sha
stringlengths 40
40
| text
stringlengths 1
13.4M
| id
stringlengths 2
117
| tags
listlengths 1
7.91k
| created_at
stringlengths 25
25
| metadata
stringlengths 2
875k
| last_modified
stringlengths 25
25
| arxiv
listlengths 0
25
| languages
listlengths 0
7.91k
| tags_str
stringlengths 17
159k
| text_str
stringlengths 1
447k
| text_lists
listlengths 0
352
| processed_texts
listlengths 1
353
| tokens_length
listlengths 1
353
| input_texts
listlengths 1
40
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
ad996008a06911e0f6f0e1387bb0a6d14d803f81
|
# Dataset Card for "bpd-twitter-plus"
I scraped my twitter timeline some time in late 2022 / v early 2023
|
boopysaur/bpd-twitter-plus
|
[
"region:us"
] |
2023-09-18T07:09:11+00:00
|
{"dataset_info": {"features": [{"name": "content", "dtype": "string"}], "splits": [{"name": "train", "num_bytes": 2872991.0, "num_examples": 42389}], "download_size": 2139467, "dataset_size": 2872991.0}, "configs": [{"config_name": "default", "data_files": [{"split": "train", "path": "data/train-*"}]}]}
|
2023-09-18T07:38:20+00:00
|
[] |
[] |
TAGS
#region-us
|
# Dataset Card for "bpd-twitter-plus"
I scraped my twitter timeline some time in late 2022 / v early 2023
|
[
"# Dataset Card for \"bpd-twitter-plus\"\n\nI scraped my twitter timeline some time in late 2022 / v early 2023"
] |
[
"TAGS\n#region-us \n",
"# Dataset Card for \"bpd-twitter-plus\"\n\nI scraped my twitter timeline some time in late 2022 / v early 2023"
] |
[
6,
30
] |
[
"passage: TAGS\n#region-us \n# Dataset Card for \"bpd-twitter-plus\"\n\nI scraped my twitter timeline some time in late 2022 / v early 2023"
] |
b7e9bed42eb6f7b135b539fb22402aa13d58f681
|
# Dataset of Yano Erika
This is the dataset of Yano Erika, containing 266 images and their tags.
Images are crawled from many sites (e.g. danbooru, pixiv, zerochan ...), the auto-crawling system is powered by [DeepGHS Team](https://github.com/deepghs)([huggingface organization](https://huggingface.co/deepghs)).
| Name | Images | Download | Description |
|:------------|---------:|:------------------------------------|:-------------------------------------------------------------------------|
| raw | 266 | [Download](dataset-raw.zip) | Raw data with meta information. |
| raw-stage3 | 580 | [Download](dataset-raw-stage3.zip) | 3-stage cropped raw data with meta information. |
| 384x512 | 266 | [Download](dataset-384x512.zip) | 384x512 aligned dataset. |
| 512x512 | 266 | [Download](dataset-512x512.zip) | 512x512 aligned dataset. |
| 512x704 | 266 | [Download](dataset-512x704.zip) | 512x704 aligned dataset. |
| 640x640 | 266 | [Download](dataset-640x640.zip) | 640x640 aligned dataset. |
| 640x880 | 266 | [Download](dataset-640x880.zip) | 640x880 aligned dataset. |
| stage3-640 | 580 | [Download](dataset-stage3-640.zip) | 3-stage cropped dataset with the shorter side not exceeding 640 pixels. |
| stage3-800 | 580 | [Download](dataset-stage3-800.zip) | 3-stage cropped dataset with the shorter side not exceeding 800 pixels. |
| stage3-1200 | 580 | [Download](dataset-stage3-1200.zip) | 3-stage cropped dataset with the shorter side not exceeding 1200 pixels. |
|
CyberHarem/yano_erika_shirobako
|
[
"task_categories:text-to-image",
"size_categories:n<1K",
"license:mit",
"art",
"not-for-all-audiences",
"region:us"
] |
2023-09-18T07:12:30+00:00
|
{"license": "mit", "size_categories": ["n<1K"], "task_categories": ["text-to-image"], "tags": ["art", "not-for-all-audiences"]}
|
2023-09-18T07:16:57+00:00
|
[] |
[] |
TAGS
#task_categories-text-to-image #size_categories-n<1K #license-mit #art #not-for-all-audiences #region-us
|
Dataset of Yano Erika
=====================
This is the dataset of Yano Erika, containing 266 images and their tags.
Images are crawled from many sites (e.g. danbooru, pixiv, zerochan ...), the auto-crawling system is powered by DeepGHS Team(huggingface organization).
|
[] |
[
"TAGS\n#task_categories-text-to-image #size_categories-n<1K #license-mit #art #not-for-all-audiences #region-us \n"
] |
[
44
] |
[
"passage: TAGS\n#task_categories-text-to-image #size_categories-n<1K #license-mit #art #not-for-all-audiences #region-us \n"
] |
ed8949577afeb3036d26072be16ff43a5eb9ea1f
|
# Dataset of Tsubaki Ando
This is the dataset of Tsubaki Ando, containing 89 images and their tags.
Images are crawled from many sites (e.g. danbooru, pixiv, zerochan ...), the auto-crawling system is powered by [DeepGHS Team](https://github.com/deepghs)([huggingface organization](https://huggingface.co/deepghs)).
| Name | Images | Download | Description |
|:------------|---------:|:------------------------------------|:-------------------------------------------------------------------------|
| raw | 89 | [Download](dataset-raw.zip) | Raw data with meta information. |
| raw-stage3 | 198 | [Download](dataset-raw-stage3.zip) | 3-stage cropped raw data with meta information. |
| 384x512 | 89 | [Download](dataset-384x512.zip) | 384x512 aligned dataset. |
| 512x512 | 89 | [Download](dataset-512x512.zip) | 512x512 aligned dataset. |
| 512x704 | 89 | [Download](dataset-512x704.zip) | 512x704 aligned dataset. |
| 640x640 | 89 | [Download](dataset-640x640.zip) | 640x640 aligned dataset. |
| 640x880 | 89 | [Download](dataset-640x880.zip) | 640x880 aligned dataset. |
| stage3-640 | 198 | [Download](dataset-stage3-640.zip) | 3-stage cropped dataset with the shorter side not exceeding 640 pixels. |
| stage3-800 | 198 | [Download](dataset-stage3-800.zip) | 3-stage cropped dataset with the shorter side not exceeding 800 pixels. |
| stage3-1200 | 198 | [Download](dataset-stage3-1200.zip) | 3-stage cropped dataset with the shorter side not exceeding 1200 pixels. |
|
CyberHarem/tsubaki_ando_shirobako
|
[
"task_categories:text-to-image",
"size_categories:n<1K",
"license:mit",
"art",
"not-for-all-audiences",
"region:us"
] |
2023-09-18T07:20:26+00:00
|
{"license": "mit", "size_categories": ["n<1K"], "task_categories": ["text-to-image"], "tags": ["art", "not-for-all-audiences"]}
|
2023-09-18T07:21:10+00:00
|
[] |
[] |
TAGS
#task_categories-text-to-image #size_categories-n<1K #license-mit #art #not-for-all-audiences #region-us
|
Dataset of Tsubaki Ando
=======================
This is the dataset of Tsubaki Ando, containing 89 images and their tags.
Images are crawled from many sites (e.g. danbooru, pixiv, zerochan ...), the auto-crawling system is powered by DeepGHS Team(huggingface organization).
|
[] |
[
"TAGS\n#task_categories-text-to-image #size_categories-n<1K #license-mit #art #not-for-all-audiences #region-us \n"
] |
[
44
] |
[
"passage: TAGS\n#task_categories-text-to-image #size_categories-n<1K #license-mit #art #not-for-all-audiences #region-us \n"
] |
889188e47998cee43bcdacf2b0da45cfc359378d
|
# Auto-ACD
Auto-ACD is a large-scale, high-quality, audio-language dataset, building on the prior of robust audio-visual correspondence in existing video datasets, VGGSound and AudioSet.
- **Homepage:** https://auto-acd.github.io/
- **Paper:** https://huggingface.co/papers/2309.11500
- **Github:** https://github.com/LoieSun/Auto-ACD
## Analysis

Auto-ACD</strong>, comprising over <strong>1.9M </strong> audio-text pairs.
As shown in figure, The text descriptions in Auto-ACD contain <strong>long texts (18 words)</strong> and <strong>diverse vocabularies (23K)</strong>, and provide information about the <strong>surrounding auditory environment</strong>(data point with <strong>shadow</strong>) in which sounds take place.
## Download
We provide a csv file. For each data pairs, we provide YouTube URLs and generated caption. Each line in the csv file has columns defined by here.
```
# YouTube ID, caption
```
## Dataset Preview

|
Loie/Auto-ACD
|
[
"arxiv:2309.11500",
"region:us"
] |
2023-09-18T07:24:55+00:00
|
{}
|
2023-11-28T07:26:42+00:00
|
[
"2309.11500"
] |
[] |
TAGS
#arxiv-2309.11500 #region-us
|
# Auto-ACD
Auto-ACD is a large-scale, high-quality, audio-language dataset, building on the prior of robust audio-visual correspondence in existing video datasets, VGGSound and AudioSet.
- Homepage: URL
- Paper: URL
- Github: URL
## Analysis

Auto-ACD</strong>, comprising over <strong>1.9M </strong> audio-text pairs.
As shown in figure, The text descriptions in Auto-ACD contain <strong>long texts (18 words)</strong> and <strong>diverse vocabularies (23K)</strong>, and provide information about the <strong>surrounding auditory environment</strong>(data point with <strong>shadow</strong>) in which sounds take place.
## Download
We provide a csv file. For each data pairs, we provide YouTube URLs and generated caption. Each line in the csv file has columns defined by here.
## Dataset Preview

|
[
"# Auto-ACD\nAuto-ACD is a large-scale, high-quality, audio-language dataset, building on the prior of robust audio-visual correspondence in existing video datasets, VGGSound and AudioSet.\n\n\n- Homepage: URL\n- Paper: URL\n- Github: URL",
"## Analysis\n\n\n\nAuto-ACD</strong>, comprising over <strong>1.9M </strong> audio-text pairs. \nAs shown in figure, The text descriptions in Auto-ACD contain <strong>long texts (18 words)</strong> and <strong>diverse vocabularies (23K)</strong>, and provide information about the <strong>surrounding auditory environment</strong>(data point with <strong>shadow</strong>) in which sounds take place.",
"## Download\n\nWe provide a csv file. For each data pairs, we provide YouTube URLs and generated caption. Each line in the csv file has columns defined by here.",
"## Dataset Preview\n\n"
] |
[
"TAGS\n#arxiv-2309.11500 #region-us \n",
"# Auto-ACD\nAuto-ACD is a large-scale, high-quality, audio-language dataset, building on the prior of robust audio-visual correspondence in existing video datasets, VGGSound and AudioSet.\n\n\n- Homepage: URL\n- Paper: URL\n- Github: URL",
"## Analysis\n\n\n\nAuto-ACD</strong>, comprising over <strong>1.9M </strong> audio-text pairs. \nAs shown in figure, The text descriptions in Auto-ACD contain <strong>long texts (18 words)</strong> and <strong>diverse vocabularies (23K)</strong>, and provide information about the <strong>surrounding auditory environment</strong>(data point with <strong>shadow</strong>) in which sounds take place.",
"## Download\n\nWe provide a csv file. For each data pairs, we provide YouTube URLs and generated caption. Each line in the csv file has columns defined by here.",
"## Dataset Preview\n\n"
] |
[
14,
68,
127,
41,
14
] |
[
"passage: TAGS\n#arxiv-2309.11500 #region-us \n# Auto-ACD\nAuto-ACD is a large-scale, high-quality, audio-language dataset, building on the prior of robust audio-visual correspondence in existing video datasets, VGGSound and AudioSet.\n\n\n- Homepage: URL\n- Paper: URL\n- Github: URL## Analysis\n\n\n\nAuto-ACD</strong>, comprising over <strong>1.9M </strong> audio-text pairs. \nAs shown in figure, The text descriptions in Auto-ACD contain <strong>long texts (18 words)</strong> and <strong>diverse vocabularies (23K)</strong>, and provide information about the <strong>surrounding auditory environment</strong>(data point with <strong>shadow</strong>) in which sounds take place.## Download\n\nWe provide a csv file. For each data pairs, we provide YouTube URLs and generated caption. Each line in the csv file has columns defined by here.## Dataset Preview\n\n"
] |
d5621b2c1e8db7e845fddb3330b98d1908afab0f
|
# Dataset Card for "qa_wikipedia_sentence_transformer"
[More Information needed](https://github.com/huggingface/datasets/blob/main/CONTRIBUTING.md#how-to-contribute-to-the-dataset-cards)
|
legacy107/sentence_transformer_wikipedia_chunked
|
[
"region:us"
] |
2023-09-18T07:27:13+00:00
|
{"dataset_info": {"features": [{"name": "id", "dtype": "string"}, {"name": "title", "dtype": "string"}, {"name": "context", "dtype": "string"}, {"name": "question", "dtype": "string"}, {"name": "answer_start", "dtype": "int64"}, {"name": "answer", "dtype": "string"}, {"name": "article", "dtype": "string"}, {"name": "chunked_article", "sequence": "string"}], "splits": [{"name": "train", "num_bytes": 3734770114, "num_examples": 27742}, {"name": "test", "num_bytes": 408448904, "num_examples": 3468}, {"name": "validation", "num_bytes": 564192755, "num_examples": 3458}], "download_size": 717817867, "dataset_size": 4707411773}, "configs": [{"config_name": "default", "data_files": [{"split": "train", "path": "data/train-*"}, {"split": "test", "path": "data/test-*"}, {"split": "validation", "path": "data/validation-*"}]}]}
|
2023-09-19T03:00:50+00:00
|
[] |
[] |
TAGS
#region-us
|
# Dataset Card for "qa_wikipedia_sentence_transformer"
More Information needed
|
[
"# Dataset Card for \"qa_wikipedia_sentence_transformer\"\n\nMore Information needed"
] |
[
"TAGS\n#region-us \n",
"# Dataset Card for \"qa_wikipedia_sentence_transformer\"\n\nMore Information needed"
] |
[
6,
19
] |
[
"passage: TAGS\n#region-us \n# Dataset Card for \"qa_wikipedia_sentence_transformer\"\n\nMore Information needed"
] |
e9425901b771e196ed3c360e8c3e68ed37cc4a3b
|
# Dataset Card for CNN Dailymail Dataset
## Table of Contents
- [Dataset Description](#dataset-description)
- [Dataset Summary](#dataset-summary)
- [Supported Tasks and Leaderboards](#supported-tasks-and-leaderboards)
- [Languages](#languages)
- [Dataset Structure](#dataset-structure)
- [Data Instances](#data-instances)
- [Data Fields](#data-fields)
- [Data Splits](#data-splits)
- [Dataset Creation](#dataset-creation)
- [Curation Rationale](#curation-rationale)
- [Source Data](#source-data)
- [Annotations](#annotations)
- [Personal and Sensitive Information](#personal-and-sensitive-information)
- [Considerations for Using the Data](#considerations-for-using-the-data)
- [Social Impact of Dataset](#social-impact-of-dataset)
- [Discussion of Biases](#discussion-of-biases)
- [Other Known Limitations](#other-known-limitations)
- [Additional Information](#additional-information)
- [Dataset Curators](#dataset-curators)
- [Licensing Information](#licensing-information)
- [Citation Information](#citation-information)
- [Contributions](#contributions)
## Dataset Description
- **Homepage:**
- **Repository:** [CNN / DailyMail Dataset repository](https://github.com/abisee/cnn-dailymail)
- **Paper:** [Abstractive Text Summarization Using Sequence-to-Sequence RNNs and Beyond](https://papers.nips.cc/paper/5945-teaching-machines-to-read-and-comprehend.pdf), [Get To The Point: Summarization with Pointer-Generator Networks](https://www.aclweb.org/anthology/K16-1028.pdf)
- **Leaderboard:** [Papers with Code leaderboard for CNN / Dailymail Dataset](https://paperswithcode.com/sota/document-summarization-on-cnn-daily-mail)
- **Point of Contact:** [Abigail See](mailto:[email protected])
### Dataset Summary
The CNN / DailyMail Dataset is an English-language dataset containing just over 300k unique news articles as written by journalists at CNN and the Daily Mail. The current version supports both extractive and abstractive summarization, though the original version was created for machine reading and comprehension and abstractive question answering.
### Supported Tasks and Leaderboards
- 'summarization': [Versions 2.0.0 and 3.0.0 of the CNN / DailyMail Dataset](https://www.aclweb.org/anthology/K16-1028.pdf) can be used to train a model for abstractive and extractive summarization ([Version 1.0.0](https://papers.nips.cc/paper/5945-teaching-machines-to-read-and-comprehend.pdf) was developed for machine reading and comprehension and abstractive question answering). The model performance is measured by how high the output summary's [ROUGE](https://huggingface.co/metrics/rouge) score for a given article is when compared to the highlight as written by the original article author. [Zhong et al (2020)](https://www.aclweb.org/anthology/2020.acl-main.552.pdf) report a ROUGE-1 score of 44.41 when testing a model trained for extractive summarization. See the [Papers With Code leaderboard](https://paperswithcode.com/sota/document-summarization-on-cnn-daily-mail) for more models.
### Languages
The BCP-47 code for English as generally spoken in the United States is en-US and the BCP-47 code for English as generally spoken in the United Kingdom is en-GB. It is unknown if other varieties of English are represented in the data.
## Dataset Structure
### Data Instances
For each instance, there is a string for the article, a string for the highlights, and a string for the id. See the [CNN / Daily Mail dataset viewer](https://huggingface.co/datasets/viewer/?dataset=cnn_dailymail&config=3.0.0) to explore more examples.
```
{'id': '0054d6d30dbcad772e20b22771153a2a9cbeaf62',
'article': '(CNN) -- An American woman died aboard a cruise ship that docked at Rio de Janeiro on Tuesday, the same ship on which 86 passengers previously fell ill, according to the state-run Brazilian news agency, Agencia Brasil. The American tourist died aboard the MS Veendam, owned by cruise operator Holland America. Federal Police told Agencia Brasil that forensic doctors were investigating her death. The ship's doctors told police that the woman was elderly and suffered from diabetes and hypertension, according the agency. The other passengers came down with diarrhea prior to her death during an earlier part of the trip, the ship's doctors said. The Veendam left New York 36 days ago for a South America tour.'
'highlights': 'The elderly woman suffered from diabetes and hypertension, ship's doctors say .\nPreviously, 86 passengers had fallen ill on the ship, Agencia Brasil says .'}
```
The average token count for the articles and the highlights are provided below:
| Feature | Mean Token Count |
| ---------- | ---------------- |
| Article | 781 |
| Highlights | 56 |
### Data Fields
- `id`: a string containing the heximal formated SHA1 hash of the url where the story was retrieved from
- `article`: a string containing the body of the news article
- `highlights`: a string containing the highlight of the article as written by the article author
### Data Splits
The CNN/DailyMail dataset has 3 splits: _train_, _validation_, and _test_. Below are the statistics for Version 3.0.0 of the dataset.
| Dataset Split | Number of Instances in Split |
| ------------- | ------------------------------------------- |
| Train | 287,113 |
| Validation | 13,368 |
| Test | 11,490 |
## Dataset Creation
### Curation Rationale
Version 1.0.0 aimed to support supervised neural methodologies for machine reading and question answering with a large amount of real natural language training data and released about 313k unique articles and nearly 1M Cloze style questions to go with the articles. Versions 2.0.0 and 3.0.0 changed the structure of the dataset to support summarization rather than question answering. Version 3.0.0 provided a non-anonymized version of the data, whereas both the previous versions were preprocessed to replace named entities with unique identifier labels.
### Source Data
#### Initial Data Collection and Normalization
The data consists of news articles and highlight sentences. In the question answering setting of the data, the articles are used as the context and entities are hidden one at a time in the highlight sentences, producing Cloze style questions where the goal of the model is to correctly guess which entity in the context has been hidden in the highlight. In the summarization setting, the highlight sentences are concatenated to form a summary of the article. The CNN articles were written between April 2007 and April 2015. The Daily Mail articles were written between June 2010 and April 2015.
The code for the original data collection is available at <https://github.com/deepmind/rc-data>. The articles were downloaded using archives of <www.cnn.com> and <www.dailymail.co.uk> on the Wayback Machine. Articles were not included in the Version 1.0.0 collection if they exceeded 2000 tokens. Due to accessibility issues with the Wayback Machine, Kyunghyun Cho has made the datasets available at <https://cs.nyu.edu/~kcho/DMQA/>. An updated version of the code that does not anonymize the data is available at <https://github.com/abisee/cnn-dailymail>.
Hermann et al provided their own tokenization script. The script provided by See uses the PTBTokenizer. It also lowercases the text and adds periods to lines missing them.
#### Who are the source language producers?
The text was written by journalists at CNN and the Daily Mail.
### Annotations
The dataset does not contain any additional annotations.
#### Annotation process
[N/A]
#### Who are the annotators?
[N/A]
### Personal and Sensitive Information
Version 3.0 is not anonymized, so individuals' names can be found in the dataset. Information about the original author is not included in the dataset.
## Considerations for Using the Data
### Social Impact of Dataset
The purpose of this dataset is to help develop models that can summarize long paragraphs of text in one or two sentences.
This task is useful for efficiently presenting information given a large quantity of text. It should be made clear that any summarizations produced by models trained on this dataset are reflective of the language used in the articles, but are in fact automatically generated.
### Discussion of Biases
[Bordia and Bowman (2019)](https://www.aclweb.org/anthology/N19-3002.pdf) explore measuring gender bias and debiasing techniques in the CNN / Dailymail dataset, the Penn Treebank, and WikiText-2. They find the CNN / Dailymail dataset to have a slightly lower gender bias based on their metric compared to the other datasets, but still show evidence of gender bias when looking at words such as 'fragile'.
Because the articles were written by and for people in the US and the UK, they will likely present specifically US and UK perspectives and feature events that are considered relevant to those populations during the time that the articles were published.
### Other Known Limitations
News articles have been shown to conform to writing conventions in which important information is primarily presented in the first third of the article [(Kryściński et al, 2019)](https://www.aclweb.org/anthology/D19-1051.pdf). [Chen et al (2016)](https://www.aclweb.org/anthology/P16-1223.pdf) conducted a manual study of 100 random instances of the first version of the dataset and found 25% of the samples to be difficult even for humans to answer correctly due to ambiguity and coreference errors.
It should also be noted that machine-generated summarizations, even when extractive, may differ in truth values when compared to the original articles.
## Additional Information
### Dataset Curators
The data was originally collected by Karl Moritz Hermann, Tomáš Kočiský, Edward Grefenstette, Lasse Espeholt, Will Kay, Mustafa Suleyman, and Phil Blunsom of Google DeepMind. Tomáš Kočiský and Phil Blunsom are also affiliated with the University of Oxford. They released scripts to collect and process the data into the question answering format.
Ramesh Nallapati, Bowen Zhou, Cicero dos Santos, and Bing Xiang of IMB Watson and Çağlar Gu̇lçehre of Université de Montréal modified Hermann et al's collection scripts to restore the data to a summary format. They also produced both anonymized and non-anonymized versions.
The code for the non-anonymized version is made publicly available by Abigail See of Stanford University, Peter J. Liu of Google Brain and Christopher D. Manning of Stanford University at <https://github.com/abisee/cnn-dailymail>. The work at Stanford University was supported by the DARPA DEFT ProgramAFRL contract no. FA8750-13-2-0040.
### Licensing Information
The CNN / Daily Mail dataset version 1.0.0 is released under the [Apache-2.0 License](http://www.apache.org/licenses/LICENSE-2.0).
### Citation Information
```
@inproceedings{see-etal-2017-get,
title = "Get To The Point: Summarization with Pointer-Generator Networks",
author = "See, Abigail and
Liu, Peter J. and
Manning, Christopher D.",
booktitle = "Proceedings of the 55th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers)",
month = jul,
year = "2017",
address = "Vancouver, Canada",
publisher = "Association for Computational Linguistics",
url = "https://www.aclweb.org/anthology/P17-1099",
doi = "10.18653/v1/P17-1099",
pages = "1073--1083",
abstract = "Neural sequence-to-sequence models have provided a viable new approach for abstractive text summarization (meaning they are not restricted to simply selecting and rearranging passages from the original text). However, these models have two shortcomings: they are liable to reproduce factual details inaccurately, and they tend to repeat themselves. In this work we propose a novel architecture that augments the standard sequence-to-sequence attentional model in two orthogonal ways. First, we use a hybrid pointer-generator network that can copy words from the source text via pointing, which aids accurate reproduction of information, while retaining the ability to produce novel words through the generator. Second, we use coverage to keep track of what has been summarized, which discourages repetition. We apply our model to the CNN / Daily Mail summarization task, outperforming the current abstractive state-of-the-art by at least 2 ROUGE points.",
}
```
```
@inproceedings{DBLP:conf/nips/HermannKGEKSB15,
author={Karl Moritz Hermann and Tomás Kociský and Edward Grefenstette and Lasse Espeholt and Will Kay and Mustafa Suleyman and Phil Blunsom},
title={Teaching Machines to Read and Comprehend},
year={2015},
cdate={1420070400000},
pages={1693-1701},
url={http://papers.nips.cc/paper/5945-teaching-machines-to-read-and-comprehend},
booktitle={NIPS},
crossref={conf/nips/2015}
}
```
### Contributions
Thanks to [@thomwolf](https://github.com/thomwolf), [@lewtun](https://github.com/lewtun), [@jplu](https://github.com/jplu), [@jbragg](https://github.com/jbragg), [@patrickvonplaten](https://github.com/patrickvonplaten) and [@mcmillanmajora](https://github.com/mcmillanmajora) for adding this dataset.
|
samyakmohelay/genai_dataset
|
[
"task_categories:summarization",
"task_ids:news-articles-summarization",
"annotations_creators:no-annotation",
"language_creators:found",
"multilinguality:monolingual",
"size_categories:100K<n<1M",
"source_datasets:original",
"language:en",
"license:apache-2.0",
"region:us"
] |
2023-09-18T07:27:28+00:00
|
{"annotations_creators": ["no-annotation"], "language_creators": ["found"], "language": ["en"], "license": ["apache-2.0"], "multilinguality": ["monolingual"], "size_categories": ["100K<n<1M"], "source_datasets": ["original"], "task_categories": ["summarization"], "task_ids": ["news-articles-summarization"], "paperswithcode_id": "cnn-daily-mail-1", "pretty_name": "CNN / Daily Mail", "dataset_info": [{"config_name": "3.0.0", "features": [{"name": "article", "dtype": "string"}, {"name": "highlights", "dtype": "string"}, {"name": "id", "dtype": "string"}], "splits": [{"name": "train", "num_bytes": 1261704133, "num_examples": 287113}, {"name": "validation", "num_bytes": 57732436, "num_examples": 13368}, {"name": "test", "num_bytes": 49925756, "num_examples": 11490}], "download_size": 585439472, "dataset_size": 1369362325}, {"config_name": "1.0.0", "features": [{"name": "article", "dtype": "string"}, {"name": "highlights", "dtype": "string"}, {"name": "id", "dtype": "string"}], "splits": [{"name": "train", "num_bytes": 1261704133, "num_examples": 287113}, {"name": "validation", "num_bytes": 57732436, "num_examples": 13368}, {"name": "test", "num_bytes": 49925756, "num_examples": 11490}], "download_size": 585439472, "dataset_size": 1369362325}, {"config_name": "2.0.0", "features": [{"name": "article", "dtype": "string"}, {"name": "highlights", "dtype": "string"}, {"name": "id", "dtype": "string"}], "splits": [{"name": "train", "num_bytes": 1261704133, "num_examples": 287113}, {"name": "validation", "num_bytes": 57732436, "num_examples": 13368}, {"name": "test", "num_bytes": 49925756, "num_examples": 11490}], "download_size": 585439472, "dataset_size": 1369362325}], "train-eval-index": [{"config": "3.0.0", "task": "summarization", "task_id": "summarization", "splits": {"eval_split": "test"}, "col_mapping": {"article": "text", "highlights": "target"}}]}
|
2023-09-18T07:45:28+00:00
|
[] |
[
"en"
] |
TAGS
#task_categories-summarization #task_ids-news-articles-summarization #annotations_creators-no-annotation #language_creators-found #multilinguality-monolingual #size_categories-100K<n<1M #source_datasets-original #language-English #license-apache-2.0 #region-us
|
Dataset Card for CNN Dailymail Dataset
======================================
Table of Contents
-----------------
* Dataset Description
+ Dataset Summary
+ Supported Tasks and Leaderboards
+ Languages
* Dataset Structure
+ Data Instances
+ Data Fields
+ Data Splits
* Dataset Creation
+ Curation Rationale
+ Source Data
+ Annotations
+ Personal and Sensitive Information
* Considerations for Using the Data
+ Social Impact of Dataset
+ Discussion of Biases
+ Other Known Limitations
* Additional Information
+ Dataset Curators
+ Licensing Information
+ Citation Information
+ Contributions
Dataset Description
-------------------
* Homepage:
* Repository: CNN / DailyMail Dataset repository
* Paper: Abstractive Text Summarization Using Sequence-to-Sequence RNNs and Beyond, Get To The Point: Summarization with Pointer-Generator Networks
* Leaderboard: Papers with Code leaderboard for CNN / Dailymail Dataset
* Point of Contact: Abigail See
### Dataset Summary
The CNN / DailyMail Dataset is an English-language dataset containing just over 300k unique news articles as written by journalists at CNN and the Daily Mail. The current version supports both extractive and abstractive summarization, though the original version was created for machine reading and comprehension and abstractive question answering.
### Supported Tasks and Leaderboards
* 'summarization': Versions 2.0.0 and 3.0.0 of the CNN / DailyMail Dataset can be used to train a model for abstractive and extractive summarization (Version 1.0.0 was developed for machine reading and comprehension and abstractive question answering). The model performance is measured by how high the output summary's ROUGE score for a given article is when compared to the highlight as written by the original article author. Zhong et al (2020) report a ROUGE-1 score of 44.41 when testing a model trained for extractive summarization. See the Papers With Code leaderboard for more models.
### Languages
The BCP-47 code for English as generally spoken in the United States is en-US and the BCP-47 code for English as generally spoken in the United Kingdom is en-GB. It is unknown if other varieties of English are represented in the data.
Dataset Structure
-----------------
### Data Instances
For each instance, there is a string for the article, a string for the highlights, and a string for the id. See the CNN / Daily Mail dataset viewer to explore more examples.
The average token count for the articles and the highlights are provided below:
### Data Fields
* 'id': a string containing the heximal formated SHA1 hash of the url where the story was retrieved from
* 'article': a string containing the body of the news article
* 'highlights': a string containing the highlight of the article as written by the article author
### Data Splits
The CNN/DailyMail dataset has 3 splits: *train*, *validation*, and *test*. Below are the statistics for Version 3.0.0 of the dataset.
Dataset Creation
----------------
### Curation Rationale
Version 1.0.0 aimed to support supervised neural methodologies for machine reading and question answering with a large amount of real natural language training data and released about 313k unique articles and nearly 1M Cloze style questions to go with the articles. Versions 2.0.0 and 3.0.0 changed the structure of the dataset to support summarization rather than question answering. Version 3.0.0 provided a non-anonymized version of the data, whereas both the previous versions were preprocessed to replace named entities with unique identifier labels.
### Source Data
#### Initial Data Collection and Normalization
The data consists of news articles and highlight sentences. In the question answering setting of the data, the articles are used as the context and entities are hidden one at a time in the highlight sentences, producing Cloze style questions where the goal of the model is to correctly guess which entity in the context has been hidden in the highlight. In the summarization setting, the highlight sentences are concatenated to form a summary of the article. The CNN articles were written between April 2007 and April 2015. The Daily Mail articles were written between June 2010 and April 2015.
The code for the original data collection is available at <URL The articles were downloaded using archives of and on the Wayback Machine. Articles were not included in the Version 1.0.0 collection if they exceeded 2000 tokens. Due to accessibility issues with the Wayback Machine, Kyunghyun Cho has made the datasets available at <URL An updated version of the code that does not anonymize the data is available at <URL
Hermann et al provided their own tokenization script. The script provided by See uses the PTBTokenizer. It also lowercases the text and adds periods to lines missing them.
#### Who are the source language producers?
The text was written by journalists at CNN and the Daily Mail.
### Annotations
The dataset does not contain any additional annotations.
#### Annotation process
[N/A]
#### Who are the annotators?
[N/A]
### Personal and Sensitive Information
Version 3.0 is not anonymized, so individuals' names can be found in the dataset. Information about the original author is not included in the dataset.
Considerations for Using the Data
---------------------------------
### Social Impact of Dataset
The purpose of this dataset is to help develop models that can summarize long paragraphs of text in one or two sentences.
This task is useful for efficiently presenting information given a large quantity of text. It should be made clear that any summarizations produced by models trained on this dataset are reflective of the language used in the articles, but are in fact automatically generated.
### Discussion of Biases
Bordia and Bowman (2019) explore measuring gender bias and debiasing techniques in the CNN / Dailymail dataset, the Penn Treebank, and WikiText-2. They find the CNN / Dailymail dataset to have a slightly lower gender bias based on their metric compared to the other datasets, but still show evidence of gender bias when looking at words such as 'fragile'.
Because the articles were written by and for people in the US and the UK, they will likely present specifically US and UK perspectives and feature events that are considered relevant to those populations during the time that the articles were published.
### Other Known Limitations
News articles have been shown to conform to writing conventions in which important information is primarily presented in the first third of the article (Kryściński et al, 2019). Chen et al (2016) conducted a manual study of 100 random instances of the first version of the dataset and found 25% of the samples to be difficult even for humans to answer correctly due to ambiguity and coreference errors.
It should also be noted that machine-generated summarizations, even when extractive, may differ in truth values when compared to the original articles.
Additional Information
----------------------
### Dataset Curators
The data was originally collected by Karl Moritz Hermann, Tomáš Kočiský, Edward Grefenstette, Lasse Espeholt, Will Kay, Mustafa Suleyman, and Phil Blunsom of Google DeepMind. Tomáš Kočiský and Phil Blunsom are also affiliated with the University of Oxford. They released scripts to collect and process the data into the question answering format.
Ramesh Nallapati, Bowen Zhou, Cicero dos Santos, and Bing Xiang of IMB Watson and Çağlar Gu̇lçehre of Université de Montréal modified Hermann et al's collection scripts to restore the data to a summary format. They also produced both anonymized and non-anonymized versions.
The code for the non-anonymized version is made publicly available by Abigail See of Stanford University, Peter J. Liu of Google Brain and Christopher D. Manning of Stanford University at <URL The work at Stanford University was supported by the DARPA DEFT ProgramAFRL contract no. FA8750-13-2-0040.
### Licensing Information
The CNN / Daily Mail dataset version 1.0.0 is released under the Apache-2.0 License.
### Contributions
Thanks to @thomwolf, @lewtun, @jplu, @jbragg, @patrickvonplaten and @mcmillanmajora for adding this dataset.
|
[
"### Dataset Summary\n\n\nThe CNN / DailyMail Dataset is an English-language dataset containing just over 300k unique news articles as written by journalists at CNN and the Daily Mail. The current version supports both extractive and abstractive summarization, though the original version was created for machine reading and comprehension and abstractive question answering.",
"### Supported Tasks and Leaderboards\n\n\n* 'summarization': Versions 2.0.0 and 3.0.0 of the CNN / DailyMail Dataset can be used to train a model for abstractive and extractive summarization (Version 1.0.0 was developed for machine reading and comprehension and abstractive question answering). The model performance is measured by how high the output summary's ROUGE score for a given article is when compared to the highlight as written by the original article author. Zhong et al (2020) report a ROUGE-1 score of 44.41 when testing a model trained for extractive summarization. See the Papers With Code leaderboard for more models.",
"### Languages\n\n\nThe BCP-47 code for English as generally spoken in the United States is en-US and the BCP-47 code for English as generally spoken in the United Kingdom is en-GB. It is unknown if other varieties of English are represented in the data.\n\n\nDataset Structure\n-----------------",
"### Data Instances\n\n\nFor each instance, there is a string for the article, a string for the highlights, and a string for the id. See the CNN / Daily Mail dataset viewer to explore more examples.\n\n\nThe average token count for the articles and the highlights are provided below:",
"### Data Fields\n\n\n* 'id': a string containing the heximal formated SHA1 hash of the url where the story was retrieved from\n* 'article': a string containing the body of the news article\n* 'highlights': a string containing the highlight of the article as written by the article author",
"### Data Splits\n\n\nThe CNN/DailyMail dataset has 3 splits: *train*, *validation*, and *test*. Below are the statistics for Version 3.0.0 of the dataset.\n\n\n\nDataset Creation\n----------------",
"### Curation Rationale\n\n\nVersion 1.0.0 aimed to support supervised neural methodologies for machine reading and question answering with a large amount of real natural language training data and released about 313k unique articles and nearly 1M Cloze style questions to go with the articles. Versions 2.0.0 and 3.0.0 changed the structure of the dataset to support summarization rather than question answering. Version 3.0.0 provided a non-anonymized version of the data, whereas both the previous versions were preprocessed to replace named entities with unique identifier labels.",
"### Source Data",
"#### Initial Data Collection and Normalization\n\n\nThe data consists of news articles and highlight sentences. In the question answering setting of the data, the articles are used as the context and entities are hidden one at a time in the highlight sentences, producing Cloze style questions where the goal of the model is to correctly guess which entity in the context has been hidden in the highlight. In the summarization setting, the highlight sentences are concatenated to form a summary of the article. The CNN articles were written between April 2007 and April 2015. The Daily Mail articles were written between June 2010 and April 2015.\n\n\nThe code for the original data collection is available at <URL The articles were downloaded using archives of and on the Wayback Machine. Articles were not included in the Version 1.0.0 collection if they exceeded 2000 tokens. Due to accessibility issues with the Wayback Machine, Kyunghyun Cho has made the datasets available at <URL An updated version of the code that does not anonymize the data is available at <URL\n\n\nHermann et al provided their own tokenization script. The script provided by See uses the PTBTokenizer. It also lowercases the text and adds periods to lines missing them.",
"#### Who are the source language producers?\n\n\nThe text was written by journalists at CNN and the Daily Mail.",
"### Annotations\n\n\nThe dataset does not contain any additional annotations.",
"#### Annotation process\n\n\n[N/A]",
"#### Who are the annotators?\n\n\n[N/A]",
"### Personal and Sensitive Information\n\n\nVersion 3.0 is not anonymized, so individuals' names can be found in the dataset. Information about the original author is not included in the dataset.\n\n\nConsiderations for Using the Data\n---------------------------------",
"### Social Impact of Dataset\n\n\nThe purpose of this dataset is to help develop models that can summarize long paragraphs of text in one or two sentences.\n\n\nThis task is useful for efficiently presenting information given a large quantity of text. It should be made clear that any summarizations produced by models trained on this dataset are reflective of the language used in the articles, but are in fact automatically generated.",
"### Discussion of Biases\n\n\nBordia and Bowman (2019) explore measuring gender bias and debiasing techniques in the CNN / Dailymail dataset, the Penn Treebank, and WikiText-2. They find the CNN / Dailymail dataset to have a slightly lower gender bias based on their metric compared to the other datasets, but still show evidence of gender bias when looking at words such as 'fragile'.\n\n\nBecause the articles were written by and for people in the US and the UK, they will likely present specifically US and UK perspectives and feature events that are considered relevant to those populations during the time that the articles were published.",
"### Other Known Limitations\n\n\nNews articles have been shown to conform to writing conventions in which important information is primarily presented in the first third of the article (Kryściński et al, 2019). Chen et al (2016) conducted a manual study of 100 random instances of the first version of the dataset and found 25% of the samples to be difficult even for humans to answer correctly due to ambiguity and coreference errors.\n\n\nIt should also be noted that machine-generated summarizations, even when extractive, may differ in truth values when compared to the original articles.\n\n\nAdditional Information\n----------------------",
"### Dataset Curators\n\n\nThe data was originally collected by Karl Moritz Hermann, Tomáš Kočiský, Edward Grefenstette, Lasse Espeholt, Will Kay, Mustafa Suleyman, and Phil Blunsom of Google DeepMind. Tomáš Kočiský and Phil Blunsom are also affiliated with the University of Oxford. They released scripts to collect and process the data into the question answering format.\n\n\nRamesh Nallapati, Bowen Zhou, Cicero dos Santos, and Bing Xiang of IMB Watson and Çağlar Gu̇lçehre of Université de Montréal modified Hermann et al's collection scripts to restore the data to a summary format. They also produced both anonymized and non-anonymized versions.\n\n\nThe code for the non-anonymized version is made publicly available by Abigail See of Stanford University, Peter J. Liu of Google Brain and Christopher D. Manning of Stanford University at <URL The work at Stanford University was supported by the DARPA DEFT ProgramAFRL contract no. FA8750-13-2-0040.",
"### Licensing Information\n\n\nThe CNN / Daily Mail dataset version 1.0.0 is released under the Apache-2.0 License.",
"### Contributions\n\n\nThanks to @thomwolf, @lewtun, @jplu, @jbragg, @patrickvonplaten and @mcmillanmajora for adding this dataset."
] |
[
"TAGS\n#task_categories-summarization #task_ids-news-articles-summarization #annotations_creators-no-annotation #language_creators-found #multilinguality-monolingual #size_categories-100K<n<1M #source_datasets-original #language-English #license-apache-2.0 #region-us \n",
"### Dataset Summary\n\n\nThe CNN / DailyMail Dataset is an English-language dataset containing just over 300k unique news articles as written by journalists at CNN and the Daily Mail. The current version supports both extractive and abstractive summarization, though the original version was created for machine reading and comprehension and abstractive question answering.",
"### Supported Tasks and Leaderboards\n\n\n* 'summarization': Versions 2.0.0 and 3.0.0 of the CNN / DailyMail Dataset can be used to train a model for abstractive and extractive summarization (Version 1.0.0 was developed for machine reading and comprehension and abstractive question answering). The model performance is measured by how high the output summary's ROUGE score for a given article is when compared to the highlight as written by the original article author. Zhong et al (2020) report a ROUGE-1 score of 44.41 when testing a model trained for extractive summarization. See the Papers With Code leaderboard for more models.",
"### Languages\n\n\nThe BCP-47 code for English as generally spoken in the United States is en-US and the BCP-47 code for English as generally spoken in the United Kingdom is en-GB. It is unknown if other varieties of English are represented in the data.\n\n\nDataset Structure\n-----------------",
"### Data Instances\n\n\nFor each instance, there is a string for the article, a string for the highlights, and a string for the id. See the CNN / Daily Mail dataset viewer to explore more examples.\n\n\nThe average token count for the articles and the highlights are provided below:",
"### Data Fields\n\n\n* 'id': a string containing the heximal formated SHA1 hash of the url where the story was retrieved from\n* 'article': a string containing the body of the news article\n* 'highlights': a string containing the highlight of the article as written by the article author",
"### Data Splits\n\n\nThe CNN/DailyMail dataset has 3 splits: *train*, *validation*, and *test*. Below are the statistics for Version 3.0.0 of the dataset.\n\n\n\nDataset Creation\n----------------",
"### Curation Rationale\n\n\nVersion 1.0.0 aimed to support supervised neural methodologies for machine reading and question answering with a large amount of real natural language training data and released about 313k unique articles and nearly 1M Cloze style questions to go with the articles. Versions 2.0.0 and 3.0.0 changed the structure of the dataset to support summarization rather than question answering. Version 3.0.0 provided a non-anonymized version of the data, whereas both the previous versions were preprocessed to replace named entities with unique identifier labels.",
"### Source Data",
"#### Initial Data Collection and Normalization\n\n\nThe data consists of news articles and highlight sentences. In the question answering setting of the data, the articles are used as the context and entities are hidden one at a time in the highlight sentences, producing Cloze style questions where the goal of the model is to correctly guess which entity in the context has been hidden in the highlight. In the summarization setting, the highlight sentences are concatenated to form a summary of the article. The CNN articles were written between April 2007 and April 2015. The Daily Mail articles were written between June 2010 and April 2015.\n\n\nThe code for the original data collection is available at <URL The articles were downloaded using archives of and on the Wayback Machine. Articles were not included in the Version 1.0.0 collection if they exceeded 2000 tokens. Due to accessibility issues with the Wayback Machine, Kyunghyun Cho has made the datasets available at <URL An updated version of the code that does not anonymize the data is available at <URL\n\n\nHermann et al provided their own tokenization script. The script provided by See uses the PTBTokenizer. It also lowercases the text and adds periods to lines missing them.",
"#### Who are the source language producers?\n\n\nThe text was written by journalists at CNN and the Daily Mail.",
"### Annotations\n\n\nThe dataset does not contain any additional annotations.",
"#### Annotation process\n\n\n[N/A]",
"#### Who are the annotators?\n\n\n[N/A]",
"### Personal and Sensitive Information\n\n\nVersion 3.0 is not anonymized, so individuals' names can be found in the dataset. Information about the original author is not included in the dataset.\n\n\nConsiderations for Using the Data\n---------------------------------",
"### Social Impact of Dataset\n\n\nThe purpose of this dataset is to help develop models that can summarize long paragraphs of text in one or two sentences.\n\n\nThis task is useful for efficiently presenting information given a large quantity of text. It should be made clear that any summarizations produced by models trained on this dataset are reflective of the language used in the articles, but are in fact automatically generated.",
"### Discussion of Biases\n\n\nBordia and Bowman (2019) explore measuring gender bias and debiasing techniques in the CNN / Dailymail dataset, the Penn Treebank, and WikiText-2. They find the CNN / Dailymail dataset to have a slightly lower gender bias based on their metric compared to the other datasets, but still show evidence of gender bias when looking at words such as 'fragile'.\n\n\nBecause the articles were written by and for people in the US and the UK, they will likely present specifically US and UK perspectives and feature events that are considered relevant to those populations during the time that the articles were published.",
"### Other Known Limitations\n\n\nNews articles have been shown to conform to writing conventions in which important information is primarily presented in the first third of the article (Kryściński et al, 2019). Chen et al (2016) conducted a manual study of 100 random instances of the first version of the dataset and found 25% of the samples to be difficult even for humans to answer correctly due to ambiguity and coreference errors.\n\n\nIt should also be noted that machine-generated summarizations, even when extractive, may differ in truth values when compared to the original articles.\n\n\nAdditional Information\n----------------------",
"### Dataset Curators\n\n\nThe data was originally collected by Karl Moritz Hermann, Tomáš Kočiský, Edward Grefenstette, Lasse Espeholt, Will Kay, Mustafa Suleyman, and Phil Blunsom of Google DeepMind. Tomáš Kočiský and Phil Blunsom are also affiliated with the University of Oxford. They released scripts to collect and process the data into the question answering format.\n\n\nRamesh Nallapati, Bowen Zhou, Cicero dos Santos, and Bing Xiang of IMB Watson and Çağlar Gu̇lçehre of Université de Montréal modified Hermann et al's collection scripts to restore the data to a summary format. They also produced both anonymized and non-anonymized versions.\n\n\nThe code for the non-anonymized version is made publicly available by Abigail See of Stanford University, Peter J. Liu of Google Brain and Christopher D. Manning of Stanford University at <URL The work at Stanford University was supported by the DARPA DEFT ProgramAFRL contract no. FA8750-13-2-0040.",
"### Licensing Information\n\n\nThe CNN / Daily Mail dataset version 1.0.0 is released under the Apache-2.0 License.",
"### Contributions\n\n\nThanks to @thomwolf, @lewtun, @jplu, @jbragg, @patrickvonplaten and @mcmillanmajora for adding this dataset."
] |
[
91,
76,
147,
69,
64,
73,
54,
126,
4,
266,
24,
17,
10,
14,
50,
91,
142,
133,
234,
26,
45
] |
[
"passage: TAGS\n#task_categories-summarization #task_ids-news-articles-summarization #annotations_creators-no-annotation #language_creators-found #multilinguality-monolingual #size_categories-100K<n<1M #source_datasets-original #language-English #license-apache-2.0 #region-us \n### Dataset Summary\n\n\nThe CNN / DailyMail Dataset is an English-language dataset containing just over 300k unique news articles as written by journalists at CNN and the Daily Mail. The current version supports both extractive and abstractive summarization, though the original version was created for machine reading and comprehension and abstractive question answering.### Supported Tasks and Leaderboards\n\n\n* 'summarization': Versions 2.0.0 and 3.0.0 of the CNN / DailyMail Dataset can be used to train a model for abstractive and extractive summarization (Version 1.0.0 was developed for machine reading and comprehension and abstractive question answering). The model performance is measured by how high the output summary's ROUGE score for a given article is when compared to the highlight as written by the original article author. Zhong et al (2020) report a ROUGE-1 score of 44.41 when testing a model trained for extractive summarization. See the Papers With Code leaderboard for more models.### Languages\n\n\nThe BCP-47 code for English as generally spoken in the United States is en-US and the BCP-47 code for English as generally spoken in the United Kingdom is en-GB. It is unknown if other varieties of English are represented in the data.\n\n\nDataset Structure\n-----------------### Data Instances\n\n\nFor each instance, there is a string for the article, a string for the highlights, and a string for the id. See the CNN / Daily Mail dataset viewer to explore more examples.\n\n\nThe average token count for the articles and the highlights are provided below:",
"passage: ### Data Fields\n\n\n* 'id': a string containing the heximal formated SHA1 hash of the url where the story was retrieved from\n* 'article': a string containing the body of the news article\n* 'highlights': a string containing the highlight of the article as written by the article author### Data Splits\n\n\nThe CNN/DailyMail dataset has 3 splits: *train*, *validation*, and *test*. Below are the statistics for Version 3.0.0 of the dataset.\n\n\n\nDataset Creation\n----------------### Curation Rationale\n\n\nVersion 1.0.0 aimed to support supervised neural methodologies for machine reading and question answering with a large amount of real natural language training data and released about 313k unique articles and nearly 1M Cloze style questions to go with the articles. Versions 2.0.0 and 3.0.0 changed the structure of the dataset to support summarization rather than question answering. Version 3.0.0 provided a non-anonymized version of the data, whereas both the previous versions were preprocessed to replace named entities with unique identifier labels.### Source Data#### Initial Data Collection and Normalization\n\n\nThe data consists of news articles and highlight sentences. In the question answering setting of the data, the articles are used as the context and entities are hidden one at a time in the highlight sentences, producing Cloze style questions where the goal of the model is to correctly guess which entity in the context has been hidden in the highlight. In the summarization setting, the highlight sentences are concatenated to form a summary of the article. The CNN articles were written between April 2007 and April 2015. The Daily Mail articles were written between June 2010 and April 2015.\n\n\nThe code for the original data collection is available at <URL The articles were downloaded using archives of and on the Wayback Machine. Articles were not included in the Version 1.0.0 collection if they exceeded 2000 tokens. Due to accessibility issues with the Wayback Machine, Kyunghyun Cho has made the datasets available at <URL An updated version of the code that does not anonymize the data is available at <URL\n\n\nHermann et al provided their own tokenization script. The script provided by See uses the PTBTokenizer. It also lowercases the text and adds periods to lines missing them.#### Who are the source language producers?\n\n\nThe text was written by journalists at CNN and the Daily Mail.### Annotations\n\n\nThe dataset does not contain any additional annotations.#### Annotation process\n\n\n[N/A]",
"passage: #### Who are the annotators?\n\n\n[N/A]### Personal and Sensitive Information\n\n\nVersion 3.0 is not anonymized, so individuals' names can be found in the dataset. Information about the original author is not included in the dataset.\n\n\nConsiderations for Using the Data\n---------------------------------### Social Impact of Dataset\n\n\nThe purpose of this dataset is to help develop models that can summarize long paragraphs of text in one or two sentences.\n\n\nThis task is useful for efficiently presenting information given a large quantity of text. It should be made clear that any summarizations produced by models trained on this dataset are reflective of the language used in the articles, but are in fact automatically generated.### Discussion of Biases\n\n\nBordia and Bowman (2019) explore measuring gender bias and debiasing techniques in the CNN / Dailymail dataset, the Penn Treebank, and WikiText-2. They find the CNN / Dailymail dataset to have a slightly lower gender bias based on their metric compared to the other datasets, but still show evidence of gender bias when looking at words such as 'fragile'.\n\n\nBecause the articles were written by and for people in the US and the UK, they will likely present specifically US and UK perspectives and feature events that are considered relevant to those populations during the time that the articles were published.### Other Known Limitations\n\n\nNews articles have been shown to conform to writing conventions in which important information is primarily presented in the first third of the article (Kryściński et al, 2019). Chen et al (2016) conducted a manual study of 100 random instances of the first version of the dataset and found 25% of the samples to be difficult even for humans to answer correctly due to ambiguity and coreference errors.\n\n\nIt should also be noted that machine-generated summarizations, even when extractive, may differ in truth values when compared to the original articles.\n\n\nAdditional Information\n----------------------"
] |
565f73fa8738c73502ef3a05db69e93c1b02e71a
|
# Dataset Card for "editorials_nyt"
[More Information needed](https://github.com/huggingface/datasets/blob/main/CONTRIBUTING.md#how-to-contribute-to-the-dataset-cards)
|
nailiamirzakhmedova/editorials_nyt
|
[
"region:us"
] |
2023-09-18T07:38:51+00:00
|
{"configs": [{"config_name": "default", "data_files": [{"split": "train", "path": "data/train-*"}]}], "dataset_info": {"features": [{"name": "id", "dtype": "string"}, {"name": "text", "dtype": "string"}], "splits": [{"name": "train", "num_bytes": 14178142, "num_examples": 5000}], "download_size": 8824350, "dataset_size": 14178142}}
|
2023-09-18T07:38:54+00:00
|
[] |
[] |
TAGS
#region-us
|
# Dataset Card for "editorials_nyt"
More Information needed
|
[
"# Dataset Card for \"editorials_nyt\"\n\nMore Information needed"
] |
[
"TAGS\n#region-us \n",
"# Dataset Card for \"editorials_nyt\"\n\nMore Information needed"
] |
[
6,
15
] |
[
"passage: TAGS\n#region-us \n# Dataset Card for \"editorials_nyt\"\n\nMore Information needed"
] |
10ad3cbd651561c5962c7637ccf8d3ba13f17ee4
|
### Dataset Summary
Articles retrieved from the [Spanish WikiHow website](https://es.wikihow.com) on September 2023.
Each article contains a tutorial about a specific topic. The format is always a "How to" question
followed by a detailed step-by-step explanation. In some cases, the response includes several methods.
The main idea is to use this data for instruction tuning of Spanish LLMs, but given its nature it
could also be used for other tasks such as text classification or summarization.
### Languages
- Spanish (ES)
### Usage
To load the full dataset:
```python
from datasets import load_dataset
all_articles = load_dataset("mapama247/wikihow_es", trust_remote_code=True)
print(all_articles.num_rows) # output: {'train': 7380}
```
To load only examples from a specific category:
```python
from datasets import load_dataset
sports_articles = load_dataset("mapama247/wikihow_es", "deportes")
print(sports_articles.num_rows) # output: {'train': 201}
```
List of available categories, with the repective number of examples:
```
computadoras-y-electrónica 821
salud 804
pasatiempos 729
cuidado-y-estilo-personal 724
carreras-y-educación 564
en-la-casa-y-el-jardín 496
finanzas-y-negocios 459
comida-y-diversión 454
relaciones 388
mascotas-y-animales 338
filosofía-y-religión 264
arte-y-entretenimiento 254
en-el-trabajo 211
adolescentes 201
deportes 201
vida-familiar 147
viajes 139
automóviles-y-otros-vehículos 100
días-de-fiesta-y-tradiciones 86
```
### Supported Tasks
This dataset can be used to train a model for...
- `instruction-tuning`
- `text-classification`
- `question-answering`
- `conversational`
- `summarization`
## Dataset Structure
### Data Instances
```python
{
'category': str,
'question': str,
'introduction': str,
'answers': List[str],
'short_answers': List[str],
'url': str,
'num_answers': int,
'num_refs': int,
'expert_author': bool,
}
```
### Data Fields
- `category`: The category (from [this list](https://es.wikihow.com/Especial:CategoryListing)) to which the example belongs to.
- `label`: Numerical representation of the category, for text classification purposes.
- `question`: The article's title, which always starts with "¿Cómo ...".
- `introduction`: Introductory text that precedes the step-by-step explanation.
- `answers`: List of complete answers, with the full explanation of each step.
- `short_answers`: List of shorter answers that only contain one-sentence steps.
- `num_answers`: The number of alternative answers provided (e.g. length of `answers`).
- `num_ref`: Number of references provided in the article.
- `expert_authors`: Whether the article's author claims to be an expert on the topic or not.
- `url`: The URL address of the original article.
### Data Splits
There is only one split (`train`) that contains a total of 7,380 examples.
## Dataset Creation
### Curation Rationale
This dataset was created for language model alignment to end tasks and user preferences.
### Source Data
How-To questions with detailed step-by-step answers, retrieved from the WikiHow website.
#### Data Collection and Normalization
All articles available in September 2023 were extracted, no filters applied.
Along with the article's content, some metadata was retrieved as well.
#### Source language producers
WikiHow users. All the content is human-generated.
### Personal and Sensitive Information
The data does not include personal or sensitive information.
## Considerations
### Social Impact
The Spanish community can benefit from the high-quality data provided by this dataset.
### Bias
No post-processing steps have been applied to mitigate potential social biases.
## Additional Information
### Curators
Marc Pàmes @ Barcelona Supercomputing Center.
### License
This dataset is licensed under a **Creative Commons CC BY-NC-SA 3.0** license.
Quote from [WikiHow's Terms of Use](https://www.wikihow.com/wikiHow:Terms-of-Use):
> All text posted by Users to the Service is sub-licensed by wikiHow to other Users under a Creative Commons license as
> provided herein. The Creative Commons license allows such user generated text content to be used freely for personal,
> non-commercial purposes, so long as it is used and attributed to the original author as specified under the terms of
> the license. Allowing free republication of our articles helps wikiHow achieve its mission by providing instruction
> on solving the problems of everyday life to more people for free. In order to support this goal, wikiHow hereby grants
> each User of the Service a license to all text content that Users contribute to the Service under the terms and
> conditions of a Creative Commons CC BY-NC-SA 3.0 License. Please be sure to read the terms of the license carefully.
> You continue to own all right, title, and interest in and to your User Content, and you are free to distribute it as
> you wish, whether for commercial or non-commercial purposes.
|
mapama247/wikihow_es
|
[
"task_categories:text-classification",
"task_categories:question-answering",
"task_categories:conversational",
"task_categories:summarization",
"multilinguality:monolingual",
"size_categories:1K<n<10K",
"language:es",
"license:cc-by-nc-sa-3.0",
"Spanish",
"WikiHow",
"Wiki Articles",
"Tutorials",
"Step-By-Step",
"Instruction Tuning",
"region:us"
] |
2023-09-18T07:39:33+00:00
|
{"language": "es", "license": "cc-by-nc-sa-3.0", "multilinguality": "monolingual", "size_categories": "1K<n<10K", "task_categories": ["text-classification", "question-answering", "conversational", "summarization"], "pretty_name": "WikiHow-ES", "tags": ["Spanish", "WikiHow", "Wiki Articles", "Tutorials", "Step-By-Step", "Instruction Tuning"]}
|
2023-12-27T09:46:57+00:00
|
[] |
[
"es"
] |
TAGS
#task_categories-text-classification #task_categories-question-answering #task_categories-conversational #task_categories-summarization #multilinguality-monolingual #size_categories-1K<n<10K #language-Spanish #license-cc-by-nc-sa-3.0 #Spanish #WikiHow #Wiki Articles #Tutorials #Step-By-Step #Instruction Tuning #region-us
|
### Dataset Summary
Articles retrieved from the Spanish WikiHow website on September 2023.
Each article contains a tutorial about a specific topic. The format is always a "How to" question
followed by a detailed step-by-step explanation. In some cases, the response includes several methods.
The main idea is to use this data for instruction tuning of Spanish LLMs, but given its nature it
could also be used for other tasks such as text classification or summarization.
### Languages
- Spanish (ES)
### Usage
To load the full dataset:
To load only examples from a specific category:
List of available categories, with the repective number of examples:
### Supported Tasks
This dataset can be used to train a model for...
- 'instruction-tuning'
- 'text-classification'
- 'question-answering'
- 'conversational'
- 'summarization'
## Dataset Structure
### Data Instances
### Data Fields
- 'category': The category (from this list) to which the example belongs to.
- 'label': Numerical representation of the category, for text classification purposes.
- 'question': The article's title, which always starts with "¿Cómo ...".
- 'introduction': Introductory text that precedes the step-by-step explanation.
- 'answers': List of complete answers, with the full explanation of each step.
- 'short_answers': List of shorter answers that only contain one-sentence steps.
- 'num_answers': The number of alternative answers provided (e.g. length of 'answers').
- 'num_ref': Number of references provided in the article.
- 'expert_authors': Whether the article's author claims to be an expert on the topic or not.
- 'url': The URL address of the original article.
### Data Splits
There is only one split ('train') that contains a total of 7,380 examples.
## Dataset Creation
### Curation Rationale
This dataset was created for language model alignment to end tasks and user preferences.
### Source Data
How-To questions with detailed step-by-step answers, retrieved from the WikiHow website.
#### Data Collection and Normalization
All articles available in September 2023 were extracted, no filters applied.
Along with the article's content, some metadata was retrieved as well.
#### Source language producers
WikiHow users. All the content is human-generated.
### Personal and Sensitive Information
The data does not include personal or sensitive information.
## Considerations
### Social Impact
The Spanish community can benefit from the high-quality data provided by this dataset.
### Bias
No post-processing steps have been applied to mitigate potential social biases.
## Additional Information
### Curators
Marc Pàmes @ Barcelona Supercomputing Center.
### License
This dataset is licensed under a Creative Commons CC BY-NC-SA 3.0 license.
Quote from WikiHow's Terms of Use:
> All text posted by Users to the Service is sub-licensed by wikiHow to other Users under a Creative Commons license as
> provided herein. The Creative Commons license allows such user generated text content to be used freely for personal,
> non-commercial purposes, so long as it is used and attributed to the original author as specified under the terms of
> the license. Allowing free republication of our articles helps wikiHow achieve its mission by providing instruction
> on solving the problems of everyday life to more people for free. In order to support this goal, wikiHow hereby grants
> each User of the Service a license to all text content that Users contribute to the Service under the terms and
> conditions of a Creative Commons CC BY-NC-SA 3.0 License. Please be sure to read the terms of the license carefully.
> You continue to own all right, title, and interest in and to your User Content, and you are free to distribute it as
> you wish, whether for commercial or non-commercial purposes.
|
[
"### Dataset Summary\n\nArticles retrieved from the Spanish WikiHow website on September 2023.\n\nEach article contains a tutorial about a specific topic. The format is always a \"How to\" question \nfollowed by a detailed step-by-step explanation. In some cases, the response includes several methods. \n\nThe main idea is to use this data for instruction tuning of Spanish LLMs, but given its nature it \ncould also be used for other tasks such as text classification or summarization.",
"### Languages\n\n- Spanish (ES)",
"### Usage\n\nTo load the full dataset:\n\n\nTo load only examples from a specific category:\n\n\nList of available categories, with the repective number of examples:",
"### Supported Tasks\n\nThis dataset can be used to train a model for...\n\n- 'instruction-tuning'\n- 'text-classification'\n- 'question-answering'\n- 'conversational'\n- 'summarization'",
"## Dataset Structure",
"### Data Instances",
"### Data Fields\n\n- 'category': The category (from this list) to which the example belongs to.\n- 'label': Numerical representation of the category, for text classification purposes.\n- 'question': The article's title, which always starts with \"¿Cómo ...\".\n- 'introduction': Introductory text that precedes the step-by-step explanation.\n- 'answers': List of complete answers, with the full explanation of each step.\n- 'short_answers': List of shorter answers that only contain one-sentence steps.\n- 'num_answers': The number of alternative answers provided (e.g. length of 'answers').\n- 'num_ref': Number of references provided in the article.\n- 'expert_authors': Whether the article's author claims to be an expert on the topic or not.\n- 'url': The URL address of the original article.",
"### Data Splits\n\nThere is only one split ('train') that contains a total of 7,380 examples.",
"## Dataset Creation",
"### Curation Rationale\n\nThis dataset was created for language model alignment to end tasks and user preferences.",
"### Source Data\n\nHow-To questions with detailed step-by-step answers, retrieved from the WikiHow website.",
"#### Data Collection and Normalization\n\nAll articles available in September 2023 were extracted, no filters applied.\n\nAlong with the article's content, some metadata was retrieved as well.",
"#### Source language producers\n\nWikiHow users. All the content is human-generated.",
"### Personal and Sensitive Information\n\nThe data does not include personal or sensitive information.",
"## Considerations",
"### Social Impact\n\nThe Spanish community can benefit from the high-quality data provided by this dataset.",
"### Bias\n\nNo post-processing steps have been applied to mitigate potential social biases.",
"## Additional Information",
"### Curators\n\nMarc Pàmes @ Barcelona Supercomputing Center.",
"### License\n\nThis dataset is licensed under a Creative Commons CC BY-NC-SA 3.0 license.\n\nQuote from WikiHow's Terms of Use:\n\n> All text posted by Users to the Service is sub-licensed by wikiHow to other Users under a Creative Commons license as \n> provided herein. The Creative Commons license allows such user generated text content to be used freely for personal, \n> non-commercial purposes, so long as it is used and attributed to the original author as specified under the terms of \n> the license. Allowing free republication of our articles helps wikiHow achieve its mission by providing instruction \n> on solving the problems of everyday life to more people for free. In order to support this goal, wikiHow hereby grants \n> each User of the Service a license to all text content that Users contribute to the Service under the terms and \n> conditions of a Creative Commons CC BY-NC-SA 3.0 License. Please be sure to read the terms of the license carefully. \n> You continue to own all right, title, and interest in and to your User Content, and you are free to distribute it as \n> you wish, whether for commercial or non-commercial purposes."
] |
[
"TAGS\n#task_categories-text-classification #task_categories-question-answering #task_categories-conversational #task_categories-summarization #multilinguality-monolingual #size_categories-1K<n<10K #language-Spanish #license-cc-by-nc-sa-3.0 #Spanish #WikiHow #Wiki Articles #Tutorials #Step-By-Step #Instruction Tuning #region-us \n",
"### Dataset Summary\n\nArticles retrieved from the Spanish WikiHow website on September 2023.\n\nEach article contains a tutorial about a specific topic. The format is always a \"How to\" question \nfollowed by a detailed step-by-step explanation. In some cases, the response includes several methods. \n\nThe main idea is to use this data for instruction tuning of Spanish LLMs, but given its nature it \ncould also be used for other tasks such as text classification or summarization.",
"### Languages\n\n- Spanish (ES)",
"### Usage\n\nTo load the full dataset:\n\n\nTo load only examples from a specific category:\n\n\nList of available categories, with the repective number of examples:",
"### Supported Tasks\n\nThis dataset can be used to train a model for...\n\n- 'instruction-tuning'\n- 'text-classification'\n- 'question-answering'\n- 'conversational'\n- 'summarization'",
"## Dataset Structure",
"### Data Instances",
"### Data Fields\n\n- 'category': The category (from this list) to which the example belongs to.\n- 'label': Numerical representation of the category, for text classification purposes.\n- 'question': The article's title, which always starts with \"¿Cómo ...\".\n- 'introduction': Introductory text that precedes the step-by-step explanation.\n- 'answers': List of complete answers, with the full explanation of each step.\n- 'short_answers': List of shorter answers that only contain one-sentence steps.\n- 'num_answers': The number of alternative answers provided (e.g. length of 'answers').\n- 'num_ref': Number of references provided in the article.\n- 'expert_authors': Whether the article's author claims to be an expert on the topic or not.\n- 'url': The URL address of the original article.",
"### Data Splits\n\nThere is only one split ('train') that contains a total of 7,380 examples.",
"## Dataset Creation",
"### Curation Rationale\n\nThis dataset was created for language model alignment to end tasks and user preferences.",
"### Source Data\n\nHow-To questions with detailed step-by-step answers, retrieved from the WikiHow website.",
"#### Data Collection and Normalization\n\nAll articles available in September 2023 were extracted, no filters applied.\n\nAlong with the article's content, some metadata was retrieved as well.",
"#### Source language producers\n\nWikiHow users. All the content is human-generated.",
"### Personal and Sensitive Information\n\nThe data does not include personal or sensitive information.",
"## Considerations",
"### Social Impact\n\nThe Spanish community can benefit from the high-quality data provided by this dataset.",
"### Bias\n\nNo post-processing steps have been applied to mitigate potential social biases.",
"## Additional Information",
"### Curators\n\nMarc Pàmes @ Barcelona Supercomputing Center.",
"### License\n\nThis dataset is licensed under a Creative Commons CC BY-NC-SA 3.0 license.\n\nQuote from WikiHow's Terms of Use:\n\n> All text posted by Users to the Service is sub-licensed by wikiHow to other Users under a Creative Commons license as \n> provided herein. The Creative Commons license allows such user generated text content to be used freely for personal, \n> non-commercial purposes, so long as it is used and attributed to the original author as specified under the terms of \n> the license. Allowing free republication of our articles helps wikiHow achieve its mission by providing instruction \n> on solving the problems of everyday life to more people for free. In order to support this goal, wikiHow hereby grants \n> each User of the Service a license to all text content that Users contribute to the Service under the terms and \n> conditions of a Creative Commons CC BY-NC-SA 3.0 License. Please be sure to read the terms of the license carefully. \n> You continue to own all right, title, and interest in and to your User Content, and you are free to distribute it as \n> you wish, whether for commercial or non-commercial purposes."
] |
[
113,
105,
9,
36,
52,
6,
6,
216,
28,
5,
27,
27,
41,
19,
18,
3,
21,
23,
5,
16,
259
] |
[
"passage: TAGS\n#task_categories-text-classification #task_categories-question-answering #task_categories-conversational #task_categories-summarization #multilinguality-monolingual #size_categories-1K<n<10K #language-Spanish #license-cc-by-nc-sa-3.0 #Spanish #WikiHow #Wiki Articles #Tutorials #Step-By-Step #Instruction Tuning #region-us \n### Dataset Summary\n\nArticles retrieved from the Spanish WikiHow website on September 2023.\n\nEach article contains a tutorial about a specific topic. The format is always a \"How to\" question \nfollowed by a detailed step-by-step explanation. In some cases, the response includes several methods. \n\nThe main idea is to use this data for instruction tuning of Spanish LLMs, but given its nature it \ncould also be used for other tasks such as text classification or summarization.### Languages\n\n- Spanish (ES)### Usage\n\nTo load the full dataset:\n\n\nTo load only examples from a specific category:\n\n\nList of available categories, with the repective number of examples:### Supported Tasks\n\nThis dataset can be used to train a model for...\n\n- 'instruction-tuning'\n- 'text-classification'\n- 'question-answering'\n- 'conversational'\n- 'summarization'## Dataset Structure### Data Instances"
] |
2f874923ea3724472eed85a88c859514e52c55eb
|
Clone from https://github.com/vietai/mTet
---
license: cc-by-4.0
task_categories:
- translation
language:
- vi
license: cc-by-4.0
---
|
hungeni/mtet_dataset
|
[
"region:us"
] |
2023-09-18T07:40:03+00:00
|
{}
|
2023-09-18T08:00:31+00:00
|
[] |
[] |
TAGS
#region-us
|
Clone from URL
---
license: cc-by-4.0
task_categories:
- translation
language:
- vi
license: cc-by-4.0
---
|
[] |
[
"TAGS\n#region-us \n"
] |
[
6
] |
[
"passage: TAGS\n#region-us \n"
] |
f5c025bd01004bc68ffc893de936424874603742
|
# Dataset of Priestess
This is the dataset of Priestess, containing 300 images and their tags.
Images are crawled from many sites (e.g. danbooru, pixiv, zerochan ...), the auto-crawling system is powered by [DeepGHS Team](https://github.com/deepghs)([huggingface organization](https://huggingface.co/deepghs)).
| Name | Images | Download | Description |
|:------------|---------:|:------------------------------------|:-------------------------------------------------------------------------|
| raw | 300 | [Download](dataset-raw.zip) | Raw data with meta information. |
| raw-stage3 | 653 | [Download](dataset-raw-stage3.zip) | 3-stage cropped raw data with meta information. |
| 384x512 | 300 | [Download](dataset-384x512.zip) | 384x512 aligned dataset. |
| 512x512 | 300 | [Download](dataset-512x512.zip) | 512x512 aligned dataset. |
| 512x704 | 300 | [Download](dataset-512x704.zip) | 512x704 aligned dataset. |
| 640x640 | 300 | [Download](dataset-640x640.zip) | 640x640 aligned dataset. |
| 640x880 | 300 | [Download](dataset-640x880.zip) | 640x880 aligned dataset. |
| stage3-640 | 653 | [Download](dataset-stage3-640.zip) | 3-stage cropped dataset with the shorter side not exceeding 640 pixels. |
| stage3-800 | 653 | [Download](dataset-stage3-800.zip) | 3-stage cropped dataset with the shorter side not exceeding 800 pixels. |
| stage3-1200 | 653 | [Download](dataset-stage3-1200.zip) | 3-stage cropped dataset with the shorter side not exceeding 1200 pixels. |
|
CyberHarem/priestess_goblinslayer
|
[
"task_categories:text-to-image",
"size_categories:n<1K",
"license:mit",
"art",
"not-for-all-audiences",
"region:us"
] |
2023-09-18T07:45:27+00:00
|
{"license": "mit", "size_categories": ["n<1K"], "task_categories": ["text-to-image"], "tags": ["art", "not-for-all-audiences"]}
|
2023-09-18T07:52:14+00:00
|
[] |
[] |
TAGS
#task_categories-text-to-image #size_categories-n<1K #license-mit #art #not-for-all-audiences #region-us
|
Dataset of Priestess
====================
This is the dataset of Priestess, containing 300 images and their tags.
Images are crawled from many sites (e.g. danbooru, pixiv, zerochan ...), the auto-crawling system is powered by DeepGHS Team(huggingface organization).
|
[] |
[
"TAGS\n#task_categories-text-to-image #size_categories-n<1K #license-mit #art #not-for-all-audiences #region-us \n"
] |
[
44
] |
[
"passage: TAGS\n#task_categories-text-to-image #size_categories-n<1K #license-mit #art #not-for-all-audiences #region-us \n"
] |
a26c3391dde6af6daefb98f50e7f3e3cadfd629c
|
# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem
## Abstract:
The sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.
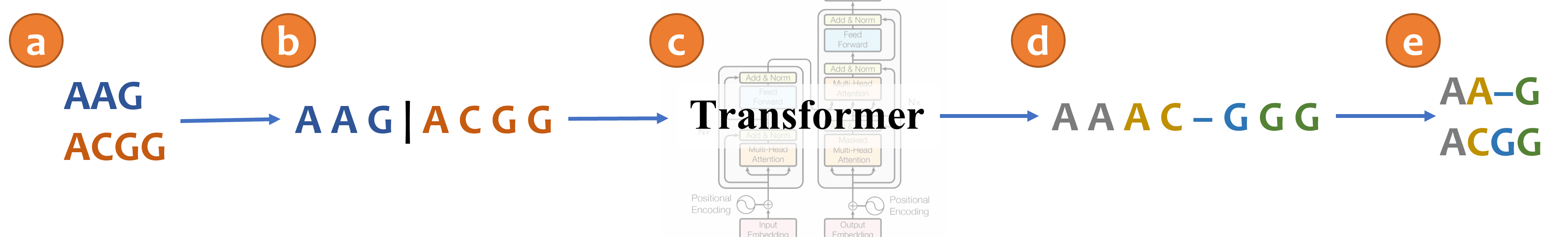
An illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences "AAG" and "ACGG". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which "AA-G" is aligned to "ACGG". The transformer architecture illustration is adapted from (Vaswani et al., 2017).
## Data:
We used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.
We generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \in (0.0,0.05)*, *A_I, A_D \in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order "T", "C", "A" and "G"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order "a", "b", "c", "d", and "e" for the substitution matrix.
## Example:
The following example correspond for the illustrated MSA in the figure above:
{"MSA": "AAAC-GGG", "unaligned_seqs": {"seq0": "AAG", "seq1": "ACGG"}}
## APA
```
Dotan, E., Belinkov, Y., Avram, O., Wygoda, E., Ecker, N., Alburquerque, M., Keren, O., Loewenthal, G., & Pupko T. (2023). Multiple sequence alignment as a sequence-to-sequence learning problem. The Eleventh International Conference on Learning Representations (ICLR 2023).
```
## BibTeX
```
@article{Dotan_multiple_2023,
author = {Dotan, Edo and Belinkov, Yonatan and Avram, Oren and Wygoda, Elya and Ecker, Noa and Alburquerque, Michael and Keren, Omri and Loewenthal, Gil and Pupko, Tal},
month = aug,
title = {{Multiple sequence alignment as a sequence-to-sequence learning problem}},
year = {2023}
}
```
|
dotan1111/MSA-amino-2-seq
|
[
"sequence-to-sequence",
"bioinformatics",
"biology",
"region:us"
] |
2023-09-18T07:45:48+00:00
|
{"tags": ["sequence-to-sequence", "bioinformatics", "biology"]}
|
2023-09-18T10:46:50+00:00
|
[] |
[] |
TAGS
#sequence-to-sequence #bioinformatics #biology #region-us
|
# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem
## Abstract:
The sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.
!image
An illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences "AAG" and "ACGG". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which "AA-G" is aligned to "ACGG". The transformer architecture illustration is adapted from (Vaswani et al., 2017).
## Data:
We used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.
We generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \in (0.0,0.05)*, *A_I, A_D \in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order "T", "C", "A" and "G"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order "a", "b", "c", "d", and "e" for the substitution matrix.
## Example:
The following example correspond for the illustrated MSA in the figure above:
{"MSA": "AAAC-GGG", "unaligned_seqs": {"seq0": "AAG", "seq1": "ACGG"}}
## APA
## BibTeX
|
[
"# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem",
"## Abstract:\nThe sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.\n!image\n\nAn illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences \"AAG\" and \"ACGG\". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which \"AA-G\" is aligned to \"ACGG\". The transformer architecture illustration is adapted from (Vaswani et al., 2017).",
"## Data:\n\nWe used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.\n\nWe generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \\in (0.0,0.05)*, *A_I, A_D \\in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order \"T\", \"C\", \"A\" and \"G\"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order \"a\", \"b\", \"c\", \"d\", and \"e\" for the substitution matrix.",
"## Example:\n\nThe following example correspond for the illustrated MSA in the figure above:\n\n{\"MSA\": \"AAAC-GGG\", \"unaligned_seqs\": {\"seq0\": \"AAG\", \"seq1\": \"ACGG\"}}",
"## APA",
"## BibTeX"
] |
[
"TAGS\n#sequence-to-sequence #bioinformatics #biology #region-us \n",
"# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem",
"## Abstract:\nThe sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.\n!image\n\nAn illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences \"AAG\" and \"ACGG\". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which \"AA-G\" is aligned to \"ACGG\". The transformer architecture illustration is adapted from (Vaswani et al., 2017).",
"## Data:\n\nWe used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.\n\nWe generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \\in (0.0,0.05)*, *A_I, A_D \\in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order \"T\", \"C\", \"A\" and \"G\"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order \"a\", \"b\", \"c\", \"d\", and \"e\" for the substitution matrix.",
"## Example:\n\nThe following example correspond for the illustrated MSA in the figure above:\n\n{\"MSA\": \"AAAC-GGG\", \"unaligned_seqs\": {\"seq0\": \"AAG\", \"seq1\": \"ACGG\"}}",
"## APA",
"## BibTeX"
] |
[
23,
23,
323,
508,
64,
2,
5
] |
[
"passage: TAGS\n#sequence-to-sequence #bioinformatics #biology #region-us \n# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem## Abstract:\nThe sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.\n!image\n\nAn illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences \"AAG\" and \"ACGG\". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which \"AA-G\" is aligned to \"ACGG\". The transformer architecture illustration is adapted from (Vaswani et al., 2017)."
] |
76cdffad389c86a38687f33bdb650165e090bee4
|
# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem
## Abstract:
The sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.

An illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences "AAG" and "ACGG". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which "AA-G" is aligned to "ACGG". The transformer architecture illustration is adapted from (Vaswani et al., 2017).
## Data:
We used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.
We generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \in (0.0,0.05)*, *A_I, A_D \in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order "T", "C", "A" and "G"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order "a", "b", "c", "d", and "e" for the substitution matrix.
## Example:
The following example correspond for the illustrated MSA in the figure above:
{"MSA": "AAAC-GGG", "unaligned_seqs": {"seq0": "AAG", "seq1": "ACGG"}}
## APA
```
Dotan, E., Belinkov, Y., Avram, O., Wygoda, E., Ecker, N., Alburquerque, M., Keren, O., Loewenthal, G., & Pupko T. (2023). Multiple sequence alignment as a sequence-to-sequence learning problem. The Eleventh International Conference on Learning Representations (ICLR 2023).
```
## BibTeX
```
@article{Dotan_multiple_2023,
author = {Dotan, Edo and Belinkov, Yonatan and Avram, Oren and Wygoda, Elya and Ecker, Noa and Alburquerque, Michael and Keren, Omri and Loewenthal, Gil and Pupko, Tal},
month = aug,
title = {{Multiple sequence alignment as a sequence-to-sequence learning problem}},
year = {2023}
}
```
|
dotan1111/MSA-amino-3-seq
|
[
"sequence-to-sequence",
"bioinformatics",
"biology",
"region:us"
] |
2023-09-18T07:46:01+00:00
|
{"tags": ["sequence-to-sequence", "bioinformatics", "biology"]}
|
2023-09-18T10:46:25+00:00
|
[] |
[] |
TAGS
#sequence-to-sequence #bioinformatics #biology #region-us
|
# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem
## Abstract:
The sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.
!image
An illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences "AAG" and "ACGG". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which "AA-G" is aligned to "ACGG". The transformer architecture illustration is adapted from (Vaswani et al., 2017).
## Data:
We used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.
We generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \in (0.0,0.05)*, *A_I, A_D \in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order "T", "C", "A" and "G"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order "a", "b", "c", "d", and "e" for the substitution matrix.
## Example:
The following example correspond for the illustrated MSA in the figure above:
{"MSA": "AAAC-GGG", "unaligned_seqs": {"seq0": "AAG", "seq1": "ACGG"}}
## APA
## BibTeX
|
[
"# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem",
"## Abstract:\nThe sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.\n!image\n\nAn illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences \"AAG\" and \"ACGG\". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which \"AA-G\" is aligned to \"ACGG\". The transformer architecture illustration is adapted from (Vaswani et al., 2017).",
"## Data:\n\nWe used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.\n\nWe generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \\in (0.0,0.05)*, *A_I, A_D \\in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order \"T\", \"C\", \"A\" and \"G\"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order \"a\", \"b\", \"c\", \"d\", and \"e\" for the substitution matrix.",
"## Example:\n\nThe following example correspond for the illustrated MSA in the figure above:\n\n{\"MSA\": \"AAAC-GGG\", \"unaligned_seqs\": {\"seq0\": \"AAG\", \"seq1\": \"ACGG\"}}",
"## APA",
"## BibTeX"
] |
[
"TAGS\n#sequence-to-sequence #bioinformatics #biology #region-us \n",
"# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem",
"## Abstract:\nThe sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.\n!image\n\nAn illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences \"AAG\" and \"ACGG\". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which \"AA-G\" is aligned to \"ACGG\". The transformer architecture illustration is adapted from (Vaswani et al., 2017).",
"## Data:\n\nWe used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.\n\nWe generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \\in (0.0,0.05)*, *A_I, A_D \\in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order \"T\", \"C\", \"A\" and \"G\"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order \"a\", \"b\", \"c\", \"d\", and \"e\" for the substitution matrix.",
"## Example:\n\nThe following example correspond for the illustrated MSA in the figure above:\n\n{\"MSA\": \"AAAC-GGG\", \"unaligned_seqs\": {\"seq0\": \"AAG\", \"seq1\": \"ACGG\"}}",
"## APA",
"## BibTeX"
] |
[
23,
23,
323,
508,
64,
2,
5
] |
[
"passage: TAGS\n#sequence-to-sequence #bioinformatics #biology #region-us \n# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem## Abstract:\nThe sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.\n!image\n\nAn illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences \"AAG\" and \"ACGG\". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which \"AA-G\" is aligned to \"ACGG\". The transformer architecture illustration is adapted from (Vaswani et al., 2017)."
] |
0ba641d2606faad334b614f06d86e4d035b5bd94
|
# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem
## Abstract:
The sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.

An illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences "AAG" and "ACGG". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which "AA-G" is aligned to "ACGG". The transformer architecture illustration is adapted from (Vaswani et al., 2017).
## Data:
We used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.
We generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \in (0.0,0.05)*, *A_I, A_D \in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order "T", "C", "A" and "G"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order "a", "b", "c", "d", and "e" for the substitution matrix.
## Example:
The following example correspond for the illustrated MSA in the figure above:
{"MSA": "AAAC-GGG", "unaligned_seqs": {"seq0": "AAG", "seq1": "ACGG"}}
## APA
```
Dotan, E., Belinkov, Y., Avram, O., Wygoda, E., Ecker, N., Alburquerque, M., Keren, O., Loewenthal, G., & Pupko T. (2023). Multiple sequence alignment as a sequence-to-sequence learning problem. The Eleventh International Conference on Learning Representations (ICLR 2023).
```
## BibTeX
```
@article{Dotan_multiple_2023,
author = {Dotan, Edo and Belinkov, Yonatan and Avram, Oren and Wygoda, Elya and Ecker, Noa and Alburquerque, Michael and Keren, Omri and Loewenthal, Gil and Pupko, Tal},
month = aug,
title = {{Multiple sequence alignment as a sequence-to-sequence learning problem}},
year = {2023}
}
```
|
dotan1111/MSA-amino-4-seq
|
[
"sequence-to-sequence",
"bioinformatics",
"biology",
"region:us"
] |
2023-09-18T07:46:34+00:00
|
{"tags": ["sequence-to-sequence", "bioinformatics", "biology"]}
|
2023-09-18T10:47:10+00:00
|
[] |
[] |
TAGS
#sequence-to-sequence #bioinformatics #biology #region-us
|
# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem
## Abstract:
The sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.
!image
An illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences "AAG" and "ACGG". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which "AA-G" is aligned to "ACGG". The transformer architecture illustration is adapted from (Vaswani et al., 2017).
## Data:
We used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.
We generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \in (0.0,0.05)*, *A_I, A_D \in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order "T", "C", "A" and "G"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order "a", "b", "c", "d", and "e" for the substitution matrix.
## Example:
The following example correspond for the illustrated MSA in the figure above:
{"MSA": "AAAC-GGG", "unaligned_seqs": {"seq0": "AAG", "seq1": "ACGG"}}
## APA
## BibTeX
|
[
"# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem",
"## Abstract:\nThe sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.\n!image\n\nAn illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences \"AAG\" and \"ACGG\". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which \"AA-G\" is aligned to \"ACGG\". The transformer architecture illustration is adapted from (Vaswani et al., 2017).",
"## Data:\n\nWe used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.\n\nWe generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \\in (0.0,0.05)*, *A_I, A_D \\in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order \"T\", \"C\", \"A\" and \"G\"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order \"a\", \"b\", \"c\", \"d\", and \"e\" for the substitution matrix.",
"## Example:\n\nThe following example correspond for the illustrated MSA in the figure above:\n\n{\"MSA\": \"AAAC-GGG\", \"unaligned_seqs\": {\"seq0\": \"AAG\", \"seq1\": \"ACGG\"}}",
"## APA",
"## BibTeX"
] |
[
"TAGS\n#sequence-to-sequence #bioinformatics #biology #region-us \n",
"# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem",
"## Abstract:\nThe sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.\n!image\n\nAn illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences \"AAG\" and \"ACGG\". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which \"AA-G\" is aligned to \"ACGG\". The transformer architecture illustration is adapted from (Vaswani et al., 2017).",
"## Data:\n\nWe used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.\n\nWe generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \\in (0.0,0.05)*, *A_I, A_D \\in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order \"T\", \"C\", \"A\" and \"G\"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order \"a\", \"b\", \"c\", \"d\", and \"e\" for the substitution matrix.",
"## Example:\n\nThe following example correspond for the illustrated MSA in the figure above:\n\n{\"MSA\": \"AAAC-GGG\", \"unaligned_seqs\": {\"seq0\": \"AAG\", \"seq1\": \"ACGG\"}}",
"## APA",
"## BibTeX"
] |
[
23,
23,
323,
508,
64,
2,
5
] |
[
"passage: TAGS\n#sequence-to-sequence #bioinformatics #biology #region-us \n# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem## Abstract:\nThe sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.\n!image\n\nAn illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences \"AAG\" and \"ACGG\". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which \"AA-G\" is aligned to \"ACGG\". The transformer architecture illustration is adapted from (Vaswani et al., 2017)."
] |
84c41cf9fa4707c3803cb5838dfd05ff1be44bce
|
# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem
## Abstract:
The sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.

An illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences "AAG" and "ACGG". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which "AA-G" is aligned to "ACGG". The transformer architecture illustration is adapted from (Vaswani et al., 2017).
## Data:
We used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.
We generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \in (0.0,0.05)*, *A_I, A_D \in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order "T", "C", "A" and "G"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order "a", "b", "c", "d", and "e" for the substitution matrix.
## Example:
The following example correspond for the illustrated MSA in the figure above:
{"MSA": "AAAC-GGG", "unaligned_seqs": {"seq0": "AAG", "seq1": "ACGG"}}
## APA
```
Dotan, E., Belinkov, Y., Avram, O., Wygoda, E., Ecker, N., Alburquerque, M., Keren, O., Loewenthal, G., & Pupko T. (2023). Multiple sequence alignment as a sequence-to-sequence learning problem. The Eleventh International Conference on Learning Representations (ICLR 2023).
```
## BibTeX
```
@article{Dotan_multiple_2023,
author = {Dotan, Edo and Belinkov, Yonatan and Avram, Oren and Wygoda, Elya and Ecker, Noa and Alburquerque, Michael and Keren, Omri and Loewenthal, Gil and Pupko, Tal},
month = aug,
title = {{Multiple sequence alignment as a sequence-to-sequence learning problem}},
year = {2023}
}
```
|
dotan1111/MSA-amino-5-seq
|
[
"sequence-to-sequence",
"bioinformatics",
"biology",
"region:us"
] |
2023-09-18T07:47:21+00:00
|
{"tags": ["sequence-to-sequence", "bioinformatics", "biology"]}
|
2023-09-18T10:47:27+00:00
|
[] |
[] |
TAGS
#sequence-to-sequence #bioinformatics #biology #region-us
|
# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem
## Abstract:
The sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.
!image
An illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences "AAG" and "ACGG". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which "AA-G" is aligned to "ACGG". The transformer architecture illustration is adapted from (Vaswani et al., 2017).
## Data:
We used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.
We generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \in (0.0,0.05)*, *A_I, A_D \in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order "T", "C", "A" and "G"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order "a", "b", "c", "d", and "e" for the substitution matrix.
## Example:
The following example correspond for the illustrated MSA in the figure above:
{"MSA": "AAAC-GGG", "unaligned_seqs": {"seq0": "AAG", "seq1": "ACGG"}}
## APA
## BibTeX
|
[
"# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem",
"## Abstract:\nThe sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.\n!image\n\nAn illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences \"AAG\" and \"ACGG\". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which \"AA-G\" is aligned to \"ACGG\". The transformer architecture illustration is adapted from (Vaswani et al., 2017).",
"## Data:\n\nWe used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.\n\nWe generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \\in (0.0,0.05)*, *A_I, A_D \\in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order \"T\", \"C\", \"A\" and \"G\"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order \"a\", \"b\", \"c\", \"d\", and \"e\" for the substitution matrix.",
"## Example:\n\nThe following example correspond for the illustrated MSA in the figure above:\n\n{\"MSA\": \"AAAC-GGG\", \"unaligned_seqs\": {\"seq0\": \"AAG\", \"seq1\": \"ACGG\"}}",
"## APA",
"## BibTeX"
] |
[
"TAGS\n#sequence-to-sequence #bioinformatics #biology #region-us \n",
"# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem",
"## Abstract:\nThe sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.\n!image\n\nAn illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences \"AAG\" and \"ACGG\". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which \"AA-G\" is aligned to \"ACGG\". The transformer architecture illustration is adapted from (Vaswani et al., 2017).",
"## Data:\n\nWe used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.\n\nWe generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \\in (0.0,0.05)*, *A_I, A_D \\in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order \"T\", \"C\", \"A\" and \"G\"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order \"a\", \"b\", \"c\", \"d\", and \"e\" for the substitution matrix.",
"## Example:\n\nThe following example correspond for the illustrated MSA in the figure above:\n\n{\"MSA\": \"AAAC-GGG\", \"unaligned_seqs\": {\"seq0\": \"AAG\", \"seq1\": \"ACGG\"}}",
"## APA",
"## BibTeX"
] |
[
23,
23,
323,
508,
64,
2,
5
] |
[
"passage: TAGS\n#sequence-to-sequence #bioinformatics #biology #region-us \n# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem## Abstract:\nThe sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.\n!image\n\nAn illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences \"AAG\" and \"ACGG\". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which \"AA-G\" is aligned to \"ACGG\". The transformer architecture illustration is adapted from (Vaswani et al., 2017)."
] |
d360b2645182eed270905bd55a566d3f6d923708
|
This dataset crawl and re-organize from site: amruta.org, extracting questions and answers for fine-tuning tasks.
By the grace of H.H. Mother Shri Mataji Nirmala Devi
|
hungeni/amrutaQA
|
[
"license:other",
"region:us"
] |
2023-09-18T07:47:41+00:00
|
{"license": "other"}
|
2023-09-20T10:47:41+00:00
|
[] |
[] |
TAGS
#license-other #region-us
|
This dataset crawl and re-organize from site: URL, extracting questions and answers for fine-tuning tasks.
By the grace of H.H. Mother Shri Mataji Nirmala Devi
|
[] |
[
"TAGS\n#license-other #region-us \n"
] |
[
11
] |
[
"passage: TAGS\n#license-other #region-us \n"
] |
83a2a5672499d808ef1a6c48dbbd8b184d8a8d8d
|
# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem
## Abstract:
The sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.

An illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences "AAG" and "ACGG". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which "AA-G" is aligned to "ACGG". The transformer architecture illustration is adapted from (Vaswani et al., 2017).
## Data:
We used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.
We generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \in (0.0,0.05)*, *A_I, A_D \in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order "T", "C", "A" and "G"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order "a", "b", "c", "d", and "e" for the substitution matrix.
## Example:
The following example correspond for the illustrated MSA in the figure above:
{"MSA": "AAAC-GGG", "unaligned_seqs": {"seq0": "AAG", "seq1": "ACGG"}}
## APA
```
Dotan, E., Belinkov, Y., Avram, O., Wygoda, E., Ecker, N., Alburquerque, M., Keren, O., Loewenthal, G., & Pupko T. (2023). Multiple sequence alignment as a sequence-to-sequence learning problem. The Eleventh International Conference on Learning Representations (ICLR 2023).
```
## BibTeX
```
@article{Dotan_multiple_2023,
author = {Dotan, Edo and Belinkov, Yonatan and Avram, Oren and Wygoda, Elya and Ecker, Noa and Alburquerque, Michael and Keren, Omri and Loewenthal, Gil and Pupko, Tal},
month = aug,
title = {{Multiple sequence alignment as a sequence-to-sequence learning problem}},
year = {2023}
}
```
|
dotan1111/MSA-amino-6-seq
|
[
"sequence-to-sequence",
"bioinformatics",
"biology",
"region:us"
] |
2023-09-18T07:50:48+00:00
|
{"tags": ["sequence-to-sequence", "bioinformatics", "biology"]}
|
2023-09-18T10:47:37+00:00
|
[] |
[] |
TAGS
#sequence-to-sequence #bioinformatics #biology #region-us
|
# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem
## Abstract:
The sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.
!image
An illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences "AAG" and "ACGG". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which "AA-G" is aligned to "ACGG". The transformer architecture illustration is adapted from (Vaswani et al., 2017).
## Data:
We used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.
We generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \in (0.0,0.05)*, *A_I, A_D \in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order "T", "C", "A" and "G"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order "a", "b", "c", "d", and "e" for the substitution matrix.
## Example:
The following example correspond for the illustrated MSA in the figure above:
{"MSA": "AAAC-GGG", "unaligned_seqs": {"seq0": "AAG", "seq1": "ACGG"}}
## APA
## BibTeX
|
[
"# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem",
"## Abstract:\nThe sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.\n!image\n\nAn illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences \"AAG\" and \"ACGG\". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which \"AA-G\" is aligned to \"ACGG\". The transformer architecture illustration is adapted from (Vaswani et al., 2017).",
"## Data:\n\nWe used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.\n\nWe generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \\in (0.0,0.05)*, *A_I, A_D \\in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order \"T\", \"C\", \"A\" and \"G\"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order \"a\", \"b\", \"c\", \"d\", and \"e\" for the substitution matrix.",
"## Example:\n\nThe following example correspond for the illustrated MSA in the figure above:\n\n{\"MSA\": \"AAAC-GGG\", \"unaligned_seqs\": {\"seq0\": \"AAG\", \"seq1\": \"ACGG\"}}",
"## APA",
"## BibTeX"
] |
[
"TAGS\n#sequence-to-sequence #bioinformatics #biology #region-us \n",
"# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem",
"## Abstract:\nThe sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.\n!image\n\nAn illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences \"AAG\" and \"ACGG\". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which \"AA-G\" is aligned to \"ACGG\". The transformer architecture illustration is adapted from (Vaswani et al., 2017).",
"## Data:\n\nWe used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.\n\nWe generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \\in (0.0,0.05)*, *A_I, A_D \\in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order \"T\", \"C\", \"A\" and \"G\"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order \"a\", \"b\", \"c\", \"d\", and \"e\" for the substitution matrix.",
"## Example:\n\nThe following example correspond for the illustrated MSA in the figure above:\n\n{\"MSA\": \"AAAC-GGG\", \"unaligned_seqs\": {\"seq0\": \"AAG\", \"seq1\": \"ACGG\"}}",
"## APA",
"## BibTeX"
] |
[
23,
23,
323,
508,
64,
2,
5
] |
[
"passage: TAGS\n#sequence-to-sequence #bioinformatics #biology #region-us \n# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem## Abstract:\nThe sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.\n!image\n\nAn illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences \"AAG\" and \"ACGG\". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which \"AA-G\" is aligned to \"ACGG\". The transformer architecture illustration is adapted from (Vaswani et al., 2017)."
] |
6fd52130ebe36874aa80b78488fa060967f76eb1
|
# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem
## Abstract:
The sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.

An illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences "AAG" and "ACGG". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which "AA-G" is aligned to "ACGG". The transformer architecture illustration is adapted from (Vaswani et al., 2017).
## Data:
We used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.
We generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \in (0.0,0.05)*, *A_I, A_D \in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order "T", "C", "A" and "G"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order "a", "b", "c", "d", and "e" for the substitution matrix.
## Example:
The following example correspond for the illustrated MSA in the figure above:
{"MSA": "AAAC-GGG", "unaligned_seqs": {"seq0": "AAG", "seq1": "ACGG"}}
## APA
```
Dotan, E., Belinkov, Y., Avram, O., Wygoda, E., Ecker, N., Alburquerque, M., Keren, O., Loewenthal, G., & Pupko T. (2023). Multiple sequence alignment as a sequence-to-sequence learning problem. The Eleventh International Conference on Learning Representations (ICLR 2023).
```
## BibTeX
```
@article{Dotan_multiple_2023,
author = {Dotan, Edo and Belinkov, Yonatan and Avram, Oren and Wygoda, Elya and Ecker, Noa and Alburquerque, Michael and Keren, Omri and Loewenthal, Gil and Pupko, Tal},
month = aug,
title = {{Multiple sequence alignment as a sequence-to-sequence learning problem}},
year = {2023}
}
```
|
dotan1111/MSA-amino-7-seq
|
[
"sequence-to-sequence",
"bioinformatics",
"biology",
"region:us"
] |
2023-09-18T07:50:50+00:00
|
{"tags": ["sequence-to-sequence", "bioinformatics", "biology"]}
|
2023-09-18T10:47:47+00:00
|
[] |
[] |
TAGS
#sequence-to-sequence #bioinformatics #biology #region-us
|
# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem
## Abstract:
The sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.
!image
An illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences "AAG" and "ACGG". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which "AA-G" is aligned to "ACGG". The transformer architecture illustration is adapted from (Vaswani et al., 2017).
## Data:
We used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.
We generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \in (0.0,0.05)*, *A_I, A_D \in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order "T", "C", "A" and "G"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order "a", "b", "c", "d", and "e" for the substitution matrix.
## Example:
The following example correspond for the illustrated MSA in the figure above:
{"MSA": "AAAC-GGG", "unaligned_seqs": {"seq0": "AAG", "seq1": "ACGG"}}
## APA
## BibTeX
|
[
"# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem",
"## Abstract:\nThe sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.\n!image\n\nAn illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences \"AAG\" and \"ACGG\". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which \"AA-G\" is aligned to \"ACGG\". The transformer architecture illustration is adapted from (Vaswani et al., 2017).",
"## Data:\n\nWe used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.\n\nWe generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \\in (0.0,0.05)*, *A_I, A_D \\in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order \"T\", \"C\", \"A\" and \"G\"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order \"a\", \"b\", \"c\", \"d\", and \"e\" for the substitution matrix.",
"## Example:\n\nThe following example correspond for the illustrated MSA in the figure above:\n\n{\"MSA\": \"AAAC-GGG\", \"unaligned_seqs\": {\"seq0\": \"AAG\", \"seq1\": \"ACGG\"}}",
"## APA",
"## BibTeX"
] |
[
"TAGS\n#sequence-to-sequence #bioinformatics #biology #region-us \n",
"# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem",
"## Abstract:\nThe sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.\n!image\n\nAn illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences \"AAG\" and \"ACGG\". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which \"AA-G\" is aligned to \"ACGG\". The transformer architecture illustration is adapted from (Vaswani et al., 2017).",
"## Data:\n\nWe used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.\n\nWe generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \\in (0.0,0.05)*, *A_I, A_D \\in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order \"T\", \"C\", \"A\" and \"G\"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order \"a\", \"b\", \"c\", \"d\", and \"e\" for the substitution matrix.",
"## Example:\n\nThe following example correspond for the illustrated MSA in the figure above:\n\n{\"MSA\": \"AAAC-GGG\", \"unaligned_seqs\": {\"seq0\": \"AAG\", \"seq1\": \"ACGG\"}}",
"## APA",
"## BibTeX"
] |
[
23,
23,
323,
508,
64,
2,
5
] |
[
"passage: TAGS\n#sequence-to-sequence #bioinformatics #biology #region-us \n# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem## Abstract:\nThe sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.\n!image\n\nAn illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences \"AAG\" and \"ACGG\". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which \"AA-G\" is aligned to \"ACGG\". The transformer architecture illustration is adapted from (Vaswani et al., 2017)."
] |
420c626215333ed4ccf7d73f9689c4fd18837d2f
|
# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem
## Abstract:
The sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.

An illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences "AAG" and "ACGG". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which "AA-G" is aligned to "ACGG". The transformer architecture illustration is adapted from (Vaswani et al., 2017).
## Data:
We used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.
We generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \in (0.0,0.05)*, *A_I, A_D \in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order "T", "C", "A" and "G"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order "a", "b", "c", "d", and "e" for the substitution matrix.
## Example:
The following example correspond for the illustrated MSA in the figure above:
{"MSA": "AAAC-GGG", "unaligned_seqs": {"seq0": "AAG", "seq1": "ACGG"}}
## APA
```
Dotan, E., Belinkov, Y., Avram, O., Wygoda, E., Ecker, N., Alburquerque, M., Keren, O., Loewenthal, G., & Pupko T. (2023). Multiple sequence alignment as a sequence-to-sequence learning problem. The Eleventh International Conference on Learning Representations (ICLR 2023).
```
## BibTeX
```
@article{Dotan_multiple_2023,
author = {Dotan, Edo and Belinkov, Yonatan and Avram, Oren and Wygoda, Elya and Ecker, Noa and Alburquerque, Michael and Keren, Omri and Loewenthal, Gil and Pupko, Tal},
month = aug,
title = {{Multiple sequence alignment as a sequence-to-sequence learning problem}},
year = {2023}
}
```
|
dotan1111/MSA-amino-9-seq
|
[
"sequence-to-sequence",
"bioinformatics",
"biology",
"region:us"
] |
2023-09-18T07:51:23+00:00
|
{"tags": ["sequence-to-sequence", "bioinformatics", "biology"]}
|
2023-09-18T10:48:30+00:00
|
[] |
[] |
TAGS
#sequence-to-sequence #bioinformatics #biology #region-us
|
# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem
## Abstract:
The sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.
!image
An illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences "AAG" and "ACGG". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which "AA-G" is aligned to "ACGG". The transformer architecture illustration is adapted from (Vaswani et al., 2017).
## Data:
We used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.
We generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \in (0.0,0.05)*, *A_I, A_D \in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order "T", "C", "A" and "G"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order "a", "b", "c", "d", and "e" for the substitution matrix.
## Example:
The following example correspond for the illustrated MSA in the figure above:
{"MSA": "AAAC-GGG", "unaligned_seqs": {"seq0": "AAG", "seq1": "ACGG"}}
## APA
## BibTeX
|
[
"# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem",
"## Abstract:\nThe sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.\n!image\n\nAn illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences \"AAG\" and \"ACGG\". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which \"AA-G\" is aligned to \"ACGG\". The transformer architecture illustration is adapted from (Vaswani et al., 2017).",
"## Data:\n\nWe used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.\n\nWe generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \\in (0.0,0.05)*, *A_I, A_D \\in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order \"T\", \"C\", \"A\" and \"G\"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order \"a\", \"b\", \"c\", \"d\", and \"e\" for the substitution matrix.",
"## Example:\n\nThe following example correspond for the illustrated MSA in the figure above:\n\n{\"MSA\": \"AAAC-GGG\", \"unaligned_seqs\": {\"seq0\": \"AAG\", \"seq1\": \"ACGG\"}}",
"## APA",
"## BibTeX"
] |
[
"TAGS\n#sequence-to-sequence #bioinformatics #biology #region-us \n",
"# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem",
"## Abstract:\nThe sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.\n!image\n\nAn illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences \"AAG\" and \"ACGG\". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which \"AA-G\" is aligned to \"ACGG\". The transformer architecture illustration is adapted from (Vaswani et al., 2017).",
"## Data:\n\nWe used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.\n\nWe generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \\in (0.0,0.05)*, *A_I, A_D \\in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order \"T\", \"C\", \"A\" and \"G\"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order \"a\", \"b\", \"c\", \"d\", and \"e\" for the substitution matrix.",
"## Example:\n\nThe following example correspond for the illustrated MSA in the figure above:\n\n{\"MSA\": \"AAAC-GGG\", \"unaligned_seqs\": {\"seq0\": \"AAG\", \"seq1\": \"ACGG\"}}",
"## APA",
"## BibTeX"
] |
[
23,
23,
323,
508,
64,
2,
5
] |
[
"passage: TAGS\n#sequence-to-sequence #bioinformatics #biology #region-us \n# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem## Abstract:\nThe sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.\n!image\n\nAn illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences \"AAG\" and \"ACGG\". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which \"AA-G\" is aligned to \"ACGG\". The transformer architecture illustration is adapted from (Vaswani et al., 2017)."
] |
fda797d55c7c506b44152131d812debbaa78192a
|
# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem
## Abstract:
The sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.

An illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences "AAG" and "ACGG". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which "AA-G" is aligned to "ACGG". The transformer architecture illustration is adapted from (Vaswani et al., 2017).
## Data:
We used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.
We generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \in (0.0,0.05)*, *A_I, A_D \in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order "T", "C", "A" and "G"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order "a", "b", "c", "d", and "e" for the substitution matrix.
## Example:
The following example correspond for the illustrated MSA in the figure above:
{"MSA": "AAAC-GGG", "unaligned_seqs": {"seq0": "AAG", "seq1": "ACGG"}}
## APA
```
Dotan, E., Belinkov, Y., Avram, O., Wygoda, E., Ecker, N., Alburquerque, M., Keren, O., Loewenthal, G., & Pupko T. (2023). Multiple sequence alignment as a sequence-to-sequence learning problem. The Eleventh International Conference on Learning Representations (ICLR 2023).
```
## BibTeX
```
@article{Dotan_multiple_2023,
author = {Dotan, Edo and Belinkov, Yonatan and Avram, Oren and Wygoda, Elya and Ecker, Noa and Alburquerque, Michael and Keren, Omri and Loewenthal, Gil and Pupko, Tal},
month = aug,
title = {{Multiple sequence alignment as a sequence-to-sequence learning problem}},
year = {2023}
}
```
|
dotan1111/MSA-amino-10-seq
|
[
"sequence-to-sequence",
"bioinformatics",
"biology",
"region:us"
] |
2023-09-18T07:51:52+00:00
|
{"tags": ["sequence-to-sequence", "bioinformatics", "biology"]}
|
2023-09-18T10:48:44+00:00
|
[] |
[] |
TAGS
#sequence-to-sequence #bioinformatics #biology #region-us
|
# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem
## Abstract:
The sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.
!image
An illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences "AAG" and "ACGG". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which "AA-G" is aligned to "ACGG". The transformer architecture illustration is adapted from (Vaswani et al., 2017).
## Data:
We used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.
We generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \in (0.0,0.05)*, *A_I, A_D \in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order "T", "C", "A" and "G"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order "a", "b", "c", "d", and "e" for the substitution matrix.
## Example:
The following example correspond for the illustrated MSA in the figure above:
{"MSA": "AAAC-GGG", "unaligned_seqs": {"seq0": "AAG", "seq1": "ACGG"}}
## APA
## BibTeX
|
[
"# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem",
"## Abstract:\nThe sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.\n!image\n\nAn illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences \"AAG\" and \"ACGG\". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which \"AA-G\" is aligned to \"ACGG\". The transformer architecture illustration is adapted from (Vaswani et al., 2017).",
"## Data:\n\nWe used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.\n\nWe generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \\in (0.0,0.05)*, *A_I, A_D \\in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order \"T\", \"C\", \"A\" and \"G\"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order \"a\", \"b\", \"c\", \"d\", and \"e\" for the substitution matrix.",
"## Example:\n\nThe following example correspond for the illustrated MSA in the figure above:\n\n{\"MSA\": \"AAAC-GGG\", \"unaligned_seqs\": {\"seq0\": \"AAG\", \"seq1\": \"ACGG\"}}",
"## APA",
"## BibTeX"
] |
[
"TAGS\n#sequence-to-sequence #bioinformatics #biology #region-us \n",
"# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem",
"## Abstract:\nThe sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.\n!image\n\nAn illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences \"AAG\" and \"ACGG\". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which \"AA-G\" is aligned to \"ACGG\". The transformer architecture illustration is adapted from (Vaswani et al., 2017).",
"## Data:\n\nWe used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.\n\nWe generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \\in (0.0,0.05)*, *A_I, A_D \\in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order \"T\", \"C\", \"A\" and \"G\"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order \"a\", \"b\", \"c\", \"d\", and \"e\" for the substitution matrix.",
"## Example:\n\nThe following example correspond for the illustrated MSA in the figure above:\n\n{\"MSA\": \"AAAC-GGG\", \"unaligned_seqs\": {\"seq0\": \"AAG\", \"seq1\": \"ACGG\"}}",
"## APA",
"## BibTeX"
] |
[
23,
23,
323,
508,
64,
2,
5
] |
[
"passage: TAGS\n#sequence-to-sequence #bioinformatics #biology #region-us \n# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem## Abstract:\nThe sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.\n!image\n\nAn illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences \"AAG\" and \"ACGG\". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which \"AA-G\" is aligned to \"ACGG\". The transformer architecture illustration is adapted from (Vaswani et al., 2017)."
] |
e0c05cfcd51f7bdef5f607da4249fb409b431ffb
|
# Dataset of Tsukimi Eiko
This is the dataset of Tsukimi Eiko, containing 299 images and their tags.
Images are crawled from many sites (e.g. danbooru, pixiv, zerochan ...), the auto-crawling system is powered by [DeepGHS Team](https://github.com/deepghs)([huggingface organization](https://huggingface.co/deepghs)).
| Name | Images | Download | Description |
|:------------|---------:|:------------------------------------|:-------------------------------------------------------------------------|
| raw | 299 | [Download](dataset-raw.zip) | Raw data with meta information. |
| raw-stage3 | 719 | [Download](dataset-raw-stage3.zip) | 3-stage cropped raw data with meta information. |
| 384x512 | 299 | [Download](dataset-384x512.zip) | 384x512 aligned dataset. |
| 512x512 | 299 | [Download](dataset-512x512.zip) | 512x512 aligned dataset. |
| 512x704 | 299 | [Download](dataset-512x704.zip) | 512x704 aligned dataset. |
| 640x640 | 299 | [Download](dataset-640x640.zip) | 640x640 aligned dataset. |
| 640x880 | 299 | [Download](dataset-640x880.zip) | 640x880 aligned dataset. |
| stage3-640 | 719 | [Download](dataset-stage3-640.zip) | 3-stage cropped dataset with the shorter side not exceeding 640 pixels. |
| stage3-800 | 719 | [Download](dataset-stage3-800.zip) | 3-stage cropped dataset with the shorter side not exceeding 800 pixels. |
| stage3-1200 | 719 | [Download](dataset-stage3-1200.zip) | 3-stage cropped dataset with the shorter side not exceeding 1200 pixels. |
|
CyberHarem/tsukimi_eiko_paripikoumei
|
[
"task_categories:text-to-image",
"size_categories:n<1K",
"license:mit",
"art",
"not-for-all-audiences",
"region:us"
] |
2023-09-18T08:02:55+00:00
|
{"license": "mit", "size_categories": ["n<1K"], "task_categories": ["text-to-image"], "tags": ["art", "not-for-all-audiences"]}
|
2023-09-18T08:07:03+00:00
|
[] |
[] |
TAGS
#task_categories-text-to-image #size_categories-n<1K #license-mit #art #not-for-all-audiences #region-us
|
Dataset of Tsukimi Eiko
=======================
This is the dataset of Tsukimi Eiko, containing 299 images and their tags.
Images are crawled from many sites (e.g. danbooru, pixiv, zerochan ...), the auto-crawling system is powered by DeepGHS Team(huggingface organization).
|
[] |
[
"TAGS\n#task_categories-text-to-image #size_categories-n<1K #license-mit #art #not-for-all-audiences #region-us \n"
] |
[
44
] |
[
"passage: TAGS\n#task_categories-text-to-image #size_categories-n<1K #license-mit #art #not-for-all-audiences #region-us \n"
] |
2f4ba3bc2aa7ad8b89cf7066472dcb0714bf248c
|
# Dataset Card for "gtzan_all_preprocessed"
[More Information needed](https://github.com/huggingface/datasets/blob/main/CONTRIBUTING.md#how-to-contribute-to-the-dataset-cards)
|
bvallegc/gtzan_all_preprocessed
|
[
"region:us"
] |
2023-09-18T08:05:43+00:00
|
{"configs": [{"config_name": "default", "data_files": [{"split": "train", "path": "data/train-*"}, {"split": "test", "path": "data/test-*"}]}], "dataset_info": {"features": [{"name": "label", "dtype": {"class_label": {"names": {"0": "blues", "1": "classical", "2": "country", "3": "disco", "4": "hiphop", "5": "jazz", "6": "metal", "7": "pop", "8": "reggae", "9": "rock"}}}}, {"name": "input_values", "sequence": "float32"}, {"name": "attention_mask", "sequence": "int32"}], "splits": [{"name": "train", "num_bytes": 3452159816, "num_examples": 899}, {"name": "test", "num_bytes": 384000696, "num_examples": 100}], "download_size": 1923103923, "dataset_size": 3836160512}}
|
2023-09-18T08:09:33+00:00
|
[] |
[] |
TAGS
#region-us
|
# Dataset Card for "gtzan_all_preprocessed"
More Information needed
|
[
"# Dataset Card for \"gtzan_all_preprocessed\"\n\nMore Information needed"
] |
[
"TAGS\n#region-us \n",
"# Dataset Card for \"gtzan_all_preprocessed\"\n\nMore Information needed"
] |
[
6,
18
] |
[
"passage: TAGS\n#region-us \n# Dataset Card for \"gtzan_all_preprocessed\"\n\nMore Information needed"
] |
75875e8fea324396baaec92d9f94c4f8427b2a07
|
Count:
- 2023-09-14 | 66371 | 2.97%
- 2023-09-15 | 595557 | 26.61%
- 2023-09-16 | 618586 | 27.64%
- 2023-09-17 | 566691 | 25.32%
- 2023-09-18 | 390878 | 17.46%
- Total items: 2238083
|
haor/OpenMid-Dataset
|
[
"task_categories:text-to-image",
"size_categories:1M<n<10M",
"language:en",
"license:cc-by-nc-4.0",
"dataset",
"Nijijourney",
"Midjourney",
"doi:10.57967/hf/1123",
"region:us"
] |
2023-09-18T08:07:40+00:00
|
{"language": ["en"], "license": "cc-by-nc-4.0", "size_categories": ["1M<n<10M"], "task_categories": ["text-to-image"], "tags": ["dataset", "Nijijourney", "Midjourney"]}
|
2023-09-18T09:46:50+00:00
|
[] |
[
"en"
] |
TAGS
#task_categories-text-to-image #size_categories-1M<n<10M #language-English #license-cc-by-nc-4.0 #dataset #Nijijourney #Midjourney #doi-10.57967/hf/1123 #region-us
|
Count:
- 2023-09-14 | 66371 | 2.97%
- 2023-09-15 | 595557 | 26.61%
- 2023-09-16 | 618586 | 27.64%
- 2023-09-17 | 566691 | 25.32%
- 2023-09-18 | 390878 | 17.46%
- Total items: 2238083
|
[] |
[
"TAGS\n#task_categories-text-to-image #size_categories-1M<n<10M #language-English #license-cc-by-nc-4.0 #dataset #Nijijourney #Midjourney #doi-10.57967/hf/1123 #region-us \n"
] |
[
70
] |
[
"passage: TAGS\n#task_categories-text-to-image #size_categories-1M<n<10M #language-English #license-cc-by-nc-4.0 #dataset #Nijijourney #Midjourney #doi-10.57967/hf/1123 #region-us \n"
] |
e7875f9ccfb0764ef40aed497a58b30ff4c4e68b
|
# Monolingual Dataset
This a a malnutrition dataset in Kinyarwanda and English, it shall be translated using translators to make it a parallel corpus.
# Source of Data
1. Rwanda Biomedical Center (RBC) (26,390 sentences)
2. GPT-4 prompting (42,576 sentences)
|
DigitalUmuganda/Monolingual_health_dataset
|
[
"size_categories:10K<n<100K",
"language:rw",
"language:en",
"license:cc-by-2.0",
"region:us"
] |
2023-09-18T08:13:20+00:00
|
{"language": ["rw", "en"], "license": "cc-by-2.0", "size_categories": ["10K<n<100K"]}
|
2023-09-18T08:37:07+00:00
|
[] |
[
"rw",
"en"
] |
TAGS
#size_categories-10K<n<100K #language-Kinyarwanda #language-English #license-cc-by-2.0 #region-us
|
# Monolingual Dataset
This a a malnutrition dataset in Kinyarwanda and English, it shall be translated using translators to make it a parallel corpus.
# Source of Data
1. Rwanda Biomedical Center (RBC) (26,390 sentences)
2. GPT-4 prompting (42,576 sentences)
|
[
"# Monolingual Dataset\n\nThis a a malnutrition dataset in Kinyarwanda and English, it shall be translated using translators to make it a parallel corpus.",
"# Source of Data\n1. Rwanda Biomedical Center (RBC) (26,390 sentences)\n2. GPT-4 prompting (42,576 sentences)"
] |
[
"TAGS\n#size_categories-10K<n<100K #language-Kinyarwanda #language-English #license-cc-by-2.0 #region-us \n",
"# Monolingual Dataset\n\nThis a a malnutrition dataset in Kinyarwanda and English, it shall be translated using translators to make it a parallel corpus.",
"# Source of Data\n1. Rwanda Biomedical Center (RBC) (26,390 sentences)\n2. GPT-4 prompting (42,576 sentences)"
] |
[
38,
39,
32
] |
[
"passage: TAGS\n#size_categories-10K<n<100K #language-Kinyarwanda #language-English #license-cc-by-2.0 #region-us \n# Monolingual Dataset\n\nThis a a malnutrition dataset in Kinyarwanda and English, it shall be translated using translators to make it a parallel corpus.# Source of Data\n1. Rwanda Biomedical Center (RBC) (26,390 sentences)\n2. GPT-4 prompting (42,576 sentences)"
] |
8690a3ca2976a61939220b5390c6fc331a954d50
|
# Dataset of High Elf Archer
This is the dataset of High Elf Archer, containing 300 images and their tags.
Images are crawled from many sites (e.g. danbooru, pixiv, zerochan ...), the auto-crawling system is powered by [DeepGHS Team](https://github.com/deepghs)([huggingface organization](https://huggingface.co/deepghs)).
| Name | Images | Download | Description |
|:------------|---------:|:------------------------------------|:-------------------------------------------------------------------------|
| raw | 300 | [Download](dataset-raw.zip) | Raw data with meta information. |
| raw-stage3 | 638 | [Download](dataset-raw-stage3.zip) | 3-stage cropped raw data with meta information. |
| 384x512 | 300 | [Download](dataset-384x512.zip) | 384x512 aligned dataset. |
| 512x512 | 300 | [Download](dataset-512x512.zip) | 512x512 aligned dataset. |
| 512x704 | 300 | [Download](dataset-512x704.zip) | 512x704 aligned dataset. |
| 640x640 | 300 | [Download](dataset-640x640.zip) | 640x640 aligned dataset. |
| 640x880 | 300 | [Download](dataset-640x880.zip) | 640x880 aligned dataset. |
| stage3-640 | 638 | [Download](dataset-stage3-640.zip) | 3-stage cropped dataset with the shorter side not exceeding 640 pixels. |
| stage3-800 | 638 | [Download](dataset-stage3-800.zip) | 3-stage cropped dataset with the shorter side not exceeding 800 pixels. |
| stage3-1200 | 638 | [Download](dataset-stage3-1200.zip) | 3-stage cropped dataset with the shorter side not exceeding 1200 pixels. |
|
CyberHarem/high_elf_archer_goblinslayer
|
[
"task_categories:text-to-image",
"size_categories:n<1K",
"license:mit",
"art",
"not-for-all-audiences",
"region:us"
] |
2023-09-18T08:13:33+00:00
|
{"license": "mit", "size_categories": ["n<1K"], "task_categories": ["text-to-image"], "tags": ["art", "not-for-all-audiences"]}
|
2023-09-18T08:16:15+00:00
|
[] |
[] |
TAGS
#task_categories-text-to-image #size_categories-n<1K #license-mit #art #not-for-all-audiences #region-us
|
Dataset of High Elf Archer
==========================
This is the dataset of High Elf Archer, containing 300 images and their tags.
Images are crawled from many sites (e.g. danbooru, pixiv, zerochan ...), the auto-crawling system is powered by DeepGHS Team(huggingface organization).
|
[] |
[
"TAGS\n#task_categories-text-to-image #size_categories-n<1K #license-mit #art #not-for-all-audiences #region-us \n"
] |
[
44
] |
[
"passage: TAGS\n#task_categories-text-to-image #size_categories-n<1K #license-mit #art #not-for-all-audiences #region-us \n"
] |
a283cef8ff18a75b56195999e83f9aeca06bd169
|
# Dataset of Kuon Nanami
This is the dataset of Kuon Nanami, containing 153 images and their tags.
Images are crawled from many sites (e.g. danbooru, pixiv, zerochan ...), the auto-crawling system is powered by [DeepGHS Team](https://github.com/deepghs)([huggingface organization](https://huggingface.co/deepghs)).
| Name | Images | Download | Description |
|:------------|---------:|:------------------------------------|:-------------------------------------------------------------------------|
| raw | 153 | [Download](dataset-raw.zip) | Raw data with meta information. |
| raw-stage3 | 358 | [Download](dataset-raw-stage3.zip) | 3-stage cropped raw data with meta information. |
| 384x512 | 153 | [Download](dataset-384x512.zip) | 384x512 aligned dataset. |
| 512x512 | 153 | [Download](dataset-512x512.zip) | 512x512 aligned dataset. |
| 512x704 | 153 | [Download](dataset-512x704.zip) | 512x704 aligned dataset. |
| 640x640 | 153 | [Download](dataset-640x640.zip) | 640x640 aligned dataset. |
| 640x880 | 153 | [Download](dataset-640x880.zip) | 640x880 aligned dataset. |
| stage3-640 | 358 | [Download](dataset-stage3-640.zip) | 3-stage cropped dataset with the shorter side not exceeding 640 pixels. |
| stage3-800 | 358 | [Download](dataset-stage3-800.zip) | 3-stage cropped dataset with the shorter side not exceeding 800 pixels. |
| stage3-1200 | 358 | [Download](dataset-stage3-1200.zip) | 3-stage cropped dataset with the shorter side not exceeding 1200 pixels. |
|
CyberHarem/kuon_nanami_paripikoumei
|
[
"task_categories:text-to-image",
"size_categories:n<1K",
"license:mit",
"art",
"not-for-all-audiences",
"region:us"
] |
2023-09-18T08:16:04+00:00
|
{"license": "mit", "size_categories": ["n<1K"], "task_categories": ["text-to-image"], "tags": ["art", "not-for-all-audiences"]}
|
2023-09-18T08:18:13+00:00
|
[] |
[] |
TAGS
#task_categories-text-to-image #size_categories-n<1K #license-mit #art #not-for-all-audiences #region-us
|
Dataset of Kuon Nanami
======================
This is the dataset of Kuon Nanami, containing 153 images and their tags.
Images are crawled from many sites (e.g. danbooru, pixiv, zerochan ...), the auto-crawling system is powered by DeepGHS Team(huggingface organization).
|
[] |
[
"TAGS\n#task_categories-text-to-image #size_categories-n<1K #license-mit #art #not-for-all-audiences #region-us \n"
] |
[
44
] |
[
"passage: TAGS\n#task_categories-text-to-image #size_categories-n<1K #license-mit #art #not-for-all-audiences #region-us \n"
] |
083ef30cfcaa446beb2f9d063544bfe753bb5f5e
|
# Dataset of Cow Girl
This is the dataset of Cow Girl, containing 119 images and their tags.
Images are crawled from many sites (e.g. danbooru, pixiv, zerochan ...), the auto-crawling system is powered by [DeepGHS Team](https://github.com/deepghs)([huggingface organization](https://huggingface.co/deepghs)).
| Name | Images | Download | Description |
|:------------|---------:|:------------------------------------|:-------------------------------------------------------------------------|
| raw | 119 | [Download](dataset-raw.zip) | Raw data with meta information. |
| raw-stage3 | 247 | [Download](dataset-raw-stage3.zip) | 3-stage cropped raw data with meta information. |
| 384x512 | 119 | [Download](dataset-384x512.zip) | 384x512 aligned dataset. |
| 512x512 | 119 | [Download](dataset-512x512.zip) | 512x512 aligned dataset. |
| 512x704 | 119 | [Download](dataset-512x704.zip) | 512x704 aligned dataset. |
| 640x640 | 119 | [Download](dataset-640x640.zip) | 640x640 aligned dataset. |
| 640x880 | 119 | [Download](dataset-640x880.zip) | 640x880 aligned dataset. |
| stage3-640 | 247 | [Download](dataset-stage3-640.zip) | 3-stage cropped dataset with the shorter side not exceeding 640 pixels. |
| stage3-800 | 247 | [Download](dataset-stage3-800.zip) | 3-stage cropped dataset with the shorter side not exceeding 800 pixels. |
| stage3-1200 | 247 | [Download](dataset-stage3-1200.zip) | 3-stage cropped dataset with the shorter side not exceeding 1200 pixels. |
|
CyberHarem/cow_girl_goblinslayer
|
[
"task_categories:text-to-image",
"size_categories:n<1K",
"license:mit",
"art",
"not-for-all-audiences",
"region:us"
] |
2023-09-18T08:24:26+00:00
|
{"license": "mit", "size_categories": ["n<1K"], "task_categories": ["text-to-image"], "tags": ["art", "not-for-all-audiences"]}
|
2023-09-18T08:25:25+00:00
|
[] |
[] |
TAGS
#task_categories-text-to-image #size_categories-n<1K #license-mit #art #not-for-all-audiences #region-us
|
Dataset of Cow Girl
===================
This is the dataset of Cow Girl, containing 119 images and their tags.
Images are crawled from many sites (e.g. danbooru, pixiv, zerochan ...), the auto-crawling system is powered by DeepGHS Team(huggingface organization).
|
[] |
[
"TAGS\n#task_categories-text-to-image #size_categories-n<1K #license-mit #art #not-for-all-audiences #region-us \n"
] |
[
44
] |
[
"passage: TAGS\n#task_categories-text-to-image #size_categories-n<1K #license-mit #art #not-for-all-audiences #region-us \n"
] |
4a415d63c24b391f4725ab348a20647a16f4bf85
|
# Dataset of Guild Girl
This is the dataset of Guild Girl, containing 96 images and their tags.
Images are crawled from many sites (e.g. danbooru, pixiv, zerochan ...), the auto-crawling system is powered by [DeepGHS Team](https://github.com/deepghs)([huggingface organization](https://huggingface.co/deepghs)).
| Name | Images | Download | Description |
|:------------|---------:|:------------------------------------|:-------------------------------------------------------------------------|
| raw | 96 | [Download](dataset-raw.zip) | Raw data with meta information. |
| raw-stage3 | 221 | [Download](dataset-raw-stage3.zip) | 3-stage cropped raw data with meta information. |
| 384x512 | 96 | [Download](dataset-384x512.zip) | 384x512 aligned dataset. |
| 512x512 | 96 | [Download](dataset-512x512.zip) | 512x512 aligned dataset. |
| 512x704 | 96 | [Download](dataset-512x704.zip) | 512x704 aligned dataset. |
| 640x640 | 96 | [Download](dataset-640x640.zip) | 640x640 aligned dataset. |
| 640x880 | 96 | [Download](dataset-640x880.zip) | 640x880 aligned dataset. |
| stage3-640 | 221 | [Download](dataset-stage3-640.zip) | 3-stage cropped dataset with the shorter side not exceeding 640 pixels. |
| stage3-800 | 221 | [Download](dataset-stage3-800.zip) | 3-stage cropped dataset with the shorter side not exceeding 800 pixels. |
| stage3-1200 | 221 | [Download](dataset-stage3-1200.zip) | 3-stage cropped dataset with the shorter side not exceeding 1200 pixels. |
|
CyberHarem/guild_girl_goblinslayer
|
[
"task_categories:text-to-image",
"size_categories:n<1K",
"license:mit",
"art",
"not-for-all-audiences",
"region:us"
] |
2023-09-18T08:31:27+00:00
|
{"license": "mit", "size_categories": ["n<1K"], "task_categories": ["text-to-image"], "tags": ["art", "not-for-all-audiences"]}
|
2023-09-18T08:36:36+00:00
|
[] |
[] |
TAGS
#task_categories-text-to-image #size_categories-n<1K #license-mit #art #not-for-all-audiences #region-us
|
Dataset of Guild Girl
=====================
This is the dataset of Guild Girl, containing 96 images and their tags.
Images are crawled from many sites (e.g. danbooru, pixiv, zerochan ...), the auto-crawling system is powered by DeepGHS Team(huggingface organization).
|
[] |
[
"TAGS\n#task_categories-text-to-image #size_categories-n<1K #license-mit #art #not-for-all-audiences #region-us \n"
] |
[
44
] |
[
"passage: TAGS\n#task_categories-text-to-image #size_categories-n<1K #license-mit #art #not-for-all-audiences #region-us \n"
] |
913bdb5524788ff6f47714e2ffb4bf3f8e9fe9c4
|
# Dataset Card for "chahieugiluon"
[More Information needed](https://github.com/huggingface/datasets/blob/main/CONTRIBUTING.md#how-to-contribute-to-the-dataset-cards)
|
maitrang/chahieugiluon
|
[
"region:us"
] |
2023-09-18T08:47:07+00:00
|
{"configs": [{"config_name": "default", "data_files": [{"split": "train", "path": "data/train-*"}]}], "dataset_info": {"features": [{"name": "id", "dtype": "string"}, {"name": "revid", "dtype": "string"}, {"name": "url", "dtype": "string"}, {"name": "title", "dtype": "string"}, {"name": "text", "dtype": "string"}], "splits": [{"name": "train", "num_bytes": 171514, "num_examples": 9}], "download_size": 88493, "dataset_size": 171514}}
|
2023-09-18T08:48:55+00:00
|
[] |
[] |
TAGS
#region-us
|
# Dataset Card for "chahieugiluon"
More Information needed
|
[
"# Dataset Card for \"chahieugiluon\"\n\nMore Information needed"
] |
[
"TAGS\n#region-us \n",
"# Dataset Card for \"chahieugiluon\"\n\nMore Information needed"
] |
[
6,
16
] |
[
"passage: TAGS\n#region-us \n# Dataset Card for \"chahieugiluon\"\n\nMore Information needed"
] |
e4886ed458e1842f391aca216ede50f3a3d77e35
|
# Dataset Card for "llama2-nuv-repeat-300"
[More Information needed](https://github.com/huggingface/datasets/blob/main/CONTRIBUTING.md#how-to-contribute-to-the-dataset-cards)
|
Luciya/llama2-nuv-repeat-300
|
[
"region:us"
] |
2023-09-18T08:47:26+00:00
|
{"dataset_info": {"features": [{"name": "text", "dtype": "string"}], "splits": [{"name": "train", "num_bytes": 59468, "num_examples": 329}], "download_size": 10788, "dataset_size": 59468}, "configs": [{"config_name": "default", "data_files": [{"split": "train", "path": "data/train-*"}]}]}
|
2023-09-18T08:47:27+00:00
|
[] |
[] |
TAGS
#region-us
|
# Dataset Card for "llama2-nuv-repeat-300"
More Information needed
|
[
"# Dataset Card for \"llama2-nuv-repeat-300\"\n\nMore Information needed"
] |
[
"TAGS\n#region-us \n",
"# Dataset Card for \"llama2-nuv-repeat-300\"\n\nMore Information needed"
] |
[
6,
20
] |
[
"passage: TAGS\n#region-us \n# Dataset Card for \"llama2-nuv-repeat-300\"\n\nMore Information needed"
] |
a594e0a8a04b02abfc2c727eef690632fc85d8d9
|
# GRAPEVISTA - GRAPE Vineyard Imaging and Segmentation Technology Archive
## Description
This dataset contains two collections of high-resolution images captured in various vineyards called **VITIGEOSS** and **WGISD_Extension**. Each image is accompanied by either ground truth annotations or produced segmentation mask, providing valuable data for vineyard-related computer vision and machine learning tasks.
## Dataset Details
- **Source**:
- **VITIGEOSS**: These images were collected by infield cameras installed in 5 different vineyards across Italy, Spain and Portugal.
- **WGISD_Extension**: These images were collected during field visits to vineyards as mentioned in the [original work](https://github.com/thsant/wgisd).
- **Citation**: Please cite the dataset as follows:
``` latex
@inproceedings{blanco23automatic,
title={On the automatic detection and monitoring of Leaves and Grapes using in-field optical cameras},
author={Blanco, Giacomo and Oldani, Federico and Salza, Dario and Rossi, Claudio},
booktitle={2023 IEEE international workshop on metrology for agriculture and forestry (MetroAgriFor)},
year={2023},
organization={IEEE}
}
```
## Dataset Content
- **Number of Images**:
- **VITIGEOSS**: 4545
- **WGISD_Extension**: 8910 training + 1107 validation
- **File Format**: JPEG
- **Ground Truth Annotation Format**: PNG
## Data Fields/Columns
The two collections are provided with the following format:
- **VITIGEOSS**:
- `image_filename`: {CompanyCode}_{VineyardCode}_{CameraCode}_{Variety}_{YYYY-MM-DDTHH:MM:SS}.jpg
- `annotation_filename`: {CompanyCode}_{VineyardCode}_{CameraCode}_{Variety}_{YYYY-MM-DDTHH:MM:SS}.png
- **WGISD_Extension**:
- `image_filename`: {WGISDOriginalName}_{N}.jpg where N is the number of augmentation of the same image
- `annotation_filename`: {WGISDOriginalName}_{N}_labelTrainIds.jpg where N is the number of augmentation of the same image
## Ground Truth Annotation
For both collections semantic segmentation annotations are reported as images where each pixel indicates class among *background, leaves and grapes* for correspondent image
- **WGISD_Extension**: Ground truth annotations are obtained together with augmented images creation
- **VITIGEOSS**: Images are not provided with ground truth annotations but with the semgnation mask produced by the model developed in the aforemention work
## Dataset extraction
In order to extract GRAPEVISTA dataset for archive files, run the following commands
``` shell
cd data
cat grapevista.tar.*.gz.part > grapevista.tar.gz
tar -xvzf grapevista.tar.gz
```
## License Information
This dataset is provided under the CC BY-NC 2.0 license. See the [LICENSE](https://creativecommons.org/licenses/by-nc/2.0/) website for details.
|
links-ads/grapevista-dataset
|
[
"task_categories:image-segmentation",
"size_categories:10K<n<100K",
"license:cc-by-2.0",
"region:us"
] |
2023-09-18T08:53:08+00:00
|
{"license": "cc-by-2.0", "size_categories": ["10K<n<100K"], "task_categories": ["image-segmentation"], "pretty_name": "GRAPEVISTA"}
|
2023-09-18T10:01:07+00:00
|
[] |
[] |
TAGS
#task_categories-image-segmentation #size_categories-10K<n<100K #license-cc-by-2.0 #region-us
|
# GRAPEVISTA - GRAPE Vineyard Imaging and Segmentation Technology Archive
## Description
This dataset contains two collections of high-resolution images captured in various vineyards called VITIGEOSS and WGISD_Extension. Each image is accompanied by either ground truth annotations or produced segmentation mask, providing valuable data for vineyard-related computer vision and machine learning tasks.
## Dataset Details
- Source:
- VITIGEOSS: These images were collected by infield cameras installed in 5 different vineyards across Italy, Spain and Portugal.
- WGISD_Extension: These images were collected during field visits to vineyards as mentioned in the original work.
- Citation: Please cite the dataset as follows:
## Dataset Content
- Number of Images:
- VITIGEOSS: 4545
- WGISD_Extension: 8910 training + 1107 validation
- File Format: JPEG
- Ground Truth Annotation Format: PNG
## Data Fields/Columns
The two collections are provided with the following format:
- VITIGEOSS:
- 'image_filename': {CompanyCode}_{VineyardCode}_{CameraCode}_{Variety}_{YYYY-MM-DDTHH:MM:SS}.jpg
- 'annotation_filename': {CompanyCode}_{VineyardCode}_{CameraCode}_{Variety}_{YYYY-MM-DDTHH:MM:SS}.png
- WGISD_Extension:
- 'image_filename': {WGISDOriginalName}_{N}.jpg where N is the number of augmentation of the same image
- 'annotation_filename': {WGISDOriginalName}_{N}_labelTrainIds.jpg where N is the number of augmentation of the same image
## Ground Truth Annotation
For both collections semantic segmentation annotations are reported as images where each pixel indicates class among *background, leaves and grapes* for correspondent image
- WGISD_Extension: Ground truth annotations are obtained together with augmented images creation
- VITIGEOSS: Images are not provided with ground truth annotations but with the semgnation mask produced by the model developed in the aforemention work
## Dataset extraction
In order to extract GRAPEVISTA dataset for archive files, run the following commands
## License Information
This dataset is provided under the CC BY-NC 2.0 license. See the LICENSE website for details.
|
[
"# GRAPEVISTA - GRAPE Vineyard Imaging and Segmentation Technology Archive",
"## Description\nThis dataset contains two collections of high-resolution images captured in various vineyards called VITIGEOSS and WGISD_Extension. Each image is accompanied by either ground truth annotations or produced segmentation mask, providing valuable data for vineyard-related computer vision and machine learning tasks.",
"## Dataset Details\n- Source: \n - VITIGEOSS: These images were collected by infield cameras installed in 5 different vineyards across Italy, Spain and Portugal.\n - WGISD_Extension: These images were collected during field visits to vineyards as mentioned in the original work.\n- Citation: Please cite the dataset as follows:",
"## Dataset Content\n- Number of Images: \n - VITIGEOSS: 4545\n - WGISD_Extension: 8910 training + 1107 validation\n- File Format: JPEG\n- Ground Truth Annotation Format: PNG",
"## Data Fields/Columns\nThe two collections are provided with the following format:\n- VITIGEOSS:\n - 'image_filename': {CompanyCode}_{VineyardCode}_{CameraCode}_{Variety}_{YYYY-MM-DDTHH:MM:SS}.jpg\n - 'annotation_filename': {CompanyCode}_{VineyardCode}_{CameraCode}_{Variety}_{YYYY-MM-DDTHH:MM:SS}.png\n- WGISD_Extension:\n - 'image_filename': {WGISDOriginalName}_{N}.jpg where N is the number of augmentation of the same image\n - 'annotation_filename': {WGISDOriginalName}_{N}_labelTrainIds.jpg where N is the number of augmentation of the same image",
"## Ground Truth Annotation\nFor both collections semantic segmentation annotations are reported as images where each pixel indicates class among *background, leaves and grapes* for correspondent image\n- WGISD_Extension: Ground truth annotations are obtained together with augmented images creation\n- VITIGEOSS: Images are not provided with ground truth annotations but with the semgnation mask produced by the model developed in the aforemention work",
"## Dataset extraction\nIn order to extract GRAPEVISTA dataset for archive files, run the following commands",
"## License Information\nThis dataset is provided under the CC BY-NC 2.0 license. See the LICENSE website for details."
] |
[
"TAGS\n#task_categories-image-segmentation #size_categories-10K<n<100K #license-cc-by-2.0 #region-us \n",
"# GRAPEVISTA - GRAPE Vineyard Imaging and Segmentation Technology Archive",
"## Description\nThis dataset contains two collections of high-resolution images captured in various vineyards called VITIGEOSS and WGISD_Extension. Each image is accompanied by either ground truth annotations or produced segmentation mask, providing valuable data for vineyard-related computer vision and machine learning tasks.",
"## Dataset Details\n- Source: \n - VITIGEOSS: These images were collected by infield cameras installed in 5 different vineyards across Italy, Spain and Portugal.\n - WGISD_Extension: These images were collected during field visits to vineyards as mentioned in the original work.\n- Citation: Please cite the dataset as follows:",
"## Dataset Content\n- Number of Images: \n - VITIGEOSS: 4545\n - WGISD_Extension: 8910 training + 1107 validation\n- File Format: JPEG\n- Ground Truth Annotation Format: PNG",
"## Data Fields/Columns\nThe two collections are provided with the following format:\n- VITIGEOSS:\n - 'image_filename': {CompanyCode}_{VineyardCode}_{CameraCode}_{Variety}_{YYYY-MM-DDTHH:MM:SS}.jpg\n - 'annotation_filename': {CompanyCode}_{VineyardCode}_{CameraCode}_{Variety}_{YYYY-MM-DDTHH:MM:SS}.png\n- WGISD_Extension:\n - 'image_filename': {WGISDOriginalName}_{N}.jpg where N is the number of augmentation of the same image\n - 'annotation_filename': {WGISDOriginalName}_{N}_labelTrainIds.jpg where N is the number of augmentation of the same image",
"## Ground Truth Annotation\nFor both collections semantic segmentation annotations are reported as images where each pixel indicates class among *background, leaves and grapes* for correspondent image\n- WGISD_Extension: Ground truth annotations are obtained together with augmented images creation\n- VITIGEOSS: Images are not provided with ground truth annotations but with the semgnation mask produced by the model developed in the aforemention work",
"## Dataset extraction\nIn order to extract GRAPEVISTA dataset for archive files, run the following commands",
"## License Information\nThis dataset is provided under the CC BY-NC 2.0 license. See the LICENSE website for details."
] |
[
39,
19,
72,
79,
47,
205,
99,
25,
26
] |
[
"passage: TAGS\n#task_categories-image-segmentation #size_categories-10K<n<100K #license-cc-by-2.0 #region-us \n# GRAPEVISTA - GRAPE Vineyard Imaging and Segmentation Technology Archive## Description\nThis dataset contains two collections of high-resolution images captured in various vineyards called VITIGEOSS and WGISD_Extension. Each image is accompanied by either ground truth annotations or produced segmentation mask, providing valuable data for vineyard-related computer vision and machine learning tasks.## Dataset Details\n- Source: \n - VITIGEOSS: These images were collected by infield cameras installed in 5 different vineyards across Italy, Spain and Portugal.\n - WGISD_Extension: These images were collected during field visits to vineyards as mentioned in the original work.\n- Citation: Please cite the dataset as follows:## Dataset Content\n- Number of Images: \n - VITIGEOSS: 4545\n - WGISD_Extension: 8910 training + 1107 validation\n- File Format: JPEG\n- Ground Truth Annotation Format: PNG## Data Fields/Columns\nThe two collections are provided with the following format:\n- VITIGEOSS:\n - 'image_filename': {CompanyCode}_{VineyardCode}_{CameraCode}_{Variety}_{YYYY-MM-DDTHH:MM:SS}.jpg\n - 'annotation_filename': {CompanyCode}_{VineyardCode}_{CameraCode}_{Variety}_{YYYY-MM-DDTHH:MM:SS}.png\n- WGISD_Extension:\n - 'image_filename': {WGISDOriginalName}_{N}.jpg where N is the number of augmentation of the same image\n - 'annotation_filename': {WGISDOriginalName}_{N}_labelTrainIds.jpg where N is the number of augmentation of the same image"
] |
69330f392ce7f726d5e952f22b0281ce6a1e44d8
|
# Dataset Card for "dataset_pfs_by_arm"
[More Information needed](https://github.com/huggingface/datasets/blob/main/CONTRIBUTING.md#how-to-contribute-to-the-dataset-cards)
|
nzindoc/dataset_pfs_by_arm
|
[
"region:us"
] |
2023-09-18T08:53:47+00:00
|
{"dataset_info": {"features": [{"name": "instruction", "dtype": "string"}, {"name": "response", "dtype": "string"}, {"name": "text", "dtype": "string"}], "splits": [{"name": "train", "num_bytes": 1015503, "num_examples": 1827}], "download_size": 93206, "dataset_size": 1015503}, "configs": [{"config_name": "default", "data_files": [{"split": "train", "path": "data/train-*"}]}]}
|
2023-09-19T00:04:38+00:00
|
[] |
[] |
TAGS
#region-us
|
# Dataset Card for "dataset_pfs_by_arm"
More Information needed
|
[
"# Dataset Card for \"dataset_pfs_by_arm\"\n\nMore Information needed"
] |
[
"TAGS\n#region-us \n",
"# Dataset Card for \"dataset_pfs_by_arm\"\n\nMore Information needed"
] |
[
6,
19
] |
[
"passage: TAGS\n#region-us \n# Dataset Card for \"dataset_pfs_by_arm\"\n\nMore Information needed"
] |
a9f96bec214903d4763ae811b4155637824055b9
|
# Dataset Card for "dataset_pfs_by_arm"
[More Information needed](https://github.com/huggingface/datasets/blob/main/CONTRIBUTING.md#how-to-contribute-to-the-dataset-cards)
|
yicozy/dataset_pfs_by_arm
|
[
"region:us"
] |
2023-09-18T09:11:25+00:00
|
{"dataset_info": {"features": [{"name": "instruction", "dtype": "string"}, {"name": "response", "dtype": "string"}, {"name": "text", "dtype": "string"}], "splits": [{"name": "train", "num_bytes": 1015503, "num_examples": 1827}], "download_size": 0, "dataset_size": 1015503}, "configs": [{"config_name": "default", "data_files": [{"split": "train", "path": "data/train-*"}]}]}
|
2023-09-19T00:51:31+00:00
|
[] |
[] |
TAGS
#region-us
|
# Dataset Card for "dataset_pfs_by_arm"
More Information needed
|
[
"# Dataset Card for \"dataset_pfs_by_arm\"\n\nMore Information needed"
] |
[
"TAGS\n#region-us \n",
"# Dataset Card for \"dataset_pfs_by_arm\"\n\nMore Information needed"
] |
[
6,
19
] |
[
"passage: TAGS\n#region-us \n# Dataset Card for \"dataset_pfs_by_arm\"\n\nMore Information needed"
] |
a3c65a7a9a9ff513e9a875959c5e06ddbbdc3ab6
|
# Bangumi Image Base of Bocchi The Rock!
This is the image base of bangumi Bocchi the Rock!, we detected 23 characters, 2223 images in total. The full dataset is [here](all.zip).
**Please note that these image bases are not guaranteed to be 100% cleaned, they may be noisy actual.** If you intend to manually train models using this dataset, we recommend performing necessary preprocessing on the downloaded dataset to eliminate potential noisy samples (approximately 1% probability).
Here is the characters' preview:
| # | Images | Download | Preview 1 | Preview 2 | Preview 3 | Preview 4 | Preview 5 | Preview 6 | Preview 7 | Preview 8 |
|:------|---------:|:---------------------------|:-------------------------------|:-------------------------------|:-------------------------------|:-------------------------------|:-------------------------------|:-------------------------------|:-------------------------------|:-------------------------------|
| 0 | 538 | [Download](0/dataset.zip) |  |  |  |  |  |  |  |  |
| 1 | 54 | [Download](1/dataset.zip) |  |  |  |  |  |  |  |  |
| 2 | 35 | [Download](2/dataset.zip) |  |  |  |  |  |  |  |  |
| 3 | 13 | [Download](3/dataset.zip) |  |  |  |  |  |  |  |  |
| 4 | 286 | [Download](4/dataset.zip) |  |  |  |  |  |  |  |  |
| 5 | 108 | [Download](5/dataset.zip) |  |  |  |  |  |  |  |  |
| 6 | 8 | [Download](6/dataset.zip) |  |  |  |  |  |  |  |  |
| 7 | 88 | [Download](7/dataset.zip) |  |  |  |  |  |  |  |  |
| 8 | 14 | [Download](8/dataset.zip) |  |  |  |  |  |  |  |  |
| 9 | 439 | [Download](9/dataset.zip) |  |  |  |  |  |  |  |  |
| 10 | 66 | [Download](10/dataset.zip) |  |  |  |  |  |  |  |  |
| 11 | 8 | [Download](11/dataset.zip) |  |  |  |  |  |  |  |  |
| 12 | 257 | [Download](12/dataset.zip) |  |  |  |  |  |  |  |  |
| 13 | 29 | [Download](13/dataset.zip) |  |  |  |  |  |  |  |  |
| 14 | 6 | [Download](14/dataset.zip) |  |  |  |  |  |  | N/A | N/A |
| 15 | 9 | [Download](15/dataset.zip) |  |  |  |  |  |  |  |  |
| 16 | 14 | [Download](16/dataset.zip) |  |  |  |  |  |  |  |  |
| 17 | 6 | [Download](17/dataset.zip) |  |  |  |  |  |  | N/A | N/A |
| 18 | 14 | [Download](18/dataset.zip) |  |  |  |  |  |  |  |  |
| 19 | 12 | [Download](19/dataset.zip) |  |  |  |  |  |  |  |  |
| 20 | 13 | [Download](20/dataset.zip) |  |  |  |  |  |  |  |  |
| 21 | 9 | [Download](21/dataset.zip) |  |  |  |  |  |  |  |  |
| noise | 197 | [Download](-1/dataset.zip) |  |  |  |  |  |  |  |  |
|
BangumiBase/bocchitherock
|
[
"size_categories:1K<n<10K",
"license:mit",
"art",
"region:us"
] |
2023-09-18T09:11:33+00:00
|
{"license": "mit", "size_categories": ["1K<n<10K"], "tags": ["art"]}
|
2023-09-29T07:50:21+00:00
|
[] |
[] |
TAGS
#size_categories-1K<n<10K #license-mit #art #region-us
|
Bangumi Image Base of Bocchi The Rock!
======================================
This is the image base of bangumi Bocchi the Rock!, we detected 23 characters, 2223 images in total. The full dataset is here.
Please note that these image bases are not guaranteed to be 100% cleaned, they may be noisy actual. If you intend to manually train models using this dataset, we recommend performing necessary preprocessing on the downloaded dataset to eliminate potential noisy samples (approximately 1% probability).
Here is the characters' preview:
|
[] |
[
"TAGS\n#size_categories-1K<n<10K #license-mit #art #region-us \n"
] |
[
25
] |
[
"passage: TAGS\n#size_categories-1K<n<10K #license-mit #art #region-us \n"
] |
339041acb23902fc795cc75c9ab813192923f547
|
所有数据都是单轮代码指令数据
325696条英语,42816条中文。
---
license: cc
---
|
nchen909/hugcode-codesft
|
[
"task_categories:text-generation",
"size_categories:100K<n<1M",
"language:code",
"license:openrail",
"code",
"region:us"
] |
2023-09-18T09:46:09+00:00
|
{"language": ["code"], "license": "openrail", "size_categories": ["100K<n<1M"], "task_categories": ["text-generation"], "pretty_name": "CodeSFT-nchen909", "tags": ["code"]}
|
2024-01-29T04:04:26+00:00
|
[] |
[
"code"
] |
TAGS
#task_categories-text-generation #size_categories-100K<n<1M #language-code #license-openrail #code #region-us
|
所有数据都是单轮代码指令数据
325696条英语,42816条中文。
---
license: cc
---
|
[] |
[
"TAGS\n#task_categories-text-generation #size_categories-100K<n<1M #language-code #license-openrail #code #region-us \n"
] |
[
41
] |
[
"passage: TAGS\n#task_categories-text-generation #size_categories-100K<n<1M #language-code #license-openrail #code #region-us \n"
] |
651c52e4dd20b7f29fce3a85696d228b74fbe8b0
|
# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem
## Abstract:
The sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.

An illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences "AAG" and "ACGG". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which "AA-G" is aligned to "ACGG". The transformer architecture illustration is adapted from (Vaswani et al., 2017).
## Data:
We used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.
We generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \in (0.0,0.05)*, *A_I, A_D \in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order "T", "C", "A" and "G"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order "a", "b", "c", "d", and "e" for the substitution matrix.
## Example:
The following example correspond for the illustrated MSA in the figure above:
{"MSA": "AAAC-GGG", "unaligned_seqs": {"seq0": "AAG", "seq1": "ACGG"}}
## APA
```
Dotan, E., Belinkov, Y., Avram, O., Wygoda, E., Ecker, N., Alburquerque, M., Keren, O., Loewenthal, G., & Pupko T. (2023). Multiple sequence alignment as a sequence-to-sequence learning problem. The Eleventh International Conference on Learning Representations (ICLR 2023).
```
## BibTeX
```
@article{Dotan_multiple_2023,
author = {Dotan, Edo and Belinkov, Yonatan and Avram, Oren and Wygoda, Elya and Ecker, Noa and Alburquerque, Michael and Keren, Omri and Loewenthal, Gil and Pupko, Tal},
month = aug,
title = {{Multiple sequence alignment as a sequence-to-sequence learning problem}},
year = {2023}
}
```
|
dotan1111/MSA-amino-8-seq
|
[
"sequence-to-sequence",
"bioinformatics",
"biology",
"region:us"
] |
2023-09-18T10:01:35+00:00
|
{"tags": ["sequence-to-sequence", "bioinformatics", "biology"]}
|
2023-09-18T10:47:59+00:00
|
[] |
[] |
TAGS
#sequence-to-sequence #bioinformatics #biology #region-us
|
# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem
## Abstract:
The sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.
!image
An illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences "AAG" and "ACGG". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which "AA-G" is aligned to "ACGG". The transformer architecture illustration is adapted from (Vaswani et al., 2017).
## Data:
We used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.
We generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \in (0.0,0.05)*, *A_I, A_D \in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order "T", "C", "A" and "G"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order "a", "b", "c", "d", and "e" for the substitution matrix.
## Example:
The following example correspond for the illustrated MSA in the figure above:
{"MSA": "AAAC-GGG", "unaligned_seqs": {"seq0": "AAG", "seq1": "ACGG"}}
## APA
## BibTeX
|
[
"# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem",
"## Abstract:\nThe sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.\n!image\n\nAn illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences \"AAG\" and \"ACGG\". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which \"AA-G\" is aligned to \"ACGG\". The transformer architecture illustration is adapted from (Vaswani et al., 2017).",
"## Data:\n\nWe used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.\n\nWe generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \\in (0.0,0.05)*, *A_I, A_D \\in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order \"T\", \"C\", \"A\" and \"G\"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order \"a\", \"b\", \"c\", \"d\", and \"e\" for the substitution matrix.",
"## Example:\n\nThe following example correspond for the illustrated MSA in the figure above:\n\n{\"MSA\": \"AAAC-GGG\", \"unaligned_seqs\": {\"seq0\": \"AAG\", \"seq1\": \"ACGG\"}}",
"## APA",
"## BibTeX"
] |
[
"TAGS\n#sequence-to-sequence #bioinformatics #biology #region-us \n",
"# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem",
"## Abstract:\nThe sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.\n!image\n\nAn illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences \"AAG\" and \"ACGG\". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which \"AA-G\" is aligned to \"ACGG\". The transformer architecture illustration is adapted from (Vaswani et al., 2017).",
"## Data:\n\nWe used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.\n\nWe generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \\in (0.0,0.05)*, *A_I, A_D \\in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order \"T\", \"C\", \"A\" and \"G\"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order \"a\", \"b\", \"c\", \"d\", and \"e\" for the substitution matrix.",
"## Example:\n\nThe following example correspond for the illustrated MSA in the figure above:\n\n{\"MSA\": \"AAAC-GGG\", \"unaligned_seqs\": {\"seq0\": \"AAG\", \"seq1\": \"ACGG\"}}",
"## APA",
"## BibTeX"
] |
[
23,
23,
323,
508,
64,
2,
5
] |
[
"passage: TAGS\n#sequence-to-sequence #bioinformatics #biology #region-us \n# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem## Abstract:\nThe sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.\n!image\n\nAn illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences \"AAG\" and \"ACGG\". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which \"AA-G\" is aligned to \"ACGG\". The transformer architecture illustration is adapted from (Vaswani et al., 2017)."
] |
bd65c0ee770380dc443ed355eea59b3ed43bb989
|
This dataset clone from amruta.org for training LLM
Contact: [email protected]
By the grace of Our H.H. Shri Mataji Nirmala Devi
|
hungeni/amrutaDB
|
[
"task_categories:text-generation",
"size_categories:1K<n<10K",
"language:en",
"language:vi",
"language:hi",
"license:other",
"region:us"
] |
2023-09-18T10:02:51+00:00
|
{"language": ["en", "vi", "hi"], "license": "other", "size_categories": ["1K<n<10K"], "task_categories": ["text-generation"]}
|
2023-09-18T10:11:08+00:00
|
[] |
[
"en",
"vi",
"hi"
] |
TAGS
#task_categories-text-generation #size_categories-1K<n<10K #language-English #language-Vietnamese #language-Hindi #license-other #region-us
|
This dataset clone from URL for training LLM
Contact: hungbui@URL
By the grace of Our H.H. Shri Mataji Nirmala Devi
|
[] |
[
"TAGS\n#task_categories-text-generation #size_categories-1K<n<10K #language-English #language-Vietnamese #language-Hindi #license-other #region-us \n"
] |
[
49
] |
[
"passage: TAGS\n#task_categories-text-generation #size_categories-1K<n<10K #language-English #language-Vietnamese #language-Hindi #license-other #region-us \n"
] |
0acd8be3fd2b222e4d3f88f4fd370310c2878120
|
# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem
## Abstract:
The sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.

An illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences "AAG" and "ACGG". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which "AA-G" is aligned to "ACGG". The transformer architecture illustration is adapted from (Vaswani et al., 2017).
## Data:
We used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.
We generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \in (0.0,0.05)*, *A_I, A_D \in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order "T", "C", "A" and "G"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order "a", "b", "c", "d", and "e" for the substitution matrix.
## Example:
The following example correspond for the illustrated MSA in the figure above:
{"MSA": "AAAC-GGG", "unaligned_seqs": {"seq0": "AAG", "seq1": "ACGG"}}
## APA
```
Dotan, E., Belinkov, Y., Avram, O., Wygoda, E., Ecker, N., Alburquerque, M., Keren, O., Loewenthal, G., & Pupko T. (2023). Multiple sequence alignment as a sequence-to-sequence learning problem. The Eleventh International Conference on Learning Representations (ICLR 2023).
```
## BibTeX
```
@article{Dotan_multiple_2023,
author = {Dotan, Edo and Belinkov, Yonatan and Avram, Oren and Wygoda, Elya and Ecker, Noa and Alburquerque, Michael and Keren, Omri and Loewenthal, Gil and Pupko, Tal},
month = aug,
title = {{Multiple sequence alignment as a sequence-to-sequence learning problem}},
year = {2023}
}
```
|
dotan1111/MSA-nuc-3-seq
|
[
"sequence-to-sequence",
"bioinformatics",
"biology",
"region:us"
] |
2023-09-18T10:04:23+00:00
|
{"tags": ["sequence-to-sequence", "bioinformatics", "biology"]}
|
2023-09-18T10:49:24+00:00
|
[] |
[] |
TAGS
#sequence-to-sequence #bioinformatics #biology #region-us
|
# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem
## Abstract:
The sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.
!image
An illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences "AAG" and "ACGG". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which "AA-G" is aligned to "ACGG". The transformer architecture illustration is adapted from (Vaswani et al., 2017).
## Data:
We used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.
We generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \in (0.0,0.05)*, *A_I, A_D \in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order "T", "C", "A" and "G"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order "a", "b", "c", "d", and "e" for the substitution matrix.
## Example:
The following example correspond for the illustrated MSA in the figure above:
{"MSA": "AAAC-GGG", "unaligned_seqs": {"seq0": "AAG", "seq1": "ACGG"}}
## APA
## BibTeX
|
[
"# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem",
"## Abstract:\nThe sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.\n!image\n\nAn illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences \"AAG\" and \"ACGG\". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which \"AA-G\" is aligned to \"ACGG\". The transformer architecture illustration is adapted from (Vaswani et al., 2017).",
"## Data:\n\nWe used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.\n\nWe generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \\in (0.0,0.05)*, *A_I, A_D \\in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order \"T\", \"C\", \"A\" and \"G\"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order \"a\", \"b\", \"c\", \"d\", and \"e\" for the substitution matrix.",
"## Example:\n\nThe following example correspond for the illustrated MSA in the figure above:\n\n{\"MSA\": \"AAAC-GGG\", \"unaligned_seqs\": {\"seq0\": \"AAG\", \"seq1\": \"ACGG\"}}",
"## APA",
"## BibTeX"
] |
[
"TAGS\n#sequence-to-sequence #bioinformatics #biology #region-us \n",
"# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem",
"## Abstract:\nThe sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.\n!image\n\nAn illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences \"AAG\" and \"ACGG\". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which \"AA-G\" is aligned to \"ACGG\". The transformer architecture illustration is adapted from (Vaswani et al., 2017).",
"## Data:\n\nWe used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.\n\nWe generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \\in (0.0,0.05)*, *A_I, A_D \\in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order \"T\", \"C\", \"A\" and \"G\"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order \"a\", \"b\", \"c\", \"d\", and \"e\" for the substitution matrix.",
"## Example:\n\nThe following example correspond for the illustrated MSA in the figure above:\n\n{\"MSA\": \"AAAC-GGG\", \"unaligned_seqs\": {\"seq0\": \"AAG\", \"seq1\": \"ACGG\"}}",
"## APA",
"## BibTeX"
] |
[
23,
23,
323,
508,
64,
2,
5
] |
[
"passage: TAGS\n#sequence-to-sequence #bioinformatics #biology #region-us \n# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem## Abstract:\nThe sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.\n!image\n\nAn illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences \"AAG\" and \"ACGG\". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which \"AA-G\" is aligned to \"ACGG\". The transformer architecture illustration is adapted from (Vaswani et al., 2017)."
] |
ffad72d3ad4b3343a1f132c041e2c58432226fee
|
# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem
## Abstract:
The sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.

An illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences "AAG" and "ACGG". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which "AA-G" is aligned to "ACGG". The transformer architecture illustration is adapted from (Vaswani et al., 2017).
## Data:
We used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.
We generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \in (0.0,0.05)*, *A_I, A_D \in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order "T", "C", "A" and "G"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order "a", "b", "c", "d", and "e" for the substitution matrix.
## Example:
The following example correspond for the illustrated MSA in the figure above:
{"MSA": "AAAC-GGG", "unaligned_seqs": {"seq0": "AAG", "seq1": "ACGG"}}
## APA
```
Dotan, E., Belinkov, Y., Avram, O., Wygoda, E., Ecker, N., Alburquerque, M., Keren, O., Loewenthal, G., & Pupko T. (2023). Multiple sequence alignment as a sequence-to-sequence learning problem. The Eleventh International Conference on Learning Representations (ICLR 2023).
```
## BibTeX
```
@article{Dotan_multiple_2023,
author = {Dotan, Edo and Belinkov, Yonatan and Avram, Oren and Wygoda, Elya and Ecker, Noa and Alburquerque, Michael and Keren, Omri and Loewenthal, Gil and Pupko, Tal},
month = aug,
title = {{Multiple sequence alignment as a sequence-to-sequence learning problem}},
year = {2023}
}
```
|
dotan1111/MSA-nuc-2-seq
|
[
"sequence-to-sequence",
"bioinformatics",
"biology",
"region:us"
] |
2023-09-18T10:04:25+00:00
|
{"tags": ["sequence-to-sequence", "bioinformatics", "biology"]}
|
2023-09-18T10:49:02+00:00
|
[] |
[] |
TAGS
#sequence-to-sequence #bioinformatics #biology #region-us
|
# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem
## Abstract:
The sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.
!image
An illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences "AAG" and "ACGG". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which "AA-G" is aligned to "ACGG". The transformer architecture illustration is adapted from (Vaswani et al., 2017).
## Data:
We used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.
We generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \in (0.0,0.05)*, *A_I, A_D \in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order "T", "C", "A" and "G"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order "a", "b", "c", "d", and "e" for the substitution matrix.
## Example:
The following example correspond for the illustrated MSA in the figure above:
{"MSA": "AAAC-GGG", "unaligned_seqs": {"seq0": "AAG", "seq1": "ACGG"}}
## APA
## BibTeX
|
[
"# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem",
"## Abstract:\nThe sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.\n!image\n\nAn illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences \"AAG\" and \"ACGG\". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which \"AA-G\" is aligned to \"ACGG\". The transformer architecture illustration is adapted from (Vaswani et al., 2017).",
"## Data:\n\nWe used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.\n\nWe generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \\in (0.0,0.05)*, *A_I, A_D \\in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order \"T\", \"C\", \"A\" and \"G\"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order \"a\", \"b\", \"c\", \"d\", and \"e\" for the substitution matrix.",
"## Example:\n\nThe following example correspond for the illustrated MSA in the figure above:\n\n{\"MSA\": \"AAAC-GGG\", \"unaligned_seqs\": {\"seq0\": \"AAG\", \"seq1\": \"ACGG\"}}",
"## APA",
"## BibTeX"
] |
[
"TAGS\n#sequence-to-sequence #bioinformatics #biology #region-us \n",
"# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem",
"## Abstract:\nThe sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.\n!image\n\nAn illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences \"AAG\" and \"ACGG\". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which \"AA-G\" is aligned to \"ACGG\". The transformer architecture illustration is adapted from (Vaswani et al., 2017).",
"## Data:\n\nWe used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.\n\nWe generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \\in (0.0,0.05)*, *A_I, A_D \\in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order \"T\", \"C\", \"A\" and \"G\"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order \"a\", \"b\", \"c\", \"d\", and \"e\" for the substitution matrix.",
"## Example:\n\nThe following example correspond for the illustrated MSA in the figure above:\n\n{\"MSA\": \"AAAC-GGG\", \"unaligned_seqs\": {\"seq0\": \"AAG\", \"seq1\": \"ACGG\"}}",
"## APA",
"## BibTeX"
] |
[
23,
23,
323,
508,
64,
2,
5
] |
[
"passage: TAGS\n#sequence-to-sequence #bioinformatics #biology #region-us \n# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem## Abstract:\nThe sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.\n!image\n\nAn illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences \"AAG\" and \"ACGG\". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which \"AA-G\" is aligned to \"ACGG\". The transformer architecture illustration is adapted from (Vaswani et al., 2017)."
] |
3e3412ae08ef71685fda34f5aeb4cfbc3b74b687
|
# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem
## Abstract:
The sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.

An illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences "AAG" and "ACGG". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which "AA-G" is aligned to "ACGG". The transformer architecture illustration is adapted from (Vaswani et al., 2017).
## Data:
We used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.
We generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \in (0.0,0.05)*, *A_I, A_D \in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order "T", "C", "A" and "G"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order "a", "b", "c", "d", and "e" for the substitution matrix.
## Example:
The following example correspond for the illustrated MSA in the figure above:
{"MSA": "AAAC-GGG", "unaligned_seqs": {"seq0": "AAG", "seq1": "ACGG"}}
## APA
```
Dotan, E., Belinkov, Y., Avram, O., Wygoda, E., Ecker, N., Alburquerque, M., Keren, O., Loewenthal, G., & Pupko T. (2023). Multiple sequence alignment as a sequence-to-sequence learning problem. The Eleventh International Conference on Learning Representations (ICLR 2023).
```
## BibTeX
```
@article{Dotan_multiple_2023,
author = {Dotan, Edo and Belinkov, Yonatan and Avram, Oren and Wygoda, Elya and Ecker, Noa and Alburquerque, Michael and Keren, Omri and Loewenthal, Gil and Pupko, Tal},
month = aug,
title = {{Multiple sequence alignment as a sequence-to-sequence learning problem}},
year = {2023}
}
```
|
dotan1111/MSA-nuc-4-seq
|
[
"sequence-to-sequence",
"bioinformatics",
"biology",
"region:us"
] |
2023-09-18T10:04:52+00:00
|
{"tags": ["sequence-to-sequence", "bioinformatics", "biology"]}
|
2023-09-18T10:49:37+00:00
|
[] |
[] |
TAGS
#sequence-to-sequence #bioinformatics #biology #region-us
|
# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem
## Abstract:
The sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.
!image
An illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences "AAG" and "ACGG". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which "AA-G" is aligned to "ACGG". The transformer architecture illustration is adapted from (Vaswani et al., 2017).
## Data:
We used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.
We generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \in (0.0,0.05)*, *A_I, A_D \in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order "T", "C", "A" and "G"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order "a", "b", "c", "d", and "e" for the substitution matrix.
## Example:
The following example correspond for the illustrated MSA in the figure above:
{"MSA": "AAAC-GGG", "unaligned_seqs": {"seq0": "AAG", "seq1": "ACGG"}}
## APA
## BibTeX
|
[
"# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem",
"## Abstract:\nThe sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.\n!image\n\nAn illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences \"AAG\" and \"ACGG\". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which \"AA-G\" is aligned to \"ACGG\". The transformer architecture illustration is adapted from (Vaswani et al., 2017).",
"## Data:\n\nWe used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.\n\nWe generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \\in (0.0,0.05)*, *A_I, A_D \\in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order \"T\", \"C\", \"A\" and \"G\"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order \"a\", \"b\", \"c\", \"d\", and \"e\" for the substitution matrix.",
"## Example:\n\nThe following example correspond for the illustrated MSA in the figure above:\n\n{\"MSA\": \"AAAC-GGG\", \"unaligned_seqs\": {\"seq0\": \"AAG\", \"seq1\": \"ACGG\"}}",
"## APA",
"## BibTeX"
] |
[
"TAGS\n#sequence-to-sequence #bioinformatics #biology #region-us \n",
"# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem",
"## Abstract:\nThe sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.\n!image\n\nAn illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences \"AAG\" and \"ACGG\". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which \"AA-G\" is aligned to \"ACGG\". The transformer architecture illustration is adapted from (Vaswani et al., 2017).",
"## Data:\n\nWe used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.\n\nWe generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \\in (0.0,0.05)*, *A_I, A_D \\in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order \"T\", \"C\", \"A\" and \"G\"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order \"a\", \"b\", \"c\", \"d\", and \"e\" for the substitution matrix.",
"## Example:\n\nThe following example correspond for the illustrated MSA in the figure above:\n\n{\"MSA\": \"AAAC-GGG\", \"unaligned_seqs\": {\"seq0\": \"AAG\", \"seq1\": \"ACGG\"}}",
"## APA",
"## BibTeX"
] |
[
23,
23,
323,
508,
64,
2,
5
] |
[
"passage: TAGS\n#sequence-to-sequence #bioinformatics #biology #region-us \n# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem## Abstract:\nThe sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.\n!image\n\nAn illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences \"AAG\" and \"ACGG\". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which \"AA-G\" is aligned to \"ACGG\". The transformer architecture illustration is adapted from (Vaswani et al., 2017)."
] |
4960274bd8044a34cfe1236478386085138d316c
|
# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem
## Abstract:
The sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.

An illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences "AAG" and "ACGG". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which "AA-G" is aligned to "ACGG". The transformer architecture illustration is adapted from (Vaswani et al., 2017).
## Data:
We used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.
We generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \in (0.0,0.05)*, *A_I, A_D \in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order "T", "C", "A" and "G"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order "a", "b", "c", "d", and "e" for the substitution matrix.
## Example:
The following example correspond for the illustrated MSA in the figure above:
{"MSA": "AAAC-GGG", "unaligned_seqs": {"seq0": "AAG", "seq1": "ACGG"}}
## APA
```
Dotan, E., Belinkov, Y., Avram, O., Wygoda, E., Ecker, N., Alburquerque, M., Keren, O., Loewenthal, G., & Pupko T. (2023). Multiple sequence alignment as a sequence-to-sequence learning problem. The Eleventh International Conference on Learning Representations (ICLR 2023).
```
## BibTeX
```
@article{Dotan_multiple_2023,
author = {Dotan, Edo and Belinkov, Yonatan and Avram, Oren and Wygoda, Elya and Ecker, Noa and Alburquerque, Michael and Keren, Omri and Loewenthal, Gil and Pupko, Tal},
month = aug,
title = {{Multiple sequence alignment as a sequence-to-sequence learning problem}},
year = {2023}
}
```
|
dotan1111/MSA-nuc-5-seq
|
[
"sequence-to-sequence",
"bioinformatics",
"biology",
"region:us"
] |
2023-09-18T10:05:24+00:00
|
{"tags": ["sequence-to-sequence", "bioinformatics", "biology"]}
|
2023-09-18T10:49:47+00:00
|
[] |
[] |
TAGS
#sequence-to-sequence #bioinformatics #biology #region-us
|
# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem
## Abstract:
The sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.
!image
An illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences "AAG" and "ACGG". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which "AA-G" is aligned to "ACGG". The transformer architecture illustration is adapted from (Vaswani et al., 2017).
## Data:
We used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.
We generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \in (0.0,0.05)*, *A_I, A_D \in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order "T", "C", "A" and "G"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order "a", "b", "c", "d", and "e" for the substitution matrix.
## Example:
The following example correspond for the illustrated MSA in the figure above:
{"MSA": "AAAC-GGG", "unaligned_seqs": {"seq0": "AAG", "seq1": "ACGG"}}
## APA
## BibTeX
|
[
"# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem",
"## Abstract:\nThe sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.\n!image\n\nAn illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences \"AAG\" and \"ACGG\". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which \"AA-G\" is aligned to \"ACGG\". The transformer architecture illustration is adapted from (Vaswani et al., 2017).",
"## Data:\n\nWe used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.\n\nWe generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \\in (0.0,0.05)*, *A_I, A_D \\in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order \"T\", \"C\", \"A\" and \"G\"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order \"a\", \"b\", \"c\", \"d\", and \"e\" for the substitution matrix.",
"## Example:\n\nThe following example correspond for the illustrated MSA in the figure above:\n\n{\"MSA\": \"AAAC-GGG\", \"unaligned_seqs\": {\"seq0\": \"AAG\", \"seq1\": \"ACGG\"}}",
"## APA",
"## BibTeX"
] |
[
"TAGS\n#sequence-to-sequence #bioinformatics #biology #region-us \n",
"# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem",
"## Abstract:\nThe sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.\n!image\n\nAn illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences \"AAG\" and \"ACGG\". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which \"AA-G\" is aligned to \"ACGG\". The transformer architecture illustration is adapted from (Vaswani et al., 2017).",
"## Data:\n\nWe used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.\n\nWe generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \\in (0.0,0.05)*, *A_I, A_D \\in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order \"T\", \"C\", \"A\" and \"G\"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order \"a\", \"b\", \"c\", \"d\", and \"e\" for the substitution matrix.",
"## Example:\n\nThe following example correspond for the illustrated MSA in the figure above:\n\n{\"MSA\": \"AAAC-GGG\", \"unaligned_seqs\": {\"seq0\": \"AAG\", \"seq1\": \"ACGG\"}}",
"## APA",
"## BibTeX"
] |
[
23,
23,
323,
508,
64,
2,
5
] |
[
"passage: TAGS\n#sequence-to-sequence #bioinformatics #biology #region-us \n# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem## Abstract:\nThe sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.\n!image\n\nAn illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences \"AAG\" and \"ACGG\". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which \"AA-G\" is aligned to \"ACGG\". The transformer architecture illustration is adapted from (Vaswani et al., 2017)."
] |
2cd2c15d08ffd0e96b6d83ab53222eff417534f8
|
# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem
## Abstract:
The sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.

An illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences "AAG" and "ACGG". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which "AA-G" is aligned to "ACGG". The transformer architecture illustration is adapted from (Vaswani et al., 2017).
## Data:
We used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.
We generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \in (0.0,0.05)*, *A_I, A_D \in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order "T", "C", "A" and "G"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order "a", "b", "c", "d", and "e" for the substitution matrix.
## Example:
The following example correspond for the illustrated MSA in the figure above:
{"MSA": "AAAC-GGG", "unaligned_seqs": {"seq0": "AAG", "seq1": "ACGG"}}
## APA
```
Dotan, E., Belinkov, Y., Avram, O., Wygoda, E., Ecker, N., Alburquerque, M., Keren, O., Loewenthal, G., & Pupko T. (2023). Multiple sequence alignment as a sequence-to-sequence learning problem. The Eleventh International Conference on Learning Representations (ICLR 2023).
```
## BibTeX
```
@article{Dotan_multiple_2023,
author = {Dotan, Edo and Belinkov, Yonatan and Avram, Oren and Wygoda, Elya and Ecker, Noa and Alburquerque, Michael and Keren, Omri and Loewenthal, Gil and Pupko, Tal},
month = aug,
title = {{Multiple sequence alignment as a sequence-to-sequence learning problem}},
year = {2023}
}
```
|
dotan1111/MSA-nuc-6-seq
|
[
"sequence-to-sequence",
"bioinformatics",
"biology",
"region:us"
] |
2023-09-18T10:05:44+00:00
|
{"tags": ["sequence-to-sequence", "bioinformatics", "biology"]}
|
2023-09-18T10:49:55+00:00
|
[] |
[] |
TAGS
#sequence-to-sequence #bioinformatics #biology #region-us
|
# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem
## Abstract:
The sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.
!image
An illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences "AAG" and "ACGG". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which "AA-G" is aligned to "ACGG". The transformer architecture illustration is adapted from (Vaswani et al., 2017).
## Data:
We used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.
We generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \in (0.0,0.05)*, *A_I, A_D \in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order "T", "C", "A" and "G"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order "a", "b", "c", "d", and "e" for the substitution matrix.
## Example:
The following example correspond for the illustrated MSA in the figure above:
{"MSA": "AAAC-GGG", "unaligned_seqs": {"seq0": "AAG", "seq1": "ACGG"}}
## APA
## BibTeX
|
[
"# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem",
"## Abstract:\nThe sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.\n!image\n\nAn illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences \"AAG\" and \"ACGG\". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which \"AA-G\" is aligned to \"ACGG\". The transformer architecture illustration is adapted from (Vaswani et al., 2017).",
"## Data:\n\nWe used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.\n\nWe generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \\in (0.0,0.05)*, *A_I, A_D \\in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order \"T\", \"C\", \"A\" and \"G\"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order \"a\", \"b\", \"c\", \"d\", and \"e\" for the substitution matrix.",
"## Example:\n\nThe following example correspond for the illustrated MSA in the figure above:\n\n{\"MSA\": \"AAAC-GGG\", \"unaligned_seqs\": {\"seq0\": \"AAG\", \"seq1\": \"ACGG\"}}",
"## APA",
"## BibTeX"
] |
[
"TAGS\n#sequence-to-sequence #bioinformatics #biology #region-us \n",
"# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem",
"## Abstract:\nThe sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.\n!image\n\nAn illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences \"AAG\" and \"ACGG\". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which \"AA-G\" is aligned to \"ACGG\". The transformer architecture illustration is adapted from (Vaswani et al., 2017).",
"## Data:\n\nWe used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.\n\nWe generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \\in (0.0,0.05)*, *A_I, A_D \\in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order \"T\", \"C\", \"A\" and \"G\"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order \"a\", \"b\", \"c\", \"d\", and \"e\" for the substitution matrix.",
"## Example:\n\nThe following example correspond for the illustrated MSA in the figure above:\n\n{\"MSA\": \"AAAC-GGG\", \"unaligned_seqs\": {\"seq0\": \"AAG\", \"seq1\": \"ACGG\"}}",
"## APA",
"## BibTeX"
] |
[
23,
23,
323,
508,
64,
2,
5
] |
[
"passage: TAGS\n#sequence-to-sequence #bioinformatics #biology #region-us \n# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem## Abstract:\nThe sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.\n!image\n\nAn illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences \"AAG\" and \"ACGG\". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which \"AA-G\" is aligned to \"ACGG\". The transformer architecture illustration is adapted from (Vaswani et al., 2017)."
] |
de2d05a999292a1578bcaa723b87dd6e1b93ba14
|
# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem
## Abstract:
The sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.

An illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences "AAG" and "ACGG". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which "AA-G" is aligned to "ACGG". The transformer architecture illustration is adapted from (Vaswani et al., 2017).
## Data:
We used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.
We generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \in (0.0,0.05)*, *A_I, A_D \in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order "T", "C", "A" and "G"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order "a", "b", "c", "d", and "e" for the substitution matrix.
## Example:
The following example correspond for the illustrated MSA in the figure above:
{"MSA": "AAAC-GGG", "unaligned_seqs": {"seq0": "AAG", "seq1": "ACGG"}}
## APA
```
Dotan, E., Belinkov, Y., Avram, O., Wygoda, E., Ecker, N., Alburquerque, M., Keren, O., Loewenthal, G., & Pupko T. (2023). Multiple sequence alignment as a sequence-to-sequence learning problem. The Eleventh International Conference on Learning Representations (ICLR 2023).
```
## BibTeX
```
@article{Dotan_multiple_2023,
author = {Dotan, Edo and Belinkov, Yonatan and Avram, Oren and Wygoda, Elya and Ecker, Noa and Alburquerque, Michael and Keren, Omri and Loewenthal, Gil and Pupko, Tal},
month = aug,
title = {{Multiple sequence alignment as a sequence-to-sequence learning problem}},
year = {2023}
}
```
|
dotan1111/MSA-nuc-7-seq
|
[
"sequence-to-sequence",
"bioinformatics",
"biology",
"region:us"
] |
2023-09-18T10:06:17+00:00
|
{"tags": ["sequence-to-sequence", "bioinformatics", "biology"]}
|
2023-09-18T10:50:02+00:00
|
[] |
[] |
TAGS
#sequence-to-sequence #bioinformatics #biology #region-us
|
# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem
## Abstract:
The sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.
!image
An illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences "AAG" and "ACGG". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which "AA-G" is aligned to "ACGG". The transformer architecture illustration is adapted from (Vaswani et al., 2017).
## Data:
We used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.
We generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \in (0.0,0.05)*, *A_I, A_D \in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order "T", "C", "A" and "G"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order "a", "b", "c", "d", and "e" for the substitution matrix.
## Example:
The following example correspond for the illustrated MSA in the figure above:
{"MSA": "AAAC-GGG", "unaligned_seqs": {"seq0": "AAG", "seq1": "ACGG"}}
## APA
## BibTeX
|
[
"# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem",
"## Abstract:\nThe sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.\n!image\n\nAn illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences \"AAG\" and \"ACGG\". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which \"AA-G\" is aligned to \"ACGG\". The transformer architecture illustration is adapted from (Vaswani et al., 2017).",
"## Data:\n\nWe used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.\n\nWe generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \\in (0.0,0.05)*, *A_I, A_D \\in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order \"T\", \"C\", \"A\" and \"G\"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order \"a\", \"b\", \"c\", \"d\", and \"e\" for the substitution matrix.",
"## Example:\n\nThe following example correspond for the illustrated MSA in the figure above:\n\n{\"MSA\": \"AAAC-GGG\", \"unaligned_seqs\": {\"seq0\": \"AAG\", \"seq1\": \"ACGG\"}}",
"## APA",
"## BibTeX"
] |
[
"TAGS\n#sequence-to-sequence #bioinformatics #biology #region-us \n",
"# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem",
"## Abstract:\nThe sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.\n!image\n\nAn illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences \"AAG\" and \"ACGG\". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which \"AA-G\" is aligned to \"ACGG\". The transformer architecture illustration is adapted from (Vaswani et al., 2017).",
"## Data:\n\nWe used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.\n\nWe generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \\in (0.0,0.05)*, *A_I, A_D \\in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order \"T\", \"C\", \"A\" and \"G\"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order \"a\", \"b\", \"c\", \"d\", and \"e\" for the substitution matrix.",
"## Example:\n\nThe following example correspond for the illustrated MSA in the figure above:\n\n{\"MSA\": \"AAAC-GGG\", \"unaligned_seqs\": {\"seq0\": \"AAG\", \"seq1\": \"ACGG\"}}",
"## APA",
"## BibTeX"
] |
[
23,
23,
323,
508,
64,
2,
5
] |
[
"passage: TAGS\n#sequence-to-sequence #bioinformatics #biology #region-us \n# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem## Abstract:\nThe sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.\n!image\n\nAn illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences \"AAG\" and \"ACGG\". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which \"AA-G\" is aligned to \"ACGG\". The transformer architecture illustration is adapted from (Vaswani et al., 2017)."
] |
139dc61980cdcd09fdaece1b791a3126ea156498
|
# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem
## Abstract:
The sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.

An illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences "AAG" and "ACGG". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which "AA-G" is aligned to "ACGG". The transformer architecture illustration is adapted from (Vaswani et al., 2017).
## Data:
We used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.
We generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \in (0.0,0.05)*, *A_I, A_D \in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order "T", "C", "A" and "G"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order "a", "b", "c", "d", and "e" for the substitution matrix.
## Example:
The following example correspond for the illustrated MSA in the figure above:
{"MSA": "AAAC-GGG", "unaligned_seqs": {"seq0": "AAG", "seq1": "ACGG"}}
## APA
```
Dotan, E., Belinkov, Y., Avram, O., Wygoda, E., Ecker, N., Alburquerque, M., Keren, O., Loewenthal, G., & Pupko T. (2023). Multiple sequence alignment as a sequence-to-sequence learning problem. The Eleventh International Conference on Learning Representations (ICLR 2023).
```
## BibTeX
```
@article{Dotan_multiple_2023,
author = {Dotan, Edo and Belinkov, Yonatan and Avram, Oren and Wygoda, Elya and Ecker, Noa and Alburquerque, Michael and Keren, Omri and Loewenthal, Gil and Pupko, Tal},
month = aug,
title = {{Multiple sequence alignment as a sequence-to-sequence learning problem}},
year = {2023}
}
```
|
dotan1111/MSA-nuc-8-seq
|
[
"sequence-to-sequence",
"bioinformatics",
"biology",
"region:us"
] |
2023-09-18T10:06:25+00:00
|
{"tags": ["sequence-to-sequence", "bioinformatics", "biology"]}
|
2023-09-18T10:50:10+00:00
|
[] |
[] |
TAGS
#sequence-to-sequence #bioinformatics #biology #region-us
|
# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem
## Abstract:
The sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.
!image
An illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences "AAG" and "ACGG". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which "AA-G" is aligned to "ACGG". The transformer architecture illustration is adapted from (Vaswani et al., 2017).
## Data:
We used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.
We generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \in (0.0,0.05)*, *A_I, A_D \in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order "T", "C", "A" and "G"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order "a", "b", "c", "d", and "e" for the substitution matrix.
## Example:
The following example correspond for the illustrated MSA in the figure above:
{"MSA": "AAAC-GGG", "unaligned_seqs": {"seq0": "AAG", "seq1": "ACGG"}}
## APA
## BibTeX
|
[
"# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem",
"## Abstract:\nThe sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.\n!image\n\nAn illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences \"AAG\" and \"ACGG\". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which \"AA-G\" is aligned to \"ACGG\". The transformer architecture illustration is adapted from (Vaswani et al., 2017).",
"## Data:\n\nWe used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.\n\nWe generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \\in (0.0,0.05)*, *A_I, A_D \\in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order \"T\", \"C\", \"A\" and \"G\"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order \"a\", \"b\", \"c\", \"d\", and \"e\" for the substitution matrix.",
"## Example:\n\nThe following example correspond for the illustrated MSA in the figure above:\n\n{\"MSA\": \"AAAC-GGG\", \"unaligned_seqs\": {\"seq0\": \"AAG\", \"seq1\": \"ACGG\"}}",
"## APA",
"## BibTeX"
] |
[
"TAGS\n#sequence-to-sequence #bioinformatics #biology #region-us \n",
"# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem",
"## Abstract:\nThe sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.\n!image\n\nAn illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences \"AAG\" and \"ACGG\". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which \"AA-G\" is aligned to \"ACGG\". The transformer architecture illustration is adapted from (Vaswani et al., 2017).",
"## Data:\n\nWe used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.\n\nWe generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \\in (0.0,0.05)*, *A_I, A_D \\in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order \"T\", \"C\", \"A\" and \"G\"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order \"a\", \"b\", \"c\", \"d\", and \"e\" for the substitution matrix.",
"## Example:\n\nThe following example correspond for the illustrated MSA in the figure above:\n\n{\"MSA\": \"AAAC-GGG\", \"unaligned_seqs\": {\"seq0\": \"AAG\", \"seq1\": \"ACGG\"}}",
"## APA",
"## BibTeX"
] |
[
23,
23,
323,
508,
64,
2,
5
] |
[
"passage: TAGS\n#sequence-to-sequence #bioinformatics #biology #region-us \n# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem## Abstract:\nThe sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.\n!image\n\nAn illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences \"AAG\" and \"ACGG\". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which \"AA-G\" is aligned to \"ACGG\". The transformer architecture illustration is adapted from (Vaswani et al., 2017)."
] |
cda0235afd488d77107ce2237cef7f5d6069bb54
|
# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem
## Abstract:
The sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.

An illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences "AAG" and "ACGG". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which "AA-G" is aligned to "ACGG". The transformer architecture illustration is adapted from (Vaswani et al., 2017).
## Data:
We used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.
We generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \in (0.0,0.05)*, *A_I, A_D \in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order "T", "C", "A" and "G"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order "a", "b", "c", "d", and "e" for the substitution matrix.
## Example:
The following example correspond for the illustrated MSA in the figure above:
{"MSA": "AAAC-GGG", "unaligned_seqs": {"seq0": "AAG", "seq1": "ACGG"}}
## APA
```
Dotan, E., Belinkov, Y., Avram, O., Wygoda, E., Ecker, N., Alburquerque, M., Keren, O., Loewenthal, G., & Pupko T. (2023). Multiple sequence alignment as a sequence-to-sequence learning problem. The Eleventh International Conference on Learning Representations (ICLR 2023).
```
## BibTeX
```
@article{Dotan_multiple_2023,
author = {Dotan, Edo and Belinkov, Yonatan and Avram, Oren and Wygoda, Elya and Ecker, Noa and Alburquerque, Michael and Keren, Omri and Loewenthal, Gil and Pupko, Tal},
month = aug,
title = {{Multiple sequence alignment as a sequence-to-sequence learning problem}},
year = {2023}
}
```
|
dotan1111/MSA-nuc-9-seq
|
[
"sequence-to-sequence",
"bioinformatics",
"biology",
"region:us"
] |
2023-09-18T10:08:12+00:00
|
{"tags": ["sequence-to-sequence", "bioinformatics", "biology"]}
|
2023-09-18T10:50:18+00:00
|
[] |
[] |
TAGS
#sequence-to-sequence #bioinformatics #biology #region-us
|
# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem
## Abstract:
The sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.
!image
An illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences "AAG" and "ACGG". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which "AA-G" is aligned to "ACGG". The transformer architecture illustration is adapted from (Vaswani et al., 2017).
## Data:
We used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.
We generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \in (0.0,0.05)*, *A_I, A_D \in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order "T", "C", "A" and "G"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order "a", "b", "c", "d", and "e" for the substitution matrix.
## Example:
The following example correspond for the illustrated MSA in the figure above:
{"MSA": "AAAC-GGG", "unaligned_seqs": {"seq0": "AAG", "seq1": "ACGG"}}
## APA
## BibTeX
|
[
"# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem",
"## Abstract:\nThe sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.\n!image\n\nAn illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences \"AAG\" and \"ACGG\". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which \"AA-G\" is aligned to \"ACGG\". The transformer architecture illustration is adapted from (Vaswani et al., 2017).",
"## Data:\n\nWe used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.\n\nWe generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \\in (0.0,0.05)*, *A_I, A_D \\in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order \"T\", \"C\", \"A\" and \"G\"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order \"a\", \"b\", \"c\", \"d\", and \"e\" for the substitution matrix.",
"## Example:\n\nThe following example correspond for the illustrated MSA in the figure above:\n\n{\"MSA\": \"AAAC-GGG\", \"unaligned_seqs\": {\"seq0\": \"AAG\", \"seq1\": \"ACGG\"}}",
"## APA",
"## BibTeX"
] |
[
"TAGS\n#sequence-to-sequence #bioinformatics #biology #region-us \n",
"# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem",
"## Abstract:\nThe sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.\n!image\n\nAn illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences \"AAG\" and \"ACGG\". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which \"AA-G\" is aligned to \"ACGG\". The transformer architecture illustration is adapted from (Vaswani et al., 2017).",
"## Data:\n\nWe used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.\n\nWe generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \\in (0.0,0.05)*, *A_I, A_D \\in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order \"T\", \"C\", \"A\" and \"G\"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order \"a\", \"b\", \"c\", \"d\", and \"e\" for the substitution matrix.",
"## Example:\n\nThe following example correspond for the illustrated MSA in the figure above:\n\n{\"MSA\": \"AAAC-GGG\", \"unaligned_seqs\": {\"seq0\": \"AAG\", \"seq1\": \"ACGG\"}}",
"## APA",
"## BibTeX"
] |
[
23,
23,
323,
508,
64,
2,
5
] |
[
"passage: TAGS\n#sequence-to-sequence #bioinformatics #biology #region-us \n# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem## Abstract:\nThe sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.\n!image\n\nAn illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences \"AAG\" and \"ACGG\". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which \"AA-G\" is aligned to \"ACGG\". The transformer architecture illustration is adapted from (Vaswani et al., 2017)."
] |
0db7deeef118375fc2a4516ad354e81a6060ad20
|
# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem
## Abstract:
The sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.

An illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences "AAG" and "ACGG". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which "AA-G" is aligned to "ACGG". The transformer architecture illustration is adapted from (Vaswani et al., 2017).
## Data:
We used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.
We generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \in (0.0,0.05)*, *A_I, A_D \in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order "T", "C", "A" and "G"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order "a", "b", "c", "d", and "e" for the substitution matrix.
## Example:
The following example correspond for the illustrated MSA in the figure above:
{"MSA": "AAAC-GGG", "unaligned_seqs": {"seq0": "AAG", "seq1": "ACGG"}}
## APA
```
Dotan, E., Belinkov, Y., Avram, O., Wygoda, E., Ecker, N., Alburquerque, M., Keren, O., Loewenthal, G., & Pupko T. (2023). Multiple sequence alignment as a sequence-to-sequence learning problem. The Eleventh International Conference on Learning Representations (ICLR 2023).
```
## BibTeX
```
@article{Dotan_multiple_2023,
author = {Dotan, Edo and Belinkov, Yonatan and Avram, Oren and Wygoda, Elya and Ecker, Noa and Alburquerque, Michael and Keren, Omri and Loewenthal, Gil and Pupko, Tal},
month = aug,
title = {{Multiple sequence alignment as a sequence-to-sequence learning problem}},
year = {2023}
}
```
|
dotan1111/MSA-nuc-10-seq
|
[
"sequence-to-sequence",
"bioinformatics",
"biology",
"region:us"
] |
2023-09-18T10:08:22+00:00
|
{"tags": ["sequence-to-sequence", "bioinformatics", "biology"]}
|
2023-09-18T10:50:27+00:00
|
[] |
[] |
TAGS
#sequence-to-sequence #bioinformatics #biology #region-us
|
# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem
## Abstract:
The sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.
!image
An illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences "AAG" and "ACGG". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which "AA-G" is aligned to "ACGG". The transformer architecture illustration is adapted from (Vaswani et al., 2017).
## Data:
We used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.
We generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \in (0.0,0.05)*, *A_I, A_D \in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order "T", "C", "A" and "G"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order "a", "b", "c", "d", and "e" for the substitution matrix.
## Example:
The following example correspond for the illustrated MSA in the figure above:
{"MSA": "AAAC-GGG", "unaligned_seqs": {"seq0": "AAG", "seq1": "ACGG"}}
## APA
## BibTeX
|
[
"# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem",
"## Abstract:\nThe sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.\n!image\n\nAn illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences \"AAG\" and \"ACGG\". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which \"AA-G\" is aligned to \"ACGG\". The transformer architecture illustration is adapted from (Vaswani et al., 2017).",
"## Data:\n\nWe used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.\n\nWe generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \\in (0.0,0.05)*, *A_I, A_D \\in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order \"T\", \"C\", \"A\" and \"G\"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order \"a\", \"b\", \"c\", \"d\", and \"e\" for the substitution matrix.",
"## Example:\n\nThe following example correspond for the illustrated MSA in the figure above:\n\n{\"MSA\": \"AAAC-GGG\", \"unaligned_seqs\": {\"seq0\": \"AAG\", \"seq1\": \"ACGG\"}}",
"## APA",
"## BibTeX"
] |
[
"TAGS\n#sequence-to-sequence #bioinformatics #biology #region-us \n",
"# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem",
"## Abstract:\nThe sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.\n!image\n\nAn illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences \"AAG\" and \"ACGG\". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which \"AA-G\" is aligned to \"ACGG\". The transformer architecture illustration is adapted from (Vaswani et al., 2017).",
"## Data:\n\nWe used SpartaABC (Loewenthal et al., 2021) to generate millions of true alignments. SpartaABC requires the following input: (1) a rooted phylogenetic tree, which includes a topology and branch lengths; (2) a substitution model (amino acids or nucleotides); (3) root sequence length; (4) the indel model parameters, which include: insertion rate (*R_I*), deletion rate (*R_D*), a parameter for the insertion Zipfian distribution (*A_I*), and a parameter for the deletion Zipfian distribution (*A_D*). MSAs were simulated along random phylogenetic tree topologies generated using the program ETE version 3.0 (Huerta-Cepas et al., 2016) with default parameters.\n\nWe generated 1,495,000, 2,000 and 3,000, protein MSAs with ten sequences that were used as training validation and testing data, respectively. We generated the same number of DNA MSAs. For each random tree, branch lengths were drawn from a uniform distribution in the range *(0.5,1.0)*. Next, the sequences were generated using SpartaABC with the following parameters: *R_I,R_D \\in (0.0,0.05)*, *A_I, A_D \\in (1.01,2.0)*. The alignment lengths as well as the sequence lengths of the tree leaves vary within and among datasets as they depend on the indel dynamics and the root length. The root length was sampled uniformly in the range *[32,44]*. Unless stated otherwise, all protein datasets were generated with the WAG+G model, and all DNA datasets were generated with the GTR+G model, with the following parameters: (1) frequencies for the different nucleotides *(0.37, 0.166, 0.307, 0.158)*, in the order \"T\", \"C\", \"A\" and \"G\"; (2) with the substitutions rate *(0.444, 0.0843, 0.116, 0.107, 0.00027)*, in the order \"a\", \"b\", \"c\", \"d\", and \"e\" for the substitution matrix.",
"## Example:\n\nThe following example correspond for the illustrated MSA in the figure above:\n\n{\"MSA\": \"AAAC-GGG\", \"unaligned_seqs\": {\"seq0\": \"AAG\", \"seq1\": \"ACGG\"}}",
"## APA",
"## BibTeX"
] |
[
23,
23,
323,
508,
64,
2,
5
] |
[
"passage: TAGS\n#sequence-to-sequence #bioinformatics #biology #region-us \n# Multiple Sequence Alignment as a Sequence-to-Sequence Learning Problem## Abstract:\nThe sequence alignment problem is one of the most fundamental problems in bioinformatics and a plethora of methods were devised to tackle it. Here we introduce BetaAlign, a methodology for aligning sequences using an NLP approach. BetaAlign accounts for the possible variability of the evolutionary process among different datasets by using an ensemble of transformers, each trained on millions of samples generated from a different evolutionary model. Our approach leads to alignment accuracy that is similar and often better than commonly used methods, such as MAFFT, DIALIGN, ClustalW, T-Coffee, PRANK, and MUSCLE.\n!image\n\nAn illustration of aligning sequences with sequence-to-sequence learning. (a) Consider two input sequences \"AAG\" and \"ACGG\". (b) The result of encoding the unaligned sequences into the source language (*Concat* representation). (c) The sentence from the source language is translated to the target language via a transformer model. (d) The translated sentence in the target language (*Spaces* representation). (e) The resulting alignment, decoded from the translated sentence, in which \"AA-G\" is aligned to \"ACGG\". The transformer architecture illustration is adapted from (Vaswani et al., 2017)."
] |
16d2d53d8276c4a6fc89a38757630ff359069ab0
|
# Dataset Card for "dataset"
[More Information needed](https://github.com/huggingface/datasets/blob/main/CONTRIBUTING.md#how-to-contribute-to-the-dataset-cards)
|
ExecrableChromosphere/dataset
|
[
"region:us"
] |
2023-09-18T10:21:10+00:00
|
{"dataset_info": {"features": [{"name": "text", "dtype": "string"}], "splits": [{"name": "train", "num_bytes": 38873, "num_examples": 228}], "download_size": 20752, "dataset_size": 38873}, "configs": [{"config_name": "default", "data_files": [{"split": "train", "path": "data/train-*"}]}]}
|
2023-09-18T10:23:57+00:00
|
[] |
[] |
TAGS
#region-us
|
# Dataset Card for "dataset"
More Information needed
|
[
"# Dataset Card for \"dataset\"\n\nMore Information needed"
] |
[
"TAGS\n#region-us \n",
"# Dataset Card for \"dataset\"\n\nMore Information needed"
] |
[
6,
12
] |
[
"passage: TAGS\n#region-us \n# Dataset Card for \"dataset\"\n\nMore Information needed"
] |
c8b3fb46cd7833811d98dba1b091a5dfaac9bcc7
|
# Dataset Card for "indian_food_images"
[More Information needed](https://github.com/huggingface/datasets/blob/main/CONTRIBUTING.md#how-to-contribute-to-the-dataset-cards)
|
RiazHussain/indian_food_images
|
[
"region:us"
] |
2023-09-18T10:36:58+00:00
|
{"configs": [{"config_name": "default", "data_files": [{"split": "train", "path": "data/train-*"}, {"split": "test", "path": "data/test-*"}]}], "dataset_info": {"features": [{"name": "image", "dtype": "image"}, {"name": "label", "dtype": {"class_label": {"names": {"0": "burger", "1": "butter_naan", "2": "chai", "3": "chapati", "4": "chole_bhature", "5": "dal_makhani", "6": "dhokla", "7": "fried_rice", "8": "idli", "9": "jalebi", "10": "kaathi_rolls", "11": "kadai_paneer", "12": "kulfi", "13": "masala_dosa", "14": "momos", "15": "paani_puri", "16": "pakode", "17": "pav_bhaji", "18": "pizza", "19": "samosa"}}}}], "splits": [{"name": "train", "num_bytes": 1370201244.9594336, "num_examples": 5328}, {"name": "test", "num_bytes": 208936489.3925666, "num_examples": 941}], "download_size": 1601617594, "dataset_size": 1579137734.3520002}}
|
2023-09-19T06:20:57+00:00
|
[] |
[] |
TAGS
#region-us
|
# Dataset Card for "indian_food_images"
More Information needed
|
[
"# Dataset Card for \"indian_food_images\"\n\nMore Information needed"
] |
[
"TAGS\n#region-us \n",
"# Dataset Card for \"indian_food_images\"\n\nMore Information needed"
] |
[
6,
17
] |
[
"passage: TAGS\n#region-us \n# Dataset Card for \"indian_food_images\"\n\nMore Information needed"
] |
9e61280a95c930eb0f72786947e375c1213aac46
|
# Bangumi Image Base of Ren`ai Flops
This is the image base of bangumi Ren`ai Flops, we detected 19 characters, 1980 images in total. The full dataset is [here](all.zip).
**Please note that these image bases are not guaranteed to be 100% cleaned, they may be noisy actual.** If you intend to manually train models using this dataset, we recommend performing necessary preprocessing on the downloaded dataset to eliminate potential noisy samples (approximately 1% probability).
Here is the characters' preview:
| # | Images | Download | Preview 1 | Preview 2 | Preview 3 | Preview 4 | Preview 5 | Preview 6 | Preview 7 | Preview 8 |
|:------|---------:|:---------------------------|:-------------------------------|:-------------------------------|:-------------------------------|:-------------------------------|:-------------------------------|:-------------------------------|:-------------------------------|:-------------------------------|
| 0 | 714 | [Download](0/dataset.zip) |  |  |  |  |  |  |  |  |
| 1 | 182 | [Download](1/dataset.zip) |  |  |  |  |  |  |  |  |
| 2 | 10 | [Download](2/dataset.zip) |  |  |  |  |  |  |  |  |
| 3 | 8 | [Download](3/dataset.zip) |  |  |  |  |  |  |  |  |
| 4 | 13 | [Download](4/dataset.zip) |  |  |  |  |  |  |  |  |
| 5 | 19 | [Download](5/dataset.zip) |  |  |  |  |  |  |  |  |
| 6 | 170 | [Download](6/dataset.zip) |  |  |  |  |  |  |  |  |
| 7 | 95 | [Download](7/dataset.zip) |  |  |  |  |  |  |  |  |
| 8 | 47 | [Download](8/dataset.zip) |  |  |  |  |  |  |  |  |
| 9 | 101 | [Download](9/dataset.zip) |  |  |  |  |  |  |  |  |
| 10 | 197 | [Download](10/dataset.zip) |  |  |  |  |  |  |  |  |
| 11 | 42 | [Download](11/dataset.zip) |  |  |  |  |  |  |  |  |
| 12 | 8 | [Download](12/dataset.zip) |  |  |  |  |  |  |  |  |
| 13 | 74 | [Download](13/dataset.zip) |  |  |  |  |  |  |  |  |
| 14 | 169 | [Download](14/dataset.zip) |  |  |  |  |  |  |  |  |
| 15 | 6 | [Download](15/dataset.zip) |  |  |  |  |  |  | N/A | N/A |
| 16 | 7 | [Download](16/dataset.zip) |  |  |  |  |  |  |  | N/A |
| 17 | 6 | [Download](17/dataset.zip) |  |  |  |  |  |  | N/A | N/A |
| noise | 112 | [Download](-1/dataset.zip) |  |  |  |  |  |  |  |  |
|
BangumiBase/renaiflops
|
[
"size_categories:1K<n<10K",
"license:mit",
"art",
"region:us"
] |
2023-09-18T10:40:24+00:00
|
{"license": "mit", "size_categories": ["1K<n<10K"], "tags": ["art"]}
|
2023-09-29T07:58:27+00:00
|
[] |
[] |
TAGS
#size_categories-1K<n<10K #license-mit #art #region-us
|
Bangumi Image Base of Ren'ai Flops
==================================
This is the image base of bangumi Ren'ai Flops, we detected 19 characters, 1980 images in total. The full dataset is here.
Please note that these image bases are not guaranteed to be 100% cleaned, they may be noisy actual. If you intend to manually train models using this dataset, we recommend performing necessary preprocessing on the downloaded dataset to eliminate potential noisy samples (approximately 1% probability).
Here is the characters' preview:
|
[] |
[
"TAGS\n#size_categories-1K<n<10K #license-mit #art #region-us \n"
] |
[
25
] |
[
"passage: TAGS\n#size_categories-1K<n<10K #license-mit #art #region-us \n"
] |
0c5eab12211ce2b057e39e97df64266e452f5d10
|
# Dataset Card for "CLM_data"
[More Information needed](https://github.com/huggingface/datasets/blob/main/CONTRIBUTING.md#how-to-contribute-to-the-dataset-cards)
|
nc33/CLM_data
|
[
"region:us"
] |
2023-09-18T10:45:44+00:00
|
{"dataset_info": [{"config_name": "default", "features": [{"name": "train", "struct": [{"name": "input", "dtype": "string"}, {"name": "instruction", "dtype": "string"}, {"name": "output", "dtype": "string"}]}], "splits": [{"name": "train", "num_bytes": 438033088, "num_examples": 227703}], "download_size": 117819233, "dataset_size": 438033088}, {"config_name": "train", "features": [{"name": "instruction", "dtype": "string"}, {"name": "input", "dtype": "string"}, {"name": "output", "dtype": "string"}], "splits": [{"name": "train", "num_bytes": 438033088, "num_examples": 227703}], "download_size": 117810940, "dataset_size": 438033088}], "configs": [{"config_name": "default", "data_files": [{"split": "train", "path": "data/train-*"}]}, {"config_name": "train", "data_files": [{"split": "train", "path": "train/train-*"}]}]}
|
2023-09-18T14:31:42+00:00
|
[] |
[] |
TAGS
#region-us
|
# Dataset Card for "CLM_data"
More Information needed
|
[
"# Dataset Card for \"CLM_data\"\n\nMore Information needed"
] |
[
"TAGS\n#region-us \n",
"# Dataset Card for \"CLM_data\"\n\nMore Information needed"
] |
[
6,
14
] |
[
"passage: TAGS\n#region-us \n# Dataset Card for \"CLM_data\"\n\nMore Information needed"
] |
67048bedce1fee9f6bc5dc82ce31dfc106362a56
|
# Dataset of Gotō Hitori
This is the dataset of Gotō Hitori, containing 300 images and their tags.
Images are crawled from many sites (e.g. danbooru, pixiv, zerochan ...), the auto-crawling system is powered by [DeepGHS Team](https://github.com/deepghs)([huggingface organization](https://huggingface.co/deepghs)).
| Name | Images | Download | Description |
|:------------|---------:|:------------------------------------|:-------------------------------------------------------------------------|
| raw | 300 | [Download](dataset-raw.zip) | Raw data with meta information. |
| raw-stage3 | 648 | [Download](dataset-raw-stage3.zip) | 3-stage cropped raw data with meta information. |
| 384x512 | 300 | [Download](dataset-384x512.zip) | 384x512 aligned dataset. |
| 512x512 | 300 | [Download](dataset-512x512.zip) | 512x512 aligned dataset. |
| 512x704 | 300 | [Download](dataset-512x704.zip) | 512x704 aligned dataset. |
| 640x640 | 300 | [Download](dataset-640x640.zip) | 640x640 aligned dataset. |
| 640x880 | 300 | [Download](dataset-640x880.zip) | 640x880 aligned dataset. |
| stage3-640 | 648 | [Download](dataset-stage3-640.zip) | 3-stage cropped dataset with the shorter side not exceeding 640 pixels. |
| stage3-800 | 648 | [Download](dataset-stage3-800.zip) | 3-stage cropped dataset with the shorter side not exceeding 800 pixels. |
| stage3-1200 | 648 | [Download](dataset-stage3-1200.zip) | 3-stage cropped dataset with the shorter side not exceeding 1200 pixels. |
|
CyberHarem/goto_hitori_bocchitherock
|
[
"task_categories:text-to-image",
"size_categories:n<1K",
"license:mit",
"art",
"not-for-all-audiences",
"region:us"
] |
2023-09-18T11:30:35+00:00
|
{"license": "mit", "size_categories": ["n<1K"], "task_categories": ["text-to-image"], "tags": ["art", "not-for-all-audiences"]}
|
2023-09-18T11:35:08+00:00
|
[] |
[] |
TAGS
#task_categories-text-to-image #size_categories-n<1K #license-mit #art #not-for-all-audiences #region-us
|
Dataset of Gotō Hitori
======================
This is the dataset of Gotō Hitori, containing 300 images and their tags.
Images are crawled from many sites (e.g. danbooru, pixiv, zerochan ...), the auto-crawling system is powered by DeepGHS Team(huggingface organization).
|
[] |
[
"TAGS\n#task_categories-text-to-image #size_categories-n<1K #license-mit #art #not-for-all-audiences #region-us \n"
] |
[
44
] |
[
"passage: TAGS\n#task_categories-text-to-image #size_categories-n<1K #license-mit #art #not-for-all-audiences #region-us \n"
] |
077c662c575282aeb21b81d0c9b00625e852fe0f
|
# Dataset Card for "banel_including_pos_training_dataset_90"
[More Information needed](https://github.com/huggingface/datasets/blob/main/CONTRIBUTING.md#how-to-contribute-to-the-dataset-cards)
|
fia24/banel_including_pos_training_dataset_90
|
[
"region:us"
] |
2023-09-18T11:36:44+00:00
|
{"configs": [{"config_name": "default", "data_files": [{"split": "train", "path": "data/train-*"}, {"split": "test", "path": "data/test-*"}]}], "dataset_info": {"features": [{"name": "id", "dtype": "string"}, {"name": "translation", "struct": [{"name": "en", "dtype": "string"}, {"name": "fr", "dtype": "string"}]}], "splits": [{"name": "train", "num_bytes": 1386207, "num_examples": 18105}, {"name": "test", "num_bytes": 155599, "num_examples": 2012}], "download_size": 621202, "dataset_size": 1541806}}
|
2023-09-18T11:36:47+00:00
|
[] |
[] |
TAGS
#region-us
|
# Dataset Card for "banel_including_pos_training_dataset_90"
More Information needed
|
[
"# Dataset Card for \"banel_including_pos_training_dataset_90\"\n\nMore Information needed"
] |
[
"TAGS\n#region-us \n",
"# Dataset Card for \"banel_including_pos_training_dataset_90\"\n\nMore Information needed"
] |
[
6,
23
] |
[
"passage: TAGS\n#region-us \n# Dataset Card for \"banel_including_pos_training_dataset_90\"\n\nMore Information needed"
] |
d9b31bbfaa54dc6cffeea89b690ed4eb9cd93442
|
# Dataset Card for "tedlium-prompted"
[More Information needed](https://github.com/huggingface/datasets/blob/main/CONTRIBUTING.md#how-to-contribute-to-the-dataset-cards)
|
distil-whisper/tedlium-prompted
|
[
"region:us"
] |
2023-09-18T11:41:46+00:00
|
{"dataset_info": {"config_name": "release3", "features": [{"name": "audio", "dtype": {"audio": {"sampling_rate": 16000}}}, {"name": "text", "dtype": "string"}, {"name": "speaker_id", "dtype": "string"}, {"name": "gender", "dtype": {"class_label": {"names": {"0": "unknown", "1": "female", "2": "male"}}}}, {"name": "file", "dtype": "string"}, {"name": "id", "dtype": "string"}, {"name": "whisper_transcript_unprompted", "dtype": "string"}, {"name": "whisper_transcript", "dtype": "string"}], "splits": [{"name": "train", "num_bytes": 52484152554.125, "num_examples": 268263}, {"name": "validation", "num_bytes": 184679438.0, "num_examples": 507}, {"name": "test", "num_bytes": 302513272.625, "num_examples": 1155}], "download_size": 52650349441, "dataset_size": 52971345264.75}, "configs": [{"config_name": "release3", "data_files": [{"split": "train", "path": "release3/train-*"}, {"split": "validation", "path": "release3/validation-*"}, {"split": "test", "path": "release3/test-*"}]}]}
|
2023-09-18T12:21:11+00:00
|
[] |
[] |
TAGS
#region-us
|
# Dataset Card for "tedlium-prompted"
More Information needed
|
[
"# Dataset Card for \"tedlium-prompted\"\n\nMore Information needed"
] |
[
"TAGS\n#region-us \n",
"# Dataset Card for \"tedlium-prompted\"\n\nMore Information needed"
] |
[
6,
16
] |
[
"passage: TAGS\n#region-us \n# Dataset Card for \"tedlium-prompted\"\n\nMore Information needed"
] |
bc7d07f99a548c3c1a11f3c836ea7af34fbf1abd
|
# Dataset Card for "cmv_op_10k"
[More Information needed](https://github.com/huggingface/datasets/blob/main/CONTRIBUTING.md#how-to-contribute-to-the-dataset-cards)
|
nailiamirzakhmedova/cmv_op_10k
|
[
"region:us"
] |
2023-09-18T11:46:07+00:00
|
{"configs": [{"config_name": "default", "data_files": [{"split": "train", "path": "data/train-*"}]}], "dataset_info": {"features": [{"name": "id", "dtype": "string"}, {"name": "title", "dtype": "string"}, {"name": "selftext", "dtype": "string"}, {"name": "text", "dtype": "string"}], "splits": [{"name": "train", "num_bytes": 33252845.121562276, "num_examples": 10000}], "download_size": 19395504, "dataset_size": 33252845.121562276}}
|
2023-09-18T12:39:33+00:00
|
[] |
[] |
TAGS
#region-us
|
# Dataset Card for "cmv_op_10k"
More Information needed
|
[
"# Dataset Card for \"cmv_op_10k\"\n\nMore Information needed"
] |
[
"TAGS\n#region-us \n",
"# Dataset Card for \"cmv_op_10k\"\n\nMore Information needed"
] |
[
6,
17
] |
[
"passage: TAGS\n#region-us \n# Dataset Card for \"cmv_op_10k\"\n\nMore Information needed"
] |
debf34de12ce3077c53f36bc0eb7ef2d84b8b6fe
|
# Dataset Card for Evaluation run of marcchew/LaMini-40k-Platypus2-7B
## Dataset Description
- **Homepage:**
- **Repository:** https://huggingface.co/marcchew/LaMini-40k-Platypus2-7B
- **Paper:**
- **Leaderboard:** https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard
- **Point of Contact:** [email protected]
### Dataset Summary
Dataset automatically created during the evaluation run of model [marcchew/LaMini-40k-Platypus2-7B](https://huggingface.co/marcchew/LaMini-40k-Platypus2-7B) on the [Open LLM Leaderboard](https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard).
The dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.
The dataset has been created from 3 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The "train" split is always pointing to the latest results.
An additional configuration "results" store all the aggregated results of the run (and is used to compute and display the aggregated metrics on the [Open LLM Leaderboard](https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard)).
To load the details from a run, you can for instance do the following:
```python
from datasets import load_dataset
data = load_dataset("open-llm-leaderboard/details_marcchew__LaMini-40k-Platypus2-7B",
"harness_gsm8k_5",
split="train")
```
## Latest results
These are the [latest results from run 2023-12-03T19:17:16.064054](https://huggingface.co/datasets/open-llm-leaderboard/details_marcchew__LaMini-40k-Platypus2-7B/blob/main/results_2023-12-03T19-17-16.064054.json)(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the "latest" split for each eval):
```python
{
"all": {
"acc": 0.0,
"acc_stderr": 0.0
},
"harness|gsm8k|5": {
"acc": 0.0,
"acc_stderr": 0.0
}
}
```
### Supported Tasks and Leaderboards
[More Information Needed]
### Languages
[More Information Needed]
## Dataset Structure
### Data Instances
[More Information Needed]
### Data Fields
[More Information Needed]
### Data Splits
[More Information Needed]
## Dataset Creation
### Curation Rationale
[More Information Needed]
### Source Data
#### Initial Data Collection and Normalization
[More Information Needed]
#### Who are the source language producers?
[More Information Needed]
### Annotations
#### Annotation process
[More Information Needed]
#### Who are the annotators?
[More Information Needed]
### Personal and Sensitive Information
[More Information Needed]
## Considerations for Using the Data
### Social Impact of Dataset
[More Information Needed]
### Discussion of Biases
[More Information Needed]
### Other Known Limitations
[More Information Needed]
## Additional Information
### Dataset Curators
[More Information Needed]
### Licensing Information
[More Information Needed]
### Citation Information
[More Information Needed]
### Contributions
[More Information Needed]
|
open-llm-leaderboard/details_marcchew__LaMini-40k-Platypus2-7B
|
[
"region:us"
] |
2023-09-18T11:51:35+00:00
|
{"pretty_name": "Evaluation run of marcchew/LaMini-40k-Platypus2-7B", "dataset_summary": "Dataset automatically created during the evaluation run of model [marcchew/LaMini-40k-Platypus2-7B](https://huggingface.co/marcchew/LaMini-40k-Platypus2-7B) on the [Open LLM Leaderboard](https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard).\n\nThe dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.\n\nThe dataset has been created from 3 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The \"train\" split is always pointing to the latest results.\n\nAn additional configuration \"results\" store all the aggregated results of the run (and is used to compute and display the aggregated metrics on the [Open LLM Leaderboard](https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard)).\n\nTo load the details from a run, you can for instance do the following:\n```python\nfrom datasets import load_dataset\ndata = load_dataset(\"open-llm-leaderboard/details_marcchew__LaMini-40k-Platypus2-7B\",\n\t\"harness_gsm8k_5\",\n\tsplit=\"train\")\n```\n\n## Latest results\n\nThese are the [latest results from run 2023-12-03T19:17:16.064054](https://huggingface.co/datasets/open-llm-leaderboard/details_marcchew__LaMini-40k-Platypus2-7B/blob/main/results_2023-12-03T19-17-16.064054.json)(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the \"latest\" split for each eval):\n\n```python\n{\n \"all\": {\n \"acc\": 0.0,\n \"acc_stderr\": 0.0\n },\n \"harness|gsm8k|5\": {\n \"acc\": 0.0,\n \"acc_stderr\": 0.0\n }\n}\n```", "repo_url": "https://huggingface.co/marcchew/LaMini-40k-Platypus2-7B", "leaderboard_url": "https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard", "point_of_contact": "[email protected]", "configs": [{"config_name": "harness_arc_challenge_25", "data_files": [{"split": "2023_09_18T12_51_11.107895", "path": ["**/details_harness|arc:challenge|25_2023-09-18T12-51-11.107895.parquet"]}, {"split": "latest", "path": ["**/details_harness|arc:challenge|25_2023-09-18T12-51-11.107895.parquet"]}]}, {"config_name": "harness_drop_3", "data_files": [{"split": "2023_10_28T08_58_28.638753", "path": ["**/details_harness|drop|3_2023-10-28T08-58-28.638753.parquet"]}, {"split": "latest", "path": ["**/details_harness|drop|3_2023-10-28T08-58-28.638753.parquet"]}]}, {"config_name": "harness_gsm8k_5", "data_files": [{"split": "2023_10_28T08_58_28.638753", "path": ["**/details_harness|gsm8k|5_2023-10-28T08-58-28.638753.parquet"]}, {"split": "2023_12_03T19_17_16.064054", "path": ["**/details_harness|gsm8k|5_2023-12-03T19-17-16.064054.parquet"]}, {"split": "latest", "path": ["**/details_harness|gsm8k|5_2023-12-03T19-17-16.064054.parquet"]}]}, {"config_name": "harness_hellaswag_10", "data_files": [{"split": "2023_09_18T12_51_11.107895", "path": ["**/details_harness|hellaswag|10_2023-09-18T12-51-11.107895.parquet"]}, {"split": "latest", "path": ["**/details_harness|hellaswag|10_2023-09-18T12-51-11.107895.parquet"]}]}, {"config_name": "harness_hendrycksTest_5", "data_files": [{"split": "2023_09_18T12_51_11.107895", "path": ["**/details_harness|hendrycksTest-abstract_algebra|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-anatomy|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-astronomy|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-business_ethics|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-clinical_knowledge|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-college_biology|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-college_chemistry|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-college_computer_science|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-college_mathematics|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-college_medicine|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-college_physics|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-computer_security|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-conceptual_physics|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-econometrics|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-electrical_engineering|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-elementary_mathematics|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-formal_logic|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-global_facts|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-high_school_biology|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-high_school_chemistry|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-high_school_computer_science|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-high_school_european_history|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-high_school_geography|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-high_school_government_and_politics|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-high_school_macroeconomics|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-high_school_mathematics|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-high_school_microeconomics|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-high_school_physics|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-high_school_psychology|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-high_school_statistics|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-high_school_us_history|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-high_school_world_history|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-human_aging|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-human_sexuality|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-international_law|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-jurisprudence|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-logical_fallacies|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-machine_learning|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-management|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-marketing|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-medical_genetics|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-miscellaneous|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-moral_disputes|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-moral_scenarios|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-nutrition|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-philosophy|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-prehistory|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-professional_accounting|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-professional_law|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-professional_medicine|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-professional_psychology|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-public_relations|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-security_studies|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-sociology|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-us_foreign_policy|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-virology|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-world_religions|5_2023-09-18T12-51-11.107895.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-abstract_algebra|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-anatomy|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-astronomy|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-business_ethics|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-clinical_knowledge|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-college_biology|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-college_chemistry|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-college_computer_science|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-college_mathematics|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-college_medicine|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-college_physics|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-computer_security|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-conceptual_physics|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-econometrics|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-electrical_engineering|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-elementary_mathematics|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-formal_logic|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-global_facts|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-high_school_biology|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-high_school_chemistry|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-high_school_computer_science|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-high_school_european_history|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-high_school_geography|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-high_school_government_and_politics|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-high_school_macroeconomics|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-high_school_mathematics|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-high_school_microeconomics|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-high_school_physics|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-high_school_psychology|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-high_school_statistics|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-high_school_us_history|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-high_school_world_history|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-human_aging|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-human_sexuality|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-international_law|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-jurisprudence|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-logical_fallacies|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-machine_learning|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-management|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-marketing|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-medical_genetics|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-miscellaneous|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-moral_disputes|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-moral_scenarios|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-nutrition|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-philosophy|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-prehistory|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-professional_accounting|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-professional_law|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-professional_medicine|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-professional_psychology|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-public_relations|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-security_studies|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-sociology|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-us_foreign_policy|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-virology|5_2023-09-18T12-51-11.107895.parquet", "**/details_harness|hendrycksTest-world_religions|5_2023-09-18T12-51-11.107895.parquet"]}]}, {"config_name": "harness_hendrycksTest_abstract_algebra_5", "data_files": [{"split": "2023_09_18T12_51_11.107895", "path": ["**/details_harness|hendrycksTest-abstract_algebra|5_2023-09-18T12-51-11.107895.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-abstract_algebra|5_2023-09-18T12-51-11.107895.parquet"]}]}, {"config_name": "harness_hendrycksTest_anatomy_5", "data_files": [{"split": "2023_09_18T12_51_11.107895", "path": ["**/details_harness|hendrycksTest-anatomy|5_2023-09-18T12-51-11.107895.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-anatomy|5_2023-09-18T12-51-11.107895.parquet"]}]}, {"config_name": "harness_hendrycksTest_astronomy_5", "data_files": [{"split": "2023_09_18T12_51_11.107895", "path": ["**/details_harness|hendrycksTest-astronomy|5_2023-09-18T12-51-11.107895.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-astronomy|5_2023-09-18T12-51-11.107895.parquet"]}]}, {"config_name": "harness_hendrycksTest_business_ethics_5", "data_files": [{"split": "2023_09_18T12_51_11.107895", "path": ["**/details_harness|hendrycksTest-business_ethics|5_2023-09-18T12-51-11.107895.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-business_ethics|5_2023-09-18T12-51-11.107895.parquet"]}]}, {"config_name": "harness_hendrycksTest_clinical_knowledge_5", "data_files": [{"split": "2023_09_18T12_51_11.107895", "path": ["**/details_harness|hendrycksTest-clinical_knowledge|5_2023-09-18T12-51-11.107895.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-clinical_knowledge|5_2023-09-18T12-51-11.107895.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_biology_5", "data_files": [{"split": "2023_09_18T12_51_11.107895", "path": ["**/details_harness|hendrycksTest-college_biology|5_2023-09-18T12-51-11.107895.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_biology|5_2023-09-18T12-51-11.107895.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_chemistry_5", "data_files": [{"split": "2023_09_18T12_51_11.107895", "path": ["**/details_harness|hendrycksTest-college_chemistry|5_2023-09-18T12-51-11.107895.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_chemistry|5_2023-09-18T12-51-11.107895.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_computer_science_5", "data_files": [{"split": "2023_09_18T12_51_11.107895", "path": ["**/details_harness|hendrycksTest-college_computer_science|5_2023-09-18T12-51-11.107895.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_computer_science|5_2023-09-18T12-51-11.107895.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_mathematics_5", "data_files": [{"split": "2023_09_18T12_51_11.107895", "path": ["**/details_harness|hendrycksTest-college_mathematics|5_2023-09-18T12-51-11.107895.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_mathematics|5_2023-09-18T12-51-11.107895.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_medicine_5", "data_files": [{"split": "2023_09_18T12_51_11.107895", "path": ["**/details_harness|hendrycksTest-college_medicine|5_2023-09-18T12-51-11.107895.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_medicine|5_2023-09-18T12-51-11.107895.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_physics_5", "data_files": [{"split": "2023_09_18T12_51_11.107895", "path": ["**/details_harness|hendrycksTest-college_physics|5_2023-09-18T12-51-11.107895.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_physics|5_2023-09-18T12-51-11.107895.parquet"]}]}, {"config_name": "harness_hendrycksTest_computer_security_5", "data_files": [{"split": "2023_09_18T12_51_11.107895", "path": ["**/details_harness|hendrycksTest-computer_security|5_2023-09-18T12-51-11.107895.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-computer_security|5_2023-09-18T12-51-11.107895.parquet"]}]}, {"config_name": "harness_hendrycksTest_conceptual_physics_5", "data_files": [{"split": "2023_09_18T12_51_11.107895", "path": ["**/details_harness|hendrycksTest-conceptual_physics|5_2023-09-18T12-51-11.107895.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-conceptual_physics|5_2023-09-18T12-51-11.107895.parquet"]}]}, {"config_name": "harness_hendrycksTest_econometrics_5", "data_files": [{"split": "2023_09_18T12_51_11.107895", "path": ["**/details_harness|hendrycksTest-econometrics|5_2023-09-18T12-51-11.107895.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-econometrics|5_2023-09-18T12-51-11.107895.parquet"]}]}, {"config_name": "harness_hendrycksTest_electrical_engineering_5", "data_files": [{"split": "2023_09_18T12_51_11.107895", "path": ["**/details_harness|hendrycksTest-electrical_engineering|5_2023-09-18T12-51-11.107895.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-electrical_engineering|5_2023-09-18T12-51-11.107895.parquet"]}]}, {"config_name": "harness_hendrycksTest_elementary_mathematics_5", "data_files": [{"split": "2023_09_18T12_51_11.107895", "path": ["**/details_harness|hendrycksTest-elementary_mathematics|5_2023-09-18T12-51-11.107895.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-elementary_mathematics|5_2023-09-18T12-51-11.107895.parquet"]}]}, {"config_name": "harness_hendrycksTest_formal_logic_5", "data_files": [{"split": "2023_09_18T12_51_11.107895", "path": ["**/details_harness|hendrycksTest-formal_logic|5_2023-09-18T12-51-11.107895.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-formal_logic|5_2023-09-18T12-51-11.107895.parquet"]}]}, {"config_name": "harness_hendrycksTest_global_facts_5", "data_files": [{"split": "2023_09_18T12_51_11.107895", "path": ["**/details_harness|hendrycksTest-global_facts|5_2023-09-18T12-51-11.107895.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-global_facts|5_2023-09-18T12-51-11.107895.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_biology_5", "data_files": [{"split": "2023_09_18T12_51_11.107895", "path": ["**/details_harness|hendrycksTest-high_school_biology|5_2023-09-18T12-51-11.107895.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_biology|5_2023-09-18T12-51-11.107895.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_chemistry_5", "data_files": [{"split": "2023_09_18T12_51_11.107895", "path": ["**/details_harness|hendrycksTest-high_school_chemistry|5_2023-09-18T12-51-11.107895.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_chemistry|5_2023-09-18T12-51-11.107895.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_computer_science_5", "data_files": [{"split": "2023_09_18T12_51_11.107895", "path": ["**/details_harness|hendrycksTest-high_school_computer_science|5_2023-09-18T12-51-11.107895.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_computer_science|5_2023-09-18T12-51-11.107895.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_european_history_5", "data_files": [{"split": "2023_09_18T12_51_11.107895", "path": ["**/details_harness|hendrycksTest-high_school_european_history|5_2023-09-18T12-51-11.107895.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_european_history|5_2023-09-18T12-51-11.107895.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_geography_5", "data_files": [{"split": "2023_09_18T12_51_11.107895", "path": ["**/details_harness|hendrycksTest-high_school_geography|5_2023-09-18T12-51-11.107895.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_geography|5_2023-09-18T12-51-11.107895.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_government_and_politics_5", "data_files": [{"split": "2023_09_18T12_51_11.107895", "path": ["**/details_harness|hendrycksTest-high_school_government_and_politics|5_2023-09-18T12-51-11.107895.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_government_and_politics|5_2023-09-18T12-51-11.107895.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_macroeconomics_5", "data_files": [{"split": "2023_09_18T12_51_11.107895", "path": ["**/details_harness|hendrycksTest-high_school_macroeconomics|5_2023-09-18T12-51-11.107895.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_macroeconomics|5_2023-09-18T12-51-11.107895.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_mathematics_5", "data_files": [{"split": "2023_09_18T12_51_11.107895", "path": ["**/details_harness|hendrycksTest-high_school_mathematics|5_2023-09-18T12-51-11.107895.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_mathematics|5_2023-09-18T12-51-11.107895.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_microeconomics_5", "data_files": [{"split": "2023_09_18T12_51_11.107895", "path": ["**/details_harness|hendrycksTest-high_school_microeconomics|5_2023-09-18T12-51-11.107895.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_microeconomics|5_2023-09-18T12-51-11.107895.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_physics_5", "data_files": [{"split": "2023_09_18T12_51_11.107895", "path": ["**/details_harness|hendrycksTest-high_school_physics|5_2023-09-18T12-51-11.107895.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_physics|5_2023-09-18T12-51-11.107895.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_psychology_5", "data_files": [{"split": "2023_09_18T12_51_11.107895", "path": ["**/details_harness|hendrycksTest-high_school_psychology|5_2023-09-18T12-51-11.107895.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_psychology|5_2023-09-18T12-51-11.107895.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_statistics_5", "data_files": [{"split": "2023_09_18T12_51_11.107895", "path": ["**/details_harness|hendrycksTest-high_school_statistics|5_2023-09-18T12-51-11.107895.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_statistics|5_2023-09-18T12-51-11.107895.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_us_history_5", "data_files": [{"split": "2023_09_18T12_51_11.107895", "path": ["**/details_harness|hendrycksTest-high_school_us_history|5_2023-09-18T12-51-11.107895.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_us_history|5_2023-09-18T12-51-11.107895.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_world_history_5", "data_files": [{"split": "2023_09_18T12_51_11.107895", "path": ["**/details_harness|hendrycksTest-high_school_world_history|5_2023-09-18T12-51-11.107895.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_world_history|5_2023-09-18T12-51-11.107895.parquet"]}]}, {"config_name": "harness_hendrycksTest_human_aging_5", "data_files": [{"split": "2023_09_18T12_51_11.107895", "path": ["**/details_harness|hendrycksTest-human_aging|5_2023-09-18T12-51-11.107895.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-human_aging|5_2023-09-18T12-51-11.107895.parquet"]}]}, {"config_name": "harness_hendrycksTest_human_sexuality_5", "data_files": [{"split": "2023_09_18T12_51_11.107895", "path": ["**/details_harness|hendrycksTest-human_sexuality|5_2023-09-18T12-51-11.107895.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-human_sexuality|5_2023-09-18T12-51-11.107895.parquet"]}]}, {"config_name": "harness_hendrycksTest_international_law_5", "data_files": [{"split": "2023_09_18T12_51_11.107895", "path": ["**/details_harness|hendrycksTest-international_law|5_2023-09-18T12-51-11.107895.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-international_law|5_2023-09-18T12-51-11.107895.parquet"]}]}, {"config_name": "harness_hendrycksTest_jurisprudence_5", "data_files": [{"split": "2023_09_18T12_51_11.107895", "path": ["**/details_harness|hendrycksTest-jurisprudence|5_2023-09-18T12-51-11.107895.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-jurisprudence|5_2023-09-18T12-51-11.107895.parquet"]}]}, {"config_name": "harness_hendrycksTest_logical_fallacies_5", "data_files": [{"split": "2023_09_18T12_51_11.107895", "path": ["**/details_harness|hendrycksTest-logical_fallacies|5_2023-09-18T12-51-11.107895.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-logical_fallacies|5_2023-09-18T12-51-11.107895.parquet"]}]}, {"config_name": "harness_hendrycksTest_machine_learning_5", "data_files": [{"split": "2023_09_18T12_51_11.107895", "path": ["**/details_harness|hendrycksTest-machine_learning|5_2023-09-18T12-51-11.107895.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-machine_learning|5_2023-09-18T12-51-11.107895.parquet"]}]}, {"config_name": "harness_hendrycksTest_management_5", "data_files": [{"split": "2023_09_18T12_51_11.107895", "path": ["**/details_harness|hendrycksTest-management|5_2023-09-18T12-51-11.107895.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-management|5_2023-09-18T12-51-11.107895.parquet"]}]}, {"config_name": "harness_hendrycksTest_marketing_5", "data_files": [{"split": "2023_09_18T12_51_11.107895", "path": ["**/details_harness|hendrycksTest-marketing|5_2023-09-18T12-51-11.107895.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-marketing|5_2023-09-18T12-51-11.107895.parquet"]}]}, {"config_name": "harness_hendrycksTest_medical_genetics_5", "data_files": [{"split": "2023_09_18T12_51_11.107895", "path": ["**/details_harness|hendrycksTest-medical_genetics|5_2023-09-18T12-51-11.107895.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-medical_genetics|5_2023-09-18T12-51-11.107895.parquet"]}]}, {"config_name": "harness_hendrycksTest_miscellaneous_5", "data_files": [{"split": "2023_09_18T12_51_11.107895", "path": ["**/details_harness|hendrycksTest-miscellaneous|5_2023-09-18T12-51-11.107895.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-miscellaneous|5_2023-09-18T12-51-11.107895.parquet"]}]}, {"config_name": "harness_hendrycksTest_moral_disputes_5", "data_files": [{"split": "2023_09_18T12_51_11.107895", "path": ["**/details_harness|hendrycksTest-moral_disputes|5_2023-09-18T12-51-11.107895.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-moral_disputes|5_2023-09-18T12-51-11.107895.parquet"]}]}, {"config_name": "harness_hendrycksTest_moral_scenarios_5", "data_files": [{"split": "2023_09_18T12_51_11.107895", "path": ["**/details_harness|hendrycksTest-moral_scenarios|5_2023-09-18T12-51-11.107895.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-moral_scenarios|5_2023-09-18T12-51-11.107895.parquet"]}]}, {"config_name": "harness_hendrycksTest_nutrition_5", "data_files": [{"split": "2023_09_18T12_51_11.107895", "path": ["**/details_harness|hendrycksTest-nutrition|5_2023-09-18T12-51-11.107895.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-nutrition|5_2023-09-18T12-51-11.107895.parquet"]}]}, {"config_name": "harness_hendrycksTest_philosophy_5", "data_files": [{"split": "2023_09_18T12_51_11.107895", "path": ["**/details_harness|hendrycksTest-philosophy|5_2023-09-18T12-51-11.107895.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-philosophy|5_2023-09-18T12-51-11.107895.parquet"]}]}, {"config_name": "harness_hendrycksTest_prehistory_5", "data_files": [{"split": "2023_09_18T12_51_11.107895", "path": ["**/details_harness|hendrycksTest-prehistory|5_2023-09-18T12-51-11.107895.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-prehistory|5_2023-09-18T12-51-11.107895.parquet"]}]}, {"config_name": "harness_hendrycksTest_professional_accounting_5", "data_files": [{"split": "2023_09_18T12_51_11.107895", "path": ["**/details_harness|hendrycksTest-professional_accounting|5_2023-09-18T12-51-11.107895.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-professional_accounting|5_2023-09-18T12-51-11.107895.parquet"]}]}, {"config_name": "harness_hendrycksTest_professional_law_5", "data_files": [{"split": "2023_09_18T12_51_11.107895", "path": ["**/details_harness|hendrycksTest-professional_law|5_2023-09-18T12-51-11.107895.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-professional_law|5_2023-09-18T12-51-11.107895.parquet"]}]}, {"config_name": "harness_hendrycksTest_professional_medicine_5", "data_files": [{"split": "2023_09_18T12_51_11.107895", "path": ["**/details_harness|hendrycksTest-professional_medicine|5_2023-09-18T12-51-11.107895.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-professional_medicine|5_2023-09-18T12-51-11.107895.parquet"]}]}, {"config_name": "harness_hendrycksTest_professional_psychology_5", "data_files": [{"split": "2023_09_18T12_51_11.107895", "path": ["**/details_harness|hendrycksTest-professional_psychology|5_2023-09-18T12-51-11.107895.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-professional_psychology|5_2023-09-18T12-51-11.107895.parquet"]}]}, {"config_name": "harness_hendrycksTest_public_relations_5", "data_files": [{"split": "2023_09_18T12_51_11.107895", "path": ["**/details_harness|hendrycksTest-public_relations|5_2023-09-18T12-51-11.107895.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-public_relations|5_2023-09-18T12-51-11.107895.parquet"]}]}, {"config_name": "harness_hendrycksTest_security_studies_5", "data_files": [{"split": "2023_09_18T12_51_11.107895", "path": ["**/details_harness|hendrycksTest-security_studies|5_2023-09-18T12-51-11.107895.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-security_studies|5_2023-09-18T12-51-11.107895.parquet"]}]}, {"config_name": "harness_hendrycksTest_sociology_5", "data_files": [{"split": "2023_09_18T12_51_11.107895", "path": ["**/details_harness|hendrycksTest-sociology|5_2023-09-18T12-51-11.107895.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-sociology|5_2023-09-18T12-51-11.107895.parquet"]}]}, {"config_name": "harness_hendrycksTest_us_foreign_policy_5", "data_files": [{"split": "2023_09_18T12_51_11.107895", "path": ["**/details_harness|hendrycksTest-us_foreign_policy|5_2023-09-18T12-51-11.107895.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-us_foreign_policy|5_2023-09-18T12-51-11.107895.parquet"]}]}, {"config_name": "harness_hendrycksTest_virology_5", "data_files": [{"split": "2023_09_18T12_51_11.107895", "path": ["**/details_harness|hendrycksTest-virology|5_2023-09-18T12-51-11.107895.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-virology|5_2023-09-18T12-51-11.107895.parquet"]}]}, {"config_name": "harness_hendrycksTest_world_religions_5", "data_files": [{"split": "2023_09_18T12_51_11.107895", "path": ["**/details_harness|hendrycksTest-world_religions|5_2023-09-18T12-51-11.107895.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-world_religions|5_2023-09-18T12-51-11.107895.parquet"]}]}, {"config_name": "harness_truthfulqa_mc_0", "data_files": [{"split": "2023_09_18T12_51_11.107895", "path": ["**/details_harness|truthfulqa:mc|0_2023-09-18T12-51-11.107895.parquet"]}, {"split": "latest", "path": ["**/details_harness|truthfulqa:mc|0_2023-09-18T12-51-11.107895.parquet"]}]}, {"config_name": "harness_winogrande_5", "data_files": [{"split": "2023_10_28T08_58_28.638753", "path": ["**/details_harness|winogrande|5_2023-10-28T08-58-28.638753.parquet"]}, {"split": "latest", "path": ["**/details_harness|winogrande|5_2023-10-28T08-58-28.638753.parquet"]}]}, {"config_name": "results", "data_files": [{"split": "2023_09_18T12_51_11.107895", "path": ["results_2023-09-18T12-51-11.107895.parquet"]}, {"split": "2023_10_28T08_58_28.638753", "path": ["results_2023-10-28T08-58-28.638753.parquet"]}, {"split": "2023_12_03T19_17_16.064054", "path": ["results_2023-12-03T19-17-16.064054.parquet"]}, {"split": "latest", "path": ["results_2023-12-03T19-17-16.064054.parquet"]}]}]}
|
2023-12-03T19:17:23+00:00
|
[] |
[] |
TAGS
#region-us
|
# Dataset Card for Evaluation run of marcchew/LaMini-40k-Platypus2-7B
## Dataset Description
- Homepage:
- Repository: URL
- Paper:
- Leaderboard: URL
- Point of Contact: clementine@URL
### Dataset Summary
Dataset automatically created during the evaluation run of model marcchew/LaMini-40k-Platypus2-7B on the Open LLM Leaderboard.
The dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.
The dataset has been created from 3 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The "train" split is always pointing to the latest results.
An additional configuration "results" store all the aggregated results of the run (and is used to compute and display the aggregated metrics on the Open LLM Leaderboard).
To load the details from a run, you can for instance do the following:
## Latest results
These are the latest results from run 2023-12-03T19:17:16.064054(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the "latest" split for each eval):
### Supported Tasks and Leaderboards
### Languages
## Dataset Structure
### Data Instances
### Data Fields
### Data Splits
## Dataset Creation
### Curation Rationale
### Source Data
#### Initial Data Collection and Normalization
#### Who are the source language producers?
### Annotations
#### Annotation process
#### Who are the annotators?
### Personal and Sensitive Information
## Considerations for Using the Data
### Social Impact of Dataset
### Discussion of Biases
### Other Known Limitations
## Additional Information
### Dataset Curators
### Licensing Information
### Contributions
|
[
"# Dataset Card for Evaluation run of marcchew/LaMini-40k-Platypus2-7B",
"## Dataset Description\n\n- Homepage: \n- Repository: URL\n- Paper: \n- Leaderboard: URL\n- Point of Contact: clementine@URL",
"### Dataset Summary\n\nDataset automatically created during the evaluation run of model marcchew/LaMini-40k-Platypus2-7B on the Open LLM Leaderboard.\n\nThe dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.\n\nThe dataset has been created from 3 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The \"train\" split is always pointing to the latest results.\n\nAn additional configuration \"results\" store all the aggregated results of the run (and is used to compute and display the aggregated metrics on the Open LLM Leaderboard).\n\nTo load the details from a run, you can for instance do the following:",
"## Latest results\n\nThese are the latest results from run 2023-12-03T19:17:16.064054(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the \"latest\" split for each eval):",
"### Supported Tasks and Leaderboards",
"### Languages",
"## Dataset Structure",
"### Data Instances",
"### Data Fields",
"### Data Splits",
"## Dataset Creation",
"### Curation Rationale",
"### Source Data",
"#### Initial Data Collection and Normalization",
"#### Who are the source language producers?",
"### Annotations",
"#### Annotation process",
"#### Who are the annotators?",
"### Personal and Sensitive Information",
"## Considerations for Using the Data",
"### Social Impact of Dataset",
"### Discussion of Biases",
"### Other Known Limitations",
"## Additional Information",
"### Dataset Curators",
"### Licensing Information",
"### Contributions"
] |
[
"TAGS\n#region-us \n",
"# Dataset Card for Evaluation run of marcchew/LaMini-40k-Platypus2-7B",
"## Dataset Description\n\n- Homepage: \n- Repository: URL\n- Paper: \n- Leaderboard: URL\n- Point of Contact: clementine@URL",
"### Dataset Summary\n\nDataset automatically created during the evaluation run of model marcchew/LaMini-40k-Platypus2-7B on the Open LLM Leaderboard.\n\nThe dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.\n\nThe dataset has been created from 3 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The \"train\" split is always pointing to the latest results.\n\nAn additional configuration \"results\" store all the aggregated results of the run (and is used to compute and display the aggregated metrics on the Open LLM Leaderboard).\n\nTo load the details from a run, you can for instance do the following:",
"## Latest results\n\nThese are the latest results from run 2023-12-03T19:17:16.064054(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the \"latest\" split for each eval):",
"### Supported Tasks and Leaderboards",
"### Languages",
"## Dataset Structure",
"### Data Instances",
"### Data Fields",
"### Data Splits",
"## Dataset Creation",
"### Curation Rationale",
"### Source Data",
"#### Initial Data Collection and Normalization",
"#### Who are the source language producers?",
"### Annotations",
"#### Annotation process",
"#### Who are the annotators?",
"### Personal and Sensitive Information",
"## Considerations for Using the Data",
"### Social Impact of Dataset",
"### Discussion of Biases",
"### Other Known Limitations",
"## Additional Information",
"### Dataset Curators",
"### Licensing Information",
"### Contributions"
] |
[
6,
24,
31,
173,
67,
10,
4,
6,
6,
5,
5,
5,
7,
4,
10,
10,
5,
5,
9,
8,
8,
7,
8,
7,
5,
6,
6,
5
] |
[
"passage: TAGS\n#region-us \n# Dataset Card for Evaluation run of marcchew/LaMini-40k-Platypus2-7B## Dataset Description\n\n- Homepage: \n- Repository: URL\n- Paper: \n- Leaderboard: URL\n- Point of Contact: clementine@URL### Dataset Summary\n\nDataset automatically created during the evaluation run of model marcchew/LaMini-40k-Platypus2-7B on the Open LLM Leaderboard.\n\nThe dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.\n\nThe dataset has been created from 3 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The \"train\" split is always pointing to the latest results.\n\nAn additional configuration \"results\" store all the aggregated results of the run (and is used to compute and display the aggregated metrics on the Open LLM Leaderboard).\n\nTo load the details from a run, you can for instance do the following:## Latest results\n\nThese are the latest results from run 2023-12-03T19:17:16.064054(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the \"latest\" split for each eval):### Supported Tasks and Leaderboards### Languages## Dataset Structure### Data Instances### Data Fields### Data Splits## Dataset Creation### Curation Rationale### Source Data#### Initial Data Collection and Normalization#### Who are the source language producers?### Annotations#### Annotation process#### Who are the annotators?### Personal and Sensitive Information## Considerations for Using the Data### Social Impact of Dataset### Discussion of Biases### Other Known Limitations## Additional Information### Dataset Curators### Licensing Information### Contributions"
] |
51b6a5ade9c53a659ae3a0992947c3be464801db
|
# Dataset of Ijichi Nijika
This is the dataset of Ijichi Nijika, containing 296 images and their tags.
Images are crawled from many sites (e.g. danbooru, pixiv, zerochan ...), the auto-crawling system is powered by [DeepGHS Team](https://github.com/deepghs)([huggingface organization](https://huggingface.co/deepghs)).
| Name | Images | Download | Description |
|:------------|---------:|:------------------------------------|:-------------------------------------------------------------------------|
| raw | 296 | [Download](dataset-raw.zip) | Raw data with meta information. |
| raw-stage3 | 684 | [Download](dataset-raw-stage3.zip) | 3-stage cropped raw data with meta information. |
| 384x512 | 296 | [Download](dataset-384x512.zip) | 384x512 aligned dataset. |
| 512x512 | 296 | [Download](dataset-512x512.zip) | 512x512 aligned dataset. |
| 512x704 | 296 | [Download](dataset-512x704.zip) | 512x704 aligned dataset. |
| 640x640 | 296 | [Download](dataset-640x640.zip) | 640x640 aligned dataset. |
| 640x880 | 296 | [Download](dataset-640x880.zip) | 640x880 aligned dataset. |
| stage3-640 | 684 | [Download](dataset-stage3-640.zip) | 3-stage cropped dataset with the shorter side not exceeding 640 pixels. |
| stage3-800 | 684 | [Download](dataset-stage3-800.zip) | 3-stage cropped dataset with the shorter side not exceeding 800 pixels. |
| stage3-1200 | 684 | [Download](dataset-stage3-1200.zip) | 3-stage cropped dataset with the shorter side not exceeding 1200 pixels. |
|
CyberHarem/ijichi_nijika_bocchitherock
|
[
"task_categories:text-to-image",
"size_categories:n<1K",
"license:mit",
"art",
"not-for-all-audiences",
"region:us"
] |
2023-09-18T11:56:02+00:00
|
{"license": "mit", "size_categories": ["n<1K"], "task_categories": ["text-to-image"], "tags": ["art", "not-for-all-audiences"]}
|
2023-09-18T12:02:38+00:00
|
[] |
[] |
TAGS
#task_categories-text-to-image #size_categories-n<1K #license-mit #art #not-for-all-audiences #region-us
|
Dataset of Ijichi Nijika
========================
This is the dataset of Ijichi Nijika, containing 296 images and their tags.
Images are crawled from many sites (e.g. danbooru, pixiv, zerochan ...), the auto-crawling system is powered by DeepGHS Team(huggingface organization).
|
[] |
[
"TAGS\n#task_categories-text-to-image #size_categories-n<1K #license-mit #art #not-for-all-audiences #region-us \n"
] |
[
44
] |
[
"passage: TAGS\n#task_categories-text-to-image #size_categories-n<1K #license-mit #art #not-for-all-audiences #region-us \n"
] |
5a23f91b986b5ca7ada07edc9c67883e6e091574
|
# Miners Detection dataset
The dataset consists of of photos captured within various mines, focusing on **miners** engaged in their work. Each photo is annotated with bounding box detection of the miners, an attribute highlights whether each miner is sitting or standing in the photo.
The dataset's diverse applications such as computer vision, safety assessment and others make it a valuable resource for *researchers, employers, and policymakers in the mining industry*.

