hexsha
stringlengths 40
40
| size
int64 6
14.9M
| ext
stringclasses 1
value | lang
stringclasses 1
value | max_stars_repo_path
stringlengths 6
260
| max_stars_repo_name
stringlengths 6
119
| max_stars_repo_head_hexsha
stringlengths 40
41
| max_stars_repo_licenses
list | max_stars_count
int64 1
191k
⌀ | max_stars_repo_stars_event_min_datetime
stringlengths 24
24
⌀ | max_stars_repo_stars_event_max_datetime
stringlengths 24
24
⌀ | max_issues_repo_path
stringlengths 6
260
| max_issues_repo_name
stringlengths 6
119
| max_issues_repo_head_hexsha
stringlengths 40
41
| max_issues_repo_licenses
list | max_issues_count
int64 1
67k
⌀ | max_issues_repo_issues_event_min_datetime
stringlengths 24
24
⌀ | max_issues_repo_issues_event_max_datetime
stringlengths 24
24
⌀ | max_forks_repo_path
stringlengths 6
260
| max_forks_repo_name
stringlengths 6
119
| max_forks_repo_head_hexsha
stringlengths 40
41
| max_forks_repo_licenses
list | max_forks_count
int64 1
105k
⌀ | max_forks_repo_forks_event_min_datetime
stringlengths 24
24
⌀ | max_forks_repo_forks_event_max_datetime
stringlengths 24
24
⌀ | avg_line_length
float64 2
1.04M
| max_line_length
int64 2
11.2M
| alphanum_fraction
float64 0
1
| cells
list | cell_types
list | cell_type_groups
list |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
cbeeef3ebddea43d68072b05a20009264dba3968
| 14,828 |
ipynb
|
Jupyter Notebook
|
docs/plotting/plotly/Range Slider and Selector in Python.ipynb
|
zhangshoug/czipline
|
6bce0abd4772443547f44669c0adb2b5c63f64db
|
[
"Apache-2.0"
] | 9 |
2019-05-18T10:44:48.000Z
|
2022-01-01T15:12:49.000Z
|
docs/plotting/plotly/Range Slider and Selector in Python.ipynb
|
yuanyichuangzhi/czipline
|
6bce0abd4772443547f44669c0adb2b5c63f64db
|
[
"Apache-2.0"
] | null | null | null |
docs/plotting/plotly/Range Slider and Selector in Python.ipynb
|
yuanyichuangzhi/czipline
|
6bce0abd4772443547f44669c0adb2b5c63f64db
|
[
"Apache-2.0"
] | 10 |
2019-05-18T10:58:55.000Z
|
2022-03-24T13:37:17.000Z
| 31.616205 | 176 | 0.383531 |
[
[
[
"Note: range sliders and range selectors are available in version 1.9.7+\n\nRun pip install plotly --upgrade to update your Plotly version",
"_____no_output_____"
]
],
[
[
"import plotly\n\nplotly.__version__",
"_____no_output_____"
]
],
[
[
"## Basic Range Slider and Range Selectors",
"_____no_output_____"
]
],
[
[
"from cswd import query_adjusted_pricing\nOHLCV = ['open','high','low','close','volume']",
"_____no_output_____"
],
[
"df = query_adjusted_pricing('000001','2007-10-1','2009-4-1',OHLCV,True)",
"_____no_output_____"
],
[
"import plotly.plotly as py\nimport plotly.graph_objs as go \n\nfrom datetime import datetime\n\ntrace = go.Scatter(x=df.index,\n y=df.high)\n\ndata = [trace]\nlayout = dict(\n title='带滑块和选择器的时间序列',\n xaxis=dict(\n rangeselector=dict(\n buttons=list([\n dict(count=1,\n label='1w',\n step='week',\n stepmode='backward'),\n dict(count=6,\n label='6m',\n step='month',\n stepmode='backward'),\n dict(count=1,\n label='YTD',\n step='year',\n stepmode='todate'),\n dict(count=1,\n label='1y',\n step='year',\n stepmode='backward'),\n dict(step='all')\n ])\n ),\n rangeslider=dict(),\n type='date'\n )\n)\n\nfig = dict(data=data, layout=layout)\npy.iplot(fig)",
"_____no_output_____"
]
],
[
[
"## Range Slider with Vertically Stacked Subplots",
"_____no_output_____"
]
],
[
[
"import plotly.plotly as py\nimport plotly.graph_objs as go\n\ntrace1 = go.Scatter(\n x = [\"2013-01-15\", \"2013-01-29\", \"2013-02-26\", \"2013-04-19\", \"2013-07-02\", \"2013-08-27\",\n \"2013-10-22\", \"2014-01-20\", \"2014-05-05\", \"2014-07-01\", \"2015-02-09\", \"2015-04-13\",\n \"2015-05-13\", \"2015-06-08\", \"2015-08-05\", \"2016-02-25\"], \n y = [\"8\", \"3\", \"2\", \"10\", \"5\", \"5\", \"6\", \"8\", \"3\", \"3\", \"7\", \"5\", \"10\", \"10\", \"9\", \"14\"], \n name = \"var0\", \n text = [\"8\", \"3\", \"2\", \"10\", \"5\", \"5\", \"6\", \"8\", \"3\", \"3\", \"7\", \"5\", \"10\", \"10\", \"9\", \"14\"], \n yaxis = \"y\", \n)\n\ntrace2 = go.Scatter(\n x = [\"2015-04-13\", \"2015-05-13\", \"2015-06-08\", \"2015-08-05\", \"2016-02-25\"], \n y = [\"53.0\", \"69.0\", \"89.0\", \"41.0\", \"41.0\"], \n name = \"var1\", \n text = [\"53.0\", \"69.0\", \"89.0\", \"41.0\", \"41.0\"], \n yaxis = \"y2\", \n)\n\ntrace3 = go.Scatter(\n x = [\"2013-01-29\", \"2013-02-26\", \"2013-04-19\", \"2013-07-02\", \"2013-08-27\", \"2013-10-22\",\n \"2014-01-20\", \"2014-04-09\", \"2014-05-05\", \"2014-07-01\", \"2014-09-30\", \"2015-02-09\",\n \"2015-04-13\", \"2015-06-08\", \"2016-02-25\"], \n y = [\"9.6\", \"4.6\", \"2.7\", \"8.3\", \"18\", \"7.3\", \"3\", \"7.5\", \"1.0\", \"0.5\", \"2.8\", \"9.2\",\n \"13\", \"5.8\", \"6.9\"], \n name = \"var2\", \n text = [\"9.6\", \"4.6\", \"2.7\", \"8.3\", \"18\", \"7.3\", \"3\", \"7.5\", \"1.0\", \"0.5\", \"2.8\", \"9.2\",\n \"13\", \"5.8\", \"6.9\"], \n yaxis = \"y3\",\n)\n\ntrace4 = go.Scatter(\n x = [\"2013-01-29\", \"2013-02-26\", \"2013-04-19\", \"2013-07-02\", \"2013-08-27\", \"2013-10-22\",\n \"2014-01-20\", \"2014-04-09\", \"2014-05-05\", \"2014-07-01\", \"2014-09-30\", \"2015-02-09\",\n \"2015-04-13\", \"2015-06-08\", \"2016-02-25\"], \n y = [\"6.9\", \"7.5\", \"7.3\", \"7.3\", \"6.9\", \"7.1\", \"8\", \"7.8\", \"7.4\", \"7.9\", \"7.9\", \"7.6\",\n \"7.2\", \"7.2\", \"8.0\"], \n name = \"var3\", \n text = [\"6.9\", \"7.5\", \"7.3\", \"7.3\", \"6.9\", \"7.1\", \"8\", \"7.8\", \"7.4\", \"7.9\", \"7.9\", \"7.6\",\n \"7.2\", \"7.2\", \"8.0\"], \n yaxis = \"y4\",\n)\n\ntrace5 = go.Scatter(\n x = [\"2013-02-26\", \"2013-07-02\", \"2013-09-26\", \"2013-10-22\", \"2013-12-04\", \"2014-01-02\",\n \"2014-01-20\", \"2014-05-05\", \"2014-07-01\", \"2015-02-09\", \"2015-05-05\"], \n y = [\"290\", \"1078\", \"263\", \"407\", \"660\", \"740\", \"33\", \"374\", \"95\", \"734\", \"3000\"], \n name = \"var4\", \n text = [\"290\", \"1078\", \"263\", \"407\", \"660\", \"740\", \"33\", \"374\", \"95\", \"734\", \"3000\"], \n yaxis = \"y5\",\n)\n\ndata = go.Data([trace1, trace2, trace3, trace4, trace5])\n\n# style all the traces\nfor k in range(len(data)):\n data[k].update(\n {\n \"type\": \"scatter\",\n \"hoverinfo\": \"name+x+text\",\n \"line\": {\"width\": 0.5}, \n \"marker\": {\"size\": 8},\n \"mode\": \"lines+markers\",\n \"showlegend\": False\n }\n )\n\nlayout = {\n \"annotations\": [\n {\n \"x\": \"2013-06-01\", \n \"y\": 0, \n \"arrowcolor\": \"rgba(63, 81, 181, 0.2)\", \n \"arrowsize\": 0.3, \n \"ax\": 0, \n \"ay\": 30, \n \"text\": \"state1\", \n \"xref\": \"x\", \n \"yanchor\": \"bottom\", \n \"yref\": \"y\"\n }, \n {\n \"x\": \"2014-09-13\", \n \"y\": 0, \n \"arrowcolor\": \"rgba(76, 175, 80, 0.1)\", \n \"arrowsize\": 0.3, \n \"ax\": 0,\n \"ay\": 30,\n \"text\": \"state2\",\n \"xref\": \"x\", \n \"yanchor\": \"bottom\", \n \"yref\": \"y\"\n }\n ], \n \"dragmode\": \"zoom\", \n \"hovermode\": \"x\", \n \"legend\": {\"traceorder\": \"reversed\"}, \n \"margin\": {\n \"t\": 100, \n \"b\": 100\n }, \n \"shapes\": [\n {\n \"fillcolor\": \"rgba(63, 81, 181, 0.2)\", \n \"line\": {\"width\": 0}, \n \"type\": \"rect\", \n \"x0\": \"2013-01-15\", \n \"x1\": \"2013-10-17\", \n \"xref\": \"x\", \n \"y0\": 0, \n \"y1\": 0.95, \n \"yref\": \"paper\"\n }, \n {\n \"fillcolor\": \"rgba(76, 175, 80, 0.1)\", \n \"line\": {\"width\": 0}, \n \"type\": \"rect\", \n \"x0\": \"2013-10-22\", \n \"x1\": \"2015-08-05\", \n \"xref\": \"x\", \n \"y0\": 0, \n \"y1\": 0.95, \n \"yref\": \"paper\"\n }\n ], \n \"xaxis\": {\n \"autorange\": True, \n \"range\": [\"2012-10-31 18:36:37.3129\", \"2016-05-10 05:23:22.6871\"], \n \"rangeslider\": {\n \"autorange\": True, \n \"range\": [\"2012-10-31 18:36:37.3129\", \"2016-05-10 05:23:22.6871\"]\n }, \n \"type\": \"date\"\n }, \n \"yaxis\": {\n \"anchor\": \"x\", \n \"autorange\": True, \n \"domain\": [0, 0.2], \n \"linecolor\": \"#673ab7\", \n \"mirror\": True, \n \"range\": [-60.0858369099, 28.4406294707], \n \"showline\": True, \n \"side\": \"right\", \n \"tickfont\": {\"color\": \"#673ab7\"}, \n \"tickmode\": \"auto\", \n \"ticks\": \"\", \n \"titlefont\": {\"color\": \"#673ab7\"}, \n \"type\": \"linear\", \n \"zeroline\": False\n }, \n \"yaxis2\": {\n \"anchor\": \"x\", \n \"autorange\": True, \n \"domain\": [0.2, 0.4], \n \"linecolor\": \"#E91E63\", \n \"mirror\": True, \n \"range\": [29.3787777032, 100.621222297], \n \"showline\": True, \n \"side\": \"right\", \n \"tickfont\": {\"color\": \"#E91E63\"}, \n \"tickmode\": \"auto\", \n \"ticks\": \"\", \n \"titlefont\": {\"color\": \"#E91E63\"}, \n \"type\": \"linear\", \n \"zeroline\": False\n }, \n \"yaxis3\": {\n \"anchor\": \"x\", \n \"autorange\": True, \n \"domain\": [0.4, 0.6], \n \"linecolor\": \"#795548\", \n \"mirror\": True, \n \"range\": [-3.73690396239, 22.2369039624], \n \"showline\": True, \n \"side\": \"right\", \n \"tickfont\": {\"color\": \"#795548\"}, \n \"tickmode\": \"auto\", \n \"ticks\": \"\", \n \"title\": \"mg/L\", \n \"titlefont\": {\"color\": \"#795548\"}, \n \"type\": \"linear\", \n \"zeroline\": False\n }, \n \"yaxis4\": {\n \"anchor\": \"x\", \n \"autorange\": True, \n \"domain\": [0.6, 0.8], \n \"linecolor\": \"#607d8b\", \n \"mirror\": True, \n \"range\": [6.63368032236, 8.26631967764], \n \"showline\": True, \n \"side\": \"right\", \n \"tickfont\": {\"color\": \"#607d8b\"}, \n \"tickmode\": \"auto\", \n \"ticks\": \"\", \n \"title\": \"mmol/L\", \n \"titlefont\": {\"color\": \"#607d8b\"}, \n \"type\": \"linear\", \n \"zeroline\": False\n }, \n \"yaxis5\": {\n \"anchor\": \"x\", \n \"autorange\": True, \n \"domain\": [0.8, 1], \n \"linecolor\": \"#2196F3\", \n \"mirror\": True, \n \"range\": [-685.336803224, 3718.33680322], \n \"showline\": True, \n \"side\": \"right\", \n \"tickfont\": {\"color\": \"#2196F3\"}, \n \"tickmode\": \"auto\",\n \"ticks\": \"\", \n \"title\": \"mg/Kg\", \n \"titlefont\": {\"color\": \"#2196F3\"}, \n \"type\": \"linear\", \n \"zeroline\": False\n }\n}\nfig = go.Figure(data=data, layout=layout)\npy.iplot(fig)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
cbef147c169c420eeda901f53400dfef033dfb93
| 15,752 |
ipynb
|
Jupyter Notebook
|
Array_creation_routines.ipynb
|
server73/numpy_excercises
|
4145f5fe8193f1110a7e83360aa6ae455cf87491
|
[
"MIT"
] | null | null | null |
Array_creation_routines.ipynb
|
server73/numpy_excercises
|
4145f5fe8193f1110a7e83360aa6ae455cf87491
|
[
"MIT"
] | null | null | null |
Array_creation_routines.ipynb
|
server73/numpy_excercises
|
4145f5fe8193f1110a7e83360aa6ae455cf87491
|
[
"MIT"
] | null | null | null | 20.9747 | 103 | 0.427501 |
[
[
[
"# Array creation routines",
"_____no_output_____"
],
[
"## Ones and zeros",
"_____no_output_____"
]
],
[
[
"import numpy as np",
"_____no_output_____"
]
],
[
[
"Create a new array of 2*2 integers, without initializing entries.",
"_____no_output_____"
],
[
"Let X = np.array([1,2,3], [4,5,6], np.int32). \nCreate a new array with the same shape and type as X.",
"_____no_output_____"
]
],
[
[
"X = np.array([[1,2,3], [4,5,6]], np.int32)\n",
"_____no_output_____"
]
],
[
[
"Create a 3-D array with ones on the diagonal and zeros elsewhere.",
"_____no_output_____"
],
[
"Create a new array of 3*2 float numbers, filled with ones.",
"_____no_output_____"
],
[
"Let x = np.arange(4, dtype=np.int64). Create an array of ones with the same shape and type as X.",
"_____no_output_____"
]
],
[
[
"x = np.arange(4, dtype=np.int64)\n",
"_____no_output_____"
]
],
[
[
"Create a new array of 3*2 float numbers, filled with zeros.",
"_____no_output_____"
],
[
"Let x = np.arange(4, dtype=np.int64). Create an array of zeros with the same shape and type as X.",
"_____no_output_____"
]
],
[
[
"x = np.arange(4, dtype=np.int64)\n",
"_____no_output_____"
]
],
[
[
"Create a new array of 2*5 uints, filled with 6.",
"_____no_output_____"
],
[
"Let x = np.arange(4, dtype=np.int64). Create an array of 6's with the same shape and type as X.",
"_____no_output_____"
]
],
[
[
"x = np.arange(4, dtype=np.int64)\n",
"_____no_output_____"
]
],
[
[
"## From existing data",
"_____no_output_____"
],
[
"Create an array of [1, 2, 3].",
"_____no_output_____"
],
[
"Let x = [1, 2]. Convert it into an array.",
"_____no_output_____"
]
],
[
[
"x = [1,2]\n",
"_____no_output_____"
]
],
[
[
"Let X = np.array([[1, 2], [3, 4]]). Convert it into a matrix.",
"_____no_output_____"
]
],
[
[
"X = np.array([[1, 2], [3, 4]])\n",
"_____no_output_____"
]
],
[
[
"Let x = [1, 2]. Conver it into an array of `float`.",
"_____no_output_____"
]
],
[
[
"x = [1, 2]\n",
"_____no_output_____"
]
],
[
[
"Let x = np.array([30]). Convert it into scalar of its single element, i.e. 30.",
"_____no_output_____"
]
],
[
[
"x = np.array([30])\n",
"_____no_output_____"
]
],
[
[
"Let x = np.array([1, 2, 3]). Create a array copy of x, which has a different id from x.",
"_____no_output_____"
]
],
[
[
"x = np.array([1, 2, 3])\n",
"70140352 [1 2 3]\n70140752 [1 2 3]\n"
]
],
[
[
"## Numerical ranges",
"_____no_output_____"
],
[
"Create an array of 2, 4, 6, 8, ..., 100.",
"_____no_output_____"
],
[
"Create a 1-D array of 50 evenly spaced elements between 3. and 10., inclusive.",
"_____no_output_____"
],
[
"Create a 1-D array of 50 element spaced evenly on a log scale between 3. and 10., exclusive.",
"_____no_output_____"
],
[
"## Building matrices",
"_____no_output_____"
],
[
"Let X = np.array([[ 0, 1, 2, 3],\n [ 4, 5, 6, 7],\n [ 8, 9, 10, 11]]).\n Get the diagonal of X, that is, [0, 5, 10].",
"_____no_output_____"
]
],
[
[
"X = np.array([[ 0, 1, 2, 3], [ 4, 5, 6, 7], [ 8, 9, 10, 11]])\n",
"_____no_output_____"
]
],
[
[
"Create a 2-D array whose diagonal equals [1, 2, 3, 4] and 0's elsewhere.",
"_____no_output_____"
],
[
"Create an array which looks like below.\narray([[ 0., 0., 0., 0., 0.],\n [ 1., 0., 0., 0., 0.],\n [ 1., 1., 0., 0., 0.]])",
"_____no_output_____"
],
[
"Create an array which looks like below.\narray([[ 0, 0, 0],\n [ 4, 0, 0],\n [ 7, 8, 0],\n [10, 11, 12]])",
"_____no_output_____"
],
[
"Create an array which looks like below. array([[ 1, 2, 3],\n [ 4, 5, 6],\n [ 0, 8, 9],\n [ 0, 0, 12]])",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
cbef16f39f95537c00fb7a39f408cd2ecea3ebff
| 5,438 |
ipynb
|
Jupyter Notebook
|
notebooks/Test.ipynb
|
slorg1/rainbowhat
|
fb7e75d62abcac81fcbd8d98015b2a5af6b26044
|
[
"Apache-2.0"
] | null | null | null |
notebooks/Test.ipynb
|
slorg1/rainbowhat
|
fb7e75d62abcac81fcbd8d98015b2a5af6b26044
|
[
"Apache-2.0"
] | null | null | null |
notebooks/Test.ipynb
|
slorg1/rainbowhat
|
fb7e75d62abcac81fcbd8d98015b2a5af6b26044
|
[
"Apache-2.0"
] | null | null | null | 23.439655 | 108 | 0.482346 |
[
[
[
"import rainbowhat as rh\nfrom enum import Enum\nimport subprocess\nimport re\nimport time\nimport itertools",
"_____no_output_____"
],
[
"class RGBColors(Enum):\n RED = (50, 0, 0)\n YELLOW = (50, 50, 0)\n PINK = (50, 10, 12)\n GREEN = (0, 50, 0)\n PURPLE = (50, 0, 50)\n ORANGE = (50, 22, 0)\n BLUE = (0, 0, 50)",
"_____no_output_____"
],
[
"def run_rainbow(it):\n for pixel, color in it:\n rh.rainbow.clear()\n rh.rainbow.set_pixel(pixel, *(color.value))\n rh.rainbow.show()\n time.sleep(0.1)\n",
"_____no_output_____"
],
[
"def open_rainbow():\n colours = list(RGBColors)\n for pixel in itertools.chain(reversed(range(4)),range(4)):\n rh.rainbow.clear()\n rh.rainbow.set_pixel(pixel, *(colours[pixel].value))\n rh.rainbow.set_pixel(-pixel+6, *(colours[-pixel+6].value))\n rh.rainbow.show()\n time.sleep(0.4)\n \n rh.rainbow.clear()\n rh.rainbow.show()\n",
"_____no_output_____"
],
[
"def right_to_left():\n run_rainbow(zip(range(7), RGBColors))",
"_____no_output_____"
],
[
"def left_to_right():\n run_rainbow(reversed(tuple(zip(range(7), RGBColors))))",
"_____no_output_____"
],
[
"\nrh.rainbow.clear()\nrh.display.clear()\n\nfor idx in range(1, 11):\n rh.display.print_str(f'{idx}')\n rh.display.show()\n\n if (idx % 2) == 1:\n left_to_right()\n else:\n right_to_left()\n \n\nrh.rainbow.clear()\nrh.rainbow.show()\nrh.display.print_str('DONE')\nrh.display.show()",
"_____no_output_____"
],
[
"TEMP_EXTRACTOR = re.compile(r\"[^=]+=(\\d+(?:\\.\\d+))'C\\n$\")",
"_____no_output_____"
],
[
"def get_temp(real_temp):\n cpu_temp_str = subprocess.check_output(\"vcgencmd measure_temp\", shell=True).decode(\"utf-8\")\n temp = rh.weather.temperature()\n cpu_temp = float(TEMP_EXTRACTOR.match(cpu_temp_str)[1])\n# print(temp, cpu_temp)\n FACTOR = -(cpu_temp - temp)/(real_temp - temp)\n print('FACTOR', FACTOR)\n FACTOR = 0.6976542896954581\n \n return temp - ((cpu_temp - temp)/FACTOR)",
"_____no_output_____"
],
[
"@rh.touch.A.press()\ndef touch_a(channel):\n rh.lights.rgb(1, 0, 0)\n rh.display.clear()\n rh.display.print_str(\"TEMP\")\n rh.display.show()\n time.sleep(1)\n rh.display.print_float(get_temp(20.5))\n rh.display.show()\n open_rainbow()\n \ndef release(channel):\n rh.display.clear()\n rh.display.show()\n rh.rainbow.clear()\n rh.rainbow.show()\n rh.lights.rgb(0, 0, 0)",
"_____no_output_____"
],
[
"@rh.touch.B.press()\ndef touch_b(channel):\n rh.lights.rgb(0, 1, 0)\n rh.display.clear()\n rh.display.print_str(\" B \")\n rh.display.show()\n time.sleep(0.5)\n right_to_left()\n",
"_____no_output_____"
],
[
"@rh.touch.C.press()\ndef touch_c(channel):\n rh.lights.rgb(0, 0, 1)\n rh.display.clear()\n rh.display.print_str(\" C\")\n rh.display.show()\n time.sleep(0.5)\n left_to_right()\n",
"_____no_output_____"
],
[
"rh.touch.A.release()(release)\nrh.touch.B.release()(release)\nrh.touch.C.release()(release)",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbef2644f29cae071d3121ab5aa701ef7a106be8
| 4,638 |
ipynb
|
Jupyter Notebook
|
Tarea3-sol.ipynb
|
diegotg2000/Tarea1-Lineal
|
70c4bcd70d9ca484b64ade59d755de4389e3e1ee
|
[
"MIT"
] | null | null | null |
Tarea3-sol.ipynb
|
diegotg2000/Tarea1-Lineal
|
70c4bcd70d9ca484b64ade59d755de4389e3e1ee
|
[
"MIT"
] | null | null | null |
Tarea3-sol.ipynb
|
diegotg2000/Tarea1-Lineal
|
70c4bcd70d9ca484b64ade59d755de4389e3e1ee
|
[
"MIT"
] | null | null | null | 18.930612 | 84 | 0.398448 |
[
[
[
"V=QQ^3\ndef f(v):\n x=v[0]\n y=v[1]\n z=v[2]\n \n a=3*x-3*y-z\n b=-y-4*z\n c=5*x-7*y+2*z\n w=vector(QQ, [a,b,c])\n return w",
"_____no_output_____"
],
[
"column_matrix([f([1,0,0]),f([0,1,0]),f([0,0,1])])\n",
"_____no_output_____"
],
[
"V=QQ^4\ndef f(v):\n x=v[0]\n y=v[1]\n z=v[2]\n w=v[3]\n \n a=x-y+z+w\n b=-x-y-z\n c=2*x-y-2*z-w\n d=2*x-y-z\n \n r=vector(QQ, [a,b,c,d])\n return r",
"_____no_output_____"
],
[
"column_matrix([f([1,0,0,0]),f([0,1,0,0]),f([0,0,1,0]),f([0,0,0,1])])\n",
"_____no_output_____"
],
[
"V=QQ^3\ncolumn_matrix([[1,0,0],[0,1,0],[0,0,0]])",
"_____no_output_____"
],
[
"B=column_matrix([[1,0,0],[0,1,0],[0,0,0]]) #rep en base B\nBaC=column_matrix([[1,0,1],[0,1,1],[1,1,0]]) #matriz de la base B a base C\nCaB=(BaC)^(-1)\n\nC=BaC*B*CaB\nC",
"_____no_output_____"
],
[
"column_matrix([[0,0,0],[0,0,0],[0,0,1]])",
"_____no_output_____"
],
[
"B=column_matrix([[0,0,0],[0,0,0],[0,0,1]]) #rep en base B\nBaC=column_matrix([[1,0,1],[0,1,1],[1,1,0]]) #matriz de la base B a base C\nCaB=(BaC)^(-1)\n\nC=BaC*B*CaB\nC",
"_____no_output_____"
],
[
"B=column_matrix([[1,0,0],[0,0,0],[0,0,0]]) #rep en base B2\nBaC=column_matrix([[2,1,1],[1,2,1],[1,1,2]]) #matriz de la base B2 a base C\nCaB=(BaC)^(-1)\n\nC=BaC*B*CaB\nC",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbef36b686ce6dd2670c07f305c7573232cb0bed
| 10,709 |
ipynb
|
Jupyter Notebook
|
Titanic (1).ipynb
|
Mhendricks85/Bemidji-DS
|
a526ce2bee6aba7bea2cff536ccd9838cc5ff883
|
[
"MIT"
] | null | null | null |
Titanic (1).ipynb
|
Mhendricks85/Bemidji-DS
|
a526ce2bee6aba7bea2cff536ccd9838cc5ff883
|
[
"MIT"
] | null | null | null |
Titanic (1).ipynb
|
Mhendricks85/Bemidji-DS
|
a526ce2bee6aba7bea2cff536ccd9838cc5ff883
|
[
"MIT"
] | null | null | null | 22.080412 | 133 | 0.545336 |
[
[
[
"## Agenda\n 1. Make sure everyone has the necesary programs:\n - Pyhton\n - Jupyter\n - Github\n 2. Intro tp Git\n 3. Clone Titanic book\n 4. Start cleaning data ",
"_____no_output_____"
],
[
"## 5 basic steps to Machine Learning:\n#### 1. Data gathering/cleaning/feature development\n 2. Model Selection\n 3. Fitting the model\n 4. Make predicitons\n 5. Validate the model",
"_____no_output_____"
]
],
[
[
"#just sticking with Pandas today to manipulate and clean the Titanic dataset \nimport pandas as pd",
"_____no_output_____"
],
[
"# you will need to map to where your files are located \ntrain = pd.read_csv(######train.csv')\ntest = pd.read_csv(#######test.csv')",
"_____no_output_____"
],
[
"train.head()\n",
"_____no_output_____"
],
[
"test.head()",
"_____no_output_____"
],
[
"# created a dataframe to hold passenger ID and eventually our submission \nSubmission = pd.DataFrame()\nSubmission['PassengerId'] = test['PassengerId']",
"_____no_output_____"
],
[
"# we are going to remove the target (survived) from train and combine train and test so that are feature is consistent\n# must be careful when doing this to not drop rows from either set during cleaning\ny = train.Survived\ncombined = pd.concat([train.drop(['Survived'], axis=1),test])",
"_____no_output_____"
],
[
"print(y.shape)\nprint(combined.shape)",
"_____no_output_____"
]
],
[
[
"#### We are going to go through each feature one by one and develop our final train and test features",
"_____no_output_____"
]
],
[
[
"combined.info()",
"_____no_output_____"
],
[
"combined.head()",
"_____no_output_____"
],
[
"# passenger ID is just an incremented count of passengers, no value so lets drop it\n\ncombined = combined.drop('PassengerId', axis= 1)",
"_____no_output_____"
],
[
"combined.head(20)",
"_____no_output_____"
],
[
"# at first glance Name appears to have no value, but notice the title within the name ex. Mrs, Mr, Master, etc\n# these may be valuable as it may signify crew or socioeconomic class \n# We need to split out the Title within the Name columnm, we can name it Title\ncombined['Title'] = combined['Name'].map(lambda name:name.split(',')[1].split('.')[0].strip())\n# and we can drop Name\ncombined = combined.drop('Name', axis=1)",
"_____no_output_____"
],
[
"combined.head(20)",
"_____no_output_____"
],
[
"# one last action to perform on the Name or now Title, we need to create dummy variables so our value is numeric\ncombined = pd.get_dummies(combined, columns=['Title'], drop_first=False)",
"_____no_output_____"
],
[
"combined.head()",
"_____no_output_____"
],
[
"# and we must do the same thing to Sex\ncombined = pd.get_dummies(combined, columns=['Sex'], drop_first=True)",
"_____no_output_____"
],
[
"combined.head()",
"_____no_output_____"
],
[
"# overall age is fine but we are missing values, we need 1309 and we only have 1046. We will do a simple imputation \n# We are also missing 1 fare value, and 2 Embarked values\n# We will do a slightly different imputation for each\n\ncombined['Age'] = combined.Age.fillna(combined.Age.mean())\ncombined['Fare'] = combined.Fare.fillna(combined.Age.median())\ncombined['Embarked'] = combined.Embarked.fillna(combined.Embarked.ffill())\n\n# and we need to create dummy variables for embarked\ncombined = pd.get_dummies(combined, columns=['Embarked'], drop_first=False)",
"_____no_output_____"
],
[
"combined.head()",
"_____no_output_____"
],
[
"combined.info()",
"_____no_output_____"
],
[
"# we just have ticket and cabin left\n# lets first look at the first 20 tickets and see if there may be any value there:\n\ncombined.Ticket[:20]",
"_____no_output_____"
],
[
"# there doesn't appear to be anything worth extracting so let's drop Ticket\ncombined = combined.drop('Ticket', axis=1)",
"_____no_output_____"
],
[
"combined.head()",
"_____no_output_____"
],
[
"# Finally we are left with cabin, lets take a look at the first 50\ncombined.Cabin[:50]",
"_____no_output_____"
],
[
"# looks like there are a lot of missing values, but they might mean they didn't have a cabin and the significance of A, B, C\n# might be something of value. Let's parse it out and assign the NAN a U for unknown\n\ncombined['Cabin'] = combined.Cabin.fillna('U')\ncombined['Cabin'] = combined['Cabin'].map(lambda c : c[0])\n\n# Finally apply dummy variables to Cabin\ncombined = pd.get_dummies(combined, columns=['Cabin'], drop_first=False)",
"_____no_output_____"
],
[
"combined.head()",
"_____no_output_____"
],
[
"combined.info()",
"_____no_output_____"
],
[
"# we are good to go. No missing values and everthing is numeric\n# lets put the test and train set back together\n\n# remebering the length of the train set\ny.shape",
"_____no_output_____"
],
[
"train = combined.iloc[:891]\ntest = combined.iloc[891:]\nprint(train.shape)\nprint(test.shape)",
"_____no_output_____"
]
],
[
[
"### We will pick up from here at our next meeting with model selction and introduction to pipelines",
"_____no_output_____"
]
],
[
[
"from sklearn.svm import LinearSVC\nfrom sklearn.ensemble import RandomForestClassifier",
"_____no_output_____"
],
[
"X_train = train.values\nX_test = test.values\ny = y.values",
"_____no_output_____"
],
[
"clf = RandomForestClassifier()",
"_____no_output_____"
],
[
"clf.fit(X_train, y)",
"_____no_output_____"
],
[
"Submission['Survived'] = clf.predict(X_test)",
"_____no_output_____"
],
[
"Submission.head()",
"_____no_output_____"
],
[
"# you need to map the submission file to your computer \nSubmission.to_csv(##submission.csv', index=False, header=True)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbef787235544a2d7d977831b9898fde142f4aeb
| 426,464 |
ipynb
|
Jupyter Notebook
|
nbs/.ipynb_checkpoints/kernel_density_estimation-checkpoint.ipynb
|
arvidl/elmed219-brainage
|
22a9815108363509736d2ea21063fe89e7d1d7a7
|
[
"MIT"
] | 1 |
2020-09-21T08:49:33.000Z
|
2020-09-21T08:49:33.000Z
|
nbs/.ipynb_checkpoints/kernel_density_estimation-checkpoint.ipynb
|
arvidl/elmed219-brainage
|
22a9815108363509736d2ea21063fe89e7d1d7a7
|
[
"MIT"
] | null | null | null |
nbs/.ipynb_checkpoints/kernel_density_estimation-checkpoint.ipynb
|
arvidl/elmed219-brainage
|
22a9815108363509736d2ea21063fe89e7d1d7a7
|
[
"MIT"
] | 3 |
2020-09-21T08:49:35.000Z
|
2021-09-19T14:49:16.000Z
| 50.612865 | 35,556 | 0.4081 |
[
[
[
"# Kernel density estimation",
"_____no_output_____"
]
],
[
[
"# Import all libraries needed for the exploration\n\n# General syntax to import specific functions in a library: \n##from (library) import (specific library function)\nfrom pandas import DataFrame, read_csv\n\n# General syntax to import a library but no functions: \n##import (library) as (give the library a nickname/alias)\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nimport pandas as pd #this is how we usually import pandas\nimport numpy as np #this is how we usually import numpy\nimport sys #only needed to determine Python version number\nimport matplotlib #only needed to determine Matplotlib version number\nimport tables # pytables is needed to read and write hdf5 files\nimport openpyxl # is used to read and write MS Excel files\n\nimport xgboost\nimport math\nfrom scipy.stats import pearsonr\nfrom sklearn.linear_model import LinearRegression\nfrom sklearn import tree, linear_model\nfrom sklearn.model_selection import cross_validate, cross_val_score\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.metrics import explained_variance_score\n\nfrom sklearn.neighbors import KernelDensity\nfrom scipy.stats import gaussian_kde\nfrom statsmodels.nonparametric.kde import KDEUnivariate\nfrom statsmodels.nonparametric.kernel_density import KDEMultivariate\n\n# Enable inline plotting\n%matplotlib inline",
"_____no_output_____"
],
[
"# Supress some warnings:\nimport warnings\nwarnings.filterwarnings('ignore')",
"_____no_output_____"
],
[
"print('Python version ' + sys.version)\nprint('Pandas version ' + pd.__version__)\nprint('Numpy version ' + np.__version__)\nprint('Matplotlib version ' + matplotlib.__version__)\nprint('Seaborn version ' + sns.__version__)",
"Python version 3.7.3 (default, Mar 27 2019, 17:13:21) [MSC v.1915 64 bit (AMD64)]\nPandas version 0.24.2\nNumpy version 1.16.2\nMatplotlib version 3.0.3\nSeaborn version 0.9.0\n"
]
],
[
[
"## Training data",
"_____no_output_____"
]
],
[
[
"data = pd.read_csv('../data/train.csv')",
"_____no_output_____"
]
],
[
[
"### Explore the data",
"_____no_output_____"
]
],
[
[
"# Check the number of data points in the data set\nprint('No observations:', len(data))\n# Check the number of features in the data set\nprint('No variables:', len(data.columns))\n# Check the data types\nprint(data.dtypes.unique())",
"No observations: 1085\nNo variables: 152\n[dtype('int64') dtype('O') dtype('float64')]\n"
],
[
"data.shape",
"_____no_output_____"
],
[
"data.columns",
"_____no_output_____"
],
[
"for i, col in enumerate(data.columns, start=0):\n print(i, col)",
"0 SubjectID\n1 Source\n2 ID\n3 Sex\n4 Height\n5 Weight\n6 FieldStrength\n7 Handedness\n8 Measure:volume\n9 Left-Lateral-Ventricle\n10 Left-Inf-Lat-Vent\n11 Left-Cerebellum-White-Matter\n12 Left-Cerebellum-Cortex\n13 Left-Thalamus-Proper\n14 Left-Caudate\n15 Left-Putamen\n16 Left-Pallidum\n17 3rd-Ventricle\n18 4th-Ventricle\n19 Brain-Stem\n20 Left-Hippocampus\n21 Left-Amygdala\n22 CSF\n23 Left-Accumbens-area\n24 Left-VentralDC\n25 Left-vessel\n26 Left-choroid-plexus\n27 Right-Lateral-Ventricle\n28 Right-Inf-Lat-Vent\n29 Right-Cerebellum-White-Matter\n30 Right-Cerebellum-Cortex\n31 Right-Thalamus-Proper\n32 Right-Caudate\n33 Right-Putamen\n34 Right-Pallidum\n35 Right-Hippocampus\n36 Right-Amygdala\n37 Right-Accumbens-area\n38 Right-VentralDC\n39 Right-vessel\n40 Right-choroid-plexus\n41 5th-Ventricle\n42 WM-hypointensities\n43 Left-WM-hypointensities\n44 Right-WM-hypointensities\n45 non-WM-hypointensities\n46 Left-non-WM-hypointensities\n47 Right-non-WM-hypointensities\n48 Optic-Chiasm\n49 CC_Posterior\n50 CC_Mid_Posterior\n51 CC_Central\n52 CC_Mid_Anterior\n53 CC_Anterior\n54 BrainSegVol\n55 BrainSegVolNotVent_x\n56 BrainSegVolNotVentSurf\n57 lhCortexVol\n58 rhCortexVol\n59 CortexVol\n60 lhCerebralWhiteMatterVol\n61 rhCerebralWhiteMatterVol\n62 CerebralWhiteMatterVol\n63 SubCortGrayVol\n64 TotalGrayVol\n65 SupraTentorialVol\n66 SupraTentorialVolNotVent\n67 SupraTentorialVolNotVentVox\n68 MaskVol\n69 BrainSegVol-to-eTIV\n70 MaskVol-to-eTIV\n71 lhSurfaceHoles\n72 rhSurfaceHoles\n73 SurfaceHoles\n74 EstimatedTotalIntraCranialVol\n75 lh.aparc.thickness\n76 lh_bankssts_thickness\n77 lh_caudalanteriorcingulate_thickness\n78 lh_caudalmiddlefrontal_thickness\n79 lh_cuneus_thickness\n80 lh_entorhinal_thickness\n81 lh_fusiform_thickness\n82 lh_inferiorparietal_thickness\n83 lh_inferiortemporal_thickness\n84 lh_isthmuscingulate_thickness\n85 lh_lateraloccipital_thickness\n86 lh_lateralorbitofrontal_thickness\n87 lh_lingual_thickness\n88 lh_medialorbitofrontal_thickness\n89 lh_middletemporal_thickness\n90 lh_parahippocampal_thickness\n91 lh_paracentral_thickness\n92 lh_parsopercularis_thickness\n93 lh_parsorbitalis_thickness\n94 lh_parstriangularis_thickness\n95 lh_pericalcarine_thickness\n96 lh_postcentral_thickness\n97 lh_posteriorcingulate_thickness\n98 lh_precentral_thickness\n99 lh_precuneus_thickness\n100 lh_rostralanteriorcingulate_thickness\n101 lh_rostralmiddlefrontal_thickness\n102 lh_superiorfrontal_thickness\n103 lh_superiorparietal_thickness\n104 lh_superiortemporal_thickness\n105 lh_supramarginal_thickness\n106 lh_frontalpole_thickness\n107 lh_temporalpole_thickness\n108 lh_transversetemporal_thickness\n109 lh_insula_thickness\n110 lh_MeanThickness_thickness\n111 BrainSegVolNotVent_y\n112 eTIV_x\n113 rh.aparc.thickness\n114 rh_bankssts_thickness\n115 rh_caudalanteriorcingulate_thickness\n116 rh_caudalmiddlefrontal_thickness\n117 rh_cuneus_thickness\n118 rh_entorhinal_thickness\n119 rh_fusiform_thickness\n120 rh_inferiorparietal_thickness\n121 rh_inferiortemporal_thickness\n122 rh_isthmuscingulate_thickness\n123 rh_lateraloccipital_thickness\n124 rh_lateralorbitofrontal_thickness\n125 rh_lingual_thickness\n126 rh_medialorbitofrontal_thickness\n127 rh_middletemporal_thickness\n128 rh_parahippocampal_thickness\n129 rh_paracentral_thickness\n130 rh_parsopercularis_thickness\n131 rh_parsorbitalis_thickness\n132 rh_parstriangularis_thickness\n133 rh_pericalcarine_thickness\n134 rh_postcentral_thickness\n135 rh_posteriorcingulate_thickness\n136 rh_precentral_thickness\n137 rh_precuneus_thickness\n138 rh_rostralanteriorcingulate_thickness\n139 rh_rostralmiddlefrontal_thickness\n140 rh_superiorfrontal_thickness\n141 rh_superiorparietal_thickness\n142 rh_superiortemporal_thickness\n143 rh_supramarginal_thickness\n144 rh_frontalpole_thickness\n145 rh_temporalpole_thickness\n146 rh_transversetemporal_thickness\n147 rh_insula_thickness\n148 rh_MeanThickness_thickness\n149 BrainSegVolNotVent\n150 eTIV_y\n151 Age\n"
],
[
"# We may have some categorical features, let's check them\ndata.select_dtypes(include=['O']).columns.tolist()",
"_____no_output_____"
],
[
"# Check any number of columns with NaN\nprint(data.isnull().any().sum(), ' / ', len(data.columns))\n# Check number of data points with any NaN\nprint(data.isnull().any(axis=1).sum(), ' / ', len(data))",
"4 / 152\n1085 / 1085\n"
]
],
[
[
"### Select features and targets",
"_____no_output_____"
]
],
[
[
"features = data.iloc[:,9:-1].columns.tolist()\ntarget = data.iloc[:,-1].name",
"_____no_output_____"
],
[
"all_lh_features = [\n 'CSF', 'CC_Posterior', 'CC_Mid_Posterior', 'CC_Central', 'CC_Mid_Anterior', 'CC_Anterior', 'EstimatedTotalIntraCranialVol',\n 'Left-Lateral-Ventricle',\n 'Left-Inf-Lat-Vent',\n 'Left-Cerebellum-White-Matter',\n 'Left-Cerebellum-Cortex',\n 'Left-Thalamus-Proper',\n 'Left-Caudate',\n 'Left-Putamen',\n 'Left-Pallidum',\n 'Left-Hippocampus',\n 'Left-Amygdala',\n 'Left-Accumbens-area',\n 'Left-VentralDC',\n 'Left-vessel',\n 'Left-choroid-plexus',\n 'Left-WM-hypointensities',\n 'Left-non-WM-hypointensities',\n 'lhCortexVol',\n 'lhCerebralWhiteMatterVol', \n 'lhSurfaceHoles', \n 'lh.aparc.thickness',\n 'lh_bankssts_thickness',\n 'lh_caudalanteriorcingulate_thickness',\n 'lh_caudalmiddlefrontal_thickness',\n 'lh_cuneus_thickness',\n 'lh_entorhinal_thickness',\n 'lh_fusiform_thickness',\n 'lh_inferiorparietal_thickness',\n 'lh_inferiortemporal_thickness',\n 'lh_isthmuscingulate_thickness',\n 'lh_lateraloccipital_thickness',\n 'lh_lateralorbitofrontal_thickness',\n 'lh_lingual_thickness',\n 'lh_medialorbitofrontal_thickness',\n 'lh_middletemporal_thickness',\n 'lh_parahippocampal_thickness',\n 'lh_paracentral_thickness',\n 'lh_parsopercularis_thickness',\n 'lh_parsorbitalis_thickness',\n 'lh_parstriangularis_thickness',\n 'lh_pericalcarine_thickness',\n 'lh_postcentral_thickness',\n 'lh_posteriorcingulate_thickness',\n 'lh_precentral_thickness',\n 'lh_precuneus_thickness',\n 'lh_rostralanteriorcingulate_thickness',\n 'lh_rostralmiddlefrontal_thickness',\n 'lh_superiorfrontal_thickness',\n 'lh_superiorparietal_thickness',\n 'lh_superiortemporal_thickness',\n 'lh_supramarginal_thickness',\n 'lh_frontalpole_thickness',\n 'lh_temporalpole_thickness',\n 'lh_transversetemporal_thickness',\n 'lh_insula_thickness',\n 'lh_MeanThickness_thickness'\n ]",
"_____no_output_____"
],
[
"data_lh = data[all_lh_features]",
"_____no_output_____"
],
[
"data_lh.describe().T",
"_____no_output_____"
],
[
"dropcolumns = [\n 'EstimatedTotalIntraCranialVol',\n 'CSF', \n 'CC_Posterior', \n 'CC_Mid_Posterior', \n 'CC_Central', \n 'CC_Mid_Anterior', \n 'CC_Anterior' \n]\n\ndf_lh = data_lh.drop(dropcolumns, axis=1)\ndf_lh",
"_____no_output_____"
],
[
"target",
"_____no_output_____"
],
[
"from sklearn.neighbors import KernelDensity\nfrom scipy.stats import gaussian_kde\nfrom statsmodels.nonparametric.kde import KDEUnivariate\nfrom statsmodels.nonparametric.kernel_density import KDEMultivariate\n\n\ndef kde_scipy(x, x_grid, bandwidth=0.2, **kwargs):\n \"\"\"Kernel Density Estimation with Scipy\"\"\"\n # Note that scipy weights its bandwidth by the covariance of the\n # input data. To make the results comparable to the other methods,\n # we divide the bandwidth by the sample standard deviation here.\n kde = gaussian_kde(x, bw_method=bandwidth / x.std(ddof=1), **kwargs)\n return kde.evaluate(x_grid)\n\n\ndef kde_statsmodels_u(x, x_grid, bandwidth=0.2, **kwargs):\n \"\"\"Univariate Kernel Density Estimation with Statsmodels\"\"\"\n kde = KDEUnivariate(x)\n kde.fit(bw=bandwidth, **kwargs)\n return kde.evaluate(x_grid)\n \n \ndef kde_statsmodels_m(x, x_grid, bandwidth=0.2, **kwargs):\n \"\"\"Multivariate Kernel Density Estimation with Statsmodels\"\"\"\n kde = KDEMultivariate(x, bw=bandwidth * np.ones_like(x),\n var_type='c', **kwargs)\n return kde.pdf(x_grid)\n\n\ndef kde_sklearn(x, x_grid, bandwidth=0.2, **kwargs):\n \"\"\"Kernel Density Estimation with Scikit-learn\"\"\"\n kde_skl = KernelDensity(bandwidth=bandwidth, **kwargs)\n kde_skl.fit(x[:, np.newaxis])\n # score_samples() returns the log-likelihood of the samples\n log_pdf = kde_skl.score_samples(x_grid[:, np.newaxis])\n return np.exp(log_pdf)\n\n\nkde_funcs = [kde_statsmodels_u, kde_statsmodels_m, kde_scipy, kde_sklearn]\nkde_funcnames = ['Statsmodels-U', 'Statsmodels-M', 'Scipy', 'Scikit-learn']\n\nprint \"Package Versions:\"\nimport sklearn; print \" scikit-learn:\", sklearn.__version__\nimport scipy; print \" scipy:\", scipy.__version__\nimport statsmodels; print \" statsmodels:\", statsmodels.__version__",
"_____no_output_____"
]
],
[
[
"### Discretization of Age variable\nQuantile-based discretization function. Discretize variable into equal-sized buckets based on rank or based on sample quantiles. For example 1000 values for 10 quantiles would produce a Categorical object indicating quantile membership for each data point.",
"_____no_output_____"
]
],
[
[
"pd.qcut(data['Age'], 8).head(1)",
"_____no_output_____"
]
],
[
[
"#### Columns with missing values",
"_____no_output_____"
]
],
[
[
"def missing(dff):\n print (round((dff.isnull().sum() * 100/ len(dff)),4).sort_values(ascending=False))",
"_____no_output_____"
],
[
"missing(df_lh)",
"lh_MeanThickness_thickness 0.0\nLeft-choroid-plexus 0.0\nlh_entorhinal_thickness 0.0\nlh_cuneus_thickness 0.0\nlh_caudalmiddlefrontal_thickness 0.0\nlh_caudalanteriorcingulate_thickness 0.0\nlh_bankssts_thickness 0.0\nlh.aparc.thickness 0.0\nlhSurfaceHoles 0.0\nlhCerebralWhiteMatterVol 0.0\nlhCortexVol 0.0\nLeft-non-WM-hypointensities 0.0\nLeft-WM-hypointensities 0.0\nLeft-vessel 0.0\nlh_inferiorparietal_thickness 0.0\nLeft-VentralDC 0.0\nLeft-Accumbens-area 0.0\nLeft-Amygdala 0.0\nLeft-Hippocampus 0.0\nLeft-Pallidum 0.0\nLeft-Putamen 0.0\nLeft-Caudate 0.0\nLeft-Thalamus-Proper 0.0\nLeft-Cerebellum-Cortex 0.0\nLeft-Cerebellum-White-Matter 0.0\nLeft-Inf-Lat-Vent 0.0\nlh_fusiform_thickness 0.0\nlh_inferiortemporal_thickness 0.0\nlh_insula_thickness 0.0\nlh_posteriorcingulate_thickness 0.0\nlh_transversetemporal_thickness 0.0\nlh_temporalpole_thickness 0.0\nlh_frontalpole_thickness 0.0\nlh_supramarginal_thickness 0.0\nlh_superiortemporal_thickness 0.0\nlh_superiorparietal_thickness 0.0\nlh_superiorfrontal_thickness 0.0\nlh_rostralmiddlefrontal_thickness 0.0\nlh_rostralanteriorcingulate_thickness 0.0\nlh_precuneus_thickness 0.0\nlh_precentral_thickness 0.0\nlh_postcentral_thickness 0.0\nlh_isthmuscingulate_thickness 0.0\nlh_pericalcarine_thickness 0.0\nlh_parstriangularis_thickness 0.0\nlh_parsorbitalis_thickness 0.0\nlh_parsopercularis_thickness 0.0\nlh_paracentral_thickness 0.0\nlh_parahippocampal_thickness 0.0\nlh_middletemporal_thickness 0.0\nlh_medialorbitofrontal_thickness 0.0\nlh_lingual_thickness 0.0\nlh_lateralorbitofrontal_thickness 0.0\nlh_lateraloccipital_thickness 0.0\nLeft-Lateral-Ventricle 0.0\ndtype: float64\n"
]
],
[
[
"#### How to remove columns with too many missing values in Python\nhttps://stackoverflow.com/questions/45515031/how-to-remove-columns-with-too-many-missing-values-in-python",
"_____no_output_____"
]
],
[
[
"def rmissingvaluecol(dff,threshold):\n l = []\n l = list(dff.drop(dff.loc[:,list((100*(dff.isnull().sum()/len(dff.index))>=threshold))].columns, 1).columns.values)\n print(\"# Columns having more than %s percent missing values:\"%threshold,(dff.shape[1] - len(l)))\n print(\"Columns:\\n\",list(set(list((dff.columns.values))) - set(l)))\n return l",
"_____no_output_____"
],
[
"#Here threshold is 10% which means we are going to drop columns having more than 10% of missing values\nrmissingvaluecol(data,10)",
"# Columns having more than 10 percent missing values: 4\nColumns:\n ['Height', 'FieldStrength', 'Weight', 'Handedness']\n"
],
[
"# Now create new dataframe excluding these columns\nl = rmissingvaluecol(data,10)\ndata1 = data[l]",
"# Columns having more than 10 percent missing values: 4\nColumns:\n ['Height', 'FieldStrength', 'Weight', 'Handedness']\n"
],
[
"# missing(data[features])",
"_____no_output_____"
]
],
[
[
"#### Correlations between features and target",
"_____no_output_____"
]
],
[
[
"correlations = {}\nfor f in features:\n data_temp = data1[[f,target]]\n x1 = data_temp[f].values\n x2 = data_temp[target].values\n key = f + ' vs ' + target\n correlations[key] = pearsonr(x1,x2)[0]",
"_____no_output_____"
],
[
"data_correlations = pd.DataFrame(correlations, index=['Value']).T\ndata_correlations.loc[data_correlations['Value'].abs().sort_values(ascending=False).index]",
"_____no_output_____"
]
],
[
[
"#### We can see that the top 5 features are the most correlated features with the target \"Age\"",
"_____no_output_____"
]
],
[
[
"y = data.loc[:,['lh_insula_thickness','rh_insula_thickness',target]].sort_values(target, ascending=True).values\nx = np.arange(y.shape[0])",
"_____no_output_____"
],
[
"%matplotlib inline\nplt.subplot(3,1,1)\nplt.plot(x,y[:,0])\nplt.title('lh_insula_thickness and rh_insula_thickness vs Age')\nplt.ylabel('lh_insula_thickness')\n\nplt.subplot(3,1,2)\nplt.plot(x,y[:,1])\nplt.ylabel('rh_insula_thickness')\n\nplt.subplot(3,1,3)\nplt.plot(x,y[:,2],'r')\nplt.ylabel(\"Age\")\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Predicting Age",
"_____no_output_____"
]
],
[
[
"# Train a simple linear regression model\nregr = linear_model.LinearRegression()\nnew_data = data[features]",
"_____no_output_____"
],
[
"X = new_data.values\ny = data.Age.values",
"_____no_output_____"
],
[
"X_train, X_test, y_train, y_test = train_test_split(X, y ,test_size=0.2)",
"_____no_output_____"
],
[
"regr.fit(X_train, y_train)\nprint(regr.predict(X_test))",
"[ 40.26215822 52.73759521 32.38092553 69.94745346 40.74674538\n 42.81256113 26.33642016 18.99090684 40.61615371 51.19628677\n 45.97370323 44.36776157 19.84677273 56.20778796 63.02045595\n 25.38893895 52.34309984 45.74582733 22.62640226 45.14721014\n 37.14349031 28.32441254 24.10945518 29.30161616 37.93751988\n 28.23567073 34.7513464 31.39147949 36.13830631 58.25515543\n 35.57164764 45.69784681 45.15737594 30.06739509 34.12306674\n 51.66018966 47.71524995 58.81758066 55.65393097 52.51658456\n 44.18512012 30.79376463 32.22232595 38.51828841 52.96832978\n 46.92018209 46.12072676 25.46805051 47.12062023 39.03335715\n 30.05147109 38.79004408 35.03470988 43.46331263 36.32689369\n 48.90200731 31.73084835 29.29195648 114.73773053 38.24888887\n 26.11832624 34.7462788 25.45241279 74.5893491 32.78911877\n 24.48251944 56.45219477 36.12498349 75.3288535 44.41877628\n 37.98615202 43.54654305 40.85339155 52.58007581 78.77423028\n 29.59495972 45.51088251 56.57490001 38.16899527 34.71658266\n 76.8004311 49.50590078 57.09155261 21.65712491 51.11620288\n 29.80038204 36.78985014 22.84257521 39.92421952 36.03288869\n 45.10387113 24.56428404 48.22150475 51.59263692 43.02033523\n 37.05562024 62.60980768 42.28242175 30.79547489 32.60328903\n 29.38755278 37.75884994 42.40022526 29.90475668 49.42087884\n 48.94888693 31.68679825 35.80977306 29.96837714 60.10826179\n 43.27746634 57.8122699 25.06753277 53.7659823 51.97374535\n 51.06587471 35.83987448 42.37056988 30.42440856 46.4567182\n 36.32989379 34.75805316 35.76178737 48.52690606 44.68705588\n 33.84440278 30.23105584 20.36428273 36.30820155 40.67482003\n 47.98653361 40.18712535 30.66578171 45.26595262 23.70375143\n 25.82974016 43.25642709 24.66856209 40.55382014 38.38516975\n 38.57785346 35.80171053 57.85234136 52.10071293 57.88729102\n 36.83679636 19.51881443 39.27125599 19.8174166 33.34619031\n 42.97007128 31.93794503 38.84718547 31.06509786 33.28890478\n 21.06786699 41.74040308 50.10704901 30.90925312 58.28471697\n 47.61880804 38.78760754 51.48352856 88.70077411 48.46445436\n 38.48157829 37.14272731 60.54347078 38.30903265 42.77007237\n 29.47713791 59.14186855 40.73437395 49.39758207 27.54173722\n 49.05241739 66.21363573 36.3753923 50.27229993 55.93518674\n 50.35393379 60.01225016 58.7745546 48.8329128 53.66858163\n 56.60071593 39.56540148 59.57448537 48.46650818 52.06455537\n 70.66170007 37.73805216 34.18627196 33.66912982 46.36351148\n 33.31082487 38.55948214 33.04736139 31.38092771 35.20967449\n 34.8712002 34.6485859 24.41518528 46.25768592 39.18291793\n 29.80731816 27.92804469 36.89325516 44.84580703 44.2700266\n 31.97058615 55.18676331 53.69868612 41.76695067 43.01728287\n 46.10134902 29.29131122]\n"
],
[
"regr.score(X_test,y_test)",
"_____no_output_____"
],
[
"# Calculate the Root Mean Squared Error\nprint(\"RMSE: %.2f\"\n % math.sqrt(np.mean((regr.predict(X_test) - y_test) ** 2)))",
"RMSE: 12.77\n"
],
[
"# Let's try XGboost algorithm to see if we can get better results\nxgb = xgboost.XGBRegressor(n_estimators=100, learning_rate=0.08, gamma=0, subsample=0.75,\n colsample_bytree=1, max_depth=7)",
"_____no_output_____"
],
[
"traindf, testdf = train_test_split(X_train, test_size = 0.3)\nxgb.fit(X_train,y_train)",
"[22:23:30] WARNING: C:/Jenkins/workspace/xgboost-win64_release_0.90/src/objective/regression_obj.cu:152: reg:linear is now deprecated in favor of reg:squarederror.\n"
],
[
"predictions = xgb.predict(X_test)\nprint(explained_variance_score(predictions,y_test))",
"0.32469110931920553\n"
]
],
[
[
"### This is worse than a simple regression model",
"_____no_output_____"
],
[
"We can use `.describe()` to calculate simple **descriptive statistics** for the dataset (rounding to 3 decimals):",
"_____no_output_____"
]
],
[
[
"new_data.describe().round(3).T",
"_____no_output_____"
]
],
[
[
"Computing the **pairwise correlation of columns** (features). Method could be ‘pearson’ (default), ‘kendall’, or ‘spearman’. ",
"_____no_output_____"
]
],
[
[
"new_data.corr().round(2)",
"_____no_output_____"
],
[
"new_data.describe()",
"_____no_output_____"
]
],
[
[
"Splitting the object (iris DataFrame) **into groups** (species)",
"_____no_output_____"
]
],
[
[
"grouped = data.groupby('Sex')",
"_____no_output_____"
],
[
"grouped.groups",
"_____no_output_____"
]
],
[
[
"Describe the group-wise `PetalLength` summary statistics",
"_____no_output_____"
]
],
[
[
"print('Age:')\ngrouped['Age'].describe()",
"Age:\n"
]
],
[
[
"Iterating through the grouped data is very natural",
"_____no_output_____"
]
],
[
[
"for name, group in grouped:\n print(name,':')\n print(group.describe().round(2).head(3))",
"F :\n SubjectID Height Weight Handedness Measure:volume \\\ncount 611.00 202.00 202.00 81.00 611.0 \nmean 764.03 163.23 65.93 94.94 0.0 \nstd 450.63 7.77 16.09 15.50 0.0 \n\n Left-Lateral-Ventricle Left-Inf-Lat-Vent \\\ncount 611.00 611.00 \nmean 7897.59 289.78 \nstd 5118.12 191.02 \n\n Left-Cerebellum-White-Matter Left-Cerebellum-Cortex \\\ncount 611.00 611.00 \nmean 14641.69 51303.14 \nstd 2275.35 5657.52 \n\n Left-Thalamus-Proper ... rh_superiortemporal_thickness \\\ncount 611.00 ... 611.00 \nmean 7344.60 ... 2.81 \nstd 895.28 ... 0.20 \n\n rh_supramarginal_thickness rh_frontalpole_thickness \\\ncount 611.00 611.00 \nmean 2.56 2.83 \nstd 0.17 0.28 \n\n rh_temporalpole_thickness rh_transversetemporal_thickness \\\ncount 611.00 611.00 \nmean 3.75 2.34 \nstd 0.30 0.26 \n\n rh_insula_thickness rh_MeanThickness_thickness BrainSegVolNotVent \\\ncount 611.00 611.00 611.00 \nmean 2.97 2.51 1072761.22 \nstd 0.20 0.14 96248.66 \n\n eTIV_y Age \ncount 611.00 611.00 \nmean 1425366.41 42.94 \nstd 126535.10 18.92 \n\n[3 rows x 146 columns]\nM :\n SubjectID Height Weight Handedness Measure:volume \\\ncount 474.00 181.00 181.00 31.00 474.0 \nmean 781.12 176.94 78.63 92.69 0.0 \nstd 444.63 7.98 17.00 26.44 0.0 \n\n Left-Lateral-Ventricle Left-Inf-Lat-Vent \\\ncount 474.00 474.00 \nmean 9587.68 347.96 \nstd 5858.83 220.57 \n\n Left-Cerebellum-White-Matter Left-Cerebellum-Cortex \\\ncount 474.00 474.00 \nmean 15906.66 57497.89 \nstd 2703.39 6289.00 \n\n Left-Thalamus-Proper ... rh_superiortemporal_thickness \\\ncount 474.00 ... 474.00 \nmean 8141.47 ... 2.84 \nstd 971.97 ... 0.18 \n\n rh_supramarginal_thickness rh_frontalpole_thickness \\\ncount 474.00 474.00 \nmean 2.57 2.79 \nstd 0.16 0.29 \n\n rh_temporalpole_thickness rh_transversetemporal_thickness \\\ncount 474.00 474.00 \nmean 3.77 2.37 \nstd 0.29 0.23 \n\n rh_insula_thickness rh_MeanThickness_thickness BrainSegVolNotVent \\\ncount 474.00 474.00 474.00 \nmean 3.02 2.52 1207838.10 \nstd 0.19 0.13 110806.96 \n\n eTIV_y Age \ncount 474.00 474.00 \nmean 1631575.33 41.10 \nstd 135881.80 18.11 \n\n[3 rows x 146 columns]\n"
]
],
[
[
"**Group-wise feature correlations**",
"_____no_output_____"
]
],
[
[
"data.groupby('Age').corr().round(3)",
"_____no_output_____"
]
],
[
[
"DataFrame has an `assign()` method that allows you to easily create new columns that are potentially derived from existing columns.",
"_____no_output_____"
]
],
[
[
"iris.assign(sepal_ratio = iris['SepalWidth'] / iris['SepalLength']).head().round(3)",
"_____no_output_____"
]
],
[
[
"In the example above, we inserted a precomputed value. <br>\nWe can also pass in a function of one argument to be evaluated on the DataFrame being assigned to.",
"_____no_output_____"
]
],
[
[
"iris.assign(sepal_ratio = lambda x: (x['SepalWidth'] /\n x['SepalLength'])).head().round(3)",
"_____no_output_____"
]
],
[
[
"`assign` always returns a copy of the data, leaving the original DataFrame untouched, e.g.",
"_____no_output_____"
]
],
[
[
"iris.head(2)",
"_____no_output_____"
]
],
[
[
"Passing a callable, as opposed to an actual value to be inserted, is useful when you don’t have a reference to the DataFrame at hand. This is common when using assign`` in a chain of operations. For example, we can limit the DataFrame to just those observations with a Sepal Length greater than 5, calculate the ratio, and plot:",
"_____no_output_____"
]
],
[
[
"(iris.query('SepalLength > 5')\n .assign(SepalRatio = lambda x: x.SepalWidth / x.SepalLength,\n PetalRatio = lambda x: x.PetalWidth / x.PetalLength)\n .plot(kind='scatter', x='SepalRatio', y='PetalRatio'))",
"_____no_output_____"
]
],
[
[
"### Classification",
"_____no_output_____"
],
[
"*Organizing data as X and y before classification*",
"_____no_output_____"
]
],
[
[
"from sklearn.preprocessing import LabelEncoder\n\n# dfX5Y = pd.read_csv('../results/02_X5Y.csv', sep=',')\n# print(dfX5Y.info())\n# print(dfX5Y.describe())\n# dfX5Y\n\n# Featuer importance XGBoost:\n# X = df.loc[:, ['CC_Mid_Anterior_w3', 'BrainSegVol-to-eTIV_w3', 'CSF_w2']] # Top three important features\n# Featuer importance RF (Strrop_3):\nX = df.loc[:, ['BrainSegVol-to-eTIV_w3', 'CC_Mid_Anterior_w3', 'ic04-ic02']] # Top three important features\n# Featuer importance RF predicrting Stroop_1_R_3:\n# X = df.loc[:, ['ic09-ic06', 'ic10-ic01', 'ic05-ic03']] # Top three important features\n# Featuer importance RF predicrting Stroop_2_R_3:\n# X = df.loc[:, ['WM-hypointensities_w3', 'ic17-ic04', 'Left-vessel_w3']] # Top three important features\n# X = df.loc[:, ['BrainSegVol-to-eTIV_w3', 'ic04-ic02']] # Two important features\n# X = df.loc[:, ['BrainSegVol-to-eTIV_w3', 'CC_Mid_Anterior_w3']] # Top two important features\nY = df.loc[:, ['Stroop_3_cat']]\ny = Y.as_matrix().ravel()\nnp.unique(y)\nX.columns",
"_____no_output_____"
],
[
"from sklearn.ensemble import VotingClassifier\nfrom sklearn.pipeline import Pipeline\nfrom sklearn.decomposition import PCA\nfrom sklearn.preprocessing import StandardScaler\nfrom sklearn.neural_network import MLPClassifier\nfrom sklearn.neighbors import KNeighborsClassifier\nfrom sklearn.linear_model import LogisticRegression\nfrom sklearn.ensemble import RandomForestClassifier\nfrom sklearn.svm import SVC\nfrom xgboost import XGBClassifier\nfrom sklearn.model_selection import LeaveOneOut\nfrom sklearn.model_selection import KFold\nfrom sklearn.model_selection import StratifiedKFold\nfrom sklearn.model_selection import cross_val_score\nfrom sklearn.metrics import accuracy_score, precision_score, recall_score, f1_score\nfrom sklearn import preprocessing\n\n# X = dfX5Y.loc[:, dfX5Y.columns != 'grp'] # Top five important connections\n# X = dfX5Y.loc[:, ['ic09-ic02', 'ic04-ic01']] # Top two important connections\n# X = df.loc[:, ['LatVent_w2', 'LatVent_w3', 'ic09-ic02', 'ic04-ic01']]\n# X = df.loc[:, ['LatVent_w3', 'ic09-ic02']]\n# X = df.loc[:, ['LatVent_w2', 'LatVent_w3']]\n# Y = df.loc[:, ['Stroop_3_cat']]\n\n# X = df.loc[:, ['BrainSegVol-to-eTIV_w3', 'CC_Mid_Anterior_w3', 'ic04-ic02']]\n# Y = df.loc[:, ['Stroop_3_cat']]\n# y = Y.as_matrix().ravel()\n\nrs = 42 # random_state (42)\nhls = 3 # MLP hidden layer size (3 or 4)\n# https://stackoverflow.com/questions/37659970/how-does-sklearn-compute-the-precision-score-metric\nmyaverage = 'weighted' # For multilabel classification 'micro', 'macro', 'samples', 'weighted'\n# see: https://stackoverflow.com/questions/37659970/how-does-sklearn-compute-the-precision-score-metric\n\n# http://scikit-learn.org/stable/modules/neural_networks_supervised.html\n# Class MLPClassifier implements a multi-layer perceptron (MLP) algorithm that \n# trains using Backpropagation.\n\n# So what about size of the hidden layer(s)--how many neurons? \n# There are some empirically-derived rules-of-thumb, of these, the most \n# commonly relied on is 'the optimal size of the hidden layer is usually \n# between the size of the input and size of the output layers'. \n# Jeff Heaton, author of Introduction to Neural Networks in Java offers a few more.\n#\n# In sum, for most problems, one could probably get decent performance (even without \n# a second optimization step) by setting the hidden layer configuration using j\n# ust two rules: \n# (i) number of hidden layers equals one; and \n# (ii) the number of neurons in that layer is the mean of the neurons in the \n# input and output layers.\n# Compute the precision\n# The precision is the ratio tp / (tp + fp) where tp is the number of true positives and \n# fp the number of false positives. The precision is intuitively the ability of the \n# classifier not to label as positive a sample that is negative.\n\n# Compute the recall\n# The recall is the ratio tp / (tp + fn) where tp is the number of true positives and \n# fn the number of false negatives. The recall is intuitively the ability of the \n# classifier to find all the positive samples.\n\n# Compute the F1 score, also known as balanced F-score or F-measure\n# The F1 score can be interpreted as a weighted average of the precision and recall, \n# where an F1 score reaches its best value at 1 and worst score at 0. \n# The relative contribution of precision and recall to the F1 score are equal. \n# The formula for the F1 score is:\n# F1 = 2 * (precision * recall) / (precision + recall)\n# In the multi-class and multi-label case, this is the weighted average of the F1 score of each class.\n\npipe_clf1 = Pipeline([\n ('scl', StandardScaler()),\n #('pca', PCA(n_components=2)),\n ('clf1', LogisticRegression(C=1., solver='saga', n_jobs=1, \n multi_class='multinomial', random_state=rs))])\n\npipe_clf2 = Pipeline([\n ('scl', StandardScaler()),\n #('pca', PCA(n_components=2)),\n ('clf2', MLPClassifier(hidden_layer_sizes=(hls, ), # =(100, ) ; =(4, )\n activation='relu', solver='adam',\n alpha=0.0001, batch_size='auto', learning_rate='constant',\n learning_rate_init=0.001, power_t=0.5, max_iter=5000,\n shuffle=True, random_state=rs, tol=0.0001, verbose=False,\n warm_start=False, momentum=0.9, nesterovs_momentum=True,\n early_stopping=False, validation_fraction=0.1,\n beta_1=0.9, beta_2=0.999, epsilon=1e-08))])\n\n# pipe_clf3 = Pipeline([\n# ('scl', StandardScaler()),\n# #('pca', PCA(n_components=2)),\n# ('clf3', RandomForestClassifier(n_estimators=100, criterion='gini', max_depth=None,\n# min_samples_split=2, min_samples_leaf=1,\n# min_weight_fraction_leaf=0.0, max_features='auto',\n# max_leaf_nodes=None, # min_impurity_split=1e-07,\n# bootstrap=True, oob_score=False, n_jobs=1,\n# random_state=rs, verbose=0, warm_start=False,\n# class_weight=None))])\n# pipe_clf3 = Pipeline([\n# ('scl', StandardScaler()),\n# #('pca', PCA(n_components=2)),\n# ('clf3', GradientBoostingClassifier(init=None, learning_rate=0.05, loss='deviance',\n# max_depth=None, max_features=None, max_leaf_nodes=None,\n# min_samples_leaf=1, min_samples_split=2,\n# min_weight_fraction_leaf=0.0, n_estimators=100,\n# presort='auto', random_state=rs, subsample=1.0, verbose=0,\n# warm_start=False)\n\npipe_clf3 = Pipeline([\n ('scl', StandardScaler()),\n #('pca', PCA(n_components=2)),\n ('clf3', XGBClassifier(base_score=0.5, colsample_bylevel=1, colsample_bytree=1,\n gamma=0, learning_rate=0.1, max_delta_step=0, max_depth=3,\n min_child_weight=1, missing=None, n_estimators=1000, nthread=-1,\n objective='multi:softprob', reg_alpha=0, reg_lambda=1,\n scale_pos_weight=1, seed=rs, silent=True, subsample=1))])\n \npipe_clf4 = Pipeline([\n ('scl', StandardScaler()),\n #('pca', PCA(n_components=2)),\n ('clf4', SVC(C=1.0, probability=True, random_state=rs))])\n# ('clf4', SVC(C=1.0, random_state=rs))])\n \npipe_clf5 = Pipeline([\n ('scl', StandardScaler()),\n #('pca', PCA(n_components=2)),\n ('clf5', KNeighborsClassifier(n_neighbors=5, weights='uniform', algorithm='kd_tree', \n leaf_size=30, p=2, metric='minkowski', \n metric_params=None, n_jobs=1))])\n\npipe_clf_vote = Pipeline([\n# ('scl', StandardScaler()),\n ('clf_vote', VotingClassifier(\n estimators=[('lr', pipe_clf1), \n ('mlp', pipe_clf2),\n ('rf', pipe_clf3),\n ('svc', pipe_clf4),\n ('knn', pipe_clf5)],\n voting = 'soft'))])\n# voting = 'hard'))])\n\nscores1_acc, scores2_acc, scores3_acc, scores4_acc, scores5_acc, scores_vote_acc = [], [], [], [], [], []\nscores1_pre, scores2_pre, scores3_pre, scores4_pre, scores5_pre, scores_vote_pre = [], [], [], [], [], []\nscores1_rec, scores2_rec, scores3_rec, scores4_rec, scores5_rec, scores_vote_rec = [], [], [], [], [], []\nscores1_f1, scores2_f1, scores3_f1, scores4_f1, scores5_f1, scores_vote_f1 = [], [], [], [], [], []\n\nn_splits = 10 # k=10\n# n_splits = X.shape[0] # i.e. Leave One Out strategy\n# for train_index, test_index in LeaveOneOut.split(X):\n\nk=1\nfor train_index, test_index in \\\nStratifiedKFold(n_splits=n_splits, shuffle=True, random_state=rs).split(X,y):\n print(\"Fold number:\", k)\n #print(\"\\nTRUE class:\\n\", list(y[test_index]))\n X_train, X_test = X.iloc[train_index], X.iloc[test_index]\n y_train, y_test = y[train_index], y[test_index]\n\n \n #clf1 = LogisticRegression\n print(\" - LogisticRegression\")\n pipe_clf1.fit(X_train, y_train)\n scores1_acc.append(accuracy_score(y_test, pipe_clf1.predict(X_test)))\n scores1_pre.append(precision_score(y_test, pipe_clf1.predict(X_test), average=myaverage))\n scores1_rec.append(recall_score(y_test, pipe_clf1.predict(X_test), average=myaverage))\n scores1_f1.append(f1_score(y_test, pipe_clf1.predict(X_test), average=myaverage))\n print(' Precision: %.2f' % (precision_score(y_test, pipe_clf1.predict(X_test), average=myaverage)))\n print(' Recall: %.2f' % (recall_score(y_test, pipe_clf1.predict(X_test), average=myaverage)))\n #print(\"LR predicted:\\n\", list(pipe_clf1.predict(X_test)))\n \n #clf2 = MLPClassifier\n print(\" - MLPClassifier\")\n pipe_clf2.fit(X_train, y_train)\n scores2_acc.append(accuracy_score(y_test, pipe_clf2.predict(X_test)))\n scores2_pre.append(precision_score(y_test, pipe_clf2.predict(X_test), average=myaverage))\n scores2_rec.append(recall_score(y_test, pipe_clf2.predict(X_test), average=myaverage))\n scores2_f1.append(f1_score(y_test, pipe_clf2.predict(X_test), average=myaverage))\n print(' Precision: %.2f' % (precision_score(y_test, pipe_clf2.predict(X_test), average=myaverage)))\n print(' Recall: %.2f' % (recall_score(y_test, pipe_clf2.predict(X_test), average=myaverage)))\n #print(\"MLP predicted:\\n\", list(pipe_clf2.predict(X_test)))\n \n #clf3 = RandomForestClassifier\n #print(\" - RandomForestClassifier\")\n #clf3 = XGBoost\n print(\" - XGBoost\")\n pipe_clf3.fit(X_train, y_train)\n scores3_acc.append(accuracy_score(y_test, pipe_clf3.predict(X_test)))\n scores3_pre.append(precision_score(y_test, pipe_clf3.predict(X_test), average=myaverage))\n scores3_rec.append(recall_score(y_test, pipe_clf3.predict(X_test), average=myaverage))\n scores3_f1.append(f1_score(y_test, pipe_clf3.predict(X_test), average=myaverage))\n print(' Precision: %.2f' % (precision_score(y_test, pipe_clf3.predict(X_test), average=myaverage)))\n print(' Recall: %.2f' % (recall_score(y_test, pipe_clf3.predict(X_test), average=myaverage)))\n #print(\"RF predicted:\\n\", list(pipe_clf3.predict(X_test)))\n #print(\"XGB predicted:\\n\", list(pipe_clf3.predict(X_test)))\n \n #clf4 = svm.SVC()\n print(\" - svm/SVC\")\n pipe_clf4.fit(X_train, y_train)\n scores4_acc.append(accuracy_score(y_test, pipe_clf4.predict(X_test)))\n scores4_pre.append(precision_score(y_test, pipe_clf4.predict(X_test), average=myaverage))\n scores4_rec.append(recall_score(y_test, pipe_clf4.predict(X_test), average=myaverage))\n scores4_f1.append(f1_score(y_test, pipe_clf4.predict(X_test), average=myaverage))\n print(' Precision: %.2f' % (precision_score(y_test, pipe_clf4.predict(X_test), average=myaverage)))\n print(' Recall: %.2f' % (recall_score(y_test, pipe_clf4.predict(X_test), average=myaverage)))\n #print(\"SVM predicted:\\n\", list(pipe_clf4.predict(X_test)))\n \n #clf5 = KNeighborsClassifier\n print(\" - KNN\")\n pipe_clf5.fit(X_train, y_train)\n scores5_acc.append(accuracy_score(y_test, pipe_clf5.predict(X_test)))\n scores5_pre.append(precision_score(y_test, pipe_clf5.predict(X_test), average=myaverage))\n scores5_rec.append(recall_score(y_test, pipe_clf5.predict(X_test), average=myaverage))\n scores5_f1.append(f1_score(y_test, pipe_clf5.predict(X_test), average=myaverage))\n #print(\"KNN predicted:\\n\", list(pipe_clf5.predict(X_test)))\n \n #clf_vote = VotingClassifier\n print(\" - VotingClassifier\")\n pipe_clf_vote.fit(X_train, y_train)\n scores_vote_acc.append(accuracy_score(y_test, pipe_clf_vote.predict(X_test)))\n scores_vote_pre.append(precision_score(y_test, pipe_clf_vote.predict(X_test), average=myaverage))\n scores_vote_rec.append(recall_score(y_test, pipe_clf_vote.predict(X_test), average=myaverage))\n scores_vote_f1.append(f1_score(y_test, pipe_clf_vote.predict(X_test), average=myaverage))\n print(' Precision: %.2f' % (precision_score(y_test, pipe_clf_vote.predict(X_test), average=myaverage)))\n print(' Recall: %.2f' % (recall_score(y_test, pipe_clf_vote.predict(X_test), average=myaverage)))\n k=k+1\n \nprint('\\nPredictors:')\nprint('X.columns = %s' % list(X.columns))\n\nprint('\\nOutcome:')\nprint(pd.qcut(df['Stroop_3_R_3'], 3).head(0))\nprint(np.unique(y))\n\nprint('\\nSome hyperparameters:')\nprint(\"MLP hidden_layer_size = %d\" % (hls))\nprint(\"random_state = %d\" % (rs))\nprint(\"score average = '%s'\" % (myaverage))\n\nprint(\"\\nLR : CV accuracy = %.3f +-%.3f (k=%d)\" % (np.mean(scores1_acc), np.std(scores1_acc), n_splits))\nprint(\"MLP: CV accuracy = %.3f +-%.3f (k=%d)\" % (np.mean(scores2_acc), np.std(scores2_acc), n_splits))\n# print(\"RF : CV accuracy = %.3f +-%.3f (k=%d)\" % (np.mean(scores3_acc), np.std(scores3_acc), n_splits))\nprint(\"XGB : CV accuracy = %.3f +-%.3f (k=%d)\" % (np.mean(scores3_acc), np.std(scores3_acc), n_splits))\nprint(\"SVM: CV accuracy = %.3f +-%.3f (k=%d)\" % (np.mean(scores4_acc), np.std(scores4_acc), n_splits))\nprint(\"KNN: CV accuracy = %.3f +-%.3f (k=%d)\" % (np.mean(scores5_acc), np.std(scores5_acc), n_splits))\nprint(\"Voting: CV accuracy = %.3f +-%.3f (k=%d)\" % (np.mean(scores_vote_acc), np.std(scores_vote_acc), n_splits))\n\nprint(\"\\nLR : CV precision = %.3f +-%.3f (k=%d)\" % (np.mean(scores1_pre), np.std(scores1_pre), n_splits))\nprint(\"MLP: CV precision = %.3f +-%.3f (k=%d)\" % (np.mean(scores2_pre), np.std(scores2_pre), n_splits))\nprint(\"XGB : CV precision = %.3f +-%.3f (k=%d)\" % (np.mean(scores3_pre), np.std(scores3_pre), n_splits))\nprint(\"SVM: CV precision = %.3f +-%.3f (k=%d)\" % (np.mean(scores4_pre), np.std(scores4_pre), n_splits))\nprint(\"KNN: CV precision = %.3f +-%.3f (k=%d)\" % (np.mean(scores5_pre), np.std(scores5_pre), n_splits))\nprint(\"Voting: CV precision = %.3f +-%.3f (k=%d)\" % (np.mean(scores_vote_pre), np.std(scores_vote_pre), n_splits))\n\nprint(\"\\nLR : CV recall = %.3f +-%.3f (k=%d)\" % (np.mean(scores1_rec), np.std(scores1_rec), n_splits))\nprint(\"MLP: CV recall = %.3f +-%.3f (k=%d)\" % (np.mean(scores2_rec), np.std(scores2_rec), n_splits))\nprint(\"XGB : CV recall = %.3f +-%.3f (k=%d)\" % (np.mean(scores3_rec), np.std(scores3_rec), n_splits))\nprint(\"SVM: CV recall = %.3f +-%.3f (k=%d)\" % (np.mean(scores4_rec), np.std(scores4_rec), n_splits))\nprint(\"KNN: CV recall = %.3f +-%.3f (k=%d)\" % (np.mean(scores5_rec), np.std(scores5_rec), n_splits))\nprint(\"Voting: CV recall = %.3f +-%.3f (k=%d)\" % (np.mean(scores_vote_rec), np.std(scores_vote_rec), n_splits))\n\nprint(\"\\nLR : CV F1-score = %.3f +-%.3f (k=%d)\" % (np.mean(scores1_f1), np.std(scores1_f1), n_splits))\nprint(\"MLP: CV F1-score = %.3f +-%.3f (k=%d)\" % (np.mean(scores2_f1), np.std(scores2_f1), n_splits))\nprint(\"XGB : CV F1-score = %.3f +-%.3f (k=%d)\" % (np.mean(scores3_f1), np.std(scores3_f1), n_splits))\nprint(\"SVM: CV F1-score = %.3f +-%.3f (k=%d)\" % (np.mean(scores4_f1), np.std(scores4_f1), n_splits))\nprint(\"KNN: CV F1-score = %.3f +-%.3f (k=%d)\" % (np.mean(scores5_f1), np.std(scores5_f1), n_splits))\nprint(\"Voting: CV F1-score = %.3f +-%.3f (k=%d)\" % (np.mean(scores_vote_f1), np.std(scores_vote_f1), n_splits))",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
]
] |
cbef7e21f8f94b3550229e133e165d8d9bb8144d
| 90,568 |
ipynb
|
Jupyter Notebook
|
nlp_ensemble_explorer.ipynb
|
nlpie/nlp-ensemble-explorer
|
b687684cf557b9badceb435485abc680face77ee
|
[
"Apache-2.0"
] | 1 |
2021-03-15T12:54:37.000Z
|
2021-03-15T12:54:37.000Z
|
nlp_ensemble_explorer.ipynb
|
nlpie/nlp-ensemble-explorer
|
b687684cf557b9badceb435485abc680face77ee
|
[
"Apache-2.0"
] | null | null | null |
nlp_ensemble_explorer.ipynb
|
nlpie/nlp-ensemble-explorer
|
b687684cf557b9badceb435485abc680face77ee
|
[
"Apache-2.0"
] | 1 |
2021-03-15T12:54:44.000Z
|
2021-03-15T12:54:44.000Z
| 43.127619 | 217 | 0.499603 |
[
[
[
" Copyright (c) 2019 Regents of the University of Minnesota.\n \n Licensed under the Apache License, Version 2.0 (the \"License\");\n you may not use this file except in compliance with the License.\n You may obtain a copy of the License at\n \n http://www.apache.org/licenses/LICENSE-2.0\n \n Unless required by applicable law or agreed to in writing, software\n distributed under the License is distributed on an \"AS IS\" BASIS,\n WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n See the License for the specific language governing permissions and\n limitations under the License.",
"_____no_output_____"
]
],
[
[
"import gevent\nimport random\nimport pandas as pd\nimport numpy as np\nimport math\nimport time \nimport functools as ft\nimport glob, os, sys \nimport operator as op\nimport shelve\nimport ipywidgets as widgets\nfrom ipywidgets import interact, interact_manual\n#from pandas.api.types import is_numeric_dtypen()\nfrom pathlib import Path\nfrom itertools import combinations, product, permutations\nfrom sqlalchemy.engine import create_engine\nfrom datetime import datetime\nfrom ast import literal_eval\nfrom scipy import stats \nfrom scipy.stats.mstats import gmean\nfrom pythonds.basic.stack import Stack\nfrom pythonds.trees.binaryTree import BinaryTree\nfrom collections import defaultdict\nimport collections\nfrom typing import List, Set, Tuple \nfrom sklearn.metrics import classification_report, confusion_matrix\nfrom scipy import sparse",
"_____no_output_____"
],
[
"#!pip install pythonds",
"_____no_output_____"
]
],
[
[
"The cell below contains the configurable parameters to ensure that our ensemble explorer runs properaly on your machine. \nPlease read carfully through steps (1-11) before running the rest of the cells.",
"_____no_output_____"
]
],
[
[
"# STEP-1: CHOOSE YOUR CORPUS\n# TODO: get working with list of corpora\n#corpora = ['mipacq','i2b2','fairview'] #options for concept extraction include 'fairview', 'mipacq' OR 'i2b2'\n\n# cross-system semantic union merge filter for cross system aggregations using custom system annotations file with corpus name and system name using 'ray_test': \n# need to add semantic type filrering when reading in sys_data\n#corpus = 'ray_test'\n#corpus = 'clinical_trial2'\ncorpus = 'fairview'\n#corpora = ['i2b2','fairview']\n\n# STEP-2: CHOOSE YOUR DATA DIRECTORY; this is where output data will be saved on your machine\ndata_directory = '/mnt/DataResearch/gsilver1/output/' \n\n# STEP-3: CHOOSE WHICH SYSTEMS YOU'D LIKE TO EVALUATE AGAINST THE CORPUS REFERENCE SET\n#systems = ['biomedicus', 'clamp', 'ctakes', 'metamap', 'quick_umls']\n#systems = ['biomedicus', 'clamp', 'metamap', 'quick_umls']\n#systems = ['biomedicus', 'quick_umls']\n#systems = ['biomedicus', 'ctakes', 'quick_umls']\nsystems = ['biomedicus', 'clamp', 'ctakes', 'metamap']\n#systems = ['biomedicus', 'clamp']\n#systems = ['ctakes', 'quick_umls', 'biomedicus', 'metamap']\n#systems = ['biomedicus', 'metamap']\n#systems = ['ray_test']\n#systems = ['metamap']\n\n# STEP-4: CHOOSE TYPE OF RUN\nrtype = 6 # OPTIONS INCLUDE: 1->Single systems; 2->Ensemble; 3->Tests; 4 -> majority vote \n # The Ensemble can include the max system set ['ctakes','biomedicus','clamp','metamap','quick_umls']\n \n# STEP-5: CHOOSE WHAT TYPE OF ANALYSIS YOU'D LIKE TO RUN ON THE CORPUS\nanalysis_type = 'full' #options include 'entity', 'cui' OR 'full'\n\n# STEP-(6A): ENTER DETAILS FOR ACCESSING MANUAL ANNOTATION DATA\ndatabase_type = 'postgresql+psycopg2' # We use mysql+pymql as default\ndatabase_username = 'gsilver1'\ndatabase_password = 'nej123' \ndatabase_url = 'd0pconcourse001' # HINT: use localhost if you're running database on your local machine\n#database_name = 'clinical_trial' # Enter database name\ndatabase_name = 'covid-19' # Enter database name\n\ndef ref_data(corpus):\n return corpus + '_all' # Enter the table within the database where your reference data is stored\n\ntable_name = ref_data(corpus)\n\n# STEP-(6B): ENTER DETAILS FOR ACCESSING SYSTEM ANNOTATION DATA\n\ndef sys_data(corpus, analysis_type):\n if analysis_type == 'entity':\n return 'analytical_'+corpus+'.csv' # OPTIONS include 'analytical_cui_mipacq_concepts.csv' OR 'analytical_cui_i2b2_concepts.csv' \n elif analysis_type in ('cui', 'full'):\n return 'analytical_'+corpus+'_cui.csv' # OPTIONS include 'analytical_cui_mipacq_concepts.csv' OR 'analytical_cui_i2b2_concepts.csv' \n \nsystem_annotation = sys_data(corpus, analysis_type)\n\n# STEP-7: CREATE A DB CONNECTION POOL\nengine_request = str(database_type)+'://'+database_username+':'+database_password+\"@\"+database_url+'/'+database_name\nengine = create_engine(engine_request, pool_pre_ping=True, pool_size=20, max_overflow=30)\n\n# STEP-(8A): FILTER BY SEMTYPE\nfilter_semtype = True #False\n\n# STEP-(8B): IF STEP-(8A) == True -> GET REFERENCE SEMTYPES\n\ndef ref_semtypes(filter_semtype, corpus):\n if filter_semtype:\n if corpus == 'fairview':\n semtypes = ['Disorders']\n else: pass\n \n return semtypes\n\nsemtypes = ref_semtypes(filter_semtype, corpus)\n\n# STEP-9: Set data directory/table for source documents for vectorization\nsrc_table = 'sofa'\n\n# STEP-10: Specificy match type from {'exact', 'overlap', 'cui' -> kludge for majority}\nrun_type = 'overlap'\n\n# for clinical trial, measurement/temoral are single system since no overlap for intersect\n# STEP-11: Specify expression type for run (TODO: run all at once; make less kludgey)\nexpression_type = 'nested' #'nested_with_singleton' # type of merge expression: nested ((A&B)|C), paired ((A&B)|(C&D)), nested_with_singleton ((A&B)|((C&D)|E)) \n# -> NB: len(systems) for pair must be >= 4, and for nested_with_singleton == 5; single-> skip merges\n\n# STEP-12: Specify type of ensemble: merge or vote\nensemble_type = 'merge'\n\n# STEP-13: run on negation modifier (TODO: negated entity)\nmodification = None #'negation' ",
"_____no_output_____"
]
],
[
[
"****** TODO \n-> add majority vote to union for analysis_type = 'full'\n-> case for multiple labels on same/overlapping span/same system; disambiguate (order by score if exists and select random for ties): done!\n-> port to command line \n----------------------->\n-> still need to validate that all semtypes in corpus!\n-> handle case where intersect merges are empty/any confusion matrix values are 0; specificallly on empty df in evaluate method: done!\n-> case when system annotations empty from semtype filter; print as 0: done!\n-> trim whitespace on CSV import -> done for semtypes\n-> eliminate rtype = 1 for expression_type = 'single'\n-> cross-system semantic union merge on aggregation\n-> negation: testing\n-> other modification, such as 'present'\n-> clean up configuration process\n-> allow iteration through all corpora and semtypes\n-> optimize vecorization (remove confusion?)",
"_____no_output_____"
]
],
[
[
"# config class for analysis\nclass AnalysisConfig():\n \"\"\"\n Configuration object:\n systems to use\n notes by corpus\n paths by output, gold and system location\n \"\"\"\n def __init__(self):\n self = self \n self.systems = systems\n self.data_dir = data_directory\n \n def corpus_config(self): \n usys_data = system_annotation\n ref_data = database_name+'.'+table_name\n return usys_data, ref_data\n\nanalysisConf = AnalysisConfig()\n#usys, ref = analysisConf.corpus_config()",
"_____no_output_____"
],
[
"class SemanticTypes(object):\n '''\n Filter semantic types based on: https://metamap.nlm.nih.gov/SemanticTypesAndGroups.shtml\n :params: semtypes list from corpus, system to query\n :return: list of equivalent system semtypes \n '''\n \n def __init__(self, semtypes, corpus):\n self = self\n \n# if corpus == 'clinical_trial2':\n# corpus = 'clinical_trial' # kludge!!\n# sql = \"SELECT st.tui, abbreviation, clamp_name, ctakes_name, biomedicus_name FROM clinical_trial.semantic_groups sg join semantic_types st on sg.tui = st.tui where \" + corpus + \"_name in ({})\"\\\n# .format(', '.join(['%s' for _ in semtypes])) \n sql = \"SELECT st.tui, abbreviation, clamp_name, ctakes_name FROM semantic_groups sg join semantic_types st on sg.tui = st.tui where group_name in ({})\"\\\n .format(', '.join(['%s' for _ in semtypes])) \n \n stypes = pd.read_sql(sql, params=[semtypes], con=engine) \n \n if len(stypes['tui'].tolist()) > 0:\n self.biomedicus_types = set(stypes['tui'].tolist())\n self.qumls_types = set(stypes['tui'].tolist())\n \n else:\n self.biomedicus_types = None\n self.qumls_types = None\n \n if stypes['clamp_name'].dropna(inplace=True) or len(stypes['clamp_name']) == 0:\n self.clamp_types = None\n else:\n self.clamp_types = set(stypes['clamp_name'].tolist()[0].split(','))\n \n if stypes['ctakes_name'].dropna(inplace=True) or len(stypes['ctakes_name']) == 0:\n self.ctakes_types = None\n else:\n self.ctakes_types = set(stypes['ctakes_name'].tolist()[0].split(','))\n \n# # # Kludge for b9 temporal\n# if stypes['biomedicus_name'].dropna(inplace=True) or len(stypes['biomedicus_name']) > 0:\n# self.biomedicus_types.update(set(stypes['biomedicus_name'].tolist()[0].split(',')))\n# #else:\n# # self.biomedicus_type = None\n \n if len(stypes['abbreviation'].tolist()) > 0:\n self.metamap_types = set(stypes['abbreviation'].tolist())\n else:\n self.metamap_types = None\n \n self.reference_types = set(semtypes)\n \n def get_system_type(self, system): \n \n if system == 'biomedicus':\n semtypes = self.biomedicus_types\n elif system == 'ctakes':\n semtypes = self.ctakes_types\n elif system == 'clamp':\n semtypes = self.clamp_types\n elif system == 'metamap':\n semtypes = self.metamap_types\n elif system == 'quick_umls':\n semtypes = self.qumls_types\n elif system == 'reference':\n semtypes = self.reference_types\n \n return semtypes\n \n# print(SemanticTypes(['Drug'], corpus).get_system_type('biomedicus'))\n#print(SemanticTypes(['Drug'], corpus).get_system_type('quick_umls'))\n#print(SemanticTypes(['drug'], corpus).get_system_type('clamp'))\n#print(SemanticTypes(['Disorders'], 'fairview').get_system_type('clamp'))",
"_____no_output_____"
],
[
"#semtypes = ['test,treatment']\n#semtypes = 'drug,drug::drug_name,drug::drug_dose,dietary_supplement::dietary_supplement_name,dietary_supplement::dietary_supplement_dose'\n#semtypes = 'demographics::age,demographics::sex,demographics::race_ethnicity,demographics::bmi,demographics::weight'\n#corpus = 'clinical_trial'\n#sys = 'quick_umls'\n\n# is semantic type in particular system\ndef system_semtype_check(sys, semtype, corpus):\n st = SemanticTypes([semtype], corpus).get_system_type(sys)\n if st:\n return sys\n else:\n return None\n\n#print(system_semtype_check(sys, semtypes, corpus))",
"_____no_output_____"
],
[
"# annotation class for systems\nclass AnnotationSystems():\n \"\"\" \n System annotations of interest for UMLS concept extraction\n NB: ctakes combines all \"mentions\" annotation types\n \n \"\"\"\n def __init__(self):\n \n \"\"\" \n annotation base types\n \"\"\" \n \n self.biomedicus_types = [\"biomedicus.v2.UmlsConcept\"]\n self.clamp_types = [\"edu.uth.clamp.nlp.typesystem.ClampNameEntityUIMA\"]\n self.ctakes_types = [\"ctakes_mentions\"]\n self.metamap_types = [\"org.metamap.uima.ts.Candidate\"]\n self.qumls_types = [\"concept_jaccard_score_False\"]\n \n def get_system_type(self, system):\n \n \"\"\"\n return system types\n \"\"\"\n \n if system == \"biomedicus\":\n view = \"Analysis\"\n else:\n view = \"_InitialView\"\n\n if system == 'biomedicus':\n types = self.biomedicus_types\n\n elif system == 'clamp':\n types = self.clamp_types\n\n elif system == 'ctakes':\n types = self.ctakes_types\n\n elif system == 'metamap':\n types = self.metamap_types\n \n elif system == \"quick_umls\":\n types = self.qumls_types\n \n return types, view\n \nannSys = AnnotationSystems()",
"_____no_output_____"
],
[
"%reload_ext Cython",
"_____no_output_____"
],
[
"#%%cython\n\n#import numpy as np # access to Numpy from Python layer\n#import math\n\nclass Metrics(object):\n \"\"\"\n metrics class:\n returns an instance with confusion matrix metrics\n \"\"\"\n def __init__(self, system_only, gold_only, gold_system_match, system_n, neither = 0): # neither: no sys or manual annotation\n\n self = self \n self.system_only = system_only\n self.gold_only = gold_only\n self.gold_system_match = gold_system_match\n self.system_n = system_n\n self.neither = neither\n \n def get_confusion_metrics(self, corpus = None, test = False):\n \n \"\"\"\n compute confusion matrix measures, as per \n https://stats.stackexchange.com/questions/51296/how-do-you-calculate-precision-and-recall-for-multiclass-classification-using-co\n \"\"\"\n# cdef:\n# int TP, FP, FN\n# double TM\n\n TP = self.gold_system_match\n FP = self.system_only\n FN = self.gold_only\n \n TM = TP/math.sqrt(self.system_n) # TigMetric\n \n if not test:\n \n if corpus == 'casi':\n recall = TP/(TP + FN)\n precision = TP/(TP + FP)\n F = 2*(precision*recall)/(precision + recall)\n else:\n if self.neither == 0:\n confusion = [[0, self.system_only],[self.gold_only,self.gold_system_match]]\n else:\n confusion = [[self.neither, self.system_only],[self.gold_only,self.gold_system_match]]\n c = np.asarray(confusion)\n \n if TP != 0 or FP != 0:\n precision = TP/(TP+FP)\n else:\n precision = 0\n \n if TP != 0 or FN != 0:\n recall = TP/(TP+FN)\n else:\n recall = 0\n \n if precision + recall != 0:\n F = 2*(precision*recall)/(precision + recall)\n else:\n F = 0\n \n# recall = np.diag(c) / np.sum(c, axis = 1)\n# precision = np.diag(c) / np.sum(c, axis = 0)\n# #print('Yo!', np.mean(precision), np.mean(recall))\n# if np.mean(precision) != 0 and np.mean(recall) != 0:\n# F = 2*(precision*recall)/(precision + recall)\n# else:\n# F = 0\n else:\n precision = TP/(TP+FP)\n recall = TP/(TP+FN)\n F = 2*(precision*recall)/(precision + recall)\n \n # Tignanelli Metric\n if FN == 0:\n TP_FN_R = TP\n elif FN > 0:\n TP_FN_R = TP/FN\n \n return F, recall, precision, TP, FP, FN, TP_FN_R, TM",
"_____no_output_____"
],
[
"def df_to_set(df, analysis_type = 'entity', df_type = 'sys', corpus = None):\n \n # get values for creation of series of type tuple\n if 'entity' in analysis_type: \n if corpus == 'casi':\n arg = df.case, df.overlap\n else: \n arg = df.begin, df.end, df.case\n \n elif 'cui' in analysis_type:\n arg = df.value, df.case\n elif 'full' in analysis_type:\n arg = df.begin, df.end, df.value, df.case\n \n return set(list(zip(*arg)))",
"_____no_output_____"
],
[
"#%%cython \n\nfrom __main__ import df_to_set, engine\nimport numpy as np \nimport pandas as pd\n\ndef get_cooccurences(ref, sys, analysis_type: str, corpus: str):\n \"\"\"\n get cooccurences between system and reference; exact match; TODO: add relaxed -> done in single system evals during ensemble run\n \"\"\"\n # cooccurences\n class Cooccurences(object):\n \n def __init__(self):\n self.ref_system_match = 0\n self.ref_only = 0\n self.system_only = 0\n self.system_n = 0\n self.ref_n = 0\n self.matches = set()\n self.false_negatives = set()\n self.corpus = corpus\n\n c = Cooccurences()\n \n if c.corpus != 'casi':\n if analysis_type in ['cui', 'full']:\n sys = sys.rename(index=str, columns={\"note_id\": \"case\", \"cui\": \"value\"})\n # do not overestimate FP\n sys = sys[~sys['value'].isnull()] \n ref = ref[~ref['value'].isnull()]\n \n if 'entity' in analysis_type: \n sys = sys.rename(index=str, columns={\"note_id\": \"case\"})\n cols_to_keep = ['begin', 'end', 'case']\n elif 'cui' in analysis_type: \n cols_to_keep = ['value', 'case']\n elif 'full' in analysis_type: \n cols_to_keep = ['begin', 'end', 'value', 'case']\n \n sys = sys[cols_to_keep].drop_duplicates()\n ref = ref[cols_to_keep].drop_duplicates()\n # matches via inner join\n tp = pd.merge(sys, ref, how = 'inner', left_on=cols_to_keep, right_on = cols_to_keep) \n # reference-only via left outer join\n fn = pd.merge(ref, sys, how = 'left', left_on=cols_to_keep, right_on = cols_to_keep, indicator=True) \n fn = fn[fn[\"_merge\"] == 'left_only']\n\n tp = tp[cols_to_keep]\n fn = fn[cols_to_keep]\n\n # use for metrics \n c.matches = c.matches.union(df_to_set(tp, analysis_type, 'ref'))\n c.false_negatives = c.false_negatives.union(df_to_set(fn, analysis_type, 'ref'))\n c.ref_system_match = len(c.matches)\n c.system_only = len(sys) - len(c.matches) # fp\n c.system_n = len(sys)\n c.ref_n = len(ref)\n c.ref_only = len(c.false_negatives)\n \n else:\n sql = \"select `case` from test.amia_2019_analytical_v where overlap = 1 and `system` = %(sys.name)s\" \n tp = pd.read_sql(sql, params={\"sys.name\":sys.name}, con=engine)\n \n sql = \"select `case` from test.amia_2019_analytical_v where (overlap = 0 or overlap is null) and `system` = %(sys.name)s\" \n fn = pd.read_sql(sql, params={\"sys.name\":sys.name}, con=engine)\n \n c.matches = df_to_set(tp, 'entity', 'sys', 'casi')\n c.fn = df_to_set(fn, 'entity', 'sys', 'casi')\n c.ref_system_match = len(c.matches)\n c.system_only = len(sys) - len(c.matches)\n c.system_n = len(tp) + len(fn)\n c.ref_n = len(tp) + len(fn)\n c.ref_only = len(fn)\n \n # sanity check\n if len(ref) - c.ref_system_match < 0:\n print('Error: ref_system_match > len(ref)!')\n if len(ref) != c.ref_system_match + c.ref_only:\n print('Error: ref count mismatch!', len(ref), c.ref_system_match, c.ref_only)\n \n return c ",
"_____no_output_____"
],
[
"def label_vector(doc: str, ann: List[int], labels: List[str]) -> np.array:\n\n v = np.zeros(doc)\n labels = list(labels)\n \n for (i, lab) in enumerate(labels):\n i += 1 # 0 is reserved for no label\n idxs = [np.arange(a.begin, a.end) for a in ann if a.label == lab]\n idxs = [j for mask in idxs for j in mask]\n v[idxs] = i\n\n return v\n\n# test confusion matrix elements for vectorized annotation set; includes TN\n# https://kawahara.ca/how-to-compute-truefalse-positives-and-truefalse-negatives-in-python-for-binary-classification-problems/\n# def confused(sys1, ann1):\n# TP = np.sum(np.logical_and(ann1 == 1, sys1 == 1))\n\n# # True Negative (TN): we predict a label of 0 (negative), and the true label is 0.\n# TN = np.sum(np.logical_and(ann1 == 0, sys1 == 0))\n\n# # False Positive (FP): we predict a label of 1 (positive), but the true label is 0.\n# FP = np.sum(np.logical_and(ann1 == 0, sys1 == 1))\n\n# # False Negative (FN): we predict a label of 0 (negative), but the true label is 1.\n# FN = np.sum(np.logical_and(ann1 == 1, sys1 == 0))\n \n# return TP, TN, FP, FN\n\ndef confused(sys1, ann1):\n TP = np.sum(np.logical_and(ann1 > 0, sys1 == ann1))\n\n # True Negative (TN): we predict a label of 0 (negative), and the true label is 0.\n TN = np.sum(np.logical_and(ann1 == 0, sys1 == ann1))\n\n # False Positive (FP): we predict a label of 1 (positive), but the true label is 0.\n FP = np.sum(np.logical_and(sys1 > 0, sys1 != ann1))\n\n # False Negative (FN): we predict a label of 0 (negative), but the true label is 1.\n FN = np.sum(np.logical_and(ann1 > 0, sys1 == 0))\n \n return TP, TN, FP, FN\n\[email protected]_cache(maxsize=None)\ndef vectorized_cooccurences(r: object, analysis_type: str, corpus: str, filter_semtype, semtype = None) -> np.int64:\n docs = get_docs(corpus)\n \n if filter_semtype:\n ann = get_ref_ann(analysis_type, corpus, filter_semtype, semtype)\n else: \n ann = get_ref_ann(analysis_type, corpus, filter_semtype)\n \n sys = get_sys_ann(analysis_type, r)\n \n #cvals = []\n if analysis_type == 'entity':\n labels = [\"concept\"]\n elif analysis_type in ['cui', 'full']:\n labels = list(set(ann[\"value\"].tolist()))\n \n sys2 = list()\n ann2 = list()\n s2 = list()\n a2 = list()\n \n for n in range(len(docs)):\n \n if analysis_type != 'cui':\n a1 = list(ann.loc[ann[\"case\"] == docs[n][0]].itertuples(index=False))\n s1 = list(sys.loc[sys[\"case\"] == docs[n][0]].itertuples(index=False))\n ann1 = label_vector(docs[n][1], a1, labels)\n sys1 = label_vector(docs[n][1], s1, labels)\n\n #TP, TN, FP, FN = confused(sys1, ann1)\n #cvals.append([TP, TN, FP, FN])\n sys2.append(list(sys1))\n ann2.append(list(ann1))\n\n else:\n a = ann.loc[ann[\"case\"] == docs[n][0]]['label'].tolist()\n s = sys.loc[sys[\"case\"] == docs[n][0]]['label'].tolist()\n x = [1 if x in a else 0 for x in labels]\n y = [1 if x in s else 0 for x in labels]\n# x_sparse = sparse.csr_matrix(x)\n# y_sparse = sparse.csr_matrix(y)\n s2.append(y)\n a2.append(x)\n \n \n #a1 = list(ann.loc[ann[\"case\"] == docs[n][0]].itertuples(index=False))\n #s1 = list(sys.loc[sys[\"case\"] == docs[n][0]].itertuples(index=False))\n \n if analysis_type != 'cui':\n a2 = [item for sublist in ann2 for item in sublist]\n s2 = [item for sublist in sys2 for item in sublist]\n report = classification_report(a2, s2, output_dict=True)\n #print('classification:', report)\n macro_precision = report['macro avg']['precision'] \n macro_recall = report['macro avg']['recall'] \n macro_f1 = report['macro avg']['f1-score']\n TN, FP, FN, TP = confusion_matrix(a2, s2).ravel()\n \n #return (np.sum(cvals, axis=0), (macro_precision, macro_recall, macro_f1))\n return ((TP, TN, FP, FN), (macro_precision, macro_recall, macro_f1))\n else:\n x_sparse = sparse.csr_matrix(a2)\n y_sparse = sparse.csr_matrix(s2)\n report = classification_report(x_sparse, y_sparse, output_dict=True)\n macro_precision = report['macro avg']['precision'] \n macro_recall = report['macro avg']['recall'] \n macro_f1 = report['macro avg']['f1-score']\n #print((macro_precision, macro_recall, macro_f1))\n return ((0, 0, 0, 0), (macro_precision, macro_recall, macro_f1))\n \n \n ",
"_____no_output_____"
],
[
"def cm_dict(ref_only: int, system_only: int, ref_system_match: int, system_n: int, ref_n: int) -> dict:\n \"\"\"\n Generate dictionary of confusion matrix params and measures\n :params: ref_only, system_only, reference_system_match -> sets\n matches, system_n, reference_n -> counts\n :return: dictionary object\n \"\"\"\n\n if ref_only + ref_system_match != ref_n:\n print('ERROR!')\n \n # get evaluation metrics\n F, recall, precision, TP, FP, FN, TP_FN_R, TM = Metrics(system_only, ref_only, ref_system_match, system_n).get_confusion_metrics()\n\n d = {\n# 'F1': F[1], \n# 'precision': precision[1], \n# 'recall': recall[1], \n 'F1': F, \n 'precision': precision, \n 'recall': recall, \n 'TP': TP, \n 'FN': FN, \n 'FP': FP, \n 'TP/FN': TP_FN_R,\n 'n_gold': ref_n, \n 'n_sys': system_n, \n 'TM': TM\n }\n \n if system_n - FP != TP:\n print('inconsistent system n!')\n\n return d",
"_____no_output_____"
],
[
"@ft.lru_cache(maxsize=None)\ndef get_metric_data(analysis_type: str, corpus: str):\n \n usys_file, ref_table = AnalysisConfig().corpus_config()\n systems = AnalysisConfig().systems\n \n sys_ann = pd.read_csv(analysisConf.data_dir + usys_file, dtype={'note_id': str})\n \n# sql = \"SELECT * FROM \" + ref_table #+ \" where semtype in('Anatomy', 'Chemicals_and_drugs')\" \n \n# ref_ann = pd.read_sql(sql, con=engine)\n sys_ann = sys_ann.drop_duplicates()\n ref_ann = None\n \n return ref_ann, sys_ann",
"_____no_output_____"
],
[
"#%%cython\n\nimport pandas as pd\nfrom scipy import stats\nfrom scipy.stats.mstats import gmean\n\ndef geometric_mean(metrics):\n \"\"\"\n 1. Get rank average of F1, TP/FN, TM\n http://www.datasciencemadesimple.com/rank-dataframe-python-pandas-min-max-dense-rank-group/\n https://stackoverflow.com/questions/46686315/in-pandas-how-to-create-a-new-column-with-a-rank-according-to-the-mean-values-o?rq=1\n 2. Take geomean of rank averages\n https://stackoverflow.com/questions/42436577/geometric-mean-applied-on-row\n \"\"\"\n \n data = pd.DataFrame() \n\n metrics['F1 rank']=metrics['F1'].rank(ascending=0,method='average')\n metrics['TP/FN rank']=metrics['TP/FN'].rank(ascending=0,method='average')\n metrics['TM rank']=metrics['TM'].rank(ascending=0,method='average')\n metrics['Gmean'] = gmean(metrics.iloc[:,-3:],axis=1)\n\n return metrics ",
"_____no_output_____"
],
[
"def generate_metrics(analysis_type: str, corpus: str, filter_semtype, semtype = None):\n start = time.time()\n\n systems = AnalysisConfig().systems\n metrics = pd.DataFrame()\n\n __, sys_ann = get_metric_data(analysis_type, corpus)\n c = None\n \n for sys in systems:\n \n if filter_semtype and semtype:\n ref_ann = get_ref_ann(analysis_type, corpus, filter_semtype, semtype)\n else:\n ref_ann = get_ref_ann(analysis_type, corpus, filter_semtype)\n \n system_annotations = sys_ann[sys_ann['system'] == sys].copy()\n\n if filter_semtype:\n st = SemanticTypes([semtype], corpus).get_system_type(sys)\n\n if st: \n system_annotations = sys_ann[sys_ann['semtypes'].isin(st)].copy()\n else:\n system_annotations = sys_ann.copy()\n\n if (filter_semtype and st) or filter_semtype is False:\n system = system_annotations.copy()\n\n if sys == 'quick_umls':\n system = system[system.score.astype(float) >= .8]\n\n if sys == 'metamap' and modification == None:\n system = system.fillna(0)\n system = system[system.score.abs().astype(int) >= 800]\n\n system = system.drop_duplicates()\n\n ref_ann = ref_ann.rename(index=str, columns={\"start\": \"begin\", \"file\": \"case\"})\n c = get_cooccurences(ref_ann, system, analysis_type, corpus) # get matches, FN, etc.\n\n \n if c.ref_system_match > 0: # compute confusion matrix metrics and write to dictionary -> df\n # get dictionary of confusion matrix metrics\n d = cm_dict(c.ref_only, c.system_only, c.ref_system_match, c.system_n, c.ref_n)\n d['system'] = sys\n\n data = pd.DataFrame(d, index=[0])\n metrics = pd.concat([metrics, data], ignore_index=True)\n metrics.drop_duplicates(keep='last', inplace=True)\n else:\n print(\"NO EXACT MATCHES FOR\", sys)\n elapsed = (time.time() - start)\n print(\"elapsed:\", sys, elapsed)\n \n if c:\n elapsed = (time.time() - start)\n print(geometric_mean(metrics))\n\n now = datetime.now()\n timestamp = datetime.timestamp(now)\n\n file_name = 'metrics_'\n\n metrics.to_csv(analysisConf.data_dir + corpus + '_' + file_name + analysis_type + '_' + str(timestamp) + '.csv')\n\n print(\"total elapsed time:\", elapsed) ",
"_____no_output_____"
],
[
"@ft.lru_cache(maxsize=None)\ndef get_ref_n(analysis_type: str, corpus: str, filter_semtype: str) -> int:\n \n ref_ann, _ = get_metric_data(analysis_type, corpus)\n \n if filter_semtype:\n ref_ann = ref_ann[ref_ann['semtype'].isin(SemanticTypes(semtypes, corpus).get_system_type('reference'))]\n \n if corpus == 'casi':\n return len(ref_ann)\n \n else:\n # do not overestimate fn\n if 'entity' in analysis_type:\n ref_ann = ref_ann[['start', 'end', 'file']].drop_duplicates()\n elif 'cui' in analysis_type:\n ref_ann = ref_ann[['value', 'file']].drop_duplicates()\n elif 'full' in analysis_type:\n ref_ann = ref_ann[['start', 'end', 'value', 'file']].drop_duplicates()\n else:\n pass\n\n ref_n = len(ref_ann.drop_duplicates())\n\n return ref_n\n \[email protected]_cache(maxsize=None)\ndef get_sys_data(system: str, analysis_type: str, corpus: str, filter_semtype, semtype = None) -> pd.DataFrame:\n \n _, data = get_metric_data(analysis_type, corpus)\n \n out = data[data['system'] == system].copy()\n \n if filter_semtype:\n st = SemanticTypes([semtype], corpus).get_system_type(system)\n print(system, 'st', st)\n \n if corpus == 'casi':\n cols_to_keep = ['case', 'overlap'] \n out = out[cols_to_keep].drop_duplicates()\n return out\n \n else:\n if filter_semtype:\n out = out[out['semtype'].isin(st)].copy()\n \n else:\n out = out[out['system']== system].copy()\n \n if modification == 'negation':\n out = out[out['modification'] == 'negation'].copy()\n \n if system == 'quick_umls':\n out = out[(out.score.astype(float) >= 0.8) & (out[\"type\"] == 'concept_jaccard_score_False')]\n # fix for leading space on semantic type field\n out = out.apply(lambda x: x.str.strip() if x.dtype == \"object\" else x) \n out['semtypes'] = out['semtypes'].str.strip()\n \n if system == 'metamap' and modification == None:\n out = out[out.score.abs().astype(int) >= 800]\n \n if 'entity' in analysis_type:\n cols_to_keep = ['begin', 'end', 'note_id']\n elif 'cui' in analysis_type:\n cols_to_keep = ['cui', 'note_id']\n elif 'full' in analysis_type:\n cols_to_keep = ['begin', 'end', 'cui', 'note_id', 'polarity']\n\n out = out[cols_to_keep]\n \n return out.drop_duplicates()",
"_____no_output_____"
]
],
[
[
"GENERATE merges",
"_____no_output_____"
]
],
[
[
"class SetTotals(object):\n \"\"\" \n returns an instance with merged match set numbers using either union or intersection of elements in set \n \"\"\"\n def __init__(self, ref_n, sys_n, match_set):\n\n self = self \n self.ref_ann = ref_n\n self.sys_n = sys_n\n self.match_set = match_set\n\n def get_ref_sys(self):\n\n ref_only = self.ref_ann - len(self.match_set)\n sys_only = self.sys_n - len(self.match_set)\n\n return ref_only, sys_only, len(self.match_set), self.match_set",
"_____no_output_____"
],
[
"def union_vote(arg):\n arg['length'] = (arg.end - arg.begin).abs()\n \n df = arg[['begin', 'end', 'note_id', 'cui', 'length', 'polarity']].copy()\n df.sort_values(by=['note_id','begin'],inplace=True)\n df = df.drop_duplicates(['begin', 'end', 'note_id', 'cui', 'polarity'])\n \n cases = set(df['note_id'].tolist())\n data = []\n out = pd.DataFrame()\n \n for case in cases:\n print(case)\n \n test = df[df['note_id']==case].copy()\n \n for row in test.itertuples():\n\n iix = pd.IntervalIndex.from_arrays(test.begin, test.end, closed='neither')\n span_range = pd.Interval(row.begin, row.end)\n fx = test[iix.overlaps(span_range)].copy()\n\n maxLength = fx['length'].max()\n minLength = fx['length'].min()\n\n if len(fx) > 1: \n #if longer span exists use as tie-breaker\n if maxLength > minLength:\n fx = fx[fx['length'] == fx['length'].max()]\n\n data.append(fx)\n\n out = pd.concat(data, axis=0)\n \n # Remaining ties on span with same or different CUIs\n # randomly reindex to keep random selected row when dropping duplicates: https://gist.github.com/cadrev/6b91985a1660f26c2742\n out.reset_index(inplace=True)\n out = out.reindex(np.random.permutation(out.index))\n \n return out.drop_duplicates(['begin', 'end', 'note_id', 'polarity']) #out.drop('length', axis=1, inplace=True) ",
"_____no_output_____"
],
[
"@ft.lru_cache(maxsize=None)\ndef process_sentence(pt, sentence, analysis_type, corpus, filter_semtype, semtype = None):\n \"\"\"\n Recursively evaluate parse tree, \n with check for existence before build\n :param sentence: to process\n :return class of merged annotations, boolean operated system df \n \"\"\"\n \n class Results(object):\n def __init__(self):\n self.results = set()\n self.system_merges = pd.DataFrame()\n \n r = Results()\n \n if 'entity' in analysis_type and corpus != 'casi': \n cols_to_keep = ['begin', 'end', 'note_id', 'polarity'] # entity only\n elif 'full' in analysis_type: \n cols_to_keep = ['cui', 'begin', 'end', 'note_id', 'polarity'] # entity only\n join_cols = ['cui', 'begin', 'end', 'note_id']\n elif 'cui' in analysis_type:\n cols_to_keep = ['cui', 'note_id', 'polarity'] # entity only\n elif corpus == 'casi':\n cols_to_keep = ['case', 'overlap']\n \n def evaluate(parseTree):\n oper = {'&': op.and_, '|': op.or_}\n \n if parseTree:\n leftC = gevent.spawn(evaluate, parseTree.getLeftChild())\n rightC = gevent.spawn(evaluate, parseTree.getRightChild())\n \n if leftC.get() is not None and rightC.get() is not None:\n system_query = pd.DataFrame()\n fn = oper[parseTree.getRootVal()]\n \n if isinstance(leftC.get(), str):\n # get system as leaf node \n if filter_semtype:\n left_sys = get_sys_data(leftC.get(), analysis_type, corpus, filter_semtype, semtype)\n else:\n left_sys = get_sys_data(leftC.get(), analysis_type, corpus, filter_semtype)\n \n elif isinstance(leftC.get(), pd.DataFrame):\n l_sys = leftC.get()\n \n if isinstance(rightC.get(), str):\n # get system as leaf node\n if filter_semtype:\n right_sys = get_sys_data(rightC.get(), analysis_type, corpus, filter_semtype, semtype)\n else:\n right_sys = get_sys_data(rightC.get(), analysis_type, corpus, filter_semtype)\n \n elif isinstance(rightC.get(), pd.DataFrame):\n r_sys = rightC.get()\n \n if fn == op.or_:\n\n if isinstance(leftC.get(), str) and isinstance(rightC.get(), str):\n frames = [left_sys, right_sys]\n\n elif isinstance(leftC.get(), str) and isinstance(rightC.get(), pd.DataFrame):\n frames = [left_sys, r_sys]\n\n elif isinstance(leftC.get(), pd.DataFrame) and isinstance(rightC.get(), str):\n frames = [l_sys, right_sys]\n\n elif isinstance(leftC.get(), pd.DataFrame) and isinstance(rightC.get(), pd.DataFrame):\n frames = [l_sys, r_sys]\n \n df = pd.concat(frames, ignore_index=True)\n \n if analysis_type == 'full':\n df = union_vote(df)\n\n if fn == op.and_:\n \n if isinstance(leftC.get(), str) and isinstance(rightC.get(), str):\n if not left_sys.empty and not right_sys.empty:\n df = left_sys.merge(right_sys, on=join_cols, how='inner')\n df = df[cols_to_keep].drop_duplicates(subset=cols_to_keep)\n else:\n df = pd.DataFrame(columns=cols_to_keep)\n\n elif isinstance(leftC.get(), str) and isinstance(rightC.get(), pd.DataFrame):\n if not left_sys.empty and not r_sys.empty:\n df = left_sys.merge(r_sys, on=join_cols, how='inner')\n df = df[cols_to_keep].drop_duplicates(subset=cols_to_keep)\n else:\n df = pd.DataFrame(columns=cols_to_keep)\n\n elif isinstance(leftC.get(), pd.DataFrame) and isinstance(rightC.get(), str):\n if not l_sys.empty and not right_sys.empty:\n df = l_sys.merge(right_sys, on=join_cols, how='inner')\n df = df[cols_to_keep].drop_duplicates(subset=cols_to_keep)\n else:\n df = pd.DataFrame(columns=cols_to_keep)\n\n elif isinstance(leftC.get(), pd.DataFrame) and isinstance(rightC.get(), pd.DataFrame):\n if not l_sys.empty and not r_sys.empty:\n df = l_sys.merge(r_sys, on=join_cols, how='inner')\n df = df[cols_to_keep].drop_duplicates(subset=cols_to_keep)\n else:\n df = pd.DataFrame(columns=cols_to_keep)\n \n # get combined system results\n r.system_merges = df\n \n if len(df) > 0:\n system_query = system_query.append(df)\n else:\n print('wtf!')\n \n return system_query\n else:\n return parseTree.getRootVal()\n \n if sentence.n_or > 0 or sentence.n_and > 0:\n evaluate(pt) \n \n # trivial case\n elif sentence.n_or == 0 and sentence.n_and == 0:\n \n if filter_semtype:\n r.system_merges = get_sys_data(sentence.sentence, analysis_type, corpus, filter_semtype, semtype)\n else:\n r.system_merges = get_sys_data(sentence.sentence, analysis_type, corpus, filter_semtype)\n \n return r",
"_____no_output_____"
],
[
"\"\"\"\nIncoming Boolean sentences are parsed into a binary tree.\n\nTest expressions to parse:\n\nsentence = '((((A&B)|C)|D)&E)'\n\nsentence = '(E&(D|(C|(A&B))))'\n\nsentence = '(((A|(B&C))|(D&(E&F)))|(H&I))'\n\n\"\"\"\n# build parse tree from passed sentence using grammatical rules of Boolean logic\ndef buildParseTree(fpexp):\n \"\"\"\n Iteratively build parse tree from passed sentence using grammatical rules of Boolean logic\n :param fpexp: sentence to parse\n :return eTree: parse tree representation\n Incoming Boolean sentences are parsed into a binary tree.\n Test expressions to parse:\n sentence = '(A&B)'\n sentence = '(A|B)'\n sentence = '((A|B)&C)'\n \n \"\"\"\n fplist = fpexp.split()\n pStack = Stack()\n eTree = BinaryTree('')\n pStack.push(eTree)\n currentTree = eTree\n\n for i in fplist:\n\n if i == '(':\n currentTree.insertLeft('')\n pStack.push(currentTree)\n currentTree = currentTree.getLeftChild()\n elif i not in ['&', '|', ')']:\n currentTree.setRootVal(i)\n parent = pStack.pop()\n currentTree = parent\n elif i in ['&', '|']:\n currentTree.setRootVal(i)\n currentTree.insertRight('')\n pStack.push(currentTree)\n currentTree = currentTree.getRightChild()\n elif i == ')':\n currentTree = pStack.pop()\n else:\n raise ValueError\n\n return eTree\n\ndef make_parse_tree(payload):\n \"\"\"\n Ensure data to create tree are in correct form\n :param sentence: sentence to preprocess\n :return pt, parse tree graph\n sentence, processed sentence to build tree\n a: order\n \"\"\"\n def preprocess_sentence(sentence):\n # prepare statement for case when a boolean AND/OR is given\n sentence = payload.replace('(', ' ( '). \\\n replace(')', ' ) '). \\\n replace('&', ' & '). \\\n replace('|', ' | '). \\\n replace(' ', ' ')\n return sentence\n\n sentence = preprocess_sentence(payload)\n print('Processing sentence:', sentence)\n \n pt = buildParseTree(sentence)\n #pt.postorder() \n \n return pt\n\nclass Sentence(object):\n '''\n Details about boolean expression -> number operators and expression\n '''\n def __init__(self, sentence):\n self = self\n self.n_and = sentence.count('&')\n self.n_or = sentence.count('|')\n self.sentence = sentence\n\[email protected]_cache(maxsize=None)\ndef get_docs(corpus):\n \n # KLUDGE!!!\n if corpus == 'ray_test':\n corpus = 'fairview'\n \n sql = 'select distinct note_id, sofa from sofas where corpus = %(corpus)s order by note_id'\n df = pd.read_sql(sql, params={\"corpus\":corpus}, con=engine)\n df.drop_duplicates()\n df['len_doc'] = df['sofa'].apply(len)\n \n subset = df[['note_id', 'len_doc']]\n docs = [tuple(x) for x in subset.to_numpy()]\n \n return docs\n\[email protected]_cache(maxsize=None)\ndef get_ref_ann(analysis_type, corpus, filter_semtype, semtype = None):\n \n if filter_semtype:\n if ',' in semtype:\n semtype = semtype.split(',')\n else:\n semtype = [semtype]\n \n ann, _ = get_metric_data(analysis_type, corpus)\n ann = ann.rename(index=str, columns={\"start\": \"begin\", \"file\": \"case\"})\n \n if filter_semtype:\n ann = ann[ann['semtype'].isin(semtype)]\n if analysis_type == 'entity': \n ann[\"label\"] = 'concept'\n elif analysis_type in ['cui','full']:\n ann[\"label\"] = ann[\"value\"]\n \n if modification == 'negation':\n ann = ann[ann['semtype'] == 'negation']\n \n \n if analysis_type == 'entity':\n cols_to_keep = ['begin', 'end', 'case', 'label']\n elif analysis_type == 'cui':\n cols_to_keep = ['value', 'case', 'label']\n elif analysis_type == 'full':\n cols_to_keep = ['begin', 'end', 'value', 'case', 'label']\n ann = ann[cols_to_keep]\n \n return ann\n\[email protected]_cache(maxsize=None)\ndef get_sys_ann(analysis_type, r):\n sys = r.system_merges \n \n sys = sys.rename(index=str, columns={\"note_id\": \"case\"})\n if analysis_type == 'entity':\n sys[\"label\"] = 'concept'\n cols_to_keep = ['begin', 'end', 'case', 'label']\n elif analysis_type == 'full':\n sys[\"label\"] = sys[\"cui\"]\n cols_to_keep = ['begin', 'end', 'case', 'value', 'label']\n elif analysis_type == 'cui':\n sys[\"label\"] = sys[\"cui\"]\n cols_to_keep = ['case', 'cui', 'label']\n \n sys = sys[cols_to_keep]\n return sys\n\[email protected]_cache(maxsize=None)\ndef get_metrics(boolean_expression: str, analysis_type: str, corpus: str, run_type: str, filter_semtype, semtype = None):\n \"\"\"\n Traverse binary parse tree representation of Boolean sentence\n :params: boolean expression in form of '(<annotator_engine_name1><boolean operator><annotator_engine_name2>)'\n analysis_type (string value of: 'entity', 'cui', 'full') used to filter set of reference and system annotations \n :return: dictionary with values needed for confusion matrix\n \"\"\"\n \n sentence = Sentence(boolean_expression) \n pt = make_parse_tree(sentence.sentence)\n \n if filter_semtype:\n r = process_sentence(pt, sentence, analysis_type, corpus, filter_semtype, semtype)\n else:\n r = process_sentence(pt, sentence, analysis_type, corpus, filter_semtype)\n \n # vectorize merges using i-o labeling\n if run_type == 'overlap':\n if filter_semtype:\n ((TP, TN, FP, FN),(p,r,f1)) = vectorized_cooccurences(r, analysis_type, corpus, filter_semtype, semtype)\n else:\n ((TP, TN, FP, FN),(p,r,f1)) = vectorized_cooccurences(r, analysis_type, corpus, filter_semtype)\n \n print('results:',((TP, TN, FP, FN),(p,r,f1)))\n # TODO: validate against ann1/sys1 where val = 1\n # total by number chars\n system_n = TP + FP\n reference_n = TP + FN\n\n if analysis_type != 'cui':\n d = cm_dict(FN, FP, TP, system_n, reference_n)\n else:\n d = dict()\n d['F1'] = 0\n d['precision'] = 0 \n d['recall'] = 0\n d['TP/FN'] = 0\n d['TM'] = 0\n \n d['TN'] = TN\n d['macro_p'] = p\n d['macro_r'] = r\n d['macro_f1'] = f1\n \n \n # return full metrics\n return d\n\n elif run_type == 'exact':\n # total by number spans\n \n if filter_semtype:\n ann = get_ref_ann(analysis_type, corpus, filter_semtype, semtype)\n else: \n ann = get_ref_ann(analysis_type, corpus, filter_semtype)\n \n c = get_cooccurences(ann, r.system_merges, analysis_type, corpus) # get matches, FN, etc.\n\n if c.ref_system_match > 0: # compute confusion matrix metrics and write to dictionary -> df\n # get dictionary of confusion matrix metrics\n d = cm_dict(c.ref_only, c.system_only, c.ref_system_match, c.system_n, c.ref_n)\n else:\n d = None\n \n return d",
"_____no_output_____"
],
[
"#get_valid_systems(['biomedicus'], 'Anatomy')",
"_____no_output_____"
],
[
"# generate all combinations of given list of annotators:\ndef partly_unordered_permutations(lst, k):\n elems = set(lst)\n for c in combinations(lst, k):\n for d in permutations(elems - set(c)):\n yield c + d\n \ndef expressions(l, n):\n for (operations, *operands), operators in product(\n combinations(l, n), product(('&', '|'), repeat=n - 1)):\n for operation in zip(operators, operands):\n operations = [operations, *operation]\n yield operations\n\n# get list of systems with a semantic type in grouping\ndef get_valid_systems(systems, semtype):\n test = []\n for sys in systems:\n st = system_semtype_check(sys, semtype, corpus)\n if st:\n test.append(sys)\n\n return test\n\n# permute system combinations and evaluate system merges for performance\ndef run_ensemble(systems, analysis_type, corpus, filter_semtype, expression_type, semtype = None):\n metrics = pd.DataFrame()\n \n # pass single system to evaluate\n if expression_type == 'single':\n for system in systems:\n if filter_semtype:\n d = get_metrics(system, analysis_type, corpus, run_type, filter_semtype, semtype)\n else:\n d = get_metrics(system, analysis_type, corpus, run_type, filter_semtype)\n d['merge'] = system\n d['n_terms'] = 1\n\n frames = [metrics, pd.DataFrame(d, index=[0])]\n metrics = pd.concat(frames, ignore_index=True, sort=False) \n \n elif expression_type == 'nested':\n for l in partly_unordered_permutations(systems, 2):\n print('processing merge combo:', l)\n for i in range(1, len(l)+1):\n test = list(expressions(l, i))\n for t in test:\n if i > 1:\n # format Boolean sentence for parse tree \n t = '(' + \" \".join(str(x) for x in t).replace('[','(').replace(']',')').replace(\"'\",\"\").replace(\",\",\"\").replace(\" \",\"\") + ')'\n\n if filter_semtype:\n d = get_metrics(t, analysis_type, corpus, run_type, filter_semtype, semtype)\n else:\n d = get_metrics(t, analysis_type, corpus, run_type, filter_semtype)\n\n d['merge'] = t\n d['n_terms'] = i\n\n frames = [metrics, pd.DataFrame(d, index=[0])]\n metrics = pd.concat(frames, ignore_index=True, sort=False) \n \n elif expression_type == 'nested_with_singleton' and len(systems) == 5:\n # form (((a&b)|c)&(d|e))\n \n nested = list(expressions(systems, 3))\n test = list(expressions(systems, 2))\n to_do_terms = []\n \n for n in nested:\n # format Boolean sentence for parse tree \n n = '(' + \" \".join(str(x) for x in n).replace('[','(').replace(']',')').replace(\"'\",\"\").replace(\",\",\"\").replace(\" \",\"\") + ')'\n\n for t in test:\n t = '(' + \" \".join(str(x) for x in t).replace('[','(').replace(']',')').replace(\"'\",\"\").replace(\",\",\"\").replace(\" \",\"\") + ')'\n\n new_and = '(' + n +'&'+ t + ')'\n new_or = '(' + n +'|'+ t + ')'\n\n if new_and.count('biomedicus') != 2 and new_and.count('clamp') != 2 and new_and.count('ctakes') != 2 and new_and.count('metamap') != 2 and new_and.count('quick_umls') != 2:\n\n if new_and.count('&') != 4 and new_or.count('|') != 4:\n #print(new_and)\n #print(new_or)\n to_do_terms.append(new_or)\n to_do_terms.append(new_and)\n \n print('nested_with_singleton', len(to_do_terms))\n for term in to_do_terms:\n if filter_semtype:\n d = get_metrics(term, analysis_type, corpus, run_type, filter_semtype, semtype)\n else:\n d = get_metrics(term, analysis_type, corpus, run_type, filter_semtype)\n \n n = term.count('&')\n m = term.count('|')\n d['merge'] = term\n d['n_terms'] = m + n + 1\n\n frames = [metrics, pd.DataFrame(d, index=[0])]\n metrics = pd.concat(frames, ignore_index=True, sort=False) \n \n elif expression_type == 'paired':\n m = list(expressions(systems, 2))\n test = list(expressions(m, 2))\n\n to_do_terms = []\n for t in test:\n # format Boolean sentence for parse tree \n t = '(' + \" \".join(str(x) for x in t).replace('[','(').replace(']',')').replace(\"'\",\"\").replace(\",\",\"\").replace(\" \",\"\") + ')'\n if t.count('biomedicus') != 2 and t.count('clamp') != 2 and t.count('ctakes') != 2 and t.count('metamap') != 2 and t.count('quick_umls') != 2:\n if t.count('&') != 3 and t.count('|') != 3:\n to_do_terms.append(t)\n if len(systems) == 5:\n for i in systems:\n if i not in t:\n #print('('+t+'&'+i+')')\n #print('('+t+'|'+i+')')\n new_and = '('+t+'&'+i+')'\n new_or = '('+t+'|'+i+')'\n to_do_terms.append(new_and)\n to_do_terms.append(new_or)\n \n print('paired', len(to_do_terms))\n for term in to_do_terms:\n if filter_semtype:\n d = get_metrics(term, analysis_type, corpus, run_type, filter_semtype, semtype)\n else:\n d = get_metrics(term, analysis_type, corpus, run_type, filter_semtype)\n \n n = term.count('&')\n m = term.count('|')\n d['merge'] = term\n d['n_terms'] = m + n + 1\n\n frames = [metrics, pd.DataFrame(d, index=[0])]\n metrics = pd.concat(frames, ignore_index=True, sort=False) \n \n return metrics\n\n# write to file\ndef generate_ensemble_metrics(metrics, analysis_type, corpus, ensemble_type, filter_semtype, semtype = None):\n now = datetime.now()\n timestamp = datetime.timestamp(now)\n \n file_name = corpus + '_all_'\n \n # drop exact matches:\n metrics = metrics.drop_duplicates()\n \n if ensemble_type == 'merge':\n metrics = metrics.sort_values(by=['n_terms', 'merge'])\n file_name += 'merge_'\n elif ensemble_type == 'vote':\n file_name += '_'\n \n #metrics = metrics.drop_duplicates(subset=['TP', 'FN', 'FP', 'n_sys', 'precision', 'recall', 'F', 'TM', 'TP/FN', 'TM', 'n_terms'])\n\n file = file_name + analysis_type + '_' + run_type +'_'\n \n if filter_semtype:\n file += semtype\n \n \n geometric_mean(metrics).to_csv(analysisConf.data_dir + file + str(timestamp) + '.csv')\n print(geometric_mean(metrics))\n \n# control ensemble run\ndef ensemble_control(systems, analysis_type, corpus, run_type, filter_semtype, semtypes = None):\n if filter_semtype:\n for semtype in semtypes:\n test = get_valid_systems(systems, semtype)\n print('SYSTEMS FOR SEMTYPE', semtype, 'ARE', test)\n metrics = run_ensemble(test, analysis_type, corpus, filter_semtype, expression_type, semtype)\n if (expression_type == 'nested_with_singleton' and len(test) == 5) or expression_type in ['nested', 'paired', 'single']:\n generate_ensemble_metrics(metrics, analysis_type, corpus, ensemble_type, filter_semtype, semtype)\n else:\n metrics = run_ensemble(systems, analysis_type, corpus, filter_semtype, expression_type)\n generate_ensemble_metrics(metrics, analysis_type, corpus, ensemble_type, filter_semtype)",
"_____no_output_____"
],
[
"# ad hoc query for performance evaluation\ndef get_merge_data(boolean_expression: str, analysis_type: str, corpus: str, run_type: str, filter_semtype, semtype = None):\n \"\"\"\n Traverse binary parse tree representation of Boolean sentence\n :params: boolean expression in form of '(<annotator_engine_name1><boolean operator><annotator_engine_name2>)'\n analysis_type (string value of: 'entity', 'cui', 'full') used to filter set of reference and system annotations \n :return: dictionary with values needed for confusion matrix\n \"\"\"\n if filter_semtype:\n ann = get_ref_ann(analysis_type, corpus, filter_semtype, semtype)\n else: \n ann = get_ref_ann(analysis_type, corpus, filter_semtype)\n \n sentence = Sentence(boolean_expression) \n\n pt = make_parse_tree(sentence.sentence)\n\n r = process_sentence(pt, sentence, analysis_type, corpus, filter_semtype, semtype)\n\n if run_type == 'overlap' and rtype != 6:\n if filter_semtype:\n ((TP, TN, FP, FN),(p,r,f1)) = vectorized_cooccurences(r, analysis_type, corpus, filter_semtype, semtype)\n else:\n ((TP, TN, FP, FN),(p,r,f1)) = vectorized_cooccurences(r, analysis_type, corpus, filter_semtype)\n\n # TODO: validate against ann1/sys1 where val = 1\n # total by number chars\n system_n = TP + FP\n reference_n = TP + FN\n\n d = cm_dict(FN, FP, TP, system_n, reference_n)\n print(d)\n \n elif run_type == 'exact':\n c = get_cooccurences(ann, r.system_merges, analysis_type, corpus) # get matches, FN, etc.\n\n if c.ref_system_match > 0: # compute confusion matrix metrics and write to dictionary -> df\n # get dictionary of confusion matrix metrics\n d = cm_dict(c.ref_only, c.system_only, c.ref_system_match, c.system_n, c.ref_n)\n\n print('cm', d)\n else:\n pass\n \n # get matched data from merge\n return r.system_merges # merge_eval(reference_only, system_only, reference_system_match, system_n, reference_n)",
"_____no_output_____"
],
[
"# ad hoc query for performance evaluation\ndef get_sys_merge(boolean_expression: str, analysis_type: str, corpus: str, run_type: str, filter_semtype, semtype = None):\n \"\"\"\n Traverse binary parse tree representation of Boolean sentence\n :params: boolean expression in form of '(<annotator_engine_name1><boolean operator><annotator_engine_name2>)'\n analysis_type (string value of: 'entity', 'cui', 'full') used to filter set of reference and system annotations \n :return: dictionary with values needed for confusion matrix\n \"\"\"\n# if filter_semtype:\n# ann = get_ref_ann(analysis_type, corpus, filter_semtype, semtype)\n# else: \n# ann = get_ref_ann(analysis_type, corpus, filter_semtype)\n \n sentence = Sentence(boolean_expression) \n\n pt = make_parse_tree(sentence.sentence)\n\n for semtype in semtypes:\n test = get_valid_systems(systems, semtype)\n r = process_sentence(pt, sentence, analysis_type, corpus, filter_semtype, semtype)\n\n# if run_type == 'overlap' and rtype != 6:\n# if filter_semtype:\n# ((TP, TN, FP, FN),(p,r,f1)) = vectorized_cooccurences(r, analysis_type, corpus, filter_semtype, semtype)\n# else:\n# ((TP, TN, FP, FN),(p,r,f1)) = vectorized_cooccurences(r, analysis_type, corpus, filter_semtype)\n\n# # TODO: validate against ann1/sys1 where val = 1\n# # total by number chars\n# system_n = TP + FP\n# reference_n = TP + FN\n\n# d = cm_dict(FN, FP, TP, system_n, reference_n)\n# print(d)\n \n# elif run_type == 'exact':\n# c = get_cooccurences(ann, r.system_merges, analysis_type, corpus) # get matches, FN, etc.\n\n# if c.ref_system_match > 0: # compute confusion matrix metrics and write to dictionary -> df\n# # get dictionary of confusion matrix metrics\n# d = cm_dict(c.ref_only, c.system_only, c.ref_system_match, c.system_n, c.ref_n)\n\n# print('cm', d)\n# else:\n# pass\n \n # get matched data from merge\n return r.system_merges # merge_eval(reference_only, system_only, reference_system_match, system_n, reference_n)",
"_____no_output_____"
],
[
"# majority vote \ndef vectorized_annotations(ann):\n \n docs = get_docs(corpus)\n labels = [\"concept\"]\n out= []\n \n for n in range(len(docs)):\n a1 = list(ann.loc[ann[\"case\"] == docs[n][0]].itertuples(index=False))\n a = label_vector(docs[n][1], a1, labels)\n out.append(a)\n\n return out\n\ndef flatten_list(l):\n return [item for sublist in l for item in sublist]\n\ndef get_reference_vector(analysis_type, corpus, filter_semtype, semtype = None):\n ref_ann = get_ref_ann(analysis_type, corpus, filter_semtype, semtype)\n\n df = ref_ann.copy()\n df = df.drop_duplicates(subset=['begin','end','case'])\n df['label'] = 'concept'\n\n cols_to_keep = ['begin', 'end', 'case', 'label']\n ref = df[cols_to_keep].copy()\n test = vectorized_annotations(ref)\n ref = np.asarray(flatten_list(test), dtype=np.int32) \n\n return ref\n\ndef majority_overlap_sys(systems, analysis_type, corpus, filter_semtype, semtype = None):\n \n d = {}\n cols_to_keep = ['begin', 'end', 'case', 'label']\n sys_test = []\n \n for system in systems:\n sys_ann = get_sys_data(system, analysis_type, corpus, filter_semtype, semtype)\n df = sys_ann.copy()\n df['label'] = 'concept'\n df = df.rename(index=str, columns={\"note_id\": \"case\"})\n sys = df[df['system']==system][cols_to_keep].copy()\n test = vectorized_annotations(sys)\n d[system] = flatten_list(test) \n sys_test.append(d[system])\n\n output = sum(np.array(sys_test))\n \n n = int(len(systems) / 2)\n #print(n)\n if ((len(systems) % 2) != 0):\n vote = np.where(output > n, 1, 0)\n else:\n vote = np.where(output > n, 1, \n (np.where(output == n, random.randint(0, 1), 0)))\n \n return vote\n\ndef majority_overlap_vote_out(ref, vote, corpus): \n TP, TN, FP, FN = confused(ref, vote)\n print(TP, TN, FP, FN)\n system_n = TP + FP\n reference_n = TP + FN\n\n d = cm_dict(FN, FP, TP, system_n, reference_n)\n\n d['TN'] = TN\n d['corpus'] = corpus\n print(d)\n \n metrics = pd.DataFrame(d, index=[0])\n \n return metrics\n\n# control vote run\ndef majority_vote(systems, analysis_type, corpus, run_type, filter_semtype, semtypes = None):\n print(semtypes, systems)\n if filter_semtype:\n for semtype in semtypes:\n test = get_valid_systems(systems, semtype)\n print('SYSYEMS FOR SEMTYPE', semtype, 'ARE', test)\n \n if run_type == 'overlap':\n ref = get_reference_vector(analysis_type, corpus, filter_semtype, semtype)\n vote = majority_overlap_sys(test, analysis_type, corpus, filter_semtype, semtype)\n metrics = majority_overlap_vote_out(ref, vote, corpus)\n #generate_ensemble_metrics(metrics, analysis_type, corpus, ensemble_type, filter_semtype, semtype)\n elif run_type == 'exact':\n sys = majority_exact_sys(test, analysis_type, corpus, filter_semtype, semtype)\n d = majority_exact_vote_out(sys, filter_semtype, semtype)\n metrics = pd.DataFrame(d, index=[0])\n elif run_type == 'cui':\n sys = majority_cui_sys(test, analysis_type, corpus, filter_semtype, semtype)\n d = majority_cui_vote_out(sys, filter_semtype, semtype)\n metrics = pd.DataFrame(d, index=[0])\n \n metrics['systems'] = ','.join(test)\n generate_ensemble_metrics(metrics, analysis_type, corpus, ensemble_type, filter_semtype, semtype)\n \n else:\n if run_type == 'overlap':\n ref = get_reference_vector(analysis_type, corpus, filter_semtype)\n vote = majority_overlap_sys(systems, analysis_type, corpus, filter_semtype)\n metrics = majority_overlap_vote_out(ref, vote, corpus)\n \n elif run_type == 'exact':\n sys = majority_exact_sys(systems, analysis_type, corpus, filter_semtype)\n d = majority_exact_vote_out(sys, filter_semtype)\n metrics = pd.DataFrame(d, index=[0])\n \n elif run_type == 'cui':\n sys = majority_cui_sys(systems, analysis_type, corpus, filter_semtype)\n d = majority_cui_vote_out(sys, filter_semtype)\n metrics = pd.DataFrame(d, index=[0])\n \n metrics['systems'] = ','.join(systems)\n generate_ensemble_metrics(metrics, analysis_type, corpus, ensemble_type, filter_semtype)\n \n print(metrics)\n \ndef majority_cui_sys(systems, analysis_type, corpus, filter_semtype, semtype = None):\n \n cols_to_keep = ['cui', 'note_id', 'system']\n \n df = pd.DataFrame()\n for system in systems:\n if filter_semtype:\n sys = get_sys_data(system, analysis_type, corpus, filter_semtype, semtype)\n else:\n sys = get_sys_data(system, analysis_type, corpus, filter_semtype)\n \n sys = sys[sys['system'] == system][cols_to_keep].drop_duplicates()\n \n frames = [df, sys]\n df = pd.concat(frames)\n \n return df\n\ndef majority_cui_vote_out(sys, filter_semtype, semtype = None):\n \n sys = sys.astype(str)\n sys['value_cui'] = list(zip(sys.cui, sys.note_id.astype(str)))\n sys['count'] = sys.groupby(['value_cui'])['value_cui'].transform('count')\n\n n = int(len(systems) / 2)\n if ((len(systems) % 2) != 0):\n sys = sys[sys['count'] > n]\n else:\n # https://stackoverflow.com/questions/23330654/update-a-dataframe-in-pandas-while-iterating-row-by-row\n for i in sys.index:\n if sys.at[i, 'count'] == n:\n sys.at[i, 'count'] = random.choice([1,len(systems)])\n sys = sys[sys['count'] > n]\n\n sys = sys.drop_duplicates(subset=['value_cui', 'cui', 'note_id'])\n ref = get_ref_ann(analysis_type, corpus, filter_semtype, semtype)\n\n c = get_cooccurences(ref, sys, analysis_type, corpus) # get matches, FN, etc.\n\n if c.ref_system_match > 0: # compute confusion matrix metrics and write to dictionary -> df\n # get dictionary of confusion matrix metrics\n print(cm_dict(c.ref_only, c.system_only, c.ref_system_match, c.system_n, c.ref_n))\n return cm_dict(c.ref_only, c.system_only, c.ref_system_match, c.system_n, c.ref_n)\n \n\ndef majority_exact_sys(systems, analysis_type, corpus, filter_semtype, semtype = None):\n \n cols_to_keep = ['begin', 'end', 'note_id', 'system']\n \n df = pd.DataFrame()\n for system in systems:\n if filter_semtype:\n sys = get_sys_data(system, analysis_type, corpus, filter_semtype, semtype)\n else:\n sys = get_sys_data(system, analysis_type, corpus, filter_semtype)\n \n sys = sys[sys['system'] == system][cols_to_keep].drop_duplicates()\n \n frames = [df, sys]\n df = pd.concat(frames)\n \n return df\n \ndef majority_exact_vote_out(sys, filter_semtype, semtype = None):\n sys['span'] = list(zip(sys.begin, sys.end, sys.note_id.astype(str)))\n sys['count'] = sys.groupby(['span'])['span'].transform('count')\n\n n = int(len(systems) / 2)\n if ((len(systems) % 2) != 0):\n sys = sys[sys['count'] > n]\n else:\n # https://stackoverflow.com/questions/23330654/update-a-dataframe-in-pandas-while-iterating-row-by-row\n for i in sys.index:\n if sys.at[i, 'count'] == n:\n sys.at[i, 'count'] = random.choice([1,len(systems)])\n sys = sys[sys['count'] > n]\n\n sys = sys.drop_duplicates(subset=['span', 'begin', 'end', 'note_id'])\n ref = get_ref_ann(analysis_type, corpus, filter_semtype, semtype)\n\n c = get_cooccurences(ref, sys, analysis_type, corpus) # get matches, FN, etc.\n\n if c.ref_system_match > 0: # compute confusion matrix metrics and write to dictionary -> df\n # get dictionary of confusion matrix metrics\n print(cm_dict(c.ref_only, c.system_only, c.ref_system_match, c.system_n, c.ref_n))\n return cm_dict(c.ref_only, c.system_only, c.ref_system_match, c.system_n, c.ref_n)\n \n#ensemble_type = 'vote' \n#filter_semtype = False\n#majority_vote(systems, analysis_type, corpus, run_type, filter_semtype, semtypes)",
