 19/30\n1/1 [==============================] - 0s 38ms/step - loss: 1.4176e-04\nEpoch 20/30\n1/1 [==============================] - 0s 40ms/step - loss: 1.3885e-04\nEpoch 21/30\n1/1 [==============================] - 0s 43ms/step - loss: 1.4064e-04\nEpoch 22/30\n1/1 [==============================] - 0s 39ms/step - loss: 1.4016e-04\nEpoch 23/30\n1/1 [==============================] - 0s 38ms/step - loss: 1.4080e-04\nEpoch 24/30\n1/1 [==============================] - 0s 42ms/step - loss: 1.3489e-04\nEpoch 25/30\n1/1 [==============================] - 0s 36ms/step - loss: 1.3025e-04\nEpoch 26/30\n1/1 [==============================] - 0s 43ms/step - loss: 1.3028e-04\nEpoch 27/30\n1/1 [==============================] - 0s 40ms/step - loss: 1.3745e-04\nEpoch 28/30\n1/1 [==============================] - 0s 40ms/step - loss: 1.4128e-04\nEpoch 29/30\n1/1 [==============================] - 0s 42ms/step - loss: 1.4748e-04\nEpoch 30/30\n1/1 [==============================] - 0s 45ms/step - loss: 1.4076e-04\n(1, 12280, 1)\nEpoch 1/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.1067\nEpoch 2/30\n1/1 [==============================] - 0s 43ms/step - loss: 0.1210\nEpoch 3/30\n1/1 [==============================] - 0s 51ms/step - loss: 3.1608e-04\nEpoch 4/30\n1/1 [==============================] - 0s 39ms/step - loss: 3.4093e-04\nEpoch 5/30\n1/1 [==============================] - 0s 42ms/step - loss: 3.3573e-04\nEpoch 6/30\n1/1 [==============================] - 0s 39ms/step - loss: 2.9036e-04\nEpoch 7/30\n1/1 [==============================] - 0s 40ms/step - loss: 2.0225e-04\nEpoch 8/30\n1/1 [==============================] - 0s 40ms/step - loss: 2.0889e-04\nEpoch 9/30\n1/1 [==============================] - 0s 44ms/step - loss: 2.3399e-04\nEpoch 10/30\n1/1 [==============================] - 0s 40ms/step - loss: 2.0975e-04\nEpoch 11/30\n1/1 [==============================] - 0s 49ms/step - loss: 2.1550e-04\nEpoch 12/30\n1/1 [==============================] - 0s 38ms/step - loss: 2.0639e-04\nEpoch 13/30\n1/1 [==============================] - 0s 40ms/step - loss: 1.7103e-04\nEpoch 14/30\n1/1 [==============================] - 0s 43ms/step - loss: 2.0255e-04\nEpoch 15/30\n1/1 [==============================] - 0s 39ms/step - loss: 2.2793e-04\nEpoch 16/30\n1/1 [==============================] - 0s 46ms/step - loss: 2.1249e-04\nEpoch 17/30\n1/1 [==============================] - 0s 43ms/step - loss: 1.9367e-04\nEpoch 18/30\n1/1 [==============================] - 0s 39ms/step - loss: 1.6115e-04\nEpoch 19/30\n1/1 [==============================] - 0s 42ms/step - loss: 1.3983e-04\nEpoch 20/30\n1/1 [==============================] - 0s 50ms/step - loss: 1.6092e-04\nEpoch 21/30\n1/1 [==============================] - 0s 40ms/step - loss: 1.8786e-04\nEpoch 22/30\n1/1 [==============================] - 0s 38ms/step - loss: 1.8291e-04\nEpoch 23/30\n1/1 [==============================] - 0s 43ms/step - loss: 1.7218e-04\nEpoch 24/30\n1/1 [==============================] - 0s 41ms/step - loss: 1.5439e-04\nEpoch 25/30\n1/1 [==============================] - 0s 42ms/step - loss: 1.2719e-04\nEpoch 26/30\n1/1 [==============================] - 0s 41ms/step - loss: 1.4595e-04\nEpoch 27/30\n1/1 [==============================] - 0s 51ms/step - loss: 1.7013e-04\nEpoch 28/30\n1/1 [==============================] - 0s 42ms/step - loss: 1.6606e-04\nEpoch 29/30\n1/1 [==============================] - 0s 42ms/step - loss: 1.6189e-04\nEpoch 30/30\n1/1 [==============================] - 0s 35ms/step - loss: 1.4434e-04\n(1, 12280, 1)\nEpoch 1/30\n1/1 [==============================] - 0s 41ms/step - loss: 1.2643e-04\nEpoch 2/30\n1/1 [==============================] - 0s 40ms/step - loss: 1.4427e-04\nEpoch 3/30\n1/1 [==============================] - 0s 42ms/step - loss: 1.7206e-04\nEpoch 4/30\n1/1 [==============================] - 0s 43ms/step - loss: 1.7132e-04\nEpoch 5/30\n1/1 [==============================] - 0s 42ms/step - loss: 1.5889e-04\nEpoch 6/30\n1/1 [==============================] - 0s 39ms/step - loss: 1.3934e-04\nEpoch 7/30\n1/1 [==============================] - 0s 49ms/step - loss: 1.1982e-04\nEpoch 8/30\n1/1 [==============================] - 0s 43ms/step - loss: 1.3871e-04\nEpoch 9/30\n1/1 [==============================] - 0s 39ms/step - loss: 1.6725e-04\nEpoch 10/30\n1/1 [==============================] - 0s 50ms/step - loss: 1.5951e-04\nEpoch 11/30\n1/1 [==============================] - 0s 38ms/step - loss: 1.5945e-04\nEpoch 12/30\n1/1 [==============================] - 0s 37ms/step - loss: 1.5013e-04\nEpoch 13/30\n1/1 [==============================] - 0s 42ms/step - loss: 1.2492e-04\nEpoch 14/30\n1/1 [==============================] - 0s 41ms/step - loss: 1.3451e-04\nEpoch 15/30\n1/1 [==============================] - 0s 45ms/step - loss: 1.5693e-04\nEpoch 16/30\n1/1 [==============================] - 0s 38ms/step - loss: 1.5830e-04\nEpoch 17/30\n1/1 [==============================] - 0s 43ms/step - loss: 1.6077e-04\nEpoch 18/30\n1/1 [==============================] - 0s 45ms/step - loss: 1.4233e-04\nEpoch 19/30\n1/1 [==============================] - 0s 46ms/step - loss: 1.2828e-04\nEpoch 20/30\n1/1 [==============================] - 0s 38ms/step - loss: 1.4369e-04\nEpoch 21/30\n1/1 [==============================] - 0s 38ms/step - loss: 1.5478e-04\nEpoch 22/30\n1/1 [==============================] - 0s 42ms/step - loss: 1.4818e-04\nEpoch 23/30\n1/1 [==============================] - 0s 37ms/step - loss: 1.4445e-04\nEpoch 24/30\n1/1 [==============================] - 0s 40ms/step - loss: 1.3406e-04\nEpoch 25/30\n1/1 [==============================] - 0s 36ms/step - loss: 1.2494e-04\nEpoch 26/30\n1/1 [==============================] - 0s 52ms/step - loss: 1.3685e-04\nEpoch 27/30\n1/1 [==============================] - 0s 44ms/step - loss: 1.5869e-04\nEpoch 28/30\n1/1 [==============================] - 0s 44ms/step - loss: 1.5938e-04\nEpoch 29/30\n1/1 [==============================] - 0s 41ms/step - loss: 1.4977e-04\nEpoch 30/30\n1/1 [==============================] - 0s 41ms/step - loss: 1.3305e-04\n(1, 12280, 1)\nEpoch 1/30\n1/1 [==============================] - 0s 43ms/step - loss: 1.1964e-04\nEpoch 2/30\n1/1 [==============================] - 0s 37ms/step - loss: 1.3340e-04\nEpoch 3/30\n1/1 [==============================] - 0s 38ms/step - loss: 1.5936e-04\nEpoch 4/30\n1/1 [==============================] - 0s 40ms/step - loss: 1.5530e-04\nEpoch 5/30\n1/1 [==============================] - 0s 42ms/step - loss: 0.0960\nEpoch 6/30\n1/1 [==============================] - 0s 37ms/step - loss: 2.1753e-04\nEpoch 7/30\n1/1 [==============================] - 0s 43ms/step - loss: 2.1764e-04\nEpoch 8/30\n1/1 [==============================] - 0s 44ms/step - loss: 1.9176e-04\nEpoch 9/30\n1/1 [==============================] - 0s 43ms/step - loss: 1.8743e-04\nEpoch 10/30\n1/1 [==============================] - 0s 36ms/step - loss: 1.9830e-04\nEpoch 11/30\n1/1 [==============================] - 0s 42ms/step - loss: 1.6774e-04\nEpoch 12/30\n1/1 [==============================] - 0s 43ms/step - loss: 1.3821e-04\nEpoch 13/30\n1/1 [==============================] - 0s 43ms/step - loss: 1.6281e-04\nEpoch 14/30\n1/1 [==============================] - 0s 40ms/step - loss: 1.8626e-04\nEpoch 15/30\n1/1 [==============================] - 0s 44ms/step - loss: 1.7820e-04\nEpoch 16/30\n1/1 [==============================] - 0s 42ms/step - loss: 1.5525e-04\nEpoch 17/30\n1/1 [==============================] - 0s 47ms/step - loss: 1.3885e-04\nEpoch 18/30\n1/1 [==============================] - 0s 42ms/step - loss: 1.5511e-04\nEpoch 19/30\n1/1 [==============================] - 0s 37ms/step - loss: 1.5708e-04\nEpoch 20/30\n1/1 [==============================] - 0s 42ms/step - loss: 1.4842e-04\nEpoch 21/30\n1/1 [==============================] - 0s 44ms/step - loss: 1.6501e-04\nEpoch 22/30\n1/1 [==============================] - 0s 41ms/step - loss: 1.7046e-04\nEpoch 23/30\n1/1 [==============================] - 0s 37ms/step - loss: 1.5307e-04\nEpoch 24/30\n1/1 [==============================] - 0s 41ms/step - loss: 1.4508e-04\nEpoch 25/30\n1/1 [==============================] - 0s 40ms/step - loss: 1.4555e-04\nEpoch 26/30\n1/1 [==============================] - 0s 46ms/step - loss: 1.5797e-04\nEpoch 27/30\n1/1 [==============================] - 0s 42ms/step - loss: 1.5756e-04\nEpoch 28/30\n1/1 [==============================] - 0s 45ms/step - loss: 1.4359e-04\nEpoch 29/30\n1/1 [==============================] - 0s 42ms/step - loss: 1.4619e-04\nEpoch 30/30\n1/1 [==============================] - 0s 45ms/step - loss: 1.5510e-04\n(1, 12280, 1)\nEpoch 1/30\n1/1 [==============================] - 0s 47ms/step - loss: 1.4887e-04\nEpoch 2/30\n1/1 [==============================] - 0s 41ms/step - loss: 1.4180e-04\nEpoch 3/30\n1/1 [==============================] - 0s 50ms/step - loss: 1.4807e-04\nEpoch 4/30\n1/1 [==============================] - 0s 42ms/step - loss: 1.5531e-04\nEpoch 5/30\n1/1 [==============================] - 0s 59ms/step - loss: 1.4723e-04\nEpoch 6/30\n1/1 [==============================] - 0s 49ms/step - loss: 1.3889e-04\nEpoch 7/30\n1/1 [==============================] - 0s 49ms/step - loss: 1.4349e-04\nEpoch 8/30\n1/1 [==============================] - 0s 43ms/step - loss: 1.5329e-04\nEpoch 9/30\n1/1 [==============================] - 0s 47ms/step - loss: 1.5005e-04\nEpoch 10/30\n1/1 [==============================] - 0s 39ms/step - loss: 1.3985e-04\nEpoch 11/30\n1/1 [==============================] - 0s 41ms/step - loss: 1.4247e-04\nEpoch 12/30\n1/1 [==============================] - 0s 42ms/step - loss: 1.5047e-04\nEpoch 13/30\n1/1 [==============================] - 0s 42ms/step - loss: 1.4618e-04\nEpoch 14/30\n1/1 [==============================] - 0s 81ms/step - loss: 1.3546e-04\nEpoch 15/30\n1/1 [==============================] - 0s 57ms/step - loss: 1.4504e-04\nEpoch 16/30\n1/1 [==============================] - 0s 47ms/step - loss: 1.5943e-04\nEpoch 17/30\n1/1 [==============================] - 0s 44ms/step - loss: 1.4646e-04\nEpoch 18/30\n1/1 [==============================] - 0s 42ms/step - loss: 1.2888e-04\nEpoch 19/30\n1/1 [==============================] - 0s 55ms/step - loss: 1.3136e-04\nEpoch 20/30\n1/1 [==============================] - 0s 40ms/step - loss: 1.5021e-04\nEpoch 21/30\n1/1 [==============================] - 0s 41ms/step - loss: 1.5386e-04\nEpoch 22/30\n1/1 [==============================] - 0s 50ms/step - loss: 1.3781e-04\nEpoch 23/30\n1/1 [==============================] - 0s 42ms/step - loss: 1.4570e-04\nEpoch 24/30\n1/1 [==============================] - 0s 40ms/step - loss: 1.5535e-04\nEpoch 25/30\n1/1 [==============================] - 0s 40ms/step - loss: 1.4146e-04\nEpoch 26/30\n1/1 [==============================] - 0s 47ms/step - loss: 1.3084e-04\nEpoch 27/30\n1/1 [==============================] - 0s 40ms/step - loss: 1.3948e-04\nEpoch 28/30\n1/1 [==============================] - 0s 46ms/step - loss: 1.5139e-04\nEpoch 29/30\n1/1 [==============================] - 0s 40ms/step - loss: 1.5128e-04\nEpoch 30/30\n1/1 [==============================] - 0s 40ms/step - loss: 1.3867e-04\n(1, 12280, 1)\nEpoch 1/30\n1/1 [==============================] - 0s 40ms/step - loss: 58.3494\nEpoch 2/30\n1/1 [==============================] - 0s 47ms/step - loss: 58.3494\nEpoch 3/30\n1/1 [==============================] - 0s 39ms/step - loss: 58.3494\nEpoch 4/30\n1/1 [==============================] - 0s 41ms/step - loss: 58.3493\nEpoch 5/30\n1/1 [==============================] - 0s 40ms/step - loss: 58.3493\nEpoch 6/30\n1/1 [==============================] - 0s 39ms/step - loss: 58.3493\nEpoch 7/30\n1/1 [==============================] - 0s 44ms/step - loss: 58.3492\nEpoch 8/30\n1/1 [==============================] - 0s 36ms/step - loss: 58.3491\nEpoch 9/30\n1/1 [==============================] - 0s 38ms/step - loss: 58.3491\nEpoch 10/30\n1/1 [==============================] - 0s 40ms/step - loss: 58.3490\nEpoch 11/30\n1/1 [==============================] - 0s 42ms/step - loss: 58.3489\nEpoch 12/30\n1/1 [==============================] - 0s 41ms/step - loss: 58.3488\nEpoch 13/30\n1/1 [==============================] - 0s 41ms/step - loss: 58.3488\nEpoch 14/30\n1/1 [==============================] - 0s 42ms/step - loss: 58.3487\nEpoch 15/30\n1/1 [==============================] - 0s 39ms/step - loss: 58.3486\nEpoch 16/30\n1/1 [==============================] - 0s 41ms/step - loss: 58.3485\nEpoch 17/30\n1/1 [==============================] - 0s 40ms/step - loss: 58.3484\nEpoch 18/30\n1/1 [==============================] - 0s 44ms/step - loss: 58.3483\nEpoch 19/30\n1/1 [==============================] - 0s 42ms/step - loss: 58.3482\nEpoch 20/30\n1/1 [==============================] - 0s 40ms/step - loss: 58.3480\nEpoch 21/30\n1/1 [==============================] - 0s 47ms/step - loss: 58.3480\nEpoch 22/30\n1/1 [==============================] - 0s 40ms/step - loss: 58.3479\nEpoch 23/30\n1/1 [==============================] - 0s 44ms/step - loss: 58.3478\nEpoch 24/30\n1/1 [==============================] - 0s 38ms/step - loss: 58.3476\nEpoch 25/30\n1/1 [==============================] - 0s 43ms/step - loss: 58.3475\nEpoch 26/30\n1/1 [==============================] - 0s 36ms/step - loss: 58.3474\nEpoch 27/30\n1/1 [==============================] - 0s 46ms/step - loss: 58.3473\nEpoch 28/30\n1/1 [==============================] - 0s 40ms/step - loss: 58.3472\nEpoch 29/30\n1/1 [==============================] - 0s 39ms/step - loss: 58.3471\nEpoch 30/30\n1/1 [==============================] - 0s 49ms/step - loss: 58.3470\n(1, 12280, 1)\nEpoch 1/30\n1/1 [==============================] - 0s 38ms/step - loss: 159.7591\nEpoch 2/30\n1/1 [==============================] - 0s 40ms/step - loss: 159.7590\nEpoch 3/30\n1/1 [==============================] - 0s 40ms/step - loss: 159.7589\nEpoch 4/30\n1/1 [==============================] - 0s 46ms/step - loss: 159.7588\nEpoch 5/30\n1/1 [==============================] - 0s 43ms/step - loss: 159.7586\nEpoch 6/30\n1/1 [==============================] - 0s 45ms/step - loss: 159.7584\nEpoch 7/30\n1/1 [==============================] - 0s 38ms/step - loss: 159.7582\nEpoch 8/30\n1/1 [==============================] - 0s 70ms/step - loss: 159.7581\nEpoch 9/30\n1/1 [==============================] - 0s 34ms/step - loss: 159.7578\nEpoch 10/30\n1/1 [==============================] - 0s 46ms/step - loss: 159.7576\nEpoch 11/30\n1/1 [==============================] - 0s 46ms/step - loss: 159.7573\nEpoch 12/30\n1/1 [==============================] - 0s 36ms/step - loss: 159.7570\nEpoch 13/30\n1/1 [==============================] - 0s 41ms/step - loss: 159.7568\nEpoch 14/30\n1/1 [==============================] - 0s 47ms/step - loss: 159.7565\nEpoch 15/30\n1/1 [==============================] - 0s 46ms/step - loss: 159.7561\nEpoch 16/30\n1/1 [==============================] - 0s 44ms/step - loss: 159.7558\nEpoch 17/30\n1/1 [==============================] - 0s 39ms/step - loss: 159.7555\nEpoch 18/30\n1/1 [==============================] - 0s 45ms/step - loss: 159.7552\nEpoch 19/30\n1/1 [==============================] - 0s 37ms/step - loss: 159.7549\nEpoch 20/30\n1/1 [==============================] - 0s 38ms/step - loss: 159.7546\nEpoch 21/30\n1/1 [==============================] - 0s 41ms/step - loss: 159.7542\nEpoch 22/30\n1/1 [==============================] - 0s 41ms/step - loss: 159.7539\nEpoch 23/30\n1/1 [==============================] - 0s 40ms/step - loss: 159.7535\nEpoch 24/30\n1/1 [==============================] - 0s 41ms/step - loss: 159.7532\nEpoch 25/30\n1/1 [==============================] - 0s 40ms/step - loss: 159.7528\nEpoch 26/30\n1/1 [==============================] - 0s 50ms/step - loss: 159.7525\nEpoch 27/30\n1/1 [==============================] - 0s 36ms/step - loss: 159.7521\nEpoch 28/30\n1/1 [==============================] - 0s 40ms/step - loss: 159.7518\nEpoch 29/30\n1/1 [==============================] - 0s 41ms/step - loss: 159.7514\nEpoch 30/30\n1/1 [==============================] - 0s 44ms/step - loss: 159.7511\n(1, 12280, 1)\nEpoch 1/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.0178\nEpoch 2/30\n1/1 [==============================] - 0s 43ms/step - loss: 0.0175\nEpoch 3/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.0172\nEpoch 4/30\n1/1 [==============================] - 0s 44ms/step - loss: 0.0168\nEpoch 5/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.0164\nEpoch 6/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.0160\nEpoch 7/30\n1/1 [==============================] - 0s 42ms/step - loss: 0.0155\nEpoch 8/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.0150\nEpoch 9/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.0145\nEpoch 10/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.0141\nEpoch 11/30\n1/1 [==============================] - 0s 50ms/step - loss: 0.0138\nEpoch 12/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.0136\nEpoch 13/30\n1/1 [==============================] - 0s 42ms/step - loss: 0.0134\nEpoch 14/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.0132\nEpoch 15/30\n1/1 [==============================] - 0s 43ms/step - loss: 0.0130\nEpoch 16/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.0127\nEpoch 17/30\n1/1 [==============================] - 0s 46ms/step - loss: 0.0123\nEpoch 18/30\n1/1 [==============================] - 0s 38ms/step - loss: 0.0119\nEpoch 19/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.0115\nEpoch 20/30\n1/1 [==============================] - 0s 57ms/step - loss: 0.0110\nEpoch 21/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.0105\nEpoch 22/30\n1/1 [==============================] - 0s 47ms/step - loss: 0.0100\nEpoch 23/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.0094\nEpoch 24/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.0089\nEpoch 25/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.0084\nEpoch 26/30\n1/1 [==============================] - 0s 42ms/step - loss: 0.0080\nEpoch 27/30\n1/1 [==============================] - 0s 37ms/step - loss: 0.0076\nEpoch 28/30\n1/1 [==============================] - 0s 48ms/step - loss: 0.0072\nEpoch 29/30\n1/1 [==============================] - 0s 44ms/step - loss: 0.0068\nEpoch 30/30\n1/1 [==============================] - 0s 44ms/step - loss: 0.0064\n(1, 12280, 1)\nEpoch 1/30\n1/1 [==============================] - 0s 46ms/step - loss: 0.0145\nEpoch 2/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.0140\nEpoch 3/30\n1/1 [==============================] - 0s 37ms/step - loss: 0.0134\nEpoch 4/30\n1/1 [==============================] - 0s 44ms/step - loss: 0.0129\nEpoch 5/30\n1/1 [==============================] - 0s 47ms/step - loss: 0.0123\nEpoch 6/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.0117\nEpoch 7/30\n1/1 [==============================] - 0s 45ms/step - loss: 0.0112\nEpoch 8/30\n1/1 [==============================] - 0s 44ms/step - loss: 0.0106\nEpoch 9/30\n1/1 [==============================] - 0s 51ms/step - loss: 0.0101\nEpoch 10/30\n1/1 [==============================] - 0s 44ms/step - loss: 0.0096\nEpoch 11/30\n1/1 [==============================] - 0s 38ms/step - loss: 0.0093\nEpoch 12/30\n1/1 [==============================] - 0s 47ms/step - loss: 0.0091\nEpoch 13/30\n1/1 [==============================] - 0s 46ms/step - loss: 0.0090\nEpoch 14/30\n1/1 [==============================] - 0s 47ms/step - loss: 0.0089\nEpoch 15/30\n1/1 [==============================] - 0s 51ms/step - loss: 0.0088\nEpoch 16/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.0086\nEpoch 17/30\n1/1 [==============================] - 0s 48ms/step - loss: 0.0084\nEpoch 18/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.0081\nEpoch 19/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.0077\nEpoch 20/30\n1/1 [==============================] - 0s 42ms/step - loss: 0.0073\nEpoch 21/30\n1/1 [==============================] - 0s 55ms/step - loss: 0.0069\nEpoch 22/30\n1/1 [==============================] - 0s 44ms/step - loss: 0.0065\nEpoch 23/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.0060\nEpoch 24/30\n1/1 [==============================] - 0s 43ms/step - loss: 0.0056\nEpoch 25/30\n1/1 [==============================] - 0s 58ms/step - loss: 0.0052\nEpoch 26/30\n1/1 [==============================] - 0s 42ms/step - loss: 0.0049\nEpoch 27/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.0046\nEpoch 28/30\n1/1 [==============================] - 0s 44ms/step - loss: 0.0044\nEpoch 29/30\n1/1 [==============================] - 0s 51ms/step - loss: 0.0042\nEpoch 30/30\n1/1 [==============================] - 0s 47ms/step - loss: 0.0039\n(1, 12280, 1)\nEpoch 1/30\n1/1 [==============================] - 0s 43ms/step - loss: 0.0575\nEpoch 2/30\n1/1 [==============================] - 0s 42ms/step - loss: 0.0576\nEpoch 3/30\n1/1 [==============================] - 0s 51ms/step - loss: 0.0576\nEpoch 4/30\n1/1 [==============================] - 0s 42ms/step - loss: 0.0574\nEpoch 5/30\n1/1 [==============================] - 0s 47ms/step - loss: 0.0572\nEpoch 6/30\n1/1 [==============================] - 0s 48ms/step - loss: 0.0570\nEpoch 7/30\n1/1 [==============================] - 0s 49ms/step - loss: 0.0566\nEpoch 8/30\n1/1 [==============================] - 0s 90ms/step - loss: 0.0562\nEpoch 9/30\n1/1 [==============================] - 0s 61ms/step - loss: 0.0557\nEpoch 10/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.0552\nEpoch 11/30\n1/1 [==============================] - 0s 46ms/step - loss: 0.0548\nEpoch 12/30\n1/1 [==============================] - 0s 53ms/step - loss: 0.0543\nEpoch 13/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.0538\nEpoch 14/30\n1/1 [==============================] - 0s 37ms/step - loss: 0.0532\nEpoch 15/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.0526\nEpoch 16/30\n1/1 [==============================] - 0s 37ms/step - loss: 0.0520\nEpoch 17/30\n1/1 [==============================] - 0s 43ms/step - loss: 0.0513\nEpoch 18/30\n1/1 [==============================] - 0s 43ms/step - loss: 0.0506\nEpoch 19/30\n1/1 [==============================] - 0s 42ms/step - loss: 0.0499\nEpoch 20/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.0491\nEpoch 21/30\n1/1 [==============================] - 0s 42ms/step - loss: 0.0483\nEpoch 22/30\n1/1 [==============================] - 0s 57ms/step - loss: 0.0475\nEpoch 23/30\n1/1 [==============================] - 0s 81ms/step - loss: 0.0467\nEpoch 24/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.0459\nEpoch 25/30\n1/1 [==============================] - 0s 38ms/step - loss: 0.0451\nEpoch 26/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.0442\nEpoch 27/30\n1/1 [==============================] - 0s 43ms/step - loss: 0.0434\nEpoch 28/30\n1/1 [==============================] - 0s 47ms/step - loss: 0.0426\nEpoch 29/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.0418\nEpoch 30/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.0410\n(1, 12280, 1)\nEpoch 1/30\n1/1 [==============================] - 0s 47ms/step - loss: 1.0196\nEpoch 2/30\n1/1 [==============================] - 0s 41ms/step - loss: 1.0187\nEpoch 3/30\n1/1 [==============================] - 0s 53ms/step - loss: 1.0179\nEpoch 4/30\n1/1 [==============================] - 0s 46ms/step - loss: 1.0171\nEpoch 5/30\n1/1 [==============================] - 0s 43ms/step - loss: 1.0162\nEpoch 6/30\n1/1 [==============================] - 0s 38ms/step - loss: 1.0154\nEpoch 7/30\n1/1 [==============================] - 0s 46ms/step - loss: 1.0145\nEpoch 8/30\n1/1 [==============================] - 0s 40ms/step - loss: 1.0137\nEpoch 9/30\n1/1 [==============================] - 0s 42ms/step - loss: 1.0131\nEpoch 10/30\n1/1 [==============================] - 0s 41ms/step - loss: 1.0126\nEpoch 11/30\n1/1 [==============================] - 0s 43ms/step - loss: 1.0127\nEpoch 12/30\n1/1 [==============================] - 0s 38ms/step - loss: 1.0128\nEpoch 13/30\n1/1 [==============================] - 0s 42ms/step - loss: 1.0132\nEpoch 14/30\n1/1 [==============================] - 0s 42ms/step - loss: 1.0135\nEpoch 15/30\n1/1 [==============================] - 0s 41ms/step - loss: 1.0137\nEpoch 16/30\n1/1 [==============================] - 0s 44ms/step - loss: 1.0138\nEpoch 17/30\n1/1 [==============================] - 0s 39ms/step - loss: 1.0138\nEpoch 18/30\n1/1 [==============================] - 0s 35ms/step - loss: 1.0138\nEpoch 19/30\n1/1 [==============================] - 0s 36ms/step - loss: 1.0136\nEpoch 20/30\n1/1 [==============================] - 0s 37ms/step - loss: 1.0134\nEpoch 21/30\n1/1 [==============================] - 0s 36ms/step - loss: 1.0131\nEpoch 22/30\n1/1 [==============================] - 0s 44ms/step - loss: 1.0128\nEpoch 23/30\n1/1 [==============================] - 0s 42ms/step - loss: 1.0124\nEpoch 24/30\n1/1 [==============================] - 0s 38ms/step - loss: 1.0121\nEpoch 25/30\n1/1 [==============================] - 0s 45ms/step - loss: 1.0121\nEpoch 26/30\n1/1 [==============================] - 0s 37ms/step - loss: 1.0120\nEpoch 27/30\n1/1 [==============================] - 0s 38ms/step - loss: 1.0121\nEpoch 28/30\n1/1 [==============================] - 0s 52ms/step - loss: 1.0122\nEpoch 29/30\n1/1 [==============================] - 0s 40ms/step - loss: 1.0122\nEpoch 30/30\n1/1 [==============================] - 0s 42ms/step - loss: 1.0122\n(1, 12280, 1)\nEpoch 1/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.0115\nEpoch 2/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.0013\nEpoch 3/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.0012\nEpoch 4/30\n1/1 [==============================] - 0s 38ms/step - loss: 0.0011\nEpoch 5/30\n1/1 [==============================] - 0s 50ms/step - loss: 0.0011\nEpoch 6/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.0010\nEpoch 7/30\n1/1 [==============================] - 0s 39ms/step - loss: 9.8082e-04\nEpoch 8/30\n1/1 [==============================] - 0s 39ms/step - loss: 9.6179e-04\nEpoch 9/30\n1/1 [==============================] - 0s 40ms/step - loss: 8.9829e-04\nEpoch 10/30\n1/1 [==============================] - 0s 39ms/step - loss: 8.8269e-04\nEpoch 11/30\n1/1 [==============================] - 0s 45ms/step - loss: 8.6078e-04\nEpoch 12/30\n1/1 [==============================] - 0s 46ms/step - loss: 8.3198e-04\nEpoch 13/30\n1/1 [==============================] - 0s 37ms/step - loss: 8.0977e-04\nEpoch 14/30\n1/1 [==============================] - 0s 39ms/step - loss: 7.6071e-04\nEpoch 15/30\n1/1 [==============================] - 0s 40ms/step - loss: 7.3309e-04\nEpoch 16/30\n1/1 [==============================] - 0s 60ms/step - loss: 7.0087e-04\nEpoch 17/30\n1/1 [==============================] - 0s 62ms/step - loss: 6.6940e-04\nEpoch 18/30\n1/1 [==============================] - 0s 42ms/step - loss: 6.2952e-04\nEpoch 19/30\n1/1 [==============================] - 0s 41ms/step - loss: 6.0895e-04\nEpoch 20/30\n1/1 [==============================] - 0s 37ms/step - loss: 5.9707e-04\nEpoch 21/30\n1/1 [==============================] - 0s 41ms/step - loss: 5.6866e-04\nEpoch 22/30\n1/1 [==============================] - 0s 39ms/step - loss: 5.5265e-04\nEpoch 23/30\n1/1 [==============================] - 0s 37ms/step - loss: 5.2490e-04\nEpoch 24/30\n1/1 [==============================] - 0s 39ms/step - loss: 4.9784e-04\nEpoch 25/30\n1/1 [==============================] - 0s 40ms/step - loss: 4.6255e-04\nEpoch 26/30\n1/1 [==============================] - 0s 37ms/step - loss: 4.3340e-04\nEpoch 27/30\n1/1 [==============================] - 0s 37ms/step - loss: 4.2730e-04\nEpoch 28/30\n1/1 [==============================] - 0s 40ms/step - loss: 4.2264e-04\nEpoch 29/30\n1/1 [==============================] - 0s 38ms/step - loss: 4.0627e-04\nEpoch 30/30\n1/1 [==============================] - 0s 36ms/step - loss: 3.7425e-04\n(1, 12280, 1)\nEpoch 1/30\n1/1 [==============================] - 0s 35ms/step - loss: 0.2697\nEpoch 2/30\n1/1 [==============================] - 0s 36ms/step - loss: 0.2697\nEpoch 3/30\n1/1 [==============================] - 0s 42ms/step - loss: 0.2696\nEpoch 4/30\n1/1 [==============================] - 0s 38ms/step - loss: 0.2696\nEpoch 5/30\n1/1 [==============================] - 0s 37ms/step - loss: 0.2696\nEpoch 6/30\n1/1 [==============================] - 0s 37ms/step - loss: 0.2695\nEpoch 7/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.2695\nEpoch 8/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.2695\nEpoch 9/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.2694\nEpoch 10/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.2694\nEpoch 11/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.2694\nEpoch 12/30\n1/1 [==============================] - 0s 44ms/step - loss: 0.2694\nEpoch 13/30\n1/1 [==============================] - 0s 47ms/step - loss: 0.2694\nEpoch 14/30\n1/1 [==============================] - 0s 37ms/step - loss: 0.2694\nEpoch 15/30\n1/1 [==============================] - 0s 50ms/step - loss: 0.2694\nEpoch 16/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.2694\nEpoch 17/30\n1/1 [==============================] - 0s 48ms/step - loss: 0.2694\nEpoch 18/30\n1/1 [==============================] - 0s 42ms/step - loss: 0.2694\nEpoch 19/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.2694\nEpoch 20/30\n1/1 [==============================] - 0s 37ms/step - loss: 0.2693\nEpoch 21/30\n1/1 [==============================] - 0s 43ms/step - loss: 0.2693\nEpoch 22/30\n1/1 [==============================] - 0s 49ms/step - loss: 0.2693\nEpoch 23/30\n1/1 [==============================] - 0s 44ms/step - loss: 0.2693\nEpoch 24/30\n1/1 [==============================] - 0s 36ms/step - loss: 0.2693\nEpoch 25/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.2693\nEpoch 26/30\n1/1 [==============================] - 0s 37ms/step - loss: 0.2692\nEpoch 27/30\n1/1 [==============================] - 0s 37ms/step - loss: 0.2692\nEpoch 28/30\n1/1 [==============================] - 0s 36ms/step - loss: 0.2692\nEpoch 29/30\n1/1 [==============================] - 0s 38ms/step - loss: 0.2692\nEpoch 30/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.2692\n(1, 12280, 1)\nEpoch 1/30\n1/1 [==============================] - 0s 37ms/step - loss: 0.1285\nEpoch 2/30\n1/1 [==============================] - 0s 37ms/step - loss: 0.1285\nEpoch 3/30\n1/1 [==============================] - 0s 38ms/step - loss: 0.1285\nEpoch 4/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.1285\nEpoch 5/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.1285\nEpoch 6/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.1285\nEpoch 7/30\n1/1 [==============================] - 0s 38ms/step - loss: 0.1285\nEpoch 8/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.1285\nEpoch 9/30\n1/1 [==============================] - 0s 38ms/step - loss: 0.1285\nEpoch 10/30\n1/1 [==============================] - 0s 37ms/step - loss: 0.1284\nEpoch 11/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.1284\nEpoch 12/30\n1/1 [==============================] - 0s 36ms/step - loss: 0.1284\nEpoch 13/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.1284\nEpoch 14/30\n1/1 [==============================] - 0s 38ms/step - loss: 0.1284\nEpoch 15/30\n1/1 [==============================] - 0s 36ms/step - loss: 0.1284\nEpoch 16/30\n1/1 [==============================] - 0s 42ms/step - loss: 0.1284\nEpoch 17/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.1284\nEpoch 18/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.1283\nEpoch 19/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.1283\nEpoch 20/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.1283\nEpoch 21/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.1283\nEpoch 22/30\n1/1 [==============================] - 0s 42ms/step - loss: 0.1283\nEpoch 23/30\n1/1 [==============================] - 0s 37ms/step - loss: 0.1283\nEpoch 24/30\n1/1 [==============================] - 0s 46ms/step - loss: 0.1283\nEpoch 25/30\n1/1 [==============================] - 0s 42ms/step - loss: 0.1283\nEpoch 26/30\n1/1 [==============================] - 0s 42ms/step - loss: 0.1282\nEpoch 27/30\n1/1 [==============================] - 0s 44ms/step - loss: 0.1282\nEpoch 28/30\n1/1 [==============================] - 0s 45ms/step - loss: 0.1282\nEpoch 29/30\n1/1 [==============================] - 0s 38ms/step - loss: 0.1282\nEpoch 30/30\n1/1 [==============================] - 0s 48ms/step - loss: 0.1282\n(1, 12280, 1)\nEpoch 1/30\n1/1 [==============================] - 0s 49ms/step - loss: 0.0612\nEpoch 2/30\n1/1 [==============================] - 0s 38ms/step - loss: 0.0611\nEpoch 3/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.0611\nEpoch 4/30\n1/1 [==============================] - 0s 38ms/step - loss: 0.0611\nEpoch 5/30\n1/1 [==============================] - 0s 35ms/step - loss: 0.0611\nEpoch 6/30\n1/1 [==============================] - 0s 37ms/step - loss: 0.0611\nEpoch 7/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.0611\nEpoch 8/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.0611\nEpoch 9/30\n1/1 [==============================] - 0s 43ms/step - loss: 0.0611\nEpoch 10/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.0610\nEpoch 11/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.0610\nEpoch 12/30\n1/1 [==============================] - 0s 45ms/step - loss: 0.0610\nEpoch 13/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.0610\nEpoch 14/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.0610\nEpoch 15/30\n1/1 [==============================] - 0s 37ms/step - loss: 0.0610\nEpoch 16/30\n1/1 [==============================] - 0s 37ms/step - loss: 0.0610\nEpoch 17/30\n1/1 [==============================] - 0s 37ms/step - loss: 0.0610\nEpoch 18/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.0610\nEpoch 19/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.0610\nEpoch 20/30\n1/1 [==============================] - 0s 37ms/step - loss: 0.0610\nEpoch 21/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.0610\nEpoch 22/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.0610\nEpoch 23/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.0610\nEpoch 24/30\n1/1 [==============================] - 0s 43ms/step - loss: 0.0610\nEpoch 25/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.0610\nEpoch 26/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.0610\nEpoch 27/30\n1/1 [==============================] - 0s 55ms/step - loss: 0.0610\nEpoch 28/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.0609\nEpoch 29/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.0609\nEpoch 30/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.0609\n(1, 12280, 1)\nEpoch 1/30\n1/1 [==============================] - 0s 40ms/step - loss: 2.1711e-04\nEpoch 2/30\n1/1 [==============================] - 0s 42ms/step - loss: 2.0158e-04\nEpoch 3/30\n1/1 [==============================] - 0s 38ms/step - loss: 2.0349e-04\nEpoch 4/30\n1/1 [==============================] - 0s 39ms/step - loss: 2.0176e-04\nEpoch 5/30\n1/1 [==============================] - 0s 39ms/step - loss: 1.9975e-04\nEpoch 6/30\n1/1 [==============================] - 0s 38ms/step - loss: 1.8624e-04\nEpoch 7/30\n1/1 [==============================] - 0s 39ms/step - loss: 1.8939e-04\nEpoch 8/30\n1/1 [==============================] - 0s 42ms/step - loss: 1.8625e-04\nEpoch 9/30\n1/1 [==============================] - 0s 39ms/step - loss: 1.8271e-04\nEpoch 10/30\n1/1 [==============================] - 0s 43ms/step - loss: 1.7140e-04\nEpoch 11/30\n1/1 [==============================] - 0s 40ms/step - loss: 1.7575e-04\nEpoch 12/30\n1/1 [==============================] - 0s 38ms/step - loss: 1.7604e-04\nEpoch 13/30\n1/1 [==============================] - 0s 39ms/step - loss: 1.8167e-04\nEpoch 14/30\n1/1 [==============================] - 0s 40ms/step - loss: 1.7355e-04\nEpoch 15/30\n1/1 [==============================] - 0s 42ms/step - loss: 1.7620e-04\nEpoch 16/30\n1/1 [==============================] - 0s 38ms/step - loss: 1.7457e-04\nEpoch 17/30\n1/1 [==============================] - 0s 39ms/step - loss: 1.7508e-04\nEpoch 18/30\n1/1 [==============================] - 0s 41ms/step - loss: 1.6652e-04\nEpoch 19/30\n1/1 [==============================] - 0s 39ms/step - loss: 1.7118e-04\nEpoch 20/30\n1/1 [==============================] - 0s 37ms/step - loss: 1.6684e-04\nEpoch 21/30\n1/1 [==============================] - 0s 38ms/step - loss: 1.6508e-04\nEpoch 22/30\n1/1 [==============================] - 0s 41ms/step - loss: 1.5456e-04\nEpoch 23/30\n1/1 [==============================] - 0s 36ms/step - loss: 1.5700e-04\nEpoch 24/30\n1/1 [==============================] - 0s 44ms/step - loss: 1.5508e-04\nEpoch 25/30\n1/1 [==============================] - 0s 37ms/step - loss: 1.5783e-04\nEpoch 26/30\n1/1 [==============================] - 0s 38ms/step - loss: 1.4620e-04\nEpoch 27/30\n1/1 [==============================] - 0s 39ms/step - loss: 1.4952e-04\nEpoch 28/30\n1/1 [==============================] - 0s 42ms/step - loss: 1.4819e-04\nEpoch 29/30\n1/1 [==============================] - 0s 42ms/step - loss: 1.5034e-04\nEpoch 30/30\n1/1 [==============================] - 0s 39ms/step - loss: 1.4108e-04\n(1, 12280, 1)\nEpoch 1/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.0166\nEpoch 2/30\n1/1 [==============================] - 0s 42ms/step - loss: 0.0166\nEpoch 3/30\n1/1 [==============================] - 0s 38ms/step - loss: 0.0166\nEpoch 4/30\n1/1 [==============================] - 0s 43ms/step - loss: 0.0166\nEpoch 5/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.0166\nEpoch 6/30\n1/1 [==============================] - 0s 42ms/step - loss: 0.0166\nEpoch 7/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.0166\nEpoch 8/30\n1/1 [==============================] - 0s 42ms/step - loss: 0.0166\nEpoch 9/30\n1/1 [==============================] - 0s 44ms/step - loss: 0.0166\nEpoch 10/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.0166\nEpoch 11/30\n1/1 [==============================] - 0s 43ms/step - loss: 0.0166\nEpoch 12/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.0166\nEpoch 13/30\n1/1 [==============================] - 0s 43ms/step - loss: 0.0166\nEpoch 14/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.0166\nEpoch 15/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.0166\nEpoch 16/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.0166\nEpoch 17/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.0166\nEpoch 18/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.0166\nEpoch 19/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.0166\nEpoch 20/30\n1/1 [==============================] - 0s 37ms/step - loss: 0.0165\nEpoch 21/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.0165\nEpoch 22/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.0166\nEpoch 23/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.0166\nEpoch 24/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.0165\nEpoch 25/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.0165\nEpoch 26/30\n1/1 [==============================] - 0s 38ms/step - loss: 0.0165\nEpoch 27/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.0165\nEpoch 28/30\n1/1 [==============================] - 0s 38ms/step - loss: 0.0165\nEpoch 29/30\n1/1 [==============================] - 0s 36ms/step - loss: 0.0165\nEpoch 30/30\n1/1 [==============================] - 0s 47ms/step - loss: 0.0165\n(1, 12280, 1)\nEpoch 1/30\n1/1 [==============================] - 0s 38ms/step - loss: 0.1058\nEpoch 2/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.1058\nEpoch 3/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.1058\nEpoch 4/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.1058\nEpoch 5/30\n1/1 [==============================] - 0s 44ms/step - loss: 0.1058\nEpoch 6/30\n1/1 [==============================] - 0s 46ms/step - loss: 0.1058\nEpoch 7/30\n1/1 [==============================] - 0s 44ms/step - loss: 0.1058\nEpoch 8/30\n1/1 [==============================] - 0s 42ms/step - loss: 0.1058\nEpoch 9/30\n1/1 [==============================] - 0s 43ms/step - loss: 0.1058\nEpoch 10/30\n1/1 [==============================] - 0s 44ms/step - loss: 0.1057\nEpoch 11/30\n1/1 [==============================] - 0s 51ms/step - loss: 0.1057\nEpoch 12/30\n1/1 [==============================] - 0s 52ms/step - loss: 0.1057\nEpoch 13/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.1057\nEpoch 14/30\n1/1 [==============================] - 0s 45ms/step - loss: 0.1057\nEpoch 15/30\n1/1 [==============================] - 0s 43ms/step - loss: 0.1057\nEpoch 16/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.1057\nEpoch 17/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.1057\nEpoch 18/30\n1/1 [==============================] - 0s 49ms/step - loss: 0.1057\nEpoch 19/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.1057\nEpoch 20/30\n1/1 [==============================] - 0s 42ms/step - loss: 0.1057\nEpoch 21/30\n1/1 [==============================] - 0s 45ms/step - loss: 0.1057\nEpoch 22/30\n1/1 [==============================] - 0s 38ms/step - loss: 0.1057\nEpoch 23/30\n1/1 [==============================] - 0s 63ms/step - loss: 0.1057\nEpoch 24/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.1057\nEpoch 25/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.1057\nEpoch 26/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.1057\nEpoch 27/30\n1/1 [==============================] - 0s 38ms/step - loss: 0.1057\nEpoch 28/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.1057\nEpoch 29/30\n1/1 [==============================] - 0s 38ms/step - loss: 0.1057\nEpoch 30/30\n1/1 [==============================] - 0s 38ms/step - loss: 0.1057\n(1, 12280, 1)\nEpoch 1/30\n1/1 [==============================] - 0s 40ms/step - loss: 2.2082e-04\nEpoch 2/30\n1/1 [==============================] - 0s 38ms/step - loss: 2.1800e-04\nEpoch 3/30\n1/1 [==============================] - 0s 41ms/step - loss: 2.1565e-04\nEpoch 4/30\n1/1 [==============================] - 0s 37ms/step - loss: 2.0505e-04\nEpoch 5/30\n1/1 [==============================] - 0s 36ms/step - loss: 2.0523e-04\nEpoch 6/30\n1/1 [==============================] - 0s 37ms/step - loss: 2.0263e-04\nEpoch 7/30\n1/1 [==============================] - 0s 38ms/step - loss: 2.0584e-04\nEpoch 8/30\n1/1 [==============================] - 0s 37ms/step - loss: 1.9676e-04\nEpoch 9/30\n1/1 [==============================] - 0s 38ms/step - loss: 1.9825e-04\nEpoch 10/30\n1/1 [==============================] - 0s 36ms/step - loss: 1.9470e-04\nEpoch 11/30\n1/1 [==============================] - 0s 37ms/step - loss: 1.9521e-04\nEpoch 12/30\n1/1 [==============================] - 0s 59ms/step - loss: 1.8783e-04\nEpoch 13/30\n1/1 [==============================] - 0s 41ms/step - loss: 1.9314e-04\nEpoch 14/30\n1/1 [==============================] - 0s 38ms/step - loss: 1.8833e-04\nEpoch 15/30\n1/1 [==============================] - 0s 39ms/step - loss: 1.8802e-04\nEpoch 16/30\n1/1 [==============================] - 0s 38ms/step - loss: 1.7895e-04\nEpoch 17/30\n1/1 [==============================] - 0s 39ms/step - loss: 1.8165e-04\nEpoch 18/30\n1/1 [==============================] - 0s 40ms/step - loss: 1.7986e-04\nEpoch 19/30\n1/1 [==============================] - 0s 42ms/step - loss: 1.8024e-04\nEpoch 20/30\n1/1 [==============================] - 0s 49ms/step - loss: 1.6930e-04\nEpoch 21/30\n1/1 [==============================] - 0s 36ms/step - loss: 1.6994e-04\nEpoch 22/30\n1/1 [==============================] - 0s 56ms/step - loss: 1.6685e-04\nEpoch 23/30\n1/1 [==============================] - 0s 42ms/step - loss: 1.6887e-04\nEpoch 24/30\n1/1 [==============================] - 0s 43ms/step - loss: 1.5840e-04\nEpoch 25/30\n1/1 [==============================] - 0s 46ms/step - loss: 1.6204e-04\nEpoch 26/30\n1/1 [==============================] - 0s 42ms/step - loss: 1.5779e-04\nEpoch 27/30\n1/1 [==============================] - 0s 41ms/step - loss: 1.5720e-04\nEpoch 28/30\n1/1 [==============================] - 0s 41ms/step - loss: 1.4777e-04\nEpoch 29/30\n1/1 [==============================] - 0s 41ms/step - loss: 1.5021e-04\nEpoch 30/30\n1/1 [==============================] - 0s 40ms/step - loss: 1.4753e-04\n(1, 12280, 1)\nEpoch 1/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.1461\nEpoch 2/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.1461\nEpoch 3/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.1461\nEpoch 4/30\n1/1 [==============================] - 0s 38ms/step - loss: 0.1461\nEpoch 5/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.1461\nEpoch 6/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.1461\nEpoch 7/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.1461\nEpoch 8/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.1461\nEpoch 9/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.1460\nEpoch 10/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.1460\nEpoch 11/30\n1/1 [==============================] - 0s 44ms/step - loss: 0.1460\nEpoch 12/30\n1/1 [==============================] - 0s 47ms/step - loss: 0.1460\nEpoch 13/30\n1/1 [==============================] - 0s 35ms/step - loss: 0.1460\nEpoch 14/30\n1/1 [==============================] - 0s 43ms/step - loss: 0.1460\nEpoch 15/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.1460\nEpoch 16/30\n1/1 [==============================] - 0s 43ms/step - loss: 0.1460\nEpoch 17/30\n1/1 [==============================] - 0s 44ms/step - loss: 0.1460\nEpoch 18/30\n1/1 [==============================] - 0s 49ms/step - loss: 0.1460\nEpoch 19/30\n1/1 [==============================] - 0s 43ms/step - loss: 0.1460\nEpoch 20/30\n1/1 [==============================] - 0s 42ms/step - loss: 0.1460\nEpoch 21/30\n1/1 [==============================] - 0s 42ms/step - loss: 0.1459\nEpoch 22/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.1459\nEpoch 23/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.1459\nEpoch 24/30\n1/1 [==============================] - 0s 44ms/step - loss: 0.1459\nEpoch 25/30\n1/1 [==============================] - 0s 43ms/step - loss: 0.1459\nEpoch 26/30\n1/1 [==============================] - 0s 43ms/step - loss: 0.1459\nEpoch 27/30\n1/1 [==============================] - 0s 38ms/step - loss: 0.1459\nEpoch 28/30\n1/1 [==============================] - 0s 42ms/step - loss: 0.1459\nEpoch 29/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.1459\nEpoch 30/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.1459\n(1, 12280, 1)\nEpoch 1/30\n1/1 [==============================] - 0s 43ms/step - loss: 0.0242\nEpoch 2/30\n1/1 [==============================] - 0s 44ms/step - loss: 0.0242\nEpoch 3/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.0242\nEpoch 4/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.0242\nEpoch 5/30\n1/1 [==============================] - 0s 43ms/step - loss: 0.0242\nEpoch 6/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.0242\nEpoch 7/30\n1/1 [==============================] - 0s 44ms/step - loss: 0.0242\nEpoch 8/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.0242\nEpoch 9/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.0242\nEpoch 10/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.0242\nEpoch 11/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.0242\nEpoch 12/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.0241\nEpoch 13/30\n1/1 [==============================] - 0s 37ms/step - loss: 0.0241\nEpoch 14/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.0241\nEpoch 15/30\n1/1 [==============================] - 0s 37ms/step - loss: 0.0241\nEpoch 16/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.0241\nEpoch 17/30\n1/1 [==============================] - 0s 38ms/step - loss: 0.0241\nEpoch 18/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.0241\nEpoch 19/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.0241\nEpoch 20/30\n1/1 [==============================] - 0s 42ms/step - loss: 0.0241\nEpoch 21/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.0241\nEpoch 22/30\n1/1 [==============================] - 0s 43ms/step - loss: 0.0241\nEpoch 23/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.0241\nEpoch 24/30\n1/1 [==============================] - 0s 43ms/step - loss: 0.0241\nEpoch 25/30\n1/1 [==============================] - 0s 38ms/step - loss: 0.0241\nEpoch 26/30\n1/1 [==============================] - 0s 42ms/step - loss: 0.0240\nEpoch 27/30\n1/1 [==============================] - 0s 44ms/step - loss: 0.0240\nEpoch 28/30\n1/1 [==============================] - 0s 44ms/step - loss: 0.0240\nEpoch 29/30\n1/1 [==============================] - 0s 43ms/step - loss: 0.0240\nEpoch 30/30\n1/1 [==============================] - 0s 42ms/step - loss: 0.0240\n(1, 12280, 1)\nEpoch 1/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.0342\nEpoch 2/30\n1/1 [==============================] - 0s 42ms/step - loss: 0.0342\nEpoch 3/30\n1/1 [==============================] - 0s 56ms/step - loss: 0.0342\nEpoch 4/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.0342\nEpoch 5/30\n1/1 [==============================] - 0s 46ms/step - loss: 0.0342\nEpoch 6/30\n1/1 [==============================] - 0s 43ms/step - loss: 0.0342\nEpoch 7/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.0342\nEpoch 8/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.0342\nEpoch 9/30\n1/1 [==============================] - 0s 42ms/step - loss: 0.0342\nEpoch 10/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.0342\nEpoch 11/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.0342\nEpoch 