# Get the dataset
### This is just an example of the data
Leave a request on [**https://trainingdata.pro/data-market**](https://trainingdata.pro/data-market?utm_source=huggingface&utm_medium=cpc&utm_campaign=miners-detection) to discuss your requirements, learn about the price and buy the dataset.
# Dataset structure
- **images** - contains of original images of miners
- **boxes** - includes bounding box labeling for the original images
- **annotations.xml** - contains coordinates of the bounding boxes and labels, created for the original photo
# Data Format
Each image from `images` folder is accompanied by an XML-annotation in the `annotations.xml` file indicating the coordinates of the bounding boxes for miners detection. For each point, the x and y coordinates are provided. The position of the miner is also provided by the attribute **is_sitting** (true, false).
# Example of XML file structure
.png?generation=1695040600108833&alt=media)
# Miners detection might be made in accordance with your requirements.
## [TrainingData](https://trainingdata.pro/data-market?utm_source=huggingface&utm_medium=cpc&utm_campaign=miners-detection) provides high-quality data annotation tailored to your needs
More datasets in TrainingData's Kaggle account: **https://www.kaggle.com/trainingdatapro/datasets**
TrainingData's GitHub: **https://github.com/Trainingdata-datamarket/TrainingData_All_datasets**
|
TrainingDataPro/miners-detection
|
[
"task_categories:image-classification",
"task_categories:object-detection",
"language:en",
"license:cc-by-nc-nd-4.0",
"code",
"region:us"
] |
2023-09-18T12:00:29+00:00
|
{"language": ["en"], "license": "cc-by-nc-nd-4.0", "task_categories": ["image-classification", "object-detection"], "tags": ["code"], "dataset_info": {"features": [{"name": "id", "dtype": "int32"}, {"name": "name", "dtype": "string"}, {"name": "image", "dtype": "image"}, {"name": "mask", "dtype": "image"}, {"name": "width", "dtype": "uint16"}, {"name": "height", "dtype": "uint16"}, {"name": "shapes", "sequence": [{"name": "label", "dtype": {"class_label": {"names": {"0": "Miner"}}}}, {"name": "type", "dtype": "string"}, {"name": "points", "sequence": {"sequence": "float32"}}, {"name": "rotation", "dtype": "float32"}, {"name": "occluded", "dtype": "uint8"}, {"name": "attributes", "sequence": [{"name": "name", "dtype": "string"}, {"name": "text", "dtype": "string"}]}]}], "splits": [{"name": "train", "num_bytes": 5907438, "num_examples": 8}], "download_size": 5795853, "dataset_size": 5907438}}
|
2023-09-29T07:29:36+00:00
|
[] |
[
"en"
] |
TAGS
#task_categories-image-classification #task_categories-object-detection #language-English #license-cc-by-nc-nd-4.0 #code #region-us
|
# Miners Detection dataset
The dataset consists of of photos captured within various mines, focusing on miners engaged in their work. Each photo is annotated with bounding box detection of the miners, an attribute highlights whether each miner is sitting or standing in the photo.
The dataset's diverse applications such as computer vision, safety assessment and others make it a valuable resource for *researchers, employers, and policymakers in the mining industry*.
.
# Example of XML file structure
.",
"# Example of XML file structure\n.",
"# Example of XML file structure\n.# Example of XML file structure\n
|
Harene/guanaco-llama2-100-rows
|
[
"region:us"
] |
2023-09-18T12:07:38+00:00
|
{"dataset_info": {"features": [{"name": "text", "dtype": "string"}], "splits": [{"name": "train", "num_bytes": 184326, "num_examples": 100}], "download_size": 111858, "dataset_size": 184326}, "configs": [{"config_name": "default", "data_files": [{"split": "train", "path": "data/train-*"}]}]}
|
2023-09-18T12:07:39+00:00
|
[] |
[] |
TAGS
#region-us
|
# Dataset Card for "guanaco-llama2-100-rows"
More Information needed
|
[
"# Dataset Card for \"guanaco-llama2-100-rows\"\n\nMore Information needed"
] |
[
"TAGS\n#region-us \n",
"# Dataset Card for \"guanaco-llama2-100-rows\"\n\nMore Information needed"
] |
[
6,
21
] |
[
"passage: TAGS\n#region-us \n# Dataset Card for \"guanaco-llama2-100-rows\"\n\nMore Information needed"
] |
709e576d9a089a5e1beffc2ab572044fda77b22a
|
# Bangumi Image Base of Futoku No Guild
This is the image base of bangumi Futoku no Guild, we detected 23 characters, 2459 images in total. The full dataset is [here](all.zip).
**Please note that these image bases are not guaranteed to be 100% cleaned, they may be noisy actual.** If you intend to manually train models using this dataset, we recommend performing necessary preprocessing on the downloaded dataset to eliminate potential noisy samples (approximately 1% probability).
Here is the characters' preview:
| # | Images | Download | Preview 1 | Preview 2 | Preview 3 | Preview 4 | Preview 5 | Preview 6 | Preview 7 | Preview 8 |
|:------|---------:|:---------------------------|:-------------------------------|:-------------------------------|:-------------------------------|:-------------------------------|:-------------------------------|:-------------------------------|:-------------------------------|:-------------------------------|
| 0 | 192 | [Download](0/dataset.zip) |  |  |  |  |  |  |  |  |
| 1 | 395 | [Download](1/dataset.zip) |  |  |  |  |  |  |  |  |
| 2 | 9 | [Download](2/dataset.zip) |  |  |  |  |  |  |  |  |
| 3 | 5 | [Download](3/dataset.zip) |  |  |  |  |  | N/A | N/A | N/A |
| 4 | 155 | [Download](4/dataset.zip) |  |  |  |  |  |  |  |  |
| 5 | 116 | [Download](5/dataset.zip) |  |  |  |  |  |  |  |  |
| 6 | 221 | [Download](6/dataset.zip) |  |  |  |  |  |  |  |  |
| 7 | 48 | [Download](7/dataset.zip) |  |  |  |  |  |  |  |  |
| 8 | 51 | [Download](8/dataset.zip) |  |  |  |  |  |  |  |  |
| 9 | 12 | [Download](9/dataset.zip) |  |  |  |  |  |  |  |  |
| 10 | 261 | [Download](10/dataset.zip) |  |  |  |  |  |  |  |  |
| 11 | 16 | [Download](11/dataset.zip) |  |  |  |  |  |  |  |  |
| 12 | 68 | [Download](12/dataset.zip) |  |  |  |  |  |  |  |  |
| 13 | 12 | [Download](13/dataset.zip) |  |  |  |  |  |  |  |  |
| 14 | 11 | [Download](14/dataset.zip) |  |  |  |  |  |  |  |  |
| 15 | 10 | [Download](15/dataset.zip) |  |  |  |  |  |  |  |  |
| 16 | 27 | [Download](16/dataset.zip) |  |  |  |  |  |  |  |  |
| 17 | 443 | [Download](17/dataset.zip) |  |  |  |  |  |  |  |  |
| 18 | 17 | [Download](18/dataset.zip) |  |  |  |  |  |  |  |  |
| 19 | 227 | [Download](19/dataset.zip) |  |  |  |  |  |  |  |  |
| 20 | 20 | [Download](20/dataset.zip) |  |  |  |  |  |  |  |  |
| 21 | 10 | [Download](21/dataset.zip) |  |  |  |  |  |  |  |  |
| noise | 133 | [Download](-1/dataset.zip) |  |  |  |  |  |  |  |  |
|
BangumiBase/futokunoguild
|
[
"size_categories:1K<n<10K",
"license:mit",
"art",
"region:us"
] |
2023-09-18T12:08:57+00:00
|
{"license": "mit", "size_categories": ["1K<n<10K"], "tags": ["art"]}
|
2023-09-29T08:08:38+00:00
|
[] |
[] |
TAGS
#size_categories-1K<n<10K #license-mit #art #region-us
|
Bangumi Image Base of Futoku No Guild
=====================================
This is the image base of bangumi Futoku no Guild, we detected 23 characters, 2459 images in total. The full dataset is here.
Please note that these image bases are not guaranteed to be 100% cleaned, they may be noisy actual. If you intend to manually train models using this dataset, we recommend performing necessary preprocessing on the downloaded dataset to eliminate potential noisy samples (approximately 1% probability).
Here is the characters' preview:
|
[] |
[
"TAGS\n#size_categories-1K<n<10K #license-mit #art #region-us \n"
] |
[
25
] |
[
"passage: TAGS\n#size_categories-1K<n<10K #license-mit #art #region-us \n"
] |
00f627806ba9ee6fd2fdfc9312d8fdd7078038ec
|
{
"data": [
{
"title": "Colliery Control Order, 2000",
"paragraphs": [
{
"context": "The Colliery Control Order, 2000 was issued by the Government of India in 2000. In exercise of the powers conferred by section 3 read with section 5 of the Essential Commodities Act, 1955 (10 of 1955) and in supersession of the Colliery Control Order, 1945, except as respects things done or omitted to be done before such supersession, the Government of India has issued a Gazette Notification on 1.1.2000 to publish the Colliery Control Order, 2000. The content of the Colliery Control Order, 2000 is given below.\n1. Short title and commencement._ (1) This Order may be called the Colliery Control Order, 2000.\n(2) It shall come into force on the 1st day of January, 2000.\n2. Definitions. _In this Order, unless there is anything repugnant in the subject or context, -\n(a) 'coal' includes anthracite, bituminous coal, lignite, peat and any other form of carbonaceous matter sold or marketed as coal and also coke;\n(b) 'Coal Controller' means the person appointed as such by the Central Government under the provisions of the Coal Controller’s Organisation (Group ‘A’ Posts) Recruitment Rules, 1986;\n(c) 'colliery' means any mine or open working where winning or extraction of coal is the principal object of the mining, quarrying or any other operation carried on therein, and includes a plant for the production of coke or for the washing of coal;\n(d) 'disposal' includes agreeing or offering to dispose of, and the disposal of ownership or any proprietary interest, the right of possession and possession whether or not accompanied by any disposal of ownership or of any proprietary interest or of the right to possession;\n(e) ‘agent’, ‘manager’, and ‘owner’ when used in relation to a colliery shall have the meanings respectively assigned to them in the Mines Act,1952;\n(f) 'size' when used in relation to coal shall have the same specification as given, from time to time, by the Bureau of Indian Standards in their specification number IS:437-1979.",
"qas": [
{
"question": "What is the short title of the Colliery Control Order, 2000?",
"id": "q1",
"answers": [
{
"text": "The Colliery Control Order, 2000",
"answer_start": 181
}
]
},
{
"question": "Under what authority was the Colliery Control Order, 2000 issued by the Government of India?",
"id": "q2",
"answers": [
{
"text": "Essential Commodities Act, 1955",
"answer_start": 85
}
]
},
{
"question": "When was the Colliery Control Order, 2000 published?",
"id": "q3",
"answers": [
{
"text": "1.1.2000",
"answer_start": 212
}
]
},
{
"question": "What is the principal objective of a colliery, as defined in the Order?",
"id": "q4",
"answers": [
{
"text": "winning or extraction of coal",
"answer_start": 299
}
]
},
{
"question": "Who is referred to as the 'Coal Controller' in the context of the Colliery Control Order, 2000?",
"id": "q5",
"answers": [
{
"text": "the person appointed as such by the Central Government under the provisions of the Coal Controller’s Organisation Recruitment Rules, 1986",
"answer_start": 377
}
]
},
{
"question": "What types of carbonaceous matter are included in the definition of 'coal' in this Order?",
"id": "q6",
"answers": [
{
"text": "anthracite, bituminous coal, lignite, peat, and any other form of carbonaceous matter sold or marketed as coal, as well as coke",
"answer_start": 424
}
]
},
{
"question": "What is the significance of size in relation to coal according to the Order?",
"id": "q7",
"answers": [
{
"text": "specified by the Bureau of Indian Standards in their specification number IS:437-1979",
"answer_start": 532
}
]
},
{
"question": "How is the categorization of coal into classes, grades, and sizes determined?",
"id": "q8",
"answers": [
{
"text": "determined by the Central Government through notifications in the Official Gazette",
"answer_start": 600
}
]
},
{
"question": "Who is responsible for laying down the procedure and method of sampling and analysis of coal for grade maintenance in a colliery?",
"id": "q9",
"answers": [
{
"text": "The Coal Controller",
"answer_start": 727
}
]
},
{
"question": "What is the procedure for resolving disputes between a consumer and the owner of a colliery regarding the declaration of grades of coal?",
"id": "q10",
"answers": [
{
"text": "Disputes regarding the declaration of grades of coal may be referred to the Coal Controller, and the decision of the Coal Controller shall be binding on the owner of the colliery. A memorandum of reference to the Coal Controller regarding such disputes should be accompanied by a fee as specified by the Coal Controller.",
"answer_start": 855
}
]
}
]
}
]
}
]
}
|
piyushghante/temp
|
[
"region:us"
] |
2023-09-18T12:12:51+00:00
|
{}
|
2023-09-18T12:17:14+00:00
|
[] |
[] |
TAGS
#region-us
|
{
"data": [
{
"title": "Colliery Control Order, 2000",
"paragraphs": [
{
"context": "The Colliery Control Order, 2000 was issued by the Government of India in 2000. In exercise of the powers conferred by section 3 read with section 5 of the Essential Commodities Act, 1955 (10 of 1955) and in supersession of the Colliery Control Order, 1945, except as respects things done or omitted to be done before such supersession, the Government of India has issued a Gazette Notification on 1.1.2000 to publish the Colliery Control Order, 2000. The content of the Colliery Control Order, 2000 is given below.\n1. Short title and commencement._ (1) This Order may be called the Colliery Control Order, 2000.\n(2) It shall come into force on the 1st day of January, 2000.\n2. Definitions. _In this Order, unless there is anything repugnant in the subject or context, -\n(a) 'coal' includes anthracite, bituminous coal, lignite, peat and any other form of carbonaceous matter sold or marketed as coal and also coke;\n(b) 'Coal Controller' means the person appointed as such by the Central Government under the provisions of the Coal Controller’s Organisation (Group ‘A’ Posts) Recruitment Rules, 1986;\n(c) 'colliery' means any mine or open working where winning or extraction of coal is the principal object of the mining, quarrying or any other operation carried on therein, and includes a plant for the production of coke or for the washing of coal;\n(d) 'disposal' includes agreeing or offering to dispose of, and the disposal of ownership or any proprietary interest, the right of possession and possession whether or not accompanied by any disposal of ownership or of any proprietary interest or of the right to possession;\n(e) ‘agent’, ‘manager’, and ‘owner’ when used in relation to a colliery shall have the meanings respectively assigned to them in the Mines Act,1952;\n(f) 'size' when used in relation to coal shall have the same specification as given, from time to time, by the Bureau of Indian Standards in their specification number IS:437-1979.",
"qas": [
{
"question": "What is the short title of the Colliery Control Order, 2000?",
"id": "q1",
"answers": [
{
"text": "The Colliery Control Order, 2000",
"answer_start": 181
}
]
},
{
"question": "Under what authority was the Colliery Control Order, 2000 issued by the Government of India?",
"id": "q2",
"answers": [
{
"text": "Essential Commodities Act, 1955",
"answer_start": 85
}
]
},
{
"question": "When was the Colliery Control Order, 2000 published?",
"id": "q3",
"answers": [
{
"text": "1.1.2000",
"answer_start": 212
}
]
},
{
"question": "What is the principal objective of a colliery, as defined in the Order?",
"id": "q4",
"answers": [
{
"text": "winning or extraction of coal",
"answer_start": 299
}
]
},
{
"question": "Who is referred to as the 'Coal Controller' in the context of the Colliery Control Order, 2000?",
"id": "q5",
"answers": [
{
"text": "the person appointed as such by the Central Government under the provisions of the Coal Controller’s Organisation Recruitment Rules, 1986",
"answer_start": 377
}
]
},
{
"question": "What types of carbonaceous matter are included in the definition of 'coal' in this Order?",
"id": "q6",
"answers": [
{
"text": "anthracite, bituminous coal, lignite, peat, and any other form of carbonaceous matter sold or marketed as coal, as well as coke",
"answer_start": 424
}
]
},
{
"question": "What is the significance of size in relation to coal according to the Order?",
"id": "q7",
"answers": [
{
"text": "specified by the Bureau of Indian Standards in their specification number IS:437-1979",
"answer_start": 532
}
]
},
{
"question": "How is the categorization of coal into classes, grades, and sizes determined?",
"id": "q8",
"answers": [
{
"text": "determined by the Central Government through notifications in the Official Gazette",
"answer_start": 600
}
]
},
{
"question": "Who is responsible for laying down the procedure and method of sampling and analysis of coal for grade maintenance in a colliery?",
"id": "q9",
"answers": [
{
"text": "The Coal Controller",
"answer_start": 727
}
]
},
{
"question": "What is the procedure for resolving disputes between a consumer and the owner of a colliery regarding the declaration of grades of coal?",
"id": "q10",
"answers": [
{
"text": "Disputes regarding the declaration of grades of coal may be referred to the Coal Controller, and the decision of the Coal Controller shall be binding on the owner of the colliery. A memorandum of reference to the Coal Controller regarding such disputes should be accompanied by a fee as specified by the Coal Controller.",
"answer_start": 855
}
]
}
]
}
]
}
]
}
|
[] |
[
"TAGS\n#region-us \n"
] |
[
6
] |
[
"passage: TAGS\n#region-us \n"
] |
5cc0c232237bfe562167c13b3ce0beb24eaf3840
|
# Dataset Card for Evaluation run of wei123602/Llama-2-13b-FINETUNE4
## Dataset Description
- **Homepage:**
- **Repository:** https://huggingface.co/wei123602/Llama-2-13b-FINETUNE4
- **Paper:**
- **Leaderboard:** https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard
- **Point of Contact:** [email protected]
### Dataset Summary
Dataset automatically created during the evaluation run of model [wei123602/Llama-2-13b-FINETUNE4](https://huggingface.co/wei123602/Llama-2-13b-FINETUNE4) on the [Open LLM Leaderboard](https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard).
The dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.
The dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The "train" split is always pointing to the latest results.
An additional configuration "results" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the [Open LLM Leaderboard](https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard)).
To load the details from a run, you can for instance do the following:
```python
from datasets import load_dataset
data = load_dataset("open-llm-leaderboard/details_wei123602__Llama-2-13b-FINETUNE4",
"harness_winogrande_5",
split="train")
```
## Latest results
These are the [latest results from run 2023-10-23T06:23:21.987505](https://huggingface.co/datasets/open-llm-leaderboard/details_wei123602__Llama-2-13b-FINETUNE4/blob/main/results_2023-10-23T06-23-21.987505.json)(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the "latest" split for each eval):
```python
{
"all": {
"em": 0.08525587248322147,
"em_stderr": 0.0028599050719363664,
"f1": 0.13560297818791875,
"f1_stderr": 0.0029877199841954003,
"acc": 0.44731455091723,
"acc_stderr": 0.010474236802343157
},
"harness|drop|3": {
"em": 0.08525587248322147,
"em_stderr": 0.0028599050719363664,
"f1": 0.13560297818791875,
"f1_stderr": 0.0029877199841954003
},
"harness|gsm8k|5": {
"acc": 0.12509476876421532,
"acc_stderr": 0.009112601439849643
},
"harness|winogrande|5": {
"acc": 0.7695343330702447,
"acc_stderr": 0.011835872164836671
}
}
```
### Supported Tasks and Leaderboards
[More Information Needed]
### Languages
[More Information Needed]
## Dataset Structure
### Data Instances
[More Information Needed]
### Data Fields
[More Information Needed]
### Data Splits
[More Information Needed]
## Dataset Creation
### Curation Rationale
[More Information Needed]
### Source Data
#### Initial Data Collection and Normalization
[More Information Needed]
#### Who are the source language producers?
[More Information Needed]
### Annotations
#### Annotation process
[More Information Needed]
#### Who are the annotators?
[More Information Needed]
### Personal and Sensitive Information
[More Information Needed]
## Considerations for Using the Data
### Social Impact of Dataset
[More Information Needed]
### Discussion of Biases
[More Information Needed]
### Other Known Limitations
[More Information Needed]
## Additional Information
### Dataset Curators
[More Information Needed]
### Licensing Information
[More Information Needed]
### Citation Information
[More Information Needed]
### Contributions
[More Information Needed]
|
open-llm-leaderboard/details_wei123602__Llama-2-13b-FINETUNE4
|
[
"region:us"
] |
2023-09-18T12:14:36+00:00
|
{"pretty_name": "Evaluation run of wei123602/Llama-2-13b-FINETUNE4", "dataset_summary": "Dataset automatically created during the evaluation run of model [wei123602/Llama-2-13b-FINETUNE4](https://huggingface.co/wei123602/Llama-2-13b-FINETUNE4) on the [Open LLM Leaderboard](https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard).\n\nThe dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.\n\nThe dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The \"train\" split is always pointing to the latest results.\n\nAn additional configuration \"results\" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the [Open LLM Leaderboard](https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard)).\n\nTo load the details from a run, you can for instance do the following:\n```python\nfrom datasets import load_dataset\ndata = load_dataset(\"open-llm-leaderboard/details_wei123602__Llama-2-13b-FINETUNE4\",\n\t\"harness_winogrande_5\",\n\tsplit=\"train\")\n```\n\n## Latest results\n\nThese are the [latest results from run 2023-10-23T06:23:21.987505](https://huggingface.co/datasets/open-llm-leaderboard/details_wei123602__Llama-2-13b-FINETUNE4/blob/main/results_2023-10-23T06-23-21.987505.json)(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the \"latest\" split for each eval):\n\n```python\n{\n \"all\": {\n \"em\": 0.08525587248322147,\n \"em_stderr\": 0.0028599050719363664,\n \"f1\": 0.13560297818791875,\n \"f1_stderr\": 0.0029877199841954003,\n \"acc\": 0.44731455091723,\n \"acc_stderr\": 0.010474236802343157\n },\n \"harness|drop|3\": {\n \"em\": 0.08525587248322147,\n \"em_stderr\": 0.0028599050719363664,\n \"f1\": 0.13560297818791875,\n \"f1_stderr\": 0.0029877199841954003\n },\n \"harness|gsm8k|5\": {\n \"acc\": 0.12509476876421532,\n \"acc_stderr\": 0.009112601439849643\n },\n \"harness|winogrande|5\": {\n \"acc\": 0.7695343330702447,\n \"acc_stderr\": 0.011835872164836671\n }\n}\n```", "repo_url": "https://huggingface.co/wei123602/Llama-2-13b-FINETUNE4", "leaderboard_url": "https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard", "point_of_contact": "[email protected]", "configs": [{"config_name": "harness_arc_challenge_25", "data_files": [{"split": "2023_09_18T13_14_12.416583", "path": ["**/details_harness|arc:challenge|25_2023-09-18T13-14-12.416583.parquet"]}, {"split": "latest", "path": ["**/details_harness|arc:challenge|25_2023-09-18T13-14-12.416583.parquet"]}]}, {"config_name": "harness_drop_3", "data_files": [{"split": "2023_10_23T06_23_21.987505", "path": ["**/details_harness|drop|3_2023-10-23T06-23-21.987505.parquet"]}, {"split": "latest", "path": ["**/details_harness|drop|3_2023-10-23T06-23-21.987505.parquet"]}]}, {"config_name": "harness_gsm8k_5", "data_files": [{"split": "2023_10_23T06_23_21.987505", "path": ["**/details_harness|gsm8k|5_2023-10-23T06-23-21.987505.parquet"]}, {"split": "latest", "path": ["**/details_harness|gsm8k|5_2023-10-23T06-23-21.987505.parquet"]}]}, {"config_name": "harness_hellaswag_10", "data_files": [{"split": "2023_09_18T13_14_12.416583", "path": ["**/details_harness|hellaswag|10_2023-09-18T13-14-12.416583.parquet"]}, {"split": "latest", "path": ["**/details_harness|hellaswag|10_2023-09-18T13-14-12.416583.parquet"]}]}, {"config_name": "harness_hendrycksTest_5", "data_files": [{"split": "2023_09_18T13_14_12.416583", "path": ["**/details_harness|hendrycksTest-abstract_algebra|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-anatomy|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-astronomy|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-business_ethics|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-clinical_knowledge|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-college_biology|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-college_chemistry|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-college_computer_science|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-college_mathematics|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-college_medicine|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-college_physics|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-computer_security|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-conceptual_physics|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-econometrics|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-electrical_engineering|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-elementary_mathematics|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-formal_logic|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-global_facts|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-high_school_biology|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-high_school_chemistry|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-high_school_computer_science|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-high_school_european_history|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-high_school_geography|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-high_school_government_and_politics|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-high_school_macroeconomics|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-high_school_mathematics|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-high_school_microeconomics|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-high_school_physics|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-high_school_psychology|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-high_school_statistics|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-high_school_us_history|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-high_school_world_history|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-human_aging|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-human_sexuality|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-international_law|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-jurisprudence|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-logical_fallacies|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-machine_learning|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-management|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-marketing|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-medical_genetics|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-miscellaneous|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-moral_disputes|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-moral_scenarios|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-nutrition|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-philosophy|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-prehistory|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-professional_accounting|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-professional_law|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-professional_medicine|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-professional_psychology|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-public_relations|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-security_studies|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-sociology|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-us_foreign_policy|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-virology|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-world_religions|5_2023-09-18T13-14-12.416583.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-abstract_algebra|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-anatomy|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-astronomy|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-business_ethics|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-clinical_knowledge|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-college_biology|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-college_chemistry|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-college_computer_science|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-college_mathematics|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-college_medicine|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-college_physics|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-computer_security|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-conceptual_physics|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-econometrics|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-electrical_engineering|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-elementary_mathematics|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-formal_logic|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-global_facts|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-high_school_biology|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-high_school_chemistry|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-high_school_computer_science|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-high_school_european_history|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-high_school_geography|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-high_school_government_and_politics|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-high_school_macroeconomics|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-high_school_mathematics|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-high_school_microeconomics|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-high_school_physics|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-high_school_psychology|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-high_school_statistics|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-high_school_us_history|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-high_school_world_history|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-human_aging|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-human_sexuality|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-international_law|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-jurisprudence|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-logical_fallacies|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-machine_learning|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-management|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-marketing|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-medical_genetics|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-miscellaneous|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-moral_disputes|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-moral_scenarios|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-nutrition|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-philosophy|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-prehistory|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-professional_accounting|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-professional_law|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-professional_medicine|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-professional_psychology|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-public_relations|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-security_studies|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-sociology|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-us_foreign_policy|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-virology|5_2023-09-18T13-14-12.416583.parquet", "**/details_harness|hendrycksTest-world_religions|5_2023-09-18T13-14-12.416583.parquet"]}]}, {"config_name": "harness_hendrycksTest_abstract_algebra_5", "data_files": [{"split": "2023_09_18T13_14_12.416583", "path": ["**/details_harness|hendrycksTest-abstract_algebra|5_2023-09-18T13-14-12.416583.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-abstract_algebra|5_2023-09-18T13-14-12.416583.parquet"]}]}, {"config_name": "harness_hendrycksTest_anatomy_5", "data_files": [{"split": "2023_09_18T13_14_12.416583", "path": ["**/details_harness|hendrycksTest-anatomy|5_2023-09-18T13-14-12.416583.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-anatomy|5_2023-09-18T13-14-12.416583.parquet"]}]}, {"config_name": "harness_hendrycksTest_astronomy_5", "data_files": [{"split": "2023_09_18T13_14_12.416583", "path": ["**/details_harness|hendrycksTest-astronomy|5_2023-09-18T13-14-12.416583.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-astronomy|5_2023-09-18T13-14-12.416583.parquet"]}]}, {"config_name": "harness_hendrycksTest_business_ethics_5", "data_files": [{"split": "2023_09_18T13_14_12.416583", "path": ["**/details_harness|hendrycksTest-business_ethics|5_2023-09-18T13-14-12.416583.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-business_ethics|5_2023-09-18T13-14-12.416583.parquet"]}]}, {"config_name": "harness_hendrycksTest_clinical_knowledge_5", "data_files": [{"split": "2023_09_18T13_14_12.416583", "path": ["**/details_harness|hendrycksTest-clinical_knowledge|5_2023-09-18T13-14-12.416583.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-clinical_knowledge|5_2023-09-18T13-14-12.416583.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_biology_5", "data_files": [{"split": "2023_09_18T13_14_12.416583", "path": ["**/details_harness|hendrycksTest-college_biology|5_2023-09-18T13-14-12.416583.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_biology|5_2023-09-18T13-14-12.416583.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_chemistry_5", "data_files": [{"split": "2023_09_18T13_14_12.416583", "path": ["**/details_harness|hendrycksTest-college_chemistry|5_2023-09-18T13-14-12.416583.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_chemistry|5_2023-09-18T13-14-12.416583.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_computer_science_5", "data_files": [{"split": "2023_09_18T13_14_12.416583", "path": ["**/details_harness|hendrycksTest-college_computer_science|5_2023-09-18T13-14-12.416583.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_computer_science|5_2023-09-18T13-14-12.416583.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_mathematics_5", "data_files": [{"split": "2023_09_18T13_14_12.416583", "path": ["**/details_harness|hendrycksTest-college_mathematics|5_2023-09-18T13-14-12.416583.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_mathematics|5_2023-09-18T13-14-12.416583.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_medicine_5", "data_files": [{"split": "2023_09_18T13_14_12.416583", "path": ["**/details_harness|hendrycksTest-college_medicine|5_2023-09-18T13-14-12.416583.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_medicine|5_2023-09-18T13-14-12.416583.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_physics_5", "data_files": [{"split": "2023_09_18T13_14_12.416583", "path": ["**/details_harness|hendrycksTest-college_physics|5_2023-09-18T13-14-12.416583.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_physics|5_2023-09-18T13-14-12.416583.parquet"]}]}, {"config_name": "harness_hendrycksTest_computer_security_5", "data_files": [{"split": "2023_09_18T13_14_12.416583", "path": ["**/details_harness|hendrycksTest-computer_security|5_2023-09-18T13-14-12.416583.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-computer_security|5_2023-09-18T13-14-12.416583.parquet"]}]}, {"config_name": "harness_hendrycksTest_conceptual_physics_5", "data_files": [{"split": "2023_09_18T13_14_12.416583", "path": ["**/details_harness|hendrycksTest-conceptual_physics|5_2023-09-18T13-14-12.416583.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-conceptual_physics|5_2023-09-18T13-14-12.416583.parquet"]}]}, {"config_name": "harness_hendrycksTest_econometrics_5", "data_files": [{"split": "2023_09_18T13_14_12.416583", "path": ["**/details_harness|hendrycksTest-econometrics|5_2023-09-18T13-14-12.416583.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-econometrics|5_2023-09-18T13-14-12.416583.parquet"]}]}, {"config_name": "harness_hendrycksTest_electrical_engineering_5", "data_files": [{"split": "2023_09_18T13_14_12.416583", "path": ["**/details_harness|hendrycksTest-electrical_engineering|5_2023-09-18T13-14-12.416583.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-electrical_engineering|5_2023-09-18T13-14-12.416583.parquet"]}]}, {"config_name": "harness_hendrycksTest_elementary_mathematics_5", "data_files": [{"split": "2023_09_18T13_14_12.416583", "path": ["**/details_harness|hendrycksTest-elementary_mathematics|5_2023-09-18T13-14-12.416583.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-elementary_mathematics|5_2023-09-18T13-14-12.416583.parquet"]}]}, {"config_name": "harness_hendrycksTest_formal_logic_5", "data_files": [{"split": "2023_09_18T13_14_12.416583", "path": ["**/details_harness|hendrycksTest-formal_logic|5_2023-09-18T13-14-12.416583.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-formal_logic|5_2023-09-18T13-14-12.416583.parquet"]}]}, {"config_name": "harness_hendrycksTest_global_facts_5", "data_files": [{"split": "2023_09_18T13_14_12.416583", "path": ["**/details_harness|hendrycksTest-global_facts|5_2023-09-18T13-14-12.416583.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-global_facts|5_2023-09-18T13-14-12.416583.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_biology_5", "data_files": [{"split": "2023_09_18T13_14_12.416583", "path": ["**/details_harness|hendrycksTest-high_school_biology|5_2023-09-18T13-14-12.416583.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_biology|5_2023-09-18T13-14-12.416583.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_chemistry_5", "data_files": [{"split": "2023_09_18T13_14_12.416583", "path": ["**/details_harness|hendrycksTest-high_school_chemistry|5_2023-09-18T13-14-12.416583.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_chemistry|5_2023-09-18T13-14-12.416583.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_computer_science_5", "data_files": [{"split": "2023_09_18T13_14_12.416583", "path": ["**/details_harness|hendrycksTest-high_school_computer_science|5_2023-09-18T13-14-12.416583.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_computer_science|5_2023-09-18T13-14-12.416583.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_european_history_5", "data_files": [{"split": "2023_09_18T13_14_12.416583", "path": ["**/details_harness|hendrycksTest-high_school_european_history|5_2023-09-18T13-14-12.416583.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_european_history|5_2023-09-18T13-14-12.416583.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_geography_5", "data_files": [{"split": "2023_09_18T13_14_12.416583", "path": ["**/details_harness|hendrycksTest-high_school_geography|5_2023-09-18T13-14-12.416583.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_geography|5_2023-09-18T13-14-12.416583.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_government_and_politics_5", "data_files": [{"split": "2023_09_18T13_14_12.416583", "path": ["**/details_harness|hendrycksTest-high_school_government_and_politics|5_2023-09-18T13-14-12.416583.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_government_and_politics|5_2023-09-18T13-14-12.416583.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_macroeconomics_5", "data_files": [{"split": "2023_09_18T13_14_12.416583", "path": ["**/details_harness|hendrycksTest-high_school_macroeconomics|5_2023-09-18T13-14-12.416583.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_macroeconomics|5_2023-09-18T13-14-12.416583.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_mathematics_5", "data_files": [{"split": "2023_09_18T13_14_12.416583", "path": ["**/details_harness|hendrycksTest-high_school_mathematics|5_2023-09-18T13-14-12.416583.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_mathematics|5_2023-09-18T13-14-12.416583.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_microeconomics_5", "data_files": [{"split": "2023_09_18T13_14_12.416583", "path": ["**/details_harness|hendrycksTest-high_school_microeconomics|5_2023-09-18T13-14-12.416583.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_microeconomics|5_2023-09-18T13-14-12.416583.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_physics_5", "data_files": [{"split": "2023_09_18T13_14_12.416583", "path": ["**/details_harness|hendrycksTest-high_school_physics|5_2023-09-18T13-14-12.416583.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_physics|5_2023-09-18T13-14-12.416583.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_psychology_5", "data_files": [{"split": "2023_09_18T13_14_12.416583", "path": ["**/details_harness|hendrycksTest-high_school_psychology|5_2023-09-18T13-14-12.416583.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_psychology|5_2023-09-18T13-14-12.416583.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_statistics_5", "data_files": [{"split": "2023_09_18T13_14_12.416583", "path": ["**/details_harness|hendrycksTest-high_school_statistics|5_2023-09-18T13-14-12.416583.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_statistics|5_2023-09-18T13-14-12.416583.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_us_history_5", "data_files": [{"split": "2023_09_18T13_14_12.416583", "path": ["**/details_harness|hendrycksTest-high_school_us_history|5_2023-09-18T13-14-12.416583.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_us_history|5_2023-09-18T13-14-12.416583.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_world_history_5", "data_files": [{"split": "2023_09_18T13_14_12.416583", "path": ["**/details_harness|hendrycksTest-high_school_world_history|5_2023-09-18T13-14-12.416583.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_world_history|5_2023-09-18T13-14-12.416583.parquet"]}]}, {"config_name": "harness_hendrycksTest_human_aging_5", "data_files": [{"split": "2023_09_18T13_14_12.416583", "path": ["**/details_harness|hendrycksTest-human_aging|5_2023-09-18T13-14-12.416583.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-human_aging|5_2023-09-18T13-14-12.416583.parquet"]}]}, {"config_name": "harness_hendrycksTest_human_sexuality_5", "data_files": [{"split": "2023_09_18T13_14_12.416583", "path": ["**/details_harness|hendrycksTest-human_sexuality|5_2023-09-18T13-14-12.416583.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-human_sexuality|5_2023-09-18T13-14-12.416583.parquet"]}]}, {"config_name": "harness_hendrycksTest_international_law_5", "data_files": [{"split": "2023_09_18T13_14_12.416583", "path": ["**/details_harness|hendrycksTest-international_law|5_2023-09-18T13-14-12.416583.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-international_law|5_2023-09-18T13-14-12.416583.parquet"]}]}, {"config_name": "harness_hendrycksTest_jurisprudence_5", "data_files": [{"split": "2023_09_18T13_14_12.416583", "path": ["**/details_harness|hendrycksTest-jurisprudence|5_2023-09-18T13-14-12.416583.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-jurisprudence|5_2023-09-18T13-14-12.416583.parquet"]}]}, {"config_name": "harness_hendrycksTest_logical_fallacies_5", "data_files": [{"split": "2023_09_18T13_14_12.416583", "path": ["**/details_harness|hendrycksTest-logical_fallacies|5_2023-09-18T13-14-12.416583.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-logical_fallacies|5_2023-09-18T13-14-12.416583.parquet"]}]}, {"config_name": "harness_hendrycksTest_machine_learning_5", "data_files": [{"split": "2023_09_18T13_14_12.416583", "path": ["**/details_harness|hendrycksTest-machine_learning|5_2023-09-18T13-14-12.416583.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-machine_learning|5_2023-09-18T13-14-12.416583.parquet"]}]}, {"config_name": "harness_hendrycksTest_management_5", "data_files": [{"split": "2023_09_18T13_14_12.416583", "path": ["**/details_harness|hendrycksTest-management|5_2023-09-18T13-14-12.416583.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-management|5_2023-09-18T13-14-12.416583.parquet"]}]}, {"config_name": "harness_hendrycksTest_marketing_5", "data_files": [{"split": "2023_09_18T13_14_12.416583", "path": ["**/details_harness|hendrycksTest-marketing|5_2023-09-18T13-14-12.416583.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-marketing|5_2023-09-18T13-14-12.416583.parquet"]}]}, {"config_name": "harness_hendrycksTest_medical_genetics_5", "data_files": [{"split": "2023_09_18T13_14_12.416583", "path": ["**/details_harness|hendrycksTest-medical_genetics|5_2023-09-18T13-14-12.416583.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-medical_genetics|5_2023-09-18T13-14-12.416583.parquet"]}]}, {"config_name": "harness_hendrycksTest_miscellaneous_5", "data_files": [{"split": "2023_09_18T13_14_12.416583", "path": ["**/details_harness|hendrycksTest-miscellaneous|5_2023-09-18T13-14-12.416583.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-miscellaneous|5_2023-09-18T13-14-12.416583.parquet"]}]}, {"config_name": "harness_hendrycksTest_moral_disputes_5", "data_files": [{"split": "2023_09_18T13_14_12.416583", "path": ["**/details_harness|hendrycksTest-moral_disputes|5_2023-09-18T13-14-12.416583.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-moral_disputes|5_2023-09-18T13-14-12.416583.parquet"]}]}, {"config_name": "harness_hendrycksTest_moral_scenarios_5", "data_files": [{"split": "2023_09_18T13_14_12.416583", "path": ["**/details_harness|hendrycksTest-moral_scenarios|5_2023-09-18T13-14-12.416583.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-moral_scenarios|5_2023-09-18T13-14-12.416583.parquet"]}]}, {"config_name": "harness_hendrycksTest_nutrition_5", "data_files": [{"split": "2023_09_18T13_14_12.416583", "path": ["**/details_harness|hendrycksTest-nutrition|5_2023-09-18T13-14-12.416583.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-nutrition|5_2023-09-18T13-14-12.416583.parquet"]}]}, {"config_name": "harness_hendrycksTest_philosophy_5", "data_files": [{"split": "2023_09_18T13_14_12.416583", "path": ["**/details_harness|hendrycksTest-philosophy|5_2023-09-18T13-14-12.416583.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-philosophy|5_2023-09-18T13-14-12.416583.parquet"]}]}, {"config_name": "harness_hendrycksTest_prehistory_5", "data_files": [{"split": "2023_09_18T13_14_12.416583", "path": ["**/details_harness|hendrycksTest-prehistory|5_2023-09-18T13-14-12.416583.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-prehistory|5_2023-09-18T13-14-12.416583.parquet"]}]}, {"config_name": "harness_hendrycksTest_professional_accounting_5", "data_files": [{"split": "2023_09_18T13_14_12.416583", "path": ["**/details_harness|hendrycksTest-professional_accounting|5_2023-09-18T13-14-12.416583.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-professional_accounting|5_2023-09-18T13-14-12.416583.parquet"]}]}, {"config_name": "harness_hendrycksTest_professional_law_5", "data_files": [{"split": "2023_09_18T13_14_12.416583", "path": ["**/details_harness|hendrycksTest-professional_law|5_2023-09-18T13-14-12.416583.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-professional_law|5_2023-09-18T13-14-12.416583.parquet"]}]}, {"config_name": "harness_hendrycksTest_professional_medicine_5", "data_files": [{"split": "2023_09_18T13_14_12.416583", "path": ["**/details_harness|hendrycksTest-professional_medicine|5_2023-09-18T13-14-12.416583.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-professional_medicine|5_2023-09-18T13-14-12.416583.parquet"]}]}, {"config_name": "harness_hendrycksTest_professional_psychology_5", "data_files": [{"split": "2023_09_18T13_14_12.416583", "path": ["**/details_harness|hendrycksTest-professional_psychology|5_2023-09-18T13-14-12.416583.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-professional_psychology|5_2023-09-18T13-14-12.416583.parquet"]}]}, {"config_name": "harness_hendrycksTest_public_relations_5", "data_files": [{"split": "2023_09_18T13_14_12.416583", "path": ["**/details_harness|hendrycksTest-public_relations|5_2023-09-18T13-14-12.416583.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-public_relations|5_2023-09-18T13-14-12.416583.parquet"]}]}, {"config_name": "harness_hendrycksTest_security_studies_5", "data_files": [{"split": "2023_09_18T13_14_12.416583", "path": ["**/details_harness|hendrycksTest-security_studies|5_2023-09-18T13-14-12.416583.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-security_studies|5_2023-09-18T13-14-12.416583.parquet"]}]}, {"config_name": "harness_hendrycksTest_sociology_5", "data_files": [{"split": "2023_09_18T13_14_12.416583", "path": ["**/details_harness|hendrycksTest-sociology|5_2023-09-18T13-14-12.416583.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-sociology|5_2023-09-18T13-14-12.416583.parquet"]}]}, {"config_name": "harness_hendrycksTest_us_foreign_policy_5", "data_files": [{"split": "2023_09_18T13_14_12.416583", "path": ["**/details_harness|hendrycksTest-us_foreign_policy|5_2023-09-18T13-14-12.416583.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-us_foreign_policy|5_2023-09-18T13-14-12.416583.parquet"]}]}, {"config_name": "harness_hendrycksTest_virology_5", "data_files": [{"split": "2023_09_18T13_14_12.416583", "path": ["**/details_harness|hendrycksTest-virology|5_2023-09-18T13-14-12.416583.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-virology|5_2023-09-18T13-14-12.416583.parquet"]}]}, {"config_name": "harness_hendrycksTest_world_religions_5", "data_files": [{"split": "2023_09_18T13_14_12.416583", "path": ["**/details_harness|hendrycksTest-world_religions|5_2023-09-18T13-14-12.416583.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-world_religions|5_2023-09-18T13-14-12.416583.parquet"]}]}, {"config_name": "harness_truthfulqa_mc_0", "data_files": [{"split": "2023_09_18T13_14_12.416583", "path": ["**/details_harness|truthfulqa:mc|0_2023-09-18T13-14-12.416583.parquet"]}, {"split": "latest", "path": ["**/details_harness|truthfulqa:mc|0_2023-09-18T13-14-12.416583.parquet"]}]}, {"config_name": "harness_winogrande_5", "data_files": [{"split": "2023_10_23T06_23_21.987505", "path": ["**/details_harness|winogrande|5_2023-10-23T06-23-21.987505.parquet"]}, {"split": "latest", "path": ["**/details_harness|winogrande|5_2023-10-23T06-23-21.987505.parquet"]}]}, {"config_name": "results", "data_files": [{"split": "2023_09_18T13_14_12.416583", "path": ["results_2023-09-18T13-14-12.416583.parquet"]}, {"split": "2023_10_23T06_23_21.987505", "path": ["results_2023-10-23T06-23-21.987505.parquet"]}, {"split": "latest", "path": ["results_2023-10-23T06-23-21.987505.parquet"]}]}]}
|
2023-10-23T05:23:36+00:00
|
[] |
[] |
TAGS
#region-us
|
# Dataset Card for Evaluation run of wei123602/Llama-2-13b-FINETUNE4
## Dataset Description
- Homepage:
- Repository: URL
- Paper:
- Leaderboard: URL
- Point of Contact: clementine@URL
### Dataset Summary
Dataset automatically created during the evaluation run of model wei123602/Llama-2-13b-FINETUNE4 on the Open LLM Leaderboard.
The dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.
The dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The "train" split is always pointing to the latest results.
An additional configuration "results" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the Open LLM Leaderboard).
To load the details from a run, you can for instance do the following:
## Latest results
These are the latest results from run 2023-10-23T06:23:21.987505(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the "latest" split for each eval):
### Supported Tasks and Leaderboards
### Languages
## Dataset Structure
### Data Instances
### Data Fields
### Data Splits
## Dataset Creation
### Curation Rationale
### Source Data
#### Initial Data Collection and Normalization
#### Who are the source language producers?
### Annotations
#### Annotation process
#### Who are the annotators?
### Personal and Sensitive Information
## Considerations for Using the Data
### Social Impact of Dataset
### Discussion of Biases
### Other Known Limitations
## Additional Information
### Dataset Curators
### Licensing Information
### Contributions
|
[
"# Dataset Card for Evaluation run of wei123602/Llama-2-13b-FINETUNE4",
"## Dataset Description\n\n- Homepage: \n- Repository: URL\n- Paper: \n- Leaderboard: URL\n- Point of Contact: clementine@URL",
"### Dataset Summary\n\nDataset automatically created during the evaluation run of model wei123602/Llama-2-13b-FINETUNE4 on the Open LLM Leaderboard.\n\nThe dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.\n\nThe dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The \"train\" split is always pointing to the latest results.\n\nAn additional configuration \"results\" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the Open LLM Leaderboard).\n\nTo load the details from a run, you can for instance do the following:",
"## Latest results\n\nThese are the latest results from run 2023-10-23T06:23:21.987505(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the \"latest\" split for each eval):",
"### Supported Tasks and Leaderboards",
"### Languages",
"## Dataset Structure",
"### Data Instances",
"### Data Fields",
"### Data Splits",
"## Dataset Creation",
"### Curation Rationale",
"### Source Data",
"#### Initial Data Collection and Normalization",
"#### Who are the source language producers?",
"### Annotations",
"#### Annotation process",
"#### Who are the annotators?",
"### Personal and Sensitive Information",
"## Considerations for Using the Data",
"### Social Impact of Dataset",
"### Discussion of Biases",
"### Other Known Limitations",
"## Additional Information",
"### Dataset Curators",
"### Licensing Information",
"### Contributions"
] |
[
"TAGS\n#region-us \n",
"# Dataset Card for Evaluation run of wei123602/Llama-2-13b-FINETUNE4",
"## Dataset Description\n\n- Homepage: \n- Repository: URL\n- Paper: \n- Leaderboard: URL\n- Point of Contact: clementine@URL",
"### Dataset Summary\n\nDataset automatically created during the evaluation run of model wei123602/Llama-2-13b-FINETUNE4 on the Open LLM Leaderboard.\n\nThe dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.\n\nThe dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The \"train\" split is always pointing to the latest results.\n\nAn additional configuration \"results\" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the Open LLM Leaderboard).\n\nTo load the details from a run, you can for instance do the following:",
"## Latest results\n\nThese are the latest results from run 2023-10-23T06:23:21.987505(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the \"latest\" split for each eval):",
"### Supported Tasks and Leaderboards",
"### Languages",
"## Dataset Structure",
"### Data Instances",
"### Data Fields",
"### Data Splits",
"## Dataset Creation",
"### Curation Rationale",
"### Source Data",
"#### Initial Data Collection and Normalization",
"#### Who are the source language producers?",
"### Annotations",
"#### Annotation process",
"#### Who are the annotators?",
"### Personal and Sensitive Information",
"## Considerations for Using the Data",
"### Social Impact of Dataset",
"### Discussion of Biases",
"### Other Known Limitations",
"## Additional Information",
"### Dataset Curators",
"### Licensing Information",
"### Contributions"
] |
[
6,
24,
31,
172,
67,
10,
4,
6,
6,
5,
5,
5,
7,
4,
10,
10,
5,
5,
9,
8,
8,
7,
8,
7,
5,
6,
6,
5
] |
[
"passage: TAGS\n#region-us \n# Dataset Card for Evaluation run of wei123602/Llama-2-13b-FINETUNE4## Dataset Description\n\n- Homepage: \n- Repository: URL\n- Paper: \n- Leaderboard: URL\n- Point of Contact: clementine@URL### Dataset Summary\n\nDataset automatically created during the evaluation run of model wei123602/Llama-2-13b-FINETUNE4 on the Open LLM Leaderboard.\n\nThe dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.\n\nThe dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The \"train\" split is always pointing to the latest results.\n\nAn additional configuration \"results\" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the Open LLM Leaderboard).\n\nTo load the details from a run, you can for instance do the following:## Latest results\n\nThese are the latest results from run 2023-10-23T06:23:21.987505(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the \"latest\" split for each eval):### Supported Tasks and Leaderboards### Languages## Dataset Structure### Data Instances### Data Fields### Data Splits## Dataset Creation### Curation Rationale### Source Data#### Initial Data Collection and Normalization#### Who are the source language producers?### Annotations#### Annotation process#### Who are the annotators?### Personal and Sensitive Information## Considerations for Using the Data### Social Impact of Dataset### Discussion of Biases### Other Known Limitations## Additional Information### Dataset Curators### Licensing Information### Contributions"
] |
7c37ffaf41d35b8173165231f48a1f241cbec9c5
|
# Dataset Card for "bus_few4_8x"
[More Information needed](https://github.com/huggingface/datasets/blob/main/CONTRIBUTING.md#how-to-contribute-to-the-dataset-cards)
|
FanChen0116/bus_few4_8x
|
[
"region:us"
] |
2023-09-18T12:15:59+00:00
|
{"dataset_info": {"features": [{"name": "id", "dtype": "int64"}, {"name": "tokens", "sequence": "string"}, {"name": "labels", "sequence": {"class_label": {"names": {"0": "O", "1": "I-from_location", "2": "B-from_location", "3": "B-leaving_date", "4": "I-leaving_date", "5": "I-to_location", "6": "B-to_location"}}}}, {"name": "request_slot", "sequence": "string"}], "splits": [{"name": "train", "num_bytes": 109163, "num_examples": 560}, {"name": "validation", "num_bytes": 6900, "num_examples": 35}, {"name": "test", "num_bytes": 70618, "num_examples": 377}], "download_size": 18363, "dataset_size": 186681}}
|
2023-09-20T12:31:17+00:00
|
[] |
[] |
TAGS
#region-us
|
# Dataset Card for "bus_few4_8x"
More Information needed
|
[
"# Dataset Card for \"bus_few4_8x\"\n\nMore Information needed"
] |
[
"TAGS\n#region-us \n",
"# Dataset Card for \"bus_few4_8x\"\n\nMore Information needed"
] |
[
6,
18
] |
[
"passage: TAGS\n#region-us \n# Dataset Card for \"bus_few4_8x\"\n\nMore Information needed"
] |
d86f32973f486cea848a737e4ce7f61c9cdba844
|
# Dataset Card for "bus_few4_16x"
[More Information needed](https://github.com/huggingface/datasets/blob/main/CONTRIBUTING.md#how-to-contribute-to-the-dataset-cards)
|
FanChen0116/bus_few4_16x
|
[
"region:us"
] |
2023-09-18T12:16:17+00:00
|
{"dataset_info": {"features": [{"name": "id", "dtype": "int64"}, {"name": "tokens", "sequence": "string"}, {"name": "labels", "sequence": {"class_label": {"names": {"0": "O", "1": "I-from_location", "2": "B-from_location", "3": "B-leaving_date", "4": "I-leaving_date", "5": "I-to_location", "6": "B-to_location"}}}}, {"name": "request_slot", "sequence": "string"}], "splits": [{"name": "train", "num_bytes": 217504, "num_examples": 1120}, {"name": "validation", "num_bytes": 6900, "num_examples": 35}, {"name": "test", "num_bytes": 70618, "num_examples": 377}], "download_size": 0, "dataset_size": 295022}}
|
2023-09-27T06:29:01+00:00
|
[] |
[] |
TAGS
#region-us
|
# Dataset Card for "bus_few4_16x"
More Information needed
|
[
"# Dataset Card for \"bus_few4_16x\"\n\nMore Information needed"
] |
[
"TAGS\n#region-us \n",
"# Dataset Card for \"bus_few4_16x\"\n\nMore Information needed"
] |
[
6,
18
] |
[
"passage: TAGS\n#region-us \n# Dataset Card for \"bus_few4_16x\"\n\nMore Information needed"
] |
3ae1d4e8c168b4d54c0421071a0d9e9c090acc72
|
# Dataset Card for "bus_few4_32x"
[More Information needed](https://github.com/huggingface/datasets/blob/main/CONTRIBUTING.md#how-to-contribute-to-the-dataset-cards)
|
FanChen0116/bus_few4_32x
|
[
"region:us"
] |
2023-09-18T12:16:31+00:00
|
{"dataset_info": {"features": [{"name": "id", "dtype": "int64"}, {"name": "tokens", "sequence": "string"}, {"name": "labels", "sequence": {"class_label": {"names": {"0": "O", "1": "I-from_location", "2": "B-from_location", "3": "B-leaving_date", "4": "I-leaving_date", "5": "I-to_location", "6": "B-to_location"}}}}, {"name": "request_slot", "sequence": "string"}], "splits": [{"name": "train", "num_bytes": 431733, "num_examples": 2240}, {"name": "validation", "num_bytes": 6900, "num_examples": 35}, {"name": "test", "num_bytes": 70618, "num_examples": 377}], "download_size": 0, "dataset_size": 509251}}
|
2023-09-27T00:49:43+00:00
|
[] |
[] |
TAGS
#region-us
|
# Dataset Card for "bus_few4_32x"
More Information needed
|
[
"# Dataset Card for \"bus_few4_32x\"\n\nMore Information needed"
] |
[
"TAGS\n#region-us \n",
"# Dataset Card for \"bus_few4_32x\"\n\nMore Information needed"
] |
[
6,
18
] |
[
"passage: TAGS\n#region-us \n# Dataset Card for \"bus_few4_32x\"\n\nMore Information needed"
] |
d9c96862af89bb033e1c96b35f8e23fbdd7b5bea
|
# Dataset Card for Evaluation run of Dampish/StellarX-4B-V0.2
## Dataset Description
- **Homepage:**
- **Repository:** https://huggingface.co/Dampish/StellarX-4B-V0.2
- **Paper:**
- **Leaderboard:** https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard
- **Point of Contact:** [email protected]
### Dataset Summary
Dataset automatically created during the evaluation run of model [Dampish/StellarX-4B-V0.2](https://huggingface.co/Dampish/StellarX-4B-V0.2) on the [Open LLM Leaderboard](https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard).
The dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.
The dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The "train" split is always pointing to the latest results.
An additional configuration "results" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the [Open LLM Leaderboard](https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard)).
To load the details from a run, you can for instance do the following:
```python
from datasets import load_dataset
data = load_dataset("open-llm-leaderboard/details_Dampish__StellarX-4B-V0.2",
"harness_winogrande_5",
split="train")
```
## Latest results
These are the [latest results from run 2023-10-25T23:58:38.907071](https://huggingface.co/datasets/open-llm-leaderboard/details_Dampish__StellarX-4B-V0.2/blob/main/results_2023-10-25T23-58-38.907071.json)(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the "latest" split for each eval):
```python
{
"all": {
"em": 0.0008389261744966443,
"em_stderr": 0.00029649629898012564,
"f1": 0.04009018456375841,
"f1_stderr": 0.0010817232514367004,
"acc": 0.30702446724546173,
"acc_stderr": 0.006841018496698701
},
"harness|drop|3": {
"em": 0.0008389261744966443,
"em_stderr": 0.00029649629898012564,
"f1": 0.04009018456375841,
"f1_stderr": 0.0010817232514367004
},
"harness|gsm8k|5": {
"acc": 0.0,
"acc_stderr": 0.0
},
"harness|winogrande|5": {
"acc": 0.6140489344909235,
"acc_stderr": 0.013682036993397402
}
}
```
### Supported Tasks and Leaderboards
[More Information Needed]
### Languages
[More Information Needed]
## Dataset Structure
### Data Instances
[More Information Needed]
### Data Fields
[More Information Needed]
### Data Splits
[More Information Needed]
## Dataset Creation
### Curation Rationale
[More Information Needed]
### Source Data
#### Initial Data Collection and Normalization
[More Information Needed]
#### Who are the source language producers?
[More Information Needed]
### Annotations
#### Annotation process
[More Information Needed]
#### Who are the annotators?
[More Information Needed]
### Personal and Sensitive Information
[More Information Needed]
## Considerations for Using the Data
### Social Impact of Dataset
[More Information Needed]
### Discussion of Biases
[More Information Needed]
### Other Known Limitations
[More Information Needed]
## Additional Information
### Dataset Curators
[More Information Needed]
### Licensing Information
[More Information Needed]
### Citation Information
[More Information Needed]
### Contributions
[More Information Needed]
|
open-llm-leaderboard/details_Dampish__StellarX-4B-V0.2
|
[
"region:us"
] |
2023-09-18T12:16:43+00:00
|
{"pretty_name": "Evaluation run of Dampish/StellarX-4B-V0.2", "dataset_summary": "Dataset automatically created during the evaluation run of model [Dampish/StellarX-4B-V0.2](https://huggingface.co/Dampish/StellarX-4B-V0.2) on the [Open LLM Leaderboard](https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard).\n\nThe dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.\n\nThe dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The \"train\" split is always pointing to the latest results.\n\nAn additional configuration \"results\" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the [Open LLM Leaderboard](https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard)).\n\nTo load the details from a run, you can for instance do the following:\n```python\nfrom datasets import load_dataset\ndata = load_dataset(\"open-llm-leaderboard/details_Dampish__StellarX-4B-V0.2\",\n\t\"harness_winogrande_5\",\n\tsplit=\"train\")\n```\n\n## Latest results\n\nThese are the [latest results from run 2023-10-25T23:58:38.907071](https://huggingface.co/datasets/open-llm-leaderboard/details_Dampish__StellarX-4B-V0.2/blob/main/results_2023-10-25T23-58-38.907071.json)(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the \"latest\" split for each eval):\n\n```python\n{\n \"all\": {\n \"em\": 0.0008389261744966443,\n \"em_stderr\": 0.00029649629898012564,\n \"f1\": 0.04009018456375841,\n \"f1_stderr\": 0.0010817232514367004,\n \"acc\": 0.30702446724546173,\n \"acc_stderr\": 0.006841018496698701\n },\n \"harness|drop|3\": {\n \"em\": 0.0008389261744966443,\n \"em_stderr\": 0.00029649629898012564,\n \"f1\": 0.04009018456375841,\n \"f1_stderr\": 0.0010817232514367004\n },\n \"harness|gsm8k|5\": {\n \"acc\": 0.0,\n \"acc_stderr\": 0.0\n },\n \"harness|winogrande|5\": {\n \"acc\": 0.6140489344909235,\n \"acc_stderr\": 0.013682036993397402\n }\n}\n```", "repo_url": "https://huggingface.co/Dampish/StellarX-4B-V0.2", "leaderboard_url": "https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard", "point_of_contact": "[email protected]", "configs": [{"config_name": "harness_arc_challenge_25", "data_files": [{"split": "2023_09_18T13_16_25.972049", "path": ["**/details_harness|arc:challenge|25_2023-09-18T13-16-25.972049.parquet"]}, {"split": "latest", "path": ["**/details_harness|arc:challenge|25_2023-09-18T13-16-25.972049.parquet"]}]}, {"config_name": "harness_drop_3", "data_files": [{"split": "2023_10_25T23_58_38.907071", "path": ["**/details_harness|drop|3_2023-10-25T23-58-38.907071.parquet"]}, {"split": "latest", "path": ["**/details_harness|drop|3_2023-10-25T23-58-38.907071.parquet"]}]}, {"config_name": "harness_gsm8k_5", "data_files": [{"split": "2023_10_25T23_58_38.907071", "path": ["**/details_harness|gsm8k|5_2023-10-25T23-58-38.907071.parquet"]}, {"split": "latest", "path": ["**/details_harness|gsm8k|5_2023-10-25T23-58-38.907071.parquet"]}]}, {"config_name": "harness_hellaswag_10", "data_files": [{"split": "2023_09_18T13_16_25.972049", "path": ["**/details_harness|hellaswag|10_2023-09-18T13-16-25.972049.parquet"]}, {"split": "latest", "path": ["**/details_harness|hellaswag|10_2023-09-18T13-16-25.972049.parquet"]}]}, {"config_name": "harness_hendrycksTest_5", "data_files": [{"split": "2023_09_18T13_16_25.972049", "path": ["**/details_harness|hendrycksTest-abstract_algebra|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-anatomy|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-astronomy|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-business_ethics|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-clinical_knowledge|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-college_biology|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-college_chemistry|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-college_computer_science|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-college_mathematics|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-college_medicine|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-college_physics|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-computer_security|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-conceptual_physics|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-econometrics|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-electrical_engineering|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-elementary_mathematics|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-formal_logic|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-global_facts|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-high_school_biology|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-high_school_chemistry|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-high_school_computer_science|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-high_school_european_history|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-high_school_geography|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-high_school_government_and_politics|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-high_school_macroeconomics|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-high_school_mathematics|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-high_school_microeconomics|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-high_school_physics|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-high_school_psychology|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-high_school_statistics|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-high_school_us_history|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-high_school_world_history|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-human_aging|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-human_sexuality|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-international_law|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-jurisprudence|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-logical_fallacies|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-machine_learning|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-management|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-marketing|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-medical_genetics|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-miscellaneous|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-moral_disputes|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-moral_scenarios|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-nutrition|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-philosophy|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-prehistory|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-professional_accounting|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-professional_law|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-professional_medicine|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-professional_psychology|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-public_relations|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-security_studies|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-sociology|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-us_foreign_policy|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-virology|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-world_religions|5_2023-09-18T13-16-25.972049.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-abstract_algebra|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-anatomy|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-astronomy|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-business_ethics|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-clinical_knowledge|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-college_biology|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-college_chemistry|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-college_computer_science|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-college_mathematics|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-college_medicine|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-college_physics|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-computer_security|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-conceptual_physics|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-econometrics|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-electrical_engineering|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-elementary_mathematics|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-formal_logic|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-global_facts|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-high_school_biology|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-high_school_chemistry|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-high_school_computer_science|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-high_school_european_history|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-high_school_geography|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-high_school_government_and_politics|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-high_school_macroeconomics|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-high_school_mathematics|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-high_school_microeconomics|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-high_school_physics|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-high_school_psychology|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-high_school_statistics|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-high_school_us_history|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-high_school_world_history|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-human_aging|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-human_sexuality|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-international_law|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-jurisprudence|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-logical_fallacies|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-machine_learning|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-management|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-marketing|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-medical_genetics|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-miscellaneous|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-moral_disputes|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-moral_scenarios|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-nutrition|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-philosophy|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-prehistory|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-professional_accounting|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-professional_law|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-professional_medicine|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-professional_psychology|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-public_relations|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-security_studies|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-sociology|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-us_foreign_policy|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-virology|5_2023-09-18T13-16-25.972049.parquet", "**/details_harness|hendrycksTest-world_religions|5_2023-09-18T13-16-25.972049.parquet"]}]}, {"config_name": "harness_hendrycksTest_abstract_algebra_5", "data_files": [{"split": "2023_09_18T13_16_25.972049", "path": ["**/details_harness|hendrycksTest-abstract_algebra|5_2023-09-18T13-16-25.972049.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-abstract_algebra|5_2023-09-18T13-16-25.972049.parquet"]}]}, {"config_name": "harness_hendrycksTest_anatomy_5", "data_files": [{"split": "2023_09_18T13_16_25.972049", "path": ["**/details_harness|hendrycksTest-anatomy|5_2023-09-18T13-16-25.972049.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-anatomy|5_2023-09-18T13-16-25.972049.parquet"]}]}, {"config_name": "harness_hendrycksTest_astronomy_5", "data_files": [{"split": "2023_09_18T13_16_25.972049", "path": ["**/details_harness|hendrycksTest-astronomy|5_2023-09-18T13-16-25.972049.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-astronomy|5_2023-09-18T13-16-25.972049.parquet"]}]}, {"config_name": "harness_hendrycksTest_business_ethics_5", "data_files": [{"split": "2023_09_18T13_16_25.972049", "path": ["**/details_harness|hendrycksTest-business_ethics|5_2023-09-18T13-16-25.972049.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-business_ethics|5_2023-09-18T13-16-25.972049.parquet"]}]}, {"config_name": "harness_hendrycksTest_clinical_knowledge_5", "data_files": [{"split": "2023_09_18T13_16_25.972049", "path": ["**/details_harness|hendrycksTest-clinical_knowledge|5_2023-09-18T13-16-25.972049.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-clinical_knowledge|5_2023-09-18T13-16-25.972049.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_biology_5", "data_files": [{"split": "2023_09_18T13_16_25.972049", "path": ["**/details_harness|hendrycksTest-college_biology|5_2023-09-18T13-16-25.972049.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_biology|5_2023-09-18T13-16-25.972049.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_chemistry_5", "data_files": [{"split": "2023_09_18T13_16_25.972049", "path": ["**/details_harness|hendrycksTest-college_chemistry|5_2023-09-18T13-16-25.972049.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_chemistry|5_2023-09-18T13-16-25.972049.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_computer_science_5", "data_files": [{"split": "2023_09_18T13_16_25.972049", "path": ["**/details_harness|hendrycksTest-college_computer_science|5_2023-09-18T13-16-25.972049.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_computer_science|5_2023-09-18T13-16-25.972049.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_mathematics_5", "data_files": [{"split": "2023_09_18T13_16_25.972049", "path": ["**/details_harness|hendrycksTest-college_mathematics|5_2023-09-18T13-16-25.972049.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_mathematics|5_2023-09-18T13-16-25.972049.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_medicine_5", "data_files": [{"split": "2023_09_18T13_16_25.972049", "path": ["**/details_harness|hendrycksTest-college_medicine|5_2023-09-18T13-16-25.972049.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_medicine|5_2023-09-18T13-16-25.972049.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_physics_5", "data_files": [{"split": "2023_09_18T13_16_25.972049", "path": ["**/details_harness|hendrycksTest-college_physics|5_2023-09-18T13-16-25.972049.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_physics|5_2023-09-18T13-16-25.972049.parquet"]}]}, {"config_name": "harness_hendrycksTest_computer_security_5", "data_files": [{"split": "2023_09_18T13_16_25.972049", "path": ["**/details_harness|hendrycksTest-computer_security|5_2023-09-18T13-16-25.972049.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-computer_security|5_2023-09-18T13-16-25.972049.parquet"]}]}, {"config_name": "harness_hendrycksTest_conceptual_physics_5", "data_files": [{"split": "2023_09_18T13_16_25.972049", "path": ["**/details_harness|hendrycksTest-conceptual_physics|5_2023-09-18T13-16-25.972049.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-conceptual_physics|5_2023-09-18T13-16-25.972049.parquet"]}]}, {"config_name": "harness_hendrycksTest_econometrics_5", "data_files": [{"split": "2023_09_18T13_16_25.972049", "path": ["**/details_harness|hendrycksTest-econometrics|5_2023-09-18T13-16-25.972049.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-econometrics|5_2023-09-18T13-16-25.972049.parquet"]}]}, {"config_name": "harness_hendrycksTest_electrical_engineering_5", "data_files": [{"split": "2023_09_18T13_16_25.972049", "path": ["**/details_harness|hendrycksTest-electrical_engineering|5_2023-09-18T13-16-25.972049.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-electrical_engineering|5_2023-09-18T13-16-25.972049.parquet"]}]}, {"config_name": "harness_hendrycksTest_elementary_mathematics_5", "data_files": [{"split": "2023_09_18T13_16_25.972049", "path": ["**/details_harness|hendrycksTest-elementary_mathematics|5_2023-09-18T13-16-25.972049.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-elementary_mathematics|5_2023-09-18T13-16-25.972049.parquet"]}]}, {"config_name": "harness_hendrycksTest_formal_logic_5", "data_files": [{"split": "2023_09_18T13_16_25.972049", "path": ["**/details_harness|hendrycksTest-formal_logic|5_2023-09-18T13-16-25.972049.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-formal_logic|5_2023-09-18T13-16-25.972049.parquet"]}]}, {"config_name": "harness_hendrycksTest_global_facts_5", "data_files": [{"split": "2023_09_18T13_16_25.972049", "path": ["**/details_harness|hendrycksTest-global_facts|5_2023-09-18T13-16-25.972049.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-global_facts|5_2023-09-18T13-16-25.972049.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_biology_5", "data_files": [{"split": "2023_09_18T13_16_25.972049", "path": ["**/details_harness|hendrycksTest-high_school_biology|5_2023-09-18T13-16-25.972049.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_biology|5_2023-09-18T13-16-25.972049.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_chemistry_5", "data_files": [{"split": "2023_09_18T13_16_25.972049", "path": ["**/details_harness|hendrycksTest-high_school_chemistry|5_2023-09-18T13-16-25.972049.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_chemistry|5_2023-09-18T13-16-25.972049.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_computer_science_5", "data_files": [{"split": "2023_09_18T13_16_25.972049", "path": ["**/details_harness|hendrycksTest-high_school_computer_science|5_2023-09-18T13-16-25.972049.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_computer_science|5_2023-09-18T13-16-25.972049.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_european_history_5", "data_files": [{"split": "2023_09_18T13_16_25.972049", "path": ["**/details_harness|hendrycksTest-high_school_european_history|5_2023-09-18T13-16-25.972049.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_european_history|5_2023-09-18T13-16-25.972049.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_geography_5", "data_files": [{"split": "2023_09_18T13_16_25.972049", "path": ["**/details_harness|hendrycksTest-high_school_geography|5_2023-09-18T13-16-25.972049.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_geography|5_2023-09-18T13-16-25.972049.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_government_and_politics_5", "data_files": [{"split": "2023_09_18T13_16_25.972049", "path": ["**/details_harness|hendrycksTest-high_school_government_and_politics|5_2023-09-18T13-16-25.972049.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_government_and_politics|5_2023-09-18T13-16-25.972049.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_macroeconomics_5", "data_files": [{"split": "2023_09_18T13_16_25.972049", "path": ["**/details_harness|hendrycksTest-high_school_macroeconomics|5_2023-09-18T13-16-25.972049.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_macroeconomics|5_2023-09-18T13-16-25.972049.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_mathematics_5", "data_files": [{"split": "2023_09_18T13_16_25.972049", "path": ["**/details_harness|hendrycksTest-high_school_mathematics|5_2023-09-18T13-16-25.972049.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_mathematics|5_2023-09-18T13-16-25.972049.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_microeconomics_5", "data_files": [{"split": "2023_09_18T13_16_25.972049", "path": ["**/details_harness|hendrycksTest-high_school_microeconomics|5_2023-09-18T13-16-25.972049.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_microeconomics|5_2023-09-18T13-16-25.972049.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_physics_5", "data_files": [{"split": "2023_09_18T13_16_25.972049", "path": ["**/details_harness|hendrycksTest-high_school_physics|5_2023-09-18T13-16-25.972049.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_physics|5_2023-09-18T13-16-25.972049.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_psychology_5", "data_files": [{"split": "2023_09_18T13_16_25.972049", "path": ["**/details_harness|hendrycksTest-high_school_psychology|5_2023-09-18T13-16-25.972049.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_psychology|5_2023-09-18T13-16-25.972049.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_statistics_5", "data_files": [{"split": "2023_09_18T13_16_25.972049", "path": ["**/details_harness|hendrycksTest-high_school_statistics|5_2023-09-18T13-16-25.972049.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_statistics|5_2023-09-18T13-16-25.972049.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_us_history_5", "data_files": [{"split": "2023_09_18T13_16_25.972049", "path": ["**/details_harness|hendrycksTest-high_school_us_history|5_2023-09-18T13-16-25.972049.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_us_history|5_2023-09-18T13-16-25.972049.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_world_history_5", "data_files": [{"split": "2023_09_18T13_16_25.972049", "path": ["**/details_harness|hendrycksTest-high_school_world_history|5_2023-09-18T13-16-25.972049.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_world_history|5_2023-09-18T13-16-25.972049.parquet"]}]}, {"config_name": "harness_hendrycksTest_human_aging_5", "data_files": [{"split": "2023_09_18T13_16_25.972049", "path": ["**/details_harness|hendrycksTest-human_aging|5_2023-09-18T13-16-25.972049.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-human_aging|5_2023-09-18T13-16-25.972049.parquet"]}]}, {"config_name": "harness_hendrycksTest_human_sexuality_5", "data_files": [{"split": "2023_09_18T13_16_25.972049", "path": ["**/details_harness|hendrycksTest-human_sexuality|5_2023-09-18T13-16-25.972049.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-human_sexuality|5_2023-09-18T13-16-25.972049.parquet"]}]}, {"config_name": "harness_hendrycksTest_international_law_5", "data_files": [{"split": "2023_09_18T13_16_25.972049", "path": ["**/details_harness|hendrycksTest-international_law|5_2023-09-18T13-16-25.972049.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-international_law|5_2023-09-18T13-16-25.972049.parquet"]}]}, {"config_name": "harness_hendrycksTest_jurisprudence_5", "data_files": [{"split": "2023_09_18T13_16_25.972049", "path": ["**/details_harness|hendrycksTest-jurisprudence|5_2023-09-18T13-16-25.972049.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-jurisprudence|5_2023-09-18T13-16-25.972049.parquet"]}]}, {"config_name": "harness_hendrycksTest_logical_fallacies_5", "data_files": [{"split": "2023_09_18T13_16_25.972049", "path": ["**/details_harness|hendrycksTest-logical_fallacies|5_2023-09-18T13-16-25.972049.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-logical_fallacies|5_2023-09-18T13-16-25.972049.parquet"]}]}, {"config_name": "harness_hendrycksTest_machine_learning_5", "data_files": [{"split": "2023_09_18T13_16_25.972049", "path": ["**/details_harness|hendrycksTest-machine_learning|5_2023-09-18T13-16-25.972049.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-machine_learning|5_2023-09-18T13-16-25.972049.parquet"]}]}, {"config_name": "harness_hendrycksTest_management_5", "data_files": [{"split": "2023_09_18T13_16_25.972049", "path": ["**/details_harness|hendrycksTest-management|5_2023-09-18T13-16-25.972049.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-management|5_2023-09-18T13-16-25.972049.parquet"]}]}, {"config_name": "harness_hendrycksTest_marketing_5", "data_files": [{"split": "2023_09_18T13_16_25.972049", "path": ["**/details_harness|hendrycksTest-marketing|5_2023-09-18T13-16-25.972049.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-marketing|5_2023-09-18T13-16-25.972049.parquet"]}]}, {"config_name": "harness_hendrycksTest_medical_genetics_5", "data_files": [{"split": "2023_09_18T13_16_25.972049", "path": ["**/details_harness|hendrycksTest-medical_genetics|5_2023-09-18T13-16-25.972049.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-medical_genetics|5_2023-09-18T13-16-25.972049.parquet"]}]}, {"config_name": "harness_hendrycksTest_miscellaneous_5", "data_files": [{"split": "2023_09_18T13_16_25.972049", "path": ["**/details_harness|hendrycksTest-miscellaneous|5_2023-09-18T13-16-25.972049.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-miscellaneous|5_2023-09-18T13-16-25.972049.parquet"]}]}, {"config_name": "harness_hendrycksTest_moral_disputes_5", "data_files": [{"split": "2023_09_18T13_16_25.972049", "path": ["**/details_harness|hendrycksTest-moral_disputes|5_2023-09-18T13-16-25.972049.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-moral_disputes|5_2023-09-18T13-16-25.972049.parquet"]}]}, {"config_name": "harness_hendrycksTest_moral_scenarios_5", "data_files": [{"split": "2023_09_18T13_16_25.972049", "path": ["**/details_harness|hendrycksTest-moral_scenarios|5_2023-09-18T13-16-25.972049.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-moral_scenarios|5_2023-09-18T13-16-25.972049.parquet"]}]}, {"config_name": "harness_hendrycksTest_nutrition_5", "data_files": [{"split": "2023_09_18T13_16_25.972049", "path": ["**/details_harness|hendrycksTest-nutrition|5_2023-09-18T13-16-25.972049.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-nutrition|5_2023-09-18T13-16-25.972049.parquet"]}]}, {"config_name": "harness_hendrycksTest_philosophy_5", "data_files": [{"split": "2023_09_18T13_16_25.972049", "path": ["**/details_harness|hendrycksTest-philosophy|5_2023-09-18T13-16-25.972049.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-philosophy|5_2023-09-18T13-16-25.972049.parquet"]}]}, {"config_name": "harness_hendrycksTest_prehistory_5", "data_files": [{"split": "2023_09_18T13_16_25.972049", "path": ["**/details_harness|hendrycksTest-prehistory|5_2023-09-18T13-16-25.972049.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-prehistory|5_2023-09-18T13-16-25.972049.parquet"]}]}, {"config_name": "harness_hendrycksTest_professional_accounting_5", "data_files": [{"split": "2023_09_18T13_16_25.972049", "path": ["**/details_harness|hendrycksTest-professional_accounting|5_2023-09-18T13-16-25.972049.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-professional_accounting|5_2023-09-18T13-16-25.972049.parquet"]}]}, {"config_name": "harness_hendrycksTest_professional_law_5", "data_files": [{"split": "2023_09_18T13_16_25.972049", "path": ["**/details_harness|hendrycksTest-professional_law|5_2023-09-18T13-16-25.972049.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-professional_law|5_2023-09-18T13-16-25.972049.parquet"]}]}, {"config_name": "harness_hendrycksTest_professional_medicine_5", "data_files": [{"split": "2023_09_18T13_16_25.972049", "path": ["**/details_harness|hendrycksTest-professional_medicine|5_2023-09-18T13-16-25.972049.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-professional_medicine|5_2023-09-18T13-16-25.972049.parquet"]}]}, {"config_name": "harness_hendrycksTest_professional_psychology_5", "data_files": [{"split": "2023_09_18T13_16_25.972049", "path": ["**/details_harness|hendrycksTest-professional_psychology|5_2023-09-18T13-16-25.972049.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-professional_psychology|5_2023-09-18T13-16-25.972049.parquet"]}]}, {"config_name": "harness_hendrycksTest_public_relations_5", "data_files": [{"split": "2023_09_18T13_16_25.972049", "path": ["**/details_harness|hendrycksTest-public_relations|5_2023-09-18T13-16-25.972049.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-public_relations|5_2023-09-18T13-16-25.972049.parquet"]}]}, {"config_name": "harness_hendrycksTest_security_studies_5", "data_files": [{"split": "2023_09_18T13_16_25.972049", "path": ["**/details_harness|hendrycksTest-security_studies|5_2023-09-18T13-16-25.972049.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-security_studies|5_2023-09-18T13-16-25.972049.parquet"]}]}, {"config_name": "harness_hendrycksTest_sociology_5", "data_files": [{"split": "2023_09_18T13_16_25.972049", "path": ["**/details_harness|hendrycksTest-sociology|5_2023-09-18T13-16-25.972049.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-sociology|5_2023-09-18T13-16-25.972049.parquet"]}]}, {"config_name": "harness_hendrycksTest_us_foreign_policy_5", "data_files": [{"split": "2023_09_18T13_16_25.972049", "path": ["**/details_harness|hendrycksTest-us_foreign_policy|5_2023-09-18T13-16-25.972049.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-us_foreign_policy|5_2023-09-18T13-16-25.972049.parquet"]}]}, {"config_name": "harness_hendrycksTest_virology_5", "data_files": [{"split": "2023_09_18T13_16_25.972049", "path": ["**/details_harness|hendrycksTest-virology|5_2023-09-18T13-16-25.972049.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-virology|5_2023-09-18T13-16-25.972049.parquet"]}]}, {"config_name": "harness_hendrycksTest_world_religions_5", "data_files": [{"split": "2023_09_18T13_16_25.972049", "path": ["**/details_harness|hendrycksTest-world_religions|5_2023-09-18T13-16-25.972049.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-world_religions|5_2023-09-18T13-16-25.972049.parquet"]}]}, {"config_name": "harness_truthfulqa_mc_0", "data_files": [{"split": "2023_09_18T13_16_25.972049", "path": ["**/details_harness|truthfulqa:mc|0_2023-09-18T13-16-25.972049.parquet"]}, {"split": "latest", "path": ["**/details_harness|truthfulqa:mc|0_2023-09-18T13-16-25.972049.parquet"]}]}, {"config_name": "harness_winogrande_5", "data_files": [{"split": "2023_10_25T23_58_38.907071", "path": ["**/details_harness|winogrande|5_2023-10-25T23-58-38.907071.parquet"]}, {"split": "latest", "path": ["**/details_harness|winogrande|5_2023-10-25T23-58-38.907071.parquet"]}]}, {"config_name": "results", "data_files": [{"split": "2023_09_18T13_16_25.972049", "path": ["results_2023-09-18T13-16-25.972049.parquet"]}, {"split": "2023_10_25T23_58_38.907071", "path": ["results_2023-10-25T23-58-38.907071.parquet"]}, {"split": "latest", "path": ["results_2023-10-25T23-58-38.907071.parquet"]}]}]}
|
2023-10-25T22:58:51+00:00
|
[] |
[] |
TAGS
#region-us
|
# Dataset Card for Evaluation run of Dampish/StellarX-4B-V0.2
## Dataset Description
- Homepage:
- Repository: URL
- Paper:
- Leaderboard: URL
- Point of Contact: clementine@URL
### Dataset Summary
Dataset automatically created during the evaluation run of model Dampish/StellarX-4B-V0.2 on the Open LLM Leaderboard.
The dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.
The dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The "train" split is always pointing to the latest results.
An additional configuration "results" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the Open LLM Leaderboard).
To load the details from a run, you can for instance do the following:
## Latest results
These are the latest results from run 2023-10-25T23:58:38.907071(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the "latest" split for each eval):
### Supported Tasks and Leaderboards
### Languages
## Dataset Structure
### Data Instances
### Data Fields
### Data Splits
## Dataset Creation
### Curation Rationale
### Source Data
#### Initial Data Collection and Normalization
#### Who are the source language producers?
### Annotations
#### Annotation process
#### Who are the annotators?
### Personal and Sensitive Information
## Considerations for Using the Data
### Social Impact of Dataset
### Discussion of Biases
### Other Known Limitations
## Additional Information
### Dataset Curators
### Licensing Information
### Contributions
|
[
"# Dataset Card for Evaluation run of Dampish/StellarX-4B-V0.2",
"## Dataset Description\n\n- Homepage: \n- Repository: URL\n- Paper: \n- Leaderboard: URL\n- Point of Contact: clementine@URL",
"### Dataset Summary\n\nDataset automatically created during the evaluation run of model Dampish/StellarX-4B-V0.2 on the Open LLM Leaderboard.\n\nThe dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.\n\nThe dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The \"train\" split is always pointing to the latest results.\n\nAn additional configuration \"results\" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the Open LLM Leaderboard).\n\nTo load the details from a run, you can for instance do the following:",
"## Latest results\n\nThese are the latest results from run 2023-10-25T23:58:38.907071(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the \"latest\" split for each eval):",
"### Supported Tasks and Leaderboards",
"### Languages",
"## Dataset Structure",
"### Data Instances",
"### Data Fields",
"### Data Splits",
"## Dataset Creation",
"### Curation Rationale",
"### Source Data",
"#### Initial Data Collection and Normalization",
"#### Who are the source language producers?",
"### Annotations",
"#### Annotation process",
"#### Who are the annotators?",
"### Personal and Sensitive Information",
"## Considerations for Using the Data",
"### Social Impact of Dataset",
"### Discussion of Biases",
"### Other Known Limitations",
"## Additional Information",
"### Dataset Curators",
"### Licensing Information",
"### Contributions"
] |
[
"TAGS\n#region-us \n",
"# Dataset Card for Evaluation run of Dampish/StellarX-4B-V0.2",
"## Dataset Description\n\n- Homepage: \n- Repository: URL\n- Paper: \n- Leaderboard: URL\n- Point of Contact: clementine@URL",
"### Dataset Summary\n\nDataset automatically created during the evaluation run of model Dampish/StellarX-4B-V0.2 on the Open LLM Leaderboard.\n\nThe dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.\n\nThe dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The \"train\" split is always pointing to the latest results.\n\nAn additional configuration \"results\" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the Open LLM Leaderboard).\n\nTo load the details from a run, you can for instance do the following:",
"## Latest results\n\nThese are the latest results from run 2023-10-25T23:58:38.907071(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the \"latest\" split for each eval):",
"### Supported Tasks and Leaderboards",
"### Languages",
"## Dataset Structure",
"### Data Instances",
"### Data Fields",
"### Data Splits",
"## Dataset Creation",
"### Curation Rationale",
"### Source Data",
"#### Initial Data Collection and Normalization",
"#### Who are the source language producers?",
"### Annotations",
"#### Annotation process",
"#### Who are the annotators?",
"### Personal and Sensitive Information",
"## Considerations for Using the Data",
"### Social Impact of Dataset",
"### Discussion of Biases",
"### Other Known Limitations",
"## Additional Information",
"### Dataset Curators",
"### Licensing Information",
"### Contributions"
] |
[
6,
21,
31,
169,
67,
10,
4,
6,
6,
5,
5,
5,
7,
4,
10,
10,
5,
5,
9,
8,
8,
7,
8,
7,
5,
6,
6,
5
] |
[
"passage: TAGS\n#region-us \n# Dataset Card for Evaluation run of Dampish/StellarX-4B-V0.2## Dataset Description\n\n- Homepage: \n- Repository: URL\n- Paper: \n- Leaderboard: URL\n- Point of Contact: clementine@URL### Dataset Summary\n\nDataset automatically created during the evaluation run of model Dampish/StellarX-4B-V0.2 on the Open LLM Leaderboard.\n\nThe dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.\n\nThe dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The \"train\" split is always pointing to the latest results.\n\nAn additional configuration \"results\" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the Open LLM Leaderboard).\n\nTo load the details from a run, you can for instance do the following:## Latest results\n\nThese are the latest results from run 2023-10-25T23:58:38.907071(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the \"latest\" split for each eval):### Supported Tasks and Leaderboards### Languages## Dataset Structure### Data Instances### Data Fields### Data Splits## Dataset Creation### Curation Rationale### Source Data#### Initial Data Collection and Normalization#### Who are the source language producers?### Annotations#### Annotation process#### Who are the annotators?### Personal and Sensitive Information## Considerations for Using the Data### Social Impact of Dataset### Discussion of Biases### Other Known Limitations## Additional Information### Dataset Curators### Licensing Information### Contributions"
] |
7e2ff1966868ddab3e0e502f63607b03414d3d5e
|
# Dataset Card for "bus_few4_80x"
[More Information needed](https://github.com/huggingface/datasets/blob/main/CONTRIBUTING.md#how-to-contribute-to-the-dataset-cards)
|
FanChen0116/bus_few4_80x
|
[
"region:us"
] |
2023-09-18T12:16:52+00:00
|
{"dataset_info": {"features": [{"name": "id", "dtype": "int64"}, {"name": "tokens", "sequence": "string"}, {"name": "labels", "sequence": {"class_label": {"names": {"0": "O", "1": "I-from_location", "2": "B-from_location", "3": "B-leaving_date", "4": "I-leaving_date", "5": "I-to_location", "6": "B-to_location"}}}}, {"name": "request_slot", "sequence": "string"}], "splits": [{"name": "train", "num_bytes": 1087354, "num_examples": 5600}, {"name": "validation", "num_bytes": 6900, "num_examples": 35}, {"name": "test", "num_bytes": 70618, "num_examples": 377}], "download_size": 0, "dataset_size": 1164872}}
|
2023-09-23T15:58:08+00:00
|
[] |
[] |
TAGS
#region-us
|
# Dataset Card for "bus_few4_80x"
More Information needed
|
[
"# Dataset Card for \"bus_few4_80x\"\n\nMore Information needed"
] |
[
"TAGS\n#region-us \n",
"# Dataset Card for \"bus_few4_80x\"\n\nMore Information needed"
] |
[
6,
18
] |
[
"passage: TAGS\n#region-us \n# Dataset Card for \"bus_few4_80x\"\n\nMore Information needed"
] |
032b99bec45ca5ac69331d2b70f00b6255aff1ab
|
# Dataset Card for "bus_few4_64x"
[More Information needed](https://github.com/huggingface/datasets/blob/main/CONTRIBUTING.md#how-to-contribute-to-the-dataset-cards)
|
FanChen0116/bus_few4_64x
|
[
"region:us"
] |
2023-09-18T12:17:34+00:00
|
{"dataset_info": {"features": [{"name": "id", "dtype": "int64"}, {"name": "tokens", "sequence": "string"}, {"name": "labels", "sequence": {"class_label": {"names": {"0": "O", "1": "I-from_location", "2": "B-from_location", "3": "B-leaving_date", "4": "I-leaving_date", "5": "I-to_location", "6": "B-to_location"}}}}, {"name": "request_slot", "sequence": "string"}], "splits": [{"name": "train", "num_bytes": 871876, "num_examples": 4480}, {"name": "validation", "num_bytes": 6900, "num_examples": 35}, {"name": "test", "num_bytes": 70618, "num_examples": 377}], "download_size": 131795, "dataset_size": 949394}}
|
2023-09-21T11:59:37+00:00
|
[] |
[] |
TAGS
#region-us
|
# Dataset Card for "bus_few4_64x"
More Information needed
|
[
"# Dataset Card for \"bus_few4_64x\"\n\nMore Information needed"
] |
[
"TAGS\n#region-us \n",
"# Dataset Card for \"bus_few4_64x\"\n\nMore Information needed"
] |
[
6,
18
] |
[
"passage: TAGS\n#region-us \n# Dataset Card for \"bus_few4_64x\"\n\nMore Information needed"
] |
28c0720f353f193fae2397a1f6afced2ea37c37a
|
# Dataset Card for "bus_few4_8x_empty"
[More Information needed](https://github.com/huggingface/datasets/blob/main/CONTRIBUTING.md#how-to-contribute-to-the-dataset-cards)
|
FanChen0116/bus_few4_8x_empty
|
[
"region:us"
] |
2023-09-18T12:18:26+00:00
|
{"dataset_info": {"features": [{"name": "id", "dtype": "int64"}, {"name": "tokens", "sequence": "string"}, {"name": "labels", "sequence": {"class_label": {"names": {"0": "O", "1": "I-from_location", "2": "B-from_location", "3": "B-leaving_date", "4": "I-leaving_date", "5": "I-to_location", "6": "B-to_location"}}}}, {"name": "request_slot", "sequence": "string"}], "splits": [{"name": "train", "num_bytes": 97021, "num_examples": 560}, {"name": "validation", "num_bytes": 6128, "num_examples": 35}, {"name": "test", "num_bytes": 70618, "num_examples": 377}], "download_size": 8558, "dataset_size": 173767}}
|
2023-09-20T12:31:29+00:00
|
[] |
[] |
TAGS
#region-us
|
# Dataset Card for "bus_few4_8x_empty"
More Information needed
|
[
"# Dataset Card for \"bus_few4_8x_empty\"\n\nMore Information needed"
] |
[
"TAGS\n#region-us \n",
"# Dataset Card for \"bus_few4_8x_empty\"\n\nMore Information needed"
] |
[
6,
21
] |
[
"passage: TAGS\n#region-us \n# Dataset Card for \"bus_few4_8x_empty\"\n\nMore Information needed"
] |
b4f8ff2fb06311192086f90a633d89529517bb47
|
# Dataset Card for "bus_few4_16x_empty"
[More Information needed](https://github.com/huggingface/datasets/blob/main/CONTRIBUTING.md#how-to-contribute-to-the-dataset-cards)
|
FanChen0116/bus_few4_16x_empty
|
[
"region:us"
] |
2023-09-18T12:18:39+00:00
|
{"dataset_info": {"features": [{"name": "id", "dtype": "int64"}, {"name": "tokens", "sequence": "string"}, {"name": "labels", "sequence": {"class_label": {"names": {"0": "O", "1": "I-from_location", "2": "B-from_location", "3": "B-leaving_date", "4": "I-leaving_date", "5": "I-to_location", "6": "B-to_location"}}}}, {"name": "request_slot", "sequence": "string"}], "splits": [{"name": "train", "num_bytes": 193483, "num_examples": 1120}, {"name": "validation", "num_bytes": 6128, "num_examples": 35}, {"name": "test", "num_bytes": 70618, "num_examples": 377}], "download_size": 0, "dataset_size": 270229}}
|
2023-09-27T06:29:11+00:00
|
[] |
[] |
TAGS
#region-us
|
# Dataset Card for "bus_few4_16x_empty"
More Information needed
|
[
"# Dataset Card for \"bus_few4_16x_empty\"\n\nMore Information needed"
] |
[
"TAGS\n#region-us \n",
"# Dataset Card for \"bus_few4_16x_empty\"\n\nMore Information needed"
] |
[
6,
21
] |
[
"passage: TAGS\n#region-us \n# Dataset Card for \"bus_few4_16x_empty\"\n\nMore Information needed"
] |
0f845c5f112b07ffe91ab20a68ef6cdc64f92941
|
# Dataset Card for "bus_few4_32x_empty"
[More Information needed](https://github.com/huggingface/datasets/blob/main/CONTRIBUTING.md#how-to-contribute-to-the-dataset-cards)
|
FanChen0116/bus_few4_32x_empty
|
[
"region:us"
] |
2023-09-18T12:18:52+00:00
|
{"dataset_info": {"features": [{"name": "id", "dtype": "int64"}, {"name": "tokens", "sequence": "string"}, {"name": "labels", "sequence": {"class_label": {"names": {"0": "O", "1": "I-from_location", "2": "B-from_location", "3": "B-leaving_date", "4": "I-leaving_date", "5": "I-to_location", "6": "B-to_location"}}}}, {"name": "request_slot", "sequence": "string"}], "splits": [{"name": "train", "num_bytes": 384184, "num_examples": 2240}, {"name": "validation", "num_bytes": 6128, "num_examples": 35}, {"name": "test", "num_bytes": 70618, "num_examples": 377}], "download_size": 0, "dataset_size": 460930}}
|
2023-09-27T00:49:53+00:00
|
[] |
[] |
TAGS
#region-us
|
# Dataset Card for "bus_few4_32x_empty"
More Information needed
|
[
"# Dataset Card for \"bus_few4_32x_empty\"\n\nMore Information needed"
] |
[
"TAGS\n#region-us \n",
"# Dataset Card for \"bus_few4_32x_empty\"\n\nMore Information needed"
] |
[
6,
21
] |
[
"passage: TAGS\n#region-us \n# Dataset Card for \"bus_few4_32x_empty\"\n\nMore Information needed"
] |
ae0c19c37fff90748fb2bff007ac662e14937ff2
|
# Dataset Card for "bus_few4_80x_empty"
[More Information needed](https://github.com/huggingface/datasets/blob/main/CONTRIBUTING.md#how-to-contribute-to-the-dataset-cards)
|
FanChen0116/bus_few4_80x_empty
|
[
"region:us"
] |
2023-09-18T12:19:18+00:00
|
{"dataset_info": {"features": [{"name": "id", "dtype": "int64"}, {"name": "tokens", "sequence": "string"}, {"name": "labels", "sequence": {"class_label": {"names": {"0": "O", "1": "I-from_location", "2": "B-from_location", "3": "B-leaving_date", "4": "I-leaving_date", "5": "I-to_location", "6": "B-to_location"}}}}, {"name": "request_slot", "sequence": "string"}], "splits": [{"name": "train", "num_bytes": 967883, "num_examples": 5600}, {"name": "validation", "num_bytes": 6128, "num_examples": 35}, {"name": "test", "num_bytes": 70618, "num_examples": 377}], "download_size": 0, "dataset_size": 1044629}}
|
2023-09-23T15:58:21+00:00
|
[] |
[] |
TAGS
#region-us
|
# Dataset Card for "bus_few4_80x_empty"
More Information needed
|
[
"# Dataset Card for \"bus_few4_80x_empty\"\n\nMore Information needed"
] |
[
"TAGS\n#region-us \n",
"# Dataset Card for \"bus_few4_80x_empty\"\n\nMore Information needed"
] |
[
6,
21
] |
[
"passage: TAGS\n#region-us \n# Dataset Card for \"bus_few4_80x_empty\"\n\nMore Information needed"
] |
73f99403e30e96b7a8bc1e4944325b4b4760f31d
|
# Dataset Card for "bus_few4_8x_pvi"
[More Information needed](https://github.com/huggingface/datasets/blob/main/CONTRIBUTING.md#how-to-contribute-to-the-dataset-cards)
|
FanChen0116/bus_few4_8x_pvi
|
[
"region:us"
] |
2023-09-18T12:20:44+00:00
|
{"dataset_info": {"features": [{"name": "id", "dtype": "int64"}, {"name": "tokens", "sequence": "string"}, {"name": "labels", "sequence": {"class_label": {"names": {"0": "O", "1": "I-from_location", "2": "B-from_location", "3": "B-leaving_date", "4": "I-leaving_date", "5": "I-to_location", "6": "B-to_location"}}}}, {"name": "request_slot", "sequence": "string"}], "splits": [{"name": "train", "num_bytes": 68839, "num_examples": 280}, {"name": "validation", "num_bytes": 6900, "num_examples": 35}, {"name": "test", "num_bytes": 70618, "num_examples": 377}], "download_size": 13438, "dataset_size": 146357}}
|
2023-09-23T16:27:11+00:00
|
[] |
[] |
TAGS
#region-us
|
# Dataset Card for "bus_few4_8x_pvi"
More Information needed
|
[
"# Dataset Card for \"bus_few4_8x_pvi\"\n\nMore Information needed"
] |
[
"TAGS\n#region-us \n",
"# Dataset Card for \"bus_few4_8x_pvi\"\n\nMore Information needed"
] |
[
6,
21
] |
[
"passage: TAGS\n#region-us \n# Dataset Card for \"bus_few4_8x_pvi\"\n\nMore Information needed"
] |
bcf1044d4b9c75191c15a2065a013028790a8620
|
# Dataset Card for "bus_few4_16x_pvi"
[More Information needed](https://github.com/huggingface/datasets/blob/main/CONTRIBUTING.md#how-to-contribute-to-the-dataset-cards)
|
FanChen0116/bus_few4_16x_pvi
|
[
"region:us"
] |
2023-09-18T12:20:57+00:00
|
{"dataset_info": {"features": [{"name": "id", "dtype": "int64"}, {"name": "tokens", "sequence": "string"}, {"name": "labels", "sequence": {"class_label": {"names": {"0": "O", "1": "I-from_location", "2": "B-from_location", "3": "B-leaving_date", "4": "I-leaving_date", "5": "I-to_location", "6": "B-to_location"}}}}, {"name": "request_slot", "sequence": "string"}], "splits": [{"name": "train", "num_bytes": 138287, "num_examples": 560}, {"name": "validation", "num_bytes": 6900, "num_examples": 35}, {"name": "test", "num_bytes": 70618, "num_examples": 377}], "download_size": 20540, "dataset_size": 215805}}
|
2023-09-27T07:41:25+00:00
|
[] |
[] |
TAGS
#region-us
|
# Dataset Card for "bus_few4_16x_pvi"
More Information needed
|
[
"# Dataset Card for \"bus_few4_16x_pvi\"\n\nMore Information needed"
] |
[
"TAGS\n#region-us \n",
"# Dataset Card for \"bus_few4_16x_pvi\"\n\nMore Information needed"
] |
[
6,
21
] |
[
"passage: TAGS\n#region-us \n# Dataset Card for \"bus_few4_16x_pvi\"\n\nMore Information needed"
] |
fb3ef2968c194ddf8ce1bcab571603de56b5153c
|
# Dataset Card for "bus_few4_32x_pvi"
[More Information needed](https://github.com/huggingface/datasets/blob/main/CONTRIBUTING.md#how-to-contribute-to-the-dataset-cards)
|
FanChen0116/bus_few4_32x_pvi
|
[
"region:us"
] |
2023-09-18T12:21:08+00:00
|
{"dataset_info": {"features": [{"name": "id", "dtype": "int64"}, {"name": "tokens", "sequence": "string"}, {"name": "labels", "sequence": {"class_label": {"names": {"0": "O", "1": "I-from_location", "2": "B-from_location", "3": "B-leaving_date", "4": "I-leaving_date", "5": "I-to_location", "6": "B-to_location"}}}}, {"name": "request_slot", "sequence": "string"}], "splits": [{"name": "train", "num_bytes": 273844, "num_examples": 1120}, {"name": "validation", "num_bytes": 6900, "num_examples": 35}, {"name": "test", "num_bytes": 70618, "num_examples": 377}], "download_size": 36847, "dataset_size": 351362}}
|
2023-09-27T02:26:08+00:00
|
[] |
[] |
TAGS
#region-us
|
# Dataset Card for "bus_few4_32x_pvi"
More Information needed
|
[
"# Dataset Card for \"bus_few4_32x_pvi\"\n\nMore Information needed"
] |
[
"TAGS\n#region-us \n",
"# Dataset Card for \"bus_few4_32x_pvi\"\n\nMore Information needed"
] |
[
6,
21
] |
[
"passage: TAGS\n#region-us \n# Dataset Card for \"bus_few4_32x_pvi\"\n\nMore Information needed"
] |
ba586440d68b0166a40f1fdeb1db06d34202abc3
|
# Dataset Card for "bus_few4_64x_pvi"
[More Information needed](https://github.com/huggingface/datasets/blob/main/CONTRIBUTING.md#how-to-contribute-to-the-dataset-cards)
|
FanChen0116/bus_few4_64x_pvi
|
[
"region:us"
] |
2023-09-18T12:31:07+00:00
|
{"dataset_info": {"features": [{"name": "id", "dtype": "int64"}, {"name": "tokens", "sequence": "string"}, {"name": "labels", "sequence": {"class_label": {"names": {"0": "O", "1": "I-from_location", "2": "B-from_location", "3": "B-leaving_date", "4": "I-leaving_date", "5": "I-to_location", "6": "B-to_location"}}}}, {"name": "request_slot", "sequence": "string"}], "splits": [{"name": "train", "num_bytes": 555679, "num_examples": 2240}, {"name": "validation", "num_bytes": 6900, "num_examples": 35}, {"name": "test", "num_bytes": 70618, "num_examples": 377}], "download_size": 67918, "dataset_size": 633197}}
|
2023-09-26T15:31:42+00:00
|
[] |
[] |
TAGS
#region-us
|
# Dataset Card for "bus_few4_64x_pvi"
More Information needed
|
[
"# Dataset Card for \"bus_few4_64x_pvi\"\n\nMore Information needed"
] |
[
"TAGS\n#region-us \n",
"# Dataset Card for \"bus_few4_64x_pvi\"\n\nMore Information needed"
] |
[
6,
21
] |
[
"passage: TAGS\n#region-us \n# Dataset Card for \"bus_few4_64x_pvi\"\n\nMore Information needed"
] |
33671ea51a08d73e1e2d80a7d74ac5a21308243c
|
# Dataset Card for "bus_few4_80x_pvi"
[More Information needed](https://github.com/huggingface/datasets/blob/main/CONTRIBUTING.md#how-to-contribute-to-the-dataset-cards)
|
FanChen0116/bus_few4_80x_pvi
|
[
"region:us"
] |
2023-09-18T12:31:17+00:00
|
{"dataset_info": {"features": [{"name": "id", "dtype": "int64"}, {"name": "tokens", "sequence": "string"}, {"name": "labels", "sequence": {"class_label": {"names": {"0": "O", "1": "I-from_location", "2": "B-from_location", "3": "B-leaving_date", "4": "I-leaving_date", "5": "I-to_location", "6": "B-to_location"}}}}, {"name": "request_slot", "sequence": "string"}], "splits": [{"name": "train", "num_bytes": 922303, "num_examples": 4480}, {"name": "validation", "num_bytes": 6900, "num_examples": 35}, {"name": "test", "num_bytes": 70618, "num_examples": 377}], "download_size": 104198, "dataset_size": 999821}}
|
2023-09-26T15:25:07+00:00
|
[] |
[] |
TAGS
#region-us
|
# Dataset Card for "bus_few4_80x_pvi"
More Information needed
|
[
"# Dataset Card for \"bus_few4_80x_pvi\"\n\nMore Information needed"
] |
[
"TAGS\n#region-us \n",
"# Dataset Card for \"bus_few4_80x_pvi\"\n\nMore Information needed"
] |
[
6,
21
] |
[
"passage: TAGS\n#region-us \n# Dataset Card for \"bus_few4_80x_pvi\"\n\nMore Information needed"
] |
07a61878508b773bb9a999a253ccc6e20dbe5b99
|
# Dataset Card for Evaluation run of PY007/TinyLlama-1.1B-intermediate-step-240k-503b
## Dataset Description
- **Homepage:**
- **Repository:** https://huggingface.co/PY007/TinyLlama-1.1B-intermediate-step-240k-503b
- **Paper:**
- **Leaderboard:** https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard
- **Point of Contact:** [email protected]
### Dataset Summary
Dataset automatically created during the evaluation run of model [PY007/TinyLlama-1.1B-intermediate-step-240k-503b](https://huggingface.co/PY007/TinyLlama-1.1B-intermediate-step-240k-503b) on the [Open LLM Leaderboard](https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard).
The dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.
The dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The "train" split is always pointing to the latest results.
An additional configuration "results" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the [Open LLM Leaderboard](https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard)).
To load the details from a run, you can for instance do the following:
```python
from datasets import load_dataset
data = load_dataset("open-llm-leaderboard/details_PY007__TinyLlama-1.1B-intermediate-step-240k-503b",
"harness_winogrande_5",
split="train")
```
## Latest results
These are the [latest results from run 2023-10-28T05:32:33.745725](https://huggingface.co/datasets/open-llm-leaderboard/details_PY007__TinyLlama-1.1B-intermediate-step-240k-503b/blob/main/results_2023-10-28T05-32-33.745725.json)(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the "latest" split for each eval):
```python
{
"all": {
"em": 0.0019924496644295304,
"em_stderr": 0.00045666764626669333,
"f1": 0.04375419463087258,
"f1_stderr": 0.0012232801051450955,
"acc": 0.2844681550025042,
"acc_stderr": 0.007722228058459302
},
"harness|drop|3": {
"em": 0.0019924496644295304,
"em_stderr": 0.00045666764626669333,
"f1": 0.04375419463087258,
"f1_stderr": 0.0012232801051450955
},
"harness|gsm8k|5": {
"acc": 0.003032600454890068,
"acc_stderr": 0.0015145735612245499
},
"harness|winogrande|5": {
"acc": 0.5659037095501184,
"acc_stderr": 0.013929882555694054
}
}
```
### Supported Tasks and Leaderboards
[More Information Needed]
### Languages
[More Information Needed]
## Dataset Structure
### Data Instances
[More Information Needed]
### Data Fields
[More Information Needed]
### Data Splits
[More Information Needed]
## Dataset Creation
### Curation Rationale
[More Information Needed]
### Source Data
#### Initial Data Collection and Normalization
[More Information Needed]
#### Who are the source language producers?
[More Information Needed]
### Annotations
#### Annotation process
[More Information Needed]
#### Who are the annotators?
[More Information Needed]
### Personal and Sensitive Information
[More Information Needed]
## Considerations for Using the Data
### Social Impact of Dataset
[More Information Needed]
### Discussion of Biases
[More Information Needed]
### Other Known Limitations
[More Information Needed]
## Additional Information
### Dataset Curators
[More Information Needed]
### Licensing Information
[More Information Needed]
### Citation Information
[More Information Needed]
### Contributions
[More Information Needed]
|
open-llm-leaderboard/details_PY007__TinyLlama-1.1B-intermediate-step-240k-503b
|
[
"region:us"
] |
2023-09-18T12:32:02+00:00
|