"_____no_output_____"
],
[
"#%%time\ndef main():\n '''\n corpora: i2b2, mipacq, fv017\n analyses: entity only (exact span), cui by document, full (aka (entity and cui on exaact span/exact cui)\n systems: ctakes, biomedicus, clamp, metamap, quick_umls\n \n TODO -> Vectorization (entity only) -> done:\n add switch for use of TN on single system performance evaluations -> done\n add switch for overlap matching versus exact span -> done\n -> Other tasks besides concept extraction\n \n ''' \n analysisConf = AnalysisConfig()\n print(analysisConf.systems, analysisConf.corpus_config())\n \n if (rtype == 1):\n print(semtypes, systems)\n if filter_semtype:\n for semtype in semtypes:\n test = get_valid_systems(systems, semtype)\n print('SYSYEMS FOR SEMTYPE', semtype, 'ARE', test)\n generate_metrics(analysis_type, corpus, filter_semtype, semtype)\n \n else:\n generate_metrics(analysis_type, corpus, filter_semtype)\n \n elif (rtype == 2):\n print('run_type:', run_type)\n if filter_semtype:\n print(semtypes)\n ensemble_control(analysisConf.systems, analysis_type, corpus, run_type, filter_semtype, semtypes)\n else:\n ensemble_control(analysisConf.systems, analysis_type, corpus, run_type, filter_semtype)\n elif (rtype == 3):\n t = ['concept_jaccard_score_false']\n test_systems(analysis_type, analysisConf.systems, corpus) \n test_count(analysis_type, corpus)\n test_ensemble(analysis_type, corpus)\n elif (rtype == 4):\n if filter_semtype:\n majority_vote(systems, analysis_type, corpus, run_type, filter_semtype, semtypes)\n else:\n majority_vote(systems, analysis_type, corpus, run_type, filter_semtype)\n elif (rtype == 5):\n \n # control filter_semtype in get_sys_data, get_ref_n and generate_metrics. TODO consolidate. \n # # run single ad hoc statement\n statement = '((ctakes&biomedicus)|metamap)'\n\n def ad_hoc(analysis_type, corpus, statement):\n sys = get_merge_data(statement, analysis_type, corpus, run_type, filter_semtype)\n sys = sys.rename(index=str, columns={\"note_id\": \"case\"})\n sys['label'] = 'concept'\n\n ref = get_reference_vector(analysis_type, corpus, filter_semtype)\n sys = vectorized_annotations(sys)\n sys = np.asarray(flatten_list(list(sys)), dtype=np.int32)\n\n return ref, sys\n\n ref, sys = ad_hoc(analysis_type, corpus, statement)\n\n elif (rtype == 6): # 5 w/o evaluation\n \n statement = '(ctakes|biomedicus)' #((((A∧C)∧D)∧E)∨B)->for covid pipeline\n\n def ad_hoc(analysis_type, corpus, statement):\n print(semtypes)\n for semtype in semtypes:\n sys = get_sys_merge(statement, analysis_type, corpus, run_type, filter_semtype, semtype)\n sys = sys.rename(index=str, columns={\"note_id\": \"case\"})\n\n return sys\n\n sys = ad_hoc(analysis_type, corpus, statement).sort_values(by=['case', 'begin'])\n sys.drop_duplicates(['cui', 'case', 'polarity'],inplace=True)\n sys.to_csv(data_directory + 'test_new.csv')\n \n test = sys.copy()\n test.drop(['begin','end','case','polarity'], axis=1, inplace=True) \n test.to_csv(data_directory + 'test_dedup_new.csv')\n\nif __name__ == '__main__':\n #%prun main()\n main()\n print('done!')\n pass",
"_____no_output_____"
]
]
] |
[
"raw",
"code",
"raw",
"code",
"markdown",
"code",
"raw",
"code"
] |
[
[
"raw"
],
[
"code",
"code"
],
[
"raw"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"raw"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbef8309189399bcaed6c7dc38df3a6cd5ba69d7
| 9,781 |
ipynb
|
Jupyter Notebook
|
.ipynb_checkpoints/Tratamento-de-dados-checkpoint.ipynb
|
repositorio-gil-rocha/Titanic
|
07a3bdce41c7ab010a4dd221f228849285792cf5
|
[
"MIT"
] | null | null | null |
.ipynb_checkpoints/Tratamento-de-dados-checkpoint.ipynb
|
repositorio-gil-rocha/Titanic
|
07a3bdce41c7ab010a4dd221f228849285792cf5
|
[
"MIT"
] | null | null | null |
.ipynb_checkpoints/Tratamento-de-dados-checkpoint.ipynb
|
repositorio-gil-rocha/Titanic
|
07a3bdce41c7ab010a4dd221f228849285792cf5
|
[
"MIT"
] | null | null | null | 31.859935 | 90 | 0.320622 |
[
[
[
"# importando as blibs\nimport pandas as pd\nimport numpy as np\nimport datetime as dt\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nimport warnings\nwarnings.filterwarnings('ignore')",
"Matplotlib is building the font cache; this may take a moment.\n"
],
[
"# Importando os dados\ndf_teste = pd.read_csv('test.csv')\ndf_treino = pd.read_csv('train.csv')\ndf_submission = pd.read_csv('gender_submission.csv')",
"_____no_output_____"
],
[
"df_teste",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code"
]
] |
cbef876145d4fb582a22b744a61ba3faf1649287
| 149,043 |
ipynb
|
Jupyter Notebook
|
sketchbook.ipynb
|
piovere/zeTorch
|
9c54f7b3fd2eee2e3cff14a97be40ca317408bf7
|
[
"MIT"
] | null | null | null |
sketchbook.ipynb
|
piovere/zeTorch
|
9c54f7b3fd2eee2e3cff14a97be40ca317408bf7
|
[
"MIT"
] | null | null | null |
sketchbook.ipynb
|
piovere/zeTorch
|
9c54f7b3fd2eee2e3cff14a97be40ca317408bf7
|
[
"MIT"
] | null | null | null | 121.173171 | 101,584 | 0.793637 |
[
[
[
"%matplotlib inline\nimport pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"def loadfile(fn):\n \"\"\"Load plasma torch spectrum from file\n \n Returns pandas dataframe with two columns. When loading, it\n checks to validate that average counts (\"Intensity\") is above 100.\n It also checks that all channels above 1000nm are less than 100.\n If either of these checks fails, it raises an error instead of\n loading the data. It also sets all channels with negative counts\n to zero.\n \"\"\"\n d = pd.read_table(\n fn,\n sep='\\s+',\n skiprows=15,\n skipfooter=240,\n engine='python',\n header=None,\n names=['Wavelength', 'Intensity']\n )\n d[d<0] = 0\n \n # Verify the mean is above 100\n mn = np.mean(d['Intensity'])\n if mn < 100:\n raise Exception('Average intensity is too low ({} < 100)'.format(np.mean(d['Intensity'])))\n \n # Verify that channels above 1000nm are < 100\n high_wavelength_values = d[d['Wavelength'] > 1000]\n mx = np.max(high_wavelength_values['Intensity'])\n if mx > 100:\n raise Exception('High-wavelength intensity too high ({} > 100)'.format(mx))\n \n # Normalize the intensity\n d['Intensity'] = d['Intensity'] / sum(d['Intensity'])\n \n return d",
"_____no_output_____"
],
[
"d = loadfile('Data/20170721-Ar-H-Torch.txt')",
"_____no_output_____"
],
[
"f = plt.figure(1, figsize=(20,20))\np = plt.subplot(111)\np.plot(d['Wavelength'], d['Intensity'])\nfig_size = [0, 0]\nfig_size[0] = 12\nfig_size[1] = 9\nplt.title(\"Spectrum\")\nplt.xlabel('Wavelength (nm)')\nplt.show()",
"_____no_output_____"
],
[
"bd1 = loadfile(\"Data/Bad/20170906_ORNL-Torch-Sil_Subt14_14-31-55-813.txt\")",
"_____no_output_____"
],
[
"bd2 = loadfile('Data/Bad/20170906_ORNL-Torch-Sil_Subt14_14-33-16-145.txt')",
"_____no_output_____"
],
[
"d",
"_____no_output_____"
],
[
"import peakutils as pk",
"_____no_output_____"
],
[
"indexes = pk.indexes(d['Intensity'], thres=0.0005, min_dist=0.1)\nprint(indexes)\nprint(len(indexes))\nprint(d['Wavelength'][indexes], d['Intensity'][indexes])",
"[ 71 85 90 92 94 97 99 104 108 113 117 119 121 124 128\n 130 134 138 140 144 147 149 151 153 155 158 160 163 168 170\n 173 175 178 181 183 186 193 197 199 201 206 212 214 218 223\n 226 229 231 233 236 238 241 247 250 252 254 257 259 262 266\n 269 271 276 279 282 286 292 294 299 301 305 308 313 319 321\n 324 329 332 344 347 351 356 360 367 372 377 379 381 384 387\n 389 392 395 403 406 415 419 422 428 430 433 436 440 444 446\n 450 457 460 467 471 478 486 494 499 501 507 517 524 530 538\n 541 544 547 552 557 562 568 573 577 580 585 589 596 600 605\n 611 636 654 659 664 667 671 674 679 682 686 688 705 707 724\n 731 733 736 738 742 744 746 748 751 755 758 764 772 777 779\n 786 793 802 812 820 823 827 829 833 837 840 844 846 850 852\n 865 870 880 897 908 911 914 919 925 936 938 946 948 951 955\n 966 972 983 1012 1015 1045 1058 1067 1070 1073 1084 1103 1105 1126 1128\n 1138 1144 1146 1160 1172 1174 1184 1199 1210 1212 1218 1227 1244 1256 1276\n 1287 1303 1306 1314 1327 1342 1351 1365 1381 1388 1394 1399 1413 1417 1432\n 1436 1453 1458 1469 1475 1478 1483 1492 1495 1500 1502 1509 1512 1515 1523\n 1526 1530 1533 1537 1541 1544 1547 1552 1554 1557 1560 1565 1570 1576 1580\n 1583 1597 1609 1614 1621 1630 1635 1638 1642 1644 1648 1651 1660 1667 1669\n 1671 1675 1678 1684 1686 1688 1690 1694 1698 1700 1703 1705 1708 1710 1713\n 1715 1722 1728 1748 1752 1765]\n306\n71 221.832\n85 228.427\n90 230.781\n92 231.722\n94 232.664\n97 234.076\n99 235.017\n104 237.369\n108 239.251\n113 241.602\n117 243.483\n119 244.423\n121 245.363\n124 246.773\n128 248.652\n130 249.592\n134 251.471\n138 253.349\n140 254.288\n144 256.166\n147 257.574\n149 258.513\n151 259.451\n153 260.389\n155 261.328\n158 262.735\n160 263.673\n163 265.080\n168 267.424\n170 268.362\n ... \n1635 927.199\n1638 928.490\n1642 930.210\n1644 931.071\n1648 932.791\n1651 934.080\n1660 937.948\n1667 940.955\n1669 941.814\n1671 942.672\n1675 944.390\n1678 945.677\n1684 948.251\n1686 949.109\n1688 949.967\n1690 950.825\n1694 952.540\n1698 954.254\n1700 955.112\n1703 956.397\n1705 957.254\n1708 958.539\n1710 959.396\n1713 960.680\n1715 961.537\n1722 964.533\n1728 967.100\n1748 975.651\n1752 977.359\n1765 982.910\nName: Wavelength, Length: 306, dtype: float64 71 0.000005\n85 0.000005\n90 0.000008\n92 0.000006\n94 0.000005\n97 0.000009\n99 0.000008\n104 0.000010\n108 0.000009\n113 0.000010\n117 0.000011\n119 0.000012\n121 0.000013\n124 0.000015\n128 0.000018\n130 0.000015\n134 0.000018\n138 0.000019\n140 0.000019\n144 0.000021\n147 0.000021\n149 0.000021\n151 0.000025\n153 0.000024\n155 0.000023\n158 0.000026\n160 0.000024\n163 0.000025\n168 0.000029\n170 0.000029\n ... \n1635 0.000025\n1638 0.000023\n1642 0.000015\n1644 0.000014\n1648 0.000014\n1651 0.000039\n1660 0.000042\n1667 0.000007\n1669 0.000007\n1671 0.000007\n1675 0.000008\n1678 0.000007\n1684 0.000006\n1686 0.000005\n1688 0.000006\n1690 0.000007\n1694 0.000005\n1698 0.000006\n1700 0.000005\n1703 0.000006\n1705 0.000006\n1708 0.000004\n1710 0.000007\n1713 0.000005\n1715 0.000006\n1722 0.000081\n1728 0.000006\n1748 0.000005\n1752 0.000023\n1765 0.000005\nName: Intensity, Length: 306, dtype: float64\n"
],
[
"peaks_x = pk.interpolate(d.as_matrix(columns=d.columns), d.as_matrix(columns=d.columns), ind=indexes)\nprint(peaks_x)",
"_____no_output_____"
],
[
"d[d.Wavelength > 500]",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbef896e04f4833ee28e914ff51b4d9af077319d
| 12,340 |
ipynb
|
Jupyter Notebook
|
Page-Rank.ipynb
|
rgaezsd/pagerank
|
6858ff1e2c19a280d3df33ac4224a88f80dbdca6
|
[
"MIT"
] | null | null | null |
Page-Rank.ipynb
|
rgaezsd/pagerank
|
6858ff1e2c19a280d3df33ac4224a88f80dbdca6
|
[
"MIT"
] | null | null | null |
Page-Rank.ipynb
|
rgaezsd/pagerank
|
6858ff1e2c19a280d3df33ac4224a88f80dbdca6
|
[
"MIT"
] | null | null | null | 31.399491 | 246 | 0.522366 |
[
[
[
"%pylab notebook\r\nimport numpy as np\r\nimport numpy.linalg as la\r\nnp.set_printoptions(suppress=True)",
"Populating the interactive namespace from numpy and matplotlib\n"
]
],
[
[
"Let's imagine a micro-internet, with just 6 websites (**A**vocado, **B**ullseye, **C**atBabel, **D**romeda, **e**Tings, and **F**aceSpace).\r\nEach website links to some of the others, and this forms a network like this.\r\n\r\n\r\n\r\nWe have 100 *Procrastinating Pat*s (see [README](https://github.com/rgaezsd/pagerank/blob/main/README.md)) on our micro-internet, each viewing a single website at a time.\r\n\r\nEach minute the Pats follow a link on their website to another site on the micro-internet.\r\n\r\nAfter a while, the websites that are most linked to will have more Pats visiting them, and in the long run, each minute for every Pat that leaves a website, another will enter keeping the total numbers of Pats on each website constant.\r\n\r\nWe represent the number of Pats on each website with the vector,\r\n$$\\mathbf{r} = \\begin{bmatrix} r_A \\\\ r_B \\\\ r_C \\\\ r_D \\\\ r_E \\\\ r_F \\end{bmatrix}$$\r\nAnd say that the number of Pats on each website in minute $i+1$ is related to those at minute $i$ by the matrix transformation\r\n\r\n$$ \\mathbf{r}^{(i+1)} = L \\,\\mathbf{r}^{(i)}$$\r\nwith the matrix $L$ taking the form,\r\n$$ L = \\begin{bmatrix}\r\nL_{A→A} & L_{B→A} & L_{C→A} & L_{D→A} & L_{E→A} & L_{F→A} \\\\\r\nL_{A→B} & L_{B→B} & L_{C→B} & L_{D→B} & L_{E→B} & L_{F→B} \\\\\r\nL_{A→C} & L_{B→C} & L_{C→C} & L_{D→C} & L_{E→C} & L_{F→C} \\\\\r\nL_{A→D} & L_{B→D} & L_{C→D} & L_{D→D} & L_{E→D} & L_{F→D} \\\\\r\nL_{A→E} & L_{B→E} & L_{C→E} & L_{D→E} & L_{E→E} & L_{F→E} \\\\\r\nL_{A→F} & L_{B→F} & L_{C→F} & L_{D→F} & L_{E→F} & L_{F→F} \\\\\r\n\\end{bmatrix}\r\n$$\r\nwhere the columns represent the probability of leaving a website for any other website, and sum to one.\r\nThe rows determine how likely you are to enter a website from any other, though these need not add to one.\r\nThe long time behaviour of this system is when $ \\mathbf{r}^{(i+1)} = \\mathbf{r}^{(i)}$, so we'll drop the superscripts here, and that allows us to write,\r\n$$ L \\,\\mathbf{r} = \\mathbf{r}$$\r\n\r\nwhich is an eigenvalue equation for the matrix $L$, with eigenvalue 1 (this is guaranteed by the probabalistic structure of the matrix $L$).",
"_____no_output_____"
]
],
[
[
"L = np.array([[0, 1/2, 1/3, 0, 0, 0 ],\r\n [1/3, 0, 0, 0, 1/2, 0 ],\r\n [1/3, 1/2, 0, 1, 0, 1/2 ],\r\n [1/3, 0, 1/3, 0, 1/2, 1/2 ],\r\n [0, 0, 0, 0, 0, 0 ],\r\n [0, 0, 1/3, 0, 0, 0 ]])",
"_____no_output_____"
],
[
"eVals, eVecs = la.eig(L)\r\norder = np.absolute(eVals).argsort()[::-1]\r\neVals = eVals[order]\r\neVecs = eVecs[:,order]\r\n\r\nr = eVecs[:, 0]\r\n100 * np.real(r / np.sum(r))",
"_____no_output_____"
]
],
[
[
"We can see from this list, the number of Procrastinating Pats that we expect to find on each website after long times.\r\nPutting them in order of *popularity* (based on this metric), the PageRank of this micro-internet is:\r\n\r\n**C**atBabel, **D**romeda, **A**vocado, **F**aceSpace, **B**ullseye, **e**Tings\r\n\r\nReferring back to the micro-internet diagram, is this what you would have expected?\r\nConvince yourself that based on which pages seem important given which others link to them, that this is a sensible ranking.\r\n\r\nLet's now try to get the same result using the Power-Iteration method.",
"_____no_output_____"
]
],
[
[
"r = 100 * np.ones(6) / 6\r\nr",
"_____no_output_____"
]
],
[
[
"Next, let's update the vector to the next minute, with the matrix $L$.\nRun the following cell multiple times, until the answer stabilises.",
"_____no_output_____"
]
],
[
[
"r = 100 * np.ones(6) / 6 \r\nfor i in np.arange(100):\r\n r = L @ r\r\nr",
"_____no_output_____"
]
],
[
[
"Or even better, we can keep running until we get to the required tolerance.",
"_____no_output_____"
]
],
[
[
"r = 100 * np.ones(6) / 6\r\nlastR = r\r\nr = L @ r\r\ni = 0\r\nwhile la.norm(lastR - r) > 0.01 :\r\n lastR = r\r\n r = L @ r\r\n i += 1\r\nprint(str(i) + \" iterations to convergence.\")\r\nr",
"18 iterations to convergence.\n"
]
],
[
[
"### Damping Parameter Case\r\n\r\nLet's consider an extension to our micro-internet where things start to go wrong.\r\n\r\nSay a new website is added to the micro-internet: *Geoff's* Website.\r\nThis website is linked to by *FaceSpace* and only links to itself.\r\n\r\n\r\n\r\nIntuitively, only *FaceSpace*, which is in the bottom half of the page rank, links to this website amongst the two others it links to,\r\nso we might expect *Geoff's* site to have a correspondingly low PageRank score.",
"_____no_output_____"
]
],
[
[
"L2 = np.array([[0, 1/2, 1/3, 0, 0, 0, 0 ],\r\n [1/3, 0, 0, 0, 1/2, 0, 0 ],\r\n [1/3, 1/2, 0, 1, 0, 1/3, 0 ],\r\n [1/3, 0, 1/3, 0, 1/2, 1/3, 0 ],\r\n [0, 0, 0, 0, 0, 0, 0 ],\r\n [0, 0, 1/3, 0, 0, 0, 0 ],\r\n [0, 0, 0, 0, 0, 1/3, 1 ]])",
"_____no_output_____"
],
[
"r = 100 * np.ones(7) / 7\r\nlastR = r\r\nr = L2 @ r\r\ni = 0\r\nwhile la.norm(lastR - r) > 0.01 :\r\n lastR = r\r\n r = L2 @ r\r\n i += 1\r\nprint(str(i) + \" iterations to convergence.\")\r\nr",
"131 iterations to convergence.\n"
]
],
[
[
"That's no good! *Geoff* seems to be taking all the traffic on the micro-internet, and somehow coming at the top of the PageRank.\r\nThis behaviour can be understood, because once a Pat get's to *Geoff's* Website, they can't leave, as all links head back to Geoff.\r\n\r\nTo combat this, we can add a small probability that the Procrastinating Pats don't follow any link on a webpage, but instead visit a website on the micro-internet at random.\r\nWe'll say the probability of them following a link is $d$ and the probability of choosing a random website is therefore $1-d$.\r\nWe can use a new matrix to work out where the Pat's visit each minute.\r\n$$ M = d \\, L + \\frac{1-d}{n} \\, J $$\r\nwhere $J$ is an $n\\times n$ matrix where every element is one.\r\n\r\nIf $d$ is one, we have the case we had previously, whereas if $d$ is zero, we will always visit a random webpage and therefore all webpages will be equally likely and equally ranked.\r\nFor this extension to work best, $1-d$ should be somewhat small - though we won't go into a discussion about exactly how small.",
"_____no_output_____"
]
],
[
[
"d = 0.5\r\nM = d * L2 + (1-d)/7 * np.ones([7, 7])",
"_____no_output_____"
],
[
"r = 100 * np.ones(7) / 7\r\nM = d * L2 + (1-d)/7 * np.ones([7, 7])\r\nlastR = r\r\nr = M @ r\r\ni = 0\r\nwhile la.norm(lastR - r) > 0.01 :\r\n lastR = r\r\n r = M @ r\r\n i += 1\r\nprint(str(i) + \" iterations to convergence.\")\r\nr",
"8 iterations to convergence.\n"
]
],
[
[
"This is certainly better, the PageRank gives sensible numbers for the Procrastinating Pats that end up on each webpage.\r\nThis method still predicts Geoff has a high ranking webpage however.\r\nThis could be seen as a consequence of using a small network.",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
cbefaa6e82549e590bbd07168f702be8be0ada12
| 44,252 |
ipynb
|
Jupyter Notebook
|
scripts/optimal_policy/4-2-75/plot.ipynb
|
ILABUTK/Integrated_PV-battery_System
|
438c363d68b959fea6773ad716726e542a7f24d4
|
[
"MIT"
] | null | null | null |
scripts/optimal_policy/4-2-75/plot.ipynb
|
ILABUTK/Integrated_PV-battery_System
|
438c363d68b959fea6773ad716726e542a7f24d4
|
[
"MIT"
] | null | null | null |
scripts/optimal_policy/4-2-75/plot.ipynb
|
ILABUTK/Integrated_PV-battery_System
|
438c363d68b959fea6773ad716726e542a7f24d4
|
[
"MIT"
] | null | null | null | 260.305882 | 39,173 | 0.91397 |
[
[
[
"import numpy as np\r\nimport pandas as pd\r\nimport matplotlib.pyplot as plt\r\nimport matplotlib.ticker as ticker",
"_____no_output_____"
],
[
"# data\r\nx = list(range(0, 60, 1))\r\ndata = pd.read_csv(\"4-2-75.csv\", index_col=False)\r\nell = data['ell'].to_list()",
"_____no_output_____"
],
[
"# font size\r\nplt.rc('axes', labelsize=18)\r\nplt.rc('xtick', labelsize=18)\r\nplt.rc('ytick', labelsize=18)\r\nplt.rc('legend', fontsize=16)\r\nplt.rc('axes', titlesize=18)\r\nplt.rc('text', usetex=True)\r\n#plt.rc('font',**{'family':'sans-serif','sans-serif':['Helvetica']})\r\nplt.rcParams['text.latex.preamble'] = [\r\n r'\\usepackage{tgheros}', # helvetica font\r\n r'\\usepackage{sansmath}', # math-font matching helvetica\r\n r'\\sansmath' # actually tell tex to use it!\r\n r'\\usepackage{siunitx}', # micro symbols\r\n r'\\sisetup{detect-all}', # force siunitx to use the fonts\r\n]",
"<ipython-input-3-b3fe6e799c48>:9: MatplotlibDeprecationWarning: Support for setting the 'text.latex.preamble' or 'pgf.preamble' rcParam to a list of strings is deprecated since 3.3 and will be removed two minor releases later; set it to a single string instead.\n plt.rcParams['text.latex.preamble'] = [\n"
],
[
"# plot\r\nfig, ax = plt.subplots(nrows=1, ncols=1, figsize=(8, 6))\r\n# threshold\r\nax.plot(\r\n x, [0.75] * len(x), linestyle='-', linewidth=3, color='red', alpha=0.5,\r\n label=r'$\\underline{\\ell}$' + ' -- minimum replacement threshold' \r\n)\r\n# optimal policy\r\nax.plot(\r\n x, ell, linestyle='-.', linewidth=3, color='blue',\r\n label=r'$\\ell^*$' + ' -- optimal replacement threshold '\r\n)\r\n# fill with shade -- Replace\r\nax.fill_between(\r\n x, [0.75] * len(x), ell,\r\n facecolor='w', hatch = '|'\r\n)\r\nax.text(\r\n 8, 0.85, 'Replace', fontsize=20,\r\n bbox = dict(facecolor = 'w', alpha = 1.0, linewidth=0)\r\n)\r\n# fill with shade -- Keep\r\nax.fill_between(\r\n x, [1.00] * len(x), ell,\r\n facecolor='w', hatch = '.'\r\n)\r\nax.text(\r\n 36, 0.85, 'Keep', fontsize=20,\r\n bbox = dict(facecolor = 'w', alpha = 1.0, linewidth=0)\r\n)\r\n# fill with shade -- Bottom\r\nax.fill_between(\r\n x, [0.70] * len(x), [0.75] * len(x),\r\n facecolor='w', hatch = '\\\\'\r\n)\r\nax.text(\r\n 14, 0.72, r'Unreachable below $\\underline{\\ell}=75\\%$', fontsize=20,\r\n bbox = dict(facecolor = 'w', alpha = 1.0, linewidth=0)\r\n)\r\nax.yaxis.set_major_formatter(ticker.PercentFormatter(xmax=1))\r\nax.set_ylim(0.70, 1.00)\r\nax.set_xlim(0, 59)\r\nax.set_xticks([0, 10, 20, 30, 40, 50, 59])\r\nax.set_xlabel(\"Day\")\r\nax.set_ylabel(\"Maximum available capacity of battery\")\r\nax.legend(loc=(0.327, 1.01))\r\nfig.tight_layout()\r\nfig.show()\r\nfig.savefig(\"4-2-75.png\", dpi=600)\r\nfig.savefig(\"4-2-75_red.png\", dpi=100)",
"<ipython-input-4-203d83566435>:48: UserWarning: Matplotlib is currently using module://ipykernel.pylab.backend_inline, which is a non-GUI backend, so cannot show the figure.\n fig.show()\n"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code"
]
] |
cbefcbc474f93a5d8818ba61b887e9916072d53b
| 74,251 |
ipynb
|
Jupyter Notebook
|
mclearn/knfst/python/test.ipynb
|
chengsoonong/mclass-sky
|
98219221c233fa490e78246eda1ead05c6cf7c17
|
[
"BSD-3-Clause"
] | 9 |
2016-06-01T12:09:47.000Z
|
2021-01-16T05:28:01.000Z
|
mclearn/knfst/python/test.ipynb
|
alasdairtran/mclearn
|
98219221c233fa490e78246eda1ead05c6cf7c17
|
[
"BSD-3-Clause"
] | 165 |
2015-01-28T10:37:34.000Z
|
2017-10-23T06:55:13.000Z
|
mclearn/knfst/python/test.ipynb
|
alasdairtran/mclearn
|
98219221c233fa490e78246eda1ead05c6cf7c17
|
[
"BSD-3-Clause"
] | 9 |
2015-01-24T16:27:54.000Z
|
2020-09-01T08:54:31.000Z
| 61.772879 | 26,121 | 0.661904 |
[
[
[
"import numpy as np\nimport scipy as sp\nimport pandas as pd\nimport urllib.request\nimport os\nimport shutil\nimport tarfile\nimport matplotlib.pyplot as plt\nfrom sklearn import datasets, cross_validation, metrics\nfrom sklearn.preprocessing import KernelCenterer\n\n%matplotlib notebook",
"_____no_output_____"
]
],
[
[
"First we need to download the Caltech256 dataset.",
"_____no_output_____"
]
],
[
[
"DATASET_URL = r\"http://homes.esat.kuleuven.be/~tuytelaa/\"\\\n\"unsup/unsup_caltech256_dense_sift_1000_bow.tar.gz\"\nDATASET_DIR = \"../../../projects/weiyen/data\"",
"_____no_output_____"
],
[
"filename = os.path.split(DATASET_URL)[1]\ndest_path = os.path.join(DATASET_DIR, filename)\n\nif os.path.exists(dest_path):\n print(\"{} exists. Skipping download...\".format(dest_path))\nelse:\n with urllib.request.urlopen(DATASET_URL) as response, open(dest_path, 'wb') as out_file:\n shutil.copyfileobj(response, out_file)\n print(\"Dataset downloaded. Extracting files...\")\n\ntar = tarfile.open(dest_path)\ntar.extractall(path=DATASET_DIR)\nprint(\"Files extracted.\")\ntar.close()\n\npath = os.path.join(DATASET_DIR, \"bow_1000_dense/\")",
"../../../projects/weiyen/data/unsup_caltech256_dense_sift_1000_bow.tar.gz exists. Skipping download...\nFiles extracted.\n"
]
],
[
[
"Calculate multi-class KNFST model for multi-class novelty detection\n \n INPUT\n K: NxN kernel matrix containing similarities of n training samples\n labels: Nx1 column vector containing multi-class labels of N training samples\n\n OUTPUT\n proj: Projection of KNFST\n target_points: The projections of training data into the null space\n",
"_____no_output_____"
],
[
"Load the dataset into memory",
"_____no_output_____"
]
],
[
[
"ds = datasets.load_files(path)\nds.data = np.vstack([np.fromstring(txt, sep='\\t') for txt in ds.data])\n",
"_____no_output_____"
],
[
"data = ds.data\ntarget = ds.target",
"_____no_output_____"
]
],
[
[
"Select a few \"known\" classes",
"_____no_output_____"
]
],
[
[
"classes = np.unique(target)\nnum_class = len(classes)\nnum_known = 5\n\nknown = np.random.choice(classes, num_known)\nmask = np.array([y in known for y in target])\n\nX_train = data[mask]\ny_train = target[mask]\n\nidx = y_train.argsort()\nX_train = X_train[idx]\ny_train = y_train[idx]\n\nprint(X_train.shape)\nprint(y_train.shape)",
"(538, 1000)\n(538,)\n"
],
[
"def _hik(x, y):\n '''\n Implements the histogram intersection kernel.\n '''\n return np.minimum(x, y).sum()\n",
"_____no_output_____"
],
[
"from scipy.linalg import svd\n\ndef nullspace(A, eps=1e-12):\n u, s, vh = svd(A)\n null_mask = (s <= eps)\n null_space = sp.compress(null_mask, vh, axis=0)\n return sp.transpose(null_space)\n\nA = np.array([[2,3,5],[-4,2,3],[0,0,0]])\nnp.array([-4,2,3]).dot(nullspace(A))",
"_____no_output_____"
]
],
[
[
"Train the model, and obtain the projection and class target points.",
"_____no_output_____"
]
],
[
[
"def learn(K, labels):\n classes = np.unique(labels)\n if len(classes) < 2:\n raise Exception(\"KNFST requires 2 or more classes\")\n n, m = K.shape\n if n != m:\n raise Exception(\"Kernel matrix must be quadratic\")\n \n centered_k = KernelCenterer().fit_transform(K)\n \n basis_values, basis_vecs = np.linalg.eigh(centered_k)\n \n basis_vecs = basis_vecs[:,basis_values > 1e-12]\n basis_values = basis_values[basis_values > 1e-12]\n \n basis_values = np.diag(1.0/np.sqrt(basis_values))\n\n basis_vecs = basis_vecs.dot(basis_values)\n\n L = np.zeros([n,n])\n for cl in classes:\n for idx1, x in enumerate(labels == cl):\n for idx2, y in enumerate(labels == cl):\n if x and y:\n L[idx1, idx2] = 1.0/np.sum(labels==cl)\n M = np.ones([m,m])/m\n H = (((np.eye(m,m)-M).dot(basis_vecs)).T).dot(K).dot(np.eye(n,m)-L)\n \n t_sw = H.dot(H.T)\n eigenvecs = nullspace(t_sw)\n if eigenvecs.shape[1] < 1:\n eigenvals, eigenvecs = np.linalg.eigh(t_sw)\n \n eigenvals = np.diag(eigenvals)\n min_idx = eigenvals.argsort()[0]\n eigenvecs = eigenvecs[:, min_idx]\n proj = ((np.eye(m,m)-M).dot(basis_vecs)).dot(eigenvecs)\n target_points = []\n for cl in classes:\n k_cl = K[labels==cl, :] \n pt = np.mean(k_cl.dot(proj), axis=0)\n target_points.append(pt)\n \n return proj, np.array(target_points)",
"_____no_output_____"
],
[
"kernel_mat = metrics.pairwise_kernels(X_train, metric=_hik)\nproj, target_points = learn(kernel_mat, y_train)",
"_____no_output_____"
],
[
"def squared_euclidean_distances(x, y):\n n = np.shape(x)[0]\n m = np.shape(y)[0]\n distmat = np.zeros((n,m))\n \n for i in range(n):\n for j in range(m):\n buff = x[i,:] - y[j,:]\n distmat[i,j] = buff.dot(buff.T)\n return distmat\n\ndef assign_score(proj, target_points, ks):\n projection_vectors = ks.T.dot(proj)\n sq_dist = squared_euclidean_distances(projection_vectors, target_points)\n scores = np.sqrt(np.amin(sq_dist, 1))\n return scores\n\n",
"_____no_output_____"
],
[
"auc_scores = []\nclasses = np.unique(target)\nnum_known = 5\nfor n in range(20):\n num_class = len(classes)\n known = np.random.choice(classes, num_known)\n mask = np.array([y in known for y in target])\n\n X_train = data[mask]\n y_train = target[mask]\n \n idx = y_train.argsort()\n X_train = X_train[idx]\n y_train = y_train[idx]\n \n sample_idx = np.random.randint(0, len(data), size=1000)\n X_test = data[sample_idx,:]\n y_labels = target[sample_idx]\n\n # Test labels are 1 if novel, otherwise 0.\n y_test = np.array([1 if cl not in known else 0 for cl in y_labels])\n \n # Train model\n kernel_mat = metrics.pairwise_kernels(X_train, metric=_hik)\n proj, target_points = learn(kernel_mat, y_train)\n \n # Test\n ks = metrics.pairwise_kernels(X_train, X_test, metric=_hik)\n scores = assign_score(proj, target_points, ks)\n auc = metrics.roc_auc_score(y_test, scores)\n print(\"AUC:\", auc)\n auc_scores.append(auc)\n\n \n",
"AUC: 0.269456410256\nAUC: 1.0\nAUC: 1.0\nAUC: 1.0\nAUC: 0.272551020408\nAUC: 0.990788126919\nAUC: 0.501006036217\nAUC: 1.0\nAUC: 0.809297709105\nAUC: 0.4522182861\nAUC: 0.502514351831\nAUC: 0.228532104602\nAUC: 0.32806368427\n"
],
[
"fpr, tpr, thresholds = metrics.roc_curve(y_test, scores)",
"_____no_output_____"
],
[
"plt.figure()\nplt.plot(fpr, tpr, label='ROC curve')\nplt.plot([0, 1], [0, 1], 'k--')\nplt.xlim([0.0, 1.0])\nplt.ylim([0.0, 1.05])\nplt.xlabel('False Positive Rate')\nplt.ylabel('True Positive Rate')\nplt.title('ROC Curve of the KNFST Novelty Classifier')\nplt.legend(loc=\"lower right\")\nplt.show()",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbefceffdab80e7aca74324e2fdebe446244caa8
| 7,393 |
ipynb
|
Jupyter Notebook
|
xgboost.ipynb
|
yashchoubey/Enlightiks-Business-Solutions
|
04496c0e5f378ed05bfdff9efe13bfeb6f5e5232
|
[
"MIT"
] | null | null | null |
xgboost.ipynb
|
yashchoubey/Enlightiks-Business-Solutions
|
04496c0e5f378ed05bfdff9efe13bfeb6f5e5232
|
[
"MIT"
] | null | null | null |
xgboost.ipynb
|
yashchoubey/Enlightiks-Business-Solutions
|
04496c0e5f378ed05bfdff9efe13bfeb6f5e5232
|
[
"MIT"
] | null | null | null | 28.766537 | 272 | 0.581767 |
[
[
[
"import pandas as pd\ndf = pd.read_excel('Data for classification.xlsx', shuffle=True)\ndf.drop('ID', axis=1, inplace=True)",
"_____no_output_____"
],
[
"from sklearn.model_selection import cross_val_score\nfrom xgboost import XGBClassifier\nfrom xgboost import plot_importance",
"_____no_output_____"
],
[
"label=df['Target'].fillna(0)\nlabel=label.astype('int')\ndf.drop('Target', axis=1, inplace=True)",
"_____no_output_____"
],
[
"model = XGBClassifier()\nfrom xgboost import plot_importance\nmodel.fit(df, label)\nplot_importance(model)",
"_____no_output_____"
],
[
"# print(model.feature_importances_).sum()\nprint model.get_booster().get_score(importance_type='weight')",
"{'feature_18': 23, 'feature_19': 2, 'feature_15': 21, 'feature_16': 12, 'feature_17': 21, 'feature_11': 51, 'feature_12': 13, 'feature_13': 104, 'feature_8': 14, 'feature_2': 71, 'feature_1': 45, 'feature_6': 6, 'feature_7': 2, 'feature_4': 14, 'feature_5': 17}\n"
],
[
"print(model.feature_importances_)",
"[0.10817308 0.17067307 0. 0.03365385 0.04086538 0.01442308\n 0.00480769 0.03365385 0. 0. 0.12259615 0.03125\n 0.25 0. 0.05048077 0.02884615 0.05048077 0.05528846\n 0.00480769]\n"
],
[
"\nresult = cross_val_score(model, df, label, scoring='f1')\nprint result.mean()",
"/usr/local/lib/python2.7/dist-packages/sklearn/preprocessing/label.py:151: DeprecationWarning: The truth value of an empty array is ambiguous. Returning False, but in future this will result in an error. Use `array.size > 0` to check that an array is not empty.\n if diff:\n/usr/local/lib/python2.7/dist-packages/sklearn/preprocessing/label.py:151: DeprecationWarning: The truth value of an empty array is ambiguous. Returning False, but in future this will result in an error. Use `array.size > 0` to check that an array is not empty.\n if diff:\n"
],
[
"df.drop('feature_3', axis=1, inplace=True)\ndf.drop('feature_14', axis=1, inplace=True)\ndf.drop('feature_9', axis=1, inplace=True)\n# df.drop('feature_10', axis=1, inplace=True)\n\nresult = cross_val_score(model, df, label, scoring='f1')\nprint result.mean()",
"/usr/local/lib/python2.7/dist-packages/sklearn/preprocessing/label.py:151: DeprecationWarning: The truth value of an empty array is ambiguous. Returning False, but in future this will result in an error. Use `array.size > 0` to check that an array is not empty.\n if diff:\n/usr/local/lib/python2.7/dist-packages/sklearn/preprocessing/label.py:151: DeprecationWarning: The truth value of an empty array is ambiguous. Returning False, but in future this will result in an error. Use `array.size > 0` to check that an array is not empty.\n if diff:\n"
],
[
"from sklearn.model_selection import train_test_split\nfrom sklearn.feature_selection import SelectFromModel\nfrom sklearn.metrics import accuracy_score,f1_score\nX_train, X_test, y_train, y_test = train_test_split(df, label, test_size=0.2)\n",
"_____no_output_____"
],
[
"model = XGBClassifier()\nmodel.fit(X_train, y_train)\n# make predictions for test data and evaluate\ny_pred = model.predict(X_test)\nf1 = f1_score(y_test, y_pred)\nacc = accuracy_score(y_test, y_pred)\nprint\"accuracy_score: \"+str(acc)\nprint\"f1_score: \"+str(f1)\n",
"accuracy_score: 0.9409422694094227\nf1_score: 0.7420289855072464\n"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbefcf4182d0c8a07b1e8dc21e6ebc9918923c29
| 33,952 |
ipynb
|
Jupyter Notebook
|
05.01-KNN-imputation.ipynb
|
sri-spirited/feature-engineering-for-ml
|
607c376cf92efd0ca9cc0f4f4959f639f793dedc
|
[
"BSD-3-Clause"
] | null | null | null |
05.01-KNN-imputation.ipynb
|
sri-spirited/feature-engineering-for-ml
|
607c376cf92efd0ca9cc0f4f4959f639f793dedc
|
[
"BSD-3-Clause"
] | null | null | null |
05.01-KNN-imputation.ipynb
|
sri-spirited/feature-engineering-for-ml
|
607c376cf92efd0ca9cc0f4f4959f639f793dedc
|
[
"BSD-3-Clause"
] | null | null | null | 29.043627 | 291 | 0.416294 |
[
[
[
"## KNN imputation\n\nThe missing values are estimated as the average value from the closest K neighbours.\n\n[KNNImputer from sklearn](https://scikit-learn.org/stable/modules/generated/sklearn.impute.KNNImputer.html#sklearn.impute.KNNImputer)\n\n- Same K will be used to impute all variables\n- Can't really optimise K to better predict the missing values\n- Could optimise K to better predict the target\n\n**Note**\n\nIf what we want is to predict, as accurately as possible the values of the missing data, then, we would not use the KNN imputer, we would build individual KNN algorithms to predict 1 variable from the remaining ones. This is a common regression problem.",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\n\nimport matplotlib.pyplot as plt\n\n# to split the datasets\nfrom sklearn.model_selection import train_test_split\n\n# multivariate imputation\nfrom sklearn.impute import KNNImputer",
"_____no_output_____"
]
],
[
[
"## Load data",
"_____no_output_____"
]
],
[
[
"# list with numerical varables\n\ncols_to_use = [\n 'MSSubClass', 'LotFrontage', 'LotArea', 'OverallQual',\n 'OverallCond', 'YearBuilt', 'YearRemodAdd', 'MasVnrArea',\n 'BsmtFinSF1', 'BsmtFinSF2', 'BsmtUnfSF', 'TotalBsmtSF',\n '1stFlrSF', '2ndFlrSF', 'LowQualFinSF', 'GrLivArea',\n 'BsmtFullBath', 'BsmtHalfBath', 'FullBath', 'HalfBath',\n 'BedroomAbvGr', 'KitchenAbvGr', 'TotRmsAbvGrd',\n 'Fireplaces', 'GarageYrBlt', 'GarageCars', 'GarageArea',\n 'WoodDeckSF', 'OpenPorchSF', 'EnclosedPorch', '3SsnPorch',\n 'ScreenPorch', 'PoolArea', 'MiscVal', 'MoSold', 'YrSold',\n 'SalePrice'\n]",
"_____no_output_____"
],
[
"# let's load the dataset with a selected variables\n\ndata = pd.read_csv('../houseprice.csv', usecols=cols_to_use)\n\n# find variables with missing data\nfor var in data.columns:\n if data[var].isnull().sum() > 1:\n print(var, data[var].isnull().sum())",
"LotFrontage 259\nMasVnrArea 8\nGarageYrBlt 81\n"
],
[
"# let's separate into training and testing set\n\n# first drop the target from the feature list\ncols_to_use.remove('SalePrice')\n\nX_train, X_test, y_train, y_test = train_test_split(\n data[cols_to_use],\n data['SalePrice'],\n test_size=0.3,\n random_state=0)\n\nX_train.shape, X_test.shape",
"_____no_output_____"
],
[
"# reset index, so we can compare values later on\n# in the demo\n\nX_train.reset_index(inplace=True, drop=True)\nX_test.reset_index(inplace=True, drop=True)",
"_____no_output_____"
]
],
[
[
"## KNN imputation",
"_____no_output_____"
]
],
[
[
"imputer = KNNImputer(\n n_neighbors=5, # the number of neighbours K\n weights='distance', # the weighting factor\n metric='nan_euclidean', # the metric to find the neighbours\n add_indicator=False, # whether to add a missing indicator\n)",
"_____no_output_____"
],
[
"imputer.fit(X_train)",
"_____no_output_____"
],
[
"train_t = imputer.transform(X_train)\ntest_t = imputer.transform(X_test)\n\n# sklearn returns a Numpy array\n# lets make a dataframe\ntrain_t = pd.DataFrame(train_t, columns=X_train.columns)\ntest_t = pd.DataFrame(test_t, columns=X_test.columns)\n\ntrain_t.head()",
"_____no_output_____"
],
[
"# variables without NA after the imputation\n\ntrain_t[['LotFrontage', 'MasVnrArea', 'GarageYrBlt']].isnull().sum()",
"_____no_output_____"
],
[
"# the obseravtions with NA in the original train set\n\nX_train[X_train['MasVnrArea'].isnull()]['MasVnrArea']",
"_____no_output_____"
],
[
"# the replacement values in the transformed dataset\n\ntrain_t[X_train['MasVnrArea'].isnull()]['MasVnrArea']",
"_____no_output_____"
],
[
"# the mean value of the variable (i.e., for mean imputation)\n\nX_train['MasVnrArea'].mean()",
"_____no_output_____"
]
],
[
[
"In some cases, the imputation values are very different from the mean value we would have used in MeanMedianImputation.",
"_____no_output_____"
],
[
"## Imputing a slice of the dataframe\n\nWe can use Feature-engine to apply the KNNImputer to a slice of the dataframe.",
"_____no_output_____"
]
],
[
[
"from feature_engine.wrappers import SklearnTransformerWrapper",
"_____no_output_____"
],
[
"data = pd.read_csv('../houseprice.csv')\n\nX_train, X_test, y_train, y_test = train_test_split(\n data.drop('SalePrice', axis=1),\n data['SalePrice'],\n test_size=0.3,\n random_state=0)\n\nX_train.shape, X_test.shape",
"_____no_output_____"
],
[
"# start the KNNimputer inside the SKlearnTransformerWrapper\n\nimputer = SklearnTransformerWrapper(\n transformer = KNNImputer(weights='distance'),\n variables = cols_to_use,\n)",
"_____no_output_____"
],
[
"# fit the wrapper + KNNImputer\nimputer.fit(X_train)\n\n# transform the data\ntrain_t = imputer.transform(X_train)\ntest_t = imputer.transform(X_test)\n\n# feature-engine returns a dataframe\ntrain_t.head()",
"_____no_output_____"
],
[
"# no NA after the imputation\n\ntrain_t['MasVnrArea'].isnull().sum()",
"_____no_output_____"
],
[
"# same imputation values as previously\n\ntrain_t[X_train['MasVnrArea'].isnull()]['MasVnrArea']",
"_____no_output_____"
]
],
[
[
"## Automatically find best imputation parameters\n\nWe can optimise the parameters of the KNN imputation to better predict our outcome.",
"_____no_output_____"
]
],
[
[
"# import extra classes for modelling\nfrom sklearn.preprocessing import StandardScaler\nfrom sklearn.linear_model import Lasso\nfrom sklearn.pipeline import Pipeline\nfrom sklearn.model_selection import GridSearchCV",
"_____no_output_____"
],
[
"# separate intro train and test set\n\nX_train, X_test, y_train, y_test = train_test_split(\n data[cols_to_use], # just the features\n data['SalePrice'], # the target\n test_size=0.3, # the percentage of obs in the test set\n random_state=0) # for reproducibility\n\nX_train.shape, X_test.shape",
"_____no_output_____"
],
[
"pipe = Pipeline(steps=[\n ('imputer', KNNImputer(\n n_neighbors=5,\n weights='distance',\n add_indicator=False)),\n \n ('scaler', StandardScaler()),\n ('regressor', Lasso(max_iter=2000)),\n])",
"_____no_output_____"
],
[
"# now we create the grid with all the parameters that we would like to test\n\nparam_grid = {\n 'imputer__n_neighbors': [3,5,10],\n 'imputer__weights': ['uniform', 'distance'],\n 'imputer__add_indicator': [True, False],\n 'regressor__alpha': [10, 100, 200],\n}\n\ngrid_search = GridSearchCV(pipe, param_grid, cv=5, n_jobs=-1, scoring='r2')\n\n# cv=3 is the cross-validation\n# no_jobs =-1 indicates to use all available cpus\n# scoring='r2' indicates to evaluate using the r squared\n\n# for more details in the grid parameters visit:\n#https://scikit-learn.org/stable/modules/generated/sklearn.model_selection.GridSearchCV.html",
"_____no_output_____"
],
[
"# and now we train over all the possible combinations \n# of the parameters above\ngrid_search.fit(X_train, y_train)\n\n# and we print the best score over the train set\nprint((\"best linear regression from grid search: %.3f\"\n % grid_search.score(X_train, y_train)))",
"best linear regression from grid search: 0.845\n"
],
[
"# let's check the performance over the test set\nprint((\"best linear regression from grid search: %.3f\"\n % grid_search.score(X_test, y_test)))",
"best linear regression from grid search: 0.730\n"
],
[
"# and find the best parameters\n\ngrid_search.best_params_",
"_____no_output_____"
]
],
[
[
"## Compare with univariate imputation",
"_____no_output_____"
]
],
[
[
"from sklearn.impute import SimpleImputer",
"_____no_output_____"
],
[
"# separate intro train and test set\n\nX_train, X_test, y_train, y_test = train_test_split(\n data[cols_to_use], # just the features\n data['SalePrice'], # the target\n test_size=0.3, # the percentage of obs in the test set\n random_state=0) # for reproducibility\n\nX_train.shape, X_test.shape",
"_____no_output_____"
],
[
"pipe = Pipeline(steps=[\n ('imputer', SimpleImputer(strategy='mean', fill_value=-1)),\n ('scaler', StandardScaler()),\n ('regressor', Lasso(max_iter=2000)),\n])\n\nparam_grid = {\n 'imputer__strategy': ['mean', 'median', 'constant'],\n 'imputer__add_indicator': [True, False],\n 'regressor__alpha': [10, 100, 200],\n}\n\ngrid_search = GridSearchCV(pipe, param_grid, cv=5, n_jobs=-1, scoring='r2')\n\n# and now we train over all the possible combinations of the parameters above\ngrid_search.fit(X_train, y_train)\n\n# and we print the best score over the train set\nprint((\"best linear regression from grid search: %.3f\"\n % grid_search.score(X_train, y_train)))",
"best linear regression from grid search: 0.845\n"
],
[
"# and finally let's check the performance over the test set\nprint((\"best linear regression from grid search: %.3f\"\n % grid_search.score(X_test, y_test)))",
"best linear regression from grid search: 0.729\n"
],
[
"# and find the best fit parameters like this\ngrid_search.best_params_",
"_____no_output_____"
]
],
[
[
"We see that imputing the values with an arbitrary value of -1, returns approximately the same performance as doing KNN imputation, so we might not want to add the additional complexity of training models to impute NA, to then go ahead and predict the real target we are interested in.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
cbf0063ee7e0583b20177784175b43bfe98bc1f7
| 29,777 |
ipynb
|
Jupyter Notebook
|
src/model.ipynb
|
GeoRouv/TP-residues-classifier
|
2810b4ee087a3c62c3bd804b36859320cf88f5c7
|
[
"MIT"
] | 1 |
2021-11-10T20:03:38.000Z
|
2021-11-10T20:03:38.000Z
|
src/model.ipynb
|
GeoRouv/TP-residues-classifier
|
2810b4ee087a3c62c3bd804b36859320cf88f5c7
|
[
"MIT"
] | null | null | null |
src/model.ipynb
|
GeoRouv/TP-residues-classifier
|
2810b4ee087a3c62c3bd804b36859320cf88f5c7
|
[
"MIT"
] | null | null | null | 14,888.5 | 29,776 | 0.76633 |
[
[
[
"import os\nimport matplotlib.pyplot as plt\nimport numpy as np\nimport pandas as pd\nfrom random import randint\nfrom numpy import array\nfrom numpy import argmax\nfrom numpy import array_equal\nimport tensorflow as tf\nfrom tensorflow.keras.utils import to_categorical\nfrom keras.models import Model\nfrom keras.layers import Input\nfrom keras.layers import LSTM\nfrom keras.layers import Dense\nfrom keras.preprocessing.sequence import pad_sequences\nfrom sklearn.model_selection import train_test_split",
"_____no_output_____"
],
[
"# from google.colab import drive\n# drive.mount('/content/drive')\n# os.chdir(\"drive/My Drive/Colab Notebooks/Structural/Project\")",
"Mounted at /content/drive\n"
]
],
[
[
"Dataset Preparation and Split",
"_____no_output_____"
]
],
[
[
"dataset = pd.read_csv('./data/dataset.ultrafltr.csv')\nprint(dataset)",
" sequence opm_class\n0 SGFEFHGYARSGVIMNDSGASTKSGAYITPAGETGGAIGRLGNQAD... 0000111111111111000000000000000000000000000001...\n1 MASMTGGQQMGRDLQVTLYGTIKAGVEVSRVKDAGTYKAQGGKSKT... 0000000000000001111111111111000000000000000000...\n2 GTMARNDGQGKAAATFMHISYNNFITEVDNLNKRMGDLRDINGEAG... 0000000000000000000000000000000000000000000011...\n3 VDFHGYARSGIGWTGSGGEQQCFQTTGAQSKYRLGNECETYAELKL... 0111111111111000000000000000000000000011111111...\n4 ANSGEAPKNFGLDVKITGESENDRDLGTAPGGTLNDIGIDLRPWAF... 0000000011111111111100000000000000011111111111...\n.. ... ...\n223 MWEANELSSTNTFSHQAEMDWPSANWWQRYQDAQLNHLIEEALQHS... 0000000000000000000000000000000000000000000000...\n224 AGLVVNDNDLRNDLAWLSDRGVIHLSLSTWPLSQEEIARALKKAKP... 0000000000000000000000000000000000000000000000...\n225 QDTSPDTLVVTANRFEQPRSTVLAPTTVVTRQDIDRWQSTSVNDVL... 0000000000000000000000000000000000000000000000...\n226 TQVFDLEGYGAISRAMGGTSSSYYTGNAALISNPATLSFAPDGNQF... 0000000000000000000000000000000000000000011111...\n227 MGSSHHHHHHSSGLVPRGSHMQQNDTSADENQQKNNAESEEEQQGD... 0000000000000000000000000000000000000000000000...\n\n[228 rows x 2 columns]\n"
]
],
[
[
"Lengths of sequences",
"_____no_output_____"
]
],
[
[
"data = dataset['sequence'].str.len()\ncounts, bins = np.histogram(data)\nplt.hist(bins[:-1], bins, weights=counts)",
"_____no_output_____"
],
[
"df_filtered = dataset[dataset['sequence'].str.len() <= 1000]\nprint(df_filtered.shape)\n\ndata = df_filtered['sequence'].str.len()\ncounts, bins = np.histogram(data)\nplt.hist(bins[:-1], bins, weights=counts)",
"(226, 2)\n"
],
[
"dataset = df_filtered\nmeasurer = np.vectorize(len)\nres1 = measurer(dataset.values.astype(str)).max(axis=0)[0]\nprint(res1)\n\ndf, df_test = train_test_split(dataset, test_size=0.1)\nprint(df)",
"913\n sequence opm_class\n18 AQAEASSAQAAQQKNFNIAAQPLQSAMLRFAEQAGMQVFFDEVKLD... 0000000000000000000000000000000000000000000000...\n25 ANVRLQHHHHHHHLEEFIADSKAELTLRNFYFDRDYKKDPYPYTAA... 0000000000000000000000111111111000000000000000...\n13 MAPKDNTWYTGAKLGWSQYHDTGLINNNGPTHENKLGAGAFGGYQV... 0000000111111111000000000000000000011111111110...\n36 DAGTVDFYGQLRTELKFLEDKDPTIGSGSSRAGVDANYTVNDSLAL... 0001111111111111000000000000000111111100000001...\n217 ENLMQVYQQARLSNPELRKSAADRDAAFEKINEARSPLLPQLGLGA... 0000000000000000000000000000000000000000111111...\n.. ... ...\n78 DNVITRVVAVRNVSVRELSPLLRQLIDNAGAGNVVHYDPANIILIT... 0000000000000000000000000000000000000000000000...\n218 MNSSRSVNPRPSFAPRALSLAIALLLGAPAFAANSGEAPKNFGLDV... 0000000000000000000000000000000000000000111111...\n110 CTMIPQYEQPKVEVAETFQNDTSVSSIRAVDLGWHDYFADPRLQKL... 0000000000000000000000000000000000000000000000...\n1 MASMTGGQQMGRDLQVTLYGTIKAGVEVSRVKDAGTYKAQGGKSKT... 0000000000000001111111111111000000000000000000...\n113 MKKRIPTLLATMIASALYSHQGLAADLASQCMLGVPSYDRPLVKGD... 0000000000000000000000000000000000000000000000...\n\n[203 rows x 2 columns]\n"
]
],
[
[
"Encoding of Aminoacids",
"_____no_output_____"
]
],
[
[
"codes = ['A', 'C', 'D', 'E', 'F', 'G', 'H', 'I', 'K', 'L',\n 'M', 'N', 'P', 'Q', 'R', 'S', 'T', 'V', 'W', 'Y']\n\ndef create_dict(codes):\n char_dict = {}\n for index, val in enumerate(codes):\n char_dict[val] = index+1\n return char_dict\nchar_dict = create_dict(codes)\n\ndef integer_encoding(data):\n \"\"\"\n - Encodes code sequence to integer values.\n - 20 common amino acids are taken into consideration\n and rest 4 are categorized as 0.\n \"\"\"\n row_encode = []\n for code in list(data):\n row_encode.append(char_dict.get(code, 0))\n return row_encode",
"_____no_output_____"
]
],
[
[
"Model",
"_____no_output_____"
]
],
[
[
"# prepare data for the LSTM\ndef get_dataset(df):\n X1, X2, y = list(), list(), list()\n\n \n for index, row in df.iterrows():\n # generate source sequence\n source = row['sequence']\n # source = source.ljust(res1, '0')\n source = integer_encoding(source)\n\n # define padded target sequence\n target = row['opm_class']\n # target = target.ljust(res1, '0')\n target = list(map(int, target))\n\n # create padded input target sequence\n target_in = [0] + target[:-1]\n\n # encode\n src_encoded = to_categorical(source, num_classes=20+1)\n tar_encoded = to_categorical(target, num_classes=2)\n tar2_encoded = to_categorical(target_in, num_classes=2)\n\n\n # store\n X1.append(src_encoded)\n X2.append(tar2_encoded)\n y.append(tar_encoded)\n\n return array(X1), array(X2), array(y)#, temp_df\n \n# Creating the first Dataframe using dictionary\nX1, X2, y = get_dataset(df)\nX1 = pad_sequences(X1, maxlen=res1, padding='post', truncating='post')\nX2 = pad_sequences(X2, maxlen=res1, padding='post', truncating='post')\ny = pad_sequences(y, maxlen=res1, padding='post', truncating='post')\n\n\n# returns train, inference_encoder and inference_decoder models\ndef define_models(n_input, n_output, n_units):\n\t# define training encoder\n\tencoder_inputs = Input(shape=(None, n_input))\n\tencoder = LSTM(n_units, return_state=True)\n\tencoder_outputs, state_h, state_c = encoder(encoder_inputs)\n\tencoder_states = [state_h, state_c]\n\t# define training decoder\n\tdecoder_inputs = Input(shape=(None, n_output))\n\tdecoder_lstm = LSTM(n_units, return_sequences=True, return_state=True)\n\tdecoder_outputs, _, _ = decoder_lstm(decoder_inputs, initial_state=encoder_states)\n\tdecoder_dense = Dense(n_output, activation='softmax')\n\tdecoder_outputs = decoder_dense(decoder_outputs)\n\tmodel = Model([encoder_inputs, decoder_inputs], decoder_outputs)\n\t# define inference encoder\n\tencoder_model = Model(encoder_inputs, encoder_states)\n\t# define inference decoder\n\tdecoder_state_input_h = Input(shape=(n_units,))\n\tdecoder_state_input_c = Input(shape=(n_units,))\n\tdecoder_states_inputs = [decoder_state_input_h, decoder_state_input_c]\n\tdecoder_outputs, state_h, state_c = decoder_lstm(decoder_inputs, initial_state=decoder_states_inputs)\n\tdecoder_states = [state_h, state_c]\n\tdecoder_outputs = decoder_dense(decoder_outputs)\n\tdecoder_model = Model([decoder_inputs] + decoder_states_inputs, [decoder_outputs] + decoder_states)\n\t# return all models\n\treturn model, encoder_model, decoder_model\n\ntrain, infenc, infdec = define_models(20+1, 2, 128)\ntrain.compile(optimizer='adam', loss='categorical_crossentropy', metrics=['accuracy'])\ntrain.summary()\n\n# train model\ntrain.fit([X1, X2], y, epochs=10)",
"/usr/local/lib/python3.7/dist-packages/ipykernel_launcher.py:41: VisibleDeprecationWarning: Creating an ndarray from ragged nested sequences (which is a list-or-tuple of lists-or-tuples-or ndarrays with different lengths or shapes) is deprecated. If you meant to do this, you must specify 'dtype=object' when creating the ndarray\n"
]
],
[
[
"",
"_____no_output_____"
],
[
"Prediction",
"_____no_output_____"
]
],
[
[
"# decode a one hot encoded string\ndef one_hot_decode(encoded_seq):\n return [argmax(vector) for vector in encoded_seq]\n\ndef compare_seqs(source, target):\n correct = 0\n for i in range(len(source)):\n if source[i] == target[i]:\n correct += 1\n \n return correct\n\n# generate target given source sequence\ndef predict_sequence(infenc, infdec, source, n_steps, cardinality):\n # encode\n state = infenc.predict(source)\n # start of sequence input\n target_seq = array([0.0 for _ in range(cardinality)]).reshape(1, 1, cardinality)\n\n # collect predictions\n output = list()\n for t in range(n_steps):\n # predict next char\n yhat, h, c = infdec.predict([target_seq] + state)\n # store prediction\n output.append(yhat[0,0,:])\n # update state\n state = [h, c]\n # update target sequence\n target_seq = yhat\n return array(output)\n\n# evaluate LSTM\nX1, X2, y = get_dataset(df_test)\nX1 = pad_sequences(X1, maxlen=res1, padding='post', truncating='post')\nX2 = pad_sequences(X2, maxlen=res1, padding='post', truncating='post')\ny = pad_sequences(y, maxlen=res1, padding='post', truncating='post')\n\naccuracies = []\nfor i in range(len(X1)):\n row = X1[i]\n row = row.reshape((1, row.shape[0], row.shape[1]))\n target = predict_sequence(infenc, infdec, row, res1, 2)\n\n curr_acc = compare_seqs(one_hot_decode(target), one_hot_decode(y[i]))/res1\n accuracies.append(curr_acc)\n\n print(f'Sequence{i} Accuracy: {curr_acc}')\n\ntotal_acc = 0\nfor i in range(len(accuracies)):\n total_acc += accuracies[i]\nprint('Total Accuracy: %.2f%%' % (float(total_acc)/float(len(X1))*100.0))",
"/usr/local/lib/python3.7/dist-packages/ipykernel_launcher.py:41: VisibleDeprecationWarning: Creating an ndarray from ragged nested sequences (which is a list-or-tuple of lists-or-tuples-or ndarrays with different lengths or shapes) is deprecated. If you meant to do this, you must specify 'dtype=object' when creating the ndarray\n"
],
[
"",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
]
] |
cbf030496e1e0af8627cd52baa94a2c9224ed9cb
| 1,713 |
ipynb
|
Jupyter Notebook
|
downloaded_kernels/university_rankings/kernel_38.ipynb
|
josepablocam/common-code-extraction
|
a6978fae73eee8ece6f1db09f2f38cf92f03b3ad
|
[
"MIT"
] | null | null | null |
downloaded_kernels/university_rankings/kernel_38.ipynb
|
josepablocam/common-code-extraction
|
a6978fae73eee8ece6f1db09f2f38cf92f03b3ad
|
[
"MIT"
] | null | null | null |
downloaded_kernels/university_rankings/kernel_38.ipynb
|
josepablocam/common-code-extraction
|
a6978fae73eee8ece6f1db09f2f38cf92f03b3ad
|
[
"MIT"
] | 2 |
2021-07-12T00:48:08.000Z
|
2021-08-11T12:53:05.000Z
| 45.078947 | 714 | 0.680677 |
[
[
[
"# This Python 3 environment comes with many helpful analytics libraries installed\n# It is defined by the kaggle/python docker image: https://github.com/kaggle/docker-python\n# For example, here's several helpful packages to load in \n\nimport numpy as np # linear algebra\nimport pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)\n\n# Input data files are available in the \"../input/\" directory.\n# For example, running this (by clicking run or pressing Shift+Enter) will list the files in the input directory\n\nfrom subprocess import check_output\nprint(check_output([\"ls\", \"../input\"]).decode(\"utf8\"))\n\n# Any results you write to the current directory are saved as output.",
"_____no_output_____"
],
[
"time_data = pd.read_csv('../input/timesData.csv')\ntime_data[20:25]",
"_____no_output_____"
],
[
"id_count_by_region = time_data.groupby('country')['world_rank'].count()\nid_count_by_region.sort_values(na_position='last', inplace=True, ascending=False)\nid_count_by_region[:10]",
"_____no_output_____"
],
[
"%matplotlib inline\nid_count_by_region[:10].plot(kind='barh', rot=0, title='Universities by Region')",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code"
]
] |
cbf038986add6c5b83ba62eec7dcbaff5719bb34
| 31,904 |
ipynb
|
Jupyter Notebook
|
Coding Bootcamps/Coding Bootcamp Part 5.ipynb
|
ds-modules/ENVECON-118-SP22
|
90d093590fc90d83b0f644fd46dcba5ef3580c89
|
[
"BSD-3-Clause"
] | 1 |
2022-03-16T04:10:52.000Z
|
2022-03-16T04:10:52.000Z
|
Coding Bootcamps/Coding Bootcamp Part 5.ipynb
|
ds-modules/ENVECON-118-SP22
|
90d093590fc90d83b0f644fd46dcba5ef3580c89
|
[
"BSD-3-Clause"
] | null | null | null |
Coding Bootcamps/Coding Bootcamp Part 5.ipynb
|
ds-modules/ENVECON-118-SP22
|
90d093590fc90d83b0f644fd46dcba5ef3580c89
|
[
"BSD-3-Clause"
] | 3 |
2022-03-17T20:54:01.000Z
|
2022-03-30T22:34:50.000Z
| 42.201058 | 686 | 0.611741 |
[
[
[
"# R Bootcamp Part 5\n\n## stargazer, xtable, robust standard errors, and fixed effects regressions\n\n\nThis bootcamp will help us get more comfortableusing **stargazer** and **xtable** to produce high-quality results and summary statistics tables, and using `felm()` from the **lfe** package for regressions (both fixed effects and regular OLS).\n\n\nFor today, let's load a few packages and read in a dataset on residential water use for residents in Alameda and Contra Costa Counties. ",
"_____no_output_____"
],
[
"## Preamble\nHere we'll load in our necessary packages and the data file",
"_____no_output_____"
]
],
[
[
"library(tidyverse)\nlibrary(haven)\nlibrary(lfe)\nlibrary(stargazer)\nlibrary(xtable)\n\n# load in wateruse data, add in measure of gallons per day \"gpd\"\nwaterdata <- read_dta(\"wateruse.dta\") %>%\n mutate(gpd = (unit*748)/num_days)\nhead(waterdata)",
"_____no_output_____"
]
],
[
[
"# Summary Statistics Tables with xtable\n\n`xtable` is a useful package for producing custom summary statistics tables. let's say we're interested in summarizing water use ($gpd$) and degree days ($degree\\_days$) according to whether a lot is less than or greater than one acre ($lotsize_1$) or more than 4 acres ($lotsize_4$):\n\n`homesize <- waterdata %>%\n select(hh, billingcycle, gpd, degree_days, lotsize) %>%\n drop_na() %>%\n mutate(lotsize_1 = ifelse((lotsize < 1), \"< 1\", \">= 1\"),\n lotsize_4 = ifelse((lotsize > 4), \"> 4\", \"<= 4\"))\nhead(homesize)`\n",
"_____no_output_____"
],
[
"We know how to create summary statistics for these two variables for both levels of $lotsize\\_1$ and $lotsize\\_4$ using `summarise()`:\n\n`sumstat_1 <- homesize %>%\n group_by(lotsize_1) %>%\n summarise(mean_gpd = mean(gpd), \n mean_degdays = mean(degree_days))\nsumstat_1`\n\n`sumstat_4 <- homesize %>%\n group_by(lotsize_4) %>%\n summarise(mean_gpd = mean(gpd), \n mean_degdays = mean(degree_days))\nsumstat_4`",
"_____no_output_____"
],
[
"And now we can use `xtable()` to put them into the same table!\n\n`full <- xtable(cbind(t(sumstat_1), t(sumstat_4)))\nrownames(full)[1] <- \"Lotsize Group\"\ncolnames(full) <- c(\"lotsize_1 = 1\", \"lotsize_1 = 0\", \"lotsize_4 = 0\", \"lotsize_4 =1\")\nfull`",
"_____no_output_____"
],
[
"We now have a table `full` that is an xtable object. \n\nWe can also spit this table out in html or latex form if needed using the `print.xtable()` function on our xtable `full`, specifying `type = \"html\":\n\n`print.xtable(full, type = \"html\")`",
"_____no_output_____"
],
[
"Copy and paste the html code here to see how it appears",
"_____no_output_____"
],
[
"# Regression Tables in Stargazer\n\n`stargazer` is a super useful package for producing professional-quality regression tables. While it defaults to producing LaTeX format tables (a typesetting language a lot of economists use), for use in our class we can also produce html code that can easily be copied into text cells and formatted perfectly.\n\nIf we run the following three regressions: \n\n\\begin{align*} GPD_{it} &= \\beta_0 + \\beta_1 degree\\_days_{it} + \\beta_2 precip_{it} ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~(1)\\\\\n GPD_{it} &= \\beta_0 + \\beta_1 degree\\_days_{it} + \\beta_2 precip_{it} + \\beta_3 lotsize_{i}~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~(2)\\\\\nGPD_{it} &= \\beta_0 + \\beta_1 degree\\_days_{it} + \\beta_2 precip_{it} + \\beta_3 lotsize_{i} + \\beta_4 Homeval_i~~~~~~~~~~~~~~~~~~(3)\n\\end{align*}\n\nWe might want to present the results side by side in the same table so that we can easily compare coefficients from one column to the other. To do that with `stargazer`, we can \n\n1. Run each regression, storing them in memory\n2. Run `stargazer(reg1, reg2, reg3, ..., type )` where the first arguments are all the regression objects we want in the table, and telling R what type of output we want\n\nIf we specify `type = \"text\"`, we'll get the table displayed directly in the output window:\n\n`reg_a <- lm(gpd ~ degree_days + precip, waterdata)\nreg_b <- lm(gpd ~ degree_days + precip + lotsize, waterdata)\nreg_c <- lm(gpd ~ degree_days + precip + lotsize + homeval, waterdata)`\n\n`stargazer(reg_a, reg_b, reg_c, type = \"text\")`",
"_____no_output_____"
],
[
"And if we specify `type = \"html\"`, we'll get html code that we need to copy and paste into a text/markdown cell:\n\n`stargazer(reg_a, reg_b, reg_c, type = \"html\")`",
"_____no_output_____"
],
[
"Now all we need to do is copy and paste that html code from the output into a text cell and we've got our table!",
"_____no_output_____"
],
[
"(copy your code here)",
"_____no_output_____"
],
[
"And we get a nice looking regression table with all three models side by side! This makes it easy to see how the coefficient on lot size falls when we add in home value, letting us quickly figure out the sign of correlation between the two.\n\n## Table Options\n\nStargazer has a ton of different options for customizing the look of our table with optional arguments, including\n* `title` lets us add a custom title\n* `column.labels` lets you add text labels to the columns\n* `covariate.labels` lets us specify custom labels for all our variables other than the variable names. Specify each label in quotations in the form of a vector with `c()`\n* `ci = TRUE` adds in confidence intervals (by default for the 10\\% level, but you can change it to the 1\\% level with `ci.level = 0.99`\n* `intercept.bottom = FALSE` will move the constant to the top of the table\n* `digits` lets you choose the number of decimal places to display\n* `notes` lets you add some notes at the bottom\n\nFor example, we could customize the above table as\n\n`stargazer(reg_a, reg_b, reg_c, type = \"text\",\n title = \"Water Use, Weather, and Home Characteristics\",\n column.labels = c(\"Weather\", \"With Lotsize\", \"With HomeVal\"),\n covariate.labels = c(\"Intercept\", \"Degree Days\", \"Precipitation (mm)\", \"Lot Size (Acres)\", \"Home Value (USD)\"),\n intercept.bottom = FALSE,\n digits = 2,\n note = \"Isn't stargazer neat?\"\n )`",
"_____no_output_____"
],
[
"# Summary Statistics Tables in Stargazer\n\nCan we use Stargazer for summary statistics tables too? You bet we can! \n\nStargazer especially comes in handy if we have a lot of variables we want to summarize and one or no variables we want to group them on. This approach works especially well with `across()` within `summarise()`.",
"_____no_output_____"
],
[
"For example, let's say we wanted to summarise the median and variance of `gpd`, `precip`, and `degree_days` by whether the home was built after 1980 or not. Rather than create separate tables for all of the variables and merge them together like with xtable, we can just summarise across with\n\n`ss_acr <- mutate(waterdata, pre_80 = ifelse(yearbuilt < 1980, \"1. Pre-1980\", \"2. 1980+\")) %>%\n group_by(pre_80) %>%\n summarise(across(.cols = c(gpd, precip, degree_days),\n .fns = list(Median = median, Variance = var)))\nss_acr`",
"_____no_output_____"
],
[
"Note that `ifelse()` is a function that follows the format\n\n`ifelse(Condition, Value if True, Value if False)`\n\nHere our condition is that the $yearbuilt$ variable is less than 1980. If it’s true, we want this\nnew variable to take on the label \"1. Pre-1980\", and otherwise\nbe \"2. 1980+\".\n\nThis table then contains everything we want, but having it displayed \"wide\" like this is a bit tough to see. If we wanted to display it \"long\" where there is one column for each or pre-1980 and post-1980 homes, we can just use the transpose function `t()`. Placing that within the `stargazer()` call and specifying that we want html code then gets us\n\n`stargazer(t(ss_acr), type = \"html\")`",
"_____no_output_____"
],
[
"(copy your html code here)",
"_____no_output_____"
],
[
"## Heteroskedasticity-Robust Standard Errors \n\n\nThere are often times where you want to use heteroskedasticity-robust standard errors in place of the normal kind to account for situations where we might be worried about violating our homoskedasticity assumption. To add robust standard errors to our table, we'll take advantage of the `lmtest` and `sandwich` packages (that we already loaded in the preamble).\n\nIf we want to see the coefficient table from Regression B with robust standard errors, we can use the `coeftest()` function as specified below:\n\n`coeftest(reg_b, vcov = vcovHC(reg_b, type = \"HC1\"))`",
"_____no_output_____"
],
[
"What the `vcovHC(reg_a, type = \"HC1\")` part is doing is telling R we want to calculate standard errors using the heteroskedasticity-robust approach (i.e. telling it a specific form of the variance-covariance matrix between our residuals). `coeftest()` then prints the nice output table. \n\nWhile this is a nice way to view the robust standard errors in a summary-style table, sometimes we want to extract the robust standard errors so we can use them elsewhere - like in stargazer!\n\nTo get a vector of robust standard errors from Regression B, we can use the following:\n\n`robust_b <- sqrt(diag(vcovHC(reg_b, type = \"HC1\")))`\n\n`robust_b`",
"_____no_output_____"
],
[
"Which matches the robust standard errors using `coeftest()` earlier. But woah there, that's a function nested in a function nested in *another function*! Let's break this down step-by-step:\n\n`vcov_b <- vcovHC(reg_b, type = \"HC1\")`\n\nThis first `vcov_b` object is getting the entire variance-covariance matrix for our regression coefficients. Since we again specified `type = \"HC1\"`, we ensure we get the heteroskedasticity-robust version of this matrix (if we had instead specified `type = \"constant\"` we would be assuming homoskedasticity and would get our usual variance estimates).\n\nWhat this looks like is\n\n\n$$VCOV_b = \\begin{matrix}{}\n\\widehat{Var}(\\hat \\beta_0) & \\widehat{Cov}(\\hat \\beta_0, \\hat\\beta_1) & \\widehat{Cov}(\\hat \\beta_0, \\hat\\beta_2) \\\\\n \\widehat{Cov}(\\hat \\beta_1, \\hat\\beta_0) & \\widehat{Var}(\\hat \\beta_1) & \\widehat{Cov}(\\hat \\beta_1, \\hat\\beta_2) \\\\\n \\widehat{Cov}(\\hat \\beta_2, \\hat\\beta_0) & \\widehat{Cov}(\\hat \\beta_2, \\hat\\beta_1) & \\widehat{Var}(\\hat \\beta_2) \n\\end{matrix}$$\n\nWhere each element is $\\hat\\sigma_i$ in the ith row mutiplied by $\\hat\\sigma_j$ in the jth column. Note that when $i = j$ in the main diagonal, we get the variance estimate for $\\hat \\beta_i$!\n\nYou can check this by running the following lines:\n\n`vcov_b <- vcovHC(reg_b, type = \"HC1\")\n vcov_b`",
"_____no_output_____"
],
[
"`var_b <- diag(vcov_b)`\n\nThe `diag()` function extracts this main diagonal, giving us a vector of our robust estimated variances\n\n`robust_b <- sqrt(var_b)`\n\nAnd taking the square root gets us our standard error estimates for our $\\hat\\beta$'s!\n\nSee the process by running the following lines:\n\n`var_b <- diag(vcov_b)\n var_b`\n\n`robust_b <- sqrt(var_b)\n robust_b`",
"_____no_output_____"
],
[
"## Stargazer and Heteroskedasticity-Robust Standard Errors \n\nNow that we know how to get our robust standard errors, we can grab them for all three of our regressions and add them to our beautiful stargazer table:\n\n`robust_a <- sqrt(diag(vcovHC(reg_a, type = \"HC1\")))\nrobust_b <- sqrt(diag(vcovHC(reg_b, type = \"HC1\")))\nrobust_c <- sqrt(diag(vcovHC(reg_c, type = \"HC1\")))`\n\n\n`stargazer(reg_a, reg_b, reg_c, \n type = \"html\",\n se = list(robust_a, robust_b, robust_c),\n omit.stat = \"f\")`\n \nHere we're adding the robust standard errors to `stargazer()` with the `se =` argument (combining them together in the right order as a list). I'm also omitting the overall F test at the bottom with `omit.stat = \"f\"` since we'd need to correct that too for heteroskedasticity. \n\nTry running this code below to see how the standard errors change when we use robust standard errors:",
"_____no_output_____"
],
[
"Copy and paste the table code here and run the cell to see it formatted.",
"_____no_output_____"
],
[
"Now that looks pretty good, though note that the less than signs in the note for significance labels don't appear right. This is because html is reading the < symbol as a piece of code and not the math symbol. To get around this, you can add dollar signs around the < signs in the html code for the note to have the signs display properly:\n\n`<sup>*</sup>p $<$ 0.1; <sup>**</sup>p $<$ 0.05; <sup>***</sup>p $<$ 0.01</td>`",
"_____no_output_____"
],
[
"\n# Fixed Effects Regression\n\n\nToday we will practice with fixed effects regressions in __R__. We have two different ways to estimate the model, and we will see how to do both and the situations in which we might favor one versus the other.\n\nLet's give this a try using the dataset `wateruse.dta`. The subset of households are high water users, people who used over 1,000 gallons per billing cycle. We have information on their water use, weather during the period, as well as information on the city and zipcode of where the home is located, and information on the size and value of the house.\n\nSuppose we are interested in running the following panel regression of residential water use:\n\n$$ GPD_{it} = \\beta_0 + \\beta_1 degree\\_days_{it} + \\beta_2 precip_{it} ~~~~~~~~~~~~~~~~~~~~~~~(1)$$\n\nWhere $GPD$ is the gallons used per day by household $i$ in billing cycle $t$, $degree\\_days$ the count of degree days experienced by the household in that billing cycle (degree days are a measure of cumulative time spent above a certain temperature threshold), and $precip$ the amount of precipitation in millimeters.\n\n`reg1 <- lm(gpd ~ degree_days + precip, data = waterdata)\nsummary(reg1)`",
"_____no_output_____"
],
[
"Here we obtain an estimate of $\\hat\\beta_1 = 0.777$, telling us that an additional degree day per billing cycle is associated with an additional $0.7769$ gallon used per day. These billing cycles are roughly two months long, so this suggests an increase of roughly 47 gallons per billing cycle. Our estimate is statistically significant at all conventional levels, suggesting residential water use does respond to increased exposure to high heat.\n\nWe estimate a statistically insignificant coefficient on additional precipitation, which tells us that on average household water use in our sample doesn't adjust to how much it rains.\n\nWe might think that characteristics of the home impact how much water is used there, so we add in some home controls:\n\n\n$$ GPD_{it} = \\beta_0 + \\beta_1 degree\\_days_{it} + \\beta_2 precip_{it} + \\beta_3 lotsize_{i} + \\beta_4 homesize_i + \\beta_5 num\\_baths_i + \\beta_6 num\\_beds_i + \\beta_7 homeval_i~~~~~~~~~~~~~~~~~~~~~~~(2)$$\n\n`reg2 <- lm(gpd ~ degree_days + precip + lotsize + homesize + num_baths + num_beds + homeval, data = waterdata)\nsummary(reg2)`",
"_____no_output_____"
],
[
"Our coefficient on $degree\\_days$ remains statistically significant and doesn't change much, so we find that $\\hat\\beta_1$ is robust to the addition of home characteristics. Of these characteristics, we obtain statistically significant coefficients on the size of the lot in acres ($lotsize$), the size of the home in square feet ($homesize$), and the number of bedrooms in the home ($num_beds$).\n\nWe get a curious result for $\\hat\\beta_6$: for each additional bedroom in the home we predict that water use will *fall* by 48 gallons per day. \n\n### Discussion: what might be driving this effect? ",
"_____no_output_____"
],
[
"Since there are likely a number of sources of omitted variable bias in the previous model, we think it might be worth including some fixed effects in our model. These will allow us to control for some of the unobserved sources of OVB without having to measure them directly!\n\n## Method 1: Fixed Effects with lm() \n\nUp to this point we have been running our regressions using the `lm()` function. We can still use `lm()` for our fixed effects models, but it takes some more work and gets increasingly time-intensive as datasets get large.\n\nRecall that we can write our general panel fixed effects model as \n\n$$ y_{it} = \\beta x_{it} + \\mathbf{a}_i + \\mathbf{d}_t + u_{it} $$\n\n* $y$ our outcome of interest, which varies in both the time and cross-sectional dimensions\n* $x_{it}$ our set of time-varying unit characteristics\n* $\\mathbf{a}_i$ our set of unit fixed effects\n* $\\mathbf{d}_t$ our time fixed effects\n\nWe can estimate this model in `lm()` provided we have variables in our dataframe that correspond to each level of $a_i$ and $d_t$. This means we'll have to generate them before we can run any regression.\n\n### Generating Dummy Variables\n\nIn order to include fixed effects for our regression, we can first generate the set of dummy variables that we want. For example, if we want to include a set of city fixed effects in our model, we need to generate them.\n\nWe can do this in a few ways.\n\n1. First, we can use `mutate()` and add a separate line for each individual city:\n\n`fe_1 <- waterdata %>%\n mutate(city_1 = as.numeric((city==1)),\n city_2 = as.numeric((city ==2)),\n city_3 = as.numeric((city ==3))) %>%\n select(n, hh, city, city_1, city_2, city_3)\nhead(fe_1)`",
"_____no_output_____"
],
[
"This can be super tedious though when we have a bunch of different levels of our variable that we want to make fixed effects for. In this case, we have 27 different cities.\n\n2. Alternatively, we can use the `spread()` function to help us out. Here we add in a constant variable `v` that is equal to one in all rows, and a copy of city that adds \"city_\" to the front of the city number. Then we pass the data to `spread`, telling it to split the variable `cty` into dummy variables for all its levels, with all the \"false\" cases filled with zeros.\n\n`fe_2 <- waterdata %>%\nselect(n, city, billingycle)`\n\n\n`fe_2 %>%\n mutate(v = 1, cty = paste0(\"city_\", city)) %>% \n spread(cty, v, fill = 0)`",
"_____no_output_____"
],
[
"That is much easier! \n\nThis is a useful approach if you want to produce summary statistics for the fixed effects (i.e. what share of the sample lives in each city), but isn't truly necessary.\n\nAlternatively, we can tell R to read our fixed effects variables as factors:\n\n`lm(gpd ~ degree_days + precip + factor(city), data = waterdata)`\n\n`factor()` around $city$ tells R to split city into dummy variables for each unique value it takes. R will then drop the first level when we run the regression - in our case making the first city our omitted group.\n\n\n`reg3 <- lm(gpd ~ degree_days + precip + factor(city), data = waterdata)\nsummary(reg3)`",
"_____no_output_____"
],
[
"Now we have everything we need to run the regression\n\n\n$$ GPD_{it} = \\beta_0 + \\beta_1 degree\\_days_{it} + \\beta_2 precip_{it} + \\mathbf{a}_i + \\mathbf{d}_t~~~~~~~~~~~~~~~~~~~~~~~(2)$$\n\nWhere $\\mathbf{a}_i$ are our city fixed effects, and $\\mathbf{d}_t$ our billing cycle fixed effects:\n\n`fe_reg1 <- lm(gpd ~ degree_days + precip + factor(city) + factor(billingcycle), data = waterdata)\nsummary(fe_reg1)`",
"_____no_output_____"
],
[
"__R__ automatically chose the first dummy variable for each set of fixed effect (city 1 and billing cycle 1) to leave out as our omitted group. \n\nNow that we account for which billing cycle we're in (i.e. whether we're in the winter or whether we're in the summer), we find that the coefficient on $degree\\_days$ is now much smaller and statistically insignificant. This makes sense, as we were falsely attributing the extra water use that comes from seasonality to temperature on its own. Now that we control for the season we're in via billing cycle fixed effects, we find that deviations in temperature exposure during a billing cycle don't result in dramatically higher water use within the sample.\n\n### Discussion: Why did we drop the home characteristics from our model?",
"_____no_output_____"
],
[
"## Method 2: Fixed Effects with felm()\n\nAlternatively, we could do everything way faster using the `felm()` function from the package __lfe__. This package doesn't require us to produce all the dummy variables by hand. Further, it performs the background math way faster so will be much quicker to estimate models using large datasets and many variables.\n\nThe syntax we use is now \n\n`felm(y ~ x1 + x2 + ... + xk | FE_1 + FE_2 + ..., data = df)`\n\n* The first section $y \\sim x1 + x2 +... xk$ is our formula, written the same way as with `lm()` - but omitting the fixed effects\n* We now add a `|` and in the second section we specify our fixed effects. Here we say $FE\\_1 + FE\\_2$ which tells __R__ to include fixed effects for each level of the variables $FE\\_1$ and $FE\\_2$.\n* we add the data source after the comma, as before.\n\nLet's go ahead and try this now with our water data model:\n\n`fe_reg2 <- felm(gpd ~ degree_days + precip | city + billingcycle, data = waterdata)\nsummary(fe_reg2)`",
"_____no_output_____"
],
[
"And we estimate the exact same coefficients on $degree\\_days$ and $precip$ as in the case where we specified everything by hand! We didn't have to mutate our data or add any variables. The one potential downside is that this approach doesn't report the fixed effects themselves by default. The tradeoff is that `felm` runs a lot faster than `lm`, especially with large datasets. \n\nWe can also recover the fixed effects with getfe(): \n\n`getfe(fe_reg2, se = TRUE, robust = TRUE)`",
"_____no_output_____"
],
[
"the argument `se = TRUE` tells it to produce standard errors too, and `robust = TRUE` further indicates that we want heteroskedasticity-robust standard errors.\n\n\nNote that this approach doesn't give you the same reference groups as before, but we get the same relative values. Note that before the coefficient on $city2$ was 301.7 and now is 73.9. But the coefficient on $city1$ is -227.8, and if we subtract $city1$ from $city2$ to get the difference in averages for city 2 relative to city 1 we get $73.9 - (-227.8) = 301.7$, the same as before!",
"_____no_output_____"
],
[
"# Fixed Effects Practice Question #1\n\n#### From a random sample of agricultural yields Y (1000 dollars per acre) for region $i$ in year $t$ for the US, we have estimated the following eqation:\n\n \\begin{align*} \\widehat{\\log(Y)}_{it} &=\t0.49\t+ .01 GE_{it} ~~~~ \tR^2 = .32\\\\\n\t&~~~~~(.11)\t ~~~~ (.01) ~~~~ n = 1526 \\end{align*}\n\n#### (a) Interpret the results on the Genetically engineered ($GE$) technology on yields. (follow SSS= Sign Size Significance)\n\n#### (b) Suppose $GE$ is used more on the West Coast, where crop yields are also higher. How would the estimated effect of GE change if we include a West Coast region dummy variable in the equation? Justify your answer.\n\n#### (c) If we include region fixed effects, would they control for the factors in (b)? Justify your answer.\n\n#### (d) If yields have been generally improving over time and GE adoption was only recently introduced in the USA, what would happen to the coefficient of GE if we included year fixed effects?\n\n\n\n# Fixed Effects Practice Question #2\n\n#### A recent paper investigates whether advertisement for Viagra causes increases in birth rates in the USA. Apparently, advertising for products, including Viagra, happens on TV and reaches households that have a TV within a Marketing region and does not happen in areas outside a designated marketing region. What the authors do is look at hospital birth rates in regions inside and near the advertising region border and collect data on dollars per 100 people (Ads) for a certain time, and compare those to the birth rates in hospitals located outside and near the advertising region designated border. They conduct a panel data analysis and estimate the following model:\n\n$$ Births_{it} = \\beta_0 + \\beta_1 Ads + \\beta_2 Ads^2 + Z_i + M_t + u_{it}$$\n\n#### Where $Z_i$ are zipcode fixed effects and $M_t$ monthly fixed effects.\n\n#### (a) Why do the authors include Zip Code Fixed Effects? In particular, what would be a variable that they are controlling for when adding Zip Code fixed effects that could cause a problem when interpreting the marginal effect of ad spending on birth rates? What would that (solved) problem be?\n\n#### (b) Why do they add month fixed effects? \n\n",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
cbf0472332b7e1bc48286b65e5739e49f5b3e23c
| 6,147 |
ipynb
|
Jupyter Notebook
|
Pytorch/Pytorch simple.ipynb
|
fastovetsilya/MyPythonScripts
|
a7f17638caeb760cba97e817b85a542458877bfb
|
[
"MIT"
] | null | null | null |
Pytorch/Pytorch simple.ipynb
|
fastovetsilya/MyPythonScripts
|
a7f17638caeb760cba97e817b85a542458877bfb
|
[
"MIT"
] | null | null | null |
Pytorch/Pytorch simple.ipynb
|
fastovetsilya/MyPythonScripts
|
a7f17638caeb760cba97e817b85a542458877bfb
|
[
"MIT"
] | null | null | null | 23.196226 | 147 | 0.496828 |
[
[
[
"import torch\nfrom torch.autograd import Variable",
"_____no_output_____"
],
[
"x = Variable(torch.ones(2, 2), requires_grad=True)\nprint(x)",
"Variable containing:\n 1 1\n 1 1\n[torch.FloatTensor of size 2x2]\n\n"
],
[
"y = x + 2 \nprint(y)",
"Variable containing:\n 3 3\n 3 3\n[torch.FloatTensor of size 2x2]\n\n"
],
[
"print(y.grad_fn)",
"<AddBackward0 object at 0x7f2723c22f60>\n"
],
[
"z = y * y * 3\nout = z.mean()\nprint(z, out)",
"Variable containing:\n 27 27\n 27 27\n[torch.FloatTensor of size 2x2]\n Variable containing:\n 27\n[torch.FloatTensor of size 1]\n\n"
],
[
"out.backward()",
"_____no_output_____"
],
[
"print(x.grad)",
"Variable containing:\n 4.5000 4.5000\n 4.5000 4.5000\n[torch.FloatTensor of size 2x2]\n\n"
],
[
"x = torch.randn(3)\nx = Variable(x, requires_grad=True)\n\ny = x * 2\n\nwhile y.data.norm() < 1000:\n y = y * 2\n\nprint(y)",
"Variable containing:\n-1293.1851\n 745.5553\n -602.8740\n[torch.FloatTensor of size 3]\n\n"
],
[
"gradients = torch.FloatTensor([0.1, 1.0, 0.0001])\nx.backward(gradients)\nprint(x.grad)",
"Variable containing:\n 102.5000\n 1025.0000\n 0.1025\n[torch.FloatTensor of size 3]\n\n"
],
[
"help(x.backward)",
"Help on method backward in module torch.autograd.variable:\n\nbackward(gradient=None, retain_graph=None, create_graph=None, retain_variables=None) method of torch.autograd.variable.Variable instance\n Computes the gradient of current variable w.r.t. graph leaves.\n \n The graph is differentiated using the chain rule. If the variable is\n non-scalar (i.e. its data has more than one element) and requires\n gradient, the function additionally requires specifying ``gradient``.\n It should be a tensor of matching type and location, that contains\n the gradient of the differentiated function w.r.t. ``self``.\n \n This function accumulates gradients in the leaves - you might need to\n zero them before calling it.\n \n Arguments:\n gradient (Tensor, Variable or None): Gradient w.r.t. the\n variable. If it is a tensor, it will be automatically converted\n to a Variable that is volatile unless ``create_graph`` is True.\n None values can be specified for scalar Variables or ones that\n don't require grad. If a None value would be acceptable then\n this argument is optional.\n retain_graph (bool, optional): If ``False``, the graph used to compute\n the grads will be freed. Note that in nearly all cases setting\n this option to True is not needed and often can be worked around\n in a much more efficient way. Defaults to the value of\n ``create_graph``.\n create_graph (bool, optional): If ``True``, graph of the derivative will\n be constructed, allowing to compute higher order derivative\n products. Defaults to ``False``, unless ``gradient`` is a volatile\n Variable.\n\n"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbf054dc80cd6508a18223dd7a604be1fb5b8166
| 6,598 |
ipynb
|
Jupyter Notebook
|
notebooks/11_Process_synchronization_example.ipynb
|
thijsreedijk/OpenCLSim
|
09f48976c584afd8860200e55e835363eadfbc27
|
[
"MIT"
] | null | null | null |
notebooks/11_Process_synchronization_example.ipynb
|
thijsreedijk/OpenCLSim
|
09f48976c584afd8860200e55e835363eadfbc27
|
[
"MIT"
] | null | null | null |
notebooks/11_Process_synchronization_example.ipynb
|
thijsreedijk/OpenCLSim
|
09f48976c584afd8860200e55e835363eadfbc27
|
[
"MIT"
] | null | null | null | 30.127854 | 542 | 0.482267 |
[
[
[
"# Demo Process Synchronization\nNext to the synchronization of activities using controll structures like **while** and **sequence** used within a single process, OpenCLSim allows to synchronize different processes using **start_events**. Start_events can be specified using the expression language as documented to **delay** the execution of a process. Control structures do not delay the execution but explicitly start the execution when possible. The difference is very well visible when you compare the activity log in this demo with the one from the sequence demo.",
"_____no_output_____"
]
],
[
[
"import datetime, time\nimport simpy\n\nimport pandas as pd\nimport openclsim.core as core\nimport openclsim.model as model\nimport openclsim.plot as plot\n\n# setup environment\nsimulation_start = 0\nmy_env = simpy.Environment(initial_time=simulation_start)\nregistry = {}",
"_____no_output_____"
]
],
[
[
"## Definition of two basic activities\nThe two activities are started in parallel, but the execution of Activity2 is delayed until Activity1 has been completed. To easier see how the two activities are interrelated a reporting activity is added.",
"_____no_output_____"
]
],
[
[
"reporting_activity = model.BasicActivity(\n env=my_env,\n name=\"Reporting activity\",\n registry=registry,\n duration=0,\n)\nactivity = model.BasicActivity(\n env=my_env,\n name=\"Activity1\",\n registry=registry,\n additional_logs=[reporting_activity],\n duration=14,\n)\nactivity2 = model.BasicActivity(\n env=my_env,\n name=\"Activity2\",\n registry=registry,\n additional_logs=[reporting_activity],\n start_event=[{\"type\": \"activity\", \"name\": \"Activity1\", \"state\": \"done\"}],\n duration=30,\n)",
"_____no_output_____"
]
],
[
[
"## Run simulation",
"_____no_output_____"
]
],
[
[
"model.register_processes([reporting_activity, activity, activity2])\nmy_env.run()",
"_____no_output_____"
],
[
"display(plot.get_log_dataframe(reporting_activity, [activity, activity2, reporting_activity]))",
"_____no_output_____"
]
],
[
[
"Both activities start at the same time. Activity2 gets into a waiting state, which ends, when Activity1 ends. Then Activity2 is executed.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
cbf05e6a2a57907e2ab8aca392933c1fc0e1e82d
| 2,265 |
ipynb
|
Jupyter Notebook
|
QS-AD/.ipynb_checkpoints/Examples_to_be_deleted-Copy1-checkpoint.ipynb
|
miguelbarretosanz/QS-AgregartorDashboard
|
59a317e16e771b190e3d27e2aff0a3da9a553aea
|
[
"MIT"
] | null | null | null |
QS-AD/.ipynb_checkpoints/Examples_to_be_deleted-Copy1-checkpoint.ipynb
|
miguelbarretosanz/QS-AgregartorDashboard
|
59a317e16e771b190e3d27e2aff0a3da9a553aea
|
[
"MIT"
] | null | null | null |
QS-AD/.ipynb_checkpoints/Examples_to_be_deleted-Copy1-checkpoint.ipynb
|
miguelbarretosanz/QS-AgregartorDashboard
|
59a317e16e771b190e3d27e2aff0a3da9a553aea
|
[
"MIT"
] | null | null | null | 28.3125 | 80 | 0.540397 |
[
[
[
"# myapp.py\n\nfrom random import random\n\nfrom bokeh.layouts import column\nfrom bokeh.models import Button\nfrom bokeh.palettes import RdYlBu3\nfrom bokeh.plotting import figure, curdoc\n\n# create a plot and style its properties\np = figure(x_range=(0, 100), y_range=(0, 100), toolbar_location=None)\np.border_fill_color = 'black'\np.background_fill_color = 'black'\np.outline_line_color = None\np.grid.grid_line_color = None\n\n# add a text renderer to our plot (no data yet)\nr = p.text(x=[], y=[], text=[], text_color=[], text_font_size=\"20pt\",\n text_baseline=\"middle\", text_align=\"center\")\n\ni = 0\n\nds = r.data_source\n\n# create a callback that will add a number in a random location\ndef callback():\n global i\n\n # BEST PRACTICE --- update .data in one step with a new dict\n new_data = dict()\n new_data['x'] = ds.data['x'] + [random()*70 + 15]\n new_data['y'] = ds.data['y'] + [random()*70 + 15]\n new_data['text_color'] = ds.data['text_color'] + [RdYlBu3[i%3]]\n new_data['text'] = ds.data['text'] + [str(i)]\n ds.data = new_data\n\n i = i + 1\n\n# add a button widget and configure with the call back\nbutton = Button(label=\"Press Me\")\nbutton.on_click(callback)\n\n# put the button and plot in a layout and add to the document\ncurdoc().add_root(column(button, p))",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code"
]
] |
cbf076a416719e25f42b7386600cd60805a5c880
| 5,981 |
ipynb
|
Jupyter Notebook
|
test/ipynb/java/JavaTest.ipynb
|
ssadedin/beakerx
|
34479b07d2dfdf1404692692f483faf0251632c3
|
[
"Apache-2.0"
] | 1 |
2018-10-16T18:59:59.000Z
|
2018-10-16T18:59:59.000Z
|
test/ipynb/java/JavaTest.ipynb
|
ssadedin/beakerx
|
34479b07d2dfdf1404692692f483faf0251632c3
|
[
"Apache-2.0"
] | 1 |
2019-10-27T19:56:51.000Z
|
2019-10-27T19:56:51.000Z
|
test/ipynb/java/JavaTest.ipynb
|
ssadedin/beakerx
|
34479b07d2dfdf1404692692f483faf0251632c3
|
[
"Apache-2.0"
] | null | null | null | 21.749091 | 100 | 0.50627 |
[
[
[
"empty"
]
]
] |
[
"empty"
] |
[
[
"empty"
]
] |
cbf08584b3f154f6487514d569a53b8e68cfef67
| 3,794 |
ipynb
|
Jupyter Notebook
|
AIDungeon_2.ipynb
|
applenick/AIDungeon
|
01c485a39c59820a8ab0361a13416edac65cddfe
|
[
"MIT"
] | 1 |
2021-01-05T16:26:25.000Z
|
2021-01-05T16:26:25.000Z
|
AIDungeon_2.ipynb
|
dyc3/AIDungeon
|
01c485a39c59820a8ab0361a13416edac65cddfe
|
[
"MIT"
] | 5 |
2020-09-26T00:29:01.000Z
|
2022-02-10T00:46:04.000Z
|
AIDungeon_2.ipynb
|
dyc3/AIDungeon
|
01c485a39c59820a8ab0361a13416edac65cddfe
|
[
"MIT"
] | null | null | null | 44.635294 | 342 | 0.591987 |
[
[
[
"\n\nSponsored by the BYU PCCL Lab.\n\n> AI Dungeon 2 is a completely AI generated text adventure built with OpenAI's largest GPT-2 model. It's a first of it's kind game that allows you to enter and will react to any action you can imagine.\n\n# Main mirrors of AI Dungeon 2 are currently down due to high download costs.\nWe are using bittorrent as a temporary solution to host game files and keep this game alive. It's not fast, but it's the best we've got right now.\n\nIf you want to help, best thing you can do is to **[download this torrent file with game files](https://github.com/nickwalton/AIDungeon/files/3935881/model_v5.torrent.zip)** and **seed it** indefinitely to the best of your ability. This will help new players download this game faster, and discover the vast worlds of AIDungeon2!\n\n- <a href=\"https://twitter.com/nickwalton00?ref_src=twsrc%5Etfw\" class=\"twitter-follow-button\" data-show-count=\"false\">Follow @nickwalton00</a> on Twitter for updates on when it will be available again.\n- **[Support AI Dungeon 2](https://www.patreon.com/AIDungeon) on Patreon to help me to continue improving the game with all the awesome ideas I have for its future!**\n\n## How to play\n1. Click \"Tools\"-> \"Settings...\" -> \"Theme\" -> \"Dark\" (optional but recommended)\n2. Click \"Runtime\" -> \"Run all\"\n3. Wait until all files are downloaded (only has to be one once, and it will take some time)\n4. It will then take a couple minutes to boot up as the model is loaded onto the GPU. \n5. If you have questions about getting it to work then please [go to github repo](https://github.com/AIDungeon/AIDungeon) to get help. \n\n## About\n* While you wait you can [read adventures others have had](https://aidungeon.io/)\n* [Read more](https://pcc.cs.byu.edu/2019/11/21/ai-dungeon-2-creating-infinitely-generated-text-adventures-with-deep-learning-language-models/) about how AI Dungeon 2 is made.- **[Support AI Dungeon 2](https://www.patreon.com/bePatron?u=19115449) on Patreon to help me to continue improving the game with all the awesome ideas I have for its future!**\n",
"_____no_output_____"
]
],
[
[
"!git clone --depth 1 --branch master https://github.com/AIDungeon/AIDungeon/\n%cd AIDungeon\n!./install.sh\nfrom IPython.display import clear_output \nclear_output()\nprint(\"Download Complete!\")",
"_____no_output_____"
],
[
"from IPython.display import Javascript\ndisplay(Javascript('''google.colab.output.setIframeHeight(0, true, {maxHeight: 5000})'''))\n!python play.py",
"_____no_output_____"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code"
]
] |
cbf0991a8f710edac475244df89161292f562079
| 3,479 |
ipynb
|
Jupyter Notebook
|
letsupgrade day 3 assignment.ipynb
|
msadithyabhat/Letsupgrade--python-assignment
|
48db90404b8740de6b1b1de1d8d06b16ecdc82d0
|
[
"Apache-2.0"
] | null | null | null |
letsupgrade day 3 assignment.ipynb
|
msadithyabhat/Letsupgrade--python-assignment
|
48db90404b8740de6b1b1de1d8d06b16ecdc82d0
|
[
"Apache-2.0"
] | null | null | null |
letsupgrade day 3 assignment.ipynb
|
msadithyabhat/Letsupgrade--python-assignment
|
48db90404b8740de6b1b1de1d8d06b16ecdc82d0
|
[
"Apache-2.0"
] | null | null | null | 20.226744 | 54 | 0.449554 |
[
[
[
"# Assignment 1",
"_____no_output_____"
]
],
[
[
"alt = int(input(\"enter the altitude\"))\nif alt<=1000:\n print(\"Safe to land\")\nelif alt>1000 and alt<5000:\n print(\"bring down to 1000\")\nelse:\n print(\"Turn around\")",
"_____no_output_____"
],
[
"alt = int(input(\"enter the altitude\"))\nif alt<=1000:\n print(\"Safe to land\")\nelif alt>1000 and alt<5000:\n print(\"bring down to 1000\")\nelse:\n print(\"Turn around\")",
"_____no_output_____"
],
[
"alt = int(input(\"enter the altitude\"))\nif alt<=1000:\n print(\"Safe to land\")\nelif alt>1000 and alt<5000:\n print(\"bring down to 1000\")\nelse:\n print(\"Turn around\")",
"_____no_output_____"
]
],
[
[
"\n# Assignment 2",
"_____no_output_____"
]
],
[
[
"lower = int(input(\"Enter lower range: \")) \nupper = int(input(\"Enter upper range: \")) \n \nfor num in range(lower,upper + 1): \n if num > 1: \n for i in range(2,num): \n if (num % i) == 0: \n break \n else: \n print(num) ",
"_____no_output_____"
]
],
[
[
"\n# Assignment 3",
"_____no_output_____"
]
],
[
[
"\n ",
"_____no_output_____"
],
[
"lower = int(input(\"Enter lower range: \")) \nupper = int(input(\"Enter upper range: \")) \n \nfor num in range(lower,upper + 1): \n sum = 0 \n temp = num \n while temp > 0: \n digit = temp % 10 \n sum += digit ** 3 \n temp //= 10 \n if num == sum: \n print(num) \n",
"Enter lower range: 1042000\nEnter upper range: 702648265\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
cbf099b9d7fbc519c00c5b6085c2d0d20f2ab358
| 40,238 |
ipynb
|
Jupyter Notebook
|
benchmarks/benchmark7.ipynb
|
stvdwtt/chimad-phase-field
|
cf0c0b923b7dcfd8eb5785fb778438387194920d
|
[
"MIT"
] | null | null | null |
benchmarks/benchmark7.ipynb
|
stvdwtt/chimad-phase-field
|
cf0c0b923b7dcfd8eb5785fb778438387194920d
|
[
"MIT"
] | 1 |
2018-01-12T21:47:30.000Z
|
2018-01-26T16:45:15.000Z
|
benchmarks/benchmark7.ipynb
|
stvdwtt/chimad-phase-field
|
cf0c0b923b7dcfd8eb5785fb778438387194920d
|
[
"MIT"
] | null | null | null | 34.959166 | 1,161 | 0.538198 |
[
[
[
"%%javascript\n MathJax.Hub.Config({\n TeX: { equationNumbers: { autoNumber: \"AMS\" } }\n });",
"_____no_output_____"
],
[
"from IPython.display import HTML\n\nHTML('''<script>\ncode_show=true; \nfunction code_toggle() {\n if (code_show){\n $('div.input').hide();\n $('div.prompt').hide();\n } else {\n $('div.input').show();\n$('div.prompt').show();\n }\n code_show = !code_show\n} \n$( document ).ready(code_toggle);\n</script>\n<form action=\"javascript:code_toggle()\"><input type=\"submit\" value=\"Code Toggle\"></form>''')",
"_____no_output_____"
],
[
"from IPython.display import HTML\n\nHTML('''\n<a href=\"{{ site.links.github }}/raw/nist-pages/benchmarks/benchmark7.ipynb\"\n download>\n<button type=\"submit\">Download Notebook</button>\n</a>\n''')",
"_____no_output_____"
]
],
[
[
"# Benchmark Problem 7: MMS Allen-Cahn",
"_____no_output_____"
]
],
[
[
"from IPython.display import HTML\n\nHTML('''{% include jupyter_benchmark_table.html num=\"[7]\" revision=0 %}''')",
"_____no_output_____"
]
],
[
[
"* [Overview](#Overview)\n* [Governing equation and manufactured solution](#Governing-equation-and-manufactured-solution)\n* [Domain geometry, boundary conditions, initial conditions, and stopping condition](#Domain-geometry,-boundary-conditions,-initial-conditions,-and-stopping-condition)\n* [Parameter values](#Parameter-values)\n* [Benchmark simulation instructions](#Benchmark-simulation-instructions)\n * [Part (a)](#Part-%28a%29)\n * [Part (b)](#Part-%28b%29)\n * [Part (c)](#Part-%28c%29)\n* [Results](#Results)\n* [Feedback](#Feedback)\n* [Appendix](#Appendix)\n * [Computer algebra systems](#Computer-algebra-systems)\n * [Source equation](#Source-equation)\n * [Code](#Code)\n\n\nSee the journal publication entitled [\"Benchmark problems for numerical implementations of phase field models\"][benchmark_paper] for more details about the benchmark problems. Furthermore, read [the extended essay][benchmarks] for a discussion about the need for benchmark problems.\n\n[benchmarks]: ../\n[benchmark_paper]: http://dx.doi.org/10.1016/j.commatsci.2016.09.022",
"_____no_output_____"
],
[
"# Overview",
"_____no_output_____"
],
[
"The Method of Manufactured Solutions (MMS) is a powerful technique for verifying the accuracy of a simulation code. In the MMS, one picks a desired solution to the problem at the outset, the \"manufactured solution\", and then determines the governing equation that will result in that solution. With the exact analytical form of the solution in hand, when the governing equation is solved using a particular simulation code, the deviation from the expected solution can be determined exactly. This deviation can be converted into an error metric to rigously quantify the error for a calculation. This error can be used to determine the order of accuracy of the simulation results to verify simulation codes. It can also be used to compare the computational efficiency of different codes or different approaches for a particular code at a certain level of error. Furthermore, the spatial/temporal distribution can give insight into the conditions resulting in the largest error (high gradients, changes in mesh resolution, etc.).\n\nAfter choosing a manufactured solution, the governing equation must be modified to force the solution to equal the manufactured solution. This is accomplished by taking the nominal equation that is to be solved (e.g. Allen-Cahn equation, Cahn-Hilliard equation, Fick's second law, Laplace equation) and adding a source term. This source term is determined by plugging the manufactured solution into the nominal governing equation and setting the source term equal to the residual. Thus, the manufactured solution satisfies the MMS governing equation (the nominal governing equation plus the source term). A more detailed discussion of MMS can be found in [the report by Salari and Knupp][mms_report].\n\nIn this benchmark problem, the objective is to use the MMS to rigorously verify phase field simulation codes and then provide a basis of comparison for the computational performance between codes and for various settings for a single code, as discussed above. To this end, the benchmark problem was chosen as a balance between two factors: simplicity, to minimize the development effort required to solve the benchmark, and transferability to a real phase field system of physical interest. \n\n[mms_report]: http://prod.sandia.gov/techlib/access-control.cgi/2000/001444.pdf",
"_____no_output_____"
],
[
"# Governing equation and manufactured solution\nFor this benchmark problem, we use a simple Allen-Cahn equation as the governing equation\n\n$$\\begin{equation}\n\\frac{\\partial \\eta}{\\partial t} = - \\left[ 4 \\eta \\left(\\eta - 1 \\right) \\left(\\eta-\\frac{1}{2} \\right) - \\kappa \\nabla^2 \\eta \\right] + S(x,y,t) \n\\end{equation}$$\n\nwhere $S(x,y,t)$ is the MMS source term and $\\kappa$ is a constant parameter (the gradient energy coefficient). \n\nThe manufactured solution, $\\eta_{sol}$ is a hyperbolic tangent function, shifted to vary between 0 and 1, with the $x$ position of the middle of the interface ($\\eta_{sol}=0.5$) given by the function $\\alpha(x,t)$:\n\n$$\\begin{equation}\n\\eta_{sol}(x,y,t) = \\frac{1}{2}\\left[ 1 - \\tanh\\left( \\frac{y-\\alpha(x,t)}{\\sqrt{2 \\kappa}} \\right) \\right] \n\\end{equation}$$\n\n$$\\begin{equation}\n\\alpha(x,t) = \\frac{1}{4} + A_1 t \\sin\\left(B_1 x \\right) + A_2 \\sin \\left(B_2 x + C_2 t \\right)\n\\end{equation}$$\n\nwhere $A_1$, $B_1$, $A_2$, $B_2$, and $C_2$ are constant parameters. \n\nThis manufactured solution is an equilbrium solution of the governing equation, when $S(x,y,t)=0$ and $\\alpha(x,t)$ is constant. The closeness of this manufactured solution to a solution of the nominal governing equation increases the likihood that the behavior of simulation codes when solving this benchmark problem is representive of the solution of the regular Allen-Cahn equation (i.e. without the source term). The form of $\\alpha(x,t)$ was chosen to yield complex behavior while still retaining a (somewhat) simple functional form. The two spatial sinusoidal terms introduce two controllable length scales to the interfacial shape. Summing them gives a \"beat\" pattern with a period longer than the period of either individual term, permitting a domain size that is larger than the wavelength of the sinusoids without a repeating pattern. The temporal sinusoidal term introduces a controllable time scale to the interfacial shape in addition to the phase transformation time scale, while the linear temporal dependence of the other term ensures that the sinusoidal term can go through multiple periods without $\\eta_{sol}$ repeating itself.\n\nInserting the manufactured solution into the governing equation and solving for $S(x,y,t)$ yields:\n\n$$\\begin{equation}\nS(x,y,t) = \\frac{\\text{sech}^2 \\left[ \\frac{y-\\alpha(x,t)}{\\sqrt{2 \\kappa}} \\right]}{4 \\sqrt{\\kappa}} \\left[-2\\sqrt{\\kappa} \\tanh \\left[\\frac{y-\\alpha(x,t)}{\\sqrt{2 \\kappa}} \\right] \\left(\\frac{\\partial \\alpha(x,t)}{\\partial x} \\right)^2+\\sqrt{2} \\left[ \\frac{\\partial \\alpha(x,t)}{\\partial t}-\\kappa \\frac{\\partial^2 \\alpha(x,t)}{\\partial x^2} \\right] \\right]\n\\end{equation}$$\n\nwhere $\\alpha(x,t)$ is given above and where:\n\n$$\\begin{equation}\n\\frac{\\partial \\alpha(x,t)}{\\partial x} = A_1 B_1 t \\cos\\left(B_1 x\\right) + A_2 B_2 \\cos \\left(B_2 x + C_2 t \\right)\n\\end{equation}$$\n\n$$\\begin{equation}\n\\frac{\\partial^2 \\alpha(x,t)}{\\partial x^2} = -A_1 B_1^2 t \\sin\\left(B_1 x\\right) - A_2 B_2^2 \\sin \\left(B_2 x + C_2 t \\right)\n\\end{equation}$$\n\n$$\\begin{equation}\n\\frac{\\partial \\alpha(x,t)}{\\partial t} = A_1 \\sin\\left(B_1 x\\right) + A_2 C_2 \\cos \\left(B_2 x + C_2 t \\right)\n\\end{equation}$$\n\n#### *N.B.*: Don't transcribe these equations. Please download the appropriate files from the [Appendix](#Appendix).",
"_____no_output_____"
],
[
"# Domain geometry, boundary conditions, initial conditions, and stopping condition\nThe domain geometry is a rectangle that spans [0, 1] in $x$ and [0, 0.5] in $y$. This elongated domain was chosen to allow multiple peaks and valleys in $\\eta_{sol}$ without stretching the interface too much in the $y$ direction (which causes the thickness of the interface to change) or having large regions where $\\eta_{sol}$ never deviates from 0 or 1. Periodic boundary conditions are applied along the $x = 0$ and the $x = 1$ boundaries to accomodate the periodicity of $\\alpha(x,t)$. Dirichlet boundary conditions of $\\eta$ = 0 and $\\eta$ = 1 are applied along the $y = 0$ and the $y = 0.5$ boundaries, respectively. These boundary conditions are chosen to be consistent with $\\eta_{sol}(x,y,t)$. The initial condition is the manufactured solution at $t = 0$:\n\n$$\n\\begin{equation}\n\\eta_{sol}(x,y,0) = \\frac{1}{2}\\left[ 1 - \\tanh\\left( \\frac{y-\\left(\\frac{1}{4}+A_2 \\sin(B_2 x) \\right)}{\\sqrt{2 \\kappa}} \\right) \\right] \n\\end{equation}\n$$\n\nThe stopping condition for all calculations is when t = 8 time units, which was chosen to let $\\alpha(x,t)$ evolve substantially, while still being slower than the characteristic time for the phase evolution (determined by the CFL condition for a uniform mesh with a reasonable level of resolution of $\\eta_{sol}$).",
"_____no_output_____"
],
[
"# Parameter values\nThe nominal parameter values for the governing equation and manufactured solution are given below. The value of $\\kappa$ will change in Part (b) in the following section and the values of $\\kappa$ and $C_2$ will change in Part (c).\n\n| Parameter | Value |\n|-----------|-------|\n| $\\kappa$ | 0.0004|\n| $A_1$ | 0.0075|\n| $B_1$ | 0.03 |\n| $A_2$ | 8.0 |\n| $B_2$ | 22.0 |\n| $C_2$ | 0.0625|",
"_____no_output_____"
],
[
"# Benchmark simulation instructions\nThis section describes three sets of tests to conduct using the MMS problem specified above. The primary purpose of the first test is provide a computationally inexpensive problem to verify a simulation code. The second and third tests are more computationally demanding and are primarily designed to serve as a basis for performance comparisons.",
"_____no_output_____"
],
[
"## Part (a)\nThe objective of this test is to verify the accuracy of your simulation code in both time and space. Here, we make use of convergence tests, where either the mesh size (or grid point spacing) or the time step size is systematically changed to determine the response of the error to these quantities. Once a convergence test is completed the order of accuracy can be calculated from the result. The order of accuracy can be compared to the theoretical order of accuracy for the numerical method employed in the simulation. If the two match (to a reasonable degree), then one can be confident that the simulation code is working as expected. The remainder of this subsection will give instructions for convergence tests for this MMS problem.\n\nImplement the MMS problem specified above using the simulation code of your choice. Perform a spatial convergence test by running the simulation for a variety of mesh sizes. For each simulation, determine the discrete $L_2$ norm of the error at $t=8$:\n\n$$\\begin{equation}\n L_2 = \\sqrt{\\sum\\limits_{x,y}\\left(\\eta^{t=8}_{x,y} - \\eta_{sol}(x,y,8)\\right)^2 \\Delta x \\Delta y}\n\\end{equation}$$\n\nFor all of these simulations, verify that the time step is small enough that any temporal error is much smaller that the total error. This can be accomplished by decreasing the time step until it has minimal effect on the error. Ensure that at least three simulation results have $L_2$ errors in the range $[5\\times10^{-3}, 1\\times10^{-4}]$, attempting to cover as much of that range as possible/practical. This maximum and minimum errors in the range roughly represent a poorly resolved simulation and a very well-resolved simulation.\n\nFor at least three simulations that have $L_2$ errors in the range $[5\\times10^{-3}, 1\\times10^{-4}]$, save the effective mesh size and $L_2$ error in a CSV or JSON file. Upload this file to the PFHub website as a 2D data set with the effective mesh size as the x-axis column and the $L_2$ error as the y-axis column. Calculate the effective element size as the square root of the area of the finest part of the mesh for nonuniform meshes. For irregular meshes with continous distributions of element sizes, approximate the effective mesh size as the average of the square root of the area of the smallest 5% of the elements.\n\nNext, confirm that the observed order of accuracy is approximately equal to the expected value. Calculate the order of accuracy, $p$, with a least squares fit of the following function:\n\n$$\\begin{equation}\n \\log(E)=p \\log(R) + b\n\\end{equation}$$\n\nwhere $E$ is the $L_2$ error, $R$ is the effective element size, and b is an intercept. Deviations of ±0.2 or more from the theoretical value are to be expected (depending on the range of errors considered and other factors).\n\nFinally, perform a similar convergence test, but for the time step, systematically changing the time step and recording the $L_2$ error. Use a time step that does not vary over the course of any single simulation. Verify that the spatial discretization error is small enough that it does not substantially contribute to the total error. Once again, ensure that at least three simulations have $L_2$ errors in the range $[5\\times10^{-3}, 1\\times10^{-4}]$, attempting to cover as much of that range as possible/practical. Save the effective mesh size and $L_2$ error for each individual simulation in a CSV or JSON file. [Upload this file to the PFHub website](https://pages.nist.gov/pfhub/simulations/upload_form/) as a 2D data set with the time step size as the x-axis column and the $L_2$ error as the y-axis column. Confirm that the observed order of accuracy is approximately equal to the expected value.",
"_____no_output_____"
],
[
"## Part (b)\nNow that your code has been verified in (a), the objective of this part is to determine the computational performance of your code at various levels of error. These results can then be used to objectively compare the performance between codes or settings within the same code. To make the problem more computationally demanding and stress solvers more than in (a), decrease $\\kappa$ by a factor of $256$ to $1.5625\\times10^{-6}$. This change will reduce the interfacial thickness by a factor of $16$.\n\nRun a series of simulations, attempting to optimize solver parameters (mesh, time step, tolerances, etc.) to minimize the required computational resources for at least three levels of $L_2$ error in range $[5\\times10^{-3}, 1\\times10^{-5}]$. Use the same CPU and processor type for all simulations. For the best of these simulations, save the wall time, number of computing cores, maximum memory usage, and $L_2$ error for each individual simulation in a CSV or JSON file. [Upload this to the PFHub website](https://pages.nist.gov/pfhub/simulations/upload_form/) as a 3D data set with the wall time as the x-axis column, the number of computing cores as the y-axis column, and the $L_2$ error as the z-axis column. (The PFHub upload system is currently limited to three columns of data. Once this constraint is relaxed, the maximum memory usage data will be incorporated as well.)\n\n<!---For the best of these simulations, submit the wall time, number of computing cores, processor speed, maximum memory usage, and $L_2$ error at $t=8$ to the CHiMaD website.--->",
"_____no_output_____"
],
[
"## Part (c)\nThis final part is designed to stress time integrators even further by increasing the rate of change of $\\alpha(x,t)$. Increase $C_2$ to $0.5$. Keep $\\kappa= 1.5625\\times10^{-6}$ from (b).\n\nRepeat the process from (b), uploading the wall time, number of computing cores, processor speed, maximum memory usage, and $L_2$ error at $t=8$ to the PFHub website.",
"_____no_output_____"
],
[
"# Results\nResults from this benchmark problem are displayed on the [simulation result page]({{ site.baseurl }}/simulations) for different codes.",
"_____no_output_____"
],
[
"# Feedback\nFeedback on this benchmark problem is appreciated. If you have questions, comments, or seek clarification, please contact the [CHiMaD phase field community](https://pages.nist.gov/chimad-phase-field/community/) through the [Gitter chat channel](https://gitter.im/usnistgov/chimad-phase-field) or by [email](https://pages.nist.gov/chimad-phase-field/mailing_list/). If you found an error, please file an [issue on GitHub](https://github.com/usnistgov/chimad-phase-field/issues/new).",
"_____no_output_____"
],
[
"# Appendix\n\n## Computer algebra systems\nRigorous verification of software frameworks using MMS requires posing the equation and manufacturing the solution with as much complexity as possible. This can be straight-forward, but interesting equations produce complicated source terms. To streamline the MMS workflow, it is strongly recommended that you use a CAS such as SymPy, Maple, or Mathematica to generate source equations and turn it into executable code automatically. For accessibility, we will use [SymPy](http://www.sympy.org/), but so long as vector calculus is supported, and CAS will do.",
"_____no_output_____"
],
[
"## Source term",
"_____no_output_____"
]
],
[
[
"from sympy import Symbol, symbols, simplify\nfrom sympy import Eq, sin, cos, cosh, sinh, tanh, sqrt\nfrom sympy.physics.vector import divergence, gradient, ReferenceFrame, time_derivative\nfrom sympy.printing import pprint\nfrom sympy.abc import kappa, S, t, x, y\n\n# Spatial coordinates: x=R[0], y=R[1], z=R[2]\nR = ReferenceFrame('R')\n\n# sinusoid amplitudes\nA1, A2 = symbols('A1 A2')\nB1, B2 = symbols('B1 B2')\nC1, C2 = symbols('C1 C2')\n\n# Define interface offset (alpha)\nalpha = (1/4 + A1 * t * sin(B1 * R[0]) \n + A2 * sin(B2 * R[0] + C2 * t)\n ).subs({R[0]: x, R[1]: y})\n\n# Define the solution equation (eta) \neta = (1/2 * (1 - tanh((R[1] - alpha) /\n sqrt(2*kappa)))\n ).subs({R[0]: x, R[1]: y})\n\n# Compute the initial condition\neta0 = eta.subs({t: 0, R[0]: x, R[1]: y})\n\n# Compute the source term from the equation of motion\nS = simplify(time_derivative(eta, R)\n + 4 * eta * (eta - 1) * (eta - 1/2)\n - divergence(kappa * gradient(eta, R), R)\n ).subs({R[0]: x, R[1]: y})",
"_____no_output_____"
],
[
"pprint(Eq(symbols('alpha'), alpha))",
"α = A₁⋅t⋅sin(B₁⋅x) + A₂⋅sin(B₂⋅x + C₂⋅t) + 0.25\n"
],
[
"pprint(Eq(symbols('eta'), eta))",
" ⎛√2⋅(-A₁⋅t⋅sin(B₁⋅x) - A₂⋅sin(B₂⋅x + C₂⋅t) + y - 0.25)⎞ \nη = - 0.5⋅tanh⎜─────────────────────────────────────────────────────⎟ + 0.5\n ⎝ 2⋅√κ ⎠ \n"
],
[
"pprint(Eq(symbols('eta0'), eta0))",
" ⎛√2⋅(-A₂⋅sin(B₂⋅x) + y - 0.25)⎞ \nη₀ = - 0.5⋅tanh⎜─────────────────────────────⎟ + 0.5\n ⎝ 2⋅√κ ⎠ \n"
],
[
"pprint(Eq(symbols('S'), S))",
" ⎛ ⎛√2⋅(A₁⋅t⋅sin(B₁⋅x) + A₂⋅sin(B₂⋅x + C₂⋅t) - y + 0.25)⎞ \n ⎜0.5⋅√κ⋅tanh⎜────────────────────────────────────────────────────⎟ - 0.25⋅\n ⎝ ⎝ 2⋅√κ ⎠ \nS = ──────────────────────────────────────────────────────────────────────────\n \n\n ⎞ ⎛ 2⎛√2⋅(A₁⋅t⋅sin(B₁⋅x) + A₂⋅sin\n√2⋅(A₁⋅sin(B₁⋅x) + A₂⋅C₂⋅cos(B₂⋅x + C₂⋅t))⎟⋅⎜tanh ⎜───────────────────────────\n ⎠ ⎝ ⎝ 2⋅√\n──────────────────────────────────────────────────────────────────────────────\n √κ \n\n(B₂⋅x + C₂⋅t) - y + 0.25)⎞ ⎞\n─────────────────────────⎟ - 1⎟\nκ ⎠ ⎠\n───────────────────────────────\n \n"
]
],
[
[
"## Code",
"_____no_output_____"
],
[
"### Python\n\nCopy the first cell under Source Term directly into your program.\nFor a performance boost, convert the expressions into lambda functions:\n```python\nfrom sympy.utilities.lambdify import lambdify\n\napy = lambdify([x, y], alpha, modules='sympy')\nepy = lambdify([x, y], eta, modules='sympy')\nipy = lambdify([x, y], eta0, modules='sympy')\nSpy = lambdify([x, y], S, modules='sympy')\n```\n#### *N.B.*: You may need to add coefficients to the variables list.",
"_____no_output_____"
],
[
"### C",
"_____no_output_____"
]
],
[
[
"from sympy.utilities.codegen import codegen\n\n[(c_name, c_code), (h_name, c_header)] = codegen([('alpha', alpha),\n ('eta', eta),\n ('eta0', eta0),\n ('S', S)],\n language='C', prefix='MMS', project='PFHub')\nprint(c_code)",
"/******************************************************************************\n * Code generated with sympy 1.1.1 *\n * *\n * See http://www.sympy.org/ for more information. *\n * *\n * This file is part of 'PFHub' *\n ******************************************************************************/\n#include \"MMS.h\"\n#include <math.h>\n\ndouble alpha(double A1, double A2, double B1, double B2, double C2, double t, double x) {\n\n double alpha_result;\n alpha_result = A1*t*sin(B1*x) + A2*sin(B2*x + C2*t) + 0.25;\n return alpha_result;\n\n}\n\ndouble eta(double A1, double A2, double B1, double B2, double C2, double kappa, double t, double x, double y) {\n\n double eta_result;\n eta_result = -0.5*tanh((1.0L/2.0L)*sqrt(2)*(-A1*t*sin(B1*x) - A2*sin(B2*x + C2*t) + y - 0.25)/sqrt(kappa)) + 0.5;\n return eta_result;\n\n}\n\ndouble eta0(double A2, double B2, double kappa, double x, double y) {\n\n double eta0_result;\n eta0_result = -0.5*tanh((1.0L/2.0L)*sqrt(2)*(-A2*sin(B2*x) + y - 0.25)/sqrt(kappa)) + 0.5;\n return eta0_result;\n\n}\n\ndouble S(double A1, double A2, double B1, double B2, double C2, double kappa, double t, double x, double y) {\n\n double S_result;\n S_result = (0.5*sqrt(kappa)*tanh((1.0L/2.0L)*sqrt(2)*(A1*t*sin(B1*x) + A2*sin(B2*x + C2*t) - y + 0.25)/sqrt(kappa)) - 0.25*sqrt(2)*(A1*sin(B1*x) + A2*C2*cos(B2*x + C2*t)))*(pow(tanh((1.0L/2.0L)*sqrt(2)*(A1*t*sin(B1*x) + A2*sin(B2*x + C2*t) - y + 0.25)/sqrt(kappa)), 2) - 1)/sqrt(kappa);\n return S_result;\n\n}\n\n"
]
],
[
[
"### C++",
"_____no_output_____"
]
],
[
[
"from sympy.printing.cxxcode import cxxcode",
"_____no_output_____"
],
[
"print(\"α:\")\ncxxcode(alpha)",
"α:\n"
],
[
"print(\"η:\")\ncxxcode(eta)",
"η:\n"
],
[
"print(\"η₀:\")\ncxxcode(eta0)",
"η₀:\n"
],
[
"print(\"S:\")\ncxxcode(S)",
"S:\n"
]
],
[
[
"### Fortran",
"_____no_output_____"
]
],
[
[
"from sympy.printing import fcode",
"_____no_output_____"
],
[
"print(\"α:\")\nfcode(alpha)",
"α:\n"
],
[
"print(\"η:\")\nfcode(eta)",
"η:\n"
],
[
"print(\"η₀:\")\nfcode(eta0)",
"η₀:\n"
],
[
"print(\"S:\")\nfcode(S)",
"S:\n"
]
],
[
[
"### Julia",
"_____no_output_____"
]
],
[
[
"from sympy.printing import julia_code",
"_____no_output_____"
],
[
"print(\"α:\")\njulia_code(alpha)",
"α:\n"
],
[
"print(\"η:\")\njulia_code(eta)",
"η:\n"
],
[
"print(\"η₀:\")\njulia_code(eta0)",
"η₀:\n"
],
[
"print(\"S:\")\njulia_code(S)",
"S:\n"
]
],
[
[
"### Mathematica",
"_____no_output_____"
]
],
[
[
"from sympy.printing import mathematica_code",
"_____no_output_____"
],
[
"print(\"α:\")\nmathematica_code(alpha)",
"α:\n"
],
[
"print(\"η:\")\nmathematica_code(eta)",
"η:\n"
],
[
"print(\"η₀:\")\nmathematica_code(eta0)",
"η₀:\n"
],
[
"print(\"S:\")\nmathematica_code(S)",
"S:\n"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
cbf0be43dd461d31e82cb4e924d498c95b533568
| 52,074 |
ipynb
|
Jupyter Notebook
|
notebooks/.ipynb_checkpoints/10-jalvaradoruiz-carga-delitos-checkpoint.ipynb
|
JALVARADORUIZ/DataAnalytic-PJUD
|
cd063efd619eca3b7bf583f1fb00383f6f774818
|
[
"MIT"
] | null | null | null |
notebooks/.ipynb_checkpoints/10-jalvaradoruiz-carga-delitos-checkpoint.ipynb
|
JALVARADORUIZ/DataAnalytic-PJUD
|
cd063efd619eca3b7bf583f1fb00383f6f774818
|
[
"MIT"
] | null | null | null |
notebooks/.ipynb_checkpoints/10-jalvaradoruiz-carga-delitos-checkpoint.ipynb
|
JALVARADORUIZ/DataAnalytic-PJUD
|
cd063efd619eca3b7bf583f1fb00383f6f774818
|
[
"MIT"
] | null | null | null | 35.740563 | 228 | 0.401774 |
[
[
[
"# EXTRACCION, LIMPIEZA Y CARGA DATA REFERENTE A DELITOS\nVERSION 0.2\n\nFECHA: 16/10/2020\n\nANALIZAR DELITOS VIGENTES Y NO VIGENTES\n",
"_____no_output_____"
]
],
[
[
"import os\nimport pandas as pd\nimport numpy as np\nfrom pyarrow import feather\nfrom tqdm import tqdm\nfrom unicodedata import normalize\n\nfrom src.data import clean_data",
"_____no_output_____"
],
[
"tqdm.pandas()",
"/Users/jalvaradoruiz/opt/anaconda3/lib/python3.7/site-packages/tqdm/std.py:658: FutureWarning: The Panel class is removed from pandas. Accessing it from the top-level namespace will also be removed in the next version\n from pandas import Panel\n"
],
[
"codigos_delitos = pd.read_excel(\"../data/external/codigos_penal_2020.xlsx\", sheet_name = \"codigos vigentes\")",
"_____no_output_____"
],
[
"codigos_delitos = codigos_delitos.drop_duplicates() # elimino filas con NaN\ncodigos_delitos = codigos_delitos.drop([0,1,2], axis = 0) # elimino 2 primeras filas que son titulos\n",