12/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.0342\nEpoch 13/30\n1/1 [==============================] - 0s 42ms/step - loss: 0.0342\nEpoch 14/30\n1/1 [==============================] - 0s 38ms/step - loss: 0.0342\nEpoch 15/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.0342\nEpoch 16/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.0342\nEpoch 17/30\n1/1 [==============================] - 0s 57ms/step - loss: 0.0342\nEpoch 18/30\n1/1 [==============================] - 0s 62ms/step - loss: 0.0342\nEpoch 19/30\n1/1 [==============================] - 0s 67ms/step - loss: 0.0342\nEpoch 20/30\n1/1 [==============================] - 0s 51ms/step - loss: 0.0341\nEpoch 21/30\n1/1 [==============================] - 0s 47ms/step - loss: 0.0341\nEpoch 22/30\n1/1 [==============================] - 0s 61ms/step - loss: 0.0341\nEpoch 23/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.0341\nEpoch 24/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.0341\nEpoch 25/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.0341\nEpoch 26/30\n1/1 [==============================] - 0s 37ms/step - loss: 0.0341\nEpoch 27/30\n1/1 [==============================] - 0s 38ms/step - loss: 0.0341\nEpoch 28/30\n1/1 [==============================] - 0s 43ms/step - loss: 0.0341\nEpoch 29/30\n1/1 [==============================] - 0s 38ms/step - loss: 0.0341\nEpoch 30/30\n1/1 [==============================] - 0s 73ms/step - loss: 0.0341\n(1, 12280, 1)\nEpoch 1/30\n1/1 [==============================] - 0s 102ms/step - loss: 0.9124\nEpoch 2/30\n1/1 [==============================] - 0s 65ms/step - loss: 0.9124\nEpoch 3/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.9124\nEpoch 4/30\n1/1 [==============================] - 0s 43ms/step - loss: 0.9123\nEpoch 5/30\n1/1 [==============================] - 0s 42ms/step - loss: 0.9123\nEpoch 6/30\n1/1 [==============================] - 0s 37ms/step - loss: 0.9123\nEpoch 7/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.9123\nEpoch 8/30\n1/1 [==============================] - 0s 42ms/step - loss: 0.9123\nEpoch 9/30\n1/1 [==============================] - 0s 42ms/step - loss: 0.9123\nEpoch 10/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.9122\nEpoch 11/30\n1/1 [==============================] - 0s 38ms/step - loss: 0.9122\nEpoch 12/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.9122\nEpoch 13/30\n1/1 [==============================] - 0s 42ms/step - loss: 0.9122\nEpoch 14/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.9122\nEpoch 15/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.9121\nEpoch 16/30\n1/1 [==============================] - 0s 124ms/step - loss: 0.9121\nEpoch 17/30\n1/1 [==============================] - 0s 51ms/step - loss: 0.9121\nEpoch 18/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.9121\nEpoch 19/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.9121\nEpoch 20/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.9121\nEpoch 21/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.9120\nEpoch 22/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.9120\nEpoch 23/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.9120\nEpoch 24/30\n1/1 [==============================] - 0s 37ms/step - loss: 0.9120\nEpoch 25/30\n1/1 [==============================] - 0s 42ms/step - loss: 0.9120\nEpoch 26/30\n1/1 [==============================] - 0s 66ms/step - loss: 0.9120\nEpoch 27/30\n1/1 [==============================] - 0s 49ms/step - loss: 0.9119\nEpoch 28/30\n1/1 [==============================] - 0s 43ms/step - loss: 0.9119\nEpoch 29/30\n1/1 [==============================] - 0s 53ms/step - loss: 0.9119\nEpoch 30/30\n1/1 [==============================] - 0s 43ms/step - loss: 0.9119\n(1, 12280, 1)\nEpoch 1/30\n1/1 [==============================] - 0s 36ms/step - loss: 0.0083\nEpoch 2/30\n1/1 [==============================] - 0s 45ms/step - loss: 7.1087e-04\nEpoch 3/30\n1/1 [==============================] - 0s 38ms/step - loss: 7.2593e-04\nEpoch 4/30\n1/1 [==============================] - 0s 40ms/step - loss: 6.9284e-04\nEpoch 5/30\n1/1 [==============================] - 0s 57ms/step - loss: 6.7323e-04\nEpoch 6/30\n1/1 [==============================] - 0s 52ms/step - loss: 6.5942e-04\nEpoch 7/30\n1/1 [==============================] - 0s 41ms/step - loss: 6.2271e-04\nEpoch 8/30\n1/1 [==============================] - 0s 60ms/step - loss: 5.8805e-04\nEpoch 9/30\n1/1 [==============================] - 0s 41ms/step - loss: 6.0568e-04\nEpoch 10/30\n1/1 [==============================] - 0s 43ms/step - loss: 6.0777e-04\nEpoch 11/30\n1/1 [==============================] - 0s 44ms/step - loss: 5.8520e-04\nEpoch 12/30\n1/1 [==============================] - 0s 42ms/step - loss: 5.4639e-04\nEpoch 13/30\n1/1 [==============================] - 0s 37ms/step - loss: 5.1920e-04\nEpoch 14/30\n1/1 [==============================] - 0s 39ms/step - loss: 5.1800e-04\nEpoch 15/30\n1/1 [==============================] - 0s 39ms/step - loss: 5.1478e-04\nEpoch 16/30\n1/1 [==============================] - 0s 41ms/step - loss: 4.9006e-04\nEpoch 17/30\n1/1 [==============================] - 0s 42ms/step - loss: 4.8532e-04\nEpoch 18/30\n1/1 [==============================] - 0s 43ms/step - loss: 4.7269e-04\nEpoch 19/30\n1/1 [==============================] - 0s 47ms/step - loss: 4.5408e-04\nEpoch 20/30\n1/1 [==============================] - 0s 38ms/step - loss: 4.3635e-04\nEpoch 21/30\n1/1 [==============================] - 0s 41ms/step - loss: 4.3163e-04\nEpoch 22/30\n1/1 [==============================] - 0s 77ms/step - loss: 4.1754e-04\nEpoch 23/30\n1/1 [==============================] - 0s 114ms/step - loss: 3.9463e-04\nEpoch 24/30\n1/1 [==============================] - 0s 52ms/step - loss: 3.6338e-04\nEpoch 25/30\n1/1 [==============================] - 0s 48ms/step - loss: 3.4862e-04\nEpoch 26/30\n1/1 [==============================] - 0s 43ms/step - loss: 3.3980e-04\nEpoch 27/30\n1/1 [==============================] - 0s 65ms/step - loss: 3.3416e-04\nEpoch 28/30\n1/1 [==============================] - 0s 78ms/step - loss: 3.0669e-04\nEpoch 29/30\n1/1 [==============================] - 0s 47ms/step - loss: 2.9818e-04\nEpoch 30/30\n1/1 [==============================] - 0s 37ms/step - loss: 2.8448e-04\n(1, 12280, 1)\nEpoch 1/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.2687\nEpoch 2/30\n1/1 [==============================] - 0s 38ms/step - loss: 0.2687\nEpoch 3/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.2687\nEpoch 4/30\n1/1 [==============================] - 0s 43ms/step - loss: 0.2687\nEpoch 5/30\n1/1 [==============================] - 0s 43ms/step - loss: 0.2686\nEpoch 6/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.2686\nEpoch 7/30\n1/1 [==============================] - 0s 38ms/step - loss: 0.2686\nEpoch 8/30\n1/1 [==============================] - 0s 42ms/step - loss: 0.2686\nEpoch 9/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.2686\nEpoch 10/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.2685\nEpoch 11/30\n1/1 [==============================] - 0s 37ms/step - loss: 0.2685\nEpoch 12/30\n1/1 [==============================] - 0s 38ms/step - loss: 0.2685\nEpoch 13/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.2685\nEpoch 14/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.2685\nEpoch 15/30\n1/1 [==============================] - 0s 38ms/step - loss: 0.2685\nEpoch 16/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.2685\nEpoch 17/30\n1/1 [==============================] - 0s 72ms/step - loss: 0.2685\nEpoch 18/30\n1/1 [==============================] - 0s 44ms/step - loss: 0.2685\nEpoch 19/30\n1/1 [==============================] - 0s 48ms/step - loss: 0.2685\nEpoch 20/30\n1/1 [==============================] - 0s 48ms/step - loss: 0.2685\nEpoch 21/30\n1/1 [==============================] - 0s 48ms/step - loss: 0.2685\nEpoch 22/30\n1/1 [==============================] - 0s 49ms/step - loss: 0.2684\nEpoch 23/30\n1/1 [==============================] - 0s 42ms/step - loss: 0.2684\nEpoch 24/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.2684\nEpoch 25/30\n1/1 [==============================] - 0s 44ms/step - loss: 0.2684\nEpoch 26/30\n1/1 [==============================] - 0s 54ms/step - loss: 0.2684\nEpoch 27/30\n1/1 [==============================] - 0s 46ms/step - loss: 0.2684\nEpoch 28/30\n1/1 [==============================] - 0s 45ms/step - loss: 0.2684\nEpoch 29/30\n1/1 [==============================] - 0s 45ms/step - loss: 0.2684\nEpoch 30/30\n1/1 [==============================] - 0s 45ms/step - loss: 0.2684\n(1, 12280, 1)\nEpoch 1/30\n1/1 [==============================] - 0s 42ms/step - loss: 743.5905\nEpoch 2/30\n1/1 [==============================] - 0s 36ms/step - loss: 0.1618\nEpoch 3/30\n1/1 [==============================] - 0s 38ms/step - loss: 46.3736\nEpoch 4/30\n1/1 [==============================] - 0s 42ms/step - loss: 0.1619\nEpoch 5/30\n1/1 [==============================] - 0s 49ms/step - loss: 0.1619\nEpoch 6/30\n1/1 [==============================] - 0s 52ms/step - loss: 0.1618\nEpoch 7/30\n1/1 [==============================] - 0s 55ms/step - loss: 0.1618\nEpoch 8/30\n1/1 [==============================] - 0s 64ms/step - loss: 0.1617\nEpoch 9/30\n1/1 [==============================] - 0s 44ms/step - loss: 0.1617\nEpoch 10/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.1617\nEpoch 11/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.1617\nEpoch 12/30\n1/1 [==============================] - 0s 47ms/step - loss: 0.1617\nEpoch 13/30\n1/1 [==============================] - 0s 58ms/step - loss: 0.1617\nEpoch 14/30\n1/1 [==============================] - 0s 53ms/step - loss: 0.1616\nEpoch 15/30\n1/1 [==============================] - 0s 45ms/step - loss: 0.1616\nEpoch 16/30\n1/1 [==============================] - 0s 43ms/step - loss: 0.1616\nEpoch 17/30\n1/1 [==============================] - 0s 36ms/step - loss: 0.1616\nEpoch 18/30\n1/1 [==============================] - 0s 43ms/step - loss: 0.1616\nEpoch 19/30\n1/1 [==============================] - 0s 92ms/step - loss: 0.1616\nEpoch 20/30\n1/1 [==============================] - 0s 56ms/step - loss: 0.1616\nEpoch 21/30\n1/1 [==============================] - 0s 75ms/step - loss: 0.1615\nEpoch 22/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.1615\nEpoch 23/30\n1/1 [==============================] - 0s 46ms/step - loss: 0.1615\nEpoch 24/30\n1/1 [==============================] - 0s 62ms/step - loss: 0.1615\nEpoch 25/30\n1/1 [==============================] - 0s 65ms/step - loss: 0.1615\nEpoch 26/30\n1/1 [==============================] - 0s 58ms/step - loss: 0.1615\nEpoch 27/30\n1/1 [==============================] - 0s 55ms/step - loss: 0.1615\nEpoch 28/30\n1/1 [==============================] - 0s 38ms/step - loss: 0.1614\nEpoch 29/30\n1/1 [==============================] - 0s 45ms/step - loss: 0.1614\nEpoch 30/30\n1/1 [==============================] - 0s 54ms/step - loss: 0.1614\n(1, 12280, 1)\nEpoch 1/30\n1/1 [==============================] - 0s 54ms/step - loss: 0.0772\nEpoch 2/30\n1/1 [==============================] - 0s 56ms/step - loss: 0.0772\nEpoch 3/30\n1/1 [==============================] - 0s 50ms/step - loss: 0.0772\nEpoch 4/30\n1/1 [==============================] - 0s 66ms/step - loss: 0.0772\nEpoch 5/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.0772\nEpoch 6/30\n1/1 [==============================] - 0s 42ms/step - loss: 0.0772\nEpoch 7/30\n1/1 [==============================] - 0s 152ms/step - loss: 0.0771\nEpoch 8/30\n1/1 [==============================] - 0s 59ms/step - loss: 0.0771\nEpoch 9/30\n1/1 [==============================] - 0s 43ms/step - loss: 0.0771\nEpoch 10/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.0771\nEpoch 11/30\n1/1 [==============================] - 0s 38ms/step - loss: 0.0771\nEpoch 12/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.0771\nEpoch 13/30\n1/1 [==============================] - 0s 44ms/step - loss: 0.0771\nEpoch 14/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.0771\nEpoch 15/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.0771\nEpoch 16/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.0771\nEpoch 17/30\n1/1 [==============================] - 0s 37ms/step - loss: 0.0771\nEpoch 18/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.0771\nEpoch 19/30\n1/1 [==============================] - 0s 52ms/step - loss: 0.0770\nEpoch 20/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.0770\nEpoch 21/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.0770\nEpoch 22/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.0770\nEpoch 23/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.0770\nEpoch 24/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.0770\nEpoch 25/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.0770\nEpoch 26/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.0770\nEpoch 27/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.0770\nEpoch 28/30\n1/1 [==============================] - 0s 38ms/step - loss: 0.0770\nEpoch 29/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.0770\nEpoch 30/30\n1/1 [==============================] - 0s 38ms/step - loss: 0.0769\n(1, 12280, 1)\nEpoch 1/30\n1/1 [==============================] - 0s 37ms/step - loss: 0.2672\nEpoch 2/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.2671\nEpoch 3/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.2671\nEpoch 4/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.2671\nEpoch 5/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.2671\nEpoch 6/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.2671\nEpoch 7/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.2671\nEpoch 8/30\n1/1 [==============================] - 0s 38ms/step - loss: 0.2671\nEpoch 9/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.2671\nEpoch 10/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.2670\nEpoch 11/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.2670\nEpoch 12/30\n1/1 [==============================] - 0s 37ms/step - loss: 0.2670\nEpoch 13/30\n1/1 [==============================] - 0s 38ms/step - loss: 0.2670\nEpoch 14/30\n1/1 [==============================] - 0s 38ms/step - loss: 0.2670\nEpoch 15/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.2670\nEpoch 16/30\n1/1 [==============================] - 0s 36ms/step - loss: 0.2670\nEpoch 17/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.2670\nEpoch 18/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.2670\nEpoch 19/30\n1/1 [==============================] - 0s 37ms/step - loss: 0.2670\nEpoch 20/30\n1/1 [==============================] - 0s 36ms/step - loss: 0.2669\nEpoch 21/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.2669\nEpoch 22/30\n1/1 [==============================] - 0s 38ms/step - loss: 0.2669\nEpoch 23/30\n1/1 [==============================] - 0s 38ms/step - loss: 0.2669\nEpoch 24/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.2669\nEpoch 25/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.2669\nEpoch 26/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.2669\nEpoch 27/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.2669\nEpoch 28/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.2669\nEpoch 29/30\n1/1 [==============================] - 0s 38ms/step - loss: 0.2668\nEpoch 30/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.2668\n(1, 12280, 1)\nEpoch 1/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.1582\nEpoch 2/30\n1/1 [==============================] - 0s 44ms/step - loss: 0.1581\nEpoch 3/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.1581\nEpoch 4/30\n1/1 [==============================] - 0s 46ms/step - loss: 0.1581\nEpoch 5/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.1581\nEpoch 6/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.1581\nEpoch 7/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.1581\nEpoch 8/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.1581\nEpoch 9/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.1581\nEpoch 10/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.1581\nEpoch 11/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.1581\nEpoch 12/30\n1/1 [==============================] - 0s 42ms/step - loss: 0.1581\nEpoch 13/30\n1/1 [==============================] - 0s 42ms/step - loss: 0.1580\nEpoch 14/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.1580\nEpoch 15/30\n1/1 [==============================] - 0s 38ms/step - loss: 0.1580\nEpoch 16/30\n1/1 [==============================] - 0s 42ms/step - loss: 0.1580\nEpoch 17/30\n1/1 [==============================] - 0s 38ms/step - loss: 0.1580\nEpoch 18/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.1580\nEpoch 19/30\n1/1 [==============================] - 0s 42ms/step - loss: 0.1580\nEpoch 20/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.1580\nEpoch 21/30\n1/1 [==============================] - 0s 38ms/step - loss: 0.1580\nEpoch 22/30\n1/1 [==============================] - 0s 37ms/step - loss: 0.1579\nEpoch 23/30\n1/1 [==============================] - 0s 36ms/step - loss: 0.1579\nEpoch 24/30\n1/1 [==============================] - 0s 38ms/step - loss: 0.1579\nEpoch 25/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.1579\nEpoch 26/30\n1/1 [==============================] - 0s 38ms/step - loss: 0.1579\nEpoch 27/30\n1/1 [==============================] - 0s 54ms/step - loss: 0.1579\nEpoch 28/30\n1/1 [==============================] - 0s 54ms/step - loss: 0.1579\nEpoch 29/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.1579\nEpoch 30/30\n1/1 [==============================] - 0s 45ms/step - loss: 0.1579\n(1, 12280, 1)\nEpoch 1/30\n1/1 [==============================] - 0s 76ms/step - loss: 0.3404\nEpoch 2/30\n1/1 [==============================] - 0s 52ms/step - loss: 0.3404\nEpoch 3/30\n1/1 [==============================] - 0s 50ms/step - loss: 0.3404\nEpoch 4/30\n1/1 [==============================] - 0s 45ms/step - loss: 0.3404\nEpoch 5/30\n1/1 [==============================] - 0s 56ms/step - loss: 0.3404\nEpoch 6/30\n1/1 [==============================] - 0s 45ms/step - loss: 0.3404\nEpoch 7/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.3403\nEpoch 8/30\n1/1 [==============================] - 0s 43ms/step - loss: 0.3403\nEpoch 9/30\n1/1 [==============================] - 0s 44ms/step - loss: 0.3403\nEpoch 10/30\n1/1 [==============================] - 0s 43ms/step - loss: 0.3403\nEpoch 11/30\n1/1 [==============================] - 0s 47ms/step - loss: 0.3403\nEpoch 12/30\n1/1 [==============================] - 0s 50ms/step - loss: 0.3403\nEpoch 13/30\n1/1 [==============================] - 0s 66ms/step - loss: 0.3403\nEpoch 14/30\n1/1 [==============================] - 0s 42ms/step - loss: 0.3403\nEpoch 15/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.3403\nEpoch 16/30\n1/1 [==============================] - 0s 45ms/step - loss: 0.3403\nEpoch 17/30\n1/1 [==============================] - 0s 42ms/step - loss: 0.3403\nEpoch 18/30\n1/1 [==============================] - 0s 38ms/step - loss: 0.3402\nEpoch 19/30\n1/1 [==============================] - 0s 38ms/step - loss: 0.3402\nEpoch 20/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.3402\nEpoch 21/30\n1/1 [==============================] - 0s 51ms/step - loss: 0.3402\nEpoch 22/30\n1/1 [==============================] - 0s 45ms/step - loss: 0.3402\nEpoch 23/30\n1/1 [==============================] - 0s 56ms/step - loss: 0.3402\nEpoch 24/30\n1/1 [==============================] - 0s 62ms/step - loss: 0.3402\nEpoch 25/30\n1/1 [==============================] - 0s 43ms/step - loss: 0.3402\nEpoch 26/30\n1/1 [==============================] - 0s 38ms/step - loss: 0.3402\nEpoch 27/30\n1/1 [==============================] - 0s 44ms/step - loss: 0.3402\nEpoch 28/30\n1/1 [==============================] - 0s 44ms/step - loss: 0.3402\nEpoch 29/30\n1/1 [==============================] - 0s 42ms/step - loss: 0.3402\nEpoch 30/30\n1/1 [==============================] - 0s 43ms/step - loss: 0.3401\n(1, 12280, 1)\nEpoch 1/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.0761\nEpoch 2/30\n1/1 [==============================] - 0s 42ms/step - loss: 0.0761\nEpoch 3/30\n1/1 [==============================] - 0s 42ms/step - loss: 0.0761\nEpoch 4/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.0761\nEpoch 5/30\n1/1 [==============================] - 0s 38ms/step - loss: 0.0761\nEpoch 6/30\n1/1 [==============================] - 0s 38ms/step - loss: 0.0761\nEpoch 7/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.0761\nEpoch 8/30\n1/1 [==============================] - 0s 45ms/step - loss: 0.0761\nEpoch 9/30\n1/1 [==============================] - 0s 57ms/step - loss: 0.0761\nEpoch 10/30\n1/1 [==============================] - 0s 38ms/step - loss: 0.0761\nEpoch 11/30\n1/1 [==============================] - 0s 53ms/step - loss: 0.0760\nEpoch 12/30\n1/1 [==============================] - 0s 61ms/step - loss: 0.0760\nEpoch 13/30\n1/1 [==============================] - 0s 40ms/step - loss: 0.0760\nEpoch 14/30\n1/1 [==============================] - 0s 85ms/step - loss: 0.0760\nEpoch 15/30\n1/1 [==============================] - 0s 39ms/step - loss: 0.0760\nEpoch 16/30\n1/1 [==============================] - 0s 54ms/step - loss: 0.0760\nEpoch 17/30\n1/1 [==============================] - 0s 44ms/step - loss: 0.0760\nEpoch 18/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.0760\nEpoch 19/30\n1/1 [==============================] - 0s 50ms/step - loss: 0.0760\nEpoch 20/30\n1/1 [==============================] - 0s 38ms/step - loss: 0.0760\nEpoch 21/30\n1/1 [==============================] - 0s 57ms/step - loss: 0.0760\nEpoch 22/30\n1/1 [==============================] - 0s 52ms/step - loss: 0.0760\nEpoch 23/30\n1/1 [==============================] - 0s 62ms/step - loss: 0.0760\nEpoch 24/30\n1/1 [==============================] - 0s 41ms/step - loss: 0.0759\nEpoch 25/30\n1/1 [==============================] - 0s 63ms/step - loss: 0.0759\nEpoch 26/30\n1/1 [==============================] - 0s 50ms/step - loss: 0.0759\nEpoch 27/30\n1/1 [==============================] - 0s 48ms/step - loss: 0.0759\nEpoch 28/30\n1/1 [==============================] - 0s 58ms/step - loss: 0.0759\nEpoch 29/30\n1/1 [==============================] - 0s 46ms/step - loss: 0.0759\nEpoch 30/30\n1/1 [==============================] - 0s 49ms/step - loss: 0.0759\n(1, 12280, 1)\nEpoch 1/30\n1/1 [==============================] - 0s 53ms/step - loss: 136.5052\nEpoch 2/30\n1/1 [==============================] - 0s 57ms/step - loss: 136.5052\nEpoch 3/30\n1/1 [==============================] - 0s 65ms/step - loss: 136.5051\nEpoch 4/30\n1/1 [==============================] - 0s 60ms/step - loss: 136.5050\nEpoch 5/30\n1/1 [==============================] - 0s 54ms/step - loss: 136.5050\nEpoch 6/30\n1/1 [==============================] - 0s 39ms/step - loss: 136.5049\nEpoch 7/30\n1/1 [==============================] - 0s 39ms/step - loss: 136.5047\nEpoch 8/30\n1/1 [==============================] - 0s 51ms/step - loss: 136.5046\nEpoch 9/30\n1/1 [==============================] - 0s 53ms/step - loss: 136.5045\nEpoch 10/30\n1/1 [==============================] - 0s 64ms/step - loss: 136.5043\nEpoch 11/30\n1/1 [==============================] - 0s 55ms/step - loss: 136.5041\nEpoch 12/30\n1/1 [==============================] - 0s 58ms/step - loss: 136.5039\nEpoch 13/30\n1/1 [==============================] - 0s 52ms/step - loss: 136.5037\nEpoch 14/30\n1/1 [==============================] - 0s 80ms/step - loss: 136.5035\nEpoch 15/30\n1/1 [==============================] - 0s 54ms/step - loss: 136.5033\nEpoch 16/30\n1/1 [==============================] - 0s 49ms/step - loss: 136.5031\nEpoch 17/30\n1/1 [==============================] - 0s 48ms/step - loss: 136.5029\nEpoch 18/30\n1/1 [==============================] - 0s 90ms/step - loss: 136.5027\nEpoch 19/30\n1/1 [==============================] - 0s 79ms/step - loss: 136.5025\nEpoch 20/30\n1/1 [==============================] - 0s 60ms/step - loss: 136.5023\nEpoch 21/30\n1/1 [==============================] - 0s 44ms/step - loss: 136.5021\nEpoch 22/30\n1/1 [==============================] - 0s 53ms/step - loss: 136.5018\nEpoch 23/30\n1/1 [==============================] - 0s 44ms/step - loss: 136.5016\nEpoch 24/30\n1/1 [==============================] - 0s 47ms/step - loss: 136.5014\nEpoch 25/30\n1/1 [==============================] - 0s 58ms/step - loss: 136.5011\nEpoch 26/30\n1/1 [==============================] - 0s 60ms/step - loss: 136.5009\nEpoch 27/30\n1/1 [==============================] - 0s 53ms/step - loss: 136.5007\nEpoch 28/30\n1/1 [==============================] - 0s 44ms/step - loss: 136.5004\nEpoch 29/30\n1/1 [==============================] - 0s 44ms/step - loss: 136.5002\nEpoch 30/30\n1/1 [==============================] - 0s 41ms/step - loss: 136.5000\n(1, 12280, 1)\nEpoch 1/30\n1/1 [==============================] - 0s 58ms/step - loss: 91.6799\nEpoch 2/30\n1/1 [==============================] - 0s 41ms/step - loss: 91.6801\nEpoch 3/30\n1/1 [==============================] - 0s 43ms/step - loss: 91.6802\nEpoch 4/30\n1/1 [==============================] - 0s 41ms/step - loss: 91.6803\nEpoch 5/30\n1/1 [==============================] - 0s 39ms/step - loss: 91.6804\nEpoch 6/30\n1/1 [==============================] - 0s 40ms/step - loss: 91.6804\nEpoch 7/30\n1/1 [==============================] - 0s 64ms/step - loss: 91.6804\nEpoch 8/30\n1/1 [==============================] - 0s 40ms/step - loss: 91.6804\nEpoch 9/30\n1/1 [==============================] - 0s 40ms/step - loss: 91.6803\nEpoch 10/30\n1/1 [==============================] - 0s 47ms/step - loss: 91.6803\nEpoch 11/30\n1/1 [==============================] - 0s 49ms/step - loss: 91.6802\nEpoch 12/30\n1/1 [==============================] - 0s 36ms/step - loss: 91.6801\nEpoch 13/30\n1/1 [==============================] - 0s 40ms/step - loss: 91.6800\nEpoch 14/30\n1/1 [==============================] - 0s 39ms/step - loss: 91.6799\nEpoch 15/30\n1/1 [==============================] - 0s 40ms/step - loss: 91.6797\nEpoch 16/30\n1/1 [==============================] - 0s 41ms/step - loss: 91.6796\nEpoch 17/30\n1/1 [==============================] - 0s 41ms/step - loss: 91.6794\nEpoch 18/30\n1/1 [==============================] - 0s 47ms/step - loss: 91.6793\nEpoch 19/30\n1/1 [==============================] - 0s 56ms/step - loss: 91.6791\nEpoch 20/30\n1/1 [==============================] - 0s 57ms/step - loss: 91.6789\nEpoch 21/30\n1/1 [==============================] - 0s 45ms/step - loss: 91.6787\nEpoch 22/30\n1/1 [==============================] - 0s 39ms/step - loss: 91.6785\nEpoch 23/30\n1/1 [==============================] - 0s 53ms/step - loss: 91.6783\nEpoch 24/30\n1/1 [==============================] - 0s 37ms/step - loss: 91.6781\nEpoch 25/30\n1/1 [==============================] - 0s 36ms/step - loss: 91.6779\nEpoch 26/30\n1/1 [==============================] - 0s 42ms/step - loss: 91.6777\nEpoch 27/30\n1/1 [==============================] - 0s 40ms/step - loss: 91.6774\nEpoch 28/30\n1/1 [==============================] - 0s 38ms/step - loss: 91.6772\nEpoch 29/30\n1/1 [==============================] - 0s 41ms/step - loss: 91.6770\nEpoch 30/30\n1/1 [==============================] - 0s 37ms/step - loss: 91.6768\n(1, 12280, 1)\nEpoch 1/30\n1/1 [==============================] - 0s 38ms/step - loss: 1617.8488\nEpoch 2/30\n1/1 [==============================] - 0s 53ms/step - loss: 1617.8486\nEpoch 3/30\n1/1 [==============================] - 0s 43ms/step - loss: 1617.8484\nEpoch 4/30\n1/1 [==============================] - 0s 45ms/step - loss: 1617.8480\nEpoch 5/30\n1/1 [==============================] - 0s 38ms/step - loss: 1617.8479\nEpoch 6/30\n1/1 [==============================] - 0s 39ms/step - loss: 1617.8474\nEpoch 7/30\n1/1 [==============================] - 0s 43ms/step - loss: 1617.8470\nEpoch 8/30\n1/1 [==============================] - 0s 39ms/step - loss: 1617.8467\nEpoch 9/30\n1/1 [==============================] - 0s 64ms/step - loss: 1617.8463\nEpoch 10/30\n1/1 [==============================] - 0s 39ms/step - loss: 1617.8459\nEpoch 11/30\n1/1 [==============================] - 0s 37ms/step - loss: 1617.8458\nEpoch 12/30\n1/1 [==============================] - 0s 42ms/step - loss: 1617.8455\nEpoch 13/30\n1/1 [==============================] - 0s 41ms/step - loss: 1617.8453\nEpoch 14/30\n1/1 [==============================] - 0s 42ms/step - loss: 1617.8451\nEpoch 15/30\n1/1 [==============================] - 0s 43ms/step - loss: 1617.8448\nEpoch 16/30\n1/1 [==============================] - 0s 42ms/step - loss: 1617.8444\nEpoch 17/30\n1/1 [==============================] - 0s 45ms/step - loss: 1617.8440\nEpoch 18/30\n1/1 [==============================] - 0s 42ms/step - loss: 1617.8436\nEpoch 19/30\n1/1 [==============================] - 0s 47ms/step - loss: 1617.8431\nEpoch 20/30\n1/1 [==============================] - 0s 43ms/step - loss: 1617.8428\nEpoch 21/30\n1/1 [==============================] - 0s 41ms/step - loss: 1617.8423\nEpoch 22/30\n1/1 [==============================] - 0s 41ms/step - loss: 1617.8418\nEpoch 23/30\n1/1 [==============================] - 0s 63ms/step - loss: 1617.8414\nEpoch 24/30\n1/1 [==============================] - 0s 47ms/step - loss: 1617.8411\nEpoch 25/30\n1/1 [==============================] - 0s 42ms/step - loss: 1617.8407\nEpoch 26/30\n1/1 [==============================] - 0s 39ms/step - loss: 1617.8403\nEpoch 27/30\n1/1 [==============================] - 0s 37ms/step - loss: 1617.8400\nEpoch 28/30\n1/1 [==============================] - 0s 43ms/step - loss: 1617.8397\nEpoch 29/30\n1/1 [==============================] - 0s 39ms/step - loss: 1617.8392\nEpoch 30/30\n1/1 [==============================] - 0s 42ms/step - loss: 1617.8387\n(1, 12280, 1)\nEpoch 1/30\n1/1 [==============================] - 0s 37ms/step - loss: 899.6222\nEpoch 2/30\n1/1 [==============================] - 0s 44ms/step - loss: 899.6220\nEpoch 3/30\n1/1 [==============================] - 0s 37ms/step - loss: 899.6217\nEpoch 4/30\n1/1 [==============================] - 0s 40ms/step - loss: 899.6215\nEpoch 5/30\n1/1 [==============================] - 0s 42ms/step - loss: 899.6214\nEpoch 6/30\n1/1 [==============================] - 0s 40ms/step - loss: 899.6211\nEpoch 7/30\n1/1 [==============================] - 0s 39ms/step - loss: 899.6208\nEpoch 8/30\n1/1 [==============================] - 0s 40ms/step - loss: 899.6204\nEpoch 9/30\n1/1 [==============================] - 0s 42ms/step - loss: 899.6201\nEpoch 10/30\n1/1 [==============================] - 0s 46ms/step - loss: 899.6198\nEpoch 11/30\n1/1 [==============================] - 0s 39ms/step - loss: 899.6194\nEpoch 12/30\n1/1 [==============================] - 0s 47ms/step - loss: 899.6189\nEpoch 13/30\n1/1 [==============================] - 0s 39ms/step - loss: 899.6184\nEpoch 14/30\n1/1 [==============================] - 0s 41ms/step - loss: 899.6181\nEpoch 15/30\n1/1 [==============================] - 0s 39ms/step - loss: 899.6177\nEpoch 16/30\n1/1 [==============================] - 0s 39ms/step - loss: 899.6172\nEpoch 17/30\n1/1 [==============================] - 0s 39ms/step - loss: 899.6167\nEpoch 18/30\n1/1 [==============================] - 0s 42ms/step - loss: 899.6161\nEpoch 19/30\n1/1 [==============================] - 0s 42ms/step - loss: 899.6157\nEpoch 20/30\n1/1 [==============================] - 0s 36ms/step - loss: 899.6151\nEpoch 21/30\n1/1 [==============================] - 0s 48ms/step - loss: 899.6146\nEpoch 22/30\n1/1 [==============================] - 0s 39ms/step - loss: 899.6142\nEpoch 23/30\n1/1 [==============================] - 0s 38ms/step - loss: 899.6137\nEpoch 24/30\n1/1 [==============================] - 0s 45ms/step - loss: 899.6130\nEpoch 25/30\n1/1 [==============================] - 0s 41ms/step - loss: 899.6126\nEpoch 26/30\n1/1 [==============================] - 0s 53ms/step - loss: 899.6121\nEpoch 27/30\n1/1 [==============================] - 0s 40ms/step - loss: 899.6116\nEpoch 28/30\n1/1 [==============================] - 0s 42ms/step - loss: 899.6110\nEpoch 29/30\n1/1 [==============================] - 0s 38ms/step - loss: 899.6106\nEpoch 30/30\n1/1 [==============================] - 0s 57ms/step - loss: 899.6102\n(1, 12280, 1)\nEpoch 1/30\n1/1 [==============================] - 0s 40ms/step - loss: 3750.1396\nEpoch 2/30\n1/1 [==============================] - 0s 40ms/step - loss: 3750.1396\nEpoch 3/30\n1/1 [==============================] - 0s 43ms/step - loss: 3750.1392\nEpoch 4/30\n1/1 [==============================] - 0s 44ms/step - loss: 3750.1392\nEpoch 5/30\n1/1 [==============================] - 0s 45ms/step - loss: 3750.1392\nEpoch 6/30\n1/1 [==============================] - 0s 42ms/step - loss: 3750.1392\nEpoch 7/30\n1/1 [==============================] - 0s 42ms/step - loss: 3750.1392\nEpoch 8/30\n1/1 [==============================] - 0s 41ms/step - loss: 3750.1392\nEpoch 9/30\n1/1 [==============================] - 0s 43ms/step - loss: 3750.1384\nEpoch 10/30\n1/1 [==============================] - 0s 42ms/step - loss: 3750.1384\nEpoch 11/30\n1/1 [==============================] - 0s 41ms/step - loss: 3750.1379\nEpoch 12/30\n1/1 [==============================] - 0s 40ms/step - loss: 3750.1379\nEpoch 13/30\n1/1 [==============================] - 0s 42ms/step - loss: 3750.1377\nEpoch 14/30\n1/1 [==============================] - 0s 40ms/step - loss: 3750.1377\nEpoch 15/30\n1/1 [==============================] - 0s 40ms/step - loss: 3750.1372\nEpoch 16/30\n1/1 [==============================] - 0s 40ms/step - loss: 3750.1372\nEpoch 17/30\n1/1 [==============================] - 0s 38ms/step - loss: 3750.1365\nEpoch 18/30\n1/1 [==============================] - 0s 39ms/step - loss: 3750.1365\nEpoch 19/30\n1/1 [==============================] - 0s 40ms/step - loss: 3750.1355\nEpoch 20/30\n1/1 [==============================] - 0s 44ms/step - loss: 3750.1353\nEpoch 21/30\n1/1 [==============================] - 0s 39ms/step - loss: 3750.1348\nEpoch 22/30\n1/1 [==============================] - 0s 38ms/step - loss: 3750.1345\nEpoch 23/30\n1/1 [==============================] - 0s 42ms/step - loss: 3750.1335\nEpoch 24/30\n1/1 [==============================] - 0s 47ms/step - loss: 3750.1335\nEpoch 25/30\n1/1 [==============================] - 0s 41ms/step - loss: 3750.1333\nEpoch 26/30\n1/1 [==============================] - 0s 40ms/step - loss: 3750.1328\nEpoch 27/30\n1/1 [==============================] - 0s 43ms/step - loss: 3750.1323\nEpoch 28/30\n1/1 [==============================] - 0s 40ms/step - loss: 3750.1321\nEpoch 29/30\n1/1 [==============================] - 0s 42ms/step - loss: 3750.1309\nEpoch 30/30\n1/1 [==============================] - 0s 37ms/step - loss: 3750.1309\n(1, 12280, 1)\nEpoch 1/30\n1/1 [==============================] - 0s 41ms/step - loss: 3900.0349\nEpoch 2/30\n1/1 [==============================] - 0s 38ms/step - loss: 3900.0349\nEpoch 3/30\n1/1 [==============================] - 0s 39ms/step - loss: 3900.0349\nEpoch 4/30\n1/1 [==============================] - 0s 39ms/step - loss: 3900.0349\nEpoch 5/30\n1/1 [==============================] - 0s 38ms/step - loss: 3900.0339\nEpoch 6/30\n1/1 [==============================] - 0s 41ms/step - loss: 3900.0337\nEpoch 7/30\n1/1 [==============================] - 0s 42ms/step - loss: 3900.0332\nEpoch 8/30\n1/1 [==============================] - 0s 42ms/step - loss: 3900.0332\nEpoch 9/30\n1/1 [==============================] - 0s 41ms/step - loss: 3900.0327\nEpoch 10/30\n1/1 [==============================] - 0s 42ms/step - loss: 3900.0315\nEpoch 11/30\n1/1 [==============================] - 0s 38ms/step - loss: 3900.0315\nEpoch 12/30\n1/1 [==============================] - 0s 40ms/step - loss: 3900.0305\nEpoch 13/30\n1/1 [==============================] - 0s 39ms/step - loss: 3900.0305\nEpoch 14/30\n1/1 [==============================] - 0s 45ms/step - loss: 3900.0295\nEpoch 15/30\n1/1 [==============================] - 0s 40ms/step - loss: 3900.0293\nEpoch 16/30\n1/1 [==============================] - 0s 38ms/step - loss: 3900.0281\nEpoch 17/30\n1/1 [==============================] - 0s 40ms/step - loss: 3900.0269\nEpoch 18/30\n1/1 [==============================] - 0s 40ms/step - loss: 3900.0256\nEpoch 19/30\n1/1 [==============================] - 0s 40ms/step - loss: 3900.0249\nEpoch 20/30\n1/1 [==============================] - 0s 43ms/step - loss: 3900.0239\nEpoch 21/30\n1/1 [==============================] - 0s 44ms/step - loss: 3900.0232\nEpoch 22/30\n1/1 [==============================] - 0s 41ms/step - loss: 3900.0220\nEpoch 23/30\n1/1 [==============================] - 0s 40ms/step - loss: 3900.0208\nEpoch 24/30\n1/1 [==============================] - 0s 40ms/step - loss: 3900.0195\nEpoch 25/30\n1/1 [==============================] - 0s 39ms/step - loss: 3900.0188\nEpoch 26/30\n1/1 [==============================] - 0s 40ms/step - loss: 3900.0183\nEpoch 27/30\n1/1 [==============================] - 0s 41ms/step - loss: 3900.0176\nEpoch 28/30\n1/1 [==============================] - 0s 43ms/step - loss: 3900.0164\nEpoch 29/30\n1/1 [==============================] - 0s 40ms/step - loss: 3900.0156\nEpoch 30/30\n1/1 [==============================] - 0s 39ms/step - loss: 3900.0156\n(1, 12280, 1)\nEpoch 1/30\n1/1 [==============================] - 0s 42ms/step - loss: 11681.9668\nEpoch 2/30\n1/1 [==============================] - 0s 37ms/step - loss: 11681.9668\nEpoch 3/30\n1/1 [==============================] - 0s 41ms/step - loss: 11681.9668\nEpoch 4/30\n1/1 [==============================] - 0s 42ms/step - loss: 11681.9658\nEpoch 5/30\n1/1 [==============================] - 0s 41ms/step - loss: 11681.9668\nEpoch 6/30\n1/1 [==============================] - 0s 39ms/step - loss: 11681.9648\nEpoch 7/30\n1/1 [==============================] - 0s 37ms/step - loss: 11681.9639\nEpoch 8/30\n1/1 [==============================] - 0s 41ms/step - loss: 11681.9639\nEpoch 9/30\n1/1 [==============================] - 0s 41ms/step - loss: 11681.9629\nEpoch 10/30\n1/1 [==============================] - 0s 40ms/step - loss: 11681.9629\nEpoch 11/30\n1/1 [==============================] - 0s 40ms/step - loss: 11681.9629\nEpoch 12/30\n1/1 [==============================] - 0s 40ms/step - loss: 11681.9619\nEpoch 13/30\n1/1 [==============================] - 0s 39ms/step - loss: 11681.9619\nEpoch 14/30\n1/1 [==============================] - 0s 40ms/step - loss: 11681.9600\nEpoch 15/30\n1/1 [==============================] - 0s 50ms/step - loss: 11681.9600\nEpoch 16/30\n1/1 [==============================] - 0s 52ms/step - loss: 11681.9600\nEpoch 17/30\n1/1 [==============================] - 0s 46ms/step - loss: 11681.9590\nEpoch 18/30\n1/1 [==============================] - 0s 46ms/step - loss: 11681.9580\nEpoch 19/30\n1/1 [==============================] - 0s 54ms/step - loss: 11681.9580\nEpoch 20/30\n1/1 [==============================] - 0s 45ms/step - loss: 11681.9580\nEpoch 21/30\n1/1 [==============================] - 0s 38ms/step - loss: 11681.9570\nEpoch 22/30\n1/1 [==============================] - 0s 40ms/step - loss: 11681.9561\nEpoch 23/30\n1/1 [==============================] - 0s 38ms/step - loss: 11681.9551\nEpoch 24/30\n1/1 [==============================] - 0s 36ms/step - loss: 11681.9541\nEpoch 25/30\n1/1 [==============================] - 0s 41ms/step - loss: 11681.9531\nEpoch 26/30\n1/1 [==============================] - 0s 39ms/step - loss: 11681.9521\nEpoch 27/30\n1/1 [==============================] - 0s 40ms/step - loss: 11681.9512\nEpoch 28/30\n1/1 [==============================] - 0s 39ms/step - loss: 11681.9492\nEpoch 29/30\n1/1 [==============================] - 0s 41ms/step - loss: 11681.9492\nEpoch 30/30\n1/1 [==============================] - 0s 44ms/step - loss: 11681.9482\n(1, 12280, 1)\nEpoch 1/30\n1/1 [==============================] - 0s 39ms/step - loss: 3362.2769\nEpoch 2/30\n1/1 [==============================] - 0s 38ms/step - loss: 3362.2769\nEpoch 3/30\n1/1 [==============================] - 0s 42ms/step - loss: 3362.2764\nEpoch 4/30\n1/1 [==============================] - 0s 50ms/step - loss: 3362.2764\nEpoch 5/30\n1/1 [==============================] - 0s 47ms/step - loss: 3362.2759\nEpoch 6/30\n1/1 [==============================] - 0s 42ms/step - loss: 3362.2759\nEpoch 7/30\n1/1 [==============================] - 0s 46ms/step - loss: 3362.2756\nEpoch 8/30\n1/1 [==============================] - 0s 42ms/step - loss: 3362.2749\nEpoch 9/30\n1/1 [==============================] - 0s 64ms/step - loss: 3362.2744\nEpoch 10/30\n1/1 [==============================] - 0s 44ms/step - loss: 3362.2737\nEpoch 11/30\n1/1 [==============================] - 0s 41ms/step - loss: 3362.2727\nEpoch 12/30\n1/1 [==============================] - 0s 37ms/step - loss: 3362.2725\nEpoch 13/30\n1/1 [==============================] - 0s 39ms/step - loss: 3362.2715\nEpoch 14/30\n1/1 [==============================] - 0s 38ms/step - loss: 3362.2705\nEpoch 15/30\n1/1 [==============================] - 0s 37ms/step - loss: 3362.2700\nEpoch 16/30\n1/1 [==============================] - 0s 40ms/step - loss: 3362.2695\nEpoch 17/30\n1/1 [==============================] - 0s 41ms/step - loss: 3362.2688\nEpoch 18/30\n1/1 [==============================] - 0s 40ms/step - loss: 3362.2681\nEpoch 19/30\n1/1 [==============================] - 0s 37ms/step - loss: 3362.2671\nEpoch 20/30\n1/1 [==============================] - 0s 45ms/step - loss: 3362.2668\nEpoch 21/30\n1/1 [==============================] - 0s 50ms/step - loss: 3362.2656\nEpoch 22/30\n1/1 [==============================] - 0s 39ms/step - loss: 3362.2649\nEpoch 23/30\n1/1 [==============================] - 0s 38ms/step - loss: 3362.2639\nEpoch 24/30\n1/1 [==============================] - 0s 46ms/step - loss: 3362.2632\nEpoch 25/30\n1/1 [==============================] - 0s 41ms/step - loss: 3362.2620\nEpoch 26/30\n1/1 [==============================] - 0s 38ms/step - loss: 3362.2612\nEpoch 27/30\n1/1 [==============================] - 0s 43ms/step - loss: 3362.2605\nEpoch 28/30\n1/1 [==============================] - 0s 38ms/step - loss: 3362.2595\nEpoch 29/30\n1/1 [==============================] - 0s 43ms/step - loss: 3362.2588\nEpoch 30/30\n1/1 [==============================] - 0s 46ms/step - loss: 3362.2581\n(1, 12280, 1)\nEpoch 1/30\n1/1 [==============================] - 0s 43ms/step - loss: 17027.5059\nEpoch 2/30\n1/1 [==============================] - 0s 42ms/step - loss: 17027.5059\nEpoch 3/30\n1/1 [==============================] - 0s 44ms/step - loss: 17027.5059\nEpoch 4/30\n1/1 [==============================] - 0s 40ms/step - loss: 17027.5059\nEpoch 5/30\n1/1 [==============================] - 0s 37ms/step - loss: 17027.5059\nEpoch 6/30\n1/1 [==============================] - 0s 42ms/step - loss: 17027.5059\nEpoch 7/30\n1/1 [==============================] - 0s 42ms/step - loss: 17027.5039\nEpoch 8/30\n1/1 [==============================] - 0s 54ms/step - loss: 17027.5039\nEpoch 9/30\n1/1 [==============================] - 0s 47ms/step - loss: 17027.5039\nEpoch 10/30\n1/1 [==============================] - 0s 40ms/step - loss: 17027.5020\nEpoch 11/30\n1/1 [==============================] - 0s 43ms/step - loss: 17027.5020\nEpoch 12/30\n1/1 [==============================] - 0s 41ms/step - loss: 17027.5020\nEpoch 13/30\n1/1 [==============================] - 0s 39ms/step - loss: 17027.5000\nEpoch 14/30\n1/1 [==============================] - 0s 40ms/step - loss: 17027.5000\nEpoch 15/30\n1/1 [==============================] - 0s 40ms/step - loss: 17027.5000\nEpoch 16/30\n1/1 [==============================] - 0s 42ms/step - loss: 17027.5000\nEpoch 17/30\n1/1 [==============================] - 0s 42ms/step - loss: 17027.4980\nEpoch 18/30\n1/1 [==============================] - 0s 43ms/step - loss: 17027.4980\nEpoch 19/30\n1/1 [==============================] - 0s 38ms/step - loss: 17027.4980\nEpoch 20/30\n1/1 [==============================] - 0s 40ms/step - loss: 17027.4980\nEpoch 21/30\n1/1 [==============================] - 0s 37ms/step - loss: 17027.4980\nEpoch 22/30\n1/1 [==============================] - 0s 54ms/step - loss: 17027.4961\nEpoch 23/30\n1/1 [==============================] - 0s 39ms/step - loss: 17027.4941\nEpoch 24/30\n1/1 [==============================] - 0s 43ms/step - loss: 17027.4941\nEpoch 25/30\n1/1 [==============================] - 0s 39ms/step - loss: 17027.4902\nEpoch 26/30\n1/1 [==============================] - 0s 40ms/step - loss: 17027.4902\nEpoch 27/30\n1/1 [==============================] - 0s 42ms/step - loss: 17027.4902\nEpoch 28/30\n1/1 [==============================] - 0s 41ms/step - loss: 17027.4902\nEpoch 29/30\n1/1 [==============================] - 0s 42ms/step - loss: 17027.4902\nEpoch 30/30\n1/1 [==============================] - 0s 42ms/step - loss: 17027.4902\n(1, 12280, 1)\nEpoch 1/30\n1/1 [==============================] - 0s 38ms/step - loss: 2077.2107\nEpoch 2/30\n1/1 [==============================] - 0s 54ms/step - loss: 2077.2109\nEpoch 3/30\n1/1 [==============================] - 0s 40ms/step - loss: 2077.2112\nEpoch 4/30\n1/1 [==============================] - 0s 40ms/step - loss: 2077.2109\nEpoch 5/30\n1/1 [==============================] - 0s 41ms/step - loss: 2077.2109\nEpoch 6/30\n1/1 [==============================] - 0s 39ms/step - loss: 2077.2107\nEpoch 7/30\n1/1 [==============================] - 0s 40ms/step - loss: 2077.2107\nEpoch 8/30\n1/1 [==============================] - 0s 37ms/step - loss: 2077.2102\nEpoch 9/30\n1/1 [==============================] - 0s 40ms/step - loss: 2077.2100\nEpoch 10/30\n1/1 [==============================] - 0s 41ms/step - loss: 2077.2095\nEpoch 11/30\n1/1 [==============================] - 0s 51ms/step - loss: 2077.2090\nEpoch 12/30\n1/1 [==============================] - 0s 39ms/step - loss: 2077.2085\nEpoch 13/30\n1/1 [==============================] - 0s 56ms/step - loss: 2077.2083\nEpoch 14/30\n1/1 [==============================] - 0s 36ms/step - loss: 2077.2075\nEpoch 15/30\n1/1 [==============================] - 0s 48ms/step - loss: 2077.2068\nEpoch 16/30\n1/1 [==============================] - 0s 43ms/step - loss: 2077.2061\nEpoch 17/30\n1/1 [==============================] - 0s 39ms/step - loss: 2077.2056\nEpoch 18/30\n1/1 [==============================] - 0s 41ms/step - loss: 2077.2048\nEpoch 19/30\n1/1 [==============================] - 0s 43ms/step - loss: 2077.2041\nEpoch 20/30\n1/1 [==============================] - 0s 46ms/step - loss: 2077.2031\nEpoch 21/30\n1/1 [==============================] - 0s 40ms/step - loss: 2077.2024\nEpoch 22/30\n1/1 [==============================] - 0s 63ms/step - loss: 2077.2019\nEpoch 23/30\n1/1 [==============================] - 0s 45ms/step - loss: 2077.2012\nEpoch 24/30\n1/1 [==============================] - 0s 39ms/step - loss: 2077.2004\nEpoch 25/30\n1/1 [==============================] - 0s 38ms/step - loss: 2077.1997\nEpoch 26/30\n1/1 [==============================] - 0s 38ms/step - loss: 2077.1990\nEpoch 27/30\n1/1 [==============================] - 0s 42ms/step - loss: 2077.1980\nEpoch 28/30\n1/1 [==============================] - 0s 52ms/step - loss: 2077.1975\nEpoch 29/30\n1/1 [==============================] - 0s 50ms/step - loss: 2077.1963\nEpoch 30/30\n1/1 [==============================] - 0s 47ms/step - loss: 2077.1953\n(1, 12280, 1)\nEpoch 1/30\n1/1 [==============================] - 0s 43ms/step - loss: 3739.3313\nEpoch 2/30\n1/1 [==============================] - 0s 44ms/step - loss: 3739.3308\nEpoch 3/30\n1/1 [==============================] - 0s 42ms/step - loss: 3739.3308\nEpoch 4/30\n1/1 [==============================] - 0s 62ms/step - loss: 3739.3303\nEpoch 5/30\n1/1 [==============================] - 0s 58ms/step - loss: 3739.3303\nEpoch 6/30\n1/1 [==============================] - 0s 67ms/step - loss: 3739.3301\nEpoch 7/30\n1/1 [==============================] - 0s 46ms/step - loss: 3739.3296\nEpoch 8/30\n1/1 [==============================] - 0s 42ms/step - loss: 3739.3291\nEpoch 9/30\n1/1 [==============================] - 0s 37ms/step - loss: 3739.3289\nEpoch 10/30\n1/1 [==============================] - 0s 39ms/step - loss: 3739.3279\nEpoch 11/30\n1/1 [==============================] - 0s 38ms/step - loss: 3739.3271\nEpoch 12/30\n1/1 [==============================] - 0s 40ms/step - loss: 3739.3269\nEpoch 13/30\n1/1 [==============================] - 0s 39ms/step - loss: 3739.3264\nEpoch 14/30\n1/1 [==============================] - 0s 41ms/step - loss: 3739.3257\nEpoch 15/30\n1/1 [==============================] - 0s 43ms/step - loss: 3739.3245\nEpoch 16/30\n1/1 [==============================] - 0s 40ms/step - loss: 3739.3240\nEpoch 17/30\n1/1 [==============================] - 0s 40ms/step - loss: 3739.3232\nEpoch 18/30\n1/1 [==============================] - 0s 39ms/step - loss: 3739.3225\nEpoch 19/30\n1/1 [==============================] - 0s 41ms/step - loss: 3739.3215\nEpoch 20/30\n1/1 [==============================] - 0s 39ms/step - loss: 3739.3208\nEpoch 21/30\n1/1 [==============================] - 0s 47ms/step - loss: 3739.3201\nEpoch 22/30\n1/1 [==============================] - 0s 45ms/step - loss: 3739.3191\nEpoch 23/30\n1/1 [==============================] - 0s 47ms/step - loss: 3739.3181\nEpoch 24/30\n1/1 [==============================] - 0s 44ms/step - loss: 3739.3176\nEpoch 25/30\n1/1 [==============================] - 0s 45ms/step - loss: 3739.3169\nEpoch 26/30\n1/1 [==============================] - 0s 55ms/step - loss: 3739.3159\nEpoch 27/30\n1/1 [==============================] - 0s 41ms/step - loss: 3739.3152\nEpoch 28/30\n1/1 [==============================] - 0s 41ms/step - loss: 3739.3145\nEpoch 29/30\n1/1 [==============================] - 0s 43ms/step - loss: 3739.3140\nEpoch 30/30\n1/1 [==============================] - 0s 45ms/step - loss: 3739.3132\n(1, 12280, 1)\nEpoch 1/30\n1/1 [==============================] - 0s 43ms/step - loss: 516.9252\nEpoch 2/30\n1/1 [==============================] - 0s 41ms/step - loss: 516.9254\nEpoch 3/30\n1/1 [==============================] - 0s 40ms/step - loss: 516.9255\nEpoch 4/30\n1/1 [==============================] - 0s 43ms/step - loss: 516.9255\nEpoch 5/30\n1/1 [==============================] - 0s 48ms/step - loss: 516.9254\nEpoch 6/30\n1/1 [==============================] - 0s 38ms/step - loss: 516.9252\nEpoch 7/30\n1/1 [==============================] - 0s 39ms/step - loss: 516.9249\nEpoch 8/30\n1/1 [==============================] - 0s 35ms/step - loss: 516.9245\nEpoch 9/30\n1/1 [==============================] - 0s 39ms/step - loss: 516.9241\nEpoch 10/30\n1/1 [==============================] - 0s 36ms/step - loss: 516.9237\nEpoch 11/30\n1/1 [==============================] - 0s 40ms/step - loss: 516.9232\nEpoch 12/30\n1/1 [==============================] - 0s 37ms/step - loss: 516.9226\nEpoch 13/30\n1/1 [==============================] - 0s 37ms/step - loss: 516.9220\nEpoch 14/30\n1/1 [==============================] - 0s 38ms/step - loss: 516.9214\nEpoch 15/30\n1/1 [==============================] - 0s 39ms/step - loss: 516.9207\nEpoch 16/30\n1/1 [==============================] - 0s 39ms/step - loss: 516.9200\nEpoch 17/30\n1/1 [==============================] - 0s 39ms/step - loss: 516.9193\nEpoch 18/30\n1/1 [==============================] - 0s 41ms/step - loss: 516.9185\nEpoch 19/30\n1/1 [==============================] - 0s 39ms/step - loss: 516.9177\nEpoch 20/30\n1/1 [==============================] - 0s 41ms/step - loss: 516.9170\nEpoch 21/30\n1/1 [==============================] - 0s 42ms/step - loss: 516.9161\nEpoch 22/30\n1/1 [==============================] - 0s 39ms/step - loss: 516.9153\nEpoch 23/30\n1/1 [==============================] - 0s 44ms/step - loss: 516.9145\nEpoch 24/30\n1/1 [==============================] - 0s 40ms/step - loss: 516.9136\nEpoch 25/30\n1/1 [==============================] - 0s 41ms/step - loss: 516.9127\nEpoch 26/30\n1/1 [==============================] - 0s 38ms/step - loss: 516.9119\nEpoch 27/30\n1/1 [==============================] - 0s 41ms/step - loss: 516.9110\nEpoch 28/30\n1/1 [==============================] - 0s 40ms/step - loss: 516.9102\nEpoch 29/30\n1/1 [==============================] - 0s 36ms/step - loss: 516.9093\nEpoch 30/30\n1/1 [==============================] - 0s 40ms/step - loss: 516.9084\n"