{"pretty_name": "Evaluation run of PY007/TinyLlama-1.1B-intermediate-step-240k-503b", "dataset_summary": "Dataset automatically created during the evaluation run of model [PY007/TinyLlama-1.1B-intermediate-step-240k-503b](https://huggingface.co/PY007/TinyLlama-1.1B-intermediate-step-240k-503b) on the [Open LLM Leaderboard](https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard).\n\nThe dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.\n\nThe dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The \"train\" split is always pointing to the latest results.\n\nAn additional configuration \"results\" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the [Open LLM Leaderboard](https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard)).\n\nTo load the details from a run, you can for instance do the following:\n```python\nfrom datasets import load_dataset\ndata = load_dataset(\"open-llm-leaderboard/details_PY007__TinyLlama-1.1B-intermediate-step-240k-503b\",\n\t\"harness_winogrande_5\",\n\tsplit=\"train\")\n```\n\n## Latest results\n\nThese are the [latest results from run 2023-10-28T05:32:33.745725](https://huggingface.co/datasets/open-llm-leaderboard/details_PY007__TinyLlama-1.1B-intermediate-step-240k-503b/blob/main/results_2023-10-28T05-32-33.745725.json)(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the \"latest\" split for each eval):\n\n```python\n{\n \"all\": {\n \"em\": 0.0019924496644295304,\n \"em_stderr\": 0.00045666764626669333,\n \"f1\": 0.04375419463087258,\n \"f1_stderr\": 0.0012232801051450955,\n \"acc\": 0.2844681550025042,\n \"acc_stderr\": 0.007722228058459302\n },\n \"harness|drop|3\": {\n \"em\": 0.0019924496644295304,\n \"em_stderr\": 0.00045666764626669333,\n \"f1\": 0.04375419463087258,\n \"f1_stderr\": 0.0012232801051450955\n },\n \"harness|gsm8k|5\": {\n \"acc\": 0.003032600454890068,\n \"acc_stderr\": 0.0015145735612245499\n },\n \"harness|winogrande|5\": {\n \"acc\": 0.5659037095501184,\n \"acc_stderr\": 0.013929882555694054\n }\n}\n```", "repo_url": "https://huggingface.co/PY007/TinyLlama-1.1B-intermediate-step-240k-503b", "leaderboard_url": "https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard", "point_of_contact": "[email protected]", "configs": [{"config_name": "harness_arc_challenge_25", "data_files": [{"split": "2023_09_18T13_31_42.519724", "path": ["**/details_harness|arc:challenge|25_2023-09-18T13-31-42.519724.parquet"]}, {"split": "latest", "path": ["**/details_harness|arc:challenge|25_2023-09-18T13-31-42.519724.parquet"]}]}, {"config_name": "harness_drop_3", "data_files": [{"split": "2023_10_28T05_32_33.745725", "path": ["**/details_harness|drop|3_2023-10-28T05-32-33.745725.parquet"]}, {"split": "latest", "path": ["**/details_harness|drop|3_2023-10-28T05-32-33.745725.parquet"]}]}, {"config_name": "harness_gsm8k_5", "data_files": [{"split": "2023_10_28T05_32_33.745725", "path": ["**/details_harness|gsm8k|5_2023-10-28T05-32-33.745725.parquet"]}, {"split": "latest", "path": ["**/details_harness|gsm8k|5_2023-10-28T05-32-33.745725.parquet"]}]}, {"config_name": "harness_hellaswag_10", "data_files": [{"split": "2023_09_18T13_31_42.519724", "path": ["**/details_harness|hellaswag|10_2023-09-18T13-31-42.519724.parquet"]}, {"split": "latest", "path": ["**/details_harness|hellaswag|10_2023-09-18T13-31-42.519724.parquet"]}]}, {"config_name": "harness_hendrycksTest_5", "data_files": [{"split": "2023_09_18T13_31_42.519724", "path": ["**/details_harness|hendrycksTest-abstract_algebra|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-anatomy|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-astronomy|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-business_ethics|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-clinical_knowledge|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-college_biology|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-college_chemistry|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-college_computer_science|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-college_mathematics|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-college_medicine|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-college_physics|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-computer_security|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-conceptual_physics|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-econometrics|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-electrical_engineering|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-elementary_mathematics|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-formal_logic|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-global_facts|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-high_school_biology|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-high_school_chemistry|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-high_school_computer_science|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-high_school_european_history|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-high_school_geography|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-high_school_government_and_politics|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-high_school_macroeconomics|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-high_school_mathematics|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-high_school_microeconomics|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-high_school_physics|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-high_school_psychology|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-high_school_statistics|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-high_school_us_history|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-high_school_world_history|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-human_aging|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-human_sexuality|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-international_law|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-jurisprudence|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-logical_fallacies|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-machine_learning|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-management|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-marketing|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-medical_genetics|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-miscellaneous|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-moral_disputes|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-moral_scenarios|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-nutrition|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-philosophy|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-prehistory|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-professional_accounting|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-professional_law|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-professional_medicine|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-professional_psychology|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-public_relations|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-security_studies|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-sociology|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-us_foreign_policy|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-virology|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-world_religions|5_2023-09-18T13-31-42.519724.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-abstract_algebra|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-anatomy|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-astronomy|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-business_ethics|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-clinical_knowledge|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-college_biology|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-college_chemistry|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-college_computer_science|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-college_mathematics|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-college_medicine|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-college_physics|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-computer_security|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-conceptual_physics|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-econometrics|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-electrical_engineering|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-elementary_mathematics|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-formal_logic|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-global_facts|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-high_school_biology|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-high_school_chemistry|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-high_school_computer_science|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-high_school_european_history|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-high_school_geography|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-high_school_government_and_politics|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-high_school_macroeconomics|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-high_school_mathematics|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-high_school_microeconomics|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-high_school_physics|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-high_school_psychology|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-high_school_statistics|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-high_school_us_history|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-high_school_world_history|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-human_aging|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-human_sexuality|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-international_law|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-jurisprudence|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-logical_fallacies|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-machine_learning|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-management|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-marketing|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-medical_genetics|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-miscellaneous|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-moral_disputes|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-moral_scenarios|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-nutrition|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-philosophy|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-prehistory|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-professional_accounting|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-professional_law|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-professional_medicine|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-professional_psychology|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-public_relations|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-security_studies|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-sociology|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-us_foreign_policy|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-virology|5_2023-09-18T13-31-42.519724.parquet", "**/details_harness|hendrycksTest-world_religions|5_2023-09-18T13-31-42.519724.parquet"]}]}, {"config_name": "harness_hendrycksTest_abstract_algebra_5", "data_files": [{"split": "2023_09_18T13_31_42.519724", "path": ["**/details_harness|hendrycksTest-abstract_algebra|5_2023-09-18T13-31-42.519724.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-abstract_algebra|5_2023-09-18T13-31-42.519724.parquet"]}]}, {"config_name": "harness_hendrycksTest_anatomy_5", "data_files": [{"split": "2023_09_18T13_31_42.519724", "path": ["**/details_harness|hendrycksTest-anatomy|5_2023-09-18T13-31-42.519724.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-anatomy|5_2023-09-18T13-31-42.519724.parquet"]}]}, {"config_name": "harness_hendrycksTest_astronomy_5", "data_files": [{"split": "2023_09_18T13_31_42.519724", "path": ["**/details_harness|hendrycksTest-astronomy|5_2023-09-18T13-31-42.519724.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-astronomy|5_2023-09-18T13-31-42.519724.parquet"]}]}, {"config_name": "harness_hendrycksTest_business_ethics_5", "data_files": [{"split": "2023_09_18T13_31_42.519724", "path": ["**/details_harness|hendrycksTest-business_ethics|5_2023-09-18T13-31-42.519724.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-business_ethics|5_2023-09-18T13-31-42.519724.parquet"]}]}, {"config_name": "harness_hendrycksTest_clinical_knowledge_5", "data_files": [{"split": "2023_09_18T13_31_42.519724", "path": ["**/details_harness|hendrycksTest-clinical_knowledge|5_2023-09-18T13-31-42.519724.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-clinical_knowledge|5_2023-09-18T13-31-42.519724.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_biology_5", "data_files": [{"split": "2023_09_18T13_31_42.519724", "path": ["**/details_harness|hendrycksTest-college_biology|5_2023-09-18T13-31-42.519724.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_biology|5_2023-09-18T13-31-42.519724.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_chemistry_5", "data_files": [{"split": "2023_09_18T13_31_42.519724", "path": ["**/details_harness|hendrycksTest-college_chemistry|5_2023-09-18T13-31-42.519724.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_chemistry|5_2023-09-18T13-31-42.519724.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_computer_science_5", "data_files": [{"split": "2023_09_18T13_31_42.519724", "path": ["**/details_harness|hendrycksTest-college_computer_science|5_2023-09-18T13-31-42.519724.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_computer_science|5_2023-09-18T13-31-42.519724.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_mathematics_5", "data_files": [{"split": "2023_09_18T13_31_42.519724", "path": ["**/details_harness|hendrycksTest-college_mathematics|5_2023-09-18T13-31-42.519724.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_mathematics|5_2023-09-18T13-31-42.519724.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_medicine_5", "data_files": [{"split": "2023_09_18T13_31_42.519724", "path": ["**/details_harness|hendrycksTest-college_medicine|5_2023-09-18T13-31-42.519724.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_medicine|5_2023-09-18T13-31-42.519724.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_physics_5", "data_files": [{"split": "2023_09_18T13_31_42.519724", "path": ["**/details_harness|hendrycksTest-college_physics|5_2023-09-18T13-31-42.519724.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_physics|5_2023-09-18T13-31-42.519724.parquet"]}]}, {"config_name": "harness_hendrycksTest_computer_security_5", "data_files": [{"split": "2023_09_18T13_31_42.519724", "path": ["**/details_harness|hendrycksTest-computer_security|5_2023-09-18T13-31-42.519724.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-computer_security|5_2023-09-18T13-31-42.519724.parquet"]}]}, {"config_name": "harness_hendrycksTest_conceptual_physics_5", "data_files": [{"split": "2023_09_18T13_31_42.519724", "path": ["**/details_harness|hendrycksTest-conceptual_physics|5_2023-09-18T13-31-42.519724.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-conceptual_physics|5_2023-09-18T13-31-42.519724.parquet"]}]}, {"config_name": "harness_hendrycksTest_econometrics_5", "data_files": [{"split": "2023_09_18T13_31_42.519724", "path": ["**/details_harness|hendrycksTest-econometrics|5_2023-09-18T13-31-42.519724.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-econometrics|5_2023-09-18T13-31-42.519724.parquet"]}]}, {"config_name": "harness_hendrycksTest_electrical_engineering_5", "data_files": [{"split": "2023_09_18T13_31_42.519724", "path": ["**/details_harness|hendrycksTest-electrical_engineering|5_2023-09-18T13-31-42.519724.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-electrical_engineering|5_2023-09-18T13-31-42.519724.parquet"]}]}, {"config_name": "harness_hendrycksTest_elementary_mathematics_5", "data_files": [{"split": "2023_09_18T13_31_42.519724", "path": ["**/details_harness|hendrycksTest-elementary_mathematics|5_2023-09-18T13-31-42.519724.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-elementary_mathematics|5_2023-09-18T13-31-42.519724.parquet"]}]}, {"config_name": "harness_hendrycksTest_formal_logic_5", "data_files": [{"split": "2023_09_18T13_31_42.519724", "path": ["**/details_harness|hendrycksTest-formal_logic|5_2023-09-18T13-31-42.519724.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-formal_logic|5_2023-09-18T13-31-42.519724.parquet"]}]}, {"config_name": "harness_hendrycksTest_global_facts_5", "data_files": [{"split": "2023_09_18T13_31_42.519724", "path": ["**/details_harness|hendrycksTest-global_facts|5_2023-09-18T13-31-42.519724.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-global_facts|5_2023-09-18T13-31-42.519724.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_biology_5", "data_files": [{"split": "2023_09_18T13_31_42.519724", "path": ["**/details_harness|hendrycksTest-high_school_biology|5_2023-09-18T13-31-42.519724.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_biology|5_2023-09-18T13-31-42.519724.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_chemistry_5", "data_files": [{"split": "2023_09_18T13_31_42.519724", "path": ["**/details_harness|hendrycksTest-high_school_chemistry|5_2023-09-18T13-31-42.519724.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_chemistry|5_2023-09-18T13-31-42.519724.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_computer_science_5", "data_files": [{"split": "2023_09_18T13_31_42.519724", "path": ["**/details_harness|hendrycksTest-high_school_computer_science|5_2023-09-18T13-31-42.519724.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_computer_science|5_2023-09-18T13-31-42.519724.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_european_history_5", "data_files": [{"split": "2023_09_18T13_31_42.519724", "path": ["**/details_harness|hendrycksTest-high_school_european_history|5_2023-09-18T13-31-42.519724.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_european_history|5_2023-09-18T13-31-42.519724.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_geography_5", "data_files": [{"split": "2023_09_18T13_31_42.519724", "path": ["**/details_harness|hendrycksTest-high_school_geography|5_2023-09-18T13-31-42.519724.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_geography|5_2023-09-18T13-31-42.519724.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_government_and_politics_5", "data_files": [{"split": "2023_09_18T13_31_42.519724", "path": ["**/details_harness|hendrycksTest-high_school_government_and_politics|5_2023-09-18T13-31-42.519724.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_government_and_politics|5_2023-09-18T13-31-42.519724.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_macroeconomics_5", "data_files": [{"split": "2023_09_18T13_31_42.519724", "path": ["**/details_harness|hendrycksTest-high_school_macroeconomics|5_2023-09-18T13-31-42.519724.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_macroeconomics|5_2023-09-18T13-31-42.519724.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_mathematics_5", "data_files": [{"split": "2023_09_18T13_31_42.519724", "path": ["**/details_harness|hendrycksTest-high_school_mathematics|5_2023-09-18T13-31-42.519724.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_mathematics|5_2023-09-18T13-31-42.519724.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_microeconomics_5", "data_files": [{"split": "2023_09_18T13_31_42.519724", "path": ["**/details_harness|hendrycksTest-high_school_microeconomics|5_2023-09-18T13-31-42.519724.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_microeconomics|5_2023-09-18T13-31-42.519724.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_physics_5", "data_files": [{"split": "2023_09_18T13_31_42.519724", "path": ["**/details_harness|hendrycksTest-high_school_physics|5_2023-09-18T13-31-42.519724.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_physics|5_2023-09-18T13-31-42.519724.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_psychology_5", "data_files": [{"split": "2023_09_18T13_31_42.519724", "path": ["**/details_harness|hendrycksTest-high_school_psychology|5_2023-09-18T13-31-42.519724.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_psychology|5_2023-09-18T13-31-42.519724.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_statistics_5", "data_files": [{"split": "2023_09_18T13_31_42.519724", "path": ["**/details_harness|hendrycksTest-high_school_statistics|5_2023-09-18T13-31-42.519724.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_statistics|5_2023-09-18T13-31-42.519724.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_us_history_5", "data_files": [{"split": "2023_09_18T13_31_42.519724", "path": ["**/details_harness|hendrycksTest-high_school_us_history|5_2023-09-18T13-31-42.519724.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_us_history|5_2023-09-18T13-31-42.519724.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_world_history_5", "data_files": [{"split": "2023_09_18T13_31_42.519724", "path": ["**/details_harness|hendrycksTest-high_school_world_history|5_2023-09-18T13-31-42.519724.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_world_history|5_2023-09-18T13-31-42.519724.parquet"]}]}, {"config_name": "harness_hendrycksTest_human_aging_5", "data_files": [{"split": "2023_09_18T13_31_42.519724", "path": ["**/details_harness|hendrycksTest-human_aging|5_2023-09-18T13-31-42.519724.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-human_aging|5_2023-09-18T13-31-42.519724.parquet"]}]}, {"config_name": "harness_hendrycksTest_human_sexuality_5", "data_files": [{"split": "2023_09_18T13_31_42.519724", "path": ["**/details_harness|hendrycksTest-human_sexuality|5_2023-09-18T13-31-42.519724.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-human_sexuality|5_2023-09-18T13-31-42.519724.parquet"]}]}, {"config_name": "harness_hendrycksTest_international_law_5", "data_files": [{"split": "2023_09_18T13_31_42.519724", "path": ["**/details_harness|hendrycksTest-international_law|5_2023-09-18T13-31-42.519724.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-international_law|5_2023-09-18T13-31-42.519724.parquet"]}]}, {"config_name": "harness_hendrycksTest_jurisprudence_5", "data_files": [{"split": "2023_09_18T13_31_42.519724", "path": ["**/details_harness|hendrycksTest-jurisprudence|5_2023-09-18T13-31-42.519724.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-jurisprudence|5_2023-09-18T13-31-42.519724.parquet"]}]}, {"config_name": "harness_hendrycksTest_logical_fallacies_5", "data_files": [{"split": "2023_09_18T13_31_42.519724", "path": ["**/details_harness|hendrycksTest-logical_fallacies|5_2023-09-18T13-31-42.519724.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-logical_fallacies|5_2023-09-18T13-31-42.519724.parquet"]}]}, {"config_name": "harness_hendrycksTest_machine_learning_5", "data_files": [{"split": "2023_09_18T13_31_42.519724", "path": ["**/details_harness|hendrycksTest-machine_learning|5_2023-09-18T13-31-42.519724.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-machine_learning|5_2023-09-18T13-31-42.519724.parquet"]}]}, {"config_name": "harness_hendrycksTest_management_5", "data_files": [{"split": "2023_09_18T13_31_42.519724", "path": ["**/details_harness|hendrycksTest-management|5_2023-09-18T13-31-42.519724.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-management|5_2023-09-18T13-31-42.519724.parquet"]}]}, {"config_name": "harness_hendrycksTest_marketing_5", "data_files": [{"split": "2023_09_18T13_31_42.519724", "path": ["**/details_harness|hendrycksTest-marketing|5_2023-09-18T13-31-42.519724.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-marketing|5_2023-09-18T13-31-42.519724.parquet"]}]}, {"config_name": "harness_hendrycksTest_medical_genetics_5", "data_files": [{"split": "2023_09_18T13_31_42.519724", "path": ["**/details_harness|hendrycksTest-medical_genetics|5_2023-09-18T13-31-42.519724.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-medical_genetics|5_2023-09-18T13-31-42.519724.parquet"]}]}, {"config_name": "harness_hendrycksTest_miscellaneous_5", "data_files": [{"split": "2023_09_18T13_31_42.519724", "path": ["**/details_harness|hendrycksTest-miscellaneous|5_2023-09-18T13-31-42.519724.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-miscellaneous|5_2023-09-18T13-31-42.519724.parquet"]}]}, {"config_name": "harness_hendrycksTest_moral_disputes_5", "data_files": [{"split": "2023_09_18T13_31_42.519724", "path": ["**/details_harness|hendrycksTest-moral_disputes|5_2023-09-18T13-31-42.519724.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-moral_disputes|5_2023-09-18T13-31-42.519724.parquet"]}]}, {"config_name": "harness_hendrycksTest_moral_scenarios_5", "data_files": [{"split": "2023_09_18T13_31_42.519724", "path": ["**/details_harness|hendrycksTest-moral_scenarios|5_2023-09-18T13-31-42.519724.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-moral_scenarios|5_2023-09-18T13-31-42.519724.parquet"]}]}, {"config_name": "harness_hendrycksTest_nutrition_5", "data_files": [{"split": "2023_09_18T13_31_42.519724", "path": ["**/details_harness|hendrycksTest-nutrition|5_2023-09-18T13-31-42.519724.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-nutrition|5_2023-09-18T13-31-42.519724.parquet"]}]}, {"config_name": "harness_hendrycksTest_philosophy_5", "data_files": [{"split": "2023_09_18T13_31_42.519724", "path": ["**/details_harness|hendrycksTest-philosophy|5_2023-09-18T13-31-42.519724.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-philosophy|5_2023-09-18T13-31-42.519724.parquet"]}]}, {"config_name": "harness_hendrycksTest_prehistory_5", "data_files": [{"split": "2023_09_18T13_31_42.519724", "path": ["**/details_harness|hendrycksTest-prehistory|5_2023-09-18T13-31-42.519724.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-prehistory|5_2023-09-18T13-31-42.519724.parquet"]}]}, {"config_name": "harness_hendrycksTest_professional_accounting_5", "data_files": [{"split": "2023_09_18T13_31_42.519724", "path": ["**/details_harness|hendrycksTest-professional_accounting|5_2023-09-18T13-31-42.519724.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-professional_accounting|5_2023-09-18T13-31-42.519724.parquet"]}]}, {"config_name": "harness_hendrycksTest_professional_law_5", "data_files": [{"split": "2023_09_18T13_31_42.519724", "path": ["**/details_harness|hendrycksTest-professional_law|5_2023-09-18T13-31-42.519724.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-professional_law|5_2023-09-18T13-31-42.519724.parquet"]}]}, {"config_name": "harness_hendrycksTest_professional_medicine_5", "data_files": [{"split": "2023_09_18T13_31_42.519724", "path": ["**/details_harness|hendrycksTest-professional_medicine|5_2023-09-18T13-31-42.519724.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-professional_medicine|5_2023-09-18T13-31-42.519724.parquet"]}]}, {"config_name": "harness_hendrycksTest_professional_psychology_5", "data_files": [{"split": "2023_09_18T13_31_42.519724", "path": ["**/details_harness|hendrycksTest-professional_psychology|5_2023-09-18T13-31-42.519724.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-professional_psychology|5_2023-09-18T13-31-42.519724.parquet"]}]}, {"config_name": "harness_hendrycksTest_public_relations_5", "data_files": [{"split": "2023_09_18T13_31_42.519724", "path": ["**/details_harness|hendrycksTest-public_relations|5_2023-09-18T13-31-42.519724.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-public_relations|5_2023-09-18T13-31-42.519724.parquet"]}]}, {"config_name": "harness_hendrycksTest_security_studies_5", "data_files": [{"split": "2023_09_18T13_31_42.519724", "path": ["**/details_harness|hendrycksTest-security_studies|5_2023-09-18T13-31-42.519724.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-security_studies|5_2023-09-18T13-31-42.519724.parquet"]}]}, {"config_name": "harness_hendrycksTest_sociology_5", "data_files": [{"split": "2023_09_18T13_31_42.519724", "path": ["**/details_harness|hendrycksTest-sociology|5_2023-09-18T13-31-42.519724.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-sociology|5_2023-09-18T13-31-42.519724.parquet"]}]}, {"config_name": "harness_hendrycksTest_us_foreign_policy_5", "data_files": [{"split": "2023_09_18T13_31_42.519724", "path": ["**/details_harness|hendrycksTest-us_foreign_policy|5_2023-09-18T13-31-42.519724.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-us_foreign_policy|5_2023-09-18T13-31-42.519724.parquet"]}]}, {"config_name": "harness_hendrycksTest_virology_5", "data_files": [{"split": "2023_09_18T13_31_42.519724", "path": ["**/details_harness|hendrycksTest-virology|5_2023-09-18T13-31-42.519724.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-virology|5_2023-09-18T13-31-42.519724.parquet"]}]}, {"config_name": "harness_hendrycksTest_world_religions_5", "data_files": [{"split": "2023_09_18T13_31_42.519724", "path": ["**/details_harness|hendrycksTest-world_religions|5_2023-09-18T13-31-42.519724.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-world_religions|5_2023-09-18T13-31-42.519724.parquet"]}]}, {"config_name": "harness_truthfulqa_mc_0", "data_files": [{"split": "2023_09_18T13_31_42.519724", "path": ["**/details_harness|truthfulqa:mc|0_2023-09-18T13-31-42.519724.parquet"]}, {"split": "latest", "path": ["**/details_harness|truthfulqa:mc|0_2023-09-18T13-31-42.519724.parquet"]}]}, {"config_name": "harness_winogrande_5", "data_files": [{"split": "2023_10_28T05_32_33.745725", "path": ["**/details_harness|winogrande|5_2023-10-28T05-32-33.745725.parquet"]}, {"split": "latest", "path": ["**/details_harness|winogrande|5_2023-10-28T05-32-33.745725.parquet"]}]}, {"config_name": "results", "data_files": [{"split": "2023_09_18T13_31_42.519724", "path": ["results_2023-09-18T13-31-42.519724.parquet"]}, {"split": "2023_10_28T05_32_33.745725", "path": ["results_2023-10-28T05-32-33.745725.parquet"]}, {"split": "latest", "path": ["results_2023-10-28T05-32-33.745725.parquet"]}]}]}
|
2023-10-28T04:32:46+00:00
|
[] |
[] |
TAGS
#region-us
|
# Dataset Card for Evaluation run of PY007/TinyLlama-1.1B-intermediate-step-240k-503b
## Dataset Description
- Homepage:
- Repository: URL
- Paper:
- Leaderboard: URL
- Point of Contact: clementine@URL
### Dataset Summary
Dataset automatically created during the evaluation run of model PY007/TinyLlama-1.1B-intermediate-step-240k-503b on the Open LLM Leaderboard.
The dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.
The dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The "train" split is always pointing to the latest results.
An additional configuration "results" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the Open LLM Leaderboard).
To load the details from a run, you can for instance do the following:
## Latest results
These are the latest results from run 2023-10-28T05:32:33.745725(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the "latest" split for each eval):
### Supported Tasks and Leaderboards
### Languages
## Dataset Structure
### Data Instances
### Data Fields
### Data Splits
## Dataset Creation
### Curation Rationale
### Source Data
#### Initial Data Collection and Normalization
#### Who are the source language producers?
### Annotations
#### Annotation process
#### Who are the annotators?
### Personal and Sensitive Information
## Considerations for Using the Data
### Social Impact of Dataset
### Discussion of Biases
### Other Known Limitations
## Additional Information
### Dataset Curators
### Licensing Information
### Contributions
|
[
"# Dataset Card for Evaluation run of PY007/TinyLlama-1.1B-intermediate-step-240k-503b",
"## Dataset Description\n\n- Homepage: \n- Repository: URL\n- Paper: \n- Leaderboard: URL\n- Point of Contact: clementine@URL",
"### Dataset Summary\n\nDataset automatically created during the evaluation run of model PY007/TinyLlama-1.1B-intermediate-step-240k-503b on the Open LLM Leaderboard.\n\nThe dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.\n\nThe dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The \"train\" split is always pointing to the latest results.\n\nAn additional configuration \"results\" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the Open LLM Leaderboard).\n\nTo load the details from a run, you can for instance do the following:",
"## Latest results\n\nThese are the latest results from run 2023-10-28T05:32:33.745725(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the \"latest\" split for each eval):",
"### Supported Tasks and Leaderboards",
"### Languages",
"## Dataset Structure",
"### Data Instances",
"### Data Fields",
"### Data Splits",
"## Dataset Creation",
"### Curation Rationale",
"### Source Data",
"#### Initial Data Collection and Normalization",
"#### Who are the source language producers?",
"### Annotations",
"#### Annotation process",
"#### Who are the annotators?",
"### Personal and Sensitive Information",
"## Considerations for Using the Data",
"### Social Impact of Dataset",
"### Discussion of Biases",
"### Other Known Limitations",
"## Additional Information",
"### Dataset Curators",
"### Licensing Information",
"### Contributions"
] |
[
"TAGS\n#region-us \n",
"# Dataset Card for Evaluation run of PY007/TinyLlama-1.1B-intermediate-step-240k-503b",
"## Dataset Description\n\n- Homepage: \n- Repository: URL\n- Paper: \n- Leaderboard: URL\n- Point of Contact: clementine@URL",
"### Dataset Summary\n\nDataset automatically created during the evaluation run of model PY007/TinyLlama-1.1B-intermediate-step-240k-503b on the Open LLM Leaderboard.\n\nThe dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.\n\nThe dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The \"train\" split is always pointing to the latest results.\n\nAn additional configuration \"results\" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the Open LLM Leaderboard).\n\nTo load the details from a run, you can for instance do the following:",
"## Latest results\n\nThese are the latest results from run 2023-10-28T05:32:33.745725(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the \"latest\" split for each eval):",
"### Supported Tasks and Leaderboards",
"### Languages",
"## Dataset Structure",
"### Data Instances",
"### Data Fields",
"### Data Splits",
"## Dataset Creation",
"### Curation Rationale",
"### Source Data",
"#### Initial Data Collection and Normalization",
"#### Who are the source language producers?",
"### Annotations",
"#### Annotation process",
"#### Who are the annotators?",
"### Personal and Sensitive Information",
"## Considerations for Using the Data",
"### Social Impact of Dataset",
"### Discussion of Biases",
"### Other Known Limitations",
"## Additional Information",
"### Dataset Curators",
"### Licensing Information",
"### Contributions"
] |
[
6,
32,
31,
180,
66,
10,
4,
6,
6,
5,
5,
5,
7,
4,
10,
10,
5,
5,
9,
8,
8,
7,
8,
7,
5,
6,
6,
5
] |
[
"passage: TAGS\n#region-us \n# Dataset Card for Evaluation run of PY007/TinyLlama-1.1B-intermediate-step-240k-503b## Dataset Description\n\n- Homepage: \n- Repository: URL\n- Paper: \n- Leaderboard: URL\n- Point of Contact: clementine@URL### Dataset Summary\n\nDataset automatically created during the evaluation run of model PY007/TinyLlama-1.1B-intermediate-step-240k-503b on the Open LLM Leaderboard.\n\nThe dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.\n\nThe dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The \"train\" split is always pointing to the latest results.\n\nAn additional configuration \"results\" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the Open LLM Leaderboard).\n\nTo load the details from a run, you can for instance do the following:## Latest results\n\nThese are the latest results from run 2023-10-28T05:32:33.745725(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the \"latest\" split for each eval):### Supported Tasks and Leaderboards### Languages## Dataset Structure### Data Instances### Data Fields### Data Splits## Dataset Creation### Curation Rationale### Source Data#### Initial Data Collection and Normalization#### Who are the source language producers?### Annotations#### Annotation process#### Who are the annotators?### Personal and Sensitive Information## Considerations for Using the Data### Social Impact of Dataset### Discussion of Biases### Other Known Limitations## Additional Information### Dataset Curators### Licensing Information### Contributions"
] |
21e773afe3dc7b5a22463832b4dbc2509a098061
|
# Dataset Card for "gtzan_all_preprocessed"
[More Information needed](https://github.com/huggingface/datasets/blob/main/CONTRIBUTING.md#how-to-contribute-to-the-dataset-cards)
|
ssahir/gtzan_all_preprocessed
|
[
"region:us"
] |
2023-09-18T12:36:57+00:00
|
{"configs": [{"config_name": "default", "data_files": [{"split": "train", "path": "data/train-*"}, {"split": "test", "path": "data/test-*"}]}], "dataset_info": {"features": [{"name": "label", "dtype": {"class_label": {"names": {"0": "blues", "1": "classical", "2": "country", "3": "disco", "4": "hiphop", "5": "jazz", "6": "metal", "7": "pop", "8": "reggae", "9": "rock"}}}}, {"name": "input_values", "sequence": "float32"}, {"name": "attention_mask", "sequence": "int32"}], "splits": [{"name": "train", "num_bytes": 3452159816, "num_examples": 899}, {"name": "test", "num_bytes": 384000696, "num_examples": 100}], "download_size": 1923103923, "dataset_size": 3836160512}}
|
2023-09-18T12:40:52+00:00
|
[] |
[] |
TAGS
#region-us
|
# Dataset Card for "gtzan_all_preprocessed"
More Information needed
|
[
"# Dataset Card for \"gtzan_all_preprocessed\"\n\nMore Information needed"
] |
[
"TAGS\n#region-us \n",
"# Dataset Card for \"gtzan_all_preprocessed\"\n\nMore Information needed"
] |
[
6,
18
] |
[
"passage: TAGS\n#region-us \n# Dataset Card for \"gtzan_all_preprocessed\"\n\nMore Information needed"
] |
bbd0e7f498e2be2f28a7227507e2bf1777907f91
|
# Dataset Card for Evaluation run of Undi95/MLewd-L2-Chat-13B
## Dataset Description
- **Homepage:**
- **Repository:** https://huggingface.co/Undi95/MLewd-L2-Chat-13B
- **Paper:**
- **Leaderboard:** https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard
- **Point of Contact:** [email protected]
### Dataset Summary
Dataset automatically created during the evaluation run of model [Undi95/MLewd-L2-Chat-13B](https://huggingface.co/Undi95/MLewd-L2-Chat-13B) on the [Open LLM Leaderboard](https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard).
The dataset is composed of 3 configuration, each one coresponding to one of the evaluated task.
The dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The "train" split is always pointing to the latest results.
An additional configuration "results" store all the aggregated results of the run (and is used to compute and display the aggregated metrics on the [Open LLM Leaderboard](https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard)).
To load the details from a run, you can for instance do the following:
```python
from datasets import load_dataset
data = load_dataset("open-llm-leaderboard/details_Undi95__MLewd-L2-Chat-13B_public",
"harness_winogrande_5",
split="train")
```
## Latest results
These are the [latest results from run 2023-11-07T04:02:20.497765](https://huggingface.co/datasets/open-llm-leaderboard/details_Undi95__MLewd-L2-Chat-13B_public/blob/main/results_2023-11-07T04-02-20.497765.json)(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the "latest" split for each eval):
```python
{
"all": {
"em": 0.039953859060402684,
"em_stderr": 0.0020056958276819816,
"f1": 0.12528313758389248,
"f1_stderr": 0.0025138994037981494,
"acc": 0.44361714795535834,
"acc_stderr": 0.010234482644867801
},
"harness|drop|3": {
"em": 0.039953859060402684,
"em_stderr": 0.0020056958276819816,
"f1": 0.12528313758389248,
"f1_stderr": 0.0025138994037981494
},
"harness|gsm8k|5": {
"acc": 0.11296436694465505,
"acc_stderr": 0.008719339028833055
},
"harness|winogrande|5": {
"acc": 0.7742699289660616,
"acc_stderr": 0.011749626260902545
}
}
```
### Supported Tasks and Leaderboards
[More Information Needed]
### Languages
[More Information Needed]
## Dataset Structure
### Data Instances
[More Information Needed]
### Data Fields
[More Information Needed]
### Data Splits
[More Information Needed]
## Dataset Creation
### Curation Rationale
[More Information Needed]
### Source Data
#### Initial Data Collection and Normalization
[More Information Needed]
#### Who are the source language producers?
[More Information Needed]
### Annotations
#### Annotation process
[More Information Needed]
#### Who are the annotators?
[More Information Needed]
### Personal and Sensitive Information
[More Information Needed]
## Considerations for Using the Data
### Social Impact of Dataset
[More Information Needed]
### Discussion of Biases
[More Information Needed]
### Other Known Limitations
[More Information Needed]
## Additional Information
### Dataset Curators
[More Information Needed]
### Licensing Information
[More Information Needed]
### Citation Information
[More Information Needed]
### Contributions
[More Information Needed]
|
open-llm-leaderboard/details_Undi95__MLewd-L2-Chat-13B
|
[
"region:us"
] |
2023-09-18T12:38:52+00:00
|
{"pretty_name": "Evaluation run of Undi95/MLewd-L2-Chat-13B", "dataset_summary": "Dataset automatically created during the evaluation run of model [Undi95/MLewd-L2-Chat-13B](https://huggingface.co/Undi95/MLewd-L2-Chat-13B) on the [Open LLM Leaderboard](https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard).\n\nThe dataset is composed of 3 configuration, each one coresponding to one of the evaluated task.\n\nThe dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The \"train\" split is always pointing to the latest results.\n\nAn additional configuration \"results\" store all the aggregated results of the run (and is used to compute and display the aggregated metrics on the [Open LLM Leaderboard](https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard)).\n\nTo load the details from a run, you can for instance do the following:\n```python\nfrom datasets import load_dataset\ndata = load_dataset(\"open-llm-leaderboard/details_Undi95__MLewd-L2-Chat-13B_public\",\n\t\"harness_winogrande_5\",\n\tsplit=\"train\")\n```\n\n## Latest results\n\nThese are the [latest results from run 2023-11-07T04:02:20.497765](https://huggingface.co/datasets/open-llm-leaderboard/details_Undi95__MLewd-L2-Chat-13B_public/blob/main/results_2023-11-07T04-02-20.497765.json)(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the \"latest\" split for each eval):\n\n```python\n{\n \"all\": {\n \"em\": 0.039953859060402684,\n \"em_stderr\": 0.0020056958276819816,\n \"f1\": 0.12528313758389248,\n \"f1_stderr\": 0.0025138994037981494,\n \"acc\": 0.44361714795535834,\n \"acc_stderr\": 0.010234482644867801\n },\n \"harness|drop|3\": {\n \"em\": 0.039953859060402684,\n \"em_stderr\": 0.0020056958276819816,\n \"f1\": 0.12528313758389248,\n \"f1_stderr\": 0.0025138994037981494\n },\n \"harness|gsm8k|5\": {\n \"acc\": 0.11296436694465505,\n \"acc_stderr\": 0.008719339028833055\n },\n \"harness|winogrande|5\": {\n \"acc\": 0.7742699289660616,\n \"acc_stderr\": 0.011749626260902545\n }\n}\n```", "repo_url": "https://huggingface.co/Undi95/MLewd-L2-Chat-13B", "leaderboard_url": "https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard", "point_of_contact": "[email protected]", "configs": [{"config_name": "harness_drop_3", "data_files": [{"split": "2023_11_05T00_36_15.205012", "path": ["**/details_harness|drop|3_2023-11-05T00-36-15.205012.parquet"]}, {"split": "2023_11_07T04_02_20.497765", "path": ["**/details_harness|drop|3_2023-11-07T04-02-20.497765.parquet"]}, {"split": "latest", "path": ["**/details_harness|drop|3_2023-11-07T04-02-20.497765.parquet"]}]}, {"config_name": "harness_gsm8k_5", "data_files": [{"split": "2023_11_05T00_36_15.205012", "path": ["**/details_harness|gsm8k|5_2023-11-05T00-36-15.205012.parquet"]}, {"split": "2023_11_07T04_02_20.497765", "path": ["**/details_harness|gsm8k|5_2023-11-07T04-02-20.497765.parquet"]}, {"split": "latest", "path": ["**/details_harness|gsm8k|5_2023-11-07T04-02-20.497765.parquet"]}]}, {"config_name": "harness_winogrande_5", "data_files": [{"split": "2023_11_05T00_36_15.205012", "path": ["**/details_harness|winogrande|5_2023-11-05T00-36-15.205012.parquet"]}, {"split": "2023_11_07T04_02_20.497765", "path": ["**/details_harness|winogrande|5_2023-11-07T04-02-20.497765.parquet"]}, {"split": "latest", "path": ["**/details_harness|winogrande|5_2023-11-07T04-02-20.497765.parquet"]}]}, {"config_name": "results", "data_files": [{"split": "2023_11_05T00_36_15.205012", "path": ["results_2023-11-05T00-36-15.205012.parquet"]}, {"split": "2023_11_07T04_02_20.497765", "path": ["results_2023-11-07T04-02-20.497765.parquet"]}, {"split": "latest", "path": ["results_2023-11-07T04-02-20.497765.parquet"]}]}]}
|
2023-12-01T14:12:01+00:00
|
[] |
[] |
TAGS
#region-us
|
# Dataset Card for Evaluation run of Undi95/MLewd-L2-Chat-13B
## Dataset Description
- Homepage:
- Repository: URL
- Paper:
- Leaderboard: URL
- Point of Contact: clementine@URL
### Dataset Summary
Dataset automatically created during the evaluation run of model Undi95/MLewd-L2-Chat-13B on the Open LLM Leaderboard.
The dataset is composed of 3 configuration, each one coresponding to one of the evaluated task.
The dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The "train" split is always pointing to the latest results.
An additional configuration "results" store all the aggregated results of the run (and is used to compute and display the aggregated metrics on the Open LLM Leaderboard).
To load the details from a run, you can for instance do the following:
## Latest results
These are the latest results from run 2023-11-07T04:02:20.497765(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the "latest" split for each eval):
### Supported Tasks and Leaderboards
### Languages
## Dataset Structure
### Data Instances
### Data Fields
### Data Splits
## Dataset Creation
### Curation Rationale
### Source Data
#### Initial Data Collection and Normalization
#### Who are the source language producers?
### Annotations
#### Annotation process
#### Who are the annotators?
### Personal and Sensitive Information
## Considerations for Using the Data
### Social Impact of Dataset
### Discussion of Biases
### Other Known Limitations
## Additional Information
### Dataset Curators
### Licensing Information
### Contributions
|
[
"# Dataset Card for Evaluation run of Undi95/MLewd-L2-Chat-13B",
"## Dataset Description\n\n- Homepage: \n- Repository: URL\n- Paper: \n- Leaderboard: URL\n- Point of Contact: clementine@URL",
"### Dataset Summary\n\nDataset automatically created during the evaluation run of model Undi95/MLewd-L2-Chat-13B on the Open LLM Leaderboard.\n\nThe dataset is composed of 3 configuration, each one coresponding to one of the evaluated task.\n\nThe dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The \"train\" split is always pointing to the latest results.\n\nAn additional configuration \"results\" store all the aggregated results of the run (and is used to compute and display the aggregated metrics on the Open LLM Leaderboard).\n\nTo load the details from a run, you can for instance do the following:",
"## Latest results\n\nThese are the latest results from run 2023-11-07T04:02:20.497765(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the \"latest\" split for each eval):",
"### Supported Tasks and Leaderboards",
"### Languages",
"## Dataset Structure",
"### Data Instances",
"### Data Fields",
"### Data Splits",
"## Dataset Creation",
"### Curation Rationale",
"### Source Data",
"#### Initial Data Collection and Normalization",
"#### Who are the source language producers?",
"### Annotations",
"#### Annotation process",
"#### Who are the annotators?",
"### Personal and Sensitive Information",
"## Considerations for Using the Data",
"### Social Impact of Dataset",
"### Discussion of Biases",
"### Other Known Limitations",
"## Additional Information",
"### Dataset Curators",
"### Licensing Information",
"### Contributions"
] |
[
"TAGS\n#region-us \n",
"# Dataset Card for Evaluation run of Undi95/MLewd-L2-Chat-13B",
"## Dataset Description\n\n- Homepage: \n- Repository: URL\n- Paper: \n- Leaderboard: URL\n- Point of Contact: clementine@URL",
"### Dataset Summary\n\nDataset automatically created during the evaluation run of model Undi95/MLewd-L2-Chat-13B on the Open LLM Leaderboard.\n\nThe dataset is composed of 3 configuration, each one coresponding to one of the evaluated task.\n\nThe dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The \"train\" split is always pointing to the latest results.\n\nAn additional configuration \"results\" store all the aggregated results of the run (and is used to compute and display the aggregated metrics on the Open LLM Leaderboard).\n\nTo load the details from a run, you can for instance do the following:",
"## Latest results\n\nThese are the latest results from run 2023-11-07T04:02:20.497765(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the \"latest\" split for each eval):",
"### Supported Tasks and Leaderboards",
"### Languages",
"## Dataset Structure",
"### Data Instances",
"### Data Fields",
"### Data Splits",
"## Dataset Creation",
"### Curation Rationale",
"### Source Data",
"#### Initial Data Collection and Normalization",
"#### Who are the source language producers?",
"### Annotations",
"#### Annotation process",
"#### Who are the annotators?",
"### Personal and Sensitive Information",
"## Considerations for Using the Data",
"### Social Impact of Dataset",
"### Discussion of Biases",
"### Other Known Limitations",
"## Additional Information",
"### Dataset Curators",
"### Licensing Information",
"### Contributions"
] |
[
6,
22,
31,
171,
66,
10,
4,
6,
6,
5,
5,
5,
7,
4,
10,
10,
5,
5,
9,
8,
8,
7,
8,
7,
5,
6,
6,
5
] |
[
"passage: TAGS\n#region-us \n# Dataset Card for Evaluation run of Undi95/MLewd-L2-Chat-13B## Dataset Description\n\n- Homepage: \n- Repository: URL\n- Paper: \n- Leaderboard: URL\n- Point of Contact: clementine@URL### Dataset Summary\n\nDataset automatically created during the evaluation run of model Undi95/MLewd-L2-Chat-13B on the Open LLM Leaderboard.\n\nThe dataset is composed of 3 configuration, each one coresponding to one of the evaluated task.\n\nThe dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The \"train\" split is always pointing to the latest results.\n\nAn additional configuration \"results\" store all the aggregated results of the run (and is used to compute and display the aggregated metrics on the Open LLM Leaderboard).\n\nTo load the details from a run, you can for instance do the following:## Latest results\n\nThese are the latest results from run 2023-11-07T04:02:20.497765(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the \"latest\" split for each eval):### Supported Tasks and Leaderboards### Languages## Dataset Structure### Data Instances### Data Fields### Data Splits## Dataset Creation### Curation Rationale### Source Data#### Initial Data Collection and Normalization#### Who are the source language producers?### Annotations#### Annotation process#### Who are the annotators?### Personal and Sensitive Information## Considerations for Using the Data### Social Impact of Dataset### Discussion of Biases### Other Known Limitations## Additional Information### Dataset Curators### Licensing Information### Contributions"
] |
3a48305689e9b774a90ff4b93b313f1dbb5284ff
|
# Dataset Card for Evaluation run of Undi95/ReMM-v2.1-L2-13B
## Dataset Description
- **Homepage:**
- **Repository:** https://huggingface.co/Undi95/ReMM-v2.1-L2-13B
- **Paper:**
- **Leaderboard:** https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard
- **Point of Contact:** [email protected]
### Dataset Summary
Dataset automatically created during the evaluation run of model [Undi95/ReMM-v2.1-L2-13B](https://huggingface.co/Undi95/ReMM-v2.1-L2-13B) on the [Open LLM Leaderboard](https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard).
The dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.
The dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The "train" split is always pointing to the latest results.
An additional configuration "results" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the [Open LLM Leaderboard](https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard)).
To load the details from a run, you can for instance do the following:
```python
from datasets import load_dataset
data = load_dataset("open-llm-leaderboard/details_Undi95__ReMM-v2.1-L2-13B",
"harness_winogrande_5",
split="train")
```
## Latest results
These are the [latest results from run 2023-10-28T01:20:40.320894](https://huggingface.co/datasets/open-llm-leaderboard/details_Undi95__ReMM-v2.1-L2-13B/blob/main/results_2023-10-28T01-20-40.320894.json)(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the "latest" split for each eval):
```python
{
"all": {
"em": 0.061556208053691275,
"em_stderr": 0.0024613859292232257,
"f1": 0.12617135067114038,
"f1_stderr": 0.002725179835134867,
"acc": 0.44332154720067884,
"acc_stderr": 0.010599334769481
},
"harness|drop|3": {
"em": 0.061556208053691275,
"em_stderr": 0.0024613859292232257,
"f1": 0.12617135067114038,
"f1_stderr": 0.002725179835134867
},
"harness|gsm8k|5": {
"acc": 0.12736921910538287,
"acc_stderr": 0.009183110326737822
},
"harness|winogrande|5": {
"acc": 0.7592738752959748,
"acc_stderr": 0.012015559212224178
}
}
```
### Supported Tasks and Leaderboards
[More Information Needed]
### Languages
[More Information Needed]
## Dataset Structure
### Data Instances
[More Information Needed]
### Data Fields
[More Information Needed]
### Data Splits
[More Information Needed]
## Dataset Creation
### Curation Rationale
[More Information Needed]
### Source Data
#### Initial Data Collection and Normalization
[More Information Needed]
#### Who are the source language producers?
[More Information Needed]
### Annotations
#### Annotation process
[More Information Needed]
#### Who are the annotators?
[More Information Needed]
### Personal and Sensitive Information
[More Information Needed]
## Considerations for Using the Data
### Social Impact of Dataset
[More Information Needed]
### Discussion of Biases
[More Information Needed]
### Other Known Limitations
[More Information Needed]
## Additional Information
### Dataset Curators
[More Information Needed]
### Licensing Information
[More Information Needed]
### Citation Information
[More Information Needed]
### Contributions
[More Information Needed]
|
open-llm-leaderboard/details_Undi95__ReMM-v2.1-L2-13B
|
[
"region:us"
] |
2023-09-18T12:44:19+00:00
|
{"pretty_name": "Evaluation run of Undi95/ReMM-v2.1-L2-13B", "dataset_summary": "Dataset automatically created during the evaluation run of model [Undi95/ReMM-v2.1-L2-13B](https://huggingface.co/Undi95/ReMM-v2.1-L2-13B) on the [Open LLM Leaderboard](https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard).\n\nThe dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.\n\nThe dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The \"train\" split is always pointing to the latest results.\n\nAn additional configuration \"results\" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the [Open LLM Leaderboard](https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard)).\n\nTo load the details from a run, you can for instance do the following:\n```python\nfrom datasets import load_dataset\ndata = load_dataset(\"open-llm-leaderboard/details_Undi95__ReMM-v2.1-L2-13B\",\n\t\"harness_winogrande_5\",\n\tsplit=\"train\")\n```\n\n## Latest results\n\nThese are the [latest results from run 2023-10-28T01:20:40.320894](https://huggingface.co/datasets/open-llm-leaderboard/details_Undi95__ReMM-v2.1-L2-13B/blob/main/results_2023-10-28T01-20-40.320894.json)(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the \"latest\" split for each eval):\n\n```python\n{\n \"all\": {\n \"em\": 0.061556208053691275,\n \"em_stderr\": 0.0024613859292232257,\n \"f1\": 0.12617135067114038,\n \"f1_stderr\": 0.002725179835134867,\n \"acc\": 0.44332154720067884,\n \"acc_stderr\": 0.010599334769481\n },\n \"harness|drop|3\": {\n \"em\": 0.061556208053691275,\n \"em_stderr\": 0.0024613859292232257,\n \"f1\": 0.12617135067114038,\n \"f1_stderr\": 0.002725179835134867\n },\n \"harness|gsm8k|5\": {\n \"acc\": 0.12736921910538287,\n \"acc_stderr\": 0.009183110326737822\n },\n \"harness|winogrande|5\": {\n \"acc\": 0.7592738752959748,\n \"acc_stderr\": 0.012015559212224178\n }\n}\n```", "repo_url": "https://huggingface.co/Undi95/ReMM-v2.1-L2-13B", "leaderboard_url": "https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard", "point_of_contact": "[email protected]", "configs": [{"config_name": "harness_arc_challenge_25", "data_files": [{"split": "2023_09_18T13_43_56.304128", "path": ["**/details_harness|arc:challenge|25_2023-09-18T13-43-56.304128.parquet"]}, {"split": "latest", "path": ["**/details_harness|arc:challenge|25_2023-09-18T13-43-56.304128.parquet"]}]}, {"config_name": "harness_drop_3", "data_files": [{"split": "2023_10_28T01_20_40.320894", "path": ["**/details_harness|drop|3_2023-10-28T01-20-40.320894.parquet"]}, {"split": "latest", "path": ["**/details_harness|drop|3_2023-10-28T01-20-40.320894.parquet"]}]}, {"config_name": "harness_gsm8k_5", "data_files": [{"split": "2023_10_28T01_20_40.320894", "path": ["**/details_harness|gsm8k|5_2023-10-28T01-20-40.320894.parquet"]}, {"split": "latest", "path": ["**/details_harness|gsm8k|5_2023-10-28T01-20-40.320894.parquet"]}]}, {"config_name": "harness_hellaswag_10", "data_files": [{"split": "2023_09_18T13_43_56.304128", "path": ["**/details_harness|hellaswag|10_2023-09-18T13-43-56.304128.parquet"]}, {"split": "latest", "path": ["**/details_harness|hellaswag|10_2023-09-18T13-43-56.304128.parquet"]}]}, {"config_name": "harness_hendrycksTest_5", "data_files": [{"split": "2023_09_18T13_43_56.304128", "path": ["**/details_harness|hendrycksTest-abstract_algebra|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-anatomy|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-astronomy|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-business_ethics|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-clinical_knowledge|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-college_biology|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-college_chemistry|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-college_computer_science|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-college_mathematics|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-college_medicine|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-college_physics|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-computer_security|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-conceptual_physics|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-econometrics|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-electrical_engineering|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-elementary_mathematics|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-formal_logic|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-global_facts|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-high_school_biology|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-high_school_chemistry|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-high_school_computer_science|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-high_school_european_history|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-high_school_geography|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-high_school_government_and_politics|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-high_school_macroeconomics|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-high_school_mathematics|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-high_school_microeconomics|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-high_school_physics|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-high_school_psychology|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-high_school_statistics|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-high_school_us_history|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-high_school_world_history|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-human_aging|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-human_sexuality|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-international_law|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-jurisprudence|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-logical_fallacies|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-machine_learning|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-management|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-marketing|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-medical_genetics|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-miscellaneous|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-moral_disputes|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-moral_scenarios|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-nutrition|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-philosophy|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-prehistory|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-professional_accounting|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-professional_law|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-professional_medicine|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-professional_psychology|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-public_relations|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-security_studies|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-sociology|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-us_foreign_policy|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-virology|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-world_religions|5_2023-09-18T13-43-56.304128.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-abstract_algebra|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-anatomy|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-astronomy|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-business_ethics|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-clinical_knowledge|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-college_biology|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-college_chemistry|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-college_computer_science|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-college_mathematics|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-college_medicine|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-college_physics|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-computer_security|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-conceptual_physics|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-econometrics|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-electrical_engineering|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-elementary_mathematics|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-formal_logic|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-global_facts|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-high_school_biology|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-high_school_chemistry|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-high_school_computer_science|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-high_school_european_history|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-high_school_geography|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-high_school_government_and_politics|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-high_school_macroeconomics|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-high_school_mathematics|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-high_school_microeconomics|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-high_school_physics|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-high_school_psychology|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-high_school_statistics|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-high_school_us_history|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-high_school_world_history|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-human_aging|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-human_sexuality|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-international_law|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-jurisprudence|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-logical_fallacies|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-machine_learning|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-management|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-marketing|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-medical_genetics|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-miscellaneous|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-moral_disputes|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-moral_scenarios|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-nutrition|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-philosophy|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-prehistory|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-professional_accounting|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-professional_law|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-professional_medicine|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-professional_psychology|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-public_relations|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-security_studies|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-sociology|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-us_foreign_policy|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-virology|5_2023-09-18T13-43-56.304128.parquet", "**/details_harness|hendrycksTest-world_religions|5_2023-09-18T13-43-56.304128.parquet"]}]}, {"config_name": "harness_hendrycksTest_abstract_algebra_5", "data_files": [{"split": "2023_09_18T13_43_56.304128", "path": ["**/details_harness|hendrycksTest-abstract_algebra|5_2023-09-18T13-43-56.304128.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-abstract_algebra|5_2023-09-18T13-43-56.304128.parquet"]}]}, {"config_name": "harness_hendrycksTest_anatomy_5", "data_files": [{"split": "2023_09_18T13_43_56.304128", "path": ["**/details_harness|hendrycksTest-anatomy|5_2023-09-18T13-43-56.304128.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-anatomy|5_2023-09-18T13-43-56.304128.parquet"]}]}, {"config_name": "harness_hendrycksTest_astronomy_5", "data_files": [{"split": "2023_09_18T13_43_56.304128", "path": ["**/details_harness|hendrycksTest-astronomy|5_2023-09-18T13-43-56.304128.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-astronomy|5_2023-09-18T13-43-56.304128.parquet"]}]}, {"config_name": "harness_hendrycksTest_business_ethics_5", "data_files": [{"split": "2023_09_18T13_43_56.304128", "path": ["**/details_harness|hendrycksTest-business_ethics|5_2023-09-18T13-43-56.304128.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-business_ethics|5_2023-09-18T13-43-56.304128.parquet"]}]}, {"config_name": "harness_hendrycksTest_clinical_knowledge_5", "data_files": [{"split": "2023_09_18T13_43_56.304128", "path": ["**/details_harness|hendrycksTest-clinical_knowledge|5_2023-09-18T13-43-56.304128.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-clinical_knowledge|5_2023-09-18T13-43-56.304128.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_biology_5", "data_files": [{"split": "2023_09_18T13_43_56.304128", "path": ["**/details_harness|hendrycksTest-college_biology|5_2023-09-18T13-43-56.304128.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_biology|5_2023-09-18T13-43-56.304128.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_chemistry_5", "data_files": [{"split": "2023_09_18T13_43_56.304128", "path": ["**/details_harness|hendrycksTest-college_chemistry|5_2023-09-18T13-43-56.304128.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_chemistry|5_2023-09-18T13-43-56.304128.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_computer_science_5", "data_files": [{"split": "2023_09_18T13_43_56.304128", "path": ["**/details_harness|hendrycksTest-college_computer_science|5_2023-09-18T13-43-56.304128.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_computer_science|5_2023-09-18T13-43-56.304128.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_mathematics_5", "data_files": [{"split": "2023_09_18T13_43_56.304128", "path": ["**/details_harness|hendrycksTest-college_mathematics|5_2023-09-18T13-43-56.304128.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_mathematics|5_2023-09-18T13-43-56.304128.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_medicine_5", "data_files": [{"split": "2023_09_18T13_43_56.304128", "path": ["**/details_harness|hendrycksTest-college_medicine|5_2023-09-18T13-43-56.304128.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_medicine|5_2023-09-18T13-43-56.304128.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_physics_5", "data_files": [{"split": "2023_09_18T13_43_56.304128", "path": ["**/details_harness|hendrycksTest-college_physics|5_2023-09-18T13-43-56.304128.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_physics|5_2023-09-18T13-43-56.304128.parquet"]}]}, {"config_name": "harness_hendrycksTest_computer_security_5", "data_files": [{"split": "2023_09_18T13_43_56.304128", "path": ["**/details_harness|hendrycksTest-computer_security|5_2023-09-18T13-43-56.304128.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-computer_security|5_2023-09-18T13-43-56.304128.parquet"]}]}, {"config_name": "harness_hendrycksTest_conceptual_physics_5", "data_files": [{"split": "2023_09_18T13_43_56.304128", "path": ["**/details_harness|hendrycksTest-conceptual_physics|5_2023-09-18T13-43-56.304128.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-conceptual_physics|5_2023-09-18T13-43-56.304128.parquet"]}]}, {"config_name": "harness_hendrycksTest_econometrics_5", "data_files": [{"split": "2023_09_18T13_43_56.304128", "path": ["**/details_harness|hendrycksTest-econometrics|5_2023-09-18T13-43-56.304128.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-econometrics|5_2023-09-18T13-43-56.304128.parquet"]}]}, {"config_name": "harness_hendrycksTest_electrical_engineering_5", "data_files": [{"split": "2023_09_18T13_43_56.304128", "path": ["**/details_harness|hendrycksTest-electrical_engineering|5_2023-09-18T13-43-56.304128.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-electrical_engineering|5_2023-09-18T13-43-56.304128.parquet"]}]}, {"config_name": "harness_hendrycksTest_elementary_mathematics_5", "data_files": [{"split": "2023_09_18T13_43_56.304128", "path": ["**/details_harness|hendrycksTest-elementary_mathematics|5_2023-09-18T13-43-56.304128.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-elementary_mathematics|5_2023-09-18T13-43-56.304128.parquet"]}]}, {"config_name": "harness_hendrycksTest_formal_logic_5", "data_files": [{"split": "2023_09_18T13_43_56.304128", "path": ["**/details_harness|hendrycksTest-formal_logic|5_2023-09-18T13-43-56.304128.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-formal_logic|5_2023-09-18T13-43-56.304128.parquet"]}]}, {"config_name": "harness_hendrycksTest_global_facts_5", "data_files": [{"split": "2023_09_18T13_43_56.304128", "path": ["**/details_harness|hendrycksTest-global_facts|5_2023-09-18T13-43-56.304128.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-global_facts|5_2023-09-18T13-43-56.304128.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_biology_5", "data_files": [{"split": "2023_09_18T13_43_56.304128", "path": ["**/details_harness|hendrycksTest-high_school_biology|5_2023-09-18T13-43-56.304128.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_biology|5_2023-09-18T13-43-56.304128.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_chemistry_5", "data_files": [{"split": "2023_09_18T13_43_56.304128", "path": ["**/details_harness|hendrycksTest-high_school_chemistry|5_2023-09-18T13-43-56.304128.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_chemistry|5_2023-09-18T13-43-56.304128.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_computer_science_5", "data_files": [{"split": "2023_09_18T13_43_56.304128", "path": ["**/details_harness|hendrycksTest-high_school_computer_science|5_2023-09-18T13-43-56.304128.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_computer_science|5_2023-09-18T13-43-56.304128.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_european_history_5", "data_files": [{"split": "2023_09_18T13_43_56.304128", "path": ["**/details_harness|hendrycksTest-high_school_european_history|5_2023-09-18T13-43-56.304128.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_european_history|5_2023-09-18T13-43-56.304128.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_geography_5", "data_files": [{"split": "2023_09_18T13_43_56.304128", "path": ["**/details_harness|hendrycksTest-high_school_geography|5_2023-09-18T13-43-56.304128.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_geography|5_2023-09-18T13-43-56.304128.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_government_and_politics_5", "data_files": [{"split": "2023_09_18T13_43_56.304128", "path": ["**/details_harness|hendrycksTest-high_school_government_and_politics|5_2023-09-18T13-43-56.304128.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_government_and_politics|5_2023-09-18T13-43-56.304128.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_macroeconomics_5", "data_files": [{"split": "2023_09_18T13_43_56.304128", "path": ["**/details_harness|hendrycksTest-high_school_macroeconomics|5_2023-09-18T13-43-56.304128.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_macroeconomics|5_2023-09-18T13-43-56.304128.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_mathematics_5", "data_files": [{"split": "2023_09_18T13_43_56.304128", "path": ["**/details_harness|hendrycksTest-high_school_mathematics|5_2023-09-18T13-43-56.304128.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_mathematics|5_2023-09-18T13-43-56.304128.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_microeconomics_5", "data_files": [{"split": "2023_09_18T13_43_56.304128", "path": ["**/details_harness|hendrycksTest-high_school_microeconomics|5_2023-09-18T13-43-56.304128.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_microeconomics|5_2023-09-18T13-43-56.304128.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_physics_5", "data_files": [{"split": "2023_09_18T13_43_56.304128", "path": ["**/details_harness|hendrycksTest-high_school_physics|5_2023-09-18T13-43-56.304128.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_physics|5_2023-09-18T13-43-56.304128.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_psychology_5", "data_files": [{"split": "2023_09_18T13_43_56.304128", "path": ["**/details_harness|hendrycksTest-high_school_psychology|5_2023-09-18T13-43-56.304128.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_psychology|5_2023-09-18T13-43-56.304128.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_statistics_5", "data_files": [{"split": "2023_09_18T13_43_56.304128", "path": ["**/details_harness|hendrycksTest-high_school_statistics|5_2023-09-18T13-43-56.304128.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_statistics|5_2023-09-18T13-43-56.304128.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_us_history_5", "data_files": [{"split": "2023_09_18T13_43_56.304128", "path": ["**/details_harness|hendrycksTest-high_school_us_history|5_2023-09-18T13-43-56.304128.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_us_history|5_2023-09-18T13-43-56.304128.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_world_history_5", "data_files": [{"split": "2023_09_18T13_43_56.304128", "path": ["**/details_harness|hendrycksTest-high_school_world_history|5_2023-09-18T13-43-56.304128.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_world_history|5_2023-09-18T13-43-56.304128.parquet"]}]}, {"config_name": "harness_hendrycksTest_human_aging_5", "data_files": [{"split": "2023_09_18T13_43_56.304128", "path": ["**/details_harness|hendrycksTest-human_aging|5_2023-09-18T13-43-56.304128.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-human_aging|5_2023-09-18T13-43-56.304128.parquet"]}]}, {"config_name": "harness_hendrycksTest_human_sexuality_5", "data_files": [{"split": "2023_09_18T13_43_56.304128", "path": ["**/details_harness|hendrycksTest-human_sexuality|5_2023-09-18T13-43-56.304128.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-human_sexuality|5_2023-09-18T13-43-56.304128.parquet"]}]}, {"config_name": "harness_hendrycksTest_international_law_5", "data_files": [{"split": "2023_09_18T13_43_56.304128", "path": ["**/details_harness|hendrycksTest-international_law|5_2023-09-18T13-43-56.304128.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-international_law|5_2023-09-18T13-43-56.304128.parquet"]}]}, {"config_name": "harness_hendrycksTest_jurisprudence_5", "data_files": [{"split": "2023_09_18T13_43_56.304128", "path": ["**/details_harness|hendrycksTest-jurisprudence|5_2023-09-18T13-43-56.304128.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-jurisprudence|5_2023-09-18T13-43-56.304128.parquet"]}]}, {"config_name": "harness_hendrycksTest_logical_fallacies_5", "data_files": [{"split": "2023_09_18T13_43_56.304128", "path": ["**/details_harness|hendrycksTest-logical_fallacies|5_2023-09-18T13-43-56.304128.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-logical_fallacies|5_2023-09-18T13-43-56.304128.parquet"]}]}, {"config_name": "harness_hendrycksTest_machine_learning_5", "data_files": [{"split": "2023_09_18T13_43_56.304128", "path": ["**/details_harness|hendrycksTest-machine_learning|5_2023-09-18T13-43-56.304128.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-machine_learning|5_2023-09-18T13-43-56.304128.parquet"]}]}, {"config_name": "harness_hendrycksTest_management_5", "data_files": [{"split": "2023_09_18T13_43_56.304128", "path": ["**/details_harness|hendrycksTest-management|5_2023-09-18T13-43-56.304128.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-management|5_2023-09-18T13-43-56.304128.parquet"]}]}, {"config_name": "harness_hendrycksTest_marketing_5", "data_files": [{"split": "2023_09_18T13_43_56.304128", "path": ["**/details_harness|hendrycksTest-marketing|5_2023-09-18T13-43-56.304128.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-marketing|5_2023-09-18T13-43-56.304128.parquet"]}]}, {"config_name": "harness_hendrycksTest_medical_genetics_5", "data_files": [{"split": "2023_09_18T13_43_56.304128", "path": ["**/details_harness|hendrycksTest-medical_genetics|5_2023-09-18T13-43-56.304128.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-medical_genetics|5_2023-09-18T13-43-56.304128.parquet"]}]}, {"config_name": "harness_hendrycksTest_miscellaneous_5", "data_files": [{"split": "2023_09_18T13_43_56.304128", "path": ["**/details_harness|hendrycksTest-miscellaneous|5_2023-09-18T13-43-56.304128.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-miscellaneous|5_2023-09-18T13-43-56.304128.parquet"]}]}, {"config_name": "harness_hendrycksTest_moral_disputes_5", "data_files": [{"split": "2023_09_18T13_43_56.304128", "path": ["**/details_harness|hendrycksTest-moral_disputes|5_2023-09-18T13-43-56.304128.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-moral_disputes|5_2023-09-18T13-43-56.304128.parquet"]}]}, {"config_name": "harness_hendrycksTest_moral_scenarios_5", "data_files": [{"split": "2023_09_18T13_43_56.304128", "path": ["**/details_harness|hendrycksTest-moral_scenarios|5_2023-09-18T13-43-56.304128.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-moral_scenarios|5_2023-09-18T13-43-56.304128.parquet"]}]}, {"config_name": "harness_hendrycksTest_nutrition_5", "data_files": [{"split": "2023_09_18T13_43_56.304128", "path": ["**/details_harness|hendrycksTest-nutrition|5_2023-09-18T13-43-56.304128.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-nutrition|5_2023-09-18T13-43-56.304128.parquet"]}]}, {"config_name": "harness_hendrycksTest_philosophy_5", "data_files": [{"split": "2023_09_18T13_43_56.304128", "path": ["**/details_harness|hendrycksTest-philosophy|5_2023-09-18T13-43-56.304128.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-philosophy|5_2023-09-18T13-43-56.304128.parquet"]}]}, {"config_name": "harness_hendrycksTest_prehistory_5", "data_files": [{"split": "2023_09_18T13_43_56.304128", "path": ["**/details_harness|hendrycksTest-prehistory|5_2023-09-18T13-43-56.304128.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-prehistory|5_2023-09-18T13-43-56.304128.parquet"]}]}, {"config_name": "harness_hendrycksTest_professional_accounting_5", "data_files": [{"split": "2023_09_18T13_43_56.304128", "path": ["**/details_harness|hendrycksTest-professional_accounting|5_2023-09-18T13-43-56.304128.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-professional_accounting|5_2023-09-18T13-43-56.304128.parquet"]}]}, {"config_name": "harness_hendrycksTest_professional_law_5", "data_files": [{"split": "2023_09_18T13_43_56.304128", "path": ["**/details_harness|hendrycksTest-professional_law|5_2023-09-18T13-43-56.304128.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-professional_law|5_2023-09-18T13-43-56.304128.parquet"]}]}, {"config_name": "harness_hendrycksTest_professional_medicine_5", "data_files": [{"split": "2023_09_18T13_43_56.304128", "path": ["**/details_harness|hendrycksTest-professional_medicine|5_2023-09-18T13-43-56.304128.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-professional_medicine|5_2023-09-18T13-43-56.304128.parquet"]}]}, {"config_name": "harness_hendrycksTest_professional_psychology_5", "data_files": [{"split": "2023_09_18T13_43_56.304128", "path": ["**/details_harness|hendrycksTest-professional_psychology|5_2023-09-18T13-43-56.304128.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-professional_psychology|5_2023-09-18T13-43-56.304128.parquet"]}]}, {"config_name": "harness_hendrycksTest_public_relations_5", "data_files": [{"split": "2023_09_18T13_43_56.304128", "path": ["**/details_harness|hendrycksTest-public_relations|5_2023-09-18T13-43-56.304128.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-public_relations|5_2023-09-18T13-43-56.304128.parquet"]}]}, {"config_name": "harness_hendrycksTest_security_studies_5", "data_files": [{"split": "2023_09_18T13_43_56.304128", "path": ["**/details_harness|hendrycksTest-security_studies|5_2023-09-18T13-43-56.304128.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-security_studies|5_2023-09-18T13-43-56.304128.parquet"]}]}, {"config_name": "harness_hendrycksTest_sociology_5", "data_files": [{"split": "2023_09_18T13_43_56.304128", "path": ["**/details_harness|hendrycksTest-sociology|5_2023-09-18T13-43-56.304128.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-sociology|5_2023-09-18T13-43-56.304128.parquet"]}]}, {"config_name": "harness_hendrycksTest_us_foreign_policy_5", "data_files": [{"split": "2023_09_18T13_43_56.304128", "path": ["**/details_harness|hendrycksTest-us_foreign_policy|5_2023-09-18T13-43-56.304128.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-us_foreign_policy|5_2023-09-18T13-43-56.304128.parquet"]}]}, {"config_name": "harness_hendrycksTest_virology_5", "data_files": [{"split": "2023_09_18T13_43_56.304128", "path": ["**/details_harness|hendrycksTest-virology|5_2023-09-18T13-43-56.304128.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-virology|5_2023-09-18T13-43-56.304128.parquet"]}]}, {"config_name": "harness_hendrycksTest_world_religions_5", "data_files": [{"split": "2023_09_18T13_43_56.304128", "path": ["**/details_harness|hendrycksTest-world_religions|5_2023-09-18T13-43-56.304128.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-world_religions|5_2023-09-18T13-43-56.304128.parquet"]}]}, {"config_name": "harness_truthfulqa_mc_0", "data_files": [{"split": "2023_09_18T13_43_56.304128", "path": ["**/details_harness|truthfulqa:mc|0_2023-09-18T13-43-56.304128.parquet"]}, {"split": "latest", "path": ["**/details_harness|truthfulqa:mc|0_2023-09-18T13-43-56.304128.parquet"]}]}, {"config_name": "harness_winogrande_5", "data_files": [{"split": "2023_10_28T01_20_40.320894", "path": ["**/details_harness|winogrande|5_2023-10-28T01-20-40.320894.parquet"]}, {"split": "latest", "path": ["**/details_harness|winogrande|5_2023-10-28T01-20-40.320894.parquet"]}]}, {"config_name": "results", "data_files": [{"split": "2023_09_18T13_43_56.304128", "path": ["results_2023-09-18T13-43-56.304128.parquet"]}, {"split": "2023_10_28T01_20_40.320894", "path": ["results_2023-10-28T01-20-40.320894.parquet"]}, {"split": "latest", "path": ["results_2023-10-28T01-20-40.320894.parquet"]}]}]}
|
2023-10-28T00:20:52+00:00
|
[] |
[] |
TAGS
#region-us
|
# Dataset Card for Evaluation run of Undi95/ReMM-v2.1-L2-13B
## Dataset Description
- Homepage:
- Repository: URL
- Paper:
- Leaderboard: URL
- Point of Contact: clementine@URL
### Dataset Summary
Dataset automatically created during the evaluation run of model Undi95/ReMM-v2.1-L2-13B on the Open LLM Leaderboard.
The dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.
The dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The "train" split is always pointing to the latest results.
An additional configuration "results" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the Open LLM Leaderboard).
To load the details from a run, you can for instance do the following:
## Latest results
These are the latest results from run 2023-10-28T01:20:40.320894(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the "latest" split for each eval):
### Supported Tasks and Leaderboards
### Languages
## Dataset Structure
### Data Instances
### Data Fields
### Data Splits
## Dataset Creation
### Curation Rationale
### Source Data
#### Initial Data Collection and Normalization
#### Who are the source language producers?
### Annotations
#### Annotation process
#### Who are the annotators?
### Personal and Sensitive Information
## Considerations for Using the Data
### Social Impact of Dataset
### Discussion of Biases
### Other Known Limitations
## Additional Information
### Dataset Curators
### Licensing Information
### Contributions
|
[
"# Dataset Card for Evaluation run of Undi95/ReMM-v2.1-L2-13B",
"## Dataset Description\n\n- Homepage: \n- Repository: URL\n- Paper: \n- Leaderboard: URL\n- Point of Contact: clementine@URL",
"### Dataset Summary\n\nDataset automatically created during the evaluation run of model Undi95/ReMM-v2.1-L2-13B on the Open LLM Leaderboard.\n\nThe dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.\n\nThe dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The \"train\" split is always pointing to the latest results.\n\nAn additional configuration \"results\" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the Open LLM Leaderboard).\n\nTo load the details from a run, you can for instance do the following:",
"## Latest results\n\nThese are the latest results from run 2023-10-28T01:20:40.320894(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the \"latest\" split for each eval):",
"### Supported Tasks and Leaderboards",
"### Languages",
"## Dataset Structure",
"### Data Instances",
"### Data Fields",
"### Data Splits",
"## Dataset Creation",
"### Curation Rationale",
"### Source Data",
"#### Initial Data Collection and Normalization",
"#### Who are the source language producers?",
"### Annotations",
"#### Annotation process",
"#### Who are the annotators?",
"### Personal and Sensitive Information",
"## Considerations for Using the Data",
"### Social Impact of Dataset",
"### Discussion of Biases",
"### Other Known Limitations",
"## Additional Information",
"### Dataset Curators",
"### Licensing Information",
"### Contributions"
] |
[
"TAGS\n#region-us \n",
"# Dataset Card for Evaluation run of Undi95/ReMM-v2.1-L2-13B",
"## Dataset Description\n\n- Homepage: \n- Repository: URL\n- Paper: \n- Leaderboard: URL\n- Point of Contact: clementine@URL",
"### Dataset Summary\n\nDataset automatically created during the evaluation run of model Undi95/ReMM-v2.1-L2-13B on the Open LLM Leaderboard.\n\nThe dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.\n\nThe dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The \"train\" split is always pointing to the latest results.\n\nAn additional configuration \"results\" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the Open LLM Leaderboard).\n\nTo load the details from a run, you can for instance do the following:",
"## Latest results\n\nThese are the latest results from run 2023-10-28T01:20:40.320894(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the \"latest\" split for each eval):",
"### Supported Tasks and Leaderboards",
"### Languages",
"## Dataset Structure",
"### Data Instances",
"### Data Fields",
"### Data Splits",
"## Dataset Creation",
"### Curation Rationale",
"### Source Data",
"#### Initial Data Collection and Normalization",
"#### Who are the source language producers?",
"### Annotations",
"#### Annotation process",
"#### Who are the annotators?",
"### Personal and Sensitive Information",
"## Considerations for Using the Data",
"### Social Impact of Dataset",
"### Discussion of Biases",
"### Other Known Limitations",
"## Additional Information",
"### Dataset Curators",
"### Licensing Information",
"### Contributions"
] |
[
6,
23,
31,
171,
67,
10,
4,
6,
6,
5,
5,
5,
7,
4,
10,
10,
5,
5,
9,
8,
8,
7,
8,
7,
5,
6,
6,
5
] |
[
"passage: TAGS\n#region-us \n# Dataset Card for Evaluation run of Undi95/ReMM-v2.1-L2-13B## Dataset Description\n\n- Homepage: \n- Repository: URL\n- Paper: \n- Leaderboard: URL\n- Point of Contact: clementine@URL### Dataset Summary\n\nDataset automatically created during the evaluation run of model Undi95/ReMM-v2.1-L2-13B on the Open LLM Leaderboard.\n\nThe dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.\n\nThe dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The \"train\" split is always pointing to the latest results.\n\nAn additional configuration \"results\" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the Open LLM Leaderboard).\n\nTo load the details from a run, you can for instance do the following:## Latest results\n\nThese are the latest results from run 2023-10-28T01:20:40.320894(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the \"latest\" split for each eval):### Supported Tasks and Leaderboards### Languages## Dataset Structure### Data Instances### Data Fields### Data Splits## Dataset Creation### Curation Rationale### Source Data#### Initial Data Collection and Normalization#### Who are the source language producers?### Annotations#### Annotation process#### Who are the annotators?### Personal and Sensitive Information## Considerations for Using the Data### Social Impact of Dataset### Discussion of Biases### Other Known Limitations## Additional Information### Dataset Curators### Licensing Information### Contributions"
] |
dabfc7f5026fef4c73d6ce99f710ed61fd8966ec
|
# Dataset Card for Evaluation run of Undi95/UndiMix-v4-13B
## Dataset Description
- **Homepage:**
- **Repository:** https://huggingface.co/Undi95/UndiMix-v4-13B
- **Paper:**
- **Leaderboard:** https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard
- **Point of Contact:** [email protected]
### Dataset Summary
Dataset automatically created during the evaluation run of model [Undi95/UndiMix-v4-13B](https://huggingface.co/Undi95/UndiMix-v4-13B) on the [Open LLM Leaderboard](https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard).
The dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.
The dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The "train" split is always pointing to the latest results.
An additional configuration "results" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the [Open LLM Leaderboard](https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard)).
To load the details from a run, you can for instance do the following:
```python
from datasets import load_dataset
data = load_dataset("open-llm-leaderboard/details_Undi95__UndiMix-v4-13B",
"harness_winogrande_5",
split="train")
```
## Latest results
These are the [latest results from run 2023-10-27T04:12:01.560692](https://huggingface.co/datasets/open-llm-leaderboard/details_Undi95__UndiMix-v4-13B/blob/main/results_2023-10-27T04-12-01.560692.json)(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the "latest" split for each eval):
```python
{
"all": {
"em": 0.14146392617449666,
"em_stderr": 0.003568960808825645,
"f1": 0.20818477348993217,
"f1_stderr": 0.0036692979641845653,
"acc": 0.4494334219138294,
"acc_stderr": 0.010726378456151354
},
"harness|drop|3": {
"em": 0.14146392617449666,
"em_stderr": 0.003568960808825645,
"f1": 0.20818477348993217,
"f1_stderr": 0.0036692979641845653
},
"harness|gsm8k|5": {
"acc": 0.1372251705837756,
"acc_stderr": 0.009477808244600401
},
"harness|winogrande|5": {
"acc": 0.7616416732438832,
"acc_stderr": 0.011974948667702308
}
}
```
### Supported Tasks and Leaderboards
[More Information Needed]
### Languages
[More Information Needed]
## Dataset Structure
### Data Instances
[More Information Needed]
### Data Fields
[More Information Needed]
### Data Splits
[More Information Needed]
## Dataset Creation
### Curation Rationale
[More Information Needed]
### Source Data
#### Initial Data Collection and Normalization
[More Information Needed]
#### Who are the source language producers?
[More Information Needed]
### Annotations
#### Annotation process
[More Information Needed]
#### Who are the annotators?
[More Information Needed]
### Personal and Sensitive Information
[More Information Needed]
## Considerations for Using the Data
### Social Impact of Dataset
[More Information Needed]
### Discussion of Biases
[More Information Needed]
### Other Known Limitations
[More Information Needed]
## Additional Information
### Dataset Curators
[More Information Needed]
### Licensing Information
[More Information Needed]
### Citation Information
[More Information Needed]
### Contributions
[More Information Needed]
|
open-llm-leaderboard/details_Undi95__UndiMix-v4-13B
|
[
"region:us"
] |
2023-09-18T12:46:18+00:00
|
{"pretty_name": "Evaluation run of Undi95/UndiMix-v4-13B", "dataset_summary": "Dataset automatically created during the evaluation run of model [Undi95/UndiMix-v4-13B](https://huggingface.co/Undi95/UndiMix-v4-13B) on the [Open LLM Leaderboard](https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard).\n\nThe dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.\n\nThe dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The \"train\" split is always pointing to the latest results.\n\nAn additional configuration \"results\" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the [Open LLM Leaderboard](https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard)).\n\nTo load the details from a run, you can for instance do the following:\n```python\nfrom datasets import load_dataset\ndata = load_dataset(\"open-llm-leaderboard/details_Undi95__UndiMix-v4-13B\",\n\t\"harness_winogrande_5\",\n\tsplit=\"train\")\n```\n\n## Latest results\n\nThese are the [latest results from run 2023-10-27T04:12:01.560692](https://huggingface.co/datasets/open-llm-leaderboard/details_Undi95__UndiMix-v4-13B/blob/main/results_2023-10-27T04-12-01.560692.json)(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the \"latest\" split for each eval):\n\n```python\n{\n \"all\": {\n \"em\": 0.14146392617449666,\n \"em_stderr\": 0.003568960808825645,\n \"f1\": 0.20818477348993217,\n \"f1_stderr\": 0.0036692979641845653,\n \"acc\": 0.4494334219138294,\n \"acc_stderr\": 0.010726378456151354\n },\n \"harness|drop|3\": {\n \"em\": 0.14146392617449666,\n \"em_stderr\": 0.003568960808825645,\n \"f1\": 0.20818477348993217,\n \"f1_stderr\": 0.0036692979641845653\n },\n \"harness|gsm8k|5\": {\n \"acc\": 0.1372251705837756,\n \"acc_stderr\": 0.009477808244600401\n },\n \"harness|winogrande|5\": {\n \"acc\": 0.7616416732438832,\n \"acc_stderr\": 0.011974948667702308\n }\n}\n```", "repo_url": "https://huggingface.co/Undi95/UndiMix-v4-13B", "leaderboard_url": "https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard", "point_of_contact": "[email protected]", "configs": [{"config_name": "harness_arc_challenge_25", "data_files": [{"split": "2023_09_18T13_45_54.862257", "path": ["**/details_harness|arc:challenge|25_2023-09-18T13-45-54.862257.parquet"]}, {"split": "latest", "path": ["**/details_harness|arc:challenge|25_2023-09-18T13-45-54.862257.parquet"]}]}, {"config_name": "harness_drop_3", "data_files": [{"split": "2023_10_27T04_12_01.560692", "path": ["**/details_harness|drop|3_2023-10-27T04-12-01.560692.parquet"]}, {"split": "latest", "path": ["**/details_harness|drop|3_2023-10-27T04-12-01.560692.parquet"]}]}, {"config_name": "harness_gsm8k_5", "data_files": [{"split": "2023_10_27T04_12_01.560692", "path": ["**/details_harness|gsm8k|5_2023-10-27T04-12-01.560692.parquet"]}, {"split": "latest", "path": ["**/details_harness|gsm8k|5_2023-10-27T04-12-01.560692.parquet"]}]}, {"config_name": "harness_hellaswag_10", "data_files": [{"split": "2023_09_18T13_45_54.862257", "path": ["**/details_harness|hellaswag|10_2023-09-18T13-45-54.862257.parquet"]}, {"split": "latest", "path": ["**/details_harness|hellaswag|10_2023-09-18T13-45-54.862257.parquet"]}]}, {"config_name": "harness_hendrycksTest_5", "data_files": [{"split": "2023_09_18T13_45_54.862257", "path": ["**/details_harness|hendrycksTest-abstract_algebra|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-anatomy|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-astronomy|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-business_ethics|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-clinical_knowledge|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-college_biology|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-college_chemistry|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-college_computer_science|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-college_mathematics|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-college_medicine|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-college_physics|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-computer_security|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-conceptual_physics|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-econometrics|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-electrical_engineering|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-elementary_mathematics|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-formal_logic|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-global_facts|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-high_school_biology|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-high_school_chemistry|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-high_school_computer_science|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-high_school_european_history|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-high_school_geography|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-high_school_government_and_politics|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-high_school_macroeconomics|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-high_school_mathematics|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-high_school_microeconomics|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-high_school_physics|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-high_school_psychology|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-high_school_statistics|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-high_school_us_history|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-high_school_world_history|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-human_aging|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-human_sexuality|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-international_law|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-jurisprudence|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-logical_fallacies|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-machine_learning|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-management|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-marketing|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-medical_genetics|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-miscellaneous|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-moral_disputes|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-moral_scenarios|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-nutrition|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-philosophy|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-prehistory|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-professional_accounting|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-professional_law|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-professional_medicine|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-professional_psychology|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-public_relations|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-security_studies|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-sociology|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-us_foreign_policy|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-virology|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-world_religions|5_2023-09-18T13-45-54.862257.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-abstract_algebra|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-anatomy|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-astronomy|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-business_ethics|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-clinical_knowledge|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-college_biology|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-college_chemistry|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-college_computer_science|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-college_mathematics|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-college_medicine|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-college_physics|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-computer_security|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-conceptual_physics|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-econometrics|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-electrical_engineering|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-elementary_mathematics|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-formal_logic|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-global_facts|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-high_school_biology|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-high_school_chemistry|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-high_school_computer_science|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-high_school_european_history|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-high_school_geography|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-high_school_government_and_politics|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-high_school_macroeconomics|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-high_school_mathematics|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-high_school_microeconomics|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-high_school_physics|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-high_school_psychology|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-high_school_statistics|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-high_school_us_history|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-high_school_world_history|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-human_aging|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-human_sexuality|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-international_law|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-jurisprudence|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-logical_fallacies|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-machine_learning|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-management|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-marketing|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-medical_genetics|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-miscellaneous|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-moral_disputes|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-moral_scenarios|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-nutrition|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-philosophy|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-prehistory|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-professional_accounting|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-professional_law|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-professional_medicine|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-professional_psychology|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-public_relations|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-security_studies|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-sociology|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-us_foreign_policy|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-virology|5_2023-09-18T13-45-54.862257.parquet", "**/details_harness|hendrycksTest-world_religions|5_2023-09-18T13-45-54.862257.parquet"]}]}, {"config_name": "harness_hendrycksTest_abstract_algebra_5", "data_files": [{"split": "2023_09_18T13_45_54.862257", "path": ["**/details_harness|hendrycksTest-abstract_algebra|5_2023-09-18T13-45-54.862257.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-abstract_algebra|5_2023-09-18T13-45-54.862257.parquet"]}]}, {"config_name": "harness_hendrycksTest_anatomy_5", "data_files": [{"split": "2023_09_18T13_45_54.862257", "path": ["**/details_harness|hendrycksTest-anatomy|5_2023-09-18T13-45-54.862257.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-anatomy|5_2023-09-18T13-45-54.862257.parquet"]}]}, {"config_name": "harness_hendrycksTest_astronomy_5", "data_files": [{"split": "2023_09_18T13_45_54.862257", "path": ["**/details_harness|hendrycksTest-astronomy|5_2023-09-18T13-45-54.862257.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-astronomy|5_2023-09-18T13-45-54.862257.parquet"]}]}, {"config_name": "harness_hendrycksTest_business_ethics_5", "data_files": [{"split": "2023_09_18T13_45_54.862257", "path": ["**/details_harness|hendrycksTest-business_ethics|5_2023-09-18T13-45-54.862257.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-business_ethics|5_2023-09-18T13-45-54.862257.parquet"]}]}, {"config_name": "harness_hendrycksTest_clinical_knowledge_5", "data_files": [{"split": "2023_09_18T13_45_54.862257", "path": ["**/details_harness|hendrycksTest-clinical_knowledge|5_2023-09-18T13-45-54.862257.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-clinical_knowledge|5_2023-09-18T13-45-54.862257.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_biology_5", "data_files": [{"split": "2023_09_18T13_45_54.862257", "path": ["**/details_harness|hendrycksTest-college_biology|5_2023-09-18T13-45-54.862257.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_biology|5_2023-09-18T13-45-54.862257.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_chemistry_5", "data_files": [{"split": "2023_09_18T13_45_54.862257", "path": ["**/details_harness|hendrycksTest-college_chemistry|5_2023-09-18T13-45-54.862257.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_chemistry|5_2023-09-18T13-45-54.862257.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_computer_science_5", "data_files": [{"split": "2023_09_18T13_45_54.862257", "path": ["**/details_harness|hendrycksTest-college_computer_science|5_2023-09-18T13-45-54.862257.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_computer_science|5_2023-09-18T13-45-54.862257.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_mathematics_5", "data_files": [{"split": "2023_09_18T13_45_54.862257", "path": ["**/details_harness|hendrycksTest-college_mathematics|5_2023-09-18T13-45-54.862257.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_mathematics|5_2023-09-18T13-45-54.862257.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_medicine_5", "data_files": [{"split": "2023_09_18T13_45_54.862257", "path": ["**/details_harness|hendrycksTest-college_medicine|5_2023-09-18T13-45-54.862257.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_medicine|5_2023-09-18T13-45-54.862257.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_physics_5", "data_files": [{"split": "2023_09_18T13_45_54.862257", "path": ["**/details_harness|hendrycksTest-college_physics|5_2023-09-18T13-45-54.862257.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_physics|5_2023-09-18T13-45-54.862257.parquet"]}]}, {"config_name": "harness_hendrycksTest_computer_security_5", "data_files": [{"split": "2023_09_18T13_45_54.862257", "path": ["**/details_harness|hendrycksTest-computer_security|5_2023-09-18T13-45-54.862257.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-computer_security|5_2023-09-18T13-45-54.862257.parquet"]}]}, {"config_name": "harness_hendrycksTest_conceptual_physics_5", "data_files": [{"split": "2023_09_18T13_45_54.862257", "path": ["**/details_harness|hendrycksTest-conceptual_physics|5_2023-09-18T13-45-54.862257.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-conceptual_physics|5_2023-09-18T13-45-54.862257.parquet"]}]}, {"config_name": "harness_hendrycksTest_econometrics_5", "data_files": [{"split": "2023_09_18T13_45_54.862257", "path": ["**/details_harness|hendrycksTest-econometrics|5_2023-09-18T13-45-54.862257.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-econometrics|5_2023-09-18T13-45-54.862257.parquet"]}]}, {"config_name": "harness_hendrycksTest_electrical_engineering_5", "data_files": [{"split": "2023_09_18T13_45_54.862257", "path": ["**/details_harness|hendrycksTest-electrical_engineering|5_2023-09-18T13-45-54.862257.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-electrical_engineering|5_2023-09-18T13-45-54.862257.parquet"]}]}, {"config_name": "harness_hendrycksTest_elementary_mathematics_5", "data_files": [{"split": "2023_09_18T13_45_54.862257", "path": ["**/details_harness|hendrycksTest-elementary_mathematics|5_2023-09-18T13-45-54.862257.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-elementary_mathematics|5_2023-09-18T13-45-54.862257.parquet"]}]}, {"config_name": "harness_hendrycksTest_formal_logic_5", "data_files": [{"split": "2023_09_18T13_45_54.862257", "path": ["**/details_harness|hendrycksTest-formal_logic|5_2023-09-18T13-45-54.862257.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-formal_logic|5_2023-09-18T13-45-54.862257.parquet"]}]}, {"config_name": "harness_hendrycksTest_global_facts_5", "data_files": [{"split": "2023_09_18T13_45_54.862257", "path": ["**/details_harness|hendrycksTest-global_facts|5_2023-09-18T13-45-54.862257.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-global_facts|5_2023-09-18T13-45-54.862257.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_biology_5", "data_files": [{"split": "2023_09_18T13_45_54.862257", "path": ["**/details_harness|hendrycksTest-high_school_biology|5_2023-09-18T13-45-54.862257.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_biology|5_2023-09-18T13-45-54.862257.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_chemistry_5", "data_files": [{"split": "2023_09_18T13_45_54.862257", "path": ["**/details_harness|hendrycksTest-high_school_chemistry|5_2023-09-18T13-45-54.862257.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_chemistry|5_2023-09-18T13-45-54.862257.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_computer_science_5", "data_files": [{"split": "2023_09_18T13_45_54.862257", "path": ["**/details_harness|hendrycksTest-high_school_computer_science|5_2023-09-18T13-45-54.862257.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_computer_science|5_2023-09-18T13-45-54.862257.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_european_history_5", "data_files": [{"split": "2023_09_18T13_45_54.862257", "path": ["**/details_harness|hendrycksTest-high_school_european_history|5_2023-09-18T13-45-54.862257.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_european_history|5_2023-09-18T13-45-54.862257.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_geography_5", "data_files": [{"split": "2023_09_18T13_45_54.862257", "path": ["**/details_harness|hendrycksTest-high_school_geography|5_2023-09-18T13-45-54.862257.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_geography|5_2023-09-18T13-45-54.862257.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_government_and_politics_5", "data_files": [{"split": "2023_09_18T13_45_54.862257", "path": ["**/details_harness|hendrycksTest-high_school_government_and_politics|5_2023-09-18T13-45-54.862257.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_government_and_politics|5_2023-09-18T13-45-54.862257.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_macroeconomics_5", "data_files": [{"split": "2023_09_18T13_45_54.862257", "path": ["**/details_harness|hendrycksTest-high_school_macroeconomics|5_2023-09-18T13-45-54.862257.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_macroeconomics|5_2023-09-18T13-45-54.862257.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_mathematics_5", "data_files": [{"split": "2023_09_18T13_45_54.862257", "path": ["**/details_harness|hendrycksTest-high_school_mathematics|5_2023-09-18T13-45-54.862257.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_mathematics|5_2023-09-18T13-45-54.862257.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_microeconomics_5", "data_files": [{"split": "2023_09_18T13_45_54.862257", "path": ["**/details_harness|hendrycksTest-high_school_microeconomics|5_2023-09-18T13-45-54.862257.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_microeconomics|5_2023-09-18T13-45-54.862257.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_physics_5", "data_files": [{"split": "2023_09_18T13_45_54.862257", "path": ["**/details_harness|hendrycksTest-high_school_physics|5_2023-09-18T13-45-54.862257.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_physics|5_2023-09-18T13-45-54.862257.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_psychology_5", "data_files": [{"split": "2023_09_18T13_45_54.862257", "path": ["**/details_harness|hendrycksTest-high_school_psychology|5_2023-09-18T13-45-54.862257.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_psychology|5_2023-09-18T13-45-54.862257.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_statistics_5", "data_files": [{"split": "2023_09_18T13_45_54.862257", "path": ["**/details_harness|hendrycksTest-high_school_statistics|5_2023-09-18T13-45-54.862257.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_statistics|5_2023-09-18T13-45-54.862257.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_us_history_5", "data_files": [{"split": "2023_09_18T13_45_54.862257", "path": ["**/details_harness|hendrycksTest-high_school_us_history|5_2023-09-18T13-45-54.862257.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_us_history|5_2023-09-18T13-45-54.862257.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_world_history_5", "data_files": [{"split": "2023_09_18T13_45_54.862257", "path": ["**/details_harness|hendrycksTest-high_school_world_history|5_2023-09-18T13-45-54.862257.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_world_history|5_2023-09-18T13-45-54.862257.parquet"]}]}, {"config_name": "harness_hendrycksTest_human_aging_5", "data_files": [{"split": "2023_09_18T13_45_54.862257", "path": ["**/details_harness|hendrycksTest-human_aging|5_2023-09-18T13-45-54.862257.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-human_aging|5_2023-09-18T13-45-54.862257.parquet"]}]}, {"config_name": "harness_hendrycksTest_human_sexuality_5", "data_files": [{"split": "2023_09_18T13_45_54.862257", "path": ["**/details_harness|hendrycksTest-human_sexuality|5_2023-09-18T13-45-54.862257.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-human_sexuality|5_2023-09-18T13-45-54.862257.parquet"]}]}, {"config_name": "harness_hendrycksTest_international_law_5", "data_files": [{"split": "2023_09_18T13_45_54.862257", "path": ["**/details_harness|hendrycksTest-international_law|5_2023-09-18T13-45-54.862257.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-international_law|5_2023-09-18T13-45-54.862257.parquet"]}]}, {"config_name": "harness_hendrycksTest_jurisprudence_5", "data_files": [{"split": "2023_09_18T13_45_54.862257", "path": ["**/details_harness|hendrycksTest-jurisprudence|5_2023-09-18T13-45-54.862257.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-jurisprudence|5_2023-09-18T13-45-54.862257.parquet"]}]}, {"config_name": "harness_hendrycksTest_logical_fallacies_5", "data_files": [{"split": "2023_09_18T13_45_54.862257", "path": ["**/details_harness|hendrycksTest-logical_fallacies|5_2023-09-18T13-45-54.862257.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-logical_fallacies|5_2023-09-18T13-45-54.862257.parquet"]}]}, {"config_name": "harness_hendrycksTest_machine_learning_5", "data_files": [{"split": "2023_09_18T13_45_54.862257", "path": ["**/details_harness|hendrycksTest-machine_learning|5_2023-09-18T13-45-54.862257.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-machine_learning|5_2023-09-18T13-45-54.862257.parquet"]}]}, {"config_name": "harness_hendrycksTest_management_5", "data_files": [{"split": "2023_09_18T13_45_54.862257", "path": ["**/details_harness|hendrycksTest-management|5_2023-09-18T13-45-54.862257.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-management|5_2023-09-18T13-45-54.862257.parquet"]}]}, {"config_name": "harness_hendrycksTest_marketing_5", "data_files": [{"split": "2023_09_18T13_45_54.862257", "path": ["**/details_harness|hendrycksTest-marketing|5_2023-09-18T13-45-54.862257.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-marketing|5_2023-09-18T13-45-54.862257.parquet"]}]}, {"config_name": "harness_hendrycksTest_medical_genetics_5", "data_files": [{"split": "2023_09_18T13_45_54.862257", "path": ["**/details_harness|hendrycksTest-medical_genetics|5_2023-09-18T13-45-54.862257.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-medical_genetics|5_2023-09-18T13-45-54.862257.parquet"]}]}, {"config_name": "harness_hendrycksTest_miscellaneous_5", "data_files": [{"split": "2023_09_18T13_45_54.862257", "path": ["**/details_harness|hendrycksTest-miscellaneous|5_2023-09-18T13-45-54.862257.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-miscellaneous|5_2023-09-18T13-45-54.862257.parquet"]}]}, {"config_name": "harness_hendrycksTest_moral_disputes_5", "data_files": [{"split": "2023_09_18T13_45_54.862257", "path": ["**/details_harness|hendrycksTest-moral_disputes|5_2023-09-18T13-45-54.862257.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-moral_disputes|5_2023-09-18T13-45-54.862257.parquet"]}]}, {"config_name": "harness_hendrycksTest_moral_scenarios_5", "data_files": [{"split": "2023_09_18T13_45_54.862257", "path": ["**/details_harness|hendrycksTest-moral_scenarios|5_2023-09-18T13-45-54.862257.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-moral_scenarios|5_2023-09-18T13-45-54.862257.parquet"]}]}, {"config_name": "harness_hendrycksTest_nutrition_5", "data_files": [{"split": "2023_09_18T13_45_54.862257", "path": ["**/details_harness|hendrycksTest-nutrition|5_2023-09-18T13-45-54.862257.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-nutrition|5_2023-09-18T13-45-54.862257.parquet"]}]}, {"config_name": "harness_hendrycksTest_philosophy_5", "data_files": [{"split": "2023_09_18T13_45_54.862257", "path": ["**/details_harness|hendrycksTest-philosophy|5_2023-09-18T13-45-54.862257.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-philosophy|5_2023-09-18T13-45-54.862257.parquet"]}]}, {"config_name": "harness_hendrycksTest_prehistory_5", "data_files": [{"split": "2023_09_18T13_45_54.862257", "path": ["**/details_harness|hendrycksTest-prehistory|5_2023-09-18T13-45-54.862257.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-prehistory|5_2023-09-18T13-45-54.862257.parquet"]}]}, {"config_name": "harness_hendrycksTest_professional_accounting_5", "data_files": [{"split": "2023_09_18T13_45_54.862257", "path": ["**/details_harness|hendrycksTest-professional_accounting|5_2023-09-18T13-45-54.862257.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-professional_accounting|5_2023-09-18T13-45-54.862257.parquet"]}]}, {"config_name": "harness_hendrycksTest_professional_law_5", "data_files": [{"split": "2023_09_18T13_45_54.862257", "path": ["**/details_harness|hendrycksTest-professional_law|5_2023-09-18T13-45-54.862257.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-professional_law|5_2023-09-18T13-45-54.862257.parquet"]}]}, {"config_name": "harness_hendrycksTest_professional_medicine_5", "data_files": [{"split": "2023_09_18T13_45_54.862257", "path": ["**/details_harness|hendrycksTest-professional_medicine|5_2023-09-18T13-45-54.862257.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-professional_medicine|5_2023-09-18T13-45-54.862257.parquet"]}]}, {"config_name": "harness_hendrycksTest_professional_psychology_5", "data_files": [{"split": "2023_09_18T13_45_54.862257", "path": ["**/details_harness|hendrycksTest-professional_psychology|5_2023-09-18T13-45-54.862257.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-professional_psychology|5_2023-09-18T13-45-54.862257.parquet"]}]}, {"config_name": "harness_hendrycksTest_public_relations_5", "data_files": [{"split": "2023_09_18T13_45_54.862257", "path": ["**/details_harness|hendrycksTest-public_relations|5_2023-09-18T13-45-54.862257.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-public_relations|5_2023-09-18T13-45-54.862257.parquet"]}]}, {"config_name": "harness_hendrycksTest_security_studies_5", "data_files": [{"split": "2023_09_18T13_45_54.862257", "path": ["**/details_harness|hendrycksTest-security_studies|5_2023-09-18T13-45-54.862257.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-security_studies|5_2023-09-18T13-45-54.862257.parquet"]}]}, {"config_name": "harness_hendrycksTest_sociology_5", "data_files": [{"split": "2023_09_18T13_45_54.862257", "path": ["**/details_harness|hendrycksTest-sociology|5_2023-09-18T13-45-54.862257.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-sociology|5_2023-09-18T13-45-54.862257.parquet"]}]}, {"config_name": "harness_hendrycksTest_us_foreign_policy_5", "data_files": [{"split": "2023_09_18T13_45_54.862257", "path": ["**/details_harness|hendrycksTest-us_foreign_policy|5_2023-09-18T13-45-54.862257.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-us_foreign_policy|5_2023-09-18T13-45-54.862257.parquet"]}]}, {"config_name": "harness_hendrycksTest_virology_5", "data_files": [{"split": "2023_09_18T13_45_54.862257", "path": ["**/details_harness|hendrycksTest-virology|5_2023-09-18T13-45-54.862257.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-virology|5_2023-09-18T13-45-54.862257.parquet"]}]}, {"config_name": "harness_hendrycksTest_world_religions_5", "data_files": [{"split": "2023_09_18T13_45_54.862257", "path": ["**/details_harness|hendrycksTest-world_religions|5_2023-09-18T13-45-54.862257.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-world_religions|5_2023-09-18T13-45-54.862257.parquet"]}]}, {"config_name": "harness_truthfulqa_mc_0", "data_files": [{"split": "2023_09_18T13_45_54.862257", "path": ["**/details_harness|truthfulqa:mc|0_2023-09-18T13-45-54.862257.parquet"]}, {"split": "latest", "path": ["**/details_harness|truthfulqa:mc|0_2023-09-18T13-45-54.862257.parquet"]}]}, {"config_name": "harness_winogrande_5", "data_files": [{"split": "2023_10_27T04_12_01.560692", "path": ["**/details_harness|winogrande|5_2023-10-27T04-12-01.560692.parquet"]}, {"split": "latest", "path": ["**/details_harness|winogrande|5_2023-10-27T04-12-01.560692.parquet"]}]}, {"config_name": "results", "data_files": [{"split": "2023_09_18T13_45_54.862257", "path": ["results_2023-09-18T13-45-54.862257.parquet"]}, {"split": "2023_10_27T04_12_01.560692", "path": ["results_2023-10-27T04-12-01.560692.parquet"]}, {"split": "latest", "path": ["results_2023-10-27T04-12-01.560692.parquet"]}]}]}
|
2023-10-27T03:12:14+00:00
|
[] |
[] |
TAGS
#region-us
|
# Dataset Card for Evaluation run of Undi95/UndiMix-v4-13B
## Dataset Description
- Homepage:
- Repository: URL
- Paper:
- Leaderboard: URL
- Point of Contact: clementine@URL
### Dataset Summary
Dataset automatically created during the evaluation run of model Undi95/UndiMix-v4-13B on the Open LLM Leaderboard.
The dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.
The dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The "train" split is always pointing to the latest results.
An additional configuration "results" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the Open LLM Leaderboard).
To load the details from a run, you can for instance do the following:
## Latest results
These are the latest results from run 2023-10-27T04:12:01.560692(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the "latest" split for each eval):
### Supported Tasks and Leaderboards
### Languages
## Dataset Structure
### Data Instances
### Data Fields
### Data Splits
## Dataset Creation
### Curation Rationale
### Source Data
#### Initial Data Collection and Normalization
#### Who are the source language producers?
### Annotations
#### Annotation process
#### Who are the annotators?
### Personal and Sensitive Information
## Considerations for Using the Data
### Social Impact of Dataset
### Discussion of Biases
### Other Known Limitations
## Additional Information
### Dataset Curators
### Licensing Information
### Contributions
|
[
"# Dataset Card for Evaluation run of Undi95/UndiMix-v4-13B",
"## Dataset Description\n\n- Homepage: \n- Repository: URL\n- Paper: \n- Leaderboard: URL\n- Point of Contact: clementine@URL",
"### Dataset Summary\n\nDataset automatically created during the evaluation run of model Undi95/UndiMix-v4-13B on the Open LLM Leaderboard.\n\nThe dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.\n\nThe dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The \"train\" split is always pointing to the latest results.\n\nAn additional configuration \"results\" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the Open LLM Leaderboard).\n\nTo load the details from a run, you can for instance do the following:",
"## Latest results\n\nThese are the latest results from run 2023-10-27T04:12:01.560692(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the \"latest\" split for each eval):",
"### Supported Tasks and Leaderboards",
"### Languages",
"## Dataset Structure",
"### Data Instances",
"### Data Fields",
"### Data Splits",
"## Dataset Creation",
"### Curation Rationale",
"### Source Data",
"#### Initial Data Collection and Normalization",
"#### Who are the source language producers?",
"### Annotations",
"#### Annotation process",
"#### Who are the annotators?",
"### Personal and Sensitive Information",
"## Considerations for Using the Data",
"### Social Impact of Dataset",
"### Discussion of Biases",
"### Other Known Limitations",
"## Additional Information",
"### Dataset Curators",
"### Licensing Information",
"### Contributions"
] |
[
"TAGS\n#region-us \n",
"# Dataset Card for Evaluation run of Undi95/UndiMix-v4-13B",
"## Dataset Description\n\n- Homepage: \n- Repository: URL\n- Paper: \n- Leaderboard: URL\n- Point of Contact: clementine@URL",
"### Dataset Summary\n\nDataset automatically created during the evaluation run of model Undi95/UndiMix-v4-13B on the Open LLM Leaderboard.\n\nThe dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.\n\nThe dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The \"train\" split is always pointing to the latest results.\n\nAn additional configuration \"results\" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the Open LLM Leaderboard).\n\nTo load the details from a run, you can for instance do the following:",
"## Latest results\n\nThese are the latest results from run 2023-10-27T04:12:01.560692(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the \"latest\" split for each eval):",
"### Supported Tasks and Leaderboards",
"### Languages",
"## Dataset Structure",
"### Data Instances",
"### Data Fields",
"### Data Splits",
"## Dataset Creation",
"### Curation Rationale",
"### Source Data",
"#### Initial Data Collection and Normalization",
"#### Who are the source language producers?",
"### Annotations",
"#### Annotation process",
"#### Who are the annotators?",
"### Personal and Sensitive Information",
"## Considerations for Using the Data",
"### Social Impact of Dataset",
"### Discussion of Biases",
"### Other Known Limitations",
"## Additional Information",
"### Dataset Curators",
"### Licensing Information",
"### Contributions"
] |
[
6,
22,
31,
170,
67,
10,
4,
6,
6,
5,
5,
5,
7,
4,
10,
10,
5,
5,
9,
8,
8,
7,
8,
7,
5,
6,
6,
5
] |
[
"passage: TAGS\n#region-us \n# Dataset Card for Evaluation run of Undi95/UndiMix-v4-13B## Dataset Description\n\n- Homepage: \n- Repository: URL\n- Paper: \n- Leaderboard: URL\n- Point of Contact: clementine@URL### Dataset Summary\n\nDataset automatically created during the evaluation run of model Undi95/UndiMix-v4-13B on the Open LLM Leaderboard.\n\nThe dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.\n\nThe dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The \"train\" split is always pointing to the latest results.\n\nAn additional configuration \"results\" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the Open LLM Leaderboard).\n\nTo load the details from a run, you can for instance do the following:## Latest results\n\nThese are the latest results from run 2023-10-27T04:12:01.560692(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the \"latest\" split for each eval):### Supported Tasks and Leaderboards### Languages## Dataset Structure### Data Instances### Data Fields### Data Splits## Dataset Creation### Curation Rationale### Source Data#### Initial Data Collection and Normalization#### Who are the source language producers?### Annotations#### Annotation process#### Who are the annotators?### Personal and Sensitive Information## Considerations for Using the Data### Social Impact of Dataset### Discussion of Biases### Other Known Limitations## Additional Information### Dataset Curators### Licensing Information### Contributions"
] |
d6ef4ec57ccae6554ea9d2e536f954376fb65bdc
|
# Dataset Card for Evaluation run of Undi95/OpenRP-13B
## Dataset Description
- **Homepage:**
- **Repository:** https://huggingface.co/Undi95/OpenRP-13B
- **Paper:**
- **Leaderboard:** https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard
- **Point of Contact:** [email protected]
### Dataset Summary
Dataset automatically created during the evaluation run of model [Undi95/OpenRP-13B](https://huggingface.co/Undi95/OpenRP-13B) on the [Open LLM Leaderboard](https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard).
The dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.
The dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The "train" split is always pointing to the latest results.
An additional configuration "results" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the [Open LLM Leaderboard](https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard)).
To load the details from a run, you can for instance do the following:
```python
from datasets import load_dataset
data = load_dataset("open-llm-leaderboard/details_Undi95__OpenRP-13B",
"harness_winogrande_5",
split="train")
```
## Latest results
These are the [latest results from run 2023-10-29T00:54:50.325458](https://huggingface.co/datasets/open-llm-leaderboard/details_Undi95__OpenRP-13B/blob/main/results_2023-10-29T00-54-50.325458.json)(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the "latest" split for each eval):
```python
{
"all": {
"em": 0.27632130872483224,
"em_stderr": 0.0045795175994957325,
"f1": 0.3337751677852358,
"f1_stderr": 0.004476795348022121,
"acc": 0.44447433030571937,
"acc_stderr": 0.010615829695443002
},
"harness|drop|3": {
"em": 0.27632130872483224,
"em_stderr": 0.0045795175994957325,
"f1": 0.3337751677852358,
"f1_stderr": 0.004476795348022121
},
"harness|gsm8k|5": {
"acc": 0.1288855193328279,
"acc_stderr": 0.009229580761400263
},
"harness|winogrande|5": {
"acc": 0.7600631412786109,
"acc_stderr": 0.012002078629485742
}
}
```
### Supported Tasks and Leaderboards
[More Information Needed]
### Languages
[More Information Needed]
## Dataset Structure
### Data Instances
[More Information Needed]
### Data Fields
[More Information Needed]
### Data Splits
[More Information Needed]
## Dataset Creation
### Curation Rationale
[More Information Needed]
### Source Data
#### Initial Data Collection and Normalization
[More Information Needed]
#### Who are the source language producers?
[More Information Needed]
### Annotations
#### Annotation process
[More Information Needed]
#### Who are the annotators?
[More Information Needed]
### Personal and Sensitive Information
[More Information Needed]
## Considerations for Using the Data
### Social Impact of Dataset
[More Information Needed]
### Discussion of Biases
[More Information Needed]
### Other Known Limitations
[More Information Needed]
## Additional Information
### Dataset Curators
[More Information Needed]
### Licensing Information
[More Information Needed]
### Citation Information
[More Information Needed]
### Contributions
[More Information Needed]
|
open-llm-leaderboard/details_Undi95__OpenRP-13B
|
[
"region:us"
] |
2023-09-18T12:49:23+00:00
|