"_____no_output_____"
],
[
"# elimino columnas con datos NaN\n\nvariables = range(2,248)\ncolumnas = []\nfor variable in variables:\n columnas.append(\"Unnamed: \" + str(variable))\n\ncodigos_delitos = codigos_delitos.drop(columns = columnas, axis = 1)",
"_____no_output_____"
],
[
"# cambio nombres columnas\n\ncodigos_delitos = codigos_delitos.rename(columns = {'VERSION AL 01/01/2018':'COD. MATERIA', 'Unnamed: 1':'MATERIA'})",
"_____no_output_____"
],
[
"codigos_delitos",
"_____no_output_____"
],
[
"delitos_vigentes = []\n\nfor item in codigos_delitos.index:\n\n if str(codigos_delitos['COD. MATERIA'][item]).isupper():\n tipologia_delito=str(codigos_delitos['COD. MATERIA'][item])\n else:\n delitos_vigentes.append([codigos_delitos['COD. MATERIA'][item],\n str(codigos_delitos['MATERIA'][item]).upper().rstrip(),\n tipologia_delito,'VIGENTE'])\n ",
"_____no_output_____"
],
[
"df_delitos_vigentes = pd.DataFrame(delitos_vigentes,columns = ['COD. MATERIA','MATERIA','TIPOLOGIA MATERIA','VIGENCIA MATERIA'])",
"_____no_output_____"
],
[
"df_delitos_vigentes\n",
"_____no_output_____"
],
[
"df_delitos_vigentes.dtypes",
"_____no_output_____"
]
],
[
[
"## Limpieza de variable MATERIA",
"_____no_output_____"
]
],
[
[
"# Elimino tildes de las columnas object\n\ncols = df_delitos_vigentes.select_dtypes(include = [\"object\"]).columns\ndf_delitos_vigentes[cols] = df_delitos_vigentes[cols].progress_apply(clean_data.elimina_tilde)\ndf_delitos_vigentes[cols] = df_delitos_vigentes[cols].progress_apply(clean_data.limpieza_caracteres)",
"100%|██████████| 4/4 [00:00<00:00, 183.12it/s]\n100%|██████████| 4/4 [00:00<00:00, 350.29it/s]\n"
],
[
"df_delitos_vigentes.tail(100)",
"_____no_output_____"
],
[
"df_delitos_vigentes['COD. MATERIA'] = df_delitos_vigentes['COD. MATERIA'].fillna(0).astype('int16')\ndf_delitos_vigentes.info()\n",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 440 entries, 0 to 439\nData columns (total 4 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 COD. MATERIA 440 non-null int16 \n 1 MATERIA 440 non-null object\n 2 TIPOLOGIA MATERIA 440 non-null object\n 3 VIGENCIA MATERIA 440 non-null object\ndtypes: int16(1), object(3)\nmemory usage: 11.3+ KB\n"
]
],
[
[
"## CARGA Y LIMPIEZA DE DATOS RELACIONADOS A DELITOS NO VIGENTES",
"_____no_output_____"
]
],
[
[
"codigos_delitos_novigentes = pd.read_excel(\"../data/external/codigos_penal_2020.xlsx\", sheet_name = \"Codigos no vigentes\")",
"_____no_output_____"
],
[
"# cambio nombres columnas\n\ncodigos_delitos_novigentes = codigos_delitos_novigentes.rename(columns = {'MATERIAS PENALES NO VIGENTES':'TIPOLOGIA MATERIA',\n 'Unnamed: 1':'COD. MATERIA','Unnamed: 2':'MATERIA'\n })\n",
"_____no_output_____"
],
[
"codigos_delitos_novigentes = codigos_delitos_novigentes.drop([0], axis = 0) # elimino primera fila que son titulos\ncodigos_delitos_novigentes = codigos_delitos_novigentes.fillna('ST') # reemplazo Nan por ST",
"_____no_output_____"
],
[
"codigos_delitos_novigentes",
"_____no_output_____"
],
[
"delitos_no_vigentes = []\nfor item in codigos_delitos_novigentes.index:\n \n tipologia_delito = codigos_delitos_novigentes['TIPOLOGIA MATERIA'][item]\n \n if tipologia_delito != 'ST':\n tipologia = codigos_delitos_novigentes['TIPOLOGIA MATERIA'][item]\n else:\n tipologia_delito = tipologia\n \n delitos_no_vigentes.append([codigos_delitos_novigentes['COD. MATERIA'][item],\n codigos_delitos_novigentes['MATERIA'][item].rstrip(),\n tipologia_delito,'NO VIGENTE'])\n ",
"_____no_output_____"
],
[
"df_delitos_no_vigentes = pd.DataFrame(delitos_no_vigentes, columns = ['COD. MATERIA','MATERIA','TIPOLOGIA MATERIA','VIGENCIA MATERIA'])",
"_____no_output_____"
],
[
"df_delitos_no_vigentes",
"_____no_output_____"
],
[
"# Elimino tildes de las columnas object\n\ncols = df_delitos_no_vigentes.select_dtypes(include = [\"object\"]).columns\ndf_delitos_no_vigentes[cols] = df_delitos_no_vigentes[cols].progress_apply(clean_data.elimina_tilde)",
"100%|██████████| 3/3 [00:00<00:00, 257.09it/s]\n"
],
[
"df_delitos_no_vigentes['COD. MATERIA'] = df_delitos_no_vigentes['COD. MATERIA'].astype('int16')\ndf_delitos_no_vigentes.dtypes",
"_____no_output_____"
]
],
[
[
"# UNION DE AMBOS DATASET CON DELITOS VIGENTES Y NO VIGENTES",
"_____no_output_____"
]
],
[
[
"df_delitos = pd.concat([df_delitos_vigentes,df_delitos_no_vigentes])\n",
"_____no_output_____"
],
[
"df_delitos",
"_____no_output_____"
],
[
"df_delitos.info()",
"<class 'pandas.core.frame.DataFrame'>\nInt64Index: 542 entries, 0 to 101\nData columns (total 4 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 COD. MATERIA 542 non-null int16 \n 1 MATERIA 542 non-null object\n 2 TIPOLOGIA MATERIA 542 non-null object\n 3 VIGENCIA MATERIA 542 non-null object\ndtypes: int16(1), object(3)\nmemory usage: 18.0+ KB\n"
],
[
"df_delitos.sort_values(\"COD. MATERIA\")",
"_____no_output_____"
]
],
[
[
"## Guardamos DF como to_feather",
"_____no_output_____"
]
],
[
[
"# Reset el index para realizar feather\n\ndf_delitos.reset_index(inplace = True)\n\n\n# Guardamos dataset como archivo feather\n\ndf_delitos.to_feather('../data/processed/Delitos_feather')",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
cbf0c8e150d6c911b9f139d60eedded4c464140a
| 6,840 |
ipynb
|
Jupyter Notebook
|
lecture_source/practical_python/class_basic.ipynb
|
nockchun/rsnet
|
24abf97265d45f12fef0267db93dcda309806057
|
[
"Apache-2.0"
] | 1 |
2021-02-22T14:07:45.000Z
|
2021-02-22T14:07:45.000Z
|
lecture_source/practical_python/class_basic.ipynb
|
nockchun/rsnet
|
24abf97265d45f12fef0267db93dcda309806057
|
[
"Apache-2.0"
] | null | null | null |
lecture_source/practical_python/class_basic.ipynb
|
nockchun/rsnet
|
24abf97265d45f12fef0267db93dcda309806057
|
[
"Apache-2.0"
] | 7 |
2020-12-13T15:22:38.000Z
|
2022-03-28T11:24:51.000Z
| 17.952756 | 69 | 0.444883 |
[
[
[
"<div class=\"alert alert-block alert-success\">\n <b><center>Class Basic</center></b>\n</div>",
"_____no_output_____"
],
[
"# Configure Environment",
"_____no_output_____"
],
[
"# Class",
"_____no_output_____"
]
],
[
[
"class Basic():\n def __init__(self):\n self.data = []\n \n def getData(self):\n return self.data\n \n def addData(self, val):\n self.data.append(val)\n \n def addDatas(self, val):\n self.addData(val)\n return self",
"_____no_output_____"
],
[
"b = Basic()",
"_____no_output_____"
],
[
"b.getData()",
"_____no_output_____"
],
[
"b.addData(3)\nb.getData()",
"_____no_output_____"
],
[
"b.addData(6)\nb.getData()",
"_____no_output_____"
],
[
"b.addDatas(4).addData(5)\nb.getData()",
"_____no_output_____"
],
[
"b",
"_____no_output_____"
]
],
[
[
"# Special Method",
"_____no_output_____"
]
],
[
[
"class Demo():\n def __init__(self, name):\n self.name = name\n self.data = [1, 2, 3]\n \n def myFunc(self, name):\n return f\"myFunc: {name}\"\n \n def __repr__(self):\n return f\"name is {self.name}\"\n \n def __call__(self, val):\n print(f\"__call__: {val}\")\n \n def __add__(self, other):\n return f\"me is {self.name}, other is {other.name}\"\n \n def __len__(self):\n return len(self.data)\n \n def __getitem__(self, position):\n return f\"{self.name}_{self.data[position]}\"",
"_____no_output_____"
],
[
"a = Demo(\"home\")\nb = Demo(\"school\")",
"_____no_output_____"
],
[
"a",
"_____no_output_____"
],
[
"a.myFunc(\"good\")",
"_____no_output_____"
],
[
"a(\"good\")",
"__call__: good\n"
],
[
"a+b",
"_____no_output_____"
],
[
"for item in a:\n print(item)",
"home_1\nhome_2\nhome_3\n"
],
[
"a[1]",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbf0ce0350c915740462178fb13a35698e93b4ff
| 138,344 |
ipynb
|
Jupyter Notebook
|
notebooks/Bayesian updating of pulldown interactions.ipynb
|
gngdb/opencast-bio
|
9fb110076295aafa696a9f8b5070b8d93c6400ce
|
[
"MIT"
] | 2 |
2016-02-24T20:44:39.000Z
|
2020-07-06T02:44:38.000Z
|
notebooks/Bayesian updating of pulldown interactions.ipynb
|
gngdb/opencast-bio
|
9fb110076295aafa696a9f8b5070b8d93c6400ce
|
[
"MIT"
] | null | null | null |
notebooks/Bayesian updating of pulldown interactions.ipynb
|
gngdb/opencast-bio
|
9fb110076295aafa696a9f8b5070b8d93c6400ce
|
[
"MIT"
] | null | null | null | 157.926941 | 21,841 | 0.872947 |
[
[
[
"empty"
]
]
] |
[
"empty"
] |
[
[
"empty"
]
] |
cbf0ce46e6be5f48763060e3c8f3a794911ee193
| 40,922 |
ipynb
|
Jupyter Notebook
|
Machine-Learning/Part 1 - Data Preprocessing/data_preprocessing.ipynb
|
SwattyX/Machine-Learning-A-Z-hands-on-Python-And-R-in-data-Science
|
dff869166dc9c9b3f64b5daa39280698612a5049
|
[
"MIT"
] | null | null | null |
Machine-Learning/Part 1 - Data Preprocessing/data_preprocessing.ipynb
|
SwattyX/Machine-Learning-A-Z-hands-on-Python-And-R-in-data-Science
|
dff869166dc9c9b3f64b5daa39280698612a5049
|
[
"MIT"
] | null | null | null |
Machine-Learning/Part 1 - Data Preprocessing/data_preprocessing.ipynb
|
SwattyX/Machine-Learning-A-Z-hands-on-Python-And-R-in-data-Science
|
dff869166dc9c9b3f64b5daa39280698612a5049
|
[
"MIT"
] | null | null | null | 30.584454 | 147 | 0.369435 |
[
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\nimport pandas as pd",
"_____no_output_____"
],
[
"name='randolph'\nname",
"_____no_output_____"
],
[
"name[:-5]",
"_____no_output_____"
],
[
"data = pd.read_excel('climate_change.xls')\ndata.head()",
"_____no_output_____"
],
[
"data.dtypes",
"_____no_output_____"
],
[
"data['Country code'].isnull().value_counts()",
"_____no_output_____"
],
[
"data['Country name'] = data['Country name'].astype(str)",
"_____no_output_____"
],
[
"data = data.loc[data['Country name'] == 'Panama',:]\ndata.head()",
"_____no_output_____"
],
[
"data['Series name'].value_counts()",
"_____no_output_____"
],
[
"df = pd.DataFrame()",
"_____no_output_____"
],
[
"#data.melt(id_vars=[\"GDP ($)\", \"Population\", \"Urban population\", \"Nationally terrestrial protected areas (% of total land area)\"],\n# var_name=\"Date\",\n# value_name=\"Value\")\n",
"_____no_output_____"
],
[
"df = data.loc[data['Series name'] == 'GDP ($)', :]\ndf.head()",
"_____no_output_____"
],
[
"#df.rename(columns={\"Series name\": \"GDP ($)\"})\ndf = df.apply(lambda x: 1 if x['Series name'] == 'GDP ($)' else 0, axis= 1)",
"_____no_output_____"
],
[
"dataset = pd.read_csv(\"Data.csv\")\ndataset.head()",
"_____no_output_____"
],
[
"X = dataset.iloc[:, :-1].values\nX",
"_____no_output_____"
],
[
"Y = dataset.iloc[:, -1]\nY",
"_____no_output_____"
],
[
"Y.dtypes",
"_____no_output_____"
],
[
"from sklearn.impute import SimpleImputer\nimputer = SimpleImputer(missing_values=np.nan, strategy='mean')\nimputer.fit(X[:,1:3])\nX[:,1:3] = imputer.transform(X[:,1:3])\nprint(X)",
"[['France' 44.0 72000.0]\n ['Spain' 27.0 48000.0]\n ['Germany' 30.0 54000.0]\n ['Spain' 38.0 61000.0]\n ['Germany' 40.0 63777.77777777778]\n ['France' 35.0 58000.0]\n ['Spain' 38.77777777777778 52000.0]\n ['France' 48.0 79000.0]\n ['Germany' 50.0 83000.0]\n ['France' 37.0 67000.0]]\n"
],
[
"print(X)",
"[['France' 44.0 72000.0]\n ['Spain' 27.0 48000.0]\n ['Germany' 30.0 54000.0]\n ['Spain' 38.0 61000.0]\n ['Germany' 40.0 63777.77777777778]\n ['France' 35.0 58000.0]\n ['Spain' 38.77777777777778 52000.0]\n ['France' 48.0 79000.0]\n ['Germany' 50.0 83000.0]\n ['France' 37.0 67000.0]]\n"
],
[
"from sklearn.compose import ColumnTransformer\n#OneHotEncoder is used for several categorical data\nfrom sklearn.preprocessing import OneHotEncoder\nct = ColumnTransformer(transformers=[('encoder', OneHotEncoder(), [0])], remainder='passthrough')\nX = np.array(ct.fit_transform(X))\n",
"_____no_output_____"
],
[
"print(X)",
"[[1.0 0.0 0.0 44.0 72000.0]\n [0.0 0.0 1.0 27.0 48000.0]\n [0.0 1.0 0.0 30.0 54000.0]\n [0.0 0.0 1.0 38.0 61000.0]\n [0.0 1.0 0.0 40.0 63777.77777777778]\n [1.0 0.0 0.0 35.0 58000.0]\n [0.0 0.0 1.0 38.77777777777778 52000.0]\n [1.0 0.0 0.0 48.0 79000.0]\n [0.0 1.0 0.0 50.0 83000.0]\n [1.0 0.0 0.0 37.0 67000.0]]\n"
],
[
"#LabelEncoder is used for two categorical data which can be directly encoded to 0 and 1\nfrom sklearn.preprocessing import LabelEncoder\nle = LabelEncoder()\nY = le.fit_transform(Y)",
"_____no_output_____"
],
[
"print(Y)",
"[0 1 0 0 1 1 0 1 0 1]\n"
],
[
"from sklearn.model_selection import train_test_split\nX_train, X_test, Y_train, Y_test = train_test_split(X,Y,test_size=0.2, random_state=1)",
"_____no_output_____"
],
[
"print(X_train)",
"[[0.0 0.0 1.0 38.77777777777778 52000.0]\n [0.0 1.0 0.0 40.0 63777.77777777778]\n [1.0 0.0 0.0 44.0 72000.0]\n [0.0 0.0 1.0 38.0 61000.0]\n [0.0 0.0 1.0 27.0 48000.0]\n [1.0 0.0 0.0 48.0 79000.0]\n [0.0 1.0 0.0 50.0 83000.0]\n [1.0 0.0 0.0 35.0 58000.0]]\n"
],
[
"print(X_test)",
"[[0.0 1.0 0.0 30.0 54000.0]\n [1.0 0.0 0.0 37.0 67000.0]]\n"
],
[
"print(Y_train)",
"[0 1 0 0 1 1 0 1]\n"
],
[
"print(Y_test)",
"[0 1]\n"
],
[
"from sklearn.preprocessing import StandardScaler\nsc = StandardScaler()\nX_train[:, 3:] = sc.fit_transform(X_train[:, 3:])\nX_test[:, 3:] = sc.transform(X_test[:, 3:])",
"_____no_output_____"
],
[
"print(X_train)",
"[[0.0 0.0 1.0 -0.19159184384578545 -1.0781259408412425]\n [0.0 1.0 0.0 -0.014117293757057777 -0.07013167641635372]\n [1.0 0.0 0.0 0.566708506533324 0.633562432710455]\n [0.0 0.0 1.0 -0.30453019390224867 -0.30786617274297867]\n [0.0 0.0 1.0 -1.9018011447007988 -1.420463615551582]\n [1.0 0.0 0.0 1.1475343068237058 1.232653363453549]\n [0.0 1.0 0.0 1.4379472069688968 1.5749910381638885]\n [1.0 0.0 0.0 -0.7401495441200351 -0.5646194287757332]]\n"
],
[
"print(X_test)",
"[[0.0 1.0 0.0 -1.4661817944830124 -0.9069571034860727]\n [1.0 0.0 0.0 -0.44973664397484414 0.2056403393225306]]\n"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbf0cfff0f33a9de835d2b67827407c2c1259adb
| 11,646 |
ipynb
|
Jupyter Notebook
|
lab4/lab4_parte1.ipynb
|
eirasf/GCED-AA2
|
123817fa5433779131f5b1011e63752b372e38f3
|
[
"MIT"
] | 2 |
2021-09-10T09:25:02.000Z
|
2021-09-18T21:38:02.000Z
|
lab4/lab4_parte1.ipynb
|
eirasf/GCED-AA2
|
123817fa5433779131f5b1011e63752b372e38f3
|
[
"MIT"
] | null | null | null |
lab4/lab4_parte1.ipynb
|
eirasf/GCED-AA2
|
123817fa5433779131f5b1011e63752b372e38f3
|
[
"MIT"
] | null | null | null | 34.557864 | 506 | 0.562081 |
[
[
[
"[](https://colab.research.google.com/github/eirasf/GCED-AA2/blob/main/lab4/lab4_parte1.ipynb)\n# Práctica 4: Redes neuronales usando Keras con Regularización\n## Parte 1. Early Stopping\n### Overfitting\nEl problema del sobreajuste (*overfitting*) consiste en que la solución aprendida se ajusta muy bien a los datos de entrenamiento, pero no generaliza adecuadamente ante la aparición de nuevos datos. \n\n# Regularización\n\nUna vez diagnosticado el sobreajuste, es hora de probar diferentes técnicas que intenten reducir la varianza, sin incrementar demasiado el sesgo y, con ello, el modelo generaliza mejor. Las técnicas de regularización que vamos a ver en este laboratorio son:\n1. *Early stopping*. Detiene el entrenamiento de la red cuando aumenta el error.\n1. Penalización basada\ten\tla\tnorma\tde\tlos\tparámetros (tanto norma L1 como L2). \n1. *Dropout*. Ampliamente utilizada en aprendizaje profundo, \"desactiva\" algunas neuronas para evitar el sobreajuste.\n\nEn esta primera parte del Laboratorio 4 nos centraremos en **Early Stopping**\n\n\n\n\n",
"_____no_output_____"
],
[
"## Pre-requisitos. Instalar paquetes\n\nPara la primera parte de este Laboratorio 4 necesitaremos TensorFlow, TensorFlow-Datasets y otros paquetes para inicializar la semilla y poder reproducir los resultados",
"_____no_output_____"
]
],
[
[
"import tensorflow as tf\nimport tensorflow_datasets as tfds\nimport os\nimport numpy as np\nimport random\n\n#Fijamos la semilla para poder reproducir los resultados\nseed=1234\nos.environ['PYTHONHASHSEED']=str(seed)\ntf.random.set_seed(seed)\nnp.random.seed(seed)\nrandom.seed(seed)\n",
"_____no_output_____"
]
],
[
[
"Además, cargamos también APIs que vamos a emplear para que el código quede más legible",
"_____no_output_____"
]
],
[
[
"#API de Keras, modelo Sequential y las capas que vamos a usar en nuestro modelo\nfrom tensorflow import keras\nfrom keras.models import Sequential\nfrom keras.layers import InputLayer\nfrom keras.layers import Dense \n#Para mostrar gráficas\nfrom matplotlib import pyplot\n\n#Necesario para el EarlyStopping\nfrom keras.callbacks import EarlyStopping\n",
"_____no_output_____"
]
],
[
[
"## Cargamos el conjunto de datos\n\nDe nuevo, seguimos empleando el conjunto *german_credit_numeric* ya empleado en los laboratorios anteriores, aunque esta vez lo dividimos para tener un subconjunto de entrenamiento, otro de validación (que nos servirá para detener el entrenamiento) y otro de test para evaluar el rendimiento del modelo.\n",
"_____no_output_____"
]
],
[
[
"# Cargamos el conjunto de datos\nds_train = tfds.load('german_credit_numeric', split='train[:40%]', as_supervised=True).batch(128)\nds_val = tfds.load('german_credit_numeric', split='train[40%:50%]', as_supervised=True).batch(128)\nds_test = tfds.load('german_credit_numeric', split='train[50%:]', as_supervised=True).batch(128)",
"_____no_output_____"
]
],
[
[
"También vamos a establecer la función de pérdida, el algoritmo que vamos a emplear para el entrenamiento y la métrica que nos servirá para evaluar el rendimiento del modelo entrenado.",
"_____no_output_____"
]
],
[
[
"#Indicamos la función de perdida, el algoritmo de optimización y la métrica para evaluar el rendimiento \nfn_perdida = tf.keras.losses.BinaryCrossentropy()\noptimizador = tf.keras.optimizers.Adam(0.001)\nmetrica = tf.keras.metrics.AUC()",
"_____no_output_____"
]
],
[
[
"## Creamos un modelo *Sequential*\nCreamos un modelo *Sequential* tal y como se ha hecho en el Laboratorio 3. Parte 2.",
"_____no_output_____"
]
],
[
[
"tamano_entrada = 24\nh0_size = 20\nh1_size = 10\nh2_size = 5\n#TODO - define el modelo Sequential\nmodel = ...\n#TODO - incluye la capa de entrada y las 4 capas Dense del modelo\n......\n\n#Construimos el modelo y mostramos \nmodel.build()\nprint(model.summary())",
"_____no_output_____"
]
],
[
[
"Completar el método *compile*.",
"_____no_output_____"
]
],
[
[
"#TODO - indicar los parametros del método compile\nmodel.compile(loss=fn_perdida,\n optimizer=optimizador,\n metrics=[metrica])",
"_____no_output_____"
]
],
[
[
"Hacemos una llamada al método *fit* usando el conjunto de entrenamiento como entrada, indicando el número de epochs y, además, incluyendo el argumento *validation_data* que permite usar un subconjunto de datos para validar. Las diferencias entre entrenamiento y validación se pueden apreciar en el gráfico.\n\n**NOTA**: Observad las diferencias de resultado entre entrenamiento, validación y test.",
"_____no_output_____"
]
],
[
[
"#Establecemos el número de epochs\nnum_epochs = 700\n\n# Guardamos los pesos antes de entrenar, para poder resetear el modelo posteriormente y hacer comparativas.\npesos_preentrenamiento = model.get_weights()\n\n#TODO - entrenar el modelo usando como entradas el conjunto de entrenamiento, \n#indicando el número de epochs y el conjunto de validación\nhistory = model.fit(....)\n\n# plot training history\npyplot.plot(history.history['loss'], label='train')\npyplot.plot(history.history['val_loss'], label='val')\npyplot.legend()\npyplot.show()",
"_____no_output_____"
],
[
"#TODO - llamar a evaluate usando el conjunto de test, guardando el resultado\nresult = model.evaluate(.....) \nprint(model.metrics_names)\nprint(result)",
"_____no_output_____"
]
],
[
[
"## Usando Early Stopping en el entrenamiento\n\nKeras nos facilita un *Callback* para realizar la parada temprana (*keras.callbacks.EarlyStopping*). De este modo, podemos parar el entrenamiento cuando una determinada medida (especificada en el argumento *monitor*) empeore su rendimiento (el argumento *mode* nos dirá si se espera que dicha medida se minimice, *min*, o maximice, *max*). Opcionalmente, el usuario puede proporcionar el argumento *patience* para especificar cuantas *epochs* debe esperar el entrenamiento antes de detenerse.\n\n**TO-DO**: Realizar varias veces el entrenamiento, cambiando los distintos parámetros para ver las diferencias en el aprendizaje. ¿Se para siempre en el mismo *epoch*? Comprobar el rendimiento en el conjunto de test.",
"_____no_output_____"
]
],
[
[
"# simple early stopping\n#TODO- indica la medida a monitorizar, el modo y la paciencia\nes = EarlyStopping(\n monitor=....\n mode=...\n patience=...\n)\n\n# Antes de entrenar, olvidamos el entrenamiento anterior restaurando los pesos iniciales\nmodel.set_weights(pesos_preentrenamiento)\n\n#TODO - entrenar el modelo usando como entradas el conjunto de entrenamiento, \n#indicando el número de epochs, el conjunto de validación y la callback para el EarlyStopping\nhistory = model.fit(...., callbacks=[es])\n\n# plot training history\npyplot.plot(history.history['loss'], label='train')\npyplot.plot(history.history['val_loss'], label='val')\npyplot.legend()\npyplot.show()",
"_____no_output_____"
]
],
[
[
"Evaluación sobre el conjunto de test (no usado para el entrenamiento).",
"_____no_output_____"
]
],
[
[
"#TODO - llamar a evaluate usando el conjunto de test, guardando el resultado\nresult = model.evaluate(....) \nprint(model.metrics_names)\nprint(result)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cbf0e88d635a102ec226c97c49720e32f43003cb
| 9,003 |
ipynb
|
Jupyter Notebook
|
Rafay notes/Samsung Course/Chapter 6/Quiz/.ipynb_checkpoints/MOhamamd Abdul Rafay BSE183009-problem_0501-checkpoint.ipynb
|
rafay99-epic/Ssmsunng-Innovation-Campus-Notes
|
19a2dfd125957d5a3d3458636d91747b48267689
|
[
"MIT"
] | null | null | null |
Rafay notes/Samsung Course/Chapter 6/Quiz/.ipynb_checkpoints/MOhamamd Abdul Rafay BSE183009-problem_0501-checkpoint.ipynb
|
rafay99-epic/Ssmsunng-Innovation-Campus-Notes
|
19a2dfd125957d5a3d3458636d91747b48267689
|
[
"MIT"
] | null | null | null |
Rafay notes/Samsung Course/Chapter 6/Quiz/.ipynb_checkpoints/MOhamamd Abdul Rafay BSE183009-problem_0501-checkpoint.ipynb
|
rafay99-epic/Ssmsunng-Innovation-Campus-Notes
|
19a2dfd125957d5a3d3458636d91747b48267689
|
[
"MIT"
] | null | null | null | 42.267606 | 1,390 | 0.610685 |
[
[
[
"## Quiz #0501",
"_____no_output_____"
],
[
"### \"Logistic Regression and Gradient Descent Algorithm\"",
"_____no_output_____"
],
[
"#### Answer the following questions by providing Python code:\n#### Objectives:\n- Code a logistic regression class using only the NumPy library.\n- Implement in Python the Sigmoid function.\n- Implement in Python the Gradient of the logarithmic likelihood.\n- Implement in Python the Gradient Descent Algorithm.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nfrom sklearn.datasets import load_breast_cancer\nfrom sklearn.model_selection import train_test_split",
"_____no_output_____"
]
],
[
[
"#### Read in data:",
"_____no_output_____"
]
],
[
[
"# Load data.\ndata = load_breast_cancer()\n# Explanatory variables.\nX = data['data']\n# Relabel such that 0 = 'benign' and 1 = malignant.\nY = 1 - data['target']",
"_____no_output_____"
],
[
"# Split the dataset into training and testing.\nX_train, X_test, Y_train, Y_test = train_test_split(X, Y, test_size=0.4, random_state=1234)",
"_____no_output_____"
]
],
[
[
"1). Define the 'sigmoid' and 'gradient' functions to produce the output shown below:",
"_____no_output_____"
]
],
[
[
"def sigmoid(x):\n x = np.linspace(-10, 10, 100)\n z = 1/(1 + np.exp(-x))\n return z\n\ndef gradient(X, Y, beta):\n size = X.shape[0]\n weight = parameters[\"data\"] \n bias = parameters[\"target\"]\n sigma = sigmoid(X)\n loss = -1/size * np.sum(Y * np.log(sigma)) + (1 - Y) * np.log(1-sigma)\n dW = 1/size * np.dot(x.T, (sigma - Y))\n db = 1/size * np.sum(sigma - Y)\n \n X[\"data\"] = weight\n X[\"target\"] = bias\n return parameters",
"_____no_output_____"
]
],
[
[
"2). Define the 'LogisticRegression' class to produce the output shown below:",
"_____no_output_____"
]
],
[
[
"class LogisticRegression:\n def __init__(self, learn_rate):\n self.lr = learning_rate\n def train(self, input_X, input_Y, n_epochs):\n parameters_out = gradient(input_X, input_Y, n_epochs)\n return parameters_out\n \n #def query(self, input_X, prob=True, cutoff=0.5):\n # <Your code goes in here>\n ",
"_____no_output_____"
]
],
[
[
"#### Sample run:",
"_____no_output_____"
]
],
[
[
"# Hyperparameter for the learner.\nlearning_rate = 0.001",
"_____no_output_____"
],
[
"# Train and predict.\nLR = LogisticRegression(learning_rate)\nLR.train(X_train, Y_train, 2000)\nY_pred = LR.query(X_test,prob=False,cutoff=0.5)",
"_____no_output_____"
],
[
"# Display the accuracy.\nacc = (Y_pred == Y_test.reshape(-1,1)).mean()\nprint('Accuracy : {}'.format(np.round(acc,3)))",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
cbf0eb9c9c183810bcdb87906fcc52b3909ebbb4
| 52,669 |
ipynb
|
Jupyter Notebook
|
notebook/milestone1.ipynb
|
williamxu7/DSCI_525_Group13_Rainfall
|
ad34c065463ac95697a01fc4bef10b97b74e1d89
|
[
"MIT"
] | 1 |
2021-04-04T00:13:08.000Z
|
2021-04-04T00:13:08.000Z
|
notebook/milestone1.ipynb
|
williamxu7/DSCI_525_Group13_Rainfall
|
ad34c065463ac95697a01fc4bef10b97b74e1d89
|
[
"MIT"
] | 17 |
2021-03-30T22:46:12.000Z
|
2021-05-04T17:48:31.000Z
|
notebook/milestone1.ipynb
|
williamxu7/DSCI_525_Group13_Rainfall
|
ad34c065463ac95697a01fc4bef10b97b74e1d89
|
[
"MIT"
] | 3 |
2021-04-02T19:42:46.000Z
|
2021-06-28T05:23:34.000Z
| 30.131007 | 450 | 0.465606 |
[
[
[
"# DSCI 525: Web and Cloud Computing\n\n## Milestone 1: Tackling Big Data on Computer\n\n### Group 13\nAuthors: Ivy Zhang, Mike Lynch, Selma Duric, William Xu",
"_____no_output_____"
],
[
"## Table of contents\n\n- [Download the data](#1)\n- [Combining data CSVs](#2)\n- [Load the combined CSV to memory and perform a simple EDA](#3)\n- [Perform a simple EDA in R](#4)\n- [Reflection](#5)",
"_____no_output_____"
],
[
"### Imports",
"_____no_output_____"
]
],
[
[
"import re\nimport os\nimport glob\nimport zipfile\nimport requests\nfrom urllib.request import urlretrieve\nimport json\nimport pandas as pd\nimport numpy as np\nimport pyarrow.feather as feather\nfrom memory_profiler import memory_usage\nimport pyarrow.dataset as ds\nimport pyarrow as pa\nimport pyarrow.parquet as pq\nimport dask.dataframe as dd",
"_____no_output_____"
],
[
"%load_ext rpy2.ipython\n%load_ext memory_profiler",
"_____no_output_____"
]
],
[
[
"## 1. Download the data <a name=\"1\"></a>",
"_____no_output_____"
],
[
"1. Download the data from figshare to local computer using the figshare API.\n2. Extract the zip file programmatically.",
"_____no_output_____"
]
],
[
[
"# Attribution: DSCI 525 lecture notebook\n# Necessary metadata\narticle_id = 14096681 # unique identifier of the article on figshare\nurl = f\"https://api.figshare.com/v2/articles/{article_id}\"\nheaders = {\"Content-Type\": \"application/json\"}\noutput_directory = \"figsharerainfall/\"",
"_____no_output_____"
],
[
"response = requests.request(\"GET\", url, headers=headers)\ndata = json.loads(response.text)\nfiles = data[\"files\"] ",
"_____no_output_____"
],
[
"%%time\nfiles_to_dl = [\"data.zip\"] \nfor file in files:\n if file[\"name\"] in files_to_dl:\n os.makedirs(output_directory, exist_ok=True)\n urlretrieve(file[\"download_url\"], output_directory + file[\"name\"])",
"CPU times: user 6.12 s, sys: 6.19 s, total: 12.3 s\nWall time: 2min 3s\n"
],
[
"%%time\nwith zipfile.ZipFile(os.path.join(output_directory, \"data.zip\"), 'r') as f:\n f.extractall(output_directory)",
"CPU times: user 20.8 s, sys: 5.39 s, total: 26.2 s\nWall time: 33.7 s\n"
]
],
[
[
"## 2. Combining data CSVs <a name=\"2\"></a>",
"_____no_output_____"
],
[
"1. Use one of the following options to combine data CSVs into a single CSV (Pandas, Dask). **We used the option of Pandas**.\n2. When combining the csv files, we added extra column called \"model\" that identifies the model (we get this column populated from the file name eg: for file name \"SAM0-UNICON_daily_rainfall_NSW.csv\", the model name is SAM0-UNICON)\n3. Compare run times and memory usages of these options on different machines within the team, and summarize observations.",
"_____no_output_____"
]
],
[
[
"%%time\n%memit\n# Shows time that regular python takes to merge file\n# Join all data together\n## here we are using a normal python way of merging the data \n# use_cols = [\"time\", \"lat_min\", \"lat_max\", \"lon_min\",\"lon_max\",\"rain (mm/day)\"]\nfiles = glob.glob('figsharerainfall/*.csv')\ndf = pd.concat((pd.read_csv(file, index_col=0)\n .assign(model=re.findall(r'[^\\/]+(?=\\_d)', file)[0])\n for file in files)\n )\ndf.to_csv(\"figsharerainfall/combined_data.csv\")",
"peak memory: 171.12 MiB, increment: 0.05 MiB\nCPU times: user 7min 49s, sys: 38.1 s, total: 8min 27s\nWall time: 10min 21s\n"
],
[
"%%time\ndf = pd.read_csv(\"figsharerainfall/combined_data.csv\")",
"CPU times: user 1min 10s, sys: 25.9 s, total: 1min 36s\nWall time: 1min 54s\n"
],
[
"%%sh\ndu -sh figsharerainfall/combined_data.csv",
"5.6G\tfigsharerainfall/combined_data.csv\n"
],
[
"print(df.shape)",
"(62513863, 7)\n"
],
[
"df.head()",
"_____no_output_____"
]
],
[
[
"**Summary of run times and memory usages:**\n\n***William***\n- Combining files: \n - peak memory: 95.41 MiB, increment: 0.26 MiB\n - CPU times: user 7min 28s, sys: 31 s, total: 7min 59s\n - Wall time: 9min 17s\n- Reading the combined file:\n - Wall time: 1min 51s\n\n***Mike***\n- Combining files: \n - peak memory: 168.59 MiB, increment: 0.12 MiB\n - CPU times: user 3min 29s, sys: 5.09 s, total: 3min 34s\n - Wall time: 3min 34s\n- Reading the combined file:\n - Wall time: 37.1 s\n\n***Selma***\n- Combining files: \n - peak memory: 150.54 MiB, increment: 0.23 MiB\n - CPU times: user 6min 46s, sys: 23.1 s, total: 7min 9s\n - Wall time: 7min 29s\n- Reading the combined file:\n - Wall time: 1min 19s\n \n***Ivy***\n- Combining files: \n - peak memory: 156.23 MiB, increment: 0.00 MiB\n - CPU times: user 5min 14s, sys: 18.2 s, total: 5min 32s\n - Wall time: 5min 45s\n- Reading the combined file:\n - Wall time: 1min 30s",
"_____no_output_____"
],
[
"## 3. Load the combined CSV to memory and perform a simple EDA <a name=\"3\"></a>",
"_____no_output_____"
],
[
"### Establish a baseline for memory usage",
"_____no_output_____"
]
],
[
[
"# First load in the dataset using default settings for dtypes\ndf_eda = pd.read_csv(\"figsharerainfall/combined_data.csv\", parse_dates=True, index_col='time')\ndf_eda.head()",
"_____no_output_____"
],
[
"# As we can see below, dtypes are float64 and object\ndf_eda.dtypes",
"_____no_output_____"
],
[
"# Measure the memory usage when representing numbers using float64 dtype\nprint(f\"Memory usage with float64: {df_eda.memory_usage().sum() / 1e6:.2f} MB\")",
"Memory usage with float64: 3500.78 MB\n"
],
[
"%%time\n%memit\n\n# Now perform a simple EDA with pandas describe function\ndf_eda.describe()",
"peak memory: 599.40 MiB, increment: 0.54 MiB\nCPU times: user 17.5 s, sys: 14.6 s, total: 32.1 s\nWall time: 43.9 s\n"
]
],
[
[
"Baseline memory and time data:\n- Memory usage with float64: 3500.78 MB\n- peak memory: 698.22 MiB, increment: 0.35 MiB\n- CPU times: user 16.2 s, sys: 13.8 s, total: 30 s\n- Wall time: 36.5 s",
"_____no_output_____"
],
[
"### Effects of changing dtypes on memory usage",
"_____no_output_____"
]
],
[
[
"# Now load in the dataset using float32 dtype to represent numbers\ncolum_dtypes = {'lat_min': np.float32, 'lat_max': np.float32, 'lon_min': np.float32, 'lon_max': np.float32, 'rain (mm/day)': np.float32, 'model': str}\ndf_eda = pd.read_csv(\"figsharerainfall/combined_data.csv\",parse_dates=True, index_col='time', dtype=colum_dtypes)\ndf_eda.head()",
"_____no_output_____"
],
[
"# As we can see below, dtypes are float32 and object\ndf_eda.dtypes",
"_____no_output_____"
],
[
"print(f\"Memory usage with float32: {df_eda.memory_usage().sum() / 1e6:.2f} MB\")",
"Memory usage with float32: 2250.50 MB\n"
],
[
"%%time\n%memit\n\n# Now perform a simple EDA with pandas describe function\ndf_eda.describe()",
"peak memory: 609.06 MiB, increment: 0.36 MiB\nCPU times: user 11.3 s, sys: 5.72 s, total: 17 s\nWall time: 22.7 s\n"
]
],
[
[
"Time and memory data when using different dtypes:\n- Memory usage with float32: 2250.50 MB\n- peak memory: 609.06 MiB, increment: 0.36 MiB\n- CPU times: user 11.3 s, sys: 5.72 s, total: 17 s\n- Wall time: 22.7 s",
"_____no_output_____"
],
[
"### Effects of loading a smaller subset of columns on memory usage",
"_____no_output_____"
]
],
[
[
"# Now load only a subset of columns from the dataset\ndf_eda = pd.read_csv(\"figsharerainfall/combined_data.csv\",parse_dates=True, index_col='time', usecols=['time', 'lat_min', 'rain (mm/day)'])\ndf_eda.head()",
"_____no_output_____"
],
[
"# As we can see below, dtypes are float64 by default\ndf_eda.dtypes",
"_____no_output_____"
],
[
"print(f\"Memory usage with reduced number of columns: {df_eda.memory_usage().sum() / 1e6:.2f} MB\")",
"Memory usage with reduced number of columns: 1500.33 MB\n"
],
[
"%%time\n%memit\n\n# Now perform a simple EDA with pandas describe function\ndf_eda.describe()",
"peak memory: 340.50 MiB, increment: 0.40 MiB\nCPU times: user 7.13 s, sys: 5.6 s, total: 12.7 s\nWall time: 18.2 s\n"
]
],
[
[
"Time and memory data when using column subset:\n- Memory usage with reduced number of columns: 1500.33 MB\n- peak memory: 340.50 MiB, increment: 0.40 MiB\n- CPU times: user 7.13 s, sys: 5.6 s, total: 12.7 s\n- Wall time: 18.2 s",
"_____no_output_____"
],
[
"### Summary",
"_____no_output_____"
],
[
"#### Using float32 vs. baseline float64 dtype to perform a simple EDA:\n- The memory usage decreased from 3500.78 MB to 2250.50 MB when representing numbers using float32 instead of float64\n- When using the pandas describe function to perform a simple EDA, we found that the peak memory increased when using float32 dtype for the numerical columns. \n- The wall time taken to perform the EDA also decreased substantially to 22.7s from the baseline of 36.5s. \n\n#### Using a reduced number of columns compared to the baseline to perform a simple EDA:\n- The memory usage decreased from 3500.78 MB to 1500.33 MB when using a subset of columns from the dataset\n- When using the pandas describe function to perform a simple EDA, we found that the peak memory increased when using fewer columns. \n- The wall time taken to perform the EDA also decreased substantially to 18.2s from the baseline of 36.5s. ",
"_____no_output_____"
],
[
"## 4. Perform a simple EDA in R <a name=\"4\"></a>",
"_____no_output_____"
],
[
"We will transform our dataframe into different formats before loading into R.\n#### I. Default memory format + feather file format",
"_____no_output_____"
]
],
[
[
"%%time\nfeather.write_feather(df, \"figsharerainfall/combined_data.feather\")",
"CPU times: user 14.1 s, sys: 20.4 s, total: 34.4 s\nWall time: 36.6 s\n"
]
],
[
[
"#### II. dask + parquet file format",
"_____no_output_____"
]
],
[
[
"ddf = dd.read_csv(\"figsharerainfall/combined_data.csv\")",
"_____no_output_____"
],
[
"%%time\ndd.to_parquet(ddf, 'figsharerainfall/combined_data.parquet')",
"CPU times: user 2min 15s, sys: 38 s, total: 2min 53s\nWall time: 1min 39s\n"
]
],
[
[
"#### III. Arrow memory format + parquet file format",
"_____no_output_____"
]
],
[
[
"%%time\n%%memit\ndataset = ds.dataset(\"figsharerainfall/combined_data.csv\", format=\"csv\")\ntable = dataset.to_table()",
"peak memory: 1529.93 MiB, increment: 945.35 MiB\nCPU times: user 23.9 s, sys: 15.5 s, total: 39.4 s\nWall time: 37.3 s\n"
],
[
"%%time\npq.write_to_dataset(table, 'figsharerainfall/rainfall.parquet')",
"CPU times: user 14.4 s, sys: 7.15 s, total: 21.5 s\nWall time: 30.9 s\n"
]
],
[
[
"#### IV. Arrow memory format + feather file format",
"_____no_output_____"
]
],
[
[
"%%time\nfeather.write_feather(table, 'figsharerainfall/rainfall.feather')",
"CPU times: user 4.5 s, sys: 5.98 s, total: 10.5 s\nWall time: 7.91 s\n"
],
[
"%%sh\ndu -sh figsharerainfall/combined_data.csv\ndu -sh figsharerainfall/combined_data.parquet\ndu -sh figsharerainfall/rainfall.parquet\ndu -sh figsharerainfall/rainfall.feather",
"5.6G\tfigsharerainfall/combined_data.csv\n2.3G\tfigsharerainfall/combined_data.parquet\n544M\tfigsharerainfall/rainfall.parquet\n1.0G\tfigsharerainfall/rainfall.feather\n"
]
],
[
[
"### Transfer different formats of data from Python to R",
"_____no_output_____"
],
[
"It is usually not efficient to directly transfer Python dataframe to R due to serialization and deserialization involved in the process. Also, we observe Arrow memory format performs better than the default memory default. Thus, our next step is to further compare the performance of transferring Arrow-feather file and Arrow-parquet file to R.\n\n#### I. Read Arrow-parquet file to R",
"_____no_output_____"
],
[
"```python\n%%time\n%%R\nlibrary(arrow)\nstart_time <- Sys.time()\nr_table <- arrow::read_parquet(\"figsharerainfall/rainfall.parquet/e5a0076fe71f4bdead893e20a935897b.parquet\")\nprint(class(r_table))\nlibrary(dplyr)\nresult <- r_table %>% count(model)\nend_time <- Sys.time()\nprint(result)\nprint(end_time - start_time)\n```",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"*Note that the code above has been commented out to ensure the workbook is reproducible. Please check Reflection for more details.*",
"_____no_output_____"
],
[
"#### II. Read Arrow-feather file to R",
"_____no_output_____"
]
],
[
[
"%%time\n%%R\nlibrary(arrow)\nstart_time <- Sys.time()\nr_table <- arrow::read_feather(\"figsharerainfall/rainfall.feather\")\nprint(class(r_table))\nlibrary(dplyr)\nresult <- r_table %>% count(model)\nend_time <- Sys.time()\nprint(result)\nprint(end_time - start_time)",
"R[write to console]: \nAttaching package: ‘arrow’\n\n\nR[write to console]: The following object is masked from ‘package:utils’:\n\n timestamp\n\n\n"
]
],
[
[
"#### Summary of format selection\nBased on the data storage and processing time comparison from above, our preferred format among all is **parquet using Arrow package**. The file with this format takes much less space to store it. Also, it takes less time to write to this format and read it in R.",
"_____no_output_____"
],
[
"## Reflection <a name=\"5\"></a>",
"_____no_output_____"
],
[
"After some trial and error, all team members were individually able to successfully run the analysis from start to finish, however during the process we did experience some problems which included the following:\n- William had issue with `%load_ext rpy2.ipython` despite the successful environment installation on his MacOS. After many hours debugging, ry2 finally worked after specifying the python version in the course yml file. The solution is to add `python=3.8.6` to the 525.yml file under `dependencies:` and reinstall the environment. \n- Even though the file sizes were only 5 GB, we actually required 10 GB of disk space since we needed to download and unzip the data.\n- We got some confusing results by accidentally re-downloading the dataset without first deleting it since we were then combining twice as many files in the next step.\n- We noticed that parquet file name under the parquet folder is generated differently every time we re-run the workbook. If we keep current file name and re-run all cells, `arrow::read_parquet` function would return an error message indicating that the file \"e5a0076fe71f4bdead893e20a935897b.parquet\" does not exist in the directory. For reproducibility reason, we decided to comment out the code but record the output for further comparison.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
cbf0ed0a02311ca486543c0d62399fdf62b4de51
| 996,668 |
ipynb
|
Jupyter Notebook
|
Lessons&CourseWorks/2.AdvanceComputerVision&DeepLearning/Project-ImageCaptioning/3_Inference.ipynb
|
imsaksham-c/Udacity-ComputerVision-Nanodegree
|
e65494dfa70ff06c8e07a6c7c29d31277cb5453f
|
[
"MIT"
] | null | null | null |
Lessons&CourseWorks/2.AdvanceComputerVision&DeepLearning/Project-ImageCaptioning/3_Inference.ipynb
|
imsaksham-c/Udacity-ComputerVision-Nanodegree
|
e65494dfa70ff06c8e07a6c7c29d31277cb5453f
|
[
"MIT"
] | null | null | null |
Lessons&CourseWorks/2.AdvanceComputerVision&DeepLearning/Project-ImageCaptioning/3_Inference.ipynb
|
imsaksham-c/Udacity-ComputerVision-Nanodegree
|
e65494dfa70ff06c8e07a6c7c29d31277cb5453f
|
[
"MIT"
] | null | null | null | 1,818.737226 | 234,920 | 0.960849 |
[
[
[
"# Computer Vision Nanodegree\n\n## Project: Image Captioning\n\n---\n\nIn this notebook, you will use your trained model to generate captions for images in the test dataset.\n\nThis notebook **will be graded**. \n\nFeel free to use the links below to navigate the notebook:\n- [Step 1](#step1): Get Data Loader for Test Dataset \n- [Step 2](#step2): Load Trained Models\n- [Step 3](#step3): Finish the Sampler\n- [Step 4](#step4): Clean up Captions\n- [Step 5](#step5): Generate Predictions!",
"_____no_output_____"
],
[
"<a id='step1'></a>\n## Step 1: Get Data Loader for Test Dataset\n\nBefore running the code cell below, define the transform in `transform_test` that you would like to use to pre-process the test images. \n\nMake sure that the transform that you define here agrees with the transform that you used to pre-process the training images (in **2_Training.ipynb**). For instance, if you normalized the training images, you should also apply the same normalization procedure to the test images.",
"_____no_output_____"
]
],
[
[
"import sys\nsys.path.append('/opt/cocoapi/PythonAPI')\nfrom pycocotools.coco import COCO\nfrom data_loader import get_loader\nfrom torchvision import transforms\n\n# TODO #1: Define a transform to pre-process the testing images.\ntransform_test = transforms.Compose([ \n transforms.Resize(256), # smaller edge of image resized to 256\n transforms.RandomCrop(224), # get 224x224 crop from random location, \n transforms.ToTensor(), # convert the PIL Image to a tensor\n transforms.Normalize((0.485, 0.456, 0.406), # normalize image for pre-trained model\n (0.229, 0.224, 0.225))])\n\n#-#-#-# Do NOT modify the code below this line. #-#-#-#\n\n# Create the data loader.\ndata_loader = get_loader(transform=transform_test, \n mode='test')",
"Vocabulary successfully loaded from vocab.pkl file!\n"
]
],
[
[
"Run the code cell below to visualize an example test image, before pre-processing is applied.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\n%matplotlib inline\n\n# Obtain sample image before and after pre-processing.\norig_image, image = next(iter(data_loader))\n\n# Visualize sample image, before pre-processing.\nplt.imshow(np.squeeze(orig_image))\nplt.title('example image')\nplt.show()",
"_____no_output_____"
]
],
[
[
"<a id='step2'></a>\n## Step 2: Load Trained Models\n\nIn the next code cell we define a `device` that you will use move PyTorch tensors to GPU (if CUDA is available). Run this code cell before continuing.",
"_____no_output_____"
]
],
[
[
"import torch\n\ndevice = torch.device(\"cuda\" if torch.cuda.is_available() else \"cpu\")",
"_____no_output_____"
]
],
[
[
"Before running the code cell below, complete the following tasks.\n\n### Task #1\n\nIn the next code cell, you will load the trained encoder and decoder from the previous notebook (**2_Training.ipynb**). To accomplish this, you must specify the names of the saved encoder and decoder files in the `models/` folder (e.g., these names should be `encoder-5.pkl` and `decoder-5.pkl`, if you trained the model for 5 epochs and saved the weights after each epoch). \n\n### Task #2\n\nPlug in both the embedding size and the size of the hidden layer of the decoder corresponding to the selected pickle file in `decoder_file`.",
"_____no_output_____"
]
],
[
[
"# Watch for any changes in model.py, and re-load it automatically.\n% load_ext autoreload\n% autoreload 2\n\nimport os\nimport torch\nfrom model import EncoderCNN, DecoderRNN\n\n# TODO #2: Specify the saved models to load.\nencoder_file = 'encoder-3.pkl' \ndecoder_file = 'decoder-3.pkl'\n\n# TODO #3: Select appropriate values for the Python variables below.\nembed_size = 250\nhidden_size = 125\n\n# The size of the vocabulary.\nvocab_size = len(data_loader.dataset.vocab)\n\n# Initialize the encoder and decoder, and set each to inference mode.\nencoder = EncoderCNN(embed_size)\nencoder.eval()\ndecoder = DecoderRNN(embed_size, hidden_size, vocab_size)\ndecoder.eval()\n\n# Load the trained weights.\nencoder.load_state_dict(torch.load(os.path.join('./models', encoder_file)))\ndecoder.load_state_dict(torch.load(os.path.join('./models', decoder_file)))\n\n# Move models to GPU if CUDA is available.\nencoder.to(device)\ndecoder.to(device)",
"Downloading: \"https://download.pytorch.org/models/resnet50-19c8e357.pth\" to /root/.torch/models/resnet50-19c8e357.pth\n100%|██████████| 102502400/102502400 [00:01<00:00, 62335034.11it/s]\n/opt/conda/lib/python3.6/site-packages/torch/nn/modules/rnn.py:38: UserWarning: dropout option adds dropout after all but last recurrent layer, so non-zero dropout expects num_layers greater than 1, but got dropout=0.5 and num_layers=1\n \"num_layers={}\".format(dropout, num_layers))\n"
]
],
[
[
"<a id='step3'></a>\n## Step 3: Finish the Sampler\n\nBefore executing the next code cell, you must write the `sample` method in the `DecoderRNN` class in **model.py**. This method should accept as input a PyTorch tensor `features` containing the embedded input features corresponding to a single image.\n\nIt should return as output a Python list `output`, indicating the predicted sentence. `output[i]` is a nonnegative integer that identifies the predicted `i`-th token in the sentence. The correspondence between integers and tokens can be explored by examining either `data_loader.dataset.vocab.word2idx` (or `data_loader.dataset.vocab.idx2word`).\n\nAfter implementing the `sample` method, run the code cell below. If the cell returns an assertion error, then please follow the instructions to modify your code before proceeding. Do **not** modify the code in the cell below. ",
"_____no_output_____"
]
],
[
[
"# Move image Pytorch Tensor to GPU if CUDA is available.\nimage = image.to(device)\n\n# Obtain the embedded image features.\nfeatures = encoder(image).unsqueeze(1)\n\n# Pass the embedded image features through the model to get a predicted caption.\noutput = decoder.sample(features)\nprint('example output:', output)\n\nassert (type(output)==list), \"Output needs to be a Python list\" \nassert all([type(x)==int for x in output]), \"Output should be a list of integers.\" \nassert all([x in data_loader.dataset.vocab.idx2word for x in output]), \"Each entry in the output needs to correspond to an integer that indicates a token in the vocabulary.\"",
"example output: [0, 3, 33, 30, 39, 3, 33, 566, 21, 3, 30, 39, 46, 18, 1]\n"
]
],
[
[
"<a id='step4'></a>\n## Step 4: Clean up the Captions\n\nIn the code cell below, complete the `clean_sentence` function. It should take a list of integers (corresponding to the variable `output` in **Step 3**) as input and return the corresponding predicted sentence (as a single Python string). ",
"_____no_output_____"
]
],
[
[
"# TODO #4: Complete the function.\ndef clean_sentence(output):\n sentence = ''\n for x in output:\n sentence = sentence + ' ' + data_loader.dataset.vocab.idx2word[x]\n sentence = sentence.strip()\n return sentence",
"_____no_output_____"
]
],
[
[
"After completing the `clean_sentence` function above, run the code cell below. If the cell returns an assertion error, then please follow the instructions to modify your code before proceeding.",
"_____no_output_____"
]
],
[
[
"sentence = clean_sentence(output)\n\nprint('example sentence:', sentence)\n\nassert type(sentence)==str, 'Sentence needs to be a Python string!'",
"example sentence: <start> a street sign on a street corner with a sign on it . <end>\n"
]
],
[
[
"<a id='step5'></a>\n## Step 5: Generate Predictions!\n\nIn the code cell below, we have written a function (`get_prediction`) that you can use to use to loop over images in the test dataset and print your model's predicted caption.",
"_____no_output_____"
]
],
[
[
"def get_prediction():\n orig_image, image = next(iter(data_loader))\n plt.imshow(np.squeeze(orig_image))\n plt.title('Sample Image')\n plt.show()\n image = image.to(device)\n features = encoder(image).unsqueeze(1)\n output = decoder.sample(features) \n sentence = clean_sentence(output)\n print(sentence)",
"_____no_output_____"
]
],
[
[
"Run the code cell below (multiple times, if you like!) to test how this function works.",
"_____no_output_____"
]
],
[
[
"get_prediction()",
"_____no_output_____"
]
],
[
[
"As the last task in this project, you will loop over the images until you find four image-caption pairs of interest:\n- Two should include image-caption pairs that show instances when the model performed well.\n- Two should highlight image-caption pairs that highlight instances where the model did not perform well.\n\nUse the four code cells below to complete this task.",
"_____no_output_____"
],
[
"### The model performed well!\n\nUse the next two code cells to loop over captions. Save the notebook when you encounter two images with relatively accurate captions.",
"_____no_output_____"
]
],
[
[
"get_prediction()",
"_____no_output_____"
],
[
"get_prediction()",
"_____no_output_____"
]
],
[
[
"### The model could have performed better ...\n\nUse the next two code cells to loop over captions. Save the notebook when you encounter two images with relatively inaccurate captions.",
"_____no_output_____"
]
],
[
[
"get_prediction()",
"_____no_output_____"
],
[
"get_prediction()",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
cbf0ed7baf256a81201a575f6cf59b1ae6128fe2
| 495,149 |
ipynb
|
Jupyter Notebook
|
data-512-project/Data-512-Analysis.ipynb
|
tharunsikhinam/data-512
|
06dd84190b42b0ea04a03419dc8f82851604dc1f
|
[
"MIT"
] | null | null | null |
data-512-project/Data-512-Analysis.ipynb
|
tharunsikhinam/data-512
|
06dd84190b42b0ea04a03419dc8f82851604dc1f
|
[
"MIT"
] | null | null | null |
data-512-project/Data-512-Analysis.ipynb
|
tharunsikhinam/data-512
|
06dd84190b42b0ea04a03419dc8f82851604dc1f
|
[
"MIT"
] | null | null | null | 232.464319 | 140,844 | 0.90268 |
[
[
[
"# FIFA Transfer Market Analysis of European Football Leagues\n## Data 512 Project\n## Tharun Sikhinam",
"_____no_output_____"
],
[
"## I. Introduction",
"_____no_output_____"
],
[
"Transfer windows are as busy a time as any other in the football world. The game attracts so much attention that even when the ball is not rolling, the eyes of the entire world are on football. Transfers occur all year round and in every corner of the globe, but activity on the transfer market is typically at its peak during the summer and winter season.\n\nThe purpose of this project is to study of the economics in play from 1990 - 2019 in the transfer market of the top european leagues. It would be interesting to know how the trends have changed over the years. And by looking closely at the data, one hopes to uncover any hidden trends and explain the rise of the elite clubs in the European continent. With each clubs bringing in vast amounts of money through TV rights and other sources, it would be interesting to know how the clubs have put this money to use.",
"_____no_output_____"
],
[
"Understanding these macroeconomics can help us build a fair game and find areas of improvement. Through this analysis we want to address the wealth inequality of the top elite european clubs and how football has almost become a ‘money-game’. \n",
"_____no_output_____"
],
[
"## II. Background",
"_____no_output_____"
],
[
"The best resource for transfmer markets is the official FIFA TMS[1]. This link holds a summary reports for each transfer window in the european leagues dating back to 2013. The analysis concentrates on a particular transfer window and not the trends over the years. While the report talks about how each country is represented, the players are not classified on their Position (Striker, Midfield etc.,) Through this analysis we wish to answer questions such as \"Which country produces the best strikers?\"\n\nKPMG also publishes reports on clubs valuations and their spendings[2]. These reports also serve as useful guidelines so that I don't replicate any existing work. Most of these papers focus on a short duration of time, and this motivated me to explore the trends dating back to 1990 and correlating some of the trends found to real-life events.\n\nThe wealth gap between the top clubs and lower level clubs has been evident for a while [3], but it would be intersting to know what the exact numbers are. And how this wealth gap has been changing over the years. This can inform football policy makers to design better laws to safeguard the values and spirit of the game.",
"_____no_output_____"
],
[
"## III. Data",
"_____no_output_____"
],
[
"The data for this analysis is scraped off of transfermarkt.co.uk\nThe scraping was done in accordance with the Terms of Use in this link: https://www.transfermarkt.co.uk/intern/anb After reading the Terms of Use, it clearly states that\n\n```4.8. With the transmission of Content, Transfermarkt grants you the irrevocable, temporally and spatially unlimited and transferable right to reproduce, distribute, publish, issue, make publicly accessible, modify, translate and store the Content. This includes the right to edit, design, adapt to the file formats required for use, and to change and / or improve the presentation quality.```\n\nFurthermore looking at the https://www.transfermarkt.co.uk/robots.txt , the pages we want to scrap off of are not disallowed and are open to web crawlers and scrapers.\n\nThis dataset consists of all transfers in the European football market from 1991-2018. The data consists of a player name, club-out, club-in along with the transfer value grouped by different leagues. The dataset also consists of free transfers and loans as well. Most of the data is updated and contributed by the users of the website, and there might be few factual inaccuracies in the transfer figures stated.",
"_____no_output_____"
],
[
"<b>Ethical considerations:</b> Player names and ages have been removed from the dataset and will not be used as part of the analysis. This analysis doesn't aim to disrespect any player or country. The results of the analysis are aimed at the governing bodies of different leagues and sport lawmakers. Individual clubs are singled out over the analysis.",
"_____no_output_____"
],
[
"## IV. Research Questions",
"_____no_output_____"
],
[
"The following research questions are posed and answered methodically following best practices:",
"_____no_output_____"
],
[
"- <b>Q1. How has the transfer spending increased in the top 5 European Leagues?</b>\n- <b>Q2. Which clubs spent the most and received the most transfer fee on players from 2010-2018?</b>\n- <b> Q3. How have transfers fees moved betwen the leagues from 2010-2018? </b>\n- <b> Q4. How has the wealth gap changed amongst the European elite? </b>\n- <b> Q5. Investigating the spending trends of Manchester City, Chelsea and Paris Saint-Germain </b>\n- <b> Q6. Which country produces the best footballing talent? </b>\n",
"_____no_output_____"
],
[
"## V. Reproducibility",
"_____no_output_____"
],
[
"Each individual section of the analysis can be reproduced in its entirety. To reproduce the analysis, the following softwares have to be installed\n- R\n- python\n- pandas\n- numpy\n- matplotlib\n- sqlite3\n- javascript/codepen\n- Tableau\n\nFor questions 1-5 of the analysis sqlite3 is used as a database to query the results from. Sqlite3 is an in-memory/local database that is easy to setup and use. An installation of sqlite3 is recommended to reproduce all parts of the analysis. https://www.sqlite.org/download.html \nJavascript is used to create a sankey diagram for research question 3. The code for the javascript visualization can be found at https://codepen.io/tharunsikhinam/pen/QWwbzKj. \nTo create annotations and vertical stacked bar chart for question 5, Tableau was used. The Tableau .twb file is stored in the images directory of the project called q5Viz.twb. Load the file into tableau desktop/online to view the visualization. \nA copy of the database used for the analysis is also stored in the cleanData table, called as transfers.db. It holds all the tables necessary for the analysis and can be directly imported into sqlite3 or any other SQL database.\n",
"_____no_output_____"
],
[
"## VI. Analysis and Code",
"_____no_output_____"
],
[
"The analysis and code section of this document is broken down into 3 main parts\n1. Data Collection\n2. Data Pre-Processing\n3. Data Analysis & Results",
"_____no_output_____"
],
[
"## 1. Data Collection\n",
"_____no_output_____"
],
[
"#### Data is scraped off of transfermarket.co.uk in accordance with their Terms of Use. \n#### The R scripts to scrape data are stored under scrapingScripts/, run the following cell to generate raw data\n``` R < scrape.R --no-save ```",
"_____no_output_____"
],
[
"### 1.1 Run R scripts to scrape data",
"_____no_output_____"
]
],
[
[
"!R < ./scrapingScripts/scrape.R --no-save",
"\nR version 3.6.1 (2019-07-05) -- \"Action of the Toes\"\nCopyright (C) 2019 The R Foundation for Statistical Computing\nPlatform: x86_64-apple-darwin15.6.0 (64-bit)\n\nR is free software and comes with ABSOLUTELY NO WARRANTY.\nYou are welcome to redistribute it under certain conditions.\nType 'license()' or 'licence()' for distribution details.\n\n Natural language support but running in an English locale\n\nR is a collaborative project with many contributors.\nType 'contributors()' for more information and\n'citation()' on how to cite R or R packages in publications.\n\nType 'demo()' for some demos, 'help()' for on-line help, or\n'help.start()' for an HTML browser interface to help.\nType 'q()' to quit R.\n\n> \n> # setup -------------------------------------------------------------------\n> getwd()\n[1] \"/Users/tharun/workspace/data-512/data-512-project\"\n> # load packages / local functions\n> source(\"./scrapingScripts/00-setup.R\")\nLoading required package: pacman\n> \n> # seasons to scrape\n> seasons <- c(2018,2017,2016,2015,2014,2013,2012,2011,2010,2009,2008,2007,2006,2005,2004,2003,2002,2001,2000)\n> \n> # scrape ---------------------------------------------------\n> \n> # epl transfers\n> epl_transfers <- map_dfr(\n+ seasons, scrape_season_transfers,\n+ league_name = \"premier-league\", league_id = \"GB1\"\n+ )\n^C\n\nExecution halted\n"
]
],
[
[
"#### scrape.R can be modified to include more seasons and leagues, by default we are considering the years 1991-2018 and the European leagues from England, Spain, Italy, France, Germany, (Top 5) , Portugal and Netherlands",
"_____no_output_____"
],
[
"### 1.2 List first 5 files in data directory",
"_____no_output_____"
]
],
[
[
"from os import walk\nf = []\nfor (dirpath, dirnames, filenames) in walk(\"./rawData\"):\n f.extend(filenames)\n break\nf[0:5]",
"_____no_output_____"
]
],
[
[
"### 1.3 Combining data into one file",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport glob\n\ninteresting_files = glob.glob(\"./rawData/*.csv\") \n\n# Combine data frames inside rawData directory\ncombinedDf = pd.concat((pd.read_csv(f, header = 0) for f in interesting_files))",
"_____no_output_____"
],
[
"combinedDf.to_csv(\"./cleanData/allSeasons.csv\")\ncombinedDf.head(5)",
"_____no_output_____"
]
],
[
[
"## 2. Data Pre-Processing",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nraw = pd.read_csv(\"./cleanData/allSeasons.csv\")",
"_____no_output_____"
]
],
[
[
"#### Shape of the dataset",
"_____no_output_____"
]
],
[
[
"raw.shape",
"_____no_output_____"
]
],
[
[
"#### Columns",
"_____no_output_____"
]
],
[
[
"raw.columns",
"_____no_output_____"
]
],
[
[
"### 2.1 Dropping unnecessary columns",
"_____no_output_____"
]
],
[
[
"raw.head(5)\nraw = raw.drop([\"player_name\",\"age\",\"fee\"],axis=1)",
"_____no_output_____"
]
],
[
[
"### 2.2 Clean up nationality and position\n#### For players belonging to more than one nation, only the first country is used\n#### Positions are generalized into Forwards, Midfield, Defense, Wingers and Goalkeepers\n",
"_____no_output_____"
]
],
[
[
"# Consider only the first country that player belongs to\ndef cleanCountry(row):\n if isinstance(row[\"nat\"],str) :\n return row[\"nat\"].split(\";\")[0]\n else:\n return row[\"nat\"]\n \n# Replace positions with a general category \ndef cleanPosition(row):\n if isinstance(row[\"position\"],str) :\n if \"Midfield\" in row[\"position\"]:\n return \"Midfield\"\n elif row[\"position\"].find(\"Back\")>-1:\n return \"Defense\"\n elif \"Forward\" in row[\"position\"]:\n return \"Forward\"\n elif \"Striker\" in row[\"position\"]:\n return \"Forward\" \n elif \"Winger\" in row[\"position\"]:\n return \"Winger\"\n else:\n return row[\"position\"]\n else:\n return row[\"position\"]",
"_____no_output_____"
]
],
[
[
"<b>Clean up country \nClean up position and add new posNew column \nRemove incoming transfers (duplicates) \nRemove rows from Championship, we're only considering top flight leagues </b>",
"_____no_output_____"
]
],
[
[
"# Clean up country\nraw[\"nat\"] = raw.apply(cleanCountry,axis=1)\n# Clean up Position aand add new posNew column\nraw[\"posNew\"] = raw.apply(cleanPosition,axis=1)\n# Remove incoming transfers (duplicates)\nraw = raw[raw['transfer_movement']=='in']\n# Remove rows from Championship, we're only considering top flights leagues\nraw = raw[raw[\"league_name\"]!=\"Championship\"]",
"_____no_output_____"
]
],
[
[
"### 2.4 Replace NA's with zero's and drop all rows with zero's\n\n#### For the purposes of this analysis we will be only considering publicly stated transfers. Transfers involving loans, free transfers and end of contract signings are not considered.",
"_____no_output_____"
]
],
[
[
"# Replace 0's with NA's and drop rows \nraw = raw.replace(0,np.nan)\nraw = raw.dropna()\nleagues = raw.league_name.unique()",
"_____no_output_____"
],
[
"raw.to_csv(\"./cleanData/allSeasonsClean.csv\")\nraw.shape",
"_____no_output_____"
]
],
[
[
"### 2.5. Load data into SQLite",
"_____no_output_____"
],
[
"<b>For all future parts of this analysis a local installation of sqlite3 is recommended. Download sqlite3 from the following link and unzip the file https://www.sqlite.org/download.html. </b>",
"_____no_output_____"
],
[
"<b>\nWe will be using an in-memory database to run SQL queries against. If you would like to create a persistent copy of the database, replace :memory: with a path in the filesystem\n</b>",
"_____no_output_____"
]
],
[
[
"import sqlite3\nimport pandas as pd\n\n# Create a new database called transfers.db\nraw = pd.read_csv(\"./cleanData/allSeasonsClean.csv\")\ncnx = sqlite3.connect(\":memory:\")\n\n# create the dataframe from a query\nraw.to_sql(\"transfer\", cnx, if_exists='append', index=False)",
"/Users/tharun/anaconda3/lib/python3.6/site-packages/pandas/core/generic.py:2531: UserWarning: The spaces in these column names will not be changed. In pandas versions < 0.14, spaces were converted to underscores.\n dtype=dtype, method=method)\n"
]
],
[
[
"#### Verifying counts in database",
"_____no_output_____"
]
],
[
[
"df = pd.read_sql_query(\"SELECT count(*) FROM transfer limit 1\", cnx)\ndf.head()",
"_____no_output_____"
]
],
[
[
"#### Loading inflation dataset",
"_____no_output_____"
],
[
"<b>British inflation data is obtained from http://inflation.iamkate.com/ under the CC0 1.0 Universal License https://creativecommons.org/publicdomain/zero/1.0/legalcode \n \n<b>Multiplier mentions by how much the price at a particular year should be multiplied by to get an inflation adjusted value</b>",
"_____no_output_____"
]
],
[
[
"inflation = pd.read_csv(\"./cleanData/inflation.csv\")\ninflation.head()",
"_____no_output_____"
],
[
"inflation.to_sql(\"inflation\", cnx, if_exists='append', index=False)",
"_____no_output_____"
]
],
[
[
"### 2.6. Adjust transfer fee for inflation",
"_____no_output_____"
],
[
"<b>Adjust transfer fees for inflation and store this data as transfers table</b>",
"_____no_output_____"
]
],
[
[
"inflation_adjusted = pd.read_sql_query(\"select transfer.*,(fee_cleaned*multiplier) as `fee_inflation_adjusted` from transfer join inflation on transfer.year = inflation.year\",cnx)\ninflation_adjusted.to_sql(\"transfers\", cnx, if_exists='append', index=False)\n",
"_____no_output_____"
]
],
[
[
"## 3. Data Analysis",
"_____no_output_____"
],
[
"#### Verifying data in the table",
"_____no_output_____"
]
],
[
[
"pd.read_sql_query(\"SELECT count(*) FROM transfers limit 1\", cnx).head()",
"_____no_output_____"
]
],
[
[
"## Q1. How has the transfer spending increased in the top 5 European Leagues?",
"_____no_output_____"
],
[
"Transfer records are being broken every summer, with player values reaching upwards for £100M pounds. English Premier League in particular seems to be setting the trend for high transfer spending with some of the richest clubs belonging to England. Through this question we want to analyze how the transfer spending has increased amongst the top flights leagues in Europe. \nThe actual inflation from 1990's to 2018's is over 2.5% for the British Pound[4], while the inflation in transfer spending appears to be extreme. We want to be able to quantify this increase in value of players.",
"_____no_output_____"
],
[
"To observe the trends over the years we write a SQL query that sums over the transfer_fee spending for each league over the years. \nSQL query - sum(transfer_fee) and group by league_name and year\n",
"_____no_output_____"
]
],
[
[
"df = pd.read_sql_query(\"SELECT league_name,year,sum(fee_inflation_adjusted) as `total_spending in £million` from transfers where\\\n league_name in ('Serie A', '1 Bundesliga', 'Premier League', 'Ligue 1','Primera Division') \\\n group by league_name,year\",cnx)\ndf.head(5)",
"_____no_output_____"
]
],
[
[
"### Plot",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nplt.rcParams[\"figure.figsize\"] = (20,10)\nfor l in ['Serie A', '1 Bundesliga', 'Premier League', 'Ligue 1','Primera Division']:\n x = df[df[\"league_name\"]==l]\n plt.plot(x[\"year\"], x[\"total_spending in £million\"],label=l)\nplt.legend(prop={'size': 15})\nplt.xlabel(\"Years\",fontsize = 20)\nplt.ylabel(\"Transfer Spending in £Million \",fontsize = 20)\nplt.title(\"Transfer Spending in the top 5 European Leagues\",fontsize = 20)\n",
"_____no_output_____"
]
],
[
[
"### Results\n- Transfer spending has been steadily increasing for all European leagues since 1991\n- The spending gap between Premier League and the others shows a steep increase since the 2010.\n- The percentage change in median price of player from 1990’s to 2018 is 521%\n- The rise TV rights revenue can be an explanation for the rise in Premier League Transfer Spending (https://www.usatoday.com/story/sports/soccer/2019/05/21/english-premier-league-broadcast-rights-rise-to-12-billion/39500789/) ",
"_____no_output_____"
],
[
"## Q 2.1. Which clubs spent the most on players from 2010-2018?",
"_____no_output_____"
],
[
"After observing the trends in transfer spending to increase by the year, it would be interesting to know who are the top spenders and which clubs receive the most in transfer fees. You would expect the top spenders to also produce the best footballing talent. The results from this question paint a different picture",
"_____no_output_____"
],
[
"### SQL Query - sum(transfer_fee) and group by club_name and order by descending",
"_____no_output_____"
]
],
[
[
"topSpenders = pd.read_sql_query(\"SELECT club_name,league_name,sum(fee_inflation_adjusted) as `total_spending in £million` from transfers where\\\n year>2010 and year<2019\\\n group by club_name,league_name order by `total_spending in £million` desc\",cnx)\ntopSpenders = topSpenders.head(10)\ntopSpenders.head(10)",
"_____no_output_____"
]
],
[
[
"### Plot",
"_____no_output_____"
]
],
[
[
"plt.rcParams[\"figure.figsize\"] = (10,5)\nplt.barh(topSpenders[\"club_name\"],topSpenders[\"total_spending in £million\"])\nplt.xlabel(\"Transfer Spending in £Million\",fontsize = 15)\nplt.ylabel(\"Club Names\",fontsize = 15)\nplt.title(\"Clubs with highest transfer fee spending (2010-2018)\",fontsize = 15)\n",
"_____no_output_____"
]
],
[
[
"## Q 2.2: Which clubs receive the highest transfer fees for their players?",
"_____no_output_____"
],
[
"### SQL Query - sum(transfer_fee) over outgoing transfers and group by club_name and order by descending",
"_____no_output_____"
]
],
[
[
"highestReceivers = pd.read_sql_query(\"SELECT club_involved_name,sum(fee_inflation_adjusted) as `total_spending in £million` from transfers where\\\n year>=2010 and year<=2019\\\n group by club_involved_name order by `total_spending in £million` desc\",cnx).head(10)\nhighestReceivers = highestReceivers.head(10)\nhighestReceivers.head(10)",
"_____no_output_____"
]
],
[
[
"### Plot",
"_____no_output_____"
]
],
[
[
"plt.rcParams[\"figure.figsize\"] = (10,5)\nplt.barh(highestReceivers[\"club_involved_name\"],highestReceivers[\"total_spending in £million\"])\nplt.xlabel(\"Transfer Spending in £Million\",fontsize = 15)\nplt.ylabel(\"Club Names\",fontsize = 15)\nplt.title(\"Clubs receiving the highest transfer fee (2010-2018)\",fontsize = 15)",
"_____no_output_____"
]
],
[
[
"### Results",
"_____no_output_____"
],
[
"- The highest spending clubs are Manchester City, Chelsea and PSG. It would be interesting to know how the transfer trends have changed for these three clubs (explored in q5)\n- The club with highest transfer fees received is Monaco a relatively small club from Ligue 1 (France). We also notice Benfica another club from Liga Nos (Portugal) that receives high transfer fees. This goes to show that the club spending the highest doesn't necessarily sell their players for a high value\n- 4 of the top 10 highest spending clubs are from the English Premier League, which leads us into the next question. ",
"_____no_output_____"
],
[
"## Q3. How have the transfers flowed betwen the leagues from 2010-2018?",
"_____no_output_____"
],
[
"### We create a new temporary table 'movements' with the following columns: from_league, fee_spent and to_league",
"_____no_output_____"
]
],
[
[
"### To find out the movement of money across the leagues, we create a temporary table\ndf = pd.read_sql_query(\"select t1.league_name as 'to_league',fee_inflation_adjusted,b.league_name as 'from_league' from transfers t1 left outer join (select club_name,league_name from transfers \\\n where league_name!='Championship' and year>=2010 \\\n group by club_name,league_name) as b on t1.club_involved_name = b.club_name where t1.year>=2010\",cnx);\n\ndf.to_sql(\"movements\", cnx, if_exists='append', index=False)\n\n",
"_____no_output_____"
],
[
"movements = pd.read_sql_query(\"select to_league,sum(fee_inflation_adjusted) as fee_spent,from_league from movements where \\\n from_league is not null and to_league!=from_league group by to_league,from_league\",cnx)\nmovements.head(5)",
"_____no_output_____"
]
],
[
[
"### The above data is loaded into a sankey diagram written in javascript, the visualization is displayed below (only visible when using notebook). \n### Code for the visualization can be found at https://codepen.io/tharunsikhinam/pen/QWwbzKj\n### Link to the visualization https://codepen.io/tharunsikhinam/full/QWwbzKj",
"_____no_output_____"
]
],
[
[
"# Display the associated webpage in a new window\nimport IPython\nurl = 'https://codepen.io/tharunsikhinam/full/QWwbzKj'\niframe = '<iframe src=' + url + ' width=1000 height=700></iframe>'\nIPython.display.HTML(iframe)",
"_____no_output_____"
]
],
[
[
"### Results\n- The league importing the maximum talent is the English Premier League. This also explains the high transfer spending in that league.\n- The league exporting most talent is La Liga (Spain). The Spanish League exports players to nearly all leagues with the highest being to English Premier League. ",
"_____no_output_____"
],
[
"## Q 4: How has the wealth gap changed amongst the European elite?",
"_____no_output_____"
],
[
"### For this part of the analysis we will be only focussing on the top 5 European leagues",
"_____no_output_____"
]
],
[
[
"leagues = ['Primera Division', 'Serie A', '1 Bundesliga', 'Premier League','Ligue 1']",
"_____no_output_____"
]
],
[
[
"### To evaluate the wealth gap\n - We collect top 5 spenders in each league and calculate their transfer spending over the years\n - We collect the bottom 15 in the league and calculate their transfer spendings over the years\n - The above two measures are plotted on a graph",
"_____no_output_____"
]
],
[
[
"i=120;\nfor l in leagues:\n plt.figure()\n # Query to get spendings of top 5 clubs in a league\n df = pd.read_sql_query(\"select '\"+l+\"' ,year,sum(fee_inflation_adjusted) from transfers where club_name in\\\n(select club_name from transfers where league_name='\"+l+\"'\\\nand year>=2010 \\\ngroup by club_name order by sum(fee_cleaned) desc limit 5) and league_name='\"+l+\"'\\\nand year>=2010 group by year\",cnx)\n plt.plot(df[\"year\"], df[\"sum(fee_inflation_adjusted)\"],label=\"Top 5\")\n \n # Query to get spendings of bottom 15 clubs in a league\n df = pd.read_sql_query(\"select '\"+l+\"' ,year,sum(fee_inflation_adjusted) from transfers where club_name not in\\\n(select club_name from transfers where league_name='\"+l+\"'\\\nand year>=2010 \\\ngroup by club_name order by sum(fee_cleaned) desc limit 5) and league_name='\"+l+\"'\\\nand year>=2010 group by year\",cnx)\n plt.title(\"Transfer spendings in \" + l )\n plt.plot(df[\"year\"], df[\"sum(fee_inflation_adjusted)\"],label=\"Bottom 15\")\n plt.xlabel(\"Years\",fontsize = 10)\n plt.ylabel(\"Transfer Spending in £Million \",fontsize = 10)\n plt.legend()\n ",
"_____no_output_____"
]
],
[
[
"### Results\n- We observe a huge wealth inequality between the top and bottom clubs in Ligue 1, Serie A and Primera Division\n- The difference is not so significant for English Premier League and the Bundesliga\n- This is still a cause for concern since the top 5 clubs hold a disproportionate share of wealth in the top flight clubs\n- These top 5 clubs in their respective leagues have won the domestic or international titles since 2010 (except for Leicester City in 2016).\n- High transfer spending for domestic and international performance can lead to inequality between leagues and clubs. ",
"_____no_output_____"
],
[
"## Q5: Investigating the spending trends of Manchester City, Chelsea and Paris Saint-Germain",
"_____no_output_____"
],
[
"We are particularly interested in the spending trends of the above 3 clubs. They have arrived into the footballing scene relatively recently and have gone on to challenge the European Elite.",
"_____no_output_____"
],
[
"#### Calculate transfer spendings over the years for the above clubs",
"_____no_output_____"
]
],
[
[
"df = pd.read_sql_query(\"select club_name,year,sum(fee_inflation_adjusted) as `transfer_fee_total` from transfers\\\n where club_name in ('Manchester City','Chelsea FC','Paris Saint-Germain') and year<=2017\\\n group by club_name,year\",cnx)\ndf.head(5)",
"_____no_output_____"
]
],
[
[
"### For this question, Tableau was used to create the visualization. The Tableau file for the visualization can be found at clubsSpending.twb. \nSteps to reproduce the visualization\n- Run the above SQL query \n- Dump the dataframe into csv file\n- Open Tableau desktop/online. \n- Load the csv file into Tableau\n- Move transfer_fee_total to rows\n- Move year to columns\n- Add club_name to color \n- Choose vertical stacked chart from the right top corner ",
"_____no_output_____"
]
],
[
[
"from IPython.display import Image\nImage(filename='./images/q5.png') ",
"_____no_output_____"
]
],
[
[
"- Chelsea, Manchester City and PSG have challenged the European elite in the past decade partly due to their huge spending\n- Chelsea’s investment grew by over 234%, while Paris Saint Germain’s by 477% and Manchester City’s by 621% \n- The huge transfer spendings can be attributed to the massive amounts of foreign investment into clubs\n\n<b>Amount & time to first title</b>\n- Chelsea - £470.5 million (2 yrs)\n- Manchester City - £761 million (5 yrs)\n- Paris SaintGermain - £421 million (2 ys)",
"_____no_output_____"
],
[
"## Q 6: Which country produces the best footballing talent?",
"_____no_output_____"
],
[
"### Following steps are performed to decide which country produces the best kind of footballing talent\n- Iterate over the years 2000-2018\n- Iterate over chosen positions\n- Sum transfer fee by nation \n- Rank countries in descending order of transfer spending\n- Compute median rank over the years\n- Sort by median rank and display for each position\n- Disregard countries that appear in the top 10 rankings only 10 times in the 18 year span",
"_____no_output_____"
]
],
[
[
"clean = raw\npositions=['Forward', 'Midfield', 'Winger', 'Defense',\n 'Goalkeeper']\n\nfinal = {}\n#Iterate over the years\nfor i in range(2000,2019):\n # Iterate over positions\n for pos in positions:\n if pos not in final:\n final[pos]=pd.DataFrame()\n # Sum over the fee by nation and rank \n x = clean[(clean[\"posNew\"]==pos)&(clean[\"year\"]==i)].groupby(['nat'])['fee_cleaned'].agg('sum').rank(ascending=False).sort_values(ascending=True)\n x = x.to_frame()\n x[\"year\"]=i\n final[pos]= final[pos].append(x)\n# Add column to maintain counts\nfor pos in positions:\n final[pos][\"count\"]=1\n \n# Compute median and display \nfor pos in positions:\n z = final[pos].groupby('nat').median().reset_index()\n z1 = final[pos].groupby(['nat']).agg({\"count\":\"count\",\"fee_cleaned\": \"median\",})\n print(pos)\n z1 = z1.rename(columns={\"fee_cleaned\": \"median_rank\",\"nat\":\"Country\"})\n print(z1[z1[\"count\"]>=10].drop(['count'],axis=1).sort_values(by=\"median_rank\").head(5))\n print(\"\\n\\n\")",
"Forward\n median_rank\nnat \nArgentina 3.0\nItaly 3.0\nSpain 4.0\nFrance 5.0\nBrazil 5.0\n\n\n\nMidfield\n median_rank\nnat \nBrazil 2.0\nFrance 3.0\nSpain 4.0\nEngland 4.0\nItaly 4.0\n\n\n\nWinger\n median_rank\nnat \nFrance 3.0\nSpain 4.0\nNetherlands 5.0\nPortugal 5.5\nEngland 6.5\n\n\n\nDefense\n median_rank\nnat \nItaly 2.0\nBrazil 3.0\nFrance 3.0\nEngland 3.0\nSpain 5.0\n\n\n\nGoalkeeper\n median_rank\nnat \nItaly 3.0\nEngland 3.5\nBrazil 5.0\nSpain 5.0\nGermany 6.0\n\n\n\n"
]
],
[
[
"## VII. Limitations\n- All of the data is collected and maintained by users of the transfermarkt.co.uk website. There might be inaccuracies in the stated transfer figures. \n- These inaccuracies might be more frequent as we go back the years (1990-2000)\n- As part of the analysis players on loan and free transfers are not being considered, this could change the results of the analysis\n- Calculation of median rank might not be the best metric for measuring which country produces the best footballing talent. Player ratings or yearly performances are a better measure for this analysis\n",
"_____no_output_____"
],
[
"## VIII. Conclusion\n\n- By analyzing the transfer market we are now aware of some of the big spenders in the European leagues and the hyper-inflation in transfer fees in the English Premier League\n\n- With clubs raking in huge amounts of revenue, checks and balances need to be put into place to prevent the sport from being dominated from a few European elite clubs. which could lead to an European Super League\n\n- High transfer spending for domestic and international performance can lead to wealth inequality between leagues and clubs. \n\n- The increase in foreign investments into European Clubs has led to the rise of super-rich clubs\n\n- Clubs chasing success are spending more and more on players, which creates an unequal playing field for all the clubs. Although, it might not be feasible to completely curb the spending of these clubs, regulations need to be put in place to prevent such clubs from taking over.\n",
"_____no_output_____"
],
[
"## IX. References\n1. FIFA TMS reports https://www.fifatms.com/data-reports/reports/\n2. Evaluation football clubs in Europe - A report by KPMG https://www.footballbenchmark.com/library/football_clubs_valuation_the_european_elite_2019\n3. Wealth gap in the top European Clubs https://www.usatoday.com/story/sports/soccer/2018/01/16/uefa-warns-of-growing-wealth-gap-in-top-clubs-finance-study/109521284/\n4. British Inflation Calculator http://inflation.iamkate.com/",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
cbf0f1238b723608d6cb9427846011a5a6f31b93
| 7,880 |
ipynb
|
Jupyter Notebook
|
simulation/7_sided_die.ipynb
|
shawlu95/Data_Science_Toolbox
|
3f5dd554667dab51b59a1668488243161786b5f1
|
[
"MIT"
] | 41 |
2019-05-04T11:02:43.000Z
|
2022-02-20T02:37:01.000Z
|
simulation/7_sided_die.ipynb
|
shawlu95/Data_Science_Toolbox
|
3f5dd554667dab51b59a1668488243161786b5f1
|
[
"MIT"
] | null | null | null |
simulation/7_sided_die.ipynb
|
shawlu95/Data_Science_Toolbox
|
3f5dd554667dab51b59a1668488243161786b5f1
|
[
"MIT"
] | 16 |
2019-04-05T00:49:16.000Z
|
2021-04-15T08:06:43.000Z
| 66.779661 | 5,148 | 0.805076 |
[
[
[
"#### Problem\n*Random number generation: Give you a dice with 7 sides, how do you generate random numbers between 1 and 10 with equal probability?*\n\n* If get 7, ignore, conditional on getting <= 6, the probability of 1~3 is 50%\n* Second row, if getting 6, 7, ignore, conditional on <= 5, each number has 20%\n* If first row get 4~6, add 5 to second row\n\n*The answer is not unique. The key concept is conditional probability.*",
"_____no_output_____"
]
],
[
[
"import random as r\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"def throw_dice():\n \"\"\"\n @return number 1 ~ 10 with equal probability.\n \"\"\"\n faces = [i + 1 for i in range(7)]\n \n first = r.choice(faces)\n while first == 7:\n first = r.choice(faces)\n \n second = r.choice(faces)\n while second >= 6:\n second = r.choice(faces)\n \n return second + 5 * (first > 3)",
"_____no_output_____"
],
[
"sample = [throw_dice() for _ in range(10000)]",
"_____no_output_____"
],
[
"# expect uniform distribution\nplt.hist(sample)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
cbf0f200017b7bc0fd0dd0cce37b9ab0633a51ac
| 63,838 |
ipynb
|
Jupyter Notebook
|
docs_src/callbacks.one_cycle.ipynb
|
ivaylojelev/fastai1
|
e201aa26c7064c6b5fff29e677a3f66950bc95ff
|
[
"Apache-2.0"
] | 59 |
2020-08-18T03:41:35.000Z
|
2022-03-23T03:51:55.000Z
|
docs_src/callbacks.one_cycle.ipynb
|
ivaylojelev/fastai1
|
e201aa26c7064c6b5fff29e677a3f66950bc95ff
|
[
"Apache-2.0"
] | 17 |
2020-08-25T14:15:32.000Z
|
2022-03-27T02:12:19.000Z
|
docs_src/callbacks.one_cycle.ipynb
|
ivaylojelev/fastai1
|
e201aa26c7064c6b5fff29e677a3f66950bc95ff
|
[
"Apache-2.0"
] | 89 |
2020-08-17T23:45:42.000Z
|
2022-03-27T20:53:43.000Z
| 116.919414 | 28,076 | 0.852188 |
[
[
[
"## The 1cycle policy",
"_____no_output_____"
]
],
[
[
"from fastai.gen_doc.nbdoc import *\nfrom fastai.vision import *\nfrom fastai.callbacks import *",
"_____no_output_____"
]
],
[
[
"## What is 1cycle?",
"_____no_output_____"
],
[
"This Callback allows us to easily train a network using Leslie Smith's 1cycle policy. To learn more about the 1cycle technique for training neural networks check out [Leslie Smith's paper](https://arxiv.org/pdf/1803.09820.pdf) and for a more graphical and intuitive explanation check out [Sylvain Gugger's post](https://sgugger.github.io/the-1cycle-policy.html).\n\nTo use our 1cycle policy we will need an [optimum learning rate](https://sgugger.github.io/how-do-you-find-a-good-learning-rate.html). We can find this learning rate by using a learning rate finder which can be called by using [`lr_finder`](/callbacks.lr_finder.html#callbacks.lr_finder). It will do a mock training by going over a large range of learning rates, then plot them against the losses. We will pick a value a bit before the minimum, where the loss still improves. Our graph would look something like this:\n\n\n\nHere anything between `3x10^-2` and `10^-2` is a good idea.\n\nNext we will apply the 1cycle policy with the chosen learning rate as the maximum learning rate. The original 1cycle policy has three steps:\n\n 1. We progressively increase our learning rate from lr_max/div_factor to lr_max and at the same time we progressively decrease our momentum from mom_max to mom_min.\n 2. We do the exact opposite: we progressively decrease our learning rate from lr_max to lr_max/div_factor and at the same time we progressively increase our momentum from mom_min to mom_max.\n 3. We further decrease our learning rate from lr_max/div_factor to lr_max/(div_factor x 100) and we keep momentum steady at mom_max.\n \nThis gives the following form:\n\n<img src=\"imgs/onecycle_params.png\" alt=\"1cycle parameteres\" width=\"500\">\n\nUnpublished work has shown even better results by using only two phases: the same phase 1, followed by a second phase where we do a cosine annealing from lr_max to 0. The momentum goes from mom_min to mom_max by following the symmetric cosine (see graph a bit below).",
"_____no_output_____"
],
[
"## Basic Training",
"_____no_output_____"
],
[
"The one cycle policy allows to train very quickly, a phenomenon termed [_superconvergence_](https://arxiv.org/abs/1708.07120). To see this in practice, we will first train a CNN and see how our results compare when we use the [`OneCycleScheduler`](/callbacks.one_cycle.html#OneCycleScheduler) with [`fit_one_cycle`](/train.html#fit_one_cycle).",
"_____no_output_____"
]
],
[
[
"path = untar_data(URLs.MNIST_SAMPLE)\ndata = ImageDataBunch.from_folder(path)\nmodel = simple_cnn((3,16,16,2))\nlearn = Learner(data, model, metrics=[accuracy])",
"_____no_output_____"
]
],
[
[
"First lets find the optimum learning rate for our comparison by doing an LR range test.",
"_____no_output_____"
]
],
[
[
"learn.lr_find()",
"LR Finder is complete, type {learner_name}.recorder.plot() to see the graph.\n"
],
[
"learn.recorder.plot()",
"_____no_output_____"
]
],
[
[
"Here 5e-2 looks like a good value, a tenth of the minimum of the curve. That's going to be the highest learning rate in 1cycle so let's try a constant training at that value.",
"_____no_output_____"
]
],
[
[
"learn.fit(2, 5e-2)",
"_____no_output_____"
]
],
[
[
"We can also see what happens when we train at a lower learning rate",
"_____no_output_____"
]
],
[
[
"model = simple_cnn((3,16,16,2))\nlearn = Learner(data, model, metrics=[accuracy])\nlearn.fit(2, 5e-3)",
"_____no_output_____"
]
],
[
[
"## Training with the 1cycle policy",
"_____no_output_____"
],
[
"Now to do the same thing with 1cycle, we use [`fit_one_cycle`](/train.html#fit_one_cycle).",
"_____no_output_____"
]
],
[
[
"model = simple_cnn((3,16,16,2))\nlearn = Learner(data, model, metrics=[accuracy])\nlearn.fit_one_cycle(2, 5e-2)",
"_____no_output_____"
]
],
[
[
"This gets the best of both world and we can see how we get a far better accuracy and a far lower loss in the same number of epochs. It's possible to get to the same amazing results with training at constant learning rates, that we progressively diminish, but it will take a far longer time.\n\nHere is the schedule of the lrs (left) and momentum (right) that the new 1cycle policy uses.",
"_____no_output_____"
]
],
[
[
"learn.recorder.plot_lr(show_moms=True)",
"_____no_output_____"
],
[
"show_doc(OneCycleScheduler)",
"_____no_output_____"
]
],
[
[
"Create a [`Callback`](/callback.html#Callback) that handles the hyperparameters settings following the 1cycle policy for `learn`. `lr_max` should be picked with the [`lr_find`](/train.html#lr_find) test. In phase 1, the learning rates goes from `lr_max/div_factor` to `lr_max` linearly while the momentum goes from `moms[0]` to `moms[1]` linearly. In phase 2, the learning rates follows a cosine annealing from `lr_max` to 0, as the momentum goes from `moms[1]` to `moms[0]` with the same annealing.",
"_____no_output_____"
]
],
[
[
"show_doc(OneCycleScheduler.steps, doc_string=False)",
"_____no_output_____"
]
],
[
[
"Build the [`Scheduler`](/callback.html#Scheduler) for the [`Callback`](/callback.html#Callback) according to `steps_cfg`.",
"_____no_output_____"
],
[
"### Callback methods",
"_____no_output_____"
],
[
"You don't call these yourself - they're called by fastai's [`Callback`](/callback.html#Callback) system automatically to enable the class's functionality.",
"_____no_output_____"
]
],
[
[
"show_doc(OneCycleScheduler.on_train_begin, doc_string=False)",
"_____no_output_____"
]
],
[
[
"Initiate the parameters of a training for `n_epochs`.",
"_____no_output_____"
]
],
[
[
"show_doc(OneCycleScheduler.on_batch_end, doc_string=False)",
"_____no_output_____"
]
],
[
[
"Prepares the hyperparameters for the next batch.",
"_____no_output_____"
],
[
"## Undocumented Methods - Methods moved below this line will intentionally be hidden",
"_____no_output_____"
],
[
"## New Methods - Please document or move to the undocumented section",
"_____no_output_____"
]
],
[
[
"show_doc(OneCycleScheduler.on_epoch_end)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
]
] |
cbf0f31e89630c0de817ea38b7fd5600c6ba956b
| 605,628 |
ipynb
|
Jupyter Notebook
|
practice/week-13/W13_2_pyspark.ipynb
|
thejungwon/bigdata-analysis-programming
|
bdff58c1f982fe9a0a4143f9610b7a56c987957e
|
[
"MIT"
] | null | null | null |
practice/week-13/W13_2_pyspark.ipynb
|
thejungwon/bigdata-analysis-programming
|
bdff58c1f982fe9a0a4143f9610b7a56c987957e
|
[
"MIT"
] | null | null | null |
practice/week-13/W13_2_pyspark.ipynb
|
thejungwon/bigdata-analysis-programming
|
bdff58c1f982fe9a0a4143f9610b7a56c987957e
|
[
"MIT"
] | null | null | null | 51.06045 | 1,111 | 0.446317 |
[
[
[
"# BIG DATA ANALYTICS PROGRAMMING : PySpark\n### PySpark 맛보기\n---",
"_____no_output_____"
]
],
[
[
"import sys\n!{sys.executable} -m pip install pyspark",
"Collecting pyspark\n Downloading pyspark-3.0.1.tar.gz (204.2 MB)\n\u001b[K |████████████████████████████████| 204.2 MB 10.4 MB/s eta 0:00:011 |██████████████ | 88.9 MB 12.2 MB/s eta 0:00:10 |██████████████████████████▌ | 169.4 MB 12.1 MB/s eta 0:00:03\n\u001b[?25hCollecting py4j==0.10.9\n Downloading py4j-0.10.9-py2.py3-none-any.whl (198 kB)\n\u001b[K |████████████████████████████████| 198 kB 11.8 MB/s eta 0:00:01\n\u001b[?25hBuilding wheels for collected packages: pyspark\n Building wheel for pyspark (setup.py) ... \u001b[?25ldone\n\u001b[?25h Created wheel for pyspark: filename=pyspark-3.0.1-py2.py3-none-any.whl size=204612244 sha256=28ff896afa1fde1de08c06e1cb91b2cb34ea0be83d0dcc8e178de48e7b544ab5\n Stored in directory: /Users/jungwons/Library/Caches/pip/wheels/5e/34/fa/b37b5cef503fc5148b478b2495043ba61b079120b7ff379f9b\nSuccessfully built pyspark\nInstalling collected packages: py4j, pyspark\nSuccessfully installed py4j-0.10.9 pyspark-3.0.1\n"
],
[
"# PYSPARK를 활용하기 위한 관련 설정\nimport os\nimport sys\n\nos.environ[\"PYSPARK_PYTHON\"]=sys.executable\nos.environ[\"PYSPARK_DRIVER_PYTHON\"]=sys.executable\n",
"_____no_output_____"
]
],
[
[
"## RDD 활용하기\n- Resilient Disributed Data",
"_____no_output_____"
]
],
[
[
"# pyspark 임포트\nfrom pyspark import SparkContext",
"_____no_output_____"
],
[
"# Spark context를 활용해 RDD를 생성 할 수 있다\nsc = SparkContext()",
"_____no_output_____"
]
],
[
[
"### 테스트 파일 생성",
"_____no_output_____"
]
],
[
[
"%%writefile example.txt\nfirst line\nsecond line\nthird line\nfourth line",
"Overwriting example.txt\n"
]
],
[
[
"### RDD 기본 동작",
"_____no_output_____"
]
],
[
[
"textFile = sc.textFile('example.txt')",
"_____no_output_____"
],
[
"textFile",
"_____no_output_____"
]
],
[
[
"### Line 수 세기",
"_____no_output_____"
]
],
[
[
"textFile.count()",
"_____no_output_____"
]
],
[
[
"### 첫번째 줄 출력",
"_____no_output_____"
]
],
[
[
"textFile.first()",
"_____no_output_____"
]
],
[
[
"## 특정 text를 포함하는 데이터 출력",
"_____no_output_____"
]
],
[
[
"secfind = textFile.filter(lambda line: 'second' in line)",
"_____no_output_____"
],
[
"# RDD, 아직까지 어떠한 연산도 이루어지지 않은 상태입니다!\nsecfind",
"_____no_output_____"
],
[
"# 이때 연산 시작\nsecfind.collect()",
"_____no_output_____"
],
[
"# 이때 연산 시작\nsecfind.count()",
"_____no_output_____"
]
],
[
[
"## RDD에서의 전처리",
"_____no_output_____"
]
],
[
[
"%%writefile example2.txt\nfirst \nsecond line\nthe third line\nthen a fourth line",
"Overwriting example2.txt\n"
],
[
"text_rdd = sc.textFile('example2.txt')",
"_____no_output_____"
],
[
"text_rdd.collect()",
"_____no_output_____"
]
],
[
[
"### Map과 Flatmap의 차이",
"_____no_output_____"
]
],
[
[
"text_rdd.map(lambda line: line.split()).collect()",
"_____no_output_____"
],
[
"# Collect everything as a single flat map\ntext_rdd.flatMap(lambda line: line.split()).collect()",
"_____no_output_____"
]
],
[
[
"### CSV 파일 전처리",
"_____no_output_____"
]
],
[
[
"rdd = sc.textFile('data.csv')",
"_____no_output_____"
],
[
"rdd.take(2)",
"_____no_output_____"
],
[
"rdd.map(lambda x: x.split(\",\")).take(3)",
"_____no_output_____"
],
[
"rdd.map(lambda x: x.replace(\" \",\"_\")).collect()",
"_____no_output_____"
],
[
"rdd.map(lambda x: x.replace(\" \",\"_\")).map(lambda x: x.replace(\"'\",\"_\")).collect()",
"_____no_output_____"
],
[
"rdd.map(lambda x: x.replace(\" \",\"_\")).map(lambda x: x.replace(\"'\",\"_\")).map(lambda x: x.replace(\"/\",\"_\")).collect()",
"_____no_output_____"
],
[
"clean_rdd = rdd.map(lambda x: x.replace(\" \",\"_\").replace(\"'\",\"_\").replace(\"/\",\"_\").replace('\"',\"\"))",
"_____no_output_____"
],
[
"clean_rdd.collect()",
"_____no_output_____"
],
[
"clean_rdd = clean_rdd.map(lambda x: x.split(\",\"))",
"_____no_output_____"
],
[
"clean_rdd.collect()",
"_____no_output_____"
]
],
[
[
"### Group BY 구현",
"_____no_output_____"
]
],
[
[
"clean_rdd.map(lambda lst: (lst[0],lst[-1])).collect()",
"_____no_output_____"
],
[
"# 첫번째 원소(lst[0])를 키로 인지\nclean_rdd.map(lambda lst: (lst[0],lst[-1]))\\\n .reduceByKey(lambda amt1,amt2 : amt1+amt2)\\\n .collect()",
"_____no_output_____"
],
[
"# 올바른 연산을 위해 Float으로 캐스팅\nclean_rdd.map(lambda lst: (lst[0],lst[-1]))\\\n .reduceByKey(lambda amt1,amt2 : float(amt1)+float(amt2))\\\n .collect()",
"_____no_output_____"
],
[
"# 최종 코드\nclean_rdd.map(lambda lst: (lst[0],lst[-1]))\\\n.reduceByKey(lambda amt1,amt2 : float(amt1)+float(amt2))\\\n.filter(lambda x: not x[0]=='gender')\\\n.sortBy(lambda stateAmount: stateAmount[1], ascending=False)\\\n.collect()",
"_____no_output_____"
]
],
[
[
"## DataFrame 활용하기",
"_____no_output_____"
]
],
[
[
"from pyspark.sql import SparkSession\n\nappName = \"Python Example - PySpark Read CSV\"\nmaster = 'local'\n\n# Create Spark session\nspark = SparkSession.builder \\\n .master(master) \\\n .appName(appName) \\\n .getOrCreate()\n\n# Convert list to data frame\ndf = spark.read.format('csv') \\\n .option('header',True) \\\n .option('multiLine', True) \\\n .load('data.csv')\ndf.show()\nprint(f'Record count is: {df.count()}')",
"+------+--------------+---------------------------+------------+-----------------------+----------+-------------+-------------+\n|gender|race/ethnicity|parental level of education| lunch|test preparation course|math score|reading score|writing score|\n+------+--------------+---------------------------+------------+-----------------------+----------+-------------+-------------+\n|female| group B| bachelor's degree| standard| none| 72| 72| 74|\n|female| group C| some college| standard| completed| 69| 90| 88|\n|female| group B| master's degree| standard| none| 90| 95| 93|\n| male| group A| associate's degree|free/reduced| none| 47| 57| 44|\n| male| group C| some college| standard| none| 76| 78| 75|\n|female| group B| associate's degree| standard| none| 71| 83| 78|\n|female| group B| some college| standard| completed| 88| 95| 92|\n| male| group B| some college|free/reduced| none| 40| 43| 39|\n| male| group D| high school|free/reduced| completed| 64| 64| 67|\n|female| group B| high school|free/reduced| none| 38| 60| 50|\n| male| group C| associate's degree| standard| none| 58| 54| 52|\n| male| group D| associate's degree| standard| none| 40| 52| 43|\n|female| group B| high school| standard| none| 65| 81| 73|\n| male| group A| some college| standard| completed| 78| 72| 70|\n|female| group A| master's degree| standard| none| 50| 53| 58|\n|female| group C| some high school| standard| none| 69| 75| 78|\n| male| group C| high school| standard| none| 88| 89| 86|\n|female| group B| some high school|free/reduced| none| 18| 32| 28|\n| male| group C| master's degree|free/reduced| completed| 46| 42| 46|\n|female| group C| associate's degree|free/reduced| none| 54| 58| 61|\n+------+--------------+---------------------------+------------+-----------------------+----------+-------------+-------------+\nonly showing top 20 rows\n\nRecord count is: 1000\n"
],
[
"df.columns",
"_____no_output_____"
],
[
"df.describe()",
"_____no_output_____"
],
[
"df.select('gender').show()",
"+------+\n|gender|\n+------+\n|female|\n|female|\n|female|\n| male|\n| male|\n|female|\n|female|\n| male|\n| male|\n|female|\n| male|\n| male|\n|female|\n| male|\n|female|\n|female|\n| male|\n|female|\n| male|\n|female|\n+------+\nonly showing top 20 rows\n\n"
],
[
"df.select('gender').distinct().show()",
"+------+\n|gender|\n+------+\n|female|\n| male|\n+------+\n\n"
],
[
"df.select('race/ethnicity').distinct().show()",
"+--------------+\n|race/ethnicity|\n+--------------+\n| group B|\n| group C|\n| group D|\n| group A|\n| group E|\n+--------------+\n\n"
],
[
"from pyspark.sql import functions as F\ndf.groupBy(\"gender\").agg(F.mean('writing score'), F.mean('math score')).show()",
"+------+------------------+------------------+\n|gender|avg(writing score)| avg(math score)|\n+------+------------------+------------------+\n|female| 72.46718146718146|63.633204633204635|\n| male| 63.31120331950208| 68.72821576763485|\n+------+------------------+------------------+\n\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbf0f4e36b0f1a32e522de3fca77d973604f3b01
| 33,383 |
ipynb
|
Jupyter Notebook
|
lessons/jk-python/python.ipynb
|
sahiggin/2015-01-07-instaar
|
7ead50a81c959cd9f38dd855645307bec6f5502e
|
[
"CC-BY-3.0"
] | null | null | null |
lessons/jk-python/python.ipynb
|
sahiggin/2015-01-07-instaar
|
7ead50a81c959cd9f38dd855645307bec6f5502e
|
[
"CC-BY-3.0"
] | null | null | null |
lessons/jk-python/python.ipynb
|
sahiggin/2015-01-07-instaar
|
7ead50a81c959cd9f38dd855645307bec6f5502e
|
[
"CC-BY-3.0"
] | null | null | null | 32.664384 | 324 | 0.563281 |
[
[
[
"empty"
]
]
] |
[
"empty"
] |
[
[
"empty"
]
] |
cbf0f5c5ebfc7b55e13f021dba1a595686371c87
| 38,820 |
ipynb
|
Jupyter Notebook
|
tutorials/91-rewrite-Wigner-symbols.ipynb
|
Joseph-33/JAC.jl
|
e866abe256e9ae4dc5293a7efe9a24a5e6213f90
|
[
"MIT"
] | 115 |
2019-03-11T11:24:12.000Z
|
2022-03-22T22:33:28.000Z
|
tutorials/91-rewrite-Wigner-symbols.ipynb
|
Joseph-33/JAC.jl
|
e866abe256e9ae4dc5293a7efe9a24a5e6213f90
|
[
"MIT"
] | 10 |
2019-04-02T22:04:24.000Z
|
2021-10-06T06:21:57.000Z
|
tutorials/91-rewrite-Wigner-symbols.ipynb
|
Joseph-33/JAC.jl
|
e866abe256e9ae4dc5293a7efe9a24a5e6213f90
|
[
"MIT"
] | 27 |
2019-03-20T10:28:48.000Z
|
2022-03-04T12:51:19.000Z
| 41.253985 | 992 | 0.540185 |
[
[
[
"empty"
]
]
] |
[
"empty"
] |
[
[
"empty"
]
] |
cbf0fa4ea60bb3fc5c17fd440deb4309df4dd281
| 41,192 |
ipynb
|
Jupyter Notebook
|
autoencoder/Simple_Autoencoder.ipynb
|
jeffaudi/deep-learning
|
d64778933ca79a09c365ed51111a2a9667408363
|
[
"MIT"
] | null | null | null |
autoencoder/Simple_Autoencoder.ipynb
|
jeffaudi/deep-learning
|
d64778933ca79a09c365ed51111a2a9667408363
|
[
"MIT"
] | null | null | null |
autoencoder/Simple_Autoencoder.ipynb
|
jeffaudi/deep-learning
|
d64778933ca79a09c365ed51111a2a9667408363
|
[
"MIT"
] | 1 |
2018-05-23T19:02:52.000Z
|
2018-05-23T19:02:52.000Z
| 137.765886 | 25,708 | 0.874684 |
[
[
[
"# A Simple Autoencoder\n\nWe'll start off by building a simple autoencoder to compress the MNIST dataset. With autoencoders, we pass input data through an encoder that makes a compressed representation of the input. Then, this representation is passed through a decoder to reconstruct the input data. Generally the encoder and decoder will be built with neural networks, then trained on example data.\n\n\n\nIn this notebook, we'll be build a simple network architecture for the encoder and decoder. Let's get started by importing our libraries and getting the dataset.",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\n\nimport numpy as np\nimport tensorflow as tf\nimport matplotlib.pyplot as plt",
"/Users/Jeff/Applications/miniconda3/envs/dlnd/lib/python3.6/importlib/_bootstrap.py:219: RuntimeWarning: compiletime version 3.5 of module 'tensorflow.python.framework.fast_tensor_util' does not match runtime version 3.6\n return f(*args, **kwds)\n"
],
[
"from tensorflow.examples.tutorials.mnist import input_data\nmnist = input_data.read_data_sets('MNIST_data', validation_size=0)",
"Successfully downloaded train-images-idx3-ubyte.gz 9912422 bytes.\nExtracting MNIST_data/train-images-idx3-ubyte.gz\nSuccessfully downloaded train-labels-idx1-ubyte.gz 28881 bytes.\nExtracting MNIST_data/train-labels-idx1-ubyte.gz\nSuccessfully downloaded t10k-images-idx3-ubyte.gz 1648877 bytes.\nExtracting MNIST_data/t10k-images-idx3-ubyte.gz\nSuccessfully downloaded t10k-labels-idx1-ubyte.gz 4542 bytes.\nExtracting MNIST_data/t10k-labels-idx1-ubyte.gz\n"
]
],
[
[
"Below I'm plotting an example image from the MNIST dataset. These are 28x28 grayscale images of handwritten digits.",
"_____no_output_____"
]
],
[
[
"img = mnist.train.images[2]\nplt.imshow(img.reshape((28, 28)), cmap='Greys_r')",
"_____no_output_____"
]
],
[
[
"We'll train an autoencoder with these images by flattening them into 784 length vectors. The images from this dataset are already normalized such that the values are between 0 and 1. Let's start by building basically the simplest autoencoder with a **single ReLU hidden layer**. This layer will be used as the compressed representation. Then, the encoder is the input layer and the hidden layer. The decoder is the hidden layer and the output layer. Since the images are normalized between 0 and 1, we need to use a **sigmoid activation on the output layer** to get values matching the input.\n\n\n\n\n> **Exercise:** Build the graph for the autoencoder in the cell below. The input images will be flattened into 784 length vectors. The targets are the same as the inputs. And there should be one hidden layer with a ReLU activation and an output layer with a sigmoid activation. Feel free to use TensorFlow's higher level API, `tf.layers`. For instance, you would use [`tf.layers.dense(inputs, units, activation=tf.nn.relu)`](https://www.tensorflow.org/api_docs/python/tf/layers/dense) to create a fully connected layer with a ReLU activation. The loss should be calculated with the cross-entropy loss, there is a convenient TensorFlow function for this `tf.nn.sigmoid_cross_entropy_with_logits` ([documentation](https://www.tensorflow.org/api_docs/python/tf/nn/sigmoid_cross_entropy_with_logits)). You should note that `tf.nn.sigmoid_cross_entropy_with_logits` takes the logits, but to get the reconstructed images you'll need to pass the logits through the sigmoid function.",
"_____no_output_____"
]
],
[
[
"# Size of the encoding layer (the hidden layer)\nencoding_dim = 32 # feel free to change this value\ntotal_pixels = mnist.train.images.shape[1]\n\n# Input and target placeholders\ninputs_ = tf.placeholder(tf.float32, (None, total_pixels), name='inputs')\ntargets_ = tf.placeholder(tf.float32, (None, total_pixels), name='targets')\n\n# Output of hidden layer, single fully connected layer here with ReLU activation\nencoded = tf.layers.dense(inputs=inputs_, units=encoding_dim, activation=None)\n\n# Output layer logits, fully connected layer with no activation\nlogits = tf.layers.dense(inputs=encoded, units=total_pixels, activation=None)\n# Sigmoid output from logits\ndecoded = tf.sigmoid(logits, name='output')\n\n# Sigmoid cross-entropy loss\nloss = tf.nn.sigmoid_cross_entropy_with_logits(labels=targets_, logits=logits)\n# Mean of the loss\ncost = tf.reduce_mean(loss)\n\n# Adam optimizer\nopt = tf.train.AdamOptimizer(0.001).minimize(cost)",
"_____no_output_____"
]
],
[
[
"## Training",
"_____no_output_____"
]
],
[
[
"# Create the session\nsess = tf.Session()",
"_____no_output_____"
]
],
[
[
"Here I'll write a bit of code to train the network. I'm not too interested in validation here, so I'll just monitor the training loss. \n\nCalling `mnist.train.next_batch(batch_size)` will return a tuple of `(images, labels)`. We're not concerned with the labels here, we just need the images. Otherwise this is pretty straightfoward training with TensorFlow. We initialize the variables with `sess.run(tf.global_variables_initializer())`. Then, run the optimizer and get the loss with `batch_cost, _ = sess.run([cost, opt], feed_dict=feed)`.",
"_____no_output_____"
]
],
[
[
"epochs = 20\nbatch_size = 200\nsess.run(tf.global_variables_initializer())\nfor e in range(epochs):\n for ii in range(mnist.train.num_examples//batch_size):\n batch = mnist.train.next_batch(batch_size)\n feed = {inputs_: batch[0], targets_: batch[0]}\n batch_cost, _ = sess.run([cost, opt], feed_dict=feed)\n\n print(\"Epoch: {}/{}: Training loss: {:.4f}\".format(e+1, epochs, batch_cost))",
"Epoch: 1/20: Training loss: 0.1783\nEpoch: 2/20: Training loss: 0.1423\nEpoch: 3/20: Training loss: 0.1252\nEpoch: 4/20: Training loss: 0.1190\nEpoch: 5/20: Training loss: 0.1013\nEpoch: 6/20: Training loss: 0.1024\nEpoch: 7/20: Training loss: 0.0988\nEpoch: 8/20: Training loss: 0.0954\nEpoch: 9/20: Training loss: 0.0945\nEpoch: 10/20: Training loss: 0.0949\nEpoch: 11/20: Training loss: 0.0928\nEpoch: 12/20: Training loss: 0.0905\nEpoch: 13/20: Training loss: 0.0927\nEpoch: 14/20: Training loss: 0.0952\nEpoch: 15/20: Training loss: 0.0918\nEpoch: 16/20: Training loss: 0.0926\nEpoch: 17/20: Training loss: 0.0945\nEpoch: 18/20: Training loss: 0.0952\nEpoch: 19/20: Training loss: 0.0930\nEpoch: 20/20: Training loss: 0.0941\n"
]
],
[
[
"## Checking out the results\n\nBelow I've plotted some of the test images along with their reconstructions. For the most part these look pretty good except for some blurriness in some parts.",
"_____no_output_____"
]
],
[
[
"fig, axes = plt.subplots(nrows=2, ncols=10, sharex=True, sharey=True, figsize=(20,4))\nin_imgs = mnist.test.images[:10]\nreconstructed, compressed = sess.run([decoded, encoded], feed_dict={inputs_: in_imgs})\n\nfor images, row in zip([in_imgs, reconstructed], axes):\n for img, ax in zip(images, row):\n ax.imshow(img.reshape((28, 28)), cmap='Greys_r')\n ax.get_xaxis().set_visible(False)\n ax.get_yaxis().set_visible(False)\n\nfig.tight_layout(pad=0.1)",
"_____no_output_____"
],
[
"sess.close()",
"_____no_output_____"
]
],
[
[
"## Up Next\n\nWe're dealing with images here, so we can (usually) get better performance using convolution layers. So, next we'll build a better autoencoder with convolutional layers.\n\nIn practice, autoencoders aren't actually better at compression compared to typical methods like JPEGs and MP3s. But, they are being used for noise reduction, which you'll also build.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
cbf102039c39459ebb3f18a978a8a47a6b1e68ce
| 127,192 |
ipynb
|
Jupyter Notebook
|
pymaceuticals_starter.ipynb
|
danchavez728/matplotlib
|
992833e5e7ff577d277088b0fc0a3f755dc46001
|
[
"ADSL"
] | null | null | null |
pymaceuticals_starter.ipynb
|
danchavez728/matplotlib
|
992833e5e7ff577d277088b0fc0a3f755dc46001
|
[
"ADSL"
] | null | null | null |
pymaceuticals_starter.ipynb
|
danchavez728/matplotlib
|
992833e5e7ff577d277088b0fc0a3f755dc46001
|
[
"ADSL"
] | null | null | null | 154.923264 | 17,756 | 0.868813 |
[
[
[
"# Observations Insights \n# As the timepoint passes the mice had seen their tumor size go down when using cap.\n# Study seems fair - there are equal parts male to female sitting at 49 - 50 % \n# there may be a correlation to weight and tumor size - possibly obesity playing a role? ",
"_____no_output_____"
]
],
[
[
"## Dependencies and starter code",
"_____no_output_____"
]
],
[
[
"# Dependencies and Setup\nimport matplotlib.pyplot as plt\nimport pandas as pd\nimport scipy.stats as st\nimport numpy as np\nfrom scipy.stats import linregress\n\n# Study data files\nmouse_metadata = \"Data/Mouse_metadata.csv\"\nstudy_results = \"Data/Study_results.csv\"\n\n# Read the mouse data and the study results\nmouse_metadata = pd.read_csv(mouse_metadata)\nstudy_results = pd.read_csv(study_results)\n\n# Combine the data into a single dataset\nmerged=pd.merge(mouse_metadata, study_results, on=\"Mouse ID\", how=\"left\")\nmerged.head()",
"_____no_output_____"
]
],
[
[
"## Summary statistics",
"_____no_output_____"
]
],
[
[
"# Generate a summary statistics table of mean, median, variance, standard deviation, and SEM of the tumor volume for each regimen\n\n# calculate stats for mean median var and stdv\nmean = merged.groupby('Drug Regimen')['Tumor Volume (mm3)'].mean()\nmedian = merged.groupby('Drug Regimen')['Tumor Volume (mm3)'].median()\nvariance = merged.groupby('Drug Regimen')['Tumor Volume (mm3)'].var()\nstdv = merged.groupby('Drug Regimen')['Tumor Volume (mm3)'].std()\nsem = merged.groupby('Drug Regimen')['Tumor Volume (mm3)'].sem()\n\n# create df \nstats_df = pd.DataFrame({\"Mean\": mean, \"Median\": median, \"Variance\": variance, \"Standard Deviation\": stdv, \"SEM\": sem})\n\n# print df\nstats_df\n\n",
"_____no_output_____"
]
],
[
[
"## Bar plots",
"_____no_output_____"
]
],
[
[
"# Generate a bar plot showing number of data points for each treatment regimen using pandas\ngroup_df = pd.DataFrame(merged.groupby([\"Drug Regimen\"]).count()).reset_index()\n\n# set to only these columns \ndrugRegimen = group_df[[\"Drug Regimen\",\"Mouse ID\"]]\ndrugRegimen = drugRegimen.rename(columns={\"Mouse ID\": \"Amount\"})\ndrugRegimen = drugRegimen.set_index(\"Drug Regimen\")\n\n# create bg \ndrugRegimen.plot(kind=\"bar\")\n\n#set bg title \nplt.title(\"Amount per Drug Regimen\")\n\n#print bg \nplt.show()\n",
"_____no_output_____"
],
[
"# Generate a bar plot showing number of data points for each treatment regimen using pyplot\n\n# convert data into lists\ndrugRegimenplt = stats_df.index.tolist()\nregCount = (merged.groupby([\"Drug Regimen\"])[\"Age_months\"].count()).tolist()\nx_axis = np.arange(len(regCount))\nx_axis = drugRegimenplt\n\n# Create a bg based upon the above data\nplt.figure(figsize=(10,4))\nplt.bar(x_axis, regCount)\n\n",
"_____no_output_____"
]
],
[
[
"## Pie plots",
"_____no_output_____"
]
],
[
[
"# Generate a pie plot showing the distribution of female versus male mice using pandas\n# generate df for sex data \nsex_df = pd.DataFrame(merged.groupby([\"Sex\"]).count()).reset_index()\n\n#set data for only these columns \nsex_df = sex_df[[\"Sex\",\"Mouse ID\"]]\nsex_df = sex_df.rename(columns={\"Mouse ID\": \"Sex Ratio\"})\n\n# generate pie plot and set ratios \nplt.figure(figsize=(10,12))\nax1 = plt.subplot(121, aspect='equal')\n\n# format to percentages and set title \nsex_df.plot(kind='pie', y = \"Sex Ratio\", ax=ax1, autopct='%1.1f%%')\n\n",
"_____no_output_____"
],
[
"# Generate a pie plot showing the distribution of female versus male mice using pyplot\nsexPie = (merged.groupby([\"Sex\"])[\"Age_months\"].count()).tolist()\n\n# format pc for title and color \nlabels = [\"Females\", \"Males\"]\ncolors = [\"orange\", \"blue\"]\n\n# generate pc and convert to percentage \nplt.pie(sexPie, labels=labels, colors=colors, autopct=\"%1.1f%%\", shadow=True, startangle=90)\n\n",
"_____no_output_____"
]
],
[
[
"## Quartiles, outliers and boxplots",
"_____no_output_____"
]
],
[
[
"# Calculate the final tumor volume of each mouse across four of the most promising treatment regimens. \n\n# get four colums for df and sort for iqr and boxplot \ntopReg = merged[merged[\"Drug Regimen\"].isin([\"Capomulin\", \"Ramicane\", \"Infubinol\", \"Ceftamin\"])]\ntopReg = topReg.sort_values([\"Timepoint\"], ascending=True)\n\n# format headers for rows \ntopRegData = topReg[[\"Drug Regimen\", \"Mouse ID\", \"Timepoint\", \"Tumor Volume (mm3)\"]]\ntopRegData.head()",
"_____no_output_____"
],
[
"#Calculate the IQR and quantitatively determine if there are any potential outliers. \n# Group data for tumor volume \ntopRegList = topRegData.groupby(['Drug Regimen', 'Mouse ID']).last()['Tumor Volume (mm3)']\n\n# create df with assigned labels\ntopRegData_df = topRegList.to_frame()\ndrugs = ['Capomulin', 'Ramicane', 'Infubinol','Ceftamin']\n\n# create box plot and format \nbox_df = topRegData_df.reset_index()\ntumors = box_df.groupby('Drug Regimen')['Tumor Volume (mm3)'].apply(list)\ntumors_df = pd.DataFrame(tumors)\ntumors_df = tumors_df.reindex(drugs)\ntumorVolumesBox = [vol for vol in tumors_df['Tumor Volume (mm3)']]\nplt.boxplot(tumorVolumesBox, labels=drugs)\nplt.ylim(0, 100)\n# display box \nplt.show()\n",
"_____no_output_____"
]
],
[
[
"## Line and scatter plots",
"_____no_output_____"
]
],
[
[
"# Generate a line plot of time point versus tumor volume for a mouse treated with Capomulin\n\n# create df for capomuline \ncapLine_df = merged.loc[merged[\"Drug Regimen\"] == \"Capomulin\"]\ncapLine_df = capLine_df.reset_index()\n\n# pull data for mouse m601\ncapMouse_df = capLine_df.loc[capLine_df[\"Mouse ID\"] == \"m601\"]\n\n# sort columns for line chart \ncapMouse_df = capMouse_df.loc[:, [\"Timepoint\", \"Tumor Volume (mm3)\"]]\n\n# create lne graph of time vs volume \ncapMouse_df.set_index('Timepoint').plot(figsize=(10, 4), lineWidth=4, color='blue')\n\n",
"_____no_output_____"
],
[
"# Generate a scatter plot of mouse weight versus average tumor volume for the Capomulin regimen\n\n# sort columns for scatter plot \nscatt_df = capLine_df.loc[:, [\"Mouse ID\",\"Weight (g)\", \"Tumor Volume (mm3)\"]]\n\n# set up df for average for mouse tumor vol \ntumAvg = pd.DataFrame(scatt_df.groupby([\"Mouse ID\", \"Weight (g)\"])[\"Tumor Volume (mm3)\"].mean()).reset_index()\n\n# set new name for Tumor volume for chart title \ntumAvg = tumAvg.rename(columns={\"Tumor Volume (mm3)\": \"Mouse Tumor Average Volume\"})\n\n# create scatter plot \ntumAvg.plot(kind=\"scatter\", x=\"Weight (g)\",y = \"Mouse Tumor Average Volume\",grid =True, figsize=(10,4), title= \"Weight (g) VS Mouse Tumor Average Volume\")\n",
"_____no_output_____"
],
[
"# Calculate the correlation coefficient and linear regression model for mouse weight and average tumor volume for the Capomulin regimen\nx = tumAvg['Weight (g)']\nx_values = tumAvg['Weight (g)']\ny_values = tumAvg['Mouse Tumor Average Volume']\n(slope, intercept, rvalue, pvalue, stderr) = linregress(x_values, y_values)\nregress_values = x_values * slope + intercept\nline_eq = \"y = \" + str(round(slope,2)) + \"x + \" + str(round(intercept,2))\nplt.scatter(x_values,y_values)\nplt.plot(x_values,regress_values,\"r-\")\nplt.annotate(line_eq,(10,4),color=\"orange\")\n# format labels for axis \nplt.xlabel('Mouse Weight')\nplt.ylabel('Mouse Tumor Average Volume')\nplt.show()\n\n",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
cbf1080e92539899a932a6e06ec9807fd0fdc60d
| 46,146 |
ipynb
|
Jupyter Notebook
|
notebooks/feature-importance.ipynb
|
samesense/mahdi_epi
|
ec002df1d6b0dbdd4be8675e48971ed604ee9014
|
[
"MIT"
] | 4 |
2018-06-05T06:13:04.000Z
|
2020-11-16T03:03:14.000Z
|
notebooks/feature-importance.ipynb
|
samesense/mahdi_epi
|
ec002df1d6b0dbdd4be8675e48971ed604ee9014
|
[
"MIT"
] | 25 |
2018-05-25T11:46:06.000Z
|
2018-05-29T10:57:07.000Z
|
notebooks/feature-importance.ipynb
|
samesense/mahdi_epi
|
ec002df1d6b0dbdd4be8675e48971ed604ee9014
|
[
"MIT"
] | null | null | null | 215.635514 | 14,116 | 0.901292 |
[
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\nimport pandas as pd\n\nfrom sklearn.datasets import make_classification\nfrom sklearn.ensemble import ExtraTreesClassifier",
"/opt/conda/lib/python3.4/site-packages/matplotlib/font_manager.py:273: UserWarning: Matplotlib is building the font cache using fc-list. This may take a moment.\n warnings.warn('Matplotlib is building the font cache using fc-list. This may take a moment.')\n/opt/conda/lib/python3.4/site-packages/matplotlib/font_manager.py:273: UserWarning: Matplotlib is building the font cache using fc-list. This may take a moment.\n warnings.warn('Matplotlib is building the font cache using fc-list. This may take a moment.')\n"
],
[
"def write_feature_importance(disease, X, indices, cols, importances, std, fout):\n for f in range(X.shape[1]):\n ls = (disease, cols[indices[f]], importances[indices[f]], std[indices[f]])\n print(\"panel\\t%s\\t%s\\t%f\\t%f\" % ls, file=fout)",
"_____no_output_____"
],
[
"afile = '../data/interim/panel.dat'\ndf = pd.read_csv(afile, sep='\\t')\ncols = ['ccr', 'fathmm', 'vest', 'missense_badness', 'missense_depletion', 'is_domain']\nfout = open('../data/interim/importance.panel', 'w')\nheader = ('eval_set', 'disease', 'feature', 'importance', 'std')\nprint('\\t'.join(header), file=fout)\n\nfor disease in set(df['Disease']):\n print(disease)\n X = df[df.Disease==disease][cols]\n y = df[df.Disease==disease]['y']\n\n # Build a forest and compute the feature importances\n forest = ExtraTreesClassifier(n_estimators=250,\n random_state=0)\n\n forest.fit(X, y)\n importances = forest.feature_importances_\n std = np.std([tree.feature_importances_ for tree in forest.estimators_],\n axis=0)\n indices = np.argsort(importances)[::-1]\n\n print(\"Feature ranking:\")\n write_feature_importance(disease, X, indices, cols, importances, std, fout)\n \n for f in range(X.shape[1]):\n print(\"%s %d. %s (%f) std %f\" % (disease, f + 1, cols[indices[f]], importances[indices[f]], std[indices[f]]))\n\n plt.figure()\n plt.title(\"Feature importances \" + disease)\n plt.bar(range(X.shape[1]), importances[indices],\n color=\"r\", yerr=std[indices], align=\"center\")\n plt.xticks(range(X.shape[1]), indices)\n plt.xlim([-1, X.shape[1]])\n plt.show()\nfout.close()",
"EPI\nFeature ranking:\nEPI 1. ccr (0.354841) std 0.181799\nEPI 2. vest (0.315044) std 0.168161\nEPI 3. fathmm (0.153860) std 0.080869\nEPI 4. missense_depletion (0.094822) std 0.056730\nEPI 5. missense_badness (0.048287) std 0.017606\nEPI 6. is_domain (0.033146) std 0.019694\n"
],
[
"!jupyter nbconvert --to=python feature-importance.ipynb --stdout > test2.py",
"[NbConvertApp] Converting notebook feature-importance.ipynb to python\r\n"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code"
]
] |
cbf1200d44c79c2db3a1e47f8384ce9357339fcd
| 4,309 |
ipynb
|
Jupyter Notebook
|
examples/ch12/snippets_ipynb/12_13_02.ipynb
|
eltechno/python_course
|
f74abac7df3f9f41864afd06479389260c29ea3a
|
[
"MIT"
] | 4 |
2019-05-04T00:33:25.000Z
|
2021-05-29T20:37:59.000Z
|
examples/ch12/snippets_ipynb/12_13_02.ipynb
|
eltechno/python_course
|
f74abac7df3f9f41864afd06479389260c29ea3a
|
[
"MIT"
] | null | null | null |
examples/ch12/snippets_ipynb/12_13_02.ipynb
|
eltechno/python_course
|
f74abac7df3f9f41864afd06479389260c29ea3a
|
[
"MIT"
] | 3 |
2020-05-05T13:14:28.000Z
|
2022-02-03T16:18:37.000Z
| 24.072626 | 101 | 0.501741 |
[
[
[
"# 12.13.2 Initiating Stream Processing\n### Authenticating",
"_____no_output_____"
]
],
[
[
"import tweepy",
"_____no_output_____"
],
[
"import keys",
"_____no_output_____"
],
[
"auth = tweepy.OAuthHandler(keys.consumer_key, \n keys.consumer_secret)",
"_____no_output_____"
],
[
"auth.set_access_token(keys.access_token, \n keys.access_token_secret)",
"_____no_output_____"
],
[
"api = tweepy.API(auth, wait_on_rate_limit=True, \n wait_on_rate_limit_notify=True)",
"_____no_output_____"
]
],
[
[
"### Creating a `TweetListener` ",
"_____no_output_____"
]
],
[
[
"from tweetlistener import TweetListener",
"_____no_output_____"
],
[
"tweet_listener = TweetListener(api)",
"_____no_output_____"
]
],
[
[
"### Creating a `Stream` ",
"_____no_output_____"
]
],
[
[
"tweet_stream = tweepy.Stream(auth=api.auth, \n listener=tweet_listener)",
"_____no_output_____"
]
],
[
[
"### Starting the Tweet Stream\n\nWe removed the is_async argument to ensure that the streamed tweets all appear below this cell.",
"_____no_output_____"
]
],
[
[
"tweet_stream.filter(track=['Mars Rover']) #, is_async=True) ",
"_____no_output_____"
]
],
[
[
"### Asynchronous vs. Synchronous Streams",
"_____no_output_____"
],
[
"### Other filter Method Parameters\n### Twitter Restrictions Note",
"_____no_output_____"
]
],
[
[
"##########################################################################\n# (C) Copyright 2019 by Deitel & Associates, Inc. and #\n# Pearson Education, Inc. All Rights Reserved. #\n# #\n# DISCLAIMER: The authors and publisher of this book have used their #\n# best efforts in preparing the book. These efforts include the #\n# development, research, and testing of the theories and programs #\n# to determine their effectiveness. The authors and publisher make #\n# no warranty of any kind, expressed or implied, with regard to these #\n# programs or to the documentation contained in these books. The authors #\n# and publisher shall not be liable in any event for incidental or #\n# consequential damages in connection with, or arising out of, the #\n# furnishing, performance, or use of these programs. #\n##########################################################################\n",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
]
] |
cbf121c130fc45c5cd16ffbf657fad2a2dfd46c3
| 59,404 |
ipynb
|
Jupyter Notebook
|
models/GAN_Mal.ipynb
|
Wapiti08/Defense_Evasion_with_GAN
|
47dac12c314e1238affc665014f13658e87d6fcb
|
[
"MIT"
] | 2 |
2021-10-09T11:44:46.000Z
|
2022-03-11T15:22:58.000Z
|
models/GAN_Mal.ipynb
|
Wapiti08/Defense_Evasion_with_GAN
|
47dac12c314e1238affc665014f13658e87d6fcb
|
[
"MIT"
] | null | null | null |