],
[
" \"\"\"Testing with random interval(DeepAnT)\"\"\"\n# Set number of test sequences \nn_test_seq = 1\n\n# Split a univariate sequence into samples\ndef generate_test_batch(raw_seq, n_test_seq):\n # Sample a portion of the raw_seq randomly\n ran_ix = random.randint(0,len(raw_seq) - n_test_seq * w - n_test_seq * p_w)\n raw_test_seq = array(raw_seq[ran_ix:ran_ix + n_test_seq * w + n_test_seq * p_w])\n batch_test_seq, batch_test_label = list(), list()\n ix = ran_ix\n for i in range(n_test_seq):\n # gather input and output parts of the pattern\n seq_x = raw_seq[ix : ix+w],\n seq_y = raw_seq[ix+w : ix+w+p_w]\n ix = ix+w+p_w\n batch_test_seq.append(seq_x)\n batch_test_label.append(seq_y)\n return array(batch_test_seq), array(batch_test_label)\n\nbatch_test_seq, batch_test_label = generate_test_batch(list(reg_data.ix[:,0]), n_test_seq)\nbatch_test_seq = batch_test_seq.reshape((batch_test_seq.shape[0], w, n_features))\nbatch_test_label = batch_test_label.reshape((batch_test_label.shape[0], p_w))\n\n# Returns the loss value & metrics values for the model in test mode\nmodel.evaluate(x=batch_test_seq,\n y=batch_test_label,\n verbose=1) ",
"C:\\Users\\smoor\\Anaconda3\\lib\\site-packages\\ipykernel_launcher.py:21: DeprecationWarning: \n.ix is deprecated. Please use\n.loc for label based indexing or\n.iloc for positional indexing\n\nSee the documentation here:\nhttp://pandas.pydata.org/pandas-docs/stable/indexing.html#ix-indexer-is-deprecated\n"
],
[
" \"\"\"Save Weights (DeepAnT)\"\"\"\n# save it to disk so we can load it back up anytime\nmodel.save_weights('ch_3_noanom_weights.h5') ",
"_____no_output_____"
],
[
" \"\"\"Predicting future sequence (DeepAnT)\"\"\"\n# Build model \nmodel = Sequential()\nmodel.add(Conv1D(filters=num_filt_1,\n kernel_size=kernel_size,\n strides=conv_strides,\n padding='valid',\n activation='relu',\n input_shape=(w, n_features)))\nmodel.add(MaxPooling1D(pool_size=pool_size_1)) \nmodel.add(Conv1D(filters=num_filt_2,\n kernel_size=kernel_size,\n strides=conv_strides,\n padding='valid',\n activation='relu'))\nmodel.add(MaxPooling1D(pool_size=pool_size_2))\nmodel.add(Flatten())\nmodel.add(Dense(units=num_nrn_dl, activation='relu')) \nmodel.add(Dropout(dropout_rate))\nmodel.add(Dense(units=num_nrn_ol))\n\n# Load the model's saved weights.\nmodel.load_weights('ch_3_noanom_weights.h5')\n \n \nraw_seq = list(reg_data.ix[:,0])\nendix = len(raw_seq) - w - p_w\ninput_seq = array(raw_seq[endix:endix+w])\ntarget_seq = array(raw_seq[endix+w:endix+w+p_w]) \ninput_seq = input_seq.reshape((1, w, n_features))\n\n# Predict the next time stampes of the sampled sequence\npredicted_seq = model.predict(input_seq, verbose=1)\n\n# Print our model's predictions.\nprint(predicted_seq)\n\n# Check our predictions against the ground truths.\nprint(target_seq) # [7, 2, 1, 0, 4]",
"C:\\Users\\smoor\\Anaconda3\\lib\\site-packages\\ipykernel_launcher.py:26: DeprecationWarning: \n.ix is deprecated. Please use\n.loc for label based indexing or\n.iloc for positional indexing\n\nSee the documentation here:\nhttp://pandas.pydata.org/pandas-docs/stable/indexing.html#ix-indexer-is-deprecated\n"
],
[
" '''Visualization of predicted time series'''\nin_seq = reg_data.ix[:,i][endix:endix+w]\ntar_seq = reg_data.ix[:,i][endix+w:endix+w+p_w]\npredicted_seq = predicted_seq.reshape((p_w))\nd = {'time': reg_data.ix[:,i][endix+w:endix+w+p_w], 'values': predicted_seq}\ndf_sine_pre = pd.DataFrame(data=d)\npre_seq = df_sine_pre['values']\n\nplt.plot(in_seq)\nplt.plot(tar_seq)\nplt.plot(pre_seq)\n\n\nplt.ylim(top = 10)\nplt.ylim(bottom=0)\n\nplt.title('Channel 27 Prediction')\nplt.ylabel('value')\nplt.xlabel('time')\nplt.legend(['input_seq', 'pre_seq', 'target_seq'], loc='upper right')\naxes = plt.gca()\naxes.set_xlim([endix,endix+w+p_w])\nfig_predict = plt.figure(figsize=(100,10))\nfig_predict.savefig('predicted_sequence.png')\nplt.show() ",
"C:\\Users\\smoor\\Anaconda3\\lib\\site-packages\\ipykernel_launcher.py:2: DeprecationWarning: \n.ix is deprecated. Please use\n.loc for label based indexing or\n.iloc for positional indexing\n\nSee the documentation here:\nhttp://pandas.pydata.org/pandas-docs/stable/indexing.html#ix-indexer-is-deprecated\n \nC:\\Users\\smoor\\Anaconda3\\lib\\site-packages\\ipykernel_launcher.py:3: DeprecationWarning: \n.ix is deprecated. Please use\n.loc for label based indexing or\n.iloc for positional indexing\n\nSee the documentation here:\nhttp://pandas.pydata.org/pandas-docs/stable/indexing.html#ix-indexer-is-deprecated\n This is separate from the ipykernel package so we can avoid doing imports until\nC:\\Users\\smoor\\Anaconda3\\lib\\site-packages\\ipykernel_launcher.py:5: DeprecationWarning: \n.ix is deprecated. Please use\n.loc for label based indexing or\n.iloc for positional indexing\n\nSee the documentation here:\nhttp://pandas.pydata.org/pandas-docs/stable/indexing.html#ix-indexer-is-deprecated\n \"\"\"\n"
],
[
"'''\n\n \"\"\"Predicting random intervals (DeepAnT)\"\"\"\n# Build model \nmodel = Sequential()\nmodel.add(Conv1D(filters=num_filt_1,\n kernel_size=kernel_size,\n strides=conv_strides,\n padding='valid',\n activation='relu',\n input_shape=(w, n_features)))\nmodel.add(MaxPooling1D(pool_size=pool_size_1)) \nmodel.add(Conv1D(filters=num_filt_2,\n kernel_size=kernel_size,\n strides=conv_strides,\n padding='valid',\n activation='relu'))\nmodel.add(MaxPooling1D(pool_size=pool_size_2))\nmodel.add(Flatten())\nmodel.add(Dense(units=num_nrn_dl, activation='relu')) \nmodel.add(Dropout(dropout_rate))\nmodel.add(Dense(units=num_nrn_ol))\n\n# Load the model's saved weights.\nmodel.load_weights('ch_1_weights.h5')\n \n# Sample a portion of the raw_seq randomly\n# 1. Choose \nran_ix = random.randint(1,len(raw_seq) - w - p_w)\ninput_seq = array(raw_seq[ran_ix : ran_ix + w])\ntarget_seq = array(raw_seq[ran_ix + w : ran_ix + w + p_w])\ninput_seq = input_seq.reshape((1, w, n_features))\n\n# Predict the next time stampes of the sampled sequence\nyhat = model.predict(input_seq, verbose=1)\n\n# Print our model's predictions.\nprint(yhat)\n\n# Check our predictions against the ground truths.\nprint(target_seq) # [7, 2, 1, 0, 4]\n'''",
"_____no_output_____"
],
[
"\"\"\"\nDetermins whether a sequence exceeds the threshold for being an anomaly\n\nreturn boolean value of whether the sequence is an anomaly or not\n\"\"\"\ndef anomaly_detector(prediction_seq, ground_truth_seq):\n # calculate Euclidean between actual seq and predicted seq\n dist = np.linalg.norm(ground_truth_seq - prediction_seq) \n if (dist > anm_det_thr):\n return true # anomaly\n else:\n return false # normal ",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb8a505bd51702c5a9698ae4cfe956253c7bfde5
| 11,096 |
ipynb
|
Jupyter Notebook
|
src/utils/.ipynb_checkpoints/split_train_data-checkpoint.ipynb
|
irebuf/audio-event-tagging
|
1fe247c1d24b9fdc02d442a9753184f7e5006cef
|
[
"MIT"
] | null | null | null |
src/utils/.ipynb_checkpoints/split_train_data-checkpoint.ipynb
|
irebuf/audio-event-tagging
|
1fe247c1d24b9fdc02d442a9753184f7e5006cef
|
[
"MIT"
] | null | null | null |
src/utils/.ipynb_checkpoints/split_train_data-checkpoint.ipynb
|
irebuf/audio-event-tagging
|
1fe247c1d24b9fdc02d442a9753184f7e5006cef
|
[
"MIT"
] | 2 |
2020-10-28T09:35:26.000Z
|
2020-12-08T12:12:31.000Z
| 28.671835 | 118 | 0.41186 |
[
[
[
"import os\nimport pandas as pd\nimport numpy as np\nimport json\nfrom collections import defaultdict",
"_____no_output_____"
],
[
"data_path = os.path.abspath(os.path.join(os.getcwd(),os.pardir,os.pardir))+'/data/'",
"_____no_output_____"
],
[
"dictionary = {'Animal': {'/m/0ch8v','/m/01280g','/m/068hy'},\n 'Humans':{'/m/02zsn','/m/01w250','/m/09hlz4','/m/0bpl036','/m/09l8g',\n '/m/0k65p','/m/01jg02','/t/dd00012','m/05zppz','/m/09x0r'},\n 'Natural':{'/m/03m9d0z','/m/0jb2l','/m/0838f','/m/02_41'}\n 'Music':{'/m/04rlf'}}",
"_____no_output_____"
]
],
[
[
"- speech is one of the most interesting feature for humans hence it's the most used \n- Music is not used atm\n- Animals: Domestic animals / Livestock / Wild animals\n- Natural: Wind / Thunderstorm / Water / Fire\n",
"_____no_output_____"
],
[
"Step 1: Take only these classes",
"_____no_output_____"
],
[
"Read dataset",
"_____no_output_____"
]
],
[
[
"lines =[]\nwith open(data_path+'unbalanced_train_segments.csv','r') as file:\n lines = file.readlines()\nlines = lines[3:]",
"_____no_output_____"
]
],
[
[
"Create dataframe for each category [it takes a while - not really efficient]",
"_____no_output_____"
]
],
[
[
"dataframe = []\n#for each category\nfor i,key in enumerate(dictionary.keys()):\n data = np.ones(shape=(1,4)) \n for line in lines:\n elements = line.rstrip().split(',')\n common = list(dictionary[key].intersection(elements[3:]))\n #no control about the subgroup\n if common != []:\n data = np.vstack([data, np.array(elements[:3] + [key]).reshape(1,4)])\n dataframe.append(data)\n ",
"_____no_output_____"
],
[
"def get_sample(arr,k=5000): #take k-sample from the dataset\n idx = np.random.choice(range(2,len(arr)),replace=False,size=k)\n return arr[idx]",
"_____no_output_____"
],
[
"temp = np.vstack([np.vstack([get_sample(dataframe[0]), get_sample(dataframe[1])]), get_sample(dataframe[2])])\ndf = pd.DataFrame(temp, columns=['url', 'start_time', 'end_time', 'class_label'])",
"_____no_output_____"
],
[
"df.class_label.value_counts() #it's balanced -> justify it",
"_____no_output_____"
],
[
"df.sample(20)",
"_____no_output_____"
],
[
"df.to_csv(data_path+'data_split.csv') #save dataset",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb8a5ac0ccd1b5b18211a8fe7302d274c3ff5f85
| 42,074 |
ipynb
|
Jupyter Notebook
|
working_on_posts/notebooks/scratch_nn.ipynb
|
a-i-dan/a-i-dan.github.io
|
ced86259bc796cc2287c17895de1130c482e38b3
|
[
"MIT"
] | null | null | null |
working_on_posts/notebooks/scratch_nn.ipynb
|
a-i-dan/a-i-dan.github.io
|
ced86259bc796cc2287c17895de1130c482e38b3
|
[
"MIT"
] | null | null | null |
working_on_posts/notebooks/scratch_nn.ipynb
|
a-i-dan/a-i-dan.github.io
|
ced86259bc796cc2287c17895de1130c482e38b3
|
[
"MIT"
] | null | null | null | 82.175781 | 13,892 | 0.742691 |
[
[
[
"# Simple Neural Networks: Revised\n\nBack in February I published a post title [<i>Simple Neural Networks with Numpy</i>](https://a-i-dan.github.io/tanh_NN). I wanted to take a deep dive into the world of neural networks and learn everything that went into making a neural net seem \"magical\". Now, a few months later, I want to rewrite that post. At the time of the original post, I was a novice in both machine learning and the Python language. While I still consider myself in the beginning stages of my growth, I have learned a lot in the recent months and have realized the mistakes that were made in my original post. I thought about deleting the post and rewriting it, but I made this blog to keep a track record of my progress, and deleting the original post goes against this idea. I am sure there will be mistakes in this post as well as I am still learning this field.\n\nThis blog post will be a revised, and way better, version of my original blog post with a similar title. Unlike the original post, in this post, building the neural network's architecture will be achieved by taking an object-oriented approach (this is mainly to help me learn). Therefore, the code will not be the most efficient code possible and will be a bit more lengthy. This neural network will contain one hidden layer and will be using the <b>sigmoid activation function</b>. The full code for this project will be posted below (including plot and predictions), then blocks of code will be seperated and explained in further detail.\n\n## What is a Neural Network?\n\nA neural network is loosely based on how the human brain works: many neurons connected to other neurons, passing information through their connections and firing when the input to a neuron surpasses a certain threshold. Our artificial neural network will consist of artificial neurons and synapses with information being passed between them. The synapses, or connections, will be weighted according to the neurons strength of influence on determining the output. These synaptic weights will go through an optimization process called <b>backpropagation</b>. For each iteration during the training process, backpropagation will be used to go backwards through the layers of the network and adjusts the weights according to their contribution to the neural net's error. \n\nNeural networks are essentially self-optimizing functions that map inputs to the correct outputs. We can then place a new input into the function, where it will predict an output based on the function it created with the training data.\n\n<img src='https://github.com/a-i-dan/a-i-dan.github.io/blob/master/images/NN_Revised/Untitled%20presentation-2.png?raw=true' style='display: block; margin: auto; width: 700px;'>\n\n## Neural Net's Goal\n\nThis neural network, like all neural networks, will have to learn what the important features are in the data to produce the output. In paticular, this neural net will be given an input matrix with six samples, each with three feature columns consisting of soley zeros and ones. For example, one sample in the training set may be [0, 1, 1]. The output to each sample will be a single one or zero. The output will be determined by the number in the first feature column of the data samples. Using the example given before, the output for [0, 1, 1] would be 0, because the first column contains a zero. An example chart will be given below to demonstrate the output for each input sample.\n\n<img src='https://github.com/a-i-dan/a-i-dan.github.io/blob/master/images/NN_Revised/Untitled%20presentation.png?raw=true' style='display: block; margin: auto; width: 600px;'>",
"_____no_output_____"
],
[
"### Full Code",
"_____no_output_____"
]
],
[
[
"import numpy as np # helps with the math\nimport matplotlib.pyplot as plt # to plot error during training\n\n# input data\ninputs = np.array([[0, 1, 0],\n [0, 1, 1],\n [0, 0, 0],\n [1, 0, 0],\n [1, 1, 1],\n [1, 0, 1]])\n# output data\noutputs = np.array([[0], [0], [0], [1], [1], [1]])\n\n# create NeuralNetwork class\nclass NeuralNetwork:\n \n # intialize variables in class\n def __init__(self, inputs, outputs):\n self.inputs = inputs\n self.outputs = outputs\n # initialize weights as .50 for simplicity\n self.weights = np.array([[.50], [.50], [.50]])\n self.error_history = []\n self.epoch_list = []\n \n #activation function ==> S(x) = 1/1+e^(-x)\n def sigmoid(self, x, deriv=False):\n if deriv == True:\n return x * (1 - x)\n return 1 / (1 + np.exp(-x))\n \n # data will flow through the neural network.\n def feed_forward(self):\n self.hidden = self.sigmoid(np.dot(self.inputs, self.weights))\n \n # going backwards through the network to update weights\n def backpropagation(self):\n self.error = self.outputs - self.hidden\n delta = self.error * self.sigmoid(self.hidden, deriv=True)\n self.weights += np.dot(self.inputs.T, delta)\n \n # train the neural net for 25,000 iterations\n def train(self, epochs=25000): \n for epoch in range(epochs):\n # flow forward and produce an output\n self.feed_forward()\n # go back though the network to make corrections based on the output\n self.backpropagation() \n # keep track of the error history over each epoch\n self.error_history.append(np.average(np.abs(self.error)))\n self.epoch_list.append(epoch)\n \n # function to predict output on new and unseen input data \n def predict(self, new_input):\n prediction = self.sigmoid(np.dot(new_input, self.weights))\n return prediction\n\n# create neural network \nNN = NeuralNetwork(inputs, outputs)\n# train neural network\nNN.train()\n\n# create two new examples to predict \nexample = np.array([[1, 1, 0]])\nexample_2 = np.array([[0, 1, 1]])\n\n# print the predictions for both examples \nprint(NN.predict(example), ' - Correct: ', example[0][0])\nprint(NN.predict(example_2), ' - Correct: ', example_2[0][0])\n\n# plot the error over the entire training duration\nplt.figure(figsize=(15,5))\nplt.plot(NN.epoch_list, NN.error_history)\nplt.xlabel('Epoch')\nplt.ylabel('Error')\nplt.show()",
"[[0.99089925]] - Correct: 1\n[[0.006409]] - Correct: 0\n"
]
],
[
[
"## Code Breakdown",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt",
"_____no_output_____"
]
],
[
[
"Before getting started, we will need to import the necessary libraries. Only two libraries will be needed for this example, without plotting the loss we would only need Numpy. Numpy is a python math library mainly used for linear algebra applications. Matplotlib is a visualization tool that we will use to create a plot to display how our error decreases over time.",
"_____no_output_____"
]
],
[
[
"inputs = np.array([[0, 1, 0],\n [0, 1, 1],\n [0, 0, 0],\n [1, 0, 0],\n [1, 1, 1],\n [1, 0, 1]])\n\noutputs = np.array([[0], [0], [0], [1], [1], [1]])",
"_____no_output_____"
]
],
[
[
"As mentioned earlier, neural networks need data to learn from. We will create our input data matrix and the corresponding outputs matrix with Numpy's `.array()` function. Each sample in the input consists of three feature columns made up of 0s and 1s that produce one output of either a 0 or 1. We want to neural network to learn that the outputs are determined by the first feature column in each sample.\n\n$$\\begin{matrix}\n[0 & 1 & 0] ==>\\\\\n[0 & 1 & 1] ==>\\\\\n[1 & 0 & 0] ==>\\\\\n[1 & 0 & 1] ==>\\end{matrix}\\begin{matrix}\n0\\\\\n0\\\\\n1\\\\\n1\\end{matrix}$$",
"_____no_output_____"
]
],
[
[
"class NeuralNetwork:\n \n def __init__(self, inputs, outputs):\n self.inputs = inputs\n self.outputs = outputs\n self.weights = np.array([[.50], [.50], [.50]])\n self.error_history = []\n self.epoch_list = []",
"_____no_output_____"
]
],
[
[
"We will take an object-oriented approach to building this paticular neural network. We can begin by creating a class called \"NeuralNetwork\" and initializing the class by defining the `__init__` function. Our `__init__` function will take the inputs and outputs as arguments. We will also need to define our weights, which, for simplicity, will start with each weight being .50. Because each feature in the data must be connected to the hidden layer, we will need a weight for each feature in the data (three weights). For plotting purposes, we will also create two empty lists: loss_history and epoch_list. This will keep track of our neural network's error at each epoch during the training process.",
"_____no_output_____"
]
],
[
[
"def sigmoid(self, x, deriv=False):\n if deriv == True:\n return x * (1 - x)\n return 1 / (1 + np.exp(-x))",
"_____no_output_____"
]
],
[
[
"This neural network will be using the sigmoid function, or logistic function, as the activation function. The sigmoid function is a popular nonlinear activation function that has a range of (0-1). The inputs to this function will always be squished down to fit inbetween the sigmoid function's two horizontal asymptotes at y=0 and y=1. The sigmoid function has some well known issues that restrict its usage. When we look at the graph below of the sigmoidal curve, we notice that as we reach the two ends of the curve, the derivatives of those points become very small. When these small derivatives are multiplied during backpropagation, they become smaller and smaller until becoming useless. Due to the derivatives, or gradients, getting smaller and smaller, the weights in the neural network will not be updated very much, if at all. This will lead the neural network to become stuck, with the situation becoming worse and worse for every additional training iteration.\n\n<img src='https://github.com/a-i-dan/a-i-dan.github.io/blob/master/images/NN_Revised/Unknown-13?raw=true' style='display: block; margin: auto; width: 600px;'>\n\nThe sigmoid function can be written as:\n$ S(x) = \\frac{1}{1 + e^{-x}} $\n\nAnd the derivative of the sigmoid function can be written as: \n$S'(x) = S(x) \\cdot (1 - S(x))$",
"_____no_output_____"
],
[
"### How to get Derivative\n\nA derivative is just a fancy word for the slope or the tangent line to a given point. Take a closer look at the sigmoid function's curve on the graph above. Where x=0, the slope is much greater than the slope where x=4 or x=-4. The amount that the weight(s) are updated is based on the derivative. If the slope is a lower value, the neural network is confident in its prediction, and less movement of the weights is needed. If the slope is of a higher value, then the neural networks predictions are closer to .50, or 50% (The highest slope value possible for the sigmoid function is at x=0 and y=.5. y is the prediction.). This means the neural network is not very confident in its prediction and is in need of a greater update to the weights. \n\nWe can find the derivative of the sigmoid function with the steps below:\n$$ S(x) = \\frac{1}{1 + e^{-x}} $$\n\n$$ S'(x) = \\frac{d}{dx}(1 + e^{-x})^{-1} $$\n\n$$ = -(1 + e^{-x})^{-2} \\cdot \\frac{d}{dx} (1 + e^{-x})$$\n\n$$ = -(1 + e^{-x})^{-2} \\cdot (\\frac{d}{dx} (1) + \\frac{d}{dx}(e^{-x}))$$\n\n$$ = -(1 + e^{-x})^{-2} \\cdot (-e^{-x}) $$\n\n$$ = \\frac{-(-e^{-x})}{(1 + e^{-x})^{2}} $$\n\n$$ = \\frac{e^{-x}}{(1 + e^{-x})^{2}} $$\n\n<center>This is the derivative of the sigmoid function, but we can simplify it further by multiplying the numerator by one:</center>\n$$ = \\frac{1}{(1 + e^{-x})} \\cdot \\frac{e^{-x}}{(1 + e^{-x})} $$\n\n<center>This will pull out another sigmoid function! We can then use a cool trick to continue the simplification: add one and subtract one to $e^{-x}$. Adding one and subtracting one will not change anything because they cancel eachother out. It is a fancy way of adding zero.</center>\n$$ = \\frac{1}{(1 + e^{-x})} \\cdot \\frac{1 + e^{-x} - 1}{(1 + e^{-x})} $$\n\n<center>By adding and subtracting one in the numerator, we can split the fraction up again and pull out another sigmoid function!</center>\n$$ = \\frac{1}{(1 + e^{-x})} \\cdot (\\frac{(1 + e^{-x})}{(1 + e^{-x})} - \\frac{1}{(1 + e^{-x})}) $$\n\n<center>Now we can simplify $\\frac{(1 + e^{-x})}{(1 + e^{-x})}$ to 1 and end up with the sigmoid functions simplified derivative.</center>\n$$ = \\frac{1}{(1 + e^{-x})} \\cdot (1 - \\frac{1}{(1 + e^{-x})}) $$\n\n<center>If we write the sigmoid function as $S(x)$, then the derivative can be written as:</center>\n$$ = S(x) \\cdot (1 - S(x)) $$",
"_____no_output_____"
]
],
[
[
"def feed_forward(self):\n self.hidden = self.sigmoid(np.dot(self.inputs, self.weights))",
"_____no_output_____"
]
],
[
[
"During our neural network's training process, the input data will be fed forward through the network's weights and functions. The result of this feed forward function will be the output of the hidden layer, or the hidden layer's best guess with the weights it is given. Each feature in the input data will have its own weight for its connection to the hidden layer. We will start by taking the sum of every feature multiplied by its corresponding weight. Once we have multiplied the input and weight matrices, we can take the results and feed it through the sigmoid function to get squished down into a probability between (0-1). The forward propagation function can be written like this, where $x_{i}$ and $w_{i}$ are individual features and weights in the matrices: \n\n$$ \\hat y = \\frac{1}{1 + e^{-(∑x_iw_i)}} $$\n\nTo reiterate, the hidden layer will be calculated with the following steps:\n* Multiply each feature column with its weight\n* Sum the products of the features and weights\n* Pass the sum into the sigmoid function to produce the output $\\hat y$\n<img src='https://github.com/a-i-dan/a-i-dan.github.io/blob/master/images/NN_Revised/Untitled%20presentation-6.png?raw=true' style='display:block; margin:auto; width:500px;'>\n\nThe above image shows the process of multiplying each feature and its corresponding weight, then taking the sum of the products. Each row in the training data will be computed this way. The resulting 4x1 matrix will be fed into the sigmoid activation function, as shown below:\n\n<img src='https://github.com/a-i-dan/a-i-dan.github.io/blob/master/images/NN_Revised/Untitled%20presentation-4.png?raw=true' style='display:block; margin:auto; width:500px;'>\n\nThe above process will result in the hidden layer's prediction. Each row in the $\\sum xw$ matrix will be entered into the sigmoid function. The colors represent the individual processes for each row in the $\\sum xw$ matrix. <b>Note:</b> this calculation only represents <b>one training iteration</b>, so the resulting $\\hat y$ matrix will not be very accurate. By computing the hidden layer this way, then using backpropagation for many iterations, the result will be much more accurate.",
"_____no_output_____"
]
],
[
[
"def backpropagation(self):\n self.error = self.outputs - self.hidden\n delta = self.error * self.sigmoid(self.hidden, deriv=True)\n self.weights += np.dot(self.inputs.T, delta)",
"_____no_output_____"
]
],
[
[
"This is the coolest part of the whole neural net: backpropagation. Backpropagation will go back through the layer(s) of the neural network, determine which weights contributed to the output and the error, then change the weights based on the gradient of the hidden layers output. This will be explained further, but for now, the whole process can be written like this, where $y$ is the correct output and $\\hat y$ is the hidden layers prediction.:\n\n$$ w_{i} = w_i + X^{T}\\cdot (y - \\hat y) \\cdot [\\frac{1}{1 + e^{-(∑x_iw_i)}} \\cdot (1 - \\frac{1}{1 + e^{-(∑x_iw_i)}})]$$\n\nTo calculate the error of the hidden layer's predictions, we will simply take the difference between the correct output matrix, $y$, and the hidden layer's matrix, $\\hat y$. This process will be shown below.\n\n<img src='https://github.com/a-i-dan/a-i-dan.github.io/blob/master/images/NN_Revised/Untitled%20presentation-5.png?raw=true' style='display:block; margin:auto; width:400px;'>",
"_____no_output_____"
],
[
"We can now multiply the error and the derivative of the hidden layer's prediction. We know that the derivative of the sigmoid function is $S(x)\\cdot (1 - S(x))$. Therefore, the derivative for each of the hidden layer's predictions would be $[\\hat y \\cdot (1 - \\hat y)]$. For example, the first row in the hidden layer's prediction matrix holds a value of $0.62$. We can substitute $\\hat y$ with $0.62$ and the result will be the derivative of the prediction. $0.62 \\cdot (1 - 0.62) = 0.2356$. Repeating this process for every row in the $\\hat y$ matrix will give you a 4x1 matrix of derivatives which you will then multiply with the error matrix.\n<img src='https://github.com/a-i-dan/a-i-dan.github.io/blob/master/images/NN_Revised/Untitled%20presentation-7.png?raw=true' style='display:block; margin:auto; width:400px;'>\n\nMultiplying the error and the derivative is used to find the change that is needed. When the sigmoid function outputs a value with a higher confidence (either close to 0 or close to 1), the derivative will be smaller, therefore the change needed will be smaller. If the sigmoid function outputs a value closer to .50, then the derivative is a larger value, which means there needs to be a larger change in order for the nerual net to become more confident.\n<img src='https://github.com/a-i-dan/a-i-dan.github.io/blob/master/images/NN_Revised/Untitled%20presentation-8.png?raw=true' style='display:block; margin:auto; width:400px;'>\n\nThis step will result with the update that will be added to the weights. We can get this update by multiplying our \"error weighted derivative\" from the above step and the inputs. If the feature in the input is a 0, then the update to the weight will be 0 and if the feature in the input is 1, the update will be added in. This will result in a (3x1) matrix that matches the shape of our weights matrix.\n<img src='https://github.com/a-i-dan/a-i-dan.github.io/blob/master/images/NN_Revised/Untitled%20presentation-9.png?raw=true' style='display:block; margin:auto; width:400px;'>\n\nOnce we have the update matrix, we can add it to our weights matrix to officially change the weights to become stronger. Even after one training iteration there is some noticeable progress! If you look at the updated weights matrix, you may notice that the first weight in the matrix has a higher value. Remember that our neural network must learn that the first feature in the inputs determines the output. We can see that our neural network is already assigning a higher value to the weight connected to the first feature in each input example!\n<img src='https://github.com/a-i-dan/a-i-dan.github.io/blob/master/images/NN_Revised/Untitled%20presentation-10.png?raw=true' style='display:block; margin:auto; width:400px;'>",
"_____no_output_____"
]
],
[
[
"def train(self, epochs=25000):\n for epoch in range(epochs):\n self.feed_forward()\n self.backpropagation()\n\n self.error_history.append(np.average(np.abs(self.error)))\n self.epoch_list.append(epoch)",
"_____no_output_____"
]
],
[
[
"The time has come to train the neural network. During the training process, the neural net will \"learn\" which features in the input data correlate with its output, and it will learn to make accurate predictions. To train our neural network, we will create the train function with the number of epochs, or iterations, to 25,000. This means the neural network will repeat the weight-updating process 25,000 times. Within the train function, we will call our `feed_forward()` function, then the `backpropagation()` function. For each iteration we will also keep track of the error produced after the `feed_forward()` function has completed. We will keep track of this by appending the error and epoch to the lists that were initialized earlier. I am sure there is an easier way to do this, but for quick prototyping, this way works just fine for now.\n\nThe training process follows the equation below for every weight in our neural net:\n* $x_i$ - Feature in Input Data\n* $w_i$ - The Weight that is Being Updated\n* $X^T$ - Transposed Input Data\n* $y$ - Correct Output\n* $\\hat y$ - Predicted Output\n* $(y - \\hat y)$ - Error\n* $∑x_iw_i$ - Sum of the Products of Input Features and Weights\n* $\\frac{1}{1 + e^{(∑x_iw_i)}}$ or $S(∑x_iw_i)$ - Sigmoid Function\n\n$$ w_i = w_i + X^{T}\\cdot(y - \\hat y) \\cdot [S(∑x_iw_i) \\cdot (1 - S(∑x_iw_i))] $$",
"_____no_output_____"
]
],
[
[
"def predict(self, new_input):\n prediction = self.sigmoid(np.dot(new_input, self.weights))\n return prediction",
"_____no_output_____"
]
],
[
[
"Now that the neural network has been trained and has learned the important features in the input data, we can begin to make predictions. The prediction function will look similar to the hidden layer, or the `feedforward()` function. The forward propagation function essentially makes a prediction as well, then backpropagation checks for the error and updates the weights. Our predict function will use the same method as the feedforward function: multiply the input matrix and the weights matrix, then feed the results through the sigmoid function to return a value between 0-1. Hopefully, our neural network will make a prediction as close as possible to the actual output.",
"_____no_output_____"
]
],
[
[
"NN = NeuralNetwork(inputs, outputs)",
"_____no_output_____"
]
],
[
[
"We will create our NN object from the NeuralNetwork class and pass in the input matrix and the output matrix.",
"_____no_output_____"
]
],
[
[
"NN.train()",
"_____no_output_____"
]
],
[
[
"We can then call the `.train()` function on our neural network object. ",
"_____no_output_____"
]
],
[
[
"example = np.array([[1, 1, 0]])\nexample_2 = np.array([[0, 1, 1]])\n\nprint(NN.predict(example), ' - Correct: ', example[0][0])\nprint(NN.predict(example_2), ' - Correct: ', example_2[0][0])",
"_____no_output_____"
]
],
[
[
"```\n[[0.99089925]] - Correct: 1\n[[0.006409]] - Correct: 0\n```\n\nNow we can create the two new examples that we want our neural network to make predictions for. We will call these \"example\" and \"example_2\". We can then call the `.predict()` function and pass through the arrays. We know that the first number, or feature, in the input determines the output. The first example, \"Example\", has a 1 in the first column, and therefore the output should be a one. The second example has a 0 in the first column, and so the output should be a 0.",
"_____no_output_____"
]
],
[
[
"plt.figure(figsize=(15,5))\nplt.plot(NN.epoch_list, NN.error_history)\nplt.xlabel('Epoch')\nplt.ylabel('Loss')",
"_____no_output_____"
]
],
[
[
"With the training complete, we can plot the error over each training iteration. The plot shows that there is a hude decrease in error during the earlier epochs, but that the error slightly plateaus after approximately 5000 iterations.\n\n<img src='https://github.com/a-i-dan/a-i-dan.github.io/blob/master/images/NN_Revised/Unknown-14?raw=true' style='display: block; margin: auto; width: 1000px;'>",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
cb8a6bc534f29683bedd887c83d83bc135de9045
| 2,653 |
ipynb
|
Jupyter Notebook
|
demo/NI_cDaq/log_server.ipynb
|
wi11dey/pylabnet
|
a6e3362f727c45aaa60e61496e858ae92e85574d
|
[
"MIT"
] | 10 |
2020-01-07T23:28:49.000Z
|
2022-02-02T19:09:17.000Z
|
demo/NI_cDaq/log_server.ipynb
|
wi11dey/pylabnet
|
a6e3362f727c45aaa60e61496e858ae92e85574d
|
[
"MIT"
] | 249 |
2019-12-28T19:38:49.000Z
|
2022-03-28T16:45:32.000Z
|
demo/NI_cDaq/log_server.ipynb
|
wi11dey/pylabnet
|
a6e3362f727c45aaa60e61496e858ae92e85574d
|
[
"MIT"
] | 5 |
2020-11-17T19:45:10.000Z
|
2022-01-04T18:07:04.000Z
| 17.925676 | 85 | 0.500188 |
[
[
[
"from pylabnet.utils.logging.logger import LogService\nfrom pylabnet.network.core.generic_server import GenericServer",
"_____no_output_____"
]
],
[
[
"# Start Logging Server",
"_____no_output_____"
]
],
[
[
"log_service = LogService()",
"_____no_output_____"
],
[
"log_server = GenericServer(\n service=log_service, \n host='localhost', \n port=14785\n)",
"_____no_output_____"
],
[
"log_server.start()",
"2020-09-03 12:59:31,027 - INFO - server started on [127.0.0.1]:14785\n"
],
[
"log_server._server.active",
"_____no_output_____"
]
],
[
[
"# Monitor Clients and Logging Output",
"_____no_output_____"
]
],
[
[
"log_server._server.clients",
"_____no_output_____"
],
[
"print (\"Log messages will be displayed below\")",
"Log messages will be displayed below\n"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
cb8a727003684a5a3563ed64e55b403c1d1bd58c
| 64,753 |
ipynb
|
Jupyter Notebook
|
Optmize_absorbates/KR opt.ipynb
|
Gregory-Cooper/TL_MOF
|
6f7f6d804962b6774ca8227a573bfc42113cdc22
|
[
"MIT"
] | null | null | null |
Optmize_absorbates/KR opt.ipynb
|
Gregory-Cooper/TL_MOF
|
6f7f6d804962b6774ca8227a573bfc42113cdc22
|
[
"MIT"
] | null | null | null |
Optmize_absorbates/KR opt.ipynb
|
Gregory-Cooper/TL_MOF
|
6f7f6d804962b6774ca8227a573bfc42113cdc22
|
[
"MIT"
] | null | null | null | 107.384743 | 24,942 | 0.854277 |
[
[
[
"# help function\nfrom transfer_learning import NeuralNet_sherpa_optimize\nfrom dataset_loader import data_loader, get_descriptors, one_filter, data_scaler\n\n# modules\nimport torch\nimport torch.nn as nn\nimport torch.optim as optim\n\nimport os\nimport numpy as np\nimport pandas as pd\n\nfrom sklearn.model_selection import train_test_split\n\n# New\nfrom transfer_learning import MyDataset\nfrom Statistics_helper import stratified_cluster_sample\nfrom ignite.engine import Engine, Events, create_supervised_evaluator\nfrom ignite.metrics import Loss\nfrom ignite.contrib.metrics.regression import R2Score\nimport time\nfrom ignite.engine import Events, create_supervised_evaluator\nimport sherpa\nfrom sklearn.metrics import r2_score\n\n\n# file name and data path\ndevice = torch.device(\"cuda\" if torch.cuda.is_available() else \"cpu\")\nbase_path = os.getcwd()\nfile_name = \"data/CrystGrowthDesign_SI.csv\"\n\n\"\"\"\nData description.\n\n Descriptors:\n 'void fraction', 'Vol. S.A.', 'Grav. S.A.', 'Pore diameter Limiting', 'Pore diameter Largest'\n Source task:\n 'H2@100 bar/243K (wt%)'\n Target tasks:\n 'H2@100 bar/130K (wt%)' 'CH4@100 bar/298 K (mg/g)' '5 bar Xe mol/kg' '5 bar Kr mol/kg'\n\"\"\"\n\ndescriptor_columns = [\n \"void fraction\",\n \"Vol. S.A.\",\n \"Grav. S.A.\",\n \"Pore diameter Limiting\",\n \"Pore diameter Largest\",\n]\none_filter_columns = [\"H2@100 bar/243K (wt%)\"]\nanother_filter_columns = ['5 bar Kr mol/kg']\n\n# load data\ndata = data_loader(base_path, file_name)\ndata = data.reset_index(drop=True)\nepochs = 10000\nbatch_size = 128\n# parameters\ninput_size = 5\noutput_size = 1\n\n# file specifics \n#filename = f\"data_epochs-{epochs}_bs-{batch_size}\"\ntrial_parameters={\n \"lr\" : 0.003014,\n \"H_l1\" : 257,\n \"activate\" : \"nn.PReLU\"\n}\n#format data\nlearning_rate = trial_parameters[\"lr\"]\ndf, t_1, t_2, y_1, y_2 = stratified_cluster_sample(\n 1, data, descriptor_columns, one_filter_columns[0], 5, net_out=True\n)\ndf = df[0]\ndf=df.drop(\"Cluster\",axis=1)\ninterest = one_filter_columns[0]\n#descriptor_columns.append(\"Cluster\")\nfeatures = descriptor_columns\n\ndf_train, df_val, y_df_train, y_df_val = train_test_split(\n df[features], df[interest], test_size=0.1\n)\ndf_train[interest] = np.array(y_df_train)\ndf_val[interest] = np.array(y_df_val)\nfirst = MyDataset(df_train, interest, features)\ntrain_loader = torch.utils.data.DataLoader(first, batch_size=batch_size)\nsecond = MyDataset(df_val, interest, features)\nval_loader = torch.utils.data.DataLoader(second, batch_size=len(df_val))\n\ntrain_loss = []\ntrain_r_2 = []\nval_loss = []\nval_r_2 = []\ntest_loss = []\ntest_r_2 = []\nnet_time = []\n#create model\nmodel = NeuralNet_sherpa_optimize(5, 1, trial_parameters).to(device)\ncriterion = nn.MSELoss()\noptimizer = optim.Adam(model.parameters(), lr=learning_rate)\n\n\ndef train_step(engine, batch):\n x, y = batch\n model.train()\n optimizer.zero_grad()\n y_pred = model(x)\n loss = criterion(y_pred, y)\n loss.backward()\n optimizer.step()\n\n return loss.item()\n\ntrainer = Engine(train_step)\n\n\nmetrics = {\"loss\": Loss(criterion), \"r_2\": R2Score()}\n\n#train_evaluator = create_supervised_evaluator(model, metrics=metrics, device=device)\n# train_evaluator.logger = setup_logger(\"Train Evaluator\")\n#validation_evaluator = create_supervised_evaluator(\n# model, metrics=metrics, device=device\n#)\n# validation_evaluator.logger = setup_logger(\"Val Evaluator\")\n\n\n\n\ntrain_loader = torch.utils.data.DataLoader(first, batch_size=batch_size,shuffle=True)\nstart = time.time()\ntrainer.logger.disabled=True\ntrainer.run(train_loader, max_epochs=epochs)\n\n\ndescriptor_columns = [\n \"void fraction\",\n \"Vol. S.A.\",\n \"Grav. S.A.\",\n \"Pore diameter Limiting\",\n \"Pore diameter Largest\",\n]\nmodel.fc1.weight.requires_grad = False\nmodel.fc1.bias.requires_grad = False\nmodel.fc2.weight.requires_grad = False\nmodel.fc2.bias.requires_grad = False\noptimizer = optim.Adam(\n filter(lambda p: p.requires_grad, model.parameters()), lr=learning_rate\n)\ndf, t_1, t_2, y_1, y_2 = stratified_cluster_sample(\n 1, data, descriptor_columns, another_filter_columns[0], 5, net_out=True\n)\ndf = df[0]\ndf=df.drop(\"Cluster\",axis=1)\ninterest = another_filter_columns[0]\n#descriptor_columns.append(\"Cluster\")\nfeatures = descriptor_columns\n\ndf_train, df_test, y_df_train, y_df_test = train_test_split(\n df[features], df[interest], test_size=0.2\n)\ny_df_train=y_df_train.reset_index(drop=False)\ndf_train, df_val, y_df_train, y_df_val = train_test_split(\n df_train[features], y_df_train[interest], test_size=0.2\n)\ndf_train[interest] = np.array(y_df_train)\ndf_val[interest] = np.array(y_df_val)\ndf_test[interest]=np.array(y_df_test)\ninterest=another_filter_columns[0]\nfirst = MyDataset(df_train, interest, features)\ntrain_loader = torch.utils.data.DataLoader(first, batch_size=batch_size)\n\nsecond = MyDataset(df_val, interest, features)\nval_loader = torch.utils.data.DataLoader(second, batch_size=len(df_val))\n\nthird = MyDataset(df_test, interest, features)\ntest_loader=torch.utils.data.DataLoader(third, batch_size=len(df_test))\n\ndef train_step_1(engine, batch):\n x, y = batch\n model.train()\n optimizer.zero_grad()\n y_pred = model(x)\n loss = criterion(y_pred, y)\n loss.backward()\n optimizer.step()\n\n return loss.item()\ntransfer_trainer = Engine(train_step_1)\nmetrics = {\"loss\": Loss(criterion), \"r_2\": R2Score()}\n@transfer_trainer.on(Events.EPOCH_COMPLETED(every=50))\ndef store_metrics(engine):\n end = time.time()\n e = engine.state.epoch\n out=float(criterion(model(train_loader.dataset.x_train),train_loader.dataset.y_train))\n out1=float(r2_score(model(train_loader.dataset.x_train).detach().numpy(),train_loader.dataset.y_train.detach().numpy()))\n out2=float(criterion(model(val_loader.dataset.x_train),val_loader.dataset.y_train))\n out3=float(r2_score(model(val_loader.dataset.x_train).detach().numpy(),val_loader.dataset.y_train.detach().numpy()))\n out4=float(criterion(model(test_loader.dataset.x_train),test_loader.dataset.y_train))\n out5=float(r2_score(model(test_loader.dataset.x_train).detach().numpy(),test_loader.dataset.y_train.detach().numpy()))\n train_loss.append(out)\n train_r_2.append(out1)\n val_loss.append(out2)\n val_r_2.append(out3)\n test_loss.append(out4)\n test_r_2.append(out5)\n net_time.append(end-start)\n print(e)\ntransfer_trainer.logger.disabled=True\ntransfer_trainer.run(train_loader, max_epochs=epochs)",
"50\n100\n150\n200\n250\n300\n350\n400\n450\n500\n550\n600\n650\n700\n750\n800\n850\n900\n950\n1000\n1050\n1100\n1150\n1200\n1250\n1300\n1350\n1400\n1450\n1500\n1550\n1600\n1650\n1700\n1750\n1800\n1850\n1900\n1950\n2000\n2050\n2100\n2150\n2200\n2250\n2300\n2350\n2400\n2450\n2500\n2550\n2600\n2650\n2700\n2750\n2800\n2850\n2900\n2950\n3000\n3050\n3100\n3150\n3200\n3250\n3300\n3350\n3400\n3450\n3500\n3550\n3600\n3650\n3700\n3750\n3800\n3850\n3900\n3950\n4000\n4050\n4100\n4150\n4200\n4250\n4300\n4350\n4400\n4450\n4500\n4550\n4600\n4650\n4700\n4750\n4800\n4850\n4900\n4950\n5000\n5050\n5100\n5150\n5200\n5250\n5300\n5350\n5400\n5450\n5500\n5550\n5600\n5650\n5700\n5750\n5800\n5850\n5900\n5950\n6000\n6050\n6100\n6150\n6200\n6250\n6300\n6350\n6400\n6450\n6500\n6550\n6600\n6650\n6700\n6750\n6800\n6850\n6900\n6950\n7000\n7050\n7100\n7150\n7200\n7250\n7300\n7350\n7400\n7450\n7500\n7550\n7600\n7650\n7700\n7750\n7800\n7850\n7900\n7950\n8000\n8050\n8100\n8150\n8200\n8250\n8300\n8350\n8400\n8450\n8500\n8550\n8600\n8650\n8700\n8750\n8800\n8850\n8900\n8950\n9000\n9050\n9100\n9150\n9200\n9250\n9300\n9350\n9400\n9450\n9500\n9550\n9600\n9650\n9700\n9750\n9800\n9850\n9900\n9950\n10000\n"
],
[
"import matplotlib.pyplot as plt\nplt.plot(val_r_2)\nplt.plot(train_r_2,label=\"t\")\nplt.plot(test_r_2,label=\"real\")\nplt.legend()",
"_____no_output_____"
],
[
"plt.plot(val_loss)\nplt.plot(train_loss,label=\"t\")\nplt.plot(test_loss,label=\"real\")\nplt.legend()",
"_____no_output_____"
],
[
"torch.save(model, \"KR.ckpt\")\npd.DataFrame()",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code"
]
] |
cb8a8277528f362e3d0447ee90eba4f6a888a359
| 124,828 |
ipynb
|
Jupyter Notebook
|
dog-breed-identification/3. Image-Similar-FCNN-Binary.ipynb
|
StudyExchange/Kaggle
|
e8306cc34da6b0ec91955d8f62124459981e605e
|
[
"MIT"
] | 2 |
2019-04-19T12:51:31.000Z
|
2019-04-19T12:51:34.000Z
|
dog-breed-identification/3. Image-Similar-FCNN-Binary.ipynb
|
StudyExchange/Kaggle
|
e8306cc34da6b0ec91955d8f62124459981e605e
|
[
"MIT"
] | null | null | null |
dog-breed-identification/3. Image-Similar-FCNN-Binary.ipynb
|
StudyExchange/Kaggle
|
e8306cc34da6b0ec91955d8f62124459981e605e
|
[
"MIT"
] | 1 |
2018-10-05T00:35:18.000Z
|
2018-10-05T00:35:18.000Z
| 156.622334 | 4,968 | 0.556534 |
[
[
[
"# 3. Image-Similar-FCNN-Binary\nFor landmark-recognition-2019 algorithm validation",
"_____no_output_____"
],
[
"## Run name",
"_____no_output_____"
]
],
[
[
"import time\n\nproject_name = 'Dog-Breed'\nstep_name = '3-Image-Similar-FCNN-Binary'\ntime_str = time.strftime(\"%Y%m%d-%H%M%S\", time.localtime())\nrun_name = project_name + '_' + step_name + '_' + time_str\nprint('run_name: ' + run_name)\nt0 = time.time()",
"run_name: Dog-Breed_3-Image-Similar-FCNN-Binary_20190507-211416\n"
]
],
[
[
"## Important params",
"_____no_output_____"
]
],
[
[
"import multiprocessing\n\ncpu_amount = multiprocessing.cpu_count()\nprint('cpu_amount: ', cpu_amount)",
"cpu_amount: 4\n"
]
],
[
[
"## Import PKGs",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport matplotlib.image as mpimg\n%matplotlib inline\nfrom IPython.display import display\n\nimport os\nimport sys\nimport gc\nimport math\nimport shutil\nimport zipfile\nimport pickle\nimport h5py\n\nfrom tqdm import tqdm\n\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.metrics import confusion_matrix, accuracy_score",
"_____no_output_____"
],
[
"import keras\nfrom keras.utils import Sequence\nfrom keras.layers import *\nfrom keras.models import *\nfrom keras.applications import *\nfrom keras.optimizers import *\nfrom keras.regularizers import *\nfrom keras.preprocessing.image import *\nfrom keras.applications.inception_v3 import preprocess_input",
"Using TensorFlow backend.\n"
]
],
[
[
"## Project folders",
"_____no_output_____"
]
],
[
[
"cwd = os.getcwd()\nfeature_folder = os.path.join(cwd, 'feature')\ninput_folder = os.path.join(cwd, 'input')\noutput_folder = os.path.join(cwd, 'output')\nmodel_folder = os.path.join(cwd, 'model')\n\norg_train_folder = os.path.join(input_folder, 'org_train')\norg_test_folder = os.path.join(input_folder, 'org_test')\ntrain_folder = os.path.join(input_folder, 'data_train')\nval_folder = os.path.join(input_folder, 'data_val')\ntest_folder = os.path.join(input_folder, 'data_test')\ntest_sub_folder = os.path.join(test_folder, 'test')\n\nvgg16_feature_file = os.path.join(feature_folder, 'feature_wrapper_171023.h5')\ntrain_csv_file = os.path.join(input_folder, 'train.csv')\ntest_csv_file = os.path.join(input_folder, 'test.csv')\nsample_submission_folder = os.path.join(input_folder, 'sample_submission.csv')\nprint(vgg16_feature_file)\nprint(train_csv_file)\nprint(test_csv_file)\nprint(sample_submission_folder)",
"D:\\Kaggle\\dog-breed-identification\\feature\\feature_wrapper_171023.h5\nD:\\Kaggle\\dog-breed-identification\\input\\train.csv\nD:\\Kaggle\\dog-breed-identification\\input\\test.csv\nD:\\Kaggle\\dog-breed-identification\\input\\sample_submission.csv\n"
]
],
[
[
"## Load feature",
"_____no_output_____"
]
],
[
[
"with h5py.File(vgg16_feature_file, 'r') as h:\n x_train = np.array(h['train'])\n y_train = np.array(h['train_label'])\n x_val = np.array(h['val'])\n y_val = np.array(h['val_label'])\n",
"_____no_output_____"
],
[
"print(x_train.shape)\nprint(y_train.shape)\nprint(x_val.shape)\nprint(y_val.shape)",
"(9199, 512)\n(9199,)\n(1023, 512)\n(1023,)\n"
],
[
"import random\nrandom.choice(list(range(10)))\nimport copy\na = list(range(10, 20))\nprint(a)\na.remove(13)\nprint(a)",
"[10, 11, 12, 13, 14, 15, 16, 17, 18, 19]\n[10, 11, 12, 14, 15, 16, 17, 18, 19]\n"
]
],
[
[
"## ImageSequence",
"_____no_output_____"
]
],
[
[