{"pretty_name": "Evaluation run of Undi95/OpenRP-13B", "dataset_summary": "Dataset automatically created during the evaluation run of model [Undi95/OpenRP-13B](https://huggingface.co/Undi95/OpenRP-13B) on the [Open LLM Leaderboard](https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard).\n\nThe dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.\n\nThe dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The \"train\" split is always pointing to the latest results.\n\nAn additional configuration \"results\" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the [Open LLM Leaderboard](https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard)).\n\nTo load the details from a run, you can for instance do the following:\n```python\nfrom datasets import load_dataset\ndata = load_dataset(\"open-llm-leaderboard/details_Undi95__OpenRP-13B\",\n\t\"harness_winogrande_5\",\n\tsplit=\"train\")\n```\n\n## Latest results\n\nThese are the [latest results from run 2023-10-29T00:54:50.325458](https://huggingface.co/datasets/open-llm-leaderboard/details_Undi95__OpenRP-13B/blob/main/results_2023-10-29T00-54-50.325458.json)(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the \"latest\" split for each eval):\n\n```python\n{\n \"all\": {\n \"em\": 0.27632130872483224,\n \"em_stderr\": 0.0045795175994957325,\n \"f1\": 0.3337751677852358,\n \"f1_stderr\": 0.004476795348022121,\n \"acc\": 0.44447433030571937,\n \"acc_stderr\": 0.010615829695443002\n },\n \"harness|drop|3\": {\n \"em\": 0.27632130872483224,\n \"em_stderr\": 0.0045795175994957325,\n \"f1\": 0.3337751677852358,\n \"f1_stderr\": 0.004476795348022121\n },\n \"harness|gsm8k|5\": {\n \"acc\": 0.1288855193328279,\n \"acc_stderr\": 0.009229580761400263\n },\n \"harness|winogrande|5\": {\n \"acc\": 0.7600631412786109,\n \"acc_stderr\": 0.012002078629485742\n }\n}\n```", "repo_url": "https://huggingface.co/Undi95/OpenRP-13B", "leaderboard_url": "https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard", "point_of_contact": "[email protected]", "configs": [{"config_name": "harness_arc_challenge_25", "data_files": [{"split": "2023_09_18T13_48_59.614981", "path": ["**/details_harness|arc:challenge|25_2023-09-18T13-48-59.614981.parquet"]}, {"split": "latest", "path": ["**/details_harness|arc:challenge|25_2023-09-18T13-48-59.614981.parquet"]}]}, {"config_name": "harness_drop_3", "data_files": [{"split": "2023_10_29T00_54_50.325458", "path": ["**/details_harness|drop|3_2023-10-29T00-54-50.325458.parquet"]}, {"split": "latest", "path": ["**/details_harness|drop|3_2023-10-29T00-54-50.325458.parquet"]}]}, {"config_name": "harness_gsm8k_5", "data_files": [{"split": "2023_10_29T00_54_50.325458", "path": ["**/details_harness|gsm8k|5_2023-10-29T00-54-50.325458.parquet"]}, {"split": "latest", "path": ["**/details_harness|gsm8k|5_2023-10-29T00-54-50.325458.parquet"]}]}, {"config_name": "harness_hellaswag_10", "data_files": [{"split": "2023_09_18T13_48_59.614981", "path": ["**/details_harness|hellaswag|10_2023-09-18T13-48-59.614981.parquet"]}, {"split": "latest", "path": ["**/details_harness|hellaswag|10_2023-09-18T13-48-59.614981.parquet"]}]}, {"config_name": "harness_hendrycksTest_5", "data_files": [{"split": "2023_09_18T13_48_59.614981", "path": ["**/details_harness|hendrycksTest-abstract_algebra|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-anatomy|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-astronomy|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-business_ethics|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-clinical_knowledge|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-college_biology|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-college_chemistry|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-college_computer_science|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-college_mathematics|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-college_medicine|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-college_physics|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-computer_security|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-conceptual_physics|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-econometrics|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-electrical_engineering|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-elementary_mathematics|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-formal_logic|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-global_facts|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-high_school_biology|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-high_school_chemistry|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-high_school_computer_science|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-high_school_european_history|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-high_school_geography|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-high_school_government_and_politics|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-high_school_macroeconomics|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-high_school_mathematics|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-high_school_microeconomics|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-high_school_physics|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-high_school_psychology|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-high_school_statistics|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-high_school_us_history|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-high_school_world_history|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-human_aging|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-human_sexuality|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-international_law|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-jurisprudence|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-logical_fallacies|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-machine_learning|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-management|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-marketing|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-medical_genetics|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-miscellaneous|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-moral_disputes|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-moral_scenarios|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-nutrition|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-philosophy|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-prehistory|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-professional_accounting|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-professional_law|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-professional_medicine|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-professional_psychology|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-public_relations|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-security_studies|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-sociology|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-us_foreign_policy|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-virology|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-world_religions|5_2023-09-18T13-48-59.614981.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-abstract_algebra|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-anatomy|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-astronomy|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-business_ethics|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-clinical_knowledge|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-college_biology|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-college_chemistry|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-college_computer_science|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-college_mathematics|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-college_medicine|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-college_physics|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-computer_security|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-conceptual_physics|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-econometrics|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-electrical_engineering|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-elementary_mathematics|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-formal_logic|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-global_facts|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-high_school_biology|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-high_school_chemistry|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-high_school_computer_science|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-high_school_european_history|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-high_school_geography|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-high_school_government_and_politics|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-high_school_macroeconomics|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-high_school_mathematics|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-high_school_microeconomics|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-high_school_physics|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-high_school_psychology|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-high_school_statistics|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-high_school_us_history|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-high_school_world_history|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-human_aging|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-human_sexuality|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-international_law|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-jurisprudence|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-logical_fallacies|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-machine_learning|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-management|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-marketing|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-medical_genetics|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-miscellaneous|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-moral_disputes|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-moral_scenarios|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-nutrition|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-philosophy|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-prehistory|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-professional_accounting|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-professional_law|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-professional_medicine|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-professional_psychology|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-public_relations|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-security_studies|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-sociology|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-us_foreign_policy|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-virology|5_2023-09-18T13-48-59.614981.parquet", "**/details_harness|hendrycksTest-world_religions|5_2023-09-18T13-48-59.614981.parquet"]}]}, {"config_name": "harness_hendrycksTest_abstract_algebra_5", "data_files": [{"split": "2023_09_18T13_48_59.614981", "path": ["**/details_harness|hendrycksTest-abstract_algebra|5_2023-09-18T13-48-59.614981.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-abstract_algebra|5_2023-09-18T13-48-59.614981.parquet"]}]}, {"config_name": "harness_hendrycksTest_anatomy_5", "data_files": [{"split": "2023_09_18T13_48_59.614981", "path": ["**/details_harness|hendrycksTest-anatomy|5_2023-09-18T13-48-59.614981.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-anatomy|5_2023-09-18T13-48-59.614981.parquet"]}]}, {"config_name": "harness_hendrycksTest_astronomy_5", "data_files": [{"split": "2023_09_18T13_48_59.614981", "path": ["**/details_harness|hendrycksTest-astronomy|5_2023-09-18T13-48-59.614981.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-astronomy|5_2023-09-18T13-48-59.614981.parquet"]}]}, {"config_name": "harness_hendrycksTest_business_ethics_5", "data_files": [{"split": "2023_09_18T13_48_59.614981", "path": ["**/details_harness|hendrycksTest-business_ethics|5_2023-09-18T13-48-59.614981.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-business_ethics|5_2023-09-18T13-48-59.614981.parquet"]}]}, {"config_name": "harness_hendrycksTest_clinical_knowledge_5", "data_files": [{"split": "2023_09_18T13_48_59.614981", "path": ["**/details_harness|hendrycksTest-clinical_knowledge|5_2023-09-18T13-48-59.614981.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-clinical_knowledge|5_2023-09-18T13-48-59.614981.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_biology_5", "data_files": [{"split": "2023_09_18T13_48_59.614981", "path": ["**/details_harness|hendrycksTest-college_biology|5_2023-09-18T13-48-59.614981.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_biology|5_2023-09-18T13-48-59.614981.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_chemistry_5", "data_files": [{"split": "2023_09_18T13_48_59.614981", "path": ["**/details_harness|hendrycksTest-college_chemistry|5_2023-09-18T13-48-59.614981.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_chemistry|5_2023-09-18T13-48-59.614981.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_computer_science_5", "data_files": [{"split": "2023_09_18T13_48_59.614981", "path": ["**/details_harness|hendrycksTest-college_computer_science|5_2023-09-18T13-48-59.614981.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_computer_science|5_2023-09-18T13-48-59.614981.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_mathematics_5", "data_files": [{"split": "2023_09_18T13_48_59.614981", "path": ["**/details_harness|hendrycksTest-college_mathematics|5_2023-09-18T13-48-59.614981.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_mathematics|5_2023-09-18T13-48-59.614981.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_medicine_5", "data_files": [{"split": "2023_09_18T13_48_59.614981", "path": ["**/details_harness|hendrycksTest-college_medicine|5_2023-09-18T13-48-59.614981.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_medicine|5_2023-09-18T13-48-59.614981.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_physics_5", "data_files": [{"split": "2023_09_18T13_48_59.614981", "path": ["**/details_harness|hendrycksTest-college_physics|5_2023-09-18T13-48-59.614981.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_physics|5_2023-09-18T13-48-59.614981.parquet"]}]}, {"config_name": "harness_hendrycksTest_computer_security_5", "data_files": [{"split": "2023_09_18T13_48_59.614981", "path": ["**/details_harness|hendrycksTest-computer_security|5_2023-09-18T13-48-59.614981.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-computer_security|5_2023-09-18T13-48-59.614981.parquet"]}]}, {"config_name": "harness_hendrycksTest_conceptual_physics_5", "data_files": [{"split": "2023_09_18T13_48_59.614981", "path": ["**/details_harness|hendrycksTest-conceptual_physics|5_2023-09-18T13-48-59.614981.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-conceptual_physics|5_2023-09-18T13-48-59.614981.parquet"]}]}, {"config_name": "harness_hendrycksTest_econometrics_5", "data_files": [{"split": "2023_09_18T13_48_59.614981", "path": ["**/details_harness|hendrycksTest-econometrics|5_2023-09-18T13-48-59.614981.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-econometrics|5_2023-09-18T13-48-59.614981.parquet"]}]}, {"config_name": "harness_hendrycksTest_electrical_engineering_5", "data_files": [{"split": "2023_09_18T13_48_59.614981", "path": ["**/details_harness|hendrycksTest-electrical_engineering|5_2023-09-18T13-48-59.614981.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-electrical_engineering|5_2023-09-18T13-48-59.614981.parquet"]}]}, {"config_name": "harness_hendrycksTest_elementary_mathematics_5", "data_files": [{"split": "2023_09_18T13_48_59.614981", "path": ["**/details_harness|hendrycksTest-elementary_mathematics|5_2023-09-18T13-48-59.614981.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-elementary_mathematics|5_2023-09-18T13-48-59.614981.parquet"]}]}, {"config_name": "harness_hendrycksTest_formal_logic_5", "data_files": [{"split": "2023_09_18T13_48_59.614981", "path": ["**/details_harness|hendrycksTest-formal_logic|5_2023-09-18T13-48-59.614981.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-formal_logic|5_2023-09-18T13-48-59.614981.parquet"]}]}, {"config_name": "harness_hendrycksTest_global_facts_5", "data_files": [{"split": "2023_09_18T13_48_59.614981", "path": ["**/details_harness|hendrycksTest-global_facts|5_2023-09-18T13-48-59.614981.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-global_facts|5_2023-09-18T13-48-59.614981.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_biology_5", "data_files": [{"split": "2023_09_18T13_48_59.614981", "path": ["**/details_harness|hendrycksTest-high_school_biology|5_2023-09-18T13-48-59.614981.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_biology|5_2023-09-18T13-48-59.614981.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_chemistry_5", "data_files": [{"split": "2023_09_18T13_48_59.614981", "path": ["**/details_harness|hendrycksTest-high_school_chemistry|5_2023-09-18T13-48-59.614981.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_chemistry|5_2023-09-18T13-48-59.614981.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_computer_science_5", "data_files": [{"split": "2023_09_18T13_48_59.614981", "path": ["**/details_harness|hendrycksTest-high_school_computer_science|5_2023-09-18T13-48-59.614981.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_computer_science|5_2023-09-18T13-48-59.614981.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_european_history_5", "data_files": [{"split": "2023_09_18T13_48_59.614981", "path": ["**/details_harness|hendrycksTest-high_school_european_history|5_2023-09-18T13-48-59.614981.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_european_history|5_2023-09-18T13-48-59.614981.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_geography_5", "data_files": [{"split": "2023_09_18T13_48_59.614981", "path": ["**/details_harness|hendrycksTest-high_school_geography|5_2023-09-18T13-48-59.614981.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_geography|5_2023-09-18T13-48-59.614981.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_government_and_politics_5", "data_files": [{"split": "2023_09_18T13_48_59.614981", "path": ["**/details_harness|hendrycksTest-high_school_government_and_politics|5_2023-09-18T13-48-59.614981.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_government_and_politics|5_2023-09-18T13-48-59.614981.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_macroeconomics_5", "data_files": [{"split": "2023_09_18T13_48_59.614981", "path": ["**/details_harness|hendrycksTest-high_school_macroeconomics|5_2023-09-18T13-48-59.614981.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_macroeconomics|5_2023-09-18T13-48-59.614981.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_mathematics_5", "data_files": [{"split": "2023_09_18T13_48_59.614981", "path": ["**/details_harness|hendrycksTest-high_school_mathematics|5_2023-09-18T13-48-59.614981.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_mathematics|5_2023-09-18T13-48-59.614981.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_microeconomics_5", "data_files": [{"split": "2023_09_18T13_48_59.614981", "path": ["**/details_harness|hendrycksTest-high_school_microeconomics|5_2023-09-18T13-48-59.614981.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_microeconomics|5_2023-09-18T13-48-59.614981.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_physics_5", "data_files": [{"split": "2023_09_18T13_48_59.614981", "path": ["**/details_harness|hendrycksTest-high_school_physics|5_2023-09-18T13-48-59.614981.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_physics|5_2023-09-18T13-48-59.614981.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_psychology_5", "data_files": [{"split": "2023_09_18T13_48_59.614981", "path": ["**/details_harness|hendrycksTest-high_school_psychology|5_2023-09-18T13-48-59.614981.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_psychology|5_2023-09-18T13-48-59.614981.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_statistics_5", "data_files": [{"split": "2023_09_18T13_48_59.614981", "path": ["**/details_harness|hendrycksTest-high_school_statistics|5_2023-09-18T13-48-59.614981.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_statistics|5_2023-09-18T13-48-59.614981.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_us_history_5", "data_files": [{"split": "2023_09_18T13_48_59.614981", "path": ["**/details_harness|hendrycksTest-high_school_us_history|5_2023-09-18T13-48-59.614981.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_us_history|5_2023-09-18T13-48-59.614981.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_world_history_5", "data_files": [{"split": "2023_09_18T13_48_59.614981", "path": ["**/details_harness|hendrycksTest-high_school_world_history|5_2023-09-18T13-48-59.614981.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_world_history|5_2023-09-18T13-48-59.614981.parquet"]}]}, {"config_name": "harness_hendrycksTest_human_aging_5", "data_files": [{"split": "2023_09_18T13_48_59.614981", "path": ["**/details_harness|hendrycksTest-human_aging|5_2023-09-18T13-48-59.614981.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-human_aging|5_2023-09-18T13-48-59.614981.parquet"]}]}, {"config_name": "harness_hendrycksTest_human_sexuality_5", "data_files": [{"split": "2023_09_18T13_48_59.614981", "path": ["**/details_harness|hendrycksTest-human_sexuality|5_2023-09-18T13-48-59.614981.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-human_sexuality|5_2023-09-18T13-48-59.614981.parquet"]}]}, {"config_name": "harness_hendrycksTest_international_law_5", "data_files": [{"split": "2023_09_18T13_48_59.614981", "path": ["**/details_harness|hendrycksTest-international_law|5_2023-09-18T13-48-59.614981.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-international_law|5_2023-09-18T13-48-59.614981.parquet"]}]}, {"config_name": "harness_hendrycksTest_jurisprudence_5", "data_files": [{"split": "2023_09_18T13_48_59.614981", "path": ["**/details_harness|hendrycksTest-jurisprudence|5_2023-09-18T13-48-59.614981.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-jurisprudence|5_2023-09-18T13-48-59.614981.parquet"]}]}, {"config_name": "harness_hendrycksTest_logical_fallacies_5", "data_files": [{"split": "2023_09_18T13_48_59.614981", "path": ["**/details_harness|hendrycksTest-logical_fallacies|5_2023-09-18T13-48-59.614981.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-logical_fallacies|5_2023-09-18T13-48-59.614981.parquet"]}]}, {"config_name": "harness_hendrycksTest_machine_learning_5", "data_files": [{"split": "2023_09_18T13_48_59.614981", "path": ["**/details_harness|hendrycksTest-machine_learning|5_2023-09-18T13-48-59.614981.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-machine_learning|5_2023-09-18T13-48-59.614981.parquet"]}]}, {"config_name": "harness_hendrycksTest_management_5", "data_files": [{"split": "2023_09_18T13_48_59.614981", "path": ["**/details_harness|hendrycksTest-management|5_2023-09-18T13-48-59.614981.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-management|5_2023-09-18T13-48-59.614981.parquet"]}]}, {"config_name": "harness_hendrycksTest_marketing_5", "data_files": [{"split": "2023_09_18T13_48_59.614981", "path": ["**/details_harness|hendrycksTest-marketing|5_2023-09-18T13-48-59.614981.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-marketing|5_2023-09-18T13-48-59.614981.parquet"]}]}, {"config_name": "harness_hendrycksTest_medical_genetics_5", "data_files": [{"split": "2023_09_18T13_48_59.614981", "path": ["**/details_harness|hendrycksTest-medical_genetics|5_2023-09-18T13-48-59.614981.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-medical_genetics|5_2023-09-18T13-48-59.614981.parquet"]}]}, {"config_name": "harness_hendrycksTest_miscellaneous_5", "data_files": [{"split": "2023_09_18T13_48_59.614981", "path": ["**/details_harness|hendrycksTest-miscellaneous|5_2023-09-18T13-48-59.614981.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-miscellaneous|5_2023-09-18T13-48-59.614981.parquet"]}]}, {"config_name": "harness_hendrycksTest_moral_disputes_5", "data_files": [{"split": "2023_09_18T13_48_59.614981", "path": ["**/details_harness|hendrycksTest-moral_disputes|5_2023-09-18T13-48-59.614981.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-moral_disputes|5_2023-09-18T13-48-59.614981.parquet"]}]}, {"config_name": "harness_hendrycksTest_moral_scenarios_5", "data_files": [{"split": "2023_09_18T13_48_59.614981", "path": ["**/details_harness|hendrycksTest-moral_scenarios|5_2023-09-18T13-48-59.614981.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-moral_scenarios|5_2023-09-18T13-48-59.614981.parquet"]}]}, {"config_name": "harness_hendrycksTest_nutrition_5", "data_files": [{"split": "2023_09_18T13_48_59.614981", "path": ["**/details_harness|hendrycksTest-nutrition|5_2023-09-18T13-48-59.614981.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-nutrition|5_2023-09-18T13-48-59.614981.parquet"]}]}, {"config_name": "harness_hendrycksTest_philosophy_5", "data_files": [{"split": "2023_09_18T13_48_59.614981", "path": ["**/details_harness|hendrycksTest-philosophy|5_2023-09-18T13-48-59.614981.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-philosophy|5_2023-09-18T13-48-59.614981.parquet"]}]}, {"config_name": "harness_hendrycksTest_prehistory_5", "data_files": [{"split": "2023_09_18T13_48_59.614981", "path": ["**/details_harness|hendrycksTest-prehistory|5_2023-09-18T13-48-59.614981.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-prehistory|5_2023-09-18T13-48-59.614981.parquet"]}]}, {"config_name": "harness_hendrycksTest_professional_accounting_5", "data_files": [{"split": "2023_09_18T13_48_59.614981", "path": ["**/details_harness|hendrycksTest-professional_accounting|5_2023-09-18T13-48-59.614981.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-professional_accounting|5_2023-09-18T13-48-59.614981.parquet"]}]}, {"config_name": "harness_hendrycksTest_professional_law_5", "data_files": [{"split": "2023_09_18T13_48_59.614981", "path": ["**/details_harness|hendrycksTest-professional_law|5_2023-09-18T13-48-59.614981.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-professional_law|5_2023-09-18T13-48-59.614981.parquet"]}]}, {"config_name": "harness_hendrycksTest_professional_medicine_5", "data_files": [{"split": "2023_09_18T13_48_59.614981", "path": ["**/details_harness|hendrycksTest-professional_medicine|5_2023-09-18T13-48-59.614981.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-professional_medicine|5_2023-09-18T13-48-59.614981.parquet"]}]}, {"config_name": "harness_hendrycksTest_professional_psychology_5", "data_files": [{"split": "2023_09_18T13_48_59.614981", "path": ["**/details_harness|hendrycksTest-professional_psychology|5_2023-09-18T13-48-59.614981.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-professional_psychology|5_2023-09-18T13-48-59.614981.parquet"]}]}, {"config_name": "harness_hendrycksTest_public_relations_5", "data_files": [{"split": "2023_09_18T13_48_59.614981", "path": ["**/details_harness|hendrycksTest-public_relations|5_2023-09-18T13-48-59.614981.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-public_relations|5_2023-09-18T13-48-59.614981.parquet"]}]}, {"config_name": "harness_hendrycksTest_security_studies_5", "data_files": [{"split": "2023_09_18T13_48_59.614981", "path": ["**/details_harness|hendrycksTest-security_studies|5_2023-09-18T13-48-59.614981.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-security_studies|5_2023-09-18T13-48-59.614981.parquet"]}]}, {"config_name": "harness_hendrycksTest_sociology_5", "data_files": [{"split": "2023_09_18T13_48_59.614981", "path": ["**/details_harness|hendrycksTest-sociology|5_2023-09-18T13-48-59.614981.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-sociology|5_2023-09-18T13-48-59.614981.parquet"]}]}, {"config_name": "harness_hendrycksTest_us_foreign_policy_5", "data_files": [{"split": "2023_09_18T13_48_59.614981", "path": ["**/details_harness|hendrycksTest-us_foreign_policy|5_2023-09-18T13-48-59.614981.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-us_foreign_policy|5_2023-09-18T13-48-59.614981.parquet"]}]}, {"config_name": "harness_hendrycksTest_virology_5", "data_files": [{"split": "2023_09_18T13_48_59.614981", "path": ["**/details_harness|hendrycksTest-virology|5_2023-09-18T13-48-59.614981.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-virology|5_2023-09-18T13-48-59.614981.parquet"]}]}, {"config_name": "harness_hendrycksTest_world_religions_5", "data_files": [{"split": "2023_09_18T13_48_59.614981", "path": ["**/details_harness|hendrycksTest-world_religions|5_2023-09-18T13-48-59.614981.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-world_religions|5_2023-09-18T13-48-59.614981.parquet"]}]}, {"config_name": "harness_truthfulqa_mc_0", "data_files": [{"split": "2023_09_18T13_48_59.614981", "path": ["**/details_harness|truthfulqa:mc|0_2023-09-18T13-48-59.614981.parquet"]}, {"split": "latest", "path": ["**/details_harness|truthfulqa:mc|0_2023-09-18T13-48-59.614981.parquet"]}]}, {"config_name": "harness_winogrande_5", "data_files": [{"split": "2023_10_29T00_54_50.325458", "path": ["**/details_harness|winogrande|5_2023-10-29T00-54-50.325458.parquet"]}, {"split": "latest", "path": ["**/details_harness|winogrande|5_2023-10-29T00-54-50.325458.parquet"]}]}, {"config_name": "results", "data_files": [{"split": "2023_09_18T13_48_59.614981", "path": ["results_2023-09-18T13-48-59.614981.parquet"]}, {"split": "2023_10_29T00_54_50.325458", "path": ["results_2023-10-29T00-54-50.325458.parquet"]}, {"split": "latest", "path": ["results_2023-10-29T00-54-50.325458.parquet"]}]}]}
|
2023-10-28T23:55:02+00:00
|
[] |
[] |
TAGS
#region-us
|
# Dataset Card for Evaluation run of Undi95/OpenRP-13B
## Dataset Description
- Homepage:
- Repository: URL
- Paper:
- Leaderboard: URL
- Point of Contact: clementine@URL
### Dataset Summary
Dataset automatically created during the evaluation run of model Undi95/OpenRP-13B on the Open LLM Leaderboard.
The dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.
The dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The "train" split is always pointing to the latest results.
An additional configuration "results" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the Open LLM Leaderboard).
To load the details from a run, you can for instance do the following:
## Latest results
These are the latest results from run 2023-10-29T00:54:50.325458(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the "latest" split for each eval):
### Supported Tasks and Leaderboards
### Languages
## Dataset Structure
### Data Instances
### Data Fields
### Data Splits
## Dataset Creation
### Curation Rationale
### Source Data
#### Initial Data Collection and Normalization
#### Who are the source language producers?
### Annotations
#### Annotation process
#### Who are the annotators?
### Personal and Sensitive Information
## Considerations for Using the Data
### Social Impact of Dataset
### Discussion of Biases
### Other Known Limitations
## Additional Information
### Dataset Curators
### Licensing Information
### Contributions
|
[
"# Dataset Card for Evaluation run of Undi95/OpenRP-13B",
"## Dataset Description\n\n- Homepage: \n- Repository: URL\n- Paper: \n- Leaderboard: URL\n- Point of Contact: clementine@URL",
"### Dataset Summary\n\nDataset automatically created during the evaluation run of model Undi95/OpenRP-13B on the Open LLM Leaderboard.\n\nThe dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.\n\nThe dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The \"train\" split is always pointing to the latest results.\n\nAn additional configuration \"results\" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the Open LLM Leaderboard).\n\nTo load the details from a run, you can for instance do the following:",
"## Latest results\n\nThese are the latest results from run 2023-10-29T00:54:50.325458(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the \"latest\" split for each eval):",
"### Supported Tasks and Leaderboards",
"### Languages",
"## Dataset Structure",
"### Data Instances",
"### Data Fields",
"### Data Splits",
"## Dataset Creation",
"### Curation Rationale",
"### Source Data",
"#### Initial Data Collection and Normalization",
"#### Who are the source language producers?",
"### Annotations",
"#### Annotation process",
"#### Who are the annotators?",
"### Personal and Sensitive Information",
"## Considerations for Using the Data",
"### Social Impact of Dataset",
"### Discussion of Biases",
"### Other Known Limitations",
"## Additional Information",
"### Dataset Curators",
"### Licensing Information",
"### Contributions"
] |
[
"TAGS\n#region-us \n",
"# Dataset Card for Evaluation run of Undi95/OpenRP-13B",
"## Dataset Description\n\n- Homepage: \n- Repository: URL\n- Paper: \n- Leaderboard: URL\n- Point of Contact: clementine@URL",
"### Dataset Summary\n\nDataset automatically created during the evaluation run of model Undi95/OpenRP-13B on the Open LLM Leaderboard.\n\nThe dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.\n\nThe dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The \"train\" split is always pointing to the latest results.\n\nAn additional configuration \"results\" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the Open LLM Leaderboard).\n\nTo load the details from a run, you can for instance do the following:",
"## Latest results\n\nThese are the latest results from run 2023-10-29T00:54:50.325458(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the \"latest\" split for each eval):",
"### Supported Tasks and Leaderboards",
"### Languages",
"## Dataset Structure",
"### Data Instances",
"### Data Fields",
"### Data Splits",
"## Dataset Creation",
"### Curation Rationale",
"### Source Data",
"#### Initial Data Collection and Normalization",
"#### Who are the source language producers?",
"### Annotations",
"#### Annotation process",
"#### Who are the annotators?",
"### Personal and Sensitive Information",
"## Considerations for Using the Data",
"### Social Impact of Dataset",
"### Discussion of Biases",
"### Other Known Limitations",
"## Additional Information",
"### Dataset Curators",
"### Licensing Information",
"### Contributions"
] |
[
6,
17,
31,
165,
67,
10,
4,
6,
6,
5,
5,
5,
7,
4,
10,
10,
5,
5,
9,
8,
8,
7,
8,
7,
5,
6,
6,
5
] |
[
"passage: TAGS\n#region-us \n# Dataset Card for Evaluation run of Undi95/OpenRP-13B## Dataset Description\n\n- Homepage: \n- Repository: URL\n- Paper: \n- Leaderboard: URL\n- Point of Contact: clementine@URL### Dataset Summary\n\nDataset automatically created during the evaluation run of model Undi95/OpenRP-13B on the Open LLM Leaderboard.\n\nThe dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.\n\nThe dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The \"train\" split is always pointing to the latest results.\n\nAn additional configuration \"results\" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the Open LLM Leaderboard).\n\nTo load the details from a run, you can for instance do the following:## Latest results\n\nThese are the latest results from run 2023-10-29T00:54:50.325458(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the \"latest\" split for each eval):### Supported Tasks and Leaderboards### Languages## Dataset Structure### Data Instances### Data Fields### Data Splits## Dataset Creation### Curation Rationale### Source Data#### Initial Data Collection and Normalization#### Who are the source language producers?### Annotations#### Annotation process#### Who are the annotators?### Personal and Sensitive Information## Considerations for Using the Data### Social Impact of Dataset### Discussion of Biases### Other Known Limitations## Additional Information### Dataset Curators### Licensing Information### Contributions"
] |
c818efd131db2ecba4abdcb7a18880e6cc85f6bf
|
# Dataset Card for Evaluation run of nicholasKluge/Aira-2-774M
## Dataset Description
- **Homepage:**
- **Repository:** https://huggingface.co/nicholasKluge/Aira-2-774M
- **Paper:**
- **Leaderboard:** https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard
- **Point of Contact:** [email protected]
### Dataset Summary
Dataset automatically created during the evaluation run of model [nicholasKluge/Aira-2-774M](https://huggingface.co/nicholasKluge/Aira-2-774M) on the [Open LLM Leaderboard](https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard).
The dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.
The dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The "train" split is always pointing to the latest results.
An additional configuration "results" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the [Open LLM Leaderboard](https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard)).
To load the details from a run, you can for instance do the following:
```python
from datasets import load_dataset
data = load_dataset("open-llm-leaderboard/details_nicholasKluge__Aira-2-774M",
"harness_winogrande_5",
split="train")
```
## Latest results
These are the [latest results from run 2023-10-23T22:37:50.525759](https://huggingface.co/datasets/open-llm-leaderboard/details_nicholasKluge__Aira-2-774M/blob/main/results_2023-10-23T22-37-50.525759.json)(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the "latest" split for each eval):
```python
{
"all": {
"em": 0.019819630872483222,
"em_stderr": 0.0014273827117585976,
"f1": 0.04258074664429536,
"f1_stderr": 0.0017104629235282784,
"acc": 0.2600631412786109,
"acc_stderr": 0.007020548332172165
},
"harness|drop|3": {
"em": 0.019819630872483222,
"em_stderr": 0.0014273827117585976,
"f1": 0.04258074664429536,
"f1_stderr": 0.0017104629235282784
},
"harness|gsm8k|5": {
"acc": 0.0,
"acc_stderr": 0.0
},
"harness|winogrande|5": {
"acc": 0.5201262825572218,
"acc_stderr": 0.01404109666434433
}
}
```
### Supported Tasks and Leaderboards
[More Information Needed]
### Languages
[More Information Needed]
## Dataset Structure
### Data Instances
[More Information Needed]
### Data Fields
[More Information Needed]
### Data Splits
[More Information Needed]
## Dataset Creation
### Curation Rationale
[More Information Needed]
### Source Data
#### Initial Data Collection and Normalization
[More Information Needed]
#### Who are the source language producers?
[More Information Needed]
### Annotations
#### Annotation process
[More Information Needed]
#### Who are the annotators?
[More Information Needed]
### Personal and Sensitive Information
[More Information Needed]
## Considerations for Using the Data
### Social Impact of Dataset
[More Information Needed]
### Discussion of Biases
[More Information Needed]
### Other Known Limitations
[More Information Needed]
## Additional Information
### Dataset Curators
[More Information Needed]
### Licensing Information
[More Information Needed]
### Citation Information
[More Information Needed]
### Contributions
[More Information Needed]
|
open-llm-leaderboard/details_nicholasKluge__Aira-2-774M
|
[
"region:us"
] |
2023-09-18T12:49:50+00:00
|
{"pretty_name": "Evaluation run of nicholasKluge/Aira-2-774M", "dataset_summary": "Dataset automatically created during the evaluation run of model [nicholasKluge/Aira-2-774M](https://huggingface.co/nicholasKluge/Aira-2-774M) on the [Open LLM Leaderboard](https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard).\n\nThe dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.\n\nThe dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The \"train\" split is always pointing to the latest results.\n\nAn additional configuration \"results\" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the [Open LLM Leaderboard](https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard)).\n\nTo load the details from a run, you can for instance do the following:\n```python\nfrom datasets import load_dataset\ndata = load_dataset(\"open-llm-leaderboard/details_nicholasKluge__Aira-2-774M\",\n\t\"harness_winogrande_5\",\n\tsplit=\"train\")\n```\n\n## Latest results\n\nThese are the [latest results from run 2023-10-23T22:37:50.525759](https://huggingface.co/datasets/open-llm-leaderboard/details_nicholasKluge__Aira-2-774M/blob/main/results_2023-10-23T22-37-50.525759.json)(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the \"latest\" split for each eval):\n\n```python\n{\n \"all\": {\n \"em\": 0.019819630872483222,\n \"em_stderr\": 0.0014273827117585976,\n \"f1\": 0.04258074664429536,\n \"f1_stderr\": 0.0017104629235282784,\n \"acc\": 0.2600631412786109,\n \"acc_stderr\": 0.007020548332172165\n },\n \"harness|drop|3\": {\n \"em\": 0.019819630872483222,\n \"em_stderr\": 0.0014273827117585976,\n \"f1\": 0.04258074664429536,\n \"f1_stderr\": 0.0017104629235282784\n },\n \"harness|gsm8k|5\": {\n \"acc\": 0.0,\n \"acc_stderr\": 0.0\n },\n \"harness|winogrande|5\": {\n \"acc\": 0.5201262825572218,\n \"acc_stderr\": 0.01404109666434433\n }\n}\n```", "repo_url": "https://huggingface.co/nicholasKluge/Aira-2-774M", "leaderboard_url": "https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard", "point_of_contact": "[email protected]", "configs": [{"config_name": "harness_arc_challenge_25", "data_files": [{"split": "2023_09_18T13_49_35.718586", "path": ["**/details_harness|arc:challenge|25_2023-09-18T13-49-35.718586.parquet"]}, {"split": "latest", "path": ["**/details_harness|arc:challenge|25_2023-09-18T13-49-35.718586.parquet"]}]}, {"config_name": "harness_drop_3", "data_files": [{"split": "2023_10_23T22_37_50.525759", "path": ["**/details_harness|drop|3_2023-10-23T22-37-50.525759.parquet"]}, {"split": "latest", "path": ["**/details_harness|drop|3_2023-10-23T22-37-50.525759.parquet"]}]}, {"config_name": "harness_gsm8k_5", "data_files": [{"split": "2023_10_23T22_37_50.525759", "path": ["**/details_harness|gsm8k|5_2023-10-23T22-37-50.525759.parquet"]}, {"split": "latest", "path": ["**/details_harness|gsm8k|5_2023-10-23T22-37-50.525759.parquet"]}]}, {"config_name": "harness_hellaswag_10", "data_files": [{"split": "2023_09_18T13_49_35.718586", "path": ["**/details_harness|hellaswag|10_2023-09-18T13-49-35.718586.parquet"]}, {"split": "latest", "path": ["**/details_harness|hellaswag|10_2023-09-18T13-49-35.718586.parquet"]}]}, {"config_name": "harness_hendrycksTest_5", "data_files": [{"split": "2023_09_18T13_49_35.718586", "path": ["**/details_harness|hendrycksTest-abstract_algebra|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-anatomy|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-astronomy|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-business_ethics|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-clinical_knowledge|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-college_biology|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-college_chemistry|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-college_computer_science|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-college_mathematics|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-college_medicine|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-college_physics|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-computer_security|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-conceptual_physics|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-econometrics|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-electrical_engineering|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-elementary_mathematics|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-formal_logic|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-global_facts|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-high_school_biology|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-high_school_chemistry|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-high_school_computer_science|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-high_school_european_history|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-high_school_geography|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-high_school_government_and_politics|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-high_school_macroeconomics|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-high_school_mathematics|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-high_school_microeconomics|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-high_school_physics|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-high_school_psychology|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-high_school_statistics|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-high_school_us_history|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-high_school_world_history|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-human_aging|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-human_sexuality|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-international_law|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-jurisprudence|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-logical_fallacies|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-machine_learning|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-management|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-marketing|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-medical_genetics|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-miscellaneous|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-moral_disputes|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-moral_scenarios|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-nutrition|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-philosophy|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-prehistory|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-professional_accounting|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-professional_law|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-professional_medicine|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-professional_psychology|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-public_relations|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-security_studies|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-sociology|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-us_foreign_policy|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-virology|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-world_religions|5_2023-09-18T13-49-35.718586.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-abstract_algebra|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-anatomy|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-astronomy|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-business_ethics|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-clinical_knowledge|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-college_biology|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-college_chemistry|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-college_computer_science|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-college_mathematics|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-college_medicine|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-college_physics|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-computer_security|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-conceptual_physics|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-econometrics|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-electrical_engineering|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-elementary_mathematics|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-formal_logic|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-global_facts|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-high_school_biology|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-high_school_chemistry|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-high_school_computer_science|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-high_school_european_history|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-high_school_geography|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-high_school_government_and_politics|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-high_school_macroeconomics|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-high_school_mathematics|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-high_school_microeconomics|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-high_school_physics|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-high_school_psychology|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-high_school_statistics|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-high_school_us_history|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-high_school_world_history|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-human_aging|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-human_sexuality|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-international_law|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-jurisprudence|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-logical_fallacies|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-machine_learning|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-management|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-marketing|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-medical_genetics|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-miscellaneous|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-moral_disputes|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-moral_scenarios|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-nutrition|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-philosophy|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-prehistory|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-professional_accounting|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-professional_law|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-professional_medicine|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-professional_psychology|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-public_relations|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-security_studies|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-sociology|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-us_foreign_policy|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-virology|5_2023-09-18T13-49-35.718586.parquet", "**/details_harness|hendrycksTest-world_religions|5_2023-09-18T13-49-35.718586.parquet"]}]}, {"config_name": "harness_hendrycksTest_abstract_algebra_5", "data_files": [{"split": "2023_09_18T13_49_35.718586", "path": ["**/details_harness|hendrycksTest-abstract_algebra|5_2023-09-18T13-49-35.718586.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-abstract_algebra|5_2023-09-18T13-49-35.718586.parquet"]}]}, {"config_name": "harness_hendrycksTest_anatomy_5", "data_files": [{"split": "2023_09_18T13_49_35.718586", "path": ["**/details_harness|hendrycksTest-anatomy|5_2023-09-18T13-49-35.718586.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-anatomy|5_2023-09-18T13-49-35.718586.parquet"]}]}, {"config_name": "harness_hendrycksTest_astronomy_5", "data_files": [{"split": "2023_09_18T13_49_35.718586", "path": ["**/details_harness|hendrycksTest-astronomy|5_2023-09-18T13-49-35.718586.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-astronomy|5_2023-09-18T13-49-35.718586.parquet"]}]}, {"config_name": "harness_hendrycksTest_business_ethics_5", "data_files": [{"split": "2023_09_18T13_49_35.718586", "path": ["**/details_harness|hendrycksTest-business_ethics|5_2023-09-18T13-49-35.718586.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-business_ethics|5_2023-09-18T13-49-35.718586.parquet"]}]}, {"config_name": "harness_hendrycksTest_clinical_knowledge_5", "data_files": [{"split": "2023_09_18T13_49_35.718586", "path": ["**/details_harness|hendrycksTest-clinical_knowledge|5_2023-09-18T13-49-35.718586.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-clinical_knowledge|5_2023-09-18T13-49-35.718586.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_biology_5", "data_files": [{"split": "2023_09_18T13_49_35.718586", "path": ["**/details_harness|hendrycksTest-college_biology|5_2023-09-18T13-49-35.718586.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_biology|5_2023-09-18T13-49-35.718586.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_chemistry_5", "data_files": [{"split": "2023_09_18T13_49_35.718586", "path": ["**/details_harness|hendrycksTest-college_chemistry|5_2023-09-18T13-49-35.718586.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_chemistry|5_2023-09-18T13-49-35.718586.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_computer_science_5", "data_files": [{"split": "2023_09_18T13_49_35.718586", "path": ["**/details_harness|hendrycksTest-college_computer_science|5_2023-09-18T13-49-35.718586.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_computer_science|5_2023-09-18T13-49-35.718586.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_mathematics_5", "data_files": [{"split": "2023_09_18T13_49_35.718586", "path": ["**/details_harness|hendrycksTest-college_mathematics|5_2023-09-18T13-49-35.718586.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_mathematics|5_2023-09-18T13-49-35.718586.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_medicine_5", "data_files": [{"split": "2023_09_18T13_49_35.718586", "path": ["**/details_harness|hendrycksTest-college_medicine|5_2023-09-18T13-49-35.718586.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_medicine|5_2023-09-18T13-49-35.718586.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_physics_5", "data_files": [{"split": "2023_09_18T13_49_35.718586", "path": ["**/details_harness|hendrycksTest-college_physics|5_2023-09-18T13-49-35.718586.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_physics|5_2023-09-18T13-49-35.718586.parquet"]}]}, {"config_name": "harness_hendrycksTest_computer_security_5", "data_files": [{"split": "2023_09_18T13_49_35.718586", "path": ["**/details_harness|hendrycksTest-computer_security|5_2023-09-18T13-49-35.718586.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-computer_security|5_2023-09-18T13-49-35.718586.parquet"]}]}, {"config_name": "harness_hendrycksTest_conceptual_physics_5", "data_files": [{"split": "2023_09_18T13_49_35.718586", "path": ["**/details_harness|hendrycksTest-conceptual_physics|5_2023-09-18T13-49-35.718586.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-conceptual_physics|5_2023-09-18T13-49-35.718586.parquet"]}]}, {"config_name": "harness_hendrycksTest_econometrics_5", "data_files": [{"split": "2023_09_18T13_49_35.718586", "path": ["**/details_harness|hendrycksTest-econometrics|5_2023-09-18T13-49-35.718586.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-econometrics|5_2023-09-18T13-49-35.718586.parquet"]}]}, {"config_name": "harness_hendrycksTest_electrical_engineering_5", "data_files": [{"split": "2023_09_18T13_49_35.718586", "path": ["**/details_harness|hendrycksTest-electrical_engineering|5_2023-09-18T13-49-35.718586.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-electrical_engineering|5_2023-09-18T13-49-35.718586.parquet"]}]}, {"config_name": "harness_hendrycksTest_elementary_mathematics_5", "data_files": [{"split": "2023_09_18T13_49_35.718586", "path": ["**/details_harness|hendrycksTest-elementary_mathematics|5_2023-09-18T13-49-35.718586.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-elementary_mathematics|5_2023-09-18T13-49-35.718586.parquet"]}]}, {"config_name": "harness_hendrycksTest_formal_logic_5", "data_files": [{"split": "2023_09_18T13_49_35.718586", "path": ["**/details_harness|hendrycksTest-formal_logic|5_2023-09-18T13-49-35.718586.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-formal_logic|5_2023-09-18T13-49-35.718586.parquet"]}]}, {"config_name": "harness_hendrycksTest_global_facts_5", "data_files": [{"split": "2023_09_18T13_49_35.718586", "path": ["**/details_harness|hendrycksTest-global_facts|5_2023-09-18T13-49-35.718586.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-global_facts|5_2023-09-18T13-49-35.718586.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_biology_5", "data_files": [{"split": "2023_09_18T13_49_35.718586", "path": ["**/details_harness|hendrycksTest-high_school_biology|5_2023-09-18T13-49-35.718586.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_biology|5_2023-09-18T13-49-35.718586.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_chemistry_5", "data_files": [{"split": "2023_09_18T13_49_35.718586", "path": ["**/details_harness|hendrycksTest-high_school_chemistry|5_2023-09-18T13-49-35.718586.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_chemistry|5_2023-09-18T13-49-35.718586.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_computer_science_5", "data_files": [{"split": "2023_09_18T13_49_35.718586", "path": ["**/details_harness|hendrycksTest-high_school_computer_science|5_2023-09-18T13-49-35.718586.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_computer_science|5_2023-09-18T13-49-35.718586.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_european_history_5", "data_files": [{"split": "2023_09_18T13_49_35.718586", "path": ["**/details_harness|hendrycksTest-high_school_european_history|5_2023-09-18T13-49-35.718586.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_european_history|5_2023-09-18T13-49-35.718586.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_geography_5", "data_files": [{"split": "2023_09_18T13_49_35.718586", "path": ["**/details_harness|hendrycksTest-high_school_geography|5_2023-09-18T13-49-35.718586.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_geography|5_2023-09-18T13-49-35.718586.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_government_and_politics_5", "data_files": [{"split": "2023_09_18T13_49_35.718586", "path": ["**/details_harness|hendrycksTest-high_school_government_and_politics|5_2023-09-18T13-49-35.718586.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_government_and_politics|5_2023-09-18T13-49-35.718586.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_macroeconomics_5", "data_files": [{"split": "2023_09_18T13_49_35.718586", "path": ["**/details_harness|hendrycksTest-high_school_macroeconomics|5_2023-09-18T13-49-35.718586.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_macroeconomics|5_2023-09-18T13-49-35.718586.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_mathematics_5", "data_files": [{"split": "2023_09_18T13_49_35.718586", "path": ["**/details_harness|hendrycksTest-high_school_mathematics|5_2023-09-18T13-49-35.718586.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_mathematics|5_2023-09-18T13-49-35.718586.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_microeconomics_5", "data_files": [{"split": "2023_09_18T13_49_35.718586", "path": ["**/details_harness|hendrycksTest-high_school_microeconomics|5_2023-09-18T13-49-35.718586.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_microeconomics|5_2023-09-18T13-49-35.718586.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_physics_5", "data_files": [{"split": "2023_09_18T13_49_35.718586", "path": ["**/details_harness|hendrycksTest-high_school_physics|5_2023-09-18T13-49-35.718586.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_physics|5_2023-09-18T13-49-35.718586.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_psychology_5", "data_files": [{"split": "2023_09_18T13_49_35.718586", "path": ["**/details_harness|hendrycksTest-high_school_psychology|5_2023-09-18T13-49-35.718586.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_psychology|5_2023-09-18T13-49-35.718586.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_statistics_5", "data_files": [{"split": "2023_09_18T13_49_35.718586", "path": ["**/details_harness|hendrycksTest-high_school_statistics|5_2023-09-18T13-49-35.718586.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_statistics|5_2023-09-18T13-49-35.718586.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_us_history_5", "data_files": [{"split": "2023_09_18T13_49_35.718586", "path": ["**/details_harness|hendrycksTest-high_school_us_history|5_2023-09-18T13-49-35.718586.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_us_history|5_2023-09-18T13-49-35.718586.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_world_history_5", "data_files": [{"split": "2023_09_18T13_49_35.718586", "path": ["**/details_harness|hendrycksTest-high_school_world_history|5_2023-09-18T13-49-35.718586.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_world_history|5_2023-09-18T13-49-35.718586.parquet"]}]}, {"config_name": "harness_hendrycksTest_human_aging_5", "data_files": [{"split": "2023_09_18T13_49_35.718586", "path": ["**/details_harness|hendrycksTest-human_aging|5_2023-09-18T13-49-35.718586.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-human_aging|5_2023-09-18T13-49-35.718586.parquet"]}]}, {"config_name": "harness_hendrycksTest_human_sexuality_5", "data_files": [{"split": "2023_09_18T13_49_35.718586", "path": ["**/details_harness|hendrycksTest-human_sexuality|5_2023-09-18T13-49-35.718586.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-human_sexuality|5_2023-09-18T13-49-35.718586.parquet"]}]}, {"config_name": "harness_hendrycksTest_international_law_5", "data_files": [{"split": "2023_09_18T13_49_35.718586", "path": ["**/details_harness|hendrycksTest-international_law|5_2023-09-18T13-49-35.718586.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-international_law|5_2023-09-18T13-49-35.718586.parquet"]}]}, {"config_name": "harness_hendrycksTest_jurisprudence_5", "data_files": [{"split": "2023_09_18T13_49_35.718586", "path": ["**/details_harness|hendrycksTest-jurisprudence|5_2023-09-18T13-49-35.718586.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-jurisprudence|5_2023-09-18T13-49-35.718586.parquet"]}]}, {"config_name": "harness_hendrycksTest_logical_fallacies_5", "data_files": [{"split": "2023_09_18T13_49_35.718586", "path": ["**/details_harness|hendrycksTest-logical_fallacies|5_2023-09-18T13-49-35.718586.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-logical_fallacies|5_2023-09-18T13-49-35.718586.parquet"]}]}, {"config_name": "harness_hendrycksTest_machine_learning_5", "data_files": [{"split": "2023_09_18T13_49_35.718586", "path": ["**/details_harness|hendrycksTest-machine_learning|5_2023-09-18T13-49-35.718586.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-machine_learning|5_2023-09-18T13-49-35.718586.parquet"]}]}, {"config_name": "harness_hendrycksTest_management_5", "data_files": [{"split": "2023_09_18T13_49_35.718586", "path": ["**/details_harness|hendrycksTest-management|5_2023-09-18T13-49-35.718586.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-management|5_2023-09-18T13-49-35.718586.parquet"]}]}, {"config_name": "harness_hendrycksTest_marketing_5", "data_files": [{"split": "2023_09_18T13_49_35.718586", "path": ["**/details_harness|hendrycksTest-marketing|5_2023-09-18T13-49-35.718586.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-marketing|5_2023-09-18T13-49-35.718586.parquet"]}]}, {"config_name": "harness_hendrycksTest_medical_genetics_5", "data_files": [{"split": "2023_09_18T13_49_35.718586", "path": ["**/details_harness|hendrycksTest-medical_genetics|5_2023-09-18T13-49-35.718586.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-medical_genetics|5_2023-09-18T13-49-35.718586.parquet"]}]}, {"config_name": "harness_hendrycksTest_miscellaneous_5", "data_files": [{"split": "2023_09_18T13_49_35.718586", "path": ["**/details_harness|hendrycksTest-miscellaneous|5_2023-09-18T13-49-35.718586.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-miscellaneous|5_2023-09-18T13-49-35.718586.parquet"]}]}, {"config_name": "harness_hendrycksTest_moral_disputes_5", "data_files": [{"split": "2023_09_18T13_49_35.718586", "path": ["**/details_harness|hendrycksTest-moral_disputes|5_2023-09-18T13-49-35.718586.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-moral_disputes|5_2023-09-18T13-49-35.718586.parquet"]}]}, {"config_name": "harness_hendrycksTest_moral_scenarios_5", "data_files": [{"split": "2023_09_18T13_49_35.718586", "path": ["**/details_harness|hendrycksTest-moral_scenarios|5_2023-09-18T13-49-35.718586.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-moral_scenarios|5_2023-09-18T13-49-35.718586.parquet"]}]}, {"config_name": "harness_hendrycksTest_nutrition_5", "data_files": [{"split": "2023_09_18T13_49_35.718586", "path": ["**/details_harness|hendrycksTest-nutrition|5_2023-09-18T13-49-35.718586.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-nutrition|5_2023-09-18T13-49-35.718586.parquet"]}]}, {"config_name": "harness_hendrycksTest_philosophy_5", "data_files": [{"split": "2023_09_18T13_49_35.718586", "path": ["**/details_harness|hendrycksTest-philosophy|5_2023-09-18T13-49-35.718586.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-philosophy|5_2023-09-18T13-49-35.718586.parquet"]}]}, {"config_name": "harness_hendrycksTest_prehistory_5", "data_files": [{"split": "2023_09_18T13_49_35.718586", "path": ["**/details_harness|hendrycksTest-prehistory|5_2023-09-18T13-49-35.718586.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-prehistory|5_2023-09-18T13-49-35.718586.parquet"]}]}, {"config_name": "harness_hendrycksTest_professional_accounting_5", "data_files": [{"split": "2023_09_18T13_49_35.718586", "path": ["**/details_harness|hendrycksTest-professional_accounting|5_2023-09-18T13-49-35.718586.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-professional_accounting|5_2023-09-18T13-49-35.718586.parquet"]}]}, {"config_name": "harness_hendrycksTest_professional_law_5", "data_files": [{"split": "2023_09_18T13_49_35.718586", "path": ["**/details_harness|hendrycksTest-professional_law|5_2023-09-18T13-49-35.718586.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-professional_law|5_2023-09-18T13-49-35.718586.parquet"]}]}, {"config_name": "harness_hendrycksTest_professional_medicine_5", "data_files": [{"split": "2023_09_18T13_49_35.718586", "path": ["**/details_harness|hendrycksTest-professional_medicine|5_2023-09-18T13-49-35.718586.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-professional_medicine|5_2023-09-18T13-49-35.718586.parquet"]}]}, {"config_name": "harness_hendrycksTest_professional_psychology_5", "data_files": [{"split": "2023_09_18T13_49_35.718586", "path": ["**/details_harness|hendrycksTest-professional_psychology|5_2023-09-18T13-49-35.718586.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-professional_psychology|5_2023-09-18T13-49-35.718586.parquet"]}]}, {"config_name": "harness_hendrycksTest_public_relations_5", "data_files": [{"split": "2023_09_18T13_49_35.718586", "path": ["**/details_harness|hendrycksTest-public_relations|5_2023-09-18T13-49-35.718586.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-public_relations|5_2023-09-18T13-49-35.718586.parquet"]}]}, {"config_name": "harness_hendrycksTest_security_studies_5", "data_files": [{"split": "2023_09_18T13_49_35.718586", "path": ["**/details_harness|hendrycksTest-security_studies|5_2023-09-18T13-49-35.718586.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-security_studies|5_2023-09-18T13-49-35.718586.parquet"]}]}, {"config_name": "harness_hendrycksTest_sociology_5", "data_files": [{"split": "2023_09_18T13_49_35.718586", "path": ["**/details_harness|hendrycksTest-sociology|5_2023-09-18T13-49-35.718586.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-sociology|5_2023-09-18T13-49-35.718586.parquet"]}]}, {"config_name": "harness_hendrycksTest_us_foreign_policy_5", "data_files": [{"split": "2023_09_18T13_49_35.718586", "path": ["**/details_harness|hendrycksTest-us_foreign_policy|5_2023-09-18T13-49-35.718586.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-us_foreign_policy|5_2023-09-18T13-49-35.718586.parquet"]}]}, {"config_name": "harness_hendrycksTest_virology_5", "data_files": [{"split": "2023_09_18T13_49_35.718586", "path": ["**/details_harness|hendrycksTest-virology|5_2023-09-18T13-49-35.718586.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-virology|5_2023-09-18T13-49-35.718586.parquet"]}]}, {"config_name": "harness_hendrycksTest_world_religions_5", "data_files": [{"split": "2023_09_18T13_49_35.718586", "path": ["**/details_harness|hendrycksTest-world_religions|5_2023-09-18T13-49-35.718586.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-world_religions|5_2023-09-18T13-49-35.718586.parquet"]}]}, {"config_name": "harness_truthfulqa_mc_0", "data_files": [{"split": "2023_09_18T13_49_35.718586", "path": ["**/details_harness|truthfulqa:mc|0_2023-09-18T13-49-35.718586.parquet"]}, {"split": "latest", "path": ["**/details_harness|truthfulqa:mc|0_2023-09-18T13-49-35.718586.parquet"]}]}, {"config_name": "harness_winogrande_5", "data_files": [{"split": "2023_10_23T22_37_50.525759", "path": ["**/details_harness|winogrande|5_2023-10-23T22-37-50.525759.parquet"]}, {"split": "latest", "path": ["**/details_harness|winogrande|5_2023-10-23T22-37-50.525759.parquet"]}]}, {"config_name": "results", "data_files": [{"split": "2023_09_18T13_49_35.718586", "path": ["results_2023-09-18T13-49-35.718586.parquet"]}, {"split": "2023_10_23T22_37_50.525759", "path": ["results_2023-10-23T22-37-50.525759.parquet"]}, {"split": "latest", "path": ["results_2023-10-23T22-37-50.525759.parquet"]}]}]}
|
2023-10-23T21:38:01+00:00
|
[] |
[] |
TAGS
#region-us
|
# Dataset Card for Evaluation run of nicholasKluge/Aira-2-774M
## Dataset Description
- Homepage:
- Repository: URL
- Paper:
- Leaderboard: URL
- Point of Contact: clementine@URL
### Dataset Summary
Dataset automatically created during the evaluation run of model nicholasKluge/Aira-2-774M on the Open LLM Leaderboard.
The dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.
The dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The "train" split is always pointing to the latest results.
An additional configuration "results" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the Open LLM Leaderboard).
To load the details from a run, you can for instance do the following:
## Latest results
These are the latest results from run 2023-10-23T22:37:50.525759(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the "latest" split for each eval):
### Supported Tasks and Leaderboards
### Languages
## Dataset Structure
### Data Instances
### Data Fields
### Data Splits
## Dataset Creation
### Curation Rationale
### Source Data
#### Initial Data Collection and Normalization
#### Who are the source language producers?
### Annotations
#### Annotation process
#### Who are the annotators?
### Personal and Sensitive Information
## Considerations for Using the Data
### Social Impact of Dataset
### Discussion of Biases
### Other Known Limitations
## Additional Information
### Dataset Curators
### Licensing Information
### Contributions
|
[
"# Dataset Card for Evaluation run of nicholasKluge/Aira-2-774M",
"## Dataset Description\n\n- Homepage: \n- Repository: URL\n- Paper: \n- Leaderboard: URL\n- Point of Contact: clementine@URL",
"### Dataset Summary\n\nDataset automatically created during the evaluation run of model nicholasKluge/Aira-2-774M on the Open LLM Leaderboard.\n\nThe dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.\n\nThe dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The \"train\" split is always pointing to the latest results.\n\nAn additional configuration \"results\" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the Open LLM Leaderboard).\n\nTo load the details from a run, you can for instance do the following:",
"## Latest results\n\nThese are the latest results from run 2023-10-23T22:37:50.525759(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the \"latest\" split for each eval):",
"### Supported Tasks and Leaderboards",
"### Languages",
"## Dataset Structure",
"### Data Instances",
"### Data Fields",
"### Data Splits",
"## Dataset Creation",
"### Curation Rationale",
"### Source Data",
"#### Initial Data Collection and Normalization",
"#### Who are the source language producers?",
"### Annotations",
"#### Annotation process",
"#### Who are the annotators?",
"### Personal and Sensitive Information",
"## Considerations for Using the Data",
"### Social Impact of Dataset",
"### Discussion of Biases",
"### Other Known Limitations",
"## Additional Information",
"### Dataset Curators",
"### Licensing Information",
"### Contributions"
] |
[
"TAGS\n#region-us \n",
"# Dataset Card for Evaluation run of nicholasKluge/Aira-2-774M",
"## Dataset Description\n\n- Homepage: \n- Repository: URL\n- Paper: \n- Leaderboard: URL\n- Point of Contact: clementine@URL",
"### Dataset Summary\n\nDataset automatically created during the evaluation run of model nicholasKluge/Aira-2-774M on the Open LLM Leaderboard.\n\nThe dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.\n\nThe dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The \"train\" split is always pointing to the latest results.\n\nAn additional configuration \"results\" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the Open LLM Leaderboard).\n\nTo load the details from a run, you can for instance do the following:",
"## Latest results\n\nThese are the latest results from run 2023-10-23T22:37:50.525759(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the \"latest\" split for each eval):",
"### Supported Tasks and Leaderboards",
"### Languages",
"## Dataset Structure",
"### Data Instances",
"### Data Fields",
"### Data Splits",
"## Dataset Creation",
"### Curation Rationale",
"### Source Data",
"#### Initial Data Collection and Normalization",
"#### Who are the source language producers?",
"### Annotations",
"#### Annotation process",
"#### Who are the annotators?",
"### Personal and Sensitive Information",
"## Considerations for Using the Data",
"### Social Impact of Dataset",
"### Discussion of Biases",
"### Other Known Limitations",
"## Additional Information",
"### Dataset Curators",
"### Licensing Information",
"### Contributions"
] |
[
6,
21,
31,
169,
67,
10,
4,
6,
6,
5,
5,
5,
7,
4,
10,
10,
5,
5,
9,
8,
8,
7,
8,
7,
5,
6,
6,
5
] |
[
"passage: TAGS\n#region-us \n# Dataset Card for Evaluation run of nicholasKluge/Aira-2-774M## Dataset Description\n\n- Homepage: \n- Repository: URL\n- Paper: \n- Leaderboard: URL\n- Point of Contact: clementine@URL### Dataset Summary\n\nDataset automatically created during the evaluation run of model nicholasKluge/Aira-2-774M on the Open LLM Leaderboard.\n\nThe dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.\n\nThe dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The \"train\" split is always pointing to the latest results.\n\nAn additional configuration \"results\" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the Open LLM Leaderboard).\n\nTo load the details from a run, you can for instance do the following:## Latest results\n\nThese are the latest results from run 2023-10-23T22:37:50.525759(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the \"latest\" split for each eval):### Supported Tasks and Leaderboards### Languages## Dataset Structure### Data Instances### Data Fields### Data Splits## Dataset Creation### Curation Rationale### Source Data#### Initial Data Collection and Normalization#### Who are the source language producers?### Annotations#### Annotation process#### Who are the annotators?### Personal and Sensitive Information## Considerations for Using the Data### Social Impact of Dataset### Discussion of Biases### Other Known Limitations## Additional Information### Dataset Curators### Licensing Information### Contributions"
] |
6b35fc9b0d099312e794bef1991b76495af88155
|
# Dataset Card for Evaluation run of PeanutJar/LLaMa-2-PeanutButter_v37_SFT-R1-DPO-R2-7B
## Dataset Description
- **Homepage:**
- **Repository:** https://huggingface.co/PeanutJar/LLaMa-2-PeanutButter_v37_SFT-R1-DPO-R2-7B
- **Paper:**
- **Leaderboard:** https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard
- **Point of Contact:** [email protected]
### Dataset Summary
Dataset automatically created during the evaluation run of model [PeanutJar/LLaMa-2-PeanutButter_v37_SFT-R1-DPO-R2-7B](https://huggingface.co/PeanutJar/LLaMa-2-PeanutButter_v37_SFT-R1-DPO-R2-7B) on the [Open LLM Leaderboard](https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard).
The dataset is composed of 61 configuration, each one coresponding to one of the evaluated task.
The dataset has been created from 1 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The "train" split is always pointing to the latest results.
An additional configuration "results" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the [Open LLM Leaderboard](https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard)).
To load the details from a run, you can for instance do the following:
```python
from datasets import load_dataset
data = load_dataset("open-llm-leaderboard/details_PeanutJar__LLaMa-2-PeanutButter_v37_SFT-R1-DPO-R2-7B",
"harness_truthfulqa_mc_0",
split="train")
```
## Latest results
These are the [latest results from run 2023-09-18T13:52:12.512549](https://huggingface.co/datasets/open-llm-leaderboard/details_PeanutJar__LLaMa-2-PeanutButter_v37_SFT-R1-DPO-R2-7B/blob/main/results_2023-09-18T13-52-12.512549.json)(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the "latest" split for each eval):
```python
{
"all": {
"acc": 0.4757356941246475,
"acc_stderr": 0.03534759162401654,
"acc_norm": 0.47972138667961756,
"acc_norm_stderr": 0.03533243222459744,
"mc1": 0.27906976744186046,
"mc1_stderr": 0.015702107090627904,
"mc2": 0.41999112300299424,
"mc2_stderr": 0.014077295047564501
},
"harness|arc:challenge|25": {
"acc": 0.5042662116040956,
"acc_stderr": 0.014610858923956955,
"acc_norm": 0.5409556313993175,
"acc_norm_stderr": 0.014562291073601233
},
"harness|hellaswag|10": {
"acc": 0.5925114519020116,
"acc_stderr": 0.004903628887264536,
"acc_norm": 0.7909778928500298,
"acc_norm_stderr": 0.004057792171893564
},
"harness|hendrycksTest-abstract_algebra|5": {
"acc": 0.31,
"acc_stderr": 0.04648231987117316,
"acc_norm": 0.31,
"acc_norm_stderr": 0.04648231987117316
},
"harness|hendrycksTest-anatomy|5": {
"acc": 0.48148148148148145,
"acc_stderr": 0.043163785995113245,
"acc_norm": 0.48148148148148145,
"acc_norm_stderr": 0.043163785995113245
},
"harness|hendrycksTest-astronomy|5": {
"acc": 0.42105263157894735,
"acc_stderr": 0.04017901275981748,
"acc_norm": 0.42105263157894735,
"acc_norm_stderr": 0.04017901275981748
},
"harness|hendrycksTest-business_ethics|5": {
"acc": 0.54,
"acc_stderr": 0.05009082659620332,
"acc_norm": 0.54,
"acc_norm_stderr": 0.05009082659620332
},
"harness|hendrycksTest-clinical_knowledge|5": {
"acc": 0.4716981132075472,
"acc_stderr": 0.0307235352490061,
"acc_norm": 0.4716981132075472,
"acc_norm_stderr": 0.0307235352490061
},
"harness|hendrycksTest-college_biology|5": {
"acc": 0.4652777777777778,
"acc_stderr": 0.04171115858181618,
"acc_norm": 0.4652777777777778,
"acc_norm_stderr": 0.04171115858181618
},
"harness|hendrycksTest-college_chemistry|5": {
"acc": 0.33,
"acc_stderr": 0.04725815626252605,
"acc_norm": 0.33,
"acc_norm_stderr": 0.04725815626252605
},
"harness|hendrycksTest-college_computer_science|5": {
"acc": 0.34,
"acc_stderr": 0.04760952285695235,
"acc_norm": 0.34,
"acc_norm_stderr": 0.04760952285695235
},
"harness|hendrycksTest-college_mathematics|5": {
"acc": 0.34,
"acc_stderr": 0.04760952285695235,
"acc_norm": 0.34,
"acc_norm_stderr": 0.04760952285695235
},
"harness|hendrycksTest-college_medicine|5": {
"acc": 0.4393063583815029,
"acc_stderr": 0.037842719328874674,
"acc_norm": 0.4393063583815029,
"acc_norm_stderr": 0.037842719328874674
},
"harness|hendrycksTest-college_physics|5": {
"acc": 0.2647058823529412,
"acc_stderr": 0.04389869956808778,
"acc_norm": 0.2647058823529412,
"acc_norm_stderr": 0.04389869956808778
},
"harness|hendrycksTest-computer_security|5": {
"acc": 0.61,
"acc_stderr": 0.04902071300001975,
"acc_norm": 0.61,
"acc_norm_stderr": 0.04902071300001975
},
"harness|hendrycksTest-conceptual_physics|5": {
"acc": 0.425531914893617,
"acc_stderr": 0.03232146916224468,
"acc_norm": 0.425531914893617,
"acc_norm_stderr": 0.03232146916224468
},
"harness|hendrycksTest-econometrics|5": {
"acc": 0.2807017543859649,
"acc_stderr": 0.042270544512322004,
"acc_norm": 0.2807017543859649,
"acc_norm_stderr": 0.042270544512322004
},
"harness|hendrycksTest-electrical_engineering|5": {
"acc": 0.47586206896551725,
"acc_stderr": 0.041618085035015295,
"acc_norm": 0.47586206896551725,
"acc_norm_stderr": 0.041618085035015295
},
"harness|hendrycksTest-elementary_mathematics|5": {
"acc": 0.2830687830687831,
"acc_stderr": 0.023201392938194974,
"acc_norm": 0.2830687830687831,
"acc_norm_stderr": 0.023201392938194974
},
"harness|hendrycksTest-formal_logic|5": {
"acc": 0.30952380952380953,
"acc_stderr": 0.04134913018303316,
"acc_norm": 0.30952380952380953,
"acc_norm_stderr": 0.04134913018303316
},
"harness|hendrycksTest-global_facts|5": {
"acc": 0.33,
"acc_stderr": 0.04725815626252605,
"acc_norm": 0.33,
"acc_norm_stderr": 0.04725815626252605
},
"harness|hendrycksTest-high_school_biology|5": {
"acc": 0.49032258064516127,
"acc_stderr": 0.028438677998909558,
"acc_norm": 0.49032258064516127,
"acc_norm_stderr": 0.028438677998909558
},
"harness|hendrycksTest-high_school_chemistry|5": {
"acc": 0.37438423645320196,
"acc_stderr": 0.03405155380561953,
"acc_norm": 0.37438423645320196,
"acc_norm_stderr": 0.03405155380561953
},
"harness|hendrycksTest-high_school_computer_science|5": {
"acc": 0.4,
"acc_stderr": 0.049236596391733084,
"acc_norm": 0.4,
"acc_norm_stderr": 0.049236596391733084
},
"harness|hendrycksTest-high_school_european_history|5": {
"acc": 0.5818181818181818,
"acc_stderr": 0.03851716319398393,
"acc_norm": 0.5818181818181818,
"acc_norm_stderr": 0.03851716319398393
},
"harness|hendrycksTest-high_school_geography|5": {
"acc": 0.51010101010101,
"acc_stderr": 0.035616254886737454,
"acc_norm": 0.51010101010101,
"acc_norm_stderr": 0.035616254886737454
},
"harness|hendrycksTest-high_school_government_and_politics|5": {
"acc": 0.6735751295336787,
"acc_stderr": 0.033840286211432945,
"acc_norm": 0.6735751295336787,
"acc_norm_stderr": 0.033840286211432945
},
"harness|hendrycksTest-high_school_macroeconomics|5": {
"acc": 0.4512820512820513,
"acc_stderr": 0.025230381238934833,
"acc_norm": 0.4512820512820513,
"acc_norm_stderr": 0.025230381238934833
},
"harness|hendrycksTest-high_school_mathematics|5": {
"acc": 0.2962962962962963,
"acc_stderr": 0.027840811495871923,
"acc_norm": 0.2962962962962963,
"acc_norm_stderr": 0.027840811495871923
},
"harness|hendrycksTest-high_school_microeconomics|5": {
"acc": 0.4411764705882353,
"acc_stderr": 0.0322529423239964,
"acc_norm": 0.4411764705882353,
"acc_norm_stderr": 0.0322529423239964
},
"harness|hendrycksTest-high_school_physics|5": {
"acc": 0.32450331125827814,
"acc_stderr": 0.038227469376587525,
"acc_norm": 0.32450331125827814,
"acc_norm_stderr": 0.038227469376587525
},
"harness|hendrycksTest-high_school_psychology|5": {
"acc": 0.636697247706422,
"acc_stderr": 0.020620603919625804,
"acc_norm": 0.636697247706422,
"acc_norm_stderr": 0.020620603919625804
},
"harness|hendrycksTest-high_school_statistics|5": {
"acc": 0.28703703703703703,
"acc_stderr": 0.030851992993257013,
"acc_norm": 0.28703703703703703,
"acc_norm_stderr": 0.030851992993257013
},
"harness|hendrycksTest-high_school_us_history|5": {
"acc": 0.553921568627451,
"acc_stderr": 0.03488845451304974,
"acc_norm": 0.553921568627451,
"acc_norm_stderr": 0.03488845451304974
},
"harness|hendrycksTest-high_school_world_history|5": {
"acc": 0.6497890295358649,
"acc_stderr": 0.031052391937584346,
"acc_norm": 0.6497890295358649,
"acc_norm_stderr": 0.031052391937584346
},
"harness|hendrycksTest-human_aging|5": {
"acc": 0.5560538116591929,
"acc_stderr": 0.03334625674242728,
"acc_norm": 0.5560538116591929,
"acc_norm_stderr": 0.03334625674242728
},
"harness|hendrycksTest-human_sexuality|5": {
"acc": 0.549618320610687,
"acc_stderr": 0.04363643698524779,
"acc_norm": 0.549618320610687,
"acc_norm_stderr": 0.04363643698524779
},
"harness|hendrycksTest-international_law|5": {
"acc": 0.6528925619834711,
"acc_stderr": 0.043457245702925335,
"acc_norm": 0.6528925619834711,
"acc_norm_stderr": 0.043457245702925335
},
"harness|hendrycksTest-jurisprudence|5": {
"acc": 0.5185185185185185,
"acc_stderr": 0.04830366024635331,
"acc_norm": 0.5185185185185185,
"acc_norm_stderr": 0.04830366024635331
},
"harness|hendrycksTest-logical_fallacies|5": {
"acc": 0.5398773006134969,
"acc_stderr": 0.039158572914369714,
"acc_norm": 0.5398773006134969,
"acc_norm_stderr": 0.039158572914369714
},
"harness|hendrycksTest-machine_learning|5": {
"acc": 0.375,
"acc_stderr": 0.04595091388086298,
"acc_norm": 0.375,
"acc_norm_stderr": 0.04595091388086298
},
"harness|hendrycksTest-management|5": {
"acc": 0.5728155339805825,
"acc_stderr": 0.048979577377811674,
"acc_norm": 0.5728155339805825,
"acc_norm_stderr": 0.048979577377811674
},
"harness|hendrycksTest-marketing|5": {
"acc": 0.6965811965811965,
"acc_stderr": 0.030118210106942638,
"acc_norm": 0.6965811965811965,
"acc_norm_stderr": 0.030118210106942638
},
"harness|hendrycksTest-medical_genetics|5": {
"acc": 0.55,
"acc_stderr": 0.04999999999999999,
"acc_norm": 0.55,
"acc_norm_stderr": 0.04999999999999999
},
"harness|hendrycksTest-miscellaneous|5": {
"acc": 0.6398467432950191,
"acc_stderr": 0.017166362471369302,
"acc_norm": 0.6398467432950191,
"acc_norm_stderr": 0.017166362471369302
},
"harness|hendrycksTest-moral_disputes|5": {
"acc": 0.5144508670520231,
"acc_stderr": 0.02690784985628254,
"acc_norm": 0.5144508670520231,
"acc_norm_stderr": 0.02690784985628254
},
"harness|hendrycksTest-moral_scenarios|5": {
"acc": 0.23798882681564246,
"acc_stderr": 0.014242630070574915,
"acc_norm": 0.23798882681564246,
"acc_norm_stderr": 0.014242630070574915
},
"harness|hendrycksTest-nutrition|5": {
"acc": 0.5032679738562091,
"acc_stderr": 0.028629305194003543,
"acc_norm": 0.5032679738562091,
"acc_norm_stderr": 0.028629305194003543
},
"harness|hendrycksTest-philosophy|5": {
"acc": 0.617363344051447,
"acc_stderr": 0.027604689028581996,
"acc_norm": 0.617363344051447,
"acc_norm_stderr": 0.027604689028581996
},
"harness|hendrycksTest-prehistory|5": {
"acc": 0.4783950617283951,
"acc_stderr": 0.02779476010500874,
"acc_norm": 0.4783950617283951,
"acc_norm_stderr": 0.02779476010500874
},
"harness|hendrycksTest-professional_accounting|5": {
"acc": 0.3617021276595745,
"acc_stderr": 0.02866382014719949,
"acc_norm": 0.3617021276595745,
"acc_norm_stderr": 0.02866382014719949
},
"harness|hendrycksTest-professional_law|5": {
"acc": 0.36114732724902215,
"acc_stderr": 0.01226793547751903,
"acc_norm": 0.36114732724902215,
"acc_norm_stderr": 0.01226793547751903
},
"harness|hendrycksTest-professional_medicine|5": {
"acc": 0.5183823529411765,
"acc_stderr": 0.030352303395351964,
"acc_norm": 0.5183823529411765,
"acc_norm_stderr": 0.030352303395351964
},
"harness|hendrycksTest-professional_psychology|5": {
"acc": 0.45098039215686275,
"acc_stderr": 0.020130388312904528,
"acc_norm": 0.45098039215686275,
"acc_norm_stderr": 0.020130388312904528
},
"harness|hendrycksTest-public_relations|5": {
"acc": 0.5545454545454546,
"acc_stderr": 0.047605488214603246,
"acc_norm": 0.5545454545454546,
"acc_norm_stderr": 0.047605488214603246
},
"harness|hendrycksTest-security_studies|5": {
"acc": 0.5142857142857142,
"acc_stderr": 0.03199615232806287,
"acc_norm": 0.5142857142857142,
"acc_norm_stderr": 0.03199615232806287
},
"harness|hendrycksTest-sociology|5": {
"acc": 0.6417910447761194,
"acc_stderr": 0.03390393042268814,
"acc_norm": 0.6417910447761194,
"acc_norm_stderr": 0.03390393042268814
},
"harness|hendrycksTest-us_foreign_policy|5": {
"acc": 0.63,
"acc_stderr": 0.04852365870939099,
"acc_norm": 0.63,
"acc_norm_stderr": 0.04852365870939099
},
"harness|hendrycksTest-virology|5": {
"acc": 0.42771084337349397,
"acc_stderr": 0.038515976837185335,
"acc_norm": 0.42771084337349397,
"acc_norm_stderr": 0.038515976837185335
},
"harness|hendrycksTest-world_religions|5": {
"acc": 0.7192982456140351,
"acc_stderr": 0.034462962170884265,
"acc_norm": 0.7192982456140351,
"acc_norm_stderr": 0.034462962170884265
},
"harness|truthfulqa:mc|0": {
"mc1": 0.27906976744186046,
"mc1_stderr": 0.015702107090627904,
"mc2": 0.41999112300299424,
"mc2_stderr": 0.014077295047564501
}
}
```
### Supported Tasks and Leaderboards
[More Information Needed]
### Languages
[More Information Needed]
## Dataset Structure
### Data Instances
[More Information Needed]
### Data Fields
[More Information Needed]
### Data Splits
[More Information Needed]
## Dataset Creation
### Curation Rationale
[More Information Needed]
### Source Data
#### Initial Data Collection and Normalization
[More Information Needed]
#### Who are the source language producers?
[More Information Needed]
### Annotations
#### Annotation process
[More Information Needed]
#### Who are the annotators?
[More Information Needed]
### Personal and Sensitive Information
[More Information Needed]
## Considerations for Using the Data
### Social Impact of Dataset
[More Information Needed]
### Discussion of Biases
[More Information Needed]
### Other Known Limitations
[More Information Needed]
## Additional Information
### Dataset Curators
[More Information Needed]
### Licensing Information
[More Information Needed]
### Citation Information
[More Information Needed]
### Contributions
[More Information Needed]
|
open-llm-leaderboard/details_PeanutJar__LLaMa-2-PeanutButter_v37_SFT-R1-DPO-R2-7B
|
[
"region:us"
] |
2023-09-18T12:52:36+00:00
|
{"pretty_name": "Evaluation run of PeanutJar/LLaMa-2-PeanutButter_v37_SFT-R1-DPO-R2-7B", "dataset_summary": "Dataset automatically created during the evaluation run of model [PeanutJar/LLaMa-2-PeanutButter_v37_SFT-R1-DPO-R2-7B](https://huggingface.co/PeanutJar/LLaMa-2-PeanutButter_v37_SFT-R1-DPO-R2-7B) on the [Open LLM Leaderboard](https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard).\n\nThe dataset is composed of 61 configuration, each one coresponding to one of the evaluated task.\n\nThe dataset has been created from 1 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The \"train\" split is always pointing to the latest results.\n\nAn additional configuration \"results\" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the [Open LLM Leaderboard](https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard)).\n\nTo load the details from a run, you can for instance do the following:\n```python\nfrom datasets import load_dataset\ndata = load_dataset(\"open-llm-leaderboard/details_PeanutJar__LLaMa-2-PeanutButter_v37_SFT-R1-DPO-R2-7B\",\n\t\"harness_truthfulqa_mc_0\",\n\tsplit=\"train\")\n```\n\n## Latest results\n\nThese are the [latest results from run 2023-09-18T13:52:12.512549](https://huggingface.co/datasets/open-llm-leaderboard/details_PeanutJar__LLaMa-2-PeanutButter_v37_SFT-R1-DPO-R2-7B/blob/main/results_2023-09-18T13-52-12.512549.json)(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the \"latest\" split for each eval):\n\n```python\n{\n \"all\": {\n \"acc\": 0.4757356941246475,\n \"acc_stderr\": 0.03534759162401654,\n \"acc_norm\": 0.47972138667961756,\n \"acc_norm_stderr\": 0.03533243222459744,\n \"mc1\": 0.27906976744186046,\n \"mc1_stderr\": 0.015702107090627904,\n \"mc2\": 0.41999112300299424,\n \"mc2_stderr\": 0.014077295047564501\n },\n \"harness|arc:challenge|25\": {\n \"acc\": 0.5042662116040956,\n \"acc_stderr\": 0.014610858923956955,\n \"acc_norm\": 0.5409556313993175,\n \"acc_norm_stderr\": 0.014562291073601233\n },\n \"harness|hellaswag|10\": {\n \"acc\": 0.5925114519020116,\n \"acc_stderr\": 0.004903628887264536,\n \"acc_norm\": 0.7909778928500298,\n \"acc_norm_stderr\": 0.004057792171893564\n },\n \"harness|hendrycksTest-abstract_algebra|5\": {\n \"acc\": 0.31,\n \"acc_stderr\": 0.04648231987117316,\n \"acc_norm\": 0.31,\n \"acc_norm_stderr\": 0.04648231987117316\n },\n \"harness|hendrycksTest-anatomy|5\": {\n \"acc\": 0.48148148148148145,\n \"acc_stderr\": 0.043163785995113245,\n \"acc_norm\": 0.48148148148148145,\n \"acc_norm_stderr\": 0.043163785995113245\n },\n \"harness|hendrycksTest-astronomy|5\": {\n \"acc\": 0.42105263157894735,\n \"acc_stderr\": 0.04017901275981748,\n \"acc_norm\": 0.42105263157894735,\n \"acc_norm_stderr\": 0.04017901275981748\n },\n \"harness|hendrycksTest-business_ethics|5\": {\n \"acc\": 0.54,\n \"acc_stderr\": 0.05009082659620332,\n \"acc_norm\": 0.54,\n \"acc_norm_stderr\": 0.05009082659620332\n },\n \"harness|hendrycksTest-clinical_knowledge|5\": {\n \"acc\": 0.4716981132075472,\n \"acc_stderr\": 0.0307235352490061,\n \"acc_norm\": 0.4716981132075472,\n \"acc_norm_stderr\": 0.0307235352490061\n },\n \"harness|hendrycksTest-college_biology|5\": {\n \"acc\": 0.4652777777777778,\n \"acc_stderr\": 0.04171115858181618,\n \"acc_norm\": 0.4652777777777778,\n \"acc_norm_stderr\": 0.04171115858181618\n },\n \"harness|hendrycksTest-college_chemistry|5\": {\n \"acc\": 0.33,\n \"acc_stderr\": 0.04725815626252605,\n \"acc_norm\": 0.33,\n \"acc_norm_stderr\": 0.04725815626252605\n },\n \"harness|hendrycksTest-college_computer_science|5\": {\n \"acc\": 0.34,\n \"acc_stderr\": 0.04760952285695235,\n \"acc_norm\": 0.34,\n \"acc_norm_stderr\": 0.04760952285695235\n },\n \"harness|hendrycksTest-college_mathematics|5\": {\n \"acc\": 0.34,\n \"acc_stderr\": 0.04760952285695235,\n \"acc_norm\": 0.34,\n \"acc_norm_stderr\": 0.04760952285695235\n },\n \"harness|hendrycksTest-college_medicine|5\": {\n \"acc\": 0.4393063583815029,\n \"acc_stderr\": 0.037842719328874674,\n \"acc_norm\": 0.4393063583815029,\n \"acc_norm_stderr\": 0.037842719328874674\n },\n \"harness|hendrycksTest-college_physics|5\": {\n \"acc\": 0.2647058823529412,\n \"acc_stderr\": 0.04389869956808778,\n \"acc_norm\": 0.2647058823529412,\n \"acc_norm_stderr\": 0.04389869956808778\n },\n \"harness|hendrycksTest-computer_security|5\": {\n \"acc\": 0.61,\n \"acc_stderr\": 0.04902071300001975,\n \"acc_norm\": 0.61,\n \"acc_norm_stderr\": 0.04902071300001975\n },\n \"harness|hendrycksTest-conceptual_physics|5\": {\n \"acc\": 0.425531914893617,\n \"acc_stderr\": 0.03232146916224468,\n \"acc_norm\": 0.425531914893617,\n \"acc_norm_stderr\": 0.03232146916224468\n },\n \"harness|hendrycksTest-econometrics|5\": {\n \"acc\": 0.2807017543859649,\n \"acc_stderr\": 0.042270544512322004,\n \"acc_norm\": 0.2807017543859649,\n \"acc_norm_stderr\": 0.042270544512322004\n },\n \"harness|hendrycksTest-electrical_engineering|5\": {\n \"acc\": 0.47586206896551725,\n \"acc_stderr\": 0.041618085035015295,\n \"acc_norm\": 0.47586206896551725,\n \"acc_norm_stderr\": 0.041618085035015295\n },\n \"harness|hendrycksTest-elementary_mathematics|5\": {\n \"acc\": 0.2830687830687831,\n \"acc_stderr\": 0.023201392938194974,\n \"acc_norm\": 0.2830687830687831,\n \"acc_norm_stderr\": 0.023201392938194974\n },\n \"harness|hendrycksTest-formal_logic|5\": {\n \"acc\": 0.30952380952380953,\n \"acc_stderr\": 0.04134913018303316,\n \"acc_norm\": 0.30952380952380953,\n \"acc_norm_stderr\": 0.04134913018303316\n },\n \"harness|hendrycksTest-global_facts|5\": {\n \"acc\": 0.33,\n \"acc_stderr\": 0.04725815626252605,\n \"acc_norm\": 0.33,\n \"acc_norm_stderr\": 0.04725815626252605\n },\n \"harness|hendrycksTest-high_school_biology|5\": {\n \"acc\": 0.49032258064516127,\n \"acc_stderr\": 0.028438677998909558,\n \"acc_norm\": 0.49032258064516127,\n \"acc_norm_stderr\": 0.028438677998909558\n },\n \"harness|hendrycksTest-high_school_chemistry|5\": {\n \"acc\": 0.37438423645320196,\n \"acc_stderr\": 0.03405155380561953,\n \"acc_norm\": 0.37438423645320196,\n \"acc_norm_stderr\": 0.03405155380561953\n },\n \"harness|hendrycksTest-high_school_computer_science|5\": {\n \"acc\": 0.4,\n \"acc_stderr\": 0.049236596391733084,\n \"acc_norm\": 0.4,\n \"acc_norm_stderr\": 0.049236596391733084\n },\n \"harness|hendrycksTest-high_school_european_history|5\": {\n \"acc\": 0.5818181818181818,\n \"acc_stderr\": 0.03851716319398393,\n \"acc_norm\": 0.5818181818181818,\n \"acc_norm_stderr\": 0.03851716319398393\n },\n \"harness|hendrycksTest-high_school_geography|5\": {\n \"acc\": 0.51010101010101,\n \"acc_stderr\": 0.035616254886737454,\n \"acc_norm\": 0.51010101010101,\n \"acc_norm_stderr\": 0.035616254886737454\n },\n \"harness|hendrycksTest-high_school_government_and_politics|5\": {\n \"acc\": 0.6735751295336787,\n \"acc_stderr\": 0.033840286211432945,\n \"acc_norm\": 0.6735751295336787,\n \"acc_norm_stderr\": 0.033840286211432945\n },\n \"harness|hendrycksTest-high_school_macroeconomics|5\": {\n \"acc\": 0.4512820512820513,\n \"acc_stderr\": 0.025230381238934833,\n \"acc_norm\": 0.4512820512820513,\n \"acc_norm_stderr\": 0.025230381238934833\n },\n \"harness|hendrycksTest-high_school_mathematics|5\": {\n \"acc\": 0.2962962962962963,\n \"acc_stderr\": 0.027840811495871923,\n \"acc_norm\": 0.2962962962962963,\n \"acc_norm_stderr\": 0.027840811495871923\n },\n \"harness|hendrycksTest-high_school_microeconomics|5\": {\n \"acc\": 0.4411764705882353,\n \"acc_stderr\": 0.0322529423239964,\n \"acc_norm\": 0.4411764705882353,\n \"acc_norm_stderr\": 0.0322529423239964\n },\n \"harness|hendrycksTest-high_school_physics|5\": {\n \"acc\": 0.32450331125827814,\n \"acc_stderr\": 0.038227469376587525,\n \"acc_norm\": 0.32450331125827814,\n \"acc_norm_stderr\": 0.038227469376587525\n },\n \"harness|hendrycksTest-high_school_psychology|5\": {\n \"acc\": 0.636697247706422,\n \"acc_stderr\": 0.020620603919625804,\n \"acc_norm\": 0.636697247706422,\n \"acc_norm_stderr\": 0.020620603919625804\n },\n \"harness|hendrycksTest-high_school_statistics|5\": {\n \"acc\": 0.28703703703703703,\n \"acc_stderr\": 0.030851992993257013,\n \"acc_norm\": 0.28703703703703703,\n \"acc_norm_stderr\": 0.030851992993257013\n },\n \"harness|hendrycksTest-high_school_us_history|5\": {\n \"acc\": 0.553921568627451,\n \"acc_stderr\": 0.03488845451304974,\n \"acc_norm\": 0.553921568627451,\n \"acc_norm_stderr\": 0.03488845451304974\n },\n \"harness|hendrycksTest-high_school_world_history|5\": {\n \"acc\": 0.6497890295358649,\n \"acc_stderr\": 0.031052391937584346,\n \"acc_norm\": 0.6497890295358649,\n \"acc_norm_stderr\": 0.031052391937584346\n },\n \"harness|hendrycksTest-human_aging|5\": {\n \"acc\": 0.5560538116591929,\n \"acc_stderr\": 0.03334625674242728,\n \"acc_norm\": 0.5560538116591929,\n \"acc_norm_stderr\": 0.03334625674242728\n },\n \"harness|hendrycksTest-human_sexuality|5\": {\n \"acc\": 0.549618320610687,\n \"acc_stderr\": 0.04363643698524779,\n \"acc_norm\": 0.549618320610687,\n \"acc_norm_stderr\": 0.04363643698524779\n },\n \"harness|hendrycksTest-international_law|5\": {\n \"acc\": 0.6528925619834711,\n \"acc_stderr\": 0.043457245702925335,\n \"acc_norm\": 0.6528925619834711,\n \"acc_norm_stderr\": 0.043457245702925335\n },\n \"harness|hendrycksTest-jurisprudence|5\": {\n \"acc\": 0.5185185185185185,\n \"acc_stderr\": 0.04830366024635331,\n \"acc_norm\": 0.5185185185185185,\n \"acc_norm_stderr\": 0.04830366024635331\n },\n \"harness|hendrycksTest-logical_fallacies|5\": {\n \"acc\": 0.5398773006134969,\n \"acc_stderr\": 0.039158572914369714,\n \"acc_norm\": 0.5398773006134969,\n \"acc_norm_stderr\": 0.039158572914369714\n },\n \"harness|hendrycksTest-machine_learning|5\": {\n \"acc\": 0.375,\n \"acc_stderr\": 0.04595091388086298,\n \"acc_norm\": 0.375,\n \"acc_norm_stderr\": 0.04595091388086298\n },\n \"harness|hendrycksTest-management|5\": {\n \"acc\": 0.5728155339805825,\n \"acc_stderr\": 0.048979577377811674,\n \"acc_norm\": 0.5728155339805825,\n \"acc_norm_stderr\": 0.048979577377811674\n },\n \"harness|hendrycksTest-marketing|5\": {\n \"acc\": 0.6965811965811965,\n \"acc_stderr\": 0.030118210106942638,\n \"acc_norm\": 0.6965811965811965,\n \"acc_norm_stderr\": 0.030118210106942638\n },\n \"harness|hendrycksTest-medical_genetics|5\": {\n \"acc\": 0.55,\n \"acc_stderr\": 0.04999999999999999,\n \"acc_norm\": 0.55,\n \"acc_norm_stderr\": 0.04999999999999999\n },\n \"harness|hendrycksTest-miscellaneous|5\": {\n \"acc\": 0.6398467432950191,\n \"acc_stderr\": 0.017166362471369302,\n \"acc_norm\": 0.6398467432950191,\n \"acc_norm_stderr\": 0.017166362471369302\n },\n \"harness|hendrycksTest-moral_disputes|5\": {\n \"acc\": 0.5144508670520231,\n \"acc_stderr\": 0.02690784985628254,\n \"acc_norm\": 0.5144508670520231,\n \"acc_norm_stderr\": 0.02690784985628254\n },\n \"harness|hendrycksTest-moral_scenarios|5\": {\n \"acc\": 0.23798882681564246,\n \"acc_stderr\": 0.014242630070574915,\n \"acc_norm\": 0.23798882681564246,\n \"acc_norm_stderr\": 0.014242630070574915\n },\n \"harness|hendrycksTest-nutrition|5\": {\n \"acc\": 0.5032679738562091,\n \"acc_stderr\": 0.028629305194003543,\n \"acc_norm\": 0.5032679738562091,\n \"acc_norm_stderr\": 0.028629305194003543\n },\n \"harness|hendrycksTest-philosophy|5\": {\n \"acc\": 0.617363344051447,\n \"acc_stderr\": 0.027604689028581996,\n \"acc_norm\": 0.617363344051447,\n \"acc_norm_stderr\": 0.027604689028581996\n },\n \"harness|hendrycksTest-prehistory|5\": {\n \"acc\": 0.4783950617283951,\n \"acc_stderr\": 0.02779476010500874,\n \"acc_norm\": 0.4783950617283951,\n \"acc_norm_stderr\": 0.02779476010500874\n },\n \"harness|hendrycksTest-professional_accounting|5\": {\n \"acc\": 0.3617021276595745,\n \"acc_stderr\": 0.02866382014719949,\n \"acc_norm\": 0.3617021276595745,\n \"acc_norm_stderr\": 0.02866382014719949\n },\n \"harness|hendrycksTest-professional_law|5\": {\n \"acc\": 0.36114732724902215,\n \"acc_stderr\": 0.01226793547751903,\n \"acc_norm\": 0.36114732724902215,\n \"acc_norm_stderr\": 0.01226793547751903\n },\n \"harness|hendrycksTest-professional_medicine|5\": {\n \"acc\": 0.5183823529411765,\n \"acc_stderr\": 0.030352303395351964,\n \"acc_norm\": 0.5183823529411765,\n \"acc_norm_stderr\": 0.030352303395351964\n },\n \"harness|hendrycksTest-professional_psychology|5\": {\n \"acc\": 0.45098039215686275,\n \"acc_stderr\": 0.020130388312904528,\n \"acc_norm\": 0.45098039215686275,\n \"acc_norm_stderr\": 0.020130388312904528\n },\n \"harness|hendrycksTest-public_relations|5\": {\n \"acc\": 0.5545454545454546,\n \"acc_stderr\": 0.047605488214603246,\n \"acc_norm\": 0.5545454545454546,\n \"acc_norm_stderr\": 0.047605488214603246\n },\n \"harness|hendrycksTest-security_studies|5\": {\n \"acc\": 0.5142857142857142,\n \"acc_stderr\": 0.03199615232806287,\n \"acc_norm\": 0.5142857142857142,\n \"acc_norm_stderr\": 0.03199615232806287\n },\n \"harness|hendrycksTest-sociology|5\": {\n \"acc\": 0.6417910447761194,\n \"acc_stderr\": 0.03390393042268814,\n \"acc_norm\": 0.6417910447761194,\n \"acc_norm_stderr\": 0.03390393042268814\n },\n \"harness|hendrycksTest-us_foreign_policy|5\": {\n \"acc\": 0.63,\n \"acc_stderr\": 0.04852365870939099,\n \"acc_norm\": 0.63,\n \"acc_norm_stderr\": 0.04852365870939099\n },\n \"harness|hendrycksTest-virology|5\": {\n \"acc\": 0.42771084337349397,\n \"acc_stderr\": 0.038515976837185335,\n \"acc_norm\": 0.42771084337349397,\n \"acc_norm_stderr\": 0.038515976837185335\n },\n \"harness|hendrycksTest-world_religions|5\": {\n \"acc\": 0.7192982456140351,\n \"acc_stderr\": 0.034462962170884265,\n \"acc_norm\": 0.7192982456140351,\n \"acc_norm_stderr\": 0.034462962170884265\n },\n \"harness|truthfulqa:mc|0\": {\n \"mc1\": 0.27906976744186046,\n \"mc1_stderr\": 0.015702107090627904,\n \"mc2\": 0.41999112300299424,\n \"mc2_stderr\": 0.014077295047564501\n }\n}\n```", "repo_url": "https://huggingface.co/PeanutJar/LLaMa-2-PeanutButter_v37_SFT-R1-DPO-R2-7B", "leaderboard_url": "https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard", "point_of_contact": "[email protected]", "configs": [{"config_name": "harness_arc_challenge_25", "data_files": [{"split": "2023_09_18T13_52_12.512549", "path": ["**/details_harness|arc:challenge|25_2023-09-18T13-52-12.512549.parquet"]}, {"split": "latest", "path": ["**/details_harness|arc:challenge|25_2023-09-18T13-52-12.512549.parquet"]}]}, {"config_name": "harness_hellaswag_10", "data_files": [{"split": "2023_09_18T13_52_12.512549", "path": ["**/details_harness|hellaswag|10_2023-09-18T13-52-12.512549.parquet"]}, {"split": "latest", "path": ["**/details_harness|hellaswag|10_2023-09-18T13-52-12.512549.parquet"]}]}, {"config_name": "harness_hendrycksTest_5", "data_files": [{"split": "2023_09_18T13_52_12.512549", "path": ["**/details_harness|hendrycksTest-abstract_algebra|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-anatomy|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-astronomy|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-business_ethics|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-clinical_knowledge|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-college_biology|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-college_chemistry|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-college_computer_science|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-college_mathematics|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-college_medicine|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-college_physics|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-computer_security|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-conceptual_physics|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-econometrics|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-electrical_engineering|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-elementary_mathematics|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-formal_logic|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-global_facts|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-high_school_biology|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-high_school_chemistry|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-high_school_computer_science|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-high_school_european_history|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-high_school_geography|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-high_school_government_and_politics|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-high_school_macroeconomics|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-high_school_mathematics|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-high_school_microeconomics|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-high_school_physics|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-high_school_psychology|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-high_school_statistics|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-high_school_us_history|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-high_school_world_history|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-human_aging|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-human_sexuality|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-international_law|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-jurisprudence|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-logical_fallacies|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-machine_learning|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-management|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-marketing|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-medical_genetics|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-miscellaneous|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-moral_disputes|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-moral_scenarios|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-nutrition|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-philosophy|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-prehistory|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-professional_accounting|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-professional_law|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-professional_medicine|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-professional_psychology|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-public_relations|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-security_studies|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-sociology|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-us_foreign_policy|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-virology|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-world_religions|5_2023-09-18T13-52-12.512549.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-abstract_algebra|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-anatomy|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-astronomy|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-business_ethics|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-clinical_knowledge|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-college_biology|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-college_chemistry|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-college_computer_science|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-college_mathematics|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-college_medicine|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-college_physics|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-computer_security|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-conceptual_physics|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-econometrics|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-electrical_engineering|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-elementary_mathematics|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-formal_logic|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-global_facts|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-high_school_biology|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-high_school_chemistry|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-high_school_computer_science|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-high_school_european_history|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-high_school_geography|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-high_school_government_and_politics|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-high_school_macroeconomics|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-high_school_mathematics|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-high_school_microeconomics|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-high_school_physics|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-high_school_psychology|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-high_school_statistics|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-high_school_us_history|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-high_school_world_history|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-human_aging|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-human_sexuality|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-international_law|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-jurisprudence|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-logical_fallacies|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-machine_learning|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-management|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-marketing|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-medical_genetics|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-miscellaneous|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-moral_disputes|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-moral_scenarios|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-nutrition|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-philosophy|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-prehistory|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-professional_accounting|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-professional_law|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-professional_medicine|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-professional_psychology|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-public_relations|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-security_studies|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-sociology|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-us_foreign_policy|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-virology|5_2023-09-18T13-52-12.512549.parquet", "**/details_harness|hendrycksTest-world_religions|5_2023-09-18T13-52-12.512549.parquet"]}]}, {"config_name": "harness_hendrycksTest_abstract_algebra_5", "data_files": [{"split": "2023_09_18T13_52_12.512549", "path": ["**/details_harness|hendrycksTest-abstract_algebra|5_2023-09-18T13-52-12.512549.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-abstract_algebra|5_2023-09-18T13-52-12.512549.parquet"]}]}, {"config_name": "harness_hendrycksTest_anatomy_5", "data_files": [{"split": "2023_09_18T13_52_12.512549", "path": ["**/details_harness|hendrycksTest-anatomy|5_2023-09-18T13-52-12.512549.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-anatomy|5_2023-09-18T13-52-12.512549.parquet"]}]}, {"config_name": "harness_hendrycksTest_astronomy_5", "data_files": [{"split": "2023_09_18T13_52_12.512549", "path": ["**/details_harness|hendrycksTest-astronomy|5_2023-09-18T13-52-12.512549.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-astronomy|5_2023-09-18T13-52-12.512549.parquet"]}]}, {"config_name": "harness_hendrycksTest_business_ethics_5", "data_files": [{"split": "2023_09_18T13_52_12.512549", "path": ["**/details_harness|hendrycksTest-business_ethics|5_2023-09-18T13-52-12.512549.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-business_ethics|5_2023-09-18T13-52-12.512549.parquet"]}]}, {"config_name": "harness_hendrycksTest_clinical_knowledge_5", "data_files": [{"split": "2023_09_18T13_52_12.512549", "path": ["**/details_harness|hendrycksTest-clinical_knowledge|5_2023-09-18T13-52-12.512549.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-clinical_knowledge|5_2023-09-18T13-52-12.512549.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_biology_5", "data_files": [{"split": "2023_09_18T13_52_12.512549", "path": ["**/details_harness|hendrycksTest-college_biology|5_2023-09-18T13-52-12.512549.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_biology|5_2023-09-18T13-52-12.512549.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_chemistry_5", "data_files": [{"split": "2023_09_18T13_52_12.512549", "path": ["**/details_harness|hendrycksTest-college_chemistry|5_2023-09-18T13-52-12.512549.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_chemistry|5_2023-09-18T13-52-12.512549.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_computer_science_5", "data_files": [{"split": "2023_09_18T13_52_12.512549", "path": ["**/details_harness|hendrycksTest-college_computer_science|5_2023-09-18T13-52-12.512549.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_computer_science|5_2023-09-18T13-52-12.512549.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_mathematics_5", "data_files": [{"split": "2023_09_18T13_52_12.512549", "path": ["**/details_harness|hendrycksTest-college_mathematics|5_2023-09-18T13-52-12.512549.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_mathematics|5_2023-09-18T13-52-12.512549.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_medicine_5", "data_files": [{"split": "2023_09_18T13_52_12.512549", "path": ["**/details_harness|hendrycksTest-college_medicine|5_2023-09-18T13-52-12.512549.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_medicine|5_2023-09-18T13-52-12.512549.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_physics_5", "data_files": [{"split": "2023_09_18T13_52_12.512549", "path": ["**/details_harness|hendrycksTest-college_physics|5_2023-09-18T13-52-12.512549.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_physics|5_2023-09-18T13-52-12.512549.parquet"]}]}, {"config_name": "harness_hendrycksTest_computer_security_5", "data_files": [{"split": "2023_09_18T13_52_12.512549", "path": ["**/details_harness|hendrycksTest-computer_security|5_2023-09-18T13-52-12.512549.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-computer_security|5_2023-09-18T13-52-12.512549.parquet"]}]}, {"config_name": "harness_hendrycksTest_conceptual_physics_5", "data_files": [{"split": "2023_09_18T13_52_12.512549", "path": ["**/details_harness|hendrycksTest-conceptual_physics|5_2023-09-18T13-52-12.512549.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-conceptual_physics|5_2023-09-18T13-52-12.512549.parquet"]}]}, {"config_name": "harness_hendrycksTest_econometrics_5", "data_files": [{"split": "2023_09_18T13_52_12.512549", "path": ["**/details_harness|hendrycksTest-econometrics|5_2023-09-18T13-52-12.512549.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-econometrics|5_2023-09-18T13-52-12.512549.parquet"]}]}, {"config_name": "harness_hendrycksTest_electrical_engineering_5", "data_files": [{"split": "2023_09_18T13_52_12.512549", "path": ["**/details_harness|hendrycksTest-electrical_engineering|5_2023-09-18T13-52-12.512549.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-electrical_engineering|5_2023-09-18T13-52-12.512549.parquet"]}]}, {"config_name": "harness_hendrycksTest_elementary_mathematics_5", "data_files": [{"split": "2023_09_18T13_52_12.512549", "path": ["**/details_harness|hendrycksTest-elementary_mathematics|5_2023-09-18T13-52-12.512549.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-elementary_mathematics|5_2023-09-18T13-52-12.512549.parquet"]}]}, {"config_name": "harness_hendrycksTest_formal_logic_5", "data_files": [{"split": "2023_09_18T13_52_12.512549", "path": ["**/details_harness|hendrycksTest-formal_logic|5_2023-09-18T13-52-12.512549.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-formal_logic|5_2023-09-18T13-52-12.512549.parquet"]}]}, {"config_name": "harness_hendrycksTest_global_facts_5", "data_files": [{"split": "2023_09_18T13_52_12.512549", "path": ["**/details_harness|hendrycksTest-global_facts|5_2023-09-18T13-52-12.512549.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-global_facts|5_2023-09-18T13-52-12.512549.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_biology_5", "data_files": [{"split": "2023_09_18T13_52_12.512549", "path": ["**/details_harness|hendrycksTest-high_school_biology|5_2023-09-18T13-52-12.512549.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_biology|5_2023-09-18T13-52-12.512549.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_chemistry_5", "data_files": [{"split": "2023_09_18T13_52_12.512549", "path": ["**/details_harness|hendrycksTest-high_school_chemistry|5_2023-09-18T13-52-12.512549.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_chemistry|5_2023-09-18T13-52-12.512549.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_computer_science_5", "data_files": [{"split": "2023_09_18T13_52_12.512549", "path": ["**/details_harness|hendrycksTest-high_school_computer_science|5_2023-09-18T13-52-12.512549.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_computer_science|5_2023-09-18T13-52-12.512549.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_european_history_5", "data_files": [{"split": "2023_09_18T13_52_12.512549", "path": ["**/details_harness|hendrycksTest-high_school_european_history|5_2023-09-18T13-52-12.512549.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_european_history|5_2023-09-18T13-52-12.512549.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_geography_5", "data_files": [{"split": "2023_09_18T13_52_12.512549", "path": ["**/details_harness|hendrycksTest-high_school_geography|5_2023-09-18T13-52-12.512549.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_geography|5_2023-09-18T13-52-12.512549.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_government_and_politics_5", "data_files": [{"split": "2023_09_18T13_52_12.512549", "path": ["**/details_harness|hendrycksTest-high_school_government_and_politics|5_2023-09-18T13-52-12.512549.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_government_and_politics|5_2023-09-18T13-52-12.512549.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_macroeconomics_5", "data_files": [{"split": "2023_09_18T13_52_12.512549", "path": ["**/details_harness|hendrycksTest-high_school_macroeconomics|5_2023-09-18T13-52-12.512549.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_macroeconomics|5_2023-09-18T13-52-12.512549.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_mathematics_5", "data_files": [{"split": "2023_09_18T13_52_12.512549", "path": ["**/details_harness|hendrycksTest-high_school_mathematics|5_2023-09-18T13-52-12.512549.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_mathematics|5_2023-09-18T13-52-12.512549.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_microeconomics_5", "data_files": [{"split": "2023_09_18T13_52_12.512549", "path": ["**/details_harness|hendrycksTest-high_school_microeconomics|5_2023-09-18T13-52-12.512549.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_microeconomics|5_2023-09-18T13-52-12.512549.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_physics_5", "data_files": [{"split": "2023_09_18T13_52_12.512549", "path": ["**/details_harness|hendrycksTest-high_school_physics|5_2023-09-18T13-52-12.512549.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_physics|5_2023-09-18T13-52-12.512549.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_psychology_5", "data_files": [{"split": "2023_09_18T13_52_12.512549", "path": ["**/details_harness|hendrycksTest-high_school_psychology|5_2023-09-18T13-52-12.512549.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_psychology|5_2023-09-18T13-52-12.512549.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_statistics_5", "data_files": [{"split": "2023_09_18T13_52_12.512549", "path": ["**/details_harness|hendrycksTest-high_school_statistics|5_2023-09-18T13-52-12.512549.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_statistics|5_2023-09-18T13-52-12.512549.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_us_history_5", "data_files": [{"split": "2023_09_18T13_52_12.512549", "path": ["**/details_harness|hendrycksTest-high_school_us_history|5_2023-09-18T13-52-12.512549.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_us_history|5_2023-09-18T13-52-12.512549.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_world_history_5", "data_files": [{"split": "2023_09_18T13_52_12.512549", "path": ["**/details_harness|hendrycksTest-high_school_world_history|5_2023-09-18T13-52-12.512549.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_world_history|5_2023-09-18T13-52-12.512549.parquet"]}]}, {"config_name": "harness_hendrycksTest_human_aging_5", "data_files": [{"split": "2023_09_18T13_52_12.512549", "path": ["**/details_harness|hendrycksTest-human_aging|5_2023-09-18T13-52-12.512549.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-human_aging|5_2023-09-18T13-52-12.512549.parquet"]}]}, {"config_name": "harness_hendrycksTest_human_sexuality_5", "data_files": [{"split": "2023_09_18T13_52_12.512549", "path": ["**/details_harness|hendrycksTest-human_sexuality|5_2023-09-18T13-52-12.512549.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-human_sexuality|5_2023-09-18T13-52-12.512549.parquet"]}]}, {"config_name": "harness_hendrycksTest_international_law_5", "data_files": [{"split": "2023_09_18T13_52_12.512549", "path": ["**/details_harness|hendrycksTest-international_law|5_2023-09-18T13-52-12.512549.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-international_law|5_2023-09-18T13-52-12.512549.parquet"]}]}, {"config_name": "harness_hendrycksTest_jurisprudence_5", "data_files": [{"split": "2023_09_18T13_52_12.512549", "path": ["**/details_harness|hendrycksTest-jurisprudence|5_2023-09-18T13-52-12.512549.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-jurisprudence|5_2023-09-18T13-52-12.512549.parquet"]}]}, {"config_name": "harness_hendrycksTest_logical_fallacies_5", "data_files": [{"split": "2023_09_18T13_52_12.512549", "path": ["**/details_harness|hendrycksTest-logical_fallacies|5_2023-09-18T13-52-12.512549.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-logical_fallacies|5_2023-09-18T13-52-12.512549.parquet"]}]}, {"config_name": "harness_hendrycksTest_machine_learning_5", "data_files": [{"split": "2023_09_18T13_52_12.512549", "path": ["**/details_harness|hendrycksTest-machine_learning|5_2023-09-18T13-52-12.512549.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-machine_learning|5_2023-09-18T13-52-12.512549.parquet"]}]}, {"config_name": "harness_hendrycksTest_management_5", "data_files": [{"split": "2023_09_18T13_52_12.512549", "path": ["**/details_harness|hendrycksTest-management|5_2023-09-18T13-52-12.512549.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-management|5_2023-09-18T13-52-12.512549.parquet"]}]}, {"config_name": "harness_hendrycksTest_marketing_5", "data_files": [{"split": "2023_09_18T13_52_12.512549", "path": ["**/details_harness|hendrycksTest-marketing|5_2023-09-18T13-52-12.512549.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-marketing|5_2023-09-18T13-52-12.512549.parquet"]}]}, {"config_name": "harness_hendrycksTest_medical_genetics_5", "data_files": [{"split": "2023_09_18T13_52_12.512549", "path": ["**/details_harness|hendrycksTest-medical_genetics|5_2023-09-18T13-52-12.512549.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-medical_genetics|5_2023-09-18T13-52-12.512549.parquet"]}]}, {"config_name": "harness_hendrycksTest_miscellaneous_5", "data_files": [{"split": "2023_09_18T13_52_12.512549", "path": ["**/details_harness|hendrycksTest-miscellaneous|5_2023-09-18T13-52-12.512549.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-miscellaneous|5_2023-09-18T13-52-12.512549.parquet"]}]}, {"config_name": "harness_hendrycksTest_moral_disputes_5", "data_files": [{"split": "2023_09_18T13_52_12.512549", "path": ["**/details_harness|hendrycksTest-moral_disputes|5_2023-09-18T13-52-12.512549.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-moral_disputes|5_2023-09-18T13-52-12.512549.parquet"]}]}, {"config_name": "harness_hendrycksTest_moral_scenarios_5", "data_files": [{"split": "2023_09_18T13_52_12.512549", "path": ["**/details_harness|hendrycksTest-moral_scenarios|5_2023-09-18T13-52-12.512549.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-moral_scenarios|5_2023-09-18T13-52-12.512549.parquet"]}]}, {"config_name": "harness_hendrycksTest_nutrition_5", "data_files": [{"split": "2023_09_18T13_52_12.512549", "path": ["**/details_harness|hendrycksTest-nutrition|5_2023-09-18T13-52-12.512549.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-nutrition|5_2023-09-18T13-52-12.512549.parquet"]}]}, {"config_name": "harness_hendrycksTest_philosophy_5", "data_files": [{"split": "2023_09_18T13_52_12.512549", "path": ["**/details_harness|hendrycksTest-philosophy|5_2023-09-18T13-52-12.512549.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-philosophy|5_2023-09-18T13-52-12.512549.parquet"]}]}, {"config_name": "harness_hendrycksTest_prehistory_5", "data_files": [{"split": "2023_09_18T13_52_12.512549", "path": ["**/details_harness|hendrycksTest-prehistory|5_2023-09-18T13-52-12.512549.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-prehistory|5_2023-09-18T13-52-12.512549.parquet"]}]}, {"config_name": "harness_hendrycksTest_professional_accounting_5", "data_files": [{"split": "2023_09_18T13_52_12.512549", "path": ["**/details_harness|hendrycksTest-professional_accounting|5_2023-09-18T13-52-12.512549.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-professional_accounting|5_2023-09-18T13-52-12.512549.parquet"]}]}, {"config_name": "harness_hendrycksTest_professional_law_5", "data_files": [{"split": "2023_09_18T13_52_12.512549", "path": ["**/details_harness|hendrycksTest-professional_law|5_2023-09-18T13-52-12.512549.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-professional_law|5_2023-09-18T13-52-12.512549.parquet"]}]}, {"config_name": "harness_hendrycksTest_professional_medicine_5", "data_files": [{"split": "2023_09_18T13_52_12.512549", "path": ["**/details_harness|hendrycksTest-professional_medicine|5_2023-09-18T13-52-12.512549.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-professional_medicine|5_2023-09-18T13-52-12.512549.parquet"]}]}, {"config_name": "harness_hendrycksTest_professional_psychology_5", "data_files": [{"split": "2023_09_18T13_52_12.512549", "path": ["**/details_harness|hendrycksTest-professional_psychology|5_2023-09-18T13-52-12.512549.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-professional_psychology|5_2023-09-18T13-52-12.512549.parquet"]}]}, {"config_name": "harness_hendrycksTest_public_relations_5", "data_files": [{"split": "2023_09_18T13_52_12.512549", "path": ["**/details_harness|hendrycksTest-public_relations|5_2023-09-18T13-52-12.512549.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-public_relations|5_2023-09-18T13-52-12.512549.parquet"]}]}, {"config_name": "harness_hendrycksTest_security_studies_5", "data_files": [{"split": "2023_09_18T13_52_12.512549", "path": ["**/details_harness|hendrycksTest-security_studies|5_2023-09-18T13-52-12.512549.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-security_studies|5_2023-09-18T13-52-12.512549.parquet"]}]}, {"config_name": "harness_hendrycksTest_sociology_5", "data_files": [{"split": "2023_09_18T13_52_12.512549", "path": ["**/details_harness|hendrycksTest-sociology|5_2023-09-18T13-52-12.512549.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-sociology|5_2023-09-18T13-52-12.512549.parquet"]}]}, {"config_name": "harness_hendrycksTest_us_foreign_policy_5", "data_files": [{"split": "2023_09_18T13_52_12.512549", "path": ["**/details_harness|hendrycksTest-us_foreign_policy|5_2023-09-18T13-52-12.512549.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-us_foreign_policy|5_2023-09-18T13-52-12.512549.parquet"]}]}, {"config_name": "harness_hendrycksTest_virology_5", "data_files": [{"split": "2023_09_18T13_52_12.512549", "path": ["**/details_harness|hendrycksTest-virology|5_2023-09-18T13-52-12.512549.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-virology|5_2023-09-18T13-52-12.512549.parquet"]}]}, {"config_name": "harness_hendrycksTest_world_religions_5", "data_files": [{"split": "2023_09_18T13_52_12.512549", "path": ["**/details_harness|hendrycksTest-world_religions|5_2023-09-18T13-52-12.512549.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-world_religions|5_2023-09-18T13-52-12.512549.parquet"]}]}, {"config_name": "harness_truthfulqa_mc_0", "data_files": [{"split": "2023_09_18T13_52_12.512549", "path": ["**/details_harness|truthfulqa:mc|0_2023-09-18T13-52-12.512549.parquet"]}, {"split": "latest", "path": ["**/details_harness|truthfulqa:mc|0_2023-09-18T13-52-12.512549.parquet"]}]}, {"config_name": "results", "data_files": [{"split": "2023_09_18T13_52_12.512549", "path": ["results_2023-09-18T13-52-12.512549.parquet"]}, {"split": "latest", "path": ["results_2023-09-18T13-52-12.512549.parquet"]}]}]}
|
2023-09-18T12:53:35+00:00
|
[] |
[] |
TAGS
#region-us
|
# Dataset Card for Evaluation run of PeanutJar/LLaMa-2-PeanutButter_v37_SFT-R1-DPO-R2-7B
## Dataset Description
- Homepage:
- Repository: URL
- Paper:
- Leaderboard: URL
- Point of Contact: clementine@URL
### Dataset Summary
Dataset automatically created during the evaluation run of model PeanutJar/LLaMa-2-PeanutButter_v37_SFT-R1-DPO-R2-7B on the Open LLM Leaderboard.
The dataset is composed of 61 configuration, each one coresponding to one of the evaluated task.
The dataset has been created from 1 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The "train" split is always pointing to the latest results.
An additional configuration "results" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the Open LLM Leaderboard).
To load the details from a run, you can for instance do the following:
## Latest results
These are the latest results from run 2023-09-18T13:52:12.512549(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the "latest" split for each eval):
### Supported Tasks and Leaderboards
### Languages
## Dataset Structure
### Data Instances
### Data Fields
### Data Splits
## Dataset Creation
### Curation Rationale
### Source Data
#### Initial Data Collection and Normalization
#### Who are the source language producers?
### Annotations
#### Annotation process
#### Who are the annotators?
### Personal and Sensitive Information
## Considerations for Using the Data
### Social Impact of Dataset
### Discussion of Biases
### Other Known Limitations
## Additional Information
### Dataset Curators
### Licensing Information
### Contributions
|
[
"# Dataset Card for Evaluation run of PeanutJar/LLaMa-2-PeanutButter_v37_SFT-R1-DPO-R2-7B",
"## Dataset Description\n\n- Homepage: \n- Repository: URL\n- Paper: \n- Leaderboard: URL\n- Point of Contact: clementine@URL",
"### Dataset Summary\n\nDataset automatically created during the evaluation run of model PeanutJar/LLaMa-2-PeanutButter_v37_SFT-R1-DPO-R2-7B on the Open LLM Leaderboard.\n\nThe dataset is composed of 61 configuration, each one coresponding to one of the evaluated task.\n\nThe dataset has been created from 1 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The \"train\" split is always pointing to the latest results.\n\nAn additional configuration \"results\" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the Open LLM Leaderboard).\n\nTo load the details from a run, you can for instance do the following:",
"## Latest results\n\nThese are the latest results from run 2023-09-18T13:52:12.512549(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the \"latest\" split for each eval):",
"### Supported Tasks and Leaderboards",
"### Languages",
"## Dataset Structure",
"### Data Instances",
"### Data Fields",
"### Data Splits",
"## Dataset Creation",
"### Curation Rationale",
"### Source Data",
"#### Initial Data Collection and Normalization",
"#### Who are the source language producers?",
"### Annotations",
"#### Annotation process",
"#### Who are the annotators?",
"### Personal and Sensitive Information",
"## Considerations for Using the Data",
"### Social Impact of Dataset",
"### Discussion of Biases",
"### Other Known Limitations",
"## Additional Information",
"### Dataset Curators",
"### Licensing Information",
"### Contributions"
] |
[
"TAGS\n#region-us \n",
"# Dataset Card for Evaluation run of PeanutJar/LLaMa-2-PeanutButter_v37_SFT-R1-DPO-R2-7B",
"## Dataset Description\n\n- Homepage: \n- Repository: URL\n- Paper: \n- Leaderboard: URL\n- Point of Contact: clementine@URL",