models/GAN_Mal.ipynb
|
Wapiti08/Defense_Evasion_with_GAN
|
47dac12c314e1238affc665014f13658e87d6fcb
|
[
"MIT"
] | 1 |
2022-03-11T15:23:02.000Z
|
2022-03-11T15:23:02.000Z
| 52.338326 | 318 | 0.412295 |
[
[
[
"from keras.layers import Input, Dense, Activation\nfrom keras.layers import Maximum, Concatenate\nfrom keras.models import Model\nfrom keras.optimizers import adam_v2\nfrom sklearn.tree import DecisionTreeClassifier\nfrom sklearn.linear_model import SGDClassifier\nfrom sklearn.ensemble import GradientBoostingClassifier\nfrom sklearn.svm import SVC\n\nfrom Ensemble_Classifiers import Ensemble_Classifier\nfrom sklearn.model_selection import train_test_split\nimport numpy as np\nglobal seed\n\nseed = 0",
"_____no_output_____"
],
[
"class MalGAN():\n def __init__(self, blackbox, X, Y, threshold):\n self.apifeature_dims = 69\n self.z_dims = 30\n self.generator_layers = [self.apifeature_dims+self.z_dims, 32, 32, 64 , self.apifeature_dims]\n # self.generator_layers = [self.apifeature_dims+self.z_dims, 64, 64, 128 , self.apifeature_dims]\n\n self.substitute_detector_layers = [self.apifeature_dims, 64, 64, 1]\n # self.substitute_detector_layers = [self.apifeature_dims, 128, 128, 1]\n self.blackbox = blackbox \n optimizer = adam_v2.Adam(learning_rate=0.0002, beta_1=0.5)\n self.X = X\n self.Y = Y\n self.threshold = threshold\n\n # Build and Train blackbox_detector\n self.blackbox_detector = self.build_blackbox_detector()\n\n # Build and compile the substitute_detector\n self.substitute_detector = self.build_substitute_detector()\n self.substitute_detector.compile(loss='binary_crossentropy', optimizer=optimizer, metrics=['accuracy'])\n\n # Build the generator\n self.generator = self.build_generator()\n\n # The generator takes malware and noise as input and generates adversarial malware examples\n example = Input(shape=(self.apifeature_dims,))\n noise = Input(shape=(self.z_dims,))\n input = [example, noise]\n malware_examples = self.generator(input)\n\n # The discriminator takes generated images as input and determines validity\n validity = self.substitute_detector(malware_examples)\n\n # The combined model (stacked generator and substitute_detector)\n # Trains the generator to fool the discriminator\n self.combined = Model(input, validity)\n self.combined.compile(loss='binary_crossentropy', optimizer=optimizer)\n \n # For the combined model we will only train the generator\n self.substitute_detector.trainable = False\n \n\n def build_blackbox_detector(self):\n if self.blackbox in ['SVM']:\n blackbox_detector = SVC(kernel = 'linear')\n \n elif self.blackbox in ['GB']:\n blackbox_detector = GradientBoostingClassifier(random_state=seed)\n \n elif self.blackbox in ['SGD']:\n blackbox_detector = SGDClassifier(random_state=seed) \n\n elif self.blackbox in ['DT']:\n blackbox_detector = DecisionTreeClassifier(random_state=seed)\n \n elif self.blackbox in ['Ensem']:\n blackbox_detector = Ensemble_Classifier()\n\n return blackbox_detector\n\n def build_generator(self):\n\n example = Input(shape=(self.apifeature_dims,))\n noise = Input(shape=(self.z_dims,))\n x = Concatenate(axis=1)([example, noise])\n for dim in self.generator_layers[1:]:\n x = Dense(dim)(x)\n x = Activation(activation='tanh')(x)\n x = Maximum()([example, x])\n generator = Model([example, noise], x, name='generator')\n generator.summary()\n return generator\n\n def build_substitute_detector(self):\n\n input = Input(shape=(self.substitute_detector_layers[0],))\n x = input\n for dim in self.substitute_detector_layers[1:]:\n x = Dense(dim)(x)\n x = Activation(activation='sigmoid')(x)\n substitute_detector = Model(input, x, name='substitute_detector')\n substitute_detector.summary()\n return substitute_detector\n\n def load_data(self):\n x_ben, x_ran,y_ben, y_ran = self.X[:self.threshold], self.X[self.threshold:], self.Y[:self.threshold], self.Y[self.threshold:]\n\n return (x_ran, y_ran), (x_ben, y_ben)\n \n \n def train(self, epochs, batch_size=32):\n\n # Load and Split the dataset\n (xmal, ymal), (xben, yben) = self.load_data()\n xtrain_mal, xtest_mal, ytrain_mal, ytest_mal = train_test_split(xmal, ymal, test_size=0.50)\n xtrain_ben, xtest_ben, ytrain_ben, ytest_ben = train_test_split(xben, yben, test_size=0.50)\n\n bl_xtrain_mal, bl_ytrain_mal, bl_xtrain_ben, bl_ytrain_ben = xtrain_mal, ytrain_mal, xtrain_ben, ytrain_ben\n\n \n self.blackbox_detector.fit(np.concatenate([xmal, xben]), np.concatenate([ymal, yben]))\n\n ytrain_ben_blackbox = self.blackbox_detector.predict(bl_xtrain_ben)\n \n Original_Train_TPR = self.blackbox_detector.score(bl_xtrain_mal, bl_ytrain_mal)\n \n Original_Test_TPR = self.blackbox_detector.score(xtest_mal, ytest_mal)\n Train_TPR, Test_TPR = [Original_Train_TPR], [Original_Test_TPR]\n\n\n for epoch in range(epochs):\n\n for step in range(xtrain_mal.shape[0] // batch_size):\n # ---------------------\n # Train substitute_detector\n # ---------------------\n\n # Select a random batch of malware examples\n idx_mal = np.random.randint(0, xtrain_mal.shape[0], batch_size)\n \n xmal_batch = xtrain_mal[idx_mal]\n \n noise = np.random.normal(0, 1, (batch_size, self.z_dims))\n \n idx_ben = np.random.randint(0, xmal_batch.shape[0], batch_size)\n \n xben_batch = xtrain_ben[idx_ben]\n yben_batch = ytrain_ben_blackbox[idx_ben]\n\n # Generate a batch of new malware examples\n gen_examples = self.generator.predict([xmal_batch, noise])\n ymal_batch = self.blackbox_detector.predict(np.ones(gen_examples.shape)*(gen_examples > 0.5))\n\n # Train the substitute_detector\n\n d_loss_real = self.substitute_detector.train_on_batch(gen_examples, ymal_batch)\n d_loss_fake = self.substitute_detector.train_on_batch(xben_batch, yben_batch)\n d_loss = 0.5 * np.add(d_loss_real, d_loss_fake)\n\n # ---------------------\n # Train Generator\n # ---------------------\n\n idx = np.random.randint(0, xtrain_mal.shape[0], batch_size)\n xmal_batch = xtrain_mal[idx]\n noise = np.random.uniform(0, 1, (batch_size, self.z_dims))\n\n # Train the generator\n g_loss = self.combined.train_on_batch([xmal_batch, noise], np.zeros((batch_size, 1)))\n\n # Compute Train TPR\n noise = np.random.uniform(0, 1, (xtrain_mal.shape[0], self.z_dims))\n gen_examples = self.generator.predict([xtrain_mal, noise])\n TPR = self.blackbox_detector.score(np.ones(gen_examples.shape) * (gen_examples > 0.5), ytrain_mal)\n Train_TPR.append(TPR)\n\n # Compute Test TPR\n noise = np.random.uniform(0, 1, (xtest_mal.shape[0], self.z_dims))\n gen_examples = self.generator.predict([xtest_mal, noise])\n TPR = self.blackbox_detector.score(np.ones(gen_examples.shape) * (gen_examples > 0.5), ytest_mal)\n Test_TPR.append(TPR)\n\n print(\"%d [D loss: %f, acc.: %.2f%%] [G loss: %f]\" % (epoch, d_loss[0], 100*d_loss[1], g_loss))\n \n if int(epoch) == int(epochs-1):\n return d_loss[0], 100*d_loss[1], g_loss\n ",
"_____no_output_____"
],
[
"# create the dict to save the D loss, acc and G loss for different classifiers\nD_loss_dict, Acc_dict, G_loss_dict = {}, {}, {}\n# get the data from Feature-Selector\nimport pandas as pd\n\ndf= pd.read_csv('../dataset/matrix/CLaMP.csv')\n\n",
"_____no_output_____"
],
[
"df.dtypes.value_counts()",
"_____no_output_____"
],
[
"df.columns",
"_____no_output_____"
],
[
"# encode categorical column\nfrom sklearn.preprocessing import LabelEncoder\ndf['packer_type'] = LabelEncoder().fit_transform(df['packer_type'])",
"_____no_output_____"
],
[
"df['packer_type'].value_counts()",
"_____no_output_____"
],
[
"Y = df['class'].values\nX = df.drop('class', axis=1).values",
"_____no_output_____"
],
[
"X.shape",
"_____no_output_____"
],
[
"from sklearn.preprocessing import MinMaxScaler\n\nX = MinMaxScaler().fit_transform(X)\n",
"_____no_output_____"
],
[
"X",
"_____no_output_____"
],
[
"from collections import Counter\nCounter(Y)",
"_____no_output_____"
],
[
"# load the classifier\nfor classifier in [ 'SVM', 'SGD', 'DT', 'GB', 'Ensem']: \n print('[+] \\nTraining the model with {} classifier\\n'.format(classifier))\n malgan = MalGAN(blackbox=classifier, X=X, Y=Y, threshold = 2488)\n d_loss, acc, g_loss = malgan.train(epochs=50, batch_size=32)\n\n D_loss_dict[classifier] = d_loss\n Acc_dict[classifier] = acc \n G_loss_dict[classifier] = g_loss\n\n\nprint('=====================')\nprint(D_loss_dict)\nprint('=====================')\nprint(Acc_dict)\nprint('=====================')\nprint(G_loss_dict)",
"[+] \nTraining the model with SVM classifier\n\nModel: \"substitute_detector\"\n_________________________________________________________________\n Layer (type) Output Shape Param # \n=================================================================\n input_26 (InputLayer) [(None, 69)] 0 \n \n dense_35 (Dense) (None, 64) 4480 \n \n dense_36 (Dense) (None, 64) 4160 \n \n dense_37 (Dense) (None, 1) 65 \n \n activation_10 (Activation) (None, 1) 0 \n \n=================================================================\nTotal params: 8,705\nTrainable params: 8,705\nNon-trainable params: 0\n_________________________________________________________________\nModel: \"generator\"\n__________________________________________________________________________________________________\n Layer (type) Output Shape Param # Connected to \n==================================================================================================\n input_27 (InputLayer) [(None, 69)] 0 [] \n \n input_28 (InputLayer) [(None, 30)] 0 [] \n \n concatenate_5 (Concatenate) (None, 99) 0 ['input_27[0][0]', \n 'input_28[0][0]'] \n \n dense_38 (Dense) (None, 32) 3200 ['concatenate_5[0][0]'] \n \n dense_39 (Dense) (None, 32) 1056 ['dense_38[0][0]'] \n \n dense_40 (Dense) (None, 64) 2112 ['dense_39[0][0]'] \n \n dense_41 (Dense) (None, 69) 4485 ['dense_40[0][0]'] \n \n activation_11 (Activation) (None, 69) 0 ['dense_41[0][0]'] \n \n maximum_5 (Maximum) (None, 69) 0 ['input_27[0][0]', \n 'activation_11[0][0]'] \n \n==================================================================================================\nTotal params: 10,853\nTrainable params: 10,853\nNon-trainable params: 0\n__________________________________________________________________________________________________\n0 [D loss: 0.539537, acc.: 71.88%] [G loss: 0.045343]\n1 [D loss: 0.514337, acc.: 73.44%] [G loss: 0.012183]\n2 [D loss: 0.479212, acc.: 79.69%] [G loss: 0.008032]\n3 [D loss: 0.321460, acc.: 92.19%] [G loss: 0.003344]\n4 [D loss: 0.327451, acc.: 92.19%] [G loss: 0.002011]\n5 [D loss: 0.294928, acc.: 90.62%] [G loss: 0.000550]\n6 [D loss: 0.215045, acc.: 95.31%] [G loss: 0.000192]\n7 [D loss: 0.189461, acc.: 93.75%] [G loss: 0.000056]\n8 [D loss: 0.161751, acc.: 96.88%] [G loss: 0.000027]\n9 [D loss: 0.183761, acc.: 95.31%] [G loss: 0.000020]\n10 [D loss: 0.250592, acc.: 92.19%] [G loss: 0.000012]\n11 [D loss: 0.142484, acc.: 96.88%] [G loss: 0.000011]\n12 [D loss: 0.253131, acc.: 92.19%] [G loss: 0.000012]\n13 [D loss: 0.179309, acc.: 95.31%] [G loss: 0.000002]\n14 [D loss: 0.124512, acc.: 96.88%] [G loss: 0.000019]\n15 [D loss: 0.298732, acc.: 87.50%] [G loss: 0.000006]\n16 [D loss: 0.381269, acc.: 85.94%] [G loss: 0.000002]\n17 [D loss: 0.178112, acc.: 93.75%] [G loss: 0.000013]\n18 [D loss: 0.213450, acc.: 89.06%] [G loss: 0.000007]\n19 [D loss: 0.196237, acc.: 95.31%] [G loss: 0.000002]\n20 [D loss: 0.255417, acc.: 92.19%] [G loss: 0.000001]\n21 [D loss: 0.096326, acc.: 96.88%] [G loss: 0.000001]\n22 [D loss: 0.175630, acc.: 93.75%] [G loss: 0.000005]\n23 [D loss: 0.118437, acc.: 93.75%] [G loss: 0.000000]\n24 [D loss: 0.056320, acc.: 100.00%] [G loss: 0.000000]\n25 [D loss: 0.148634, acc.: 95.31%] [G loss: 0.000001]\n26 [D loss: 0.168127, acc.: 95.31%] [G loss: 0.000000]\n27 [D loss: 0.106564, acc.: 96.88%] [G loss: 0.000000]\n28 [D loss: 0.082563, acc.: 96.88%] [G loss: 0.000000]\n29 [D loss: 0.196098, acc.: 95.31%] [G loss: 0.000000]\n30 [D loss: 0.204871, acc.: 92.19%] [G loss: 0.000001]\n31 [D loss: 0.178122, acc.: 95.31%] [G loss: 0.000000]\n32 [D loss: 0.085559, acc.: 98.44%] [G loss: 0.000000]\n33 [D loss: 0.107219, acc.: 98.44%] [G loss: 0.000000]\n34 [D loss: 0.270811, acc.: 90.62%] [G loss: 0.000001]\n35 [D loss: 0.070889, acc.: 100.00%] [G loss: 0.000002]\n36 [D loss: 0.151385, acc.: 95.31%] [G loss: 0.000000]\n37 [D loss: 0.103830, acc.: 96.88%] [G loss: 0.000001]\n38 [D loss: 0.122855, acc.: 96.88%] [G loss: 0.000001]\n39 [D loss: 0.104025, acc.: 95.31%] [G loss: 0.000001]\n40 [D loss: 0.045923, acc.: 100.00%] [G loss: 0.000000]\n41 [D loss: 0.054304, acc.: 100.00%] [G loss: 0.000001]\n42 [D loss: 0.267024, acc.: 92.19%] [G loss: 0.000000]\n43 [D loss: 0.145469, acc.: 96.88%] [G loss: 0.000000]\n44 [D loss: 0.234447, acc.: 95.31%] [G loss: 0.000001]\n45 [D loss: 0.121235, acc.: 95.31%] [G loss: 0.000000]\n46 [D loss: 0.098315, acc.: 95.31%] [G loss: 0.000000]\n47 [D loss: 0.156507, acc.: 95.31%] [G loss: 0.000000]\n48 [D loss: 0.118286, acc.: 96.88%] [G loss: 0.000000]\n49 [D loss: 0.214374, acc.: 92.19%] [G loss: 0.000000]\n[+] \nTraining the model with SGD classifier\n\nModel: \"substitute_detector\"\n_________________________________________________________________\n Layer (type) Output Shape Param # \n=================================================================\n input_31 (InputLayer) [(None, 69)] 0 \n \n dense_42 (Dense) (None, 64) 4480 \n \n dense_43 (Dense) (None, 64) 4160 \n \n dense_44 (Dense) (None, 1) 65 \n \n activation_12 (Activation) (None, 1) 0 \n \n=================================================================\nTotal params: 8,705\nTrainable params: 8,705\nNon-trainable params: 0\n_________________________________________________________________\nModel: \"generator\"\n__________________________________________________________________________________________________\n Layer (type) Output Shape Param # Connected to \n==================================================================================================\n input_32 (InputLayer) [(None, 69)] 0 [] \n \n input_33 (InputLayer) [(None, 30)] 0 [] \n \n concatenate_6 (Concatenate) (None, 99) 0 ['input_32[0][0]', \n 'input_33[0][0]'] \n \n dense_45 (Dense) (None, 32) 3200 ['concatenate_6[0][0]'] \n \n dense_46 (Dense) (None, 32) 1056 ['dense_45[0][0]'] \n \n dense_47 (Dense) (None, 64) 2112 ['dense_46[0][0]'] \n \n dense_48 (Dense) (None, 69) 4485 ['dense_47[0][0]'] \n \n activation_13 (Activation) (None, 69) 0 ['dense_48[0][0]'] \n \n maximum_6 (Maximum) (None, 69) 0 ['input_32[0][0]', \n 'activation_13[0][0]'] \n \n==================================================================================================\nTotal params: 10,853\nTrainable params: 10,853\nNon-trainable params: 0\n__________________________________________________________________________________________________\n0 [D loss: 0.560470, acc.: 79.69%] [G loss: 0.070669]\n1 [D loss: 0.631458, acc.: 76.56%] [G loss: 0.063685]\n2 [D loss: 0.526189, acc.: 78.12%] [G loss: 0.026516]\n3 [D loss: 0.434511, acc.: 76.56%] [G loss: 0.016415]\n4 [D loss: 0.375236, acc.: 84.38%] [G loss: 0.012311]\n5 [D loss: 0.426508, acc.: 76.56%] [G loss: 0.008566]\n6 [D loss: 0.382926, acc.: 82.81%] [G loss: 0.011727]\n7 [D loss: 0.302153, acc.: 81.25%] [G loss: 0.006293]\n8 [D loss: 0.267416, acc.: 87.50%] [G loss: 0.000785]\n9 [D loss: 0.281879, acc.: 84.38%] [G loss: 0.000301]\n10 [D loss: 0.210437, acc.: 92.19%] [G loss: 0.000570]\n11 [D loss: 0.230503, acc.: 92.19%] [G loss: 0.000295]\n12 [D loss: 0.224950, acc.: 90.62%] [G loss: 0.000174]\n13 [D loss: 0.241756, acc.: 90.62%] [G loss: 0.000258]\n14 [D loss: 0.132709, acc.: 96.88%] [G loss: 0.000145]\n15 [D loss: 0.189781, acc.: 95.31%] [G loss: 0.000062]\n16 [D loss: 0.130304, acc.: 96.88%] [G loss: 0.000039]\n17 [D loss: 0.233896, acc.: 90.62%] [G loss: 0.000340]\n18 [D loss: 0.187010, acc.: 95.31%] [G loss: 0.000132]\n19 [D loss: 0.220850, acc.: 92.19%] [G loss: 0.000070]\n20 [D loss: 0.255564, acc.: 87.50%] [G loss: 0.000020]\n21 [D loss: 0.272240, acc.: 90.62%] [G loss: 0.000057]\n22 [D loss: 0.170161, acc.: 93.75%] [G loss: 0.000018]\n23 [D loss: 0.156027, acc.: 95.31%] [G loss: 0.000056]\n24 [D loss: 0.183404, acc.: 93.75%] [G loss: 0.000006]\n25 [D loss: 0.176130, acc.: 95.31%] [G loss: 0.000039]\n26 [D loss: 0.128596, acc.: 95.31%] [G loss: 0.000022]\n27 [D loss: 0.253872, acc.: 90.62%] [G loss: 0.000012]\n28 [D loss: 0.174997, acc.: 92.19%] [G loss: 0.000007]\n29 [D loss: 0.097610, acc.: 98.44%] [G loss: 0.000027]\n30 [D loss: 0.131244, acc.: 95.31%] [G loss: 0.000004]\n31 [D loss: 0.137328, acc.: 96.88%] [G loss: 0.000013]\n32 [D loss: 0.144717, acc.: 90.62%] [G loss: 0.000003]\n33 [D loss: 0.298392, acc.: 90.62%] [G loss: 0.000032]\n34 [D loss: 0.163233, acc.: 96.88%] [G loss: 0.000002]\n35 [D loss: 0.079941, acc.: 96.88%] [G loss: 0.000004]\n36 [D loss: 0.078496, acc.: 98.44%] [G loss: 0.000001]\n37 [D loss: 0.121086, acc.: 93.75%] [G loss: 0.000000]\n38 [D loss: 0.103405, acc.: 95.31%] [G loss: 0.000001]\n39 [D loss: 0.084932, acc.: 96.88%] [G loss: 0.000001]\n40 [D loss: 0.127238, acc.: 95.31%] [G loss: 0.000005]\n41 [D loss: 0.100307, acc.: 96.88%] [G loss: 0.000003]\n42 [D loss: 0.137738, acc.: 95.31%] [G loss: 0.000002]\n43 [D loss: 0.151586, acc.: 90.62%] [G loss: 0.000001]\n44 [D loss: 0.135681, acc.: 93.75%] [G loss: 0.000001]\n45 [D loss: 0.063676, acc.: 98.44%] [G loss: 0.000002]\n46 [D loss: 0.213562, acc.: 93.75%] [G loss: 0.000001]\n47 [D loss: 0.208648, acc.: 95.31%] [G loss: 0.000001]\n48 [D loss: 0.137129, acc.: 93.75%] [G loss: 0.000000]\n49 [D loss: 0.136670, acc.: 95.31%] [G loss: 0.000000]\n[+] \nTraining the model with DT classifier\n\nModel: \"substitute_detector\"\n_________________________________________________________________\n Layer (type) Output Shape Param # \n=================================================================\n input_36 (InputLayer) [(None, 69)] 0 \n \n dense_49 (Dense) (None, 64) 4480 \n \n dense_50 (Dense) (None, 64) 4160 \n \n dense_51 (Dense) (None, 1) 65 \n \n activation_14 (Activation) (None, 1) 0 \n \n=================================================================\nTotal params: 8,705\nTrainable params: 8,705\nNon-trainable params: 0\n_________________________________________________________________\nModel: \"generator\"\n__________________________________________________________________________________________________\n Layer (type) Output Shape Param # Connected to \n==================================================================================================\n input_37 (InputLayer) [(None, 69)] 0 [] \n \n input_38 (InputLayer) [(None, 30)] 0 [] \n \n concatenate_7 (Concatenate) (None, 99) 0 ['input_37[0][0]', \n 'input_38[0][0]'] \n \n dense_52 (Dense) (None, 32) 3200 ['concatenate_7[0][0]'] \n \n dense_53 (Dense) (None, 32) 1056 ['dense_52[0][0]'] \n \n dense_54 (Dense) (None, 64) 2112 ['dense_53[0][0]'] \n \n dense_55 (Dense) (None, 69) 4485 ['dense_54[0][0]'] \n \n activation_15 (Activation) (None, 69) 0 ['dense_55[0][0]'] \n \n maximum_7 (Maximum) (None, 69) 0 ['input_37[0][0]', \n 'activation_15[0][0]'] \n \n==================================================================================================\nTotal params: 10,853\nTrainable params: 10,853\nNon-trainable params: 0\n__________________________________________________________________________________________________\n0 [D loss: 0.470500, acc.: 76.56%] [G loss: 0.015478]\n1 [D loss: 0.281149, acc.: 92.19%] [G loss: 0.007004]\n2 [D loss: 0.249665, acc.: 90.62%] [G loss: 0.003139]\n3 [D loss: 0.274744, acc.: 90.62%] [G loss: 0.002155]\n4 [D loss: 0.277593, acc.: 84.38%] [G loss: 0.001984]\n5 [D loss: 0.268764, acc.: 85.94%] [G loss: 0.001235]\n6 [D loss: 0.264095, acc.: 85.94%] [G loss: 0.000876]\n7 [D loss: 0.234939, acc.: 90.62%] [G loss: 0.001125]\n8 [D loss: 0.259193, acc.: 90.62%] [G loss: 0.001170]\n9 [D loss: 0.195469, acc.: 95.31%] [G loss: 0.001250]\n10 [D loss: 0.177671, acc.: 90.62%] [G loss: 0.000567]\n11 [D loss: 0.119328, acc.: 93.75%] [G loss: 0.002052]\n12 [D loss: 0.203810, acc.: 93.75%] [G loss: 0.000603]\n13 [D loss: 0.196677, acc.: 89.06%] [G loss: 0.001937]\n14 [D loss: 0.250313, acc.: 89.06%] [G loss: 0.000496]\n15 [D loss: 0.307570, acc.: 89.06%] [G loss: 0.001236]\n16 [D loss: 0.148317, acc.: 92.19%] [G loss: 0.000761]\n17 [D loss: 0.183912, acc.: 93.75%] [G loss: 0.001219]\n18 [D loss: 0.395257, acc.: 82.81%] [G loss: 0.000799]\n19 [D loss: 0.209102, acc.: 90.62%] [G loss: 0.000840]\n20 [D loss: 0.309130, acc.: 87.50%] [G loss: 0.001170]\n21 [D loss: 0.137122, acc.: 92.19%] [G loss: 0.000931]\n22 [D loss: 0.206550, acc.: 92.19%] [G loss: 0.000514]\n23 [D loss: 0.144322, acc.: 96.88%] [G loss: 0.000513]\n24 [D loss: 0.248394, acc.: 90.62%] [G loss: 0.001212]\n25 [D loss: 0.215556, acc.: 90.62%] [G loss: 0.000886]\n26 [D loss: 0.195571, acc.: 90.62%] [G loss: 0.000503]\n27 [D loss: 0.274772, acc.: 82.81%] [G loss: 0.000395]\n28 [D loss: 0.384415, acc.: 87.50%] [G loss: 0.000267]\n29 [D loss: 0.165094, acc.: 95.31%] [G loss: 0.000276]\n30 [D loss: 0.195638, acc.: 89.06%] [G loss: 0.000270]\n31 [D loss: 0.293304, acc.: 90.62%] [G loss: 0.000317]\n32 [D loss: 0.314830, acc.: 87.50%] [G loss: 0.000181]\n33 [D loss: 0.132081, acc.: 96.88%] [G loss: 0.000239]\n34 [D loss: 0.202171, acc.: 95.31%] [G loss: 0.000273]\n35 [D loss: 0.228417, acc.: 89.06%] [G loss: 0.000298]\n36 [D loss: 0.219766, acc.: 89.06%] [G loss: 0.000439]\n37 [D loss: 0.166166, acc.: 93.75%] [G loss: 0.000247]\n38 [D loss: 0.164932, acc.: 92.19%] [G loss: 0.000438]\n39 [D loss: 0.209162, acc.: 92.19%] [G loss: 0.000281]\n40 [D loss: 0.218597, acc.: 87.50%] [G loss: 0.000259]\n41 [D loss: 0.300279, acc.: 87.50%] [G loss: 0.000495]\n42 [D loss: 0.216715, acc.: 89.06%] [G loss: 0.000433]\n43 [D loss: 0.237669, acc.: 90.62%] [G loss: 0.000547]\n44 [D loss: 0.190998, acc.: 93.75%] [G loss: 0.000509]\n45 [D loss: 0.276852, acc.: 90.62%] [G loss: 0.000604]\n46 [D loss: 0.213999, acc.: 92.19%] [G loss: 0.000429]\n47 [D loss: 0.167948, acc.: 90.62%] [G loss: 0.000252]\n48 [D loss: 0.218098, acc.: 90.62%] [G loss: 0.000273]\n49 [D loss: 0.182178, acc.: 92.19%] [G loss: 0.000247]\n[+] \nTraining the model with GB classifier\n\nModel: \"substitute_detector\"\n_________________________________________________________________\n Layer (type) Output Shape Param # \n=================================================================\n input_41 (InputLayer) [(None, 69)] 0 \n \n dense_56 (Dense) (None, 64) 4480 \n \n dense_57 (Dense) (None, 64) 4160 \n \n dense_58 (Dense) (None, 1) 65 \n \n activation_16 (Activation) (None, 1) 0 \n \n=================================================================\nTotal params: 8,705\nTrainable params: 8,705\nNon-trainable params: 0\n_________________________________________________________________\nModel: \"generator\"\n__________________________________________________________________________________________________\n Layer (type) Output Shape Param # Connected to \n==================================================================================================\n input_42 (InputLayer) [(None, 69)] 0 [] \n \n input_43 (InputLayer) [(None, 30)] 0 [] \n \n concatenate_8 (Concatenate) (None, 99) 0 ['input_42[0][0]', \n 'input_43[0][0]'] \n \n dense_59 (Dense) (None, 32) 3200 ['concatenate_8[0][0]'] \n \n dense_60 (Dense) (None, 32) 1056 ['dense_59[0][0]'] \n \n dense_61 (Dense) (None, 64) 2112 ['dense_60[0][0]'] \n \n dense_62 (Dense) (None, 69) 4485 ['dense_61[0][0]'] \n \n activation_17 (Activation) (None, 69) 0 ['dense_62[0][0]'] \n \n maximum_8 (Maximum) (None, 69) 0 ['input_42[0][0]', \n 'activation_17[0][0]'] \n \n==================================================================================================\nTotal params: 10,853\nTrainable params: 10,853\nNon-trainable params: 0\n__________________________________________________________________________________________________\n0 [D loss: 0.573416, acc.: 71.88%] [G loss: 0.042877]\n1 [D loss: 0.313247, acc.: 90.62%] [G loss: 0.014194]\n2 [D loss: 0.265982, acc.: 90.62%] [G loss: 0.009463]\n3 [D loss: 0.392246, acc.: 81.25%] [G loss: 0.010947]\n4 [D loss: 0.259212, acc.: 87.50%] [G loss: 0.011197]\n5 [D loss: 0.248394, acc.: 89.06%] [G loss: 0.005363]\n6 [D loss: 0.250565, acc.: 89.06%] [G loss: 0.004234]\n7 [D loss: 0.222129, acc.: 90.62%] [G loss: 0.004072]\n8 [D loss: 0.223532, acc.: 89.06%] [G loss: 0.001930]\n9 [D loss: 0.300477, acc.: 85.94%] [G loss: 0.001843]\n10 [D loss: 0.146298, acc.: 93.75%] [G loss: 0.001027]\n11 [D loss: 0.284012, acc.: 87.50%] [G loss: 0.001909]\n12 [D loss: 0.184279, acc.: 90.62%] [G loss: 0.000594]\n13 [D loss: 0.106002, acc.: 95.31%] [G loss: 0.000729]\n14 [D loss: 0.122617, acc.: 96.88%] [G loss: 0.000205]\n15 [D loss: 0.145742, acc.: 89.06%] [G loss: 0.000101]\n16 [D loss: 0.215438, acc.: 90.62%] [G loss: 0.000074]\n17 [D loss: 0.243511, acc.: 89.06%] [G loss: 0.000081]\n18 [D loss: 0.151225, acc.: 90.62%] [G loss: 0.000060]\n19 [D loss: 0.219926, acc.: 93.75%] [G loss: 0.000134]\n20 [D loss: 0.203309, acc.: 90.62%] [G loss: 0.000166]\n21 [D loss: 0.158108, acc.: 92.19%] [G loss: 0.000054]\n22 [D loss: 0.174401, acc.: 93.75%] [G loss: 0.000076]\n23 [D loss: 0.179505, acc.: 92.19%] [G loss: 0.000153]\n24 [D loss: 0.172350, acc.: 93.75%] [G loss: 0.000060]\n25 [D loss: 0.202041, acc.: 93.75%] [G loss: 0.000059]\n26 [D loss: 0.311082, acc.: 89.06%] [G loss: 0.000049]\n27 [D loss: 0.173305, acc.: 90.62%] [G loss: 0.000071]\n28 [D loss: 0.146377, acc.: 92.19%] [G loss: 0.000034]\n29 [D loss: 0.109577, acc.: 96.88%] [G loss: 0.000011]\n30 [D loss: 0.176321, acc.: 92.19%] [G loss: 0.000047]\n31 [D loss: 0.106688, acc.: 96.88%] [G loss: 0.000028]\n32 [D loss: 0.204070, acc.: 90.62%] [G loss: 0.000030]\n33 [D loss: 0.276153, acc.: 87.50%] [G loss: 0.000033]\n34 [D loss: 0.130390, acc.: 95.31%] [G loss: 0.000022]\n35 [D loss: 0.107560, acc.: 93.75%] [G loss: 0.000018]\n36 [D loss: 0.244759, acc.: 92.19%] [G loss: 0.000029]\n37 [D loss: 0.207966, acc.: 89.06%] [G loss: 0.000065]\n38 [D loss: 0.129487, acc.: 92.19%] [G loss: 0.000009]\n39 [D loss: 0.128798, acc.: 93.75%] [G loss: 0.000041]\n40 [D loss: 0.200801, acc.: 89.06%] [G loss: 0.000046]\n41 [D loss: 0.112566, acc.: 93.75%] [G loss: 0.000020]\n42 [D loss: 0.192431, acc.: 87.50%] [G loss: 0.000037]\n43 [D loss: 0.102662, acc.: 96.88%] [G loss: 0.000028]\n44 [D loss: 0.072342, acc.: 95.31%] [G loss: 0.000033]\n45 [D loss: 0.140514, acc.: 96.88%] [G loss: 0.000014]\n46 [D loss: 0.182096, acc.: 92.19%] [G loss: 0.000027]\n47 [D loss: 0.126825, acc.: 95.31%] [G loss: 0.000007]\n48 [D loss: 0.172399, acc.: 90.62%] [G loss: 0.000026]\n49 [D loss: 0.085044, acc.: 98.44%] [G loss: 0.000014]\n[+] \nTraining the model with Ensem classifier\n\nModel: \"substitute_detector\"\n_________________________________________________________________\n Layer (type) Output Shape Param # \n=================================================================\n input_46 (InputLayer) [(None, 69)] 0 \n \n dense_63 (Dense) (None, 64) 4480 \n \n dense_64 (Dense) (None, 64) 4160 \n \n dense_65 (Dense) (None, 1) 65 \n \n activation_18 (Activation) (None, 1) 0 \n \n=================================================================\nTotal params: 8,705\nTrainable params: 8,705\nNon-trainable params: 0\n_________________________________________________________________\nModel: \"generator\"\n__________________________________________________________________________________________________\n Layer (type) Output Shape Param # Connected to \n==================================================================================================\n input_47 (InputLayer) [(None, 69)] 0 [] \n \n input_48 (InputLayer) [(None, 30)] 0 [] \n \n concatenate_9 (Concatenate) (None, 99) 0 ['input_47[0][0]', \n 'input_48[0][0]'] \n \n dense_66 (Dense) (None, 32) 3200 ['concatenate_9[0][0]'] \n \n dense_67 (Dense) (None, 32) 1056 ['dense_66[0][0]'] \n \n dense_68 (Dense) (None, 64) 2112 ['dense_67[0][0]'] \n \n dense_69 (Dense) (None, 69) 4485 ['dense_68[0][0]'] \n \n activation_19 (Activation) (None, 69) 0 ['dense_69[0][0]'] \n \n maximum_9 (Maximum) (None, 69) 0 ['input_47[0][0]', \n 'activation_19[0][0]'] \n \n==================================================================================================\nTotal params: 10,853\nTrainable params: 10,853\nNon-trainable params: 0\n__________________________________________________________________________________________________\n0 [D loss: 0.725446, acc.: 64.06%] [G loss: 0.106034]\n1 [D loss: 0.472506, acc.: 82.81%] [G loss: 0.044921]\n2 [D loss: 0.493638, acc.: 78.12%] [G loss: 0.046864]\n3 [D loss: 0.429565, acc.: 81.25%] [G loss: 0.024836]\n4 [D loss: 0.472476, acc.: 79.69%] [G loss: 0.011504]\n5 [D loss: 0.355399, acc.: 82.81%] [G loss: 0.006065]\n6 [D loss: 0.352146, acc.: 89.06%] [G loss: 0.005009]\n7 [D loss: 0.261766, acc.: 87.50%] [G loss: 0.005003]\n8 [D loss: 0.435049, acc.: 81.25%] [G loss: 0.003219]\n9 [D loss: 0.323544, acc.: 87.50%] [G loss: 0.002419]\n10 [D loss: 0.309001, acc.: 85.94%] [G loss: 0.002507]\n11 [D loss: 0.217984, acc.: 90.62%] [G loss: 0.002454]\n12 [D loss: 0.318078, acc.: 89.06%] [G loss: 0.002410]\n13 [D loss: 0.295441, acc.: 85.94%] [G loss: 0.001993]\n14 [D loss: 0.294457, acc.: 92.19%] [G loss: 0.001727]\n15 [D loss: 0.305529, acc.: 87.50%] [G loss: 0.001971]\n16 [D loss: 0.293821, acc.: 90.62%] [G loss: 0.001392]\n17 [D loss: 0.263283, acc.: 92.19%] [G loss: 0.001976]\n18 [D loss: 0.332522, acc.: 87.50%] [G loss: 0.001959]\n19 [D loss: 0.396115, acc.: 82.81%] [G loss: 0.000624]\n20 [D loss: 0.432034, acc.: 81.25%] [G loss: 0.001168]\n21 [D loss: 0.357209, acc.: 84.38%] [G loss: 0.001341]\n22 [D loss: 0.266621, acc.: 87.50%] [G loss: 0.001281]\n23 [D loss: 0.277092, acc.: 89.06%] [G loss: 0.000914]\n24 [D loss: 0.200488, acc.: 95.31%] [G loss: 0.000854]\n25 [D loss: 0.319952, acc.: 87.50%] [G loss: 0.000716]\n26 [D loss: 0.314751, acc.: 87.50%] [G loss: 0.000990]\n27 [D loss: 0.355572, acc.: 85.94%] [G loss: 0.001957]\n28 [D loss: 0.273745, acc.: 89.06%] [G loss: 0.000668]\n29 [D loss: 0.184017, acc.: 92.19%] [G loss: 0.000616]\n30 [D loss: 0.247044, acc.: 89.06%] [G loss: 0.000610]\n31 [D loss: 0.436773, acc.: 79.69%] [G loss: 0.000571]\n32 [D loss: 0.354982, acc.: 84.38%] [G loss: 0.001340]\n33 [D loss: 0.240508, acc.: 90.62%] [G loss: 0.001228]\n34 [D loss: 0.235917, acc.: 92.19%] [G loss: 0.000923]\n35 [D loss: 0.287815, acc.: 87.50%] [G loss: 0.000744]\n36 [D loss: 0.320785, acc.: 87.50%] [G loss: 0.001186]\n37 [D loss: 0.263758, acc.: 87.50%] [G loss: 0.001270]\n38 [D loss: 0.356739, acc.: 85.94%] [G loss: 0.001088]\n39 [D loss: 0.353877, acc.: 82.81%] [G loss: 0.001489]\n40 [D loss: 0.248005, acc.: 90.62%] [G loss: 0.000971]\n41 [D loss: 0.206246, acc.: 90.62%] [G loss: 0.000793]\n42 [D loss: 0.189100, acc.: 93.75%] [G loss: 0.000844]\n43 [D loss: 0.330642, acc.: 90.62%] [G loss: 0.000715]\n44 [D loss: 0.334137, acc.: 85.94%] [G loss: 0.000845]\n45 [D loss: 0.271033, acc.: 85.94%] [G loss: 0.002023]\n46 [D loss: 0.249067, acc.: 89.06%] [G loss: 0.001103]\n47 [D loss: 0.311791, acc.: 84.38%] [G loss: 0.000498]\n48 [D loss: 0.384985, acc.: 84.38%] [G loss: 0.000987]\n49 [D loss: 0.319834, acc.: 89.06%] [G loss: 0.001135]\n=====================\n{'SVM': 0.2143743298947811, 'SGD': 0.1366698555648327, 'DT': 0.18217830266803503, 'GB': 0.08504400122910738, 'Ensem': 0.31983402371406555}\n=====================\n{'SVM': 92.1875, 'SGD': 95.3125, 'DT': 92.1875, 'GB': 98.4375, 'Ensem': 89.0625}\n=====================\n{'SVM': 3.931344494390032e-08, 'SGD': 2.905404699049541e-07, 'DT': 0.00024743395624682307, 'GB': 1.3990780644235201e-05, 'Ensem': 0.0011351570719853044}\n"
],
[
"\nmatrix_dict = {}\n\nfor key, value in D_loss_dict.items():\n matrix_dict[key] = []\n\n\nfor key, value in D_loss_dict.items():\n matrix_dict[key].append(D_loss_dict[key])\n matrix_dict[key].append(Acc_dict[key])\n matrix_dict[key].append(G_loss_dict[key])",
"_____no_output_____"
],
[
"import pandas as pd\n\ndf = pd.DataFrame.from_dict(matrix_dict, orient='columns') \ndf.index= list([ 'D_Loss', 'Acc', 'G_Loss'])\ndf",
"_____no_output_____"
],
[
"import dataframe_image as dfi\ndfi.export(df, '64_mal_matrix.png')",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbf15b7a24a514aaf48bb9a8adcc25579bee26de
| 6,305 |
ipynb
|
Jupyter Notebook
|
ner/ner.ipynb
|
v-smwang/AI-NLP-Tutorial
|
3dbfdc7e19a025e00febab97f4948da8a3710f34
|
[
"Apache-2.0"
] | null | null | null |
ner/ner.ipynb
|
v-smwang/AI-NLP-Tutorial
|
3dbfdc7e19a025e00febab97f4948da8a3710f34
|
[
"Apache-2.0"
] | null | null | null |
ner/ner.ipynb
|
v-smwang/AI-NLP-Tutorial
|
3dbfdc7e19a025e00febab97f4948da8a3710f34
|
[
"Apache-2.0"
] | null | null | null | 34.453552 | 226 | 0.57161 |
[
[
[
"<font color=\"red\">训练直接将样本的分词+词性 改为 分词+命名实体类型即可</font>\n\n## 目录\n- [8. 命名实体识别](#8-命名实体识别)\n- [8.1 概述](#81-概述)\n- [8.2 基于隐马尔可夫模型序列标注的命名实体识别](#82-基于隐马尔可夫模型序列标注的命名实体识别)\n- [8.3 基于感知机序列标注的命名实体识别](#83-基于感知机序列标注的命名实体识别)\n- [8.4 基于条件随机场序列标注的命名实体识别](#84-基于条件随机场序列标注的命名实体识别)\n- [8.5 命名实体识别标准化评测](#85-命名实体识别标准化评测)\n- [8.6 自定义领域命名实体识别](#86-自定义领域命名实体识别)\n\n## 8. 命名实体识别\n\n### 8.1 概述\n\n1. **命名实体**\n\n 文本中有一些描述实体的词汇。比如人名、地名、组织机构名、股票基金、医学术语等,称为**命名实体**。具有以下共性:\n\n - 数量无穷。比如宇宙中的恒星命名、新生儿的命名不断出现新组合。\n - 构词灵活。比如中国工商银行,既可以称为工商银行,也可以简称工行。\n - 类别模糊。有一些地名本身就是机构名,比如“国家博物馆”\n\n2. **命名实体识别**\n\n 识别出句子中命名实体的边界与类别的任务称为**命名实体识别**。由于上述难点,命名实体识别也是一个统计为主、规则为辅的任务。\n\n 对于规则性较强的命名实体,比如网址、E-mail、IBSN、商品编号等,完全可以通过正则表达式处理,未匹配上的片段交给统计模型处理。\n\n 命名实体识别也可以转化为一个序列标注问题。具体做法是将命名实体识别附着到{B,M,E,S}标签,比如, 构成地名的单词标注为“B/ME/S- 地名”,以此类推。对于那些命名实体边界之外的单词,则统一标注为0 ( Outside )。具体实施时,HanLP做了一个简化,即所有非复合词的命名实体都标注为S,不再附着类别。这样标注集更精简,模型更小巧。\n\n命名实体识别实际上可以看作分词与词性标注任务的集成: 命名实体的边界可以通过{B,M,E,S}确定,其类别可以通过 B-nt 等附加类别的标签来确定。\n\nHanLP内部提供了语料库转换工序,用户无需关心,只需要传入 PKU 格式的语料库路径即可。\n\n\n\n### 8.2 基于隐马尔可夫模型序列标注的命名实体识别\n\n之前我们就介绍过隐马尔可夫模型,详细见: [4.隐马尔可夫模型与序列标注](https://github.com/NLP-LOVE/Introduction-NLP/blob/master/chapter/4.%E9%9A%90%E9%A9%AC%E5%B0%94%E5%8F%AF%E5%A4%AB%E6%A8%A1%E5%9E%8B%E4%B8%8E%E5%BA%8F%E5%88%97%E6%A0%87%E6%B3%A8.md)\n\n隐马尔可夫模型命名实体识别代码见(**自动下载 PKU 语料库**): hmm_ner.py\n\n[https://github.com/NLP-LOVE/Introduction-NLP/tree/master/code/ch08/hmm_ner.py](https://github.com/NLP-LOVE/Introduction-NLP/tree/master/code/ch08/hmm_ner.py)\n\n运行代码后结果如下:\n\n```\n华北电力公司/nt 董事长/n 谭旭光/nr 和/c 秘书/n 胡花蕊/nr 来到/v 美国纽约/ns 现代/ntc 艺术/n 博物馆/n 参观/v\n```\n\n其中机构名“华北电力公司”、人名“谭旭光”“胡花蕊”全部识别正确。但是地名“美国纽约现代艺术博物馆”则无法识别。有以下两个原因:\n\n- PKU 语料库中没有出现过这个样本。\n- 隐马尔可夫模型无法利用词性特征。\n\n对于第一个原因,只能额外标注一些语料。对于第二个原因可以通过切换到更强大的模型来解决。\n\n\n\n### 8.3 基于感知机序列标注的命名实体识别\n\n之前我们就介绍过感知机模型,详细见: [5.感知机分类与序列标注](https://github.com/NLP-LOVE/Introduction-NLP/blob/master/chapter/5.%E6%84%9F%E7%9F%A5%E6%9C%BA%E5%88%86%E7%B1%BB%E4%B8%8E%E5%BA%8F%E5%88%97%E6%A0%87%E6%B3%A8.md)\n\n感知机模型词性标注代码见(**自动下载 PKU 语料库**): perceptron_ner.py\n\n[https://github.com/NLP-LOVE/Introduction-NLP/tree/master/code/ch08/perceptron_ner.py](https://github.com/NLP-LOVE/Introduction-NLP/tree/master/code/ch08/perceptron_ner.py)\n\n运行会有些慢,结果如下:\n\n```\n华北电力公司/nt 董事长/n 谭旭光/nr 和/c 秘书/n 胡花蕊/nr 来到/v [美国纽约/ns 现代/ntc 艺术/n 博物馆/n]/ns 参观/v\n```\n\n与隐马尔可夫模型相比,已经能够正确识别地名了。\n\n\n\n### 8.4 基于条件随机场序列标注的命名实体识别\n\n之前我们就介绍过条件随机场模型,详细见: [6.条件随机场与序列标注](https://github.com/NLP-LOVE/Introduction-NLP/blob/master/chapter/6.%E6%9D%A1%E4%BB%B6%E9%9A%8F%E6%9C%BA%E5%9C%BA%E4%B8%8E%E5%BA%8F%E5%88%97%E6%A0%87%E6%B3%A8.md)\n\n条件随机场模型词性标注代码见(**自动下载 PKU 语料库**): crf_ner.py\n\n[https://github.com/NLP-LOVE/Introduction-NLP/tree/master/code/ch08/crf_ner.py](https://github.com/NLP-LOVE/Introduction-NLP/tree/master/code/ch08/crf_ner.py)\n\n运行时间会比较长,结果如下:\n\n```\n华北电力公司/nt 董事长/n 谭旭光/nr 和/c 秘书/n 胡花蕊/nr 来到/v [美国纽约/ns 现代/ntc 艺术/n 博物馆/n]/ns 参观/v\n```\n\n得到了结果是一样的。\n\n\n\n### 8.5 命名实体识别标准化评测\n\n各个命名实体识别模块的准确率如何,并非只能通过几个句子主观感受。任何监督学习任务都有一套标准化评测方案,对于命名实体识别,按照惯例引入P、R 和 F1 评测指标。\n\n在1998年1月《人民日报》语料库上的标准化评测结果如下:\n\n| 模型 | P | R | F1 |\n| -------------- | ----- | ----- | ----- |\n| 隐马尔可夫模型 | 79.01 | 30.14 | 43.64 |\n| 感知机 | 87.33 | 78.98 | 82.94 |\n| 条件随机场 | 87.93 | 73.75 | 80.22 |\n\n值得一提的是,准确率与评测策略、特征模板、语料库规模息息相关。通常而言,当语料库较小时,应当使用简单的特征模板,以防止模型过拟合;当语料库较大时,则建议使用更多特征,以期更高的准确率。当特征模板固定时,往往是语料库越大,准确率越高。\n\n\n\n### 8.6 自定义领域命名实体识别\n\n以上我们接触的都是通用领域上的语料库,所含的命名实体仅限于人名、地名、机构名等。假设我们想要识别专门领域中的命名实体,这时,我们就要自定义领域的语料库了。\n\n1. **标注领域命名实体识别语料库**\n\n 首先我们需要收集一些文本, 作为标注语料库的原料,称为**生语料**。由于我们的目标是识别文本中的战斗机名称或型号,所以生语料的来源应当是些军事网站的报道。在实际工程中,求由客户提出,则应当由该客户提供生语料。语料的量级越大越好,一般最低不少于数千个句子。\n\n 生语料准备就绪后,就可以开始标注了。对于命名实体识别语料库,若以词语和词性为特征的话,还需要标注分词边界和词性。不过我们不必从零开始标注,而可以在HanLP的标注基础上进行校正,这样工作量更小。\n\n 样本标注了数千个之后,生语料就被标注成了**熟语料**。下面代码自动下载语料库。\n\n2. **训练领域模型**\n\n 选择感知机作为训练算法(**自动下载 战斗机 语料库**): plane_ner.py\n\n [https://github.com/NLP-LOVE/Introduction-NLP/tree/master/code/ch08/plane_ner.py](https://github.com/NLP-LOVE/Introduction-NLP/tree/master/code/ch08/plane_ner.py)\n\n 运行结果如下:\n\n ```\n 下载 http://file.hankcs.com/corpus/plane-re.zip 到 /usr/local/lib/python3.7/site-packages/pyhanlp/static/data/test/plane-re.zip\n 100.00%, 0 MB, 552 KB/s, 还有 0 分 0 秒 \n 米高扬/nrf 设计/v [米格/nr -/w 17/m PF/nx]/np :/w [米格/nr -/w 17/m]/np PF/n 型/k 战斗机/n 比/p [米格/nr -/w 17/m P/nx]/np 性能/n 更好/l 。/w\n [米格/nr -/w 阿帕奇/nrf -/w 666/m S/q]/np 横空出世/l 。/w\n ```\n\n 这句话已经在语料库中出现过,能被正常识别并不意外。我们可以伪造一款“米格-阿帕奇-666S”战斗机,试试模型的繁华能力,发现依然能够正确识别。\n\n\n\n\n",
"_____no_output_____"
]
]
] |
[
"markdown"
] |
[
[
"markdown"
]
] |
cbf1685c22d50494e98d526dc703db899c3d63d2
| 5,030 |
ipynb
|
Jupyter Notebook
|
Functions 2.ipynb
|
sumathi-kanthakumar/PythonITMFCS
|
720874976907b79a688377d794c0f14dcf77979c
|
[
"MIT"
] | null | null | null |
Functions 2.ipynb
|
sumathi-kanthakumar/PythonITMFCS
|
720874976907b79a688377d794c0f14dcf77979c
|
[
"MIT"
] | null | null | null |
Functions 2.ipynb
|
sumathi-kanthakumar/PythonITMFCS
|
720874976907b79a688377d794c0f14dcf77979c
|
[
"MIT"
] | null | null | null | 17.900356 | 92 | 0.452883 |
[
[
[
"# *args",
"_____no_output_____"
]
],
[
[
"def myfunc(a,b):\n #return a+b\n return sum((a,b))",
"_____no_output_____"
],
[
"myfunc(1,5)",
"_____no_output_____"
],
[
"# problem in the above function is we cannot change the number of arguments. Always 2.",
"_____no_output_____"
],
[
"def myNewFunc(*numbers):\n return sum(numbers)",
"_____no_output_____"
],
[
"myNewFunc(1,3,4)",
"_____no_output_____"
],
[
"myNewFunc(1)\nmyNewFunc(2)",
"_____no_output_____"
],
[
"myNewFunc()",
"_____no_output_____"
]
],
[
[
"# **kwargs",
"_____no_output_____"
]
],
[
[
"def my_sec_func(**kwargs):\n for k in kwargs:\n print (k)\n if 'fruit' in kwargs:\n print(f\"My Fav fruit is {kwargs['fruit']}\")\n elif 'veggie' in kwargs:\n print(f\"My Fav veggie is {kwargs['veggie']}\")\n else:\n print(\"No fruits in the list\")\n \n",
"_____no_output_____"
],
[
"my_sec_func(fruit='Apple')",
"My Fav fruit is Apple\n"
],
[
"my_sec_func(fruit='Apple',veggie='potato')",
"fruit\nveggie\nMy Fav fruit is Apple\n"
]
],
[
[
"# Exercise: Take name of the student and marks and print in a formatted Report Card",
"_____no_output_____"
]
],
[
[
"\ndef print_report_card(**markList):\n print(f\"Name | Mark\")\n print(f\"=============\")\n for name,mark in markList.items():\n print(f\"{name} | {mark}\")\n ",
"_____no_output_____"
],
[
"print_report_card(kayal='85', sai='50')",
"Name | Mark\n=============\nkayal | 85\nsai | 50\n"
],
[
"#print_report_card({'kayal':'85', 'sai':'80'}) => TO DO Why not Dictionary",
"_____no_output_____"
],
[
"#python -m pip install <package-name>",
"_____no_output_____"
],
[
"! set PATH=%PATH%;c:\\\\ProgramData\\Anaconda3",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
cbf16c7da25b9b3e78847f9680a07c727c0a56f3
| 74,205 |
ipynb
|
Jupyter Notebook
|
week-4/week-4-web-scraping.ipynb
|
pg2455/accessing-research-data
|
38711b74e0f881d29d00fc558c17380dadcfc90e
|
[
"CC0-1.0"
] | null | null | null |
week-4/week-4-web-scraping.ipynb
|
pg2455/accessing-research-data
|
38711b74e0f881d29d00fc558c17380dadcfc90e
|
[
"CC0-1.0"
] | null | null | null |
week-4/week-4-web-scraping.ipynb
|
pg2455/accessing-research-data
|
38711b74e0f881d29d00fc558c17380dadcfc90e
|
[
"CC0-1.0"
] | null | null | null | 39.491751 | 3,072 | 0.634957 |
[
[
[
"empty"
]
]
] |
[
"empty"
] |
[
[
"empty"
]
] |
cbf16ca932d7272c026fb11c800430f8d1b317d0
| 3,875 |
ipynb
|
Jupyter Notebook
|
notebooks/Misc.ipynb
|
gngdb/opencast-bio
|
9fb110076295aafa696a9f8b5070b8d93c6400ce
|
[
"MIT"
] | 2 |
2016-02-24T20:44:39.000Z
|
2020-07-06T02:44:38.000Z
|
notebooks/Misc.ipynb
|
gngdb/opencast-bio
|
9fb110076295aafa696a9f8b5070b8d93c6400ce
|
[
"MIT"
] | null | null | null |
notebooks/Misc.ipynb
|
gngdb/opencast-bio
|
9fb110076295aafa696a9f8b5070b8d93c6400ce
|
[
"MIT"
] | null | null | null | 21.527778 | 226 | 0.481032 |
[
[
[
"empty"
]
]
] |
[
"empty"
] |
[
[
"empty"
]
] |
cbf16f6e1ea64a8b2fadb7857b7aea4b8ac71c6c
| 9,719 |
ipynb
|
Jupyter Notebook
|
vPythonSetup.ipynb
|
smithrockmaker/PH213
|
1f5796337c1f17f649532f6ccdfb59f02e397963
|
[
"MIT"
] | null | null | null |
vPythonSetup.ipynb
|
smithrockmaker/PH213
|
1f5796337c1f17f649532f6ccdfb59f02e397963
|
[
"MIT"
] | null | null | null |
vPythonSetup.ipynb
|
smithrockmaker/PH213
|
1f5796337c1f17f649532f6ccdfb59f02e397963
|
[
"MIT"
] | null | null | null | 35.863469 | 383 | 0.569091 |
[
[
[
"# Setting up vPython in Jupyterlab\n\n## Version Compatibility\n\nAt this time (10/20) vPython appears to be compatible with verisons of Jupyterlab through 1.2.6. You may need to remove your current install of Jupyterlab to do this. To install specific versions of Jupyterlab (and remove the application) go to the settings icon in the top right corner of the Jupyterlab frame on the Home tab in Anaconda Navigator.\n\n\n \n\nThe most effective instructions I have found for installing vpython in a Jupyterlab environment are [here - thanks to Bruce Sherwood](https://www.vpython.org/presentation2018/install.html). Here are the principle steps in that process.\n\nPreparation: Launch Juypterlab from Anaconda Navigator which should open a window in your browser.\n\ni: Update python by going to the Environments tab on Anaconda Navigator. Choose to display the Installed environments and scroll down to find your python package. To the right is a column which indicates the version number. If there is a blue arrow there indicating an update to the package you can click to install. At the time of this document my python version was 3.8.3.\n\n \n\nii: From the File drop down menu (in the Jupyterlab tab on your browser) select New Launcher and launch a Terminal window.\n\n\n\n\n\niii: In that terminal window you will execute the following commands (from the document linked above). Anaconda will sort out what needs to be installed and updated. For the two installs it will ask you if you want to proceed at some point and you need to enter y (y/n are options).\n\n```conda install -c vpython vpython``` \n```conda install -c anaconda nodejs``` \n\n\n\niv: Assuming that you get no error messages in the previous installs then the next terminal command is connects vpython to the Jupyterlab environment. \n\n```jupyter labextension install vpython```\n\nIf this labextension install gives you conflicts it may be that your installed version of Jupyterlab is too new.\n\nv: From the File drop down menu select Shut Down to exit from Jupyterlab and close the tab in your browser. \n\nvi: Relaunch Jupyterlab and the sample vPython code below should execute correctly.",
"_____no_output_____"
],
[
"## Downloading this Notebook\n\nIf you are reading this notebook you probably found the [github](https://github.com/smithrockmaker/PH213) where it is hosted. Generally one clones the entire github repository but for physics classes we often wish to download only one of the notebooks hosted on the github. Here is the process that appears to work.\n\ni: Select the notebook of interest which will be rendered so that you can explore it. \nii At the top right of the menu bar is a 'raw' button. 'Right click' on the raw button and select 'Save Link As..'\n\n \n\n\niii: The downloaded file will have an .ipynb suffix. To correctly display this page on your computer you will need to got to the images folder on the github and download the relevant images to an appropiate folder.",
"_____no_output_____"
]
],
[
[
"from vpython import *",
"_____no_output_____"
],
[
"scene=canvas(title=\"Constant Velocity\",x=0, y=0, width=800,\n height=600, autoscale=0, center=vector(0,0,0))\nt = 0.0\ndt = .1\ns = vector(0.0,0.0,0.0)\nv = vector(5.0,0.0,0.0)\na = vector(2.0, 0.0,0.0)\ncart = sphere(pos=s,radius = .3, color = color.blue)\nwhile t < 1.5:\n rate(10)\n s = s+v*dt\n v = v+a*dt\n print ('t =',t, 's =',s, 'v =',v)\n cart.pos=s\n ballghost = sphere(pos=s,radius=.1, color = color.yellow)\n t = t + dt",
"_____no_output_____"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code"
]
] |
cbf171eb9abfb967b58d7567c0a1b6844686a0a6
| 21,749 |
ipynb
|
Jupyter Notebook
|
m04_machine_learning/m04_c04_metrics_and_model_selection/m04_c04_lab.ipynb
|
simonmasnu/mat281_portfolio
|
9c7c0b5747c8db33a75d700fbfdb3ad61017e65c
|
[
"MIT",
"BSD-3-Clause"
] | null | null | null |
m04_machine_learning/m04_c04_metrics_and_model_selection/m04_c04_lab.ipynb
|
simonmasnu/mat281_portfolio
|
9c7c0b5747c8db33a75d700fbfdb3ad61017e65c
|
[
"MIT",
"BSD-3-Clause"
] | null | null | null |
m04_machine_learning/m04_c04_metrics_and_model_selection/m04_c04_lab.ipynb
|
simonmasnu/mat281_portfolio
|
9c7c0b5747c8db33a75d700fbfdb3ad61017e65c
|
[
"MIT",
"BSD-3-Clause"
] | null | null | null | 30.849645 | 262 | 0.444204 |
[
[
[
"<img src=\"https://upload.wikimedia.org/wikipedia/commons/4/47/Logo_UTFSM.png\" width=\"200\" alt=\"utfsm-logo\" align=\"left\"/>\n\n# MAT281\n### Aplicaciones de la Matemática en la Ingeniería",
"_____no_output_____"
],
[
"## Módulo 04\n## Laboratorio Clase 04: Métricas y selección de modelos",
"_____no_output_____"
],
[
"### Instrucciones\n\n\n* Completa tus datos personales (nombre y rol USM) en siguiente celda.\n* La escala es de 0 a 4 considerando solo valores enteros.\n* Debes _pushear_ tus cambios a tu repositorio personal del curso.\n* Como respaldo, debes enviar un archivo .zip con el siguiente formato `mXX_cYY_lab_apellido_nombre.zip` a [email protected], debe contener todo lo necesario para que se ejecute correctamente cada celda, ya sea datos, imágenes, scripts, etc.\n* Se evaluará:\n - Soluciones\n - Código\n - Que Binder esté bien configurado.\n - Al presionar `Kernel -> Restart Kernel and Run All Cells` deben ejecutarse todas las celdas sin error.",
"_____no_output_____"
],
[
"__Nombre__: Simón Masnú\n\n__Rol__: 201503026-K",
"_____no_output_____"
],
[
"En este laboratorio utilizaremos el conjunto de datos _Abolone_. ",
"_____no_output_____"
],
[
"**Recuerdo**\n\nLa base de datos contiene mediciones a 4177 abalones, donde las mediciones posibles son sexo ($S$), peso entero $W_1$, peso sin concha $W_2$, peso de visceras $W_3$, peso de concha $W_4$, largo ($L$), diametro $D$, altura $H$, y el número de anillos $A$. ",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np",
"_____no_output_____"
],
[
"abalone = pd.read_csv(\n \"data/abalone.data\",\n header=None,\n names=[\"sex\", \"length\", \"diameter\", \"height\", \"whole_weight\", \"shucked_weight\", \"viscera_weight\", \"shell_weight\", \"rings\"]\n)\n\nabalone_data = (\n abalone.assign(sex=lambda x: x[\"sex\"].map({\"M\": 1, \"I\": 0, \"F\": -1}))\n .loc[lambda x: x.drop(columns=\"sex\").gt(0).all(axis=1)]\n .astype(np.float)\n)\nabalone_data.head()",
"_____no_output_____"
]
],
[
[
"#### Modelo A\nConsideramos 9 parámetros, llamados $\\alpha_i$, para el siguiente modelo:\n$$ \\log(A) = \\alpha_0 + \\alpha_1 W_1 + \\alpha_2 W_2 +\\alpha_3 W_3 +\\alpha_4 W_4 + \\alpha_5 S + \\alpha_6 \\log L + \\alpha_7 \\log D+ \\alpha_8 \\log H$$",
"_____no_output_____"
]
],
[
[
"def train_model_A(data):\n y = np.log(data.loc[:, \"rings\"].values.ravel())\n X = (\n data.assign(\n intercept=1.,\n length=lambda x: x[\"length\"].apply(np.log),\n diameter=lambda x: x[\"diameter\"].apply(np.log),\n height=lambda x: x[\"height\"].apply(np.log),\n )\n .loc[: , [\"intercept\", \"whole_weight\", \"shucked_weight\", \"viscera_weight\", \"shell_weight\", \"sex\", \"length\", \"diameter\", \"height\"]]\n .values\n )\n coeffs = np.linalg.lstsq(X, y, rcond=None)[0]\n return coeffs\n\ndef test_model_A(data, coeffs):\n X = (\n data.assign(\n intercept=1.,\n length=lambda x: x[\"length\"].apply(np.log),\n diameter=lambda x: x[\"diameter\"].apply(np.log),\n height=lambda x: x[\"height\"].apply(np.log),\n )\n .loc[: , [\"intercept\", \"whole_weight\", \"shucked_weight\", \"viscera_weight\", \"shell_weight\", \"sex\", \"length\", \"diameter\", \"height\"]]\n .values\n )\n ln_anillos = np.dot(X, coeffs)\n return np.exp(ln_anillos)",
"_____no_output_____"
]
],
[
[
"#### Modelo B\nConsideramos 6 parámetros, llamados $\\beta_i$, para el siguiente modelo:\n$$ \\log(A) = \\beta_0 + \\beta_1 W_1 + \\beta_2 W_2 +\\beta_3 W_3 +\\beta W_4 + \\beta_5 \\log( L D H ) $$",
"_____no_output_____"
]
],
[
[
"def train_model_B(data):\n y = np.log(data.loc[:, \"rings\"].values.ravel())\n X = (\n data.assign(\n intercept=1.,\n ldh=lambda x: (x[\"length\"] * x[\"diameter\"] * x[\"height\"]).apply(np.log),\n )\n .loc[: , [\"intercept\", \"whole_weight\", \"shucked_weight\", \"viscera_weight\", \"shell_weight\", \"ldh\"]]\n .values\n )\n coeffs = np.linalg.lstsq(X, y, rcond=None)[0]\n return coeffs\n\ndef test_model_B(data, coeffs):\n X = (\n data.assign(\n intercept=1.,\n ldh=lambda x: (x[\"length\"] * x[\"diameter\"] * x[\"height\"]).apply(np.log),\n )\n .loc[: , [\"intercept\", \"whole_weight\", \"shucked_weight\", \"viscera_weight\", \"shell_weight\", \"ldh\"]]\n .values\n )\n ln_anillos = np.dot(X, coeffs)\n return np.exp(ln_anillos)",
"_____no_output_____"
]
],
[
[
"#### Modelo C\nConsideramos 12 parámetros, llamados $\\theta_i^{k}$, con $k \\in \\{M, F, I\\}$, para el siguiente modelo:\n\nSi $S=male$:\n$$ \\log(A) = \\theta_0^M + \\theta_1^M W_2 + \\theta_2^M W_4 + \\theta_3^M \\log( L D H ) $$\n\nSi $S=female$\n$$ \\log(A) = \\theta_0^F + \\theta_1^F W_2 + \\theta_2^F W_4 + \\theta_3^F \\log( L D H ) $$\n\nSi $S=indefined$\n$$ \\log(A) = \\theta_0^I + \\theta_1^I W_2 + \\theta_2^I W_4 + \\theta_3^I \\log( L D H ) $$",
"_____no_output_____"
]
],
[
[
"def train_model_C(data):\n df = (\n data.assign(\n intercept=1.,\n ldh=lambda x: (x[\"length\"] * x[\"diameter\"] * x[\"height\"]).apply(np.log),\n )\n .loc[: , [\"intercept\", \"shucked_weight\", \"shell_weight\", \"ldh\", \"sex\", \"rings\"]]\n )\n coeffs_dict = {}\n for sex, df_sex in df.groupby(\"sex\"):\n X = df_sex.drop(columns=[\"sex\", \"rings\"])\n y = np.log(df_sex[\"rings\"].values.ravel())\n coeffs_dict[sex] = np.linalg.lstsq(X, y, rcond=None)[0]\n return coeffs_dict\n\ndef test_model_C(data, coeffs_dict):\n df = (\n data.assign(\n intercept=1.,\n ldh=lambda x: (x[\"length\"] * x[\"diameter\"] * x[\"height\"]).apply(np.log),\n )\n .loc[: , [\"intercept\", \"shucked_weight\", \"shell_weight\", \"ldh\", \"sex\", \"rings\"]]\n )\n pred_dict = {}\n for sex, df_sex in df.groupby(\"sex\"):\n X = df_sex.drop(columns=[\"sex\", \"rings\"])\n ln_anillos = np.dot(X, coeffs_dict[sex])\n pred_dict[sex] = np.exp(ln_anillos)\n return pred_dict",
"_____no_output_____"
]
],
[
[
"### 1. Split Data (1 pto)\n\nCrea dos dataframes, uno de entrenamiento (80% de los datos) y otro de test (20% restante de los datos) a partir de `abalone_data`.\n\n_Hint:_ `sklearn.model_selection.train_test_split` funciona con dataframes!",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import train_test_split\n\nabalone_train, abalone_test = train_test_split(abalone_data, test_size=0.20,random_state=42) #la misma seed de la clase\n\nabalone_train.head()",
"_____no_output_____"
]
],
[
[
"### 2. Entrenamiento (1 pto)\n\nUtilice las funciones de entrenamiento definidas más arriba con tal de obtener los coeficientes para los datos de entrenamiento. Recuerde que para el modelo C se retorna un diccionario donde la llave corresponde a la columna `sex`.",
"_____no_output_____"
]
],
[
[
"coeffs_A = train_model_A(abalone_train)\ncoeffs_B = train_model_B(abalone_train)\ncoeffs_C = train_model_C(abalone_train)",
"_____no_output_____"
]
],
[
[
"### 3. Predicción (1 pto)\n\nCon los coeficientes de los modelos realize la predicción utilizando el conjunto de test. El resultado debe ser un array de shape `(835, )` por lo que debes concatenar los resultados del modelo C. \n\n**Hint**: Usar `np.concatenate`.",
"_____no_output_____"
]
],
[
[
"y_pred_A = test_model_A(abalone_test,coeffs_A)\ny_pred_B = test_model_B(abalone_test,coeffs_B)\ny_pred_C = np.concatenate([test_model_C(abalone_test,coeffs_C)[-1],test_model_C(abalone_test,coeffs_C)[0],test_model_C(abalone_test,coeffs_C)[1]])",
"_____no_output_____"
]
],
[
[
"### 4. Cálculo del error (1 pto)\n\nSe utilizará el Error Cuadrático Medio (MSE) que se define como \n\n$$\\textrm{MSE}(y,\\hat{y}) =\\dfrac{1}{n}\\sum_{t=1}^{n}\\left | y_{t}-\\hat{y}_{t}\\right |^2$$\n\nDefina una la función `MSE` y el vectores `y_test_A`, `y_test_B` e `y_test_C` para luego calcular el error para cada modelo. \n\n**Ojo:** Nota que al calcular el error cuadrático medio se realiza una resta elemento por elemento, por lo que el orden del vector es importante, en particular para el modelo que separa por `sex`.",
"_____no_output_____"
]
],
[
[
"def MSE(y_real, y_pred):\n return sum(np.absolute(y_real-y_pred)**2)/len(y_real) ",
"_____no_output_____"
],
[
"y_test_A = abalone_test.loc[:,'rings']\ny_test_B = abalone_test.loc[:,'rings']\ny_test_C = np.concatenate([abalone_test[ abalone_test['sex'] == -1].loc[:,'rings']\n ,abalone_test[ abalone_test['sex'] == 0].loc[:,'rings']\n ,abalone_test[ abalone_test['sex'] == 1].loc[:,'rings']]\n ,axis=None) #perdon por el Hard-Coding",
"_____no_output_____"
],
[
"error_A = MSE(y_test_A,y_pred_A)\nerror_B = MSE(y_test_B,y_pred_B)\nerror_C = MSE(y_test_C,y_pred_C)",
"_____no_output_____"
],
[
"print(f\"Error modelo A: {error_A:.2f}\")\nprint(f\"Error modelo B: {error_B:.2f}\")\nprint(f\"Error modelo C: {error_C:.2f}\")",
"Error modelo A: 5.13\nError modelo B: 5.12\nError modelo C: 5.22\n"
]
],
[
[
"**¿Cuál es el mejor modelo considerando esta métrica?**",
"_____no_output_____"
],
[
"El mejor modelo considerando como métrica el `MSE` es el modelo **B**.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
]
] |
cbf1a0022f030e8efe43ee003e2492b5f1078215
| 114,507 |
ipynb
|
Jupyter Notebook
|
Chapter05/Exercise45.ipynb
|
JDhyeok/Data-Science-with-Python
|
8d82bb3aa224baf5cf867fb04a047b7be446d63b
|
[
"MIT"
] | null | null | null |
Chapter05/Exercise45.ipynb
|
JDhyeok/Data-Science-with-Python
|
8d82bb3aa224baf5cf867fb04a047b7be446d63b
|
[
"MIT"
] | null | null | null |
Chapter05/Exercise45.ipynb
|
JDhyeok/Data-Science-with-Python
|
8d82bb3aa224baf5cf867fb04a047b7be446d63b
|
[
"MIT"
] | null | null | null | 57.025398 | 7,400 | 0.637192 |
[
[
[
"import numpy as np\nimport pandas as pd",
"_____no_output_____"
],
[
"data = pd.read_csv(\"./data/adult-data.csv\", names=['age', 'workclass', \n 'fnlwgt', 'education-num', 'occupation', 'capital-gain', 'capital-loss', \n 'hours-per-week', 'income'])",
"_____no_output_____"
],
[
"from sklearn.preprocessing import LabelEncoder\ndata['workclass'] = LabelEncoder().fit_transform(data['workclass'])\ndata['occupation'] = LabelEncoder().fit_transform(data['occupation'])\ndata['income'] = LabelEncoder().fit_transform(data['income'])\ndata['education-num'] = LabelEncoder().fit_transform(data['education-num'])\ndata['hours-per-week'] = LabelEncoder().fit_transform(data['hours-per-week'])\n",
"_____no_output_____"
],
[
"data.head(10)",
"_____no_output_____"
],
[
"import xgboost as xgb\nX = data.copy()\nX.drop(\"income\", inplace = True, axis = 1)\nY = data['income']\n\nX_train, X_test = X[:int(X.shape[0]*0.8)].values,X[int(X.shape[0]*0.8):].values\nY_train, Y_test = Y[:int(Y.shape[0]*0.8)].values,Y[int(Y.shape[0]*0.8):].values",
"_____no_output_____"
],
[
"train = xgb.DMatrix(X_train,label=Y_train)\ntest = xgb.DMatrix(X_test,label=Y_test)",
"_____no_output_____"
],
[
"test_error = {}\nfor i in range(20):\n param = {'max_depth':i, 'eta':0.1, 'silent':1, 'objective':'binary:hinge'}\n num_round = 50\n model_metrics = xgb.cv(param,train,num_round,nfold=10)\n test_error[i] = model_metrics.iloc[-1]['test-error-mean']",
"[20:32:15] WARNING: C:/Users/Administrator/workspace/xgboost-win64_release_1.3.0/src/learner.cc:541: \nParameters: { silent } might not be used.\n\n This may not be accurate due to some parameters are only used in language bindings but\n passed down to XGBoost core. Or some parameters are not used but slip through this\n verification. Please open an issue if you find above cases.\n\n\n[20:32:15] WARNING: C:/Users/Administrator/workspace/xgboost-win64_release_1.3.0/src/learner.cc:541: \nParameters: { silent } might not be used.\n\n This may not be accurate due to some parameters are only used in language bindings but\n passed down to XGBoost core. Or some parameters are not used but slip through this\n verification. Please open an issue if you find above cases.\n\n\n[20:32:15] WARNING: C:/Users/Administrator/workspace/xgboost-win64_release_1.3.0/src/learner.cc:541: \nParameters: { silent } might not be used.\n\n This may not be accurate due to some parameters are only used in language bindings but\n passed down to XGBoost core. Or some parameters are not used but slip through this\n verification. Please open an issue if you find above cases.\n\n\n[20:32:15] WARNING: C:/Users/Administrator/workspace/xgboost-win64_release_1.3.0/src/learner.cc:541: \nParameters: { silent } might not be used.\n\n This may not be accurate due to some parameters are only used in language bindings but\n passed down to XGBoost core. Or some parameters are not used but slip through this\n verification. Please open an issue if you find above cases.\n\n\n[20:32:15] WARNING: C:/Users/Administrator/workspace/xgboost-win64_release_1.3.0/src/learner.cc:541: \nParameters: { silent } might not be used.\n\n This may not be accurate due to some parameters are only used in language bindings but\n passed down to XGBoost core. Or some parameters are not used but slip through this\n verification. Please open an issue if you find above cases.\n\n\n[20:32:15] WARNING: C:/Users/Administrator/workspace/xgboost-win64_release_1.3.0/src/learner.cc:541: \nParameters: { silent } might not be used.\n\n This may not be accurate due to some parameters are only used in language bindings but\n passed down to XGBoost core. Or some parameters are not used but slip through this\n verification. Please open an issue if you find above cases.\n\n\n[20:32:15] WARNING: C:/Users/Administrator/workspace/xgboost-win64_release_1.3.0/src/learner.cc:541: \nParameters: { silent } might not be used.\n\n This may not be accurate due to some parameters are only used in language bindings but\n passed down to XGBoost core. Or some parameters are not used but slip through this\n verification. Please open an issue if you find above cases.\n\n\n[20:32:15] WARNING: C:/Users/Administrator/workspace/xgboost-win64_release_1.3.0/src/learner.cc:541: \nParameters: { silent } might not be used.\n\n This may not be accurate due to some parameters are only used in language bindings but\n passed down to XGBoost core. Or some parameters are not used but slip through this\n verification. Please open an issue if you find above cases.\n\n\n[20:32:15] WARNING: C:/Users/Administrator/workspace/xgboost-win64_release_1.3.0/src/learner.cc:541: \nParameters: { silent } might not be used.\n\n This may not be accurate due to some parameters are only used in language bindings but\n passed down to XGBoost core. Or some parameters are not used but slip through this\n verification. Please open an issue if you find above cases.\n\n\n[20:32:15] WARNING: C:/Users/Administrator/workspace/xgboost-win64_release_1.3.0/src/learner.cc:541: \nParameters: { silent } might not be used.\n\n This may not be accurate due to some parameters are only used in language bindings but\n passed down to XGBoost core. Or some parameters are not used but slip through this\n verification. Please open an issue if you find above cases.\n\n\n[20:32:16] WARNING: C:/Users/Administrator/workspace/xgboost-win64_release_1.3.0/src/learner.cc:541: \nParameters: { silent } might not be used.\n\n This may not be accurate due to some parameters are only used in language bindings but\n passed down to XGBoost core. Or some parameters are not used but slip through this\n verification. Please open an issue if you find above cases.\n\n\n[20:32:16] WARNING: C:/Users/Administrator/workspace/xgboost-win64_release_1.3.0/src/learner.cc:541: \nParameters: { silent } might not be used.\n\n This may not be accurate due to some parameters are only used in language bindings but\n passed down to XGBoost core. Or some parameters are not used but slip through this\n verification. Please open an issue if you find above cases.\n\n\n[20:32:16] WARNING: C:/Users/Administrator/workspace/xgboost-win64_release_1.3.0/src/learner.cc:541: \nParameters: { silent } might not be used.\n\n This may not be accurate due to some parameters are only used in language bindings but\n passed down to XGBoost core. Or some parameters are not used but slip through this\n verification. Please open an issue if you find above cases.\n\n\n[20:32:16] WARNING: C:/Users/Administrator/workspace/xgboost-win64_release_1.3.0/src/learner.cc:541: \nParameters: { silent } might not be used.\n\n This may not be accurate due to some parameters are only used in language bindings but\n passed down to XGBoost core. Or some parameters are not used but slip through this\n verification. Please open an issue if you find above cases.\n\n\n[20:32:16] WARNING: C:/Users/Administrator/workspace/xgboost-win64_release_1.3.0/src/learner.cc:541: \nParameters: { silent } might not be used.\n\n This may not be accurate due to some parameters are only used in language bindings but\n passed down to XGBoost core. Or some parameters are not used but slip through this\n verification. Please open an issue if you find above cases.\n\n\n[20:32:16] WARNING: C:/Users/Administrator/workspace/xgboost-win64_release_1.3.0/src/learner.cc:541: \nParameters: { silent } might not be used.\n\n This may not be accurate due to some parameters are only used in language bindings but\n passed down to XGBoost core. Or some parameters are not used but slip through this\n verification. Please open an issue if you find above cases.\n\n\n[20:32:16] WARNING: C:/Users/Administrator/workspace/xgboost-win64_release_1.3.0/src/learner.cc:541: \nParameters: { silent } might not be used.\n\n This may not be accurate due to some parameters are only used in language bindings but\n passed down to XGBoost core. Or some parameters are not used but slip through this\n verification. Please open an issue if you find above cases.\n\n\n[20:32:16] WARNING: C:/Users/Administrator/workspace/xgboost-win64_release_1.3.0/src/learner.cc:541: \nParameters: { silent } might not be used.\n\n This may not be accurate due to some parameters are only used in language bindings but\n passed down to XGBoost core. Or some parameters are not used but slip through this\n verification. Please open an issue if you find above cases.\n\n\n[20:32:16] WARNING: C:/Users/Administrator/workspace/xgboost-win64_release_1.3.0/src/learner.cc:541: \nParameters: { silent } might not be used.\n\n This may not be accurate due to some parameters are only used in language bindings but\n passed down to XGBoost core. Or some parameters are not used but slip through this\n verification. Please open an issue if you find above cases.\n\n\n[20:32:16] WARNING: C:/Users/Administrator/workspace/xgboost-win64_release_1.3.0/src/learner.cc:541: \nParameters: { silent } might not be used.\n\n This may not be accurate due to some parameters are only used in language bindings but\n passed down to XGBoost core. Or some parameters are not used but slip through this\n verification. Please open an issue if you find above cases.\n\n\n[20:32:17] WARNING: C:/Users/Administrator/workspace/xgboost-win64_release_1.3.0/src/learner.cc:541: \nParameters: { silent } might not be used.\n\n This may not be accurate due to some parameters are only used in language bindings but\n passed down to XGBoost core. Or some parameters are not used but slip through this\n verification. Please open an issue if you find above cases.\n\n\n[20:32:17] WARNING: C:/Users/Administrator/workspace/xgboost-win64_release_1.3.0/src/learner.cc:541: \nParameters: { silent } might not be used.\n\n This may not be accurate due to some parameters are only used in language bindings but\n passed down to XGBoost core. Or some parameters are not used but slip through this\n verification. Please open an issue if you find above cases.\n\n\n[20:32:17] WARNING: C:/Users/Administrator/workspace/xgboost-win64_release_1.3.0/src/learner.cc:541: \nParameters: { silent } might not be used.\n\n This may not be accurate due to some parameters are only used in language bindings but\n passed down to XGBoost core. Or some parameters are not used but slip through this\n verification. Please open an issue if you find above cases.\n\n\n[20:32:17] WARNING: C:/Users/Administrator/workspace/xgboost-win64_release_1.3.0/src/learner.cc:541: \nParameters: { silent } might not be used.\n\n This may not be accurate due to some parameters are only used in language bindings but\n passed down to XGBoost core. Or some parameters are not used but slip through this\n verification. Please open an issue if you find above cases.\n\n\n[20:32:17] WARNING: C:/Users/Administrator/workspace/xgboost-win64_release_1.3.0/src/learner.cc:541: \nParameters: { silent } might not be used.\n\n This may not be accurate due to some parameters are only used in language bindings but\n passed down to XGBoost core. Or some parameters are not used but slip through this\n verification. Please open an issue if you find above cases.\n\n\n[20:32:17] WARNING: C:/Users/Administrator/workspace/xgboost-win64_release_1.3.0/src/learner.cc:541: \nParameters: { silent } might not be used.\n\n This may not be accurate due to some parameters are only used in language bindings but\n passed down to XGBoost core. Or some parameters are not used but slip through this\n verification. Please open an issue if you find above cases.\n\n\n[20:32:17] WARNING: C:/Users/Administrator/workspace/xgboost-win64_release_1.3.0/src/learner.cc:541: \nParameters: { silent } might not be used.\n\n This may not be accurate due to some parameters are only used in language bindings but\n passed down to XGBoost core. Or some parameters are not used but slip through this\n verification. Please open an issue if you find above cases.\n\n\n[20:32:17] WARNING: C:/Users/Administrator/workspace/xgboost-win64_release_1.3.0/src/learner.cc:541: \nParameters: { silent } might not be used.\n\n This may not be accurate due to some parameters are only used in language bindings but\n passed down to XGBoost core. Or some parameters are not used but slip through this\n verification. Please open an issue if you find above cases.\n\n\n[20:32:17] WARNING: C:/Users/Administrator/workspace/xgboost-win64_release_1.3.0/src/learner.cc:541: \nParameters: { silent } might not be used.\n\n This may not be accurate due to some parameters are only used in language bindings but\n passed down to XGBoost core. Or some parameters are not used but slip through this\n verification. Please open an issue if you find above cases.\n\n\n[20:32:17] WARNING: C:/Users/Administrator/workspace/xgboost-win64_release_1.3.0/src/learner.cc:541: \nParameters: { silent } might not be used.\n\n This may not be accurate due to some parameters are only used in language bindings but\n passed down to XGBoost core. Or some parameters are not used but slip through this\n verification. Please open an issue if you find above cases.\n\n\n"
],
[
"import matplotlib.pyplot as plt\nplt.scatter(test_error.keys(), test_error.values())\nplt.xlabel('Max Depth')\nplt.ylabel('Test Error')\nplt.show()",
"_____no_output_____"
],
[
"lr = [0.001*(i) for i in test_error.keys()]\nplt.scatter(temp,test_error.values())\nplt.xlabel('Learning Rate')\nplt.ylabel('Error')\nplt.show()",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbf1ae802f7585984d7acd29750b8686dd3d3a61
| 5,433 |
ipynb
|
Jupyter Notebook
|
codes/mlmodels/NaiveBayes.ipynb
|
NilLau/NilLau.github.io
|
e55768be0be4d6549b24c702554c11e64958d4c7
|
[
"MIT"
] | null | null | null |
codes/mlmodels/NaiveBayes.ipynb
|
NilLau/NilLau.github.io
|
e55768be0be4d6549b24c702554c11e64958d4c7
|
[
"MIT"
] | 6 |
2019-06-20T10:05:10.000Z
|
2019-07-08T04:53:01.000Z
|
codes/mlmodels/NaiveBayes.ipynb
|
GaoangLiu/GaoangLiu.github.io
|
3552e9bfd96d4d2e2a213f9758dea44173aab70c
|
[
"MIT"
] | null | null | null | 34.826923 | 141 | 0.515185 |
[
[
[
"# Naive Bayes\n## Bayes Theorem\n$P(A|B) = \\frac{P(B|A) P(A)}{P(B)} $\n\nIn our case, given features $X = (x_1, ..., x_n)$, the class probability $P(y|X)$:\n\n$P(y|X) = \\frac{P(X|y) P(y)}{P(X)}$\n\nWe're making an assumption all features are **mutually independent**.\n\n$P(y|X) = \\frac{P(x_1|y) \\cdot P(x_2|y) \\cdot P(x_3|y) \\cdots P(x_n|y) \\cdot P(y)} {P(X)}$\n\nNote that $P(y|X)$ called the posterior probability, $P(x_i|y)$ class conditional probability, and $P(y)$ prior probability of $y$. \n\n## Select class with highest probability \n\n$y = argmax_yP(y|X) = argmax_y \\frac{P(x_1|y) \\cdot P(x_2|y) \\cdot P(x_3|y) \\cdots P(x_n|y) \\cdot P(y)} {P(X)}$\n\nSince $P(X)$ is certain, \n\n$y = argmax_y P(x_1|y) \\cdot P(x_2|y) \\cdot P(x_3|y) \\cdots P(x_n|y) \\cdot P(y)$\n\nTo avoid overfollow problem, we use a little trick:\n\n$y = argmax_y (\\log(P(x_1|y)) + \\log(P(x_2|y)) + \\log(P(x_3|y)) \\cdots \\log(P(x_n|y)) + \\log(P(y)) )$\n\n## Model class conditional probability $P(x_i|y)$ by Gaussian\n\n$P(x_i|y) = \\frac{1}{\\sqrt{2\\pi \\sigma^2}} \\cdot e^{-\\frac{(x_i - \\mu_y)^2}{2 \\sigma_y^2}}$\n",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\nclass NaiveBayes:\n def fit(self, X, y):\n n_samples, n_features = X.shape\n self._classes = np.unique(y)\n n_classes = len(self._classes)\n \n self._mean = np.zeros((n_classes, n_features), dtype=np.float64)\n self._var = np.zeros((n_classes, n_features), dtype=np.float64)\n self._priors = np.zeros(n_classes, dtype=np.float64)\n \n for idx, c in enumerate(self._classes):\n X_c = X[y==c] \n self._mean[idx, :] = X_c.mean(axis=0)\n self._var[idx, :] = X_c.var(axis=0)\n \n # prior probability of y, or frequency, how often this class C occur\n self._priors[idx] = X_c.shape[0] / float(n_samples)\n print(self._classes)\n print(self._mean, self._var, self._priors)\n \n\n def predict(self, X):\n y_pred = [self._predict(x) for x in X]\n return y_pred \n \n def _predict(self, x):\n '''Make prediction on a single instance.'''\n posteriors = []\n for idx, c in enumerate(self._classes):\n prior = np.log(self._priors[idx])\n class_conditional = np.sum(np.log(self._probability_dense_function(idx, x)))\n _posterior = prior + class_conditional\n posteriors.append(_posterior)\n \n return self._classes[np.argmax(posteriors)]\n \n def _probability_dense_function(self, class_idx, x):\n mean = self._mean[class_idx]\n var = self._var[class_idx]\n numerator = np.exp(-(x - mean) ** 2 / (2 * var))\n denominator = np.sqrt(2 * np.pi * var)\n return numerator / denominator\n \n ",
"_____no_output_____"
],
[
"from sklearn import datasets\nimport xgboost\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.metrics import accuracy_score\nfrom sklearn.naive_bayes import MultinomialNB\n\ndata = datasets.load_breast_cancer()\ndata = datasets.load_iris()\nX, y = data.data, data.target \ny[y == 0] = -1\nn_estimators=10\n\nX_train, X_val, y_train, y_val = train_test_split(X, y, test_size=0.2, random_state=0)\n\nnb = NaiveBayes()\n# nb = MultinomialNB()\nnb.fit(X_train, y_train)\ny_pred = nb.predict(X_val)\nprint(\"Accuracy score \", accuracy_score(y_val, y_pred))",
"[-1 1 2]\n[[5.02051282 3.4025641 1.46153846 0.24102564]\n [5.88648649 2.76216216 4.21621622 1.32432432]\n [6.63863636 2.98863636 5.56590909 2.03181818]] [[0.12932281 0.1417883 0.02031558 0.01113741]\n [0.26387144 0.1039737 0.2300073 0.04075968]\n [0.38918905 0.10782541 0.29451963 0.06444215]] [0.325 0.30833333 0.36666667]\nAccuracy score 0.9666666666666667\n"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code"
]
] |
cbf1c12a4bfca83c2a78e8e7a95aa65187165b45
| 24,843 |
ipynb
|
Jupyter Notebook
|
I Python Basics & Pandas/#02. Data Manipulation & Visualization to Enhance the Discipline/.ipynb_checkpoints/02practice-checkpoint.ipynb
|
iamoespana92/machine-learning-program
|
a6c6dac18d41b3795685fcb3dd358ab1f64a4ee4
|
[
"MIT"
] | null | null | null |
I Python Basics & Pandas/#02. Data Manipulation & Visualization to Enhance the Discipline/.ipynb_checkpoints/02practice-checkpoint.ipynb
|
iamoespana92/machine-learning-program
|
a6c6dac18d41b3795685fcb3dd358ab1f64a4ee4
|
[
"MIT"
] | null | null | null |
I Python Basics & Pandas/#02. Data Manipulation & Visualization to Enhance the Discipline/.ipynb_checkpoints/02practice-checkpoint.ipynb
|
iamoespana92/machine-learning-program
|
a6c6dac18d41b3795685fcb3dd358ab1f64a4ee4
|
[
"MIT"
] | null | null | null | 27.180525 | 134 | 0.381395 |
[
[
[
"<font size=\"+5\">#03. Data Manipulation & Visualization to Enhance the Discipline</font>",
"_____no_output_____"
],
[
"- Book + Private Lessons [Here ↗](https://sotastica.com/reservar)\n- Subscribe to my [Blog ↗](https://blog.pythonassembly.com/)\n- Let's keep in touch on [LinkedIn ↗](www.linkedin.com/in/jsulopz) 😄",
"_____no_output_____"
],
[
"# Load the Data",
"_____no_output_____"
],
[
"> - By executing the below lines of code,\n> - You will see a list of possible datasets that we can load to python by just typing the name in the function",
"_____no_output_____"
]
],
[
[
"import seaborn as sns\nsns.get_dataset_names()",
"_____no_output_____"
]
],
[
[
"> - For example, `mpg`:\n\n**PS**: It will be more challenging & fun for your learning to try other dataset than `mpg`",
"_____no_output_____"
]
],
[
[
"your_dataset = 'mpg'\n\ndf = sns.load_dataset(name='mpg')\ndf.head()",
"_____no_output_____"
]
],
[
[
"# Scatterplot with 2 Variables",
"_____no_output_____"
],
[
"> - Variable in X Axis\n> - Variable in Y Axis",
"_____no_output_____"
],
[
"# Scatterplot with 3 variables",
"_____no_output_____"
],
[
"> - Variable in X Axis\n> - Variable in Y Axis\n> - Color each point regarding a different value in a column",
"_____no_output_____"
],
[
"# Other Data Visualization Figures",
"_____no_output_____"
],
[
"> We'll head over the 3 main libraries used in Python to visualize data: `matplotlib`, `seaborn` and `plotly`.\n>\n> We'll reproduce at least one example from the links provided below. Therefore, we need to:\n>\n> 1. Click in the link\n> 2. Pick up an example\n> 3. Copy-paste the lines of code\n> 4. Run the code",
"_____no_output_____"
],
[
"## Seaborn",
"_____no_output_____"
],
[
"### Seaborn Example Gallery",
"_____no_output_____"
],
[
"> - https://seaborn.pydata.org/examples/index.html",
"_____no_output_____"
],
[
"### Repeat the same Visualization Figures ",
"_____no_output_____"
],
[
"> - Now with you `DataFrame` ↓",
"_____no_output_____"
]
],
[
[
"df = sns.load_dataset(name=your_dataset)\ndf.head()",
"_____no_output_____"
]
],
[
[
"## Matplotlib",
"_____no_output_____"
],
[
"## Matplotlib Example Gallery",
"_____no_output_____"
],
[
"> - https://towardsdatascience.com/matplotlib-tutorial-with-code-for-pythons-powerful-data-visualization-tool-8ec458423c5e",
"_____no_output_____"
],
[
"### Repeat the same Visualization Figures ",
"_____no_output_____"
],
[
"> - Now with you `DataFrame` ↓",
"_____no_output_____"
]
],
[
[
"df = sns.load_dataset(name=your_dataset)\ndf.head()",
"_____no_output_____"
]
],
[
[
"## Plotly",
"_____no_output_____"
],
[
"## Plotly Example Gallery",
"_____no_output_____"
],
[
"> - https://plotly.com/python/",
"_____no_output_____"
],
[
"### Repeat the same Visualization Figures ",
"_____no_output_____"
],
[
"> - Now with you `DataFrame` ↓",
"_____no_output_____"
]
],
[
[
"df = sns.load_dataset(name=your_dataset)\ndf.head()",
"_____no_output_____"
]
],
[
[
"# Achieved Goals",
"_____no_output_____"
],
[
"_Double click on **this cell** and place an `X` inside the square brackets (i.e., [X]) if you think you understand the goal:_\n\n- [ ] All roads lead to Rome; We can achieve the **the same result with different lines of code**.\n - We reproduced a scatterplot using 3 different lines of code.\n- [ ] Different **`libraries`** may have `functions()` that do the **same thing**.\n - `matplotlib`, `seaborn` & `plotly` makes outstanding Visualization Figures\n - But you probably wouldn't use `plotly` on a paper. Unless we are on Harry Potter 😜\n- [ ] Understand that **coding is a matter of necessity**. Not a serie of mechanical steps to achieve a goal.\n - You need to create art (code) by solving one problem at a time.\n- [ ] Understand that **there isn't a unique solution**.\n - We can achieve the same result with different approaches.\n - For example, changing the colors of the points.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
cbf1da05c7c90bd30a1978614ffe6f6636ee324c
| 154,420 |
ipynb
|
Jupyter Notebook
|
test.ipynb
|
MinHee-L/oss-test
|
56dc1d331d2bd05949d8a279effbdcdbd0f22c4c
|
[
"MIT"
] | null | null | null |
test.ipynb
|
MinHee-L/oss-test
|
56dc1d331d2bd05949d8a279effbdcdbd0f22c4c
|
[
"MIT"
] | 1 |
2022-03-30T02:40:08.000Z
|
2022-03-30T02:40:08.000Z
|
test.ipynb
|
minhe8564/oss-test
|
56dc1d331d2bd05949d8a279effbdcdbd0f22c4c
|
[
"MIT"
] | null | null | null | 190.641975 | 73,753 | 0.826816 |
[
[
[
"<a href=\"https://colab.research.google.com/github/minhe8564/oss-test/blob/main/test.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport matplotlib.pyplot as plt\nimport seaborn as sns",
"_____no_output_____"
],
[
"train = pd.read_csv('/content/sample_data/california_housing_test.csv')\ntest = pd.read_csv('/content/sample_data/california_housing_train.csv')\ntrain.head()",
"_____no_output_____"
],
[
"test.head()",
"_____no_output_____"
],
[
"train.describe()",
"_____no_output_____"
],
[
"train.hist(figsize=(15,13), grid=False, bins=50)\nplt.show()",
"_____no_output_____"
],
[
"correlation = train.corr()",
"_____no_output_____"
],
[
"plt.figure(figsize=(10,10))\nsns.heatmap(correlation, annot=True)\nplt.show()",
"_____no_output_____"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbf1f49b8f620e48528a3b6dd227b428ed571e95
| 291,711 |
ipynb
|
Jupyter Notebook
|
monthly_update/templates/discourse_report.ipynb
|
choldgraf/jupyter-activity-snapshot
|
080f8c34e1e3e5081c4b733592b114b01b09b8c0
|
[
"BSD-3-Clause"
] | 7 |
2019-08-26T13:19:05.000Z
|
2021-11-18T16:34:01.000Z
|
monthly_update/templates/discourse_report.ipynb
|
choldgraf/jupyter-activity-snapshot
|
080f8c34e1e3e5081c4b733592b114b01b09b8c0
|
[
"BSD-3-Clause"
] | 3 |
2019-11-27T19:25:27.000Z
|
2021-03-13T01:19:45.000Z
|
monthly_update/templates/discourse_report.ipynb
|
choldgraf/jupyter-activity-snapshot
|
080f8c34e1e3e5081c4b733592b114b01b09b8c0
|
[
"BSD-3-Clause"
] | 4 |
2019-06-20T17:49:53.000Z
|
2021-05-21T21:06:18.000Z
| 80.516423 | 39,874 | 0.780375 |
[
[
[
"import requests\nfrom IPython.display import Markdown\nfrom tqdm import tqdm, tqdm_notebook\nimport pandas as pd\nfrom matplotlib import pyplot as plt\nimport numpy as np\nimport altair as alt\nfrom requests.utils import quote\nimport os\nfrom datetime import timedelta\nfrom mod import alt_theme",
"_____no_output_____"
],
[
"fmt = \"{:%Y-%m-%d}\"\n\n# Can optionally use number of days to choose dates\nn_days = 60\nend_date = fmt.format(pd.datetime.today())\nstart_date = fmt.format(pd.datetime.today() - timedelta(days=n_days))\n\nrenderer = \"kaggle\"\ngithub_orgs = [\"jupyterhub\", \"jupyter\", \"jupyterlab\", \"jupyter-widgets\", \"ipython\", \"binder-examples\", \"nteract\"]\nbot_names = [\"stale\", \"codecov\", \"jupyterlab-dev-mode\", \"henchbot\"]",
"<ipython-input-2-149463e7ffff>:5: FutureWarning: The pandas.datetime class is deprecated and will be removed from pandas in a future version. Import from datetime module instead.\n end_date = fmt.format(pd.datetime.today())\n<ipython-input-2-149463e7ffff>:6: FutureWarning: The pandas.datetime class is deprecated and will be removed from pandas in a future version. Import from datetime module instead.\n start_date = fmt.format(pd.datetime.today() - timedelta(days=n_days))\n"
],
[
"alt.renderers.enable(renderer);\nalt.themes.register('my_theme', alt_theme)\nalt.themes.enable(\"my_theme\")",
"_____no_output_____"
],
[
"# Discourse API key\napi = {'Api-Key': os.environ['DISCOURSE_API_KEY'],\n 'Api-Username': os.environ['DISCOURSE_API_USERNAME']}",
"_____no_output_____"
],
[
"# Discourse\ndef topics_to_markdown(topics, n_list=10):\n body = []\n for _, topic in topics.iterrows():\n title = topic['fancy_title']\n slug = topic['slug']\n posts_count = topic['posts_count']\n url = f'https://discourse.jupyter.org/t/{slug}'\n body.append(f'* [{title}]({url}) ({posts_count} posts)')\n body = body[:n_list]\n return '\\n'.join(body)\n\ndef counts_from_activity(activity):\n counts = activity.groupby('category_id').count()['bookmarked'].reset_index()\n counts['parent_category'] = None\n for ii, irow in counts.iterrows():\n if parent_categories[irow['category_id']] is not None:\n counts.loc[ii, 'parent_category'] = parent_categories[irow['category_id']]\n\n counts['category_id'] = counts['category_id'].map(lambda a: category_mapping[a])\n counts['parent_category'] = counts['parent_category'].map(lambda a: category_mapping[a] if a is not None else 'parent')\n is_parent = counts['parent_category'] == 'parent'\n counts.loc[is_parent, 'parent_category'] = counts.loc[is_parent, 'category_id'] \n counts['parent/category'] = counts.apply(lambda a: a['parent_category']+'/'+a['category_id'], axis=1)\n counts = counts.sort_values(['parent_category', 'bookmarked'], ascending=False)\n return counts",
"_____no_output_____"
]
],
[
[
"# Community forum activity\n\nThe [Jupyter Community Forum](https://discourse.jupyter.org) is a place for Jovyans across the\ncommunity to talk about Jupyter tools in interactive computing and how they fit into their\nworkflows. It's also a place for developers to share ideas, tools, tips, and help one another.\n\nBelow are a few updates from activity in the Discourse. For more detailed information about\nthe activity on the Community Forum, check out these links:\n\n* [The users page](https://discourse.jupyter.org/u) has information about user activity\n* [The top posts page](https://discourse.jupyter.org/top) contains a list of top posts, sorted\n by various metrics.",
"_____no_output_____"
]
],
[
[
"# Get categories for IDs\nurl = \"https://discourse.jupyter.org/site.json\"\nresp = requests.get(url, headers=api)\ncategory_mapping = {cat['id']: cat['name'] for cat in resp.json()['categories']}\nparent_categories = {cat['id']: cat.get(\"parent_category_id\", None) for cat in resp.json()['categories']}",
"_____no_output_____"
],
[
"# Base URL to use\nurl = \"https://discourse.jupyter.org/latest.json\"",
"_____no_output_____"
]
],
[
[
"## Topics with lots of likes\n\n\"Likes\" are a way for community members to say thanks for a helpful post, show their\nsupport for an idea, or generally to share a little positivity with somebody else.\nThese are topics that have generated lots of likes in recent history.",
"_____no_output_____"
]
],
[
[
"params = {\"order\": \"likes\", \"ascending\": \"False\"}\nresp = requests.get(url, headers=api, params=params)\n\n# Topics with the most likes in recent history\nliked = pd.DataFrame(resp.json()['topic_list']['topics'])\nMarkdown(topics_to_markdown(liked))",
"_____no_output_____"
]
],
[
[
"## Active topics on the Community Forum\n\nThese are topics with lots of activity in recent history.",
"_____no_output_____"
]
],
[
[
"params = {\"order\": \"posts\", \"ascending\": \"False\"}\nresp = requests.get(url, headers=api, params=params)\n\n# Topics with the most posts in recent history\nposts = pd.DataFrame(resp.json()['topic_list']['topics'])\nMarkdown(topics_to_markdown(posts))",
"_____no_output_____"
],
[
"counts = counts_from_activity(posts)\nalt.Chart(data=counts, width=700, height=300, title=\"Activity by category\").mark_bar().encode(\n x=alt.X(\"parent/category\", sort=alt.Sort(counts['category_id'].values.tolist())),\n y=\"bookmarked\",\n color=\"parent_category\"\n)",
"_____no_output_____"
]
],
[
[
"## Recently-created topics\n\nThese are topics that were recently created, sorted by the amount of activity\nin each one.",
"_____no_output_____"
]
],
[
[
"params = {\"order\": \"created\", \"ascending\": \"False\"}\nresp = requests.get(url, headers=api, params=params)\n\n# Sort created by the most posted for recently-created posts\ncreated = pd.DataFrame(resp.json()['topic_list']['topics'])\ncreated = created.sort_values('posts_count', ascending=False)\nMarkdown(topics_to_markdown(created))",
"_____no_output_____"
],
[
"counts = counts_from_activity(created)\nalt.Chart(data=counts, width=700, height=300, title=\"Activity by category\").mark_bar().encode(\n x=alt.X(\"parent/category\", sort=alt.Sort(counts['category_id'].values.tolist())),\n y=\"bookmarked\",\n color=\"parent_category\"\n)",
"_____no_output_____"
]
],
[
[
"## User activity in the Community Forum\n\n**Top posters**\n\nThese people have posted lots of comments, replies, answers, etc in the community forum.",
"_____no_output_____"
]
],
[
[
"def plot_user_data(users, column, sort=False):\n plt_data = users.sort_values(column, ascending=False).head(50)\n x = alt.X(\"username\", sort=plt_data['username'].tolist()) if sort is True else 'username'\n ch = alt.Chart(data=plt_data).mark_bar().encode(\n x=x,\n y=column\n )\n return ch",
"_____no_output_____"
],
[
"url = \"https://discourse.jupyter.org/directory_items.json\"\nparams = {\"period\": \"quarterly\", \"order\": \"post_count\"}\nresp = requests.get(url, headers=api, params=params)\n\n# Topics with the most likes in recent history\nusers = pd.DataFrame(resp.json()['directory_items'])\nusers['username'] = users['user'].map(lambda a: a['username'])",
"_____no_output_____"
],
[
"plot_user_data(users.head(50), 'post_count')",
"_____no_output_____"
]
],
[
[
"**Forum users, sorted by likes given**\n\nThese are Community Forum members that \"liked\" other people's posts. We appreciate\nanybody taking the time to tell someone else they like what they're shared!",
"_____no_output_____"
]
],
[
[
"plot_user_data(users.head(50), 'likes_given')",
"_____no_output_____"
]
],
[
[
"**Forum users, sorted by likes received**\n\nThese are folks that posted things other people in the Community Forum liked.",
"_____no_output_____"
]
],
[
[
"plot_user_data(users.head(50), 'likes_received')",
"_____no_output_____"
],
[
"%%html\n<script src=\"https://cdn.rawgit.com/parente/4c3e6936d0d7a46fd071/raw/65b816fb9bdd3c28b4ddf3af602bfd6015486383/code_toggle.js\"></script>",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
cbf209a09b157c0d938ec490cdaacbcc601ed71f
| 108,447 |
ipynb
|
Jupyter Notebook
|
python/examples/notebooks/Fluorescent Bead - Synthetic Deconvolution.ipynb
|
russellb/flowdec
|
0239b7aedf497133c8073476cc868cc455a87428
|
[
"Apache-2.0"
] | 86 |
2018-04-02T16:25:11.000Z
|
2022-03-17T18:19:30.000Z
|
python/examples/notebooks/Fluorescent Bead - Synthetic Deconvolution.ipynb
|
russellb/flowdec
|
0239b7aedf497133c8073476cc868cc455a87428
|
[
"Apache-2.0"
] | 38 |
2018-09-25T15:21:16.000Z
|
2020-11-19T19:42:43.000Z
|
python/examples/notebooks/Fluorescent Bead - Synthetic Deconvolution.ipynb
|
russellb/flowdec
|
0239b7aedf497133c8073476cc868cc455a87428
|
[
"Apache-2.0"
] | 28 |
2018-03-27T21:14:36.000Z
|
2021-12-16T17:46:15.000Z
| 753.104167 | 63,900 | 0.953507 |
[
[
[
"# Deconvolution of Downsampled \"Bead\" Synthetic Dataset\n\nDeconvolution of [Single Fluorescent Bead](http://bigwww.epfl.ch/deconvolution/bead/) dataset.\n\n**Reference**<br>\nD. Sage, L. Donati, F. Soulez, D. Fortun, G. Schmit, A. Seitz, R. Guiet, C. Vonesch, M. Unser<br>\nDeconvolutionLab2: An Open-Source Software for Deconvolution Microscopy<br>\nMethods - Image Processing for Biologists, 115, 2017.",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nimport numpy as np\nimport matplotlib.pyplot as plt\nfrom flowdec.nb import utils as nbutils \nfrom flowdec import data as fd_data",
"_____no_output_____"
],
[
"# Load volume downsampled to 25%\nacq = fd_data.bead_25pct()\nacq.shape()",
"_____no_output_____"
],
[
"nbutils.plot_rotations(acq.data)",
"_____no_output_____"
],
[
"import tensorflow as tf\nfrom flowdec import restoration as fd_restoration\n\nres = fd_restoration.richardson_lucy(\n acq, niter=25, \n # Disable GPUs for this since on windows at least, repeat runs of TF graphs\n # on GPUs in a jupyter notebook do not go well (crashes with mem allocation errors)\n session_config=tf.ConfigProto(device_count={'GPU': 0})\n)\nres.shape",
"_____no_output_____"
],
[
"nbutils.plot_rotations(res)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
cbf20a92bded4cecb1addd6a23d4be76af5fb52d
| 21,225 |
ipynb
|
Jupyter Notebook
|
Cats and Dogs Classification with Augmentation and Dropout.ipynb
|
sproboticworks/ml-course
|
dabd81bd9062a9e10f22738497b03bb47446c13b
|
[
"Apache-2.0"
] | null | null | null |
Cats and Dogs Classification with Augmentation and Dropout.ipynb
|
sproboticworks/ml-course
|
dabd81bd9062a9e10f22738497b03bb47446c13b
|
[
"Apache-2.0"
] | null | null | null |
Cats and Dogs Classification with Augmentation and Dropout.ipynb
|
sproboticworks/ml-course
|
dabd81bd9062a9e10f22738497b03bb47446c13b
|
[
"Apache-2.0"
] | null | null | null | 29.93653 | 295 | 0.459176 |
[
[
[
"<a href=\"https://colab.research.google.com/github/sproboticworks/ml-course/blob/master/Cats%20and%20Dogs%20Classification%20with%20Augmentation%20and%20Dropout.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# Import Packages",
"_____no_output_____"
]
],
[
[
"import tensorflow as tf\nimport numpy as np\nimport matplotlib.pyplot as plt",
"_____no_output_____"
]
],
[
[
"# Download Data",
"_____no_output_____"
]
],
[
[
"url = 'https://storage.googleapis.com/mledu-datasets/cats_and_dogs_filtered.zip'\nzip_dir = tf.keras.utils.get_file('cats_and_dogs_filtered.zip', origin=url, extract=True)",
"_____no_output_____"
]
],
[
[
"The dataset we have downloaded has the following directory structure.\n\n<pre style=\"font-size: 10.0pt; font-family: Arial; line-height: 2; letter-spacing: 1.0pt;\" >\n<b>cats_and_dogs_filtered</b>\n|__ <b>train</b>\n |______ <b>cats</b>: [cat.0.jpg, cat.1.jpg, cat.2.jpg ...]\n |______ <b>dogs</b>: [dog.0.jpg, dog.1.jpg, dog.2.jpg ...]\n|__ <b>validation</b>\n |______ <b>cats</b>: [cat.2000.jpg, cat.2001.jpg, cat.2002.jpg ...]\n |______ <b>dogs</b>: [dog.2000.jpg, dog.2001.jpg, dog.2002.jpg ...]\n</pre>\n",
"_____no_output_____"
],
[
"## List the directories with the following terminal command:",
"_____no_output_____"
]
],
[
[
"import os\nzip_dir_base = os.path.dirname(zip_dir)\n!find $zip_dir_base -type d -print",
"_____no_output_____"
]
],
[
[
"## Assign Directory Variables",
"_____no_output_____"
]
],
[
[
"base_dir = os.path.join(os.path.dirname(zip_dir), 'cats_and_dogs_filtered')\n\ntrain_dir = os.path.join(base_dir, 'train')\nvalidation_dir = os.path.join(base_dir, 'validation')\n\n# Directory with our training cat/dog pictures\ntrain_cats_dir = os.path.join(train_dir, 'cats')\ntrain_dogs_dir = os.path.join(train_dir, 'dogs')\n\n# Directory with our validation cat/dog pictures\nvalidation_cats_dir = os.path.join(validation_dir, 'cats')\nvalidation_dogs_dir = os.path.join(validation_dir, 'dogs')",
"_____no_output_____"
]
],
[
[
"## Print Filenames",
"_____no_output_____"
]
],
[
[
"train_cat_fnames = os.listdir( train_cats_dir )\ntrain_dog_fnames = os.listdir( train_dogs_dir )\n\nprint(train_cat_fnames[:10])\nprint(train_dog_fnames[:10])",
"_____no_output_____"
]
],
[
[
"## Print number of Training and Validation images",
"_____no_output_____"
]
],
[
[
"num_cats_tr = len(os.listdir(train_cats_dir))\nnum_dogs_tr = len(os.listdir(train_dogs_dir))\n\nnum_cats_val = len(os.listdir(validation_cats_dir))\nnum_dogs_val = len(os.listdir(validation_dogs_dir))\n\ntotal_train = num_cats_tr + num_dogs_tr\ntotal_val = num_cats_val + num_dogs_val\n\nprint('total training cat images :', len(os.listdir( train_cats_dir ) ))\nprint('total training dog images :', len(os.listdir( train_dogs_dir ) ))\n\nprint('total validation cat images :', len(os.listdir( validation_cats_dir ) ))\nprint('total validation dog images :', len(os.listdir( validation_dogs_dir ) ))",
"_____no_output_____"
]
],
[
[
"# Data Preparation",
"_____no_output_____"
]
],
[
[
"BATCH_SIZE = 20\nIMG_SHAPE = 150\nfrom tensorflow.keras.preprocessing.image import ImageDataGenerator\n\n# All images will be rescaled by 1./255.\ntrain_datagen = ImageDataGenerator( rescale = 1.0/255. )\nvalidation_datagen = ImageDataGenerator( rescale = 1.0/255. )\n\n# --------------------\n# Flow training images in batches of 20 using train_datagen generator\n# --------------------\ntrain_generator = train_datagen.flow_from_directory(train_dir,\n batch_size=BATCH_SIZE,\n class_mode='binary',\n target_size=(IMG_SHAPE, IMG_SHAPE))\n \n# --------------------\n# Flow validation images in batches of 20 using test_datagen generator\n# --------------------\nvalidation_generator = validation_datagen.flow_from_directory(validation_dir,\n batch_size=BATCH_SIZE,\n class_mode = 'binary',\n target_size = (IMG_SHAPE, IMG_SHAPE))\n",
"_____no_output_____"
]
],
[
[
"## Visualizing Training images",
"_____no_output_____"
]
],
[
[
"def plotImages(images_arr):\n fig, axes = plt.subplots(1, 5, figsize=(20,20))\n axes = axes.flatten()\n for img, ax in zip(images_arr, axes):\n ax.imshow(img)\n plt.tight_layout()\n plt.show()",
"_____no_output_____"
],
[
"sample_training_images, _ = next(train_generator)",
"_____no_output_____"
],
[
"plotImages(sample_training_images[:5]) # Plot images 0-4",
"_____no_output_____"
]
],
[
[
"# Image Augmentation",
"_____no_output_____"
],
[
"## Flipping the image horizontally",
"_____no_output_____"
]
],
[
[
"train_datagen = ImageDataGenerator(rescale=1./255, horizontal_flip=True)\n\ntrain_generator = train_datagen.flow_from_directory(batch_size=BATCH_SIZE,\n directory=train_dir,\n shuffle=True,\n target_size=(IMG_SHAPE,IMG_SHAPE))",
"_____no_output_____"
],
[
"augmented_images = [train_generator[0][0][0] for i in range(5)]\nplotImages(augmented_images)",
"_____no_output_____"
]
],
[
[
"## Rotating the image",
"_____no_output_____"
]
],
[
[
"train_datagen = ImageDataGenerator(rescale=1./255, rotation_range=45)\n\ntrain_generator = train_datagen.flow_from_directory(batch_size=BATCH_SIZE,\n directory=train_dir,\n shuffle=True,\n target_size=(IMG_SHAPE, IMG_SHAPE))",
"_____no_output_____"
],
[
"augmented_images = [train_generator[0][0][0] for i in range(5)]\nplotImages(augmented_images)",
"_____no_output_____"
]
],
[
[
"## Applying Zoom",
"_____no_output_____"
]
],
[
[
"train_datagen = ImageDataGenerator(rescale=1./255, zoom_range=0.5)\n\ntrain_generator = train_datagen.flow_from_directory(batch_size=BATCH_SIZE,\n directory=train_dir,\n shuffle=True,\n target_size=(IMG_SHAPE, IMG_SHAPE))",
"_____no_output_____"
],
[
"augmented_images = [train_generator[0][0][0] for i in range(5)]\nplotImages(augmented_images)",
"_____no_output_____"
]
],
[
[
"## Putting it all together",
"_____no_output_____"
]
],
[
[
"train_datagen = ImageDataGenerator(\n rescale=1./255,\n rotation_range=40,\n width_shift_range=0.2,\n height_shift_range=0.2,\n shear_range=0.2,\n zoom_range=0.2,\n horizontal_flip=True,\n fill_mode='nearest')\n\ntrain_generator = train_datagen.flow_from_directory(batch_size=BATCH_SIZE,\n directory=train_dir,\n shuffle=True,\n target_size=(IMG_SHAPE,IMG_SHAPE),\n class_mode='binary')",
"_____no_output_____"
],
[
"augmented_images = [train_generator[0][0][0] for i in range(5)]\nplotImages(augmented_images)",
"_____no_output_____"
]
],
[
[
"# Build Model",
"_____no_output_____"
]
],
[
[
"model = tf.keras.models.Sequential([\n # Note the input shape is the desired size of the image 150x150 with 3 bytes color\n tf.keras.layers.Conv2D(16, (3,3), padding = 'same', activation='relu', input_shape=(150, 150, 3)),\n tf.keras.layers.MaxPooling2D(2,2),\n tf.keras.layers.Conv2D(32, (3,3), padding = 'same', activation='relu'),\n tf.keras.layers.MaxPooling2D(2,2), \n tf.keras.layers.Conv2D(64, (3,3), padding = 'same', activation='relu'), \n tf.keras.layers.MaxPooling2D(2,2),\n # Dropout\n tf.keras.layers.Dropout(0.5),\n # Flatten the results to feed into a DNN\n tf.keras.layers.Flatten(), \n # 512 neuron hidden layer\n tf.keras.layers.Dense(512, activation='relu'), \n # Our last layer (our classifier) consists of a Dense layer with 2 output units and a softmax activation function\n # tf.keras.layers.Dense(2, activation='softmax') \n # Another popular approach when working with binary classification problems, is to use a classifier that consists of a Dense layer with 1 output unit and a sigmoid activation function\n # It will contain a value from 0-1 where 0 for 1 class ('cats') and 1 for the other ('dogs')\n tf.keras.layers.Dense(1, activation='sigmoid') \n])",
"_____no_output_____"
],
[
"model.summary()",
"_____no_output_____"
],
[
"from tensorflow.keras.optimizers import RMSprop\n\nmodel.compile(optimizer=RMSprop(lr=0.001),\n loss='binary_crossentropy',\n metrics = ['accuracy'])",
"_____no_output_____"
]
],
[
[
"# Training Model",
"_____no_output_____"
]
],
[
[
"EPOCHS = 100\nhistory = model.fit(train_generator,\n validation_data=validation_generator,\n steps_per_epoch=int(np.ceil(total_train / float(BATCH_SIZE))),\n epochs=EPOCHS,\n validation_steps=int(np.ceil(total_val / float(BATCH_SIZE))),\n verbose=2)",
"_____no_output_____"
]
],
[
[
"# Visualizing results of the training",
"_____no_output_____"
]
],
[
[
"acc = history.history['accuracy']\nval_acc = history.history['val_accuracy']\n\nloss = history.history['loss']\nval_loss = history.history['val_loss']\n\nepochs_range = range(EPOCHS)\n\nplt.figure(figsize=(8, 8))\nplt.subplot(1, 2, 1)\nplt.plot(epochs_range, acc, label='Training Accuracy')\nplt.plot(epochs_range, val_acc, label='Validation Accuracy')\nplt.legend(loc='lower right')\nplt.title('Training and Validation Accuracy')\n\nplt.subplot(1, 2, 2)\nplt.plot(epochs_range, loss, label='Training Loss')\nplt.plot(epochs_range, val_loss, label='Validation Loss')\nplt.legend(loc='upper right')\nplt.title('Training and Validation Loss')\n#plt.savefig('./foo.png')\nplt.show()",
"_____no_output_____"
]
],
[
[
"# Prediction using the Model\n\nLet's now take a look at actually running a prediction using the model. ",
"_____no_output_____"
]
],
[
[
"test_images, test_labels = next(validation_generator)\nclasses = model.predict(test_images, 10)\nclasses = classes.flatten()\nprint(classes)\nprint(test_labels)",
"_____no_output_____"
],
[
"fig, axes = plt.subplots(4, 5, figsize=(20,20))\naxes = axes.flatten()\ni = 0\nfor img, ax in zip(test_images, axes):\n ax.imshow(img)\n ax.axis('off')\n color = 'blue'\n if round(classes[i]) != test_labels[i] :\n color = 'red'\n if classes[i]>0.5:\n ax.set_title(\"Dog\",fontdict = {'size' : 20, 'color' : color});\n else :\n ax.set_title(\"Cat\",fontdict = {'size' : 20, 'color' : color});\n i+=1\nplt.tight_layout()\nplt.show()",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
cbf20c1e9c631249226e6f9c3b4bd20601abb963
| 534,888 |
ipynb
|
Jupyter Notebook
|
Course2-Build Better Generative Adversarial Networks (GANs)/week3/StyleGAN2.ipynb
|
shreyansh26/GANs-Specialization-Deeplearning.ai
|
1c9f5324f67360209a76e0040c607991bd8f4655
|
[
"MIT"
] | 1 |
2020-12-30T13:50:59.000Z
|
2020-12-30T13:50:59.000Z
|
Course2-Build Better Generative Adversarial Networks (GANs)/week3/StyleGAN2.ipynb
|
shreyansh26/GANs-Specialization-Deeplearning.ai
|
1c9f5324f67360209a76e0040c607991bd8f4655
|
[
"MIT"
] | null | null | null |
Course2-Build Better Generative Adversarial Networks (GANs)/week3/StyleGAN2.ipynb
|
shreyansh26/GANs-Specialization-Deeplearning.ai
|
1c9f5324f67360209a76e0040c607991bd8f4655
|
[
"MIT"
] | null | null | null | 1,473.520661 | 513,920 | 0.955469 |
[
[
[
"# StyleGAN2\n*Please note that this is an optional notebook that is meant to introduce more advanced concepts, if you're up for a challenge. So, don't worry if you don't completely follow every step! We provide external resources for extra base knowledge required to grasp some components of the advanced material.*\n\nIn this notebook, you're going to learn about StyleGAN2, from the paper [Analyzing and Improving the Image Quality of StyleGAN](https://arxiv.org/abs/1912.04958) (Karras et al., 2019), and how it builds on StyleGAN. This is the V2 of StyleGAN, so be prepared for even more extraordinary outputs. Here's the quick version: \n\n1. **Demodulation.** The instance normalization of AdaIN in the original StyleGAN actually was producing “droplet artifacts” that made the output images clearly fake. AdaIN is modified a bit in StyleGAN2 to make this not happen. Below, *Figure 1* from the StyleGAN2 paper is reproduced, showing the droplet artifacts in StyleGAN. \n\n\n\n2. **Path length regularization.** “Perceptual path length” (or PPL, which you can explore in [another optional notebook](https://www.coursera.org/learn/build-better-generative-adversarial-networks-gans/ungradedLab/BQjUq/optional-ppl)) was introduced in the original StyleGAN paper, as a metric for measuring the disentanglement of the intermediate noise space W. PPL measures the change in the output image, when interpolating between intermediate noise vectors $w$. You'd expect a good model to have a smooth transition during interpolation, where the same step size in $w$ maps onto the same amount of perceived change in the resulting image. \n\n Using this intuition, you can make the mapping from $W$ space to images smoother, by encouraging a given change in $w$ to correspond to a constant amount of change in the image. This is known as path length regularization, and as you might expect, included as a term in the loss function. This smoothness also made the generator model \"significantly easier to invert\"! Recall that inversion means going from a real or fake image to finding its $w$, so you can easily adapt the image's styles by controlling $w$.\n\n\n3. **No progressive growing.** While progressive growing was seemingly helpful for training the network more efficiently and with greater stability at lower resolutions before progressing to higher resolutions, there's actually a better way. Instead, you can replace it with 1) a better neural network architecture with skip and residual connections (which you also see in Course 3 models, Pix2Pix and CycleGAN), and 2) training with all of the resolutions at once, but gradually moving the generator's _attention_ from lower-resolution to higher-resolution dimensions. So in a way, still being very careful about how to handle different resolutions to make training eaiser, from lower to higher scales.\n\nThere are also a number of performance optimizations, like calculating the regularization less frequently. We won't focus on those in this notebook, but they are meaningful technical contributions. \n",
"_____no_output_____"
],
[
"But first, some useful imports:",
"_____no_output_____"
]
],
[
[
"import torch\nimport torch.nn as nn\nimport torch.nn.functional as F\nfrom torchvision.utils import make_grid\nimport matplotlib.pyplot as plt\n\ndef show_tensor_images(image_tensor, num_images=16, size=(3, 64, 64), nrow=3):\n '''\n Function for visualizing images: Given a tensor of images, number of images,\n size per image, and images per row, plots and prints the images in an uniform grid.\n '''\n image_tensor = (image_tensor + 1) / 2\n image_unflat = image_tensor.detach().cpu().clamp_(0, 1)\n image_grid = make_grid(image_unflat[:num_images], nrow=nrow, padding=2)\n plt.imshow(image_grid.permute(1, 2, 0).squeeze())\n plt.axis('off')\n plt.show()",
"_____no_output_____"
]
],
[
[
"## Fixing Instance Norm\nOne issue with instance normalization is that it can lose important information that is typically communicated by relative magnitudes. In StyleGAN2, it was proposed that the droplet artifects are a way for the network to \"sneak\" this magnitude information with a single large spike. This issue was also highlighted in the paper which introduced GauGAN, [Semantic Image Synthesis with Spatially-Adaptive Normalization](https://arxiv.org/abs/1903.07291) (Park et al.), earlier in 2019. In that more extreme case, instance normalization could sometimes eliminate all semantic information, as shown in their paper's *Figure 3*: \n\n\n\n\nWhile removing normalization is technically possible, it reduces the controllability of the model, a major feature of StyleGAN. Here's one solution from the paper:\n\n### Output Demodulation\nThe first solution notes that the scaling the output of a convolutional layer by style has a consistent and numerically reproducible impact on the standard deviation of its output. By scaling down the standard deviation of the output to 1, the droplet effect can be reduced. \n\nMore specifically, the style $s$, when applied as a multiple to convolutional weights $w$, resulting in weights $w'_{ijk}=s_i \\cdot w_{ijk}$ will have standard deviation $\\sigma_j = \\sqrt{\\sum_{i,k} w'^2_{ijk}}$. One can simply divide the output of the convolution by this factor. \n\nHowever, the authors note that dividing by this factor can also be incorporated directly into the the convolutional weights (with an added $\\epsilon$ for numerical stability):\n\n$$w''_{ijk}=\\frac{w'_{ijk}}{\\sqrt{\\sum_{i,k} w'^2_{ijk} + \\epsilon}}$$\n\nThis makes it so that this entire operation can be baked into a single convolutional layer, making it easier to work with, implement, and integrate into the existing architecture of the model.\n",
"_____no_output_____"
]
],
[
[
"class ModulatedConv2d(nn.Module):\n '''\n ModulatedConv2d Class, extends/subclass of nn.Module\n Values:\n channels: the number of channels the image has, a scalar\n w_dim: the dimension of the intermediate tensor, w, a scalar \n '''\n\n def __init__(self, w_dim, in_channels, out_channels, kernel_size, padding=1):\n super().__init__()\n self.conv_weight = nn.Parameter(\n torch.randn(out_channels, in_channels, kernel_size, kernel_size)\n )\n self.style_scale_transform = nn.Linear(w_dim, in_channels)\n self.eps = 1e-6\n self.padding = padding\n\n def forward(self, image, w):\n # There is a more efficient (vectorized) way to do this using the group parameter of F.conv2d,\n # but for simplicity and readibility you will go through one image at a time.\n images = []\n for i, w_cur in enumerate(w):\n # Calculate the style scale factor\n style_scale = self.style_scale_transform(w_cur)\n # Multiply it by the corresponding weight to get the new weights\n w_prime = self.conv_weight * style_scale[None, :, None, None]\n # Demodulate the new weights based on the above formula\n w_prime_prime = w_prime / torch.sqrt(\n (w_prime ** 2).sum([1, 2, 3])[:, None, None, None] + self.eps\n )\n images.append(F.conv2d(image[i][None], w_prime_prime, padding=self.padding))\n return torch.cat(images)\n \n def forward_efficient(self, image, w):\n # Here's the more efficient approach. It starts off mostly the same\n style_scale = self.style_scale_transform(w)\n w_prime = self.conv_weight[None] * style_scale[:, None, :, None, None]\n w_prime_prime = w_prime / torch.sqrt(\n (w_prime ** 2).sum([2, 3, 4])[:, :, None, None, None] + self.eps\n )\n # Now, the trick is that we'll make the images into one image, and \n # all of the conv filters into one filter, and then use the \"groups\"\n # parameter of F.conv2d to apply them all at once\n batchsize, in_channels, height, width = image.shape\n out_channels = w_prime_prime.shape[2]\n # Create an \"image\" where all the channels of the images are in one sequence\n efficient_image = image.view(1, batchsize * in_channels, height, width)\n efficient_filter = w_prime_prime.view(batchsize * out_channels, in_channels, *w_prime_prime.shape[3:])\n efficient_out = F.conv2d(efficient_image, efficient_filter, padding=self.padding, groups=batchsize)\n return efficient_out.view(batchsize, out_channels, *image.shape[2:])\n\nexample_modulated_conv = ModulatedConv2d(w_dim=128, in_channels=3, out_channels=3, kernel_size=3)\nnum_ex = 2\nimage_size = 64\nrand_image = torch.randn(num_ex, 3, image_size, image_size) # A 64x64 image with 3 channels\nrand_w = torch.randn(num_ex, 128)\nnew_image = example_modulated_conv(rand_image, rand_w)\nsecond_modulated_conv = ModulatedConv2d(w_dim=128, in_channels=3, out_channels=3, kernel_size=3)\nsecond_image = second_modulated_conv(new_image, rand_w)\n\nprint(\"Original noise (left), noise after modulated convolution (middle), noise after two modulated convolutions (right)\")\nplt.rcParams['figure.figsize'] = [8, 8]\nshow_tensor_images(torch.stack([rand_image, new_image, second_image], 1).view(-1, 3, image_size, image_size))",
"Original noise (left), noise after modulated convolution (middle), noise after two modulated convolutions (right)\n"
]
],
[
[
"## Path Length Regularization\n\nPath length regularization was introduced based on the usefulness of PPL, or perceptual path length, a metric used of evaluating disentanglement proposed in the original StyleGAN paper -- feel free to check out the [optional notebook](https://www.coursera.org/learn/build-better-generative-adversarial-networks-gans/ungradedLab/BQjUq/optional-ppl) for a detailed overview! In essence, for a fixed-size step in any direction in $W$ space, the metric attempts to make the change in image space to have a constant magnitude $a$. This is accomplished (in theory) by first taking the Jacobian of the generator with respect to $w$, which is $\\mathop{\\mathrm{J}_{\\mathrm{w}}}={\\partial g(\\mathrm{w})} / {\\partial \\mathrm{w}}$. \n\nThen, you take the L2 norm of Jacobian matrix and you multiply that by random images (that you sample from a normal distribution, as you often do): \n$\\Vert \\mathrm{J}_{\\mathrm{w}}^T \\mathrm{y} \\Vert_2$. This captures the expected magnitude of the change in pixel space. From this, you get a loss term, which penalizes the distance between this magnitude and $a$. The paper notes that this has similarities to spectral normalization (discussed in [another optional notebook](https://www.coursera.org/learn/build-basic-generative-adversarial-networks-gans/ungradedLab/c2FPs/optional-sn-gan) in Course 1), because it constrains multiple norms. \n\nAn additional optimization is also possible and ultimately used in the StyleGAN2 model: instead of directly computing $\\mathrm{J}_{\\mathrm{w}}^T \\mathrm{y}$, you can more efficiently calculate the gradient \n$\\nabla_{\\mathrm{w}} (g(\\mathrm{w}) \\cdot \\mathrm{y})$.\n\nFinally, a bit of talk on $a$: $a$ is not a fixed constant, but an exponentially decaying average of the magnitudes over various runs -- as with most times you see (decaying) averages being used, this is to smooth out the value of $a$ across multiple iterations, not just dependent on one. Notationally, with decay rate $\\gamma$, $a$ at the next iteration $a_{t+1} = {a_t} * (1 - \\gamma) + \\Vert \\mathrm{J}_{\\mathrm{w}}^T \\mathrm{y} \\Vert_2 * \\gamma$. \n\nHowever, for your one example iteration you can treat $a$ as a constant for simplicity. There is also an example of an update of $a$ after the calculation of the loss, so you can see what $a_{t+1}$ looks like with exponential decay.",
"_____no_output_____"
]
],
[
[
"# For convenience, we'll define a very simple generator here:\nclass SimpleGenerator(nn.Module):\n '''\n SimpleGenerator Class, for path length regularization demonstration purposes\n Values:\n channels: the number of channels the image has, a scalar\n w_dim: the dimension of the intermediate tensor, w, a scalar \n '''\n\n def __init__(self, w_dim, in_channels, hid_channels, out_channels, kernel_size, padding=1, init_size=64):\n super().__init__()\n self.w_dim = w_dim\n self.init_size = init_size\n self.in_channels = in_channels\n self.c1 = ModulatedConv2d(w_dim, in_channels, hid_channels, kernel_size)\n self.activation = nn.ReLU()\n self.c2 = ModulatedConv2d(w_dim, hid_channels, out_channels, kernel_size)\n\n def forward(self, w):\n image = torch.randn(len(w), self.in_channels, self.init_size, self.init_size).to(w.device)\n y = self.c1(image, w)\n y = self.activation(y)\n y = self.c2(y, w)\n return y",
"_____no_output_____"
],
[
"from torch.autograd import grad\ndef path_length_regulization_loss(generator, w, a):\n # Generate the images from w\n fake_images = generator(w)\n # Get the corresponding random images\n random_images = torch.randn_like(fake_images)\n # Output variation that we'd like to regularize\n output_var = (fake_images * random_images).sum()\n # Calculate the gradient with respect to the inputs\n cur_grad = grad(outputs=output_var, inputs=w)[0]\n # Calculate the distance from a\n penalty = (((cur_grad - a) ** 2).sum()).sqrt()\n return penalty, output_var\nsimple_gen = SimpleGenerator(w_dim=128, in_channels=3, hid_channels=64, out_channels=3, kernel_size=3)\nsamples = 10\ntest_w = torch.randn(samples, 128).requires_grad_()\na = 10\npenalty, variation = path_length_regulization_loss(simple_gen, test_w, a=a)\n\ndecay = 0.001 # How quickly a should decay\nnew_a = a * (1 - decay) + variation * decay\nprint(f\"Old a: {a}; new a: {new_a.item()}\")",
"Old a: 10; new a: 9.948847770690918\n"
]
],
[
[
"## No More Progressive Growing\nWhile the concepts behind progressive growing remain, you get to see how that is revamped and beefed up in StyleGAN2. This starts with generating all resolutions of images from the very start of training. You might be wondering why they didn't just do this in the first place: in the past, this has generally been unstable to do. However, by using residual or skip connections (there are two variants that both do better than without them), StyleGAN2 manages to replicate many of the dynamics of progressive growing in a less explicit way. Three architectures were considered for StyleGAN2 to replace the progressive growing. \n\nNote that in the following figure, *tRGB* and *fRGB* refer to the $1 \\times 1$ convolutions which transform the noise with some number channels at a given layer into a three-channel image for the generator, and vice versa for the discriminator.\n\n\n\n*The set of architectures considered for StyleGAN2 (from the paper). Ultimately, the skip generator and residual discriminator (highlighted in green) were chosen*.\n\n### Option a: MSG-GAN\n[MSG-GAN](https://arxiv.org/abs/1903.06048) (from Karnewar and Wang 2019), proposed a somewhat natural approach: generate all resolutions of images, but also directly pass each corresponding resolution to a block of the discriminator responsible for dealing with that resolution. \n\n### Option b: Skip Connections\n\nIn the skip-connection approach, each block takes the previous noise as input and generates the next resolution of noise. For the generator, each noise is converted to an image, upscaled to the maximum size, and then summed together. For the discriminator, the images are downsampled to each block's size and converted to noises.\n\n### Option c: Residual Nets\n\nIn the residual network approach, each block adds residual detail to the noise, and the image conversion happens at the end for the generator and at the start for the discriminator.\n\n### StyleGAN2: Skip Generator, Residual Discriminator\nBy experiment, the skip generator and residual discriminator were chosen. One interesting effect is that, as the images for the skip generator are additive, you can explicitly see the contribution from each of them, and measure the magnitude of each block's contribution. If you're not 100% sure how to implement skip and residual models yet, don't worry - you'll get a lot of practice with that in Course 3!\n\n\n\n*Figure 8 from StyleGAN2 paper, showing generator contributions by different resolution blocks of the generator over time. The y-axis is the standard deviation of the contributions, and the x-axis is the number of millions of images that the model has been trained on (training progress).*",
"_____no_output_____"
],
[
"Now, you've seen the primary changes, and you understand the current state-of-the-art in image generation, StyleGAN2, congratulations! \n\nIf you're the type of person who reads through the optional notebooks for fun, maybe you'll make the next state-of-the-art! Can't wait to cover your GAN in a new notebook :)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
]
] |
cbf2261b9a785deb50e1fb316337af1a4fdc9b1f
| 351,167 |
ipynb
|
Jupyter Notebook
|
Visual_Analysis.ipynb
|
mohantechis/analysis-data
|
dfe4f38d791a9be875003ea1de83f31ad76ac43b
|
[
"MIT"
] | null | null | null |
Visual_Analysis.ipynb
|
mohantechis/analysis-data
|
dfe4f38d791a9be875003ea1de83f31ad76ac43b
|
[
"MIT"
] | null | null | null |
Visual_Analysis.ipynb
|
mohantechis/analysis-data
|
dfe4f38d791a9be875003ea1de83f31ad76ac43b
|
[
"MIT"
] | null | null | null | 816.667442 | 318,008 | 0.952328 |
[
[
[
"# Visual Analysis\n\nThis notebook has been created to support two main purposes:\n* Based on an input image and a set of models, display the action-space probability distribution.\n* Based on an input image and a set of models, visualize which parts of the image the model looks at.\n\n## Usage\n\nThe workbook requires the following:\n* A set of raw images captured from the front camera of the car\n* One or more static model files (`model_*.pb`)\n* The `model_metadata.json`\n\n## Contributions\n\nAs usual, your ideas are very welcome and encouraged so if you have any suggestions either bring them to [the AWS DeepRacer Community](http://join.deepracing.io) or share as code contributions.\n\n## Requirements\n\nBefore you start using the notebook, you will need to install some dependencies. If you haven't yet done so, have a look at [The README.md file](/edit/README.md#running-the-notebooks) to find what you need to install.\n\nThis workbook will require `tensorflow` and `cv2` to work.\n\n## Imports\n\nRun the imports block below:",
"_____no_output_____"
]
],
[
[
"import json\nimport os\nimport glob\n\nimport numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\n\nimport cv2\n\nimport tensorflow as tf\nfrom tensorflow.gfile import GFile\n\nfrom deepracer.model import load_session, visualize_gradcam_discrete_ppo, rgb2gray",
"_____no_output_____"
]
],
[
[
"## Configure and load files\n\nProvide the paths where the image and models are stored. Also define which iterations you would like to review.",
"_____no_output_____"
]
],
[
[
"img_selection = 'logs/sample-model/pictures/*.png'\nmodel_path = 'logs/sample-model/model'\niterations = [15, 30, 48]",
"_____no_output_____"
]
],
[
[
"Load the model metadata in, and define which sensor is in use.",
"_____no_output_____"
]
],
[
[
"with open(\"{}/model_metadata.json\".format(model_path),\"r\") as jsonin:\n model_metadata=json.load(jsonin)\nmy_sensor = [sensor for sensor in model_metadata['sensor'] if sensor != \"LIDAR\"][0]\ndisplay(model_metadata)",
"_____no_output_____"
]
],
[
[
"Load in the pictures from the pre-defined path.",
"_____no_output_____"
]
],
[
[
"picture_files = sorted(glob.glob(img_selection))\ndisplay(picture_files)",
"_____no_output_____"
],
[
"action_names = []\ndegree_sign= u'\\N{DEGREE SIGN}'\nfor action in model_metadata['action_space']:\n action_names.append(str(action['steering_angle'])+ degree_sign + \" \"+\"%.1f\"%action[\"speed\"])\ndisplay(action_names)",
"_____no_output_____"
]
],
[
[
"## Load the model files and process pictures\nWe will now load in the models and process the pictures. Output is a nested list with size `n` models as the outer and `m` picture as the inner list. The inner list will contain a number of values equal to the ",
"_____no_output_____"
]
],
[
[
"model_inference = []\nmodels_file_path = []\n\nfor n in iterations:\n models_file_path.append(\"{}/model_{}.pb\".format(model_path,n))\ndisplay(models_file_path)\n\nfor model_file in models_file_path:\n model, obs, model_out = load_session(model_file, my_sensor)\n arr = []\n for f in picture_files[:]:\n img = cv2.imread(f)\n img = cv2.resize(img, dsize=(160, 120), interpolation=cv2.INTER_CUBIC)\n img_arr = np.array(img)\n img_arr = rgb2gray(img_arr)\n img_arr = np.expand_dims(img_arr, axis=2)\n current_state = {\"observation\": img_arr} #(1, 120, 160, 1)\n y_output = model.run(model_out, feed_dict={obs:[img_arr]})[0]\n arr.append (y_output)\n \n model_inference.append(arr)\n model.close()\n tf.reset_default_graph()",
"_____no_output_____"
]
],
[
[
"## Simulation Image Analysis - Probability distribution on decisions (actions)\n\nWe will now show the probabilities per action for the selected picture and iterations. The higher the probability of one single action the more mature is the model. Comparing different models enables the developer to see how the model is becoming more certain over time.",
"_____no_output_____"
]
],
[
[
"PICTURE_INDEX=1\ndisplay(picture_files[PICTURE_INDEX])",
"_____no_output_____"
],
[