"# class ImageSequence(Sequence):\n# def __init__(self, x, y, batch_size, times_for_1_image, positive_rate):\n# self.x = x\n# self.y = y\n# self.batch_size = batch_size\n# self.times_for_1_image = times_for_1_image\n# self.positive_rate = positive_rate\n \n# self.len_x = self.x.shape[0]\n# self.index = list(range(self.len_x))\n# self.group = {}\n# self.classes = list(set(self.y))\n# self.classes.sort()\n# for c in self.classes:\n# self.group[c] = []\n# for i, y_i in enumerate(self.y):\n# temp_arr = self.group[y_i]\n# temp_arr.append(i)\n# self.group[y_i] = temp_arr\n \n# def __len__(self):\n# # times_for_1_image: the times to train one image\n# # 2: positive example and negative example\n# return self.times_for_1_image * 2 * (math.ceil(self.len_x/self.batch_size))\n# def __getitem__(self, idx):\n# batch_main_x = []\n# batch_libary_x = []\n# batch_x = {}\n# batch_y = [] # 0 or 1\n# for i in range(self.batch_size):\n# # prepare main image\n# item_main_image_idx = random.choice(self.index) # random choice one image from all train images\n# item_main_image_y = self.y[item_main_image_idx]\n \n# # prepare libary image\n# is_positive = random.random() < self.positive_rate\n# if is_positive: # chioce a positive image as libary_x\n# # choice one image from itself group\n# item_libary_image_idx = random.choice(self.group[item_main_image_y]) # don't exclude item_main_image_idx, so it could choice a idx same to item_main_image_idx.\n# else: # chioce a negative image as libary_x\n# # choice group\n# new_class = copy.deepcopy(self.classes)\n# new_class.remove(item_main_image_y)\n# item_libary_image_group_num = random.choice(new_class)\n# # choice one image from group\n# item_libary_image_idx = random.choice(self.group[item_libary_image_group_num])\n# # add item data to batch\n# batch_main_x.append(self.x[item_main_image_idx])\n# batch_libary_x.append(self.x[item_libary_image_idx])\n# batch_y.append(int(is_positive))\n# # concatenate array to np.array\n# batch_x = {\n# 'main_input': np.array(batch_main_x),\n# 'library_input': np.array(batch_libary_x)\n# }\n# batch_y = np.array(batch_y)\n# return batch_x, batch_y\n\n# demo_sequence = ImageSequence(x_train[:200], y_train[:200], 128, 3, 0.1)\n# print(len(demo_sequence))\n# print(type(demo_sequence))\n\n# batch_index = 0\n# demo_batch = demo_sequence[batch_index]\n# demo_batch_x = demo_batch[0]\n# demo_batch_y = demo_batch[1]\n# print(type(demo_batch_x))\n# print(type(demo_batch_y))\n\n# demo_main_input = demo_batch_x['main_input']\n# demo_library_input = demo_batch_x['library_input']\n# print(demo_main_input.shape)\n# print(demo_library_input.shape)\n# print(demo_batch_y.shape)\n\n# # print(demo_main_input[0])\n# print(demo_batch_y)",
"_____no_output_____"
],
[
"class ImageSequence(Sequence):\n def __init__(self, x, y, batch_size, times_for_1_image, positive_rate):\n self.x = x\n self.y = y\n self.batch_size = batch_size\n self.times_for_1_image = times_for_1_image\n self.positive_rate = positive_rate\n \n self.len_x = self.x.shape[0]\n self.index = list(range(self.len_x))\n self.group = {}\n self.classes = list(set(self.y))\n self.classes.sort()\n for c in self.classes:\n self.group[c] = []\n for i, y_i in enumerate(self.y):\n temp_arr = self.group[y_i]\n temp_arr.append(i)\n self.group[y_i] = temp_arr\n \n def __len__(self):\n # times_for_1_image: the times to train one image\n # 2: positive example and negative example\n return self.times_for_1_image * 2 * (math.ceil(self.len_x/self.batch_size))\n def __getitem__(self, idx):\n batch_main_x = np.zeros((self.batch_size, self.x.shape[1]))\n batch_libary_x = np.zeros((self.batch_size, self.x.shape[1]))\n batch_x = {}\n batch_y = [] # 0 or 1\n for i in range(self.batch_size):\n # prepare main image\n item_main_image_idx = random.choice(self.index) # random choice one image from all train images\n item_main_image_y = self.y[item_main_image_idx]\n \n # prepare libary image\n is_positive = random.random() < self.positive_rate\n if is_positive: # chioce a positive image as libary_x\n # choice one image from itself group\n item_libary_image_idx = random.choice(self.group[item_main_image_y]) # don't exclude item_main_image_idx, so it could choice a idx same to item_main_image_idx.\n else: # chioce a negative image as libary_x\n # choice group\n new_class = copy.deepcopy(self.classes)\n new_class.remove(item_main_image_y)\n item_libary_image_group_num = random.choice(new_class)\n # choice one image from group\n item_libary_image_idx = random.choice(self.group[item_libary_image_group_num])\n # add item data to batch\n batch_main_x[i] = self.x[item_main_image_idx]\n batch_libary_x[i] = self.x[item_libary_image_idx]\n batch_y.append(int(is_positive))\n # concatenate array to np.array\n batch_x = {\n 'main_input': batch_main_x,\n 'library_input': batch_libary_x\n }\n batch_y = np.array(batch_y)\n return batch_x, batch_y\n\ndemo_sequence = ImageSequence(x_train[:200], y_train[:200], 128, 3, 0.1)\nprint(len(demo_sequence))\nprint(type(demo_sequence))\n\nbatch_index = 0\ndemo_batch = demo_sequence[batch_index]\ndemo_batch_x = demo_batch[0]\ndemo_batch_y = demo_batch[1]\nprint(type(demo_batch_x))\nprint(type(demo_batch_y))\n\ndemo_main_input = demo_batch_x['main_input']\ndemo_library_input = demo_batch_x['library_input']\nprint(demo_main_input.shape)\nprint(demo_library_input.shape)\nprint(demo_batch_y.shape)\n\n# print(demo_main_input[0])\nprint(demo_batch_y)",
"12\n<class '__main__.ImageSequence'>\n<class 'dict'>\n<class 'numpy.ndarray'>\n(128, 512)\n(128, 512)\n(128,)\n[0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 1 0 0 0 0 1 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 0 1 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 1 0 0 0 0 1 0 0 0 0 0 0 0 1 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 1 1 0 0 0 0]\n"
],
[
"train_sequence = ImageSequence(x_train, y_train, 32, 3, 0.5)\nval_sequence = ImageSequence(x_val, y_val, 32, 3, 0.5)",
"_____no_output_____"
]
],
[
[
"## Model",
"_____no_output_____"
]
],
[
[
"main_input = Input((x_train.shape[1],), dtype='float32', name='main_input')\nlibrary_input = Input((x_train.shape[1],), dtype='float32', name='library_input')\n\nx = keras.layers.concatenate([main_input, library_input])\nx = Dense(x_train.shape[1]*2, activation='sigmoid')(x)\n\nx = Dense(1024, activation='sigmoid')(x)\nx = Dense(1024, activation='sigmoid')(x)\noutput = Dense(1, activation='sigmoid')(x)\n\nmodel = Model(inputs=[main_input, library_input], outputs=[output])\nmodel.compile(optimizer=Adam(lr=1e-4), loss='binary_crossentropy', metrics=['accuracy'])",
"WARNING:tensorflow:From C:\\Users\\study\\Anaconda3\\lib\\site-packages\\tensorflow\\python\\framework\\op_def_library.py:263: colocate_with (from tensorflow.python.framework.ops) is deprecated and will be removed in a future version.\nInstructions for updating:\nColocations handled automatically by placer.\n"
],
[
"model.summary()",
"__________________________________________________________________________________________________\nLayer (type) Output Shape Param # Connected to \n==================================================================================================\nmain_input (InputLayer) (None, 512) 0 \n__________________________________________________________________________________________________\nlibrary_input (InputLayer) (None, 512) 0 \n__________________________________________________________________________________________________\nconcatenate_1 (Concatenate) (None, 1024) 0 main_input[0][0] \n library_input[0][0] \n__________________________________________________________________________________________________\ndense_1 (Dense) (None, 1024) 1049600 concatenate_1[0][0] \n__________________________________________________________________________________________________\ndense_2 (Dense) (None, 1024) 1049600 dense_1[0][0] \n__________________________________________________________________________________________________\ndense_3 (Dense) (None, 1024) 1049600 dense_2[0][0] \n__________________________________________________________________________________________________\ndense_4 (Dense) (None, 1) 1025 dense_3[0][0] \n==================================================================================================\nTotal params: 3,149,825\nTrainable params: 3,149,825\nNon-trainable params: 0\n__________________________________________________________________________________________________\n"
],
[
"hist = model.fit_generator(\n train_sequence, \n steps_per_epoch=128, \n epochs=300, \n verbose=1, \n callbacks=None, \n validation_data=val_sequence, \n validation_steps=128, \n class_weight=None, \n max_queue_size=10, \n workers=1, \n use_multiprocessing=False, \n shuffle=True, \n initial_epoch=0\n)",
"WARNING:tensorflow:From C:\\Users\\study\\Anaconda3\\lib\\site-packages\\tensorflow\\python\\ops\\math_ops.py:3066: to_int32 (from tensorflow.python.ops.math_ops) is deprecated and will be removed in a future version.\nInstructions for updating:\nUse tf.cast instead.\nEpoch 1/300\n128/128 [==============================] - ETA: 2:43 - loss: 0.7173 - acc: 0.437 - ETA: 1:27 - loss: 0.7095 - acc: 0.484 - ETA: 1:02 - loss: 0.6981 - acc: 0.520 - ETA: 49s - loss: 0.7058 - acc: 0.515 - ETA: 41s - loss: 0.7066 - acc: 0.50 - ETA: 36s - loss: 0.7042 - acc: 0.51 - ETA: 32s - loss: 0.7079 - acc: 0.50 - ETA: 30s - loss: 0.7043 - acc: 0.51 - ETA: 28s - loss: 0.7039 - acc: 0.52 - ETA: 26s - loss: 0.7075 - acc: 0.52 - ETA: 24s - loss: 0.7068 - acc: 0.52 - ETA: 23s - loss: 0.7026 - acc: 0.53 - ETA: 22s - loss: 0.7027 - acc: 0.52 - ETA: 21s - loss: 0.7017 - acc: 0.52 - ETA: 20s - loss: 0.7013 - acc: 0.51 - ETA: 19s - loss: 0.7007 - acc: 0.51 - ETA: 19s - loss: 0.7004 - acc: 0.50 - ETA: 18s - loss: 0.7001 - acc: 0.50 - ETA: 18s - loss: 0.6996 - acc: 0.50 - ETA: 17s - loss: 0.6988 - acc: 0.51 - ETA: 17s - loss: 0.6981 - acc: 0.51 - ETA: 16s - loss: 0.6983 - acc: 0.51 - ETA: 16s - loss: 0.6973 - acc: 0.51 - ETA: 15s - loss: 0.6991 - acc: 0.51 - ETA: 15s - loss: 0.6983 - acc: 0.51 - ETA: 15s - loss: 0.7007 - acc: 0.51 - ETA: 14s - loss: 0.7009 - acc: 0.50 - ETA: 14s - loss: 0.7010 - acc: 0.50 - ETA: 14s - loss: 0.7016 - acc: 0.50 - ETA: 13s - loss: 0.7014 - acc: 0.50 - ETA: 13s - loss: 0.7023 - acc: 0.50 - ETA: 13s - loss: 0.7010 - acc: 0.50 - ETA: 13s - loss: 0.7008 - acc: 0.50 - ETA: 12s - loss: 0.7009 - acc: 0.50 - ETA: 12s - loss: 0.7007 - acc: 0.50 - ETA: 12s - loss: 0.7004 - acc: 0.50 - ETA: 12s - loss: 0.7002 - acc: 0.50 - ETA: 11s - loss: 0.6993 - acc: 0.50 - ETA: 11s - loss: 0.6981 - acc: 0.51 - ETA: 11s - loss: 0.6992 - acc: 0.51 - ETA: 11s - loss: 0.7010 - acc: 0.50 - ETA: 11s - loss: 0.7012 - acc: 0.51 - ETA: 10s - loss: 0.7014 - acc: 0.51 - ETA: 10s - loss: 0.7007 - acc: 0.51 - ETA: 10s - loss: 0.7005 - acc: 0.51 - ETA: 10s - loss: 0.7001 - acc: 0.51 - ETA: 10s - loss: 0.6998 - acc: 0.51 - ETA: 10s - loss: 0.6997 - acc: 0.51 - ETA: 9s - loss: 0.6997 - acc: 0.5159 - ETA: 9s - loss: 0.7003 - acc: 0.511 - ETA: 9s - loss: 0.7002 - acc: 0.510 - ETA: 9s - loss: 0.6999 - acc: 0.512 - ETA: 9s - loss: 0.7005 - acc: 0.510 - ETA: 9s - loss: 0.7003 - acc: 0.511 - ETA: 9s - loss: 0.6999 - acc: 0.512 - ETA: 8s - loss: 0.7002 - acc: 0.512 - ETA: 8s - loss: 0.6998 - acc: 0.513 - ETA: 8s - loss: 0.6995 - acc: 0.514 - ETA: 8s - loss: 0.6996 - acc: 0.514 - ETA: 8s - loss: 0.6993 - acc: 0.515 - ETA: 8s - loss: 0.6988 - acc: 0.517 - ETA: 8s - loss: 0.6991 - acc: 0.516 - ETA: 7s - loss: 0.6989 - acc: 0.516 - ETA: 7s - loss: 0.6987 - acc: 0.516 - ETA: 7s - loss: 0.6985 - acc: 0.517 - ETA: 7s - loss: 0.6988 - acc: 0.516 - ETA: 7s - loss: 0.6988 - acc: 0.516 - ETA: 7s - loss: 0.6987 - acc: 0.517 - ETA: 7s - loss: 0.6986 - acc: 0.517 - ETA: 6s - loss: 0.6977 - acc: 0.520 - ETA: 6s - loss: 0.6980 - acc: 0.520 - ETA: 6s - loss: 0.6984 - acc: 0.519 - ETA: 6s - loss: 0.6984 - acc: 0.519 - ETA: 6s - loss: 0.6984 - acc: 0.518 - ETA: 6s - loss: 0.6986 - acc: 0.516 - ETA: 6s - loss: 0.6987 - acc: 0.515 - ETA: 6s - loss: 0.6994 - acc: 0.512 - ETA: 6s - loss: 0.6997 - acc: 0.510 - ETA: 5s - loss: 0.6995 - acc: 0.510 - ETA: 5s - loss: 0.6995 - acc: 0.508 - ETA: 5s - loss: 0.6993 - acc: 0.509 - ETA: 5s - loss: 0.6989 - acc: 0.510 - ETA: 5s - loss: 0.6992 - acc: 0.509 - ETA: 5s - loss: 0.6996 - acc: 0.509 - ETA: 5s - loss: 0.6996 - acc: 0.509 - ETA: 4s - loss: 0.6994 - acc: 0.510 - ETA: 4s - loss: 0.6993 - acc: 0.510 - ETA: 4s - loss: 0.6996 - acc: 0.509 - ETA: 4s - loss: 0.6995 - acc: 0.509 - ETA: 4s - loss: 0.6995 - acc: 0.508 - ETA: 4s - loss: 0.6995 - acc: 0.507 - ETA: 4s - loss: 0.6997 - acc: 0.506 - ETA: 4s - loss: 0.6997 - acc: 0.506 - ETA: 4s - loss: 0.6996 - acc: 0.505 - ETA: 3s - loss: 0.6996 - acc: 0.506 - ETA: 3s - loss: 0.6995 - acc: 0.505 - ETA: 3s - loss: 0.6994 - acc: 0.505 - ETA: 3s - loss: 0.6994 - acc: 0.505 - ETA: 3s - loss: 0.6997 - acc: 0.503 - ETA: 3s - loss: 0.6996 - acc: 0.503 - ETA: 3s - loss: 0.6994 - acc: 0.505 - ETA: 3s - loss: 0.6994 - acc: 0.504 - ETA: 2s - loss: 0.6996 - acc: 0.504 - ETA: 2s - loss: 0.6994 - acc: 0.505 - ETA: 2s - loss: 0.6992 - acc: 0.505 - ETA: 2s - loss: 0.6995 - acc: 0.504 - ETA: 2s - loss: 0.6996 - acc: 0.503 - ETA: 2s - loss: 0.6996 - acc: 0.504 - ETA: 2s - loss: 0.6996 - acc: 0.503 - ETA: 2s - loss: 0.6994 - acc: 0.504 - ETA: 1s - loss: 0.6992 - acc: 0.505 - ETA: 1s - loss: 0.6994 - acc: 0.505 - ETA: 1s - loss: 0.6996 - acc: 0.504 - ETA: 1s - loss: 0.6994 - acc: 0.505 - ETA: 1s - loss: 0.6990 - acc: 0.506 - ETA: 1s - loss: 0.6992 - acc: 0.505 - ETA: 1s - loss: 0.6989 - acc: 0.506 - ETA: 1s - loss: 0.6988 - acc: 0.507 - ETA: 1s - loss: 0.6989 - acc: 0.505 - ETA: 0s - loss: 0.6989 - acc: 0.505 - ETA: 0s - loss: 0.6990 - acc: 0.505 - ETA: 0s - loss: 0.6990 - acc: 0.505 - ETA: 0s - loss: 0.6991 - acc: 0.504 - ETA: 0s - loss: 0.6991 - acc: 0.503 - ETA: 0s - loss: 0.6991 - acc: 0.504 - ETA: 0s - loss: 0.6991 - acc: 0.503 - ETA: 0s - loss: 0.6991 - acc: 0.502 - 16s 127ms/step - loss: 0.6991 - acc: 0.5029 - val_loss: 0.6899 - val_acc: 0.5381\nEpoch 2/300\n"
],
[
"import matplotlib.pyplot as plt\n%matplotlib inline\n\nplt.plot(hist.history['loss'], color='b')\nplt.plot(hist.history['val_loss'], color='r')\nplt.show()\nplt.plot(hist.history['acc'], color='b')\nplt.plot(hist.history['val_acc'], color='r')\nplt.show()",
"_____no_output_____"
],
[
"def saveModel(model, run_name):\n cwd = os.getcwd()\n modelPath = os.path.join(cwd, 'model')\n if not os.path.isdir(modelPath):\n os.mkdir(modelPath)\n weigths_file = os.path.join(modelPath, run_name + '.h5')\n print(weigths_file)\n model.save(weigths_file)\nsaveModel(model, run_name)",
"_____no_output_____"
],
[
"print('Time elapsed: %.1fs' % (time.time() - t0))\nprint(run_name)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb8aab194d042d350b1d77337aa59b4cfb9870f7
| 925,069 |
ipynb
|
Jupyter Notebook
|
Basic_ML/A3-SVM and kernel.ipynb
|
PinmanHuang/CrashCourseML
|
b59ebf138d42fc9a1669735c6363d50938200e69
|
[
"MIT"
] | 3 |
2019-02-16T05:57:09.000Z
|
2019-09-16T07:07:18.000Z
|
Basic_ML/A3-SVM and kernel.ipynb
|
PinmanHuang/CrashCourseML
|
b59ebf138d42fc9a1669735c6363d50938200e69
|
[
"MIT"
] | null | null | null |
Basic_ML/A3-SVM and kernel.ipynb
|
PinmanHuang/CrashCourseML
|
b59ebf138d42fc9a1669735c6363d50938200e69
|
[
"MIT"
] | 8 |
2019-02-14T02:51:26.000Z
|
2019-10-07T07:44:24.000Z
| 2,301.166667 | 275,018 | 0.949927 |
[
[
[
"# Support Vector Machine",
"_____no_output_____"
]
],
[
[
"from PIL import Image\nimport numpy as np\n%matplotlib inline\nimport matplotlib\nimport matplotlib.pyplot as plt\nfrom sklearn import datasets, svm, linear_model\nmatplotlib.style.use('bmh')\nmatplotlib.rcParams['figure.figsize']=(10,10)",
"_____no_output_____"
]
],
[
[
"### 2D Linear",
"_____no_output_____"
]
],
[
[
"# Random 2d X\nX0 = np.random.normal(-2, size=(30,2))\nX1 = np.random.normal(2, size=(30,2))\nX = np.concatenate([X0,X1], axis=0)\n\ny = X @ [1,1] > 0\n\nclf=svm.SVC(kernel='linear', C=1000)\nclf.fit(X, y)\n\n# 邊界\nx_min, y_min = X.min(axis=0)-1\nx_max, y_max = X.max(axis=0)+1\n\n# 座標點\ngrid = np.mgrid[x_min:x_max:200j, y_min:y_max:200j]\n# grid.shape = (2, 200, 200)\n\n# 在座標點 算出 svm 的判斷函數\nZ = clf.decision_function(grid.reshape(2, -1).T)\nZ = Z.reshape(grid.shape[1:])\n\n# 畫出顏色和邊界\nplt.pcolormesh(grid[0], grid[1], Z > 0, cmap=plt.cm.rainbow, alpha=0.1)\nplt.contour(grid[0], grid[1], Z, colors=['k', 'k', 'k'], linestyles=['--', '-', '--'],\n levels=[-1, 0, 1])\n# 標出 sample 點\nplt.scatter(X[:,0], X[:, 1], c=y, cmap=plt.cm.rainbow, zorder=10, s=50);\n",
"_____no_output_____"
]
],
[
[
"3D view",
"_____no_output_____"
]
],
[
[
"from mpl_toolkits.mplot3d import Axes3D\nax = plt.gca(projection='3d')\nax.plot_surface(grid[0], grid[1], Z, cmap=plt.cm.rainbow, alpha=0.2)\nax.plot_wireframe(grid[0], grid[1], Z, alpha=0.2, rstride=20, cstride=20)\nax.scatter(X[:, 0], X[:, 1], y, c=y, cmap=plt.cm.rainbow, s=30);\nax.set_zlim3d(-2,2)\nax.set_xlim3d(-3,3)\nax.set_ylim3d(-3,3)\nax.view_init(15, -75)",
"_____no_output_____"
]
],
[
[
"Linear Nonseparable",
"_____no_output_____"
]
],
[
[
"# Random 2d X\nX = np.random.uniform(-1.5, 1.5, size=(100,2))\n\ny = (X**2).sum(axis=1) > 1\n\nclf=svm.SVC(kernel='linear', C=1000)\nclf.fit(X, y)\n\n# 邊界\nx_min, y_min = X.min(axis=0)-1\nx_max, y_max = X.max(axis=0)+1\n\n# 座標點\ngrid = np.mgrid[x_min:x_max:200j, y_min:y_max:200j]\n# grid.shape = (2, 200, 200)\n\n# 在座標點 算出 svm 的判斷函數\nZ = clf.decision_function(grid.reshape(2, -1).T)\nZ = Z.reshape(grid.shape[1:])\n\n# 畫出顏色和邊界\nplt.pcolormesh(grid[0], grid[1], Z > 0, cmap=plt.cm.rainbow, alpha=0.1)\nplt.contour(grid[0], grid[1], Z, colors=['k', 'k', 'k'], linestyles=['--', '-', '--'],\n levels=[-1, 0, 1])\n# 標出 sample 點\nplt.scatter(X[:,0], X[:, 1], c=y, cmap=plt.cm.rainbow, zorder=10, s=20);\n",
"_____no_output_____"
],
[
"(np.linspace(-1.5,1.5, 10)[:, None] @ np.linspace(-1.5,1.5, 10)[None, :]).shape\n",
"_____no_output_____"
],
[
"# Random 2d X\nX = np.random.uniform(-1.5, 1.5, size=(100,2))\n# more feature (x**2, y**2, x*y)\nX2 = np.concatenate([X, X**2, (X[:, 0]*X[:, 1])[:, None]], axis=1)\ny = (X**2).sum(axis=1) > 1\n\nclf=svm.SVC(kernel='linear', C=1000)\nclf.fit(X2, y)\n\n# 邊界\nx_min, y_min = X.min(axis=0)-1\nx_max, y_max = X.max(axis=0)+1\n\n# 座標點\ngrid = np.mgrid[x_min:x_max:200j, y_min:y_max:200j]\n# grid.shape = (2, 200, 200)\nG = grid.reshape(2, -1).T\nG = np.concatenate([G, G**2, (G[:, 0]*G[:, 1])[:, None]], axis=1)\n\n# 在座標點 算出 svm 的判斷函數\nZ = clf.decision_function(G)\nZ = Z.reshape(grid.shape[1:])\n\n# 畫出顏色和邊界\nplt.pcolormesh(grid[0], grid[1], Z > 0, cmap=plt.cm.rainbow, alpha=0.1)\nplt.contour(grid[0], grid[1], Z, colors=['k', 'k', 'k'], linestyles=['--', '-', '--'],\n levels=[-1, 0, 1])\n# 標出 sample 點\nplt.scatter(X[:,0], X[:, 1], c=y, cmap=plt.cm.rainbow, zorder=10, s=20);\n",
"_____no_output_____"
],
[
"#%matplotlib qt\nax = plt.gca(projection='3d')\nax.plot_surface(grid[0], grid[1], Z, cmap=plt.cm.rainbow, alpha=0.2)\nax.plot_wireframe(grid[0], grid[1], Z, alpha=0.2, rstride=20, cstride=20)\nax.scatter(X[:, 0], X[:, 1], y, c=y, cmap=plt.cm.rainbow, s=30);\n#plt.show()",
"_____no_output_____"
]
],
[
[
"With kernel",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nmatplotlib.rcParams['figure.figsize']=(10,10)",
"_____no_output_____"
],
[
"# Random 2d X\nX = np.random.uniform(-1.5, 1.5, size=(100,2))\n# more feature (x**2, y**2, x*y)\nX2 = np.concatenate([X, X**2, (X[:, 0]*X[:, 1])[:, None]], axis=1)\ny = (X**2).sum(axis=1) > 1\n\nclf=svm.SVC(kernel='rbf', C=1000)\nclf.fit(X2, y)\n\n# 邊界\nx_min, y_min = X.min(axis=0)-1\nx_max, y_max = X.max(axis=0)+1\n\n# 座標點\ngrid = np.mgrid[x_min:x_max:200j, y_min:y_max:200j]\n# grid.shape = (2, 200, 200)\nG = grid.reshape(2, -1).T\nG = np.concatenate([G, G**2, (G[:, 0]*G[:, 1])[:, None]], axis=1)\n\n# 在座標點 算出 svm 的判斷函數\nZ = clf.decision_function(G)\nZ = Z.reshape(grid.shape[1:])\n\n# 畫出顏色和邊界\nplt.pcolormesh(grid[0], grid[1], Z > 0, cmap=plt.cm.rainbow, alpha=0.1)\nplt.contour(grid[0], grid[1], Z, colors=['k', 'k', 'k'], linestyles=['--', '-', '--'],\n levels=[-1, 0, 1])\n# 標出 sample 點\nplt.scatter(X[:,0], X[:, 1], c=y, cmap=plt.cm.rainbow, zorder=10, s=20);\n",
"_____no_output_____"
],
[
"#%matplotlib qt\nax = plt.gca(projection='3d')\nax.plot_surface(grid[0], grid[1], Z, cmap=plt.cm.rainbow, alpha=0.2)\nax.plot_wireframe(grid[0], grid[1], Z, alpha=0.2, rstride=20, cstride=20)\nax.scatter(X[:, 0], X[:, 1], y, c=y, cmap=plt.cm.rainbow, s=30);\n#plt.show()",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
cb8ab2e7cf7291d219739a5409c7cb646edfda05
| 21,583 |
ipynb
|
Jupyter Notebook
|
exercise/Question_SQL_Programming_Hands_on-1.ipynb
|
iitmsofteng/prework-sql
|
471209e9a2c4225d309f1028acdae98b1765fe7f
|
[
"MIT"
] | null | null | null |
exercise/Question_SQL_Programming_Hands_on-1.ipynb
|
iitmsofteng/prework-sql
|
471209e9a2c4225d309f1028acdae98b1765fe7f
|
[
"MIT"
] | null | null | null |
exercise/Question_SQL_Programming_Hands_on-1.ipynb
|
iitmsofteng/prework-sql
|
471209e9a2c4225d309f1028acdae98b1765fe7f
|
[
"MIT"
] | null | null | null | 29.404632 | 214 | 0.470741 |
[
[
[
"<img src=\"http://drive.google.com/uc?export=view&id=1tpOCamr9aWz817atPnyXus8w5gJ3mIts\" width=500px>\n\nProprietary content. © Great Learning. All Rights Reserved. Unauthorized use or distribution prohibited.",
"_____no_output_____"
],
[
"---\n# Hands-on - Advanced Certificate in Software Engineering - IIT Madras\n---",
"_____no_output_____"
],
[
"# Instructions\n\n- You need to add the code where ever you see \"`#### Add your code here ####`\"\n- Marks are mentioned along with the cells\n- **Do not edit any of the prefilled text/code part**",
"_____no_output_____"
],
[
"## Run below cells before starting the test - Mandatory",
"_____no_output_____"
]
],
[
[
"#### Please run this cell. Don't edit anything ####\n\n!pip install --upgrade pip\n!pip install ipython-sql\n\n%load_ext sql\n%sql sqlite://",
"Collecting pip\n Downloading pip-21.1.2-py3-none-any.whl (1.5 MB)\nInstalling collected packages: pip\n Attempting uninstall: pip\n Found existing installation: pip 20.1.1\n Uninstalling pip-20.1.1:\n Successfully uninstalled pip-20.1.1\n"
]
],
[
[
"## <font color='blue'> Please run the following cell. Don't edit anything </font>",
"_____no_output_____"
]
],
[
[
"%%sql\n\n\nCREATE TABLE if not exists `user`(\n `user_id` VARCHAR(50) NOT NULL,\n `user_name` VARCHAR(50) NOT NULL,\n `email` VARCHAR(50) NOT NULL UNIQUE,\n `access_level` VARCHAR(50)\n);\n \nINSERT INTO `user` (`user_id`,`user_name`,`email`,`access_level`) VALUES ('U101','Alex','[email protected]','Normal');\nINSERT INTO `user` (`user_id`,`user_name`,`email`,`access_level`) VALUES ('U102','Dwight','[email protected]','Admin');\nINSERT INTO `user` (`user_id`,`user_name`,`email`,`access_level`) VALUES ('U103','Michael','[email protected]','Admin');\nINSERT INTO `user` (`user_id`,`user_name`,`email`,`access_level`) VALUES ('U104','Jim','[email protected]','Admin');\nINSERT INTO `user` (`user_id`,`user_name`,`email`,`access_level`) VALUES ('U105','Pam','[email protected]','Normal');\n\n\n\nCREATE TABLE if not exists `products`(\n `product_id` VARCHAR(50) NOT NULL,\n `product_name` VARCHAR(50) NOT NULL,\n `brand_name` VARCHAR(50) NOT NULL,\n `price` INT\n);\n \nINSERT INTO `products` (`product_id`,`product_name`,`brand_name`,`price`) VALUES ('1','soap','vivel',10);\nINSERT INTO `products` (`product_id`,`product_name`,`brand_name`,`price`) VALUES ('2','box','naman',15);\nINSERT INTO `products` (`product_id`,`product_name`,`brand_name`,`price`) VALUES ('3','shampoo','vivel',30);\nINSERT INTO `products` (`product_id`,`product_name`,`brand_name`,`price`) VALUES ('4','detergent','quarts',20);\nINSERT INTO `products` (`product_id`,`product_name`,`brand_name`,`price`) VALUES ('5','brush','pepsod',100);\nINSERT INTO `products` (`product_id`,`product_name`,`brand_name`,`price`) VALUES ('6','fridge','kelvinator',35);\nINSERT INTO `products` (`product_id`,`product_name`,`brand_name`,`price`) VALUES ('7','cooler','kelvinator',40);\nINSERT INTO `products` (`product_id`,`product_name`,`brand_name`,`price`) VALUES ('8','shirt','raymond',60);\nINSERT INTO `products` (`product_id`,`product_name`,`brand_name`,`price`) VALUES ('9','heater','kelvinator',70);\nINSERT INTO `products` (`product_id`,`product_name`,`brand_name`,`price`) VALUES ('10','trouser','raymond',21);\n\n",
" * sqlite://\nDone.\n1 rows affected.\n1 rows affected.\n1 rows affected.\n1 rows affected.\n1 rows affected.\nDone.\n1 rows affected.\n1 rows affected.\n1 rows affected.\n1 rows affected.\n1 rows affected.\n1 rows affected.\n1 rows affected.\n1 rows affected.\n1 rows affected.\n1 rows affected.\n"
]
],
[
[
"## <font color='blue'> Please run the following cell. Don't edit anything </font>",
"_____no_output_____"
]
],
[
[
"%%sql\n\nselect * from user;",
" * sqlite://\nDone.\n"
]
],
[
[
"## <font color='blue'> Please run the following cell. Don't edit anything </font>",
"_____no_output_____"
]
],
[
[
"%%sql\n\nselect * from products;",
" * sqlite://\nDone.\n"
]
],
[
[
"## Question 1 (4 Marks)\n\nCount the number of `user_id` with `Normal` access_level in the `user` table.",
"_____no_output_____"
]
],
[
[
"%%sql\n\nselect count(user_id) from user where access_level = 'Normal';",
" * sqlite://\nDone.\n"
]
],
[
[
"## Question 2 (3 Marks)\n\nPrint the all the product information for `brand_name` `vivel` in the `products` table.",
"_____no_output_____"
]
],
[
[
"%%sql\n\nselect * from products where brand_name = 'vivel';",
" * sqlite://\nDone.\n"
]
],
[
[
"## Question 3 (3 Marks)\n\nPrint the all the unique `brand_name` from the `products` table. ",
"_____no_output_____"
]
],
[
[
"%%sql\n\nselect distinct(brand_name) from products; ",
" * sqlite://\nDone.\n"
],
[
"%%sql\nselect count(brand_name), brand_name from products group by brand_name;",
" * sqlite://\nDone.\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
cb8ab5ed8af016aa7209e4406052f1a16141d98c
| 63,771 |
ipynb
|
Jupyter Notebook
|
notebooks/03_numpy/03-Numpy-PyConES2018_EXTENDED.ipynb
|
PyDataMallorca/PyConES2018_Introduccion_a_data_science_en_Python
|
71d0c6f86afdc992e3b2ba1d6862eb34dead5698
|
[
"MIT"
] | 15 |
2018-09-03T16:45:42.000Z
|
2020-02-16T13:50:06.000Z
|
notebooks/03_numpy/03-Numpy-PyConES2018_EXTENDED.ipynb
|
NachoAG76/PyConES2018_Introduccion_a_data_science_en_Python
|
71d0c6f86afdc992e3b2ba1d6862eb34dead5698
|
[
"MIT"
] | null | null | null |
notebooks/03_numpy/03-Numpy-PyConES2018_EXTENDED.ipynb
|
NachoAG76/PyConES2018_Introduccion_a_data_science_en_Python
|
71d0c6f86afdc992e3b2ba1d6862eb34dead5698
|
[
"MIT"
] | 9 |
2018-09-08T10:06:00.000Z
|
2020-05-09T05:09:38.000Z
| 23.671492 | 441 | 0.536702 |
[
[
[
"import sys",
"_____no_output_____"
],
[
"import numpy as np",
"_____no_output_____"
]
],
[
[
"# Numpy",
"_____no_output_____"
],
[
"Numpy proporciona un nuevo contenedor de datos a Python, los `ndarray`s, además de funcionalidad especializada para poder manipularlos de forma eficiente.\n\nHablar de manipulación de datos en Python es sinónimo de Numpy y prácticamente todo el ecosistema científico de Python está construido sobre Numpy. Digamos que Numpy es el ladrillo que ha permitido levantar edificios tan sólidos como Pandas, Matplotlib, Scipy, scikit-learn,...\n\n**Índice**\n\n* [¿Por qué un nuevo contenedor de datos?](#%C2%BFPor-qu%C3%A9-un-nuevo-contenedor-de-datos?)\n* [Tipos de datos](#Tipos-de-datos)\n* [Creación de `numpy` arrays](#Creaci%C3%B3n-de-numpy-arrays)\n* [Operaciones disponibles más típicas](#Operaciones-disponibles-m%C3%A1s-t%C3%ADpicas)\n* [Metadatos y anatomía de un `ndarray`](#Metadatos-y-anatom%C3%ADa-de-un-ndarray)\n* [Indexación](#Indexaci%C3%B3n)\n* [Manejo de valores especiales](#Manejo-de-valores-especiales)\n* [Subarrays, vistas y copias](#Subarrays,-vistas-y-copias)\n* [¿Cómo funcionan los ejes de un `ndarray`?](#%C2%BFC%C3%B3mo-funcionan-los-ejes-en-un-ndarray?)\n* [Reformateo de `ndarray`s](#Reformateo-de-ndarrays)\n* [Broadcasting](#Broadcasting)\n\n\n* [`ndarrays` estructurados y `recarray`s](#ndarrays-estructurados-y-recarrays)\n* [Concatenación y partición de `ndarray`s](#Concatenaci%C3%B3n-y-partici%C3%B3n-de-ndarrays)\n* [Funciones matemáticas, funciones universales *ufuncs* y vectorización](#Funciones-matem%C3%A1ticas,-funciones-universales-ufuncs-y-vectorizaci%C3%B3n)\n* [Estadística](#Estad%C3%ADstica)\n* [Ordenando, buscando y contando](#Ordenando,-buscando-y-contando)\n* [Polinomios](#Polinomios)\n* [Álgebra lineal](#%C3%81lgebra-lineal)\n* [Manipulación de `ndarray`s](#Manipulaci%C3%B3n-de-ndarrays)\n* [Módulos de interés dentro de numpy](#M%C3%B3dulos-de-inter%C3%A9s-dentro-de-numpy)\n* [Cálculo matricial](#C%C3%A1lculo-matricial)",
"_____no_output_____"
],
[
"## ¿Por qué un nuevo contenedor de datos?",
"_____no_output_____"
],
[
"En Python, disponemos, de partida, de diversos contenedores de datos, listas, tuplas, diccionarios, conjuntos,..., ¿por qué añadir uno más?.\n\n¡Por conveniencia!, a pesar de la pérdida de flexibilidad. Es una solución de compromiso.\n\n* Uso de memoria más eficiente: Por ejemplo, una lista puede contener distintos tipos de objetos lo que provoca que Python deba guardar información del tipo de cada elemento contenido en la lista. Por otra parte, un `ndarray` contiene tipos homogéneos, es decir, todos los elementos son del mismo tipo, por lo que la información del tipo solo debe guardarse una vez independientemente del número de elementos que tenga el `ndarray`.\n\n\n\n***(imagen por Jake VanderPlas y extraída [de GitHub](https://github.com/jakevdp/PythonDataScienceHandbook)).***\n\n* Más rápido: Por ejemplo, en una lista que consta de elementos con diferentes tipos Python debe realizar trabajos extra para saber si los tipos son compatibles con las operaciones que estamos realizando. Cuando trabajamos con un `ndarray` ya podemos saber eso de partida y podemos tener operaciones más eficientes (además de que mucha funcionalidad está programada en C, C++, Cython, Fortran).\n\n\n* Operaciones vectorizadas\n\n\n* Funcionalidad extra: Muchas operaciones de álgebra lineal, transformadas rápidas de Fourier, estadística básica, histogramas,...\n\n\n* Acceso a los elementos más conveniente: Indexación más avanzada que con los tipos normales de Python\n\n\n* ...",
"_____no_output_____"
],
[
"Uso de memoria",
"_____no_output_____"
]
],
[
[
"# AVISO: SYS.GETSYZEOF NO ES FIABLE\n\nlista = list(range(5_000_000))\narr = np.array(lista, dtype=np.uint32)\nprint(\"5 millones de elementos\")\nprint(sys.getsizeof(lista))\nprint(sys.getsizeof(arr))\n\nprint()\n\nlista = list(range(100))\narr = np.array(lista, dtype=np.uint8)\nprint(\"100 elementos\")\nprint(sys.getsizeof(lista))\nprint(sys.getsizeof(arr))",
"_____no_output_____"
]
],
[
[
"Velocidad de operaciones",
"_____no_output_____"
]
],
[
[
"a = list(range(1000000))\n%timeit sum(a)\nprint(sum(a))",
"_____no_output_____"
],
[
"a = np.array(a)\n%timeit np.sum(a)\nprint(np.sum(a))",
"_____no_output_____"
]
],
[
[
"Operaciones vectorizadas",
"_____no_output_____"
]
],
[
[
"# Suma de dos vectores elemento a elemento\na = [1, 1, 1]\nb = [3, 4, 3]\nprint(a + b)\nprint('Fail')",
"_____no_output_____"
],
[
"# Suma de dos vectores elemento a elemento\na = np.array([1, 1, 1])\nb = np.array([3, 4, 3])\nprint(a + b)\nprint('\\o/')",
"_____no_output_____"
]
],
[
[
"Funcionalidad más conveniente",
"_____no_output_____"
]
],
[
[
"# suma acumulada\na = list(range(100))\nprint([sum(a[:i+1]) for i in a])\n\na = np.array(a)\nprint(a.cumsum())",
"_____no_output_____"
]
],
[
[
"Acceso a elementos más conveniente",
"_____no_output_____"
]
],
[
[
"a = [[11, 12, 13],\n [21, 22, 23],\n [31, 32, 33]]\nprint('acceso a la primera fila: ', a[0])\nprint('acceso a la primera columna: ', a[:][0], ' Fail!!!')",
"_____no_output_____"
],
[
"a = np.array(a)\nprint('acceso a la primera fila: ', a[0])\nprint('acceso a la primera columna: ', a[:,0], ' \\o/')",
"_____no_output_____"
]
],
[
[
"...",
"_____no_output_____"
],
[
"Recapitulando un poco.\n\n***Los `ndarray`s son contenedores multidimensionales, homogéneos con elementos de tamaño fijo, de dimensión predefinida.***",
"_____no_output_____"
],
[
"## Tipos de datos",
"_____no_output_____"
],
[
"Como los arrays deben ser homogéneos tenemos tipos de datos. Algunos de ellos se pueden ver en la siguiente tabla:\n\n| Data type\t | Descripción |\n|---------------|-------------|\n| ``bool_`` | Booleano (True o False) almacenado como un Byte |\n| ``int_`` | El tipo entero por defecto (igual que el `long` de C; normalmente será `int64` o `int32`)| \n| ``intc`` | Idéntico al ``int`` de C (normalmente `int32` o `int64`)| \n| ``intp`` | Entero usado para indexación (igual que `ssize_t` en C; normalmente `int32` o `int64`)| \n| ``int8`` | Byte (de -128 a 127)| \n| ``int16`` | Entero (de -32768 a 32767)|\n| ``int32`` | Entero (de -2147483648 a 2147483647)|\n| ``int64`` | Entero (de -9223372036854775808 a 9223372036854775807)| \n| ``uint8`` | Entero sin signo (de 0 a 255)| \n| ``uint16`` | Entero sin signo (de 0 a 65535)| \n| ``uint32`` | Entero sin signo (de 0 a 4294967295)| \n| ``uint64`` | Entero sin signo (de 0 a 18446744073709551615)| \n| ``float_`` | Atajo para ``float64``.| \n| ``float16`` | Half precision float: un bit para el signo, 5 bits para el exponente, 10 bits para la mantissa| \n| ``float32`` | Single precision float: un bit para el signo, 8 bits para el exponente, 23 bits para la mantissa|\n| ``float64`` | Double precision float: un bit para el signo, 11 bits para el exponente, 52 bits para la mantissa|\n| ``complex_`` | Atajo para `complex128`.| \n| ``complex64`` | Número complejo, represantedo por dos *floats* de 32-bits| \n| ``complex128``| Número complejo, represantedo por dos *floats* de 64-bits| \n\nEs posible tener una especificación de tipos más detallada, pudiendo especificar números con *big endian* o *little endian*. No vamos a ver esto en este momento.\n\nEl tipo por defecto que usa `numpy` al crear un *ndarray* es `np.float_`, siempre que no específiquemos explícitamente el tipo a usar.",
"_____no_output_____"
],
[
"Por ejemplo, un array de tipo `np.uint8` puede tener los siguientes valores:",
"_____no_output_____"
]
],
[
[
"import itertools\n\nfor i, bits in enumerate(itertools.product((0, 1), repeat=8)):\n print(i, bits)",
"_____no_output_____"
]
],
[
[
"Es decir, puede contener valores que van de 0 a 255 ($2^8$).",
"_____no_output_____"
],
[
"¿Cuántos bytes tendrá un `ndarray` de 10 elementos cuyo tipo de datos es un `np.int8`?",
"_____no_output_____"
]
],
[
[
"a = np.arange(10, dtype=np.int8)\nprint(a.nbytes)\nprint(sys.getsizeof(a))",
"_____no_output_____"
],
[
"a = np.repeat(1, 100000).astype(np.int8)\nprint(a.nbytes)\nprint(sys.getsizeof(a))",
"_____no_output_____"
]
],
[
[
"## Creación de numpy arrays",
"_____no_output_____"
],
[
"Podemos crear numpy arrays de muchas formas.\n\n* Rangos numéricos\n\n`np.arange`, `np.linspace`, `np.logspace`\n\n* Datos homogéneos\n\n`np.zeros`, `np.ones`\n\n* Elementos diagonales\n\n`np.diag`, `np.eye`\n\n* A partir de otras estructuras de datos ya creadas\n\n`np.array`\n\n* A partir de otros numpy arrays\n\n`np.empty_like`\n\n* A partir de ficheros\n\n`np.loadtxt`, `np.genfromtxt`,...\n\n\n* A partir de un escalar\n\n`np.full`, `np.tile`,...\n\n* A partir de valores aleatorios\n\n`np.random.randint`, `np.random.randint`, `np.random.randn`,...\n\n...",
"_____no_output_____"
]
],
[
[
"a = np.arange(10) # similar a range pero devuelve un ndarray en lugar de un objeto range\nprint(a)",
"_____no_output_____"
],
[
"a = np.linspace(0, 1, 101)\nprint(a)",
"_____no_output_____"
],
[
"a_i = np.zeros((2, 3), dtype=np.int)\na_f = np.zeros((2, 3))\nprint(a_i)\nprint(a_f)",
"_____no_output_____"
],
[
"a = np.eye(3)\nprint(a)",
"_____no_output_____"
],
[
"a = np.array(\n (\n (1, 2, 3, 4, 5, 6),\n (10, 20, 30, 40, 50, 60)\n ), \n dtype=np.float\n)\nprint(a)",
"_____no_output_____"
],
[
"np.full((5, 5), -999)",
"_____no_output_____"
],
[
"np.random.randint(0, 50, 15)",
"_____no_output_____"
]
],
[
[
"<div class=\"alert alert-success\">\n <p>Referencias:</p>\n <p><a href=\"https://docs.scipy.org/doc/numpy/user/basics.creation.html#arrays-creation\">array creation</a></p>\n <p><a href=\"https://docs.scipy.org/doc/numpy/reference/routines.array-creation.html#routines-array-creation\">routines for array creation</a></p>\n</div>",
"_____no_output_____"
],
[
"**Practicando**\n\nRecordad que siempre podéis usar `help`, `?`, `np.lookfor`,..., para obtener más información.",
"_____no_output_____"
]
],
[
[
"help(np.sum)",
"_____no_output_____"
],
[
"np.rad2deg?",
"_____no_output_____"
],
[
"np.lookfor(\"create array\")",
"_____no_output_____"
]
],
[
[
"Ved un poco como funciona `np.repeat`, `np.empty_like`,... ",
"_____no_output_____"
]
],
[
[
"# Play area\n\n",
"_____no_output_____"
],
[
"%load ../../solutions/03_01_np_array_creacion.py",
"_____no_output_____"
]
],
[
[
"## Operaciones disponibles más típicas",
"_____no_output_____"
]
],
[
[
"a = np.random.rand(5, 2)\nprint(a)",
"_____no_output_____"
],
[
"a.sum()",
"_____no_output_____"
],
[
"a.sum(axis=0)",
"_____no_output_____"
],
[
"a.sum(axis=1)",
"_____no_output_____"
],
[
"a.ravel()",
"_____no_output_____"
],
[
"a.reshape(2, 5)",
"_____no_output_____"
],
[
"a.T",
"_____no_output_____"
],
[
"a.transpose()",
"_____no_output_____"
],
[
"a.mean()",
"_____no_output_____"
],
[
"a.mean(axis=1)",
"_____no_output_____"
],
[
"a.cumsum(axis=1)",
"_____no_output_____"
]
],
[
[
"<div class=\"alert alert-success\">\n <p>Referencias:</p>\n <p><a href=\"https://docs.scipy.org/doc/numpy/user/quickstart.html\">Quick start tutorial</a></p>\n</div>",
"_____no_output_____"
],
[
"**Practicando**\n\nMirad más métodos de un `ndarray` y toquetead. Si no entendéis algo, preguntad:",
"_____no_output_____"
]
],
[
[
"dir(a)",
"_____no_output_____"
],
[
"# Play area\n\n",
"_____no_output_____"
],
[
"%load ../../solutions/03_02_np_operaciones_tipicas.py",
"_____no_output_____"
]
],
[
[
"## Metadatos y anatomía de un `ndarray`",
"_____no_output_____"
],
[
"En realidad, un `ndarray` es un bloque de memoria con información extra sobre como interpretar su contenido. La memoria dinámica (RAM) se puede considerar como un 'churro' lineal y es por ello que necesitamos esa información extra para saber como formar ese `ndarray`, sobre todo la información de `shape` y `strides`.\n\nEsta parte va a ser un poco más esotérica para los no iniciados pero considero que es necesaria para poder entender mejor nuestra nueva estructura de datos y poder sacarle mejor partido.",
"_____no_output_____"
]
],
[
[
"a = np.random.randn(5000, 5000)",
"_____no_output_____"
]
],
[
[
"El número de dimensiones del `ndarray`",
"_____no_output_____"
]
],
[
[
"a.ndim",
"_____no_output_____"
]
],
[
[
"El número de elementos en cada una de las dimensiones",
"_____no_output_____"
]
],
[
[
"a.shape",
"_____no_output_____"
]
],
[
[
"El número de elementos",
"_____no_output_____"
]
],
[
[
"a.size",
"_____no_output_____"
]
],
[
[
"El tipo de datos de los elementos",
"_____no_output_____"
]
],
[
[
"a.dtype",
"_____no_output_____"
]
],
[
[
"El número de bytes de cada elemento",
"_____no_output_____"
]
],
[
[
"a.itemsize",
"_____no_output_____"
]
],
[
[
"El número de bytes que ocupa el `ndarray` (es lo mismo que `size` por `itemsize`)",
"_____no_output_____"
]
],
[
[
"a.nbytes",
"_____no_output_____"
]
],
[
[
"El *buffer* que contiene los elementos del `ndarray`",
"_____no_output_____"
]
],
[
[
"a.data",
"_____no_output_____"
]
],
[
[
"Pasos a dar en cada dimensión cuando nos movemos entre elementos",
"_____no_output_____"
]
],
[
[
"a.strides",
"_____no_output_____"
]
],
[
[
"\n***(imagen extraída [de GitHub](https://github.com/btel/2016-erlangen-euroscipy-advanced-numpy)).***",
"_____no_output_____"
],
[
"Más cosas",
"_____no_output_____"
]
],
[
[
"a.flags",
"_____no_output_____"
]
],
[
[
"Pequeño ejercicio, ¿por qué tarda menos en sumar elementos en una dimensión que en otra si es un array regular?",
"_____no_output_____"
]
],
[
[
"%timeit a.sum(axis=0)\n%timeit a.sum(axis=1)",
"_____no_output_____"
]
],
[
[
"Pequeño ejercicio, ¿por qué ahora el resultado es diferente?",
"_____no_output_____"
]
],
[
[
"aT = a.T\n%timeit aT.sum(axis=0)\n%timeit aT.sum(axis=1)",
"_____no_output_____"
],
[
"print(aT.strides)\nprint(aT.flags)",
"_____no_output_____"
],
[
"print(np.repeat((1,2,3), 3))\nprint()\na = np.repeat((1,2,3), 3).reshape(3, 3)\nprint(a)\nprint()\nprint(a.sum(axis=0))\nprint()\nprint(a.sum(axis=1))",
"_____no_output_____"
]
],
[
[
"<div class=\"alert alert-success\">\n <p>Referencias:</p>\n <p><a href=\"https://docs.scipy.org/doc/numpy/reference/arrays.ndarray.html#internal-memory-layout-of-an-ndarray\">Internal memory layout of an ndarray</a></p>\n <p><a href=\"https://docs.scipy.org/doc/numpy/reference/internals.html#multidimensional-array-indexing-order-issues\">multidimensional array indexing order issues</a></p>\n</div>",
"_____no_output_____"
],
[
"## Indexación",
"_____no_output_____"
],
[
"Si ya has trabajado con indexación en estructuras de Python, como listas, tuplas o strings, la indexación en Numpy te resultará muy familiar. \n\nPor ejemplo, por hacer las cosas sencillas, vamos a crear un `ndarray` de 1D:",
"_____no_output_____"
]
],
[
[
"a = np.arange(10, dtype=np.uint8)\nprint(a)",
"_____no_output_____"
],
[
"print(a[:]) # para acceder a todos los elementos\nprint(a[:-1]) # todos los elementos menos el último\nprint(a[1:]) # todos los elementos menos el primero\nprint(a[::2]) # el primer, el tercer, el quinto,..., elemento\nprint(a[3]) # el cuarto elemento\nprint(a[-1:-5:-1]) # ¿?",
"_____no_output_____"
],
[
"# Practicad vosotros\n",
"_____no_output_____"
]
],
[
[
"Para *ndarrays* de una dimensión es exactamente igual que si usásemos listas o tuplas de Python:\n\n* Primer elemento tiene índice 0\n* Los índices negativos empiezan a contar desde el final\n* slices/rebanadas con `[start:stop:step]`",
"_____no_output_____"
],
[
"Con un `ndarray` de más dimensiones las cosas ya cambian con respecto a Python puro:",
"_____no_output_____"
]
],
[
[
"a = np.random.randn(10, 2)\nprint(a)",
"_____no_output_____"
],
[
"a[1] # ¿Qué nos dará esto?\na[1, 1] # Si queremos acceder a un elemento específico hay que dar su posición completa en el ndarray\na[::3, 1]",
"_____no_output_____"
]
],
[
[
"Si tenemos dimensiones mayores a 1 es parecido a las listas pero los índices se separan por comas para las nuevas dimensiones.\n<img src=\"../../images/03_03_arraygraphics_0.png\" width=400px />\n(imagen extraída de [aquí](https://github.com/gertingold/euroscipy-numpy-tutorial))",
"_____no_output_____"
]
],
[
[
"a = np.arange(40).reshape(5, 8)\nprint(a)",
"_____no_output_____"
],
[
"a[2, -3]",
"_____no_output_____"
]
],
[
[
"Para obtener más de un elemento hacemos *slicing* para cada eje:\n<img src=\"../../images/03_04_arraygraphics_1.png\" width=400px />\n(imagen extraída de [aquí](https://github.com/gertingold/euroscipy-numpy-tutorial))",
"_____no_output_____"
]
],
[
[
"a[:3, :5]",
"_____no_output_____"
]
],
[
[
"¿Cómo podemos conseguir los elementos señalados en esta imagen?\n<img src=\"../../images/03_06_arraygraphics_2_wo.png\" width=400px />\n\n(imagen extraída de [aquí](https://github.com/gertingold/euroscipy-numpy-tutorial))",
"_____no_output_____"
]
],
[
[
"a[x:x ,x:x]",
"_____no_output_____"
]
],
[
[
"¿Cómo podemos conseguir los elementos señalados en esta imagen?\n<img src=\"../../images/03_08_arraygraphics_3_wo.png\" width=400px />\n\n(imagen extraída de [aquí](https://github.com/gertingold/euroscipy-numpy-tutorial))",
"_____no_output_____"
]
],
[
[
"a[x:x ,x:x]",
"_____no_output_____"
]
],
[
[
"¿Cómo podemos conseguir los elementos señalados en esta imagen?\n<img src=\"../../images/03_10_arraygraphics_4_wo.png\" width=400px />\n\n(imagen extraída de [aquí](https://github.com/gertingold/euroscipy-numpy-tutorial))",
"_____no_output_____"
]
],
[
[
"a[x:x ,x:x]",
"_____no_output_____"
]
],
[
[
"¿Cómo podemos conseguir los elementos señalados en esta imagen?\n<img src=\"../../images/03_12_arraygraphics_5_wo.png\" width=400px />\n\n(imagen extraída de [aquí](https://github.com/gertingold/euroscipy-numpy-tutorial))",
"_____no_output_____"
]
],
[
[
"a[x:x ,x:x]",
"_____no_output_____"
]
],
[
[
"Soluciones a lo anterior:\n\n<img src=\"../../images/03_05_arraygraphics_2.png\" width=200px />\n<img src=\"../../images/03_07_arraygraphics_3.png\" width=200px />\n<img src=\"../../images/03_09_arraygraphics_4.png\" width=200px />\n<img src=\"../../images/03_11_arraygraphics_5.png\" width=200px />\n\n(imágenes extraídas de [aquí](https://github.com/gertingold/euroscipy-numpy-tutorial))",
"_____no_output_____"
],
[
"**Fancy indexing**",
"_____no_output_____"
],
[
"Con *fancy indexing* podemos hacer cosas tan variopintas como:",
"_____no_output_____"
],
[
"<img src=\"../../images/03_13_arraygraphics_6.png\" width=300px />\n<img src=\"../../images/03_14_arraygraphics_7.png\" width=300px />\n\n(imágenes extraídas de [aquí](https://github.com/gertingold/euroscipy-numpy-tutorial))",
"_____no_output_____"
],
[
"Es decir, podemos indexar usando `ndarray`s de booleanos ó usando listas de índices para extraer elementos concretos de una sola vez.\n\n**WARNING: En el momento que usamos *fancy indexing* nos devuelve un nuevo *ndarray* que no tiene porque conservar la estructura original.**",
"_____no_output_____"
],
[
"Por ejemplo, en el siguiente caso no devuelve un *ndarray* de dos dimensiones porque la máscara no tiene porqué ser regular y, por tanto, devuelve solo los valores que cumplen el criterio en un vector (*ndarray* de una dimensión).",
"_____no_output_____"
]
],
[
[
"a = np.arange(10).reshape(2, 5)\nprint(a)",
"_____no_output_____"
],
[
"bool_indexes = (a % 2 == 0)\nprint(bool_indexes)",
"_____no_output_____"
],
[
"a[bool_indexes]",
"_____no_output_____"
]
],
[
[
"Sin embargo, sí que lo podríamos usar para modificar el *ndarray* original en base a un criterio y seguir manteniendo la misma forma.",
"_____no_output_____"
]
],
[
[
"a[bool_indexes] = 999\nprint(a)",
"_____no_output_____"
]
],
[
[
"<div class=\"alert alert-success\">\n <p>Referencias:</p>\n <p><a href=\"https://docs.scipy.org/doc/numpy/reference/arrays.indexing.html#arrays-indexing\">array indexing</a></p>\n <p><a href=\"https://docs.scipy.org/doc/numpy/reference/arrays.ndarray.html#indexing-arrays\">indexing arrays</a></p>\n</div>",
"_____no_output_____"
],
[
"## Manejo de valores especiales",
"_____no_output_____"
],
[
"`numpy` provee de varios valores especiales: `np.nan`, `np.Inf`, `np.Infinity`, `np.inf`, `np.infty`,...",
"_____no_output_____"
]
],
[
[
"a = 1 / np.arange(10)\nprint(a)",
"_____no_output_____"
],
[
"a[0] == np.inf",
"_____no_output_____"
],
[
"a.max() # Esto no es lo que queremos",
"_____no_output_____"
],
[
"a.mean() # Esto no es lo que queremos",
"_____no_output_____"
],
[
"a[np.isfinite(a)].max()",
"_____no_output_____"
],
[
"a[-1] = np.nan\nprint(a)",
"_____no_output_____"
],
[
"a.mean()",
"_____no_output_____"
],
[
"np.isnan(a)",
"_____no_output_____"
],
[
"np.isfinite(a)",
"_____no_output_____"
],
[
"np.isinf(a) # podéis mirar también np.isneginf, np.isposinf",
"_____no_output_____"
]
],
[
[
"`numpy` usa el estándar IEEE de números flotantes para aritmética (IEEE 754). Esto significa que *Not a\nNumber* no es equivalente a *infinity*. También, *positive infinity* no es equivalente a *negative infinity*. Pero *infinity* es equivalente a *positive infinity*.",
"_____no_output_____"
]
],
[
[
"1 < np.inf",
"_____no_output_____"
],
[
"1 < -np.inf",
"_____no_output_____"
],
[
"1 > -np.inf",
"_____no_output_____"
],
[
"1 == np.inf",
"_____no_output_____"
],
[
"1 < np.nan",
"_____no_output_____"
],
[
"1 > np.nan",
"_____no_output_____"
],
[
"1 == np.nan",
"_____no_output_____"
]
],
[
[
"## Subarrays, vistas y copias",
"_____no_output_____"
],
[
"**¡IMPORTANTE!**\n\nVistas y copias: `numpy`, por defecto, siempre devuelve vistas para evitar incrementos innecesarios de memoria. Este comportamiento difiere del de Python puro donde una rebanada (*slicing*) de una lista devuelve una copia. Si queremos una copia de un `ndarray` debemos obtenerla de forma explícita:",
"_____no_output_____"
]
],
[
[
"a = np.arange(10)\nb = a[2:5]\nprint(a)\nprint(b)",
"_____no_output_____"
],
[
"b[0] = 222\nprint(a)\nprint(b)",
"_____no_output_____"
]
],
[
[
"Este comportamiento por defecto es realmente muy útil, significa que, trabajando con grandes conjuntos de datos, podemos acceder y procesar piezas de estos conjuntos de datos sin necesidad de copiar el buffer de datos original.",
"_____no_output_____"
],
[
"A veces, es necesario crear una copia. Esto se puede realizar fácilmente usando el método `copy` de los *ndarrays*. El ejemplo anterior usando una copia en lugar de una vista:",
"_____no_output_____"
]
],
[
[
"a = np.arange(10)\nb = a[2:5].copy()\nprint(a)\nprint(b)\nb[0] = 222\nprint(a)\nprint(b)",
"_____no_output_____"
]
],
[
[
"## ¿Cómo funcionan los ejes en un `ndarray`?",
"_____no_output_____"
],
[
"Por ejemplo, cuando hacemos `a.sum()`, `a.sum(axis=0)`, `a.sum(axis=1)`.\n\n¿Qué pasa si tenemos más de dos dimensiones?\n\nVamos a ver ejemplos:",
"_____no_output_____"
]
],
[
[
"a = np.arange(10).reshape(5,2)",
"_____no_output_____"
],
[
"a.shape",
"_____no_output_____"
],
[
"a.sum()",
"_____no_output_____"
],
[
"a.sum(axis=0)",
"_____no_output_____"
],
[
"a.sum(axis=1)",
"_____no_output_____"
]
],
[
[
"\n(imagen extraída de [aquí](https://github.com/gertingold/euroscipy-numpy-tutorial))",
"_____no_output_____"
]
],
[
[
"a = np.arange(9).reshape(3, 3)\nprint(a)\nprint(a.sum(axis=0))\nprint(a.sum(axis=1))",
"_____no_output_____"
]
],
[
[
"\n(imagen extraída de [aquí](https://github.com/gertingold/euroscipy-numpy-tutorial))",
"_____no_output_____"
]
],
[
[
"a = np.arange(24).reshape(2, 3, 4)\nprint(a)\nprint(a.sum(axis=0))\nprint(a.sum(axis=1))\nprint(a.sum(axis=2))",
"_____no_output_____"
]
],
[
[