"### Dataset Summary\n\nDataset automatically created during the evaluation run of model PeanutJar/LLaMa-2-PeanutButter_v37_SFT-R1-DPO-R2-7B on the Open LLM Leaderboard.\n\nThe dataset is composed of 61 configuration, each one coresponding to one of the evaluated task.\n\nThe dataset has been created from 1 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The \"train\" split is always pointing to the latest results.\n\nAn additional configuration \"results\" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the Open LLM Leaderboard).\n\nTo load the details from a run, you can for instance do the following:",
"## Latest results\n\nThese are the latest results from run 2023-09-18T13:52:12.512549(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the \"latest\" split for each eval):",
"### Supported Tasks and Leaderboards",
"### Languages",
"## Dataset Structure",
"### Data Instances",
"### Data Fields",
"### Data Splits",
"## Dataset Creation",
"### Curation Rationale",
"### Source Data",
"#### Initial Data Collection and Normalization",
"#### Who are the source language producers?",
"### Annotations",
"#### Annotation process",
"#### Who are the annotators?",
"### Personal and Sensitive Information",
"## Considerations for Using the Data",
"### Social Impact of Dataset",
"### Discussion of Biases",
"### Other Known Limitations",
"## Additional Information",
"### Dataset Curators",
"### Licensing Information",
"### Contributions"
] |
[
6,
40,
31,
188,
66,
10,
4,
6,
6,
5,
5,
5,
7,
4,
10,
10,
5,
5,
9,
8,
8,
7,
8,
7,
5,
6,
6,
5
] |
[
"passage: TAGS\n#region-us \n# Dataset Card for Evaluation run of PeanutJar/LLaMa-2-PeanutButter_v37_SFT-R1-DPO-R2-7B## Dataset Description\n\n- Homepage: \n- Repository: URL\n- Paper: \n- Leaderboard: URL\n- Point of Contact: clementine@URL### Dataset Summary\n\nDataset automatically created during the evaluation run of model PeanutJar/LLaMa-2-PeanutButter_v37_SFT-R1-DPO-R2-7B on the Open LLM Leaderboard.\n\nThe dataset is composed of 61 configuration, each one coresponding to one of the evaluated task.\n\nThe dataset has been created from 1 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The \"train\" split is always pointing to the latest results.\n\nAn additional configuration \"results\" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the Open LLM Leaderboard).\n\nTo load the details from a run, you can for instance do the following:## Latest results\n\nThese are the latest results from run 2023-09-18T13:52:12.512549(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the \"latest\" split for each eval):### Supported Tasks and Leaderboards### Languages## Dataset Structure### Data Instances### Data Fields### Data Splits## Dataset Creation### Curation Rationale### Source Data#### Initial Data Collection and Normalization#### Who are the source language producers?### Annotations#### Annotation process#### Who are the annotators?### Personal and Sensitive Information## Considerations for Using the Data### Social Impact of Dataset### Discussion of Biases### Other Known Limitations## Additional Information### Dataset Curators### Licensing Information### Contributions"
] |
8242f208742d032e9ad1741eb25c0457ed4bcc9f
|
# Dataset Card for Evaluation run of Undi95/Unholy-v1-12L-13B
## Dataset Description
- **Homepage:**
- **Repository:** https://huggingface.co/Undi95/Unholy-v1-12L-13B
- **Paper:**
- **Leaderboard:** https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard
- **Point of Contact:** [email protected]
### Dataset Summary
Dataset automatically created during the evaluation run of model [Undi95/Unholy-v1-12L-13B](https://huggingface.co/Undi95/Unholy-v1-12L-13B) on the [Open LLM Leaderboard](https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard).
The dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.
The dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The "train" split is always pointing to the latest results.
An additional configuration "results" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the [Open LLM Leaderboard](https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard)).
To load the details from a run, you can for instance do the following:
```python
from datasets import load_dataset
data = load_dataset("open-llm-leaderboard/details_Undi95__Unholy-v1-12L-13B",
"harness_winogrande_5",
split="train")
```
## Latest results
These are the [latest results from run 2023-10-29T08:07:07.360378](https://huggingface.co/datasets/open-llm-leaderboard/details_Undi95__Unholy-v1-12L-13B/blob/main/results_2023-10-29T08-07-07.360378.json)(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the "latest" split for each eval):
```python
{
"all": {
"em": 0.022651006711409395,
"em_stderr": 0.0015237307803438198,
"f1": 0.09728712248322129,
"f1_stderr": 0.00210132435826052,
"acc": 0.44169065680213837,
"acc_stderr": 0.010210392359241776
},
"harness|drop|3": {
"em": 0.022651006711409395,
"em_stderr": 0.0015237307803438198,
"f1": 0.09728712248322129,
"f1_stderr": 0.00210132435826052
},
"harness|gsm8k|5": {
"acc": 0.1106899166034875,
"acc_stderr": 0.008642172551392465
},
"harness|winogrande|5": {
"acc": 0.7726913970007893,
"acc_stderr": 0.011778612167091087
}
}
```
### Supported Tasks and Leaderboards
[More Information Needed]
### Languages
[More Information Needed]
## Dataset Structure
### Data Instances
[More Information Needed]
### Data Fields
[More Information Needed]
### Data Splits
[More Information Needed]
## Dataset Creation
### Curation Rationale
[More Information Needed]
### Source Data
#### Initial Data Collection and Normalization
[More Information Needed]
#### Who are the source language producers?
[More Information Needed]
### Annotations
#### Annotation process
[More Information Needed]
#### Who are the annotators?
[More Information Needed]
### Personal and Sensitive Information
[More Information Needed]
## Considerations for Using the Data
### Social Impact of Dataset
[More Information Needed]
### Discussion of Biases
[More Information Needed]
### Other Known Limitations
[More Information Needed]
## Additional Information
### Dataset Curators
[More Information Needed]
### Licensing Information
[More Information Needed]
### Citation Information
[More Information Needed]
### Contributions
[More Information Needed]
|
open-llm-leaderboard/details_Undi95__Unholy-v1-12L-13B
|
[
"region:us"
] |
2023-09-18T12:52:43+00:00
|
{"pretty_name": "Evaluation run of Undi95/Unholy-v1-12L-13B", "dataset_summary": "Dataset automatically created during the evaluation run of model [Undi95/Unholy-v1-12L-13B](https://huggingface.co/Undi95/Unholy-v1-12L-13B) on the [Open LLM Leaderboard](https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard).\n\nThe dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.\n\nThe dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The \"train\" split is always pointing to the latest results.\n\nAn additional configuration \"results\" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the [Open LLM Leaderboard](https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard)).\n\nTo load the details from a run, you can for instance do the following:\n```python\nfrom datasets import load_dataset\ndata = load_dataset(\"open-llm-leaderboard/details_Undi95__Unholy-v1-12L-13B\",\n\t\"harness_winogrande_5\",\n\tsplit=\"train\")\n```\n\n## Latest results\n\nThese are the [latest results from run 2023-10-29T08:07:07.360378](https://huggingface.co/datasets/open-llm-leaderboard/details_Undi95__Unholy-v1-12L-13B/blob/main/results_2023-10-29T08-07-07.360378.json)(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the \"latest\" split for each eval):\n\n```python\n{\n \"all\": {\n \"em\": 0.022651006711409395,\n \"em_stderr\": 0.0015237307803438198,\n \"f1\": 0.09728712248322129,\n \"f1_stderr\": 0.00210132435826052,\n \"acc\": 0.44169065680213837,\n \"acc_stderr\": 0.010210392359241776\n },\n \"harness|drop|3\": {\n \"em\": 0.022651006711409395,\n \"em_stderr\": 0.0015237307803438198,\n \"f1\": 0.09728712248322129,\n \"f1_stderr\": 0.00210132435826052\n },\n \"harness|gsm8k|5\": {\n \"acc\": 0.1106899166034875,\n \"acc_stderr\": 0.008642172551392465\n },\n \"harness|winogrande|5\": {\n \"acc\": 0.7726913970007893,\n \"acc_stderr\": 0.011778612167091087\n }\n}\n```", "repo_url": "https://huggingface.co/Undi95/Unholy-v1-12L-13B", "leaderboard_url": "https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard", "point_of_contact": "[email protected]", "configs": [{"config_name": "harness_arc_challenge_25", "data_files": [{"split": "2023_09_18T13_52_19.375562", "path": ["**/details_harness|arc:challenge|25_2023-09-18T13-52-19.375562.parquet"]}, {"split": "latest", "path": ["**/details_harness|arc:challenge|25_2023-09-18T13-52-19.375562.parquet"]}]}, {"config_name": "harness_drop_3", "data_files": [{"split": "2023_10_29T08_07_07.360378", "path": ["**/details_harness|drop|3_2023-10-29T08-07-07.360378.parquet"]}, {"split": "latest", "path": ["**/details_harness|drop|3_2023-10-29T08-07-07.360378.parquet"]}]}, {"config_name": "harness_gsm8k_5", "data_files": [{"split": "2023_10_29T08_07_07.360378", "path": ["**/details_harness|gsm8k|5_2023-10-29T08-07-07.360378.parquet"]}, {"split": "latest", "path": ["**/details_harness|gsm8k|5_2023-10-29T08-07-07.360378.parquet"]}]}, {"config_name": "harness_hellaswag_10", "data_files": [{"split": "2023_09_18T13_52_19.375562", "path": ["**/details_harness|hellaswag|10_2023-09-18T13-52-19.375562.parquet"]}, {"split": "latest", "path": ["**/details_harness|hellaswag|10_2023-09-18T13-52-19.375562.parquet"]}]}, {"config_name": "harness_hendrycksTest_5", "data_files": [{"split": "2023_09_18T13_52_19.375562", "path": ["**/details_harness|hendrycksTest-abstract_algebra|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-anatomy|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-astronomy|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-business_ethics|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-clinical_knowledge|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-college_biology|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-college_chemistry|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-college_computer_science|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-college_mathematics|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-college_medicine|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-college_physics|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-computer_security|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-conceptual_physics|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-econometrics|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-electrical_engineering|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-elementary_mathematics|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-formal_logic|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-global_facts|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-high_school_biology|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-high_school_chemistry|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-high_school_computer_science|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-high_school_european_history|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-high_school_geography|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-high_school_government_and_politics|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-high_school_macroeconomics|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-high_school_mathematics|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-high_school_microeconomics|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-high_school_physics|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-high_school_psychology|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-high_school_statistics|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-high_school_us_history|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-high_school_world_history|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-human_aging|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-human_sexuality|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-international_law|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-jurisprudence|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-logical_fallacies|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-machine_learning|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-management|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-marketing|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-medical_genetics|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-miscellaneous|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-moral_disputes|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-moral_scenarios|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-nutrition|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-philosophy|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-prehistory|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-professional_accounting|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-professional_law|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-professional_medicine|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-professional_psychology|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-public_relations|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-security_studies|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-sociology|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-us_foreign_policy|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-virology|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-world_religions|5_2023-09-18T13-52-19.375562.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-abstract_algebra|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-anatomy|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-astronomy|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-business_ethics|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-clinical_knowledge|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-college_biology|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-college_chemistry|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-college_computer_science|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-college_mathematics|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-college_medicine|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-college_physics|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-computer_security|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-conceptual_physics|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-econometrics|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-electrical_engineering|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-elementary_mathematics|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-formal_logic|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-global_facts|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-high_school_biology|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-high_school_chemistry|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-high_school_computer_science|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-high_school_european_history|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-high_school_geography|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-high_school_government_and_politics|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-high_school_macroeconomics|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-high_school_mathematics|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-high_school_microeconomics|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-high_school_physics|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-high_school_psychology|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-high_school_statistics|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-high_school_us_history|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-high_school_world_history|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-human_aging|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-human_sexuality|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-international_law|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-jurisprudence|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-logical_fallacies|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-machine_learning|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-management|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-marketing|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-medical_genetics|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-miscellaneous|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-moral_disputes|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-moral_scenarios|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-nutrition|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-philosophy|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-prehistory|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-professional_accounting|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-professional_law|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-professional_medicine|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-professional_psychology|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-public_relations|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-security_studies|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-sociology|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-us_foreign_policy|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-virology|5_2023-09-18T13-52-19.375562.parquet", "**/details_harness|hendrycksTest-world_religions|5_2023-09-18T13-52-19.375562.parquet"]}]}, {"config_name": "harness_hendrycksTest_abstract_algebra_5", "data_files": [{"split": "2023_09_18T13_52_19.375562", "path": ["**/details_harness|hendrycksTest-abstract_algebra|5_2023-09-18T13-52-19.375562.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-abstract_algebra|5_2023-09-18T13-52-19.375562.parquet"]}]}, {"config_name": "harness_hendrycksTest_anatomy_5", "data_files": [{"split": "2023_09_18T13_52_19.375562", "path": ["**/details_harness|hendrycksTest-anatomy|5_2023-09-18T13-52-19.375562.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-anatomy|5_2023-09-18T13-52-19.375562.parquet"]}]}, {"config_name": "harness_hendrycksTest_astronomy_5", "data_files": [{"split": "2023_09_18T13_52_19.375562", "path": ["**/details_harness|hendrycksTest-astronomy|5_2023-09-18T13-52-19.375562.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-astronomy|5_2023-09-18T13-52-19.375562.parquet"]}]}, {"config_name": "harness_hendrycksTest_business_ethics_5", "data_files": [{"split": "2023_09_18T13_52_19.375562", "path": ["**/details_harness|hendrycksTest-business_ethics|5_2023-09-18T13-52-19.375562.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-business_ethics|5_2023-09-18T13-52-19.375562.parquet"]}]}, {"config_name": "harness_hendrycksTest_clinical_knowledge_5", "data_files": [{"split": "2023_09_18T13_52_19.375562", "path": ["**/details_harness|hendrycksTest-clinical_knowledge|5_2023-09-18T13-52-19.375562.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-clinical_knowledge|5_2023-09-18T13-52-19.375562.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_biology_5", "data_files": [{"split": "2023_09_18T13_52_19.375562", "path": ["**/details_harness|hendrycksTest-college_biology|5_2023-09-18T13-52-19.375562.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_biology|5_2023-09-18T13-52-19.375562.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_chemistry_5", "data_files": [{"split": "2023_09_18T13_52_19.375562", "path": ["**/details_harness|hendrycksTest-college_chemistry|5_2023-09-18T13-52-19.375562.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_chemistry|5_2023-09-18T13-52-19.375562.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_computer_science_5", "data_files": [{"split": "2023_09_18T13_52_19.375562", "path": ["**/details_harness|hendrycksTest-college_computer_science|5_2023-09-18T13-52-19.375562.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_computer_science|5_2023-09-18T13-52-19.375562.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_mathematics_5", "data_files": [{"split": "2023_09_18T13_52_19.375562", "path": ["**/details_harness|hendrycksTest-college_mathematics|5_2023-09-18T13-52-19.375562.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_mathematics|5_2023-09-18T13-52-19.375562.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_medicine_5", "data_files": [{"split": "2023_09_18T13_52_19.375562", "path": ["**/details_harness|hendrycksTest-college_medicine|5_2023-09-18T13-52-19.375562.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_medicine|5_2023-09-18T13-52-19.375562.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_physics_5", "data_files": [{"split": "2023_09_18T13_52_19.375562", "path": ["**/details_harness|hendrycksTest-college_physics|5_2023-09-18T13-52-19.375562.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_physics|5_2023-09-18T13-52-19.375562.parquet"]}]}, {"config_name": "harness_hendrycksTest_computer_security_5", "data_files": [{"split": "2023_09_18T13_52_19.375562", "path": ["**/details_harness|hendrycksTest-computer_security|5_2023-09-18T13-52-19.375562.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-computer_security|5_2023-09-18T13-52-19.375562.parquet"]}]}, {"config_name": "harness_hendrycksTest_conceptual_physics_5", "data_files": [{"split": "2023_09_18T13_52_19.375562", "path": ["**/details_harness|hendrycksTest-conceptual_physics|5_2023-09-18T13-52-19.375562.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-conceptual_physics|5_2023-09-18T13-52-19.375562.parquet"]}]}, {"config_name": "harness_hendrycksTest_econometrics_5", "data_files": [{"split": "2023_09_18T13_52_19.375562", "path": ["**/details_harness|hendrycksTest-econometrics|5_2023-09-18T13-52-19.375562.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-econometrics|5_2023-09-18T13-52-19.375562.parquet"]}]}, {"config_name": "harness_hendrycksTest_electrical_engineering_5", "data_files": [{"split": "2023_09_18T13_52_19.375562", "path": ["**/details_harness|hendrycksTest-electrical_engineering|5_2023-09-18T13-52-19.375562.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-electrical_engineering|5_2023-09-18T13-52-19.375562.parquet"]}]}, {"config_name": "harness_hendrycksTest_elementary_mathematics_5", "data_files": [{"split": "2023_09_18T13_52_19.375562", "path": ["**/details_harness|hendrycksTest-elementary_mathematics|5_2023-09-18T13-52-19.375562.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-elementary_mathematics|5_2023-09-18T13-52-19.375562.parquet"]}]}, {"config_name": "harness_hendrycksTest_formal_logic_5", "data_files": [{"split": "2023_09_18T13_52_19.375562", "path": ["**/details_harness|hendrycksTest-formal_logic|5_2023-09-18T13-52-19.375562.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-formal_logic|5_2023-09-18T13-52-19.375562.parquet"]}]}, {"config_name": "harness_hendrycksTest_global_facts_5", "data_files": [{"split": "2023_09_18T13_52_19.375562", "path": ["**/details_harness|hendrycksTest-global_facts|5_2023-09-18T13-52-19.375562.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-global_facts|5_2023-09-18T13-52-19.375562.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_biology_5", "data_files": [{"split": "2023_09_18T13_52_19.375562", "path": ["**/details_harness|hendrycksTest-high_school_biology|5_2023-09-18T13-52-19.375562.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_biology|5_2023-09-18T13-52-19.375562.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_chemistry_5", "data_files": [{"split": "2023_09_18T13_52_19.375562", "path": ["**/details_harness|hendrycksTest-high_school_chemistry|5_2023-09-18T13-52-19.375562.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_chemistry|5_2023-09-18T13-52-19.375562.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_computer_science_5", "data_files": [{"split": "2023_09_18T13_52_19.375562", "path": ["**/details_harness|hendrycksTest-high_school_computer_science|5_2023-09-18T13-52-19.375562.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_computer_science|5_2023-09-18T13-52-19.375562.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_european_history_5", "data_files": [{"split": "2023_09_18T13_52_19.375562", "path": ["**/details_harness|hendrycksTest-high_school_european_history|5_2023-09-18T13-52-19.375562.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_european_history|5_2023-09-18T13-52-19.375562.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_geography_5", "data_files": [{"split": "2023_09_18T13_52_19.375562", "path": ["**/details_harness|hendrycksTest-high_school_geography|5_2023-09-18T13-52-19.375562.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_geography|5_2023-09-18T13-52-19.375562.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_government_and_politics_5", "data_files": [{"split": "2023_09_18T13_52_19.375562", "path": ["**/details_harness|hendrycksTest-high_school_government_and_politics|5_2023-09-18T13-52-19.375562.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_government_and_politics|5_2023-09-18T13-52-19.375562.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_macroeconomics_5", "data_files": [{"split": "2023_09_18T13_52_19.375562", "path": ["**/details_harness|hendrycksTest-high_school_macroeconomics|5_2023-09-18T13-52-19.375562.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_macroeconomics|5_2023-09-18T13-52-19.375562.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_mathematics_5", "data_files": [{"split": "2023_09_18T13_52_19.375562", "path": ["**/details_harness|hendrycksTest-high_school_mathematics|5_2023-09-18T13-52-19.375562.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_mathematics|5_2023-09-18T13-52-19.375562.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_microeconomics_5", "data_files": [{"split": "2023_09_18T13_52_19.375562", "path": ["**/details_harness|hendrycksTest-high_school_microeconomics|5_2023-09-18T13-52-19.375562.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_microeconomics|5_2023-09-18T13-52-19.375562.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_physics_5", "data_files": [{"split": "2023_09_18T13_52_19.375562", "path": ["**/details_harness|hendrycksTest-high_school_physics|5_2023-09-18T13-52-19.375562.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_physics|5_2023-09-18T13-52-19.375562.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_psychology_5", "data_files": [{"split": "2023_09_18T13_52_19.375562", "path": ["**/details_harness|hendrycksTest-high_school_psychology|5_2023-09-18T13-52-19.375562.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_psychology|5_2023-09-18T13-52-19.375562.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_statistics_5", "data_files": [{"split": "2023_09_18T13_52_19.375562", "path": ["**/details_harness|hendrycksTest-high_school_statistics|5_2023-09-18T13-52-19.375562.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_statistics|5_2023-09-18T13-52-19.375562.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_us_history_5", "data_files": [{"split": "2023_09_18T13_52_19.375562", "path": ["**/details_harness|hendrycksTest-high_school_us_history|5_2023-09-18T13-52-19.375562.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_us_history|5_2023-09-18T13-52-19.375562.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_world_history_5", "data_files": [{"split": "2023_09_18T13_52_19.375562", "path": ["**/details_harness|hendrycksTest-high_school_world_history|5_2023-09-18T13-52-19.375562.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_world_history|5_2023-09-18T13-52-19.375562.parquet"]}]}, {"config_name": "harness_hendrycksTest_human_aging_5", "data_files": [{"split": "2023_09_18T13_52_19.375562", "path": ["**/details_harness|hendrycksTest-human_aging|5_2023-09-18T13-52-19.375562.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-human_aging|5_2023-09-18T13-52-19.375562.parquet"]}]}, {"config_name": "harness_hendrycksTest_human_sexuality_5", "data_files": [{"split": "2023_09_18T13_52_19.375562", "path": ["**/details_harness|hendrycksTest-human_sexuality|5_2023-09-18T13-52-19.375562.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-human_sexuality|5_2023-09-18T13-52-19.375562.parquet"]}]}, {"config_name": "harness_hendrycksTest_international_law_5", "data_files": [{"split": "2023_09_18T13_52_19.375562", "path": ["**/details_harness|hendrycksTest-international_law|5_2023-09-18T13-52-19.375562.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-international_law|5_2023-09-18T13-52-19.375562.parquet"]}]}, {"config_name": "harness_hendrycksTest_jurisprudence_5", "data_files": [{"split": "2023_09_18T13_52_19.375562", "path": ["**/details_harness|hendrycksTest-jurisprudence|5_2023-09-18T13-52-19.375562.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-jurisprudence|5_2023-09-18T13-52-19.375562.parquet"]}]}, {"config_name": "harness_hendrycksTest_logical_fallacies_5", "data_files": [{"split": "2023_09_18T13_52_19.375562", "path": ["**/details_harness|hendrycksTest-logical_fallacies|5_2023-09-18T13-52-19.375562.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-logical_fallacies|5_2023-09-18T13-52-19.375562.parquet"]}]}, {"config_name": "harness_hendrycksTest_machine_learning_5", "data_files": [{"split": "2023_09_18T13_52_19.375562", "path": ["**/details_harness|hendrycksTest-machine_learning|5_2023-09-18T13-52-19.375562.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-machine_learning|5_2023-09-18T13-52-19.375562.parquet"]}]}, {"config_name": "harness_hendrycksTest_management_5", "data_files": [{"split": "2023_09_18T13_52_19.375562", "path": ["**/details_harness|hendrycksTest-management|5_2023-09-18T13-52-19.375562.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-management|5_2023-09-18T13-52-19.375562.parquet"]}]}, {"config_name": "harness_hendrycksTest_marketing_5", "data_files": [{"split": "2023_09_18T13_52_19.375562", "path": ["**/details_harness|hendrycksTest-marketing|5_2023-09-18T13-52-19.375562.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-marketing|5_2023-09-18T13-52-19.375562.parquet"]}]}, {"config_name": "harness_hendrycksTest_medical_genetics_5", "data_files": [{"split": "2023_09_18T13_52_19.375562", "path": ["**/details_harness|hendrycksTest-medical_genetics|5_2023-09-18T13-52-19.375562.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-medical_genetics|5_2023-09-18T13-52-19.375562.parquet"]}]}, {"config_name": "harness_hendrycksTest_miscellaneous_5", "data_files": [{"split": "2023_09_18T13_52_19.375562", "path": ["**/details_harness|hendrycksTest-miscellaneous|5_2023-09-18T13-52-19.375562.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-miscellaneous|5_2023-09-18T13-52-19.375562.parquet"]}]}, {"config_name": "harness_hendrycksTest_moral_disputes_5", "data_files": [{"split": "2023_09_18T13_52_19.375562", "path": ["**/details_harness|hendrycksTest-moral_disputes|5_2023-09-18T13-52-19.375562.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-moral_disputes|5_2023-09-18T13-52-19.375562.parquet"]}]}, {"config_name": "harness_hendrycksTest_moral_scenarios_5", "data_files": [{"split": "2023_09_18T13_52_19.375562", "path": ["**/details_harness|hendrycksTest-moral_scenarios|5_2023-09-18T13-52-19.375562.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-moral_scenarios|5_2023-09-18T13-52-19.375562.parquet"]}]}, {"config_name": "harness_hendrycksTest_nutrition_5", "data_files": [{"split": "2023_09_18T13_52_19.375562", "path": ["**/details_harness|hendrycksTest-nutrition|5_2023-09-18T13-52-19.375562.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-nutrition|5_2023-09-18T13-52-19.375562.parquet"]}]}, {"config_name": "harness_hendrycksTest_philosophy_5", "data_files": [{"split": "2023_09_18T13_52_19.375562", "path": ["**/details_harness|hendrycksTest-philosophy|5_2023-09-18T13-52-19.375562.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-philosophy|5_2023-09-18T13-52-19.375562.parquet"]}]}, {"config_name": "harness_hendrycksTest_prehistory_5", "data_files": [{"split": "2023_09_18T13_52_19.375562", "path": ["**/details_harness|hendrycksTest-prehistory|5_2023-09-18T13-52-19.375562.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-prehistory|5_2023-09-18T13-52-19.375562.parquet"]}]}, {"config_name": "harness_hendrycksTest_professional_accounting_5", "data_files": [{"split": "2023_09_18T13_52_19.375562", "path": ["**/details_harness|hendrycksTest-professional_accounting|5_2023-09-18T13-52-19.375562.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-professional_accounting|5_2023-09-18T13-52-19.375562.parquet"]}]}, {"config_name": "harness_hendrycksTest_professional_law_5", "data_files": [{"split": "2023_09_18T13_52_19.375562", "path": ["**/details_harness|hendrycksTest-professional_law|5_2023-09-18T13-52-19.375562.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-professional_law|5_2023-09-18T13-52-19.375562.parquet"]}]}, {"config_name": "harness_hendrycksTest_professional_medicine_5", "data_files": [{"split": "2023_09_18T13_52_19.375562", "path": ["**/details_harness|hendrycksTest-professional_medicine|5_2023-09-18T13-52-19.375562.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-professional_medicine|5_2023-09-18T13-52-19.375562.parquet"]}]}, {"config_name": "harness_hendrycksTest_professional_psychology_5", "data_files": [{"split": "2023_09_18T13_52_19.375562", "path": ["**/details_harness|hendrycksTest-professional_psychology|5_2023-09-18T13-52-19.375562.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-professional_psychology|5_2023-09-18T13-52-19.375562.parquet"]}]}, {"config_name": "harness_hendrycksTest_public_relations_5", "data_files": [{"split": "2023_09_18T13_52_19.375562", "path": ["**/details_harness|hendrycksTest-public_relations|5_2023-09-18T13-52-19.375562.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-public_relations|5_2023-09-18T13-52-19.375562.parquet"]}]}, {"config_name": "harness_hendrycksTest_security_studies_5", "data_files": [{"split": "2023_09_18T13_52_19.375562", "path": ["**/details_harness|hendrycksTest-security_studies|5_2023-09-18T13-52-19.375562.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-security_studies|5_2023-09-18T13-52-19.375562.parquet"]}]}, {"config_name": "harness_hendrycksTest_sociology_5", "data_files": [{"split": "2023_09_18T13_52_19.375562", "path": ["**/details_harness|hendrycksTest-sociology|5_2023-09-18T13-52-19.375562.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-sociology|5_2023-09-18T13-52-19.375562.parquet"]}]}, {"config_name": "harness_hendrycksTest_us_foreign_policy_5", "data_files": [{"split": "2023_09_18T13_52_19.375562", "path": ["**/details_harness|hendrycksTest-us_foreign_policy|5_2023-09-18T13-52-19.375562.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-us_foreign_policy|5_2023-09-18T13-52-19.375562.parquet"]}]}, {"config_name": "harness_hendrycksTest_virology_5", "data_files": [{"split": "2023_09_18T13_52_19.375562", "path": ["**/details_harness|hendrycksTest-virology|5_2023-09-18T13-52-19.375562.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-virology|5_2023-09-18T13-52-19.375562.parquet"]}]}, {"config_name": "harness_hendrycksTest_world_religions_5", "data_files": [{"split": "2023_09_18T13_52_19.375562", "path": ["**/details_harness|hendrycksTest-world_religions|5_2023-09-18T13-52-19.375562.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-world_religions|5_2023-09-18T13-52-19.375562.parquet"]}]}, {"config_name": "harness_truthfulqa_mc_0", "data_files": [{"split": "2023_09_18T13_52_19.375562", "path": ["**/details_harness|truthfulqa:mc|0_2023-09-18T13-52-19.375562.parquet"]}, {"split": "latest", "path": ["**/details_harness|truthfulqa:mc|0_2023-09-18T13-52-19.375562.parquet"]}]}, {"config_name": "harness_winogrande_5", "data_files": [{"split": "2023_10_29T08_07_07.360378", "path": ["**/details_harness|winogrande|5_2023-10-29T08-07-07.360378.parquet"]}, {"split": "latest", "path": ["**/details_harness|winogrande|5_2023-10-29T08-07-07.360378.parquet"]}]}, {"config_name": "results", "data_files": [{"split": "2023_09_18T13_52_19.375562", "path": ["results_2023-09-18T13-52-19.375562.parquet"]}, {"split": "2023_10_29T08_07_07.360378", "path": ["results_2023-10-29T08-07-07.360378.parquet"]}, {"split": "latest", "path": ["results_2023-10-29T08-07-07.360378.parquet"]}]}]}
|
2023-10-29T08:07:19+00:00
|
[] |
[] |
TAGS
#region-us
|
# Dataset Card for Evaluation run of Undi95/Unholy-v1-12L-13B
## Dataset Description
- Homepage:
- Repository: URL
- Paper:
- Leaderboard: URL
- Point of Contact: clementine@URL
### Dataset Summary
Dataset automatically created during the evaluation run of model Undi95/Unholy-v1-12L-13B on the Open LLM Leaderboard.
The dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.
The dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The "train" split is always pointing to the latest results.
An additional configuration "results" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the Open LLM Leaderboard).
To load the details from a run, you can for instance do the following:
## Latest results
These are the latest results from run 2023-10-29T08:07:07.360378(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the "latest" split for each eval):
### Supported Tasks and Leaderboards
### Languages
## Dataset Structure
### Data Instances
### Data Fields
### Data Splits
## Dataset Creation
### Curation Rationale
### Source Data
#### Initial Data Collection and Normalization
#### Who are the source language producers?
### Annotations
#### Annotation process
#### Who are the annotators?
### Personal and Sensitive Information
## Considerations for Using the Data
### Social Impact of Dataset
### Discussion of Biases
### Other Known Limitations
## Additional Information
### Dataset Curators
### Licensing Information
### Contributions
|
[
"# Dataset Card for Evaluation run of Undi95/Unholy-v1-12L-13B",
"## Dataset Description\n\n- Homepage: \n- Repository: URL\n- Paper: \n- Leaderboard: URL\n- Point of Contact: clementine@URL",
"### Dataset Summary\n\nDataset automatically created during the evaluation run of model Undi95/Unholy-v1-12L-13B on the Open LLM Leaderboard.\n\nThe dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.\n\nThe dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The \"train\" split is always pointing to the latest results.\n\nAn additional configuration \"results\" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the Open LLM Leaderboard).\n\nTo load the details from a run, you can for instance do the following:",
"## Latest results\n\nThese are the latest results from run 2023-10-29T08:07:07.360378(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the \"latest\" split for each eval):",
"### Supported Tasks and Leaderboards",
"### Languages",
"## Dataset Structure",
"### Data Instances",
"### Data Fields",
"### Data Splits",
"## Dataset Creation",
"### Curation Rationale",
"### Source Data",
"#### Initial Data Collection and Normalization",
"#### Who are the source language producers?",
"### Annotations",
"#### Annotation process",
"#### Who are the annotators?",
"### Personal and Sensitive Information",
"## Considerations for Using the Data",
"### Social Impact of Dataset",
"### Discussion of Biases",
"### Other Known Limitations",
"## Additional Information",
"### Dataset Curators",
"### Licensing Information",
"### Contributions"
] |
[
"TAGS\n#region-us \n",
"# Dataset Card for Evaluation run of Undi95/Unholy-v1-12L-13B",
"## Dataset Description\n\n- Homepage: \n- Repository: URL\n- Paper: \n- Leaderboard: URL\n- Point of Contact: clementine@URL",
"### Dataset Summary\n\nDataset automatically created during the evaluation run of model Undi95/Unholy-v1-12L-13B on the Open LLM Leaderboard.\n\nThe dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.\n\nThe dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The \"train\" split is always pointing to the latest results.\n\nAn additional configuration \"results\" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the Open LLM Leaderboard).\n\nTo load the details from a run, you can for instance do the following:",
"## Latest results\n\nThese are the latest results from run 2023-10-29T08:07:07.360378(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the \"latest\" split for each eval):",
"### Supported Tasks and Leaderboards",
"### Languages",
"## Dataset Structure",
"### Data Instances",
"### Data Fields",
"### Data Splits",
"## Dataset Creation",
"### Curation Rationale",
"### Source Data",
"#### Initial Data Collection and Normalization",
"#### Who are the source language producers?",
"### Annotations",
"#### Annotation process",
"#### Who are the annotators?",
"### Personal and Sensitive Information",
"## Considerations for Using the Data",
"### Social Impact of Dataset",
"### Discussion of Biases",
"### Other Known Limitations",
"## Additional Information",
"### Dataset Curators",
"### Licensing Information",
"### Contributions"
] |
[
6,
23,
31,
171,
66,
10,
4,
6,
6,
5,
5,
5,
7,
4,
10,
10,
5,
5,
9,
8,
8,
7,
8,
7,
5,
6,
6,
5
] |
[
"passage: TAGS\n#region-us \n# Dataset Card for Evaluation run of Undi95/Unholy-v1-12L-13B## Dataset Description\n\n- Homepage: \n- Repository: URL\n- Paper: \n- Leaderboard: URL\n- Point of Contact: clementine@URL### Dataset Summary\n\nDataset automatically created during the evaluation run of model Undi95/Unholy-v1-12L-13B on the Open LLM Leaderboard.\n\nThe dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.\n\nThe dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The \"train\" split is always pointing to the latest results.\n\nAn additional configuration \"results\" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the Open LLM Leaderboard).\n\nTo load the details from a run, you can for instance do the following:## Latest results\n\nThese are the latest results from run 2023-10-29T08:07:07.360378(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the \"latest\" split for each eval):### Supported Tasks and Leaderboards### Languages## Dataset Structure### Data Instances### Data Fields### Data Splits## Dataset Creation### Curation Rationale### Source Data#### Initial Data Collection and Normalization#### Who are the source language producers?### Annotations#### Annotation process#### Who are the annotators?### Personal and Sensitive Information## Considerations for Using the Data### Social Impact of Dataset### Discussion of Biases### Other Known Limitations## Additional Information### Dataset Curators### Licensing Information### Contributions"
] |
a110c44de50f860e540d22b28735dd18437d2705
|
# Dataset of Yamada Ryō
This is the dataset of Yamada Ryō, containing 282 images and their tags.
Images are crawled from many sites (e.g. danbooru, pixiv, zerochan ...), the auto-crawling system is powered by [DeepGHS Team](https://github.com/deepghs)([huggingface organization](https://huggingface.co/deepghs)).
| Name | Images | Download | Description |
|:------------|---------:|:------------------------------------|:-------------------------------------------------------------------------|
| raw | 282 | [Download](dataset-raw.zip) | Raw data with meta information. |
| raw-stage3 | 631 | [Download](dataset-raw-stage3.zip) | 3-stage cropped raw data with meta information. |
| 384x512 | 282 | [Download](dataset-384x512.zip) | 384x512 aligned dataset. |
| 512x512 | 282 | [Download](dataset-512x512.zip) | 512x512 aligned dataset. |
| 512x704 | 282 | [Download](dataset-512x704.zip) | 512x704 aligned dataset. |
| 640x640 | 282 | [Download](dataset-640x640.zip) | 640x640 aligned dataset. |
| 640x880 | 282 | [Download](dataset-640x880.zip) | 640x880 aligned dataset. |
| stage3-640 | 631 | [Download](dataset-stage3-640.zip) | 3-stage cropped dataset with the shorter side not exceeding 640 pixels. |
| stage3-800 | 631 | [Download](dataset-stage3-800.zip) | 3-stage cropped dataset with the shorter side not exceeding 800 pixels. |
| stage3-1200 | 631 | [Download](dataset-stage3-1200.zip) | 3-stage cropped dataset with the shorter side not exceeding 1200 pixels. |
|
CyberHarem/yamada_ryo_bocchitherock
|
[
"task_categories:text-to-image",
"size_categories:n<1K",
"license:mit",
"art",
"not-for-all-audiences",
"region:us"
] |
2023-09-18T12:53:59+00:00
|
{"license": "mit", "size_categories": ["n<1K"], "task_categories": ["text-to-image"], "tags": ["art", "not-for-all-audiences"]}
|
2023-09-18T12:56:26+00:00
|
[] |
[] |
TAGS
#task_categories-text-to-image #size_categories-n<1K #license-mit #art #not-for-all-audiences #region-us
|
Dataset of Yamada Ryō
=====================
This is the dataset of Yamada Ryō, containing 282 images and their tags.
Images are crawled from many sites (e.g. danbooru, pixiv, zerochan ...), the auto-crawling system is powered by DeepGHS Team(huggingface organization).
|
[] |
[
"TAGS\n#task_categories-text-to-image #size_categories-n<1K #license-mit #art #not-for-all-audiences #region-us \n"
] |
[
44
] |
[
"passage: TAGS\n#task_categories-text-to-image #size_categories-n<1K #license-mit #art #not-for-all-audiences #region-us \n"
] |
f4991d8a4d0363bae5c1dfdb923ede27e4dc28ad
|
# Dataset Card for Evaluation run of Undi95/MLewdBoros-L2-13B
## Dataset Description
- **Homepage:**
- **Repository:** https://huggingface.co/Undi95/MLewdBoros-L2-13B
- **Paper:**
- **Leaderboard:** https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard
- **Point of Contact:** [email protected]
### Dataset Summary
Dataset automatically created during the evaluation run of model [Undi95/MLewdBoros-L2-13B](https://huggingface.co/Undi95/MLewdBoros-L2-13B) on the [Open LLM Leaderboard](https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard).
The dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.
The dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The "train" split is always pointing to the latest results.
An additional configuration "results" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the [Open LLM Leaderboard](https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard)).
To load the details from a run, you can for instance do the following:
```python
from datasets import load_dataset
data = load_dataset("open-llm-leaderboard/details_Undi95__MLewdBoros-L2-13B",
"harness_winogrande_5",
split="train")
```
## Latest results
These are the [latest results from run 2023-10-28T22:12:00.775103](https://huggingface.co/datasets/open-llm-leaderboard/details_Undi95__MLewdBoros-L2-13B/blob/main/results_2023-10-28T22-12-00.775103.json)(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the "latest" split for each eval):
```python
{
"all": {
"em": 0.41820469798657717,
"em_stderr": 0.005051486654118123,
"f1": 0.4659270134228202,
"f1_stderr": 0.0048870842597281815,
"acc": 0.4397330497800048,
"acc_stderr": 0.010226033876351036
},
"harness|drop|3": {
"em": 0.41820469798657717,
"em_stderr": 0.005051486654118123,
"f1": 0.4659270134228202,
"f1_stderr": 0.0048870842597281815
},
"harness|gsm8k|5": {
"acc": 0.10993176648976498,
"acc_stderr": 0.008616195587865394
},
"harness|winogrande|5": {
"acc": 0.7695343330702447,
"acc_stderr": 0.01183587216483668
}
}
```
### Supported Tasks and Leaderboards
[More Information Needed]
### Languages
[More Information Needed]
## Dataset Structure
### Data Instances
[More Information Needed]
### Data Fields
[More Information Needed]
### Data Splits
[More Information Needed]
## Dataset Creation
### Curation Rationale
[More Information Needed]
### Source Data
#### Initial Data Collection and Normalization
[More Information Needed]
#### Who are the source language producers?
[More Information Needed]
### Annotations
#### Annotation process
[More Information Needed]
#### Who are the annotators?
[More Information Needed]
### Personal and Sensitive Information
[More Information Needed]
## Considerations for Using the Data
### Social Impact of Dataset
[More Information Needed]
### Discussion of Biases
[More Information Needed]
### Other Known Limitations
[More Information Needed]
## Additional Information
### Dataset Curators
[More Information Needed]
### Licensing Information
[More Information Needed]
### Citation Information
[More Information Needed]
### Contributions
[More Information Needed]
|
open-llm-leaderboard/details_Undi95__MLewdBoros-L2-13B
|
[
"region:us"
] |
2023-09-18T12:57:01+00:00
|
{"pretty_name": "Evaluation run of Undi95/MLewdBoros-L2-13B", "dataset_summary": "Dataset automatically created during the evaluation run of model [Undi95/MLewdBoros-L2-13B](https://huggingface.co/Undi95/MLewdBoros-L2-13B) on the [Open LLM Leaderboard](https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard).\n\nThe dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.\n\nThe dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The \"train\" split is always pointing to the latest results.\n\nAn additional configuration \"results\" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the [Open LLM Leaderboard](https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard)).\n\nTo load the details from a run, you can for instance do the following:\n```python\nfrom datasets import load_dataset\ndata = load_dataset(\"open-llm-leaderboard/details_Undi95__MLewdBoros-L2-13B\",\n\t\"harness_winogrande_5\",\n\tsplit=\"train\")\n```\n\n## Latest results\n\nThese are the [latest results from run 2023-10-28T22:12:00.775103](https://huggingface.co/datasets/open-llm-leaderboard/details_Undi95__MLewdBoros-L2-13B/blob/main/results_2023-10-28T22-12-00.775103.json)(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the \"latest\" split for each eval):\n\n```python\n{\n \"all\": {\n \"em\": 0.41820469798657717,\n \"em_stderr\": 0.005051486654118123,\n \"f1\": 0.4659270134228202,\n \"f1_stderr\": 0.0048870842597281815,\n \"acc\": 0.4397330497800048,\n \"acc_stderr\": 0.010226033876351036\n },\n \"harness|drop|3\": {\n \"em\": 0.41820469798657717,\n \"em_stderr\": 0.005051486654118123,\n \"f1\": 0.4659270134228202,\n \"f1_stderr\": 0.0048870842597281815\n },\n \"harness|gsm8k|5\": {\n \"acc\": 0.10993176648976498,\n \"acc_stderr\": 0.008616195587865394\n },\n \"harness|winogrande|5\": {\n \"acc\": 0.7695343330702447,\n \"acc_stderr\": 0.01183587216483668\n }\n}\n```", "repo_url": "https://huggingface.co/Undi95/MLewdBoros-L2-13B", "leaderboard_url": "https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard", "point_of_contact": "[email protected]", "configs": [{"config_name": "harness_arc_challenge_25", "data_files": [{"split": "2023_09_18T13_56_38.282478", "path": ["**/details_harness|arc:challenge|25_2023-09-18T13-56-38.282478.parquet"]}, {"split": "latest", "path": ["**/details_harness|arc:challenge|25_2023-09-18T13-56-38.282478.parquet"]}]}, {"config_name": "harness_drop_3", "data_files": [{"split": "2023_10_28T22_12_00.775103", "path": ["**/details_harness|drop|3_2023-10-28T22-12-00.775103.parquet"]}, {"split": "latest", "path": ["**/details_harness|drop|3_2023-10-28T22-12-00.775103.parquet"]}]}, {"config_name": "harness_gsm8k_5", "data_files": [{"split": "2023_10_28T22_12_00.775103", "path": ["**/details_harness|gsm8k|5_2023-10-28T22-12-00.775103.parquet"]}, {"split": "latest", "path": ["**/details_harness|gsm8k|5_2023-10-28T22-12-00.775103.parquet"]}]}, {"config_name": "harness_hellaswag_10", "data_files": [{"split": "2023_09_18T13_56_38.282478", "path": ["**/details_harness|hellaswag|10_2023-09-18T13-56-38.282478.parquet"]}, {"split": "latest", "path": ["**/details_harness|hellaswag|10_2023-09-18T13-56-38.282478.parquet"]}]}, {"config_name": "harness_hendrycksTest_5", "data_files": [{"split": "2023_09_18T13_56_38.282478", "path": ["**/details_harness|hendrycksTest-abstract_algebra|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-anatomy|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-astronomy|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-business_ethics|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-clinical_knowledge|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-college_biology|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-college_chemistry|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-college_computer_science|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-college_mathematics|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-college_medicine|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-college_physics|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-computer_security|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-conceptual_physics|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-econometrics|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-electrical_engineering|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-elementary_mathematics|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-formal_logic|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-global_facts|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-high_school_biology|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-high_school_chemistry|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-high_school_computer_science|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-high_school_european_history|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-high_school_geography|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-high_school_government_and_politics|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-high_school_macroeconomics|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-high_school_mathematics|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-high_school_microeconomics|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-high_school_physics|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-high_school_psychology|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-high_school_statistics|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-high_school_us_history|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-high_school_world_history|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-human_aging|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-human_sexuality|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-international_law|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-jurisprudence|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-logical_fallacies|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-machine_learning|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-management|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-marketing|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-medical_genetics|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-miscellaneous|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-moral_disputes|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-moral_scenarios|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-nutrition|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-philosophy|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-prehistory|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-professional_accounting|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-professional_law|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-professional_medicine|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-professional_psychology|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-public_relations|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-security_studies|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-sociology|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-us_foreign_policy|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-virology|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-world_religions|5_2023-09-18T13-56-38.282478.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-abstract_algebra|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-anatomy|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-astronomy|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-business_ethics|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-clinical_knowledge|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-college_biology|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-college_chemistry|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-college_computer_science|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-college_mathematics|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-college_medicine|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-college_physics|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-computer_security|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-conceptual_physics|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-econometrics|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-electrical_engineering|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-elementary_mathematics|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-formal_logic|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-global_facts|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-high_school_biology|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-high_school_chemistry|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-high_school_computer_science|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-high_school_european_history|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-high_school_geography|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-high_school_government_and_politics|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-high_school_macroeconomics|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-high_school_mathematics|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-high_school_microeconomics|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-high_school_physics|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-high_school_psychology|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-high_school_statistics|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-high_school_us_history|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-high_school_world_history|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-human_aging|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-human_sexuality|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-international_law|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-jurisprudence|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-logical_fallacies|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-machine_learning|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-management|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-marketing|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-medical_genetics|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-miscellaneous|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-moral_disputes|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-moral_scenarios|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-nutrition|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-philosophy|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-prehistory|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-professional_accounting|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-professional_law|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-professional_medicine|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-professional_psychology|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-public_relations|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-security_studies|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-sociology|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-us_foreign_policy|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-virology|5_2023-09-18T13-56-38.282478.parquet", "**/details_harness|hendrycksTest-world_religions|5_2023-09-18T13-56-38.282478.parquet"]}]}, {"config_name": "harness_hendrycksTest_abstract_algebra_5", "data_files": [{"split": "2023_09_18T13_56_38.282478", "path": ["**/details_harness|hendrycksTest-abstract_algebra|5_2023-09-18T13-56-38.282478.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-abstract_algebra|5_2023-09-18T13-56-38.282478.parquet"]}]}, {"config_name": "harness_hendrycksTest_anatomy_5", "data_files": [{"split": "2023_09_18T13_56_38.282478", "path": ["**/details_harness|hendrycksTest-anatomy|5_2023-09-18T13-56-38.282478.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-anatomy|5_2023-09-18T13-56-38.282478.parquet"]}]}, {"config_name": "harness_hendrycksTest_astronomy_5", "data_files": [{"split": "2023_09_18T13_56_38.282478", "path": ["**/details_harness|hendrycksTest-astronomy|5_2023-09-18T13-56-38.282478.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-astronomy|5_2023-09-18T13-56-38.282478.parquet"]}]}, {"config_name": "harness_hendrycksTest_business_ethics_5", "data_files": [{"split": "2023_09_18T13_56_38.282478", "path": ["**/details_harness|hendrycksTest-business_ethics|5_2023-09-18T13-56-38.282478.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-business_ethics|5_2023-09-18T13-56-38.282478.parquet"]}]}, {"config_name": "harness_hendrycksTest_clinical_knowledge_5", "data_files": [{"split": "2023_09_18T13_56_38.282478", "path": ["**/details_harness|hendrycksTest-clinical_knowledge|5_2023-09-18T13-56-38.282478.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-clinical_knowledge|5_2023-09-18T13-56-38.282478.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_biology_5", "data_files": [{"split": "2023_09_18T13_56_38.282478", "path": ["**/details_harness|hendrycksTest-college_biology|5_2023-09-18T13-56-38.282478.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_biology|5_2023-09-18T13-56-38.282478.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_chemistry_5", "data_files": [{"split": "2023_09_18T13_56_38.282478", "path": ["**/details_harness|hendrycksTest-college_chemistry|5_2023-09-18T13-56-38.282478.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_chemistry|5_2023-09-18T13-56-38.282478.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_computer_science_5", "data_files": [{"split": "2023_09_18T13_56_38.282478", "path": ["**/details_harness|hendrycksTest-college_computer_science|5_2023-09-18T13-56-38.282478.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_computer_science|5_2023-09-18T13-56-38.282478.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_mathematics_5", "data_files": [{"split": "2023_09_18T13_56_38.282478", "path": ["**/details_harness|hendrycksTest-college_mathematics|5_2023-09-18T13-56-38.282478.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_mathematics|5_2023-09-18T13-56-38.282478.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_medicine_5", "data_files": [{"split": "2023_09_18T13_56_38.282478", "path": ["**/details_harness|hendrycksTest-college_medicine|5_2023-09-18T13-56-38.282478.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_medicine|5_2023-09-18T13-56-38.282478.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_physics_5", "data_files": [{"split": "2023_09_18T13_56_38.282478", "path": ["**/details_harness|hendrycksTest-college_physics|5_2023-09-18T13-56-38.282478.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_physics|5_2023-09-18T13-56-38.282478.parquet"]}]}, {"config_name": "harness_hendrycksTest_computer_security_5", "data_files": [{"split": "2023_09_18T13_56_38.282478", "path": ["**/details_harness|hendrycksTest-computer_security|5_2023-09-18T13-56-38.282478.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-computer_security|5_2023-09-18T13-56-38.282478.parquet"]}]}, {"config_name": "harness_hendrycksTest_conceptual_physics_5", "data_files": [{"split": "2023_09_18T13_56_38.282478", "path": ["**/details_harness|hendrycksTest-conceptual_physics|5_2023-09-18T13-56-38.282478.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-conceptual_physics|5_2023-09-18T13-56-38.282478.parquet"]}]}, {"config_name": "harness_hendrycksTest_econometrics_5", "data_files": [{"split": "2023_09_18T13_56_38.282478", "path": ["**/details_harness|hendrycksTest-econometrics|5_2023-09-18T13-56-38.282478.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-econometrics|5_2023-09-18T13-56-38.282478.parquet"]}]}, {"config_name": "harness_hendrycksTest_electrical_engineering_5", "data_files": [{"split": "2023_09_18T13_56_38.282478", "path": ["**/details_harness|hendrycksTest-electrical_engineering|5_2023-09-18T13-56-38.282478.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-electrical_engineering|5_2023-09-18T13-56-38.282478.parquet"]}]}, {"config_name": "harness_hendrycksTest_elementary_mathematics_5", "data_files": [{"split": "2023_09_18T13_56_38.282478", "path": ["**/details_harness|hendrycksTest-elementary_mathematics|5_2023-09-18T13-56-38.282478.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-elementary_mathematics|5_2023-09-18T13-56-38.282478.parquet"]}]}, {"config_name": "harness_hendrycksTest_formal_logic_5", "data_files": [{"split": "2023_09_18T13_56_38.282478", "path": ["**/details_harness|hendrycksTest-formal_logic|5_2023-09-18T13-56-38.282478.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-formal_logic|5_2023-09-18T13-56-38.282478.parquet"]}]}, {"config_name": "harness_hendrycksTest_global_facts_5", "data_files": [{"split": "2023_09_18T13_56_38.282478", "path": ["**/details_harness|hendrycksTest-global_facts|5_2023-09-18T13-56-38.282478.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-global_facts|5_2023-09-18T13-56-38.282478.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_biology_5", "data_files": [{"split": "2023_09_18T13_56_38.282478", "path": ["**/details_harness|hendrycksTest-high_school_biology|5_2023-09-18T13-56-38.282478.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_biology|5_2023-09-18T13-56-38.282478.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_chemistry_5", "data_files": [{"split": "2023_09_18T13_56_38.282478", "path": ["**/details_harness|hendrycksTest-high_school_chemistry|5_2023-09-18T13-56-38.282478.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_chemistry|5_2023-09-18T13-56-38.282478.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_computer_science_5", "data_files": [{"split": "2023_09_18T13_56_38.282478", "path": ["**/details_harness|hendrycksTest-high_school_computer_science|5_2023-09-18T13-56-38.282478.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_computer_science|5_2023-09-18T13-56-38.282478.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_european_history_5", "data_files": [{"split": "2023_09_18T13_56_38.282478", "path": ["**/details_harness|hendrycksTest-high_school_european_history|5_2023-09-18T13-56-38.282478.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_european_history|5_2023-09-18T13-56-38.282478.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_geography_5", "data_files": [{"split": "2023_09_18T13_56_38.282478", "path": ["**/details_harness|hendrycksTest-high_school_geography|5_2023-09-18T13-56-38.282478.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_geography|5_2023-09-18T13-56-38.282478.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_government_and_politics_5", "data_files": [{"split": "2023_09_18T13_56_38.282478", "path": ["**/details_harness|hendrycksTest-high_school_government_and_politics|5_2023-09-18T13-56-38.282478.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_government_and_politics|5_2023-09-18T13-56-38.282478.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_macroeconomics_5", "data_files": [{"split": "2023_09_18T13_56_38.282478", "path": ["**/details_harness|hendrycksTest-high_school_macroeconomics|5_2023-09-18T13-56-38.282478.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_macroeconomics|5_2023-09-18T13-56-38.282478.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_mathematics_5", "data_files": [{"split": "2023_09_18T13_56_38.282478", "path": ["**/details_harness|hendrycksTest-high_school_mathematics|5_2023-09-18T13-56-38.282478.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_mathematics|5_2023-09-18T13-56-38.282478.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_microeconomics_5", "data_files": [{"split": "2023_09_18T13_56_38.282478", "path": ["**/details_harness|hendrycksTest-high_school_microeconomics|5_2023-09-18T13-56-38.282478.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_microeconomics|5_2023-09-18T13-56-38.282478.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_physics_5", "data_files": [{"split": "2023_09_18T13_56_38.282478", "path": ["**/details_harness|hendrycksTest-high_school_physics|5_2023-09-18T13-56-38.282478.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_physics|5_2023-09-18T13-56-38.282478.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_psychology_5", "data_files": [{"split": "2023_09_18T13_56_38.282478", "path": ["**/details_harness|hendrycksTest-high_school_psychology|5_2023-09-18T13-56-38.282478.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_psychology|5_2023-09-18T13-56-38.282478.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_statistics_5", "data_files": [{"split": "2023_09_18T13_56_38.282478", "path": ["**/details_harness|hendrycksTest-high_school_statistics|5_2023-09-18T13-56-38.282478.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_statistics|5_2023-09-18T13-56-38.282478.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_us_history_5", "data_files": [{"split": "2023_09_18T13_56_38.282478", "path": ["**/details_harness|hendrycksTest-high_school_us_history|5_2023-09-18T13-56-38.282478.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_us_history|5_2023-09-18T13-56-38.282478.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_world_history_5", "data_files": [{"split": "2023_09_18T13_56_38.282478", "path": ["**/details_harness|hendrycksTest-high_school_world_history|5_2023-09-18T13-56-38.282478.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_world_history|5_2023-09-18T13-56-38.282478.parquet"]}]}, {"config_name": "harness_hendrycksTest_human_aging_5", "data_files": [{"split": "2023_09_18T13_56_38.282478", "path": ["**/details_harness|hendrycksTest-human_aging|5_2023-09-18T13-56-38.282478.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-human_aging|5_2023-09-18T13-56-38.282478.parquet"]}]}, {"config_name": "harness_hendrycksTest_human_sexuality_5", "data_files": [{"split": "2023_09_18T13_56_38.282478", "path": ["**/details_harness|hendrycksTest-human_sexuality|5_2023-09-18T13-56-38.282478.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-human_sexuality|5_2023-09-18T13-56-38.282478.parquet"]}]}, {"config_name": "harness_hendrycksTest_international_law_5", "data_files": [{"split": "2023_09_18T13_56_38.282478", "path": ["**/details_harness|hendrycksTest-international_law|5_2023-09-18T13-56-38.282478.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-international_law|5_2023-09-18T13-56-38.282478.parquet"]}]}, {"config_name": "harness_hendrycksTest_jurisprudence_5", "data_files": [{"split": "2023_09_18T13_56_38.282478", "path": ["**/details_harness|hendrycksTest-jurisprudence|5_2023-09-18T13-56-38.282478.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-jurisprudence|5_2023-09-18T13-56-38.282478.parquet"]}]}, {"config_name": "harness_hendrycksTest_logical_fallacies_5", "data_files": [{"split": "2023_09_18T13_56_38.282478", "path": ["**/details_harness|hendrycksTest-logical_fallacies|5_2023-09-18T13-56-38.282478.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-logical_fallacies|5_2023-09-18T13-56-38.282478.parquet"]}]}, {"config_name": "harness_hendrycksTest_machine_learning_5", "data_files": [{"split": "2023_09_18T13_56_38.282478", "path": ["**/details_harness|hendrycksTest-machine_learning|5_2023-09-18T13-56-38.282478.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-machine_learning|5_2023-09-18T13-56-38.282478.parquet"]}]}, {"config_name": "harness_hendrycksTest_management_5", "data_files": [{"split": "2023_09_18T13_56_38.282478", "path": ["**/details_harness|hendrycksTest-management|5_2023-09-18T13-56-38.282478.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-management|5_2023-09-18T13-56-38.282478.parquet"]}]}, {"config_name": "harness_hendrycksTest_marketing_5", "data_files": [{"split": "2023_09_18T13_56_38.282478", "path": ["**/details_harness|hendrycksTest-marketing|5_2023-09-18T13-56-38.282478.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-marketing|5_2023-09-18T13-56-38.282478.parquet"]}]}, {"config_name": "harness_hendrycksTest_medical_genetics_5", "data_files": [{"split": "2023_09_18T13_56_38.282478", "path": ["**/details_harness|hendrycksTest-medical_genetics|5_2023-09-18T13-56-38.282478.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-medical_genetics|5_2023-09-18T13-56-38.282478.parquet"]}]}, {"config_name": "harness_hendrycksTest_miscellaneous_5", "data_files": [{"split": "2023_09_18T13_56_38.282478", "path": ["**/details_harness|hendrycksTest-miscellaneous|5_2023-09-18T13-56-38.282478.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-miscellaneous|5_2023-09-18T13-56-38.282478.parquet"]}]}, {"config_name": "harness_hendrycksTest_moral_disputes_5", "data_files": [{"split": "2023_09_18T13_56_38.282478", "path": ["**/details_harness|hendrycksTest-moral_disputes|5_2023-09-18T13-56-38.282478.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-moral_disputes|5_2023-09-18T13-56-38.282478.parquet"]}]}, {"config_name": "harness_hendrycksTest_moral_scenarios_5", "data_files": [{"split": "2023_09_18T13_56_38.282478", "path": ["**/details_harness|hendrycksTest-moral_scenarios|5_2023-09-18T13-56-38.282478.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-moral_scenarios|5_2023-09-18T13-56-38.282478.parquet"]}]}, {"config_name": "harness_hendrycksTest_nutrition_5", "data_files": [{"split": "2023_09_18T13_56_38.282478", "path": ["**/details_harness|hendrycksTest-nutrition|5_2023-09-18T13-56-38.282478.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-nutrition|5_2023-09-18T13-56-38.282478.parquet"]}]}, {"config_name": "harness_hendrycksTest_philosophy_5", "data_files": [{"split": "2023_09_18T13_56_38.282478", "path": ["**/details_harness|hendrycksTest-philosophy|5_2023-09-18T13-56-38.282478.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-philosophy|5_2023-09-18T13-56-38.282478.parquet"]}]}, {"config_name": "harness_hendrycksTest_prehistory_5", "data_files": [{"split": "2023_09_18T13_56_38.282478", "path": ["**/details_harness|hendrycksTest-prehistory|5_2023-09-18T13-56-38.282478.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-prehistory|5_2023-09-18T13-56-38.282478.parquet"]}]}, {"config_name": "harness_hendrycksTest_professional_accounting_5", "data_files": [{"split": "2023_09_18T13_56_38.282478", "path": ["**/details_harness|hendrycksTest-professional_accounting|5_2023-09-18T13-56-38.282478.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-professional_accounting|5_2023-09-18T13-56-38.282478.parquet"]}]}, {"config_name": "harness_hendrycksTest_professional_law_5", "data_files": [{"split": "2023_09_18T13_56_38.282478", "path": ["**/details_harness|hendrycksTest-professional_law|5_2023-09-18T13-56-38.282478.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-professional_law|5_2023-09-18T13-56-38.282478.parquet"]}]}, {"config_name": "harness_hendrycksTest_professional_medicine_5", "data_files": [{"split": "2023_09_18T13_56_38.282478", "path": ["**/details_harness|hendrycksTest-professional_medicine|5_2023-09-18T13-56-38.282478.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-professional_medicine|5_2023-09-18T13-56-38.282478.parquet"]}]}, {"config_name": "harness_hendrycksTest_professional_psychology_5", "data_files": [{"split": "2023_09_18T13_56_38.282478", "path": ["**/details_harness|hendrycksTest-professional_psychology|5_2023-09-18T13-56-38.282478.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-professional_psychology|5_2023-09-18T13-56-38.282478.parquet"]}]}, {"config_name": "harness_hendrycksTest_public_relations_5", "data_files": [{"split": "2023_09_18T13_56_38.282478", "path": ["**/details_harness|hendrycksTest-public_relations|5_2023-09-18T13-56-38.282478.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-public_relations|5_2023-09-18T13-56-38.282478.parquet"]}]}, {"config_name": "harness_hendrycksTest_security_studies_5", "data_files": [{"split": "2023_09_18T13_56_38.282478", "path": ["**/details_harness|hendrycksTest-security_studies|5_2023-09-18T13-56-38.282478.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-security_studies|5_2023-09-18T13-56-38.282478.parquet"]}]}, {"config_name": "harness_hendrycksTest_sociology_5", "data_files": [{"split": "2023_09_18T13_56_38.282478", "path": ["**/details_harness|hendrycksTest-sociology|5_2023-09-18T13-56-38.282478.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-sociology|5_2023-09-18T13-56-38.282478.parquet"]}]}, {"config_name": "harness_hendrycksTest_us_foreign_policy_5", "data_files": [{"split": "2023_09_18T13_56_38.282478", "path": ["**/details_harness|hendrycksTest-us_foreign_policy|5_2023-09-18T13-56-38.282478.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-us_foreign_policy|5_2023-09-18T13-56-38.282478.parquet"]}]}, {"config_name": "harness_hendrycksTest_virology_5", "data_files": [{"split": "2023_09_18T13_56_38.282478", "path": ["**/details_harness|hendrycksTest-virology|5_2023-09-18T13-56-38.282478.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-virology|5_2023-09-18T13-56-38.282478.parquet"]}]}, {"config_name": "harness_hendrycksTest_world_religions_5", "data_files": [{"split": "2023_09_18T13_56_38.282478", "path": ["**/details_harness|hendrycksTest-world_religions|5_2023-09-18T13-56-38.282478.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-world_religions|5_2023-09-18T13-56-38.282478.parquet"]}]}, {"config_name": "harness_truthfulqa_mc_0", "data_files": [{"split": "2023_09_18T13_56_38.282478", "path": ["**/details_harness|truthfulqa:mc|0_2023-09-18T13-56-38.282478.parquet"]}, {"split": "latest", "path": ["**/details_harness|truthfulqa:mc|0_2023-09-18T13-56-38.282478.parquet"]}]}, {"config_name": "harness_winogrande_5", "data_files": [{"split": "2023_10_28T22_12_00.775103", "path": ["**/details_harness|winogrande|5_2023-10-28T22-12-00.775103.parquet"]}, {"split": "latest", "path": ["**/details_harness|winogrande|5_2023-10-28T22-12-00.775103.parquet"]}]}, {"config_name": "results", "data_files": [{"split": "2023_09_18T13_56_38.282478", "path": ["results_2023-09-18T13-56-38.282478.parquet"]}, {"split": "2023_10_28T22_12_00.775103", "path": ["results_2023-10-28T22-12-00.775103.parquet"]}, {"split": "latest", "path": ["results_2023-10-28T22-12-00.775103.parquet"]}]}]}
|
2023-10-28T21:12:12+00:00
|
[] |
[] |
TAGS
#region-us
|
# Dataset Card for Evaluation run of Undi95/MLewdBoros-L2-13B
## Dataset Description
- Homepage:
- Repository: URL
- Paper:
- Leaderboard: URL
- Point of Contact: clementine@URL
### Dataset Summary
Dataset automatically created during the evaluation run of model Undi95/MLewdBoros-L2-13B on the Open LLM Leaderboard.
The dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.
The dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The "train" split is always pointing to the latest results.
An additional configuration "results" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the Open LLM Leaderboard).
To load the details from a run, you can for instance do the following:
## Latest results
These are the latest results from run 2023-10-28T22:12:00.775103(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the "latest" split for each eval):
### Supported Tasks and Leaderboards
### Languages
## Dataset Structure
### Data Instances
### Data Fields
### Data Splits
## Dataset Creation
### Curation Rationale
### Source Data
#### Initial Data Collection and Normalization
#### Who are the source language producers?
### Annotations
#### Annotation process
#### Who are the annotators?
### Personal and Sensitive Information
## Considerations for Using the Data
### Social Impact of Dataset
### Discussion of Biases
### Other Known Limitations
## Additional Information
### Dataset Curators
### Licensing Information
### Contributions
|
[
"# Dataset Card for Evaluation run of Undi95/MLewdBoros-L2-13B",
"## Dataset Description\n\n- Homepage: \n- Repository: URL\n- Paper: \n- Leaderboard: URL\n- Point of Contact: clementine@URL",
"### Dataset Summary\n\nDataset automatically created during the evaluation run of model Undi95/MLewdBoros-L2-13B on the Open LLM Leaderboard.\n\nThe dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.\n\nThe dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The \"train\" split is always pointing to the latest results.\n\nAn additional configuration \"results\" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the Open LLM Leaderboard).\n\nTo load the details from a run, you can for instance do the following:",
"## Latest results\n\nThese are the latest results from run 2023-10-28T22:12:00.775103(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the \"latest\" split for each eval):",
"### Supported Tasks and Leaderboards",
"### Languages",
"## Dataset Structure",
"### Data Instances",
"### Data Fields",
"### Data Splits",
"## Dataset Creation",
"### Curation Rationale",
"### Source Data",
"#### Initial Data Collection and Normalization",
"#### Who are the source language producers?",
"### Annotations",
"#### Annotation process",
"#### Who are the annotators?",
"### Personal and Sensitive Information",
"## Considerations for Using the Data",
"### Social Impact of Dataset",
"### Discussion of Biases",
"### Other Known Limitations",
"## Additional Information",
"### Dataset Curators",
"### Licensing Information",
"### Contributions"
] |
[
"TAGS\n#region-us \n",
"# Dataset Card for Evaluation run of Undi95/MLewdBoros-L2-13B",
"## Dataset Description\n\n- Homepage: \n- Repository: URL\n- Paper: \n- Leaderboard: URL\n- Point of Contact: clementine@URL",
"### Dataset Summary\n\nDataset automatically created during the evaluation run of model Undi95/MLewdBoros-L2-13B on the Open LLM Leaderboard.\n\nThe dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.\n\nThe dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The \"train\" split is always pointing to the latest results.\n\nAn additional configuration \"results\" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the Open LLM Leaderboard).\n\nTo load the details from a run, you can for instance do the following:",
"## Latest results\n\nThese are the latest results from run 2023-10-28T22:12:00.775103(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the \"latest\" split for each eval):",
"### Supported Tasks and Leaderboards",
"### Languages",
"## Dataset Structure",
"### Data Instances",
"### Data Fields",
"### Data Splits",
"## Dataset Creation",
"### Curation Rationale",
"### Source Data",
"#### Initial Data Collection and Normalization",
"#### Who are the source language producers?",
"### Annotations",
"#### Annotation process",
"#### Who are the annotators?",
"### Personal and Sensitive Information",
"## Considerations for Using the Data",
"### Social Impact of Dataset",
"### Discussion of Biases",
"### Other Known Limitations",
"## Additional Information",
"### Dataset Curators",
"### Licensing Information",
"### Contributions"
] |
[
6,
23,
31,
171,
66,
10,
4,
6,
6,
5,
5,
5,
7,
4,
10,
10,
5,
5,
9,
8,
8,
7,
8,
7,
5,
6,
6,
5
] |
[
"passage: TAGS\n#region-us \n# Dataset Card for Evaluation run of Undi95/MLewdBoros-L2-13B## Dataset Description\n\n- Homepage: \n- Repository: URL\n- Paper: \n- Leaderboard: URL\n- Point of Contact: clementine@URL### Dataset Summary\n\nDataset automatically created during the evaluation run of model Undi95/MLewdBoros-L2-13B on the Open LLM Leaderboard.\n\nThe dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.\n\nThe dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The \"train\" split is always pointing to the latest results.\n\nAn additional configuration \"results\" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the Open LLM Leaderboard).\n\nTo load the details from a run, you can for instance do the following:## Latest results\n\nThese are the latest results from run 2023-10-28T22:12:00.775103(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the \"latest\" split for each eval):### Supported Tasks and Leaderboards### Languages## Dataset Structure### Data Instances### Data Fields### Data Splits## Dataset Creation### Curation Rationale### Source Data#### Initial Data Collection and Normalization#### Who are the source language producers?### Annotations#### Annotation process#### Who are the annotators?### Personal and Sensitive Information## Considerations for Using the Data### Social Impact of Dataset### Discussion of Biases### Other Known Limitations## Additional Information### Dataset Curators### Licensing Information### Contributions"
] |
27216281f0cb4c481968766647b9c58ab12fb8ca
|
# Dataset Card for Evaluation run of Undi95/ReMM-v2-L2-13B
## Dataset Description
- **Homepage:**
- **Repository:** https://huggingface.co/Undi95/ReMM-v2-L2-13B
- **Paper:**
- **Leaderboard:** https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard
- **Point of Contact:** [email protected]
### Dataset Summary
Dataset automatically created during the evaluation run of model [Undi95/ReMM-v2-L2-13B](https://huggingface.co/Undi95/ReMM-v2-L2-13B) on the [Open LLM Leaderboard](https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard).
The dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.
The dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The "train" split is always pointing to the latest results.
An additional configuration "results" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the [Open LLM Leaderboard](https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard)).
To load the details from a run, you can for instance do the following:
```python
from datasets import load_dataset
data = load_dataset("open-llm-leaderboard/details_Undi95__ReMM-v2-L2-13B",
"harness_winogrande_5",
split="train")
```
## Latest results
These are the [latest results from run 2023-10-24T07:00:18.944945](https://huggingface.co/datasets/open-llm-leaderboard/details_Undi95__ReMM-v2-L2-13B/blob/main/results_2023-10-24T07-00-18.944945.json)(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the "latest" split for each eval):
```python
{
"all": {
"em": 0.056312919463087245,
"em_stderr": 0.0023607917437880183,
"f1": 0.12075503355704631,
"f1_stderr": 0.002645290783284543,
"acc": 0.4452013645505283,
"acc_stderr": 0.010675124517934693
},
"harness|drop|3": {
"em": 0.056312919463087245,
"em_stderr": 0.0023607917437880183,
"f1": 0.12075503355704631,
"f1_stderr": 0.002645290783284543
},
"harness|gsm8k|5": {
"acc": 0.13191811978771797,
"acc_stderr": 0.009321265253857515
},
"harness|winogrande|5": {
"acc": 0.7584846093133386,
"acc_stderr": 0.012028983782011872
}
}
```
### Supported Tasks and Leaderboards
[More Information Needed]
### Languages
[More Information Needed]
## Dataset Structure
### Data Instances
[More Information Needed]
### Data Fields
[More Information Needed]
### Data Splits
[More Information Needed]
## Dataset Creation
### Curation Rationale
[More Information Needed]
### Source Data
#### Initial Data Collection and Normalization
[More Information Needed]
#### Who are the source language producers?
[More Information Needed]
### Annotations
#### Annotation process
[More Information Needed]
#### Who are the annotators?
[More Information Needed]
### Personal and Sensitive Information
[More Information Needed]
## Considerations for Using the Data
### Social Impact of Dataset
[More Information Needed]
### Discussion of Biases
[More Information Needed]
### Other Known Limitations
[More Information Needed]
## Additional Information
### Dataset Curators
[More Information Needed]
### Licensing Information
[More Information Needed]
### Citation Information
[More Information Needed]
### Contributions
[More Information Needed]
|
open-llm-leaderboard/details_Undi95__ReMM-v2-L2-13B
|
[
"region:us"
] |
2023-09-18T12:59:09+00:00
|
{"pretty_name": "Evaluation run of Undi95/ReMM-v2-L2-13B", "dataset_summary": "Dataset automatically created during the evaluation run of model [Undi95/ReMM-v2-L2-13B](https://huggingface.co/Undi95/ReMM-v2-L2-13B) on the [Open LLM Leaderboard](https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard).\n\nThe dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.\n\nThe dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The \"train\" split is always pointing to the latest results.\n\nAn additional configuration \"results\" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the [Open LLM Leaderboard](https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard)).\n\nTo load the details from a run, you can for instance do the following:\n```python\nfrom datasets import load_dataset\ndata = load_dataset(\"open-llm-leaderboard/details_Undi95__ReMM-v2-L2-13B\",\n\t\"harness_winogrande_5\",\n\tsplit=\"train\")\n```\n\n## Latest results\n\nThese are the [latest results from run 2023-10-24T07:00:18.944945](https://huggingface.co/datasets/open-llm-leaderboard/details_Undi95__ReMM-v2-L2-13B/blob/main/results_2023-10-24T07-00-18.944945.json)(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the \"latest\" split for each eval):\n\n```python\n{\n \"all\": {\n \"em\": 0.056312919463087245,\n \"em_stderr\": 0.0023607917437880183,\n \"f1\": 0.12075503355704631,\n \"f1_stderr\": 0.002645290783284543,\n \"acc\": 0.4452013645505283,\n \"acc_stderr\": 0.010675124517934693\n },\n \"harness|drop|3\": {\n \"em\": 0.056312919463087245,\n \"em_stderr\": 0.0023607917437880183,\n \"f1\": 0.12075503355704631,\n \"f1_stderr\": 0.002645290783284543\n },\n \"harness|gsm8k|5\": {\n \"acc\": 0.13191811978771797,\n \"acc_stderr\": 0.009321265253857515\n },\n \"harness|winogrande|5\": {\n \"acc\": 0.7584846093133386,\n \"acc_stderr\": 0.012028983782011872\n }\n}\n```", "repo_url": "https://huggingface.co/Undi95/ReMM-v2-L2-13B", "leaderboard_url": "https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard", "point_of_contact": "[email protected]", "configs": [{"config_name": "harness_arc_challenge_25", "data_files": [{"split": "2023_09_18T13_58_45.934639", "path": ["**/details_harness|arc:challenge|25_2023-09-18T13-58-45.934639.parquet"]}, {"split": "latest", "path": ["**/details_harness|arc:challenge|25_2023-09-18T13-58-45.934639.parquet"]}]}, {"config_name": "harness_drop_3", "data_files": [{"split": "2023_10_24T07_00_18.944945", "path": ["**/details_harness|drop|3_2023-10-24T07-00-18.944945.parquet"]}, {"split": "latest", "path": ["**/details_harness|drop|3_2023-10-24T07-00-18.944945.parquet"]}]}, {"config_name": "harness_gsm8k_5", "data_files": [{"split": "2023_10_24T07_00_18.944945", "path": ["**/details_harness|gsm8k|5_2023-10-24T07-00-18.944945.parquet"]}, {"split": "latest", "path": ["**/details_harness|gsm8k|5_2023-10-24T07-00-18.944945.parquet"]}]}, {"config_name": "harness_hellaswag_10", "data_files": [{"split": "2023_09_18T13_58_45.934639", "path": ["**/details_harness|hellaswag|10_2023-09-18T13-58-45.934639.parquet"]}, {"split": "latest", "path": ["**/details_harness|hellaswag|10_2023-09-18T13-58-45.934639.parquet"]}]}, {"config_name": "harness_hendrycksTest_5", "data_files": [{"split": "2023_09_18T13_58_45.934639", "path": ["**/details_harness|hendrycksTest-abstract_algebra|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-anatomy|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-astronomy|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-business_ethics|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-clinical_knowledge|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-college_biology|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-college_chemistry|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-college_computer_science|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-college_mathematics|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-college_medicine|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-college_physics|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-computer_security|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-conceptual_physics|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-econometrics|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-electrical_engineering|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-elementary_mathematics|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-formal_logic|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-global_facts|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-high_school_biology|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-high_school_chemistry|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-high_school_computer_science|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-high_school_european_history|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-high_school_geography|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-high_school_government_and_politics|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-high_school_macroeconomics|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-high_school_mathematics|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-high_school_microeconomics|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-high_school_physics|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-high_school_psychology|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-high_school_statistics|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-high_school_us_history|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-high_school_world_history|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-human_aging|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-human_sexuality|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-international_law|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-jurisprudence|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-logical_fallacies|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-machine_learning|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-management|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-marketing|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-medical_genetics|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-miscellaneous|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-moral_disputes|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-moral_scenarios|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-nutrition|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-philosophy|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-prehistory|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-professional_accounting|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-professional_law|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-professional_medicine|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-professional_psychology|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-public_relations|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-security_studies|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-sociology|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-us_foreign_policy|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-virology|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-world_religions|5_2023-09-18T13-58-45.934639.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-abstract_algebra|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-anatomy|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-astronomy|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-business_ethics|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-clinical_knowledge|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-college_biology|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-college_chemistry|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-college_computer_science|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-college_mathematics|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-college_medicine|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-college_physics|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-computer_security|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-conceptual_physics|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-econometrics|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-electrical_engineering|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-elementary_mathematics|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-formal_logic|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-global_facts|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-high_school_biology|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-high_school_chemistry|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-high_school_computer_science|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-high_school_european_history|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-high_school_geography|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-high_school_government_and_politics|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-high_school_macroeconomics|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-high_school_mathematics|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-high_school_microeconomics|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-high_school_physics|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-high_school_psychology|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-high_school_statistics|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-high_school_us_history|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-high_school_world_history|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-human_aging|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-human_sexuality|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-international_law|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-jurisprudence|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-logical_fallacies|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-machine_learning|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-management|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-marketing|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-medical_genetics|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-miscellaneous|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-moral_disputes|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-moral_scenarios|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-nutrition|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-philosophy|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-prehistory|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-professional_accounting|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-professional_law|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-professional_medicine|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-professional_psychology|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-public_relations|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-security_studies|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-sociology|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-us_foreign_policy|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-virology|5_2023-09-18T13-58-45.934639.parquet", "**/details_harness|hendrycksTest-world_religions|5_2023-09-18T13-58-45.934639.parquet"]}]}, {"config_name": "harness_hendrycksTest_abstract_algebra_5", "data_files": [{"split": "2023_09_18T13_58_45.934639", "path": ["**/details_harness|hendrycksTest-abstract_algebra|5_2023-09-18T13-58-45.934639.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-abstract_algebra|5_2023-09-18T13-58-45.934639.parquet"]}]}, {"config_name": "harness_hendrycksTest_anatomy_5", "data_files": [{"split": "2023_09_18T13_58_45.934639", "path": ["**/details_harness|hendrycksTest-anatomy|5_2023-09-18T13-58-45.934639.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-anatomy|5_2023-09-18T13-58-45.934639.parquet"]}]}, {"config_name": "harness_hendrycksTest_astronomy_5", "data_files": [{"split": "2023_09_18T13_58_45.934639", "path": ["**/details_harness|hendrycksTest-astronomy|5_2023-09-18T13-58-45.934639.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-astronomy|5_2023-09-18T13-58-45.934639.parquet"]}]}, {"config_name": "harness_hendrycksTest_business_ethics_5", "data_files": [{"split": "2023_09_18T13_58_45.934639", "path": ["**/details_harness|hendrycksTest-business_ethics|5_2023-09-18T13-58-45.934639.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-business_ethics|5_2023-09-18T13-58-45.934639.parquet"]}]}, {"config_name": "harness_hendrycksTest_clinical_knowledge_5", "data_files": [{"split": "2023_09_18T13_58_45.934639", "path": ["**/details_harness|hendrycksTest-clinical_knowledge|5_2023-09-18T13-58-45.934639.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-clinical_knowledge|5_2023-09-18T13-58-45.934639.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_biology_5", "data_files": [{"split": "2023_09_18T13_58_45.934639", "path": ["**/details_harness|hendrycksTest-college_biology|5_2023-09-18T13-58-45.934639.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_biology|5_2023-09-18T13-58-45.934639.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_chemistry_5", "data_files": [{"split": "2023_09_18T13_58_45.934639", "path": ["**/details_harness|hendrycksTest-college_chemistry|5_2023-09-18T13-58-45.934639.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_chemistry|5_2023-09-18T13-58-45.934639.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_computer_science_5", "data_files": [{"split": "2023_09_18T13_58_45.934639", "path": ["**/details_harness|hendrycksTest-college_computer_science|5_2023-09-18T13-58-45.934639.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_computer_science|5_2023-09-18T13-58-45.934639.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_mathematics_5", "data_files": [{"split": "2023_09_18T13_58_45.934639", "path": ["**/details_harness|hendrycksTest-college_mathematics|5_2023-09-18T13-58-45.934639.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_mathematics|5_2023-09-18T13-58-45.934639.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_medicine_5", "data_files": [{"split": "2023_09_18T13_58_45.934639", "path": ["**/details_harness|hendrycksTest-college_medicine|5_2023-09-18T13-58-45.934639.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_medicine|5_2023-09-18T13-58-45.934639.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_physics_5", "data_files": [{"split": "2023_09_18T13_58_45.934639", "path": ["**/details_harness|hendrycksTest-college_physics|5_2023-09-18T13-58-45.934639.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_physics|5_2023-09-18T13-58-45.934639.parquet"]}]}, {"config_name": "harness_hendrycksTest_computer_security_5", "data_files": [{"split": "2023_09_18T13_58_45.934639", "path": ["**/details_harness|hendrycksTest-computer_security|5_2023-09-18T13-58-45.934639.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-computer_security|5_2023-09-18T13-58-45.934639.parquet"]}]}, {"config_name": "harness_hendrycksTest_conceptual_physics_5", "data_files": [{"split": "2023_09_18T13_58_45.934639", "path": ["**/details_harness|hendrycksTest-conceptual_physics|5_2023-09-18T13-58-45.934639.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-conceptual_physics|5_2023-09-18T13-58-45.934639.parquet"]}]}, {"config_name": "harness_hendrycksTest_econometrics_5", "data_files": [{"split": "2023_09_18T13_58_45.934639", "path": ["**/details_harness|hendrycksTest-econometrics|5_2023-09-18T13-58-45.934639.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-econometrics|5_2023-09-18T13-58-45.934639.parquet"]}]}, {"config_name": "harness_hendrycksTest_electrical_engineering_5", "data_files": [{"split": "2023_09_18T13_58_45.934639", "path": ["**/details_harness|hendrycksTest-electrical_engineering|5_2023-09-18T13-58-45.934639.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-electrical_engineering|5_2023-09-18T13-58-45.934639.parquet"]}]}, {"config_name": "harness_hendrycksTest_elementary_mathematics_5", "data_files": [{"split": "2023_09_18T13_58_45.934639", "path": ["**/details_harness|hendrycksTest-elementary_mathematics|5_2023-09-18T13-58-45.934639.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-elementary_mathematics|5_2023-09-18T13-58-45.934639.parquet"]}]}, {"config_name": "harness_hendrycksTest_formal_logic_5", "data_files": [{"split": "2023_09_18T13_58_45.934639", "path": ["**/details_harness|hendrycksTest-formal_logic|5_2023-09-18T13-58-45.934639.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-formal_logic|5_2023-09-18T13-58-45.934639.parquet"]}]}, {"config_name": "harness_hendrycksTest_global_facts_5", "data_files": [{"split": "2023_09_18T13_58_45.934639", "path": ["**/details_harness|hendrycksTest-global_facts|5_2023-09-18T13-58-45.934639.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-global_facts|5_2023-09-18T13-58-45.934639.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_biology_5", "data_files": [{"split": "2023_09_18T13_58_45.934639", "path": ["**/details_harness|hendrycksTest-high_school_biology|5_2023-09-18T13-58-45.934639.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_biology|5_2023-09-18T13-58-45.934639.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_chemistry_5", "data_files": [{"split": "2023_09_18T13_58_45.934639", "path": ["**/details_harness|hendrycksTest-high_school_chemistry|5_2023-09-18T13-58-45.934639.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_chemistry|5_2023-09-18T13-58-45.934639.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_computer_science_5", "data_files": [{"split": "2023_09_18T13_58_45.934639", "path": ["**/details_harness|hendrycksTest-high_school_computer_science|5_2023-09-18T13-58-45.934639.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_computer_science|5_2023-09-18T13-58-45.934639.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_european_history_5", "data_files": [{"split": "2023_09_18T13_58_45.934639", "path": ["**/details_harness|hendrycksTest-high_school_european_history|5_2023-09-18T13-58-45.934639.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_european_history|5_2023-09-18T13-58-45.934639.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_geography_5", "data_files": [{"split": "2023_09_18T13_58_45.934639", "path": ["**/details_harness|hendrycksTest-high_school_geography|5_2023-09-18T13-58-45.934639.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_geography|5_2023-09-18T13-58-45.934639.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_government_and_politics_5", "data_files": [{"split": "2023_09_18T13_58_45.934639", "path": ["**/details_harness|hendrycksTest-high_school_government_and_politics|5_2023-09-18T13-58-45.934639.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_government_and_politics|5_2023-09-18T13-58-45.934639.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_macroeconomics_5", "data_files": [{"split": "2023_09_18T13_58_45.934639", "path": ["**/details_harness|hendrycksTest-high_school_macroeconomics|5_2023-09-18T13-58-45.934639.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_macroeconomics|5_2023-09-18T13-58-45.934639.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_mathematics_5", "data_files": [{"split": "2023_09_18T13_58_45.934639", "path": ["**/details_harness|hendrycksTest-high_school_mathematics|5_2023-09-18T13-58-45.934639.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_mathematics|5_2023-09-18T13-58-45.934639.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_microeconomics_5", "data_files": [{"split": "2023_09_18T13_58_45.934639", "path": ["**/details_harness|hendrycksTest-high_school_microeconomics|5_2023-09-18T13-58-45.934639.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_microeconomics|5_2023-09-18T13-58-45.934639.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_physics_5", "data_files": [{"split": "2023_09_18T13_58_45.934639", "path": ["**/details_harness|hendrycksTest-high_school_physics|5_2023-09-18T13-58-45.934639.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_physics|5_2023-09-18T13-58-45.934639.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_psychology_5", "data_files": [{"split": "2023_09_18T13_58_45.934639", "path": ["**/details_harness|hendrycksTest-high_school_psychology|5_2023-09-18T13-58-45.934639.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_psychology|5_2023-09-18T13-58-45.934639.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_statistics_5", "data_files": [{"split": "2023_09_18T13_58_45.934639", "path": ["**/details_harness|hendrycksTest-high_school_statistics|5_2023-09-18T13-58-45.934639.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_statistics|5_2023-09-18T13-58-45.934639.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_us_history_5", "data_files": [{"split": "2023_09_18T13_58_45.934639", "path": ["**/details_harness|hendrycksTest-high_school_us_history|5_2023-09-18T13-58-45.934639.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_us_history|5_2023-09-18T13-58-45.934639.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_world_history_5", "data_files": [{"split": "2023_09_18T13_58_45.934639", "path": ["**/details_harness|hendrycksTest-high_school_world_history|5_2023-09-18T13-58-45.934639.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_world_history|5_2023-09-18T13-58-45.934639.parquet"]}]}, {"config_name": "harness_hendrycksTest_human_aging_5", "data_files": [{"split": "2023_09_18T13_58_45.934639", "path": ["**/details_harness|hendrycksTest-human_aging|5_2023-09-18T13-58-45.934639.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-human_aging|5_2023-09-18T13-58-45.934639.parquet"]}]}, {"config_name": "harness_hendrycksTest_human_sexuality_5", "data_files": [{"split": "2023_09_18T13_58_45.934639", "path": ["**/details_harness|hendrycksTest-human_sexuality|5_2023-09-18T13-58-45.934639.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-human_sexuality|5_2023-09-18T13-58-45.934639.parquet"]}]}, {"config_name": "harness_hendrycksTest_international_law_5", "data_files": [{"split": "2023_09_18T13_58_45.934639", "path": ["**/details_harness|hendrycksTest-international_law|5_2023-09-18T13-58-45.934639.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-international_law|5_2023-09-18T13-58-45.934639.parquet"]}]}, {"config_name": "harness_hendrycksTest_jurisprudence_5", "data_files": [{"split": "2023_09_18T13_58_45.934639", "path": ["**/details_harness|hendrycksTest-jurisprudence|5_2023-09-18T13-58-45.934639.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-jurisprudence|5_2023-09-18T13-58-45.934639.parquet"]}]}, {"config_name": "harness_hendrycksTest_logical_fallacies_5", "data_files": [{"split": "2023_09_18T13_58_45.934639", "path": ["**/details_harness|hendrycksTest-logical_fallacies|5_2023-09-18T13-58-45.934639.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-logical_fallacies|5_2023-09-18T13-58-45.934639.parquet"]}]}, {"config_name": "harness_hendrycksTest_machine_learning_5", "data_files": [{"split": "2023_09_18T13_58_45.934639", "path": ["**/details_harness|hendrycksTest-machine_learning|5_2023-09-18T13-58-45.934639.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-machine_learning|5_2023-09-18T13-58-45.934639.parquet"]}]}, {"config_name": "harness_hendrycksTest_management_5", "data_files": [{"split": "2023_09_18T13_58_45.934639", "path": ["**/details_harness|hendrycksTest-management|5_2023-09-18T13-58-45.934639.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-management|5_2023-09-18T13-58-45.934639.parquet"]}]}, {"config_name": "harness_hendrycksTest_marketing_5", "data_files": [{"split": "2023_09_18T13_58_45.934639", "path": ["**/details_harness|hendrycksTest-marketing|5_2023-09-18T13-58-45.934639.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-marketing|5_2023-09-18T13-58-45.934639.parquet"]}]}, {"config_name": "harness_hendrycksTest_medical_genetics_5", "data_files": [{"split": "2023_09_18T13_58_45.934639", "path": ["**/details_harness|hendrycksTest-medical_genetics|5_2023-09-18T13-58-45.934639.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-medical_genetics|5_2023-09-18T13-58-45.934639.parquet"]}]}, {"config_name": "harness_hendrycksTest_miscellaneous_5", "data_files": [{"split": "2023_09_18T13_58_45.934639", "path": ["**/details_harness|hendrycksTest-miscellaneous|5_2023-09-18T13-58-45.934639.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-miscellaneous|5_2023-09-18T13-58-45.934639.parquet"]}]}, {"config_name": "harness_hendrycksTest_moral_disputes_5", "data_files": [{"split": "2023_09_18T13_58_45.934639", "path": ["**/details_harness|hendrycksTest-moral_disputes|5_2023-09-18T13-58-45.934639.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-moral_disputes|5_2023-09-18T13-58-45.934639.parquet"]}]}, {"config_name": "harness_hendrycksTest_moral_scenarios_5", "data_files": [{"split": "2023_09_18T13_58_45.934639", "path": ["**/details_harness|hendrycksTest-moral_scenarios|5_2023-09-18T13-58-45.934639.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-moral_scenarios|5_2023-09-18T13-58-45.934639.parquet"]}]}, {"config_name": "harness_hendrycksTest_nutrition_5", "data_files": [{"split": "2023_09_18T13_58_45.934639", "path": ["**/details_harness|hendrycksTest-nutrition|5_2023-09-18T13-58-45.934639.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-nutrition|5_2023-09-18T13-58-45.934639.parquet"]}]}, {"config_name": "harness_hendrycksTest_philosophy_5", "data_files": [{"split": "2023_09_18T13_58_45.934639", "path": ["**/details_harness|hendrycksTest-philosophy|5_2023-09-18T13-58-45.934639.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-philosophy|5_2023-09-18T13-58-45.934639.parquet"]}]}, {"config_name": "harness_hendrycksTest_prehistory_5", "data_files": [{"split": "2023_09_18T13_58_45.934639", "path": ["**/details_harness|hendrycksTest-prehistory|5_2023-09-18T13-58-45.934639.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-prehistory|5_2023-09-18T13-58-45.934639.parquet"]}]}, {"config_name": "harness_hendrycksTest_professional_accounting_5", "data_files": [{"split": "2023_09_18T13_58_45.934639", "path": ["**/details_harness|hendrycksTest-professional_accounting|5_2023-09-18T13-58-45.934639.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-professional_accounting|5_2023-09-18T13-58-45.934639.parquet"]}]}, {"config_name": "harness_hendrycksTest_professional_law_5", "data_files": [{"split": "2023_09_18T13_58_45.934639", "path": ["**/details_harness|hendrycksTest-professional_law|5_2023-09-18T13-58-45.934639.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-professional_law|5_2023-09-18T13-58-45.934639.parquet"]}]}, {"config_name": "harness_hendrycksTest_professional_medicine_5", "data_files": [{"split": "2023_09_18T13_58_45.934639", "path": ["**/details_harness|hendrycksTest-professional_medicine|5_2023-09-18T13-58-45.934639.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-professional_medicine|5_2023-09-18T13-58-45.934639.parquet"]}]}, {"config_name": "harness_hendrycksTest_professional_psychology_5", "data_files": [{"split": "2023_09_18T13_58_45.934639", "path": ["**/details_harness|hendrycksTest-professional_psychology|5_2023-09-18T13-58-45.934639.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-professional_psychology|5_2023-09-18T13-58-45.934639.parquet"]}]}, {"config_name": "harness_hendrycksTest_public_relations_5", "data_files": [{"split": "2023_09_18T13_58_45.934639", "path": ["**/details_harness|hendrycksTest-public_relations|5_2023-09-18T13-58-45.934639.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-public_relations|5_2023-09-18T13-58-45.934639.parquet"]}]}, {"config_name": "harness_hendrycksTest_security_studies_5", "data_files": [{"split": "2023_09_18T13_58_45.934639", "path": ["**/details_harness|hendrycksTest-security_studies|5_2023-09-18T13-58-45.934639.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-security_studies|5_2023-09-18T13-58-45.934639.parquet"]}]}, {"config_name": "harness_hendrycksTest_sociology_5", "data_files": [{"split": "2023_09_18T13_58_45.934639", "path": ["**/details_harness|hendrycksTest-sociology|5_2023-09-18T13-58-45.934639.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-sociology|5_2023-09-18T13-58-45.934639.parquet"]}]}, {"config_name": "harness_hendrycksTest_us_foreign_policy_5", "data_files": [{"split": "2023_09_18T13_58_45.934639", "path": ["**/details_harness|hendrycksTest-us_foreign_policy|5_2023-09-18T13-58-45.934639.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-us_foreign_policy|5_2023-09-18T13-58-45.934639.parquet"]}]}, {"config_name": "harness_hendrycksTest_virology_5", "data_files": [{"split": "2023_09_18T13_58_45.934639", "path": ["**/details_harness|hendrycksTest-virology|5_2023-09-18T13-58-45.934639.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-virology|5_2023-09-18T13-58-45.934639.parquet"]}]}, {"config_name": "harness_hendrycksTest_world_religions_5", "data_files": [{"split": "2023_09_18T13_58_45.934639", "path": ["**/details_harness|hendrycksTest-world_religions|5_2023-09-18T13-58-45.934639.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-world_religions|5_2023-09-18T13-58-45.934639.parquet"]}]}, {"config_name": "harness_truthfulqa_mc_0", "data_files": [{"split": "2023_09_18T13_58_45.934639", "path": ["**/details_harness|truthfulqa:mc|0_2023-09-18T13-58-45.934639.parquet"]}, {"split": "latest", "path": ["**/details_harness|truthfulqa:mc|0_2023-09-18T13-58-45.934639.parquet"]}]}, {"config_name": "harness_winogrande_5", "data_files": [{"split": "2023_10_24T07_00_18.944945", "path": ["**/details_harness|winogrande|5_2023-10-24T07-00-18.944945.parquet"]}, {"split": "latest", "path": ["**/details_harness|winogrande|5_2023-10-24T07-00-18.944945.parquet"]}]}, {"config_name": "results", "data_files": [{"split": "2023_09_18T13_58_45.934639", "path": ["results_2023-09-18T13-58-45.934639.parquet"]}, {"split": "2023_10_24T07_00_18.944945", "path": ["results_2023-10-24T07-00-18.944945.parquet"]}, {"split": "latest", "path": ["results_2023-10-24T07-00-18.944945.parquet"]}]}]}
|
2023-10-24T06:00:31+00:00
|
[] |
[] |
TAGS
#region-us
|
# Dataset Card for Evaluation run of Undi95/ReMM-v2-L2-13B
## Dataset Description
- Homepage:
- Repository: URL
- Paper:
- Leaderboard: URL
- Point of Contact: clementine@URL
### Dataset Summary
Dataset automatically created during the evaluation run of model Undi95/ReMM-v2-L2-13B on the Open LLM Leaderboard.
The dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.
The dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The "train" split is always pointing to the latest results.
An additional configuration "results" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the Open LLM Leaderboard).
To load the details from a run, you can for instance do the following:
## Latest results
These are the latest results from run 2023-10-24T07:00:18.944945(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the "latest" split for each eval):
### Supported Tasks and Leaderboards
### Languages
## Dataset Structure
### Data Instances
### Data Fields
### Data Splits
## Dataset Creation
### Curation Rationale
### Source Data
#### Initial Data Collection and Normalization
#### Who are the source language producers?
### Annotations
#### Annotation process
#### Who are the annotators?
### Personal and Sensitive Information
## Considerations for Using the Data
### Social Impact of Dataset
### Discussion of Biases
### Other Known Limitations
## Additional Information
### Dataset Curators
### Licensing Information
### Contributions
|
[
"# Dataset Card for Evaluation run of Undi95/ReMM-v2-L2-13B",
"## Dataset Description\n\n- Homepage: \n- Repository: URL\n- Paper: \n- Leaderboard: URL\n- Point of Contact: clementine@URL",
"### Dataset Summary\n\nDataset automatically created during the evaluation run of model Undi95/ReMM-v2-L2-13B on the Open LLM Leaderboard.\n\nThe dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.\n\nThe dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The \"train\" split is always pointing to the latest results.\n\nAn additional configuration \"results\" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the Open LLM Leaderboard).\n\nTo load the details from a run, you can for instance do the following:",
"## Latest results\n\nThese are the latest results from run 2023-10-24T07:00:18.944945(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the \"latest\" split for each eval):",
"### Supported Tasks and Leaderboards",
"### Languages",
"## Dataset Structure",
"### Data Instances",
"### Data Fields",
"### Data Splits",
"## Dataset Creation",
"### Curation Rationale",
"### Source Data",
"#### Initial Data Collection and Normalization",
"#### Who are the source language producers?",
"### Annotations",
"#### Annotation process",
"#### Who are the annotators?",
"### Personal and Sensitive Information",
"## Considerations for Using the Data",
"### Social Impact of Dataset",
"### Discussion of Biases",
"### Other Known Limitations",
"## Additional Information",
"### Dataset Curators",
"### Licensing Information",
"### Contributions"
] |
[
"TAGS\n#region-us \n",
"# Dataset Card for Evaluation run of Undi95/ReMM-v2-L2-13B",
"## Dataset Description\n\n- Homepage: \n- Repository: URL\n- Paper: \n- Leaderboard: URL\n- Point of Contact: clementine@URL",
"### Dataset Summary\n\nDataset automatically created during the evaluation run of model Undi95/ReMM-v2-L2-13B on the Open LLM Leaderboard.\n\nThe dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.\n\nThe dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The \"train\" split is always pointing to the latest results.\n\nAn additional configuration \"results\" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the Open LLM Leaderboard).\n\nTo load the details from a run, you can for instance do the following:",
"## Latest results\n\nThese are the latest results from run 2023-10-24T07:00:18.944945(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the \"latest\" split for each eval):",
"### Supported Tasks and Leaderboards",
"### Languages",
"## Dataset Structure",
"### Data Instances",
"### Data Fields",
"### Data Splits",
"## Dataset Creation",
"### Curation Rationale",
"### Source Data",
"#### Initial Data Collection and Normalization",
"#### Who are the source language producers?",
"### Annotations",
"#### Annotation process",
"#### Who are the annotators?",
"### Personal and Sensitive Information",
"## Considerations for Using the Data",
"### Social Impact of Dataset",
"### Discussion of Biases",
"### Other Known Limitations",
"## Additional Information",
"### Dataset Curators",
"### Licensing Information",
"### Contributions"
] |
[
6,
22,
31,
170,
67,
10,
4,
6,
6,
5,
5,
5,
7,
4,
10,
10,
5,
5,
9,
8,
8,
7,
8,
7,
5,
6,
6,
5
] |
[
"passage: TAGS\n#region-us \n# Dataset Card for Evaluation run of Undi95/ReMM-v2-L2-13B## Dataset Description\n\n- Homepage: \n- Repository: URL\n- Paper: \n- Leaderboard: URL\n- Point of Contact: clementine@URL### Dataset Summary\n\nDataset automatically created during the evaluation run of model Undi95/ReMM-v2-L2-13B on the Open LLM Leaderboard.\n\nThe dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.\n\nThe dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The \"train\" split is always pointing to the latest results.\n\nAn additional configuration \"results\" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the Open LLM Leaderboard).\n\nTo load the details from a run, you can for instance do the following:## Latest results\n\nThese are the latest results from run 2023-10-24T07:00:18.944945(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the \"latest\" split for each eval):### Supported Tasks and Leaderboards### Languages## Dataset Structure### Data Instances### Data Fields### Data Splits## Dataset Creation### Curation Rationale### Source Data#### Initial Data Collection and Normalization#### Who are the source language producers?### Annotations#### Annotation process#### Who are the annotators?### Personal and Sensitive Information## Considerations for Using the Data### Social Impact of Dataset### Discussion of Biases### Other Known Limitations## Additional Information### Dataset Curators### Licensing Information### Contributions"
] |
bbc3a7b808ac2444c97fc4d0e14548b23baa3a5f
|
# Dataset Card for Evaluation run of Doctor-Shotgun/CalliopeDS-L2-13B
## Dataset Description
- **Homepage:**
- **Repository:** https://huggingface.co/Doctor-Shotgun/CalliopeDS-L2-13B
- **Paper:**
- **Leaderboard:** https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard
- **Point of Contact:** [email protected]
### Dataset Summary
Dataset automatically created during the evaluation run of model [Doctor-Shotgun/CalliopeDS-L2-13B](https://huggingface.co/Doctor-Shotgun/CalliopeDS-L2-13B) on the [Open LLM Leaderboard](https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard).
The dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.
The dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The "train" split is always pointing to the latest results.
An additional configuration "results" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the [Open LLM Leaderboard](https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard)).
To load the details from a run, you can for instance do the following:
```python
from datasets import load_dataset
data = load_dataset("open-llm-leaderboard/details_Doctor-Shotgun__CalliopeDS-L2-13B",
"harness_winogrande_5",
split="train")
```
## Latest results
These are the [latest results from run 2023-10-26T04:36:21.549191](https://huggingface.co/datasets/open-llm-leaderboard/details_Doctor-Shotgun__CalliopeDS-L2-13B/blob/main/results_2023-10-26T04-36-21.549191.json)(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the "latest" split for each eval):
```python
{
"all": {
"em": 0.02307046979865772,
"em_stderr": 0.0015374446489046481,
"f1": 0.08979446308724821,
"f1_stderr": 0.0020360011017500185,
"acc": 0.4351997070321265,
"acc_stderr": 0.010043960065261932
},
"harness|drop|3": {
"em": 0.02307046979865772,
"em_stderr": 0.0015374446489046481,
"f1": 0.08979446308724821,
"f1_stderr": 0.0020360011017500185
},
"harness|gsm8k|5": {
"acc": 0.10007581501137225,
"acc_stderr": 0.008266274528685632
},
"harness|winogrande|5": {
"acc": 0.7703235990528808,
"acc_stderr": 0.011821645601838232
}
}
```
### Supported Tasks and Leaderboards
[More Information Needed]
### Languages
[More Information Needed]
## Dataset Structure
### Data Instances
[More Information Needed]
### Data Fields
[More Information Needed]
### Data Splits
[More Information Needed]
## Dataset Creation
### Curation Rationale
[More Information Needed]
### Source Data
#### Initial Data Collection and Normalization
[More Information Needed]
#### Who are the source language producers?
[More Information Needed]
### Annotations
#### Annotation process
[More Information Needed]
#### Who are the annotators?
[More Information Needed]
### Personal and Sensitive Information
[More Information Needed]
## Considerations for Using the Data
### Social Impact of Dataset
[More Information Needed]
### Discussion of Biases
[More Information Needed]
### Other Known Limitations
[More Information Needed]
## Additional Information
### Dataset Curators
[More Information Needed]
### Licensing Information
[More Information Needed]
### Citation Information
[More Information Needed]
### Contributions
[More Information Needed]
|
open-llm-leaderboard/details_Doctor-Shotgun__CalliopeDS-L2-13B
|
[
"region:us"
] |
2023-09-18T13:01:15+00:00
|