"x = list(range(1,len(action_names)+1))\n\nnum_plots = len(iterations)\nfig, ax = plt.subplots(num_plots,1,figsize=(20,3*num_plots),sharex=True,squeeze=False)\n\nfor p in range(0, num_plots):\n ax[p][0].bar(x,model_inference[p][PICTURE_INDEX][::-1])\n plt.setp(ax[p, 0], ylabel=os.path.basename(models_file_path[p]))\n \nplt.xticks(x,action_names[::-1],rotation='vertical')\nplt.show()",
"_____no_output_____"
]
],
[
[
"## What is the model looking at?\n\nGradcam: visual heatmap of where the model is looking to make its decisions. based on https://arxiv.org/pdf/1610.02391.pdf",
"_____no_output_____"
]
],
[
[
"heatmaps = []\nview_models = models_file_path[1:3]\n\nfor model_file in view_models:\n model, obs, model_out = load_session(model_file, my_sensor)\n arr = []\n for f in picture_files:\n img = cv2.imread(f)\n img = cv2.resize(img, dsize=(160, 120), interpolation=cv2.INTER_CUBIC)\n heatmap = visualize_gradcam_discrete_ppo(model, img, category_index=0, num_of_actions=len(action_names))\n heatmaps.append(heatmap) \n\n tf.reset_default_graph()",
"Device mapping:\n/job:localhost/replica:0/task:0/device:XLA_CPU:0 -> device: XLA_CPU device\n\nload graph: model/Demo-Reinvent-1/model/model_30.pb\nDevice mapping:\n/job:localhost/replica:0/task:0/device:XLA_CPU:0 -> device: XLA_CPU device\n\nload graph: model/Demo-Reinvent-1/model/model_48.pb\n"
],
[
"fig, ax = plt.subplots(len(view_models),len(picture_files),\n figsize=(7*len(view_models),2.5*len(picture_files)), sharex=True, sharey=True, squeeze=False)\n\nfor i in list(range(len(view_models))):\n plt.setp(ax[i, 0], ylabel=os.path.basename(view_models[i]))\n for j in list(range(len(picture_files))):\n ax[i][j].imshow(heatmaps[i * len(picture_files) + j])\n plt.setp(ax[-1:, j], xlabel=os.path.basename(picture_files[j]))\n \nplt.show()",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
cbf23504740c121c927e620667800e696ad65a4c
| 68,572 |
ipynb
|
Jupyter Notebook
|
TwoStateProblem.ipynb
|
youhom/CS332-project
|
d5b4c68917dbde0b346cfb23ae56fb060392e45b
|
[
"MIT"
] | null | null | null |
TwoStateProblem.ipynb
|
youhom/CS332-project
|
d5b4c68917dbde0b346cfb23ae56fb060392e45b
|
[
"MIT"
] | null | null | null |
TwoStateProblem.ipynb
|
youhom/CS332-project
|
d5b4c68917dbde0b346cfb23ae56fb060392e45b
|
[
"MIT"
] | null | null | null | 211.641975 | 18,016 | 0.888876 |
[
[
[
"import numpy as np\nimport pandas as pd\nfrom copy import deepcopy\nfrom matplotlib import pyplot as plt\nimport calculations\nimport agent\nimport environment",
"_____no_output_____"
],
[
"def train(env, agent, num_iterations, algo, learning_rate, lam):\n weights = []\n new_weight = np.zeros(agent.num_features)\n z = np.zeros(agent.num_features)\n \n cur_st = env.state\n cur_sib_st = cur_st\n \n for ctr in range(1, num_iterations+1):\n # Perform action in environment using current weights self.w\n act = agent.get_action(cur_st)\n next_st, reward = env.step(act)\n\n # Get next sibling state\n next_sib_st, _ = calculations.sibling(cur_st, act, next_st, env.transition_matrix)\n\n # Sample reward from current sibling state\n env_clone = deepcopy(env)\n env_clone.state = cur_sib_st\n _, reward_sib = env_clone.step(agent.get_action(cur_sib_st))\n\n # Update eligibility traces and weights based on chosen algorithm\n if algo == 'TD':\n featdiff = env.gamma * agent.features[next_st] - agent.features[cur_st]\n d = reward + sum(featdiff * new_weight)\n new_weight = new_weight + learning_rate * d * z / ctr\n z = env.gamma * lam * z + agent.features[next_st]\n\n elif algo == 'STD-99':\n featdiff = env.gamma * (agent.features[next_st] - agent.features[next_sib_st]) - (\n agent.features[cur_st] - agent.features[cur_sib_st])\n d = reward + sum(featdiff * new_weight)\n new_weight = new_weight + learning_rate * d * z / ctr\n z = env.gamma * lam * z + agent.features[next_st] - agent.features[next_sib_st]\n\n elif algo == 'STD-01':\n featdiff = gamma * (agent.features[next_st] - agent.features[next_sib_st]) - (\n agent.features[cur_st] - agent.features[cur_sib_st])\n d = reward - reward_sib + sum(featdiff * new_weight)\n new_weight = new_weight + learning_rate * d * z / ctr\n z = gamma * lam * z + (agent.features[next_st] - agent.features[next_sib_st]) / \\\n env.transition_matrix[cur_st][act][next_st]\n\n # update historical sibling states\n cur_st = next_st\n cur_sib_st = next_sib_st\n weights.append(new_weight)\n agent.weight = new_weight\n return weights",
"_____no_output_____"
],
[
"num_states = 2\nnum_actions = 2\n\n# Randomly initialize transition_matrix, rewards, alpha, and features\ntransition_matrix = np.random.rand(num_states,num_actions,num_states)\nfor i in range(num_states):\n for j in range(num_actions):\n transition_matrix[i][j] = transition_matrix[i][j] / sum(transition_matrix[i][j])\nrewards = np.random.rand(num_states,num_states)*2 - 1\ngamma = np.random.rand(1)[0]\nfeatures = np.random.rand(num_states,num_states-1)*2\n\n# Calculate value function for each policy\npolicy_values = calculations.policy_values(transition_matrix, rewards, gamma)\noptimal_policy = max(list(policy_values.items()), key=lambda x: sum(x[1]))[0]",
"_____no_output_____"
],
[
"# Set up environment\nenv = environment.Environment(transition_matrix, rewards, gamma)\n\n# Set up and run policy iteration on agent\ntd_agent = agent.Agent(features, optimal_policy)\ntd_weights = train(env, td_agent, num_iterations=10000, algo=\"TD\", learning_rate=2.0, lam=1.0)\ntd_agent.policy = calculations.make_new_policy(td_agent, td_weights[-1], transition_matrix, rewards, gamma)\n\n# Plot weight values at each iteration\nplt.plot(td_weights[50:])\nplt.xlabel(\"Number of Transitions\")\nplt.ylabel(\"w\")\nplt.title(\"TD Weights\")\nplt.show()\nprint()\n\n# Plot value differences at each iteration\nstdAB = calculations.value_difference([1,-1], features, td_weights, optimal_policy, policy_values)\nplt.plot(stdAB[50:])\nplt.xlabel(\"Number of Transitions\")\nplt.title(\"TD Difference in Approximation and Real V(A) - V(B)\")\nplt.show()\n",
"_____no_output_____"
],
[
"# Set up environment\nenv = environment.Environment(transition_matrix, rewards, gamma)\n\n# Set up and run policy iteration on agent\nstd_agent = agent.Agent(features, optimal_policy)\nstd_weights = train(env, std_agent, num_iterations=10000, algo=\"STD-01\", learning_rate=2.0, lam=1.0)\nstd_agent.policy = calculations.make_new_policy(std_agent, std_weights[-1], transition_matrix, rewards, gamma)\n\n# Plot weight values at each iteration\nplt.plot(std_weights[50:])\nplt.xlabel(\"Number of Transitions\")\nplt.ylabel(\"w\")\nplt.title(\"STD Weights\")\nplt.show()\nprint()\n\n# Plot value differences at each iteration\nstdAB = calculations.value_difference([1,-1], features, std_weights, optimal_policy, policy_values)\nplt.plot(stdAB[50:])\nplt.xlabel(\"Number of Transitions\")\nplt.title(\"STD Difference in Approximation and Real V(A) - V(B)\")\nplt.show()\n",
"_____no_output_____"
],
[
"pd.DataFrame(np.array([[optimal_policy, td_agent.policy, std_agent.policy],\nlist(map(lambda x: sum(policy_values[x]), [optimal_policy, td_agent.policy, std_agent.policy]))]),\n [\"Policy\", \"Policy Value\"], [\"Optimal\", \"TD\", \"STD\"])",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbf237157778e3d717733353ced7419b26d74871
| 74,621 |
ipynb
|
Jupyter Notebook
|
Traffic_Sign_Classifier.ipynb
|
atadkase/udacitysdcnd-p2
|
cb12e691822d87566b21e353c2670dad0ea92d66
|
[
"MIT"
] | null | null | null |
Traffic_Sign_Classifier.ipynb
|
atadkase/udacitysdcnd-p2
|
cb12e691822d87566b21e353c2670dad0ea92d66
|
[
"MIT"
] | null | null | null |
Traffic_Sign_Classifier.ipynb
|
atadkase/udacitysdcnd-p2
|
cb12e691822d87566b21e353c2670dad0ea92d66
|
[
"MIT"
] | null | null | null | 87.276023 | 19,876 | 0.791399 |
[
[
[
"# Self-Driving Car Engineer Nanodegree\n\n## Deep Learning\n\n## Project: Build a Traffic Sign Recognition Classifier\n\nIn this notebook, a template is provided for you to implement your functionality in stages, which is required to successfully complete this project. If additional code is required that cannot be included in the notebook, be sure that the Python code is successfully imported and included in your submission if necessary. \n\n> **Note**: Once you have completed all of the code implementations, you need to finalize your work by exporting the iPython Notebook as an HTML document. Before exporting the notebook to html, all of the code cells need to have been run so that reviewers can see the final implementation and output. You can then export the notebook by using the menu above and navigating to \\n\",\n \"**File -> Download as -> HTML (.html)**. Include the finished document along with this notebook as your submission. \n\nIn addition to implementing code, there is a writeup to complete. The writeup should be completed in a separate file, which can be either a markdown file or a pdf document. There is a [write up template](https://github.com/udacity/CarND-Traffic-Sign-Classifier-Project/blob/master/writeup_template.md) that can be used to guide the writing process. Completing the code template and writeup template will cover all of the [rubric points](https://review.udacity.com/#!/rubrics/481/view) for this project.\n\nThe [rubric](https://review.udacity.com/#!/rubrics/481/view) contains \"Stand Out Suggestions\" for enhancing the project beyond the minimum requirements. The stand out suggestions are optional. If you decide to pursue the \"stand out suggestions\", you can include the code in this Ipython notebook and also discuss the results in the writeup file.\n\n\n>**Note:** Code and Markdown cells can be executed using the **Shift + Enter** keyboard shortcut. In addition, Markdown cells can be edited by typically double-clicking the cell to enter edit mode.",
"_____no_output_____"
],
[
"---\n## Step 0: Load The Data",
"_____no_output_____"
]
],
[
[
"# Load pickled data\nimport pickle\n\n# TODO: Fill this in based on where you saved the training and testing data\n\ntraining_file = './traffic-signs-data/train.p'\nvalidation_file= './traffic-signs-data/valid.p'\ntesting_file = './traffic-signs-data/test.p'\n\nwith open(training_file, mode='rb') as f:\n train = pickle.load(f)\nwith open(validation_file, mode='rb') as f:\n valid = pickle.load(f)\nwith open(testing_file, mode='rb') as f:\n test = pickle.load(f)\n \nX_train, y_train = train['features'], train['labels']\nX_valid, y_valid = valid['features'], valid['labels']\nX_test, y_test = test['features'], test['labels']",
"_____no_output_____"
]
],
[
[
"---\n\n## Step 1: Dataset Summary & Exploration\n\nThe pickled data is a dictionary with 4 key/value pairs:\n\n- `'features'` is a 4D array containing raw pixel data of the traffic sign images, (num examples, width, height, channels).\n- `'labels'` is a 1D array containing the label/class id of the traffic sign. The file `signnames.csv` contains id -> name mappings for each id.\n- `'sizes'` is a list containing tuples, (width, height) representing the original width and height the image.\n- `'coords'` is a list containing tuples, (x1, y1, x2, y2) representing coordinates of a bounding box around the sign in the image. **THESE COORDINATES ASSUME THE ORIGINAL IMAGE. THE PICKLED DATA CONTAINS RESIZED VERSIONS (32 by 32) OF THESE IMAGES**\n\nComplete the basic data summary below. Use python, numpy and/or pandas methods to calculate the data summary rather than hard coding the results. For example, the [pandas shape method](http://pandas.pydata.org/pandas-docs/stable/generated/pandas.DataFrame.shape.html) might be useful for calculating some of the summary results. ",
"_____no_output_____"
],
[
"### Provide a Basic Summary of the Data Set Using Python, Numpy and/or Pandas",
"_____no_output_____"
]
],
[
[
"### Replace each question mark with the appropriate value. \n### Use python, pandas or numpy methods rather than hard coding the results\n\nimport pandas as pd\nimport numpy as np\nimport tensorflow as tf\n\n\n\n# TODO: Number of training examples\nn_train = len(X_train)\n\n# TODO: Number of validation examples\nn_validation = len(X_valid)\n\n# TODO: Number of testing examples.\nn_test = len(X_test)\n\n# TODO: What's the shape of an traffic sign image?\nimage_shape = np.shape(X_train[0]) \n\n# TODO: How many unique classes/labels there are in the dataset.\nn_classes = np.shape(np.unique(y_train))[0]\n\nprint(\"Number of training examples =\", n_train)\nprint(\"Number of validation examples =\", n_validation)\nprint(\"Number of testing examples =\", n_test)\nprint(\"Image data shape =\", image_shape)\nprint(\"Number of classes =\", n_classes)",
"/home/ashu/udacity/miniconda3/envs/IntroToTensorFlow/lib/python3.6/site-packages/h5py/__init__.py:36: FutureWarning: Conversion of the second argument of issubdtype from `float` to `np.floating` is deprecated. In future, it will be treated as `np.float64 == np.dtype(float).type`.\n from ._conv import register_converters as _register_converters\n"
]
],
[
[
"### Include an exploratory visualization of the dataset",
"_____no_output_____"
],
[
"Visualize the German Traffic Signs Dataset using the pickled file(s). This is open ended, suggestions include: plotting traffic sign images, plotting the count of each sign, etc. \n\nThe [Matplotlib](http://matplotlib.org/) [examples](http://matplotlib.org/examples/index.html) and [gallery](http://matplotlib.org/gallery.html) pages are a great resource for doing visualizations in Python.\n\n**NOTE:** It's recommended you start with something simple first. If you wish to do more, come back to it after you've completed the rest of the sections. It can be interesting to look at the distribution of classes in the training, validation and test set. Is the distribution the same? Are there more examples of some classes than others?",
"_____no_output_____"
]
],
[
[
"### Data exploration visualization code goes here.\n### Feel free to use as many code cells as needed.\nimport matplotlib.pyplot as plt\nimport random\n# Visualizations will be shown in the notebook.\n%matplotlib inline\n\nindex = random.randint(0, len(X_train))\nimage = X_train[index].squeeze()\n\nplt.figure(figsize=(1,1))\nplt.imshow(image)\nprint(y_train[index])\n\n[n_classes, counts] = np.unique(y_train, return_counts=True)\nplt.figure(figsize=(5,5))\nplt.bar(n_classes,counts)\nplt.xlabel('Traffic Sign Class ID')\nplt.title('Distribution of traffic sign classes in training input')\nplt.ylabel('Number of images')\nplt.show()",
"5\n"
]
],
[
[
"----\n\n## Step 2: Design and Test a Model Architecture\n\nDesign and implement a deep learning model that learns to recognize traffic signs. Train and test your model on the [German Traffic Sign Dataset](http://benchmark.ini.rub.de/?section=gtsrb&subsection=dataset).\n\nThe LeNet-5 implementation shown in the [classroom](https://classroom.udacity.com/nanodegrees/nd013/parts/fbf77062-5703-404e-b60c-95b78b2f3f9e/modules/6df7ae49-c61c-4bb2-a23e-6527e69209ec/lessons/601ae704-1035-4287-8b11-e2c2716217ad/concepts/d4aca031-508f-4e0b-b493-e7b706120f81) at the end of the CNN lesson is a solid starting point. You'll have to change the number of classes and possibly the preprocessing, but aside from that it's plug and play! \n\nWith the LeNet-5 solution from the lecture, you should expect a validation set accuracy of about 0.89. To meet specifications, the validation set accuracy will need to be at least 0.93. It is possible to get an even higher accuracy, but 0.93 is the minimum for a successful project submission. \n\nThere are various aspects to consider when thinking about this problem:\n\n- Neural network architecture (is the network over or underfitting?)\n- Play around preprocessing techniques (normalization, rgb to grayscale, etc)\n- Number of examples per label (some have more than others).\n- Generate fake data.\n\nHere is an example of a [published baseline model on this problem](http://yann.lecun.com/exdb/publis/pdf/sermanet-ijcnn-11.pdf). It's not required to be familiar with the approach used in the paper but, it's good practice to try to read papers like these.",
"_____no_output_____"
],
[
"### Pre-process the Data Set (normalization, grayscale, etc.)",
"_____no_output_____"
],
[
"Minimally, the image data should be normalized so that the data has mean zero and equal variance. For image data, `(pixel - 128)/ 128` is a quick way to approximately normalize the data and can be used in this project. \n\nOther pre-processing steps are optional. You can try different techniques to see if it improves performance. \n\nUse the code cell (or multiple code cells, if necessary) to implement the first step of your project.",
"_____no_output_____"
]
],
[
[
"### Preprocess the data here. It is required to normalize the data. Other preprocessing steps could include \n### converting to grayscale, etc.\n### Feel free to use as many code cells as needed.\nfrom sklearn.utils import shuffle\n\nX_train, y_train = shuffle(X_train, y_train)\nX_train_norm = (X_train-128.0)/128.0\nX_valid_norm = (X_valid - 128.0)/128.0\nX_test_norm = (X_test - 128.0)/128.0\n",
"_____no_output_____"
]
],
[
[
"### Model Architecture",
"_____no_output_____"
]
],
[
[
"### Define your architecture here.\n### Feel free to use as many code cells as needed.\nfrom tensorflow.contrib.layers import flatten\n\ndef LeNet(x, keep_probability):\n # Hyperparameters\n mu = 0\n sigma = 0.1\n\n # Layer 1: Convolutional. Input = 32x32x1. Output = 28x28x12.\n conv1_W = tf.Variable(tf.truncated_normal(shape=(5, 5, 3, 12), mean = mu, stddev = sigma))\n conv1_b = tf.Variable(tf.zeros(12))\n conv1 = tf.nn.conv2d(x, conv1_W, strides=[1, 1, 1, 1], padding='VALID') + conv1_b\n\n # Activation.\n conv1 = tf.nn.relu(conv1)\n\n #Pooling. Input = 28x28x12. Output = 14x14x12.\n conv1 = tf.nn.max_pool(conv1, ksize=[1, 2, 2, 1], strides=[1, 2, 2, 1], padding='VALID')\n\n #Layer 2: Convolutional. Output = 10x10x28.\n conv2_W = tf.Variable(tf.truncated_normal(shape=(5, 5, 12, 28), mean = mu, stddev = sigma))\n conv2_b = tf.Variable(tf.zeros(28))\n conv2 = tf.nn.conv2d(conv1, conv2_W, strides=[1, 1, 1, 1], padding='VALID') + conv2_b\n\n #Activation.\n conv2 = tf.nn.relu(conv2)\n\n \n #Layer 3: Convolutional. Output = 6x6x36.\n conv3_W = tf.Variable(tf.truncated_normal(shape=(5, 5, 28, 36), mean = mu, stddev = sigma))\n conv3_b = tf.Variable(tf.zeros(36))\n conv3 = tf.nn.conv2d(conv2, conv3_W, strides=[1, 1, 1, 1], padding='VALID') + conv3_b\n\n #Activation.\n conv3 = tf.nn.relu(conv3)\n\n # Pooling. Input = 6x6x36. Output = 3x3x36.\n conv3 = tf.nn.max_pool(conv3, ksize=[1, 2, 2, 1], strides=[1, 2, 2, 1], padding='VALID')\n \n \n\n #Flatten. Input = 3x3x36. Output = 324.\n fc0 = flatten(conv3)\n\n #Layer 3: Fully Connected. Input = 324. Output = 240.\n fc1_W = tf.Variable(tf.truncated_normal(shape=(324, 240), mean = mu, stddev = sigma))\n fc1_b = tf.Variable(tf.zeros(240))\n fc1 = tf.matmul(fc0, fc1_W) + fc1_b\n\n # Activation.\n fc1 = tf.nn.relu(fc1)\n \n fc1 = tf.nn.dropout(fc1, keep_prob=keep_probability)\n\n # Layer 4: Fully Connected. Input = 240. Output = 98.\n fc2_W = tf.Variable(tf.truncated_normal(shape=(240, 98), mean = mu, stddev = sigma))\n fc2_b = tf.Variable(tf.zeros(98))\n fc2 = tf.matmul(fc1, fc2_W) + fc2_b\n\n # Activation.\n fc2 = tf.nn.relu(fc2)\n\n # Layer 5: Fully Connected. Input = 98. Output = 43.\n fc3_W = tf.Variable(tf.truncated_normal(shape=(98, 43), mean = mu, stddev = sigma))\n fc3_b = tf.Variable(tf.zeros(43))\n logits = tf.matmul(fc2, fc3_W) + fc3_b\n\n return logits\n",
"_____no_output_____"
]
],
[
[
"### Train, Validate and Test the Model",
"_____no_output_____"
],
[
"A validation set can be used to assess how well the model is performing. A low accuracy on the training and validation\nsets imply underfitting. A high accuracy on the training set but low accuracy on the validation set implies overfitting.",
"_____no_output_____"
]
],
[
[
"### Train your model here.\n### Calculate and report the accuracy on the training and validation set.\n### Once a final model architecture is selected, \n### the accuracy on the test set should be calculated and reported as well.\n### Feel free to use as many code cells as needed.\n\nEPOCHS = 10\nBATCH_SIZE = 128\nx = tf.placeholder(tf.float32, (None, 32, 32, 3))\ny = tf.placeholder(tf.int32, (None))\nkeep_probability = tf.placeholder(tf.float32)\none_hot_y = tf.one_hot(y, 43)\n\n\nrate = 0.001\n\nlogits = LeNet(x, keep_probability)\ncross_entropy = tf.nn.softmax_cross_entropy_with_logits(labels=one_hot_y, logits=logits)\nloss_operation = tf.reduce_mean(cross_entropy)\noptimizer = tf.train.AdamOptimizer(learning_rate = rate)\ntraining_operation = optimizer.minimize(loss_operation)\n\n\ncorrect_prediction = tf.equal(tf.argmax(logits, 1), tf.argmax(one_hot_y, 1))\naccuracy_operation = tf.reduce_mean(tf.cast(correct_prediction, tf.float32))\nsaver = tf.train.Saver()\n\ndef evaluate(X_data, y_data):\n num_examples = len(X_data)\n total_accuracy = 0\n sess = tf.get_default_session()\n for offset in range(0, num_examples, BATCH_SIZE):\n batch_x, batch_y = X_data[offset:offset+BATCH_SIZE], y_data[offset:offset+BATCH_SIZE]\n accuracy = sess.run(accuracy_operation, feed_dict={x: batch_x, y: batch_y, keep_probability: 1.0})\n total_accuracy += (accuracy * len(batch_x))\n return total_accuracy / num_examples\n",
"WARNING:tensorflow:From <ipython-input-6-f6278b7cdf0f>:18: softmax_cross_entropy_with_logits (from tensorflow.python.ops.nn_ops) is deprecated and will be removed in a future version.\nInstructions for updating:\n\nFuture major versions of TensorFlow will allow gradients to flow\ninto the labels input on backprop by default.\n\nSee @{tf.nn.softmax_cross_entropy_with_logits_v2}.\n\n"
],
[
"with tf.Session() as sess:\n sess.run(tf.global_variables_initializer())\n num_examples = len(X_train)\n \n print(\"Training...\")\n print()\n for i in range(EPOCHS):\n X_train_norm, y_train = shuffle(X_train_norm, y_train)\n for offset in range(0, num_examples, BATCH_SIZE):\n end = offset + BATCH_SIZE\n batch_x, batch_y = X_train_norm[offset:end], y_train[offset:end]\n sess.run(training_operation, feed_dict={x: batch_x, y: batch_y, keep_probability: 0.7})\n \n validation_accuracy = evaluate(X_valid_norm, y_valid)\n training_accuracy = evaluate(X_train_norm, y_train)\n print(\"EPOCH {} ...\".format(i+1))\n print(\"Training Accuracy = {:.3f}\".format(training_accuracy))\n print(\"Validation Accuracy = {:.3f}\".format(validation_accuracy))\n print()\n \n saver.save(sess, './lenet')\n print(\"Model saved\")",
"Training...\n\nEPOCH 1 ...\nTraining Accuracy = 0.943\nValidation Accuracy = 0.884\n\nEPOCH 2 ...\nTraining Accuracy = 0.982\nValidation Accuracy = 0.945\n\nEPOCH 3 ...\nTraining Accuracy = 0.989\nValidation Accuracy = 0.944\n\nEPOCH 4 ...\nTraining Accuracy = 0.993\nValidation Accuracy = 0.956\n\nEPOCH 5 ...\nTraining Accuracy = 0.993\nValidation Accuracy = 0.959\n\nEPOCH 6 ...\nTraining Accuracy = 0.990\nValidation Accuracy = 0.957\n\nEPOCH 7 ...\nTraining Accuracy = 0.996\nValidation Accuracy = 0.962\n\nEPOCH 8 ...\nTraining Accuracy = 0.995\nValidation Accuracy = 0.954\n\nEPOCH 9 ...\nTraining Accuracy = 0.998\nValidation Accuracy = 0.964\n\nEPOCH 10 ...\nTraining Accuracy = 0.998\nValidation Accuracy = 0.970\n\nModel saved\n"
],
[
"with tf.Session() as sess:\n saver.restore(sess, tf.train.latest_checkpoint('.'))\n\n test_accuracy = evaluate(X_test_norm, y_test)\n print(\"Test Accuracy = {:.3f}\".format(test_accuracy))\n",
"INFO:tensorflow:Restoring parameters from ./lenet\nTest Accuracy = 0.951\n"
]
],
[
[
"---\n\n## Step 3: Test a Model on New Images\n\nTo give yourself more insight into how your model is working, download at least five pictures of German traffic signs from the web and use your model to predict the traffic sign type.\n\nYou may find `signnames.csv` useful as it contains mappings from the class id (integer) to the actual sign name.",
"_____no_output_____"
],
[
"### Load and Output the Images",
"_____no_output_____"
]
],
[
[
"### Load the images and plot them here.\n### Feel free to use as many code cells as needed.\nfrom skimage.transform import resize\n\nPATH = \"./test-traffic-signs/test{0:1d}.jpg\"\nnum_images = 5\ntest_images = np.ndarray((num_images,32,32,3))\nfig = plt.figure(figsize=(5, 5)) \ntest_outputs = [33, 13, 36, 4, 11]\nfor i in range(1,num_images+1):\n p = PATH.format(i)\n image = plt.imread(p)\n image = (image-128.0)/128.0\n resized_image = resize(image, (32,32))\n test_images[i-1] = resized_image\n sub = fig.add_subplot(num_images, 1, i)\n sub.imshow(test_images[i-1,:,:,:].squeeze())",
"/home/ashu/udacity/miniconda3/envs/IntroToTensorFlow/lib/python3.6/site-packages/skimage/transform/_warps.py:84: UserWarning: The default mode, 'constant', will be changed to 'reflect' in skimage 0.15.\n warn(\"The default mode, 'constant', will be changed to 'reflect' in \"\nClipping input data to the valid range for imshow with RGB data ([0..1] for floats or [0..255] for integers).\nClipping input data to the valid range for imshow with RGB data ([0..1] for floats or [0..255] for integers).\nClipping input data to the valid range for imshow with RGB data ([0..1] for floats or [0..255] for integers).\nClipping input data to the valid range for imshow with RGB data ([0..1] for floats or [0..255] for integers).\nClipping input data to the valid range for imshow with RGB data ([0..1] for floats or [0..255] for integers).\n"
]
],
[
[
"### Predict the Sign Type for Each Image",
"_____no_output_____"
]
],
[
[
"### Run the predictions here and use the model to output the prediction for each image.\n### Make sure to pre-process the images with the same pre-processing pipeline used earlier.\n### Feel free to use as many code cells as needed.\nwith tf.Session() as sess:\n saver.restore(sess, tf.train.latest_checkpoint('.'))\n test_predictions = sess.run(tf.argmax(logits,1), feed_dict={x: test_images, y: test_outputs, keep_probability: 1.0})\n print(test_predictions)\n",
"INFO:tensorflow:Restoring parameters from ./lenet\n[33 13 36 3 11]\n"
]
],
[
[
"### Analyze Performance",
"_____no_output_____"
]
],
[
[
"### Calculate the accuracy for these 5 new images. \n### For example, if the model predicted 1 out of 5 signs correctly, it's 20% accurate on these new images.\nwith tf.Session() as sess:\n saver.restore(sess, tf.train.latest_checkpoint('.'))\n\n test_accuracy = evaluate(test_images, test_outputs)\n print(\"Test Accuracy = {:.3f}\".format(test_accuracy))",
"INFO:tensorflow:Restoring parameters from ./lenet\nTest Accuracy = 0.800\n"
]
],
[
[
"### Output Top 5 Softmax Probabilities For Each Image Found on the Web",
"_____no_output_____"
],
[
"For each of the new images, print out the model's softmax probabilities to show the **certainty** of the model's predictions (limit the output to the top 5 probabilities for each image). [`tf.nn.top_k`](https://www.tensorflow.org/versions/r0.12/api_docs/python/nn.html#top_k) could prove helpful here. \n\nThe example below demonstrates how tf.nn.top_k can be used to find the top k predictions for each image.\n\n`tf.nn.top_k` will return the values and indices (class ids) of the top k predictions. So if k=3, for each sign, it'll return the 3 largest probabilities (out of a possible 43) and the correspoding class ids.\n\nTake this numpy array as an example. The values in the array represent predictions. The array contains softmax probabilities for five candidate images with six possible classes. `tf.nn.top_k` is used to choose the three classes with the highest probability:\n\n```\n# (5, 6) array\na = np.array([[ 0.24879643, 0.07032244, 0.12641572, 0.34763842, 0.07893497,\n 0.12789202],\n [ 0.28086119, 0.27569815, 0.08594638, 0.0178669 , 0.18063401,\n 0.15899337],\n [ 0.26076848, 0.23664738, 0.08020603, 0.07001922, 0.1134371 ,\n 0.23892179],\n [ 0.11943333, 0.29198961, 0.02605103, 0.26234032, 0.1351348 ,\n 0.16505091],\n [ 0.09561176, 0.34396535, 0.0643941 , 0.16240774, 0.24206137,\n 0.09155967]])\n```\n\nRunning it through `sess.run(tf.nn.top_k(tf.constant(a), k=3))` produces:\n\n```\nTopKV2(values=array([[ 0.34763842, 0.24879643, 0.12789202],\n [ 0.28086119, 0.27569815, 0.18063401],\n [ 0.26076848, 0.23892179, 0.23664738],\n [ 0.29198961, 0.26234032, 0.16505091],\n [ 0.34396535, 0.24206137, 0.16240774]]), indices=array([[3, 0, 5],\n [0, 1, 4],\n [0, 5, 1],\n [1, 3, 5],\n [1, 4, 3]], dtype=int32))\n```\n\nLooking just at the first row we get `[ 0.34763842, 0.24879643, 0.12789202]`, you can confirm these are the 3 largest probabilities in `a`. You'll also notice `[3, 0, 5]` are the corresponding indices.",
"_____no_output_____"
]
],
[
[
"### Print out the top five softmax probabilities for the predictions on the German traffic sign images found on the web. \n### Feel free to use as many code cells as needed.\nwith tf.Session() as sess:\n saver.restore(sess, tf.train.latest_checkpoint('.'))\n test_top_5 = sess.run(tf.nn.top_k(tf.nn.softmax(logits),k=5), feed_dict = {x:test_images, y:test_outputs, keep_probability: 1.0})\n print(test_top_5)",
"INFO:tensorflow:Restoring parameters from ./lenet\nTopKV2(values=array([[9.9999464e-01, 5.2261566e-06, 7.3018022e-08, 5.4251206e-08,\n 2.3633202e-08],\n [1.0000000e+00, 2.3822607e-22, 4.8681307e-23, 9.0203673e-24,\n 2.0791327e-26],\n [9.9999976e-01, 1.8436631e-07, 1.2037944e-10, 9.2007055e-11,\n 7.8432115e-11],\n [9.9997795e-01, 2.1118827e-05, 8.9822697e-07, 9.7055183e-11,\n 8.1984232e-11],\n [9.9999583e-01, 4.1915223e-06, 5.3396528e-12, 1.2637004e-12,\n 8.3137639e-14]], dtype=float32), indices=array([[33, 35, 4, 40, 5],\n [13, 15, 25, 9, 35],\n [36, 38, 41, 35, 20],\n [ 3, 5, 31, 35, 4],\n [11, 30, 23, 27, 20]], dtype=int32))\n"
]
],
[
[
"### Project Writeup\n\nOnce you have completed the code implementation, document your results in a project writeup using this [template](https://github.com/udacity/CarND-Traffic-Sign-Classifier-Project/blob/master/writeup_template.md) as a guide. The writeup can be in a markdown or pdf file. ",
"_____no_output_____"
],
[
"> **Note**: Once you have completed all of the code implementations and successfully answered each question above, you may finalize your work by exporting the iPython Notebook as an HTML document. You can do this by using the menu above and navigating to \\n\",\n \"**File -> Download as -> HTML (.html)**. Include the finished document along with this notebook as your submission.",
"_____no_output_____"
],
[
"---\n\n## Step 4 (Optional): Visualize the Neural Network's State with Test Images\n\n This Section is not required to complete but acts as an additional excersise for understaning the output of a neural network's weights. While neural networks can be a great learning device they are often referred to as a black box. We can understand what the weights of a neural network look like better by plotting their feature maps. After successfully training your neural network you can see what it's feature maps look like by plotting the output of the network's weight layers in response to a test stimuli image. From these plotted feature maps, it's possible to see what characteristics of an image the network finds interesting. For a sign, maybe the inner network feature maps react with high activation to the sign's boundary outline or to the contrast in the sign's painted symbol.\n\n Provided for you below is the function code that allows you to get the visualization output of any tensorflow weight layer you want. The inputs to the function should be a stimuli image, one used during training or a new one you provided, and then the tensorflow variable name that represents the layer's state during the training process, for instance if you wanted to see what the [LeNet lab's](https://classroom.udacity.com/nanodegrees/nd013/parts/fbf77062-5703-404e-b60c-95b78b2f3f9e/modules/6df7ae49-c61c-4bb2-a23e-6527e69209ec/lessons/601ae704-1035-4287-8b11-e2c2716217ad/concepts/d4aca031-508f-4e0b-b493-e7b706120f81) feature maps looked like for it's second convolutional layer you could enter conv2 as the tf_activation variable.\n\nFor an example of what feature map outputs look like, check out NVIDIA's results in their paper [End-to-End Deep Learning for Self-Driving Cars](https://devblogs.nvidia.com/parallelforall/deep-learning-self-driving-cars/) in the section Visualization of internal CNN State. NVIDIA was able to show that their network's inner weights had high activations to road boundary lines by comparing feature maps from an image with a clear path to one without. Try experimenting with a similar test to show that your trained network's weights are looking for interesting features, whether it's looking at differences in feature maps from images with or without a sign, or even what feature maps look like in a trained network vs a completely untrained one on the same sign image.\n\n<figure>\n <img src=\"visualize_cnn.png\" width=\"380\" alt=\"Combined Image\" />\n <figcaption>\n <p></p> \n <p style=\"text-align: center;\"> Your output should look something like this (above)</p> \n </figcaption>\n</figure>\n <p></p> \n",
"_____no_output_____"
]
],
[
[
"### Visualize your network's feature maps here.\n### Feel free to use as many code cells as needed.\n\n# image_input: the test image being fed into the network to produce the feature maps\n# tf_activation: should be a tf variable name used during your training procedure that represents the calculated state of a specific weight layer\n# activation_min/max: can be used to view the activation contrast in more detail, by default matplot sets min and max to the actual min and max values of the output\n# plt_num: used to plot out multiple different weight feature map sets on the same block, just extend the plt number for each new feature map entry\n\ndef outputFeatureMap(image_input, tf_activation, activation_min=-1, activation_max=-1 ,plt_num=1):\n # Here make sure to preprocess your image_input in a way your network expects\n # with size, normalization, ect if needed\n # image_input =\n # Note: x should be the same name as your network's tensorflow data placeholder variable\n # If you get an error tf_activation is not defined it may be having trouble accessing the variable from inside a function\n activation = tf_activation.eval(session=sess,feed_dict={x : image_input})\n featuremaps = activation.shape[3]\n plt.figure(plt_num, figsize=(15,15))\n for featuremap in range(featuremaps):\n plt.subplot(6,8, featuremap+1) # sets the number of feature maps to show on each row and column\n plt.title('FeatureMap ' + str(featuremap)) # displays the feature map number\n if activation_min != -1 & activation_max != -1:\n plt.imshow(activation[0,:,:, featuremap], interpolation=\"nearest\", vmin =activation_min, vmax=activation_max, cmap=\"gray\")\n elif activation_max != -1:\n plt.imshow(activation[0,:,:, featuremap], interpolation=\"nearest\", vmax=activation_max, cmap=\"gray\")\n elif activation_min !=-1:\n plt.imshow(activation[0,:,:, featuremap], interpolation=\"nearest\", vmin=activation_min, cmap=\"gray\")\n else:\n plt.imshow(activation[0,:,:, featuremap], interpolation=\"nearest\", cmap=\"gray\")",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
]
] |
cbf24e0f979f77bdc9f3e1b92d3b94a3bf251e58
| 103,038 |
ipynb
|
Jupyter Notebook
|
notebooks/results.ipynb
|
btaba/handwriting-generation
|
7ae4d03dec0482a18c60148598f559a697637e8e
|
[
"MIT"
] | null | null | null |
notebooks/results.ipynb
|
btaba/handwriting-generation
|
7ae4d03dec0482a18c60148598f559a697637e8e
|
[
"MIT"
] | null | null | null |
notebooks/results.ipynb
|
btaba/handwriting-generation
|
7ae4d03dec0482a18c60148598f559a697637e8e
|
[
"MIT"
] | 2 |
2020-10-27T16:13:36.000Z
|
2021-05-04T10:40:09.000Z
| 730.765957 | 54,048 | 0.948893 |
[
[
[
"# Results\n",
"_____no_output_____"
]
],
[
[
"from handwriting_gen.plotting import plot_stroke\nfrom handwriting_gen.models import generate_unconditionally, generate_conditionally",
"_____no_output_____"
]
],
[
[
"\n### Unconditional generation:",
"_____no_output_____"
]
],
[
[
"stroke = generate_unconditionally(random_seed=5)\nplot_stroke(stroke)",
"Loading: {'save_path': '/home/trex/handwriting-generation/models/unconditional-stroke-model', 'learning_rate': 0.0001, 'mixture_components': 20, 'num_layers': 3, 'hidden_size': 200, 'dropout': 0.0, 'max_grad_norm': 5, 'decay': 0.95, 'momentum': 0.9, 'rnn_steps': 1, 'batch_size': 1, 'is_train': False}\nINFO:tensorflow:Restoring parameters from /home/trex/handwriting-generation/models/unconditional-stroke-model/-91500\n"
]
],
[
[
"### Conditional generation:",
"_____no_output_____"
]
],
[
[
"stroke = generate_conditionally(\n text='Baruch was here ', random_seed=42,\n std_bias=5)\nplot_stroke(stroke)",
"Loading: {'save_path': '/home/trex/handwriting-generation/models/conditional-stroke-model', 'learning_rate': 0.0001, 'mixture_components': 20, 'vocab_size': 60, 'char_dict': {' ': 0, '<UNK>': 1, 'D': 2, \"'\": 3, 'P': 4, 'Q': 5, 'v': 6, 'z': 7, 'M': 8, 'A': 9, 'c': 10, 'b': 11, 'k': 12, 'J': 13, 'Y': 14, 'O': 15, 'x': 16, 'o': 17, 'N': 18, 'l': 19, 'a': 20, 'S': 21, 'r': 22, '-': 23, 'i': 24, 'R': 25, 'U': 26, '.': 27, 'X': 28, 'C': 29, 'G': 30, 't': 31, 'q': 32, ',': 33, 'B': 34, 'W': 35, '\"': 36, 'H': 37, 'w': 38, 'y': 39, 'Z': 40, 'm': 41, 'F': 42, 's': 43, 'f': 44, 'K': 45, 'n': 46, 'g': 47, '?': 48, 'L': 49, 'e': 50, 'u': 51, 'I': 52, 'h': 53, 'j': 54, 'V': 55, 'E': 56, 'T': 57, 'p': 58, 'd': 59}, 'window_components': 10, 'char_seq_len': 17, 'num_layers': 3, 'hidden_size': 200, 'dropout': 0.0, 'max_grad_norm': 10, 'decay': 0.95, 'momentum': 0.9, 'rnn_steps': 1, 'batch_size': 1, 'is_train': False}\nINFO:tensorflow:Restoring parameters from /home/trex/handwriting-generation/models/conditional-stroke-model/-9078\n"
]
],
[
[
"### Handwriting recognition:\n\nNA",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
cbf25017478356ee401a5771b1de11e502b55349
| 4,911 |
ipynb
|
Jupyter Notebook
|
v1/notebooks/AzureStorage.ipynb
|
Muflhi01/HealthyCountryAI
|
ba4b033b5ba0d20a90d477feea19db587631e331
|
[
"MIT"
] | 39 |
2019-11-21T00:31:21.000Z
|
2022-03-19T06:29:31.000Z
|
v1/notebooks/AzureStorage.ipynb
|
Muflhi01/HealthyCountryAI
|
ba4b033b5ba0d20a90d477feea19db587631e331
|
[
"MIT"
] | 82 |
2019-11-18T05:37:02.000Z
|
2020-05-08T01:47:19.000Z
|
v1/notebooks/AzureStorage.ipynb
|
Muflhi01/HealthyCountryAI
|
ba4b033b5ba0d20a90d477feea19db587631e331
|
[
"MIT"
] | 15 |
2019-12-19T00:09:45.000Z
|
2022-01-31T21:10:10.000Z
| 39.288 | 109 | 0.671961 |
[
[
[
"from azure.common import AzureMissingResourceHttpError\nfrom azure.storage.blob import BlockBlobService, PublicAccess\nfrom azure.storage.file import FileService\nfrom azure.storage.table import TableService, Entity",
"_____no_output_____"
],
[
"#Blob Service...\ndef get_block_blob_service(account_name, storage_key):\n return BlockBlobService(account_name=account_name, account_key=storage_key)\n\ndef blob_service_create_container(account_name, storage_key, container_name):\n containers = blob_service_list_containers(account_name, storage_key)\n if container_name not in containers:\n block_blob_service = get_block_blob_service(account_name, storage_key)\n block_blob_service.create_container(container_name)\n block_blob_service.set_container_acl(container_name, public_access=PublicAccess.Container)\n\ndef blob_service_create_blob_from_bytes(account_name, storage_key, container_name, blob_name, blob):\n block_blob_service = get_block_blob_service(account_name, storage_key)\n block_blob_service.create_blob_from_bytes(container_name, blob_name, blob)\n\ndef blob_service_get_blob_to_path(account_name, storage_key, container_name, blob_name, file_path):\n block_blob_service = get_block_blob_service(account_name, storage_key)\n block_blob_service.get_blob_to_path(container_name, blob_name, file_path)\n\ndef blob_service_insert(account_name, storage_key, container_name, blob_name, text):\n block_blob_service = get_block_blob_service(account_name, storage_key)\n block_blob_service.create_blob_from_text(container_name, blob_name, text)\n\ndef blob_service_list_blobs(account_name, storage_key, container_name):\n blobs = []\n block_blob_service = get_block_blob_service(account_name, storage_key)\n generator = block_blob_service.list_blobs(container_name)\n for blob in generator:\n blobs.append(blob.name)\n return blobs\n\ndef blob_service_list_containers(account_name, storage_key):\n containers = []\n block_blob_service = get_block_blob_service(account_name, storage_key)\n generator = block_blob_service.list_containers()\n for container in generator:\n containers.append(container.name)\n return containers\n\n# File Service...\ndef get_file_service(account_name, storage_key):\n return FileService(account_name=account_name, account_key=storage_key)\n\ndef file_service_list_directories_and_files(account_name, storage_key, share_name, directory_name):\n file_or_dirs = []\n file_service = get_file_service(account_name, storage_key)\n generator = file_service.list_directories_and_files(share_name, directory_name)\n for file_or_dir in generator:\n file_or_dirs.append(file_or_dir.name)\n return file_or_dirs\n\n# Table Service...\ndef get_table_service(account_name, storage_key):\n return TableService(account_name=account_name, account_key=storage_key)\n\ndef table_service_get_entity(account_name, storage_key, table, partition_key, row_key):\n table_service = get_table_service(account_name, storage_key)\n return table_service.get_entity(table, partition_key, row_key)\n\ndef table_service_insert(account_name, storage_key, table, entity):\n table_service = get_table_service(account_name, storage_key)\n table_service.insert_entity(table, entity)\n \ndef table_service_query_entities(account_name, storage_key, table, filter):\n table_service = get_table_service(account_name, storage_key)\n return table_service.query_entities(table, filter)",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code"
]
] |
cbf25bf82b0971d00d83a1a7a1ddf2d436ec3a85
| 133,904 |
ipynb
|
Jupyter Notebook
|
notebooks/2018-08-23-lab-ddqnpre.ipynb
|
ucaiado/banana-rl
|
4e4295eca4b4ea157c907f1b21397322f8f9a4e9
|
[
"MIT"
] | 8 |
2018-08-24T03:48:59.000Z
|
2022-01-28T14:39:35.000Z
|
notebooks/2018-08-23-lab-ddqnpre.ipynb
|
ucaiado/banana-rl
|
4e4295eca4b4ea157c907f1b21397322f8f9a4e9
|
[
"MIT"
] | null | null | null |
notebooks/2018-08-23-lab-ddqnpre.ipynb
|
ucaiado/banana-rl
|
4e4295eca4b4ea157c907f1b21397322f8f9a4e9
|
[
"MIT"
] | 4 |
2018-08-26T16:10:44.000Z
|
2020-10-22T14:38:52.000Z
| 151.303955 | 67,776 | 0.864455 |
[
[
[
"# Project 1: Navigation\n### Test 3 - DDQN model with Prioritized Experience Replay\n\n<sub>Uirá Caiado. August 23, 2018<sub>\n\n#### Abstract\n\n\n_In this notebook, I will use the Unity ML-Agents environment to train a DDQN model with PER for the first project of the [Deep Reinforcement Learning Nanodegree](https://www.udacity.com/course/deep-reinforcement-learning-nanodegree--nd893)._",
"_____no_output_____"
],
[
"## 1. What we are going to test\n\nQuoting the seminal [Prioritized Experience Replay](https://arxiv.org/abs/1511.05952) paper, from the Deep Mind team, experience replay lets online reinforcement learning agents remember and reuse experiences from the past. Bellow, I am going to test my implementation of the PER buffer in conjunction to Double DQN. Thus, let's begin by checking the environment where I am going to run these tests.",
"_____no_output_____"
]
],
[
[
"%load_ext version_information\n%version_information numpy, unityagents, torch, matplotlib, pandas, gym",
"The version_information extension is already loaded. To reload it, use:\n %reload_ext version_information\n"
]
],
[
[
"Now, let's define some meta variables to use in this notebook",
"_____no_output_____"
]
],
[
[
"import os\nfig_prefix = 'figures/2018-08-23-'\ndata_prefix = '../data/2018-08-23-'\ns_currentpath = os.getcwd()",
"_____no_output_____"
]
],
[
[
"Also, let's import some of the necessary packages for this experiment.",
"_____no_output_____"
]
],
[
[
"from unityagents import UnityEnvironment",
"_____no_output_____"
],
[
"import sys\nimport os\nsys.path.append(\"../\") # include the root directory as the main\nimport eda\nimport pandas as pd\nimport numpy as np",
"_____no_output_____"
]
],
[
[
"## 2. Training the agent\n\nThe environment used for this project is the Udacity version of the Banana Collector environment, from [Unity](https://youtu.be/heVMs3t9qSk). The goal of the agent is to collect as many yellow bananas as possible while avoiding blue bananas. Bellow, we are going to start this environment.",
"_____no_output_____"
]
],
[
[
"env = UnityEnvironment(file_name=\"../Banana_Linux_NoVis/Banana.x86_64\")",
"INFO:unityagents:\n'Academy' started successfully!\nUnity Academy name: Academy\n Number of Brains: 1\n Number of External Brains : 1\n Lesson number : 0\n Reset Parameters :\n\t\t\nUnity brain name: BananaBrain\n Number of Visual Observations (per agent): 0\n Vector Observation space type: continuous\n Vector Observation space size (per agent): 37\n Number of stacked Vector Observation: 1\n Vector Action space type: discrete\n Vector Action space size (per agent): 4\n Vector Action descriptions: , , , \n"
]
],
[
[
"Unity Environments contain brains which are responsible for deciding the actions of their associated agents. Here we check for the first brain available, and set it as the default brain we will be controlling from Python.",
"_____no_output_____"
]
],
[
[
"# get the default brain\nbrain_name = env.brain_names[0]\nbrain = env.brains[brain_name]",
"_____no_output_____"
]
],
[
[
"Now, we are going to collect some basic information about the environment.",
"_____no_output_____"
]
],
[
[
"# reset the environment\nenv_info = env.reset(train_mode=True)[brain_name]\n\n# number of actions\naction_size = brain.vector_action_space_size\n\n# examine the state space \nstate = env_info.vector_observations[0]\nstate_size = len(state)",
"_____no_output_____"
]
],
[
[
"And finally, we are going to train the model. We will consider that this environment is solved if the agent is able to receive an average reward (over 100 episodes) of at least +13.",
"_____no_output_____"
]
],
[
[
"%%time\n\nimport gym\nimport pickle\nimport random\nimport torch\nimport numpy as np\nfrom collections import deque\nfrom drlnd.dqn_agent import DQNAgent, DDQNAgent, DDQNPREAgent\n\nn_episodes = 2000\neps_start = 1.\neps_end=0.01\neps_decay=0.995\nmax_t = 1000\ns_model = 'ddqnpre'\n\nagent = DDQNPREAgent(state_size=state_size, action_size=action_size, seed=0)\n\nscores = [] # list containing scores from each episode\nscores_std = [] # List containing the std dev of the last 100 episodes\nscores_avg = [] # List containing the mean of the last 100 episodes\nscores_window = deque(maxlen=100) # last 100 scores\neps = eps_start # initialize epsilon\n\nfor i_episode in range(1, n_episodes+1):\n env_info = env.reset(train_mode=True)[brain_name] # reset the environment\n state = env_info.vector_observations[0] # get the current state\n score = 0 # initialize the score\n for t in range(max_t):\n # action = np.random.randint(action_size) # select an action\n action = agent.act(state, eps)\n env_info = env.step(action)[brain_name] # send the action to the environment\n next_state = env_info.vector_observations[0] # get the next state\n reward = env_info.rewards[0] # get the reward\n done = env_info.local_done[0] # see if episode has finished\n agent.step(state, action, reward, next_state, done)\n score += reward # update the score\n state = next_state # roll over the state to next time step\n if done: # exit loop if episode finished\n break\n scores_window.append(score) # save most recent score\n scores.append(score) # save most recent score\n scores_std.append(np.std(scores_window)) # save most recent std dev\n scores_avg.append(np.mean(scores_window)) # save most recent std dev\n eps = max(eps_end, eps_decay*eps) # decrease epsilon\n print('\\rEpisode {}\\tAverage Score: {:.2f}'.format(i_episode, np.mean(scores_window)), end=\"\")\n if i_episode % 100 == 0:\n print('\\rEpisode {}\\tAverage Score: {:.2f}'.format(i_episode, np.mean(scores_window)))\n if np.mean(scores_window)>=13.0:\n s_msg = '\\nEnvironment solved in {:d} episodes!\\tAverage Score: {:.2f}'\n print(s_msg.format(i_episode, np.mean(scores_window)))\n torch.save(agent.qnet.state_dict(), '%scheckpoint_%s.pth' % (data_prefix, s_model))\n break\n \n# save data to use latter\nd_data = {'episodes': i_episode,\n 'scores': scores,\n 'scores_std': scores_std,\n 'scores_avg': scores_avg,\n 'scores_window': scores_window}\npickle.dump(d_data, open('%ssim-data-%s.data' % (data_prefix, s_model), 'wb'))",
"Episode 100\tAverage Score: 0.44\nEpisode 200\tAverage Score: 2.84\nEpisode 300\tAverage Score: 6.03\nEpisode 400\tAverage Score: 9.25\nEpisode 500\tAverage Score: 11.74\nEpisode 562\tAverage Score: 13.06\nEnvironment solved in 562 episodes!\tAverage Score: 13.06\nCPU times: user 16min 37s, sys: 20.7 s, total: 16min 58s\nWall time: 19min 18s\n"
]
],
[
[
"## 3. Results\n\nThe agent using Double DQN with Prioritized Experience Replay was able to solve the Banana Collector environment in 562 episodes of 1000 steps, each.",
"_____no_output_____"
]
],
[
[
"import pickle\n\nd_data = pickle.load(open('../data/2018-08-23-sim-data-ddqnpre.data', 'rb'))\ns_msg = 'Environment solved in {:d} episodes!\\tAverage Score: {:.2f} +- {:.2f}'\nprint(s_msg.format(d_data['episodes'], np.mean(d_data['scores_window']), np.std(d_data['scores_window'])))",
"Environment solved in 562 episodes!\tAverage Score: 13.06 +- 3.47\n"
]
],
[
[
"Now, let's plot the rewards per episode. In the right panel, we will plot the rolling average score over 100 episodes $\\pm$ its standard deviation, as well as the goal of this project (13+ on average over the last 100 episodes).",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nimport seaborn as sns\nimport numpy as np\n\n%matplotlib inline\n\n#recover data\nna_raw = np.array(d_data['scores'])\nna_mu = np.array(d_data['scores_avg'])\nna_sigma = np.array(d_data['scores_std'])\n\n# plot the scores\nf, (ax1, ax2) = plt.subplots(1, 2, figsize=(12, 5), sharex=True, sharey=True)\n\n# plot the sores by episode\nax1.plot(np.arange(len(na_raw)), na_raw)\nax1.set_xlim(0, len(na_raw)+1)\nax1.set_ylabel('Score')\nax1.set_xlabel('Episode #')\nax1.set_title('raw scores')\n\n# plot the average of these scores\nax2.axhline(y=13., xmin=0.0, xmax=1.0, color='r', linestyle='--', linewidth=0.7, alpha=0.9)\nax2.plot(np.arange(len(na_mu)), na_mu)\nax2.fill_between(np.arange(len(na_mu)), na_mu+na_sigma, na_mu-na_sigma, facecolor='gray', alpha=0.1)\nax2.set_ylabel('Average Score')\nax2.set_xlabel('Episode #')\nax2.set_title('average scores')\n\nf.tight_layout()",
"_____no_output_____"
],
[
"# f.savefig(fig_prefix + 'ddqnpre-learning-curve.eps', format='eps', dpi=1200)\nf.savefig(fig_prefix + 'ddqnpre-learning-curve.jpg', format='jpg')",
"_____no_output_____"
],
[
"env.close()",
"_____no_output_____"
]
],
[
[
"## 4. Conclusion\n\n\nThe Double Deep Q-learning agent using Prioritized Experience Replay was able to solve the environment in 562 episodes and was the worst performance among all implementations. However, something that is worth noting that this implementation is seen to present the most smooth learning curve.",
"_____no_output_____"
]
],
[
[
"import pickle\n\nd_ddqnper = pickle.load(open('../data/2018-08-23-sim-data-ddqnpre.data', 'rb'))\nd_ddqn = pickle.load(open('../data/2018-08-24-sim-data-ddqn.data', 'rb'))\nd_dqn = pickle.load(open('../data/2018-08-24-sim-data-dqn.data', 'rb'))",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt\nimport seaborn as sns\nimport numpy as np\n\n%matplotlib inline\n\ndef recover_data(d_data):\n #recover data\n na_raw = np.array(d_data['scores'])\n na_mu = np.array(d_data['scores_avg'])\n na_sigma = np.array(d_data['scores_std'])\n return na_raw, na_mu, na_sigma\n\n# plot the scores\nf, ax2 = plt.subplots(1, 1, figsize=(8, 4), sharex=True, sharey=True)\n\n\nfor s_model, d_data in zip(['DQN', 'DDQN', 'DDQN with PER'], [d_ddqnper, d_ddqn, d_dqn]):\n\n na_raw, na_mu, na_sigma = recover_data(d_data)\n if s_model == 'DDQN with PER':\n ax2.set_xlim(0, 572)\n # plot the average of these scores\n ax2.axhline(y=13., xmin=0.0, xmax=1.0, color='r', linestyle='--', linewidth=0.7, alpha=0.9)\n ax2.plot(np.arange(len(na_mu)), na_mu, label=s_model)\n# ax2.fill_between(np.arange(len(na_mu)), na_mu+na_sigma, na_mu-na_sigma, alpha=0.15)\n\n# format axis\nax2.legend()\nax2.set_title('Learning Curves')\nax2.set_ylabel('Average Score in 100 episodes')\nax2.set_xlabel('Episode #')\n\n# Shrink current axis's height by 10% on the bottom\nbox = ax2.get_position()\nax2.set_position([box.x0, box.y0 + box.height * 0.1,\n box.width, box.height * 0.9])\n\n# Put a legend below current axis\nlgd = ax2.legend(loc='upper center', bbox_to_anchor=(0.5, -0.10),\n fancybox=False, shadow=False, ncol=3)\n\nf.tight_layout()",
"_____no_output_____"
],
[
"f.savefig(fig_prefix + 'final-comparition-2.eps', format='eps',\n bbox_extra_artists=(lgd,), bbox_inches='tight', dpi=1200)",
"_____no_output_____"
]
],
[
[
"Finally, let's compare the score distributions generated by the agents. I am going to perform the one-sided Welch's unequal variances t-test for the null hypothesis that the DDQN model has the expected score higher than the other agents on the final 100 episodes of each experiment. As the implementation of the t-test in the [Scipy](https://goo.gl/gs222c) assumes a two-sided t-test, to perform the one-sided test, we will divide the p-value by 2 to compare to a critical value of 0.05 and requires that the t-value is greater than zero.",
"_____no_output_____"
]
],
[
[
"import pandas as pd\n\ndef extract_info(s, d_data):\n return {'model': s,\n 'episodes': d_data['episodes'],\n 'mean_score': np.mean(d_data['scores_window']),\n 'std_score': np.std(d_data['scores_window'])}\n\nl_data = [extract_info(s, d) for s, d in zip(['DDQN with PER', 'DDQN', 'DQN'],\n [d_ddqnper, d_ddqn, d_dqn])]\n\ndf = pd.DataFrame(l_data)\ndf.index = df.model\ndf.drop('model', axis=1, inplace=True)\nprint(df.sort_values(by='episodes'))",
" episodes mean_score std_score\nmodel \nDDQN 462 13.01 3.479928\nDQN 529 13.01 3.421973\nDDQN with PER 562 13.06 3.466468\n"
],
[
"import scipy\n#performs t-test\na = [float(pd.DataFrame(d_dqn['scores']).iloc[-1].values)] * 2\nb = list(pd.DataFrame(d_rtn_test_1r['pnl']['test']).fillna(method='ffill').iloc[-1].values)\ntval, p_value = scipy.stats.ttest_ind(a, b, equal_var=False)",
"_____no_output_____"
],
[
"import scipy\ntval, p_value = scipy.stats.ttest_ind(d_ddqn['scores'], d_dqn['scores'], equal_var=False)\nprint(\"DDQN vs. DQN: t-value = {:0.6f}, p-value = {:0.8f}\".format(tval, p_value))\ntval, p_value = scipy.stats.ttest_ind(d_ddqn['scores'], d_ddqnper['scores'], equal_var=False)\nprint(\"DDQN vs. DDQNPRE: t-value = {:0.6f}, p-value = {:0.8f}\".format(tval, p_value))",
"DDQN vs. DQN: t-value = -0.625024, p-value = 0.53210301\nDDQN vs. DDQNPRE: t-value = 0.504778, p-value = 0.61382809\n"
]
],
[
[
"There was no significant difference between the performances of the agents.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
]
] |
cbf26a70e8d3cfda32f1e52a65017ab23d4199a7
| 2,382 |
ipynb
|
Jupyter Notebook
|
Python Data Science Toolbox -Part 1/Writing your own functions/05.A brief introduction to tuples.ipynb
|
nazmusshakib121/Python-Programming
|
3ea852641cd5fe811228f27a780109a44174e8e5
|
[
"MIT"
] | null | null | null |
Python Data Science Toolbox -Part 1/Writing your own functions/05.A brief introduction to tuples.ipynb
|
nazmusshakib121/Python-Programming
|
3ea852641cd5fe811228f27a780109a44174e8e5
|
[
"MIT"
] | null | null | null |
Python Data Science Toolbox -Part 1/Writing your own functions/05.A brief introduction to tuples.ipynb
|
nazmusshakib121/Python-Programming
|
3ea852641cd5fe811228f27a780109a44174e8e5
|
[
"MIT"
] | null | null | null | 25.340426 | 248 | 0.592779 |
[
[
[
"Alongside learning about functions, you've also learned about tuples! Here, you will practice what you've learned about tuples: how to construct, unpack, and access tuple elements. Recall how Hugo unpacked the tuple even_nums in the video:\n\na, b, c = even_nums\n\nA three-element tuple named nums has been preloaded for this exercise. Before completing the script, perform the following:\n\nPrint out the value of nums in the IPython shell. Note the elements in the tuple.\nIn the IPython shell, try to change the first element of nums to the value 2 by doing an assignment: nums[0] = 2. What happens?",
"_____no_output_____"
],
[
"Unpack nums to the variables num1, num2, and num3.",
"_____no_output_____"
],
[
"Construct a new tuple, even_nums composed of the same elements in nums, but with the 1st element replaced with the value, 2.",
"_____no_output_____"
]
],
[
[
"# Unpack nums into num1, num2, and num3\nnums = (3, 4, 6)\nnum1 , num2 , num3 = nums\n\n# Construct even_nums\n\neven_nums=(2 , num2 , num3)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code"
]
] |
cbf26e2c4ca0f294413d97aa11aa25bbf9127b36
| 113,170 |
ipynb
|
Jupyter Notebook
|
Convolutional_Autoencoder.ipynb
|
Meirtz/Convolutional_Autoencoder
|
470e898cb646f2310846194b528e5b3e98d3a78e
|
[
"MIT"
] | null | null | null |
Convolutional_Autoencoder.ipynb
|
Meirtz/Convolutional_Autoencoder
|
470e898cb646f2310846194b528e5b3e98d3a78e
|
[
"MIT"
] | null | null | null |
Convolutional_Autoencoder.ipynb
|
Meirtz/Convolutional_Autoencoder
|
470e898cb646f2310846194b528e5b3e98d3a78e
|
[
"MIT"
] | null | null | null | 141.994981 | 49,972 | 0.835955 |
[
[
[
"# Convolutional Autoencoder\n\nSticking with the MNIST dataset, let's improve our autoencoder's performance using convolutional layers. Again, loading modules and the data.",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\n\nimport numpy as np\nimport tensorflow as tf\nimport matplotlib.pyplot as plt\n#from tqdm import tqdm",
"_____no_output_____"
],
[
"from tensorflow.examples.tutorials.mnist import input_data\nmnist = input_data.read_data_sets('MNIST_data', validation_size=0)",
"Extracting MNIST_data/train-images-idx3-ubyte.gz\nExtracting MNIST_data/train-labels-idx1-ubyte.gz\nExtracting MNIST_data/t10k-images-idx3-ubyte.gz\nExtracting MNIST_data/t10k-labels-idx1-ubyte.gz\n"
],
[
"img = mnist.train.images[2]\nplt.imshow(img.reshape((28, 28)), cmap='Greys_r')",
"_____no_output_____"
]
],
[
[
"## Network Architecture\n\nThe encoder part of the network will be a typical convolutional pyramid. Each convolutional layer will be followed by a max-pooling layer to reduce the dimensions of the layers. The decoder though might be something new to you. The decoder needs to convert from a narrow representation to a wide reconstructed image. For example, the representation could be a 4x4x8 max-pool layer. This is the output of the encoder, but also the input to the decoder. We want to get a 28x28x1 image out from the decoder so we need to work our way back up from the narrow decoder input layer. A schematic of the network is shown below.\n\n\n\nHere our final encoder layer has size 4x4x8 = 128. The original images have size 28x28 = 784, so the encoded vector is roughly 16% the size of the original image. These are just suggested sizes for each of the layers. Feel free to change the depths and sizes, but remember our goal here is to find a small representation of the input data.\n\n### What's going on with the decoder\n\nOkay, so the decoder has these \"Upsample\" layers that you might not have seen before. First off, I'll discuss a bit what these layers *aren't*. Usually, you'll see **deconvolutional** layers used to increase the width and height of the layers. They work almost exactly the same as convolutional layers, but it reverse. A stride in the input layer results in a larger stride in the deconvolutional layer. For example, if you have a 3x3 kernel, a 3x3 patch in the input layer will be reduced to one unit in a convolutional layer. Comparatively, one unit in the input layer will be expanded to a 3x3 path in a deconvolutional layer. Deconvolution is often called \"transpose convolution\" which is what you'll find with the TensorFlow API, with [`tf.nn.conv2d_transpose`](https://www.tensorflow.org/api_docs/python/tf/nn/conv2d_transpose). \n\nHowever, deconvolutional layers can lead to artifacts in the final images, such as checkerboard patterns. This is due to overlap in the kernels which can be avoided by setting the stride and kernel size equal. In [this Distill article](http://distill.pub/2016/deconv-checkerboard/) from Augustus Odena, *et al*, the authors show that these checkerboard artifacts can be avoided by resizing the layers using nearest neighbor or bilinear interpolation (upsampling) followed by a convolutional layer. In TensorFlow, this is easily done with [`tf.image.resize_images`](https://www.tensorflow.org/versions/r1.1/api_docs/python/tf/image/resize_images), followed by a convolution. Be sure to read the Distill article to get a better understanding of deconvolutional layers and why we're using upsampling.\n\n> **Exercise:** Build the network shown above. Remember that a convolutional layer with strides of 1 and 'same' padding won't reduce the height and width. That is, if the input is 28x28 and the convolution layer has stride = 1 and 'same' padding, the convolutional layer will also be 28x28. The max-pool layers are used the reduce the width and height. A stride of 2 will reduce the size by 2. Odena *et al* claim that nearest neighbor interpolation works best for the upsampling, so make sure to include that as a parameter in `tf.image.resize_images` or use [`tf.image.resize_nearest_neighbor`]( https://www.tensorflow.org/api_docs/python/tf/image/resize_nearest_neighbor).",
"_____no_output_____"
]
],
[
[
"learning_rate = 0.001\ninputs_ = tf.placeholder(tf.float32, shape=(None,28,28,1), name='inputs')\ntargets_ = tf.placeholder(tf.float32, shape=(None,28,28,1), name='targets')\n\n### Encoder\nconv1 = tf.layers.conv2d(inputs_, 16, (3,3), padding='same', activation=tf.nn.relu)\n# Now 28x28x16\nmaxpool1 = tf.layers.max_pooling2d(conv1, (2,2), (2,2))\n# Now 14x14x16\nconv2 = tf.layers.conv2d(maxpool1, 8, (3,3), padding='same', activation=tf.nn.relu)\n# Now 14x14x8\nmaxpool2 = tf.layers.max_pooling2d(conv2, (2,2), (2,2))\n# Now 7x7x8\nconv3 = tf.layers.conv2d(maxpool2, 8, (3,3), padding='same', activation=tf.nn.relu)\n# Now 7x7x8\nencoded = tf.layers.max_pooling2d(conv3, (2,2), (2,2), padding='same')\n# Now 4x4x8\n\n### Decoder\nupsample1 = tf.image.resize_nearest_neighbor(conv3, (7,7))\n# Now 7x7x8\nconv4 = tf.layers.conv2d(upsample1, 8, (3,3), padding='same', activation=tf.nn.relu)\n# Now 7x7x8\nupsample2 = tf.image.resize_nearest_neighbor(conv4, (14,14))\n# Now 14x14x8\nconv5 = tf.layers.conv2d(upsample2, 8, (3,3), padding='same', activation=tf.nn.relu)\n# Now 14x14x8\nupsample3 = tf.image.resize_nearest_neighbor(conv5, (28,28))\n# Now 28x28x8\nconv6 = tf.layers.conv2d(upsample3, 16, (3,3), padding='same', activation=tf.nn.relu)\n# Now 28x28x16\n\nlogits = tf.layers.conv2d(upsample3, 1, (3,3), padding='same', activation=None)\n#Now 28x28x1\n\n# Pass logits through sigmoid to get reconstructed image\ndecoded = tf.nn.sigmoid(logits, name='decoded')\n\n# Pass logits through sigmoid and calculate the cross-entropy loss\nloss = tf.nn.sigmoid_cross_entropy_with_logits(labels=targets_, logits=logits)\n\n# Get cost and define the optimizer\ncost = tf.reduce_mean(loss)\nopt = tf.train.AdamOptimizer(learning_rate).minimize(cost)",
"_____no_output_____"
]
],
[
[
"## Training\n\nAs before, here wi'll train the network. Instead of flattening the images though, we can pass them in as 28x28x1 arrays.",
"_____no_output_____"
]
],
[
[
"sess = tf.Session()",
"_____no_output_____"
],
[
"epochs = 20\nbatch_size = 256\nsess.run(tf.global_variables_initializer())\nfor e in range(epochs):\n for ii in range(mnist.train.num_examples//batch_size):\n batch = mnist.train.next_batch(batch_size)\n imgs = batch[0].reshape((-1, 28, 28, 1))\n batch_cost, _ = sess.run([cost, opt], feed_dict={inputs_: imgs,\n targets_: imgs})\n if ii % 100 == 0:\n print(\"Epoch: {}/{}...\".format(e+1, epochs),\n \"Training loss: {:.4f}\".format(batch_cost))",
"Epoch: 1/20... Training loss: 0.6935\nEpoch: 1/20... Training loss: 0.1773\nEpoch: 1/20... Training loss: 0.1284\nEpoch: 2/20... Training loss: 0.1167\nEpoch: 2/20... Training loss: 0.1019\nEpoch: 2/20... Training loss: 0.0972\nEpoch: 3/20... Training loss: 0.0967\nEpoch: 3/20... Training loss: 0.0915\nEpoch: 3/20... Training loss: 0.0907\nEpoch: 4/20... Training loss: 0.0847\nEpoch: 4/20... Training loss: 0.0869\nEpoch: 4/20... Training loss: 0.0846\nEpoch: 5/20... Training loss: 0.0845\nEpoch: 5/20... Training loss: 0.0858\nEpoch: 5/20... Training loss: 0.0846\nEpoch: 6/20... Training loss: 0.0820\nEpoch: 6/20... Training loss: 0.0828\nEpoch: 6/20... Training loss: 0.0803\nEpoch: 7/20... Training loss: 0.0814\nEpoch: 7/20... Training loss: 0.0806\nEpoch: 7/20... Training loss: 0.0824\nEpoch: 8/20... Training loss: 0.0813\nEpoch: 8/20... Training loss: 0.0792\nEpoch: 8/20... Training loss: 0.0788\nEpoch: 9/20... Training loss: 0.0784\nEpoch: 9/20... Training loss: 0.0801\nEpoch: 9/20... Training loss: 0.0801\nEpoch: 10/20... Training loss: 0.0770\nEpoch: 10/20... Training loss: 0.0778\nEpoch: 10/20... Training loss: 0.0801\nEpoch: 11/20... Training loss: 0.0797\nEpoch: 11/20... Training loss: 0.0768\nEpoch: 11/20... Training loss: 0.0759\nEpoch: 12/20... Training loss: 0.0751\nEpoch: 12/20... Training loss: 0.0765\nEpoch: 12/20... Training loss: 0.0783\nEpoch: 13/20... Training loss: 0.0762\nEpoch: 13/20... Training loss: 0.0774\nEpoch: 13/20... Training loss: 0.0790\nEpoch: 14/20... Training loss: 0.0760\nEpoch: 14/20... Training loss: 0.0768\nEpoch: 14/20... Training loss: 0.0753\nEpoch: 15/20... Training loss: 0.0772\nEpoch: 15/20... Training loss: 0.0751\nEpoch: 15/20... Training loss: 0.0749\nEpoch: 16/20... Training loss: 0.0745\nEpoch: 16/20... Training loss: 0.0768\nEpoch: 16/20... Training loss: 0.0781\nEpoch: 17/20... Training loss: 0.0730\nEpoch: 17/20... Training loss: 0.0756\nEpoch: 17/20... Training loss: 0.0743\nEpoch: 18/20... Training loss: 0.0743\nEpoch: 18/20... Training loss: 0.0738\nEpoch: 18/20... Training loss: 0.0766\nEpoch: 19/20... Training loss: 0.0743\nEpoch: 19/20... Training loss: 0.0747\nEpoch: 19/20... Training loss: 0.0742\nEpoch: 20/20... Training loss: 0.0752\nEpoch: 20/20... Training loss: 0.0741\nEpoch: 20/20... Training loss: 0.0749\n"
],
[
"fig, axes = plt.subplots(nrows=2, ncols=10, sharex=True, sharey=True, figsize=(20,4))\nin_imgs = mnist.test.images[:10]\nreconstructed = sess.run(decoded, feed_dict={inputs_: in_imgs.reshape((10, 28, 28, 1))})\n\nfor images, row in zip([in_imgs, reconstructed], axes):\n for img, ax in zip(images, row):\n ax.imshow(img.reshape((28, 28)), cmap='Greys_r')\n ax.get_xaxis().set_visible(False)\n ax.get_yaxis().set_visible(False)\n\n\nfig.tight_layout(pad=0.1)",
"_____no_output_____"
],
[
"sess.close()",
"_____no_output_____"
]
],
[
[
"## Denoising\n\nAs I've mentioned before, autoencoders like the ones you've built so far aren't too useful in practive. However, they can be used to denoise images quite successfully just by training the network on noisy images. We can create the noisy images ourselves by adding Gaussian noise to the training images, then clipping the values to be between 0 and 1. We'll use noisy images as input and the original, clean images as targets. Here's an example of the noisy images I generated and the denoised images.\n\n\n\n\nSince this is a harder problem for the network, we'll want to use deeper convolutional layers here, more feature maps. I suggest something like 32-32-16 for the depths of the convolutional layers in the encoder, and the same depths going backward through the decoder. Otherwise the architecture is the same as before.\n\n> **Exercise:** Build the network for the denoising autoencoder. It's the same as before, but with deeper layers. I suggest 32-32-16 for the depths, but you can play with these numbers, or add more layers.",
"_____no_output_____"
]
],
[
[
"learning_rate = 0.001\ninputs_ = tf.placeholder(tf.float32, shape=(None,28,28,1), name='inputs')\ntargets_ = tf.placeholder(tf.float32, shape=(None,28,28,1), name='targets')\n\n### Encoder\nconv1 = tf.layers.conv2d(inputs_, 16, (3,3), padding='same', activation=tf.nn.relu)\n# Now 28x28x16\nmaxpool1 = tf.layers.max_pooling2d(conv1, (2,2), (2,2))\n# Now 14x14x16\nconv2 = tf.layers.conv2d(maxpool1, 8, (3,3), padding='same', activation=tf.nn.relu)\n# Now 14x14x8\nmaxpool2 = tf.layers.max_pooling2d(conv2, (2,2), (2,2))\n# Now 7x7x8\nconv3 = tf.layers.conv2d(maxpool2, 8, (3,3), padding='same', activation=tf.nn.relu)\n# Now 7x7x8\nencoded = tf.layers.max_pooling2d(conv3, (2,2), (2,2), padding='same')\n# Now 4x4x8\n\n### Decoder\nupsample1 = tf.image.resize_nearest_neighbor(conv3, (7,7))\n# Now 7x7x8\nconv4 = tf.layers.conv2d(upsample1, 8, (3,3), padding='same', activation=tf.nn.relu)\n# Now 7x7x8\nupsample2 = tf.image.resize_nearest_neighbor(conv4, (14,14))\n# Now 14x14x8\nconv5 = tf.layers.conv2d(upsample2, 8, (3,3), padding='same', activation=tf.nn.relu)\n# Now 14x14x8\nupsample3 = tf.image.resize_nearest_neighbor(conv5, (28,28))\n# Now 28x28x8\nconv6 = tf.layers.conv2d(upsample3, 16, (3,3), padding='same', activation=tf.nn.relu)\n# Now 28x28x16\n\nlogits = tf.layers.conv2d(upsample3, 1, (3,3), padding='same', activation=None)\n#Now 28x28x1\n\n# Pass logits through sigmoid to get reconstructed image\ndecoded = tf.nn.sigmoid(logits, name='decoded')\n\n# Pass logits through sigmoid and calculate the cross-entropy loss\nloss = tf.nn.sigmoid_cross_entropy_with_logits(labels=targets_, logits=logits)\n\n# Get cost and define the optimizer\ncost = tf.reduce_mean(loss)\nopt = tf.train.AdamOptimizer(learning_rate).minimize(cost)",
"_____no_output_____"
],
[
"sess = tf.Session()",
"_____no_output_____"
],
[
"epochs = 100\nbatch_size = 256\n# Set's how much noise we're adding to the MNIST images\nnoise_factor = 0.5\nsess.run(tf.global_variables_initializer())\nfor e in range(epochs):\n for ii in range(mnist.train.num_examples//batch_size):\n batch = mnist.train.next_batch(batch_size)\n # Get images from the batch\n imgs = batch[0].reshape((-1, 28, 28, 1))\n \n # Add random noise to the input images\n noisy_imgs = imgs + noise_factor * np.random.randn(*imgs.shape)\n # Clip the images to be between 0 and 1\n noisy_imgs = np.clip(noisy_imgs, 0., 1.)\n \n # Noisy images as inputs, original images as targets\n batch_cost, _ = sess.run([cost, opt], feed_dict={inputs_: noisy_imgs,\n targets_: imgs})\n if ii % 100 == 0:\n print(\"Epoch: {}/{}...\".format(e+1, epochs),\n \"Training loss: {:.4f}\".format(batch_cost))",
"Epoch: 1/100... Training loss: 0.6973\nEpoch: 1/100... Training loss: 0.2817\nEpoch: 1/100... Training loss: 0.1699\nEpoch: 2/100... Training loss: 0.1646\nEpoch: 2/100... Training loss: 0.1476\nEpoch: 2/100... Training loss: 0.1344\nEpoch: 3/100... Training loss: 0.1350\nEpoch: 3/100... Training loss: 0.1319\nEpoch: 3/100... Training loss: 0.1276\nEpoch: 4/100... Training loss: 0.1267\nEpoch: 4/100... Training loss: 0.1234\nEpoch: 4/100... Training loss: 0.1236\nEpoch: 5/100... Training loss: 0.1219\nEpoch: 5/100... Training loss: 0.1208\nEpoch: 5/100... Training loss: 0.1223\nEpoch: 6/100... Training loss: 0.1201\nEpoch: 6/100... Training loss: 0.1173\nEpoch: 6/100... Training loss: 0.1194\nEpoch: 7/100... Training loss: 0.1200\nEpoch: 7/100... Training loss: 0.1183\nEpoch: 7/100... Training loss: 0.1175\nEpoch: 8/100... Training loss: 0.1157\nEpoch: 8/100... Training loss: 0.1131\nEpoch: 8/100... Training loss: 0.1142\nEpoch: 9/100... Training loss: 0.1144\nEpoch: 9/100... Training loss: 0.1132\nEpoch: 9/100... Training loss: 0.1147\nEpoch: 10/100... Training loss: 0.1137\nEpoch: 10/100... Training loss: 0.1127\nEpoch: 10/100... Training loss: 0.1133\nEpoch: 11/100... Training loss: 0.1128\nEpoch: 11/100... Training loss: 0.1141\nEpoch: 11/100... Training loss: 0.1142\nEpoch: 12/100... Training loss: 0.1149\nEpoch: 12/100... Training loss: 0.1137\nEpoch: 12/100... Training loss: 0.1119\nEpoch: 13/100... Training loss: 0.1131\nEpoch: 13/100... Training loss: 0.1141\nEpoch: 13/100... Training loss: 0.1106\nEpoch: 14/100... Training loss: 0.1140\nEpoch: 14/100... Training loss: 0.1113\nEpoch: 14/100... Training loss: 0.1098\nEpoch: 15/100... Training loss: 0.1116\nEpoch: 15/100... Training loss: 0.1110\nEpoch: 15/100... Training loss: 0.1111\nEpoch: 16/100... Training loss: 0.1135\nEpoch: 16/100... Training loss: 0.1124\nEpoch: 16/100... Training loss: 0.1122\nEpoch: 17/100... Training loss: 0.1121\nEpoch: 17/100... Training loss: 0.1114\nEpoch: 17/100... Training loss: 0.1137\nEpoch: 18/100... Training loss: 0.1129\nEpoch: 18/100... Training loss: 0.1084\nEpoch: 18/100... Training loss: 0.1090\nEpoch: 19/100... Training loss: 0.1111\nEpoch: 19/100... Training loss: 0.1083\nEpoch: 19/100... Training loss: 0.1143\nEpoch: 20/100... Training loss: 0.1103\nEpoch: 20/100... Training loss: 0.1110\nEpoch: 20/100... Training loss: 0.1113\nEpoch: 21/100... Training loss: 0.1065\nEpoch: 21/100... Training loss: 0.1108\nEpoch: 21/100... Training loss: 0.1074\nEpoch: 22/100... Training loss: 0.1065\nEpoch: 22/100... Training loss: 0.1075\nEpoch: 22/100... Training loss: 0.1125\nEpoch: 23/100... Training loss: 0.1094\nEpoch: 23/100... Training loss: 0.1101\nEpoch: 23/100... Training loss: 0.1096\nEpoch: 24/100... Training loss: 0.1072\nEpoch: 24/100... Training loss: 0.1112\nEpoch: 24/100... Training loss: 0.1090\nEpoch: 25/100... Training loss: 0.1095\nEpoch: 25/100... Training loss: 0.1093\nEpoch: 25/100... Training loss: 0.1092\nEpoch: 26/100... Training loss: 0.1095\nEpoch: 26/100... Training loss: 0.1104\nEpoch: 26/100... Training loss: 0.1085\nEpoch: 27/100... Training loss: 0.1093\nEpoch: 27/100... Training loss: 0.1093\nEpoch: 27/100... Training loss: 0.1076\nEpoch: 28/100... Training loss: 0.1089\nEpoch: 28/100... Training loss: 0.1078\nEpoch: 28/100... Training loss: 0.1055\nEpoch: 29/100... Training loss: 0.1067\nEpoch: 29/100... Training loss: 0.1081\nEpoch: 29/100... Training loss: 0.1078\nEpoch: 30/100... Training loss: 0.1066\nEpoch: 30/100... Training loss: 0.1098\nEpoch: 30/100... Training loss: 0.1064\nEpoch: 31/100... Training loss: 0.1110\nEpoch: 31/100... Training loss: 0.1076\nEpoch: 31/100... Training loss: 0.1103\nEpoch: 32/100... Training loss: 0.1083\nEpoch: 32/100... Training loss: 0.1092\nEpoch: 32/100... Training loss: 0.1079\nEpoch: 33/100... Training loss: 0.1062\nEpoch: 33/100... Training loss: 0.1055\nEpoch: 33/100... Training loss: 0.1049\nEpoch: 34/100... Training loss: 0.1094\nEpoch: 34/100... Training loss: 0.1086\nEpoch: 34/100... Training loss: 0.1034\nEpoch: 35/100... Training loss: 0.1083\nEpoch: 35/100... Training loss: 0.1098\nEpoch: 35/100... Training loss: 0.1078\nEpoch: 36/100... Training loss: 0.1055\nEpoch: 36/100... Training loss: 0.1077\nEpoch: 36/100... Training loss: 0.1070\nEpoch: 37/100... Training loss: 0.1051\nEpoch: 37/100... Training loss: 0.1073\nEpoch: 37/100... Training loss: 0.1050\nEpoch: 38/100... Training loss: 0.1091\nEpoch: 38/100... Training loss: 0.1059\nEpoch: 38/100... Training loss: 0.1067\nEpoch: 39/100... Training loss: 0.1080\nEpoch: 39/100... Training loss: 0.1054\nEpoch: 39/100... Training loss: 0.1067\nEpoch: 40/100... Training loss: 0.1063\nEpoch: 40/100... Training loss: 0.1091\nEpoch: 40/100... Training loss: 0.1074\nEpoch: 41/100... Training loss: 0.1090\nEpoch: 41/100... Training loss: 0.1059\nEpoch: 41/100... Training loss: 0.1068\nEpoch: 42/100... Training loss: 0.1079\nEpoch: 42/100... Training loss: 0.1075\nEpoch: 42/100... Training loss: 0.1070\nEpoch: 43/100... Training loss: 0.1093\nEpoch: 43/100... Training loss: 0.1064\nEpoch: 43/100... Training loss: 0.1052\nEpoch: 44/100... Training loss: 0.1052\nEpoch: 44/100... Training loss: 0.1068\nEpoch: 44/100... Training loss: 0.1080\nEpoch: 45/100... Training loss: 0.1059\nEpoch: 45/100... Training loss: 0.1067\nEpoch: 45/100... Training loss: 0.1062\nEpoch: 46/100... Training loss: 0.1078\nEpoch: 46/100... Training loss: 0.1104\nEpoch: 46/100... Training loss: 0.1080\nEpoch: 47/100... Training loss: 0.1043\nEpoch: 47/100... Training loss: 0.1100\nEpoch: 47/100... Training loss: 0.1070\nEpoch: 48/100... Training loss: 0.1060\nEpoch: 48/100... Training loss: 0.1056\nEpoch: 48/100... Training loss: 0.1048\nEpoch: 49/100... Training loss: 0.1071\nEpoch: 49/100... Training loss: 0.1065\nEpoch: 49/100... Training loss: 0.1055\nEpoch: 50/100... Training loss: 0.1036\nEpoch: 50/100... Training loss: 0.1050\nEpoch: 50/100... Training loss: 0.1060\nEpoch: 51/100... Training loss: 0.1038\nEpoch: 51/100... Training loss: 0.1071\nEpoch: 51/100... Training loss: 0.1066\nEpoch: 52/100... Training loss: 0.1082\nEpoch: 52/100... Training loss: 0.1058\nEpoch: 52/100... Training loss: 0.1063\nEpoch: 53/100... Training loss: 0.1063\nEpoch: 53/100... Training loss: 0.1069\nEpoch: 53/100... Training loss: 0.1072\nEpoch: 54/100... Training loss: 0.1034\nEpoch: 54/100... Training loss: 0.1076\nEpoch: 54/100... Training loss: 0.1056\nEpoch: 55/100... Training loss: 0.1082\nEpoch: 55/100... Training loss: 0.1057\nEpoch: 55/100... Training loss: 0.1070\nEpoch: 56/100... Training loss: 0.1058\nEpoch: 56/100... Training loss: 0.1062\nEpoch: 56/100... Training loss: 0.1093\nEpoch: 57/100... Training loss: 0.1065\nEpoch: 57/100... Training loss: 0.1055\nEpoch: 57/100... Training loss: 0.1061\nEpoch: 58/100... Training loss: 0.1059\nEpoch: 58/100... Training loss: 0.1053\nEpoch: 58/100... Training loss: 0.1098\nEpoch: 59/100... Training loss: 0.1065\nEpoch: 59/100... Training loss: 0.1066\nEpoch: 59/100... Training loss: 0.1033\nEpoch: 60/100... Training loss: 0.1062\nEpoch: 60/100... Training loss: 0.1073\nEpoch: 60/100... Training loss: 0.1077\nEpoch: 61/100... Training loss: 0.1059\nEpoch: 61/100... Training loss: 0.1042\nEpoch: 61/100... Training loss: 0.1078\nEpoch: 62/100... Training loss: 0.1073\nEpoch: 62/100... Training loss: 0.1045\nEpoch: 62/100... Training loss: 0.1050\nEpoch: 63/100... Training loss: 0.1054\nEpoch: 63/100... Training loss: 0.1070\nEpoch: 63/100... Training loss: 0.1059\nEpoch: 64/100... Training loss: 0.1087\nEpoch: 64/100... Training loss: 0.1067\nEpoch: 64/100... Training loss: 0.1059\nEpoch: 65/100... Training loss: 0.1054\nEpoch: 65/100... Training loss: 0.1048\nEpoch: 65/100... Training loss: 0.1027\nEpoch: 66/100... Training loss: 0.1015\nEpoch: 66/100... Training loss: 0.1071\nEpoch: 66/100... Training loss: 0.1071\nEpoch: 67/100... Training loss: 0.1022\nEpoch: 67/100... Training loss: 0.1077\nEpoch: 67/100... Training loss: 0.1051\nEpoch: 68/100... Training loss: 0.1057\nEpoch: 68/100... Training loss: 0.1029\nEpoch: 68/100... Training loss: 0.1042\nEpoch: 69/100... Training loss: 0.1057\nEpoch: 69/100... Training loss: 0.1026\nEpoch: 69/100... Training loss: 0.1065\nEpoch: 70/100... Training loss: 0.1073\nEpoch: 70/100... Training loss: 0.1027\nEpoch: 70/100... Training loss: 0.1039\nEpoch: 71/100... Training loss: 0.1064\n"
]
],
[
[
"## Checking out the performance\n\nHere I'm adding noise to the test images and passing them through the autoencoder. It does a suprisingly great job of removing the noise, even though it's sometimes difficult to tell what the original number is.",
"_____no_output_____"
]
],
[
[
"fig, axes = plt.subplots(nrows=2, ncols=10, sharex=True, sharey=True, figsize=(20,4))\nin_imgs = mnist.test.images[50:60]\nnoisy_imgs = in_imgs + noise_factor * np.random.randn(*in_imgs.shape)\nnoisy_imgs = np.clip(noisy_imgs, 0., 1.)\n\nreconstructed = sess.run(decoded, feed_dict={inputs_: noisy_imgs.reshape((10, 28, 28, 1))})\n\nfor images, row in zip([noisy_imgs, reconstructed], axes):\n for img, ax in zip(images, row):\n ax.imshow(img.reshape((28, 28)), cmap='Greys_r')\n ax.get_xaxis().set_visible(False)\n ax.get_yaxis().set_visible(False)\n\nfig.tight_layout(pad=0.1)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
cbf27593b7bfa829ccd81cb83008d294a44c28b5
| 166,754 |
ipynb
|
Jupyter Notebook
|
site/en/r2/guide/keras/training_and_evaluation.ipynb
|
angersson/docs
|
ead810deac521543f71a27c75739f93a81601061
|
[
"Apache-2.0"
] | 2 |
2021-07-05T19:07:31.000Z
|
2021-11-17T11:09:30.000Z
|
site/en/r2/guide/keras/training_and_evaluation.ipynb
|
angersson/docs
|
ead810deac521543f71a27c75739f93a81601061
|
[
"Apache-2.0"
] | null | null | null |
site/en/r2/guide/keras/training_and_evaluation.ipynb
|
angersson/docs
|
ead810deac521543f71a27c75739f93a81601061
|
[
"Apache-2.0"
] | 1 |
2019-11-10T04:01:29.000Z
|
2019-11-10T04:01:29.000Z
| 67.429842 | 68,444 | 0.701914 |
[
[
[
"##### Copyright 2019 The TensorFlow Authors.",
"_____no_output_____"
]
],
[
[
"#@title Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# https://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.",
"_____no_output_____"
]
],
[
[
"# Training and Evaluation with TensorFlow Keras\n",
"_____no_output_____"
],
[
"\n<table class=\"tfo-notebook-buttons\" align=\"left\">\n <td>\n <a target=\"_blank\" href=\"https://www.tensorflow.org/alpha/guide/keras/training_and_evaluation\"><img src=\"https://www.tensorflow.org/images/tf_logo_32px.png\" />View on TensorFlow.org</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://colab.research.google.com/github/tensorflow/docs/blob/master/site/en/r2/guide/keras/training_and_evaluation.ipynb\"><img src=\"https://www.tensorflow.org/images/colab_logo_32px.png\" />Run in Google Colab</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://github.com/tensorflow/docs/blob/master/site/en/r2/guide/keras/training_and_evaluation.ipynb\"><img src=\"https://www.tensorflow.org/images/GitHub-Mark-32px.png\" />View source on GitHub</a>\n </td>\n</table>\n",
"_____no_output_____"
],
[
"\nThis guide covers training, evaluation, and prediction (inference) models in TensorFlow 2.0 in two broad situations:\n\n- When using built-in APIs for training & validation (such as `model.fit()`, `model.evaluate()`, `model.predict()`). This is covered in the section **\"Using build-in training & evaluation loops\"**.\n- When writing custom loops from scratch using eager execution and the `GradientTape` object. This is covered in the section **\"Writing your own training & evaluation loops from scratch\"**.\n\nIn general, whether you are using built-in loops or writing your own, model training & evaluation works strictly in the same way across every kind of Keras model -- Sequential models, models built with the Functional API, and models written from scratch via model subclassing.\n\nThis guide doesn't cover distributed training.",
"_____no_output_____"
],
[
"## Setup",
"_____no_output_____"
]
],
[
[
"!pip install pydot\n!apt-get install graphviz",
"Requirement already satisfied: pydot in /usr/local/lib/python3.6/dist-packages (1.3.0)\nRequirement already satisfied: pyparsing>=2.1.4 in /usr/local/lib/python3.6/dist-packages (from pydot) (2.3.1)\nReading package lists... Done\nBuilding dependency tree \nReading state information... Done\ngraphviz is already the newest version (2.40.1-2).\n0 upgraded, 0 newly installed, 0 to remove and 10 not upgraded.\n"
],
[
"from __future__ import absolute_import, division, print_function\n\n!pip install tensorflow-gpu==2.0.0-alpha0\nimport tensorflow as tf\n\ntf.keras.backend.clear_session() # For easy reset of notebook state.",
"_____no_output_____"
]
],
[
[
"## Part I: Using build-in training & evaluation loops\n\nWhen passing data to the built-in training loops of a model, you should either use **Numpy arrays** (if your data is small and fits in memory) or **tf.data Dataset** objects. In the next few paragraphs, we'll use the MNIST dataset as Numpy arrays, in order to demonstrate how to use optimizers, losses, and metrics.",
"_____no_output_____"
],
[
"### API overview: a first end-to-end example\n\nLet's consider the following model (here, we build in with the Functional API, but it could be a Sequential model or a subclassed model as well):\n\n\n\n",
"_____no_output_____"
]
],
[
[
"from tensorflow import keras\nfrom tensorflow.keras import layers\n\ninputs = keras.Input(shape=(784,), name='digits')\nx = layers.Dense(64, activation='relu', name='dense_1')(inputs)\nx = layers.Dense(64, activation='relu', name='dense_2')(x)\noutputs = layers.Dense(10, activation='softmax', name='predictions')(x)\n\nmodel = keras.Model(inputs=inputs, outputs=outputs)",
"_____no_output_____"
]
],
[
[
"Here's what the typical end-to-end workflow looks like, consisting of training, validation on a holdout set generated from the original training data, and finally evaluation on the test data:\n",
"_____no_output_____"
]
],
[
[
"# Load a toy dataset for the sake of this example\n(x_train, y_train), (x_test, y_test) = keras.datasets.mnist.load_data()\n\n# Preprocess the data (these are Numpy arrays)\nx_train = x_train.reshape(60000, 784).astype('float32') / 255\nx_test = x_test.reshape(10000, 784).astype('float32') / 255\n\n# Reserve 10,000 samples for validation\nx_val = x_train[-10000:]\ny_val = y_train[-10000:]\nx_train = x_train[:-10000]\ny_train = y_train[:-10000]\n\n# Specify the training configuration (optimizer, loss, metrics)\nmodel.compile(optimizer=keras.optimizers.RMSprop(), # Optimizer\n # Loss function to minimize\n loss=keras.losses.SparseCategoricalCrossentropy(),\n # List of metrics to monitor\n metrics=[keras.metrics.SparseCategoricalAccuracy()])\n\n# Train the model by slicing the data into \"batches\"\n# of size \"batch_size\", and repeatedly iterating over\n# the entire dataset for a given number of \"epochs\"\nprint('# Fit model on training data')\nhistory = model.fit(x_train, y_train,\n batch_size=64,\n epochs=3,\n # We pass some validation for\n # monitoring validation loss and metrics\n # at the end of each epoch\n validation_data=(x_val, y_val))\n\n# The returned \"history\" object holds a record\n# of the loss values and metric values during training\nprint('\\nhistory dict:', history.history)\n\n# Evaluate the model on the test data using `evaluate`\nprint('\\n# Evaluate on test data')\nresults = model.evaluate(x_test, y_test, batch_size=128)\nprint('test loss, test acc:', results)\n\n# Generate predictions (probabilities -- the output of the last layer)\n# on new data using `predict`\nprint('\\n# Generate predictions for 3 samples')\npredictions = model.predict(x_test[:3])\nprint('predictions shape:', predictions.shape)",
"Downloading data from https://storage.googleapis.com/tensorflow/tf-keras-datasets/mnist.npz\n11493376/11490434 [==============================] - 0s 0us/step\n# Fit model on training data\nTrain on 50000 samples, validate on 10000 samples\nEpoch 1/3\n50000/50000 [==============================] - 5s 110us/sample - loss: 0.3377 - sparse_categorical_accuracy: 0.9053 - val_loss: 0.2020 - val_sparse_categorical_accuracy: 0.9402\nEpoch 2/3\n50000/50000 [==============================] - 5s 107us/sample - loss: 0.1576 - sparse_categorical_accuracy: 0.9528 - val_loss: 0.1379 - val_sparse_categorical_accuracy: 0.9616\nEpoch 3/3\n50000/50000 [==============================] - 5s 106us/sample - loss: 0.1143 - sparse_categorical_accuracy: 0.9650 - val_loss: 0.1128 - val_sparse_categorical_accuracy: 0.9681\n\nhistory dict: {'loss': [0.33772996835231783, 0.15758442388363184, 0.11431736122608185], 'sparse_categorical_accuracy': [0.90532, 0.95276, 0.96504], 'val_loss': [0.2019659897595644, 0.13788076196610927, 0.1128087827205658], 'val_sparse_categorical_accuracy': [0.9402, 0.9616, 0.9681]}\n\n# Evaluate on test data\n10000/10000 [==============================] - 0s 36us/sample - loss: 0.1238 - sparse_categorical_accuracy: 0.9606\ntest loss, test acc: [0.12378974738866091, 0.9606]\n\n# Generate predictions for 3 samples\npredictions shape: (3, 10)\n"
]
],
[
[
"### Specifying a loss, metrics, and an optimizer\n\nTo train a model with `fit`, you need to specify a loss function, an optimizer, and optionally, some metrics to monitor.\n\nYou pass these to the model as arguments to the `compile()` method:\n\n",
"_____no_output_____"
]
],
[
[
"model.compile(optimizer=keras.optimizers.RMSprop(learning_rate=1e-3),\n loss=keras.losses.SparseCategoricalCrossentropy(),\n metrics=[keras.metrics.SparseCategoricalAccuracy()])",
"_____no_output_____"
]
],
[
[
"The `metrics` argument should be a list -- you model can have any number of metrics.\n\nIf your model has multiple outputs, you can specify different losses and metrics for each output,\nand you can modulate to contribution of each output to the total loss of the model. You will find more details about this in the section \"**Passing data to multi-input, multi-output models**\".\n\nNote that in many cases, the loss and metrics are specified via string identifiers, as a shortcut:\n",
"_____no_output_____"
]
],
[
[
"model.compile(optimizer=keras.optimizers.RMSprop(learning_rate=1e-3),\n loss='sparse_categorical_crossentropy',\n metrics=['sparse_categorical_accuracy'])",
"_____no_output_____"
]
],
[
[
"For later reuse, let's put our model definition and compile step in functions; we will call them several times across different examples in this guide.",
"_____no_output_____"
]
],
[
[
"def get_uncompiled_model():\n inputs = keras.Input(shape=(784,), name='digits')\n x = layers.Dense(64, activation='relu', name='dense_1')(inputs)\n x = layers.Dense(64, activation='relu', name='dense_2')(x)\n outputs = layers.Dense(10, activation='softmax', name='predictions')(x)\n model = keras.Model(inputs=inputs, outputs=outputs)\n return model\n\ndef get_compiled_model():\n model = get_uncompiled_model()\n model.compile(optimizer=keras.optimizers.RMSprop(learning_rate=1e-3),\n loss='sparse_categorical_crossentropy',\n metrics=['sparse_categorical_accuracy'])\n return model",
"_____no_output_____"
]
],
[
[
"#### Many built-in optimizers, losses, and metrics are available\n\nIn general, you won't have to create from scratch your own losses, metrics, or optimizers, because what you need is likely already part of the Keras API:\n\nOptimizers:\n- `SGD()` (with or without momentum)\n- `RMSprop()`\n- `Adam()`\n- etc.\n\nLosses:\n- `MeanSquaredError()`\n- `KLDivergence()`\n- `CosineSimilarity()`\n- etc.\n\nMetrics:\n- `AUC()`\n- `Precision()`\n- `Recall()`\n- etc.",
"_____no_output_____"
],
[
"#### Writing custom losses and metrics\n\nIf you need a metric that isn't part of the API, you can easily create custom metrics by subclassing the `Metric` class. You will need to implement 4 methods:\n\n- `__init__(self)`, in which you will create state variables for your metric.\n- `update_state(self, y_true, y_pred, sample_weight=None)`, which uses the targets `y_true` and the model predictions `y_pred` to update the state variables.\n- `result(self)`, which uses the state variables to compute the final results.\n- `reset_states(self)`, which reinitializes the state of the metric.\n\nState update and results computation are kept separate (in `update_state()` and `result()`, respectively) because in some cases, results computation might be very expensive, and would only be done periodically.\n\nHere's a simple example showing how to implement a `CatgoricalTruePositives` metric, that counts how many samples where correctly classified as belonging to a given class:",
"_____no_output_____"
]
],
[
[
"class CatgoricalTruePositives(keras.metrics.Metric):\n \n def __init__(self, name='binary_true_positives', **kwargs):\n super(CatgoricalTruePositives, self).__init__(name=name, **kwargs)\n self.true_positives = self.add_weight(name='tp', initializer='zeros')\n\n def update_state(self, y_true, y_pred, sample_weight=None):\n y_pred = tf.argmax(y_pred)\n values = tf.equal(tf.cast(y_true, 'int32'), tf.cast(y_pred, 'int32'))\n values = tf.cast(values, 'float32')\n if sample_weight is not None:\n sample_weight = tf.cast(sample_weight, 'float32')\n values = tf.multiply(values, sample_weight)\n return self.true_positives.assign_add(tf.reduce_sum(values)) # TODO: fix\n\n def result(self):\n return tf.identity(self.true_positives) # TODO: fix\n \n def reset_states(self):\n # The state of the metric will be reset at the start of each epoch.\n self.true_positives.assign(0.)\n\n\nmodel.compile(optimizer=keras.optimizers.RMSprop(learning_rate=1e-3),\n loss=keras.losses.SparseCategoricalCrossentropy(),\n metrics=[CatgoricalTruePositives()])\nmodel.fit(x_train, y_train,\n batch_size=64,\n epochs=3)\n",
"_____no_output_____"
]
],
[
[
"#### Handling losses and metrics that don't fit the standard signature\n\nThe overwhelming majority of losses and metrics can be computed from `y_true` and `y_pred`, where `y_pred` is an output of your model. But not all of them. For instance, a regularization loss may only require the activation of a layer (there are no targets in this case), and this activation may not be a model output.\n\nIn such cases, you can call `self.add_loss(loss_value)` from inside the `call` method of a custom layer. Here's a simple example that adds activity regularization (note that activity regularization is built-in in all Keras layers -- this layer is just for the sake of providing a concrete example):\n",
"_____no_output_____"
]
],
[
[
"class ActivityRegularizationLayer(layers.Layer):\n \n def call(self, inputs):\n self.add_loss(tf.reduce_sum(inputs) * 0.1)\n return inputs # Pass-through layer.\n \ninputs = keras.Input(shape=(784,), name='digits')\nx = layers.Dense(64, activation='relu', name='dense_1')(inputs)\n\n# Insert activity regularization as a layer\nx = ActivityRegularizationLayer()(x)\n\nx = layers.Dense(64, activation='relu', name='dense_2')(x)\noutputs = layers.Dense(10, activation='softmax', name='predictions')(x)\n\nmodel = keras.Model(inputs=inputs, outputs=outputs)\nmodel.compile(optimizer=keras.optimizers.RMSprop(learning_rate=1e-3),\n loss='sparse_categorical_crossentropy')\n\n# The displayed loss will be much higher than before\n# due to the regularization component.\nmodel.fit(x_train, y_train,\n batch_size=64,\n epochs=1)",
"50000/50000 [==============================] - 4s 75us/sample - loss: 2.5322\n"
]
],
[
[
"You can do the same for logging metric values:",
"_____no_output_____"
]
],
[
[
"class MetricLoggingLayer(layers.Layer):\n \n def call(self, inputs):\n # The `aggregation` argument defines\n # how to aggregate the per-batch values\n # over each epoch:\n # in this case we simply average them.\n self.add_metric(keras.backend.std(inputs),\n name='std_of_activation',\n aggregation='mean')\n return inputs # Pass-through layer.\n\n \ninputs = keras.Input(shape=(784,), name='digits')\nx = layers.Dense(64, activation='relu', name='dense_1')(inputs)\n\n# Insert std logging as a layer.\nx = MetricLoggingLayer()(x)\n\nx = layers.Dense(64, activation='relu', name='dense_2')(x)\noutputs = layers.Dense(10, activation='softmax', name='predictions')(x)\n\nmodel = keras.Model(inputs=inputs, outputs=outputs)\nmodel.compile(optimizer=keras.optimizers.RMSprop(learning_rate=1e-3),\n loss='sparse_categorical_crossentropy')\nmodel.fit(x_train, y_train,\n batch_size=64,\n epochs=1)",
"50000/50000 [==============================] - 4s 76us/sample - loss: 0.3366 - std_of_activation: 0.9773\n"
]
],
[
[
"In the [Functional API](functional.ipynb), you can also call `model.add_loss(loss_tensor)`, or `model.add_metric(metric_tensor, name, aggregation)`. \n\nHere's a simple example:",
"_____no_output_____"
]
],
[
[
"inputs = keras.Input(shape=(784,), name='digits')\nx1 = layers.Dense(64, activation='relu', name='dense_1')(inputs)\nx2 = layers.Dense(64, activation='relu', name='dense_2')(x1)\noutputs = layers.Dense(10, activation='softmax', name='predictions')(x2)\nmodel = keras.Model(inputs=inputs, outputs=outputs)\n\nmodel.add_loss(tf.reduce_sum(x1) * 0.1)\n\nmodel.add_metric(keras.backend.std(x1),\n name='std_of_activation',\n aggregation='mean')\n\nmodel.compile(optimizer=keras.optimizers.RMSprop(1e-3),\n loss='sparse_categorical_crossentropy')\nmodel.fit(x_train, y_train,\n batch_size=64,\n epochs=1)",
"50000/50000 [==============================] - 4s 80us/sample - loss: 2.5158 - std_of_activation: 0.0020\n"
]
],
[
[
"\n#### Automatically setting apart a validation holdout set\n\nIn the first end-to-end example you saw, we used the `validation_data` argument to pass a tuple\nof Numpy arrays `(x_val, y_val)` to the model for evaluating a validation loss and validation metrics at the end of each epoch.\n\nHere's another option: the argument `validation_split` allows you to automatically reserve part of your training data for validation. The argument value represents the fraction of the data to be reserved for validation, so it should be set to a number higher than 0 and lower than 1. For instance, `validation_split=0.2` means \"use 20% of the data for validation\", and `validation_split=0.6` means \"use 60% of the data for validation\".\n\nThe way the validation is computed is by *taking the last x% samples of the arrays received by the `fit` call, before any shuffling*.\n\nYou can only use `validation_split` when training with Numpy data.",
"_____no_output_____"
]
],
[
[
"model = get_compiled_model()\nmodel.fit(x_train, y_train, batch_size=64, validation_split=0.2, epochs=3)",
"Train on 40000 samples, validate on 10000 samples\nEpoch 1/3\n40000/40000 [==============================] - 3s 82us/sample - loss: 0.3735 - sparse_categorical_accuracy: 0.8951 - val_loss: 0.2413 - val_sparse_categorical_accuracy: 0.9272\nEpoch 2/3\n40000/40000 [==============================] - 3s 82us/sample - loss: 0.1688 - sparse_categorical_accuracy: 0.9499 - val_loss: 0.1781 - val_sparse_categorical_accuracy: 0.9468\nEpoch 3/3\n40000/40000 [==============================] - 3s 79us/sample - loss: 0.1232 - sparse_categorical_accuracy: 0.9638 - val_loss: 0.1518 - val_sparse_categorical_accuracy: 0.9539\n"
]
],
[
[
"### Training & evaluation from tf.data Datasets\n\nIn the past few paragraphs, you've seen how to handle losses, metrics, and optimizers, and you've seen how to use the `validation_data` and `validation_split` arguments in `fit`, when your data is passed as Numpy arrays.\n\nLet's now take a look at the case where your data comes in the form of a tf.data Dataset.\n\nThe tf.data API is a set of utilities in TensorFlow 2.0 for loading and preprocessing data in a way that's fast and scalable. \n\nFor a complete guide about creating Datasets, see [the tf.data documentation](https://www.tensorflow.org/versions/r2.0/api_docs/python/tf).\n\nYou can pass a Dataset instance directly to the methods `fit()`, `evaluate()`, and `predict()`:",
"_____no_output_____"
]
],
[
[
"model = get_compiled_model()\n\n# First, let's create a training Dataset instance.\n# For the sake of our example, we'll use the same MNIST data as before.\ntrain_dataset = tf.data.Dataset.from_tensor_slices((x_train, y_train))\n# Shuffle and slice the dataset.\ntrain_dataset = train_dataset.shuffle(buffer_size=1024).batch(64)\n\n# Now we get a test dataset.\ntest_dataset = tf.data.Dataset.from_tensor_slices((x_test, y_test))\ntest_dataset = test_dataset.batch(64)\n\n# Since the dataset already takes care of batching,\n# we don't pass a `batch_size` argument.\nmodel.fit(train_dataset, epochs=3)\n\n# You can also evaluate or predict on a dataset.\nprint('\\n# Evaluate')\nmodel.evaluate(test_dataset)",
"Epoch 1/3\n782/782 [==============================] - 5s 7ms/step - loss: 0.3250 - sparse_categorical_accuracy: 0.9074\nEpoch 2/3\n782/782 [==============================] - 4s 6ms/step - loss: 0.1484 - sparse_categorical_accuracy: 0.9559\nEpoch 3/3\n782/782 [==============================] - 4s 5ms/step - loss: 0.1074 - sparse_categorical_accuracy: 0.9685\n\n# Evaluate\n157/157 [==============================] - 1s 3ms/step - loss: 0.1137 - sparse_categorical_accuracy: 0.9665\n"
]
],
[
[
"Note that the Dataset is reset at the end of each epoch, so it can be reused of the next epoch.\n\nIf you want to run training only on a specific number of batches from this Dataset, you can pass the `steps_per_epoch` argument, which specifies how many training steps the model should run using this Dataset before moving on to the next epoch.\n\nIf you do this, the dataset is not reset at the end of each epoch, instead we just keep drawing the next batches. The dataset will eventually run out of data (unless it is an infinitely-looping dataset).",
"_____no_output_____"
]
],
[
[
"model = get_compiled_model()\n\n# Prepare the training dataset\ntrain_dataset = tf.data.Dataset.from_tensor_slices((x_train, y_train))\ntrain_dataset = train_dataset.shuffle(buffer_size=1024).batch(64)\n\n# Only use the 100 batches per epoch (that's 64 * 100 samples)\nmodel.fit(train_dataset, epochs=3, steps_per_epoch=100)",
"Epoch 1/3\n100/100 [==============================] - 1s 11ms/step - loss: 0.7733 - sparse_categorical_accuracy: 0.8067\nEpoch 2/3\n100/100 [==============================] - 0s 5ms/step - loss: 0.3706 - sparse_categorical_accuracy: 0.8922\nEpoch 3/3\n100/100 [==============================] - 1s 5ms/step - loss: 0.3379 - sparse_categorical_accuracy: 0.9011\n"
]
],
[
[
"#### Using a validation dataset\n\nYou can pass a Dataset instance as the `validation_data` argument in `fit`:",
"_____no_output_____"
]
],
[
[
"model = get_compiled_model()\n\n# Prepare the training dataset\ntrain_dataset = tf.data.Dataset.from_tensor_slices((x_train, y_train))\ntrain_dataset = train_dataset.shuffle(buffer_size=1024).batch(64)\n\n# Prepare the validation dataset\nval_dataset = tf.data.Dataset.from_tensor_slices((x_val, y_val))\nval_dataset = val_dataset.batch(64)\n\nmodel.fit(train_dataset, epochs=3, validation_data=val_dataset)",
"Epoch 1/3\n782/782 [==============================] - 7s 8ms/step - loss: 0.3440 - sparse_categorical_accuracy: 0.9020 - val_loss: 0.1838 - val_sparse_categorical_accuracy: 0.9490\nEpoch 2/3\n782/782 [==============================] - 7s 9ms/step - loss: 0.1649 - sparse_categorical_accuracy: 0.9515 - val_loss: 0.1391 - val_sparse_categorical_accuracy: 0.9603\nEpoch 3/3\n782/782 [==============================] - 8s 10ms/step - loss: 0.1216 - sparse_categorical_accuracy: 0.9645 - val_loss: 0.1208 - val_sparse_categorical_accuracy: 0.9672\n"
]
],
[
[
"At the end of each epoch, the model will iterate over the validation Dataset and compute the validation loss and validation metrics.\n\nIf you want to run validation only on a specific number of batches from this Dataset, you can pass the `validation_steps` argument, which specifies how many validation steps the model should run with the validation Dataset before interrupting validation and moving on to the next epoch:",
"_____no_output_____"
]
],
[
[
"model = get_compiled_model()\n\n# Prepare the training dataset\ntrain_dataset = tf.data.Dataset.from_tensor_slices((x_train, y_train))\ntrain_dataset = train_dataset.shuffle(buffer_size=1024).batch(64)\n\n# Prepare the validation dataset\nval_dataset = tf.data.Dataset.from_tensor_slices((x_val, y_val))\nval_dataset = val_dataset.batch(64)\n\nmodel.fit(train_dataset, epochs=3,\n # Only run validation using the first 10 batches of the dataset\n # using the `validation_steps` argument\n validation_data=val_dataset, validation_steps=10)",
"Epoch 1/3\n782/782 [==============================] - 9s 12ms/step - loss: 0.3359 - sparse_categorical_accuracy: 0.9053 - val_loss: 0.3095 - val_sparse_categorical_accuracy: 0.9187\nEpoch 2/3\n782/782 [==============================] - 7s 9ms/step - loss: 0.1593 - sparse_categorical_accuracy: 0.9528 - val_loss: 0.2196 - val_sparse_categorical_accuracy: 0.9438\nEpoch 3/3\n782/782 [==============================] - 7s 9ms/step - loss: 0.1158 - sparse_categorical_accuracy: 0.9661 - val_loss: 0.1840 - val_sparse_categorical_accuracy: 0.9469\n"
]
],
[
[
"Note that the validation Dataset will be reset after each use (so that you will always be evaluating on the same samples from epoch to epoch).\n\nThe argument `validation_split` (generating a holdout set from the training data) is not supported when training from Dataset objects, since this features requires the ability to index the samples of the datasets, which is not possible in general with the Dataset API.",
"_____no_output_____"
],
[
"\n### Other input formats supported\n\nBesides Numpy arrays and TensorFlow Datasets, it's possible to train a Keras model using Pandas dataframes, or from Python generators that yield batches.\n\nIn general, we recommend that you use Numpy input data if your data is small and fits in memory, and Datasets otherwise.",
"_____no_output_____"
],
[
"### Using sample weighting and class weighting\n\nBesides input data and target data, it is possible to pass sample weights or class weights to a model when using `fit`:\n\n- When training from Numpy data: via the `sample_weight` and `class_weight` arguments.\n- When training from Datasets: by having the Dataset return a tuple `(input_batch, target_batch, sample_weight_batch)` .\n\nA \"sample weights\" array is an array of numbers that specify how much weight each sample in a batch should have in computing the total loss. It is commonly used in imbalanced classification problems (the idea being to give more weight to rarely-seen classes). When the weights used are ones and zeros, the array can be used as a *mask* for the loss function (entirely discarding the contribution of certain samples to the total loss).\n\nA \"class weights\" dict is a more specific instance of the same concept: it maps class indices to the sample weight that should be used for samples belonging to this class. For instance, if class \"0\" is twice less represented than class \"1\" in your data, you could use `class_weight={0: 1., 1: 0.5}`.",
"_____no_output_____"
],
[
"Here's a Numpy example where we use class weights or sample weights to give more importance to the correct classification of class #5 (which is the digit \"5\" in the MNIST dataset).",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\nclass_weight = {0: 1., 1: 1., 2: 1., 3: 1., 4: 1.,\n # Set weight \"2\" for class \"5\",\n # making this class 2x more important\n 5: 2.,\n 6: 1., 7: 1., 8: 1., 9: 1.}\nmodel.fit(x_train, y_train,\n class_weight=class_weight,\n batch_size=64,\n epochs=4)\n\n# Here's the same example using `sample_weight` instead:\nsample_weight = np.ones(shape=(len(y_train),))\nsample_weight[y_train == 5] = 2.\n\nmodel = get_compiled_model()\nmodel.fit(x_train, y_train,\n sample_weight=sample_weight,\n batch_size=64,\n epochs=4)",
"Epoch 1/4\n50000/50000 [==============================] - 4s 89us/sample - loss: 0.1040 - sparse_categorical_accuracy: 0.9715\nEpoch 2/4\n50000/50000 [==============================] - 4s 83us/sample - loss: 0.0872 - sparse_categorical_accuracy: 0.9751\nEpoch 3/4\n50000/50000 [==============================] - 4s 85us/sample - loss: 0.0734 - sparse_categorical_accuracy: 0.9789\nEpoch 4/4\n50000/50000 [==============================] - 4s 81us/sample - loss: 0.0657 - sparse_categorical_accuracy: 0.9818\nEpoch 1/4\n50000/50000 [==============================] - 4s 87us/sample - loss: 0.3647 - sparse_categorical_accuracy: 0.9063\nEpoch 2/4\n50000/50000 [==============================] - 5s 91us/sample - loss: 0.1703 - sparse_categorical_accuracy: 0.9525\nEpoch 3/4\n50000/50000 [==============================] - 4s 81us/sample - loss: 0.1276 - sparse_categorical_accuracy: 0.9647\nEpoch 4/4\n50000/50000 [==============================] - 4s 83us/sample - loss: 0.1016 - sparse_categorical_accuracy: 0.9719\n"
]
],
[
[
"Here's a matching Dataset example:",
"_____no_output_____"
]
],
[
[
"sample_weight = np.ones(shape=(len(y_train),))\nsample_weight[y_train == 5] = 2.\n\n# Create a Dataset that includes sample weights\n# (3rd element in the return tuple).\ntrain_dataset = tf.data.Dataset.from_tensor_slices(\n (x_train, y_train, sample_weight))\n\n# Shuffle and slice the dataset.\ntrain_dataset = train_dataset.shuffle(buffer_size=1024).batch(64)\n\nmodel = get_compiled_model()\nmodel.fit(train_dataset, epochs=3)",
"Epoch 1/3\n782/782 [==============================] - 9s 11ms/step - loss: 0.3666 - sparse_categorical_accuracy: 0.9046\nEpoch 2/3\n782/782 [==============================] - 7s 9ms/step - loss: 0.1646 - sparse_categorical_accuracy: 0.9539\nEpoch 3/3\n782/782 [==============================] - 7s 9ms/step - loss: 0.1178 - sparse_categorical_accuracy: 0.9677\n"
]
],
[
[
"### Passing data to multi-input, multi-output models\n\nIn the previous examples, we were considering a model with a single input (a tensor of shape `(764,)`) and a single output (a prediction tensor of shape `(10,)`). But what about models that have multiple inputs or outputs?\n\nConsider the following model, which has an image input of shape `(32, 32, 3)` (that's `(height, width, channels)`) and a timeseries input of shape `(None, 10)` (that's `(timesteps, features)`). Our model will have two outputs computed from the combination of these inputs: a \"score\" (of shape `(1,)`) and a probability distribution over 5 classes (of shape `(10,)`).\n",
"_____no_output_____"
]
],
[
[
"from tensorflow import keras\nfrom tensorflow.keras import layers\n\nimage_input = keras.Input(shape=(32, 32, 3), name='img_input')\ntimeseries_input = keras.Input(shape=(None, 10), name='ts_input')\n\nx1 = layers.Conv2D(3, 3)(image_input)\nx1 = layers.GlobalMaxPooling2D()(x1)\n\nx2 = layers.Conv1D(3, 3)(timeseries_input)\nx2 = layers.GlobalMaxPooling1D()(x2)\n\nx = layers.concatenate([x1, x2])\n\nscore_output = layers.Dense(1, name='score_output')(x)\nclass_output = layers.Dense(5, activation='softmax', name='class_output')(x)\n\nmodel = keras.Model(inputs=[image_input, timeseries_input],\n outputs=[score_output, class_output])",
"_____no_output_____"
]
],
[
[
"Let's plot this model, so you can clearly see what we're doing here (note that the shapes shown in the plot are batch shapes, rather than per-sample shapes).",
"_____no_output_____"
]
],
[
[
"keras.utils.plot_model(model, 'multi_input_and_output_model.png', show_shapes=True)",
"_____no_output_____"
]
],
[
[
"At compilation time, we can specify different losses to different ouptuts, by passing the loss functions as a list:",
"_____no_output_____"
]
],
[
[
"model.compile(\n optimizer=keras.optimizers.RMSprop(1e-3),\n loss=[keras.losses.MeanSquaredError(),\n keras.losses.CategoricalCrossentropy()])",
"_____no_output_____"
]
],
[
[
"If we only passed a single loss function to the model, the same loss function would be applied to every output, which is not appropriate here.\n\nLikewise for metrics:",
"_____no_output_____"
]
],
[
[
"model.compile(\n optimizer=keras.optimizers.RMSprop(1e-3),\n loss=[keras.losses.MeanSquaredError(),\n keras.losses.CategoricalCrossentropy()],\n metrics=[[keras.metrics.MeanAbsolutePercentageError(),\n keras.metrics.MeanAbsoluteError()],\n [keras.metrics.CategoricalAccuracy()]])",
"_____no_output_____"
]
],
[
[
"Since we gave names to our output layers, we coud also specify per-output losses and metrics via a dict:",
"_____no_output_____"
]
],
[
[
"model.compile(\n optimizer=keras.optimizers.RMSprop(1e-3),\n loss={'score_output': keras.losses.MeanSquaredError(),\n 'class_output': keras.losses.CategoricalCrossentropy()},\n metrics={'score_output': [keras.metrics.MeanAbsolutePercentageError(),\n keras.metrics.MeanAbsoluteError()],\n 'class_output': [keras.metrics.CategoricalAccuracy()]})",
"_____no_output_____"
]
],
[
[
"We recommend the use of explicit names and dicts if you have more than 2 outputs.",
"_____no_output_____"
],
[
"It's possible to give different weights to different output-specific losses (for instance, one might wish to privilege the \"score\" loss in our example, by giving to 2x the importance of the class loss), using the `loss_weight` argument:",
"_____no_output_____"
]
],
[
[
"model.compile(\n optimizer=keras.optimizers.RMSprop(1e-3),\n loss={'score_output': keras.losses.MeanSquaredError(),\n 'class_output': keras.losses.CategoricalCrossentropy()},\n metrics={'score_output': [keras.metrics.MeanAbsolutePercentageError(),\n keras.metrics.MeanAbsoluteError()],\n 'class_output': [keras.metrics.CategoricalAccuracy()]},\n loss_weight={'score_output': 2., 'class_output': 1.})",
"_____no_output_____"
]
],
[
[
"You could also chose not to compute a loss for certain outputs, if these outputs meant for prediction but not for training:",
"_____no_output_____"
]
],
[
[
"# List loss version\nmodel.compile(\n optimizer=keras.optimizers.RMSprop(1e-3),\n loss=[None, keras.losses.CategoricalCrossentropy()])\n\n# Or dict loss version\nmodel.compile(\n optimizer=keras.optimizers.RMSprop(1e-3),\n loss={'class_output': keras.losses.CategoricalCrossentropy()})",
"WARNING: Logging before flag parsing goes to stderr.\nW0305 23:50:32.918388 140053718652800 training_utils.py:1152] Output score_output missing from loss dictionary. We assume this was done on purpose. The fit and evaluate APIs will not be expecting any data to be passed to score_output.\n"
]
],
[
[
"Passing data to a multi-input or multi-output model in `fit` works in a similar way as specifying a loss function in `compile`:\nyou can pass *lists of Numpy arrays (with 1:1 mapping to the outputs that received a loss function)* or *dicts mapping output names to Numpy arrays of training data*.",
"_____no_output_____"
]
],
[
[
"model.compile(\n optimizer=keras.optimizers.RMSprop(1e-3),\n loss=[keras.losses.MeanSquaredError(),\n keras.losses.CategoricalCrossentropy()])\n\n# Generate dummy Numpy data\nimg_data = np.random.random_sample(size=(100, 32, 32, 3))\nts_data = np.random.random_sample(size=(100, 20, 10))\nscore_targets = np.random.random_sample(size=(100, 1))\nclass_targets = np.random.random_sample(size=(100, 5))\n\n# Fit on lists\nmodel.fit([img_data, ts_data], [score_targets, class_targets],\n batch_size=32,\n epochs=3)\n\n# Alernatively, fit on dicts\nmodel.fit({'img_input': img_data, 'ts_input': ts_data},\n {'score_output': score_targets, 'class_output': class_targets},\n batch_size=32,\n epochs=3)",
"Epoch 1/3\n100/100 [==============================] - 1s 6ms/sample - loss: 7.6847 - score_output_loss: 0.7406 - class_output_loss: 6.9441\nEpoch 2/3\n100/100 [==============================] - 0s 1ms/sample - loss: 7.0638 - score_output_loss: 0.3140 - class_output_loss: 6.7499\nEpoch 3/3\n100/100 [==============================] - 0s 1ms/sample - loss: 6.7368 - score_output_loss: 0.1928 - class_output_loss: 6.5440\nEpoch 1/3\n100/100 [==============================] - 0s 4ms/sample - loss: 6.4485 - score_output_loss: 0.1420 - class_output_loss: 6.3065\nEpoch 2/3\n100/100 [==============================] - 0s 4ms/sample - loss: 6.1095 - score_output_loss: 0.1428 - class_output_loss: 5.9667\nEpoch 3/3\n100/100 [==============================] - 0s 4ms/sample - loss: 5.8362 - score_output_loss: 0.1219 - class_output_loss: 5.7143\n"
]
],
[
[
"Here's the Dataset use case: similarly as what we did for Numpy arrays, the Dataset should return\na tuple of dicts.",
"_____no_output_____"
]
],
[
[
"train_dataset = tf.data.Dataset.from_tensor_slices(\n ({'img_input': img_data, 'ts_input': ts_data},\n {'score_output': score_targets, 'class_output': class_targets}))\ntrain_dataset = train_dataset.shuffle(buffer_size=1024).batch(64)\n\nmodel.fit(train_dataset, epochs=3)",
"Epoch 1/3\n2/2 [==============================] - 0s 152ms/step - loss: 5.6598 - score_output_loss: 0.1304 - class_output_loss: 5.5127\nEpoch 2/3\n2/2 [==============================] - 0s 107ms/step - loss: 5.5597 - score_output_loss: 0.1229 - class_output_loss: 5.4204\nEpoch 3/3\n2/2 [==============================] - 0s 145ms/step - loss: 5.4660 - score_output_loss: 0.1176 - class_output_loss: 5.3324\n"
]
],
[
[
"### Using callbacks\n\nCallbacks in Keras are objects that are called at different point during training (at the start of an epoch, at the end of a batch, at the end of an epoch, etc.) and which can be used to implement behaviors such as:\n\n- Doing validation at different points during training (beyond the built-in per-epoch validation)\n- Checkpointing the model at regular intervals or when it exceeds a certain accuracy threshold\n- Changing the learning rate of the model when training seems to be plateauing\n- Doing fine-tuning of the top layers when training seems to be plateauing\n- Sending email or instant message notifications when training ends or where a certain performance threshold is exceeded\n- Etc.\n\nCallbacks can be passed as a list to your call to `fit`:",
"_____no_output_____"
]
],
[
[
"model = get_compiled_model()\n\ncallbacks = [\n keras.callbacks.EarlyStopping(\n # Stop training when `val_loss` is no longer improving\n monitor='val_loss',\n # \"no longer improving\" being defined as \"no better than 1e-2 less\"\n min_delta=1e-2,\n # \"no longer improving\" being further defined as \"for at least 2 epochs\"\n patience=2,\n verbose=1)\n]\nmodel.fit(x_train, y_train,\n epochs=20,\n batch_size=64,\n callbacks=callbacks,\n validation_split=0.2)",
"Train on 40000 samples, validate on 10000 samples\nEpoch 1/20\n40000/40000 [==============================] - 4s 102us/sample - loss: 0.3712 - sparse_categorical_accuracy: 0.8955 - val_loss: 0.2237 - val_sparse_categorical_accuracy: 0.9325\nEpoch 2/20\n40000/40000 [==============================] - 4s 93us/sample - loss: 0.1754 - sparse_categorical_accuracy: 0.9483 - val_loss: 0.1784 - val_sparse_categorical_accuracy: 0.9440\nEpoch 3/20\n40000/40000 [==============================] - 3s 84us/sample - loss: 0.1255 - sparse_categorical_accuracy: 0.9619 - val_loss: 0.1583 - val_sparse_categorical_accuracy: 0.9514\nEpoch 4/20\n40000/40000 [==============================] - 4s 90us/sample - loss: 0.1003 - sparse_categorical_accuracy: 0.9703 - val_loss: 0.1404 - val_sparse_categorical_accuracy: 0.9587\nEpoch 5/20\n40000/40000 [==============================] - 4s 88us/sample - loss: 0.0829 - sparse_categorical_accuracy: 0.9757 - val_loss: 0.1332 - val_sparse_categorical_accuracy: 0.9617\nEpoch 6/20\n40000/40000 [==============================] - 4s 97us/sample - loss: 0.0705 - sparse_categorical_accuracy: 0.9789 - val_loss: 0.1341 - val_sparse_categorical_accuracy: 0.9641\nEpoch 00006: early stopping\n"
]
],
[
[
"#### Many built-in callbacks are available\n\n- `ModelCheckpoint`: Periodically save the model.\n- `EarlyStopping`: Stop training when training is no longer improving the validation metrics.\n- `TensorBoard`: periodically write model logs that can be visualized in TensorBoard (more details in the section \"Visualization\").\n- `CSVLogger`: streams loss and metrics data to a CSV file.\n- etc.\n\n\n\n#### Writing your own callback\n\nYou can create a custom callback by extending the base class keras.callbacks.Callback. A callback has access to its associated model through the class property `self.model`.\n\nHere's a simple example saving a list of per-batch loss values during training:\n\n```python\nclass LossHistory(keras.callbacks.Callback):\n\n def on_train_begin(self, logs):\n self.losses = []\n\n def on_batch_end(self, batch, logs):\n self.losses.append(logs.get('loss'))\n```",
"_____no_output_____"
],
[
"### Checkpointing models\n\nWhen you're training model on relatively large datasets, it's crucial to save checkpoints of your model at frequent intervals.\n\nThe easiest way to achieve this is with the `ModelCheckpoint` callback:",
"_____no_output_____"
]
],
[
[
"model = get_compiled_model()\n\ncallbacks = [\n keras.callbacks.ModelCheckpoint(\n filepath='mymodel_{epoch}.h5',\n # Path where to save the model\n # The two parameters below mean that we will overwrite\n # the current checkpoint if and only if\n # the `val_loss` score has improved.\n save_best_only=True,\n monitor='val_loss',\n verbose=1)\n]\nmodel.fit(x_train, y_train,\n epochs=3,\n batch_size=64,\n callbacks=callbacks,\n validation_split=0.2)",
"Train on 40000 samples, validate on 10000 samples\nEpoch 1/3\n39936/40000 [============================>.] - ETA: 0s - loss: 0.3635 - sparse_categorical_accuracy: 0.8971\nEpoch 00001: val_loss improved from inf to 0.21655, saving model to mymodel_1.h5\n40000/40000 [==============================] - 4s 108us/sample - loss: 0.3631 - sparse_categorical_accuracy: 0.8972 - val_loss: 0.2166 - val_sparse_categorical_accuracy: 0.9347\nEpoch 2/3\n39360/40000 [============================>.] - ETA: 0s - loss: 0.1669 - sparse_categorical_accuracy: 0.9506\nEpoch 00002: val_loss improved from 0.21655 to 0.17676, saving model to mymodel_2.h5\n40000/40000 [==============================] - 4s 97us/sample - loss: 0.1669 - sparse_categorical_accuracy: 0.9505 - val_loss: 0.1768 - val_sparse_categorical_accuracy: 0.9456\nEpoch 3/3\n39424/40000 [============================>.] - ETA: 0s - loss: 0.1232 - sparse_categorical_accuracy: 0.9624\nEpoch 00003: val_loss improved from 0.17676 to 0.15663, saving model to mymodel_3.h5\n40000/40000 [==============================] - 4s 99us/sample - loss: 0.1236 - sparse_categorical_accuracy: 0.9624 - val_loss: 0.1566 - val_sparse_categorical_accuracy: 0.9536\n"
]
],
[
[