"Por ejemplo, en el primer caso, `axis=0`, lo que sucede es que cogemos todos los elementos del primer índice y aplicamos la operación para cada uno de los elementos de los otros dos ejes. Hecho de uno en uno sería lo siguiente:",
"_____no_output_____"
]
],
[
[
"print(a[:,0,0].sum(), a[:,0,1].sum(), a[:,0,2].sum(), a[:,0,3].sum())\nprint(a[:,1,0].sum(), a[:,1,1].sum(), a[:,1,2].sum(), a[:,1,3].sum())\nprint(a[:,2,0].sum(), a[:,2,1].sum(), a[:,2,2].sum(), a[:,2,3].sum())",
"_____no_output_____"
]
],
[
[
"Sin contar el eje que estamos usando, las dimensiones que quedan son 3 x 4 (segunda y tercera dimensiones) por lo que el resultado son 12 elementos.",
"_____no_output_____"
],
[
"Para el caso de `axis=1`:",
"_____no_output_____"
]
],
[
[
"print(a[0,:,0].sum(), a[0,:,1].sum(), a[0,:,2].sum(), a[0,:,3].sum())\nprint(a[1,:,0].sum(), a[1,:,1].sum(), a[1,:,2].sum(), a[1,:,3].sum())",
"_____no_output_____"
]
],
[
[
"Sin contar el eje que estamos usando, las dimensiones que quedan son 2 x 4 (primera y tercera dimensiones) por lo que el resultado son 8 elementos.",
"_____no_output_____"
],
[
"Para el caso de `axis=2`:",
"_____no_output_____"
]
],
[
[
"print(a[0,0,:].sum(), a[0,1,:].sum(), a[0,2,:].sum())\nprint(a[1,0,:].sum(), a[1,1,:].sum(), a[1,2,:].sum())",
"_____no_output_____"
]
],
[
[
"Sin contar el eje que estamos usando, las dimensiones que quedan son 2 x 3 (primera y segunda dimensiones) por lo que el resultado son 3 elementos.",
"_____no_output_____"
],
[
"## Reformateo de `ndarray`s",
"_____no_output_____"
],
[
"Podemos cambiar la forma de los `ndarray`s usando el método `reshape`. Por ejemplo, si queremos colocar los números del 1 al 9 en un grid $3 \\times 3$ lo podemos hacer de la siguiente forma:",
"_____no_output_____"
]
],
[
[
"a = np.arange(1, 10).reshape(3, 3)",
"_____no_output_____"
]
],
[
[
"Para que el cambio de forma no dé errores hemos de tener cuidado en que los tamaños del `ndarray` inicial y del `ndarray` final sean compatibles.",
"_____no_output_____"
]
],
[
[
"# Por ejemplo, lo siguiente dará error?\na = np.arange(1, 10). reshape(5, 2)",
"_____no_output_____"
]
],
[
[
"Otro patrón común de cambio de forma sería la conversion de un `ndarray` de 1D en uno de 2D añadiendo un nuevo eje. Lo podemos hacer usando, nuevamente, el método `reshape` o usando `numpy.newaxis`.",
"_____no_output_____"
]
],
[
[
"# Por ejemplo un array 2D de una fila\na = np.arange(3)\na1_2D = a.reshape(1,3)\na2_2D = a[np.newaxis, :]\nprint(a1_2D)\nprint(a1_2D.shape)\nprint(a2_2D)\nprint(a2_2D.shape)",
"_____no_output_____"
],
[
"# Por ejemplo un array 2D de una columna\na = np.arange(3)\na1_2D = a.reshape(3,1)\na2_2D = a[:, np.newaxis]\nprint(a1_2D)\nprint(a1_2D.shape)\nprint(a2_2D)\nprint(a2_2D.shape)",
"_____no_output_____"
]
],
[
[
"## Broadcasting",
"_____no_output_____"
],
[
"Es poible realizar operaciones en *ndarrays* de diferentes tamaños. En algunos casos `numpy` puede transformar estos *ndarrays* automáticamente de forma que todos tienen la misma forma. Esta conversión automática se llama **broadcasting**.",
"_____no_output_____"
],
[
"Normas del Broadcasting\n\nPara determinar la interacción entre dos `ndarray`s en Numpy se sigue un conjunto de reglas estrictas:\n\n* Regla 1: Si dos `ndarray`s difieren en su número de dimensiones la forma de aquel con menos dimensiones se rellena con 1's a su derecha.\n- Regla 2: Si la forma de dos `ndarray`s no es la misma en ninguna de sus dimensiones, el `ndarry` con forma igual a 1 en esa dimensión se 'alarga' para tener simulares dimensiones que los del otros `ndarray`.\n- Regla 3: Si en cualquier dimensión el tamaño no es igual y ninguno de ellos es igual a 1 entonces obtendremos un error.\n\nResumiendo, cuando se opera en dos *ndarrays*, `numpy` compara sus formas (*shapes*) elemento a elemento. Empieza por las dimensiones más a la izquierda y trabaja hacia las siguientes dimensiones. Dos dimensiones son compatibles cuando\n\n ambas son iguales o\n una de ellas es 1\n\nSi estas condiciones no se cumplen se lanzará una excepción `ValueError: frames are not aligned` indicando que los *ndarrays* tienen formas incompatibles. El tamaño del *ndarray* resultante es el tamaño máximo a lo largo de cada dimensión de los *ndarrays* de partida.",
"_____no_output_____"
],
[
"De forma más gráfica:\n\n\n(imagen extraída de [aquí](https://github.com/btel/2016-erlangen-euroscipy-advanced-numpy))\n\n```\na: 4 x 3 a: 4 x 3 a: 4 x 1\nb: 4 x 3 b: 3 b: 3\nresult: 4 x 3 result: 4 x 3 result: 4 x 3\n```\n\nIntentemos reproducir los esquemas de la imagen anterior.",
"_____no_output_____"
]
],
[
[
"a = np.repeat((0, 10, 20, 30), 3).reshape(4, 3)\nb = np.repeat((0, 1, 2), 4).reshape(3,4).T\nprint(a)\nprint(b)\nprint(a + b)",
"_____no_output_____"
],
[
"a = np.repeat((0, 10, 20, 30), 3).reshape(4, 3)\nb = np.array((0, 1, 2))\nprint(a)\nprint(b)\nprint(a + b)",
"_____no_output_____"
],
[
"a = np.array((0, 10, 20, 30)).reshape(4,1)\nb = np.array((0, 1, 2))\nprint(a)\nprint(b)\nprint(a + b)",
"_____no_output_____"
]
],
[
[
"<div class=\"alert alert-success\">\n <p>Referencias:</p>\n <p><a href=\"https://docs.scipy.org/doc/numpy/user/basics.broadcasting.html\">Basic broadcasting</a></p>\n <p><a href=\"http://scipy.github.io/old-wiki/pages/EricsBroadcastingDoc\">Broadcasting more in depth</a></p>\n</div>",
"_____no_output_____"
],
[
"## `ndarrays` estructurados y `recarray`s",
"_____no_output_____"
],
[
"Antes hemos comentado que los `ndarray`s deben ser homogéneos pero era un poco inexacto, en realidad, podemos tener `ndarray`s que tengan diferentes tipos. Estos se llaman `ndarray`s estructurados y `recarray`s.\n\nVeamos ejemplos:",
"_____no_output_____"
]
],
[
[
"nombre = ['paca', 'pancracio', 'nemesia', 'eulogio']\nedad = [72, 68, 86, 91]\na = np.array(np.zeros(4), dtype=[('name', '<S10'), ('age', np.int)])\na['name'] = nombre\na['age'] = edad\nprint(a)",
"_____no_output_____"
]
],
[
[
"Podemos acceder a las columnas por nombre",
"_____no_output_____"
]
],
[
[
"a['name']",
"_____no_output_____"
]
],
[
[
"A todos los elementos menos el primero",
"_____no_output_____"
]
],
[
[
"a['age'][1:]",
"_____no_output_____"
]
],
[
[
"Un `recarray` es similar pero podemos acceder a los campos con notación de punto (*dot notation*).",
"_____no_output_____"
]
],
[
[
"ra = a.view(np.recarray)",
"_____no_output_____"
],
[
"ra.name",
"_____no_output_____"
]
],
[
[
"Esto introduce un poco de *overhead* para acceder ya que se realizan algunas operaciones de más.",
"_____no_output_____"
],
[
"## Concatenación y partición de `ndarrays`",
"_____no_output_____"
],
[
"Podemos combinar múltiples *ndarrays* en uno o separar uno en varios.\n\nPara concatenar podemos usar `np.concatenate`, `np.hstack`, `np.vstack`, `np.dstack`. Ejemplos:",
"_____no_output_____"
]
],
[
[
"a = np.array([1, 1, 1, 1])\nb = np.array([2, 2, 2, 2])",
"_____no_output_____"
]
],
[
[
"Podemos concatenar esos dos arrays usando `np.concatenate`:",
"_____no_output_____"
]
],
[
[
"np.concatenate([a, b])",
"_____no_output_____"
]
],
[
[
"No solo podemos concatenar *ndarrays* de una sola dimensión:",
"_____no_output_____"
]
],
[
[
"np.concatenate([a.reshape(2, 2), b.reshape(2, 2)])",
"_____no_output_____"
]
],
[
[
"Podemos elegir sobre qué eje concatenamos:",
"_____no_output_____"
]
],
[
[
"np.concatenate([a.reshape(2, 2), b.reshape(2, 2)], axis=1)",
"_____no_output_____"
]
],
[
[
"Podemos concatenar más de dos arrays:",
"_____no_output_____"
]
],
[
[
"c = [3, 3, 3, 3]\nnp.concatenate([a, b, c])",
"_____no_output_____"
]
],
[
[
"Si queremos ser más explícitos podemos usar `np.hstack` o `np.vstack`. La `h` y la `v` son para horizontal y vertical, respectivamente.",
"_____no_output_____"
]
],
[
[
"np.hstack([a, b])",
"_____no_output_____"
],
[
"np.vstack([a, b])",
"_____no_output_____"
]
],
[
[
"Podemos concatenar en la tercera dimensión usamos `np.dstack`.",
"_____no_output_____"
],
[
"De la misma forma que podemos concatenar, podemos partir *ndarrays* usando `np.split`, `np.hsplit`, `np.vsplit`, `np.dsplit`.",
"_____no_output_____"
]
],
[
[
"# Intentamos entender como funciona la partición probando...\n",
"_____no_output_____"
]
],
[
[
"## Funciones matemáticas, funciones universales *ufuncs* y vectorización",
"_____no_output_____"
],
[
"¿Qué es eso de *ufunc*? \n\nDe la [documentación oficial de Numpy](http://docs.scipy.org/doc/numpy/reference/ufuncs.html):\n \n> A universal function (or ufunc for short) is a function that operates on ndarrays in an element-by-element fashion, supporting array broadcasting, type casting, and several other standard features. That is, a ufunc is a “**vectorized**” wrapper for a function that takes a **fixed number of scalar inputs** and produces a **fixed number of scalar outputs**.\n\nUna *ufunc* es una *Universal function* o función universal que actúa sobre todos los elementos de un `ndarray`, es decir aplica la funcionalidad sobre cada uno de los elementos del `ndarray`. Esto se conoce como vectorización.\n\nPor ejemplo, veamos la operación de elevar al cuadrado una lista en python puro o en `numpy`:",
"_____no_output_____"
]
],
[
[
"# En Python puro\na_list = list(range(10000))\n\n%timeit [i ** 2 for i in a_list]",
"_____no_output_____"
],
[
"# En numpy\nan_arr = np.arange(10000)\n\n%timeit np.power(an_arr, 2)",
"_____no_output_____"
],
[
"a = np.arange(10)\n\nnp.power(a, 2)",
"_____no_output_____"
]
],
[
[
"La función anterior eleva al cuadrado cada uno de los elementos del `ndarray` anterior.\n\nDentro de `numpy` hay muchísimas *ufuncs* y `scipy` (no lo vamos a ver) dispone de muchas más *ufuns* mucho más especializadas.\n\nEn `numpy` tenemos, por ejemplo:",
"_____no_output_____"
],
[
"* Funciones trigonométricas: `sin`, `cos`, `tan`, `arcsin`, `arccos`, `arctan`, `hypot`, `arctan2`, `degrees`, `radians`, `unwrap`, `deg2rad`, `rad2deg`",
"_____no_output_____"
]
],
[
[
"# juguemos un poco con ellas\n",
"_____no_output_____"
]
],
[
[
"* Funciones hiperbólicas: `sinh`, `cosh`, `tanh`, `arcsinh`, `arccosh`, `arctanh`",
"_____no_output_____"
]
],
[
[
"# juguemos un poco con ellas\n",
"_____no_output_____"
]
],
[
[
"* Redondeo: `around`, `round_`, `rint`, `fix`, `floor`, `ceil`, `trunc`",
"_____no_output_____"
]
],
[
[
"# juguemos un poco con ellas\n",
"_____no_output_____"
]
],
[
[
"* Sumas, productos, diferencias: `prod`, `sum`, `nansum`, `cumprod`, `cumsum`, `diff`, `ediff1d`, `gradient`, `cross`, `trapz`",
"_____no_output_____"
]
],
[
[
"# juguemos un poco con ellas\n",
"_____no_output_____"
]
],
[
[
"* Exponentes y logaritmos: `exp`, `expm1`, `exp2`, `log`, `log10`, `log2`, `log1p`, `logaddexp`, `logaddexp2`",
"_____no_output_____"
]
],
[
[
"# juguemos un poco con ellas\n",
"_____no_output_____"
]
],
[
[
"* Otras funciones especiales: `i0`, `sinc`",
"_____no_output_____"
]
],
[
[
"# juguemos un poco con ellas\n",
"_____no_output_____"
]
],
[
[
"* Trabajo con decimales: `signbit`, `copysign`, `frexp`, `ldexp`",
"_____no_output_____"
]
],
[
[
"# juguemos un poco con ellas\n",
"_____no_output_____"
]
],
[
[
"* Operaciones aritméticas: `add`, `reciprocal`, `negative`, `multiply`, `divide`, `power`, `subtract`, `true_divide`, `floor_divide`, `fmod`, `mod`, `modf`, `remainder`",
"_____no_output_____"
]
],
[
[
"# juguemos un poco con ellas\n",
"_____no_output_____"
]
],
[
[
"* Manejo de números complejos: `angle`, `real`, `imag`, `conj`",
"_____no_output_____"
]
],
[
[
"# juguemos un poco con ellas\n",
"_____no_output_____"
]
],
[
[
"* Miscelanea: `convolve`, `clip`, `sqrt`, `square`, `absolute`, `fabs`, `sign`, `maximum`, `minimum`, `fmax`, `fmin`, `nan_to_num`, `real_if_close`, `interp`\n\n\n...",
"_____no_output_____"
]
],
[
[
"# juguemos un poco con ellas\n",
"_____no_output_____"
]
],
[
[
"<div class=\"alert alert-success\">\n <p>Referencias:</p>\n <p><a href=\"http://docs.scipy.org/doc/numpy/reference/ufuncs.html\">Ufuncs</a></p>\n</div>",
"_____no_output_____"
],
[
"## Estadística",
"_____no_output_____"
],
[
"* Orden: `amin`, `amax`, `nanmin`, `nanmax`, `ptp`, `percentile`, `nanpercentile`\n\n\n* Medias y varianzas: `median`, `average`, `mean`, `std`, `var`, `nanmedian`, `nanmean`, `nanstd`, `nanvar`\n\n\n* Correlacionando: `corrcoef`, `correlate`, `cov`\n\n\n* Histogramas: `histogram`, `histogram2d`, `histogramdd`, `bincount`, `digitize`\n\n...",
"_____no_output_____"
]
],
[
[
"# juguemos un poco con ellas\n",
"_____no_output_____"
]
],
[
[
"## Ordenando, buscando y contando",
"_____no_output_____"
],
[
"* Ordenando: `sort`, `lexsort`, `argsort`, `ndarray.sort`, `msort`, `sort_complex`, `partition`, `argpartition`\n\n\n* Buscando: `argmax`, `nanargmax`, `argmin`, `nanargmin`, `argwhere`, `nonzero`, `flatnonzero`, `where`, `searchsorted`, `extract`\n\n\n* Contando: `count_nonzero`\n\n...",
"_____no_output_____"
]
],
[
[
"# juguemos un poco con ellas\n",
"_____no_output_____"
]
],
[
[
"## Polinomios",
"_____no_output_____"
],
[
"* Series de potencias: `numpy.polynomial.polynomial`\n\n\n* Clase Polynomial: `np.polynomial.Polynomial`\n\n\n* Básicos: `polyval`, `polyval2d`, `polyval3d`, `polygrid2d`, `polygrid3d`, `polyroots`, `polyfromroots`\n\n\n* Ajuste: `polyfit`, `polyvander`, `polyvander2d`, `polyvander3d`\n\n\n* Cálculo: `polyder`, `polyint`\n\n\n* Álgebra: `polyadd`, `polysub`, `polymul`, `polymulx`, `polydiv`, `polypow`\n\n\n* Miscelánea: `polycompanion`, `polydomain`, `polyzero`, `polyone`, `polyx`, `polytrim`, `polyline`\n\n\n* Otras funciones polinómicas: `Chebyshev`, `Legendre`, `Laguerre`, `Hermite`\n\n...",
"_____no_output_____"
]
],
[
[
"# juguemos un poco con ellas\n",
"_____no_output_____"
]
],
[
[
"## Álgebra lineal",
"_____no_output_____"
],
[
"Lo siguiente que se encuentra dentro de `numpy.linalg` vendrá precedido por `LA`.\n\n* Productos para vectores y matrices: `dot`, `vdot`, `inner`, `outer`, `matmul`, `tensordot`, `einsum`, `LA.matrix_power`, `kron`\n\n\n* Descomposiciones: `LA.cholesky`, `LA.qr`, `LA.svd`\n\n\n* Eigenvalores: `LA.eig`, `LA.eigh`, `LA.eigvals`, `LA.eigvalsh`\n\n\n* Normas y otros números: `LA.norm`, `LA.cond`, `LA.det`, `LA.matrix_rank`, `LA.slogdet`, `trace`\n\n\n* Resolución de ecuaciones e inversión de matrices: `LA.solve`, `LA.tensorsolve`, `LA.lstsq`, `LA.inv`, `LA.pinv`, `LA.tensorinv`\n\n\nDentro de `scipy` tenemos más cosas relacionadas.",
"_____no_output_____"
]
],
[
[
"# juguemos un poco con ellas\n",
"_____no_output_____"
]
],
[
[
"## Manipulación de `ndarrays`",
"_____no_output_____"
],
[
"`tile`, `hstack`, `vstack`, `dstack`, `hsplit`, `vsplit`, `dsplit`, `repeat`, `reshape`, `ravel`, `resize`,...",
"_____no_output_____"
]
],
[
[
"# juguemos un poco con ellas\n",
"_____no_output_____"
]
],
[
[
"## Módulos de interés dentro de `numpy`",
"_____no_output_____"
],
[
"Dentro de `numpy` podemos encontrar módulos para:\n\n* Usar números aleatorios: `np.random`\n\n\n* Usar FFT: `np.fft`\n\n\n* Usar *masked arrays*: `np.ma`\n\n\n* Usar polinomios: `np.polynomial`\n\n\n* Usar álgebra lineal: `np.linalg`\n\n\n* Usar matrices: `np.matlib`\n\n\n* ...\n\nToda esta funcionalidad se puede ampliar y mejorar usando `scipy`.",
"_____no_output_____"
],
[
"## Cálculo matricial",
"_____no_output_____"
]
],
[
[
"a1 = np.repeat(2, 9).reshape(3, 3)\na2 = np.tile(2, (3, 3))\na3 = np.ones((3, 3), dtype=np.int) * 2\nprint(a1)\nprint(a2)\nprint(a3)",
"_____no_output_____"
],
[
"b = np.arange(1,4)\nprint(b)",
"_____no_output_____"
],
[
"print(a1.dot(b))\nprint(np.dot(a2, b))\nprint(a3 @ b) # only python version >= 3.5",
"_____no_output_____"
]
],
[
[
"Lo anterior lo hemos hecho usando *ndarrays* pero `numpy` también ofrece una estructura de datos `matrix`.",
"_____no_output_____"
]
],
[
[
"a_mat = np.matrix(a1)\na_mat",
"_____no_output_____"
],
[
"b_mat = np.matrix(b)",
"_____no_output_____"
],
[
"a_mat @ b_mat",
"_____no_output_____"
],
[
"a_mat @ b_mat.T",
"_____no_output_____"
]
],
[
[
"Como vemos, con los *ndarrays* no hace falta que seamos rigurosos con las dimensiones, en cambio, si usamos `np.matrix` como tipos hemos de realizar operaciones matriciales válidas (por ejemplo, que las dimensiones sean correctas).",
"_____no_output_____"
],
[
"A efectos prácticos, en general, los *ndarrays* se pueden usar como `matrix` conociendo estas pequeñas cosas.",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
]
] |
cb8ac962a3f50ad3cbaf31985d11d706056a33e6
| 125,418 |
ipynb
|
Jupyter Notebook
|
notebooks/OOI_demo.ipynb
|
SvenGastauer/echopype
|
a7c32f4aa7d567e1dc441d99822db104d7ba1e84
|
[
"Apache-2.0"
] | 1 |
2021-09-04T07:19:35.000Z
|
2021-09-04T07:19:35.000Z
|
notebooks/OOI_demo.ipynb
|
SvenGastauer/echopype
|
a7c32f4aa7d567e1dc441d99822db104d7ba1e84
|
[
"Apache-2.0"
] | null | null | null |
notebooks/OOI_demo.ipynb
|
SvenGastauer/echopype
|
a7c32f4aa7d567e1dc441d99822db104d7ba1e84
|
[
"Apache-2.0"
] | null | null | null | 216.611399 | 79,148 | 0.922156 |
[
[
[
"### Processing Echosounder Data from Ocean Observatories Initiative with `echopype`.",
"_____no_output_____"
],
[
"Downloading a file from the OOI website. We pick August 21, 2017 since this was the day of the solar eclipse which affected the traditional patterns of the marine life.",
"_____no_output_____"
]
],
[
[
"# downloading the file\n!wget https://rawdata.oceanobservatories.org/files/CE04OSPS/PC01B/ZPLSCB102_10.33.10.143/OOI-D20170821-T163049.raw ",
"--2019-02-18 10:54:47-- https://rawdata.oceanobservatories.org/files/CE04OSPS/PC01B/ZPLSCB102_10.33.10.143/OOI-D20170821-T163049.raw\nResolving rawdata.oceanobservatories.org (rawdata.oceanobservatories.org)... 128.6.240.153\nConnecting to rawdata.oceanobservatories.org (rawdata.oceanobservatories.org)|128.6.240.153|:443... connected.\nHTTP request sent, awaiting response... 200 OK\nLength: 52431936 (50M) [text/plain]\nSaving to: ‘OOI-D20170821-T163049.raw’\n\nOOI-D20170821-T1630 100%[===================>] 50.00M 19.5MB/s in 2.6s \n\n2019-02-18 10:54:50 (19.5 MB/s) - ‘OOI-D20170821-T163049.raw’ saved [52431936/52431936]\n\n"
],
[
"filename = 'OOI-D20170821-T163049.raw'",
"_____no_output_____"
]
],
[
[
"**Converting from Raw to Standartized Netcdf Format**",
"_____no_output_____"
]
],
[
[
"# import as part of a submodule\nfrom echopype.convert import ConvertEK60\ndata_tmp = ConvertEK60(filename)\ndata_tmp.raw2nc()\nos.remove(filename)",
"10:55:03 converting file: OOI-D20170821-T163049.raw\n"
]
],
[
[
"**Calibrating, Denoising, Mean Volume Backscatter Strength**",
"_____no_output_____"
]
],
[
[
"from echopype.model import EchoData\ndata = EchoData(filename[:-4]+'.nc')\ndata.calibrate() # Calibration and echo-integration\ndata.remove_noise(save=True) # Save denoised Sv to FILENAME_Sv_clean.nc\ndata.get_MVBS(save=True)",
"10:55:08 saving calibrated Sv to OOI-D20170821-T163049_Sv.nc\n10:55:19 saving denoised Sv to OOI-D20170821-T163049_Sv_clean.nc\n10:55:20 saving MVBS to OOI-D20170821-T163049_MVBS.nc\n"
]
],
[
[
"**Visualizing the Result**",
"_____no_output_____"
]
],
[
[
"%matplotlib inline",
"_____no_output_____"
],
[
"data.MVBS.MVBS.sel(frequency=200000).plot(x='ping_time',cmap = 'jet')",
"_____no_output_____"
]
],
[
[
"**Processing Multiple Files**",
"_____no_output_____"
],
[
"To process multiple file from the OOI website we need to scrape the names of the existing files there. We will use the `Beautiful Soup`\npackage for that. ",
"_____no_output_____"
]
],
[
[
"!conda install --yes beautifulsoup4",
"_____no_output_____"
],
[
"from bs4 import BeautifulSoup\nfrom urllib.request import urlopen",
"_____no_output_____"
],
[
"path = 'https://rawdata.oceanobservatories.org/files/CE04OSPS/PC01B/ZPLSCB102_10.33.10.143/'",
"_____no_output_____"
],
[
"response = urlopen(path)\nsoup = BeautifulSoup(response.read(), \"html.parser\")",
"_____no_output_____"
],
[
"# urls = []\n# for item in soup.find_all(text=True):\n# if '.raw' in item:\n# urls.append(path+'/'+item) ",
"_____no_output_____"
],
[
"urls = [path+'/'+item for item in soup.find_all(text=True) if '.raw' in item]",
"_____no_output_____"
],
[
"# urls",
"_____no_output_____"
],
[
"from datetime import datetime",
"_____no_output_____"
]
],
[
[
"Specify range:",
"_____no_output_____"
]
],
[
[
"start_time = '20170821-T000000'\nend_time = '20170822-T235959'",
"_____no_output_____"
],
[
"# convert the times to datetime format\nstart_datetime = datetime.strptime(start_time,'%Y%m%d-T%H%M%S')\nend_datetime = datetime.strptime(end_time,'%Y%m%d-T%H%M%S')",
"_____no_output_____"
],
[
"# function to check if a date is in the date range\ndef in_range(date_str, start_time, end_time):\n date_str = datetime.strptime(date_str,'%Y%m%d-T%H%M%S')\n true = date_str >= start_datetime and date_str <= end_datetime\n return(true)\n ",
"_____no_output_____"
],
[
"# identify the list of urls in range\nrange_urls = []\nfor url in urls: \n date_str = url[-20:-4]\n if in_range(date_str, start_time, end_time):\n range_urls.append(url)\n ",
"_____no_output_____"
],
[
"range_urls",
"_____no_output_____"
],
[
"rawnames = [url.split('//')[-1] for url in range_urls]",
"_____no_output_____"
],
[
"ls",
"_____no_output_____"
],
[
"import os",
"_____no_output_____"
]
],
[
[
"**Downloading the Files**",
"_____no_output_____"
]
],
[
[
"# Download the files\nimport requests\nrawnames = []\nfor url in range_urls:\n r = requests.get(url, allow_redirects=True)\n rawnames.append(url.split('//')[-1])\n open(url.split('//')[-1], 'wb').write(r.content)",
"_____no_output_____"
],
[
"!pip install echopype\n ",
"Requirement already satisfied: echopype in /srv/conda/lib/python3.6/site-packages (0.1.2)\r\nRequirement already satisfied: click in /srv/conda/lib/python3.6/site-packages (from echopype) (7.0)\r\n"
],
[
"ls",
"_____no_output_____"
]
],
[
[
"**Converting from Raw to Standartized Netcdf Format**",
"_____no_output_____"
]
],
[
[
"# import as part of a submodule\nfrom echopype.convert import ConvertEK60\nfor filename in rawnames:\n data_tmp = ConvertEK60(filename)\n data_tmp.raw2nc()\n os.remove(filename)",
"_____no_output_____"
],
[
"#ls",
"_____no_output_____"
]
],
[
[
"**Calibrating, Denoising, Mean Volume Backscatter Strength**",
"_____no_output_____"
]
],
[
[
"# calibrate and denoise\nfrom echopype.model import EchoData\n\nfor filename in rawnames:\n\n data = EchoData(filename[:-4]+'.nc')\n data.calibrate() # Calibration and echo-integration\n data.remove_noise(save=False) # Save denoised Sv to FILENAME_Sv_clean.nc\n data.get_MVBS(save=True)\n os.remove(filename[:-4]+'.nc')\n os.remove(filename[:-4]+'_Sv.nc')",
"_____no_output_____"
]
],
[
[
"**Opening and Visualizing the Results in Parallel**",
"_____no_output_____"
],
[
"No that all files are in an appropriate format, we can open them and visualize them in parallel. For that we will need to install the `dask` parallelization library.",
"_____no_output_____"
]
],
[
[
"!conda install --yes dask",
"Solving environment: done\n\n\n==> WARNING: A newer version of conda exists. <==\n current version: 4.5.11\n latest version: 4.6.4\n\nPlease update conda by running\n\n $ conda update -n base conda\n\n\n\n# All requested packages already installed.\n\n"
],
[
"import xarray as xr",
"_____no_output_____"
],
[
"res = xr.open_mfdataset('*MVBS.nc')",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"plt.figure(figsize = (15,5))\nres.MVBS.sel(frequency=200000).plot(x='ping_time',cmap = 'jet')",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
cb8acbfc8244bf4f6d612dcad41d6ae9da569be2
| 1,040 |
ipynb
|
Jupyter Notebook
|
index.ipynb
|
toson77/binder_octave_signal
|
0bf61ba6e32fe3f967168614a25315669e720979
|
[
"BSD-3-Clause"
] | null | null | null |
index.ipynb
|
toson77/binder_octave_signal
|
0bf61ba6e32fe3f967168614a25315669e720979
|
[
"BSD-3-Clause"
] | null | null | null |
index.ipynb
|
toson77/binder_octave_signal
|
0bf61ba6e32fe3f967168614a25315669e720979
|
[
"BSD-3-Clause"
] | null | null | null | 23.111111 | 273 | 0.572115 |
[
[
[
"empty"
]
]
] |
[
"empty"
] |
[
[
"empty"
]
] |
cb8ae5e2dd622706236b42a960bbaf92da1d342d
| 320,123 |
ipynb
|
Jupyter Notebook
|
_notebooks/2021-07-01-book-crossing-surprise-svd-nmf.ipynb
|
recohut/notebook
|
610670666a1c3d8ef430d42f712ff72ecdbd8f86
|
[
"Apache-2.0"
] | null | null | null |
_notebooks/2021-07-01-book-crossing-surprise-svd-nmf.ipynb
|
recohut/notebook
|
610670666a1c3d8ef430d42f712ff72ecdbd8f86
|
[
"Apache-2.0"
] | 1 |
2022-01-12T05:40:57.000Z
|
2022-01-12T05:40:57.000Z
|
_notebooks/2021-07-01-book-crossing-surprise-svd-nmf.ipynb
|
recohut/notebook
|
610670666a1c3d8ef430d42f712ff72ecdbd8f86
|
[
"Apache-2.0"
] | 1 |
2021-08-13T19:00:26.000Z
|
2021-08-13T19:00:26.000Z
| 60.083146 | 47,486 | 0.643821 |
[
[
[
"# Book-Crossing Recommendation System\n> Book recommender system on book crossing dataset using surprise SVD and NMF models\n\n- toc: true\n- badges: true\n- comments: true\n- categories: [Surprise, SVD, NMF, Book]\n- author: \"<a href='https://github.com/tttgm/fellowshipai'>Tom McKenzie</a>\"\n- image:",
"_____no_output_____"
],
[
"## Setup",
"_____no_output_____"
]
],
[
[
"!pip install -q git+https://github.com/sparsh-ai/recochef.git",
" Installing build dependencies ... \u001b[?25l\u001b[?25hdone\n Getting requirements to build wheel ... \u001b[?25l\u001b[?25hdone\n Preparing wheel metadata ... \u001b[?25l\u001b[?25hdone\n\u001b[K |████████████████████████████████| 4.3MB 5.4MB/s \n\u001b[?25h Building wheel for recochef (PEP 517) ... \u001b[?25l\u001b[?25hdone\n"
],
[
"import pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport pickle\n\nfrom sklearn import model_selection\n\nimport warnings\nwarnings.filterwarnings('ignore')\n\nplt.style.use('seaborn-white')\nplt.rcParams.update({'font.size': 15})\n%matplotlib inline",
"_____no_output_____"
],
[
"from recochef.datasets.bookcrossing import BookCrossing",
"_____no_output_____"
]
],
[
[
"## Load the dataset",
"_____no_output_____"
]
],
[
[
"bookcrossing = BookCrossing()\nusers = bookcrossing.load_users()\nbooks = bookcrossing.load_items()\nbook_ratings = bookcrossing.load_interactions()",
"_____no_output_____"
],
[
"users.head()",
"_____no_output_____"
],
[
"books.head()",
"_____no_output_____"
],
[
"book_ratings.head()",
"_____no_output_____"
],
[
"print(f'Users: {len(users)}\\nBooks: {len(books)}\\nRatings: {len(book_ratings)}')",
"Users: 278858\nBooks: 271360\nRatings: 1149780\n"
]
],
[
[
"## EDA and Data cleaning",
"_____no_output_____"
],
[
"### Users",
"_____no_output_____"
]
],
[
[
"users.describe(include='all').T",
"_____no_output_____"
]
],
[
[
"The age range goes from 0 to 244 years old! Obviously this cannot be correct; I'll set all ages less than 5 and older than 100 to NaN to try keep them realistic.",
"_____no_output_____"
]
],
[
[
"users.loc[(users.AGE<5) | (users.AGE>100), 'AGE'] = np.nan",
"_____no_output_____"
],
[
"u = users.AGE.value_counts().sort_index()\nplt.figure(figsize=(20, 10))\nplt.bar(u.index, u.values)\nplt.xlabel('Age')\nplt.ylabel('counts')\nplt.show()",
"_____no_output_____"
]
],
[
[
"Next, can we expand the 'Location' field to break it up into 'City', 'State', and 'Country'.",
"_____no_output_____"
],
[
"> Note: Used Pandas Series.str.split method as it has an 'expand' parameter which can handle None cases",
"_____no_output_____"
]
],
[
[
"user_location_expanded = users.LOCATION.str.split(',', 2, expand=True)\nuser_location_expanded.columns = ['CITY', 'STATE', 'COUNTRY']\nusers = users.join(user_location_expanded)\nusers.COUNTRY.replace('', np.nan, inplace=True)\nusers.drop(columns=['LOCATION'], inplace=True)\nusers.head()",
"_____no_output_____"
]
],
[
[
"### Books",
"_____no_output_____"
]
],
[
[
"books.head(2)",
"_____no_output_____"
],
[
"books.describe(include='all').T",
"_____no_output_____"
],
[
"# Convert years to float\nbooks.YEAR = pd.to_numeric(books.YEAR, errors='coerce')",
"_____no_output_____"
],
[
"# Replace all years of zero with NaN\nbooks.YEAR.replace(0, np.nan, inplace=True)",
"_____no_output_____"
],
[
"yr = books.YEAR.value_counts().sort_index()\nyr = yr.where(yr>5) # filter out counts less than 5\nplt.figure(figsize=(20, 10))\nplt.bar(yr.index, yr.values)\nplt.xlabel('Year of Publication')\nplt.ylabel('counts')\nplt.show()",
"_____no_output_____"
]
],
[
[
"Note that in the plot above we filtered out counts less than 5, as there are a few books in the dataset with publication years in the 1300s, and a few in the future (?!). The plot above show the general trend that more recent books are much more frequent.\n\nLet's take a look at some of those 'outlier' books. Maybe we'll even keep them as a separate dataset so we can filter them out if we need to later in the analysis. We'll leave them in for now, and then figure out how to handle them once we have more info later on.",
"_____no_output_____"
]
],
[
[
"historical_books = books[books.YEAR<1900] # create df of old books\nbooks_from_the_future = books[books.YEAR>2018] # create df of books with publication yrs in the future!\n\nhist_books_mini = historical_books[['TITLE', 'YEAR']]\nfuture_books_mini = books_from_the_future[['TITLE', 'YEAR']]\nprint(f'Historical books:\\n{hist_books_mini}')\nprint('\\n')\nprint(f'Future books:\\n{future_books_mini}')",
"Historical books:\n TITLE YEAR\n171817 Complete Works 10 Volumes [2,6,7,8,9] (Notable... 1806.0\n227531 Dalan-i bihisht (Dastan-i Irani) 1378.0\n245607 Hugh Wynne, Free Quaker (2 Volumes (BCL1-PS Am... 1897.0\n253750 Tasht-i khun 1376.0\n\n\nFuture books:\n TITLE YEAR\n30010 The Royals 2020.0\n37487 MY TEACHER FRIED MY BRAINS (RACK SIZE) (MY TEA... 2030.0\n55676 MY TEACHER FLUNKED THE PLANET (RACK SIZE) (MY ... 2030.0\n78168 Crossing America 2030.0\n80264 Alice's Adventures in Wonderland and Through t... 2050.0\n92156 Edgar Allen Poe Collected Poems 2020.0\n97826 Outline of European Architecture (Pelican S.) 2050.0\n116053 Three Plays of Eugene Oneill 2038.0\n118294 Das gro��e B�¶se- M�¤dchen- Lesebuch. 2026.0\n183769 Heidi 2021.0\n192993 Field Guide to the Birds of North America, 3rd... 2030.0\n228173 FOREST PEOPLE (Touchstone Books (Hardcover)) 2030.0\n238817 LOOK HOMEWARD ANGEL 2020.0\n240169 In Our Time: Stories (Scribner Classic) 2030.0\n246842 CLOUT 2024.0\n255409 To Have and Have Not 2037.0\n260974 FOOTBALL SUPER TEAMS : FOOTBALL SUPER TEAMS 2030.0\n"
]
],
[
[
"I think we can probably omit the 'historical_books' as they may potentially skew the model and do not seem to have much relevance to the wider userbase.\n\nSome of the 'future' books actually appear to be errors (e.g. Alice in Wonderland, Edgar Allen Poe, etc.)... Perhaps they were supposed to be e.g. 1950 instead of 2050? However, instead of investigating this further, since there are <20 books here I will simply remove them from the 'books' table.",
"_____no_output_____"
]
],
[
[
"print(f'Length of books dataset before removal: {len(books)}')\nbooks = books.loc[~(books.ITEMID.isin(historical_books.ITEMID))] # remove historical books\nbooks = books.loc[~(books.ITEMID.isin(books_from_the_future.ITEMID))] # remove historical books\nprint(f'Length of books dataset after removal: {len(books)}')",
"Length of books dataset before removal: 271360\nLength of books dataset after removal: 271339\n"
]
],
[
[
"We clean up the ampersand formatting in the Publisher field.",
"_____no_output_____"
]
],
[
[
"books.PUBLISHER = books.PUBLISHER.str.replace('&', '&', regex=False)\nbooks.head()",
"_____no_output_____"
]
],
[
[
"Check that there are no duplicated book entries.",
"_____no_output_____"
]
],
[
[
"uniq_books = books.ITEMID.nunique()\nall_books = books.ITEMID.count()\nprint(f'No. of unique books: {uniq_books} | All book entries: {all_books}')",
"No. of unique books: 271339 | All book entries: 271339\n"
]
],
[
[
"Let's look at the most frequent Publishing houses in the dataset.",
"_____no_output_____"
]
],
[
[
"top_publishers = books.PUBLISHER.value_counts()[:10]\nprint(f'The 10 publishers with the most entries in the books table are:\\n{top_publishers}')",
"The 10 publishers with the most entries in the books table are:\nHarlequin 7535\nSilhouette 4220\nPocket 3905\nBallantine Books 3783\nBantam Books 3646\nScholastic 3159\nSimon &; Schuster 2969\nPenguin Books 2844\nBerkley Publishing Group 2771\nWarner Books 2727\nName: PUBLISHER, dtype: int64\n"
]
],
[
[
"What about authors with the most entries?",
"_____no_output_____"
]
],
[
[
"top_authors = books.AUTHOR.value_counts()[:10]\nprint(f'The 10 authors with the most entries in the books table are:\\n{top_authors}')",
"The 10 authors with the most entries in the books table are:\nAgatha Christie 632\nWilliam Shakespeare 567\nStephen King 524\nAnn M. Martin 423\nCarolyn Keene 373\nFrancine Pascal 372\nIsaac Asimov 330\nNora Roberts 315\nBarbara Cartland 307\nCharles Dickens 302\nName: AUTHOR, dtype: int64\n"
]
],
[
[
"We should search for empty or NaN values in these fields too.",
"_____no_output_____"
]
],
[
[
"empty_string_publisher = books[books.PUBLISHER == ''].PUBLISHER.count()\nnan_publisher = books.PUBLISHER.isnull().sum()\nprint(f'There are {empty_string_publisher} entries with empty strings, and {nan_publisher} NaN entries in the Publisher field')",
"There are 0 entries with empty strings, and 2 NaN entries in the Publisher field\n"
]
],
[
[
"Great - no empty strings in the Publisher field, and only 2 NaNs.",
"_____no_output_____"
]
],
[
[
"empty_string_author = books[books.AUTHOR == ''].AUTHOR.count()\nnan_author = books.AUTHOR.isnull().sum()\nprint(f'There are {empty_string_author} entries with empty strings, and {nan_author} NaN entries in the Author field')",
"There are 0 entries with empty strings, and 1 NaN entries in the Author field\n"
]
],
[
[
"Cool, only 1 NaN in the Author field.\n\nLet's look at the titles.",
"_____no_output_____"
]
],
[
[
"top_titles = books.TITLE.value_counts()[:10]\nprint(f'The 10 book titles with the most entries in the books table are:\\n{top_titles}')",
"The 10 book titles with the most entries in the books table are:\nSelected Poems 27\nLittle Women 24\nWuthering Heights 21\nThe Secret Garden 20\nAdventures of Huckleberry Finn 20\nDracula 20\nJane Eyre 19\nThe Night Before Christmas 18\nPride and Prejudice 18\nGreat Expectations 17\nName: TITLE, dtype: int64\n"
]
],
[
[
"This is actually quite an important observation. Although all of the ISBN entries are *unique* in the 'books' dataframe, different *forms* of the **same** book will have different ISBNs - i.e. paperback, e-book, etc. Therefore, we can see that some books have multiple ISBN entries (e.g. Jane Eyre has 19 different ISBNs, each corresponding to a different version of the book).\n\nLet's take a look at, for example, the entries for 'Jane Eyre'.",
"_____no_output_____"
]
],
[
[
"books[books.TITLE=='Jane Eyre']",
"_____no_output_____"
]
],
[
[
"It looks like each ISBN assigned to the book 'Jane Eyre' has different Publisher and Year of Publication values also.\n\nIt might be more useful for our model if we simplified this to give each book a *unique* identifier, independent of the book format, as our recommendations will be for a book, not a specific version of a book. Therefore, all values in the Jane Eyre example above would stay the same, except all of the Jane Eyre entries would additionally be assigned a *unique ISBN* code as a new field.\n\n**Will create this more unique identifier under the field name 'UNIQUE_ITEMIDS'. Note that entries with only a single ISBN number will be left the same. However, will need to do this after joining to the other tables in the dataset, as some ISBNs in the 'book-rating' table may be removed if done prior.**",
"_____no_output_____"
],
[
"### Interactions",
"_____no_output_____"
]
],
[
[
"book_ratings.head()",
"_____no_output_____"
],
[
"book_ratings.describe(include='all').T",
"_____no_output_____"
],
[
"book_ratings.dtypes",
"_____no_output_____"
]
],
[
[
"The data types already look good. Remember that the ISBN numbers may contain letters, and so should be left as strings.\n\nWhich users contribute the most ratings?",
"_____no_output_____"
]
],
[
[
"super_users = book_ratings.groupby('USERID').ITEMID.count().sort_values(ascending=False)\nprint(f'The 20 users with the most ratings:\\n{super_users[:20]}')",
"The 20 users with the most ratings:\nUSERID\n11676 13602\n198711 7550\n153662 6109\n98391 5891\n35859 5850\n212898 4785\n278418 4533\n76352 3367\n110973 3100\n235105 3067\n230522 2991\n16795 2948\n234623 2674\n36836 2529\n52584 2512\n245963 2507\n204864 2504\n55492 2459\n185233 2448\n171118 2421\nName: ITEMID, dtype: int64\n"
]
],
[
[
"Wow! User \\#11676 has almost twice as many ratings as the next highest user! All of the top 20 users have thousands of ratings, which seems like a lot, although maybe I'm just a slow reader...\n\nLet's see how they are distributed.",
"_____no_output_____"
]
],
[
[
"# user distribution - users with more than 50 ratings removed\nuser_hist = super_users.where(super_users<50)\nuser_hist.hist(bins=30)\nplt.xlabel('No. of ratings')\nplt.ylabel('count')\nplt.show()",
"_____no_output_____"
]
],
[
[
"It looks like **_by far_** the most frequent events are users with only 1 or 2 rating entries. We can see that the 'super users' with thousands of ratings are significant outliers.\n\nThis becomes clear if we make the same histogram with a cutoff for users with a minimum of 1000 ratings.",
"_____no_output_____"
]
],
[
[
"# only users with more than 1000 ratings\nsuper_user_hist = super_users.where(super_users>1000)\nsuper_user_hist.hist(bins=30)\nplt.xlabel('No. of ratings (min. 1000)')\nplt.ylabel('count')\nplt.show()",
"_____no_output_____"
]
],
[
[
"Let's see what the distribution of **ratings** looks like.",
"_____no_output_____"
]
],
[
[
"rtg = book_ratings.RATING.value_counts().sort_index()\n\nplt.figure(figsize=(10, 5))\nplt.bar(rtg.index, rtg.values)\nplt.xlabel('Rating')\nplt.ylabel('counts')\nplt.show()",
"_____no_output_____"
]
],
[
[
"Seems like most of the entries have a rating of zero!\n\nAfter doing some research on the internet regarding this (and similar) datasets, it appears that the rating scale is actually from 1 to 10, and a 0 indicates an 'implicit' rather than an 'explicit' rating. An implicit rating represents an interaction (may be positive or negative) between the user and the item. Implicit interactions usually need to be handled differently from explicit ones.\n\nFor the modeling step we'll only be looking at *explicit* ratings, and so the 0 rating entry rows will be removed.",
"_____no_output_____"
]
],
[
[
"print(f'Size of book_ratings before removing zero ratings: {len(book_ratings)}')\nbook_ratings = book_ratings[book_ratings.RATING != 0]\nprint(f'Size of book_ratings after removing zero ratings: {len(book_ratings)}')",
"Size of book_ratings before removing zero ratings: 1149780\nSize of book_ratings after removing zero ratings: 433671\n"
]
],
[
[
"By removing the implicit ratings we have reduced our sample size by more than half.\n\nLet's look at how the ratings are distributed again.",
"_____no_output_____"
]
],
[
[
"rtg = book_ratings.RATING.value_counts().sort_index()\n\nplt.figure(figsize=(10, 5))\nplt.bar(rtg.index, rtg.values)\nplt.xlabel('Rating')\nplt.ylabel('counts')\nplt.show()",
"_____no_output_____"
]
],
[
[
"This is much more clear! Now we can see that 8 is the most frequent rating, while users tend to give ratings > 5, with very few low ratings given.",
"_____no_output_____"
],
[
"### Merge",
"_____no_output_____"
],
[
"First, we'll join the 'books' table to the 'book_ratings' table on the ISBN field.",
"_____no_output_____"
]
],
[
[
"print(f'Books table size: {len(books)}')\nprint(f'Ratings table size: {len(book_ratings)}')\nbooks_with_ratings = book_ratings.join(books.set_index('ITEMID'), on='ITEMID')\nprint(f'New table size: {len(books_with_ratings)}')",
"Books table size: 271339\nRatings table size: 433671\nNew table size: 433671\n"
]
],
[
[
"Let's take a look at the new table.",
"_____no_output_____"
]
],
[
[
"books_with_ratings.head()",
"_____no_output_____"
],
[
"print(f'There are {books_with_ratings.TITLE.isnull().sum()} books with no title/author information.')\nprint(f'This represents {len(books_with_ratings)/books_with_ratings.TITLE.isnull().sum():.2f}% of the ratings dataset.')",
"There are 49861 books with no title/author information.\nThis represents 8.70% of the ratings dataset.\n"
]
],
[
[
"There seems to be quite a few ISBNs in the ratings table that did not match an ISBN in the books table, almost 9% of all entries!\n\nThere isn't really anything we can do about that, but we should really remove them from the dataset as we won't be able to access the title of the book to make a recommendation even if the model can use them.",
"_____no_output_____"
]
],
[
[
"books_with_ratings.info()",
"<class 'pandas.core.frame.DataFrame'>\nInt64Index: 433671 entries, 1 to 1149779\nData columns (total 10 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 USERID 433671 non-null int64 \n 1 ITEMID 433671 non-null object \n 2 RATING 433671 non-null int64 \n 3 TITLE 383810 non-null object \n 4 AUTHOR 383809 non-null object \n 5 YEAR 378026 non-null float64\n 6 PUBLISHER 383808 non-null object \n 7 URLSMALL 383810 non-null object \n 8 URLMEDIUM 383810 non-null object \n 9 URLLARGE 383809 non-null object \ndtypes: float64(1), int64(2), object(7)\nmemory usage: 36.4+ MB\n"
]
],
[
[
"It looks like the ```year_of_publication``` field contains the most NaN entries, while ```USERID```, ```isbn```, and ```book_rating``` are full. The ```book_title```, ```book_author```, and ```publisher``` fields contain approximately the same number of missing entries.\n\nWe'll choose to remove rows for which the ```book_title``` is empty, as this is the most crucial piece of data needed to identify the book.",
"_____no_output_____"
]
],
[
[
"books_with_ratings.dropna(subset=['TITLE'], inplace=True) # remove rows with missing title/author data",
"_____no_output_____"
]
],
[
[
"Let's see which books have the highest **cumulative** book rating values.",
"_____no_output_____"
]
],
[
[
"cm_rtg = books_with_ratings.groupby('TITLE').RATING.sum()\ncm_rtg = cm_rtg.sort_values(ascending=False)[:10]\nidx = cm_rtg.index.tolist() # Get sorted book titles\nvals = cm_rtg.values.tolist() # Get corresponding cm_rtg values\n\nplt.figure(figsize=(10, 5))\nplt.bar(range(len(idx)), vals)\nplt.xticks(range(len(idx)), idx, rotation='vertical')\nplt.ylabel('cumulative rating score')\nplt.show()",
"_____no_output_____"
]
],
[
[
"This seems about right as it combines the total number of ratings with the score given, so these are all really popular book titles.\n\nWhat about the highest **average ratings** (with a minimum of at least 50 ratings recieved)?",
"_____no_output_____"
]
],
[
[
"cutoff = books_with_ratings.TITLE.value_counts()\nmean_rtg = books_with_ratings[books_with_ratings.TITLE.isin(cutoff[cutoff>50].index)].groupby('TITLE')['RATING'].mean()\nmean_rtg.sort_values(ascending=False)[:10] # show only top 10",
"_____no_output_____"
]
],
[
[
"This looks perfectly reasonable. The Harry Potter and Lord of the Rings books rate extremely highly, as expected.\n\nHow about the **lowest-rated** books?",
"_____no_output_____"
]
],
[
[
"mean_rtg.sort_values(ascending=False)[-10:] # bottom 10 only",
"_____no_output_____"
]
],
[
[
"Seems like the *lowest average* rating in the dataset is only a 4.39 - and all the rest of the books have average ratings higher than 5.\n\nI haven't heard of any of these books, so I can't really comment on if they seem correct here.\n\n**Now I'd like to tackle the challenge of the same book potentially having multiple ISBN numbers (for the different formats it is available in). We should clean that up here before we add the 'user' table.**",
"_____no_output_____"
],
[
"### Single ISBN per book\nRestrict books to a \"single ISBN per book\" (regardless of format)",
"_____no_output_____"
],
[
"Let's look again at the book titles which have the most associated ISBN numbers.",
"_____no_output_____"
]
],
[
[
"books_with_ratings.groupby('TITLE').ITEMID.nunique().sort_values(ascending=False)[:10]",
"_____no_output_____"
],
[
"multiple_isbns = books_with_ratings.groupby('TITLE').ITEMID.nunique()\nmultiple_isbns.value_counts()",
"_____no_output_____"
]
],
[
[
"We can see that the vast majority of books have less only 1 associated ISBN number, however quite a few multiple ISBNs. We want to create a ```UNIQUE_ITEMIDS``` such that a single book will only have 1 identifier when fed to the recommendation model.",
"_____no_output_____"
]
],
[
[
"has_mult_isbns = multiple_isbns.where(multiple_isbns>1)\nhas_mult_isbns.dropna(inplace=True) # remove NaNs, which in this case is books with a single ISBN number",
"_____no_output_____"
],
[
"print(f'There are {len(has_mult_isbns)} book titles with multiple ISBN numbers which we will try to re-assign to a unique identifier')",
"There are 10231 book titles with multiple ISBN numbers which we will try to re-assign to a unique identifier\n"
],
[
"# Check to see that our friend Jane Eyre still has multiple ISBN values\nhas_mult_isbns['Jane Eyre']",
"_____no_output_____"
]
],
[
[
"**Note:** Created the dictionary below and pickled it, just need to load it again (or run it if the first time on a new system).",
"_____no_output_____"
]
],
[
[
"# Create dictionary for books with multiple isbns\ndef make_isbn_dict(df):\n title_isbn_dict = {}\n for title in has_mult_isbns.index:\n isbn_series = df.loc[df.TITLE==title].ITEMID.unique() # returns only the unique ISBNs\n title_isbn_dict[title] = isbn_series.tolist()\n return title_isbn_dict\n\n%time dict_UNIQUE_ITEMIDS = make_isbn_dict(books_with_ratings)\n\n# As the loop takes a while to run (8 min on the full dataset), pickle this dict for future use\nwith open('multiple_isbn_dict.pickle', 'wb') as handle:\n pickle.dump(dict_UNIQUE_ITEMIDS, handle, protocol=pickle.HIGHEST_PROTOCOL)\n\n# LOAD isbn_dict back into namespace\nwith open('multiple_isbn_dict.pickle', 'rb') as handle:\n multiple_isbn_dict = pickle.load(handle)",
"CPU times: user 8min 32s, sys: 1.61 s, total: 8min 34s\nWall time: 8min 33s\n"
],
[
"print(f'There are now {len(multiple_isbn_dict)} books in the ISBN dictionary that have multiple ISBN numbers')",
"There are now 10231 books in the ISBN dictionary that have multiple ISBN numbers\n"
]
],
[
[
"Let's take a quick look in the dict we just created for the 'Jane Eyre' entry - it should contain a list of 14 ISBN numbers.",
"_____no_output_____"
]
],
[
[
"print(f'Length of Jane Eyre dict entry: {len(multiple_isbn_dict[\"Jane Eyre\"])}\\n')\nmultiple_isbn_dict['Jane Eyre']",
"Length of Jane Eyre dict entry: 14\n\n"
]
],
[
[
"Looking good!\n\nAs I don't really know what each of the different ISBN numbers refers to (from what I understand the code actually signifies various things including publisher, year, type of print, etc, but decoding this is outside the scope of this analysis), I'll just select the **first** ISBN number that appears in the list of values to set as our ```UNIQUE_ITEMIDS``` for that particular book.\n\n_**Note**_: ISBN numbers are currently 13 digits long, but used to be 10. Any ISBN that isn't 10 or 13 digits long is probably an error that should be handled somehow. Any that are 9 digits long might actually be SBN numbers (pre-1970), and can be converted into ISBN's by just pre-fixing with a zero.",
"_____no_output_____"
]
],
[
[
"# Add 'UNIQUE_ITEMIDS' column to 'books_with_ratings' dataframe that includes the first ISBN if multiple ISBNS,\n# or just the ISBN if only 1 ISBN present anyway.\ndef add_UNIQUE_ITEMIDS_col(df):\n df['UNIQUE_ITEMIDS'] = df.apply(lambda row: multiple_isbn_dict[row.TITLE][0] if row.TITLE in multiple_isbn_dict.keys() else row.ITEMID, axis=1)\n return df\n\n%time books_with_ratings = add_UNIQUE_ITEMIDS_col(books_with_ratings)",
"CPU times: user 7.77 s, sys: 53.9 ms, total: 7.82 s\nWall time: 7.82 s\n"
],
[
"books_with_ratings.head()",
"_____no_output_____"
]
],
[
[
"The table now includes our ```UNIQUE_ITEMIDS``` field.\n\nLet's check to see if the 'Jane Eyre' entries have been assigned the ISBN '1590071212', which was the first val in the dictionary for this title.",
"_____no_output_____"
]
],
[
[
"books_with_ratings[books_with_ratings.TITLE=='Jane Eyre'].head()",
"_____no_output_____"
]
],
[
[
"Great! Seems to have worked well.\n\nWe won't replace the original ISBN column with the 'UNIQUE_ITEMIDS' column, but just note that the recommendation model should be based on the 'UNIQUE_ITEMIDS' field.",
"_____no_output_____"
],
[
"### Remove Small and Large book-cover URL columns",
"_____no_output_____"
]
],
[
[
"books_users_ratings.drop(['URLSMALL', 'URLLARGE'], axis=1, inplace=True)",
"_____no_output_____"
]
],
[
[
"## Join the 'users' table on the 'USERID' field",
"_____no_output_____"
]
],
[
[
"print(f'Books+Ratings table size: {len(books_with_ratings)}')\nprint(f'Users table size: {len(users)}')\nbooks_users_ratings = books_with_ratings.join(users.set_index('USERID'), on='USERID')\nprint(f'New \"books_users_ratings\" table size: {len(books_users_ratings)}')",
"Books+Ratings table size: 383810\nUsers table size: 278858\nNew \"books_users_ratings\" table size: 383810\n"
]
],
[
[
"Inspect the new table.",
"_____no_output_____"
]
],
[
[
"books_users_ratings.head()",
"_____no_output_____"
],
[
"books_users_ratings.info()",
"<class 'pandas.core.frame.DataFrame'>\nInt64Index: 383810 entries, 1 to 1149778\nData columns (total 13 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 USERID 383810 non-null int64 \n 1 ITEMID 383810 non-null object \n 2 RATING 383810 non-null int64 \n 3 TITLE 383810 non-null object \n 4 AUTHOR 383809 non-null object \n 5 YEAR 378026 non-null float64\n 6 PUBLISHER 383808 non-null object \n 7 URLMEDIUM 383810 non-null object \n 8 UNIQUE_ITEMIDS 383810 non-null object \n 9 AGE 267996 non-null float64\n 10 CITY 383810 non-null object \n 11 STATE 383810 non-null object \n 12 COUNTRY 373330 non-null object \ndtypes: float64(2), int64(2), object(9)\nmemory usage: 41.0+ MB\n"
]
],
[
[
"There are a few missing ```age```, ```year_of_publication```, ```publisher```, and ```country``` entries, but the primary fields of ```USERID```, ```UNIQUE_ITEMIDS```, and ```book_rating``` are all full, which is good.\n\nIn terms of the data types, ```USERID``` and ```book_rating``` are integers, while the ```UNIQUE_ITEMIDS``` are strings (which is expected as the ISBN numbers may also contain letters).",