{"pretty_name": "Evaluation run of Doctor-Shotgun/CalliopeDS-L2-13B", "dataset_summary": "Dataset automatically created during the evaluation run of model [Doctor-Shotgun/CalliopeDS-L2-13B](https://huggingface.co/Doctor-Shotgun/CalliopeDS-L2-13B) on the [Open LLM Leaderboard](https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard).\n\nThe dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.\n\nThe dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The \"train\" split is always pointing to the latest results.\n\nAn additional configuration \"results\" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the [Open LLM Leaderboard](https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard)).\n\nTo load the details from a run, you can for instance do the following:\n```python\nfrom datasets import load_dataset\ndata = load_dataset(\"open-llm-leaderboard/details_Doctor-Shotgun__CalliopeDS-L2-13B\",\n\t\"harness_winogrande_5\",\n\tsplit=\"train\")\n```\n\n## Latest results\n\nThese are the [latest results from run 2023-10-26T04:36:21.549191](https://huggingface.co/datasets/open-llm-leaderboard/details_Doctor-Shotgun__CalliopeDS-L2-13B/blob/main/results_2023-10-26T04-36-21.549191.json)(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the \"latest\" split for each eval):\n\n```python\n{\n \"all\": {\n \"em\": 0.02307046979865772,\n \"em_stderr\": 0.0015374446489046481,\n \"f1\": 0.08979446308724821,\n \"f1_stderr\": 0.0020360011017500185,\n \"acc\": 0.4351997070321265,\n \"acc_stderr\": 0.010043960065261932\n },\n \"harness|drop|3\": {\n \"em\": 0.02307046979865772,\n \"em_stderr\": 0.0015374446489046481,\n \"f1\": 0.08979446308724821,\n \"f1_stderr\": 0.0020360011017500185\n },\n \"harness|gsm8k|5\": {\n \"acc\": 0.10007581501137225,\n \"acc_stderr\": 0.008266274528685632\n },\n \"harness|winogrande|5\": {\n \"acc\": 0.7703235990528808,\n \"acc_stderr\": 0.011821645601838232\n }\n}\n```", "repo_url": "https://huggingface.co/Doctor-Shotgun/CalliopeDS-L2-13B", "leaderboard_url": "https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard", "point_of_contact": "[email protected]", "configs": [{"config_name": "harness_arc_challenge_25", "data_files": [{"split": "2023_09_18T14_00_51.912601", "path": ["**/details_harness|arc:challenge|25_2023-09-18T14-00-51.912601.parquet"]}, {"split": "latest", "path": ["**/details_harness|arc:challenge|25_2023-09-18T14-00-51.912601.parquet"]}]}, {"config_name": "harness_drop_3", "data_files": [{"split": "2023_10_26T04_36_21.549191", "path": ["**/details_harness|drop|3_2023-10-26T04-36-21.549191.parquet"]}, {"split": "latest", "path": ["**/details_harness|drop|3_2023-10-26T04-36-21.549191.parquet"]}]}, {"config_name": "harness_gsm8k_5", "data_files": [{"split": "2023_10_26T04_36_21.549191", "path": ["**/details_harness|gsm8k|5_2023-10-26T04-36-21.549191.parquet"]}, {"split": "latest", "path": ["**/details_harness|gsm8k|5_2023-10-26T04-36-21.549191.parquet"]}]}, {"config_name": "harness_hellaswag_10", "data_files": [{"split": "2023_09_18T14_00_51.912601", "path": ["**/details_harness|hellaswag|10_2023-09-18T14-00-51.912601.parquet"]}, {"split": "latest", "path": ["**/details_harness|hellaswag|10_2023-09-18T14-00-51.912601.parquet"]}]}, {"config_name": "harness_hendrycksTest_5", "data_files": [{"split": "2023_09_18T14_00_51.912601", "path": ["**/details_harness|hendrycksTest-abstract_algebra|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-anatomy|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-astronomy|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-business_ethics|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-clinical_knowledge|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-college_biology|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-college_chemistry|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-college_computer_science|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-college_mathematics|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-college_medicine|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-college_physics|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-computer_security|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-conceptual_physics|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-econometrics|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-electrical_engineering|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-elementary_mathematics|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-formal_logic|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-global_facts|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-high_school_biology|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-high_school_chemistry|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-high_school_computer_science|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-high_school_european_history|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-high_school_geography|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-high_school_government_and_politics|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-high_school_macroeconomics|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-high_school_mathematics|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-high_school_microeconomics|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-high_school_physics|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-high_school_psychology|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-high_school_statistics|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-high_school_us_history|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-high_school_world_history|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-human_aging|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-human_sexuality|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-international_law|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-jurisprudence|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-logical_fallacies|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-machine_learning|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-management|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-marketing|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-medical_genetics|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-miscellaneous|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-moral_disputes|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-moral_scenarios|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-nutrition|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-philosophy|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-prehistory|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-professional_accounting|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-professional_law|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-professional_medicine|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-professional_psychology|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-public_relations|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-security_studies|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-sociology|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-us_foreign_policy|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-virology|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-world_religions|5_2023-09-18T14-00-51.912601.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-abstract_algebra|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-anatomy|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-astronomy|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-business_ethics|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-clinical_knowledge|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-college_biology|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-college_chemistry|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-college_computer_science|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-college_mathematics|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-college_medicine|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-college_physics|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-computer_security|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-conceptual_physics|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-econometrics|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-electrical_engineering|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-elementary_mathematics|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-formal_logic|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-global_facts|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-high_school_biology|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-high_school_chemistry|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-high_school_computer_science|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-high_school_european_history|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-high_school_geography|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-high_school_government_and_politics|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-high_school_macroeconomics|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-high_school_mathematics|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-high_school_microeconomics|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-high_school_physics|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-high_school_psychology|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-high_school_statistics|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-high_school_us_history|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-high_school_world_history|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-human_aging|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-human_sexuality|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-international_law|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-jurisprudence|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-logical_fallacies|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-machine_learning|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-management|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-marketing|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-medical_genetics|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-miscellaneous|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-moral_disputes|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-moral_scenarios|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-nutrition|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-philosophy|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-prehistory|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-professional_accounting|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-professional_law|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-professional_medicine|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-professional_psychology|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-public_relations|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-security_studies|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-sociology|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-us_foreign_policy|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-virology|5_2023-09-18T14-00-51.912601.parquet", "**/details_harness|hendrycksTest-world_religions|5_2023-09-18T14-00-51.912601.parquet"]}]}, {"config_name": "harness_hendrycksTest_abstract_algebra_5", "data_files": [{"split": "2023_09_18T14_00_51.912601", "path": ["**/details_harness|hendrycksTest-abstract_algebra|5_2023-09-18T14-00-51.912601.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-abstract_algebra|5_2023-09-18T14-00-51.912601.parquet"]}]}, {"config_name": "harness_hendrycksTest_anatomy_5", "data_files": [{"split": "2023_09_18T14_00_51.912601", "path": ["**/details_harness|hendrycksTest-anatomy|5_2023-09-18T14-00-51.912601.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-anatomy|5_2023-09-18T14-00-51.912601.parquet"]}]}, {"config_name": "harness_hendrycksTest_astronomy_5", "data_files": [{"split": "2023_09_18T14_00_51.912601", "path": ["**/details_harness|hendrycksTest-astronomy|5_2023-09-18T14-00-51.912601.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-astronomy|5_2023-09-18T14-00-51.912601.parquet"]}]}, {"config_name": "harness_hendrycksTest_business_ethics_5", "data_files": [{"split": "2023_09_18T14_00_51.912601", "path": ["**/details_harness|hendrycksTest-business_ethics|5_2023-09-18T14-00-51.912601.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-business_ethics|5_2023-09-18T14-00-51.912601.parquet"]}]}, {"config_name": "harness_hendrycksTest_clinical_knowledge_5", "data_files": [{"split": "2023_09_18T14_00_51.912601", "path": ["**/details_harness|hendrycksTest-clinical_knowledge|5_2023-09-18T14-00-51.912601.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-clinical_knowledge|5_2023-09-18T14-00-51.912601.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_biology_5", "data_files": [{"split": "2023_09_18T14_00_51.912601", "path": ["**/details_harness|hendrycksTest-college_biology|5_2023-09-18T14-00-51.912601.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_biology|5_2023-09-18T14-00-51.912601.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_chemistry_5", "data_files": [{"split": "2023_09_18T14_00_51.912601", "path": ["**/details_harness|hendrycksTest-college_chemistry|5_2023-09-18T14-00-51.912601.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_chemistry|5_2023-09-18T14-00-51.912601.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_computer_science_5", "data_files": [{"split": "2023_09_18T14_00_51.912601", "path": ["**/details_harness|hendrycksTest-college_computer_science|5_2023-09-18T14-00-51.912601.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_computer_science|5_2023-09-18T14-00-51.912601.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_mathematics_5", "data_files": [{"split": "2023_09_18T14_00_51.912601", "path": ["**/details_harness|hendrycksTest-college_mathematics|5_2023-09-18T14-00-51.912601.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_mathematics|5_2023-09-18T14-00-51.912601.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_medicine_5", "data_files": [{"split": "2023_09_18T14_00_51.912601", "path": ["**/details_harness|hendrycksTest-college_medicine|5_2023-09-18T14-00-51.912601.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_medicine|5_2023-09-18T14-00-51.912601.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_physics_5", "data_files": [{"split": "2023_09_18T14_00_51.912601", "path": ["**/details_harness|hendrycksTest-college_physics|5_2023-09-18T14-00-51.912601.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_physics|5_2023-09-18T14-00-51.912601.parquet"]}]}, {"config_name": "harness_hendrycksTest_computer_security_5", "data_files": [{"split": "2023_09_18T14_00_51.912601", "path": ["**/details_harness|hendrycksTest-computer_security|5_2023-09-18T14-00-51.912601.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-computer_security|5_2023-09-18T14-00-51.912601.parquet"]}]}, {"config_name": "harness_hendrycksTest_conceptual_physics_5", "data_files": [{"split": "2023_09_18T14_00_51.912601", "path": ["**/details_harness|hendrycksTest-conceptual_physics|5_2023-09-18T14-00-51.912601.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-conceptual_physics|5_2023-09-18T14-00-51.912601.parquet"]}]}, {"config_name": "harness_hendrycksTest_econometrics_5", "data_files": [{"split": "2023_09_18T14_00_51.912601", "path": ["**/details_harness|hendrycksTest-econometrics|5_2023-09-18T14-00-51.912601.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-econometrics|5_2023-09-18T14-00-51.912601.parquet"]}]}, {"config_name": "harness_hendrycksTest_electrical_engineering_5", "data_files": [{"split": "2023_09_18T14_00_51.912601", "path": ["**/details_harness|hendrycksTest-electrical_engineering|5_2023-09-18T14-00-51.912601.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-electrical_engineering|5_2023-09-18T14-00-51.912601.parquet"]}]}, {"config_name": "harness_hendrycksTest_elementary_mathematics_5", "data_files": [{"split": "2023_09_18T14_00_51.912601", "path": ["**/details_harness|hendrycksTest-elementary_mathematics|5_2023-09-18T14-00-51.912601.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-elementary_mathematics|5_2023-09-18T14-00-51.912601.parquet"]}]}, {"config_name": "harness_hendrycksTest_formal_logic_5", "data_files": [{"split": "2023_09_18T14_00_51.912601", "path": ["**/details_harness|hendrycksTest-formal_logic|5_2023-09-18T14-00-51.912601.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-formal_logic|5_2023-09-18T14-00-51.912601.parquet"]}]}, {"config_name": "harness_hendrycksTest_global_facts_5", "data_files": [{"split": "2023_09_18T14_00_51.912601", "path": ["**/details_harness|hendrycksTest-global_facts|5_2023-09-18T14-00-51.912601.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-global_facts|5_2023-09-18T14-00-51.912601.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_biology_5", "data_files": [{"split": "2023_09_18T14_00_51.912601", "path": ["**/details_harness|hendrycksTest-high_school_biology|5_2023-09-18T14-00-51.912601.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_biology|5_2023-09-18T14-00-51.912601.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_chemistry_5", "data_files": [{"split": "2023_09_18T14_00_51.912601", "path": ["**/details_harness|hendrycksTest-high_school_chemistry|5_2023-09-18T14-00-51.912601.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_chemistry|5_2023-09-18T14-00-51.912601.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_computer_science_5", "data_files": [{"split": "2023_09_18T14_00_51.912601", "path": ["**/details_harness|hendrycksTest-high_school_computer_science|5_2023-09-18T14-00-51.912601.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_computer_science|5_2023-09-18T14-00-51.912601.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_european_history_5", "data_files": [{"split": "2023_09_18T14_00_51.912601", "path": ["**/details_harness|hendrycksTest-high_school_european_history|5_2023-09-18T14-00-51.912601.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_european_history|5_2023-09-18T14-00-51.912601.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_geography_5", "data_files": [{"split": "2023_09_18T14_00_51.912601", "path": ["**/details_harness|hendrycksTest-high_school_geography|5_2023-09-18T14-00-51.912601.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_geography|5_2023-09-18T14-00-51.912601.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_government_and_politics_5", "data_files": [{"split": "2023_09_18T14_00_51.912601", "path": ["**/details_harness|hendrycksTest-high_school_government_and_politics|5_2023-09-18T14-00-51.912601.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_government_and_politics|5_2023-09-18T14-00-51.912601.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_macroeconomics_5", "data_files": [{"split": "2023_09_18T14_00_51.912601", "path": ["**/details_harness|hendrycksTest-high_school_macroeconomics|5_2023-09-18T14-00-51.912601.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_macroeconomics|5_2023-09-18T14-00-51.912601.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_mathematics_5", "data_files": [{"split": "2023_09_18T14_00_51.912601", "path": ["**/details_harness|hendrycksTest-high_school_mathematics|5_2023-09-18T14-00-51.912601.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_mathematics|5_2023-09-18T14-00-51.912601.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_microeconomics_5", "data_files": [{"split": "2023_09_18T14_00_51.912601", "path": ["**/details_harness|hendrycksTest-high_school_microeconomics|5_2023-09-18T14-00-51.912601.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_microeconomics|5_2023-09-18T14-00-51.912601.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_physics_5", "data_files": [{"split": "2023_09_18T14_00_51.912601", "path": ["**/details_harness|hendrycksTest-high_school_physics|5_2023-09-18T14-00-51.912601.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_physics|5_2023-09-18T14-00-51.912601.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_psychology_5", "data_files": [{"split": "2023_09_18T14_00_51.912601", "path": ["**/details_harness|hendrycksTest-high_school_psychology|5_2023-09-18T14-00-51.912601.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_psychology|5_2023-09-18T14-00-51.912601.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_statistics_5", "data_files": [{"split": "2023_09_18T14_00_51.912601", "path": ["**/details_harness|hendrycksTest-high_school_statistics|5_2023-09-18T14-00-51.912601.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_statistics|5_2023-09-18T14-00-51.912601.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_us_history_5", "data_files": [{"split": "2023_09_18T14_00_51.912601", "path": ["**/details_harness|hendrycksTest-high_school_us_history|5_2023-09-18T14-00-51.912601.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_us_history|5_2023-09-18T14-00-51.912601.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_world_history_5", "data_files": [{"split": "2023_09_18T14_00_51.912601", "path": ["**/details_harness|hendrycksTest-high_school_world_history|5_2023-09-18T14-00-51.912601.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_world_history|5_2023-09-18T14-00-51.912601.parquet"]}]}, {"config_name": "harness_hendrycksTest_human_aging_5", "data_files": [{"split": "2023_09_18T14_00_51.912601", "path": ["**/details_harness|hendrycksTest-human_aging|5_2023-09-18T14-00-51.912601.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-human_aging|5_2023-09-18T14-00-51.912601.parquet"]}]}, {"config_name": "harness_hendrycksTest_human_sexuality_5", "data_files": [{"split": "2023_09_18T14_00_51.912601", "path": ["**/details_harness|hendrycksTest-human_sexuality|5_2023-09-18T14-00-51.912601.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-human_sexuality|5_2023-09-18T14-00-51.912601.parquet"]}]}, {"config_name": "harness_hendrycksTest_international_law_5", "data_files": [{"split": "2023_09_18T14_00_51.912601", "path": ["**/details_harness|hendrycksTest-international_law|5_2023-09-18T14-00-51.912601.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-international_law|5_2023-09-18T14-00-51.912601.parquet"]}]}, {"config_name": "harness_hendrycksTest_jurisprudence_5", "data_files": [{"split": "2023_09_18T14_00_51.912601", "path": ["**/details_harness|hendrycksTest-jurisprudence|5_2023-09-18T14-00-51.912601.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-jurisprudence|5_2023-09-18T14-00-51.912601.parquet"]}]}, {"config_name": "harness_hendrycksTest_logical_fallacies_5", "data_files": [{"split": "2023_09_18T14_00_51.912601", "path": ["**/details_harness|hendrycksTest-logical_fallacies|5_2023-09-18T14-00-51.912601.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-logical_fallacies|5_2023-09-18T14-00-51.912601.parquet"]}]}, {"config_name": "harness_hendrycksTest_machine_learning_5", "data_files": [{"split": "2023_09_18T14_00_51.912601", "path": ["**/details_harness|hendrycksTest-machine_learning|5_2023-09-18T14-00-51.912601.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-machine_learning|5_2023-09-18T14-00-51.912601.parquet"]}]}, {"config_name": "harness_hendrycksTest_management_5", "data_files": [{"split": "2023_09_18T14_00_51.912601", "path": ["**/details_harness|hendrycksTest-management|5_2023-09-18T14-00-51.912601.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-management|5_2023-09-18T14-00-51.912601.parquet"]}]}, {"config_name": "harness_hendrycksTest_marketing_5", "data_files": [{"split": "2023_09_18T14_00_51.912601", "path": ["**/details_harness|hendrycksTest-marketing|5_2023-09-18T14-00-51.912601.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-marketing|5_2023-09-18T14-00-51.912601.parquet"]}]}, {"config_name": "harness_hendrycksTest_medical_genetics_5", "data_files": [{"split": "2023_09_18T14_00_51.912601", "path": ["**/details_harness|hendrycksTest-medical_genetics|5_2023-09-18T14-00-51.912601.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-medical_genetics|5_2023-09-18T14-00-51.912601.parquet"]}]}, {"config_name": "harness_hendrycksTest_miscellaneous_5", "data_files": [{"split": "2023_09_18T14_00_51.912601", "path": ["**/details_harness|hendrycksTest-miscellaneous|5_2023-09-18T14-00-51.912601.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-miscellaneous|5_2023-09-18T14-00-51.912601.parquet"]}]}, {"config_name": "harness_hendrycksTest_moral_disputes_5", "data_files": [{"split": "2023_09_18T14_00_51.912601", "path": ["**/details_harness|hendrycksTest-moral_disputes|5_2023-09-18T14-00-51.912601.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-moral_disputes|5_2023-09-18T14-00-51.912601.parquet"]}]}, {"config_name": "harness_hendrycksTest_moral_scenarios_5", "data_files": [{"split": "2023_09_18T14_00_51.912601", "path": ["**/details_harness|hendrycksTest-moral_scenarios|5_2023-09-18T14-00-51.912601.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-moral_scenarios|5_2023-09-18T14-00-51.912601.parquet"]}]}, {"config_name": "harness_hendrycksTest_nutrition_5", "data_files": [{"split": "2023_09_18T14_00_51.912601", "path": ["**/details_harness|hendrycksTest-nutrition|5_2023-09-18T14-00-51.912601.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-nutrition|5_2023-09-18T14-00-51.912601.parquet"]}]}, {"config_name": "harness_hendrycksTest_philosophy_5", "data_files": [{"split": "2023_09_18T14_00_51.912601", "path": ["**/details_harness|hendrycksTest-philosophy|5_2023-09-18T14-00-51.912601.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-philosophy|5_2023-09-18T14-00-51.912601.parquet"]}]}, {"config_name": "harness_hendrycksTest_prehistory_5", "data_files": [{"split": "2023_09_18T14_00_51.912601", "path": ["**/details_harness|hendrycksTest-prehistory|5_2023-09-18T14-00-51.912601.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-prehistory|5_2023-09-18T14-00-51.912601.parquet"]}]}, {"config_name": "harness_hendrycksTest_professional_accounting_5", "data_files": [{"split": "2023_09_18T14_00_51.912601", "path": ["**/details_harness|hendrycksTest-professional_accounting|5_2023-09-18T14-00-51.912601.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-professional_accounting|5_2023-09-18T14-00-51.912601.parquet"]}]}, {"config_name": "harness_hendrycksTest_professional_law_5", "data_files": [{"split": "2023_09_18T14_00_51.912601", "path": ["**/details_harness|hendrycksTest-professional_law|5_2023-09-18T14-00-51.912601.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-professional_law|5_2023-09-18T14-00-51.912601.parquet"]}]}, {"config_name": "harness_hendrycksTest_professional_medicine_5", "data_files": [{"split": "2023_09_18T14_00_51.912601", "path": ["**/details_harness|hendrycksTest-professional_medicine|5_2023-09-18T14-00-51.912601.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-professional_medicine|5_2023-09-18T14-00-51.912601.parquet"]}]}, {"config_name": "harness_hendrycksTest_professional_psychology_5", "data_files": [{"split": "2023_09_18T14_00_51.912601", "path": ["**/details_harness|hendrycksTest-professional_psychology|5_2023-09-18T14-00-51.912601.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-professional_psychology|5_2023-09-18T14-00-51.912601.parquet"]}]}, {"config_name": "harness_hendrycksTest_public_relations_5", "data_files": [{"split": "2023_09_18T14_00_51.912601", "path": ["**/details_harness|hendrycksTest-public_relations|5_2023-09-18T14-00-51.912601.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-public_relations|5_2023-09-18T14-00-51.912601.parquet"]}]}, {"config_name": "harness_hendrycksTest_security_studies_5", "data_files": [{"split": "2023_09_18T14_00_51.912601", "path": ["**/details_harness|hendrycksTest-security_studies|5_2023-09-18T14-00-51.912601.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-security_studies|5_2023-09-18T14-00-51.912601.parquet"]}]}, {"config_name": "harness_hendrycksTest_sociology_5", "data_files": [{"split": "2023_09_18T14_00_51.912601", "path": ["**/details_harness|hendrycksTest-sociology|5_2023-09-18T14-00-51.912601.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-sociology|5_2023-09-18T14-00-51.912601.parquet"]}]}, {"config_name": "harness_hendrycksTest_us_foreign_policy_5", "data_files": [{"split": "2023_09_18T14_00_51.912601", "path": ["**/details_harness|hendrycksTest-us_foreign_policy|5_2023-09-18T14-00-51.912601.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-us_foreign_policy|5_2023-09-18T14-00-51.912601.parquet"]}]}, {"config_name": "harness_hendrycksTest_virology_5", "data_files": [{"split": "2023_09_18T14_00_51.912601", "path": ["**/details_harness|hendrycksTest-virology|5_2023-09-18T14-00-51.912601.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-virology|5_2023-09-18T14-00-51.912601.parquet"]}]}, {"config_name": "harness_hendrycksTest_world_religions_5", "data_files": [{"split": "2023_09_18T14_00_51.912601", "path": ["**/details_harness|hendrycksTest-world_religions|5_2023-09-18T14-00-51.912601.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-world_religions|5_2023-09-18T14-00-51.912601.parquet"]}]}, {"config_name": "harness_truthfulqa_mc_0", "data_files": [{"split": "2023_09_18T14_00_51.912601", "path": ["**/details_harness|truthfulqa:mc|0_2023-09-18T14-00-51.912601.parquet"]}, {"split": "latest", "path": ["**/details_harness|truthfulqa:mc|0_2023-09-18T14-00-51.912601.parquet"]}]}, {"config_name": "harness_winogrande_5", "data_files": [{"split": "2023_10_26T04_36_21.549191", "path": ["**/details_harness|winogrande|5_2023-10-26T04-36-21.549191.parquet"]}, {"split": "latest", "path": ["**/details_harness|winogrande|5_2023-10-26T04-36-21.549191.parquet"]}]}, {"config_name": "results", "data_files": [{"split": "2023_09_18T14_00_51.912601", "path": ["results_2023-09-18T14-00-51.912601.parquet"]}, {"split": "2023_10_26T04_36_21.549191", "path": ["results_2023-10-26T04-36-21.549191.parquet"]}, {"split": "latest", "path": ["results_2023-10-26T04-36-21.549191.parquet"]}]}]}
|
2023-10-26T03:36:34+00:00
|
[] |
[] |
TAGS
#region-us
|
# Dataset Card for Evaluation run of Doctor-Shotgun/CalliopeDS-L2-13B
## Dataset Description
- Homepage:
- Repository: URL
- Paper:
- Leaderboard: URL
- Point of Contact: clementine@URL
### Dataset Summary
Dataset automatically created during the evaluation run of model Doctor-Shotgun/CalliopeDS-L2-13B on the Open LLM Leaderboard.
The dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.
The dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The "train" split is always pointing to the latest results.
An additional configuration "results" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the Open LLM Leaderboard).
To load the details from a run, you can for instance do the following:
## Latest results
These are the latest results from run 2023-10-26T04:36:21.549191(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the "latest" split for each eval):
### Supported Tasks and Leaderboards
### Languages
## Dataset Structure
### Data Instances
### Data Fields
### Data Splits
## Dataset Creation
### Curation Rationale
### Source Data
#### Initial Data Collection and Normalization
#### Who are the source language producers?
### Annotations
#### Annotation process
#### Who are the annotators?
### Personal and Sensitive Information
## Considerations for Using the Data
### Social Impact of Dataset
### Discussion of Biases
### Other Known Limitations
## Additional Information
### Dataset Curators
### Licensing Information
### Contributions
|
[
"# Dataset Card for Evaluation run of Doctor-Shotgun/CalliopeDS-L2-13B",
"## Dataset Description\n\n- Homepage: \n- Repository: URL\n- Paper: \n- Leaderboard: URL\n- Point of Contact: clementine@URL",
"### Dataset Summary\n\nDataset automatically created during the evaluation run of model Doctor-Shotgun/CalliopeDS-L2-13B on the Open LLM Leaderboard.\n\nThe dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.\n\nThe dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The \"train\" split is always pointing to the latest results.\n\nAn additional configuration \"results\" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the Open LLM Leaderboard).\n\nTo load the details from a run, you can for instance do the following:",
"## Latest results\n\nThese are the latest results from run 2023-10-26T04:36:21.549191(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the \"latest\" split for each eval):",
"### Supported Tasks and Leaderboards",
"### Languages",
"## Dataset Structure",
"### Data Instances",
"### Data Fields",
"### Data Splits",
"## Dataset Creation",
"### Curation Rationale",
"### Source Data",
"#### Initial Data Collection and Normalization",
"#### Who are the source language producers?",
"### Annotations",
"#### Annotation process",
"#### Who are the annotators?",
"### Personal and Sensitive Information",
"## Considerations for Using the Data",
"### Social Impact of Dataset",
"### Discussion of Biases",
"### Other Known Limitations",
"## Additional Information",
"### Dataset Curators",
"### Licensing Information",
"### Contributions"
] |
[
"TAGS\n#region-us \n",
"# Dataset Card for Evaluation run of Doctor-Shotgun/CalliopeDS-L2-13B",
"## Dataset Description\n\n- Homepage: \n- Repository: URL\n- Paper: \n- Leaderboard: URL\n- Point of Contact: clementine@URL",
"### Dataset Summary\n\nDataset automatically created during the evaluation run of model Doctor-Shotgun/CalliopeDS-L2-13B on the Open LLM Leaderboard.\n\nThe dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.\n\nThe dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The \"train\" split is always pointing to the latest results.\n\nAn additional configuration \"results\" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the Open LLM Leaderboard).\n\nTo load the details from a run, you can for instance do the following:",
"## Latest results\n\nThese are the latest results from run 2023-10-26T04:36:21.549191(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the \"latest\" split for each eval):",
"### Supported Tasks and Leaderboards",
"### Languages",
"## Dataset Structure",
"### Data Instances",
"### Data Fields",
"### Data Splits",
"## Dataset Creation",
"### Curation Rationale",
"### Source Data",
"#### Initial Data Collection and Normalization",
"#### Who are the source language producers?",
"### Annotations",
"#### Annotation process",
"#### Who are the annotators?",
"### Personal and Sensitive Information",
"## Considerations for Using the Data",
"### Social Impact of Dataset",
"### Discussion of Biases",
"### Other Known Limitations",
"## Additional Information",
"### Dataset Curators",
"### Licensing Information",
"### Contributions"
] |
[
6,
24,
31,
172,
67,
10,
4,
6,
6,
5,
5,
5,
7,
4,
10,
10,
5,
5,
9,
8,
8,
7,
8,
7,
5,
6,
6,
5
] |
[
"passage: TAGS\n#region-us \n# Dataset Card for Evaluation run of Doctor-Shotgun/CalliopeDS-L2-13B## Dataset Description\n\n- Homepage: \n- Repository: URL\n- Paper: \n- Leaderboard: URL\n- Point of Contact: clementine@URL### Dataset Summary\n\nDataset automatically created during the evaluation run of model Doctor-Shotgun/CalliopeDS-L2-13B on the Open LLM Leaderboard.\n\nThe dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.\n\nThe dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The \"train\" split is always pointing to the latest results.\n\nAn additional configuration \"results\" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the Open LLM Leaderboard).\n\nTo load the details from a run, you can for instance do the following:## Latest results\n\nThese are the latest results from run 2023-10-26T04:36:21.549191(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the \"latest\" split for each eval):### Supported Tasks and Leaderboards### Languages## Dataset Structure### Data Instances### Data Fields### Data Splits## Dataset Creation### Curation Rationale### Source Data#### Initial Data Collection and Normalization#### Who are the source language producers?### Annotations#### Annotation process#### Who are the annotators?### Personal and Sensitive Information## Considerations for Using the Data### Social Impact of Dataset### Discussion of Biases### Other Known Limitations## Additional Information### Dataset Curators### Licensing Information### Contributions"
] |
f8573dd2cdf6d1fbebcc74ebf1c2c2a93a9a6117
|
# Dataset Card for "iCliniq_data"
[More Information needed](https://github.com/huggingface/datasets/blob/main/CONTRIBUTING.md#how-to-contribute-to-the-dataset-cards)
|
DaisyStar004/iCliniq_data
|
[
"region:us"
] |
2023-09-18T13:02:16+00:00
|
{"dataset_info": {"features": [{"name": "text", "dtype": "string"}], "splits": [{"name": "train", "num_bytes": 7579290, "num_examples": 7321}], "download_size": 4355411, "dataset_size": 7579290}, "configs": [{"config_name": "default", "data_files": [{"split": "train", "path": "data/train-*"}]}]}
|
2023-09-18T13:14:49+00:00
|
[] |
[] |
TAGS
#region-us
|
# Dataset Card for "iCliniq_data"
More Information needed
|
[
"# Dataset Card for \"iCliniq_data\"\n\nMore Information needed"
] |
[
"TAGS\n#region-us \n",
"# Dataset Card for \"iCliniq_data\"\n\nMore Information needed"
] |
[
6,
16
] |
[
"passage: TAGS\n#region-us \n# Dataset Card for \"iCliniq_data\"\n\nMore Information needed"
] |
7021998cc3d1270cf54ddcb2280ad6d339aa4477
|
# Dataset Card for "gtzan_all_preprocessed_kaggle_version"
[More Information needed](https://github.com/huggingface/datasets/blob/main/CONTRIBUTING.md#how-to-contribute-to-the-dataset-cards)
|
barto17/gtzan_all_preprocessed_kaggle_version
|
[
"region:us"
] |
2023-09-18T13:07:54+00:00
|
{"configs": [{"config_name": "default", "data_files": [{"split": "train", "path": "data/train-*"}, {"split": "test", "path": "data/test-*"}]}], "dataset_info": {"features": [{"name": "label", "dtype": {"class_label": {"names": {"0": "blues", "1": "classical", "2": "country", "3": "disco", "4": "hiphop", "5": "jazz", "6": "metal", "7": "pop", "8": "reggae", "9": "rock"}}}}, {"name": "input_values", "sequence": "float32"}, {"name": "attention_mask", "sequence": "int32"}], "splits": [{"name": "train", "num_bytes": 3452159816, "num_examples": 899}, {"name": "test", "num_bytes": 384000696, "num_examples": 100}], "download_size": 1923103931, "dataset_size": 3836160512}}
|
2023-09-18T13:56:06+00:00
|
[] |
[] |
TAGS
#region-us
|
# Dataset Card for "gtzan_all_preprocessed_kaggle_version"
More Information needed
|
[
"# Dataset Card for \"gtzan_all_preprocessed_kaggle_version\"\n\nMore Information needed"
] |
[
"TAGS\n#region-us \n",
"# Dataset Card for \"gtzan_all_preprocessed_kaggle_version\"\n\nMore Information needed"
] |
[
6,
23
] |
[
"passage: TAGS\n#region-us \n# Dataset Card for \"gtzan_all_preprocessed_kaggle_version\"\n\nMore Information needed"
] |
c619a36f74a5344aef0bc9cfe2bca7a0e90b69ce
|
# Dataset Card for Evaluation run of teknium/OpenHermes-7B
## Dataset Description
- **Homepage:**
- **Repository:** https://huggingface.co/teknium/OpenHermes-7B
- **Paper:**
- **Leaderboard:** https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard
- **Point of Contact:** [email protected]
### Dataset Summary
Dataset automatically created during the evaluation run of model [teknium/OpenHermes-7B](https://huggingface.co/teknium/OpenHermes-7B) on the [Open LLM Leaderboard](https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard).
The dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.
The dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The "train" split is always pointing to the latest results.
An additional configuration "results" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the [Open LLM Leaderboard](https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard)).
To load the details from a run, you can for instance do the following:
```python
from datasets import load_dataset
data = load_dataset("open-llm-leaderboard/details_teknium__OpenHermes-7B",
"harness_winogrande_5",
split="train")
```
## Latest results
These are the [latest results from run 2023-10-26T05:03:25.636029](https://huggingface.co/datasets/open-llm-leaderboard/details_teknium__OpenHermes-7B/blob/main/results_2023-10-26T05-03-25.636029.json)(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the "latest" split for each eval):
```python
{
"all": {
"em": 0.2645763422818792,
"em_stderr": 0.004517352215857921,
"f1": 0.33702810402684713,
"f1_stderr": 0.004480224621998652,
"acc": 0.3975524975571051,
"acc_stderr": 0.009127124661977076
},
"harness|drop|3": {
"em": 0.2645763422818792,
"em_stderr": 0.004517352215857921,
"f1": 0.33702810402684713,
"f1_stderr": 0.004480224621998652
},
"harness|gsm8k|5": {
"acc": 0.050037907505686124,
"acc_stderr": 0.006005442354577731
},
"harness|winogrande|5": {
"acc": 0.745067087608524,
"acc_stderr": 0.012248806969376422
}
}
```
### Supported Tasks and Leaderboards
[More Information Needed]
### Languages
[More Information Needed]
## Dataset Structure
### Data Instances
[More Information Needed]
### Data Fields
[More Information Needed]
### Data Splits
[More Information Needed]
## Dataset Creation
### Curation Rationale
[More Information Needed]
### Source Data
#### Initial Data Collection and Normalization
[More Information Needed]
#### Who are the source language producers?
[More Information Needed]
### Annotations
#### Annotation process
[More Information Needed]
#### Who are the annotators?
[More Information Needed]
### Personal and Sensitive Information
[More Information Needed]
## Considerations for Using the Data
### Social Impact of Dataset
[More Information Needed]
### Discussion of Biases
[More Information Needed]
### Other Known Limitations
[More Information Needed]
## Additional Information
### Dataset Curators
[More Information Needed]
### Licensing Information
[More Information Needed]
### Citation Information
[More Information Needed]
### Contributions
[More Information Needed]
|
open-llm-leaderboard/details_teknium__OpenHermes-7B
|
[
"region:us"
] |
2023-09-18T13:09:24+00:00
|
{"pretty_name": "Evaluation run of teknium/OpenHermes-7B", "dataset_summary": "Dataset automatically created during the evaluation run of model [teknium/OpenHermes-7B](https://huggingface.co/teknium/OpenHermes-7B) on the [Open LLM Leaderboard](https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard).\n\nThe dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.\n\nThe dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The \"train\" split is always pointing to the latest results.\n\nAn additional configuration \"results\" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the [Open LLM Leaderboard](https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard)).\n\nTo load the details from a run, you can for instance do the following:\n```python\nfrom datasets import load_dataset\ndata = load_dataset(\"open-llm-leaderboard/details_teknium__OpenHermes-7B\",\n\t\"harness_winogrande_5\",\n\tsplit=\"train\")\n```\n\n## Latest results\n\nThese are the [latest results from run 2023-10-26T05:03:25.636029](https://huggingface.co/datasets/open-llm-leaderboard/details_teknium__OpenHermes-7B/blob/main/results_2023-10-26T05-03-25.636029.json)(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the \"latest\" split for each eval):\n\n```python\n{\n \"all\": {\n \"em\": 0.2645763422818792,\n \"em_stderr\": 0.004517352215857921,\n \"f1\": 0.33702810402684713,\n \"f1_stderr\": 0.004480224621998652,\n \"acc\": 0.3975524975571051,\n \"acc_stderr\": 0.009127124661977076\n },\n \"harness|drop|3\": {\n \"em\": 0.2645763422818792,\n \"em_stderr\": 0.004517352215857921,\n \"f1\": 0.33702810402684713,\n \"f1_stderr\": 0.004480224621998652\n },\n \"harness|gsm8k|5\": {\n \"acc\": 0.050037907505686124,\n \"acc_stderr\": 0.006005442354577731\n },\n \"harness|winogrande|5\": {\n \"acc\": 0.745067087608524,\n \"acc_stderr\": 0.012248806969376422\n }\n}\n```", "repo_url": "https://huggingface.co/teknium/OpenHermes-7B", "leaderboard_url": "https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard", "point_of_contact": "[email protected]", "configs": [{"config_name": "harness_arc_challenge_25", "data_files": [{"split": "2023_09_18T14_09_00.502210", "path": ["**/details_harness|arc:challenge|25_2023-09-18T14-09-00.502210.parquet"]}, {"split": "latest", "path": ["**/details_harness|arc:challenge|25_2023-09-18T14-09-00.502210.parquet"]}]}, {"config_name": "harness_drop_3", "data_files": [{"split": "2023_10_26T05_03_25.636029", "path": ["**/details_harness|drop|3_2023-10-26T05-03-25.636029.parquet"]}, {"split": "latest", "path": ["**/details_harness|drop|3_2023-10-26T05-03-25.636029.parquet"]}]}, {"config_name": "harness_gsm8k_5", "data_files": [{"split": "2023_10_26T05_03_25.636029", "path": ["**/details_harness|gsm8k|5_2023-10-26T05-03-25.636029.parquet"]}, {"split": "latest", "path": ["**/details_harness|gsm8k|5_2023-10-26T05-03-25.636029.parquet"]}]}, {"config_name": "harness_hellaswag_10", "data_files": [{"split": "2023_09_18T14_09_00.502210", "path": ["**/details_harness|hellaswag|10_2023-09-18T14-09-00.502210.parquet"]}, {"split": "latest", "path": ["**/details_harness|hellaswag|10_2023-09-18T14-09-00.502210.parquet"]}]}, {"config_name": "harness_hendrycksTest_5", "data_files": [{"split": "2023_09_18T14_09_00.502210", "path": ["**/details_harness|hendrycksTest-abstract_algebra|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-anatomy|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-astronomy|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-business_ethics|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-clinical_knowledge|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-college_biology|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-college_chemistry|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-college_computer_science|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-college_mathematics|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-college_medicine|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-college_physics|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-computer_security|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-conceptual_physics|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-econometrics|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-electrical_engineering|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-elementary_mathematics|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-formal_logic|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-global_facts|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-high_school_biology|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-high_school_chemistry|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-high_school_computer_science|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-high_school_european_history|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-high_school_geography|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-high_school_government_and_politics|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-high_school_macroeconomics|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-high_school_mathematics|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-high_school_microeconomics|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-high_school_physics|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-high_school_psychology|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-high_school_statistics|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-high_school_us_history|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-high_school_world_history|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-human_aging|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-human_sexuality|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-international_law|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-jurisprudence|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-logical_fallacies|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-machine_learning|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-management|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-marketing|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-medical_genetics|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-miscellaneous|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-moral_disputes|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-moral_scenarios|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-nutrition|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-philosophy|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-prehistory|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-professional_accounting|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-professional_law|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-professional_medicine|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-professional_psychology|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-public_relations|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-security_studies|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-sociology|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-us_foreign_policy|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-virology|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-world_religions|5_2023-09-18T14-09-00.502210.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-abstract_algebra|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-anatomy|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-astronomy|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-business_ethics|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-clinical_knowledge|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-college_biology|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-college_chemistry|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-college_computer_science|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-college_mathematics|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-college_medicine|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-college_physics|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-computer_security|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-conceptual_physics|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-econometrics|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-electrical_engineering|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-elementary_mathematics|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-formal_logic|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-global_facts|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-high_school_biology|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-high_school_chemistry|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-high_school_computer_science|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-high_school_european_history|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-high_school_geography|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-high_school_government_and_politics|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-high_school_macroeconomics|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-high_school_mathematics|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-high_school_microeconomics|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-high_school_physics|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-high_school_psychology|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-high_school_statistics|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-high_school_us_history|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-high_school_world_history|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-human_aging|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-human_sexuality|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-international_law|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-jurisprudence|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-logical_fallacies|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-machine_learning|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-management|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-marketing|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-medical_genetics|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-miscellaneous|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-moral_disputes|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-moral_scenarios|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-nutrition|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-philosophy|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-prehistory|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-professional_accounting|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-professional_law|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-professional_medicine|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-professional_psychology|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-public_relations|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-security_studies|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-sociology|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-us_foreign_policy|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-virology|5_2023-09-18T14-09-00.502210.parquet", "**/details_harness|hendrycksTest-world_religions|5_2023-09-18T14-09-00.502210.parquet"]}]}, {"config_name": "harness_hendrycksTest_abstract_algebra_5", "data_files": [{"split": "2023_09_18T14_09_00.502210", "path": ["**/details_harness|hendrycksTest-abstract_algebra|5_2023-09-18T14-09-00.502210.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-abstract_algebra|5_2023-09-18T14-09-00.502210.parquet"]}]}, {"config_name": "harness_hendrycksTest_anatomy_5", "data_files": [{"split": "2023_09_18T14_09_00.502210", "path": ["**/details_harness|hendrycksTest-anatomy|5_2023-09-18T14-09-00.502210.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-anatomy|5_2023-09-18T14-09-00.502210.parquet"]}]}, {"config_name": "harness_hendrycksTest_astronomy_5", "data_files": [{"split": "2023_09_18T14_09_00.502210", "path": ["**/details_harness|hendrycksTest-astronomy|5_2023-09-18T14-09-00.502210.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-astronomy|5_2023-09-18T14-09-00.502210.parquet"]}]}, {"config_name": "harness_hendrycksTest_business_ethics_5", "data_files": [{"split": "2023_09_18T14_09_00.502210", "path": ["**/details_harness|hendrycksTest-business_ethics|5_2023-09-18T14-09-00.502210.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-business_ethics|5_2023-09-18T14-09-00.502210.parquet"]}]}, {"config_name": "harness_hendrycksTest_clinical_knowledge_5", "data_files": [{"split": "2023_09_18T14_09_00.502210", "path": ["**/details_harness|hendrycksTest-clinical_knowledge|5_2023-09-18T14-09-00.502210.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-clinical_knowledge|5_2023-09-18T14-09-00.502210.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_biology_5", "data_files": [{"split": "2023_09_18T14_09_00.502210", "path": ["**/details_harness|hendrycksTest-college_biology|5_2023-09-18T14-09-00.502210.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_biology|5_2023-09-18T14-09-00.502210.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_chemistry_5", "data_files": [{"split": "2023_09_18T14_09_00.502210", "path": ["**/details_harness|hendrycksTest-college_chemistry|5_2023-09-18T14-09-00.502210.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_chemistry|5_2023-09-18T14-09-00.502210.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_computer_science_5", "data_files": [{"split": "2023_09_18T14_09_00.502210", "path": ["**/details_harness|hendrycksTest-college_computer_science|5_2023-09-18T14-09-00.502210.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_computer_science|5_2023-09-18T14-09-00.502210.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_mathematics_5", "data_files": [{"split": "2023_09_18T14_09_00.502210", "path": ["**/details_harness|hendrycksTest-college_mathematics|5_2023-09-18T14-09-00.502210.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_mathematics|5_2023-09-18T14-09-00.502210.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_medicine_5", "data_files": [{"split": "2023_09_18T14_09_00.502210", "path": ["**/details_harness|hendrycksTest-college_medicine|5_2023-09-18T14-09-00.502210.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_medicine|5_2023-09-18T14-09-00.502210.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_physics_5", "data_files": [{"split": "2023_09_18T14_09_00.502210", "path": ["**/details_harness|hendrycksTest-college_physics|5_2023-09-18T14-09-00.502210.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_physics|5_2023-09-18T14-09-00.502210.parquet"]}]}, {"config_name": "harness_hendrycksTest_computer_security_5", "data_files": [{"split": "2023_09_18T14_09_00.502210", "path": ["**/details_harness|hendrycksTest-computer_security|5_2023-09-18T14-09-00.502210.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-computer_security|5_2023-09-18T14-09-00.502210.parquet"]}]}, {"config_name": "harness_hendrycksTest_conceptual_physics_5", "data_files": [{"split": "2023_09_18T14_09_00.502210", "path": ["**/details_harness|hendrycksTest-conceptual_physics|5_2023-09-18T14-09-00.502210.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-conceptual_physics|5_2023-09-18T14-09-00.502210.parquet"]}]}, {"config_name": "harness_hendrycksTest_econometrics_5", "data_files": [{"split": "2023_09_18T14_09_00.502210", "path": ["**/details_harness|hendrycksTest-econometrics|5_2023-09-18T14-09-00.502210.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-econometrics|5_2023-09-18T14-09-00.502210.parquet"]}]}, {"config_name": "harness_hendrycksTest_electrical_engineering_5", "data_files": [{"split": "2023_09_18T14_09_00.502210", "path": ["**/details_harness|hendrycksTest-electrical_engineering|5_2023-09-18T14-09-00.502210.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-electrical_engineering|5_2023-09-18T14-09-00.502210.parquet"]}]}, {"config_name": "harness_hendrycksTest_elementary_mathematics_5", "data_files": [{"split": "2023_09_18T14_09_00.502210", "path": ["**/details_harness|hendrycksTest-elementary_mathematics|5_2023-09-18T14-09-00.502210.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-elementary_mathematics|5_2023-09-18T14-09-00.502210.parquet"]}]}, {"config_name": "harness_hendrycksTest_formal_logic_5", "data_files": [{"split": "2023_09_18T14_09_00.502210", "path": ["**/details_harness|hendrycksTest-formal_logic|5_2023-09-18T14-09-00.502210.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-formal_logic|5_2023-09-18T14-09-00.502210.parquet"]}]}, {"config_name": "harness_hendrycksTest_global_facts_5", "data_files": [{"split": "2023_09_18T14_09_00.502210", "path": ["**/details_harness|hendrycksTest-global_facts|5_2023-09-18T14-09-00.502210.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-global_facts|5_2023-09-18T14-09-00.502210.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_biology_5", "data_files": [{"split": "2023_09_18T14_09_00.502210", "path": ["**/details_harness|hendrycksTest-high_school_biology|5_2023-09-18T14-09-00.502210.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_biology|5_2023-09-18T14-09-00.502210.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_chemistry_5", "data_files": [{"split": "2023_09_18T14_09_00.502210", "path": ["**/details_harness|hendrycksTest-high_school_chemistry|5_2023-09-18T14-09-00.502210.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_chemistry|5_2023-09-18T14-09-00.502210.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_computer_science_5", "data_files": [{"split": "2023_09_18T14_09_00.502210", "path": ["**/details_harness|hendrycksTest-high_school_computer_science|5_2023-09-18T14-09-00.502210.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_computer_science|5_2023-09-18T14-09-00.502210.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_european_history_5", "data_files": [{"split": "2023_09_18T14_09_00.502210", "path": ["**/details_harness|hendrycksTest-high_school_european_history|5_2023-09-18T14-09-00.502210.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_european_history|5_2023-09-18T14-09-00.502210.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_geography_5", "data_files": [{"split": "2023_09_18T14_09_00.502210", "path": ["**/details_harness|hendrycksTest-high_school_geography|5_2023-09-18T14-09-00.502210.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_geography|5_2023-09-18T14-09-00.502210.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_government_and_politics_5", "data_files": [{"split": "2023_09_18T14_09_00.502210", "path": ["**/details_harness|hendrycksTest-high_school_government_and_politics|5_2023-09-18T14-09-00.502210.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_government_and_politics|5_2023-09-18T14-09-00.502210.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_macroeconomics_5", "data_files": [{"split": "2023_09_18T14_09_00.502210", "path": ["**/details_harness|hendrycksTest-high_school_macroeconomics|5_2023-09-18T14-09-00.502210.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_macroeconomics|5_2023-09-18T14-09-00.502210.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_mathematics_5", "data_files": [{"split": "2023_09_18T14_09_00.502210", "path": ["**/details_harness|hendrycksTest-high_school_mathematics|5_2023-09-18T14-09-00.502210.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_mathematics|5_2023-09-18T14-09-00.502210.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_microeconomics_5", "data_files": [{"split": "2023_09_18T14_09_00.502210", "path": ["**/details_harness|hendrycksTest-high_school_microeconomics|5_2023-09-18T14-09-00.502210.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_microeconomics|5_2023-09-18T14-09-00.502210.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_physics_5", "data_files": [{"split": "2023_09_18T14_09_00.502210", "path": ["**/details_harness|hendrycksTest-high_school_physics|5_2023-09-18T14-09-00.502210.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_physics|5_2023-09-18T14-09-00.502210.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_psychology_5", "data_files": [{"split": "2023_09_18T14_09_00.502210", "path": ["**/details_harness|hendrycksTest-high_school_psychology|5_2023-09-18T14-09-00.502210.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_psychology|5_2023-09-18T14-09-00.502210.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_statistics_5", "data_files": [{"split": "2023_09_18T14_09_00.502210", "path": ["**/details_harness|hendrycksTest-high_school_statistics|5_2023-09-18T14-09-00.502210.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_statistics|5_2023-09-18T14-09-00.502210.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_us_history_5", "data_files": [{"split": "2023_09_18T14_09_00.502210", "path": ["**/details_harness|hendrycksTest-high_school_us_history|5_2023-09-18T14-09-00.502210.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_us_history|5_2023-09-18T14-09-00.502210.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_world_history_5", "data_files": [{"split": "2023_09_18T14_09_00.502210", "path": ["**/details_harness|hendrycksTest-high_school_world_history|5_2023-09-18T14-09-00.502210.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_world_history|5_2023-09-18T14-09-00.502210.parquet"]}]}, {"config_name": "harness_hendrycksTest_human_aging_5", "data_files": [{"split": "2023_09_18T14_09_00.502210", "path": ["**/details_harness|hendrycksTest-human_aging|5_2023-09-18T14-09-00.502210.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-human_aging|5_2023-09-18T14-09-00.502210.parquet"]}]}, {"config_name": "harness_hendrycksTest_human_sexuality_5", "data_files": [{"split": "2023_09_18T14_09_00.502210", "path": ["**/details_harness|hendrycksTest-human_sexuality|5_2023-09-18T14-09-00.502210.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-human_sexuality|5_2023-09-18T14-09-00.502210.parquet"]}]}, {"config_name": "harness_hendrycksTest_international_law_5", "data_files": [{"split": "2023_09_18T14_09_00.502210", "path": ["**/details_harness|hendrycksTest-international_law|5_2023-09-18T14-09-00.502210.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-international_law|5_2023-09-18T14-09-00.502210.parquet"]}]}, {"config_name": "harness_hendrycksTest_jurisprudence_5", "data_files": [{"split": "2023_09_18T14_09_00.502210", "path": ["**/details_harness|hendrycksTest-jurisprudence|5_2023-09-18T14-09-00.502210.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-jurisprudence|5_2023-09-18T14-09-00.502210.parquet"]}]}, {"config_name": "harness_hendrycksTest_logical_fallacies_5", "data_files": [{"split": "2023_09_18T14_09_00.502210", "path": ["**/details_harness|hendrycksTest-logical_fallacies|5_2023-09-18T14-09-00.502210.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-logical_fallacies|5_2023-09-18T14-09-00.502210.parquet"]}]}, {"config_name": "harness_hendrycksTest_machine_learning_5", "data_files": [{"split": "2023_09_18T14_09_00.502210", "path": ["**/details_harness|hendrycksTest-machine_learning|5_2023-09-18T14-09-00.502210.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-machine_learning|5_2023-09-18T14-09-00.502210.parquet"]}]}, {"config_name": "harness_hendrycksTest_management_5", "data_files": [{"split": "2023_09_18T14_09_00.502210", "path": ["**/details_harness|hendrycksTest-management|5_2023-09-18T14-09-00.502210.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-management|5_2023-09-18T14-09-00.502210.parquet"]}]}, {"config_name": "harness_hendrycksTest_marketing_5", "data_files": [{"split": "2023_09_18T14_09_00.502210", "path": ["**/details_harness|hendrycksTest-marketing|5_2023-09-18T14-09-00.502210.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-marketing|5_2023-09-18T14-09-00.502210.parquet"]}]}, {"config_name": "harness_hendrycksTest_medical_genetics_5", "data_files": [{"split": "2023_09_18T14_09_00.502210", "path": ["**/details_harness|hendrycksTest-medical_genetics|5_2023-09-18T14-09-00.502210.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-medical_genetics|5_2023-09-18T14-09-00.502210.parquet"]}]}, {"config_name": "harness_hendrycksTest_miscellaneous_5", "data_files": [{"split": "2023_09_18T14_09_00.502210", "path": ["**/details_harness|hendrycksTest-miscellaneous|5_2023-09-18T14-09-00.502210.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-miscellaneous|5_2023-09-18T14-09-00.502210.parquet"]}]}, {"config_name": "harness_hendrycksTest_moral_disputes_5", "data_files": [{"split": "2023_09_18T14_09_00.502210", "path": ["**/details_harness|hendrycksTest-moral_disputes|5_2023-09-18T14-09-00.502210.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-moral_disputes|5_2023-09-18T14-09-00.502210.parquet"]}]}, {"config_name": "harness_hendrycksTest_moral_scenarios_5", "data_files": [{"split": "2023_09_18T14_09_00.502210", "path": ["**/details_harness|hendrycksTest-moral_scenarios|5_2023-09-18T14-09-00.502210.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-moral_scenarios|5_2023-09-18T14-09-00.502210.parquet"]}]}, {"config_name": "harness_hendrycksTest_nutrition_5", "data_files": [{"split": "2023_09_18T14_09_00.502210", "path": ["**/details_harness|hendrycksTest-nutrition|5_2023-09-18T14-09-00.502210.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-nutrition|5_2023-09-18T14-09-00.502210.parquet"]}]}, {"config_name": "harness_hendrycksTest_philosophy_5", "data_files": [{"split": "2023_09_18T14_09_00.502210", "path": ["**/details_harness|hendrycksTest-philosophy|5_2023-09-18T14-09-00.502210.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-philosophy|5_2023-09-18T14-09-00.502210.parquet"]}]}, {"config_name": "harness_hendrycksTest_prehistory_5", "data_files": [{"split": "2023_09_18T14_09_00.502210", "path": ["**/details_harness|hendrycksTest-prehistory|5_2023-09-18T14-09-00.502210.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-prehistory|5_2023-09-18T14-09-00.502210.parquet"]}]}, {"config_name": "harness_hendrycksTest_professional_accounting_5", "data_files": [{"split": "2023_09_18T14_09_00.502210", "path": ["**/details_harness|hendrycksTest-professional_accounting|5_2023-09-18T14-09-00.502210.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-professional_accounting|5_2023-09-18T14-09-00.502210.parquet"]}]}, {"config_name": "harness_hendrycksTest_professional_law_5", "data_files": [{"split": "2023_09_18T14_09_00.502210", "path": ["**/details_harness|hendrycksTest-professional_law|5_2023-09-18T14-09-00.502210.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-professional_law|5_2023-09-18T14-09-00.502210.parquet"]}]}, {"config_name": "harness_hendrycksTest_professional_medicine_5", "data_files": [{"split": "2023_09_18T14_09_00.502210", "path": ["**/details_harness|hendrycksTest-professional_medicine|5_2023-09-18T14-09-00.502210.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-professional_medicine|5_2023-09-18T14-09-00.502210.parquet"]}]}, {"config_name": "harness_hendrycksTest_professional_psychology_5", "data_files": [{"split": "2023_09_18T14_09_00.502210", "path": ["**/details_harness|hendrycksTest-professional_psychology|5_2023-09-18T14-09-00.502210.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-professional_psychology|5_2023-09-18T14-09-00.502210.parquet"]}]}, {"config_name": "harness_hendrycksTest_public_relations_5", "data_files": [{"split": "2023_09_18T14_09_00.502210", "path": ["**/details_harness|hendrycksTest-public_relations|5_2023-09-18T14-09-00.502210.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-public_relations|5_2023-09-18T14-09-00.502210.parquet"]}]}, {"config_name": "harness_hendrycksTest_security_studies_5", "data_files": [{"split": "2023_09_18T14_09_00.502210", "path": ["**/details_harness|hendrycksTest-security_studies|5_2023-09-18T14-09-00.502210.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-security_studies|5_2023-09-18T14-09-00.502210.parquet"]}]}, {"config_name": "harness_hendrycksTest_sociology_5", "data_files": [{"split": "2023_09_18T14_09_00.502210", "path": ["**/details_harness|hendrycksTest-sociology|5_2023-09-18T14-09-00.502210.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-sociology|5_2023-09-18T14-09-00.502210.parquet"]}]}, {"config_name": "harness_hendrycksTest_us_foreign_policy_5", "data_files": [{"split": "2023_09_18T14_09_00.502210", "path": ["**/details_harness|hendrycksTest-us_foreign_policy|5_2023-09-18T14-09-00.502210.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-us_foreign_policy|5_2023-09-18T14-09-00.502210.parquet"]}]}, {"config_name": "harness_hendrycksTest_virology_5", "data_files": [{"split": "2023_09_18T14_09_00.502210", "path": ["**/details_harness|hendrycksTest-virology|5_2023-09-18T14-09-00.502210.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-virology|5_2023-09-18T14-09-00.502210.parquet"]}]}, {"config_name": "harness_hendrycksTest_world_religions_5", "data_files": [{"split": "2023_09_18T14_09_00.502210", "path": ["**/details_harness|hendrycksTest-world_religions|5_2023-09-18T14-09-00.502210.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-world_religions|5_2023-09-18T14-09-00.502210.parquet"]}]}, {"config_name": "harness_truthfulqa_mc_0", "data_files": [{"split": "2023_09_18T14_09_00.502210", "path": ["**/details_harness|truthfulqa:mc|0_2023-09-18T14-09-00.502210.parquet"]}, {"split": "latest", "path": ["**/details_harness|truthfulqa:mc|0_2023-09-18T14-09-00.502210.parquet"]}]}, {"config_name": "harness_winogrande_5", "data_files": [{"split": "2023_10_26T05_03_25.636029", "path": ["**/details_harness|winogrande|5_2023-10-26T05-03-25.636029.parquet"]}, {"split": "latest", "path": ["**/details_harness|winogrande|5_2023-10-26T05-03-25.636029.parquet"]}]}, {"config_name": "results", "data_files": [{"split": "2023_09_18T14_09_00.502210", "path": ["results_2023-09-18T14-09-00.502210.parquet"]}, {"split": "2023_10_26T05_03_25.636029", "path": ["results_2023-10-26T05-03-25.636029.parquet"]}, {"split": "latest", "path": ["results_2023-10-26T05-03-25.636029.parquet"]}]}]}
|
2023-10-26T04:03:37+00:00
|
[] |
[] |
TAGS
#region-us
|
# Dataset Card for Evaluation run of teknium/OpenHermes-7B
## Dataset Description
- Homepage:
- Repository: URL
- Paper:
- Leaderboard: URL
- Point of Contact: clementine@URL
### Dataset Summary
Dataset automatically created during the evaluation run of model teknium/OpenHermes-7B on the Open LLM Leaderboard.
The dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.
The dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The "train" split is always pointing to the latest results.
An additional configuration "results" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the Open LLM Leaderboard).
To load the details from a run, you can for instance do the following:
## Latest results
These are the latest results from run 2023-10-26T05:03:25.636029(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the "latest" split for each eval):
### Supported Tasks and Leaderboards
### Languages
## Dataset Structure
### Data Instances
### Data Fields
### Data Splits
## Dataset Creation
### Curation Rationale
### Source Data
#### Initial Data Collection and Normalization
#### Who are the source language producers?
### Annotations
#### Annotation process
#### Who are the annotators?
### Personal and Sensitive Information
## Considerations for Using the Data
### Social Impact of Dataset
### Discussion of Biases
### Other Known Limitations
## Additional Information
### Dataset Curators
### Licensing Information
### Contributions
|
[
"# Dataset Card for Evaluation run of teknium/OpenHermes-7B",
"## Dataset Description\n\n- Homepage: \n- Repository: URL\n- Paper: \n- Leaderboard: URL\n- Point of Contact: clementine@URL",
"### Dataset Summary\n\nDataset automatically created during the evaluation run of model teknium/OpenHermes-7B on the Open LLM Leaderboard.\n\nThe dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.\n\nThe dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The \"train\" split is always pointing to the latest results.\n\nAn additional configuration \"results\" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the Open LLM Leaderboard).\n\nTo load the details from a run, you can for instance do the following:",
"## Latest results\n\nThese are the latest results from run 2023-10-26T05:03:25.636029(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the \"latest\" split for each eval):",
"### Supported Tasks and Leaderboards",
"### Languages",
"## Dataset Structure",
"### Data Instances",
"### Data Fields",
"### Data Splits",
"## Dataset Creation",
"### Curation Rationale",
"### Source Data",
"#### Initial Data Collection and Normalization",
"#### Who are the source language producers?",
"### Annotations",
"#### Annotation process",
"#### Who are the annotators?",
"### Personal and Sensitive Information",
"## Considerations for Using the Data",
"### Social Impact of Dataset",
"### Discussion of Biases",
"### Other Known Limitations",
"## Additional Information",
"### Dataset Curators",
"### Licensing Information",
"### Contributions"
] |
[
"TAGS\n#region-us \n",
"# Dataset Card for Evaluation run of teknium/OpenHermes-7B",
"## Dataset Description\n\n- Homepage: \n- Repository: URL\n- Paper: \n- Leaderboard: URL\n- Point of Contact: clementine@URL",
"### Dataset Summary\n\nDataset automatically created during the evaluation run of model teknium/OpenHermes-7B on the Open LLM Leaderboard.\n\nThe dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.\n\nThe dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The \"train\" split is always pointing to the latest results.\n\nAn additional configuration \"results\" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the Open LLM Leaderboard).\n\nTo load the details from a run, you can for instance do the following:",
"## Latest results\n\nThese are the latest results from run 2023-10-26T05:03:25.636029(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the \"latest\" split for each eval):",
"### Supported Tasks and Leaderboards",
"### Languages",
"## Dataset Structure",
"### Data Instances",
"### Data Fields",
"### Data Splits",
"## Dataset Creation",
"### Curation Rationale",
"### Source Data",
"#### Initial Data Collection and Normalization",
"#### Who are the source language producers?",
"### Annotations",
"#### Annotation process",
"#### Who are the annotators?",
"### Personal and Sensitive Information",
"## Considerations for Using the Data",
"### Social Impact of Dataset",
"### Discussion of Biases",
"### Other Known Limitations",
"## Additional Information",
"### Dataset Curators",
"### Licensing Information",
"### Contributions"
] |
[
6,
17,
31,
165,
67,
10,
4,
6,
6,
5,
5,
5,
7,
4,
10,
10,
5,
5,
9,
8,
8,
7,
8,
7,
5,
6,
6,
5
] |
[
"passage: TAGS\n#region-us \n# Dataset Card for Evaluation run of teknium/OpenHermes-7B## Dataset Description\n\n- Homepage: \n- Repository: URL\n- Paper: \n- Leaderboard: URL\n- Point of Contact: clementine@URL### Dataset Summary\n\nDataset automatically created during the evaluation run of model teknium/OpenHermes-7B on the Open LLM Leaderboard.\n\nThe dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.\n\nThe dataset has been created from 2 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The \"train\" split is always pointing to the latest results.\n\nAn additional configuration \"results\" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the Open LLM Leaderboard).\n\nTo load the details from a run, you can for instance do the following:## Latest results\n\nThese are the latest results from run 2023-10-26T05:03:25.636029(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the \"latest\" split for each eval):### Supported Tasks and Leaderboards### Languages## Dataset Structure### Data Instances### Data Fields### Data Splits## Dataset Creation### Curation Rationale### Source Data#### Initial Data Collection and Normalization#### Who are the source language producers?### Annotations#### Annotation process#### Who are the annotators?### Personal and Sensitive Information## Considerations for Using the Data### Social Impact of Dataset### Discussion of Biases### Other Known Limitations## Additional Information### Dataset Curators### Licensing Information### Contributions"
] |
85418d04e4f63e38370b1bb3653f7eef711bfeba
|
# Dataset Card for "flower_arrangement"
[More Information needed](https://github.com/huggingface/datasets/blob/main/CONTRIBUTING.md#how-to-contribute-to-the-dataset-cards)
|
Falah/flower_arrangement
|
[
"region:us"
] |
2023-09-18T13:11:09+00:00
|
{"dataset_info": {"features": [{"name": "prompts", "dtype": "string"}], "splits": [{"name": "train", "num_bytes": 367584, "num_examples": 1000}], "download_size": 41547, "dataset_size": 367584}}
|
2023-09-18T13:11:11+00:00
|
[] |
[] |
TAGS
#region-us
|
# Dataset Card for "flower_arrangement"
More Information needed
|
[
"# Dataset Card for \"flower_arrangement\"\n\nMore Information needed"
] |
[
"TAGS\n#region-us \n",
"# Dataset Card for \"flower_arrangement\"\n\nMore Information needed"
] |
[
6,
13
] |
[
"passage: TAGS\n#region-us \n# Dataset Card for \"flower_arrangement\"\n\nMore Information needed"
] |
275410f4ad7b46a51d4c42b66fd6c00301adfa3a
|
# Dataset Card for "stories_0_prompts"
[More Information needed](https://github.com/huggingface/datasets/blob/main/CONTRIBUTING.md#how-to-contribute-to-the-dataset-cards)
|
Falah/stories_0_prompts
|
[
"region:us"
] |
2023-09-18T13:11:51+00:00
|
{"dataset_info": {"features": [{"name": "prompts", "dtype": "string"}], "splits": [{"name": "train", "num_bytes": 3009, "num_examples": 11}], "download_size": 4074, "dataset_size": 3009}}
|
2023-09-18T14:21:27+00:00
|
[] |
[] |
TAGS
#region-us
|
# Dataset Card for "stories_0_prompts"
More Information needed
|
[
"# Dataset Card for \"stories_0_prompts\"\n\nMore Information needed"
] |
[
"TAGS\n#region-us \n",
"# Dataset Card for \"stories_0_prompts\"\n\nMore Information needed"
] |
[
6,
17
] |
[
"passage: TAGS\n#region-us \n# Dataset Card for \"stories_0_prompts\"\n\nMore Information needed"
] |
ec578758cc1702f3b6bfe206a8698457fc1fb86c
|
# Dataset Card for "stories_1_prompts"
[More Information needed](https://github.com/huggingface/datasets/blob/main/CONTRIBUTING.md#how-to-contribute-to-the-dataset-cards)
|
Falah/stories_1_prompts
|
[
"region:us"
] |
2023-09-18T13:11:55+00:00
|
{"dataset_info": {"features": [{"name": "prompts", "dtype": "string"}], "splits": [{"name": "train", "num_bytes": 3381, "num_examples": 11}], "download_size": 5022, "dataset_size": 3381}}
|
2023-09-18T14:21:33+00:00
|
[] |
[] |
TAGS
#region-us
|
# Dataset Card for "stories_1_prompts"
More Information needed
|
[
"# Dataset Card for \"stories_1_prompts\"\n\nMore Information needed"
] |
[
"TAGS\n#region-us \n",
"# Dataset Card for \"stories_1_prompts\"\n\nMore Information needed"
] |
[
6,
16
] |
[
"passage: TAGS\n#region-us \n# Dataset Card for \"stories_1_prompts\"\n\nMore Information needed"
] |
5fefdd0add89c6b884c845a8b24a96e803bbb31b
|
# Dataset Card for "stories_2_prompts"
[More Information needed](https://github.com/huggingface/datasets/blob/main/CONTRIBUTING.md#how-to-contribute-to-the-dataset-cards)
|
Falah/stories_2_prompts
|
[
"region:us"
] |
2023-09-18T13:12:00+00:00
|
{"dataset_info": {"features": [{"name": "prompts", "dtype": "string"}], "splits": [{"name": "train", "num_bytes": 3595, "num_examples": 12}], "download_size": 5193, "dataset_size": 3595}}
|
2023-09-18T14:21:40+00:00
|
[] |
[] |
TAGS
#region-us
|
# Dataset Card for "stories_2_prompts"
More Information needed
|
[
"# Dataset Card for \"stories_2_prompts\"\n\nMore Information needed"
] |
[
"TAGS\n#region-us \n",
"# Dataset Card for \"stories_2_prompts\"\n\nMore Information needed"
] |
[
6,
17
] |
[
"passage: TAGS\n#region-us \n# Dataset Card for \"stories_2_prompts\"\n\nMore Information needed"
] |
fd593d761df438a09d5bc5cfcd5b3651a34a883a
|
# Dataset Card for Evaluation run of lgaalves/llama-2-13b-hf-platypus
## Dataset Description
- **Homepage:**
- **Repository:** https://huggingface.co/lgaalves/llama-2-13b-hf-platypus
- **Paper:**
- **Leaderboard:** https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard
- **Point of Contact:** [email protected]
### Dataset Summary
Dataset automatically created during the evaluation run of model [lgaalves/llama-2-13b-hf-platypus](https://huggingface.co/lgaalves/llama-2-13b-hf-platypus) on the [Open LLM Leaderboard](https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard).
The dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.
The dataset has been created from 3 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The "train" split is always pointing to the latest results.
An additional configuration "results" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the [Open LLM Leaderboard](https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard)).
To load the details from a run, you can for instance do the following:
```python
from datasets import load_dataset
data = load_dataset("open-llm-leaderboard/details_lgaalves__llama-2-13b-hf-platypus",
"harness_winogrande_5",
split="train")
```
## Latest results
These are the [latest results from run 2023-10-28T02:33:59.939371](https://huggingface.co/datasets/open-llm-leaderboard/details_lgaalves__llama-2-13b-hf-platypus/blob/main/results_2023-10-28T02-33-59.939371.json)(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the "latest" split for each eval):
```python
{
"all": {
"em": 0.0017827181208053692,
"em_stderr": 0.00043200973460388544,
"f1": 0.05985213926174496,
"f1_stderr": 0.0013641672120704657,
"acc": 0.4325617395685546,
"acc_stderr": 0.009923090021448928
},
"harness|drop|3": {
"em": 0.0017827181208053692,
"em_stderr": 0.00043200973460388544,
"f1": 0.05985213926174496,
"f1_stderr": 0.0013641672120704657
},
"harness|gsm8k|5": {
"acc": 0.09401061410159212,
"acc_stderr": 0.00803881981887246
},
"harness|winogrande|5": {
"acc": 0.771112865035517,
"acc_stderr": 0.011807360224025398
}
}
```
### Supported Tasks and Leaderboards
[More Information Needed]
### Languages
[More Information Needed]
## Dataset Structure
### Data Instances
[More Information Needed]
### Data Fields
[More Information Needed]
### Data Splits
[More Information Needed]
## Dataset Creation
### Curation Rationale
[More Information Needed]
### Source Data
#### Initial Data Collection and Normalization
[More Information Needed]
#### Who are the source language producers?
[More Information Needed]
### Annotations
#### Annotation process
[More Information Needed]
#### Who are the annotators?
[More Information Needed]
### Personal and Sensitive Information
[More Information Needed]
## Considerations for Using the Data
### Social Impact of Dataset
[More Information Needed]
### Discussion of Biases
[More Information Needed]
### Other Known Limitations
[More Information Needed]
## Additional Information
### Dataset Curators
[More Information Needed]
### Licensing Information
[More Information Needed]
### Citation Information
[More Information Needed]
### Contributions
[More Information Needed]
|
open-llm-leaderboard/details_lgaalves__llama-2-13b-hf-platypus
|
[
"region:us"
] |
2023-09-18T13:16:10+00:00
|
{"pretty_name": "Evaluation run of lgaalves/llama-2-13b-hf-platypus", "dataset_summary": "Dataset automatically created during the evaluation run of model [lgaalves/llama-2-13b-hf-platypus](https://huggingface.co/lgaalves/llama-2-13b-hf-platypus) on the [Open LLM Leaderboard](https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard).\n\nThe dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.\n\nThe dataset has been created from 3 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The \"train\" split is always pointing to the latest results.\n\nAn additional configuration \"results\" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the [Open LLM Leaderboard](https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard)).\n\nTo load the details from a run, you can for instance do the following:\n```python\nfrom datasets import load_dataset\ndata = load_dataset(\"open-llm-leaderboard/details_lgaalves__llama-2-13b-hf-platypus\",\n\t\"harness_winogrande_5\",\n\tsplit=\"train\")\n```\n\n## Latest results\n\nThese are the [latest results from run 2023-10-28T02:33:59.939371](https://huggingface.co/datasets/open-llm-leaderboard/details_lgaalves__llama-2-13b-hf-platypus/blob/main/results_2023-10-28T02-33-59.939371.json)(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the \"latest\" split for each eval):\n\n```python\n{\n \"all\": {\n \"em\": 0.0017827181208053692,\n \"em_stderr\": 0.00043200973460388544,\n \"f1\": 0.05985213926174496,\n \"f1_stderr\": 0.0013641672120704657,\n \"acc\": 0.4325617395685546,\n \"acc_stderr\": 0.009923090021448928\n },\n \"harness|drop|3\": {\n \"em\": 0.0017827181208053692,\n \"em_stderr\": 0.00043200973460388544,\n \"f1\": 0.05985213926174496,\n \"f1_stderr\": 0.0013641672120704657\n },\n \"harness|gsm8k|5\": {\n \"acc\": 0.09401061410159212,\n \"acc_stderr\": 0.00803881981887246\n },\n \"harness|winogrande|5\": {\n \"acc\": 0.771112865035517,\n \"acc_stderr\": 0.011807360224025398\n }\n}\n```", "repo_url": "https://huggingface.co/lgaalves/llama-2-13b-hf-platypus", "leaderboard_url": "https://huggingface.co/spaces/HuggingFaceH4/open_llm_leaderboard", "point_of_contact": "[email protected]", "configs": [{"config_name": "harness_arc_challenge_25", "data_files": [{"split": "2023_09_18T14_15_46.670153", "path": ["**/details_harness|arc:challenge|25_2023-09-18T14-15-46.670153.parquet"]}, {"split": "latest", "path": ["**/details_harness|arc:challenge|25_2023-09-18T14-15-46.670153.parquet"]}]}, {"config_name": "harness_drop_3", "data_files": [{"split": "2023_10_28T00_17_42.072889", "path": ["**/details_harness|drop|3_2023-10-28T00-17-42.072889.parquet"]}, {"split": "2023_10_28T02_33_59.939371", "path": ["**/details_harness|drop|3_2023-10-28T02-33-59.939371.parquet"]}, {"split": "latest", "path": ["**/details_harness|drop|3_2023-10-28T02-33-59.939371.parquet"]}]}, {"config_name": "harness_gsm8k_5", "data_files": [{"split": "2023_10_28T00_17_42.072889", "path": ["**/details_harness|gsm8k|5_2023-10-28T00-17-42.072889.parquet"]}, {"split": "2023_10_28T02_33_59.939371", "path": ["**/details_harness|gsm8k|5_2023-10-28T02-33-59.939371.parquet"]}, {"split": "latest", "path": ["**/details_harness|gsm8k|5_2023-10-28T02-33-59.939371.parquet"]}]}, {"config_name": "harness_hellaswag_10", "data_files": [{"split": "2023_09_18T14_15_46.670153", "path": ["**/details_harness|hellaswag|10_2023-09-18T14-15-46.670153.parquet"]}, {"split": "latest", "path": ["**/details_harness|hellaswag|10_2023-09-18T14-15-46.670153.parquet"]}]}, {"config_name": "harness_hendrycksTest_5", "data_files": [{"split": "2023_09_18T14_15_46.670153", "path": ["**/details_harness|hendrycksTest-abstract_algebra|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-anatomy|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-astronomy|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-business_ethics|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-clinical_knowledge|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-college_biology|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-college_chemistry|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-college_computer_science|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-college_mathematics|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-college_medicine|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-college_physics|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-computer_security|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-conceptual_physics|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-econometrics|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-electrical_engineering|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-elementary_mathematics|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-formal_logic|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-global_facts|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-high_school_biology|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-high_school_chemistry|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-high_school_computer_science|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-high_school_european_history|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-high_school_geography|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-high_school_government_and_politics|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-high_school_macroeconomics|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-high_school_mathematics|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-high_school_microeconomics|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-high_school_physics|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-high_school_psychology|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-high_school_statistics|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-high_school_us_history|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-high_school_world_history|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-human_aging|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-human_sexuality|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-international_law|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-jurisprudence|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-logical_fallacies|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-machine_learning|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-management|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-marketing|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-medical_genetics|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-miscellaneous|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-moral_disputes|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-moral_scenarios|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-nutrition|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-philosophy|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-prehistory|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-professional_accounting|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-professional_law|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-professional_medicine|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-professional_psychology|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-public_relations|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-security_studies|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-sociology|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-us_foreign_policy|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-virology|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-world_religions|5_2023-09-18T14-15-46.670153.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-abstract_algebra|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-anatomy|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-astronomy|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-business_ethics|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-clinical_knowledge|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-college_biology|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-college_chemistry|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-college_computer_science|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-college_mathematics|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-college_medicine|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-college_physics|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-computer_security|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-conceptual_physics|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-econometrics|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-electrical_engineering|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-elementary_mathematics|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-formal_logic|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-global_facts|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-high_school_biology|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-high_school_chemistry|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-high_school_computer_science|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-high_school_european_history|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-high_school_geography|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-high_school_government_and_politics|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-high_school_macroeconomics|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-high_school_mathematics|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-high_school_microeconomics|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-high_school_physics|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-high_school_psychology|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-high_school_statistics|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-high_school_us_history|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-high_school_world_history|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-human_aging|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-human_sexuality|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-international_law|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-jurisprudence|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-logical_fallacies|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-machine_learning|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-management|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-marketing|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-medical_genetics|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-miscellaneous|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-moral_disputes|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-moral_scenarios|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-nutrition|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-philosophy|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-prehistory|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-professional_accounting|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-professional_law|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-professional_medicine|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-professional_psychology|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-public_relations|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-security_studies|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-sociology|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-us_foreign_policy|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-virology|5_2023-09-18T14-15-46.670153.parquet", "**/details_harness|hendrycksTest-world_religions|5_2023-09-18T14-15-46.670153.parquet"]}]}, {"config_name": "harness_hendrycksTest_abstract_algebra_5", "data_files": [{"split": "2023_09_18T14_15_46.670153", "path": ["**/details_harness|hendrycksTest-abstract_algebra|5_2023-09-18T14-15-46.670153.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-abstract_algebra|5_2023-09-18T14-15-46.670153.parquet"]}]}, {"config_name": "harness_hendrycksTest_anatomy_5", "data_files": [{"split": "2023_09_18T14_15_46.670153", "path": ["**/details_harness|hendrycksTest-anatomy|5_2023-09-18T14-15-46.670153.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-anatomy|5_2023-09-18T14-15-46.670153.parquet"]}]}, {"config_name": "harness_hendrycksTest_astronomy_5", "data_files": [{"split": "2023_09_18T14_15_46.670153", "path": ["**/details_harness|hendrycksTest-astronomy|5_2023-09-18T14-15-46.670153.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-astronomy|5_2023-09-18T14-15-46.670153.parquet"]}]}, {"config_name": "harness_hendrycksTest_business_ethics_5", "data_files": [{"split": "2023_09_18T14_15_46.670153", "path": ["**/details_harness|hendrycksTest-business_ethics|5_2023-09-18T14-15-46.670153.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-business_ethics|5_2023-09-18T14-15-46.670153.parquet"]}]}, {"config_name": "harness_hendrycksTest_clinical_knowledge_5", "data_files": [{"split": "2023_09_18T14_15_46.670153", "path": ["**/details_harness|hendrycksTest-clinical_knowledge|5_2023-09-18T14-15-46.670153.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-clinical_knowledge|5_2023-09-18T14-15-46.670153.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_biology_5", "data_files": [{"split": "2023_09_18T14_15_46.670153", "path": ["**/details_harness|hendrycksTest-college_biology|5_2023-09-18T14-15-46.670153.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_biology|5_2023-09-18T14-15-46.670153.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_chemistry_5", "data_files": [{"split": "2023_09_18T14_15_46.670153", "path": ["**/details_harness|hendrycksTest-college_chemistry|5_2023-09-18T14-15-46.670153.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_chemistry|5_2023-09-18T14-15-46.670153.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_computer_science_5", "data_files": [{"split": "2023_09_18T14_15_46.670153", "path": ["**/details_harness|hendrycksTest-college_computer_science|5_2023-09-18T14-15-46.670153.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_computer_science|5_2023-09-18T14-15-46.670153.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_mathematics_5", "data_files": [{"split": "2023_09_18T14_15_46.670153", "path": ["**/details_harness|hendrycksTest-college_mathematics|5_2023-09-18T14-15-46.670153.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_mathematics|5_2023-09-18T14-15-46.670153.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_medicine_5", "data_files": [{"split": "2023_09_18T14_15_46.670153", "path": ["**/details_harness|hendrycksTest-college_medicine|5_2023-09-18T14-15-46.670153.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_medicine|5_2023-09-18T14-15-46.670153.parquet"]}]}, {"config_name": "harness_hendrycksTest_college_physics_5", "data_files": [{"split": "2023_09_18T14_15_46.670153", "path": ["**/details_harness|hendrycksTest-college_physics|5_2023-09-18T14-15-46.670153.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-college_physics|5_2023-09-18T14-15-46.670153.parquet"]}]}, {"config_name": "harness_hendrycksTest_computer_security_5", "data_files": [{"split": "2023_09_18T14_15_46.670153", "path": ["**/details_harness|hendrycksTest-computer_security|5_2023-09-18T14-15-46.670153.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-computer_security|5_2023-09-18T14-15-46.670153.parquet"]}]}, {"config_name": "harness_hendrycksTest_conceptual_physics_5", "data_files": [{"split": "2023_09_18T14_15_46.670153", "path": ["**/details_harness|hendrycksTest-conceptual_physics|5_2023-09-18T14-15-46.670153.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-conceptual_physics|5_2023-09-18T14-15-46.670153.parquet"]}]}, {"config_name": "harness_hendrycksTest_econometrics_5", "data_files": [{"split": "2023_09_18T14_15_46.670153", "path": ["**/details_harness|hendrycksTest-econometrics|5_2023-09-18T14-15-46.670153.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-econometrics|5_2023-09-18T14-15-46.670153.parquet"]}]}, {"config_name": "harness_hendrycksTest_electrical_engineering_5", "data_files": [{"split": "2023_09_18T14_15_46.670153", "path": ["**/details_harness|hendrycksTest-electrical_engineering|5_2023-09-18T14-15-46.670153.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-electrical_engineering|5_2023-09-18T14-15-46.670153.parquet"]}]}, {"config_name": "harness_hendrycksTest_elementary_mathematics_5", "data_files": [{"split": "2023_09_18T14_15_46.670153", "path": ["**/details_harness|hendrycksTest-elementary_mathematics|5_2023-09-18T14-15-46.670153.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-elementary_mathematics|5_2023-09-18T14-15-46.670153.parquet"]}]}, {"config_name": "harness_hendrycksTest_formal_logic_5", "data_files": [{"split": "2023_09_18T14_15_46.670153", "path": ["**/details_harness|hendrycksTest-formal_logic|5_2023-09-18T14-15-46.670153.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-formal_logic|5_2023-09-18T14-15-46.670153.parquet"]}]}, {"config_name": "harness_hendrycksTest_global_facts_5", "data_files": [{"split": "2023_09_18T14_15_46.670153", "path": ["**/details_harness|hendrycksTest-global_facts|5_2023-09-18T14-15-46.670153.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-global_facts|5_2023-09-18T14-15-46.670153.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_biology_5", "data_files": [{"split": "2023_09_18T14_15_46.670153", "path": ["**/details_harness|hendrycksTest-high_school_biology|5_2023-09-18T14-15-46.670153.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_biology|5_2023-09-18T14-15-46.670153.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_chemistry_5", "data_files": [{"split": "2023_09_18T14_15_46.670153", "path": ["**/details_harness|hendrycksTest-high_school_chemistry|5_2023-09-18T14-15-46.670153.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_chemistry|5_2023-09-18T14-15-46.670153.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_computer_science_5", "data_files": [{"split": "2023_09_18T14_15_46.670153", "path": ["**/details_harness|hendrycksTest-high_school_computer_science|5_2023-09-18T14-15-46.670153.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_computer_science|5_2023-09-18T14-15-46.670153.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_european_history_5", "data_files": [{"split": "2023_09_18T14_15_46.670153", "path": ["**/details_harness|hendrycksTest-high_school_european_history|5_2023-09-18T14-15-46.670153.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_european_history|5_2023-09-18T14-15-46.670153.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_geography_5", "data_files": [{"split": "2023_09_18T14_15_46.670153", "path": ["**/details_harness|hendrycksTest-high_school_geography|5_2023-09-18T14-15-46.670153.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_geography|5_2023-09-18T14-15-46.670153.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_government_and_politics_5", "data_files": [{"split": "2023_09_18T14_15_46.670153", "path": ["**/details_harness|hendrycksTest-high_school_government_and_politics|5_2023-09-18T14-15-46.670153.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_government_and_politics|5_2023-09-18T14-15-46.670153.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_macroeconomics_5", "data_files": [{"split": "2023_09_18T14_15_46.670153", "path": ["**/details_harness|hendrycksTest-high_school_macroeconomics|5_2023-09-18T14-15-46.670153.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_macroeconomics|5_2023-09-18T14-15-46.670153.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_mathematics_5", "data_files": [{"split": "2023_09_18T14_15_46.670153", "path": ["**/details_harness|hendrycksTest-high_school_mathematics|5_2023-09-18T14-15-46.670153.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_mathematics|5_2023-09-18T14-15-46.670153.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_microeconomics_5", "data_files": [{"split": "2023_09_18T14_15_46.670153", "path": ["**/details_harness|hendrycksTest-high_school_microeconomics|5_2023-09-18T14-15-46.670153.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_microeconomics|5_2023-09-18T14-15-46.670153.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_physics_5", "data_files": [{"split": "2023_09_18T14_15_46.670153", "path": ["**/details_harness|hendrycksTest-high_school_physics|5_2023-09-18T14-15-46.670153.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_physics|5_2023-09-18T14-15-46.670153.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_psychology_5", "data_files": [{"split": "2023_09_18T14_15_46.670153", "path": ["**/details_harness|hendrycksTest-high_school_psychology|5_2023-09-18T14-15-46.670153.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_psychology|5_2023-09-18T14-15-46.670153.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_statistics_5", "data_files": [{"split": "2023_09_18T14_15_46.670153", "path": ["**/details_harness|hendrycksTest-high_school_statistics|5_2023-09-18T14-15-46.670153.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_statistics|5_2023-09-18T14-15-46.670153.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_us_history_5", "data_files": [{"split": "2023_09_18T14_15_46.670153", "path": ["**/details_harness|hendrycksTest-high_school_us_history|5_2023-09-18T14-15-46.670153.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_us_history|5_2023-09-18T14-15-46.670153.parquet"]}]}, {"config_name": "harness_hendrycksTest_high_school_world_history_5", "data_files": [{"split": "2023_09_18T14_15_46.670153", "path": ["**/details_harness|hendrycksTest-high_school_world_history|5_2023-09-18T14-15-46.670153.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-high_school_world_history|5_2023-09-18T14-15-46.670153.parquet"]}]}, {"config_name": "harness_hendrycksTest_human_aging_5", "data_files": [{"split": "2023_09_18T14_15_46.670153", "path": ["**/details_harness|hendrycksTest-human_aging|5_2023-09-18T14-15-46.670153.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-human_aging|5_2023-09-18T14-15-46.670153.parquet"]}]}, {"config_name": "harness_hendrycksTest_human_sexuality_5", "data_files": [{"split": "2023_09_18T14_15_46.670153", "path": ["**/details_harness|hendrycksTest-human_sexuality|5_2023-09-18T14-15-46.670153.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-human_sexuality|5_2023-09-18T14-15-46.670153.parquet"]}]}, {"config_name": "harness_hendrycksTest_international_law_5", "data_files": [{"split": "2023_09_18T14_15_46.670153", "path": ["**/details_harness|hendrycksTest-international_law|5_2023-09-18T14-15-46.670153.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-international_law|5_2023-09-18T14-15-46.670153.parquet"]}]}, {"config_name": "harness_hendrycksTest_jurisprudence_5", "data_files": [{"split": "2023_09_18T14_15_46.670153", "path": ["**/details_harness|hendrycksTest-jurisprudence|5_2023-09-18T14-15-46.670153.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-jurisprudence|5_2023-09-18T14-15-46.670153.parquet"]}]}, {"config_name": "harness_hendrycksTest_logical_fallacies_5", "data_files": [{"split": "2023_09_18T14_15_46.670153", "path": ["**/details_harness|hendrycksTest-logical_fallacies|5_2023-09-18T14-15-46.670153.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-logical_fallacies|5_2023-09-18T14-15-46.670153.parquet"]}]}, {"config_name": "harness_hendrycksTest_machine_learning_5", "data_files": [{"split": "2023_09_18T14_15_46.670153", "path": ["**/details_harness|hendrycksTest-machine_learning|5_2023-09-18T14-15-46.670153.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-machine_learning|5_2023-09-18T14-15-46.670153.parquet"]}]}, {"config_name": "harness_hendrycksTest_management_5", "data_files": [{"split": "2023_09_18T14_15_46.670153", "path": ["**/details_harness|hendrycksTest-management|5_2023-09-18T14-15-46.670153.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-management|5_2023-09-18T14-15-46.670153.parquet"]}]}, {"config_name": "harness_hendrycksTest_marketing_5", "data_files": [{"split": "2023_09_18T14_15_46.670153", "path": ["**/details_harness|hendrycksTest-marketing|5_2023-09-18T14-15-46.670153.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-marketing|5_2023-09-18T14-15-46.670153.parquet"]}]}, {"config_name": "harness_hendrycksTest_medical_genetics_5", "data_files": [{"split": "2023_09_18T14_15_46.670153", "path": ["**/details_harness|hendrycksTest-medical_genetics|5_2023-09-18T14-15-46.670153.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-medical_genetics|5_2023-09-18T14-15-46.670153.parquet"]}]}, {"config_name": "harness_hendrycksTest_miscellaneous_5", "data_files": [{"split": "2023_09_18T14_15_46.670153", "path": ["**/details_harness|hendrycksTest-miscellaneous|5_2023-09-18T14-15-46.670153.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-miscellaneous|5_2023-09-18T14-15-46.670153.parquet"]}]}, {"config_name": "harness_hendrycksTest_moral_disputes_5", "data_files": [{"split": "2023_09_18T14_15_46.670153", "path": ["**/details_harness|hendrycksTest-moral_disputes|5_2023-09-18T14-15-46.670153.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-moral_disputes|5_2023-09-18T14-15-46.670153.parquet"]}]}, {"config_name": "harness_hendrycksTest_moral_scenarios_5", "data_files": [{"split": "2023_09_18T14_15_46.670153", "path": ["**/details_harness|hendrycksTest-moral_scenarios|5_2023-09-18T14-15-46.670153.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-moral_scenarios|5_2023-09-18T14-15-46.670153.parquet"]}]}, {"config_name": "harness_hendrycksTest_nutrition_5", "data_files": [{"split": "2023_09_18T14_15_46.670153", "path": ["**/details_harness|hendrycksTest-nutrition|5_2023-09-18T14-15-46.670153.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-nutrition|5_2023-09-18T14-15-46.670153.parquet"]}]}, {"config_name": "harness_hendrycksTest_philosophy_5", "data_files": [{"split": "2023_09_18T14_15_46.670153", "path": ["**/details_harness|hendrycksTest-philosophy|5_2023-09-18T14-15-46.670153.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-philosophy|5_2023-09-18T14-15-46.670153.parquet"]}]}, {"config_name": "harness_hendrycksTest_prehistory_5", "data_files": [{"split": "2023_09_18T14_15_46.670153", "path": ["**/details_harness|hendrycksTest-prehistory|5_2023-09-18T14-15-46.670153.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-prehistory|5_2023-09-18T14-15-46.670153.parquet"]}]}, {"config_name": "harness_hendrycksTest_professional_accounting_5", "data_files": [{"split": "2023_09_18T14_15_46.670153", "path": ["**/details_harness|hendrycksTest-professional_accounting|5_2023-09-18T14-15-46.670153.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-professional_accounting|5_2023-09-18T14-15-46.670153.parquet"]}]}, {"config_name": "harness_hendrycksTest_professional_law_5", "data_files": [{"split": "2023_09_18T14_15_46.670153", "path": ["**/details_harness|hendrycksTest-professional_law|5_2023-09-18T14-15-46.670153.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-professional_law|5_2023-09-18T14-15-46.670153.parquet"]}]}, {"config_name": "harness_hendrycksTest_professional_medicine_5", "data_files": [{"split": "2023_09_18T14_15_46.670153", "path": ["**/details_harness|hendrycksTest-professional_medicine|5_2023-09-18T14-15-46.670153.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-professional_medicine|5_2023-09-18T14-15-46.670153.parquet"]}]}, {"config_name": "harness_hendrycksTest_professional_psychology_5", "data_files": [{"split": "2023_09_18T14_15_46.670153", "path": ["**/details_harness|hendrycksTest-professional_psychology|5_2023-09-18T14-15-46.670153.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-professional_psychology|5_2023-09-18T14-15-46.670153.parquet"]}]}, {"config_name": "harness_hendrycksTest_public_relations_5", "data_files": [{"split": "2023_09_18T14_15_46.670153", "path": ["**/details_harness|hendrycksTest-public_relations|5_2023-09-18T14-15-46.670153.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-public_relations|5_2023-09-18T14-15-46.670153.parquet"]}]}, {"config_name": "harness_hendrycksTest_security_studies_5", "data_files": [{"split": "2023_09_18T14_15_46.670153", "path": ["**/details_harness|hendrycksTest-security_studies|5_2023-09-18T14-15-46.670153.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-security_studies|5_2023-09-18T14-15-46.670153.parquet"]}]}, {"config_name": "harness_hendrycksTest_sociology_5", "data_files": [{"split": "2023_09_18T14_15_46.670153", "path": ["**/details_harness|hendrycksTest-sociology|5_2023-09-18T14-15-46.670153.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-sociology|5_2023-09-18T14-15-46.670153.parquet"]}]}, {"config_name": "harness_hendrycksTest_us_foreign_policy_5", "data_files": [{"split": "2023_09_18T14_15_46.670153", "path": ["**/details_harness|hendrycksTest-us_foreign_policy|5_2023-09-18T14-15-46.670153.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-us_foreign_policy|5_2023-09-18T14-15-46.670153.parquet"]}]}, {"config_name": "harness_hendrycksTest_virology_5", "data_files": [{"split": "2023_09_18T14_15_46.670153", "path": ["**/details_harness|hendrycksTest-virology|5_2023-09-18T14-15-46.670153.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-virology|5_2023-09-18T14-15-46.670153.parquet"]}]}, {"config_name": "harness_hendrycksTest_world_religions_5", "data_files": [{"split": "2023_09_18T14_15_46.670153", "path": ["**/details_harness|hendrycksTest-world_religions|5_2023-09-18T14-15-46.670153.parquet"]}, {"split": "latest", "path": ["**/details_harness|hendrycksTest-world_religions|5_2023-09-18T14-15-46.670153.parquet"]}]}, {"config_name": "harness_truthfulqa_mc_0", "data_files": [{"split": "2023_09_18T14_15_46.670153", "path": ["**/details_harness|truthfulqa:mc|0_2023-09-18T14-15-46.670153.parquet"]}, {"split": "latest", "path": ["**/details_harness|truthfulqa:mc|0_2023-09-18T14-15-46.670153.parquet"]}]}, {"config_name": "harness_winogrande_5", "data_files": [{"split": "2023_10_28T00_17_42.072889", "path": ["**/details_harness|winogrande|5_2023-10-28T00-17-42.072889.parquet"]}, {"split": "2023_10_28T02_33_59.939371", "path": ["**/details_harness|winogrande|5_2023-10-28T02-33-59.939371.parquet"]}, {"split": "latest", "path": ["**/details_harness|winogrande|5_2023-10-28T02-33-59.939371.parquet"]}]}, {"config_name": "results", "data_files": [{"split": "2023_09_18T14_15_46.670153", "path": ["results_2023-09-18T14-15-46.670153.parquet"]}, {"split": "2023_10_28T00_17_42.072889", "path": ["results_2023-10-28T00-17-42.072889.parquet"]}, {"split": "2023_10_28T02_33_59.939371", "path": ["results_2023-10-28T02-33-59.939371.parquet"]}, {"split": "latest", "path": ["results_2023-10-28T02-33-59.939371.parquet"]}]}]}
|
2023-10-28T01:34:08+00:00
|
[] |
[] |
TAGS
#region-us
|
# Dataset Card for Evaluation run of lgaalves/llama-2-13b-hf-platypus
## Dataset Description
- Homepage:
- Repository: URL
- Paper:
- Leaderboard: URL
- Point of Contact: clementine@URL
### Dataset Summary
Dataset automatically created during the evaluation run of model lgaalves/llama-2-13b-hf-platypus on the Open LLM Leaderboard.
The dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.
The dataset has been created from 3 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The "train" split is always pointing to the latest results.
An additional configuration "results" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the Open LLM Leaderboard).
To load the details from a run, you can for instance do the following:
## Latest results
These are the latest results from run 2023-10-28T02:33:59.939371(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the "latest" split for each eval):
### Supported Tasks and Leaderboards
### Languages
## Dataset Structure
### Data Instances
### Data Fields
### Data Splits
## Dataset Creation
### Curation Rationale
### Source Data
#### Initial Data Collection and Normalization
#### Who are the source language producers?
### Annotations
#### Annotation process
#### Who are the annotators?
### Personal and Sensitive Information
## Considerations for Using the Data
### Social Impact of Dataset
### Discussion of Biases
### Other Known Limitations
## Additional Information
### Dataset Curators
### Licensing Information
### Contributions
|
[
"# Dataset Card for Evaluation run of lgaalves/llama-2-13b-hf-platypus",
"## Dataset Description\n\n- Homepage: \n- Repository: URL\n- Paper: \n- Leaderboard: URL\n- Point of Contact: clementine@URL",
"### Dataset Summary\n\nDataset automatically created during the evaluation run of model lgaalves/llama-2-13b-hf-platypus on the Open LLM Leaderboard.\n\nThe dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.\n\nThe dataset has been created from 3 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The \"train\" split is always pointing to the latest results.\n\nAn additional configuration \"results\" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the Open LLM Leaderboard).\n\nTo load the details from a run, you can for instance do the following:",
"## Latest results\n\nThese are the latest results from run 2023-10-28T02:33:59.939371(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the \"latest\" split for each eval):",
"### Supported Tasks and Leaderboards",
"### Languages",
"## Dataset Structure",
"### Data Instances",
"### Data Fields",
"### Data Splits",
"## Dataset Creation",
"### Curation Rationale",
"### Source Data",
"#### Initial Data Collection and Normalization",
"#### Who are the source language producers?",
"### Annotations",
"#### Annotation process",
"#### Who are the annotators?",
"### Personal and Sensitive Information",
"## Considerations for Using the Data",
"### Social Impact of Dataset",
"### Discussion of Biases",
"### Other Known Limitations",
"## Additional Information",
"### Dataset Curators",
"### Licensing Information",
"### Contributions"
] |
[
"TAGS\n#region-us \n",
"# Dataset Card for Evaluation run of lgaalves/llama-2-13b-hf-platypus",
"## Dataset Description\n\n- Homepage: \n- Repository: URL\n- Paper: \n- Leaderboard: URL\n- Point of Contact: clementine@URL",
"### Dataset Summary\n\nDataset automatically created during the evaluation run of model lgaalves/llama-2-13b-hf-platypus on the Open LLM Leaderboard.\n\nThe dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.\n\nThe dataset has been created from 3 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The \"train\" split is always pointing to the latest results.\n\nAn additional configuration \"results\" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the Open LLM Leaderboard).\n\nTo load the details from a run, you can for instance do the following:",
"## Latest results\n\nThese are the latest results from run 2023-10-28T02:33:59.939371(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the \"latest\" split for each eval):",
"### Supported Tasks and Leaderboards",
"### Languages",
"## Dataset Structure",
"### Data Instances",
"### Data Fields",
"### Data Splits",
"## Dataset Creation",
"### Curation Rationale",
"### Source Data",
"#### Initial Data Collection and Normalization",
"#### Who are the source language producers?",
"### Annotations",
"#### Annotation process",
"#### Who are the annotators?",
"### Personal and Sensitive Information",
"## Considerations for Using the Data",
"### Social Impact of Dataset",
"### Discussion of Biases",
"### Other Known Limitations",
"## Additional Information",
"### Dataset Curators",
"### Licensing Information",
"### Contributions"
] |
[
6,
26,
31,
174,
67,
10,
4,
6,
6,
5,
5,
5,
7,
4,
10,
10,
5,
5,
9,
8,
8,
7,
8,
7,
5,
6,
6,
5
] |
[
"passage: TAGS\n#region-us \n# Dataset Card for Evaluation run of lgaalves/llama-2-13b-hf-platypus## Dataset Description\n\n- Homepage: \n- Repository: URL\n- Paper: \n- Leaderboard: URL\n- Point of Contact: clementine@URL### Dataset Summary\n\nDataset automatically created during the evaluation run of model lgaalves/llama-2-13b-hf-platypus on the Open LLM Leaderboard.\n\nThe dataset is composed of 64 configuration, each one coresponding to one of the evaluated task.\n\nThe dataset has been created from 3 run(s). Each run can be found as a specific split in each configuration, the split being named using the timestamp of the run.The \"train\" split is always pointing to the latest results.\n\nAn additional configuration \"results\" store all the aggregated results of the run (and is used to compute and display the agregated metrics on the Open LLM Leaderboard).\n\nTo load the details from a run, you can for instance do the following:## Latest results\n\nThese are the latest results from run 2023-10-28T02:33:59.939371(note that their might be results for other tasks in the repos if successive evals didn't cover the same tasks. You find each in the results and the \"latest\" split for each eval):### Supported Tasks and Leaderboards### Languages## Dataset Structure### Data Instances### Data Fields### Data Splits## Dataset Creation### Curation Rationale### Source Data#### Initial Data Collection and Normalization#### Who are the source language producers?### Annotations#### Annotation process#### Who are the annotators?### Personal and Sensitive Information## Considerations for Using the Data### Social Impact of Dataset### Discussion of Biases### Other Known Limitations## Additional Information### Dataset Curators### Licensing Information### Contributions"
] |
6ea21f81bb8e58b70bab0737d3fb49e63a733be7
|
# Dataset Card for "l_cls_labelled_from_distilbert_masking_heaps"
[More Information needed](https://github.com/huggingface/datasets/blob/main/CONTRIBUTING.md#how-to-contribute-to-the-dataset-cards)
|
johannes-garstenauer/l_cls_labelled_from_distilbert_masking_heaps
|
[
"region:us"
] |
2023-09-18T13:16:27+00:00
|
{"dataset_info": {"features": [{"name": "last_cls", "sequence": "float32"}, {"name": "label", "dtype": "int64"}], "splits": [{"name": "train", "num_bytes": 3084000, "num_examples": 1000}], "download_size": 0, "dataset_size": 3084000}}
|
2023-09-18T13:16:59+00:00
|
[] |
[] |
TAGS
#region-us
|
# Dataset Card for "l_cls_labelled_from_distilbert_masking_heaps"
More Information needed
|
[
"# Dataset Card for \"l_cls_labelled_from_distilbert_masking_heaps\"\n\nMore Information needed"
] |
[
"TAGS\n#region-us \n",
"# Dataset Card for \"l_cls_labelled_from_distilbert_masking_heaps\"\n\nMore Information needed"
] |
[
6,
29
] |
[
"passage: TAGS\n#region-us \n# Dataset Card for \"l_cls_labelled_from_distilbert_masking_heaps\"\n\nMore Information needed"
] |
ea7a32be60d0d75053264f457e08c814cbf45623
|
# Dataset Card for "amazon_product_reviews_datafiniti"
[More Information needed](https://github.com/huggingface/datasets/blob/main/CONTRIBUTING.md#how-to-contribute-to-the-dataset-cards)
|
m-ric/amazon_product_reviews_datafiniti
|
[
"task_categories:text-classification",
"task_categories:question-answering",
"task_categories:feature-extraction",
"size_categories:1K<n<10K",
"language:en",
"region:us"
] |
2023-09-18T13:16:55+00:00
|
{"language": ["en"], "size_categories": ["1K<n<10K"], "task_categories": ["text-classification", "question-answering", "feature-extraction"], "pretty_name": "Amazon Product Reviews by Datafiniti", "configs": [{"config_name": "default", "data_files": [{"split": "train", "path": "data/train-*"}, {"split": "test", "path": "data/test-*"}]}], "dataset_info": {"features": [{"name": "brand", "dtype": {"class_label": {"names": {"0": "Amazon", "1": "AmazonBasics", "2": "Amazonbasics"}}}}, {"name": "primaryCategories", "dtype": "string"}, {"name": "reviews.numHelpful", "dtype": "float64"}, {"name": "reviews.rating", "dtype": "int64"}, {"name": "reviews.text", "dtype": "string"}], "splits": [{"name": "train", "num_bytes": 1107781.5, "num_examples": 6000}, {"name": "test", "num_bytes": 369260.5, "num_examples": 2000}], "download_size": 704792, "dataset_size": 1477042}}
|
2023-09-26T13:12:40+00:00
|
[] |
[
"en"
] |
TAGS
#task_categories-text-classification #task_categories-question-answering #task_categories-feature-extraction #size_categories-1K<n<10K #language-English #region-us
|
# Dataset Card for "amazon_product_reviews_datafiniti"
More Information needed
|
[
"# Dataset Card for \"amazon_product_reviews_datafiniti\"\n\nMore Information needed"
] |
[
"TAGS\n#task_categories-text-classification #task_categories-question-answering #task_categories-feature-extraction #size_categories-1K<n<10K #language-English #region-us \n",
"# Dataset Card for \"amazon_product_reviews_datafiniti\"\n\nMore Information needed"
] |
[
57,
21
] |
[
"passage: TAGS\n#task_categories-text-classification #task_categories-question-answering #task_categories-feature-extraction #size_categories-1K<n<10K #language-English #region-us \n# Dataset Card for \"amazon_product_reviews_datafiniti\"\n\nMore Information needed"
] |
40607285f73f84168de9346019ee04ff94188434
|
# Dataset of Kita Ikuyo
This is the dataset of Kita Ikuyo, containing 296 images and their tags.
Images are crawled from many sites (e.g. danbooru, pixiv, zerochan ...), the auto-crawling system is powered by [DeepGHS Team](https://github.com/deepghs)([huggingface organization](https://huggingface.co/deepghs)).
| Name | Images | Download | Description |
|:------------|---------:|:------------------------------------|:-------------------------------------------------------------------------|
| raw | 296 | [Download](dataset-raw.zip) | Raw data with meta information. |
| raw-stage3 | 650 | [Download](dataset-raw-stage3.zip) | 3-stage cropped raw data with meta information. |
| 384x512 | 296 | [Download](dataset-384x512.zip) | 384x512 aligned dataset. |
| 512x512 | 296 | [Download](dataset-512x512.zip) | 512x512 aligned dataset. |
| 512x704 | 296 | [Download](dataset-512x704.zip) | 512x704 aligned dataset. |
| 640x640 | 296 | [Download](dataset-640x640.zip) | 640x640 aligned dataset. |
| 640x880 | 296 | [Download](dataset-640x880.zip) | 640x880 aligned dataset. |
| stage3-640 | 650 | [Download](dataset-stage3-640.zip) | 3-stage cropped dataset with the shorter side not exceeding 640 pixels. |
| stage3-800 | 650 | [Download](dataset-stage3-800.zip) | 3-stage cropped dataset with the shorter side not exceeding 800 pixels. |
| stage3-1200 | 650 | [Download](dataset-stage3-1200.zip) | 3-stage cropped dataset with the shorter side not exceeding 1200 pixels. |
|
CyberHarem/kita_ikuyo_bocchitherock
|
[
"task_categories:text-to-image",
"size_categories:n<1K",
"license:mit",
"art",
"not-for-all-audiences",
"region:us"
] |
2023-09-18T13:17:41+00:00
|
{"license": "mit", "size_categories": ["n<1K"], "task_categories": ["text-to-image"], "tags": ["art", "not-for-all-audiences"]}
|
2023-09-18T13:22:56+00:00
|
[] |
[] |
TAGS
#task_categories-text-to-image #size_categories-n<1K #license-mit #art #not-for-all-audiences #region-us
|
Dataset of Kita Ikuyo
=====================
This is the dataset of Kita Ikuyo, containing 296 images and their tags.
Images are crawled from many sites (e.g. danbooru, pixiv, zerochan ...), the auto-crawling system is powered by DeepGHS Team(huggingface organization).
|
[] |
[
"TAGS\n#task_categories-text-to-image #size_categories-n<1K #license-mit #art #not-for-all-audiences #region-us \n"
] |
[
44
] |
[
"passage: TAGS\n#task_categories-text-to-image #size_categories-n<1K #license-mit #art #not-for-all-audiences #region-us \n"
] |
b9e4f50df4fedbd0f4b27acefbc5d7e9817529ef
|
# Dataset Card for "iCliniq-llama2-7k"
[More Information needed](https://github.com/huggingface/datasets/blob/main/CONTRIBUTING.md#how-to-contribute-to-the-dataset-cards)
|
DaisyStar004/iCliniq-llama2-7k
|
[
"region:us"
] |
2023-09-18T13:18:34+00:00
|
{"dataset_info": {"features": [{"name": "text", "dtype": "string"}], "splits": [{"name": "train", "num_bytes": 7229044, "num_examples": 7000}], "download_size": 4177341, "dataset_size": 7229044}, "configs": [{"config_name": "default", "data_files": [{"split": "train", "path": "data/train-*"}]}]}
|
2023-09-18T13:18:37+00:00
|
[] |
[] |
TAGS
#region-us
|
# Dataset Card for "iCliniq-llama2-7k"
More Information needed
|
[
"# Dataset Card for \"iCliniq-llama2-7k\"\n\nMore Information needed"
] |
[
"TAGS\n#region-us \n",
"# Dataset Card for \"iCliniq-llama2-7k\"\n\nMore Information needed"
] |
[
6,
20
] |
[
"passage: TAGS\n#region-us \n# Dataset Card for \"iCliniq-llama2-7k\"\n\nMore Information needed"
] |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.