"You call also write your own callback for saving and restoring models.\n\nFor a complete guide on serialization and saving, see [Guide to Saving and Serializing Models](./saving_and_serializing.ipynb).",
"_____no_output_____"
],
[
"### Using learning rate schedules\n\nA common pattern when training deep learning models is to gradually reduce the learning as training progresses. This is generally known as \"learning rate decay\".\n\nThe learning decay schedule could be static (fixed in advance, as a function of the current epoch or the current batch index), or dynamic (responding to the current behavior of the model, in particular the validation loss).\n\n#### Passing a schedule to an optimizer\n\nYou can easily use a static learning rate decay schedule by passing a schedule object as the `learning_rate` argument in your optimizer:\n",
"_____no_output_____"
]
],
[
[
"initial_learning_rate = 0.1\nlr_schedule = keras.optimizers.schedules.ExponentialDecay(\n initial_learning_rate,\n decay_steps=100000,\n decay_rate=0.96,\n staircase=True)\n\noptimizer = keras.optimizers.RMSprop(learning_rate=lr_schedule)",
"_____no_output_____"
]
],
[
[
"Several built-in schedules are available: `ExponentialDecay`, `PiecewiseConstantDecay`, `PolynomialDecay`, and `InverseTimeDecay`.",
"_____no_output_____"
],
[
"#### Using callbacks to implement a dynamic learning rate schedule\n\nA dynamic learning rate schedule (for instance, decreasing the learning rate when the validation loss is no longer improving) cannot be achieved with these schedule objects since the optimizer does not have access to validation metrics.\n\nHowever, callbacks do have access to all metrics, including validation metrics! You can thus achieve this pattern by using a callback that modifies the current learning rate on the optimizer. In fact, this is even built-in as the `ReduceLROnPlateau` callback.",
"_____no_output_____"
],
[
"### Visualizing loss and metrics during training\n\nThe best way to keep an eye on your model during training is to use [TensorBoard](https://www.tensorflow.org/tensorboard), a browser-based application that you can run locally that provides you with:\n\n- Live plots of the loss and metrics for training and evaluation\n- (optionally) Visualizations of the histograms of your layer activations\n- (optionally) 3D visualizations of the embedding spaces learned by your `Embedding` layers\n\nIf you have installed TensorFlow with pip, you should be able to launch TensorBoard from the command line:\n\n```\ntensorboard --logdir=/full_path_to_your_logs\n```\n\n#### Using the TensorBoard callback\n\nThe easiest way to use TensorBoard with a Keras model and the `fit` method is the `TensorBoard` callback.\n\nIn the simplest case, just specify where you want te callback to write logs, and you're good to go:\n\n```python\ntensorboard_cbk = keras.callbacks.TensorBoard(log_dir='/full_path_to_your_logs')\nmodel.fit(dataset, epochs=10, callbacks=[tensorboard_cbk])\n```\n\nThe `TensorBoard` callback has many useful options, including whether to log embeddings, histograms, and how often to write logs:\n\n```python\nkeras.callbacks.TensorBoard(\n log_dir='/full_path_to_your_logs',\n histogram_freq=0, # How often to log histogram visualizations\n embeddings_freq=0, # How often to log embedding visualizations\n update_freq='epoch') # How often to write logs (default: once per epoch)\n```\n\n",
"_____no_output_____"
],
[
"## Part II: Writing your own training & evaluation loops from scratch\n\nIf you want lower-level over your training & evaluation loops than what `fit()` and `evaluate()` provide, you should write your own. It's actually pretty simple! But you should be ready to have a lot more debugging to do on your own.",
"_____no_output_____"
],
[
"### Using the GradientTape: a first end-to-end example\n\nCalling a model inside a `GradientTape` scope enables you to retrieve the gradients of the trainable weights of the layer with respect to a loss value. Using an optimizer instance, you can use these gradients to update these variables (which you can retrieve using `model.trainable_variables`).\n\nLet's reuse our initial MNIST model from Part I, and let's train it using mini-batch gradient with a custom training loop. ",
"_____no_output_____"
]
],
[
[
"# Get the model.\ninputs = keras.Input(shape=(784,), name='digits')\nx = layers.Dense(64, activation='relu', name='dense_1')(inputs)\nx = layers.Dense(64, activation='relu', name='dense_2')(x)\noutputs = layers.Dense(10, activation='softmax', name='predictions')(x)\nmodel = keras.Model(inputs=inputs, outputs=outputs)\n\n# Instantiate an optimizer.\noptimizer = keras.optimizers.SGD(learning_rate=1e-3)\n# Instantiate a loss function.\nloss_fn = keras.losses.SparseCategoricalCrossentropy()\n\n# Prepare the training dataset.\nbatch_size = 64\ntrain_dataset = tf.data.Dataset.from_tensor_slices((x_train, y_train))\ntrain_dataset = train_dataset.shuffle(buffer_size=1024).batch(batch_size)\n\n# Iterate over epochs.\nfor epoch in range(3):\n print('Start of epoch %d' % (epoch,))\n \n # Iterate over the batches of the dataset.\n for step, (x_batch_train, y_batch_train) in enumerate(train_dataset):\n\n # Open a GradientTape to record the operations run\n # during the forward pass, which enables autodifferentiation.\n with tf.GradientTape() as tape:\n\n # Run the forward pass of the layer.\n # The operations that the layer applies\n # to its inputs are going to be recorded\n # on the GradientTape.\n logits = model(x_batch_train) # Logits for this minibatch\n\n # Compute the loss value for this minibatch.\n loss_value = loss_fn(y_batch_train, logits)\n\n # Use the gradient tape to automatically retrieve\n # the gradients of the trainable variables with respect to the loss.\n grads = tape.gradient(loss_value, model.trainable_variables)\n\n # Run one step of gradient descent by updating\n # the value of the variables to minimize the loss.\n optimizer.apply_gradients(zip(grads, model.trainable_variables))\n\n # Log every 200 batches.\n if step % 200 == 0:\n print('Training loss (for one batch) at step %s: %s' % (step, float(loss_value)))\n print('Seen so far: %s samples' % ((step + 1) * 64))",
"Start of epoch 0\nTraining loss (for one batch) at step 0: 2.295337200164795\nSeen so far: 64 samples\nTraining loss (for one batch) at step 200: 2.267664909362793\nSeen so far: 12864 samples\nTraining loss (for one batch) at step 400: 2.1268270015716553\nSeen so far: 25664 samples\nTraining loss (for one batch) at step 600: 2.0609934329986572\nSeen so far: 38464 samples\nStart of epoch 1\nTraining loss (for one batch) at step 0: 1.9627395868301392\nSeen so far: 64 samples\nTraining loss (for one batch) at step 200: 1.9132888317108154\nSeen so far: 12864 samples\nTraining loss (for one batch) at step 400: 1.7715450525283813\nSeen so far: 25664 samples\nTraining loss (for one batch) at step 600: 1.680647611618042\nSeen so far: 38464 samples\nStart of epoch 2\nTraining loss (for one batch) at step 0: 1.554194450378418\nSeen so far: 64 samples\nTraining loss (for one batch) at step 200: 1.5058209896087646\nSeen so far: 12864 samples\nTraining loss (for one batch) at step 400: 1.3611259460449219\nSeen so far: 25664 samples\nTraining loss (for one batch) at step 600: 1.2863078117370605\nSeen so far: 38464 samples\n"
]
],
[
[
"### Low-level handling of metrics\n\nLet's add metrics to the mix. You can readily reuse the built-in metrics (or custom ones you wrote) in such training loops written from scratch. Here's the flow:\n\n- Instantiate the metric at the start of the loop\n- Call `metric.update_state()` after each batch\n- Call `metric.result()` when you need to display the current value of the metric\n- Call `metric.reset_states()` when you need to clear the state of the metric (typically at the end of an epoch)\n\nLet's use this knowledge to compute `SparseCategoricalAccuracy` on validation data at the end of each epoch:",
"_____no_output_____"
]
],
[
[
"# Get model\ninputs = keras.Input(shape=(784,), name='digits')\nx = layers.Dense(64, activation='relu', name='dense_1')(inputs)\nx = layers.Dense(64, activation='relu', name='dense_2')(x)\noutputs = layers.Dense(10, activation='softmax', name='predictions')(x)\nmodel = keras.Model(inputs=inputs, outputs=outputs)\n\n# Instantiate an optimizer to train the model.\noptimizer = keras.optimizers.SGD(learning_rate=1e-3)\n# Instantiate a loss function.\nloss_fn = keras.losses.SparseCategoricalCrossentropy()\n\n# Prepare the metrics.\ntrain_acc_metric = keras.metrics.SparseCategoricalAccuracy() \nval_acc_metric = keras.metrics.SparseCategoricalAccuracy()\n\n# Prepare the training dataset.\nbatch_size = 64\ntrain_dataset = tf.data.Dataset.from_tensor_slices((x_train, y_train))\ntrain_dataset = train_dataset.shuffle(buffer_size=1024).batch(batch_size)\n\n# Prepare the validation dataset.\nval_dataset = tf.data.Dataset.from_tensor_slices((x_val, y_val))\nval_dataset = val_dataset.batch(64)\n\n\n# Iterate over epochs.\nfor epoch in range(3):\n print('Start of epoch %d' % (epoch,))\n \n # Iterate over the batches of the dataset.\n for step, (x_batch_train, y_batch_train) in enumerate(train_dataset):\n with tf.GradientTape() as tape:\n logits = model(x_batch_train)\n loss_value = loss_fn(y_batch_train, logits)\n grads = tape.gradient(loss_value, model.trainable_variables)\n optimizer.apply_gradients(zip(grads, model.trainable_variables))\n \n # Update training metric.\n train_acc_metric(y_batch_train, logits)\n\n # Log every 200 batches.\n if step % 200 == 0:\n print('Training loss (for one batch) at step %s: %s' % (step, float(loss_value)))\n print('Seen so far: %s samples' % ((step + 1) * 64))\n\n # Display metrics at the end of each epoch.\n train_acc = train_acc_metric.result()\n print('Training acc over epoch: %s' % (float(train_acc),))\n # Reset training metrics at the end of each epoch\n train_acc_metric.reset_states()\n\n # Run a validation loop at the end of each epoch.\n for x_batch_val, y_batch_val in val_dataset:\n val_logits = model(x_batch_val)\n # Update val metrics\n val_acc_metric(y_batch_val, val_logits)\n val_acc = val_acc_metric.result()\n val_acc_metric.reset_states()\n print('Validation acc: %s' % (float(val_acc),))",
"Start of epoch 0\nTraining loss (for one batch) at step 0: 2.3286547660827637\nSeen so far: 64 samples\nTraining loss (for one batch) at step 200: 2.297130823135376\nSeen so far: 12864 samples\nTraining loss (for one batch) at step 400: 2.168592929840088\nSeen so far: 25664 samples\nTraining loss (for one batch) at step 600: 2.037825107574463\nSeen so far: 38464 samples\nTraining acc over epoch: 0.2502399981021881\nValidation acc: 0.4449000060558319\nStart of epoch 1\nTraining loss (for one batch) at step 0: 1.9728939533233643\nSeen so far: 64 samples\nTraining loss (for one batch) at step 200: 1.9893989562988281\nSeen so far: 12864 samples\nTraining loss (for one batch) at step 400: 1.7468760013580322\nSeen so far: 25664 samples\nTraining loss (for one batch) at step 600: 1.6020689010620117\nSeen so far: 38464 samples\nTraining acc over epoch: 0.5704200267791748\nValidation acc: 0.6780999898910522\nStart of epoch 2\nTraining loss (for one batch) at step 0: 1.476192831993103\nSeen so far: 64 samples\nTraining loss (for one batch) at step 200: 1.558509349822998\nSeen so far: 12864 samples\nTraining loss (for one batch) at step 400: 1.267077922821045\nSeen so far: 25664 samples\nTraining loss (for one batch) at step 600: 1.1969627141952515\nSeen so far: 38464 samples\nTraining acc over epoch: 0.7189000248908997\nValidation acc: 0.7734000086784363\n"
]
],
[
[
"### Low-level handling of extra losses\n\nYou saw in the previous section that it is possible for regularization losses to be added by a layer by calling `self.add_loss(value)` in the `call` method.\n\nIn the general case, you will want to take these losses into account in your custom training loops (unless you've written the model yourself and you already know that it creates no such losses).\n\nRecall this example from the previous section, featuring a layer that creates a regularization loss:\n",
"_____no_output_____"
]
],
[
[
"class ActivityRegularizationLayer(layers.Layer):\n \n def call(self, inputs):\n self.add_loss(1e-2 * tf.reduce_sum(inputs))\n return inputs\n \ninputs = keras.Input(shape=(784,), name='digits')\nx = layers.Dense(64, activation='relu', name='dense_1')(inputs)\n# Insert activity regularization as a layer\nx = ActivityRegularizationLayer()(x)\nx = layers.Dense(64, activation='relu', name='dense_2')(x)\noutputs = layers.Dense(10, activation='softmax', name='predictions')(x)\n\nmodel = keras.Model(inputs=inputs, outputs=outputs)\n",
"_____no_output_____"
]
],
[
[
"When you call a model, like this:\n\n```python\nlogits = model(x_train)\n```\n\nthe losses it creates during the forward pass are added to the `model.losses` attribute:",
"_____no_output_____"
]
],
[
[
"logits = model(x_train[:64])\nprint(model.losses)",
"[<tf.Tensor: id=999790, shape=(), dtype=float32, numpy=6.8533154>]\n"
]
],
[
[
"The tracked losses are first cleared at the start of the model `__call__`, so you will only see the losses created during this one forward pass. For instance, calling the model repeatedly and then querying `losses` only displays the latest losses, created during the last call:",
"_____no_output_____"
]
],
[
[
"logits = model(x_train[:64])\nlogits = model(x_train[64: 128])\nlogits = model(x_train[128: 192])\nprint(model.losses)",
"[<tf.Tensor: id=999851, shape=(), dtype=float32, numpy=6.88884>]\n"
]
],
[
[
"To take these losses into account during training, all you have to do is to modify your training loop to add `sum(model.losses)` to your total loss:",
"_____no_output_____"
]
],
[
[
"optimizer = keras.optimizers.SGD(learning_rate=1e-3)\n\nfor epoch in range(3):\n print('Start of epoch %d' % (epoch,))\n\n for step, (x_batch_train, y_batch_train) in enumerate(train_dataset):\n with tf.GradientTape() as tape:\n logits = model(x_batch_train)\n loss_value = loss_fn(y_batch_train, logits)\n\n # Add extra losses created during this forward pass:\n loss_value += sum(model.losses)\n \n grads = tape.gradient(loss_value, model.trainable_variables)\n optimizer.apply_gradients(zip(grads, model.trainable_variables))\n\n # Log every 200 batches.\n if step % 200 == 0:\n print('Training loss (for one batch) at step %s: %s' % (step, float(loss_value)))\n print('Seen so far: %s samples' % ((step + 1) * 64))",
"Start of epoch 0\nTraining loss (for one batch) at step 0: 9.747203826904297\nSeen so far: 64 samples\nTraining loss (for one batch) at step 200: 2.5395843982696533\nSeen so far: 12864 samples\nTraining loss (for one batch) at step 400: 2.427178144454956\nSeen so far: 25664 samples\nTraining loss (for one batch) at step 600: 2.324587821960449\nSeen so far: 38464 samples\nStart of epoch 1\nTraining loss (for one batch) at step 0: 2.322904586791992\nSeen so far: 64 samples\nTraining loss (for one batch) at step 200: 2.334357976913452\nSeen so far: 12864 samples\nTraining loss (for one batch) at step 400: 2.3377459049224854\nSeen so far: 25664 samples\nTraining loss (for one batch) at step 600: 2.3055613040924072\nSeen so far: 38464 samples\nStart of epoch 2\nTraining loss (for one batch) at step 0: 2.3104405403137207\nSeen so far: 64 samples\nTraining loss (for one batch) at step 200: 2.317152261734009\nSeen so far: 12864 samples\nTraining loss (for one batch) at step 400: 2.319432020187378\nSeen so far: 25664 samples\nTraining loss (for one batch) at step 600: 2.303823471069336\nSeen so far: 38464 samples\n"
]
],
[
[
"That was the last piece of the puzzle! You've reached the end of this guide.\n\nNow you know everything there is to know about using built-in training loops and writing your own from scratch.\n",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
cbf279d33208b9b567ed64506d3bac5ed5021761
| 44,149 |
ipynb
|
Jupyter Notebook
|
CS229_PS/.ipynb_checkpoints/PS4_Q6_ReinforcementLearning-checkpoint.ipynb
|
meichen91/MachineLearning-Snippets
|
f423661e0d54ee4d6808e6ba4a8ee8e267bf5964
|
[
"MIT"
] | 6 |
2019-01-07T12:41:09.000Z
|
2019-11-05T09:37:35.000Z
|
CS229_PS/.ipynb_checkpoints/PS4_Q6_ReinforcementLearning-checkpoint.ipynb
|
meichen91/MachineLearning-Snippets
|
f423661e0d54ee4d6808e6ba4a8ee8e267bf5964
|
[
"MIT"
] | null | null | null |
CS229_PS/.ipynb_checkpoints/PS4_Q6_ReinforcementLearning-checkpoint.ipynb
|
meichen91/MachineLearning-Snippets
|
f423661e0d54ee4d6808e6ba4a8ee8e267bf5964
|
[
"MIT"
] | 5 |
2018-09-26T17:35:32.000Z
|
2022-02-18T10:40:29.000Z
| 96.185185 | 27,992 | 0.811321 |
[
[
[
"# Inverted Pendulum: Reinforcement learning\nMeichen Lu ([email protected]) 26th April 2018\n\nSource: CS229: PS4Q6\n\nStarting code: http://cs229.stanford.edu/ps/ps4/q6/\nReference: https://github.com/zyxue/stanford-cs229/blob/master/Problem-set-4/6-reinforcement-learning-the-inverted-pendulum/control.py",
"_____no_output_____"
]
],
[
[
"from cart_pole import CartPole, Physics\nimport numpy as np\nfrom scipy.signal import lfilter\nimport matplotlib.pyplot as plt\n%matplotlib inline",
"_____no_output_____"
],
[
"# Simulation parameters\npause_time = 0.0001\nmin_trial_length_to_start_display = 100\ndisplay_started = min_trial_length_to_start_display == 0\n\nNUM_STATES = 163\nNUM_ACTIONS = 2\nGAMMA = 0.995\nTOLERANCE = 0.01\nNO_LEARNING_THRESHOLD = 20",
"_____no_output_____"
],
[
"# Time cycle of the simulation\ntime = 0\n\n# These variables perform bookkeeping (how many cycles was the pole\n# balanced for before it fell). Useful for plotting learning curves.\ntime_steps_to_failure = []\nnum_failures = 0\ntime_at_start_of_current_trial = 0\n\n# You should reach convergence well before this\nmax_failures = 500\n\n# Initialize a cart pole\ncart_pole = CartPole(Physics())",
"_____no_output_____"
],
[
"# Starting `state_tuple` is (0, 0, 0, 0)\n# x, x_dot, theta, theta_dot represents the actual continuous state vector\nx, x_dot, theta, theta_dot = 0.0, 0.0, 0.0, 0.0\nstate_tuple = (x, x_dot, theta, theta_dot)\n\n# `state` is the number given to this state, you only need to consider\n# this representation of the state\nstate = cart_pole.get_state(state_tuple)\n# if min_trial_length_to_start_display == 0 or display_started == 1:\n# cart_pole.show_cart(state_tuple, pause_time)",
"_____no_output_____"
],
[
"# Perform all your initializations here:\n# Assume no transitions or rewards have been observed.\n# Initialize the value function array to small random values (0 to 0.10,\n# say).\n# Initialize the transition probabilities uniformly (ie, probability of\n# transitioning for state x to state y using action a is exactly\n# 1/NUM_STATES).\n# Initialize all state rewards to zero.\n\n###### BEGIN YOUR CODE ######\nV_s = np.random.rand(NUM_STATES)\nP_sa = np.ones((NUM_STATES,NUM_ACTIONS, NUM_STATES))/NUM_STATES\nR_s = np.zeros((NUM_STATES))\n# Initialise intermediate variables\nstate_transition_count = np.zeros((NUM_STATES,NUM_ACTIONS, NUM_STATES))\nnew_state_count = np.zeros(NUM_STATES)\nR_new_state = np.zeros(NUM_STATES)\n###### END YOUR CODE ######",
"_____no_output_____"
],
[
"# This is the criterion to end the simulation.\n# You should change it to terminate when the previous\n# 'NO_LEARNING_THRESHOLD' consecutive value function computations all\n# converged within one value function iteration. Intuitively, it seems\n# like there will be little learning after this, so end the simulation\n# here, and say the overall algorithm has converged.\nconsecutive_no_learning_trials = 0\n\nwhile consecutive_no_learning_trials < NO_LEARNING_THRESHOLD:\n\n # Write code to choose action (0 or 1).\n # This action choice algorithm is just for illustration. It may\n # convince you that reinforcement learning is nice for control\n # problems!Replace it with your code to choose an action that is\n # optimal according to the current value function, and the current MDP\n # model.\n ###### BEGIN YOUR CODE ######\n # TODO:\n action = np.argmax(np.sum(P_sa[state]*V_s, axis = 1))\n ###### END YOUR CODE ######\n # Get the next state by simulating the dynamics\n state_tuple = cart_pole.simulate(action, state_tuple)\n\n # Increment simulation time\n time = time + 1\n\n # Get the state number corresponding to new state vector\n new_state = cart_pole.get_state(state_tuple)\n # if display_started == 1:\n # cart_pole.show_cart(state_tuple, pause_time)\n\n # reward function to use - do not change this!\n if new_state == NUM_STATES - 1:\n R = -1\n else:\n R = 0\n\n # Perform model updates here.\n # A transition from `state` to `new_state` has just been made using\n # `action`. The reward observed in `new_state` (note) is `R`.\n # Write code to update your statistics about the MDP i.e. the\n # information you are storing on the transitions and on the rewards\n # observed. Do not change the actual MDP parameters, except when the\n # pole falls (the next if block)!\n\n ###### BEGIN YOUR CODE ######\n # record the number of times `state, action, new_state` occurs\n state_transition_count[state, action, new_state] += 1\n # record the rewards for every `new_state`\n R_new_state[new_state] += R\n # record the number of time `new_state` was reached\n new_state_count[new_state] += 1\n ###### END YOUR CODE ######\n \n # Recompute MDP model whenever pole falls\n # Compute the value function V for the new model\n if new_state == NUM_STATES - 1:\n\n # Update MDP model using the current accumulated statistics about the\n # MDP - transitions and rewards.\n # Make sure you account for the case when a state-action pair has never\n # been tried before, or the state has never been visited before. In that\n # case, you must not change that component (and thus keep it at the\n # initialized uniform distribution).\n\n ###### BEGIN YOUR CODE ######\n # TODO:\n sum_state = np.sum(state_transition_count, axis = 2)\n mask = sum_state > 0\n P_sa[mask] = state_transition_count[mask]/sum_state[mask].reshape(-1, 1)\n \n # Update reward function\n mask = new_state_count>0\n R_s[mask] = R_new_state[mask]/new_state_count[mask]\n \n ###### END YOUR CODE ######\n\n # Perform value iteration using the new estimated model for the MDP.\n # The convergence criterion should be based on `TOLERANCE` as described\n # at the top of the file.\n # If it converges within one iteration, you may want to update your\n # variable that checks when the whole simulation must end.\n\n ###### BEGIN YOUR CODE ######\n iter = 0\n tol = 1\n while tol > TOLERANCE:\n V_old = V_s\n V_s = R_s + GAMMA * np.max(np.sum(P_sa*V_s, axis = 2), axis = 1)\n tol = np.max(np.abs(V_s - V_old))\n iter = iter + 1\n \n if iter == 1:\n consecutive_no_learning_trials += 1\n else:\n # Reset\n consecutive_no_learning_trials = 0\n ###### END YOUR CODE ######\n\n # Do NOT change this code: Controls the simulation, and handles the case\n # when the pole fell and the state must be reinitialized.\n if new_state == NUM_STATES - 1:\n num_failures += 1\n if num_failures >= max_failures:\n break\n print('[INFO] Failure number {}'.format(num_failures))\n time_steps_to_failure.append(time - time_at_start_of_current_trial)\n # time_steps_to_failure[num_failures] = time - time_at_start_of_current_trial\n time_at_start_of_current_trial = time\n\n if time_steps_to_failure[num_failures - 1] > min_trial_length_to_start_display:\n display_started = 1\n\n # Reinitialize state\n # x = 0.0\n x = -1.1 + np.random.uniform() * 2.2\n x_dot, theta, theta_dot = 0.0, 0.0, 0.0\n state_tuple = (x, x_dot, theta, theta_dot)\n state = cart_pole.get_state(state_tuple)\n else:\n state = new_state",
"[INFO] Failure number 1\n[INFO] Failure number 2\n[INFO] Failure number 3\n[INFO] Failure number 4\n[INFO] Failure number 5\n[INFO] Failure number 6\n[INFO] Failure number 7\n[INFO] Failure number 8\n[INFO] Failure number 9\n[INFO] Failure number 10\n[INFO] Failure number 11\n[INFO] Failure number 12\n[INFO] Failure number 13\n[INFO] Failure number 14\n[INFO] Failure number 15\n[INFO] Failure number 16\n[INFO] Failure number 17\n[INFO] Failure number 18\n[INFO] Failure number 19\n[INFO] Failure number 20\n[INFO] Failure number 21\n[INFO] Failure number 22\n[INFO] Failure number 23\n[INFO] Failure number 24\n[INFO] Failure number 25\n[INFO] Failure number 26\n[INFO] Failure number 27\n[INFO] Failure number 28\n[INFO] Failure number 29\n[INFO] Failure number 30\n[INFO] Failure number 31\n[INFO] Failure number 32\n[INFO] Failure number 33\n[INFO] Failure number 34\n[INFO] Failure number 35\n[INFO] Failure number 36\n[INFO] Failure number 37\n[INFO] Failure number 38\n[INFO] Failure number 39\n[INFO] Failure number 40\n[INFO] Failure number 41\n[INFO] Failure number 42\n[INFO] Failure number 43\n[INFO] Failure number 44\n[INFO] Failure number 45\n[INFO] Failure number 46\n[INFO] Failure number 47\n[INFO] Failure number 48\n[INFO] Failure number 49\n[INFO] Failure number 50\n[INFO] Failure number 51\n[INFO] Failure number 52\n[INFO] Failure number 53\n[INFO] Failure number 54\n[INFO] Failure number 55\n[INFO] Failure number 56\n[INFO] Failure number 57\n[INFO] Failure number 58\n[INFO] Failure number 59\n[INFO] Failure number 60\n[INFO] Failure number 61\n[INFO] Failure number 62\n[INFO] Failure number 63\n[INFO] Failure number 64\n[INFO] Failure number 65\n[INFO] Failure number 66\n[INFO] Failure number 67\n[INFO] Failure number 68\n[INFO] Failure number 69\n[INFO] Failure number 70\n[INFO] Failure number 71\n[INFO] Failure number 72\n[INFO] Failure number 73\n[INFO] Failure number 74\n[INFO] Failure number 75\n[INFO] Failure number 76\n[INFO] Failure number 77\n[INFO] Failure number 78\n[INFO] Failure number 79\n[INFO] Failure number 80\n[INFO] Failure number 81\n[INFO] Failure number 82\n[INFO] Failure number 83\n[INFO] Failure number 84\n[INFO] Failure number 85\n[INFO] Failure number 86\n[INFO] Failure number 87\n[INFO] Failure number 88\n[INFO] Failure number 89\n[INFO] Failure number 90\n[INFO] Failure number 91\n[INFO] Failure number 92\n[INFO] Failure number 93\n[INFO] Failure number 94\n[INFO] Failure number 95\n[INFO] Failure number 96\n[INFO] Failure number 97\n[INFO] Failure number 98\n[INFO] Failure number 99\n[INFO] Failure number 100\n[INFO] Failure number 101\n[INFO] Failure number 102\n[INFO] Failure number 103\n[INFO] Failure number 104\n[INFO] Failure number 105\n[INFO] Failure number 106\n[INFO] Failure number 107\n[INFO] Failure number 108\n[INFO] Failure number 109\n[INFO] Failure number 110\n[INFO] Failure number 111\n[INFO] Failure number 112\n[INFO] Failure number 113\n[INFO] Failure number 114\n[INFO] Failure number 115\n[INFO] Failure number 116\n[INFO] Failure number 117\n[INFO] Failure number 118\n[INFO] Failure number 119\n[INFO] Failure number 120\n[INFO] Failure number 121\n[INFO] Failure number 122\n[INFO] Failure number 123\n[INFO] Failure number 124\n[INFO] Failure number 125\n[INFO] Failure number 126\n[INFO] Failure number 127\n[INFO] Failure number 128\n[INFO] Failure number 129\n[INFO] Failure number 130\n[INFO] Failure number 131\n[INFO] Failure number 132\n[INFO] Failure number 133\n[INFO] Failure number 134\n[INFO] Failure number 135\n[INFO] Failure number 136\n"
],
[
"# plot the learning curve (time balanced vs. trial)\nlog_tstf = np.log(np.array(time_steps_to_failure))\nplt.plot(np.arange(len(time_steps_to_failure)), log_tstf, 'k')\nwindow = 30\nw = np.array([1/window for _ in range(window)])\nweights = lfilter(w, 1, log_tstf)\nx = np.arange(window//2, len(log_tstf) - window//2)\nplt.plot(x, weights[window:len(log_tstf)], 'r--')\nplt.xlabel('Num failures')\nplt.ylabel('Num steps to failure')\nplt.show()",
"_____no_output_____"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbf27c6784dcdf0a654906caa340588c55d3352a
| 19,818 |
ipynb
|
Jupyter Notebook
|
lab5/ex1.ipynb
|
evalevanto/qiskit-gss-labs
|
da804f1db38e8b8c83303ba8a05291abe2eb2527
|
[
"Unlicense"
] | null | null | null |
lab5/ex1.ipynb
|
evalevanto/qiskit-gss-labs
|
da804f1db38e8b8c83303ba8a05291abe2eb2527
|
[
"Unlicense"
] | null | null | null |
lab5/ex1.ipynb
|
evalevanto/qiskit-gss-labs
|
da804f1db38e8b8c83303ba8a05291abe2eb2527
|
[
"Unlicense"
] | null | null | null | 27.952045 | 399 | 0.585528 |
[
[
[
"",
"_____no_output_____"
],
[
"# Lab 5: Quantum error correction\n\nYou can do actual insightful science with IBMQ devices and the knowledge you have about quantum error correction. All you need are a few tools from Qiskit.",
"_____no_output_____"
]
],
[
[
"!pip install -U -r grading_tools/requirements.txt\nfrom qiskit import *\nfrom IPython.display import clear_output\nclear_output()",
"_____no_output_____"
]
],
[
[
"## Using a noise model\n\nIn this lab we are going to deal with noisy quantum systems, or at least simulations of them. To deal with this in Qiskit, we need to import some things.",
"_____no_output_____"
]
],
[
[
"from qiskit.providers.aer.noise import NoiseModel\nfrom qiskit.providers.aer.noise.errors import pauli_error, depolarizing_error\nfrom qiskit.providers.aer.noise import thermal_relaxation_error",
"_____no_output_____"
]
],
[
[
"The following function is designed to create a noise model which will be good for what we are doing here. It has two types of noise:\n* Errors on `cx` gates in which an `x`, `y` or `z` is randomly applied to each qubit.\n* Errors in measurement which simulated a thermal process happening over time.",
"_____no_output_____"
]
],
[
[
"def make_noise(p_cx=0,T1T2Tm=(1,1,0)):\n '''\n Returns a noise model specified by the inputs\n - p_cx: probability of depolarizing noise on each\n qubit during a cx\n - T1T2Tm: tuple with (T1,T2,Tm), the T1 and T2 times\n and the measurement time\n '''\n \n noise_model = NoiseModel()\n \n # depolarizing error for cx\n error_cx = depolarizing_error(p_cx, 1)\n error_cx = error_cx.tensor(error_cx)\n noise_model.add_all_qubit_quantum_error(error_cx, [\"cx\"])\n \n # thermal error for measurement\n (T1,T2,Tm) = T1T2Tm\n error_meas = thermal_relaxation_error(T1, T2, Tm)\n noise_model.add_all_qubit_quantum_error(error_meas, \"measure\")\n \n return noise_model",
"_____no_output_____"
]
],
[
[
"Let's check it out on a simple four qubit circuit. One qubit has an `x` applied. Two others has a `cx`. One has nothing. Then all are measured.",
"_____no_output_____"
]
],
[
[
"qc = QuantumCircuit(4)\nqc.x(0)\nqc.cx(1,2)\nqc.measure_all()\n\nqc.draw(output='mpl')",
"_____no_output_____"
]
],
[
[
"This is a simple circuit with a simple output, as we'll see when we run it.",
"_____no_output_____"
]
],
[
[
"execute( qc, Aer.get_backend('qasm_simulator'), shots=8192).result().get_counts()",
"_____no_output_____"
]
],
[
[
"Now let's run it with noise on the `cx` gates only.",
"_____no_output_____"
]
],
[
[
"noise_model = make_noise(p_cx=0.1)\n\nexecute( qc, Aer.get_backend('qasm_simulator'), noise_model=noise_model, shots=8192).result().get_counts()",
"_____no_output_____"
]
],
[
[
"The measurement noise depends on three numbers: $T_1$, $T_2$ and $T_m$. The first two describe the timescale for certain noise processes. The last describes how long measurements take. For simplicity we'll set $T_1=T_2=1$ and vary $T_m$. For $T_m=0$, the measurement is too fast to see any noise. The longer it takes, the more noise we'll see.",
"_____no_output_____"
]
],
[
[
"for Tm in (0.01,0.1,1,10):\n\n noise_model = make_noise(p_cx=0, T1T2Tm=(1,1,Tm))\n\n counts = execute( qc, Aer.get_backend('qasm_simulator'), noise_model=noise_model, shots=8192).result().get_counts()\n print('Tm =',Tm,', counts =',counts)",
"_____no_output_____"
]
],
[
[
"The most notable effect of this noise is that it causes `1` values to relax down to `0`.",
"_____no_output_____"
],
[
"# Running repetition codes\n\nQiskit has tools to make it easy to set up, run and analyze repetition codes.",
"_____no_output_____"
]
],
[
[
"from qiskit.ignis.verification.topological_codes import RepetitionCode\nfrom qiskit.ignis.verification.topological_codes import GraphDecoder\nfrom qiskit.ignis.verification.topological_codes import lookuptable_decoding, postselection_decoding",
"_____no_output_____"
]
],
[
[
"Here's one with four repetitions and a single measurement round.",
"_____no_output_____"
]
],
[
[
"d = 4\nT = 1\ncode = RepetitionCode(d,T)",
"_____no_output_____"
]
],
[
[
"The repetition code object contains a couple of circuits: for encoded logical values of `0` and `1`.",
"_____no_output_____"
]
],
[
[
"code.circuit",
"_____no_output_____"
]
],
[
[
"Here's the one for `0`.",
"_____no_output_____"
]
],
[
[
"code.circuit['0'].draw(output='text')",
"_____no_output_____"
]
],
[
[
"And for `1`.",
"_____no_output_____"
]
],
[
[
"code.circuit['1'].draw(output='text')",
"_____no_output_____"
]
],
[
[
"We can run both circuits at once by first converting them into a list.",
"_____no_output_____"
]
],
[
[
"circuits = code.get_circuit_list()\n\njob = execute( circuits, Aer.get_backend('qasm_simulator'), noise_model=noise_model, shots=8192)",
"_____no_output_____"
]
],
[
[
"Once they've run, we can extract the results and convert them into a form that allows us to more easily look at syndrome changes.",
"_____no_output_____"
]
],
[
[
"raw_results = {}\nfor log in ['0','1']:\n raw_results[log] = job.result().get_counts(log)\n\nresults = code.process_results( raw_results )",
"_____no_output_____"
]
],
[
[
"It's easiest to just package this up into a function.",
"_____no_output_____"
]
],
[
[
"def get_results(code, noise_model, shots=8192):\n \n circuits = code.get_circuit_list()\n\n job = execute( circuits, Aer.get_backend('qasm_simulator'), noise_model=noise_model, shots=shots)\n\n raw_results = {}\n for log in ['0','1']:\n raw_results[log] = job.result().get_counts(log)\n\n results = code.process_results( raw_results )\n \n return results",
"_____no_output_____"
]
],
[
[
"First let's look at an example without any noise, to keep things simple.",
"_____no_output_____"
]
],
[
[
"noise_model = make_noise() # noise model with no noise\n\nresults = get_results(code, noise_model)\n\nresults",
"_____no_output_____"
]
],
[
[
"Here's an example with some `cx` noise.",
"_____no_output_____"
]
],
[
[
"noise_model = make_noise(p_cx=0.01)\n\nresults = get_results(code, noise_model)\n\nfor log in results:\n print('\\nMost common results for a stored',log)\n for output in results[log]:\n if results[log][output]>100:\n print(output,'ocurred for',results[log][output],'samples.')",
"_____no_output_____"
]
],
[
[
"The main thing we need to know is the probability of a logical error. By setting up and using a decoder, we can find out!",
"_____no_output_____"
]
],
[
[
"decoder = GraphDecoder(code)\n\ndecoder.get_logical_prob(results)",
"_____no_output_____"
]
],
[
[
"By calculating these value for different sizes of code and noise models, we can learn more about how the noise will affect large circuits. This is important for error correction, but also for the applications that we'll try to run before error correction is possible.\n\nEven more importantly, running these codes on real devices allows us to see the effects of real noise. Small-scale quantum error correction experiments like these will allow us to study the devices we have access to, understand what they do and why they do it, and test their abilities.\n\nThis is the most important exercise that you can try: doing real and insightful experiments on cutting-edge quantum hardware. It's the kind of thing that professional researchers do and write papers about. I know this because I'm one of those researchers.\n\nSee the following examples:\n* [\"A repetition code of 15 qubits\", James R. Wootton and Daniel Loss, Phys. Rev. A 97, 052313 (2018)](https://arxiv.org/abs/1709.00990)\n* [\"Benchmarking near-term devices with quantum error correction\", James R. Wootton, Quantum Science and Technology (2020)](https://arxiv.org/abs/2004.11037)\n\nAs well as the relevant chapter of the Qiskit textbook: [5.1 Introduction to Quantum Error Correction using Repetition Codes](https://qiskit.org/textbook/ch-quantum-hardware/error-correction-repetition-code.html).\n\nBy running repetition codes on the IBM quantum devices available to you, looking at the results and figuring out why they look like they do, you could soon know things about them that no-one else does!",
"_____no_output_____"
],
[
"## Transpiling for real devices\n\nThe first step toward using a real quantum device is to load your IBMQ account and set up the provider.",
"_____no_output_____"
]
],
[
[
"# IBMQ.save_account(\"d25b4b26f7725b7768cc6394319cf4d7528c7d037bd8b2752f51b9be9da98ff1cff30053a2c2ef65bef631496cd60c7b15b658be2953a7eb14a13fe71e8eafeb\")\nIBMQ.load_account()\n\nprovider = IBMQ.get_provider(hub='ibm-q')",
"_____no_output_____"
]
],
[
[
"Now you can set up a backend object for your device of choice. We'll go for the biggest device on offer: Melbourne.",
"_____no_output_____"
]
],
[
[
"backend = provider.get_backend('ibmq_16_melbourne')",
"_____no_output_____"
]
],
[
[
"Using the Jupyter tools, we can take a closer look.",
"_____no_output_____"
]
],
[
[
"import qiskit.tools.jupyter\n%matplotlib inline\n\nbackend",
"_____no_output_____"
]
],
[
[
"This has enough qubits to run a $d=8$ repetition code. Let's set this up and get the circuits to run.",
"_____no_output_____"
]
],
[
[
"d = 8\ncode = RepetitionCode(8,1)\n\nraw_circuits = code.get_circuit_list()",
"_____no_output_____"
]
],
[
[
"Rather than show such a big circuit, let's just look at how many of each type of gate there are. For example, repetition codes should have $2(d-1)$ `cx` gates in, which means 14 in this case.",
"_____no_output_____"
]
],
[
[
"raw_circuits[1].count_ops()",
"_____no_output_____"
]
],
[
[
"Before running on a real device we need to transpile. This is the process of turning the circuits into ones that the device can actually run. It is usually done automatically before running, but we can also do it ourself using the code below.",
"_____no_output_____"
]
],
[
[
"circuits = []\nfor qc in raw_circuits:\n circuits.append( transpile(qc, backend=backend) )",
"_____no_output_____"
]
],
[
[
"Let's check what this process did to the gates in the circuit.",
"_____no_output_____"
]
],
[
[
"circuits[1].count_ops()",
"_____no_output_____"
]
],
[
[
"Note that this has `u3` gates (which the circuit previously didn't) and the `x` gates have disappeared. The solution to this is simple. The `x` gates have just been described as specific forms of `u3` gates, which is the way that the hardware understands single qubit operations.\n\nMore concerning is what has happened to the `cx` gates. There are now 74!.\n\nThis is due to connectivity. If you ask for a combination of `cx` gates that cannot be directly implemented, the transpiler will do some fancy tricks to make a circuit which is effectively the same as the one you want. This comes at the cost of inserting `cx` gates. For more information, see [2.4 More Circuit-Identities](https://qiskit.org/textbook/ch-gates/more-circuit-identities.html).\n\nHowever, here our circuit *is* something that can be directly implemented. The transpiler just didn't realize (and figuring it out is a hard problem). We can solve the problem by telling the transpiler exactly which qubits on the device should be used as the qubits in our code.\n\nThis is done by setting up an `initial_layout` as follows.",
"_____no_output_____"
]
],
[
[
"def get_initial_layout(code,line):\n initial_layout = {}\n for j in range(code.d):\n initial_layout[code.code_qubit[j]] = line[2*j]\n for j in range(code.d-1):\n initial_layout[code.link_qubit[j]] = line[2*j+1]\n return initial_layout\n \nline = [6,5,4,3,2,1,0,14,13,12,11,10,9,8,7]\n \ninitial_layout = get_initial_layout(code,line)\n\ninitial_layout",
"_____no_output_____"
]
],
[
[
"With this, let's try transpilation again.",
"_____no_output_____"
]
],
[
[
"circuits = []\nfor qc in raw_circuits:\n circuits.append( transpile(qc, backend=backend, initial_layout=initial_layout) )\n \ncircuits[1].count_ops()",
"_____no_output_____"
]
],
[
[
"Perfect!\n\nNow try for yourself on one of the devices that we've now retired: Tokyo.",
"_____no_output_____"
]
],
[
[
"from qiskit.test.mock import FakeTokyo\n\nbackend = FakeTokyo()\n\nbackend",
"_____no_output_____"
]
],
[
[
"The largest repetition code this can handle is one with $d=10$.",
"_____no_output_____"
]
],
[
[
"d = 10\ncode = RepetitionCode(d,1)\nraw_circuits = code.get_circuit_list()\n\nraw_circuits[1].count_ops()",
"_____no_output_____"
]
],
[
[
"For this we need to find a line of 19 qubits across the coupling map.",
"_____no_output_____"
]
],
[
[
"line = [0,1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19]\ninitial_layout = get_initial_layout(code,line)\n\ncircuits = []\nfor qc in raw_circuits:\n circuits.append(transpile(qc, backend=backend, initial_layout=initial_layout) )\n \ncircuits[1].count_ops()",
"_____no_output_____"
]
],
[
[
"Clearly, the line chosen in the cell above was not a good example. Find a line such that the transpiled circuit `circuits[1]` has exactly 18 `cx` gates.",
"_____no_output_____"
]
],
[
[
"line = None\n# define line variable so the transpiled circuit has exactly 18 CNOTs.\n### WRITE YOUR CODE BETWEEN THESE LINES - START\nline = 0,1,2,3,9,4,8,7,6,5,10,15,16,17,11,12,13,18,14,19\n### WRITE YOUR CODE BETWEEN THESE LINES - END\n\ninitial_layout = get_initial_layout(code,line)\n\ncircuits = []\nfor qc in raw_circuits:\n circuits.append(transpile(qc, backend=backend, initial_layout=initial_layout) )\n \ncircuits[1].count_ops()",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cbf299a58d460bf5a560055f34245913573d2031
| 6,365 |
ipynb
|
Jupyter Notebook
|
JupyterNotebooks/Labs/Lab 3.ipynb
|
CJP-12/CMPT-120L-910-20F
|
ec74b248ed1957ceabd3f96e176e0e244220a9cd
|
[
"MIT"
] | null | null | null |
JupyterNotebooks/Labs/Lab 3.ipynb
|
CJP-12/CMPT-120L-910-20F
|
ec74b248ed1957ceabd3f96e176e0e244220a9cd
|
[
"MIT"
] | null | null | null |
JupyterNotebooks/Labs/Lab 3.ipynb
|
CJP-12/CMPT-120L-910-20F
|
ec74b248ed1957ceabd3f96e176e0e244220a9cd
|
[
"MIT"
] | null | null | null | 25.873984 | 186 | 0.536528 |
[
[
[
"# Lab Three\n---\n\nFor this lab we're going to be making and using a bunch of functions. \n\nOur Goals are:\n- Searching our Documentation\n- Using built in functions\n- Making our own functions\n- Combining functions\n- Structuring solutions",
"_____no_output_____"
]
],
[
[
"# For the following built in functions we didn't touch on them in class. I want you to look for them in the python documentation and implement them.",
"_____no_output_____"
],
[
"# I want you to find a built in function to SWAP CASE on a string. Print it.\n\n# For example the string \"HeY thERe HowS iT GoING\" turns into \"hEy THerE hOWs It gOing\"\nsample_string = \"HeY thERe HowS iT GoING\"\nprint(sample_string.swapcase())",
"hEy THerE hOWs It gOing\n"
],
[
"# I want you to find a built in function to CENTER a string and pad the sides with 4 dashes(-) a side. Print it.\n\n# For example the string \"Hey There\" becomes \"----Hey There----\"\n\nsample_string = \"Hey There\"\nprint(sample_string.center(17,\"-\"))",
"----Hey There----\n"
],
[
"# I want you to find a built in function to PARTITION a string. Print it.\n\n# For example the string \"abcdefg.hijklmnop\" would come out to be [\"abcdefg\",\".\",\"hijklmnop\"]\n\nsample_string = \"abcdefg.hijklmnop\"\nprint(sample_string.partition(\".\"))",
"('abcdefg', '.', 'hijklmnop')\n"
],
[
"# I want you to write a function that will take in a number and raise it to the power given. \n\n# For example if given the numbers 2 and 3. The math that the function should do is 2^3 and should print out or return 8. Print the output.\ndef power(number,exponent) -> int:\n return number ** exponent\n\nexample = power(2,3)\nprint(example)",
"8\n"
],
[
"# I want you to write a function that will take in a list and see how many times a given number is in the list. \n\n# For example if the array given is [2,3,5,2,3,6,7,8,2] and the number given is 2 the function should print out or return 3. Print the output.\narray = [2,3,4,2,3,6,7,8,2]\ndef multiplicity(array,target):\n count = 0\n for number in array:\n if number == target:\n count += 1\n return count\nexample = multiplicity(array, 2)\nprint(example)",
"3\n"
],
[
"# Use the functions given to create a slope function. The function should be named slope and have 4 parameters.\n\n# If you don't remember the slope formula is (y2 - y1) / (x2 - x1) If this doesn't make sense look up `Slope Formula` on google.\n\ndef division(x, y):\n return x / y\n\ndef subtraction(x, y):\n return x - y\n\ndef slope(x1, x2, y1, y2):\n return division(subtraction(y2,y1), subtraction(x2,x1))\n\nexample = slope(1, 3, 2, 6)\nprint(example)",
"2.0\n"
],
[
"# Use the functions given to create a distance function. The function should be named function and have 4 parameters.\n\n# HINT: You'll need a built in function here too. You'll also be able to use functions written earlier in the notebook as long as you've run those cells.\n\n# If you don't remember the distance formula it is the square root of the following ((x2 - x1)^2 + (y2 - y1)^2). If this doesn't make sense look up `Distance Formula` on google.\n\nimport math\n\ndef addition(x, y):\n return x + y\n\ndef distance(x1, x2, y1, y2):\n x_side = power(subtraction(x2, x1), 2)\n y_side = power(subtraction(y2, y1), 2)\n combined_sides = addition(x_side, y_side)\n\n return math.sqrt(combined_sides)\n\nprint(distance(1, 3, 2, 6))",
"4.47213595499958\n"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbf2a4989824c02f18d73c9415b2ef66c2aea751
| 209,284 |
ipynb
|
Jupyter Notebook
|
07 - Inferential Stats with Python/notebooks/03_VariableRelationshipTests(CorrelationTest).ipynb
|
hossainlab/PY4R
|
cd905ce424de67b4b9fea2b371f286db9d147cb6
|
[
"CC0-1.0"
] | 2 |
2021-09-06T14:05:27.000Z
|
2021-09-11T14:42:25.000Z
|
07 - Inferential Stats with Python/notebooks/03_VariableRelationshipTests(CorrelationTest).ipynb
|
hossainlab/PY4R
|
cd905ce424de67b4b9fea2b371f286db9d147cb6
|
[
"CC0-1.0"
] | null | null | null |
07 - Inferential Stats with Python/notebooks/03_VariableRelationshipTests(CorrelationTest).ipynb
|
hossainlab/PY4R
|
cd905ce424de67b4b9fea2b371f286db9d147cb6
|
[
"CC0-1.0"
] | 3 |
2021-09-06T13:05:27.000Z
|
2021-09-11T14:42:27.000Z
| 205.381747 | 54,092 | 0.898621 |
[
[
[
"# Variable Relationship Tests (correlation)\n- Pearson’s Correlation Coefficient\n- Spearman’s Rank Correlation\n- Kendall’s Rank Correlation\n- Chi-Squared Test",
"_____no_output_____"
],
[
"## Correlation Test\nCorrelation Measures whether greater values of one variable correspond to greater values in the other. Scaled to always lie between +1 and −1\n\n- Correlation is Positive when the values increase together.\n- Correlation is Negative when one value decreases as the other increases.\n- A correlation is assumed to be linear.\n- 1 is a perfect positive correlation\n- 0 is no correlation (the values don’t seem linked at all)\n- -1 is a perfect negative correlation",
"_____no_output_____"
],
[
"## Correlation Methods\n- **Pearson's Correlation Test:** assumes the data is normally distributed and measures linear correlation.\n- **Spearman's Correlation Test:** does not assume normality and measures non-linear correlation.\n- **Kendall's Correlation Test:** similarly does not assume normality and measures non-linear correlation, but it less commonly used.\n",
"_____no_output_____"
],
[
"## Difference Between Pearson's and Spearman's \nPearson's Test | Spearman's Test\n---------------|----------------\nParamentric Correlation | Non-parametric \nLinear relationship | Non-linear relationship\nContinuous variables | continuous or ordinal variables\nPropotional change | Change not at constant rate",
"_____no_output_____"
]
],
[
[
"import statsmodels.api as sm\nimport matplotlib.pyplot as plt\nimport pandas as pd\nimport numpy as np\nimport seaborn as sns \nsns.set(font_scale=2, palette= \"viridis\")\nfrom sklearn.preprocessing import scale\nimport researchpy as rp\nfrom scipy import stats",
"_____no_output_____"
],
[
"data = pd.read_csv('../data/pulse_data.csv')\ndata.head() ",
"_____no_output_____"
]
],
[
[
"## Pearson’s Correlation Coefficient\nTests whether two samples have a linear relationship.\n\n### Assumptions\n\n- Observations in each sample are independent and identically distributed (iid).\n- Observations in each sample are normally distributed.\n- Observations in each sample have the same variance.\n\n### Interpretation\n- H0: There is a relationship between two variables \n- Ha: There is no relationship between two variables \n\n\n__Question: Is there any relationship between height and weight?__",
"_____no_output_____"
]
],
[
[
"data.Height.corr(data.Weight)",
"_____no_output_____"
],
[
"data.Height.corr(data.Weight, method=\"pearson\")",
"_____no_output_____"
],
[
"data.Height.corr(data.Weight, method=\"spearman\")",
"_____no_output_____"
],
[
"plt.figure(figsize=(10,8))\nsns.scatterplot(data=data, x='Height', y=\"Weight\")\nplt.show()",
"_____no_output_____"
],
[
"plt.figure(figsize=(10,8))\nsns.regplot(data=data, x='Height', y=\"Weight\")\nplt.show()",
"_____no_output_____"
],
[
"stat, p_value = stats.shapiro(data['Height'])\nprint(f'statistic = {stat}, p-value = {p_value}')\n\nalpha = 0.05 \nif p_value > alpha: \n print(\"The sample has normal distribution(Fail to reject the null hypothesis, the result is not significant)\")\nelse: \n print(\"The sample does not have a normal distribution(Reject the null hypothesis, the result is significant)\")",
"statistic = 0.9885410666465759, p-value = 0.4920051395893097\nThe sample has normal distribution(Fail to reject the null hypothesis, the result is not significant)\n"
],
[
"stat, p_value = stats.shapiro(data['Weight'])\nprint(f'statistic = {stat}, p-value = {p_value}')\n\nalpha = 0.05 \nif p_value > alpha: \n print(\"The sample has normal distribution(Fail to reject the null hypothesis, the result is not significant)\")\nelse: \n print(\"The sample does not have a normal distribution(Reject the null hypothesis, the result is significant)\")",
"statistic = 0.9418081641197205, p-value = 0.00013631694309879094\nThe sample does not have a normal distribution(Reject the null hypothesis, the result is significant)\n"
],
[
"# Checking for normality by Q-Q plot graph\nplt.figure(figsize=(12, 8))\nstats.probplot(data['Height'], plot=plt, dist='norm')\nplt.show()",
"_____no_output_____"
],
[
"# Checking for normality by Q-Q plot graph\nplt.figure(figsize=(12, 8))\nstats.probplot(data['Weight'], plot=plt, dist='norm')\nplt.show()",
"_____no_output_____"
],
[
"stats.levene(data['Height'], data['Weight'])",
"_____no_output_____"
],
[
"stat, p, = stats.levene(data['Height'], data['Weight'])\nprint(f'stat={stat}, p-value={p}')\nalpha = 0.05 \nif p > alpha:\n print('The variances are equal between two variables(reject H0, not significant)')\nelse:\n print('The variances are not equal between two variables(reject H0, significant)')",
"stat=135.08608012666895, p-value=1.5839821691245638e-24\nThe variances are not equal between two variables(reject H0, significant)\n"
],
[
"stats.pearsonr(data['Height'], data['Weight'])",
"_____no_output_____"
],
[
"stat, p, = stats.pearsonr(data['Height'], data['Weight'])\nprint(f'stat={stat}, p-value={p}')\nalpha = 0.05 \nif p > alpha:\n print('There is a relationship between two variables(fail to reject H0, not significant)')\nelse:\n print('There is a no relationship between two variables(reject H0, significant)')",
"stat=0.7413041988574969, p-value=4.5627070950916043e-20\nThere is a no relationship between two variables(reject H0, significant)\n"
]
],
[
[
"## Spearman’s Rank Correlation Test \nTests whether two samples have a monotonic relationship.\n\n### Assumptions\n- Observations in each sample are independent and identically distributed (iid).\n- Observations in each sample can be ranked.\n\n\n\n### Interpretation\n- **H0 hypothesis:** There is is relationship between variable 1 and variable 2\n- **H1 hypothesis:** There is no relationship between variable 1 and variable 2\n",
"_____no_output_____"
]
],
[
[
"stats.spearmanr(data['Height'], data['Weight'])",
"_____no_output_____"
],
[
"stat, p = stats.spearmanr(data['Height'], data['Weight'])\nprint(f'stat={stat}, p-value={p}')\n\nalpha = 0.05 \nif p > alpha:\n print('There is a relationship between two variables(fail to reject H0, not significant)')\nelse:\n print('There is a no relationship between two variables(reject H0, significant)')",
"stat=0.7535418080977205, p-value=4.989916471360893e-21\nThere is a no relationship between two variables(reject H0, significant)\n"
]
],
[
[
"## Kendall’s Rank Correlation Test \n\n### Assumptions\n\n- Observations in each sample are independent and identically distributed (iid).\n- Observations in each sample can be ranked.\n\n### Interpretation\n\n\n### Interpretation\n- **H0 hypothesis:** There is a relationship between variable 1 and variable 2\n- **H1 hypothesis:** There is no relationship between variable 1 and variable 2",
"_____no_output_____"
]
],
[
[
"stats.spearmanr(data['Height'], data['Weight'])",
"_____no_output_____"
],
[
"stat, p, = stats.kendalltau(data['Height'], data['Weight'])\nprint(f'stat={stat}, p-value={p}')\nalpha = 0.05 \nif p > alpha:\n print('Accept null hypothesis; there is a relationship between Height and Weight(fail to reject H0, not significant)')\nelse:\n print('Reject the null hypothesis; there is no relationship between Height and Weight (reject H0, significant)')",
"stat=0.57668142211864, p-value=7.200475725682383e-18\nReject the null hypothesis; there is no relationship between Height and Weight (reject H0, significant)\n"
]
],
[
[
"## Chi-Squared Test\n- The Chi-square test of independence tests if there is a significant relationship between two categorical variables\n- The test is comparing the observed observations to the expected observations.\n- The data is usually displayed in a cross-tabulation format with each row representing a category for one variable and each column representing a category for another variable.\n- Chi-square test of independence is an omnibus test. Meaning it tests the data as a whole. This means that one will not be able to tell which levels (categories) of the variables are responsible for the relationship if the Chi-square table is larger than 2×2\n- If the test is larger than 2×2, it requires post hoc testing. If this doesn’t make much sense right now, don’t worry. Further explanation will be provided when we start working with the data.\n\n### Assumptions\n- It should be two categorical variables(e.g; Gender)\n- Each variables should have at leats two groups(e.g; Gender = Female or Male)\n- There should be independence of observations(between and within subjects)\n- Large sample size \n - The expected frequencies should be at least 1 for each cell. \n - The expected frequencies for the majority(80%) of the cells should be at least 5.\n \nIf the sample size is small, we have to use **Fisher's Exact Test**\n\n**Fisher's Exact Test** is similar to Chi-squared test, but it is used for small-sized samples.\n\n## Interpretation \n- The H0 (Null Hypothesis): There is a relationship between variable one and variable two.\n- The Ha (Alternative Hypothesis): There is no relationship between variable 1 and variable 2.",
"_____no_output_____"
],
[
"### Contingency Table\nContingency table is a table with at least two rows and two columns(2x2) and its use to present categorical data in terms of frequency counts.",
"_____no_output_____"
]
],
[
[
"data = pd.read_csv('../data/KosteckiDillon.csv', usecols=['id', 'time', 'dos', 'hatype', 'age', 'airq',\n 'medication', 'headache', 'sex'])\ndata.head() ",
"_____no_output_____"
],
[
"table = pd.crosstab(data['sex'], data['headache'])\ntable",
"_____no_output_____"
],
[
"stats.chi2_contingency(table)",
"_____no_output_____"
],
[
"stat, p, dof, expected = stats.chi2_contingency(table)\nprint(f'stat={stat}, p-value={p}') \nalpha = 0.05 \nif p > alpha:\n print('There is a relationship between sex and headache(fail to reject Ho, not significant)')\nelse:\n print('There is no relationship between sex and headache.(reject H0, significant)')",
"stat=0.042688484136431136, p-value=0.8363129528687885\nThere is a relationship between sex andheadache(fail to reject Ho, not significant)\n"
]
],
[
[
"## Fisher’s Test",
"_____no_output_____"
]
],
[
[
"stat, p, = stats.fisher_exact(table)\nprint(f'stat={stat}, p-value={p}') \nif p > 0.05:\n print('Probably independent')\nelse:\n print('Probably dependent')",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
cbf2ab920e77c66ea53ee6764e9c2e49579ba8d5
| 94,787 |
ipynb
|
Jupyter Notebook
|
Pandas_profiling.ipynb
|
wlazlod/CalcRisk
|
2983a5a5fcbd80f47bf9a67b101aaabb66437c46
|
[
"MIT"
] | null | null | null |
Pandas_profiling.ipynb
|
wlazlod/CalcRisk
|
2983a5a5fcbd80f47bf9a67b101aaabb66437c46
|
[
"MIT"
] | null | null | null |
Pandas_profiling.ipynb
|
wlazlod/CalcRisk
|
2983a5a5fcbd80f47bf9a67b101aaabb66437c46
|
[
"MIT"
] | null | null | null | 38.112988 | 231 | 0.391045 |
[
[
[
"<a href=\"https://colab.research.google.com/github/wlazlod/CalcRisk/blob/master/Pandas_profiling.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"!pip install --upgrade --force-reinstall --quiet pandas_profiling",
"\u001b[31mERROR: tensorflow 2.3.0 has requirement numpy<1.19.0,>=1.16.0, but you'll have numpy 1.19.1 which is incompatible.\u001b[0m\n\u001b[31mERROR: tensorflow 2.3.0 has requirement scipy==1.4.1, but you'll have scipy 1.5.2 which is incompatible.\u001b[0m\n\u001b[31mERROR: kaggle 1.5.6 has requirement urllib3<1.25,>=1.21.1, but you'll have urllib3 1.25.10 which is incompatible.\u001b[0m\n\u001b[31mERROR: jupyter-console 5.2.0 has requirement prompt-toolkit<2.0.0,>=1.0.0, but you'll have prompt-toolkit 3.0.5 which is incompatible.\u001b[0m\n\u001b[31mERROR: google-colab 1.0.0 has requirement ipykernel~=4.10, but you'll have ipykernel 5.3.4 which is incompatible.\u001b[0m\n\u001b[31mERROR: google-colab 1.0.0 has requirement ipython~=5.5.0, but you'll have ipython 7.16.1 which is incompatible.\u001b[0m\n\u001b[31mERROR: google-colab 1.0.0 has requirement notebook~=5.3.0; python_version >= \"3.0\", but you'll have notebook 6.1.1 which is incompatible.\u001b[0m\n\u001b[31mERROR: google-colab 1.0.0 has requirement pandas~=1.0.0; python_version >= \"3.0\", but you'll have pandas 1.1.0 which is incompatible.\u001b[0m\n\u001b[31mERROR: google-colab 1.0.0 has requirement requests~=2.23.0, but you'll have requests 2.24.0 which is incompatible.\u001b[0m\n\u001b[31mERROR: google-colab 1.0.0 has requirement tornado~=5.1.0; python_version >= \"3.0\", but you'll have tornado 6.0.4 which is incompatible.\u001b[0m\n\u001b[31mERROR: datascience 0.10.6 has requirement folium==0.2.1, but you'll have folium 0.8.3 which is incompatible.\u001b[0m\n\u001b[31mERROR: basketball-reference-web-scraper 4.9.4 has requirement certifi==2018.10.15, but you'll have certifi 2020.6.20 which is incompatible.\u001b[0m\n\u001b[31mERROR: basketball-reference-web-scraper 4.9.4 has requirement idna==2.7, but you'll have idna 2.10 which is incompatible.\u001b[0m\n\u001b[31mERROR: basketball-reference-web-scraper 4.9.4 has requirement pytz==2018.6, but you'll have pytz 2020.1 which is incompatible.\u001b[0m\n\u001b[31mERROR: basketball-reference-web-scraper 4.9.4 has requirement requests==2.20.0, but you'll have requests 2.24.0 which is incompatible.\u001b[0m\n\u001b[31mERROR: basketball-reference-web-scraper 4.9.4 has requirement urllib3==1.24.3, but you'll have urllib3 1.25.10 which is incompatible.\u001b[0m\n\u001b[31mERROR: albumentations 0.1.12 has requirement imgaug<0.2.7,>=0.2.5, but you'll have imgaug 0.2.9 which is incompatible.\u001b[0m\n"
],
[
"!pip install --quiet basketball_reference_web_scraper",
"\u001b[K |████████████████████████████████| 61kB 2.5MB/s \n\u001b[K |████████████████████████████████| 5.5MB 6.7MB/s \n\u001b[K |████████████████████████████████| 153kB 26.5MB/s \n\u001b[K |████████████████████████████████| 61kB 5.4MB/s \n\u001b[K |████████████████████████████████| 122kB 29.6MB/s \n\u001b[K |████████████████████████████████| 512kB 44.2MB/s \n\u001b[31mERROR: tensorboard 2.3.0 has requirement requests<3,>=2.21.0, but you'll have requests 2.20.0 which is incompatible.\u001b[0m\n\u001b[31mERROR: pandas-profiling 2.8.0 has requirement requests>=2.23.0, but you'll have requests 2.20.0 which is incompatible.\u001b[0m\n\u001b[31mERROR: google-colab 1.0.0 has requirement ipykernel~=4.10, but you'll have ipykernel 5.3.4 which is incompatible.\u001b[0m\n\u001b[31mERROR: google-colab 1.0.0 has requirement ipython~=5.5.0, but you'll have ipython 7.16.1 which is incompatible.\u001b[0m\n\u001b[31mERROR: google-colab 1.0.0 has requirement notebook~=5.3.0; python_version >= \"3.0\", but you'll have notebook 6.1.1 which is incompatible.\u001b[0m\n\u001b[31mERROR: google-colab 1.0.0 has requirement pandas~=1.0.0; python_version >= \"3.0\", but you'll have pandas 1.1.0 which is incompatible.\u001b[0m\n\u001b[31mERROR: google-colab 1.0.0 has requirement requests~=2.23.0, but you'll have requests 2.20.0 which is incompatible.\u001b[0m\n\u001b[31mERROR: google-colab 1.0.0 has requirement tornado~=5.1.0; python_version >= \"3.0\", but you'll have tornado 6.0.4 which is incompatible.\u001b[0m\n\u001b[31mERROR: datascience 0.10.6 has requirement folium==0.2.1, but you'll have folium 0.8.3 which is incompatible.\u001b[0m\n\u001b[?25h"
],
[
"import numpy as np\nimport pandas as pd\nfrom pandas_profiling import ProfileReport",
"/usr/local/lib/python3.6/dist-packages/statsmodels/tools/_testing.py:19: FutureWarning: pandas.util.testing is deprecated. Use the functions in the public API at pandas.testing instead.\n import pandas.util.testing as tm\n"
]
],
[
[
"# Pobranie danych",
"_____no_output_____"
]
],
[
[
"from basketball_reference_web_scraper import client\n\nplayers = pd.DataFrame(client.players_season_totals(season_end_year=2020))\nplayers['team'] = players['team'].astype(str).str[5:].str.replace(\"_\", \" \")\nplayers['positions'] = players['positions'].astype(str).str.extract(\"'([^']*)'\")\n\nplayers",
"_____no_output_____"
]
],
[
[
"# Dodanie średnich i średnich per 36",
"_____no_output_____"
]
],
[
[
"players.columns",
"_____no_output_____"
],
[
"columns = ['made_field_goals', 'attempted_field_goals', 'made_three_point_field_goals',\n 'attempted_three_point_field_goals', 'made_free_throws',\n 'attempted_free_throws', 'offensive_rebounds', 'defensive_rebounds',\n 'assists', 'steals', 'blocks', 'turnovers', 'personal_fouls', 'points']",
"_____no_output_____"
]
],
[
[
"2 pętle dodające statystyki na mecz i na 36 minut",
"_____no_output_____"
]
],
[
[
"for stat in columns:\n players[stat+\"_per_game\"] = players[stat] / players[\"games_played\"]\n\nfor stat in columns:\n players[stat+\"_per_36\"] = players[stat] / players[\"minutes_played\"] * 36.",
"_____no_output_____"
],
[
"players",
"_____no_output_____"
],
[
"profile = ProfileReport(players, title='NBA Players Profiling Report')",
"_____no_output_____"
],
[
"profile.to_notebook_iframe()",
"_____no_output_____"
],
[
"profile.to_file(\"nba_players.html\")",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbf2c3816a2d62b111bc6e0c6f1ebee19035981a
| 13,406 |
ipynb
|
Jupyter Notebook
|
BIG_GAME - Hackerearth/6. VotingClassifier.ipynb
|
phileinSophos/ML-DL_Problems
|
033fc8a0086883fbe6748f2bf4725de7e8376e4b
|
[
"MIT"
] | null | null | null |
BIG_GAME - Hackerearth/6. VotingClassifier.ipynb
|
phileinSophos/ML-DL_Problems
|
033fc8a0086883fbe6748f2bf4725de7e8376e4b
|
[
"MIT"
] | null | null | null |
BIG_GAME - Hackerearth/6. VotingClassifier.ipynb
|
phileinSophos/ML-DL_Problems
|
033fc8a0086883fbe6748f2bf4725de7e8376e4b
|
[
"MIT"
] | null | null | null | 32.381643 | 140 | 0.460764 |
[
[
[
"## VotingClassifier - [ BaggingClassifier, RandomForestClassifier, XGBClassifier ]",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nfrom xgboost import XGBClassifier, XGBRegressor\nfrom sklearn.preprocessing import LabelEncoder\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.metrics import f1_score\nfrom sklearn.ensemble import BaggingClassifier, RandomForestClassifier, VotingClassifier\nfrom sklearn.multiclass import OneVsOneClassifier,OneVsRestClassifier\nfrom sklearn.svm import LinearSVC, SVC\nfrom sklearn.linear_model import LinearRegression, LogisticRegression, RidgeClassifier\nfrom sklearn.tree import DecisionTreeClassifier, DecisionTreeRegressor\nfrom sklearn.neighbors import KNeighborsClassifier\nfrom imblearn.ensemble import BalancedBaggingClassifier, BalancedRandomForestClassifier",
"_____no_output_____"
],
[
"training_data = pd.read_csv('train.csv')\nprint(training_data.shape)\ntraining_data.columns",
"(6500, 10)\n"
]
],
[
[
"### Dropping columns -- [ 'ID','Team_Value','Playing_Style','Won_Championship','Previous_SB_Wins' ]",
"_____no_output_____"
]
],
[
[
"y = training_data.Won_Championship\ntraining_data = training_data.drop(columns=['Won_Championship','ID','Team_Value','Playing_Style','Previous_SB_Wins'],axis=1)\n\nle_Number_Of_Injured_Players = LabelEncoder()\ntraining_data['Number_Of_Injured_Players'] = le_Number_Of_Injured_Players.fit_transform(training_data['Number_Of_Injured_Players'])\n\nle_Coach_Experience_Level = LabelEncoder()\ntraining_data['Coach_Experience_Level'] = le_Coach_Experience_Level.fit_transform(training_data['Coach_Experience_Level'])\ntraining_data.head()",
"_____no_output_____"
],
[
"x_train,x_test, y_train, y_test = train_test_split(training_data,y,test_size=0.2)",
"_____no_output_____"
],
[
"bags = BalancedBaggingClassifier(n_estimators=100,oob_score=True,bootstrap_features=True,replacement=True)\nbags.fit(x_train,y_train)\n#bags.fit(training_data,y)\nprediction = bags.predict(x_test)\nacc = 100 * (f1_score(y_test,prediction,average='binary'))\nacc",
"_____no_output_____"
],
[
"bal_rfc = BalancedRandomForestClassifier(class_weight='balanced_subsample',criterion='entropy')\nbal_rfc.fit(x_train,y_train)\n#bal_rfc.fit(training_data,y)\nprediction = bal_rfc.predict(x_test)\nacc = 100 * (f1_score(y_test,prediction,average='binary'))\nacc",
"_____no_output_____"
],
[
"xgb = XGBClassifier(n_estimators=500,learning_rate=0.1,max_depth=10,reg_lambda=0.1,importance_type='total_gain')\nxgb.fit(x_train,y_train)\n#xgb.fit(training_data,y)\nprediction = xgb.predict(x_test)\nacc = 100 * (f1_score(y_test,prediction,average='binary'))\nacc",
"_____no_output_____"
],
[
"bag = BalancedBaggingClassifier(n_estimators=100,oob_score=True,bootstrap_features=True,replacement=True)\nxgb = XGBClassifier(n_estimators=500,learning_rate=0.1,max_depth=10,reg_lambda=0.1,importance_type='total_gain')\nbal_rfc = BalancedRandomForestClassifier(class_weight='balanced_subsample',criterion='entropy')\n\nvoting = VotingClassifier(estimators=[\n ('bag', bag), ('rfc', bal_rfc), ('xgb', xgb)], voting='hard')\nvoting.fit(training_data, y)",
"_____no_output_____"
],
[
"prediction = voting.predict(x_test)\nacc = 100 * (f1_score(y_test,prediction,average='binary'))\nacc",
"_____no_output_____"
],
[
"cols = training_data.columns\ntest_data = pd.read_csv('test.csv')\nevent_id = test_data['ID']\n\nprint(test_data.shape)\ntest_data = test_data.drop(columns=['ID','Team_Value','Playing_Style','Previous_SB_Wins'],axis=1)\n\ntest_data['Number_Of_Injured_Players'] = le_Number_Of_Injured_Players.fit_transform(test_data['Number_Of_Injured_Players'])\n\ntest_data['Coach_Experience_Level'] = le_Coach_Experience_Level.fit_transform(test_data['Coach_Experience_Level'])\n\n\npredictions = voting.predict(test_data)\nresult_df = pd.DataFrame({'ID':event_id,'Won_Championship':predictions})\nresult_df.to_csv('Prediction.csv',index=False)",
"(3500, 9)\n"
]
],
[
[
"#### Online ACCURACY - 76.4, when local accuracy - 78.9 on whole data (VotingClassifier)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
cbf2ce31fc219231955f415ea65918b1b324953d
| 9,091 |
ipynb
|
Jupyter Notebook
|
notebooks/4-Views.ipynb
|
digitronik/widgetastic_notebooks
|
3d0d07b3df2017d2fc3582eaafb09da073db2cf4
|
[
"Apache-2.0"
] | 5 |
2020-03-27T09:08:02.000Z
|
2022-02-21T17:24:50.000Z
|
notebooks/4-Views.ipynb
|
digitronik/widgetastic_notebooks
|
3d0d07b3df2017d2fc3582eaafb09da073db2cf4
|
[
"Apache-2.0"
] | null | null | null |
notebooks/4-Views.ipynb
|
digitronik/widgetastic_notebooks
|
3d0d07b3df2017d2fc3582eaafb09da073db2cf4
|
[
"Apache-2.0"
] | 3 |
2018-07-15T16:57:10.000Z
|
2020-01-20T06:27:16.000Z
| 21.390588 | 104 | 0.509295 |
[
[
[
"# Views",
"_____no_output_____"
],
[
"- Views are nothing but widget only but having capability to hold widgets.",
"_____no_output_____"
]
],
[
[
"from webdriver_kaifuku import BrowserManager\nfrom widgetastic.widget import Browser\n\ncommand_executor = \"http://localhost:4444/wd/hub\"\n\nconfig = {\n \"webdriver\": \"Remote\",\n \"webdriver_options\":\n {\"desired_capabilities\": {\"browserName\": \"firefox\"},\n \"command_executor\": command_executor,\n }\n}\nmgr = BrowserManager.from_conf(config)\nsel = mgr.ensure_open()\n\n\nclass MyBrowser(Browser):\n pass\n\nbrowser = MyBrowser(selenium=sel)\nbrowser.url = \"http://0.0.0.0:8000/test_page.html\"",
"_____no_output_____"
],
[
"from widgetastic.widget import View, Text, TextInput, Checkbox, ColourInput, Select",
"_____no_output_____"
],
[
"# Example-1",
"_____no_output_____"
],
[
"class BasicWidgetView(View):\n text_input = TextInput(id=\"text_input\")\n checkbox = Checkbox(id=\"checkbox_input\")\n button = Text(locator=\".//button[@id='a_button']\")\n color_input = ColourInput(id=\"color_input\")",
"_____no_output_____"
],
[
"view = BasicWidgetView(browser)",
"_____no_output_____"
]
],
[
[
"### Nested Views",
"_____no_output_____"
]
],
[
[
"# Example-2",
"_____no_output_____"
],
[
"class MyNestedView(View):\n @View.nested\n class basic(View): #noqa\n text_input = TextInput(id=\"text_input\")\n checkbox = Checkbox(id=\"checkbox_input\")\n \n @View.nested\n class conditional(View): \n select_input = Select(id=\"select_lang\")\n ",
"_____no_output_____"
],
[
"view = MyNestedView(browser)",
"_____no_output_____"
],
[
"view.fill({'basic': {'text_input': 'hi', 'checkbox': True},\n 'conditional': {'select_input': 'Go'}})",
"_____no_output_____"
],
[
"# Example-3",
"_____no_output_____"
],
[
"class Basic(View):\n text_input = TextInput(id=\"text_input\")\n checkbox = Checkbox(id=\"checkbox_input\")\n \nclass Conditional(View):\n select_input = Select(id=\"select_lang\")",
"_____no_output_____"
],
[
"class MyNestedView(View):\n basic = View.nested(Basic)\n conditional = View.nested(Conditional)",
"_____no_output_____"
],
[
"view = MyNestedView(browser)",
"_____no_output_____"
],
[
"view.read()",
"_____no_output_____"
]
],
[
[
"### Switchable Conditional Views",
"_____no_output_____"
]
],
[
[
"from widgetastic.widget import ConditionalSwitchableView",
"_____no_output_____"
],
[
"# Example-4: Switchable widgets",
"_____no_output_____"
],
[
"class MyConditionalWidgetView(View):\n select_input = Select(id=\"select_lang\")\n \n lang_label = ConditionalSwitchableView(reference=\"select_input\")\n \n lang_label.register(\"Python\", default=True, widget=Text(locator=\".//h3[@id='lang-1']\"))\n lang_label.register(\"Go\", widget=Text(locator=\".//h3[@id='lang-2']\"))",
"_____no_output_____"
],
[
"view = MyConditionalWidgetView(browser)",
"_____no_output_____"
],
[
"# Example-5: Switchable Views",
"_____no_output_____"
],
[
"class MyConditionalView(View):\n select_input = Select(id=\"select_lang\")\n \n lang = ConditionalSwitchableView(reference=\"select_input\")\n \n @lang.register(\"Python\", default=True)\n class PythonView(View):\n # some more widgets\n lang_label = Text(locator=\".//h3[@id='lang-1']\")\n \n @lang.register(\"Go\")\n class GoView(View):\n lang_label = Text(locator=\".//h3[@id='lang-2']\")\n ",
"_____no_output_____"
],
[
"view = MyConditionalView(browser)",
"_____no_output_____"
]
],
[
[
"### Parametrized Views",
"_____no_output_____"
]
],
[
[
"from widgetastic.widget import ParametrizedView\nfrom widgetastic.utils import ParametrizedLocator",
"_____no_output_____"
],
[
"# Example-6",
"_____no_output_____"
],
[
"class MyParametrizedView(ParametrizedView):\n PARAMETERS = ('name',)\n \n ROOT = ParametrizedLocator(\".//div[contains(label, {name|quote})]\")\n \n widget = Checkbox(locator=\".//input\")\n ",
"_____no_output_____"
],
[
"view = MyParametrizedView(browser, additional_context={'name': 'widget 1'})",
"_____no_output_____"
],
[
"# Example-7: Nested Parametrized View",
"_____no_output_____"
],
[
"class MyNestedParametrizedView(View):\n\n @View.nested\n class widget_selector(ParametrizedView):\n PARAMETERS = ('name',)\n\n ROOT = ParametrizedLocator(\".//div[contains(label, {name|quote})]\")\n\n widget = Checkbox(locator=\".//input\")",
"_____no_output_____"
],
[
"view = MyNestedParametrizedView(browser)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbf2d40cd00d6265e44099df9560f9f38e2ca6c9
| 6,921 |
ipynb
|
Jupyter Notebook
|
Section 5/Deploy_chatbot.ipynb
|
PacktPublishing/Interactive-Chatbots-with-TensorFlow-
|
0e3a665e9a22cb852e7325420af3d0fd37fc7c34
|
[
"MIT"
] | 26 |
2019-02-02T13:32:17.000Z
|
2022-02-23T20:07:00.000Z
|
Section 5/Deploy_chatbot.ipynb
|
PacktPublishing/Interactive-Chatbots-with-TensorFlow-
|
0e3a665e9a22cb852e7325420af3d0fd37fc7c34
|
[
"MIT"
] | 1 |
2019-04-20T07:46:34.000Z
|
2019-04-20T07:46:34.000Z
|
Section 5/Deploy_chatbot.ipynb
|
PacktPublishing/Interactive-Chatbots-with-TensorFlow-
|
0e3a665e9a22cb852e7325420af3d0fd37fc7c34
|
[
"MIT"
] | 21 |
2019-02-08T00:35:28.000Z
|
2021-06-10T17:47:16.000Z
| 28.717842 | 269 | 0.526947 |
[
[
[
"import tensorflow as tf\nimport numpy as np\nimport pandas as pd\n\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.preprocessing import LabelEncoder\nfrom sklearn.feature_extraction.text import CountVectorizer\n\nfrom nltk.stem import PorterStemmer\nfrom autocorrect import spell\n\nimport os\nfrom six.moves import cPickle\nimport re\n",
"/home/omar/anaconda2/envs/py35/lib/python3.5/site-packages/h5py/__init__.py:36: FutureWarning: Conversion of the second argument of issubdtype from `float` to `np.floating` is deprecated. In future, it will be treated as `np.float64 == np.dtype(float).type`.\n from ._conv import register_converters as _register_converters\n"
],
[
"MAX_LEN = 25\nBATCH_SIZE = 64\n\nstemmer = PorterStemmer()\ndef process_str(string, bot_input=False, bot_output=False):\n string = string.strip().lower()\n string = re.sub(r\"[^A-Za-z0-9(),!?\\'\\`:]\", \" \", string)\n string = re.sub(r\"\\'s\", \" \\'s\", string)\n string = re.sub(r\"\\'ve\", \" \\'ve\", string)\n string = re.sub(r\"n\\'t\", \" n\\'t\", string)\n string = re.sub(r\"\\'re\", \" \\'re\", string)\n string = re.sub(r\"\\'d\", \" \\'d\", string)\n string = re.sub(r\"\\'ll\", \" \\'ll\", string)\n string = re.sub(r\",\", \" , \", string)\n string = re.sub(r\"!\", \" ! \", string)\n string = re.sub(r\"\\s{2,}\", \" \", string)\n string = string.split(\" \")\n string = [re.sub(r\"[0-9]+\", \"NUM\", token) for token in string]\n string = [stemmer.stem(re.sub(r'(.)\\1+', r'\\1\\1', token)) for token in string]\n string = [spell(token).lower() for token in string]\n # Truncate string\n while True:\n try:\n string.remove(\"\")\n except:\n break\n if(not bot_input and not bot_output):\n string = string[0:MAX_LEN]\n elif(bot_input):\n string = string[0:MAX_LEN-1]\n string.insert(0, \"</start>\")\n else:\n string = string[0:MAX_LEN-1]\n string.insert(len(string), \"</end>\")\n old_len = len(string)\n for i in range((MAX_LEN) - len(string)):\n string.append(\" </pad> \")\n string = re.sub(\"\\s+\", \" \", \" \".join(string)).strip()\n return string, old_len",
"_____no_output_____"
],
[
"imported_graph = tf.train.import_meta_graph('checkpoints/best_validation.meta')\nsess = tf.InteractiveSession()\nimported_graph.restore(sess, \"checkpoints/best_validation\")\n\nsess.run(tf.tables_initializer())\ngraph = tf.get_default_graph()",
"INFO:tensorflow:Restoring parameters from checkpoints/best_validation\n"
],
[
"def test(text):\n text, text_len = process_str(text)\n text = [text] + [\"hi\"] * (BATCH_SIZE-1)\n text_len = [text_len] + [1] * (BATCH_SIZE-1)\n return text, text_len",
"_____no_output_____"
],
[
"test_init_op = graph.get_operation_by_name('data/dataset_init')\n\nuser_ph = graph.get_tensor_by_name(\"user_placeholder:0\")\nbot_inp_ph = graph.get_tensor_by_name(\"bot_inp_placeholder:0\")\nbot_out_ph = graph.get_tensor_by_name(\"bot_out_placeholder:0\")\n\nuser_lens_ph = graph.get_tensor_by_name(\"user_len_placeholder:0\")\nbot_inp_lens_ph = graph.get_tensor_by_name(\"bot_inp_lens_placeholder:0\")\nbot_out_lens_ph = graph.get_tensor_by_name(\"bot_out_lens_placeholder:0\")\n\nwords = graph.get_tensor_by_name(\"inference/words:0\")\n",
"_____no_output_____"
],
[
"def chat(text):\n user, user_lens = test(text)\n sess.run(test_init_op, feed_dict={\n user_ph: user,\n bot_inp_ph: [\"hi\"] * BATCH_SIZE,\n bot_out_ph: [\"hi\"] * BATCH_SIZE,\n user_lens_ph: user_lens,\n bot_inp_lens_ph: [1] * BATCH_SIZE,\n bot_out_lens_ph: [1] * BATCH_SIZE\n })\n translations_text = sess.run(words)\n output = [item.decode() for item in translations_text[0]]\n if(\"</end>\" in output):\n end_idx = output.index(\"</end>\")\n output = output[0:end_idx]\n output = \" \".join(output)\n print(\"BOT: \" + output)",
"_____no_output_____"
],
[
"while True:\n chat(input())",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbf2da30e9112eeaed974b9f8d39d2ddf82fd8a4
| 19,539 |
ipynb
|
Jupyter Notebook
|
Week-4/linreg.ipynb
|
MLunov/Introduction-to-Machine-Learning-HSE-Yandex
|
a7366f96e3a15e08eda88baefd9c4c140659845d
|
[
"MIT"
] | null | null | null |
Week-4/linreg.ipynb
|
MLunov/Introduction-to-Machine-Learning-HSE-Yandex
|
a7366f96e3a15e08eda88baefd9c4c140659845d
|
[
"MIT"
] | null | null | null |
Week-4/linreg.ipynb
|
MLunov/Introduction-to-Machine-Learning-HSE-Yandex
|
a7366f96e3a15e08eda88baefd9c4c140659845d
|
[
"MIT"
] | null | null | null | 32.510815 | 775 | 0.49977 |
[
[
[
"# Programming Assignment: Линейная регрессия: прогноз оклада по описанию вакансии\n\n## Введение\nЛинейные методы хорошо подходят для работы с разреженными данными — к таковым относятся, например, тексты. Это можно объяснить высокой скоростью обучения и небольшим количеством параметров, благодаря чему удается избежать переобучения.\n\nЛинейная регрессия имеет несколько разновидностей в зависимости от того, какой регуляризатор используется. Мы будем работать с гребневой регрессией, где применяется квадратичный, или `L2`-регуляризатор.\n\n## Реализация в Scikit-Learn\nДля извлечения `TF-IDF`-признаков из текстов воспользуйтесь классом `sklearn.feature_extraction.text.TfidfVectorizer`.\n\nДля предсказания целевой переменной мы будем использовать гребневую регрессию, которая реализована в классе `sklearn.linear_model.Ridge`.\n\nОбратите внимание, что признаки `LocationNormalized` и `ContractTime` являются строковыми, и поэтому с ними нельзя работать напрямую. Такие нечисловые признаки с неупорядоченными значениями называют категориальными или номинальными. Типичный подход к их обработке — кодирование категориального признака с m возможными значениями с помощью m бинарных признаков. Каждый бинарный признак соответствует одному из возможных значений категориального признака и является индикатором того, что на данном объекте он принимает данное значение. Данный подход иногда называют `one-hot`-кодированием. Воспользуйтесь им, чтобы перекодировать признаки `LocationNormalized` и `ContractTime`. Он уже реализован в классе `sklearn.feature_extraction.DictVectorizer`. Пример использования:",
"_____no_output_____"
]
],
[
[
"from sklearn.feature_extraction import DictVectorizer\nenc = DictVectorizer()\nX_train_categ = enc.fit_transform(data_train[['LocationNormalized', 'ContractTime']].to_dict('records'))\nX_test_categ = enc.transform(data_test[['LocationNormalized', 'ContractTime']].to_dict('records'))",
"_____no_output_____"
]
],
[
[
"Вам понадобится производить замену пропущенных значений на специальные строковые величины (например, `'nan'`). Для этого подходит следующий код:",
"_____no_output_____"
]
],
[
[
"data_train['LocationNormalized'].fillna('nan', inplace=True)\ndata_train['ContractTime'].fillna('nan', inplace=True)",
"_____no_output_____"
]
],
[
[
"## Инструкция по выполнению\n\n### Шаг 1:\nЗагрузите данные об описаниях вакансий и соответствующих годовых зарплатах из файла `salary-train.csv` (либо его заархивированную версию `salary-train.zip`).",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nfrom sklearn.feature_extraction.text import TfidfVectorizer\nfrom sklearn.feature_extraction import DictVectorizer\nfrom sklearn.linear_model import Ridge\nfrom scipy.sparse import hstack",
"_____no_output_____"
],
[
"train = pd.read_csv('salary-train.csv')\ntest = pd.read_csv('salary-test-mini.csv')",
"_____no_output_____"
],
[
"train.head()",
"_____no_output_____"
],
[
"test.head()",
"_____no_output_____"
]
],
[
[
"### Шаг 2:\nПроведите предобработку:\n* Приведите тексты к нижнему регистру (`text.lower()`).\n* Замените все, кроме букв и цифр, на пробелы — это облегчит дальнейшее разделение текста на слова. Для такой замены в строке `text` подходит следующий вызов: `re.sub('[^a-zA-Z0-9]', ' ', text)`. Также можно воспользоваться методом `replace` у `DataFrame`, чтобы сразу преобразовать все тексты:",
"_____no_output_____"
]
],
[
[
"train['FullDescription'] = train['FullDescription'].replace('[^a-zA-Z0-9]', ' ', regex = True)",
"_____no_output_____"
]
],
[
[
"* Примените `TfidfVectorizer` для преобразования текстов в векторы признаков. Оставьте только те слова, которые встречаются хотя бы в 5 объектах (параметр `min_df` у `TfidfVectorizer`).\n* Замените пропуски в столбцах `LocationNormalized` и `ContractTime` на специальную строку `'nan'`. Код для этого был приведен выше.\n* Примените `DictVectorizer` для получения `one-hot`-кодирования признаков `LocationNormalized` и `ContractTime`.\n* Объедините все полученные признаки в одну матрицу \"объекты-признаки\". Обратите внимание, что матрицы для текстов и категориальных признаков являются разреженными. Для объединения их столбцов нужно воспользоваться функцией `scipy.sparse.hstack`.",
"_____no_output_____"
]
],
[
[
"train['FullDescription'] = train['FullDescription'].replace('[^a-zA-Z0-9]', ' ', regex = True).str.lower()\ntest['FullDescription'] = test['FullDescription'].replace('[^a-zA-Z0-9]', ' ', regex = True).str.lower()",
"_____no_output_____"
],
[
"train.head()",
"_____no_output_____"
],
[
"test.head()",
"_____no_output_____"
],
[
"vectorizer = TfidfVectorizer(min_df = 5)\n\nX_train = vectorizer.fit_transform(train['FullDescription'])\nX_test = vectorizer.transform(test['FullDescription'])",
"_____no_output_____"
],
[
"train['LocationNormalized'].fillna('nan', inplace=True)\ntrain['ContractTime'].fillna('nan', inplace=True)",
"_____no_output_____"
],
[
"enc = DictVectorizer()\n\nX_train_categ = enc.fit_transform(train[['LocationNormalized', 'ContractTime']].to_dict('records'))\nX_test_categ = enc.transform(test[['LocationNormalized', 'ContractTime']].to_dict('records'))",
"_____no_output_____"
],
[
"X = hstack([X_train, X_train_categ])\ny = train['SalaryNormalized']",
"_____no_output_____"
]
],
[
[
"### Шаг 3:\n\nОбучите гребневую регрессию с параметрами `alpha=1` и `random_state=241`. Целевая переменная записана в столбце `SalaryNormalized`.",
"_____no_output_____"
]
],
[
[
"clf = Ridge(alpha=1, random_state=241)\nclf.fit(X, y)",
"_____no_output_____"
]
],
[
[
"### Шаг 4: \n\nПостройте прогнозы для двух примеров из файла `salary-test-mini.csv`. Значения полученных прогнозов являются ответом на задание. Укажите их через пробел.",
"_____no_output_____"
]
],
[
[
"X = hstack([X_test, X_test_categ])\n\nans = clf.predict(X)\nans",
"_____no_output_____"
],
[
"def write_answer(ans, n):\n with open(\"ans{}.txt\".format(n), \"w\") as fout:\n fout.write(str(ans))\n \nwrite_answer(str(ans)[1:-1], 1)",
"_____no_output_____"
]
],
[
[
"Если ответом является нецелое число, то целую и дробную часть необходимо разграничивать точкой, например, 0.42. При необходимости округляйте дробную часть до двух знаков.",
"_____no_output_____"
]
]
] |
[
"markdown",
"raw",
"markdown",
"raw",
"markdown",
"code",
"markdown",
"raw",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"raw"
],
[
"markdown"
],
[
"raw"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"raw"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
cbf2db78c157612bc4950cbfb07a6068032bdd95
| 2,356 |
ipynb
|
Jupyter Notebook
|
Business Problem.ipynb
|
scarlioD/AppliedDataScienceCapstone
|
4abebc71b3572258af20b52b9d53d5add7fc4d6f
|
[
"MIT"
] | null | null | null |
Business Problem.ipynb
|
scarlioD/AppliedDataScienceCapstone
|
4abebc71b3572258af20b52b9d53d5add7fc4d6f
|
[
"MIT"
] | null | null | null |
Business Problem.ipynb
|
scarlioD/AppliedDataScienceCapstone
|
4abebc71b3572258af20b52b9d53d5add7fc4d6f
|
[
"MIT"
] | null | null | null | 29.08642 | 509 | 0.629457 |
[
[
[
"<h1>Business Problem</h1>",
"_____no_output_____"
],
[
"In this section I am going to discuss my idea for the Capstone project",
"_____no_output_____"
],
[
"<h2>Location of Interest</h2>",
"_____no_output_____"
],
[
"First of all I would like to introduce the location of Interest I chose for my project. The business idea will be developed on the city of Salerno (Italy) and its neighborhoods. Salerno is my hometown, I chose it because I know it well and I can help with results interpretation. The city is quite small so I am going to take in account some near places. This area is suitable for my goal because tourism is a great source of income and the amount of places will provide an interesting scenario to study",
"_____no_output_____"
],
[
"<h2>A tourist guide made with unsupervised machine learning tecnicques</h2>",
"_____no_output_____"
],
[
"The idea is to create a tourist guide using geodata and machine learning. I am going to cluster the area in order to find the places more suitable for every need, cluster them and then make a comparison between the logical clusters and the geographical area. \n\nThe goal is to create list of clusters grouped by most common venuetypes and where they are located.\n\n<i>Example</i>\n\n<blockquote>Suppose you are visiting the city and you want to eat a pizza you can check the clusters in order to find how the Pizza Places are distributed</blockquote> ",
"_____no_output_____"
]
]
] |
[
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
cbf2eee56a595298f5220942bdf6cfe77604932f
| 363,512 |
ipynb
|
Jupyter Notebook
|
first-neural-network/Your_first_neural_network.ipynb
|
BB-Y/cn-deep-learning
|
0d6c2ba5ec33ce437d046c6a631dadcc031252fd
|
[
"MIT"
] | null | null | null |
first-neural-network/Your_first_neural_network.ipynb
|
BB-Y/cn-deep-learning
|
0d6c2ba5ec33ce437d046c6a631dadcc031252fd
|
[
"MIT"
] | null | null | null |
first-neural-network/Your_first_neural_network.ipynb
|
BB-Y/cn-deep-learning
|
0d6c2ba5ec33ce437d046c6a631dadcc031252fd
|
[
"MIT"
] | null | null | null | 355.339198 | 233,052 | 0.918946 |
[
[
[
"# 你的第一个神经网络\n\n在此项目中,你将构建你的第一个神经网络,并用该网络预测每日自行车租客人数。我们提供了一些代码,但是需要你来实现神经网络(大部分内容)。提交此项目后,欢迎进一步探索该数据和模型。",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\n%config InlineBackend.figure_format = 'retina'\n\nimport numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt",
"_____no_output_____"
]
],
[
[
"## 加载和准备数据\n\n构建神经网络的关键一步是正确地准备数据。不同尺度级别的变量使网络难以高效地掌握正确的权重。我们在下方已经提供了加载和准备数据的代码。你很快将进一步学习这些代码!",
"_____no_output_____"
]
],
[
[
"data_path = 'Bike-Sharing-Dataset/hour.csv'\n\nrides = pd.read_csv(data_path)",
"_____no_output_____"
],
[
"rides.head()",
"_____no_output_____"
]
],
[
[
"## 数据简介\n\n此数据集包含的是从 2011 年 1 月 1 日到 2012 年 12 月 31 日期间每天每小时的骑车人数。骑车用户分成临时用户和注册用户,cnt 列是骑车用户数汇总列。你可以在上方看到前几行数据。\n\n下图展示的是数据集中前 10 天左右的骑车人数(某些天不一定是 24 个条目,所以不是精确的 10 天)。你可以在这里看到每小时租金。这些数据很复杂!周末的骑行人数少些,工作日上下班期间是骑行高峰期。我们还可以从上方的数据中看到温度、湿度和风速信息,所有这些信息都会影响骑行人数。你需要用你的模型展示所有这些数据。",
"_____no_output_____"
]
],
[
[
"rides[:24*10].plot(x='dteday', y='cnt')",
"_____no_output_____"
]
],
[
[
"### 虚拟变量(哑变量)\n\n下面是一些分类变量,例如季节、天气、月份。要在我们的模型中包含这些数据,我们需要创建二进制虚拟变量。用 Pandas 库中的 `get_dummies()` 就可以轻松实现。",
"_____no_output_____"
]
],
[
[
"dummy_fields = ['season', 'weathersit', 'mnth', 'hr', 'weekday']\nfor each in dummy_fields:\n dummies = pd.get_dummies(rides[each], prefix=each, drop_first=False)\n rides = pd.concat([rides, dummies], axis=1)\n\nfields_to_drop = ['instant', 'dteday', 'season', 'weathersit', \n 'weekday', 'atemp', 'mnth', 'workingday', 'hr']\ndata = rides.drop(fields_to_drop, axis=1)\ndata.head()",
"_____no_output_____"
]
],
[
[
"### 调整目标变量\n\n为了更轻松地训练网络,我们将对每个连续变量标准化,即转换和调整变量,使它们的均值为 0,标准差为 1。\n\n我们会保存换算因子,以便当我们使用网络进行预测时可以还原数据。",
"_____no_output_____"
]
],
[
[
"quant_features = ['casual', 'registered', 'cnt', 'temp', 'hum', 'windspeed']\n# Store scalings in a dictionary so we can convert back later\nscaled_features = {}\nfor each in quant_features:\n mean, std = data[each].mean(), data[each].std()\n scaled_features[each] = [mean, std]\n data.loc[:, each] = (data[each] - mean)/std",
"_____no_output_____"
]
],
[
[
"### 将数据拆分为训练、测试和验证数据集\n\n我们将大约最后 21 天的数据保存为测试数据集,这些数据集会在训练完网络后使用。我们将使用该数据集进行预测,并与实际的骑行人数进行对比。",
"_____no_output_____"
]
],
[
[
"# Save data for approximately the last 21 days \ntest_data = data[-21*24:]\n\n# Now remove the test data from the data set \ndata = data[:-21*24]\n\n# Separate the data into features and targets\ntarget_fields = ['cnt', 'casual', 'registered']\nfeatures, targets = data.drop(target_fields, axis=1), data[target_fields]\ntest_features, test_targets = test_data.drop(target_fields, axis=1), test_data[target_fields]",
"_____no_output_____"
]
],
[
[
"我们将数据拆分为两个数据集,一个用作训练,一个在网络训练完后用来验证网络。因为数据是有时间序列特性的,所以我们用历史数据进行训练,然后尝试预测未来数据(验证数据集)。",
"_____no_output_____"
]
],
[
[
"# Hold out the last 60 days or so of the remaining data as a validation set\ntrain_features, train_targets = features[:-60*24], targets[:-60*24]\nval_features, val_targets = features[-60*24:], targets[-60*24:]",
"_____no_output_____"
]
],
[
[
"## 开始构建网络\n\n下面你将构建自己的网络。我们已经构建好结构和反向传递部分。你将实现网络的前向传递部分。还需要设置超参数:学习速率、隐藏单元的数量,以及训练传递数量。\n\n<img src=\"assets/neural_network.png\" width=300px>\n\n该网络有两个层级,一个隐藏层和一个输出层。隐藏层级将使用 S 型函数作为激活函数。输出层只有一个节点,用于递归,节点的输出和节点的输入相同。即激活函数是 $f(x)=x$。这种函数获得输入信号,并生成输出信号,但是会考虑阈值,称为激活函数。我们完成网络的每个层级,并计算每个神经元的输出。一个层级的所有输出变成下一层级神经元的输入。这一流程叫做前向传播(forward propagation)。\n\n我们在神经网络中使用权重将信号从输入层传播到输出层。我们还使用权重将错误从输出层传播回网络,以便更新权重。这叫做反向传播(backpropagation)。\n\n> **提示**:你需要为反向传播实现计算输出激活函数 ($f(x) = x$) 的导数。如果你不熟悉微积分,其实该函数就等同于等式 $y = x$。该等式的斜率是多少?也就是导数 $f(x)$。\n\n\n你需要完成以下任务:\n\n1. 实现 S 型激活函数。将 `__init__` 中的 `self.activation_function` 设为你的 S 型函数。\n2. 在 `train` 方法中实现前向传递。\n3. 在 `train` 方法中实现反向传播算法,包括计算输出错误。\n4. 在 `run` 方法中实现前向传递。\n\n ",
"_____no_output_____"
]
],
[
[
"class NeuralNetwork(object):\n def __init__(self, input_nodes, hidden_nodes, output_nodes, learning_rate):\n # Set number of nodes in input, hidden and output layers.\n self.input_nodes = input_nodes\n self.hidden_nodes = hidden_nodes\n self.output_nodes = output_nodes\n\n # Initialize weights\n self.weights_input_to_hidden = np.random.normal(0.0, self.input_nodes**-0.5, \n (self.input_nodes, self.hidden_nodes))\n\n self.weights_hidden_to_output = np.random.normal(0.0, self.hidden_nodes**-0.5, \n (self.hidden_nodes, self.output_nodes))\n self.lr = learning_rate\n \n #### TODO: Set self.activation_function to your implemented sigmoid function ####\n #\n # Note: in Python, you can define a function with a lambda expression,\n # as shown below.\n self.activation_function = lambda x : 1/(1 + np.exp(-x)) # Replace 0 with your sigmoid calculation.\n \n ### If the lambda code above is not something you're familiar with,\n # You can uncomment out the following three lines and put your \n # implementation there instead.\n #\n #def sigmoid(x):\n # return 0 # Replace 0 with your sigmoid calculation here\n #self.activation_function = sigmoid\n \n \n def train(self, features, targets):\n ''' Train the network on batch of features and targets. \n \n Arguments\n ---------\n \n features: 2D array, each row is one data record, each column is a feature\n targets: 1D array of target values\n \n '''\n n_records = features.shape[0]\n delta_weights_i_h = np.zeros(self.weights_input_to_hidden.shape)\n delta_weights_h_o = np.zeros(self.weights_hidden_to_output.shape)\n for X, y in zip(features, targets):\n #### Implement the forward pass here ####\n ### Forward pass ###\n # TODO: Hidden layer - Replace these values with your calculations.\n hidden_inputs = np.dot(X, self.weights_input_to_hidden) # signals into hidden layer\n hidden_outputs = self.activation_function(hidden_inputs) # signals from hidden layer\n \n # TODO: Output layer - Replace these values with your calculations.\n final_inputs = np.dot(hidden_outputs, self.weights_hidden_to_output) # signals into final output layer\n final_outputs = final_inputs # signals from final output layer\n \n #### Implement the backward pass here ####\n ### Backward pass ###\n\n # TODO: Output error - Replace this value with your calculations.\n error = y - final_outputs # Output layer error is the difference between desired target and actual output.\n\n # TODO: Calculate the hidden layer's contribution to the error\n hidden_error = np.dot(error[0], self.weights_hidden_to_output.T[0])\n\n # TODO: Backpropagated error terms - Replace these values with your calculations.\n output_error_term = error\n hidden_error_term = hidden_error * hidden_outputs * (1 - hidden_outputs)\n\n # Weight step (input to hidden)\n delta_weights_i_h += hidden_error_term * X[:, None]\n \n # Weight step (hidden to output)\n s = output_error_term * hidden_outputs\n delta_weights_h_o += s[:,None]\n \n\n # TODO: Update the weights - Replace these values with your calculations.\n self.weights_hidden_to_output += self.lr * delta_weights_h_o / n_records # update hidden-to-output weights with gradient descent step\n self.weights_input_to_hidden += self.lr * delta_weights_i_h / n_records # update input-to-hidden weights with gradient descent step\n \n def run(self, features):\n ''' Run a forward pass through the network with input features \n \n Arguments\n ---------\n features: 1D array of feature values\n '''\n \n #### Implement the forward pass here ####\n # TODO: Hidden layer - replace these values with the appropriate calculations.\n hidden_inputs = np.dot(features, self.weights_input_to_hidden) # signals into hidden layer\n hidden_outputs = self.activation_function(hidden_inputs) # signals from hidden layer\n \n # TODO: Output layer - Replace these values with the appropriate calculations.\n final_inputs = np.dot(hidden_outputs, self.weights_hidden_to_output) # signals into final output layer\n final_outputs = final_inputs # signals from final output layer \n \n return final_outputs",
"_____no_output_____"
],
[
"def MSE(y, Y):\n return np.mean((y-Y)**2)",
"_____no_output_____"
]
],
[
[
"## 单元测试\n\n运行这些单元测试,检查你的网络实现是否正确。这样可以帮助你确保网络已正确实现,然后再开始训练网络。这些测试必须成功才能通过此项目。",
"_____no_output_____"
]
],
[
[
"import unittest\n\ninputs = np.array([[0.5, -0.2, 0.1]])\ntargets = np.array([[0.4]])\ntest_w_i_h = np.array([[0.1, -0.2],\n [0.4, 0.5],\n [-0.3, 0.2]])\ntest_w_h_o = np.array([[0.3],[-0.1]])\n\nclass TestMethods(unittest.TestCase):\n \n ##########\n # Unit tests for data loading\n ##########\n \n def test_data_path(self):\n # Test that file path to dataset has been unaltered\n self.assertTrue(data_path.lower() == 'bike-sharing-dataset/hour.csv')\n \n def test_data_loaded(self):\n # Test that data frame loaded\n self.assertTrue(isinstance(rides, pd.DataFrame))\n \n ##########\n # Unit tests for network functionality\n ##########\n\n def test_activation(self):\n network = NeuralNetwork(3, 2, 1, 0.5)\n # Test that the activation function is a sigmoid\n self.assertTrue(np.all(network.activation_function(0.5) == 1/(1+np.exp(-0.5))))\n\n def test_train(self):\n # Test that weights are updated correctly on training\n network = NeuralNetwork(3, 2, 1, 0.5)\n network.weights_input_to_hidden = test_w_i_h.copy()\n network.weights_hidden_to_output = test_w_h_o.copy()\n \n network.train(inputs, targets)\n self.assertTrue(np.allclose(network.weights_hidden_to_output, \n np.array([[ 0.37275328], \n [-0.03172939]])))\n self.assertTrue(np.allclose(network.weights_input_to_hidden,\n np.array([[ 0.10562014, -0.20185996], \n [0.39775194, 0.50074398], \n [-0.29887597, 0.19962801]])))\n\n def test_run(self):\n # Test correctness of run method\n network = NeuralNetwork(3, 2, 1, 0.5)\n network.weights_input_to_hidden = test_w_i_h.copy()\n network.weights_hidden_to_output = test_w_h_o.copy()\n\n self.assertTrue(np.allclose(network.run(inputs), 0.09998924))\n\nsuite = unittest.TestLoader().loadTestsFromModule(TestMethods())\nunittest.TextTestRunner().run(suite)",
".....\n----------------------------------------------------------------------\nRan 5 tests in 0.005s\n\nOK\n"
]
],
[
[
"## 训练网络\n\n现在你将设置网络的超参数。策略是设置的超参数使训练集上的错误很小但是数据不会过拟合。如果网络训练时间太长,或者有太多的隐藏节点,可能就会过于针对特定训练集,无法泛化到验证数据集。即当训练集的损失降低时,验证集的损失将开始增大。\n\n你还将采用随机梯度下降 (SGD) 方法训练网络。对于每次训练,都获取随机样本数据,而不是整个数据集。与普通梯度下降相比,训练次数要更多,但是每次时间更短。这样的话,网络训练效率更高。稍后你将详细了解 SGD。\n\n\n### 选择迭代次数\n\n也就是训练网络时从训练数据中抽样的批次数量。迭代次数越多,模型就与数据越拟合。但是,如果迭代次数太多,模型就无法很好地泛化到其他数据,这叫做过拟合。你需要选择一个使训练损失很低并且验证损失保持中等水平的数字。当你开始过拟合时,你会发现训练损失继续下降,但是验证损失开始上升。\n\n### 选择学习速率\n\n速率可以调整权重更新幅度。如果速率太大,权重就会太大,导致网络无法与数据相拟合。建议从 0.1 开始。如果网络在与数据拟合时遇到问题,尝试降低学习速率。注意,学习速率越低,权重更新的步长就越小,神经网络收敛的时间就越长。\n\n\n### 选择隐藏节点数量\n\n隐藏节点越多,模型的预测结果就越准确。尝试不同的隐藏节点的数量,看看对性能有何影响。你可以查看损失字典,寻找网络性能指标。如果隐藏单元的数量太少,那么模型就没有足够的空间进行学习,如果太多,则学习方向就有太多的选择。选择隐藏单元数量的技巧在于找到合适的平衡点。",
"_____no_output_____"
]
],
[
[
"import sys\n\n### TODO:Set the hyperparameters here, you need to change the defalut to get a better solution ###\niterations = 5000\nlearning_rate = 0.8\nhidden_nodes = 15\noutput_nodes = 1\n\nN_i = train_features.shape[1]\nnetwork = NeuralNetwork(N_i, hidden_nodes, output_nodes, learning_rate)\n\nlosses = {'train':[], 'validation':[]}\nfor ii in range(iterations):\n # Go through a random batch of 128 records from the training data set\n batch = np.random.choice(train_features.index, size=128)\n X, y = train_features.ix[batch].values, train_targets.ix[batch]['cnt']\n \n network.train(X, y)\n \n # Printing out the training progress\n train_loss = MSE(network.run(train_features).T, train_targets['cnt'].values)\n val_loss = MSE(network.run(val_features).T, val_targets['cnt'].values)\n sys.stdout.write(\"\\rProgress: {:2.1f}\".format(100 * ii/float(iterations)) \\\n + \"% ... Training loss: \" + str(train_loss)[:5] \\\n + \" ... Validation loss: \" + str(val_loss)[:5])\n sys.stdout.flush()\n \n losses['train'].append(train_loss)\n losses['validation'].append(val_loss)",
"Progress: 0.2% ... Training loss: 0.945 ... Validation loss: 1.365"
],
[
"plt.plot(losses['train'], label='Training loss')\nplt.plot(losses['validation'], label='Validation loss')\nplt.legend()\n_ = plt.ylim()",
"_____no_output_____"
]
],
[
[
"## 检查预测结果\n\n使用测试数据看看网络对数据建模的效果如何。如果完全错了,请确保网络中的每步都正确实现。",
"_____no_output_____"
]
],
[
[
"fig, ax = plt.subplots(figsize=(15,4))\n\nmean, std = scaled_features['cnt']\npredictions = network.run(test_features).T*std + mean\nax.plot(predictions[0], label='Prediction')\nax.plot((test_targets['cnt']*std + mean).values, label='Data')\nax.set_xlim(right=len(predictions))\nax.legend()\n\ndates = pd.to_datetime(rides.ix[test_data.index]['dteday'])\ndates = dates.apply(lambda d: d.strftime('%b %d'))\nax.set_xticks(np.arange(len(dates))[12::24])\n_ = ax.set_xticklabels(dates[12::24], rotation=45)",
"/Users/hzx/anaconda3/lib/python3.7/site-packages/ipykernel_launcher.py:10: DeprecationWarning: \n.ix is deprecated. Please use\n.loc for label based indexing or\n.iloc for positional indexing\n\nSee the documentation here:\nhttp://pandas.pydata.org/pandas-docs/stable/indexing.html#ix-indexer-is-deprecated\n # Remove the CWD from sys.path while we load stuff.\n"
]
],
[
[
"## 可选:思考下你的结果(我们不会评估这道题的答案)\n\n \n请针对你的结果回答以下问题。模型对数据的预测效果如何?哪里出现问题了?为何出现问题呢?\n\n> **注意**:你可以通过双击该单元编辑文本。如果想要预览文本,请按 Control + Enter\n\n#### 请将你的答案填写在下方\n预测效果大体符合实际情况,但是22-26日圣诞节前后的预测与实际值相差较大。由于一年只有一次圣诞节,第二年圣诞节作为测试集,训练数据过少所以在这个日期预测的不准确",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
cbf2f7b51565831f4b06fe196d8555284031041f
| 43,040 |
ipynb
|
Jupyter Notebook
|
Copy_of_keras_wide_deep.ipynb
|
Amro-source/Deep-Learning
|
1e3e1fdffbf3b8495b245066eb33ce7a8beb5682
|
[
"CC0-1.0"
] | null | null | null |
Copy_of_keras_wide_deep.ipynb
|
Amro-source/Deep-Learning
|
1e3e1fdffbf3b8495b245066eb33ce7a8beb5682
|
[
"CC0-1.0"
] | null | null | null |
Copy_of_keras_wide_deep.ipynb
|
Amro-source/Deep-Learning
|
1e3e1fdffbf3b8495b245066eb33ce7a8beb5682
|
[
"CC0-1.0"
] | null | null | null | 49.757225 | 525 | 0.522955 |
[
[
[
"<a href=\"https://colab.research.google.com/github/Amro-source/Deep-Learning/blob/main/Copy_of_keras_wide_deep.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"To run this model directly in the browser with zero setup, open it in [Colab here](https://colab.research.google.com/github/sararob/keras-wine-model/blob/master/keras-wide-deep.ipynb).",
"_____no_output_____"
]
],
[
[
"from __future__ import absolute_import\nfrom __future__ import division\nfrom __future__ import print_function",
"_____no_output_____"
],
[
"# Install the latest version of TensorFlow\n!pip install -q -U tensorflow==1.7.0",
"_____no_output_____"
],
[
"import itertools\nimport os\nimport math\nimport numpy as np\nimport pandas as pd\nimport tensorflow as tf\n\nfrom sklearn.preprocessing import LabelEncoder\nfrom tensorflow import keras\nlayers = keras.layers\n\n# This code was tested with TensorFlow v1.7\nprint(\"You have TensorFlow version\", tf.__version__)",
"You have TensorFlow version 1.7.0\n"
],
[
"# Get the data: original source is here: https://www.kaggle.com/zynicide/wine-reviews/data\nURL = \"https://storage.googleapis.com/sara-cloud-ml/wine_data.csv\"\npath = tf.keras.utils.get_file(URL.split('/')[-1], URL)\n",
"_____no_output_____"
],
[
"# Convert the data to a Pandas data frame\ndata = pd.read_csv(path)",
"_____no_output_____"
],
[
"# Shuffle the data\ndata = data.sample(frac=1)\n\n# Print the first 5 rows\ndata.head()",
"_____no_output_____"
],
[
"# Do some preprocessing to limit the # of wine varities in the dataset\ndata = data[pd.notnull(data['country'])]\ndata = data[pd.notnull(data['price'])]\ndata = data.drop(data.columns[0], axis=1) \n\nvariety_threshold = 500 # Anything that occurs less than this will be removed.\nvalue_counts = data['variety'].value_counts()\nto_remove = value_counts[value_counts <= variety_threshold].index\ndata.replace(to_remove, np.nan, inplace=True)\ndata = data[pd.notnull(data['variety'])]",
"_____no_output_____"
],
[
"# Split data into train and test\ntrain_size = int(len(data) * .8)\nprint (\"Train size: %d\" % train_size)\nprint (\"Test size: %d\" % (len(data) - train_size))",
"Train size: 95646\nTest size: 23912\n"
],
[
"# Train features\ndescription_train = data['description'][:train_size]\nvariety_train = data['variety'][:train_size]\n\n# Train labels\nlabels_train = data['price'][:train_size]\n\n# Test features\ndescription_test = data['description'][train_size:]\nvariety_test = data['variety'][train_size:]\n\n# Test labels\nlabels_test = data['price'][train_size:]",
"_____no_output_____"
],
[
"# Create a tokenizer to preprocess our text descriptions\nvocab_size = 12000 # This is a hyperparameter, experiment with different values for your dataset\ntokenize = keras.preprocessing.text.Tokenizer(num_words=vocab_size, char_level=False)\ntokenize.fit_on_texts(description_train) # only fit on train",
"_____no_output_____"
],
[
"# Wide feature 1: sparse bag of words (bow) vocab_size vector \ndescription_bow_train = tokenize.texts_to_matrix(description_train)\ndescription_bow_test = tokenize.texts_to_matrix(description_test)",
"_____no_output_____"
],
[
"# Wide feature 2: one-hot vector of variety categories\n\n# Use sklearn utility to convert label strings to numbered index\nencoder = LabelEncoder()\nencoder.fit(variety_train)\nvariety_train = encoder.transform(variety_train)\nvariety_test = encoder.transform(variety_test)\nnum_classes = np.max(variety_train) + 1\n\n# Convert labels to one hot\nvariety_train = keras.utils.to_categorical(variety_train, num_classes)\nvariety_test = keras.utils.to_categorical(variety_test, num_classes)",
"_____no_output_____"
],
[
"# Define our wide model with the functional API\nbow_inputs = layers.Input(shape=(vocab_size,))\nvariety_inputs = layers.Input(shape=(num_classes,))\nmerged_layer = layers.concatenate([bow_inputs, variety_inputs])\nmerged_layer = layers.Dense(256, activation='relu')(merged_layer)\npredictions = layers.Dense(1)(merged_layer)\nwide_model = keras.Model(inputs=[bow_inputs, variety_inputs], outputs=predictions)",
"_____no_output_____"
],
[
"wide_model.compile(loss='mse', optimizer='adam', metrics=['accuracy'])\nprint(wide_model.summary())",
"__________________________________________________________________________________________________\nLayer (type) Output Shape Param # Connected to \n==================================================================================================\ninput_1 (InputLayer) (None, 12000) 0 \n__________________________________________________________________________________________________\ninput_2 (InputLayer) (None, 40) 0 \n__________________________________________________________________________________________________\nconcatenate_1 (Concatenate) (None, 12040) 0 input_1[0][0] \n input_2[0][0] \n__________________________________________________________________________________________________\ndense_1 (Dense) (None, 256) 3082496 concatenate_1[0][0] \n__________________________________________________________________________________________________\ndense_2 (Dense) (None, 1) 257 dense_1[0][0] \n==================================================================================================\nTotal params: 3,082,753\nTrainable params: 3,082,753\nNon-trainable params: 0\n__________________________________________________________________________________________________\nNone\n"
],
[
"# Deep model feature: word embeddings of wine descriptions\ntrain_embed = tokenize.texts_to_sequences(description_train)\ntest_embed = tokenize.texts_to_sequences(description_test)\n\nmax_seq_length = 170\ntrain_embed = keras.preprocessing.sequence.pad_sequences(\n train_embed, maxlen=max_seq_length, padding=\"post\")\ntest_embed = keras.preprocessing.sequence.pad_sequences(\n test_embed, maxlen=max_seq_length, padding=\"post\")",
"_____no_output_____"
],
[
"# Define our deep model with the Functional API\ndeep_inputs = layers.Input(shape=(max_seq_length,))\nembedding = layers.Embedding(vocab_size, 8, input_length=max_seq_length)(deep_inputs)\nembedding = layers.Flatten()(embedding)\nembed_out = layers.Dense(1)(embedding)\ndeep_model = keras.Model(inputs=deep_inputs, outputs=embed_out)\nprint(deep_model.summary())",
"_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\ninput_3 (InputLayer) (None, 170) 0 \n_________________________________________________________________\nembedding_1 (Embedding) (None, 170, 8) 96000 \n_________________________________________________________________\nflatten_1 (Flatten) (None, 1360) 0 \n_________________________________________________________________\ndense_3 (Dense) (None, 1) 1361 \n=================================================================\nTotal params: 97,361\nTrainable params: 97,361\nNon-trainable params: 0\n_________________________________________________________________\nNone\n"
],
[
"deep_model.compile(loss='mse',\n optimizer='adam',\n metrics=['accuracy'])",
"_____no_output_____"
],
[
"# Combine wide and deep into one model\nmerged_out = layers.concatenate([wide_model.output, deep_model.output])\nmerged_out = layers.Dense(1)(merged_out)\ncombined_model = keras.Model(wide_model.input + [deep_model.input], merged_out)\nprint(combined_model.summary())\n\ncombined_model.compile(loss='mse',\n optimizer='adam',\n metrics=['accuracy'])",
"__________________________________________________________________________________________________\nLayer (type) Output Shape Param # Connected to \n==================================================================================================\ninput_1 (InputLayer) (None, 12000) 0 \n__________________________________________________________________________________________________\ninput_2 (InputLayer) (None, 40) 0 \n__________________________________________________________________________________________________\ninput_3 (InputLayer) (None, 170) 0 \n__________________________________________________________________________________________________\nconcatenate_1 (Concatenate) (None, 12040) 0 input_1[0][0] \n input_2[0][0] \n__________________________________________________________________________________________________\nembedding_1 (Embedding) (None, 170, 8) 96000 input_3[0][0] \n__________________________________________________________________________________________________\ndense_1 (Dense) (None, 256) 3082496 concatenate_1[0][0] \n__________________________________________________________________________________________________\nflatten_1 (Flatten) (None, 1360) 0 embedding_1[0][0] \n__________________________________________________________________________________________________\ndense_2 (Dense) (None, 1) 257 dense_1[0][0] \n__________________________________________________________________________________________________\ndense_3 (Dense) (None, 1) 1361 flatten_1[0][0] \n__________________________________________________________________________________________________\nconcatenate_2 (Concatenate) (None, 2) 0 dense_2[0][0] \n dense_3[0][0] \n__________________________________________________________________________________________________\ndense_4 (Dense) (None, 1) 3 concatenate_2[0][0] \n==================================================================================================\nTotal params: 3,180,117\nTrainable params: 3,180,117\nNon-trainable params: 0\n__________________________________________________________________________________________________\nNone\n"
],
[
"# Run training\ncombined_model.fit([description_bow_train, variety_train] + [train_embed], labels_train, epochs=10, batch_size=128)",
"Epoch 1/10\n95646/95646 [==============================] - 52s 546us/step - loss: 1165.0368 - acc: 0.0276\nEpoch 2/10\n95646/95646 [==============================] - 57s 593us/step - loss: 915.0286 - acc: 0.0352\nEpoch 3/10\n95646/95646 [==============================] - 58s 608us/step - loss: 785.9011 - acc: 0.0396\nEpoch 4/10\n95646/95646 [==============================] - 56s 590us/step - loss: 668.5673 - acc: 0.0445\nEpoch 5/10\n95646/95646 [==============================] - 55s 576us/step - loss: 550.9641 - acc: 0.0483\nEpoch 6/10\n95646/95646 [==============================] - 55s 571us/step - loss: 440.3762 - acc: 0.0563\nEpoch 7/10\n95646/95646 [==============================] - 57s 594us/step - loss: 342.1391 - acc: 0.0651\nEpoch 8/10\n95646/95646 [==============================] - 59s 615us/step - loss: 260.5558 - acc: 0.0734\nEpoch 9/10\n95646/95646 [==============================] - 55s 579us/step - loss: 199.9219 - acc: 0.0853\nEpoch 10/10\n95646/95646 [==============================] - 57s 601us/step - loss: 152.7983 - acc: 0.0962\n"
],
[
"combined_model.evaluate([description_bow_test, variety_test] + [test_embed], labels_test, batch_size=128)",
"23912/23912 [==============================] - 6s 252us/step\n"
],
[
"# Generate predictions\npredictions = combined_model.predict([description_bow_test, variety_test] + [test_embed])",
"_____no_output_____"
],
[
"# Compare predictions with actual values for the first few items in our test dataset\nnum_predictions = 40\ndiff = 0\n\nfor i in range(num_predictions):\n val = predictions[i]\n print(description_test.iloc[i])\n print('Predicted: ', val[0], 'Actual: ', labels_test.iloc[i], '\\n')\n diff += abs(val[0] - labels_test.iloc[i])",
"Not a whole lot to say about the soft, flat nose. It's o.k., but that's really about it. The palate, meanwhile, turns sweet, nearly to the point of candied. And the finish is downright sugary. Decent weight and feel help it along.\nPredicted: 16.776217 Actual: 14.0 \n\nThis 70-30 blend of Chardonnay and Trebbiano is driven by a strong mineral vein that imparts dusty notes of slate, vitamin and granite over fuller aromas of exotic fruit and peach. The wine is simple and tart in the mouth with a creamy, soft feel overall.\nPredicted: 19.51367 Actual: 18.0 \n\nLight, fresh aromas of tomato and plum are gritty. This feels round for the most part, with rubbery tannins. Medicinal, slightly minty plum and berry flavors end with acceptable rawness and a latex note.\nPredicted: 21.455168 Actual: 10.0 \n\nA densely fruity wine, with berry and plum fruits, pushed by some firm tannins. It is the ripe fruit, brimming over the glass, that makes this wine so delicious and approachable now.\nPredicted: 25.588463 Actual: 17.0 \n\nWild, briary, brambly and semi-sweet from this hot vintage, this is a Zin made for easy drinking with roasts and cheeses. It's a big, hot wine, with some raisiny flavors mixed in with the cherries and spices.\nPredicted: 30.262243 Actual: 38.0 \n\nThe nose of this Chard is so lively and welcoming with accents of lilac, jasmine, white peach slices and clementine. Matured in barrel for 10 months, the wine's palate offers more intense, mature flavors of peaches and cream, ginger and supporting tropical acidity. Well structured with a defined and evolving finish, it takes you from citrus to fresh peach to vanilla wooded spice.\nPredicted: 13.833609 Actual: 15.0 \n\nFull bodied with a velvety heft on the palate, this Reserve Merlot from Hudson-Chatham offers a striking balance of intensity and elegance. From the nose to the palate, there's a bounty of rich dark fruit, bramble and smoke layered with silky oak and firm tannins.\nPredicted: 18.991003 Actual: 22.0 \n\nCranberry, Bing cherry and red plum flavors dominate the fruit profile, enlivened by a bright spice note. The tannins turn quite chalky and drying, which lessens the pleasure of the finish.\nPredicted: 11.892209 Actual: 19.0 \n\nA wine that is all about crispness, structure and bright fruits. The creamy texture from lees aging gives a rounder dimension, but the main thrust here is minerality coupled with delicious acidity.\nPredicted: 28.631481 Actual: 20.0 \n\nAt its best Rubicon stuns with power and richness. In lesser vintages (which are usually hotter ones) the wine, which is largely Cabernet Sauvignon, can be raisiny. While 2004 was a warm year, diligent viticulture paid off, resulting in an opulent wine with the purest expression of crushed cherries and blackberries, and oak-inspired hints of nougat and caramel. Fairly aggressive in tannins now, it should begin to open by 2008 and drink well for a decade.\nPredicted: 262.89868 Actual: 125.0 \n\nA bit unripe, with a slight vegetal intrusion into the blackberries and cherries. Teeter-totters just on the edge of ripeness, but falls short. Otherwise, the tannins are pretty, and the oak is deftly applied.\nPredicted: 48.683483 Actual: 50.0 \n\nA good everyday wine. It's dry, full-bodied and has enough berry-cherry flavors to get by, wrapped into a smooth texture.\nPredicted: 9.694958 Actual: 10.0 \n\nCombines dusky peach and passion fruit aromas and flavors in a wine that's a bit softer and sweeter-tasting than many Marlborough Sauvignon Blancs, yet still carries a refreshing touch of acid on the finish. Imported by New Zealand Wine Imports.\nPredicted: 17.764606 Actual: 17.0 \n\nA light-pink color is backed by a simple nose that's nondescript but fresh and clean. A slightly foamy but firm palate isn't all that deep or rich, while mild grapefruit and blood orange flavors finish steady.\nPredicted: 12.843844 Actual: 12.0 \n\nA nice, clean, attractive version of affordable Carmenère? Yep, that's what Odfjell delivers pretty much every year with its Armador bottling. Aromas are floral, minerally and deep, and so is the palate, which is pure, spicy and loaded with blackberry fruit. Smooth on the finish, too. One of Chile's best value-priced Carmenères.\nPredicted: 15.080006 Actual: 12.0 \n\nPlum, red berry, cinnamon and mint aromas give this savory-style red from Lebanon a serious start. Soft and elegant on the palate but with a touch of spice and lively acid, the wine is balanced and unfolding, and will pair well with dishes that offer complex spice. An interesting red with exotic appeal.\nPredicted: 30.442371 Actual: 50.0 \n\nPowerful vanilla scents rise from the glass, but the fruit, even in this difficult vintage, comes out immediately. It's tart and sharp, with a strong herbal component, and the wine snaps into focus quickly with fruit, acid, tannin, herb and vanilla in equal proportion. Firm and tight, still quite young, this wine needs decanting and/or further bottle age to show its best.\nPredicted: 46.233624 Actual: 45.0 \n\nThe bouquet delivers touches of graphite and rubber in front of traditional dried cherry and raspberry. Nice in the mouth, with softly fading fruit riding a wave of persistent acids and tannins. Snappy, old-school and succulent. Drink now through 2010.\nPredicted: 24.337423 Actual: 26.0 \n\nOak powers the nose on this entry-level bottling from the Ballard Canyon-based winery, with peach custard, cinnamon and other brown spices. It's wide and ripe on the palate at first, with cooked Meyer lemons, nectarine and pineapple, but also boasts a line of brisker acidity trending toward lime.\nPredicted: 25.915466 Actual: 24.0 \n\nShows plenty of weight on the palate, but also an overall lack of intensity to its slightly vegetal—green pea—flavors. Still, there's enough passion fruit and grapefruit to provide a sense of balance and a crisp, clean finish.\nPredicted: 16.476027 Actual: 11.0 \n\nFresh, vivid fruit flavors pop from this light-colored, well-balanced, mouthwatering wine. Ripe pear, crisp apple and a touch of baking spices mingle on the palate and linger on the finish.\nPredicted: 16.757448 Actual: 16.0 \n\nDark mineral, toasted French oak and black fruit carry the nose. This is a sturdy, nicely made high-end Rioja, but due to the heat of the year its range of flavors is narrow as it settles on baked plum and molasses. Medium long on the finish, with a lasting taste of chocolate.\nPredicted: 81.83065 Actual: 100.0 \n\nThis medium-bodied dry Riesling is nicely balanced between apple and lime, with hints of petrol and spice to bring home its Rieslingness. Drink now for the vibrance and freshness of the fruit, but it should age well for at least a few years.\nPredicted: 15.825063 Actual: 18.0 \n\nPretty good, but a little on the rustic side, with baked blackberry, cherry and currant flavors wrapped into some rugged, edgy tannins. Could just be going through an awkward phase, but its future is hard to predict.\nPredicted: 26.417683 Actual: 40.0 \n\nQuite woody for this weight class, with balsam shavings and berry aromas that are distant. Chewy, lactic and creamy across the tongue, with a flavor profile that brings chocolate and berry syrup flavors. Finishes with a final wave of oaky spice and bitterness; good but needs more fruit.\nPredicted: 13.144117 Actual: 15.0 \n\nFrom an organic, dry farmed vineyard, this wine is as feral as the forest—a study in wild boar, wildflowers and dried herbs. Bright acidity buoys that meaty quality in the form of tart rhubarb and strawberry tea flavors. Floral on the nose and palate, it's spicy too, with bold seasonings of clove and pepper.\nPredicted: 67.539925 Actual: 95.0 \n\nThis is a very enjoyable Chardonnay from Friuli in northeastern Italy with vibrant luminosity and fresh tones of stone fruit, yellow rose and honey. The mouthfeel is naturally creamy but the wine ends on a bright, crisp note.\nPredicted: 17.144892 Actual: 16.0 \n\nA powerful, fat, still young wine, full of yeast flavors, green apples, plums and with a fine, tight structure—this is a food wine that could certainly age, maybe over 3 to 4 years.\nPredicted: 25.042484 Actual: 23.0 \n\nPast its prime, with herbal, leafy notes dominating the nectarine and melon fruit. Soft and even a bit sweet, with a chalky finish.\nPredicted: 16.498108 Actual: 15.0 \n\nSoft, creamy and buttery, this will surely please those who like a banana cream pie style of Chardonnay. There's peaches in there too, and papaya—a lot of ripe fruit, nicely matched to the oak flavors. Drink now.\nPredicted: 26.30731 Actual: 24.0 \n\nPutting your nose into a glass of Oddero Barolo Villero is like sniffing a handful of Langhe soil. This wine speaks highly of its territory with aromas of smoke, earth, forest berry, white truffle, hazelnut, cola, root beer, humus and dried apple skins. In the mouth, the tannins are impressively polished and fine.\nPredicted: 60.76769 Actual: 65.0 \n\nA reasonably good buy in vintage Port, Silval is something of a second label for Quinta do Noval. It's less concentrated and suppler in the mouth than its sister wine, but still boasts compelling floral aromas allied to prune, berry and chocolate flavors. Worth trying a bottle before 2010.\nPredicted: 44.748398 Actual: 37.0 \n\nRustic tomato and cherry aromas make for a scratchy opening. This is wiry and grating in feel, with sour plum and red-currant flavors. A fresh but scratchy and herbal tasting finish isn't exactly friendly.\nPredicted: 10.840133 Actual: 13.0 \n\nFragrant and spicy, this opens with green pepper notes and boatloads of prickly/brambly raspberry/strawberry fruit. From a cool vintage, the blend includes 15% Syrah. It's nicely handled, with great penetration and a dancer's precision.\nPredicted: 27.572311 Actual: 25.0 \n\nThis is a cheerful, agreeable wine, with loose-knit fruit flavors of strawberry and sweet cranberry, interwoven with generous streaks of butter and toast. Drink up.\nPredicted: 20.560844 Actual: 15.0 \n\nThis terroir-driven Pinot, from the Oregon side of the Columbia River, shows a dramatically different profile than the Willamette Valley Pinots commonly associated with the state. Kudos to winemaker Peter Rosback for finding great Pinot all over Oregon. This unique vineyard shows classic varietal fruit laced with mineral, kissed with toast, and finished with delicious mocha flavors from new oak barrels. The tart, tangy spine keeps it balanced and extends the fruit flavors well into a long, crisp finish.\nPredicted: 38.203552 Actual: 42.0 \n\nHere's a modern, round and velvety Barolo (from Monforte d'Alba) that will appeal to those who love a thick and juicy style of wine. The aromas include lavender, allspice, cinnamon, white chocolate and vanilla. Tart berry flavors backed by crisp acidity and firm tannins give the mouthfeel determination and grit.\nPredicted: 41.028854 Actual: 49.0 \n\nJammy and simple, with ripe cherry, blackberry and pepper flavors. Nothing terribly complex, but an easy everyday Zin at a decent price.\nPredicted: 9.812495 Actual: 11.0 \n\nMade in a sweeter, more candied style, with lemon, pineapple and vanilla flavors, although crisp acidity provides much needed and drying balance. But it's still a fairly simple sipper.\nPredicted: 19.652079 Actual: 21.0 \n\nIt's obvious from the alcohol that the Merlot here was very ripe. But, happily, it doesn't taste that way. The balance of rich raisin and red plum flavors are well balanced by acidity. There is some smokiness from wood, the ripeness finishing freshly with a black cherry taste.\nPredicted: 41.17104 Actual: 26.0 \n\n"