"_____no_output_____"
]
],
[
[
"books_users_ratings.shape",
"_____no_output_____"
]
],
[
[
"## Recommender model\n\nCollaborative filtering use similarities of the 'user' and 'item' fields, with values of 'rating' predicted based on either user-item, or item-item similarity:\n - Item-Item CF: \"Users who liked this item also liked...\"\n - User-Item CF: \"Users who are similar to you also liked...\"\n \nIn both cases, we need to create a user-item matrix built from the entire dataset. We'll create a matrix for each of the training and testing sets, with the users as the rows, the books as the columns, and the rating as the matrix value. Note that this will be a very sparse matrix, as not every user will have watched every movie etc.\n\nWe'll first create a new dataframe that contains only the relevant columns (```USERID```, ```UNIQUE_ITEMIDS```, and ```book_rating```).",
"_____no_output_____"
]
],
[
[
"user_item_rating = books_users_ratings[['USERID', 'UNIQUE_ITEMIDS', 'RATING']]\nuser_item_rating.head()",
"_____no_output_____"
]
],
[
[
"We know what the distribution of ratings should look like (as we plotted it earlier) - let's plot it again on this new dataframe to just quickly check that it looks right.",
"_____no_output_____"
]
],
[
[
"rtg = user_item_rating.RATING.value_counts().sort_index()\n\nplt.figure(figsize=(10, 5))\nplt.bar(rtg.index, rtg.values)\nplt.xlabel('Rating')\nplt.ylabel('counts')\nplt.show()",
"_____no_output_____"
]
],
[
[
"Looks perfect! Continue.",
"_____no_output_____"
],
[
"### Using ```sklearn``` to generate training and testing subsets",
"_____no_output_____"
]
],
[
[
"train_data, test_data = model_selection.train_test_split(user_item_rating, test_size=0.20)",
"_____no_output_____"
],
[
"print(f'Training set size: {len(train_data)}')\nprint(f'Testing set size: {len(test_data)}')\nprint(f'Test set is {(len(test_data)/(len(train_data)+len(test_data))*100):.0f}% of the full dataset.')",
"Training set size: 307048\nTesting set size: 76762\nTest set is 20% of the full dataset.\n"
]
],
[
[
"### Map the ```USERID``` and ```UNIQUE_ITEMIDS``` fields to sequential integers for matrix processing",
"_____no_output_____"
]
],
[
[
"### TRAINING SET\n# Get int mapping for USERID\nu_unique_train = train_data.USERID.unique() # create a 'set' (i.e. all unique) list of vals\ntrain_data_user2idx = {o:i for i, o in enumerate(u_unique_train)}\n# Get int mapping for UNIQUE_ITEMIDS\nb_unique_train = train_data.UNIQUE_ITEMIDS.unique() # create a 'set' (i.e. all unique) list of vals\ntrain_data_book2idx = {o:i for i, o in enumerate(b_unique_train)}\n\n### TESTING SET\n# Get int mapping for USERID\nu_unique_test = test_data.USERID.unique() # create a 'set' (i.e. all unique) list of vals\ntest_data_user2idx = {o:i for i, o in enumerate(u_unique_test)}\n# Get int mapping for UNIQUE_ITEMIDS\nb_unique_test = test_data.UNIQUE_ITEMIDS.unique() # create a 'set' (i.e. all unique) list of vals\ntest_data_book2idx = {o:i for i, o in enumerate(b_unique_test)}",
"_____no_output_____"
],
[
"### TRAINING SET\ntrain_data['USER_UNIQUE'] = train_data['USERID'].map(train_data_user2idx)\ntrain_data['ITEM_UNIQUE'] = train_data['UNIQUE_ITEMIDS'].map(train_data_book2idx)\n\n### TESTING SET\ntest_data['USER_UNIQUE'] = test_data['USERID'].map(test_data_user2idx)\ntest_data['ITEM_UNIQUE'] = test_data['UNIQUE_ITEMIDS'].map(test_data_book2idx)\n\n### Convert back to 3-column df\ntrain_data = train_data[['USER_UNIQUE', 'ITEM_UNIQUE', 'RATING']]\ntest_data = test_data[['USER_UNIQUE', 'ITEM_UNIQUE', 'RATING']]",
"_____no_output_____"
],
[
"train_data.tail()",
"_____no_output_____"
],
[
"train_data.dtypes",
"_____no_output_____"
]
],
[
[
"This dataset is now ready to be processed via a collaborative filtering approach!\n\n**Note:** When we need to identify the user or book from the model we'll need to refer back to the ```train_data_user2idx``` and ```train_data_book2idx``` dictionaries to locate the ```USERID``` and ```UNIQUE_ITEMIDS```, respectively.",
"_____no_output_____"
]
],
[
[
"### TRAINING SET\n# Create user-item matrices\nn_users = train_data['USER_UNIQUE'].nunique()\nn_books = train_data['ITEM_UNIQUE'].nunique()\n\n# First, create an empty matrix of size USERS x BOOKS (this speeds up the later steps)\ntrain_matrix = np.zeros((n_users, n_books))\n\n# Then, add the appropriate vals to the matrix by extracting them from the df with itertuples\nfor entry in train_data.itertuples(): # entry[1] is the user-id, entry[2] is the book-isbn\n train_matrix[entry[1]-1, entry[2]-1] = entry[3] # -1 is to counter 0-based indexing",
"_____no_output_____"
],
[
"train_matrix.shape",
"_____no_output_____"
]
],
[
[
"Now do the same for the test set.",
"_____no_output_____"
]
],
[
[
"### TESTING SET\n# Create user-item matrices\nn_users = test_data['u_unique'].nunique()\nn_books = test_data['b_unique'].nunique()\n\n# First, create an empty matrix of size USERS x BOOKS (this speeds up the later steps)\ntest_matrix = np.zeros((n_users, n_books))\n\n# Then, add the appropriate vals to the matrix by extracting them from the df with itertuples\nfor entry in test_data.itertuples(): # entry[1] is the user-id, entry[2] is the book-isbn\n test_matrix[entry[1]-1, entry[2]-1] = entry[3] # -1 is to counter 0-based indexing",
"_____no_output_____"
],
[
"test_matrix.shape",
"_____no_output_____"
]
],
[
[
"Now the matrix is in the correct format, with the user and book entries encoded from the mapping dict created above!",
"_____no_output_____"
],
[
"### Calculating cosine similarity with the 'pairwise distances' function\n\nTo determine the similarity between users/items we'll use the 'cosine similarity' which is a common n-dimensional distance metric.\n\n**Note:** since all of the rating values are positive (1-10 scale), the cosine distances will all fall between 0 and 1.",
"_____no_output_____"
]
],
[
[
"# It may take a while to calculate, so I'll perform on a subset initially\ntrain_matrix_small = train_matrix[:10000, :10000]\ntest_matrix_small = test_matrix[:10000, :10000]\n\nfrom sklearn.metrics.pairwise import pairwise_distances\nuser_similarity = pairwise_distances(train_matrix_small, metric='cosine')\nitem_similarity = pairwise_distances(train_matrix_small.T, metric='cosine') # .T transposes the matrix (NumPy)",
"_____no_output_____"
]
],
[
[
"If we are looking at similarity between users we need to account for the average behaviour of that individual user. For example, one user may give all movies quite high ratings, whereas one might give all ratings between 3 and 7. These users might otherwise have quite similar preferences.\n\nTo do this, we use the users average rating as a 'weighting' factor.\n\nIf we are looking at item-based similarity we don't need to add this weighting factor.\n\nWe can incorporate this into a ```predict()``` function, like so:",
"_____no_output_____"
]
],
[
[
"def predict(ratings, similarity, type='user'): # default type is 'user'\n if type == 'user':\n mean_user_rating = ratings.mean(axis=1)\n # Use np.newaxis so that mean_user_rating has the same format as ratings\n ratings_diff = (ratings - mean_user_rating[:, np.newaxis])\n pred = mean_user_rating[:, np.newaxis] + similarity.dot(ratings_diff) / np.array([np.abs(similarity).sum(axis=1)]).T\n elif type == 'item':\n pred = ratings.dot(similarity) / np.array([np.abs(similarity).sum(axis=1)])\n return pred",
"_____no_output_____"
]
],
[
[
"Then can make our predictions!",
"_____no_output_____"
]
],
[
[
"item_prediction = predict(train_matrix_small, item_similarity, type='item')\nuser_prediction = predict(train_matrix_small, user_similarity, type='user')",
"_____no_output_____"
]
],
[
[
"### Evaluation\n\nHow do we know if this is making good ```rating``` predictions?\n\nWe'll start by just taking the root mean squared error (RMSE) (from ```sklearn```) of predicted values in the ```test_set``` (i.e. where we know what the answer should be).\n\nSince we want to compare only predicted ratings that are in the test set, we can filter out all other predictions that aren't in the test matrix.",
"_____no_output_____"
]
],
[
[
"from sklearn.metrics import mean_squared_error\nfrom math import sqrt\n\ndef rmse(prediction, test_matrix):\n prediction = prediction[test_matrix.nonzero()].flatten()\n test_matrix = test_matrix[test_matrix.nonzero()].flatten()\n return sqrt(mean_squared_error(prediction, test_matrix))\n\n# Call on test set to get error from each approach ('user' or 'item')\nprint(f'User-based CF RMSE: {rmse(user_prediction, test_matrix_small)}')\nprint(f'Item-based CF RMSE: {rmse(item_prediction, test_matrix_small)}')",
"User-based CF RMSE: 7.929926397008533\nItem-based CF RMSE: 7.930326958653732\n"
]
],
[
[
"For the user-item and the item-item recommendations we get RMSE = 7.85 (MSE > 60) for both. This is pretty bad, but we only trained over a small subset of the data.\n\nAlthough this collaborative filtering setup is relatively simple to write, it doesn't scale very well at all, as it is all stored in memory! (Hence why we only used a subset of the training/testing data).\n\n----------------\n\nInstead, we should really use a model-based (based on matrix factorization) recommendation algorithm. These are inherently more scalable and can deal with higher sparsity level than memory-based models, and are considered more powerful due to their ability to pick up on \"latent factors\" in the relationships between what sets of items users like. However, they still suffer from the \"cold start\" problem (where a new user has no history).\n\nFortunately, there is a Python library called ```surprise``` that was built specifically for the implementation of model-based recommendation systems! This library comes with many of the leading algorithms in this space already built-in. Let's try use it for our book recommender system.",
"_____no_output_____"
],
[
"# Using the ```surprise``` library for building a recommender system\nSeveral common model-based algorithms including SVD, KNN, and non-negative matrix factorization are built-in! \nSee [here](http://surprise.readthedocs.io/en/stable/getting_started.html#basic-usage) for the docs.",
"_____no_output_____"
]
],
[
[
"from surprise import Reader, Dataset",
"_____no_output_____"
],
[
"user_item_rating.head() # take a look at our data",
"_____no_output_____"
],
[
"# First need to create a 'Reader' object to set the scale/limit of the ratings field\nreader = Reader(rating_scale=(1, 10))\n\n# Load the data into a 'Dataset' object directly from the pandas df.\n# Note: The fields must be in the order: user, item, rating\ndata = Dataset.load_from_df(user_item_rating, reader)",
"_____no_output_____"
],
[
"# Load the models and 'evaluation' method\nfrom surprise import SVD, NMF, model_selection, accuracy",
"_____no_output_____"
]
],
[
[
"Where: SVD = Singular Value Decomposition (orthogonal factorization), NMF = Non-negative Matrix Factorization.\n\n**Note** that when using the ```surprise``` library we don't need to manually create the mapping of USERID and UNIQUE_ITEMIDS to integers in a custom dict. See [here](http://surprise.readthedocs.io/en/stable/FAQ.html#raw-inner-note) for details. ",
"_____no_output_____"
],
[
"### SVD model",
"_____no_output_____"
],
[
"**_Using cross-validation (5 folds)_**",
"_____no_output_____"
]
],
[
[
"# Load SVD algorithm\nmodel = SVD()\n\n# Train on books dataset\n%time model_selection.cross_validate(model, data, measures=['RMSE'], cv=5, verbose=True)",
"Evaluating RMSE of algorithm SVD on 5 split(s).\n\n Fold 1 Fold 2 Fold 3 Fold 4 Fold 5 Mean Std \nRMSE (testset) 1.6379 1.6342 1.6399 1.6333 1.6383 1.6367 0.0025 \nFit time 27.50 29.12 28.19 28.50 27.17 28.09 0.70 \nTest time 1.26 1.28 1.25 1.07 1.22 1.22 0.07 \nCPU times: user 2min 32s, sys: 1.44 s, total: 2min 33s\nWall time: 2min 34s\n"
]
],
[
[
"The SVD model gives an average RMSE of ca. 1.64 after 5-folds, with a fit time of ca. 28 s for each fold.",
"_____no_output_____"
],
[
"**_Using test-train split_**",
"_____no_output_____"
]
],
[
[
"# set test set to 20%.\ntrainset, testset = model_selection.train_test_split(data, test_size=0.2)\n\n# Instantiate the SVD model.\nmodel = SVD()\n\n# Train the algorithm on the training set, and predict ratings for the test set\nmodel.fit(trainset)\npredictions = model.test(testset)\n\n# Then compute RMSE\naccuracy.rmse(predictions)",
"RMSE: 1.6355\n"
]
],
[
[
"Using a 80% train-test split, the SVD model gave a RMSE of 1.6426.\n\n-----------\n\nWe can see that using the SVD algorithm has already far out-performed the memory-based collaborative filtering approach (RMSE of 1.64 vs 7.92)!",
"_____no_output_____"
],
[
"### NMF model",
"_____no_output_____"
]
],
[
[
"# Load NMF algorithm\nmodel = NMF()\n# Train on books dataset\n%time model_selection.cross_validate(model, data, measures=['RMSE'], cv=5, verbose=True)",
"Evaluating RMSE of algorithm NMF on 5 split(s).\n\n Fold 1 Fold 2 Fold 3 Fold 4 Fold 5 Mean Std \nRMSE (testset) 2.4719 2.4753 2.4991 2.4583 2.4690 2.4747 0.0134 \nFit time 48.55 48.36 48.67 48.21 49.12 48.58 0.31 \nTest time 1.00 1.22 1.24 0.89 1.27 1.12 0.15 \nCPU times: user 4min 12s, sys: 2.77 s, total: 4min 15s\nWall time: 4min 15s\n"
]
],
[
[
"The NMF model gave a mean RMSE of ca. 2.47, with a fit time of ca. 48 s.\n\nIt seems like the SVD algorithm is the best choice for this dataset.",
"_____no_output_____"
],
[
"## Optimizing the SVD algorithm with parameter tuning\nSince it seems like the SVD algorithm is our best choice, let's see if we can improve the predictions even further by optimizing some of the algorithm hyperparameters.\n\nOne way of doing this is to use the handy ```GridSearchCV``` method from the ```surprise``` library. When passed a range of hyperparameter values, ```GridSearchCV``` will automatically search through the parameter-space to find the best-performing set of hyperparameters.",
"_____no_output_____"
]
],
[
[
"# We'll remake the training set, keeping 20% for testing\ntrainset, testset = model_selection.train_test_split(data, test_size=0.2)",
"_____no_output_____"
],
[
"### Fine-tune Surprise SVD model useing GridSearchCV\nfrom surprise.model_selection import GridSearchCV\n\nparam_grid = {'n_factors': [80, 100, 120], 'lr_all': [0.001, 0.005, 0.01], 'reg_all': [0.01, 0.02, 0.04]}\n\n# Optimize SVD algorithm for both root mean squared error ('rmse') and mean average error ('mae')\ngs = GridSearchCV(SVD, param_grid, measures=['rmse', 'mae'], cv=3)",
"_____no_output_____"
],
[
"# Fit the gridsearch result on the entire dataset\n%time gs.fit(data)",
"CPU times: user 34min 9s, sys: 10.8 s, total: 34min 20s\nWall time: 34min 29s\n"
],
[
"# Return the best version of the SVD algorithm\nmodel = gs.best_estimator['rmse']\n\nprint(gs.best_score['rmse'])\nprint(gs.best_params['rmse'])",
"1.6376613092691847\n{'n_factors': 80, 'lr_all': 0.005, 'reg_all': 0.04}\n"
],
[
"model_selection.cross_validate(model, data, measures=['rmse', 'mae'], cv=5, verbose=True)",
"Evaluating RMSE, MAE of algorithm SVD on 5 split(s).\n\n Fold 1 Fold 2 Fold 3 Fold 4 Fold 5 Mean Std \nRMSE (testset) 1.6326 1.6292 1.6314 1.6323 1.6296 1.6310 0.0014 \nMAE (testset) 1.2594 1.2612 1.2611 1.2587 1.2563 1.2593 0.0018 \nFit time 25.22 24.25 24.24 24.00 24.18 24.38 0.43 \nTest time 1.08 1.46 1.46 1.05 1.04 1.22 0.20 \n"
]
],
[
[
"The mean RSME using the optimized parameters was 1.6351 over 5 folds, with an average fit time of ca. 24s.",
"_____no_output_____"
]
],
[
[
"### Use the new parameters with the training set\nmodel = SVD(n_factors=80, lr_all=0.005, reg_all=0.04)\nmodel.fit(trainset) # re-fit on only the training data using the best hyperparameters\ntest_pred = model.test(testset)\nprint(\"SVD : Test Set\")\naccuracy.rmse(test_pred, verbose=True)",
"SVD : Test Set\nRMSE: 1.6287\n"
]
],
[
[
"Using the optimized hyperparameters we see a slight improvement in the resulting RMSE (1.629) compared with the unoptimized SVD algorithm (1.635)1",
"_____no_output_____"
],
[
"## Testing some of the outputs (ratings and recommendations)\nWould like to do an intuitive check of some of the recommendations being made.\n\nLet's just choose a random user/book pair (represented in the ```suprise``` library as ```uid``` and ```iid```, respectively).\n\n**Note:** The ```model``` being used here is the optimized SVD algorithm that has been fit on the training set.",
"_____no_output_____"
]
],
[
[
"# get a prediction for specific users and items.\nuid = 276744 # the USERID int\niid = '038550120X' # the UNIQUE_ITEMIDS string\n# This pair has an actual rating of 7!\n\npred = model.predict(uid, iid, verbose=True)",
"user: 276744 item: 038550120X r_ui = None est = 7.18 {'was_impossible': False}\n"
]
],
[
[
"Can access the attributes of the ```predict``` method to get a nicer output.",
"_____no_output_____"
]
],
[
[
"print(f'The estimated rating for the book with the \"UNIQUE_ITEMIDS\" code {pred.iid} from user #{pred.uid} is {pred.est:.2f}.\\n')\nactual_rtg = user_item_rating[(user_item_rating.USERID==pred.uid) & (user_item_rating.UNIQUE_ITEMIDS==pred.iid)].RATING.values[0]\nprint(f'The real rating given for this was {actual_rtg:.2f}.')",
"The estimated rating for the book with the \"unique_isbn\" code 038550120X from user #276744 is 7.18.\n\nThe real rating given for this was 7.00.\n"
],
[
"# get a prediction for specific users and items.\nuid = 95095 # the USERID int\niid = '0140079963' # the UNIQUE_ITEMIDS string\n# This pair has an actual rating of 6.0!\n\npred = model.predict(uid, iid, verbose=True)",
"user: 95095 item: 0140079963 r_ui = None est = 6.87 {'was_impossible': False}\n"
],
[
"print(f'The estimated rating for the book with the \"UNIQUE_ITEMIDS\" code {pred.iid} from user #{pred.uid} is {pred.est:.2f}.\\n')\nactual_rtg = user_item_rating[(user_item_rating.USERID==pred.uid) & (user_item_rating.UNIQUE_ITEMIDS==pred.iid)].RATING.values[0]\nprint(f'The real rating given for this was {actual_rtg:.2f}.')",
"The estimated rating for the book with the \"unique_isbn\" code 0140079963 from user #95095 is 6.87.\n\nThe real rating given for this was 6.00.\n"
]
],
[
[
"The following function was adapted from the ```surprise``` docs, and can be used to get the top book recommendations for each user.",
"_____no_output_____"
]
],
[
[
"from collections import defaultdict\n\ndef get_top_n(predictions, n=10):\n '''Return the top-N recommendation for each user from a set of predictions.\n\n Args:\n predictions(list of Prediction objects): The list of predictions, as\n returned by the test method of an algorithm.\n n(int): The number of recommendation to output for each user. Default\n is 10.\n\n Returns:\n A dict where keys are user (raw) ids and values are lists of tuples:\n [(raw item id, rating estimation), ...] of size n.\n '''\n\n # First map the predictions to each user.\n top_n = defaultdict(list)\n for uid, iid, true_r, est, _ in predictions:\n top_n[uid].append((iid, est))\n\n # Then sort the predictions for each user and retrieve the k highest ones.\n for uid, user_ratings in top_n.items():\n user_ratings.sort(key=lambda x: x[1], reverse=True)\n top_n[uid] = user_ratings[:n]\n \n return top_n",
"_____no_output_____"
]
],
[
[
"Let's get the Top 10 recommended books for each USERID in the test set.",
"_____no_output_____"
]
],
[
[
"pred = model.test(testset)\ntop_n = get_top_n(pred)",
"_____no_output_____"
],
[
"def get_reading_list(userid):\n \"\"\"\n Retrieve full book titles from full 'books_users_ratings' dataframe\n \"\"\"\n reading_list = defaultdict(list)\n top_n = get_top_n(predictions, n=10)\n for n in top_n[userid]:\n book, rating = n\n title = books_users_ratings.loc[books_users_ratings.UNIQUE_ITEMIDS==book].TITLE.unique()[0]\n reading_list[title] = rating\n return reading_list",
"_____no_output_____"
],
[
"# Just take a random look at USERID=60337\nexample_reading_list = get_reading_list(userid=60337)\nfor book, rating in example_reading_list.items():\n print(f'{book}: {rating}')",
"A Natural History of the Senses: 9.124985146110324\nGrace For The Moment: 9.004232452533614\nEva Luna: 8.957717816714606\nA Slender Thread : Rediscovering Hope at the Heart of Crisis: 8.81756023683983\nTell Me the Truth About Love: Ten Poems: 8.729204207356146\nPapa, My Father: A Celebration of Dads: 8.708985984501158\nLet the Dead Bury Their Dead (Harvest American Writing Series): 8.646273705729296\nThe Doctor's Book of Home Remedies : Thousands of Tips and Techniques Anyone Can Use to Heal Everyday Health Problems: 8.635645117795315\nWhy We Cant Wait: 8.535174763005822\nLump It or Leave It: 8.535174763005822\n"
]
],
[
[
"Have tried out a few different ```userid``` entries (from the ```testset```) to see what the top 10 books that user would like are and they seem pretty well related, indicating that the recommendation engine is performing reasonably well!",
"_____no_output_____"
],
[
"# Summary",
"_____no_output_____"
],
[
"In this notebook a dataset from the 'Book-Crossing' website was used to create a recommendation system. A few different approaches were investigated, including memory-based correlations, and model-based matrix factorization algorithms[2]. Of these, the latter - and particularly the Singular Value Decomposition (SVD) algorithm - gave the best performance as assessed by comparing the predicted book ratings for a given user with the actual rating in a test set that the model was not trained on.\n\nThe only fields that were used for the model were the \"user ID\", \"book ID\", and \"rating\". There were others available in the dataset, such as \"age\", \"location\", \"publisher\", \"year published\", etc, however for these types of recommendation systems it has often been found that additional data fields do not increase the accuracy of the models significantly[1]. A \"Grid Search Cross Validation\" method was used to optimize some of the hyperparameters for the model, resulting in a slight improvement in model performance from the default values.\n\nFinally, we were able to build a recommender that could predict the 10 most likely book titles to be rated highly by a given user.\n\nIt should be noted that this approach still suffers from the \"cold start problem\"[3] - that is, for users with no ratings or history the model will not make accurate predictions. One way we could tackle this problem may be to initially start with popularity-based recommendations, before building up enough user history to implement the model. Another piece of data that was not utilised in the current investigation was the \"implicit\" ratings - denoted as those with a rating of \"0\" in the dataset. Although more information about these implicit ratings (for example, does it represent a positive or negative interaction), these might be useful for supplementing the \"explicit\" ratings recommender.",
"_____no_output_____"
],
[
"# References",
"_____no_output_____"
],
[
"1. http://blog.ethanrosenthal.com/2015/11/02/intro-to-collaborative-filtering/\n2. https://cambridgespark.com/content/tutorials/implementing-your-own-recommender-systems-in-Python/index.html\n3. https://towardsdatascience.com/building-a-recommendation-system-for-fragrance-5b00de3829da",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
cb8ae72200b842a65ff04b6bd53dd99db1640a49
| 21,579 |
ipynb
|
Jupyter Notebook
|
code/chapter03_DL-basics/3.2_linear-regression-scratch.ipynb
|
tainenko/Dive-into-DL-TensorFlow2.0
|
8a7c4fe2cd4a3dd2da52f45f9e218b776033aef6
|
[
"Apache-2.0"
] | 8 |
2019-12-07T19:12:55.000Z
|
2021-04-11T07:47:30.000Z
|
Codes (for executing)/ch3_DL_basics/3.2 linear regression from scratch.ipynb
|
ShirleyGao1023/Dive-into-DL-TensorFlow2.0
|
b4db6aa4f23f29f4bb78080876bdc606b5b15d40
|
[
"Apache-2.0"
] | null | null | null |
Codes (for executing)/ch3_DL_basics/3.2 linear regression from scratch.ipynb
|
ShirleyGao1023/Dive-into-DL-TensorFlow2.0
|
b4db6aa4f23f29f4bb78080876bdc606b5b15d40
|
[
"Apache-2.0"
] | 4 |
2019-12-14T00:24:07.000Z
|
2021-12-12T14:58:38.000Z
| 65.390909 | 14,232 | 0.805783 |
[
[
[
"import tensorflow as tf\nprint(tf.__version__)\n# from Ipython import display\nfrom matplotlib import pyplot as plt\nimport random\n%matplotlib inline",
"2.0.0\n"
]
],
[
[
"## 3.2.1 gen_dataset",
"_____no_output_____"
]
],
[
[
"num_inputs = 2\nnum_examples = 1000\ntrue_w = [2, -3.4]\ntrue_b = 4.2\nfeatures = tf.random.normal((num_examples, num_inputs),stddev = 1)\nlabels = true_w[0] * features[:,0] + true_w[1] * features[:,1] + true_b\nlabels += tf.random.normal(labels.shape,stddev=0.01)\n\nfeatures[0], labels[0]",
"_____no_output_____"
],
[
"def use_svg_display():\n display.set_matplotlib_formats('svg')\ndef set_figsize(figsize=(3.5, 2.5)):\n# use_svg_display()\n plt.rcParams['figure.figsize'] = figsize\n\nset_figsize()\nplt.scatter(features[:, 1], labels, 1)",
"_____no_output_____"
]
],
[
[
"## 3.2.2 read data",
"_____no_output_____"
]
],
[
[
"import numpy as np\ndef data_iter(batch_size, features, labels):\n features = np.array(features)\n labels = np.array(labels)\n num_examples = len(features)\n indices = list(range(num_examples))\n random.shuffle(indices)\n for i in range(0, num_examples, batch_size):\n j = np.array(indices[i:min(i + batch_size, num_examples)])\n yield features[j], labels[j]",
"_____no_output_____"
],
[
"batch_size = 10\n\nfor X, y in data_iter(batch_size, features, labels):\n print(X, y)\n break",
"[[-0.15791896 -1.368419 ]\n [-1.2912872 -2.9434786 ]\n [-1.7658867 -1.4701791 ]\n [ 0.17313462 0.61840177]\n [ 0.98486614 1.8532189 ]\n [ 0.98916334 1.760945 ]\n [-0.854792 -1.3276836 ]\n [ 0.8285209 1.9348817 ]\n [ 1.3753697 1.2648998 ]\n [-0.7374796 -0.71223205]] [ 8.530758 11.613669 5.662739 2.4489765 -0.1518561 0.18878557\n 6.994077 -0.7146609 2.6620245 5.160786 ]\n"
]
],
[
[
"## 3.2.3 initialize weight",
"_____no_output_____"
]
],
[
[
"w = tf.Variable(tf.random.normal((num_inputs, 1), stddev=0.01))\nb = tf.Variable(tf.zeros((1,)))",
"_____no_output_____"
]
],
[
[
"## 3.2.4 define model",
"_____no_output_____"
]
],
[
[
"def linreg(X, w, b):\n return tf.matmul(X, w) + b",
"_____no_output_____"
]
],
[
[
"## 3.2.5 define loss",
"_____no_output_____"
]
],
[
[
"def squared_loss(y_hat, y):\n return (y_hat - tf.reshape(y, y_hat.shape)) ** 2 /2",
"_____no_output_____"
]
],
[
[
"## 3.2.6 define optimization",
"_____no_output_____"
]
],
[
[
"def sgd(params, lr, batch_size):\n for param in params:\n# param[:] = param - lr * t.gradient(l, param) / batch_size\n param.assign_sub(lr * t.gradient(l, param) / batch_size)",
"_____no_output_____"
]
],
[
[
"## 3.2.7 training",
"_____no_output_____"
]
],
[
[
"lr = 0.03\nnum_epochs = 3\nnet = linreg\nloss = squared_loss\n\nfor epoch in range(num_epochs):\n for X, y in data_iter(batch_size, features, labels):\n with tf.GradientTape(persistent=True) as t:\n t.watch([w,b])\n l = loss(net(X, w, b), y)\n sgd([w, b], lr, batch_size)\n train_l = loss(net(features, w, b), labels)\n print('epoch %d, loss %f' % (epoch + 1, tf.reduce_mean(train_l)))",
"epoch 1, loss 0.052947\nepoch 2, loss 0.000244\nepoch 3, loss 0.000044\n"
],
[
"true_w, w",
"_____no_output_____"
],
[
"true_b, b",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
cb8af60c4c0af525a7aff2867290c1529ada9f90
| 13,494 |
ipynb
|
Jupyter Notebook
|
tp2/.ipynb_checkpoints/experimentacion_calidad_de_los_resultados_obtenidos_al_combinar_KNN_con_y_sin_PCA-checkpoint.ipynb
|
sebasbocaccio/tp2-metodos
|
916f569e75cb570e9ccc193427df9618a508322b
|
[
"MIT"
] | null | null | null |
tp2/.ipynb_checkpoints/experimentacion_calidad_de_los_resultados_obtenidos_al_combinar_KNN_con_y_sin_PCA-checkpoint.ipynb
|
sebasbocaccio/tp2-metodos
|
916f569e75cb570e9ccc193427df9618a508322b
|
[
"MIT"
] | null | null | null |
tp2/.ipynb_checkpoints/experimentacion_calidad_de_los_resultados_obtenidos_al_combinar_KNN_con_y_sin_PCA-checkpoint.ipynb
|
sebasbocaccio/tp2-metodos
|
916f569e75cb570e9ccc193427df9618a508322b
|
[
"MIT"
] | null | null | null | 40.890909 | 410 | 0.52616 |
[
[
[
"%load_ext autoreload\n%autoreload 2\nimport pandas as pd\nfrom tqdm import tqdm\nimport metnum\nimport numpy\nfrom sklearn.utils import shuffle\nfrom sklearn.metrics import accuracy_score,precision_score,recall_score,f1_score\nimport csv\nimport time\n",
"The autoreload extension is already loaded. To reload it, use:\n %reload_ext autoreload\n"
],
[
"def correr_Knn_con_k_aumentando_en(porcentage_para_entrenar,cant_muestras=42000,semilla = 2,intervalo_k):\n df_train = pd.read_csv(\"../data/train.csv\")\n df_train = df_train.sample(frac=1, random_state=semilla)\n X= df_train[df_train.columns[1:]].values\n y = df_train[\"label\"].values.reshape(-1, 1)\n\n accuracy_of_all_k = []\n precision_df = pd.DataFrame(columns=(0,1,2,3,4,5,6,7,8,9))\n recall_df = pd.DataFrame(columns=(0,1,2,3,4,5,6,7,8,9))\n f1_df = pd.DataFrame(columns=(0,1,2,3,4,5,6,7,8,9))\n times = [] \n \n setup_time_start_time = time.time()\n \n limit = int(0.8 * X.shape[0]) \n X_train = X[:limit]\n X_val = X[limit:]\n y_train = y[:limit]\n y_val = y[limit:]\n alpha = 0\n # Hago el fit generico que guarda los datos \n clf.fit(X_train, y_train)\n setup_time_end_time = time.time()\n setup_time = setup_time_end_time-setup_time_start_time\n for k in tqdm(range(1,X_train.shape[0]+1,intervalo_k)):\n\n # Correr knn \n clf = metnum.KNNClassifier(k)\n knn_start_time = time.time()\n \n y_pred = clf.predict(X_val)\n print(accuracy_score(y_val, y_pred))\n knn_end_time = time.time()\n knn_time = knn_end_time-knn_start_time\n # Calcular metricas de interes\n labels= [0,1,2,3,4,5,6,7,8,9]\n precision = precision_score(y_val, y_pred,labels=labels, average=None)\n accuracy = accuracy_score(y_val, y_pred)\n recall = recall_score(y_val,y_pred,labels=labels, average=None)\n f1 = f1_score(y_val,y_pred,labels=labels, average=None)\n\n # Escribir los resultados\n accuracy_of_all_k.append([alpha, k ,accuracy])\n times.append(setup_time+knn_time)\n\n # Agregar una fila al dataframe de precision\n digit = 0 \n precision_dict={}\n for i in range(0,10,1):\n precision_dict[digit]=precision[i]\n digit += 1 \n precision_dict['k']=k\n precision_dict['alpha']=alpha\n precision_df = precision_df.append(precision_dict,ignore_index=True)\n\n # Agregar una fila al dataframe de recall\n digit = 0 \n recall_dict={}\n for i in range(0,10,1):\n recall_dict[digit]=recall[i]\n digit += 1 \n recall_dict['k']=k\n recall_dict['alpha']=alpha\n recall_df = recall_df.append(recall_dict,ignore_index=True)\n \n # Agregar una fila al dataframe de f1\n \n digit = 0 \n f1_dict={}\n for i in range(0,10,1):\n f1_dict[digit]=f1[i]\n digit += 1 \n f1_dict['k']=k\n f1_dict['alpha']=alpha\n f1_df = f1_df.append(f1_dict,ignore_index=True)\n \n\n # Escribo los resultados a un archivo para no tener que correr devuelta los resultados.\n precision_df.to_csv('knn_solo_precision.csv', index=False) \n recall_df.to_csv('knn_solo_recall.csv', index=False) \n f1_df.to_csv('knn_solo_f1.csv', index=False) \n\n with open('knn_solo_acuracy.csv', 'w', newline='') as myfile:\n wr = csv.writer(myfile, quoting=csv.QUOTE_ALL)\n wr.writerow(accuracy_of_all_k)\n\n with open('knn_solo_time.csv', 'w', newline='') as myfile:\n wr = csv.writer(myfile, quoting=csv.QUOTE_ALL)\n wr.writerow(times)\n \n \n with open('knn_solo_predicciones.csv', 'w', newline='') as myfile:\n wr = csv.writer(myfile, quoting=csv.QUOTE_ALL)\n wr.writerow(y_pred)\n\n\n\n ",
"_____no_output_____"
],
[
"correr_Knn_con_todos_los_k_posibles(0.8)",
"_____no_output_____"
],
[
"def correr_Knn_pca(porcentage_para_entrenar,cant_muestras=42000,semilla = 2,intervalo_k,intervalo_alpha):\n df_train = pd.read_csv(\"../data/train.csv\")\n df_train = df_train.sample(frac=1, random_state=semilla)\n X_original = df_train[df_train.columns[1:]].values\n y = df_train[\"label\"].values.reshape(-1, 1)\n\n valores_k = [1,5,10,25,50,75,100,200,500,1000,2000,5000,10000,20000]\n accuracy_of_all_k = []\n precision_df = pd.DataFrame(columns=('alpha','k',0,1,2,3,4,5,6,7,8,9))\n recall_df = pd.DataFrame(columns=('alpha','k',0,1,2,3,4,5,6,7,8,9))\n f1_df = pd.DataFrame(columns=(0,1,2,3,4,5,6,7,8,9))\n times = [] \n for alpha in tqdm(range(1,29,intervalo_alpha)):\n \n setup_time_start_time = time.time()\n pca = metnum.PCA(alpha)\n X = pca.transform(X_original)\n limit = int(0.8 * X.shape[0]) \n X_train = X[:limit]\n X_val = X[limit:]\n y_train = y[:limit]\n y_val = y[limit:]\n\n \n # Hago el fit generico que guarda los datos \n\n setup_time_end_time = time.time()\n setup_time = setup_time_end_time-setup_time_start_time\n \n for k in tqdm(valores_k):\n\n # Correr knn \n clf = metnum.KNNClassifier(k)\n \n knn_start_time = time.time()\n clf.fit(X_train, y_train)\n y_pred = clf.predict(X_val)\n print(accuracy_score(y_val, y_pred))\n knn_end_time = time.time()\n knn_time = knn_end_time-knn_start_time\n # Calcular metricas de interes\n labels= [0,1,2,3,4,5,6,7,8,9]\n precision = precision_score(y_val, y_pred,labels=labels, average=None)\n accuracy = accuracy_score(y_val, y_pred)\n recall = recall_score(y_val,y_pred,labels=labels, average=None)\n f1 = f1_score(y_val,y_pred,labels=labels, average=None)\n\n # Escribir los resultados\n accuracy_of_all_k.append([alpha, k ,accuracy])\n times.append(setup_time+knn_time)\n\n # Agregar una fila al dataframe de precision\n digit = 0 \n precision_dict={}\n for i in range(0,10,1):\n precision_dict[digit]=precision[i]\n digit += 1 \n precision_dict['k']=k\n precision_dict['alpha']=alpha\n precision_df = precision_df.append(precision_dict,ignore_index=True)\n\n # Agregar una fila al dataframe de recall\n digit = 0 \n recall_dict={}\n for i in range(0,10,1):\n recall_dict[digit]=recall[i]\n digit += 1 \n recall_dict['k']=k\n recall_dict['alpha']=alpha\n recall_df = recall_df.append(recall_dict,ignore_index=True)\n \n \n # Agregar una fila al dataframe de f1\n \n digit = 0 \n f1_dict={}\n for i in range(0,10,1):\n f1_dict[digit]=f1[i]\n digit += 1 \n f1_dict['k']=k\n f1_dict['alpha']=alpha\n f1_df = f1_df.append(f1_dict,ignore_index=True)\n\n\n # Escribo los resultados a un archivo para no tener que correr devuelta los resultados.\n precision_df.to_csv('knn_pca_precision_re_do.csv', index=False) \n recall_df.to_csv('knn_pca_recall.csv_re_do', index=False) \n\n with open('knn_pca_acuracy_re_do.csv', 'w', newline='') as myfile:\n wr = csv.writer(myfile, quoting=csv.QUOTE_ALL)\n wr.writerow(accuracy_of_all_k)\n\n with open('knn_pca_time_re_do.csv', 'w', newline='') as myfile:\n wr = csv.writer(myfile, quoting=csv.QUOTE_ALL)\n wr.writerow(times)\n\n with open('knn_pca_predicciones.csv', 'w', newline='') as myfile:\n wr = csv.writer(myfile, quoting=csv.QUOTE_ALL)\n wr.writerow(y_pred)\n\n \n\n ",
"_____no_output_____"
],
[
"correr_Knn_pca(0.8)",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code"
]
] |
cb8b050734a3dc69e1e6608cebbd299b247597ea
| 18,018 |
ipynb
|
Jupyter Notebook
|
tutorial7.ipynb
|
dschwen/learn_python
|
9c49fc732dd99a632984a537c1b00d5ba1ffabaa
|
[
"CC-BY-3.0"
] | 1 |
2022-02-02T03:21:37.000Z
|
2022-02-02T03:21:37.000Z
|
tutorial7.ipynb
|
jrincayc/learn_python
|
9c49fc732dd99a632984a537c1b00d5ba1ffabaa
|
[
"CC-BY-3.0"
] | null | null | null |
tutorial7.ipynb
|
jrincayc/learn_python
|
9c49fc732dd99a632984a537c1b00d5ba1ffabaa
|
[
"CC-BY-3.0"
] | 1 |
2021-08-29T16:05:02.000Z
|
2021-08-29T16:05:02.000Z
| 33.553073 | 649 | 0.534133 |
[
[
[
"\nVariables with more than one value\n==================================\n\n\nYou have already seen ordinary variables that store a single value. However other variable types can hold more than one value. The simplest type is called a list. Here is a example of a list being used:\n",
"_____no_output_____"
]
],
[
[
"which_one = int(input(\"What month (1-12)? \"))\nmonths = ['January', 'February', 'March', 'April', 'May', 'June', 'July',\\\n 'August', 'September', 'October', 'November', 'December']\nif 1 <= which_one <= 12:\n print(\"The month is\", months[which_one - 1])",
"_____no_output_____"
]
],
[
[
"and an output example:",
"_____no_output_____"
]
],
[
[
"What month (1-12)? 3\nThe month is March",
"_____no_output_____"
]
],
[
[
"\n\nIn this example the months is a list. months is defined with the lines months = ['January', 'February', 'March', 'April', 'May', 'June', 'July', 'August', 'September', 'October', 'November', 'December'] (Note that a `\\` can be used to split a long line). The `[` and `]` start and end the list with comma's (`,`) separating the list items. The list is used in `months[which_one - 1]`. A list consists of items that are numbered starting at 0. In other words if you wanted January you would type in 1 and that would have 1 subtracted off to use `months[0]`. Give a list a number and it will return the value that is stored at that location.\n\n\nThe statement `if 1 <= which_one <= 12:` will only be true if `which_one` is between one and twelve inclusive (in other words it is what you would expect if you have seen that in algebra). Since 1 is subtracted from `which_one` we get list locations from 0 to 11.\n\n\nLists can be thought of as a series of boxes. For example, the boxes created by demolist = ['life', 42, 'the', 'universe', 6, 'and', 7] would look like this:\n\n\n\n\n| | | | | | | | | \n| --- | --- | --- | --- | --- | --- | --- | --- | \n| box number | 0 | 1 | 2 | 3 | 4 | 5 | 6 | \n| demolist | 'life' | 42 | 'the' | 'universe' | 6 | 'and' | 7 | \n\n\n\nEach box is referenced by its number so the statement `demolist[0]` would get 'life', `demolist[1]` would get 42 and so on up to `demolist[6]` getting 7.\n\n\nMore features of lists\n======================\n\n\nThe next example is just to show a lot of other stuff lists can do (for once, I don't expect you to type it in, but you should probably play around with lists until you are comfortable with them. Also, there will be another program that uses most of these features soon.). Here goes:\n",
"_____no_output_____"
]
],
[
[
"demolist = ['life', 42, 'the', 'universe', 6, 'and', 7]\nprint('demolist = ', demolist)\ndemolist.append('everything')\nprint(\"after 'everything' was appended demolist is now:\")\nprint(demolist)\nprint('len(demolist) =', len(demolist))\nprint('demolist.index(42) =', demolist.index(42))\nprint('demolist[1] =', demolist[1])\n#Next we will loop through the list\nc = 0\nwhile c < len(demolist):\n print('demolist[', c, ']=', demolist[c])\n c = c + 1\ndel demolist[2]\nprint(\"After 'the universe' was removed demolist is now:\")\nprint(demolist)\nif 'life' in demolist:\n print(\"'life' was found in demolist\")\nelse:\n print(\"'life' was not found in demolist\")\nif 'amoeba' in demolist:\n print(\"'amoeba' was found in demolist\")\nif 'amoeba' not in demolist:\n print(\"'amoeba' was not found in demolist\")\nint_list = []\nc = 0\nwhile c < len(demolist):\n if type(0) == type(demolist[c]):\n int_list.append(demolist[c])\n c = c + 1\nprint('int_list is', int_list)\nint_list.sort()\nprint('The sorted int_list is ', int_list)",
"_____no_output_____"
]
],
[
[
"\n\nThe output is:\n",
"_____no_output_____"
]
],
[
[
"demolist = ['life', 42, 'the', 'universe', 6, 'and', 7]\nafter 'everything' was appended demolist is now:\n['life', 42, 'the', 'universe', 6, 'and', 7, 'everything']\nlen(demolist) = 8\ndemolist.index(42) = 1\ndemolist[1] = 42\ndemolist[ 0 ]= life\ndemolist[ 1 ]= 42\ndemolist[ 2 ]= the\ndemolist[ 3 ]= universe\ndemolist[ 4 ]= 6\ndemolist[ 5 ]= and\ndemolist[ 6 ]= 7\ndemolist[ 7 ]= everything\nAfter 'the universe' was removed demolist is now:\n['life', 42, 'universe', 6, 'and', 7, 'everything']\n'life' was found in demolist\n'amoeba' was not found in demolist\nint_list is [42, 6, 7]\nThe sorted int_list is [6, 7, 42]",
"_____no_output_____"
]
],
[
[
"\n\nThis example uses a whole bunch of new functions. Notice that you can\njust print a whole list. Next the append function is used\nto add a new item to the end of the list. `len` returns how many\nitems are in a list. The valid indexes (as in numbers that can be\nused inside of the []) of a list range from 0 to len - 1. The\nindex function tell where the first location of an item is\nlocated in a list. Notice how `demolist.index(42)` returns 1 and\nwhen `demolist[1]` is run it returns 42. The line\n`#Next we will loop through the list` is a just a reminder to the\nprogrammer (also called a comment). Python will ignore any lines that\nstart with a `#`. Next the lines:\n",
"_____no_output_____"
]
],
[
[
"c = 0\nwhile c < len(demolist):\n print('demolist[', c, ']=', demolist[c])\n c = c + 1",
"_____no_output_____"
]
],
[
[
"\nThis creates a variable c which starts at 0 and is incremented until it reaches the last index of the list. Meanwhile the print function prints out each element of the list.\n\n\nThe `del` command can be used to remove a given element in a list. The next few lines use the in operator to test if a element is in or is not in a list.\n\n\nThe `sort` function sorts the list. This is useful if you need a\nlist in order from smallest number to largest or alphabetical. Note\nthat this rearranges the list. Note also that the numbers were put in\na new list, and that was sorted, instead of trying to sort a mixed\nlist. Sorting numbers and strings does not really make sense and results\nin an error.\n\n\nIn summary for a list the following operations exist:\n\n\n\n\n| | | | |\n| --- | --- | --- | --- |\n| example | explanation | | |\n| list[2] | accesses the element at index 2 | | |\n| list[2] = 3 | sets the element at index 2 to be 3 | | |\n| del list[2] | removes the element at index 2 | | |\n| len(list) | returns the length of list | | |\n| \"value\" in list | is true if \"value\" is an element in list | | |\n| \"value\" not in list | is true if \"value\" is not an element in list | | |\n| list.sort() | sorts list | | |\n| list.index(\"value\") | returns the index of the first place that \"value\" occurs | | |\n| list.append(\"value\") | adds an element \"value\" at the end of the list | | |\n\n\n\nThis next example uses these features in a more useful way:\n",
"_____no_output_____"
]
],
[
[
"menu_item = 0\nlist = []\nwhile menu_item != 9:\n print(\"--------------------\")\n print(\"1. Print the list\")\n print(\"2. Add a name to the list\")\n print(\"3. Remove a name from the list\")\n print(\"4. Change an item in the list\")\n print(\"9. Quit\")\n menu_item = int(input(\"Pick an item from the menu: \"))\n if menu_item == 1:\n current = 0\n if len(list) > 0:\n while current < len(list):\n print(current, \". \", list[current])\n current = current + 1\n else:\n print(\"List is empty\")\n elif menu_item == 2:\n name = input(\"Type in a name to add: \")\n list.append(name)\n elif menu_item == 3:\n del_name = input(\"What name would you like to remove: \")\n if del_name in list:\n item_number = list.index(del_name)\n del list[item_number]\n #The code above only removes the first occurance of\n # the name. The code below from Gerald removes all.\n #while del_name in list:\n # item_number = list.index(del_name)\n # del list[item_number]\n else:\n print(del_name, \" was not found\")\n elif menu_item == 4:\n old_name = input(\"What name would you like to change: \")\n if old_name in list:\n item_number = list.index(old_name)\n new_name = input(\"What is the new name: \")\n list[item_number] = new_name\n else:\n print(old_name, \" was not found\")\nprint(\"Goodbye\")",
"_____no_output_____"
]
],
[
[
"\n\nAnd here is part of the output:\n",
"_____no_output_____"
]
],
[
[
"--------------------\n1. Print the list\n2. Add a name to the list\n3. Remove a name from the list\n4. Change an item in the list\n9. Quit",
"_____no_output_____"
]
],
[
[
"\n",
"_____no_output_____"
]
],
[
[
"Pick an item from the menu: 2\nType in a name to add: Jack\n\nPick an item from the menu: 2\nType in a name to add: Jill\n\nPick an item from the menu: 1\n0 . Jack\n1 . Jill\n\nPick an item from the menu: 3\nWhat name would you like to remove: Jack\n\nPick an item from the menu: 4\nWhat name would you like to change: Jill\nWhat is the new name: Jill Peters\n\nPick an item from the menu: 1\n0 . Jill Peters\n\nPick an item from the menu: 9\nGoodbye",
"_____no_output_____"
]
],
[
[
"\n\nThat was a long program. Let's take a look at the source code. The line `list = []` makes the variable list a list with no items (or elements). The next important line is `while menu_item != 9:` . This line starts a loop that allows the menu system for this program. The next few lines display a menu and decide which part of the program to run.\n\n\nThe section:\n",
"_____no_output_____"
]
],
[
[
"current = 0\nif len(list) > 0:\n while current < len(list):\n print(current, \". \", list[current])\n current = current + 1\nelse:\n print(\"List is empty\")",
"_____no_output_____"
]
],
[
[
"\ngoes through the list and prints each name. `len(list_name)` tell how many items are in a list. If len returns `0` then the list is empty.\n\n\nThen a few lines later the statement `list.append(name)` appears. It uses the append function to add a item to the end of the list. Jump down another two lines and notice this section of code:\n",
"_____no_output_____"
]
],
[
[
"item_number = list.index(del_name)\ndel list[item_number]",
"_____no_output_____"
]
],
[
[
"\nHere the index function is used to find the index value that will be used later to remove the item. `del list[item_number]` is used to remove a element of the list.\n\n\nThe next section\n",
"_____no_output_____"
]
],
[
[
"old_name = input(\"What name would you like to change: \")\nif old_name in list:\n item_number = list.index(old_name)\n new_name = input(\"What is the new name: \")\n list[item_number] = new_name\nelse:\n print(old_name, \" was not found\")",
"_____no_output_____"
]
],
[
[
"\nuses index to find the `item_number` and then puts `new_name` where the `old_name` was.\n\n\nCongratulations, with lists under your belt, you now know enough of the language\nthat you could do any computations that a computer can do (this is technically known as Turing-Completeness). Of course, there are still many features that\nare used to make your life easier. \n\n\nExamples\n========\n\n\n\ntest.py\n",
"_____no_output_____"
]
],
[
[
"## This program runs a test of knowledge\n\n# First get the test questions\n# Later this will be modified to use file io.\ndef get_questions():\n # notice how the data is stored as a list of lists\n return [[\"What color is the daytime sky on a clear day?\", \"blue\"],\\\n [\"What is the answer to life, the universe and everything?\", \"42\"],\\\n [\"What is a three letter word for mouse trap?\", \"cat\"]]\n\n# This will test a single question\n# it takes a single question in\n# it returns true if the user typed the correct answer, otherwise false\ndef check_question(question_and_answer):\n #extract the question and the answer from the list\n question = question_and_answer[0]\n answer = question_and_answer[1]\n # give the question to the user\n given_answer = input(question)\n # compare the user's answer to the tester's answer\n if answer == given_answer:\n print(\"Correct\")\n return True\n else:\n print(\"Incorrect, correct was:\", answer)\n return False\n\n# This will run through all the questions\ndef run_test(questions):\n if len(questions) == 0:\n print(\"No questions were given.\")\n # the return exits the function\n return\n index = 0\n right = 0\n while index < len(questions):\n #Check the question\n if check_question(questions[index]):\n right = right + 1\n #go to the next question\n index = index + 1\n #notice the order of the computation, first multiply, then divide\n print(\"You got \", right*100//len(questions), \"% right out of\", len(questions))\n\n#now lets run the questions\nrun_test(get_questions())",
"_____no_output_____"
]
],
[
[
"\n\nSample Output:\n",
"_____no_output_____"
]
],
[
[
"What color is the daytime sky on a clear day?green\nIncorrect, correct was: blue\nWhat is the answer to life, the universe and everything?42\nCorrect\nWhat is a three letter word for mouse trap?cat\nCorrect\nYou got 66 % right out of 3",
"_____no_output_____"
]
],
[
[
"\n\nExercises\n=========\n\n\n\nExpand the test.py program so it has menu giving the option of taking\nthe test, viewing the list of questions and answers, and an option to\nQuit. Also, add a new question to ask, “What noise does a truly\nadvanced machine make?” with the answer of “ping”.\n",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"raw",
"markdown",
"code",
"markdown",
"raw",
"markdown",
"code",
"markdown",
"code",
"markdown",
"raw",
"markdown",
"raw",
"markdown",
"raw",
"markdown",
"raw",
"markdown",
"raw",
"markdown",
"code",
"markdown",
"raw",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"raw"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"raw"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"raw"
],
[
"markdown"
],
[
"raw"
],
[
"markdown"
],
[
"raw"
],
[
"markdown"
],
[
"raw"
],
[
"markdown"
],
[
"raw"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"raw"
],
[
"markdown"
]
] |
cb8b1091e7b54ce77f4f78f5ff3bf19c9cced9b6
| 37,088 |
ipynb
|
Jupyter Notebook
|
polynomial-design/notebooks/RST-example-hydropower-dam.ipynb
|
kjartan-at-tec/mr2007-computerized-control
|
16e35f5007f53870eaf344eea1165507505ab4aa
|
[
"MIT"
] | 2 |
2020-11-07T05:20:37.000Z
|
2020-12-22T09:46:13.000Z
|
polynomial-design/notebooks/RST-example-hydropower-dam.ipynb
|
alfkjartan/control-computarizado
|
5b9a3ae67602d131adf0b306f3ffce7a4914bf8e
|
[
"MIT"
] | 4 |
2020-06-12T20:44:41.000Z
|
2020-06-12T20:49:00.000Z
|
polynomial-design/notebooks/RST-example-hydropower-dam.ipynb
|
alfkjartan/control-computarizado
|
5b9a3ae67602d131adf0b306f3ffce7a4914bf8e
|
[
"MIT"
] | 1 |
2019-09-25T20:02:23.000Z
|
2019-09-25T20:02:23.000Z
| 92.258706 | 8,892 | 0.82005 |
[
[
[
"# Control of a hydropower dam\nConsider a hydropower plant with a dam. We want to control the flow through the dam gates in order to keep the amount of water at a desired level. \n<p><img src=\"hydropowerdam-wikipedia.png\" alt=\"Hydro power from Wikipedia\" width=\"400\"></p>\nThe system is a typical integrator, and is given by the difference equation\n$$ y(k+1) = y(k) + b_uu(k) - b_vv(k), $$\nwhere $x$ is the deviation of the water level from a reference level, $u$ is the change in the flow through the dam gates. A positive value of $u$ corresponds to less flow through the gates, relative to an operating point. The flow $v$ corresponds to changes in the flow in (from river) or out (through power plant). \nThe pulse transfer function of the dam is thus $$H(z) = \\frac{b_u}{z-1}.$$\nWe want to control the system using a two-degree-of-freedom controller, including an anti-aliasing filter modelled as a delay of one sampling period. This gives the block diagram <p><img src=\"2dof-block-integrator.png\" alt=\"Block diagram\" width=\"700\"></p>\nThe desired closed-loop system from the command signal $u_c$ to the output $y$ should have poles in $z=0.7$, and any observer poles should be chosen faster than the closed-loop poles, say in $z=0.5$.",
"_____no_output_____"
],
[
"## The closed-loop pulse-transfer functions\nWith $F_b(z) = \\frac{S(z)}{R(z)}$ and $F_f(z) = \\frac{T(z)}{R(z)}$, and using Mason's rule, we get that the closed-loop pulse-transfer function from command signal $u_c$ to output $y$ becomes\n$$G_c(z) = \\frac{\\frac{T(z)}{R(z)}\\frac{b_u}{z-1}}{1 + \\frac{S(z)}{R(z)} \\frac{b_u}{(z-1)z}} = \\frac{b_uzT(z)}{z(z-1)R(z) + b_uS(z)}.$$\nThe closed-loop transfer function from disturbance to output becomes\n$$G_{cv}(z) = \\frac{\\frac{b_v}{z-1}}{1 + \\frac{S(z)}{R(z)} \\frac{b_u}{(z-1)z}} = \\frac{b_vzR(z)}{z(z-1)R(z) + b_uS(z)}.$$",
"_____no_output_____"
],
[
"## The Diophantine equation\nThe diophantine equation becomes\n$$z(z-1)R(z) + b_uS(z) = A_c(z)A_o(z)$$ We want to find the smallest order controller that can satisfy the Diophantine equation. Since the feedback controller is given by \n$$ F_b(z) = \\frac{s_0z^n + s_1z^{n-1} + \\cdots + s_n}{z^n + r_1z^{n-1} + \\cdots + r_n}$$ and has $2\\deg R + 1$ unknown parameters, and since we should choose the order of the Diphantine equation to be the same as the number of unknown parameters, we get \n$$ \\deg \\big((z(z-1)R(z) + b_uS(z)\\big) = \\deg R + 2 = 2\\deg R + 1 \\quad \\Rightarrow \\quad \\deg R = n = 1.$$",
"_____no_output_____"
],
[
"The Diophantine equation thus becomes\n$$ z(z-1)(z+r_1) + b_u(s_0z+s_1) = (z-0.7)^2(z-0.5), $$\nwhere $A_o(z) = z-0.5$ is the observer polynomial. Working out the expressions on both sides gives\n$$ z^3-(1-r_1)z^2 -r_1 z + b_us_0z + b_us_1 = (z^2 - 1.4z + 0.49)(z-0.5)$$\n$$ z^3 -(1-r_1)z^2 +(b_us_0-r_1)z + b_us_1 = z^3 - (1.4+0.5)z^2 + (0.49+0.7)z -0.245$$\nFrom the Diophantine equation we get the following equations in the unknowns\n\\begin{align}\nz^2: &\\quad 1-r_1 = 1.9\\\\\nz^1: &\\quad b_us_0 - r_1 = 1.19\\\\\nz^0: &\\quad b_us_1 = -0.245\n\\end{align}\nThis is a linear system of equations in the unknown, and can be solved in many different ways. Here we see that with simple substitution we find\n\\begin{align}\nr_1 &= 1-1.9 = -0.9\\\\\ns_0 &= \\frac{1}{b_u}(1.19+r_1) = \\frac{0.29}{b_u}\\\\\ns_1 &= -\\frac{0.245}{b_u}\n\\end{align}",
"_____no_output_____"
],
[
"## The feedforward \nWe set $T(z) = t_0A_o(z)$ which gives the closed-loop pulse-transfer function\n$$G_c(z) = \\frac{b_uzT(z)}{z(z-1)R(z) + b_uS(z)}= \\frac{b_ut_0zA_o(z)}{A_c(z)A_o(z)} = \\frac{b_u t_0z}{A_c(z)}$$\nIn order for this pulse-transfer function to have unit DC-gain (static gain) we must have $G_c(1) = 1$, or \n$$ \\frac{b_ut_0}{A_c(1)} = 1. $$\nThe solution is \n$$ t_0 = \\frac{A_c(1)}{b_u} = \\frac{(1-0.7)^2}{b_u} = \\frac{0.3^2}{b_u}. $$",
"_____no_output_____"
],
[
"## Verify by symbolic computer algebra",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport sympy as sy",
"_____no_output_____"
],
[
"z = sy.symbols('z', real=False)\nbu,r1,s0,s1 = sy.symbols('bu,r1,s0,s1', real=True)\npc,po = sy.symbols('pc,po', real=True) # Closed-loop pole and observer pole",
"_____no_output_____"
],
[
"# The polynomials\nAp = sy.Poly(z*(z-1), z)\nBp = sy.Poly(bu,z)\nRp = sy.Poly(z+r1, z)\nSp = sy.Poly(s0*z+s1, z)\nAc = sy.Poly((z-pc)**2, z)\nAo = sy.Poly(z-po, z)",
"_____no_output_____"
],
[
"# The diophantine eqn\ndioph = Ap*Rp + Bp*Sp - Ac*Ao\n# Form system of eqs from coefficients, then solve\ndioph_coeffs = dioph.all_coeffs()\n\n# Solve for r1, s0 and s1, \nsol = sy.solve(dioph_coeffs, (r1,s0,s1))\nprint('r_1 = %s' % sol[r1])\nprint('s_0 = %s' % sol[s0])\nprint('s_1 = %s' % sol[s1])",
"r_1 = -2*pc - po + 1\ns_0 = (pc**2 + 2*pc*po - 2*pc - po + 1)/bu\ns_1 = -pc**2*po/bu\n"
],
[
"# Substitute values for the desired closed-loop pole and observer pole\nsubstitutions = [(pc, 0.7), (po, 0.5)]\nprint('r_1 = %s' % sol[r1].subs(substitutions))\nprint('s_0 = %s' % sol[s0].subs(substitutions))\nprint('s_1 = %s' % sol[s1].subs(substitutions))",
"r_1 = -0.900000000000000\ns_0 = 0.29/bu\ns_1 = -0.245/bu\n"
],
[
"# The forward controller\nt0 = (Ac.eval(1)/Bp.eval(1))\nprint('t_0 = %s' % t0)\nprint('t_0 = %s' % t0.subs(substitutions))",
"t_0 = (pc**2 - 2*pc + 1)/bu\nt_0 = 0.0900000000000001/bu\n"
]
],
[
[
"## Requirements on the closed-loop poles and observer poles in order to obtain stable controller\nNotice the solution for the controller denominator\n$$ R(z) = z+r_1 = z -2p_c -p_o + 1, $$\nwhere $0\\ge p_c<1$ is the desired closed-loop pole and $0 \\ge p_o<1$ is the observer pole. Sketch in the $(p_c, p_o)$-plane the region which will give a stable controller $F_b(z) = \\frac{S(z)}{R(z)}$!",
"_____no_output_____"
],
[
"## Simulate a particular case\nLet $b_u=1$, $p_c = p_o = \\frac{2}{3}$. Analyze the closed-loop system by simulation",
"_____no_output_____"
]
],
[
[
"import control\nimport control.matlab as cm\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"sbs = [(bu, 1), (pc, 2.0/3.0), (po, 2.0/3.0)]\nRcoeffs = [1, float(sol[r1].subs(sbs))]\nScoeffs = [float(sol[s0].subs(sbs)), float(sol[s1].subs(sbs))]\nTcoeffs = float(t0.subs(sbs))*np.array([1, float(pc.subs(sbs))])\nAcoeffs = [1, -1]\n\nH = cm.tf(float(bu.subs(sbs)), Acoeffs, 1)\nFf = cm.tf(Tcoeffs, Rcoeffs, 1)\nFb = cm.tf(Scoeffs, Rcoeffs, 1)\nHaa = cm.tf(1, [1, 0], 1) # The delay due to the anti-aliasing filter\nGc = cm.minreal(Ff*cm.feedback(H, Haa*Fb ))\nGcv = cm.feedback(H, Haa*Fb)\n\n# Pulse trf fcn from command signal to control signal\nGcu = Ff*cm.feedback(1, H*Haa*Fb)",
"1 states have been removed from the model\n"
],
[
"cm.pzmap(Fb)",
"_____no_output_____"
],
[
"tvec = np.arange(40)\n(t1, y1) = control.step_response(Gc,tvec)\nplt.figure(figsize=(14,4))\nplt.step(t1, y1[0])\nplt.xlabel('k')\nplt.ylabel('y')\nplt.title('Output')",
"_____no_output_____"
],
[
"(t1, y1) = control.step_response(Gcv,tvec)\nplt.figure(figsize=(14,4))\nplt.step(t1, y1[0])\nplt.xlabel('k')\nplt.ylabel('y')\nplt.title('Output')",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
cb8b187d11965a744d3fe0bc4c9c003bf989e069
| 2,813 |
ipynb
|
Jupyter Notebook
|
30septiembre.ipynb
|
pandemicbat801/daa_2021_1
|
4b912d0ca5631882f8137583dfbc25280ec8c574
|
[
"MIT"
] | null | null | null |
30septiembre.ipynb
|
pandemicbat801/daa_2021_1
|
4b912d0ca5631882f8137583dfbc25280ec8c574
|
[
"MIT"
] | null | null | null |
30septiembre.ipynb
|
pandemicbat801/daa_2021_1
|
4b912d0ca5631882f8137583dfbc25280ec8c574
|
[
"MIT"
] | null | null | null | 28.13 | 236 | 0.453964 |
[
[
[
"<a href=\"https://colab.research.google.com/github/pandemicbat801/daa_2021_1/blob/master/30septiembre.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"#Palindromos\nEs una palabra que se lee de igual formade un sentido y de sentido inverso, ejemplo:\n1. sugus\n2. reconocer\n3. oso\n4. 10101\n5. 10:01\netc.\n\n###Planteamiento del problema\nSe deseada encontrar todos los palindromos quese encuentran en un dia completo tomando como horario inicial las 00:00 horas y como horario final las 23:59 horas. \n\nEl algortimodebe mostrar en pantalla todos los algoritmos exixtentes en ese rango, al final debe mostrar el conteo de todos los palindromos existentes.\n",
"_____no_output_____"
]
],
[
[
"horas=\"0000\"\ncont=0\nwhile horas!=\"2359\":\n\n inv=horas[::-1]\n if horas==inv and int(horas[2])<=5:\n cont+=1\n print(horas[0:2],\":\",horas[2:4])\n new=int(horas)\n new+=1\n horas=str(new).zfill(4)\nprint(\"son \",cont,\"palindromos\")",
"00 : 00\n01 : 10\n02 : 20\n03 : 30\n04 : 40\n05 : 50\n10 : 01\n11 : 11\n12 : 21\n13 : 31\n14 : 41\n15 : 51\n20 : 02\n21 : 12\n22 : 22\n23 : 32\nson 16 palindromos\n"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
]
] |
cb8b297522ff26a64fb105aac013d81c09ba5ca9
| 329,478 |
ipynb
|
Jupyter Notebook
|
I Python Basics & Pandas/06_Applied For Loops vs DataFrame Functionalities/.ipynb_checkpoints/06session_pandas-vs-for_loop-checkpoint.ipynb
|
iamoespana92/machine-learning-program
|
a6c6dac18d41b3795685fcb3dd358ab1f64a4ee4
|
[
"MIT"
] | null | null | null |
I Python Basics & Pandas/06_Applied For Loops vs DataFrame Functionalities/.ipynb_checkpoints/06session_pandas-vs-for_loop-checkpoint.ipynb
|
iamoespana92/machine-learning-program
|
a6c6dac18d41b3795685fcb3dd358ab1f64a4ee4
|
[
"MIT"
] | null | null | null |
I Python Basics & Pandas/06_Applied For Loops vs DataFrame Functionalities/.ipynb_checkpoints/06session_pandas-vs-for_loop-checkpoint.ipynb
|
iamoespana92/machine-learning-program
|
a6c6dac18d41b3795685fcb3dd358ab1f64a4ee4
|
[
"MIT"
] | null | null | null | 44.463968 | 21,980 | 0.596887 |
[
[
[
"import pandas as pd",
"_____no_output_____"
],
[
"df = pd.read_csv(filepath_or_buffer='https://raw.githubusercontent.com/justmarkham/DAT8/master/data/chipotle.tsv', sep='\\t').iloc[:100,:]",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
]
],
[
[
"## Cuantos pedidos por cada orden?",
"_____no_output_____"
]
],
[
[
"mask = df['order_id'] == 1",
"_____no_output_____"
],
[
"df[mask]",
"_____no_output_____"
],
[
"df[mask].quantity",
"_____no_output_____"
],
[
"df[mask].quantity.sum()",
"_____no_output_____"
],
[
"mask = df['order_id'] == 2",
"_____no_output_____"
],
[
"df[mask]",
"_____no_output_____"
],
[
"df[mask].quantity",
"_____no_output_____"
],
[
"df[mask].quantity.sum()",
"_____no_output_____"
],
[
"mask = df['order_id'] == 3\n\ndf[mask]\n\ndf[mask].quantity\n\ndf[mask].quantity.sum()",
"_____no_output_____"
],
[
"mask = df['order_id'] == pepa\n\ndf[mask]\n\ndf[mask].quantity\n\ndf[mask].quantity.sum()",
"_____no_output_____"
],
[
"for pepa in [1,2,3]:\n\n mask = df['order_id'] == pepa\n\n df[mask]\n\n df[mask].quantity\n\n df[mask].quantity.sum()",
"_____no_output_____"
],
[
"for pepa in [1,2,3, 4, 5, 6,7,8,9]: # pepa = 1, # pepa = 2\n\n mask = df['order_id'] == pepa # mask = df['order_id'] == 1, mask = df['order_id'] == 2\n\n print(df[mask].quantity.sum())",
"4\n2\n2\n2\n2\n2\n2\n2\n3\n"
],
[
"n_productos_pedidos = []",
"_____no_output_____"
],
[
"for pepa in [1,2,3,4,5,6,7,8,9]: # pepa = 1, # pepa = 2\n\n mask = df['order_id'] == pepa # mask = df['order_id'] == 1, mask = df['order_id'] == 2\n\n n_productos_pedidos.append(df[mask].quantity.sum())",
"_____no_output_____"
],
[
"n_productos_pedidos",
"_____no_output_____"
],
[
"n_productos_pedidos = []",
"_____no_output_____"
],
[
"for pepa in [1,2,3,4,5,6,7,8,9]: # pepa = 1, # pepa = 2\n\n mask = df['order_id'] == pepa # mask = df['order_id'] == 1, mask = df['order_id'] == 2\n\n n_productos_pedidos.append(df[mask])",
"_____no_output_____"
],
[
"n_productos_pedidos[0]",
"_____no_output_____"
],
[
"n_productos_pedidos[1]",
"_____no_output_____"
],
[
"n_productos_pedidos[2]",
"_____no_output_____"
],
[
"n_productos_pedidos[3]",
"_____no_output_____"
],
[
"n_productos_pedidos[4]",
"_____no_output_____"
],
[
"n_productos_pedidos",
"_____no_output_____"
],
[
"dic['order 1'] = 5",
"_____no_output_____"
],
[
"dic",
"_____no_output_____"
],
[
"dic['order 2'] = 3",
"_____no_output_____"
],
[
"dic",
"_____no_output_____"
],
[
"dic_pedidos = {}",
"_____no_output_____"
],
[
"dic_pedidos",
"_____no_output_____"
],
[
"dic_pedidos['order 1'] = 4",
"_____no_output_____"
],
[
"dic_pedidos",
"_____no_output_____"
],
[
"dic_pedidos[1] = 4",
"_____no_output_____"
],
[
"dic_pedidos",
"_____no_output_____"
],
[
"dic_pedidos['clave'] = 89",
"_____no_output_____"
],
[
"dic_pedidos = {}",
"_____no_output_____"
],
[
"for pepa in [1,2,3,4,5,6,7,8,9]: # pepa = 1, # pepa = 2\n\n mask = df['order_id'] == pepa # mask = df['order_id'] == 1, mask = df['order_id'] == 2\n \n dic_pedidos[pepa] = df[mask].quantity.sum()",
"_____no_output_____"
],
[
"dic_pedidos",
"_____no_output_____"
],
[
"df",
"_____no_output_____"
],
[
"dic_pedidos = {}",
"_____no_output_____"
],
[
"for pepa in [1,2,3,4,5,6,7,8,9]: # pepa = 1, # pepa = 2\n\n mask = df['order_id'] == pepa # mask = df['order_id'] == 1, mask = df['order_id'] == 2\n \n dic_pedidos[pepa] = df[mask].quantity.sum()",
"_____no_output_____"
],
[
"dic_pedidos",
"_____no_output_____"
],
[
"df",
"_____no_output_____"
],
[
"df['order_id']",
"_____no_output_____"
],
[
"pepas = df['order_id'].unique()",
"_____no_output_____"
],
[
"for pepa in pepas:",
"_____no_output_____"
],
[
"dic_pedidos = {}",
"_____no_output_____"
],
[
"for pepa in array([ 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17,\n 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34,\n 35, 36, 37, 38, 39, 40, 41, 42, 43, 44]): # pepa = 1, # pepa = 2\n\n mask = df['order_id'] == pepa # mask = df['order_id'] == 1, mask = df['order_id'] == 2\n \n dic_pedidos[pepa] = df[mask].quantity.sum()",
"_____no_output_____"
],
[
"dic_pedidos",
"_____no_output_____"
],
[
"dic_pedidos = {}",
"_____no_output_____"
],
[
"for pepa in df['order_id'].unique(): # pepa = 1, # pepa = 2\n\n mask = df['order_id'] == pepa # mask = df['order_id'] == 1, mask = df['order_id'] == 2\n \n dic_pedidos[pepa] = df[mask].quantity.sum()",
"_____no_output_____"
],
[
"dic_pedidos",
"_____no_output_____"
],
[
"dfg = df.groupby('order_id')",
"_____no_output_____"
],
[
"dfg.get_group(1).quantity.sum()",
"_____no_output_____"
],
[
"dfg.get_group(2)",
"_____no_output_____"
],
[
"df.quantity",
"_____no_output_____"
],
[
"for i in ...",
"_____no_output_____"
],
[
"import seaborn as sns",
"_____no_output_____"
],
[
"df = sns.load_dataset('mpg')",
"_____no_output_____"
],
[
"df",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"plt.scatter(x='weight', y='mpg', data=df)",
"_____no_output_____"
],
[
"mask_usa = df.origin == 'usa'",
"_____no_output_____"
],
[
"mask_japan = df.origin == 'japan'",
"_____no_output_____"
],
[
"plt.scatter(x='weight', y='mpg', data=df[mask_usa])\nplt.scatter(x='weight', y='mpg', data=df[mask_japan])",
"_____no_output_____"
],
[
"'x'",
"_____no_output_____"
],
[
"x",
"_____no_output_____"
],
[
"mask = df.origin == x\n\nplt.scatter(x='weight', y='mpg', data=df[mask])",
"_____no_output_____"
],
[
"for x in ['usa','japan']:\n\n mask = df.origin == x\n\n plt.scatter(x='weight', y='mpg', data=df[mask])",
"_____no_output_____"
],
[
"df",
"_____no_output_____"
],
[
"paises = df.origin.unique()",
"_____no_output_____"
],
[
"paises",
"_____no_output_____"
],
[
"for x in paises:\n\n mask = df.origin == x\n\n plt.scatter(x='weight', y='mpg', data=df[mask])",
"_____no_output_____"
],
[
"for x in df.cylinders.unique():\n\n mask = df.cylinders == x\n\n plt.scatter(x='weight', y='mpg', data=df[mask])",
"_____no_output_____"
],
[
"dic = {}",
"_____no_output_____"
],
[
"for x in df.cylinders.unique():\n\n mask = df.cylinders == x\n\n plt.scatter(x='weight', y='mpg', data=df[mask])\n \n dic[x] = len(df[mask].cylinders.unique())",
"_____no_output_____"
],
[
"dic",
"_____no_output_____"
],
[
"dfsel = df[df['order_id'] == 1]",
"_____no_output_____"
],
[
"n_pedidos = dfsel.shape[0]",
"_____no_output_____"
],
[
"dfsel",
"_____no_output_____"
],
[
"dfsel = df[df['order_id'] == 2]",
"_____no_output_____"
],
[
"n_pedidos = dfsel.shape[0]",
"_____no_output_____"
],
[
"dfsel",
"_____no_output_____"
],
[
"dfsel = df[df['order_id'] == 3]",
"_____no_output_____"
],
[
"n_pedidos = dfsel.shape[0]",
"_____no_output_____"
],
[
"dfsel",
"_____no_output_____"
],
[
"dic_pedidos = {}",
"_____no_output_____"
],
[
"dfsel = df[df['order_id'] == 1]",
"_____no_output_____"
],
[
"n_pedidos = dfsel.shape[0]",
"_____no_output_____"
],
[
"dfsel",
"_____no_output_____"
],
[
"dic_pedidos[1] = n_pedidos",
"_____no_output_____"
],
[
"dic_pedidos",
"_____no_output_____"
],
[
"dfsel = df[df['order_id'] == 2]",
"_____no_output_____"
],
[
"n_pedidos = dfsel.shape[0]",
"_____no_output_____"
],
[
"dfsel",
"_____no_output_____"
],
[
"dic_pedidos[2] = n_pedidos",
"_____no_output_____"
],
[
"dic_pedidos",
"_____no_output_____"
],
[
"dfsel = df[df['order_id'] == 3]",
"_____no_output_____"
],
[
"n_pedidos = dfsel.shape[0]",
"_____no_output_____"
],
[
"dfsel",
"_____no_output_____"
],
[
"dic_pedidos[3] = n_pedidos",
"_____no_output_____"
],
[
"dic_pedidos",
"_____no_output_____"
],
[
"dic_pedidos = {}",
"_____no_output_____"
],
[
"dfsel = df[df['order_id'] == 3]\n\nn_pedidos = dfsel.shape[0]\n\ndfsel\n\ndic_pedidos[3] = n_pedidos\n\ndic_pedidos",
"_____no_output_____"
],
[
"dic_pedidos = {}",
"_____no_output_____"
],
[
"dfsel = df[df['order_id'] == pepa]\n\nn_pedidos = dfsel.shape[0]\n\ndfsel\n\ndic_pedidos[pepa] = n_pedidos\n\ndic_pedidos",
"_____no_output_____"
],
[
"dic_pedidos = {}",
"_____no_output_____"
],
[
"for pepa in [1,2,3]:\n dfsel = df[df['order_id'] == pepa]\n\n n_pedidos = dfsel.shape[0]\n\n dfsel\n\n dic_pedidos[pepa] = n_pedidos\n\n dic_pedidos",
"_____no_output_____"
],
[
"dic_pedidos",
"_____no_output_____"
],
[
"df",
"_____no_output_____"
],
[
"df.order_id",
"_____no_output_____"
],
[
"pedidos = df.order_id.unique()",
"_____no_output_____"
],
[
"dic_pedidos = {}",
"_____no_output_____"
],
[
"for pepa in pedidos:\n dfsel = df[df['order_id'] == pepa]\n\n n_pedidos = dfsel.shape[0]\n\n dfsel\n\n dic_pedidos[pepa] = n_pedidos\n\n dic_pedidos",
"_____no_output_____"
],
[
"dic_pedidos",
"_____no_output_____"
],
[
"pd.Series(dic_pedidos)",
"_____no_output_____"
],
[
"df.order_id.value_counts()",
"_____no_output_____"
],
[
"df.order_id.value_counts(sort=False)",
"_____no_output_____"
],
[
"df.order_id.value_counts(sort=False)",
"_____no_output_____"
],
[
"dic_pedidos = {}\n\nfor pepa in pedidos:\n dfsel = df[df['order_id'] == pepa]\n n_pedidos = dfsel.shape[0]\n dic_pedidos[pepa] = n_pedidos\n\npd.Series(dic_pedidos)",
"_____no_output_____"
],
[
"df.item_price.sum()",
"_____no_output_____"
],
[
"for i in df.item_price:\n print(i)",
"$2.39 \n$3.39 \n$3.39 \n$2.39 \n$16.98 \n$10.98 \n$1.69 \n$11.75 \n$9.25 \n$9.25 \n$4.45 \n$8.75 \n$8.75 \n$11.25 \n$4.45 \n$2.39 \n$8.49 \n$8.49 \n$2.18 \n$8.75 \n$4.45 \n$8.99 \n$3.39 \n$10.98 \n$3.39 \n$2.39 \n$8.49 \n$8.99 \n$1.09 \n$8.49 \n$2.39 \n$8.99 \n$1.69 \n$8.99 \n$1.09 \n$8.75 \n$8.75 \n$4.45 \n$2.95 \n$11.75 \n$2.15 \n$4.45 \n$11.25 \n$11.75 \n$8.75 \n$10.98 \n$8.99 \n$3.39 \n$8.99 \n$3.99 \n$8.99 \n$2.18 \n$10.98 \n$1.09 \n$8.99 \n$2.39 \n$9.25 \n$11.25 \n$11.75 \n$2.15 \n$4.45 \n$9.25 \n$11.25 \n$8.75 \n$8.99 \n$8.99 \n$3.39 \n$8.99 \n$10.98 \n$8.99 \n$1.69 \n$8.99 \n$3.99 \n$8.75 \n$4.45 \n$8.75 \n$8.75 \n$2.15 \n$8.75 \n$11.25 \n$2.15 \n$9.25 \n$8.75 \n$8.75 \n$9.25 \n$8.49 \n$8.99 \n$1.09 \n$9.25 \n$2.95 \n$11.75 \n$11.75 \n$9.25 \n$11.75 \n$4.45 \n$9.25 \n$4.45 \n$11.75 \n$8.75 \n$8.75 \n"
],
[
"i = '$2.39 '",
"_____no_output_____"
],
[
"clean_i = i.replace('$', '').replace(' ', '')",
"_____no_output_____"
],
[
"float_i = float(clean_i)",
"_____no_output_____"
],
[
"float_i",
"_____no_output_____"
],
[
"i = '$2.39 '\n\nclean_i = i.replace('$', '').replace(' ', '')\n\nfloat_i = float(clean_i)\n\nfloat_i",
"_____no_output_____"
],
[
" i = '$2.39 '\n\n clean_i = i.replace('$', '').replace(' ', '')\n\n float_i = float(clean_i)\n\n float_i",
"_____no_output_____"
],
[
"for i in df.item_price:\n \n clean_i = i.replace('$', '').replace(' ', '')\n\n float_i = float(clean_i)\n\n float_i",
"_____no_output_____"
],
[
"float_i",
"_____no_output_____"
],
[
"lista_precios = []",
"_____no_output_____"
],
[
"for i in df.item_price:\n \n clean_i = i.replace('$', '').replace(' ', '')\n\n float_i = float(clean_i)\n\n lista_precios.append(float_i)",
"_____no_output_____"
],
[
"lista_precios",
"_____no_output_____"
],
[
"df['precio'] = lista_precios",
"_____no_output_____"
],
[
"df",
"_____no_output_____"
],
[
"df.item_price.sum()",
"_____no_output_____"
],
[
"df.precio.sum()",
"_____no_output_____"
],
[
"df",
"_____no_output_____"
],
[
"for item in df.choice_description:\n print(item)",
"nan\n[Clementine]\n[Apple]\nnan\n[Tomatillo-Red Chili Salsa (Hot), [Black Beans, Rice, Cheese, Sour Cream]]\n[Fresh Tomato Salsa (Mild), [Rice, Cheese, Sour Cream, Guacamole, Lettuce]]\nnan\n[Tomatillo Red Chili Salsa, [Fajita Vegetables, Black Beans, Pinto Beans, Cheese, Sour Cream, Guacamole, Lettuce]]\n[Tomatillo Green Chili Salsa, [Pinto Beans, Cheese, Sour Cream, Lettuce]]\n[Fresh Tomato Salsa, [Rice, Black Beans, Pinto Beans, Cheese, Sour Cream, Lettuce]]\nnan\n[Roasted Chili Corn Salsa, [Fajita Vegetables, Rice, Black Beans, Cheese, Sour Cream]]\n[Roasted Chili Corn Salsa, [Rice, Black Beans, Cheese, Sour Cream]]\n[Fresh Tomato Salsa, [Fajita Vegetables, Rice, Cheese, Sour Cream, Guacamole]]\nnan\nnan\n[Tomatillo-Green Chili Salsa (Medium), [Pinto Beans, Cheese, Sour Cream]]\n[Fresh Tomato Salsa (Mild), [Black Beans, Rice, Cheese, Sour Cream, Lettuce]]\n[Sprite]\n[Tomatillo Red Chili Salsa, [Fajita Vegetables, Black Beans, Sour Cream, Cheese, Lettuce]]\nnan\n[[Fresh Tomato Salsa (Mild), Tomatillo-Green Chili Salsa (Medium), Tomatillo-Red Chili Salsa (Hot)], [Rice, Cheese, Sour Cream, Lettuce]]\n[Pomegranate Cherry]\n[[Tomatillo-Green Chili Salsa (Medium), Tomatillo-Red Chili Salsa (Hot)], [Pinto Beans, Rice, Cheese, Sour Cream, Guacamole, Lettuce]]\n[Grapefruit]\nnan\n[Roasted Chili Corn Salsa (Medium), [Pinto Beans, Rice, Fajita Veggies, Cheese, Sour Cream, Lettuce]]\n[[Tomatillo-Green Chili Salsa (Medium), Roasted Chili Corn Salsa (Medium)], [Black Beans, Rice, Sour Cream, Lettuce]]\n[Dr. Pepper]\n[Tomatillo-Green Chili Salsa (Medium), [Pinto Beans, Rice, Cheese, Sour Cream]]\nnan\n[[Roasted Chili Corn Salsa (Medium), Fresh Tomato Salsa (Mild)], [Rice, Black Beans, Sour Cream]]\nnan\n[Tomatillo-Red Chili Salsa (Hot), [Black Beans, Rice, Fajita Veggies, Cheese, Sour Cream, Lettuce]]\nnan\n[Roasted Chili Corn Salsa, Rice]\n[Roasted Chili Corn Salsa, [Cheese, Lettuce]]\nnan\nnan\n[Roasted Chili Corn Salsa, [Fajita Vegetables, Rice, Black Beans, Cheese, Sour Cream, Guacamole, Lettuce]]\nnan\nnan\n[Roasted Chili Corn Salsa, [Rice, Black Beans, Cheese, Sour Cream, Guacamole, Lettuce]]\n[Fresh Tomato Salsa, [Rice, Pinto Beans, Cheese, Sour Cream, Guacamole, Lettuce]]\n[Fresh Tomato Salsa, [Fajita Vegetables, Pinto Beans, Lettuce]]\n[Tomatillo-Red Chili Salsa (Hot), [Black Beans, Fajita Veggies, Cheese, Sour Cream, Guacamole, Lettuce]]\n[Tomatillo-Red Chili Salsa (Hot), [Rice, Fajita Veggies, Cheese]]\n[Blackberry]\n[[Fresh Tomato Salsa (Mild), Roasted Chili Corn Salsa (Medium), Tomatillo-Red Chili Salsa (Hot)], [Black Beans, Rice, Sour Cream]]\nnan\n[Roasted Chili Corn Salsa (Medium), [Rice, Fajita Veggies, Cheese, Sour Cream, Lettuce]]\n[Mountain Dew]\n[Roasted Chili Corn Salsa (Medium), [Black Beans, Rice, Fajita Veggies, Cheese, Sour Cream, Guacamole, Lettuce]]\n[Sprite]\n[Fresh Tomato Salsa (Mild), [Black Beans, Rice, Fajita Veggies, Cheese, Sour Cream]]\nnan\n[Fresh Tomato Salsa, [Fajita Vegetables, Black Beans, Lettuce]]\n[Tomatillo Red Chili Salsa, [Fajita Vegetables, Rice, Black Beans, Cheese, Sour Cream, Guacamole, Lettuce]]\n[Roasted Chili Corn Salsa, [Fajita Vegetables, Rice, Black Beans, Cheese, Sour Cream, Guacamole, Lettuce]]\nnan\nnan\n[Fresh Tomato Salsa, Cheese]\n[Fresh Tomato Salsa, [Fajita Vegetables, Rice, Black Beans, Cheese, Sour Cream, Guacamole, Lettuce]]\n[Fresh Tomato Salsa, [Rice, Black Beans, Cheese]]\n[Tomatillo-Red Chili Salsa (Hot), [Black Beans, Rice, Fajita Veggies, Cheese, Sour Cream, Lettuce]]\n[Tomatillo-Red Chili Salsa (Hot), [Rice, Cheese, Sour Cream, Lettuce]]\n[Blackberry]\n[Tomatillo-Red Chili Salsa (Hot), [Rice, Cheese]]\n[Tomatillo-Red Chili Salsa (Hot), [Black Beans, Fajita Veggies, Cheese, Sour Cream, Guacamole, Lettuce]]\n[Roasted Chili Corn Salsa (Medium), [Rice, Black Beans, Sour Cream]]\nnan\n[Tomatillo-Red Chili Salsa (Hot), [Black Beans, Rice, Cheese, Sour Cream, Lettuce]]\nnan\n[Tomatillo Red Chili Salsa, [Rice, Black Beans, Cheese, Sour Cream]]\nnan\n[Tomatillo Red Chili Salsa, [Rice, Cheese, Sour Cream, Lettuce]]\n[Fresh Tomato Salsa, [Rice, Black Beans, Pinto Beans, Cheese, Lettuce]]\nnan\n[Fresh Tomato Salsa, [Rice, Black Beans, Cheese, Sour Cream, Lettuce]]\n[Roasted Chili Corn Salsa, [Rice, Cheese, Lettuce, Guacamole]]\nnan\n[Fresh Tomato Salsa, [Rice, Pinto Beans, Cheese, Sour Cream, Lettuce]]\n[Fresh Tomato Salsa, [Rice, Black Beans, Cheese, Sour Cream, Lettuce]]\n[Tomatillo Green Chili Salsa, [Rice, Black Beans, Sour Cream, Cheese]]\n[Tomatillo Red Chili Salsa]\n[Tomatillo-Red Chili Salsa (Hot), [Pinto Beans, Black Beans, Rice, Fajita Veggies, Lettuce]]\n[Tomatillo-Red Chili Salsa (Hot), [Pinto Beans, Rice, Fajita Veggies, Cheese, Lettuce]]\nnan\n[Fresh Tomato Salsa, [Rice, Pinto Beans, Sour Cream, Cheese]]\nnan\n[Fresh Tomato Salsa, [Rice, Black Beans, Cheese, Sour Cream, Guacamole]]\n[Fresh Tomato Salsa, [Fajita Vegetables, Cheese, Sour Cream, Guacamole]]\n[Fresh Tomato Salsa, Sour Cream]\n[Roasted Chili Corn Salsa, [Sour Cream, Guacamole]]\nnan\n[Fresh Tomato Salsa, [Fajita Vegetables, Rice, Cheese, Sour Cream]]\nnan\n[Fresh Tomato Salsa, [Fajita Vegetables, Rice, Black Beans, Cheese, Guacamole, Lettuce]]\n[Tomatillo Red Chili Salsa, [Rice, Black Beans, Cheese]]\n[Tomatillo Red Chili Salsa, [Rice, Fajita Vegetables, Sour Cream, Lettuce]]\n"
],
[
"item = '[Tomatillo-Red Chili Salsa (Hot), [Black Beans, Rice, Cheese, Sour Cream]]'",
"_____no_output_____"
],
[
"item = item.replace('[', '').replace(']', '')",
"_____no_output_____"
],
[
"item",
"_____no_output_____"
],
[
"lista_item = item.split(', ')",
"_____no_output_____"
],
[
"lista_item",
"_____no_output_____"
],
[
"item = '[Tomatillo-Red Chili Salsa (Hot), [Black Beans, Rice, Cheese, Sour Cream]]'\n\nitem = item.replace('[', '').replace(']', '')\n\nitem\n\nlista_item = item.split(', ')\n\nlista_item",
"_____no_output_____"
],
[
" item = '[Tomatillo-Red Chili Salsa (Hot), [Black Beans, Rice, Cheese, Sour Cream]]'\n\n item = item.replace('[', '').replace(']', '')\n\n item\n\n lista_item = item.split(', ')\n\n lista_item",
"_____no_output_____"
],
[
"for item in df.choice_description:\n \n item = '[Tomatillo-Red Chili Salsa (Hot), [Black Beans, Rice, Cheese, Sour Cream]]'\n\n item = item.replace('[', '').replace(']', '')\n\n item\n\n lista_item = item.split(', ')\n\n lista_item",
"_____no_output_____"
],
[
"for item in df.choice_description:\n\n item = item.replace('[', '').replace(']', '')\n\n item\n\n lista_item = item.split(', ')\n\n lista_item",
"_____no_output_____"
],
[
"for item in df.choice_description:\n \n print(item)\n \n item = item.replace('[', '').replace(']', '')\n\n item\n\n lista_item = item.split(', ')\n\n lista_item",
"nan\n"
],
[
"import numpy as np",
"_____no_output_____"
],
[
"np.nan",
"_____no_output_____"
],
[
"np.nan.replace()",
"_____no_output_____"
],
[
"if type(item) == float:\n print('bingo')\nelse:\n print('nanai')",
"bingo\n"
],
[
"for item in df.choice_description:\n if type(item) == float:\n \n item\n \n else:\n \n item = item.replace('[', '').replace(']', '')\n\n item\n\n lista_item = item.split(', ')\n\n lista_item",
"_____no_output_____"
],
[
"for item in df.choice_description:\n if type(item) == float:\n \n item\n \n else:\n \n item = item.replace('[', '').replace(']', '')\n\n item\n\n lista_item = item.split(', ')\n\n print(lista_item)",
"['Clementine']\n['Apple']\n['Tomatillo-Red Chili Salsa (Hot)', 'Black Beans', 'Rice', 'Cheese', 'Sour Cream']\n['Fresh Tomato Salsa (Mild)', 'Rice', 'Cheese', 'Sour Cream', 'Guacamole', 'Lettuce']\n['Tomatillo Red Chili Salsa', 'Fajita Vegetables', 'Black Beans', 'Pinto Beans', 'Cheese', 'Sour Cream', 'Guacamole', 'Lettuce']\n['Tomatillo Green Chili Salsa', 'Pinto Beans', 'Cheese', 'Sour Cream', 'Lettuce']\n['Fresh Tomato Salsa', 'Rice', 'Black Beans', 'Pinto Beans', 'Cheese', 'Sour Cream', 'Lettuce']\n['Roasted Chili Corn Salsa', 'Fajita Vegetables', 'Rice', 'Black Beans', 'Cheese', 'Sour Cream']\n['Roasted Chili Corn Salsa', 'Rice', 'Black Beans', 'Cheese', 'Sour Cream']\n['Fresh Tomato Salsa', 'Fajita Vegetables', 'Rice', 'Cheese', 'Sour Cream', 'Guacamole']\n['Tomatillo-Green Chili Salsa (Medium)', 'Pinto Beans', 'Cheese', 'Sour Cream']\n['Fresh Tomato Salsa (Mild)', 'Black Beans', 'Rice', 'Cheese', 'Sour Cream', 'Lettuce']\n['Sprite']\n['Tomatillo Red Chili Salsa', 'Fajita Vegetables', 'Black Beans', 'Sour Cream', 'Cheese', 'Lettuce']\n['Fresh Tomato Salsa (Mild)', 'Tomatillo-Green Chili Salsa (Medium)', 'Tomatillo-Red Chili Salsa (Hot)', 'Rice', 'Cheese', 'Sour Cream', 'Lettuce']\n['Pomegranate Cherry']\n['Tomatillo-Green Chili Salsa (Medium)', 'Tomatillo-Red Chili Salsa (Hot)', 'Pinto Beans', 'Rice', 'Cheese', 'Sour Cream', 'Guacamole', 'Lettuce']\n['Grapefruit']\n['Roasted Chili Corn Salsa (Medium)', 'Pinto Beans', 'Rice', 'Fajita Veggies', 'Cheese', 'Sour Cream', 'Lettuce']\n['Tomatillo-Green Chili Salsa (Medium)', 'Roasted Chili Corn Salsa (Medium)', 'Black Beans', 'Rice', 'Sour Cream', 'Lettuce']\n['Dr. Pepper']\n['Tomatillo-Green Chili Salsa (Medium)', 'Pinto Beans', 'Rice', 'Cheese', 'Sour Cream']\n['Roasted Chili Corn Salsa (Medium)', 'Fresh Tomato Salsa (Mild)', 'Rice', 'Black Beans', 'Sour Cream']\n['Tomatillo-Red Chili Salsa (Hot)', 'Black Beans', 'Rice', 'Fajita Veggies', 'Cheese', 'Sour Cream', 'Lettuce']\n['Roasted Chili Corn Salsa', 'Rice']\n['Roasted Chili Corn Salsa', 'Cheese', 'Lettuce']\n['Roasted Chili Corn Salsa', 'Fajita Vegetables', 'Rice', 'Black Beans', 'Cheese', 'Sour Cream', 'Guacamole', 'Lettuce']\n['Roasted Chili Corn Salsa', 'Rice', 'Black Beans', 'Cheese', 'Sour Cream', 'Guacamole', 'Lettuce']\n['Fresh Tomato Salsa', 'Rice', 'Pinto Beans', 'Cheese', 'Sour Cream', 'Guacamole', 'Lettuce']\n['Fresh Tomato Salsa', 'Fajita Vegetables', 'Pinto Beans', 'Lettuce']\n['Tomatillo-Red Chili Salsa (Hot)', 'Black Beans', 'Fajita Veggies', 'Cheese', 'Sour Cream', 'Guacamole', 'Lettuce']\n['Tomatillo-Red Chili Salsa (Hot)', 'Rice', 'Fajita Veggies', 'Cheese']\n['Blackberry']\n['Fresh Tomato Salsa (Mild)', 'Roasted Chili Corn Salsa (Medium)', 'Tomatillo-Red Chili Salsa (Hot)', 'Black Beans', 'Rice', 'Sour Cream']\n['Roasted Chili Corn Salsa (Medium)', 'Rice', 'Fajita Veggies', 'Cheese', 'Sour Cream', 'Lettuce']\n['Mountain Dew']\n['Roasted Chili Corn Salsa (Medium)', 'Black Beans', 'Rice', 'Fajita Veggies', 'Cheese', 'Sour Cream', 'Guacamole', 'Lettuce']\n['Sprite']\n['Fresh Tomato Salsa (Mild)', 'Black Beans', 'Rice', 'Fajita Veggies', 'Cheese', 'Sour Cream']\n['Fresh Tomato Salsa', 'Fajita Vegetables', 'Black Beans', 'Lettuce']\n['Tomatillo Red Chili Salsa', 'Fajita Vegetables', 'Rice', 'Black Beans', 'Cheese', 'Sour Cream', 'Guacamole', 'Lettuce']\n['Roasted Chili Corn Salsa', 'Fajita Vegetables', 'Rice', 'Black Beans', 'Cheese', 'Sour Cream', 'Guacamole', 'Lettuce']\n['Fresh Tomato Salsa', 'Cheese']\n['Fresh Tomato Salsa', 'Fajita Vegetables', 'Rice', 'Black Beans', 'Cheese', 'Sour Cream', 'Guacamole', 'Lettuce']\n['Fresh Tomato Salsa', 'Rice', 'Black Beans', 'Cheese']\n['Tomatillo-Red Chili Salsa (Hot)', 'Black Beans', 'Rice', 'Fajita Veggies', 'Cheese', 'Sour Cream', 'Lettuce']\n['Tomatillo-Red Chili Salsa (Hot)', 'Rice', 'Cheese', 'Sour Cream', 'Lettuce']\n['Blackberry']\n['Tomatillo-Red Chili Salsa (Hot)', 'Rice', 'Cheese']\n['Tomatillo-Red Chili Salsa (Hot)', 'Black Beans', 'Fajita Veggies', 'Cheese', 'Sour Cream', 'Guacamole', 'Lettuce']\n['Roasted Chili Corn Salsa (Medium)', 'Rice', 'Black Beans', 'Sour Cream']\n['Tomatillo-Red Chili Salsa (Hot)', 'Black Beans', 'Rice', 'Cheese', 'Sour Cream', 'Lettuce']\n['Tomatillo Red Chili Salsa', 'Rice', 'Black Beans', 'Cheese', 'Sour Cream']\n['Tomatillo Red Chili Salsa', 'Rice', 'Cheese', 'Sour Cream', 'Lettuce']\n['Fresh Tomato Salsa', 'Rice', 'Black Beans', 'Pinto Beans', 'Cheese', 'Lettuce']\n['Fresh Tomato Salsa', 'Rice', 'Black Beans', 'Cheese', 'Sour Cream', 'Lettuce']\n['Roasted Chili Corn Salsa', 'Rice', 'Cheese', 'Lettuce', 'Guacamole']\n['Fresh Tomato Salsa', 'Rice', 'Pinto Beans', 'Cheese', 'Sour Cream', 'Lettuce']\n['Fresh Tomato Salsa', 'Rice', 'Black Beans', 'Cheese', 'Sour Cream', 'Lettuce']\n['Tomatillo Green Chili Salsa', 'Rice', 'Black Beans', 'Sour Cream', 'Cheese']\n['Tomatillo Red Chili Salsa']\n['Tomatillo-Red Chili Salsa (Hot)', 'Pinto Beans', 'Black Beans', 'Rice', 'Fajita Veggies', 'Lettuce']\n['Tomatillo-Red Chili Salsa (Hot)', 'Pinto Beans', 'Rice', 'Fajita Veggies', 'Cheese', 'Lettuce']\n['Fresh Tomato Salsa', 'Rice', 'Pinto Beans', 'Sour Cream', 'Cheese']\n['Fresh Tomato Salsa', 'Rice', 'Black Beans', 'Cheese', 'Sour Cream', 'Guacamole']\n['Fresh Tomato Salsa', 'Fajita Vegetables', 'Cheese', 'Sour Cream', 'Guacamole']\n['Fresh Tomato Salsa', 'Sour Cream']\n['Roasted Chili Corn Salsa', 'Sour Cream', 'Guacamole']\n['Fresh Tomato Salsa', 'Fajita Vegetables', 'Rice', 'Cheese', 'Sour Cream']\n['Fresh Tomato Salsa', 'Fajita Vegetables', 'Rice', 'Black Beans', 'Cheese', 'Guacamole', 'Lettuce']\n['Tomatillo Red Chili Salsa', 'Rice', 'Black Beans', 'Cheese']\n['Tomatillo Red Chili Salsa', 'Rice', 'Fajita Vegetables', 'Sour Cream', 'Lettuce']\n"
],
[
"for item in df.choice_description:\n if type(item) == float:\n \n lista_item = []\n \n else:\n \n item = item.replace('[', '').replace(']', '')\n\n item\n\n lista_item = item.split(', ')\n\n print(lista_item)",
"['Clementine']\n['Apple']\n['Tomatillo-Red Chili Salsa (Hot)', 'Black Beans', 'Rice', 'Cheese', 'Sour Cream']\n['Fresh Tomato Salsa (Mild)', 'Rice', 'Cheese', 'Sour Cream', 'Guacamole', 'Lettuce']\n['Tomatillo Red Chili Salsa', 'Fajita Vegetables', 'Black Beans', 'Pinto Beans', 'Cheese', 'Sour Cream', 'Guacamole', 'Lettuce']\n['Tomatillo Green Chili Salsa', 'Pinto Beans', 'Cheese', 'Sour Cream', 'Lettuce']\n['Fresh Tomato Salsa', 'Rice', 'Black Beans', 'Pinto Beans', 'Cheese', 'Sour Cream', 'Lettuce']\n['Roasted Chili Corn Salsa', 'Fajita Vegetables', 'Rice', 'Black Beans', 'Cheese', 'Sour Cream']\n['Roasted Chili Corn Salsa', 'Rice', 'Black Beans', 'Cheese', 'Sour Cream']\n['Fresh Tomato Salsa', 'Fajita Vegetables', 'Rice', 'Cheese', 'Sour Cream', 'Guacamole']\n['Tomatillo-Green Chili Salsa (Medium)', 'Pinto Beans', 'Cheese', 'Sour Cream']\n['Fresh Tomato Salsa (Mild)', 'Black Beans', 'Rice', 'Cheese', 'Sour Cream', 'Lettuce']\n['Sprite']\n['Tomatillo Red Chili Salsa', 'Fajita Vegetables', 'Black Beans', 'Sour Cream', 'Cheese', 'Lettuce']\n['Fresh Tomato Salsa (Mild)', 'Tomatillo-Green Chili Salsa (Medium)', 'Tomatillo-Red Chili Salsa (Hot)', 'Rice', 'Cheese', 'Sour Cream', 'Lettuce']\n['Pomegranate Cherry']\n['Tomatillo-Green Chili Salsa (Medium)', 'Tomatillo-Red Chili Salsa (Hot)', 'Pinto Beans', 'Rice', 'Cheese', 'Sour Cream', 'Guacamole', 'Lettuce']\n['Grapefruit']\n['Roasted Chili Corn Salsa (Medium)', 'Pinto Beans', 'Rice', 'Fajita Veggies', 'Cheese', 'Sour Cream', 'Lettuce']\n['Tomatillo-Green Chili Salsa (Medium)', 'Roasted Chili Corn Salsa (Medium)', 'Black Beans', 'Rice', 'Sour Cream', 'Lettuce']\n['Dr. Pepper']\n['Tomatillo-Green Chili Salsa (Medium)', 'Pinto Beans', 'Rice', 'Cheese', 'Sour Cream']\n['Roasted Chili Corn Salsa (Medium)', 'Fresh Tomato Salsa (Mild)', 'Rice', 'Black Beans', 'Sour Cream']\n['Tomatillo-Red Chili Salsa (Hot)', 'Black Beans', 'Rice', 'Fajita Veggies', 'Cheese', 'Sour Cream', 'Lettuce']\n['Roasted Chili Corn Salsa', 'Rice']\n['Roasted Chili Corn Salsa', 'Cheese', 'Lettuce']\n['Roasted Chili Corn Salsa', 'Fajita Vegetables', 'Rice', 'Black Beans', 'Cheese', 'Sour Cream', 'Guacamole', 'Lettuce']\n['Roasted Chili Corn Salsa', 'Rice', 'Black Beans', 'Cheese', 'Sour Cream', 'Guacamole', 'Lettuce']\n['Fresh Tomato Salsa', 'Rice', 'Pinto Beans', 'Cheese', 'Sour Cream', 'Guacamole', 'Lettuce']\n['Fresh Tomato Salsa', 'Fajita Vegetables', 'Pinto Beans', 'Lettuce']\n['Tomatillo-Red Chili Salsa (Hot)', 'Black Beans', 'Fajita Veggies', 'Cheese', 'Sour Cream', 'Guacamole', 'Lettuce']\n['Tomatillo-Red Chili Salsa (Hot)', 'Rice', 'Fajita Veggies', 'Cheese']\n['Blackberry']\n['Fresh Tomato Salsa (Mild)', 'Roasted Chili Corn Salsa (Medium)', 'Tomatillo-Red Chili Salsa (Hot)', 'Black Beans', 'Rice', 'Sour Cream']\n['Roasted Chili Corn Salsa (Medium)', 'Rice', 'Fajita Veggies', 'Cheese', 'Sour Cream', 'Lettuce']\n['Mountain Dew']\n['Roasted Chili Corn Salsa (Medium)', 'Black Beans', 'Rice', 'Fajita Veggies', 'Cheese', 'Sour Cream', 'Guacamole', 'Lettuce']\n['Sprite']\n['Fresh Tomato Salsa (Mild)', 'Black Beans', 'Rice', 'Fajita Veggies', 'Cheese', 'Sour Cream']\n['Fresh Tomato Salsa', 'Fajita Vegetables', 'Black Beans', 'Lettuce']\n['Tomatillo Red Chili Salsa', 'Fajita Vegetables', 'Rice', 'Black Beans', 'Cheese', 'Sour Cream', 'Guacamole', 'Lettuce']\n['Roasted Chili Corn Salsa', 'Fajita Vegetables', 'Rice', 'Black Beans', 'Cheese', 'Sour Cream', 'Guacamole', 'Lettuce']\n['Fresh Tomato Salsa', 'Cheese']\n['Fresh Tomato Salsa', 'Fajita Vegetables', 'Rice', 'Black Beans', 'Cheese', 'Sour Cream', 'Guacamole', 'Lettuce']\n['Fresh Tomato Salsa', 'Rice', 'Black Beans', 'Cheese']\n['Tomatillo-Red Chili Salsa (Hot)', 'Black Beans', 'Rice', 'Fajita Veggies', 'Cheese', 'Sour Cream', 'Lettuce']\n['Tomatillo-Red Chili Salsa (Hot)', 'Rice', 'Cheese', 'Sour Cream', 'Lettuce']\n['Blackberry']\n['Tomatillo-Red Chili Salsa (Hot)', 'Rice', 'Cheese']\n['Tomatillo-Red Chili Salsa (Hot)', 'Black Beans', 'Fajita Veggies', 'Cheese', 'Sour Cream', 'Guacamole', 'Lettuce']\n['Roasted Chili Corn Salsa (Medium)', 'Rice', 'Black Beans', 'Sour Cream']\n['Tomatillo-Red Chili Salsa (Hot)', 'Black Beans', 'Rice', 'Cheese', 'Sour Cream', 'Lettuce']\n['Tomatillo Red Chili Salsa', 'Rice', 'Black Beans', 'Cheese', 'Sour Cream']\n['Tomatillo Red Chili Salsa', 'Rice', 'Cheese', 'Sour Cream', 'Lettuce']\n['Fresh Tomato Salsa', 'Rice', 'Black Beans', 'Pinto Beans', 'Cheese', 'Lettuce']\n['Fresh Tomato Salsa', 'Rice', 'Black Beans', 'Cheese', 'Sour Cream', 'Lettuce']\n['Roasted Chili Corn Salsa', 'Rice', 'Cheese', 'Lettuce', 'Guacamole']\n['Fresh Tomato Salsa', 'Rice', 'Pinto Beans', 'Cheese', 'Sour Cream', 'Lettuce']\n['Fresh Tomato Salsa', 'Rice', 'Black Beans', 'Cheese', 'Sour Cream', 'Lettuce']\n['Tomatillo Green Chili Salsa', 'Rice', 'Black Beans', 'Sour Cream', 'Cheese']\n['Tomatillo Red Chili Salsa']\n['Tomatillo-Red Chili Salsa (Hot)', 'Pinto Beans', 'Black Beans', 'Rice', 'Fajita Veggies', 'Lettuce']\n['Tomatillo-Red Chili Salsa (Hot)', 'Pinto Beans', 'Rice', 'Fajita Veggies', 'Cheese', 'Lettuce']\n['Fresh Tomato Salsa', 'Rice', 'Pinto Beans', 'Sour Cream', 'Cheese']\n['Fresh Tomato Salsa', 'Rice', 'Black Beans', 'Cheese', 'Sour Cream', 'Guacamole']\n['Fresh Tomato Salsa', 'Fajita Vegetables', 'Cheese', 'Sour Cream', 'Guacamole']\n['Fresh Tomato Salsa', 'Sour Cream']\n['Roasted Chili Corn Salsa', 'Sour Cream', 'Guacamole']\n['Fresh Tomato Salsa', 'Fajita Vegetables', 'Rice', 'Cheese', 'Sour Cream']\n['Fresh Tomato Salsa', 'Fajita Vegetables', 'Rice', 'Black Beans', 'Cheese', 'Guacamole', 'Lettuce']\n['Tomatillo Red Chili Salsa', 'Rice', 'Black Beans', 'Cheese']\n['Tomatillo Red Chili Salsa', 'Rice', 'Fajita Vegetables', 'Sour Cream', 'Lettuce']\n"
],
[
"lista_todos = []",
"_____no_output_____"
],
[
"for item in df.choice_description:\n if type(item) == float:\n \n lista_item = []\n lista_todos.append(lista_item)\n else:\n \n item = item.replace('[', '').replace(']', '')\n\n item\n\n lista_item = item.split(', ')\n\n lista_todos.append(lista_item)",
"_____no_output_____"
],
[
"lista_todos",
"_____no_output_____"
],
[
"from sklearn.feature_extraction.text import TfidfVectorizer\n\nbag_of_words = CountVectorizer(tokenizer=lambda doc: doc, lowercase=False).fit_transform(lista_todos)",
"_____no_output_____"
],
[
"bag_of_words.get",
"_____no_output_____"
],
[
"vec = TfidfVectorizer()",
"_____no_output_____"
],
[
"lista_todos = [','.join(i) for i in lista_todos]",
"_____no_output_____"
],
[
"vec.fit(lista_todos)",
"_____no_output_____"
],
[
"data = vec.transform(lista_todos).toarray()",
"_____no_output_____"
],
[
"vec.get_feature_names_out()",
"_____no_output_____"
],
[
"pd.DataFrame(data)",
"_____no_output_____"
],
[
"pd.DataFrame(bag_of_words.toarray())",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code"
] |
[
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb8b4820846b9fed1cfbaeec1536938fc0a68567
| 57,635 |
ipynb
|
Jupyter Notebook
|
mz685_pricing_vocram.ipynb
|
GeweiWang/notebooks
|
bc5e928bc662ffd364bd9423fef037c63b116a3e
|
[
"MIT"
] | null | null | null |
mz685_pricing_vocram.ipynb
|
GeweiWang/notebooks
|
bc5e928bc662ffd364bd9423fef037c63b116a3e
|
[
"MIT"
] | null | null | null |
mz685_pricing_vocram.ipynb
|
GeweiWang/notebooks
|
bc5e928bc662ffd364bd9423fef037c63b116a3e
|
[
"MIT"
] | null | null | null | 112.78865 | 15,136 | 0.846777 |
[
[
[
"### Simulate the flight reservation process (MZ685 Case Vocram)",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nfrom random import random",
"_____no_output_____"
],
[
"# Constants\nFLIGHTS = 1000 # The number of flights for simulation (can be changed)\nCALLS = 10 # The number of calls for each flight\nSEATS = 3 # The number of available seats for each flight\nWTP_PUBLIC = 100 # The WTP upper bound of public employee\nWTP_PRIVATE = 150 # The WTP upper bound of private individual\nP = 0.5 # P: The probability of a call being public employee \n # 1-P: The probability of a call being private individual",
"_____no_output_____"
]
],
[
[
"### Replicate the Status Quo\n\nAssumptions:\n\n* Single price: $65\n* The WTP of public employee follows a discrete uniform distribution [1, 100]\n* The WTP of private individual follows a discrete uniform distribution [1, 150]\n* A strict first-call first-serve system: whoever called and made a reservation for a specific departure would be given a seat, provided one was available.\n\nSimulation results are expected to be in line with the fact:\n\nHistorically, 40% of the clients were scientists/researchers/public employees and 60% were private individuals.",
"_____no_output_____"
]
],
[
[
"def one_flight(flight_no, price, reservations):\n \"\"\"Simulate the reservation process for one flight.\n \n Parameters\n ----------\n flight_no: flight sequence number for bookkeeping\n \"\"\"\n seats_left = SEATS\n \n for i in range(CALLS):\n whocall = [0, 1][random()>P]\n if whocall == 0: # 0-public employee\n wtp = np.random.randint(1, WTP_PUBLIC+1)\n if wtp >= price:\n seats_left -= 1\n reservations.append((flight_no, whocall, wtp))\n if seats_left == 0:\n break\n elif whocall == 1: # 1-private individual\n wtp = np.random.randint(1, WTP_PRIVATE+1)\n if wtp >= price:\n seats_left -= 1\n reservations.append((flight_no, whocall, wtp))\n if seats_left == 0:\n break ",
"_____no_output_____"
],
[
"def revenue(price, more_info=False):\n \"\"\"Simulate FLIGHT times and return the average revenue per flight.\n \"\"\"\n reservations = []\n \n for i in range(1, FLIGHTS+1):\n one_flight(i, price, reservations)\n \n avg_revenue = price * len(reservations) / FLIGHTS\n \n if more_info == True:\n print('The # of availabe seats in {} flights is {}. The # of seats sold is {}.' \\\n .format(FLIGHTS, FLIGHTS*SEATS, len(reservations)))\n pcnt = ((pd.DataFrame(reservations, columns=['flight_no', 'whocall', 'wtp'])). \\\n groupby('whocall').size()) / len(reservations) * 100\n title = 'Price: \\$' + str(price) + ', Average revenue per flight: \\$' + str(round(avg_revenue)) + \\\n '\\n' + ' %' + str(round(len(reservations) / (FLIGHTS*SEATS) * 100)) + ' seats were sold'\n ax = pcnt.plot(kind='bar', title=title, figsize=(8,5))\n ax.set_xlabel(\"0-public employee 1-private individual\")\n ax.set_ylabel(\"Percent (%)\")\n \n return avg_revenue",
"_____no_output_____"
],
[
"revenue(65, True)",
"The # of availabe seats in 1000 flights is 3000. The # of seats sold is 2895.\n"
]
],
[
[
"### Get the Optimal Single Price",
"_____no_output_____"
]
],
[
[
"# Step 1: Get a rough range of the optimal single price\n\noptimal_price, max_revenue = 1, 1\n\nfor p in range(60, 150):\n r = revenue(p)\n if max_revenue < r:\n optimal_price, max_revenue = p, r\n \nprint(optimal_price, max_revenue)",
"80 210.88\n"
],
[
"# Step 2: Run simulation many times within a smaller price range\n\noptima = []\n\nfor i in range(100):\n optimal_price, max_revenue = 1, 1\n \n for p in range(75, 90):\n r = revenue(p)\n if max_revenue < r:\n optimal_price, max_revenue = p, r\n \n optima.append((optimal_price, max_revenue)) ",
"_____no_output_____"
],
[
"# Step 3: Average the results from Step 2 to get the optimal single price\n\n(pd.DataFrame(optima, columns=['price', 'revenue'])).mean()",
"_____no_output_____"
],
[
"revenue(82, True)",
"The # of availabe seats in 1000 flights is 3000. The # of seats sold is 2545.\n"
]
],
[
[
"### Two-tier Pricing\n\nAssumptions:\n* Stu will ask a customer calling in to know what type the person belongs to. For public employee, he will quote the high price; for private individual, he will quote the low price.\n* Stu doesn't set a booking limit for public employee.\n",
"_____no_output_____"
]
],
[
[
"def one_flight_two_tier(flight_no, high_price, low_price, reservations, booking_limit=SEATS):\n \"\"\" Simulate the reservation process for one flight with two-tier pricing system\n \n Parameters\n ----------\n flight_no: flight sequence number for bookkeeping\n high_price: price for private individual\n low_price: price for public employee\n booking_limit: the max number of seats allocated to public employee per flight\n \"\"\"\n\n seats_left = SEATS\n seats_for_public = booking_limit\n \n for i in range(CALLS):\n whocall = [0, 1][random()>P]\n if whocall == 0: # 0-public employee\n wtp = np.random.randint(1, WTP_PUBLIC+1)\n if seats_for_public == 0:\n continue\n if wtp >= low_price:\n seats_left -= 1\n seats_for_public -= 1\n reservations.append((flight_no, whocall, wtp))\n if seats_left == 0:\n break\n elif whocall == 1: # 1-private individual\n wtp = np.random.randint(1, WTP_PRIVATE+1)\n if wtp >= high_price:\n seats_left -= 1\n reservations.append((flight_no, whocall, wtp))\n if seats_left == 0:\n break",
"_____no_output_____"
],
[
"def revenue_two_tier(high_price, low_price, booking_limit=SEATS, more_info=False):\n \"\"\"Return the average revenue per flight\n \"\"\"\n reservations = []\n \n for i in range(1, FLIGHTS+1):\n one_flight_two_tier(i, high_price, low_price, reservations, booking_limit)\n \n df = pd.DataFrame(reservations, columns=['flight_no', 'whocall', 'wtp'])\n avg_revenue = (high_price * len(df[df.whocall==1]) + low_price * len(df[df.whocall==0])) / FLIGHTS\n \n if more_info == True:\n print('The # of availabe seats in {} flights is {}. The # of seats sold is {}.' \\\n .format(FLIGHTS, FLIGHTS*SEATS, len(reservations)))\n pcnt = df.groupby('whocall').size() / len(reservations) * 100\n title = 'High Price: \\$' + str(high_price) + ', Low Price: \\$' + str(low_price) + \\\n '\\nAverage revenue per flight: \\$' + str(round(avg_revenue)) + \\\n '\\n' + ' %' + str(round(len(reservations) / (FLIGHTS*SEATS) * 100)) + ' seats were sold'\n ax = pcnt.plot(kind='bar', title=title, figsize=(8,5))\n ax.set_xlabel(\"0-public employee 1-private individual\")\n ax.set_ylabel(\"Percent (%)\")\n \n return avg_revenue",
"_____no_output_____"
]
],
[
[
"### Get the Optimal Two-tier System",
"_____no_output_____"
]
],
[
[
"# Step 1: Get rough ranges of the optimal two prices\n\noptimal_high_price, optimal_low_price, max_revenue = 1, 1, 1\n\nfor high in range(70, 150):\n for low in range(50, 100):\n r = revenue_two_tier(high, low)\n if max_revenue < r:\n max_revenue = r\n optimal_high_price, optimal_low_price = high, low\n \nprint(optimal_high_price, optimal_low_price, max_revenue)",
"103 73 224.178\n"
],
[
"# Step 2: Run simulation many times within smaller price ranges\n\nN = 100\noptima = []\n\nfor i in range(N):\n optimal_high_price, optimal_low_price, max_revenue = 1, 1, 1\n \n for high in range(90, 110):\n for low in range(65, 80):\n r = revenue_two_tier(high, low)\n if max_revenue < r:\n max_revenue = r\n optimal_high_price, optimal_low_price = high, low\n \n optima.append((optimal_high_price, optimal_low_price, max_revenue)) \n",
"_____no_output_____"
],
[
"# Step 3: Average the results from Step 2 to get the optimal two prices\n\n(pd.DataFrame(optima, columns=['high_price', 'low_price', 'revenue'])).mean()",
"_____no_output_____"
],
[
"revenue_two_tier(98, 73, more_info=True)",
"The # of availabe seats in 1000 flights is 3000. The # of seats sold is 2523.\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
cb8b4b20e0ce39ac7e8b22e682f3f04bd1f6e61f
| 34,521 |
ipynb
|
Jupyter Notebook
|
notebooks/4_Tokenization_Lemmatization_V3.ipynb
|
RichardScottOZ/dh2loop
|
1d19b26c058833976cca93a9e3553240ff6c5c61
|
[
"MIT"
] | 20 |
2020-06-11T02:19:46.000Z
|
2022-01-27T13:37:15.000Z
|
notebooks/4_Tokenization_Lemmatization_V3.ipynb
|
RichardScottOZ/dh2loop
|
1d19b26c058833976cca93a9e3553240ff6c5c61
|
[
"MIT"
] | 2 |
2020-03-20T02:58:23.000Z
|
2021-04-30T03:09:50.000Z
|
notebooks/.ipynb_checkpoints/4_Tokenization_Lemmatization_V3-checkpoint.ipynb
|
Loop3D/dh2loop
|
1d19b26c058833976cca93a9e3553240ff6c5c61
|
[
"MIT"
] | 3 |
2020-10-14T08:23:56.000Z
|
2021-06-18T15:53:08.000Z
| 38.271619 | 129 | 0.473972 |
[
[
[