],
[
"# Compare the average difference between actual price and the model's predicted price\nprint('Average prediction difference: ', diff / num_predictions)",
"Average prediction difference: 8.719513726234435\n"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbf2ff2159e99137c0773b6a8e310acf8232eee7
| 81,646 |
ipynb
|
Jupyter Notebook
|
nlp.ipynb
|
joeytai/Machine-Learning
|
26e9e93b6b37c36d903177906b9e82c4db95ae23
|
[
"MIT"
] | null | null | null |
nlp.ipynb
|
joeytai/Machine-Learning
|
26e9e93b6b37c36d903177906b9e82c4db95ae23
|
[
"MIT"
] | null | null | null |
nlp.ipynb
|
joeytai/Machine-Learning
|
26e9e93b6b37c36d903177906b9e82c4db95ae23
|
[
"MIT"
] | null | null | null | 41.63488 | 1,783 | 0.508035 |
[
[
[
"import requests\n# 585247235\n# 245719505、\n# 观视频工作室 房价 543149108\n# https://api.bilibili.com/x/v2/reply/reply?callback=jQuery17206010832908607249_1608646550339&jsonp=jsonp&pn=1&type=1&oid=585247235&\nheader = {\"User-Agent\": \"Mozilla/5.0 (X11; Ubuntu; Linux x86_64; rv:22.0) Gecko/20100101 Firefox/22.0\",\n\"Cookie\": \"\"}\ncomments = []\noriginal_url = \"https://api.bilibili.com/x/v2/reply?jsonp=jsonp&type=1&oid=543149108&sort=2&pn=\"\n\nfor page in range(1, 60): # 页码这里就简单处理了\n url = original_url + str(page)\n print(url)\n try:\n html = requests.get(url, headers=header)\n data = html.json()\n if data['data']['replies']:\n for i in data['data']['replies']:\n comments.append(i['content']['message'])\n except Exception as err:\n print(url)\n print(err)",
"https://api.bilibili.com/x/v2/reply?jsonp=jsonp&type=1&oid=543149108&sort=2&pn=1\nhttps://api.bilibili.com/x/v2/reply?jsonp=jsonp&type=1&oid=543149108&sort=2&pn=2\nhttps://api.bilibili.com/x/v2/reply?jsonp=jsonp&type=1&oid=543149108&sort=2&pn=3\nhttps://api.bilibili.com/x/v2/reply?jsonp=jsonp&type=1&oid=543149108&sort=2&pn=4\nhttps://api.bilibili.com/x/v2/reply?jsonp=jsonp&type=1&oid=543149108&sort=2&pn=5\nhttps://api.bilibili.com/x/v2/reply?jsonp=jsonp&type=1&oid=543149108&sort=2&pn=6\nhttps://api.bilibili.com/x/v2/reply?jsonp=jsonp&type=1&oid=543149108&sort=2&pn=7\nhttps://api.bilibili.com/x/v2/reply?jsonp=jsonp&type=1&oid=543149108&sort=2&pn=8\nhttps://api.bilibili.com/x/v2/reply?jsonp=jsonp&type=1&oid=543149108&sort=2&pn=9\nhttps://api.bilibili.com/x/v2/reply?jsonp=jsonp&type=1&oid=543149108&sort=2&pn=10\nhttps://api.bilibili.com/x/v2/reply?jsonp=jsonp&type=1&oid=543149108&sort=2&pn=11\nhttps://api.bilibili.com/x/v2/reply?jsonp=jsonp&type=1&oid=543149108&sort=2&pn=12\nhttps://api.bilibili.com/x/v2/reply?jsonp=jsonp&type=1&oid=543149108&sort=2&pn=13\nhttps://api.bilibili.com/x/v2/reply?jsonp=jsonp&type=1&oid=543149108&sort=2&pn=14\nhttps://api.bilibili.com/x/v2/reply?jsonp=jsonp&type=1&oid=543149108&sort=2&pn=15\nhttps://api.bilibili.com/x/v2/reply?jsonp=jsonp&type=1&oid=543149108&sort=2&pn=16\nhttps://api.bilibili.com/x/v2/reply?jsonp=jsonp&type=1&oid=543149108&sort=2&pn=17\nhttps://api.bilibili.com/x/v2/reply?jsonp=jsonp&type=1&oid=543149108&sort=2&pn=18\nhttps://api.bilibili.com/x/v2/reply?jsonp=jsonp&type=1&oid=543149108&sort=2&pn=19\nhttps://api.bilibili.com/x/v2/reply?jsonp=jsonp&type=1&oid=543149108&sort=2&pn=20\nhttps://api.bilibili.com/x/v2/reply?jsonp=jsonp&type=1&oid=543149108&sort=2&pn=21\nhttps://api.bilibili.com/x/v2/reply?jsonp=jsonp&type=1&oid=543149108&sort=2&pn=22\nhttps://api.bilibili.com/x/v2/reply?jsonp=jsonp&type=1&oid=543149108&sort=2&pn=23\nhttps://api.bilibili.com/x/v2/reply?jsonp=jsonp&type=1&oid=543149108&sort=2&pn=24\nhttps://api.bilibili.com/x/v2/reply?jsonp=jsonp&type=1&oid=543149108&sort=2&pn=25\nhttps://api.bilibili.com/x/v2/reply?jsonp=jsonp&type=1&oid=543149108&sort=2&pn=26\nhttps://api.bilibili.com/x/v2/reply?jsonp=jsonp&type=1&oid=543149108&sort=2&pn=27\nhttps://api.bilibili.com/x/v2/reply?jsonp=jsonp&type=1&oid=543149108&sort=2&pn=28\nhttps://api.bilibili.com/x/v2/reply?jsonp=jsonp&type=1&oid=543149108&sort=2&pn=29\nhttps://api.bilibili.com/x/v2/reply?jsonp=jsonp&type=1&oid=543149108&sort=2&pn=30\nhttps://api.bilibili.com/x/v2/reply?jsonp=jsonp&type=1&oid=543149108&sort=2&pn=31\nhttps://api.bilibili.com/x/v2/reply?jsonp=jsonp&type=1&oid=543149108&sort=2&pn=32\nhttps://api.bilibili.com/x/v2/reply?jsonp=jsonp&type=1&oid=543149108&sort=2&pn=33\nhttps://api.bilibili.com/x/v2/reply?jsonp=jsonp&type=1&oid=543149108&sort=2&pn=34\nhttps://api.bilibili.com/x/v2/reply?jsonp=jsonp&type=1&oid=543149108&sort=2&pn=35\nhttps://api.bilibili.com/x/v2/reply?jsonp=jsonp&type=1&oid=543149108&sort=2&pn=36\nhttps://api.bilibili.com/x/v2/reply?jsonp=jsonp&type=1&oid=543149108&sort=2&pn=37\nhttps://api.bilibili.com/x/v2/reply?jsonp=jsonp&type=1&oid=543149108&sort=2&pn=38\nhttps://api.bilibili.com/x/v2/reply?jsonp=jsonp&type=1&oid=543149108&sort=2&pn=39\nhttps://api.bilibili.com/x/v2/reply?jsonp=jsonp&type=1&oid=543149108&sort=2&pn=40\nhttps://api.bilibili.com/x/v2/reply?jsonp=jsonp&type=1&oid=543149108&sort=2&pn=41\nhttps://api.bilibili.com/x/v2/reply?jsonp=jsonp&type=1&oid=543149108&sort=2&pn=42\nhttps://api.bilibili.com/x/v2/reply?jsonp=jsonp&type=1&oid=543149108&sort=2&pn=43\nhttps://api.bilibili.com/x/v2/reply?jsonp=jsonp&type=1&oid=543149108&sort=2&pn=44\nhttps://api.bilibili.com/x/v2/reply?jsonp=jsonp&type=1&oid=543149108&sort=2&pn=45\nhttps://api.bilibili.com/x/v2/reply?jsonp=jsonp&type=1&oid=543149108&sort=2&pn=46\nhttps://api.bilibili.com/x/v2/reply?jsonp=jsonp&type=1&oid=543149108&sort=2&pn=47\nhttps://api.bilibili.com/x/v2/reply?jsonp=jsonp&type=1&oid=543149108&sort=2&pn=48\nhttps://api.bilibili.com/x/v2/reply?jsonp=jsonp&type=1&oid=543149108&sort=2&pn=49\nhttps://api.bilibili.com/x/v2/reply?jsonp=jsonp&type=1&oid=543149108&sort=2&pn=50\nhttps://api.bilibili.com/x/v2/reply?jsonp=jsonp&type=1&oid=543149108&sort=2&pn=51\nhttps://api.bilibili.com/x/v2/reply?jsonp=jsonp&type=1&oid=543149108&sort=2&pn=52\nhttps://api.bilibili.com/x/v2/reply?jsonp=jsonp&type=1&oid=543149108&sort=2&pn=53\nhttps://api.bilibili.com/x/v2/reply?jsonp=jsonp&type=1&oid=543149108&sort=2&pn=54\nhttps://api.bilibili.com/x/v2/reply?jsonp=jsonp&type=1&oid=543149108&sort=2&pn=55\nhttps://api.bilibili.com/x/v2/reply?jsonp=jsonp&type=1&oid=543149108&sort=2&pn=56\nhttps://api.bilibili.com/x/v2/reply?jsonp=jsonp&type=1&oid=543149108&sort=2&pn=57\nhttps://api.bilibili.com/x/v2/reply?jsonp=jsonp&type=1&oid=543149108&sort=2&pn=58\nhttps://api.bilibili.com/x/v2/reply?jsonp=jsonp&type=1&oid=543149108&sort=2&pn=59\n"
],
[
"url = 'https://m.weibo.cn/comments/hotflow?id=4595898757681897&mid=4595898757681897&max_id_type=0'\n# url = 'https://m.weibo.cn/profile/info?uid=5393135816'\nheader = {\"User-Agent\": \"Mozilla/5.0 (X11; Ubuntu; Linux x86_64; rv:22.0) Gecko/20100101 Firefox/22.0\",\n\"Cookie\": \"\"}\nhtml = requests.get(url, headers=header)",
"_____no_output_____"
],
[
"html.json()",
"_____no_output_____"
],
[
"for i in range(100):\n print(data['data']['replies'][i]['member']['vip']['vipType'])",
"0\n2\n1\n0\n2\n0\n0\n1\n2\n2\n1\n1\n2\n2\n2\n1\n1\n1\n0\n2\n"
],
[
"comments[:5]",
"_____no_output_____"
],
[
"type(comments)",
"_____no_output_____"
],
[
"len(comments)",
"_____no_output_____"
],
[
"from gensim.models import word2vec\nimport jieba\nfrom gensim.models.keyedvectors import KeyedVectors",
"_____no_output_____"
],
[
"name = 'fangjia'\nwith open(f'comments_{name}.txt', 'w') as f:\n for item in comments:\n f.write(\"%s\\n\" % item)",
"_____no_output_____"
],
[
"stopwords = stopwordslist('hit.txt')",
"_____no_output_____"
],
[
"# stopwords",
"_____no_output_____"
],
[
" #打开文件夹,读取内容,并进行分词\nfinal = []\nstopwords = stopwordslist('hit.txt')\nwith open(f'comments_{name}.txt') as f:\n for line in f.readlines():\n word = jieba.cut(line)\n for i in word:\n if i not in stopwords:\n# final = final + i +\" \"\n final.append(i)\n \n",
"_____no_output_____"
],
[
"# final\nwith open(f'fenci_comments_{name}.txt', 'w') as f:\n for item in final:\n f.write(\"%s\\n\" % item)",
"_____no_output_____"
],
[
"with open(f'fenci_comments_{name}.txt', 'r') as f:\n sentences = f.readlines()\n sentences = [s.split() for s in sentences]\n # size表示词向量维度 iter表示迭代次数\n model = word2vec.Word2Vec(sentences, window=5,size=300, min_count=2, iter=500, workers=3)\n #model = Word2Vec.load(\"word2vec.model\")\n #model.wv.save('vectors_300d_word2vec') # 保存训练过程\n #model.save('vectors_300d_word2vec') # 保存训练过程\n model.wv.save_word2vec_format('vectors_chenping_word2vec.txt') # 仅保留词向量\n",
"_____no_output_____"
],
[
"sentences=word2vec.Text8Corpus('comments_chenping.txt')",
"_____no_output_____"
],
[
"word_vectors = KeyedVectors.load_word2vec_format('', binary=False) ",
"_____no_output_____"
],
[
"word_vectors.most_similar('房价')",
"_____no_output_____"
],
[
"model.wv.similar_by_word('内卷', topn =100)",
"_____no_output_____"
],
[
"model.wv.similar_by_word('战争', topn =10)",
"_____no_output_____"
],
[
"model.wv.similar_by_word('战争', topn =10)",
"_____no_output_____"
],
[
"model.wv.similar_by_word('中国', topn =10)",
"_____no_output_____"
],
[
"model.wv.similar_by_word('中国', topn =10)",
"_____no_output_____"
],
[
"model.wv.similar_by_word('美国', topn =10)",
"_____no_output_____"
],
[
"model.wv.similar_by_word('美国', topn =10)",
"_____no_output_____"
],
[
"model.wv.similar_by_word('欧洲', topn =10)",
"_____no_output_____"
],
[
"model.wv.similar_by_word('联合国', topn =10)",
"_____no_output_____"
],
[
"model.wv.similar_by_word('历史', topn =10)",
"_____no_output_____"
],
[
"model.wv.similar_by_word('朝鲜', topn =10)",
"_____no_output_____"
],
[
"model.wv.similar_by_word('高晓松', topn =100)",
"_____no_output_____"
],
[
"model.wv.similar_by_word('公知', topn =10)",
"_____no_output_____"
],
[
"from collections import Counter\nimport jieba\njieba.load_userdict('userdict.txt')\n\n# 创建停用词list\ndef stopwordslist(filepath):\n stopwords = [line.strip() for line in open(filepath, 'r').readlines()]\n return stopwords\n\n# 对句子进行分词\ndef seg_sentence(sentence):\n sentence_seged = jieba.cut(sentence.strip())\n stopwords = stopwordslist('G:\\\\哈工大停用词表.txt') # 这里加载停用词的路径\n outstr = ''\n for word in sentence_seged:\n if word not in stopwords:\n if word != '\\t':\n outstr += word\n outstr += \" \"\n return outstr\n\n\ninputs = open('hebing_wenben\\\\wenben.txt', 'r') #加载要处理的文件的路径\noutputs = open('output.txt', 'w') #加载处理后的文件路径\nfor line in inputs:\n line_seg = seg_sentence(line) # 这里的返回值是字符串\n outputs.write(line_seg)\noutputs.close()\ninputs.close()\n# WordCount\nwith open('output.txt', 'r') as fr: #读入已经去除停用词的文件\n data = jieba.cut(fr.read())\ndata = dict(Counter(data))\n\nwith open('cipin.txt', 'w') as fw: #读入存储wordcount的文件路径\n for k, v in data.items():\n fw.write('%s,%d\\n' % (k, v))",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbf30fe0cba8da68f9e3dfa981a9088389315e8a
| 35,648 |
ipynb
|
Jupyter Notebook
|
k_pro_memo.ipynb
|
chun1182/start_ai_project
|
69c63d73d929f1bbf76c1d292a705fd5c3aa7fb3
|
[
"MIT"
] | null | null | null |
k_pro_memo.ipynb
|
chun1182/start_ai_project
|
69c63d73d929f1bbf76c1d292a705fd5c3aa7fb3
|
[
"MIT"
] | null | null | null |
k_pro_memo.ipynb
|
chun1182/start_ai_project
|
69c63d73d929f1bbf76c1d292a705fd5c3aa7fb3
|
[
"MIT"
] | 2 |
2020-01-01T23:47:18.000Z
|
2020-01-07T01:27:23.000Z
| 24.233855 | 297 | 0.37946 |
[
[
[
"#入力\na,b=map(int, input().split())\nc=list(map(int, input().split()))\nprint(a,b,c)",
"1 5\n1 5 6 2\n1 5 [1, 5, 6, 2]\n"
],
[
"#初期化\na=[0]*5 \nb=a\nb2=a[:]\na[1]=3\nprint('b:{}, b2:{}'.format(b,b2))\n\nimport copy\na= [[0]*3 for i in range(5)] #2次元配列はこう準備、[[0]*5]*5\nb=copy.deepcopy(a)\na[1][0]=5\nprint(b)",
"b:[0, 3, 0, 0, 0], b2:[0, 0, 0, 0, 0]\n[[0, 0, 0], [0, 0, 0], [0, 0, 0], [0, 0, 0], [0, 0, 0]]\n"
],
[
"#内包表記奇数のみ\nodd=[i for i in range(20) if i%2==1]\nprint(a)\n#ソート\nw=[[1, 2], [2, 6] , [3, 6], [4, 5], [5, 7]]\nw.sort()\nprint(w)\nw.sort(key=lambda x:x[1],reverse=True)\nprint(w)",
"[1, 3, 5, 7, 9, 11, 13, 15, 17, 19]\n[[1, 2], [2, 6], [3, 6], [4, 5], [5, 7]]\n[[5, 7], [2, 6], [3, 6], [4, 5], [1, 2]]\n"
],
[
"#二重ループ\nn,y=1000, 1234000\nfor i in range(n+1):\n for j in range(n-i+1):\n if y-10000*i-5000*j==1000*(n-i-j):\n print(i, j, n-i-j)\n break\n else:\n continue\n break\nelse:\n print('-1 -1 -1')",
"2 54 944\n"
],
[
"#二部探索\nimport bisect\na = [1, 2, 3, 5, 6, 7, 8, 9]\nb=bisect.bisect_left(a, 8)\nbisect.insort_left(a, 4)\nprint(a,b)",
"[1, 2, 3, 4, 5, 6, 7, 8, 9] 6\n"
],
[
"%%time\n#素数\nn = 10**6\nprimes = set(range(2, n+1))\nfor i in range(2, int(n**0.5+1)):\n primes.difference_update(range(i*2, n+1, i))\nprimes=list(primes)\n#print(primes)",
"Wall time: 695 ms\n"
],
[
"#combinations、組み合わせ、順列\nfrom itertools import permutations, combinations,combinations_with_replacement,product\na=['a','b','C']\nprint(list(permutations(a)))\nprint(list(combinations(a,2)))\nprint(list(combinations_with_replacement(a,3)))\nprint(list(product(['a','b','C'],repeat=2)))\n",
"[('a', 'b', 'C'), ('a', 'C', 'b'), ('b', 'a', 'C'), ('b', 'C', 'a'), ('C', 'a', 'b'), ('C', 'b', 'a')]\n[('a', 'b'), ('a', 'C'), ('b', 'C')]\n[('a', 'a', 'a'), ('a', 'a', 'b'), ('a', 'a', 'C'), ('a', 'b', 'b'), ('a', 'b', 'C'), ('a', 'C', 'C'), ('b', 'b', 'b'), ('b', 'b', 'C'), ('b', 'C', 'C'), ('C', 'C', 'C')]\n[('a', 'a'), ('a', 'b'), ('a', 'C'), ('b', 'a'), ('b', 'b'), ('b', 'C'), ('C', 'a'), ('C', 'b'), ('C', 'C')]\n"
],
[
"# 0埋め\na=100\nb=0.987654321\nprint('{0:06d}-{1:6f}'.format(a,b))",
"000100-0.987654\n"
],
[
"#最大公約数、最小公倍数、階乗\nimport fractions, math\na,b=map(int, input().split())\nf=fractions.gcd(a,b)\nf2=a*b//f\nprint(f,f2)\nprint(math.factorial(5))",
"72 50\n2 1800\n120\n"
],
[
"#文字式を評価\na=eval('1*2*3')\nprint(a)",
"6\n"
],
[
"from collections import Counter\na=[2,2,2,3,4,3,1,2,1,3,1,2,1,2,2,1,2,1]\na=Counter(a)\nfor i in a.most_common(3):print(i)",
"(2, 8)\n(1, 6)\n(3, 3)\n"
],
[
"import numpy as np\ns=[list(input()) for i in range(4)]\ns=np.array(s)\ns=s[::-1,:].T",
"1234\n1234\n1234\n1234\n"
],
[
"i=0\nj=1\nnp.sum(s[:3,:2]=='5')\n",
"_____no_output_____"
],
[
"#最短経路\nys,xs=2,2\nyg,xg=4,5\na=['########', '#......#', '#.######', '#..#...#', '#..##..#', '##.....#', '########']\nn=[(ys-1,xs-1)]\nroute={n[0]:0}\np=[[1,0],[0,1],[-1,0],[0,-1]]\ncount=1\nwhile route.get((yg-1,xg-1),0)==0 and count != 10000:\n n2=[]\n for i in n:\n for j in p:\n np=(i[0]+j[0],i[1]+j[1])\n if a[np[0]][np[1]]=='.' and route.get(np,-1)==-1:\n n2.append(np)\n route[np]=count\n n=n2\n count+=1\nprint(n,route)",
"[(3, 6), (3, 4)] {(1, 1): 0, (2, 1): 1, (1, 2): 1, (3, 1): 2, (1, 3): 2, (4, 1): 3, (3, 2): 3, (1, 4): 3, (4, 2): 4, (1, 5): 4, (5, 2): 5, (1, 6): 5, (5, 3): 6, (5, 4): 7, (5, 5): 8, (5, 6): 9, (4, 5): 9, (4, 6): 10, (3, 5): 10, (3, 6): 11, (3, 4): 11}\n"
],
[
"a",
"_____no_output_____"
],
[
"#最短経路\nW,H=4,5\nstep={(2,2):0}\nroute=[[(2,2)]]\n#map=[[0 for i in range(W)] for j in range(H)]\nmap=[[0, 0, 0, 0], [1, 1, 0, 0], [0, 0, 0, 1], [0, 0, 1, 1], [0, 0, 0, 0]]\nprint(map)\n\nfor i in range(15):\n next_list=[]\n for j in range(len(route[i])):\n next_list.append((route[i][j][0]+1,route[i][j][1]))\n next_list.append((route[i][j][0] ,route[i][j][1]+1))\n next_list.append((route[i][j][0]-1,route[i][j][1]))\n next_list.append((route[i][j][0] ,route[i][j][1]-1))\n s=set(next_list)\n #print(s) \n\n n_list=[]\n for l, (j,k) in enumerate(next_list):\n if W>j>=0 and H>k>=0 :\n #print(map[j][k])\n if map[k][j]==0:\n n_list.append(next_list[l])\n n_list=sorted(set(n_list),key=n_list.index)\n\n remove=[]\n for l in n_list:\n if step.setdefault(l, i+1) <i+1:\n remove.append(l)\n for l in remove:\n n_list.remove(l)\n route.append(n_list)\n print(i)\n if (3,4) in step:\n break\nprint(step)\nprint(route) \nprint(len(step.keys()))\nprint(step.get((3,4),-1))",
"[[0, 0, 0, 0], [1, 1, 0, 0], [0, 0, 0, 1], [0, 0, 1, 1], [0, 0, 0, 0]]\n0\n1\n2\n3\n4\n{(2, 2): 0, (1, 2): 1, (2, 1): 1, (1, 3): 2, (0, 2): 2, (3, 1): 2, (2, 0): 2, (1, 4): 3, (0, 3): 3, (3, 0): 3, (1, 0): 3, (2, 4): 4, (0, 4): 4, (0, 0): 4, (3, 4): 5}\n[[(2, 2)], [(1, 2), (2, 1)], [(1, 3), (0, 2), (3, 1), (2, 0)], [(1, 4), (0, 3), (3, 0), (1, 0)], [(2, 4), (0, 4), (0, 0)], [(3, 4)]]\n15\n5\n"
],
[
"#深さ探索\n#q=list()\n#for i in range(5):\n# q.append(set(map(int, input().split())))\nq=[{1, 3}, {1, 4}, {9, 5}, {5, 2}, {6, 5},{3,5},{8,9},{7,9}]\ncount=0\nwhile count!=10000:\n a=q.pop()\n for j in q:\n if len(j&a) != 0:\n j |=a\n count=0\n break\n else:q=[a]+q\n if count>len(q): break\n count+=1\n print(count,q)",
"1 [{1, 3}, {1, 4}, {9, 5, 7}, {2, 5}, {5, 6}, {3, 5}, {8, 9}]\n1 [{1, 3}, {1, 4}, {5, 7, 8, 9}, {2, 5}, {5, 6}, {3, 5}]\n1 [{1, 3, 5}, {1, 4}, {5, 7, 8, 9}, {2, 5}, {5, 6}]\n1 [{1, 3, 5, 6}, {1, 4}, {5, 7, 8, 9}, {2, 5}]\n1 [{1, 2, 3, 5, 6}, {1, 4}, {5, 7, 8, 9}]\n1 [{1, 2, 3, 5, 6, 7, 8, 9}, {1, 4}]\n1 [{1, 2, 3, 4, 5, 6, 7, 8, 9}]\n2 [{1, 2, 3, 4, 5, 6, 7, 8, 9}]\n"
],
[
"#深さ探索2\n#n=int(input())\n#pt=[[] for i in range(n)]\n#for i in range(n-1):\n# a,b=map(int,input().split())\n# pt[a-1].append(b-1)\n# pt[b-1].append(a-1)\nn=7\npt=[[1, 2, 3], [0], [5, 0], [6, 0], [6], [2], [3, 4]]\ndef dfs(v):\n d=[-1 for i in range(n)]\n q=[]\n d[v]=0\n q.append(v)\n while q:\n v=q.pop()\n for i in pt[v]:\n if d[i]==-1:\n d[i]=d[v]+1\n q.append(i)\n print(d,q)\n return d\nprint(dfs(0))",
"[0, 1, 1, 1, -1, -1, -1] [1, 2, 3]\n[0, 1, 1, 1, -1, -1, 2] [1, 2, 6]\n[0, 1, 1, 1, 3, -1, 2] [1, 2, 4]\n[0, 1, 1, 1, 3, -1, 2] [1, 2]\n[0, 1, 1, 1, 3, 2, 2] [1, 5]\n[0, 1, 1, 1, 3, 2, 2] [1]\n[0, 1, 1, 1, 3, 2, 2] []\n[0, 1, 1, 1, 3, 2, 2]\n"
],
[
"#幅優先探索\nn=7\npt=[[1, 2, 3], [0], [5, 0], [6, 0], [6], [2], [3, 4]]\ndef bfs(v):\n d=[-1]*n\n d[v]=0\n q=[v]\n c=1\n while q:\n q1=[]\n for i in q:\n for j in pt[i]:\n if d[j]==-1:\n d[j]=c\n q1.append(j)\n q=q1\n c+=1\n print(d,q)\n return d\nprint(bfs(0))",
"[0, 1, 1, 1, -1, -1, -1] [1, 2, 3]\n[0, 1, 1, 1, -1, 2, 2] [5, 6]\n[0, 1, 1, 1, 3, 2, 2] [4]\n[0, 1, 1, 1, 3, 2, 2] []\n[0, 1, 1, 1, 3, 2, 2]\n"
],
[
"#周辺埋め\nh,w=map(int, input().split())\ns = [\".\"*(w+2)]+[\".\"+input()+\".\" for i in range(h)]+[\".\"*(w+2)]\nprint(s)",
"2 3\n...\n###\n['.....', '.....', '.###.', '.....']\n"
],
[
"h,w=map(int, input().split())\ns = [\".\"*(w+2)]+[\".\"+input()+\".\" for i in range(h)]+[\".\"*(w+2)]\ns2 = [[0 for i in range(w+2)] for i in range(h+2)]\nprint(s)\nfor i in range(h+2):\n for j in range(w+2):\n if s[i][j]=='#':\n s2[i][j]=1\nprint(s2)",
"2 3\n...\n###\n['.....', '.....', '.###.', '.....']\n[[0, 0, 0, 0, 0], [0, 0, 0, 0, 0], [0, 1, 1, 1, 0], [0, 0, 0, 0, 0]]\n"
],
[
"import random\nprint([random.randint(0,1) for i in range(10)])",
"[0, 0, 1, 1, 1, 0, 1, 0, 1, 0]\n"
],
[
"import random\ncount=0\nfor i in range(1000000):\n a=1\n while a==1:\n a=random.randint(0,1)\n #print(a,end=(' '))\n if a==1:\n count+=1\n \nprint(count)",
"999729\n"
],
[
"#しゃくとり方\nn=int(input())\na=list(map(int, input().split()))\ncount=0\nright=0\nfor left in range(n):\n if right==left: right+=1\n while right<n and a[right-1]<a[right]:\n right+=1\n count+=right-left\nprint(count)",
"6\n5 6 1 2 3 4\n13\n"
],
[
"#しゃくとり、同じものを保存\nn=int(input())\na=list(map(int, input().split()))\n\ncount=0\nright=0\nm=dict()\nfor left in range(n):\n while right<n and m.get(a[right],0)==0:\n m[a[right]]=m.get(a[right],0)+1\n right+=1\n count=max(count,right-left)\n m[a[left]]=m.get(a[left],1)-1\nprint(count)",
"_____no_output_____"
],
[
"# 累積和\nb=list(range(1,30))\nimport numpy\nb2=numpy.cumsum([0]+b)\na2=[0]\nfor i in b:a2.append(a2[-1]+i)\nprint(b2)\nprint(a2)",
"[ 0 1 3 6 10 15 21 28 36 45 55 66 78 91 105 120 136 153\n 171 190 210 231 253 276 300 325 351 378 406 435]\n[0, 1, 3, 6, 10, 15, 21, 28, 36, 45, 55, 66, 78, 91, 105, 120, 136, 153, 171, 190, 210, 231, 253, 276, 300, 325, 351, 378, 406, 435]\n"
],
[
"#DP1\nn=6\nw=8\nweight=[2,1,3,2,1,5]\nvalue=[3,2,6,1,3,85]\n\ndp=[[0 for i in range(w+1)] for j in range(n+1)]\nfor i in range(n):\n for j in range(w+1):\n if j>=weight[i] : dp[i+1][j]=max(dp[i][j-weight[i]]+value[i],dp[i][j])\n else: dp[i+1][j]=dp[i][j]\n print(dp[:i+2])\nprint(dp[n][w])",
"[[0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 3, 3, 3, 3, 3, 3, 3]]\n[[0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 3, 3, 3, 3, 3, 3, 3], [0, 2, 3, 5, 5, 5, 5, 5, 5]]\n[[0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 3, 3, 3, 3, 3, 3, 3], [0, 2, 3, 5, 5, 5, 5, 5, 5], [0, 2, 3, 6, 8, 9, 11, 11, 11]]\n[[0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 3, 3, 3, 3, 3, 3, 3], [0, 2, 3, 5, 5, 5, 5, 5, 5], [0, 2, 3, 6, 8, 9, 11, 11, 11], [0, 2, 3, 6, 8, 9, 11, 11, 12]]\n[[0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 3, 3, 3, 3, 3, 3, 3], [0, 2, 3, 5, 5, 5, 5, 5, 5], [0, 2, 3, 6, 8, 9, 11, 11, 11], [0, 2, 3, 6, 8, 9, 11, 11, 12], [0, 3, 5, 6, 9, 11, 12, 14, 14]]\n[[0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 3, 3, 3, 3, 3, 3, 3], [0, 2, 3, 5, 5, 5, 5, 5, 5], [0, 2, 3, 6, 8, 9, 11, 11, 11], [0, 2, 3, 6, 8, 9, 11, 11, 12], [0, 3, 5, 6, 9, 11, 12, 14, 14], [0, 3, 5, 6, 9, 85, 88, 90, 91]]\n91\n"
],
[
"#DP2\nn=5\na=[7,5,3,1,8]\nA=12\ndp=[[30 for i in range(A+1)] for j in range(n+1)]\ndp[0][0]=0\n\nfor i in range(n):\n for j in range(A+1):\n if j>=a[i] : dp[i+1][j]=min(dp[i][j-a[i]]+1,dp[i][j])\n else: dp[i+1][j]=dp[i][j]\n print(dp[:i+2])\nprint(dp[n][A])",
"[[0, 30, 30, 30, 30, 30, 30, 30, 30, 30, 30, 30, 30], [0, 30, 30, 30, 30, 30, 30, 1, 30, 30, 30, 30, 30]]\n[[0, 30, 30, 30, 30, 30, 30, 30, 30, 30, 30, 30, 30], [0, 30, 30, 30, 30, 30, 30, 1, 30, 30, 30, 30, 30], [0, 30, 30, 30, 30, 1, 30, 1, 30, 30, 30, 30, 2]]\n[[0, 30, 30, 30, 30, 30, 30, 30, 30, 30, 30, 30, 30], [0, 30, 30, 30, 30, 30, 30, 1, 30, 30, 30, 30, 30], [0, 30, 30, 30, 30, 1, 30, 1, 30, 30, 30, 30, 2], [0, 30, 30, 1, 30, 1, 30, 1, 2, 30, 2, 30, 2]]\n[[0, 30, 30, 30, 30, 30, 30, 30, 30, 30, 30, 30, 30], [0, 30, 30, 30, 30, 30, 30, 1, 30, 30, 30, 30, 30], [0, 30, 30, 30, 30, 1, 30, 1, 30, 30, 30, 30, 2], [0, 30, 30, 1, 30, 1, 30, 1, 2, 30, 2, 30, 2], [0, 1, 30, 1, 2, 1, 2, 1, 2, 3, 2, 3, 2]]\n[[0, 30, 30, 30, 30, 30, 30, 30, 30, 30, 30, 30, 30], [0, 30, 30, 30, 30, 30, 30, 1, 30, 30, 30, 30, 30], [0, 30, 30, 30, 30, 1, 30, 1, 30, 30, 30, 30, 2], [0, 30, 30, 1, 30, 1, 30, 1, 2, 30, 2, 30, 2], [0, 1, 30, 1, 2, 1, 2, 1, 2, 3, 2, 3, 2], [0, 1, 30, 1, 2, 1, 2, 1, 1, 2, 2, 2, 2]]\n2\n"
],
[
"#ビット演算、式を計算\nfrom itertools import permutations, combinations,combinations_with_replacement,product\na=list(product(['+','-'],repeat=3))\ns=['5', '5', '3', '4']\nfor i in a:\n ans=s[0]+i[0]+s[1]+i[1]+s[2]+i[2]+s[3]\n if eval(ans)==7:\n print(ans+'=7')\n break",
"5-5+3+4=7\n"
],
[
"#ワーシャルフロイト法\nimport random\nn=int(input())\n#c=[list(map(int, input().split())) for i in range(n)]\nc=[[random.randint(1, 10) for i in range(n)] for i in range(n)]\nc[0][4]=0\nc[0][3]=0\nc[0][2]=0\nprint(c)\nfor k in range(n):\n for i in range(n):\n for j in range(n):\n c[i][j]=min(c[i][j],c[i][k]+c[k][j])\nfor i in c:\n print(i)\n\nfrom scipy.sparse.csgraph import floyd_warshall \ncost=floyd_warshall(c)\ncost",
"5\n[[7, 10, 0, 0, 0], [3, 5, 7, 6, 10], [5, 8, 9, 9, 9], [1, 10, 6, 5, 2], [10, 6, 5, 5, 6]]\n[1, 6, 0, 0, 0]\n[3, 5, 3, 3, 3]\n[5, 8, 5, 5, 5]\n[1, 7, 1, 1, 1]\n[6, 6, 5, 5, 6]\n"
],
[
"#ベルマンフォード法\ndef BF(p,n,s):\n inf=float(\"inf\")\n d=[inf for i in range(n)]\n d[s-1]=0\n for i in range(n+1):\n for e in p:\n if e[0]!=inf and d[e[1]-1]>d[e[0]-1]+e[2]:\n d[e[1]-1] = d[e[0]-1] + e[2]\n if i==n-1:t=d[-1]\n if i==n and t!=d[-1]:\n return [0,'inf']\n return list(map(lambda x:-x, d))\n\nn,m=map(int, input().split())\na=[list(map(int, input().split())) for i in range(m)]\na=[[x,y,-z] for x,y,z in a]\nprint(BF(a, n, 1)[-1])",
"2 2\n1 2 1\n2 1 1\ninf\n"
],
[
"# ダイクストラ法\nmp2=[[2, 4, 2], [3, 4, 5], [3, 2, 1], [1, 3, 2], [2, 0, 8], [0, 2, 8], [1, 2, 4], [0, 1, 3]]\n\nfrom heapq import heappop, heappush\ninf=float('inf')\nd=[inf for i in range(5)]\nd[0]=0\nprev=[None]*5\np=dict()\nfor i,j,k in mp2: p[i]=p.get(i,[])+[(j,k)]\nprint(p)\n\nq=[]\nheappush(q,(d[0],0))\nwhile q:\n print(q,d,prev)\n du, u = heappop(q)\n if d[u]<du: continue\n for v,weight in p.get(u,[]):\n alt=du+weight\n if d[v]>alt:\n d[v]=alt\n prev[v]=u\n heappush(q, (alt,v))\nprint('p',p)\np=[4]\nwhile prev[p[-1]]!=None: p.append(prev[p[-1]])\nprint('最短経路',*p[::-1])\nprint('最短距離',d)",
"{2: [(4, 2), (0, 8)], 3: [(4, 5), (2, 1)], 1: [(3, 2), (2, 4)], 0: [(2, 8), (1, 3)]}\n[(0, 0)] [0, inf, inf, inf, inf] [None, None, None, None, None]\n[(3, 1), (8, 2)] [0, 3, 8, inf, inf] [None, 0, 0, None, None]\n[(5, 3), (8, 2), (7, 2)] [0, 3, 7, 5, inf] [None, 0, 1, 1, None]\n[(6, 2), (7, 2), (10, 4), (8, 2)] [0, 3, 6, 5, 10] [None, 0, 3, 1, 3]\n[(7, 2), (8, 2), (10, 4), (8, 4)] [0, 3, 6, 5, 8] [None, 0, 3, 1, 2]\n[(8, 2), (8, 4), (10, 4)] [0, 3, 6, 5, 8] [None, 0, 3, 1, 2]\n[(8, 4), (10, 4)] [0, 3, 6, 5, 8] [None, 0, 3, 1, 2]\n[(10, 4)] [0, 3, 6, 5, 8] [None, 0, 3, 1, 2]\np {2: [(4, 2), (0, 8)], 3: [(4, 5), (2, 1)], 1: [(3, 2), (2, 4)], 0: [(2, 8), (1, 3)]}\n最短経路 0 1 3 2 4\n最短距離 [0, 3, 6, 5, 8]\n"
],
[
"#ダイクストラscipy\nfrom scipy.sparse import csr_matrix\nfrom scipy.sparse.csgraph import dijkstra\nimport numpy as np\nmp2=[[2, 4, 2], [3, 4, 5], [3, 2, 1], [1, 3, 2], [2, 0, 8], [0, 2, 8], [1, 2, 4], [0, 1, 3]]\nin_,out,weight=zip(*mp2)\ngraph = csr_matrix((weight, (in_, out)), shape=(5, 5), dtype=np.int64)\ndists = dijkstra(graph, indices=0)\nprint(dists)",
"_____no_output_____"
],
[
"p",
"_____no_output_____"
],
[
"#複数の文字列を変換\nS='54IZSB'\nS = S.translate(str.maketrans(\"ODIZSB\",\"001258\"))\nprint(S)",
"541258\n"
],
[
"#素因数分解\npf={}\nm=341555136\nfor i in range(2,int(m**0.5)+1):\n while m%i==0:\n pf[i]=pf.get(i,0)+1\n m//=i\nif m>1:pf[m]=1\nprint(pf)",
"{2: 6, 3: 1, 13: 1, 136841: 1}\n"
],
[
"#組み合わせのmod対応\ndef framod(n, mod, a=1):\n for i in range(1,n+1):\n a=a * i % mod\n return a\n\ndef power(n, r, mod):\n if r == 0: return 1\n if r%2 == 0:\n return power(n*n % mod, r//2, mod) % mod\n if r%2 == 1:\n return n * power(n, r-1, mod) % mod\n \ndef comb(n, k, mod):\n a=framod(n, mod)\n b=framod(k, mod)\n c=framod(n-k, mod)\n return (a * power(b, mod-2, mod) * power(c, mod-2, mod)) % mod\n\nprint(comb(10**5,5000,10**9+7))\nprint(comb(100000, 50000,10**9+7))\n",
"434941573\n149033233\n"
],
[
"#ヒープ 最小値の取り出し\nfrom heapq import heappop,heappush\nan=[[1,3],[2,1],[3,4]]\nplus,h=[],[]\n\nfor i,(a,b) in enumerate(an):\n plus.append(b)\n heappush(h,(a,i))\n\nans,k=0,7\nfor i in range(k):\n x,i=heappop(h)\n ans+=x\n heappush(h,(x+plus[i],i))\n print(ans,x)",
"1 1\n3 2\n6 3\n9 3\n13 4\n17 4\n22 5\n"
],
[
"#アルファベット\nal=[chr(ord('a') + i) for i in range(26)]\nprint(''.join(al))",
"abcdefghijklmnopqrstuvwxyz\n"
],
[
"#2で何回割れるか\nn=8896\nprint(bin(n),len(bin(n)),bin(n).rfind(\"1\"))\nprint(len(bin(n)) - bin(n).rfind(\"1\") - 1)\nwhile not n%2:\n n/=2\n print(n)",
"_____no_output_____"
],
[
"#union find\nclass UnionFind(object):\n def __init__(self, n=1):\n self.par = [i for i in range(n)]\n self.rank = [0 for _ in range(n)]\n\n def find(self, x):\n if self.par[x] == x:\n return x\n else:\n self.par[x] = self.find(self.par[x])\n return self.par[x]\n\n def union(self, x, y):\n x = self.find(x)\n y = self.find(y)\n if x != y:\n if self.rank[x] < self.rank[y]:\n x, y = y, x\n if self.rank[x] == self.rank[y]:\n self.rank[x] += 1\n self.par[y] = x\n\n def is_same(self, x, y):\n return self.find(x) == self.find(y)\n \nn,m=map(int, input().split())\np=list(map(int, input().split()))\nuf1=UnionFind(n)\nfor i in range(m):\n a,b=map(int, input().split())\n uf1.union(a-1,b-1)\ncount=0\nfor i in range(n):\n if uf1.is_same(i,p[i]-1):count+=1\nprint(count)\nprint(uf1.par)",
"5 2\n1 3 5 2 4\n1 3\n2 4\n2\n[0, 1, 0, 1, 4]\n"
],
[
"#組み合わせの数\nfrom scipy.special import comb\ncomb(10**5, 100, exact=True)%(10**9+7)",
"_____no_output_____"
],
[
"#マラソンマッチ\nfrom time import time\nst=time()\nnow=0\nwhile time() - st<2:\n now=(now+1)%10**10\nprint(now)",
"6332049\n"
],
[
"#n進数\nn=64\nk=-3\nbi=''\nwhile n!=0:\n bi+=str(n%abs(k))\n if k<0:n=-(-n//k)\n else:n=n//k\nprint(bi[::-1])",
"11101\n"
],
[
"#マンハッタン距離重心&cost\na=[5,2,7,2,12,5]\nimport numpy as np\nb=np.int64([0]+a).cumsum().cumsum()[:-1]\nc=np.int64([0]+a[::-1]).cumsum().cumsum()[:-1]\nprint(b+c[::-1], (b+c[::-1]).min(), (b+c[::-1]).argmin())\n\nb=sum(a)\nc=0\nfor j,i in enumerate(a):\n c+=i\n if c>b//2:break\nprint(sum([a[i]*abs(i-j) for i in range(len(a))]),j)",
"[95 72 53 48 47 70] 47 4\n47 4\n"
],
[
"#入力\nN, x, *A = map(int, open(0).read().split())",
"_____no_output_____"
],
[
"#メモ化再帰\nfrom functools import lru_cache\nimport sys\nsys.setrecursionlimit(10000)\n@lru_cache(maxsize=10000)\ndef fibm(n):\n if n<2:return n\n else:return (fibm(n-1) + fibm(n-2))%(10**9+7)\nprint(fibm(1200)%(10**9+7))",
"_____no_output_____"
],
[
"#上下左右 幅優先探索\nh,w=5,5\ns=[['#', '.', '.', '.', '#'],\n ['#', '.', '.', '#', '#'],\n ['.', '.', '.', '.', '.'],\n ['#', '#', '#', '.', '#'],\n ['.', '.', '.', '.', '#']]\nsta=(2,2)\np=[[-1]*w for i in range(h)]\nnp=[(1,0),(-1,0),(0,1),(0,-1)]\nq={sta}\np[sta[0]][sta[1]]=0\nstep=0\nwhile step<k and q:\n step+=1\n nq=set()\n while q:\n now=q.pop()\n for i,j in np:\n nx,ny=now[0]+i,now[1]+j\n if nx<0 or nx==h or ny<0 or ny==w:continue\n if s[nx][ny]=='.' and p[nx][ny]==-1:\n p[nx][ny]=step\n nq.add((nx,ny))\n q=nq.copy()\n print(q)\nfor i in range(h):\n for j in range(w):\n print('{:2d}'.format(p[i][j]),end=' ')\n print()",
"{(1, 2), (2, 3), (2, 1)}\n{(2, 0), (1, 1), (3, 3), (2, 4), (0, 2)}\n{(0, 1), (0, 3), (4, 3)}\n{(4, 2)}\n-1 3 2 3 -1 \n-1 2 1 -1 -1 \n 2 1 0 1 2 \n-1 -1 -1 2 -1 \n-1 -1 4 3 -1 \n"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbf3152c6954b8cf9eedbbfb0ca69ba655147a0c
| 63,962 |
ipynb
|
Jupyter Notebook
|
PythonSolution/Main.ipynb
|
A2Amir/Behavior-Planning-by-Finite-State-Machine
|
8f654556c3229875239054184ebd3193593137b9
|
[
"MIT"
] | 1 |
2021-05-12T13:13:55.000Z
|
2021-05-12T13:13:55.000Z
|
PythonSolution/Main.ipynb
|
A2Amir/Behavior-Planning-by-Finite-State-Machine
|
8f654556c3229875239054184ebd3193593137b9
|
[
"MIT"
] | null | null | null |
PythonSolution/Main.ipynb
|
A2Amir/Behavior-Planning-by-Finite-State-Machine
|
8f654556c3229875239054184ebd3193593137b9
|
[
"MIT"
] | 1 |
2020-11-05T16:14:33.000Z
|
2020-11-05T16:14:33.000Z
| 40.610794 | 54 | 0.053172 |
[
[
[
"import simulate_behavior",
"_____no_output_____"
],
[
"simulate_behavior.run_simulation(VISUALIZE=True)",
"000 - | | | | |\n | | | | |\n | | | | |\n | | | *** | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | 014 |\n | | | | |\n | | 006 | | 015 |\n | | | | |\n | | | | |\n | | | 010 | |\n | 001 | 007 | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n020 - | | | | 016 |\n | | | | |\n | 002 | | | |\n | | | 011 | |\n | | | | |\n | | | | |\n | 003 | | | |\n | | | | 017 |\n | | | | |\n | | | | 018 |\n | | | | |\n | | 008 | | |\n | | | | |\n | | | | |\n | | | 012 | 019 |\n | | 009 | | |\n | 004 | | | 020 |\n | | | | |\n | | | 013 | |\n | | | | |\n\n000 - | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | *** |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | 006 | | 014 |\n | | | | |\n020 - | | | | 015 |\n | 001 | | | |\n | | 007 | 010 | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | 002 | | | |\n | | | | 016 |\n | | | | |\n | | | 011 | |\n | 003 | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | 017 |\n | | | | |\n | | 008 | | 018 |\n | | | | |\n\n000 - | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n020 - | | | | *** |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | 006 | | |\n | | | | |\n | 001 | | | 014 |\n | | | | |\n | | 007 | | 015 |\n | | | 010 | |\n | | | | |\n | | | | |\n | | | | |\n | 002 | | | |\n | | | | |\n | | | | |\n | | | | |\n | 003 | | | 016 |\n | | | 011 | |\n\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n020 - | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | *** |\n | | | | |\n | | | | |\n | | 006 | | |\n | 001 | | | |\n | | | | |\n | | | | |\n | | 007 | | 014 |\n | | | | |\n | | | 010 | 015 |\n | | | | |\n040 - | 002 | | | |\n | | | | |\n | | | | |\n | | | | |\n | 003 | | | |\n | | | | |\n | | | | |\n | | | 011 | 016 |\n | | | | |\n\n | | | | |\n020 - | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | 001 | 006 | | *** |\n040 - | | | | |\n | | | | |\n | | | | |\n | | 007 | | |\n | | | | |\n | | | | 014 |\n | 002 | | 010 | |\n | | | | 015 |\n | | | | |\n | | | | |\n | 003 | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | 011 | |\n | | | | 016 |\n | | | | |\n | | | | |\n\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n040 - | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | 001 | | | |\n | | 006 | | *** |\n | | | | |\n | | | | |\n | | | | |\n | | 007 | | |\n | | | | |\n | 002 | | | |\n | | | | |\n | | | 010 | 014 |\n | | | | |\n | 003 | | | 015 |\n | | | | |\n | | | | |\n | | | | |\n060 - | | | | |\n | | | | |\n | | | | |\n | | | 011 | |\n | | | | |\n | | | | 016 |\n\n | | | | |\n | | | | |\n | | | | |\n040 - | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | 001 | | | |\n | | | | |\n | | 006 | | |\n | | | | |\n | | | | |\n | | | | |\n | | 007 | | *** |\n | 002 | | | |\n | | | | |\n060 - | | | | |\n | | | | |\n | 003 | | 010 | |\n | | | | 014 |\n | | | | |\n | | | | 015 |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | | |\n | | | 011 | |\n | 004 | | | |\n | | 008 | | |\n | | | | 016 |\n | | | | |\n | | | | |\n\n"
]
]
] |
[
"code"
] |
[
[
"code",
"code"
]
] |
cbf325cfb2ec5343292aa1ac12d9255719f1261f
| 3,388 |
ipynb
|
Jupyter Notebook
|
db/cleanup_raw_location_data.ipynb
|
danishabdullah/lunatic
|
0345d34ba402e0d33bc2028e09cf9be259ce1ec0
|
[
"MIT"
] | null | null | null |
db/cleanup_raw_location_data.ipynb
|
danishabdullah/lunatic
|
0345d34ba402e0d33bc2028e09cf9be259ce1ec0
|
[
"MIT"
] | null | null | null |
db/cleanup_raw_location_data.ipynb
|
danishabdullah/lunatic
|
0345d34ba402e0d33bc2028e09cf9be259ce1ec0
|
[
"MIT"
] | null | null | null | 19.033708 | 98 | 0.475207 |
[
[
[
"import pandas as pd",
"_____no_output_____"
],
[
"filepath = 'src/rawdata/location/cities1000-clean.tsv'\ndata = pd.read_csv(filepath, encoding='utf-8', header=None, sep='\\t', quotechar='\"' )",
"_____no_output_____"
],
[
"# to_be_lowered = [6]\n# for col in to_be_lowered:\n# data[col] = data[col].str.lower()\n",
"_____no_output_____"
],
[
"# fill_n_a = [4, 6, 7]\n# for col in fill_n_a:\n# data[col].fillna('N-A', inplace=True)",
"_____no_output_____"
],
[
"data.fillna(\"NULL\", inplace=True)\ndata.loc[data[6].isna(), 6] = 'na'",
"_____no_output_____"
],
[
"data.to_csv(filepath, encoding='utf-8', header=None, sep='\\t', quotechar='\"', index=False)",
"_____no_output_____"
],
[
"def date_reformat(value):\n day, month, year = value.split(\"/\")\n return f\"{year}-{month}-{day}\"\ndata[9] = data[9].apply(lambda x: date_reformat(x))",
"_____no_output_____"
],
[
"data.loc[data[3]=='NULL', 3] = ''",
"_____no_output_____"
],
[
"data[3].isna().describe()",
"_____no_output_____"
],
[
"len(data[3].unique())",
"_____no_output_____"
],
[
"data[3].shape",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbf32618af4091094c9813f4ede57085f6e418d4
| 31,186 |
ipynb
|
Jupyter Notebook
|
LinALgPython_JOCSON.ipynb
|
domsjcsn/Linear-ALgebra---Python
|
9f43c5cc6f0484cbe9a0d57c82685222a0242313
|
[
"Apache-2.0"
] | null | null | null |
LinALgPython_JOCSON.ipynb
|
domsjcsn/Linear-ALgebra---Python
|
9f43c5cc6f0484cbe9a0d57c82685222a0242313
|
[
"Apache-2.0"
] | null | null | null |
LinALgPython_JOCSON.ipynb
|
domsjcsn/Linear-ALgebra---Python
|
9f43c5cc6f0484cbe9a0d57c82685222a0242313
|
[
"Apache-2.0"
] | null | null | null | 23.952381 | 367 | 0.424453 |
[
[
[
"<a href=\"https://colab.research.google.com/github/domsjcsn/Linear-ALgebra---Python/blob/main/LinALgPython_JOCSON.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# **WELCOME TO PYTHON FUNDAMENTALS**\n\nIn this module, we are going to establish our skills in Python programming. In this notebook we are going to cover:\n\n* Variable and Data Types\n* Operations\n* Input and Output Operations\n* Logic Control\n* Iterables\n* Functions",
"_____no_output_____"
],
[
"## **Variables and Data Types**\n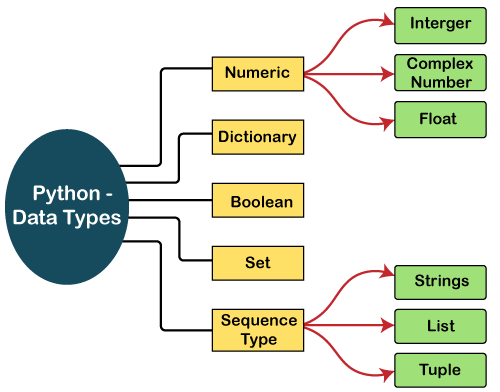 \n\n<blockquote> Variable and data types may be utilized by the user for giving definition, declaring, and executing mathematical terms and functions. Also, variables and data types are the given different values. You can store a value in a variable, which is also a memory location. And the variable and data types will depend on what the user will input. [1]",
"_____no_output_____"
]
],
[
[
"x = 1\na,b = 3, -2",
"_____no_output_____"
],
[
"type(x)",
"_____no_output_____"
],
[
"y = 3.0\ntype(y)",
"_____no_output_____"
],
[
"x = float(x)\ntype(x)\nx",
"_____no_output_____"
],
[
"s, t, u = \"1\", '3', 'three'\ntype(s) ",
"_____no_output_____"
]
],
[
[
"## **Operations**\n\n<blockquote>\nIn order to solve and calculate for a mathematical problem, mathematical process is applied. There are numerous operations that one can use, including the basics, which are the Addition, Multiplication, Subtradction, and Division. [2]",
"_____no_output_____"
],
[
"## **Arithmetic** \n\n=========================================\n\nArithmetic are symbols that indicate that a mathematical operation is required. \n",
"_____no_output_____"
]
],
[
[
"w, x, y,z = 4.0, -3.0, 2, -32",
"_____no_output_____"
],
[
"### Addition\nS = w + x\nS",
"_____no_output_____"
],
[
"### Subtraction",
"_____no_output_____"
],
[
"D = y - z\nD",
"_____no_output_____"
],
[
"### Multiplication\nP = w*z\nP",
"_____no_output_____"
],
[
"### Division\nQ = y/x\nQ",
"_____no_output_____"
]
],
[
[
" \n\n<blockquote>\nFloor division returns the highest possible integer. The floor division is indicated by the \"⌊ ⌋\" symbol. [3]",
"_____no_output_____"
]
],
[
[
"### Floor Division\nQf = w//x\nQf",
"_____no_output_____"
],
[
"### Exponentiation\nE = w**w\nE",
"_____no_output_____"
]
],
[
[
"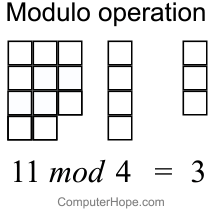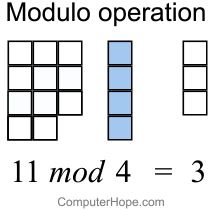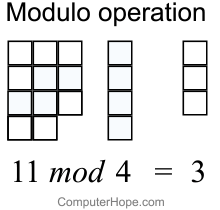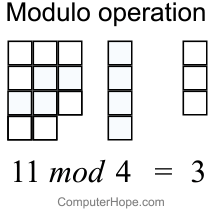 \n\n<blockquote>\nModelo operations is also a mathematical operation like the Floor division. The modelo operator is utilized to find the remainder of two divided numbers. It is indicated by the symbol \"%\". [4]",
"_____no_output_____"
]
],
[
[
"### Modulo\nmod = z%x\nmod",
"_____no_output_____"
]
],
[
[
"## ***Assignment*** \n\nThe Assignment operator is the operator used to assign a new value to a variable, property, event or indexer element. ",
"_____no_output_____"
]
],
[
[
"A, B, C, D, E = 0, 100, 2, 1, 2",
"_____no_output_____"
],
[
"A += w\nA",
"_____no_output_____"
],
[
"B -= x\nB",
"_____no_output_____"
],
[
"C *= w\nC",
"_____no_output_____"
],
[
"D /= x\nD",
"_____no_output_____"
],
[
"E **= y\nE",
"_____no_output_____"
]
],
[
[
"## ***Comparators***\n\n\nComparators are used to compare different types of values or objects based on one's liking. [6]",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
],
[
[
"size_1, size_2, size_3 = 1,2.0, \"1\"\ntrue_size = 1.0",
"_____no_output_____"
],
[
"## Equality \nsize_1 == true_size",
"_____no_output_____"
],
[
"## Non-Equality\nsize_2 != true_size",
"_____no_output_____"
],
[
"## Inequality\ns1 = size_1 > size_2\ns2 = size_1 < size_2/2\ns3 = true_size >= size_1\ns4 = size_2 <= true_size",
"_____no_output_____"
]
],
[
[
"## ***Logical***\n\nThis is a set of principles that depicts how components should be lay out so your device can execute tasks without failures. ",
"_____no_output_____"
]
],
[
[
"size_1 == true_size\nsize_1",
"_____no_output_____"
],
[
"size_1 is true_size",
"_____no_output_____"
],
[
"size_1 is not true_size",
"_____no_output_____"
],
[
"P, Q = True, False\nconj = P and Q\nconj",
"_____no_output_____"
],
[
"disj = P or Q\ndisj",
"_____no_output_____"
],
[
"nand = not(P and Q)\nnand",
"_____no_output_____"
],
[
"xor = (not P and Q) or (P and not Q)\nxor",
"_____no_output_____"
]
],
[
[
"## **Input & Output**\n\nInput refers to what the user loads to the program. And Output refers to what the program gives back to the user. ",
"_____no_output_____"
]
],
[
[
"print(\"Hello World!\")",
"Hello World!\n"
],
[
"cnt = 14000",
"_____no_output_____"
],
[
"string= \"Hello World!\"\nprint(string, \", Current COVID count is:\", cnt)\ncnt += 10000",
"Hello World! , Current COVID count is: 14000\n"
],
[
"print(f\"{string}, current count is: {cnt}\")",
"Hello World!, current count is: 24000\n"
],
[
"sem_grade = 86.25\nname = \"Franz\"\nprint(\"Hello {}, your semestral grade is: {}\".format(name, sem_grade))",
"Hello Franz, your semestral grade is: 86.25\n"
],
[
"pg, mg, fg = 0.3, 0.3, 0.4\nprint(\"The weights of your semestral grades are:\\\n\\n\\t {:.2%} for Prelim\\\n\\n\\t {:.2%} for Midterms, and\\\n\\n\\t {:.2%} for Finals.\".format(pg, mg, fg))",
"The weights of your semestral grades are:\n\t 30.00% for Prelim\n\t 30.00% for Midterms, and\n\t 40.00% for Finals.\n"
],
[
"e = input(\"Enter an number: \")\ne",
"Enter an number: 1\n"
],
[
"name = input(\"Enter your name: \")\nprelimsgrade =int(input(\"Enter your prelim grade: \"))\nmidtermsgrade =int(input(\"Enter your midterm grade: \"))\nfinalsgrade =int(input(\"Enter your finals grade: \"))\nsum = int(prelimsgrade + midtermsgrade + finalsgrade)\nsemester_grade = sum/3\nprint(\"Hello {}, your semestral grade is: {}\".format(name, semester_grade))",
"_____no_output_____"
]
],
[
[
"## ***Looping Statements*** \n<blockquote> \nLooping statements performs for a number of repetitions until specific conditions are met. \n\n",
"_____no_output_____"
],
[
"## ***While Statement*** \n\nWhile statement executes for a number of repetition until it reaches a specific condition that is false. ",
"_____no_output_____"
]
],
[
[
"## while loops \ni, j = 0, 10\nwhile(i<=j):\n print(f\"{i}\\t|\\t{j}\")\n i += 1",
"_____no_output_____"
]
],
[
[
"### ***For Statement***\n\nFor statement enables the looping of sets for a number of repetition until it achieves a false condition. ",
"_____no_output_____"
]
],
[
[
"# for(int = 0; i<10; i++){\n# printf(i)\n# }\n\ni = 0\nfor i in range(11):\n print(i)",
"_____no_output_____"
],
[
"playlist = [\"Beside You\", \"Meet me at our spot\", \"Sandali\"]\nprint('Now playing:\\n')\nfor song in playlist:\n print(song)",
"_____no_output_____"
]
],
[
[
"## **FLOW CONTROL**",
"_____no_output_____"
],
[
"## ***Conditional Statement***\n\nThe conditional statement decides whether certain statements need to be executed or not based on a given condition. ",
"_____no_output_____"
]
],
[
[
"num_1, num_2 = 12, 12\nif(num_1 == num_2):\n print(\"HAHA\")\n elif(num_1>num_2):\n print(\"HOHO\")\n else:\n print(\"HUHU\")",
"_____no_output_____"
]
],
[
[
"## ***FUNCTIONS***\n\nA function is a block of code that is utilized to reach a specific result or to execute a single action. ",
"_____no_output_____"
]
],
[
[
"# void DeleteUser (int userid){\n# delete(userid);\n# }\n\ndef delete_user (userid):\n print(\"Successfully delete user: {}\".format(userid))",
"_____no_output_____"
],
[
"user = 202014736 \ndelete_user(202014736)",
"_____no_output_____"
],
[
"def add(addend1, addend2):\n sum = addend1 + addend2\n return sum",
"_____no_output_____"
],
[
"add(3, 4)",
"_____no_output_____"
]
],
[
[
"## **REFERENCES**\n\n[1] Priya Pedamkar (2020). [Variable Types](https://www.educba.com/python-variable-types)\n\n[2] w3Schools (2021). [Operations](https://www.w3schools.com/python/python_operators.asp)\n\n[3] Python Tutorial (2021). [Floor Division](https://www.pythontutorial.net/advanced-python/python-floor-division/)\n\n[4] Tutorials Point (2021). [Comparators](https://www.tutorialspoint.com/python/python_basic_operators.htm)",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
]
] |
cbf340bcba62460b3393113983009a1982a5c52a
| 19,651 |
ipynb
|
Jupyter Notebook
|
onestop_data_clean.ipynb
|
ppiont/tensor-flow-state
|
5d94f1080aef6dab1214518eb6a9f92db33a58a9
|
[
"MIT"
] | null | null | null |
onestop_data_clean.ipynb
|
ppiont/tensor-flow-state
|
5d94f1080aef6dab1214518eb6a9f92db33a58a9
|
[
"MIT"
] | null | null | null |
onestop_data_clean.ipynb
|
ppiont/tensor-flow-state
|
5d94f1080aef6dab1214518eb6a9f92db33a58a9
|
[
"MIT"
] | null | null | null | 34.536028 | 241 | 0.420233 |
[
[
[
"<a href=\"https://colab.research.google.com/github/ppiont/tensor-flow-state/blob/master/onestop_data_clean.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"from google.colab import drive\ndrive.mount(\"/gdrive\", force_remount = True)",
"Mounted at /gdrive\n"
],
[
"%cd \"/gdrive/My Drive/tensor-flow-state/tensor-flow-state\"",
"/gdrive/My Drive/tensor-flow-state/tensor-flow-state\n"
],
[
"import pandas as pd\nimport numpy as np",
"_____no_output_____"
],
[
"data_dir = \"data/\"\n# Define sensors to process\nsensor_name_list = [\"RWS01_MONIBAS_0021hrl0414ra\", \"RWS01_MONIBAS_0021hrl0403ra\", \"RWS01_MONIBAS_0021hrl0409ra\", \"RWS01_MONIBAS_0021hrl0420ra\", \"RWS01_MONIBAS_0021hrl0426ra\"]",
"_____no_output_____"
]
],
[
[
"### ------------------------------------------------------------ START OF MESSING AROUND ------------------------------------------------------------",
"_____no_output_____"
]
],
[
[
"",
"_____no_output_____"
]
],
[
[
"### ------------------------------------------------------------ END OF MESSING AROUND ------------------------------------------------------------",
"_____no_output_____"
],
[
"### Clean sensor data",
"_____no_output_____"
]
],
[
[
"import datetime\ndef dateparse (time_in_secs):\n # Unix/epoch time to \"YYYY-MM-DD HH:MM:SS\"\n return datetime.datetime.fromtimestamp(float(time_in_secs))\n\ndef repair_datetime_index(df, freq = \"T\"):\n df = df.loc[~df.index.duplicated(keep = \"first\")] # remove duplicate date time indexes\n df = df.reindex(pd.date_range(start = df.index.min(), end = df.index.max(), freq = freq)) # add missing date time indexes\n df.index = df.index.tz_localize(\"UTC\").tz_convert(\"Europe/Amsterdam\")\n\n return df\n\ndef fix_values(df):\n # The order of these operations is currently important! Pay attention when making changes\n df[\"speed_limit\"] = np.where((df.index.hour < 19) & (df.index.hour >= 6), 100, 130)\n df.loc[df.flow < 0, \"flow\"] = np.nan # flow is either -2 (missing data) or 0 or positive. -2 to nan\n df.loc[df.speed < -1, \"speed\"] = np.nan # -2 (missing data) as well as oddities (-1.33, an average over -2 and -1 lanes?) to nan \n df.speed.mask(df.speed == -1, df.speed_limit, inplace = True) # -1 means no cars, setting it to speed limit\n df.loc[(df.speed < 0) & (df.speed > -1), \"speed\"] = 0 # anything else below zero is between 0 and -1, occuring when some lanes have non-moving cars while others have have no cars.\n df.speed.mask(df.speed > df.speed_limit, df.speed_limit, inplace = True) # cap speed at speed_limit, since higher speed dosn't add to representation\n \n return df\n\nimport os\ndef reduce_cols(sensors, path_in = \"data/ndw_raw/\", path_out = \"data/\"):\n sensor_df_list = list()\n for sensor in sensors:\n df = pd.read_csv(os.path.join(path_in, sensor + \".csv\"), header = None, \\\n usecols = [0, 86, 87], names = [\"timestamp\", \"speed\", \"flow\"], \\\n index_col = \"timestamp\", parse_dates = True, date_parser = dateparse)\n df.flow /= 60 # change flow unit to min^-1\n df = repair_datetime_index(df)\n df = fix_values(df) \n #df.to_csv(path_out + sensor)\n sensor_df_list.append(df)\n\n return sensor_df_list",
"_____no_output_____"
],
[
"sensor_df_list = reduce_cols(sensor_name_list)",
"_____no_output_____"
]
],
[
[
"### Join Sensors",
"_____no_output_____"
]
],
[
[
"def join_sensors(sensor_df_list, sensor_name_list):\n combined_df = pd.DataFrame({\"timestamp\": pd.date_range(start = \"2011-01-01\", end = \"2019-12-31\", freq = \"T\")})\n combined_df.set_index(\"timestamp\", drop = True, inplace = True)\n combined_df.index = combined_df.index.tz_localize(\"UTC\").tz_convert(\"Europe/Amsterdam\")\n d = {}\n for i, sensor in enumerate(sensor_df_list):\n # only add speed limit on the final sensor\n if i == len(sensor_df_list) - 1:\n d[sensor_name_list[i]] = sensor_df_list[i]\n combined_df = combined_df.join(d[sensor_name_list[i]], how = \"outer\", rsuffix = '_' + sensor_name_list[i])\n else:\n d[sensor_name_list[i]] = sensor_df_list[i].iloc[:, :2]\n combined_df = combined_df.join(d[sensor_name_list[i]], how = \"outer\", rsuffix = \"_\" + sensor_name_list[i])\n combined_df.dropna(how = \"all\", axis = 0, inplace = True) # this works in all cases because speed_limit is never NA on a sensor df\n \n return combined_df",
"_____no_output_____"
],
[
"# Join sensors to one table\ndf = join_sensors(sensor_df_list, sensor_name_list)\n\n# Rename and reorder columns\ndf.rename({\"speed_RWS01_MONIBAS_0021hrl0403ra\": \"speed_-2\", \"speed_RWS01_MONIBAS_0021hrl0409ra\": \"speed_-1\",\\\n \"speed_RWS01_MONIBAS_0021hrl0420ra\": \"speed_+1\", \"speed_RWS01_MONIBAS_0021hrl0426ra\": \"speed_+2\",\\\n \"flow_RWS01_MONIBAS_0021hrl0403ra\": \"flow_-2\", \"flow_RWS01_MONIBAS_0021hrl0409ra\": \"flow_-1\",\\\n \"flow_RWS01_MONIBAS_0021hrl0420ra\": \"flow_+1\", \"flow_RWS01_MONIBAS_0021hrl0426ra\": \"flow_+2\"\\\n }, axis = 1, inplace = True)\ncol_order = [\"speed\", \"flow\", \"speed_-2\", \"speed_-1\",\"speed_+1\", \"speed_+2\", \"flow_-2\", \"flow_-1\", \"flow_+1\", \"flow_+2\", \"speed_limit\"]\ndf = df[col_order]\n\n# Save table to csv\n#df.to_csv(data_dir + \"combined_df.csv\")",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
]
],
[
[
"### Impute data",
"_____no_output_____"
]
],
[
[
"cols = col_order\nspeed_cols = [\"speed\", \"speed_-2\", \"speed_-1\",\"speed_+1\", \"speed_+2\"]\nflow_cols = [\"flow\", \"flow_-2\", \"flow_-1\", \"flow_+1\", \"flow_+2\"]",
"_____no_output_____"
],
[
"# Where values are missing in one or more sensors, but are present in others, impute with mean of others\ndef fill_na_row_mean(df):\n row_avgs = df.mean(axis = 1).values.reshape(-1, 1)\n df = df.fillna(0) + df.isna().values * row_avgs\n return df",
"_____no_output_____"
],
[
"speed_df = fill_na_row_mean(df[speed_cols])\nflow_df = fill_na_row_mean(df[flow_cols])",
"_____no_output_____"
],
[
"df = speed_df.join(flow_df, how = \"inner\").join(df[[\"speed_limit\"]], how = \"inner\")",
"_____no_output_____"
],
[
"# Interpolate null vals for the first week of data of speed and flow cols\ndef interpolate_week(df, cols):\n week = 7 * 24 * 60\n for col in cols: \n df.iloc[:week, df.columns.get_loc(col)] = df[col][:week].interpolate(method = \"time\")\n return df\n\n# Replace remaining nulls with value from 1 week previous\ndef shift_week(df, cols):\n # Use RangeIndex for the this operation\n df[\"timestamp\"] = df.index\n df.reset_index(drop = True, inplace = True)\n week = 7 * 24 * 60\n for col in cols:\n col_index = df.columns.get_loc(col)\n for row in df.itertuples():\n if np.isnan(row[col_index + 1]):\n df.iat[row[0], col_index] = df.iat[(row[0] - week), col_index]\n # Return to DateTimeIndex again\n df.set_index(pd.to_datetime(df.timestamp.values), inplace = True) \n df.drop(\"timestamp\", axis = 1, inplace = True)\n return df",
"_____no_output_____"
],
[
"df = interpolate_week(df, cols)\ndf = shift_week(df, cols)\n#df.to_csv(\"data/df_imputed_week_shift.csv\")",
"_____no_output_____"
],
[
"import holidays\ndf[\"density\"] = (df.flow * 60) / df.speed\ndf[\"weekend\"] = np.where(df.index.weekday > 4, 1, 0).astype(np.int16)\ndf[\"holiday\"] = np.array([int(x in holidays.NL()) for x in df.index]).astype(np.int16)\ndf[\"speed_limit\"] = np.where(df.speed_limit > 115, 1, 0)",
"_____no_output_____"
],
[
"df.to_csv(\"data/df_imputed_week_shift_added_holiday_weekends_speed_limit_130.csv\")",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbf352708c6af36cbe7215a4efc4913cad423f5a
| 19,945 |
ipynb
|
Jupyter Notebook
|
DeepLearning.AI Tensorflow Developer/Course 1 - Introduction to TensorFlow for Artificial Intelligence, Machine Learning, and Deep Learning/Resources/Copy of Course 1 - Part 8 - Lesson 3 - Notebook.ipynb
|
lucigrigo/CourseraCourses
|
a53a7ccc35861b1c23a536c50e10e17b8d9742e6
|
[
"MIT"
] | null | null | null |
DeepLearning.AI Tensorflow Developer/Course 1 - Introduction to TensorFlow for Artificial Intelligence, Machine Learning, and Deep Learning/Resources/Copy of Course 1 - Part 8 - Lesson 3 - Notebook.ipynb
|
lucigrigo/CourseraCourses
|
a53a7ccc35861b1c23a536c50e10e17b8d9742e6
|
[
"MIT"
] | null | null | null |
DeepLearning.AI Tensorflow Developer/Course 1 - Introduction to TensorFlow for Artificial Intelligence, Machine Learning, and Deep Learning/Resources/Copy of Course 1 - Part 8 - Lesson 3 - Notebook.ipynb
|
lucigrigo/CourseraCourses
|
a53a7ccc35861b1c23a536c50e10e17b8d9742e6
|
[
"MIT"
] | null | null | null | 19,945 | 19,945 | 0.711156 |
[
[
[
"##### Copyright 2019 The TensorFlow Authors.",
"_____no_output_____"
]
],
[
[
"#@title Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# https://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.",
"_____no_output_____"
],
[
"!wget --no-check-certificate \\\n https://storage.googleapis.com/laurencemoroney-blog.appspot.com/horse-or-human.zip \\\n -O /tmp/horse-or-human.zip",
"_____no_output_____"
],
[
"!wget --no-check-certificate \\\n https://storage.googleapis.com/laurencemoroney-blog.appspot.com/validation-horse-or-human.zip \\\n -O /tmp/validation-horse-or-human.zip",
"_____no_output_____"
]
],
[
[
"The following python code will use the OS library to use Operating System libraries, giving you access to the file system, and the zipfile library allowing you to unzip the data. ",
"_____no_output_____"
]
],
[
[
"import os\nimport zipfile\n\nlocal_zip = '/tmp/horse-or-human.zip'\nzip_ref = zipfile.ZipFile(local_zip, 'r')\nzip_ref.extractall('/tmp/horse-or-human')\nlocal_zip = '/tmp/validation-horse-or-human.zip'\nzip_ref = zipfile.ZipFile(local_zip, 'r')\nzip_ref.extractall('/tmp/validation-horse-or-human')\nzip_ref.close()",
"_____no_output_____"
]
],
[
[
"The contents of the .zip are extracted to the base directory `/tmp/horse-or-human`, which in turn each contain `horses` and `humans` subdirectories.\n\nIn short: The training set is the data that is used to tell the neural network model that 'this is what a horse looks like', 'this is what a human looks like' etc. \n\nOne thing to pay attention to in this sample: We do not explicitly label the images as horses or humans. If you remember with the handwriting example earlier, we had labelled 'this is a 1', 'this is a 7' etc. Later you'll see something called an ImageGenerator being used -- and this is coded to read images from subdirectories, and automatically label them from the name of that subdirectory. So, for example, you will have a 'training' directory containing a 'horses' directory and a 'humans' one. ImageGenerator will label the images appropriately for you, reducing a coding step. \n\nLet's define each of these directories:",
"_____no_output_____"
]
],
[
[
"# Directory with our training horse pictures\ntrain_horse_dir = os.path.join('/tmp/horse-or-human/horses')\n\n# Directory with our training human pictures\ntrain_human_dir = os.path.join('/tmp/horse-or-human/humans')\n\n# Directory with our training horse pictures\nvalidation_horse_dir = os.path.join('/tmp/validation-horse-or-human/horses')\n\n# Directory with our training human pictures\nvalidation_human_dir = os.path.join('/tmp/validation-horse-or-human/humans')",
"_____no_output_____"
]
],
[
[
"Now, let's see what the filenames look like in the `horses` and `humans` training directories:",
"_____no_output_____"
]
],
[
[
"train_horse_names = os.listdir(train_horse_dir)\nprint(train_horse_names[:10])\n\ntrain_human_names = os.listdir(train_human_dir)\nprint(train_human_names[:10])\n\nvalidation_horse_hames = os.listdir(validation_horse_dir)\nprint(validation_horse_hames[:10])\n\nvalidation_human_names = os.listdir(validation_human_dir)\nprint(validation_human_names[:10])",
"_____no_output_____"
]
],
[
[
"Let's find out the total number of horse and human images in the directories:",
"_____no_output_____"
]
],
[
[
"print('total training horse images:', len(os.listdir(train_horse_dir)))\nprint('total training human images:', len(os.listdir(train_human_dir)))\nprint('total validation horse images:', len(os.listdir(validation_horse_dir)))\nprint('total validation human images:', len(os.listdir(validation_human_dir)))",
"_____no_output_____"
]
],
[
[
"Now let's take a look at a few pictures to get a better sense of what they look like. First, configure the matplot parameters:",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\n\nimport matplotlib.pyplot as plt\nimport matplotlib.image as mpimg\n\n# Parameters for our graph; we'll output images in a 4x4 configuration\nnrows = 4\nncols = 4\n\n# Index for iterating over images\npic_index = 0",
"_____no_output_____"
]
],
[
[
"Now, display a batch of 8 horse and 8 human pictures. You can rerun the cell to see a fresh batch each time:",
"_____no_output_____"
]
],
[
[
"# Set up matplotlib fig, and size it to fit 4x4 pics\nfig = plt.gcf()\nfig.set_size_inches(ncols * 4, nrows * 4)\n\npic_index += 8\nnext_horse_pix = [os.path.join(train_horse_dir, fname) \n for fname in train_horse_names[pic_index-8:pic_index]]\nnext_human_pix = [os.path.join(train_human_dir, fname) \n for fname in train_human_names[pic_index-8:pic_index]]\n\nfor i, img_path in enumerate(next_horse_pix+next_human_pix):\n # Set up subplot; subplot indices start at 1\n sp = plt.subplot(nrows, ncols, i + 1)\n sp.axis('Off') # Don't show axes (or gridlines)\n\n img = mpimg.imread(img_path)\n plt.imshow(img)\n\nplt.show()\n",
"_____no_output_____"
]
],
[
[
"## Building a Small Model from Scratch\n\nBut before we continue, let's start defining the model:\n\nStep 1 will be to import tensorflow.",
"_____no_output_____"
]
],
[
[
"import tensorflow as tf",
"_____no_output_____"
]
],
[
[
"We then add convolutional layers as in the previous example, and flatten the final result to feed into the densely connected layers.",
"_____no_output_____"
],
[
"Finally we add the densely connected layers. \n\nNote that because we are facing a two-class classification problem, i.e. a *binary classification problem*, we will end our network with a [*sigmoid* activation](https://wikipedia.org/wiki/Sigmoid_function), so that the output of our network will be a single scalar between 0 and 1, encoding the probability that the current image is class 1 (as opposed to class 0).",
"_____no_output_____"
]
],
[
[
"model = tf.keras.models.Sequential([\n # Note the input shape is the desired size of the image 300x300 with 3 bytes color\n # This is the first convolution\n tf.keras.layers.Conv2D(16, (3,3), activation='relu', input_shape=(300, 300, 3)),\n tf.keras.layers.MaxPooling2D(2, 2),\n # The second convolution\n tf.keras.layers.Conv2D(32, (3,3), activation='relu'),\n tf.keras.layers.MaxPooling2D(2,2),\n # The third convolution\n tf.keras.layers.Conv2D(64, (3,3), activation='relu'),\n tf.keras.layers.MaxPooling2D(2,2),\n # The fourth convolution\n tf.keras.layers.Conv2D(64, (3,3), activation='relu'),\n tf.keras.layers.MaxPooling2D(2,2),\n # The fifth convolution\n tf.keras.layers.Conv2D(64, (3,3), activation='relu'),\n tf.keras.layers.MaxPooling2D(2,2),\n # Flatten the results to feed into a DNN\n tf.keras.layers.Flatten(),\n # 512 neuron hidden layer\n tf.keras.layers.Dense(512, activation='relu'),\n # Only 1 output neuron. It will contain a value from 0-1 where 0 for 1 class ('horses') and 1 for the other ('humans')\n tf.keras.layers.Dense(1, activation='sigmoid')\n])",
"_____no_output_____"
]
],
[
[
"The model.summary() method call prints a summary of the NN ",
"_____no_output_____"
]
],
[
[
"model.summary()",
"_____no_output_____"
]
],
[
[
"The \"output shape\" column shows how the size of your feature map evolves in each successive layer. The convolution layers reduce the size of the feature maps by a bit due to padding, and each pooling layer halves the dimensions.",
"_____no_output_____"
],
[
"Next, we'll configure the specifications for model training. We will train our model with the `binary_crossentropy` loss, because it's a binary classification problem and our final activation is a sigmoid. (For a refresher on loss metrics, see the [Machine Learning Crash Course](https://developers.google.com/machine-learning/crash-course/descending-into-ml/video-lecture).) We will use the `rmsprop` optimizer with a learning rate of `0.001`. During training, we will want to monitor classification accuracy.\n\n**NOTE**: In this case, using the [RMSprop optimization algorithm](https://wikipedia.org/wiki/Stochastic_gradient_descent#RMSProp) is preferable to [stochastic gradient descent](https://developers.google.com/machine-learning/glossary/#SGD) (SGD), because RMSprop automates learning-rate tuning for us. (Other optimizers, such as [Adam](https://wikipedia.org/wiki/Stochastic_gradient_descent#Adam) and [Adagrad](https://developers.google.com/machine-learning/glossary/#AdaGrad), also automatically adapt the learning rate during training, and would work equally well here.)",
"_____no_output_____"
]
],
[
[
"from tensorflow.keras.optimizers import RMSprop\n\nmodel.compile(loss='binary_crossentropy',\n optimizer=RMSprop(lr=0.001),\n metrics=['accuracy'])",
"_____no_output_____"
]
],
[
[
"### Data Preprocessing\n\nLet's set up data generators that will read pictures in our source folders, convert them to `float32` tensors, and feed them (with their labels) to our network. We'll have one generator for the training images and one for the validation images. Our generators will yield batches of images of size 300x300 and their labels (binary).\n\nAs you may already know, data that goes into neural networks should usually be normalized in some way to make it more amenable to processing by the network. (It is uncommon to feed raw pixels into a convnet.) In our case, we will preprocess our images by normalizing the pixel values to be in the `[0, 1]` range (originally all values are in the `[0, 255]` range).\n\nIn Keras this can be done via the `keras.preprocessing.image.ImageDataGenerator` class using the `rescale` parameter. This `ImageDataGenerator` class allows you to instantiate generators of augmented image batches (and their labels) via `.flow(data, labels)` or `.flow_from_directory(directory)`. These generators can then be used with the Keras model methods that accept data generators as inputs: `fit`, `evaluate_generator`, and `predict_generator`.",
"_____no_output_____"
]
],
[
[
"from tensorflow.keras.preprocessing.image import ImageDataGenerator\n\n# All images will be rescaled by 1./255\ntrain_datagen = ImageDataGenerator(rescale=1/255)\nvalidation_datagen = ImageDataGenerator(rescale=1/255)\n\n# Flow training images in batches of 128 using train_datagen generator\ntrain_generator = train_datagen.flow_from_directory(\n '/tmp/horse-or-human/', # This is the source directory for training images\n target_size=(300, 300), # All images will be resized to 300x300\n batch_size=128,\n # Since we use binary_crossentropy loss, we need binary labels\n class_mode='binary')\n\n# Flow training images in batches of 128 using train_datagen generator\nvalidation_generator = validation_datagen.flow_from_directory(\n '/tmp/validation-horse-or-human/', # This is the source directory for training images\n target_size=(300, 300), # All images will be resized to 300x300\n batch_size=32,\n # Since we use binary_crossentropy loss, we need binary labels\n class_mode='binary')",
"_____no_output_____"
]
],
[
[
"### Training\nLet's train for 15 epochs -- this may take a few minutes to run.\n\nDo note the values per epoch.\n\nThe Loss and Accuracy are a great indication of progress of training. It's making a guess as to the classification of the training data, and then measuring it against the known label, calculating the result. Accuracy is the portion of correct guesses. ",
"_____no_output_____"
]
],
[
[
"history = model.fit(\n train_generator,\n steps_per_epoch=8, \n epochs=15,\n verbose=1,\n validation_data = validation_generator,\n validation_steps=8)",
"_____no_output_____"
]
],
[
[
"###Running the Model\n\nLet's now take a look at actually running a prediction using the model. This code will allow you to choose 1 or more files from your file system, it will then upload them, and run them through the model, giving an indication of whether the object is a horse or a human.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nfrom google.colab import files\nfrom keras.preprocessing import image\n\nuploaded = files.upload()\n\nfor fn in uploaded.keys():\n \n # predicting images\n path = '/content/' + fn\n img = image.load_img(path, target_size=(300, 300))\n x = image.img_to_array(img)\n x = np.expand_dims(x, axis=0)\n\n images = np.vstack([x])\n classes = model.predict(images, batch_size=10)\n print(classes[0])\n if classes[0]>0.5:\n print(fn + \" is a human\")\n else:\n print(fn + \" is a horse\")\n ",
"_____no_output_____"
]
],
[
[
"### Visualizing Intermediate Representations\n\nTo get a feel for what kind of features our convnet has learned, one fun thing to do is to visualize how an input gets transformed as it goes through the convnet.\n\nLet's pick a random image from the training set, and then generate a figure where each row is the output of a layer, and each image in the row is a specific filter in that output feature map. Rerun this cell to generate intermediate representations for a variety of training images.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport random\nfrom tensorflow.keras.preprocessing.image import img_to_array, load_img\n\n# Let's define a new Model that will take an image as input, and will output\n# intermediate representations for all layers in the previous model after\n# the first.\nsuccessive_outputs = [layer.output for layer in model.layers[1:]]\n#visualization_model = Model(img_input, successive_outputs)\nvisualization_model = tf.keras.models.Model(inputs = model.input, outputs = successive_outputs)\n# Let's prepare a random input image from the training set.\nhorse_img_files = [os.path.join(train_horse_dir, f) for f in train_horse_names]\nhuman_img_files = [os.path.join(train_human_dir, f) for f in train_human_names]\nimg_path = random.choice(horse_img_files + human_img_files)\n\nimg = load_img(img_path, target_size=(300, 300)) # this is a PIL image\nx = img_to_array(img) # Numpy array with shape (150, 150, 3)\nx = x.reshape((1,) + x.shape) # Numpy array with shape (1, 150, 150, 3)\n\n# Rescale by 1/255\nx /= 255\n\n# Let's run our image through our network, thus obtaining all\n# intermediate representations for this image.\nsuccessive_feature_maps = visualization_model.predict(x)\n\n# These are the names of the layers, so can have them as part of our plot\nlayer_names = [layer.name for layer in model.layers[1:]]\n\n# Now let's display our representations\nfor layer_name, feature_map in zip(layer_names, successive_feature_maps):\n if len(feature_map.shape) == 4:\n # Just do this for the conv / maxpool layers, not the fully-connected layers\n n_features = feature_map.shape[-1] # number of features in feature map\n # The feature map has shape (1, size, size, n_features)\n size = feature_map.shape[1]\n # We will tile our images in this matrix\n display_grid = np.zeros((size, size * n_features))\n for i in range(n_features):\n # Postprocess the feature to make it visually palatable\n x = feature_map[0, :, :, i]\n x -= x.mean()\n x /= x.std()\n x *= 64\n x += 128\n x = np.clip(x, 0, 255).astype('uint8')\n # We'll tile each filter into this big horizontal grid\n display_grid[:, i * size : (i + 1) * size] = x\n # Display the grid\n scale = 20. / n_features\n plt.figure(figsize=(scale * n_features, scale))\n plt.title(layer_name)\n plt.grid(False)\n plt.imshow(display_grid, aspect='auto', cmap='viridis')",
"_____no_output_____"
]
],
[
[
"As you can see we go from the raw pixels of the images to increasingly abstract and compact representations. The representations downstream start highlighting what the network pays attention to, and they show fewer and fewer features being \"activated\"; most are set to zero. This is called \"sparsity.\" Representation sparsity is a key feature of deep learning.\n\n\nThese representations carry increasingly less information about the original pixels of the image, but increasingly refined information about the class of the image. You can think of a convnet (or a deep network in general) as an information distillation pipeline.",
"_____no_output_____"
],
[
"## Clean Up\n\nBefore running the next exercise, run the following cell to terminate the kernel and free memory resources:",
"_____no_output_____"
]
],
[
[
"import os, signal\nos.kill(os.getpid(), signal.SIGKILL)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
]
] |
cbf359ac70eed9e54da952a237857ac14ed71160
| 38,328 |
ipynb
|
Jupyter Notebook
|
docs/test/testinnsending/.ipynb_checkpoints/upersonlig-as-checkpoint.ipynb
|
Skatteetaten/skattemeldingen
|
b06506e3c9f853b850d63ec6ceae1217af9219c1
|
[
"Apache-2.0"
] | 14 |
2020-10-31T21:41:47.000Z
|
2022-01-31T07:36:56.000Z
|
docs/test/testinnsending/.ipynb_checkpoints/upersonlig-as-checkpoint.ipynb
|
Skatteetaten/skattemeldingen
|
b06506e3c9f853b850d63ec6ceae1217af9219c1
|
[
"Apache-2.0"
] | 6 |
2020-09-03T05:47:07.000Z
|
2021-11-16T13:44:37.000Z
|
docs/test/testinnsending/.ipynb_checkpoints/upersonlig-as-checkpoint.ipynb
|
Skatteetaten/skattemeldingen
|
b06506e3c9f853b850d63ec6ceae1217af9219c1
|
[
"Apache-2.0"
] | 9 |
2020-09-03T06:07:52.000Z
|
2021-11-08T10:14:57.000Z
| 76.502994 | 2,384 | 0.694871 |
[
[
[
"# Testinnsening av upersonlig skattemelding med næringspesifikasjon\nDenne demoen er ment for å vise hvordan flyten for et sluttbrukersystem kan hente et utkast, gjøre endringer, validere/kontrollere det mot Skatteetatens apier, for å sende det inn via Altinn3. ",
"_____no_output_____"
]
],
[
[
"try: \n from altinn3 import *\n from skatteetaten_api import main_relay, base64_decode_response, decode_dokument\n import requests\n import base64\n import xmltodict\n import xml.dom.minidom\n from pathlib import Path\nexcept ImportError as e:\n print(\"Mangler en avhengighet, installer dem via pip\")\n !pip install python-jose\n !pip install xmltodict\n !pip install pathlib\n import xmltodict\n from skatteetaten_api import main_relay, base64_decode_response, decode_dokument\n\n \n#hjelpe metode om du vil se en request printet som curl \ndef print_request_as_curl(r):\n command = \"curl -X {method} -H {headers} -d '{data}' '{uri}'\"\n method = r.request.method\n uri = r.request.url\n data = r.request.body\n headers = ['\"{0}: {1}\"'.format(k, v) for k, v in r.request.headers.items()]\n headers = \" -H \".join(headers)\n print(command.format(method=method, headers=headers, data=data, uri=uri))",
"_____no_output_____"
],
[
"idporten_header = main_relay()",
"https://oidc-ver2.difi.no/idporten-oidc-provider/authorize?scope=skatteetaten%3Aformueinntekt%2Fskattemelding%20openid&acr_values=Level3&client_id=8d7adad7-b497-40d0-8897-9a9d86c95306&redirect_uri=http%3A%2F%2Flocalhost%3A12345%2Ftoken&response_type=code&state=5_1plE-p4OF9rQEFdIXkag&nonce=1633506921962760&resource=https%3A%2F%2Fmp-test.sits.no%2Fapi%2Feksterntapi%2Fformueinntekt%2Fskattemelding%2F&code_challenge=-FL7Q5j47R2ZDcJ-3i1g7tijySrDk3hzlzi3LQGk26M=&code_challenge_method=S256&ui_locales=nb\nAuthorization token received\n{'code': ['X2pgIYVfyCWuBnLEeZd0GOFXCQaZc1nsGGemqigmll0'], 'state': ['5_1plE-p4OF9rQEFdIXkag']}\nJS : \n{'access_token': 'eyJraWQiOiJjWmswME1rbTVIQzRnN3Z0NmNwUDVGSFpMS0pzdzhmQkFJdUZiUzRSVEQ0IiwiYWxnIjoiUlMyNTYifQ.eyJzdWIiOiItMHp1cDZaNWJqSFB2LUQ1eXpyUWJaQ1JXem4xVnVlRnpTQXdBUTBITjRvPSIsImlzcyI6Imh0dHBzOlwvXC9vaWRjLXZlcjIuZGlmaS5ub1wvaWRwb3J0ZW4tb2lkYy1wcm92aWRlclwvIiwiY2xpZW50X2FtciI6Im5vbmUiLCJwaWQiOiIwMTAxNDcwMTM3NyIsInRva2VuX3R5cGUiOiJCZWFyZXIiLCJjbGllbnRfaWQiOiI4ZDdhZGFkNy1iNDk3LTQwZDAtODg5Ny05YTlkODZjOTUzMDYiLCJhdWQiOiJodHRwczpcL1wvbXAtdGVzdC5zaXRzLm5vXC9hcGlcL2Vrc3Rlcm50YXBpXC9mb3JtdWVpbm50ZWt0XC9za2F0dGVtZWxkaW5nXC8iLCJhY3IiOiJMZXZlbDMiLCJzY29wZSI6Im9wZW5pZCBza2F0dGVldGF0ZW46Zm9ybXVlaW5udGVrdFwvc2thdHRlbWVsZGluZyIsImV4cCI6MTYzMzU5MzM0NCwiaWF0IjoxNjMzNTA2OTQ0LCJjbGllbnRfb3Jnbm8iOiI5NzQ3NjEwNzYiLCJqdGkiOiJuNVJuZERaSzNWakN1bGlrMFc5T2FoRkdYbGFnYmpYUmNCdFVnU3R6ZGpVIiwiY29uc3VtZXIiOnsiYXV0aG9yaXR5IjoiaXNvNjUyMy1hY3RvcmlkLXVwaXMiLCJJRCI6IjAxOTI6OTc0NzYxMDc2In19.N9cyh-kI5EFmLYnnHr-vbQk5O8WYwH7ITUe5m8qlEXDNULXqn6a6hPRme-zq6RDxkCx4apr5NBNy1AcFUhCjOVZLJYm6LmcH4JckIoftVTnCBQLD8V8NOMXrJXSw5X59Im1XGligjZOd2LwhGU0b2oi3Ma6kFeux-H_6m8KWzJhid7wqH-je8DXaNwxI1l2FhQLpawvC-F4s2odZdVZB7AaN1o8h-N8vPGQ79hCucgi49xYjT6j-_coEt_ggfs-P8qgzbWu_wFNdpYsaye5WS2T5Y2xZzR5iWRlQt43ye8q_Vemea0gNGU5bzknWEvR17GaVPcPHJC_aaBybFc-hOQ', 'id_token': 'eyJraWQiOiJjWmswME1rbTVIQzRnN3Z0NmNwUDVGSFpMS0pzdzhmQkFJdUZiUzRSVEQ0IiwiYWxnIjoiUlMyNTYifQ.eyJhdF9oYXNoIjoia0plbzFsS2Z5RVpNQks1T2Mtc3hUdyIsInN1YiI6Ii0wenVwNlo1YmpIUHYtRDV5enJRYlpDUld6bjFWdWVGelNBd0FRMEhONG89IiwiYW1yIjpbIk1pbmlkLVBJTiJdLCJpc3MiOiJodHRwczpcL1wvb2lkYy12ZXIyLmRpZmkubm9cL2lkcG9ydGVuLW9pZGMtcHJvdmlkZXJcLyIsInBpZCI6IjAxMDE0NzAxMzc3IiwibG9jYWxlIjoibmIiLCJub25jZSI6IjE2MzM1MDY5MjE5NjI3NjAiLCJzaWQiOiJmUFNtd3pHMWVycG5XZ0l2ekZIU1Q5ZlBZb1JxSmRKYS1OUVdwTl9TQmpZIiwiYXVkIjoiOGQ3YWRhZDctYjQ5Ny00MGQwLTg4OTctOWE5ZDg2Yzk1MzA2IiwiYWNyIjoiTGV2ZWwzIiwiYXV0aF90aW1lIjoxNjMzNTA2OTQ0LCJleHAiOjE2MzM1MDcwNjQsImlhdCI6MTYzMzUwNjk0NCwianRpIjoiQmJEdVZhODBaZXlPbHdtT3ZxV1Z3aThLUlZGMVhwckpKc25aNlBJX1FpbyJ9.TA0Ck2z7_p5fjLBU_AZa2MOhWu_-kRKd06cgFsEVouEk7UPuDCbjcwZRahxnPAWuYO6hWw6ZN8uyb1TmEQVzy9B4RV4FoYsNzyXM-adGmicNukKjAHMMh3IvezS8DTfKlwGWwxfpXE6iuM17cbm7yOiSKdRH0QRiXenNlS7RQ_rQKR4tS3MIFP2it-QybIGXee4Nh3AM9CTKs2OZFsuaQ6XQNSLoHFsxq3ErVQ5IjFknJQry1Ll8v42pVMyLIefQrshnHojIvoHTF0zgqMx0YlEb50yWWFdR1z17dGEwVDgTeKeZ-GbUD9f_u4ZIHLHbJwobiMcxftsaEQAsCf_pjw', 'token_type': 'Bearer', 'expires_in': 86399, 'scope': 'openid skatteetaten:formueinntekt/skattemelding'}\nThe token is good, expires in 86399 seconds\n\nBearer eyJraWQiOiJjWmswME1rbTVIQzRnN3Z0NmNwUDVGSFpMS0pzdzhmQkFJdUZiUzRSVEQ0IiwiYWxnIjoiUlMyNTYifQ.eyJzdWIiOiItMHp1cDZaNWJqSFB2LUQ1eXpyUWJaQ1JXem4xVnVlRnpTQXdBUTBITjRvPSIsImlzcyI6Imh0dHBzOlwvXC9vaWRjLXZlcjIuZGlmaS5ub1wvaWRwb3J0ZW4tb2lkYy1wcm92aWRlclwvIiwiY2xpZW50X2FtciI6Im5vbmUiLCJwaWQiOiIwMTAxNDcwMTM3NyIsInRva2VuX3R5cGUiOiJCZWFyZXIiLCJjbGllbnRfaWQiOiI4ZDdhZGFkNy1iNDk3LTQwZDAtODg5Ny05YTlkODZjOTUzMDYiLCJhdWQiOiJodHRwczpcL1wvbXAtdGVzdC5zaXRzLm5vXC9hcGlcL2Vrc3Rlcm50YXBpXC9mb3JtdWVpbm50ZWt0XC9za2F0dGVtZWxkaW5nXC8iLCJhY3IiOiJMZXZlbDMiLCJzY29wZSI6Im9wZW5pZCBza2F0dGVldGF0ZW46Zm9ybXVlaW5udGVrdFwvc2thdHRlbWVsZGluZyIsImV4cCI6MTYzMzU5MzM0NCwiaWF0IjoxNjMzNTA2OTQ0LCJjbGllbnRfb3Jnbm8iOiI5NzQ3NjEwNzYiLCJqdGkiOiJuNVJuZERaSzNWakN1bGlrMFc5T2FoRkdYbGFnYmpYUmNCdFVnU3R6ZGpVIiwiY29uc3VtZXIiOnsiYXV0aG9yaXR5IjoiaXNvNjUyMy1hY3RvcmlkLXVwaXMiLCJJRCI6IjAxOTI6OTc0NzYxMDc2In19.N9cyh-kI5EFmLYnnHr-vbQk5O8WYwH7ITUe5m8qlEXDNULXqn6a6hPRme-zq6RDxkCx4apr5NBNy1AcFUhCjOVZLJYm6LmcH4JckIoftVTnCBQLD8V8NOMXrJXSw5X59Im1XGligjZOd2LwhGU0b2oi3Ma6kFeux-H_6m8KWzJhid7wqH-je8DXaNwxI1l2FhQLpawvC-F4s2odZdVZB7AaN1o8h-N8vPGQ79hCucgi49xYjT6j-_coEt_ggfs-P8qgzbWu_wFNdpYsaye5WS2T5Y2xZzR5iWRlQt43ye8q_Vemea0gNGU5bzknWEvR17GaVPcPHJC_aaBybFc-hOQ\n"
]
],
[
[
"# Hent utkast og gjeldende\nHer legger vi inn fødselsnummeret vi logget oss inn med, Dersom du velger et annet fødselsnummer så må den du logget på med ha tilgang til skattemeldingen du ønsker å hente\n\n#### Parten nedenfor er brukt for demostrasjon, pass på bruk deres egne testparter når dere tester\n\n\n01014701377 er daglig leder for 811422762 ",
"_____no_output_____"
]
],
[
[
"s = requests.Session()\ns.headers = dict(idporten_header)\nfnr=\"01014701377\" #oppdater med test fødselsnummerene du har fått tildelt\norgnr_as = \"811423262\"",
"_____no_output_____"
]
],
[
[
"### Utkast",
"_____no_output_____"
]
],
[
[
"url_utkast = f'https://mp-test.sits.no/api/skattemelding/v2/utkast/2021/{orgnr_as}'\n\nr = s.get(url_utkast)\nr",
"_____no_output_____"
]
],
[
[
"### Gjeldende",
"_____no_output_____"
]
],
[
[
"url_gjeldende = f'https://mp-test.sits.no/api/skattemelding/v2/2021/{orgnr_as}'\nr_gjeldende = s.get(url_gjeldende)\nr_gjeldende",
"_____no_output_____"
]
],
[
[
"## Fastsatt\nHer får en _http 404_ om vedkommende ikke har noen fastsetting, rekjørt denne etter du har sendt inn og fått tilbakemdling i Altinn at den har blitt behandlet, du skal nå ha en fastsatt skattemelding om den har blitt sent inn som Komplett",
"_____no_output_____"
]
],
[
[
"url_fastsatt = f'https://mp-test.sits.no/api/skattemelding/v2/fastsatt/2021/{orgnr_as}'\nr_fastsatt = s.get(url_fastsatt)\nr_fastsatt",
"_____no_output_____"
],
[
"r_fastsatt.headers",
"_____no_output_____"
]
],
[
[
"## Svar fra hent gjeldende \n\n### Gjeldende dokument referanse: \nI responsen på alle api kallene, være seg utkast/fastsatt eller gjeldene, så følger det med en dokumentreferanse. \nFor å kalle valider tjenesten, er en avhengig av å bruke korrekt referanse til gjeldende skattemelding. \n\nCellen nedenfor henter ut gjeldende dokumentrefranse printer ut responsen fra hent gjeldende kallet ",
"_____no_output_____"
]
],
[
[
"sjekk_svar = r_gjeldende\n\nsme_og_naering_respons = xmltodict.parse(sjekk_svar.text)\nskattemelding_base64 = sme_og_naering_respons[\"skattemeldingOgNaeringsspesifikasjonforespoerselResponse\"][\"dokumenter\"][\"skattemeldingdokument\"]\nsme_base64 = skattemelding_base64[\"content\"]\ndokref = sme_og_naering_respons[\"skattemeldingOgNaeringsspesifikasjonforespoerselResponse\"][\"dokumenter\"]['skattemeldingdokument']['id']\ndecoded_sme_xml = decode_dokument(skattemelding_base64)\nsme_utkast = xml.dom.minidom.parseString(decoded_sme_xml[\"content\"]).toprettyxml()\n\nprint(f\"Responsen fra hent gjeldende ser slik ut, gjeldende dokumentrerefanse er {dokref}\")\nprint(sjekk_svar.request.method ,sjekk_svar.request.url)\nprint(xml.dom.minidom.parseString(sjekk_svar.text).toprettyxml())\n",
"_____no_output_____"
],
[
"#Legg merge til dokumenter.dokument.type = skattemeldingUpersonlig\n\nwith open(\"../../../src/resources/eksempler/v2/Naeringspesifikasjon-enk-v2.xml\", 'r') as f:\n naering_as_xml = f.read()\n \ninnsendingstype = \"komplett\"\nnaeringsspesifikasjoner_as_b64 = base64.b64encode(naering_as_xml.encode(\"utf-8\"))\nnaeringsspesifikasjoner_as_b64 = str(naeringsspesifikasjoner_as_b64.decode(\"utf-8\"))\nnaeringsspesifikasjoner_base64=naeringsspesifikasjoner_as_b64\ndok_ref=dokref\n\nvalider_konvlutt_v2 = \"\"\"\n<?xml version=\"1.0\" encoding=\"utf-8\" ?>\n<skattemeldingOgNaeringsspesifikasjonRequest xmlns=\"no:skatteetaten:fastsetting:formueinntekt:skattemeldingognaeringsspesifikasjon:request:v2\">\n <dokumenter>\n <dokument>\n <type>skattemeldingUpersonlig</type>\n <encoding>utf-8</encoding>\n <content>{sme_base64}</content>\n </dokument>\n <dokument>\n <type>naeringsspesifikasjon</type>\n <encoding>utf-8</encoding>\n <content>{naeringsspeifikasjon_base64}</content>\n </dokument>\n </dokumenter>\n <dokumentreferanseTilGjeldendeDokument>\n <dokumenttype>skattemeldingPersonlig</dokumenttype>\n <dokumentidentifikator>{dok_ref}</dokumentidentifikator>\n </dokumentreferanseTilGjeldendeDokument>\n <inntektsaar>2021</inntektsaar>\n <innsendingsinformasjon>\n <innsendingstype>{innsendingstype}</innsendingstype>\n <opprettetAv>TurboSkatt</opprettetAv>\n </innsendingsinformasjon>\n</skattemeldingOgNaeringsspesifikasjonRequest>\n\"\"\".replace(\"\\n\",\"\")\n\n\nnaering_enk = valider_konvlutt_v2.format(sme_base64=sme_base64,\n naeringsspeifikasjon_base64=naeringsspesifikasjoner_base64,\n dok_ref=dok_ref,\n innsendingstype=innsendingstype)",
"_____no_output_____"
]
],
[
[
"# Valider utkast sme med næringsopplysninger",
"_____no_output_____"
]
],
[
[
"def valider_sme(payload):\n url_valider = f'https://mp-test.sits.no/api/skattemelding/v2/valider/2021/{orgnr_as}'\n header = dict(idporten_header)\n header[\"Content-Type\"] = \"application/xml\"\n return s.post(url_valider, headers=header, data=payload)\n\n\nvalider_respons = valider_sme(naering_enk)\nresultatAvValidering = xmltodict.parse(valider_respons.text)[\"skattemeldingerOgNaeringsspesifikasjonResponse\"][\"resultatAvValidering\"]\n\nif valider_respons:\n print(resultatAvValidering)\n print()\n print(xml.dom.minidom.parseString(valider_respons.text).toprettyxml())\nelse:\n print(valider_respons.status_code, valider_respons.headers, valider_respons.text)",
"_____no_output_____"
]
],
[
[
"# Altinn 3",
"_____no_output_____"
],
[
"1. Hent Altinn Token\n2. Oppretter en ny instans av skjemaet\n3. lasteropp metadata til skjemaet\n4. last opp vedlegg til skattemeldingen\n5. oppdater skattemelding xml med referanse til vedlegg_id fra altinn3. \n6. laster opp skattemeldingen og næringsopplysninger som et vedlegg",
"_____no_output_____"
]
],
[
[
"#1\naltinn3_applikasjon = \"skd/formueinntekt-skattemelding-v2\"\naltinn_header = hent_altinn_token(idporten_header)\n#2\ninstans_data = opprett_ny_instans(altinn_header, fnr, appnavn=altinn3_applikasjon)",
"_____no_output_____"
]
],
[
[
"### 3 Last opp metadata (skattemelding_V1)\n",
"_____no_output_____"
]
],
[
[
"print(f\"innsendingstypen er satt til: {innsendingstype}\")\nreq_metadata = last_opp_metadata_json(instans_data, altinn_header, inntektsaar=2021, appnavn=altinn3_applikasjon)\nreq_metadata",
"_____no_output_____"
]
],
[
[
"## Last opp skattemelding",
"_____no_output_____"
]
],
[
[
"#Last opp skattemeldingen\nreq_send_inn = last_opp_skattedata(instans_data, altinn_header, \n xml=naering_enk, \n data_type=\"skattemeldingOgNaeringspesifikasjon\",\n appnavn=altinn3_applikasjon)\nreq_send_inn",
"_____no_output_____"
]
],
[
[
"### Sett statusen klar til henting av skatteetaten. ",
"_____no_output_____"
]
],
[
[
"req_bekreftelse = endre_prosess_status(instans_data, altinn_header, \"next\", appnavn=altinn3_applikasjon)\nreq_bekreftelse = endre_prosess_status(instans_data, altinn_header, \"next\", appnavn=altinn3_applikasjon)\nreq_bekreftelse",
"_____no_output_____"
]
],
[
[
"### Framtidig: Sjekk status på altinn3 instansen om skatteetaten har hentet instansen. ",
"_____no_output_____"
],
[
"### Se innsending i Altinn\n\nTa en slurk av kaffen og klapp deg selv på ryggen, du har nå sendt inn, la byråkratiet gjøre sin ting... og det tar litt tid. Pt så sjekker skatteeaten hos Altinn3 hvert 5 min om det har kommet noen nye innsendinger. ",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
cbf3676e87baf7789fef58296dc8dd8bcfdaaf6e
| 60,829 |
ipynb
|
Jupyter Notebook
|
courses/coursera/DeepLearning/ConvolutionalNeuralNetworks/Keras+-+Tutorial+-+Happy+House+v2.ipynb
|
sshh12/MLCode
|
692730c83d2ed72ce6e1b0c5e734dffab4c5b53b
|
[
"MIT"
] | 2 |
2017-08-18T12:51:43.000Z
|
2017-10-29T03:56:23.000Z
|
courses/coursera/DeepLearning/ConvolutionalNeuralNetworks/Keras+-+Tutorial+-+Happy+House+v2.ipynb
|
sshh12/MLCode
|
692730c83d2ed72ce6e1b0c5e734dffab4c5b53b
|
[
"MIT"
] | null | null | null |
courses/coursera/DeepLearning/ConvolutionalNeuralNetworks/Keras+-+Tutorial+-+Happy+House+v2.ipynb
|
sshh12/MLCode
|
692730c83d2ed72ce6e1b0c5e734dffab4c5b53b
|
[
"MIT"
] | null | null | null | 85.674648 | 27,832 | 0.732973 |
[
[
[
"# Keras tutorial - the Happy House\n\nWelcome to the first assignment of week 2. In this assignment, you will:\n1. Learn to use Keras, a high-level neural networks API (programming framework), written in Python and capable of running on top of several lower-level frameworks including TensorFlow and CNTK. \n2. See how you can in a couple of hours build a deep learning algorithm.\n\nWhy are we using Keras? Keras was developed to enable deep learning engineers to build and experiment with different models very quickly. Just as TensorFlow is a higher-level framework than Python, Keras is an even higher-level framework and provides additional abstractions. Being able to go from idea to result with the least possible delay is key to finding good models. However, Keras is more restrictive than the lower-level frameworks, so there are some very complex models that you can implement in TensorFlow but not (without more difficulty) in Keras. That being said, Keras will work fine for many common models. \n\nIn this exercise, you'll work on the \"Happy House\" problem, which we'll explain below. Let's load the required packages and solve the problem of the Happy House!",
"_____no_output_____"
]
],
[
[
"import numpy as np\nfrom keras import layers\nfrom keras.layers import Input, Dense, Activation, ZeroPadding2D, BatchNormalization, Flatten, Conv2D\nfrom keras.layers import AveragePooling2D, MaxPooling2D, Dropout, GlobalMaxPooling2D, GlobalAveragePooling2D\nfrom keras.models import Model\nfrom keras.preprocessing import image\nfrom keras.utils import layer_utils\nfrom keras.utils.data_utils import get_file\nfrom keras.applications.imagenet_utils import preprocess_input\nimport pydot\nfrom IPython.display import SVG\nfrom keras.utils.vis_utils import model_to_dot\nfrom keras.utils import plot_model\nfrom kt_utils import *\n\nimport keras.backend as K\nK.set_image_data_format('channels_last')\nimport matplotlib.pyplot as plt\nfrom matplotlib.pyplot import imshow\n\n%matplotlib inline",
"Using TensorFlow backend.\n"
]
],
[
[
"**Note**: As you can see, we've imported a lot of functions from Keras. You can use them easily just by calling them directly in the notebook. Ex: `X = Input(...)` or `X = ZeroPadding2D(...)`.",
"_____no_output_____"
],
[
"## 1 - The Happy House \n\nFor your next vacation, you decided to spend a week with five of your friends from school. It is a very convenient house with many things to do nearby. But the most important benefit is that everybody has commited to be happy when they are in the house. So anyone wanting to enter the house must prove their current state of happiness.\n\n<img src=\"images/happy-house.jpg\" style=\"width:350px;height:270px;\">\n<caption><center> <u> <font color='purple'> **Figure 1** </u><font color='purple'> : **the Happy House**</center></caption>\n\n\nAs a deep learning expert, to make sure the \"Happy\" rule is strictly applied, you are going to build an algorithm which that uses pictures from the front door camera to check if the person is happy or not. The door should open only if the person is happy. \n\nYou have gathered pictures of your friends and yourself, taken by the front-door camera. The dataset is labbeled. \n\n<img src=\"images/house-members.png\" style=\"width:550px;height:250px;\">\n\nRun the following code to normalize the dataset and learn about its shapes.",
"_____no_output_____"
]
],
[
[
"X_train_orig, Y_train_orig, X_test_orig, Y_test_orig, classes = load_dataset()\n\n# Normalize image vectors\nX_train = X_train_orig/255.\nX_test = X_test_orig/255.\n\n# Reshape\nY_train = Y_train_orig.T\nY_test = Y_test_orig.T\n\nprint (\"number of training examples = \" + str(X_train.shape[0]))\nprint (\"number of test examples = \" + str(X_test.shape[0]))\nprint (\"X_train shape: \" + str(X_train.shape))\nprint (\"Y_train shape: \" + str(Y_train.shape))\nprint (\"X_test shape: \" + str(X_test.shape))\nprint (\"Y_test shape: \" + str(Y_test.shape))",
"number of training examples = 600\nnumber of test examples = 150\nX_train shape: (600, 64, 64, 3)\nY_train shape: (600, 1)\nX_test shape: (150, 64, 64, 3)\nY_test shape: (150, 1)\n"
]
],
[
[
"**Details of the \"Happy\" dataset**:\n- Images are of shape (64,64,3)\n- Training: 600 pictures\n- Test: 150 pictures\n\nIt is now time to solve the \"Happy\" Challenge.",
"_____no_output_____"
],
[
"## 2 - Building a model in Keras\n\nKeras is very good for rapid prototyping. In just a short time you will be able to build a model that achieves outstanding results.\n\nHere is an example of a model in Keras:\n\n```python\ndef model(input_shape):\n # Define the input placeholder as a tensor with shape input_shape. Think of this as your input image!\n X_input = Input(input_shape)\n\n # Zero-Padding: pads the border of X_input with zeroes\n X = ZeroPadding2D((3, 3))(X_input)\n\n # CONV -> BN -> RELU Block applied to X\n X = Conv2D(32, (7, 7), strides = (1, 1), name = 'conv0')(X)\n X = BatchNormalization(axis = 3, name = 'bn0')(X)\n X = Activation('relu')(X)\n\n # MAXPOOL\n X = MaxPooling2D((2, 2), name='max_pool')(X)\n\n # FLATTEN X (means convert it to a vector) + FULLYCONNECTED\n X = Flatten()(X)\n X = Dense(1, activation='sigmoid', name='fc')(X)\n\n # Create model. This creates your Keras model instance, you'll use this instance to train/test the model.\n model = Model(inputs = X_input, outputs = X, name='HappyModel')\n \n return model\n```\n\nNote that Keras uses a different convention with variable names than we've previously used with numpy and TensorFlow. In particular, rather than creating and assigning a new variable on each step of forward propagation such as `X`, `Z1`, `A1`, `Z2`, `A2`, etc. for the computations for the different layers, in Keras code each line above just reassigns `X` to a new value using `X = ...`. In other words, during each step of forward propagation, we are just writing the latest value in the commputation into the same variable `X`. The only exception was `X_input`, which we kept separate and did not overwrite, since we needed it at the end to create the Keras model instance (`model = Model(inputs = X_input, ...)` above). \n\n**Exercise**: Implement a `HappyModel()`. This assignment is more open-ended than most. We suggest that you start by implementing a model using the architecture we suggest, and run through the rest of this assignment using that as your initial model. But after that, come back and take initiative to try out other model architectures. For example, you might take inspiration from the model above, but then vary the network architecture and hyperparameters however you wish. You can also use other functions such as `AveragePooling2D()`, `GlobalMaxPooling2D()`, `Dropout()`. \n\n**Note**: You have to be careful with your data's shapes. Use what you've learned in the videos to make sure your convolutional, pooling and fully-connected layers are adapted to the volumes you're applying it to.",
"_____no_output_____"
]
],
[
[
"# GRADED FUNCTION: HappyModel\n\ndef HappyModel(input_shape):\n \"\"\"\n Implementation of the HappyModel.\n \n Arguments:\n input_shape -- shape of the images of the dataset\n\n Returns:\n model -- a Model() instance in Keras\n \"\"\"\n \n ### START CODE HERE ###\n # Feel free to use the suggested outline in the text above to get started, and run through the whole\n # exercise (including the later portions of this notebook) once. The come back also try out other\n # network architectures as well. \n X_input = Input(input_shape)\n\n X = ZeroPadding2D((3, 3))(X_input)\n\n X = Conv2D(32, (7, 7), strides = (1, 1), name = 'conv0')(X)\n X = BatchNormalization(axis = 3, name = 'bn0')(X)\n X = Activation('relu')(X)\n\n X = MaxPooling2D((2, 2), name='max_pool')(X)\n\n X = Flatten()(X)\n X = Dense(1, activation='sigmoid', name='fc')(X)\n\n model = Model(inputs = X_input, outputs = X, name='HappyModel')\n \n ### END CODE HERE ###\n \n return model",
"_____no_output_____"
]
],
[
[
"You have now built a function to describe your model. To train and test this model, there are four steps in Keras:\n1. Create the model by calling the function above\n2. Compile the model by calling `model.compile(optimizer = \"...\", loss = \"...\", metrics = [\"accuracy\"])`\n3. Train the model on train data by calling `model.fit(x = ..., y = ..., epochs = ..., batch_size = ...)`\n4. Test the model on test data by calling `model.evaluate(x = ..., y = ...)`\n\nIf you want to know more about `model.compile()`, `model.fit()`, `model.evaluate()` and their arguments, refer to the official [Keras documentation](https://keras.io/models/model/).\n\n**Exercise**: Implement step 1, i.e. create the model.",
"_____no_output_____"
]
],
[
[
"### START CODE HERE ### (1 line)\nhappyModel = HappyModel((64, 64, 3))\n### END CODE HERE ###",
"_____no_output_____"
]
],
[
[
"**Exercise**: Implement step 2, i.e. compile the model to configure the learning process. Choose the 3 arguments of `compile()` wisely. Hint: the Happy Challenge is a binary classification problem.",
"_____no_output_____"
]
],
[
[
"### START CODE HERE ### (1 line)\nhappyModel.compile(optimizer='adam', loss='binary_crossentropy', metrics = [\"accuracy\"])\n### END CODE HERE ###",
"_____no_output_____"
]
],
[
[
"**Exercise**: Implement step 3, i.e. train the model. Choose the number of epochs and the batch size.",
"_____no_output_____"
]
],
[
[
"### START CODE HERE ### (1 line)\nhappyModel.fit(X_train, Y_train, epochs=45, batch_size=16)\n### END CODE HERE ###",
"Epoch 1/45\n600/600 [==============================] - 13s - loss: 1.4679 - acc: 0.6800 \nEpoch 2/45\n600/600 [==============================] - 13s - loss: 0.2057 - acc: 0.9217 \nEpoch 3/45\n600/600 [==============================] - 13s - loss: 0.1417 - acc: 0.9533 \nEpoch 4/45\n600/600 [==============================] - 13s - loss: 0.1116 - acc: 0.9617 \nEpoch 5/45\n600/600 [==============================] - 13s - loss: 0.1223 - acc: 0.9467 \nEpoch 6/45\n600/600 [==============================] - 13s - loss: 0.0845 - acc: 0.9633 \nEpoch 7/45\n600/600 [==============================] - 13s - loss: 0.0827 - acc: 0.9700 \nEpoch 8/45\n600/600 [==============================] - 13s - loss: 0.1012 - acc: 0.9667 \nEpoch 9/45\n600/600 [==============================] - 13s - loss: 0.0752 - acc: 0.9750 \nEpoch 10/45\n600/600 [==============================] - 13s - loss: 0.0867 - acc: 0.9700 \nEpoch 11/45\n600/600 [==============================] - 13s - loss: 0.0507 - acc: 0.9850 \nEpoch 12/45\n600/600 [==============================] - 13s - loss: 0.1187 - acc: 0.9500 \nEpoch 13/45\n600/600 [==============================] - 12s - loss: 0.0848 - acc: 0.9750 \nEpoch 14/45\n600/600 [==============================] - 12s - loss: 0.1569 - acc: 0.9467 \nEpoch 15/45\n600/600 [==============================] - 12s - loss: 0.0560 - acc: 0.9767 \nEpoch 16/45\n600/600 [==============================] - 12s - loss: 0.0365 - acc: 0.9917 \nEpoch 17/45\n600/600 [==============================] - 12s - loss: 0.0357 - acc: 0.9883 \nEpoch 18/45\n600/600 [==============================] - 12s - loss: 0.0407 - acc: 0.9850 \nEpoch 19/45\n600/600 [==============================] - 12s - loss: 0.1026 - acc: 0.9583 \nEpoch 20/45\n600/600 [==============================] - 12s - loss: 0.0465 - acc: 0.9833 \nEpoch 21/45\n600/600 [==============================] - 12s - loss: 0.1568 - acc: 0.9433 \nEpoch 22/45\n600/600 [==============================] - 12s - loss: 0.0750 - acc: 0.9767 \nEpoch 23/45\n600/600 [==============================] - 12s - loss: 0.0551 - acc: 0.9750 \nEpoch 24/45\n600/600 [==============================] - 12s - loss: 0.0709 - acc: 0.9733 \nEpoch 25/45\n600/600 [==============================] - 12s - loss: 0.0983 - acc: 0.9700 \nEpoch 26/45\n600/600 [==============================] - 12s - loss: 0.0733 - acc: 0.9767 \nEpoch 27/45\n600/600 [==============================] - 12s - loss: 0.0366 - acc: 0.9867 \nEpoch 28/45\n600/600 [==============================] - 12s - loss: 0.0820 - acc: 0.9683 \nEpoch 29/45\n600/600 [==============================] - 12s - loss: 0.1024 - acc: 0.9617 \nEpoch 30/45\n600/600 [==============================] - 13s - loss: 0.1453 - acc: 0.9583 \nEpoch 31/45\n600/600 [==============================] - 13s - loss: 0.0346 - acc: 0.9917 \nEpoch 32/45\n600/600 [==============================] - 13s - loss: 0.0797 - acc: 0.9767 \nEpoch 33/45\n600/600 [==============================] - 13s - loss: 0.0187 - acc: 0.9933 \nEpoch 34/45\n600/600 [==============================] - 13s - loss: 0.0751 - acc: 0.9817 \nEpoch 35/45\n600/600 [==============================] - 13s - loss: 0.0688 - acc: 0.9800 \nEpoch 36/45\n600/600 [==============================] - 14s - loss: 0.0400 - acc: 0.9883 \nEpoch 37/45\n600/600 [==============================] - 14s - loss: 0.0396 - acc: 0.9867 \nEpoch 38/45\n600/600 [==============================] - 14s - loss: 0.0169 - acc: 0.9950 \nEpoch 39/45\n600/600 [==============================] - 14s - loss: 0.1753 - acc: 0.9583 \nEpoch 40/45\n600/600 [==============================] - 14s - loss: 0.0873 - acc: 0.9700 \nEpoch 41/45\n600/600 [==============================] - 14s - loss: 0.0373 - acc: 0.9900 \nEpoch 42/45\n600/600 [==============================] - 14s - loss: 0.0159 - acc: 0.9983 \nEpoch 43/45\n600/600 [==============================] - 14s - loss: 0.0057 - acc: 0.9967 \nEpoch 44/45\n600/600 [==============================] - 14s - loss: 0.0136 - acc: 0.9967 \nEpoch 45/45\n600/600 [==============================] - 14s - loss: 0.0264 - acc: 0.9900 \n"
]
],
[
[
"Note that if you run `fit()` again, the `model` will continue to train with the parameters it has already learnt instead of reinitializing them.\n\n**Exercise**: Implement step 4, i.e. test/evaluate the model.",
"_____no_output_____"
]
],
[
[
"### START CODE HERE ### (1 line)\npreds = happyModel.evaluate(X_test, Y_test)\n### END CODE HERE ###\nprint()\nprint (\"Loss = \" + str(preds[0]))\nprint (\"Test Accuracy = \" + str(preds[1]))",
"150/150 [==============================] - 2s \n\nLoss = 0.72265175422\nTest Accuracy = 0.88666667064\n"
]
],
[
[
"If your `happyModel()` function worked, you should have observed much better than random-guessing (50%) accuracy on the train and test sets.\n\nTo give you a point of comparison, our model gets around **95% test accuracy in 40 epochs** (and 99% train accuracy) with a mini batch size of 16 and \"adam\" optimizer. But our model gets decent accuracy after just 2-5 epochs, so if you're comparing different models you can also train a variety of models on just a few epochs and see how they compare. \n\nIf you have not yet achieved a very good accuracy (let's say more than 80%), here're some things you can play around with to try to achieve it:\n\n- Try using blocks of CONV->BATCHNORM->RELU such as:\n```python\nX = Conv2D(32, (3, 3), strides = (1, 1), name = 'conv0')(X)\nX = BatchNormalization(axis = 3, name = 'bn0')(X)\nX = Activation('relu')(X)\n```\nuntil your height and width dimensions are quite low and your number of channels quite large (≈32 for example). You are encoding useful information in a volume with a lot of channels. You can then flatten the volume and use a fully-connected layer.\n- You can use MAXPOOL after such blocks. It will help you lower the dimension in height and width.\n- Change your optimizer. We find Adam works well. \n- If the model is struggling to run and you get memory issues, lower your batch_size (12 is usually a good compromise)\n- Run on more epochs, until you see the train accuracy plateauing. \n\nEven if you have achieved a good accuracy, please feel free to keep playing with your model to try to get even better results. \n\n**Note**: If you perform hyperparameter tuning on your model, the test set actually becomes a dev set, and your model might end up overfitting to the test (dev) set. But just for the purpose of this assignment, we won't worry about that here.\n",
"_____no_output_____"
],
[
"## 3 - Conclusion\n\nCongratulations, you have solved the Happy House challenge! \n\nNow, you just need to link this model to the front-door camera of your house. We unfortunately won't go into the details of how to do that here. ",
"_____no_output_____"
],
[
"<font color='blue'>\n**What we would like you to remember from this assignment:**\n- Keras is a tool we recommend for rapid prototyping. It allows you to quickly try out different model architectures. Are there any applications of deep learning to your daily life that you'd like to implement using Keras? \n- Remember how to code a model in Keras and the four steps leading to the evaluation of your model on the test set. Create->Compile->Fit/Train->Evaluate/Test.",
"_____no_output_____"
],
[
"## 4 - Test with your own image (Optional)\n\nCongratulations on finishing this assignment. You can now take a picture of your face and see if you could enter the Happy House. To do that:\n 1. Click on \"File\" in the upper bar of this notebook, then click \"Open\" to go on your Coursera Hub.\n 2. Add your image to this Jupyter Notebook's directory, in the \"images\" folder\n 3. Write your image's name in the following code\n 4. Run the code and check if the algorithm is right (0 is unhappy, 1 is happy)!\n \nThe training/test sets were quite similar; for example, all the pictures were taken against the same background (since a front door camera is always mounted in the same position). This makes the problem easier, but a model trained on this data may or may not work on your own data. But feel free to give it a try! ",
"_____no_output_____"
]
],
[
[
"### START CODE HERE ###\nimg_path = 'images/my_image.jpg'\n### END CODE HERE ###\nimg = image.load_img(img_path, target_size=(64, 64))\nimshow(img)\n\nx = image.img_to_array(img)\nx = np.expand_dims(x, axis=0)\nx = preprocess_input(x)\n\nprint(happyModel.predict(x))",
"[[ 0.]]\n"
]
],
[
[
"## 5 - Other useful functions in Keras (Optional)\n\nTwo other basic features of Keras that you'll find useful are:\n- `model.summary()`: prints the details of your layers in a table with the sizes of its inputs/outputs\n- `plot_model()`: plots your graph in a nice layout. You can even save it as \".png\" using SVG() if you'd like to share it on social media ;). It is saved in \"File\" then \"Open...\" in the upper bar of the notebook.\n\nRun the following code.",
"_____no_output_____"
]
],
[
[
"happyModel.summary()",
"_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\ninput_1 (InputLayer) (None, 64, 64, 3) 0 \n_________________________________________________________________\nzero_padding2d_1 (ZeroPaddin (None, 70, 70, 3) 0 \n_________________________________________________________________\nconv0 (Conv2D) (None, 64, 64, 32) 4736 \n_________________________________________________________________\nbn0 (BatchNormalization) (None, 64, 64, 32) 128 \n_________________________________________________________________\nactivation_1 (Activation) (None, 64, 64, 32) 0 \n_________________________________________________________________\nmax_pool (MaxPooling2D) (None, 32, 32, 32) 0 \n_________________________________________________________________\nflatten_1 (Flatten) (None, 32768) 0 \n_________________________________________________________________\nfc (Dense) (None, 1) 32769 \n=================================================================\nTotal params: 37,633\nTrainable params: 37,569\nNon-trainable params: 64\n_________________________________________________________________\n"
],
[
"plot_model(happyModel, to_file='HappyModel.png')\nSVG(model_to_dot(happyModel).create(prog='dot', format='svg'))",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
cbf367cd52802afe168796669d0cec4d3c3d1d16
| 5,649 |
ipynb
|
Jupyter Notebook
|
1_2_Convolutional_Filters_Edge_Detection/7. Haar Cascade, Face Detection.ipynb
|
schase15/CV_Udacity
|
2d8859580e76fc94b3f154a5b52e1029a8217f5c
|
[
"MIT"
] | null | null | null |
1_2_Convolutional_Filters_Edge_Detection/7. Haar Cascade, Face Detection.ipynb
|
schase15/CV_Udacity
|
2d8859580e76fc94b3f154a5b52e1029a8217f5c
|
[
"MIT"
] | null | null | null |
1_2_Convolutional_Filters_Edge_Detection/7. Haar Cascade, Face Detection.ipynb
|
schase15/CV_Udacity
|
2d8859580e76fc94b3f154a5b52e1029a8217f5c
|
[
"MIT"
] | null | null | null | 32.653179 | 401 | 0.615861 |
[
[
[
"## Face detection using OpenCV\n\nOne older (from around 2001), but still popular scheme for face detection is a Haar cascade classifier; these classifiers in the OpenCV library and use feature-based classification cascades that learn to isolate and detect faces in an image. You can read [the original paper proposing this approach here](https://www.cs.cmu.edu/~efros/courses/LBMV07/Papers/viola-cvpr-01.pdf).\n\nLet's see how face detection works on an exampe in this notebook.",
"_____no_output_____"
]
],
[
[
"# import required libraries for this section\n%matplotlib inline\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport cv2 ",
"_____no_output_____"
],
[
"# load in color image for face detection\nimage = cv2.imread('images/multi_faces.jpg')\n\n# convert to RBG\nimage = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)\n\nplt.figure(figsize=(20,10))\nplt.imshow(image)",
"_____no_output_____"
]
],
[
[
"To use a face detector, we'll first convert the image from color to grayscale. For face detection this is perfectly fine to do as there is plenty non-color specific structure in the human face for our detector to learn on.",
"_____no_output_____"
]
],
[
[
"# convert to grayscale\ngray = cv2.cvtColor(image, cv2.COLOR_BGR2GRAY) \n\nplt.figure(figsize=(20,10))\nplt.imshow(gray, cmap='gray')",
"_____no_output_____"
]
],
[
[
"Next we load in the fully trained architecture of the face detector, found in the file `detector_architectures/ haarcascade_frontalface_default.xml`,and use it on our image to find faces!\n\n**A note on parameters** \n\nHow many faces are detected is determined by the function, `detectMultiScale` which aims to detect faces of varying sizes. The inputs to this function are: `(image, scaleFactor, minNeighbors)`; you will often detect more faces with a smaller scaleFactor, and lower value for minNeighbors, but raising these values often produces better matches. Modify these values depending on your input image.",
"_____no_output_____"
]
],
[
[
"# load in cascade classifier\nface_cascade = cv2.CascadeClassifier('detector_architectures/haarcascade_frontalface_default.xml')\n\n# run the detector on the grayscale image\nfaces = face_cascade.detectMultiScale(gray, 4, 6)",
"_____no_output_____"
]
],
[
[
"The output of the classifier is an array of detections; coordinates that define the dimensions of a bounding box around each face. Note that this always outputs a bounding box that is square in dimension.",
"_____no_output_____"
]
],
[
[
"# print out the detections found\nprint ('We found ' + str(len(faces)) + ' faces in this image')\nprint (\"Their coordinates and lengths/widths are as follows\")\nprint ('=============================')\nprint (faces)",
"_____no_output_____"
]
],
[
[
"Let's plot the corresponding detection boxes on our original image to see how well we've done. \n- The coordinates are in form (x,y,w,h)\n- To get the four coordinates of the bounding box (x, y, x+w, y+h)",
"_____no_output_____"
]
],
[
[
"# make a copy of the original image to plot rectangle detections ontop of\nimg_with_detections = np.copy(image) \n\n# loop over our detections and draw their corresponding boxes on top of our original image\nfor (x,y,w,h) in faces:\n # draw next detection as a red rectangle on top of the original image. \n # Note: the fourth element (255,0,0) determines the color of the rectangle, \n # and the final argument (here set to 5) determines the width of the lines that draw the rectangle\n cv2.rectangle(img_with_detections,(x,y),(x+w,y+h),(255,0,0),5) \n\n# display the result\nplt.figure(figsize=(20,10))\nplt.imshow(img_with_detections)",
"_____no_output_____"
]
],
[
[
"- Article about a GAN model that detects its own bias (racial/gender) and corrects its predictions (https://godatadriven.com/blog/fairness-in-machine-learning-with-pytorch/)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
cbf3754e4fdf4b9cf8de961c674e6a86af68922e
| 404,671 |
ipynb
|
Jupyter Notebook
|
docs/example-analyses/vote-fatigue.ipynb
|
htshnr/caption-contest-data
|
13d61b1448bca8d19464faca79d16d16f2fe8fc6
|
[
"CC-BY-4.0",
"BSD-3-Clause"
] | 62 |
2016-04-12T14:00:47.000Z
|
2022-03-18T19:09:47.000Z
|
docs/example-analyses/vote-fatigue.ipynb
|
htshnr/caption-contest-data
|
13d61b1448bca8d19464faca79d16d16f2fe8fc6
|
[
"CC-BY-4.0",
"BSD-3-Clause"
] | 28 |
2016-02-26T17:39:14.000Z
|
2022-02-10T00:24:51.000Z
|
docs/example-analyses/vote-fatigue.ipynb
|
blakemas/NEXT_data
|
7cbe8080b441fc91e2e8198ec47c750e6517f83f
|
[
"CC-BY-4.0",
"BSD-3-Clause"
] | 12 |
2017-10-20T08:46:04.000Z
|
2021-12-25T01:35:12.000Z
| 473.299415 | 228,580 | 0.929254 |
[
[
[
"# How do ratings behave after users have seen many captions?",
"_____no_output_____"
],
[
"This notebook looks at the \"vote decay\" of users. The New Yorker caption contest organizer, Bob Mankoff, has received many emails like the one below (name/personal details left out for anonymity)\n\n> Here's my issue. \n>\n> First time I encounter something, I might say it's funny. \n>\n> Then it comes back in many forms over and over and it's no longer funny and I wish I could go back to the first one and say it's not funny. \n>\n> But it's funny, and then I can't decide whether to credit everyone with funny or keep hitting unfunny. What I really like to find out is who submitted it first, but often it's slightly different and there may be a best version. Auggh!\n>\n> How should we do this???\n\nWe can investigate this: we have all the data at hand. We record the timestamp, participant ID and their rating for a given caption. So let's see how votes go after a user has seen $n$ captions!",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nimport pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nplt.style.use('seaborn')",
"_____no_output_____"
],
[
"import caption_contest_data as ccd",
"_____no_output_____"
]
],
[
[
"## Reading in data\nLet's read in the data. As the last column can contain a non-escaped comma, we have to fix that before doing any analysis.\n\nNote that two versions of this notebook exist (the previous notebook can be found in [43bc5d]). This highlights some of the differences required to read in the earlier datasets.\n\n[43bc5d]:https://github.com/nextml/caption-contest-data/commit/43bc5d23ee287b8b34cc4eb0181484bd21bbd341",
"_____no_output_____"
]
],
[
[
"contest = 540\n\nresponses = ccd.responses(contest)\n\nprint(len(responses))\nresponses.head()",
"452547\n"
]
],
[
[
"## Seeing how many captions a user has seen\nThis is the workhorse of the notebook: it sees how many captions one participant has seen. I sorted by timestamp (and with an actual timestamp, not a str) to collect the ratings in the order a user has seen. I do not assume that only one user answers at a time.",
"_____no_output_____"
]
],
[
[
"last_id = None\ni = 0\nnum_responses = []\ncaptions_seen = []\nresponses = responses.sort_values(by='timestamp_query_generated')\n# responses = responses[0:1000] # debug",
"_____no_output_____"
],
[
"captions_seen_by = {}\ncaptions_seen = []\nfor _, response in responses.iterrows():\n id_, rating = response['participant_uid'], response['target_reward']\n if id_ not in captions_seen_by:\n captions_seen_by[id_] = 0\n captions_seen_by[id_] += 1\n captions_seen += [captions_seen_by[id_]]\n num_responses += [i]",
"_____no_output_____"
],
[
"responses['number of captions seen'] = captions_seen\nresponses.head()",
"_____no_output_____"
]
],
[
[
"## Viewing the data\nNow let's format the data to view it. We can view the data in two ways: as we only have three rating values, we can view the probability of a person rating 1, 2 or 3, and can also view the mean.\n\nIn this, we rely on `pd.pivot_table`. This can take DataFrame that looks like a list of dictionaries and compute `aggfunc` (by default `np.mean`) for all items that contain common keys (indicated by `index` and `columns`). It's similar to Excel's pivot table functionality.",
"_____no_output_____"
],
[
"### Probability of rating {1, 2, 3}",
"_____no_output_____"
]
],
[
[
"def prob(x):\n n = len(x)\n ret = {'n': n}\n ret.update({name: np.sum(x == i) for name, i in [('unfunny', 1),\n ('somewhat funny', 2),\n ('funny', 3)]})\n return ret\n\nprobs = responses.pivot_table(index='number of captions seen',\n columns='alg_label', values='target_reward',\n aggfunc=prob)\nprobs.head()",
"_____no_output_____"
],
[
"d = {label: dict(probs[label]) for label in ['RandomSampling']}\nfor label in d.keys():\n for n in d[label].keys():\n if d[label][n] is None:\n continue\n for rating in ['unfunny', 'somewhat funny', 'funny']:\n d[label][n][rating] = d[label][n][rating] / d[label][n]['n']",
"_____no_output_____"
],
[
"df = pd.DataFrame(d['RandomSampling']).T\ndf = pd.concat({'RandomSampling': df}, axis=1)\ndf.head()",
"_____no_output_____"
],
[
"plt.style.use(\"default\")\n\nfig, axs = plt.subplots(figsize=(8, 4), ncols=2)\n\nalg = \"RandomSampling\"\nshow = df[alg].copy()\nshow[\"captions seen\"] = show.index\n\nfor y in [\"funny\", \"somewhat funny\", \"unfunny\"]:\n show.plot(x=\"captions seen\", y=y, ax=axs[0])\nshow.plot(x=\"captions seen\", y=\"n\", ax=axs[1])\n\nfor ax in axs:\n ax.set_xlim(0, 100)\n ax.grid(linestyle='--', alpha=0.5)\n",
"_____no_output_____"
],
[
"plt.style.use(\"default\")\n\ndef plot(alg):\n fig = plt.figure(figsize=(10, 5))\n ax = plt.subplot(1, 2, 1)\n df[alg][['unfunny', 'somewhat funny', 'funny']].plot(ax=ax)\n plt.xlim(0, 100)\n plt.title('{} ratings\\nfor contest {}'.format(alg, contest))\n plt.ylabel('Probability of rating')\n plt.xlabel('Number of captions seen')\n plt.grid(linestyle=\"--\", alpha=0.6)\n\n ax = plt.subplot(1, 2, 2)\n df[alg]['n'].plot(ax=ax, logy=False)\n plt.ylabel('Number of users')\n plt.xlabel('Number of captions seen, $n$')\n plt.title('Number of users that have\\nseen $n$ captions')\n plt.xlim(0, 100)\n plt.grid(linestyle=\"--\", alpha=0.6)\n \nfor alg in ['RandomSampling']:\n fig = plot(alg)\n plt.show()",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
cbf380208f0463afcd4a9e91d5708ddf8b021a63
| 18,596 |
ipynb
|
Jupyter Notebook
|
sagemaker-experiments/mnist-handwritten-digits-classification-experiment/mnist-handwritten-digits-classification-experiment.ipynb
|
fhirschmann/amazon-sagemaker-examples
|
bb4a4ed78cd4f3673bd6894f0b92ab08aa7f8f29
|
[
"Apache-2.0"
] | null | null | null |
sagemaker-experiments/mnist-handwritten-digits-classification-experiment/mnist-handwritten-digits-classification-experiment.ipynb
|
fhirschmann/amazon-sagemaker-examples
|
bb4a4ed78cd4f3673bd6894f0b92ab08aa7f8f29
|
[
"Apache-2.0"
] | null | null | null |
sagemaker-experiments/mnist-handwritten-digits-classification-experiment/mnist-handwritten-digits-classification-experiment.ipynb
|
fhirschmann/amazon-sagemaker-examples
|
bb4a4ed78cd4f3673bd6894f0b92ab08aa7f8f29
|
[
"Apache-2.0"
] | null | null | null | 33.207143 | 533 | 0.590826 |
[
[
[
"## MNIST Handwritten Digits Classification Experiment\n\nThis demo shows how you can use SageMaker Experiment Management Python SDK to organize, track, compare, and evaluate your machine learning (ML) model training experiments.\n\nYou can track artifacts for experiments, including data sets, algorithms, hyper-parameters, and metrics. Experiments executed on SageMaker such as SageMaker Autopilot jobs and training jobs will be automatically tracked. You can also track artifacts for additional steps within an ML workflow that come before/after model training e.g. data pre-processing or post-training model evaluation.\n\nThe APIs also let you search and browse your current and past experiments, compare experiments, and identify best performing models.\n\nNow we will demonstrate these capabilities through an MNIST handwritten digits classification example. The experiment will be organized as follow:\n\n1. Download and prepare the MNIST dataset.\n2. Train a Convolutional Neural Network (CNN) Model. Tune the hyper parameter that configures the number of hidden channels in the model. Track the parameter configurations and resulting model accuracy using SageMaker Experiments Python SDK.\n3. Finally use the search and analytics capabilities of Python SDK to search, compare and evaluate the performance of all model versions generated from model tuning in Step 2.\n4. We will also see an example of tracing the complete linage of a model version i.e. the collection of all the data pre-processing and training configurations and inputs that went into creating that model version.\n\nMake sure you selected `Python 3 (Data Science)` kernel.",
"_____no_output_____"
],
[
"### Install Python SDKs",
"_____no_output_____"
]
],
[
[
"import sys",
"_____no_output_____"
],
[
"!{sys.executable} -m pip install sagemaker-experiments==0.1.24",
"_____no_output_____"
]
],
[
[
"### Install PyTroch",
"_____no_output_____"
]
],
[
[
"# pytorch version needs to be the same in both the notebook instance and the training job container \n# https://github.com/pytorch/pytorch/issues/25214\n!{sys.executable} -m pip install torch==1.1.0\n!{sys.executable} -m pip install torchvision==0.3.0\n!{sys.executable} -m pip install pillow==6.2.2\n!{sys.executable} -m pip install --upgrade sagemaker",
"_____no_output_____"
]
],
[
[
"### Setup",
"_____no_output_____"
]
],
[
[
"import time\n\nimport boto3\nimport numpy as np\nimport pandas as pd\nfrom IPython.display import set_matplotlib_formats\nfrom matplotlib import pyplot as plt\nfrom torchvision import datasets, transforms\n\nimport sagemaker\nfrom sagemaker import get_execution_role\nfrom sagemaker.session import Session\nfrom sagemaker.analytics import ExperimentAnalytics\n\nfrom smexperiments.experiment import Experiment\nfrom smexperiments.trial import Trial\nfrom smexperiments.trial_component import TrialComponent\nfrom smexperiments.tracker import Tracker\n\nset_matplotlib_formats('retina')",
"_____no_output_____"
],
[
"sess = boto3.Session()\nsm = sess.client('sagemaker')\nrole = get_execution_role()",
"_____no_output_____"
]
],
[
[
"### Create a S3 bucket to hold data",
"_____no_output_____"
]
],
[
[
"# create a s3 bucket to hold data, note that your account might already created a bucket with the same name\naccount_id = sess.client('sts').get_caller_identity()[\"Account\"]\nbucket = 'sagemaker-experiments-{}-{}'.format(sess.region_name, account_id)\nprefix = 'mnist'\n\ntry:\n if sess.region_name == \"us-east-1\":\n sess.client('s3').create_bucket(Bucket=bucket)\n else:\n sess.client('s3').create_bucket(Bucket=bucket, \n CreateBucketConfiguration={'LocationConstraint': sess.region_name})\nexcept Exception as e:\n print(e)",
"_____no_output_____"
]
],
[
[
"### Dataset\nWe download the MNIST hand written digits dataset, and then apply transformation on each of the image.",
"_____no_output_____"
]
],
[
[
"# TODO: can be removed after upgrade to torchvision==0.9.1\n# see github.com/pytorch/vision/issues/1938 and github.com/pytorch/vision/issues/3549\ndatasets.MNIST.urls = [\n 'https://ossci-datasets.s3.amazonaws.com/mnist/train-images-idx3-ubyte.gz',\n 'https://ossci-datasets.s3.amazonaws.com/mnist/train-labels-idx1-ubyte.gz',\n 'https://ossci-datasets.s3.amazonaws.com/mnist/t10k-images-idx3-ubyte.gz',\n 'https://ossci-datasets.s3.amazonaws.com/mnist/t10k-labels-idx1-ubyte.gz'\n]\n\n# download the dataset\n# this will not only download data to ./mnist folder, but also load and transform (normalize) them\ntrain_set = datasets.MNIST('mnist', train=True, transform=transforms.Compose([\n transforms.ToTensor(),\n transforms.Normalize((0.1307,), (0.3081,))]), \n download=True)\n \ntest_set = datasets.MNIST('mnist', train=False, transform=transforms.Compose([\n transforms.ToTensor(),\n transforms.Normalize((0.1307,), (0.3081,))]),\n download=False)",
"_____no_output_____"
],
[
"plt.imshow(train_set.data[2].numpy())",
"_____no_output_____"
]
],
[
[
"After transforming the images in the dataset, we upload it to s3.",