"import warnings\nwarnings.filterwarnings('ignore')\n\nimport nltk\nnltk.download('stopwords')\nnltk.download('punkt')\nnltk.download('wordnet')\nfrom nltk.corpus import stopwords\n\nimport pandas as pd\nimport numpy as np\nfrom glove import Glove\nfrom sklearn.preprocessing import LabelEncoder\nfrom sklearn import metrics\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.preprocessing import OneHotEncoder\nfrom keras.models import Sequential\nfrom keras.layers import Dense\n\nimport geopandas as gpd\nimport os\n\nimport json\nimport h5py\n\nlabelEncoder = LabelEncoder()\none_enc = OneHotEncoder()\nlemma = nltk.WordNetLemmatizer()",
"[nltk_data] Downloading package stopwords to /home/ranee/nltk_data...\n[nltk_data] Package stopwords is already up-to-date!\n[nltk_data] Downloading package punkt to /home/ranee/nltk_data...\n[nltk_data] Package punkt is already up-to-date!\n[nltk_data] Downloading package wordnet to /home/ranee/nltk_data...\n[nltk_data] Package wordnet is already up-to-date!\n"
]
],
[
[
"## Manual Classification",
"_____no_output_____"
]
],
[
[
"#Dir = '/mnt/d/Dropbox/Ranee_Joshi_PhD_Local/04_PythonCodes/dh2loop_old/shp_NSW'\n#DF=litho_Dataframe(Dir)\n#DF.to_csv('export.csv')\nDF = pd.read_csv('/mnt/d/Dropbox/Ranee_Joshi_PhD_Local/04_PythonCodes/dh2loop/notebooks/Upscaled_Litho_Test2.csv')\nDF['FromDepth'] = pd.to_numeric(DF.FromDepth)\nDF['ToDepth'] = pd.to_numeric(DF.ToDepth)\nDF['TopElev'] = pd.to_numeric(DF.TopElev)\nDF['BottomElev'] = pd.to_numeric(DF.BottomElev)\nDF['x'] = pd.to_numeric(DF.x)\nDF['y'] = pd.to_numeric(DF.y)\nprint('number of original litho classes:', len(DF.MajorLithCode.unique()))\nprint('number of litho classes :',\n len(DF['reclass'].unique()))\nprint('unclassified descriptions:',\n len(DF[DF['reclass'].isnull()]))",
"number of original litho classes: 340\nnumber of litho classes : 78\nunclassified descriptions: 0\n"
],
[
"def save_file(DF, name):\n '''Function to save manually reclassified dataframe\n Inputs:\n -DF: reclassified pandas dataframe\n -name: name (string) to save dataframe file\n '''\n DF.to_pickle('{}.pkl'.format(name))",
"_____no_output_____"
],
[
"save_file(DF, 'manualTest_ygsb')",
"_____no_output_____"
]
],
[
[
"## MLP Classification",
"_____no_output_____"
]
],
[
[
"def load_geovec(path):\n instance = Glove()\n with h5py.File(path, 'r') as f:\n v = np.zeros(f['vectors'].shape, f['vectors'].dtype)\n f['vectors'].read_direct(v)\n dct = f['dct'][()].tostring().decode('utf-8')\n dct = json.loads(dct)\n instance.word_vectors = v\n instance.no_components = v.shape[1]\n instance.word_biases = np.zeros(v.shape[0])\n instance.add_dictionary(dct)\n return instance",
"_____no_output_____"
],
[
"# Stopwords\nextra_stopwords = [\n 'also',\n]\nstop = stopwords.words('english') + extra_stopwords",
"_____no_output_____"
],
[
"def tokenize(text, min_len=1):\n '''Function that tokenize a set of strings\n Input:\n -text: set of strings\n -min_len: tokens length\n Output:\n -list containing set of tokens'''\n\n tokens = [word.lower() for sent in nltk.sent_tokenize(text)\n for word in nltk.word_tokenize(sent)]\n filtered_tokens = []\n\n for token in tokens:\n if token.isalpha() and len(token) >= min_len:\n filtered_tokens.append(token)\n\n return [x.lower() for x in filtered_tokens if x not in stop]\n\n\ndef tokenize_and_lemma(text, min_len=0):\n '''Function that retrieves lemmatised tokens\n Inputs:\n -text: set of strings\n -min_len: length of text\n Outputs:\n -list containing lemmatised tokens'''\n filtered_tokens = tokenize(text, min_len=min_len)\n\n lemmas = [lemma.lemmatize(t) for t in filtered_tokens]\n return lemmas\n\n\ndef get_vector(word, model, return_zero=False):\n '''Function that retrieves word embeddings (vector)\n Inputs:\n -word: token (string)\n -model: trained MLP model\n -return_zero: boolean variable\n Outputs:\n -wv: numpy array (vector)'''\n epsilon = 1.e-10\n\n unk_idx = model.dictionary['unk']\n idx = model.dictionary.get(word, unk_idx)\n wv = model.word_vectors[idx].copy()\n\n if return_zero and word not in model.dictionary:\n n_comp = model.word_vectors.shape[1]\n wv = np.zeros(n_comp) + epsilon\n\n return wv",
"_____no_output_____"
],
[
"def mean_embeddings(dataframe_file, model):\n '''Function to retrieve sentence embeddings from dataframe with\n lithological descriptions.\n Inputs:\n -dataframe_file: pandas dataframe containing lithological descriptions\n and reclassified lithologies\n -model: word embeddings model generated using GloVe\n Outputs:\n -DF: pandas dataframe including sentence embeddings'''\n DF = pd.read_pickle(dataframe_file)\n DF = DF.drop_duplicates(subset=['x', 'y', 'z'])\n DF['tokens'] = DF['Description'].apply(lambda x: tokenize_and_lemma(x))\n DF['length'] = DF['tokens'].apply(lambda x: len(x))\n DF = DF.loc[DF['length']> 0]\n DF['vectors'] = DF['tokens'].apply(lambda x: np.asarray([get_vector(n, model) for n in x]))\n DF['mean'] = DF['vectors'].apply(lambda x: np.mean(x[~np.all(x == 1.e-10, axis=1)], axis=0))\n DF['reclass'] = pd.Categorical(DF.reclass)\n DF['code'] = DF.reclass.cat.codes\n DF['drop'] = DF['mean'].apply(lambda x: (~np.isnan(x).any()))\n DF = DF[DF['drop']]\n return DF",
"_____no_output_____"
],
[
"# loading word embeddings model\n# (This can be obtained from https://github.com/spadarian/GeoVec )\n#modelEmb = Glove.load('/home/ignacio/Documents/chapter2/best_glove_300_317413_w10_lemma.pkl')\nmodelEmb = load_geovec('geovec_300d_v1.h5')\n\n# getting the mean embeddings of descriptions\nDF = mean_embeddings('manualTest_ygsb.pkl', modelEmb)",
"_____no_output_____"
],
[
"DF2 = DF[DF['code'].isin(DF['code'].value_counts()[DF['code'].value_counts()>2].index)]\nprint(DF2)",
" Description HydroCode FromDepth ToDepth TopElev \\\n0 Transported hardpan 124897 0 1.0 400.000000 \n1 Transported hardpan 124897 1 2.0 399.133975 \n2 Regolith Clay 124897 2 3.0 398.267949 \n3 Regolith Clay 124897 3 4.0 397.401924 \n4 Regolith Clay 124897 4 5.0 396.535898 \n... ... ... ... ... ... \n159942 Basalt 4167632 25 28.0 375.000000 \n159943 Basalt 4167632 28 29.0 372.000000 \n159944 Banded Iron Formation BIF 4167633 1 4.0 399.000000 \n159945 Basalt 4167633 4 5.0 396.000000 \n159946 Banded Iron Formation BIF 4167634 46 47.0 354.000000 \n\n BottomElev MajorLithCode geometry z \\\n0 399.133975 NHO NaN 399.566987 \n1 398.267949 NHO NaN 398.700962 \n2 397.401924 RCY NaN 397.834937 \n3 396.535898 RCY NaN 396.968911 \n4 395.669873 RCY NaN 396.102886 \n... ... ... ... ... \n159942 372.000000 MBA NaN 373.500000 \n159943 371.000000 MBA NaN 371.500000 \n159944 396.000000 SBI NaN 397.500000 \n159945 395.000000 MBA NaN 395.500000 \n159946 353.000000 SBI NaN 353.500000 \n\n OWN reclass x y \\\n0 Transported hardpan colluvium 513822.0449 6952747.766 \n1 Transported hardpan colluvium 513822.5449 6952747.766 \n2 Regolith Clay mud 513823.0449 6952747.766 \n3 Regolith Clay mud 513823.5449 6952747.766 \n4 Regolith Clay mud 513824.0449 6952747.766 \n... ... ... ... ... \n159942 Basalt basalt 576956.0811 6882354.387 \n159943 Basalt basalt 576956.0811 6882354.387 \n159944 Banded Iron Formation BIF Other 577054.2428 6882353.752 \n159945 Basalt basalt 577054.2428 6882353.752 \n159946 Banded Iron Formation BIF Other 577152.4045 6882353.117 \n\n tokens length \\\n0 [transported, hardpan] 2 \n1 [transported, hardpan] 2 \n2 [regolith, clay] 2 \n3 [regolith, clay] 2 \n4 [regolith, clay] 2 \n... ... ... \n159942 [basalt] 1 \n159943 [basalt] 1 \n159944 [banded, iron, formation, bif] 4 \n159945 [basalt] 1 \n159946 [banded, iron, formation, bif] 4 \n\n vectors \\\n0 [[-0.18778053, -0.0063699493, -0.016271375, -0... \n1 [[-0.18778053, -0.0063699493, -0.016271375, -0... \n2 [[0.029590942, 0.08967461, -0.16049796, -0.342... \n3 [[0.029590942, 0.08967461, -0.16049796, -0.342... \n4 [[0.029590942, 0.08967461, -0.16049796, -0.342... \n... ... \n159942 [[-0.2746381, -0.23468062, -0.37358257, 0.0047... \n159943 [[-0.2746381, -0.23468062, -0.37358257, 0.0047... \n159944 [[0.02457981, -0.39863607, 0.04441907, 0.09330... \n159945 [[-0.2746381, -0.23468062, -0.37358257, 0.0047... \n159946 [[0.02457981, -0.39863607, 0.04441907, 0.09330... \n\n mean code drop \n0 [-0.10188103, -0.009773923, -0.030616358, -0.1... 14 True \n1 [-0.10188103, -0.009773923, -0.030616358, -0.1... 14 True \n2 [0.09962925, -0.10881818, -0.29363656, -0.2486... 44 True \n3 [0.09962925, -0.10881818, -0.29363656, -0.2486... 44 True \n4 [0.09962925, -0.10881818, -0.29363656, -0.2486... 44 True \n... ... ... ... \n159942 [-0.2746381, -0.23468062, -0.37358257, 0.00474... 4 True \n159943 [-0.2746381, -0.23468062, -0.37358257, 0.00474... 4 True \n159944 [0.010618463, -0.33650404, -0.120667905, -0.02... 0 True \n159945 [-0.2746381, -0.23468062, -0.37358257, 0.00474... 4 True \n159946 [0.010618463, -0.33650404, -0.120667905, -0.02... 0 True \n\n[122502 rows x 19 columns]\n"
],
[
"def split_stratified_dataset(Dataframe, test_size, validation_size):\n '''Function that split dataset into test, training and validation subsets\n Inputs:\n -Dataframe: pandas dataframe with sentence mean_embeddings\n -test_size: decimal number to generate the test subset\n -validation_size: decimal number to generate the validation subset\n Outputs:\n -X: numpy array with embeddings\n -Y: numpy array with lithological classes\n -X_test: numpy array with embeddings for test subset\n -Y_test: numpy array with lithological classes for test subset\n -Xt: numpy array with embeddings for training subset\n -yt: numpy array with lithological classes for training subset\n -Xv: numpy array with embeddings for validation subset\n -yv: numpy array with lithological classes for validation subset\n '''\n #df2 = Dataframe[Dataframe['code'].isin(Dataframe['code'].value_counts()[Dataframe['code'].value_counts()>2].index)]\n #X = np.vstack(df2['mean'].values)\n #Y = df2.code.values.reshape(len(df2.code), 1)\n X = np.vstack(Dataframe['mean'].values)\n Y = Dataframe.code.values.reshape(len(Dataframe.code), 1)\n #print(X.shape)\n #print (Dataframe.code.values.shape)\n #print (len(Dataframe.code))\n #print (Y.shape)\n X_train, X_test, y_train, y_test = train_test_split(X,\n Y, stratify=Y,\n test_size=test_size,\n random_state=42)\n #print(X_train.shape)\n #print(Y_train.shape)\n Xt, Xv, yt, yv = train_test_split(X_train,\n y_train,\n test_size=validation_size,\n stratify=None,\n random_state=1)\n return X, Y, X_test, y_test, Xt, yt, Xv, yv",
"_____no_output_____"
],
[
"# subseting dataset for training classifier\nX, Y, X_test, Y_test, X_train, Y_train, X_validation, Y_validation = split_stratified_dataset(DF2, 0.1, 0.1)\n\n# encoding lithological classes\nencodes = one_enc.fit_transform(Y_train).toarray()\n\n# MLP model generation\nmodel = Sequential()\nmodel.add(Dense(100, input_dim=300, activation='relu'))\nmodel.add(Dense(100, activation='relu'))\nmodel.add(Dense(100, activation='relu'))\nmodel.add(Dense(units=len(DF2.code.unique()), activation='softmax'))\nmodel.compile(loss='categorical_crossentropy',\n optimizer='adam',\n metrics=['accuracy'])\n\n# training MLP model\nmodel.fit(X_train, encodes, epochs=30, batch_size=100, verbose=2)\n\n# saving MLP model\nmodel.save('mlp_prob_model.h5')",
"Epoch 1/30\n - 3s - loss: 0.2120 - accuracy: 0.9601\nEpoch 2/30\n - 3s - loss: 0.0060 - accuracy: 0.9990\nEpoch 3/30\n - 5s - loss: 0.0037 - accuracy: 0.9994\nEpoch 4/30\n - 5s - loss: 0.0040 - accuracy: 0.9991\nEpoch 5/30\n - 5s - loss: 0.0020 - accuracy: 0.9997\nEpoch 6/30\n - 5s - loss: 0.0036 - accuracy: 0.9993\nEpoch 7/30\n - 7s - loss: 0.0019 - accuracy: 0.9997\nEpoch 8/30\n - 9s - loss: 0.0016 - accuracy: 0.9998\nEpoch 9/30\n - 6s - loss: 0.0016 - accuracy: 0.9998\nEpoch 10/30\n - 4s - loss: 0.0028 - accuracy: 0.9995\nEpoch 11/30\n - 4s - loss: 0.0032 - accuracy: 0.9994\nEpoch 12/30\n - 3s - loss: 0.0024 - accuracy: 0.9996\nEpoch 13/30\n - 4s - loss: 0.0020 - accuracy: 0.9996\nEpoch 14/30\n - 3s - loss: 0.0035 - accuracy: 0.9994\nEpoch 15/30\n - 3s - loss: 0.0018 - accuracy: 0.9997\nEpoch 16/30\n - 3s - loss: 0.0015 - accuracy: 0.9998\nEpoch 17/30\n - 3s - loss: 0.0015 - accuracy: 0.9998\nEpoch 18/30\n - 3s - loss: 0.0015 - accuracy: 0.9998\nEpoch 19/30\n - 3s - loss: 0.0016 - accuracy: 0.9998\nEpoch 20/30\n - 3s - loss: 0.0015 - accuracy: 0.9998\nEpoch 21/30\n - 3s - loss: 0.0015 - accuracy: 0.9998\nEpoch 22/30\n - 3s - loss: 0.0015 - accuracy: 0.9998\nEpoch 23/30\n - 3s - loss: 0.0038 - accuracy: 0.9993\nEpoch 24/30\n - 3s - loss: 0.0019 - accuracy: 0.9997\nEpoch 25/30\n - 3s - loss: 0.0015 - accuracy: 0.9998\nEpoch 26/30\n - 3s - loss: 0.0015 - accuracy: 0.9998\nEpoch 27/30\n - 3s - loss: 0.0014 - accuracy: 0.9998\nEpoch 28/30\n - 3s - loss: 0.0015 - accuracy: 0.9998\nEpoch 29/30\n - 3s - loss: 0.0015 - accuracy: 0.9998\nEpoch 30/30\n - 3s - loss: 0.0015 - accuracy: 0.9998\n"
],
[
"def retrieve_predictions(classifier, x):\n '''Function that retrieves lithological classes using the trained classifier\n Inputs:\n -classifier: trained MLP classifier\n -x: numpy array containing embbedings\n Outputs:\n -codes_pred: numpy array containing lithological classes predicted'''\n preds = classifier.predict(x, verbose=0)\n new_onehot = np.zeros((x.shape[0], 72))\n new_onehot[np.arange(len(preds)), preds.argmax(axis=1)] = 1\n codes_pred = one_enc.inverse_transform(new_onehot)\n return codes_pred\n\n\ndef classifier_assess(classifier, x, y):\n '''Function that prints the performance of the classifier\n Inputs:\n -classifier: trained MLP classifier\n -x: numpy array with embeddings\n -y: numpy array with lithological classes predicted'''\n Y2 = retrieve_predictions(classifier, x)\n print('f1 score: ', metrics.f1_score(y, Y2, average='macro'),\n 'accuracy: ', metrics.accuracy_score(y, Y2),\n 'balanced_accuracy:', metrics.balanced_accuracy_score(y, Y2))\n\n\ndef save_predictions(Dataframe, classifier, x, name):\n '''Function that saves dataframe predictions as a pickle file\n Inputs:\n -Dataframe: pandas dataframe with mean_embeddings\n -classifier: trained MLP model,\n -x: numpy array with embeddings,\n -name: string name to save dataframe\n Outputs:\n -save dataframe'''\n preds = classifier.predict(x, verbose=0)\n Dataframe['predicted_probabilities'] = preds.tolist()\n Dataframe['pred'] = retrieve_predictions(classifier, x).astype(np.int32)\n Dataframe[['x', 'y', 'FromDepth', 'ToDepth', 'TopElev', 'BottomElev',\n 'mean', 'predicted_probabilities', 'pred', 'reclass', 'code']].to_pickle('{}.pkl'.format(name))",
"_____no_output_____"
],
[
"# assessment of model performance\nclassifier_assess(model, X_validation, Y_validation)\n\n# save lithological prediction likelihoods dataframe\nsave_predictions(DF2, model, X, 'YGSBpredictions')",
"f1 score: 1.0 accuracy: 1.0 balanced_accuracy: 1.0\n"
],
[
"import pickle\n\nwith open('YGSBpredictions.pkl', 'rb') as f:\n data = pickle.load(f)",
"_____no_output_____"
],
[
"print(data)",
" x y FromDepth ToDepth TopElev BottomElev \\\n0 513822.0449 6952747.766 0 1.0 400.000000 399.133975 \n1 513822.5449 6952747.766 1 2.0 399.133975 398.267949 \n2 513823.0449 6952747.766 2 3.0 398.267949 397.401924 \n3 513823.5449 6952747.766 3 4.0 397.401924 396.535898 \n4 513824.0449 6952747.766 4 5.0 396.535898 395.669873 \n... ... ... ... ... ... ... \n159942 576956.0811 6882354.387 25 28.0 375.000000 372.000000 \n159943 576956.0811 6882354.387 28 29.0 372.000000 371.000000 \n159944 577054.2428 6882353.752 1 4.0 399.000000 396.000000 \n159945 577054.2428 6882353.752 4 5.0 396.000000 395.000000 \n159946 577152.4045 6882353.117 46 47.0 354.000000 353.000000 \n\n mean \\\n0 [-0.10188103, -0.009773923, -0.030616358, -0.1... \n1 [-0.10188103, -0.009773923, -0.030616358, -0.1... \n2 [0.09962925, -0.10881818, -0.29363656, -0.2486... \n3 [0.09962925, -0.10881818, -0.29363656, -0.2486... \n4 [0.09962925, -0.10881818, -0.29363656, -0.2486... \n... ... \n159942 [-0.2746381, -0.23468062, -0.37358257, 0.00474... \n159943 [-0.2746381, -0.23468062, -0.37358257, 0.00474... \n159944 [0.010618463, -0.33650404, -0.120667905, -0.02... \n159945 [-0.2746381, -0.23468062, -0.37358257, 0.00474... \n159946 [0.010618463, -0.33650404, -0.120667905, -0.02... \n\n predicted_probabilities pred reclass \\\n0 [2.280496902429263e-09, 8.17267023641658e-15, ... 14 colluvium \n1 [2.280496902429263e-09, 8.17267023641658e-15, ... 14 colluvium \n2 [3.0045320837842383e-28, 3.419642554591617e-34... 44 mud \n3 [3.0045320837842383e-28, 3.419642554591617e-34... 44 mud \n4 [3.0045320837842383e-28, 3.419642554591617e-34... 44 mud \n... ... ... ... \n159942 [0.0, 0.0, 0.0, 0.0, 1.0, 0.0, 0.0, 0.0, 0.0, ... 4 basalt \n159943 [0.0, 0.0, 0.0, 0.0, 1.0, 0.0, 0.0, 0.0, 0.0, ... 4 basalt \n159944 [1.0, 3.293394082603164e-20, 5.359187797035615... 0 Other \n159945 [0.0, 0.0, 0.0, 0.0, 1.0, 0.0, 0.0, 0.0, 0.0, ... 4 basalt \n159946 [1.0, 3.293394082603164e-20, 5.359187797035615... 0 Other \n\n code \n0 14 \n1 14 \n2 44 \n3 44 \n4 44 \n... ... \n159942 4 \n159943 4 \n159944 0 \n159945 4 \n159946 0 \n\n[122502 rows x 11 columns]\n"
],
[
"len(data)",
"_____no_output_____"
],
[
"data.head()",
"_____no_output_____"
],
[
"tmp = data['predicted_probabilities'][0]",
"_____no_output_____"
],
[
"len(tmp)",
"_____no_output_____"
],
[
"data.to_csv('YGSBpredictions.csv')",
"_____no_output_____"
],
[
"import base64\nwith open('a.csv', 'a', encoding='utf8') as csv_file:\n wr = csv.writer(csv_file, delimiter='|')\n pickle_bytes = pickle.dumps(obj) # unsafe to write\n b64_bytes = base64.b64encode(pickle_bytes) # safe to write but still bytes\n b64_str = b64_bytes.decode('utf8') # safe and in utf8\n wr.writerow(['col1', 'col2', b64_str])\n\n\n# the file contains\n# col1|col2|gANdcQAu\n\nwith open('a.csv', 'r') as csv_file:\n for line in csv_file:\n line = line.strip('\\n')\n b64_str = line.split('|')[2] # take the pickled obj\n obj = pickle.loads(base64.b64decode(b64_str)) #",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb8b5227a1d231559d8275cb4e88fcbc08e237e0
| 11,305 |
ipynb
|
Jupyter Notebook
|
concurrent_task_progress_bar.ipynb
|
mns1yash/Work_Yash
|
80dccc7dcbcc52d0760bb70442dbcf5e9eb066fc
|
[
"MIT"
] | null | null | null |
concurrent_task_progress_bar.ipynb
|
mns1yash/Work_Yash
|
80dccc7dcbcc52d0760bb70442dbcf5e9eb066fc
|
[
"MIT"
] | null | null | null |
concurrent_task_progress_bar.ipynb
|
mns1yash/Work_Yash
|
80dccc7dcbcc52d0760bb70442dbcf5e9eb066fc
|
[
"MIT"
] | null | null | null | 22.037037 | 944 | 0.507917 |
[
[
[
"# Displaying Progress Bar for Concurrent Tasks",
"_____no_output_____"
],
[
"`pip install tqdm`",
"_____no_output_____"
]
],
[
[
"import time\nfrom tqdm.notebook import tqdm",
"_____no_output_____"
],
[
"N = 30",
"_____no_output_____"
],
[
"def foo(a):\n time.sleep(0.2)\n return a**2",
"_____no_output_____"
],
[
"x = []\nfor i in tqdm(range(N)):\n x.append(foo(i))",
"_____no_output_____"
],
[
"x = [foo(i) for i in tqdm(range(N))]",
"_____no_output_____"
],
[
"x = list(tqdm(map(foo, range(N)), total=N))",
"_____no_output_____"
]
],
[
[
"## multiprocessing",
"_____no_output_____"
]
],
[
[
"from multiprocessing.pool import Pool\nfrom concurrent.futures import ProcessPoolExecutor\nfrom tqdm.contrib.concurrent import process_map",
"_____no_output_____"
],
[
"with Pool() as p:\n x = list(tqdm(p.imap(foo, range(N)), total=N))",
"_____no_output_____"
],
[
"with ProcessPoolExecutor() as executor:\n x = list(tqdm(executor.map(foo, range(N)), total=N))",
"_____no_output_____"
],
[
"x = process_map(foo, range(N))",
"_____no_output_____"
]
],
[
[
"## multithreading",
"_____no_output_____"
]
],
[
[
"from multiprocessing.pool import ThreadPool\nfrom concurrent.futures import ThreadPoolExecutor\nfrom tqdm.contrib.concurrent import thread_map",
"_____no_output_____"
],
[
"with ThreadPool(processes=10) as p:\n x = list(tqdm(p.imap(foo, range(N)), total=N))",
"_____no_output_____"
],
[
"with ThreadPoolExecutor(max_workers=10) as executor:\n x = list(tqdm(executor.map(foo, range(N)), total=N))",
"_____no_output_____"
],
[
"x = thread_map(foo, range(N), max_workers=10)",
"_____no_output_____"
]
],
[
[
"## asyncio",
"_____no_output_____"
],
[
"`pip install asyncio`\n\n`pip install nest_asyncio`",
"_____no_output_____"
]
],
[
[
"import asyncio\nimport nest_asyncio\nnest_asyncio.apply()",
"_____no_output_____"
],
[
"async def afoo(a):\n await asyncio.sleep(0.2)\n return a**2",
"_____no_output_____"
],
[
"asyncio.run(afoo(2))",
"_____no_output_____"
],
[
"tasks = list(map(afoo, range(N)))\nx = asyncio.run(asyncio.gather(*tasks))",
"_____no_output_____"
],
[
"async def do1():\n tasks = list(map(afoo, range(N)))\n return [await t for t in tqdm(asyncio.as_completed(tasks), total=N)]",
"_____no_output_____"
],
[
"x = asyncio.run(do1())",
"_____no_output_____"
],
[
"async def do2():\n tasks = list(map(asyncio.create_task, map(afoo, range(N))))\n for t in tqdm(asyncio.as_completed(tasks), total=N):\n await t\n return [t.result() for t in tasks]",
"_____no_output_____"
],
[
"x = asyncio.run(do2())",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb8b5fadf61f6098b90a9b14d631461b7eb0d782
| 112,995 |
ipynb
|
Jupyter Notebook
|
lastfm/lastfm_dash.ipynb
|
mcwaage1/qs
|
6ca4e96dac753d2cc886e788b2a4e92f41a0b514
|
[
"MIT"
] | 7 |
2021-12-13T15:10:58.000Z
|
2022-02-27T22:58:56.000Z
|
lastfm/lastfm_dash.ipynb
|
mcwaage1/qs
|
6ca4e96dac753d2cc886e788b2a4e92f41a0b514
|
[
"MIT"
] | null | null | null |
lastfm/lastfm_dash.ipynb
|
mcwaage1/qs
|
6ca4e96dac753d2cc886e788b2a4e92f41a0b514
|
[
"MIT"
] | null | null | null | 32.733198 | 233 | 0.36237 |
[
[
[
"# https://community.plotly.com/t/different-colors-for-bars-in-barchart-by-their-value/6527/7\n%reset",
"Once deleted, variables cannot be recovered. Proceed (y/[n])? y\n"
],
[
"# Run this app with `python app.py` ando\n# visit http://127.0.0.1:8050/ in your web browser.\n\nimport dash\nimport dash_core_components as dcc\nimport dash_html_components as html\nimport plotly.express as px\nimport jupyter_dash\nimport pandas as pd\nfrom dash.dependencies import Input, Output\nimport plotly.graph_objects as go\nfrom plotly.subplots import make_subplots\n\nexternal_stylesheets = ['https://codepen.io/chriddyp/pen/bWLwgP.css']",
"_____no_output_____"
],
[
"top_artists_df = pd.read_csv('~/qs/lastfm/data/lastfm_top_artists.csv')\ntop_tracks_df = pd.read_csv('~/qs/lastfm/data/lastfm_top_tracks.csv')\ntop_albums_df = pd.read_csv('~/qs/lastfm/data/lastfm_top_albums.csv')",
"_____no_output_____"
],
[
"new_top_artists_df = pd.read_csv('~/qs/lastfm/data/lastfm_top_artists_with_tags.csv', usecols=[1, 2, 3])\nnew_top_artists_df\ntop_artists_df = new_top_artists_df\ntop_artists_df",
"_____no_output_____"
],
[
"print('Top Artists')\nprint(f\"{top_artists_df.head(5)} \\n\")\n# print('Top Tracks')\n# print(f\"{top_tracks_df.head(5)} \\n\")\n# print('Top Albums')\n# print(top_albums_df.head(5))",
"Top Artists\n artist play_count tags\n0 Opeth 668 progressive metal\n1 Between the Buried and Me 186 progressive metal\n2 King Gizzard & The Lizard Wizard 165 psychedelic rock\n3 Lucifer 154 doom metal\n4 Gojira 139 death metal \n\n"
],
[
"top_artists_df.tail(5)",
"_____no_output_____"
],
[
"df = top_artists_df",
"_____no_output_____"
],
[
"# total_songs = top_artists_df['play_count'].sum()\n# total_songs",
"_____no_output_____"
],
[
"# total_artists = len(top_artists_df['artist'])\n# total_artists",
"_____no_output_____"
],
[
"# def num_unique_tags(df):\n# unique_tags = []\n# for tags, num_artists_in_tags in df.groupby('tags'):\n# unique_tags.append(len(num_artists_in_tags))\n# unique_tags.sort(reverse=True)\n# return len(unique_tags)",
"_____no_output_____"
],
[
"# num_unique_tags(top_artists_df)",
"_____no_output_____"
],
[
"metal = 'metal|core|sludge'\nrock = 'rock'\nlen(df.loc[df['tags'].str.contains(metal)])",
"_____no_output_____"
],
[
"# if you want custom colors for certain words in the tags\n\n#https://stackoverflow.com/questions/23400743/pandas-modify-column-values-in-place-based-on-boolean-array\n#px.colors.qualitative.Plotly\n\ncustom_colors = ['#EF553B'] * 200\ndf['colors'] = custom_colors\n\nmetal = 'metal|core|sludge'\ndf.loc[df['tags'].str.contains(metal), 'colors'] = '#636EFA'\n\nrock = 'rock|blues'\ndf.loc[df['tags'].str.contains(rock), 'colors'] = '#00CC96'\n\npunk = 'punk'\ndf.loc[df['tags'].str.contains(punk), 'colors'] = '#AB63FA'\n\nalternative = 'alternative'\ndf.loc[df['tags'].str.contains(alternative), 'colors'] = '#FFA15A'\n\nindie = 'indie'\ndf.loc[df['tags'].str.contains(indie), 'colors'] = '#19D3F3'\n\nbilly = 'billy'\ndf.loc[df['tags'].str.contains(billy), 'colors'] = '#FF6692'\n\nrap = 'rap|hip|rnb'\ndf.loc[df['tags'].str.contains(rap), 'colors'] = '#B6E880'\n\npop = 'pop|soul'\ndf.loc[df['tags'].str.contains(pop), 'colors'] = '#FF97FF'\n\nelectronic = 'electronic|synthwave'\ndf.loc[df['tags'].str.contains(electronic), 'colors'] = '#FECB52'\n\ndf",
"_____no_output_____"
],
[
"title = f'Last.fm Dashboard'\n\n#https://stackoverflow.com/questions/22291395/sorting-the-grouped-data-as-per-group-size-in-pandas\ndf_grouped = sorted(df.groupby('tags'), key=lambda x: len(x[1]), reverse=True)\n\nartist = df['artist']\nplay_count = df['play_count']\ntags = df['tags']\n\ntotal_songs = df['play_count'].sum()\ntotal_artists = len(df['artist'])\nnum_unique_tags = len(df.groupby('tags'))\n\nfig = go.Figure()\n\ndef make_traces(df):\n for tags, df in df_grouped:\n num_tags = len(df)\n fig.add_trace(go.Bar(y=df['artist'],\n x=df['play_count'],\n orientation='h',\n text=df['play_count'],\n textposition='outside',\n name=f\"{tags} ({num_tags})\",\n customdata = df['tags'],\n hovertemplate = \n \"Artist: %{y}<br>\" +\n \"Play Count: %{x}<br>\" +\n \"Tag: %{customdata}\" +\n \"<extra></extra>\",\n \n # for custom colors\n marker_color=df['colors'],\n \n #https://community.plotly.com/t/different-colors-for-bars-in-barchart-by-their-value/6527/4\n #marker={'color': colors[tags]},\n showlegend=True,\n ))\n \nmake_traces(top_artists_df)\n\nfig.update_layout(title=dict(text=title, \n yanchor=\"top\",\n y=.95,\n xanchor=\"left\",\n x=.075),\n legend_title=f'Unique Tags: ({num_unique_tags})',\n legend_itemclick='toggle',\n legend_itemdoubleclick='toggleothers',\n margin_l = 240,\n xaxis=dict(fixedrange=True),\n \n # https://towardsdatascience.com/4-ways-to-improve-your-plotly-graphs-517c75947f7e\n yaxis=dict(categoryorder='total descending'),\n \n dragmode='pan',\n annotations=[\n #https://plotly.com/python/text-and-annotations/#adding-annotations-with-xref-and-yref-as-paper\n #https://community.plotly.com/t/how-to-add-a-text-area-with-custom-text-as-a-kind-of-legend-in-a-plotly-plot/24349/3\n go.layout.Annotation(\n text=f'Total Songs Tracked: {total_songs}<br>Total Artists Tracked: {total_artists}',\n align='left',\n showarrow=False,\n xref='paper',\n yref='paper',\n yanchor=\"bottom\",\n y=1.02,\n xanchor=\"right\",\n x=1,\n )])\n\nfig.update_yaxes(title_text=\"Artist\",\n type='category',\n range=[25.5, -.5],\n \n # https://plotly.com/python/setting-graph-size/#adjusting-height-width--margins\n automargin=False\n )\nfig.update_xaxes(title_text=\"Play Count \",\n range=[0, 700],\n dtick=100,\n )\n \napp = jupyter_dash.JupyterDash(__name__,\n external_stylesheets=external_stylesheets,\n title=f\"{title}\")\n\ndef make_footer():\n return html.Div(html.Footer([\n 'Matthew Waage',\n html.A('github.com/mcwaage1',\n href='http://www.github.com/mcwaage1',\n target='_blank',\n style = {'margin': '.5em'}),\n html.A('[email protected]',\n href=\"mailto:[email protected]\",\n target='_blank',\n style = {'margin': '.5em'}),\n html.A('waage.dev',\n href='http://www.waage.dev', \n target='_blank',\n style = {'margin': '.5em'})\n ], style={'position': 'fixed',\n 'text-align': 'right',\n 'left': '0px',\n 'bottom': '0px',\n 'margin-right': '10%',\n 'color': 'black',\n 'display': 'inline-block',\n 'background': '#f2f2f2',\n 'border-top': 'solid 2px #e4e4e4',\n 'width': '100%'}))\n\napp.layout = html.Div([\n dcc.Graph(\n figure=fig,\n \n #https://plotly.com/python/setting-graph-size/\n #https://stackoverflow.com/questions/46287189/how-can-i-change-the-size-of-my-dash-graph\n style={'height': '95vh'}\n ),\n make_footer(),\n])\n\nif __name__ == '__main__':\n app.run_server(mode ='external', port=8070, debug=True)",
"Dash app running on http://127.0.0.1:8070/\n"
],
[
"# title = '2020 Last.fm Dashboard'\n\n# fig = go.Figure()\n\n# fig.add_trace(go.Bar(\n# x=top_artists_df['artist'],\n# y=top_artists_df['play_count'],\n# text=top_artists_df['play_count'],\n# ))\n\n# fig.update_traces(textposition='outside')\n\n# fig.update_layout(title=dict(text=title, \n# yanchor=\"top\",\n# y=.95,\n# xanchor=\"left\",\n# x=.075),\n# dragmode='pan',\n# annotations=[\n# #https://plotly.com/python/text-and-annotations/#adding-annotations-with-xref-and-yref-as-paper\n# #https://community.plotly.com/t/how-to-add-a-text-area-with-custom-text-as-a-kind-of-legend-in-a-plotly-plot/24349/3\n# go.layout.Annotation(\n# text=f'Total Songs Played: {total_songs}',\n# align='left',\n# showarrow=False,\n# xref='paper',\n# yref='paper',\n# yanchor=\"bottom\",\n# y=1.02,\n# xanchor=\"right\",\n# x=1,\n# )])\n\n# # https://stackoverflow.com/questions/61782622/plotly-how-to-add-a-horizontal-scrollbar-to-a-plotly-express-figure\n# # https://community.plotly.com/t/freeze-y-axis-while-scrolling-along-x-axis/4898/5\n# fig.update_layout(\n# xaxis=dict(\n# rangeslider=dict(\n# visible=True,\n# )))\n\n# fig.update_xaxes(title_text=\"Artist\",\n# type='category',\n# range=[-.5, 25.5],\n# )\n# fig.update_yaxes(title_text=\"Play Count \",\n# range=[0, 750],\n# dtick=100,\n# )\n \n# app = jupyter_dash.JupyterDash(__name__,\n# external_stylesheets=external_stylesheets,\n# title=f\"{title}\")\n\n# app.layout = html.Div([\n# dcc.Graph(\n# figure=fig,\n \n# #https://plotly.com/python/setting-graph-size/\n# #https://stackoverflow.com/questions/46287189/how-can-i-change-the-size-of-my-dash-graph\n# style={'height': '95vh'}\n# )\n# ])\n\n# if __name__ == '__main__':\n# app.run_server(mode ='external', debug=True, port=8080)",
"_____no_output_____"
],
[
"import requests\nimport json\n\nheaders = {\n 'user-agent': 'mcwaage1'\n}\n\nwith open(\"data/credentials.json\", \"r\") as file:\n credentials = json.load(file)\n last_fm_cr = credentials['last_fm']\n key = last_fm_cr['KEY']\n username = last_fm_cr['USERNAME']\n\nlimit = 20 #api lets you retrieve up to 200 records per call\nextended = 0 #api lets you retrieve extended data for each track, 0=no, 1=yes\npage = 1 #page of results to start retrieving at",
"_____no_output_____"
]
],
[
[
"### Testing out api calls of artists",
"_____no_output_____"
]
],
[
[
"artist = \"death from above 1979\"\nartist = artist.replace(' ', '+')\nartist",
"_____no_output_____"
],
[
"method = 'artist.gettoptags'\nrequest_url = f'http://ws.audioscrobbler.com/2.0/?method={method}&artist={artist}&api_key={key}&limit={limit}&extended={extended}&page={page}&format=json'\nresponse = requests.get(request_url, headers=headers)\nresponse.status_code",
"_____no_output_____"
],
[
"artist_tags = [tag['name'] for tag in response.json()['toptags']['tag'][:3]]\nartist_tags",
"_____no_output_____"
],
[
"method = 'tag.gettoptags'\nrequest_url = f'http://ws.audioscrobbler.com/2.0/?method={method}&api_key={key}&limit={limit}&extended={extended}&page={page}&format=json'\nresponse = requests.get(request_url, headers=headers)\nresponse.status_code",
"_____no_output_____"
],
[
"top_tags = artist_tags\ntop_tags",
"_____no_output_____"
],
[
"method = 'user.getinfo'\nrequest_url = f'http://ws.audioscrobbler.com/2.0/?method={method}&user={username}&api_key={key}&limit={limit}&extended={extended}&page={page}&format=json'\nresponse = requests.get(request_url, headers=headers)\nresponse.status_code",
"_____no_output_____"
],
[
"response.json()",
"_____no_output_____"
],
[
"user_info = [response.json()['user']['name']]\nuser_info",
"_____no_output_____"
],
[
"user_info.append(response.json()['user']['url'])",
"_____no_output_____"
],
[
"user_info.append(response.json()['user']['image'][0]['#text'])\nuser_info",
"_____no_output_____"
],
[
"# def make_user_info(user_info):\n# return html.Div(children=[html.Img(src=f'{user_info[2]}'),\n# html.A(f'{user_info[0]}',\n# href=f'{user_info[1]}',\n# target='_blank',\n# style={'margin': '.5em'}\n# ),\n# ])",
"_____no_output_____"
]
],
[
[
"### End of testing",
"_____no_output_____"
]
],
[
[
"artists = []\nartists",
"_____no_output_____"
],
[
"def get_artists():\n artists = []\n for artist in top_artists_df['artist']:\n artist = artist.replace(' ', '+')\n artists.append(artist)\n return artists",
"_____no_output_____"
],
[
"artists_to_parse = get_artists()\nartists_to_parse",
"_____no_output_____"
]
],
[
[
"# To start the api calling process and get new data",
"_____no_output_____"
]
],
[
[
"# replace the [:1] with [:3] or whatever for more tags of artist\nartist_genre = []\nfor artist in artists_to_parse:\n request_url = f'http://ws.audioscrobbler.com/2.0/?method=artist.gettoptags&artist={artist}&api_key={key}&limit={limit}&extended={extended}&page={page}&format=json'\n response = requests.get(request_url, headers=headers)\n artist_tags = [tag['name'] for tag in response.json()['toptags']['tag'][:1]]\n artist_genre.append(artist_tags)",
"_____no_output_____"
],
[
"artist_genre",
"_____no_output_____"
],
[
"# https://stackoverflow.com/questions/12555323/adding-new-column-to-existing-dataframe-in-python-pandas\ntop_artists_df['tags'] = artist_genre",
"_____no_output_____"
],
[
"top_artists_df.head(5)",
"_____no_output_____"
],
[
"top_artists_df['tags'] = top_artists_df['tags'].astype(str)",
"_____no_output_____"
],
[
"top_artists_df['tags'] = top_artists_df['tags'].str.strip(\"['']\")",
"_____no_output_____"
],
[
"top_artists_df['tags'] = top_artists_df['tags'].str.lower()\ntop_artists_df.head(5)",
"_____no_output_____"
]
],
[
[
"### To replace tags that you don't want",
"_____no_output_____"
]
],
[
[
"tags_to_replace = 'seen live|vocalists'\n\ndef get_new_artists(tags_to_replace):\n artists_to_replace = []\n for artist in df.loc[df['tags'].str.contains(tags_to_replace)]['artist']:\n artists_to_replace.append(artist)\n return artists_to_replace\n\nget_new_artists(tags_to_replace)",
"_____no_output_____"
],
[
"tags_to_replace = 'seen live|vocalists'\n\ndef get_artists_to_replace(tags_to_replace):\n artists_to_replace = []\n for artist in df.loc[df['tags'].str.contains(tags_to_replace)]['artist']:\n artist = artist.replace(' ', '+')\n artists_to_replace.append(artist)\n return artists_to_replace\n\nget_artists_to_replace(tags_to_replace)",
"_____no_output_____"
],
[
"new_artists_to_parse = get_artists_to_replace(tags_to_replace)\nnew_artists_tags = []\nfor artist in new_artists_to_parse:\n request_url = f'http://ws.audioscrobbler.com/2.0/?method=artist.gettoptags&artist={artist}&api_key={key}&limit={limit}&extended={extended}&page={page}&format=json'\n response = requests.get(request_url, headers=headers)\n artist_tags = [tag['name'] for tag in response.json()['toptags']['tag'][1:2]]\n new_artists_tags.append(artist_tags)\nnew_artists_tags",
"_____no_output_____"
],
[
"new_artists_tags = [str(x) for x in new_artists_tags]\nnew_artists_tags = [x.strip(\"['']\") for x in new_artists_tags]\nnew_artists_tags = [x.lower() for x in new_artists_tags]\n\nnew_artists_tags",
"_____no_output_____"
],
[
"for artist in get_new_artists(tags_to_replace):\n print(artist)",
"Demob Happy\nLana Del Rey\nCRX\n"
],
[
"for k, v in zip(get_new_artists(tags_to_replace), new_artists_tags):\n df.loc[df['artist'].str.contains(k), 'tags'] = v",
"_____no_output_____"
]
],
[
[
"### End of replacing tags",
"_____no_output_____"
]
],
[
[
"top_artists_df.to_csv('~/qs/lastfm/data/lastfm_top_artists_with_tags.csv')",
"_____no_output_____"
],
[
"from IPython.display import display\nwith pd.option_context('display.max_rows', 205, 'display.max_columns', 5):\n display(top_artists_df)",
"_____no_output_____"
],
[
"# unique_tags = top_artists_df['tags'].unique()\n# unique_tags = pd.Series(unique_tags)\n# print(\"Type: \", type(unique_tags))\n# print('')\n# for tag in unique_tags:\n# print(tag)",
"_____no_output_____"
],
[
"# len(unique_tags)",
"_____no_output_____"
],
[
"# def get_sorted_tags(df):\n# unique_tags = df['tags'].unique()\n# unique_tags = pd.Series(unique_tags) \n\n# sorted_tags = []\n# for tag in unique_tags:\n# #sorted_tags.append((top_artists_df['tags'].str.count(tag).sum(), tag))\n# #sorted_tags.append(top_artists_df['tags'].str.count(tag).sum())\n# sorted_tags.sort(reverse=True)\n# return sorted_tags",
"_____no_output_____"
],
[
"# get_sorted_tags(top_artists_df)",
"_____no_output_____"
],
[
"# unique_tags = unique_tags.str.split()",
"_____no_output_____"
],
[
"# type(unique_tags)",
"_____no_output_____"
],
[
"# unique_tags",
"_____no_output_____"
],
[
"# for tag in unique_tags:\n# print(tag, unique_tags.str.count(tag).sum())",
"_____no_output_____"
],
[
"# px.colors.qualitative.Plotly",
"_____no_output_____"
],
[
"# https://plotly.com/python/discrete-color/\n#fig = px.colors.qualitative.swatches()\n\n#https://plotly.com/python/renderers/\n#fig.show(renderer='iframe')",
"_____no_output_____"
],
[
"# One way to replace value in one series based on another, better version below\n# top_artists_df['colors'][top_artists_df['tags'].str.contains('metal')] = '#636EFA'",
"_____no_output_____"
],
[
"# top_tags",
"_____no_output_____"
],
[
"#https://stackoverflow.com/questions/23400743/pandas-modify-column-values-in-place-based-on-boolean-array\npx.colors.qualitative.Plotly\n# custom_colors = ['#EF553B'] * 200\n# df['colors'] = custom_colors\n\n# df.loc[df['tags'].str.contains('metal'), 'colors'] = '#636EFA'\n# df.loc[df['tags'].str.contains('rock'), 'colors'] = '#00CC96'\n# df.loc[df['tags'].str.contains('punk'), 'colors'] = '#AB63FA'\n# df.loc[df['tags'].str.contains('alternative'), 'colors'] = '#FFA15A'\n# df.loc[df['tags'].str.contains('indie'), 'colors'] = '#19D3F3'\n# df.loc[df['tags'].str.contains('billy'), 'colors'] = '#FF6692'\n\n# df",
"_____no_output_____"
],
[
"# colors = {'Ugly': 'red',\n# 'Bad': 'orange',\n# 'Good': 'lightgreen',\n# 'Great': 'darkgreen'\n# }",
"_____no_output_____"
],
[
"# from IPython.display import display\n# with pd.option_context('display.max_rows', 265, 'display.max_columns', 5):\n# display(top_artists_df)",
"_____no_output_____"
],
[
"# for tag, artist in top_artists_df.groupby('tags'):\n# print(tag, len(artist))\n# print(artist)\n# print('')",
"_____no_output_____"
],
[
"# top_artists_df.loc[top_artists_df['tags'].str.contains('metal')]",
"_____no_output_____"
],
[
"# type(top_artists_df.loc[top_artists_df['tags'].str.contains('metal')])",
"_____no_output_____"
],
[
"# len(top_artists_df.loc[top_artists_df['tags'].str.contains('metal')])",
"_____no_output_____"
],
[
"# def print_tags(df):\n# printed_tags = []\n# for tags, top_artists in top_artists_df.groupby('tags'):\n# printed_tags.append([len(top_artists), tags])\n# printed_tags.sort(reverse=True)\n# return printed_tags",
"_____no_output_____"
],
[
"# print_tags(top_artists_df)",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb8b614532fdc2df40f299ed8649a674d718a3a1
| 314,675 |
ipynb
|
Jupyter Notebook
|
.ipynb_checkpoints/hawaii_climate_analysis_sql_alchemy-checkpoint.ipynb
|
jhustles/mysql_sql_alchemy_sakila_db
|
51433887bf767a7711ebe5f9c34202868cdbef0a
|
[
"MIT"
] | 1 |
2020-04-04T08:15:59.000Z
|
2020-04-04T08:15:59.000Z
|
hawaii_climate_analysis_sql_alchemy.ipynb
|
jhustles/mysql_sql_alchemy_sakila_db
|
51433887bf767a7711ebe5f9c34202868cdbef0a
|
[
"MIT"
] | null | null | null |
hawaii_climate_analysis_sql_alchemy.ipynb
|
jhustles/mysql_sql_alchemy_sakila_db
|
51433887bf767a7711ebe5f9c34202868cdbef0a
|
[
"MIT"
] | null | null | null | 115.817078 | 27,968 | 0.606232 |
[
[
[
"# Hawaii - A Climate Analysis And Exploration\n### For data between August 23, 2016 - August 23, 2017\n---",
"_____no_output_____"
]
],
[
[
"# Import dependencies\n\n%matplotlib inline\nfrom matplotlib import style\nstyle.use('fivethirtyeight')\nimport matplotlib.pyplot as plt\nimport numpy as np\nimport pandas as pd\nimport datetime as dt\n# Python SQL toolkit and Object Relational Mapper\nimport sqlalchemy\nfrom sqlalchemy.ext.automap import automap_base\nfrom sqlalchemy.orm import Session\nfrom sqlalchemy import create_engine, func, inspect",
"_____no_output_____"
]
],
[
[
"## Reflect Tables into SQLAlchemy ORM",
"_____no_output_____"
]
],
[
[
"# Set up query engine. 'echo=True is the default - will keep a log of activities'\n\nengine = create_engine(\"sqlite:///Resources/hawaii.sqlite\")",
"_____no_output_____"
],
[
"# Reflect an existing database into a new model\nBase = automap_base()\n# Reflect the tables\nBase.prepare(engine, reflect=True)",
"_____no_output_____"
],
[
"# We can view all of the classes that automap found\nBase.classes.keys()",
"_____no_output_____"
],
[
"# Another way to get table names from SQL-lite\ninspector = inspect(engine)\ninspector.get_table_names()",
"_____no_output_____"
]
],
[
[
"## Exploratory Climate Analysis",
"_____no_output_____"
]
],
[
[
"# Save references to each table\nMeasurement = Base.classes.measurement\nStation = Base.classes.station",
"_____no_output_____"
],
[
"# Create our session (link) from Python to the DB\nsession = Session(engine)",
"_____no_output_____"
],
[
"# Display details of 'measurement' table\ncolumns = inspector.get_columns('measurement')\nfor c in columns:\n print(c['name'], c['type'])",
"id INTEGER\nstation TEXT\ndate TEXT\nprcp FLOAT\ntobs FLOAT\n"
],
[
"# DISPLY number of line items measurement, and remove tuple form\nresult, = engine.execute('SELECT COUNT(*) FROM measurement').fetchall()[0]\nprint(result,)",
"19550\n"
],
[
"# Display details of 'station' table\ncolumns = inspector.get_columns('station')\nfor c in columns:\n print(c['name'], c['type'])",
"id INTEGER\nstation TEXT\nname TEXT\nlatitude FLOAT\nlongitude FLOAT\nelevation FLOAT\n"
],
[
"# DISPLY number of line items station, and remove tuple form\n\nresult, = engine.execute('SELECT COUNT(*) FROM station').fetchall()[0]\nprint(result,)",
"9\n"
],
[
"# FULL INNTER JOIN BOTH THE MEASUREMENT AND STATION TABLE\n\n# engine.execute('SELECT measurement.*, station.name, station.latitude FROM measurement INNER JOIN station ON measurement.station = station.station;').fetchall()\njoin_result = engine.execute('SELECT * FROM measurement INNER JOIN station ON measurement.station = station.station;').fetchall()\njoin_result",
"_____no_output_____"
],
[
"# Another way to PERFORM AN INNER JOIN ON THE MEASUREMENT AND STATION TABLES\n\nengine.execute('SELECT measurement.*, station.* FROM measurement, station WHERE measurement.station=station.station;').fetchall()",
"_____no_output_____"
],
[
"# Query last date of the measurement file\n\nlast_date = session.query(Measurement.date).order_by(Measurement.date.desc()).first()[0]\n\nprint(last_date) \n\nlast_date_measurement = dt.date(2017, 8 ,23)",
"2017-08-23\n"
],
[
"# Calculate the date 1 year delta of the \"last date measurement\"\n\none_year_ago = last_date_measurement - dt.timedelta(days=365)\nprint(one_year_ago)",
"2016-08-23\n"
],
[
"# Plotting precipitation data from 1 year ago\n\ndate = dt.date(2016, 8, 23)\n\n#sel = [Measurement.id, Measurement.station, Measurement.date, Measurement.prcp, Measurement.tobs]\n\nsel = [Measurement.date, Measurement.prcp]\n\nprint(date)\n\n# date = \"2016-08-23\"\n\nresult = session.query(Measurement.date, Measurement.prcp).\\\n filter(Measurement.date >= date).all()\n\n# get the count / length of the list of tuples\nprint(len(result))\n",
"2016-08-23\n2230\n"
],
[
"# Created a line plot and saved the figure\n\ndf = pd.DataFrame(result, columns=['Date', 'Precipitation'])\ndf.sort_values(by=['Date'])\ndf.set_index('Date', inplace=True)\ns = df['Precipitation']\nax = s.plot(figsize=(8,6), use_index=True, title='Precipitation Data Between 8/23/2016 - 8/23/2017')\nfig = ax.get_figure()\nfig.savefig('./Images/precipitation_line.png')",
"_____no_output_____"
],
[
"# Use Pandas to calcualte the summary statistics for the precipitation data\n\ndf.describe()",
"_____no_output_____"
],
[
"# Design a query to show how many stations are available in this dataset?\n\nsession.query(Measurement.station).\\\n group_by(Measurement.station).count()",
"_____no_output_____"
],
[
"# Querying for the most active stations (i.e. what stations have the most rows)?\n# List the stations and the counts in descending order.\n\nengine.execute('SELECT DISTINCT station, COUNT(id) FROM measurement GROUP BY station ORDER BY COUNT(id) DESC').fetchall()",
"_____no_output_____"
],
[
"# Query for stations from the measurement table\n\nsession.query(Measurement.station).\\\n group_by(Measurement.station).all()",
"_____no_output_____"
],
[
"# Using the station id from the previous query, calculate the lowest temperature recorded, \n# highest temperature recorded, and average temperature most active station?\n\nsel = [func.min(Measurement.tobs), func.max(Measurement.tobs), func.avg(Measurement.tobs)]\n\nsession.query(*sel).\\\n filter(Measurement.station == 'USC00519281').all()\n",
"_____no_output_____"
],
[
"# Query the dates of the last 12 months of the most active station\nlast_date = session.query(Measurement.date).\\\n filter(Measurement.station == 'USC00519281').\\\n order_by(Measurement.date.desc()).first()[0]\n\nprint(last_date) ",
"2017-08-18\n"
],
[
"last_date_USC00519281 = dt.date(2017, 8 ,18)\n\nlast_year_USC00519281 = last_date_USC00519281 - dt.timedelta(days=365)\nprint(last_year_USC00519281)",
"2016-08-18\n"
],
[
"# SET UP HISTOGRAM QUERY AND PLOT\n\nsel_two = [Measurement.tobs]\nresults_tobs_hist = session.query(*sel_two).\\\n filter(Measurement.date >= last_year_USC00519281).\\\n filter(Measurement.station == 'USC00519281').all()",
"_____no_output_____"
],
[
"# HISTOGRAM Plot\n\ndf = pd.DataFrame(results_tobs_hist, columns=['tobs'])\nax = df.plot.hist(figsize=(8,6), bins=12, use_index=False, title='Hawaii - Temperature Histogram Between 8/23/2016 - 8/23/2017')\nfig = ax.get_figure()\nfig.savefig('./Images/temperature_histogram.png')",
"_____no_output_____"
],
[
"# Created a function called `calc_temps` that accepts a 'start date' and 'end date' in the format 'YYYY-MM-DD' \n# and return the minimum, average, and maximum temperatures for that range of dates\n\ndef calc_temps(start_date, end_date):\n \"\"\"Temp MIN,Temp AVG, and Temp MAX for a list of dates.\n \n Args are:\n start_date (string): A date string in the format YYYY-MM-DD\n end_date (string): A date string in the format YYYY-MM-DD\n \n Returns:\n T-MIN, T-AVG, and T-MAX\n \"\"\"\n \n return session.query(func.min(Measurement.tobs), func.avg(Measurement.tobs), func.max(Measurement.tobs)).\\\n filter(Measurement.date >= start_date).filter(Measurement.date <= end_date).all()\n\n\nprint(calc_temps('2017-08-01', '2017-08-07'))",
"[(72.0, 79.25, 83.0)]\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb8b6324a0057dcd0ab1e9795103b68764a9cf93
| 52,405 |
ipynb
|
Jupyter Notebook
|
module2-wrangle-ml-datasets/Baseline_github.ipynb
|
ddodds42/DS-Unit-2-Applied-Modeling
|
e83efdf31051977da62beb8de62ed38d696e612b
|
[
"MIT"
] | null | null | null |
module2-wrangle-ml-datasets/Baseline_github.ipynb
|
ddodds42/DS-Unit-2-Applied-Modeling
|
e83efdf31051977da62beb8de62ed38d696e612b
|
[
"MIT"
] | null | null | null |
module2-wrangle-ml-datasets/Baseline_github.ipynb
|
ddodds42/DS-Unit-2-Applied-Modeling
|
e83efdf31051977da62beb8de62ed38d696e612b
|
[
"MIT"
] | null | null | null | 35.053512 | 90 | 0.338994 |
[
[
[
"import pandas as pd\nimport numpy as np\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.metrics import f1_score",
"_____no_output_____"
],
[
"df = pd.read_excel('Master_MELT.xlsx')\ndf",
"_____no_output_____"
],
[
"df.describe()",
"_____no_output_____"
],
[
"df.head(1)",
"_____no_output_____"
],
[
"State_fail_yr_5 = df[df.year0 < 2014].iloc[:,4:5]\nState_fail_yr_5.columns=['predicted']\nninety_19 = df[df.year0 > 1989]\nninety_14 = ninety_19[ninety_19['year0'] < 2015]\nState_fail_yr_5['actual'] = np.array(ninety_14.iloc[:,6:7])\nState_fail_yr_5",
"_____no_output_____"
],
[
"f1_score(State_fail_yr_5['actual'], State_fail_yr_5['predicted'])",
"_____no_output_____"
],
[
"# BASELINE T_State_Fail_yr_1\n# From the other notebook, For state failure on year one,\n# predicted by state failure in year 0, an F-Score of 0.80547\n\n# BASELINE T_State_Fail_yr_5\n# For state failure on year five, predicted by state failure in year 0,\n# an F-Score of 0.49036",
"_____no_output_____"
],
[
"State_fail_nxt_5 = df[df.year0 < 2014].iloc[:,4:5]\nState_fail_nxt_5.columns=['predicted']\nninety_19 = df[df.year0 > 1989]\nninety_14 = ninety_19[ninety_19['year0'] < 2015]\nState_fail_nxt_5['actual'] = np.array(ninety_14.iloc[:,5:6])\nState_fail_nxt_5",
"_____no_output_____"
],
[
"f1_score(State_fail_nxt_5['actual'], State_fail_nxt_5['predicted'])",
"_____no_output_____"
],
[
"# BASELINE T_State_Fail_plus_5\n# For state failure in any of the next 5 years,\n# predicted by state failure in year 0, an F-Score of 0.6685",
"_____no_output_____"
],
[
"HDI_Mean = df[df.year0 < 2012].iloc[:,2:3]\nHDI_Mean.columns=['predicted']\n\neighty9_16 = df[df.year0 < 2017]\nninety4_16 = eighty9_16[eighty9_16.year0 > 1993]\n\nHDI_Mean['actual'] = np.array(ninety4_16.iloc[:,2:3])\nHDI_Mean",
"_____no_output_____"
],
[
"from sklearn.metrics import mean_squared_error\nfrom sklearn.metrics import r2_score",
"_____no_output_____"
],
[
"(mean_squared_error(HDI_Mean['actual'], HDI_Mean['predicted']))**0.5",
"_____no_output_____"
],
[
"# BASELINE T_HDI_Mean_plus_5\n# For HDI Mean in the next 5 years, predicted by HDI Mean in the last 5 years,\n# an RMSE of 0.03012",
"_____no_output_____"
],
[
"HDI_Trend = df[df.year0 < 2012].iloc[:,3:4]\nHDI_Trend.columns=['predicted']\n\neighty9_16 = df[df.year0 < 2017]\nninety4_16 = eighty9_16[eighty9_16.year0 > 1993]\n\nHDI_Trend['actual'] = np.array(ninety4_16.iloc[:,3:4])\nHDI_Trend",
"_____no_output_____"
],
[
"(mean_squared_error(HDI_Trend['actual'], HDI_Trend['predicted']))**0.5",
"_____no_output_____"
],
[
"# BASELINE T_HDI_Trend_plus_5\n# For HDI Trend in the next 5 years, predicted by HDI Trend in the last 5 years,\n# an RMSE of 0.004824",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.