"_____no_output_____"
]
],
[
[
"inputs = sagemaker.Session().upload_data(path='mnist', bucket=bucket, key_prefix=prefix)\nprint('input spec: {}'.format(inputs))",
"_____no_output_____"
]
],
[
[
"Now lets track the parameters from the data pre-processing step.",
"_____no_output_____"
]
],
[
[
"with Tracker.create(display_name=\"Preprocessing\", sagemaker_boto_client=sm) as tracker:\n tracker.log_parameters({\n \"normalization_mean\": 0.1307,\n \"normalization_std\": 0.3081,\n })\n # we can log the s3 uri to the dataset we just uploaded\n tracker.log_input(name=\"mnist-dataset\", media_type=\"s3/uri\", value=inputs)",
"_____no_output_____"
]
],
[
[
"### Step 1 - Set up the Experiment\n\nCreate an experiment to track all the model training iterations. Experiments are a great way to organize your data science work. You can create experiments to organize all your model development work for : [1] a business use case you are addressing (e.g. create experiment named “customer churn prediction”), or [2] a data science team that owns the experiment (e.g. create experiment named “marketing analytics experiment”), or [3] a specific data science and ML project. Think of it as a “folder” for organizing your “files”.",
"_____no_output_____"
],
[
"### Create an Experiment",
"_____no_output_____"
]
],
[
[
"mnist_experiment = Experiment.create(\n experiment_name=f\"mnist-hand-written-digits-classification-{int(time.time())}\", \n description=\"Classification of mnist hand-written digits\", \n sagemaker_boto_client=sm)\nprint(mnist_experiment)",
"_____no_output_____"
]
],
[
[
"### Step 2 - Track Experiment\n### Now create a Trial for each training run to track the it's inputs, parameters, and metrics.\nWhile training the CNN model on SageMaker, we will experiment with several values for the number of hidden channel in the model. We will create a Trial to track each training job run. We will also create a TrialComponent from the tracker we created before, and add to the Trial. This will enrich the Trial with the parameters we captured from the data pre-processing stage.\n\nNote the execution of the following code takes a while.",
"_____no_output_____"
]
],
[
[
"from sagemaker.pytorch import PyTorch, PyTorchModel",
"_____no_output_____"
],
[
"hidden_channel_trial_name_map = {}",
"_____no_output_____"
]
],
[
[
"If you want to run the following training jobs asynchronously, you may need to increase your resource limit. Otherwise, you can run them sequentially.",
"_____no_output_____"
]
],
[
[
"preprocessing_trial_component = tracker.trial_component",
"_____no_output_____"
],
[
"for i, num_hidden_channel in enumerate([2, 5, 10, 20, 32]):\n # create trial\n trial_name = f\"cnn-training-job-{num_hidden_channel}-hidden-channels-{int(time.time())}\"\n cnn_trial = Trial.create(\n trial_name=trial_name, \n experiment_name=mnist_experiment.experiment_name,\n sagemaker_boto_client=sm,\n )\n hidden_channel_trial_name_map[num_hidden_channel] = trial_name\n \n # associate the proprocessing trial component with the current trial\n cnn_trial.add_trial_component(preprocessing_trial_component)\n \n # all input configurations, parameters, and metrics specified in estimator \n # definition are automatically tracked\n estimator = PyTorch(\n py_version='py3',\n entry_point='./mnist.py',\n role=role,\n sagemaker_session=sagemaker.Session(sagemaker_client=sm),\n framework_version='1.1.0',\n instance_count=1,\n instance_type='ml.c4.xlarge',\n hyperparameters={\n 'epochs': 2,\n 'backend': 'gloo',\n 'hidden_channels': num_hidden_channel,\n 'dropout': 0.2,\n 'kernel_size': 5,\n 'optimizer': 'sgd'\n },\n metric_definitions=[\n {'Name':'train:loss', 'Regex':'Train Loss: (.*?);'},\n {'Name':'test:loss', 'Regex':'Test Average loss: (.*?),'},\n {'Name':'test:accuracy', 'Regex':'Test Accuracy: (.*?)%;'}\n ],\n enable_sagemaker_metrics=True\n )\n \n cnn_training_job_name = \"cnn-training-job-{}\".format(int(time.time()))\n \n # Now associate the estimator with the Experiment and Trial\n estimator.fit(\n inputs={'training': inputs}, \n job_name=cnn_training_job_name,\n experiment_config={\n \"TrialName\": cnn_trial.trial_name,\n \"TrialComponentDisplayName\": \"Training\",\n },\n wait=True,\n )\n \n # give it a while before dispatching the next training job\n time.sleep(2)",
"_____no_output_____"
]
],
[
[
"### Compare the model training runs for an experiment\n\nNow we will use the analytics capabilities of Python SDK to query and compare the training runs for identifying the best model produced by our experiment. You can retrieve trial components by using a search expression.",
"_____no_output_____"
],
[
"### Some Simple Analyses",
"_____no_output_____"
]
],
[
[
"search_expression = {\n \"Filters\":[\n {\n \"Name\": \"DisplayName\",\n \"Operator\": \"Equals\",\n \"Value\": \"Training\",\n }\n ],\n}",
"_____no_output_____"
],
[
"trial_component_analytics = ExperimentAnalytics(\n sagemaker_session=Session(sess, sm), \n experiment_name=mnist_experiment.experiment_name,\n search_expression=search_expression,\n sort_by=\"metrics.test:accuracy.max\",\n sort_order=\"Descending\",\n metric_names=['test:accuracy'],\n parameter_names=['hidden_channels', 'epochs', 'dropout', 'optimizer']\n)",
"_____no_output_____"
],
[
"trial_component_analytics.dataframe()",
"_____no_output_____"
]
],
[
[
"To isolate and measure the impact of change in hidden channels on model accuracy, we vary the number of hidden channel and fix the value for other hyperparameters.\n\nNext let's look at an example of tracing the lineage of a model by accessing the data tracked by SageMaker Experiments for `cnn-training-job-2-hidden-channels` trial",
"_____no_output_____"
]
],
[
[
"lineage_table = ExperimentAnalytics(\n sagemaker_session=Session(sess, sm), \n search_expression={\n \"Filters\":[{\n \"Name\": \"Parents.TrialName\",\n \"Operator\": \"Equals\",\n \"Value\": hidden_channel_trial_name_map[2]\n }]\n },\n sort_by=\"CreationTime\",\n sort_order=\"Ascending\",\n)",
"_____no_output_____"
],
[
"lineage_table.dataframe()",
"_____no_output_____"
]
],
[
[
"## Deploy endpoint for the best training-job / trial component\n\nNow we'll take the best (as sorted) and create an endpoint for it.",
"_____no_output_____"
]
],
[
[
"#Pulling best based on sort in the analytics/dataframe so first is best....\nbest_trial_component_name = trial_component_analytics.dataframe().iloc[0]['TrialComponentName']\nbest_trial_component = TrialComponent.load(best_trial_component_name)\n\nmodel_data = best_trial_component.output_artifacts['SageMaker.ModelArtifact'].value\nenv = {'hidden_channels': str(int(best_trial_component.parameters['hidden_channels'])), \n 'dropout': str(best_trial_component.parameters['dropout']), \n 'kernel_size': str(int(best_trial_component.parameters['kernel_size']))}\nmodel = PyTorchModel(\n model_data, \n role, \n './mnist.py', \n py_version='py3',\n env=env, \n sagemaker_session=sagemaker.Session(sagemaker_client=sm),\n framework_version='1.1.0',\n name=best_trial_component.trial_component_name)\n\npredictor = model.deploy(\n instance_type='ml.m5.xlarge',\n initial_instance_count=1)",
"_____no_output_____"
]
],
[
[
"## Cleanup\n\nOnce we're doing don't forget to clean up the endpoint to prevent unnecessary billing.\n\n> Trial components can exist independent of trials and experiments. You might want keep them if you plan on further exploration. If so, comment out tc.delete()",
"_____no_output_____"
]
],
[
[
"predictor.delete_endpoint()",
"_____no_output_____"
],
[
"mnist_experiment.delete_all(action='--force')",
"_____no_output_____"
]
],
[
[
"## Contact\nSubmit any questions or issues to https://github.com/aws/sagemaker-experiments/issues or mention @aws/sagemakerexperimentsadmin ",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
cbf380f4269bc61722fef5538fbee57237a4382e
| 1,065 |
ipynb
|
Jupyter Notebook
|
Ch01_Introduction/install.ipynb
|
dltech-xyz/d2l-en-read
|
b8f191111ed861c2e0830f0564ed5732b9b2dbb9
|
[
"MIT"
] | 1 |
2021-08-01T02:38:30.000Z
|
2021-08-01T02:38:30.000Z
|
Ch01_Introduction/install.ipynb
|
StevenJokess/d2l-en-read
|
71b0f35971063b9fe5f21319b8072d61c9e5a298
|
[
"MIT"
] | null | null | null |
Ch01_Introduction/install.ipynb
|
StevenJokess/d2l-en-read
|
71b0f35971063b9fe5f21319b8072d61c9e5a298
|
[
"MIT"
] | 1 |
2021-05-05T13:54:26.000Z
|
2021-05-05T13:54:26.000Z
| 18.050847 | 72 | 0.524883 |
[
[
[
"import torch",
"_____no_output_____"
],
[
"torch.__version__",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code"
]
] |
cbf382ffe10cd758c3874d79a6d3dad2918f7234
| 14,061 |
ipynb
|
Jupyter Notebook
|
04_codage_information/07_nbs_flottants.ipynb
|
efloti/cours-nsi-premiere
|
5b05bc81e5f8d7df47bf5785068b4bf4d1e357bb
|
[
"CC0-1.0"
] | null | null | null |
04_codage_information/07_nbs_flottants.ipynb
|
efloti/cours-nsi-premiere
|
5b05bc81e5f8d7df47bf5785068b4bf4d1e357bb
|
[
"CC0-1.0"
] | null | null | null |
04_codage_information/07_nbs_flottants.ipynb
|
efloti/cours-nsi-premiere
|
5b05bc81e5f8d7df47bf5785068b4bf4d1e357bb
|
[
"CC0-1.0"
] | null | null | null | 30.435065 | 265 | 0.523647 |
[
[
[
"# Codage des nombres réels",
"_____no_output_____"
],
[
"Il repose sur l'**écriture scientifique** des nombres réels:\n$${\\Large \\pm\\ \\text{Mantisse}\\times\\text{Base}^{\\text{Exposant}} }$$\n\n$$\\text{où Mantisse}\\in[1;\\text{Base}[\\\\ \\text{ et Exposant est un entier «signé»}$$\n",
"_____no_output_____"
],
[
"Exemples en base 10:\n- $-0,\\!000000138$ s'écrit $-1,\\!38\\times 10^{-7}$,\n- $299\\,790\\,000$ s'écrit $+2,9979\\times 10^{8}$.\n- $-5,\\!29$ s'écrit $-5,\\!29\\cdot 10^{0}$.",
"_____no_output_____"
],
[
"**Note**: en toute rigueur, 0 ne peut pas être représenté dans cette écriture.",
"_____no_output_____"
],
[
"## Codage des nombres purement fractionnaires (strictement inférieurs à 1)",
"_____no_output_____"
],
[
"Les puissances négatives de deux sont **un-demi**, **un-quart**, **un-huitième**, **un-seizième**, **un-trente-deuxième**, etc.",
"_____no_output_____"
],
[
"| **puissances négatives de deux** | $2^{-1}$ | $2^{-2}$ | $2^{-3}$ | $2^{-4}$ | $2^{-5}$ |\n|------------------------|:-----:|:-----:|:-----:|:------:|:---------:|\n| **décimal** | $0,\\!5$ | $0,\\!25$ | $0,\\!125$ | $0,\\!0625$ | $0,\\!03125$ | \n| **binaire** | $0,\\!1$ | $0,\\!01$ | $0,\\!001$ | $0,\\!0001$ | $0,\\!00001$ |",
"_____no_output_____"
],
[
"**Méthode1**: Pour trouver l'écriture binaire d'un nombre de l'intervalle $[0;1[$ (purement fractionnaire), on peut procèder de manière analogue à la «méthode des différences» pour les entiers.",
"_____no_output_____"
],
[
"**Exemple**: Soit à coder le nombre (en base 10) $0,696$ sur **4 bits** - on ne s'intéresse qu'aux bits situés après la virgule. ",
"_____no_output_____"
],
[
"$$\\begin{array}{r|c|l}\n\\text{puissances de 2} & \\text{différences}&\\text{bits}\\cr\n\\hline & 0,\\!696 & \\cr\n0,\\!5 & 0,\\!196& 1\\cr\n0,\\!25 & & 0\\cr\n0,\\!125 & 0,\\!071 & 1\\cr\n0,\\!0625 & 0,\\!0085& 1\\cr\n\\hline\n\\end{array}\n\\\\\\text{donc, sur 4 bits: }0,\\!696 \\text{ correspond environ à }0,\\!1011\\text{ en base } 2$$",
"_____no_output_____"
],
[
"**Notes**: \n\n- En observant les puissances négatives de deux, observer que sur 5 bits, le dernier chiffre du motif binaire serait 0 et qu'il s'agirait toujours d'une **approximation**.\n\n En fait, même avec un très grand nombre de bits, la dernière différence ne serait probablement pas nulle pour cet exemple même si elle tendrait bien sûr à diminuer.\n\n- Il n'est pas difficile de comprendre qu'avec 4 bits l'approximation effectuée ne peut excédé un-seizieme ($2^{-4}$), avec 10 bits elle serait au pire de $1/2^{10}<$ un-millième, avec 20 bits au pire d'un-millionième, etc. ",
"_____no_output_____"
],
[
"**méthode 2**: On peut aussi utiliser des multiplications successives par 2:\n\n**Tant que** la *partie fractionnaire pure n'est pas nulle*:\n- la multiplier **par 2**\n- placer la partie entière (0 ou 1) et la partie fractionnaire pure du résultat dans la colonne correspondante.\n- arrêter la boucle si le nombre d'itération dépasse une «certaine valeur»...\n\nLe codage binaire du nombre fractionnaire pure initial est donné par la colonne parties entières lue de haut en bas.",
"_____no_output_____"
],
[
"$$\\begin{array}{r|l}\n\\text{partie entière}&\\text{partie fractionnaire pure}\\cr\n\\hline\n& 0,\\!696 \\cr\n1 & 0,\\!392 \\cr\n0 & 0,\\!784\\cr\n1 & 0,\\!568\\cr\n1 & 0,\\!136\\cr\n\\dots & \\dots\\cr\n\\end{array}\n\\\\\\text{donc: }0,\\!696 \\text{ correspond environ à }0,\\!1011\\dots\\text{ en base 2}$$",
"_____no_output_____"
],
[
"**Inversement**, Quelle est la valeur en base 10 de $0,\\!0110$ (en base 2)?\n\nLe premier chiffre après la virgule correspond à un-demi, le second à un-quart, etc.\n\ndonc on a un-quart + un-huitième soit $0,25+0,125={\\bf 0,\\!375}$",
"_____no_output_____"
],
[
"## Norme IEEE 754",
"_____no_output_____"
],
[
"L'exemple précédent doit vous faire sentir la difficulté du codage des réels; il a donc été décidé de normaliser cette représentation; il y a (entre autre)\n- la **simple précision** avec 32 bits,\n- la **double précision** avec 64 bits.",
"_____no_output_____"
],
[
"Dans tous les cas, le codage utilise l'écriture scientifique où la «Base» est 2.\n\n$${\\Large \\pm\\ \\text{Mantisse}\\times\\text{2}^{\\text{Exposant}} }$$\n\n$$\\text{où Mantisse}\\in[1;2[\\\\ \\text{ et Exposant est un entier «signé»}$$",
"_____no_output_____"
],
[
"En **simple précision**, Le motif binaire correspondant est organisé comme suit:",
"_____no_output_____"
],
[
"$$\\begin{array}{ccc}\n1\\text{ bit}& 8 \\text{ bits}& 23 \\text{ bits}\\cr\n\\hline\n\\text{signe}& \\text{exposant} & \\text{mantisse}\\cr\n\\hline\n\\end{array}$$",
"_____no_output_____"
],
[
"**Signe**: 0 signifie + et 1 signifie -",
"_____no_output_____"
],
[
"**Mantisse**: Comme elle est toujours de la forme: $$1,\\!b_1b_2b_3\\dots\\\\ \\text{les } b_i \\text{représentent un bit}$$les 23 bits de la mantisse correspondent aux bits situés après la virgule; le bit obligatoirement à 1 est omis - on parle de bit caché.",
"_____no_output_____"
],
[
"*Exemple*: Si la mantisse en mémoire est $0110\\dots 0$, elle correspond à $$\\underbrace{1}_{\\text{bit caché}}+1/4+1/8=1,\\!375$$",
"_____no_output_____"
],
[
"**Exposant**: il peut-être positif ou négatif et on utilise le codage par «valeur biaisée (ou décalée)» pour le représenter.",
"_____no_output_____"
],
[
"*Exemple*: si l'exposant est $0111\\,1010$, qui correspond à soixante-quatre + trente-deux + seize + huit + deux soit à $122$,\n\non lui soustrait le **biais** soit $127$ (pour un octet) ce qui donne $-5$",
"_____no_output_____"
],
[
"*Exemple récapitulatif*: Le mot de 32 bits qui suit interprété comme un flottant en simple précision,\n\n$$\\large\\overbrace{\\color{red}{1}}^{\\text{signe}}\\ \\overbrace{\\color{green}{0111\\,1010}}^{\\text{exposant}}\\ \\overbrace{\\color{blue}{0110\\,0000\\,0000\\,0000\\,0000\\,000}}^{\\text{mantisse}}$$",
"_____no_output_____"
],
[
"correspond au nombre: \n\n$$\\Large\\overbrace{\\color{red}{-}}^{\\text{signe}}\\overbrace{\\color{blue}{1,\\!375}}^{\\text{mantisse}} \\times 2^{\\overbrace{\\color{green}{-5}}^{\\text{exposant}}}\\\\ =-4,\\!296875\\times 10^{-2}$$",
"_____no_output_____"
],
[
"**Notes**: \n- en **simple précision**, on peut représenter approximativement les nombres décimaux positifs de l'intervalle $[10^{-38}; 10^{38}]$ ainsi que les nombres négatifs correspondants. Voici comment on le voit:\n\n$$2^{128}=2^8\\times (2^{10})^{12}\\approx 100 \\times(10^3)^{12}=10^2\\times 10^{36}=10^{38}$$\n\n- en **double précision** (64 bits), la méthode est la même et:\n - l'**exposant** est codé sur **11 bits** (avec un décalage ou biais de 1023),\n - et la **mantisse** sur **52 bits**.",
"_____no_output_____"
],
[
"## Valeurs spéciales",
"_____no_output_____"
],
[
"La norme précise que les valeurs $0000\\,0000$ et $1111\\,1111$ de l'exposant sont **réservées**:\n - le nombre $0$ est représenté conventionnellement avec un bit de signe arbitraires et tous les autres à $0$: on distingue donc $+0$ et $-0$,\n - les **infinis** sont représentés par l'exposant $1111\\,1111$ et une mantisse nulle: ils servent à indiquer le dépassement de capacité,\n - une valeur spéciale `NaN` (pour *Not a Number*) est représentée par un signe à 0, l'exposant $1111\\,1111$ et une mantisse non nulle: elle sert à représenter le résultat d'opérations invalides comme $0/0$, $\\sqrt{-1}$, $0\\times +\\infty$ etc.\n - enfin, lorsque l'exposant est nulle et la mantisse non, on convient que le nombre représenté est:\n $${\\large \\pm~ {\\bf 0},\\!m \\times 2^{-126}}\\qquad \\text{nombre dénormalisé sur 32 bits}$$ où $m$ est la représentation décimale de la mantisse non nulle.",
"_____no_output_____"
],
[
"| Signe | Exposant | mantisse | valeur spéciale |\n|:------:|:--------:|:--------:|:--------------------------------:|\n| 0 | 0...0 | 0 | $+0$ |\n| 1 | 0...0 | 0 | $-0$ |\n| 0 ou 1 | 0...0 | $\\neq 0$ | $\\pm {\\bf 0},\\!m\\times 2^{-127}$ |\n| 0 | 1...1 | 0 | $+\\infty$ |\n| 1 | 1...1 | 0 | $-\\infty$ |\n| 0 | 1...1 | $\\neq 0$ | `NaN` |",
"_____no_output_____"
],
[
"## Avec Python",
"_____no_output_____"
],
[
"Les nombres flottants suivent la norme IEEE 754 en **double précision** (64 bits).\n\nOn peut utiliser la notation décimale ou scientifique pour les définir:",
"_____no_output_____"
]
],
[
[
"x = 1.6\ny = 1.2e-4 # 1,2x10^{-4}\nprint(f\"x={x}, y={y}\")",
"_____no_output_____"
]
],
[
[
"la fonction `float` convertie un entier en un flottant tandis que `int` fait le contraire:",
"_____no_output_____"
]
],
[
[
"x = float(-4)\ny = int(5.9) # simple troncature\nprint(f\"x={x}, y={y}\")",
"_____no_output_____"
]
],
[
[
"L'opérateur `/` de division produit toujours un flottant quelque soit le type de ses arguments.\n\nNote: `isinstance(valeur, type)` renvoie `True` ou `False` selon que `valeur` est du type `type` ou non.",
"_____no_output_____"
]
],
[
[
"x = 4 / 2\nprint(f\"x est un entier? {isinstance(x, int)}, x est un flottant? {isinstance(x, float)}\")",
"_____no_output_____"
]
],
[
[
"Certaines expression peuvent générer des valeurs spéciales:",
"_____no_output_____"
]
],
[
[
"x = 1e200\ny = x * x\nz = y * 0\nprint(f\"x={x}, y={y}, z={z}\")",
"_____no_output_____"
],
[
"x = 10 ** 400\n# conversion implicite en flottant pour effectuer l'addition ...\nx + 0.5 # erreur...",
"_____no_output_____"
]
],
[
[
"Un simple calcul peut donner un résultat inattendu ...",
"_____no_output_____"
]
],
[
[
"1.2 * 3",
"_____no_output_____"
]
],
[
[
"On prendra donc garde à éviter tout test d'égalité `==` avec les flottants.\n\nÀ la place, on peut vérifier que la valeur absolue `abs` de leur différence est petite (très petite); par exemple:",
"_____no_output_____"
]
],
[
[
"x = 0.1+0.2\ny = 0.3\nprint(f\"x et y sont identiques? {x==y}\")\nprint(f\"x et y sont très proches? {abs(x-y) < 1e-10}\")",
"_____no_output_____"
]
],
[
[
"### Complément",
"_____no_output_____"
],
[
"À faire ...",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
cbf390e1c6155289ea92f955ce25af6daf16eb41
| 30,483 |
ipynb
|
Jupyter Notebook
|
docs/practices/pretrained_word_embeddings/pretrained_word_embeddings.ipynb
|
Aurelius84/docs
|
2f9bddedb631b41622eaa4f0b4804baa95b99249
|
[
"Apache-2.0"
] | 104 |
2018-09-04T08:16:05.000Z
|
2021-05-06T20:45:26.000Z
|
docs/practices/pretrained_word_embeddings/pretrained_word_embeddings.ipynb
|
Aurelius84/docs
|
2f9bddedb631b41622eaa4f0b4804baa95b99249
|
[
"Apache-2.0"
] | 1,582 |
2018-06-25T06:14:11.000Z
|
2021-05-14T16:00:43.000Z
|
docs/practices/pretrained_word_embeddings/pretrained_word_embeddings.ipynb
|
Aurelius84/docs
|
2f9bddedb631b41622eaa4f0b4804baa95b99249
|
[
"Apache-2.0"
] | 387 |
2018-06-20T07:42:32.000Z
|
2021-05-14T08:35:28.000Z
| 45.361607 | 2,228 | 0.579438 |
[
[
[
"# 使用预训练的词向量完成文本分类任务\n\n**作者**: [fiyen](https://github.com/fiyen)<br>\n**日期**: 2021.10<br>\n**摘要**: 本示例教程将会演示如何使用飞桨内置的Imdb数据集,并使用预训练词向量进行文本分类。",
"_____no_output_____"
],
[
"## 一、环境设置\n本教程基于Paddle 2.2.0-rc0 编写,如果你的环境不是本版本,请先参考官网[安装](https://www.paddlepaddle.org.cn/install/quick) Paddle 2.2.0-rc0。",
"_____no_output_____"
]
],
[
[
"import paddle\r\nfrom paddle.io import Dataset\r\nimport numpy as np\r\nimport paddle.text as text\r\nimport random\r\n\r\nprint(paddle.__version__)",
"2.2.0-rc0\n"
]
],
[
[
"## 二、数据载入\n\n在这个示例中,将使用 Paddle 2.2.0-rc0 完成针对 Imdb 数据集(电影评论情感二分类数据集)的分类训练和测试。Imdb 将直接调用自 Paddle 2.2.0-rc0,同时,\n利用预训练的词向量([GloVe embedding](http://nlp.stanford.edu/projects/glove/))完成任务。",
"_____no_output_____"
]
],
[
[
"print('自然语言相关数据集:', paddle.text.__all__)",
"自然语言相关数据集: ['Conll05st', 'Imdb', 'Imikolov', 'Movielens', 'UCIHousing', 'WMT14', 'WMT16']\n"
]
],
[
[
"\n由于 Paddle 2.2.0-rc0 提供了经过处理的Imdb数据集,可以方便地调用所需要的数据实例,省去了数据预处理的麻烦。目前, Paddle 2.2.0-rc0 以及内置的高质量\n数据集包括 Conll05st、Imdb、Imikolov、Movielens、HCIHousing、WMT14 和 WMT16 等,未来还将提供更多常用数据集的调用接口。\n\n以下定义了调用 imdb 训练集合测试集的方法。其中,cutoff 定义了构建词典的截止大小,即数据集中出现频率在 cutoff 以下的不予考虑;mode 定义了返回的数据用于何种用途(test: 测试集,train: 训练集)。\n\n### 2.1 定义数据集",
"_____no_output_____"
]
],
[
[
"imdb_train = text.Imdb(mode='train', cutoff=150)\r\nimdb_test = text.Imdb(mode='test', cutoff=150)",
"_____no_output_____"
]
],
[
[
"调用 Imdb 得到的是经过编码的数据集,每个 term 对应一个唯一 id,映射关系可以通过 imdb_train.word_idx 查看。将每一个样本即一条电影评论,表示成 id 序列。可以检查一下以上生成的数据内容:",
"_____no_output_____"
]
],
[
[
"print(\"训练集样本数量: %d; 测试集样本数量: %d\" % (len(imdb_train), len(imdb_test)))\r\nprint(f\"样本标签: {set(imdb_train.labels)}\")\r\nprint(f\"样本字典: {list(imdb_train.word_idx.items())[:10]}\")\r\nprint(f\"单个样本: {imdb_train.docs[0]}\")\r\nprint(f\"最小样本长度: {min([len(x) for x in imdb_train.docs])};最大样本长度: {max([len(x) for x in imdb_train.docs])}\")",
"训练集样本数量: 25000; 测试集样本数量: 25000\n样本标签: {0, 1}\n样本字典: [(b'the', 0), (b'and', 1), (b'a', 2), (b'of', 3), (b'to', 4), (b'is', 5), (b'in', 6), (b'it', 7), (b'i', 8), (b'this', 9)]\n单个样本: [5146, 43, 71, 6, 1092, 14, 0, 878, 130, 151, 5146, 18, 281, 747, 0, 5146, 3, 5146, 2165, 37, 5146, 46, 5, 71, 4089, 377, 162, 46, 5, 32, 1287, 300, 35, 203, 2136, 565, 14, 2, 253, 26, 146, 61, 372, 1, 615, 5146, 5, 30, 0, 50, 3290, 6, 2148, 14, 0, 5146, 11, 17, 451, 24, 4, 127, 10, 0, 878, 130, 43, 2, 50, 5146, 751, 5146, 5, 2, 221, 3727, 6, 9, 1167, 373, 9, 5, 5146, 7, 5, 1343, 13, 2, 5146, 1, 250, 7, 98, 4270, 56, 2316, 0, 928, 11, 11, 9, 16, 5, 5146, 5146, 6, 50, 69, 27, 280, 27, 108, 1045, 0, 2633, 4177, 3180, 17, 1675, 1, 2571]\n最小样本长度: 10;最大样本长度: 2469\n"
]
],
[
[
"对于训练集,将数据的顺序打乱,以优化将要进行的分类模型训练的效果。",
"_____no_output_____"
]
],
[
[
"shuffle_index = list(range(len(imdb_train)))\r\nrandom.shuffle(shuffle_index)\r\ntrain_x = [imdb_train.docs[i] for i in shuffle_index]\r\ntrain_y = [imdb_train.labels[i] for i in shuffle_index]\r\n\r\ntest_x = imdb_test.docs\r\ntest_y = imdb_test.labels",
"_____no_output_____"
]
],
[
[
"从样本长度上可以看到,每个样本的长度是不相同的。然而,在模型的训练过程中,需要保证每个样本的长度相同,以便于构造矩阵进行批量运算。\n因此,需要先对所有样本进行填充或截断,使样本的长度一致。",
"_____no_output_____"
]
],
[
[
"def vectorizer(input, label=None, length=2000):\r\n if label is not None:\r\n for x, y in zip(input, label):\r\n yield np.array((x + [0]*length)[:length]).astype('int64'), np.array([y]).astype('int64')\r\n else:\r\n for x in input:\r\n yield np.array((x + [0]*length)[:length]).astype('int64')",
"_____no_output_____"
]
],
[
[
"### 2.2 载入预训练向量\n以下给出的文件较小,可以直接完全载入内存。对于大型的预训练向量,无法一次载入内存的,可以采用分批载入,并行处理的方式进行匹配。\n此外,AIStudio 中提供了 glove.6B 数据集挂载,用户可在 AIStudio 中直接载入数据集并解压。",
"_____no_output_____"
]
],
[
[
"# 下载并解压预训练向量\n!wget http://nlp.stanford.edu/data/glove.6B.zip\n!unzip -q glove.6B.zip",
"_____no_output_____"
],
[
"glove_path = \"./glove.6B.100d.txt\"\r\nembeddings = {}",
"_____no_output_____"
]
],
[
[
"观察上述GloVe预训练向量文件一行的数据:",
"_____no_output_____"
]
],
[
[
"# 使用utf8编码解码\r\nwith open(glove_path, encoding='utf-8') as gf:\r\n line = gf.readline()\r\n print(\"GloVe单行数据:'%s'\" % line)",
"GloVe单行数据:'the -0.038194 -0.24487 0.72812 -0.39961 0.083172 0.043953 -0.39141 0.3344 -0.57545 0.087459 0.28787 -0.06731 0.30906 -0.26384 -0.13231 -0.20757 0.33395 -0.33848 -0.31743 -0.48336 0.1464 -0.37304 0.34577 0.052041 0.44946 -0.46971 0.02628 -0.54155 -0.15518 -0.14107 -0.039722 0.28277 0.14393 0.23464 -0.31021 0.086173 0.20397 0.52624 0.17164 -0.082378 -0.71787 -0.41531 0.20335 -0.12763 0.41367 0.55187 0.57908 -0.33477 -0.36559 -0.54857 -0.062892 0.26584 0.30205 0.99775 -0.80481 -3.0243 0.01254 -0.36942 2.2167 0.72201 -0.24978 0.92136 0.034514 0.46745 1.1079 -0.19358 -0.074575 0.23353 -0.052062 -0.22044 0.057162 -0.15806 -0.30798 -0.41625 0.37972 0.15006 -0.53212 -0.2055 -1.2526 0.071624 0.70565 0.49744 -0.42063 0.26148 -1.538 -0.30223 -0.073438 -0.28312 0.37104 -0.25217 0.016215 -0.017099 -0.38984 0.87424 -0.72569 -0.51058 -0.52028 -0.1459 0.8278 0.27062\n'\n"
]
],
[
[
"可以看到,每一行都以单词开头,其后接上该单词的向量值,各个值之间用空格隔开。基于此,可以用如下方法得到所有词向量的字典。",
"_____no_output_____"
]
],
[
[
"with open(glove_path, encoding='utf-8') as gf:\r\n for glove in gf:\r\n word, embedding = glove.split(maxsplit=1)\r\n embedding = [float(s) for s in embedding.split(' ')]\r\n embeddings[word] = embedding\r\nprint(\"预训练词向量总数:%d\" % len(embeddings))\r\nprint(f\"单词'the'的向量是:{embeddings['the']}\")",
"预训练词向量总数:400000\n单词'the'的向量是:[-0.038194, -0.24487, 0.72812, -0.39961, 0.083172, 0.043953, -0.39141, 0.3344, -0.57545, 0.087459, 0.28787, -0.06731, 0.30906, -0.26384, -0.13231, -0.20757, 0.33395, -0.33848, -0.31743, -0.48336, 0.1464, -0.37304, 0.34577, 0.052041, 0.44946, -0.46971, 0.02628, -0.54155, -0.15518, -0.14107, -0.039722, 0.28277, 0.14393, 0.23464, -0.31021, 0.086173, 0.20397, 0.52624, 0.17164, -0.082378, -0.71787, -0.41531, 0.20335, -0.12763, 0.41367, 0.55187, 0.57908, -0.33477, -0.36559, -0.54857, -0.062892, 0.26584, 0.30205, 0.99775, -0.80481, -3.0243, 0.01254, -0.36942, 2.2167, 0.72201, -0.24978, 0.92136, 0.034514, 0.46745, 1.1079, -0.19358, -0.074575, 0.23353, -0.052062, -0.22044, 0.057162, -0.15806, -0.30798, -0.41625, 0.37972, 0.15006, -0.53212, -0.2055, -1.2526, 0.071624, 0.70565, 0.49744, -0.42063, 0.26148, -1.538, -0.30223, -0.073438, -0.28312, 0.37104, -0.25217, 0.016215, -0.017099, -0.38984, 0.87424, -0.72569, -0.51058, -0.52028, -0.1459, 0.8278, 0.27062]\n"
]
],
[
[
"### 3.3 给数据集的词表匹配词向量\n接下来,提取数据集的词表,需要注意的是,词表中的词编码的先后顺序是按照词出现的频率排列的,频率越高的词编码值越小。",
"_____no_output_____"
]
],
[
[
"word_idx = imdb_train.word_idx\r\nvocab = [w for w in word_idx.keys()]\r\nprint(f\"词表的前5个单词:{vocab[:5]}\")\r\nprint(f\"词表的后5个单词:{vocab[-5:]}\")",
"词表的前5个单词:[b'the', b'and', b'a', b'of', b'to']\n词表的后5个单词:[b'troubles', b'virtual', b'warriors', b'widely', '<unk>']\n"
]
],
[
[
"观察词表的后5个单词,发现最后一个词是\"\\<unk\\>\",这个符号代表所有词表以外的词。另外,对于形式b'the',是字符串'the'\n的二进制编码形式,使用中注意使用b'the'.decode()来进行转换('\\<unk\\>'并没有进行二进制编码,注意区分)。\n接下来,给词表中的每个词匹配对应的词向量。预训练词向量可能没有覆盖数据集词表中的所有词,对于没有的词,设该词的词\n向量为零向量。",
"_____no_output_____"
]
],
[
[
"# 定义词向量的维度,注意与预训练词向量保持一致\r\ndim = 100\r\n\r\nvocab_embeddings = np.zeros((len(vocab), dim))\r\nfor ind, word in enumerate(vocab):\r\n if word != '<unk>':\r\n word = word.decode()\r\n embedding = embeddings.get(word, np.zeros((dim,)))\r\n vocab_embeddings[ind, :] = embedding",
"_____no_output_____"
]
],
[
[
"## 四、组网\n\n### 4.1 构建基于预训练向量的Embedding\n对于预训练向量的Embedding,一般期望它的参数不再变动,所以要设置trainable=False。如果希望在此基础上训练参数,则需要\n设置trainable=True。",
"_____no_output_____"
]
],
[
[
"pretrained_attr = paddle.ParamAttr(name='embedding',\r\n initializer=paddle.nn.initializer.Assign(vocab_embeddings),\r\n trainable=False)\r\nembedding_layer = paddle.nn.Embedding(num_embeddings=len(vocab),\r\n embedding_dim=dim,\r\n padding_idx=word_idx['<unk>'],\r\n weight_attr=pretrained_attr)",
"_____no_output_____"
]
],
[
[
"### 4.2 构建分类器\n这里,构建简单的基于一维卷积的分类模型,其结构为:Embedding->Conv1D->Pool1D->Linear。在定义Linear时,由于需要知\n道输入向量的维度,可以按照公式[官方文档](https://www.paddlepaddle.org.cn/documentation/docs/zh/2.0-beta/api/paddle/nn/layer/conv/Conv2d_cn.html)\n来进行计算。这里给出计算的函数如下:",
"_____no_output_____"
]
],
[
[
"def cal_output_shape(input_shape, out_channels, kernel_size, stride, padding=0, dilation=1):\r\n return out_channels, int((input_shape + 2*padding - (dilation*(kernel_size - 1) + 1)) / stride) + 1\r\n\r\n\r\n# 定义每个样本的长度\r\nlength = 2000\r\n\r\n# 定义卷积层参数\r\nkernel_size = 5\r\nout_channels = 10\r\nstride = 2\r\npadding = 0\r\n\r\noutput_shape = cal_output_shape(length, out_channels, kernel_size, stride, padding)\r\noutput_shape = cal_output_shape(output_shape[1], output_shape[0], 2, 2, 0)\r\nsim_model = paddle.nn.Sequential(embedding_layer,\r\n paddle.nn.Conv1D(in_channels=dim, out_channels=out_channels, kernel_size=kernel_size,\r\n stride=stride, padding=padding, data_format='NLC', bias_attr=True),\r\n paddle.nn.ReLU(),\r\n paddle.nn.MaxPool1D(kernel_size=2, stride=2),\r\n paddle.nn.Flatten(),\r\n paddle.nn.Linear(in_features=np.prod(output_shape), out_features=2, bias_attr=True),\r\n paddle.nn.Softmax())\r\n\r\npaddle.summary(sim_model, input_size=(-1, length), dtypes='int64')",
"---------------------------------------------------------------------------\n Layer (type) Input Shape Output Shape Param # \n===========================================================================\n Embedding-1 [[1, 2000]] [1, 2000, 100] 514,700 \n Conv1D-1 [[1, 2000, 100]] [1, 998, 10] 5,010 \n ReLU-1 [[1, 998, 10]] [1, 998, 10] 0 \n MaxPool1D-1 [[1, 998, 10]] [1, 998, 5] 0 \n Flatten-1 [[1, 998, 5]] [1, 4990] 0 \n Linear-1 [[1, 4990]] [1, 2] 9,982 \n Softmax-1 [[1, 2]] [1, 2] 0 \n===========================================================================\nTotal params: 529,692\nTrainable params: 14,992\nNon-trainable params: 514,700\n---------------------------------------------------------------------------\nInput size (MB): 0.01\nForward/backward pass size (MB): 1.75\nParams size (MB): 2.02\nEstimated Total Size (MB): 3.78\n---------------------------------------------------------------------------\n\n"
]
],
[
[
"### 4.3 读取数据,进行训练\n可以利用飞桨2.0的io.Dataset模块来构建一个数据的读取器,方便地将数据进行分批训练。",
"_____no_output_____"
]
],
[
[
"class DataReader(Dataset):\r\n def __init__(self, input, label, length):\r\n self.data = list(vectorizer(input, label, length=length))\r\n\r\n def __getitem__(self, idx):\r\n return self.data[idx]\r\n\r\n def __len__(self):\r\n return len(self.data)\r\n \r\n\r\n# 定义输入格式\r\ninput_form = paddle.static.InputSpec(shape=[None, length], dtype='int64', name='input')\r\nlabel_form = paddle.static.InputSpec(shape=[None, 1], dtype='int64', name='label')\r\n\r\nmodel = paddle.Model(sim_model, input_form, label_form)\r\nmodel.prepare(optimizer=paddle.optimizer.Adam(learning_rate=0.001, parameters=model.parameters()),\r\n loss=paddle.nn.loss.CrossEntropyLoss(),\r\n metrics=paddle.metric.Accuracy())\r\n\r\n# 分割训练集和验证集\r\neval_length = int(len(train_x) * 1/4)\r\nmodel.fit(train_data=DataReader(train_x[:-eval_length], train_y[:-eval_length], length),\r\n eval_data=DataReader(train_x[-eval_length:], train_y[-eval_length:], length),\r\n batch_size=32, epochs=10, verbose=1)",
"The loss value printed in the log is the current step, and the metric is the average value of previous steps.\nEpoch 1/10\nstep 586/586 [==============================] - loss: 0.5608 - acc: 0.7736 - 5ms/step \nEval begin...\nstep 196/196 [==============================] - loss: 0.4902 - acc: 0.8000 - 4ms/step \nEval samples: 6250\nEpoch 2/10\nstep 586/586 [==============================] - loss: 0.4298 - acc: 0.8138 - 5ms/step \nEval begin...\nstep 196/196 [==============================] - loss: 0.4801 - acc: 0.8142 - 4ms/step \nEval samples: 6250\nEpoch 3/10\nstep 586/586 [==============================] - loss: 0.4947 - acc: 0.8298 - 6ms/step \nEval begin...\nstep 196/196 [==============================] - loss: 0.4568 - acc: 0.8230 - 4ms/step \nEval samples: 6250\nEpoch 4/10\nstep 586/586 [==============================] - loss: 0.4202 - acc: 0.8455 - 5ms/step \nEval begin...\nstep 196/196 [==============================] - loss: 0.4503 - acc: 0.8266 - 4ms/step \nEval samples: 6250\nEpoch 5/10\nstep 586/586 [==============================] - loss: 0.4847 - acc: 0.8564 - 5ms/step \nEval begin...\nstep 196/196 [==============================] - loss: 0.4647 - acc: 0.8280 - 4ms/step \nEval samples: 6250\nEpoch 6/10\nstep 586/586 [==============================] - loss: 0.4952 - acc: 0.8667 - 5ms/step \nEval begin...\nstep 196/196 [==============================] - loss: 0.4855 - acc: 0.8272 - 4ms/step \nEval samples: 6250\nEpoch 7/10\nstep 586/586 [==============================] - loss: 0.4016 - acc: 0.8704 - 5ms/step \nEval begin...\nstep 196/196 [==============================] - loss: 0.4764 - acc: 0.8248 - 4ms/step \nEval samples: 6250\nEpoch 8/10\nstep 586/586 [==============================] - loss: 0.4262 - acc: 0.8807 - 5ms/step \nEval begin...\nstep 196/196 [==============================] - loss: 0.4970 - acc: 0.8104 - 4ms/step \nEval samples: 6250\nEpoch 9/10\nstep 586/586 [==============================] - loss: 0.3585 - acc: 0.8862 - 6ms/step \nEval begin...\nstep 196/196 [==============================] - loss: 0.4614 - acc: 0.8272 - 4ms/step \nEval samples: 6250\nEpoch 10/10\nstep 586/586 [==============================] - loss: 0.3333 - acc: 0.8935 - 5ms/step \nEval begin...\nstep 196/196 [==============================] - loss: 0.4986 - acc: 0.8272 - 4ms/step \nEval samples: 6250\n"
]
],
[
[
"## 五、评估效果并用模型预测",
"_____no_output_____"
]
],
[
[
"# 评估\r\nmodel.evaluate(eval_data=DataReader(test_x, test_y, length), batch_size=32, verbose=1)\r\n\r\n# 预测\r\ntrue_y = test_y[100:105] + test_y[-110:-105]\r\npred_y = model.predict(DataReader(test_x[100:105] + test_x[-110:-105], None, length), batch_size=1)\r\ntest_x_doc = test_x[100:105] + test_x[-110:-105]\r\n\r\n# 标签编码转文字\r\nlabel_id2text = {0: 'positive', 1: 'negative'}\r\n\r\nfor index, y in enumerate(pred_y[0]):\r\n print(\"原文本:%s\" % ' '.join([vocab[i].decode() for i in test_x_doc[index] if i < len(vocab) - 1]))\r\n print(\"预测的标签是:%s, 实际标签是:%s\" % (label_id2text[np.argmax(y)], label_id2text[true_y[index]]))",
"Eval begin...\nstep 782/782 [==============================] - loss: 0.4462 - acc: 0.8262 - 4ms/step \nEval samples: 25000\nPredict begin...\nstep 10/10 [==============================] - 4ms/step \nPredict samples: 10\n原文本:albert and tom are brilliant as sir and his of course the play is brilliant to begin with and nothing can compare with the and of theatre and i think you listen better in theatre but on the screen we become more intimate were more than we are in the theatre we witness subtle changes in expression we see better as well as listen both the play and the movie are moving intelligent the story of the company of historical context of the two main characters and of the parallel characters in itself if you cannot get to see it in a theatre i dont imagine its produced much these days then please do yourself a favor and get the video\n预测的标签是:positive, 实际标签是:positive\n原文本:this film has its and may some folks who frankly need a good the head but the film is top notch in every way engaging poignant relevant naturally is larger than life makes an ideal i thought the performances to be terribly strong in both leads and character provides plenty of dark humor the period is well captured the supporting cast well chosen this is to be seen and like a fine i only wish it were out on dvd\n预测的标签是:positive, 实际标签是:positive\n原文本:this is a movie that deserves another you havent seen it for a while or a first you were too young when it came out 1983 based on a play by the same name it is the story of an older actor who heads a company in england during world war ii it deals with his stress of trying to perform a shakespeare each night while facing problems such as theaters and a company made up of older or physically young able ones being taken for military service it also deals with his relationship with various members of his company especially with his so far it all sounds rather dull but nothing could be further from the truth while tragic overall the story is told with a lot of humor and emotions run high throughout the two male leads both received oscar for best actor and so i strongly recommend this movie to anyone who enjoys human drama shakespeare or who has ever worked in any the make up another of the movie that will be fascinating to most viewers\n预测的标签是:positive, 实际标签是:positive\n原文本:sir has played over tonight he cant remember his opening at the eyes reflect the kings madness his him the is an air of desperation about both these great actor knowing his powers are major wife aware of his into madness and knowing he is to do more than ease his passing the is really a love story between the the years they have become on one another to the extent that neither can a future without the other set during the second world concerns the of a frankly second rate company an equal number of has and led by sir a theatrical knight of what might be called the old part he is playing he stage and out over the his audience into inside most of the time deep beneath the he still remains an occasional of his earlier is to catch a glimpse of this that his audiences hope for mr very cleverly on the to the point of when you are ready to his performance as mere and he will produce a moment of subtlety and that makes you realise that a great actor is playing a great actor the same goes for mr easy to write off his br of norman as an exercise in we have a middle aged rather than camp theatrical his way through the company of the girls and loving the wicked in the were and i strongly suspect still are many men just like norman in the kind and more about the plays than many of the run with wisdom and believe the vast majority of them would with laughter at mr portrait i saw the on the london stage the norman was rather more than in the was played by the great mr jones to huge from the was a memorable performance that mr him rather to an also ran as opposed to an actor on level idea that sir and norman might be almost without each other went right out of the window norman was reduced to being his im not sure was what made for breathtaking theatre and the balance in the to the relationship both men have come a long way since their early appearances in the british new wave pictures when they became the of the vaguely class and ashamed of it the british cinema virtually committed in the 1970s they on the theatre apart from a few roles to keep the wolf from the the of more in the bright br the their with energy and talent to the world at large were still not a big movie but is a great one\n预测的标签是:positive, 实际标签是:positive\n原文本:anyone who fine acting and dialogue will br this film taken from taking sides its a funnybr br and ultimately of a relationship between br very types albert is as the br actor who barely the world war br around him so intent is he on the of his br company and his own psychological and emotional br tom is as norman the of the br whose apparent turns out to be anything but br really a must see\n预测的标签是:positive, 实际标签是:positive\n原文本:well i guess i know the answer to that question for the money we have been so with cat in the hat advertising and that we almost believe there has to be something good about this movie i admit i thought the trailers looked bad but i still had to give it a chance well i should have went with my it was a complete piece hollywood trash once again that the average person can be into believing anything they say is good must be good aside from the insulting fact that the film is only about 80 minutes long it obviously started with a eaten script its full of failed attempts at senseless humor and awful it jumps all over the universe with no nor direction this is then with yes ill say it bad acting i couldnt help but feel like i was watching coffee talk on every time mike myers opened his mouth was the cat intended to be a middle aged jewish woman and were no prize either but mr myers should disappear under a rock somewhere until hes ready to make another austin powers movie f no stars 0 on a scale of 110 save your money\n预测的标签是:negative, 实际标签是:negative\n原文本:when my own child is me to leave the opening show of this film i know it is bad i wanted to my eyes out i wanted to reach through the screen and slap mike myers for the last of dignity he had this is one of the few films in my life i have watched and immediately wished to if only it were possible the other films being 2 and fast and both which are better than this crap in the br i may drink myself to sleep tonight in a attempt to forget i ever witnessed this on the good br to mike myers i say stick with austin or even world just because it worked for jim carrey doesnt mean is a success for all br\n预测的标签是:negative, 实际标签是:negative\n原文本:holy what a piece of this movie is i didnt how these filmmakers could take a word book and turn it into a movie i guess they didnt know either i dont remember any or in the book do youbr br they took this all times childrens classic added some and sexual and it into a joke this should give you a good idea of what these hollywood producers think like i have to say visually it was interesting but the brilliant visual story is ruined by toilet humor if you even think that kind of thing is funny i dont want the kids that i know to think it isbr br dont take your kids to see dont rent the dvd i hope the ghost of doctor ghost comes and the people that made this movie\n预测的标签是:negative, 实际标签是:negative\n原文本:i was so looking forward to seeing this when it was in it turned out to be the the biggest let down a far cry from the world of dr it was and i dont think dr would have the stole christmas was much better i understand it had some subtle adult jokes in it but my children have yet to catch on whereas the cat in the hat they caught a lot more than i would have up with dr it really bothered me to see how this timeless classic got on the big screen lets see what they do with a hope this one does dr some justice\n预测的标签是:positive, 实际标签是:negative\n原文本:ive seen some bad things in my time a half dead trying to get out of high a head on between two cars a thousand on a kitchen floor human beings living like br but never in my life have i seen anything as bad as the cat in the br this film is worse than 911 worse than hitler worse than the worse than people who put in br it is the most disturbing film of all time br i used to think it was a joke some elaborate joke and that mike myers was maybe a high drug who lost a bet or br i\n预测的标签是:negative, 实际标签是:negative\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cbf3a0ff35ee6d1f8e48ce7b919bea5d5e55744e
| 106,706 |
ipynb
|
Jupyter Notebook
|
chapter9-4.ipynb
|
cutelittletiantian/python-ML-principles-and-practice
|
199dee9a7ff83cae793597171ddbdd92a63eeb2e
|
[
"MIT"
] | 2 |
2021-07-07T10:29:49.000Z
|
2021-07-12T12:56:51.000Z
|
chapter9-4.ipynb
|
cutelittletiantian/python-ML-principles-and-practice
|
199dee9a7ff83cae793597171ddbdd92a63eeb2e
|
[
"MIT"
] | null | null | null |
chapter9-4.ipynb
|
cutelittletiantian/python-ML-principles-and-practice
|
199dee9a7ff83cae793597171ddbdd92a63eeb2e
|
[
"MIT"
] | 1 |
2022-01-09T02:18:39.000Z
|
2022-01-09T02:18:39.000Z
| 472.150442 | 43,804 | 0.942974 |
[
[
[
"#本章需导入的模块\nimport numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nfrom pylab import *\nimport matplotlib.cm as cm\nimport warnings\nwarnings.filterwarnings(action = 'ignore')\n%matplotlib inline\nplt.rcParams['font.sans-serif']=['SimHei'] #解决中文显示乱码问题\nplt.rcParams['axes.unicode_minus']=False\nfrom sklearn import svm\nimport sklearn.linear_model as LM\nimport scipy.stats as st\nfrom scipy.optimize import root,fsolve\nfrom sklearn.feature_selection import VarianceThreshold,SelectKBest,f_classif,chi2\nfrom sklearn.feature_selection import RFE,RFECV,SelectFromModel\nfrom sklearn.linear_model import Lasso,LassoCV,lasso_path,Ridge,RidgeCV\nfrom sklearn.linear_model import enet_path,ElasticNetCV,ElasticNet",
"_____no_output_____"
],
[
"data=pd.read_table('邮政编码数据.txt',sep=' ',header=None)\ntmp=data.loc[(data[0]==1) | (data[0]==3)]\nX=tmp.iloc[:,1:-1]\nY=tmp.iloc[:,0]\nfig,axes=plt.subplots(nrows=1,ncols=2,figsize=(12,5))\nalphas=list(np.linspace(0,1,20))\nalphas.extend([2,3])\ncoef=np.zeros((len(alphas),X.shape[1]))\nerr=[]\nfor i,alpha in enumerate(alphas):\n modelLasso = Lasso(alpha=alpha)\n modelLasso.fit(X,Y)\n if i==0:\n coef[i]=modelLasso.coef_\n else:\n coef[i]=(modelLasso.coef_/coef[0])\n err.append(1-modelLasso.score(X,Y)) \nprint('前5个变量的回归系数(alpha=0):%s'%coef[0,][0:5])\nfor i in np.arange(0,X.shape[1]):\n axes[0].plot(coef[1:-1,i])\naxes[0].set_title(\"Lasso回归中的收缩参数alpha和回归系数\")\naxes[0].set_xlabel(\"收缩参数alpha变化\")\naxes[0].set_xticks(np.arange(len(alphas)))\naxes[0].set_ylabel(\"Beta(alpha)/Beta(alpha=0)\")\n\naxes[1].plot(err)\naxes[1].set_title(\"Lasso回归中的收缩参数alpha和训练误差\")\naxes[1].set_xlabel(\"收缩参数alpha变化\")\naxes[1].set_xticks(np.arange(len(alphas)))\naxes[1].set_ylabel(\"错判率\")\n",
"前5个变量的回归系数(alpha=0):[ 0.10647164 0.08368913 -0.04294124 0.00500405 -0.03054124]\n"
],
[
"alphas_lasso, coefs_lasso, _ = lasso_path(X, Y)\nl1 = plt.plot(-np.log10(alphas_lasso), coefs_lasso.T)\nplt.xlabel('-Log(alpha)')\nplt.ylabel('回归系数')\nplt.title('Lasso回归中的收缩参数alpha和回归系数')\nplt.show()",
"_____no_output_____"
],
[
"model = LassoCV() #默认采用3-折交叉验证确定的alpha\nmodel.fit(X,Y)\nprint('Lasso剔除的变量:%d'%sum(model.coef_==0))\nprint('Lasso的最佳的alpha:',model.alpha_) # 只有在使用LassoCV有效\nlassoAlpha=model.alpha_\n\nestimator = Lasso(alpha=lassoAlpha) \nselector=SelectFromModel(estimator=estimator)\nselector.fit(X,Y)\nprint(\"阈值:%s\"%selector.threshold_)\nprint(\"保留的特征个数:%d\"%len(selector.get_support(indices=True)))\nXtmp=selector.inverse_transform(selector.transform(X))\nplt.figure(figsize=(8,8))\nnp.random.seed(1)\nids=np.random.choice(len(Y),25)\nfor i,item in enumerate(ids):\n img=np.array(Xtmp[item,]).reshape((16,16))\n plt.subplot(5,5,i+1)\n plt.imshow(img,cmap=cm.gray)\nplt.show()\n",
"Lasso剔除的变量:159\nLasso的最佳的alpha: 0.0016385673057918155\n阈值:1e-05\n保留的特征个数:97\n"
],
[
"modelLasso = Lasso(alpha=lassoAlpha)\nmodelLasso.fit(X,Y)\nprint(\"lasso训练误差:%.2f\"%(1-modelLasso.score(X,Y)))\nmodelRidge = RidgeCV() # RidgeCV自动调节alpha可以实现选择最佳的alpha。\nmodelRidge.fit(X,Y)\nprint('岭回归剔除的变量:%d'%sum(modelRidge.coef_==0))\nprint('岭回归最优alpha:',modelRidge.alpha_) \nprint(\"岭回归训练误差:%.2f\"%(1-modelRidge.score(X,Y)))",
"lasso训练误差:0.02\n岭回归剔除的变量:0\n岭回归最优alpha: 10.0\n岭回归训练误差:0.02\n"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code"
]
] |
cbf3a783798cb5b593c30ee354a20722579f89f0
| 7,864 |
ipynb
|
Jupyter Notebook
|
minitorch/operators.ipynb
|
chris-tng/minitorch
|
f5facd366aac6a4d3a437796c43ac02a9b7069ff
|
[
"MIT"
] | null | null | null |
minitorch/operators.ipynb
|
chris-tng/minitorch
|
f5facd366aac6a4d3a437796c43ac02a9b7069ff
|
[
"MIT"
] | null | null | null |
minitorch/operators.ipynb
|
chris-tng/minitorch
|
f5facd366aac6a4d3a437796c43ac02a9b7069ff
|
[
"MIT"
] | null | null | null | 22.927114 | 205 | 0.46147 |
[
[
[
"Basic math operations as well as a few warmup problems for testing out your functional programming chops in python. (Please ignore the @jit decorator for now. It will come back in later assignments.)",
"_____no_output_____"
]
],
[
[
"try:\n from .util import jit\nexcept:\n from util import jit\nimport math",
"_____no_output_____"
],
[
"@jit\ndef mul(x, y):\n \":math:`f(x, y) = x * y`\"\n return x * y",
"_____no_output_____"
],
[
"@jit\ndef id(x):\n \":math:`f(x) = x`\"\n return x",
"_____no_output_____"
],
[
"@jit\ndef add(x, y):\n \":math:`f(x, y) = x + y`\"\n return float(x + y)",
"_____no_output_____"
],
[
"@jit\ndef neg(x):\n \":math:`f(x) = -x`\"\n return -float(x)",
"_____no_output_____"
],
[
"@jit\ndef lt(x, y):\n \":math:`f(x) =` 1.0 if x is greater then y else 0.0\"\n return 1.0 if x > y else 0.",
"_____no_output_____"
],
[
"EPS = 1e-6\n\n@jit\ndef log(x):\n \":math:`f(x) = log(x)`\"\n return math.log(x + EPS)\n\n@jit\ndef exp(x):\n \":math:`f(x) = e^{x}`\"\n return math.exp(x)\n\n@jit\ndef log_back(a, b):\n return b / (a + EPS)",
"_____no_output_____"
],
[
"@jit\ndef sigmoid(x):\n r\"\"\"\n :math:`f(x) = \\frac{1.0}{(1.0 + e^{-a})}`\n (See https://en.wikipedia.org/wiki/Sigmoid_function .)\n \"\"\"\n return 1.0 / add(1.0, exp(-x))",
"_____no_output_____"
],
[
"@jit\ndef relu(x):\n \"\"\"\n :math:`f(x) =` x if x is greater then y else 0\n (See https://en.wikipedia.org/wiki/Rectifier_(neural_networks).)\n \"\"\"\n return x if x > 0. else 0.",
"_____no_output_____"
],
[
"@jit\ndef relu_back(x, y):\n \":math:`f(x) =` y if x is greater then 0 else 0\"\n return y if x > 0. else 0.",
"_____no_output_____"
],
[
"def inv(x):\n \":math:`f(x) = 1/x`\"\n return 1.0 / x",
"_____no_output_____"
],
[
"def inv_back(a, b):\n return -(1.0 / a ** 2) * b",
"_____no_output_____"
],
[
"def eq(x, y):\n \":math:`f(x) =` 1.0 if x is equal to y else 0.0\"\n return 1. if x == y else 0.",
"_____no_output_____"
]
],
[
[
"### Higher-order functions",
"_____no_output_____"
]
],
[
[
"def map(fn):\n \"\"\"\n Higher-order map.\n .. image:: figs/Ops/maplist.png\n See https://en.wikipedia.org/wiki/Map_(higher-order_function)\n Args:\n fn (one-arg function): process one value\n Returns:\n function : a function that takes a list and applies `fn` to each element\n \"\"\"\n def _fn(ls):\n return [fn(e) for e in ls]\n return _fn",
"_____no_output_____"
],
[
"def negList(ls):\n \"Use :func:`map` and :func:`neg` negate each element in `ls`\"\n return map(neg)(ls)",
"_____no_output_____"
],
[
"def zipWith(fn):\n \"\"\"\n Higher-order zipwith (or map2).\n .. image:: figs/Ops/ziplist.png\n See https://en.wikipedia.org/wiki/Map_(higher-order_function)\n Args:\n fn (two-arg function): combine two values\n Returns:\n function : takes two equally sized lists `ls1` and `ls2`, produce a new list by\n applying fn(x, y) one each pair of elements.\n \"\"\"\n def _fn(ls1, ls2):\n return [fn(e1, e2) for e1, e2 in zip(ls1, ls2)]\n return _fn",
"_____no_output_____"
],
[
"def addLists(ls1, ls2):\n \"Add the elements of `ls1` and `ls2` using :func:`zipWith` and :func:`add`\"\n return zipWith(add)(ls1, ls2)",
"_____no_output_____"
],
[
"def reduce(fn, start):\n r\"\"\"\n Higher-order reduce.\n .. image:: figs/Ops/reducelist.png\n Args:\n fn (two-arg function): combine two values\n start (float): start value :math:`x_0`\n Returns:\n function : function that takes a list `ls` of elements\n :math:`x_1 \\ldots x_n` and computes the reduction :math:`fn(x_3, fn(x_2,\n fn(x_1, x_0)))`\n \"\"\"\n def _fn(ls):\n r = start\n for e in ls:\n r = fn(r, e)\n return r\n return _fn",
"_____no_output_____"
],
[
"def sum(ls):\n \"\"\"\n Sum up a list using :func:`reduce` and :func:`add`.\n \"\"\"\n return reduce(add, 0)(ls)",
"_____no_output_____"
],
[
"def prod(ls):\n \"\"\"\n Product of a list using :func:`reduce` and :func:`mul`.\n \"\"\"\n return reduce(mul, 1)(ls)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbf3acaf63d69901c50d9e2811899332fe9a4563
| 30,833 |
ipynb
|
Jupyter Notebook
|
notebooks/Insurance Premium Predictor - P8 notebook.ipynb
|
Anughag/NLP_Python_Flask
|
0c2c353e2c68175cce95863ce2ab90bd63343c87
|
[
"Apache-2.0"
] | 26 |
2020-06-08T16:31:10.000Z
|
2021-11-23T14:58:30.000Z
|
notebooks/Insurance Premium Predictor - P8 notebook.ipynb
|
dadberg/autoai
|
aff82d659faa818ec760743c1207355873295bb5
|
[
"Apache-2.0"
] | 4 |
2020-07-13T23:23:28.000Z
|
2021-04-27T10:47:16.000Z
|
notebooks/Insurance Premium Predictor - P8 notebook.ipynb
|
dadberg/autoai
|
aff82d659faa818ec760743c1207355873295bb5
|
[
"Apache-2.0"
] | 37 |
2020-06-08T18:19:57.000Z
|
2022-03-28T04:53:20.000Z
| 30,833 | 30,833 | 0.688807 |
[
[
[
"################################################################################\n#Licensed Materials - Property of IBM\n#(C) Copyright IBM Corp. 2019\n#US Government Users Restricted Rights - Use, duplication disclosure restricted\n#by GSA ADP Schedule Contract with IBM Corp.\n################################################################################\n\nThe auto-generated notebooks are subject to the International License Agreement for Non-Warranted Programs (or equivalent) and License Information document for Watson Studio Auto-generated Notebook (\"License Terms\"), such agreements located in the link below.\nSpecifically, the Source Components and Sample Materials clause included in the License Information document for\nWatson Studio Auto-generated Notebook applies to the auto-generated notebooks. \nBy downloading, copying, accessing, or otherwise using the materials, you agree to the License Terms.\nhttp://www14.software.ibm.com/cgi-bin/weblap/lap.pl?li_formnum=L-AMCU-BHU2B7&title=IBM%20Watson%20Studio%20Auto-generated%20Notebook%20V2.1\n",
"_____no_output_____"
],
[
"## IBM AutoAI Auto-Generated Notebook v1.11.7\n### Representing Pipeline: P8 from run 25043980-e0c9-476a-8385-755ecd49aa48\n\n**Note**: Notebook code generated using AutoAI will execute successfully.\nIf code is modified or reordered, there is no guarantee it will successfully execute.\nThis pipeline is optimized for the original dataset. The pipeline may fail or produce sub-optimium results if used with different data.\nFor different data, please consider returning to AutoAI Experiements to generate a new pipeline.\nPlease read our documentation for more information: \n(IBM Cloud Platform) https://dataplatform.cloud.ibm.com/docs/content/wsj/analyze-data/autoai-notebook.html . \n(IBM Cloud Pak For Data) https://www.ibm.com/support/knowledgecenter/SSQNUZ_3.0.0/wsj/analyze-data/autoai-notebook.html . \n\nBefore modifying the pipeline or trying to re-fit the pipeline, consider: \nThe notebook converts dataframes to numpy arrays before fitting the pipeline (a current restriction of the preprocessor pipeline).\nThe known_values_list is passed by reference and populated with categorical values during fit of the preprocessing pipeline. Delete its members before re-fitting.\n",
"_____no_output_____"
],
[
"### 1. Set Up",
"_____no_output_____"
]
],
[
[
"#attempt import of autoai_libs and install if missing\ntry:\n import autoai_libs\nexcept Exception as e:\n print('attempting to install missing autoai_libs from pypi, this may take tens of seconds to complete.')\n import subprocess\n try:\n # attempt to install missing autoai-libs from pypi\n out = subprocess.check_output('pip install autoai-libs', shell=True)\n for line in out.splitlines():\n print(line)\n except Exception as e:\n print(str(e))\ntry:\n import autoai_libs\nexcept Exception as e:\n print('attempting to install missing autoai_libs from local filesystem, this may take tens of seconds to complete.')\n import subprocess\n # attempt to install missing autoai-libs from local filesystem\n try:\n out = subprocess.check_output('pip install .', shell=True, cwd='software/autoai_libs')\n for line in out.splitlines():\n print(line)\n import autoai_libs\n except Exception as e:\n print(str(e))\nimport sklearn\ntry:\n import xgboost\nexcept:\n print('xgboost, if needed, will be installed and imported later')\ntry:\n import lightgbm\nexcept:\n print('lightgbm, if needed, will be installed and imported later')\nfrom sklearn.cluster import FeatureAgglomeration\nimport numpy\nfrom numpy import inf, nan, dtype, mean\nfrom autoai_libs.sklearn.custom_scorers import CustomScorers\nfrom autoai_libs.cognito.transforms.transform_utils import TExtras, FC\nfrom autoai_libs.transformers.exportable import *\nfrom autoai_libs.utils.exportable_utils import *\nfrom sklearn.pipeline import Pipeline\nknown_values_list=[]\n",
"lightgbm, if needed, will be installed and imported later\n"
],
[
"# compose a decorator to assist pipeline instantiation via import of modules and installation of packages\ndef decorator_retries(func):\n def install_import_retry(*args, **kwargs):\n retries = 0\n successful = False\n failed_retries = 0\n while retries < 100 and failed_retries < 10 and not successful:\n retries += 1\n failed_retries += 1\n try:\n result = func(*args, **kwargs)\n successful = True\n except Exception as e:\n estr = str(e)\n if estr.startswith('name ') and estr.endswith(' is not defined'):\n try:\n import importlib\n module_name = estr.split(\"'\")[1]\n module = importlib.import_module(module_name)\n globals().update({module_name: module})\n print('import successful for ' + module_name)\n failed_retries -= 1\n except Exception as import_failure:\n print('import of ' + module_name + ' failed with: ' + str(import_failure))\n import subprocess\n print('attempting pip install of ' + module_name)\n process = subprocess.Popen('pip install ' + module_name, shell=True)\n process.wait()\n try:\n print('re-attempting import of ' + module_name)\n module = importlib.import_module(module_name)\n globals().update({module_name: module})\n print('import successful for ' + module_name)\n failed_retries -= 1\n except Exception as import_or_installation_failure:\n print('failure installing and/or importing ' + module_name + ' error was: ' + str(\n import_or_installation_failure))\n raise (ModuleNotFoundError('Missing package in environment for ' + module_name +\n '? Try import and/or pip install manually?'))\n elif type(e) is AttributeError:\n if 'module ' in estr and ' has no attribute ' in estr:\n pieces = estr.split(\"'\")\n if len(pieces) == 5:\n try:\n import importlib\n print('re-attempting import of ' + pieces[3] + ' from ' + pieces[1])\n module = importlib.import_module('.' + pieces[3], pieces[1])\n failed_retries -= 1\n except:\n print('failed attempt to import ' + pieces[3])\n raise (e)\n else:\n raise (e)\n else:\n raise (e)\n if successful:\n print('Pipeline successfully instantiated')\n else:\n raise (ModuleNotFoundError(\n 'Remaining missing imports/packages in environment? Retry cell and/or try pip install manually?'))\n return result\n return install_import_retry\n",
"_____no_output_____"
]
],
[
[
"### 2. Compose Pipeline",
"_____no_output_____"
]
],
[
[
"# metadata necessary to replicate AutoAI scores with the pipeline\n_input_metadata = {'run_uid': '25043980-e0c9-476a-8385-755ecd49aa48', 'pn': 'P8', 'data_source': '', 'target_label_name': 'charges', 'learning_type': 'regression', 'optimization_metric': 'neg_root_mean_squared_error', 'random_state': 33, 'cv_num_folds': 3, 'holdout_fraction': 0.1, 'pos_label': None}\n\n# define a function to compose the pipeline, and invoke it\n@decorator_retries\ndef compose_pipeline():\n import numpy\n from numpy import nan, dtype, mean\n #\n # composing steps for toplevel Pipeline\n #\n _input_metadata = {'run_uid': '25043980-e0c9-476a-8385-755ecd49aa48', 'pn': 'P8', 'data_source': '', 'target_label_name': 'charges', 'learning_type': 'regression', 'optimization_metric': 'neg_root_mean_squared_error', 'random_state': 33, 'cv_num_folds': 3, 'holdout_fraction': 0.1, 'pos_label': None}\n steps = []\n #\n # composing steps for preprocessor Pipeline\n #\n preprocessor__input_metadata = None\n preprocessor_steps = []\n #\n # composing steps for preprocessor_features FeatureUnion\n #\n preprocessor_features_transformer_list = []\n #\n # composing steps for preprocessor_features_categorical Pipeline\n #\n preprocessor_features_categorical__input_metadata = None\n preprocessor_features_categorical_steps = []\n preprocessor_features_categorical_steps.append(('cat_column_selector', autoai_libs.transformers.exportable.NumpyColumnSelector(columns=[0, 1, 3, 4, 5])))\n preprocessor_features_categorical_steps.append(('cat_compress_strings', autoai_libs.transformers.exportable.CompressStrings(activate_flag=True, compress_type='hash', dtypes_list=['int_num', 'char_str', 'int_num', 'char_str', 'char_str'], missing_values_reference_list=['', '-', '?', nan], misslist_list=[[], [], [], [], []])))\n preprocessor_features_categorical_steps.append(('cat_missing_replacer', autoai_libs.transformers.exportable.NumpyReplaceMissingValues(filling_values=nan, missing_values=[])))\n preprocessor_features_categorical_steps.append(('cat_unknown_replacer', autoai_libs.transformers.exportable.NumpyReplaceUnknownValues(filling_values=nan, filling_values_list=[nan, nan, nan, nan, nan], known_values_list=known_values_list, missing_values_reference_list=['', '-', '?', nan])))\n preprocessor_features_categorical_steps.append(('boolean2float_transformer', autoai_libs.transformers.exportable.boolean2float(activate_flag=True)))\n preprocessor_features_categorical_steps.append(('cat_imputer', autoai_libs.transformers.exportable.CatImputer(activate_flag=True, missing_values=nan, sklearn_version_family='20', strategy='most_frequent')))\n preprocessor_features_categorical_steps.append(('cat_encoder', autoai_libs.transformers.exportable.CatEncoder(activate_flag=True, categories='auto', dtype=numpy.float64, encoding='ordinal', handle_unknown='error', sklearn_version_family='20')))\n preprocessor_features_categorical_steps.append(('float32_transformer', autoai_libs.transformers.exportable.float32_transform(activate_flag=True)))\n # assembling preprocessor_features_categorical_ Pipeline\n preprocessor_features_categorical_pipeline = sklearn.pipeline.Pipeline(steps=preprocessor_features_categorical_steps)\n preprocessor_features_transformer_list.append(('categorical', preprocessor_features_categorical_pipeline))\n #\n # composing steps for preprocessor_features_numeric Pipeline\n #\n preprocessor_features_numeric__input_metadata = None\n preprocessor_features_numeric_steps = []\n preprocessor_features_numeric_steps.append(('num_column_selector', autoai_libs.transformers.exportable.NumpyColumnSelector(columns=[2])))\n preprocessor_features_numeric_steps.append(('num_floatstr2float_transformer', autoai_libs.transformers.exportable.FloatStr2Float(activate_flag=True, dtypes_list=['float_num'], missing_values_reference_list=[])))\n preprocessor_features_numeric_steps.append(('num_missing_replacer', autoai_libs.transformers.exportable.NumpyReplaceMissingValues(filling_values=nan, missing_values=[])))\n preprocessor_features_numeric_steps.append(('num_imputer', autoai_libs.transformers.exportable.NumImputer(activate_flag=True, missing_values=nan, strategy='median')))\n preprocessor_features_numeric_steps.append(('num_scaler', autoai_libs.transformers.exportable.OptStandardScaler(num_scaler_copy=None, num_scaler_with_mean=None, num_scaler_with_std=None, use_scaler_flag=False)))\n preprocessor_features_numeric_steps.append(('float32_transformer', autoai_libs.transformers.exportable.float32_transform(activate_flag=True)))\n # assembling preprocessor_features_numeric_ Pipeline\n preprocessor_features_numeric_pipeline = sklearn.pipeline.Pipeline(steps=preprocessor_features_numeric_steps)\n preprocessor_features_transformer_list.append(('numeric', preprocessor_features_numeric_pipeline))\n # assembling preprocessor_features_ FeatureUnion\n preprocessor_features_pipeline = sklearn.pipeline.FeatureUnion(transformer_list=preprocessor_features_transformer_list)\n preprocessor_steps.append(('features', preprocessor_features_pipeline))\n preprocessor_steps.append(('permuter', autoai_libs.transformers.exportable.NumpyPermuteArray(axis=0, permutation_indices=[0, 1, 3, 4, 5, 2])))\n # assembling preprocessor_ Pipeline\n preprocessor_pipeline = sklearn.pipeline.Pipeline(steps=preprocessor_steps)\n steps.append(('preprocessor', preprocessor_pipeline))\n #\n # composing steps for cognito Pipeline\n #\n cognito__input_metadata = None\n cognito_steps = []\n cognito_steps.append(('0', autoai_libs.cognito.transforms.transform_utils.TA2(fun=numpy.add, name='sum', datatypes1=['intc', 'intp', 'int_', 'uint8', 'uint16', 'uint32', 'uint64', 'int8', 'int16', 'int32', 'int64', 'short', 'long', 'longlong', 'float16', 'float32', 'float64'], feat_constraints1=[autoai_libs.utils.fc_methods.is_not_categorical], datatypes2=['intc', 'intp', 'int_', 'uint8', 'uint16', 'uint32', 'uint64', 'int8', 'int16', 'int32', 'int64', 'short', 'long', 'longlong', 'float16', 'float32', 'float64'], feat_constraints2=[autoai_libs.utils.fc_methods.is_not_categorical], tgraph=None, apply_all=True, col_names=['age', 'sex', 'bmi', 'children', 'smoker', 'region'], col_dtypes=[dtype('float32'), dtype('float32'), dtype('float32'), dtype('float32'), dtype('float32'), dtype('float32')], col_as_json_objects=None)))\n cognito_steps.append(('1', autoai_libs.cognito.transforms.transform_utils.FS1(cols_ids_must_keep=range(0, 6), additional_col_count_to_keep=8, ptype='regression')))\n # assembling cognito_ Pipeline\n cognito_pipeline = sklearn.pipeline.Pipeline(steps=cognito_steps)\n steps.append(('cognito', cognito_pipeline))\n steps.append(('estimator', sklearn.ensemble.forest.RandomForestRegressor(bootstrap=True, criterion='friedman_mse', max_depth=4, max_features=0.9832410473940374, max_leaf_nodes=None, min_impurity_decrease=0.0, min_impurity_split=None, min_samples_leaf=3, min_samples_split=2, min_weight_fraction_leaf=0.0, n_estimators=29, n_jobs=4, oob_score=False, random_state=33, verbose=0, warm_start=False)))\n # assembling Pipeline\n pipeline = sklearn.pipeline.Pipeline(steps=steps)\n return pipeline\npipeline = compose_pipeline()\n",
"re-attempting import of ensemble from sklearn\nPipeline successfully instantiated\n"
]
],
[
[
"### 3. Extract needed parameter values from AutoAI run metadata",
"_____no_output_____"
]
],
[
[
"\n# Metadata used in retrieving data and computing metrics. Customize as necessary for your environment.\n#data_source='replace_with_path_and_csv_filename'\ntarget_label_name = _input_metadata['target_label_name']\nlearning_type = _input_metadata['learning_type']\noptimization_metric = _input_metadata['optimization_metric']\nrandom_state = _input_metadata['random_state']\ncv_num_folds = _input_metadata['cv_num_folds']\nholdout_fraction = _input_metadata['holdout_fraction']\nif 'data_provenance' in _input_metadata:\n data_provenance = _input_metadata['data_provenance']\nelse:\n data_provenance = None\nif 'pos_label' in _input_metadata and learning_type == 'classification':\n pos_label = _input_metadata['pos_label']\nelse:\n pos_label = None\n",
"_____no_output_____"
]
],
[
[
"### 4. Create dataframe from dataset in IBM Cloud Object Storage or IBM Cloud Pak For Data",
"_____no_output_____"
]
],
[
[
"\n# @hidden_cell\n# The following code contains the credentials for a file in your IBM Cloud Object Storage.\n# You might want to remove those credentials before you share your notebook.\ncredentials_0 = {\n \n }\n",
"_____no_output_____"
],
[
"# Read the data as a dataframe\nimport pandas as pd\n\ncsv_encodings=['UTF-8','Latin-1'] # supplement list of encodings as necessary for your data\ndf = None\nreadable = None # if automatic detection fails, you can supply a filename here\n\n# First, obtain a readable object\n# IBM Cloud Object Storage data access\n# Assumes COS credentials are in a dictionary named 'credentials_0'\ncos_credentials = df = globals().get('credentials_0') \nif readable is None and cos_credentials is not None:\n print('accessing data via IBM Cloud Object Storage')\n try:\n import types\n from botocore.client import Config\n import ibm_boto3\n\n def __iter__(self): return 0\n\n if 'SERVICE_NAME' not in cos_credentials: # in case of Studio-supplied credentials for a different dataset\n cos_credentials['SERVICE_NAME'] = 's3'\n client = ibm_boto3.client(service_name=cos_credentials['SERVICE_NAME'],\n ibm_api_key_id=cos_credentials['IBM_API_KEY_ID'],\n ibm_auth_endpoint=cos_credentials['IBM_AUTH_ENDPOINT'],\n config=Config(signature_version='oauth'),\n endpoint_url=cos_credentials['ENDPOINT'])\n\n try:\n readable = client.get_object(Bucket=cos_credentials['BUCKET'],Key=cos_credentials['FILE'])['Body']\n # add missing __iter__ method, so pandas accepts readable as file-like object\n if not hasattr(readable, \"__iter__\"): readable.__iter__ = types.MethodType( __iter__, readable )\n except Exception as cos_access_exception:\n print('unable to access data object in cloud object storage with credentials supplied')\n except Exception as cos_exception:\n print('unable to create client for cloud object storage')\n\n# IBM Cloud Pak for Data data access\nproject_filename = globals().get('project_filename') \nif readable is None and 'credentials_0' in globals() and 'ASSET_ID' in credentials_0:\n project_filename = credentials_0['ASSET_ID']\nif project_filename is not None:\n print('attempting project_lib access to ' + str(project_filename))\n try:\n from project_lib import Project\n project = Project.access()\n storage_credentials = project.get_storage_metadata()\n readable = project.get_file(project_filename)\n except Exception as project_exception:\n print('unable to access data using the project_lib interface and filename supplied')\n\n# Use data_provenance as filename if other access mechanisms are unsuccessful\nif readable is None and type(data_provenance) is str:\n print('attempting to access local file using path and name ' + data_provenance)\n readable = data_provenance\n\n# Second, use pd.read_csv to read object, iterating over list of csv_encodings until successful\nif readable is not None:\n for encoding in csv_encodings:\n try:\n df = pd.read_csv(readable, encoding=encoding)\n print('successfully loaded dataframe using encoding = ' + str(encoding))\n break\n except Exception as exception_csv:\n print('unable to read csv using encoding ' + str(encoding))\n print('handled error was ' + str(exception_csv))\n if df is None:\n print('unable to read file/object as a dataframe using supplied csv_encodings ' + str(csv_encodings))\n print(\"Please use 'insert to code' on data panel to load dataframe.\")\n raise(ValueError('unable to read file/object as a dataframe using supplied csv_encodings ' + str(csv_encodings)))\n\nif df is None:\n print('Unable to access bucket/file in IBM Cloud Object Storage or asset in IBM Cloud Pak for Data with the parameters supplied.')\n print('This is abnormal, but proceeding assuming the notebook user will supply a dataframe by other means.')\n print(\"Please use 'insert to code' on data panel to load dataframe.\")\n\n",
"accessing data via IBM Cloud Object Storage\nsuccessfully loaded dataframe using encoding = UTF-8\n"
]
],
[
[
"### 5. Preprocess Data",
"_____no_output_____"
]
],
[
[
"# Drop rows whose target is not defined\ntarget = target_label_name # your target name here\nif learning_type == 'regression':\n df[target] = pd.to_numeric(df[target], errors='coerce')\ndf.dropna('rows', how='any', subset=[target], inplace=True)\n",
"_____no_output_____"
],
[
"# extract X and y\ndf_X = df.drop(columns=[target])\ndf_y = df[target]\n",
"_____no_output_____"
],
[
"# Detach preprocessing pipeline (which needs to see all training data)\npreprocessor_index = -1\npreprocessing_steps = [] \nfor i, step in enumerate(pipeline.steps):\n preprocessing_steps.append(step)\n if step[0]=='preprocessor':\n preprocessor_index = i\n break\nif len(pipeline.steps) > preprocessor_index+1 and pipeline.steps[preprocessor_index + 1][0] == 'cognito':\n preprocessor_index += 1\n preprocessing_steps.append(pipeline.steps[preprocessor_index])\nif preprocessor_index >= 0:\n preprocessing_pipeline = Pipeline(memory=pipeline.memory, steps=preprocessing_steps)\n pipeline = Pipeline(steps=pipeline.steps[preprocessor_index+1:])",
"_____no_output_____"
],
[
"# Preprocess X\n# preprocessor should see all data for cross_validate on the remaining steps to match autoai scores\nknown_values_list.clear() # known_values_list is filled in by the preprocessing_pipeline if needed\npreprocessing_pipeline.fit(df_X.values, df_y.values)\nX_prep = preprocessing_pipeline.transform(df_X.values)",
"_____no_output_____"
]
],
[
[
"### 6. Split data into Training and Holdout sets",
"_____no_output_____"
]
],
[
[
"# determine learning_type and perform holdout split (stratify conditionally)\nif learning_type is None:\n # When the problem type is not available in the metadata, use the sklearn type_of_target to determine whether to stratify the holdout split\n # Caution: This can mis-classify regression targets that can be expressed as integers as multiclass, in which case manually override the learning_type\n from sklearn.utils.multiclass import type_of_target\n if type_of_target(df_y.values) in ['multiclass', 'binary']:\n learning_type = 'classification'\n else:\n learning_type = 'regression'\n print('learning_type determined by type_of_target as:',learning_type)\nelse:\n print('learning_type specified as:',learning_type)\n \nfrom sklearn.model_selection import train_test_split\nif learning_type == 'classification':\n X, X_holdout, y, y_holdout = train_test_split(X_prep, df_y.values, test_size=holdout_fraction, random_state=random_state, stratify=df_y.values)\nelse:\n X, X_holdout, y, y_holdout = train_test_split(X_prep, df_y.values, test_size=holdout_fraction, random_state=random_state)\n",
"learning_type specified as: regression\n"
]
],
[
[
"### 7. Additional setup: Define a function that returns a scorer for the target's positive label",
"_____no_output_____"
]
],
[
[
"# create a function to produce a scorer for a given positive label\ndef make_pos_label_scorer(scorer, pos_label):\n kwargs = {'pos_label':pos_label}\n for prop in ['needs_proba', 'needs_threshold']:\n if prop+'=True' in scorer._factory_args():\n kwargs[prop] = True\n if scorer._sign == -1:\n kwargs['greater_is_better'] = False\n from sklearn.metrics import make_scorer\n scorer=make_scorer(scorer._score_func, **kwargs)\n return scorer",
"_____no_output_____"
]
],
[
[
"### 8. Fit pipeline, predict on Holdout set, calculate score, perform cross-validation",
"_____no_output_____"
]
],
[
[
"# fit the remainder of the pipeline on the training data\npipeline.fit(X,y)",
"_____no_output_____"
],
[
"# predict on the holdout data\ny_pred = pipeline.predict(X_holdout)",
"_____no_output_____"
],
[
"# compute score for the optimization metric\n# scorer may need pos_label, but not all scorers take pos_label parameter\nfrom sklearn.metrics import get_scorer\nscorer = get_scorer(optimization_metric)\nscore = None\n#score = scorer(pipeline, X_holdout, y_holdout) # this would suffice for simple cases\npos_label = None # if you want to supply the pos_label, specify it here\nif pos_label is None and 'pos_label' in _input_metadata:\n pos_label=_input_metadata['pos_label']\ntry:\n score = scorer(pipeline, X_holdout, y_holdout)\nexcept Exception as e1:\n if pos_label is None or str(pos_label)=='':\n print('You may have to provide a value for pos_label in order for a score to be calculated.')\n raise(e1)\n else:\n exception_string=str(e1)\n if 'pos_label' in exception_string:\n try:\n scorer = make_pos_label_scorer(scorer, pos_label=pos_label)\n score = scorer(pipeline, X_holdout, y_holdout)\n print('Retry was successful with pos_label supplied to scorer')\n except Exception as e2:\n print('Initial attempt to use scorer failed. Exception was:')\n print(e1)\n print('')\n print('Retry with pos_label failed. Exception was:')\n print(e2)\n else:\n raise(e1)\n\nif score is not None:\n print(score)",
"-4436.65923852591\n"
],
[
"# cross_validate pipeline using training data\nfrom sklearn.model_selection import cross_validate\nfrom sklearn.model_selection import StratifiedKFold, KFold\nif learning_type == 'classification':\n fold_generator = StratifiedKFold(n_splits=cv_num_folds, random_state=random_state)\nelse:\n fold_generator = KFold(n_splits=cv_num_folds, random_state=random_state)\ncv_results = cross_validate(pipeline, X, y, cv=fold_generator, scoring={optimization_metric:scorer}, return_train_score=True)\nimport numpy as np\nnp.mean(cv_results['test_' + optimization_metric])",
"_____no_output_____"
],
[
"cv_results",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
cbf3ae800a76966abd23c947e8c4b17382815e6f
| 2,255 |
ipynb
|
Jupyter Notebook
|
code/week8_dataingestion/Kafka Sample Consumer.ipynb
|
kaopanboonyuen/2110446_DataScience_2021s2
|
511611edf2c84a07e94cc9496858f805e85a71c3
|
[
"MIT"
] | 15 |
2022-01-11T06:07:28.000Z
|
2022-02-02T17:58:55.000Z
|
code/week8_dataingestion/Kafka Sample Consumer.ipynb
|
kaopanboonyuen/2110446_DataScience_2021s2
|
511611edf2c84a07e94cc9496858f805e85a71c3
|
[
"MIT"
] | null | null | null |
code/week8_dataingestion/Kafka Sample Consumer.ipynb
|
kaopanboonyuen/2110446_DataScience_2021s2
|
511611edf2c84a07e94cc9496858f805e85a71c3
|
[
"MIT"
] | 7 |
2022-02-18T18:35:48.000Z
|
2022-03-26T06:54:22.000Z
| 20.87963 | 114 | 0.53969 |
[
[
[
"# import required libraries\nfrom kafka import KafkaConsumer",
"_____no_output_____"
],
[
"# Connect to kafka broker running in your local host (docker). Change this to your kafka broker if needed\nkafka_broker = '34.87.22.17:9092'",
"_____no_output_____"
],
[
"consumer = KafkaConsumer(\n 'sample',\n bootstrap_servers=[kafka_broker],\n enable_auto_commit=True,\n value_deserializer=lambda x: x.decode('utf-8'))",
"_____no_output_____"
],
[
"print('Running Consumer')\nfor message in consumer:\n print('[{}:{}] {}'.format(message.timestamp, message.offset, message.value))",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code"
]
] |
cbf3bedd21f2a09a7aa48ab84537d494cea43eab
| 15,270 |
ipynb
|
Jupyter Notebook
|
research/Tutorial.ipynb
|
dreamshark/SteganoGAN
|
67c73783a8f5cb416b94cafdab80319f304e0cd7
|
[
"MIT"
] | null | null | null |
research/Tutorial.ipynb
|
dreamshark/SteganoGAN
|
67c73783a8f5cb416b94cafdab80319f304e0cd7
|
[
"MIT"
] | null | null | null |
research/Tutorial.ipynb
|
dreamshark/SteganoGAN
|
67c73783a8f5cb416b94cafdab80319f304e0cd7
|
[
"MIT"
] | null | null | null | 31.878914 | 291 | 0.544335 |
[
[
[
"## Requirements",
"_____no_output_____"
],
[
"\nBefore using this tutorial, ensure that the following are on your system:\n\n- <b>SteganoGAN is installed</b>. Install via pip or source code. \n- <b>Training and Validation Dataset are available </b>. Download via data/download.sh or retrieve your own.\n\nIt is also suggested that you have the following:\n\n- <b>CUDA-enabled machine</b>. SteganoGAN takes very long to train without a GPU. Use AWS to have access to CUDA machines.\n\n\nNow, we retrieve each of the imports required by steganoGAN\n\n## Imports\n",
"_____no_output_____"
]
],
[
[
"import numpy as np #numpy is used for a parameter input",
"_____no_output_____"
]
],
[
[
"This imports the SteganoGAN class which has the wrapper functions for SteganoGAN usage:\n\n- <b>Create a SteganoGAN architecture</b> \n- <b>Train a SteganoGAN architecture</b>\n- <b>Load a SteganoGAN model</b>\n- <b>Encode and decode operations for SteganoGAN models</b>\n\nWe retrieve each of these functions later in the tutorial. ",
"_____no_output_____"
]
],
[
[
"from steganogan import SteganoGAN",
"_____no_output_____"
]
],
[
[
"The DataLoader is used to do the following:\n\n- <b>Load images</b> from a selected database\n- <b>Specify hyperparameters</b> for database loading\n",
"_____no_output_____"
]
],
[
[
"from steganogan.loader import DataLoader",
"_____no_output_____"
]
],
[
[
"The encoders are the architectural models that are used to encode the messages inside the image. There are two types of encoders that can be imported:\n\n- <b>Basic Encoder</b>: This is memory-efficient but not as robust as the other model\n- <b>Dense Encoder</b>: This is a more robust model with a denser architecture\n\nPlease review the SteganoGAN paper for images of the two architectures. A steganoGAN model can only use one of these encoders. You may select which one to use in your model. \n",
"_____no_output_____"
]
],
[
[
"from steganogan.encoders import BasicEncoder, DenseEncoder",
"_____no_output_____"
]
],
[
[
"The decoders are the architectural models that are used to decode the messages inside the image. There are two types of decoders that can be imported:\n\n- <b>Basic Decoder</b>: This is memory-efficient but not as robust as the other model\n- <b>Dense Decoder</b>: This is a more robust model with a denser architecture\n\nPlease review the SteganoGAN paper for images of the two architectures. A steganoGAN model can only use one of these dector. You may select which one to use in your model. \n",
"_____no_output_____"
]
],
[
[
"from steganogan.decoders import BasicDecoder, DenseDecoder",
"_____no_output_____"
]
],
[
[
"The Critic checks if an image is steganographic or not. At the current moment, we have the following Critic:\n\n- <b>Basic Critic</b>: This is a GAN discriminator that ensures images are well hid. \n\nSteganoGAN currently only uses a BasicCritic. This parameter will never be changed \n",
"_____no_output_____"
]
],
[
[
"from steganogan.critics import BasicCritic",
"_____no_output_____"
]
],
[
[
"## Loading Data\n\n\nIn the next cell, we load in the data for our training and validation dataset. The training dataset is used to train the model while the validation dataset is used to ensure that the model is working correctly. There are several parameters that can you choose to tune.\n\n- <b>path:str</b> - This can be a relative path or an absolute path from the notebook file. \n\n- <b>limit:int</b> - The number of images you wish to use. If limit is set as np.inf, all the images in the directory will be used.\n\n- <b>shuffle:bool</b> - If true, your images will be randomly shuffled before being used for training.\n\n- <b>batch_size:int</b> - The number of images to use in a batch. A batch represents the number of images that are trained in a single training cycle (i.e. batch_size=10, means 10 images are sent through the network at once during training)",
"_____no_output_____"
]
],
[
[
"# Load the data\ntrain = DataLoader('D:/dataset/train/', limit=np.inf, shuffle=True, batch_size=4)\n\nvalidation = DataLoader('D:/dataset/val/', limit=np.inf, shuffle=True, batch_size=4)",
"_____no_output_____"
]
],
[
[
"## Selecting an Architecture\n\nBelow we are deciding on the architecture that we want to use for SteganoGAN. There are several parameters that you can tune here to decide on the architecture. Let us go over them:\n\n- <b>data_depth:int</b> - Represents how many layers we want to represent the data with. Currently, data is representated as a N x data_depth x H x W. Usually, we set this to 1 since that suffices for our needs. For more robustness set this data depth to a higher number.\n- <b>encoder:EncoderInstance</b> - You can choose either a BasicEncoder or DenseEncoder.\n- <b>decoder:DecoderInstance</b> - You can choose either a DenseEncoder or DenseDecoder.\n- <b>critic:CritcInstance</b> - The only option is the BasicCritic\n- <b>hidden_size:int</b> - The number of channels we wish to use in the hidden layers of our architecture. You can tune this parameter. We chose 32 as we find relatively good models with these number of channels.\n- <b>cuda:bool</b> - If true and the machine is CUDA-enabled, CUDA will be used for training/execution\n- <b>verbose:bool</b> - If true, the system will print more output to console",
"_____no_output_____"
]
],
[
[
"# Create the SteganoGAN instance\nsteganogan = SteganoGAN(1, BasicEncoder, BasicDecoder, BasicCritic, hidden_size=32, cuda=True, verbose=True)",
"Using CUDA device\n"
]
],
[
[
"## Training and Saving the Model\n\n\nOnce the architecture has been decided and the training and validation data are we loaded, we can begin training. To train call the fit function with the following parameter options:\n\n- <b>train:DataLoaderInstance</b> - This is the training set that you loaded earlier.\n- <b>validation:DataLoaderInstance</b> - This is the validation that you loaded earlier.\n- <b>epochs:int</b> - This is the number of epochs you wish to train for. A larger number of epochs will lead to a more precise model. \n",
"_____no_output_____"
]
],
[
[
"# Fit on the given data\nsteganogan.fit(train, validation, epochs=5)",
"\r 0%| | 0/300 [00:00<?, ?it/s]"
]
],
[
[
"Once the model is trained, we save the model to a .steg file. In this file, we save all the model weights and the architectures that these weights compose. Both the encoder and decoder are saved in the same file.\n\nThe arguments taken are:\n- <b>path:str</b> - This is the path to save the model. Make sure that the directory exists. ",
"_____no_output_____"
]
],
[
[
"# Save the fitted model\nsteganogan.save('demo.steg')",
"_____no_output_____"
]
],
[
[
"## Loading and Executing a Model\n\nThe next command loads a previously generated model. It takes a couple of different parameters. \n\n- <b>architecture:str</b> - You can select either 'basic' or 'dense' architectures. \n- <b>path:str</b> - The path to a model that you have previously generated. \n- <b>cuda:bool</b> - If true and the machine is CUDA-enabled, CUDA will be used for training/execution\n- <b>verbose:bool</b> - If true, the system will print more output to console\n\nNote: <b>either architectures or path but not both must not be None</b>",
"_____no_output_____"
]
],
[
[
"# Load the model\nsteganogan = SteganoGAN.load(architecture='demo', path=None, cuda=True, verbose=True)",
"Using CUDA device\n"
]
],
[
[
"This function encodes an input image with a message and outputs a steganographic image. Note that since SteganoGAN only works on spatial-domains, the output image must be a PNG image. \n\nThe function takes the following arguments:\n- <b>input_image:str</b>: The path to the input image\n- <b>output_image:str</b>: The path to the output image\n- <b>secret_message:str</b>: The secret message you wish to embed.\n",
"_____no_output_____"
]
],
[
[
"# Encode a message in input.png\nsteganogan.encode('input.png', 'output.png', 'This is asuper secret message!')",
"Encoding completed.\n"
]
],
[
[
"This function decode a steganographic image with a message and outputs a message. If no message is found, an error will be thrown by the function. Since steganoGAN encoders and decoders come in pairs, you <b>must</b> use the decoder that was trained with its corresponding encoder. \n\nThe function takes the following arguments:\n- <b>stego_image:str</b>: The path to the steganographic image \n",
"_____no_output_____"
]
],
[
[
"# Decode the message from output.png\nsteganogan.decode('test.png')",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cbf3c8a06426d45170f704c9ddcd3d48a8e50001
| 880,915 |
ipynb
|
Jupyter Notebook
|
MARKETING CAMPAIGN ANALYSIS.ipynb
|
PrasannaDataBus/MARKETING-CAMPAIGN-ANALYSIS
|
920fbc84fe069e0b3da2b314e2b37148fde45890
|
[
"Apache-2.0"
] | null | null | null |
MARKETING CAMPAIGN ANALYSIS.ipynb
|
PrasannaDataBus/MARKETING-CAMPAIGN-ANALYSIS
|
920fbc84fe069e0b3da2b314e2b37148fde45890
|
[
"Apache-2.0"
] | null | null | null |
MARKETING CAMPAIGN ANALYSIS.ipynb
|
PrasannaDataBus/MARKETING-CAMPAIGN-ANALYSIS
|
920fbc84fe069e0b3da2b314e2b37148fde45890
|
[
"Apache-2.0"
] | null | null | null | 579.549342 | 408,684 | 0.937594 |
[
[
[
"import pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport seaborn as sns\n%matplotlib inline",
"_____no_output_____"
],
[
"df = pd.read_csv(r'C:\\Users\\prasa\\Desktop\\advertising.csv')",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
],
[
"df.columns",
"_____no_output_____"
],
[
"df.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 1000 entries, 0 to 999\nData columns (total 10 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 Daily Time Spent on Site 1000 non-null float64\n 1 Age 1000 non-null int64 \n 2 Area Income 1000 non-null float64\n 3 Daily Internet Usage 1000 non-null float64\n 4 Ad Topic Line 1000 non-null object \n 5 City 1000 non-null object \n 6 Male 1000 non-null int64 \n 7 Country 1000 non-null object \n 8 Timestamp 1000 non-null object \n 9 Clicked on Ad 1000 non-null int64 \ndtypes: float64(3), int64(3), object(4)\nmemory usage: 78.2+ KB\n"
],
[
"df.describe()",
"_____no_output_____"
],
[
"sns.set_style('darkgrid')\ndf['Age'].hist(bins=35)\nplt.xlabel('Age')",
"_____no_output_____"
],
[
"pd.crosstab(df['Country'], df['Clicked on Ad']).sort_values( 1,ascending = False).tail(15)",
"_____no_output_____"
],
[
"df[df['Clicked on Ad']==1]['Country'].value_counts().head(15)",
"_____no_output_____"
],
[
"df['Country'].value_counts().head(15)",
"_____no_output_____"
],
[
"pd.crosstab(index=df['Country'],columns='count').sort_values(['count'], ascending=False).head(15)",
"_____no_output_____"
],
[
"df.isnull().sum()",
"_____no_output_____"
],
[
"type(df['Timestamp'][1])",
"_____no_output_____"
],
[
"df['Timestamp'] = pd.to_datetime(df['Timestamp']) \n",
"_____no_output_____"
],
[
"df['Month'] = df['Timestamp'].dt.month ",
"_____no_output_____"
],
[
"df['Day'] = df['Timestamp'].dt.day",
"_____no_output_____"
],
[
"df['Hour'] = df['Timestamp'].dt.hour ",
"_____no_output_____"
],
[
"df[\"Weekday\"] = df['Timestamp'].dt.dayofweek ",
"_____no_output_____"
],
[
"df = df.drop(['Timestamp'], axis=1)",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
],
[
"sns.countplot(x = 'Clicked on Ad', data = df)\nsns.set_style('darkgrid')\n",
"_____no_output_____"
],
[
"sns.jointplot(x = \"Age\", y= \"Daily Time Spent on Site\", data = df)",
"_____no_output_____"
],
[
"sns.scatterplot(x = \"Age\", y= \"Daily Time Spent on Site\",hue='Clicked on Ad', data = df) ",
"_____no_output_____"
],
[
"sns.lmplot(x = \"Age\", y= \"Daily Time Spent on Site\",hue='Clicked on Ad', data = df) ",
"_____no_output_____"
],
[
"sns.pairplot(df, hue = 'Clicked on Ad', vars = ['Daily Time Spent on Site', 'Age', 'Area Income', 'Daily Internet Usage'],palette = 'rocket')",
"_____no_output_____"
],
[
"plots = ['Daily Time Spent on Site', 'Age', 'Area Income','Daily Internet Usage']\nfor i in plots:\n plt.figure(figsize = (12, 6))\n \n plt.subplot(2,3,1)\n sns.boxplot(data= df, y=df[i],x='Clicked on Ad')\n plt.subplot(2,3,2)\n sns.boxplot(data= df, y=df[i])\n plt.subplot(2,3,3)\n sns.distplot(df[i],bins= 20,) \n plt.tight_layout()\n plt.title(i) \n plt.show()",
"_____no_output_____"
],
[
"print('oldest person didn\\'t clicked on the ad was of was of:', df['Age'].max(), 'Years')\nprint('oldest person who clicked on the ad was of:', df[df['Clicked on Ad']==0]['Age'].max(), 'Years')",
"oldest person didn't clicked on the ad was of was of: 61 Years\noldest person who clicked on the ad was of: 53 Years\n"
],
[
"print('Youngest person was of:', df['Age'].min(), 'Years')\nprint('Youngest person who clicked on the ad was of:', df[df['Clicked on Ad']==0]['Age'].min(), 'Years')\n",
"Youngest person was of: 19 Years\nYoungest person who clicked on the ad was of: 19 Years\n"
],
[
"print('Average age was of:', df['Age'].mean(), 'Years')",
"Average age was of: 36.009 Years\n"
],
[
"fig = plt.figure(figsize = (12,10))\nsns.heatmap(df.corr(), cmap='viridis', annot = True)",
"_____no_output_____"
],
[
"f,ax=plt.subplots(1,2,figsize=(14,5))\ndf['Month'][df['Clicked on Ad']==1].value_counts().sort_index().plot(ax=ax[0])\nax[0].set_ylabel('Count of Clicks')\npd.crosstab(df[\"Clicked on Ad\"], df[\"Month\"]).T.plot(kind = 'Bar',ax=ax[1]\nplt.show()",
"_____no_output_____"
],
[
"from sklearn.model_selection import train_test_split",
"_____no_output_____"
],
[
"X = df[['Daily Time Spent on Site', 'Age', 'Area Income','Daily Internet Usage', 'Male']]\ny = df['Clicked on Ad']",
"_____no_output_____"
],
[
"X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.33, random_state=101)",
"_____no_output_____"
],
[
"print(X_train.shape, y_train.shape)\nprint(X_test.shape, y_test.shape)",
"(670, 5) (670,)\n(330, 5) (330,)\n"
],
[
"from sklearn.linear_model import LogisticRegression",
"_____no_output_____"
],
[
"logmodel = LogisticRegression(solver='lbfgs')\nlogmodel.fit(X_train,y_train)",
"C:\\Users\\prasa\\anaconda3\\lib\\site-packages\\sklearn\\linear_model\\_logistic.py:940: ConvergenceWarning: lbfgs failed to converge (status=1):\nSTOP: TOTAL NO. of ITERATIONS REACHED LIMIT.\n\nIncrease the number of iterations (max_iter) or scale the data as shown in:\n https://scikit-learn.org/stable/modules/preprocessing.html\nPlease also refer to the documentation for alternative solver options:\n https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression\n extra_warning_msg=_LOGISTIC_SOLVER_CONVERGENCE_MSG)\n"
],
[
"predictions = logmodel.predict(X_test)",
"_____no_output_____"
],
[
"from sklearn.metrics import classification_report",
"_____no_output_____"
],
[
"print(classification_report(y_test,predictions))",
" precision recall f1-score support\n\n 0 0.97 0.99 0.98 171\n 1 0.99 0.97 0.98 159\n\n accuracy 0.98 330\n macro avg 0.98 0.98 0.98 330\nweighted avg 0.98 0.98 0.98 330\n\n"
],
[
"# Importing a pure confusion matrix from sklearn.metrics family\nfrom sklearn.metrics import confusion_matrix\n\n# Printing the confusion_matrix\nprint(confusion_matrix(y_test, predictions))",
"[[170 1]\n [ 5 154]]\n"
],
[
"logmodel.coef_",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbf3cc78f1b136463bb62ce21e37f19432fb4d04
| 285,991 |
ipynb
|
Jupyter Notebook
|
P1.ipynb
|
auto-ctec/ComputerVision-Finding-LaneLines-Project
|
b3a88d2399c09e7500034777eeb67abbf3761fb3
|
[
"MIT"
] | null | null | null |
P1.ipynb
|
auto-ctec/ComputerVision-Finding-LaneLines-Project
|
b3a88d2399c09e7500034777eeb67abbf3761fb3
|
[
"MIT"
] | null | null | null |
P1.ipynb
|
auto-ctec/ComputerVision-Finding-LaneLines-Project
|
b3a88d2399c09e7500034777eeb67abbf3761fb3
|
[
"MIT"
] | null | null | null | 235.771641 | 118,256 | 0.892668 |
[
[
[
"# Self-Driving Car Engineer Nanodegree\n\n\n## Project: **Finding Lane Lines on the Road** \n***\nIn this project, you will use the tools you learned about in the lesson to identify lane lines on the road. You can develop your pipeline on a series of individual images, and later apply the result to a video stream (really just a series of images). Check out the video clip \"raw-lines-example.mp4\" (also contained in this repository) to see what the output should look like after using the helper functions below. \n\nOnce you have a result that looks roughly like \"raw-lines-example.mp4\", you'll need to get creative and try to average and/or extrapolate the line segments you've detected to map out the full extent of the lane lines. You can see an example of the result you're going for in the video \"P1_example.mp4\". Ultimately, you would like to draw just one line for the left side of the lane, and one for the right.\n\nIn addition to implementing code, there is a brief writeup to complete. The writeup should be completed in a separate file, which can be either a markdown file or a pdf document. There is a [write up template](https://github.com/udacity/CarND-LaneLines-P1/blob/master/writeup_template.md) that can be used to guide the writing process. Completing both the code in the Ipython notebook and the writeup template will cover all of the [rubric points](https://review.udacity.com/#!/rubrics/322/view) for this project.\n\n---\nLet's have a look at our first image called 'test_images/solidWhiteRight.jpg'. Run the 2 cells below (hit Shift-Enter or the \"play\" button above) to display the image.\n\n**Note: If, at any point, you encounter frozen display windows or other confounding issues, you can always start again with a clean slate by going to the \"Kernel\" menu above and selecting \"Restart & Clear Output\".**\n\n---",
"_____no_output_____"
],
[
"**The tools you have are color selection, region of interest selection, grayscaling, Gaussian smoothing, Canny Edge Detection and Hough Tranform line detection. You are also free to explore and try other techniques that were not presented in the lesson. Your goal is piece together a pipeline to detect the line segments in the image, then average/extrapolate them and draw them onto the image for display (as below). Once you have a working pipeline, try it out on the video stream below.**\n\n---\n\n<figure>\n <img src=\"examples/line-segments-example.jpg\" width=\"380\" alt=\"Combined Image\" />\n <figcaption>\n <p></p> \n <p style=\"text-align: center;\"> Your output should look something like this (above) after detecting line segments using the helper functions below </p> \n </figcaption>\n</figure>\n <p></p> \n<figure>\n <img src=\"examples/laneLines_thirdPass.jpg\" width=\"380\" alt=\"Combined Image\" />\n <figcaption>\n <p></p> \n <p style=\"text-align: center;\"> Your goal is to connect/average/extrapolate line segments to get output like this</p> \n </figcaption>\n</figure>",
"_____no_output_____"
],
[
"## Import Packages",
"_____no_output_____"
],
[
"**Run the cell below to import some packages. If you get an `import error` for a package you've already installed, try changing your kernel (select the Kernel menu above --> Change Kernel). Still have problems? Try relaunching Jupyter Notebook from the terminal prompt. Also, consult the forums for more troubleshooting tips.** ",
"_____no_output_____"
]
],
[
[
"#importing some useful packages\nimport matplotlib.pyplot as plt\nimport matplotlib.image as mpimg\nimport numpy as np\nimport cv2\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"# Read in an Image",
"_____no_output_____"
]
],
[
[
"#reading in an image\nimage = mpimg.imread('test_images/solidWhiteRight.jpg')\n\n#printing out some stats and plotting\nprint('This image is:', type(image), 'with dimensions:', image.shape)\nplt.imshow(image) # if you wanted to show a single color channel image called 'gray', for example, call as plt.imshow(gray, cmap='gray')",
"This image is: <class 'numpy.ndarray'> with dimensions: (540, 960, 3)\n"
]
],
[
[
"## Ideas for Lane Detection Pipeline",
"_____no_output_____"
],
[
"**Some OpenCV functions (beyond those introduced in the lesson) that might be useful for this project are:**\n\n`cv2.inRange()` for color selection \n`cv2.fillPoly()` for regions selection \n`cv2.line()` to draw lines on an image given endpoints \n`cv2.addWeighted()` to coadd / overlay two images\n`cv2.cvtColor()` to grayscale or change color\n`cv2.imwrite()` to output images to file \n`cv2.bitwise_and()` to apply a mask to an image\n\n**Check out the OpenCV documentation to learn about these and discover even more awesome functionality!**",
"_____no_output_____"
],
[
"## Helper Functions",
"_____no_output_____"
],
[
"Below are some helper functions to help get you started. They should look familiar from the lesson!",
"_____no_output_____"
],
[
"## Test Images\n\nBuild your pipeline to work on the images in the directory \"test_images\" \n**You should make sure your pipeline works well on these images before you try the videos.**",
"_____no_output_____"
]
],
[
[
"import math\n\ndef grayscale(img):\n \"\"\"Applies the Grayscale transform\n This will return an image with only one color channel\n but NOTE: to see the returned image as grayscale\n (assuming your grayscaled image is called 'gray')\n you should call plt.imshow(gray, cmap='gray')\"\"\"\n return cv2.cvtColor(img, cv2.COLOR_RGB2GRAY)\n # Or use BGR2GRAY if you read an image with cv2.imread()\n # return cv2.cvtColor(img, cv2.COLOR_BGR2GRAY)\n \ndef canny(img, low_threshold, high_threshold):\n \"\"\"Applies the Canny transform\"\"\"\n return cv2.Canny(img, low_threshold, high_threshold)\n\ndef gaussian_blur(img, kernel_size):\n \"\"\"Applies a Gaussian Noise kernel\"\"\"\n return cv2.GaussianBlur(img, (kernel_size, kernel_size), 0)\n\ndef region_of_interest(img, vertices):\n \"\"\"\n Applies an image mask.\n \n Only keeps the region of the image defined by the polygon\n formed from `vertices`. The rest of the image is set to black.\n `vertices` should be a numpy array of integer points.\n \"\"\"\n #defining a blank mask to start with\n mask = np.zeros_like(img) \n \n #defining a 3 channel or 1 channel color to fill the mask with depending on the input image\n if len(img.shape) > 2:\n channel_count = img.shape[2] # i.e. 3 or 4 depending on your image\n ignore_mask_color = (255,) * channel_count\n else:\n ignore_mask_color = 255\n \n #filling pixels inside the polygon defined by \"vertices\" with the fill color \n cv2.fillPoly(mask, vertices, ignore_mask_color)\n \n #returning the image only where mask pixels are nonzero\n masked_image = cv2.bitwise_and(img, mask)\n return masked_image\n\n\ndef draw_lines(img, lines, color=[255, 0, 0], thickness=2):\n \"\"\"\n NOTE: this is the function you might want to use as a starting point once you want to \n average/extrapolate the line segments you detect to map out the full\n extent of the lane (going from the result shown in raw-lines-example.mp4\n to that shown in P1_example.mp4). \n \n Think about things like separating line segments by their \n slope ((y2-y1)/(x2-x1)) to decide which segments are part of the left\n line vs. the right line. Then, you can average the position of each of \n the lines and extrapolate to the top and bottom of the lane.\n \n This function draws `lines` with `color` and `thickness`. \n Lines are drawn on the image inplace (mutates the image).\n If you want to make the lines semi-transparent, think about combining\n this function with the weighted_img() function below\n \"\"\"\n #for line in lines:\n # for x1,y1,x2,y2 in line:\n # cv2.line(img, (x1, y1), (x2, y2), color, thickness)\n\ndef hough_lines(img, rho, theta, threshold, min_line_len, max_line_gap):\n \"\"\"\n `img` should be the output of a Canny transform.\n \n Returns an image with hough lines drawn.\n \"\"\"\n lines = cv2.HoughLinesP(img, rho, theta, threshold, np.array([]), minLineLength=min_line_len, maxLineGap=max_line_gap)\n line_img = np.zeros((img.shape[0], img.shape[1], 3), dtype=np.uint8)\n draw_lines(line_img, lines)\n return line_img\n\n# Python 3 has support for cool math symbols.\n\ndef weighted_img(img, initial_img, α=0.8, β=1., γ=0.):\n \"\"\"\n `img` is the output of the hough_lines(), An image with lines drawn on it.\n Should be a blank image (all black) with lines drawn on it.\n \n `initial_img` should be the image before any processing.\n \n The result image is computed as follows:\n \n initial_img * α + img * β + γ\n NOTE: initial_img and img must be the same shape!\n \"\"\"\n return cv2.addWeighted(initial_img, α, img, β, γ)",
"_____no_output_____"
],
[
"import os\nos.listdir(\"test_images/\")",
"_____no_output_____"
]
],
[
[
"## Build a Lane Finding Pipeline\n\n",
"_____no_output_____"
],
[
"Build the pipeline and run your solution on all test_images. Make copies into the `test_images_output` directory, and you can use the images in your writeup report.\n\nTry tuning the various parameters, especially the low and high Canny thresholds as well as the Hough lines parameters.",
"_____no_output_____"
],
[
"SOLUTION FOR IMAGES",
"_____no_output_____"
]
],
[
[
"######## SOLUTION FOR IMAGES ########\n\nimport matplotlib.pyplot as plt\nimport matplotlib.image as mpimg\nimport numpy as np\nimport cv2\n%matplotlib inline\n\n# Read in the image\nchoose_picture_name = \"whiteCarLaneSwitch\"\nimage_name = 'test_images/'+choose_picture_name+'.jpg'\nimage = mpimg.imread(image_name)\noriginal = image\n\n\n# Grab the x and y size and make a copy of the image\nysize = image.shape[0]\nxsize = image.shape[1]\ncolor_select = np.copy(image)\nline_image = np.copy(color_select)\n\n######## COLOR SELECTION ########\nred_threshold = 200\ngreen_threshold = 100\nblue_threshold = 100\n\nrgb_threshold = [red_threshold, green_threshold, blue_threshold]\n\ncolor_thresholds = (image[:,:,0] < rgb_threshold[0]) \\\n | (image[:,:,1] < rgb_threshold[1]) \\\n | (image[:,:,2] < rgb_threshold[2])\n\n######## MASKING #############\nleft_bottom = [50, 539]\nright_bottom = [900, 539]\napex = [475, 280]\n# Perform a linear fit (y=Ax+B) to each of the three sides of the triangle\n# np.polyfit returns the coefficients [A, B] of the fit\nfit_left = np.polyfit((left_bottom[0], apex[0]), (left_bottom[1], apex[1]), 1)\nfit_right = np.polyfit((right_bottom[0], apex[0]), (right_bottom[1], apex[1]), 1)\nfit_bottom = np.polyfit((left_bottom[0], right_bottom[0]), (left_bottom[1], right_bottom[1]), 1)\n# Find the region inside the lines\nXX, YY = np.meshgrid(np.arange(0, xsize), np.arange(0, ysize))\nregion_thresholds = (YY > (XX*fit_left[0] + fit_left[1])) & \\\n (YY > (XX*fit_right[0] + fit_right[1])) & \\\n (YY < (XX*fit_bottom[0] + fit_bottom[1]))\n# Mask color and region selection\ncolor_select[color_thresholds | ~region_thresholds] = [0, 0, 0]\n# Color pixels red where both color and region selections met\nline_image[~color_thresholds & region_thresholds] = [255, 0, 0]\n\n######## CANNY EDGES ########\ngray = cv2.cvtColor(color_select,cv2.COLOR_RGB2GRAY)\n# Define a kernel size for Gaussian smoothing / blurring\nkernel_size = 5 # Must be an odd number (3, 5, 7...)\nblur_gray = cv2.GaussianBlur(gray,(kernel_size, kernel_size),0)\n# Define our parameters for Canny and run it\nlow_threshold = 50\nhigh_threshold = 150\nedges = cv2.Canny(blur_gray, low_threshold, high_threshold)\n\n######## REGION OF INTEREST ########\n# Next we'll create a masked edges image using cv2.fillPoly()\nmask = np.zeros_like(edges) \nignore_mask_color = 255 \nimshape = image.shape\nvertices = np.array([[(0,imshape[0]),(0, 0), (imshape[1], 0), (imshape[1],imshape[0])]], dtype=np.int32)\ncv2.fillPoly(mask, vertices, ignore_mask_color)\nmasked_edges = cv2.bitwise_and(edges, mask)\n\n######## HOUGH TRANSFORM ########\n# Hough transform parameters\nrho = 1 # distance resolution in pixels of the Hough grid\ntheta = np.pi/180 # angular resolution in radians of the Hough grid\nthreshold = 35 # minimum number of votes (intersections in Hough grid cell)\nmin_line_length = 5 #minimum number of pixels making up a line\nmax_line_gap = 2 # maximum gap in pixels between connectable line segments\nline_image = np.copy(image)*0 # creating a blank to draw lines on\n# Hough on edge detected image\nlines = cv2.HoughLinesP(masked_edges, rho, theta, threshold, np.array([]),\n min_line_length, max_line_gap)\n\n######## IMPROVED DRAWN LINES ####\n# IMPROVED DRAWN LINES A)\n# In case of error, don't draw the line\ndraw_right = True\ndraw_left = True\n\n# IMPROVED DRAWN LINES B)\n# Find slopes of all lines\n# But only care about lines where abs(slope) > slope_threshold\nslope_threshold = 0.5\nconsidered_lane_height = 0.4\nslopes = []\nnew_lines = []\nfor line in lines:\n x1, y1, x2, y2 = line[0] # line = [[x1, y1, x2, y2]]\n\n # Calculate slope\n if x2 - x1 == 0.: # corner case, avoiding division by 0\n slope = 999. # practically infinite slope\n else:\n slope = (y2 - y1) / (x2 - x1)\n\n # Filter lines based on slope\n if abs(slope) > slope_threshold:\n slopes.append(slope)\n new_lines.append(line)\n\nlines = new_lines\n\n# IMPROVED DRAWN LINES C)\n# Split lines into right_lines and left_lines, representing the right and left lane lines\n# Right/left lane lines must have positive/negative slope, and be on the right/left half of the image\nright_lines = []\nleft_lines = []\nfor i, line in enumerate(lines):\n x1, y1, x2, y2 = line[0]\n img_x_center = image.shape[1] / 2 # x coordinate of center of image\n if slopes[i] > 0 and x1 > img_x_center and x2 > img_x_center:\n right_lines.append(line)\n elif slopes[i] < 0 and x1 < img_x_center and x2 < img_x_center:\n left_lines.append(line)\n\n# IMPROVED DRAWN LINES D)\n# Run linear regression to find best fit line for right and left lane lines\n# Right lane lines\nright_lines_x = []\nright_lines_y = []\n\nfor line in right_lines:\n x1, y1, x2, y2 = line[0]\n\n right_lines_x.append(x1)\n right_lines_x.append(x2)\n\n right_lines_y.append(y1)\n right_lines_y.append(y2)\n\nif len(right_lines_x) > 0:\n right_m, right_b = np.polyfit(right_lines_x, right_lines_y, 1) # y = m*x + b\nelse:\n right_m, right_b = 1, 1\n draw_right = False\n\n# Left lane lines\nleft_lines_x = []\nleft_lines_y = []\n\nfor line in left_lines:\n x1, y1, x2, y2 = line[0]\n\n left_lines_x.append(x1)\n left_lines_x.append(x2)\n\n left_lines_y.append(y1)\n left_lines_y.append(y2)\n\nif len(left_lines_x) > 0:\n left_m, left_b = np.polyfit(left_lines_x, left_lines_y, 1) # y = m*x + b\nelse:\n left_m, left_b = 1, 1\n draw_left = False\n\n# IMPROVED DRAWN LINES E)\n# Find 2 end points for right and left lines, used for drawing the line\n# y = m*x + b --> x = (y - b)/m\ny1 = image.shape[0]\ny2 = image.shape[0] * (1 - considered_lane_height)\n\nright_x1 = (y1 - right_b) / right_m\nright_x2 = (y2 - right_b) / right_m\n\nleft_x1 = (y1 - left_b) / left_m\nleft_x2 = (y2 - left_b) / left_m\n\n# IMPROVED DRAWN LINES F)\n# Convert calculated end points from float to int\ny1 = int(y1)\ny2 = int(y2)\nright_x1 = int(right_x1)\nright_x2 = int(right_x2)\nleft_x1 = int(left_x1)\nleft_x2 = int(left_x2)\n\n# IMPROVED DRAWN LINES G)\n# Draw the right and left lines on image\nif draw_right:\n cv2.line(line_image, (right_x1, y1), (right_x2, y2), (255,0,0), 10)\nif draw_left:\n cv2.line(line_image, (left_x1, y1), (left_x2, y2), (255,0,0), 10)\n\n\n# Combine the lines with the original image\nresult = cv2.addWeighted(image, 0.8, line_image, 1, 0)\n\nplt.imshow(image)\n\n# Save result image with lines\nmpimg.imsave(\"test_images_output/\"+ choose_picture_name +\"_original.png\", original)\nmpimg.imsave(\"test_images_output/\"+ choose_picture_name +\"_result.png\", result)\nmpimg.imsave(\"test_images_output/\"+ choose_picture_name +\"_color_select.png\", color_select)\nmpimg.imsave(\"test_images_output/\"+ choose_picture_name +\"_edges.png\", edges)\nmpimg.imsave(\"test_images_output/\"+ choose_picture_name +\"_line_image.png\", line_image)",
"_____no_output_____"
]
],
[
[
"## Test on Videos\n\nYou know what's cooler than drawing lanes over images? Drawing lanes over video!\n\nWe can test our solution on two provided videos:\n\n`solidWhiteRight.mp4`\n\n`solidYellowLeft.mp4`\n\n**Note: if you get an import error when you run the next cell, try changing your kernel (select the Kernel menu above --> Change Kernel). Still have problems? Try relaunching Jupyter Notebook from the terminal prompt. Also, consult the forums for more troubleshooting tips.**\n\n**If you get an error that looks like this:**\n```\nNeedDownloadError: Need ffmpeg exe. \nYou can download it by calling: \nimageio.plugins.ffmpeg.download()\n```\n**Follow the instructions in the error message and check out [this forum post](https://discussions.udacity.com/t/project-error-of-test-on-videos/274082) for more troubleshooting tips across operating systems.**",
"_____no_output_____"
],
[
"SOLUTION FOR VIDEO",
"_____no_output_____"
]
],
[
[
"# Import everything needed to edit/save/watch video clips\nfrom moviepy.editor import VideoFileClip\nfrom IPython.display import HTML",
"_____no_output_____"
],
[
"def process_image(image):\n # Grab the x and y size and make a copy of the image\n ysize = image.shape[0]\n xsize = image.shape[1]\n color_select = np.copy(image)\n line_image = np.copy(color_select)\n\n ######## COLOR SELECTION ########\n red_threshold = 200\n green_threshold = 100\n blue_threshold = 100\n\n rgb_threshold = [red_threshold, green_threshold, blue_threshold]\n\n color_thresholds = (image[:,:,0] < rgb_threshold[0]) \\\n | (image[:,:,1] < rgb_threshold[1]) \\\n | (image[:,:,2] < rgb_threshold[2])\n\n ######## MASKING #############\n left_bottom = [50, 539]\n right_bottom = [900, 539]\n apex = [475, 280]\n # Perform a linear fit (y=Ax+B) to each of the three sides of the triangle\n # np.polyfit returns the coefficients [A, B] of the fit\n fit_left = np.polyfit((left_bottom[0], apex[0]), (left_bottom[1], apex[1]), 1)\n fit_right = np.polyfit((right_bottom[0], apex[0]), (right_bottom[1], apex[1]), 1)\n fit_bottom = np.polyfit((left_bottom[0], right_bottom[0]), (left_bottom[1], right_bottom[1]), 1)\n # Find the region inside the lines\n XX, YY = np.meshgrid(np.arange(0, xsize), np.arange(0, ysize))\n region_thresholds = (YY > (XX*fit_left[0] + fit_left[1])) & \\\n (YY > (XX*fit_right[0] + fit_right[1])) & \\\n (YY < (XX*fit_bottom[0] + fit_bottom[1]))\n # Mask color and region selection\n color_select[color_thresholds | ~region_thresholds] = [0, 0, 0]\n # Color pixels red where both color and region selections met\n line_image[~color_thresholds & region_thresholds] = [255, 0, 0]\n\n ######## CANNY EDGES ########\n gray = cv2.cvtColor(color_select,cv2.COLOR_RGB2GRAY)\n # Define a kernel size for Gaussian smoothing / blurring\n kernel_size = 5 # Must be an odd number (3, 5, 7...)\n blur_gray = cv2.GaussianBlur(gray,(kernel_size, kernel_size),0)\n # Define our parameters for Canny and run it\n low_threshold = 50\n high_threshold = 150\n edges = cv2.Canny(blur_gray, low_threshold, high_threshold)\n\n ######## REGION OF INTEREST ########\n # Next we'll create a masked edges image using cv2.fillPoly()\n mask = np.zeros_like(edges) \n ignore_mask_color = 255 \n imshape = image.shape\n vertices = np.array([[(0,imshape[0]),(0, 0), (imshape[1], 0), (imshape[1],imshape[0])]], dtype=np.int32)\n cv2.fillPoly(mask, vertices, ignore_mask_color)\n masked_edges = cv2.bitwise_and(edges, mask)\n\n ######## HOUGH TRANSFORM ########\n # Hough transform parameters\n rho = 1 # distance resolution in pixels of the Hough grid\n theta = np.pi/180 # angular resolution in radians of the Hough grid\n threshold = 35 # minimum number of votes (intersections in Hough grid cell)\n min_line_length = 5 #minimum number of pixels making up a line\n max_line_gap = 2 # maximum gap in pixels between connectable line segments\n line_image = np.copy(image)*0 # creating a blank to draw lines on\n # Hough on edge detected image\n lines = cv2.HoughLinesP(masked_edges, rho, theta, threshold, np.array([]),\n min_line_length, max_line_gap)\n\n ######## IMPROVED DRAWN LINES ####\n # IMPROVED DRAWN LINES A)\n # In case of error, don't draw the line\n draw_right = True\n draw_left = True\n\n # IMPROVED DRAWN LINES B)\n # Find slopes of all lines\n # But only care about lines where abs(slope) > slope_threshold\n slope_threshold = 0.5\n considered_lane_height = 0.4\n slopes = []\n new_lines = []\n for line in lines:\n x1, y1, x2, y2 = line[0] # line = [[x1, y1, x2, y2]]\n\n # Calculate slope\n if x2 - x1 == 0.: # corner case, avoiding division by 0\n slope = 999. # practically infinite slope\n else:\n slope = (y2 - y1) / (x2 - x1)\n\n # Filter lines based on slope\n if abs(slope) > slope_threshold:\n slopes.append(slope)\n new_lines.append(line)\n\n lines = new_lines\n\n # IMPROVED DRAWN LINES C)\n # Split lines into right_lines and left_lines, representing the right and left lane lines\n # Right/left lane lines must have positive/negative slope, and be on the right/left half of the image\n right_lines = []\n left_lines = []\n for i, line in enumerate(lines):\n x1, y1, x2, y2 = line[0]\n img_x_center = image.shape[1] / 2 # x coordinate of center of image\n if slopes[i] > 0 and x1 > img_x_center and x2 > img_x_center:\n right_lines.append(line)\n elif slopes[i] < 0 and x1 < img_x_center and x2 < img_x_center:\n left_lines.append(line)\n\n # IMPROVED DRAWN LINES D)\n # Run linear regression to find best fit line for right and left lane lines\n # Right lane lines\n right_lines_x = []\n right_lines_y = []\n\n for line in right_lines:\n x1, y1, x2, y2 = line[0]\n\n right_lines_x.append(x1)\n right_lines_x.append(x2)\n\n right_lines_y.append(y1)\n right_lines_y.append(y2)\n\n if len(right_lines_x) > 0:\n right_m, right_b = np.polyfit(right_lines_x, right_lines_y, 1) # y = m*x + b\n else:\n right_m, right_b = 1, 1\n draw_right = False\n\n # Left lane lines\n left_lines_x = []\n left_lines_y = []\n\n for line in left_lines:\n x1, y1, x2, y2 = line[0]\n\n left_lines_x.append(x1)\n left_lines_x.append(x2)\n\n left_lines_y.append(y1)\n left_lines_y.append(y2)\n\n if len(left_lines_x) > 0:\n left_m, left_b = np.polyfit(left_lines_x, left_lines_y, 1) # y = m*x + b\n else:\n left_m, left_b = 1, 1\n draw_left = False\n\n # IMPROVED DRAWN LINES E)\n # Find 2 end points for right and left lines, used for drawing the line\n # y = m*x + b --> x = (y - b)/m\n y1 = image.shape[0]\n y2 = image.shape[0] * (1 - considered_lane_height)\n\n right_x1 = (y1 - right_b) / right_m\n right_x2 = (y2 - right_b) / right_m\n\n left_x1 = (y1 - left_b) / left_m\n left_x2 = (y2 - left_b) / left_m\n\n # IMPROVED DRAWN LINES F)\n # Convert calculated end points from float to int\n y1 = int(y1)\n y2 = int(y2)\n right_x1 = int(right_x1)\n right_x2 = int(right_x2)\n left_x1 = int(left_x1)\n left_x2 = int(left_x2)\n\n # IMPROVED DRAWN LINES G)\n # Draw the right and left lines on image\n if draw_right:\n cv2.line(line_image, (right_x1, y1), (right_x2, y2), (255,0,0), 10)\n if draw_left:\n cv2.line(line_image, (left_x1, y1), (left_x2, y2), (255,0,0), 10)\n result = cv2.addWeighted(image, 0.8, line_image, 1, 0) \n\n return result",
"_____no_output_____"
]
],
[
[
"Let's try the one with the solid white lane on the right first ...",
"_____no_output_____"
]
],
[
[
"white_output = 'test_videos_output/solidWhiteRight.mp4'\n## To speed up the testing process you may want to try your pipeline on a shorter subclip of the video\n## To do so add .subclip(start_second,end_second) to the end of the line below\n## Where start_second and end_second are integer values representing the start and end of the subclip\n## You may also uncomment the following line for a subclip of the first 5 seconds\n#clip1 = VideoFileClip(\"test_videos/solidWhiteRight.mp4\").subclip(0,3)\nclip1 = VideoFileClip(\"test_videos/solidWhiteRight.mp4\")\nwhite_clip = clip1.fl_image(process_image) #NOTE: this function expects color images!!\n%time white_clip.write_videofile(white_output, audio=False)",
"[MoviePy] >>>> Building video test_videos_output/solidWhiteRight.mp4\n[MoviePy] Writing video test_videos_output/solidWhiteRight.mp4\n"
]
],
[
[
"Play the video inline, or if you prefer find the video in your filesystem (should be in the same directory) and play it in your video player of choice.",
"_____no_output_____"
]
],
[
[
"HTML(\"\"\"\n<video width=\"960\" height=\"540\" controls>\n <source src=\"{0}\">\n</video>\n\"\"\".format(white_output))",
"_____no_output_____"
]
],
[
[
"## Improve the draw_lines() function\n\n**At this point, if you were successful with making the pipeline and tuning parameters, you probably have the Hough line segments drawn onto the road, but what about identifying the full extent of the lane and marking it clearly as in the example video (P1_example.mp4)? Think about defining a line to run the full length of the visible lane based on the line segments you identified with the Hough Transform. As mentioned previously, try to average and/or extrapolate the line segments you've detected to map out the full extent of the lane lines. You can see an example of the result you're going for in the video \"P1_example.mp4\".**\n\n**Go back and modify your draw_lines function accordingly and try re-running your pipeline. The new output should draw a single, solid line over the left lane line and a single, solid line over the right lane line. The lines should start from the bottom of the image and extend out to the top of the region of interest.**",
"_____no_output_____"
],
[
"Now for the one with the solid yellow lane on the left. This one's more tricky!",
"_____no_output_____"
]
],
[
[
"yellow_output = 'test_videos_output/solidYellowLeft.mp4'\n## To speed up the testing process you may want to try your pipeline on a shorter subclip of the video\n## To do so add .subclip(start_second,end_second) to the end of the line below\n## Where start_second and end_second are integer values representing the start and end of the subclip\n## You may also uncomment the following line for a subclip of the first 5 seconds\n##clip2 = VideoFileClip('test_videos/solidYellowLeft.mp4').subclip(0,5)\nclip2 = VideoFileClip('test_videos/solidYellowLeft.mp4')\nyellow_clip = clip2.fl_image(process_image)\n%time yellow_clip.write_videofile(yellow_output, audio=False)",
"[MoviePy] >>>> Building video test_videos_output/solidYellowLeft.mp4\n[MoviePy] Writing video test_videos_output/solidYellowLeft.mp4\n"
],
[
"HTML(\"\"\"\n<video width=\"960\" height=\"540\" controls>\n <source src=\"{0}\">\n</video>\n\"\"\".format(yellow_output))",
"_____no_output_____"
]
],
[
[
"## Writeup and Submission\n\nIf you're satisfied with your video outputs, it's time to make the report writeup in a pdf or markdown file. Once you have this Ipython notebook ready along with the writeup, it's time to submit for review! Here is a [link](https://github.com/udacity/CarND-LaneLines-P1/blob/master/writeup_template.md) to the writeup template file.\n",
"_____no_output_____"
],
[
"## Optional Challenge\n\nTry your lane finding pipeline on the video below. Does it still work? Can you figure out a way to make it more robust? If you're up for the challenge, modify your pipeline so it works with this video and submit it along with the rest of your project!",
"_____no_output_____"
]
],
[
[
"challenge_output = 'test_videos_output/challenge.mp4'\n## To speed up the testing process you may want to try your pipeline on a shorter subclip of the video\n## To do so add .subclip(start_second,end_second) to the end of the line below\n## Where start_second and end_second are integer values representing the start and end of the subclip\n## You may also uncomment the following line for a subclip of the first 5 seconds\nclip3 = VideoFileClip('test_videos/challenge.mp4').subclip(0,5)\n#clip3 = VideoFileClip('test_videos/challenge.mp4')\nchallenge_clip = clip3.fl_image(process_image)\n%time challenge_clip.write_videofile(challenge_output, audio=False)",
"[MoviePy] >>>> Building video test_videos_output/challenge.mp4\n[MoviePy] Writing video test_videos_output/challenge.mp4\n"
],
[
"HTML(\"\"\"\n<video width=\"960\" height=\"540\" controls>\n <source src=\"{0}\">\n</video>\n\"\"\".format(challenge_output))",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
]
] |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.